Analysis Report: HEK293_OSMI2_6hA vs HEK293_DMSO_6hA
WANG Ziyi
Date: 18 5月 2022
Summary: HEK293_OSMI2_6hA vs HEK293_DMSO_6hA
1. Differentially Expressed Genes
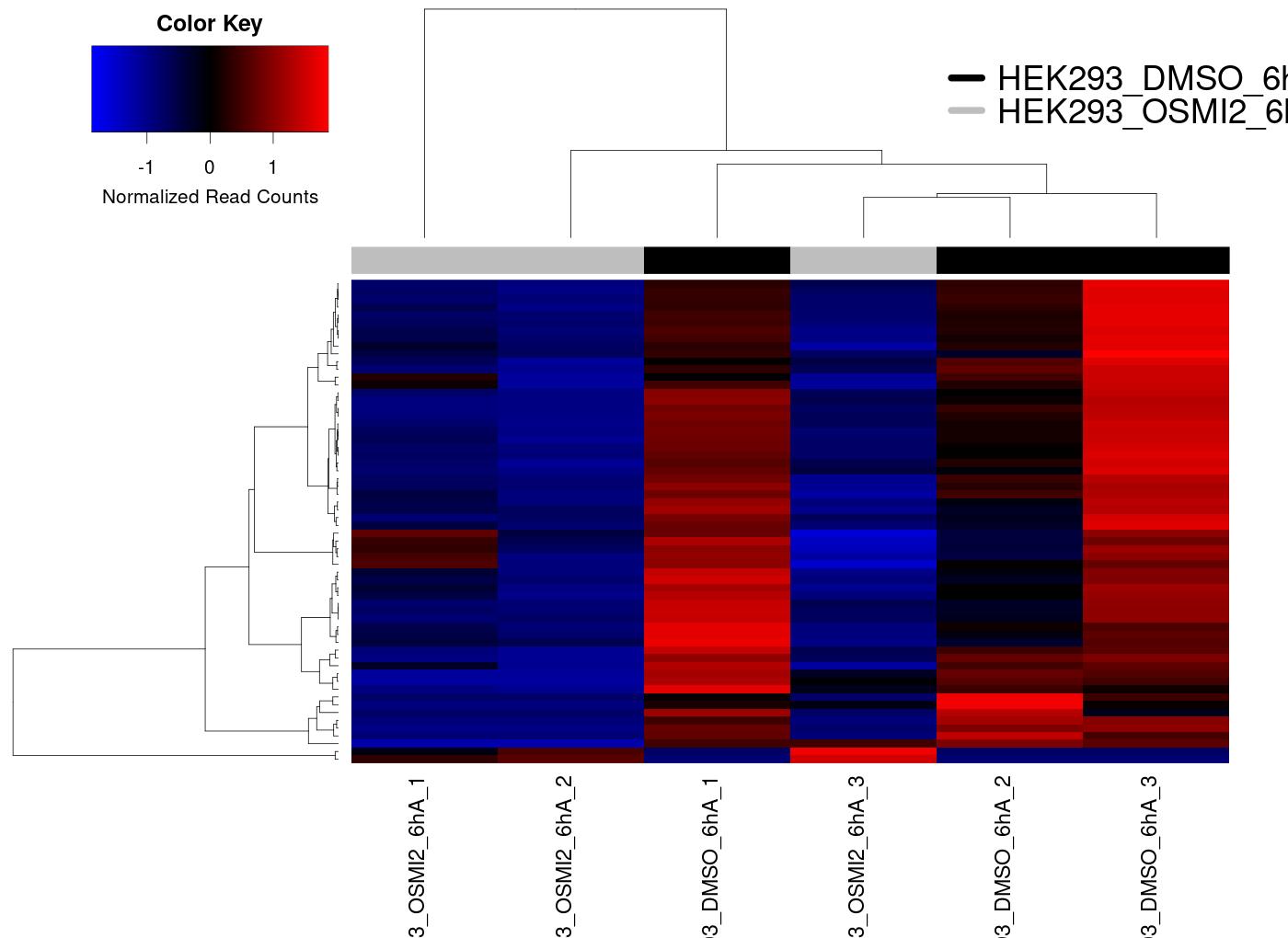
Heatmap
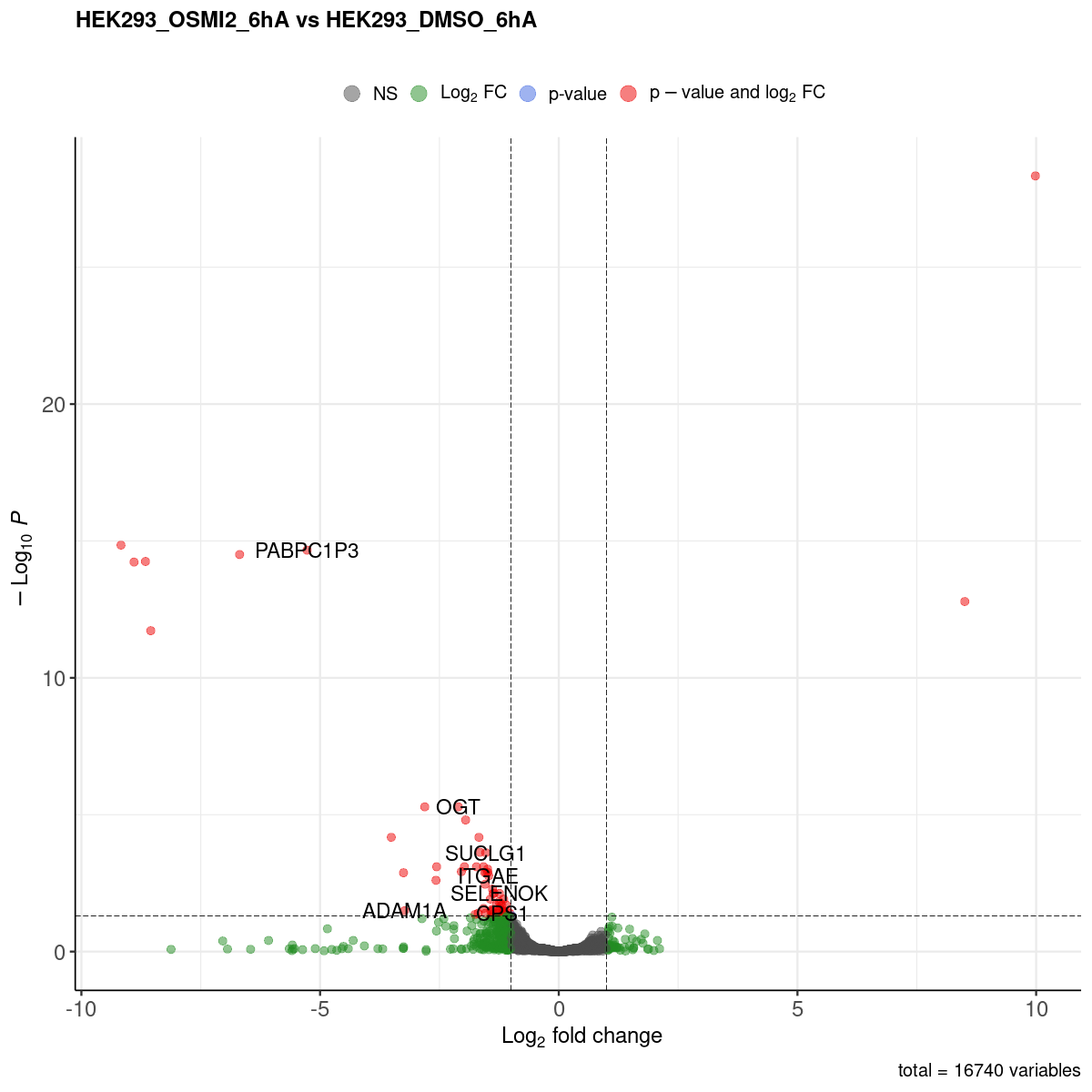
Volcano plot
DEGs list
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_OSMI2_6hA | SE_Intercept_HEK293_OSMI2_6hA | Intercept_HEK293_DMSO_6hA | SE_Intercept_HEK293_DMSO_6hA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000021826 | 1373 | CPS1 | protein_coding | P31327 | FUNCTION: Involved in the urea cycle of ureotelic animals where the enzyme plays an important role in removing excess ammonia from the cell. | 3D-structure;ATP-binding;Acetylation;Allosteric enzyme;Alternative splicing;Disease variant;Glycoprotein;Ligase;Mitochondrion;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transit peptide;Urea cycle | The mitochondrial enzyme encoded by this gene catalyzes synthesis of carbamoyl phosphate from ammonia and bicarbonate. This reaction is the first committed step of the urea cycle, which is important in the removal of excess urea from cells. The encoded protein may also represent a core mitochondrial nucleoid protein. Three transcript variants encoding different isoforms have been found for this gene. The shortest isoform may not be localized to the mitochondrion. Mutations in this gene have been associated with carbamoyl phosphate synthetase deficiency, susceptibility to persistent pulmonary hypertension, and susceptibility to venoocclusive disease after bone marrow transplantation.[provided by RefSeq, May 2010]. | hsa:1373; | cytoplasm [GO:0005737]; mitochondrial inner membrane [GO:0005743]; mitochondrial matrix [GO:0005759]; mitochondrial nucleoid [GO:0042645]; nucleolus [GO:0005730]; protein-containing complex [GO:0032991]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; carbamoyl-phosphate synthase (ammonia) activity [GO:0004087]; carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity [GO:0004088]; endopeptidase activity [GO:0004175]; glutamate binding [GO:0016595]; modified amino acid binding [GO:0072341]; phospholipid binding [GO:0005543]; protein-containing complex binding [GO:0044877]; 'de novo' pyrimidine nucleobase biosynthetic process [GO:0006207]; anion homeostasis [GO:0055081]; carbamoyl phosphate biosynthetic process [GO:0070409]; cellular response to ammonium ion [GO:0071242]; cellular response to cAMP [GO:0071320]; cellular response to fibroblast growth factor stimulus [GO:0044344]; cellular response to glucagon stimulus [GO:0071377]; cellular response to oleic acid [GO:0071400]; citrulline biosynthetic process [GO:0019240]; glutamine metabolic process [GO:0006541]; hepatocyte differentiation [GO:0070365]; homocysteine metabolic process [GO:0050667]; midgut development [GO:0007494]; nitric oxide metabolic process [GO:0046209]; nitrogen compound metabolic process [GO:0006807]; response to amine [GO:0014075]; response to amino acid [GO:0043200]; response to dexamethasone [GO:0071548]; response to food [GO:0032094]; response to growth hormone [GO:0060416]; response to lipopolysaccharide [GO:0032496]; response to starvation [GO:0042594]; response to toxic substance [GO:0009636]; response to xenobiotic stimulus [GO:0009410]; response to zinc ion [GO:0010043]; triglyceride catabolic process [GO:0019433]; urea cycle [GO:0000050]; vasodilation [GO:0042311] | 12655559_The entire DNA sequence of the human CPS1 gene is presented, including all exon-intron boundaries. 14718356_Observational study of gene-disease association. (HuGE Navigator) 17188582_CPSI T1405N genotype appears to be an important new factor in predicting susceptibility to pulmonry hypertension following surgical repair of congenital cardiac defects in children. 17188582_Observational study of gene-disease association. (HuGE Navigator) 17597649_CPPS1 T1404N polymorphism may be associated with the risk of necrotizing enterocolitis in preterm infants. 17597649_Observational study of gene-disease association. (HuGE Navigator) 19754428_Tight hydrogen binding mode is supported by the observation of reduced NAG affinity upon mutation of N-acetyl-L-glutamate-interacting residues of CPSI 19793055_Allelic imbalance may explain clinical variability in CPS1 deficiency in some families. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19926579_The differential expression of Hep Par 1(carbamoyl phosphate synthetase I) in dysplastic vs malignant tumors of the small intestine may be diagnostically useful in difficult cases. 20031578_CPS1, MUT, NOX4, and DPEP1 is associated with plasma homocysteine in healthy Women. 20031578_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20154341_Observational study and genome-wide association study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20154341_These data confirm a recent finding that CPS1 is a locus influencing homocysteine levels in women and suggest that genetic effects on Hcy may differ across developmental stages. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20383146_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20456087_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20520828_Observational study of gene-disease association. (HuGE Navigator) 20520828_The present study in preterm infants did not confirm the earlier reported association between CPS1 genotype and L-arginine levels in term infants. 20578160_Data reported five of the CPS1 mutations (p.T471N, p.Q678P, p.P774L, p.R1453Q, and p.R1453W) in severe CPS1D patients. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20659789_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20800523_structure-function analysis and pathogenicity-testing of mutations in CPS1 20877624_Observational study of gene-disease association. (HuGE Navigator) 21120950_This is the first large-scale report of CPS1 mutations spanning a wide variety of molecular defects highlighting important regions in this protein. 21281797_DNA methylation is a key mechanism of silencing CPS1 expression in human hepatocellular carcinoma cells. 21837743_Case Report: Late-onset carbamoyl phosphate synthetase 1 deficiency in an adult cured by liver transplantation. 22173106_Mutation analysis in these patients identified 17 genetic lesions, 9 of which were new confirming their 'private' nature. 22521883_the human CPS unknown function domains are spatially located in a region that corresponds to the a/b subunits interface in Escherichia coli CPS. [Review] 22692827_Data show that carbamoyl phosphate synthetase 1, an enzyme involved in the urea cycle, 8-oxoguanine DNA glycosylase 1 and DNA polymerase beta, enzymes involved in DNA repair, were expressed at higher levels in Batten disease cells than in normal cells. 24763545_More HCC cells could be identified by the antibody cocktail for CPS1 and P-CK compared with a single antibody. 24813853_Findings support the disease-causing role of the mutations reported to affect the CPS1 deficiency, revealing a key role of the small CPS1 domain of unknown function (UFSD) for proper enzyme folding. 24888247_study examined patient characteristics, including genetic polymorphism, to identify risk factors associated with development of hyperammonemia during valproic acid-based therapy; found CPS1 4217C>A polymorphism may not be associated with development of hyperammonemia in Japanese population 24924744_CPS1 becomes readily detectable upon hepatocyte apoptotic and necrotic death. Its abundance and short serum half-life suggest that it may be a useful prognostic biomarker in acute liver injury. 25099619_Overexpression of CPS1 is associated with rectal cancers. 25410056_characterized the only currently known recurrent CPS1 mutation, p.Val1013del found in eleven unrelated patients of Turkish descent; mutation p.V1013del inactivates CPS1 but does not render the enzyme grossly unstable or insoluble 26059772_Mechanism for Switching On/Off the Urea Cycle 26499888_CPS1 and CPS1IT1 may be potential prognostic indicators for patients with intrahepatic cholangiocarcinoma. 26592762_Molecular structure of CPS1 has been deciphered. 26822151_These results suggest that glycine metabolism and/or the urea cycle represent potentially novel sex-specific mechanisms for the development of atherosclerosis. 26938218_CPS1 is involved in the urea cycle in weight maintenance. 27425868_These results may offer an increasing understand that CPS1 might have a function in differentiation. 27833157_Allele and genotype frequencies of the p.Thr1406Asn polymorphism did not differ between the infants with and without NEC, but the minor A-allele was less frequent in the group of 64 infants with the combined outcome NEC or death before 34 weeks of corrected gestational age than in the infants without this outcome. A significant negative association of the A-allele with the combined outcome NEC or death was found. 28187035_Hepatocyte Antigen Expression in Barrett Esophagus and Associated Neoplasia. 28272778_HNF3beta plays a vital role in regulation of CPS1 gene and could promote the metabolism of ammonia by regulating CPS1 expression. 28376202_CPS1 knockdown reduced cell growth, decreased levels of metabolites associated with nucleic acid biosynthesis. 28538732_CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells 29441491_In silico analysis potentially links CPS1 SNPs with major depression disorder. 29801986_To investigate the efficacy of gene therapy for CPS deficiency following knock-down of hepatic endogenous CPS1 expression, we injected these mice with a helper-dependent adenoviral vector (HDAd) expressing the large murine CPS1 cDNA under control of the phosphoenolpyruvate carboxykinase promoter 30698308_Serum CPS1 appears to be a promising marker for the identi fi cation of mitochondrial damage and the progress of liver pathologies in HCV infected patients, particularly in early stages of the disease. 30802674_ureagenesis can be improved in HepaRG cells by CPS1 overexpression, however, only in combination with DMF-culturing, suggesting that both the low CPS1 level and static-culturing, possibly due to insufficient mitochondria, are limiting urea cycle. 30843237_A liver-humanized mouse model of carbamoyl phosphate synthetase 1-deficiency. 31386258_Non-alcoholic fatty liver disease alters expression of genes governing hepatic nitrogen conversion. 31492588_Expression and clinical significance of CPS1 in glioblastoma multiforme. 31565867_Caspase recruitment domain family member 10 regulates carbamoyl phosphate synthase 1 and promotes cancer growth in bladder cancer cells. 31749211_Molecular, biochemical, and clinical analyses of five patients with carbamoyl phosphate synthetase 1 deficiency. 32273051_Neonatal factors related to survival and intellectual and developmental outcome of patients with early-onset urea cycle disorders. 32934962_The Application of Next-Generation Sequencing (NGS) in Neonatal-Onset Urea Cycle Disorders (UCDs): Clinical Course, Metabolomic Profiling, and Genetic Findings in Nine Chinese Hyperammonemia Patients. 33317798_CPS1: Looking at an ancient enzyme in a modern light. 33387664_Mitochondrial dysfunction as a mechanistic biomarker in patients with non-alcoholic fatty liver disease (NAFLD). 33493519_Suppression of the NTS-CPS1 regulatory axis by AFF1 in lung adenocarcinoma cells. 33772623_Association between genetic variations in carbamoyl-phosphate synthetase gene and persistent neonatal pulmonary hypertension. 33851512_Variants associated with urea cycle disorders in Japanese patients: Nationwide study and literature review. 34343359_Discovery of a Carbamoyl Phosphate Synthetase 1-Deficient HCC Subtype With Therapeutic Potential Through Integrative Genomic and Experimental Analysis. 34973183_Unfavorable clinical outcomes in patients with carbamoyl phosphate synthetase 1 deficiency. 35008510_The Role of TRIP6, ABCC3 and CPS1 Expression in Resistance of Ovarian Cancer to Taxanes. | ENSMUSG00000025991 | Cps1 | 861.06504 | 4.412809e-01 | -1.180231 | 0.3043776 | 14.39607 | 1.481115e-04 | 4.083909e-02 | Yes | No | 6.718742e+02 | 1.519860e+02 | 1.085048e+03 | 2.512831e+02 | |
| ENSG00000070718 | 10947 | AP3M2 | protein_coding | P53677 | FUNCTION: Part of the AP-3 complex, an adaptor-related complex which is not clathrin-associated. The complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and may be directly involved in trafficking to lysosomes. In concert with the BLOC-1 complex, AP-3 is required to target cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals. | Alternative splicing;Cytoplasmic vesicle;Golgi apparatus;Membrane;Protein transport;Reference proteome;Transport | This gene encodes a subunit of the heterotetrameric adaptor-related protein comlex 3 (AP-3), which belongs to the adaptor complexes medium subunits family. The AP-3 complex plays a role in protein trafficking to lysosomes and specialized organelles. Multiple alternatively spliced variants, encoding the same protein, have been identified. [provided by RefSeq, Aug 2008]. | hsa:10947; | AP-type membrane coat adaptor complex [GO:0030119]; axon cytoplasm [GO:1904115]; clathrin adaptor complex [GO:0030131]; cytoplasmic vesicle [GO:0031410]; cytoplasmic vesicle membrane [GO:0030659]; Golgi apparatus [GO:0005794]; anterograde axonal transport [GO:0008089]; anterograde synaptic vesicle transport [GO:0048490]; endocytosis [GO:0006897]; intracellular protein transport [GO:0006886]; vesicle-mediated transport [GO:0016192] | 17293072_Observational study of genotype prevalence. (HuGE Navigator) 17293072_some AP3M2 mutations still remain candidates for unmapped disorders including epilepsy, febrile seizure, and other neuronal developmental disorders associated with functional abnormalities of GABAergic transmission 19481122_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000031539 | Ap3m2 | 1223.79459 | 4.374965e-01 | -1.192657 | 0.2886117 | 16.40391 | 5.117951e-05 | 2.035957e-02 | Yes | No | 7.449327e+02 | 1.248815e+02 | 1.493492e+03 | 2.558296e+02 | |
| ENSG00000079332 | 56681 | SAR1A | protein_coding | Q9NR31 | FUNCTION: Involved in transport from the endoplasmic reticulum to the Golgi apparatus (By similarity). Required to maintain SEC16A localization at discrete locations on the ER membrane perhaps by preventing its dissociation. SAR1A-GTP-dependent assembly of SEC16A on the ER membrane forms an organized scaffold defining endoplasmic reticulum exit sites (ERES). {ECO:0000250, ECO:0000269|PubMed:17005010}. | 3D-structure;Alternative splicing;ER-Golgi transport;Endoplasmic reticulum;GTP-binding;Golgi apparatus;Nucleotide-binding;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:56681; | COPII vesicle coat [GO:0030127]; endoplasmic reticulum exit site [GO:0070971]; Golgi apparatus [GO:0005794]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; COPII-coated vesicle cargo loading [GO:0090110]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; intracellular protein transport [GO:0006886]; membrane organization [GO:0061024]; positive regulation of protein exit from endoplasmic reticulum [GO:0070863]; regulation of COPII vesicle coating [GO:0003400]; vesicle organization [GO:0016050] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 17373700_Observational study of gene-disease association. (HuGE Navigator) 17981133_The Sec31 fragment stimulates GAP activity of Sec23/24, and a convergence of Sec31 and Sec23 residues at the Sar1 GTPase active site explains how GTP hydrolysis is triggered leading to COPII coat disassembly. 18318767_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19135528_Sar 1 H79G mutants efficiently blocked the plasma membrane trafficking of the Kir3.1/Kir3.4 complex however they did not block the Gbeta1gamma2/Kir3.1 interaction. 19357197_Kir6.2 contains a di-acidic endoplasmic reticulum exit signal, which promotes endoplasmic reticulum exit via a process that requires Sar1. 20457124_the intracellular trafficking of hCaR from the endoplasmic reticulum is Sar1-dependent via coat protein complex-II vesicles. 20624903_lipid-directed and tether-assisted Sar1 organization controls membrane constriction to regulate ER export. 20634891_Observational study of gene-disease association. (HuGE Navigator) 22605651_Sar1 mutant proteins added to metaphase-arrested Xenopus laevis egg extracts cause dramatic effects on membrane organization. 22974979_the behavior of the human of Sar1A and Sar1B, a key component of the COPII family of vesicle coat proteins, was examined. 23019651_Sedlin bound and promoted efficient cycling of Sar1, a guanosine triphosphatase that can constrict membranes, and thus allowed nascent carriers to grow and incorporate procollagen prefibrils. 24338480_although Sar1A antagonizes the lipoprotein secretion-promoting activity of Sar1B, both isoforms modulate the expression of genes encoding cholesterol biosynthetic enzymes and the synthesis of cholesterol de novo. 24914133_Data indicate that hydroxyurea (HU) induces SAR1 protein expression, which in turn activates gamma-globin expression, predominantly through the Gialpha/JNK/Jun pathway. 26607390_SAR1A gene silencing in hepatocytes mimics the effect of ethanol: dedimerization of giantin, arresting PDIA3 in the endoplasmic reticulum (ER) and large-scale alterations in Golgi architecture. 27101143_Sar1 is a novel regulator of endoplasmic reticulum-mitochondrial contact sites in the metazoans. 28499394_No associations were observed between any of the secretion associated Ras related GTPase 1A (SAR1a) promoter variants and baseline fetal hemoglobin HbF among sickle cell disease (SCD) patients. 29718541_Study shows that the bona fide nonglycoprotein Nox5, a transmembrane superoxide-producing NADPH oxidase, is transported to the cell surface in a manner resistant to co-expression of Sar1 (H79G), a GTP-fixed mutant of the small GTPase Sar1, which blocks COPII vesicle fission from the endoplasmic reticulum. 30078678_The Sar1 and PREB are upregulated following the induction of morphological differentiation, suggesting the potential role of signaling through Sar1a during morphological differentiation. 30251687_Study data suggest that SAR1A and SAR1B are the critical regulators of trafficking of Nav1.5. Moreover, SAR1A and SAR1B interact with MOG1, and are required for MOG1-mediated cell surface expression and function of Nav1.5. 30485159_FABP5 promotes the budding of particles approximately 150 nm in diameter and modulates the kinetics of the SAR1 GTPase cycle. 32358066_Small sequence variations between two mammalian paralogs of the small GTPase SAR1 underlie functional differences in coat protein complex II assembly. | ENSMUSG00000020088 | Sar1a | 2098.95921 | 4.456507e-01 | -1.166015 | 0.2963991 | 14.86748 | 1.153341e-04 | 3.491909e-02 | Yes | No | 1.290814e+03 | 2.676371e+02 | 2.601689e+03 | 5.524341e+02 | ||
| ENSG00000083457 | 3682 | ITGAE | protein_coding | P38570 | FUNCTION: Integrin alpha-E/beta-7 is a receptor for E-cadherin. It mediates adhesion of intra-epithelial T-lymphocytes to epithelial cell monolayers. | Calcium;Cell adhesion;Cleavage on pair of basic residues;Direct protein sequencing;Disulfide bond;Glycoprotein;Integrin;Magnesium;Membrane;Metal-binding;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | Integrins are heterodimeric integral membrane proteins composed of an alpha chain and a beta chain. This gene encodes an I-domain-containing alpha integrin that undergoes post-translational cleavage in the extracellular domain, yielding disulfide-linked heavy and light chains. In combination with the beta 7 integrin, this protein forms the E-cadherin binding integrin known as the human mucosal lymphocyte-1 antigen. This protein is preferentially expressed in human intestinal intraepithelial lymphocytes (IEL), and in addition to a role in adhesion, it may serve as an accessory molecule for IEL activation. [provided by RefSeq, Jul 2008]. | hsa:3682; | external side of plasma membrane [GO:0009897]; integrin complex [GO:0008305]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; cell adhesion [GO:0007155]; integrin-mediated signaling pathway [GO:0007229] | 12370249_has a role in mucosal immunity and graft rejection 12414996_Involvement of the adhesive ligand pair CD103-E-cadherin in human thymocyte cell proliferation 14532999_Differential up-regulation of CD103 expression on lamina propria lymphocytes in Crohn's disease may indicate differential humoral or cellular regulation in inducing CD103 molecules on lymphocytes in patients with this disease 15196058_CD103 promotes destruction of renal allografts by CD8 T cells 16177076_Integrin/activation marker CD103 (alphaEbeta7) is expressed on Epstein-Barr virus-specific tonsil-resident (but not peripheral blood mononuclear leukocyte-derived) cytotoxic T lymphocytes. 16216886_CD103 expression on dendritic cells influences the balance between effector and regulatory T cell activity in the intestine. 16920912_human alloantigen-induced CD103+ CD8+ T cells possess functional features of regulatory T cells 17325197_CD8+/CD103+ tumor-reactive T lymphocytes infiltrating epithelial tumors most likely play a major role in antitumor cytotoxic response through alphaEbeta7-E-cadherin interactions. 17975119_Observational study of gene-disease association. (HuGE Navigator) 18182176_An investigation to evaluate the diagnostic value of relative number of CD103 expressing CD4(+) T-lymphocytes in the BAL fluid of patients with a variety of interstitial lung diseases, including pulmonary sarcoidosis, is reported. 18492951_integrin alpha(E)(CD103)beta(7) is not only involved in epithelial retention, but also in shaping and proper intraepithelial morphogenesis of some leukocytes. 19023099_Observational study of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19150377_Phenotypic analysis of peripheral blood and decidual CD8+ T cells showed the presence of activated CD8+ T cells as well as a population of CD8+CD103+ T cells in decidual tissue 19604725_A higher proportion of CD103+CD4+ T cells and ITGAE -1088 AA genotype might be associated with fibrosis formation in pulmonary sarcoidosis. 19604725_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20417566_Observational study of gene-disease association. (HuGE Navigator) 20588308_Observational study of gene-disease association. (HuGE Navigator) 20875079_results show that binding of the human alphaE I domain to oral and skin keratinocytes is independent of E-cadherin 21739671_Studies indicate that the mechanisms that regulate the retention of tissue-resident memory T cells include TGF-beta-mediated induction of CD103 and downregulation of CCR7. 21917686_These findings support the usefulness of CD123 and CD103 to aid in the differential diagnosis of B-cell lymphoproliferative disorders 21948982_Lymphoid tissue (LT)-resident CD8alpha-positive dendritic cells (DCs) and non(LT)-derived CD103-positive DCs express transgenic XCR1 and are characterized by a unique transcriptional fingerprint, irrespective of their tissue of origin. 22922816_Memory CD103+ CD8 T cells that persist within the brain after an acute infection with vesicular stomatitis virus are bona fide tissue resident memory cells. 24190978_Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. 24477908_Using a human CTL system model based on a CD8(+)/CD103(-) T cell clone specific of a lung tumor-associated Ag, we demonstrated that the transcription factors Smad2/3 and NFAT-1 are two critical regulators of this process. 25025448_Report CD103 expression occurs most frequently in gastrointestinal T-cell lymphomas. 25584868_Cell-sorting experiments demonstrated that IDO1 expression is found in a subset of CD1a(+)CD14(-)langerin(+) cells, expressing CD103 25637255_This report highlights the diagnostic difficulties and the need of consensus in categorizing clonal CD103(+) lymphocytosis in patients with refractory celiac disease. 25957117_PD-1(+)CD103(+) CD8 TIL might regain functional antitumor activity, an effect that potentially could be augmented by immune modulation. 26522261_Levels of GZMA and ITGAE mRNAs in colon tissues can identify patients with UC who are most likely to benefit from etrolizumab; expression levels decrease with etrolizumab administration in biomarker(high) patients. 26813744_detected CD103 tumor cells in 41% (16/39) of lymph node-involved adult T-cell leukemia/lymphoma 27230039_The CD103+CD4+/CD4+ ratio does not accurately discriminate between sarcoidosis and other causes of lymphocytic alveolitis, neither alone nor in combination with the CD4+/CD8+ ratio, and is not a powerful marker for the diagnosis of sarcoidosis. 27543429_AEbeta7 is of key relevance for gut trafficking of inflammatory bowel disease (IBD) CD8(+) T cells and CD4(+) Th9 cells in vivo and mainly retention might account for this effect. These findings indicate that blockade of alphaEbeta7 in addition to alpha4beta7 may be particularly effective in intestinal disorders with expansion of CD8(+) and Th9 cells such as IBD. 28087652_CD103 expression in human dendritic cells is regulated differentially by retinoic acid and Toll-like receptor ligands. 29021139_Pxn binding to the CD103 cytoplasmic tail triggers alphaEbeta7 integrin outside-in signaling that promotes CD8(+) T-cell migratory behavior and effector functions. 29603342_Indeterminate pediatric acute liver failure (iPALF) cases are characterized by a perforin+CD103+CD8+ T-cell predominant hepatic infiltrate consistent with expansion of a Trm CD8+ T-cell phenotype, which is clearly different from noninflammatory causes of PALF. 29912405_alphaE gene expression levels were significantly higher in ileal compared with colonic biopsies in both Crohn's disease and Ulcerative colitis, as well as in healthy subjects. 30006565_Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. 30100393_ITGAE+ lymphocytes tended to associate with the epithelial-mesenchymal transition (EMT) with decreased Snail and increased E-cadherin expression accompanied. 30622531_Study data indicate that CD103 engages in dendritic cell (DC)-epithelial cell interactions upon contact with epithelial E-cadherin but is not a major driver of DC adhesion to gastrointestinal epithelia. 30683885_FGL2 promotes tumor progression in the brain by suppressing CD103-expressing dendritic cell differentiation. 30975145_CD103-expressing cancer stem cells exosomes acted to guide them to target cancer cells and organs, conferring the higher metastatic capacity of clear cell renal cell carcinoma to lungs. 31771983_Self-Maintaining CD103(+) Cancer-Specific T Cells Are Highly Energetic with Rapid Cytotoxic and Effector Responses. 31844672_Intravenous CBD-CCL4 administration recruits CD103(+) DCs and CD8(+) T cells and improves the antitumor effect of CPI immunotherapy in multiple tumor models, including poor responders to CPI. 32477365_KLRG1 and CD103 Expressions Define Distinct Intestinal Tissue-Resident Memory CD8 T Cell Subsets Modulated in Crohn's Disease. 33479027_CD8+CD103+ tissue-resident memory T cells convey reduced protective immunity in cutaneous squamous cell carcinoma. 33827905_Intratumoral CD103+ CD8+ T cells predict response to PD-L1 blockade. 34298400_The differentiation of new human CD303(+) Plasmacytoid dendritic cell subpopulations expressing CD205 and/or CD103 regulated by Non-Small-Cell lung cancer cells. 34658046_CD103 integrin identifies a high IL-10-producing FoxP3(+) regulatory T-cell population suppressing allergic airway inflammation. | ENSMUSG00000005947 | Itgae | 786.09113 | 3.612954e-01 | -1.468749 | 0.3063683 | 21.90032 | 2.871856e-06 | 1.705551e-03 | Yes | No | 3.928554e+02 | 8.244991e+01 | 1.043322e+03 | 2.232780e+02 | |
| ENSG00000087995 | 339175 | METTL2A | protein_coding | Q96IZ6 | FUNCTION: S-adenosyl-L-methionine-dependent methyltransferase that mediates N(3)-methylcytidine modification of residue 32 of the tRNA anticodon loop of tRNA(Thr)(UGU) and tRNA(Arg)(CCU). {ECO:0000269|PubMed:28655767}. | Acetylation;Alternative splicing;Methyltransferase;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine;Transferase;tRNA processing | hsa:339175; | tRNA (cytosine) methyltransferase activity [GO:0016427]; tRNA (cytosine-3-)-methyltransferase activity [GO:0052735]; tRNA methylation [GO:0030488] | 28655767_Data, including data from studies using mutant/knockout mice and cell lines from such mice, suggest that METTL2 and METTL6 (but not METTL8) contribute to post-transcriptional methylation of cytidine (formation of 3-methylcytidine) in tRNA; METTL2 methylates tRNA(Thr) and tRNA(Arg); METTL6 interacts with seryl-tRNA synthetase and methylates tRNA(Ser); METTL8 catalyzes post-transcriptional methylation of cytidine in mRNA. 34268557_Mutually exclusive substrate selection strategy by human m3C RNA transferases METTL2A and METTL6. | ENSMUSG00000020691 | Mettl2 | 1116.37482 | 4.922717e-01 | -1.022473 | 0.2628951 | 14.45681 | 1.434102e-04 | 4.030565e-02 | Yes | Yes | 7.945312e+02 | 1.114610e+02 | 1.438012e+03 | 2.059553e+02 | ||
| ENSG00000090263 | 51650 | MRPS33 | protein_coding | Q9Y291 | 3D-structure;Acetylation;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. The 28S subunit of the mammalian mitoribosome may play a crucial and characteristic role in translation initiation. This gene encodes a 28S subunit protein that is one of the more highly conserved mitochondrial ribosomal proteins among mammals, Drosophila and C. elegans. Splice variants that differ in the 5' UTR have been found for this gene; all variants encode the same protein. Pseudogenes corresponding to this gene are found on chromosomes 1q, 4p, 4q, and 20q [provided by RefSeq, Jul 2008]. | hsa:51650; | mitochondrial inner membrane [GO:0005743]; mitochondrial small ribosomal subunit [GO:0005763]; mitochondrion [GO:0005739]; structural constituent of ribosome [GO:0003735]; translation [GO:0006412] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000029918 | Mrps33 | 1276.63037 | 4.304075e-01 | -1.216225 | 0.2918316 | 16.86394 | 4.015720e-05 | 1.722409e-02 | Yes | No | 8.206603e+02 | 1.283017e+02 | 1.694868e+03 | 2.704945e+02 | ||
| ENSG00000100372 | 10478 | SLC25A17 | protein_coding | O43808 | FUNCTION: Peroxisomal transporter for multiple cofactors like coenzyme A (CoA), flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN) and nucleotide adenosine monophosphate (AMP), and to a lesser extent for nicotinamide adenine dinucleotide (NAD(+)), adenosine diphosphate (ADP) and adenosine 3',5'-diphosphate (PAP). May catalyze the transport of free CoA, FAD and NAD(+) from the cytosol into the peroxisomal matrix by a counter-exchange mechanism. Inhibited by pyridoxal 5'-phosphate and bathophenanthroline in vitro. {ECO:0000269|PubMed:12445829, ECO:0000269|PubMed:22185573}. | Cytoplasm;Membrane;Peroxisome;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene encodes a peroxisomal membrane protein that belongs to the family of mitochondrial solute carriers. It is expressed in the liver, and is likely involved in transport. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:10478; | cytosol [GO:0005829]; integral component of peroxisomal membrane [GO:0005779]; membrane [GO:0016020]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; adenine nucleotide transmembrane transporter activity [GO:0000295]; ADP transmembrane transporter activity [GO:0015217]; AMP transmembrane transporter activity [GO:0080122]; ATP transmembrane transporter activity [GO:0005347]; chaperone binding [GO:0051087]; coenzyme A transmembrane transporter activity [GO:0015228]; FAD transmembrane transporter activity [GO:0015230]; FMN transmembrane transporter activity [GO:0044610]; NAD transmembrane transporter activity [GO:0051724]; ATP transport [GO:0015867]; fatty acid alpha-oxidation [GO:0001561]; fatty acid beta-oxidation [GO:0006635]; fatty acid transport [GO:0015908] | 12445829_Results describe the identification of PMP34, a peroxisomal membrane protein belonging to the mitochondrial solute carrier family, as an adenine nucleotide transporter. 22185573_SLC25A17 is a transporter for CoA and FAD, and to a lesser extent NAD 23266187_Compares and contrasts all the known human SLC25A* genes and includes functional information. 25002403_PEX16 mediates the peroxisomal trafficking of two distinct peroxisomal membrane proteins, PEX3 and PMP34, via the endoplasmic reticulum | ENSMUSG00000022404 | Slc25a17 | 1683.10903 | 4.038913e-01 | -1.307961 | 0.3196048 | 16.39491 | 5.142304e-05 | 2.035957e-02 | Yes | No | 8.561869e+02 | 1.505079e+02 | 2.148830e+03 | 3.859809e+02 | |
| ENSG00000100814 | 57820 | CCNB1IP1 | protein_coding | Q9NPC3 | FUNCTION: Ubiquitin E3 ligase that acts as a limiting factor for crossing-over during meiosis: required during zygonema to limit the colocalization of RNF212 with MutS-gamma-associated recombination sites and thereby establish early differentiation of crossover and non-crossover sites. Later, it is directed by MutL-gamma to stably accumulate at designated crossover sites. Probably promotes the dissociation of RNF212 and MutS-gamma to allow the progression of recombination and the implementation of the final steps of crossing over (By similarity). Modulates cyclin-B levels and participates in the regulation of cell cycle progression through the G2 phase. Overexpression causes delayed entry into mitosis. {ECO:0000250, ECO:0000269|PubMed:12612082, ECO:0000269|PubMed:17297447}.; FUNCTION: E3 ubiquitin-protein ligase. Modulates cyclin B levels and participates in the regulation of cell cycle progression through the G2 phase. Overexpression causes delayed entry into mitosis. | Chromosome;Coiled coil;Meiosis;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | HEI10 is a member of the E3 ubiquitin ligase family and functions in progression of the cell cycle through G(2)/M.[supplied by OMIM, Apr 2004]. | hsa:57820; | synaptonemal complex [GO:0000795]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; blastocyst formation [GO:0001825]; chiasma assembly [GO:0051026]; reciprocal meiotic recombination [GO:0007131]; spermatid development [GO:0007286] | 12612082_HEI10 defines a divergent class of E3 ubiquitin ligase, functioning in progression through G(2)/M. 16532029_The cyclin B-binding protein and cell cycle regulator HEI10 is identified as a novel merlin-binding partner; interaction with merlin affects HEI10 protein levels. | ENSMUSG00000071470 | Ccnb1ip1 | 841.63102 | 4.232968e-01 | -1.240258 | 0.3113885 | 15.50234 | 8.240329e-05 | 2.959045e-02 | Yes | No | 4.983871e+02 | 9.210481e+01 | 1.137567e+03 | 2.144405e+02 |
| ENSG00000102158 | 84061 | MAGT1 | protein_coding | Q9H0U3 | FUNCTION: Accessory component of the STT3B-containing form of the N-oligosaccharyl transferase (OST) complex which catalyzes the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains (PubMed:31831667). Involved in N-glycosylation of STT3B-dependent substrates (PubMed:31831667). Specifically required for the glycosylation of a subset of acceptor sites that are near cysteine residues; in this function seems to act redundantly with TUSC3. In its oxidized form proposed to form transient mixed disulfides with a glycoprotein substrate to facilitate access of STT3B to the unmodified acceptor site. Has also oxidoreductase-independent functions in the STT3B-containing OST complex possibly involving substrate recognition. {ECO:0000269|PubMed:25135935, ECO:0000269|PubMed:26864433, ECO:0000269|PubMed:31036665, ECO:0000269|PubMed:31831667, ECO:0000305}.; FUNCTION: May be involved in Mg(2+) transport in epithelial cells. {ECO:0000305|PubMed:15804357, ECO:0000305|PubMed:19717468}. | 3D-structure;Alternative splicing;Cell membrane;Congenital disorder of glycosylation;Disease variant;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Magnesium;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix;Transport | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:31831667}. | This gene encodes a ubiquitously expressed magnesium cation transporter protein that localizes to the cell membrane. This protein also associates with N-oligosaccharyl transferase and therefore may have a role in N-glycosylation. Mutations in this gene cause a form of X-linked intellectual disability (XLID). This gene may have multiple in-frame translation initiation sites, one of which would encode a shorter protein with an N-terminus containing a signal peptide at amino acids 1-29. [provided by RefSeq, Jul 2017]. | hsa:84061; | azurophil granule membrane [GO:0035577]; endoplasmic reticulum [GO:0005783]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; oligosaccharyltransferase complex [GO:0008250]; plasma membrane [GO:0005886]; magnesium ion transmembrane transporter activity [GO:0015095]; cognition [GO:0050890]; magnesium ion transport [GO:0015693]; protein N-linked glycosylation [GO:0006487]; protein N-linked glycosylation via asparagine [GO:0018279]; transmembrane transport [GO:0055085] | 15804357_identifed a novel gene that encodes a protein involved with Mg2+-evoked transport, MagT1; results suggest that MagT1 may provide a selective and regulated pathway for Mg2+ transport in epithelial cells[MagT1] 18455129_study shows that mutations in two OTase-subunit genes, N33/TUSC3 and IAP result in autosomal-recessive nonsyndromic mental retardation 19717468_MagT1 is universally expressed in all tissues and its expression level is up-regulated in low extracellular Mg(2+). Knockdown of either MagT1 or TUSC3 protein significantly lowers the total and free intracellular Mg(2+) concentrations in cell lines. 21627970_Data show that gene expression of the Mg(2+) selective transporter MagT1 is upregulated in TRPM7(-/-) cells. 21983175_A deficiency in magnesium transporter 1 (MAGT1), an Mg(2+)-specific transporter, leads to the absence of a T cell antigen receptor-stimulated Mg(2+)flux and an attenuation of T cell activation. 24130152_A 0.8 kb intronic duplication in MAGT1 and a single base pair deletion in the last exon of ATRX were identified in a family with five males demonstrating intellectual disability (ID) and unusual skin findings (e.g., generalized pruritus). 26617690_evaluation of MAGT1 and AKAP13 expression in clinical hepatocellular carcinoma tissues by immunohistochemistry suggested that both proteins were strongly expressed in tumor tissues with significantly higher average immunoreactive scores of Remmele and Stegner (IRS) than in non-tumor tissues 29051561_MAGT1 expression is defective in CD8+positive T cells of chronic hepatitis B patients. 30385806_TRPM7 and MagT1 are upregulated in osteogenic differentiation and silencing either one accelerates osteogenic differentiation, partly through the activation of autophagy. .. These results underpin the contribution of magnesium, TRPM7 and MagT1 to autophagy and osteoblastogenesis. 30952425_This study demonstrates that the simultaneous downregulation of TRPM7 and MagT1 inhibits cell growth and activates autophagy, which is required in the early phases of osteoblastogenesis. 31038761_microRNA-199a-5p suppresses glioma progression by inhibiting MAGT1. 31337704_Authors observed that MAGT1-dependent glycosylation is sensitive to Mg(2+) levels and that reduced Mg(2+) impairs immune-cell function via the loss of specific glycoproteins. Findings reveal that defects in protein glycosylation and gene expression underlie immune defects in an inherited disease due to MAGT1 deficiency. 32633723_TRPM7 and MagT1 regulate the proliferation of osteoblast-like SaOS-2 cells through different mechanisms. 33210605_miR-199a-5p targeted regulation of MAGT1 expression in the functional depletion of CD8(+)T cells in HBV infection. 34086870_CRISPR-targeted MAGT1 insertion restores XMEN patient hematopoietic stem cells and lymphocytes. 34499581_MAGT1 is required for HeLa cell proliferation through regulating p21 expression, S-phase progress, and ERK/p38 MAPK MYC axis. 35264785_Identification of a novel MAGT1 mutation supports a diagnosis of XMEN disease. | ENSMUSG00000031232 | Magt1 | 1989.74941 | 4.387586e-01 | -1.188501 | 0.3137127 | 13.86630 | 1.962871e-04 | 4.860935e-02 | Yes | No | 1.236909e+03 | 2.675257e+02 | 2.624718e+03 | 5.812886e+02 |
| ENSG00000111843 | 51522 | TMEM14C | protein_coding | Q9P0S9 | FUNCTION: Required for normal heme biosynthesis. {ECO:0000250}. | 3D-structure;Heme biosynthesis;Membrane;Mitochondrion;Reference proteome;Transmembrane;Transmembrane helix | hsa:51522; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrial membrane [GO:0031966]; erythrocyte differentiation [GO:0030218]; heme biosynthetic process [GO:0006783]; mitochondrial transport [GO:0006839]; regulation of heme biosynthetic process [GO:0070453] | 19656490_Required for functional heme biosynthesis in erythroid cells. 20877624_Observational study of gene-disease association. (HuGE Navigator) 34172893_Phase I First-in-Human Dose Escalation Study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. 34861039_Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. | ENSMUSG00000021361 | Tmem14c | 1898.30372 | 4.487538e-01 | -1.156004 | 0.2916352 | 15.14794 | 9.940666e-05 | 3.265826e-02 | Yes | No | 1.069420e+03 | 1.719345e+02 | 2.402569e+03 | 3.948362e+02 | ||
| ENSG00000113811 | 58515 | SELENOK | protein_coding | Q9Y6D0 | FUNCTION: Required for Ca(2+) flux in immune cells and plays a role in T-cell proliferation and in T-cell and neutrophil migration (By similarity). Involved in endoplasmic reticulum-associated degradation (ERAD) of soluble glycosylated proteins (PubMed:22016385). Required for palmitoylation and cell surface expression of CD36 and involved in macrophage uptake of low-density lipoprotein and in foam cell formation (By similarity). Together with ZDHHC6, required for palmitoylation of ITPR1 in immune cells, leading to regulate ITPR1 stability and function (PubMed:25368151). Plays a role in protection of cells from ER stress-induced apoptosis (PubMed:20692228). Protects cells from oxidative stress when overexpressed in cardiomyocytes (PubMed:16962588). {ECO:0000250|UniProtKB:Q9JLJ1, ECO:0000269|PubMed:16962588, ECO:0000269|PubMed:20692228, ECO:0000269|PubMed:22016385, ECO:0000269|PubMed:25368151}. | 3D-structure;Calcium;Calcium transport;Cell membrane;Direct protein sequencing;Endoplasmic reticulum;Ion transport;Membrane;Reference proteome;Selenocysteine;Transmembrane;Transmembrane helix;Transport;Ubl conjugation | The protein encoded by this gene belongs to the selenoprotein K family. It is a transmembrane protein that is localized in the endoplasmic reticulum (ER), and is involved in ER-associated degradation (ERAD) of misfolded, glycosylated proteins. It also has a role in the protection of cells from ER stress-induced apoptosis. Knockout studies in mice show the importance of this gene in promoting Ca(2+) flux in immune cells and mounting effective immune response. This protein is a selenoprotein, containing the rare amino acid selenocysteine (Sec). Sec is encoded by the UGA codon, which normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, designated the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon, rather than as a stop signal. Pseudogenes of this locus have been identified on chromosomes 6 and 19.[provided by RefSeq, Aug 2017]. | hsa:58515; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; calcium ion transport [GO:0006816]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress [GO:0070059]; macrophage derived foam cell differentiation [GO:0010742]; positive regulation of defense response to virus by host [GO:0002230]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of monocyte chemotactic protein-1 production [GO:0071639]; positive regulation of T cell migration [GO:2000406]; positive regulation of T cell proliferation [GO:0042102]; positive regulation of tumor necrosis factor production [GO:0032760]; protein palmitoylation [GO:0018345]; regulation of calcium-mediated signaling [GO:0050848]; regulation of protein transport [GO:0051223]; respiratory burst after phagocytosis [GO:0045728]; response to oxidative stress [GO:0006979] | 16962588_SelK is a novel antioxidant in cardiomyocytes and is related to the regulation of cellular redox balance. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21220695_Human lymphocyte SelK colocalizes with endoplasmic reticulum biomarker KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum receptor. 22016385_Data suggest that SelK is involved in the Derlin-dependent ERAD of glycosylated misfolded proteins and that the function defined by the prototypic SelK is the widespread function of selenium in eukaryotes. 23133653_Polymorphisms in thioredoxin reductase and selenoprotein K genes and selenium status modulate risk of prostate cancer 25117454_SelK reduces hydrophobic substrates, such as phospholipid hydroperoxides, which are known to damage cell membranes. 25368151_IP3R palmitoylation is a critical regulator of Ca(2+) flux in immune cells and define a previously unidentified DHHC/Selk complex responsible for this process. 26567579_overexpression of SelK can inhibit human cancer cell Matrigel adhesion and migration 26735936_SELK has the dynamic structural features to be defined as a HUB protein able to interact with multiple members. 27275761_miR-544a expression was found to be modulated by selenium treatment, suggesting a possible role in SELK induction by selenium. 28130108_These results hereby imply SelK may regulate the release of Ca(2+) by CHERP and play an important role in the proliferation and differentiation of T cell by TCR stimulation. 30082770_The levels of SelS and SelK were decreased during adipocyte differentiation and were inversely related to the levels of peroxisome proliferator-activated receptor gamma (PPARgamma), a central regulator of adipogenesis. 30526872_Three crystal structures have been reported of a CRL2 substrate receptor, KLHDC2, in complex with the diglycine-ending C-end degrons of an early terminated selenoprotein, SelK, and the N-terminal proteolytic fragment of USP1. | ENSMUSG00000042682 | Selenok | 1130.77022 | 4.229073e-01 | -1.241587 | 0.2812352 | 18.72742 | 1.507983e-05 | 7.511217e-03 | Yes | No | 6.296759e+02 | 9.757948e+01 | 1.510604e+03 | 2.385919e+02 | |
| ENSG00000117748 | 6118 | RPA2 | protein_coding | P15927 | FUNCTION: As part of the heterotrimeric replication protein A complex (RPA/RP-A), binds and stabilizes single-stranded DNA intermediates, that form during DNA replication or upon DNA stress. It prevents their reannealing and in parallel, recruits and activates different proteins and complexes involved in DNA metabolism. Thereby, it plays an essential role both in DNA replication and the cellular response to DNA damage. In the cellular response to DNA damage, the RPA complex controls DNA repair and DNA damage checkpoint activation. Through recruitment of ATRIP activates the ATR kinase a master regulator of the DNA damage response. It is required for the recruitment of the DNA double-strand break repair factors RAD51 and RAD52 to chromatin in response to DNA damage. Also recruits to sites of DNA damage proteins like XPA and XPG that are involved in nucleotide excision repair and is required for this mechanism of DNA repair. Plays also a role in base excision repair (BER) probably through interaction with UNG. Also recruits SMARCAL1/HARP, which is involved in replication fork restart, to sites of DNA damage. May also play a role in telomere maintenance. {ECO:0000269|PubMed:15205463, ECO:0000269|PubMed:17765923, ECO:0000269|PubMed:17959650, ECO:0000269|PubMed:19116208, ECO:0000269|PubMed:20154705, ECO:0000269|PubMed:21504906, ECO:0000269|PubMed:2406247, ECO:0000269|PubMed:24332808, ECO:0000269|PubMed:7697716, ECO:0000269|PubMed:7700386, ECO:0000269|PubMed:8702565, ECO:0000269|PubMed:9430682, ECO:0000269|PubMed:9765279}. | 3D-structure;Acetylation;Alternative splicing;DNA damage;DNA recombination;DNA repair;DNA replication;DNA-binding;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes a subunit of the heterotrimeric Replication Protein A (RPA) complex, which binds to single-stranded DNA (ssDNA), forming a nucleoprotein complex that plays an important role in DNA metabolism, being involved in DNA replication, repair, recombination, telomere maintenance, and co-ordinating the cellular response to DNA damage through activation of the ataxia telangiectasia and Rad3-related protein (ATR) kinase. The RPA complex protects single-stranded DNA from nucleases, prevents formation of secondary structures that would interfere with repair, and co-ordinates the recruitment and departure of different genome maintenance factors. The heterotrimeric complex has two different modes of ssDNA binding, a low-affinity and high-affinity mode, determined by which oligonucleotide/oligosaccharide-binding (OB) domains of the complex are utilized, and differing in the length of DNA bound. This subunit contains a single OB domain that participates in high-affinity DNA binding and also contains a winged helix domain at its carboxy terminus, which interacts with many genome maintenance protein. Post-translational modifications of the RPA complex also plays a role in co-ordinating different damage response pathways. [provided by RefSeq, Sep 2017]. | hsa:6118; | chromatin [GO:0000785]; chromosome, telomeric region [GO:0000781]; DNA replication factor A complex [GO:0005662]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; damaged DNA binding [GO:0003684]; enzyme binding [GO:0019899]; G-rich strand telomeric DNA binding [GO:0098505]; protein N-terminus binding [GO:0047485]; protein phosphatase binding [GO:0019903]; single-stranded DNA binding [GO:0003697]; ubiquitin protein ligase binding [GO:0031625]; base-excision repair [GO:0006284]; DNA replication [GO:0006260]; double-strand break repair via homologous recombination [GO:0000724]; mismatch repair [GO:0006298]; mitotic G1 DNA damage checkpoint signaling [GO:0031571]; nucleotide-excision repair [GO:0006289]; protein localization to chromosome [GO:0034502]; regulation of DNA damage checkpoint [GO:2000001]; regulation of double-strand break repair via homologous recombination [GO:0010569]; telomere maintenance [GO:0000723] | 11731442_Phosphorylation of the RPA2 subunit is observed after exposure of cells to ionizing radiation (IR) and other DNA-damaging agents, which implicates the modified protein in the regulation of DNA replication after DNA damage or in DNA repair. 12509449_RPA2 binds to menin and has a role in multiple endocrine neoplasia 15793585_C-terminal domain of hRPA32 subunit (RPA32C) facilitates initiation of SV40 replication. 17035231_in response to UV-induced DNA damage, ATR rapidly phosphorylates RPA2, disrupting its association with replication centers in the S-phase and contributing to the inhibition of DNA replication 17583916_Determination at single-nucleotide resolution the relative positions of the single-stranded DNA with interacting intrinsic tryptophans of RPA32. 17928296_RPA phosphorylation facilitates chromosomal DNA repair. 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19386720_RPA32 is extensively phosphorylated after the induction of EBV lytic replication. Rad51 and RPA32 are necessary for the completion of EBV lytic infection. 19586055_The N-terminus of RPA1 and phosphorylation of RPA2 regulate RPA interactions with the MRE11-RAD50-NBS1 (MRN) complex and are important in the response to DNA damage. 19671522_mitotic phosphorylation of RPA2 starts at the onset of mitosis, and dephosphorylation occurs during late cytokinesis. 19704001_These data indicate that PP2A-mediated RPA32 dephosphorylation is required for the efficient DNA damage repair. 19834905_RPA32, critical for cell proliferation and maintenance of genome stability, are markedly down-regulated, Data hypothesized that their DNA-related functions could be partially limited in TRAIL-resistant HL-60 cells. 20130019_data suggest that RPA2 hyperphosphorylation plays a critical role in maintenance of genomic stability and cell survival after a DNA replication block via promotion of homologus recombination 20154705_Data suggest that PP4-mediated dephosphorylation of RPA2 is necessary for an efficient DNA-damage response. 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20679368_At the subunit level, 13 proteins out of 30 examined may interact with RPA2. 21062395_RPA1 and RPA2 overexpression seems to be more important during early T-categories of bladder carcinogenesis, showing similar kinetics with cyclin D1 21137066_RPA2 up-regulation may be involved in the growth and/or survival of BRCA1 tumor cells and useful in immunohistochemical discrimination of triple-negative BRCA1 tumors. 21496876_Replication protein A1, replication protein A2, and cyclins D2 and D3 seem to have a parallel role in the promotion of cell cycle in astrocytic tumors being implicated in the malignant progression of these neoplasms. 21731742_RPA2 hyperphosphorylation by DNA-PK in response to DNA double-strand breaks blocks unscheduled homologous recombination and delays mitotic entry. 22521144_Data show that the R88C variant impairs binding of the R88C variant impairs binding of uracil-DNA glycosylase UNG2 to replication protein A RPA2. 22684010_4E-BP3 regulates eIF4E-mediated nuclear mRNA export and interacts with replication protein A2 23047005_this study has explored the role of RPA32 phosphorylation at CDK and ATR sites and propose that phosphorylation of the RPA32 subunit is dispensable for checkpoint activation induced by replication stress with aphidicolin. 23393223_study concludes RPA2 expression is translationally regulated via internal ribosome entry site and by eIF3a and that this regulation is partly accountable for cellular response to DNA damage and survival. 24730652_study reports the characterization of the RPA32C-SMARCAL1 interface at the molecular level; implications of results are discussed with respect to the recruitment of SMARCAL1 and other DNA damage response and repair proteins to stalled replication forks 24819595_RPA32 phosphorylation regulates replication arrest, recombination, late origin firing, and mitotic catastrophe 24910198_Conserved motifs are required for RPA32 binding the the N-terminus of SMARCAL1. 25113031_Expression of mutant RPA2 or loss of PALB2 expression led to significant DNA damage after replication stress, a defect accentuated by poly-ADP (adenosine diphosphate) ribose polymerase inhibitors. 27068393_The authors show that Vpr can form a trimolecular complex with UNG2 and RPA32 and the positive effect of UNG2 and RPA32 on the reverse transcription process leading to optimal virus replication and dissemination between the primary target cells of HIV-1. 28007956_knockdown of RPA2 promoted formation of the menin-p65 complex and repressed the expression of NF-kappaB-mediated genes. RPA2 expression was induced via an E2F1-dependent mechanism in MCF7 and MDA-MB-231 cells treated with NF-kappaB activators, TNF-alpha or lipopolysaccharide (LPS). 28107649_RPA, best known for its role in DNA replication and repair, recruits HIRA to promoters and enhancers and regulates deposition of newly synthesized H3.3 to these regulatory elements for gene regulation. 28575657_Single point mutations in the RPA32 subunit of RPA that abolish interaction with RFWD3 also inhibit interstrand crossling repair, demonstrating that RPA-mediated RFWD3 recruitment to stalled replication forks is important for ICL repair. 28575658_E3 ligase RFWD3 functions in timely removal and degradation of RPA and RAD51 to allow homologous recombination progression to subsequent steps following mitomycin C damage. 31582797_HERC2 regulates RPA2 by mediating ATR-induced Ser33 phosphorylation and ubiquitin-dependent degradation. 32856505_Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair. 33784377_RPA2 winged-helix domain facilitates UNG-mediated removal of uracil from ssDNA; implications for repair of mutagenic uracil at the replication fork. 34642383_hSSB2 (NABP1) is required for the recruitment of RPA during the cellular response to DNA UV damage. | ENSMUSG00000028884 | Rpa2 | 5539.65371 | 4.787433e-01 | -1.062676 | 0.2701359 | 14.91182 | 1.126551e-04 | 3.479016e-02 | Yes | No | 3.885396e+03 | 5.487399e+02 | 6.830313e+03 | 9.883553e+02 | |
| ENSG00000117862 | 51060 | TXNDC12 | protein_coding | O95881 | FUNCTION: Possesses significant protein thiol-disulfide oxidase activity. {ECO:0000269|PubMed:12761212}. | 3D-structure;Direct protein sequencing;Disulfide bond;Endoplasmic reticulum;Oxidoreductase;Redox-active center;Reference proteome;Signal | This gene encodes a member of the thioredoxin superfamily. Members of this family are characterized by a conserved active motif called the thioredoxin fold that catalyzes disulfide bond formation and isomerization. This protein localizes to the endoplasmic reticulum and has a single atypical active motif. The encoded protein is mainly involved in catalyzing native disulfide bond formation and displays activity similar to protein-disulfide isomerases. This protein may play a role in defense against endoplasmic reticulum stress. Alternate splicing results in both coding and non-coding variants. [provided by RefSeq, Mar 2012]. | hsa:51060; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum lumen [GO:0005788]; protein-disulfide reductase (glutathione) activity [GO:0019153]; protein-disulfide reductase activity [GO:0015035]; negative regulation of cell death [GO:0060548]; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902236] | 12761212_putative physiological role for endoplasmic reticulum thioredoxin superfamily member p18(ERp18) in native disulfide bond formation is discussed 18628206_ERp16 mediates disulfide bond formation in the ER and plays an important role in cellular defense against prolonged ER stress 19361226_the solution structure of oxidized ERp18 as determined using NMR spectroscopy 19887585_ERp18 shows specificity towards a component of the complement cascade, pentraxin-related protein PTX3. 25940440_We conclude that ERp19 contributes to tumorigenicity and metastasis of gastric cancer 31368601_Data demonstrate that ERp18 monitors ATF6a endoplasmic reticulum quality control to ensure optimal processing following trafficking to the Golgi. 31570854_TXNDC12 promotes EMT and metastasis of hepatocellular carcinoma cells via activation of beta-catenin. 34629960_Clinical Value of TXNDC12 Combined With IDH and 1p19q as Biomarkers for Prognosis of Glioma. | ENSMUSG00000028567 | Txndc12 | 1779.62930 | 4.575736e-01 | -1.127924 | 0.2966619 | 14.17871 | 1.662407e-04 | 4.350716e-02 | Yes | No | 1.020235e+03 | 1.475523e+02 | 2.316591e+03 | 3.420346e+02 | |
| ENSG00000125037 | 55831 | EMC3 | protein_coding | Q9P0I2 | FUNCTION: Part of the endoplasmic reticulum membrane protein complex (EMC) that enables the energy-independent insertion into endoplasmic reticulum membranes of newly synthesized membrane proteins (PubMed:30415835, PubMed:29809151, PubMed:29242231, PubMed:32459176, PubMed:32439656). Preferentially accommodates proteins with transmembrane domains that are weakly hydrophobic or contain destabilizing features such as charged and aromatic residues (PubMed:30415835, PubMed:29809151, PubMed:29242231). Involved in the cotranslational insertion of multi-pass membrane proteins in which stop-transfer membrane-anchor sequences become ER membrane spanning helices (PubMed:30415835, PubMed:29809151). It is also required for the post-translational insertion of tail-anchored/TA proteins in endoplasmic reticulum membranes (PubMed:29809151, PubMed:29242231). By mediating the proper cotranslational insertion of N-terminal transmembrane domains in an N-exo topology, with translocated N-terminus in the lumen of the ER, controls the topology of multi-pass membrane proteins like the G protein-coupled receptors (PubMed:30415835). By regulating the insertion of various proteins in membranes, it is indirectly involved in many cellular processes (Probable). {ECO:0000269|PubMed:29242231, ECO:0000269|PubMed:29809151, ECO:0000269|PubMed:30415835, ECO:0000269|PubMed:32439656, ECO:0000269|PubMed:32459176, ECO:0000305}. | 3D-structure;Alternative splicing;Direct protein sequencing;Endoplasmic reticulum;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:55831; | EMC complex [GO:0072546]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of membrane [GO:0016021]; protein insertion into ER membrane by stop-transfer membrane-anchor sequence [GO:0045050]; tail-anchored membrane protein insertion into ER membrane [GO:0071816] | Mouse_homologues 29083321_EMC3 was essential for the processing and routing of surfactant proteins, SP-B and SP-C, and the biogenesis of the phospholipid transport protein ABCA3. 32886670_Loss of the ER membrane protein complex subunit Emc3 leads to retinal bipolar cell degeneration in aged mice. 33785873_Emc3 maintains intestinal homeostasis by preserving secretory lineages. | ENSMUSG00000030286 | Emc3 | 2001.38732 | 4.553054e-01 | -1.135093 | 0.2994960 | 14.05246 | 1.777812e-04 | 4.500197e-02 | Yes | No | 1.253721e+03 | 1.823277e+02 | 2.587790e+03 | 3.846656e+02 | ||
| ENSG00000129315 | 904 | CCNT1 | protein_coding | O60563 | FUNCTION: Regulatory subunit of the cyclin-dependent kinase pair (CDK9/cyclin-T1) complex, also called positive transcription elongation factor B (P-TEFb), which is proposed to facilitate the transition from abortive to productive elongation by phosphorylating the CTD (C-terminal domain) of the large subunit of RNA polymerase II (RNA Pol II). {ECO:0000269|PubMed:16109376, ECO:0000269|PubMed:16109377}.; FUNCTION: (Microbial infection) In case of HIV or SIV infections, binds to the transactivation domain of the viral nuclear transcriptional activator, Tat, thereby increasing Tat's affinity for the transactivating response RNA element (TAR RNA). Serves as an essential cofactor for Tat, by promoting RNA Pol II activation, allowing transcription of viral genes. {ECO:0000269|PubMed:10329125, ECO:0000269|PubMed:10329126}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Coiled coil;Cyclin;Direct protein sequencing;Host-virus interaction;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a member of the highly conserved cyclin C subfamily. The encoded protein tightly associates with cyclin-dependent kinase 9, and is a major subunit of positive transcription elongation factor b (p-TEFb). In humans, there are multiple forms of positive transcription elongation factor b, which may include one of several different cyclins along with cyclin-dependent kinase 9. The complex containing the encoded cyclin and cyclin-dependent kinase 9 acts as a cofactor of human immunodeficiency virus type 1 (HIV-1) Tat protein, and is both necessary and sufficient for full activation of viral transcription. This cyclin and its kinase partner are also involved in triggering transcript elongation through phosphorylation of the carboxy-terminal domain of the largest RNA polymerase II subunit. Overexpression of this gene is implicated in tumor growth. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2013]. | hsa:904; | cyclin/CDK positive transcription elongation factor complex [GO:0008024]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; P-TEFb complex [GO:0070691]; 7SK snRNA binding [GO:0097322]; chromatin binding [GO:0003682]; cyclin-dependent protein serine/threonine kinase activator activity [GO:0061575]; cyclin-dependent protein serine/threonine kinase regulator activity [GO:0016538]; DNA binding [GO:0003677]; DNA-binding transcription factor binding [GO:0140297]; protein kinase binding [GO:0019901]; RNA polymerase binding [GO:0070063]; transcription cis-regulatory region binding [GO:0000976]; cell cycle [GO:0007049]; cell division [GO:0051301]; negative regulation of mRNA polyadenylation [GO:1900364]; phosphorylation of RNA polymerase II C-terminal domain serine 2 residues involved in positive regulation of transcription elongation from RNA polymerase II promoter [GO:1903655]; phosphorylation of RNA polymerase II C-terminal domain serine 5 residues involved in positive regulation of transcription elongation from RNA polymerase II promoter [GO:1903654]; positive regulation by host of viral transcription [GO:0043923]; positive regulation of DNA-templated transcription, elongation [GO:0032786]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription elongation from RNA polymerase II promoter [GO:0032968]; protein phosphorylation [GO:0006468]; regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0000079]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription by RNA polymerase II [GO:0006366] | 11713533_Activity inhibited by 7SK small nuclear RNA. 11884399_P-TEFb containing cyclin K and Cdk9 can activate transcription via RNA. 12115727_CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. 12177005_P-TEFb is a key mediator of Myc activated transcription by stimulating elongation 12368300_fluctuation in CycT1 levels during human macrophage differentiation may be involved in the regulation of HIV-1 replication 12368330_chimeras between kinase-inactive mutant Cdk9 and truncated cyclin T1 proteins efficiently inhibit Tat transactivation and human immunodeficiency virus gene expression 12458222_Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by this protein requires Sp1 12486002_Cyclin T1 binding to HIV-1 tat is regulated by the differential acetylation of tat 12588988_cyclin T1 binds with granulin to inhibit transcription 12651893_the transcriptional repressor PIE-1 from Caenorhabditis elegans binds Cyclin T1 via an alanine-containing heptapeptide repeat and inhibits transcriptional elongation {cyclin T1) 12727882_recruitment to nuclear bodies via direct interaction with PML protein 12832472_Since Tat also binds to this cyclin T1 domain and the association between 7SK RNA/MAQ1 and P-TEFb competes with the binding of Tat to cyclin T1, we speculate that the TAR RNA/Tat lentivirus system has evolved to subvert the cellular 7SK RNA/MAQ1 system 14627702_major portion of nuclear P-TEFb is sequestered and inactivated by the coordinated actions of the 7SK snRNA 14963154_RNAi-mediated gene silencing of P-TEFb in HeLa cells was not lethal and inhibited Tat transactivation and HIV-1 replication in host cells 15049426_review of work indicating under some circumstances TAK/P-TEFb is likely to be limiting for HIV replication in CD4+ T cells and macrophages; review of mechanisms of regulation of the TAK/P-TEFb subunits in these cell types 15107825_NUFIP and P-TEFb with BRCA1 activate transcription by RNA polymerase II 15563843_a histidine-rich stretch in CycT1 was found to direct the transcriptional activity of this P-TEFb complex when tethered artificially to DNA. 15713661_HEXIM1 and HEXIM2 maintain the balance between active and inactive forms of P-TEFb, a heterodimeric complex composed of cyclin-dependent kinase 9 and cyclin T. 15913611_downregulation of hCycT1 did not cause apoptotic cell death 15940264_increased estrogen down regulated gene 1 expression results in inhibition of cyclin T1 recruitment and estrogen receptor 1 DNA binding 15994294_analysis of positive transcription elongation factor - HEXIM1 - 7SK RNA complex 16308568_Role of P-TEFb as an activator of transcription elongation can be separated from its role in RNA processing; neither function is universally required for expression of mammalian pol II-dependent genes. 16362050_the interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct its binding to P-TEFb and subcellular localization that culminates in the inhibition of transcription 16439541_human cyclin T1 expression is sufficient to support Tat-mediated transactivation in primary mouse CD4 T lymphocytes and monocytes/macrophages and increases in vitro and in vivo HIV-1 production by these stimulated cells. 16615932_Finally, two inhibitors of cyclin-dependent kinase 9 (a dominant negative CDK9 and flavopiridol) repressed activity from the MR and BMPR2 promoters 16764723_Using DNA microarray technology, we found that more than 20% of genes induced by PMA require cyclin T1 for their normal level of induction, and approximately 15% of genes repressed by PMA require cyclin T1 for their normal level of repression. 16980611_These data link the P-TEFb equilibrium to the intracellular transcriptional demand and proliferative/differentiated states of cells. 17014716_Prostratin induces Cyclin T1 expression and P-TEFb function and this is likely to be involved in prostratin reactivation of latent HIV-1 proviruses 17317667_WRN is a novel cellular cofactor for HIV-1 replication, and the WRN helicase participates in the recruitment of PCAF/P-TEFb-containing transcription complexes 17352406_Cdk9/Cyclin T1 complex may be important in the activation/differentiation program of lymphoid cells and that its upregulation may contribute to malignant transformation. 17452463_These results suggest that acetylation of CDK9 is an important posttranslational modification that is involved in regulating P-TEFb transcriptional elongation function. 17502349_Upon induction of NF-kappaB, a subset of target genes is regulated differentially by either P-TEFb or DSIF.[P-TEFb, DSIF] 17540406_Regions important for the interactions with partners as the TRM give insights into the spatial arrangment of critical residues that build up the interface for its regulatory factors. 17686863_Tax may compete and functionally substitute for Brd4 in P-TEFb regulation.Tax and Brd4 compete for binding to P-TEFb through direct interaction with cyclin T1 17690245_Overexpression of the BRD4 P-TEFb-interacting domain disrupts the interaction between the HIV transactivator Tat and P-TEFb and suppresses the ability of Tat to transactivate the HIV promoter 17724342_provide structural insights how Hexim1 recognizes the Cyclin T1 subunit of positive transcription elongation factor b 17855633_Identification of ENL-associated proteins by mass spectrometry revealed enzymes with a known role in transcriptional elongation: pTEFb and DOT1L. 18039861_while the P-TEFb level remains constant, the Brd4-P-TEFb interaction increases dramatically in cells progressing from late mitosis to early G(1). 18205180_This data suggests an active role for the Cdk9/Cyclin T1 complex during lymphoid differentiation through germinal center reaction. 18353948_HIV infection increases the sensitivity of microglia and astrocytes to inflammatory stimulation and support the use of these mice as a model to investigate various aspects of the in vivo mechanism of HIV-induced neuronal dysfunction 18391197_TAF7 interacts with the transcription factors, TFIIH and P-TEFb, resulting in the inhibition of their Pol II CTD kinase activities 18566585_Study shows that CDK9/CycT1 autophosphorylates on Thr186 in the activation segment and three C-terminal phosphorylation sites; autophosphorylation on all sites occurs in cis. 18620576_CycT1-U7 mutant inhibits HIV transcription by promoting a rapid degradation of Tat. 18773076_Cyclin T1-dependent genes in activated CD4 T and macrophage cell lines appear enriched in HIV-1 co-factors 18780834_functional and physical interaction of GATA-1 with components of the positive transcriptional elongation factor P-TEFb 18931076_CycT1 may potentially serve as a target for the development of anti-HIV therapies by inactivating viral Tat protein. 19179338_a direct impact of CDK9-cyclin T1 on the nuclear localization and activity of the viral regulator pUL69. 19297489_binding of Brd4 to the cdk9 complex is not required but that efficient binding to cyclin T1 is essential 19387490_Cyclin T1 acetylation triggers dissociation of Hexim1 and 7SK snRNA from cyclin T1/CDK9 and activates the transcriptional activity of P-TEFb. 19883659_The tripartite protein-RNA complex formation between Hexim, Cyclin T and 7SK snRNA, was analyzed. 20210365_binding to Cyclin T1 induces an asymmetry or sterical hindrance in the first coiled coil segment of dimeric Hexim1 that disallows formation of a 2:2 complex. 20618343_study showed that P-TEFb and Dengue virus (DENV) core protein work in concert to enhance IL-8 gene expression by DENV infection 20714219_Cyclin T1 oncogenic function and suggest a role for this protein in controlling cell cycle probably via Rb/E2F1 pathway. 20943989_results suggest that cyclin T1/CDK9 serves as an adapter to mediate the interaction of influenza A virus RNA-dependent RNA polymerase and RNA Pol II and promote viral transcription 21450947_HCMV inter-regulation with CDK9/cyclin T1 is at least partly based on a pUL69-cylin T1 interaction. 21555514_Cyclin K inhibits HIV-1 gene expression and replication by interfering with cyclin-dependent kinase 9 (CDK9)-cyclin T1 interaction in Nef-dependent manner. 21763494_The role of the CycT1 N-terminal region in Tat action, was investigated. 21763496_This investigation reveals a novel mechanism of cyclin T1 regulation and establishes NF90 as a regulator of HIV-1 replication during both productive infection and induction from latency. 22205749_MiR-27b, miR-29b, miR-223, and miR-150 are regulators of cyclin T1 protein levels and HIV-1 replication in resting CD4+ T cells. 22342181_N-terminal residues in cyclin T1 are specifically involved in the binding of cyclin T1 to HEXIM1 but not to Tat. 22959624_Structural determination of a short-lived conformational state between CCNT1 and CDK9 that is adopted during the phosphorylation cycle. 23152527_The authors find that resting CD4(+) T cells, whether naive or memory and independent of their HIV infection status, have low levels of Cyclin T1 and T-loop-phosphorylated CDK9, which increase upon activation. 23471103_AFF4 is positioned to make unexpected direct contacts with HIV Tat, and Tat enhances P-TEFb/CCNT1 affinity for AFF4. 23539624_positive transcription elongation factor b (P-TEFb, made of CDK9 and CycT1) is released from its inhibitory complex by histone deacetylase inhibitors, which also activate HIV transcription 23569210_CYCT1b is a newly identified cyclin T1 splice variant whose expression leads to direct inhibition of TAT-transactivated transcription of the HIV-1 LTR promoter 23691059_All of the hCD4/R5/cT1 mice developed disseminated infection of tissues that included the spleen, small intestine, lymph nodes and lungs after intravenous injection with an HIV-1 infectious molecular clone 23977272_Plk1 negatively regulates the RNA polymerase II-dependent transcription through inhibiting the activity of cyclin T1/Cdk9 complex. 24351800_The sequence domain of human cytomegalovirus pUL97 responsible for the interaction with cyclin T1 was between amino acids 231-280. 24515107_the release of P-TEFb from the 7SK snRNP led to increased synthesis of HEXIM1 but not HEXIM2 25909811_the introduction of HIV-1 Tat and Mg12+ ion resulted in the active site architectural characteristics of phosphorylated CDK9/cyclin T1 complex 27193293_Quantitative measurement of the molecular interactions among Tat, CycT1 and CDK9 has showed that any third molecule enhances the binding between the other two molecules. These findings suggest that each component of the Tat:P-TEFb complex stabilizes the overall complex, thereby supporting the efficient transcriptional elongation during viral RNA synthesis. 27731797_The TAR central loop contacts the CycT1 Tat-TAR recognition motif (TRM) and the second Tat Zn(2+)-binding loop. Hydrogen-deuterium exchange (HDX) shows that AFF4 helix 2 is stabilized in the TAR complex despite not touching the RNA, explaining how it enhances TAR binding to the SEC 50-fold. 28422315_ASF/SF2 is identified as a splicing regulator of cyclin T1, which contributes to the control of the subsequent transcription events. 30782840_Cyclins B1, T1, and H differ in their molecular mode of interaction with cytomegalovirus protein kinase pUL97. 30786885_CycT1 is most highly upregulated in maximally activated memory CD4 T cells as expected, but may become less associated with T cell activation during HIV replication. 31603123_CDK9/Cyclin T1 kinase is a protein kinase, indirectly involved in the cell cycle progression. 32108986_Genome-wide association study of word reading: Overlap with risk genes for neurodevelopmental disorders. | ENSMUSG00000011960 | Ccnt1 | 1051.82505 | 3.430746e-01 | -1.543406 | 0.3293660 | 20.40484 | 6.267118e-06 | 3.456092e-03 | Yes | No | 5.853558e+02 | 1.225196e+02 | 1.533988e+03 | 3.278068e+02 | |
| ENSG00000131238 | 5538 | PPT1 | protein_coding | P50897 | FUNCTION: Removes thioester-linked fatty acyl groups such as palmitate from modified cysteine residues in proteins or peptides during lysosomal degradation. Prefers acyl chain lengths of 14 to 18 carbons (PubMed:8816748). {ECO:0000269|PubMed:8816748}. | 3D-structure;Alternative splicing;Disease variant;Disulfide bond;Glycoprotein;Hydrolase;Lysosome;Neurodegeneration;Neuronal ceroid lipofuscinosis;Reference proteome;Secreted;Sensory transduction;Signal;Vision | The protein encoded by this gene is a small glycoprotein involved in the catabolism of lipid-modified proteins during lysosomal degradation. The encoded enzyme removes thioester-linked fatty acyl groups such as palmitate from cysteine residues. Defects in this gene are a cause of infantile neuronal ceroid lipofuscinosis 1 (CLN1, or INCL) and neuronal ceroid lipofuscinosis 4 (CLN4). Two transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Dec 2008]. | hsa:5538; | axon [GO:0030424]; cytosol [GO:0005829]; dendrite [GO:0030425]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; membrane [GO:0016020]; membrane raft [GO:0045121]; neuronal cell body [GO:0043025]; nucleus [GO:0005634]; synaptic vesicle [GO:0008021]; lysophosphatidic acid binding [GO:0035727]; palmitoyl-(protein) hydrolase activity [GO:0008474]; palmitoyl-CoA hydrolase activity [GO:0016290]; sulfatide binding [GO:0120146]; thiolester hydrolase activity [GO:0016790]; adult locomotory behavior [GO:0008344]; associative learning [GO:0008306]; brain development [GO:0007420]; cellular protein catabolic process [GO:0044257]; fatty-acyl-CoA biosynthetic process [GO:0046949]; grooming behavior [GO:0007625]; lipid catabolic process [GO:0016042]; lysosomal lumen acidification [GO:0007042]; membrane raft organization [GO:0031579]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell growth [GO:0030308]; negative regulation of neuron apoptotic process [GO:0043524]; nervous system development [GO:0007399]; neuron development [GO:0048666]; neurotransmitter secretion [GO:0007269]; pinocytosis [GO:0006907]; positive regulation of pinocytosis [GO:0048549]; positive regulation of receptor-mediated endocytosis [GO:0048260]; protein catabolic process [GO:0030163]; protein depalmitoylation [GO:0002084]; protein transport [GO:0015031]; receptor-mediated endocytosis [GO:0006898]; regulation of phospholipase A2 activity [GO:0032429]; regulation of synapse structure or activity [GO:0050803]; response to stimulus [GO:0050896]; sphingolipid catabolic process [GO:0030149]; visual perception [GO:0007601] | 8816748_The function of PPT1 is to hydrolyze long-chain fatty acids from cysteine residues of fatty acylated proteins in the lysosome. 12025857_mutated in neuronal ceroid lipofuscinosis 12125808_The clinical, biochemical, and molecular genetic aspects of lysosomal storage disorders are discussed in this review 12855696_The crystal structure of palmitoyl protein thioesterase-2 (PPT2) reveals the basis for divergent substrate specificities of the two lysosomal thioesterases, PPT1 and PPT2. 16518810_there is a close correlation between CLN2 and CLN1 expression and colorectal carcinoma progression and metastasis and suggest that they may be potential molecular targets 16542649_Results show that PPT1-deficiency causes a defect in fluid-phase and receptor-mediated endocytosis. 16571600_ER stress due to PPT1-deficiency increases ROS and disrupts calcium homeostasis activating caspase-9 and caspase-9 activation mediates caspase-3 activation and apoptosis contributing to rapid neurodegeneration in INCL. 17261688_Adult neuronal ceroid lipofuscinosis caused by deficiency in palmitoyl protein thioesterase 1. 18704195_Palmitoyl protein thioesterase-1 deficiency impairs synaptic vesicle recycling at nerve terminals in humans and mice 19054571_Observational study of gene-disease association. (HuGE Navigator) 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19201763_Observational study of gene-disease association. (HuGE Navigator) 19302939_study presents clinical and diagnostic investigations in six children with variant late infantile neuronal ceroid lipofuscinosis and mutations in CLN1 including a new large-scale deletion on one allele 19793631_Results describe the correlation between the three-dimensional structural changes in mutant palmitoyl protein thioesterase 1 and biochemical phenotypes. 21704547_Stop codon read-through with PTC124 induces palmitoyl-protein thioesterase-1 activity, reduces thioester load and suppresses apoptosis in cultured cells from Infantile neuronal ceroid lipofuscinosis patients. 22520356_This neuroimaging finding in PPT1-related neuronal ceroid lipofuscinosis was not previously reported. 24083319_Data suggest that human monocytes and macrophages express PPT1; PPT1 appears to contribute 32-40% of 2-arachidonylglycerol hydrolysis activity in THP1 monocyte cell line. 25233404_Data (including data from knockout mice) suggest that deficiency of PPT1 leads to accumulation of granular osmiophilic deposits in many cell types, especially in astrocytes. [review-like article] 25865307_analysis of the palmitoyl protein thioesterase 1 interactome in SH-SY5Y human neuroblastoma cells 26731412_we reveal the existence of a positive feedback loop, where palmitoylation of PPT1 results in decreased activity and subsequent elevation in the amount of palmitoylated proteins. 28334871_Proteomics analysis on isolated cilia revealed 660 proteins, which differed in their abundance levels between wild type and Ppt1 knock out. 28899863_Targeting PPT1 blocks mTOR signaling in a manner distinct from catalytic inhibitors, while concurrently inhibiting autophagy, thereby providing a new strategy for cancer therapy. 28978646_the combination of elevated glycolysis and deficient MRPL13 activity was closely linked to CLN1-mediated tumor activity in human hepatoma cells 29631617_The results demonstrate that these patient iPSC derived NCL NSCs are valid cell- based disease models with characteristic disease phenotypes that can be used for study of disease pathophysiology and drug development. 30442709_High PPT1 expression is associated with cancer. 30541466_the study contributes four novel variants to the spectrum of PPT1 gene mutations and eight novel variants to the TPP1 gene mutation data in neuronal ceroid lipofuscinoses type I and type II 33561134_Human INCL fibroblasts display abnormal mitochondrial and lysosomal networks and heightened susceptibility to ROS-induced cell death. | ENSMUSG00000028657 | Ppt1 | 4739.09420 | 4.510100e-01 | -1.148769 | 0.2930297 | 15.07401 | 1.033771e-04 | 3.325513e-02 | Yes | No | 2.668961e+03 | 4.020733e+02 | 5.891035e+03 | 9.087143e+02 | |
| ENSG00000132196 | 51478 | HSD17B7 | protein_coding | P56937 | FUNCTION: Bifunctional enzyme involved in steroid-hormone metabolism and cholesterol biosynthesis (PubMed:12574203, PubMed:12732193, PubMed:12829805, PubMed:20659585, PubMed:19772289, PubMed:11165030). Catalyzes the NADP(H)-dependent reduction of estrogens and androgens and regulates the biological potency of these steroids. Converts estrone (E1) to a more potent estrogen, 17beta-estradiol (E2) (PubMed:12574203, PubMed:12732193, PubMed:19772289). Converts dihydrotestosterone (DHT) to its inactive form 5a-androstane-3b,17b-diol (PubMed:12574203, PubMed:12732193, PubMed:19772289). Converts moderately progesterone to 3beta-hydroxypregn-4-ene-20-one, leading to its inactivation (PubMed:12574203, PubMed:12732193). Additionally, participates in the post-squalene cholesterol biosynthesis, as a 3-ketosteroid reductase (PubMed:12829805, PubMed:20659585, PubMed:11165030). {ECO:0000269|PubMed:11165030, ECO:0000269|PubMed:12574203, ECO:0000269|PubMed:12732193, ECO:0000269|PubMed:12829805, ECO:0000269|PubMed:19772289, ECO:0000269|PubMed:20659585}.; FUNCTION: [Isoform 3]: Does not have enzymatic activities toward E1 and DHT. {ECO:0000269|PubMed:12732193}. | Alternative splicing;Endoplasmic reticulum;Glycoprotein;Lipid biosynthesis;Lipid metabolism;Membrane;NAD;NADP;Oxidoreductase;Reference proteome;Steroid biosynthesis;Transmembrane;Transmembrane helix | PATHWAY: Steroid biosynthesis; estrogen biosynthesis. {ECO:0000269|PubMed:12574203, ECO:0000269|PubMed:12732193, ECO:0000269|PubMed:19772289}.; PATHWAY: Steroid biosynthesis; zymosterol biosynthesis; zymosterol from lanosterol: step 5/6. {ECO:0000269|PubMed:11165030, ECO:0000269|PubMed:12829805}. | HSD17B7 encodes an enzyme that functions both as a 17-beta-hydroxysteroid dehydrogenase (EC 1.1.1.62) in the biosynthesis of sex steroids and as a 3-ketosteroid reductase (EC 1.1.1.270) in the biosynthesis of cholesterol (Marijanovic et al., 2003 [PubMed 12829805]).[supplied by OMIM, May 2010]. | hsa:51478; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; 17-beta-hydroxysteroid dehydrogenase (NADP+) activity [GO:0072582]; 3-keto sterol reductase activity [GO:0000253]; 5alpha-androstane-3beta,17beta-diol dehydrogenase activity [GO:0047024]; cycloeucalenone reductase activity [GO:0102176]; estradiol 17-beta-dehydrogenase activity [GO:0004303]; androgen metabolic process [GO:0008209]; cholesterol biosynthetic process [GO:0006695]; estrogen biosynthetic process [GO:0006703] | 12829805_HSD17B7 is a novel candidate for inborn errors of cholesterol metabolism 15862973_Comparison of the promoter region of the human and murine gene. 16356630_The identified proximal promoter regions of both human and murine HSD17B7 genes contain multiple transcription factor binding sites and show strong similarity to cholesterogenic genes. 16901934_Results provide unequivocal evidence for a role of 17beta-hydroxysteroid dehydrogenase type-7 in cholesterol biosynthesis. 17454161_17beta-hydroxysteroid dehydrogenase type 7(17beta-HSD type 7)was significantly upregulated in ovarian tissue of patients with ovarian endometriosis. 17498944_17-beta hydroxysteroid dehydrogenase type 7 (HSD17B7) -shRNA sequences were designed and tested for their effectiveness. 18660489_Observational study of gene-disease association. (HuGE Navigator) 19019335_Observational study of gene-disease association. (HuGE Navigator) 19429442_determined the activity and expression levels of known estrogenic 17beta-HSDs, namely types 1, 7 and 12 17beta-HSD in preadipocytes before and after differentiation into mature adipocytes 19460435_There is no difference in catalytic properties between variants of 17beta-HSD types 7 and 12 and wild-type enzymes, while variants p.Glu77Gly and p.Lys183Arg in 17beta-HSD type 5 showed a slightly decreased activity. 19536175_Observational study of gene-disease association. (HuGE Navigator) 19735314_The transcriptional activity of the HSD17B7 gene containing the G allele is higher than that of the C allele. This difference in HSD17B7 expression may regulate the risk of peripheral edema as an adverse reaction induced by estramustine phosphate sodium 20140262_Observational study of gene-disease association. (HuGE Navigator) 20214802_Observational study of gene-disease association. (HuGE Navigator) 20215536_increased expression of HSD17B7 is associated with breast cancer. 21086175_Data show that apicidin significantly lowers HSD17B1 transcript and protein levels in endometrial adenocarcinoma cells, with no significant effect on HSD17B1 transcript stability. 21372145_estradiol stimulates HSD17B7 transcriptional activity in breast cancer cells through a novel mechanism requiring NF1 and strongly suggest a positive feedback mechanism, increasing estradiol synthesis causing growth of estrogen-dependent breast cancers 23300627_17betaHSD7 is not the key enzyme responsible for androstenone and testosterone metabolism in porcine liver cells 25257817_17beta-HSD1 and 17beta-HSD7 are principal reductive 17beta-hydroxysteroid dehydrogenases and major players in the viability of estrogen-dependent breast cancer cells. 25966904_the dual functional 17beta-HSD7 is proposed as a novel target for estrogen-dependent breast cancer by regulating the balance of estradiol and dihydrotestosterone. 28554725_Substrate inhibition of 17beta-HSD1 in tumor epithelial cells and regulation of 17beta-HSD7 by 17beta-HSD1 knockdown has been demonstrated. 28645527_Inhibition of 17beta-HSD 7 modulates breast cancer protein profile and enhances apoptosis by down-regulating GRP78. 30227243_Substrate affinity of 17beta-hydroxysteroid dehydrogenase type 7 toward estrone and 5alpha-dihydrotestosterone are similar. 31207361_Inhibition of 17beta-HSD7can provide a new basis for the design of combined endocrine therapy for breast cancer by controlling E2 and DHT, while up-regulating AR expression and stably controlling expression of ERalpha. 34185380_HSD17B7 gene in self-renewal and oncogenicity of keratinocytes from Black versus White populations. | ENSMUSG00000026675 | Hsd17b7 | 356.65624 | 4.197525e-01 | -1.252389 | 0.2951172 | 17.17664 | 3.405983e-05 | 1.502622e-02 | Yes | No | 2.091397e+02 | 3.290914e+01 | 4.485588e+02 | 7.117178e+01 |
| ENSG00000134419 | 6210 | RPS15A | protein_coding | P62244 | FUNCTION: Structural component of the ribosome (PubMed:23636399). Required for proper erythropoiesis (PubMed:27909223). {ECO:0000269|PubMed:23636399, ECO:0000269|PubMed:27909223}. | 3D-structure;Diamond-Blackfan anemia;Direct protein sequencing;Reference proteome;Ribonucleoprotein;Ribosomal protein | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 40S subunit. The protein belongs to the S8P family of ribosomal proteins. It is located in the cytoplasm. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome. [provided by RefSeq, Jul 2008]. | hsa:6210; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; cytosolic ribosome [GO:0022626]; cytosolic small ribosomal subunit [GO:0022627]; extracellular exosome [GO:0070062]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; cytoplasmic translation [GO:0002181]; positive regulation of cell cycle [GO:0045787]; positive regulation of cell population proliferation [GO:0008284]; response to virus [GO:0009615]; translation [GO:0006412] | 24334120_RPS15A may play a prominent role in heptocarcinogenesis and serve as a potential therapeutic target in hepatocellular carcinoma. 25833696_we identify RPS15A as a novel potential oncogenic gene involved in lung carcinogenesis. 26537582_Our findings indicate that RPS15A promotes cell proliferation and migration in glioblastoma for the first time. RPS15A might play a distinct role in glioblastoma and serve as a potential target for therapy 26847263_Results show that RPS15A is overexpressed in colorectal cancer (CRC) tissue and predicts a worse prognosis and outcome of CRC patients through misregulation of cell cycle progression and promotion of metastasis. 27035327_the present study provides novel evidence indicating that RPS15A modulates AML cell growth in vitro, and may be considered a novel therapeutic target for the treatment of AML. 27130037_High RPS15A expression is associated with glioblastoma. 27909223_RPS15A is a novel causative gene for Diamond-Blackfan anemia. 29242604_High RPS15A expression is associated with angiogenesis in hepatocellular carcinoma. 30450850_The present study found RPS15a expression to be higher in gastric tumors and its silencing in gastric cancer cells to inhibit the proliferation, growth, and migration thereof. Accordingly, RPS15a may be considered as a potential therapeutic target in gastric cancer. 30569143_A preliminary experimental basis for RPS15A as a novel oncogene and potential therapeutic target in RCC. 30661291_Ribosomal protein S15a (RPS15A) is up-regulated in gastric cancer (GC) patients and associated with poor prognosis. 31303129_miR-29a-3p inhibits CRC cell function possibly by targeting RPS15A 31535410_Ribosomal protein small subunit 15A (RPS15A) inhibits the apoptosis of breast cancer MDA-MB-231 cells via upregulating phosphorylated ERK1/2, Bad, and Chk1. 31665807_MicroRNA-147b suppresses the proliferation and invasion of non-small-cell lung cancer cells through downregulation of Wnt/beta-catenin signalling via targeting of RPS15A. 34156695_Ribosomal protein RPS15A augments proliferation of colorectal cancer RKO cells via regulation of BIRC3, p38 MAPK and Chk1. | 22061.15520 | 3.853507e-01 | -1.375756 | 0.3535788 | 14.28484 | 1.571256e-04 | 4.183064e-02 | Yes | No | 1.468258e+04 | 3.407659e+03 | 2.921342e+04 | 6.953978e+03 | |||
| ENSG00000139131 | 51067 | YARS2 | protein_coding | Q9Y2Z4 | FUNCTION: Catalyzes the attachment of tyrosine to tRNA(Tyr) in a two-step reaction: tyrosine is first activated by ATP to form Tyr-AMP and then transferred to the acceptor end of tRNA(Tyr). {ECO:0000269|PubMed:15779907, ECO:0000269|PubMed:17997975}. | 3D-structure;ATP-binding;Acetylation;Aminoacyl-tRNA synthetase;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Primary mitochondrial disease;Protein biosynthesis;Reference proteome;Transit peptide | This gene encodes a mitochondrial protein that catalyzes the attachment of tyrosine to tRNA(Tyr). Mutations in this gene are associated with myopathy with lactic acidosis and sideroblastic anemia type 2 (MLASA2). [provided by RefSeq, Jan 2011]. | hsa:51067; | cytosol [GO:0005829]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; nuclear body [GO:0016604]; ATP binding [GO:0005524]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; tRNA binding [GO:0000049]; tyrosine binding [GO:0072545]; tyrosine-tRNA ligase activity [GO:0004831]; mitochondrial tyrosyl-tRNA aminoacylation [GO:0070184]; translation [GO:0006412]; tRNA aminoacylation [GO:0043039] | 15779907_The gene for mitochondrial tyrosyl-tRNA synthetase is described and the initial characterization of the enzyme is reported. Genes for the remaining missing synthetases have also been found with the exception of human glutaminyl-tRNA synthetase. 15840810_first example of a TyrRS lacking specificity toward N1-N72 and thus of a TyrRS disobeying the identity rules 17672918_the apparent occurrence of an unusual TG 3' splice site in intron 5 is discussed 17997975_the structure of a strictly mitochondrial human synthetase, namely tyrosyl-tRNA synthetase (mt-TyrRS), in complex with an adenylate analog at 2.2 A resolution 20598274_The YARS2 mutation reported here is an alternative cause of MLASA. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22504945_The study confirms mutations in YARS2 as a cause of MLASA and shows that, like some of the cytoplasmic ARSs, mitochondrial ARSs occur in high-molecular-weight complexes. 24344687_Data identified novel YARS2 mutations and noted marked phenotypic variability among YARS2 MLASA patients, with phenotypes ranging from mild to lethal, suggesting that the background mtDNA haplotype may be contributing to the phenotypic variability. 26647310_The mutation in YARS2 gene is a nuclear modifier for the phenotypic manifestation of Leber's hereditary optic neuropathy-associated mitochondrial DNA mutation. 28395030_The p.Leu392Ser variant is likely a newly identified founder YARS2 mutation in mitochondrial myopathy. 29976739_tyrosyl-tRNA synthetase 2 tyrosine 30026338_Data suggest that biallelic tyrosyl-tRNA synthetase 2 (YARS2) variants, including severe loss-of-function alleles should be considered as a cause of isolated congenital sideroblastic anemia, as well as the myopathy, lactic acidosis and sideroblastic anemia 2 (MLASA2) syndromic phenotype. 31685661_findings provide molecular-level insights into the pathophysiology of maternally transmitted deafness arising from the synergy between tRNA(Ser(UCN)) and mitochondrial YARS mutations | ENSMUSG00000022792 | Yars2 | 213.79214 | 3.352900e-01 | -1.576519 | 0.3894140 | 15.80742 | 7.012693e-05 | 2.647013e-02 | Yes | Yes | 9.304249e+01 | 2.489372e+01 | 2.836681e+02 | 7.754668e+01 | |
| ENSG00000142396 | 105372481 | ERVK3-1 | protein_coding | A0A0U1RQV1 | Proteomics identification;Reference proteome | 1241.63639 | 4.669699e-01 | -1.098599 | 0.2638419 | 16.53945 | 4.764834e-05 | 1.988481e-02 | Yes | No | 7.534418e+02 | 1.064949e+02 | 1.504350e+03 | 2.169944e+02 | ||||||||
| ENSG00000143106 | 5686 | PSMA5 | protein_coding | P28066 | FUNCTION: Component of the 20S core proteasome complex involved in the proteolytic degradation of most intracellular proteins. This complex plays numerous essential roles within the cell by associating with different regulatory particles. Associated with two 19S regulatory particles, forms the 26S proteasome and thus participates in the ATP-dependent degradation of ubiquitinated proteins. The 26S proteasome plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins that could impair cellular functions, and by removing proteins whose functions are no longer required. Associated with the PA200 or PA28, the 20S proteasome mediates ubiquitin-independent protein degradation. This type of proteolysis is required in several pathways including spermatogenesis (20S-PA200 complex) or generation of a subset of MHC class I-presented antigenic peptides (20S-PA28 complex). {ECO:0000269|PubMed:15244466, ECO:0000269|PubMed:27176742, ECO:0000269|PubMed:8610016}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Glycoprotein;Nucleus;Phosphoprotein;Proteasome;Reference proteome | The proteasome is a multicatalytic proteinase complex with a highly ordered ring-shaped 20S core structure. The core structure is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides. This gene encodes a member of the peptidase T1A family, that is a 20S core alpha subunit. Multiple alternatively spliced transcript variants encoding two distinct isoforms have been found for this gene. [provided by RefSeq, Dec 2010]. | hsa:5686; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; proteasome complex [GO:0000502]; proteasome core complex [GO:0005839]; proteasome core complex, alpha-subunit complex [GO:0019773]; secretory granule lumen [GO:0034774]; proteasomal protein catabolic process [GO:0010498]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161] | 11771738_Selective upregulation of the ubiquitin-proteasome proteolytic pathway proteins, proteasome zeta chain and isopeptidase T in fetal Down syndrome. 30738879_increased ubiquitin B mRNA expression followed by significant downregulation of proteasome subunits; 26S proteasome non-ATPase regulatory subunit 1, and proteasome subunit alpha-type 5 was found in pseudoexfoliation syndrome (PEXS) individuals. Decrease in chymotrypsin-like proteasome activity and increased apoptosis were also observed in PEX subjects. 30807553_PSMA5 promotes the tumorigenic process of prostate cancer and is related to bortezomib resistance. 34831298_Conserved Mitotic Phosphorylation of a Proteasome Subunit Regulates Cell Proliferation. | ENSMUSG00000068749 | Psma5 | 3859.04079 | 3.561063e-01 | -1.489620 | 0.3005030 | 23.49188 | 1.254422e-06 | 9.684766e-04 | Yes | No | 2.056692e+03 | 4.079561e+02 | 5.448448e+03 | 1.106975e+03 | |
| ENSG00000146530 | 221806 | VWDE | protein_coding | Q8N2E2 | Alternative splicing;Disulfide bond;EGF-like domain;Glycoprotein;Reference proteome;Repeat;Secreted;Signal | hsa:221806; | cell surface [GO:0009986]; extracellular region [GO:0005576]; signaling receptor binding [GO:0005102]; anatomical structure development [GO:0048856] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | 85.59903 | 2.964665e-01 | -1.754059 | 0.4560492 | 14.13174 | 1.704437e-04 | 4.386369e-02 | Yes | Yes | 3.639919e+01 | 9.809313e+00 | 1.258631e+02 | 3.426037e+01 | |||||
| ENSG00000147162 | 8473 | OGT | protein_coding | O15294 | FUNCTION: Catalyzes the transfer of a single N-acetylglucosamine from UDP-GlcNAc to a serine or threonine residue in cytoplasmic and nuclear proteins resulting in their modification with a beta-linked N-acetylglucosamine (O-GlcNAc) (PubMed:26678539, PubMed:23103939, PubMed:21240259, PubMed:21285374, PubMed:15361863). Glycosylates a large and diverse number of proteins including histone H2B, AKT1, ATG4B, EZH2, PFKL, KMT2E/MLL5, MAPT/TAU and HCFC1 (PubMed:19451179, PubMed:20200153, PubMed:21285374, PubMed:22923583, PubMed:23353889, PubMed:24474760, PubMed:26678539, PubMed:27527864). Can regulate their cellular processes via cross-talk between glycosylation and phosphorylation or by affecting proteolytic processing (PubMed:21285374). Probably by glycosylating KMT2E/MLL5, stabilizes KMT2E/MLL5 by preventing its ubiquitination (PubMed:26678539). Involved in insulin resistance in muscle and adipocyte cells via glycosylating insulin signaling components and inhibiting the 'Thr-308' phosphorylation of AKT1, enhancing IRS1 phosphorylation and attenuating insulin signaling (By similarity). Involved in glycolysis regulation by mediating glycosylation of 6-phosphofructokinase PFKL, inhibiting its activity (PubMed:22923583). Component of a THAP1/THAP3-HCFC1-OGT complex that is required for the regulation of the transcriptional activity of RRM1. Plays a key role in chromatin structure by mediating O-GlcNAcylation of 'Ser-112' of histone H2B: recruited to CpG-rich transcription start sites of active genes via its interaction with TET proteins (TET1, TET2 or TET3) (PubMed:22121020, PubMed:23353889). As part of the NSL complex indirectly involved in acetylation of nucleosomal histone H4 on several lysine residues (PubMed:20018852). O-GlcNAcylation of 'Ser-75' of EZH2 increases its stability, and facilitating the formation of H3K27me3 by the PRC2/EED-EZH2 complex (PubMed:24474760). Regulates circadian oscillation of the clock genes and glucose homeostasis in the liver. Stabilizes clock proteins ARNTL/BMAL1 and CLOCK through O-glycosylation, which prevents their ubiquitination and subsequent degradation. Promotes the CLOCK-ARNTL/BMAL1-mediated transcription of genes in the negative loop of the circadian clock such as PER1/2 and CRY1/2 (PubMed:12150998, PubMed:19451179, PubMed:20018868, PubMed:20200153, PubMed:21285374, PubMed:15361863). O-glycosylates HCFC1 and regulates its proteolytic processing and transcriptional activity (PubMed:21285374, PubMed:28584052, PubMed:28302723). Regulates mitochondrial motility in neurons by mediating glycosylation of TRAK1 (By similarity). Glycosylates HOXA1 (By similarity). O-glycosylates FNIP1 (PubMed:30699359). Promotes autophagy by mediating O-glycosylation of ATG4B (PubMed:27527864). {ECO:0000250|UniProtKB:P56558, ECO:0000250|UniProtKB:Q8CGY8, ECO:0000269|PubMed:12150998, ECO:0000269|PubMed:15361863, ECO:0000269|PubMed:19451179, ECO:0000269|PubMed:20018852, ECO:0000269|PubMed:20018868, ECO:0000269|PubMed:20200153, ECO:0000269|PubMed:21240259, ECO:0000269|PubMed:21285374, ECO:0000269|PubMed:22121020, ECO:0000269|PubMed:22923583, ECO:0000269|PubMed:23103939, ECO:0000269|PubMed:23353889, ECO:0000269|PubMed:24474760, ECO:0000269|PubMed:26678539, ECO:0000269|PubMed:27527864, ECO:0000269|PubMed:28302723, ECO:0000269|PubMed:28584052, ECO:0000269|PubMed:30699359}.; FUNCTION: [Isoform 2]: The mitochondrial isoform (mOGT) is cytotoxic and triggers apoptosis in several cell types including INS1, an insulinoma cell line. {ECO:0000269|PubMed:20824293}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Biological rhythms;Cell membrane;Cell projection;Chromatin regulator;Cytoplasm;Direct protein sequencing;Disease variant;Glycoprotein;Glycosyltransferase;Host-virus interaction;Lipid-binding;Membrane;Mental retardation;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:15361863, ECO:0000269|PubMed:21240259, ECO:0000269|PubMed:21285374, ECO:0000269|PubMed:23103939, ECO:0000269|PubMed:26678539}. | This gene encodes a glycosyltransferase that catalyzes the addition of a single N-acetylglucosamine in O-glycosidic linkage to serine or threonine residues. Since both phosphorylation and glycosylation compete for similar serine or threonine residues, the two processes may compete for sites, or they may alter the substrate specificity of nearby sites by steric or electrostatic effects. The protein contains multiple tetratricopeptide repeats that are required for optimal recognition of substrates. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Oct 2009]. | hsa:8473; | cell projection [GO:0042995]; cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; mitochondrial membrane [GO:0031966]; NSL complex [GO:0044545]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein N-acetylglucosaminyltransferase complex [GO:0017122]; protein-containing complex [GO:0032991]; acetylglucosaminyltransferase activity [GO:0008375]; phosphatidylinositol-3,4,5-trisphosphate binding [GO:0005547]; protein N-acetylglucosaminyltransferase activity [GO:0016262]; protein O-GlcNAc transferase activity [GO:0097363]; apoptotic process [GO:0006915]; chromatin organization [GO:0006325]; circadian regulation of gene expression [GO:0032922]; histone H3-K4 trimethylation [GO:0080182]; histone H4-K16 acetylation [GO:0043984]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; negative regulation of protein ubiquitination [GO:0031397]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of histone H3-K27 methylation [GO:0061087]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of proteolysis [GO:0045862]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein O-linked glycosylation [GO:0006493]; protein processing [GO:0016485]; regulation of dosage compensation by inactivation of X chromosome [GO:1900095]; regulation of gluconeogenesis [GO:0006111]; regulation of glycolytic process [GO:0006110]; regulation of insulin receptor signaling pathway [GO:0046626]; regulation of necroptotic process [GO:0060544]; regulation of Rac protein signal transduction [GO:0035020]; regulation of transcription by RNA polymerase II [GO:0006357]; response to insulin [GO:0032868]; response to nutrient [GO:0007584]; signal transduction [GO:0007165] | 11773972_We have delineated the complete genomic structure of human OGT spanning approx 43 kb of genomic DNA in Xq13.1 11846551_homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily 12136128_O-linked GlcNAc transferase participates in a hexosamine-dependent signaling pathway that is linked to insulin resistance and leptin production 14601650_a novel HLA-A0201-restricted cytotoxic T lymphocyte (CTL)-epitope (28-SLYKFSPFPL; FSP06) derived from a mutant OGT-protein 15336570_OGT can respond rapidly to heat stress through the enhancement of nucleocytoplasmic protein O-GlcNAcylation. 15561949_Staining of OGT in streptozotocin diabetic rat liver is clearly diminished, but it was substantially restored after 6 days of insulin treatment 15795231_By using a series of 4-methylumbelliferyl 2-deoxy-2-N-fluoroacetyl-beta-D-glucopyranoside substrates, Taft-like linear free energy analyses of these enzymes indicates that O-GlcNAcase uses a catalytic mechanism involving anchimeric assistance 15896326_Thus, stably transfected HeLa cells provide an abundant source of enzyme that can be used to study the structure, function, and regulation of OGT. 16105839_analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase 16966374_Overall, transcriptional inhibition is related to the integrated effect of O-GlcNAc by direct modification of critical elements of the transcriptome and indirectly through O-GlcNAc modification of the proteasome. 18174169_O-GlcNAc modification stimulated by glucose deprivation results from increased OGT and decreased O-GlcNAcase levels and that these changes affect cell metabolism, thus inactivating glycogen synthase. 18536723_The structure of an intact OGT homolog and kinetic analysis of human OGT variants reveal a contiguous superhelical groove that directs substrates to the active site. 18653473_the O-GlcNAc cycling enzymes associate with kinases and phosphatases at M phase to regulate the posttranslational status of vimentin 19073609_Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. 20068230_Data show that forced overexpression of OGT increased the inhibitory phosphorylation of CDK1 and reduced the phosphorylation of CDK1 target proteins. 20190804_OGT regulates breast cancer tumorignenesis and cancer growth through targeting FixM1. 20200153_THAP1 was found to bind HCF-1 in vitro and to associate with HCF-1 and OGT in vivo. 20206135_OGT could be a co-regulatory subunit shared by functionally distinct complexes supporting epigenetic regulation of MIP-1alpha gene promoter. 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20805223_regulating the amount of OGT during mitosis is important in ensuring correct chromosomal segregation during mitosis. 20824293_Enhanced OGT expression efficiently triggered programmed cell death. 20845477_Observational study of gene-disease association. (HuGE Navigator) 20876116_OGT deletion in infarcted mice significantly exacerbated cardiac dysfunction. 21240259_two crystal structures of human OGT, as a binary complex with UDP (2.8 A resolution) and as a ternary complex with UDP and a peptide substrate (1.95 A). 21327254_Data identify Tau as potential substrates for the O-beta-N-acetylglucosaminyltransferase (OGT). 21567137_Decrease in MGEA5 and increase in O-GlcNAc transferease expression in higher grade tumors suggests that increased O-GlcNAc modification may be implicated in breast tumor progression and metastasis. 22275356_as prostate cancer cells alter glucose and glutamine levels, O-GlcNAc modifications and OGT levels become elevated and are required for regulation of malignant properties 22294689_a 154-amino acid region of MIBP1 was necessary for its O-GlcNAc transferase binding and O-GlcNAcylation. 22311971_Data show that the interplay between O-GlcNAc and phosphorylation on proteins and indicate that these effects can be mediated by changes in hOGT and hOGA kinetic activity. 22371499_These studies identify a molecular mechanism of GR transrepression, and highlight the function of O-GlcNAc in hormone signaling. 22384635_O-GlcNAcylation may be an important regulatory modification involved in endometrial cancer pathogenesis but the actual significance of this modification for endometrial cancer progression needs to be investigated further. 22496241_Hsp90 is involved in the regulation of OGT and O-GlcNAc modification and that Hsp90 inhibitors might be used to modulate O-GlcNAc modification and reverse its adverse effects in human diseases. 22574218_AMPK functions as a physiological suppressor of 26S proteasomes through OGT 22783592_Analysis of urinary content of MGEA5 and OGT may be useful for bladder cancer diagnostics. 22883232_O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. 23103939_we describe structural snapshots of all species along the kinetic pathway for human O-linked beta-N-acetylglucosamine transferase (O-GlcNAc transferase), an intracellular enzyme that catalyzes installation of a dynamic post-translational modification 23103942_we define how human OGT recognizes the sugar donor and acceptor peptide and uses a new catalytic mechanism of glycosyl transfer, involving the sugar donor alpha-phosphate as the catalytic base as well as an essential lysine 23152511_The human respiratory syncytial virus-induced sequestration of p38-P in IBs resulted in a substantial reduction in the accumulation of a downstream signaling substrate, MAPK-activated protein kinase 2 (MK2). 23222540_The double epigenetic modifications on both DNA and histones by TET2 and OGT coordinate together for the regulation of gene transcription. 23487789_These studies identified OGT as a promising placental biomarker of maternal stress exposure that may relate to sex-biased outcomes in neurodevelopment. 23642195_Data suggest that changes in OGT (O-linked N-acetylglucosamine transferase) and OGA (peptide O-linked N-acetylglucosamine-beta-N-acetylglucosaminidase) expression are correlated with cancer prognosis. [REVIEW] 23700425_The backbone carbonyl oxygen of Leu653 and the hydroxyl group of Thr560 in OGT contribute to the recognition of sugar moieties via hydrogen bonds. 23720054_Expression of c-MYC and OGT was tightly correlated in human prostate cancer samples. 24256146_Data suggest that with multi-substrate enzymes, such as OGT, specific inhibition can rarely be achieved with ligands that compete solely with one of the substrates; OGT is inhibited by bisubstrate UDP-oligopeptide conjugates. 24311690_study reports the tetratricopeptide-repeat domain of O-GlcNAc transferase binds the carboxyl-terminal portion of an HCF-1 proteolytic repeat such that the cleavage region lies in the glycosyltransferase active site above uridine diphosphate-GlcNAc; protein glycosylation and HCF-1 cleavage occur in the same active site 24365779_Estrogen replacement therapy and plyometric training influence muscle OGT and OGA gene expression, which may be one of the mechanisms by which HRT and PT prevent aging-related loss of muscle mass. 24394411_OGT catalyzes the O-GlcNAcylation of TET3, promotes TET3 nuclear export, and, consequently, inhibits the formation of 5-hydroxymethylcytosine catalyzed by TET3. 24580054_Endogenous OTX2 from a medulloblastoma cell line is O-GlcNAcylated at several sites. 24928395_Instead, an adipogenesis-dependent increase in O-linked beta-N-acetylglucosamine (O-GlcNAc) glycosylation of EWS was observed. 25173736_Amino acid composition of splice variants, post-translational modifications, and stable associations with regulatory proteins influence subcellular distribution/substrate specificity of OGT and OGA (O-GlcNAcase beta-N-acetylglucosaminidase). [REVIEW] 25419848_O-linked beta-N-acetylglucosamine transferase mediates O-GlcNAcylation of the SNARE protein SNAP-29 and regulates autophagy in a nutrient-dependent manner. 25568311_Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). 25663381_Hexosamine biosynthetic pathway flux is increased in idiopathic pulmonary artery hypertension and drives OGT-facilitated pulmonary artery smooth muscle cell proliferation via specific proteolysis and direct activation of host cell factor-1. 25773598_Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. 25776937_miR4235p was associated with congestive heart failure and the expression levels of proBNP; in addition, OGT was found to be a direct target of miR4235p. 26041297_Inhibition of O-Linked N-Acetylglucosamine Transferase Reduces Replication of Herpes Simplex Virus and Human Cytomegalovirus. 26237509_This work reveals that although the N-terminal TPR repeats of OGT may have roles in substrate recognition, the sequence restriction imposed by the peptide-binding site makes a substantial contribution to O-GlcNAc site specificity. 26240142_Use of OGT(C917A) enhances O-GlcNDAz production, yielding improved cross-linking of O-GlcNDAz-modified molecules 26252736_These results suggested roles of O-GlcNAcylation in modulating serine phosphorylation, as well as in regulating PKM2 activity and expression. 26305326_These results demonstrate that distinct OGT-binding sites in HCF-1 promote proteolysis, and provide novel insights into the mechanism of this unusual protease activity. 26397041_We concluded that OGT plays a key role in gastric cancer proliferation and survival, and could be a potential target for therapy. 26399441_OGT expression is increased under hypoxic conditions. 26408091_a new function of histone O-GlcNAcylation in DNA damage response 26527687_E2F1 negatively regulates both Ogt and Mgea5 expression in an Rb1 protein-dependent manner. 26707622_OGT inhibited the formation of the Ecadherin/catenin complex through reducing the interaction between p120 and Ecadherin. 26807597_Data suggest RNA polymerase II (POLR2A) is extensively modified on its unique C-terminal domain (CTD) by O-GlcNAc transferase (OGT); efficient O-GlcNAcylation requires a minimum of 20 heptad CTD repeats in POLR2A and more than half of NTD of OGT. 26854602_Together, these findings suggest that induction of SNO-OGT by Ab exposure is a pathogenic mechanism to cause cellular hypo-O-GlcNAcylation by which Ab neurotoxicity is executed 27060025_The findings suggest that OGT promotes the O-GlcNAc modification of HDAC1 in the development of progression hepatocellular carcinoma. 27129214_data indicate that O-GlcNAc-transferase activity is essential for RNA pol II promoter recruitment and that pol II goes through a cycling of O-GlcNAcylation at the promoter 27131860_O-GlcNAcylation expression and its nuclear expression were associated with the carcinogenesis and progression of gastric carcinoma. 27217568_These results support a model in which OGT modifies HIRA to regulate HIRA-H3.3 complex formation and H3.3 nucleosome assembly and reveal the mechanism by which OGT functions in cellular senescence. 27231347_the O-linked N-acetylglucosamine (O-GlcNAc) processing enzymes, O-GlcNAc-transferase (OGT) and O-GlcNAcase (OGA), interact with the (A)gamma-globin promoter at the -566 GATA repressor site 27294441_Beyond its well-known role in adding beta-O-GlcNAc to serine and threonine residues of nuclear and cytoplasmic proteins, OGT also acts as a protease in the maturation of the cell cycle regulator, HCF-1, and serves as an integral member of several protein complexes, many of them linked to gene expression. (Review) 27331873_Findings indicate O-linked N-acetylglucosamine transferase (OGT) as a cellular factor involved in human papillomaviruses type 16/18 E6 and E7 expressions and cervical cancer tumorigenesis, suggesting that targeting OGT in cervical cancer may have potential therapeutic benefit. 27505673_This work uncovers that URI-regulated OGT confers c-MYC-dependent survival functions in response to glucose fluctuations. 27705803_We identified two human PRC2 complexes and two PR-DUB deubiquitination complexes, which contain the O-linked N-acetylglucosamine transferase OGT1 and several transcription factors. 27845045_OGT functions in metastatic spread of HPV E6/E7-positive HeLa cells to xenografted lungs through E6/E7, HCF-1 and CXCR4 28100784_Reducing endogenous mitochondrial OGT expression leads to alterations in mitochondrial structure and function, including Drp1-dependent mitochondrial fragmentation, reduction in mitochondrial membrane potential, and a significant loss of mitochondrial content in the absence of mitochondrial reactive oxygen species. 28115479_The results of this study showed that the OGT is essential for sensory neuron survival and target innervation. 28232487_Fatty acid synthase fine-tunes the cell's response to stress and injury by remodeling cellular O-GlcNAcylation 28302723_Thus, a single amino acid substitution in the regulatory domain (the tetratricopeptide repeat domain) of OGT, which catalyzes the O-GlcNAc post-translational modification of nuclear and cytosolic proteins, appears causal for X-linked intellectual disability. 28347804_OGT, a unique glycosyltransferase enzyme, was identified to be upregulated in non-alcoholic fatty liver disease-associated hepatocellular carcinoma tissues by transcriptome sequencing. Here, we found that OGT plays a role in cancer by promoting tumor growth and metastasis in cell models. This effect is mediated by the induction of palmitic acid. 28450392_Data suggest that O-GlcNAc transferase 1 (OGT1) specifically binds to, O-GlcNAcylates, and stabilizes nonspecific lethal protein3 (NSL3); stabilization of NSL3 by OGT1 up-regulates global acetylation levels of histone 4 at Lys-5, Lys-8, and Lys-16. 28455227_conclusion, our results demonstrated that miR24 inhibits breast cancer cells invasion by targeting OGT and reducing FOXA1 stability. These results also indicated that OGT might be a potential target for the diagnosis and therapy of breast cancer metastasis. 28584052_Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability 28625484_Nrf1 is regulated by O-GlcNAc transferase. 28663241_The authors show that O-GlcNAcylation of KEAP1 by OGT at serine 104 is required for the efficient ubiquitination and degradation of NRF2. 28742148_Tax interacts with the host OGT/OGA complex and inhibits the activity of OGT-bound OGA. 28929346_These data predict that under conditions where O-GlcNAc levels are high (breast cancer) progesterone receptor (PR) through an interaction with the modifying enzyme OGT, will exhibit increased O-GlcNAcylation and potentiated transcriptional activity. Therapeutic strategies aimed at altering cellular O-GlcNAc levels may have profound effects on PR transcriptional activity in breast cancer 29059153_High OGT expression promotes cancer lipid metabolism via SREBP-1 regulation. 29208956_Findings demonstrate a novel role of Poleta O-GlcNAcylation by OGT in translesion DNA synthesis regulation and genome stability maintenance. 29465778_The O-GlcNAc transferase OGT interacts with and post-translationally modifies the transcription factor HOXA1. 29556021_a novel ASXL1-OGT axis and raise the possibility that this axis has a tumor-suppressor role in myeloid malignancies. 29577901_LXRalpha interacts with OGT in its N-terminal domain and ligand-binding domain (LBD) in a ligand-independent fashion. 29606577_The intellectual disability L254F mutation in OGT affects activity. The L254F mutation leads to shifts up to 12 A in the OGT structure. Thermal denaturing studies reveal reduction in tetratricopeptide repeat domain stability caused by L254F. L254F OGT mutation leads to conformational changes of the tetratricopeptide repeats and reduced activity, revealing the molecular mechanisms contributing to pathogenesis. 29769320_O-GlcNAc transferase missense mutations is associated with X-linked intellectual disability. 29788742_miR-483 inhibited the expression level of OGT mRNA by direct binding to its 3'-untranslated region. Expression of miR-483 was negatively correlated with OGT in gastric cancer tissues. 29967448_Study found that levels of placental Ogt determine sex differences in fetally derived placental trophoblast transcriptome profiles associated with key developmental processes and shape genome-wide patterning of the ubiquitous epigenetic transcriptional repressive mark, H3K27me3. 30069701_O-GlcNAc transferase (OGT) is a partner of the MCM2-7 complex and O-GlcNAcylation might regulate MCM2-7 complex by regulating the chromatin loading of MCM6 and MCM7 and stabilizing MCM/MCM interactions. 30106436_High OGT expression is associated with breast cancer. 30453909_O-GlcNAcylation and the expression of O-linked-beta-N-acetylglucosamine transferase (OGT) were upregulated in bladder cancer cell lines and tissue specimens. 30543776_these findings provide a mechanistic link between the OGT-mediated glucose metabolic pathway and antiviral innate immune signaling by targeting MAVS, and expand our current understanding of importance of glucose metabolic regulation in viral infection-associated diseases. 30550897_The ligand transport mechanism in the OGT enzymatic process is described here and is a great resource for designing inhibitors based on UDP or UDP-GlcNAc. 30555541_These findings suggest that down-regulation of OGT enhances cisplatin-induced autophagy via SNAP-29, resulting in cisplatin-resistant ovarian cancer. 30587575_novel regulatory mechanism for O-GlcNAcylation during FA complex formation, which thereby affects integrin activation and integrin-mediated functions such as cell adhesion and migration 30677218_our findings indicate that OGT promotes the stem-like cell potential of hepatoma cell through O-GlcNAcylation of eIF4E 30953348_HNF1A regulates ogt transcription in a time-dependent manner and that O-GlcNAcylation of HNF1A represses ogt transcription. 14 O-GlcNAc sites on HNF1A were revealed, six of which are predominantly modified, including Ser(303/304) , Ser(471) , Ser(560) and Thr(563/564). Loss of O-GlcNAcylation at Ser(303/304) or Thr(563/564) significantly elevates ogt transcription. 31149037_OGT is essential for proliferation of prostate cancer cells. OGT activity is required for the interaction between MYC and HCF-1 and expression of MYC-regulated mitotic proteins. 31296563_Pathogenic N567K mutation leads to loss of O-GlcNAcase activity and delayed differentiation down the neuronal lineage in mouse and Drosophila model. 31373491_Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. 31373757_silencing of OGT in HT29 cells upregulates E-cadherin (a major actor of epithelial-to-mesenchymal transition) and changes its glycosylation. On the other hand, OGT silencing perturbs biosynthesis of glycosphingolipids resulting in a decrease in gangliosides and an increase in globosides. 31527085_O-GlcNAcylation does not affect sOGT activity but does affect sOGT-interacting proteins. S56A bound to and hence glycosylated more proteins in contrast to T12A and WT sOGT. 31567281_Two cases of hand myoepithelioma showing unusual clinicopathologic features and novel OGT-FOXO3 gene fusions are described. 31627256_A missense mutation in the catalytic domain of O-GlcNAc transferase links perturbations in protein O-GlcNAcylation to X-linked intellectual disability. 31628985_Disease related single point mutations alter the global dynamics of a tetratricopeptide (TPR) alpha-solenoid domain. 31847126_findings in this report are the first to describe a role for the OGT/O-GlcNAc axis in modulating VEGF expression and vascularization in Idiopathic pulmonary arterial hypertension. 31974291_O-GlcNAc Transferase Regulates Cancer Stem-like Potential of Breast Cancer Cells. 32272438_Epigenetic activation of O-linked beta-N-acetylglucosamine transferase overrides the differentiation blockage in acute leukemia. 32310828_SIRT1 regulates O-GlcNAcylation of tau through OGT. 32471715_O-GlcNAc transferase affects the signal transduction of beta1 adrenoceptor in adult rat cardiomyocytes by increasing the O-GlcNAcylation of beta1 adrenoceptor. 32663610_Glucosamine regulates hepatic lipid accumulation by sensing glucose levels or feeding states of normal and excess. 32994395_Mutual regulation between OGT and XIAP to control colon cancer cell growth and invasion. 33006972_The O-GlcNAc transferase OGT is a conserved and essential regulator of the cellular and organismal response to hypertonic stress. 33215629_Inhibition of mechanistic target of rapamycin signaling decreases levels of O-GlcNAc transferase and increases serotonin release in the human placenta. 33307156_Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8(+) T cells. 33333092_Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. 33419956_Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. 33801653_Feedback Regulation of O-GlcNAc Transferase through Translation Control to Maintain Intracellular O-GlcNAc Homeostasis. 33909326_The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. 34046694_Dual regulation of fatty acid synthase (FASN) expression by O-GlcNAc transferase (OGT) and mTOR pathway in proliferating liver cancer cells. 34288245_Upregulation of OGT by Caveolin-1 promotes hepatocellular carcinoma cell migration and invasion. 34502531_OGT Protein Interaction Network (OGT-PIN): A Curated Database of Experimentally Identified Interaction Proteins of OGT. 34510715_P53 suppresses the progression of hepatocellular carcinoma via miR-15a by decreasing OGT expression and EZH2 stabilization. 34608265_CEMIP, a novel adaptor protein of OGT, promotes colorectal cancer metastasis through glutamine metabolic reprogramming via reciprocal regulation of beta-catenin. 34638625_Regulation of O-Linked N-Acetyl Glucosamine Transferase (OGT) through E6 Stimulation of the Ubiquitin Ligase Activity of E6AP. 34943826_Inhibition of O-GlcNAc Transferase Alters the Differentiation and Maturation Process of Human Monocyte Derived Dendritic Cells. 34948036_TET3- and OGT-Dependent Expression of Genes Involved in Epithelial-Mesenchymal Transition in Endometrial Cancer. | ENSMUSG00000034160 | Ogt | 4537.39302 | 2.325027e-01 | -2.104680 | 0.3301933 | 35.08574 | 3.155033e-09 | 5.155453e-06 | Yes | Yes | 1.731133e+03 | 3.656385e+02 | 6.400889e+03 | 1.384251e+03 |
| ENSG00000151779 | 51594 | NBAS | protein_coding | A2RRP1 | FUNCTION: Involved in Golgi-to-endoplasmic reticulum (ER) retrograde transport; the function is proposed to depend on its association in the NRZ complex which is believed to play a role in SNARE assembly at the ER (PubMed:19369418). Required for normal embryonic development (By similarity). May play a role in the nonsense-mediated decay pathway of mRNAs containing premature stop codons (By similarity). {ECO:0000250|UniProtKB:Q5TYW4, ECO:0000269|PubMed:19369418}. | Acetylation;Alternative splicing;Cytoplasm;Disease variant;Dwarfism;Endoplasmic reticulum;Membrane;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport;WD repeat | This gene encodes a protein with two leucine zipper domains, a ribosomal protein S14 signature domain and a Sec39 like domain. The protein is thought to be involved in Golgi-to-ER transport. Mutations in this gene are associated with short stature, optic nerve atrophy, and Pelger-Huet anomaly. [provided by RefSeq, Oct 2012]. | hsa:51594; | cytosol [GO:0005829]; Dsl1/NZR complex [GO:0070939]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; membrane [GO:0016020]; SNARE binding [GO:0000149]; negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:2000623]; nuclear-transcribed mRNA catabolic process [GO:0000956]; protein transport [GO:0015031]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890] | 17028906_There may be a subset of NB in which enhanced DDX1 and low-NAG expression consequent to DDX1 co-amplification without NAG amplification contributes to susceptibility to intensive therapy. 19369418_Results together suggest that NAG links between p31 and ZW10-RINT-1 and is involved in Golgi-to-ER transport. 19407829_Defects in NAG-ELMO1 is associated with leukemic progression. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20577004_These findings suggest that function of NBAS may associate with the pathogenesis of short stature syndrome as well as optic atrophy and Pelger-Huet anomaly. 23828042_DHX34 and NBAS act in concert with core nonsense-mediated mRNA decay factors to co-regulate a large number of endogenous RNA targets. 23991058_Data indicate that expression of several predicted chimeric genes and genes with disrupted exon structure including ALK, NBAS, FHIT, PTPRD and ODZ4 in neuroblastoma. 26073778_Biallelic Mutations in NBAS Cause Recurrent Acute Liver Failure with Onset in Infancy. 26286438_NBAS mutations cause a multisystem disorder involving bone, connective tissue, liver, immune system, and retina. 27789416_variants in NBAS, are reported as a cause of bone fragility in humans, and expand the phenotypic spectrum associated with NBAS. 28576691_A novel compound heterozygous mutations of NBAS (NM_015909.3): c.680A > C (p.His227Pro) were identified in two siblings with acute liver failure. 28629372_NBAS was the only candidate gene mutated in more than one patient. All NBAS mutations were novel and predictedly pathogenic. Of these mutations, 3 lay in distal (C-terminal) regions of NBAS, a novel distribution. Unlike the 2 patients without NBAS mutations, the 3 patients with confirmed NBAS mutations all suffered from a febrile illness before each episode of liver crisis (fever-related recurrent acute liver failure) 29369590_The age of the mutation in Yakutia was estimated to be about 804 +/- 140 years. The frequency of heterozygous carriers of mutation G5741-->A (R1914H) in gene NBAS was found, which averaged 13 per 1000 healthy Yakuts 29929043_A novel homozygous truncating mutation in the NBAS gene was identified in a family with Acrofrontofacionasal Dysostosis type 1. 30825388_Data report two unrelated subjects with a trait associated with defective NBAS function sharing a previously unreported pathogenic 'synonymous' change demonstrated to affect proper NBAS transcript processing. Assessing the clinical features and signs of affected subjects with biallelic NBAS variants documents the occurrence of a recognizable facial profile for these patients. 32146038_Mutations in NBAS and SCYL1, genetic causes of recurrent liver failure in children: Three case reports and a literature review. 32297715_Severe SOPH syndrome due to a novel NBAS mutation in a 27-year-old woman-Review of this pleiotropic, autosomal recessive disorder: Mystery solved after two decades. 33542026_Novel compound heterozygous variants in the NBAS gene in a child with osteogenesis imperfecta and recurrent acute liver failure. 33707149_Characterization of a complex phenotype (fever-dependent recurrent acute liver failure and osteogenesis imperfecta) due to NBAS and P4HB variants. 34386911_NBAS Variants Are Associated with Quantitative and Qualitative NK and B Cell Deficiency. | ENSMUSG00000020576 | Nbas | 947.81878 | 3.702086e-01 | -1.433590 | 0.3326742 | 17.65022 | 2.654851e-05 | 1.206112e-02 | Yes | No | 7.072649e+02 | 1.529872e+02 | 1.464994e+03 | 3.241442e+02 | |
| ENSG00000155561 | 23165 | NUP205 | protein_coding | Q92621 | FUNCTION: Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance (PubMed:9348540). May anchor NUP62 and other nucleoporins, but not NUP153 and TPR, to the NPC (PubMed:15229283). {ECO:0000269|PubMed:15229283, ECO:0000269|PubMed:9348540}. | 3D-structure;Acetylation;Direct protein sequencing;Disease variant;Membrane;Nuclear pore complex;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Translocation;Transport;mRNA transport | This gene encodes a nucleoporin, which is a subunit of the nuclear pore complex that functions in active transport of proteins, RNAs and ribonucleoprotein particles between the nucleus and cytoplasm. Mutations in this gene are associated with steroid-resistant nephrotic syndrome. [provided by RefSeq, Jul 2016]. | hsa:23165; | cytosol [GO:0005829]; membrane [GO:0016020]; nuclear envelope [GO:0005635]; nuclear inner membrane [GO:0005637]; nuclear membrane [GO:0031965]; nuclear outer membrane [GO:0005640]; nuclear periphery [GO:0034399]; nuclear pore [GO:0005643]; nuclear pore inner ring [GO:0044611]; structural constituent of nuclear pore [GO:0017056]; mRNA transport [GO:0051028]; nuclear pore complex assembly [GO:0051292]; nuclear pore organization [GO:0006999]; nucleocytoplasmic transport [GO:0006913]; protein transport [GO:0015031] | 25210169_These data show for first time that adenovirus type 5 E4orf4 interacts with and modifies the nuclear pore complex and that Nup205-E4orf4 binding is required for normal regulation of viral gene expression and viral replication. 26878725_NUP93 knockdown reduced the presence of NUP205 in the nuclear pore compex, and, reciprocally, a NUP205 alteration abrogated NUP93 interaction. 31298340_LncRNA SNHG1 overexpression regulates the proliferation of acute myeloid leukemia cells through miR-488-5p/NUP205 axis. 31306055_NUP205 and NUP210 mutations are associated with defects in cardiac patterning. 32964975_LINC00887 regulates the proliferation of nasopharyngeal carcinoma via targeting miRNA-203b-3p to upregulate NUP205. 33107391_LncRNA HOTAIR Influences the Growth, Migration, and Invasion of Papillary Thyroid Carcinoma via Affection on the miR-488-5p/NUP205 Axis. 33230144_Biallelic loss of function NEK3 mutations deacetylate alpha-tubulin and downregulate NUP205 that predispose individuals to cilia-related abnormal cardiac left-right patterning. | ENSMUSG00000038759 | Nup205 | 6790.72123 | 4.489497e-01 | -1.155374 | 0.2686878 | 17.64955 | 2.655775e-05 | 1.206112e-02 | Yes | No | 6.515043e+03 | 1.425519e+03 | 9.504918e+03 | 2.132764e+03 | |
| ENSG00000158164 | 11013 | TMSB15A | protein_coding | P0CG34 | FUNCTION: Plays an important role in the organization of the cytoskeleton. Binds to and sequesters actin monomers (G actin) and therefore inhibits actin polymerization (By similarity). {ECO:0000250}. | Actin-binding;Cytoplasm;Cytoskeleton;Direct protein sequencing;Reference proteome | hsa:11013;hsa:286527; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; actin monomer binding [GO:0003785]; actin filament organization [GO:0007015]; regulation of cell migration [GO:0030334]; sequestering of actin monomers [GO:0042989] | 8946830_Identification of thymosin beta15, the rat homolog of human thymosin beta 15/thymosin beta NB/thymosin-like 8. Thymosin beta15 levels increase in human prostate cancer, correlating positively with Gleason tumor grade. 17567946_TMSL8 is well conserved in many mammalian species. Two isoforms, thymosin beta15a and b, exist at different locations, 1.4Mb apart, on human chromosome X, with 98% identity across the coding sequences. 17888914_Human thymosin beta neuroblastoma has a higher affinity for actin in comparison to thymosin beta4 and promotes cell migration. 18452710_Thymosin beta15 (Tbeta15) was translocated into nuclei following kainic acid (KA) treatments in a subset of entorhinal cortex neurons which are the population that undergo delayed apoptosis after KA treatment. 18754322_findings suggest that increased thymosin beta15 expression correlates with the progression and metastasis of non-small cell lung cancer 19296525_Our data show that the TMSB15A and TMSB15B isoforms have distinct expression patterns in different tumor cell lines and tissues. 23079573_Thymosin beta 15A (TMSB15A) is a predictor of chemotherapy response in triple-negative breast cancer. | 273.13976 | 3.026425e-01 | -1.724314 | 0.3440410 | 24.04342 | 9.418740e-07 | 7.956840e-04 | Yes | No | 1.306848e+02 | 2.594166e+01 | 3.886784e+02 | 7.749281e+01 | ||||
| ENSG00000162813 | 10380 | BPNT1 | protein_coding | O95861 | FUNCTION: Converts adenosine 3'-phosphate 5'-phosphosulfate (PAPS) to adenosine 5'-phosphosulfate (APS) and 3'(2')-phosphoadenosine 5'- phosphate (PAP) to AMP. Has 1000-fold lower activity towards inositol 1,4-bisphosphate (Ins(1,4)P2) and inositol 1,3,4-trisphosphate (Ins(1,3,4)P3), but does not hydrolyze Ins(1)P, Ins(3,4)P2, Ins(1,3,4,5)P4 or InsP6. {ECO:0000269|PubMed:10224133}. | 3D-structure;Acetylation;Alternative splicing;Hydrolase;Lithium;Magnesium;Metal-binding;Phosphoprotein;Reference proteome | BPNT1, also called bisphosphate 3-prime-nucleotidase, or BPntase, is a member of a magnesium-dependent phosphomonoesterase family. Lithium, a major drug used to treat manic depression, acts as an uncompetitive inhibitor of BPntase. The predicted human protein is 92% identical to mouse BPntase. BPntase's physiologic role in nucleotide metabolism may be regulated by inositol signaling pathways. The inhibition of human BPntase may account for lithium-induced nephrotoxicity. [provided by RefSeq, Jul 2008]. | hsa:10380; | cytosol [GO:0005829]; 3'(2'),5'-bisphosphate nucleotidase activity [GO:0008441]; metal ion binding [GO:0046872]; 3'-phosphoadenosine 5'-phosphosulfate metabolic process [GO:0050427]; inositol phosphate dephosphorylation [GO:0046855]; nervous system development [GO:0007399]; nucleobase-containing compound metabolic process [GO:0006139]; phosphatidylinositol phosphate biosynthetic process [GO:0046854] | 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) | ENSMUSG00000026617 | Bpnt1 | 427.46646 | 2.429551e-01 | -2.041239 | 0.4129010 | 22.98642 | 1.631496e-06 | 1.199616e-03 | Yes | No | 1.352894e+02 | 3.884560e+01 | 6.045504e+02 | 1.762034e+02 | |
| ENSG00000163541 | 8802 | SUCLG1 | protein_coding | P53597 | FUNCTION: Succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of either ATP or GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The alpha subunit of the enzyme binds the substrates coenzyme A and phosphate, while succinate binding and specificity for either ATP or GTP is provided by different beta subunits. {ECO:0000255|HAMAP-Rule:MF_03222}. | 3D-structure;Acetylation;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Primary mitochondrial disease;Reference proteome;Transit peptide;Tricarboxylic acid cycle | PATHWAY: Carbohydrate metabolism; tricarboxylic acid cycle; succinate from succinyl-CoA (ligase route): step 1/1. {ECO:0000255|HAMAP-Rule:MF_03222}. | This gene encodes the alpha subunit of the heterodimeric enzyme succinate coenzyme A ligase. This enzyme is targeted to the mitochondria and catalyzes the conversion of succinyl CoA and ADP or GDP to succinate and ATP or GTP. Mutations in this gene are the cause of the metabolic disorder fatal infantile lactic acidosis and mitochondrial DNA depletion. [provided by RefSeq, Feb 2010]. | hsa:8802; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; succinate-CoA ligase complex (ADP-forming) [GO:0009361]; succinate-CoA ligase complex (GDP-forming) [GO:0045244]; nucleotide binding [GO:0000166]; RNA binding [GO:0003723]; succinate-CoA ligase (ADP-forming) activity [GO:0004775]; succinate-CoA ligase (GDP-forming) activity [GO:0004776]; tricarboxylic acid cycle [GO:0006099] | 20197121_This report enlarges the phenotypic spectrum of SUCLG1 mutations and confirms that a characteristic metabolic profile (presence of MMA and C4-DC carnitine in urines) and basal ganglia MRI lesions are the hallmarks of SCS defects. 20227526_A novel mutation in the SUCLG1 gene was found in two newborns having lethal lactic acidosis, multi-organ failure and congenital malformations including interrupted aortic arch. 20693550_Our results suggest that SUCLG1 mutations that lead to complete absence of SUCLG1 protein are responsible for a very severe disorder with antenatal manifestations, whereas a SUCLA2-like phenotype is found in patients with residual SUCLG1 protein. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22980518_3 novel mutations have been identified in patients with the mitochondrial DNA depletion syndrome (c.1048G>A and c.1049G>T in SUCLA2 and c.531+4A>T in SUCLG1). 26028457_First Chinese report of succinyl-CoA ligase deficiency caused by novel SUCLG1 mutations; five novel pathogenic mutations in SUCLG1 were identified 26475597_Long survival, to age 20 years or older, was reported in 12% of SUCLA2 and in 10% of SUCLG1 patients. 27484306_These abnormal phenotypes are rescued upon ectopic expression of wild-type SUCLG1 in the patient's fibroblasts, thus functionally confirming the pathogenic nature of the SUCLG1 VUS identified in this patient and expanding the phenotypic spectrum for SUCLG1 deficiency 29217198_We report two Tunisian patients belonging to a consanguineous family with mitochondrial encephalomyopathy. Mutational analysis of SUCLG1 gene showed the presence of c.41T > C in exon 1 in a homozygous state. This mutation substitutes a conserved methionine residue to a threonine at position 14 (p.M14T) located at the SUCLG1 protein mitochondrial targeting sequence. | ENSMUSG00000052738 | Suclg1 | 2172.93042 | 3.464061e-01 | -1.529464 | 0.2874271 | 26.70605 | 2.368785e-07 | 2.438427e-04 | Yes | No | 1.207706e+03 | 2.595885e+02 | 3.024852e+03 | 6.656675e+02 |
| ENSG00000163682 | 6133 | RPL9 | protein_coding | P32969 | 3D-structure;Acetylation;Direct protein sequencing;Reference proteome;Ribonucleoprotein;Ribosomal protein | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 60S subunit. The protein belongs to the L6P family of ribosomal proteins. It is located in the cytoplasm. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:6133; | cytosol [GO:0005829]; cytosolic large ribosomal subunit [GO:0022625]; cytosolic ribosome [GO:0022626]; focal adhesion [GO:0005925]; membrane [GO:0016020]; nucleus [GO:0005634]; ribosome [GO:0005840]; RNA binding [GO:0003723]; rRNA binding [GO:0019843]; structural constituent of ribosome [GO:0003735]; cytoplasmic translation [GO:0002181]; translation [GO:0006412] | 26655239_The authors propose that during rabies virus infection, viral phosphoprotein P binds to ribosomal protein L9 that translocates from the nucleus to the cytoplasm, inhibiting the initial stage of viral transcription. 27633352_High RPL9 expression is associated with colorectal carcinoma. 30266104_UNR could bind to the 3'UTR of PTEN and RPL9 in glioma cell lines, therefore promoting glioma cell migration and regulating the expression of RPL9. 31799629_Here we report an individual diagnosed with DBA carrying a variant in the 5'UTR of RPL9 (uL6). Additionally, we report two individuals from a family with multiple cancer incidences carrying a RPL9 missense variant 33576138_Investigating the folding mechanism of the N-terminal domain of ribosomal protein L9. 34483448_Genetic Variants of RPL5 and RPL9 Genes among Saudi Patients Diagnosed with Thrombosis. | 8115.43887 | 2.540973e-01 | -1.976547 | 0.3968435 | 24.13831 | 8.965870e-07 | 7.956840e-04 | Yes | No | 3.616498e+03 | 1.347320e+03 | 1.237116e+04 | 4.729519e+03 | ||||
| ENSG00000164032 | 3015 | H2AZ1 | protein_coding | P0C0S5 | FUNCTION: Variant histone H2A which replaces conventional H2A in a subset of nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. May be involved in the formation of constitutive heterochromatin. May be required for chromosome segregation during cell division. {ECO:0000269|PubMed:15878876}. | 3D-structure;Acetylation;Chromosome;DNA-binding;Isopeptide bond;Methylation;Nucleosome core;Nucleus;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene encodes a replication-independent member of the histone H2A family that is distinct from other members of the family. Studies in mice have shown that this particular histone is required for embryonic development and indicate that lack of functional histone H2A leads to embryonic lethality. [provided by RefSeq, Jul 2008]. | hsa:3015; | euchromatin [GO:0000791]; extracellular exosome [GO:0070062]; heterochromatin [GO:0000792]; nucleosome [GO:0000786]; nucleus [GO:0005634]; chromatin DNA binding [GO:0031490]; DNA binding [GO:0003677]; nucleosomal DNA binding [GO:0031492]; protein heterodimerization activity [GO:0046982]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II core promoter sequence-specific DNA binding [GO:0000979]; cellular response to estradiol stimulus [GO:0071392]; heterochromatin assembly [GO:0031507]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 15878876_chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z 16809769_neither H2AZ itself nor other features of the H2AZ-containing nucleosome spread to the neighboring nucleosomes in vivo, arguing against a role for H2AZ as a self-perpetuating epigenetic mark 17194760_identify the essential histone variant H2A.Z as a new structural component of the centromere 17636032_Monoubiquitylation of H2A.z distinguishes its association with euchromatin or facultative heterochromatin. 17671089_Upon DNA damage, histone H2A.Z is first evicted from the p21 promoter, followed by the recruitment of the Tip60 histone acetyltransferase to activate p21 transcription. 18414489_histone variant H2A.Z is associated with breast cancer progression 19246569_Results show that H2A.Z nucleosomes protect only approximately 120 bp of DNA from MNase digestion and exhibit specific sequence preferences, suggesting a novel mechanism of nucleosome organization for the H2A.Z variant. 19261190_Both genetic and epigenetic features are likely to participate in targeting H2A.Z to distinct chromatin loci. 19385636_The nucleosome destabilizing effect of H2A.Z acetylation takes place synergistically with the acetylation of the rest of the core histones. 19515975_H2A.Z is incorporated into the promoter regions of estrogen receptor (ERalpha) target genes only upon gene induction, and that, in a cyclic pattern 19834540_show that upon gene induction, human H2A.Z associates with gene promoters and helps in recruiting the transcriptional machinery. 19856965_Both H2A.Z and H3.3 affect nucleosome positioning, either creating new positions or altering the relative occupancy of the existing nucleosome position space. Only H2A.Z-containing nucleosomes exhibit altered linker histone binding. 20023423_Eesterogen Receptor alpha directly associates to the H2A.Z promoter, and consequently modulates its expression. 20364108_This review provides a brief overview of H2A.Z biology and presents hypotheses that could reconcile contradictory reports that are found in the literature regarding the influence of H2A.Z on nucleosome stability. 20864037_H2A.Z is maintained during mitosis and marks the +1 nucleosome of active genes, which shifts during mitosis, resulting in occupancy at the transcriptional start site and a reduced nucleosome-depleted region. 21788347_acetylation of H2A.Z is a key modification associated with gene activity in normal cells and epigenetic gene deregulation in tumorigenesis. 22393239_nucleosomes containing H2AZ are primarily composed of H4 K12ac and H3 K4me3 but not H3 K36me3 22493515_The short forms of H2A.Z in both yeast and human cells are more loosely associated with chromatin than the full-length proteins, indicating a conserved function for the H2A.Z C-terminal tail in regulating the association of H2A.Z with nucleosomes. 22678762_ZNF24 may be implicated in transcriptional regulation of genes associated with oncogenesis via interaction with H2A.Z. 23034477_Study mapped H2A.Z genome-wide in embryonic stem cells and neural progenitors; H2A.Z is deposited at promoters and enhancers, and correlates strongly with H3K4 methylation. H2A.Z is present at poised promoters with bivalent chromatin and at active promoters with H3K4 methylation, but is absent from stably repressed promoters that are enriched for H3K27 trimethylation. 23064015_incorporation of the histone variant H2A.Z at the promoter regions of PPARgamma target genes by p400/Brd8 is essential to allow fat cell differentiation 23108396_H2A.Z-dependent crosstalk between enhancer and promoter regulates cyclin D1 expression. 23122415_H2A.Z exchange promotes specific patterns of histone modification and reorganization of the chromatin architecture, leading to the assembly of a chromatin template that is an efficient substrate for the DNA double-strand break repair machinery. 23146670_age-dependent p400 downregulation and loss of H2A.Z localisation may contribute to the onset of replicative senescence through a sustained high rate of p21 transcription 23245330_Data indicate that histone H2A.Z as a protein capable of binding ST1926 specifically. 23324626_SETD6 monomethylates H2AZ on lysine 7. 23349794_Data show that histone deacetylase inhibitors (HDACi) induce p21 transcription and reduce cell proliferation of MDA-MB231, an ERalpha-negative mammary tumor cell line, in a H2A.Z dependent manner. 24127549_Sirt1 and H2A.Z deregulation in prostate cancer are related. Epigenetic mechanisms, mostly histone post-translational modifications, are likely involved and impair sirt1-mediated downregulation of H2A.Z via proteasome-mediated degradation. 24240188_Depletion of H2A.Z in the osteosarcoma U2OS cell line and in immortalized human fibroblasts does not change parameters of DNA double-strand breaks repair while affecting clonogenic ability and cell cycle distribution. 24311584_A mutational analysis revealed that the amino-acid difference at position 38 is at least partially responsible for the structural polymorphism in the L1 loop region of H2A.Z.1 and H2A.Z.2. 24397596_the predictive values regarding low expressions of H2AFZ and CASP8AP2 and high white blood cell count suggest that these features could help to identify more accurately patients at greater risk of relapse. 24613878_Anp32e may help to resolve the non-nucleosomal H2A.Z aggregates and also facilitate the removal of H2A.Z at the +1 nucleosomes, and the latter may help RNA polymerase II to pass the first nucleosomal barrier. 25392085_Results demonstrated male-selective association of the H2AFZ gene with schizophrenia, and that modification of the H2AFZ signaling pathway warrants further study in terms of the pathophysiology of schizophrenia 26034280_Dynamic modulation of H2A.Z exchange and removal by Anp32e reveals the importance of the nucleosome surface and nucleosome dynamics in processing the damaged chromatin template during DSB repair. 26051178_The findings implicate H2A.Z.2 as a mediator of cell proliferation and drug sensitivity in malignant melanoma. 26142279_H2A.Z removal from chromatin is the primary function of INO80 and ANP32E in promoting homologous recombination. 26246156_the H2AFZ gene may confer a risk for schizophrenia and contribute to the impairment of executive function in Han Chinese patients with schizophrenia. 26833946_Results suggest that the N-terminal tail of H2A.Z makes distinctively different contributions to epigenetic events. 26863632_Findings suggest the oncogenic potential of H2A.Z.1 in liver tumorigenesis and that it plays established role in accelerating cell cycle transition and EMT during hepatocarcinogenesis. 26974126_The 2.7-A-resolution crystal structure of the human YL1-H2A.Z-H2B complex shows that YL1 binding, similarly to ANP32E binding, triggers an extension of the H2A.Z alphaC helix. 27358293_Crystal structure results show that the flexible character of the H2A.Z L1 loop plays an essential role in forming the stable heterotypic H2A.Z/H2A nucleosome. 27569210_SMYD3-mediated H2A.Z.1K101 dimethylation activates cyclin A1 expression and contributes to driving the proliferation of breast cancer cells. 27692985_Monoubiquitination of histone H2B blocks eviction of histone variant H2A.Z from inducible enhancers. 28301306_Two possible modes of pioneering associated with combinations of H2A.Z and p300/CBP at nucleosome-occupied enhancers. 28645917_Thus, PWWP2A is a novel H2A.Z-specific multivalent chromatin binder providing a surprising link between H2A.Z, chromosome segregation, and organ development. 29036442_Results indicate that accumulation of H2A.Z within repressed genes can also be a consequence of the repression of gene transcription rather than an active mechanism required to establish the repression. 29116202_H2A.Z associates with epigenetic gene activation in prostate cancer.Acetylated H2A.Z role in activation of newly formed enhancers in prostate cancer. 29437725_study identifies GAS41 as a histone acetylation reader that promotes histone H2A.Z deposition in non-small cell lung cancer. 29532867_The present study demonstrated that H2A.Z is overexpressed in ICC and expression of H2A.Z correlated with poor prognosis in patients with ICC. H2A.Z regulated cell proliferation in vitro and in vivo via H2A.Z/S-phase kinase-associated protein 2/p27/p21 signaling. 29985131_FBXL10-RNF68-RNF2 ubiquitin ligase complex (FRRUC) is rapidly and transiently recruited to sites of DNA damage in a PARP1- and TIMELESS-dependent manner to promote mono-ubiquitylation of H2A at Lys119. 29986055_Data show that loading and acetylation of H2A.Z are required to assure tight control of canonical notch receptor (Notch) activation. 31491386_these results illuminate the mechanism underlying a human syndrome and uncover selective functions of H2A.Z subtypes during development. 31682939_Conclusively, chromatin immunoprecipitation-deep sequencing, confirmed the predominant association of OGDH and ACAA2 with H2A.Z-occupied transcription start sites and enhancers, the former of which we confirmed is conserved in both mouse and human tissue. 31875854_results suggest that the histone variant H2A.Z epigenetically regulates the licensing and activation of early replication origins and maintains replication timing through the SUV420H1-H4K20me2-ORC1 axis 32109204_Integrated analysis of H2A.Z isoforms function reveals a complex interplay in gene regulation. 32500630_The N-terminal and C-terminal halves of histone H2A.Z independently function in nucleosome positioning and stability. 32708675_VPS72/YL1-Mediated H2A.Z Deposition Is Required for Nuclear Reassembly after Mitosis. 33073403_Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2. 33953180_Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells. 34120148_H2A.Z acetylation by lincZNF337-AS1 via KAT5 implicated in the transcriptional misregulation in cancer signaling pathway in hepatocellular carcinoma. 34349258_Deficient H2A.Z deposition is associated with genesis of uterine leiomyoma. 34537242_Linking nuclear matrix-localized PIAS1 to chromatin SUMOylation via direct binding of histones H3 and H2A.Z. 34840626_Characterization of Cancer Stem Cell Characteristics and Development of a Prognostic Stemness Index Cell-Related Signature in Oral Squamous Cell Carcinoma. 35027526_H2A Histone Family Member Z (H2AFZ) Serves as a Prognostic Biomarker in Lung Adenocarcinoma: Bioinformatic Analysis and Experimental Validation. | ENSMUSG00000037894 | H2az1 | 20262.29694 | 3.862425e-01 | -1.372421 | 0.3132699 | 18.64825 | 1.571918e-05 | 7.584996e-03 | Yes | No | 1.034222e+04 | 1.847107e+03 | 2.689316e+04 | 4.924376e+03 | |
| ENSG00000164199 | 84059 | ADGRV1 | protein_coding | Q8WXG9 | FUNCTION: G-protein coupled receptor which has an essential role in the development of hearing and vision. Couples to G-alpha(i)-proteins, GNAI1/2/3, G-alpha(q)-proteins, GNAQ, as well as G-alpha(s)-proteins, GNAS, inhibiting adenylate cyclase (AC) activity and cAMP production. Required for the hair bundle ankle formation, which connects growing stereocilia in developing cochlear hair cells of the inner ear. In response to extracellular calcium, activates kinases PKA and PKC to regulate myelination by inhibiting the ubiquitination of MAG, thus enhancing the stability of this protein in myelin-forming cells of the auditory pathway. In retina photoreceptors, the USH2 complex is required for the maintenance of periciliary membrane complex that seems to play a role in regulating intracellular protein transport. Involved in the regulation of bone metabolism. {ECO:0000250|UniProtKB:Q8VHN7}.; FUNCTION: [ADGRV1 subunit beta]: Cleaved ADGRV1 beta-subunit couples with G-alpha(i)-proteins, GNAI1/2/3, and constitutively inhibits adenylate cyclase (AC) activity with a stronger effect than full ADGRV1. {ECO:0000250|UniProtKB:Q8VHN7}. | Alternative splicing;Calcium;Cell membrane;Cell projection;Deafness;G-protein coupled receptor;Hydrolase;Membrane;Receptor;Reference proteome;Repeat;Retinitis pigmentosa;Sensory transduction;Signal;Transducer;Transmembrane;Transmembrane helix;Usher syndrome;Vision | This gene encodes a member of the G-protein coupled receptor superfamily. The encoded protein contains a 7-transmembrane receptor domain, binds calcium and is expressed in the central nervous system. Mutations in this gene are associated with Usher syndrome 2 and familial febrile seizures. Several alternatively spliced transcripts have been described. [provided by RefSeq, Jul 2008]. | hsa:84059; | cell surface [GO:0009986]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; periciliary membrane compartment [GO:1990075]; photoreceptor inner segment [GO:0001917]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; stereocilia ankle link [GO:0002141]; stereocilia ankle link complex [GO:0002142]; stereocilium membrane [GO:0060171]; synapse [GO:0045202]; USH2 complex [GO:1990696]; adenylate cyclase inhibitor activity [GO:0010855]; calcium ion binding [GO:0005509]; G protein-coupled receptor activity [GO:0004930]; G-protein alpha-subunit binding [GO:0001965]; hydrolase activity [GO:0016787]; cell surface receptor signaling pathway [GO:0007166]; cell-cell adhesion [GO:0098609]; cellular response to calcium ion [GO:0071277]; detection of mechanical stimulus involved in sensory perception of sound [GO:0050910]; establishment of protein localization [GO:0045184]; G protein-coupled receptor signaling pathway [GO:0007186]; inner ear development [GO:0048839]; inner ear receptor cell stereocilium organization [GO:0060122]; maintenance of animal organ identity [GO:0048496]; negative regulation of adenylate cyclase activity [GO:0007194]; nervous system development [GO:0007399]; nervous system process [GO:0050877]; photoreceptor cell maintenance [GO:0045494]; positive regulation of bone mineralization [GO:0030501]; positive regulation of protein kinase A signaling [GO:0010739]; positive regulation of protein kinase C signaling [GO:0090037]; regulation of protein stability [GO:0031647]; self proteolysis [GO:0097264]; sensory perception of light stimulus [GO:0050953]; sensory perception of sound [GO:0007605]; visual perception [GO:0007601] | 12217514_has seven copies of the EPTP repeat, a unifying protein sequence motif of a heterogenous group of proteins linked to epileptic diseases. The EPTP repeat probably forms a beta-propeller structure. 12402266_A nonsense mutation (S2652X) causing a deletion of the C-terminal 126 amino acid residues was identified in one family with febrile and afebrile seizures. 15671307_USH2C and USH2A manifest photoreceptor disease with rod- and cone-mediated visual losses and thinning of the outer nuclear layer. 18463160_Humans with PCDH15 (USH1F), USH2A or GPR98 (USH2C) had a similar retinal phenotype to MYO7A 18854872_GPR98 genes and the phenotypic heterogeneity and particularly the severe ocular affection first observed in one Usher syndrome patient. 19357117_study describes for the first time two male patients with Usher syndrome type 2 with novel GPR98 mutations 19683999_Observational study of gene-disease association and genetic testing. (HuGE Navigator) 19730683_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20195266_Clinical trial and genome-wide association study of gene-disease association. (HuGE Navigator) 20200978_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20801516_Observational study of genetic testing. (HuGE Navigator) 22147658_Mutation found in USH2A, GPR98, or DFNB31 account for the vast majority of USH2 patients and their analysis provide a robust pathway for routine molecular diagnosis. 22419726_genetic association studies in postmenopausal Japanese women: Study found association between an SNP in GPR98 (rs10514346) and bone mineral density in this population; data suggest that Gpr98 signaling pathway regulates bone metabolism. 23441107_In Spain, USH2A and GPR98 are responsible for 95.8% and 5.2% of Usher syndrome 2 mutated cases, respectively. DFNB31 plays a minor role in the Spanish population. There was a group of patients in whom no mutation was found. 24962568_identified an independent Galphai signaling pathway of the VLGR1 beta-subunit and its regulatory mechanisms that may have a role in the development of Usher syndrome 25511798_our results suggest that low expression of VLGR1 is a significant risk factor of epileptic seizures in patients with low-grade glioma 25572244_Our findings also expand the spectrum of GPR98 mutations in USH and demonstrate that the long isoform of GPR98 might carry even more mutations of the GPR98 gene. 25743181_Diagnosis of Usher Syndrome 2 caused by GPR98 mutations in advance of visual defects in the cohort of nonsyndromic HL patients highlights importane of genetic testing in the diagnosis. 26432996_We identified two novel truncation mutations in GPR98 causing Usher syndrome. 28653555_7 patients clinically classified as having USH2, genetic tests confirmed the USH2 diagnosis in 5 cases. Of these, 4 patients showed mutations in the USH2A gene and 1 patient in the ADGRV1 gene. The mutation of the ADGRV1/GPR98 gene has an extremely rare incidence and is associated with a diagnosis of USH type 2C. 28951997_we found new causative mutations in heterozygous compound state, one missense and one nonsense mutation in the GPR98 gene in three deaf sibs. The first mutation located in exon7, corresponds to c.1054C > A, which causes a proline to threonine change at position 352 of the protein (p.Pro352Thr). The second mutation located within exon77 is c.16544delT that leads to a stop codon at position 5515of the protein (p.Leu5515*). 29142287_Likely causative mutations were found in all patients: 25 pathogenic variants, 18 previously reported and 7 novel, were identified in three genes (USH2A, MYO7A, ADGRV1). All USH1 presented biallelic MYO7A mutations, one USH2 exhibited ADGRV1 mutations, whereas 16 USH2 displayed USH2A mutations 29266188_Data suggest that the ADGRV1 variation contributes to epilepsy with myoclonic seizures, although the inheritance pattern may be complex in many cases. In patients with 5q14.3 deletion and epilepsy, ADGRV1 haploinsufficiency likely contributes to seizure development. 29883260_The mutations found in our study not only broaden the mutation spectrum of ADGRV1, but also provide assistances for future genetic diagnosis and treatment for Usher syndrome patients. 29890953_GPR98 missense mutation is associated with usher syndrome type IIC. 31792237_Genomewide Gene-by-Sex Interaction Scans Identify ADGRV1 for Sex Differences in Opioid Dependent African Americans. 34160719_Biallelic ADGRV1 variants are associated with Rolandic epilepsy. 34638692_Characteristics of Retinitis Pigmentosa Associated with ADGRV1 and Comparison with USH2A in Patients from a Multicentric Usher Syndrome Study Treatrush. | ENSMUSG00000069170 | Adgrv1 | 387.83392 | 4.156785e-01 | -1.266460 | 0.3289431 | 14.45476 | 1.435665e-04 | 4.030565e-02 | Yes | No | 2.614599e+02 | 4.488774e+01 | 5.426387e+02 | 9.441585e+01 | |
| ENSG00000165775 | 65991 | FUNDC2 | protein_coding | Q9BWH2 | FUNCTION: Binds directly and specifically 1,2-Diacyl-sn-glycero-3-phospho-(1'-myo-inositol-3',4',5'-bisphosphate) (PIP3) leading to the recruitment of PIP3 to mitochondria and may play a role in the regulation of the platelet activation via AKT/GSK3B/cGMP signaling pathways (PubMed:29786068). May act as transcription factor that regulates SREBP1 (isoform SREBP-1C) expression in order to modulate triglyceride (TG) homeostasis in hepatocytes (PubMed:29187281, PubMed:25855506). {ECO:0000269|PubMed:25855506, ECO:0000269|PubMed:29187281, ECO:0000269|PubMed:29786068}. | Membrane;Mitochondrion;Mitochondrion outer membrane;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA | hsa:65991; | integral component of mitochondrial outer membrane [GO:0031307]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; phosphatidylinositol-3,4,5-trisphosphate binding [GO:0005547]; autophagy of mitochondrion [GO:0000422]; cellular triglyceride homeostasis [GO:0035356]; regulation of platelet activation [GO:0010543] | 25855506_our results provide new evidence that miR-122-regulated HCBP6 functions as a sensor protein to maintain intrahepatocyte Triglyceride levels. 29187281_These results indicate that HCBP6 upregulates human SREBP1c expression by binding to the C/EBPbeta-binding site in the SREBP1c promoter. 29786068_This data indicate that FUNDC2 directly and selectively binds to PIP3 via its PB motif. Fractionation assays with HeLa cell lysates biochemically confirmed the localization of endogenous FUNDC2 to mitochondria. | ENSMUSG00000031198+ENSMUSG00000074619 | Fundc2+1700034I23Rik | 3096.07423 | 3.447600e-01 | -1.536336 | 0.3963230 | 14.53805 | 1.373569e-04 | 4.001749e-02 | Yes | No | 1.543109e+03 | 4.040434e+02 | 4.586538e+03 | 1.231203e+03 | |
| ENSG00000167447 | 55181 | SMG8 | protein_coding | Q8ND04 | FUNCTION: Involved in nonsense-mediated decay (NMD) of mRNAs containing premature stop codons. Is recruited by release factors to stalled ribosomes together with SMG1 and SMG9 (forming the SMG1C protein kinase complex) and, in the SMG1C complex, is required to mediate the recruitment of SMG1 to the ribosome:SURF complex and to suppress SMG1 kinase activity until the ribosome:SURF complex locates the exon junction complex (EJC). Acts as a regulator of kinase activity. {ECO:0000269|PubMed:19417104}. | 3D-structure;Alternative splicing;Cataract;Disease variant;Mental retardation;Methylation;Nonsense-mediated mRNA decay;Phosphoprotein;Reference proteome | hsa:55181; | cytosol [GO:0005829]; nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184]; regulation of protein kinase activity [GO:0045859] | 21245168_large-scale conformational changes induced by SMG-8 after SMG-9-mediated recruitment tune SMG-1 kinase activity to modulate nonsense-mediated mRNA decay 23983263_knockdown of SMG-8 produced the best effect for restoring defective mRNA and protein levels without affecting cell growth, cell-cycle progression, or endoplasmic reticulum stress in Ullrich congenital muscular dystrophy fibroblasts 31729466_Cryo-EM structure of SMG1-SMG8-SMG9 complex. 31792449_This study reports the 3.45-A resolution cryo-EM structure of human SMG1-SMG8-SMG9, a phosphatidylinositol-3-kinase (PI(3)K)-related protein kinase (PIKK) complex central to messenger RNA surveillance. 32469312_Structure of substrate-bound SMG1-8-9 kinase complex reveals molecular basis for phosphorylation specificity. 33242396_Recessive, Deleterious Variants in SMG8 Expand the Role of Nonsense-Mediated Decay in Developmental Disorders in Humans. 34761517_Expanding the phenotypic and allelic spectrum of SMG8: Clinical observations reveal overlap with SMG9-associated disease trait. | ENSMUSG00000020495 | Smg8 | 1095.58319 | 4.263719e-01 | -1.229816 | 0.3037154 | 15.80315 | 7.028531e-05 | 2.647013e-02 | Yes | No | 6.288958e+02 | 1.124333e+02 | 1.368348e+03 | 2.498479e+02 | ||
| ENSG00000168036 | 1499 | CTNNB1 | protein_coding | P35222 | FUNCTION: Key downstream component of the canonical Wnt signaling pathway (PubMed:17524503, PubMed:18077326, PubMed:18086858, PubMed:18957423, PubMed:21262353, PubMed:22155184, PubMed:22647378, PubMed:22699938). In the absence of Wnt, forms a complex with AXIN1, AXIN2, APC, CSNK1A1 and GSK3B that promotes phosphorylation on N-terminal Ser and Thr residues and ubiquitination of CTNNB1 via BTRC and its subsequent degradation by the proteasome (PubMed:17524503, PubMed:18077326, PubMed:18086858, PubMed:18957423, PubMed:21262353, PubMed:22155184, PubMed:22647378, PubMed:22699938). In the presence of Wnt ligand, CTNNB1 is not ubiquitinated and accumulates in the nucleus, where it acts as a coactivator for transcription factors of the TCF/LEF family, leading to activate Wnt responsive genes (PubMed:17524503, PubMed:18077326, PubMed:18086858, PubMed:18957423, PubMed:21262353, PubMed:22155184, PubMed:22647378, PubMed:22699938). Involved in the regulation of cell adhesion, as component of an E-cadherin:catenin adhesion complex (By similarity). Acts as a negative regulator of centrosome cohesion (PubMed:18086858). Involved in the CDK2/PTPN6/CTNNB1/CEACAM1 pathway of insulin internalization (PubMed:21262353). Blocks anoikis of malignant kidney and intestinal epithelial cells and promotes their anchorage-independent growth by down-regulating DAPK2 (PubMed:18957423). Disrupts PML function and PML-NB formation by inhibiting RANBP2-mediated sumoylation of PML (PubMed:22155184). Promotes neurogenesis by maintaining sympathetic neuroblasts within the cell cycle (By similarity). Involved in chondrocyte differentiation via interaction with SOX9: SOX9-binding competes with the binding sites of TCF/LEF within CTNNB1, thereby inhibiting the Wnt signaling (By similarity). {ECO:0000250|UniProtKB:Q02248, ECO:0000269|PubMed:17524503, ECO:0000269|PubMed:18077326, ECO:0000269|PubMed:18086858, ECO:0000269|PubMed:18957423, ECO:0000269|PubMed:21262353, ECO:0000269|PubMed:22155184, ECO:0000269|PubMed:22647378, ECO:0000269|PubMed:22699938}. | 3D-structure;Acetylation;Activator;Cell adhesion;Cell junction;Cell membrane;Cell projection;Chromosomal rearrangement;Cytoplasm;Cytoskeleton;Disease variant;Glycoprotein;Host-virus interaction;Membrane;Mental retardation;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Repeat;S-nitrosylation;Synapse;Transcription;Transcription regulation;Ubl conjugation;Wnt signaling pathway | The protein encoded by this gene is part of a complex of proteins that constitute adherens junctions (AJs). AJs are necessary for the creation and maintenance of epithelial cell layers by regulating cell growth and adhesion between cells. The encoded protein also anchors the actin cytoskeleton and may be responsible for transmitting the contact inhibition signal that causes cells to stop dividing once the epithelial sheet is complete. Finally, this protein binds to the product of the APC gene, which is mutated in adenomatous polyposis of the colon. Mutations in this gene are a cause of colorectal cancer (CRC), pilomatrixoma (PTR), medulloblastoma (MDB), and ovarian cancer. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2016]. | hsa:1499; | adherens junction [GO:0005912]; apical part of cell [GO:0045177]; apicolateral plasma membrane [GO:0016327]; basolateral plasma membrane [GO:0016323]; beta-catenin destruction complex [GO:0030877]; beta-catenin-ICAT complex [GO:1990711]; beta-catenin-TCF complex [GO:1990907]; beta-catenin-TCF7L2 complex [GO:0070369]; bicellular tight junction [GO:0005923]; catenin complex [GO:0016342]; cell cortex [GO:0005938]; cell junction [GO:0030054]; cell periphery [GO:0071944]; cell-cell junction [GO:0005911]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; euchromatin [GO:0000791]; extracellular exosome [GO:0070062]; fascia adherens [GO:0005916]; flotillin complex [GO:0016600]; focal adhesion [GO:0005925]; lamellipodium [GO:0030027]; lateral plasma membrane [GO:0016328]; membrane [GO:0016020]; microvillus membrane [GO:0031528]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; postsynaptic density, intracellular component [GO:0099092]; postsynaptic membrane [GO:0045211]; presynaptic active zone cytoplasmic component [GO:0098831]; presynaptic membrane [GO:0042734]; protein-containing complex [GO:0032991]; protein-DNA complex [GO:0032993]; Schaffer collateral - CA1 synapse [GO:0098685]; Scrib-APC-beta-catenin complex [GO:0034750]; spindle pole [GO:0000922]; synapse [GO:0045202]; transcription regulator complex [GO:0005667]; Wnt signalosome [GO:1990909]; Z disc [GO:0030018]; alpha-catenin binding [GO:0045294]; beta-catenin binding [GO:0008013]; cadherin binding [GO:0045296]; chromatin binding [GO:0003682]; disordered domain specific binding [GO:0097718]; DNA-binding transcription factor binding [GO:0140297]; enzyme binding [GO:0019899]; estrogen receptor binding [GO:0030331]; histone methyltransferase binding [GO:1990226]; I-SMAD binding [GO:0070411]; kinase binding [GO:0019900]; nuclear receptor binding [GO:0016922]; protein C-terminus binding [GO:0008022]; protein kinase binding [GO:0019901]; protein phosphatase binding [GO:0019903]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; SMAD binding [GO:0046332]; transcription coactivator activity [GO:0003713]; transcription coregulator binding [GO:0001221]; transmembrane transporter binding [GO:0044325]; adherens junction assembly [GO:0034333]; anterior/posterior axis specification [GO:0009948]; astrocyte-dopaminergic neuron signaling [GO:0036520]; bone resorption [GO:0045453]; branching involved in blood vessel morphogenesis [GO:0001569]; branching involved in ureteric bud morphogenesis [GO:0001658]; canonical Wnt signaling pathway [GO:0060070]; canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation [GO:0044338]; canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation [GO:1904954]; canonical Wnt signaling pathway involved in negative regulation of apoptotic process [GO:0044336]; canonical Wnt signaling pathway involved in osteoblast differentiation [GO:0044339]; canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation [GO:0061324]; canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition [GO:0044334]; cell adhesion [GO:0007155]; cell fate specification [GO:0001708]; cell maturation [GO:0048469]; cell morphogenesis involved in differentiation [GO:0000904]; cell-cell adhesion [GO:0098609]; cell-matrix adhesion [GO:0007160]; cellular response to growth factor stimulus [GO:0071363]; cellular response to indole-3-methanol [GO:0071681]; central nervous system vasculogenesis [GO:0022009]; chemical synaptic transmission [GO:0007268]; cranial ganglion development [GO:0061550]; cranial skeletal system development [GO:1904888]; detection of muscle stretch [GO:0035995]; dorsal root ganglion development [GO:1990791]; dorsal/ventral axis specification [GO:0009950]; ectoderm development [GO:0007398]; embryonic axis specification [GO:0000578]; embryonic brain development [GO:1990403]; embryonic digit morphogenesis [GO:0042733]; embryonic foregut morphogenesis [GO:0048617]; embryonic forelimb morphogenesis [GO:0035115]; embryonic heart tube development [GO:0035050]; embryonic hindlimb morphogenesis [GO:0035116]; embryonic skeletal limb joint morphogenesis [GO:0036023]; endodermal cell fate commitment [GO:0001711]; endothelial tube morphogenesis [GO:0061154]; epithelial cell differentiation involved in prostate gland development [GO:0060742]; epithelial to mesenchymal transition [GO:0001837]; epithelial tube branching involved in lung morphogenesis [GO:0060441]; fungiform papilla formation [GO:0061198]; gastrulation with mouth forming second [GO:0001702]; genitalia morphogenesis [GO:0035112]; glial cell fate determination [GO:0007403]; hair cell differentiation [GO:0035315]; hair follicle morphogenesis [GO:0031069]; hair follicle placode formation [GO:0060789]; hindbrain development [GO:0030902]; in utero embryonic development [GO:0001701]; layer formation in cerebral cortex [GO:0021819]; lens morphogenesis in camera-type eye [GO:0002089]; lung cell differentiation [GO:0060479]; lung induction [GO:0060492]; lung-associated mesenchyme development [GO:0060484]; male genitalia development [GO:0030539]; mesenchymal cell proliferation involved in lung development [GO:0060916]; mesenchymal stem cell differentiation [GO:0072497]; metanephros morphogenesis [GO:0003338]; midbrain dopaminergic neuron differentiation [GO:1904948]; negative regulation of angiogenesis [GO:0016525]; negative regulation of apoptotic process [GO:0043066]; negative regulation of apoptotic signaling pathway [GO:2001234]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of chondrocyte differentiation [GO:0032331]; negative regulation of gene expression [GO:0010629]; negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis [GO:0003340]; negative regulation of mitotic cell cycle, embryonic [GO:0045976]; negative regulation of oligodendrocyte differentiation [GO:0048715]; negative regulation of osteoclast differentiation [GO:0045671]; negative regulation of oxidative stress-induced neuron death [GO:1903204]; negative regulation of protein sumoylation [GO:0033234]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; nephron tubule formation [GO:0072079]; neural plate development [GO:0001840]; neuron migration [GO:0001764]; neuron projection extension [GO:1990138]; odontogenesis of dentin-containing tooth [GO:0042475]; oocyte development [GO:0048599]; osteoclast differentiation [GO:0030316]; oviduct development [GO:0060066]; pancreas development [GO:0031016]; positive regulation of apoptotic process [GO:0043065]; positive regulation of branching involved in lung morphogenesis [GO:0061047]; positive regulation of core promoter binding [GO:1904798]; positive regulation of determination of dorsal identity [GO:2000017]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of endothelial cell differentiation [GO:0045603]; positive regulation of epithelial cell proliferation involved in prostate gland development [GO:0060769]; positive regulation of epithelial to mesenchymal transition [GO:0010718]; positive regulation of fibroblast growth factor receptor signaling pathway [GO:0045743]; positive regulation of gene expression [GO:0010628]; positive regulation of heparan sulfate proteoglycan biosynthetic process [GO:0010909]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of mesenchymal cell proliferation [GO:0002053]; positive regulation of neuroblast proliferation [GO:0002052]; positive regulation of neuron apoptotic process [GO:0043525]; positive regulation of osteoblast differentiation [GO:0045669]; positive regulation of skeletal muscle tissue development [GO:0048643]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomere maintenance via telomerase [GO:0032212]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription elongation from RNA polymerase II promoter [GO:0032968]; positive regulation of transcription, DNA-templated [GO:0045893]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein localization to cell surface [GO:0034394]; protein polyubiquitination [GO:0000209]; proximal/distal pattern formation [GO:0009954]; regulation of angiogenesis [GO:0045765]; regulation of calcium ion import [GO:0090279]; regulation of centriole-centriole cohesion [GO:0030997]; regulation of centromeric sister chromatid cohesion [GO:0070602]; regulation of fibroblast proliferation [GO:0048145]; regulation of myelination [GO:0031641]; regulation of nephron tubule epithelial cell differentiation [GO:0072182]; regulation of neurogenesis [GO:0050767]; regulation of protein localization to cell surface [GO:2000008]; regulation of secondary heart field cardioblast proliferation [GO:0003266]; regulation of smooth muscle cell proliferation [GO:0048660]; regulation of T cell proliferation [GO:0042129]; regulation of timing of anagen [GO:0051884]; renal inner medulla development [GO:0072053]; renal outer medulla development [GO:0072054]; renal vesicle formation [GO:0072033]; response to estradiol [GO:0032355]; response to xenobiotic stimulus [GO:0009410]; smooth muscle cell differentiation [GO:0051145]; stem cell population maintenance [GO:0019827]; sympathetic ganglion development [GO:0061549]; synapse organization [GO:0050808]; synaptic vesicle clustering [GO:0097091]; synaptic vesicle transport [GO:0048489]; T cell differentiation in thymus [GO:0033077]; thymus development [GO:0048538]; trachea formation [GO:0060440] | 11747475_Loss of beta-catenin may result in the disruption of the function of the cell-cell adhesion complex, which may cause weak cell-cell adhesion and confer invasive properties on a tumor. 11790773_aardvark gene product 11819825_Abnormal beta-catenin gene expression with invasiveness of primary hepatocellular carcinoma in China. 11831984_Beta-catenin might be related to the occurrence and development of kidney tumor. 11834740_Proof for the regulated phosphorylation of the Ser/Thr residues of beta-catenin by Wnt signaling. 11836379_No germline mutations of CTNNB1 have been identified nor linkage to chromosome 3p21 been demonstrated in 8 subjects with Birt-Hogg-Dube syndrome, suggesting that CTNNB1 should be excluded as a candidate gene for BHD. 11836555_Immunohistochemistry of cyclin D1 and beta-catenin, and mutational analysis of exon 3 of beta-catenin gene in parathyroid adenomas 11839663_Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival 11856748_The minimal necessary components of the androgen receptor and beta-catenin required for binding nuclear accumulation of beta-catenin nuclear import appears to be the DNA/ligand binding regions and the Armadillo repeats of beta-catenin 11857309_increased expression predicts favorable prognosis in resected nonsmall cell lung carcinoma 11893906_findings indicate that alterations of beta-catenin are frequent in cancer of the uterine cervix and suggest that they may play an important role in the development of these tumors 11920497_mutations rare in ulcerative colitis-related colorectal carcinomas 11921277_CTNNB1 signaling plays a critical role in the development of a significant fraction of prostate cancers. 11930117_One out of 62 melanoma cell lines was found to carry a mutation in exon 3 of the beta-catenin gene indicating that aberration of the Wnt-1/wingless pathway through activation of beta-catenin is a rare event 11939410_results imply that claudin-1 is involved in the beta-catenin-Tcf/LEF signaling pathway 11940574_Activation of AXIN2 expression by beta-catenin-T cell factor 11950845_ErbB-beta-catenin complexes are associated with human infiltrating ductal breast and murine mammary tumor virus (MMTV)-Wnt-1 and MMTV-c-Neu transgenic carcinomas 11953860_nuclear beta-catenin expression significantly related to ulcerative growth of colorectal cancer 11957146_beta-catenin plays a role in endometrial carcinogenesis, particularly in endometrioid carcinomas 11967263_Arg(96) mutant has a dominant-negative effect on GSK-3beta-dependent phosphorylation of beta-catenin and targeting of beta-catenin for degradation requires prior priming through phosphorylation of Ser(45) 12037680_Protein kinase CK2 dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with beta-TRCp and enhances beta-catenin degradation [UBC3B] 12045208_results indicate selection for APC genotypes that are likely to retain some activity in downregulating beta-catenin signaling 12052822_wnt3a-beta catenin signaling regulates LEF-1 gene expression 12060769_targeted inactivation reveals effects of beta-catenin mutation 12111402_Mutation in exon 3 of the beta-catenin gene was found in 2 of the 20 endometrial cancer samples; however, it was not found in the 25 endometrial hyperplasias or the 20 associated hyperplasias. 12124804_Molecular genetic analysis of malignant melanomas for aberrations of the WNT signaling pathway genes CTNNB1, APC, ICAT and BTRC. 12127563_Anticancer-drug-induced apoptotic cell death in leukemia cells is associated with proteolysis of beta-catenin. beta-Catenin plays a role in promoting Jurkat survival. 12130512_Regulation of leukemic cell adhesion, proliferation, and survival by beta-catenin. 12169098_oxidative stress induces tyrosine phosphorylation and cellular redistribution of occludin-ZO-1 and E-cadherin-beta-catenin complexes by a tyrosine-kinase-dependent mechanism 12183361_Nr-CAM is the gene most extensively induced by beta catenin 12209953_data support the notion that upregulation of cyclin D1 and Fra-1 in human colorectal adenocarcinomas is driven by abnormally expressed beta-catenin; however, the regulation of c-myc expression in colorectal tumors appears to be more complex 12209999_Role of Wnt pathway in medulloblastoma oncogenesis: accumulation of beta-catenin in tumor cells was immunohistochemically proven in 5 cases; 2 cases showed positive immunoreactivity for Wnt-1 and another 2 showed mutation of either CTNNB1 or AXIN1 12219004_Restoration of E-cadherin/beta-catenin expression in pancreatic cancer cells inhibits growth by induction of apoptosis. 12235125_Beta catenin induced human melanoma growth requires the downstream target Microphthalmia-associated transcription factor. 12297048_Presenilin couples beta-catenin phosphorylation through two sequential kinase activities independent of the Wnt-regulated Axin/CK1alpha complex. 12297840_beta-catenin, which participates in the Wnt signaling pathway, might play a more important role in the formation of hepatic adenoma than in that of focal nodular hyperplasia. 12389996_found a pattern of beta-catenin immunostaining in typical carcinoid tumors of the appendix that was different from the pattern seen in non-appendiceal carcinoid tumors 12398896_beta-catenin plays an important role in oncogenesis through the crossregulation of NF-kappa B in breast and colonic neoplasms 12452049_abnormal expression of beta-catenin in gastric carcinoma and survival 12478897_The aberrant expression of beta-catenin protein was statistically correlated to the lymph node metastasis in esophageal cancer. 12482967_These results suggest that the TAK1-NLK MAPK cascade is activated by the noncanonical Wnt-5a/Ca(2+) pathway and antagonizes canonical Wnt/beta-catenin signaling. 12515622_beta-catenin, p53 and PCNA may play important roles in the carcinogenesis of colorectal adenoma. 12532436_Expression of e-cadherin and beta-catenin in human esophageal squamous cell carcinoma: relationships with prognosis. 12532469_Abnormal E-cadherin and alpha-catenin and beta-catenin in pancreatic carcinoma tissues. Abnormal E-cadherin and alpha-catenin with differentiation, lymph node and liver metastases. Aberrant beta-catenin with lymph node and liver metastases. 12556519_CKI epsilon-dependent phosphorylation of Dvl enhances the formation of a complex of Dvl-1 with Frat-1 and this complex leads to the activation of Wnt-3a-induced accumulation of beta-catenin 12575848_In human pilomatricoma, the frequency of beta-catenin gene mutations was remarkably low (30%),in Exon 3 of the beta-catenin gene . 12587534_matrix metalloproteinase-2 and 9 and membrane-type 1 matrix metalloproteinase mRNA expression in endometriosis was higher than in normal endometrium whereas E-cadherin, alpha- and beta-catenin mRNA expression was not suppressed in endometriosis 12592400_Data show that beta-catenin is overexpressed in Kaposi sarcoma and primary effusion lymphoma, and that this overexpression is regulated by the Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen LANA. 12603528_Mutations of beta-catenin, as well as overexpression of beta-catenin and the Tcf-4 gene, independently activate the Wnt pathway in HCC, with the target gene most likely to be C-myc. 12606575_Data indicate that the rate of nuclear export of adenomatous polyposis coli protein (APC), rather than its nuclear import or steady-state levels, determines the transcriptional activity of beta-catenin. 12606944_Pulse-labeled beta-catenin replaces the beta-catenin bound to the cell surface prebiotinylated E-cadherin immediately after synthesis or arrives at the plasma membrane in a complex with the E-cadherin precursor. 12618757_The MUC1/beta-catenin interaction occurs in primary tumors, & is dramatically increased in metastatic lesions. 12679314_dysregulation of beta-catenin may contribute to pancreatic duct adenocarcinoma progression through distinct mechanisms 12692418_These results indicate that the altered expression of beta-catenin, but not cyclin D1, in hepatocellular carcinoma may play an important role in tumor progression by stimulating tumor cell proliferation. 12694354_Aberrant beta-catenin expression may play an important role in the histologic differentiation and tumor staging of mucoepidermoid carcinoma. 12708483_There is a possible role of progesterone in regulation of beta-catenin expression in endometrial tumors. Nuclear beta-catenin accumulation, like gene abnormalities, is associated with the alteration of tumor morphology due to progesterone. 12729800_Data show that beta-catenin and poly(ADP-ribose) polymerase are cleaved during rhodostomin-induced apoptosis, indicating that cell detachment is a prerequisite for apoptosis. 12737446_CTNNB1 mutations were found in 2/19 adenomas without APC mutation. 12748295_Data show that both UBF1 and UBF2 activate RNA polymerase II-regulated, beta-catenin-responsive promoters. 12771132_beta-catenin is degraded via a retinoid X receptor-mediated pathway 12810642_When colon cancer cells with high beta-catenin levels were treated with beta-catenin antisense ODNs, VEGF-A expression was reduced by more than 50%. There is a close link between beta-catenin signaling & VEGF-A expression regulation in colon cancer. 12820959_CTNNB is ubiquinated at specific lysines by the F-box protein beta-TrCP1. 12824925_types of mutations in sinonasal NK/T cell lymphoma in northeast district of China 12839684_In nasopharyngeal cancer, methylation of promoter is a major cause of down-regulation of E-cadherin which may finally lead to detachment and metastasis of neoplastic cells. 12857869_a role for beta-catenin in the control of cell cycle and apoptosis at G2/M 12883680_Mutations of CTNNB1 may not be a factor in tumorigenesis of cervical cancer 12907143_in breast and lung tumor cells, MDA-7 protein expression modulates cell-cell adhesion and intracellular signaling via coordinate regulation of the beta-catenin and PI3K pathways 12917489_model of the interactions between beta-catenin and hedgehog signaling in the epidermis in which SHH promotes proliferation of progenitors of the hair lineages whereas IHH stimulates proliferation of sebocyte precursors 12927518_IpaC interacts with beta-catenin and destabilizes the cadherin-mediated cell adhesion complex. 12937339_provides support that E-cadherin induction by WNT/beta-catenin signaling is an evolutionarily conserved pathway operative in lung cancer cells and that loss of expression may be important in lung cancer development or progression 12952940_Results report a new noncanonical pathway through which Wnt-5a antagonizes the canonical Wnt pathway by promoting the degradation of beta-catenin. 12969793_Overexpression of beta-catenin is associated with hepatocellular carcinoma 12970740_our data indicate that inactivation of beta-catenin by a 3p21.3 homozygous deletion might be a crucial event in the development of the mesothelioma NCI-H28 cells. 12972427_interaction with the co-activator, p300, underlies the trans-repression of beta-catenin signaling by nuclear receptors and their ligands 14520463_Increased expression of beta-catenin is associated with ovarian epithelial cell transformation and in tumour progression 14563838_the beta-catenin pathway is activated by histamine 14633602_Beta-catenin gene mutations are a peculiar feature of skin tumors with matrical differentiation and correlate with a pattern of intense and diffuse beta-catenin nuclear expression. 14637149_We conclude, therefore, that two major components of cell-cell interaction synergistically regulate cell cycle progression in HEK293 cells by regulating p21 expression in a beta-catenin/TCF-dependent manner. 14660579_beta-catenin interferes with transforming growth factor-beta-mediated growth arrest by inducing the expression of BAMBI 14742711_FAM associates with E-cadherin and beta-catenin during trafficking to the plasma membrane 14766232_beta-catenin binds to MUC1 and has a role in T cell receptor signaling 14977843_beta-catenin has a role in progression of colorectal neoplasms 14985333_beta-catenin phosphorylation/degradation is regulated by CDK2 14993280_alpha-catenin and alpha-catulin have distinct activities that downregulate, respectively, beta-catenin and Ras signals converging on the cyclin D1 promoter 15064706_p53-mediated reduction in beta-catenin involves enhanced phosphorylation of beta-catenin on key NH(2)-terminal serines. 15077166_Cyr61 activated the beta-catenin/TCF4 complex, which promoted the expression of c-myc and the latter induced expression of p53 15082903_Wnt signaling pathway associated with beta-catenin regulation in breast cancer tissue 15102686_MUC1 and beta-catenin have a role in progression and invasiveness of colorectal carcinomas 15111320_Beta-catenin simultaneously induces activation of the p53-p21WAF1 pathway and overexpression of cyclin D1 during tumor cell differentiation 15126105_High nuclear expression of beta-catenin is correlated with locally advanced colorectal cancer 15138556_Translocalization of beta-catenin is associated with invasion in gastric cancer 15149841_CTNNB strongly increased activity of the eN/Nt5 promoter and this increase depended on the presence of TCF-1; demonstrated a link between Wnt signaling and the regulation of eN and ADA, which control the metabolism of adenosine 15194432_E-cadherin-mediated cell adhesion is required for keratinocyte-mediated control of melanocytic cells, which can override proliferative activity of beta-catenin. 15215241_beta-catenin/TCF transcriptional activity is blocked by Cdx1 and Cdx2, which then inhibits colon cancer cell proliferation 15239100_LECT2, which encodes a protein with chemotactic properties for human neutrophils, is a direct target gene of Wnt/beta-catenin signaling in the liver. 15254684_beta-catenin nuclear accumulation plays a role in Dukes' D human colorectal cancers 15279902_beta-catenin is downregulated by H2O2 which negatively modulates the Wnt signal pathway 15289833_abnormal immunhistochemical E-cadherin and beta-catenin expression is associated with changes in pit pattern in invasive colorectal neoplasms 15304487_there is cross-talk between Wnt and estrogen signaling pathways via functional interaction between beta-catenin and ERalpha 15331612_In HT29 and HCT116 colorectal cancer cells, beta-catenin/TCF transcriptional activity is inhibited by AP-2alpha due to formation of AP-2alpha/APC/beta-catenin complex. 15381698_terminal tail is responsible for discerning among binding of factors to the armadillo domain 15381903_negative immunoreactivity of beta-catenin in serous carcinomas and the presence of residual tumor seem to be useful markers in selecting patients likely to have an unfavorable course 15455387_Lower levels of nuclear beta-catenin is associated with prostate cancer progression 15492040_beta-catenin is targeted to adhesive or transcriptional complexes, depending on its molecular form 15500294_The beta-catenin, the main system of adherens junction, present in the tight junctions in HepG2 cells. 15514031_IGF-I modulates androgen signaling through beta-catenin 15520370_Observational study of gene-disease association. (HuGE Navigator) 15523694_Observational study of gene-disease association. (HuGE Navigator) 15557107_beta-catenin accumulates in the nucleus of epithelial cells of juvenile polyps 15572674_The interaction and functional cooperation between FHL2, CITED4, and CTNNB were studied. 15591320_identify selective beta-catenin binding hot spots of Tcf4, E-cadherin, and APC 15654359_beta-catenin plays a central role in mesenchymal cells during the healing process, and is an appealing therapeutic target for disorders of wound healing. 15660698_Loss of expression of E-cadherin and beta-catenin may play an important role in the progression of pulmonary adenocarcinoma. 15665104_Induction of the Wnt/beta-catenin pathway by LiCl also elevated PPARgamma levels and induced PPARgamma-dependent reporter and endogenous target genes. 15668893_Alterations in beta-catenin and PTEN genes, as well as MSI, are frequent in low-stage ovarian carcinomas of endometrioid type that have a favorable prognosis. 15670774_Together, we suggest that quercetin is an excellent inhibitor of beta-catenin/Tcf signaling in SW480 cell lines, and the reduced beta-catenin/Tcf transcriptional activity is due to the decreased nuclear beta-catenin and Tcf-4 proteins. 15684397_Suppression of beta-catenin expression by small interfering RNA decreased the apoptotic response to TGF-beta. 15694380_Here, we generated a fusion in which XWnt8 was fused to the N-terminus of LRP6 and show it synergizes with both Fz4 and Fz5 to potently transactivate beta-catenin-dependent Wnt signaling 15695815_ubiquitin-independent degradation of alpha-catenin regulates beta-catenin signaling and maintenance of the differentiated phenotype of articular chondrocytes 15696778_All 61 meningothelial meningiomas, 10 of 12 invasive meningiomas, and 3 of 5 anaplastic meningiomas were positive for both ECAD and beta-catenin, while these were both negative in all of the fibrous meningiomas. 15698401_Beta-catenin stabilization because of either beta-catenin or AXIN I mutation might be a late event for malignant progression rather than an early genetic event involving the initiation of HCC development. 15728254_LEF-1 expression is regulated through PITX2, LEF-1 and beta-catenin direct physical interactions 15737630_In conclusion, prolonged CPB time entails neutrophil-mediated decrease in MVEC beta-catenin expression, and thus may be an important trigger for BBB disruption. 15781969_Findings suggest that phospho-beta-catenin accumulation in Alzheimer's disease might result from impaired proteasome function. 15790758_crosstalk between the beta-catenin and NF-kappaB signaling pathways is an important regulator of intestinal inflammation 15791567_data indicate that altered expression of beta-catenin may play an important role in oral cancer progression through increased proliferation and invasiveness under epidermal growth factor receptor (EGFR) activation but not mutation or cyclin D1 expression 15806138_TCF4 expression mediated by beta-catenin/p300 may be important for initial steps during trans-differentiation of endometrial carcinoma cells. 15817486_Gas2DN can increase the activity of calpain and induce degradation of stabilized/mutated beta-catenin 15832407_The beta-catenin expression in hepatocellular carcinoma cells was heterogeneous among types of hepatitis viral infection 15837931_Lysophosphatidic acid induced colon cancer cell proliferation requires the beta-catenin signaling pathway. 15853773_The regulation of GLCE expression by 2 cis-acting elements of the beta-catenin-TCF4 complex located in the enhancer region of the promoter are reported. 15888491_Tumor sections frmo colorectal cancer patients showed elevated expression levels of AKT1, correlating with enhanced cytoplasmic/nuclear expression of beta-catenin. 15896469_Wnt/beta-catenin signalling pathway is activated in most of gastric cancers, which may play pivotal roles either in gastric cancer formation or in tumour invasion and dissemination 15899904_cyclooxygenase-2/PGE2 may exert pro-oncogenic actions through stimulating the beta-catenin/T cell factor-mediated transcription, which plays critical roles in colorectal carcinogenesis 15905404_results demonstrate a role for beta-catenin in regulating FOXO function that is particularly important under conditions of oxidative stress 15916880_significantly reduced rates of lymph-node metastases were observed in beta-catenin-positive T1 and T2 squamous cell carcinomas of the mouth floor 15927956_analysis of conformation of the oncogenic protein beta-catenin containing the phosphorylated motif DpSGXXpS bound to the beta-TrCP protein 15958533_ADAM10 has a role in E-cadherin shedding and epithelial cell-cell adhesion, migration, and beta-catenin translocation 15972952_The presence of activated beta-catenin and c-myc in the epidermis of chronic wounds may serve as a molecular marker of impaired healing 15987741_Transendothelial migration is compromised in melanoma cells expressing a dominant-negative form of beta-catenin, thus supporting a regulatory role of beta-catenin signaling in this process. 16012954_beta-Catenin transgenic mice show an in vivo hepatotrophic effect secondary to increased basal hepatocyte proliferation 16038041_Both cell surface reduction and intranuclear accumulation of beta-catenin were detected in intestinal metaplasia 16082250_nuclear beta-catenin is a rare phenomenon in colorectal SRCC, but the involvement of it may indicate a worse prognosis with shorter survival than colorectal SRCC without nuclear beta-catenin expression 16084063_results suggest that the Wnt/beta-catenin signaling pathway plays dual functions in head and neck squamous cell carcinoma (HNSCC) development: promoting both cell survival and invasive growth of HNSCC cells. 16114033_Splice variants of CTNNB1 ands downstream targets were used as markers for neoplastic progression of esophageal cancer. 16124054_Overexpression of Pin1 and beta-catenin may be closely related with the development and/or progression of colorectal carcinoma and further supports that Pin1 overexpression might contribute to the upregulation of beta-catenin. 16126725_cadherins mediate both the association of PS1 and beta-catenin and the effects of PS1 on the cellular levels of beta-catenin 16132582_Altered beta-catenin distribution in gastric cancer may result from the imbalance of E-cadherin production and Wnt expression, which confers on gastric cancer cells more aggressive behaviors. 16133456_The expression of beta-catenin, p63 and CD34 in the course of androgenetic alopecia is reported. 16163548_The cytoplasmic accumulation of beta-catenin is a common characteristic of oral SCC, but is not closely associated with mutational alterations in the APC, beta-catenin and Axin1 genes. 16199882_PKA inhibits the ubiquitination of beta-catenin by phosphorylating beta-catenin, thereby causing beta-catenin to accumulate and the Wnt signaling pathway to be activated. 16204248_TIS7, a negative regulator of transcriptional activity, represses expression of OPN and beta-catenin/Tcf-4 target genes 16239965_Data suggest a beta-catenin-dependent, stage-specific role for Notch1 signaling in promoting the progression of primary melanoma. 16254206_Beta-catenin may be an important modulator of angiogenesis and myocyte regeneration 16291872_The positive inter-regulation between beta-cat/Tcf-4 signaling and ET-1 signaling potentiates proliferation and survival of prostate cancer (CaP) cells, thereby representing a novel mechanism that contributes to CaP progression. 16311123_results suggest an established Wnt signaling pathway in most gastric cancers, a close correlation of beta-catenin/TCF4-mediated signaling with tumor dissemination, and the unlikelihood of a direct effect of activated Wnt signaling on CD44 expression 16328013_Results showed that inhibition of PI-3 kinase with wortmannin was accompanied by a considerably reduced expression of beta-catenin. 16329837_Wnt-1, beta-catenin and APC expressions were related to the differentiation of oral squamous cell carcinoma. 16343437_These results indicate that CIP4 is critical for beta-catenin-mediated cell-cell adhesion. 16344550_CoCoA uses different combinations of functional domains in its synergistic coactivator function with beta-catenin or GRIP1 16356174_APC and K-ras, but not CTNNB1 mutations have roles in regulation of expression of hMLH1 in sporadic colorectal carcinomas 16356174_Observational study of gene-disease association. (HuGE Navigator) 16371504_The data suggest a novel role for tyrosine phosphorylation of N-cadherin by Src family kinases in the regulation of beta-catenin association during transendothelial migration of melanoma cells. 16378715_beta-catenin mutation and its nuclear localization are frequent causes of Wnt signaling pathway activation suggesting that beta-catenin activation mutations contribute to tumorigenesis of pilomatricomas 16378739_colon cancer cells retain significant amounts of LEF-1 induced nuclear beta-catenin compared to LEF-1 transfected normal epithelial cells; beta-Catenin binds directly to CRM1 & overexpression of CRM1 reduces nuclear beta-catenin-mediated transactivation 16382042_sulindac sulfone can modulate the APC/beta-catenin pathway in vitro but its efficacy is dependent upon the mutational status of APC and beta-catenin 16407829_In esophageal adenocarcinomas, nuclear translocation of beta-catenin was observed regardless of the expression of APC. 16428447_High Pin1 expression in primary prostate cancer markedly inhibits the beta-catenin interaction with androgen receptor. 16442529_Expressed Bcr is able to bind the transcription factor Tcf1 to disrupt the Tcf1/beta-catenin complex. Phosphorylation of Bcr by the tyrosine kinase pp60(src) can lead to dissociation of the transcriptionally inactive Bcr/Tcf1 complex 16462762_These results unravel a novel pathway in the control of beta-catenin cellular transport and strongly suggest that SYT-SSX2 contributes to tumor development, in part through beta-catenin signaling 16465411_Immunohistological examination of nuclear accumulation of beta-catenin may be useful for diagnosing malignant immunohistological examination of nuclear accumulation of beta-catenin may thus be useful for diagnosing malignant PLTs. 16474376_Phospho-beta-catenin may have a different involvement in invasive breast carcinomas, according to its subcellular distribution. 16476742_analysis of a novel, noncanonical mechanism of modulation of beta-catenin signaling through direct phosphorylation of beta-catenin by PKA, promoting its interaction with CREB-binding protein 16496348_beta-catenin is activated by HBxAg, in part, through the upregulated expression of the HBxAg effector URG11; URG11 stimulates the beta-catenin promoter and hepatocellular growth and survival 16501564_Nuclear localization of beta-catenin, an indirect evidence of deregulated Wnt signaling pathway, was observed in 5 (19%) small intestinal adenocarcinomas and 36 (71%) colorectal adenocarcinomas. 16507986_the cross talk of KLF4 and beta-catenin plays a critical role in homeostasis of the normal intestine as well as in tumorigenesis of colorectal cancers. 16513652_active CKIepsilon generation may induce a negative feedback loop by phosphorylation of sites on LRP5/6 that modulate axin binding and hence beta-catenin degradation 16565090_the MED12 interface within Mediator is a new component in the Wnt/beta-catenin pathway 16574648_PCP-2 may play an important role in the maintenance of epithelial integrity, and a loss of its regulatory function may be an alternative mechanism for activating beta-catenin signaling. 16621789_BMP-2 antagonizes Wnt-3a signaling in osteoblast progenitors by promoting an interaction between Smad1 and Dvl-1 that restricts beta-catenin activation 16628468_The results indicate that estrogen plus overexpressed ERalpha induce LoVo cell apoptosis might mediate through the increase of hTNF-alpha gene expression, which in turn activate caspase-8, -9 and caspase-3 and lead to the DNA fragmentation and apoptosis. 16644723_GR can bind beta-catenin in vitro, suggesting that GC and Wnt signaling pathways are linked directly through their effectors 16688229_variations in beta-catenin protein levels were dependent on post-transcriptional mechanisms involving the Wnt/beta-catenin pathway only in leukemic cells 16696969_Results suggest that alternative splicing and AU-rich elements can act together in regulating beta-catenin mRNA stability and thereby provide a step of controlling the cellular beta-catenin concentration. 16707106_beta-catenin is regulated via epsilon-cleavage of N-cadherin 16712787_STMN2 is required for maintaining the anchorage-independent growth state of beta-catenin/TCF-activated hepatoma cells 16717102_histone deacetylase inhibitor sodium butyrate induces G1/S phase arrest in E1A + Ras-transformed cells through down-regulation of E2F1 activity and stabilization of beta-catenin 16724116_These data indicate that the intracellular amounts of HIC1 protein can modulate the level of the transcriptional stimulation of the genes regulated by canonical Wnt/beta-catenin signaling. 16730693_When beta-catenin is activated in transgenic En1 expressing cells, it induces Dermo1 expression in all cells of the En1 domain and disrupts muscle gene expression. 16753179_the central region of APC is unstructured in the absence of beta-catenin and Axin; beta-catenin may interact with each of the APC 15aa and 20aa repeats independently 16756720_Wnt/beta-catenin activation was observed in 65% of pancreatic adenocarcinomas, independently of beta-catenin gene mutations in most tumors 16760136_beta-catenin level depends on the way and level of Wnt pathway activation 16772034_Splice forms of crucial genes of the Wnt-pathway, beta-Catenin, LRP5, GSK3beta, Axin-1 and CtBP1 are expressed in human colorectal tissue. 16786128_beta-catenin might be involved in the Hh signaling pathway via enhancement of the transcriptional activity of GLI 16798748_APC regulates beta-catenin phosphorylation and ubiquitination by distinct domains and by separate molecular mechanisms 16815294_In conclusion, the canonical Wnt/beta-catenin pathway enhanced monocyte-endothelial cell adhesion without changing expression levels of adhesion molecules. 16830381_Comparative analysis of nuclear and membrane/cytoplasmic beta-catenin can predict local tumor infiltration. 16843107_Observational study of gene-disease association. (HuGE Navigator) 16843107_These data indicate that somatic mutations affecting APC and CTNNB1 do not play a major role in the pathogenesis of sporadic ependymomas. 16865250_Reduced membranous expression of beta-catenin was associated with metastasis in salivary adenoid cystic carcinoma 16927799_There was no statistical significance for beta-catenin expression between the invasive group and noninvasive group in pituitary adenoma. 16930546_in vivo modulation of PRA1 may be involved in TCF/beta-catenin signaling, as well as cellular proliferation and tumorigenesis 16942611_These data indicate that changes in Wnt expression per se are unlikely to be the cause of the observed dysregulation of beta-catenin expression in DD. 16945989_These findings suggest that the participation of beta-catenin in adhesion and signaling may represent a novel mechanism through which gonadotropins may regulate the cellular fate of human ovarian surface epithelial cells. 16952352_Data suggest that intact 654 and 670 tyrosine residues in beta-catenin are crucial in HGF-mediated Met-beta-catenin dissociation, beta-catenin translocation, activation and mitogenesis. 16953230_A functional crosstalk between hepatocyte growth factor receptor (MET) and beta-catenin signaling sustains and increases colorectal carcinoma cell invasive properties. 17008323_vascular endothelial growth factor expression is induced through the glucocorticoid receptor-related phosphatidylinositol 3-kinase/Akt and beta-catenin/T-cell factor-dependent pathway in human endothelial cells 17018282_Data show that P68 RNA helicase mediates platelet-derived growth factor-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. 17030184_Exposure of colon cancer cells to nitric oxide unraveled a so-far-unidentified mechanism of beta-catenin regulation. 17050667_These data reveal a potentially important role for transcriptionally active beta-catenin in the regulation of Rad6B gene expression, and link aberrant beta-catenin signaling with transcriptional deregulation of Rad6B and breast cancer development. 17052462_crystallographic analysis of how beta-catenin, BCL9, BCL9-2 and Tcf4 interact 17090192_Alterations in adenomatous polyposis coli/beta-catenin pathway and cyclin D1 dysregulation may contribute to pathogenesis of pleuropulmonary desmoid tumors and solitary fibrous tumors. 17090604_Beta-catenin relieves I-mfa-mediated suppression of LEF-1 in mammalian cells. 17121828_Wnt/beta-catenin signaling may contribute to colorectal carcinogenesis by reducing the level of the E2F4 cell cycle repressor via an antisense mechanism 17128412_COX-2 and beta-Catenin may have roles in regulating intracellular Survivin levels in mouse and human colon cancer 17129455_The Pin1 and beta-catenin signalling pathways were activated in salivary adenoid cystic carcinoma. 17143299_Wnt/beta-cat | ENSMUSG00000006932 | Ctnnb1 | 8686.18110 | 3.419246e-01 | -1.548250 | 0.3202994 | 22.48946 | 2.112994e-06 | 1.313953e-03 | Yes | No | 3.977480e+03 | 8.181180e+02 | 1.182550e+04 | 2.493135e+03 | |
| ENSG00000173915 | 84833 | ATP5MK | protein_coding | Q96IX5 | FUNCTION: Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation (PubMed:29917077). ATP5MK is a minor subunit of the mitochondrial membrane ATP synthase required for dimerization of the ATP synthase complex and as such regulates ATP synthesis in the mitochondria (PubMed:21345788, PubMed:29917077). {ECO:0000269|PubMed:21345788, ECO:0000269|PubMed:29917077, ECO:0000303|PubMed:29917077}. | Acetylation;Membrane;Mitochondrion;Primary mitochondrial disease;Reference proteome;Transmembrane;Transmembrane helix | hsa:84833; | integral component of membrane [GO:0016021]; mitochondrial proton-transporting ATP synthase complex [GO:0005753]; mitochondrion [GO:0005739] | 21345788_a critical role of DAPIT in maintaining the ATP synthase population in mitochondria and raise an intriguing possibility of active role of DAPIT in cellular energy metabolism. 22688299_confirm the mitochondrial presence of DAPIT on cellular level. We also show that DAPIT is localized in lysosomes of HUVEC and HEK 293T cells 26161955_DAPIT over-expression thus appears to modulate mitochondrial functions and alter cellular regulations, promote anaerobic metabolism and induce EMT-like transition. 29234032_To determine whether USMG5 is related to the development of heart failure, authors performed clinical and experimental studies. Microarray analysis showed that the expression levels of USMG5 were positively correlated with those of natriuretic peptide precursor A in the human failed myocardium. | ENSMUSG00000071528 | Atp5md | 1187.57369 | 4.517137e-01 | -1.146519 | 0.2995631 | 14.32923 | 1.534637e-04 | 4.157251e-02 | Yes | No | 7.653172e+02 | 1.520069e+02 | 1.587919e+03 | 3.223989e+02 | ||
| ENSG00000176225 | 25914 | RTTN | protein_coding | Q86VV8 | FUNCTION: Involved in the genetic cascade that governs left-right specification. Plays a role in the maintenance of a normal ciliary structure. Required for correct asymmetric expression of NODAL, LEFTY and PITX2. {ECO:0000269|PubMed:22939636}. | Acetylation;Alternative splicing;Cell projection;Cilium;Cytoplasm;Cytoskeleton;Developmental protein;Disease variant;Phosphoprotein;Reference proteome | This gene encodes a large protein whose specific function is unknown. Absence of the orthologous protein in mouse results in embryonic lethality with deficient axial rotation, abnormal differentiation of the neural tube, and randomized looping of the heart tube during development. In human, mutations in this gene are associated with polymicrogyria with seizures. In human fibroblasts this protein localizes at the ciliary basal bodies. Given the intracellular localization of this protein and the phenotypic effects of mutations, this gene is suspected of playing a role in the maintenance of normal ciliary structure which in turn effects the developmental process of left-right organ specification, axial rotation, and perhaps notochord development. [provided by RefSeq, Jan 2013]. | hsa:25914; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; centriole replication [GO:0007099]; centriole-centriole cohesion [GO:0010457]; ciliary basal body organization [GO:0032053]; determination of left/right symmetry [GO:0007368] | 11900971_Study characterizing mouse rotatin gene. 22939636_RTTN mutations therefore link aberrant ciliary function to abnormal development and organization of the cortex in human individuals. 26608784_RTTN mutations cause primary microcephaly and primordial dwarfism in humans. 26940245_We found a novel homozygous mutation in RTTN associated with microcephalic PD as well as complex brain malformations and congenital dermatitis, thus expanding the phenotypic spectrum of both RTTN-associated diseases and ciliary dysfunction. 28811500_RTTN directly interacts with STIL and acts downstream of STIL-mediated centriole assembly, contributing to building full-length centrioles. 29883675_We here report three individuals from two unrelated families with novel mutations in the RTTN gene. The phenotype consisted of microcephaly, short stature, pachygyria or polymicrogyria, colpocephaly, hypoplasia of the corpus callosum and superior vermis. These findings provide further confirmation of the phenotype related to pathogenic variants in RTTN. 30121372_We identified, by trio based whole exome sequencing, a homozygous missense mutation in the RTTN gene (c.2953A>G; p.(Arg985Gly)) in one Moroccan patient from a consanguineous family. 30168418_Data show that PPP1R35 acts downstream of, and forms a complex with, RTTN, a microcephaly protein required for distal centriole elongation. 30927481_Study reports on a consanguineous family with three adult members with primary microcephaly, developmental delay, primordial dwarfism, and brachydactyly segregating a homozygous splice site variant NM_173630.3:c.5648-5T>A in RTTN. The variant RTTN allele results in a nonhypomorphic skipping of exon 42 and a frameshift [(NP_775901.3:p.Ala1883Glyfs*6)]. 34207628_Human Microcephaly Protein RTTN Is Required for Proper Mitotic Progression and Correct Spindle Position. | ENSMUSG00000023066 | Rttn | 594.32117 | 3.915917e-01 | -1.352578 | 0.2991330 | 18.85117 | 1.413241e-05 | 7.273952e-03 | Yes | No | 3.795043e+02 | 6.389836e+01 | 8.268628e+02 | 1.415424e+02 | |
| ENSG00000181610 | 51649 | MRPS23 | protein_coding | Q9Y3D9 | 3D-structure;Acetylation;Disease variant;Mitochondrion;Primary mitochondrial disease;Reference proteome;Ribonucleoprotein;Ribosomal protein | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein. A pseudogene corresponding to this gene is found on chromosome 7p. [provided by RefSeq, Jul 2008]. | hsa:51649; | mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nuclear membrane [GO:0031965]; ribosome [GO:0005840]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; translation [GO:0006412] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 31950385_MRPS23 amplification and gene expression in breast cancer; association with proliferation and the non-basal subtypes. 33927350_Arginine and lysine methylation of MRPS23 promotes breast cancer metastasis through regulating OXPHOS. 33964376_Mitochondrial ribosomal small subunit proteins (MRPS) MRPS6 and MRPS23 show dysregulation in breast cancer affecting tumorigenic cellular processes. | ENSMUSG00000023723 | Mrps23 | 1152.04196 | 3.815118e-01 | -1.390200 | 0.3096225 | 19.57308 | 9.682391e-06 | 5.155372e-03 | Yes | No | 5.904447e+02 | 1.109229e+02 | 1.500092e+03 | 2.876866e+02 | ||
| ENSG00000186567 | 56971 | CEACAM19 | protein_coding | Q7Z692 | Alternative splicing;Glycoprotein;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:56971; | integral component of membrane [GO:0016021] | 20634891_Observational study of gene-disease association. (HuGE Navigator) 23525470_Our results suggest that CEACAM19 mRNA expression represents a promising, novel and clinically useful tissue biomarker for breast cancer management. 27909883_In conclusion, CEACAM19 showed high expression in tumor samples compared to normal mammary tissue. 30217308_High CEACAM19 expression is associated with gastric cancer. 32659328_Identification and expression analysis of novel splice variants of the human carcinoembryonic antigen-related cell adhesion molecule 19 (CEACAM19) gene using a high-throughput sequencing approach. | ENSMUSG00000049848 | Ceacam19 | 142.98237 | 1.428803e-01 | -2.807121 | 0.4530886 | 34.97550 | 3.338808e-09 | 5.155453e-06 | Yes | Yes | 2.784475e+01 | 7.794590e+00 | 2.190474e+02 | 5.932509e+01 | |||
| ENSG00000189043 | 4697 | NDUFA4 | protein_coding | O00483 | FUNCTION: Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules unsing 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix (PubMed:22902835). NDUFA4 is required for complex IV maintenance (PubMed:22902835). {ECO:0000269|PubMed:22902835}. | 3D-structure;Acetylation;Electron transport;Leigh syndrome;Membrane;Mitochondrion;Mitochondrion inner membrane;Phosphoprotein;Primary mitochondrial disease;Reference proteome;Respiratory chain;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene belongs to the complex I 9kDa subunit family. Mammalian complex I of mitochondrial respiratory chain is composed of 45 different subunits. This protein has NADH dehydrogenase activity and oxidoreductase activity. It transfers electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone. [provided by RefSeq, Jul 2008]. | hsa:4697; | mitochondrial inner membrane [GO:0005743]; mitochondrial membrane [GO:0031966]; mitochondrial respiratory chain complex I [GO:0005747]; mitochondrial respiratory chain complex IV [GO:0005751]; mitochondrion [GO:0005739]; NADH dehydrogenase (ubiquinone) activity [GO:0008137]; protein-containing complex binding [GO:0044877]; mitochondrial electron transport, NADH to ubiquinone [GO:0006120]; positive regulation of cytochrome-c oxidase activity [GO:1904960] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 23746447_Findings support reassignment of the NDUFA4 protein to complex IV and suggest that patients with unexplained COX deficiency should be screened for NDUFA4 mutations. 30238562_Results found that NDUFA4 mRNAs are upregulated and associated with oxidative phosphorylation pathway in gastric cancer. Also, NDUFA4 seems to promote the proliferation, reduce apoptosis, and regulate the oxidative phosphorylation pathway in gastric cancer cells. 30348529_NDUFA4 was identified as a direct target of miR-147b and knockdown of NDUFA4 abolished the oncogenic role of miR-147b inhibitor. 33837217_Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. | ENSMUSG00000029632 | Ndufa4 | 2960.42773 | 3.490145e-01 | -1.518641 | 0.3128244 | 22.54706 | 2.050582e-06 | 1.313953e-03 | Yes | No | 1.500479e+03 | 2.567854e+02 | 4.206271e+03 | 7.363778e+02 | |
| ENSG00000198408 | 10724 | OGA | protein_coding | O60502 | FUNCTION: [Isoform 1]: Cleaves GlcNAc but not GalNAc from O-glycosylated proteins. Can use p-nitrophenyl-beta-GlcNAc and 4-methylumbelliferone-GlcNAc as substrates but not p-nitrophenyl-beta-GalNAc or p-nitrophenyl-alpha-GlcNAc (in vitro) (PubMed:11148210). Does not bind acetyl-CoA and does not have histone acetyltransferase activity (PubMed:24088714). {ECO:0000269|PubMed:11148210, ECO:0000269|PubMed:11788610, ECO:0000269|PubMed:20673219, ECO:0000269|PubMed:22365600, ECO:0000269|PubMed:24088714}.; FUNCTION: [Isoform 3]: Cleaves GlcNAc but not GalNAc from O-glycosylated proteins. Can use p-nitrophenyl-beta-GlcNAc as substrate but not p-nitrophenyl-beta-GalNAc or p-nitrophenyl-alpha-GlcNAc (in vitro), but has about six times lower specific activity than isoform 1. {ECO:0000269|PubMed:20673219}. | 3D-structure;Alternative splicing;Cytoplasm;Glycosidase;Host-virus interaction;Hydrolase;Nucleus;Phosphoprotein;Reference proteome | The dynamic modification of cytoplasmic and nuclear proteins by O-linked N-acetylglucosamine (O-GlcNAc) addition and removal on serine and threonine residues is catalyzed by OGT (MIM 300255), which adds O-GlcNAc, and MGEA5, a glycosidase that removes O-GlcNAc modifications (Gao et al., 2001 [PubMed 11148210]).[supplied by OMIM, Mar 2008]. | hsa:10724; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine O-N-acetyl-alpha-D-glucosaminase activity [GO:0102167]; [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine/L-threonine O-N-acetyl-alpha-D-glucosaminase activity [GO:0102571]; [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-threonine O-N-acetyl-alpha-D-glucosaminase activity [GO:0102166]; beta-N-acetylglucosaminidase activity [GO:0016231]; hyalurononglucosaminidase activity [GO:0004415]; glycoprotein catabolic process [GO:0006516]; glycoprotein metabolic process [GO:0009100]; N-acetylglucosamine metabolic process [GO:0006044]; protein deglycosylation [GO:0006517]; protein O-linked glycosylation [GO:0006493] | 11148210_This protein is a cytosolic, neutral, O-GlcNAc specific hexosaminidase termed O-GlcNAcase. 12359146_Investigated this locus in Pima Indians who have the world's highest prevalence of NIDDM. Concluded that mutations in MGEA5 are unlikely to contribute to NIDDM in this population 12359146_Observational study of gene-disease association. (HuGE Navigator) 17546623_In type 2 Diabetes patients in Mexico City, the frequency of the T allele of MGEAT5 was higher (2.6%) in the cases than in controls (1.8%), but not a significant deviation from Hardy_Weinberg proportions. 17546623_Observational study of gene-disease association. (HuGE Navigator) 18641620_review of modifications, phosphorylation and a specific form of glycosylation, O-linked -N-acetylglucosaminylation by O-GlcNAc, relevant to pathological tau phosphorylation 19423084_the short nuclear variant of O-GlcNAcase, which has the identical catalytic domain as the full-length enzyme, has similar trends in a pH-rate profile and Taft linear free energy analysis as the full-length enzyme 19715310_characterization of O-GlcNAcase transition states using several series of substrates to generate multiple simultaneous free-energy relationships 19913121_Observational study of gene-disease association. (HuGE Navigator) 20198314_This study analyzes the activity of the enzyme involved in the removal of these sugar residues, i.e. beta-N-acetylglucosaminidase (O-GlcNAcase) as well as the level of N-acetylglucosamine in benign and malignant thyroid lesions. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20673219_N-terminal region of OGA contains the catalytic site and the C-terminal region stabilizes the protein structure and affects substrate affinity. 20863279_Results provide evidence that OGA may possess a substrate-recognition mechanism tinvolving interactions with O-GlcNAcylated proteins beyond the GlcNAc-binding site. 21178104_Direct evidence links muscle atrophy and the disruption of O-GlcNAcase activity in male bitransgenic mice. 21471514_Reducing ChREBP(OG) levels via OGA overexpression decreased lipogenic protein content (ACC, FAS), prevented hepatic steatosis, and improved the lipidic profile of OGA-treated db/db mice. 21567137_Decrease in MGEA5 and increase in O-GlcNAc transferease expression in higher grade tumors suggests that increased O-GlcNAc modification may be implicated in breast tumor progression and metastasis. 21717526_Chromosomal translocation t(1;10) is consistent with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. 22311971_Data show that the interplay between O-GlcNAc and phosphorylation on proteins and indicate that these effects can be mediated by changes in hOGT and hOGA kinetic activity. 22730328_O-linked beta-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-beta-D-glucosaminidase silencing on cell phenotype and transcriptome. 22783592_Analysis of urinary content of MGEA5 and OGT may be useful for bladder cancer diagnostics. 24365779_Estrogen replacement therapy and plyometric training influence muscle OGT and OGA gene expression, which may be one of the mechanisms by which HRT and PT prevent aging-related loss of muscle mass. 24705316_Report the presence of TGFBR3 and/or MGEA5 rearrangements in pleomorphic hyalinizing angiectatic tumors and the spectrum of related neoplasms. 25173736_Amino acid composition of splice variants, post-translational modifications, and stable associations with regulatory proteins influence subcellular distribution/substrate specificity of OGA and OGT (O-linked N-acetylglucosamine transferase). [REVIEW] 25183011_This work identifies the first target of miR-539 in the heart and the first miRNA that regulates OGA. 26269457_OGA overexpression in endothelial cells improves endothelial function and may have a beneficial effect on coronary vascular complications in diabetes. 26527687_E2F1 negatively regulates both Ogt and Mgea5 expression in an Rb1 protein-dependent manner. 26980036_TGFBR3 and/or MGEA5 rearrangements are much more common in hybrid hemosiderotic fibrolipomatous tumor-myxoinflammatory fibroblastic sarcomas than in classical myxoinflammatory fibroblastic sarcomas. 27231347_the O-linked N-acetylglucosamine (O-GlcNAc) processing enzymes, O-GlcNAc-transferase (OGT) and O-GlcNAcase (OGA), interact with the (A)gamma-globin promoter at the -566 GATA repressor site 27601472_OGA is physically associated with the known RNA polymerase II (pol II) pausing/elongation factors SPT5 and TRIM28-KAP1-TIF1beta, and a purified OGA-SPT5-TIF1beta complex has elongation properties. 28319083_hOGA forms an unusual arm-in-arm homodimer in which the catalytic domain of one monomer is covered by the stalk domain of the sister monomer to create a substrate-binding cleft. 28627871_Data suggest that the substrate specificity of O-GlcNAcase/OGA does not extend to proteins/peptides modified with S-GlcNAc (an analog of O-GlcNAc); proteins modified with S-GlcNAc appear to be stable against O-GlcNAcase/OGA hydrolysis. 28742148_Tax interacts with the host OGT/OGA complex and inhibits the activity of OGT-bound OGA. 28939839_Beta-N-acetylhexosaminidase substrate recognition and specificity 30052810_OGT promotes carcinogenesis and metastasis of cervical cancer cells. OGT's expression is significantly upregulated in cervical cancer, and low OGT level is correlated with improved prognosis 30920848_role of the tubular biomarkers NAG, kidney injury molecule-1 and neutrophil gelatinase-associated lipocalin in patients with chest pain before contrast media exposition 31501520_Study reports that OGA is upregulated in a wide range of human cancers and drives aerobic glycolysis and tumor growth by inhibiting pyruvate kinase M2 (PKM2). PKM2 is dynamically O-GlcNAcylated in response to changes in glucose availability. Under high glucose conditions, PKM2 is a target of OGA-associated acetyltransferase activity, which facilitates O-GlcNAcylation of PKM2 by O-GlcNAc transferase (OGT). 32365408_Pharmacological inhibition and knockdown of O-GlcNAcase reduces cellular internalization of alpha-synuclein preformed fibrils. 33086728_Involvement of NDPK-B in Glucose Metabolism-Mediated Endothelial Damage via Activation of the Hexosamine Biosynthesis Pathway and Suppression of O-GlcNAcase Activity. 33310751_Bacterial O-GlcNAcase genes abundance decreases in ulcerative colitis patients and its administration ameliorates colitis in mice. 33333092_Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. 34135314_OGA is associated with deglycosylation of NONO and the KU complex during DNA damage repair. | ENSMUSG00000025220 | Oga | 3874.04451 | 3.354435e-01 | -1.575858 | 0.3072666 | 24.19871 | 8.689042e-07 | 7.956840e-04 | Yes | Yes | 1.705042e+03 | 2.590085e+02 | 5.594516e+03 | 8.688342e+02 | |
| ENSG00000198522 | 11321 | GPN1 | protein_coding | Q9HCN4 | FUNCTION: Small GTPase required for proper nuclear import of RNA polymerase II (RNAPII) (PubMed:20855544, PubMed:21768307). May act at an RNAP assembly step prior to nuclear import (PubMed:21768307). Forms an interface between the RNA polymerase II enzyme and chaperone/scaffolding proteins, suggesting that it is required to connect RNA polymerase II to regulators of protein complex formation (PubMed:17643375). May be involved in nuclear localization of XPA (PubMed:11058119). {ECO:0000269|PubMed:17643375, ECO:0000269|PubMed:20855544, ECO:0000269|PubMed:21768307, ECO:0000305|PubMed:11058119}. | Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;GTP-binding;Hydrolase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a guanosine triphosphatase enzyme. The encoded protein may play a role in DNA repair and may function in activation of transcription. Alternatively spliced transcript variants have been described. [provided by RefSeq, Feb 2009]. | hsa:11321; | cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; GTP binding [GO:0005525]; GTPase activity [GO:0003924] | 12588985_relieves MBD2-mediated transcriptional repression and reactivates transcription from methylated promoters 20855544_RPAP4/GPN1 is a member of a newly discovered GTPase family that contains a highly conserved GPN loop motif that is essential, along with its GTP-binding motifs, for nuclear localization of POLR2A/RPB1 in a process that also requires microtubule assembly 21768307_Data show that GPN1/GPN3 define a new family of small GTPases that are specialized for the transport of RNA polymerase II into the nucleus. 22796641_A highly active, evolutionarily conserved nuclear export sequences in XAB1/Gpn1, was identified. 25241168_Gpn1-Gpn3 interaction was essential to maintain steady-state protein levels of both GTPases 27749845_GPN1 gene variation is associated with oral cavity and pharyngeal cancer. 28940195_an acquired PDZ-binding motif in Gpn3 Q279* caused Gpn3 nuclear entry, and inhibited Gpn1 nuclear export and Gpn3-mediated RNAPII nuclear targeting. 29029378_Results show that Gpn1-inhibitable, nuclear polyubiquitination on lysine 216 regulates the half-life of Gpn3 by tagging it for proteasomal degradation. | ENSMUSG00000064037 | Gpn1 | 942.45401 | 2.586587e-01 | -1.950878 | 0.3292254 | 32.64238 | 1.107726e-08 | 1.554946e-05 | Yes | No | 3.905056e+02 | 9.000665e+01 | 1.415623e+03 | 3.327060e+02 | |
| ENSG00000198743 | 6526 | SLC5A3 | protein_coding | P53794 | FUNCTION: Prevents intracellular accumulation of high concentrations of myo-inositol (an osmolyte) that result in impairment of cellular function. | Glycoprotein;Ion transport;Membrane;Phosphoprotein;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | Mouse_homologues mmu:53881; | integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; voltage-gated potassium channel complex [GO:0008076]; fucose transmembrane transporter activity [GO:0015150]; glucose:sodium symporter activity [GO:0005412]; myo-inositol transmembrane transporter activity [GO:0005365]; myo-inositol:sodium symporter activity [GO:0005367]; pentose transmembrane transporter activity [GO:0015146]; polyol transmembrane transporter activity [GO:0015166]; potassium channel regulator activity [GO:0015459]; transmembrane transporter binding [GO:0044325]; fucose transmembrane transport [GO:0015756]; glucose transmembrane transport [GO:1904659]; inositol metabolic process [GO:0006020]; myo-inositol import across plasma membrane [GO:1904679]; myo-inositol transport [GO:0015798]; pentose transmembrane transport [GO:0015750]; peripheral nervous system development [GO:0007422]; polyol transport [GO:0015791]; positive regulation of catalytic activity [GO:0043085]; positive regulation of protein localization to membrane [GO:1905477]; positive regulation of reactive oxygen species biosynthetic process [GO:1903428]; regulation of respiratory gaseous exchange [GO:0043576]; regulation of transporter activity [GO:0032409]; transport across blood-brain barrier [GO:0150104] | 17178845_Tumor cells silence SLC5A8 and convert pyruvate into lactate as complementary mechanisms to avoid pyruvate-induced cell death. 19032932_Expression and/or function of SMIT2 may be reduced in diabetes mellitus, insulin resistance and polycystic ovary syndrome causing abnormal inositol meteabolism 19198609_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20738937_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20971364_Observational study of gene-disease association. (HuGE Navigator) 23207989_JAK2 contributes to the regulation of the myoinositol transporter SMIT. 24944204_A kinetic model has been generated for the transport mechanism of SMIT2 allowing insight into the transport of members of the LeuT structural family at the millisecond timescale. 25756525_Hypotonic stress causes a significant upregulation of SLC5A3 gene expression as detected by semiquantitative RT-PCR and Western blot analysis. 27217553_overexpression of the Na(+)/myo-inositol cotransporter (SMIT1) and myo-inositol supplementation enlarged intracellular PI(4,5)P2 pools, modulated several PI(4,5)P2-dependent ion channels including KCNQ2/3 channels, and attenuated the action potential firing of superior cervical ganglion neurons 28128227_Results show that SMIT1 is expressed in the heart. SMIT1 is able to detect increased extracellular glucose levels and triggers signaling leading to NOX2 activation and ROS production. 28793216_SMIT1 Modifies KCNQ Channel Function and Pharmacology by Physical Interaction with the Pore 32235474_Myo-Inositol Transporter SLC5A3 Associates with Degenerative Changes and Inflammation in Sporadic Inclusion Body Myositis. | ENSMUSG00000089774 | Slc5a3 | 625.18818 | 3.765412e-01 | -1.409120 | 0.3501852 | 15.23795 | 9.477949e-05 | 3.229504e-02 | Yes | No | 3.686736e+02 | 7.700405e+01 | 8.391216e+02 | 1.785787e+02 | ||
| ENSG00000198843 | 51714 | SELENOT | protein_coding | P62341 | FUNCTION: Selenoprotein with thioredoxin reductase-like oxidoreductase activity (By similarity). Protects dopaminergic neurons against oxidative stress and cell death (PubMed:26866473). Involved in ADCYAP1/PACAP-induced calcium mobilization and neuroendocrine secretion (By similarity). Plays a role in fibroblast anchorage and redox regulation (By similarity). In gastric smooth muscle, modulates the contraction processes through the regulation of calcium release and MYLK activation (By similarity). In pancreatic islets, involved in the control of glucose homeostasis, contributes to prolonged ADCYAP1/PACAP-induced insulin secretion (By similarity). {ECO:0000250|UniProtKB:P62342, ECO:0000250|UniProtKB:Q1H5H1, ECO:0000269|PubMed:26866473}. | Endoplasmic reticulum;Membrane;NADP;Oxidoreductase;Redox-active center;Reference proteome;Selenocysteine;Signal;Transmembrane;Transmembrane helix | This gene encodes a selenoprotein, containing a selenocysteine (Sec) residue at the active site. Sec is encoded by the UGA codon that normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon rather than as a stop signal. This protein is localized in the endoplasmic reticulum. It belongs to the SelWTH family that possesses a thioredoxin-like fold and a conserved CxxU (C is cysteine, U is Sec) motif found in several redox active proteins. Studies in mice indicate a crucial role for this gene in the protection of dopaminergic neurons against oxidative stress in Parkinson's disease, and in the control of glucose homeostasis in pancreatic beta-cells. Pseudogenes of this locus have been identified on chromosomes 9 and 5. [provided by RefSeq, Sep 2017]. | hsa:51714; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; selenium binding [GO:0008430]; thioredoxin-disulfide reductase activity [GO:0004791]; cell redox homeostasis [GO:0045454]; cellular oxidant detoxification [GO:0098869]; glucose homeostasis [GO:0042593]; insulin secretion involved in cellular response to glucose stimulus [GO:0035773]; pancreas development [GO:0031016]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of growth hormone secretion [GO:0060124]; response to glucose [GO:0009749]; selenocysteine incorporation [GO:0001514] | 12775843_The paper reported that the full-length SELT protein contains 182 amino acids. 23913443_Data suggest SELT is expressed in pancreatic beta- and delta-cells; inactivation of SELT in beta-cells leads to glucose intolerance via impaired glucose homeostasis and PACAP (pituitary adenylate cyclase-activating polypeptide) insulinotropic effect. | ENSMUSG00000075700 | Selenot | 1073.57923 | 3.777741e-01 | -1.404404 | 0.3553021 | 15.55743 | 8.003633e-05 | 2.942478e-02 | Yes | No | 5.041251e+02 | 9.420788e+01 | 1.387382e+03 | 2.643160e+02 | |
| ENSG00000215695 | 6248 | RSC1A1 | protein_coding | Q92681 | FUNCTION: Mediates transcriptional and post-transcriptional regulation of SLC5A1. Inhibits a dynamin and PKC-dependent exocytotic pathway of SLC5A1. Also involved in transcriptional regulation of SLC22A2. Exhibits glucose-dependent, short-term inhibition of SLC5A1 and SLC22A2 by inhibiting the release of vesicles from the trans-Golgi network. {ECO:0000269|PubMed:14724758, ECO:0000269|PubMed:16788146, ECO:0000269|PubMed:8836035}. | Cell membrane;Golgi apparatus;Membrane;Nucleus;Reference proteome;Transcription;Transcription regulation | Mouse_homologues NA; + ;NA | The protein encoded by this intronless gene inhibits the expression of the solute carrier family 5 (sodium/glucose cotransporter), member 1 gene (SLC5A1) and downregulates exocytosis of the SLC5A1 protein. The encoded protein is sometimes found coating the trans-Golgi network and other times is localized to the nucleus, depending on the cell cycle stage. This protein also inhibits the expression of solute carrier family 22 (organic cation transporter), member 2 (SLC22A2). [provided by RefSeq, Dec 2015]. | hsa:6248; | cell junction [GO:0030054]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; sodium channel inhibitor activity [GO:0019871]; negative regulation of exocytosis [GO:0045920]; negative regulation of glucose transmembrane transport [GO:0010829]; negative regulation of Golgi to plasma membrane protein transport [GO:0042997]; negative regulation of nucleoside transport [GO:0032243]; negative regulation of protein localization to plasma membrane [GO:1903077]; negative regulation of transport [GO:0051051]; regulation of transcription, DNA-templated [GO:0006355] | 14724758_Taken together, these data indicate that RSC1A1 modulates dynamin-dependent trafficking of intracellular vesicles containing hSGLT1 in Xenopus oocytes, and modulates PKC-dependent short-term regulation of this transporter. 16788146_hRS1 protein exhibits glucose-dependent, short-term inhibition of hSGLT1 and hOCT2 by inhibiting the release of vesicles from the trans-Golgi network 16788147_RS1 inhibits the dynamin-dependent release of SGLT1-containing vesicles from the trans-golgi network 17686765_RSC1A1 codes for a 67-kDa protein named RS1 that mediates transcriptional and post-transcriptional regulation of Na(+)-D-glucose cotransporter SGLT1. 20800603_Observational study of gene-disease association. (HuGE Navigator) 27555600_Binding of RS1-Reg to ODC1 inhibits the enzymatic activity at low intracellular glucose, which is blunted at high intracellular glucose. 30355744_A domain of protein RS1 (RSC1A1) called RS1-Reg down-regulates the plasma membrane abundance of Na(+)-d-glucose cotransporter. | ENSMUSG00000078515+ENSMUSG00000040715 | Ddi2+Rsc1a1 | 990.43725 | 3.569049e-01 | -1.486388 | 0.2975965 | 22.79627 | 1.801147e-06 | 1.264160e-03 | Yes | No | 7.293338e+02 | 9.626960e+02 | 1.301908e+03 | 1.778408e+03 |
| ENSG00000229186 | ADAM1A | unitary_pseudogene | 25.22808 | 1.064682e-01 | -3.231506 | 0.8158208 | 15.20967 | 9.620956e-05 | 3.229504e-02 | Yes | No | 2.196288e+00 | 1.035361e+01 | 1.508625e+01 | 7.372824e+01 | |||||||||||
| ENSG00000230673 | PABPC1P3 | processed_pseudogene | 267.77978 | 2.584702e-02 | -5.273858 | 0.6870919 | 79.76349 | 4.220193e-19 | 2.172133e-15 | Yes | No | 1.690179e+02 | 5.586506e+02 | 4.515820e+02 | 1.594383e+03 | |||||||||||
| ENSG00000237550 | 254948 | transcribed_processed_pseudogene | 79.56404 | 1.698905e-01 | -2.557323 | 0.5230489 | 23.96883 | 9.790814e-07 | 7.956840e-04 | Yes | No | 2.922232e+01 | 1.837545e+01 | 1.358782e+02 | 8.664996e+01 | |||||||||||
| ENSG00000248101 | lncRNA | 29.12489 | 3.633203e+02 | 8.505098 | 1.3058096 | 69.56435 | 7.396159e-17 | 1.631487e-13 | Yes | No | 4.831114e+01 | 1.552371e+02 | 1.486664e-01 | 6.038730e-01 | ||||||||||||
| ENSG00000249967 | protein_coding | E9PAM4 | Mouse_homologues FUNCTION: Membrane-bound phosphatidylinositol-4 kinase (PI4-kinase) that catalyzes the phosphorylation of phosphatidylinositol (PI) to phosphatidylinositol 4-phosphate (PI4P), a lipid that plays important roles in endocytosis, Golgi function, protein sorting and membrane trafficking and is required for prolonged survival of neurons. Besides, phosphorylation of phosphatidylinositol (PI) to phosphatidylinositol 4-phosphate (PI4P) is the first committed step in the generation of phosphatidylinositol 4,5-bisphosphate (PIP2), a precursor of the second messenger inositol 1,4,5-trisphosphate (InsP3). {ECO:0000269|PubMed:19581584, ECO:0000305}. | ATP-binding;Kinase;Membrane;Nucleotide-binding;Reference proteome;Transferase | Mouse_homologues mmu:84095; | plasma membrane [GO:0005886]; 1-phosphatidylinositol 4-kinase activity [GO:0004430]; ATP binding [GO:0005524] | Mouse_homologues 15944223_PI4KIIalpha is present on adaptor protein (AP) -3 organelles where it regulates AP-3 function 16772528_The unique N terminus of PI4K type IIalpha is required for the targeting of the enzyme to cellugyrin-positive vesicles. 19010779_AP-3 and BLOC-1 act, either in concert or sequentially, to specify sorting of PI4KIIalpha along the endocytic route. 20154717_PI4KIIalpha is a novel regulator of tumor growth by its action on angiogenesis and HIF-1alpha regulation. 23146885_Itch directly associates with and ubiquitinates PI4KIIalpha, and both proteins colocalize on endosomes containing Wnt-activated frizzled 4 (Fz4) receptor 33618608_Mouse models for hereditary spastic paraplegia uncover a role of PI4K2A in autophagic lysosome reformation. | ENSMUSG00000025178 | Pi4k2a | 49.25765 | 2.477119e-03 | -8.657121 | 1.3825839 | 76.87618 | 1.820195e-18 | 5.621127e-15 | Yes | No | 1.502818e-01 | 4.968985e-01 | 7.643456e+01 | 2.187136e+02 | ||||
| ENSG00000255513 | transcribed_processed_pseudogene | 74.54457 | 1.049727e-01 | -3.251914 | 0.6702016 | 22.47643 | 2.127377e-06 | 1.313953e-03 | Yes | No | 1.581684e+01 | 5.180942e+01 | 6.214905e+01 | 2.114860e+02 | ||||||||||||
| ENSG00000259494 | 26589 | MRPL46 | protein_coding | Q9H2W6 | 3D-structure;Acetylation;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transit peptide | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 39S subunit protein. [provided by RefSeq, Jul 2008]. | hsa:26589; | cell junction [GO:0030054]; mitochondrial inner membrane [GO:0005743]; mitochondrial large ribosomal subunit [GO:0005762]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; structural constituent of ribosome [GO:0003735] | 11761714_Expression is enriched in testis. Positioned to chromosome 15q24 by radiation hybrid mapping. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000030612 | Mrpl46 | 724.52752 | 4.259679e-01 | -1.231183 | 0.3119551 | 15.30770 | 9.134342e-05 | 3.205531e-02 | Yes | No | 4.131772e+02 | 6.886869e+01 | 9.265116e+02 | 1.570554e+02 | ||
| ENSG00000259522 | protein_coding | H3BMG7 | Mouse_homologues FUNCTION: Plays an essential role in autophagy. {ECO:0000250}.; + ;NA | Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA | Mouse_homologues mmu:74140;;NA | integral component of membrane [GO:0016021]; membrane [GO:0016020]; protein localization to membrane [GO:0072657] | Mouse_homologues NA; + ;NA | ENSMUSG00000002320+ENSMUSG00000115232 | Tm9sf1+Gm49378 | 129.37786 | 9.711637e-03 | -6.686070 | 0.9076039 | 78.47406 | 8.105550e-19 | 3.128945e-15 | Yes | No | 7.459291e+01 | 8.234683e+01 | 2.260936e+02 | 2.571598e+02 | |||
| ENSG00000260566 | lncRNA | 95.85831 | 1.734872e-03 | -9.170955 | 1.3808913 | 81.40087 | 1.842766e-19 | 1.422708e-15 | Yes | No | 1.042490e-03 | 3.262009e-02 | 2.056014e+06 | 6.608698e+07 | ||||||||||||
| ENSG00000261326 | LINC01355 | lncRNA | 169.57675 | 3.101990e-01 | -1.688734 | 0.4245461 | 14.55354 | 1.362326e-04 | 4.001749e-02 | Yes | No | 5.643139e+01 | 1.815087e+01 | 2.258379e+02 | 7.288787e+01 | |||||||||||
| ENSG00000261740 | 107282092 | BOLA2-SMG1P6 | protein_coding | H3BVE0 | Coiled coil;Proteomics identification;Reference proteome | This gene represents naturally-occurring readthrough transcription between the upstream BOLA2 (bolA family member 2) and downstream SMG1 pseudogene 6 (SMG1P6) loci. Alternative splicing results in multiple transcript variants, some of which may encode proteins with an N-terminus similar to BOLA2 and a C-terminus related to SMG1 phosphatidylinositol 3-kinase-related kinase. [provided by RefSeq, Feb 2016]. | 360.84238 | 3.737526e-01 | -1.419845 | 0.3648863 | 14.91507 | 1.124609e-04 | 3.479016e-02 | Yes | No | 1.721299e+02 | 3.230597e+01 | 4.589176e+02 | 8.703476e+01 | |||||||
| ENSG00000263244 | lncRNA | 71.06895 | 2.096910e-03 | -8.897519 | 1.3973692 | 76.41628 | 2.297490e-18 | 5.912591e-15 | Yes | No | 1.670115e-01 | 5.641044e+00 | 1.222259e+02 | 4.249388e+03 | ||||||||||||
| ENSG00000264112 | lncRNA | 583.84465 | 3.136977e-01 | -1.672553 | 0.2946065 | 29.51460 | 5.549702e-08 | 6.679136e-05 | Yes | No | 2.216360e+02 | 3.219089e+01 | 7.542965e+02 | 1.098101e+02 | ||||||||||||
| ENSG00000270149 | protein_coding | R4GMV9 | Reference proteome | 76.18064 | 1.012964e+03 | 9.984367 | 1.2530628 | 144.33353 | 3.003792e-33 | 4.638155e-29 | Yes | No | 1.231317e+03 | 2.269176e+03 | 1.362813e+00 | 3.333793e+00 | ||||||||||
| ENSG00000273373 | lncRNA | 678.93544 | 3.181941e-01 | -1.652021 | 0.3056007 | 26.90727 | 2.134544e-07 | 2.354249e-04 | Yes | No | 2.335895e+02 | 6.218905e+01 | 8.878349e+02 | 2.407622e+02 | ||||||||||||
| ENSG00000274290 | 8344 | H2BC6 | protein_coding | P62807 | FUNCTION: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.; FUNCTION: Has broad antibacterial activity. May contribute to the formation of the functional antimicrobial barrier of the colonic epithelium, and to the bactericidal activity of amniotic fluid. | 3D-structure;Acetylation;Antibiotic;Antimicrobial;Chromosome;DNA-binding;Direct protein sequencing;Glycoprotein;Hydroxylation;Isopeptide bond;Methylation;Nucleosome core;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. The protein has antibacterial and antifungal antimicrobial activity. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H2B family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6. [provided by RefSeq, Aug 2015]. | hsa:3017;hsa:8339;hsa:8343;hsa:8344;hsa:8346;hsa:8347; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; protein heterodimerization activity [GO:0046982]; antibacterial humoral response [GO:0019731]; antimicrobial humoral immune response mediated by antimicrobial peptide [GO:0061844]; defense response to Gram-positive bacterium [GO:0050830]; innate immune response in mucosa [GO:0002227]; nucleosome assembly [GO:0006334] | 12860195_Histone H2B antimicrobial peptides participate in the colonic defense against bacteria and fungi. | ENSMUSG00000018102 | H2bc4 | 36.94501 | 1.681209e-01 | -2.572429 | 0.5262775 | 21.10427 | 4.349569e-06 | 2.487470e-03 | Yes | Yes | 6.452495e+00 | 4.289412e+00 | 4.160097e+01 | 2.840941e+01 | |
| ENSG00000277991 | lncRNA | 54.91756 | 8.784113e-02 | -3.508960 | 0.6563757 | 29.48908 | 5.623260e-08 | 6.679136e-05 | Yes | No | 5.603382e+00 | 2.081577e+01 | 4.015423e+01 | 1.568841e+02 | ||||||||||||
| ENSG00000280893 | protein_coding | A0A0G2JLL6 | Reference proteome | integral component of membrane [GO:0016021] | 37.05979 | 2.672953e-03 | -8.547350 | 1.4076022 | 64.46667 | 9.817910e-16 | 1.894979e-12 | Yes | No | 4.742503e-10 | 3.422587e-09 | 7.121100e+01 | 4.798296e+02 |
- Note:
- The spreadsheet above contains the genes with significantly changes in expression (P-value < 0.05 and |log2FC| > 1). Please Click HERE to check all mapped genes in a Microsoft .excel file.
- The comparisons made can be identified from the website name. “HEK293_OSMI2_6hA vs HEK293_DMSO_6hA” means that “HEK293_DMSO_6hA” is considered the ground state and “HEK293_OSMI2_6hA” the changed state. This also means that a positive log2FC value indicates that the gene expression is increased in “HEK293_OSMI2_6hA” compared to “HEK293_DMSO_6hA”.
- Click HERE to check all parameters of DESeq2 model for all detected genes of the current samples set in a Microsoft .excel file. For more detials of the DESeq2 model, please Click HERE.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| lncRNA | 7 |
| processed_pseudogene | 1 |
| protein_coding | 51 |
| transcribed_processed_pseudogene | 2 |
| unitary_pseudogene | 1 |
AS in non-DEGs
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_OSMI2_6hA | SE_Intercept_HEK293_OSMI2_6hA | Intercept_HEK293_DMSO_6hA | SE_Intercept_HEK293_DMSO_6hA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000000460 | 55732 | C1orf112 | protein_coding | Q9NSG2 | Acetylation;Alternative splicing;Phosphoprotein;Reference proteome | hsa:55732; | 19064610_Observational study of gene-disease association. (HuGE Navigator) 19536175_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23039964_Knockdown of C1orf112 by siRNA in HeLa cells significantly suppressed their growth. | ENSMUSG00000041406 | BC055324 | 5.415265e+02 | 0.4268943 | -1.228049073 | 0.3433095 | 1.237578e+01 | 0.0004349386 | 0.07261768 | No | Yes | 2.815087e+02 | 56.100987 | 6.313732e+02 | 128.143481 | ||||
| ENSG00000002586 | 4267 | CD99 | protein_coding | P14209 | FUNCTION: Involved in T-cell adhesion processes and in spontaneous rosette formation with erythrocytes. Plays a role in a late step of leukocyte extravasation helping leukocytes to overcome the endothelial basement membrane. Acts at the same site as, but independently of, PECAM1. Involved in T-cell adhesion processes (By similarity). {ECO:0000250}. | Alternative splicing;Cell adhesion;Direct protein sequencing;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a cell surface glycoprotein involved in leukocyte migration, T-cell adhesion, ganglioside GM1 and transmembrane protein transport, and T-cell death by a caspase-independent pathway. In addition, the encoded protein may have the ability to rearrange the actin cytoskeleton and may also act as an oncosuppressor in osteosarcoma. This gene is found in the pseudoautosomal region of chromosomes X and Y and escapes X-chromosome inactivation. There is a related pseudogene located immediately adjacent to this locus. [provided by RefSeq, Mar 2016]. | hsa:4267; | cytoplasm [GO:0005737]; focal adhesion [GO:0005925]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; homotypic cell-cell adhesion [GO:0034109]; positive regulation of neutrophil extravasation [GO:2000391]; T cell extravasation [GO:0072683] | 12172043_Results suggest that type I is the major isoform of CD99 expressed in non-neoplastic gastric mucosa and gastric adenocarcinomas. 12216109_Functional involvement of src and focal adhesion kinase in a CD99 splice variant-induced motility of human breast cancer cells 12681511_engagement triggers the exocytic transport of ganglioside GM1 and the reorganization of actin cytoskeleton 12832073_inhibition by MHC class I engagement or apoptosis and upregulation of T cell receptor and MHC molecules in human thymocytes and T cell line induced by CD99 14623115_CD99 epitopes plays a distinct role in T cell biology, especially in T cell apoptosis. 14646598_CD99 type II acts as a negative regulator in the neural differentiation of precursor cells that might occur during nerve system development 14744088_Solitary sclerotic fibroma is a fibrotic lesion with cells positive for CD34 and O13. O13 expression in SF has not been previously described. 15184883_our findings indicate that CD99 functions occur through reorganization of cytoskeleton and identify actin and zyxin as the early signaling events driven by CD99 engagement. 15359120_solution structure of cytoplasmic domain of human CD99 type I shows that it has a hairpin shape anchored by two flexible loops 15388255_cyclophilin A binds CD99 and may be either a signaling mediator or a signaling regulator for CD99 15469477_loss of CD99 expression in PETs was found to be associated with ominous prognostic indicators 15528994_Cooperation of CD99 engagement with suboptimal TCR/CD3 signals resulted in enhanced CD4+ T cell proliferation, elevated expression of CD25 and GM1, increased apoptosis, augmented activation of JNK, and increased AP-1 activation 15716602_CD99 was found to be widely expressed in both sarcomatous and carcinoma component in pleomorphic carcinomas of the lung 16361803_High expression of CD99 in ALK-positive anaplastic large cell lymphomas is an unexpected finding and its biologic and clinical significance has yet to be clarified. 16421247_The findings point to an antioncogenic role for CD99 in osteosarcoma, likely through the regulation of caveolin-1 and inhibition of c-Src kinase activity. 16614706_LMP-1 induced down-regulation of the CD99 pathway is important in nasopharyngeal carcinogenesis, and the expression of CD99 in lymphoid stroma may regulate immune response to nasopharyngeal carcinoma. 16614706_LMP-1 induced down-regulation of the CD99 pathway is important in nasopharyngeal carcinogenesis, and the expression of CD99 in lymphoid stroma may regulate immune response to nasopharyngeal carcinoma. (LMP-1= EBV latent membrane protein 1) 16825498_subsets of CD34+ cells with high or low levels of CD99 expression produce different numbers of erythroid, natural killer (NK), or dendritic cells in the in vitro differentiation assays 16984917_The results of this study suggest that expression of a splice variant of CD99 contributes to the invasive ability of human breast cancer cells by up-regulating AP-1-mediated gene expression through the Akt-dependent ERK and JNK signaling pathways. 17202377_PECAM (CD31) and CD99 regulate distinct, sequential steps in the transendothelial migration of neutrophils during inflammation 17464179_These data demonstrate the unique properties of CD99 co-stimulation that distinguish this molecule from CD28 and other raft-resident co-stimulatory factors. 17471235_Our findings highlight the involvement of CD99 in crucial processes of cancer malignancy. 17652531_CK19/CD99 immunoexpression did not correlate with extent of tumor invasion, mitotic activity, Ki-67 labeling index, presence of extracellular mucinous pools dissecting muscle, and angiolymphatic and perineural/neural invasion in goblet cell carcinoids. 18552083_CD99-immunohistochemistry is unique in that it is helpful for the diagnosis of clear cell brain tumors through the visualization of CD99-negative clear cells 18849489_CD99 also binds p230/golgin-245, a coiled-coil protein that recycles between the cytosol and buds/vesicles of the TGN and which plays a fundamental role in trafficking transport vesicles. 19289593_CD99 is frequently expressed in anaplastic large cell lymphoma 19307784_CD99 plays a critical role in human monocyte and lymphocyte transendothelial migration across pocine endothelial cells. 19723899_Observational study of gene-disease association. (HuGE Navigator) 19901887_Type B3 thymic epithelial tumor in an adolescent detected by immunohistochemical staining for CD5, CD99, and KIT (CD117): a case report. 20184665_CD99 showed no preferential staining of Atypical fibroxanthoma, spindle cell malignant melanoma or sarcomatoid carcinoma/spindle cell squamous cell carcinoma 20197622_Analysis of a panel of human EWS cells revealed an inverse correlation between CD99 and H-neurofilament expression, as well as an inverse correlation between neural differentiation and oncogenic transformation. 20217126_Protein expression and gene promoter hypermethylation of CD99 in transitional cell carcinoma of urinary bladder 20453109_CD99 may play a physiologic role in the clonal deletion processes necessary for B-lymphoid selection 21196719_CD99 positivity was significantly associated with advanced stage (p < 0.01), higher risk group according to the International Prognostic Index risk score (p < 0.01) and non-germinal center B cell-like type (p = 0.01). 21267687_High CD99 is associated with central primitive neuroectodermal tumors and Ewing's sarcoma 21566515_Our study has identified for the first time that pancreatic solid-pseudopapillary neoplasm exhibits a unique dot-like staining pattern for CD99. 21775056_Data show that CD99 combined with E-cadherin/beta-catenin and CD10 can be used as a relatively specific expression profile of SPTs. 22020966_upregulation of CD99 in H/RS cells induces terminal B-cell differentiation, which may provide a novel therapeutic strategies for cHL 22083306_NF-kappaB2 exhibits the major inhibitory role in the transcription at the CD99 promoter 22189791_Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31) and CD99 are critical in lymphatic transmigration of human dendritic cells. 22335486_The new antibody scFvC7 recognizes the CD99 extracellular domain. 22392539_the membrane protein CD99 was identified as a novel stromal factor with clinical relevance. The results support the concept that stromal properties have an important impact on tumour progression. 22781601_CD99 is highly expressed in anaplastic large-cell lymphoma, and showed high rate of co-expression with ALK. 22864685_Data suggest that CD99 expression in lymph node/germinal center biopsies at time of diagnosis is not prognostic marker for survival in patients with diffuse large B-cell lymphoma treated with rituximab-CHOP immunochemotherapy. 22986530_we show that miR-30a-5p provides an essential link between EWS-FLI1 and CD99, two major biomarkers in Ewing sarcoma 23145531_We report an unusual pattern of paranuclear dot-like expression of CD99 in 14 cases of Merkel cell carcinoma, two of which did not express CK20. 23152061_CD99-induced HSP70 expression in B and T lymphocytes. 23333754_PVR resides in the recently identified lateral border recycling compartment, similar to PECAM and CD99. 23644663_we have shown that forced expression of wild-type CD99 in osteosarcoma cells induces downregulation of genes such as ROCK2 23743627_CXCL16, inversely correlated with CD99 expression in Hodgkin Reed-Sternberg (H/RS) cells. 24094957_Solid pseudopapillary neoplasm of the pancreas exhibits a very unique immunostaining pattern for CD99 that is present in all cases. 24247571_dermatofibromastrongly expresses CD99 in a diffuse pattern that may serve as evidence in distinction from dermatofibrosarcoma 24341229_It is assumed that the expression of CD99 plays an important role in the pathogenesis of psoriasis, in the processes of emigration of leukocytes, and in their tropism toward to the epidermis 24677094_CD99 induces ERK activation, increasing its membrane-bound/cytoplasmic form rather than affecting its nuclear localization. 24702676_CD99 expression in nasal lobular capillary haemangioma. 24797829_CD99 expression in astrocytomas of different malignant grades might contribute to the infiltrative ability and support the importance of CD99 as a potential target to reduce infiltrative astrocytoma capacity in migration and invasion. 24798881_Report purification of human antibody scFv anti-CD99 for blocking monocyte transendothelial migration. 25501132_The dAbd C7 induced rapid and massive EWS cell death through Mdm2 degradation and p53 reactivation. Mdm2 overexpression as well as silencing of p53 in p53wt EWS cells decreased CD99-induced EWS cell death 25957287_Our results suggest that the CD34/CD25/CD123/CD99(+) LAIP is strictly associated with FLT3-ITD-positive cells. 26000312_The results highlight an important role of CD99 in the differentiation and activity of human osteoblasts in physiological and pathological conditions. 26000982_These results suggest that SEPTIN2-mediated cytoskeletal rearrangement and STATHMIN-mediated differentiation may contribute to changes in cell morphology and differentiation of H/RS cells with CD99 upregulation in Hodgkin lymphoma. 26172215_CD99 can suppress CD98-mediated assembly of pro-tumorigenic signaling complexes through its ability to dephosphorylate focal adhesion kinase. 26279084_The pseudoautosomal region of human X and Y chromosome encoded CD99 protein was predicted to interact with at least 14 proteins encoded on the somatic chromosomes. 26522646_Data show that CD99 antigen expression increased during in vitro differentiation of germinal center B cells and was highest in plasma cells (PCs). 26616853_CD99 counteracts EWS-FLI1 in controlling NF-kappaB signaling through the miR-34a, which is increased and secreted into exosomes released by CD99-silenced Ewing sarcoma cells. 26782152_CD99 was also paranuclear positive in 4 of 11 (36%) small cell lung carcinomas 27002769_High expression of EMMPRIN plays a crucial role in breast cancer cell proliferation, matrigel invasion and tumor formation. 27094031_these observations suggest that CD99 is involved in the regulation of CD1a transcription and expression by increasing ATF-2. 27792997_Besides confirming the importance of CD99, findings indicate that polymorphic variations in this gene may affect either development or progression of EWS. 27866429_CD99 expression is increased in patients with active Inflammatory bowel disease, and positively correlated with disease activity. 28123069_CD99 is a marker of acute myeloid leukemia and myelodysplastic syndrome stem cells 29403080_In summary, we established that tumor suppressor CD99 is markedly downregulated in multiple myeloma. 30061310_The present findings identify the genetic basis of the erythroid-specific Xg(a)/CD99 blood group phenotypes and reveal the molecular background of their formation. 30396687_Authors demonstrated that ligation of CD99 on monocytes by anti-CD99 mAb MT99/3 could mediate T cell hypo-responsiveness. These findings provide the first evidence of the role of CD99 on monocytes that contributes to T cell activation. 30484860_findings demonstrate that high expression levels of CD99 are mainly found in high-risk BCP-ALL, e.g. BCR-ABL1 and CRLF2(Re/Hi) , and that high CD99 mRNA levels are strongly associated with a high frequency of relapse, high proportion of positive for minimal residual disease at day 29 and poor overall survival in paediatric cohorts 30760844_A new gene regulatory function for MMP-9 in chronic lymphocytic leukemia (CLL). It also identifies CD99 as an MMP-9 target and a novel contributor to CLL cell adhesion, migration and arrest. CD99 thus constitutes a new therapeutic target in CLL, complementary to MMP-9. 30845661_Study showed that isoform 1, of CD99, is exclusively present in human astrocytomas and cell line and regulates functions, such as cytoskeleton remodeling, cell migration, invasion, and adhesion. The present transcriptome data of CD99 silenced GBM cells, suggest that CD99 modulates FAK1/c-Src signaling pathways, related to actin cytoskeleton dynamics. 31120992_CD99 and its ligand interaction induced the upregulation of IL-6 and TNF-alpha expression in anti-CD3 monoclonal antibody activated T cells. 31128035_Most cases stained positive for CD99 and negative for lymphocytic markers. Markers of epithelial differentiation displayed variable results. In all cases tested, characteristic translocations were displayed supporting the diagnosis. 31371417_Clinical and preclinical characterization of CD99 isoforms in acute myeloid leukemia. 31472965_Data show that nuclear factor erythroid 2 like 2 (Nrf2) induced cisplatin resistance in ovarian cancer cells by promoting T-cell surface glycoprotein E2 (CD99) expression. 32535112_Anti-CD99 scFv-ELP nanoworms for the treatment of acute myeloid leukemia. 32823905_Association of Maternal Regulatory Single Nucleotide Polymorphic CD99 Genotype with Preeclampsia in Pregnancies Carrying Male Fetuses in Ethiopian Women. 33147457_GDF6-CD99 Signaling Regulates Src and Ewing Sarcoma Growth. 33215253_Anti-human CD99 antibody exerts potent antitumor effects in mantle cell lymphoma. 34312421_Leukocytes with chromosome Y loss have reduced abundance of the cell surface immunoprotein CD99. 34374417_Tumour cell CD99 regulates transendothelial migration via CDC42 and actin remodelling. 34383378_CD99 and polymeric immunoglobulin receptor peptides deregulation in critical COVID-19: A potential link to molecular pathophysiology? 34920053_CD99 in malignant hematopoiesis. 35339749_Role of CD99 in regulating homeostasis and differentiation in normal human epidermal keratinocytes. | 9.419730e+03 | 1.1809451 | 0.239941898 | 0.3013582 | 6.357988e-01 | 0.4252362160 | 0.85078492 | No | Yes | 9.376166e+03 | 789.409621 | 7.389865e+03 | 638.454321 | |||
| ENSG00000005486 | 57414 | RHBDD2 | protein_coding | Q6NTF9 | Alternative splicing;Golgi apparatus;Membrane;Reference proteome;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the rhomboid family of membrane-bound proteases and is overexpressed in some breast cancers. Members of this family are involved in intramembrane proteolysis. In mouse, the orthologous protein associates with the Golgi body. [provided by RefSeq, Sep 2016]. | hsa:57414; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; Hrd1p ubiquitin ligase ERAD-L complex [GO:0000839]; integral component of endoplasmic reticulum membrane [GO:0030176]; nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; misfolded protein binding [GO:0051787]; serine-type endopeptidase activity [GO:0004252]; ubiquitin-specific protease binding [GO:1990381]; endoplasmic reticulum unfolded protein response [GO:0030968]; ubiquitin-dependent ERAD pathway [GO:0030433] | 19616622_RHBDD2 over-expression behaves as an indicator of poor prognosis and may play a role facilitating breast cancer progression. 22965880_Increased RHBDD2 expression in response to the chemotherapy is associated with advanced stages of colorectal cancer. 23386608_RHBDD2 protein plays important roles in the development and normal function of the retina 24078384_RHBDD2 overexpression in breast cancer could represent an adaptive phenotype to the stressful tumor microenvironment by modulating the endoplasmic reticulum stress response. 24185965_Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis showed a significant RHBDD2 mRNA overexpression in advanced breast cancer compared with normal tissue samples (p = 0.012). 29901166_The RHBDD2 subcellular localization was corroborated at the Golgi apparatus. 32339641_RHBDD2 overexpression promotes a chemoresistant and invasive phenotype to rectal cancer tumors via modulating UPR and focal adhesion genes. 34109992_RHBDD2WWOX protein interaction during proliferative and differentiated stages in normal and breast cancer cells. | ENSMUSG00000039917 | Rhbdd2 | 3.270492e+03 | 1.2542090 | 0.326777785 | 0.3317526 | 9.642406e-01 | 0.3261207663 | 0.82378328 | No | Yes | 3.359139e+03 | 399.468112 | 2.462053e+03 | 300.717028 | ||
| ENSG00000005801 | 7748 | ZNF195 | protein_coding | O14628 | FUNCTION: May be involved in transcriptional regulation. | Acetylation;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein belonging to the Krueppel C2H2-type zinc-finger protein family. These family members are transcription factors that are implicated in a variety of cellular processes. This gene is located near the centromeric border of chromosome 11p15.5, next to an imprinted domain that is associated with maternal-specific loss of heterozygosity in Wilms' tumors. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2012]. | hsa:7748; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; regulation of transcription, DNA-templated [GO:0006355] | 24817947_ZNF195 and SBF1 are potential biomarkers for gemcitabine sensitivity in head and neck squamous cell carcinoma cell lines. | 4.799524e+02 | 0.8129100 | -0.298832541 | 0.3257264 | 8.050906e-01 | 0.3695756983 | 0.83664500 | No | Yes | 3.212882e+02 | 50.611778 | 4.191213e+02 | 67.195755 | |||
| ENSG00000005884 | 3675 | ITGA3 | protein_coding | P26006 | FUNCTION: Integrin alpha-3/beta-1 is a receptor for fibronectin, laminin, collagen, epiligrin, thrombospondin and CSPG4. Integrin alpha-3/beta-1 provides a docking site for FAP (seprase) at invadopodia plasma membranes in a collagen-dependent manner and hence may participate in the adhesion, formation of invadopodia and matrix degradation processes, promoting cell invasion. Alpha-3/beta-1 may mediate with LGALS3 the stimulation by CSPG4 of endothelial cells migration. {ECO:0000269|PubMed:10455171, ECO:0000269|PubMed:15181153}.; FUNCTION: (Microbial infection) Integrin ITGA3:ITGB1 may act as a receptor for R.delemar CotH7 in alveolar epithelial cells, which may be an early step in pulmonary mucormycosis disease progression. {ECO:0000269|PubMed:32487760}. | Alternative splicing;Calcium;Cell adhesion;Cell junction;Cell membrane;Cell projection;Cleavage on pair of basic residues;Direct protein sequencing;Disease variant;Disulfide bond;Epidermolysis bullosa;Glycoprotein;Integrin;Lipoprotein;Membrane;Metal-binding;Palmitate;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The gene encodes a member of the integrin alpha chain family of proteins. Integrins are heterodimeric integral membrane proteins composed of an alpha chain and a beta chain that function as cell surface adhesion molecules. The encoded preproprotein is proteolytically processed to generate light and heavy chains that comprise the alpha 3 subunit. This subunit joins with a beta 1 subunit to form an integrin that interacts with extracellular matrix proteins including members of the laminin family. Expression of this gene may be correlated with breast cancer metastasis. [provided by RefSeq, Oct 2015]. | hsa:3675; | basolateral plasma membrane [GO:0016323]; cell periphery [GO:0071944]; cell surface [GO:0009986]; excitatory synapse [GO:0060076]; external side of plasma membrane [GO:0009897]; extracellular exosome [GO:0070062]; filopodium membrane [GO:0031527]; focal adhesion [GO:0005925]; growth cone filopodium [GO:1990812]; integrin alpha3-beta1 complex [GO:0034667]; integrin complex [GO:0008305]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; synaptic membrane [GO:0097060]; collagen binding [GO:0005518]; fibronectin binding [GO:0001968]; integrin binding [GO:0005178]; laminin binding [GO:0043236]; metal ion binding [GO:0046872]; protease binding [GO:0002020]; protein domain specific binding [GO:0019904]; protein heterodimerization activity [GO:0046982]; cell-matrix adhesion [GO:0007160]; dendritic spine maintenance [GO:0097062]; exploration behavior [GO:0035640]; heart development [GO:0007507]; integrin-mediated signaling pathway [GO:0007229]; lung development [GO:0030324]; maternal process involved in female pregnancy [GO:0060135]; memory [GO:0007613]; mesodermal cell differentiation [GO:0048333]; negative regulation of cell projection organization [GO:0031345]; negative regulation of Rho protein signal transduction [GO:0035024]; nephron development [GO:0072006]; neuron migration [GO:0001764]; positive regulation of cell-substrate adhesion [GO:0010811]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of gene expression [GO:0010628]; positive regulation of neuron projection development [GO:0010976]; positive regulation of protein localization to plasma membrane [GO:1903078]; regulation of BMP signaling pathway [GO:0030510]; regulation of transforming growth factor beta receptor signaling pathway [GO:0017015]; regulation of Wnt signaling pathway [GO:0030111]; renal filtration [GO:0097205]; response to gonadotropin [GO:0034698]; response to xenobiotic stimulus [GO:0009410]; skin development [GO:0043588] | 11907260_Data demonstrate that multiple tetraspanin (transmembrane 4 superfamily) proteins such as CD151 are palmitoylated, and that palmitoylation is not required for cd151-alpha3beta1 integrin association. 11988844_Human prostate tumors display on their cell surface the alpha6beta1 and/or alpha3beta1 integrins 12372459_initial adhesion of endometrial cells to mesothelium is not mediated by alpha(3)beta(1)integrin 12452046_Integrin-alpha 3 sub-unit participates the process of regulating growth of meningiomas. 12615788_Observational study of gene-disease association. (HuGE Navigator) 12699087_expression of alpha3 integrin subunit is responsible for differentiation-associated changes in cells behavior in terminally differentiated oral keratinocytes 12717361_signaling through both constitutively expressed alpha2 integrin and Matrigel-induced alpha3 integrin expression is required to acquire a differentiated phenotype in Caco-2 cells. 12919677_Data show that macrophage stimulating protein (MSP) and its receptor (Ron) induce phosphorylation of both Ron and alpha6beta4 integrin, and result in activation of alpha3beta1 integrin. 14596610_Disulfide bonding patterns of the integrin alpha 3 chain are described; the alpha chain displays differences in the disulfide patterns of those bonds near their C-terminal regions linking the heavy and light chains. 14602071_Data demonstrate that alpha6beta4 integrin mediates pancreatic epithelial cell adhesion to Netrin-1, whereas recruitment of alpha6beta4 and alpha3beta1 regulate the migration of putative pancreatic progenitors on Netrin-1. 14607975_ligand-binding specificities of integrin alpha3beta1 and alpha6beta1 14612440_alpha3beta1 integrin binding to laminin-5 depends on its proteolytic processing 14645603_results suggest that a direct interaction occurs between the Adenovirus penton base protein and the integrin receptor alpha3beta1 in vitro and in vivo 14662754_results show how laterally associated EWI-2 might regulate alpha3beta1 function in disease and development, and demonstrate how tetraspanin proteins can assemble multiple nontetraspanin proteins into functional complexes 14666169_alpha3beta1, alpha4beta1 and alphaVbeta1 integrins may play an important role in the implantation process 14706682_Observational study of gene-disease association. (HuGE Navigator) 15024036_Data suggest that the early arrest of tumor cells in the pulmonary vasculature through interaction of alpha3beta1 integrin with laminin-5 in exposed basement membrane provides a basis for cell arrest during pulmonary metastasis. 15109703_Observational study of gene-disease association. (HuGE Navigator) 15194015_Observational study of gene-environment interaction. (HuGE Navigator) 15280429_Alpha 3 beta 1 integrin is necessary and sufficient for maximal keratinocyte survival on laminin-5. 15302884_cAMP-Epac-Rap1 pathway regulates cell spreading and cell adhesion to laminin-5 through the alpha3beta1 integrin but not the alpha6beta4 integrin 15346842_Observational study of gene-disease association. (HuGE Navigator) 15351855_Observational study of gene-disease association. (HuGE Navigator) 15677332_Results indicate that CD151 association modulates the ligand-binding activity of integrin alpha3beta1 through stabilizing its activated conformation not only with purified proteins but also in a physiological context. 15805105_Beta ig-h3 induces keratinocyte differentiation via modulation of involucrin and transglutaminase expression through the integrin alpha3beta1 and the phosphatidylinositol 3-kinase/Akt signaling pathway 15878864_KSHV gB can activate VEGFR-3 on the microvascular endothelium and modulate endothelial cell migration and proliferation via an interaction between the alpha3beta1 integrin and the VEGFR-3 receptor 15919367_alpha3beta1 integrin binding results in a suppression of the interleukin-1 signaling pathway leading to the activation of NF-(kappa)B 15983209_alpha3, alpha5, and alpha6 beta1 integrins are expressed in ductal cells at 8 weeks, before glucagon- and insulin-immunoreactive cells bud off in fetal pancrea. 16055706_the signaling network involving Smad-dependent TGFbeta, PKCdelta, and integrin alpha2beta1/alpha3beta1, regulates cell spreading, motility, and invasion of the SNU16mAd gastric carcinoma cell variant 16103120_phenotypic conversion and reversion of bladder cancer cells is controlled by a glycosynapse 3 microdomain through GM3-mediated interaction of alpha3beta1 integrin with CD9 16273094_localized expression of the integrin alpha3 protein is regulated at the level of RNA localization by MBNL1; integrin alpha3 transcripts are physically associated with MLP1 in cells and MLP1 binds to a specific ACACCC motif in the integrin alpha3 3' UTR 16307893_Thus, the integrin/Src pathway negatively regulates the induction of long-term plasticity at inhibitory synapses on a cerebellar Purkinje cells. 16339173_Data show that alpha6 and alpha3 integrin subunits interact with laminin 5 to increase expression of E-cadherin, and suggest that phosphoinositide 3-kinase (PI 3-kinase) activation plays a key role in this cross-talk. 16373174_Our data show that alpha3beta1 integrin function may be altered by glycosylation, that both subunits contribute to these changes, and glycosylation may be considered a newly found mechanism in the regulation of integrin function. 16390868_Results suggest that loss of E-cadherin function is linked to regulation of cell-cell and cell-matrix adhesion, based in part on cell surface expression of alpha2, alpha3 and beta1 integrins. 16459165_decreased podocyte expression of alpha3beta1 integrins is closely related with podocyte depletion, glomerular sclerosis, and daily protein loss in patients with primary focal glomerulosclerosis 16475024_TGF-beta1 stimulates hepatoma cells to express a higher level of alpha3 integrin by transcriptional upregulation via Ets transcription factors and to exhibit a more invasive phenotype. 16510444_This study supports matrix-induced clustering of alpha3beta1 integrin promotes uPAR/alpha3beta1 interaction; potentiating cellular signal transduction pathways culminating in activation of uPA expression and enhanced uPA-dependent invasive behavior. 16537545_EWI proteins EWI-2 and EWI-F, alpha3beta1 and alpha6beta4 integrins, and protein palmitoylation have contrasting effects on cell surface CD9 organization 16571677_Both integrin alpha3beta1- and alpha6beta4-dependent cell adhesion to laminin-5 were also impaired in CD151-silenced cells. 16707493_Slug regulates integrin alpha3, beta1, and beta4 expression and cell proliferation in human epidermal keratinocytes 16731529_integrin alpha3beta1 is a receptor for the alpha3NC1 domain and transdominantly inhibits integrin alphavbeta3 activation 16732726_Acquisition of multiple antitumor drugs was accompanied by a drastically reduced expression of alpha3beta1 in the adenocarcinoma cells. 16785564_Binding of Borrelia burgdorferi to integrin alpha 3 beta 1 results in release of inflammatory mediators from human chondrocytes and is proposed as a Toll-like receptor-independent pathway for activation of the innate immune response. 17265493_Chondrocytes with low chondrogenic capacity expressed higher levels of IGF-1, MMP-2, aggrecanase 2, while chondrocytes with high chondrogenic capacity expressed higher levels of CD44, CD151, and CD49c. 17475774_Integrin-mediated adhesion via alpha3beta1, but not alpha6beta4 integrin, supports cell survival through EGFR by signaling downstream to Erk. 17481915_These results indicate that binding of the alpha3beta1 integrin results in a suppression in the activation of the IL-1 induced intracellular signaling pathway from JNK to AP-1. 17498594_findings suggest an important role of integrin alpha2beta1, alpha3beta1, and alpha5beta1 in the architectural characteristics of ameloblastomas and adenomatoid odentogenic tumor 17630833_Beta1 and alpha3 integrins are functionally important receptors for type 1 pili-expressing bacteria within the urinary tract and possibly at other sites within the host. 17888902_identification of alpha3beta1 as the predominant integrin and laminin receptor in glioma cells, and as a brain invasion-mediating integrin 17997226_These results strongly suggest that enhanced expression of alpha3beta1 integrin on hepatocellular carcinoma cells is directly involved in their malignant phenotypes such as invasion and metastasis. 18224668_Increase in ITGA3 is associated with lymph node metastasis in squamous cell carcinoma of the tongue 18384059_Observational study of gene-disease association. (HuGE Navigator) 18464290_Both integrins, v3 and v5, are involved in the glioma cell radioresistance through the integrin-linked kinase (ILK) and the small GTPase RhoB, at least by regulating the radiation-induced mitotic cell death. 18492066_CD151 regulates integrin alpha3beta1 functions in two independent aspects: potentiation of integrin alpha3beta1-mediated cell adhesion and promotion of integrin alpha3beta1-stimulated signaling events involving tyrosine phosphorylation 18550570_Observational study of gene-disease association. (HuGE Navigator) 18695939_Results suggest that HAb18G/CD147 enhances the invasion and metastatic potentials of human hepatoma cells via integrin alpha3beta1-mediated FAK-paxillin and FAKPI3K-Ca(2+) signal pathways. 18839319_Src overexpression downregulates alpha3 integrin total protein expression and localization to the cell surface of HCT116 colon cancer cells. This indicates that Src activity may enhance metastasis by altering alpha3 integrin expression. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19130304_alpha 3, alpha 6A, and beta 1 integrin expression in cancer cells at the invasive front are related to the mode of invasion and prognosis in oral squamous cell carcinoma 19224172_Proliferation activity and malignant grade of meningiomas were increased with decreased expression of integrin-alpha(3), and down-regulation of integrin-alpha(3) mRNA was associated with the invasive biological behaviors in meningiomas. 19230171_Glomerular deposition of C5b-9 and a positive correlation with the intensity of tubular alpha3beta1-integrin suggests a possible implication in the development of tubulointerstitial fibrosis. 19297475_a cellular protein that appeared to be degraded by E1B55K in combination with the E4orf6 protein was a species of molecular mass approximately 130 kDa that was identified as the integrin alpha3 subunit 19339270_Observational study of gene-disease association. (HuGE Navigator) 19411307_alpha(2)-, alpha(3)-, and beta(1)-integrins and E-cadherin expression in normal human lung and bronchopulmonary sequestration and congenital cystic adenomatoid malformation were evaluated using Western blot and immunohistochemistry. 19451223_Nox1 knockdown led to a loss of directional migration which takes place through a RhoA-dependent alpha2/alpha3 integrin switch. 19730683_Observational study of gene-disease association. (HuGE Navigator) 19752234_For the first time, alpha3- and alpha5-laminins are shown to promote both adhesion and migration of mast cells, and alpha1 integrin is shown to mediate cell migration. 19755493_Gal-3 promotes epithelial cell migration by cross-linking MGAT5-modified complex N-glycans on alpha3beta1 integrin. 19759146_Integrin alpha3 is a bona fide substrate of the adenovirus serotype 5 E4orf6 and E1B55K protein-formed cullin 5-based E3 ubiquitin ligase complex. 20352300_The results suggest that the production of MMP-9 by MKN1 cells was potentiated by the alpha3beta1 integrin-laminin-5 interaction, which facilitated their invasion via degradation of the matrix. 20412552_down-regulation of alpha3beta1 integrin recapitulates crucial events governing keratinocyte migration associated with wound healing and tissue repair. 20877569_U937 macrophage responses to the TLR2 ligand, Pam3CSK4, are dependent upon integrin alpha3beta1. 20926544_Observational study of gene-disease association. (HuGE Navigator) 20927591_CD151, c-Met, and integrin alpha3/alpha6 were all overexpressed in pancreatic ductal adenocarcinoma. CD151 and c-Met might be new molecular markers to predict the prognosis of pancreatic ductal adenocarcinoma patients. 21182210_Studies have shown that ITGA3 plays a key role during cortical development, involved in neuronal migration and placement, as well as cortical layering 21195710_Data suggest that the alpha5 laminins emerge as putative primary extracellular matrix mediators of melanoma invasion and metastasis via alpha3/alpha6beta1 and other integrin receptors. 21530503_With increased tension cytoskeletal stress fibers develop that contain alphaSMA and alphavbeta3 integrin that replaces alpha2beta1 integrin, consistent with cell switching from collagen to non-collagen proteins interactions. 22512483_We identified three patients with homozygous mutations in the integrin alpha(3) gene that were associated with disrupted basement-membrane structures and compromised barrier functions in kidney, lung, and skin. 22684562_Taken together, it was concluded that CD151 promotes the proliferation and migration of PC3 cells through the formation of CD151-integrin complex and the activation of phosphorylated ERK. 22843693_CD151 links alpha(3)beta(1)/alpha(6)beta(1) integrins to Ras, Rac1, and Cdc42 by promoting the formation of multimolecular complexes in the membrane, which leads to the up-regulation of adhesion-dependent small GTPase activation 22917688_Most actinic cheilitis cases showed reduced expression of integrin alpha 3 and superficially invasive squamous cell carcinoma lacked intergrin alpha 3 in the invasive front. 22986527_Data indicate that alpha3beta1 and the tetraspanin CD151 directly associate at the front and retracting rear of polarised migrating breast carcinoma cells. 23011394_Loss of alpha3 integrin-adenomatous poliposis coli interaction promotes endothelial apoptosis. 23074279_Data show that TIMP-2-mediated inhibition of vascular endothelial cell permeability involves an integrin alpha3beta1-Shp-1-cAMP/protein kinase A-dependent vascular endothelial cadherin cytoskeletal association. 23094960_These results suggest that Ets-1 is involved in transcriptional activation of the alpha3 integrin gene through its binding to the Ets-consensus sequence at -133 bp 23233127_Collectively, these findings suggest that integrin alpha3beta1 plays pivotal roles in regulating cell proliferation and migration that enhance the invasive type of p53-deficient NSCLC cells. 23248240_Data indicate that Slug siRNA suppressed the TGF-beta1-induced integrin alpha3beta1-mediated cell migration ability of squamous cell carcinoma HSC-4 cells. 23533596_CD151 is positively associated with the invasiveness of HGC, and CD151 or the combination of CD151 and integrin alpha3 is a novel marker for predicting the prognosis of HGC patient. 23590307_Suggest that fibronectin fine tunes LM332-mediated migration by boosting bronchiolar cell adhesion to substrate via integrin alpha3beta1 integrin. 23613949_Both CD9/CD81-silenced cells and CD151-silenced cells showed delayed alpha3beta1-dependent cell spreading on laminin-332. 23652300_Integrin alpha3 contributes to the invasive nature of glioma stem-like cell via ERK1/2, which renders integrin alpha3 a prime candidate for anti-invasion therapy for glioblastoma. 23686814_The interaction between ephrin-As, Eph receptors and integrin alpha3 is plausibly important for the crosstalk between Eph and integrin signalling pathways at the membrane protrusions and in the migration of brain cancer cells. 23687274_These results showed evidence of Borrelia burgdorferi BB0172 localization in the outer membrane, the orientation of the vWFA domain to the extracellular environment, and its function as a metal ion-dependent integrin-binding protein. 23786209_study concluded that ITGA3 is a potential molecular marker for cells undergoing enhanced epithelial-mesenchymal transition as well as for cancer cells with aggressive phenotypes; integrin alpha3 likely plays a crucial role in the progression of both cancer cells and fibroblastic cells in cancer microenvironments 24006899_Two types of metastatic trait were found in OSCC: locoregional dissemination, which was reflected by high-ITGA3/CD9, and distant metastasis through hematogenous dissemination, uniquely distinguished by high-ITGB4/JUP. 24084442_High integrin alpha 3 expression is associated with malignant pleural mesothelioma. 24220332_the calf-1 domain is required for the transport of alpha3 from the ER to the Golgi apparatus to maintain the integrity of epithelial tissues, and hence the impairment of the calf-1 domain by the R628P mutation leads to severe diseases of the kidneys, lungs, and skin. 24289209_Integrin alpha2beta1 and integrin alpha3beta1 suppress anoikis in undifferentiated cells. 24381140_ITGA3 gene polymorphism is associated with with susceptibility of osteosarcoma. 24434582_Integrin alpha3beta1 mediates regulation of COX2 mRNA stability in human breast cancer cells. 24616281_Exposure to magnesium ions facilitated hBMSC proliferation via integrin alpha2 and alpha3 expression and partly promoted differentiation into osteoblasts via the alteration of ALP expression and activity 24621570_integrin-alpha3 mutation confers major renal developmental defects 24675526_endothelial integrin alpha3beta1 stabilizes tumor/endothelial cell adhesion and induces the formation of macromolecular signaling complex activating several major signaling pathways in endothelial cells. 24950714_COX2 and alpha3 are correlated in invasive ductal carcinoma independently of hormone receptor status or other clinicopathologic features, supporting the hypothesis that integrin alpha3beta1 is a determinant of COX2 expression in human breast cancer. 24958723_Molecular modeling indicated that galactosylation occurred on the periphery of alpha2beta1 integrin interaction with alpha1(IV)382-393 but right in the middle of alpha3beta1 integrin interaction with alpha1(IV)531-543 24985492_CTGF blocks integrin alpha3beta1-dependent adhesion of cancer cells. 25078904_ITGA3 translocation to the plasma membrane suppressed by hypoxia through inhibition of glycosylation facilitated cell invasion in A431. 25096929_NOX4 is highly predictive of relapse in stage II left-side colon cancer, whereas integrin alpha 3 beta 1 (ITGA3) is predictive of relapse in stage II right-side colon cancer. 25356755_our study demonstrates that CD151-alpha3beta1 integrin complexes regulate ovarian tumor growth by repressing Slug-mediated epithelial to mesenchymal transition and Wnt signaling 25472585_ITGA3, ITGA6, ITGB3,ITGB4 and ITGB5 are associated with GC susceptibility (rs2675), ITGA3, ITGA6, ITGB3, ITGB4 and ITGB5 are associated with gastric cancer susceptibility tumor stage and lymphatic metastasis in Chinese Han population 25497870_Findings show that alpha3 integrin is essentially involved in head and neck squamous cell carcinoma (HNSCC) cell radioresistance. 26318455_studies suggest that the presence and spacing of the RGD and synergy sites modulate integrin alpha3beta1 binding to Fn 26350464_based on the interaction motifs in Sdc1 and Sdc4, called synstatins (SSTN210-240 and SSTN87-131) competitively displace the receptor tyrosine kinase and alpha3beta1 integrin from the syndecan with an IC50 of 100-300 nm 26377974_Data show that CD151 protein (CD151)-alpha3beta1 integrin complexes cooperated with epidermal growth factor receptor (EGFR) to drive tumor cell motility. 26418968_the role of the CD151-alpha3beta1 complex in carcinoma progression is context dependent, and may depend on the mode of tumor cell invasion. 26464707_In high grade DCIS, when stratified according to the HER2 status, in the HER2-negative subgroup, CD151 assessed in combination with alpha3beta1 was significantly correlated with prognosis. 26674523_Genetic and Immunohistochemical Expression of Integrins ITGAV, ITGA6, and ITGA3 As Prognostic Factor for Colorectal Cancer 26788840_Expression of the alpha3-Integrin splice variants in the brain 26854491_ITGA3 mutation is responsible in the development of epidermolysis bullosa. 26992919_these findings identify CD151 and its interactions with integrins a3 and a6 as potential therapeutic targets for inhibiting stemness-driving mechanisms and stem cell populations in Glioblastoma. 27340677_The results suggested that podocyte detachment during early stage of diabetic nephropathy is mediated through upregulation of alpha3beta1-integrin. 27509031_Results provide evidence that alpha3 and alpha6 integrins have significantly different internalization kinetics and that coordination exists between them for internalization in prostate cancer cells. 27562932_During the past decades, many studies have provided evidence for a role of laminin-binding integrins in tumorigenesis, and both tumor-promoting and suppressive activities have been identified. (Review) 27717396_This study reports a variant of ILNEB syndrome in two siblings differing from the previously reported patients in the lack of nephrotic impairment and survival beyond childhood; they are the first reported compound heterozygous for ITGA3 mutations. 27821163_our studies clearly provide evidence that aberrant expression of sFRP2 can contribute to the invasive and metastatic potential for osteosarcoma. 27974569_Competition binding and infection experiments and biochemical assays pointed out alphaVbeta1 and alpha3beta1 to be of importance for human adenovirus-37 infection of corneal tissue. 28102841_Our study elucidates the molecular mechanisms of miR-101/ITGA3 pathway in regulating nasopharyngeal carcinoma (NPC)metastasis and angiogenesis, and the systemic delivery of miR-101 provides a potent evidence for the development of a novel microRNA-targeting anticancer strategy for NPC patients. 28324890_High ITGA3 expression is associated with bladder cancer. 28416479_findings identify a novel physiological context for combinatorial integrin signaling, laying the foundation for therapeutic strategies that manipulate alpha9beta1 and/or alpha3beta1 during wound healing 28612520_Results showed that ITGA3 was directly regulated by miR-199 family in head and neck squamous cell carcinoma cells (HNSCC). Its knockdown significantly inhibited cancer cell migration and invasion by HNSCC cells. 28618934_Both in cell lines and in mouse model, the extracellular matrix receptors including the integrin ( ITGA3 and ITGA2B), collagen ( COL5A1), and laminin ( LAMA5) were significantly inhibited by curcumin at messenger RNA and protein levels. 29511671_these findings suggest that ITGA3 may play a role as a potential oncogene in intrahepatic cholangiocarcinoma (ICC)and suppression of ITGA3 expression may establish a novel target for guiding the therapy of ICC patients. 29758195_ITGA3 is a novel target of miR-124 in colorectal cancer cells. 30128883_Bioinformatics analysis identifies ITGA3 as an oncogene in human tongue cancer. 30466509_Results show that in the absence of integrin alpha3, keratinocytes of patients with interstitial lung disease, nephrotic syndrome and epidermolysis bullosa (ILNEB) produce a fibronectin-rich microenvironment and make use of fibronectin-binding integrin subunits alphav and alpha5. 30614803_Up-regulation of integrin subunit alpha 3 (ITGA3) is observed in bladder cancer (BC) clinical samples and tumor cell lines. Silencing ITGA3 enhances apoptosis, inhibits tumor cell migration and invasion, and therefore decreases viability and proliferation of BC cells. 30659093_both integrin alpha3beta1 and PI4KIIalpha co-localized to the trans-Golgi network, where they physically interacted with each other, and PI4KIIalpha specifically associated with integrin alpha3 but not alpha5 30941771_MiR-524-5p targeting on FOXE1 and ITGA3 prevents thyroid cancer progression through different pathways including cell cycling and autophagy. 30944668_Elevated serum sFRP2 levels are associated with primary tumor size, tumor stage, and lymph node metastases. 30971252_Levels of antibodies that inhibit the binding of children's IE to the receptors ICAM-1, integrin alpha3beta1 and laminin increased with age. The breadth of antibodies that inhibit ICAM-1 and laminin adhesion (defined as the proportion of IE isolates whose binding was reduced by >/= 50%) also significantly increased with age. 31101121_Downregulation of alpha3beta1 in a HER2-driven mouse model and in HER2-overexpressing human mammary carcinoma cells promotes progression and invasiveness of tumors. The invasion-suppressive role of alpha3beta1 was not observed in triple-negative mammary carcinoma cells, illustrating the tumor type-specific and complex function of alpha3beta1 in breast cancer. 31340144_Integrin-alpha3 Is a Functional Marker of Ex Vivo Expanded Human Long-Term Hematopoietic Stem Cells. 31421623_The results strongly suggest that ITGA3 is a key molecule for cellular attachment and entry of non-enveloped hepatitis E virus. 31987909_findings indicate that integrin alpha3 interacts with VASP and regulates its expression 32210347_Anatomy of a viral entry platform differentially functionalized by integrins alpha3 and alpha6. 32356431_Alterations in the immunoreactivity of laminin, type IV collagen and alpha3beta1 integrin in diabetic rat ovarian follicles. 32705201_MicroRNA199a5p suppresses cell proliferation, migration and invasion by targeting ITGA3 in colorectal cancer. 32851057_TMT-Based Quantitative Proteomic Analysis Identification of Integrin Alpha 3 and Integrin Alpha 5 as Novel Biomarkers in Pathogenesis of Acute Aortic Dissection. 33099569_Expression and Prognostic Analyses of ITGA3, ITGA5, and ITGA6 in Head and Neck Squamous Cell Carcinoma. 33173103_Paracoccidioides brasiliensis downmodulates alpha3 integrin levels in human lung epithelial cells in a TLR2-dependent manner. 33428143_Key Role of CD151-integrin Complex in Lung Cancer Metastasis and Mechanisms Involved. 33768705_Skin fragility, renal malformation and interstitial lung disease due to compound heterozygous ITGA3 mutations. 34023363_Homozygous ITGA3 Missense Mutation in Adults in a Family with Syndromic Epidermolysis Bullosa (ILNEB) without Pulmonary Involvement. 34102169_Detection of bladder cancer with aberrantly fucosylated ITGA3. 34242748_CD9 and ITGA3 are regulated during HIV-1 infection in macrophages to support viral replication. 34938358_miRNA-144-5p/ITGA3 Suppressed the Tumor-Promoting Behaviors of Thyroid Cancer Cells by Downregulating ITGA3. | ENSMUSG00000001507 | Itga3 | 2.837039e+02 | 1.0547524 | 0.076904402 | 0.3011719 | 6.539008e-02 | 0.7981710358 | 0.96139092 | No | Yes | 2.299958e+02 | 45.473075 | 2.222474e+02 | 45.397319 | |
| ENSG00000007264 | 4145 | MATK | protein_coding | P42679 | FUNCTION: Could play a significant role in the signal transduction of hematopoietic cells. May regulate tyrosine kinase activity of SRC-family members in brain by specifically phosphorylating their C-terminal regulatory tyrosine residue which acts as a negative regulatory site. It may play an inhibitory role in the control of T-cell proliferation. {ECO:0000269|PubMed:9171348}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Reference proteome;SH2 domain;SH3 domain;Transferase;Tyrosine-protein kinase | The protein encoded by this gene has amino acid sequence similarity to Csk tyrosine kinase and has the structural features of the CSK subfamily: SRC homology SH2 and SH3 domains, a catalytic domain, a unique N terminus, lack of myristylation signals, lack of a negative regulatory phosphorylation site, and lack of an autophosphorylation site. This protein is thought to play a significant role in the signal transduction of hematopoietic cells. It is able to phosphorylate and inactivate Src family kinases, and may play an inhibitory role in the control of T-cell proliferation. This protein might be involved in signaling in some cases of breast cancer. Three alternatively spliced transcript variants that encode different isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:4145; | cytosol [GO:0005829]; membrane [GO:0016020]; ATP binding [GO:0005524]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; positive regulation of cell population proliferation [GO:0008284]; protein phosphorylation [GO:0006468] | 12122014_ERBB2 binds to the SH2 domain of CHK and inhibits cell growth in human breast tumor cell lines 12429987_Overexpression of the Csk homologous kinase facilitates phosphorylation of akt/protein kinase b in breast neoplasms 12782282_CHK Leu223 stabilizes the movement of the alphaC-helix of the protein tyrosine kinase 15329911_Loss of CHK expression is associated with human brain tumors 15748901_Nuclear multi-lobulation in late S phase, which is dependent on polymerization and depolymerization of microtubules, may be involved in nuclear Chk-induced inhibition of proliferation. 15890649_striking functional differences between the Csk and Chk SH2 domains and revealed functional similarities between the Chk and Src SH2 domains 16168623_This study describes for the first time the Src-independent actions of CHK and provides novel insights into CHK function in neural cells. 16574955_Matk/CHK is not functionally redundant with Csk, and this tyrosine kinase plays an important role as a regulator of immunologic responses 16707123_Findings indicate that the importance of the N-terminal domain to Chk-induced tyrosine phosphorylation in the nucleus, and implicate that nuclear tyrosine-phosphorylated proteins may contribute to inhibition of cell proliferation. 17492661_Progesterone increases MATK in mast cells and reducs cell proliferation. 17934522_These results reveal a potentially important role for CHK in Src activation and tumorigenicity in colon cancer cells. 18292939_CHK is capable of inhibiting the CXCL12-CXCR4 pathway in neuroblastoma. 21799000_After IGF-I stimulation, CTK is recruited to IGF-IR and its recruitment facilitates CTK's subsequent association with phospho-SHPS-1. | ENSMUSG00000004933 | Matk | 2.355806e+03 | 1.5026662 | 0.587524583 | 0.3214395 | 3.314026e+00 | 0.0686910072 | 0.71206859 | No | Yes | 2.835000e+03 | 414.524735 | 1.568186e+03 | 235.773247 | |
| ENSG00000010244 | 7756 | ZNF207 | protein_coding | O43670 | FUNCTION: Kinetochore- and microtubule-binding protein that plays a key role in spindle assembly (PubMed:24462186, PubMed:24462187, PubMed:26388440). ZNF207/BuGZ is mainly composed of disordered low-complexity regions and undergoes phase transition or coacervation to form temperature-dependent liquid droplets. Coacervation promotes microtubule bundling and concentrates tubulin, promoting microtubule polymerization and assembly of spindle and spindle matrix by concentrating its building blocks (PubMed:26388440). Also acts as a regulator of mitotic chromosome alignment by mediating the stability and kinetochore loading of BUB3 (PubMed:24462186, PubMed:24462187). Mechanisms by which BUB3 is protected are unclear: according to a first report, ZNF207/BuGZ may act by blocking ubiquitination and proteasomal degradation of BUB3 (PubMed:24462186). According to another report, the stabilization is independent of the proteasome (PubMed:24462187). {ECO:0000269|PubMed:24462186, ECO:0000269|PubMed:24462187, ECO:0000269|PubMed:26388440}. | Alternative splicing;Cell cycle;Cell division;Centromere;Chromosome;Chromosome partition;Cytoplasm;Cytoskeleton;DNA-binding;Kinetochore;Metal-binding;Microtubule;Mitosis;Nucleus;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:7756; | cytoplasm [GO:0005737]; kinetochore [GO:0000776]; microtubule [GO:0005874]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spindle [GO:0005819]; spindle matrix [GO:1990047]; DNA binding [GO:0003677]; heparin binding [GO:0008201]; metal ion binding [GO:0046872]; microtubule binding [GO:0008017]; RNA binding [GO:0003723]; attachment of spindle microtubules to kinetochore [GO:0008608]; cell division [GO:0051301]; microtubule bundle formation [GO:0001578]; microtubule polymerization [GO:0046785]; mitotic sister chromatid segregation [GO:0000070]; mitotic spindle assembly [GO:0090307]; mitotic spindle assembly checkpoint signaling [GO:0007094]; protein stabilization [GO:0050821]; regulation of chromosome segregation [GO:0051983] | 26388440_BuGZ forms temperature-dependent liquid droplets alone or on microtubules in physiological buffers. Coacervation in vitro or in spindle and spindle matrix depends on hydrophobic residues in BuGZ. BuGZ coacervation and its binding to microtubules and tubulin are required to promote assembly of spindle and spindle matrix in Xenopus egg extract and in mammalian cells. 29074706_the two zinc fingers of BuGZ directly bind to AurA and BuGZ coacervation appears to promote AurA activation during spindle assembly. 30349051_a distinct isoform of ZNF207 functions in hESCs at the nexus that balances pluripotency and differentiation to ectoderm. 31113916_ZNF207 expression is elevated in hepatocellular carcinoma 32820050_BuGZ facilitates loading of spindle assembly checkpoint proteins to kinetochores in early mitosis. | ENSMUSG00000017421 | Zfp207 | 4.095497e+03 | 1.1090628 | 0.149341012 | 0.3059635 | 2.319987e-01 | 0.6300457647 | 0.91646120 | No | Yes | 4.320387e+03 | 717.928577 | 3.359592e+03 | 572.623680 | ||
| ENSG00000010256 | 7384 | UQCRC1 | protein_coding | P31930 | FUNCTION: Component of the ubiquinol-cytochrome c oxidoreductase, a multisubunit transmembrane complex that is part of the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. The cytochrome b-c1 complex catalyzes electron transfer from ubiquinol to cytochrome c, linking this redox reaction to translocation of protons across the mitochondrial inner membrane, with protons being carried across the membrane as hydrogens on the quinol. In the process called Q cycle, 2 protons are consumed from the matrix, 4 protons are released into the intermembrane space and 2 electrons are passed to cytochrome c (By similarity). The 2 core subunits UQCRC1/QCR1 and UQCRC2/QCR2 are homologous to the 2 mitochondrial-processing peptidase (MPP) subunits beta-MPP and alpha-MPP respectively, and they seem to have preserved their MPP processing properties (By similarity). May be involved in the in situ processing of UQCRFS1 into the mature Rieske protein and its mitochondrial targeting sequence (MTS)/subunit 9 when incorporated into complex III (Probable). Seems to play an important role in the maintenance of proper mitochondrial function in nigral dopaminergic neurons (PubMed:33141179). {ECO:0000250|UniProtKB:P07256, ECO:0000250|UniProtKB:P31800, ECO:0000269|PubMed:33141179, ECO:0000305|PubMed:29243944}. | 3D-structure;Acetylation;Direct protein sequencing;Disease variant;Electron transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Neuropathy;Parkinsonism;Phosphoprotein;Reference proteome;Respiratory chain;Transit peptide;Transport | hsa:7384; | mitochondrial inner membrane [GO:0005743]; mitochondrial respirasome [GO:0005746]; mitochondrial respiratory chain complex III [GO:0005750]; mitochondrion [GO:0005739]; metal ion binding [GO:0046872]; protein-containing complex binding [GO:0044877]; ubiquinol-cytochrome-c reductase activity [GO:0008121]; ubiquitin protein ligase binding [GO:0031625]; aerobic respiration [GO:0009060]; mitochondrial electron transport, ubiquinol to cytochrome c [GO:0006122]; oxidative phosphorylation [GO:0006119]; response to activity [GO:0014823]; response to alkaloid [GO:0043279] | 16775426_UQCRC1 was highly expressed in breast and ovarian tumors 19110265_This protein has been found differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 20877624_Observational study of gene-disease association. (HuGE Navigator) 26943237_Gene expression level of UQCRC1 is significantly higher in AD patients when compared to normal controls. 27845902_dysregulated UQCRC1 and UQCRFS1 are involved in impaired mitochondrial electron transport chain function. 31276465_High miR-214-3p expression may promote osteosarcoma cell proliferation by targeting UQCRC1. 32666668_The lack of association between ubiquinol-cytochrome c reductase core protein I (UQCRC1) variants and Parkinson's disease in an eastern Chinese population. 33141179_Mitochondrial UQCRC1 mutations cause autosomal dominant parkinsonism with polyneuropathy. 33248804_Lack of evidence for association of UQCRC1 with Parkinson's disease in Europeans. | ENSMUSG00000025651 | Uqcrc1 | 2.106576e+04 | 1.1661035 | 0.221695831 | 0.3265526 | 4.576709e-01 | 0.4987145125 | 0.87548575 | No | Yes | 2.235372e+04 | 3040.305846 | 1.601398e+04 | 2234.023055 | ||
| ENSG00000010318 | 51533 | PHF7 | protein_coding | Q9BWX1 | FUNCTION: May play a role in spermatogenesis. | Alternative splicing;Metal-binding;Nucleus;Reference proteome;Zinc;Zinc-finger | Spermatogenesis is a complex process regulated by extracellular and intracellular factors as well as cellular interactions among interstitial cells of the testis, Sertoli cells, and germ cells. This gene is expressed in the testis in Sertoli cells but not germ cells. The protein encoded by this gene contains plant homeodomain (PHD) finger domains, also known as leukemia associated protein (LAP) domains, believed to be involved in transcriptional regulation. The protein, which localizes to the nucleus of transfected cells, has been implicated in the transcriptional regulation of spermatogenesis. Alternate splicing results in multiple transcript variants of this gene. [provided by RefSeq, May 2013]. | hsa:51533; | cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872] | 22894908_PHF7 promoter binds H4K12ac in mature spermatozoa. | ENSMUSG00000021902 | Phf7 | 2.944493e+02 | 0.9300140 | -0.104675701 | 0.3281851 | 1.034193e-01 | 0.7477643224 | 0.94694596 | No | Yes | 2.178393e+02 | 40.418693 | 2.748402e+02 | 52.081706 | |
| ENSG00000010319 | 56920 | SEMA3G | protein_coding | Q9NS98 | FUNCTION: Has chemorepulsive activities for sympathetic axons. Ligand of NRP2 (By similarity). {ECO:0000250}. | Disulfide bond;Glycoprotein;Immunoglobulin domain;Reference proteome;Secreted;Signal | The transcription of this gene is activated by PPAR-gamma, and the resulting protein product plays a role in endothelial cell migration. Expression of this gene also inhibits tumor cell migration and invasion. [provided by RefSeq, Jul 2016]. | hsa:56920; | extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; chemorepellent activity [GO:0045499]; semaphorin receptor binding [GO:0030215]; signaling receptor binding [GO:0005102]; axon guidance [GO:0007411]; negative chemotaxis [GO:0050919]; negative regulation of axon extension [GO:0030517]; negative regulation of axon extension involved in axon guidance [GO:0048843]; neural crest cell migration [GO:0001755]; positive regulation of cell migration [GO:0030335]; semaphorin-plexin signaling pathway [GO:0071526] | 18781179_In gliomas, SEMA3B, SEMA3G and NRP2 expressions were related to prolonged survival 18818766_Sema3G inhibits tumor progression from MDA-MB-435 but not from MDA-MB-231 cancer cells. It inhibits tumor angiogenesis in MDA-MB-231 cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20947821_Identify Sema3G as a primarily endothelial cell-expressed class 3 semaphorin that controls endothelial and smooth muscle cell functions in autocrine and paracrine manners. 22562223_overexpressed SEMA3G inhibited the migratory and invasive behavior of U251MG cancer cells 25335934_Results identify Sema3g as one of the downstream effectors of PPAR-gamma, which is centrally involved in regulating endothelial cell migration. 27180624_Results indicate that Sema3G protein is secreted by podocytes and protects them from inflammatory kidney diseases and diabetic nephropathy. 32452055_Identification of a four immune-related genes signature based on an immunogenomic landscape analysis of clear cell renal cell carcinoma. 33586674_Endothelium-derived semaphorin 3G attenuates ischemic retinopathy by coordinating beta-catenin-dependent vascular remodeling. | ENSMUSG00000021904 | Sema3g | 3.848774e+02 | 1.0475750 | 0.067053588 | 0.3188295 | 4.366182e-02 | 0.8344840653 | 0.96761594 | No | Yes | 2.678606e+02 | 37.985912 | 2.783086e+02 | 40.332848 | |
| ENSG00000011143 | 54903 | MKS1 | protein_coding | Q9NXB0 | FUNCTION: Component of the tectonic-like complex, a complex localized at the transition zone of primary cilia and acting as a barrier that prevents diffusion of transmembrane proteins between the cilia and plasma membranes. Involved in centrosome migration to the apical cell surface during early ciliogenesis. Required for ciliary structure and function, including a role in regulating length and appropriate number through modulating centrosome duplication. Required for cell branching morphology. {ECO:0000269|PubMed:17185389, ECO:0000269|PubMed:19515853, ECO:0000269|PubMed:26490104}. | Alternative splicing;Bardet-Biedl syndrome;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Disease variant;Joubert syndrome;Meckel syndrome;Mental retardation;Obesity;Reference proteome | The protein encoded by this gene localizes to the basal body and is required for formation of the primary cilium in ciliated epithelial cells. Mutations in this gene result in Meckel syndrome type 1 and in Bardet-Biedl syndrome type 13. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2009]. | hsa:54903; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; MKS complex [GO:0036038]; branching morphogenesis of an epithelial tube [GO:0048754]; cardiac septum morphogenesis [GO:0060411]; cilium assembly [GO:0060271]; common bile duct development [GO:0061009]; determination of left/right symmetry [GO:0007368]; embryonic brain development [GO:1990403]; embryonic digit morphogenesis [GO:0042733]; embryonic skeletal system development [GO:0048706]; epithelial structure maintenance [GO:0010669]; head development [GO:0060322]; inner ear receptor cell stereocilium organization [GO:0060122]; motile cilium assembly [GO:0044458]; neural tube closure [GO:0001843]; non-motile cilium assembly [GO:1905515]; regulation of canonical Wnt signaling pathway [GO:0060828]; regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning [GO:1901620]; regulation of Wnt signaling pathway, planar cell polarity pathway [GO:2000095]; smoothened signaling pathway involved in regulation of secondary heart field cardioblast proliferation [GO:0003271] | 16415886_identification of a gene, MKS1,(Meckel syndrome) mutated in MKS families linked to 17q. 17185389_The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. 17377820_Study concluded that MKS1 and MKS3 account for the majority of Meckel-Gruber syndrome; polydactyly is usually found in MKS1 but rare in MKS3; cases with no, or milder, CNS phenotypes were only found in MKS3. 17397051_Observational study of genotype prevalence. (HuGE Navigator) 17397051_genotyping of MKS1 & MKS3 genes in a large, multiethnic cohort of 120 independent cases of Meckel syndrome; first results indicate that the MKS1 & MKS3 genes are each responsible for about 7% of MKS cases with various mutations in different populations 17437276_Our results indicate that MKS1 mutations are not restricted to the Caucasian gene pool and suggest splicing defects are a crucial mutational mechanism in MKS1, and further genetic heterogeneity for MKS. 17935508_Observational study of genotype prevalence. (HuGE Navigator) 18327255_Mutations in MKS1 is associated with Bardet-Biedl syndrome 19208769_MKS-1 and MKS-1-related proteins 1 and 2 (MKSR-1, MKSR-2), localize to transition zones/basal bodies of sensory cilia; subcellular localization is largely co-dependent, pointing to a functional relationship between the proteins 19515853_Kidney tissue and cells from MKS1 and MKS3 patients showed defects in centrosome and cilia number, including multi-ciliated respiratory-like epithelia, and longer cilia. 21068128_Observational study of gene-disease association. (HuGE Navigator) 24886560_describe four patients with mild Joubert phenotypes who carry pathogenic mutations in either MKS1 or B9D1, two genes previously implicated only in Meckel syndrome 26490104_MKS1 functions in the transition zone at the base of the cilium to regulate ciliary INPP5E content. 27340223_Dnah11(avc)(4) did not disrupt SHF Hh signaling and caused Atrioventricular septal defects (AVSDs) only concurrently with heterotaxy, a left/right axis abnormality. In contrast, Mks1(avc)(6) disrupted SHF Hh signaling and caused AVSDs without heterotaxy.We speculate that cilia gene mutations contribute to both syndromic and non-syndromic AVSDs in humans 27570071_we have described a pathogenic variant in the MKS1 resulting in a mild Joubert syndrome phenotype, which broadens the spectrum of mutations in the MKS1. 32726168_Formation of the B9-domain protein complex MKS1-B9D2-B9D1 is essential as a diffusion barrier for ciliary membrane proteins. | ENSMUSG00000034121 | Mks1 | 2.158727e+03 | 1.0472502 | 0.066606154 | 0.2733139 | 6.030418e-02 | 0.8060158200 | 0.96141221 | No | Yes | 1.829517e+03 | 120.193957 | 1.811220e+03 | 121.787632 | |
| ENSG00000011260 | 51096 | UTP18 | protein_coding | Q9Y5J1 | FUNCTION: Involved in nucleolar processing of pre-18S ribosomal RNA. {ECO:0000250}. | 3D-structure;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;WD repeat;rRNA processing | hsa:51096; | nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; Pwp2p-containing subcomplex of 90S preribosome [GO:0034388]; small-subunit processome [GO:0032040]; RNA binding [GO:0003723]; rRNA processing [GO:0006364] | ENSMUSG00000054079 | Utp18 | 8.456820e+02 | 0.7400619 | -0.434282244 | 0.3158344 | 1.855808e+00 | 0.1731093970 | 0.77611497 | No | Yes | 7.188856e+02 | 122.013307 | 9.435777e+02 | 163.916481 | |||
| ENSG00000011376 | 23395 | LARS2 | protein_coding | Q15031 | FUNCTION: Catalyzes the attachment of leucine to its cognate tRNA. {ECO:0000269|PubMed:26537577}. | ATP-binding;Acetylation;Aminoacyl-tRNA synthetase;Deafness;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Phosphoprotein;Protein biosynthesis;Reference proteome;Transit peptide | This gene encodes a class 1 aminoacyl-tRNA synthetase, mitochondrial leucyl-tRNA synthetase. Each of the twenty aminoacyl-tRNA synthetases catalyzes the aminoacylation of a specific tRNA or tRNA isoaccepting family with the cognate amino acid. [provided by RefSeq, Jul 2008]. | hsa:23395; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; aminoacyl-tRNA editing activity [GO:0002161]; ATP binding [GO:0005524]; leucine-tRNA ligase activity [GO:0004823]; leucyl-tRNA aminoacylation [GO:0006429]; mitochondrial translation [GO:0032543]; tRNA aminoacylation for protein translation [GO:0006418] | 15737668_Upregulation of LARS2 is a hallmark of 324A>G mutation. The accumulation of 3243A>G mutation in the brain may have a pathophysiologic role in bipolar disorder and schizophrenia. 15919814_In this study, we provide evidence that the LARS2 gene may represent a novel type 2 diabetes susceptibility gene. 15919814_Observational study of gene-disease association. (HuGE Navigator) 18796578_There was investigated whether overexpression of human mitochondrial LeuRS suppressed translation and respiratory chain defects associated with the pathogenic A3243G mutation in human cells. 19129950_data indicate that inactivation of LARS2 by both genetic and epigenetic mechanisms may be a common and important event in the carcinogenesis of nasopharyngeal carcinoma 19847392_No evidence to support previous data indicating a role in type 2 diabetes susceptibility in humans with LARS2 single nucleotide polymorphisms 19847392_Observational study of gene-disease association. (HuGE Navigator) 20194621_The alteration of aminoacylation tRNA(Leu(UUR)) caused by the A3243G mutation led to mitochondrial translational defects and thereby reduced the aminoacylated efficiencies of tRNA(Leu(UUR)) as well as tRNA(Ala) and tRNA(Met). 20877624_Observational study of gene-disease association. (HuGE Navigator) 23541342_Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. 24413189_Leucyl tRNA synthetase is able to partially rescue defects caused by mutations in non-cognate itochondrial-tRNAs. 26272616_analysis of the CP1 domain in human mitochondrial leucyl-tRNA synthetase 26657938_This represents the first independent replication of the involvement of LARS2 mutations in Perrault syndrome, contributing valuable information for the further understanding of this disease 26970254_ere we present eight families affected by Perrault syndrome. In five families we identified novel or previously reported variants in HSD17B4, LARS2, CLPP and C10orf2 29071539_IARS2 knockdown inhibits proliferation, suppresses colony formation, and causes cell cycle arrest in AGS cells. 29205794_We concluded that Perrault syndrome patients with LARS2 mutations are at risk for neurologic problems, despite previous notions otherwise. 29762635_We report novel associations between methylation at MSI2 and LARS2 and obesity-related traits. These results provide further insight into mechanisms underlying obesity-related traits, which can enable identification of new biomarkers in obesity-related chronic diseases. 30737337_This study adds LARS2 and KARS pathogenic variants as gene defects that may underlie deafness, ovarian failure, and leukodystrophy with mitochondrial signature. 32442335_The expanding LARS2 phenotypic spectrum: HLASA, Perrault syndrome with leukodystrophy, and mitochondrial myopathy. | ENSMUSG00000035202 | Lars2 | 6.939135e+03 | 0.9879597 | -0.017475861 | 0.2779326 | 3.989538e-03 | 0.9496368777 | 0.99022718 | No | Yes | 6.051497e+03 | 506.967433 | 6.158160e+03 | 528.939874 | |
| ENSG00000011426 | 54443 | ANLN | protein_coding | Q9NQW6 | FUNCTION: Required for cytokinesis (PubMed:16040610). Essential for the structural integrity of the cleavage furrow and for completion of cleavage furrow ingression. Plays a role in bleb assembly during metaphase and anaphase of mitosis (PubMed:23870127). May play a significant role in podocyte cell migration (PubMed:24676636). {ECO:0000269|PubMed:10931866, ECO:0000269|PubMed:12479805, ECO:0000269|PubMed:15496454, ECO:0000269|PubMed:16040610, ECO:0000269|PubMed:16357138, ECO:0000269|PubMed:23870127, ECO:0000269|PubMed:24676636}. | 3D-structure;Acetylation;Actin-binding;Alternative splicing;Cell cycle;Cell division;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Isopeptide bond;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes an actin-binding protein that plays a role in cell growth and migration, and in cytokinesis. The encoded protein is thought to regulate actin cytoskeletal dynamics in podocytes, components of the glomerulus. Mutations in this gene are associated with focal segmental glomerulosclerosis 8. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Oct 2014]. | hsa:54443; | actin cytoskeleton [GO:0015629]; actomyosin contractile ring [GO:0005826]; bleb [GO:0032059]; cell cortex [GO:0005938]; cytosol [GO:0005829]; midbody [GO:0030496]; nucleoplasm [GO:0005654]; actin binding [GO:0003779]; cadherin binding [GO:0045296]; actomyosin contractile ring assembly [GO:0000915]; glomerular visceral epithelial cell migration [GO:0090521]; hematopoietic progenitor cell differentiation [GO:0002244]; mitotic cytokinesis [GO:0000281]; positive regulation of bleb assembly [GO:1904172]; regulation of exit from mitosis [GO:0007096]; septin ring assembly [GO:0000921]; septin ring organization [GO:0031106] | 15496454_anillin has a role in spatially regulating the contractile activity of myosin II during cytokinesis 16040610_Anillin is a substrate of APC/C that controls spatial contractility of myosin during late cytokinesis. 16203764_anillin is overexpressed in diverse common human tumors, but not simply because of its role in cell proliferation 16357138_Over-expression of ANLN is associated with pulmonary carcinogenesis 18158243_Human anillin contains a conserved C-terminal domain that is essential for its function and localization. 19995712_This study was to establish if ANLN was a molecular marker for pancreatic cancer. 21109967_Cytoplasmic anillin expression is a marker of favourable prognosis in renal cell carcinoma patients. 21849473_CIT-K is capable of physically and functionally interacting with the actin-binding protein anillin. 22514687_Data supports an analogous function for the anillin-Ect2 complex in human cells and one hypothesis is that this complex has functionally replaced the Drosophila anillin-RacGAP50C complex. 23547718_Antibody screening revealed 3 candidate prognostic markers in breast cancer: the Anillin (ANLN); PDZ-Binding Kinase (PBK); and, PDZ-Domain Containing 1 (PDZK1). 24088567_Supervillin concentrates activated and total myosin II at the furrow, and simultaneous knockdown of supervillin and anillin additively increases cell division failure. 24676636_Identified a missense mutation R431C in anillin (ANLN), an F-actin binding cell cycle gene, as a cause of FSGS. We screened 250 additional families with FSGS and found another variant, G618C, that segregates with disease in a second family with FSGS. 24809965_ANLN is positively associated with Wnt/beta-catenin signaling in gastric cancer.Elevated expression of ANLN is a predictor of intestinal type gastric cancer. ER-alpha is negatively associated with the expression of ANLN in gastric cancer. 24994938_The sequestration of anillin by astral microtubules might alter the organization of cortical proteins to polarize cells for cytokinesis 25223638_Data suggest that anillin (ANLN) could be involved in breast cancer progression and a potential target candidate in breast cancer. 25359885_these data demonstrate that a complex of p190RhoGAP-A and anillin modulates RhoA-GTP levels in the cytokinetic furrow to ensure progression of cytokinesis. 25809162_Anillin regulates intercellular adhesion in model human epithelia by mechanisms involving the suppression of JNK activity and controlling the assembly of the perijunctional cytoskeleton. 26135313_High nuclear expression of anillin (ANLN) is an independent predictor of poor disease-specific survival and it is a useful prognostic marker of urothelial carcinoma of the upper urinary tract (UCUT). 26319989_Identification 4 novel and 18 known exonic ANLN variants associated with carotid intima-media thickness at bifurcation. 26486082_Findings indicate that miR-497 is a potent tumor suppressor that inhibits cancer phenotypes by targeting ANLN and HSPA4L in NPC. 27062703_ANLN expression is elevated in colorectal cancer and has a strong potential to act as a biomarker for the prognosis of colorectal cancer. 27863473_Anillin (ANLN) expression in tumor cells is correlated to poor prognosis in breast cancer patients, independent of Ki-67 or tumor size. 28081137_S635 phosphorylation is essential for cytokinesis. 28600503_Bladder urothelial carcinoma patients with elevated ANLN expression had poorer survival prospects. 28931593_Authors found that decreasing Ran-GTP levels or tethering active Ran to the equatorial membrane affects anillin's localization and causes cytokinesis phenotypes. 29274368_Knockdown of ANLN in liver cells blocks cytokinesis and inhibits development of liver tumors. 30530503_High ANLN expression is associated with glioma. 30548429_Missense mutation in ANLN was identified as a cause of branchio-otic syndrome in a specific Chinese family. 31268619_Study suggests that the expression of ANLN in lung adenocarcinoma is associated with metastasis of cancer cells. ANLN may be involved in the metastasis of lung adenocarcinoma by promoting epithelial mesenchymal transformation of tumor cells. 31578580_concomitantly high expression of ANLN and KDR is associated with breast cancer. 31853940_Visualizing dynamic actin cross-linking processes driven by the actin-binding protein anillin. 31910867_Anillin regulates breast cancer cell migration, growth, and metastasis by non-canonical mechanisms involving control of cell stemness and differentiation. 32198770_LncRNA XIST promotes chemoresistance of breast cancer cells to doxorubicin by sponging miR-200c-3p to upregulate ANLN. 32413362_High mobility group AT-hook 2 promotes tumorigenicity of pancreatic cancer cells via upregulating ANLN. 32880660_Anillin is an emerging regulator of tumorigenesis, acting as a cortical cytoskeletal scaffold and a nuclear modulator of cancer cell differentiation. 33089886_Actin-binding protein Anillin promotes the progression of gastric cancer in vitro and in mice. 33862101_Molecular basis of functional exchangeability between ezrin and other actin-membrane associated proteins during cytokinesis. 34082790_ANLN promotes carcinogenesis in oral cancer by regulating the PI3K/mTOR signaling pathway. 34321459_Anillin propels myosin-independent constriction of actin rings. 34344861_Alternatively spliced ANLN isoforms synergistically contribute to the progression of head and neck squamous cell carcinoma. 34777569_ANLN Regulated by miR-30a-5p Mediates Malignant Progression of Lung Adenocarcinoma. 35340220_Comprehensive Analysis of ANLN in Human Tumors: A Prognostic Biomarker Associated with Cancer Immunity. | ENSMUSG00000036777 | Anln | 8.130994e+02 | 0.9040604 | -0.145508883 | 0.3377848 | 1.815271e-01 | 0.6700641177 | 0.92913032 | No | Yes | 7.890795e+02 | 136.675426 | 8.245494e+02 | 146.426898 | |
| ENSG00000012061 | 2067 | ERCC1 | protein_coding | P07992 | FUNCTION: [Isoform 1]: Non-catalytic component of a structure-specific DNA repair endonuclease responsible for the 5'-incision during DNA repair. Responsible, in conjunction with SLX4, for the first step in the repair of interstrand cross-links (ICL). Participates in the processing of anaphase bridge-generating DNA structures, which consist in incompletely processed DNA lesions arising during S or G2 phase, and can result in cytokinesis failure. Also required for homology-directed repair (HDR) of DNA double-strand breaks, in conjunction with SLX4. {ECO:0000269|PubMed:17273966, ECO:0000269|PubMed:23623389, ECO:0000269|PubMed:24036546}.; FUNCTION: [Isoform 2]: Not functional in the nucleotide excision repair pathway. {ECO:0000305|PubMed:24036546}.; FUNCTION: [Isoform 3]: Not functional in the nucleotide excision repair pathway. {ECO:0000305|PubMed:24036546}.; FUNCTION: [Isoform 4]: Not functional in the nucleotide excision repair pathway. {ECO:0000305|PubMed:24036546}. | 3D-structure;Acetylation;Alternative splicing;Cataract;Cytoplasm;DNA damage;DNA repair;DNA-binding;Isopeptide bond;Nucleus;Reference proteome;Ubl conjugation | The product of this gene functions in the nucleotide excision repair pathway, and is required for the repair of DNA lesions such as those induced by UV light or formed by electrophilic compounds including cisplatin. The encoded protein forms a heterodimer with the XPF endonuclease (also known as ERCC4), and the heterodimeric endonuclease catalyzes the 5' incision in the process of excising the DNA lesion. The heterodimeric endonuclease is also involved in recombinational DNA repair and in the repair of inter-strand crosslinks. Mutations in this gene result in cerebrooculofacioskeletal syndrome, and polymorphisms that alter expression of this gene may play a role in carcinogenesis. Multiple transcript variants encoding different isoforms have been found for this gene. The last exon of this gene overlaps with the CD3e molecule, epsilon associated protein gene on the opposite strand. [provided by RefSeq, Oct 2009]. | hsa:2067; | chromosome, telomeric region [GO:0000781]; cytoplasm [GO:0005737]; ERCC4-ERCC1 complex [GO:0070522]; nucleoplasm [GO:0005654]; nucleotide-excision repair complex [GO:0000109]; nucleotide-excision repair factor 1 complex [GO:0000110]; damaged DNA binding [GO:0003684]; endonuclease activity [GO:0004519]; promoter-specific chromatin binding [GO:1990841]; protein C-terminus binding [GO:0008022]; protein domain specific binding [GO:0019904]; single-stranded DNA binding [GO:0003697]; TFIID-class transcription factor complex binding [GO:0001094]; cell population proliferation [GO:0008283]; DNA recombination [GO:0006310]; DNA repair [GO:0006281]; double-strand break repair via nonhomologous end joining [GO:0006303]; embryonic organ development [GO:0048568]; interstrand cross-link repair [GO:0036297]; isotype switching [GO:0045190]; male gonad development [GO:0008584]; meiotic mismatch repair [GO:0000710]; mitotic recombination [GO:0006312]; multicellular organism aging [GO:0010259]; multicellular organism growth [GO:0035264]; negative regulation of protection from non-homologous end joining at telomere [GO:1905765]; negative regulation of telomere maintenance [GO:0032205]; nucleotide-excision repair [GO:0006289]; nucleotide-excision repair, DNA incision [GO:0033683]; nucleotide-excision repair, DNA incision, 3'-to lesion [GO:0006295]; nucleotide-excision repair, DNA incision, 5'-to lesion [GO:0006296]; oogenesis [GO:0048477]; positive regulation of t-circle formation [GO:1904431]; positive regulation of transcription initiation from RNA polymerase II promoter [GO:0060261]; post-embryonic hemopoiesis [GO:0035166]; pyrimidine dimer repair by nucleotide-excision repair [GO:0000720]; replicative senescence [GO:0090399]; response to cadmium ion [GO:0046686]; response to immobilization stress [GO:0035902]; response to nutrient [GO:0007584]; response to oxidative stress [GO:0006979]; response to sucrose [GO:0009744]; response to X-ray [GO:0010165]; spermatogenesis [GO:0007283]; syncytium formation [GO:0006949]; t-circle formation [GO:0090656]; telomeric DNA-containing double minutes formation [GO:0061819]; UV protection [GO:0009650]; UV-damage excision repair [GO:0070914] | 11062157_Observational study of genotype prevalence. (HuGE Navigator) 11936875_Lactacystin enhances cisplatin sensitivity in resistant human ovarian cancer cell lines via inhibition of DNA repair and ERCC-1 expression. 12164325_Observational study of genotype prevalence. (HuGE Navigator) 12189194_Inter-individual variation, seasonal variation and close correlation of OGG1 and ERCC1 mRNA levels in full blood 12220217_Observational study of gene-disease association. (HuGE Navigator) 12643788_Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 and ERCC1 in DU145 and LNCaP prostate carcinoma through MAPK signaling. 12645871_The results show a significant association between BCC and the A-allele of a polymorphism in ERCCI exon4 (Odds ratio 12;95% Confidence Interval 1.17-124; p(chi2, two-side) = 0.019) and to a lesser extent with XPD exon6 (p = 0.06). 12800797_ERCC1 gene is a useful independent prognostic tumor marker in patients with early stage non-small-cell lung cancer. 12865926_Observational study of gene-disease association. (HuGE Navigator) 12869411_Observational study of gene-disease association. (HuGE Navigator) 14690602_Data reveal an unanticipated involvement of the ERCC1/XPF NER endonuclease in the regulation of telomere integrity. 14734547_the ternary complex of hRad52 and XPF/ERCC1 is the active species that processes recombination intermediates generated during the repair of DNA double strand breaks and in homology-dependent gene targeting events. 15095299_XPA, ERCC1 and XPF DNA repair protein expression is reduced in testis neoplasms 15140544_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15173087_ERCC1 has a role as a nucleotide excision repair protein involved in the repair of therapeutic radiation-induced DNA damage 15213713_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15277258_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15297394_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15297394_the ERCC1 C8092A polymorphism may have a role in progression of non-small cell lung cancer 15688021_This alternate transcript is ubiquitous in human tissues and cancer cell lines, but is absent in mouse and thus does not appear to be cancer related. 15709194_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15734977_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15746040_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15746057_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15788669_High ERCC1 gene expression is associated with chemotherapy resistance in esophageal cancer 15849729_Observational study of gene-disease association. (HuGE Navigator) 15885892_Observational study of gene-disease association. (HuGE Navigator) 15922480_Overexpression of ERCC1 is associated with liver fibrogenesis and cancer and could be related to the well recognized resistance of HCC to chemotherapeutics. 15932882_The XPF binding sites of ERCC1 were located in helices H1 and H3 and in the C-terminal region, similar to the involved surface of XPF. 15936590_Observational study of gene-disease association. (HuGE Navigator) 15958648_Observational study of gene-disease association. (HuGE Navigator) 16034668_A nuclear magnetic resonance examination of the DNA binding site. 16076955_XPF-ERCC1 recognizes a branched DNA substrate by binding the two ssDNA arms with the two HhH2 domains of XPF and ERCC1 and by binding the 5'-ssDNA arm with the central domain of ERCC1. 16144923_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16195237_Observational study of gene-disease association. (HuGE Navigator) 16212814_Observational study of gene-disease association. (HuGE Navigator) 16224397_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16284373_Observational study of gene-disease association. (HuGE Navigator) 16284380_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16308313_Observational study of gene-disease association. (HuGE Navigator) 16315315_The half-life of ERCC1 protein in a testis tumor cell line was not significantly different to that in a prostate cancer cell line. These results suggest that constitutive levels of these DNA repair proteins are controlled at the level of translation. 16338413_The ERCC1 domain folds properly only in the presence of the XPF domain, which implies a role for XPF as a scaffold for the folding of ERCC1. 16341770_Low variant A-allele frequency of ERCC1 G19007A in the Chinese population may suggest that the genetic contribution to cancer risk differs substantially between ethnic groups. 16351803_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16364765_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16393248_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16407418_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16507781_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16510122_Observational study of gene-disease association. (HuGE Navigator) 16537713_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16609022_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16622263_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16690207_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16723154_These results suggest that the ERCC1 8092C>A polymorphism may be related to the occurrence of childhood ALL in a Chinese population. 16817948_Observational study of gene-disease association. (HuGE Navigator) 16819291_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16875718_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16896002_Observational study of gene-disease association. (HuGE Navigator) 16899630_The conflicting information regarding the ERCC1 codon 118 polymorphism might be explained by the coexistence of major confounding factors(smoking or diet), which might mask the relatively minor gene effect associated with a single genetic polymorphism. 16979838_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16985021_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17064812_ERCC1 expression may results in chemoresistance. 17078101_Observational study of gene-disease association. (HuGE Navigator) 17131345_Observational study of gene-disease association. (HuGE Navigator) 17151930_data indicated that ERCC1 mRNA level is associated with CDDP, BCNU & VCR cytotoxicity, while ERCC2 expression is associated with BCNU cytotoxicity in gliomas; however, there was no association between ERCC1, ERCC2 SNP and chemotherapy drug response 17197435_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17210993_Observational study of gene-disease association. (HuGE Navigator) 17222938_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17229776_results of the study indicate that ERCC1 may predict survival in bladder cancer treated by platinum-based therapy 17273966_ERCC1 deficiency is associated with cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure 17299578_Meta-analysis of gene-disease association. (HuGE Navigator) 17303907_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17313739_Observational study of gene-disease association. (HuGE Navigator) 17314486_Observational study of gene-disease association. (HuGE Navigator) 17314486_Our findings suggest that the C19007T ERCC1 polymorphism is unlikely to play an important role in epithelial ovarian or endometrial cancer in Korean women. 17401013_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17438655_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17502833_Observational study of gene-disease association. (HuGE Navigator) 17502833_variant genotypes of the rs3212948 C allele were associated with significantly decreased risk of lung cancer 17522621_Meta-analysis of gene-disease association. (HuGE Navigator) 17522621_meta-analysis suggests that the ERCC1 C8092A and T19007C polymorphisms had no association to cancer risk 17534174_the benefit of adjuvant cisplatin-based chemotherapy was more profound in patients with low ERCC1 expression. 17549067_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17606717_ERCC1 has a role in head and neck squamous cell carcinomaresponse to Cisplatin-based induction chemotherapy 17617021_Observational study of gene-disease association. (HuGE Navigator) 17685459_Observational study of gene-disease association. (HuGE Navigator) 17720715_Identification of two distinct interaction surfaces of ERCC1 that mediate XPA and DNA binding. 17912758_The XPF HhH homodimer has a larger interaction interface, aromatic stacking interactions, and additional hydrogen bond contacts as compared to the XPF/ERCC1 HhH complex, which accounts for its higher stability 17925548_Observational study of gene-disease association. (HuGE Navigator) 17932351_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17961161_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17961161_Patients with ERCC1-negative tumors appear to have significantly better response to platinum-based chemotherapy compared to patients with ERCC1-positive tumors;the TT genotype seems to be favorable toward better response to platinum-based chemotherapy. 17987380_ERCC1 expression could be useful in identifying patients who may benefit from platinum-containing treatments. 18006494_These results implicate the XPF-ERCC1 complex in initiating interstrand cross-links (ICL) repair by unhooking the ICL, which simultaneously induces a double strand break at a stalled fork. 18024864_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18026184_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18036700_ERCC1 expression was identified as a positive prognostic marker in resected non-small cell lung cancer 18081788_Upregulation of ERCC1 and XPD protein expression was associated with resistance process to platinum-based chemotherapy in advanced epithelial ovarian cancer 18204222_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18204222_in our study ERCC1 was not associated with any clinical characteristic of colorectal cancer 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18289367_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18289367_The previously identified haplotype of ERCC1 and its present polymorphisms are not associated with an increased risk for colorectal cancer. 18332046_Observational study of gene-disease association. (HuGE Navigator) 18332046_Significantly lower ERCC1 expression is associated with prostate cancer 18347182_Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. 18347182_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18358500_Observational study of gene-disease association. (HuGE Navigator) 18386788_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18395930_loss of ERCC1 and class III beta-tubulin protein expression were predictors of better survival in non-small-cell lung cancer patients who received a platinum-based plus taxane chemotherapy. 18396111_To determine whether a human flap endonuclease could recognize and process this potential intermediate, the genetic requirement for the ERCC1/XPF heterodimer during LINE-1 retrotransposition was characterized 18425336_ERCC1 polymorphism may be a useful prognostic marker in patients with pancreatic cancer treated with platinum-based chemotherapy. 18430179_demonstrates increased survival for neoadjuvant treated esophageal adenocarcinoma with TS, ERCC1, or GSTP-1 mRNA level at or below the median 18451256_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18451256_study identified 2 functional variants in the ERCC1 5'-flanking region, -433T>C & 262G>T, which cooperatively influence transcriptional regulation of ERCC1; the -433C & 262G alleles are associated with susceptibility to lung cancer 18478337_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18478337_Polymorphisms of ERCC1 are associated with esophageal adenocarcinoma risk. 18483312_Observational study of genotype prevalence. (HuGE Navigator) 18494946_Significantly higher ERCC1 mRNA expression was found in SCLC compared weith NSCLC. There was no correlation between mRNA expression of the ERCC1 gene and chemosensitivity to cisplatin, carboplatin, or gemcitabine. 18505590_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18509181_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18520795_ERCC1 was confirmed to be an independent prognostic factor for survival in LD patients. No significant role was found for ERCC1, RRM1 and TopoIIalpha in ED patients. 18564169_expression of ERCC1 to b-actin was significantly higher (P < 0.01) in ZNRD1-espressing cells 18594541_ERCC1 expression might be a predictive marker of survival in locally advanced SCCHN in patients treated with cisplatin-based CCRT. 18604718_High expression of ERCC1, FLT1, NME4 and PCNA in myelodysplastic syndrome was associated with poor prognosis and disease progression along the sequence of 5q-syndrome/RCMD/RAEB/de novo AML 18615480_ERCC1 polymorphisms are associated with nasopharyngeal carcinoma. 18615480_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18616887_ERCC1 and ERCC2 polymorphisms may have a role in idiopathic azoospermia in a Chinese population 18616887_Observational study of gene-disease association. (HuGE Navigator) 18623378_ERCC1 overexpression is associated with resistance to platinum based chemotherapy in patients with nonsmall-cell lung cancer 18635523_Haplotypes of ERCC1 are significantly associated with high levels of DNA adducts compared with their most common haplotypes in normal human lymphocytes. 18635523_Observational study of gene-disease association. (HuGE Navigator) 18640939_Although the ERCC1 codon 118 polymorphism does not seem to be associated with clinical outcome, the C8092A polymorphism was an independent predictor of survivalin women with optimally resected stage III epithelial ovarian cancer 18672388_Following ERCC1 depletion, FANCD2-Ub formation is reduced and FANCD2 foci are eliminated. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18683754_By down-regulating ERCC1, BRCA1, hMLH1 genes, blocking G0/G1 phase, and increasing apoptosis rate, mifepristone could enhance anti-tumor effect of cisplatin. 18756932_ERCC1 gene is associated with the resistance to cisplatin, and the sensitivity to cisplatin can be enhanced by RNA interfering ERCC1 in ovarian cancer. 18797464_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18823676_High ERCC1 expression was associated with drug resistance to platinum-based chemotherapy in non-small-cell lung cancer. 18830263_Observational study of gene-disease association. (HuGE Navigator) 18854777_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18977553_the level of ERCC1 expression may predict survival in NSCLC patients receiving carboplatin and paclitaxel 18990028_Observational study of gene-disease association. (HuGE Navigator) 18990748_Meta-analysis of gene-disease association. (HuGE Navigator) 19009659_ERCC1 polymorphism may be a useful predictive parameter for the relapse and survival of gastric cancer patients receiving oxaliplatin-based adjuvant chemotherapy. 19009659_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19020759_Measurement of TS and ERCC1 mRNA levels in primary colorectal cancer can predict those in synchronous liver metastases, but not in metachronous ones. 19029193_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19035454_DNA repair proteins BRCA1 and ERCC1 have roles in inhibiting progression of ovarian cancer 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19068092_ERCC1 codon 118 C-->T polymorphism is associated with FOLFOX-4 treatment in metastatic colorectal carcinoma. 19068092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19074750_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19116388_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19124499_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19124499_The SNP of ERCC1 3' untranslated region (UTR) showed a significant association with glioma risk. 19150580_high expression is an independent prognostic factor for poor overall survival in advanced non-small cell lung cancer patients 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19173905_Decreased expression and mutation of ERCC1 gene may be a possible carcinogenic mechanism of CdCl2. 19194123_ERCC1 positives combined with thymidylate synthase, and glutathione S-transferase pi positives are associated with drug resistance in advanced colorectal cancer. 19203783_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19235214_lower levels of ERCC1 were found in patients with early aseptic loosening compared to patients with aseptic loosening later than 10 years 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19240185_ERCC1 expression is the first factor for predicting survival in adrenocortical carcinoma patients treated with platinum-based chemotherapy. 19247656_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19253035_platinum-based chemotherapy affects the expression of tissue marker (ERCC1)in lung cancer 19257887_Observational study of gene-disease association. (HuGE Navigator) 19258314_a model for the mechanism of ICL repair in mammalian cells that implicates the DNA glycosylase activity of NEIL1 downstream of XPF/ERCC1 and translesion DNA synthesis repair steps. 19270000_Observational study of gene-disease association. (HuGE Navigator) 19289372_examined ERCC1, RRM1, XPG, and BRCA1 mRNA levels by real-time quantitative polymerase chain reaction in 54 patients with stage IB-IIB resected NSCLC. 19297315_ERCC1 and XPF are correlated and expressed at a higher level in NS(Never Smoking) compared with ES(Ever Smoking) 19318434_Single nucleotide polymorphisms in ERCC2, ERCC1, and GLTSCR1 are associated with a 23% reduction in glioblastoma risk. 19332728_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19339270_Observational study of gene-disease association. (HuGE Navigator) 19361884_Observational study of gene-disease association. (HuGE Navigator) 19361884_The ERCC1 C8092A polymorphism may influence the non-small cell lung cancer prognosis regardless of the ERCC1 protein expression and platinum sensitivity. 19362955_ERCC1 gene polymorphisms is associated with favorable clinical outcome in advanced non-small-cell lung cancer. 19362955_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19421825_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19421825_Single-nucleotide polymorphisms of ERCC1 and XRCC1 could be applied to further individualize treatment strategies. 19434073_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19470925_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19488864_Tumor ERCC1 expression is associated with tumor response and patient's survival in Chinese advanced non-small-cell lung cancer patients treated with platinum compounds. 19513514_Examine role of ERCC1 polymorphisms in patient response to gastric cancers treated with FOLFOX or FOLFIRI. 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19538866_Intratumoral ERCC1 mRNA expression is an independent predictive marker for survival of the patients with non-small cell lung cancer receiving platinum-based chemotherapy. 19543324_Data show that the combination of RRM1 and ERCC1 expression is prognostic in pancreatic cancer patients after a complete resection. 19549713_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19549713_No differences in survival or response rates of patients with recurrent Nasopharyngeal carcinoma treated with the GEMOX regimen were found between ERCC1 genotypes. 19552012_The analysis of ERCC1 is useful to individualize therapy accordingly to individual levels which are modified by genetic mutations, and Polymorphisms in codons 118 C/T and C8092A, seem to influence the carcinogenesis, cytostatic resistance, survival. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19574766_Overexpression of ERCC1 is associated with platinum resistance in epithelial ovarian cancer. 19580345_maintained expression and even up-regulation of some (PNPT1, PMPCB, HMMR/RHAMM, BSG and ERCC1) tumor associated antigens in CD40-activated leukemic cells. 19620936_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19626585_aberrant CpG island methylation in ERCC1 promoter region exists in human glioma cell lines as well as clinical glioma samples 19636001_Observational study of genotype prevalence, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19661089_Observational study of gene-disease association. (HuGE Navigator) 19661343_Elevated ERCC1 gene expression in blood cells associated with exposure to arsenic from drinking water. 19664717_methylation status of ERCC1 is associated with radiosensitivity in glioma cell lines 19667277_Data show that women who were ERCC1 negative did not have a survival advantage over ERCC1-positive women. 19672255_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19687761_overexpression of ERCC1 is associated with brain and adrenal metastases in non-small cell lung cancer. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 19786980_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19789190_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19805513_Data demonstrate that the unhooking of an interstrand cross-link by XPF-ERCC1 is necessary for the stable localization of FANCD2 to the chromatin and subsequent homologous recombination-mediated DSB repair. 19822419_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19832035_Results indicate that ERCC1 exon VIII alternative splicing does exist in some ovarian cancer cell lines, and regulates cisplatin-resistance in ovarian cancer cells. 19850635_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19875192_mRNA expression predicts overall survival in patients with non-small cell lung cancer who received neoadjuvant cisplatin-based chemotherapy 19878615_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19922504_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19940136_Data show that mutation of two conserved residues (Asn-110 and Tyr-145) located in the XPA-binding site of ERCC1 dramatically affected NER. 19956886_ERCC1 and XPF are upregulated during testicular germ cell tumor progression 19960344_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20003391_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20003391_Study aimed to investigate the association between the ERCC2 751, 312 and ERCC1 118 polymorphisms and the risk of lung adenocarcinoma in Chinese non-smoking females. 20003463_Genetic polymorphisms in ERCC1 and XRCC1 genes might be prognostic factors in non-smoking female patients with lung adenocarcinoma. 20003463_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20009541_Data suggested that ERCC-1 was involved in the resistance of cetuximab combined with DDP. 20021611_determination of the expression of excision repair cross-complementation group 1 and class IIIbeta tubulin is useful to predict the effects of platinum-based anticancer drugs. 20047592_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20061190_A single SNP analysis showed a significantly increased risk given by XRCC1-rs915927 and a protective effect of the rare alleles of 3 ERCC1 SNPs: rs967591, rs735482 and rs2336219. 20061190_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20070981_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20070981_Two polymorphisms, C8092A and C118T, affected the prognosis of non-small-cell lung cancer patients who received adjuvant and/or neoadjuvant platinum-based chemotherapy. 20085902_Tau expression but not excision repair cross-complementing 1 in advanced breast cancer predicts poor response to combination chemotherapy of paclitaxel and cisplatin. 20104194_High expression of ERCC1 in typical carcinoid is associated with the failure of platinum-based therapy. 20143185_Platinum-based chemotherapy sensitivity was significantly associated with polymorphism of ERCC1 C118T. 20189873_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20189873_There was no significant association between the ERCC1 C118T polymorphism and treatment response in advanced NSCLC patients. 20211060_Compared with their parental cells, the expressions of CDA, RRM1, PTEN and ERCC1 increase in human gemcitabine-resistant non-small cell lung cancer cell lines. 20216541_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20221251_This demonstrates that at least part of the DNA repair defect and symptoms associated with mutations in XPF are due to mislocalization of XPF-ERCC1 into the cytoplasm of cells. 20232390_Observational study of gene-disease association. (HuGE Navigator) 20331623_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20332140_High ERCC1 is associated with non-small-cell lung cancer. 20351547_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20351547_The C->T change of ERCC1 Asn118Asn polymorphism and the C->A change of ERCC1 Gln504Lys polymorphism have statistically significant association with elevated or descendent platinum-based chemotherapy response respectively. 20354815_Observational study of gene-disease association. (HuGE Navigator) 20354815_results suggest that polymorphism Asn118Asn in ERCC1, A67T in iASPP and Asn148Glu in APE1 may associated with early onset of lung cancer as well as some specific subtype of lung cancer 20368715_Observational study of gene-disease association. (HuGE Navigator) 20378615_Observational study of gene-disease association. (HuGE Navigator) 20385995_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20391138_Observational study of gene-disease association. (HuGE Navigator) 20395129_Numerically more toxicity was observed in the entire population of ERCC1-negative tumours and reached significance in patients with adenocarcinomas regarding leukopenia (P=0.015), nausea/vomiting (P=0.040) and neurotoxicity (P=0.037). 20429839_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20462983_Data show that overall survival was significantly improved in the patients with ERCC1 118 T/T or C/T treated by platinum-based chemotherapy. 20462983_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20467918_ERCC1, RRM1 and BRCA1 are promising predictive and prognostic biomarkers in advanced non-small cell lung cancer. 20470393_Over-expression of Fas reverses drug resistance of H446/CDDP cells, possibly due to the increased cell sensitivity to apoptosis and the decreased expressions of GST-pi and ERCC1 20495366_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20504223_High level of ERCC1 mRNA may serve as a useful prognostic factor for poor outcome in advanced NSCLC patients treated w | ENSMUSG00000003549 | Ercc1 | 2.956345e+03 | 1.1762448 | 0.234188372 | 0.3209743 | 5.278239e-01 | 0.4675231830 | 0.86798431 | No | Yes | 3.145844e+03 | 412.768626 | 2.201275e+03 | 296.559999 | |
| ENSG00000012211 | 4007 | PRICKLE3 | protein_coding | O43900 | FUNCTION: Involved in the planar cell polarity (PCP) pathway that is essential for the polarization of epithelial cells during morphogenetic processes, including gastrulation and neurulation (By similarity). PCP is maintained by two molecular modules, the global and the core modules, PRICKLE3 being part of the core module (By similarity). Distinct complexes of the core module segregate to opposite sides of the cell, where they interact with the opposite complex in the neighboring cell at or near the adherents junctions (By similarity). Involved in the organization of the basal body (By similarity). Involved in cilia growth and positioning (By similarity). Required for proper assembly, stability, and function of mitochondrial membrane ATP synthase (mitochondrial complex V) (PubMed:32516135). {ECO:0000250|UniProtKB:A8WH69, ECO:0000269|PubMed:32516135}. | Alternative splicing;Cell membrane;Cilium biogenesis/degradation;Cytoplasm;Developmental protein;Disease variant;LIM domain;Leber hereditary optic neuropathy;Membrane;Metal-binding;Mitochondrion;Phosphoprotein;Primary mitochondrial disease;Reference proteome;Repeat;Zinc | LIM domain only 6 is a three LIM domain-containing protein. The LIM domain is a cysteine-rich sequence motif that binds zinc atoms to form a specific protein-binding interface for protein-protein interactions. [provided by RefSeq, Jul 2008]. | hsa:4007; | mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; zinc ion binding [GO:0008270]; cell projection organization [GO:0030030] | 32516135_PRICKLE3 linked to ATPase biogenesis manifested Leber's hereditary optic neuropathy. | ENSMUSG00000031145 | Prickle3 | 6.569277e+02 | 1.1993130 | 0.262208207 | 0.3113028 | 6.983429e-01 | 0.4033410670 | 0.84651196 | No | Yes | 6.848587e+02 | 87.934562 | 5.253305e+02 | 69.748485 | |
| ENSG00000013573 | 1663 | DDX11 | protein_coding | Q96FC9 | FUNCTION: DNA-dependent ATPase and ATP-dependent DNA helicase that participates in various functions in genomic stability, including DNA replication, DNA repair and heterochromatin organization as well as in ribosomal RNA synthesis (PubMed:10648783, PubMed:21854770, PubMed:23797032, PubMed:26089203, PubMed:26503245). Its double-stranded DNA helicase activity requires either a minimal 5'-single-stranded tail length of approximately 15 nt (flap substrates) or 10 nt length single-stranded gapped DNA substrates of a partial duplex DNA structure for helicase loading and translocation along DNA in a 5' to 3' direction (PubMed:18499658, PubMed:22102414). The helicase activity is capable of displacing duplex regions up to 100 bp, which can be extended up to 500 bp by the replication protein A (RPA) or the cohesion CTF18-replication factor C (Ctf18-RFC) complex activities (PubMed:18499658). Shows also ATPase- and helicase activities on substrates that mimic key DNA intermediates of replication, repair and homologous recombination reactions, including forked duplex, anti-parallel G-quadruplex and three-stranded D-loop DNA molecules (PubMed:22102414, PubMed:26503245). Plays a role in DNA double-strand break (DSB) repair at the DNA replication fork during DNA replication recovery from DNA damage (PubMed:23797032). Recruited with TIMELESS factor upon DNA-replication stress response at DNA replication fork to preserve replication fork progression, and hence ensure DNA replication fidelity (PubMed:26503245). Cooperates also with TIMELESS factor during DNA replication to regulate proper sister chromatid cohesion and mitotic chromosome segregation (PubMed:17105772, PubMed:18499658, PubMed:20124417, PubMed:23116066, PubMed:23797032). Stimulates 5'-single-stranded DNA flap endonuclease activity of FEN1 in an ATP- and helicase-independent manner; and hence it may contribute in Okazaki fragment processing at DNA replication fork during lagging strand DNA synthesis (PubMed:18499658). Its ability to function at DNA replication fork is modulated by its binding to long non-coding RNA (lncRNA) cohesion regulator non-coding RNA DDX11-AS1/CONCR, which is able to increase both DDX11 ATPase activity and binding to DNA replicating regions (PubMed:27477908). Plays also a role in heterochromatin organization (PubMed:21854770). Involved in rRNA transcription activation through binding to active hypomethylated rDNA gene loci by recruiting UBTF and the RNA polymerase Pol I transcriptional machinery (PubMed:26089203). Plays a role in embryonic development and prevention of aneuploidy (By similarity). Involved in melanoma cell proliferation and survival (PubMed:23116066). Associates with chromatin at DNA replication fork regions (PubMed:27477908). Binds to single- and double-stranded DNAs (PubMed:9013641, PubMed:18499658, PubMed:22102414). {ECO:0000250|UniProtKB:Q6AXC6, ECO:0000269|PubMed:10648783, ECO:0000269|PubMed:17105772, ECO:0000269|PubMed:18499658, ECO:0000269|PubMed:20124417, ECO:0000269|PubMed:21854770, ECO:0000269|PubMed:22102414, ECO:0000269|PubMed:23116066, ECO:0000269|PubMed:23797032, ECO:0000269|PubMed:26089203, ECO:0000269|PubMed:26503245, ECO:0000269|PubMed:27477908}.; FUNCTION: (Microbial infection) Required for bovine papillomavirus type 1 regulatory protein E2 loading onto mitotic chromosomes during DNA replication for the viral genome to be maintained and segregated. {ECO:0000269|PubMed:17189189}. | 4Fe-4S;ATP-binding;Activator;Alternative splicing;Chromosome;Cytoplasm;Cytoskeleton;DNA damage;DNA repair;DNA replication;DNA-binding;Developmental protein;Disease variant;Helicase;Host-virus interaction;Hydrolase;Iron;Iron-sulfur;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transcription;Transcription regulation | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which is an enzyme that possesses both ATPase and DNA helicase activities. This gene is a homolog of the yeast CHL1 gene, and may function to maintain chromosome transmission fidelity and genome stability. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jul 2008]. | hsa:1663; | centrosome [GO:0005813]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; fibrillar center [GO:0001650]; midbody [GO:0030496]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; spindle pole [GO:0000922]; 4 iron, 4 sulfur cluster binding [GO:0051539]; 5'-3' DNA helicase activity [GO:0043139]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; ATP-dependent activity, acting on DNA [GO:0008094]; ATP-dependent activity, acting on RNA [GO:0008186]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; DNA replication origin binding [GO:0003688]; double-stranded DNA binding [GO:0003690]; G-quadruplex DNA binding [GO:0051880]; helicase activity [GO:0004386]; metal ion binding [GO:0046872]; single-stranded DNA binding [GO:0003697]; single-stranded RNA binding [GO:0003727]; triplex DNA binding [GO:0045142]; cellular response to bleomycin [GO:1904976]; cellular response to cisplatin [GO:0072719]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to hydroxyurea [GO:0072711]; DNA duplex unwinding [GO:0032508]; DNA repair [GO:0006281]; establishment of sister chromatid cohesion [GO:0034085]; G-quadruplex DNA unwinding [GO:0044806]; negative regulation of protein binding [GO:0032091]; nucleolar chromatin organization [GO:1990700]; positive regulation of chromatin binding [GO:0035563]; positive regulation of double-strand break repair [GO:2000781]; positive regulation of endodeoxyribonuclease activity [GO:0032079]; positive regulation of sister chromatid cohesion [GO:0045876]; positive regulation of transcription of nucleolar large rRNA by RNA polymerase I [GO:1901838]; replication fork processing [GO:0031297]; sister chromatid cohesion [GO:0007062] | 15520935_We present preliminary analysis of a strong candidate gene in the region, the DNA helicase DDX11. In conclusion, we report mapping of the first locus that determines mean telomere length in humans. 17105772_human ChlR1 is required for sister chromatid cohesion and, hence, normal mitotic progression. 17189189_These studies provide compelling evidence that ChlR1 association is required for loading the papillomavirus E2 protein onto mitotic chromosomes and represents a kinetochore-independent mechanism for viral genome maintenance and segregation. 18499658_hChlR1 has a role in the establishment of sister chromatid cohesion, with Ctf18-RFC and Fen1 20124417_Timeless associates with the cohesion-promoting DNA helicase ChlR1, which, when overexpressed, partially alleviates the cohesion defect of cells depleted of Timeless-Tipin. 21489590_These data provide evidence that the association of bovine papillomavirus E2 with human ChlR1 contributes to a loading mechanism during DNA replication rather than direct tethering during mitotic division. 21854770_The ChlR1 is involved in the proper formation of heterochromatin, which in turn contributes to global nuclear organization and pleiotropic effects. 22102414_Wild-type ChlR1 required a minimal 5' single-stranded DNA tail of 15 nucleotides to efficiently unwind a simple duplex DNA substrate. 22678773_The results identify novel roles of Ddx11 during embryo morphogenesis and demonstrate that the activity of its motif V is essential for DDX11 function. 23033317_Data indicate a homozygous mutation (c.788G>A [p.R263Q]) in DDX11 in three affected siblings with severe intellectual disability and many of the congenital abnormalities reported in the Warsaw breakage syndrome (WABS) original case. 23116066_helicase DDX11 is expressed at high levels in primary and metastatic melanoma, and that interfering with its expression leads to severe chromosome segregation defects, telomere shortening, and massive melanoma cell apoptosis. 24487782_ChlR1 plays a critically important role in cellular replication and/or DNA repair. [review] 25561740_A distinct triplex DNA unwinding activity of ChlR1 helicase. 26032365_In this study, we found that cohesion establishment factors, like CHlR1, cooperatively stimulate endonuclease activity of hFen1 in in vivo mimic condition, including replication protein-A-coated DNA and high salt. 26089203_our results indicate that DDX11 functions as a positive regulator of rRNA transcription and provides a novel insight into the pathogenesis of WABS. 26474416_Q Motif Is Involved in DNA Binding but Not ATP Binding in ChlR1 Helicase 26503245_DDX11 and Tim proteins physically and functionally interact and act in concert to preserve replication fork progression in perturbed conditions. 27795438_A mutation has been identified in HPV16 E2 that abrogates interaction with ChlR1, and it was shown that ChlR1 regulates the chromatin association of HPV16 E2 and that this virus-host interaction is essential for viral episome maintenance. 28960803_We present two new cases of Warsaw Breakage Syndrome (WABS), an autosomal recessive cohesinopathy, in sisters aged 13 and 11 years who both had compound heterozygous mutations in DDX11 30061412_DDX11 orchestrates jointly with 9-1-1 and its loader, RAD17, DNA damage tolerance at sites of bulky lesions, and endogenous abasic sites. These functions may explain the essential roles of DDX11 and its similarity with 9-1-1 during development. 30303954_Results indicate a role for the DDX11-Timeless interaction in coordinating DNA replication with sister chromatid cohesion, and suggest implications for understanding the molecular basis of Warsaw breakage syndrome (WABS). 31097223_The DDX11-AS1 may be a novel oncogene in hepatocarcinogenesis by repressing LATS2, providing a potential therapeutic target for hepatocellular carcinoma treatment. 31935221_DDX11, ESCO1 and ESCO2 control different fractions of cohesin that are spatially and mechanistically separated. 32014424_Final rescue tests in vitro and in vivo further elucidated that DDX11 overexpression could reversed the DDX11-AS1 downregulation-mediated effect on osteosarcoma progression 32071282_The iron-sulfur helicase DDX11 promotes the generation of single-stranded DNA for CHK1 activation. 32271422_The long non-coding RNA DDX11-AS1 facilitates cell progression and oxaliplatin resistance via regulating miR-326/IRS1 axis in gastric cancer. 32332880_E2F1 mediated DDX11 transcriptional activation promotes hepatocellular carcinoma progression through PI3K/AKT/mTOR pathway. 32422563_DDX11-AS1exacerbates bladder cancer progression by enhancing CDK6 expression via suppressing miR-499b-5p. 32705708_Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication. 32772230_LncRNA DDX11-AS1: a novel oncogene in human cancer. 32855419_Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. 33591602_Finding underlying genetic mechanisms of two patients with autism spectrum disorder carrying familial apparently balanced chromosomal translocations. 34299244_Expression of DDX11 and DNM1L at the 12p11 Locus Modulates Systemic Lupus Erythematosus Susceptibility. | ENSMUSG00000035842 | Ddx11 | 7.266455e+03 | 0.7854853 | -0.348343889 | 0.2573871 | 1.829255e+00 | 0.1762156250 | 0.77663851 | No | Yes | 4.710619e+03 | 688.055335 | 6.900779e+03 | 1033.414867 | |
| ENSG00000013588 | 9052 | GPRC5A | protein_coding | Q8NFJ5 | FUNCTION: Orphan receptor. Could be involved in modulating differentiation and maintaining homeostasis of epithelial cells. This retinoic acid-inducible GPCR provide evidence for a possible interaction between retinoid and G-protein signaling pathways. Functions as a negative modulator of EGFR signaling (By similarity). May act as a lung tumor suppressor (PubMed:18000218). {ECO:0000250|UniProtKB:Q8BHL4, ECO:0000269|PubMed:18000218}. | Cell membrane;Cytoplasmic vesicle;G-protein coupled receptor;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Transducer;Transmembrane;Transmembrane helix;Tumor suppressor | This gene encodes a member of the type 3 G protein-coupling receptor family, characterized by the signature 7-transmembrane domain motif. The encoded protein may be involved in interaction between retinoid acid and G protein signalling pathways. Retinoic acid plays a critical role in development, cellular growth, and differentiation. This gene may play a role in embryonic development and epithelial cell differentiation. [provided by RefSeq, Jul 2008]. | hsa:9052; | cytoplasmic vesicle membrane [GO:0030659]; extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; nucleolus [GO:0005730]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; vesicle [GO:0031982]; cadherin binding [GO:0045296]; G protein-coupled receptor activity [GO:0004930]; protein kinase activator activity [GO:0030295]; negative regulation of epidermal growth factor-activated receptor activity [GO:0007175]; signal transduction [GO:0007165] | 15659406_RAI3 is a cell growth-promoting gene and a novel P53 transcriptional target 17055459_This evidence explains one of the mechanisms of the GPRC5A-regulated cell growth in some cancer cell lines. 18000218_Gprc5a functions as a tumor suppressor in mouse lung, and human GPRC5A may share this property. 19279407_Mechanisms underlying the induction of the putative human tumor suppressor GPRC5A are reported. 19552806_analysis of RAI3 expression in normal and cancerous human breast tissue at both the mRNA and protein levels 20563252_Loss of GPRC5A is associated with lung adenocarcinomas. 23154545_Decreased GPRC5A expression is associated with non-small cell lung cancers and lung inflammation. 23632812_RAI3 may contribute to the malignant progression of hepatocellular carcinoma 24470238_Data indicate that in mammary tumors, the mRNA expression of GPRC5A significantly correlated with that of BRCA1. 24984703_The interaction of miR-103a-3p with each of the two 5' UTR targets reduces the expression levels of both GPRC5A mRNA and GPRC5A protein in one normal epithelial and two pancreatic cancer cell lines. 25311788_EGFR interacted with GPRC5A and phosphorylated it in two conserved double-tyrosine motifs, Y317/Y320 and Y347/ Y350, at the C-terminal tail of GPRC5A. 25744720_Results show how GPRC5A deficiency leads to dysregulated EGFR and STAT3 signaling and lung tumorigenesis. 26227221_elevated levels of GPRC5A played significant roles in gastric cancer progression 27273304_under-expressed GPRC5A during lung tumorigenesis enhances any transcriptional stimulation through an active translational status. 27387124_Mutations of ARID1A, GPRC5A and MLL2 grant bladder cancer non-stem cells the capability of self-renewal. 27415424_GPRC5A is a potential oncogene in pancreatic ductal adenocarcinoma cells that is upregulated by gemcitabine with help from HuR. 27599526_our results implicate GPRC5A as a tumor suppressor in breast cancer cells, and GPRC5A exerts its tumor-suppressive function by inhibiting EGFR and its downstream pathway 27715394_findings reveal an unprecedented role for GPRC5A in regulation of the ITGB1-mediated cell adhesion and it's downstream signaling, thus indicating a potential novel role for GPRC5A in human epithelial cancers. 28114355_Suppression of GPRC5a results in decreased cell growth, proliferation and migration in pancreatic cancer cell lines via a STAT3 modulated pathway, independent from ERK activation 28316092_Data show that G protein-coupled receptor, family C, group 5, member A protein (GPRC5A) regulates oxidative stress through vanin 1 protein (VNN1). 28653505_Study underscores genomic alterations that represent early events in the development of Kras mutant LUAD following Gprc5a loss and tobacco carcinogen exposure. 28849235_p53 overexpression and GPRC5A induction markedly inhibited tumor cell viability and induced apoptosis. 28870805_These results suggest RAI3 plays an important role in adipogenesis of hASCs and may have a potential use in the future application. 29283424_miR-204 inhibits cell proliferation in gastric cancer by targeting CKS1B, CXCL1 and GPRC5A. 29636387_On the basis of our findings in human DN and in Gprc5a deficient mice, we believe that chronic reduction in Gprc5a expression plays a pathogenic role in the progression of DN. Results in cell culture support our other findings in mice showing that Gprc5a modulates EGFR and TGF-b signaling in podocytes. 29949874_GPRC5a was upregulated in pancreatic cancer (PaCa) leading to an enhanced drug resistance in PaCa cells. 30707896_RAI3 overexpression is associated with poor prognosis in esophageal cancers 30901718_The lung tumor suppressor gene GPRC5A played a protective role in cigarette smoke-induced lung tumor initiation. 31115523_Our findings demonstrated that miR342 regulates the cell proliferation of glioma by targeting GPRC5A, which indicates that miR342 is a target of interest regarding the treatment of refractory glioma, and it may provide a promising prognostic and therapeutic strategy for glioma treatment. 31276604_RNA-seq revealed that GPRC5A KO PC3 cells had dysregulated expression of cell cycle-related genes, leading to cell cycle arrest at the G2/M phase. Furthermore, the registered gene expression profile data set showed that the expression level of GPRC5A in original lesions of prostate cancer patients with bone metastasis was higher than that without bone metastasis. 32014253_RAI3 knockdown enhances osteogenic differentiation of bone marrow mesenchymal stem cells via STAT3 signaling pathway. 32115889_Adaptive RSK-EphA2-GPRC5A signaling switch triggers chemotherapy resistance in ovarian cancer. 32157214_Identification of Sca-1(+)Abcg1(+) bronchioalveolar epithelial cells as the origin of lung adenocarcinoma in Gprc5a-knockout mouse model through the interaction between lung progenitor AT2 and Lgr5 cells. 32410473_GPRC5a suppresses the proliferation of non-small cell lung cancer under wild type p53 background. 32719397_RhoA/C inhibits proliferation by inducing the synthesis of GPRC5A. 32758941_GPRC5A reduction contributes to pollutant benzo[a]pyrene injury via aggravating murine fibrosis, leading to poor prognosis of IIP patients. 33788883_Prognostic and clinicopathological significance of GPRC5A in various cancers: A systematic review and meta-analysis. 34350749_Knockdown of GPRC5A inhibits cell proliferation, migration and invasion in osteosarcoma. | ENSMUSG00000046733 | Gprc5a | 1.123381e+02 | 1.7573593 | 0.813409199 | 0.4447002 | 3.306796e+00 | 0.0689938734 | 0.71206859 | No | Yes | 1.033317e+02 | 38.134921 | 4.962617e+01 | 18.835678 | |
| ENSG00000015479 | 9782 | MATR3 | protein_coding | P43243 | FUNCTION: May play a role in transcription or may interact with other nuclear matrix proteins to form the internal fibrogranular network. In association with the SFPQ-NONO heteromer may play a role in nuclear retention of defective RNAs. Plays a role in the regulation of DNA virus-mediated innate immune response by assembling into the HDP-RNP complex, a complex that serves as a platform for IRF3 phosphorylation and subsequent innate immune response activation through the cGAS-STING pathway (PubMed:28712728). May bind to specific miRNA hairpins (PubMed:28431233). {ECO:0000269|PubMed:11525732, ECO:0000269|PubMed:28431233, ECO:0000269|PubMed:28712728}. | Acetylation;Alternative splicing;Amyotrophic lateral sclerosis;Direct protein sequencing;Disease variant;Immunity;Innate immunity;Isopeptide bond;Metal-binding;Neurodegeneration;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ubl conjugation;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA | This gene encodes a nuclear matrix protein, which is proposed to stabilize certain messenger RNA species. Mutations of this gene are associated with distal myopathy 2, which often includes vocal cord and pharyngeal weakness. Alternatively spliced transcript variants, including read-through transcripts composed of the upstream small nucleolar RNA host gene 4 (non-protein coding) and matrin 3 gene sequence, have been identified. Pseudogenes of this gene are located on chromosomes 1 and X. [provided by RefSeq, Aug 2013]. | hsa:9782; | membrane [GO:0016020]; nuclear inner membrane [GO:0005637]; nuclear matrix [GO:0016363]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; miRNA binding [GO:0035198]; RNA binding [GO:0003723]; structural molecule activity [GO:0005198]; zinc ion binding [GO:0008270]; activation of innate immune response [GO:0002218]; heart valve development [GO:0003170]; innate immune response [GO:0045087]; posttranscriptional regulation of gene expression [GO:0010608]; ventricular septum development [GO:0003281] | 12469345_Data show that the nuclear matrix protein matrin 3, cytoskeletal motor protein HMP, and the circadian clock protein lark were significantly decreased in fetal Down syndrome brain. 17658460_These results suggest that the functions of matrin 3 could be regulated by both Ca(2+)-dependent interaction with calmodulin and caspase-mediated cleavage. 18618731_the spatial proximities among a constellation of functionally related sites that are found within euchromatic regions of the cell nucleus including: HP1gamma, RNA polymerase II, matrin 3, and SAF-A sites 18676680_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19240791_Observational study of gene-disease association. (HuGE Navigator) 19344878_nonconservative S85C missense mutation in vocal cord and pharyngeal weakness with distal myopathy 19562669_study of the association of matr3 with chromosome territories and identification of potential interacting proteins 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19836620_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20421735_MATR3 to be a novel ATM target in response to DNA damage. 21182838_matrin 3 plays a significant role in controlling cell growth and proliferation 21683594_This study identified no pathogenic mutations in BAG3, MATR3, PTRF or TCAP in Australian muscular dystrophy. 21771346_MATR3 binds viral RNA and is required for the Rev/RRE mediated nuclear export of unspliced HIV-1 RNAs. 21771347_Matrin 3 binds Rev RNA to stabilize HIV-1 transcripts leading to increased cytoplasmic expression. 21771348_Rev needs MATR3 to promote the cytoplasmic accumulation and translation of unspliced RRE-containing mRNA. 21858232_Data suggest that the cellular level of MATR3, known to be highly regulated, modulates the stability of a group of gene transcripts. 24558381_the pY RNA1-s2/Matr3 interaction could play a role in vision 24686783_This study identified mutations in MATR3 in ALS kindreds. also observed MATR3 pathology in ALS-affected spinal cords with and without MATR3 mutations. 25154462_Sixteen patients from 6 families with late onset distal myopathy associated with the p.S85C MATR3 mutation were characterized. 25158920_Mutations in MATR3 are rare in French familial ALS and ALS with FTLD patients. 25523636_No mutations were identified, indicating that MATR3 mutations are not a common cause of Amyotrophic lateral sclerosis in Australian familial cases with predominately European ancestry 25574029_MATR3 gene disruption is associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. 25599992_Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. 25677933_This study shows a high risk of abnormal respiratory function with progressive worsening in MATR3 myopathy. 25771394_MATR3 mutation is identified to be a possible cause of amyotrophic lateral sclerosis. 25948554_Three-dimensional mapping of the lamin A-matrin-3 interface showed that the LMNA truncating mutation Delta303, which lacks the matrin-3 binding domain, was associated with an increased distance between lamin A and matrin-3. 26129669_Suppressing Matrin 3 powers a heightened and broader ZAP restriction of HIV-1 gene expression. 26493020_Its mutations strengthens the role of RNA metabolism in amyotrophic lateral sclerosis etiology. 26528920_Our findings indicate that mutations in Matrin 3 that are associated with ALS and myopathy do not dramatically alter the normal localization of the protein or readily induce inclusion formation. 26708275_It may not be a common genetic factor in Chinese amyotrophic lateral sclerosis patients. 26899464_A missense mutation in MATR3 was identified in myopathy patients undergoing a needle electromyography. 27731383_Depletion of SAFB1 reduced FUS's localization to chromatin-bound fraction and splicing activity, suggesting SAFB1 could tether FUS to chromatin compartment thorough N-terminal DNA-binding motif. Moreover, FUS interacts with another nuclear matrix-associated protein, Matrin3. 28580901_Using a novel RNA pulldown approach that utilized endogenous S1-tagged PINCR (a p53-regulated long noncoding RNA), the authors show that PINCR associates with the enhancer region of these genes by binding to RNA-binding protein Matrin 3 that, in turn, associates with p53. 28695676_Matrin3 physically interacts with intronic pyrimidine-rich sequences and controls alternative splicing. 29511296_The intra-nuclear localization and interaction network of MATR3 is strongly modulated by its RRM2 domain. 29763601_This might contribute to cellular damage and progression of dystrophy in muscle of Matrin-3 myopathy 30015619_cytoplasmic MATR3 redistribution mitigated neurodegeneration, suggesting that nuclear MATR3 mediates toxicity. 30078707_PTBP1 and MATR3 co-bind and repress RNA processing within and around young LINEs 30398641_FUS, EWSR1, TAF15 and MATR3 within the RNAP II/U1 snRNP machinery play distinct roles in the development of amyotrophic lateral sclerosis and spinal muscular atrophy that are more intimately tied to one another than previously thought. 30425153_In this work we identify the cellular protein MATR3 as an essential cofactor of viral RNA processing. Reactivation of HIV-1 transcription is not sufficient to allow completion of a full life cycle of the virus if MATR3 is depleted. 30516047_identified MATR3 as binding to SNHG1 and the interaction might be involved in splicing events that enhance neuroblastoma progression. 30563574_The data of this study suggest that MATR3 may have regulatory mechanisms that are influenced by tissue type and/or mutation status 30565205_Data shows the expression profile of Gle1, MATR3 and FUS genes in Spinal muscular atrophy (SMA), and suggest a critical role of FUS protein in SMA pathogenesis. Note that the authors appear to erroneously refer to the human MATR3 gene (GeneID: 9782) as MART3 (GeneID: 203430) in this publication. 30790622_MATR3 regulates miR-138 in neurons, with important implications for miR-138 regulation during neuronal development, synaptic plasticity and memory-related processes. 31019288_N-terminal sequences in MATR3 can mediate phase separation into intranuclear droplet-like structures that can recruit TDP43 under conditions of low RNA binding. 31056746_Mutant MATR3 causes multisystem proteinopathy in mice. 31441370_MATR3 is a splicing activator during PDCD1 exon 3 splicing that operates through binding to enhancer elements ESE3a and ESE3b. 32166857_Effect of up-regulation of circMATR3 on the proliferation, metastasis, progression and survival of hypopharyngeal carcinoma. 32361838_Whole-body muscle MRI of patients with MATR3-associated distal myopathy reveals a distinct pattern of muscular involvement and highlights the value of whole-body examination. 32515490_Knockdown of genes involved in axonal transport enhances the toxicity of human neuromuscular disease-linked MATR3 mutations in Drosophila. 32943284_Matrin-3 plays an important role in cell cycle and apoptosis for survival in malignant melanoma. 33129345_RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3. 33357424_Intronic Determinants Coordinate Charme lncRNA Nuclear Activity through the Interaction with MATR3 and PTBP1. 33427209_Matrin 3 in neuromuscular disease: physiology and pathophysiology. 34173818_MATR3 haploinsufficiency and early-onset neurodegeneration. | ENSMUSG00000117694+ENSMUSG00000037236 | Snhg4+Matr3 | 2.886604e+03 | 0.7937811 | -0.333186825 | 0.3150586 | 1.097810e+00 | 0.2947471721 | 0.81312088 | No | Yes | 2.155082e+03 | 354.178887 | 2.587034e+03 | 435.765373 |
| ENSG00000015568 | 84220 | RGPD5 | protein_coding | Q99666 | Alternative splicing;Cytoplasm;Phosphoprotein;Reference proteome;Repeat;TPR repeat | RAN is a small GTP-binding protein of the RAS superfamily that is associated with the nuclear membrane and is thought to control a variety of cellular functions through its interactions with other proteins. This gene shares a high degree of sequence identity with RANBP2, a large RAN-binding protein localized at the cytoplasmic side of the nuclear pore complex. It is believed that this RANBP2 gene family member arose from a duplication event 3 Mb distal to RANBP2. Alternative splicing has been observed for this locus and two variants are described. Additional splicing is suggested but complete sequence for further transcripts has not been determined. [provided by RefSeq, Jul 2008]. | hsa:729540;hsa:84220; | cytoplasm [GO:0005737]; nuclear pore [GO:0005643]; NLS-bearing protein import into nucleus [GO:0006607]; regulation of catalytic activity [GO:0050790] | 2.900335e+02 | 0.4590952 | -1.123134860 | 0.3432627 | 9.933997e+00 | 0.0016225382 | 0.15496288 | No | Yes | 1.795457e+02 | 40.969761 | 3.890955e+02 | 90.152987 | |||||
| ENSG00000017483 | 92745 | SLC38A5 | protein_coding | Q8WUX1 | FUNCTION: Functions as a sodium-dependent amino acid transporter which countertransport protons. Mediates the saturable, pH-sensitive, and electrogenic cotransport of several neutral amino acids including glycine, asparagine, alanine, serine, glutamine and histidine with sodium. {ECO:0000269|PubMed:11243884}. | Acetylation;Alternative splicing;Cell membrane;Disulfide bond;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | The protein encoded by this gene is a system N sodium-coupled amino acid transporter. The encoded protein transports glutamine, asparagine, histidine, serine, alanine, and glycine across the cell membrane, but does not transport charged amino acids, imino acids, or N-alkylated amino acids. Alternative splicing results in multiple transcript variants, but the full-length nature of some of these variants has not been determined. [provided by RefSeq, Aug 2013]. | hsa:92745; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; alanine transmembrane transporter activity [GO:0022858]; amino acid transmembrane transporter activity [GO:0015171]; glycine transmembrane transporter activity [GO:0015187]; L-asparagine transmembrane transporter activity [GO:0015182]; L-glutamine transmembrane transporter activity [GO:0015186]; L-histidine transmembrane transporter activity [GO:0005290]; L-serine transmembrane transporter activity [GO:0015194]; serine transmembrane transporter activity [GO:0022889]; amino acid transmembrane transport [GO:0003333]; amino acid transport [GO:0006865]; asparagine transmembrane transport [GO:1903713]; glutamine transport [GO:0006868]; glycine transport [GO:0015816]; L-alanine transmembrane transport [GO:1904557]; L-histidine transmembrane transport [GO:0089709]; L-serine transport [GO:0015825]; serine transport [GO:0032329]; transport across blood-brain barrier [GO:0150104] | 17333282_A 50kb deletion at Xp11.23 including the two genes, SLC38A5 and FTSJ1 was found in 3 brothers with moderate to severe mental retardation. 29499643_Confirmed by mRNA and protein expression, the amino acid transporters SLC7A7 and SLC38A5 showed marked differences between controls and intrauterine growth restriction/pre-eclampsia and were regulated by both diseases. In contrast, ABCA1 may play an exclusive role in the development of pre-eclempsia. 34704597_Expression and function of SLC38A5, an amino acid-coupled Na+/H+ exchanger, in triple-negative breast cancer and its relevance to macropinocytosis. | ENSMUSG00000031170 | Slc38a5 | 1.960546e+01 | 0.5770168 | -0.793314885 | 0.6846348 | 1.269733e+00 | 0.2598167592 | 0.79684153 | No | Yes | 8.590246e+00 | 3.646114 | 1.325095e+01 | 5.752680 | |
| ENSG00000026297 | 8635 | RNASET2 | protein_coding | O00584 | FUNCTION: Ribonuclease that plays an essential role in innate immune response by recognizing and degrading RNAs from microbial pathogens that are subsequently sensed by TLR8 (PubMed:31778653). Cleaves preferentially single-stranded RNA molecules between purine and uridine residues, which critically contributes to the supply of catabolic uridine and the generation of purine-2',3'-cyclophosphate-terminated oligoribonucleotides (PubMed:31778653). In turn, RNase T2 degradation products promote the RNA-dependent activation of TLR8 (PubMed:31778653). Plays also a key role in degradation of mitochondrial RNA and processing of non-coding RNA imported from the cytosol into mitochondria (PubMed:28730546, PubMed:30184494). Participates as well in degradation of mitochondrion-associated cytosolic rRNAs (PubMed:30385512). {ECO:0000269|PubMed:16620762, ECO:0000269|PubMed:19525954, ECO:0000269|PubMed:22735700, ECO:0000269|PubMed:28730546, ECO:0000269|PubMed:30184494, ECO:0000269|PubMed:30385512, ECO:0000269|PubMed:31778653}. | 3D-structure;Alternative splicing;Disease variant;Disulfide bond;Endonuclease;Endoplasmic reticulum;Glycoprotein;Hydrolase;Immunity;Innate immunity;Lyase;Lysosome;Mitochondrion;Nuclease;Reference proteome;Secreted;Signal | Mouse_homologues NA; + ;NA; + ;NA; + ;NA | This ribonuclease gene is a novel member of the Rh/T2/S-glycoprotein class of extracellular ribonucleases. It is a single copy gene that maps to 6q27, a region associated with human malignancies and chromosomal rearrangement. [provided by RefSeq, Jul 2008]. | hsa:8635; | azurophil granule lumen [GO:0035578]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; mitochondrial intermembrane space [GO:0005758]; endoribonuclease activity [GO:0004521]; ribonuclease activity [GO:0004540]; ribonuclease T2 activity [GO:0033897]; RNA binding [GO:0003723]; innate immune response [GO:0045087]; RNA catabolic process [GO:0006401] | 15809705_RNASET2 found to significantly decrease the metastatic potential of ovarian cancer cell line in vivo; tumor suppression by RNASET2 is suggested to not be mediated by its ribonuclease activity 16620762_The results presented herein represent a further advancement toward the molecular understanding of the tumour suppressive properties of the human RNASET2 protein. 18543608_Loss of RNASET2 is associated with melanoma 19382914_RNaseT2 is a cell growth regulator and it does not induce senescence in SV40 immortalized cell lines. 19525954_Study shows that loss-of-function mutations in the gene encoding the RNASET2 glycoprotein lead to cystic leukoencephalopathy, an autosomal recessive disorder with an indistinguishable clinical and neuroradiological phenotype. 20526339_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20601676_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 21199949_Familial cystic leukoencephalopathy arising in RNASET2-deficient humans is a manifestation of an lysosomal storage disorders in which rRNA is the best candidate for the noxious storage material. 21446958_The expression of the human tumor suppressor protein RNASET2 was studied in baculovirus-insect cell and Pichia pastoris heterologous systems. 21646684_Molecular signature induced by RNASET2, a tumor antagonizing gene, in ovarian cancer cells. 21994792_Tax represses expression of RNase T2. 22188480_a possible involvement of RNASET2 in P-body formation in mammalian cells. 22732457_RNASET2--an autoantigen in anaplastic large cell lymphoma identified by protein array analysis. 22735700_The catalytic features of RNase T2 in presence of bivalent cations were analyzed and the structural consequences of known clinical mutations were investigated. 23258633_Higher expression of RNASET2 in the semen of asthenozoospermia individuals may contribute to sperm motility impairment. 23630276_Ribonuclease T2, an ancient and phylogenetically conserved RNase, has a role in development of ovarian neoplasms 24327149_Genotypes of single nucleotide polymorphisms (SNPs) in RNASET2 gene were determined. 24457966_RNASET2 contributes to vitiligo pathogenesis by inhibiting TRAF2 expression. 24842157_Results show that downregulation of RNASET2 and GGNBP2 in drug-resistant ovarian cancer tissues/cells contributes to the regulation of drug resistance in ovarian cancer. 25426551_RNASET2 has antitumorigenic and antiangiogenic activities; a truncated version of human RNASET2, starting at E50 (trT2-50) and devoid of ribonuclease activity, has actin binding and anticancer-related biological activities 25663099_describe a multi-step strategy that allows production of highly pure, catalytically competent recombinant RNASET2 in both wild-type and mutant forms 25797262_Biological features allow to put forward the hypothesis that the RNASET2 protein can act as a molecular barrier for limiting the damages and tissue remodeling events occurring during the earlier step of cell transformation. 25928629_RNASET2 tag SNP but not CCR6 polymorphisms is associated with autoimmune thyroid diseases in the Chinese Han population 26067323_RNASET2 may contribute to the development of vitiligo by inhibiting TNF Receptor-Associated Factor 2 expression and lead directly to apoptosis of melanocytes 26293343_inhibits melanocyte outgrowth through interacting with shootin1; this effect may be associated with vitiligo pathogenesis 28400196_RNASET2, an IBD susceptibility gene, is a component of TL1A-mediated pathways regulating cytokine production. 28568286_Studied association of and RNASET2, GPR174, and PTPN22 gene polymorphisms and liver damage(LD) due to Graves' disease (GD) hyperthyroidism. Found GPR174 rs3827440, PTPN22 rs3789604, and RNASET2 rs9355610 were significantly associated with altered GD-derived LD risk. 28730546_RNASET2 is the enzyme that degrades the RNAs 29581387_High RNASET2 expression is associated with reduced sperm motility. 29763721_In poorly differentiated lung neuroendocrine carcinomas, RNASET2 expression may be upregulated as a consequence of the activation of the hypoxia-induced HIF-1alpha pathway, thus behaving as an alarmin-like molecule. 30218741_The human RNASET2 protein affects the polarization pattern of human macrophages in vitro. 30385512_Regulation of mitochondrion-associated cytosolic ribosomes by mammalian mitochondrial ribonuclease T2 31778653_the lysosomal endoribonuclease RNase T2 is a non-redundant upstream component of TLR8-dependent RNA recognition. 32294405_Immune Sensing of Synthetic, Bacterial, and Protozoan RNA by Toll-like Receptor 8 Requires Coordinated Processing by RNase T2 and RNase 2. 32295832_Zebrafish disease model of human RNASET2-deficient cystic leukoencephalopathy displays abnormalities in early microglia. 33767133_FBXO6-mediated RNASET2 ubiquitination and degradation governs the development of ovarian cancer. 34299186_Hypoxia Enhances the Expression of RNASET2 in Human Monocyte-Derived Dendritic Cells: Role of PI3K/AKT Pathway. | ENSMUSG00000116988+ENSMUSG00000094724+ENSMUSG00000116876+ENSMUSG00000095687 | Gm49673+Rnaset2b+Gm49721+Rnaset2a | 1.460166e+03 | 0.9158964 | -0.126743701 | 0.2619498 | 2.336579e-01 | 0.6288246955 | 0.91646120 | No | Yes | 1.165896e+03 | 73.339610 | 1.265368e+03 | 81.365621 |
| ENSG00000027697 | 3459 | IFNGR1 | protein_coding | P15260 | FUNCTION: Receptor subunit for interferon gamma/INFG that plays crucial roles in antimicrobial, antiviral, and antitumor responses by activating effector immune cells and enhancing antigen presentation (PubMed:20015550). Associates with transmembrane accessory factor IFNGR2 to form a functional receptor (PubMed:7615558, PubMed:2971451, PubMed:7617032, PubMed:10986460, PubMed:7673114). Upon ligand binding, the intracellular domain of IFNGR1 opens out to allow association of downstream signaling components JAK1 and JAK2. In turn, activated JAK1 phosphorylates IFNGR1 to form a docking site for STAT1. Subsequent phosphorylation of STAT1 leads to dimerization, translocation to the nucleus, and stimulation of target gene transcription (PubMed:28883123). STAT3 can also be activated in a similar manner although activation seems weaker. IFNGR1 intracellular domain phosphorylation also provides a docking site for SOCS1 that regulates the JAK-STAT pathway by competing with STAT1 binding to IFNGR1 (By similarity). {ECO:0000250|UniProtKB:P15261, ECO:0000269|PubMed:10986460, ECO:0000269|PubMed:20015550, ECO:0000269|PubMed:28883123, ECO:0000269|PubMed:2971451, ECO:0000269|PubMed:7615558, ECO:0000269|PubMed:7617032, ECO:0000269|PubMed:7673114}. | 3D-structure;Alternative splicing;Cell membrane;Direct protein sequencing;Disease variant;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix;Ubl conjugation | This gene (IFNGR1) encodes the ligand-binding chain (alpha) of the gamma interferon receptor. Human interferon-gamma receptor is a heterodimer of IFNGR1 and IFNGR2. A genetic variation in IFNGR1 is associated with susceptibility to Helicobacter pylori infection. In addition, defects in IFNGR1 are a cause of mendelian susceptibility to mycobacterial disease, also known as familial disseminated atypical mycobacterial infection. [provided by RefSeq, Jul 2008]. | hsa:3459; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; cytokine binding [GO:0019955]; cytokine receptor activity [GO:0004896]; interferon-gamma receptor activity [GO:0004906]; astrocyte activation [GO:0048143]; cytokine-mediated signaling pathway [GO:0019221]; defense response to virus [GO:0051607]; microglial cell activation [GO:0001774]; negative regulation of amyloid-beta clearance [GO:1900222]; positive regulation of amyloid-beta formation [GO:1902004]; positive regulation of gene expression [GO:0010628]; positive regulation of tumor necrosis factor production [GO:0032760]; response to virus [GO:0009615]; signal transduction [GO:0007165] | 11240951_Observational study of gene-disease association. (HuGE Navigator) 11857344_IL-12R beta 1- and IFN-gamma R1 signals co-ordinately regulate IFN-gamma production, but only IL-12 negatively controls IL-4 production. IL-12 and IFN-gamma signals are each sufficient for IFN-gamma production but both are needed for optimal production. 11865431_partial IFNGR1 mutations in Japanese patients with BCG osteomyelitis 12020266_Observational study of gene-disease association. (HuGE Navigator) 12023780_IFNGR1 gene promoter polymorphisms may be assocaited with susceptibility to cerebral malaria 12023780_Observational study of gene-disease association. (HuGE Navigator) 12027427_Mutations in interferon-gamma receptor 1. 12034035_This study identified a further role of IFN-gamma on IL-4 responses, including reduced IL-4R surface expression by human monocytes. 12165521_Lipid microdomains are required sites for the selective endocytosis and nuclear translocation of IFN-gamma receptor-1. 12244188_Partial deficiency of IFN-gamma receptor 1 results in abrogation of IFN-gamma-induced killing of Salmonella typhimurium and Toxoplasma gondii due to IFN-gamma unresponsiveness of patients' cells of the monocyte/macrophage lineage. 12438563_FRET was used to demonstrate that the IFNGR chains were preassembled on the cell membrane. 12454749_suppressed by 2- to 3-fold in B-cell chronic lymphocytic leukemia cells, which is expected to increase CLL cell survival 12516030_Genome analysis identified polymorphism in the human interferon gamma receptor affecting Helicobacter pylori infection. 12543882_MHC Class II proteins, interferon-alpha, interferon-gamma receptor and the capacity to present antigen may be crucial in HIV-associated nephropathy pathogenesis. 12743658_Mutations have no association with the susceptibility to lepromatous leprosy in the Korean population. 12743658_Observational study of gene-disease association. (HuGE Navigator) 12753505_Observational study of gene-disease association. (HuGE Navigator) 12753505_Unidentified allelic variations in the IFNGR1 gene might elevate or decrease the risk in the Croatian population, as a part of the multigenic predisposition to tuberculosis. 12851715_In this study, although IFN-gamma production in the allergic patients with L467P was equivalent to that in the non-allergic subjects, their serum IgE levels were high and they had allergic diseases 12851715_Observational study of gene-disease association. (HuGE Navigator) 14734726_IFN-gamma receptor deficiency alters the epitope hierarchy of the pool of lymphocytic choriomeningitis virus-specific memory CD8 T cells without significantly affecting the immunodominance of the primary CD8 T cell response in an acute infection. 14763782_Observational study of gene-disease association. (HuGE Navigator) 15004750_Observational study of gene-disease association. (HuGE Navigator) 15047947_Observational study of gene-disease association. (HuGE Navigator) 15182327_Observational study of gene-disease association. (HuGE Navigator) 15207788_Observational study of gene-disease association. (HuGE Navigator) 15527154_Observational study of gene-disease association. (HuGE Navigator) 15756299_disease susceptibility in Schistosoma mansoni infection to hepatic fibrosis is linked to a SNP in the interferon gamma receptor locus (P=0.000001). 15952126_Observational study of gene-disease association. (HuGE Navigator) 15952126_The IFN-GammaR2 Arg64/Arg64 genotype does not determine susceptibility to SLE in Chinese people, and the combination of IFN-Gamma R2 Arg64/Arg64 genotype and IFN-Gamma R1 Val14/Val14 genotype does not, either. 16115485_IFNG T874A, IFNGR1 C-56T and IFNGR2 A839G genotypes were not associated with the incidence of angiographic and clinical restenosis (P>0.23). 16115485_Observational study of gene-disease association. (HuGE Navigator) 16233916_Observational study of gene-disease association. (HuGE Navigator) 16233916_The relationship between polymorphisms at IFNGR1 and susceptibility to pulmonary tuberculosis is reported in Iranian patients. 16476014_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16491350_Observational study of gene-disease association. (HuGE Navigator) 16563189_IFNGR1 does not contribute to susceptibility to rheumatoid arthritis in Caucasians, although a single nucleotide polymorphisms exist in this disease. 16563189_Observational study of gene-disease association. (HuGE Navigator) 16690980_Novel tuberculosis association was found with the 56CC genotype of the IFNGR1 promotor. 16690980_Observational study of gene-disease association. (HuGE Navigator) 16785527_Interferon (IFN)-gamma and its receptor subunit IFNGR1 bind to the IFN-gamma-activated sequence (GAS) response element in the promoter region of IFN-gamma-activated genes. This binding results in enhanced activation of IFN-gamma-induced genes. 16867043_Observational study of gene-disease association. (HuGE Navigator) 16944293_Observational study of gene-disease association. (HuGE Navigator) 16944293_no statistically significant association with susceptibility to persistent HBV infection was observed with the IFN-gamma, IFNGR-1 and 2, and IRF-1 gene polymorphisms under the codominant, dominant, and recessive models 17023216_Observational study of gene-disease association. (HuGE Navigator) 17030574_Infection of HEK 293 cells with C. psittaci increased IFN-gammaR expression only in cells expressing either TLR2 or TLR4 and the adaptor protein MD-2. 17136124_Observational study of gene-disease association. (HuGE Navigator) 17136124_The results suggest a complex pattern of haplotypic variation at the IFNGR1 promoter locus associated with post-kala-azar dermal leishmanaisis susceptibility. 17152005_Observational study of gene-disease association. (HuGE Navigator) 17166914_The K3 and K5 proteins of Kaposi's sarcoma-associated herpesvirus (KSHV) both specifically target IFN-gammaR1 and induce its ubiquitination, endocytosis, and degradation. 17251453_Observational study of gene-disease association. (HuGE Navigator) 17251453_The -56T allele in the IFNGR1 promoter results in higher IFNGR1 transcriptional activity and represents a genetic risk factor for atopic cataracts. 17339358_expression downregulated by Mycobacterium tuberculosis in human cells as immune evasion mechanism. 17348823_Observational study of gene-disease association. (HuGE Navigator) 17392024_Observational study of gene-disease association. (HuGE Navigator) 17431682_Observational study of gene-disease association. (HuGE Navigator) 17431682_analyzed candidate genes related to TNFalpha regulation and found that interleukin (IL)-10, interferon-gamma receptor 1 (IFNGR1), and TNFalpha receptor 1 (TNFR1) genes were linked and associated to both tuberculosis and TNFalpha 17477815_Observational study of gene-disease association. (HuGE Navigator) 17513528_A deletion in this receptor produces a truncated form of IFNgammaR1, which has a dominant-negative effect on IFNgamma signal transduction through altered receptor stability. 17546485_Variation in transcriptional activity of genes encoding INF-gamma receptor subunits may affect function of microvasculature and thereby participate in the pathology of cardiac syndrome X 17572155_Observational study of genotype prevalence. (HuGE Navigator) 17618444_IFNGR polymorphisms (Val14Met and Gln64Arg) are protective in systemic lupus erythematosus in Chinese patients 17618444_Observational study of gene-disease association. (HuGE Navigator) 17697357_expressed IFNgamma and IFNgamma-Ralpha together with the nuclear localization of IFNgamma-Rbeta, could be a tumoral cell response 17986123_Observational study of gene-disease association. (HuGE Navigator) 17986123_The IFN-gamma receptor 1 gene polymorphism does not appear to be responsible for host susceptibility to pulmonary tuberculosis in the Korean population. 18287876_Observational study of gene-disease association. (HuGE Navigator) 18414508_Frequent mutations in microsatellite-instable (MSI-H) tumours and cell lines of a conserved A14 repeat within the 3'-untranslated region of the interferon-gamma receptor 1 gene (IFNGR1). 18548239_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18548239_The polymorphisms in the IFNG and IFNGR1 genes were studied with the aim of clarifying the relationships among these polymorphisms, penicillin allergy and anti-penicillin antibodies. 18555234_Regulation of the IFN-gamma R and its signaling response in human endometrial stromal cells during decidualization. 18593809_Observational study of gene-disease association. (HuGE Navigator) 18593809_results indicate that the IFNGR1 -56C/T polymorphism is a relevant host susceptibility factor for Gastric Carcinoma development. 18620489_with progression of HIV-1 infection, interferon-gamma production declines whereas expression of interferon-gamma receptors (R1 and R2) increases 18633131_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18702743_allele (CA)(25)of the interferon gamma receptor 1 gene appeared to be susceptible to TB, while the allele (CA)(26) was protective towards TB 18809513_The urine evaluation of the balance between IFN-gammaR1 and IL4R receptors might be informative for the immune states of HUS patients. 18931463_Finds lack of association between functional polymorphisms in intron 1 of IFN-gamma gene at position +874 and silicosis and pulmonary tuberculosis in Chinese iron miners 18953482_Observational study of gene-disease association. (HuGE Navigator) 19141860_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19172849_the transcriptional of IFNgRb/IFNgRa in the heart bioptates appeared to be an early and sensitive marker of inflammatory status of patients with myocarditis 19247692_Observational study of gene-disease association. (HuGE Navigator) 19269302_Results show a significant down regulation in expression of the alpha-chain of the IFN-gamma receptor on surfaces of alveolar macrophages acquired from smokers when compared with non-smokers. 19279332_Tat impaired the IFNgamma-receptor signaling pathway at the level of STAT1 activation, possibly via Tat-dependent induction of SOCS-2 activity. 19488747_Observational study of gene-disease association. (HuGE Navigator) 19488747_Results show that that a functional -56C/T SNP in IFNGR1 promoter is associated with the clinical outcome of HBV infection in this Chinese population. 19575238_Strong association with this gene and pulmonary tuberculosis susceptibility in African Americans. 19625176_Observational study of gene-disease association. (HuGE Navigator) 19681704_The interleukin-12/interferon-gamma axis appears to be critical for control of coccidioidomycosis and mycobacteriosis in interferon-gamma receptor 1 deficiency with a IFNGR1 mutation 19692168_Observational study of gene-disease association. (HuGE Navigator) 19712753_A case-control association analysis failed to detect significant association between the IFNGR1 polymorphisms and cerebral malaria in the Thai population 19723394_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19734231_A putative SOCS-1 binding site, tyrosine 441 on IFN-gamma receptor subunit 1, contributes to the regulation of interferon-gamma signaling through recruitment of SOCS-1 in wild-type but not mutant cells. 19739012_The urinary soluble IFN-gamma receptor levels did not correlate with the clinical severity of vesicoureteral reflux 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20015550_Functional analysis of naturally occurring amino acid substitutions in human IFN-gammaR1. 20055726_Observational study of gene-disease association. (HuGE Navigator) 20070287_Observational study of gene-disease association. (HuGE Navigator) 20070287_study showed a positive association between -56C/C genotype of IFNGR1 (OR = 1.7; 95% CI = 1.1-2.7) and pre-eclampsia. 20196868_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20331378_Observational study of gene-disease association. (HuGE Navigator) 20399512_Clinical trial of gene-disease association. (HuGE Navigator) 20412699_Observational study of gene-disease association. (HuGE Navigator) 20412699_results do not show an implication of IFNGR1gene polymorphisms in the susceptibility to and clinical expression of giant cell arteritis 20452482_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20500698_Observational study of gene-disease association. (HuGE Navigator) 20587546_Observational study of gene-disease association. (HuGE Navigator) 20588308_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20673868_Observational study of gene-disease association. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 20799037_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20959405_Observational study of gene-disease association. (HuGE Navigator) 20980339_Observational study of gene-disease association. (HuGE Navigator) 21190998_IL-29 up-regulated, whereas IFNalpha down-regulated, the surface expression of the IFNgamma receptor 1 chain on macrophages, thereby resulting in differential responsiveness of TLR-challenged macrophages to IFNgamma. 21221749_The Japanese patients with a genetic mutation in the IFN-gamma-R1 gene were more susceptible to developing recurrent disseminated mycobacterial infections. 21460021_The autosomal recessive disorder, because of a single mutation in interferon-gamma receptor-1(IFNGR1) at position -56, was found to be associated with susceptibility to leprosy in children of the same family. 21722521_CD74 gene over-expression in TEC can increase IFN-gammaR mRNA expression 21731057_IFNGR1 is a modifier gene of cystic fibrosis disease. 21859832_Single nucleotide polymorphism in IFNGR1 gene is associated with rectal cancer. 22564607_Monocyte-derived CD40 expression is regulated by interferon-gamma/interferon-gamma receptor-1 pathway when acting as a bridge during their interaction with T cells and allogeneic endothelial cells. 22595141_A novel endocytosis motif shares characteristics of tyrosine-based and dileucine-based internalization sequences and is highly conserved in IFN-gamma receptors across species. 22644879_Interaction of IFNgammaR1 with TRAF6 regulates NF-kappaB activation and IFNlambdaR1 stability. 22711893_Absence of early innate IFN-gamma production in transgenic mice with combined defects in both IL-12 and type I IFN receptor negates the impact of programmed death ligand-1 blockade. 23040881_An association study of functional polymorphic genes .... IFNGR-1, ...... with disease progression, aspartate aminotransferase, alanine aminotransferase, and viral load in chronic hepatitis B and C. 23935197_These data suggest that type I IFN stimulation induces a rapid recruitment of a repressive Egr3/Nab1 complex that silences transcription from the ifngr1 promoter. 24199198_Increasing affinity of interferon-gamma receptor 1 to interferon-gamma by computer-aided design. 24220318_A novel heterozygous frameshift mutation (805delT) encoding the IFN-gamma receptor 1 (IFNGR1) was identified, presenting in a case of Mycobacterium intracellulare infection. 24254535_Intact IFN-gammaR1 expression and function distinguishes Langerhans cell histiocytosis from mendelian susceptibility to mycobacterial disease. 24453034_work aimed to evaluate single nucleotide polymorphisms (SNPs) of IFNGR1, GSTT1, and GSTP1 genes samples in gastric cancer 24680779_Genetic polymorphisms in IFNGR1 gene are involved in the risk of tuberculosis in the Chinese population. 25108563_FcgammaRIIa cross-talk with TLRs, IL-1R, and IFNgammaR selectively modulates cytokine production in human myeloid cells. 25242146_The study identifies a mechanism of cellular signaling regulated by extracellular vesicles -associated IFN-gamma/Ifngr1 complexes, which grafted stem cells may use to communicate with the host immune system. 25382336_The study did not provide enough powerful evidence to identify a significant association between IFNGR1 -56C/T polymorphism and tuberculosis susceptibility (meta-analysis). 25466928_Uniformly low expression of IFN and IFNGR1 in post Kala-azar dermal leishmaniasis skin biopsies could explain parasite persistence and is consistent with prior demonstration of genetic association with IFNGR1 polymorphisms. 25708927_In an African-American population, a significant difference in IFNGR1 expression between patients with RA and controls. However, IFNGR1 expression levels were not statistically significantly associated with erosion status or other radiographic outcomes. 25815589_Statistical analyses revealed that four genetic variants in IFNGR1 were marginally associated with the risk of Tuberculosis (P = 0.02-0.04), while other single nucleotide polymorphisms in IFNGR1 and IFNGR2 did not exhibit any associations 26251056_Mendelian susceptibility to mycobacteria due to a partial dominant mutation of the interferon gamma receptor 1 gene. 26343451_Targeted deep sequencing identifies rare loss-of-function variants in IFNGR1 for risk of atopic dermatitis complicated by eczema herpeticum 26931784_The deletion of IFNGR1 causes complete IFN-gammaR1 deficiency. Despite the deletion ending very close to the IL22RA2 gene, it does not appear to affect IL22RA2 transcription. 27020872_A significant association of IFN-gammaR1 and P2X7 genes polymorphisms with risk of developing TB in Iranian population. 27069113_B cell type 1 IFN receptor signals accelerate, but are not required for, lupus development. 27356097_All patients tested were positive for mycobacteria; one was heterozygous for the IFNGR1 exon 5 single-nucleotide-missense substitution 27941164_the cause of active TB in the patients seems to be due to the lack of effective IFNgamma function or the lack of effective signaling connection between IFNgamma and its receptor in presence of -56 C/T polymorphism in promoter region of IFNgammaR1 gene 28652404_Data suggest IFNG plays various roles in dynamics of inflammation in subjects with underlying autoimmunity modeled as 'canonical' and 'non-canonical' pathways; in canonical pathway, IFNG dimerizes and binds to IFNGR1 in IFNGR1/IFNGR2 hetero-multimer; STAT transcription factors are involved in non-canonical pathway. (IFNG = interferon gamma; IFNGR = IFNG receptor; STAT = signal transducers and activators of transcription) 28719321_results showed a significant correlation between IFNGR1- T-56CSNP and Nontuberculous mycobacteria infection among studied populations 28744922_All known mutations, as well as 287 other variations, have been deposited in the online IFNGR1 variation database . In this article, we review the function of IFN-gammaR1 and molecular genetics of human IFNGR1. 29209098_Positive reaction in interferon-gamma release tests is not associated with IFNGR1 SNPs. 29249666_Impaired IFNgamma-Signaling and Mycobacterial Clearance in IFNgammaR1-Deficient Human iPSC-Derived Macrophages. 29343571_cellular kinase, casein kinase 1alpha (CK1alpha), is crucial for IAV HA-induced degradation of both IFNGR1 and IFNAR1. 29441481_discovered that a rare variant c.G40A in interferon gamma receptor 1 potentially contributes to the myasthenia gravis pathogenesis 29516882_The two genetic variants were found to have potential risk in association with active disease development among Sudanese patients. 31310896_Genetic variants in IFNG and IFNGR1 and tuberculosis susceptibility. 31585982_Data data demonstrate that there are multiple mechanisms by which IFNGR1 availability is regulated in myeloid cells, which is counter to the accepted dogma that IFNGR is constitutively expressed. 31687049_IFNG and IFNGR1 polymorphisms are associated with latent tuberculosis infection 31760574_Mutual alteration of NOD2-associated Blau syndrome and IFNgammaR1 deficiency. 32271156_Regulation of Interferon-gamma receptor (IFN-gammaR) expression in macrophages during Mycobacterium tuberculosis infection. 32284542_A negative correlation between ELF5, FBXW7 and IFNGR1 expression in the tumours of patients with TNBC. 32770644_Risk factors for recurrence of Helicobacter pylori infection after successful eradication in Chinese children: A prospective, nested case-control study. 33017569_Promoting effect of long non-coding RNA SNHG1 on osteogenic differentiation of fibroblastic cells from the posterior longitudinal ligament by the microRNA-320b/IFNGR1 network. 33393726_Genetic Association of a Gain-of-Function IFNGR1 Polymorphism and the Intergenic Region LNCAROD/DKK1 With Behcet's Disease. 33501617_Complete IFN-gammaR1 Deficiency in a Boy Due to UPD(6)mat with IFNGR1 Novel Splicing Variant. 33667716_Paraspeckle Promotes Hepatocellular Carcinoma Immune Escape by Sequestering IFNGR1 mRNA. 34148879_PD_BiBIM: Biclustering-based biomarker identification in ESCC microarray data. 34181338_Predictive Value of Interferon gamma Receptor Gene Polymorphisms for Hepatocellular Carcinoma Susceptibility. 34646402_Genetic Polymorphisms of IFNG, IFNGR1, and Androgen Receptor and Chronic Prostatitis/Chronic Pelvic Pain Syndrome in a Chinese Han Population. 35074548_Early diagnosis of partial interferon-gamma receptor 1 deficiency prevents the development of Bacille de Calmette et Guerin osteomyelitis. | ENSMUSG00000020009 | Ifngr1 | 3.376675e+02 | 0.7727255 | -0.371972062 | 0.3939713 | 8.709687e-01 | 0.3506872660 | 0.82969965 | No | Yes | 2.940090e+02 | 65.383361 | 3.376471e+02 | 76.876184 | |
| ENSG00000029993 | 3149 | HMGB3 | protein_coding | O15347 | FUNCTION: Multifunctional protein with various roles in different cellular compartments. May act in a redox sensitive manner. Associates with chromatin and binds DNA with a preference to non-canonical DNA structures such as single-stranded DNA. Can bent DNA and enhance DNA flexibility by looping thus providing a mechanism to promote activities on various gene promoters (By similarity). Proposed to be involved in the innate immune response to nucleic acids by acting as a cytoplasmic promiscuous immunogenic DNA/RNA sensor (By similarity). Negatively regulates B-cell and myeloid cell differentiation. In hematopoietic stem cells may regulate the balance between self-renewal and differentiation. Involved in negative regulation of canonical Wnt signaling (By similarity). {ECO:0000250|UniProtKB:O54879, ECO:0000250|UniProtKB:P09429, ECO:0000250|UniProtKB:P40618}. | 3D-structure;Acetylation;Chromosome;Cytoplasm;DNA-binding;Disulfide bond;Immunity;Innate immunity;Microphthalmia;Nucleus;Oxidation;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | This gene encodes a member of a family of proteins containing one or more high mobility group DNA-binding motifs. The encoded protein plays an important role in maintaining stem cell populations, and may be aberrantly expressed in tumor cells. A mutation in this gene was associated with microphthalmia, syndromic 13. There are numerous pseudogenes of this gene on multiple chromosomes. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2014]. | hsa:3149; | chromosome [GO:0005694]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; DNA binding, bending [GO:0008301]; double-stranded DNA binding [GO:0003690]; four-way junction DNA binding [GO:0000400]; RNA binding [GO:0003723]; DNA geometric change [GO:0032392]; DNA recombination [GO:0006310]; innate immune response [GO:0045087]; negative regulation of B cell differentiation [GO:0045578]; negative regulation of myeloid cell differentiation [GO:0045638]; regulation of transcription by RNA polymerase II [GO:0006357] | 22912879_Data indicate that myomiR-206 modulates Hmgb3 expression during myogenesis. 23326140_HMGB3 has a role in gastric cancer disease onset and progression by regulating the cell cycle 23609034_HMGB3 plays an important role in non-small cell lung cancer progression. 24098490_regulation of HMGB3 by miR-205 reduced both proliferation and invasion of breast cancer cells. 24338397_Association between co-expression networks involving HMGB3 and differentially-expressed genes in gastric cancer. 24993872_In this family, microphthalmia, microcephaly, intellectual disability, and short stature are associated with a mutation on the X chromosome in the HMGB3 gene. 25095979_High expression of HMGB3 is associated with lymph node metastases in gastric cancer. 25647262_our data suggests HMGB3 may serve as an important oncoprotein and indicate that overexpression of HMGB3 in urinary bladder cancer could be used as a potential prognostic marker. 25755721_these findings demonstrate that HMBG3 may be a potential molecular marker for predicting the prognosis of esophageal squamous cell carcinoma patients 27363334_Therefore, we infer that HMGB3 is a functional target of miR-27b in modulation of tamoxifen resistance and EMT. 28678825_HMGB3 regulates the genes expression of WNT/beta-catenin pathway 29196733_Luciferase reporter assays showed that HMGB3 was directly regulated by miR-205-5p in prostate cancer cells. 30166860_HMGB3 is involved in malignant transformation of hepatocytes and could be a useful biomarker for diagnosis and a potential target for therapy of liver cancer. 30232806_The study strongly suggests that HMGB3 promotes GBM oncogenesis through the MAPK signalling pathway while miR-200b-3p and miR-200c-3p inhibit its expression. 30343649_HMGB3 overexpression or miR-200b downregulation was associated with poor prognosis. Our findings suggest HMGB3 may serve as an important oncoprotein whose expression is negatively regulated by miR-200b in hepatocellular carcinoma. 30446524_HMGB3 has higher expression in non-small cell lung cancer tissues.HMGB3 is the target gene of miR-758. 30543931_HMGB3 might be of very great significance to the pathogenesis of FGR and might play the role by leading the dysfunction of placental villous trophoblast cells and through the interaction with some other proteins 31061066_High HMGB3 expression is associated with Chemo-resistance in Ovarian Cancer. 31422060_HOTTIP was upregulated and miR-615-3p was downregulated in non-small cell lung cancer (NSCLC) tissues and cells. Hypoxia induced glycolysis, increased HOTTIP and HMGB3 mRNA levels and repressed miR-615-3p expression in NSCLC cells. HOTTIP deficiency or miR-615-3p expression restoration repressed hypoxia-induced glycolysis. HOTTIP acted as a molecular sponge for miR-615-3p and HMGB3 was a direct target of miR-615-3p. 31621076_Bioinformatics analysis of the prognosis and biological significance of HMGB1, HMGB2, and HMGB3 in gastric cancer. 31773719_HMGB3 small interfere RNA suppresses mammosphere formation of MDA-MB-231 cells by down-regulating expression of HIF1alpha. 31858556_Circ-0001801 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) in glioblastoma by regulating miR-628-5p/HMGB3 axis. 32131767_LncRNA SNHG5 promotes nasopharyngeal carcinoma progression by regulating miR-1179/HMGB3 axis. 32386184_High-mobility group box 3 (HMGB3) silencing inhibits non-small cell lung cancer development through regulating Wnt/beta-catenin pathway. 32407168_Long Noncoding RNA SOX2-OT Knockdown Inhibits Proliferation and Metastasis of Prostate Cancer Cells Through Modulating the miR-452-5p/HMGB3 Axis and Inactivating Wnt/beta-Catenin Pathway. 32871048_LncRNA PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA. 32911346_CircRNA_102179 promotes the proliferation, migration and invasion in non-small cell lung cancer cells by regulating miR-330-5p/HMGB3 axis. 33078596_High mobility group box 3 promotes cervical cancer proliferation by regulating Wnt/beta-catenin pathway. 33428061_The role of high mobility group protein B3 (HMGB3) in tumor proliferation and drug resistance. 33727043_HDAC3 increases HMGB3 expression to facilitate the immune escape of breast cancer cells via down-regulating microRNA-130a-3p. 33773992_MiR-93/HMGB3 regulatory axis exerts tumor suppressive effects in colorectal carcinoma cells. 34408262_LINC00319 promotes cancer stem cell-like properties in laryngeal squamous cell carcinoma via E2F1-mediated upregulation of HMGB3. 34753396_LINC00857 promotes colorectal cancer progression by sponging miR-150-5p and upregulating HMGB3 (high mobility group box 3) expression. 34973275_miR-142-3p simultaneously targets HMGA1, HMGA2, HMGB1, and HMGB3 and inhibits tumorigenic properties and in-vivo metastatic potential of human cervical cancer cells. 35332131_HMGB3 promotes PARP inhibitor resistance through interacting with PARP1 in ovarian cancer. | ENSMUSG00000015217 | Hmgb3 | 6.348696e+03 | 1.1983187 | 0.261011669 | 0.2498639 | 1.102860e+00 | 0.2936393639 | 0.81275304 | No | Yes | 7.237353e+03 | 728.869737 | 5.412071e+03 | 559.020463 | |
| ENSG00000035664 | 23604 | DAPK2 | protein_coding | Q9UIK4 | FUNCTION: Calcium/calmodulin-dependent serine/threonine kinase involved in multiple cellular signaling pathways that trigger cell survival, apoptosis, and autophagy. Regulates both type I apoptotic and type II autophagic cell death signals, depending on the cellular setting. The former is caspase-dependent, while the latter is caspase-independent and is characterized by the accumulation of autophagic vesicles. Acts as a mediator of anoikis and a suppressor of beta-catenin-dependent anchorage-independent growth of malignant epithelial cells. May play a role in granulocytic maturation (PubMed:17347302). Regulates granulocytic motility by controlling cell spreading and polarization (PubMed:24163421). {ECO:0000269|PubMed:17347302, ECO:0000269|PubMed:24163421, ECO:0000269|PubMed:26047703}.; FUNCTION: Isoform 2 is not regulated by calmodulin. It can phosphorylate MYL9. It can induce membrane blebbing and autophagic cell death. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Calmodulin-binding;Cytoplasm;Cytoplasmic vesicle;Kinase;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a protein that belongs to the serine/threonine protein kinase family. This protein contains a N-terminal protein kinase domain followed by a conserved calmodulin-binding domain with significant similarity to that of death-associated protein kinase 1 (DAPK1), a positive regulator of programmed cell death. Overexpression of this gene was shown to induce cell apoptosis. It uses multiple polyadenylation sites. [provided by RefSeq, Jul 2008]. | hsa:23604; | autophagosome lumen [GO:0034423]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; identical protein binding [GO:0042802]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; anoikis [GO:0043276]; apoptotic process [GO:0006915]; intracellular signal transduction [GO:0035556]; positive regulation of apoptotic process [GO:0043065]; positive regulation of eosinophil chemotaxis [GO:2000424]; positive regulation of neutrophil chemotaxis [GO:0090023]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of apoptotic process [GO:0042981]; regulation of autophagy [GO:0010506]; regulation of intrinsic apoptotic signaling pathway [GO:2001242] | 11839660_High frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patients 11839665_distinct methylation pattern in bladder cancer with frequent methylation of RARbeta, DAPK, E-cadherin, and p16. 12087472_gene expression in colorectal and gastric cancer silenced by DNA methylation and histone deacetylation 17347302_Results implicate a novel role for DAPK2 in the regulation of normal myelopoiesis. 18521079_DAPK2 as a novel Sp1-dependent target gene for E2F1 and KLF6 in cell death response. 18957423_beta-catenin-induced down-regulation of DAPk-2 represents a novel signaling mechanism by which beta-catenin promotes the survival of malignant epithelial cells 19221469_Results showed that the inactivation of RASSF1A, RARbeta2 and DAP-Kinase by hypermethylation is a key step in NPC tumorigenesis and progression. 21408167_DRP-1 and ZIPk most likely evolved from their ancient ancestor gene DAPk by two gene duplication events that occurred close to the emergence of vertebrates 22160140_Sodium butyrate induced DAPK1/2 expression in human gastric cancer cells and this expression prompted apoptosis by decreasing FAK levels. 23700042_DAPK2 is upregulated in uterosacral ligaments in pelvic organ prolapse 24038216_The tumor suppressor gene DAPK2 is induced by the myeloid transcription factors PU.1 and C/EBPalpha during granulocytic differentiation but repressed by PML-RARalpha in APL. 24163421_The defect in chemotaxis in DAPK2-inactive granulocytes is likely a result of reduced polarization of the cells, mediated by a lack of MLC phosphorylation, resulting in radial F-actin and pseudopod formation. 25361081_DAPK2 is a novel kinase of mTORC1 and is a potential new member of this multiprotein complex, modulating mTORC1 activity and autophagy levels under stress and steady-state conditions. 25741596_DAPK2 regulates oxidative stress in cancer cells by preserving mitochondrial function 25982274_miR-520h suppresses Death-associated protein kinase 2 (DAPK2) expression, as restoring DAPK2 abolished miR-520h-promoted drug resistance, and knockdown of DAPK2 mitigated cell death caused by the depletion of miR-520h. 26038578_This study links adipocyte expression of an autophagy-regulating kinase, lysosome-mediated clearance and fat cell lipid accumulation; it demonstrates obesity-related attenuated autophagy in adipocytes, and identifies DAPK2 dependence in this regulation. 26047703_DAPK2-induced apoptosis is negatively regulated by Akt and 14-3-3 proteins. 26483415_that Death-associated protein kinase 2 effector functions are influenced by the protein's subcellular localization 27049921_This study suggests that miR-520g contributes to tumor progression and drug resistance by post-transcriptionally downregulating DAPK2 in patients with epithelial ovarian cancer 27653365_Thyroid hormone promotes selective autophagy via induction of DAPK2-SQSTM1 cascade, which in turn protects hepatocytes from diethylnitrosamine-induced hepatotoxicity or carcinogenesis. 29717115_study reveals a unique calmodulin-independent mechanism for DAPK2 activation, critical to its function as a novel downstream effector of AMPK in autophagy 30243997_remission effect of DAPK2 on placental cell oxidative damage and apoptosis in HDCP via mTOR activation 31116076_The intracellular signaling pathways that lead to Ser289 phosphorylation are mutually-exclusive and different for each kinase. In addition, Ser289 phosphorylation in fact enhances DAPK1 catalytic activity, similar to the effect on DAPK2. Thus, Ser289 phosphorylation activates both DAPK1 and DAPK2, but in response to different intracellular signaling pathways. 32244500_miR-1285-3p Controls Colorectal Cancer Proliferation and Escape from Apoptosis through DAPK2. 34298122_Cigarette smoking induces aberrant N(6)-methyladenosine of DAPK2 to promote non-small cell lung cancer progression by activating NF-kappaB pathway. 34413451_14-3-3 proteins inactivate DAPK2 by promoting its dimerization and protecting key regulatory phosphosites. | ENSMUSG00000032380 | Dapk2 | 3.734378e+01 | 1.8287689 | 0.870872761 | 0.4979961 | 3.107037e+00 | 0.0779546656 | 0.72626643 | No | Yes | 2.816655e+01 | 7.632030 | 1.780902e+01 | 4.932377 | |
| ENSG00000035681 | 8439 | NSMAF | protein_coding | Q92636 | FUNCTION: Couples the p55 TNF-receptor (TNF-R55 / TNFR1) to neutral sphingomyelinase (N-SMASE). Specifically binds to the N-smase activation domain of TNF-R55. May regulate ceramide production by N-SMASE. | Alternative splicing;Reference proteome;Repeat;WD repeat | This gene encodes a WD-repeat protein that binds the cytoplasmic sphingomyelinase activation domain of the 55kD tumor necrosis factor receptor. This protein is required for TNF-mediated activation of neutral sphingomyelinase and may play a role in regulating TNF-induced cellular responses such as inflammation. Alternative splicing results in multiple transcript variants.[provided by RefSeq, Jan 2009]. | hsa:8439; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; sphingomyelin phosphodiesterase activator activity [GO:0016230]; ceramide metabolic process [GO:0006672]; positive regulation of apoptotic process [GO:0043065]; signal transduction [GO:0007165] | 12391233_The interaction of FAN with receptor for activated C-kinase 1 (RACK1) appears to be dependent on the folding of the WD repeats into a secondary structure, because no linear binding motifs are identified in the WD-repeat region of FAN. 15653433_sphingolipid activator proteins like domain may stabilize the fold of acid sphingomyelinase 18653803_Regulation of NSMAF by TNF-alpha involves PRKCD in lung epithelial cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 29115933_The rs1050504 C > T genotype was observed to be significantly associated with an increased risk for developing pulmonary tuberculosis. 29621545_these data identify a novel caspase-2-interacting factor, FAN, and expand the role for the enzyme in seemingly non-apoptotic cellular mechanisms. | ENSMUSG00000028245 | Nsmaf | 3.615213e+02 | 0.6866149 | -0.542426884 | 0.3329039 | 2.628963e+00 | 0.1049302201 | 0.75783482 | No | Yes | 3.009416e+02 | 51.444030 | 4.630638e+02 | 80.734906 | |
| ENSG00000037241 | 51121 | RPL26L1 | protein_coding | Q9UNX3 | Direct protein sequencing;Isopeptide bond;Reference proteome;Ribonucleoprotein;Ribosomal protein;Ubl conjugation | This gene encodes a protein that shares high sequence similarity with ribosomal protein L26. Alternative splicing results in multiple transcript variants encoding the same protein. [provided by RefSeq, Dec 2015]. | hsa:51121; | cytosolic large ribosomal subunit [GO:0022625]; extracellular exosome [GO:0070062]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; cytoplasmic translation [GO:0002181]; ribosomal large subunit biogenesis [GO:0042273] | 1.027141e+03 | 0.8022079 | -0.317951863 | 0.2727353 | 1.352889e+00 | 0.2447736988 | 0.78892886 | No | Yes | 9.073460e+02 | 108.247344 | 1.084539e+03 | 132.322485 | |||||
| ENSG00000038219 | 259282 | BOD1L1 | protein_coding | Q8NFC6 | FUNCTION: Component of the fork protection machinery required to protect stalled/damaged replication forks from uncontrolled DNA2-dependent resection. Acts by stabilizing RAD51 at stalled replication forks and protecting RAD51 nucleofilaments from the antirecombinogenic activities of FBH1 and BLM (PubMed:26166705, PubMed:29937342). Does not regulate spindle orientation (PubMed:26166705). {ECO:0000269|PubMed:26166705, ECO:0000269|PubMed:29937342}. | Acetylation;Chromosome;DNA damage;DNA repair;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:259282; | centrosome [GO:0005813]; nucleoplasm [GO:0005654]; outer kinetochore [GO:0000940]; spindle microtubule [GO:0005876]; spindle pole [GO:0000922]; protein phosphatase 2A binding [GO:0051721]; protein phosphatase inhibitor activity [GO:0004864]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; negative regulation of phosphoprotein phosphatase activity [GO:0032515]; replication fork processing [GO:0031297] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 26166705_Results show that BOD1L protects against severe genome instability caused by replication stress; it maintains fork stability by stabilizing RAD51 nucleofilaments. 26889944_Protection or resection: BOD1L as a novel replication fork protection factor. | ENSMUSG00000061755 | Bod1l | 9.376479e+02 | 1.0530628 | 0.074591543 | 0.3355076 | 4.952759e-02 | 0.8238873587 | 0.96654947 | No | Yes | 1.161826e+03 | 267.772588 | 9.182206e+02 | 217.225522 | ||
| ENSG00000038358 | 23644 | EDC4 | protein_coding | Q6P2E9 | FUNCTION: In the process of mRNA degradation, seems to play a role in mRNA decapping. Component of a complex containing DCP2 and DCP1A which functions in decapping of ARE-containing mRNAs. Promotes complex formation between DCP1A and DCP2. Enhances the catalytic activity of DCP2 (in vitro). {ECO:0000269|PubMed:16364915}. | Acetylation;Alternative splicing;Coiled coil;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:23644; | cytoplasm [GO:0005737]; cytoplasmic ribonucleoprotein granule [GO:0036464]; cytosol [GO:0005829]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; P-body [GO:0000932]; deadenylation-independent decapping of nuclear-transcribed mRNA [GO:0031087] | 22982864_EDC4 might contribute to regulation of CoA biosynthesis in addition to its scaffold function in processing bodies 24510189_The data indicates that DCP2 activation by DCP1 occurs preferentially on the EDC4 scaffold, which may serve to couple DCP2 activation by DCP1 with 5'-to-3' mRNA degradation by XRN1 in human cells. 24755989_LMKB is the first protein identified to date that interacts with this portion of Ge-1. LMKB was expressed in human B and T lymphocyte cell lines; depletion of LMKB increased expression of IFI44L. 24858563_EDC4 is a processing body (P-body) component. 25970328_The assembly of EDC4 and Dcp1a into processing bodies is critical for the translational regulation of IL-6. 29511213_EDC4 plays a key role in homologous recombination by stimulating end resection at double-strand breaks. 32510323_A non-canonical role for the EDC4 decapping factor in regulating MARF1-mediated mRNA decay. 33054858_Enhancer of mRNA Decapping protein 4 (EDC4) interacts with replication protein a (RPA) and contributes to Cisplatin resistance in cervical Cancer by alleviating DNA damage. | ENSMUSG00000036270 | Edc4 | 3.621383e+03 | 1.1602924 | 0.214488368 | 0.2807008 | 5.845137e-01 | 0.4445485326 | 0.85798841 | No | Yes | 3.150982e+03 | 330.150981 | 2.852914e+03 | 307.051771 | ||
| ENSG00000040341 | 27067 | STAU2 | protein_coding | Q9NUL3 | FUNCTION: RNA-binding protein required for the microtubule-dependent transport of neuronal RNA from the cell body to the dendrite. As protein synthesis occurs within the dendrite, the localization of specific mRNAs to dendrites may be a prerequisite for neurite outgrowth and plasticity at sites distant from the cell body (By similarity). {ECO:0000250|UniProtKB:Q68SB1}. | Alternative splicing;Cytoplasm;Endoplasmic reticulum;Microtubule;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Transport | Staufen homolog 2 is a member of the family of double-stranded RNA (dsRNA)-binding proteins involved in the transport and/or localization of mRNAs to different subcellular compartments and/or organelles. These proteins are characterized by the presence of multiple dsRNA-binding domains which are required to bind RNAs having double-stranded secondary structures. Staufen homolog 2 shares 48.5% and 59.9% similarity with drosophila and human staufen, respectively. The exact function of Staufen homolog 2 is not known, but since it contains 3 copies of conserved dsRNA binding domain, it could be involved in double-stranded RNA binding events. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2009]. | hsa:27067; | axon [GO:0030424]; cytoplasmic stress granule [GO:0010494]; dendrite cytoplasm [GO:0032839]; dendritic shaft [GO:0043198]; endoplasmic reticulum [GO:0005783]; membrane [GO:0016020]; messenger ribonucleoprotein complex [GO:1990124]; microtubule [GO:0005874]; neuronal cell body [GO:0043025]; nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; double-stranded RNA binding [GO:0003725]; Hsp70 protein binding [GO:0030544]; kinesin binding [GO:0019894]; mitogen-activated protein kinase binding [GO:0051019]; ribosome binding [GO:0043022]; RNA binding [GO:0003723]; anterograde dendritic transport of messenger ribonucleoprotein complex [GO:0098964]; cellular response to oxidative stress [GO:0034599]; eye morphogenesis [GO:0048592]; positive regulation of dendritic spine morphogenesis [GO:0061003]; positive regulation of long-term synaptic depression [GO:1900454]; positive regulation of synapse assembly [GO:0051965]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of filopodium assembly [GO:0051489]; regulation of gene expression [GO:0010468] | 12140260_Data show that Stau2 is localized to the neuronal soma and dendrites, but it does not colocalize with Stau1-containing particles. 18094122_Stau1- and Stau2-mRNPs associate with distinct but overlapping sets of cellular mRNAs. 21508097_We suggest a role for Stau2 in the generation and regulation of Map1b mRNA containing granules that are required for mGluR-long-term depression 23263869_Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. 24520823_Authors establish that human Staufen-2, a host factor which is up-regulated upon HIV-1 infection, interacts with HIV-1 Rev, thereby promoting its RNA export activity and progeny virus formation. 26843428_during genotoxic stress, Stau2 is downregulated at the promoter level in an ATR- and E2F1-dependent manner, leading to increased levels of DNA damage and apoptosis. 33441653_Quantitative STAU2 measurement in lymphocytes for breast cancer risk assessment. 33663378_STAU2 protein level is controlled by caspases and the CHK1 pathway and regulates cell cycle progression in the non-transformed hTERT-RPE1 cells. 34960728_Encapsidation of Staufen-2 Enhances Infectivity of HIV-1. | ENSMUSG00000025920 | Stau2 | 3.940098e+02 | 1.0676395 | 0.094424654 | 0.3495594 | 7.313943e-02 | 0.7868194403 | 0.95754084 | No | Yes | 4.301755e+02 | 80.049868 | 3.608153e+02 | 68.922049 | |
| ENSG00000040487 | 54896 | SLC66A1 | protein_coding | Q6ZP29 | FUNCTION: Amino acid transporter that specifically mediates the pH-dependent export of the cationic amino acids arginine, histidine and lysine from lysosomes. {ECO:0000269|PubMed:22822152, ECO:0000269|PubMed:23169667}. | Alternative splicing;Amino-acid transport;Glycoprotein;Lysosome;Membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | hsa:54896; | integral component of membrane [GO:0016021]; integral component of organelle membrane [GO:0031301]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal membrane [GO:0005765]; arginine transmembrane transporter activity [GO:0015181]; basic amino acid transmembrane transporter activity [GO:0015174]; L-lysine transmembrane transporter activity [GO:0015189]; amino acid homeostasis [GO:0080144]; arginine transport [GO:0015809]; lysine transport [GO:0015819]; transmembrane transport [GO:0055085] | 23169667_PQLC2 and Ypq1-3 proteins are lysosomal/vacuolar exporters of CAAs and suggest that small-molecule transport is a conserved feature of the PQ-loop protein family 30729615_knockdown caused growth arrest and cell death of cancer cells and suppressed tumor growth in a mouse xenograft model. These results suggest that targeting PQLC2 is an effective strategy for GC treatment. 33597295_Receptor-like role for PQLC2 amino acid transporter in the lysosomal sensing of cationic amino acids. | ENSMUSG00000028744 | Slc66a1 | 1.286153e+03 | 1.1757430 | 0.233572724 | 0.3188671 | 5.330369e-01 | 0.4653328580 | 0.86741238 | No | Yes | 1.278945e+03 | 160.088617 | 9.798896e+02 | 126.223814 | ||
| ENSG00000048028 | 57646 | USP28 | protein_coding | Q96RU2 | FUNCTION: Deubiquitinase involved in DNA damage response checkpoint and MYC proto-oncogene stability. Involved in DNA damage induced apoptosis by specifically deubiquitinating proteins of the DNA damage pathway such as CLSPN. Also involved in G2 DNA damage checkpoint, by deubiquitinating CLSPN, and preventing its degradation by the anaphase promoting complex/cyclosome (APC/C). In contrast, it does not deubiquitinate PLK1. Specifically deubiquitinates MYC in the nucleoplasm, leading to prevent MYC degradation by the proteasome: acts by specifically interacting with isoform 1 of FBXW7 (FBW7alpha) in the nucleoplasm and counteracting ubiquitination of MYC by the SCF(FBW7) complex. In contrast, it does not interact with isoform 4 of FBXW7 (FBW7gamma) in the nucleolus, allowing MYC degradation and explaining the selective MYC degradation in the nucleolus. Deubiquitinates ZNF304, hence preventing ZNF304 degradation by the proteasome and leading to the activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) in a subset of colorectal cancers (CRC) cells (PubMed:24623306). {ECO:0000269|PubMed:16901786, ECO:0000269|PubMed:17558397, ECO:0000269|PubMed:17873522, ECO:0000269|PubMed:18662541, ECO:0000269|PubMed:24623306}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;Hydrolase;Isopeptide bond;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation;Ubl conjugation pathway | The protein encoded by this gene is a deubiquitinase involved in the DNA damage pathway and DNA damage-induced apoptosis. Overexpression of this gene is seen in several cancers. [provided by RefSeq, Oct 2016]. | hsa:57646; | cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; cysteine-type endopeptidase activity [GO:0004197]; thiol-dependent deubiquitinase [GO:0004843]; cell population proliferation [GO:0008283]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to UV [GO:0034644]; DNA damage checkpoint signaling [GO:0000077]; DNA repair [GO:0006281]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; protein deubiquitination [GO:0016579]; Ras protein signal transduction [GO:0007265]; regulation of protein stability [GO:0031647]; response to ionizing radiation [GO:0010212]; ubiquitin-dependent protein catabolic process [GO:0006511] | 11597335_molecular cloning of USP28 & characterization of alternatively spliced products and tissue-specific isoforms 16901786_Using a human cell line that faithfully recapitulated the Chk2-p53-PUMA pathway, we show that USP28 is required to stabilize Chk2 and 53BP1 in response to DNA damage. 17558397_High expression levels of USP28 are found in colon and breast carcinomas, and stabilization of MYC by USP28 is essential for tumour-cell proliferation. 17873522_Usp28 dissociates from Fbw7alpha in response to UV irradiation, providing a mechanism how Fbw7-mediated degradation of Myc is enhanced upon DNA damage. 22144179_A new pathway that could be targeted at the level of GSK-3, Fbw7, or USP28 to influence HIF-1alpha-dependent processes like angiogenesis and metastasis. 23832602_USP28 gene expression is down regulated by oxidative stress through the mediation of reactive oxygen species 24075993_Study reveals a critical mechanism underlying the epigenetic regulation by USP28. 24347490_Usp28 expression was indentified as a independent predictors of survival (P = 0.001) and potentially valuable in prognostic evaluation of bladder cancer 24687851_USP28 is not a critical factor in double-strand break metabolism and is unlikely to be an attractive target for therapeutic intervention aimed at chemotherapy sensitization. 24960159_identified Usp28 as a c-MYC target gene highly expressed in colorectal cancers, which indicates that USP28 and c-MYC form a positive feedback loop that maintains high c-MYC protein levels in tumors 25359778_USP28 has a chain preference activity for Lys(11), Lys(48), and Lys(63) diubiquitin linkages 25437563_Dual regulation of Fbw7 activity by Usp28 is a safeguard mechanism for maintaining physiological levels of proto-oncogenic Fbw7 substrates, which is equivalently disrupted by loss or overexpression of Usp28. 25656529_Data indicte that deubiquitinating enzyme USP28 was targeted by microRNA miR-4295. 26209720_These results showed that USP28 is overexpressed in human glioblastomas and it contributes to glioma tumorigenicity. 26268556_findings provide a first insight into understanding how the enzymatic activity of Usp28 is regulated by its non-catalytic UBR and endogenous ligands. 27371829_The authors identified 53BP1 and USP28 as essential components acting upstream of p53, evoking p21-dependent cell cycle arrest in response not only to centrosome loss, but also to other distinct defects causing prolonged mitosis. 27432896_USP28-53BP1-p53-p21 signaling pathway is also required to arrest cell growth after a prolonged prometaphase. 27432897_analysis of centrinone resistance identified a 53BP1-USP28 module as critical for communicating mitotic challenges to the p53 circuit and TRIM37 as an enforcer of the singularity of centrosome assembly. 27546791_53BP1-USP28 cooperation is essential for normal p53-promoter element interactions and gene transactivation-associated events, yet dispensable for 53BP1-dependent DNA double-strand repair regulation. 29089421_USP28 controls activation of both the TP53 branch and the GATA4/NFkB branch that controls the senescence-associated secretory phenotype (SASP) 29545478_Lack of USP28 promotes a more malignant state of breast cancer cells, indicated by an epithelial-to-mesenchymal (EMT) transition, elevated proliferation, migration, and angiogenesis as well as a decreased adhesion. 29880484_USP28 enhances MAPK activity through the stabilization of RAF family members and is a key factor in BRAF inhibitor resistance. 30206969_Knockdown of USP28 enhanced the radiosensitivity of esophageal cancer cells by destabilizing c-Myc and enhancing the accumulation of HIF-1alpha. Therefore, USP28 may serve as a novel therapeutic target to overcome esophageal cancer radioresistance. 30543854_consequential impacts of USP28-mediated stabilization of LIN28A protein on enhancing cancer cell viability, migration and ultimately augmenting LIN28A-mediated tumor progression. Overall, these data suggest that a synergistic, combinatorial approach of targeting LIN28A with USP28 would contribute to effective cancer therapeutics. 30910399_The effect of USP28 on cell proliferation was mediated by regulating the expression of p53, p21 and p16(INK4a). 30926242_We confirm oligomeric states of USP25 and USP28 in cells and show that modulating oligomerization affects substrate stabilization in accordance with in vitro activity data 30926243_Our work led to the identification of significant differences between USP25 and USP28 and provided the molecular basis for the development of new and highly specific anti-cancer drugs 31604991_The MTH1 inhibitor TH588 is a microtubule-modulating agent that eliminates cancer cells by activating the mitotic surveillance pathway. 31938050_Deubiquitinase USP28 inhibits ubiquitin ligase KLHL2-mediated uridine-cytidine kinase 1 degradation and confers sensitivity to 5'-azacytidine-resistant human leukemia cells. 31982308_A non-canonical role of caspase-8 exploited by cancer cells to override the p53-dependent G2/M cell-cycle checkpoint through cleavage of USP28. 32053284_MicroRNA-216b suppresses the cell growth of hepatocellular carcinoma by inhibiting Ubiquitin-specific peptidase 28 expression. 32128997_Maintaining protein stability of Np63 via USP28 is required by squamous cancer cells. 32578360_USP28 and USP25 are downregulated by Vismodegib in vitro and in colorectal cancer cell lines. 33664871_Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. 34106567_USP28 promotes aerobic glycolysis of colorectal cancer by increasing stability of FOXC1. 34584067_USP28 facilitates pancreatic cancer progression through activation of Wnt/beta-catenin pathway via stabilising FOXM1. 34962618_Loss of USP28 and SPINT2 expression promotes cancer cell survival after whole genome doubling. | ENSMUSG00000032267 | Usp28 | 7.661708e+02 | 0.7602928 | -0.395372924 | 0.3231584 | 1.449953e+00 | 0.2285355103 | 0.78792184 | No | Yes | 7.262021e+02 | 122.570128 | 9.328009e+02 | 161.286073 | |
| ENSG00000049768 | 50943 | FOXP3 | protein_coding | Q9BZS1 | FUNCTION: Transcriptional regulator which is crucial for the development and inhibitory function of regulatory T-cells (Treg) (PubMed:17377532, PubMed:21458306, PubMed:30513302, PubMed:23947341, PubMed:24354325, PubMed:24722479, PubMed:24835996). Plays an essential role in maintaining homeostasis of the immune system by allowing the acquisition of full suppressive function and stability of the Treg lineage, and by directly modulating the expansion and function of conventional T-cells (PubMed:23169781). Can act either as a transcriptional repressor or a transcriptional activator depending on its interactions with other transcription factors, histone acetylases and deacetylases (PubMed:17377532, PubMed:21458306, PubMed:23947341, PubMed:24354325, PubMed:24722479). The suppressive activity of Treg involves the coordinate activation of many genes, including CTLA4 and TNFRSF18 by FOXP3 along with repression of genes encoding cytokines such as interleukin-2 (IL2) and interferon-gamma (IFNG) (PubMed:17377532, PubMed:21458306, PubMed:23947341, PubMed:24354325, PubMed:24722479). Inhibits cytokine production and T-cell effector function by repressing the activity of two key transcription factors, RELA and NFATC2 (PubMed:15790681). Mediates transcriptional repression of IL2 via its association with histone acetylase KAT5 and histone deacetylase HDAC7 (PubMed:17360565). Can activate the expression of TNFRSF18, IL2RA and CTLA4 and repress the expression of IL2 and IFNG via its association with transcription factor RUNX1 (PubMed:17377532). Inhibits the differentiation of IL17 producing helper T-cells (Th17) by antagonizing RORC function, leading to down-regulation of IL17 expression, favoring Treg development (PubMed:18368049). Inhibits the transcriptional activator activity of RORA (PubMed:18354202). Can repress the expression of IL2 and IFNG via its association with transcription factor IKZF4 (By similarity). {ECO:0000250|UniProtKB:Q99JB6, ECO:0000269|PubMed:15790681, ECO:0000269|PubMed:17360565, ECO:0000269|PubMed:17377532, ECO:0000269|PubMed:18354202, ECO:0000269|PubMed:18368049, ECO:0000269|PubMed:21458306, ECO:0000269|PubMed:23169781, ECO:0000269|PubMed:24835996, ECO:0000269|PubMed:30513302, ECO:0000303|PubMed:23947341, ECO:0000303|PubMed:24354325, ECO:0000303|PubMed:24722479}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cytoplasm;DNA-binding;Diabetes mellitus;Disease variant;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a member of the forkhead/winged-helix family of transcriptional regulators. Defects in this gene are the cause of immunodeficiency polyendocrinopathy, enteropathy, X-linked syndrome (IPEX), also known as X-linked autoimmunity-immunodeficiency syndrome. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:50943; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; histone acetyltransferase binding [GO:0035035]; histone deacetylase binding [GO:0042826]; metal ion binding [GO:0046872]; NF-kappaB binding [GO:0051059]; NFAT protein binding [GO:0051525]; protein homodimerization activity [GO:0042803]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription corepressor activity [GO:0003714]; B cell homeostasis [GO:0001782]; CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment [GO:0002362]; chromatin remodeling [GO:0006338]; establishment of endothelial blood-brain barrier [GO:0014045]; gene expression [GO:0010467]; myeloid cell homeostasis [GO:0002262]; negative regulation of activated T cell proliferation [GO:0046007]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of chronic inflammatory response [GO:0002677]; negative regulation of CREB transcription factor activity [GO:0032792]; negative regulation of cytokine production [GO:0001818]; negative regulation of defense response to virus [GO:0050687]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of histone acetylation [GO:0035067]; negative regulation of histone deacetylation [GO:0031064]; negative regulation of immune response [GO:0050777]; negative regulation of interferon-gamma production [GO:0032689]; negative regulation of interleukin-10 production [GO:0032693]; negative regulation of interleukin-17 production [GO:0032700]; negative regulation of interleukin-2 production [GO:0032703]; negative regulation of interleukin-4 production [GO:0032713]; negative regulation of interleukin-5 production [GO:0032714]; negative regulation of interleukin-6 production [GO:0032715]; negative regulation of isotype switching to IgE isotypes [GO:0048294]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; negative regulation of T cell cytokine production [GO:0002725]; negative regulation of T cell proliferation [GO:0042130]; negative regulation of T-helper 17 cell differentiation [GO:2000320]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of tumor necrosis factor production [GO:0032720]; positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation [GO:0032831]; positive regulation of histone acetylation [GO:0035066]; positive regulation of immature T cell proliferation in thymus [GO:0033092]; positive regulation of interleukin-4 production [GO:0032753]; positive regulation of peripheral T cell tolerance induction [GO:0002851]; positive regulation of regulatory T cell differentiation [GO:0045591]; positive regulation of T cell anergy [GO:0002669]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of transforming growth factor beta1 production [GO:0032914]; regulation of isotype switching to IgG isotypes [GO:0048302]; regulation of T cell anergy [GO:0002667]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; response to lipopolysaccharide [GO:0032496]; response to radiation [GO:0009314]; response to rapamycin [GO:1901355]; response to virus [GO:0009615]; T cell activation [GO:0042110]; T cell homeostasis [GO:0043029]; T cell mediated immunity [GO:0002456]; T cell receptor signaling pathway [GO:0050852]; tolerance induction to self antigen [GO:0002513] | 11483607_The ability of scurfin to bind DNA, and presumably repress transcription, plays a paramount role in determining the amplitude of the response of CD4 T cells to activation. 11768393_novel mutations of FOXP3 in two Japanese patients with immune dysregulation, polyendocrinopathy, enteropathy, X linked syndrome (IPEX) 12296863_Random X inactivation was found in the T lymphocytes of a carrier of a FOXP3 mutation (Ala384Thr)with an affected son. 12750858_Observational study of gene-disease association. (HuGE Navigator) 14597769_FoxP3 is expressed predominantly in Tr cells and that it may serve as a master regulator of this cell population. 14671208_characterization of the underlying FOXP3 mutation in two families with features consistent with IPEX; analysis elucidated the molecular basis of IPEX in one family and provided evidence for an autosomal locus, suggesting genetic heterogeneity 14997560_impact of disease-causing missense mutations on the three-dimensional structure, stability, and surface electrostatic charge distribution of the forkhead domains is examined 15100250_TGF-beta-mediated induction of Foxp3 in CD4+CD25- T cells results in the development of T cells with regulatory functions. 15172973_Foxp3 regulatory gene expression may have a role in graft-versus-host disease 15220219_Allelic variation in or near the coding regions of the FOXP3 gene does not have a major role in the inherited susceptibility to the common form of type 1 diabetes. 15220219_Observational study of gene-disease association. (HuGE Navigator) 15246158_Human cord blood CD25+CD4+ T cells preferentially transcribed Foxp3, which governs the regulatory function of this subset i 15374887_FoxP3 protein, but not mRNA, was specifically expressed by cord blood CD4-CD25 derived T-cells 15466453_FOXP3 is a crucial regulatory gene for the development and function of CD25(+)CD4(+) regulatory T cells 15620457_FOXP3 expression in humans, unlike mice, may not be specific for cells with a regulatory phenotype and may be only a consequence of activation status. 15674359_The relative expression of FOXP3 protein mrNA was estimated by the ratio of the Foxp3 value to the HPRT value. 15952173_Multiple sclerosis patients have abnormalities in FOXP3 message and protein expression levels in peripheral CD4+ CD25+ T cells that are quantitatively related to a reduction in functional suppression induced during suboptimal T-cell receptor ligation. 15972448_Gene expression of regulatory T cells transcription factor FOXP3 was reduced in chronic graft-versus-host disease patients 16003241_FOXP3 mRNA expression levels in allostimulated CD25 cells and CD25 cells and in peripheral blood mononuclear cells. 16091206_Initiation and suppressive function of FOXP3 and regulatory T cells in the context of allergic asthma. Review. 16211090_Ectopic expression of FOXP3 in human CD4+ T cells does not act as a molecular switch to induce regulatory T cells in vitro 16278306_The determination of FOXP3+/CD8+ ratio can be a helpful tool to discriminate graft versus hosst disease from infectious inflammation after allogeneic stem cell transplantation. 16322292_High expression levels of FoxP3 is associated with ovarian cancer 16339542_Costimulation of effector T cells with anti-CD3 and TLR5 ligand flagellin resulted in enhanced proliferation and production of IL-2, and potently increased their suppressive capacity and enhanced expression of FOXP3. 16339919_Increased expression of interleukin-10 and transforming growth factor-beta1 mRNA was also detected in patients with TB but did not correlate with regulatory T-cell markers. 16368541_analysis of transcriptional regulation by FOXP3 [review] 16410445_Increase of functional FOXP3 in T cells in cancer patients is a response to the process of malignant transformation. 16493082_Fox3p expression is inherent feature of the function of type 1 regulatory T cells specific for desmoglein-3, the autoantigen of pemphigus vulgaris. 16517728_reveals the structure of the human FOXP3 promoter and provides new insights in mechanisms of addressing T regulatory cell-inducing signals useful for promoting immune tolerance 16551363_Increased IL-10 and Foxp3 induction in cord blood mononuclear cells of non-atopic compared to atopic mothers, may indicate an increased capacity to respond to microbial stimuli. 16557241_The data support the conclusion that the neoplastic cells in CTCL do not express the T(reg) marker FOXP3. 16574699_The finding of reduced endometrial Foxp3 implicates impaired differentiation of uterine T cells into the Treg phenotype as a key determinant of fertility in women. 16583400_These data are the first to demonstrate differences in function and FOXP3 expression of CD4(+)CD25(+) T cells from patients with RR- and SP-MS. 16596204_Expression of Foxp3 in non-small cell lung cancer patients is significantly higher in tumor tissues than in normal tissues, especially in tumors smaller than 30 mm 16617117_Most patients lacked FOXP3-expressing cells, further solidifying the association between FOXP3 deficiency and immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. 16645171_IL-2 treatment resulted in anincrease in FOXP3 expression in CD3+ T cells. CD56+CD3- natural killer (NK) cells in cancer. 16652285_HTLV-1-associated myelopathy/tropical spastic paraparesis was associated with lower expression of Foxp3 in leukocytes; suggests impaired Foxp3 expression may contribute to development of inflammatory disease in HTLV-1 infection 16728694_Regulatory T cells (T(regs)) containing FOXP3+ were increased in HIV patients after HAART therapy. 16741580_These findings indicate that forkhead box protein P3 mutations in immune dysregulation, polyendocrinopathy, enteropathy, X-linked patients result in heterogeneous biological abnormalities, leading to a dysfunction in regulatory and effector T cells. 16764698_Measurement of mRNA expression levels of specific marker FOXP3 in synovial fluid of rheumatic joints confirms the presence of regulatory CD25(bright)CD4+ T cells. 16818738_The orderly regulation of trafficking receptors in FOXP3-positive T cells according to stages of differentiation and activation is potentially important for their tissue-specific migration and regulation of immune responses in humans. 16825494_Prognostic significance of FOXP3-positive regulatory T cells in overall survival in follicular lymphoma. 16881731_Differences in the level of FOXP3 expression in the conjunctiva may indicate a role for regulatory T cells in the resolution of the conjunctival immune response, which is important in protection from immunopathology of trachoma. 16901927_Observational study of gene-disease association. (HuGE Navigator) 16920951_polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) mutations in these domains correlate with deficiencies in FOXP3 repressor function, corroborating their in vivo relevance 16955142_CD4+ CD45RO+ Foxp3+ CD25hi T lymphocytes are highly proliferative, with a doubling time of 8 days, compared with memory CD4+ CD45RO+ Foxp3- CD25- (24 days). 16956389_Here we show that levels of Foxp3 mRNA are increased in CD4+ T cells from HIV-infected patients responding to antiretroviral therapy. 16996248_Observational study of gene-disease association. (HuGE Navigator) 17028180_Foxp3 may regulate transcription through direct chromatin remodeling, and Foxp3 function is influenced by signals from the TCR 17033038_Samples from the long-survival group were more likely than those from the short-survival group to have CD4+ staining cells and to have FOXP3-positive cells in a perifollicular location. 17154262_Expression of endogenous FOXP3 in humans is not sufficient to induce regulatory T cell activity or to identify regulatory T (Treg) cells. 17161353_Data showed that autologous bone marrow graft recipients whithout graft versus host disease (GVHD) had significantly higher FOXP3 compared to those with GVHD. 17175222_assessment of Foxp3 mRNA inside an allograft may distinguish vascular from nonvascular rejection. 17183612_In this review, a new role is suggested for FOXP3 in human CD4+ T cells: down-modulating responses to T cell receptor-mediated stimulation. 17216339_Heme oxygenase-1 mRNA expression is linked to the induction of Foxp3 in CD4+ CD25+ glioma infiltrating regulatory T-cells. 17230494_Mucosal Foxp3 mRNA levels and Foxp3+CD3+ cells were significantly reduced in transplant recipients using prednisone/azathioprine/tacrolimus compared with controls but no direct relationship between Foxp3 expression and 1 specific drug was detected. 17262084_Abundant intraepithelial infiltrations of regulatory T cells with very high levels of Foxp3 expression was detected in five primary tumours. 17286616_Regulatory T cells concentrate in tubules in kidney transplantation, contributing to FOXP3 mRNA in urine. 17289884_FOXP3+ regulatory T cells affect the development and progression of hepatocarcinogenesis 17299718_BCG vaccine-receiving tuberculosis patients showed increase in FOXP3 mRNA levels. 17309822_The current study suggests that activated NK interfere with CD28-mediated Foxp3 protein and RNA expression in CD4(+)CD25(-) T lymphocytes. 17311282_regulatory roles of T-cell receptor (TCR), co-stimulatory molecules, interleukin-2 (IL-2), transforming growth factor-beta (TGF-beta) and beyond pathways on Foxp3 expression 17327427_This study reports that altered peripheral blood frequencies of Tregs, as defined by the expression of FOXP3, are not specifically associated with type 1 diabetes. 17329235_Expression of FOXP3 is a normal consequence of CD4(+) T cell activation and can no longer be used as an exclusive marker of T regulatory cells. 17360565_FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression 17377532_In natural T(R) cells, Foxp3 interacts physically with AML1 17378693_The immunohistochemistry of FOXP3 is used in the rapid screening for IPEX syndrome. 17389235_implicate Interleukin-10-producing CD25(-)Foxp3(-) T cells in the pathogenesis of human visceral leishmaniasis 17393103_Analysis of cd4+ CD25+ regulatory T cells and FOXP3 mRNA in the peripheral blood of patients with asthma. 17414320_These findings indicate that induction of FOXP3 is intrinsic to CD8 T cells that are activated in the presence of IL-2 or IL-15. 17414718_Intrahepatic FOXP3 levels are associated with HCV reinfection, a history of acute rejection, and increased within the first year after liver transplantation. 17418529_Observational study of gene-disease association. (HuGE Navigator) 17418529_results suggest that polymorphisms of the FOXP3 gene may play a role in the genetic susceptibility to autoimmune thyroid diseases in Caucasians, perhaps by altering FOXP3 function and/or expression 17445472_In Dermatophagoides farinae-stimulated PBMCs from patients with asthma, expression of GATA-3 and T-bet and FOPX3 expression is decreased. 17463169_NFAT1 could explain low FOXP3 expression and diminished Treg frequency in aplastic anemia. 17508019_Phenotypic Foxp3+ Tregs, in conjunction with immature dendritic cells and Th2 cytokines, suggests an attenuated state of immunity to human basal cell carcinoma. 17526924_Observational study of gene-disease association. (HuGE Navigator) 17526924_study failed to find an association between SNPs in the FOXP3 gene region and Juvenile Idiopathic Arthritis 17565321_Posttransplantation, the FOXP3 mRNA levels were higher in endomyocardial biopsies assigned to a higher International Society for Heart and Lung Transplantation rejection grade. 17570480_Our data demonstrate that FOXP3 is an X-linked breast cancer suppressor gene and an important regulator of the HER-2/ErbB2 oncogene. 17574040_Exposure of peripheral blood mononuclear cells to melanoma results in induction of FOXP3(+) T(reg) cells. 17589345_Peritransplanat donor vertebral body bone marrow cells have higher 10-year renal transplant acceptance, chimerism, and FoxP3 mRNA in unclarified regulatory cell phenotypes. 17612516_This study showed no association between microsatellite polymorphism in the promoter region of FOXP3 gene and susceptibility to type 1 diabetes in the Japanese population, though examination in larger samples is preferred to confirm this result. 17615291_reveal a novel role for non-Hodgkin lymphoma B cells in the development of intratumoral regulatory T cells 17641056_The T regulatory (Treg) cell-defining transcription factor FOXP3 in cooperation with NF-AT promotes TLR1- gene expression. 17644307_Observational study of gene-disease association. (HuGE Navigator) 17644307_We have identified the FOXP3 gene in juvenile Graves' disease. 17666212_Essential transcription factor not only for differentiation and function of naturally occurring Treg cells but also for regulation of intracellular molecules related to effector T-cell responses. Review. 17694575_FOXP3 DNA demethylation constitutes the most reliable criterion for natural regulatory T cells available at present 17703412_Observational study of gene-disease association. (HuGE Navigator) 17706604_analysis of FOXP3-transduced Jurkat-T cells as regulatory T (Treg)-like cells 17712427_Data suggest that lLow frequencies of Foxp3-Treg in all developmental stages of human plaque formation could explain the smoldering chronic inflammatory process that takes place throughout the longstanding course of atherosclerosis. 17712989_FOXP3 has a role in IPEX syndrome [review] 17712998_CD4+CD25+CTLA-4+FoxP3+ T cells have roles in patients with systemic lupus erythematosus and are impaired after conventional treatments 17713426_Regulatory T cells (Treg) suppress immune responses, making them an exciting therapeutic target for achieving operational transplant tolerance. FOXP3 is a master gene required for the development and function of Treg. 17804750_Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. 17851585_proportions of FOXP3-bearing lymphocytes were higher in vertical than in radial growth phase, as well as in late than in early melanoma stages 17878390_FOXP3+ CD4+ CD25(+high) regulatory T cells both in peripheral blood and in renal infiltrates were higher in donor-specific hyporesponders than in nonhyporesponders 17903368_Fibrosing cholestatic hepatitis has specific pathological characteristics which might be caused by high expression of FOXP3 in liver tissues. 17916446_patients with FOXP3 gene mutations revealed a paucity of CD4(+)CD25(+)FOXP3(+) T cells. 17921346_mechanism for IL-21-mediated expansion of antigen-specific CTLs that involves suppression of Foxp3-expressing cells and reversal of inhibition to tumor-associated antigen-specific cytotoxic T cell generation 17932340_FoxP3 as a biomarker should be explored for human tumors to enable physicians to make better decisions in oncologic care and to prepare the field for novel therapeutic agents directed at the elimination of regulatory T cells within tumors. 17954183_Foxp3+ cells can be detected in kidney transplant biopsies with acute rejection. 17970785_Despite the expression of CTLA-4 and Foxp3, tumors were incapable of suppressing the proliferation of autologous T8 cells. ATL cells are phenotypically Treg cells in at least some patients, but lack immunoregulatory functions toward T8 cells. 17975141_CD4+CD25highFoxp3+ T cells have a role in squamous cell carcinoma of the head and neck 18008005_downregulation of SKP2 was critical for FOXP3-mediated growth inhibition in breast cancer cells that do not overexpress ERBB2/HER2. 18024321_The ratio of FOXP3+ regulatory T-cells to granzyme B+ cytotoxic T/NK Cells predicts prognosis in classical Hodgkin lymphoma. 18034969_Our results show the presence of CD25 FoxP3 T regulatory cells in chronic atopic dermatitis lesions 18047933_in contrast to intragraft FOXP3 gene expression, the peripheral FOXP3 mRNA levels lack correlation with anti-donor immune responses in the graft. FOXP3 does not appear to be a candidate gene for non-invasive diagnosis of non-responsiveness or rejection. 18089323_CD8+FOXP3+ T cells play an important role in unresponsiveness related to upregulated interleukin receptors on dendritic cells. 18092263_There is an accumulation of FoxP3-expressing regulatory T cells in RA SF, and such recruitment may be dependent on the distinct chemokine receptors expressed on regulatory T cells. 18155891_Homozygous loss of function of foxp3 gene results in the absence of development of a crucial subpopulation of lymphocytes with CD4+CD25+ phenotype, called regulatory T-cells. [REVIEW] 18156149_LPS-activated monocytes suppress T cell immune responses and induce FOXP3 expression in resting CD4+CD25- T cells through a COX-2-PGE2-dependent mechanism. 18162042_interleukin 4 (IL-4) present at the time of T cell priming inhibits FOXP3 18171284_Forkhead transcription factor 3 (FoxP3) expression and T(reg) cell suppression were greater in patients with acute HCV infection , 6 months later, the resolution of disease was associated with a relative loss of functional suppression 18173798_FOXP3 expression was found in some CD4(+) T cells and CD25(+) cells but not in CD8(+) T cells. FOXP3(+) T cells in periodontitis lesions can be considered to be effector T cells. 18178814_TNreg cells are precursors for human Treg cells; these cells require a high level of FoxP3 expression to maintain their suppressive function. 18231910_higher percentages of FOXP3-positive Tregs on initial tumor biopsy evidenced a significantly longer OS 18240282_Tissue samples at different points showed a significant reduction in IL-1beta mRNA expression, and a significant increase in Foxp3 mRNA expression in ulcerative colitis. 18246047_FOXP3 expression in adult T-cell leukemia/lymphoma and reflects morphological features and is clinically and pathologically associated with an immunosuppressive state. 18261176_Campath-1H (Alemtuzumab) is an effective immunodepletion agent used in renal transplantation which increass FOXP3 regulatory T cells. 18270250_After stimulation of CD4+CD25- T cells, the cells express FOXP3 display potent immunosuppressive properties. 18270368_activation of STAT5 sustains FOXP3 expression in both Treg and Teff cells and contribute to our understanding of how cytokines affect the expression of FOXP3 18279718_After culture with mesenchymal stromal cells (MSCs) both immunoselected fractions registered increases in the CD4(+)/CD25(bright)/FoxP3 expression was downregulated. 18286169_The unique FOXP3 promoter methylation profile in Tregs suggests that a demethylated pattern is a prerequisite for stable FOXP3 expression and suppressive phenotype 18294387_B7-H1, PD-1 and FOXP3 may have roles in progression of breast neoplasms 18304876_Higher FoxP3 expression at diagnosis predicted worse future glycemic control while higher mean numbers of IL-10 cells were associated with better future glucose control. 18317533_Immunodysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome is a rare disorder caused by mutations in the FOXP3 gene 18324310_IL2/FOXP3-dependent negative feedback loop that normally regulates the T cell immune response 18331599_FOXP3 expression in cutaneous and systemic CD30-positive lymphoproliferations is generally confined to tumour infiltrating rgulatory T-lymphocytes 18354202_expand the possible targets of FOXP3-mediated repression and demonstrate functional differences between FOXP3 isoforms 18368049_the decision of antigen-stimulated cells to differentiate into either T(H)17 or T(reg) cells depends on the cytokine-regulated balance of RORgammat and Foxp3 18394345_The low expression of Foxp3 in CD4+CD25high Treg cells in cord blood suggests Treg cells in neonates are not mature in suppression. 18395862_FOXP3-independent mechanisms, mediated by IL-10, contribute to the induction and suppressor functions of type 1 regulatory T cells 18412171_Both Foxp3 isoforms possess similar capacities to induce Treg. However, unnaturally high expression levels are required to convey Treg functions to CD4(+)CD25(-) cells. 18413409_The level of expression of FOXP3 was normal in Treg cells from patients with SLE. 18413770_FoxP3, a key regulator of immune suppression, is not only expressed by regulatory T cells but also by melanoma cells, Epstein Barr virus-transformed B cells, and a wide variety of tumor cell lines. 18424697_Foxp3 can regulate pim 2 expression. 18430198_Foxp3 mRNA as well as Foxp3 protein was detected in all tumor cell lines, correlating with the expression levels of IL-10 and TGFb1 18434325_Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat 18496979_Although the FOXP3 gene polymorphism has no effect on susceptibility to sarcoidosis, the (GT)15 allele may exert protective effects against skin involvement. 18496979_Observational study of gene-disease association. (HuGE Navigator) 18510637_FOXP3 expression did not correlate with favorable graft outcomes, even when the analysis was restricted to biopsies with rejection 18510697_Foxp3 in normal T cells, but not mRNA, is basically potent at discriminating a subset of Treg cells from CD25(+) T-cell populations, in leukemic T cells could be implicated in oncogenesis and has a potentially useful clinical role. 18533240_The correlation between the presence of FOXP3(+) Tregs and high vessel density in endometrial adenocarcinomas suggests a link between immunity, intratumoral angiogenesis and poor prognosis. 18544681_FoxP3(+) Treg cells are productively infected and play an important role in acute HIV-1 infection in vivo 18579608_Preservation of FoxP3+ regulatory T cells in the peripheral blood of HIV-1-infected elite suppressors correlates with low CD4+ T-cell activation. 18606654_results, demonstrating that FOXP3 expression driven solely by the CD28/B7 interaction inhibited T cell activation, support the role of CD28 in the regulation of peripheral tolerance and suggest a new mechanism through which it could occur 18628982_GARP is a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression 18641303_IRF-1 is a key negative regulator of CD4(+)CD25(+) Treg cells through direct repression of Foxp3 expression 18641304_acquisition of suppressive behavior by activated CD4(+)CD25(-) T cells requires the expression of CTLA-4, a feature that appears to be facilitated by, but is not dependent on, expression of FoxP3 18665940_H. pylori-positive patients revealed a high induction of FOXP3 transcript which correlated positively with inflammation and TGF-beta1 levels. 18667728_Foxp3 has a role in preventing progression toward acute rejection of kidney transplants 18681861_There were fewer numbers of Foxp3+ T cells in the monoclonal variants of pityriasis lichenoides chronica and pigmented purpuric dermatosis. 18690669_The results from this study will allow the assessment of FoxP3 by flow cytometry on samples from clinical sites that are analyzed in real time on fresh blood or frozen PBMC. 18769452_malignant T cells express low molecular splice forms of FOXP3 and function as Tregs 18794055_FoxP3+ peripheral T-cell lymphomas not otherwise specified presumably derived from bona fide Treg cells occurs but seems rare in the Western population 18802341_Foxp3-positive Tregs detected in skin biopsies of atopic dermatitis patients: controversy 18818377_Foxp3+ T-regulatory cell frequency in the minor salivary glands lesions of Sjogren's syndrome patients correlates with inflammation grade and certain risk factors for lymphoma development. 18825388_The greatest upregulation of IL-23p19, Foxp3 and survivin mRNA was seen in colorectal carcinomas than normal mucosa. 18829063_FOXP3 decreases binding of NFAT2 to the HIV-1 LTR in vivo 18836525_Effects of natalizumab treatment on FOXP3 T regulatory cells is reported. 18844067_In active phase AA patients, the levels of FOXP3 and Notchl mRNA were lower than that in the control group. 18924611_The kinetics of FOXP3 T-cell accumulation during a human cutaneous antigen-specific memory response in vivo are reported. 18931102_FOXP3 sequencing should be performed in any male patient with the diagnosis of diabetes in the first 6 months who develops other possible autoimmune-associated conditions. 18941119_CD49d provides access to highly pure populations of untouched Foxp3(+) Treg cells conferring maximal safety for future clinical applications 18951619_FOXP3 protein expression did not correlate with disease severity. 18973208_Children with type 1 diabetes show decreased induction of FOXP3 in T cells. 19022313_The presence of Foxp3(+) cells may help to control the inflammatory process through the management of lymphocyte migration and, consequently, prevent neuronal destruction and chagasic megacolon development. 19034005_Evidence is provided that the increase of CD4+CD25 high T cells and FoxP3 transcripts is associated with operational tolerance in liver transplanted patients during immunosuppression withdrawal. 19039775_FOXP3 acts as a quantitative regulator rather than a simple molecular switch for Treg 19085184_Observational study of gene-disease association. (HuGE Navigator) 19085184_the (GT)n microsatelloite polymorphism in the FOXP3/Scrufin gene was associated with Japanese adult onset type 1 diabetes, especially in females, and slowly progressive type 1 diabetes. 19089920_cervical mRNA expression of IFN-gamma, IL-10, and IL-12, as well as the Treg transcription factor Forkhead box P3 (Foxp3), in a cohort of young women showed elevated Foxp3 expression. 19096978_pronounced expression of FOXP3 mRNA was found in small bowel mucosal specimens from children with coexisting celiac and type i diabetes 19104431_First report showing that Foxp3 cells are present within grafts in a subset of tolerant patients after human liver transplantation. 19108017_The prevalence of Foxp3-positive Tregs and dendritic cells was significantly higher in squamous cell carcinoma and Bowen's disease than in actinic keratosis. 19111574_both mRNA and protein expression level in CD4(+)CD25(+) Tregs are down-regulated in myasthenia gravis patients 19120312_Children with only T1 diabtes mellitus generally showed a lower FOXP3 mRNA expression than did children with celiac disease (CD), or with T1D in combination with CD. 19124747_Signal transducer and activator of transcription (STAT)1 binding elements found within the proximal part of the human foxp3 promoter function as a key regulatory unit for promoting foxp3 gene expression. 19125672_in leishmaniasis due to Leishmania guyanensis, high intralesional expression of Foxp3 might be an indicator of poor response to treatment, depending on the duration of lesion 19132983_This study shows the possible role of FoxP3 in the development of tissue characteristic humoral immunity in type 1 diabetes. 19136904_demonstrates an association between donor-specific transfusions and FoxP3+ T cells 19141582_Observational study of gene-disease association. (HuGE Navigator) 19150256_The autoimmune susceptibility in Turner Syndrome may be due to an alteration in the expression of the X-linked FOXP3 gene. 19155519_Phenotypically, CD4+CD25-Foxp3+ T cells resemble regulatory T cells (Treg) rather than activated T cells; their selective functional defects might contribute to failure of Treg to control autoimmune dysregulation in systemic lupus erythematosus patients. 19168733_Levels of Foxp3 mRNA transcripts were significantly higher in lesions from patients with chronic cutaneous leishmaniasis (CCL) CCL than in the acute disease(ACL), suggesting a critical role of intralesional Treg cells in CCL. 19178794_Our data indicate that FOXP3 is functional in nonallergic healthy donors as well as allergic patients, and FOXP3-expressing T cells may be responsible for the down-regulation of allergen-specific T-helper responses in individuals. 19189134_Report describes a case (5-year-old boy)with IPEX syndrome with a 3 bp deletion in the FOXP3 gene and a paucity of CD4(+)CD25(+)FOXP3(+) T-lymphocytes. 19192224_There is a role for lung Foxp3(+)CD4(+)CD25(+) T cells in the regulation of asthma phenotypes, presumably through an IL-10-mediated mechanism. 19199536_For HIV-1 infected persons, the TST depends on memory T cells and is more strongly associated with the numbers of circulating FoxP3(+)CD4(+) T cells than with the total number of CD4(+) T cells. 19201288_reduced FoxP3 gene expression or impaired FoxP3 functions are involved in the development of allergic disease in humans. 19203731_Utilizing FOXP3 as a Treg marker,patients who received CD4+DLI with IL-2 had greater expansion of Treg compared to patients who received IL-2 (P = .03) or CD4(+)DLI alone (P = .001). FOXP3 expression correlated with absolute CD4(+)CD25(+) cell counts. 19237575_during clinical remission, increases in CD58 expression, mediated by the protective allele, up-regulate the expression of FoxP3 through engagement of the CD58 receptor, CD2, leading to the enhanced function of TREG cells that are defective in MS 19242793_In contrast to H. pylori-induced carditis, FOXP3-expressing T cells are not associated with gastroesophageal reflux disease-associated carditis. 19255314_FOXP3 expression was associated with overall and distant metastasis-free survival but not with local relapse 19255331_FOXP3 expression in breast tumors was associated with worse overall survival probability,the risk increased with increasing FOXP3 immunostaining.FOXP3 was a strong prognostic factor for distant metastases-free survival not for local recurrence. 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19264232_Although FOXP3 is a biologically relevant gene in the pathogenesis of breast cancer, germline variation was not meaningfully associated with risk of the disease. 19264232_Observational study of gene-disease association. (HuGE Navigator) 19264985_Observational study of gene-disease association. (HuGE Navigator) 19273860_IL-17-producing Treg cells may play critical roles in antimicrobial defense, while controlling autoimmunity and inflammation. 19276356_Data provides a novel mechanism for transcription activation by FOXP3 and a genetic mechanism for lack of p21 in a large proportion of breast cancer. 19283780_CpG methylation in a conserved region within the FOXP3 gene locus increased in CD4(+)CD25(+)CD127(low) Treg, correlating with loss of FOXP3 expression and emergence of pro-inflammatory cytokines. 19328914_Biopsies from human hand transplants have been investigated for expression of Foxp3 as a marker of skin transplantation rejection. 19358983_histone deacetylase inhibitors induce the generation of regulatory T cells from human CD4(+)CD25(-) T cells. They express regulatory T cell-associated transcription factor Foxp3 and display suppressive activity against CD4(+)CD25(-) T cell proliferation. 19363449_Expression of FoxP3 is associated with cellular activation in both CD4 and CD8 T cells. 19364305_Seems likely that FOXP3mRNA expression is associated with graft acceptance after living donor liver transplantation. 19373655_FoxP3+ T-reg cells was identified as an independent predictor of failure-free survival. 19383912_Data reinforce the fact that FOXP3 remains an accurate marker to define primary Tregs in patients with cancer and autoimmune disease. 19392991_Asthmatic patients have decreased FOXP3 protein expression within their CD4(+)CD25(high) T(reg). 19394278_Study shows for the first time that CVID patients with autoimmune disease have a significantly reduced frequency of CD4(+)CD25(HIGH)FOXP3(+) cells in their peripheral blood accompanied by a decreased intensity of FOXP3 expression. 19408243_miR-31 negatively regulates FOXP3 expression in Treg by binding directly to its potential target site in the 3' UTR of FOXP3 mRNA. miR-21 acts as a positive, though indirect, regulator of FOXP3 expression in Treg. 19419438_in active periodontal lesions, Foxp3 was over-expressed; speculate Foxp3(+) cells may have role in pathogenesis of periodontal lesions by downregulating TGF-beta1 & IL-10 synthesis leading to overexpression of Th17-associated cytokines 19424039_FOXP3 does not predict response to kidney transplantation antirejection therapy but does correlates with renal fibrosis, TGF-beta, and poor late renal outcome. 19439651_human memory Tregs secrete IL-17 ex vivo and constitutively express RORgamma t 19453521_GARP is a key receptor controlling FOXP3 in T(reg) cells following T-cell activation in a positive feedback loop assisted by LGALS3 and LGMN 19464196_the dissection of FoxP3+ cells into subsets enables on | ENSMUSG00000039521 | Foxp3 | 4.130829e+01 | 2.0945894 | 1.066667491 | 0.4894542 | 4.906956e+00 | 0.0267487447 | 0.55439915 | No | Yes | 3.898062e+01 | 9.899211 | 1.937614e+01 | 5.261415 | |
| ENSG00000049860 | 3074 | HEXB | protein_coding | P07686 | FUNCTION: Hydrolyzes the non-reducing end N-acetyl-D-hexosamine and/or sulfated N-acetyl-D-hexosamine of glycoconjugates, such as the oligosaccharide moieties from proteins and neutral glycolipids, or from certain mucopolysaccharides (PubMed:11707436, PubMed:9694901, PubMed:8672428, PubMed:8123671). The isozyme B does not hydrolyze each of these substrates, however hydrolyzes efficiently neutral oligosaccharide (PubMed:11707436). Only the isozyme A is responsible for the degradation of GM2 gangliosides in the presence of GM2A (PubMed:9694901, PubMed:8672428, PubMed:8123671). During fertilization is responsible, at least in part, for the zona block to polyspermy. Present in the cortical granules of non-activated oocytes, is exocytosed during the cortical reaction in response to oocyte activation and inactivates the sperm galactosyltransferase-binding site, accounting for the block in sperm binding to the zona pellucida (By similarity). {ECO:0000250|UniProtKB:P20060, ECO:0000269|PubMed:11707436, ECO:0000269|PubMed:8123671, ECO:0000269|PubMed:8672428, ECO:0000269|PubMed:9694901}. | 3D-structure;Cytoplasmic vesicle;Direct protein sequencing;Disease variant;Disulfide bond;Gangliosidosis;Glycoprotein;Glycosidase;Hydrolase;Lipid metabolism;Lysosome;Neurodegeneration;Reference proteome;Signal;Zymogen | Hexosaminidase B is the beta subunit of the lysosomal enzyme beta-hexosaminidase that, together with the cofactor GM2 activator protein, catalyzes the degradation of the ganglioside GM2, and other molecules containing terminal N-acetyl hexosamines. Beta-hexosaminidase is composed of two subunits, alpha and beta, which are encoded by separate genes. Both beta-hexosaminidase alpha and beta subunits are members of family 20 of glycosyl hydrolases. Mutations in the alpha or beta subunit genes lead to an accumulation of GM2 ganglioside in neurons and neurodegenerative disorders termed the GM2 gangliosidoses. Beta subunit gene mutations lead to Sandhoff disease (GM2-gangliosidosis type II). Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2014]. | hsa:3074; | acrosomal vesicle [GO:0001669]; azurophil granule [GO:0042582]; azurophil granule lumen [GO:0035578]; beta-N-acetylhexosaminidase complex [GO:1905379]; cortical granule [GO:0060473]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; membrane [GO:0016020]; acetylglucosaminyltransferase activity [GO:0008375]; beta-N-acetylhexosaminidase activity [GO:0004563]; identical protein binding [GO:0042802]; N-acetyl-beta-D-galactosaminidase activity [GO:0102148]; astrocyte cell migration [GO:0043615]; cellular calcium ion homeostasis [GO:0006874]; cellular protein metabolic process [GO:0044267]; ganglioside catabolic process [GO:0006689]; glycosaminoglycan metabolic process [GO:0030203]; lipid storage [GO:0019915]; locomotory behavior [GO:0007626]; lysosome organization [GO:0007040]; male courtship behavior [GO:0008049]; myelination [GO:0042552]; neuromuscular process controlling balance [GO:0050885]; oligosaccharide catabolic process [GO:0009313]; oogenesis [GO:0048477]; penetration of zona pellucida [GO:0007341]; phospholipid biosynthetic process [GO:0008654]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of cell shape [GO:0008360]; sensory perception of sound [GO:0007605]; single fertilization [GO:0007338]; skeletal system development [GO:0001501] | 12413610_plasma activity of total Hex does not appear to be a reliable marker of erosion and cartilage degradation in rheumatoid arthritis patients; liver function appears to be the major determinant for the plasma Hex activity in these patients 12706724_The X-ray crystal structure of beta-hexosaminidase B provides new insights into mutations that cause Sandhoff disease. 15485660_alpha-subunit loop structure is required for GM2 activator protein binding by beta-hexosaminidase A 15953731_Therefore, our bicistronic lentivector supplying both alpha- and beta-subunits of beta-hexosaminidases may provide a potential therapeutic tool for the treatment of Sandhoff disease. 16352452_Observational study of genetic testing. (HuGE Navigator) 16710745_The highest activities of exoglycosidases were observed in high-grade gliomas, and a positive correlation of enzyme activities and degree of malignancy was noted. 17251047_novel c.1556A>G transition in exon 12 of the HEXB gene associated with chronic Sandhoff's disease 18180457_Beta-hexosaminidase is a peptidoglycan hydrolase that surprisingly exerts its mycobactericidal effect at the macrophage plasma membrane during mycobacteria-induced secretion of lysosomes 18204279_Elevated activity of beta-hexosaminidase observed in subjects with asthma suggests that the beta-hex isozyme could take part in airway inflammation and remodeling in asthma. 18217416_following HIV infection, there is an increased rate of catabolism of glycoconjugates in saliva resulting from changes in the proportions of the activity of isoenzymes A and B of N-acetyl-beta-hexosaminidase, beta-galactosidase and alpha-fucosidase 18552385_Lysosome-related genes, such as CLN2, CLN3, and HEXB, may be involved in the pathogenesis of adipose tissue hypertrophy in TED. 18588514_These results reveal a new aspect of beta-hexosaminidase biology and suggest that a fully processed membrane-associated form of Hex is translocated from the lysosomal membrane to the PM by an as yet unknown mechanism. W 18758829_Results describe the molecular genetics of Sandhoff disease in Italy and provide new insights into the molecular basis of the disease through HEXB mutation. 18930675_A new D459A missense HEXB mutation was discovered in six juvenile patients with Sandhoff disease. 19278737_Gene therapy reduced GM(2) storage and ameliorated neuroinflammation in the brain of HexB(-/-) mice, as well as attenuated behavioral deficits. 19615986_Data suggest that cigarette smoking can inhibit, by the influence on N-acetyl-beta-hexosaminidase activity, catabolism of oligosaccharide chains in cancer tissues. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 21307379_Urinary NAG (ninefold), NGAL (1.5-fold), and H-FABP (3.5-fold) were significantly elevated in normoalbuminuric diabetic patients compared with nondiabetic control subjects. 21321400_Plasma beta-hexosaminidase and beta-galactosidase) levels are higher in patients with Alzheimer's disease-type 2 diabetes mellitus (T2DM) compared to those with T2DM alone. 21637923_Down-regulation of beta-N-acetyl-D-glucosaminidase increases Akt1 activity in thyroid anaplastic cancer cells 21978926_The non-random distribution of plasma membrane-associated beta-hexosaminidase and beta-galactosidase and their localization within lipid microdomains, suggest a role of these enzymes in the local reorganization of glycosphingolipid-based signalling units. 21997228_The absence of beta-N-acetyl-hexosaminidase activity does not alter the differentiation of i-DCs from HSCs, but it is critical for the activation of CD4(+)T cells because knock-down of HEXA or HEXB gene causes a loss of function of i-DCs. 22191674_We describe a novel HEXB mutation that is shared among 4 patients with Sandhoff disease. 22789865_identified 27 different mutations, 14 of which were novel, in the HEXA gene and 14 different mutations, 8 of which unreported until now, in the HEXB gene, and attempted to correlate these mutations with the clinical presentation of the patients 22848519_minigene studies revealed the presence of a novel alternative spliced HEXB mRNA variant also present in normal cells 22863301_Expression of beta-hexosaminidase in the neurons of Sandhoff disease patients rescues transgenic mice from neurodegeneration. 23046579_Characterization of seven novel mutations on the HEXB gene in French Sandhoff patients. 23370522_GM2 gangliosidosis is caused by the gene mutation. (review) 23886397_A patient with Sandhoff disease also is found to have a compound macro-deletion in HEXB. 23906468_A highly significant correlation of HEX-7 and %CDT has been found. Because of exclusion of the P isoform, HEX-7 could be a useful supplementary marker for detecting chronic alcohol abuse. 23911049_Concentration and specific activity of N-acetyl-B-hexosaminidase in palatine tonsils in patients with tonsillar hypertrophy and chronic tonsillitis both in childhood and adulthood significantly increase in comparison to healthy individuals. 24461908_A total of 19 HEXB variants were found in the 1092 genomes of which 5 are suspected of having a deleterious effect on hexosaminidase activity. 24518553_DNA from Iranian Tay-Sachs patients reveals a novel mutation in HEXB predicting a termination codon or nonsense mutation. 26582265_report on the heterogeneity of the mutational spectrum of the HEXB gene in Indian patients with Sandhoff disease 27018595_a modified human hexosaminidase subunit beta (HexB), which we have termed mod2B, composed of homodimeric beta subunits that contain amino acid sequences from the alpha subunit that confer GM2 ganglioside-degrading activity and protease resistance. 27682710_Mutations of the HEXB gene is associated with maple syrup urine disease or Sandhoff disease. 28846871_Reported data present, for the first time, reference values for urinary activities of HEX and its isoenzymes HEX A and HEX B in children and adolescent. 29448188_Direct sequencing of HEXA and HEXB genes showed recurrent homozygous variants at c.509G>A (p.Arg170Gln) and c.850C>T (p.Arg284Ter) in gangliosidosis, respectively 31682993_Novel bicistronic lentiviral vectors correct beta-Hexosaminidase deficiency in neural and hematopoietic stem cells and progeny: implications for in vivo and ex vivo gene therapy of GM2 gangliosidosis. 31852446_identified a homozygous splice site variant (NM_000521:c.445 + 1G > T) in the hexosaminidase B (HEXB) gene confirming a diagnosis of Sandhoff disease (SD; type II GM2-gangliosidosis), an autosomal recessive lysosomal storage disorder caused by deficiency of hexosaminidases in a single family 31919734_Clinical and Molecular Characteristics of Two Chinese Children with Infantile Sandhoff Disease and Review of the Literature. 33407268_Infantile onset Sandhoff disease: clinical manifestation and a novel common mutation in Thai patients. 34554397_Analysis of the HEXA, HEXB, ARSA, and SMPD1 Genes in 68 Iranian Patients. | ENSMUSG00000021665 | Hexb | 8.358556e+02 | 0.9576657 | -0.062406034 | 0.3064820 | 4.152527e-02 | 0.8385273671 | 0.96857761 | No | Yes | 7.887542e+02 | 119.954312 | 8.154542e+02 | 127.095760 | |
| ENSG00000050820 | 9564 | BCAR1 | protein_coding | P56945 | FUNCTION: Docking protein which plays a central coordinating role for tyrosine kinase-based signaling related to cell adhesion (PubMed:12832404, PubMed:12432078). Implicated in induction of cell migration and cell branching (PubMed:12432078, PubMed:12832404, PubMed:17038317). Involved in the BCAR3-mediated inhibition of TGFB signaling (By similarity). {ECO:0000250|UniProtKB:Q61140, ECO:0000269|PubMed:12432078, ECO:0000269|PubMed:12832404, ECO:0000269|PubMed:17038317}. | 3D-structure;Acetylation;Alternative splicing;Cell adhesion;Cell junction;Cell projection;Cytoplasm;Diabetes mellitus;Direct protein sequencing;Phosphoprotein;Reference proteome;SH3 domain;SH3-binding | The protein encoded by this gene is a member of the Crk-associated substrate (CAS) family of scaffold proteins, characterized by the presence of multiple protein-protein interaction domains and many serine and tyrosine phosphorylation sites. The encoded protein contains a Src-homology 3 (SH3) domain, a proline-rich domain, a substrate domain which contains 15 repeat of the YxxP consensus phosphorylation motif for Src family kinases, a serine-rich domain, and a bipartite Src-binding domain, which can bind both SH2 and SH3 domains. This adaptor protein functions in multiple cellular pathways, including in cell motility, apoptosis and cell cycle control. Dysregulation of this gene can have a wide range of effects, affecting different pathways, including cardiac development, vascular smooth muscle cells, liver and kidney function, endothelial migration, and cancer. [provided by RefSeq, Sep 2017]. | hsa:9564; | actin cytoskeleton [GO:0015629]; axon [GO:0030424]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; lamellipodium [GO:0030027]; membrane [GO:0016020]; ruffle [GO:0001726]; protein kinase binding [GO:0019901]; SH3 domain binding [GO:0017124]; actin filament organization [GO:0007015]; actin filament reorganization [GO:0090527]; antigen receptor-mediated signaling pathway [GO:0050851]; B cell receptor signaling pathway [GO:0050853]; cell adhesion [GO:0007155]; cell chemotaxis [GO:0060326]; cell division [GO:0051301]; cell migration [GO:0016477]; cellular response to hepatocyte growth factor stimulus [GO:0035729]; endothelin receptor signaling pathway [GO:0086100]; epidermal growth factor receptor signaling pathway [GO:0007173]; G protein-coupled receptor signaling pathway [GO:0007186]; hepatocyte growth factor receptor signaling pathway [GO:0048012]; insulin receptor signaling pathway [GO:0008286]; integrin-mediated signaling pathway [GO:0007229]; neurotrophin TRK receptor signaling pathway [GO:0048011]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of cell migration [GO:0030335]; positive regulation of endothelial cell migration [GO:0010595]; regulation of apoptotic process [GO:0042981]; regulation of cell growth [GO:0001558]; T cell receptor signaling pathway [GO:0050852]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11779709_Tyrosine phosphorylation of p130CAS regulates localization and downstream signaling with profound affects on cell movement. 11820787_Binding of the adapter protein p130Cas to the C-terminal of Pyk2 in cultured human umbilical vein endothelial cells is phosphorylation-independent and is not affected by acute exposure to thrombin. 12135674_The association of Cas with Wiskott-Aldrich syndrome protein is associated with cell migration in stromal cell-derived factor-1alpha-stimulated Jurkat cells 12397603_phosphorylation of p130(Cas) can prevent cells from anoikis and contribute to tumor cell anchorage independence and metastasis 12529399_R-Ras promotes focal adhesion formation by signaling to FAK and p130(Cas) through a novel mechanism that differs from but synergizes with the alpha2beta1 integrin. 12738793_Requirement of the p130CAS-Crk coupling for metastasis suppressor KAI1/CD82-mediated inhibition of cell migration in a metastatic prostate cancer cell line. 15121874_endogenous Sin influences T-lymphocyte signaling by sequestering signaling substrates and regulating their availability 15448007_BCAR1 has a role in progression of primary breast cancer 15784259_crystal structure of p130cas SH3 domain 15923424_Our studies suggested that pRb2/p130-complexes bind to the ER-alpha promoter and could be involved in the transcriptional regulation of the ER-alpha gene by altering chromatin structure and DNA methylation pattern. 16040804_p130Cas and paxillin function as effectors of GD3-mediated signaling, leading to such malignant properties as rapid cell growth and invasion in melanoma cells 16440329_LOX regulates cell motility/migration through changes in actin filament polymerization, which involve the regulation of the p130(Cas)/Crk/DOCK180 16597701_Fibronectin rigidity response involves force-dependent Fyn phosphorylation of p130Cas with rigidity-dependent displacement. 16644720_PTP1B mediates of RhoA-dependent phosphorylation of p130Cas. 17038317_The interaction between Ack1 and p130(Cas) occurred through their respective SH3 domains, while the substrate domain of p130(Cas) was the major site of Ack1-dependent phosphorylation. 17129785_Cas acts as a primary force sensor, transducing force into mechanical extension and thereby priming phosphorylation and activation of downstream signaling. 17251438_p130CAS is an important component in the netrin signaling pathway acting between tyrosine kinases and small GTPase Rac1 and is essential for commissural axon guidance in vivo. 17616674_the spatial and temporal regulation of BCAR3/p130(Cas) interactions within the cell is important for controlling breast cancer cell motility 17982677_Activation of the FAK-src molecular scaffolds and p130Cas-JNK signaling cascades by alpha1-integrins during colon cancer cell invasion. 18078823_Focal adhesion kinase as well as p130Cas and paxillin should be a crucial molecule undergoing stronger tyrosine phosphorylation in GD3-expressing melanoma cells. 18095869_focal adhesion kinase positively regulates Caco-2 spreading on collagen IV via p130(Cas) phosphorylation 18164686_The interaction of MT1-MMP with p130Cas at the cell periphery suggests the existence of a close interplay between pericellular proteolysis and signaling pathways involved in endothelial cell migration. 18321991_p130Cas-mediated control of TGF-beta/Smad signaling may provide an additional clue to the mechanism underlying resistance to TGF-beta-induced growth inhibition. 18725541_uPAR cooperates with integrin complexes containing beta(3) integrin to drive formation of the p130Cas-CrkII signaling complex and activation of Rac, resulting in a Rac-driven elongated-mesenchymal morphology, cell motility, and invasion. 18842495_In hepatocellular carcinoma (HCC), there is a negative correlation between the positive expression of p130Cas and the normal expression of E-cadherin/beta-catenin. p130Cas plays important roles in the invasion, metastasis and prognosis of HCC. 19029090_The lysyl oxidase pro-peptide attenuates fibronectin-mediated activation of focal adhesion kinase and p130Cas in breast cancer cells. 19086031_Data suggest that endothelin-1 stimulates the GTPase Rap1 by a mechanism involving Pyk2 activation and recruitment of the p130Cas/BCAR3 complex in human glomerular mesangial cells. 19329671_Evidence for the role of CAS in the regulation of vascular smooth muscle contractility, cell migration, hypertrophy, and growth is presented 19330798_targeting the product of the BCAR1 gene by a peptide which mimics the phosphorylated substrate domain may provide a new molecular avenue for treatment of antiestrogen resistant breast cancers. 19331827_BCAR1 is essential for the rapid estrogen effect on osteoclast differentiation, through estrogen receptor alpha and possibly Traf6. 19357231_Propofol inhibits pressure-stimulated macrophage phagocytosis via the GABAA receptor and dysregulation of p130cas phosphorylation. 19412734_The crucial interactions required for anti-estrogen resistance occur within the substrate domain of BCAR1 19822523_p130Cas is required for mammary tumor growth and transforming growth factor-beta-mediated metastasis through regulation of Smad2/3 activity 19844255_Observational study of gene-disease association. (HuGE Navigator) 19940159_the c-Src/Cas/BCAR3 signaling axis is a prominent regulator of c-Src activity, which in turn controls cell behaviors that lead to aggressive and invasive breast tumor phenotypes 20688056_CAS plays a role in regulating the extension of cell protrusions and promotes the migration of cancer cells. 20961652_Analyses indicate that p130Cas expression in ErbB2 positive human breast cancers significantly correlates with higher risk to develop distant metastasis, thus underlying the value of the p130Cas/ErbB2 synergism in regulating breast cancer invasion. 21047529_BCAR-1 is a physiological substrate of Syk. 21245381_Knockdown of NRP1 or P130Cas or expression of either NRP1DeltaC or a non-tyrosine-phosphorylatable substrate domain mutant protein (p130(Cas15F)) was sufficient to inhibit growth factor-mediated migration of glioma and endothelial cells. 21291860_p130Cas, Src and talin function in both oral carcinoma invasion and resistance to cisplatin. 21306301_These results indicate a role for NRP1 and NRP1 glycosylation in mediating PDGF-induced VSMC migration, possibly by acting as a co-receptor for PDGFRalpha and via selective mobilization of a novel p130Cas tyrosine phosphorylation pathway. 21630091_Immunohistochemical analysis of microarrayed human oral squamous cell carcinoma revealed a significant correlation between uPAR and p130cas expression. 21765937_Cas proteins do not affect E-cadherin transcription, but rather, BCAR1 and NEDD9 signal through SRC to promote E-cadherin removal from the cell membrane and lysosomal degradation. 21937722_These findings reveal an important role of CAS Y12 phosphorylation in the regulation of focal adhesion assembly, cell migration, and invasiveness of Src-transformed cells. 21957230_Increased p130cas expression is associated with poor clinical outcome in human ovarian carcinoma, and p130cas gene silencing decreases tumor growth through stimulation of apoptotic and autophagic cell death. 22081014_The structure of the NSP3-p130Cas complex reveals that this closed conformation is instrumental for interaction of NSP proteins with a focal adhesion-targeting domain present in Cas proteins. 22084245_a novel function for PTK6 at the plasma membrane 22106368_Data show that phosphorylation of Src family kinases and the adaptor protein p130CAS, resulting in actin recruitment and CD36 clustering by 50-60% of adherent beads. 22144090_CrkI and p130(Cas) complex regulates the migration and invasion of prostate cancer cells. 22241677_BCAR1 is an independent predictor of recurrence following radical prostatectomy for 'low risk' prostate cancer. 22395610_Zyxin phenotype is mediated by partners alpha-actinin and p130Cas and ensures that motile cells in a three-dimensional matrix explore the largest space possible in minimum time. 22431919_p130Cas signaling induces the expression of EGR1 and NAB2 22476538_Ezrin and BCAR1/p130Cas mediate breast cancer growth as 3-D spheroids 22558353_Overexpression of BCAR1 is a predictor of poor prognosis in non-small-cell lung cancer and plays important carcinogenic roles in carcinogenesis. 22711540_BCAR3 expression may regulate Src signaling in a BCAR3-p130(cas) complex-dependent fashion by altering the ability of the Src SH3 domain to bind the p130(cas) SBD 22892392_Study shows that BCAR4 expression identifies a subgroup of ER-positive breast cancer patients without overexpression of ERBB2 who have a poor outcome and might benefit from combined ERBB2-targeted and antioestrogen therapy. 23042269_The aim of this study is to evaluate the role of p130cas, E-cadherin, and beta-catenin expression in patients with non-small cell lung cancer. 23098208_these data identify a new p130Cas/Cyclooxygenase-2 axis as a crucial element in the control of breast tumor plasticity. 23152477_This study identified rs4888378 in the BCAR1-CFDP1-TMEM170A locus as a novel genetic determinant of carotid intima-media thickness and coronary artery disease risk. 23239970_These results suggest that alteration of morphogenetic pathways due to p130Cas over-expression might prime mammary epithelium to tumorigenesis 23287717_p130Cas acts as survival factor by limiting PMA-mediated cell cluster disruption and resulting cell death in HL-60 cells. 23345605_Disruption of p130Cas attenuates both invasion and migration of the metastatic variant 23457408_BCAR1 rs7202877 may mediate its diabetogenic impact through impaired beta-cell function. 23740246_Data indicate that Abi1 is activated by the c-Abl-Crk-associated substrate (CAS) pathway, and Abi1 reciprocally controls the activation of its upstream regulator c-Abl. 23839042_P130Cas overexpression synergizes with ErbB2 in mammary cell transformation and promotes ErbB2-dependent invasion. 23872147_Our results suggest that elevated expression and tyrosine phosphorylation of p130Cas contributes to the resistance to TGF-beta-induced growth inhibition. 23904007_Increased BCAR1 expression is associated with non-small cell lung cancer. 24284072_Our results show that endogenous Cul5 suppresses epithelial cell transformation by several pathways, including inhibition of Src-Cas-induced ruffling through SOCS6 24494199_Data introduce hitherto unappreciated paradigms whereby reactive oxygen species can reciprocally regulate the cellular localization of pro- and anti-migratory signaling molecules, p130cas and PTEN, respectively. 24584939_BCAR1 and BCAR3 scaffolding proteins have roles in cell signaling and antiestrogen resistance 24928898_Cas promotes cell migration by linking actomyosin contractions to the adhesion complexes through interaction with Src and the actin cytoskeleton. 25253349_Collectively, these studies demonstrate that p130Cas acts as a bridging molecule between the Kaposi's sarcoma-associated herpesvirus-induced entry signal complex and the downstream trafficking signalosome in endothelial cells. 25727852_BCAR1 has a pivotal role in the regulation of tissue homeostasis in pathological conditions such as cancer. (Review) 25805500_p130(Cas) exon 1 variants display altered functional properties; shorter 1B isoform exhibited diminished FAK binding activity + reduced cell migration + invasion; longest variant 1B1 exhibited the most efficient FAK binding + greatly enhanced migration 26276885_expression quantitative trait loci studies implicate BCAR1 as the causal gene of coronary artery disease Carotid intima-media thickness 26716506_Elevated levels of p130Cas is associated with trastuzumab resistance in breast cancer. 26867768_Full-length and truncated p130Cas phosphorylated substrate domain molecules were expressed in breast cancer cells. Breast cancer cells expressing the full-length SD and the functional smaller SD fragment (spanning SD motifs 6-10) were injected into the mammary fat pads of mice. Both the complete and truncated SD significantly increased the occurrence of metastases to multiple organs. 27068854_blockade of GD3-mediated growth signaling pathways by siRNAs might be a novel and promising therapeutic strategy against malignant melanomas, provided signaling molecules such as p130Cas and paxillin are significantly expressed in individual cases. 27293031_Silencing of p130Cas and inhibition of FAK activity both strongly reduced imatinib and nilotinib stimulated invasion. 27400161_Tyrosine phosphorylation of focal adhesion kinase (FAK) and p130 Crk-associated substrate (CAS) was found to be correlated with pancreatic cancer cell invasiveness. 28223315_the p130Cas FAT domain uniquely confers a mechanosensing function. 28337997_These results suggest that miR-24-3p functions as a tumor suppressor and the miR-24-3p/p130Cas axis is a novel factor of cancer progression by regulating cell migration and invasion. 28442738_The authors herein prove, for the first time, that the transcriptional repressor Blimp1 is a novel mediator of p130Cas/ErbB2-mediated invasiveness. Indeed, high Blimp1 expression levels are detected in invasive p130Cas/ErbB2 cells and correlate with metastatic status in human breast cancer patients. 29304771_the results of our study identify BCAR1 as a prognostic biomarker with potential clinical value for risk stratification of ERG-negative prostate cancer. 30188962_study showed that RBMS1 gene rs7593730 and BCAR1 gene rs7202877 were significantly associated with type 2 diabetes in the Chinese population 30422386_a novel interaction of p130Cas with Ser/Thr kinase PKN3, is reported. 30531837_a feedforward autocrine loop to promote invasion through a FAK>p130Cas>c-Jun>MMP-9 signaling axis. 30639111_p130Cas (BCAR1) is recruited to filopodia tips via its C-terminal Cas family homology domain (CCHD) and acts as a mechanosensitive regulator of filopodia stability. 33001583_BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A. 33042270_CSRP2 suppresses colorectal cancer progression via p130Cas/Rac1 axis-meditated ERK, PAK, and HIPPO signaling pathways. 33144694_Mutant TP53 interacts with BCAR1 to contribute to cancer cell invasion. 33334252_High Expression of BCAR1 by Circulating Tumor Cells and Tumor Tissues Is Predictive of a Poor Prognosis of Early-Stage Lung Adenocarcinoma Potentially Due to Regulation of Epithelial-Mesenchymal Transition. 34169835_A Cas-BCAR3 co-regulatory circuit controls lamellipodia dynamics. 34192548_Targeting p130Cas- and microtubule-dependent MYC regulation sensitizes pancreatic cancer to ERK MAPK inhibition. 34326687_BCAR1 plays critical roles in the formation and immunoevasion of invasive circulating tumor cells in lung adenocarcinoma. 34830244_p130Cas Is Correlated with EREG Expression and a Prognostic Factor Depending on Colorectal Cancer Stage and Localization Reducing FOLFIRI Efficacy. 35031902_Comprehensive immunohistochemical analysis of RET, BCAR1, and BCAR3 expression in patients with Luminal A and B breast cancer subtypes. | ENSMUSG00000031955 | Bcar1 | 3.025132e+03 | 1.3544509 | 0.437708073 | 0.3227200 | 1.830336e+00 | 0.1760878655 | 0.77663851 | No | Yes | 3.220287e+03 | 452.343015 | 2.119231e+03 | 305.853251 | |
| ENSG00000051009 | 84067 | FHIP1B | protein_coding | Q8N612 | FUNCTION: Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). FHF complex promotes the distribution of AP-4 complex to the perinuclear area of the cell (PubMed:32073997). {ECO:0000269|PubMed:18799622, ECO:0000269|PubMed:32073997}. | Alternative splicing;Phosphoprotein;Protein transport;Reference proteome;Transport | The protein encoded by this gene is part of the FTS/Hook/FHIP (FHF) complex, which can interact with members of the homotypic vesicular protein sorting (HOPS) complex. This interaction suggests that the encoded protein is involved in vesicle trafficking. [provided by RefSeq, Dec 2016]. | hsa:84067; | cytosol [GO:0005829]; FHF complex [GO:0070695]; early endosome to late endosome transport [GO:0045022]; endosome organization [GO:0007032]; endosome to lysosome transport [GO:0008333]; lysosome organization [GO:0007040]; protein localization to perinuclear region of cytoplasm [GO:1905719]; protein transport [GO:0015031] | ENSMUSG00000044465 | Fhip1b | 2.287205e+03 | 1.0495630 | 0.069788807 | 0.2846924 | 6.052980e-02 | 0.8056605168 | 0.96141221 | No | Yes | 2.030866e+03 | 218.648943 | 2.054887e+03 | 227.071328 | ||
| ENSG00000051341 | 10721 | POLQ | protein_coding | O75417 | FUNCTION: DNA polymerase that promotes microhomology-mediated end-joining (MMEJ), an alternative non-homologous end-joining (NHEJ) machinery triggered in response to double-strand breaks in DNA (PubMed:25642963, PubMed:25643323). MMEJ is an error-prone repair pathway that produces deletions of sequences from the strand being repaired and promotes genomic rearrangements, such as telomere fusions, some of them leading to cellular transformation (PubMed:25642963, PubMed:25643323). POLQ acts as an inhibitor of homology-recombination repair (HR) pathway by limiting RAD51 accumulation at resected ends (PubMed:25642963). POLQ-mediated MMEJ may be required to promote the survival of cells with a compromised HR repair pathway, thereby preventing genomic havoc by resolving unrepaired lesions (By similarity). The polymerase acts by binding directly the 2 ends of resected double-strand breaks, allowing microhomologous sequences in the overhangs to form base pairs. It then extends each strand from the base-paired region using the opposing overhang as a template. Requires partially resected DNA containing 2 to 6 base pairs of microhomology to perform MMEJ (PubMed:25643323). The polymerase activity is highly promiscuous: unlike most polymerases, promotes extension of ssDNA and partial ssDNA (pssDNA) substrates (PubMed:18503084, PubMed:21050863, PubMed:22135286). Also exhibits low-fidelity DNA synthesis, translesion synthesis and lyase activity, and it is implicated in interstrand-cross-link repair, base excision repair and DNA end-joining (PubMed:14576298, PubMed:18503084, PubMed:19188258, PubMed:24648516). Involved in somatic hypermutation of immunoglobulin genes, a process that requires the activity of DNA polymerases to ultimately introduce mutations at both A/T and C/G base pairs (By similarity). {ECO:0000250|UniProtKB:Q8CGS6, ECO:0000269|PubMed:14576298, ECO:0000269|PubMed:18503084, ECO:0000269|PubMed:19188258, ECO:0000269|PubMed:21050863, ECO:0000269|PubMed:22135286, ECO:0000269|PubMed:24648516, ECO:0000269|PubMed:25642963, ECO:0000269|PubMed:25643323}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Chromosome;Coiled coil;DNA damage;DNA repair;DNA-directed DNA polymerase;Nucleotide-binding;Nucleotidyltransferase;Nucleus;Reference proteome;Transferase | hsa:10721; | chromosome [GO:0005694]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; 5'-deoxyribose-5-phosphate lyase activity [GO:0051575]; ATP binding [GO:0005524]; chromatin binding [GO:0003682]; damaged DNA binding [GO:0003684]; DNA-directed DNA polymerase activity [GO:0003887]; identical protein binding [GO:0042802]; single-stranded DNA helicase activity [GO:0017116]; base-excision repair [GO:0006284]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; DNA-dependent DNA replication [GO:0006261]; double-strand break repair [GO:0006302]; double-strand break repair via alternative nonhomologous end joining [GO:0097681]; negative regulation of double-strand break repair via homologous recombination [GO:2000042]; protein homooligomerization [GO:0051260]; somatic hypermutation of immunoglobulin genes [GO:0016446] | 12051913_DNA polymerase theta purified from human cells is a high-fidelity enzyme. 14576298_the isolation of the full-length human DNA POLQ gene, and an initial characterization of its gene product, DNA polymerase theta 14735462_DNA Pol theta has a specialized function in lymphocytes and in tumor progression 15496986_POLQ has a high efficiency in by-passing DNA damage. 16188888_analysis of human DNA polymerase eta error-prone synthesis on DNA deoxyadenosine adducts 16520097_The results demonstrate that activation of a UV-induced DNA damage response pathway, involving phosphorylation of RPA p34 by DNA-PK, is enhanced in cells lacking poleta. 16603639_Pol eta can play an important role in determining the cellular sensitivity to therapeutic agents. 16824193_DNA polymerase eta (Poleta) is responsible for efficient translesion synthesis (TLS) past cis-syn cyclobutane thymine dimers (TT dimers), the major DNA lesions induced by UV irradiation. 16835218_Human DNA polymerase eta selectively produces a two-base deletion in copying the N2-guanyl adduct of 2-amino-3-methylimidazo[4,5-f]quinoline but not the C8 adduct at the NarI G3 site 17150533_The enzymatic reactions with human DNA polymerase eta on oxidative products of guanine and 8-oxoG, is investigated. 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18366182_Evolutionary conservation of efficient T[CPD]T bypass by HsPoleta and AtPoleta may reflect a high degree of exposure of human skin and plants to solar UV-B radiation 18503084_When copying undamaged DNA, DNA polymerase theta generates single base errors at rates 10- to more than 100-fold higher than for other family A members. 19188258_Domain mapping of the 98-kDa enzyme by limited proteolysis and NaBH(4) cross-linking with a base excision repair intermediate revealed that the dRP lyase active site resides in a 24-kDa domain of Pol theta. 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19661089_Observational study of gene-disease association. (HuGE Navigator) 20624954_Data show that POLQ overexpression may be a promising genetic instability and prognostic marker for breast cancer. 20700469_overexpression in breast cancer confers an adverse prognosis and is associated with key cancer pathways 22135286_POLQ possesses a DNA polymerase activity that appears to be template independent and allows efficient extension of single-stranded DNA as well as duplex DNA with either protruding or multiply mismatched 3'-OH termini. 23577232_Use fluorescence resonance energy transfer to monitor assembly of the human replicative polymerase holoenzyme. 24648516_A role for DNA polymerase theta; in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. 25409685_A DNA repair variant in POLQ (c.-1060A > G) is associated to hereditary breast cancer patients. 25417172_These results suggest that variants in the POLQ gene may be associated with the risk of Luminal breast cancer 25642960_depletion of Poltheta; has a synergistic effect on cell survival in the absence of BRCA genes, suggesting that the inhibition of this mutagenic polymerase represents a valid therapeutic avenue for tumours carrying mutations in homology-directed repair genes 25642963_results reveal a synthetic lethal relationship between the homologous-recombination pathway and Poltheta;-mediated repair in epithelial ovarian cancers, and identify Poltheta; as a novel druggable target for cancer therapy 25643323_Microhomology-mediated end-joining is dependent on Poltheta; in human cells. 25775267_Polymerase theta; uses a specialized thumb subdomain to establish unique upstream contacts to the primer DNA strand. 27533083_Our results indicate that there is a synthetic lethal relationship between pol theta;-mediated DNA repair and homologous recombination pathways 28668117_This article summarizes work on the expression and purification of the full-length protein, and then focus on the design, expression, and purification of an active C-terminal polymerase fragment. Strategies to obtain and improve crystals of a ternary POLQ complex (enzyme:DNA:nucleotide) are also presented, along with key elements of the structure. 28695890_Cells doubly deficient in Pol theta; and Lig4 exhibit 100% gene-targeting efficiency because of virtually no random integration events. 28939775_Data suggest that error-free DNA replication through 3-deaza-3-methyladenine adduct is mediated via three different pathways dependent upon POL-iota/POL-kappa, POL-theta, and POL-zeta. 29243925_results suggest that bypass of Tg by Pol theta; results in mutations opposite the lesion, as well as frameshift mutations 29330301_Data suggest that translesion DNA synthesis mediated by (1) POLI-dependent pathway (2) REV1- and POLN-dependent pathway, or (3) POLtheta-dependent pathway occur in predominantly error-free manner in human cells. (POLI = DNA polymerase iota; REV1 = DNA repair protein-REV1; POLN = DNA polymerase nu; POLtheta = DNA polymerase theta) 29334356_Importantly, p53 defects or depletion unexpectedly allow mutagenic RAD52 and POLtheta; pathways to hijack stalled replication forks, which the authors find reflected in p53 defective breast-cancer patient COSMIC mutational signatures. These data uncover p53 as a keystone regulator of replication homeostasis within a DNA restart network. 29509408_Heightened Poltheta; expression levels were also associated with elevated mtDNA mutation rates in a selected panel of human tumor tissues, suggesting that this protein can influence mutational frequencies in tumors. T 30099721_Based on previous studies and gene ontology database, we found that POLQ encoding DNA polymerase theta enzyme and FNIP1 encoding tumor suppressor folliculin-interacting protein might have contributed to the Interdigitating dendritic cell sarcoma (IDCS). Our study provides potential causative genetic factors of IDCS and plays a role in advancing the understanding of IDCS pathogenesis 30299084_Expanded Substrate Scope of DNA Polymerase theta; and DNA Polymerase beta: Lyase Activity on 5'-Overhangs and Clustered Lesions. 30808656_Rev1 polymerase and Poltheta; conduct translesion synthesis opposite 1,N(6)-ethenodeoxyadenosine via alternative error-prone pathways 30818397_Poltheta; shows a strong preference for adding deoxyribonucleotides to RNA, but can also add ribonucleotides with relatively high efficiency in particular sequence contexts; this unique activity of Poltheta; will become invaluable for applications requiring 3' terminal modification of RNA and potentially enzymatic synthesis of RNA 31381562_RAD52 and POLQ are both synthetic lethal with loss of the BRCA1 and BRCA2 tumor suppressor genes. Furthermore, RAD52 and POLQ have been implicated in chromosomal break repair events that use flanking repeats to restore the chromosome. Combined disruption of RAD52 and POLQ causes at least additive hypersensitivity to cisplatin and a synthetic reduction in replication fork restart velocity. 32098870_The roles of polymerases nu and theta in replicative bypass of O (6)- and N (2)-alkyl-2'-deoxyguanosine lesions in human cells. 32169903_Genetic evidence for reconfiguration of DNA polymerase theta active site for error-free translesion synthesis in human cells. 32302896_DNA polymerase theta (Poltheta) - an error-prone polymerase necessary for genome stability. 32838755_Mutant POLQ and POLZ/REV3L DNA polymerases may contribute to the favorable survival of patients with tumors with POLE mutations outside the exonuclease domain. 32873648_POLQ suppresses interhomolog recombination and loss of heterozygosity at targeted DNA breaks. 33577776_Human DNA polymerase theta harbors DNA end-trimming activity critical for DNA repair. 33911081_Recurrent deletions in clonal hematopoiesis are driven by microhomology-mediated end joining. 33920223_Translesion Synthesis or Repair by Specialized DNA Polymerases Limits Excessive Genomic Instability upon Replication Stress. 34473992_DNA polymerase theta promotes CAG*CTG repeat expansions in Huntington's disease via insertion sequences of its catalytic domain. | ENSMUSG00000034206 | Polq | 5.468507e+02 | 0.6776187 | -0.561454420 | 0.3365073 | 2.782012e+00 | 0.0953283594 | 0.75783482 | No | Yes | 5.201203e+02 | 95.614291 | 7.020630e+02 | 131.863436 | ||
| ENSG00000054523 | 23095 | KIF1B | protein_coding | O60333 | FUNCTION: Motor for anterograde transport of mitochondria. Has a microtubule plus end-directed motility. Isoform 2 is required for induction of neuronal apoptosis. {ECO:0000269|PubMed:18334619}.; FUNCTION: Isoform 1 mediates the transport of synaptic vesicles in neuronal cells. {ECO:0000250|UniProtKB:O88658}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Apoptosis;Cell junction;Cell projection;Charcot-Marie-Tooth disease;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Disease variant;Microtubule;Mitochondrion;Motor protein;Neurodegeneration;Neuropathy;Nucleotide-binding;Phosphoprotein;Reference proteome;Synapse | This gene encodes a motor protein that transports mitochondria and synaptic vesicle precursors. Mutations in this gene cause Charcot-Marie-Tooth disease, type 2A1. [provided by RefSeq, Jul 2008]. | hsa:23095; | axon [GO:0030424]; axon cytoplasm [GO:1904115]; cytoplasmic vesicle [GO:0031410]; cytoplasmic vesicle membrane [GO:0030659]; dendrite [GO:0030425]; kinesin complex [GO:0005871]; microtubule [GO:0005874]; microtubule associated complex [GO:0005875]; mitochondrion [GO:0005739]; neuron projection [GO:0043005]; synaptic vesicle [GO:0008021]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; kinesin binding [GO:0019894]; microtubule binding [GO:0008017]; microtubule motor activity [GO:0003777]; plus-end-directed microtubule motor activity [GO:0008574]; anterograde axonal transport [GO:0008089]; apoptotic process [GO:0006915]; cytoskeleton-dependent intracellular transport [GO:0030705]; microtubule-based movement [GO:0007018]; neuromuscular synaptic transmission [GO:0007274]; neuron-neuron synaptic transmission [GO:0007270]; vesicle-mediated transport [GO:0016192] | 11835375_Observational study of genotype prevalence. (HuGE Navigator) 12888911_KIF1Ba in addition to KIF1Bbeta may not be a candidate tumor suppressor gene for neuroblastoma 16225668_KBP is a new binding partner for KIF1Balpha that is a regulator of its transport function and thus represents a new type of kinesin interacting protein. 17418584_We detected ALS-specific down-regulation of KIF1Bbeta and novel KIF3Abeta, two isoforms we show to be enriched in the brain, and also of SOD1, a key enzyme linked to familial ALS. 18334619_Study identified inherited loss-of-function KIF1Bbeta missense mutations in neuroblastomas and pheochromocytomas and an acquired loss-of-function mutation in a medulloblastoma. 18614535_KIF1Bbeta may act as a haploinsufficient tumor suppressor, and its allelic loss may be involved in the pathogenesis of neuroblastoma and other cancers. 18726616_a germline mutation in the KIF1B beta gene on 1p36 may have a role in neural and nonneural tumors 18997785_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18997785_Study reports a genome wide association study identifying a new locus replicated in 2,679 cases and 3,125 controls; an rs10492972[C] variant located in the KIF1B gene was associated with MS with an odds ratio of 1.35 (P = 2.5 x 10(-10)). 19776380_Observational study of gene-disease association. (HuGE Navigator) 20067515_KIF1B rs10492972 allelic variant does not act as a risk factor as well as a disease modifier in a Italian cohort of patients with progressive relapsing multiple sclerosis. 20067515_Observational study of gene-disease association. (HuGE Navigator) 20190806_Bmi1 is a MYCN target gene that regulates tumorigenesis through repression of KIF1Bbeta and TSLC1 in neuroblastoma. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20502484_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20676096_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 21424745_No association is found between rs10492972 KIF1B polymorphism and the progression of multiple sclerosis in Greek subjects. 21594895_analysis of the KIF1B rs10492972*C allelic association in multiple sclerosis 21606458_Data show that no evidence could be found for a determining influence of carriership of the risk allele or genotype of the KIF1B gene on any of the multiple sclerosis neurodegenerative phenotypic markers studied. 21680216_Polymorphic locus rs10492972 of the KIF1B gene associates with multiple sclerosis in Russia. 22363396_This study showed the new locus identified for hepatocellular carcinoma, KIF1B, was not associated with progression to chronic hepatitis B. 22945799_activity-dependent synaptic recruitment of KIF1Bbeta, its interaction with Ca(2 ) sensor Calmodulin and its role as a dendritic motor of ribonucleoprotein complexes provide a novel basis for understanding the coordination of motor protein mobilization and synaptic signaling pathways 23028799_Polymorphisms at KIF1B gene locus investigated in this study showed no significant association with Hepatitis B virus infection. 23354307_Leptin, MT1-MMP and KIF1B are overexpressed in GC tissues. 23528589_the TT genotype of rs1535045 was associated with a slower progression of MS and early MS onset. 23634229_KIF1B may play a critical role in the development of hepatocellular carcinoma 23803045_the KIF1B gene SNP (rs174019660) showed no significant association with HBV-related hepatocellular carcinoma in Thai patients infected with HBV, indicating that there must be other mechanisms or pathways involved in hepatocellular carcinoma. 23831022_analysis of the recognition sequence for DJ-1 protease and its interactions with KIF1B and ABL1 24469107_Results show that KIF1Bbeta has neuroblastoma tumor-suppressor properties and promotes and requires nuclear-localized DHX9 for its apoptotic function by activating XAF1 expression. 24952890_The meta-analysis showed a significant association between kinesin family member 1B (KIF1B) single nucleotide polymorphism (rs17401966) and hepatocellular carcinoma (HCC). 25153661_The variant G allele of rs17401966 may be a favorable biomarker for the prognosis of intermediate or advanced hepatitis B virus-related hepatocellular carcinoma patients in this Chinese population 25355294_Results from targeted sequencing in patients with acute lymphoblastic leukemia identified KMT2D and KIF1B as novel putative driver genes and a putative regulatory non-coding variant that coincided with overexpression of the growth factor MDK. 25518808_The tumor suppressor DLC2 and Kif1B are central components of a signaling network that guides spindle positioning, cell-cell adhesion and mitotic fidelity. 25854172_the rs17401966 polymorphism likely regulates KIF1B mRNA expression and thus may be associated with epithelial ovarian cancer risk in Eastern Chinese women. 26217094_Downregulation of KIF1B in hepatocellular carcinoma tissues is associated with poor prognosis. 26576027_Increased KIF1B was associated with worse WHO pathological classification, Karnofsky performance status, and prognosis. Silencing KIF1B inhibited expression of membranal MT1-MMP. 26812016_that KIF1Bbeta affects mitochondrial dynamics through calcineurin-dependent dephosphorylation of Dynamin-related protein 1 (DRP1), causing mitochondrial fission and apoptosis 26954557_Study found that in peripheral blood mononuclear cells the median expression of KIFC3, KIF1B, and KIF5C was much lower than the expression of dynactin subunits DCTN1 and DCTN3, in both sporadic amyotrophic lateral sclerosis and healthy cases 27122668_The gene-environment interaction between the KIF1B rs17401966 variant and alcohol consumption may contribute to the development of hepatocellular carcinoma in Chinese individuals. 27851960_BORC and Arl8 function upstream of two structurally distinct kinesin types: kinesin-1 (KIF5B) and kinesin-3 (KIF1Bbeta and KIF1A). 28427253_The rs17401966 polymorphism reduced the risk for HCC under the allele, heterozygous, homozygous, and dominant models but not under the additive or recessive models. 28515046_In conclusion, using a panel including 17 susceptibility genes, we documented the presence of somatic mutations in over 50% of pheochromocytomas and paragangliomas (PPGL). We confirmed the high frequency of NF1 somatic mutations and identified KIF1B as the second most frequently mutated gene in PPGL tissues. 30859632_The authors' findings indicate that tumor suppressor KIF1Bbeta plays an important role in intrinsic mitochondria-mediated apoptosis through the regulation of structural and functional dynamics of mitochondria in collaboration with YME1L1. 30947687_results revealed a significant association between KIF1B rs17401966 polymorphism and susceptibility to HCC under a random-effect allelic model 31541015_The Kif1bbeta and Fignl1 loss of function similarly altered zebrafish motor axon pathfinding and increased dynein-based transport velocity of Rab3 vesicles in these navigating axons, pinpointing Fignl1/Kif1bbeta as a dynein speed limiter complex. 31934989_associations of single nucleotide polymorphisms of KIF1B gene with the severity of clinical manifestations of multiple sclerosis 33397043_Whole Exome Sequencing Identifies Novel Genetic Alterations in Patients with Pheochromocytoma/Paraganglioma. 34112167_USP9X promotes apoptosis in cholangiocarcinoma by modulation expression of KIF1Bbeta via deubiquitinating EGLN3. | ENSMUSG00000063077 | Kif1b | 2.639842e+03 | 1.0297070 | 0.042233853 | 0.2572397 | 2.651406e-02 | 0.8706512036 | 0.97573306 | No | Yes | 2.428446e+03 | 270.537238 | 2.152021e+03 | 245.899068 | |
| ENSG00000055332 | 5610 | EIF2AK2 | protein_coding | P19525 | FUNCTION: IFN-induced dsRNA-dependent serine/threonine-protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (EIF2S1/eIF-2-alpha) and plays a key role in the innate immune response to viral infection (PubMed:18835251, PubMed:19507191, PubMed:19189853, PubMed:21123651, PubMed:21072047, PubMed:22948139, PubMed:23229543, PubMed:22381929). Inhibits viral replication via the integrated stress response (ISR): EIF2S1/eIF-2-alpha phosphorylation in response to viral infection converts EIF2S1/eIF-2-alpha in a global protein synthesis inhibitor, resulting to a shutdown of cellular and viral protein synthesis, while concomitantly initiating the preferential translation of ISR-specific mRNAs, such as the transcriptional activator ATF4 (PubMed:19189853, PubMed:21123651, PubMed:22948139, PubMed:23229543). Exerts its antiviral activity on a wide range of DNA and RNA viruses including hepatitis C virus (HCV), hepatitis B virus (HBV), measles virus (MV) and herpes simplex virus 1 (HHV-1) (PubMed:11836380, PubMed:19189853, PubMed:20171114, PubMed:19840259, PubMed:21710204, PubMed:23115276, PubMed:23399035). Also involved in the regulation of signal transduction, apoptosis, cell proliferation and differentiation: phosphorylates other substrates including p53/TP53, PPP2R5A, DHX9, ILF3, IRS1 and the HHV-1 viral protein US11 (PubMed:11836380, PubMed:22214662, PubMed:19229320). In addition to serine/threonine-protein kinase activity, also has tyrosine-protein kinase activity and phosphorylates CDK1 at 'Tyr-4' upon DNA damage, facilitating its ubiquitination and proteosomal degradation (PubMed:20395957). Either as an adapter protein and/or via its kinase activity, can regulate various signaling pathways (p38 MAP kinase, NF-kappa-B and insulin signaling pathways) and transcription factors (JUN, STAT1, STAT3, IRF1, ATF3) involved in the expression of genes encoding proinflammatory cytokines and IFNs (PubMed:22948139, PubMed:23084476, PubMed:23372823). Activates the NF-kappa-B pathway via interaction with IKBKB and TRAF family of proteins and activates the p38 MAP kinase pathway via interaction with MAP2K6 (PubMed:10848580, PubMed:15121867, PubMed:15229216). Can act as both a positive and negative regulator of the insulin signaling pathway (ISP) (PubMed:20685959). Negatively regulates ISP by inducing the inhibitory phosphorylation of insulin receptor substrate 1 (IRS1) at 'Ser-312' and positively regulates ISP via phosphorylation of PPP2R5A which activates FOXO1, which in turn up-regulates the expression of insulin receptor substrate 2 (IRS2) (PubMed:20685959). Can regulate NLRP3 inflammasome assembly and the activation of NLRP3, NLRP1, AIM2 and NLRC4 inflammasomes (PubMed:22801494). Plays a role in the regulation of the cytoskeleton by binding to gelsolin (GSN), sequestering the protein in an inactive conformation away from actin (By similarity). {ECO:0000250|UniProtKB:Q03963, ECO:0000269|PubMed:10848580, ECO:0000269|PubMed:11836380, ECO:0000269|PubMed:15121867, ECO:0000269|PubMed:15229216, ECO:0000269|PubMed:18835251, ECO:0000269|PubMed:19189853, ECO:0000269|PubMed:19229320, ECO:0000269|PubMed:19507191, ECO:0000269|PubMed:19840259, ECO:0000269|PubMed:20171114, ECO:0000269|PubMed:20395957, ECO:0000269|PubMed:20685959, ECO:0000269|PubMed:21072047, ECO:0000269|PubMed:21123651, ECO:0000269|PubMed:21710204, ECO:0000269|PubMed:22214662, ECO:0000269|PubMed:22381929, ECO:0000269|PubMed:22801494, ECO:0000269|PubMed:22948139, ECO:0000269|PubMed:23084476, ECO:0000269|PubMed:23115276, ECO:0000269|PubMed:23229543, ECO:0000269|PubMed:23372823, ECO:0000269|PubMed:23399035, ECO:0000269|PubMed:32197074}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Antiviral defense;Cytoplasm;Direct protein sequencing;Host-virus interaction;Immunity;Innate immunity;Isopeptide bond;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase;Tyrosine-protein kinase;Ubl conjugation | The protein encoded by this gene is a serine/threonine protein kinase that is activated by autophosphorylation after binding to dsRNA. The activated form of the encoded protein can phosphorylate translation initiation factor EIF2S1, which in turn inhibits protein synthesis. This protein is also activated by manganese ions and heparin. The encoded protein plays an important role in the innate immune response against multiple DNA and RNA viruses. [provided by RefSeq, Jul 2021]. | hsa:5610; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; ribosome [GO:0005840]; ATP binding [GO:0005524]; double-stranded RNA binding [GO:0003725]; eukaryotic translation initiation factor 2alpha kinase activity [GO:0004694]; identical protein binding [GO:0042802]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase activity [GO:0004672]; protein phosphatase regulator activity [GO:0019888]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; RNA binding [GO:0003723]; cellular response to amino acid starvation [GO:0034198]; defense response to virus [GO:0051607]; endoplasmic reticulum unfolded protein response [GO:0030968]; innate immune response [GO:0045087]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of osteoblast proliferation [GO:0033689]; negative regulation of translation [GO:0017148]; negative regulation of viral genome replication [GO:0045071]; positive regulation of chemokine production [GO:0032722]; positive regulation of cytokine production [GO:0001819]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of stress-activated MAPK cascade [GO:0032874]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of hematopoietic progenitor cell differentiation [GO:1901532]; regulation of hematopoietic stem cell differentiation [GO:1902036]; regulation of hematopoietic stem cell proliferation [GO:1902033]; regulation of NLRP3 inflammasome complex assembly [GO:1900225]; response to interferon-alpha [GO:0035455]; response to virus [GO:0009615]; translation [GO:0006412] | 11991642_The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR (TCTP PROTEIN) 12051728_IL-1beta and IL-10 interact with IFN-alpha to up- and down-regulate PKR gene expression, respectively, by modulating STAT1 activation induced by IFN-alpha 12055262_Protein kinase R (PKR) regulates double-stranded RNA induction of TNF-alpha but not IL-1 beta mRNA expression in airway epithelial cells. 12060652_PKR phosphorylates small delta antigen 12138106_PKR protein kinase phosphorylation and dimerization is controlled byt PP1C 12231563_PKR activated by direct binding in heparin-treated vascular smooth muscle cells (VSMCs)blocks G1- to S-phase transition. 12349906_The induction of anti-HIV CTL activity by p9-RNA could be mediated by PKR through NF-kappaB activation 12368306_PKR may assume multiple roles in modulating hepatitis C virus replication and protein synthesis 12368348_Us11 protein of herpes simplex virus type 1 can block PKR activation by PACT both in vitro and in vivo 12396729_The promoter-proximal KCS element of the PKR kinase gene enhances transcription irrespective of orientation and position relative to the ISRE element. 12397061_PRK activity is suppressed by HSP70 in cooperation with FANCC 12447867_Observational study of gene-environment interaction. (HuGE Navigator) 12473108_PKR binds to ASK1 and is involved in apoptosis signalling pathways 12483527_PKR upregulation occurs at defined steps in cancer progression, probably as a cellular response to neoplasia 12882984_overexpression of NPM suppressed PKR activity, enhanced protein synthesis, and inhibited apoptosis. Fanconi anemia lymphoblasts expressed low levels of NPM, which correlated with high ground-state activation of PKR and hypersensitivity to apoptotic cues 12944978_Observational study of gene-disease association. (HuGE Navigator) 12944978_Polymorphisms in the interferon-inducible elF2alpha kinase gene is associated with outcome of hepatitis C virus infection 13678666_In contrast to nondemented controls, Alzheimer cases show prominent protein kinase PKR activation in association with neuritic plaques and pyramidal neurons in the hippocampus and neocortex. 14638359_PKR is overexpressed in hepatitis C virus related hepatocellular carcinoma. 14698665_viral-induced PKR activation may play a significant role in pathogenesis by mediating the host response to poliovirus encephalitis 14765129_PKR might play a role in ER stress-induced apoptosis and in Alzheimer's disease. 14961569_Loss of PKR activity may contribute to the formation and/or maintenance of B-cell chronic lymphocytic leukemia 15121867_TRAF-PKR interaction model in which the C-terminal domain of TRAF binds to a predicted TRAF interaction motif present in the PKR kinase domain. 15122791_level of PKR reduced in hepatocellular carcinoma tumor tissues, suggesting a role in promoting tumor growth; hepatitis B virus may participate in altering the level of PKR, but other factors should play a more determining role in regulation of PKR in HCC 15299030_Inappropriaate activation of PKR may cause mutations in fanconi anemia proteins. 15535414_upregulated in liver and PMBCs of chronic hepatitis C patients, and nonresponsivenes to IFN-alpha treatment is associated with this upregulation. The PKR gene response to exogenous IFN was similar in responders and non-responders in PBMCs. 15596837_phosphyorylation inhibited by Epstein-Barr virus BILF1 protein 15607693_first detection of a mutation in the RNA-binding domains of PKR gene from tumor cells taken directly from patients 15625311_double-stranded RNA-dependent protein kinase/eukaryotic initiation factor-2alpha has a role in okadaic acid-induced apoptosis in human osteoblastic MG63 cells 15737233_results support the hypothesis that PKR phosphorylation of Tat may be important for its function in HIV-1 LTR transactivation 15880455_hepatocarcinoma core positive cells are characterized by a significant PKR phosphorylation in Thr 446 residue, which leads deregulation of mitosis 15907845_PKR may play a critical role in mediating the subversive effects of HIV Tat resulting in IL-10 induction 16156900_Hepatitis c virus polyprotein expression caused a severe cytopathological effect in human cells as a result of inhibition of protein synthesis and apoptosis induction, triggered by the activation of the IFN-induced enzymes PKR and RNase L systems. 16271080_results do not support the concept of PKR as a tumor suppressor in small-size peripheral adenocarcinomas of the lung 16339759_in the absence of IFN treatment, activation of PKR basal expression is mediated by Sp1 and Sp3 proteins in a cooperative manner 16446363_HCV core expression leads to deregulation of the mitotic checkpoint via a p38/PKR-dependent pathway 16466763_We demonstrated that the direct binding of the influenza A virus NS1A protein to the N-terminal 230 amino acid region of PKR inhibits PKR activation by PACT and dsRNA 16580685_interaction between PKR and dsRNAs represents a crucial host cell defense mechanism against viral infection 16785445_These observations suggest a model for PKR activation upon binding of dsRNA or PACT. 16861340_RAX may function as a negative regulator of growth that is required to activate PKR in response to a broad range of apoptosis-inducing stress. 16861808_DRBP-120 is a double-stranded RNA (dsRNA)-binding protein, and it was detected in both the cytoplasm and the nucleus of HeLa cells associated with PKR. 16924232_PKR differentially regulates TNF signaling; IKK, Akt and JNK were positively regulated, whereas p44/p42 MAPK and p38 MAPK were negatively regulated. 16954686_levels of PKR in lymphocytes could follow the cognitive decline in Alzheimer's disease 16989899_Protein kinase R is involved in NF-kappaB mediated gene transcription of pro-inflammatory cytokines via IkappaB kinase-beta. 17202131_the salt bridge interaction and dimer interface observed in the PKR structure is critical for the activity of PKR, GCN2, and PERK 17284445_kinase domains is critical in the propagation of the activation signal and for PKR dimerization 17290288_PKR tyrosine phosphorylation provides an important link between IFN signalling and translational control through the regulation of eIF2alpha phosphorylation. 17307214_the effects of PKR on the replication cycle of parainfluenza virus type 5 17318221_Phosphorylation of translation initiation factor 2-alpha (eukaryotic initiation factor 2-alpha (eIF2-alpha)) and its kinase, PKR, occur in dsRNA-induced apoptosis but not in necrosis. 17420072_The present work analyses another gene involved in the host cell response to HSV-1, EIF2AK2 (eukaryotic translation initiation factor 2-alpha kinase 2. 17451862_PKR can function to control cell growth and cell differentiation, and its activity can be controlled by the action of several oncogenes [review] 17522227_Data suggest that protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. 17541283_The putative dsRNA receptor PKR may not play a pivotal role in the dsRNA-stimulated expression of inflammatory chemokines in airway epithelial cells. 17596833_it appears that HCV protein expression is directly dependent on PKR expression; PKR is antiviral toward HCV and responsible for IFN's effect against HCV. 17597457_The effects of ribavirin plus interferon alpha (IFN-alpha) on EIF2AK2 activity were greater than those observed for treatment with either ribavirin alone or IFN- alpha alone. 17612505_These results suggest that a novel mechanism other than endoplasmic reticulum(ER) stress element-dependent mRNA transcription is required for induction of GRP94 and phosphorylation of PKR contributes to the induction of GRP94 under ER stress. 17686861_PKR was not necessary for Sendai virus-induced IFN synthesis, suggesting that PKR is particularly important for recognition of WNV infection 17716668_These results indicate that the endoplasmic reticulum-stress-mediated eIF2alpha/ATF4/CHOP/cell death pathway is, to some degree, dependent on PACT-mediated PKR activation apart from the PERK pathway. 17785458_PKR activates glycogen synthase kinase 3 to promote the proteasomal degradation of p53. 17851256_Airway smooth muscle cells expressed PKR but it did not seem to be involved in poly I:C effects on muscarinic receptors. 17928244_this study suggests that the role played by non-coding T cells transcript in the activation of lymphocytes can be mediated by PKR through NF-kappaB activation 17928446_study provides evidence that double-stranded RNA-activated protein kinase (PKR), a stress kinase, is involved in HIV/gp120-associated neurodegeneration 17959656_the single-cycle yield of delta-E3L virus was increased by nearly 2 log10 in PKR-deficient cells over the impaired growth in PKR-sufficient cells. 17975119_Observational study of gene-disease association. (HuGE Navigator) 17977969_the paramyxovirus P/V gene products limit induction of PKR by limiting the synthesis of aberrant viral mRNAs and double-stranded RNA and thus prevent the shutdown of translation 18023289_PrP aggresomes initiate a cell stress response by activating the RNA-dependent protein kinase (PKR). 18048689_study reports that RNAs with very limited secondary structures activate PKR in a 5'-triphosphate-dependent fashion in vitro and in vivo 18063576_PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2alpha phosphorylation 18087277_Phosphorylation of PKR may be an important initiator of muscle wasting in cancer patients. 18426922_PKR activation by ssRNA and dsRNA containing internal nucleobase, sugar, and phosphodiester modifications was analyzed. 18496558_this protein kinase could be involved in the pathogenesis of myelodysplastic sysdromes 18599499_Double-stranded RNA-activated protein kinase mediates induction of interleukin-8 expression by deoxynivalenol, Shiga toxin 1, and ricin in monocytes. 18684815_Results demonstrate that the Sendai virus C protein limits generation of double stranded RNA, thereby keeping protein kinase R inactive to maintain intracellular protein synthesis. 18684960_Epidermal keratinocytes efficiently respond to viral double-stranded dsRNA equivalent poly(I:C) by expressing protein kinase R (PKR) dsRNA sensing molecules together with their downstream signaling pathway in a functional and differentially regulated way. 18728014_NS5A protein down-regulates the expression of spindle gene Aspm through PKR-p38 signaling pathway 18835251_These data provide the first evidence identifying Nck-1 as a novel endogenous regulator of PKR and support the notion that Nck-1-PKR interaction could be a way to limit PKR activation. 18927075_PKR facilitates the host innate immune response and apoptosis in virus-infected cells by mediating IRF-3 activation through the mitochondrial IPS-1 signal transduction pathway 18936160_TRBP controls PACT activation of PKR, an activity that is reversed by stress. 18957415_PKR regulates B56(alpha)-mediated BCL2 phosphatase activity in acute lymphoblastic leukemia-derived REH cells. 18976633_These results indicated that oxidative stress induces PKR expression essentially via the IFN-gamma activation signal, and causes apoptosis in Jurkat T cells. 19004947_These results indicate that PKR plays an important antiviral role during measles virus infection but that the virus growth restriction by PKR is not dependent upon the induction of apoptosis. 19023099_Observational study of gene-disease association. (HuGE Navigator) 19028691_PKR, in addition to IPS-1 and IRF3 but not TRIF, was required for maximal type I IFN-beta induction and the induction of apoptosis by both transfected PRNAs and polyinosinic-polycytidylic acid. 19106640_PKR expression is high in various cancer cells/low in normal cells. Knockdown of PKR protein expression induced cell death. PKR signaling via either siRNA or Ad-Delta6PKR sensitizes cancer cells to etoposide or cisplatin-mediated cell death. 19109397_Inhibition of PKR in SARS-CoV-infected cells does not affect viral replication. SARS-CoV infection activates PKR and PERK, leading to sustained eIF2alpha phosphorylation. Viral activation of PKR can lead to apoptosis. 19151623_These findings suggest that neuronal apoptosis in human CJD may result from PKR(p)-mediated cell stress and are consistent with recent studies supporting a pathogenic role for intracellular or transmembrane PrP. 19189853_Expression of PKR inhibited HCV replication with concomitant increase of eIF2alpha phosphorylation. 19210572_PKR represents a cognitive decline biomarker able to dysregulate translation via two consecutive targets p53 and Redd1 in Alzheimer disease lymphocytes 19229320_protein kinase R (PKR) is shown to interact with RNA helicase A; RHA is identified as a substrate for PKR, with phosphorylation perturbing the association of the helicase with double-stranded RNA 19259124_The eukaryotic translation initiation factor 2-alpha kinase 2 mediates the regulation of the protein level and the phosphorylation status of Proto-Oncogene Proteins c-bcl-2, providing a novel mechanism of palmitate-induced apoptosis in HepG2 cells. 19349624_results suggested that PKR is the primary target for HSV-1 early RNA during induction of FUT3, FUT5, and FUT6 19353519_Induction of IDO1 by TLR involved an autocrine interferon (IFN)-beta signaling loop, which was dependent on protein kinase R 19416861_p53-mediated tumor suppression can be attributed at least in part to the biological functions of PKR induced by p53 in genotoxic conditions 19434718_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19445956_These studies support that dimerization of a 15- to 30-bp hairpin RNA, which effectively doubles its length, is a key step in driving activation of PKR and provide a model for how RNA folding can be related to human disease. 19507191_Constitutive basal activity of PKR is required for leukemic cell homeostasis and growth and functions as a negative regulator of AKT, thereby increasing the pool of potentially active GSK-3. 19515768_The authors show that the IRF inhibitory domain of VP35 also mediates the inhibition of PKR and enhances the synthesis of coexpressed proteins. 19605474_two interferon-induced proteins, ADAR1 and PKR, have antagonistic functions on HIV production 19605483_PKR is activated in adenovirus-infected cells with multiple layers of regulation imposed on eIF2alpha phosphorylation by the E1B-55K/E4orf6 complex, which also regulates viral late protein synthesis 19642004_The expression of intrahepatic PKR protein at the ultra-structural level was characterised in patients with chronic hepatitis C disease compared to normal human peripheral blood mononuclear cells, HepG2 cells and a normal human liver biopsy. 19710021_the antiapoptotic activity of ADAR1 is achieved through suppression of activation of proapoptotic and double-stranded RNA-dependent activities, as exemplified by PKR and IRF-3. 19733181_Results indicate that the ADAR1 deaminase increases exogenous gene expression at the translational level by decreasing PKR-dependent eIF-2alpha phosphorylation. 19744687_Virus-induced eIF2alpha phosphorylation no longer occurred in PKR-deficient HeLa cells, suggesting PKR was responsible for vaccinia virus-induced eIF2alpha modification. 19776135_Infectious bronchitis virus has developed a combination of two mechanisms, i.e., blocking PKR activation and inducing GADD34 expression, to maintain de novo protein synthesis in IBV-infected cells and, meanwhile, to enhance viral replication. 19812373_antiviral PKR-dependent response also modulates the protozoan parasitic L. amazonensis infection. This effect depends on PKR-induced production of the suppressor cytokine IL-10. 19846517_PKR and mitochondrial adapter IPS-1 were required for maximal measles virus protein C(ko)-mediated IFN-beta induction, which correlated with the PKR-mediated enhancement of MAPK and NF-kappaB activation 19846675_EIF2alpha phosphorylation in vaccinia virus infected cells is mediated by protein kinase R(PKR). 20006836_Hepatitis C virus activated cellular protein kinase R to block translation of interferon-stimulated genes. 20006840_HCV infection triggers phosphorylation and activation of PKR which inhibits eIF2 alpha and attenuates interferon-stimulated protein expression. 20171114_PKR plays an important role in mediating the cytokine gene expression induced by LMP1 through NF-kappaB activation and histone H3 Ser 10 phosphorylation. 20181660_Taken together, PKR and Hck were critical for DON-induced ribosomal recruitment of p38, its subsequent phosphorylation, and, ultimately, p38-driven proinflammatory cytokine expression. 20309637_These findings indicate that the expression of MxA, 2',5'-OAS and PKR are up-regulate by PI3K-AKT signal pathway, and Raf-MEK-ERK signal pathway has a negative regulatory effect on the expression of MxA and no significant effect on 2',5'-OAS and PKR. 20331378_Observational study of gene-disease association. (HuGE Navigator) 20353946_response to lethal stress, PKR, without being phosphorylated, activates the FADD/caspase-8/caspase-3 pathway to trigger HuR cleavage, and the HuR-CPs are then capable of promoting apoptosis 20395957_Cdc2 was downregulated under genotoxic stresses and that double-stranded RNA-activated protein kinase (PKR) was involved in the process. 20447405_While it has been demonstrated that domains III-IV activate protein kinase R, the authors report here that domain II of the IRES of hepatitis C virus mRNA also potently activates. 20485506_The first evidence that Hepatitis C virus uses PKR to restrain its ability to induce IFN through the RIG-I/MAVS pathway, is provided. 20587610_Observational study of gene-disease association. (HuGE Navigator) 20588308_Observational study of gene-disease association. (HuGE Navigator) 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20685959_PKR regulates the upstream central transmitters of insulin signaling, IRS1 and IRS2, through different mechanisms. 20930042_The findings first indicate that PKR expression is an independent prognostic variable in non-small cell lung cancer patients. 21029237_PKR could represent a crucial signaling point relaying stress signals to neuronal pathways leading to cellular degeneration in Alzheimer's disease 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21072047_status of nuclear PKR in acute leukemia cell lines 21123378_Human parainfluenza virus type 1 C protein suppresses dsRNA intracellular accumulation and activation of MDA5 and PKR. 21123651_Activation of the dsRNA-activated protein kinase PKR by viral RNA enabled phosphorylation of NFAR1 and NFAR2 on Thr 188 and Thr 315 21338484_These data demonstrate that the PKR- and NF-kappaB-dependent induction of pro-inflammatory molecules that accompanies reovirus-mediated killing. 21377708_Activation of PKP is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. 21385567_MSK2 functions as an adaptor in mediating PKR activation, apparently independent of its catalytic activity. 21468538_PKR activation led to selective apoptosis induction in K562 cells, which correlated with caspase-8 activity and enhanced expression of BAX. 21504114_Specific inhibition of PKR at the peripheral level might decrease the inflammatory response in Alzheimer's disease. 21526770_results demonstrate for the first time that stress-induced PACT phosphorylation functions to free PACT from the inhibitory interaction with TRBP and also to enhance its interaction with PKR 21698289_Cellular localization studies showed that NP and Hsp40 co-localize primarily in the nucleus. During IAV infection in mammalian cells, expression of NP coincided with the dissociation of P58(IPK) from Hsp40 and decrease PKR phosphorylation 21710204_functions as a key mediator of IFN-alpha in opposing HBV replication 21715487_Mammalian orthoreovirus escape from host cell translational shutoff correlates with virus-induced cellular stress granules disruption and occurs in the presence of phosphorylated eIF2alpha in a PKR-independent manner. 21790829_The PACT-PKR pathway represents a potential link between Abeta accumulation, PKR activation and tau phosphorylation. 21846836_Leishmania infection increased the expression of PKR and IFN-beta on induction of PKR-promoter activity. The observed effects required the engagement of TLR2. 21882225_NF-kappa-B is involved in the protein activator of PKR-RNA-dependent protein kinase interaction and the production of pro-inflammatory cytokines in periodontitis. 22102852_Reduced PKR is associated with non-small cell lung cancer. 22174754_PKR is involved in the innate immune effects mediated by HBx-siRNAs and further contributes to hepatitis B virus inhibition. 22278235_Cytomegalovirus TRS1 protein antagonize protein kinase R in a host-specific manner. 22281122_The evaluation of CSF T-PKR and pPKR can discriminate between AD patients and NDC and could help to improve the biochemical diagnosis of AD. 22306812_findings provide a new translational regulation of BACE1, under the control of double-stranded RNA dependant protein kinase in oxidative stress, where eIF2-alpha phosphorylation regulates 22381929_This study used complementation of human cell lines stably deficient in PKR to probe the basis of PCR-mediated activation of MAPK and interferon beta induction by measles virus. 22473766_Data demonstrate that PACT-PACT interaction is essential for efficient PKR activation. 22787234_Japanese encephalitis virus nonstructural protein 2A blocks double-stranded RNA-activated protein kinase PKR 22801494_results show a crucial role for PKR in inflammasome activation, and indicate that it should be possible to pharmacologically target this molecule to treat inflammation 22894766_Suggest that PKR exerts a positive role in cell growth control of HCV-4 related hepatocellular carcinoma. 22896602_The restriction of Bunyamwera virus replication mediated by interferon is an accumulated effect of at least three interferon-stimulated genes viperin, MTAP44 and PKR. 22912486_5'-triphosphate-containing, weakly structured RNAs activate PKR via interactions with both the dsRBD and a distinct triphosphate binding site that lacks 5'-nucleobase specificity. 22948139_an eIF-2alpha-dependent translation inhibition mechanism is sufficient to explain the PKR-mediated amplification of IPS-1-dependent IFN-beta induction by foreign RNA. 22986343_By binding to PKR with a comparable affinity, nc886 competes with dsRNA and attenuates PKR activation by dsRNA. 23064357_evidence that type 1 diabetes (DMT1) is response to viral infection: Data suggest that presence of immunoreactive enteroviral VP1 within pancreatic beta cells in DMT1 is associated with induction of PKR (and down-regulation of bcl-2-like protein 3). 23084476_Report mechanism of autophagy control that involves STAT3 and PKR as interacting partners. 23115276_Measles virus-induced stress granule formation was PKR dependent but impaired by ADAR1. 23115300_PKR expression and the PKR binding domain of herpes simplex virus 1 Us11 are required for the antiautophagic activity of Us11. 23148689_The substrate interacting residues in ck1alpha have been identified using the structural model of kinase - substrate peptide. The PKR interacting NS5A 1b residues have also been predicted. 23221979_STAT3 and EIF2AK2 interact and have roles in controlling fatty acid-induced autophagy 23229543_Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation 23302873_The results suggest that early in infection the herpes simplex virus 1 VHS RNase degrades RNAs that activate PKR. 23314571_PKR is crucial for the integrity of newly synthesized IFN mRNA, thereby generating an optimal host antiviral immune response. 23320095_At the time of Alzheimer's disease diagnosis, a higher level of cerebrospinal fluid PKR can predict a faster rate of cognitive decline 23370317_Low PKR and high EphA2 ratio is associated with non-small-cell lung cancer. 23372823_PKR acts as a negative regulator of IFN induction triggered by DENVs and poly(IC), and this regulation relies on its dsRNA binding activity. 23399035_Authors conclude that host cells can employ PKR activation to restrict hepatitis C virus 1a replication through regulation of NF-kB expression. 23527187_RNA-dependent protein kinase is essential for 2-methoxyestradiol-induced autophagy in osteosarcoma cells. 23682076_Low EIF2AK2 expression is associated with low response to therapy in endometrial cancer. 23706307_These results suggest that PKR and PKZ function by distinguishable mechanisms to modulate host responses including protein synthesis inhibition and stress granule formation. 23853588_Data suggest that M029 (an RNA-binding protein) plays pivotal role in determining cellular tropism of Myxoma virus in all mammalian cells tested; human protein kinase R/EIF2AK2 and RNA helicase A/DHX9 are two binding partners of M029. 24020926_In contrast to its previously described activity, PACT contributes to PKR dephosphorylation during HIV-1 replication. 24089560_IFNA2 inhibits viral protein expression through PKR activation, leading to a decrease of viral protein synthesis. 24334130_PKR activation plays a major regulatory role in induction of apoptosis in response to ER stress and indicates the potential of PKR as possible target for neuroprotective therapeutics. 24347309_This study identified 120 proteins present in active and inactive PKR nuclear complexes. 24463368_PKR is a maladaptive factor upregulated in hemodynamic overload that contributes to myocardial inflammation, cardiomyocyte apoptosis, and the development of congestive heart failure. 24623135_NF90 exerts antiviral activity through regulation of PKR phosphorylation and stress granules in infected cells. 24684861_PKR is activated by dsRNA generated by Newcastle disease virus infection and inhibits Newcastle disease virus replication by eIF2alpha phosphorylation. 24786893_Cyclophilin inhibitors reduce phosphorylation of PKR and eIF2alpha during HCV infection to allow for translation of ISG products. 24992036_G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. 25074923_SUMO potentiates the inhibition of protein synthesis induced by PKR in response to dsRNA. 25297997_Early dsRNA induced transient activation of the cellular dsRNA sensor protein kinase R (PKR), resulting in enhanced production of interferons and cytokines in cells and mice. 25389016_This study demonstrates that two interferon stimulated genes, i.e. PKR and ADAR1 have opposite effects on HTLV replication in vivo. 25410857_Further studies revealed that Andes virus nucleocapsid protein inhibited PKR dimerization, a critical step required for PKR autophosphorylation to attain activity. 25520508_Authors show that the PXXP domain within G3BP1 is essential for the recruitment of PKR to stress granules, for eIF2alpha phosphorylation driven by PKR, and for nucleating stress granules of normal composition. 25592072_The results from this study indicate an important role of RAX/PKR association in regulating PKR activity as well as ethanol neurotoxicity 25607115_No significant association was determined between the rs2254958 EIF2AK2 polymorphism and the development of IBD, or clinical outcome. 25653431_PKR directly interacts with HIV-1 Tat and phosphorylates the first exon of Tat exclusively at five Ser/Thr residues, which inhibits Tat-mediated provirus transcription. 25704249_Protein levels of PRKR were significantly increased in prefrontal cortex in chronic excessive alcohol use. 25715336_these data indicate a pivotal role for PKR's protein-binding function on the proliferation of pancreatic beta cells through TRAF2/RIP1/NF-kappaB/c-Myc pathways. 25784705_The G3BP1-Caprin1-PKR complex represents a new mode of PKR activation and is important for antiviral activity of G3BP1 and PKR during infection with mengovirus. 25899421_Classical swine fever virus (CSFV) infection increased the phosphorylation of eukaryotic translation initiation factor (eIF)2alpha and its kinase PKR. The activation of PKR during CSFV infection is beneficial to the virus. 26202421_PKR expression correlates with inferior survival and shorter remission duration for acute myeloid leukemia patients. 26216977_Mechanism by which PK2 inhibits the model eIF2alpha kinase human RNA-dependent protein kinase (PKR) as well as native insect eIF2alpha kinases, is reported. 26231208_the affinity of PACT-PACT and PACT-PKR interactions is enhanced in dystonia patient lymphoblasts, thereby leading to intensified PKR activation and enhanced cellular death. 26321373_Tyrosine phosphorylated EIF2AK2 plays a role in the regulation of insulin induced protein synthesis and in maintaining insulin sensitivity. 26488609_The data support a model in which activating RNAs induce formation of a back-to-back parallel PKR kinase dimer whereas nonactivating RNAs either fail to induce dimerization or produce an alternative, inactive dimer configuration. 26608746_the expression of a Tat construct containing mutations in the basic region (49-57aa), which is responsible for the interaction with PKR, favored neither parasite growth nor IL-10 expression in infected macrophages. 26698173_results show that ceramide acts at two distinct levels of the insulin signaling pathway (IRS1 and Akt). PKR, which is induced by both inflammation signals and ceramide, could play a major role in the development of insulin resistance in muscle cells. 26716879_The sole essential function of cytomegalovirus TRS1 is to antagonize host PKR. 26725122_This study provides insight into the molecular pathology of Cornelia de Lange syndrome by establishing a relationship between NIPBL and HDAC8 mutations and PKR activation. 26831644_The PKR is a key constituent of the metaflammasome and interacts directly with several inflammatory kinases, such as inhibitor kappaB (IkappaB) kinase (IKK) and c-Jun N-terminal kinase (JNK), insulin receptor substrate 1 (IRS1), and component of the translational machinery such as eIF2alpha. 26869028_The Newcastle disease virus-induced translation shutoff at late infection times was attributed to sustaining phosphorylation of eIF2a, which is mediated by continual activation of PKR and degradation of PP1. 27021640_Crucially, Chlamydia trachomatis infection resulted in robust IRE1alpha RNAse activity that was dependent on TLR4 signalling and inhibition of IRE1alpha RNAse activity prevented PKR activation. 27203671_Findings suggest a novel role for PKR in lung cancer cells as a mediator of radiation resistance possibly through translocation of the protein product to the nucleus. 27422384_In insulitic islets from living patients with recent-onset T1D, most of the overexpressed ISGs, including GBP1, TLR3, OAS1, EIF2AK2, HLA-E, IFI6, and STAT1, showed higher expression in the islet core compared with the peri-islet area containing the surrounding immune cells 27423063_NF90 exerts its antiviral activity by antagonizing the inhibitory role of NS1 on PKR phosphorylation 27560627_Protein kinase R (PKR) was required for induction of stress granules (SGs) by mumps virus (MuV) infection and regulated type III IFN (IFN-lambda1) mRNA stability. 27833096_Gene silencing studies showed that the suppression of immunoproteasome induction is essentially dependent on protein kinase R (PKR). Indeed, the generation of a strictly immunoproteasome-dependent cytotoxic T lymphocyte epitope was impaired in in vitro processing experiments using isolated 20S proteasomes from HCV-infected cells and was restored by the silencing of PKR expression. 28069888_The stem-loop of noncoding RNA 886 is structural feature not only critical for inhibiting PKR autophosphorylation, but also the phosphorylation of its cellular substrate, EIF-2alpha. 28112215_The finding that zebularine upregulates CYP gene expression through DNMT1 and PKR modulation sheds light on the mechanisms controlling hepatocyte function and thus may aid in the development of new in-vitro systems using high-functioning hepatocytes 28167698_It was established in this report that interactions between PACT, ADAR1 and HIV-1-encoded Tat protein dimin | ENSMUSG00000024079 | Eif2ak2 | 6.924911e+02 | 1.2499531 | 0.321873985 | 0.3631507 | 7.752246e-01 | 0.3786056154 | 0.84115767 | No | Yes | 7.089469e+02 | 152.722283 | 4.790193e+02 | 105.509772 | |
| ENSG00000057252 | 6646 | SOAT1 | protein_coding | P35610 | FUNCTION: Catalyzes the formation of fatty acid-cholesterol esters, which are less soluble in membranes than cholesterol (PubMed:16154994, PubMed:16647063, PubMed:9020103, PubMed:32433614, PubMed:32433613, PubMed:32944968). Plays a role in lipoprotein assembly and dietary cholesterol absorption (PubMed:16154994, PubMed:9020103). Utilizes oleoyl-CoA ((9Z)-octadecenoyl-CoA) preferentially as susbstrate: shows a higher activity towards an acyl-CoA substrate with a double bond at the delta-9 position (9Z) than towards saturated acyl-CoA or an unsaturated acyl-CoA with a double bond at the delta-7 (7Z) or delta-11 (11Z) positions (PubMed:11294643, PubMed:32433614). {ECO:0000269|PubMed:11294643, ECO:0000269|PubMed:16154994, ECO:0000269|PubMed:16647063, ECO:0000269|PubMed:32433613, ECO:0000269|PubMed:32433614, ECO:0000269|PubMed:32944968, ECO:0000269|PubMed:9020103}. | 3D-structure;Acetylation;Acyltransferase;Alternative splicing;Cholesterol metabolism;Disulfide bond;Endoplasmic reticulum;Lipid metabolism;Membrane;Phosphoprotein;Reference proteome;Steroid metabolism;Sterol metabolism;Transferase;Transmembrane;Transmembrane helix | The protein encoded by this gene belongs to the acyltransferase family. It is located in the endoplasmic reticulum, and catalyzes the formation of fatty acid-cholesterol esters. This gene has been implicated in the formation of beta-amyloid and atherosclerotic plaques by controlling the equilibrium between free cholesterol and cytoplasmic cholesteryl esters. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Nov 2011]. | hsa:6646; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; cholesterol binding [GO:0015485]; cholesterol O-acyltransferase activity [GO:0034736]; fatty-acyl-CoA binding [GO:0000062]; identical protein binding [GO:0042802]; O-acyltransferase activity [GO:0008374]; sterol O-acyltransferase activity [GO:0004772]; cholesterol efflux [GO:0033344]; cholesterol esterification [GO:0034435]; cholesterol homeostasis [GO:0042632]; cholesterol metabolic process [GO:0008203]; cholesterol storage [GO:0010878]; low-density lipoprotein particle clearance [GO:0034383]; macrophage derived foam cell differentiation [GO:0010742]; positive regulation of amyloid precursor protein biosynthetic process [GO:0042986]; very-low-density lipoprotein particle assembly [GO:0034379] | 12533546_allosteric activation by cholesterol 12851640_polymorphism of the gene encoding acyl-coenzyme A: cholesterol acyltransferase 1 (SOAT1) is involved in the regulation of beta-amyloid peptide generation, is associated with low brain amyloid load and with low cerebrospinal fluid levels of cholesterol 14729857_ACAT-1 transcripts predominate in human liver and ACAT-2 transcripts predominate in human duodenum and support the notion that ACAT-2 has an important regulatory role in liver and intestine. 15158756_Observational study of gene-disease association. (HuGE Navigator) 15219857_expression in monocytes infiltrating from the circulation to vascular walls may be enhanced by pre-existing transforming growth factor-beta1 15253151_A stable upstream stem-loop structure enhances selection of the first 5'-ORF-AUG as a main start codon for translation initiation of ACAT1 mRNA. 15308631_increasing DGAT1, ACAT1, or ACAT2 expression stimulates the assembly and secretion of VLDL from liver cells 15353128_a glucocorticoid response element (GRE) located within human ACAT1 gene P1 promoter to response to the elevation of human ACAT1 gene expression by dexamethasone could be functionally bound with glucocorticoid receptor (GR) proteins. 15768051_The results of a comprehensive genetic assessment of SOAT1 variants in the NIMH AD Genetics Initiative study sample are presented. 15850387_Disulfide linkage map shows that cysteine(C)92 is located on the cytoplasmic side of the endoplasmic reticulum (ER) membrane and the disulfide is located in the ER lumen, while all other free Cs are located within the hydrophobic region(s) of the enzyme. 15992359_The structural features of various sterols as substrates and/or activators of ACAT1 and ACAT2 in vitro are reported. 16013913_Observational study of gene-disease association. (HuGE Navigator) 16043284_Observational study of gene-disease association. (HuGE Navigator) 16043284_Our results indicate that the polymorphism rs1044925 in the 3'UTR of SOAT1 gene does not affect the risk of SAD in the northern Han-Chinese. 16154994_results led us to construct a revised, nine-transmembrane domain model, with the active site His-460 located within a hitherto undisclosed transmembrane domain, between Arg-443 and Tyr-462 16230498_Serum-induced depletion of cellular cholesterol available for esterification by ACAT was a strong, independent predictor of major adverse cardiovascular events and death. 16474185_The new ACAT inhibitor VULM1457 in concentration 0.03 and 0.1 micromol/l significantly down-regulated specific AM receptors on HepG2 cells, reduced AM secretion of HepG2 cells exposed to hypoxia. 16647063_histidine residues located at the active site are very crucial both for the catalytic activity of the enzyme and for distinguishing ACAT1 from ACAT2 with respect to enzyme catalysis and substrate specificity 16763159_Observational study of gene-disease association. (HuGE Navigator) 17412327_These data show that even a modest decrease in ACAT activity can have robust suppressive effects on Abeta generation. 17593314_Observational study of gene-disease association. (HuGE Navigator) 17593314_Our study confirmed cognitive decline and highly frequent delirium after bypass heart surgery and excluded the possible role of SOAT-1 genotype polymorphisms in their genesis 17622762_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17622762_SOAT1 gene may possibly be only a minor risk factor in Alzheimer's disease 17691824_Several residues in one subunit closely interact with the same residues in the other subunit; mutating these res. to Cys does not lead to loss in enzyme activity. Mutating residues F453, A457, or H460 to Cys causes large loss in enzyme activity. 18000807_increase in the in vitro ACAT1 activity in PC-3 prostate cancer cells treated with androgen 18269457_In clear cell type renal cell carcinoma upregulation of ACAT-1 leads to high ACAT enzymatic activity, which accelerates the accumulation of cholesterol ester. 18393248_Observational study of gene-disease association. (HuGE Navigator) 18393248_The results suggest that rs1044925 polymorphism in ACAT1 gene is not only associated with serum LDL-C and nHDLC levels in healthy Chinese subjects in Chengdu area, but also with HDL-C level in subjects with endogenous hypertriglyceridemia. 18542101_RNA secondary structures located in the vicinity of the GGC(1274-1276) codon are required for production of the 56-kDa isoform. 18779653_ACAT inhibition may stimulate cholesterol-catabolic (cytochrome P450) pathway in lesion-macrophages. 18971559_Angiotensin II enhances foam cell formation by upregulating ACAT1 expression predominantly through the actions of AT(1) receptor via the G protein/c-Src/PKC/MAPK pathway in human monocyte-macrophages 19217763_Docosahexaenoic acid can act as a substrate for ACAT1. In the manner of a poor substrate, docosahexaenoic acid also inhibited the activity of ACAT1. 19269342_Results suggest that the ERK, p38MAPK and JNK signaling pathways may be involved in insulin-mediated regulation of ACAT1, but no PI3K and PLC-gamma signaling pathways were involved in the present study. 19502590_Results show that signaling through ACAT/cholesterol esterification is a novel pathway for the CCK2R that contributes to tumor cell proliferation and invasion. 19625677_Leptin accelerates cholesteryl ester accumulation in human monocyte-derived macrophages by increasing ACAT-1 expression. 19851860_High ACAT1 expression is associated with estrogen receptor negative basal-like breast cancer. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20031551_Observational study of gene-disease association. (HuGE Navigator) 20460577_Macrophages cope with cholesterol loading by using a novel mechanism: they produce more ER-derived vesicles with elevated ACAT1 enzyme activity without having to produce more ACAT1 protein. 20602615_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20843517_the plaque-modulating effects of K-604 can be explained by stimulation of procollagen production independent of ACAT inhibition in addition to potent inhibition of macrophage ACAT-1 20945045_Visfatin may down-regulate the ABCA1 expression and up-regulate the ACAT1 expression via PPARgamma signal transduction pathway, which decreases the outflow of free cholesterol, increases the content of cholesterol esters, and then induces foam cell formation. 21143839_the polymorphism of rs1044925 in the ACAT-1 gene is mainly associated with female serum total cholesterol, LDL-C and ApoB levels in the Bai Ku Yao population 21177161_In THP-1-derived macrophages and foam cells, the expression level of ACAT-1 and cellular cholesterol content increased significantly in response to asymmetric dimethylarginine treatment in a time- and concentration-dependent manner. 21239516_SF-1-dependent up-regulation of SOAT1 may be important for maintaining readily-releasable cholesterol reserves needed for active steroidogenesis and during episodes of recurrent stress. 21266192_These findings suggest the potential involvement of MAPK and STAT pathways in norcantharidin-induced apoptogenesis. 21803834_These studies demonstrate that both SIAE and SOAT activities seem to be responsible for the enhanced level of Neu5,9Ac(2) in lymphoblasts, which is a hallmark in acute lymphoblastic leukemia 22243772_The present study shows that the C allele carriers of ACAT-1 rs1044925 SNP in male hyperlipidemic subjects had higher serum total cholesterol, HDL-cholesterol and ApoAI levels than the C allele noncarriers. 22275303_Essential oil of Pinus koraiensis leaves significantly inhibited hACAT1 levels in HepG2 cells. 23109900_Several lipid-related gene polymorphisms interact with overweight/obesity to modulate blood pressure levels. 23305686_ABCA1 and ACAT1 DNA methylation induced by homocysteine may play a potential role in ABCA1 and ACAT1 expression and the accumulation of cholesterol in monocyte-derived foam cells 23564383_The molecular mechanism of insulin action is mediated via interaction of the functional IRE upstream of the ACAT1 P1 promoter with C/EBPalpha and is MAPK-dependent. 23835473_The exo-endo trans-splicing is dependent on the interchromosomal region of the 4.3-kb human ACAT1 chimeric mRNA, and that the chimeric mRNA is necessary for the production of the ACAT1 56-kDa isoform. 23994608_PLA/AT-1 is at least partly responsible for the generation of N-acylphosphatidylethanolamine in mammalian cells. 24407243_Induction of apoptosis and necroptosis by 24(S)-hydroxycholesterol is dependent on activity ACAT1. 24517390_the enzyme activity of ACAT1 with Gln526 is less active than that of ACAT1 with Arg526 by 40%; Pro347 located near transmembrane domain 5 plays an important role in modulating enzyme catalysis 24577316_Data show that the C allele of acyl-CoA acyltransferase-1 (ACAT-1) rs1044925 was associated with a decreased risk of coronary artery disease and ischemic stroke patients. 25339759_Acat1 gene knock-out increases phagocytic uptake of amyloid beta-protein (1-42). 25617738_Suggest retinal pigment epithelium metabolism of 7-ketocholesterol occurs by esterification to fatty acids via cPLA2alpha and SOAT1 followed by selective efflux to HDL. 26252415_ACAT1 regulates glioblastoma-cell proliferation via modification of the Akt and/or the ERK1/2 pathway. 26305886_Data indicate that mitotane confers adrenal-specific cytotoxicity and down-regulates steroidogenesis by inhibition of sterol-O-acyl-transferase 1 (SOAT1) leading to lipid-induced endoplasmic reticulum (ER) stress. 26474739_ACAT1 has a role in regulating the dynamics of free cholesterols in plasma membrane which leads to the APP-alpha-processing alteration 27177773_TLR4 siRNA inhibits cell proliferation, migration and invasion by suppressing ACAT1 expression, suggesting that TLR4 may be a potential therapeutic target for the treatment of colorectal cancer 27281560_Intracranial GBM xenografts were used to determine the effects of genetically silencing SOAT1 and SREBP-1 on tumor growth. 27346255_Our results demonstrated the contrasting effects of STC1 and STC2-derived peptides on human macrophage foam cell formation associated with ACAT1 expression and on HASMC migration. 27647838_these results illustrate that ACAT1-catalyzed esterification of 24S-OHC with long-chain unsaturated fatty acid followed by formation of atypical LD-like structures at the ER membrane is a critical requirement for 24S-OHC-induced cell death. 28595267_Higher Gleason grade was associated with lower LDLR expression, lower SOAT1 and higher SQLE expression. Besides high SQLE expression, cancers that became lethal despite primary treatment were characterized by low LDLR expression (odds ratio for highest versus lowest quintile, 0.37; 95% CI 0.18-0.76) and by low SOAT1 expression (odds ratio, 0.41; 95% CI 0.21-0.83). 29567472_Soat-1 role in atherosclerosis 29593722_the transcription factor signal transducer and activator of transcription 3 was activated by TB-PE, and its chemical inhibition prevented the accumulation of lipid bodies and ACAT expression in macrophages. In terms of the host immune response, TB-PE-treated macrophages displayed immunosuppressive properties and bore higher bacillary loads. 31236660_There was a significant difference and a higher frequency of AA and AC genotypes (p = 0.047 and p = 0.016, respectively) of the SOAT1 (ACAT-1) gene in asymptomatic chagasic individuals. 31978092_the expression and contribution of ACAT-1 (SOAT1) in ovarian cancer progression, was examined. 32101051_Association Between LOX-1, LAL, and ACAT1 Gene Single Nucleotide Polymorphisms and Carotid Plaque in a Northern Chinese Population. 32433613_cryo-electron microscopy structure of human ACAT1 (SOAT1) in complex with nevanimibe, an inhibitor that is in clinical trials for the treatment of congenital adrenal hyperplasia 32433614_cryo-electron microscopy structure of human ACAT1(SOAT1) as a dimer of dimers; structural and biochemical characterization helps to rationalize the preference of ACAT1 for unsaturated acyl chains, and provides insight into the catalytic mechanism of enzymes within the MBOAT family 32633781_SOAT1 promotes mevalonate pathway dependency in pancreatic cancer. 32646003_A Dipeptidyl Peptidase-4 Inhibitor Inhibits Foam Cell Formation of Macrophages in Type 1 Diabetes via Suppression of CD36 and ACAT-1 Expression. 33057949_Relationship Between Genetic Variants of ACAT1 and APOE with the Susceptibility to Dementia (SADEM Study). 34039309_Impacts of the SOAT1 genetic variants and protein expression on HBV-related hepatocellular carcinoma. 35354120_piRNA-6426 increases DNMT3B-mediated SOAT1 methylation and improves heart failure. 35409086_SOAT1: A Suitable Target for Therapy in High-Grade Astrocytic Glioma? | ENSMUSG00000026600 | Soat1 | 8.493588e+02 | 0.9992978 | -0.001013470 | 0.3467300 | 8.490876e-06 | 0.9976750389 | 0.99948746 | No | Yes | 7.094304e+02 | 136.756891 | 6.943278e+02 | 137.172178 | |
| ENSG00000058668 | 493 | ATP2B4 | protein_coding | P23634 | FUNCTION: Calcium/calmodulin-regulated and magnesium-dependent enzyme that catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell (PubMed:8530416). By regulating sperm cell calcium homeostasis, may play a role in sperm motility (By similarity). {ECO:0000250|UniProtKB:Q6Q477, ECO:0000269|PubMed:8530416}. | 3D-structure;ATP-binding;Alternative splicing;Calcium;Calcium transport;Calmodulin-binding;Cell membrane;Cell projection;Cilium;Direct protein sequencing;Flagellum;Ion transport;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Translocase;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene belongs to the family of P-type primary ion transport ATPases characterized by the formation of an aspartyl phosphate intermediate during the reaction cycle. These enzymes remove bivalent calcium ions from eukaryotic cells against very large concentration gradients and play a critical role in intracellular calcium homeostasis. The mammalian plasma membrane calcium ATPase isoforms are encoded by at least four separate genes and the diversity of these enzymes is further increased by alternative splicing of transcripts. The expression of different isoforms and splice variants is regulated in a developmental, tissue- and cell type-specific manner, suggesting that these pumps are functionally adapted to the physiological needs of particular cells and tissues. This gene encodes the plasma membrane calcium ATPase isoform 4. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:493; | basolateral plasma membrane [GO:0016323]; caveola [GO:0005901]; glutamatergic synapse [GO:0098978]; integral component of plasma membrane [GO:0005887]; integral component of presynaptic active zone membrane [GO:0099059]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; membrane raft [GO:0045121]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; sperm flagellum [GO:0036126]; sperm principal piece [GO:0097228]; T-tubule [GO:0030315]; Z disc [GO:0030018]; ATP binding [GO:0005524]; ATPase-coupled cation transmembrane transporter activity [GO:0019829]; calcium ion transmembrane transporter activity [GO:0015085]; calcium-dependent protein binding [GO:0048306]; calmodulin binding [GO:0005516]; metal ion binding [GO:0046872]; nitric-oxide synthase binding [GO:0050998]; nitric-oxide synthase inhibitor activity [GO:0036487]; P-type calcium transporter activity [GO:0005388]; PDZ domain binding [GO:0030165]; protein kinase binding [GO:0019901]; protein phosphatase 2B binding [GO:0030346]; scaffold protein binding [GO:0097110]; sodium channel regulator activity [GO:0017080]; calcium ion export [GO:1901660]; calcium ion import across plasma membrane [GO:0098703]; calcium ion transmembrane import into cytosol [GO:0097553]; calcium ion transmembrane transport [GO:0070588]; cellular calcium ion homeostasis [GO:0006874]; cellular response to acetylcholine [GO:1905145]; cellular response to epinephrine stimulus [GO:0071872]; flagellated sperm motility [GO:0030317]; hippocampus development [GO:0021766]; ion transmembrane transport [GO:0034220]; negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process [GO:0140199]; negative regulation of angiogenesis [GO:0016525]; negative regulation of arginine catabolic process [GO:1900082]; negative regulation of blood vessel endothelial cell migration [GO:0043537]; negative regulation of calcineurin-NFAT signaling cascade [GO:0070885]; negative regulation of cardiac muscle hypertrophy in response to stress [GO:1903243]; negative regulation of cellular response to vascular endothelial growth factor stimulus [GO:1902548]; negative regulation of citrulline biosynthetic process [GO:1903249]; negative regulation of gene expression [GO:0010629]; negative regulation of nitric oxide biosynthetic process [GO:0045019]; negative regulation of nitric oxide mediated signal transduction [GO:0010751]; negative regulation of nitric-oxide synthase activity [GO:0051001]; negative regulation of peptidyl-cysteine S-nitrosylation [GO:1902083]; negative regulation of the force of heart contraction [GO:0098736]; neural retina development [GO:0003407]; positive regulation of cAMP-dependent protein kinase activity [GO:2000481]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of protein localization to plasma membrane [GO:1903078]; regulation of cardiac conduction [GO:1903779]; regulation of cell cycle G1/S phase transition [GO:1902806]; regulation of cytosolic calcium ion concentration [GO:0051480]; regulation of sodium ion transmembrane transport [GO:1902305]; regulation of transcription by RNA polymerase II [GO:0006357]; response to hydrostatic pressure [GO:0051599]; spermatogenesis [GO:0007283]; transport across blood-brain barrier [GO:0150104]; urinary bladder smooth muscle contraction [GO:0014832] | 11751908_We demonstrate here that, in the early phase of apoptosis, hPMCA4b is cleaved at aspartic acid Asp(1080) in hPMCA4b-transfected COS-7 cells or in HeLa cells that naturally express this protein 12511555_plasma membrane Ca2+ pump 4b/CI binds to Ca2+/calmodulin-dependent membrane-associated kinase CASK 12540962_co-immunoprecipitation, treatment with tyrosine kinase inhibitors and integrin inhibition experiments suggest that FAK is responsible for PMCA4b tyrosine phosphorylation during platelet activation. 12784250_role in the intracellular Ca(2+) extrusion of syncytiotrophoblast-like structure originating from the differentiation of cultured trophoblast cells isolated from human term placenta 12944246_results suggest that a decrease in PMCA4b expression in Meg-01 cells is compensated to maintain normal intracellular calcium levels. 15145946_functional association with RASSF1 indicates a role for PMCA4b in the modulation of Ras-mediated signaling 15292209_residue Asp(170), in the putative 'A' domain of human PMCA isoform 4xb, plays a critical role in autoinhibition 15955804_a novel functional interaction between PMCA and calcineurin, suggesting a role for PMCA as a negative modulator of calcineurin-mediated signaling pathways in mammalian cells. 16080782_The characteristics of the fragment of hPMCA4b produced by caspase-3 are reported. 16216224_Although differences in PMCA4 mRNA levels were observed between breast cell lines, they were not of the magnitude observed for PMCA2. 16412504_PMCA activity is influenced by membrane lipid composition and structure. The naturally high degree of lipid order in plasma membranes such as those found in human lens may serve to support PMCA activity. 16973504_found abnormal expression of both PMCA and SERCA-type CA2+-ATPases in platelets of patients with adolescent idiopathic scoliosis 16978418_PMCA4 is a good housekeeping gene for normalization of gene expression for polytopic membrane proteins including transporters and receptors. 17242280_physiological relevance of the interaction between PMCA4b and nNOS and suggests its signaling role in the heart 17393022_PMCA4b is localized in non-filamentous actin complexes in resting platelets by means of PDZ domain interactions and then associates with the actin cytoskeleton during cytoskeletal rearrangement upon platelet activation. 17883705_Compared with the expression of PMCA4b upon platelet maturation, platelets from diabetic patients exhibit similarities with immature megakaryocytes 17957572_platelets in hypertensive patients demonstrated a major increase in plasma membrane Ca(2+)ATPase 4b (PMCA4b) expression when compared with normal controls 18657858_PMCA4 seems to be preferentially distributed in both human syncytiotrophoblast plasma membranes. 19034380_This protein has been found differentially expressed in patients with schizophrenia. 19073225_Results suggest that PSD-95 promotes the formation of high-density PMCA4b microdomains in the plasma membrane and that the membrane cytoskeleton plays an important role in the regulation of this process. 19287093_Pmca4b likely reduces the local Ca2+ signals involved in reactive cardiomyocyte hypertrophy via calcineurin regulation. 19536175_Observational study of gene-disease association. (HuGE Navigator) 19755660_PMCA4 is significantly (P < 0.000001) downregulated early in the progression of some colon cancers as these cells become less differentiated 19996092_Data show that the anchors correspond to Phe-1110 and Trp-1093, respectively, in full-length PMCA4b, and the peptide and CaM are oriented in an anti-parallel manner. 20211863_Report a novel interaction between endogenous plasma membrane calcium ATPase (PMCA) and eNOS in endothelial cells. PMCA may negatively modulate eNOS activity, and NO-dependent signal transduction pathways. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21073872_regulatory domain works independently of its terminal localization and if auto-inhibitory domains 21139678_PMCA4 is strongly expressed in rabbit CE and its immunolocalization exhibits marked changes in distribution during the wound healing process. 21167220_The recently emerging role of the plasma membrane calcium/calmodulin dependent ATPase isoform 4 (PMCA4) in regulating calcium signalling, is reviewed. 22480210_The results of this study suggest posttranscriptional regulation of PMCA4 during carcinogenesis. 22525089_GUSB and ATP2B4 have been validated as a reliable gene combination for Cystic Fibrosis Transmembrane Conductance Regulator gene qPCR data normalization. 22767601_Alternative pathways for association and dissociation of the calmodulin-binding domain of plasma membrane Ca(2+)-ATPase isoform 4b (PMCA4b). 22895189_Genome-wide association study for severe malaria identified a locus on chromosome 1q32 within the ATP2B4 gene, and another locus on chromosome 16q22.2, possibly linked to a neighbouring gene encoding the tight-junction protein MARVELD3 23266958_High PMCA4 gene expression correlates with high peak bone mass in humans. 23444010_An ATP2B4 variant reduces the odds of P. falciparum infection in pregnancy and mitigates the odds of associated maternal anemia. 23549614_The PMCA4b enriched in lipid rafts decreases the local [Ca2+] levels, downregulating the nNOS activity dynamically associated to PMCA4b via Ca2+-dependent PDZ domain-mediated protein- protein interactions. 23830917_a di-leucine-like internalization signal at the C-tail of PMCA4b, is reported. 24583174_A significant relationship between ATP2B4 gene expression and the tumor location was detected in patients with rectum tumors. 24583174_No significant relationship was detected in the level of expression of the ATP2B4 and ATP5B genes in cancerous and healthy tissues of colorectal cancer patients. 25119969_Calcium dysregulation resulting from a novel missense mutation (c.803G>A, p.R268Q) in the PMCA4 (ATP2B4) gene may be associated with the pathogenesis of familial spastic paraplegia. 25147342_PMCA4 inhibits the activation of the calcineurin/NFAT pathway on VEGF stimulation of endothelial cells, leading to a significant attenuation of VEGF-mediated angiogenesis. 25233416_Mechanism of PMCA4 that creates lipid asymmetry is well understood in terms of ATP hydrolysis; molecular models exist for trajectory taken by phospholipid substrates through the enzyme. [review-like article] 25690014_The slowly activating PMCA4b isoform produced long-lasting Ca2+ oscillations in response to store-operated Ca2+ entry. 25798335_p.R268Q mutation in PMCA4 resulted in functional changes in calcium homeostasis in human neuronal cells. This suggests that calcium dysregulation may be associated with the pathogenesis of Familial spastic paraplegia 26116539_While PMCA1b has a housekeeping function in colon cancer cells, PMCA4b participates in the reorganization of the calcium signaling machinery during cell differentiation. 26448358_We also nominate a new candidate gene in congenital arrhythmia, ATP2B4, and provide experimental evidence of a regulatory role for variants discovered using this framework. 26676968_RNA sequencing identified a novel ATPase, Ca2+ transporting, plasma membrane 4(ATP2B4)-protein kinase C-alpha (PRKCA) fusion transcript. 26729804_our data show that CD147 interacts via its immunomodulatory domains with PMCA4 to bypass TCR proximal signaling and inhibit IL-2 expression. 27813079_Increased plasma membrane abundance of PMCA4b in vemurafenib-treated BRAF mutant cells is associated with enhanced Ca2+ clearance. 28216081_data suggest that an altered regulation of gene expression is responsible for the reduced RBC-PMCA4b levels that is probably linked to the development of human disease-related phenotypes. 28327142_The role of heterozygous variants of ATP2B4 is regulated by HSPG2 protein with transcription factors in bone formation through modulation of calcium signaling. 28684310_study provides evidence for the therapeutic potential of targeting PMCA4 to improve VEGF-based pro-angiogenic interventions. This goal will require the development of refined, highly selective versions of ATA, or the identification of novel PMCA4 inhibitors. 28714864_the ATP2B4 enhancer mediates red blood cell hydration and malaria susceptibility 28787189_Na+, K+-ATPase and Ca2+-ATPase activity and Na+, K+-ATPase alpha4 and PMCA4 isoform expressions examined in asthenozoospermic men 29042438_Data (including data from studies in knockout mice) suggest that renalase functions as a protective plasma protein that reduces pancreatic acinar cell injury and prevents pancreatitis via interactions with plasma membrane calcium ATPase PMCA4b. 29487197_PMCA4b overexpression preserves cardiac function following ischemia reperfusion injury, heightens cardiac performance and limits infarct progression, cardiac hypertrophy and heart failure. 31594854_The Ca(2+) export pump PMCA clears near-membrane Ca(2+) to facilitate store-operated Ca(2+) entry and NFAT activation. 32002807_Molecular and Electrophysiological Analyses of ATP2B4 Gene Variants in Bilateral Adrenal Hyperaldosteronism. 32414111_P38 MAPK Promotes Migration and Metastatic Activity of BRAF Mutant Melanoma Cells by Inducing Degradation of PMCA4b. 32860837_The calcium pump PMCA4 prevents epithelial-mesenchymal transition by inhibiting NFATc1-ZEB1 pathway in gastric cancer. 32926932_CircATP2B4 promotes hypoxia-induced proliferation and migration of pulmonary arterial smooth muscle cells via the miR-223/ATR axis. 34093590_Plasma Membrane Calcium ATPase Regulates Stoichiometry of CD4(+) T-Cell Compartments. 34174923_Replicative verification of susceptibility genes previously identified from families with segregating developmental dysplasia of the hip. 35322081_Renalase and its receptor, PMCA4b, are expressed in the placenta throughout the human gestation. | ENSMUSG00000026463 | Atp2b4 | 1.746692e+03 | 0.9324853 | -0.100847099 | 0.2538602 | 1.571186e-01 | 0.6918233382 | 0.93376105 | No | Yes | 1.651363e+03 | 139.715698 | 1.744513e+03 | 151.209134 | |
| ENSG00000058673 | 9877 | ZC3H11A | protein_coding | O75152 | FUNCTION: RNA-binding protein that interacts with purine-rich sequences and is involved in nuclear mRNA export; probably mediated by association with the TREX complex. {ECO:0000269|PubMed:22928037, ECO:0000269|PubMed:29610341}.; FUNCTION: (Microbial infection) Plays a role in efficient growth of several nuclear-replicating viruses such as HIV-1, influenza virus or herpes simplex virus 1/HHV-1. Required for efficient viral mRNA Export. {ECO:0000269|PubMed:29610341}. | Coiled coil;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transport;Ubl conjugation;Zinc;Zinc-finger;mRNA transport | Mouse_homologues NA; + ;NA | hsa:9877; | nucleoplasm [GO:0005654]; metal ion binding [GO:0046872]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; poly(A)+ mRNA export from nucleus [GO:0016973] | 19536175_Observational study of gene-disease association. (HuGE Navigator) 22928037_PDIP3 and ZC11A associate with the human TREX complex in an ATP-dependent manner and function in mRNA export. 29610341_Knockout of ZC3H11A in HeLa cells demonstrated that several nuclear-replicating viruses are dependent on ZC3H11A for efficient growth whereas cytoplasmic replicating viruses are not. ZC3H11A binds to short purine-rich ribonucleotide stretches in cellular and adenoviral transcripts. ZC3H11A is important for maintaining nuclear export of mRNAs during stress. Several nuclear-replicating viruses take advantage of this. | ENSMUSG00000102976+ENSMUSG00000116275 | Zc3h11a+Zc3h11a | 3.105477e+03 | 0.7662076 | -0.384192847 | 0.3215692 | 1.417167e+00 | 0.2338703300 | 0.78818582 | No | Yes | 2.343379e+03 | 400.676962 | 2.981539e+03 | 522.479027 | |
| ENSG00000061455 | 93166 | PRDM6 | protein_coding | Q9NQX0 | FUNCTION: Putative histone methyltransferase that acts as a transcriptional repressor of smooth muscle gene expression. Promotes the transition from differentiated to proliferative smooth muscle by suppressing differentiation and maintaining the proliferative potential of vascular smooth muscle cells. Also plays a role in endothelial cells by inhibiting endothelial cell proliferation, survival and differentiation. It is unclear whether it has histone methyltransferase activity in vivo. According to some authors, it does not act as a histone methyltransferase by itself and represses transcription by recruiting EHMT2/G9a. According to others, it possesses histone methyltransferase activity when associated with other proteins and specifically methylates 'Lys-20' of histone H4 in vitro. 'Lys-20' methylation represents a specific tag for epigenetic transcriptional repression. {ECO:0000250|UniProtKB:Q3UZD5}. | Alternative splicing;Chromatin regulator;Disease variant;Metal-binding;Methyltransferase;Nucleus;Reference proteome;Repeat;Repressor;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Zinc;Zinc-finger | The protein encoded by this gene is a transcriptional repressor and a member of the PRDM family. Family members contain a PR domain and multiple zinc-finger domains. The encoded protein is involved in regulation of vascular smooth muscle cells (VSMC) contractile proteins. Mutations in this gene result in patent ductus arteriosus 3 (PDA3). [provided by RefSeq, Apr 2017]. | hsa:93166; | nucleus [GO:0005634]; histone methyltransferase activity [GO:0042054]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; negative regulation of smooth muscle cell differentiation [GO:0051151]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neurogenesis [GO:0022008]; regulation of gene expression [GO:0010468] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25035420_Three novel loci were identified in East Asians with cardiac arrhythmias: rs2483280 (PRDM16 locus) and rs335206 (PRDM6 locus) were associated with QRS duration; and rs17026156 (SLC8A1 locus) correlated with PR interval. 27181681_Mutations in the Histone Modifier PRDM6 Are Associated with Isolated Nonsyndromic Patent Ductus Arteriosus. 30677029_we detected a novel obesity and BMI-associated locus at PKHD1 and novel variants driving associations at previously established signals (e.g. rs205262 at the SNRPC/C6orf106 locus and rs112446794 at the PRDM6-CEP120 locus). | ENSMUSG00000069378 | Prdm6 | 3.088411e+02 | 1.3102706 | 0.389864796 | 0.2921449 | 1.775649e+00 | 0.1826844640 | 0.77830220 | No | Yes | 3.661405e+02 | 45.086874 | 2.529632e+02 | 32.029338 | |
| ENSG00000061794 | 60488 | MRPS35 | protein_coding | P82673 | 3D-structure;Alternative splicing;Coiled coil;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transit peptide | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein that has had confusing nomenclature in the literature. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. Pseudogenes corresponding to this gene are found on chromosomes 3p, 5q, and 10q. [provided by RefSeq, Jul 2010]. | hsa:60488; | mitochondrial inner membrane [GO:0005743]; mitochondrial small ribosomal subunit [GO:0005763]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; mitochondrial translation [GO:0032543] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000040112 | Mrps35 | 7.438595e+02 | 1.4759974 | 0.561690176 | 0.3421729 | 2.667098e+00 | 0.1024426801 | 0.75783482 | No | Yes | 9.563038e+02 | 192.683814 | 4.968537e+02 | 103.003445 | ||
| ENSG00000062194 | 65056 | GPBP1 | protein_coding | Q86WP2 | FUNCTION: Functions as a GC-rich promoter-specific transactivating transcription factor. {ECO:0000250|UniProtKB:Q6NXH3}. | Activator;Alternative splicing;Cytoplasm;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | Mouse_homologues NA; + ;NA | This gene was originally isolated by subtractive hybridization of cDNAs expressed in atherosclerotic plaques with a thrombus, and was found to be expressed only in vascular smooth muscle cells. However, a shorter splice variant was found to be more ubiquitously expressed. This protein is suggested to play a role in the development of atherosclerosis. Studies in mice suggest that it may also function as a GC-rich promoter-specific trans-activating transcription factor. Several alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Feb 2011]. | hsa:65056; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; RNA binding [GO:0003723]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription, DNA-templated [GO:0006355]; transcription, DNA-templated [GO:0006351] | 11256614_This publication discusses both human and mouse GC-rich promoter binding protein. 12842993_Vasculin is a novel vascular protein differentially expressed in human atherogenesis 17672918_occurrence of an unusual TG 3' splice site in intron 9 | ENSMUSG00000032745+ENSMUSG00000116016 | Gpbp1+Gm49496 | 8.057942e+02 | 0.7820857 | -0.354601472 | 0.3655416 | 8.969433e-01 | 0.3436026443 | 0.82878860 | No | Yes | 7.663680e+02 | 160.621159 | 8.527412e+02 | 183.217127 |
| ENSG00000063241 | 79763 | ISOC2 | protein_coding | Q96AB3 | Mouse_homologues NA; + ;NA | Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome | Mouse_homologues NA; + ;NA | hsa:79763; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; protein destabilization [GO:0031648] | 17658461_ISOC2 is a novel functional protein, which is able to bind and co-localize with a tumor suppressor gene p16(INK4a). | ENSMUSG00000052605+ENSMUSG00000086784 | Isoc2b+Isoc2a | 4.087475e+03 | 1.3556499 | 0.438984598 | 0.3417657 | 1.626740e+00 | 0.2021545407 | 0.78445471 | No | Yes | 4.607386e+03 | 732.965556 | 2.754525e+03 | 450.020911 | |
| ENSG00000063854 | 3029 | HAGH | protein_coding | Q16775 | FUNCTION: Thiolesterase that catalyzes the hydrolysis of S-D-lactoyl-glutathione to form glutathione and D-lactic acid. | 3D-structure;Acetylation;Alternative initiation;Alternative splicing;Cytoplasm;Hydrolase;Metal-binding;Mitochondrion;Reference proteome;Transit peptide;Zinc | PATHWAY: Secondary metabolite metabolism; methylglyoxal degradation; (R)-lactate from methylglyoxal: step 2/2. | The enzyme encoded by this gene is classified as a thiolesterase and is responsible for the hydrolysis of S-lactoyl-glutathione to reduced glutathione and D-lactate. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2013]. | hsa:3029; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; hydroxyacylglutathione hydrolase activity [GO:0004416]; metal ion binding [GO:0046872]; glutathione biosynthetic process [GO:0006750]; glutathione metabolic process [GO:0006749]; methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione [GO:0019243] | 15117945_hydroxyacylglutathione hydrolase (HAGH) gene encodes both cytosolic and mitochondrial forms of glyoxalase II 16803681_Overexpression of glyoxalase II is associated with kidney tumor 16831876_Data show that the GLX2 gene, which encodes glyoxalase II enzyme, is up-regulated by p63 and p73. 18344682_In androgen-dependent prostate cancer cells, testosterone upregulates GLO2 mRNA levels. In androgen-independent prostate cancer cells, it downregulates GLO2 mRNA. 19413286_Human glyoxalase II contains an Fe(II)Zn(II) center but is active as a mononuclear Zn(II) enzyme 20237496_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 24671236_No association between genetic variants of the HAGH gene and autism spectrum disorder was found. 25645869_This study suggested that HAGH, rs11859266 and rs3743852 showed significant associations with schizophrenia in males at allelic and genotype levels. 26552067_Studies indicate that the most extensively investigated The most extensively investigated glyoxalase enzymes are glyoxalase I and glyoxalase II (Glo1 and Glo2). 26914966_epidermal expression stronger in older skin donors 27696457_Glo2, together with Glo1, represents a novel mechanism in prostate cancer progression as part of a pathway driven by PTEN/PI3K/AKT/mTOR signaling. 27935136_This article reports for the first time a possible additional role of Glo2 that, after interacting with a target protein, is able to promote S-glutathionylation. 29385039_The critical role of glyoxalases as regulators of tumorigenesis in the prostate through modulation of various critical signaling pathways, and an overview of the current knowledge on glyoxalases in bladder, kidney and testis cancers is reviewed. (GLO1, GLO2) 29950256_Down regulation of Glyoxalase II was observed in cases of diabetic retinopathy as compared to controls. 32427856_CDH6 and HAGH protein levels in plasma associate with Alzheimer's disease in APOE epsilon4 carriers. 33725370_Sirtuin 2 Regulates Protein LactoylLys Modifications. | ENSMUSG00000024158 | Hagh | 2.959410e+03 | 1.1424697 | 0.192155915 | 0.3085277 | 3.895232e-01 | 0.5325501554 | 0.88844615 | No | Yes | 2.879605e+03 | 317.872030 | 2.323947e+03 | 263.571796 |
| ENSG00000064547 | 9170 | LPAR2 | protein_coding | Q9HBW0 | FUNCTION: Receptor for lysophosphatidic acid (LPA), a mediator of diverse cellular activities. Seems to be coupled to the G(i)/G(o), G(12)/G(13), and G(q) families of heteromeric G proteins. Plays a key role in phospholipase C-beta (PLC-beta) signaling pathway. Stimulates phospholipase C (PLC) activity in a manner that is independent of RALA activation. {ECO:0000269|PubMed:15143197, ECO:0000269|PubMed:19306925}. | 3D-structure;Cell membrane;G-protein coupled receptor;Glycoprotein;Lipoprotein;Membrane;Palmitate;Receptor;Reference proteome;Transducer;Transmembrane;Transmembrane helix | This gene encodes a member of family I of the G protein-coupled receptors, as well as the EDG family of proteins. This protein functions as a lysophosphatidic acid (LPA) receptor and contributes to Ca2+ mobilization, a critical cellular response to LPA in cells, through association with Gi and Gq proteins. An alternative splice variant has been described but its full length sequence has not been determined. [provided by RefSeq, Jul 2008]. | hsa:9170; | cell surface [GO:0009986]; cytoplasm [GO:0005737]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; G protein-coupled receptor activity [GO:0004930]; lipid binding [GO:0008289]; lysophosphatidic acid receptor activity [GO:0070915]; activation of phospholipase C activity [GO:0007202]; adenylate cyclase-activating G protein-coupled receptor signaling pathway [GO:0007189]; G protein-coupled receptor signaling pathway [GO:0007186]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; regulation of metabolic process [GO:0019222] | 12123830_demonstrate that two biological fluids, blood plasma and seminal plasma, differentially activate LPA receptors 12668280_results suggested that LPA(2) and LPA(3) may be involved in VEGF expression mediated by LPA signals in human ovarian oncogenesis 12759391_LPA may directly increase the level of cyclin D1 in ovarian cancer cells, increasing their proliferation. 15535846_Upregulation of LPA2 may play a role in carcinogenesis, particularly in postmenopausal breast cancer. 15728708_LPA2 is the major LPA receptor in colon cancer cells and cellular signals by LPA2 are largely mediated through its ability to interact with NHERF2. 15755723_formation of the LPA receptor/PDZ domain-containing RhoGEF complex plays a pivotal role in LPA-induced RhoA activation 16904289_These results demonstrate that MAGI-3 interacts directly with LPA(2) and regulates the ability of LPA(2) to activate Erk and RhoA. 17496233_EDG4 and EDG2 cooperate to promote LPA-stimulated chemotaxis in breast tumor cell lines. 17765657_data suggest that LPA receptor-dependent expression of CTGF and CYR61 represents a common host response after interaction with bacteria. 17965021_lysophosphatidic acid 2 receptor mediates down-regulation of Siva-1 to promote cell survival 18703779_A role for the transgenic lysophosphatidic acid (LPA)2 receptor is identified in regulating smooth muscle cell migratory responses in the context of vascular injury. 18754873_LPA and LPA receptors, LPA(2) as well as LPA(1), represent potential therapeutic targets for patients with MPM 19001604_Expression of LPA2 during ovarian carcinogenesis contributes to ovarian cancer aggressiveness, suggesting that the targeting of LPA production and action may have potential for the treatment of ovarian cancer. 19025891_Switching of LPA receptor expression from LPA3 to LPA1, may be involved in prostate cancer progression and/or androgen independence 19081821_LPA(1) receptor, LPA(2) and LPA(3) receptors-induced VASP phosphorylation is a critical mediator of tumor cell migration initiation 19116446_LPA2 and Gi/Src pathways are significant for LPA-induced COX-2 expression and cell migration that could be a promising drug target for ovarian cancer cell metastasis. 19860625_Data show that CLL cells express LPA receptors LPA(1-5) and VEGF receptors, and the plasma levels of VEGF are elevated in CLL patients. 19899077_show that human microglia express LPA receptor subtypes LPA(1), LPA(2), and LPA(3) on mRNA and protein level. LPA activation of C13NJ cells induced Rho and extracellular signal-regulated kinase activation and enhanced cellular ATP production. 20890765_LPA2 gene mutation may play some role in the pathogenesis of colon cancer. 21134377_MAGI-3 competes with NHERF-2 to negatively regulate LPA2 receptor signaling in colon cancer cells. 21234797_This work shows for the first time that key components of the LPA pathway are modulated following traumatic brain injuries in humans. 21964883_found that LPA receptor 2/3-mediated IL-8 expression occurs through Gi/PI3K/AKT, Gi/PKC and IkappaB/NF-kappaB signaling 23084965_LPA2 and LPA6 receptor subtypes are predominant in both HPAECs and HMVECs 23569130_Lysophosphatidic acid (LPA) increased hepatocellular carcinoma cells cell invasion, which was LPA-receptor dependent. 24061591_LPA1 and LPA2 are major LPA receptor subtypes compared with low-expressed LPA3 in PANC-1 tumor cells. 24613836_Crystal structure of NHERF2 PDZ1 domain complex with C-terminal LPA2 sequence. The PDZ1-LPA2 binding specificity is achieved by hydrogen bonds and hydrophobic contacts with the last four LPA2 residues contributing to specific interactions. 24950964_the RhoA-regulated formin Dia1 is involved in entosis downstream of LPAR2 25463482_Suggest that LPA2 and LPA3 may function as a molecular switch and play opposing roles during megakaryopoiesis of K562 cells. 26327335_Data show high expression levels of LPAR2 and LPAR1 in endometrial cancer tissue with positive correlations with FIGO stage suggesting them as potential biomarkers for endometrial cancer progression. 26937138_LPAR2 mRNA is up-regulation in colorectal cancer. 27124742_epithelial dysplasia was observed in founder mouse intestine, correlating LPA2 overexpression with epithelial dysplasia. The current study demonstrates that overexpression of LPA2 alone can lead to intestinal dysplasia. 27244685_The results indicate that LPA2 and LPA3 receptors play opposing roles during red blood cells differentiation. 27583415_LPA2 mRNA levels were associated with poorer differentiation, and higher LPA6 levels were associated with microvascular invasion in HCC; both became a risk factor for recurrence after surgical treatment when combined with increased serum ATX levels 27805252_LPA2 expression was associated with HIF-1alpha expression and that a high level of LPA2 was associated with shorter overall survival and was an independent prognostic predictor for breast cancer in Chinese women. 28205098_These results suggest that LPA signaling via LPA2 may play an important role in the regulation of cellular functions in HT1080 cells treated with cisplatin. 29621954_Study shows that due to the high LPAR2 and LPAR4 transcript and protein expression in endometriotic ovarian cysts and positive correlations of both these receptors with the PR-B and ERbeta, respectively, those receptors seem to be the most promising predictors of the endometriotic cysts. 29859140_Investigated the roles of LPA receptors in the regulation of cellular functions during tumor progression in osteosarcoma cell lines. MG63-R7-C cell activities were inhibited by LPA2 knockdown, suggesting that LPA signaling via LPA2 plays an important role in the acquisition of malignant properties during tumor progression in MG-63 cells. 30093116_LPAR2 and LPAR5 regulate cellular functions during tumor progression in fibrosarcoma HT1080 cells. 31115486_It was revealed that LPA may stimulate the expression of Notch1 and Hes family bHLH transcription factor 1, and the phosphorylation of protein kinase B which belongs to the Notch pathway. 31235682_The positive expression rate of LPA2 and KLF5 were statistical different in gastric adenocarcinoma, GIN, and normal gastric tissue (P<0.05). LPA2 positive expression was associated with tumor invasion depth, Lauren type, vascular invasion, local lymph node metastasis, and clinical stage (P<0.05). 32006610_Effects of lysophosphatidic acid (LPA) receptor-2 (LPA2) and LPA3 on the regulation of chemoresistance to anticancer drug in lung cancer cells. 33347862_Roles of endothelial cells in the regulation of cell motility via lysophosphatidic acid receptor-2 (LPA2) and LPA3 in osteosarcoma cells. 33406026_Transcriptional regulation of lysophosphatidic acid receptors 2 and 3 regulates myeloid commitment of hematopoietic stem cells. 33959968_BRET analysis reveals interaction between the lysophosphatidic acid receptor LPA2 and the lysophosphatidylinositol receptor GPR55 in live cells. 35241179_LPAR2 correlated with different prognosis and immune cell infiltration in head and neck squamous cell carcinoma and kidney renal clear cell carcinoma. | ENSMUSG00000031861 | Lpar2 | 1.352827e+02 | 0.7714266 | -0.374399209 | 0.3487280 | 1.149754e+00 | 0.2836006227 | 0.80801941 | No | Yes | 9.488784e+01 | 15.888637 | 1.297419e+02 | 21.994766 | |
| ENSG00000065150 | 3843 | IPO5 | protein_coding | O00410 | FUNCTION: Functions in nuclear protein import as nuclear transport receptor. Serves as receptor for nuclear localization signals (NLS) in cargo substrates. Is thought to mediate docking of the importin/substrate complex to the nuclear pore complex (NPC) through binding to nucleoporin and the complex is subsequently translocated through the pore by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to the importin, the importin/substrate complex dissociates and importin is re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus (By similarity). Mediates the nuclear import of ribosomal proteins RPL23A, RPS7 and RPL5. Binds to a beta-like import receptor binding (BIB) domain of RPL23A. In vitro, mediates nuclear import of H2A, H2B, H3 and H4 histones. Binds to CPEB3 and mediates its nuclear import following neuronal stimulation (By similarity). In case of HIV-1 infection, binds and mediates the nuclear import of HIV-1 Rev. {ECO:0000250|UniProtKB:Q8BKC5, ECO:0000269|PubMed:9687515}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Host-virus interaction;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | Nucleocytoplasmic transport, a signal- and energy-dependent process, takes place through nuclear pore complexes embedded in the nuclear envelope. The import of proteins containing a nuclear localization signal (NLS) requires the NLS import receptor, a heterodimer of importin alpha and beta subunits also known as karyopherins. Importin alpha binds the NLS-containing cargo in the cytoplasm and importin beta docks the complex at the cytoplasmic side of the nuclear pore complex. In the presence of nucleoside triphosphates and the small GTP binding protein Ran, the complex moves into the nuclear pore complex and the importin subunits dissociate. Importin alpha enters the nucleoplasm with its passenger protein and importin beta remains at the pore. Interactions between importin beta and the FG repeats of nucleoporins are essential in translocation through the pore complex. The protein encoded by this gene is a member of the importin beta family. [provided by RefSeq, Jul 2008]. | hsa:3843; | cytoplasm [GO:0005737]; membrane [GO:0016020]; nuclear pore [GO:0005643]; nucleolus [GO:0005730]; nucleus [GO:0005634]; GTPase inhibitor activity [GO:0005095]; nuclear import signal receptor activity [GO:0061608]; nuclear localization sequence binding [GO:0008139]; RNA binding [GO:0003723]; small GTPase binding [GO:0031267]; cellular response to amino acid stimulus [GO:0071230]; negative regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0045736]; NLS-bearing protein import into nucleus [GO:0006607]; positive regulation of protein import into nucleus [GO:0042307]; protein import into nucleus [GO:0006606]; ribosomal protein import into nucleus [GO:0006610] | 12620808_L1 major capsid protein of human papillomavirus type 11 interacts with Kap beta3 nuclear import receptors 15364420_The KPNB3 locus may contain a disease-causing variant for schizophrenia. 15507604_HPV16 L2 interacts via its NLSs with a network of karyopherins and can enter the nucleus via several import pathways mediated by Kapalpha(2)beta(1) heterodimers, Kapbeta(2), and Kapbeta(3). 16644122_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16644122_The present work suggests that the combination of the KPNA3 gene and the KPNB3 gene may increase a genetic risk for schizophrenia. 17005651_RanBP5 acts as an import factor for the newly synthesized influenza A virus RNA-dependent-RNA polymerase by targeting the PB1-PA dimer to the nucleus. 17458142_Observational study of gene-disease association. (HuGE Navigator) 18295457_Lack of association of the KPNB3 locus in schizophrenia. 18455505_HPV-16 E5 protein binds to karyopherin beta3. 18562802_These results suggest that karyopherin beta3 plays a crucial role in apo A-I secretion. 19581934_Impaired p53 binding to importin: a novel mechanism of cytoplasmic sequestration identified in oxaliplatin-resistant cells. 20542336_Observational study of gene-disease association. (HuGE Navigator) 20542336_The results of this study suggested that abnormal expression and alternative splicing of the IPO5 gene may be involved in the pathophysiology of schizophrenia. 20828572_importin beta3 is essential for the nuclear import of RPL7. The import is mediated via the multifaceted basic amino acid clusters present in the NH(2)-region of RPL7, and is RanGTP-dependent 21562121_The N-terminal of influenza A virus PB1 mediates its binding to host RanBP5. 23266416_In case of L7, importin beta2 or importin beta3 are preferentially used by clusters with a high import efficiency. 24196961_IQGAP1 interacts with human importin-beta5 in HEK 293T cells. 26488411_Single nucleotide polymorphism in IPO5 gene is associated with Peripheral Arterial Disease. 27094387_importin-alpha5, which is a key regulator of interferon signaling following Ebola virus infection, as one putative target of miRNA. 27528606_Importins, Impbeta, Kapbeta2, Imp4, Imp5, Imp7, Imp9, and Impalpha, show the H3 tail binding more tightly than the H4 tail. The H3 tail binds Kapbeta2 and Imp5 with KD values of 77 and 57 nm, respectively, and binds the other five Importins more weakly. 27703004_The nuclear importin IPO5 was identified as a novel interacting protein of SMAD1. Overexpression of IPO5 in various cell lines specifically increases nuclear localization of BMP receptor-activated SMADs (R-SMADs) confirming a functional relationship between IPO5 and BMP but not TGF-beta R-SMADs. 29127199_Results indicate that the interaction between FLIL33 and IPO5 is localized to a specific segment of the FLIL33 protein, is not required for nuclear localization of FLIL33, and protects FLIL33 from proteasome-dependent degradation. 31288861_IPO5 is an oncogene involved in CRC cell proliferation and migration. This highlights the significance of IPO5 in 5-fluorouracil-resistant CRC cells. The oncogenic function of IPO5 was mediated by promoting RAS signalling by increasing the nuclear translocation of RASAL2 31340999_DDX56 inhibits type I interferon by disrupting assembly of IRF3-IPO5 to inhibit IRF3 nucleus import. 32222384_X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking. 32373960_IPO5 promotes malignant progression of esophageal cancer through activating MMP7. | ENSMUSG00000030662 | Ipo5 | 3.560845e+03 | 0.7237434 | -0.466449844 | 0.3194550 | 2.149849e+00 | 0.1425839264 | 0.76841160 | No | Yes | 3.144668e+03 | 615.733621 | 4.050888e+03 | 813.253355 | |
| ENSG00000065357 | 1606 | DGKA | protein_coding | P23743 | FUNCTION: Diacylglycerol kinase that converts diacylglycerol/DAG into phosphatidic acid/phosphatidate/PA and regulates the respective levels of these two bioactive lipids (PubMed:2175712, PubMed:15544348). Thereby, acts as a central switch between the signaling pathways activated by these second messengers with different cellular targets and opposite effects in numerous biological processes (PubMed:2175712, PubMed:15544348). Also plays an important role in the biosynthesis of complex lipids (Probable). Can also phosphorylate 1-alkyl-2-acylglycerol in vitro as efficiently as diacylglycerol provided it contains an arachidonoyl group (PubMed:15544348). Also involved in the production of alkyl-lysophosphatidic acid, another bioactive lipid, through the phosphorylation of 1-alkyl-2-acetyl glycerol (PubMed:22627129). {ECO:0000269|PubMed:15544348, ECO:0000269|PubMed:2175712, ECO:0000269|PubMed:22627129, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Calcium;Cytoplasm;Kinase;Lipid metabolism;Metal-binding;Nucleotide-binding;Reference proteome;Repeat;Transferase;Zinc;Zinc-finger | PATHWAY: Lipid metabolism; glycerolipid metabolism. {ECO:0000305|PubMed:15544348, ECO:0000305|PubMed:2175712, ECO:0000305|PubMed:22627129}. | The protein encoded by this gene belongs to the eukaryotic diacylglycerol kinase family. It acts as a modulator that competes with protein kinase C for the second messenger diacylglycerol in intracellular signaling pathways. It also plays an important role in the resynthesis of phosphatidylinositols and phosphorylating diacylglycerol to phosphatidic acid. Several transcript variants encoding different isoforms have been identified for this gene. [provided by RefSeq, Apr 2017]. | hsa:1606; | cytosol [GO:0005829]; membrane [GO:0016020]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; diacylglycerol kinase activity [GO:0004143]; kinase activity [GO:0016301]; lipid binding [GO:0008289]; NAD+ kinase activity [GO:0003951]; phospholipid binding [GO:0005543]; diacylglycerol metabolic process [GO:0046339]; glycerolipid metabolic process [GO:0046486]; intracellular signal transduction [GO:0035556]; lipid phosphorylation [GO:0046834]; phosphatidic acid biosynthetic process [GO:0006654]; platelet activation [GO:0030168]; protein kinase C-activating G protein-coupled receptor signaling pathway [GO:0007205] | 14734770_Defects in both polymorphonuclear neutrophil (PMN) transendothelial migration and PMN diacylglycerol kinase alpha signaling are implicated as disordered functions in subjects with localized aggressive periodontitis. 15117825_PPARgamma agonists upregulate DGKalpha production.This suppresses the diacylglycerol/protein-kinase-C signaling pathway. 15870081_DGKalpha is crucial for the control of cell activation and also for the regulation of the secretion of lethal exosomes, which in turn controls cell death. 15928040_ALK-mediated alphaDGK activation is dependent on p60src tyrosine kinase, with which alphaDGK forms a complex; alphaDGK activation is involved in the control of ALK-mediated mitogenic properties. 17276726_These results strongly suggest that DGKalpha is a novel positive regulator of NF-kappaB, which suppresses TNF-alpha-induced melanoma cell apoptosis. 17911109_diacylglycerol kinase alpha-conserved domains have a role in membrane targeting in intact T cells 18004883_2,3-dioleoylglycerol binds to a site on the alpha and zeta isoforms of diacylglycerol kinase that is exposed as a consequence of the substrate binding to the active site. 18424699_Lck-dependent tyrosine phosphorylation of diacylglycerol kinase alpha regulates its membrane association in T cells.( 19751727_These results strongly suggest that DGKalpha positively regulates TNF-alpha-dependent NF-kappaB activation via the PKCzeta-mediated Ser311 phosphorylation of p65/RelA. 21252909_Diacylglycerol kinase alpha is a key regulator of the polarised secretion of exosomes. 21493725_findings further suggest that DGL-alpha and -beta may regulate neurite outgrowth by engaging temporally and spatially distinct molecular pathways 22048771_SAP-mediated inhibition of DGKalpha sustains diacylglycerol signaling, thereby regulating T cell activation 22271650_Antigen-specific CD8-positive T cells from DGKalpha-deficient transgenic mice show enhanced expansion and increased cytokine production after lymphocytic choriomeningitis virus infection, yet DGK-deficient memory CD8+ T cells exhibit impaired expansion. 22425622_DGKalpha is involved in hepatocellular carcinoma progression by activation of the MAPK pathway. 22573804_DGK-alpha was more highly expressed in CD8-tumor-infiltrating T cellscompared with that in CD8non-tumor kidney-infiltrating lymphocytes. 24158111_High diacylglycerol kinase alpha expression is associated with glioblastoma. 24887021_These data indicates the existence of a SDF-1alpha induced DGKalpha - atypical PKC - beta1 integrin signaling pathway, which is essential for matrix invasion of carcinoma cells. 25248744_DGKalpha generates phosphatidic acid to drive its own recruitment to tubular recycling endosomes via its interaction with MICAL-L1 25921290_Redundant and specialized roles for diacylglycerol kinases alpha and zeta in the control of T cell functions. 26420856_An abandoned compound that also inhibits serotonin receptors may have more translational potential as a DGKa inhibitor, but more potent and specific DGKa inhibitors are sorely needed 26964756_Decreased DNA methylation at this enhancer enables recruitment of the profibrotic transcription factor early growth response 1 (EGR1) and facilitates radiation-induced DGKA transcription in cells from patients later developing fibrosis. 27498782_LIPFDGKA might serve as a potential possible biomarkers for diagnosis of gastric cancer, and their downregulation may bring new perspective into the investigation of gastric cancer prognosis 27697466_Diacylglycerol kinases alpha and zeta are up-regulated in cancer in cancer, and contribute towards tumor immune evasion and T cells clonal anergy. (Review) 27731506_DGKalpha isoform is highly expressed in the nuclei of human erythroleukemia cell line K562, and its nuclear activity drives K562 cells through the G1/S transition during cell cycle progression. 29967261_This novel study demonstrates efficient ablation of diacylglycerol kinase in human CAR-T cells that leads to improved antitumor immunity and may have significant impact in human cancer immunotherapy 30653270_This study presents the first crystal structure of EF-hand domains of diacylglycerol kinase alpha in its Ca(2+) bound form and characterize Ca(2+) -induced conformational changes, which likely regulates intra-molecular interactions. 31766109_Upon neutrophil stimulation, DGK-alpha activation is necessary for migration and a productive response. This paper focuses on the role of DGK-alpha in obstructive respiratory diseases, including asthma and chronic obstructive pulmonary disease, but also rare genetic diseases such as alpha-1-antitrypsin deficiency. [review] 32341033_DGKA Provides Platinum Resistance in Ovarian Cancer Through Activation of c-JUN-WEE1 Signaling. 32345612_Diacylglycerol kinases regulate TRPV1 channel activity. 33608256_DGKA Mediates Resistance to PD-1 Blockade. 34293268_Diacylglycerol Kinase Inhibition Reduces Airway Contraction by Negative Feedback Regulation of Gq-Signaling. 35131384_DGKA interacts with SRC/FAK to promote the metastasis of non-small cell lung cancer. | ENSMUSG00000025357 | Dgka | 2.776790e+02 | 0.9497850 | -0.074327081 | 0.3239883 | 5.272962e-02 | 0.8183798265 | 0.96462618 | No | Yes | 2.798229e+02 | 44.182581 | 3.276369e+02 | 53.513294 |
| ENSG00000065548 | 55854 | ZC3H15 | protein_coding | Q8WU90 | FUNCTION: Protects DRG1 from proteolytic degradation (PubMed:19819225). Stimulates DRG1 GTPase activity likely by increasing the affinity for the potassium ions (PubMed:23711155). {ECO:0000269|PubMed:19819225, ECO:0000269|PubMed:23711155}. | Alternative splicing;Coiled coil;Cytoplasm;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:55854; | cytosol [GO:0005829]; nucleus [GO:0005634]; cadherin binding [GO:0045296]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; cytokine-mediated signaling pathway [GO:0019221]; cytoplasmic translation [GO:0002181]; positive regulation of GTPase activity [GO:0043547] | 20925576_down-regulation of LEREPO4 expression caused a significant inhibition of HIV replication 23624947_it is hypothesized that ZC3H15 may interact with TRAF-2 functionally within the NF-kappaB pathway, and may be explored as a potential target in acute myeloid leukemia 23711155_Lerepo4 action leaves Drg1 affinity for nucleotides unaffected, feasibly favoring a switch I reorientation, mainly via the TGS domain. 27191988_ZC3H15 is an independent prognostic marker in hepatocellular carcinoma patients that is clinicopathologically associated with tumor invasion and serum a-fetoprotein levels. 34988227_ZC3H15 Correlates with a Poor Prognosis and Tumor Progression in Melanoma. 35027542_ZC3H15 promotes glioblastoma progression through regulating EGFR stability. | ENSMUSG00000027091 | Zc3h15 | 1.184228e+03 | 0.8797286 | -0.184869559 | 0.3539270 | 2.695881e-01 | 0.6036081959 | 0.90922991 | No | Yes | 1.060481e+03 | 194.956194 | 1.204320e+03 | 226.872054 | ||
| ENSG00000065559 | 6416 | MAP2K4 | protein_coding | P45985 | FUNCTION: Dual specificity protein kinase which acts as an essential component of the MAP kinase signal transduction pathway. Essential component of the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway. With MAP2K7/MKK7, is the one of the only known kinase to directly activate the stress-activated protein kinase/c-Jun N-terminal kinases MAPK8/JNK1, MAPK9/JNK2 and MAPK10/JNK3. MAP2K4/MKK4 and MAP2K7/MKK7 both activate the JNKs by phosphorylation, but they differ in their preference for the phosphorylation site in the Thr-Pro-Tyr motif. MAP2K4 shows preference for phosphorylation of the Tyr residue and MAP2K7/MKK7 for the Thr residue. The phosphorylation of the Thr residue by MAP2K7/MKK7 seems to be the prerequisite for JNK activation at least in response to proinflammatory cytokines, while other stimuli activate both MAP2K4/MKK4 and MAP2K7/MKK7 which synergistically phosphorylate JNKs. MAP2K4 is required for maintaining peripheral lymphoid homeostasis. The MKK/JNK signaling pathway is also involved in mitochondrial death signaling pathway, including the release cytochrome c, leading to apoptosis. Whereas MAP2K7/MKK7 exclusively activates JNKs, MAP2K4/MKK4 additionally activates the p38 MAPKs MAPK11, MAPK12, MAPK13 and MAPK14. {ECO:0000269|PubMed:7716521}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Apoptosis;Cytoplasm;Kinase;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Stress response;Transferase;Tyrosine-protein kinase | This gene encodes a member of the mitogen-activated protein kinase (MAPK) family. Members of this family act as an integration point for multiple biochemical signals and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development. They form a three-tiered signaling module composed of MAPKKKs, MAPKKs, and MAPKs. This protein is phosphorylated at serine and threonine residues by MAPKKKs and subsequently phosphorylates downstream MAPK targets at threonine and tyrosine residues. A similar protein in mouse has been reported to play a role in liver organogenesis. A pseudogene of this gene is located on the long arm of chromosome X. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:6416; | cytosol [GO:0005829]; nucleus [GO:0005634]; ATP binding [GO:0005524]; JUN kinase kinase activity [GO:0008545]; MAP kinase kinase activity [GO:0004708]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein tyrosine kinase activity [GO:0004713]; apoptotic process [GO:0006915]; cellular response to mechanical stimulus [GO:0071260]; cellular senescence [GO:0090398]; Fc-epsilon receptor signaling pathway [GO:0038095]; JNK cascade [GO:0007254]; MAPK cascade [GO:0000165]; negative regulation of motor neuron apoptotic process [GO:2000672]; positive regulation of smooth muscle cell apoptotic process [GO:0034393]; response to wounding [GO:0009611]; signal transduction [GO:0007165] | 11754110_There appears to be consistent rate of genetic inactivation of MAP2K4 among most tumor types, including breast cancer. 12223490_JNK-dependent phosphorylation and thus inactivation of Mcl-1 may be one of the mechanisms through which oxidative stress induces cellular damage 12368275_Jun N-terminal kinase has a role in IL-4 induction 12456688_Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways 12714585_in the setting of wild-type PTEN, PI3K- and MKK4/JNK-dependent pathways cooperate to maintain cell survival. 12730213_regulation of fibroblast functions important for wound healing by basal JNK activity 12788955_docking site in MKK4 mediates high affinity binding to JNK MAPKs and competes with similar docking sites in JNK substrates 13130464_JNK, MKK-4, and MKK-7 form an active signaling complex in rheumatoid arthritis and this novel JNK signalsome is activated in response to IL-1 and migrates to the nucleus 14511403_JNK and p38 MAPK activities in UVA-induced signaling pathways leading to AP-1 activation and c-Fos expression 14661062_MMP-9 inhibition activity of resveratrol and its inhibition of JNK and PKC-delta may have a therapeutic potential in cancer. 14716300_Apoptosis signaling triggered by ascididemin is routed via CASP2 and JNKK to mitochondria. 15256484_Curcumin induces c-jun N-terminal kinase-dependent apoptosis in HCT116 human colon cancer cells. 15262961_AP-1 and JNK have roles in reactive oxygen species activation in tobacco-induced mucin production in lung cells 15328343_PP5 plays an important role in the survival of cells in a low oxygen environment by suppressing a hypoxia-induced ASK-1/MKK4/JNK signaling cascade that promotes an apoptotic response 15496400_CYLD has a role in megative regulation of JNK signaling 15592684_Our investigations revealed significantly reduced mRNA expression of metastases suppressor gene Mkk4 in breast cancer brain metastasis. 15623633_Loss of Mkk4 protein expression is associated with pancreatic carcinoma progression 15670787_Taken together, our data suggested that the JNK/c-Jun signaling cascade plays a crucial role in Cd-induced neuronal cell apoptosis and provides a molecular linkage between oxidative stress and neuronal apoptosis. 15737736_Pharmacological inhibition of oxidative stress diminished the elevated p38, JNK activity and PARP cleavage, and enhanced PD cybrid viability. 15911620_JIP1 and JIP3, have a cross-talk that leads to the regulation of the ASK1-SEK1-JNK signal during glucose deprivation; cross-talk between JIP3 and JIP1 is mediated through SEK1-JNK2 and Akt1. 16167336_Our results demonstrate that radioresistance of Saos2 osteosarcoma cells is due to NFkappaB-mediated inhibition of JNK 16197369_an independent role for p38 MAPK and JNK in LPA-induced IL-8 expression and secretion via NF-kappaB and AP-1 transcription respectively in human bronchial epithelial cells. 16339111_These results indicate that SPC stimulates the proliferation of hADSCs through the Gi/Go-PLC-JNK pathway and that LPA receptors may be responsible in part for the SPC-induced proliferation 16388335_The present study demonstrates that HSP cells compared to controls are more sensitive to DNA damages induced by H2O2 treatment, and that JNK phosphorylation levels are increased in HSP fibroblasts after hydrogen peroxide and serum stimuli. 16627982_Homozygous deletion or reduced expression of MKK4 may contribute to the development of ovarian serous carcinoma. 16709574_Bax is phosphorylated by stress-activated JNK and/or p38 kinase and phosphorylation of Bax leads to mitochondrial translocation prior to apoptosis 16802349_JNK (c-Jun N-terminal kinase) function might be modulated by targeting MKK-7 to suppress cytokine-mediated fibroblast-like synoviocytes (FLS) activation while leaving other stress responses intact. 16964394_p38 MAPK and JNK pathways play an important role in VEGF secretion from malignant glioma cells under normoxic conditions. 17009014_Phosphorylated forms of MKK4, JNK, and c-Jun were detected in salivary infiltrating mononuclear cells in Sjogren's syndrome patients 17158870_MKK4 decreases phosphatase and tensin homologue deleted from chromosome 10 (PTEN) and promotes survival in non-small-cell lung cancer (NSCLC) cells. 17178861_LMP1-mediated DNA methyltransferase-1 (DNMT1) activation involves JN kinase. 17322004_The results suggest that JNK regulates human iNOS expression by stabilizing iNOS mRNA possibly by a tristetraprolin -dependent mechanism. 17389591_TNF-induced TRAF2-RIP1-AIP1-ASK1 complex formation and for the activation of ASK1-JNK/p38 apoptotic signaling. 17675521_PF4-stimulated immediate monocyte functions (oxygen radical formation) are regulated by p38 MAPK, Syk, and PI3K, whereas delayed functions (survival and cytokine expression) are controlled by Erk and JNK. 18337456_Cytosolic mRNA is shifted toward active polysomes in prostate tumor cells with higher levels of MKK4 protein, suggesting that MKK4 mRNA is translated more efficiently in these cells. 18408005_JNK3 recruits MKK4 to the beta-arrestin-2 scaffold complex by binding to the MAPK docking domain (D-domain) located within the N terminus of MKK4. 18436711_Disruption of signaling through MKK4 yields differential response in hypoxic colon cancer cells treated with oxaliplatin. 18713996_JNK is differentially regulated by MKK4 and MKK7 depending on the stimulus. 19001375_the molecular interactions of arrestin2 and arrestin3 and their individual domains with the components of the two MAPK cascades, ASK1-MKK4-JNK3 and c-Raf-1-MEK1-ERK2 19012245_multiple copy number alterations in chromosome regions implicated in malignancy progression and indicated a strong expression of MAP2K4 and MCL1 genes in rhabdomyosarcoma 19265040_Cardiac-specific deletion of mkk4 reveals its role in pathological hypertrophic remodeling but not in physiological cardiac growth. 19351724_An Ask1-MKK4-p38MAPK/JNK pathway reflects adipocyte stress associated with adipose tissue inflammation, linking visceral adiposity to whole-body insulin resistance in obesity. 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19513509_Overexpression of mitogen-activated protein kinase kinase 4 and nuclear factor-kappaB in laryngeal squamous cell carcinoma: a potential indicator for poor prognosis. 19610067_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19610067_Polymorphism in the promoter of MKK4 gene is associated with colorectal cancer. 19638505_We identified MEK4 as a proinvasion protein in six human prostate cancer cell lines and the target for genistein. 19861690_Elevated MKK4 abundance inhibited cell proliferation and increased the phosphorylation and activity of p38 and PRAK. Thus, multiple microRNAs acting on a single target, the MKK4 mRNA, collectively influence MKK4 abundance during replicative senescence 20156194_Filamin A is a scaffold protein whose function is to link MKK4 and MKK7 together and promote JNK1 activation. 20309881_Data indicate that MKK4 gene knockdown in MDAH2774 cells over-expressing MKK4 increased invasion activity. 20554746_Functional -1304G variant in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. 20554746_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20732303_the crystal structures of human non-phosphorylated MKK4 kinase domain (npMKK4) complexed with AMP-PNP (npMKK4/AMP) and a ternary complex of npMKK4, AMP-PNP and p38alpha peptide (npMKK4/AMP/p38) were determined. 21030692_These results suggest that JNK affects the association of alpha-catenin with the adherens junction complex and regulates adherens junctions. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21372598_Loss of MKK4 expression is associated with endometrial cancer. 21487811_These results indicated that MKK4 acts as a tumor suppressor and may represent an important therapeutic target for the treatment of ovarian cancer 21518142_Compared with the -1304TT genotype, patients with the -1304TG genotype had a significantly decreased risk of acute myeloid leukemia 21575258_MAP2K4 is targeted by genetic inactivation in ovarian cancer and restricted to high grade serous and endometrioid carcinomas in our cohort. 21638028_MKK4 was identified as playing a key role in Tau-S422 phosphorylation in human neuroblastoma cells. 21702039_protective role of genetic variant MKK4 -1304T>G is restrained in nasopharyngeal carcinoma (NPC) with Epstein-Barr Virus (EBV) infection. These findings implicate the role of EBV and MKK4 -1304 T>G interaction as a causative factor for the NPC. 21896780_8 out of 11 cancer-associated MAP2K4 mutations reduce MKK4 protein stability or impair its kinase activity 21915030_The purpose of the study was to investigate the potential contribution of HPK1, MEKK1, TAK1, p-MKK4 to the development of extramammary Paget disease 21925502_Building on the foundation of daring hypotheses: using the MKK4 metastasis suppressor to develop models of dormancy and metastatic colonization. 22154052_Overexpression of MAP2K4 in osteosarcoma was correlated with poor treatment response, disease progression and poor overall survival. 22154358_Data suggest aberrant MAP2K protein (MKK3, MKK4, MKK6, and MKK7) expression indicates that altered cellular signal transduction mediated via JNK and p38 may be common in bladder cancer. 22158075_Suggest that in pancreatic ductal adenocarcinomas, the MKK4 protein was directly related to high cell proliferation. 22335172_the functional -1304G variant in the MKK4 promoter contributes to a decreased risk of cervical cancer 22526163_Results suggest that the functional -1304G variant in the MKK4 promoter decreases the risk of PCa by increasing the promoter activity. 22730327_SEPW1 silencing increases MKK4, which activates p38gamma, p38delta, and JNK2 to phosphorylate p53 on Ser-33 and cause a transient G(1) arrest. 22828509_Crystal structures combined with small-angle X-ray scattering experiments revealed that the apo form of non-phosphorylated MAP2K4 (npMAP2K4) exists in a transient state which has a longer conformation compared with the typical kinase folding. 23299404_Rs12939944 located in the MAP2K4 intron was associated with decreased risk. 23355465_MicroRNA-92a negatively regulates Toll-like receptor (TLR)-triggered inflammatory response in macrophages by targeting MKK4 kinase 23653187_If both p53 and the SAPKK MKK4 are simultaneously inactivated, persistent polo-like kinase 4 activity combined with the lack of SAPK-mediated inhibition of centrosome duplication conspire to induce supernumerary centrosomes under stress. 23921907_Single nucleotide polymorphisms in MAP2K4 gene is associated with gastric cancer. 23960075_Arrestin-3 directly interacts with MKK7 and promotes JNK3alpha2 phosphorylation by both MKK4 and MKK7 in vitro as well as in intact cells. 24532253_Lipopolysaccharide induced miR-181a promotes pancreatic cancer cell migration via targeting PTEN and MAP2K4. 24556602_MicroRNA-27a promotes proliferation, migration and invasion by targeting MAP2K4 in human osteosarcoma cells. 24721794_Demonstrate that Mkk4 is a negative regulator of the TGF-beta1 signaling associated with atrial remodeling and arrhythmogenesis with age. 25019290_MAP2K4 increases human prostate cancer metastasis, and prolonged over expression induces long term changes in cell signaling pathways leading to independence from p38 MAPK and JNK. 25244576_knockdown of Sec8 enhances the binding of JIP4 to MAPK kinase 4, thereby decreasing the phosphorylation of MAPK kinase 4, JNK, and p38. 25732927_Data suggest a genetic interaction between MAP2K4 and HLA-DRB1, and the importance of rs10468473 and MAP2K4 splice variants in the development of autoantibody-positive RA. 26028649_MKK4 is activated in vitro by reduced Trx but not oxidized Trx in the absence of an upstream kinase, suggesting that autophosphorylation of this protein occurs due to reduction of Cys-246 and Cys-266 by Trx. 26080319_The plasma level of protein MAP2K4 was found to suggestively associate negatively with the volume of the left entorhinal cortex in asymptomatic older twins. 26554761_the presence of the -1304T > G polymorphism is likely to decrease risk of cancer (Meta-Analysis) 26856463_In Chinese Han ischemic stroke patients rs3826392 C/A genotype carriers showed significantly higher IL-1b serum levels. 27173611_These results suggest that ROCK may be important in IL-1-induced signaling through MKK4 to JNK and the activation of p38 MAPK. 27353001_Manipulating the expression of both miR-222 and miR-25 influenced diverse gene expression changes in thyroid cells. Increased expression of miR-25 reduced MEK4 and TRAIL protein expression, and cell adhesion and apoptosis are important aspects of miR-25 functioning in thyroid cells. 27509166_Association between MKK4 promoter polymorphism and breast cancer risk in Kashmiri population 27594411_Androgen-induced miR-27A acted as a tumor suppressor by targeting MAP2K4 and mediated prostate cancer progression 28319306_the expression level of MAP2K4 was inversely associated with the expression of miR-802 in tongue squamous cell carcinoma (TSCC) tissues; demonstration that the MAP2K4 expression was upregulated in TSCC cell lines; elevated expression of miR-802 inhibited TSCC cell viability and invasion through inhibiting MAP2K4 expression 28423721_Study provides evidence that phosphorylated MKK4 (pMKK4) might function as a tumor suppressor in colorectal cancer (CRC). Downregulation of pMKK4 was associated with a more aggressive phenotype and with increases in local invasion and metastasis. pMKK4 was also strongly associated with disease-free survival. 28733031_MKK4 activates non-canonical NFkappaB signaling by promoting NFkappaB2-p100 processing. 29072705_MKK4 overexpression enhanced TNF-alpha-mediated signaling activation and transcription of downstream catabolic genes, and consequently worsened cartilage degradation. 29248490_These results provide valuable insights into the role of acetylation in MKK4-JNK signaling in T cells. 29276882_This study demonstrates that MKK4 employs a subtle combination of interaction modes in order to bind to p38 alpha, leading to a complex displaying significantly different dynamics across the bound regions. 29472518_This study supports a role for NEK9 and MAP2K4 in mediating buparlisib resistance and demonstrates the value of unbiased omic analyses in uncovering resistance mechanisms to targeted therapy in Triple-Negative Breast Cancers. 30537492_high MKK4 expression and its combination with high MKK7 expression both predicted favorable prognosis in resectable pancreatic ductal adenocarcinoma. 30548122_The initial characterization of tumor-suppressing kinases- in particular members of the protein kinase C (PKC) family, MKK4 of the mitogen-activated protein kinase kinase family, and DAPK3 of the death-associated protein kinase family- laid the foundation for bioinformatic approaches that enable the identification of other tumor-suppressing kinases. 31635803_The ensemble structures provided more detailed mechanisms for regulating MAP2K4 in addition to those delineated only by the crystal structures in three states. 31761784_MAP2K4 interacts with Vimentin to activate the PI3K/AKT pathway and promotes breast cancer pathogenesis. 32495394_microRNA-124-3p inhibits tumourigenesis by targeting mitogen-activated protein kinase 4 in papillary thyroid carcinoma. 32552144_Is there any correlation among MKK4 (mitogen-activated protein kinase kinase 4) expression, clinicopathological features, and KRAS/NRAS mutation in colorectal cancer. 32779194_Long noncoding RNA HNF1A-AS1 regulates proliferation and apoptosis of glioma through activation of the JNK signaling pathway via miR-363-3p/MAP2K4. 33341545_A new prognosis prediction model combining TNM stage with MAP2K4 and JNK in postoperative pancreatic cancer patients. 34219538_Role of miR-27a in the regulation of cellular function via the inhibition of MAP2K4 in patients with asthma. | ENSMUSG00000033352 | Map2k4 | 3.957182e+02 | 0.9282527 | -0.107410536 | 0.3750244 | 8.072992e-02 | 0.7763106733 | 0.95540959 | No | Yes | 4.045896e+02 | 86.248703 | 3.657584e+02 | 79.989327 | |
| ENSG00000066117 | 6602 | SMARCD1 | protein_coding | Q96GM5 | FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner (PubMed:8804307, PubMed:29374058). Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). Has a strong influence on vitamin D-mediated transcriptional activity from an enhancer vitamin D receptor element (VDRE). May be a link between mammalian SWI-SNF-like chromatin remodeling complexes and the vitamin D receptor (VDR) heterodimer (PubMed:14698202). Mediates critical interactions between nuclear receptors and the BRG1/SMARCA4 chromatin-remodeling complex for transactivation (PubMed:12917342). {ECO:0000250|UniProtKB:Q61466, ECO:0000269|PubMed:12917342, ECO:0000269|PubMed:14698202, ECO:0000269|PubMed:29374058, ECO:0000269|PubMed:8804307, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. | 3D-structure;Acetylation;Alternative splicing;Chromatin regulator;Coiled coil;Disease variant;Isopeptide bond;Mental retardation;Methylation;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene is a member of the SWI/SNF family of proteins, whose members display helicase and ATPase activities and which are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. The encoded protein is part of the large ATP-dependent chromatin remodeling complex SNF/SWI and has sequence similarity to the yeast Swp73 protein. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:6602; | intracellular membrane-bounded organelle [GO:0043231]; nBAF complex [GO:0071565]; npBAF complex [GO:0071564]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; SWI/SNF complex [GO:0016514]; chromatin binding [GO:0003682]; molecular adaptor activity [GO:0060090]; signaling receptor binding [GO:0005102]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; cellular response to fatty acid [GO:0071398]; chromatin remodeling [GO:0006338]; epigenetic maintenance of chromatin in transcription-competent conformation [GO:0045815]; nervous system development [GO:0007399]; nucleosome disassembly [GO:0006337]; regulation of transcription by RNA polymerase II [GO:0006357] | 12917342_In addition to previously identified BAF250, BAF60a may provide another critical and direct link between nuclear receptors and the BRG1 complex that is required for promoter recruitment and subsequent chromatin remodeling 19762545_SMARCD1/BAF60a is an androgen receptor cofactor that modulates TMPRSS2 expression 25396734_the association of EGFR, CALM3 and SMARCD1 gene polymorphisms with bone mineral density in white women, as conducted. 28716547_BAF57, BAF60a and SNF5 might act as novel pro-senescence factors in both normal and tumor human skin cells 29079174_miR-223 targets the expression of SWI/SNF complex protein SMARCD1 in atypical proliferative serous tumor and high-grade ovarian serous carcinomas. 30515787_MITF promoted BAF60A recruitment to melanocyte-specific promoters, and BAF60A was required to promote BRG1 recruitment and chromatin remodeling. 30879640_identification of a human neurodevelopmental disorder caused by SMARCD1 mutations 31097748_Modulation of chromatin remodeling proteins SMYD1 and SMARCD1 promotes contractile function of human pluripotent stem cell-derived ventricular cardiomyocyte in 3D-engineered cardiac tissues. 31637714_SMARCD1 is a transcriptional target of specific non-hotspot mutant p53 forms. 31992292_Expression of SMARCD1 interacts with age in association with asthma control on inhaled corticosteroid therapy. 32299427_hsa-miR-100-5p, an overexpressed miRNA in human ovarian endometriotic stromal cells, promotes invasion through attenuation of SMARCD1 expression. 32514535_Enhanced SMARCD1, a subunit of the SWI/SNF complex, promotes liver cancer growth through the mTOR pathway. 32787523_BAF60a Deficiency in Vascular Smooth Muscle Cells Prevents Abdominal Aortic Aneurysm by Reducing Inflammation and Extracellular Matrix Degradation. 33168186_NMR spectroscopy uncovers direct interaction between BAF60A and p53. 33190170_Smarcd1 Inhibits the Malignant Phenotypes of Human Glioblastoma Cells via Crosstalk with Notch1. 35148461_microRNA-99a-5p induces cellular senescence in gemcitabine-resistant bladder cancer by targeting SMARCD1. 35158202_Assembly and interaction of core subunits of BAF complexes and crystal study of the SMARCC1/SMARCE1 binary complex. | ENSMUSG00000023018 | Smarcd1 | 8.873424e+03 | 1.2111849 | 0.276419135 | 0.2971149 | 8.668072e-01 | 0.3518407379 | 0.82969965 | No | Yes | 9.455317e+03 | 802.696318 | 7.150839e+03 | 623.067113 | |
| ENSG00000066427 | 4287 | ATXN3 | protein_coding | P54252 | FUNCTION: Deubiquitinating enzyme involved in protein homeostasis maintenance, transcription, cytoskeleton regulation, myogenesis and degradation of misfolded chaperone substrates (PubMed:12297501, PubMed:17696782, PubMed:23625928, PubMed:28445460, PubMed:16118278). Binds long polyubiquitin chains and trims them, while it has weak or no activity against chains of 4 or less ubiquitins (PubMed:17696782). Involved in degradation of misfolded chaperone substrates via its interaction with STUB1/CHIP: recruited to monoubiquitinated STUB1/CHIP, and restricts the length of ubiquitin chain attached to STUB1/CHIP substrates and preventing further chain extension (By similarity). Interacts with key regulators of transcription and represses transcription: acts as a histone-binding protein that regulates transcription (PubMed:12297501). Regulates autophagy via the deubiquitination of 'Lys-402' of BECN1 leading to the stabilization of BECN1 (PubMed:28445460). {ECO:0000250|UniProtKB:Q9CVD2, ECO:0000269|PubMed:12297501, ECO:0000269|PubMed:16118278, ECO:0000269|PubMed:17696782, ECO:0000269|PubMed:23625928, ECO:0000269|PubMed:28445460}. | 3D-structure;Alternative splicing;Hydrolase;Isopeptide bond;Neurodegeneration;Nucleus;Phosphoprotein;Protease;Reference proteome;Repeat;Spinocerebellar ataxia;Thiol protease;Transcription;Transcription regulation;Triplet repeat expansion;Ubl conjugation;Ubl conjugation pathway | Machado-Joseph disease, also known as spinocerebellar ataxia-3, is an autosomal dominant neurologic disorder. The protein encoded by this gene contains (CAG)n repeats in the coding region, and the expansion of these repeats from the normal 12-44 to 52-86 is one cause of Machado-Joseph disease. There is a negative correlation between the age of onset and CAG repeat numbers. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jul 2016]. | hsa:4287; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrial matrix [GO:0005759]; mitochondrial membrane [GO:0031966]; nuclear inclusion body [GO:0042405]; nuclear matrix [GO:0016363]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; synapse [GO:0045202]; ATPase binding [GO:0051117]; identical protein binding [GO:0042802]; Lys48-specific deubiquitinase activity [GO:1990380]; Lys63-specific deubiquitinase activity [GO:0061578]; thiol-dependent deubiquitinase [GO:0004843]; ubiquitin protein ligase binding [GO:0031625]; actin cytoskeleton organization [GO:0030036]; cellular response to heat [GO:0034605]; cellular response to misfolded protein [GO:0071218]; chemical synaptic transmission [GO:0007268]; intermediate filament cytoskeleton organization [GO:0045104]; microtubule cytoskeleton organization [GO:0000226]; monoubiquitinated protein deubiquitination [GO:0035520]; nervous system development [GO:0007399]; nucleotide-excision repair [GO:0006289]; positive regulation of ERAD pathway [GO:1904294]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein deubiquitination [GO:0016579]; protein K48-linked deubiquitination [GO:0071108]; protein K63-linked deubiquitination [GO:0070536]; protein localization to cytosolic proteasome complex involved in ERAD pathway [GO:1904379]; protein quality control for misfolded or incompletely synthesized proteins [GO:0006515]; regulation of cell-substrate adhesion [GO:0010810]; ubiquitin-dependent protein catabolic process [GO:0006511] | 11121205_Observational study of gene-disease association. (HuGE Navigator) 11804332_Observational study of genotype prevalence. (HuGE Navigator) 12084819_Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. 12166658_Observational study of gene-disease association. (HuGE Navigator) 12766160_examination of structural instability and fibrillar aggregation of non-expanded form under high pressure and temperature 12832059_expression of mutant form is involved in down regulation of heat shock protein 27 in neuronal and non-neuronal cells 12857950_functional ubiquitin-binding motifs in Atx-3 and p62 proteins are required for the localization of both proteins into aggregates 12873751_Spinocerebellar ataxia type 3 presenting as an L-DOPA responsive dystonia phenotype in a Chinese family.and SCA 3 should be considered a differential diagnosis in adult patients who present with DRD phenotype and with a positive family history. 12914917_ataxin-3 human and mouse protein sequences share a highly conserved N-terminus and differ only in the length of the glutamine repeats and in the C-terminus; the domain architecture of both is detailed 12940846_Observational study of gene-disease association. (HuGE Navigator) 12940846_The significance of SCA3 mutation in either sporadic or familial young-onset dopa-responsive parkinsonism suggestive of Parkinson's disease is not proven in a population of 85 Serbian patients. 12944474_ataxin-3 interacts with ubiquitinated proteins, can bind the proteasome, and, when the gene harbors an expanded repeat length, can interfere with the degradation of a well-characterized substrate. Additionally, ataxin-3 associates with Rad23 and VCP/p97 14559776_Ataxin-3 binds polyubiquitylated proteins and has deubiquitylating activity. 14602712_ataxin-3 is a poly-ubiquitin-binding protein 14659761_ataxin-3 folds reversibly via a single intermediate; partial destabilization of ataxin-3 by chemical denaturation leads to the formation of fibrillar aggregates by the non-pathological variant 14661975_A soluble human ataxin-3 variant with a moderately expanded polyQ tract of 36 glutamine residues gives rise to amyloid fibrils on increasing temperature above 40 degrees C. 14679302_metabolic changes associated with transgenic mouse model of spinocerebellar ataxia 3 14746390_Observational study of gene-disease association. (HuGE Navigator) 14756671_Observational study of gene-disease association. (HuGE Navigator) 15026782_These results strongly suggest that ADCA families can be traced back to common ancestors in particular parts of the Netherlands. 15128861_Transgenic rats expressing a human ataxin-3 fragment with an elongated polyglutamyl stretch under control of the human prepro-orexin promoter exhibit narcolepsy-cataplexy and postnatal loss of orexin-positive neurons 15140190_Caspase-mediated cleavage of expanded ataxin-3 resulted in increased ataxin-3 aggregation, suggesting a potential role for caspase-mediated proteolysis in spinocerebellar ataxia type-3 pathogenesis 15210524_Observational study of healthcare-related. (HuGE Navigator) 15223312_The molecular architecture of CAG repeats in mutant SCA3 transcripts was studied. 15236410_Data report sequence-independent discrimination of mutant and wild-type ataxin-3 alleles by small interfering RNA. 15265035_exploration of protein architectures of ataxin-2 and ataxin-3 15316156_The results of this study indicate that the clinical entity of Machado-Joseph disease can occur with 51 trinucleotide repeats, and that the clinical features of Machado-Joseph disease might cover a wider spectrum than previously believed. 15345714_Results show no difference in the un/folding transitions of three ataxin-3 variants, which indicates that the stability of the native conformation was not affected by polyglutamine tract extension. 15537899_An ataxin-3 fragment containing residues C terminal to amino acid 221 including the polyglutamine expansion is toxic to neuroblastoma cells and is found in vivo in affected brain regions. 15544810_Data support a mechanism in which the thermodynamic stability of ataxin-3 is governed by the properties of the Josephin domain, but the presence of an expanded polyQ tract increases dramatically the protein's tendency to aggregate. 15630566_NMR structure of ataxin 3 15767577_the deubiquitylating activity of AT3 and its ubiquitin interacting motifs play essential roles in CFTRDeltaF508 aggresome formation 15808507_Normal human ataxin-3 is a striking suppressor of polyglutamine neurodegeneration in vivo. 15952105_Findings suggest that the interaction of small ubiquitin-like modifier 1 with N-terminus of ataxin-3 first and the relevant sumoylation probably participate in the post-translation modifying of ataxin-3 and in the pathogenesis of SCA3/MJD. 16040601_Through its ubiquitin interaction motifs, normal or expanded ataxin-3 binds a broad range of ubiquitinated proteins that accumulate when the proteasome is inhibited. 16118278_ataxin-3 may function as a polyubiquitin chain-editing enzyme 16126176_cells surviving under conditions of polyglutamine cytotoxicity unfold and remove mutant ataxin-3 in presence of increased level of Hsp27 16389595_Observational study of gene-disease association. (HuGE Navigator) 16525503_VCP-Atx-3 association is a potential target for therapeutic intervention and suggest that it might influence the progression of spinocerebellar ataxia type 3. 16624810_ataxin-3 has an inherent capacity to aggregate through its non-polyglutamine domains 16687213_Observational study of gene-disease association. (HuGE Navigator) 16724006_The main mutation mechanism occurring in the evolution of the polymorphic CAG region at MJD/SCA3 locus is a multistep one, either by gene conversion or DNA slippage; repeats with 14, 21, 23 and 27 CAGs are the main alleles involved in this process. 16791428_ATX3 aggregation is noticeably reduced by deletion or replacement of regions other than the polyglutamine tract. The nature of the amino acid homo-sequences also has a strong influence on aggregation. 16822850_Normal function of AT3 might be to regulate flow through the ERAD pathway by modulating VCP-dependent extraction of proteins from the ER. 16967484_linkage and association for three CAG triplet repeat markers (Spinocerebellar Ataxia Type 1, SCA1; Machado-Joseph Disease, MJD; Dentatorubro-pallidoluysian Atrophy, DRPLA) to assess their contribution to variation in cognitive ability 17000876_We present evidence that atx3 may promote p97-associated deubiquitination to facilitate the transfer of polypeptides from p97 to the proteasome. 17079677_Normal AT3 binds target promoter regions and represses transcription of a GATA-2-dependent target gene via formation of histone-deacetylating repressor complexes requiring its ubiquitin-interacting motif-associated function. 17302910_The N-terminus of ataxin-3, which serves as a recognition site by p45, is necessary for the proteolytic process of ataxin-3. 17434145_Our results imply that phosphorylation of serine 256 in ataxin-3 by GSK 3beta regulates ataxin-3 aggregation. 17440947_Observational study of genotype prevalence. (HuGE Navigator) 17440947_To investigate the role of SCA2 and SCA3 mutations in Chinese familial and early-onset Parkinson's disease (PD) patients, CAG triplet repeat expansions of SCA2 and SCA3 genes in 73 Taiwanese/Ethnic Chinese PD patients was analyzed. 17488727_ataxin-3 proteolysis in neuroblastoma cells and in vitro and show that calcium-dependent calpain proteases generate aggregation-competent ataxin-3 fragments 17626202_Nuclear localization of ataxin-3(SCA3) is required for the manifestation of symptoms in spinocerebellar ataxia type 3 in vivo. Severity of symptoms in SCA3 is linked to the number of expanded polyglutamine repeats. 17632007_results support the toxic fragment hypothesis and narrow the mutant ataxin-3 cleavage site to the N-terminus of amino acid 190 17683516_Machado-Joseph disease enhances genetic fitness: a comparison between affected and unaffected women and between MJD and he general population is reported. 17693639_ataxin-3 cellular turnover is regulated by its catalytic activity 17696782_findings show that the enzyme activity of human AT3 is not affected by the length of polyQ in its C-terminus, even when it is in the range associated with spinocerebellar ataxia type 3 17935801_A new molecular interaction between wild-type ataxin-3 and NEDD8, using in vitro and in situ approaches, is reported. 17948873_assessed the influence of other cis and inter-allelic acting factors, at the ATXN3 locus, through the analysis of MJD lineages, flanking STR-based haplotypes, the initial repeat size and parental age 17956866_the pathogenic ataxin-3 protein of the human disease spinocerebellar ataxia type 3 (SCA3) and the yeast prion Sup35 mediate protein aggregation in Drosophila 18160752_Data show that SCA1, 2 and 3 accounted for more than one third of the ataxia cases seen in the clinic, and in cases with established family history and autosomal dominant inheritance SCA1 was most prevalent followed by SCA2 and SCA3. 18182848_Observational study of gene-disease association. (HuGE Navigator) 18353661_All ataxins-3 (AT-3) undergo the same proteolytic cleavage, removing the first 27 amino acids; while normal ataxins are further cleaved at a number of caspase sites, pathological AT-3 is proteolyzed to a much lesser extent. 18358414_homozygosity of atxn3 aggravates the clinical phenotype of spinocerebellar ataxia type 3 18385100_Overexpression of mutant human ataxin-3 in rats led to apomorphine-induced turning behavior, ataxin-3 aggregations and loss of dopaminergic markers. 18449188_a Drosophila screen for modifiers of polyQ degeneration induced by the spinocerebellar ataxia type 3 (SCA3) protein ataxin-3 18599482_The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains 18927607_No pathogenic mutations were found in PRKN protein and are not a frequent cause of Parkinson disease in Nigeria. 18927607_Observational study of gene-disease association. (HuGE Navigator) 18990604_Two SCA3 and one SCA2 cases have been identified which show autosomal dominant inheritance in Parkinson disease. 19049837_rate of clinical disease progression at presentation, especially in SCA2, is dependent on the CAG repeat size, and may commence linearly from birth 19065528_Observational study of gene-disease association. (HuGE Navigator) 19185026_Therefore, we suggest that the cell damage caused by greater oxidative stress in SCA3 mutant cells plays an important role, at least in part, in the disease progression. 19235102_Observational study of gene-disease association. (HuGE Navigator) 19259763_Observational study of gene-disease association. (HuGE Navigator) 19382171_Data show that the presence of two ubiquitin-binding sites explains how ataxin-3 binds poly-ubiquitin chains and provides new insights into the molecular mechanism of ubiquitin recognition. 19429074_findings provide the first evidence that expanded ataxin-3 interferes with Hsp27 synthesis, which may contribute to the impairment of the cells' ability to respond to stresses and trigger the progression of Machado-Joseph disease 19492089_Conformation-specific intrabody co-localizes with intracellular aggregates of misfolded ataxin-3 and a pathological fragment of huntingtin, and enhances the aggregation propensity of both disease-linked polyglutamine proteins. 19503814_analysis of the nucleocytoplasmic shuttling activity of ataxin-3 19542537_CK2-dependent phosphorylation controls the nuclear localization, aggregation and stability of ataxin-3. 19575245_Results describe the mode of interaction of ataxin-3 Q36 (AT-3 Q36) with selected endogenous and exogenous metal ions, and effects on protein structure. 19660550_Relevant signals for intracellular localization of the amino- and carboxyl-termini of ataxin-3 demonstrate its distribution during the potential pathogenesis of spinocerebellar ataxia type 3. 19661182_Gp78 promotes SOD1 and ataxin-3 degradation in endoplasmic reticulum. 19672991_This study suggested that a mutation in SCA2 or SCA3/MJD may be one of the genetic causes of Parkinson's disease in china. 19714377_Data demonstrate high variability in the ATXN3 gene transcripts, providing a basis for further investigation on the contribution of alternative splicing to the MJD pathogenic process, as well as to the larger group of the polyglutamine disorders. 19783548_comparison of flies expressing either human wild-type or caspase-site mutant proteins indicates that Ataxin-3 cleavage enhances neuronal loss in vivo. 19843543_Oxidative stress and heat shock induced nuclear localization of Atx3. 19935829_The CAG repeat size in genomic DNA was not always directly transcribed into the mRNA. 20017304_Observational study of gene-disease association. (HuGE Navigator) 20069235_Observational study of gene-disease association. (HuGE Navigator) 20140862_Autophagy is involved in the degradation of mutant ataxin-3. 20219685_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20334689_CAG repeats of ATXN3 have found a much higher frequency (72.5%) of Machado-Joseph disease in southeastern Chinese kindreds patients compared to previous reports. 20376803_the shortest expanded allele associated with a disease phenotype in the Machado-Joseph disease gene reported to date 20510362_Transgenic CMVMJD94 mice are a useful model to study the early stages in the pathogenesis of Machado-Joseph disease and to explore the molecular mechanisms involved in ataxin-3 CAG repeat instability. 20533274_It was confirmed that the numbers of CAG repeat in the SCA3/MJD was abnormal by gene diagnosis in the pedigree with spinocerebellar ataxia. 20546612_Observational study of gene-disease association. (HuGE Navigator) 20625959_A striking feature of the Polish population is the lack of SCA3 - the most frequent type in Western Europe. 20637808_Data show that the absence of ATXN3 leads to an overt cytoskeletal/adhesion defect raising the possibility that this protein may play a role in the cytoskeleton. 20810784_Results provide evidence that protein nonpathologic function can play an active role in preventing aberrant fibrillization and suggest the molecular mechanism whereby this occurs in ataxin-3. 20865150_Ub-binding site 1 and 2 are necessary for ataxin-3 to cleave polyubiquitin chains. 20940148_The interaction between ataxin-3 and parkin is direct, involves multiple domains and is greatly enhanced by parkin self-ubiquitination. 20943656_Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117 21060878_Regardless of polyglutamine expansion, 3UIM ataxin-3 is the predominant isoform in brain. Although 2UIM and 3UIM ataxin-3 display similar in vitro deubiquitinating activity, 2UIM ataxin-3 is prone to aggregate and more rapidly degraded by the proteasome 21092747_Polyglutamine-expanded ataxin-3 upregulates mRNA expression of Bax and causes apoptotic death of affected neurons by enhancing phosphorylation and transcriptional activity of p53. 21118805_Crystal structure of a Josephin-ubiquitin complex: evolutionary restraints on ataxin-3 deubiquitinating activity. 21334959_In Korean population, the mutation frequencies of SCA3 to parkinsonism was insignificant. 21386698_HAP1/stigmoid body interacts with the normal ataxin-3 through Josephin domain 21504740_a key role of Josephin in ataxin-3 fibrillar aggregation 21533208_monitored the aggregation of a normal (AT3Q24) ataxin-3, an expanded (AT3Q55) ataxin-3, and the JD in isolation. We observed that all of them aggregated, although the latter did so at a much slower rate 21536589_Findings support that ATXN3 plays an important role in regulating the FOXO4-dependent antioxidant stress response via SOD2. 21827905_This sudy showed that ATXN3 CAG trinucleotide repeat mutation related to Machado-Joseph disease. 21869492_identification of SCA3 gene mutations in a slowly progressive isolated form of ataxia in a chinese cohort 22037589_Transiently transfected HEK cell lines with expanded (Q84) ataxin-3 exhibited a higher susceptibility to 3-nitropropionic acid (3-NP), an irreversible inhibitor of mitochondrial complex II. 22113611_-glutamate-induced excitation of patient-specific induced pluripotent stem cell (iPSC)-derived neurons initiates Ca(2+)-dependent proteolysis of ATXN3 followed by the formation of SDS-insoluble aggregates 22129356_the central flexible region enhances protein aggregation and can populate conformational states with different degrees of compactness 22234302_These results underscore ataxin 3 capability of undergoing multiple aggregation pathways that lead to end products endowed with substantially different molecular structures. 22422287_no functional effect could be predicted for ATXN3 gene variant. 22491195_This study demonistrated that Disease progressed of autosomal dominant cerebellar ataxia and spastic paraplegia faster in SCA s with polyglutamine expansions in SCA1, 2, and 3. 22706685_10 SNPs (none in core splicing signals) were found from exonic & flanking intronic regions in genomic DNA from Machado-Joseph disease patients & controls. The SNPs implied losses & gains of splicing-factor-recognition motifs. 22761419_the sequestration of misfolded SOD1 into aggresomes, which is driven by ataxin-3, plays an important role in attenuating protein misfolding-induced cell toxicity. 22777171_Data present that galectin-1, ataxin-3 and sprouty-related EVH1 domain-containing protein 2 (SPRED2) may be associated with the progression and development of Down syndrome. 22843411_ATX3 proteolysis in transgenic mice by human calpains mediates ATX3 translocation to the nucleus, aggregation and toxicity. 22970133_VCP/p97 was shown to be an activator specifically of wild-type ataxin-3. 23100324_Ataxin-3 is cleaved by calpains. Increased proteolytic cleavage of ataxin-3 results in a more severe and faster progressing neurological phenotype of spinocerebellar ataxia type 3. 23184527_This study demonistrated that Altered expression of carbonic anhydrase-related protein XI in neuronal cells expressing mutant ataxin-3. 23349684_the efficacy of gene silencing in blocking the MJD-associated motor-behavior and neuropathological abnormalities 23382880_our study demonstrated that SUMOylation on K166, the first described residue of SUMO-1 modification of ataxin-3, partially increased the stability of mutant-type ataxin-3, and the rate of apoptosis arisen from the cytotoxicity of the modified protein 23562578_Our data suggest that ataxin-3 plays an important role in regulating the Bcl-XL-Bax-mediated anti-oxidative response by modulating the interaction between Bcl-XL and Bax. 23926002_The cloned A3IP gene encodes A3IP, a novel ataxin-3 interacting protein. 24106274_Our work also suggests that ataxin-3 suppresses degeneration by regulating toxic protein aggregation rather than stability. 24196352_interaction between UbD2 and p97/Atx3 mediates retranslocation of UbD2 to the cytoplasm for terminal degradation in the proteasomes. 24292675_In ATXN3-depleted cells, under conditions of transcriptional inhibition, PTEN and PTENP1 mRNAs rapidly decay. 24685680_Authors have investigated the interaction of AT3 with tubulin and HDAC6. 24817148_ataxin-3 fragment aggregates in a polyQ length-dependent manner in C. elegans muscle cells and that this aggregation is associated with cellular dysfunction 25139423_Results suggest that polyglutamine-expanded ataxin-3-Q79 impairs histone acetyltransferase activity, leading to impaired induction of cerebellar long-term depression in the spinocerebellar ataxia type 3 transgenic mouse 25143392_Data support the importance of ATXN3 in neuronal cells and indicate that an expanded polyQ tract leads to a partial loss of the cellular function of ATXN3 that may be relevant to neurodegeneration. 25144244_Ubiquitination of ataxin-3 is not necessary for its proteasomal degradation.Ataxin-3 is regulated by ubiquitin-binding site 2 on its N terminus.Ubiquitin-binding site 2 of ataxin-3 prevents its proteasomal degradation by interacting with Rad23. 25268243_The At3 N-terminal Josephin domain aggregation might be a multistep process. 25448680_substrate recognition by the Josephin domain of ataxin-3 25451224_miR-25 reduced both wild-type and polyQ-expanded mutant ataxin-3 protein levels by interacting with the 3'UTR of ATXN3 mRNA 25566755_Data show that homozygosity for Machado-Joseph disease (MJD)/SCA3 protein enhances the clinical severity of the disease. 25590633_We now report that the mutant ATXN3 protein interacts with and inactivates PNKP (polynucleotide kinase 3'-phosphatase), an essential DNA strand break repair enzyme 25633985_Here we report that purified wild-type (WT) ATXN3 stimulates, and by contrast the mutant form specifically inhibits, PNKP's 3' phosphatase activity in vitro. ATXN3-deficient cells also show decreased PNKP activity 25689313_Machado-Joseph disease patients carrying the rs709930 A allele and rs910369 T allele of ATXN3 experienced an earlier age of onset of approximately 2 to 4 years. 25700012_A multistage aggregation mechanism for ataxin-3 is described in which flanking domain self-assembly precedes polyglutamine aggregation yet is influenced by polyglutamine expansion. 25869926_South American cohort did not confirm the effect of the four candidate loci as modifier of onset age: mithocondrial A10398G polymorphism and CAGn at RAI1, CACNA1A, ATXN3, and ATXN7 genes 26037349_Wide type and polyQ-expanded ataxin-3 both showed partial co-localization to endoplasmic reticulum. 26067219_This study did not find relationship between CAG expansion length and psychiatric disorders. 26210447_Evaluated the level of truncated pathological recombinant Ataxin-3 in a Drosophila model, in presence or absence of two suppressors and during aging. Suppressing truncated Ataxin-3-induced toxicity lowered the level of aggregated polyglutamine protein. 26215704_Results suggest that the aggregation of Josephin proceeds from the monomer state to the formation of spheroidal intermediates with a native structure. Only successively, these intermediates evolve into misfolded aggregates and into the final fibrils. 26260925_polyglutamine expansion increases the molecular mobility of two juxtaposed helices critical to ataxin-3 deubiquitinase activity 26266536_Based on these data and other related studies, we presumed that de novo mutations of ATXN3 emerging from large ANs are at least one survival mechanisms of mutational ATXN3 and we can redefine the range of CAG repeats as: ANs=44, 45 =AIs =49 and AEs>/=50 26693702_Segregation patterns and factors influencing instability of expanded ATXN3 CAG transmissions in Machado-Joseph disease have been analyzed. 26808260_USP19_b up-regulates the protein levels of the polyglutamine (polyQ)-containing proteins, ataxin-3 (Atx3) and huntingtin (Htt), and thus promotes aggregation of their polyQ-expanded species in cell models 26880203_Ataxin-3 phosphorylation decreases neuronal defects in spinocerebellar ataxia type 3 models. 27731380_Low exon skipping efficiencies combined with reduction in important ataxin-3 protein functions suggest that skipping of exon 8 and 9 is not a viable therapeutic option for spinocerebellar ataxia type-3. The modified protein was incapable of binding poly-ubiquitin chains, which may interfere with its normal deubiquitinating function. 27847820_we demonstrated that neural differentiation in these iPS cells was accompanied by autophagy and that rapamycin promoted autophagy through degradation of mutant ATXN3 proteins in neurally differentiated spinocerebellar ataxia-3 human induced pluripotent stem cells (p < 0.05). In conclusion, patient-derived iPS cells are a good model for studying the mechanisms of SCA3 and may provide a tool for drug discovery in vitro. 28094059_DNA methylation levels in the ATXN3 promoter were significantly higher in SCA3/MJD patients 28158474_Our data reveal a previously unrecognized balance between pathogenic and potentially therapeutic properties of the ataxin-3-Rad23 interaction; they highlight this interaction as critical for the toxicity of the SCA3 protein, and emphasize the importance of considering protein context when pursuing suppressive avenues. 28180282_The findings reveal ATXN3 to be a novel deubiquitinase of Chk1, providing a new mechanism of Chk1 stabilization in genome integrity maintenance. 28275011_the opposing activities of RNF4 and ataxin-3 consolidate robust MDC1-dependent signaling and repair ofDNA double-strand break. 28334907_data elucidate the important role of ataxin-3 proteolysis in the pathogenesis of Machado-Joseph disease. 28939772_Data suggest ATXN3 binds with low-micromolar affinity to both wild-type p97/VCP and mutants linked to proteostasis deficiency multisystem proteinopathy 1 (MSP1; also called hereditary inclusion body myopathy); stoichiometry of binding is one ATXN3 molecule per p97/VCP hexamer in presence of ATP; MSP1 mutants of p97/VCP bind ATXN3 irrespective of nucleotide state. (VCP = valosin-containing protein/ATPase; ATXN3 = ataxin-3) 29401586_polyQ-expanded Htt-N552 and Atx-3 sequester endogenous Ub adaptors, human RAD23 homolog B (hHR23B) and ubiquilin (UBQLN)-2, into inclusions. This sequestration effect is dependent on the UBA domains of Ub adaptors and the conjugated Ub of the aggregated proteins. 29476013_Studied the role of KPNA3 (karyopherin subunit alpha 3) in nuclear transport of ataxin-3 in spinocerebellar ataxia type 3. 29726932_Coexpression of the chaperon HSP40 and HSP70 effectively rescues the effects of Atxn3-84Q, indicating that polyQ protein aggregation in glia is deleterious. Intriguingly, coexpression of wild-type Atxn3-27Q can also rescue BBB/BRB impairment, suggesting that normal polyQ protein may have a protective function. 29902454_Functionally, Ataxin-3 overexpression promoted cell proliferation, and Ataxin-3 knockdown inhibited cell proliferation in testicular cancer cell. In addition, up-regulation of Ataxin-3 inhibited the expression of PTEN and activated the AKT/mTOR pathway. 30036587_Overexpression, purification, and preliminary biophysical and structural characterization of the Machado-Joseph disease-associated expanded form of ataxin-3 has been reported. 30430184_Impaired eyeblink conditioning is an early marker of cerebellar dysfunction in preclinical SCA3 mutation carriers. 30455355_At the physiological level, the study shows that alternative splicing and the premature stop codon alter ataxin-3 stability and that ataxin-3 isoforms differ in their enzymatic deubiquitination activity, subcellular distribution, and interaction with other proteins. 30942397_miR-25 promoted human colon cancer cell growth and migration, and inhibited apoptosis via ATXN3 expression. The miR-25/ATXN3 axis provides novel insight into the pathogenesis of human colon cancer, particularly with respect to promote proliferation and metastasis of CRC 30994454_The authors report that HTT forms a transcription-coupled DNA repair (TCR) complex with RNA polymerase II subunit A (POLR2A), ataxin-3, the DNA repair enzyme polynucleotide-kinase-3'-phosphatase (PNKP), and cyclic AMP-response element-binding (CREB) protein (CBP). This complex senses and facilitates DNA damage repair during transcriptional elongation, but its functional integrity is impaired by mutant HTT. 31107713_Divalproex sodium regulates ataxin-3 translocation likely by an importin alpha1-dependent pathway. 31189928_Machado-Joseph disease (MJD), also known as spinocerebellar ataxia type 3 (SCA3), is the most frequent dominant ataxia worldwide. In 1994, a kindred from Yemen was described as the first Jewish family with Machado-Joseph disease (MJD/SCA3), a dominant ataxia caused by the expansion of a (CAG)n above 61 repeats, in ATXN3. 31310802_In a drosophila transgenic model, ataxin-3 isoforms are toxic, isoform 1 contributes to Spinocerebellar Ataxia Type 3 due to its higher level and isoform 2 rapidly degrades. 31379806_Ataxin-3 Links NOD2 and TLR2 Mediated Innate Immune Sensing and Metabolism in Myeloid Cells. 31563563_Study demonstrated that ATXN3 significantly correlated with KLF4. High ATXN3/KLF4 expression was associated with a poor prognosis in breast cancer patients. ATXN3 is the deubiquitinating enzyme of KLF4 in breast cancer. ATXN3 promotes breast cancer metastasis by deubiquitinating and stabilizing KLF4, revealing the role of the KLF4 deubiquitinating process in breast cancer metastasis. 31613024_Inactivation of the p97-ATX3 complex affects the non-homologous end joining DNA repair pathway and hypersensitises human cancer cells to ionising radiation 31783119_Druggable genome screen identifies new regulators of the abundance and toxicity of ATXN3, the Spinocerebellar Ataxia type 3 disease protein. 32205441_Study provides evidence that wild-type ATXN3 plays an important role in error-free repair of DNA double-strand breaks in the transcribed genes. In contrast, mutant ATXN3 blocks the activity of a DNA end-processing enzyme, polynucleotide kinase 3'-phosphatase (PNKP), leading to progressive accumulation of double-strand breaks and abrogation of global transcription. 32274760_REVIEW: intimate link between ataxin-3 and cellular stress and its relevance for therapeutic intervention in Machado-Joseph disease 32643267_Clinical features and genetic characteristics of homozygous spinocerebellar ataxia type 3. 32643791_Degron capability of the hydrophobic C-terminus of the polyglutamine disease protein, ataxin-3. 32789747_Antisense Oligonucleotide Therapy Targeted Against ATXN3 Improves Potassium Channel-Mediated Purkinje Neuron Dysfunction in Spinocerebellar Ataxia Type 3. 32978817_CAG Repeat Size Influences the Progression Rate of Spinocerebellar Ataxia Type 3. 33065068_Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3. 33157084_RAN Translation of the Expanded CAG Repeats in the SCA3 Disease Context. 33159825_Expanded CAG Repeats in ATXN1, ATXN2, ATXN3, and HTT in the 1000 Genomes Project. 33306823_[Detection and analysis of dynamic variant in a pedigree affected with spinocerebellar ataxia type 3]. 33411688_Polyglutamine-expanded ataxin3 alter specific gene expressions through changing DNA methylation status in SCA3/MJD. 33477953_Pathological ATX3 Expression Induces Cell Perturbations in E. coli as Revealed by Biochemical and Biophysical Investigations. 33798946_PML-II recruits ataxin-3 to PML-NBs and inhibits its deubiquitinating activity. 34428509_The deubiquitinating enzyme ATXN3 promotes the progression of anaplastic thyroid carcinoma by stabilizing EIF5A2. 35163312_Anti-Excitotoxic Effects of N-Butylidenephthalide Revealed by Chemically Insulted Purkinje Progenitor Cells Derived from SCA3 iPSCs. | ENSMUSG00000021189 | Atxn3 | 1.525982e+02 | 0.7467567 | -0.421289731 | 0.3906354 | 1.098326e+00 | 0.2946337358 | 0.81312088 | No | Yes | 1.098004e+02 | 21.793760 | 1.634601e+02 | 32.868364 | |
| ENSG00000067369 | 7158 | TP53BP1 | protein_coding | Q12888 | FUNCTION: Double-strand break (DSB) repair protein involved in response to DNA damage, telomere dynamics and class-switch recombination (CSR) during antibody genesis (PubMed:12364621, PubMed:22553214, PubMed:23333306, PubMed:17190600, PubMed:21144835, PubMed:27153538, PubMed:28241136). Plays a key role in the repair of double-strand DNA breaks (DSBs) in response to DNA damage by promoting non-homologous end joining (NHEJ)-mediated repair of DSBs and specifically counteracting the function of the homologous recombination (HR) repair protein BRCA1 (PubMed:22553214, PubMed:23727112, PubMed:23333306, PubMed:27153538). In response to DSBs, phosphorylation by ATM promotes interaction with RIF1 and dissociation from NUDT16L1/TIRR, leading to recruitment to DSBs sites (PubMed:28241136). Recruited to DSBs sites by recognizing and binding histone H2A monoubiquitinated at 'Lys-15' (H2AK15Ub) and histone H4 dimethylated at 'Lys-20' (H4K20me2), two histone marks that are present at DSBs sites (PubMed:23760478, PubMed:27153538, PubMed:28241136, PubMed:17190600). Required for immunoglobulin class-switch recombination (CSR) during antibody genesis, a process that involves the generation of DNA DSBs (PubMed:23345425). Participates in the repair and the orientation of the broken DNA ends during CSR (By similarity). In contrast, it is not required for classic NHEJ and V(D)J recombination (By similarity). Promotes NHEJ of dysfunctional telomeres via interaction with PAXIP1 (PubMed:23727112). {ECO:0000250|UniProtKB:P70399, ECO:0000269|PubMed:12364621, ECO:0000269|PubMed:17190600, ECO:0000269|PubMed:21144835, ECO:0000269|PubMed:22553214, ECO:0000269|PubMed:23333306, ECO:0000269|PubMed:23345425, ECO:0000269|PubMed:23727112, ECO:0000269|PubMed:23760478, ECO:0000269|PubMed:27153538, ECO:0000269|PubMed:28241136}. | 3D-structure;Activator;Alternative splicing;Centromere;Chromosomal rearrangement;Chromosome;DNA damage;DNA repair;DNA-binding;Isopeptide bond;Kinetochore;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a protein that functions in the DNA double-strand break repair pathway choice, promoting non-homologous end joining (NHEJ) pathways, and limiting homologous recombination. This protein plays multiple roles in the DNA damage response, including promoting checkpoint signaling following DNA damage, acting as a scaffold for recruitment of DNA damage response proteins to damaged chromatin, and promoting NHEJ pathways by limiting end resection following a double-strand break. These roles are also important during V(D)J recombination, class switch recombination and at unprotected telomeres. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Aug 2017]. | hsa:7158; | chromosome, telomeric region [GO:0000781]; cytoplasm [GO:0005737]; DNA repair complex [GO:1990391]; kinetochore [GO:0000776]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; replication fork [GO:0005657]; site of double-strand break [GO:0035861]; damaged DNA binding [GO:0003684]; histone binding [GO:0042393]; methylated histone binding [GO:0035064]; p53 binding [GO:0002039]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; telomeric DNA binding [GO:0042162]; transcription coregulator activity [GO:0003712]; ubiquitin modification-dependent histone binding [GO:0061649]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to X-ray [GO:0071481]; DNA damage checkpoint signaling [GO:0000077]; double-strand break repair via nonhomologous end joining [GO:0006303]; negative regulation of double-strand break repair via homologous recombination [GO:2000042]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of isotype switching [GO:0045830]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein homooligomerization [GO:0051260] | 11877378_Structure of the human 53BP1 BRCT region bound to p53 and its comparison to the rat Brca1 BRCT structure is presented. 12110597_crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor 12170762_Pts with p53 gene mutations were 3-times more likely to have an early onset breast cancer. Pts with both missense & silent mutations were 7-times & Pts with mutations in exon 8 of the p53 gene 6 times more likely to have early onset breast cancer. 12351827_Purification, crystallization and preliminary X-ray analysis of the BRCT domains of human 53BP1 bound to the p53 tumour suppressor 12364621_demonstrated that 53BP1 is a key transducer of the DNA damage checkpoint signal; required for p53 accumulation, G2-M checkpoint arrest, and the intra-S-phase checkpoint in response to ionizing radiation 12446782_Results suggest that 53BP1 and Snm1 may cooperate in the cellular response to genotoxic damage. 12447382_53BP1 functions as a DNA damage checkpoint protein 12551934_53BP1 is involved in a pathway with NFBD1, H2AX, and Chk2 in recruitment of repair and signaling proteins to sites of DNA damage 12646190_P202 inhibits E2F1-mediated apoptosis in prostate cancer cells 12668657_This protein interacts with HDAC4 protein to mediate DNA damage response. 12824158_53BP1 has the potential to participate directly in the repair of DNA double-strand breaks 14695167_In cells with wild-type Nbs1, suppression of 53BP1 expression had no effect on ATM activation but was associated with increased recruitment of NFBD1/MDC1 and Nbs1 to sites of DNA breaks. 15279780_53BP1 is an activator of ATM in response to DNA damage [review] 15525939_53BP1 senses DNA double-stranded breaks indirectly through changes in higher-order chromatin structure that expose the 53BP1 binding site 15539948_Thus, optimal repair of damaged replication fork lesions likely requires both ATR and ATM. BLM recruits 53BP1 to these lesions independent of its helicase activity, and optimal activation of ATM requires both p53 and BLM helicase activities. 15542852_Snm1 and 53BP1 are components of a mitotic stress checkpoint that negatively targets the APC prior to chromosome condensation 15611139_LC8 binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation 15840649_RNA has a role in the binding of 53BP1 to chromatin damaged by ionizing radiation 15970701_the BRCA1 promoter is positively regulated by 53BP1 16187115_Jun activation domain-binding protein 1 (Jab1) was identified as a 53BP1-binding protein, and the interaction between them was confirmed to occur in mammalian cells. 16294045_GAR (Glycine Arginine rich) motif is a region required for 53BP1 DNA binding activity and as the site of methylation by PRMT1. 16294047_a glycine-arginine rich (GAR) stretch of 53BP1 lying upstream of the Tudor motifs is methylated by PRMT1 16314399_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17043355_53BP1 only has limited checkpoint functions but rather acts as an adaptor in the repair of DNA double strand breaks 17161371_analysis of nonconserved residues that enforced p53 core domain binding with BRCA1-BRCT in a way similar to p53-53BP1 binding 17190600_Using X-ray crystallography and NMR spectroscopy, we show that, despite low amino acid sequence conservation, both 53BP1 and Crb2 contain tandem tudor domains that interact with histone H4 specifically dimethylated at Lys20 (H4-K20me2). 17546051_Contrary to carcinomas, almost no activation or loss of MDC1 or 53BP1 were found among testicular germ-cell tumours (TGCTs), a tumour type with unique biology and exceptionally low incidence of p53 mutations. 17591918_BLM helicase-dependent and -independent roles of TP53BP1 during replication stress-mediated homologous recombination were studied. 17634560_Genetic variation increases had and neck cancer risk. 17634560_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17637749_These studies highlight the role of 53BP1 copy loss in primary human DLBCLs and the value of integrative analyses in detecting this genetic lesion in human tumors 17695720_53BP1 oligomerization is not dependent on the presence of disulfide bridges 17884766_MDC1 and 53BP1 expressions were observed for the first time in human esophageal carcinoma cell lines TE-1,TE-13 and Eca109 cells, at both the mRNA and protein levels. 17927872_53BP1 T885G mutation may not be correlated to the susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. 17927872_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17940005_These data indicate the existence of a DNA double-strand break-repair protein that functions upstream of 53BP1 and contributes to the normal development of the human immune system. 17984114_the phosphorylation-mediated interactions between BLM, 53BP1 and RAD51 are required for their regulatory roles during homologous recombination 17985346_analysis of 53BP1 expression can be a useful tool to estimate the level of genomic instability and, simultaneously, the malignant potency of human thyroid tumors. 18337245_RNF8 ubiquitylation pathways are essential for 53BP1 regulation in response to ionizing radiation, whereas RNF8-independent pathways contribute to 53BP1 targeting and phosphorylation in response to UV light and other forms of DNA replication stress 18380789_a number of nuclear p53-binding protein 1 foci in human skin tumorigenesis 18664457_Nbs1 has a function in ATR signalling in a manner distinct to any role at stalled replication forks. Replication-independent ATR signalling also requires the mediator proteins, 53BP1 and MDC1, providing direct evidence for their role in ATR signalling. 18804090_The two major roles of 53BP1, the checkpoint signaling and repair for DNA damage, can be functionally separated. 18840612_analysis of the 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling 18931659_53BP1 (also known as TRP53BP1), a component of DNA damage foci, changes the dynamic behaviour of chromatin to promote DNA repair 18986980_the interaction between 53BP1 and MDC1 plays a role in the regulation of mitosis 18996087_ATM phosphorylates the S1219 residue of 53BP1 in vitro; this residue is phosphorylated in cells exposed to ionizing radiation 19001859_NFBD1, 53BP1 and BRCA1 have both unique and redundant functions in radiation-induced phosphorylation and localization events in the ATM-Chk2 pathway. 19064641_Additional elements in 53BP1 that facilitate recognition of DNA double-strand breaks, were identified. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19116388_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19176521_PP5 plays an important role in the regulation of 53BP1 phosphorylation and activity in vivo 19414588_PTIP regulates 53BP1-dependent signaling pathway following DNA damage 19422301_Quantitative proteomic analysis integrated with microarray data reveals that TP53BP1 is important for chemotherapy response in ovarian cancers. 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19626602_Observational study of gene-disease association. (HuGE Navigator) 19650074_MSL1 plays an important role in mediating irradiation-induced DNA repair through formation of HAT complexes and interaction with 53BP1. P8 acts as a negative regulator of this process by interacting with MSL1 and preventing its role on HAT activity. 19656881_Data suggest that the association between Zta and 53BP1 is involved in the viral replication cycle. 19690177_Observational study of gene-disease association. (HuGE Navigator) 19806024_The recruitment of tumor protein p53 binding protein 1 into gamma H2AFX protein-containing DNA damage foci was inhibited at G(2)/M phase. 19876065_Depletion of 53BP1 triggers the demise of HIV-1-elicited syncytia through mitotic catastrophe. 20010693_53BP1 promotes ATM activity through direct interactions with the MRN complex. 20081839_53BP1 amplifies Mre11-NBS1 accumulation at late-repairing DSBs, concentrating active ATM and leading to robust, localized pKAP-1. 20106900_Observational study of gene-disease association. (HuGE Navigator) 20307547_Binding of the 53BP1 Tudor domain to p53 dimethylated at K382 may facilitate p53 accumulation at DNA damage sites and promote DNA repair as suggested by chromatin immunoprecipitation and DNA repair assays. 20351298_analysis of formation of gammaH2AX and 53BP1 that coincide at sites of double-strand breaks (DSBs) after ionizing radiation 20372852_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20372852_Polymorphism-polymorphism interaction (both multiplicative and additive) between TP53BP1 and TP53 SNPs was suggested for lung cancer risk in a Japanese population. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20385987_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20453858_find reduced 53BP1 expression in subsets of sporadic triple-negative and BRCA1-associated breast cancers 20596616_53BP1 expression plays an important role in cisplatin resistance and predicts the prognosis for lung adneocarcinomas. 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20646866_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20686496_Observational study of gene-disease association. (HuGE Navigator) 20686496_three single nucleotide polymorphisms for 53BP1 were significantly associated with skin cancer risk 20724228_53Bp1 is an inhibitor of homologous recombination. Loss of 53BP1 correlates with high grade human breast cancers. 21293379_a pathway involving gammaH2AX-MDC1-MMSET regulates the induction of H4K20 methylation on histones around DSBs, which, in turn, facilitates 53BP1 recruitment 21317883_53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress 21361779_an interaction between endogenous p53 and ATM or 53BP1 21412056_The E3 Ubiquitin ligases, RNF8 and RNF168, are recruited to DNA damage foci in late mitosis, presumably to prime sites for the DNA damage response, 53BP1, recruitment in early G1. 21444690_The findings invoke a model wherein incomplete DNA synthesis during S phase leads to a DNA damage response and formation of 53BP1-OPT domains in the subsequent G1. 21635553_Glu353Asp and T-885G polymorphic variants might not have an influence on breast cancer risk, thus might not be candidates for cancer susceptibility; Glu353Asp variant may be associated with tumour aggressiveness 21840613_Constitutive gamma-H2AX and 53BP1 staining reveals a subset of patients with triple negative breast tumors that have a significantly poorer prognosis. 21914189_both 53BP1 and SRC3 have a common function that converge at the BRCA1 promoter and possibly other genes important for DNA repair and genomic stability 21931182_Data suggest a link between recruitment of 53BP1, resolution of DNA damage, and increased species lifespan. 22034884_Immunofluorescence analysis of 53BP1 expression can be a useful tool with which to estimate the level of genomic instability 22075984_Data show that the C-terminal part of NUP153 is required for effective 53BP1 nuclear import, and that 53BP1 is imported to the nucleus through the NUP153-importin-beta interplay. 22088370_53BP1 mutations may be involved in the carcinogenesis of prostatic adenocarcinoma. There was no obvious correlation between 53BP1 gene mutations and the various clinicopathologic parameters. 22266878_53BP1 is involved in several important steps in controlling cell proliferation and growth and preventing ovarian tumor development. 22323184_survival analysis revealed that ATM inhibition, but not 53BP1 depletion, strongly radiosensitised cervical cancer cells 22373579_The authors propose that the RNF8-dependent degradation of JMJD2A regulates DNA repair by controlling the recruitment of 53BP1 at DNA damage sites. 22421153_results uncouple 53Bp1 phosphorylation from double-strand breaks localization and support parallel pathways for 53Bp1 biology during the DNA damage response 22553214_Authors propose that BRCA1 antagonises 53BP1-dependent DNA repair in S phase by inhibiting its interaction with chromatin proximal to damage sites. 22621922_Data show that vaccinia-related kinase 1 (VRK1) in resting cells plays an important role in the formation of ionizing radiation-induced foci that assemble on the 53BP1 scaffold protein during the DNA damage response. 22677490_distinct dynamics of MDC1 and 53BP1 at different types of nuclear structures 22860035_proteins accumulate into foci with characteristic mean recruitment times tau(1). Mdc1 accumulates faster than 53BP1 after high LET irradiation 22865450_Data indicate that RNF8-dependent chromatin ubiquitination is required for RAD51 assembly in BRCA1/53BP1-depleted cells. 22886457_Clinically evident local recurrence of residual cholangiocarcinoma in situ at ductal stumps is closely associated with 53BP1 inactivation and decreased apoptosis. 23027628_53BP1 functions as a tumor suppressor in breast cancer via the inhibition of NF-kappaB through miR-146a. 23206244_repair signaling in CD133 + UCBC is different from CD133 - umbilical cord blood cells 23246380_BRCA1-mutated ovarian carcinomas have higher 53BP1 protein expression than wildtype or BRCA2-mutated carcinomas. 23329852_Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair 23333306_RIF1 as the critical effector of 53BP1 during DSB repair 23337117_BRCA1 loss activates cathepsin L (CTSL)-mediated degradation of 53BP1. 23377543_acetylation regulates relative BRCA1 and 53BP1 DNA double strand breaks chromatin occupancy to direct DNA repair mechanism 23680719_Single nucleotide polymorphisms in TP53BP1 is associated with radiation-dose response of glycophorin A somatic mutation in survivors with subsequent cancer development. 23728321_We found that beam shielding resulted in increased induction of 53BP1 foci and micronuclei in a cell-type-dependent manner compared with the unshielded 200 MeV/u Fe ion beam. 23910218_53BP1 plays a vital role in inhibiting angiogenesis. 24004175_Abnormal/high DDR type of 53BP1 expression might be associated with genomic instability and papillary/trabecular morphology at an early stage of papillary thyroid carcinoma carcinogenesis through BRAF(V) (600E) mutation. 24091731_CTSL-mediated degradation of 53BP1 is an endogenous mechanism of non-cycling cells to balance NHEJ and HR. 24164899_Studies indicate that the HIN (hematopoietic expression, interferon-inducible nature, and nuclear localization) domains exhibit either absent in melanoma2 (Aim2) HIN-like or p202 HINa-like modes of DNA binding. 24191908_BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. 24316395_53BP1 and TP53 gene interactions were not associated with lung cancer risk. 24326623_53BP1 recruitment to DNA repair foci requires the direct recognition of a double strand break-specific histone code. [Review] 24603722_The Glu353Asp (rs560191) polymorphism in TP53BP1 gene was not associated with risk of cancer. (Meta-analysis) 24681733_The level of 53BP1 NF was significantly increased during cervical cancer initiation and progression, whereas the relative level of 53BP1 mRNA was significantly decreased in the cervical cancer compared with that in the precancerous cervical lesion tissue. 24703952_Protein phosphatase complex PP4C/R3beta dephosphorylates T1609 and S1618 to allow the recruitment of 53BP1 to chromatin in G1 phase. 25049398_Binding of 53BP1 to methyl K810 occurs on E2 promoter binding factor target genes and allows pRb activity to be effectively integrated with the DNA damage response. 25262557_ATMIN is required for the ATM-mediated signaling and recruitment of 53BP1 to DNA damage sites upon replication stress. 25294876_SMCHD1 recruitment to DNA damage foci is regulated by 53BP1. 25337968_Depletion of RNF8 or RNF168 blocks the degradation of diffusely localized nuclear 53BP1. 25422456_UbcH7 has a role in regulating 53BP1 stability and DSB repair 25532599_Studies performed genotyping analyses of eight functional CDKN1A p21, oncoprotein p53, TP53BP1 protein and tumor protein p73 single nucleotide polymorphisms (SNPs) in 330 gastric cardia adenocarcinoma (GCA) cases and 608 controls in a Chinese population. 25586219_Depletion of both 53BP1 and BRCA1 increases repair needing microhomology usage and augments loss of DNA sequence, suggesting that MMEJ is a highly regulated DSB repair process 25645366_53BP1 binds to lamins A/C via its Tudor domain, and this is abrogated by DNA damage. Lamins A/C regulate 53BP1 levels and consequently lamin A/C-null HDF display a 53BP1 null-like phenotype. 25976740_we generated two HEP-2 cell lines with a stable knockdown of MDC1 or 53BP1 with short hairpin RNA (shRNA), respectively, and investigated the effect of MDC1 and 53BP1 on cell radiosensitivity 25999347_CK2/WIP1-mediated modulation of LSD1 phosphorylation facilitates RNF168-dependent ubiquitination and recruitment of 53BP1 to the DNA damage sites. 26011542_53BP1 functioned as a tumor suppressor gene by its novel negative control of Epithelial-mesenchymal transition through regulating the expression of miR-200b/429 and their target gene ZEB1. 26199937_After nanogold treatment, an inverse correlation between the formation of 53BP1 foci and micronuclei generation was observed. The robust 53BP1 recruitment resulted in reduced micronuclei production 26243567_the formation of 53BP1, gammaH2AX foci and their co-localization induced by gamma-rays (2, 5, 10, 50, 200 cGy) in human lymphocytes, was analyzed. 26261485_Suggest 53BP1 may serve as a candidate biomarker for predicting prognosis and disease development in colorectal cancer. 26446986_Impaired TIP60-mediated H4K16 acetylation accounts for the aberrant chromatin accumulation of 53BP1 and RAP80 in Fanconi anemia pathway-deficient cells. 26482424_kinetics of the accumulation of selected DNA repair-related proteins is protein specific at locally induced DNA lesions, and that the formation of gH2AX- and NBS1-positive foci, but not 53BP1-positive NBs, is cell cycle dependent in HeLa cells 26541290_The study shows higher expression of gamma-H2AX and 53BP1 foci in rectal cancer patients compared with healthy individuals. Yet the data in vitro were not predictive in regard to the radiotherapy outcome. 26615718_interplay between 53BP1/NHEJ and BRCA1/HR is of great relevance for tumor treatment, as the 53BP1 status would be highly important for the treatment response of BRCA1-associated tumors. 26628370_the interaction of 53BP1 with gammaH2AX is required for sustaining the 53BP1-dependent focal concentration of activated ATM that facilitates repair of DNA double-strand breaks in heterochromatin in G1. 26637364_we show that XIST and 53BP1 can be used to identify BRCA1-like breast cancer patients that have higher event rates and poor outcome after HD chemotherapy. 26684013_Depleting 53BP1, which slows cNHEJ, fully rescues the nuclear and cytogenetic abnormalities seen with EEPD1 depletion 26766492_Combined effect of dynamic recruitment of RNF4 to KAP1 regulates the relative occupancy of 53BP1 and BRCA1 at double-strand break sites to direct DNA repair in a cell cycle-dependent manner. 26820970_For both, gamma-H2AX and 53BP1, the cellular focus number as well as the percentage of positive cells did not differ between patients with clinically isolated syndrome/early relapsing-remitting multiple sclerosis and healthy controls. 26854228_5-Hydroxymethylcytosine (5hmC) accumulates at DNA damage foci and colocalizes with major DNA damage response proteins 53BP1 and gH2AX, revealing 5hmC as an epigenetic marker of DNA damage. 26935218_Increased 53BP1 expression (i.e., 'unstable' expression) in nuclear foci of oncocytic follicular adenoma (FA) of the thyroid correlates with a higher incidence of DNA copy numbers compared with conventional FA. 27041576_Ras-induced senescent cells are hindered in their ability to recruit BRCA1 and 53BP1 to DNA damage sites. Whereas BRCA1 is downregulated at transcripts levels, 53BP1 loss is caused by activation of cathepsin L-mediated degradation of 53BP1 protein. we discovered a marked downregulation of vitamin D receptor (VDR) during OIS, and a role for the vitamin D/VDR axis regulating the levels of these DNA repair 27153538_TIP60 complex regulates bivalent chromatin recognition/modification by 53BP1 through direct H4K20me binding and H2AK15 acetylation. 27348077_Exhaustion of 53BP1 by increasing the load of double strand breaks suppresses RAD51 accumulation in repair foci during S and G2. 27371829_The authors identified 53BP1 and USP28 as essential components acting upstream of p53, evoking p21-dependent cell cycle arrest in response not only to centrosome loss, but also to other distinct defects causing prolonged mitosis. 27432896_USP28-53BP1-p53-p21 signaling pathway is also required to arrest cell growth after a prolonged prometaphase. 27432897_analysis of centrinone resistance identified a 53BP1-USP28 module as critical for communicating mitotic challenges to the p53 circuit and TRIM37 as an enforcer of the singularity of centrosome assembly. 27462807_cryo-EM structure of a dimerized human 53BP1 fragment bound to a H4K20me2-containing and H2AK15ub-containing nucleosome core particle at 4.5 A resolution 27465548_High 53BP1 mRNA is associated with head and neck cancer. 27466483_During live-cell imaging, 53BP1-GFP focus formation was observed within 10 minutes after UVC irradiation. Most 53BP1 foci resolved by 100 minutes. To block UVC-induced double-strand break repair in cancer cells, poly(ADP-ribose) polymerase (PARP) was targeted with ABT-888 (veliparib). PARP inhibition markedly enhanced UVC-irradiation-induced persistence of 53BP1-foci 27494840_53BP1/RIF1 has a role in limiting BRCA1/CtIP-mediated end resection to control double strand break repair pathway choice 27499037_Deficiency of 53BP1 inhibits the radiosensitivity of colorectal cancer. 27546791_53BP1-USP28 cooperation is essential for normal p53-promoter element interactions and gene transactivation-associated events, yet dispensable for 53BP1-dependent DNA double-strand repair regulation. 27613518_Results provide evidence that 53BP1 is involved in breast cancer cells resistance for PARP inhibitor; its depletion causes resistance in ATM-deficient tumor cells. 27716486_53BP1 embodies two distinct activities: it can act as a DNA repair factor on the chromatin that flanks double-strand DNA repairs, and it promotes optimal p53 activity. 27798638_his shows that 53BP1 protects both close and distant DSEs from degradation and that the association of unprotection with distance between DSEs favors ECS capture. Reciprocally, silencing CtIP lessens ECS capture both in control and 53BP1-depleted cells. We propose that close ends are immediately/rapidly tethered and ligated, whereas distant ends first require synapsis of the distant DSEs prior to ligation 27838786_hese findings confirmed that 53BP1 loss might be a negative factor for chemotherapy efficacy, promoting cell proliferation and inhibiting apoptosis by suppressing ATM-CHK2-P53 signaling, and finally inducing 5-FU resistance. 27922110_Gamma-H2AX, phosphorylated KAP-1 and 53BP1 play an important role in the repair of heterochromatic radon-induced DNA double-strand breaks. 28076794_BRCA1 promotes PP4C-dependent 53BP1 dephosphorylation and RIF1 release, directing repair toward homologous recombination. 28213517_TIRR is a novel 53BP1-interacting protein that participates in the DNA damage response 28228263_A reciprocal regulation between 53BP1 and APC/C that is required for response to mitotic stress. 28241136_findings identify TIRR as a new factor that influences double-strand break repair using a unique mechanism of masking the histone methyl-lysine binding function of 53BP1 28255090_These data suggest that multiple pathways collectively fine-tune the cellular levels of 53BP1 protein to ensure proper DSB repair and cell survival. 28359030_Co-localization of gammaH2AX and 53BP1 indicates promotion of (in)effective nonhomologous end-joining repair mechanisms at sites of DSB. Moreover, gammaH2AX/53BP1 foci distribution presumably reveals a non-random spatial organization of the genome in MDS and AML. 28397142_It observed a distinct accumulation of 53BP1 protein to UV-induced DNA lesions: in R273C mutants, 53BP1 appeared transiently at DNA lesions, during 10-30 min after irradiation; the mutation R282W was responsible for accumulation of 53BP1 immediately after UVA-damage; and in L194F mutants, the first appearance of 53BP1 protein at the lesions occurred during 60-70 min. 28475402_PAXIP1 and 53BP1 protein levels followed gene expression results, i.e., are intrinsically correlated, and also reduced in more advanced breast cancer tumors. 28498430_UVA-induced progerinlamin A complex formation was largely responsible for suppressing 53BP1-mediated NHEJ DSB repair activity. The present study is the first to demonstrate that UVA-induced progerin upregulation adversely affects 53BP1-mediated NHEJ DSB repair in human keratinocytes via progerinlamin A complex formation. 28506460_Ubiquitin ligases RNF168, RNF169, and RAD18 specifically bind histone H2A Lys13/15-ubiquitylated nucleosomes. 53BP1 chromatin recruitment may be activated by RNF168 and blocked by RNF169 and RAD18. 28564601_premature maturation of post-replicative chromatin restores Histone h4 lysine 20 methylation and rescues 53BP1 accumulation on replicated chromatin. 28576968_Despite the requirement of all three nucleoporins for accurate NHEJ, only Nup153 is needed for proper nuclear import of 53BP1 and SENP1-dependent sumoylation of 53BP1. Data support the role of Nup153 as an important regulator of 53BP1 activity and efficient NHEJ. 28751496_results further highlight the antagonistic relationship between 53BP1 and BRCA1, and place Nup153 and Nup50 in a molecular pathway that regulates 53BP1 function by counteracting BRCA1-mediated events. 28930533_Data show that the expression of tumor protein p53 binding protein 1 (53BP1) varies at different stages of cell cycle, with high-level expression observed in mitosis. 28958991_Results indicate that 53BP1 is a biomarker of response to anti-PARP therapy in the laboratory, and our DNA damage response gene signature may be used to identify patients who are most likely to respond to PARP inhibition. 29106372_These results reveal two distinct fork restart pathways, which are antagonistically controlled by 53BP1 and BRCA1 in a double-strand DNA break repair-independent manner. 29115375_the two cell lines exhibited relatively low protein expression levels of p53, lower levels of p53 and TPp53BP1 transcripts were detected in the K562/G cells. Taken together, these findings suggest that the resistance of CML to the tyrosine kinase inhibitor, imatinib, may be associated with persistent STAT5-mediated ROS production, and the abnormality of the p53 pathway 29160738_the differential H4K20 methylation status between pre-replicative and post-replicative DNA represents an intrinsic mechanism that locally ensures appropriate recruitment of the 53BP1-RIF1-MAD2L2 complex at DNA double strand breaks. 29176614_Inhibition of 53BP1 is a robust method to increase efficiency of HDR-based precise genome editing. 29190394_Results indicate that the acetylation status of P53-binding protein 1 (53BP1) plays a key role in its recruitment to DNA double strand break (DSB) repair by promoting non-homologous end joining (NHEJ). 29274141_Study demonstrates a consistent resistance profile to PARPi and a unique cross-resistance profile to non-PARPi drugs in different PARPi-resistant U251 glioblastoma cells and reveals 53BP1 loss and SAMHD1 overexpression as the primary mechanisms responsible for their resistance to PARPi and Ara-C, respectively. 29309426_number of gammaH2AX foci did not significantly change following cardiac MR (median foci per cell pre-MR = 0.11, post-MR = 0.11, p = .90), but the number of 53BP1 foci significantly increased following MR 29328365_As shown in xenograft model of glioblastoma phosphorylation of 53BP1 by GSK3beta was indispensable for DNA double-strand break repair. 29445165_Data indicate that p53-binding protein 1 (53BP1) is required to prevent excessive chromosome missegregation and probably genome hyper-instability, and also for optimal growth in cancer cells. 29603287_Results indicate that integrity of the nuclear localization signal is important for 53BP1 nuclear localization. 29651020_GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. 29844495_This study elucidates the mechanism by which TIRR recognizes 53BP1 Tudor and functions as a cellular inhibitor of the histone methyl-lysine readers. 29967538_X-ray crystal structures of TIRR and a designer protein bound to 53BP1 now reveal a mechanism in which an intricate binding area centered on an essential TIRR arginine residue blocks the methylated-chromatin-binding surface of 53BP1. A 53BP1 separation-of-function mutation that abolishes TIRR-mediated regulation in cells renders 53BP1 hyperactive in response to DSBs, highlighting the key inhibitory function of TIRR. 29990989_results suggest that there is a direct interaction between 53BP1 and MCMs, which is essential for 53BP1 chromatin fraction and foci formation in hepatoma HepG2 cells. 30002377_Data indicate the molecular mechanism underlying Tudor interacting repair regulator (TIRR)-mediated suppression of tumor protein p53 binding protein 1 (53BP1)-dependent DNA damage repair. 30022158_data suggest that CST-Polalpha-mediated fill-in helps to control the repair of double-strand breaks by 53BP1, RIF1 and shieldin 30022168_binding of single-stranded DNA by SHLD2 is critical for shieldin function, consistent with a model in which shieldin protects DNA ends to mediate 53BP1-dependent DNA repair 30082159_The results may suggest that TP53BP1 and MFN1 frameshift mutations and their intratumoral heterogeneity (ITH) could contribute to cancer development by inhibiting the TSG activities. 30104380_results highlight the interplay of RNF169 with 53BP1 in fine-tuning choice of DSB repair pathways. 30257212_53BP1 loss of function promotes homologous recombination and PARPi resistance in a BRCA1 mutation-specific fashion. 30312172_H3K9me3 and H4K20me3 represent epigenetic markers that are important for the function of the 53BP1 protein in non-homologous end joining (NHEJ) repair. 30341375_this study identified clear differences in the 53BP1 expression patterns in urothelial carcinogenesis, and their close association with genomic instability 30445982_HPV16 E6 and E7 oncoproteins interact with 53BP1 inside nuclear foci after DNA damage 30497961_structure and function of 53BP1 and its contribution to cancer 30602538_Mitotic regulators TPX2 and Aurora A protect DNA forks during replication stress by counteracting 53BP1 function. 30620991_53BP1 deficiency promotes pathological neovascularization in proliferative retinopathy attributed to an increased homologous recombination rate. 30668775_CTD Tyr1-phosphorylated RNAPII (CTD Y1P) synthetizes strand-specific, damage-responsive transcripts (DARTs), which trigger formation of double-stranded (ds)RNA intermediates via DNA-RNA hybrid intermediates to promote recruitment of p53-binding protein 1 (53BP1) and Mediator of DNA damage checkpoint 1 (MDC1) to endogenous DSBs. 30685087_analysis of recognition of the 53BP1 nuclear localization signal by importin-alpha 30686551_53BP1 deletion increased HR in BRCA1-mutant COV362 cells and decreased PARPi sensitivity in vitro. In 36 women with relapsed ovarian cancer, responses to the PARPi ABT-767 were observed exclusively in cancers with HR deficiency. 30718900_Study shows that UTX and 53BP1 directly interact and co-occupy promoters in human embryonic stem cells and differentiating neural progenitor cells. Data suggests that the 53BP1-UTX interaction supports the activation of key genes required for human neurod | ENSMUSG00000043909 | Trp53bp1 | 1.708226e+03 | 0.5882048 | -0.765609574 | 0.2883997 | 6.981591e+00 | 0.0082352385 | 0.33086870 | No | Yes | 1.321243e+03 | 169.348938 | 2.134506e+03 | 279.695554 | |
| ENSG00000067840 | 57595 | PDZD4 | protein_coding | Q76G19 | Alternative splicing;Coiled coil;Cytoplasm;Phosphoprotein;Reference proteome | hsa:57595; | cell cortex [GO:0005938] | 15077175_Might play an important role in the proliferation of SS cells and might be a suitable target for SS drugs. 15138636_PDZRN4L showed 49.9% total-amino-acid identity with PDZRN4 short isoform (PDZRN4S). PDZ, PR34H1 and PR34H2 domains were conserved between PDZRN4L and PDZRN4S. | ENSMUSG00000002006 | Pdzd4 | 7.579572e+02 | 1.2665581 | 0.340913290 | 0.2712748 | 1.602447e+00 | 0.2055567317 | 0.78688920 | No | Yes | 7.850588e+02 | 94.712514 | 6.301609e+02 | 78.868153 | |||
| ENSG00000067900 | 6093 | ROCK1 | protein_coding | Q13464 | FUNCTION: Protein kinase which is a key regulator of the actin cytoskeleton and cell polarity (PubMed:10436159, PubMed:10652353, PubMed:11018042, PubMed:11283607, PubMed:17158456, PubMed:18573880, PubMed:19131646, PubMed:8617235, PubMed:9722579). Involved in regulation of smooth muscle contraction, actin cytoskeleton organization, stress fiber and focal adhesion formation, neurite retraction, cell adhesion and motility via phosphorylation of DAPK3, GFAP, LIMK1, LIMK2, MYL9/MLC2, TPPP, PFN1 and PPP1R12A (PubMed:10436159, PubMed:10652353, PubMed:11018042, PubMed:11283607, PubMed:17158456, PubMed:18573880, PubMed:19131646, PubMed:8617235, PubMed:9722579, PubMed:23093407, PubMed:23355470). Phosphorylates FHOD1 and acts synergistically with it to promote SRC-dependent non-apoptotic plasma membrane blebbing (PubMed:18694941). Phosphorylates JIP3 and regulates the recruitment of JNK to JIP3 upon UVB-induced stress (PubMed:19036714). Acts as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability (By similarity). Acts as a negative regulator of VEGF-induced angiogenic endothelial cell activation (PubMed:19181962). Required for centrosome positioning and centrosome-dependent exit from mitosis (By similarity). Plays a role in terminal erythroid differentiation (PubMed:21072057). Inhibits podocyte motility via regulation of actin cytoskeletal dynamics and phosphorylation of CFL1 (By similarity). Promotes keratinocyte terminal differentiation (PubMed:19997641). Involved in osteoblast compaction through the fibronectin fibrillogenesis cell-mediated matrix assembly process, essential for osteoblast mineralization (By similarity). May regulate closure of the eyelids and ventral body wall by inducing the assembly of actomyosin bundles (By similarity). {ECO:0000250|UniProtKB:P70335, ECO:0000250|UniProtKB:Q8MIT6, ECO:0000269|PubMed:10436159, ECO:0000269|PubMed:10652353, ECO:0000269|PubMed:11018042, ECO:0000269|PubMed:11283607, ECO:0000269|PubMed:17158456, ECO:0000269|PubMed:18573880, ECO:0000269|PubMed:18694941, ECO:0000269|PubMed:19036714, ECO:0000269|PubMed:19131646, ECO:0000269|PubMed:19181962, ECO:0000269|PubMed:19997641, ECO:0000269|PubMed:21072057, ECO:0000269|PubMed:23093407, ECO:0000269|PubMed:23355470, ECO:0000269|PubMed:8617235, ECO:0000269|PubMed:9722579}. | 3D-structure;ATP-binding;Acetylation;Apoptosis;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Direct protein sequencing;Golgi apparatus;Kinase;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Zinc;Zinc-finger | This gene encodes a protein serine/threonine kinase that is activated when bound to the GTP-bound form of Rho. The small GTPase Rho regulates formation of focal adhesions and stress fibers of fibroblasts, as well as adhesion and aggregation of platelets and lymphocytes by shuttling between the inactive GDP-bound form and the active GTP-bound form. Rho is also essential in cytokinesis and plays a role in transcriptional activation by serum response factor. This protein, a downstream effector of Rho, phosphorylates and activates LIM kinase, which in turn, phosphorylates cofilin, inhibiting its actin-depolymerizing activity. A pseudogene, related to this gene, is also located on chromosome 18. [provided by RefSeq, Aug 2015]. | hsa:6093; | bleb [GO:0032059]; centriole [GO:0005814]; cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; extracellular region [GO:0005576]; Golgi membrane [GO:0000139]; lamellipodium [GO:0030027]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; secretory granule lumen [GO:0034774]; aspartic-type endopeptidase inhibitor activity [GO:0019828]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; Rho-dependent protein serine/threonine kinase activity [GO:0072518]; small GTPase binding [GO:0031267]; tau protein binding [GO:0048156]; tau-protein kinase activity [GO:0050321]; actin cytoskeleton organization [GO:0030036]; actomyosin structure organization [GO:0031032]; aortic valve morphogenesis [GO:0003180]; apical constriction [GO:0003383]; apoptotic process [GO:0006915]; bleb assembly [GO:0032060]; blood vessel diameter maintenance [GO:0097746]; cortical actin cytoskeleton organization [GO:0030866]; embryonic morphogenesis [GO:0048598]; epithelial to mesenchymal transition [GO:0001837]; glomerular visceral epithelial cell migration [GO:0090521]; I-kappaB kinase/NF-kappaB signaling [GO:0007249]; leukocyte cell-cell adhesion [GO:0007159]; leukocyte migration [GO:0050900]; leukocyte tethering or rolling [GO:0050901]; membrane to membrane docking [GO:0022614]; mitotic cytokinesis [GO:0000281]; mRNA destabilization [GO:0061157]; myoblast migration [GO:0051451]; negative regulation of amyloid precursor protein catabolic process [GO:1902992]; negative regulation of amyloid-beta formation [GO:1902430]; negative regulation of angiogenesis [GO:0016525]; negative regulation of bicellular tight junction assembly [GO:1903347]; negative regulation of biomineral tissue development [GO:0070168]; negative regulation of membrane protein ectodomain proteolysis [GO:0051045]; negative regulation of myosin-light-chain-phosphatase activity [GO:0035509]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of phosphorylation [GO:0042326]; negative regulation of protein binding [GO:0032091]; neuron projection arborization [GO:0140058]; neuron projection development [GO:0031175]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of amyloid-beta clearance [GO:1900223]; positive regulation of autophagy [GO:0010508]; positive regulation of cardiac muscle hypertrophy [GO:0010613]; positive regulation of connective tissue replacement [GO:1905205]; positive regulation of dephosphorylation [GO:0035306]; positive regulation of focal adhesion assembly [GO:0051894]; positive regulation of gene expression [GO:0010628]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of phosphatase activity [GO:0010922]; protein localization to plasma membrane [GO:0072659]; protein phosphorylation [GO:0006468]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of amyloid-beta formation [GO:1902003]; regulation of angiotensin-activated signaling pathway [GO:0110061]; regulation of autophagy [GO:0010506]; regulation of cell adhesion [GO:0030155]; regulation of cell junction assembly [GO:1901888]; regulation of cell migration [GO:0030334]; regulation of cell motility [GO:2000145]; regulation of establishment of cell polarity [GO:2000114]; regulation of establishment of endothelial barrier [GO:1903140]; regulation of focal adhesion assembly [GO:0051893]; regulation of keratinocyte differentiation [GO:0045616]; regulation of microtubule cytoskeleton organization [GO:0070507]; regulation of neuron differentiation [GO:0045664]; regulation of stress fiber assembly [GO:0051492]; regulation of synaptic vesicle endocytosis [GO:1900242]; response to angiotensin [GO:1990776]; response to transforming growth factor beta [GO:0071559]; Rho protein signal transduction [GO:0007266]; signal transduction [GO:0007165]; smooth muscle contraction [GO:0006939] | 11751986_role in the control of cell shape and lymphocyte chemotaxis 11818523_Expressed in myometrium 11893932_ROCK-1 may contribute to pancreatic cancer cell invasion and/or metastasis by facilitating cancer cell migration 11992112_ROCK and Dia have opposing effects on adherens junctions downstream of Rho 12034773_The Rho-associated protein kinase p160ROCK is required for centrosome positioning. 12193698_Activation of RhoA kinase promotes beta1 and beta2 integrin-dependent leukocyte de-adhesion by inhibiting cytoskeletal-dependent spreading, a new mechanism for regulation of integrin receptor avidity. 12749591_dual function in human parturition: in uterine muscle its expression and reversion contributes to contractions throughout labor; irreversible activation contributes to involution of uterus post-partum 12850840_Continued disruption of the cytoskeletal microtubule ring, appears to be a Rhokinase-dependent event involved in the transformation of discoid platelets into spheres. 14610060_Breast epithelial cells sense the rigidity or density of their environment via ROCK-mediated contractility and a subsequent down-regulation of Rho and FAK function, which is necessary for breast epithelial tubulogenesis to occur. 14660612_ROCKI interacts with the switch regions of RhoA 14716844_invasiveness of hepatocellular carcinoma is facilitated by the Rho/Rho-kinase pathway is likely to be relevant to tumor progression 15044207_results indicate that remnant-like particles from sudden cardiac death patients upregulate Rho-kinase in coronary vascular smooth muscle cells and markedly enhance coronary vasospastic activity 15096506_RhoA, mDia, and ROCK have roles in cell shape-dependent control of the Skp2-p27kip1 pathway and the G1/S transition 15208091_Potassium chloride acts in part through stimulation of Rho and ROCK, possibly secondary to voltage-dependent Ca2+ influx. 15494373_a novel nuclear export is activated by the ROCK signaling pathway to exclude hnRNP C1/C2 from nucleus, by which the compartmentalization of specific hnRNP components is disturbed in apoptotic c 15611088_epithelial cell motility is modulated by integrin engagment through RhoA/ROCK and PAK1 15647781_data suggest that statins exert their effects on shedding of sAPP(alpha) from cultured cells, at least in part, by modulation of the isoprenoid pathway and ROCK1 15665056_Enhanced activation of Rho kinase contributes to augmented contraction of the basilar artery to serotonin after subarachnoid hemorrhage. 15713824_Rho-kinase inhibition may be of value in treating age-related erectile dysfunction. 15723050_a Cdc42-MRCK signal mediates myosin-dependent cell motility and there is convergence between Rho and Cdc42 signalling 15843433_a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module is regulated by Protein Kinase A gating and then regulates invasion in breast cancer cell lines 15855233_CD95 capping and the subsequent cellular polarization is a ROCK signaling-regulated process. 15910744_These results suggest that Rho-kinase regulates the association of alpha-adducin and spectrin with the actin cytoskeleton in platelet activation. 15956119_Hyperglycemia stimulates Rho-kinase activity via PKC- and oxidative stress-dependent pathways, leading to increased PAI-1 gene transcription 16141422_ROCK has a role in ischemia-induced endothelial dysfunction during stroke 16249185_two N-terminal regions interact to form a dimerization domain linking two kinase domains together 16371346_Our data suggested that p160ROCK was involved in ovarian cancer cell invasion and metastasis by facilitating cancer cell migration, and that p160ROCK might be a potential new effective target for preventing metastasis of ovarian cancer. 16407310_Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1 16413470_ERK-MAPK promotes endothelial cell survival and sprouting by downregulating Rho-kinase signaling. 16543245_there is an alternative mechanism for the hypoxic induction of VEGF in colon cancer that does not depend upon HIF-1alpha but instead requires the activation of PI3K/Rho/ROCK and c-Myc 16687572_Rho kinase, myosin-II, and p42/44 MAPK are potentially important players in different aspects of bile canalicular lumen morphogenesis. 16712513_A constitutively active dimeric human ROCK I (3-543) kinase domain was expressed in Sf-9 cells. We have identified a minimal functional subdomain of ROCK I that can be used in crystallization studies. 16760434_Vascular endothelial growth factor (VEGF) elicits the activation of the VEGFR2-ROCK pathway, leading to phosphorylation of Ser732 within FAK which changes the conformation of FAK, making it accessible to Pyk2. 16772336_Ability of MSCs to differentiate into neuron-like cells in response to CoCl(2), an effect that might act, in part, through HIF-1 activation and cell-cycle arrest, and which is potentiated by inhibition of ROCK. 16784244_burst in presteady-state kinetics & viscosity effect on k(cat) of 1.3 +/- 0.2 characterize phosphoryl transfer step to be fast & release of product &/or enzyme isomerization step accompanying it as rate-limiting at V(max) conditions. 16825579_cholesterol potentiates the Ca(2+) sensitization of VSM mediated by a SPC/Src-TK/Rho-kinase pathway 16891369_These results suggest that ROCK plays an essential role in the regulation of the intracellular mechanical response to VEGF of endothelial cells in a 3D matrix. 16978606_Data suggest that Rho kinase-dependent contractile force generation leads to co-alignment of cells and collagen fibrils along the plane of greatest resistance, and that this process contributes to global matrix contraction 16983089_ROCK-1-dependent caspase-3 activation was coupled with the activation of PTEN and the subsequent inhibition of protein kinase B (Akt) activity, all of which was attenuated by siRNA directed against ROCK-1 expression. 17012370_These results indicate that 2ME-induced barrier disruption is governed by microtubule depolymerization and p38- and ROCK-dependent mechanisms. 17028776_The inhibition of Rho-Associated coiled-coil-containing protein kinase (ROCK), an activator of FAK, suppressed CAGE-promoted motility. 17044646_ROCK1 is associated with the apoptosis-inducing effect of endogenous nitric oxide suppression in cancer cells. 17138966_Adhesive and protrusive events involved in organized epithelial motility downstream of ROCK are separately coordinated through the phosphorylation of (respectively) myosin light chain and cofilin. 17167780_Dual regulatory role for Rho kinase in fibroblast regulation in which lipid-induced Rho kinase activation suppresses fibroblast morphogenesis while TGF-beta1-induced Rho kinase activation culminates in transformation into myofibroblasts. 17172270_Rho kinase mediates both early- and late-phase cold-induced vasoconstriction (VC) providing a putative mechanism through which both adrenergic and nonadrenergic cold-induced VC occurs in an in vivo human thermoregulatory model 17244674_MLC2 phosphorylation is regulated by both ROCK and MLC kinase and plays an important role in platelet biogenesis by controlling proplatelet formation and fragmentation. 17344417_findings report that the Notch1 gene is a p53 target in human keratinocytes with a role in tumor suppression of this cell type through negative regulation of the ROCK1/2 and MRCKalpha kinases 17346310_These data demonstrate a role for Anaplasma phagocytophilum-mediated ROCK1 phosphorylation in infection induced by PSGL-1 and mediated by Syk. 17651694_These results indicate that Rho-kinase can directly phosphorylate eNOS at Thr495 to suppress nitric oxide production in endothelium. 17673200_These results suggest the existence of a feedback loop between cytoskeletal tension, adhesion maturation, and ROCK signaling that likely contributes to numerous mechanochemical processes. 17728102_ROCK I cooperates with RhoB to activate NF-kappaB, and suppression of ROCK I activity by genetic or pharmacological inhibitors blocks NF-kappaB activation. 17853893_that ALIX and TSG101/ESCRT-I also bind a series of proteins involved in cytokinesis, including CEP55, CD2AP, ROCK1, and IQGAP1. 17855350_plexin-B1 promotes endothelial cell motility through RhoA and ROK by regulating the integrin-dependent signaling networks that result in the activation of PI3K and Akt 18079197_results suggest that ROCK and ERK1/2 play more important roles than RhoA in PGE(2)-mediated migration stimulation of first-trimester trophoblasts. 18187541_Members of the ROCK/Rho pathway act upstream from the kinase that phosphorylates P450c17 in a fashion that augments 17,20 lyase activity, possibly acting to catalyze a priming phosphorylation. 18204440_PDK1 competes directly with RhoE for binding to ROCK1. In the absence of PDK1, negative regulation by RhoE predominates, causing reduced acto-myosin contractility and motility. 18239683_ROCK, activated by G protein-coupled receptor ligands such as thrombin, directly phosphorylates FHOD1 at the C-terminal region, which renders this formin in the active form, leading to stress fibre formation. 18331468_Inhibition of ROCK activity in human-derived cells enhanced either bacterial adhesion or adhesion and entry in an InlF-independent manner, further suggesting a host species or cell type-specific role for InlF. 18353901_Permeability of cations in the alkali metal series was increased by PRLprolactin, and such increases were abolished by ROCK inhibitors. 18396681_The expression of ROCK-1 was closely related to the invasion capability and migratory activity of ovarian cancer cells. 18477564_RhoA/Rho-associated kinase signaling plays positive and negative roles in myogenic differentiation, mediated by MRTF-A/Smad-dependent transcription of the Id3 gene in a differentiation stage-specific manner 18495812_Rock1 and 2 are activated in response to wounding. ROCK activities enhance cell proliferation and epithelial differentiation, but negatively modulate cell migration and adhesion and therefore play a role in regulating corneal epithelial wound healing. 18499753_Even though ROCK signaling plays a role in human trophoblast cell invasion, extravillous cytotrophoblastic cell migration can still occur in the absence of ROCK activity. 18615591_Placenta growth factor directly regulates the motility of human lung cancer cells and that this regulation critically dependent on ROCK-1 18648664_DLC1 negatively regulates Rho/ROCK/MLC2 18668558_Rock potently stimulates the differentiation of resting fibroblasts into myofibroblasts and the production of extracellular matrix in scleroderma fibroblasts. 18684864_the late failure of cytokinesis responsible for the endomitotic process is related to a partial defect in the Rho/Rock pathway activation 18694941_The Diaphanous-related Formin FHOD1 associates with ROCK1 and promotes Src-dependent plasma membrane blebbing. 18695890_ROCKI and possibly ROCKII are key factors in regulation of motility/invasion of breast cancer cells. 18713748_the signal initiated by thrombin bifurcates at the level of RhoA to promote changes in the cytoskeletal architecture through ROCK, and the remodeling of focal adhesion components through protein kinase C-related kinase 18780293_Prolonged androgen receptor (AR) localization to the regulatory regions of AR targeted genes and the recruitment of p160 coactivators are a potential mechanism leading to androgen-independent activation of the AR in prostate cancer. 18852895_direct interaction between ROCK1 and LIMK2 in polarised but not blebbing cells; results point to a specific role for the ROCK1:LIMK2 pathway in mesenchymal-mode migration 18922892_a feedback loop between Rho/ROCK, Src, and phosphorylated Cav1 in tumor cell protrusions, identifying a novel function for Cav1 in tumor metastasis that may contribute to the poor prognosis of some Cav1-expressing tumors. 18941185_Phosphorylation of ezrin/moesin by ROCK is involved in the early steps of apoptotic signaling following Fas receptor triggering and regulates apoptosis induction in Jurkat cells. 18946488_mutation of the RhoE effector region attenuates RhoE-mediated disruption of the actin cytoskeleton, indicating that RhoE exerts its inhibitory effects on ROCK-I through protein(s) binding to its effector region. 18948190_Rho-Associated Kinases is involved in Proton-Translocating ATPases /P2RY13 protein-mediated Lipoproteins, HDL endocytosis pathway. 18952846_ROCK, a key downstream mediator of TGF-beta and other factors might become a unique therapeutic target in the treatment of proliferative vitreoretinal diseases. 19001122_Rap1 as another downstream target of the Abl-CrkII signaling module and show that Abl-CrkII collaborates with Rho-ROCK1 to stimulate cell retraction. 19036714_Upon UVB-induced stress in keratinocytes, ROCK1 was activated, bound to JIP-3, and activated the JNK pathway 19131646_ROCK1 and ROCK2 regulated myosin light chain phosphatase. ROCK isoforms had distinct and opposing effects on vascular smooth muscle cell (VSMC) morphology and ROCK2, but not ROCK1, had a predominant role in VSMC contractility. 19160018_Rho mediates various phenotypes of malignant transformation by Ras and Src through its effectors, ROCK and mDia [review] 19167712_Rho kinase activity is inhibited by HMG-CoA statins in patients with atherosclerosis 19181962_Inhibition of Rho-dependent kinases ROCK I/II activates VEGF-driven retinal neovascularization and sprouting angiogenesis. 19403695_Both receptor-stimulated and steady-state Kv1.2 trafficking modulated by RhoA/ROCK required the activation of dynamin as well as the ROCK effector Lim-kinase, indicating a key role for actin remodeling in RhoA-dependent Kv1.2 regulation. 19404162_Observational study of gene-disease association. (HuGE Navigator) 19404162_We did not find association between studied SNPs of ROCK1 and schizophrenia in the Japanese population 19406912_Rho kinase-1 mediates cardiac fibrosis by regulating fibroblast precursor cell differentiation. 19420982_Caov-3 cell growth was dependent on the Rho effector p160 Rho kinase 19549780_alpha2-integrin expression and Rho kinase activity are regulated by protein kinase signaling cascades upon activation of alpha7 nicotinic receptor 19573309_The increased expression of RhoA and Rho kinase in uterine smooth muscle may play an important role in the initiation of labor. 19590140_Provide evidence for Rho-kinase activation in patients with pulmonary arterial hypertension. 19667069_ROCK-mediated up-regulation of p21(Cip1) and the cytoskeleton in PKCdelta-dependent apoptosis in prostate cancer cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19997641_These data suggest that ROCK1 and ROCK2 play distinct roles in regulating keratinocyte adhesion and terminal differentiation. 20039424_These results demonstrate a new interaction that directly links ROCK to increased cartilage matrix production via activation of SOX9 in response to mechanical and growth factor stimulation 20053635_Results demonstrate a biphasic regulation of ROCK in response to adhesion, inhibition of ROCK initially and subsequently, re-activation is required for maturation of adhesion complexes and enhanced tumor cell migration. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20056645_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20064505_Data show that both lysophosphatidic acid and ROCK inhibition enhanced cell motility and that their combined effects significantly increased migration rate. 20068229_Stduies indicate that cGMP-dependent protein kinase I modulates the RhoA-ROCK pathway and insulin receptor signaling to elicit BAT differentiation and stimulate thermogenesis. 20070312_Helicobacter pylori disrupts gastric epithelial barrier structure via IL-1RI-dependent activation of ROCK, which in turn mediates tight junctional claudin-4 disruption. 20126978_Rho-kinase regulates negatively EGF-induced cell proliferation upstream of Akt/GSK-3beta in colon cancer cells 20130732_[review] Rho-kinase is constitutively active in regulating vasoconstrictor tone in the pulmonary and systemic vascular bed and thus may have an important role in the pathogenesis of pulmonary hypertension. 20140017_somatic ROCK1 mutations lead to elevated kinase activity and drive actin cytoskeleton rearrangements that promote increased motility and decreased adhesion, characteristics of cancer progression. 20223287_activation of the RhoA-ROCK-p38MAPK-MSK signaling pathway is essential for the regulation of the phenotype and of the CST5/cystatin D candidate tumor suppressor and other target genes by 1,25(OH)2D3 in colon cancer cells 20364104_Data show that the Rho/ROCK signaling pathway may prove to be a promising target in anti-tumor therapy. 20379197_Mutant PIK3CA interferes with apoptosis downstream of death receptor-signal complex, allowing caspase-8 activation without apoptosis. Cleavage of ROCK-1 by caspase-8 triggers amoeboid shape and invasiveness of HCT116 cells in response to TRAIL and CD95L. 20558775_Heparin inhibits pulmonary artery smooth muscle cell proliferation through GEF-H1/RhoA/ROCK/p27 signaling pathway. 20682776_Polarization and migration of hematopoietic stem and progenitor cells rely on the RhoA/ROCK I pathway and an active reorganization of the microtubule network. 20734064_Observational study of gene-disease association. (HuGE Navigator) 20801872_hHS-M(21) is a heart-specific effector of ROCK and plays a regulatory role in the MYPT1 phosphorylation at Thr-696 by ROCK 20801873_PHD2 affects cell migration and F-actin formation via RhoA/rho-associated kinase-dependent cofilin phosphorylation 21035804_the phospho myosin-binding subunit of ROCK has a role in cardiovascular disease 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21051874_Rho proteins and ROK play a pivotal role in this signaling cascade 21085597_Studies indicate that inhibition of Rho-associated kinase (ROCK) decreased PTHrP expression. 21119662_miR-584 is a new tumour suppressor miR in ccRCC and inhibits cell motility through downregulation of ROCK-1 21206971_The beneficial effects of simvastatin may be mediated by ROCK. 21265091_In embryonic fibroblasts, protein levels of RhoA, ROCK1 and CTGF were up-regulated after 1.5 h of hypoxia and increased further with longer exposure to hypoxia. 21305317_The genes identified KTN1, ROCK1, and ZAK may be responsible for loss of cellular homeostasis in giant cells tumors of bone 21387396_highly expressed in osteosarcoma; knockdown inhibits proliferation and induces apoptosis 21417762_ROCK and cAMP both promote lymphatic endothelial barrier function, however ROCK appears to also serve as a mediator of histamine and thrombin-induced barrier dysfunction 21445309_present the crystal structure of a large section of the central coiled-coil domain of human ROCK I (amino acids 535-700). The structure forms a parallel alpha-helical coiled-coil dimer that is structurally similar to tropomyosin 21575520_Expression levels of RhoC and ROCK-1 in prostate carcinoma are higher than those in corresponding paracancerous tissues, showing a significant positive correlation with distant metastasis. 21739156_These data suggest that osteogenic growth peptide not only acts on bone marrow mesenchymal stem cells to stimulate osteogenic differentiation, but also in a dose-dependent manner, and this effect is mediated via the activation of RhoA/ROCK pathway. 21747168_p120-catenin regulates growth of metastatic lobular carcinoma through Rock1-mediated anoikis resistance 21840487_show that cytokine signaling through the receptor subunit GP130-IL6ST and the kinase JAK1 generates actomyosin contractility through Rho-kinase dependent signaling 21847509_Rho-kinase signaling pathway plays a suppressive role in the intracellular vesicle trafficking of pEGFR via the endocytic pathway. 21907926_We show constitutive activation of Rho kinase in cells bearing oncogenic forms of KIT, FLT3, and BCR-ABL, which is dependent on PI3K and Rho GTPase. 21908715_Inhibition of the ROCK pathway restored the ability of gamma-actin-knockdown cells to migrate. This demonstrates gamma-actin as a potential upstream regulator of ROCK mediated cell migration. 21986457_ROCK1 may play a role in the development of albuminuria in diabetic kidney disease by downregulating the endocytosis receptors complex - megalin/cubilin. 21994419_Results show an inverse correlation between miR-148a and ROCK1 expression in gastric cancer tissues. 22010828_14-3-3 proteins mediate CaR-dependent Rho signalling and may modulate the plasma membrane expression of the CaR. 22031287_p120 dynamically regulates Rho-GTPase activity at the E-cadherin complex through transient interaction with several of its up- and downstream effectors, including ROCK1 22041269_Fasudil can reduce the secretion of downstream inflammatory factors and cytokines by inhibiting high glucose-activated HMCs Rho/ROCK signaling pathway in mesangial cells. 22085929_MYBPH inhibited assembly competence-conferring phosphorylation of the myosin regulatory light chain (RLC) as well as activating phosphorylation of LIM domain kinase, unexpectedly through its direct physical interaction with Rho kinase 1. 22155728_ROCK1 has a role in regulating myometrial contractility. 22206673_These results clearly suggested that the Rho/ROCK is a potent primary effector of mechanical stress in the pluripotent stem cells and it participates to pluripotency-related signaling cascades as an upper stream regulator. 22232555_Results demonstrate differential effects of Snail and Slug in pancreatic cancer and identify the interplay between Rho signaling and beta1-integrin that regulates the scattering and migration of Snail- and Slug-expressing pancreatic cancer cells. 22328514_LIMK1 and LIMK2, the downstream effectors of RhoA and ROCK, regulate centrosome integrity and astral microtubule organization, respectively. 22348083_ROCK1 and ROCK2 have distinct and separate roles in adhesion complex assembly and turnover in human epidermal keratinocytes. 22363817_Matrix-associated PAI-1 plays a role in the cell behavior needed for amoeboid migration by maintaining cell blebbing, localizing PDK1 and ROCK1 at the cell membrane and maintaining the RhoA/ROCK1/MLC-P pathway activation. 22462535_We conclude that ROCK proteins are necessary for acquisition of elongated and geometric cell shape, two key events for epithelial differentiation. 22493320_The structure of Shrm domain 2 (SD2), which mediates the interaction with Rock and is required for Shrm function, is reported. 22563075_analysis of the mechanism for negatively regulating Rho-kinase (ROCK) signaling through Coronin1B protein in neuregulin 1 (NRG-1)-induced tumor cell motility 22584506_Inhibition of ROCK by Y-27632 inhibited the activity of RhoA and promoted the apoptosis of gastric cancer cells. 22798677_our overall results reveal a model whereby CD1d-mediated Ag presentation is negatively regulated by ROCK1 and ROCK2 via its effects on the actin cytoskeleton. 22813839_Rho-kinase activation is substantially involved in the pathogenesis of the DES-induced coronary hyperconstricting responses in patients with CAD, suggesting the therapeutic importance of Rho-kinase pathway. 22846914_Crystal structures of the RKI-1447/ROCK1 complex revealed that RKI-1447 is a Type I kinase inhibitor. 22913419_ROCK1-positive fibroblasts were also identified by immunohistochemistry in the alveolar parenchyma in lung tissue sections from COPD patients 22921817_Increased Rho kinase activity predicted worse cardiovascular outcome in patients with acute coronary syndrome. 22942252_suppression of ROCK1 or ROCK2 expression alone was sufficient to impair anchorage-independent growth, supporting their nonoverlapping roles in oncogenesis 22975682_Inhibition of ROCK1 during the starvation period led to a more rapid response with the production of larger early autophagosomes that matured into enlarged late degradative autolysosomes. 22986902_the two isoforms of Rho-kinase distribute differentially to accomplish their specific functions 23066013_RhoA is a PKGIalpha target and direct binding of activated PKGIalpha to RhoA is central to cGMP-mediated inhibition of the VSMC Rho kinase contractile pathway 23094262_DEX interferes with the adhesion function of U937 cells through the inhibition of ROCK1 activity 23097497_Data indicate that phosphorylation of FilGAP by ROCK appears to promote amoeboid morphology. 23168335_Rho-associated protein kinase and Wnt signaling regulates Mypt1 and Nkx2.5 colocalization outside the cell nucleus. 23188716_NDRG1 inhibited an important regulatory pathway mediated by ROCK1/pMLC2 pathway that modulates stress fiber assembly. 23233531_miR-148b controls malignancy by coordinating a novel pathway involving over 130 genes and, in particular, it directly targets players of the integrin signaling, such as ITGA5, ROCK1, PIK3CA/p110alpha, and NRAS, as well as CSF1 23298303_Positive rates of Rac1, Pak1 and Rock1 expression in normal tissue, dysplasia and gastric carcinoma show an increasing trend and are correlated with tumor lymph node metastasis and TNM stage. 23328676_Our data showed that the ROCK1 and ROCK2 genes might be a risk factor for colorectal cancer development 23350987_Rapid secretion of TSP-1 from MDA-MB-231 cells depends on ROCK1 inhibition. 23355470_dual Rock and Cdk phosphorylation of Tppp1 inhibits its regulation of the cell cycle to increase cell proliferation 23479079_Sulfide induces both ROCK-1 and ROCK-2 cleavage and causes ROCKdependent cell blebbing during apoptosis. 23615712_Results suggest miR-126 function as a potential tumor suppressor in colon cancer progression and miR-126/RhoA/ROCK may be a novel candidate for developing rational therapeutic strategies. 23649168_CCR4 mediates CCL17 induced cell migration via increased total RhoA and active RhoA protein levels in colon cancer cells. 23665024_These findings indicate that Rho-kinase regulates ER stress-mediated VCAM-1 induction by ATF4- and p38MAPK-dependent signaling pathways. 23670799_Data indicate that knockdown of ROCK1 reversed epithelial-to-mesenchymal transition (EMT) resembling that of miR-148a overexpression. 23720748_Data indicate that Epac1-Rap1A-RhoA-ROCK signaling affects Cl- secretion via effects on the apical expression of KCNN4c channels. 23726972_Exposure of smooth muscle cells to apo(a) in migration assays induced a potent, concentration-dependent chemorepulsion that was RhoA and integrin alphaVbeta3-dependent, but transforming growth factor beta-independent. 23732111_Ostf1b could constitutively activate the Rho kinase 1 (ROCK1) and myosin light chain 2 (MLC2) signalling pathway that promotes cell migration, epithelial mesenchymal transition (EMT) and cytoskeletal dynamics through stress fibre formation. 23775364_Androgen receptor might play a negative role during prostrate cancer progression via influencing entosis by modulating Rho/ROCK. pathway. 23782575_Low frequency intermediate penetrance (LFIP) variants in the ROCK1 gene predispose to the risk of TOF 23789633_Immunohistochemical and western blot analyses showed no change in ROCK1 and ROCK2 protein expressions in Barrett's esophagus 23828571_The RhoA/ROCK1 signalling pathway has important roles in the determination of cell fate. 23872151_miR-340 acts as a tumor suppressor and its downregulation in tumor tissues may contribute to the progression and metastasis of OS through a mechanism involving ROCK1 23877263_ROCK1 acts as a prominent upstream regulator of Beclin1-mediated autophagy and maintains a homeostatic balance between apoptosis and autophagy. 23936026_These results revealed a novel mechanism that miR-124 inhibits glioma cells migration and invasion via ROCK1 downregulation. 23951055_Results suggest that Aurora B, but not Rho/MLCK (myosin-light-chain kinase) signaling, is essential for the localization of 2P-MRLC (myosin regulatory light chains) to the midzone in dividing HeLa cells. 23975506_ROCK1 was negatively regulated by miR-335 at the posttranscriptional level, via a specific target site within the 3'UTR by luciferase reporter assay. 24019071_Intracellular forms of NOTCH1 trigger the immediate activation of ROCK1 independent of its transcriptional activity, promoting differentiation and resulting in decreased clonogenicity of normal human keratinocytes. 24149105_it is suggested ROCK1 phosphorylates and activates SOX9 in Sertoli cells. Testes formation might be initiated by an alternative signaling pathway attributed to ROCK1, not SRY, activation in XX testes. 24168723_Active ROCKI is phosphorylated at Ser1333 site. Antibodies that recognize phospho-Ser1333 of ROCKI and phospho-S1366 residues of ROCKII offer a means to discriminate their individual active status in cells and tissues. 24170433_ROCK1 deletion altered the substrate-dependent cell migration in glioblastoma cells. 24180482_miR-124-3p can repress the migration and invasion of bladder cancer cells via regulating ROCK1 24240172_IKKgamma facilitates RhoA activation via a guanine nucletotide exchange factor, which in turn activates ROCK to phosphorylate IKKbeta, leading to NF-kappaB activation that induced the chemokine expression and cell migration upon TGF-beta1. 24296361_This study demonistrated by Gene expression profile that ROCK1 upregulaion in fibroblasts of Huntington's disease patients. 24297045_A systematic review of RhoA/ROCK blockade approaches analyzes the impact of bias that may lead to inflated effect sizes and functional locomotor recovery after experimental thoracic spinal cord injury. 24317511_ROCK has a crucial role in regulating prostate tumor growth through interaction with c-Myc 24323043_RACK1, by acting as a component of the IKK signalosome, plays a key role in inhibiting IKK activity by interfering with TRAF2-mediated recruitment of the IKK complex. 24324133_coordinated RhoA-ROCK1 expression and signaling are mediated by hypoxia-inducible factors in breast cancer cells 24331979_results demonstrated that p38 and ERK1/2 played crucial positive roles in adipogenesis, and FAK, RhoA/ROCK and cytoskeleton played negative roles 24398981_ROCK1 upregulation occurred more frequently in osteosarcoma tissues with positive metastasis and poor response to pre-operative chemotherapy. 24457551_RhoC/ROCK1 signaling pathways are likely involved in the progression of cervical squamous cell carcinoma 24465504_MicroRNA 135a suppresses lymph node metastasis through down-regulation of ROCK1 in early gastric cancer. 24528629_ROCK-1 protein plays a key role in regulating metastasis and invasion of retinoblastoma cells. 24566943_This is the first study to show that ROCK1 gene polymorphisms may have a significant impact on susceptibility to BD. 24573110_ROCK1, as a novel target of miR-145, acts as a positive regulator of glioma cell invasion. Therefore, ROCK1 may constitute a promising target for glioma treatment. 24587343_An mDia2/ROCK signaling axis regulates invasive egress from epithelial ovarian cancer spheroids. 24617500_This is the first study to examine the involvement of ROCK1 and ROCK2 gene variations in the risk of primary open-angle glaucoma development. 24696479_ROCK1- and LIMK1-regulated phosphorylation of cofilin played a role in retrovirus release from host cells and in cell-cell transmission events. 24717605_It regulates relaxation of vascular smooth muscle. 24801908_miR-145 may act as a tumor suppressor and contributes to the progression of OS through targeting ROCK1. 24894676_Data indicate that Rho-associated protein kinase 1 (ROCK1) was inversely correlated with miR-186 expression in non-small cell lung cancer (NSCLC). 24902944_Data indicate that dexamethasone inhibition of Rho-associated coiled-coil-containing protein kinase 1 (ROCK1) activity involved Ras homolog family member A (RhoA)-independent signaling pathways. 25011614_Data suggests that the rho-associated protein kinase (ROCK)-focal adhesion kinase (p-FAK cascade plays an important role in the effect of caffeine in glioma cells. 25169484_silencing of ROCK1 suppressed cell proliferation, migration, and invasion, while supplementation of ROCK1 remarkably restored the cell growth and invasion inhibited by miR-124. 25218053_The differential expression of ROCK-1 and MMP-9 proteins suggests their potential use as prognostic markers in breast cancer. 25237195_aberrant modulation of Rock1 expression and activity induced | ENSMUSG00000024290 | Rock1 | 1.158218e+03 | 1.3168969 | 0.397142443 | 0.3300203 | 1.435292e+00 | 0.2309027770 | 0.78818582 | No | Yes | 1.311145e+03 | 257.532439 | 8.835442e+02 | 178.230120 | |
| ENSG00000068097 | 63897 | HEATR6 | protein_coding | Q6AI08 | FUNCTION: Amplification-dependent oncogene. | Phosphoprotein;Reference proteome;Repeat | hsa:63897; | RNA binding [GO:0003723] | ENSMUSG00000000976 | Heatr6 | 1.613369e+03 | 0.7706220 | -0.375904745 | 0.2743937 | 1.863102e+00 | 0.1722672143 | 0.77611497 | No | Yes | 1.452078e+03 | 177.686922 | 1.858657e+03 | 232.977110 | |||
| ENSG00000068400 | 56850 | GRIPAP1 | protein_coding | Q4V328 | FUNCTION: Regulates the endosomal recycling back to the neuronal plasma membrane, possibly by connecting early and late recycling endosomal domains and promoting segregation of recycling endosomes from early endosomal membranes. Involved in the localization of recycling endosomes to dendritic spines, thereby playing a role in the maintenance of dendritic spine morphology. Required for the activity-induced AMPA receptor recycling to dendrite membranes and for long-term potentiation and synaptic plasticity (By similarity). {ECO:0000250|UniProtKB:Q9JHZ4}.; FUNCTION: [GRASP-1 C-terminal chain]: Functions as a scaffold protein to facilitate MAP3K1/MEKK1-mediated activation of the JNK1 kinase by phosphorylation, possibly by bringing MAP3K1/MEKK1 and JNK1 in close proximity. {ECO:0000269|PubMed:17761173}. | Acetylation;Alternative splicing;Cell junction;Cell projection;Coiled coil;Direct protein sequencing;Endosome;Membrane;Phosphoprotein;Protein transport;Reference proteome;Synapse;Transport | This gene encodes a guanine nucleotide exchange factor for the Ras family of small G proteins (RasGEF). The encoded protein interacts in a complex with glutamate receptor interacting protein 1 (GRIP1) and plays a role in the regulation of AMPA receptor function. [provided by RefSeq, Aug 2013]. | hsa:56850; | axon [GO:0030424]; blood microparticle [GO:0072562]; cytosol [GO:0005829]; dendrite [GO:0030425]; extrinsic component of postsynaptic early endosome membrane [GO:0098998]; glutamatergic synapse [GO:0098978]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; postsynaptic recycling endosome [GO:0098837]; recycling endosome membrane [GO:0055038]; identical protein binding [GO:0042802]; neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0098887]; regulation of modification of synaptic structure [GO:1905244]; regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0099152]; regulation of recycling endosome localization within postsynapse [GO:0099158] | 15897011_GRASP-1 was identified as an soluble autoantigen that has numerous coiled-coil domains throughout the protein. 17761173_Results suggest that GRASP-1 serves as a scaffold protein to facilitate MEKK-1 activation of JNK signaling in neurons. 28106924_We identified a new TFE3 fusion partner, GRIPAP1, in translocation renal cell carcinoma 28285821_results demonstrate a requirement for normal recycling endosome function in AMPAR-dependent synaptic function and neuronal connectivity in vivo, and suggest a potential role for GRASP1 in the pathophysiology of human cognitive disorders. | ENSMUSG00000031153 | Gripap1 | 2.397408e+03 | 1.0136421 | 0.019548355 | 0.2500190 | 6.136384e-03 | 0.9375615170 | 0.98859743 | No | Yes | 1.903783e+03 | 166.765061 | 1.989562e+03 | 178.788392 | |
| ENSG00000070061 | 8518 | ELP1 | protein_coding | O95163 | FUNCTION: Component of the elongator complex which is required for multiple tRNA modifications, including mcm5U (5-methoxycarbonylmethyl uridine), mcm5s2U (5-methoxycarbonylmethyl-2-thiouridine), and ncm5U (5-carbamoylmethyl uridine) (PubMed:29332244). The elongator complex catalyzes the formation of carboxymethyluridine in the wobble base at position 34 in tRNAs (PubMed:29332244). Regulates the migration and branching of projection neurons in the developing cerebral cortex, through a process depending on alpha-tubulin acetylation (By similarity). ELP1 binds to tRNA, mediating interaction of the elongator complex with tRNA (By similarity). May act as a scaffold protein that assembles active IKK-MAP3K14 complexes (IKKA, IKKB and MAP3K14/NIK) (PubMed:9751059). {ECO:0000250|UniProtKB:Q06706, ECO:0000250|UniProtKB:Q7TT37, ECO:0000269|PubMed:9751059, ECO:0000303|PubMed:29332244}. | 3D-structure;Cytoplasm;Direct protein sequencing;Disease variant;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Reference proteome;tRNA processing | PATHWAY: tRNA modification; 5-methoxycarbonylmethyl-2-thiouridine-tRNA biosynthesis. {ECO:0000303|PubMed:29332244}. | The protein encoded by this gene is a scaffold protein and a regulator for three different kinases involved in proinflammatory signaling. The encoded protein can bind NF-kappa-B-inducing kinase and I-kappa-B kinases through separate domains and assemble them into an active kinase complex. Mutations in this gene have been associated with familial dysautonomia. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jan 2016]. | hsa:8518; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; elongator holoenzyme complex [GO:0033588]; nucleus [GO:0005634]; protein self-association [GO:0043621]; tRNA binding [GO:0000049]; tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation [GO:0002926] | 12058026_novel role for the I kappa B kinase complex-associated protein (IKAP) in the regulation of activation of the mammalian stress response via the c-Jun N-terminal kinase (JNK)-signaling pathway 12102458_Genetics of familial dysautonomia; tissue-specific expression of a splicing mutation (REVIEW) 12116234_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 12577200_Tissue-specific reduction in splicing efficiency of this protein is due to the major mutation associated with familial dysautonomia. 12774215_Observational study of gene-disease association. (HuGE Navigator) 12774215_The study results suggest that the polymorphisms in the coding region of the IKAP gene are unlikely to contribute to atopic disease risk in the Czech population. 16713582_whereas IKBKAP (Elongator) is recruited to both target and nontarget genes, only target genes display histone H3 hypoacetylation and progressively lower RNAPII density through the coding region in familial dysautonomia cells 16964593_Neurodevelopmental disease familial dysautonomia (FD)caused by a single-base change in the 5' splice site (5'ss) of intron 20 in the IKBKAP gene (c.2204+6T>C). 17137217_Observational study of gene-disease association. (HuGE Navigator) 17206408_investigated the nature of the FD splicing defect and the mechanism by which kinetin improves exon inclusion 17591626_IKAP/hELP1 may play a role in oligodendrocyte differentiation and/or myelin formation. 17644305_description of a humanized IKBKAP transgenic mouse that models a tissue-specific human splicing defect 17703412_Observational study of gene-disease association. (HuGE Navigator) 18091349_IKBKAP may have a role in familial dysautonomia 18264947_Observational study of genotype prevalence. (HuGE Navigator) 18303054_Evidence for the role of the cytosolic interactions of IKAP in cell adhesion and migration, and support the notion that cell-motility deficiencies could contribute to familial dysautonomia. 18434448_Observational study of gene-disease association. (HuGE Navigator) 19015235_IKAP is crucial for both vascular and neural development during embryogenesis and that protein function is conserved between mouse and human. 19247692_Observational study of gene-disease association. (HuGE Navigator) 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19262425_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19651702_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20098615_Observational study of gene-disease association. (HuGE Navigator) 20361209_IKBKAP is a candidate gene for Hirschsprung's disease and was mapped to chromosome 9q31 locus. 20361209_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20503287_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20671422_IKAP regulates contactin levels for appropriate cell-cell adhesion that could modulate neuronal growth of neurons during development 21098405_IKAP is critical for the development of afferent baroreflex pathways and has therapeutic implications in the management of these patients. 21209961_Phosphatidylserine increases IKBKAP levels in familial dysautonomia cells 21273291_IKK complex-associated protein deficiency upregulates the microtubule destabilizing protein SCG10 and, in parallel, disorganizes the cytoskeleton 21559466_IKAP/hELP1 deficiency has an effect on gene expression in differentiating neuroblastoma cells, and possibly on familial dysautonomia 22384137_IKAP plays pleiotropic roles in both the peripheral and central nervous systems 22495984_Combined treatment with epigallocatechin gallate and genistein synergistically upregulates wild-type IKBKAP-encoded RNA and protein levels in familial dysautonomia-derived cells. 23515154_Phosphatidylserine increases IKBKAP levels in a humanized knock-in IKBKAP mouse model for Familial dysautonomia. 23711097_Digoxin-mediated repression of SRSF3 expression plays a role in the digoxin-mediated inclusion of exon 20 in the IKBKAP transcript generated from the familial dysautonomia mutant allele. 24268683_IKBKAP mRNA levels decreased during a familial dysautonomia crisis and returned to baseline after recovery. The cause-and-effect relationship is unclear. 26261306_The formation of the Elp1 dimer contributes to its stability in vitro and in vivo and is required for the assembly of human Elongator complexes. 26437462_IKAP might be a vesicular like protein that might be involved in neuronal transport in hESC derived PNS neurons 27483351_overexpression of miR-203a-3p leads to a decrease of NOVA1, counter-balanced by an increase of IKAP, supporting a potential interaction between NOVA1 and IKAP. 29289840_The founder mutation in the IKBKAP gene affects the development of vestibular afferent pathways, leading to attenuated cVEMPs. 29701768_This splice-switching class of molecules is the first to specifically correct the ELP1 exon 20 splicing defect. Our data provide proof of principle of ExSpeU1s-adeno-associated virus particles as a novel therapeutic strategy for FD. 29762696_Data indicate that heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is a negative regulator of IKK-complex-associated protein (IKBKAP) exon 20 splicing. 30085848_A secondary assay that measures ELP1 splicing in FD patient-derived fibroblasts. 30905397_postnatal correction of the underlying ELP1 splicing defect can rescue devastating disease phenotypes in a humanized ELP1 mouse model 30989732_The authors identified two potentially functional single nucleotide polymorphisms (IKBKAP rs4978754 C > T and TNFRSF1B rs677844 T > C) to be associated with survival of patients with non-small cell lung cancer. 32296180_Tumours from patients with ELP1-associated medulloblastoma subgroup Sonic Hedgehog were characterized by a destabilized Elongator complex, loss of Elongator-dependent tRNA modifications, codon-dependent translational reprogramming | ENSMUSG00000028431 | Elp1 | 2.707605e+03 | 0.7622029 | -0.391753040 | 0.3059077 | 1.638251e+00 | 0.2005656701 | 0.78383913 | No | Yes | 2.343531e+03 | 385.900616 | 2.863153e+03 | 483.251426 |
| ENSG00000070081 | 4925 | NUCB2 | protein_coding | P80303 | FUNCTION: Calcium-binding protein which may have a role in calcium homeostasis (By similarity). Acts as a non-receptor guanine nucleotide exchange factor which binds to and activates guanine nucleotide-binding protein (G-protein) alpha subunit GNAI3 (By similarity). {ECO:0000250|UniProtKB:P81117, ECO:0000250|UniProtKB:Q9JI85}.; FUNCTION: [Nesfatin-1]: Anorexigenic peptide, seems to play an important role in hypothalamic pathways regulating food intake and energy homeostasis, acting in a leptin-independent manner. May also exert hypertensive roles and modulate blood pressure through directly acting on peripheral arterial resistance. {ECO:0000250|UniProtKB:Q9JI85}. | Alternative splicing;Calcium;Cleavage on pair of basic residues;Cytoplasm;DNA-binding;Direct protein sequencing;Endoplasmic reticulum;Golgi apparatus;Guanine-nucleotide releasing factor;Membrane;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal | This gene encodes a protein with a suggested role in calcium level maintenance, eating regulation in the hypothalamus, and release of tumor necrosis factor from vascular endothelial cells. This protein binds calcium and has EF-folding domains. [provided by RefSeq, Oct 2011]. | hsa:4925; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; nuclear envelope [GO:0005635]; plasma membrane [GO:0005886]; calcium ion binding [GO:0005509]; DNA binding [GO:0003677]; G-protein alpha-subunit binding [GO:0001965]; guanyl-nucleotide exchange factor activity [GO:0005085]; negative regulation of appetite [GO:0032099]; small GTPase mediated signal transduction [GO:0007264] | 16407280_Calcium-dependent ARTS-1-nucleobindin 2 complexes associate with TNFR1 prior to the commitment of TNFR1 to pathways that result in the constitutive release of TNFR1 exosome-like vesicles or the inducible proteolytic cleavage of TNFR1 ectodomains. 16407280_NUCB2-ARTS-1 complexes play an important role in mediating tumor necrosis factor receptor 1 release to the extracellular compartment. 18154733_results suggest that the functions of nucleobindins could be modulated by caspase-mediated cleavage in apoptosis 19262995_low ghrelin and especially the dramatically elevated nesfatin-1 levels might contribute to the pathophyisology of epilepsy[Nesfatin-1] 19351608_NUCB2 may be implicated in gastric secretion by establishing an agonist-releasable Ca2+ store in ER or Golgi apparatus, signalling via heterotrimeric Galpha proteins and/or mediating the exocytosis of the secretory granules. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20030931_nucb2 is the direct target gene of AML1 and AML1-ETO, the transcription regulation of AML1, AML1-ETO on nucb2 is carried out via repressing its promoter activity. 20032201_The release of NUCB2 from isolated islets was significantly elevated following glucose challenge. 20427481_Data report that nesfatin-1 is a novel depot specific adipokine preferentially produced by sc tissue, with obesity- and food deprivation-regulated expression. 20625762_lack of nesfatin-1 response to the exercise protocols may be partially due to the fasting condition 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20813143_Apelins and nesfatin-1 concentrations were higher in mature milk than in colostrum (P<0.05). The concentration of apelins, ghrelins and nesfatin-1 in serum and milk in gestational diabetic lactating women was lower than in control samples 20937336_nesfatin-1 levels may be regulated by nutrition status and response to starvation. 21252251_Potential weight-reducing actions of nesfatin-1/NUCB-2 treatment. 21459029_polymorphisms in the NUCB2 gene could play an important role in the protection against the development of obesity in male subjects 21554911_Increased serum prolactin and nesfatin-1 concentrations, decreased ghrelin concentrations could be used as markers to identify patients that have suffered a recent epileptic seizure or other paroxysmal event (psychogenic). 21653697_structural basis for the properties of Calnuc and NUCB2 binding to Galpha subunits and its regulation by calcium ions. 21803054_This study demonistrated thatsex difference in NUCB2 dynamic expressions in the midbrain of drug-free depressed suicide victims. 21988594_High NUCB2 is associated with recurrence and metastasis in breast carcinoma. 22020667_Plasma nesfatin-1 concentrations were found to be elevated in subjects with both impaired glucose tolerance and newly diagnosed T2 diabetes mellitus. 22036920_nesfatin-1 levels did not differ between hemodialysis patients and healthy subjects and negatively correlated with protein intake 22108805_Human islet NUCB2 mRNA was reduced in T2D subjects but upregulated after culture in glucolipotoxic conditions. Furthermore, a positive correlation between NUCB2 and glucagon and insulin gene expression, as well as insulin secretory capacity, was evident. 22203468_Data suggest that maternal serum levels of nesfatin-1 (derived from NUCB2) are lower in patients with gestational diabetes compared with control pregnant women; however, the cord blood nesfatin-1 levels are similar. 22225987_the details of nesfatin-1 physiology ought to be clarified, and it may be considered suitable in the future, as a potential drug in the pharmacotherapy of obesity--{REVIEW} 22270345_We have demonstrated that the Wa antigen is NEFA or nucleobindin-2, which binds specific tRNA species, and is distributed in specific human tissues. 22334726_NUCB2 may operate directly at the testicular level to link energy homeostasis, puberty onset, and gonadal function 22367584_Lower nesfatin-1 concentration may plays a very important role in the development of polycystic ovary syndrome 22950627_Nesfatin-1 concentrations were reduced in non-alcoholic fatty liver disease. 23141462_we have identified the first rare genetic variants in the NUCB2 gene in obese individuals (frequency 0.2%), suggesting that NUCB2 might indeed be involved in the regulation of energy homeostasis and food intake. 23155701_Nesfatin-1 is involved in the physiological regulation of intrauterine and postnatal growth and development in small for gestational ages infants. 23178145_nesfatin-1 levels are decreased in large for gestational age compared to appropriate for gestational age fetuses. 23266808_The NUCB2 variant c.1012C>G (Q338E) is associated with childhood adiposity. The GG genotype may be protective against excessive weight gain. 23269222_Loss of fat mass may decrease serum nesfatin-1 level in lung cancer patients with weight loss. 23306670_In advanced cystic fibrosis and low fat mass, nesfatin-1 plasma levels are significantly increased and inversely correlated with the fat mass. 23346951_Data suggest that serum nesfatin-1 are significantly lower in obese youth than in healthy control youth; no association was found with insulin resistance in obese subjects. 23515787_Ghrelin and NUCB2/nesfatin-1 are expressed in the same gastric cell and differentially correlated with body mass index in obese subjects. 23524986_A statistically significant correlation was found between the serum nesfatin-1 level and supraventricular tachycardia. 23613885_pro-osteogenic activity of NUCB2(1-83) 23739643_Nesfatin-1 levels were lower in children with untreated autoimmune thyroid diseases, however, the mechanism is also unknown. 23764403_This study provides a novel regulatory signaling pathway of nesfatin-1-regulated ovarian epithelial carcinoma growth and may contribute to ovarian cancer prevention and therapy, especially in obese patients. 23765387_Increased nucleobindin-2 (NUCB2) transcriptional activity links the regulation of insulin sensitivity in Type 2 diabetes mellitus. 23768444_Circulating nesfatin-1 may modulate glucose homeostasis during diabetes remission in gastric bypass and sleeve gastrectomy, and participate in regulating body weight in sleeve gastrectomy 23796625_plasma NUCB2/nesfatin-1 levels were altered under conditions of perceived anxiety, stress and depression in obese women. 23806888_In acute myocardial infarction patients, plasma nesfatin-1 levels were negatively correlated with high-sensitivity C-reactive protein, neutrophil% or Gensini scores. 23955480_Data suggest that NUCB2 expression is widespread in including adipose tissue, pancreas, and digestive tract; NUCB2 appears to participate in regulation of energy homeostasis/appetite; genetic polymorphisms in NUCB2 may predispose to obesity. [REVIEW] 23958433_Kaplan-Meier survival analysis showed that patients with high NUCB2 expression have shorter biochemical recurrence (BCR)-free survival time compared to patients with low NUCB2 expression. 23980879_Review highlights evidence of changes in circulating levels of NUCB2/nesfatin-1 in disease states, occurrence of NUCB2 polymorphisms and independence of leptin signalling known to be blunted under conditions of chronically increased body weight. 24048879_Role of NUCB2/Nesfatin-1 in the hypothalamic control of energy homeostasis. 24092574_we demonstrated that high NUCB2 mRNA expression correlated with poor overall survival in patients with prostate cancer 24259949_Nesfatin-1 is present in articular tissues and may contribute to the physiopathologic changes in OA. Nesfatin-1, accompanied with hsCRP and IL-18, could have roles in the progression of knee osteoarthritis 24333832_Nesfatin-1 might have an important role in regulation of food intake and pathogenesis of insulin resistance in obese children and young adolescents. 24422979_NUCB2 protein expression showed a strong association with the potencies of BCR and progression of PCa, and that may be applied as a novel biomarker. 24464950_study demonstrated cytoplasmic localization of NUCB2/nesfatin-1 peptide in both human and murine chondrocytes. 24825087_there was no significant increase in postprandial level of nesfatin-1. This suggests that oral glucose load in obese children may not be sufficient for nesfatin-1 response and that nesfatin-1 may not have an effect as short-term regulator of food intake. 24857385_Data indicate that patients with severe preeclampsia showed significantly decreased levels of serum nesfatin-1 compared with those with mild preeclampsia. 24891763_NUCB2/nesfatin-1 and visfatin are novel factors associated with systemic inflammation in COPD; visfatin may mediate impaired pulmonary diffusing capacity 24920281_Data suggest that plasma nesfatin-1/NUCB2 levels are up-regulated in pregnant women recently diagnosed with polycystic ovary syndrome (PCOS) as compared to healthy women; many of those with PCOS were also overweight and exhibited insulin resistance. 25068306_Serum nesfatin-1 levels were measured in patients with subclinical or overt hyperthyroidism to see whether nesfatin-1 is involved in the regulation of appetite and body weight in hyperthyroidism. No significant difference from normal controls was found. 25322808_Our results show that NUCB2 plays an important role in tumorigenesis and progression and is a potential molecular biomarker for the diagnosis and targeted therapy of ccRCC. 25432328_Nesfatin-1 levels were significantly lower in metabolic syndrome (MS) group compared to non-MS group 25581765_Plasma IL-6 and TNF-alpha did not significantly change after training, but nesfatin increased significantly only with high-intensity interval training compared with the control group. 25637303_intensity of exercise may have an important role in changes of nesfatin-1, leptin, FFA, and epinephrine concentrations even though this was not the case for IL-6 and insulin resistance 25833360_Serum nesfatin-1 levels are significantly higher in girls with premature thelarche compared to prepubertal controls. 25841171_This study shows an association of serum nesfatin-1 concentrations and the development and severity of peripheral arterial disease in type 2 diabetes mellitus patients. 25869615_nesfatin-1 inhibits the growth of adrenocortical H295R cells and promotes apoptosis, potentially via the involvement of Bax, BCL-XL and BCL-2 genes as well as ERK1/2, p38 and JNK1/2 signalling cascades 25872767_This investigation indicates a marked association of serum and synovial fluid nesfatin-1 concentrations with osteoarthritis disease severity. 26043295_serum level elevated in fetal growth retardation 26143537_These results corroborate the suggestion of NUCB2/nesfatin-1 being relevantly involved in the regulation of mood and stress in a sex-specific way 26162003_Circulating NUCB2/nesfatin-1 levels correlated positively with perceived anxiety, whereas no association with body mass index or eating disorder symptoms was observed. 26369257_NUCB2 protein levels in the obese PCOS group were significantly lower than those in the non-obese PCOS and control groups. There was no statistically significant difference in the distribution of NUCB2 genotypes among the groups. 26624852_1012C>G polymorphism of NUCB2 is correlated with a reduced risk of developing MetS in a Chinese Han population. 26662184_we summarize nesfatin-1 main functions, focusing on its cardiovascular implications--{REVIEW} 26744860_nesfatin-1 might have an important role in regulation of food intake and pathogenesis of loss of appetite in children. 26831553_Data indicate a positive correlation of nesfatin-1 and a negative correlation of orexin-A with body mass index. 26842712_Data suggest that NUCB2 (almost negligible in normal endometrium) plays pivotal roles in cell proliferation and migration/invasiveness of endometrial carcinoma; NUCB2 may serve as a prognostic biomarker for endometrial carcinoma. Expression of NUCB2 appears almost negligible in normal endometrium. 27049252_Nesfatin-1 hormone levels increased significantly in morbidly obese patients following laparoscopic sleeve gastrectomy. 27150059_Data indicate that nesfatin-1/NUCB-2 enhanced migration, invasion and epithelial-mesenchymal transition (EMT) in colon cancer cells through LKB1/AMPK/TORC1/ZEB1 pathways in vitro and in vivo. 27155585_Increased plasma nesfatin-1 concentrations are associated closely with inflammation, trauma severity and clinical outcomes, indicating that nesfatin-1 might be involved in inflammation and become a good prognostic biomarker following traumatic brain injury 27235705_Based on the obtained results, it was proposed that ghrelin may be considered as playing a role in the etiopathogenesis of hyperemesis gravidarum that may result in disruption of the relationship between nesfatin-1 and ghrelin. 27539099_This study supports the role of insulin resistance in decreased nesfatin-1 levels in patients with Type 2 diabetes mellitus and metabolic syndrome 27681383_Nesfatin-1 levels decrease following biliopancreatic diversion with duodenal switch-induced weight loss and are significantly associated with parameters of metabolic health. 27806328_Study demonstrated that high expression level of NUCB2 was significantly associated with poor clinical outcome of patient with non-metastatic clear cell renal cell carcinoma. 28137218_The multifunctional biological actions of nesfatin-1 propelled this peptide as a therapeutic target, and as a potential biomarker of diseases. However, a better and comprehensive understanding of tissue specific effects of nesfatin-1 is critical prior to exploring its possible use in the detection and treatment of diseases. 28273586_women and men showed an inverse association between NUCB2/nesfatin-1 and anxiety with a positive correlation in women and a negative correlation in men. However, no significant change of NUCB2/nesfatin-1 following improvement of anxiety has been observed. 28361552_Data suggest that levels of nesfatin-1 in maternal serum and cord blood are up-regulated in women with gestational diabetes (GD) as compared to control pregnant women; nesfatin-1 also appears to be up-regulated in subcutaneous abdominal adipose tissue in GD. This study was conducted in China in women of the Han ethnicity. 28714371_nucleobindin 2 might play a crucial role in gastric cancer development and could serve as an independent predictor of prognosis of gastric cancer patients. 28720397_expressions of p-Akt/Akt and p-GSK-3beta/GSK-3beta in the myocardium of MI group rats were significantly increased by nesfatin-1 administration, suggesting that nesfatin-1, which appears to possess anti-apoptotic and anti-inflammatory properties, may confer protection against ISO-induced MI via an Akt/GSK-3beta-dependent mechanism. 28814086_Our study was the first to investigate the nesfatin 1 levels in patients with the first episode psychosis. Based on our study results, nesfatin 1 might be related to some central nervous system pathologies, including the severity of a psychiatric disorder. 28855042_In all trimesters of human pregnancy, NUCB2/nesfatin-1 was highly expressed in syncytiotrophoblast. In addition, there was a significant increase in NUCB2 expression in human primary trophoblast cells induced to syncytialise. 29278806_Patients with schizophrenia had significantly higher serum nesfatin-1 levels compared to healthy controls. 29445751_Data suggest that circulating nesfatin-1 levels are up-regulated in patients newly diagnosed with type 2 diabetes; non-newly diagnosed patients treated with antidiabetic medications exhibit lower circulating nesfatin-1 levels. [META-ANALYSIS, REVIEW] 29452603_Nesfatin-1 is involved in cardiovascular regulation, stress-related responses. 29627363_Study data identified nesfatin-1 as a key modulator in hypertension and vascular remodeling by facilitating vascular smooth muscle cells phenotypic switching and proliferation. 29747779_serum nesfatin-1 concentrations were inversely correlated with atrial fibrillation development. 29940459_Serum nesfatin-1 levels were significantly lower in major depressive disorder patients with suicidal ideation compared to healthy volunteers. 30055641_Authors demonstrated that KLF4 was induced by ER stress in melanoma cells, and increased KLF4 inhibited cell apoptosis and promoted cell metastasis. Further mechanistic studies revealed that KLF4 directly bound to the promoter of NUCB2, facilitating its transcription. 30084805_This study finds that soccer matches performed different workout times have strong stimulatory effects on irisin levels in all subjects but nesfatin-1 response varied among the subjects and it did not change significantly in afternoon match. 30292960_The present review will describe the nesfatin-1 's relations to anxiety, depressiveness and stress in animal models and humans and also discuss existing gaps in knowledge in order to stimulate further research. [review] 30327690_NUCB2 played an important role in bladder cancer and could be considered a potent prognostic factor 30599062_plasma nesfatin-1 levels were found to be high in patients with coronary artery disease (CAD) and were associated with CAD independent of atherosclerotic risk factors 30599786_Nesfatin-1 levels are inversely associated with severity of carotid artery stenosis and plaque morphology. 30621139_nesfatin-1 is potential novel biomarker for the prediction and early diagnosis of Gestational Diabetes Mellitus. 30625474_findings of the present study are suggestive of a potential role for NUCB2/nesfatin-1 as an integral regulator of food intake and energy homeostasis in the human hypothalamus. 30959145_Serum nesfatin-1 concentrations are closely related to the severity of acute myocardial infarction. 31219502_Nesfatin is a potent anorectic neuropeptide which plays an important role in food intake and energy expenditure. 31383892_Does nesfatin-1 influence the hypothalamic-pituitary-gonadal axis in adult males with obstructive sleep apnoea? 31686413_Studied the expression levels of nesfatin 1 and other tumor markers in two types of endometrial cancer. 31697592_Nucleobindin-2 (NUCB2) over-expression is correlated with the high degree of recurrence of patients with glioblastoma. NUCB2 promotes cell proliferation and invasion of glioblastoma in vitro and promotes the growth and metastasis of glioblastoma in mice. 32116029_Plasma levels of nesfatin-1 as a new biomarker in depression in Asians: evidence from meta-analysis. 32162872_The Performance of Nesfatin-1 in Distinguishing Irritable Bowel Syndrome Presenting Predominantly with Diarrhea from Celiac Disease. 32184136_Calcium ions modulate the structure of the intrinsically disordered Nucleobindin-2 protein. 32265847_Evidence of a Relationship Between Plasma Leptin, Not Nesfatin-1, and Craving in Male Alcohol-Dependent Patients After Abstinence. 32311618_Relationship of spontaneous subarachnoid haemorrhage and cerebral aneurysm to serum Visfatin and Nesfatin-1 levels. 32356443_Nesfatin 1: a promising biomarker predicting successful reperfusion after coronary artery bypass surgery. 32427366_Serum nesfatin-1 is associated with testosterone and the severity of erectile dysfunction. 32432771_Nesfatin-1 alleviates acute lung injury through reducing inflammation and oxidative stress via the regulation of HMGB1. 32902020_Nucleobindin 2/nesfatin-1 expression and colocalisation with neuropeptide Y and cocaine- and amphetamine-regulated transcript in the human brainstem. 32974855_Nesfatin-1 Promotes Proliferation, Migration and Invasion of HTR-8/SVneo Trophoblast Cells and Inhibits Oxidative Stress via Activation of PI3K/AKT/mTOR and AKT/GSK3beta Pathway. 33019423_Increased serum nesfatin-1 levels in patients with acromegaly. 33110207_circ_001504 promotes the development of renal cell carcinoma by sponging microRNA-149 to increase NUCB2. 33318307_Detection of NUCB2/nesfatin-1 in cerebrospinal fluid of multiple sclerosis patients. 34073612_The Multifaceted Nature of Nucleobindin-2 in Carcinogenesis. 34082080_Localization of nucleobindin2/nesfatin-1-like immunoreactivity in human lungs and neutrophils. 34389003_Plasma nesfatin-1 and DDP-4 levels in patients with coronary artery disease: Kozani study. 35480963_Serum Cortisol, Nesfatin-1, and IL-1beta: Potential Diagnostic Biomarkers in Elderly Patients with Treatment-Resistant Depression. | ENSMUSG00000030659 | Nucb2 | 7.037578e+02 | 1.0988001 | 0.135928992 | 0.3192426 | 1.796892e-01 | 0.6716404884 | 0.92978311 | No | Yes | 6.759820e+02 | 111.203265 | 5.587969e+02 | 94.339611 | |
| ENSG00000070501 | 5423 | POLB | protein_coding | P06746 | FUNCTION: Repair polymerase that plays a key role in base-excision repair. Has 5'-deoxyribose-5-phosphate lyase (dRP lyase) activity that removes the 5' sugar phosphate and also acts as a DNA polymerase that adds one nucleotide to the 3' end of the arising single-nucleotide gap. Conducts 'gap-filling' DNA synthesis in a stepwise distributive fashion rather than in a processive fashion as for other DNA polymerases. {ECO:0000269|PubMed:11805079, ECO:0000269|PubMed:21362556, ECO:0000269|PubMed:9207062, ECO:0000269|PubMed:9572863}. | 3D-structure;Acetylation;Cytoplasm;DNA damage;DNA repair;DNA replication;DNA synthesis;DNA-binding;DNA-directed DNA polymerase;Isopeptide bond;Lyase;Magnesium;Metal-binding;Methylation;Nucleotidyltransferase;Nucleus;Reference proteome;Sodium;Transferase;Ubl conjugation | hsa:5423; | cytoplasm [GO:0005737]; microtubule [GO:0005874]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; spindle microtubule [GO:0005876]; damaged DNA binding [GO:0003684]; DNA-(apurinic or apyrimidinic site) endonuclease activity [GO:0003906]; DNA-directed DNA polymerase activity [GO:0003887]; enzyme binding [GO:0019899]; lyase activity [GO:0016829]; metal ion binding [GO:0046872]; microtubule binding [GO:0008017]; aging [GO:0007568]; base-excision repair [GO:0006284]; base-excision repair, base-free sugar-phosphate removal [GO:0006286]; base-excision repair, gap-filling [GO:0006287]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; DNA-dependent DNA replication [GO:0006261]; double-strand break repair via nonhomologous end joining [GO:0006303]; homeostasis of number of cells [GO:0048872]; immunoglobulin heavy chain V-D-J recombination [GO:0071707]; in utero embryonic development [GO:0001701]; inflammatory response [GO:0006954]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; lymph node development [GO:0048535]; neuron apoptotic process [GO:0051402]; pyrimidine dimer repair [GO:0006290]; response to ethanol [GO:0045471]; response to gamma radiation [GO:0010332]; response to hyperoxia [GO:0055093]; salivary gland morphogenesis [GO:0007435]; somatic hypermutation of immunoglobulin genes [GO:0016446]; spleen development [GO:0048536] | Mouse_homologues 12713817_beta-pol has a role in DNA replication fidelity and/or base excision repair capacity and in lens epithelial cell differentiation 15195508_polymerase beta was not involved in thermal radiosensitization 15485914_proliferation and early differentiation of neuronal progenitors in Polbeta(-/-) p53(-/-) mice appeared normal, but their brains obviously displayed cytoarchitectural abnormalities 15647757_Results suggest a genetic interaction between DNA polymerase beta and DNA-PKcs in embryogenesis and neurogenesis. 16165404_DNA polymerase beta is not an essential component of the machinery that maintains mtDNA integrity. 16179390_Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. 16188889_cell-free extracts incubated with Ape1-incised 2-deoxyribonolactone substrates under non-repair conditions give rise to DNA-protein cross-links, with a major species dependent on the presence of polbeta 17126614_role for DNA polymerase beta in radiosensitivity in vivo 17591858_Polbeta attempts to faithfully repair immunoglobulin switch region lesions but fails to repair them all. 18650495_Normal POLB abundance is necessary for normal base excision repair activity, which is critical in maintaining a low germline mutant frequency. 19674974_Data show that disruption of DNA polymerase beta and FEN1 coordination during long-patch base excision repair results in CAG repeat expansion. 19748837_results highlight that Pol beta and poly(ADP-ribose) polymerase (PARP) function in the same repair pathway, but also suggest that there is repair independent of both Pol beta and PARP activities. 19838217_Data provide the first link between oxaliplatin sensitivity and DNA repair involving Pol beta. 19997493_The stoichiometry of base excision repair enzymes is one critical factor underlying the tissue selectivity of somatic CAG expansion. 20006562_This was the first evidence of Pol beta recruitment in long patch base excision repair in vivo. 20019666_Pol beta-deficient spermatocytes are defective in meiotic chromosome synapsis and undergo apoptosis during Prophase I. 20404327_Folate deficiency provides protection against colon carcinogenesis in DNA polymerase beta haploinsufficient mice 20510258_Pol beta deficiency could enable ROS accumulation. 20805875_both pol lambda and pol beta interact with the upstream DNA glycosylases for repair of alkylated and oxidized DNA bases 21333614_Pol beta is critical for genome stability in the germline 21351627_Overexpression of Pol beta could reduce oxidative damage and protect cells from hydroquinone genotoxicity. 22070286_Key residues for the rat DNA polymerase beta conformational transition was studied using elastic network model. 22162999_SAF-A interacts with BRG1 and both components are required for RNA Polymerase II Mediated Transcription 22357513_Polbeta could play a role in protecting cell lines from genotoxicity and genetic instability induced by benzo(a)pyrene. 22357534_Polbeta could protect cells from apoptosis induced by hydroquinone through decrease of ROS level and depolarization of mitochondria. 22493258_data clearly demonstrate that the DNA polymerase activity of pol beta is essential for survival and genome stability 23154201_Data indicate an association of ovarian stimulation with a downregulation of mRNAs encoding the base excision repair proteins APEX1 and POLB as well as the 5-methyl-CpG-binding domain protein MBD3 in individual morula embryos. 24084171_Results show that deficiency of either DNA polymerases beta or lambda or both results in a modest but significant decrease in V region somatic hypermutation (SHM) with no effect on mutation specificity, suggesting no direct role in SHM. 24121118_Transient OGG1, APE1, PARP1 and Polbeta expression in an Alzheimer's disease mouse model. 24388753_Y265C POLB mutation leads to several pathologies associated with SLE and short CDR3 regions during VDJ recombination in B cells. 25184665_The aberrant DNA replication mediated by the PCNA-DNA pol-b complex induces p53-dependent neuronal cell death 25886163_we show here that heterozygosity for a Y265C mutation in Polbeta, a key polymerase in the BER pathway, is enough to significantly reduce both the number of expansions seen in paternal gametes and the extent of somatic expansion in some tissues 26123757_pol beta plays an important role in bypassing a 5',8-cyclo-dA during DNA replication and repair. 27686631_Analyses of DNA damage and apoptosis revealed significantly greater degeneration of olfactory bulb neurons in transgenic Alzheimer's (3xTgAD)/Polbeta(+/-) mice compared to 3xTgAD mice. 27956495_pol beta contains a specific NLS sequence in the N-terminal lyase domain that promotes transport of the protein independent of its interaction partners. Active nuclear uptake allows development of a nuclear/cytosolic concentration gradient against a background of passive diffusion. 29100041_Mitochondria from pol beta-deficient mouse fibroblasts had compromised DNA repair and showed elevated levels of superoxide radicals after hydrogen peroxide treatment. Mitochondria in pol beta-deficient fibroblasts displayed altered morphology by electron microscopy. 29968395_To evaluate whether Polb might be causative in senescence induction, the study evaluated the impact of murine Polb nullizygosity on senescence. Unexposed DNA polymerase beta-null primary fibroblasts exhibit a robust increase in the number of senescent cells compared to wild-type (11-fold, P < 0.001), demonstrating that loss DNA polymerase betais sufficient to induce senescence. 30274781_The Polb/p53 double knockout animals exclusively developed medulloblastoma and no other type of brain tumor. 30763888_DNA polymerases beta and lambda in single-nucleotide and multinucleotide pathways of mammalian base excision DNA repair 33540226_DNA polymerase beta outperforms DNA polymerase gamma in key mitochondrial base excision repair activities. 34144266_DNA polymerase beta deficiency promotes the occurrence of esophageal precancerous lesions in mice. 34186496_DNA glycosylase deficiency leads to decreased severity of lupus in the Polb-Y265C mouse model. | ENSMUSG00000031536 | Polb | 2.744906e+02 | 0.8062449 | -0.310709958 | 0.3554364 | 7.284018e-01 | 0.3934017197 | 0.84317192 | No | Yes | 2.684458e+02 | 51.274577 | 3.081509e+02 | 60.317222 | ||
| ENSG00000070614 | 3340 | NDST1 | protein_coding | P52848 | FUNCTION: Essential bifunctional enzyme that catalyzes both the N-deacetylation and the N-sulfation of glucosamine (GlcNAc) of the glycosaminoglycan in heparan sulfate. Modifies the GlcNAc-GlcA disaccharide repeating sugar backbone to make N-sulfated heparosan, a prerequisite substrate for later modifications in heparin biosynthesis (PubMed:10758005, PubMed:12634318). Plays a role in determining the extent and pattern of sulfation of heparan sulfate. Compared to other NDST enzymes, its presence is absolutely required. Participates in biosynthesis of heparan sulfate that can ultimately serve as L-selectin ligands, thereby playing a role in inflammatory response (PubMed:10758005, PubMed:12634318). Required for the exosomal release of SDCBP, CD63 and syndecan (PubMed:22660413). {ECO:0000269|PubMed:10758005, ECO:0000269|PubMed:12634318, ECO:0000269|PubMed:22660413}. | 3D-structure;Alternative splicing;Disease variant;Disulfide bond;Glycoprotein;Golgi apparatus;Hydrolase;Inflammatory response;Membrane;Mental retardation;Multifunctional enzyme;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Glycan metabolism; heparan sulfate biosynthesis.; PATHWAY: Glycan metabolism; heparin biosynthesis. | This gene encodes a member of the heparan sulfate/heparin GlcNAc N-deacetylase/ N-sulfotransferase family. The encoded enzyme is a type II transmembrane protein that resides in the Golgi apparatus. The encoded protein catalyzes the transfer of sulfate from 3'-phosphoadenosine 5'-phosphosulfate to nitrogen of glucosamine in heparan sulfate. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2014]. | hsa:3340; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; [heparan sulfate]-glucosamine N-sulfotransferase activity [GO:0015016]; deacetylase activity [GO:0019213]; heparan sulfate N-deacetylase activity [GO:0102140]; N-acetylglucosamine deacetylase activity [GO:0050119]; aorta development [GO:0035904]; cardiac septum development [GO:0003279]; cell population proliferation [GO:0008283]; coronary vasculature development [GO:0060976]; embryonic neurocranium morphogenesis [GO:0048702]; embryonic viscerocranium morphogenesis [GO:0048703]; fibroblast growth factor receptor signaling pathway [GO:0008543]; forebrain development [GO:0030900]; glycosaminoglycan biosynthetic process [GO:0006024]; heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process [GO:0015014]; heparin biosynthetic process [GO:0030210]; inflammatory response [GO:0006954]; midbrain development [GO:0030901]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of smoothened signaling pathway [GO:0045880]; protein sulfation [GO:0006477]; respiratory gaseous exchange by respiratory system [GO:0007585] | 14966466_Altered expression of NDST-1 mRNA in glomerular basement membrane in puromycin aminonucleoside nephrosis. 19336475_Observational study of gene-disease association. (HuGE Navigator) 19710461_effect of targeted inactivation of the Ndst1 gene on the inflammatory response associated with allergic inflammation 19851296_Observational study of gene-disease association. (HuGE Navigator) 19915053_the expression of the NDST-1 isoform was approximately equal at all stages of mast cell maturation 20129923_Inhibitory peptides of the sulfotransferase domain of the heparan sulfate enzyme, N-deacetylase-N-sulfotransferase-1. 20198315_Observational study of gene-disease association. (HuGE Navigator) 21947487_MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803 23884416_These findings establish NDST1 as a target of miR-24 and demonstrate how such NDST1 suppression in endothelial cells results in reduced responsiveness to VEGFA 25125150_Our data confirm NDST1 mutations as a cause of autosomal recessive intellectual disability with a distinctive phenotype, and support an important function of NDST1 in human development. 25156775_Upregulation of NDST1 is associated with chemoresistance in breast cancer. 26109066_demonstrate the essential role of domain cooperation within NDST-1 in producing HS with specific domain structures 28211985_Compound heterozygous mutations in NDST1 were identified, in the heparan sulfate N deacetylatase domain of one allele and the sulfotransferase domain of the other allele. This report expands the phenotypic spectrum of Ndst1 deficiency in humans. 28404855_Among the genes enriched in this screening, the authors found that TM9SF2 is critical for N-sulfation of heparan sulfate and therefore for chikungunya virus infection because it is involved in the proper localization and stability of NDST1. 31364126_MiRNA-191 functions as an oncogene in primary glioblastoma by directly targeting NDST1. 32878022_Two Cases of Recessive Intellectual Disability Caused by NDST1 and METTL23 Variants. | ENSMUSG00000054008 | Ndst1 | 5.933138e+03 | 1.2967716 | 0.374924445 | 0.3040822 | 1.505476e+00 | 0.2198306806 | 0.78763590 | No | Yes | 5.927917e+03 | 661.821049 | 4.515454e+03 | 517.321310 |
| ENSG00000071282 | 29995 | LMCD1 | protein_coding | Q9NZU5 | FUNCTION: Transcriptional cofactor that restricts GATA6 function by inhibiting DNA-binding, resulting in repression of GATA6 transcriptional activation of downstream target genes. Represses GATA6-mediated trans activation of lung- and cardiac tissue-specific promoters. Inhibits DNA-binding by GATA4 and GATA1 to the cTNC promoter (By similarity). Plays a critical role in the development of cardiac hypertrophy via activation of calcineurin/nuclear factor of activated T-cells signaling pathway. {ECO:0000250, ECO:0000269|PubMed:20026769}. | Alternative splicing;Cytoplasm;LIM domain;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc | This gene encodes a member of the LIM-domain family of zinc finger proteins. The encoded protein contains an N-terminal cysteine-rich domain and two C-terminal LIM domains. The presence of LIM domains suggests involvement in protein-protein interactions. The protein may act as a co-regulator of transcription along with other transcription factors. Alternate splicing results in multiple transcript variants of this gene. [provided by RefSeq, May 2013]. | hsa:29995; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription corepressor activity [GO:0003714]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of calcineurin-NFAT signaling cascade [GO:0070886]; regulation of cardiac muscle hypertrophy [GO:0010611] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21996735_Together, our results suggest that LMCD1 mutations are potential oncogenic events in HCC metastasis to promote cell migration through the Rac1-signaling pathway. 26879886_The rs1800775 [1.31 (1.22-1.42); p = 3.41E-12] in the CETP gene and rs359027 [1.26 (1.16-1.36); p = 2.55E-08] in the LMCD1 gene were significantly associated with LHDLC levels 29326163_LIM and cysteine-rich domains 1 is required for thrombin-induced smooth muscle cell proliferation and promotes atherogenesis 31501411_LMCD1 promotes osteogenic differentiation of human bone marrow stem cells by regulating BMP signaling. 31666122_Skeletal muscle LMCD1 expression is reduced in patients with skeletal muscle disease. We analyzed gene expression data from muscle of patients with diverse muscle pathologies and identified LMCD1 as a gene strongly associated with skeletal muscle function. 32160773_LMCD1 acts as an activator of E2F1 in humans in the induction of CDC6 and IL-33 expression during development of vascular lesions. | ENSMUSG00000057604 | Lmcd1 | 9.327204e+01 | 0.5063904 | -0.981678159 | 0.4915552 | 4.010720e+00 | 0.0452118340 | 0.64941017 | No | Yes | 3.767468e+01 | 11.708935 | 6.355733e+01 | 20.250858 | |
| ENSG00000071994 | 5134 | PDCD2 | protein_coding | Q16342 | FUNCTION: May be a DNA-binding protein with a regulatory function. May play an important role in cell death and/or in regulation of cell proliferation. | Alternative splicing;Apoptosis;DNA-binding;Metal-binding;Nucleus;Reference proteome;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a nuclear protein expressed in a variety of tissues. Expression of this gene has been shown to be repressed by B-cell CLL/lymphoma 6 (BCL6), a transcriptional repressor required for lymph node germinal center development, suggesting that BCL6 regulates apoptosis by its effects on this protein. Alternative splicing results in multiple transcript variants and pseudogenes have been identified on chromosomes 9 and 12. [provided by RefSeq, Dec 2010]. | hsa:5134; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; DNA binding [GO:0003677]; enzyme binding [GO:0019899]; metal ion binding [GO:0046872]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic process [GO:0006915]; positive regulation of apoptotic process [GO:0043065]; positive regulation of hematopoietic stem cell proliferation [GO:1902035]; regulation of hematopoietic progenitor cell differentiation [GO:1901532] | 15848047_Observational study of gene-disease association. (HuGE Navigator) 16311922_To study the role of PDCD2_C domain in apoptosis, the cDNAs of two isoforms of PDCD2 and MGC13096 were cloned. PDCD2 (NM_002598) was over expressed when endothelial cells treated with leukotriene D4 or natural killer cells were activated by IL-2. 17468402_repression of PDCD2 by BCL6 is likely important in the pathogenesis of certain human lymphomas 19146857_parkin interacts with programmed cell death-2 isoform 1 (PDCD2-1) 20605493_Transfection of a construct expressing PDCD2 induces apoptosis in human cell lines through activation of the caspase cascade. 22922207_PDCD2 has a novel regulatory role in human hematopoiesis and is essential for erythroid development. 23760497_PDCD2 RNA expression in acute leukemia patients correlates with disease status and is a significant predictor of clinical relapse. 24381196_Expression of human PDCD2 fully rescues the Drosophila Zfrp8 phenotype, underlining the functional conservation of Zfrp8/PDCD2. 25111461_Low PDCD2 expression is associated with gastric cancer. 25334010_Loss of PDCD2 expression could contribute to gastric cancer development and progression. 26589942_PDCD2 and NCoR1 may act as tumor suppressors in Gastrointestinal stromal tumors cells through the Smad signaling pathway. 26617804_PDCD2 may play an important role in tumor suppression in osteosarcoma. These mechanisms might be related to immune response induced by CD4(+) and CD8(+) T cells. 30664177_the present study elucidated for the first time, to the best of our knowledge, that PDCD2 sensitizes sorafenibresistant HepG2 cells to sorafenib by the downregulation of EMT. PDCD2 may serve as a potential therapeutic target in the treatment of sorafenibresistant liver cancer. 33245768_PDCD2 functions as an evolutionarily conserved chaperone dedicated for the 40S ribosomal protein uS5 (RPS2). | ENSMUSG00000014771 | Pdcd2 | 2.940140e+03 | 0.8037993 | -0.315092837 | 0.2601768 | 1.479313e+00 | 0.2238819691 | 0.78763590 | No | Yes | 2.712241e+03 | 259.764775 | 3.270992e+03 | 320.798808 | |
| ENSG00000072135 | 26469 | PTPN18 | protein_coding | Q99952 | FUNCTION: Differentially dephosphorylate autophosphorylated tyrosine kinases which are known to be overexpressed in tumor tissues. | 3D-structure;Alternative splicing;Cytoplasm;Hydrolase;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, the mitotic cycle, and oncogenic transformation. This PTP contains a PEST motif, which often serves as a protein-protein interaction domain, and may be related to protein intracellular half-live. This protein can differentially dephosphorylate autophosphorylated tyrosine kinases that are overexpressed in tumor tissues, and it appears to regulate HER2, a member of the epidermal growth factor receptor family of receptor tyrosine kinases. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2008]. | hsa:26469; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; non-membrane spanning protein tyrosine phosphatase activity [GO:0004726]; protein tyrosine phosphatase activity [GO:0004725]; blastocyst formation [GO:0001825]; negative regulation of ERBB signaling pathway [GO:1901185]; peptidyl-tyrosine dephosphorylation [GO:0035335]; protein dephosphorylation [GO:0006470] | 14660651_BDP1 has a role in negative regulation of HER2 signaling 16303740_novel isoform of PTPN18 based on analysis of expressed sequence tags was discovered; deletion of 4 exons in the catalytic domain of the isoform may alter enzymatic activity; 2 of the novel isoform predictions were experimentally validated through RT-PCR 25081058_PTPN18 regulates HER2-mediated cellular functions through defining both its phosphorylation and ubiquitination barcodes. 29742497_PTPN12, PTPRN and PTPN18 were independent prognostic factors in Hepatocellular Carcinoma. 31034093_Downregulation of PTPN18 can inhibit proliferation and metastasis and promote apoptosis of endometrial cancer 32027948_Phospho-PTM proteomic discovery of novel EPO- modulated kinases and phosphatases, including PTPN18 as a positive regulator of EPOR/JAK2 Signaling. | ENSMUSG00000026126 | Ptpn18 | 1.643262e+03 | 1.1811648 | 0.240210311 | 0.2862844 | 6.984540e-01 | 0.4033036741 | 0.84651196 | No | Yes | 1.633491e+03 | 149.567393 | 1.316537e+03 | 124.210003 | |
| ENSG00000072195 | 10290 | SPEG | protein_coding | Q15772 | FUNCTION: Isoform 3 may have a role in regulating the growth and differentiation of arterial smooth muscle cells. | 3D-structure;ATP-binding;Alternative promoter usage;Alternative splicing;Differentiation;Disease variant;Disulfide bond;Immunoglobulin domain;Kinase;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | This gene encodes a protein with similarity to members of the myosin light chain kinase family. This protein family is required for myocyte cytoskeletal development. Along with the desmin gene, expression of this gene may be controlled by the desmin locus control region. Mutations in this gene are associated with centronuclear myopathy 5. [provided by RefSeq, Jun 2016]. | hsa:10290; | nucleus [GO:0005634]; ATP binding [GO:0005524]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; muscle cell differentiation [GO:0042692]; muscle organ development [GO:0007517]; negative regulation of cell population proliferation [GO:0008285] | 15784173_Genomic rearrangement on APEG1 in arteriosclerosis was studied. 16354304_the RGD motif might play a role not only in the adhesion of Aortic Preferentially Expressed Protein-1 and extracellular proteins but also in intracellular protein-protein interactions 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25087613_SPEG is present in cardiac muscle, where it plays a critical role; therefore, individuals with SPEG mutations additionally present with dilated cardiomyopathy. 30412272_Clinicians should consider evaluating a centronuclear myopathies patient for SPEG mutations even in the absence of centronuclear myopathies features 31625632_Novel SPEG variant cause centronuclear myopathy in China. 31790338_Striated muscle-specific serine/threonine-protein kinase beta segregates with high versus low responsiveness to endurance exercise training. 32683896_Loss of SPEG Inhibitory Phosphorylation of Ryanodine Receptor Type-2 Promotes Atrial Fibrillation. 32925938_A Novel Recessive Mutation in SPEG Causes Early Onset Dilated Cardiomyopathy. 33794647_Homozygous SPEG Mutation Is Associated With Isolated Dilated Cardiomyopathy. | ENSMUSG00000026207 | Speg | 4.486755e+02 | 1.0319841 | 0.045420798 | 0.2998959 | 2.290720e-02 | 0.8796986483 | 0.97741608 | No | Yes | 4.115805e+02 | 69.381039 | 4.027582e+02 | 69.920573 | |
| ENSG00000072422 | 9886 | RHOBTB1 | protein_coding | O94844 | GTP-binding;Nucleotide-binding;Reference proteome;Repeat | The protein encoded by this gene belongs to the Rho family of the small GTPase superfamily. It contains a GTPase domain, a proline-rich region, a tandem of 2 BTB (broad complex, tramtrack, and bric-a-brac) domains, and a conserved C-terminal region. The protein plays a role in small GTPase-mediated signal transduction and the organization of the actin filament system. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Dec 2008]. | hsa:9886; | cell cortex [GO:0005938]; cell projection [GO:0042995]; cytoplasmic vesicle [GO:0031410]; cytoskeleton [GO:0005856]; endosome membrane [GO:0010008]; plasma membrane [GO:0005886]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; protein kinase binding [GO:0019901]; actin filament organization [GO:0007015]; cortical cytoskeleton organization [GO:0030865]; engulfment of apoptotic cell [GO:0043652]; establishment or maintenance of cell polarity [GO:0007163]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of cell shape [GO:0008360]; small GTPase mediated signal transduction [GO:0007264] | 16170569_Frequent allelic loss and decreased expression of RHOBTB1 suggested that this gene has a role in tumorigenesis of a subset of HNSCC (Head-Neck Squamous Cell Carcinoma). 16385451_Observational study of gene-disease association. (HuGE Navigator) 23258531_we found that miR-31 acts as an oncogene in colon cancer and identified RhoBTB1 as a new target of miR-31 further study demonstrated that miR-31 contributed to the development of colon cancer at least partly by targeting RhoBTB1. 28219369_RhoBTB1 regulates the integrity of the Golgi complex through METTL7B. RhoBTB1 is required for expression of METTL7B and silencing of either protein leads to fragmentation of the Golgi. Loss of RhoBTB1 expression is linked to Golgi fragmentation in breast cancer cells. Restoration of normal RhoBTB1 expression rescues Golgi morphology and dramatically inhibits breast cancer cell invasion. 31431478_RhoBTB1 associates with ROCK1 and ROCK2 and its association with ROCK1 is via its Rho domain. 32354068_RNA Interference Screening Identifies Novel Roles for RhoBTB1 and RhoBTB3 in Membrane Trafficking Events in Mammalian Cells. | ENSMUSG00000019944 | Rhobtb1 | 1.871100e+02 | 0.5858045 | -0.771508781 | 0.3403747 | 5.013348e+00 | 0.0251526168 | 0.54724823 | No | Yes | 1.315726e+02 | 20.359046 | 2.099761e+02 | 32.704641 | ||
| ENSG00000072736 | 4775 | NFATC3 | protein_coding | Q12968 | FUNCTION: Acts as a regulator of transcriptional activation. Plays a role in the inducible expression of cytokine genes in T-cells, especially in the induction of the IL-2 (PubMed:18815128). Along with NFATC4, involved in embryonic heart development (By similarity). {ECO:0000250|UniProtKB:P97305, ECO:0000269|PubMed:18815128}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cytoplasm;DNA-binding;Developmental protein;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | The product of this gene is a member of the nuclear factors of activated T cells DNA-binding transcription complex. This complex consists of at least two components: a preexisting cytosolic component that translocates to the nucleus upon T cell receptor (TCR) stimulation and an inducible nuclear component. Other members of this family participate to form this complex also. The product of this gene plays a role in the regulation of gene expression in T cells and immature thymocytes. Several transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Nov 2010]. | hsa:4775; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; calcineurin-NFAT signaling cascade [GO:0033173]; inflammatory response [GO:0006954]; negative regulation of pri-miRNA transcription by RNA polymerase II [GO:1902894]; negative regulation of vascular associated smooth muscle cell differentiation [GO:1905064]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 11997392_transactivation activity and role in inducing differentiation of CD4(+)CD8(+) T cells 15173172_NFAT and thyroid transcription factor-1 have roles in regulating transcription of the surfactant protein D gene 16260608_NFATc3 is negatively regulated by class II histone deacetylases through the DnaJ (heat shock protein-40) superfamily member Mrj 16445977_We found that [Ca(2+)](i) oscillations were associated with NFAT translocation into the nucleus in undifferentiated hMSCs. As the hMSCs differentiated to adipocytes, the [Ca(2+)](i) oscillations disappeared and the translocation of NFAT ceased. 17035332_Calcium signaling and the activation of NFAT in glial cells are required for JC Virus infection of the CNS. 18676376_NFAT and MyoD cooperation regulates myogenin expression and myogenesis 18815128_CHP2 has a role in tumorigenesis and as an activator of the calcineurin/NFAT signaling pathway 19913121_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21047202_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21642596_NFATc3 is specifically required for IL2 and cyclooxygenase-2 (COX2) gene expression in T cells and for T-cell proliferation and NFATc3 regulates COX2 in endothelial cells 22749879_The authors report that NFAT4 and NF-kappaB interact at the KB element to co-operatively activate both human polyomavirus JC early and late transcription and viral DNA replication. 22846573_Abeta-activated NFAT4 proteins were associated with astrocytic BACE1 gene expression via direct interaction with the BACE1 promoter region. 22977251_Data indicate that NFATc3 undergoes rapid dephosphorylation and nuclear translocation that are essentially complete within 20 min, although NFATc4 remains phosphorylated and localized to the cytosol. 23219532_The closely related transcription factors NFAT1 and NFAT4 possess distinct nuclear localization dynamics in response to cell stimulation. 23929433_AP-1 and NFAT4 complex promotes miR-23a expression. 24582564_Microvesicles from tumor cells transferred TrpC5 to endothelial cells, inducing the expression of P-glycoprotein by activation of the transcription factor NFATc3 (nuclear factor of activated T cells isoform c3). 25215946_NFATc3 interacted in a SUMO-dependent manner with Trim17, an E3 ubiquitin ligase necessary for neuronal apoptosis 25422138_Suggest nuclear NF-AT3 and NF-AT4 participates in atrial structural remodeling, and that PICP and TGF-beta1 levels may be sensitive serum biomarkers to estimate atrial structural remodeling with atrial fibrillation. 25818645_Results show that two protein isoforms NFAT1 and NFAT4 are both cytosolic and stimulated by the same Ca2+ messenger but require distinct subcellular Ca2+ signals for activity. 26374065_two NFAT isoforms (NFAT4 and NFAT1) have shifted band-pass windows for the same receptor in the GPCR signaling pathway 26527057_Data indicate that RNA interference of NFAT isoforms NFATc1, NFATc2, NFATc3 and NFATc4 regulate gene expression differentially in human retinal microvascular endothelial cells (HRMEC). 27123462_Calcineurin together with its upstream molecule, calpain 2, and its downstream effector, NFAT-c3, might contribute to the development of atrial fibrillation in patients with heart valve disease and diabetes. 27697837_the transcription factor NFATC3 binds to IRF7 and functions synergistically to enhance IRF7-mediated IFN expression in Plasmacytoid dendritic cells. 27863227_NFAT1 is stimulated by subplasmalemmal Ca2+ microdomains, whereas NFAT4 additionally requires Ca2+ mobilization from the inner nuclear envelope by nuclear InsP3 receptors. 28125639_we found that VIP inhibits NFAT nuclear translocation in primary human pulmonary artery smooth muscle cells (PASMC). Early activation of NFATc3 in IPF patients may contribute to disease progression and the increase in VIP expression could be a protective compensatory mechanism 28627449_MicroRNA-214 regulates immunity-related genes in bovine mammary epithelial cells by targeting NFATc3 and TRA 28724635_The NFATc3 first induced the expression of its interaction partner FosB before forming the heterodimeric NFATc3-FosB transcription factor complex, which bound the proximal AP-1 site in the TF gene promoter and activated TF expression. 29330284_Expression of the NFATC3-PLA2G15 chimera correlated with aggressive disease biology in murine patient-derived T-ALL xenografts, and poor prognosis in human T-ALL patients. 30895498_data indicated that NRON alleviates atrial fibrosis via promoting NFATc3 phosphorylation. 32653643_Blocking NFATc3 ameliorates azoxymethane/dextran sulfate sodium induced colitis-associated colorectal cancer in mice via the inhibition of inflammatory responses and epithelial-mesenchymal transition. 32667692_circNFATC3 sponges miR-548I acts as a ceRNA to protect NFATC3 itself and suppressed hepatocellular carcinoma progression. 33520407_NFATc3 inhibits hepatocarcinogenesis and HBV replication via positively regulating RIG-I-mediated interferon transcription. 33840648_Fusobacterium nucleatum Promotes Cisplatin-Resistance and Migration of Oral Squamous Carcinoma Cells by Up-Regulating Wnt5a-Mediated NFATc3 Expression. 34570211_Macrophage NFATc3 prevents foam cell formation and atherosclerosis: evidence and mechanisms. 35484132_Hypoxia-induced NFATc3 deSUMOylation enhances pancreatic carcinoma progression. | ENSMUSG00000031902 | Nfatc3 | 3.125617e+03 | 1.0125725 | 0.018025143 | 0.2574713 | 4.888649e-03 | 0.9442582312 | 0.98943389 | No | Yes | 2.998415e+03 | 262.600362 | 2.696404e+03 | 242.338643 | |
| ENSG00000073008 | 5817 | PVR | protein_coding | P15151 | FUNCTION: Mediates NK cell adhesion and triggers NK cell effector functions. Binds two different NK cell receptors: CD96 and CD226. These interactions accumulates at the cell-cell contact site, leading to the formation of a mature immunological synapse between NK cell and target cell. This may trigger adhesion and secretion of lytic granules and IFN-gamma and activate cytotoxicity of activated NK cells. May also promote NK cell-target cell modular exchange, and PVR transfer to the NK cell. This transfer is more important in some tumor cells expressing a lot of PVR, and may trigger fratricide NK cell activation, providing tumors with a mechanism of immunoevasion. Plays a role in mediating tumor cell invasion and migration. {ECO:0000269|PubMed:15471548, ECO:0000269|PubMed:15607800}.; FUNCTION: (Microbial infection) Acts as a receptor for poliovirus. May play a role in axonal transport of poliovirus, by targeting virion-PVR-containing endocytic vesicles to the microtubular network through interaction with DYNLT1. This interaction would drive the virus-containing vesicle to the axonal retrograde transport. {ECO:0000269|PubMed:2538245}.; FUNCTION: (Microbial infection) Acts as a receptor for Pseudorabies virus. {ECO:0000269|PubMed:9616127}.; FUNCTION: (Microbial infection) Is prevented to reach cell surface upon infection by Human cytomegalovirus /HHV-5, presumably to escape immune recognition of infected cell by NK cells. {ECO:0000269|PubMed:15640804}. | 3D-structure;Alternative splicing;Cell adhesion;Cell membrane;Disulfide bond;Glycoprotein;Host cell receptor for virus entry;Host-virus interaction;Immunoglobulin domain;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a transmembrane glycoprotein belonging to the immunoglobulin superfamily. The external domain mediates cell attachment to the extracellular matrix molecule vitronectin, while its intracellular domain interacts with the dynein light chain Tctex-1/DYNLT1. The gene is specific to the primate lineage, and serves as a cellular receptor for poliovirus in the first step of poliovirus replication. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2008]. | hsa:5817; | adherens junction [GO:0005912]; cell surface [GO:0009986]; cytoplasm [GO:0005737]; extracellular space [GO:0005615]; focal adhesion [GO:0005925]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; cell adhesion molecule binding [GO:0050839]; signaling receptor activity [GO:0038023]; virus receptor activity [GO:0001618]; heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules [GO:0007157]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; positive regulation of natural killer cell mediated cytotoxicity [GO:0045954]; positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target [GO:0002860]; susceptibility to natural killer cell mediated cytotoxicity [GO:0042271]; susceptibility to T cell mediated cytotoxicity [GO:0060370] | 11751937_We propose that the cytoplasmic domain may target CD155-containing endocytic vesicles to the microtubular network 11983699_activation of expression of sonic hedgehog protein 12913096_Data show that both PVR and Nectin-2 represent specific ligands for the DNAM-1 triggering receptor. 12943679_CD155 may have an important role in cellular function 14652024_These data indicate that Tage4 represents the functional orthologue of CD155 in mouse. 15076773_Observational study of gene-disease association. (HuGE Navigator) 15136589_that DNAM-1 regulates monocyte extravasation via its interaction with CD226 expressed at endothelial junctions on normal cells. 15194502_These results suggest that CD155alpha plays a role in the regulation of cell adhesion and cell motility. 15194795_cytoplasmic domain of PVR directly interacts with Tctex-1 and plays an important role in retrograde transport of poliovirus-containing vesicles along microtubules in vivo 15279713_Upregulation of the molecular target CD155 renders explant cultures of high-grade malignant gliomas highly susceptible to a prototype oncolytic poliovirus recombinant. 15536144_Analysis of the ligands for triggering NK receptors revealed the consistent expression of cd155 and cd112 in myeloid leukemias, and less frequent expression in lymphoblastic leukemias 15640804_Evasion of NK cell killing was mediated by human cytomegalovirus UL141 blocking surface expression of CD155 17446174_Necl-5 has a critical role in integrin alphavbeta3 clustering and focal complex formation 17507470_Results describe the establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. 17534374_no statistically significant association between this marker allele and non-syndromic clefting 17893876_CD155, at least in part, enhances the proliferation of ras-mutated cells 19011098_crystal structure of C155 D1D2 has been determined to 3.5-A resolution and fitted into approximately 8.5-A resolution cryoelectron microscopy reconstructions of the virus-receptor complexes for the 3 PV serotypes 19056733_CD96-driven adhesion to CD155 may be crucial in developmental processes 19319949_The Ala67Thr mutation in the poliovirus receptor is a possible risk factor for the development of vaccine-associated or paralytic poliomyelitis associated with wild-type virus. 19801517_evidence provided for the contribution of DNAM-1/CD155 interactions to the reduction of DNAM-1 expression, suggesting that chronic receptor-ligand interactions in the tumor environment may induce loss of DNAM-1 on tumor-associated NK cells. 19815499_TIGIT is expressed by all NK cells, it binds PVR and PVRL2 but not PVRL3, and it inhibits NK cytotoxicity 20331633_Necl-5 plays a role in mediating tumor cell invasion and that the overexpression of Necl-5 in cancer cells has clinical significance for prognostic evaluation of patients with primary pulmonary adenocarcinoma. 20634891_Observational study of gene-disease association. (HuGE Navigator) 21330602_CD155 is an IFNgamma-inducible immune regulatory protein on the surface of human endothelial cells that attenuates the acquisition of effector functions in CD8 T cells. 21383766_Data show that a high expression of CD112 and CD155 (DNAM-1 ligands) on leukemic blasts. 21998457_The host TICAM-1 pathway, particularly in macrophages, serves as a source of type I interferon induction that protects poliovirus (PV) receptor-bearing transgenic mice from PV infection and paralytic death. 22169283_Expression of PVR in B-ALL cells is modulated by epigenetic mechanisms. 22301152_the PVR downmodulation by Nef and Vpu is a strategy evolved by HIV-1 to prevent NK cell-mediated lysis of infected cells. 22363471_This investigation has enhanced understanding of cell invasion and confirmed CD44 to play a more significant role in this biological process than CD155. 22692919_we demonstrated the expression of both CD155 mRNA and protein in bone and soft tissue sarcoma cell lines 22929570_The concordant computational and experimental data of the present study indicate that the extent of NECL-5 expression correlates with melanoma progression. 23276719_findings suggest Necl-5 expression in lung cancer cells is crucial for their invasiveness in the cancer-stromal interaction 23333754_PVR resides in the recently identified lateral border recycling compartment, similar to PECAM and CD99. 23980210_The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. 24045107_Vpr upregulates PVR during Hiv-1 infection by activating ATR kinase that triggers the DNA damage rsponse pathway and G2 arrest. 24598754_UL141 can inhibit cell-surface expression of both natural killer (NK) cell-activating ligand CD155 as well as TRAIL death receptors (TRAIL-R1 and TRAIL-R2). 24817116_TIGIT/PVR ligation signaling mediates suppression of IFN-gamma production via the NF-kappaB pathway. 24828608_In granulosa cells, there are significant changes in expression during follicular maturation. 25113908_Ala residues 10, 14 and 18 in the TM domain of Vpu are required for CD155 downregulation. 25209846_UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. 25320021_Our findings suggest that loss of CD155 expression may play an important role in the immune escape of HCC cells and thus CD155 may serve as a prognostic marker as well as a potential therapeutic target for HCC. 25609078_The present study provides evidence that regulation of the PVR/CD155 DNAM-1 ligand expression by nitric oxide may represent an additional immune-mediated mechanism and supports the anti-myeloma activity of nitric oxide donors. 25631086_The cell-surface receptor (Pvr) catalyzes a large structural change in the poliovirus that exposes membrane-binding protein chains. 25862891_CD155 may play a critical role through both immunological and non-immuno logical mechanisms in pancreatic cancer and may be a therapeutic target for this intractable malignancy. 25972481_CD155 (PVR/Necl5) mediates a costimulatory signal in CD4+ T cells and regulates allergic inflammation. 26842126_implying that TIGIT exerts immunosuppressive effects by competing with DNAM-1 for the same ligand, CD155 27049654_sCD155 levels were significantly decreased after surgical resection of cancers. Thus, sCD155 level in serum may be potentially useful as a biomarker for cancer development and progression 27296670_The authors demonstrate that HIV and specifically Nef and/or Vpu do not modulate CD155 on infected primary T cells and both CD155 and NKG2D ligands synergize as a natural killer cell receptor to trigger natural killer cell lysis of the infected cell. 27733551_data reveal that MICA and PVR are directly regulated by human cytomegalovirus immediate early proteins, and this may be crucial for the onset of an early host antiviral response 27834324_The SNP detection assay was successfully developed for identification of Ala67Thr polymorphism in human PVR/CD155 gene. The SNP assay will be useful for large scale screening of DNA samples. 28084312_These findings highlight the importance of the TIGIT/CD226/PVR axis as an immune checkpoint barrier that could hinder future 'cure' strategies requiring potent HIV-specific CD8(+) T cells 28395975_soluble CD226 elevated in sera of CTCL patients would be important for tumor immunity by interacting with CD155 on tumor cells. 28730595_Studies showed that CD155 was frequently overexpressed in human malignant tumors. Its overexpression promotes tumor cell invasion and migration, and is associated with tumor progression. [review] 28816021_CD155 is one of key molecules promoting the growth and metastasis of colorectal cancer. 28883004_Data show that gastric cancer cells inhibit T-cell metabolism through CD155/TIGIT signaling. 29381645_Studied association of poliovirus receptor (PVR/CD155) mutation and cleft lip and cleft palate. Validated previous findings that PVR/CD155 markers are associated with cleft lip and palate. 29431243_Study investigated the more detailed mechanism for this cis-interaction of Necl-5 with the PDGF receptor beta. Necl-5 contains three Ig-like domains and the PDGF receptor beta contains five Ig-like domains at their extracellular regions; showed that the third Ig-like domain of Necl-5 cis-interacted with the fifth Ig-like domain of the PDGF receptor beta. 29878245_CD155 was found to be expressed in pediatric brain tumors and in medulloblastoma and pleomorphic xanthoastrocytoma tumor cell lines. 30039180_Our results also suggest that CD155 upregulation may be a mechanism underlying Adr resistance by breast cancer cells. 30528596_Results show the structural binding of CD155 ectodomain with the first immunoglobulin domain (D1) of CD96. 30591568_The hybrid complex structure of CD226-ecto binding to the first domain of CD155 (hCD155-D1) reveals a conserved binding interface with the first domain of CD226 (D1), whereas the second domain of CD226 (D2) both provides structural supports for the unique architecture of CD226 and forms direct interactions with CD155. 30880756_The results of real-time PCR indicated that poliovirus receptor (CD155) was significantly overexpressed in human colorectal adenocarcinoma cell lines in comparison with the human normal cell line. 31035013_These results suggest an interaction between CD155 expression and tumor-infiltrating lymphocytes (TILs) in breast cancer (BC), and they also suggest that CD155 could be an effective prognostic biomarker for BC. 31036057_The increased MIF secretion by the A549R and H460R cells could be suppressed by a multiple kinase inhibitor, dasatinib, which resulted in the decreased of oncogenic network of Src, CD155 and MIF expression 31253644_The DNAM-1/Necl-5 interaction was underpinned by conserved lock-and-key motifs located within their respective D1 domains, but also included a distinct interface derived from DNAM-1 D2. Mutation of the signature DNAM-1 'key' motif within the D1 domain attenuated Necl-5 binding and natural killer cell-mediated cytotoxicity. 31372841_High expression of soluble CD155 in estrogen receptor-negative breast cancer. 31377744_Large-scale analysis reveals the specific clinical and immune features of CD155 in glioma. 31383549_Poliovirus receptor CD155 is up-regulated in muscle-invasive bladder cancer and predicts poor prognosis. 31387897_Blockade of TIGIT/CD155 Signaling Reverses T-cell Exhaustion and Enhances Antitumor Capability in Head and Neck Squamous Cell Carcinoma. 31485637_Expression of TIGIT/CD155 in cancerous tissue is significantly elevated in patients with hepatocellular carcinoma. 31830330_Study identified for the first time that CD155 is overexpressed in triple-negative breast cancer (TNBC), is associated with poor prognosis and contributes to the aggressive phenotype. Mechanistically, IL-6/Stat3 and TGF-beta/Smad3 pathways may be involved in CD155-associated TNBC epithelial-to-mesenchymal transition and metastasis. CD155 knockdown suppressed TNBC cell in vitro motility and in vivo metastasis. 31883911_Tumor intrinsic and extrinsic immune functions of CD155. 31954274_Combined evaluation of the expression status of CD155 and TIGIT plays an important role in the prognosis of LUAD (lung adenocarcinoma). 32040157_Tumor-derived soluble CD155 inhibits DNAM-1-mediated antitumor activity of natural killer cells. 32321756_Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155. 32345648_Tumor CD155 Expression Is Associated with Resistance to Anti-PD1 Immunotherapy in Metastatic Melanoma. 32411795_Overexpression of an Immune Checkpoint (CD155) in Breast Cancer Associated with Prognostic Significance and Exhausted Tumor-Infiltrating Lymphocytes: A Cohort Study. 32446718_The immune suppressive factors CD155 and PD-L1 show contrasting expression patterns and immune correlates in ovarian and other cancers. 32554931_Combination of PD-L1 and PVR determines sensitivity to PD-1 blockade. 32727790_Mutant KRAS Promotes NKG2D(+) T Cell Infiltration and CD155 Dependent Immune Evasion. 32804915_TIGIT Can Exert Immunosuppressive Effects on CD8+ T Cells by the CD155/TIGIT Signaling Pathway for Hepatocellular Carcinoma In Vitro. 32902876_The risk of unintentional propagation of poliovirus can be minimized by using human cell lines lacking the functional CD155 gene. 32917981_Direct interaction between CD155 and CD96 promotes immunosuppression in lung adenocarcinoma. 33053330_CD155 on Tumor Cells Drives Resistance to Immunotherapy by Inducing the Degradation of the Activating Receptor CD226 in CD8(+) T Cells. 33185939_Aberrant expression of junctional adhesion molecule-A contributes to the malignancy of cervical adenocarcinoma by interaction with poliovirus receptor/CD155. 33399495_Targeting CD155 by rediocide-A overcomes tumour immuno-resistance to natural killer cells. 33504618_Aryl Hydrocarbon Receptor Signaling Controls CD155 Expression on Macrophages and Mediates Tumor Immunosuppression. 33576304_CD155-Prognostic and Immunotherapeutic Implications Based on Multiple Analyses of Databases Across 33 Human Cancers. 33616718_Overexpression of poliovirus receptor is associated with poor prognosis in head and neck squamous cell carcinoma patients. 33879814_Overexpression of PVR and PD-L1 and its association with prognosis in surgically resected squamous cell lung carcinoma. 34030043_The N-linked glycosylations of TIGIT Asn(32) and Asn(101) facilitate PVR/TIGIT interaction. 34115802_Immunohistochemical analysis of CD155 expression in triple-negative breast cancer patients. 34353426_Acetate decreases PVR/CD155 expression via PI3K/AKT pathway in cancer cells. 34504191_DNAM-1/CD226 is functionally expressed on acute myeloid leukemia (AML) cells and is associated with favorable prognosis. 34608548_Immune checkpoint CD155 promoter methylation profiling reveals cancer-associated behaviors within breast neoplasia. 35324958_Prognostic value of CD155/TIGIT expression in patients with colorectal cancer. 35384345_CD155/SRC complex promotes hepatocellular carcinoma progression via inhibiting the p38 MAPK signalling pathway and correlates with poor prognosis. | ENSMUSG00000040511 | Pvr | 3.557480e+03 | 1.2216752 | 0.288860800 | 0.2838757 | 1.038838e+00 | 0.3080917781 | 0.81529480 | No | Yes | 3.501822e+03 | 242.331685 | 2.825383e+03 | 200.794979 | |
| ENSG00000073536 | 54475 | NLE1 | protein_coding | Q9NVX2 | FUNCTION: Plays a role in regulating Notch activity. Plays a role in regulating the expression of CDKN1A and several members of the Wnt pathway, probably via its effects on Notch activity. Required during embryogenesis for inner mass cell survival (By similarity). {ECO:0000250}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Notch signaling pathway;Nucleus;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:54475; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; hematopoietic stem cell homeostasis [GO:0061484]; inner cell mass cell differentiation [GO:0001826]; kidney development [GO:0001822]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway [GO:2001268]; negative regulation of mitotic cell cycle [GO:0045930]; Notch signaling pathway [GO:0007219]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; ribosomal large subunit assembly [GO:0000027]; skeletal system morphogenesis [GO:0048705]; somitogenesis [GO:0001756] | Mouse_homologues 16611995_mNle is mainly required in inner cell mass cells, being instrumental for their survival, and raise the possibility that the death of mNle-deficient embryos might result from abnormal Notch signaling during the first steps of development. 24062412_These indings establish an essential role for Nle in hematopoietic stem cells. 24875805_These data demonstrate an essential role for Nle1 during organogenesis and in particular during axial development. 30478226_activated skeletal muscle satellite cells in conditional Nle mutant mice are arrested in proliferation; however, deletion of Nle in myofibres does not impair myogenesis. 34019907_Knockdown of NLE1 inhibits development of malignant melanoma in vitro and in vivo NLE1 promotes development of malignant melanoma. | ENSMUSG00000020692 | Nle1 | 3.278927e+03 | 1.6286233 | 0.703652921 | 0.3218145 | 4.814525e+00 | 0.0282208500 | 0.56656291 | No | Yes | 4.499237e+03 | 690.116813 | 2.211922e+03 | 348.525449 | ||
| ENSG00000073605 | 55876 | GSDMB | protein_coding | Q8TAX9 | FUNCTION: [Gasdermin-B]: Precursor of a pore-forming protein that acts as a downstream mediator of granzyme-mediated cell death (PubMed:32299851). This form constitutes the precursor of the pore-forming protein: upon cleavage, the released N-terminal moiety (Gasdermin-B, N-terminal) binds to membranes and forms pores, triggering pyroptosis (PubMed:32299851). {ECO:0000269|PubMed:32299851}.; FUNCTION: [Gasdermin-B, N-terminal]: Pore-forming protein produced by cleavage by granzyme A (GZMA), which causes membrane permeabilization and pyroptosis in target cells of cytotoxic T and natural killer (NK) cells (PubMed:27281216, PubMed:32299851). Key downstream mediator of granzyme-mediated cell death: (1) granzyme A (GZMA), delivered to target cells from cytotoxic T- and NK-cells, (2) specifically cleaves Gasdermin-B to generate this form (PubMed:32299851). After cleavage, moves to the plasma membrane, homooligomerizes within the membrane and forms pores of 10-15 nanometers (nm) of inner diameter, triggering pyroptosis (PubMed:32299851). Binds to membrane inner leaflet lipids, such as phosphatidylinositol 4-phosphate, phosphatidylinositol 5-phosphate, bisphosphorylated phosphatidylinositols, such as phosphatidylinositol (4,5)-bisphosphate, and more weakly to phosphatidic acid (PubMed:28154144). Also binds sufatide, a component of the apical membrane of epithelial cells (PubMed:28154144). {ECO:0000269|PubMed:27281216, ECO:0000269|PubMed:28154144, ECO:0000269|PubMed:32299851}. | 3D-structure;Alternative splicing;Cell membrane;Coiled coil;Cytolysis;Cytoplasm;Direct protein sequencing;Membrane;Necrosis;Reference proteome;Transmembrane;Transmembrane beta strand | This gene encodes a member of the gasdermin-domain containing protein family. Other gasdermin-family genes are implicated in the regulation of apoptosis in epithelial cells, and are linked to cancer. Alternative splicing and the use of alternative promoters results in multiple transcript variants. Additional variants have been described, but they are candidates for nonsense-mediated mRNA decay (NMD) and are unlikely to be protein-coding. [provided by RefSeq, Nov 2016]. | hsa:55876; | cytoplasm [GO:0005737]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; phosphatidylinositol-4-phosphate binding [GO:0070273]; phosphatidylserine binding [GO:0001786]; wide pore channel activity [GO:0022829]; cytolysis [GO:0019835]; cytotoxic T cell pyroptotic process [GO:1902483]; defense response to bacterium [GO:0042742]; granzyme-mediated programmed cell death signaling pathway [GO:0140507]; pyroptosis [GO:0070269] | 15010812_Evolutionary recombination hotspot around the GSDML-GSDM locus is closely linked to oncogenomic recombination hotspot around the PPP1R1B-STARD3-TCAP-PNMT-PERLD1-ERBB2-C17orf37-GRB7 amplicon at human chromosome 17q12. 18038310_GSDML may be a secretory or metabolic product involved in a secretory pathway, and changes in the regulation of GSDML splicing variant transcription and translation may be seen in the development and/or progression of gastrointestinal and hepatic cancers. 19029000_Observational study of gene-disease association. (HuGE Navigator) 19051310_Study investigated the expression pattern of the GSDM family genes in the upper gastrointestinal epithelium and cancers 19068216_Observational study of gene-disease association. (HuGE Navigator) 19133921_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19133921_polymorphisms in ORMDL3 and the adjacent GSDML may contribute to childhood asthma. 19732864_Observational study of gene-disease association. (HuGE Navigator) 19732864_The disease-linked haplotype and putative causal DNA variants of ZPBP2/GSDMB/ORMDL3 locus via a combination of genetic and functional analyses, were identified. 19760754_Observational study of gene-disease association. (HuGE Navigator) 20378605_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20601676_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20816195_Observational study of gene-disease association. (HuGE Navigator) 20860503_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21337730_The GSDMA (rs7212938) and GSDMB (rs7216389) polymorphisms are associated with asthma susceptibility and intermediate asthma phenotypes. 22295569_Five markers on chromosome 17q12-21 showed statistically significant association with bronchial asthma .SNP rs7216389 with the strongest evidence for association is located within the first intron of the GSDMB gene. 22370936_Significant associations between two SNPs, rs2305480 and rs8067378 in the GSDML gene, and asthma were found in this study. 22732088_GSDMB/ORMDL3 variants contribute to asthma susceptibility and eosinophil-mediated bronchial hyperresponsiveness. 24044605_rs11078928 is associated with the production of a novel GSDMB transcript lacking an internal segment 24066901_GSDMB SNP rs2305480 (Ser311Pro) was associated with asthma diagnosis (p = 8.9x10-4), bronchial hyperresponsiveness to methacholine (p = 8.2x10-4) and severity (p = 1.5x10-4) with supporting evidence from a second GSDMB SNP rs11078927 (intronic). 24315451_3 SNPs associated with the fraction of exhaled nitric oxide in childhood asthma are rs3751972 in LYRM9; rs944722 in NOS2; and rs8069176 near GSDMB; all at 17q12-q21. 24675552_Gasdermin-B promotes invasion and metastasis in breast cancer cells 24848122_No association between GSDMB polymorphisms and rheumatoid arthritis was observed. 25256354_Results confirmed the genetic association between GSDMB/ ORMDL3 and childhood asthma and show significant differences in the DNA methylation levels of ORMDL3 promoter of asthmatic children. 26016667_The GSDMB-driven HSVtk expression vector had a therapeutic effect on the occult peritoneal dissemination model mice. 26270739_GSDMB variants have been shown to be associated with Asthma in children with Rhinovirus infections -induced wheezing illnesses. 26484354_Based on our results and published findings on GSDMA, GSDMB, LRRC3C, and related proteins, we propose that this locus in part affects IBD susceptibility via effects on apoptosis and cell proliferation 26534891_childhood asthma is associated with gene polymorphism; meta-analysis 26886240_we investigated the association between GSDMA and GSDMB variants and the incidence of adult and childhood asthma among Jordanians.An association between the GSDMB T/C single nucleotide polymorphism (SNP) genotype and the incidence of childhood asthma was found 27462779_Data identifies the ERBB2 co-amplified and co-expressed gene GSDMB as a critical determinant of poor prognosis and therapeutic response in HER2-positive breast cancer. 27799535_These studies demonstrate that GSDMB, a gene highly linked to asthma but whose function in asthma is previously unknown, regulates AHR and airway remodeling without airway inflammation through a previously unrecognized pathway in which GSDMB induces 5-LO to induce TGF-beta1 in bronchial epithelium. 28052796_We found strong associations among GSDMB polymorphisms and the presence of AERD and FEV1 in Korean patients with asthma. Our findings indicated that genetic variations of GSDMB may be associated with the development of AERD and aspirin-induced bronchospasm. 28120299_The rs8067378 SNP variants may increase the expression of GSDMB and the risk of the development and progression of cervical SCC. 28154144_Full-length GSDMB and its N-terminal domain bind to phosphoinositides or sulfatide, but not cardiolipin. The GSDMB N-terminal domain binds liposomes containing sulfatide, suggesting a role in sulfatide transport. A loop with SNP AAs from controls (Gly299:Pro306) shows high conformational flexibility, but one with AAs (Arg299:Ser306) from people at risk for IBD and asthma is less so and has higher positive surface charge. 28272342_GSDMB levels are significantly modulated by NMD. Importantly, both AS isoforms and the identified ecircRNA were significantly dysregulated in peripheral blood mononuclear cells of relapsing-remitting MS patients compared to controls. 28588209_These results showed that the PBC susceptibility allele of rs12946510 disrupted the enhancer region for ORMDL3 and GSDMB gene expression (Fig. 6). Chromatin interaction between the sequence that contains rs12946510 and the upstream sequence of ORMDL3 and GSDMB also supported this enhancer model 28826527_Chromosome 17q21 genes ORMDL3 and GSDMB are linked to asthma and other immune diseases. (Review) 29330013_Our study identified a functional asthma variant in the GSDMB gene of the 17q21 locus and implicates GSDMB-mediated epithelial cell pyroptosis in pathogenesis. 29374573_we show that local CpG methylation mediates a proportion, but not all, of the functional effects of cis-acting asthma-susceptibility regulatory variants on GSDMB and ORMDL3 gene expression 30321352_GSDMB promotes caspase-4 activity, which is required for the cleavage of GSDMD in non-canonical pyroptosis, by directly binding to the CARD domain of caspase-4. 32299851_this study demonstrated that GZMA from cytotoxic lymphocytes cleaves and activates GSDMB to induce target cell pyroptosis. 32496997_Association between Gasdermin A, Gasdermin B Polymorphisms and Allergic Rhinitis Amongst Jordanians. 32795586_Genetic analyses identify GSDMB associated with asthma severity, exacerbations, and antiviral pathways. 33810791_Household mold exposure interacts with inflammation-related genetic variants on childhood asthma: a case-control study. 33941434_ORMDL3/GSDMB genotype is associated with distinct phenotypes of adult asthma. 33963941_Association of Gasdermin B Gene GSDMB Polymorphisms with Risk of Allergic Diseases. 34076350_The GSDMB rs7216389 SNP is associated with chronic rhinosinusitis in a multi-institutional cohort. 34326684_USP24-GSDMB complex promotes bladder cancer proliferation via activation of the STAT3 pathway. 34957313_Upregulated GSDMB in Clear Cell Renal Cell Carcinoma Is Associated with Immune Infiltrates and Poor Prognosis. 35021065_GSDMB is increased in IBD and regulates epithelial restitution/repair independent of pyroptosis. | 2.128639e+02 | 0.6516765 | -0.617772083 | 0.3087774 | 3.934963e+00 | 0.0472921680 | 0.65905990 | No | Yes | 1.114282e+02 | 24.099101 | 2.015380e+02 | 44.212685 | |||
| ENSG00000074266 | 8726 | EED | protein_coding | O75530 | FUNCTION: Polycomb group (PcG) protein. Component of the PRC2/EED-EZH2 complex, which methylates 'Lys-9' and 'Lys-27' of histone H3, leading to transcriptional repression of the affected target gene. Also recognizes 'Lys-26' trimethylated histone H1 with the effect of inhibiting PRC2 complex methyltransferase activity on nucleosomal histone H3 'Lys-27', whereas H3 'Lys-27' recognition has the opposite effect, enabling the propagation of this repressive mark. The PRC2/EED-EZH2 complex may also serve as a recruiting platform for DNA methyltransferases, thereby linking two epigenetic repression systems. Genes repressed by the PRC2/EED-EZH2 complex include HOXC8, HOXA9, MYT1 and CDKN2A. {ECO:0000269|PubMed:10581039, ECO:0000269|PubMed:14532106, ECO:0000269|PubMed:15225548, ECO:0000269|PubMed:15231737, ECO:0000269|PubMed:15385962, ECO:0000269|PubMed:16357870, ECO:0000269|PubMed:18285464, ECO:0000269|PubMed:20974918, ECO:0000269|PubMed:28229514, ECO:0000269|PubMed:9584199}. | 3D-structure;Acetylation;Alternative initiation;Alternative splicing;Chromatin regulator;Chromosome;Disease variant;Host-virus interaction;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;WD repeat | This gene encodes a member of the Polycomb-group (PcG) family. PcG family members form multimeric protein complexes, which are involved in maintaining the transcriptional repressive state of genes over successive cell generations. This protein interacts with enhancer of zeste 2, the cytoplasmic tail of integrin beta7, immunodeficiency virus type 1 (HIV-1) MA protein, and histone deacetylase proteins. This protein mediates repression of gene activity through histone deacetylation, and may act as a specific regulator of integrin function. Two transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:8726; | chromosome [GO:0005694]; cytosol [GO:0005829]; ESC/E(Z) complex [GO:0035098]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; enzyme activator activity [GO:0008047]; histone methyltransferase activity [GO:0042054]; identical protein binding [GO:0042802]; transcription corepressor binding [GO:0001222]; heterochromatin assembly [GO:0031507]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; spinal cord development [GO:0021510] | 14759364_EED is a nuclear factor and repressor of transcription and is recruited to the plasma membrane by HIV-1 nef 17547741_EED exerted an antiviral activity at the late stage of HIV-1 replication, which included genomic RNA packaging and virus assembly, resulting possibly from a mistrafficking of viral genomic RNA (gRNA) or gRNA/Gag complex. 17804093_NIPP1 is present in a complex with EED and EZH2 in vivo and has distinct binding sites for these proteins. 18302803_Identification of antibody-, MA-, IN- and EZH2-binding sites at EED's surface provides a global picture of the immunogenic and protein-protein interacting regions in the EED C-terminal domain, organized as a seven-bladed beta-propeller protein. 19578722_histone modification including PRC2-mediated repressive histone marker H3K27me3 and active histone marker acH4 may involve in CD11b transcription during HL-60 leukemia cells reprogramming to terminal differentiation[polycomb repressive complex 2 ] 19767730_the carboxy-terminal domain of EED specifically binds to histone tails carrying trimethyl-lysine residues associated with repressive chromatin marks, and this leads to the allosteric activation of the methyltransferase activity of PRC2. 20974918_molecular basis of EED-methyllysine recognition, and the biochemical characterization of how the activity of a histone methyltransferase is oppositely regulated by two histone marks. 21540835_The authors found that Sox2 and Eed positively regulate each other's expression and contribute to the maintenance of self-renewal in embryonic stem cells by controlling histone methylation and acetylation. 22271413_Promoter polymorphism of the EED gene is associated with the susceptibility to ulcerative colitis. 22308284_Genetic defects in PRC2 components other than EZH2 are not common in myeloid malignancies. 22733077_EED mutants impair polycomb repressive complex 2 and is associated with myelodysplastic syndrome and related neoplasms 23709348_These results suggest that the SNPs of the EED gene might not be associated with susceptibility to CRC. 23982173_Although inactivating mutations in PRC2-encoding genes EZH2, EED, and SUZ12 are present in T-cell acute lymphoblastic leukemia and in myeloid malignancies, gain-of-function mutations in EZH2 are frequently observed in B-cell lymphoma. 24320048_EZH2-EED is necessary and sufficient for binding to the lncRNA HOTAIR. 24457600_An integral role for EED as an epigenetic exchange factor coordinating the activities of PRC1 and 2, is reported. 25240281_Polycomb repressive complex 2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. 25264103_EED, a component of Polycomb repressive complex-2 (PRC2) that catalyzes methylation of lysine 27 of histone H3 (H3K27), was involved in epithelial-mesenchymal transition (EMT) of cancer cells induced by Transforming Growth Factor-beta (TGF-beta). 25326896_Data show that overall enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 homolog (SUZ12) expression in the colorectal cancer (CRC) tissues was significantly increased than in the non-cancerous tissue. 27000413_Mutations of SUZ12 and EED are reported to have tumor suppressive functions. (Review) 27193220_we have found two unrelated families of different ethnicities, with a similar rare phenotype, both associated with de novo mutations in this member of the PRC2 complex, we are confident that EED is indeed a novel overgrowth gene. 27868325_These findings support that Weaver syndrome is a disorder with locus heterogeneity and can be due to pathogenic variants in either EZH2 or EED. This case highlights the utility of exome sequencing as a clinical diagnostic tool for novel gene discovery as well as the importance of re-examination of exome data as new information about gene-disease associations becomes available. 28229514_we analyzed eight probands with clinically suspected Weaver syndrome by whole-exome sequencing and identified three mutations: a 25.4-kb deletion partially involving EZH2 and CUL1 ,a missense mutation , and a missense mutation in SUZ12 inherited from her father .In vitro functional analyses demonstrated that the identified EED and SUZ12 missense mutations cause decreased decreased trimethylation of lysine 27 of H3 29410511_we present a patient with a clinical diagnosis of Weaver syndrome and novel de novo sequence variant in EED. Our observation together with previous reports [2, 3, 5] suggests that EED gene testing is warranted in patients with the overgrowth syndrome features and suspicion of Weaver syndrome with normal results for EZH2 gene sequencing. 29415665_Data suggest the polycomb repressive complex 2 subunits EZH2, SUZ12, and EED protein axis as promising therapeutic target for treating sarcoma. 29754954_These findings identify Eed/PRC2 as necessary for maintenance of global gene silencing and terminal differentiation in b cells, and suggest a ''two-hit'' (chromatin and hyperglycemia) model of b cell dedifferentiation. 30595380_PICOT knock-down in Jurkat T cells resulted in a reduced histone H3 lysine 27 trimethylation (H3K27me3) at the PRC2 target gene, myelin transcription factor 1 (MYT1), suggesting that PICOT binding to EED alters PRC2-regulated transcriptional repression, and potentially contributes to the epigenetic regulation of chromatin silencing and remodeling. 30628724_Results report that EED presents a direct interaction with androgen receptor (AR). In the context of AR-positive prostate cancer, EED along with EZH2 regulate AR expression levels and its downstream targets. 30793471_Mutation in the EED gene is associated with Cohen-Gibson syndrome. 30858506_In silico modeling and calculations of the free energy changes resulting from these variants suggested that they not only destabilize the EED protein structure but also adversely affect interactions between EED, EZH2, and/or H3K27me3. 31023785_Identification of mutations in the PRC2 components EED and SUZ12 in the majority of MPNSTs may imply noncanonical oncogenic activities of the intact component, EZH2, and provide new opportunities for therapeutic intervention. 31395608_BR-001, a potent inhibitor of the EED subunit of the PRC2 complex, suppresses tumor progression by modulating the tumor microenvironment. 31527584_PICOT binding to chromatin-associated EED negatively regulates cyclin D2 expression by increasing H3K27me3 at the CCND2 gene promoter. 31724824_review the PRC2 complex and clinical syndromes of overgrowth and intellectual disability associated with core components EZH2, EED, and SUZ12 33882457_Methylation of microRNA-338-5p by EED promotes METTL3-mediated translation of oncogene CDCP1 in gastric cancer. 34544341_Embryonic Ectoderm Development (EED) as a Novel Target for Cancer Treatment. | ENSMUSG00000030619 | Eed | 7.759024e+02 | 0.8541059 | -0.227513131 | 0.2836693 | 6.395524e-01 | 0.4238729381 | 0.85075211 | No | Yes | 6.639944e+02 | 88.303616 | 8.097716e+02 | 110.159858 | |
| ENSG00000075643 | 55034 | MOCOS | protein_coding | Q96EN8 | FUNCTION: Sulfurates the molybdenum cofactor. Sulfation of molybdenum is essential for xanthine dehydrogenase (XDH) and aldehyde oxidase (ADO) enzymes in which molybdenum cofactor is liganded by 1 oxygen and 1 sulfur atom in active form. In vitro, the C-terminal domain is able to reduce N-hydroxylated prodrugs, such as benzamidoxime. {ECO:0000255|HAMAP-Rule:MF_03050, ECO:0000269|PubMed:16973608}. | Disease variant;Molybdenum cofactor biosynthesis;Phosphoprotein;Pyridoxal phosphate;Reference proteome;Transferase | This gene encodes an enzyme that sulfurates the molybdenum cofactor which is required for activation of the xanthine dehydrogenase (XDH) and aldehyde oxidase (AO) enzymes. XDH catalyzes the conversion of hypoxanthine to uric acid via xanthine, as well as the conversion of allopurinol to oxypurinol, and pyrazinamide to 5-hydroxy pyrazinamide. Mutations in this gene cause the metabolic disorder classical xanthinuria type II which is characterized by the loss of XDH/XO and AO enzyme activity, decreased levels of uric acid in the urine, increased levels of xanthine and hypoxanthine in the serum and urine, formation of xanthine stones in the urinary tract, and myositis due to tissue deposition of xanthine. [provided by RefSeq, Apr 2017]. | hsa:55034; | cytosol [GO:0005829]; lyase activity [GO:0016829]; Mo-molybdopterin cofactor sulfurase activity [GO:0008265]; molybdenum cofactor sulfurtransferase activity [GO:0102867]; molybdenum ion binding [GO:0030151]; pyridoxal phosphate binding [GO:0030170]; Mo-molybdopterin cofactor biosynthetic process [GO:0006777]; molybdopterin cofactor biosynthetic process [GO:0032324]; molybdopterin cofactor metabolic process [GO:0043545] | 17368066_Two mutations, both in the N-terminal domain of the Human Molybdenum Cofactor Sulfurase (HMCS), were reported in patients with type II xanthinuria. 19500084_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22495427_MOCOS sulfurates the molybdenum cofactor of xanthine oxidase (XDH) and aldehyde oxidase 1 (AOX1), which is required for their activities. 31900757_The rs594445 in MOCOS gene is associated with risk of autism spectrum disorder. 32327736_Impaired expression of the COSMOC/MOCOS gene unit in ASD patient stem cells. | ENSMUSG00000039616 | Mocos | 5.851917e+02 | 0.8497209 | -0.234939042 | 0.3168065 | 5.640536e-01 | 0.4526315999 | 0.86206872 | No | Yes | 4.682257e+02 | 56.127504 | 5.501711e+02 | 67.493613 | |
| ENSG00000075914 | 23016 | EXOSC7 | protein_coding | Q15024 | FUNCTION: Non-catalytic component of the RNA exosome complex which has 3'->5' exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. In the nucleus, the RNA exosome complex is involved in proper maturation of stable RNA species such as rRNA, snRNA and snoRNA, in the elimination of RNA processing by-products and non-coding 'pervasive' transcripts, such as antisense RNA species and promoter-upstream transcripts (PROMPTs), and of mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm. The RNA exosome may be involved in Ig class switch recombination (CSR) and/or Ig variable region somatic hypermutation (SHM) by targeting AICDA deamination activity to transcribed dsDNA substrates. In the cytoplasm, the RNA exosome complex is involved in general mRNA turnover and specifically degrades inherently unstable mRNAs containing AU-rich elements (AREs) within their 3' untranslated regions, and in RNA surveillance pathways, preventing translation of aberrant mRNAs. It seems to be involved in degradation of histone mRNA. The catalytic inactive RNA exosome core complex of 9 subunits (Exo-9) is proposed to play a pivotal role in the binding and presentation of RNA for ribonucleolysis, and to serve as a scaffold for the association with catalytic subunits and accessory proteins or complexes. | 3D-structure;Acetylation;Cytoplasm;Direct protein sequencing;Exosome;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;rRNA processing | hsa:23016; | cytoplasmic exosome (RNase complex) [GO:0000177]; cytosol [GO:0005829]; exosome (RNase complex) [GO:0000178]; nuclear exosome (RNase complex) [GO:0000176]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 3'-5'-exoribonuclease activity [GO:0000175]; RNA binding [GO:0003723]; exonucleolytic catabolism of deadenylated mRNA [GO:0043928]; exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [GO:0000467]; nuclear mRNA surveillance [GO:0071028]; nuclear polyadenylation-dependent mRNA catabolic process [GO:0071042]; nuclear polyadenylation-dependent rRNA catabolic process [GO:0071035]; nuclear polyadenylation-dependent tRNA catabolic process [GO:0071038]; nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5' [GO:0034427]; RNA catabolic process [GO:0006401]; RNA processing [GO:0006396]; rRNA catabolic process [GO:0016075]; rRNA processing [GO:0006364]; U1 snRNA 3'-end processing [GO:0034473]; U4 snRNA 3'-end processing [GO:0034475]; U5 snRNA 3'-end processing [GO:0034476] | 11812149_The association of hCsl4p with the exosome is mediated by protein-protein interactions with hRrp42p and hRrp46p. 12419256_Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. 21509594_EXOSC7 gene expression is decreased in both classic and follicular variants of papillary thyroid carcinoma. | ENSMUSG00000025785 | Exosc7 | 2.335838e+03 | 0.8652462 | -0.208817477 | 0.3019410 | 4.850570e-01 | 0.4861406007 | 0.87430359 | No | Yes | 1.986692e+03 | 183.268427 | 2.193572e+03 | 207.324137 | ||
| ENSG00000076356 | 5362 | PLXNA2 | protein_coding | O75051 | FUNCTION: Coreceptor for SEMA3A and SEMA6A. Necessary for signaling by SEMA6A and class 3 semaphorins and subsequent remodeling of the cytoskeleton. Plays a role in axon guidance, invasive growth and cell migration. Class 3 semaphorins bind to a complex composed of a neuropilin and a plexin. The plexin modulates the affinity of the complex for specific semaphorins, and its cytoplasmic domain is required for the activation of down-stream signaling events in the cytoplasm (By similarity). {ECO:0000250, ECO:0000269|PubMed:10520995}. | 3D-structure;Alternative splicing;Cell membrane;Coiled coil;Disulfide bond;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of the plexin-A family of semaphorin co-receptors. Semaphorins are a large family of secreted or membrane-bound proteins that mediate repulsive effects on axon pathfinding during nervous system development. A subset of semaphorins are recognized by plexin-A/neuropilin transmembrane receptor complexes, triggering a cellular signal transduction cascade that leads to axon repulsion. This plexin-A family member is thought to transduce signals from semaphorin-3A and -3C. [provided by RefSeq, Jul 2008]. | hsa:5362; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; semaphorin receptor complex [GO:0002116]; identical protein binding [GO:0042802]; semaphorin receptor activity [GO:0017154]; centrosome localization [GO:0051642]; cerebellar granule cell precursor tangential migration [GO:0021935]; limb bud formation [GO:0060174]; negative regulation of cell adhesion [GO:0007162]; neural tube development [GO:0021915]; pharyngeal system development [GO:0060037]; positive regulation of axonogenesis [GO:0050772]; regulation of cell migration [GO:0030334]; regulation of cell shape [GO:0008360]; regulation of GTPase activity [GO:0043087]; semaphorin-plexin signaling pathway [GO:0071526]; semaphorin-plexin signaling pathway involved in axon guidance [GO:1902287]; somitogenesis [GO:0001756] | 16402134_An association is identified between variants in the PLXNA2 gene and schizophrenia in two collections of schizophrenia cases and controls. High-density SNP mapping showed that the region of association spans approximately 60 kb of the PLXNA2 gene. 16402134_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 16932874_Observational study of gene-disease association. (HuGE Navigator) 17339520_Observational study of gene-disease association. (HuGE Navigator) 17346868_Observational study of gene-disease association. (HuGE Navigator) 17346868_results suggest that PLXNA2 may not play a major role in the development of schizophrenia in our Japanese sample 18065206_Observational study of gene-disease association. (HuGE Navigator) 18065206_PLXNA2 confers a varying genetic risk for schizophrenia among different populations. 18096369_Analysis of 3 SNPs at the PLXN A2 locus; we failed to replicate previously reported association of this locus and schizophrenia. 18583979_Meta-analysis of gene-disease association. (HuGE Navigator) 18628988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19480842_Data show that the expression of Sema3A receptors (neuropilin-1 (NRP-1), NRP-2, plexin A1, plexin A2, and plexin A3) significantly increased during M-CSF-mediated differentiation of monocytes into macrophages. 19666519_mutations in GATA6 are genetic causes of congenital heart diseases involving outflow tract defects, as a result of the disruption of the direct regulation of semaphorin 3C-plexin A2 signaling 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20455246_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20633049_Observational study of gene-disease association. (HuGE Navigator) 21925246_in vitro analysis of PLXNA2 revealed that the gene has higher expression in more aggressive breast cancer cell types. 22011406_PLXNA2 polymorphisms show association with ankylosing spondylitis. 22095611_results of our study reveal that PlxnA2 has a pro-osteogenic function by modulating BMP2 signaling 22912587_PLXNA2 has been identified as a new rare copy number variations gene for tetralogy of Fallot. 23708657_PLXNA2 upregulation contributes to TMPRSS2:ERG-mediated enhancements of PC3c cell migration and invasion. 25335892_although plexin-A4 overexpression restored Sema3A signaling in plexin-A1-silenced cells, it failed to restore Sema3B signaling in plexin-A2-silenced cells. 25518740_Data indicate that plexin A1-4 (PLXNA1-4) mediation of neuroanatomical traits can be detected using in vivo neuroimaging techniques. 29091353_Data suggest that Fyn tyrosine kinase (Fyn)-dependent phosphorylation at two critical tyrosines is a key feature of vertebrate plexin A1 (PlxnA1) and plexin A2 (PlxnA2) signal transduction. 33749153_PLXNA2 and LRRC40 as candidate genes in autism spectrum disorder. 34327814_PLXNA2 as a candidate gene in patients with intellectual disability. | ENSMUSG00000026640 | Plxna2 | 4.117788e+02 | 1.6549608 | 0.726797011 | 0.3328819 | 4.929529e+00 | 0.0264015173 | 0.55176060 | No | Yes | 4.646729e+02 | 82.012987 | 2.898454e+02 | 52.717429 | |
| ENSG00000077092 | 5915 | RARB | protein_coding | P10826 | FUNCTION: Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RXR/RAR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence or presence of hormone ligand, acts mainly as an activator of gene expression due to weak binding to corepressors (PubMed:12554770). The RXRA/RARB heterodimer can act as a repressor on the DR1 element and as an activator on the DR5 element (PubMed:29021580). In concert with RARG, required for skeletal growth, matrix homeostasis and growth plate function (By similarity). {ECO:0000250|UniProtKB:P22605, ECO:0000269|PubMed:12554770, ECO:0000269|PubMed:29021580}. | 3D-structure;Alternative splicing;Cytoplasm;DNA-binding;Disease variant;Metal-binding;Microphthalmia;Nucleus;Phosphoprotein;Proto-oncogene;Receptor;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes retinoic acid receptor beta, a member of the thyroid-steroid hormone receptor superfamily of nuclear transcriptional regulators. This receptor localizes to the cytoplasm and to subnuclear compartments. It binds retinoic acid, the biologically active form of vitamin A which mediates cellular signalling in embryonic morphogenesis, cell growth and differentiation. It is thought that this protein limits growth of many cell types by regulating gene expression. The gene was first identified in a hepatocellular carcinoma where it flanks a hepatitis B virus integration site. Alternate promoter usage and differential splicing result in multiple transcript variants. [provided by RefSeq, Mar 2014]. | hsa:5915; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; nuclear receptor activity [GO:0004879]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; zinc ion binding [GO:0008270]; cell differentiation [GO:0030154]; embryonic digestive tract development [GO:0048566]; embryonic eye morphogenesis [GO:0048048]; embryonic hindlimb morphogenesis [GO:0035116]; glandular epithelial cell development [GO:0002068]; growth plate cartilage development [GO:0003417]; hormone-mediated signaling pathway [GO:0009755]; multicellular organism growth [GO:0035264]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of chondrocyte differentiation [GO:0032331]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neurogenesis [GO:0022008]; positive regulation of apoptotic process [GO:0043065]; positive regulation of transcription by RNA polymerase II [GO:0045944]; retinoic acid receptor signaling pathway [GO:0048384]; signal transduction [GO:0007165]; striatum development [GO:0021756]; ureteric bud development [GO:0001657]; ventricular cardiac muscle cell differentiation [GO:0055012] | 11769677_The expression of exogenous hRAR beta gene in HL-60R cells could resume their sensitivity to retionoids in inhibiting cell proliferation and inducing granulocytic differentiation. 11834837_PML-RAR fusion protein induces gene hypermethylation and silencing by recruiting DNA methyltransferases to the RARbeta2 promoter 11839665_distinct methylation pattern in bladder cancer with frequent methylation of RARbeta, DAPK, E-cadherin, and p16. 11925591_Results indicate that loss of RAR-beta expression and accumulation of p 53 and Ki67 proteins may serve as biomarkers for early identification of esophageal cancer in the high-risk populations. 11945179_Methylation of the 5' region of RAR-beta 2 gene may contribute to gene silencing and may be an important and early event in cervical carcinogenesis. 11980632_Endogenous reactivation of the RARbeta2 tumor suppressor gene epigenetically silenced in breast cancer. 12009305_Results show that both RARalpha and RARbeta are mediators in the anticancer function of All-trans retinoic acid via AP-1 activity inhibition. 12118004_downstream codons in mRNAs initiate translation of a protein isoform that disrupts retinoid-activated transcription 12124324_Loss of retinoic acid receptor beta gene expression is linked to aberrant histone H3 acetylation in lung cancer cell lines. 12191570_STAT-1, IRF-1, and RAR-beta expression were enhanced by IFN-gamma and ATRA in combination, and to a greater degree in BALM-3 cells than in BALM-1 cells, suggesting that these IFN-gamma related genes were involved in the induction of apoptosis. 12399530_Present only in basal epithelial nuclei. RAR-beta and -gamma were increased in basal and luminal epithelial nuclei in glands with benign prostatic hyperplasia. 12529350_RARbeta expression may be an indicator of increased risk of lung cancer in heavy smokers. 12554770_RARbeta and RARgamma interact only weakly with SMRT. 12576329_results suggest that oxidized phospholipids inhibit transcription of the thrombomodulin gene in vascular endothelium by inhibiting the binding of retinoic acid receptor beta-retinoid x receptor alpha heterodimer and Sp1 and Sp3 to thrombomodulin promoter 12579317_ATRA increased RARbeta2 mRNA in non-metastatic breast cancer cells. The same treatment of metastatic cells resulted in an increase in RARbeta4 & a decrease in RARbeta2 mRNA. RARbeta4 may contribute to metastatic properties of breast cancer cell lines. 12665583_RAR beta and RAR gamma receptors appear to adopt a constitutively closed helix 12 conformation in the absence of hormone that may approximate the conformation of RAR alpha when bound to hormone agonist. 12789467_the role of RAR-beta2 induction with respect to clonogenic survival of different human tumor cells under retinoid treatment alone or in combination with irradiation 12805409_c-myc, FOG1, GATA6, glutamate dehydrogenase, glutathione S-transferase homologue (p28), Foxq1, Hic5, Meis1a, Dab2, midkine, and the PDGF-alpha receptor are genes regulated specifically by RARbeta(2) in F9 cells 12839938_The regulatin oexpression of this receptor in cancer cells is affected by ligands of ppargamma. 12839965_Promoter hypermethylation of this gene is demonstrated in esophageal squamous cell carcinoma. 14601057_hypermethylated in invasive and in in situ lobular breast cancer 14614007_Hypermethylation-associated inactivation of retinoic acid receptor beta is associated with esophageal squamous cell carcinoma 14691372_In a 9-cis retinoic acid-dependent fashion in cells in vitro, retinoic acid receptor beta isoform stimulates the expression of reporter constructs containing the site that binds aldehyde dehydrogenase-2. 14726683_The adult RARbeta2 isoform also shows age-related methylation in normal tissues but more variable methylation in colorectal cancer. 14726690_Isoforms are involved in colon cancer cell growth. 15014026_RAR-beta(2) silencing by methylation is an early event in head and neck carcinogenesis. 15217922_Retinoic acid receptor beta2 hypermethylation has a role in prostate cancer [editorial] 15217932_RARbeta2 methylation has a role in development of prostate neoplasms 15255287_Loss of expression of RARbeta is associated with esophageal squamous cell carcinomas 15361842_RARbeta2 acts as a tumor suppressor gene in myelofibrosis with myeloid metaplasia and epigenetic changes are the most significant determinants of RARbeta2 gene activity in these patients. 15375805_Review loss or abnormality of RAR-b in lung cancer cell lines. It may have tumor suppression function. 15383624_(RAR)beta functions as a tumor suppressor gene in various contexts where its absence is associated with tumorigenicity and its presence causes cell cycle arrest. 15467435_Increased acetylation of RARbeta1 is associated with head and neck cancer 15502323_crystal structure of retinoic acid receptor beta 15635645_Observational study of gene-disease association. (HuGE Navigator) 16128742_results show that inactivation of the retinoic acid signaling-associated genes RAR-beta, CRBP1, and TIG1 by DNA methylation occurs frequently in esophageal squamous cell carcinoma 16134180_Hypermethylation of retinoid acid receptor beta is associated with gastric carcinogenesis 16244585_Study demonstrate for the first time a significant and specific overexpression of RAR-beta(1) in chromophobe renal cell carcinoma 16255778_RARbeta2 induces a number of tumor suppressor functions and metastasis suppressors 16287870_RARbeta2 silencing and retinoic acid (RA) resistance are consequent to an impaired integration of RA signal at RARbeta2 chromatin 16288117_In H358 lung cancer cells transiently transfected with RARbeta1', RA treatment restored target gene expression compared with that in vector-transfected cells and suppressed cell growth compared with that in untreated cells. 16389900_Genetically deregulated in cerebral glioma. (review) 16478744_RARbeta2 was methylated in the tumor epithelium of all five prostate cancer patients and in the tumor-associated stroma in four of the five patients. 16613851_MOZ-TIF2 associates with the RARbeta2 promoter in vivo, resulting in altered recruitment of CBP/p300, aberrant histone modification, and down-regulation of the RARbeta2 gene 16858683_Aberrant methylation and hence silencing of TIMP3, SLC5A8, DAPK and RARbeta2, in association with BRAF mutation, may be an important step in PTC tumorigenesis and progression. 17001699_Results suggest that the truncated form of retinoic acid receptor beta induces retinoid resistance rather than sensitivity, and alternative pathways of cell death are mediated by different isoforms in breast cancer cells. 17065567_Immunohistochemistry (demonstrated no overt difference between CDH, hypoplastic, and control lungs, either in the localization nor the timing of the first expression of glucocorticoid, retinoid, and thyroid hormone receptors 17244680_RARbeta2 is commonly silenced by hypermethylation in primary AML blasts but not in normal hematopoietic precursors. 17252005_Biological properties of the other RAR isoforms (RARbeta and RARgamma), through the generation pf fusion protein isoforms in acutte promyelocytic leukemia. 17549354_Hypopharynx tumors showed significantly lower hypermethylation and higher mRNA expression levels of RAR beta2 compared to the tumors located at other sites of the head and neck 17556535_Shows a higher methylation in all six patients with familial partial lipodystrophy when compared with progeria patients with other LMNA mutations as well as the healthy controls. 17615267_RARss is downregulated upon TGF-ss3-driven chondrogenic differentiation of adult bone marrow mesenchymal stem cells. RARss chondrogenic pathway is independent of upregulation in SOX9 and does not lead to hypertrophy. 17686773_ectopic expression of RAR beta2 is able to down-regulate HPV-18 transcription by selectively abrogating the binding of AP-1 to the viral regulatory region in a ligand-independent manner. 17804711_HOXA5 acts directly downstream of RARbeta and may contribute to retinoid-induced anticancer and chemopreventive effects. 17900311_Phosphorylation of RXRalpha abolishes its ability to form homodimers and heterodimers with RXR and retinoic acid receptor beta. 18036402_Tumor-specific RARB hypermethylation was observed in four papillomas. 18058812_No field effect, defined as absence of epigenetically transformed cells, for GSTP1 was observed, whereas APC, RARbeta2, and RASSF1A showed a field effect up to 3 mm from the malignant core in three prostatectomy samples. 18166136_endogenous expression of retinoids receptor RARbeta gene determines the susceptibility of HCC cells to fenretinide-induced apoptosis 18212063_TopoIIbeta interacts with the 5' RARE region of the RARbeta gene. 18254975_Observational study of gene-disease association. (HuGE Navigator) 18271925_the expression level of RAR beta in endometrial carcinoma is significantly lower than that in endometrial hyperplasia 18349282_Methylation of RARB was present in 80% of NSCLC tissues but only 14% of noncancerous tissues. 18483325_Methylation of RARB was significantly positvely correlated with breast cytologic atypia, increasing between ages 35 and 45. 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18538349_The promoter methylation status of a panel of critical growth regulatory genes, RASSF1A, RARbeta2, BRCA1 and HOXA5, in 54 breast cancers and 5 distant normal breast tissues of Indian patients, was analyzed. 18547710_reduced expresion in cervical cancer 18663751_methylation pattern of the promoter region. We show that hypo- and hypermethylated alleles coexist in 5/11 cell lines in which RARB2 is inactivated. 18673301_the increase in RAR-beta expression is not associated with telomere shortening, a typical biomarker of cellular senescence 18783461_Promoter region methylation of RARb2 genes is an early event in endometrial carcinogenesis. 18825962_Methylation of promoter region of RAR-beta2 gene in renal cell, breast, and ovarian carcinomas 18852534_The 5-Aza-deoxycytidine treatment induced RARbetamRNA expression not only in ARO but also in FRO and TT cell lines, whose RARbeta2 promoter was unmethylated. 18957410_bexarotene increased the occupancy of the identified enhancer element in IGFBP-6 gene by RXRalpha, RARbeta, cJun, cFos, and p300 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19081515_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19127267_ATP7A expression is regulated by retinoic acid receptor beta and it has effects on intracellular copper levels, revealing a link between the anticancer action of retinoids and copper metabolism. 19221469_Results showed that the inactivation of RASSF1A, RARbeta2 and DAP-Kinase by hypermethylation is a key step in NPC tumorigenesis and progression. 19258476_The combination of RARB M4, INK4a/ARF, PRB, and HIN-1 CpG island promoter methylation may predict non-BRCA1/2-associated mammary carcinogenesis and tumor progression. 19258542_retinoic acid receptor-beta2 has a role in suppression of cyclooxygenase-2 in esophageal neoplasms 19469866_Our data indicated that RAR beta is expressed in dermatofibrosarcoma protuberans and correlated with COX-2 expression. 19618401_Data show that methylation of CCND-2, p16, RAR-beta and RASSF-1a was significantly more prevalent in tumor than in normal tissue specimens. 19657672_Findings suggested that aberrant methylation of RARbeta2 occurs frequently in Tunisian breast cancer patients compared with others. 19809523_RARbeta and MGMT genes were found to be more frequently methylated in 70.58% and 58.8% of glioblastomas, respectively. 19828754_HBx induced promoter hypermethylation of RARbeta2, resulting in downregulation of its expression in hepatocellular carcinoma cells. 19829046_DNA hypermethylation of RARbeta2 is associated with inflammatory breast cancer. 19902255_Data show significant associations between SNPs in RARA, RARB, TOP2B and RARG, RXRA, TLR3, TRIM5 and RIG-I genes and rubella virus-specific cytokine immune responses. 19902255_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19940364_Promoter hypermethylation was observed in PRB 67%, ERalpha 64%, RASSF1A 63%, p16INK4A 51%, RARbeta2 22%, GSTP1 25% and BRCA1 27% of the breast cancers, respectively. 19945765_hypermethylation in tumors from non-small cell lung cancer patients is associated with gender and histologic type 19961602_RAR-beta2 mRNAs increase signs the normal differentiation of the human bronchial epithelium while a decrease is observed in most lung cancer cell lines 20302734_The expression of RAR-beta decreases significantly in cancer tissues in patients with colorectal cancer. Reduced expression is correlated negatively with the lymph node metastasis and advanced clinicopathological stage. 20346360_Observational study of gene-disease association. (HuGE Navigator) 20374520_Promoter methylation is responsible for silencing of RAR-beta2 in some melanoma cells and pan-acetylation of H3 likely plays a permissive role in expression of RAR-beta2. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20496080_HIN-1, RAR-beta, RASSF1A, and Twist had the ability to distinguish high-grade squamous intraepithelial lesions/squamous cell carcinomas from low-grade squamous intraepithelial lesions/negative cervical scrapings 20505321_Data provide evidence that RAR-beta promoter hypermethylation is a major mechanism involved in RAR-beta gene silencing, resulting in impaired RAR-beta function during breast cancer development. 20508731_Five single nucleotide polymorphism variants in RARbeta were detected in Chinese subjects with high myopia, none of them were associated significantly with high myopia. 20508731_Observational study of gene-disease association. (HuGE Navigator) 20535589_We investigated the utility of a set of four independent genes (P16, TSHR, RASSF1A and RARbeta2) promoter hypermethylation analysis as a diagnostic or prognostic tool in papillary thyroid tumors 20571967_RARbeta2 was frequently methylated in colorectal cancer and correlated with a worse prognosis and high expression of COX-2 suggesting a link between these two proteins in colorectal carcinogenesis. 20602615_Observational study of gene-disease association. (HuGE Navigator) 20606036_The higher methylation of RARbeta2 genes in prostate tissue samples from AA in comparison with Cau and may potentially contribute to the racial differences that are observed in prostate cancer pathogenesis. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20704749_Combined effects of cigarette smoke and hypermethylations of retinoic acid receptor beta are associated with non small cell lung cancer. 20711807_New insights into the possible effect of some lifestyle-related factors on the aberrant methylation drift of ERalpha and RARbeta2 genes in breast cancer. 20864173_Data show that methylation of at least one gene was detected in 18 patients (45%), the genes methylated were CDKN2B (20%), RASSF1 (18%), RARB (10%), CDH13 (7.5%) and FHIT (5%). 20865461_The methylation rate of RAR-beta2 in breast cancer and precancerous lesions of breast cancer were higher than that of normal tissues. 21162886_Chemotherapy response in esophageal squamous cell carcinoma is much more effective in RAR-beta positive patients than in RAR-beta negative patients. 21254357_One variant in the RARA gene (rs12051734), three variants in the RARB gene (rs6799734, rs12630816, rs17016462), and one variant in the RARG gene (rs3741434) were found to be statistically significant at p < 0.05 as risk factors for meningomyelocele. 21287504_The p16, DAPK and RAR beta genes methylation are strongly associated with clinical data of non-small cell lung cancer, and methylation of p16 and DAPK genes are associated with tobacco smoking. 21638515_This work demonstrates a dual role for RARB2 tumor suppression and tumor promotion 21680534_Methylated RARbeta(2) and APC genes might be valuable urinary molecular markers for early detection of bilharzial and nonbilharzial bladder cancer. 21795979_Circulating DNA-based testing verifies RARbeta2 methylation level is significantly increased in plasma from patients with non-small cell lung cancer compared with healthy individuals. 21822036_A significant association between high level of RARB methylation and Gleason score (P=0.01) in prostate cancer 21916701_Dietary intakes of folate, vitamins B2, B6, B12, and methionine were not associated with likelihood of promoter methylation of E- cadherin, p16, and RAR-beta2. 22116316_trend toward increasing methylation with severity of cervical squamous intraepithelial lesion cytology 22286019_A significant association was observed between urinary retinoic acid receptor-beta2 gene promoter methylation and hyaluronidase activity and advanced grade tumor. 22451234_Significant correlation was observed between promoter hypermethylation of estrogen receptor beta and RARb2 in tumors and paired sera. Breast cancer patients with RARb2 hypermethylation showed higher recurrence and shorter overall survival. 22487818_RARbeta gene promoter methylation and P53 mutation were detected in 58.1% and 36.4% of tumors, respectively. 22659417_LMP1 overcomes all-trans retinoic acid-induced apoptosis by repressing RAR-b2 expression via DNA methylation. 22693611_nucleolin plays a coregulatory role in transcriptional regulation of the tumor suppressor RARB2 by COUP-TFII 22694820_In papillary thyroid cancer, significant correlations between the methylation status of the RARbeta2 gene and the V600E BRAF mutation were found. 22901127_Low RARbeta expression is associated with advanced gastric cancer. 22964004_HIF-1alpha up-regulation is dependent on intracrine PGE2-RARbeta signalling. 22967446_RARbeta2 and p16(INK4alpha) hypermethylation was significantly related with pathological stage, lymph node metastasis, and invasion of nerves and vessels in patients with esophageal squamous cell carcinoma. 23102699_Results define PARG as a coactivator regulating chromatin remodeling during retinoic acid receptor-dependent gene expression. 23156677_Significant correlation was found between alterations of methylation status of miR-9-1, miR-9-3, miR-34b/c, miR-193a and the expression level of RAR-beta2 target gene in the primary lung tumors. 23299856_Second RARbeta2 loss of expression was found associated with different epigenetic profiles in LNCaP and DU145 cells. 23376149_Methylation of the RARB gene increases prostate cancer risk in black Americans. 23456936_HHIP, HDAC4, NCR3 and RARB polymorphisms may have a role in impaired lung function that begins in early life 23474497_findings showed that hepatitis C virus Core induced promoter hypermethylation of RAR-beta2 to inhibit its expression; accordingly, Core-expressing cells exhibited resistance to ATRA-induced growth inhibition 23504373_The highest loss of heterozygosity and/or microsatellite imbalance is revealed in RARB locus in non-small cell lung cancer. 23644172_EGFR transactivation by intracellular PGE2-activated EP receptors results in the sequential activation of RARbeta and HIF-1alpha 23675444_RARbeta2 methylation is associated with the risk of developing prostate cancer. (Meta-analysis) 23716670_Data show that TLR3 activation by poly I-C induces up-regulation of microRNA-29b, -29c, -148b, and -152 targeting DNA methyltransferases, leading to demethylation and reexpression of the oncosuppressor retinoic acid receptor beta (RARbeta). 23851445_The data suggest that RARbeta and p14ARF may be linked in a signaling pathway that is operating in melanocytic cells and is lost in the majority of melanomas. 23946634_All-trans retinoic acid inhibits human scleral fibroblasts proliferation by a mechanism associated with modulation of ERK 1/2 and JNK activation and depends on stimulation of retinoic acid receptor beta. 23969901_Differences in RAR and RXR subtype mRNA expression patterns in various PTCs may contribute to the immunochemistry data available, and may thus find exploitation in clinical oncology, particularly in the differential diagnosis of thyroid neoplasms 24050824_The current status of knowledge indicates that there might be inter- or overlapping actions between PPARg and RARs, and there might be an association of PPARg/RARs(RARa, RARb, and RARg) with renal diseases 24075189_Both recessive and dominant mutations in RARB cause anophthalmia. 24414001_RARB and STMN2 polymorphisms were not associated with sporadic CJD in the Korean population. 24492483_RARbeta promoter is hypermethylated in all studied prostate tumors and methylation levels are positively correlated with H3K27me3 enrichments. 24796328_promoter methylation may play an important role in carcinogenesis of the NSCLC. 24851851_Data indicate a differential sensitivity of chondral versus endochondral differentiation pathways to retinoic Acid receptor-Beta (RARbeta) signaling. 24913706_Results suggest that epigenetic alterations of P16INK4A (p16) and retinoic acid receptor beta (RAR-beta) have an important role in ovarian carcinogenesis. 24953041_These results highlight the relevance of MSK1 in the up-regulation of RARbeta by prostaglandin E2. 25040980_The total frequency of FHIT, RASSF1A and RARbeta gene methylation was significantly higher in lung cancer. 25053593_Our meta-analyses identified strong associations of four aberrantly methylated genes (GSTP1, RASSF1, p16, and RARB) with Prostate cancer 25069511_Analysis indicates that the genes BIRC5, HOXA1 and RARB are critical targets that play an important regulatory role in cervical cancer pathogenesis. 25110096_RARbeta2 circulating DNA in lung cancer patients is at greater methylation rate than in healthy donors. 25413479_RAR-beta, whereas ATRA alone had no effect on methylation. The RAR-beta gene was reexpressed following DAC-ATRA combination treatment, and both agents had no effect on p16 expression. 25422594_in the absence of retinoid signaling via RARbeta, reduced IGF-1 signaling results in suppression of epithelial-mesenchymal transition and delays tumorigenesis induced by the Wnt1 oncogene 25514805_Hypermethylation of RARB was associated with esophageal squamous cell carcinoma. 25605205_Methylation of the RAR-beta gene with cigarette smoking is associated with non-small cell lung cancer. 25684670_Methylated APC and RARbeta2 genes might be valuable serum-based molecular markers for early detection of breast cancer. 25805039_Hypermethylation at promoters of RARB, Dietary folate and cobalamin intake is inversely associated with methylated RARB and BRCA1. High dietary intake of riboflavin and pyridoxine is associated with increased methylation in the RARB promoter. 25970467_Retinoic acid receptor beta and angiopoietin-like protein 1 are involved in the regulation of human androgen biosynthesis 25981383_Curcumin upregulates RARbeta by inhibiting DNMT3b expression which reverses RARbeta promoter methylation. 26080448_revealed that HSP70 could directly regulate retinoid acid (RA)-induced retinoid acid receptor beta2 (RARbeta2) gene transcription through its binding to chromatin, with R469me1 being essential in this process 26352270_Whole-exome sequencing in a South American cohort links ALDH1A3, FOXN1 and RARB/retinoic acid regulation pathways to autism spectrum disorders. 26451736_RARbeta2 methylation is significantly increased in breast cancer samples when compared to non-cancerous controls 26681652_shows that RAR-beta methylation detected in lung tissue may be used as a predictive marker for non-small cell lung cancer diagnosis and that APC methylation in tumor sample may be a useful marker 26823825_methylated promoters of DAPK1 combined with MGMT, MGMT combined with RARB, DAPK1 combined with RARB were positive correlated with cervical disease grade 27011326_Overexpression of miR-146a-5p and miR-146b-5p caused a 31% and 33% decrease in endogenous RARB mRNA levels. 27120018_Study identified a novel mutation in RARB gene in patients with intellectual disability and progressive motor impairment which confers gain-of-function further promoting the retinoic acid (RA) ligand-induced transcriptional activity by twofold to threefold over the wild-type receptor. These results providing novel insight into the role of RA in neural networks in humans. 27306217_Lack of RARbeta nuclear expression is associated with Non-small cell lung cancer development and associated with a worse prognosis. 27342319_Data shows that miR-106a directly targets RARB 3'-UTR and the miR-106a-RARB complex promotes the viability of thyroid cancer. 27599527_Upregulation of RARB enhances the sensitivity of cholangiocarcinoma cells to chemotherapeutic agents in vitro. 27670690_mRNAs expression and methylation pattern of RARB, NR4A1 and HSD3B2 genes in human adrenal tissues (HAT) and in pediatric virilizing adrenocortical tumors (VAT) were analyzed. 28008143_Results indicate that RARbeta hypermethylation correlates well with an increased risk in non-small cell lung cancer (NSCLC) patients. RARbeta gene inactivation caused by RARbeta methylation contributes the NSCLC tumorigenesis and may serve as a potential risk factor, diagnostic marker and drug target of NSCLC. [review] 28639889_RARb and FHIT promoter methylation may be associated with the carcinogenesis of cervical cancer. FHIT promoter methylation may play a crucial role in cervical cancer progression. Additional studies with large sample sizes are essential to confirm our findings. 28759951_Hypermethylation frequency of CpG islands in the promoters of retinoic acid receptor beta gene (RAR-beta) gene was respectively 61 and 33% for diffuse and intestinal type n gastric cancer tissues. 29021580_retinoic acid receptor beta-retinoic X receptor alpha heterodimer quaternary architecture variable 29077166_RAR-beta expression state is one of the independent factors for the prognosis of lung squamous cell carcinoma patients 29329300_DBP(n) are non-toxic to the cells and have a weak overall demethylation effect on genomic DNA. DBP(n) demethylate the promoter regions of the tumor suppressor genes PTEN and RARB. DBP(n) promotes expression of the genes RARB, PTEN, CDKN2A, RUNX3, Apaf-1 and APC 'silent' in the MCF-7 because of the hypermethylation of their promoter regions 29421320_Knockdown of RARbeta by RARbeta siRNA increased the human adenoviruses production and the overexpression of RARbeta decreased the virus production. 29538221_There were significant associations between RARB promoter hypermethylation and Oral Cancer risk. 29554659_Although studies reported different rates for RAR beta promoter methylation in prostate can tissues, the total meta-analysis demonstrated that RAR beta promoter methylation may be correlated with prostate cancer carcinogenesis and that the RAR beta gene is particularly susceptible. 29851970_Hypermethylation of RARB gene is associated with high grade lung cancer. 29898214_Hypermethylation-induced RARbeta down-expression was associated with LS-VSCC and correlates with the upregulation of c-Jun. The degree of methylation of RARbeta promoter increased with the malignancy of LS-VSCC. Therefore, RARbeta gene dysregulation may play a role in progression of LS-VSCC. 29921692_Our results demonstrate that the majority of RARA-negative acute promyelocytic leukemia (APL) have RARB translocations, thereby forming a novel, distinct subgroup of APL. TBL1XR1-RARB as an oncogenic protein exerts effects similar to those of PML-RARA, underpinning the importance of retinoic acid pathway alterations in the pathogenesis of APL. 30055117_expression significantly reduced in cirrhotic and hepatocellular carcinoma tissues 30251476_A genetic variant of the RARB gene was associated with carotid intima-media thickness (CIMT) at the genome-wide level of significance. The present findings strongly suggest that genetic variation within RARB contributes to the development of subclinical atherosclerosis in patients with Rheumatoid Arthritis. 30252536_Results from this study revealed a novel anticancer mechanism of RARbeta via miR-22 induction to epigenetically regulate itself and NUR77, providing a promising cancer treatment modality using miR-22 and its inducers 30601638_The results of study demonstrate that increased level of expression in the first instance of isoform RARa in combination with hyper-expression of isoform RARbeta but not RARa can have unfavorable significance in case of evaluation of response to medicinal therapy and prognosis of total survival in patients with multiple melanoma. 30803219_High expression of RARB is associated with high grade of Cervical Pre-Neoplastic Lesions. 30944670_High expression of RARbeta was associated with overall survival in colorectal cancer patients. 31202904_Data provide a strong evidence for association involving RARB methylation on prostate cancer risk. [review] 31391070_our results defined NR1B2 as a tumor suppressor in KIRC that restricted EMT by the LATS1/2-YAP pathway. 31816153_High-throughput custom capture sequencing identifies novel mutations in coloboma-associated genes: Mutation in DNA-binding domain of retinoic acid receptor beta affects nuclear localization causing ocular coloboma. 31870161_By using two longitudinal cohort studies, we consistently found that rs1529672 in RARB could significantly modify and enhance the effect of polycyclic aromatic hydrocarbon (PAH) exposure on increased annual FEV1/FVC decline. Specifically, PAH exposure may lead to RARB hypermethylation, and the rs1529672-A allele was associated with decreased RARB mRNA expression. mRNA expression. 32278009_Retinoic acid receptor-beta prevents cisplatin-induced proximal tubular cell death. 32530576_DNA methylation patterns of RAR-beta2 and RASSF1A gene promoters in FNAB samples from Greek population with benign or malignant breast lesions. 32865619_Protein Kinase C Alpha (PKCalpha) overexpression leads to a better response to retinoid acid therapy through Retinoic Acid Receptor Beta (RARbeta) activation in mammary cancer cells. 34086186_MiR-141-3p and miR-200a-3p are involved in Th17 cell differentiation by negatively regulating RARB expression. 34470463_Polymorphisms in Vitamin A-Related Genes and Their Functions in Autoimmune Thyroid Disease. 35248720_Aberrant epigenetic regulation of RARbeta by TET2 is involved in cutaneous squamous cell carcinoma resistance to retinoic acid. | ENSMUSG00000017491 | Rarb | 8.819041e+01 | 0.7729477 | -0.371557315 | 0.4538572 | 6.485325e-01 | 0.4206378445 | 0.84954815 | No | Yes | 7.339930e+01 | 16.771551 | 8.401960e+01 | 19.598341 | |
| ENSG00000077463 | 51548 | SIRT6 | protein_coding | Q8N6T7 | FUNCTION: NAD-dependent protein deacetylase involved in various processes including telomere maintenance and gene expression, and consequently has roles in genomic stability, cell senescence and apoptosis (PubMed:18337721, PubMed:19135889, PubMed:19625767, PubMed:21362626). Has very weak deacetylase activity and can bind NAD(+) in the absence of acetylated substrate (PubMed:21362626). Has deacetylase activity towards histone H3K9Ac and H3K56Ac (PubMed:19625767, PubMed:21362626). Modulates acetylation of histone H3 in telomeric chromatin during the S-phase of the cell cycle (PubMed:19625767). May also be required for the association of WRN with telomeres during S-phase and for normal telomere maintenance (PubMed:18337721). Deacetylates histone H3K9Ac at NF-kappa-B target promoters and may down-regulate the expression of a subset of NF-kappa-B target genes (PubMed:21362626). Deacetylation of nucleosomes interferes with RELA binding to target DNA (PubMed:19135889). Acts as a corepressor of the transcription factor Hif1a to control the expression of multiple glycolytic genes to regulate glucose homeostasis (By similarity). Required for normal IGF1 serum levels and normal glucose homeostasis (By similarity). Regulates the production of TNF protein (By similarity). Has a role in the regulation of life span (By similarity). {ECO:0000250|UniProtKB:P59941, ECO:0000269|PubMed:18337721, ECO:0000269|PubMed:19135889, ECO:0000269|PubMed:19625767, ECO:0000269|PubMed:21362626}. | 3D-structure;Acetylation;Alternative splicing;Disease variant;Metal-binding;NAD;Nucleus;Phosphoprotein;Reference proteome;Transferase;Zinc | This gene encodes a member of the sirtuin family of NAD-dependent enzymes that are implicated in cellular stress resistance, genomic stability, aging and energy homeostasis. The encoded protein is localized to the nucleus, exhibits ADP-ribosyl transferase and histone deacetylase activities, and plays a role in DNA repair, maintenance of telomeric chromatin, inflammation, lipid and glucose metabolism. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Mar 2016]. | hsa:51548; | chromosome, subtelomeric region [GO:0099115]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; deacetylase activity [GO:0019213]; histone deacetylase activity [GO:0004407]; NAD(P)+-protein-arginine ADP-ribosyltransferase activity [GO:0003956]; NAD+ ADP-ribosyltransferase activity [GO:0003950]; NAD+ binding [GO:0070403]; NAD-dependent histone deacetylase activity [GO:0017136]; NAD-dependent histone deacetylase activity (H3-K9 specific) [GO:0046969]; NAD-dependent protein deacetylase activity [GO:0034979]; transcription corepressor activity [GO:0003714]; zinc ion binding [GO:0008270]; base-excision repair [GO:0006284]; glucose homeostasis [GO:0042593]; histone H3 deacetylation [GO:0070932]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of gene expression, epigenetic [GO:0045814]; negative regulation of glucose import [GO:0046325]; negative regulation of glycolytic process [GO:0045820]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of blood vessel branching [GO:1905555]; positive regulation of chondrocyte proliferation [GO:1902732]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of stem cell proliferation [GO:2000648]; positive regulation of telomere maintenance [GO:0032206]; positive regulation of transcription factor catabolic process [GO:1901485]; positive regulation of vascular endothelial cell proliferation [GO:1905564]; protein ADP-ribosylation [GO:0006471]; protein deacetylation [GO:0006476]; protein destabilization [GO:0031648]; regulation of double-strand break repair via homologous recombination [GO:0010569]; subtelomeric heterochromatin assembly [GO:0031509] | 18242175_Levels of the mammalian sirtuin, SIRT6, increased upon nutrient deprivation in cultured cells, in mice after fasting, and in rats fed a calorie-restricted diet. 18337721_the first identification of a physiological enzymatic activity of SIRT6 (as histone H3 lysine 9 deacetylase), and linking chromatin regulation by SIRT6 to telomere maintenance and a human premature ageing syndrome 18388907_suggests similarities in the molecular mechanisms by which SIRT6 and WRN may prevent premature aging-like phenotypes 19625767_Study identifies H3K56Ac as a novel substrate for SIRT6, a role for SIRT6 in modulating this chromatin mark at telomeric chromatin. H3K56Ac levels in human cells are differentially regulated at telomeres and globally in response to cell cycle arrest. 20157594_SIRT6 associates dynamically with chromatin in response to DNA damage, and stabilizes the DNA double-strand break (DSB) repair factor, DNA-dependent protein kinase (DNA-PK), at DSBs. 21224216_These findings suggest that accelerated epithelial senescence plays a role in idiopathic pulmonary fibrosis pathogenesis through perpetuating abnormal epithelial-mesenchymal interactions, which can be antagonized by SIRT6. 21349544_This study suggested that altered SIRT1, 2 and 6 expression is state-dependent and might be associated with the pathogenesis and/or pathophysiology of mood disorders. 21362626_SIRT6 could act as an NAD(+) metabolite sensor 21502801_The 25% CR diet increased the expression of both SIRT1 and SIRT6 in the ovary. 21680843_results indicate that SIRT6 physically associates with PARP1 and mono-ADP-ribosylates PARP1 on lysine residue 521, stimulating PARP1 poly-ADP-ribosylase activity and enhancing DSB repair under oxidative stress 21847107_SIRT6 is important for maintaining telomeric chromatin structure that is required for silencing of nearby genes. 21900744_SIRT6 overexpression selectively kills cancer cells. 21981042_SIRT6 knockdown in HDFs influence the synthesis and degradation of collagen by hyperactive NF-kappaB signalling, which leads to a decrease in dermal collagen fibrils. SIRT6 may therefore play an important role in the process of skin anti-aging. 22087257_observed significant associations between genetic variants in the SIRT6 and UCP5 genes and atherosclerotic plaque 22700961_in TLR4-stimulated promonocytes SirT1 and SirT 6 support a switch from increased glycolysis to increased fatty acid oxidation as early inflammation converts to late inflammation. 22743824_Sirt6 was enriched in the nucleolus in the G 1 phase of the cell cycle, while S phase nucleoli were almost entirely free of Sirt6. 22753495_Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. 22792191_We observed that overexpression of SIRT6 had little influence on NFkappaB-dependent genes, but overexpression of the catalytically inactive mutant affected gene expression in developmental pathways. 23041974_Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. 23075334_Studies indicate that SIRT1, SIRT3 and SIRT6 are induced by calorie restriction conditions and are considered anti-aging molecules. 23086953_NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses 23132960_Results demonstrate that the loss of SIRT6 in endothelial cells is associated with upregulation of genes involved in inflammation, vascular remodelling, and angiogenesis. 23136298_SIRT6 plays a role in regulating the terminal effector pathways of human labor and delivery via the NFKB pathway. 23201774_SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. 23217706_By using a combination of in vitro and in vivo studies, as well as data from several human cancer databases, study demonstrated that loss of SIRT6 leads to tumorigenesis, and its expression is selectively downregulated in several human cancers. 23514751_SIRT6 is upregulated in both paclitaxel-resistant and epirubicin-resistant MCF-7 cells. 23526469_Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. 23552949_the crystal structure of SIRT6 reveals a large hydrophobic pocket that can accommodate long-chain fatty acyl groups; SIRT6 is an enzyme that controls protein lysine fatty acylation 23653361_Data indicate that miR-766 regulates SIRT6 expression posttranscriptionally. 23860128_SIRT6 is a direct target of miR-34a in primary human keratinocytes and its down-modulation is sufficient to reproduce miR-34a pro-differentiation effects. 23928404_Endotoxin and tumor necrosis factor-alpha suppressed SIRT1, SIRT3, and SIRT6 expression in human monocytes 23982738_Tissue micro-array studies confirmed the higher levels of SIRT6 in both prostate tumor tissues and prostate cancer cells than in their normal counterparts. Knockdown of SIRT6 in human prostate cancer cells led to sub-G1 phase arrest of cell cycle, increased apoptosis, elevated DNA damage level and decrease in BCL2 gene expression. 24043303_CHIP noncanonically ubiquitinates SirT6 at K170, which stabilizes SirT6 and prevents SirT6 canonical ubiquitination by other ubiquitin ligases. 24052263_Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. 24163442_our study reveals putative means of regulation of SIRT6 functions via interactions and modifications, providing an important resource for future studies on the molecular mechanisms underlying sirtuin functions. 24169447_Extended analysis of the SIRT6 interaction with G3BP1, a master stress response factor, uncovers an unexpected role and mechanism of SIRT6 in regulating stress granule assembly and cellular stress resistance. 24171769_REVIEW: current information regarding the molecular and physiological relevance of SIRT6 in the context of epigenetics, metabolism and disease 24366394_Overexpression of Sirt6 in the HepG2 HCC cell line exhibited antitumor effects through the induction of apoptosis and the inhibition of the ERK1/2 signaling pathway. 24367027_SIRT6 expression decreased in COPD lung homogenates correlating with lung function tests. Overexpression induced autophagy. SIRT6 affects CSE-induced HBEC senescence via autophagy regulation attributed to attenuation of IGF-Akt-mTOR signaling. 24438746_SIRT6 impacts upon cellular homeostasis by regulating DNA repair, telomere maintenance, and glucose and lipid metabolism, thus affecting diseases such as diabetes, obesity, heart disease, and cancer. (Review) 24607900_SIRT6 inhibits glioma cell proliferation and colony formation in vitro and glioma cell growth in vivo in a PCBP2 dependent manner. SIRT6 inhibits PCBP2 expression through deacetylating H3K9ac and SIRT6 acts as a tumor suppressor in glioma. 24638860_SIRT6 regulates the cigarette smoke-induced signalling in rheumatoid arthritis synovial fibroblasts 24782448_Interactions of two highly acetylated proteins, nucleophosmin (NPM1) and nucleolin, with SIRT6 and SIRT7 were confirmed by co-immunoprecipitation. 24842653_The methylation status of CTCF and SIRT6 promoter regions was not statistically different in cancer lesions compared with matched normal tissues. 25009184_p53 directly activates expression of the NAD(+)-dependent histone deacetylase sirtuin 6 (SIRT6), whose interaction with FoxO1 leads to FoxO1 deacetylation and export to the cytoplasm. 25074979_MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 has a role in promoting tumorigenesis and trastuzumab resistance in breast cancer 25083875_Study reports that SIRT6 defines the circadian oscillation of a distinct group of hepatic genes, different from the ones under SIRT1 control. 25162034_In conclusion, these results suggest that Sirt6 is a critical regulator of endothelial senescence and oxidative stress-induced downregulation of Sirt6 is likely involved in the pathogenesis of diabetic retinopathy. 25175731_Newly discovered S-sulfenylations suggested novel redox mechanisms, including S-sulfenyl-mediated redox regulation of the transcription factor HIF1A by SIRT6. 25181338_The histone acetyltransferase hMOF suppresses hepatocellular carcinoma growth by targeting the expression of SIRT6. 25197348_these results demonstrated for the first time that SIRT6 suppressed non-small cell lung cancer cells proliferation via down-regulation of Twist1 expression 25227832_The over-expression of adenovirus-mediated SIRT6, which has radiosensitization effect on A549 cells of NSCLC, can inhibit the proliferation of A549 cells and cause G0/G1 phase retardation as well as induce apoptosis of cells. 25304127_review describes the protective effects of SIRT1 and SIRT6 against COPD and their target proteins involved in the pathophysiology of COPD. 25320180_SIRT6 functions as an oncogene in the epidermis. 25325735_These findings establish the involvement of SIRT6 in the inflammatory pathways of diabetic atherosclerotic lesions and suggest its possible positive modulation by incretin. 25361925_This study demonstrated that SIRT6 downregulation in the liver of non-alcoholic fatty liver disease patients. 25400728_The differential expression patterns of SIRT6 amongst different T stages of muscle invasive bladder cancers indicate less reliance on glycolysis when urothelial carcinoma invades deeper through the bladder and into the adjacent tissues. 25503141_In the cancer group, the expression level of SIRT6 and SIRT7 were significantly up-regulated and are potential circulating prognostic markers for head and neck squamous cell carcinoma. 25541994_CC or CT genotype at rs107251 within SIRT6 displayed >5-year mean survival advantages compared to the TT genotype in age cohort. 25607651_SIRT6 regulates base excision repair in a PARP1-depdendent manner. 25660418_SIRT2 and SIRT6 are expressed in retinoblastoma, as well as in some normal ocular structures. 25683165_Sirtuin 6 promotes transforming growth factor-beta1/H2O2/HOCl-mediated enhancement of hepatocellular carcinoma cell tumorigenicity by suppressing cellular senescence. 25713071_Data indicate that cyclic AMP signaling reduces the expression of sirtuin 6 deacetylase via inhibition of the c-Raf-MEK1-ERK1/2 pathway. 25777360_ITCH modulates SIRT6 and SREBP2 to influence lipid metabolism and atherosclerosis in ApoE null mice 25807436_SIRT6 expression was significantly reduced in human ovarian cancer tissues compared to the normal tissues. 25808624_These results support a hypothesis whereby RUNX2-mediated repression of the SIRT6 tumor suppressor regulates metabolic pathways that promote breast cancer progression 25816777_Indicate a novel oncogenic role of E2F1 by enhancing glycolysis via suppression Sirt6 transcription in bladder and prostate tumor cell lines. 26114387_associations between SIRT6 rs107251 and pulse wave velocity, as well as an interaction between SIRT6 rs107251 and soybean intake for different levels of pulse wave velocity were found. 26180037_SIRT6 expression in NSCLC could be a useful prognostic marker and that SIRT6 might represent a novel target gene for predicting sensitivity of chemotherapy in lung adenocarcinoma. 26183563_Data show that SIRT6, a member of the sirtuin family, induces apoptosis, and its induction is attributed to the repression of survivin protein, which could be a target for endometrial cancer treatment. 26332421_Findings suggest that polymorphisms in SIRT6/UCP1 genes may be important for increased carotid plaque burden and echodensity 26456828_SIRT6 mutations identified in human cancers alter either SIRT6 stability, localization, or enzymatic activity. Deacetylase, not demyristoylase, activity is critical for SIRT6 tumor-suppressor. 26549451_Lamin A promotes SIRT6-dependent DNA-PKcs (DNA-PK catalytic subunit) recruitment to chromatin. 26639398_These data strongly suggest that overexpression of Sirt6 can prevent osteoarthritis development by reducing both the inflammatory response and chondrocytes senescence. 26648570_our data suggest that SIRT6 suppresses glioma cell growth via induction of apoptosis, inhibition of oxidative stress and inhibition of the activation of the JAK2/STAT3 signaling pathway. 26657850_EGF stimulates loss of FOXO3/SIRT6, which is blockaded by the inhibition of upstream pathways as well as lipid synthesis, revealing existence of a negative regulatory loop between LDs and FOXO3/SIRT6. 26675349_These findings therefore provide insights into the functional interplay between SIRT6 and DNA repair mechanisms, with implications for both tumorigenesis and the treatment of multiple myeloma. 26701732_Data indicate that compared to non-neoplastic endometria (NNE), endometrial cancer (EC) showed SIRT7 mRNA overexpression, whereas SIRT1, SIRT2, SIRT4 and SIRT5 were underexpressed, and no significant differences were observed for SIRT3 and SIRT6. 26711340_Results reveal a pathway controlled by ATM, SIRT6, and SNF2H to block HUWE1, which stabilizes H2AX and induces its incorporation into chromatin only when cells are damaged. 26768768_SIRT6 in a protein complex with both nuclear factor erythroid 2-related factor 2 (NRF2) and RNA polymerase II. 26787900_demonstrate that PKM2 deacetylation is integral to SIRT6-mediated tumor suppression and inhibition of metastasis. Additionally, reduced SIRT6 levels correlate with elevated nuclear acetylated PKM2 levels in increasing grades of hepatocellular carcinoma 26802595_Results find no significant association between common polymorphisms and haplotypes in the SIRT6 gene and longevity. 26803106_this study shoes that through the antiglycolytic activity of SIRT6, the autophagy is suppressed, which is beneficial to nasal polyp formation 26861461_in vitro and in vivo studies showed that gene silencing of SIRT6 suppressed cell proliferation and promoted cellular apoptosis by activating the Bax-dependent apoptotic signal pathway in Hepatocellular Carcinoma cells. Furthermore, SIRT6 knockdown could increase liver cancer cell sensitivity to chemotherapy drug doxorubicin. SIRT6 is an important protumorigenic factor in liver carcinogenesis. 26886147_The promoter regions of the SIRT6 gene were genetically analyzed in large cohorts of MI patients (n = 371) and ethnically-matched controls (n = 383). Results: A total of 15 DNA sequence variants (DSVs) were identified, including seven single-nucleotide polymorphisms (SNPs). Two novel heterozygous DSVs, g.4183823G>C and g.4183742G>A, were identified in two MI patients but in none of the controls. 26898756_Thus, these results reveal that SUMOylation has an important role in regulation of Sirt6 deacetylation on H3K56, as well as its tumor suppressive activity. 26983852_SIRT6 induced autophagy via attenuation of AKT signaling and treatment with autophagy inhibitor 3-MA or knockdown of autophagy-related protein Atg5 rescued H2O2-induced neuronal injury. 26987016_we examined the role and mechanisms of SIRT6 in suppressing postoperative epidural scar formation. We showed that SIRT6 promoted the expression of miR-21 and then suppressed TGF-beta2 expression in a targeted manner. 27041572_HBx increased signs of DNA damage such as accumulation of 8-hydroxy-2'-deoxyguanosine and comet formation, which were reversed by overexpression of PARP1 and/or Sirt6..physical interaction of HBx and PARP1 accelerates DNA damage by inhibiting recruitment of the DNA repair complex to the damaged DNA sites, which may lead to the onset of hepatocarcinogenesis 27043296_SIRT6 promotes deacetylation of a new substrate, residue K18 of histone H3 (H3K18), and inactivation of SIRT6 in cells leads to H3K18 hyperacetylation and aberrant accumulation of pericentric transcripts. 27101740_These data provide the first evidence of the Egt ability to interfere with endothelial senescence linked to hyperglycaemia through the regulation of SIRT1 and SIRT6 signaling, thus further strengthening the already assessed role of these two histone deacetylases in type 2 diabetes. 27118880_Data provided evidence that SIRT6 tagSNPs rs352493 and rs3760908 play significant roles in the severity of coronary artery disease (CAD) in Chinese Han subjects, which might be useful predictors of the severity of CAD. 27156849_p53-dependent SIRT6 expression protects cells from Abeta42-induced DNA damage. 27180906_SIRT6 as an important pancreatic ductal adenocarcinoma tumor suppressor 27249230_Loss- and gain-of-SIRT6 function studies in cultured human endothelial cells (ECs) showed that SIRT6 attenuated monocyte adhesion to ECs. 27322069_The data indicate that distinct activities of SIRT6 regulate different pathways and that the G60A mutant is a useful tool to study the contribution of defatty-acylase activity to SIRT6's various functions. 27496874_SIRT6, TNF-alpha, and IFN-gamma were significantly higher in lesional than perilesional and control skin. 27534902_In the model of CIA, forced expression of SIRT6 ameliorated disease progression, osteoblastic synthesis of Cyr61, and macrophage recruitment. More importantly, expression of LDHA and oxidative lesions were decreased in osteoblasts of SIRT6-treated joints. Our findings suggest that SIRT6 suppresses inflammatory response in osteoblasts via modulation of glucose metabolism and redox homeostasis. 27536992_positive regulator of aldose reductase expression in U937 and HeLa cells under osmotic stress 27568560_post-translational modification facilitates the mobilization of SIRT6 to DNA damage sites and is required for efficient recruitment of poly (ADP-ribose) polymerase 1 (PARP1) to DNA break sites and for efficient repair of double-strand break. 27746184_This study demonstrates that CSNK2A1 and SIRT6 are indicators of poor prognosis for breast carcinomas and that CSNK2A1-mediated phosphorylation of SIRT6 might be involved in the progression of breast carcinoma. 27766571_Strong correlation has been proved between the expression levels of HDAC4 and SIRT6. 27777384_SIRT6 is upregulated in non-small cell lung cancer; it may have a functional role in promoting migration and invasion through ERK1/2/MMP9 signaling 27794562_The expression of SIRT6 was reduced during cellular senescence, whereas enforced SIRT6 expression promoted cell proliferation and antagonized cellular senescence.Furthermore, SIRT6 directly interacted with p27. Finally, SIRT6 markedly rescued senescence induced by p27. 27824900_acts as a tumor promoter by preventing DNA damage and cellular senescence in hepatocellular carcinoma 27923994_our findings describe TRF2 as a novel SIRT6 substrate and demonstrate that acetylation of TRF2 plays a crucial role in the regulation of TRF2 protein stability, thus providing a new route for modulating its expression level during oncogenesis and damage response. 28215636_SIRT6 prevents matrix degradation of nucleus pulposus via the NF-kappaB signaling pathway in intervertebral disc degeneration. SIRT6 physically interacted with nuclear factor-kappaB (NF-kappaB). 28228253_SIRT6 overexpression suppresses PI3K signaling. 28238784_Data show that the increased acetylation of Ku autoantigen 70kDa (Ku70) in sirtuin 6 protein (SIRT6)-depleted cells disrupt its interaction with Bax apoptosis regulator protein (Bax), which finally resulted in Bax mitochondrial translocalization. 28250020_In obese patients, the expression of Sirt6 expression is reduced. 28296196_Data suggest, in macrophages, SIRT6 plays role in preventing atherosclerosis by reducing foam cell formation via autophagy-dependent pathway involving regulation of expression of ATG5, LC3B, LAMP1, ABCA1, ABCG1, and MIRN33. (ATG5 = autophagy-related protein 5; LC3B = microtubule-associated protein 1 light chain 3 beta; LAMP1 = lysosomal-associated membrane protein 1; ABCA1/ABCG1 = ATP-binding cassette transporters 1/8) 28355558_SIRT6 and its downstream signaling could be targeted in Alzheimer's disease and age-related neurodegeneration. 28393212_these results suggest that SIRT6 enhances cell aggressiveness in PTC via BRAF/ERK/Mcl1 pathway, and thus may be a promising target in the treatment of the disease. 28399814_The single nucleotide polymorphisms rs117385980 (C;T) in sirtuin 6 , situated 23 bases downstream of the exon 2 exon/intron border was found in heterozygous form in 1/43 longer-living healthy men (Minor allele frequency (MAF) 0,0116) and in 9/92 controls. 28443459_Taken together, these data demonstrated that astragaloside IV sensitized tumor cells to gefitinib via regulation of SIRT6, suggesting that astragaloside IV may serve as potential therapeutic approach for lung cance 28478957_SIRT6 over-expression establishes a condition whereby reconfiguration of the Hexokinase 2 promoter chromatin structure makes it receptive to interaction with MZF1/SIRT6 complex, thereby favouring a regulatory state conducive to diminished transcription 28653878_our studies shed insights into the crucial functions of sirtuin 6 in esophageal carcinoma cells and provide evidence supporting sirtuin 6-based personalized therapies in esophageal carcinoma cell patients 28656307_Low SIRT6 expression is associated with gastric cancer. 28661724_We provide a comprehensive overview of recent developments on the molecular signaling pathways controlled by SIRT1 and SIRT6, two post-translational modifiers proven to be valuable tools to dampen inflammation and oxidative stress at the cardiovascular level 28677777_Low SIRT6 expression is associated with glioma. 28685526_These findings reveal a previously unknown role for nasal mucosa steady-state conditions in the control of Sirt6 activity, and provide evidence for a relationship between HMGB1 and Sirt6 in chronic rhinosinusitis with nasal polyps (CRSwNP), and promising benefits of glycyrrhetinic acid for CRSwNP patients. 28723567_SIRT6 interacts with and promotes phospho-ATF2 binding to the PGC-1alpha gene promoter to activate its expression. The present study reveals a critical role for SIRT6 in regulating thermogenesis of fat. 28786706_Low SIRT6 expression is associated with Hodgkin lymphoma. 28871079_Sirt6 inhibits Notch1 and Notch4 transcription by deacetylating histone H3K9. 28887543_These results suggest that direct and reversible cysteine thiol 144 may play a functional role in SIRT6-dependent control over monocyte glycolysis, an important determinant of effector innate immune responses. 28921546_Low SIRT6 expression is associated with bone marrow metastasis in neuroblastoma. 28935467_Previously unknown reciprocal influence of SIRT6 and HK2 in regulating autophagy driven monocyte differentiation. 28983623_miR378b represses the mRNA expression levels of COL1A1 via interference with SIRT6 in human dermal fibroblasts. 29025907_our data suggest that inactivation of SIRT6 in leukemia cells leads to disruption of DNA-repair mechanisms, genomic instability and aggressive AML. This synthetic lethal approach, enhancing DNA damage while concomitantly blocking repair responses, provides the rationale for the clinical evaluation of SIRT6 modulators in the treatment of leukemia 29197589_Study shows that SIRT6 protein levels are lower in patients with prediabetes (PreDM) and type 2 diabetes mellitus (T2DM) and implies that SIRT6 may take play in development T2DM by altering the expression of genes involved in glucose metabolism through histone modification rather than its role in DNA repair. 29215322_that the bimodal expression of SIRT6 at different melanoma stages plays a critical role in regulating melanoma growth through an autophagy-dependent manner, which indicates the potential of SIRT6 to be a biomarker and a therapeutic target in melanoma 29217762_Low SIRT6 expression is associated with Liver Tumorigenesis. 29227545_experiments revealed the mechanism for SIRT6 in facilitating tumorigenesis and metastasis of colon cancer cells, suggesting that SIRT6 might be a potential therapeutic target for treating colon cancer. 29233643_this study not only suggests potential roles of SIRT6 in regulating apoptosis and stress resistance via direct deacetylation of p53, but also provides lead compound for the development of potent and selective SIRT6 inhibitors. 29317652_SirT6 promotes cysteine ubiquitination in the PRE-SET domain of Suv39h1. 29352194_Data showed that sirt6 attenuated cell senescence, and reduced apoptosis, by triggering autophagy that ultimately ameliorated IDD. 29363378_Downregulation of SIRT6 expression may promote non-small cell lung cancer malignancy in the Chinese Han population. 29387864_Sirt6 plays an important role in protecting intestinal epithelial cells against inflammatory injury in a mechanism associated with preserving Rspo1 levels in the cells. 29465379_nf-kappab was was increases duto to SIRT6 silencing in the absence of UV-B 29476161_The results demonstrate the separation of function between SIRT6 catalytic activities and suggest that SIRT6 deacylation activity in cells is important for glucose metabolism and can be mediated by still unknown acylated cellular proteins. 29555651_SIRT6 is a key factor in human development and identifying the first mutation in a chromatin factor behind a human syndrome of perinatal lethality. 29599436_SIRT6 recruits the chromatin-remodeling protein SNF2H to damaged telomeres, which appears to promote chromatin decondensation independent of its deacetylase activity. 29659670_SIRT6 may suppress cell proliferation, migration, and invasion via inhibition of the NOTCH3 signaling pathway in glioma 29686974_SIRT6 negatively regulates Dengue virus-induced inflammatory response via RIG-I-like receptor and Toll-like receptor 3 signaling pathways. 29957460_SIRT6 inhibited proliferation, migration, and invasion of colon cancer cells by up-regulating PTEN expression and down-regulating AKT1 expression. 29966233_The role of SIRT6 as a regulator or even a nutrient detector in cells has diversified its impact on aging-related processes and major human diseases such as cancer, diabetes, neurodegenerative diseases, and heart disease. SIRT6 has been shown to be involved in gene expression in the nucleus with regards to chromatin and, more recently, has also been shown to take part SG formation in the cytosol. [review] 30032073_Overexpression of SIRT6 reduced acetylation of Ku70 and promoted interaction of Ku70 with the proapoptotic protein Bax. 30230565_SIRT6 prevents lung fibroblast-to-myofibroblast differentiation through inhibition of the TGF-beta1/Smad2 and NF-kappaB signaling pathways. 30250025_These results clearly demonstrate that pharmacological activation of SIRT6 triggers an increase of autophagy that results in apoptosis in different human cancer cells. 30256390_SIRT6 overexpression inhibits cementogenesis by suppressing the activity of glucose transporter 1. 30402853_We showed that sirtuin 6 facilitates CP-induced myeloma cell KM-HM_(31) aging via suppressing Hippo 30409187_This study showed that the SIRT6 plays a pathogenic and pro-inflammatory role in Parkinson's and that nicotine can provide neuroprotection by accelerating its degradation. Inhibition of SIRT6 may be a promising strategy to ameliorate Parkinson's and neurodegeneration. 30429089_Our data identify a role of myeloid Sirt6 in clinical and experimental RA and suggest that myeloid Sirt6 may be an intriguing therapeutic target. 30483801_FOXN3 was notably downregulated in osteosarcoma (OS) tissues and the expression of FOXN3 was negatively correlated with tumor size, metastasis and TNM stage. Also, FOXN3 suppressed the proliferation, migration and invasion of osteosarcoma cells. Furthermore, FOXN3 was demonstrated to transcriptionally suppress SIRT6 expression, thereby inhibiting MMP9 secretion. 30514106_SIRT6 affects intracellular NAMPT activity. 30542728_Study findings present a novel mechanism that controls FOXO3 activation and revealed that SIRT6 is a pivotal regulatory factor in determining liver cancer chemosensitivity. SIRT6 interacts with FOXO3 and this interaction increases FOXO3 ubiquitination and decreases its stability. 30577808_Findings provide mechanistic insights into the pro-metastatic activity of SIRT6 and highlight the role of the SIRT6/Snail/KLF4 axis in regulating EMT and invasion of NSCLC cells. 30587342_The findings unveiled that Long non-coding RNA LINC00319 contributed to acute myeloid leukemia (AML) leukemogenesis via elevating SIRT6 expression, indicating a possible molecular target of LINC00319 for AML treatment. 30641770_SIRT6 gene expression is altered in adipose tissue from overweight and obese subjects compared to normal weight subjects. 30651359_SIRT6 interacts with KSHV DNA. 30700227_Overexpression of SIRT6 can protect kidney epithelial cell damage of acute kidney injury by inhibiting apoptosis and inducing autophagy, which might become a new approach to the treatment of acute kidney injury. 30787391_Sirtuin 6 in preosteoclasts suppresses age- and estrogen deficiency-related bone loss by stabilizing estrogen receptor alpha. 30858544_Sirtuin-mediated hnRNP A1 deacetylation inhibits Hepatocellular carcinoma cell proliferation and tumorigenesis in a PKM2-dependent manner. These findings point to the metabolic reprogramming induced by hnRNP A1 acetylation in order to adapt to the nutritional status of the tumor microenvironment. 30987683_Hyperglycemia appeared to promote the mRNA expression of SIRT6, TET2 and TE3, which in turn might cause the dynamic changes of 5mC and 5hmC in white blood cells from type 2 diabetes mellitus patients. 30989475_research findings demonstrate that SIRT6 serves as a tumor suppressor via regulation of the NF-kappaB pathway, which could offer an innovative strategy to treat adrenocortical carcinoma. 31004738_Both in vitro and in vivo experiments confirmed that FKHRL1-SIRT6 axis played a pivotal role in cell metabolism and tumor growth. The results indicate that FKHRL1-SIRT6 axis regulates cell metabolism and may provide clues for glioblastoma treatment. 31113929_Adipocyte Sirt6 regulates body weight gain and insulin sensitivity independent of diet, and the increased IL-4 production by Sirt6 and resultant M2 polarization of adipose tissue macrophages may attenuate proinflammatory responses in adipose tissue. 31149050_The expression level of SIRT6 in patients' specimens is lower than that of normal controls, and patients with higher SIRT6 level have a better prognosis. Transcriptional factor FoxO3a is a direct up-stream of SIRT6 and positively regulated SIRT6 expression, which in turn, promotes apoptosis by activating Bax and mitochondrial pathway. 31257493_SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/ERK2 signaling pathway. 31265839_SIRT6 protects vascular endothelial cells from angiotensin II-induced apoptosis and oxidative stress by promoting the activation of Nrf2/ARE signaling 31295533_The epigenetic regulator SIRT6 protects the liver from alcohol-induced tissue injury by reducing oxidative stress in mice. 31321952_Study shows that mammalian sterile 20-like 1 suppresses viability and promotes apoptosis of glioma cells via upregulating Sirtuin 6 expression. 31399344_results identified SIRT6 as a Pol II promoter-proximal pausing-dedicated histone deacetylase. 31437090_The multiple roles of SIRT6, a chromatin deacylase with unique and important functions in maintaining cellular homeostasis are REVIEWED. The ability of SIRT6 to interconnect chromatin dynamics with metabolism and DNA repair is discussed. 31442424_SIRT6 is the FoxO1 deacetylase suppressed by mTORC2 and show an endogenous interaction between SIRT6 and mTORC2 in both mouse and human cells. 31477582_MiR-128 inhibits the osteogenic differentiation in osteoporosis by down-regulating SIRT6 expression. 31541078_in liver tissues obtained from patients with nonalcoholic fatty liver disease (NAFLD), the extent of XBP1s acetylation correlated positively with the NAFLD activity score but negatively with the Sirt6 level. 31551254_These results established a relationship between SIRT6 and hepatocellular carcinoma (HCC) epithelial-mesenchymal transition (EMT) and further elucidated the mechanisms underlying HCC metastasis, helping provide a promising approach for the treatment of HCC. 31591350_Evidence shows that SIRT6 overexpression or knockout in cells can result in different biological responses as tumor suppressor or promoter, depending on cell and tumor type. [review] 31603249_Sirt6 attenuates hypoxia-induced tubular epithelial cell injury via targeting G2/M phase arrest. 31626739_Long noncoding RNA HIF1A-AS2 facilitates cell survival and migration by sponging miR-33b-5p to modulate SIRT6 expression in osteosarcoma. 31696978_T | ENSMUSG00000034748 | Sirt6 | 1.571370e+03 | 1.1496358 | 0.201176915 | 0.3364923 | 3.562128e-01 | 0.5506171155 | 0.89250744 | No | Yes | 1.507679e+03 | 210.800122 | 1.177399e+03 | 169.209701 | |
| ENSG00000077684 | 79960 | JADE1 | protein_coding | Q6IE81 | FUNCTION: Scaffold subunit of some HBO1 complexes, which have a histone H4 acetyltransferase activity (PubMed:16387653, PubMed:19187766, PubMed:20129055, PubMed:24065767). Plays a key role in HBO1 complex by directing KAT7/HBO1 specificity towards histone H4 acetylation (H4K5ac, H4K8ac and H4K12ac), regulating DNA replication initiation, regulating DNA replication initiation (PubMed:20129055, PubMed:24065767). May also promote acetylation of nucleosomal histone H4 by KAT5 (PubMed:15502158). Promotes apoptosis (PubMed:16046545). May act as a renal tumor suppressor (PubMed:16046545). Negatively regulates canonical Wnt signaling; at least in part, cooperates with NPHP4 in this function (PubMed:22654112). {ECO:0000269|PubMed:15502158, ECO:0000269|PubMed:16046545, ECO:0000269|PubMed:16387653, ECO:0000269|PubMed:19187766, ECO:0000269|PubMed:20129055, ECO:0000269|PubMed:22654112, ECO:0000269|PubMed:24065767}. | Acetylation;Activator;Alternative splicing;Apoptosis;Cell projection;Chromosome;Cytoplasm;Cytoskeleton;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:79960; | chromosome [GO:0005694]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; transcription coactivator activity [GO:0003713]; apoptotic process [GO:0006915]; histone H3 acetylation [GO:0043966]; histone H4-K12 acetylation [GO:0043983]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; histone modification [GO:0016570]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of cell growth [GO:0030308]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; regulation of cell cycle [GO:0051726]; regulation of cell growth [GO:0001558]; regulation of DNA biosynthetic process [GO:2000278]; regulation of DNA replication [GO:0006275]; regulation of transcription, DNA-templated [GO:0006355] | 12169691_Jade-1 protein is a novel candidate regulatory factor in Von Hippel Lindau-mediated renal tumor suppression 15502158_Jade-1 is a novel candidate transcriptional co-activator associated with HAT activity and may play a key role in the pathogenesis of renal cancer and von Hippel-Lindau disease 16046545_Jade-1 may suppress renal cancer cell growth in part by increasing apoptosis. 18684714_Jade-1/1L are crucial co-factors for HBO1-mediated histone H4 acetylation 18806787_The pVHL tumour suppressor and the Wnt tumorigenesis pathway are therefore directly linked through Jade-1. 22516360_RCC with a low expression of Jade-1 is associated with a poor outcome and decreased survival 22654112_The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate beta-catenin signaling. 23824745_Reduced Jade-1 expression is associated with clear-cell renal cell carcinomas. 24097061_lncRNA-JADE is a key functional link that connects the DDR to histone H4 acetylation, and that dysregulation of lncRNA-JADE may contribute to breast tumorigenesis. 24739512_Increase in the number of cells with cytoplasmic JADE1S correlated with activation of tubular cell proliferation and inversely correlated with the number of cells with nuclear JADE1S, supporting role of HBO1-JADE1 shuttling during organ regeneration. 25100726_Casein kinase 1 alpha specifically phosphorylates Jade-1 at an unprimed SLS phosphorylation site. 26151225_JADE1S protein localized to centrosomes in interphase and mitotic cells, and to the midbody during cytokinesis. 26919559_Interactome analyses revealed that the Jade-1S mutant unable to be phosphorylated by CK1alpha has an increased binding affinity to proteins involved in chromatin remodelling, histone deacetylation, transcriptional repression, and ribosome biogenesis. 27155521_JADE1 expression has a role in colon cancers, renal carcinomas, and in acute kidney injuries and regeneration. (Review) 27160456_identification of SNPs within the IQCJ, NXPH1, PHF17 and MYB genes partly explaining the large interindividual variability observed in plasma triglyceride levels in response to an n-3 fatty acid supplementation 28134766_A genome-wide association study (GWAS) identified loci associated with the plasma triglyceride (TG) response to omega-3 fatty acid (FA) supplementation in IQCJ, NXPH1, PHF17 and MYB. 29382722_results indicate that the N-terminal region of JADE1 functions as a platform that brings together the catalytic HBO1 subunit with its cognate H3-H4 substrate for histone acetylation 34719765_Genome-wide association study and functional validation implicates JADE1 in tauopathy. | ENSMUSG00000025764 | Jade1 | 1.133361e+03 | 0.9056742 | -0.142935884 | 0.2652212 | 2.898181e-01 | 0.5903371375 | 0.90610296 | No | Yes | 1.027027e+03 | 113.804735 | 1.016080e+03 | 115.343605 | ||
| ENSG00000077782 | 2260 | FGFR1 | protein_coding | P11362 | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of embryonic development, cell proliferation, differentiation and migration. Required for normal mesoderm patterning and correct axial organization during embryonic development, normal skeletogenesis and normal development of the gonadotropin-releasing hormone (GnRH) neuronal system. Phosphorylates PLCG1, FRS2, GAB1 and SHB. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. Promotes phosphorylation of SHC1, STAT1 and PTPN11/SHP2. In the nucleus, enhances RPS6KA1 and CREB1 activity and contributes to the regulation of transcription. FGFR1 signaling is down-regulated by IL17RD/SEF, and by FGFR1 ubiquitination, internalization and degradation. {ECO:0000250|UniProtKB:P16092, ECO:0000269|PubMed:10830168, ECO:0000269|PubMed:11353842, ECO:0000269|PubMed:12181353, ECO:0000269|PubMed:1379697, ECO:0000269|PubMed:1379698, ECO:0000269|PubMed:15117958, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17623664, ECO:0000269|PubMed:18480409, ECO:0000269|PubMed:19224897, ECO:0000269|PubMed:19261810, ECO:0000269|PubMed:19665973, ECO:0000269|PubMed:20133753, ECO:0000269|PubMed:20139426, ECO:0000269|PubMed:21765395, ECO:0000269|PubMed:8622701, ECO:0000269|PubMed:8663044}. | 3D-structure;ATP-binding;Alternative splicing;Cell membrane;Chromosomal rearrangement;Craniosynostosis;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Disease variant;Disulfide bond;Dwarfism;Glycoprotein;Heparin-binding;Holoprosencephaly;Hypogonadotropic hypogonadism;Immunoglobulin domain;Kallmann syndrome;Kinase;Membrane;Mental retardation;Nucleotide-binding;Nucleus;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transcription;Transcription regulation;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase;Ubl conjugation | The protein encoded by this gene is a member of the fibroblast growth factor receptor (FGFR) family, where amino acid sequence is highly conserved between members and throughout evolution. FGFR family members differ from one another in their ligand affinities and tissue distribution. A full-length representative protein consists of an extracellular region, composed of three immunoglobulin-like domains, a single hydrophobic membrane-spanning segment and a cytoplasmic tyrosine kinase domain. The extracellular portion of the protein interacts with fibroblast growth factors, setting in motion a cascade of downstream signals, ultimately influencing mitogenesis and differentiation. This particular family member binds both acidic and basic fibroblast growth factors and is involved in limb induction. Mutations in this gene have been associated with Pfeiffer syndrome, Jackson-Weiss syndrome, Antley-Bixler syndrome, osteoglophonic dysplasia, and autosomal dominant Kallmann syndrome 2. Chromosomal aberrations involving this gene are associated with stem cell myeloproliferative disorder and stem cell leukemia lymphoma syndrome. Alternatively spliced variants which encode different protein isoforms have been described; however, not all variants have been fully characterized. [provided by RefSeq, Jul 2008]. | hsa:2260; | cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; extracellular region [GO:0005576]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; fibroblast growth factor binding [GO:0017134]; fibroblast growth factor-activated receptor activity [GO:0005007]; heparin binding [GO:0008201]; identical protein binding [GO:0042802]; protein homodimerization activity [GO:0042803]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; receptor-receptor interaction [GO:0090722]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; cell migration [GO:0016477]; chordate embryonic development [GO:0043009]; epithelial to mesenchymal transition [GO:0001837]; fibroblast growth factor receptor signaling pathway [GO:0008543]; MAPK cascade [GO:0000165]; neuron migration [GO:0001764]; peptidyl-tyrosine phosphorylation [GO:0018108]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of blood vessel endothelial cell migration [GO:0043536]; positive regulation of cell differentiation [GO:0045597]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of endothelial cell chemotaxis to fibroblast growth factor [GO:2000546]; positive regulation of kinase activity [GO:0033674]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of phospholipase activity [GO:0010518]; positive regulation of phospholipase C activity [GO:0010863]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of vascular endothelial cell proliferation [GO:1905564]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of cell differentiation [GO:0045595]; regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001239]; skeletal system development [GO:0001501]; skeletal system morphogenesis [GO:0048705]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11693202_vitronectin increased the presence of all four growth factor receptors and most notably, VEGFR-1; in contrast, fibrin decreased all four receptors, especially FGFR-1 and FGFR-2 11746971_In the fusion of the FGFR1 and BCR genes in myeloproliferative disorder, it is likely that the dimerization properties of BCR lead to aberrant FGFR1 signaling and neoplastic transformation. 11759058_distribution in normal endocrine cells and related tumors of the gastroenteropancreatic system; immunoreactive in rare duodenal endocrine cells and in pancreatic A cells 11919391_REVIEW; The 8p11 myeloproliferative syndrome is a distinct clinical entity caused by constitutive activation of FGFR1. 12031912_overexpressed in acute myeloid luekemia while translocations associated with this gene are absent, and more frequently in patients with CD56 immunophenoytpe 12080186_Inhibiting expression of bFGF or FGFR-1 in only the melanoma cells is as effective in blocking tumor growth as simultaneously inhibiting bFGF or FGFR-1 synthesis in the melanoma cells and the melanoma cell-interspersing vasculature. 12121226_CD56 molecules on NK cells interact with fibroblast growth factor receptor 1 on Jurkat T cells to trigger IL-2 production. 12141425_Review of FGFR1 isoforms and structure-activity analysis [review] 12373339_Differences in spatial patterns of FGFR expression in normal skin may generate functional diversity in response to FGFs, and in wounded skin FGFs may function in wound healing via induced FGFRs. 12397010_Our results suggest an autocrine role of the FGF-FGFR-1 system in the pathogenesis of COPD-associated vascular remodeling. 12411316_Fibroblast growth factor receptor-1 is expressed by endothelial progenitor cells. 12440521_Recombinant FGFR1 was expressed on the surface of Sf9 insect cells. The peptide ValTyrMetSerProPhe can specifically bind to the hydrophobic surface of FGFR1. 12573278_alternatively spliced FGFR-1 isoforms induce differential signal transduction pathways 12594223_ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. 12604616_FGFR1 tyrosine phosphorylation is inhibited by sef protein 12614330_cAMP-induced nuclear accumulation of FGFR1 provides a signal that triggers molecular events leading to neuronal differentiation of neuronal progenitor cells 12627230_FGFR1 has a role in autosomal dominant Kallmann syndrome 12651930_expression system is involved in angiogenesis in inflamed synovial tissue in the temporomandibular joint 12746216_TSH stimulates FGFR1 but not FGF-2 expression and PKC activation stimulates FGF-2 synthesis and secretion, and TSH synergizes with PKC activators so increases in FGFR1 or FGF-2 or in both may contribute to goitrogenesis. 12791257_Results describe a direct interaction between neural cell adhesion molecule (fibronectin type III [F3] modules 1 and 2) and fibroblast growth factor receptor R1 (Ig modules 2 and 3) by surface plasmon resonance analysis. 12794748_HFGFR1 was expressed primarily in the ventricular zone embryologically 12799194_Tyrosine 463 is phosphorylated and able to transduce signals in response to FGF-2 treatment alone. Furthermore, FGFR-1 dimerization/kinase activation is stabilized by heparin. 14587039_involvement of a nuclear matrix bound FGFR1 in transcriptional and RNA processing events in the cell nucleus 14636241_Here we show that the TCR and fibroblast growth factor receptors co-localize during combined stimulation [which] synergistically enhances the activation of nuclear factors of activated T cells. 15001591_two novel intragenic FGFR1 mutations in two sporadic male cases in Kallmann syndrome 15096041_The weak binding affinity of the fibroblast growth factor receptor (FGFR) 1 interaction with heparin suggests that in this model, FGFR and heparan sulfate proteoglycan are unbound in the absence of FGF ligand on the cell surface. 15117958_Results suggest that active fibroblast growth factor receptor 1 kinase regulates the functions of nuclear 90-kDa ribosomal S6 kinase. 15273729_Although FGFR-1 dimerization achieved by fgfr-2 injection led to highly differentiated and smaller bladder tumors, no sign of reduction of tumor angiogenesis was observed, thus suggesting that endothelial cells are resistant to FGF. 15297314_fibrinogen binding of FGF-2 enhances EC proliferation through the coordinated effects of colocalized alpha(v)beta(3) and FGFR1 15316024_insulin receptor substrate-4 and ShcA have roles in signaling by the fibroblast growth factor receptor 15509650_conjoint endocytosis and trafficking is a novel mechanism for the coregulation of E-cadherin and FGFR1 during cell signaling and morphogenesis 15558020_The reciprocal relationship in gene expression between FGFR1 and FGFR3 in colorectal tissue plays an important role in the progression of the carcinomas to malignancy. 15564375_Recruitment of SRC to FRS2 leads to activation of signal attenuation pathways. 15613419_the reversal of hypogonadotropic hypogonadism in a proband carrying an FGFR1 mutation suggests a role of FGFR1 beyond embryonic GnRH neuron migration, and a loss of function mutation in the FGFR1 gene causing delayed puberty. 15618886_In 'undifferentiated' neurospheres of embryonic brain and spinal cord, transcripts from FGFR1 and FGFR2 were consistently detected. 15680705_Fibroblast growth factor trophic signaling to differentiated neurons could involve the release of astrocytic basic FGF acting on neuronal FGFR1 expression. 15774903_FGFR-1 is expressed in early hematopoietic/endothelial precursor cells, as well as in a subpool of endothelial cells in tumor vessels, and that it is critical for hematopoietic but not for vascular development 15817662_When used individually, FGFR1 partially prevented goiter and sVEGFR1 partially reduced vascular volume. 15929978_The interaction of FGFR1 with CREB binding protein allows activation of gene transcription and may play a role in cell differentiation. 15955231_involvement of a nuclear matrix bound FGFR1 in transcriptional and RNA processing events in the cell nucleus 16091423_Data indicate that after endocytosis, fibroblast growth factor receptor (FGFR)4 and its bound ligand, FGF1, are sorted mainly to the recycling compartment, whereas FGFR1-3 with ligand are sorted mainly to degradation in the lysosomes. 16186508_Two somatic mutations in kinase domain found in glioblastomas (N546K, R576W). 16188231_Stimulation of FGFR-1 results in a Ca2+ channel-independent change of gene expression in retinal pigment epithelial cells. 16305343_data describe percentage of bone marrow cells expressing receptors for interleukin-1, platelet-derived growth factor, fibroblast growth factor, transforming growth factor-beta, epidermal growth factor and c-Fos and c-Myc in untreated lung & breast cancer 16316338_the initial stimulus for renal inflammation, whether immune complex, hypersensitivity or rejection, did not alter expression patterns of FGF-1 or its receptor 16365308_FGF-receptor-mediated mitogen-activated protein kinase stimulation is potentiated in cells costimulated with ephrin-A1 16424058_up-regulation of the secreted FGF-BP1 protein during initiation of pancreas and colon neoplasia could make this protein a possible serum marker indicating the presence of high-risk premalignant lesions 16598308_The activation of FGF-2/FGFR1beta supports progression and chemoresistance in subsets of AML. Therefore, FGFR1 targeting may be of therapeutic benefit in subsets of AML. 16606836_Mutations leading to FGFR1 loss-of-function were found. 16685373_These results suggest that constitutive levels of both FGFR1 and FGFR3, but not FGFR4 are essential for FGF-stimulated anchorage-independent growth of SW-13 cells. 16757108_Paediatric phenotypic expression of FGFR1 loss of function mutations is highly variable, the severity of the oro-facial malformations at birth does not predict gonadotropic function at the puberty. 16764984_Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. 16807070_Analysis of FGFR1 protein revealed that a high FGFR1 gene expression is a distinct molecular feature of early OSCC indicating a participation in initial oral carcinogenesis. 16807244_analysis of heparan sulfate-related oligosaccharide binding to fibroblast growth factors 1 and 2 and their receptors 16876430_Data suggest that the relative concentrations of Anosmin-1 and FGF-2 modulate the migration of oligodendrocyte precursors during development through their interaction with FGFR1. 16949906_Immunohistochemical expression of FGFR1 in tumors was confirmed by real-time polymerase chain reaction. 17121884_FGFR1 signaling contributes to the survival of MDA-MB-134 cells. 17154279_Identification of 15 new FGFR1 sequence variants in 17 patients with Kallman Syndrome. 17255109_PP2A binding to Sprouty2 and phosphorylation changes are a prerequisite for ERK inhibition downstream of FGFR stimulation 17318851_Data suggest that preferential premolar agenesis is associated with FGFR and IRF6. 17318851_Observational study of gene-disease association. (HuGE Navigator) 17343269_Observational study of gene-disease association. (HuGE Navigator) 17360555_FGF3, FGF7, FGF10, FGF18, and FGFR1 may have roles in nonsyndromic cleft lip and palate 17366557_results of analysis of the genome-wide scan data was a 20 cM region at 8p11-23 in which markers had LODs > or =1.0. Linkage and association analyses of these SNPs yield suggestive results for markers in FGFR1 and BAG4. 17389761_ZNF198-FGFR1 activated both the AKT and mitogen activated protein kinase (MAPK) prosurvival signaling pathways, resulting in elevated phosphorylation of the AKT target FOXO3a at T32 and BAD at S112, respectively. 17397528_FGFR1 has a role in preventing progression of breast neoplasms 17414280_no evidence that mosaicism for mutations, normally associated with syndromal forms of craniosynostosis, occur in single suture craniosynostosis 17545628_FGF receptor 1 (FGFR1), which is expressed mainly in neoplastic thyroid cells, propagates MAPK activation and promotes tumor progression. In contrast, FGFR2 is down-regulated in neoplastic thyroid cells through DNA promoter methylation. 17671674_analysis of EGFR, HER2 and HER3 expression in esophageal primary tumours and corresponding metastases 17696196_data suggest an important role for FGFR1 and FGFR1-downstream genes in rhabdomyosarcoma (RMS) tumorigenesis and a possible association with the deregulation of proliferation and differentiation of skeletal myoblasts in RMS 17893707_Observational study of gene-disease association. (HuGE Navigator) 17963255_FGF and FGFR may have a role in cleft lip and cleft palate 18034870_Current evidence supports a heparan sulphate -dependent interaction between anosmin-1 and FGFR1, where anosmin-1 serves as a co-ligand activator enhancing the signal activity. (Review) 18042549_the involvement of FGFR-1 through FGF2 in eliciting PGE(2) angiogenic responses 18059337_Molecular analyses in salivary gland tumors revealed that ring formation consistently generated novel FGFR1-PLAG1 gene fusions in which the 5'-part of FGFR1 is linked to the coding sequence of PLAG1 18160472_KAL1 mutations result in a more severe reproductive phenotype than FGFR1/KAL2 mutations. 18171671_Basic fibroblast growth factor-induced neuronal differentiation of mouse bone marrow stromal cells requires FGFR-1, MAPK/ERK, and transcription factor AP-1 18216705_No sequence variation was found, indicating that mutations in the 'hot spot' exons are not associated with nonsynostotic plagiocephaly. 18216705_Observational study of genotype prevalence. (HuGE Navigator) 18231572_In human uterine leiomyomas, FGFR1 were also overexpressed. 18315732_Observational study of gene-disease association. (HuGE Navigator) 18411303_p38alpha MAPK has a critical role in the regulated translocation of exogenous FGF1 into the cytosol/nucleus 18415014_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18463157_12% of Kallman syndrome males have KAL1 deletions, but intragenic deletions of the FGFR1, GNRH1, GNRHR, GPR54 and NELF genes are uncommon in Idiopathic hypogonadotropic hypogonadism/Kallman syndrome. 18469019_Identification of two previously unreported SNPs in FGFR1 and FABP3 associated with BMD and a third SNP in TIMP2 related to risk for non-vertebral osteoporotic fractures. 18469019_Observational study of gene-disease association. (HuGE Navigator) 18480409_fibroblast growth factor receptor 1 ubiquitination is required for its intracellular sorting but not for its endocytosis 18563556_BRCA2-associated cancers were characterized by the higher relative expression of FGF1 and FGFR2 18665077_FGFR1-IIIb and FGFR1-IIIc are coexpressed, and the FGFR1-III isoforms are differentially regulated by growth factors and cyclin D1. 18669637_the FGF binding domain and the heparin binding domain are necessary for the hBP3 interaction with endogenous FGF and the activation of FGFR signaling in vivo 18682503_Observational study of gene-disease association. (HuGE Navigator) 18719360_Temporal-mediatd FGFR1 indepepndence: implications for targeting candidate molecules in prostate cancer are reported. 18723471_Observational study of gene-disease association. (HuGE Navigator) 18723471_no mutations found in Kallmann syndrome 18790757_These results indicate that EphA4 plays an important role in malignant phenotypes of glioblastoma by enhancing cell proliferation and migration through accelerating a canonical FGFR signaling pathway. 18829480_bFGF, FGFR1, and FGFR2 are frequently overexpressed in squamous cell carcinoma and adenocarcinoma of the lung and may have a role in neoplasm pathogenesis 18940940_Fibroblast growth factor receptor 1 activity is required for FGF-1 stimulated cell proliferation and priming in early adipogenic events. 18978678_Observational study of gene-disease association. (HuGE Navigator) 19082464_Overexpression of the FGFR-1 gene may thus be a useful predictor of liver metastasis in patients with colorectal cancer. 19096867_FGF-2 and FGFR1 expression is preserved in the motor system in end stage ALS. 19211868_Signaling of transgenic Fgfr1 and Fgfr2 is important for their potential ability to mediate axon-glial interaction in the peripheral sensory pain pathway, via influencing myelinating and nonmyelinating Schwann cell function. 19224897_sequential autophosphorylation of five tyrosines in the FGFR1 kinase domain is under kinetic control, mediated by both the amino acid sequence surrounding the tyrosines and their locations within the kinase structure 19258500_Results show the transforming activity of FGFR1 in mammary epithelial cells and identify RSK as a critical component of FGFR1 signaling in lobular carcinomas. 19330026_Data show that FGFR1 and DDHD2 at 8p12 cooperated functionally with MYC, whereas CCND1 and ZNF703 cooperated with a dominant negative form of TP53. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19458078_FGFR1 has significant effects on urothelial cell phenotype and may represent a useful therapeutic target in some cases of urothelial carcinoma. 19489874_Observational study of gene-disease association. (HuGE Navigator) 19574212_that the main role of heparin in FGF-induced signaling is to protect this naturally unstable protein against heat and/or proteolytic degradation and heparin is not essential for a direct FGF1-FGFR interaction and receptor activation 19658100_ZNF198-FGFR1 is associated with phosphorylation of several proteins including SSBP2, ABL, FLJ14235, CALM and TRIM4 proteins. 19663702_monoclonal antibody analysis of multiple FGFR1 isoforms, generated by alternative splicing and post-translational modifications through glycosylation 19665973_Study describes the crystal structure of the activated tyrosine kinase domain of FGFR1 in complex with a phospholipase Cgamma fragment. 19666467_a neurofascin intracellular domain activates FGFR1 for neurite outgrowth, whereas the extracellular domain functions as an additional, regulatory FGFR1 interaction domain in the course of development 19696444_binding of anosmin-1 to FGFR1 and heparin can play a dual role in assembly and activity of the ternary FGFR1.FGF2.heparin complex. 19727229_Observational study of gene-disease association. (HuGE Navigator) 19727229_There was no association among gene FGFR1 rs13317, p. E467K, p. M369I, p. S393S and gene FGF10 rs1448037 and nonsyndromic cleft lip with or without palate in Chinese population. 19728793_The aim of this study was to investigate and compare FGFR expression in in vivo embryonic limb development and in vitro chondrogenesis of mesenchymal stem cells. 19730683_Observational study of gene-disease association. (HuGE Navigator) 19802384_syndecan-1 and FGF-2, but not FGFR-1 share a common transport route and co-localize with heparanase in the nucleus, and this transport is mediated by the RMKKK motif in syndecan-1 19820032_Loss-of-function mutations in FGFR1 underlie 7% of normosmic idiopathic hypogonadotropic hypogonadism with different degrees of impairment in vitro. 19822079_Data report that the bFGF, FGFR1/2 and syndecan 1-4 expressions are altered in bladder tumours. 19890272_depressed expression of the Klotho-FGFR1 complex in hyperplastic glands underlies the pathogenesis of secondary hyperparathyroidism and its resistance to extremely high FGF23 levels in uremic patients. 19920251_Data show that oncogenic forms of fibroblast growth factor receptor type 1 inhibit the pyruvate kinase M2 (PKM2) isoform by direct phosphorylation of PKM2 tyrosine residue 105 (Y(105)). 19920824_Endogenous SPARC expression can be modulated by FGFR1-III isoform expression. Endogenous SPARC expression in PANC-1 cells was increased in FGFR1-IIIb over-expressing cells, but decreased in FGFR1-IIIc over-expressing cells. 19923290_Loss of FGFR1 signaling provides evidence that extracellular signals regulate not simply the proliferation or survival of radial glial cells, but specifically their progression to intermediate progenitor cells during neurogenesis in vivo. 20024612_FGFR-1 amplification or protein overexpression in breast cancers may be an indicator for brivanib treatment, where it may have direct anti-proliferative effects in addition to its' anti-angiogenic effects. 20036812_Studies indicate communication between tumor cells and their microenvironment is through polypeptide growth factors EGF, FGF, PDGF and receptors for these growth factors. 20068287_Decreased expression of alpha-Klotho and FGFR1c in parallel with CaR expression and parathyroid cell growth may be involved in the pathogenesis of secondary hyperparathyroidism 20103604_brivanib is a dual inhibitor of vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1 kinases 20117945_Recent advances in interactions of Kallman Syndrome (KS)-associated molecules within the FGFR1 signalling complex are covered in this review, and linkage of autosomal dominant and sex-linked modes of inheritance are discussed [review] 20179196_Data suggest that amplification and overexpression of FGFR1 may be a major contributor to poor prognosis in luminal-type breast cancers, driving anchorage-independent proliferation and endocrine therapy resistance. 20362962_Mutations of FGFR1 underlie an autosomal dominant form of Kallmann syndrome. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20388777_amplified FGFR expression engages the STAT3 pathway 20389085_Role of FGFR1 mutations in Kallmann syndrome (Review) 20389169_Based on our results, it is possible that a subtle dysfunction (expression) of the FGFR1 gene is involved in the development of the most common male reproductive tract disorder - unilateral or bilateral cryptorchidism 20421966_the phosphorylation state of FLRT1, which is itself FGFR1 dependent, may play a critical role in the potentiation of FGFR1 signalling and may also depend on a SFK-dependent phosphorylation mechanism acting via the FGFR 20536592_Dental agenesis may be a clinical feature of Kallmann syndrome caused by a mutation in the FGFR1 gene. 20538960_Observational study of gene-disease association. (HuGE Navigator) 20544801_Observational study of gene-disease association. (HuGE Navigator) 20606682_The bFGF-FGFR1-PI3K-Rac1 pathway in the bone microenvironment may have a significant role in the invasion and metastasis of Ewing sarcoma. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20643727_Observational study of gene-disease association. (HuGE Navigator) 20672350_Observational study of gene-disease association. (HuGE Navigator) 20686451_FGFR1 expression levels in parathyroid glands were found to be positively correlated to renal function and significantly decreased over chronic kidney disase stages. 20717167_Observational study of gene-disease association. (HuGE Navigator) 20734064_Observational study of gene-disease association. (HuGE Navigator) 20855522_Ginsenoside-Rg1 induces angiogenesis via non-genomic crosstalk of glucocorticoid receptor and fibroblast growth factor receptor-1. 20889570_Data suggest that an FGFR1alpha-to-FGFR1beta isoform switch and increased FGF1-induced activation of FGFR1beta may result in a proliferative advantage that plays a key role during bladder tumor progression. 20932831_Grb14 was recruited to FGFR1 into a trimeric complex containing also phospholipase C gamma 21041608_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21160078_Focal FGFR1 amplification is common in squamous cell lung cancer and associated with tumor growth and survival, suggesting that FGFR inhibitors may be a viable therapeutic option in this cohort of patients. 21329481_FGFR1 is highly expressed in renal cell carcinoma patients 21330321_we identified a novel CUX1-FGFR1 fusion oncogene in a patient with the 8p11 myeloproliferative syndrome and demonstrated its transforming potential in the Ba/F3 cell line. 21430024_FGFR1 is a novel obesity gene that may promote obesity by influencing adipose tissue and the hypothalamic control of appetite. 21451043_FGF signaling, through cooperation between Fgfr1 and Fgfr2 (but not Fgfr3), is required for initial generation of oligodendrocyte precursors in transgenic mouse ventral forebrain, with Fgfr1 being a stronger inducer than Fgfr2. 21515689_amitriptyline-induced FGFR activation might occur by the MMP-dependent shedding of FGFR ligands, such as FGF-2, thus resulting in GDNF production 21527526_FGFR1/2 are expressed by bone-derived mesenchymal stem cells in vivo and in vitro and are developmentally regulated during their differentiation. 21621521_homodimerization of FGFR1 appear to be a fundamental mechanism for the agonist activity of all FGF ligands at least in the case of the MAPK signaling. 21666686_Case Report: identify a novel fusion partner FGFR1 for the known anchor gene FOXO1 in alveolar rhabdomyosarcoma. 21666749_These studies show that FGFR1 amplification is common in squamous cell lung cancer, and that FGFR1 may represent a promising therapeutic target in non-small cell lung cancer. 21682876_Comprehensive mutation analysis of all 7 known KS genes (KAL1, FGFR1, FGF8, PROK2, PROKR2, CHD7, and WDR11) in 30 well-phenotyped probands revealed mutations in KAL1 (3 men) and FGFR1 (all 5 women vs. 4/25 men), but not in other genes in Finland patients. 21712446_Data show that the majority of TN cell lines only modest sensitive to FGFR inhibition in growth but were highly sensitive in anchorage-independent conditions. 21739604_These results point to NCAM-mediated stimulation of FGFR as a novel mechanism underlying epithelial ovarian carcinoma malignancy and indicate that this interplay may represent a valuable therapeutic target. 21765395_Nedd4-1 binds directly to and ubiquitylates activated FGFR1, by interacting primarily via its WW3 domain with a novel non-canonical sequence on FGFR1, regulating endocytosis & signalling during neuronal differentiation & embryonic development. 21779335_Data indicate that clathrin-mediated endocytosis is required for efficient internalization and downregulation of FGFR1 while FGFR3, however, is internalized by both clathrin-dependent and clathrin-independent mechanisms. 21835001_FGFR1 is the major mediator with the degenerative potential in the presence of FGF-2 in human adult articular chondrocytes 21868365_inductiion of mammary tumorigenesis requires activation of the epidermal growth factor receptor 21885851_MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. 21942441_Levels of FGF2 and FGFR1 in saliva and serum from patients with salivary gland tumors were significantly higher than those from healthy control subjects suggesting their potential use as cancer biomarkers. 21952621_For the first time, our gene expression profiling experiment on archival tumour materials has identified upregulated FGFR1 expression to be associated with PC progression to the CR state. 21969607_LDH-A is tyrosine phosphorylated and activated by FGFR1 in cancer cells. 22030335_FGFR1 gene rearrangement is associated with systemic mastocytosis and myeloid/lymphoid neoplasm in blast crisis. 22179561_An increased FGFR1 gene copy number was found in 32 (32%) lung cancer patients 22247553_Fibronectin induces endothelial cell migration through beta1 integrin and Src-dependent phosphorylation of fibroblast growth factor receptor-1 at tyrosines 653/654 and 766. 22248288_Deletion of the D1 and the D1-D2 linker (the D1/linker region) from FGFR1c led to beta-Klotho-independent receptor activation by FGF21, suggesting that there may be a direct interaction between FGF21 and the D1/linker region-deficient FGFR1c. 22319038_genetic association studies in 103 patients from US and UK: Mutations in FGFR1, FGF8, or PROKR2 contributed to 7.8% of patients with combined pituitary hormone deficiency or septo-optic dysplasia. Data suggest genetic overlap with Kallmann syndrome. 22438050_The expression of FGFR1 in patients' biopsies may serve as a marker of response to chemoradiotherapy. 22442730_Differential specificity of endocrine FGF19 and FGF21 to FGFR1 and FGFR4 in complex with KLB. 22499828_FGFR1 amplification is a common genetic event occurring at a frequency of 16% in L-SCCs. Moreover, lymph node metastases derived from FGFR1-amplified L-SCCs also exhibit FGFR1 amplification. 22514272_INFS/Nurr1 nuclear partnership provides a novel mechanism for TH gene regulation in mDA neurons and a potential therapeutic target in neurodevelopmental and neurodegenerative disorders. 22523080_KLB and FGFR1 form a 1:1 heterocomplex independent of the galectin lattice that transitions to a 1:2 complex upon the addition of FGF21. 22569333_FGFR1 initiates MAPK signaling, whereas S4-dependent FGFR1 macropinocytosis modulates the kinetics of MAPK activation 22573348_Targeting FGFR signaling is a promising new approach to treating aggressive prostate cancer 22648708_There were no significant associations between FGFR1 and clinicopathological parameters in lung cancer 22665522_FGFR1 localized to the nucleus specifically in invading cells in both clinical material and a three-dimensional model of breast cancer 22673519_proteins with LRR, IG, and FNIII are candidate regulators of the FGFRs. Here we identify leucine-rich repeat, immunoglobulin-like and transmembrane domain 3 (LRIT3) as a regulator of the FGFRs 22684217_Conclude that FGFR1 amplification is one of the most frequent therapeutically tractable genetic lesions in pulmonary carcinomas. 22701738_FGFR1 activation in urothelial carcinoma cell lines promotes epithelial-mesenchymal transition via coordinated activation of multiple signalling pathways and by promoting activation of prostaglandin synthesis. 22724017_The data shows that patients with congenital hypogonadotropic hypogonadism and a splice-site mutation in FGFR1 can undergo reversal. 22781593_Mutations in FGFR1 gene is associated with the development of myeloid and lymphoid malignancies. 22791819_results of the present study suggest that MUC4 promotes invasion and metastasis by FGFR1 stabilization through the N-cadherin upregulation. 22837387_study reports that a small subset of glioblastoma multiforme tumors harbors oncogenic chromosomal translocations that fuse in-frame the tyrosine kinase coding domains of fibroblast growth factor receptor genes(FGFR1 or FGFR3) to the transforming acidic coiled-coil coding domains of TACC1 or TACC3; the FGFR-TACC fusion protein displays oncogenic activity 22842457_The novel findings reported in this study are expected to provide valuable clues toward a complete understanding of the other genetic diseases linked to mutations in the FGFR. 22863309_FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. 22903848_Report recycling/degradation pathways of FRG receptor 1 in human glioma cell line. 22955284_Nectin-1 Ig3 induced phosphorylation of FGFR1c in the same manner as the whole nectin-1 ectodomain, and promoted survival of cerebellar granule neurons induced to undergo apoptosis. 22989054_ErbB3 but not Fgfr1 mRNA levels are reduced in leukocytes of patients with major depressive disorders compared to healthy subjects. 23029290_Fibroblast growth factor receptor 1 activation leads to induction of CX3CL1 in a tumor setting. 23070782_FGFR1 polymorphisms, especially rs4647905, can have an important role in the normal human skull variation, primarily due to their influence in head length. 23082000_The incidence of FGFR1 amplification within Chinese patient non-small cell lung carcinoma tumors was 6 of 48 squamous origin and 5 of 76 adenocarcinoma. 23154428_demonstrate skeletal phenotypic characterization of patients presenting with Kallmann syndrome and FGFR1 mutations 23154548_FGFR1 amplification is associated with squamous cell carcinoma of the lung. 23171834_A novel mRNA in-frame fusion between exon 4 of the breakpoint cluster region (BCR) gene at chromosome 22q11 and exon 9 of FGFR1 gene on chromosome 8p11-12 was identified by reverse transcription polymerase chain reaction 23182986_Data indicate that FGFR1 amplification is an independent negative prognostic factor in surgically resected squamous cell carcinoma of the lung (SCCL) and is associated with cigarette smoking in a dose-dependent manner. 23243019_Data suggest for therapeutic targeting of the FGF-2/FGFR1/CEP57 axis in prostate cancer. 23276709_2 patients with congenital isolated hypogonadotropic hypogonadism were found to have mutations in FGFR1 (R254W and R254Q); both are loss-of-function mutations demonstrated by their reduced overall and cell surface expression suggesting a deleterious effect on receptor folding and stability 23296701_Upregulation of FGFR1 is associated with gastric cancer. 23334987_The role of the FGFR/klotho-axis remains still unclear in primary hyperparathyroidism. 23343422_the combination of dovitinib + NVP-BEZ235 or dovitinib + AEE788 results in strong inhibition of tumor growth and a block in metastatic spread. Only these combinations strongly down-regulate the FGFR/FRS2/Erk and PI3K/Akt/mTOR signaling pathways 23348274_In Thai Pfeiffer syndrome patients, FGFR1 mutations in exon 5 were identified. 23401445_FGFR1 amplification is much more common in squamous cell lung cancers (21%) than in lung adenocarcinoma (3.4%). Survival of FGFR1-amplified lung cancer cell lines was additionally shown to be dependent on overexpression of the FGFR1 kinase. 23434054_Copy number variations of the FGFR1 gene occur in a subset of OTSCC with approximately 10% of cases showing amplification of the gene. FGFR1 amplification may represent a therapeutic target in OTSCC 23468956_data demonstrate that FGFR1 and FGFR3 have largely non-overlapping roles in regulating invasion/metastasis and proliferation in distinct 'mesenchymal' and 'epithelial' subsets of human BC cells 23536707_FGF2 -FGFR1 activation through an autocrine loop is a novel mechanism of acquired resistance to EGFR-tyrosine kinase inhibitors. 23563700_Growth inhibition induced by ponatinib was associated with inactivation of FGFR1 and its downstream targets. 23564461_Phosphorylation of serine 779 in fibroblast growth factor receptor 1 and 2 by protein kinase C(epsilon) regulates Ras/mitogen-activated protein kinase signaling and neuronal differentiation. 23569208_KLF10 is an effective repressor of myoblast proliferation and | ENSMUSG00000031565 | Fgfr1 | 4.654232e+03 | 1.0409137 | 0.057850397 | 0.2828731 | 4.180609e-02 | 0.8379898426 | 0.96856922 | No | Yes | 3.967650e+03 | 342.705673 | 3.998047e+03 | 354.219621 | |
| ENSG00000078124 | 55331 | ACER3 | protein_coding | Q9NUN7 | FUNCTION: Endoplasmic reticulum and Golgi ceramidase that catalyzes the hydrolysis of unsaturated long-chain C18:1-, C20:1- and C20:4-ceramides, dihydroceramides and phytoceramides into sphingoid bases like sphingosine and free fatty acids at alkaline pH (PubMed:20068046, PubMed:26792856, PubMed:20207939, PubMed:11356846, PubMed:30575723). Ceramides, sphingosine, and its phosphorylated form sphingosine-1-phosphate are bioactive lipids that mediate cellular signaling pathways regulating several biological processes including cell proliferation, apoptosis and differentiation (PubMed:20068046). Controls the generation of sphingosine in erythrocytes, and thereby sphingosine-1-phosphate in plasma (PubMed:20207939). Through the regulation of ceramides and sphingosine-1-phosphate homeostasis in the brain may play a role in neurons survival and function (By similarity). By regulating the levels of proinflammatory ceramides in immune cells and tissues, may modulate the inflammatory response (By similarity). {ECO:0000250|UniProtKB:Q9D099, ECO:0000269|PubMed:11356846, ECO:0000269|PubMed:20068046, ECO:0000269|PubMed:20207939, ECO:0000269|PubMed:26792856, ECO:0000269|PubMed:30575723, ECO:0000303|PubMed:20068046}. | 3D-structure;Alternative splicing;Calcium;Disease variant;Endoplasmic reticulum;Golgi apparatus;Hydrolase;Leukodystrophy;Lipid metabolism;Membrane;Metal-binding;Reference proteome;Sphingolipid metabolism;Transmembrane;Transmembrane helix;Zinc | PATHWAY: Lipid metabolism; sphingolipid metabolism. {ECO:0000269|PubMed:20068046, ECO:0000269|PubMed:20207939, ECO:0000269|PubMed:30575723}. | hsa:55331; | endoplasmic reticulum membrane [GO:0005789]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of Golgi membrane [GO:0030173]; integral component of membrane [GO:0016021]; calcium ion binding [GO:0005509]; ceramidase activity [GO:0102121]; dihydroceramidase activity [GO:0071633]; N-acylsphingosine amidohydrolase activity [GO:0017040]; phytoceramidase activity [GO:0070774]; zinc ion binding [GO:0008270]; ceramide catabolic process [GO:0046514]; inflammatory response [GO:0006954]; myelination [GO:0042552]; phytosphingosine biosynthetic process [GO:0071602]; positive regulation of cell population proliferation [GO:0008284]; regulation of programmed cell death [GO:0043067]; sphingolipid biosynthetic process [GO:0030148]; sphingosine biosynthetic process [GO:0046512] | 19834535_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20068046_ACER3 catalyzes the hydrolysis of unsaturated long chain ceramides and dihydroceramides and coordinates with ACER2 to regulate cell proliferation and survival 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 26792856_Homozygosity for the p.E33G mutation in the ACER3 gene results in inactivation of ACER3. 27470583_The ACER3 deficiency resulted in decreased cell growth and colony formation, elevated apoptosis, and lower AKT signaling of leukemia cells. This study indicates that ACER3 contributes to AML pathogenesis, and suggests that alkaline ceramidase inhibition is an option to treat acute myeloid leukemia. 30097213_Our study suggests that Acer3 contributes to hepatocellular carcinoma propagation 34281620_ACER3-related leukoencephalopathy: expanding the clinical and imaging findings spectrum due to novel variants. 34817752_LINC01087 indicates a poor prognosis of glioma patients with preoperative MRI. 35034572_Circular RNA circ_0001955 promotes hepatocellular carcinoma tumorigenesis by up-regulating alkaline ceramidase 3 expression through microRNA-655-3p. | ENSMUSG00000030760 | Acer3 | 2.405501e+02 | 0.5758043 | -0.796349423 | 0.3610307 | 4.804192e+00 | 0.0283905772 | 0.56784832 | No | Yes | 1.684847e+02 | 36.855973 | 2.834443e+02 | 62.950684 | |
| ENSG00000078142 | 5289 | PIK3C3 | protein_coding | Q8NEB9 | FUNCTION: Catalytic subunit of the PI3K complex that mediates formation of phosphatidylinositol 3-phosphate; different complex forms are believed to play a role in multiple membrane trafficking pathways: PI3KC3-C1 is involved in initiation of autophagosomes and PI3KC3-C2 in maturation of autophagosomes and endocytosis (PubMed:14617358, PubMed:7628435, PubMed:33637724). As part of PI3KC3-C1, promotes endoplasmic reticulum membrane curvature formation prior to vesicle budding (PubMed:32690950). Involved in regulation of degradative endocytic trafficking and required for the abcission step in cytokinesis, probably in the context of PI3KC3-C2 (PubMed:20208530, PubMed:20643123). Involved in the transport of lysosomal enzyme precursors to lysosomes (By similarity). Required for transport from early to late endosomes (By similarity). {ECO:0000250|UniProtKB:O88763, ECO:0000269|PubMed:14617358, ECO:0000269|PubMed:20208530, ECO:0000269|PubMed:20643123, ECO:0000269|PubMed:32690950, ECO:0000269|PubMed:33637724, ECO:0000269|PubMed:7628435}.; FUNCTION: (Microbial infection) Kinase activity is required for SARS coronavirus-2/SARS-CoV-2 replication. {ECO:0000269|PubMed:34320401}. | 3D-structure;ATP-binding;Autophagy;Cell cycle;Cell division;Cytoplasmic vesicle;Endosome;Kinase;Lipid metabolism;Manganese;Nucleotide-binding;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation | hsa:5289; | autolysosome [GO:0044754]; axoneme [GO:0005930]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endosome [GO:0005768]; late endosome [GO:0005770]; membrane [GO:0016020]; midbody [GO:0030496]; peroxisome [GO:0005777]; phagocytic vesicle membrane [GO:0030670]; phagophore assembly site [GO:0000407]; phosphatidylinositol 3-kinase complex, class III [GO:0035032]; phosphatidylinositol 3-kinase complex, class III, type I [GO:0034271]; phosphatidylinositol 3-kinase complex, class III, type II [GO:0034272]; 1-phosphatidylinositol-3-kinase activity [GO:0016303]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; phosphatidylinositol kinase activity [GO:0052742]; protein kinase activity [GO:0004672]; autophagosome assembly [GO:0000045]; autophagosome maturation [GO:0097352]; autophagy [GO:0006914]; autophagy of peroxisome [GO:0030242]; cell cycle [GO:0007049]; cell division [GO:0051301]; cellular response to glucose starvation [GO:0042149]; early endosome to late endosome transport [GO:0045022]; endocytosis [GO:0006897]; endosome organization [GO:0007032]; macroautophagy [GO:0016236]; phosphatidylinositol phosphate biosynthetic process [GO:0046854]; phosphatidylinositol-3-phosphate biosynthetic process [GO:0036092]; phosphatidylinositol-mediated signaling [GO:0048015]; protein lipidation [GO:0006497]; protein localization to phagophore assembly site [GO:0034497]; protein phosphorylation [GO:0006468]; protein processing [GO:0016485]; regulation of autophagy [GO:0010506]; regulation of cytokinesis [GO:0032465]; regulation of macroautophagy [GO:0016241]; regulation of protein secretion [GO:0050708]; response to leucine [GO:0043201] | 12925680_Results describe how Mycobacterium tuberculosis toxin lipoarabinomannan causes phagosome maturation arrest, interfering with a calcium/calmodulin phosphatidylinositol (PI)3 kinase hVPS34 cascade. 14617358_Data identify rab7 as an important regulator of late endosomal VPS34 function and link rab7 to the regulation of phosphatidylinositol 3'-kinase cycling between early and late endosomes. 15121481_A promoter mutation in a PI regulator affecting the binding of a POU-type transcription factor may be involved in BD and SZ in a subset of patients. 15121481_Observational study of gene-disease association. (HuGE Navigator) 16049009_hVps34 is a nutrient-regulated lipid kinase that integrates amino acid and glucose inputs to mTOR and S6K1 16176982_Amino acids mediate mTOR activation by signaling through class 3 PI3K, hVps34. 16390869_Results argue against a role for Beclin 1 as an essential chaperone or adaptor for hVps34 in normal vesicular trafficking, and they support the hypothesis that Beclin 1 functions mainly to engage hVps34 in the autophagic pathway. 16874027_There is a connection between Beclin 1-associated Class III PI3K/Vps34-dependent autophagy, but not VPS, function and the mechanism of Beclin 1 tumor suppressor action in human breast cancer cells. 17319803_study indicates that hVps34 and its product PI(3)P are involved in endosome to Golgi transport of ricin, and that SNX2 and SNX4 are likely to be effectors in this pathway 18048384_the lipid kinase activity of Vps34 has a role in resveratrol-induced apoptosis and in the formation of autophagolysosomes 18077426_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18420347_Observational study of gene-disease association. (HuGE Navigator) 18420347_our results do support PIK3C3 play a significant role in the etiology of schizophrenia 18649358_Observational study of gene-disease association. (HuGE Navigator) 18725540_SopB mediates PI(3)P production on the SCV indirectly through recruitment of Rab5 and its effector Vps34. 18755982_Class III PI3K Vps34 is responsible for the synthesis of PtdIns(3)P on phagosomes containing either S aureus or E coli. PtdIns(3)P binding to p40(phox) is important for CD18-dependent activation oxidase activation in response to S aureus and E coli 18774718_A phylogenetic study revealing co-evolution of myotubularins phosphoinositides phosphatases with PI 3-kinase class III complex 18957027_hVps34 activity is regulated through its interactions with hVps15, but is independent of Ca2+/CaM. 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19359265_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19887755_was expressed in cancer tissues at 11 times the level of that found in normal tissue; findings suggest that activation of the PI3K-AKT signal pathway is associated with oral carcinogenesis 20453000_Observational study of gene-disease association. (HuGE Navigator) 20529838_Data show that knockdown of Vps34 reduces gossypol-induced autophagy in both MCF-7 human breast adenocarcinoma and HeLa cell lines. 20602615_Observational study of gene-disease association. (HuGE Navigator) 20643123_A specific sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates both receptor degradation and cytokinesis, whereas ATG14L, a PI3K-III subunit involved in autophagy, is not required. 20671926_A PIK3C3 promoter variant (rs3813065/-442 C/T) in an independent multiancestral cohort of 478 systemic lupus erythematosus cases and 522 controls, was examined. 20671926_Observational study of gene-disease association. (HuGE Navigator) 20885446_14-3-3zeta proteins are shown as a negative regulator of autophagy through regulation of a key component of early stages of the autophagy pathway, such as hVps34. 20955765_Pik3c3 is essential for central nervous system neuronal homeostasis and Pik3c3 deletion; CaMKII-Cre transgenic mouse is a useful model for studying pathological changes in progressive forebrain neurodegeneration. 21062745_a critical role of the Rubicon RUN domain in PI3KC3 and autophagy regulation 21702994_Activation of mTOR by leucine or insulin upregulated hVps34. 21835792_Coimmunoprecipitation assay indicated that hepatitis C virus NS4B formed a complex with human Rab5 and Vps34, supporting the notion that Rab5 and Vps34 are involved in NS4B-induced autophagy. 22021616_Though dispensable for autophagy induction, transgenic Vps34 is a critical regulator of naive T cell homeostasis, modulating interleukin (IL)-7 receptor alpha trafficking, signaling, and recycling. 22024166_Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway 22095288_Results describe PKD as a novel Vps34 kinase that functions as an effecter of autophagy under oxidative stress. 22493499_conclude that Slamf1 recruits a subset of Vps34-associated proteins, which is involved in membrane fusion and NOX2 regulation 22719072_Via direct interaction with the class III PI-3-kinase (PI3KC3)/Beclin1, DEDD activated autophagy and induced the degradation of Snail and Twist, two master regulators of EMT 24013218_A mechanistic link between amino acid starvation and autophagy induction via the direct activation of the autophagy-specific PIK3C3 kinase. 24466196_Ric-8A co-localized with Vps34 at the midbody. 24582588_Insulin can spatially regulate VPS34 activity through Src-mediated tyrosine phosphorylation. 24785657_NRBF2 regulates macroautophagy as a component of Vps34 Complex I. 25046113_reveal a novel function of GABP in the regulation of autophagy via transcriptional activation of the BECN1-PIK3C3 complex 25395352_VPS34-IN1 will provide a useful tool to decipher the kinase-dependent functions of Vps34, with acute changes in SGK3 phosphorylation and subcellular localization being new biomarkers of Vps34 activity. 25402320_Data indicte that Compound 31 constitutes an optimized class III phosphoinositide 3-kinase Vps34 inhibitor that could be used to investigate cancer biology. 25593308_DNA damage regulates Vps34 complexes and its downstream mechanisms, including autophagy and receptor endocytosis, through SCF (Skp1-Cul1-F-box)-mediated ubiquitination and degradation 25714112_cis-unsaturated fatty acids require neither BECN1 nor PIK3C3 to stimulate the autophagic flux 25840748_High expression of PI3K core complex genes is associated with poor prognosis in chronic lymphocytic leukemia. 26139536_tubulation requires mTOR activity, and we identified two direct mTOR phosphorylation sites on UVRAG (S550 and S571) that activate VPS34. 26686095_Arf tumor suppressor as a new transcriptional target of nuclear EGFR and highlight Vps34 as an important regulator of the nuclear EGFR/Arf survival pathway. 26687681_Study identifies a key role of Cul3-KLHL20 in autophagy termination by controlling autophagy-dependent turnover of ULK1 and VPS34 complex subunits and reveals the pathophysiological functions of this autophagy termination mechanism. 27385829_Here we show that a putative fifth subunit, nuclear receptor binding factor 2 (NRBF2), is a tightly bound component of the class III phosphatidylinositol 3-kinase complex I that profoundly affects its activity and architecture. NRBF2 is a homodimer and drives the dimerization of the larger PI3KC3-C1 complex, with implications for the higher-order organization of the preautophagosomal structure. 27630019_Atg38 and its human ortholog NRBF2, accessory components of complex I consisting of Vps15-Vps34-Vps30/Atg6-Atg14 (yeast) and PIK3R4/VPS15-PIK3C3/VPS34-BECN1/Beclin 1-ATG14 (human), were characterized. 27793976_Vps34 has a previously unknown role in regulating Rab7 activity and late endosomal trafficking. 27821547_These results establish Vps34 as an essential determinant of both short-term and long-term canonical GPCR signaling. 28038917_High expression of VPS34 promoted GRP78 transcription by modulating ATF6. VPS34 could enhance GRP78 protein stability. 28059666_This study reveals NRBF2 as a critical molecular switch of PtdIns3K and autophagy activation, and its on/off state is precisely controlled by MTORC1 through phosphorylation. 28157699_Low VPS34 expression is associated with cancer. 28844862_p300-dependent VPS34 acetylation/deacetylation is the physiological key to VPS34 activation, which controls the initiation of canonical autophagy and of non-canonical autophagy in which the upstream kinases of VPS34 can be bypassed. 28846113_Vps34 stimulates tumor development mainly through PKC-delta- activation of p62. 29230017_Ribophagic flux was not induced upon inhibition of translational elongation or nascent chain uncoupling, but was induced in a comparatively selective manner under proteotoxic stress induced by arsenite (10) or chromosome mis-segregation (11) , dependent upon VPS34 and ATG8 conjugation 30517873_ULK1 O-GlcNAcylation is crucial for binding and phosphorylation of ATG14L, allowing the activation of lipid kinase VPS34. 30982765_MiR-338-5p induces migration, invasion and metastasis of CRC in part through PIK3C3-related autophagy pathway. The miR-338-5p/PIK3C3 ratio may become a prognostic biomarker for CRC patients. 31177902_Group A Streptococcus modulates RAB1- and PIK3C3 complex-dependent autophagy. 31885313_Binding of Avibirnavirus VP3 to the PIK3C3-PDPK1 complex inhibits autophagy by activating the AKT-MTOR pathway. 32101753_Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. 32207410_PIK3C3 Acts as a Tumor Suppressor in Esophageal Squamous Cell Carcinoma and Was Regulated by MiR-340-5p. 32248620_The Warburg Micro Syndrome-associated Rab3GAP-Rab18 module promotes autolysosome maturation through the Vps34 Complex I. 32513919_PIK3C3 regulates the expansion of liver CSCs and PIK3C3 inhibition counteracts liver cancer stem cell activity induced by PI3K inhibitor. 32602837_Membrane characteristics tune activities of endosomal and autophagic human VPS34 complexes. 32833964_An iron-dependent metabolic vulnerability underlies VPS34-dependence in RKO cancer cells. 33400087_Targeting the Autophagy Specific Lipid Kinase VPS34 for Cancer Treatment: An Integrative Repurposing Strategy. 33637724_VPS34 K29/K48 branched ubiquitination governed by UBE3C and TRABID regulates autophagy, proteostasis and liver metabolism. 33664238_SRSF1 inhibits autophagy through regulating Bcl-x splicing and interacting with PIK3C3 in lung cancer. 33692360_Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. | ENSMUSG00000033628 | Pik3c3 | 1.836282e+02 | 0.5270877 | -0.923885010 | 0.4927593 | 3.458861e+00 | 0.0629136542 | 0.70014047 | No | Yes | 8.950216e+01 | 26.947745 | 1.787429e+02 | 54.969639 | ||
| ENSG00000078304 | 5527 | PPP2R5C | protein_coding | Q13362 | FUNCTION: The B regulatory subunit might modulate substrate selectivity and catalytic activity, and also might direct the localization of the catalytic enzyme to a particular subcellular compartment. The PP2A-PPP2R5C holoenzyme may specifically dephosphorylate and activate TP53 and play a role in DNA damage-induced inhibition of cell proliferation. PP2A-PPP2R5C may also regulate the ERK signaling pathway through ERK dephosphorylation. {ECO:0000269|PubMed:16456541, ECO:0000269|PubMed:17245430}. | 3D-structure;Acetylation;Alternative splicing;Centromere;Chromosome;Nucleus;Phosphoprotein;Reference proteome | The product of this gene belongs to the phosphatase 2A regulatory subunit B family. Protein phosphatase 2A is one of the four major Ser/Thr phosphatases, and it is implicated in the negative control of cell growth and division. It consists of a common heteromeric core enzyme, which is composed of a catalytic subunit and a constant regulatory subunit, that associates with a variety of regulatory subunits. The B regulatory subunit might modulate substrate selectivity and catalytic activity. This gene encodes a gamma isoform of the regulatory subunit B56 subfamily. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:5527; | chromosome, centromeric region [GO:0000775]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein phosphatase type 2A complex [GO:0000159]; protein phosphatase activator activity [GO:0072542]; protein phosphatase regulator activity [GO:0019888]; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest [GO:0006977]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; negative regulation of cell population proliferation [GO:0008285]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein dephosphorylation [GO:0006470]; signal transduction [GO:0007165] | 15778281_B56gamma1 alters the dynamic assembly/disassembly process of nuclear speckles in heart cells. 16524888_Dihydroxyphenylethanol induces apoptosis by activating serine/threonine protein phosphatase PP2A 17245430_B56gamma, mediates DNA damage-induced dephosphorylation of p53 at Thr55. 17663574_PP2A-B56gamma1 phosphatase complexes have a role in cardiac local signaling 17967874_show an important link between ATM activity and the tumor-suppressive function of B56gamma-protein phosphatase 2A 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20448040_the dynamic nuclear distribution of the B56gamma3 regulatory subunit controls nuclear PP2A activity, which regulates cell cycle controllers, such as p27, to restrain cell cycle progression, and may be responsible for the tumor suppressor function of PP2A 20473327_a mechanistic basis for the p53-dependent and -independent functions of B56gamma-PP2A 21460856_ataxia-telangiectasia mutated (ATM) directly phosphorylates and specifically regulates B56gamma3, B56gamma2 and B56delta, after DNA damage. 21548944_Overexpression of PPP2R5C in T-cell malignancy as well as in myeloid leukemia cells might relate to its proliferation and differentiation. 22315229_structural insight into the PP2A holoenzyme assembly and emphasizes the importance of HEAT repeat 1 in B56gamma-PP2A tumor-suppressive function 23723076_Results show that mutations lost B56gamma tumor-suppressive activity by two distinct mechanisms: one is by disrupting interactions with the PP2A AC core and the other with B56gamma-PP2A substrates (p53 and unknown proteins). 23941244_PPP2R5C-siRNA treatment altered gene expression profiles in malignant T cells 24719332_Data indicate that the regulatory PP2A subunit B56gamma mediates suppression of NF-kappaB resulting in increased NF-kappaB target gene expression in T cells. 25512391_KIF4A and PP2A-B56G and -B56E create a spatially restricted negative feedback loop counteracting Aurora B in anaphase. 25884766_Differential expression of the transcripts PPP2R5C connects ubiquitin-proteasome system with infection-inflammation in preterm births and preterm premature rupture of membranes. 25888193_propose that the mechanism for proliferation inhibition and increased apoptosis of K562 cells following PPP2R5C suppression may be related to the alteration of expression profiles of BRAF, AKT2, AKT3, NFKB2 and STAT3 genes 25972378_Mutations in the PP2A regulatory subunit B family genes PPP2R5B, PPP2R5C and PPP2R5D cause human overgrowth 26440364_our data suggest that hepatic PPP2R5C represents an important factor in the functional wiring of energy metabolism and the maintenance of a metabolically healthy state. 26986830_PPP2R5C deletion or EGFR mutation that could be responsible for IER3/pERK overexpression was found in at least 8 cases (67% or more) of lung adenocarcinoma. 32071084_Kelch-like protein 42 is a profibrotic ubiquitin E3 ligase involved in systemic sclerosis. 32195664_A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment. 32561747_Structural basis of host protein hijacking in human T-cell leukemia virus integration. 33028863_Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56gamma. 35123608_[The Relationship between PPP2R5C and Molt-4 Cell Viability, HSP90-GR Signal in Childhood Acute T Lymphocytic Leukemia]. | ENSMUSG00000017843 | Ppp2r5c | 1.590839e+03 | 0.9023604 | -0.148224348 | 0.2950044 | 2.514514e-01 | 0.6160549614 | 0.91334267 | No | Yes | 1.514546e+03 | 217.279967 | 1.599369e+03 | 235.112345 | |
| ENSG00000078319 | 5379 | PMS2P1 | transcribed_unprocessed_pseudogene | 6.985660e+02 | 0.9077240 | -0.139674384 | 0.2752059 | 2.560977e-01 | 0.6128138718 | 0.91285485 | No | Yes | 5.757774e+02 | 50.593552 | 5.876039e+02 | 52.973343 | ||||||||||
| ENSG00000078399 | 3205 | HOXA9 | protein_coding | P31269 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. Required for induction of E-selectin and VCAM-1, on the endothelial cells surface at sites of inflammation. {ECO:0000269|PubMed:22269951}. | Chromosomal rearrangement;DNA-binding;Developmental protein;Homeobox;Methylation;Nucleus;Proto-oncogene;Reference proteome;Transcription;Transcription regulation | In vertebrates, the genes encoding the class of transcription factors called homeobox genes are found in clusters named A, B, C, and D on four separate chromosomes. Expression of these proteins is spatially and temporally regulated during embryonic development. This gene is part of the A cluster on chromosome 7 and encodes a DNA-binding transcription factor which may regulate gene expression, morphogenesis, and differentiation. This gene is highly similar to the abdominal-B (Abd-B) gene of Drosophila. A specific translocation event which causes a fusion between this gene and the NUP98 gene has been associated with myeloid leukemogenesis. Read-through transcription exists between this gene and the upstream homeobox A10 (HOXA10) gene.[provided by RefSeq, Mar 2011]. | hsa:3205; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; enzyme binding [GO:0019899]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; embryonic skeletal system morphogenesis [GO:0048704]; endothelial cell activation [GO:0042118]; multicellular organism development [GO:0007275]; negative regulation of myeloid cell differentiation [GO:0045638]; proximal/distal pattern formation [GO:0009954]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription, DNA-templated [GO:0006351] | 9058712_demonstrates a physiological role of the mouse protein in blood cell differentiation, with the greatest apparent influence at the level of the commited progenitor 11830496_the fusion gene NUP98-HOXA9 is an important gene in myeloid leukemogenesis. 12082612_Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1. 12112533_The HOXA9 cluster gene is frequently expressed in cell lines from acute myeloid leukemia cases with 11p15 translocations; in some cases HOX9 is not fused to NUP98. 12138901_The NUP98/HOXA9 FUSION transcript was detected by PC at exon A and not exon B of NUP98. 12923056_HOXA9 complexed with Pbx1 and DNA, so that the posterior Hox hexapeptide adopts an altered conformation. 14561764_Nup98-HOXA9 has a role in inducing gene transcription in myeloid cells 14562113_B-lineage development can proceed in t(4;11) leukemic blasts in the absence of HOX-A gene expression. 14701735_Hoxa9 and Hoxa7 as well as the Hox coregulators Meis1 and Pbx3 among the targets upregulated by MLL-ENL-ERtm in conditionally transformed cells 14738146_Lower expression of HOXA9 is associated with acute myeloid leukemia-M2 14764452_HoxA9 binds to the EphB4 promoter & stimulates its expression resulting in an increase of endothelial cell migration and tube forming activity. Thus, modulation of EphB4 expression may contribute to the proangiogenic effect of HoxA9 in endothelial cells. 15160920_suggest that MLL aberrations may regulate MEIS1 and HOXA9 gene expression in ALL-derived cell lines, while AML-derived cell lines express these genes independently of the MLL status 15254242_HOXA9 nuclear transport is induced by thrombopoietin in immature hematopoietic cells 15454493_Dnalc4, Fcgr2b, Fcrl, and Con1 genes cooperated with NUP98-HOXA9 in transforming NIH 3T3 cells 15479723_advantage to Meis1-HoxA9 coexpressing cells in vivo, leading to leukemogenesis. 15657436_HOXA9, is a positive regulator of eIF4E. HOXA9 stimulates eIF4E-dependent export of cyclin D1 and ornithine decarboxylase (ODC) mRNAs in the nucleus, as well as increases the translation efficiency of ODC mRNA in the cytoplasm. 15681849_CYBB is a common target gene repressed by HoxA10 and activated by HoxA9, and Meis1 and Nup98-hoxA9 have roles in repressing myeloid-specific gene transcription 15960975_The HOXA9 protein expression was significantly downregulated in the MOF knockdown cells compared to control siRNA-treated cells. 16105979_90% of meningioma 1-ets variant gene 6 +/HOXA9+ mice developed AML much more rapidly than control HOXA9+ mice 16630659_The relative HOXA9 expression was higher in patients in the accelerated phase of the disease (p<0.005). Interestingly, a patient with poorer prognosis (high Sokal's score), showing the highest HOXA9/ABL ratio, quickly entered a blast crisis and died. 16803519_the expression of abd-b genes may be an imortant step involved in cervical cancer. 17178874_The t(7;11)(p15;p15) translocation, observed in acute myelogenous leukemia and myelodysplastic syndrome, generates a chimeric gene where the 5' portion of the sequence encoding the human nucleoporin NUP98 protein is fused to the 3' region of HOXA9. 17327400_Pim1 appears to be a direct transcriptional target of HOXA9 and a mediator of its antiapoptotic and proproliferative effects in early cells 17452460_HOXA9 participates in the transcriptional activation of SELE in endothelial cells. 17586512_HOXA9 inhibits endothelial cell activation downstream of NF-kappaB nuclear localization by interfering with NF-kappaB DNA binding, but not transactivation capacity 17623056_DNA hypermethylation of tumour suppressor genes seems to play an important role in ovarian carcinogenesis and HOXA9, HOXB5, SCGB3A1, and CRABP1 are identified as novel hypermethylated target genes in this tumour type 17761289_In this expt. mice with a compound deficiency in hoxa9, hoxb3 and hoxb4 (hoxa9/b3/b4) were investigated for evidence of synergy between these genes in hematopoiesis. 17885552_In hypertensive patients, downregulation of HOXA9 expression in peripheral CD34+ cells may have a role in the loss of circulating endothelial progenitor cells, potentially impairing postnatal neovascularization and vascular repair. 18068911_nuclear factor-kappaB (NF-kappaB) activation is an essential step for HOXA9 downregulation. And HOXA9 regulates its own expression by positive feedback mechanism. 18337761_HoxA9 overexpression induces IGF-1R expression and subsequently promotes leukemic cell growth 18410541_analysis of the role of deregulated PcG genes in acute myeloid leukemia, and the downstream PcG targets HOXA4, HOXA9 and MEIS1 18474618_miR-126 may function in normal hematopoietic cells to modulate HOXA9 protein. 18668134_HOXA9 overexpression is associated with acute myeloid leukemias. 19056693_An important role for HOXA9 in human MLL-rearranged leukemias. 19157684_Enforced overexpression of HOXA6 or HOXA9 in FDCP-Mix resulted in increased proliferation and colony formation but had negligible effect on differentiation in early multipotential and later committed precursor cells 19346496_Gfi1 integrates 2 events during normal myeloid differentiation; the suppression of a HoxA9-Pbx1-Meis1 progenitor program and the induction of a granulopoietic transcription program. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19620287_CREB1 may mediate HOXA9 modulation of Meis1 expression. 19696924_effects of NUP98-HOXA9 on gene transcription and cell transformation are mediated by two distinct mechanisms: promoter binding through homeodomain with direct transcriptional activation, and another depending on NUP98 moiety not involving DNA binding 19938081_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20068170_HOXA9 activation is a novel, independent, and negative prognostic marker in GBM that is reversible through a PI3K-associated epigenetic mechanism. Findings suggest a transcriptional pathway through which PI3K activates oncogenic HOXA expression. 20141430_Increased expression of HoxA9 is associated with chronic myeloid leukemia. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20389018_HOXA9 restricts breast tumor aggression by modulating expression of the tumor suppressor gene BRCA1 20565746_major differentiation factors of the NK-cell lineage, including HOXA9, HOXA10 and ID2, were (de)regulated via PRC2 which therefore contributes to T-cell leukemogenesis. 20661663_knockdown of BMI-1 expression can induce cell-cycle arrest and up-regulate p16INK4a, HOXA9 and HOXC13 in HeLa cells 20727230_Studies indicate that HOXA9 restrains breast cancer progression by upregulation of BRCA1. 20935218_Data show an in vitro link between phosphoinositide 3-kinase-mediated HOXA9/HOXA10 expression, and a drug-resistant, progenitor cell phenotype in MGMT-independent pediatric glioblastoma. 21259057_the aim of this study was to investigate the prognosis and clinicopathologic roles of beta-catenin, Wnt1, Smad4, Hoxa9, and Bmi-1 in esophageal squamous cell carcinoma 21294585_proposed two possible model structures of Hoxa9 and Smad4 complex. 21467841_NUP98 and leukemogenic fusion protein NUP98-HOXA9 interaction partners disrupt the leukemic cell cycle. 21480815_prevalence of HOXA9 promoter methylation in tissue and induced sputum samples from Korean patients with lung cancer 21518888_Several targets of MLL fusions, MEIS1, HOXA7, HOXA9, and HOXA10 are functionally related and have been implicated in leukemias. Each of the four genes was knocked down separately in the precursor B-cell leukemic line RS4;11 expressing MLL-AF4. 21558411_HOXA9 promoter hypermethylation is associated with oral cavity squamous cell carcinoma. 21971947_Hoxa9 and Hoxa13 are involved in the early and organised patterning of ENS development in the zebrafish model. 22251480_Data show that up-regulation of the HOXA7, HOXA9, HOXA11, and PBX3 resulting from the down-regulation of miR-181 family members probably contribute to the poor prognosis of patients with nonfavorable cytogenetically abnormal AML (CA-AML). 22269951_HOXA9 is required for TNF-alpha-dependent binding of PRMT5 to the E-selectin promoter in endothelial cells. 22353710_findings show that HOXA9 and MEIS1 are direct targets of miRNA-196b, a microRNA located adjacent to and co-expressed with HOXA9, in MLL-rearranged leukaemic cells 22541086_All-trans-Retinoic Acid can up-regulate the expression of HOXA9 mRNA and protein in HL-60 cells. 22633751_Both truncated and full-length forms of HOXA9 are highly expressed in human MLL-rearranged leukemia and cooperate with each other in cell transformation and leukemogenesis. 22713874_PHD12 facilitates the localization of MOZ onto the promoter locus of the HOXA9 gene, thereby promoting the H3 acetylation around the promoter region. 22902925_HOXA9-DACH1 complex mediated by the carboxyl terminus of DACH1 in t(9;11) leukemia cells was identified. 22944570_Inhibition of HoxA9EC induced by high glucose contributed to endothelial cell dysfunction, which could be rescued by augmentation of HoxA9EC. 22945634_HOXA9 expression is associated with poor outcomes in epithelial ovarian cancer & and in mouse EOC xenografts. HOXA9 expression in EOC cells promotes a microenvironment permissive for tumor growth, via paracrine TGF-beta-2 action on fibroblasts. 23028966_Data identified the homeodomain protein HOXA9 as a positive regulator of SHOX expression in U2OS cells. 23056278_Methylation levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 in urine specimens are promising diagnostic biomarkers for bladder cancer recurrence surveillance 23264595_Collectively, our data suggest that PBX3 is a critical cofactor of HOXA9 in leukemogenesis. 23326393_Data indicate that retroviral transduction-mediated overexpression or siRNA-mediated knock-down of Hoxa9 respectively down-regulated or up-regulated Geminin in hematopoietic cells. 23349797_Data indicate that genes with hypermethylated CpG islands in malignant meningiomas, such as HOXA6 and HOXA9, tend to coincide with the binding sites of polycomb repressive complexes (PRC) in early developmental stage. 23403989_The shRNA for HMGB1 and siRNA for HOXA9 successfully decreased the expression levels of HMGB1 and HOXA9, respectively 23436614_The methylation status of Homeobox A9 (HOXA9), ISL LIM homeobox 1 (ISL1) and Aldehyde dehydrogenase 1 family, member A3 (ALDH1A3) was significantly associated with decreased gene expression levels 23466061_The competition for this binding site inhibits the expression of HOXA9 and induces different transcriptional outcomes, which suggests a new direction for investigation of the mechanism underlying targeted therapy of malignant gliomas. 23539541_Cell growth and function are dependent on maintained levels of signature gene HOXA9. 23575938_The complete remission rate of the patients who expressed the gene was significantly lower than that in patients who did not express the gene after chemotherapy. 23607948_High expression of HOXA9 is associated with the occurrence of acute leukemia, and its expression level is significantly higher in children with AML than in those with ALL. 23660297_Hoxa7, Hoxa9 and Hox cofactor Meis1 were identified as AP-2alpha target genes, which are involved in myeloid leukemogenesis. 23716660_Data indicate that the genes comprising the HMGA2-TET1-HOXA9 pathway are coordinately regulated in breast cancer and together encompass a prognostic signature for patient survival. 23840580_DYNLT1 interacts with nucleoporins and plays a role in the dysregulation of gene expression and induction of hematopoietic cell proliferation by the leukemogenic nucleoporin fusion, NUP98-HOXA9 23894305_A low level of expression of miR-196b can cause up-regulation of BCR-ABL1 and HOXA9 expression, which leads to the development of chronic myeloid leukemia. 23994619_Symplekin interacts and co-localizes with both MOZ and MLL in immature hematopoietic cells. Its inhibition leads to a decrease of the HOXA9 protein level but not of Hoxa9 mRNA. 24051379_SALL4 binds a specific region of the HOXA9 promoter in leukemic cells. 24081945_hypermethylation of HIST1H4F, PCDHGB6, NPBWR1, ALX1, and HOXA9 was significantly associated with shorter survival in stage 1 Non-small-cell lung cancer 24308157_HOXA9 acts as an oncogene in laryngeal squamous cell carcinoma, promoting tumor cell proliferation and migration. 24332016_HOXA9 may, therefore, stimulate ovarian cancer progression by promoting an immunosuppressive microenvironment via paracrine effects on peritoneal macrophages. 24486589_we define a three-gene panel, CDO1, HOXA9, and TAC1, which we subsequently validate in two independent cohorts of primary NSCLC samples 24535405_Hoxa9T on its own induced acute myeloid leukemia after a similar latency as Hoxa9FLim, despite its inability to bind DNA 24681432_HOXA9 methylation could be detected in plasma from HCC patients (n=40) but not in normal plasma (n=34) (p<0.0005). 24817037_present study suggests that HOXA9 inhibits migration of lung cancer cells and its hypermethylation is an independent prognostic factor for recurrence-free survival in never-smokers with NSCLC 24886209_HOXA9 methylation is frequent in oral cancers and levels are higher in tumors with greater risk of metastasis. Expression of HOXA9 is low in cells with high levels of methylation and reduced expression appears to confer a growth advantage. 25023983_These findings indicate that HOXA9 contributes to poor outcomes in Epithelial ovarian cancer in part by promoting intraperitoneal dissemination via its induction of P-cadherin. 25031718_these findings indicate that homeobox A9 regulates the expression of VEGF in epidermal stem cells 25185710_Different gain-of-function and loss-of-function studies reveal that HOXA9 enhances hematopoietic differentiation of embryonic stem cells by specifically promoting the commitment of hemogenic precursors into primitive and total CD45(+) blood cells. 25246116_variants in the HOXA7 and HOXA9 genes are not common in Chinese women with Mullerian duct abnormalities 25762636_Data establish HOXA9 as a driver of glioma initiation, aggressiveness and resistance to therapy. 25987065_Increased expression of HOXA9 is associated with acute myeloid leukemia. 26028034_With continued advances in understanding HOXA9-mediated transformation, there is a wealth of opportunity for developing novel therapeutics that would be applicable for greater than 50% of AML with overexpression of HOXA9 26059450_HOXA9 and MEIS1 overexpression are inversely correlated with relapse and overall survival, so the genes could become useful predictive markers of the clinical course of pediatric acute leukemias. 26130510_Pbx3 contributes to Hoxa9 leukemogenesis through stabilization of the Meis1 protein. 26332997_Concurrent ISL1/HOXA9 methylation in HG-NMIBC reliably predicted tumour recurrence and progression within one year, and was associated with disease-specific mortality 26586336_This study identified HOXA9 as a target gene of miR-196b and determined that the mechanism of miR-196b-mediated epithelial-to-mesenchymal transition and invasion processes involves the regulation of HOXA9 expression in non-small cell lung cancer cells. 26679037_Study found that hypermethylation of CpG probes in the promoter regions of HOXA9 and PCDH10 was associated with mature B-cell neoplasms. 26740045_The results indicate that highly selective targeting of Nup98-fusion proteins to Hox cluster regions via prebound Crm1 induces the formation of higher order chromatin structures that causes aberrant Hox gene regulation. 27020427_Promoter variants in HOXA9, TPM1, and TPM2, alter promoter expression suggesting that they have a functional role in clubfoot. 27039722_Controlling HOXA9 expression appears to be a necessary step during CC development. 27409175_HOXA9 expression is upregulated in three-dimensional organotypic culture of claudin-low breast cancer cells. 27598218_In the present study, we found in a translational stepwise approach that promoter DNA methylation of the homeobox (HOX) gene HOXA9 could help predict resistance or response to cisplatin-based chemotherapy in patients with bladder cancer 27838340_NUP98-HOXA9 ability to induce blood cell expansion is evolutionarily conserved. 27880728_Low HOXA9 expression is associated with Non Small Cell Lung Cancer. 28210005_The HOXA9 moiety of NUP98-HOXA9 is essential for binding to the Hoxa gene locus. MLL is important for the recruitment of NUP98-HOXA9 to the HOXA locus and for NUP98-HOXA9-induced HOXA gene expression. 28292467_HOXA9 transcriptionally regulated EPHB4 expression to modulate trophoblasts migration and invasion, which may suggest a contribution of HOXA9-EPHB4 in the poor placentation in the pathogenesis of preeclampsia. 28387653_Homeobox A9 (HOXA9) expression was inversely correlated with microRNA miR-196b levels in recurrent epithelial ovarian cancer (EOC). 28399410_HOXA9 role in acute myeloid leukemia. 28464221_Overexpression of HOXA4 and HOXA9 contributes to self-renewal and overpopulation of stem cells in colorectal cancers. 28630438_NHA9 (NUP98-HOXA9 fusion protein) deregulates the expression of key leukemic genes, including MEIS1-HOXA9-PBX3 complex, through the enhancer binding and the direct interaction of the fusion protein with HDAC and p300 transcriptional regulators 28707666_This is the first time the protein partners of either E2A-PBX1 or HOXA9 oncoproteins were identified using an unbiased biochemical approach. The identification of translation initiation factors associated with HOXA9 might indicate a novel function for HOX proteins independent of their transcriptional activity. 28713930_knockdown of HOXA9 abrogated NAC1induced drug resistance. 28723696_Advanced glycation end products could induce endothelial cell dysfunction through NF-kappaB dependent HoxA9 downregulation. 28748001_Results show that a combination of HOXA9 and HOXA10 promoter methylation markers is significantly associated with the prognosis of breast cancer patients a subtype-independent manner. 28780773_The present study demonstrated that miR-139-5p, functioning as a suppressor in the tumorigenesis process, suppressed oral squamous carcinoma cells mobility by targeting HOXA9 directly. 28947139_HOTTIP/WDR5/HOXA9/Wnt axis contributes to pancreatic tumor stem cell stemness. 28978671_Beta-catenin regulates expression of downstream targets of a key transcriptional memory gene, Hoxa9 that is highly enriched in myeloid leukemia cancer stem cells and helps sustain leukemic self-renewal. 29054094_MiR-182/HOXA9 was involved in the process of RUNX3-mediated GC tumor growth. 29384529_Downregulation of HOXA9 in myeloid leukaemia cells led to increased differentiation capacity in vitro. 29496663_we show that HOXA9 and JAK3/STAT5 are bona fide cooperating factors in driving leukemia development and that the cooperation is situated at the transcriptional level where STAT5 and HOXA9 co-occupy similar genomic regions 29572507_High HOXA9 expression is associated with hematogical malignancy. 29662084_HOXA9 inhibits HIF1A-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. 30032824_this study shows HOXA9 methylation and blood vessel invasion in formalin-fixed paraffin-embedded tissues for prognostic stratification of stage I lung adenocarcinoma patients 30214626_These data indicate that NPMc+ leukemic cell survival requires upregulation of PBX3 and HOXA9. 30270123_that therapeutic targeting of HOXA9-dependent enhancer reorganization can be an effective therapeutic strategy in acute leukemia with HOXA9 overexpression 30418048_HOXA9, RASSF1A and SCGB3A1 promoter methylation significantly associated with testicular germ cell tumor stage. 30552679_The results indicated that completely different sets of transcription factors coregulate HOXA4 and HOXD10 (but not HOXA9) and their expression-correlated genes. 30575897_miR-1294 functions as a tumor suppressor in OS progression by targeting HOXA9. 30622285_High HOXA9 expression is associated with increased metabolic dysregulation and leukemogenesis. 30674274_Based on the human, animal and cellular data, the transcription factor HOXA9 may promote the expression of pro-myopia genes and retinal pigment epithelial proliferation, which eventually contribute to myopia development. 30816476_Results identified HOXA9 as a potential target of miR873. 3'UTR of HOXA9 was recognized and targeted by miR873 in osteosarcoma (OS) cells. HOXA9 expression was upregulated in OS tissues, and inversely correlated with miR873 expression. 30839116_MicroRNA-652-3p promotes the proliferation and invasion of the trophoblast HTR-8/SVneo cell line by targeting homeobox A9 to modulate the expression of ephrin receptor B4 30843252_HOXA9 promoter hypermethylation is associated with the risk for head and neck squamous cell carcinoma and its progression and metastasis. 31041828_Comparative transcriptome analysis of sinonasal inverted papilloma and associated squamous cell carcinoma: Out-HOXing developmental genes. 31043660_Transcription Factor HOXA9 is Linked to the Calcification and Invasion of Papillary Thyroid Carcinoma. 31099823_HOXA9 drives growth in premalignant colonic polyps, but simultaneously prevents further transformation 31112908_A novel, so far overlooked, protein encoded by HoxA9 in humans and mice, has been described. 31424148_HOXA9 rs3801776 G>A polymorphism increases congenital talipes equinovarus risk in a Chinese population. 31670914_Correlation of miR-181a and three HOXA genes as useful biomarkers in acute myeloid leukemia. 31683603_Loss of HOXA9 in cSCC significantly upregulates RELA expression and thus enhances NF-kappaB pathway. RELA transcriptionally promotes not only anti-apoptotic factor BCL-XL but also autophagic genes including ATG1, ATG3, and ATG12. 31740654_The direct regulation of HOXA9 by miR-652 was experimentally validated in uveal melanoma cells by dual luciferase assay and Western blotting. Authors also observed that miR-652 promoted HIF-1alpha signaling via repression of HOXA9 in uveal melanoma cells. 31768018_MicroRNA-708 is a novel regulator of the Hoxa9 program in myeloid cells. 31865042_Longitudinal monitoring of HOXA9 meth-ctDNA is clinically feasible and is strongly correlated to clinical outcomes (PFS, OS). 31875764_MiR-210 inhibited the expression of HOXA9 to activate the NF-kappaB signaling pathway and mediated the occurrence of epithelial-mesenchymal transition (EMT) of pancreatic cancer cells induced by HIF-1a under hypoxia. 31881106_miR-647 inhibits glioma cell proliferation, colony formation and invasion by regulating HOXA9. 31906958_MiR-182-5p and its target HOXA9 in non-small cell lung cancer: a clinical and in-silico exploration with the combination of RT-qPCR, miRNA-seq and miRNA-chip. 31923345_A novel molecular link between HOXA9 and WNT6 in glioblastoma identifies a subgroup of patients with particular poor prognosis. 31986484_Generation and characterization of a human iPSC line derived from congenital clubfoot amniotic fluid cells. 32191343_Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer. 32430478_Detection of Promoter DNA Methylation in Urine and Plasma Aids the Detection of Non-Small Cell Lung Cancer. 32730594_Trib1 promotes acute myeloid leukemia progression by modulating the transcriptional programs of Hoxa9. 32820015_Entospletinib in Combination with Induction Chemotherapy in Previously Untreated Acute Myeloid Leukemia: Response and Predictive Significance of HOXA9 and MEIS1 Expression. 33001025_Functional interrogation of HOXA9 regulome in MLLr leukemia via reporter-based CRISPR/Cas9 screen. 33057942_Disruption of CTCF Boundary at HOXA Locus Promote BET Inhibitors' Therapeutic Sensitivity in Acute Myeloid Leukemia. 33069783_HOXA9/IRX1 expression pattern defines two subgroups of infant MLL-AF4-driven acute lymphoblastic leukemia. 33202249_Gene- and Species-Specific Hox mRNA Translation by Ribosome Expansion Segments. 33461426_Homeobox A5 and A9 expression and beta-thalassemia. 33924850_Nuclear FGFR2 Interacts with the MLL-AF4 Oncogenic Chimera and Positively Regulates HOXA9 Gene Expression in t(4;11) Leukemia Cells. 34163069_Phase separation drives aberrant chromatin looping and cancer development. 34416888_MicroRNA-638 inhibits the progression of breast cancer through targeting HOXA9 and suppressing Wnt/beta-cadherin pathway. 34514844_HOXA9-methylated DNA as a diagnostic biomarker of ovarian malignancy. 34831074_The Retinoblastoma Tumor Suppressor Is Required for the NUP98-HOXA9-Induced Aberrant Nuclear Envelope Phenotype. 34850551_ALKBH5-mediated m6A modification of lncRNA KCNQ1OT1 triggers the development of LSCC via upregulation of HOXA9. 35372588_Comprehensive Landscape of HOXA2, HOXA9, and HOXA10 as Potential Biomarkers for Predicting Progression and Prognosis in Prostate Cancer. | ENSMUSG00000038227 | Hoxa9 | 1.252749e+03 | 1.0914483 | 0.126243859 | 0.2872917 | 1.981172e-01 | 0.6562448830 | 0.92574148 | No | Yes | 1.055613e+03 | 133.929486 | 1.031059e+03 | 134.107130 | |
| ENSG00000078668 | 7419 | VDAC3 | protein_coding | Q9Y277 | FUNCTION: Forms a channel through the mitochondrial outer membrane that allows diffusion of small hydrophilic molecules. {ECO:0000250|UniProtKB:P21796}. | Acetylation;Alternative splicing;Ion transport;Isopeptide bond;Membrane;Mitochondrion;Mitochondrion outer membrane;NAD;Nucleotide-binding;Phosphoprotein;Porin;Reference proteome;Transmembrane;Transmembrane beta strand;Transport;Ubl conjugation | This gene encodes a voltage-dependent anion channel (VDAC), and belongs to the mitochondrial porin family. VDACs are small, integral membrane proteins that traverse the outer mitochondrial membrane and conduct ATP and other small metabolites. They are known to bind several kinases of intermediary metabolism, thought to be involved in translocation of adenine nucleotides, and are hypothesized to form part of the mitochondrial permeability transition pore, which results in the release of cytochrome c at the onset of apoptotic cell death. Alternatively transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Oct 2011]. | hsa:7419; | extracellular exosome [GO:0070062]; membrane [GO:0016020]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; pore complex [GO:0046930]; nucleotide binding [GO:0000166]; porin activity [GO:0015288]; voltage-gated anion channel activity [GO:0008308]; adenine transport [GO:0015853]; regulation of cilium assembly [GO:1902017] | 14739283_VDAC2 and VDAC3 might have an alternative structural organization and different functions in outer dense fibers than in mitochondria 17683036_VDAC3 is a novel target for protein S-nigrosylation in spermatozoa. 20138821_VDAC3 has a limited ability to support mitochondrial respiration and has no influence in the control of ROS production. 20434446_The substitution of the VDAC3 N-terminus with the VDAC1 N-terminus caused the chimaera to become more active than VDAC1. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22935710_VDAC3 is present at the mother centriole and modulates centriole assembly by recruiting Mps1 to centrosomes. 23060438_VDAC 1, 2, and 3 recruit Parkin to defective mitochondria to promote mitochondrial autophagy. 23388454_a VDAC3-Mps1 module at the centrosome promotes ciliary disassembly during cell cycle entry. 25171321_observed electrophysiological properties of hVDAC3 are surprisingly different from the other isoforms and are discussed in relation to the proposed physiological role of the protein in mammalian cells 25341036_These data suggest that an interaction between Mcl-1 and VDAC promotes lung cancer cell migration by a mechanism that involves Ca(2+)-dependent reactive oxygen species production. 26407725_channel gating of VDAC3 might be controlled by redox sensing under physiological conditions 26806159_VDAC3 is able to modulate its pore size and current by exploiting the mobility of the N-terminal and forming, upon external stimuli, disulfide bridges with cysteine residues located on the barrel and exposed to the inter-membrane space. 26947058_The works available on VDAC cysteines support the notion that VDAC1, VDAC2, and VDAC3 proteins are paralogs with a similar pore-function and slightly different, but important, ancillary biological functions. (Review) 28431403_The AG genotype of rs16891278 showed a significantly lower sperm concentration compared with the AA genotype (P = 0.044). Findings suggest that VDAC3 genetic variants may be associated with human sperm count. 28916538_This cohort study showed that the VDAC3 gene was down regulation between patients with idiopathic Parkinson disease and controls 29378599_miR-3928v is induced by HBx through the NF-kappaB/EGR1 signaling pathway and down-regulates the tumor suppressor gene VDAC3 to accelerate the progression of HCC. 31794266_Data suggest that the hub genes NADH:ubiquinone oxidoreductase subunit B5 (NDUFB5), translocase of inner mitochondrial membrane domain containing 1 (TIMMDC1), and voltage-dependent anion channel 3 (VDAC3) might serve as potential biomarkers for diagnosis and/or therapeutic targets for precise treatment of septic cardiomyopathy (SC) . 31935282_A lower affinity to cytosolic proteins reveals VDAC3 isoform-specific role in mitochondrial biology. 31974380_VDAC3 role in the melanoma drug resistance to erastin.Erastin-induced resistance mediated by FOXM1-Nedd4-VDAC2/VDAC3 negative feedback loop in melanoma.Nedd4 ubiquitinates and degrades VDAC3. 32098132_A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (hVDACs). 32829673_Expression of voltage-dependent anion channels in endometrial cancer and its potential prognostic significance. | ENSMUSG00000008892 | Vdac3 | 2.128502e+03 | 0.5335620 | -0.906272063 | 0.3222853 | 7.937378e+00 | 0.0048423950 | 0.24781502 | No | Yes | 1.546154e+03 | 298.108020 | 2.695569e+03 | 532.314897 | |
| ENSG00000079435 | 3991 | LIPE | protein_coding | Q05469 | FUNCTION: Lipase with broad substrate specificity, catalyzing the hydrolysis of triacylglycerols (TAGs), diacylglycerols (DAGs), monoacylglycerols (MAGs), cholesteryl esters and retinyl esters (PubMed:8812477, PubMed:15955102, PubMed:15716583, PubMed:19800417). Shows a preferential hydrolysis of DAGs over TAGs and MAGs and preferentially hydrolyzes the fatty acid (FA) esters at the sn-3 position of the glycerol backbone in DAGs (PubMed:19800417). Preferentially hydrolyzes FA esters at the sn-1 and sn-2 positions of the glycerol backbone in TAGs (By similarity). Catalyzes the hydrolysis of 2-arachidonoylglycerol, an endocannabinoid and of 2-acetyl monoalkylglycerol ether, the penultimate precursor of the pathway for de novo synthesis of platelet-activating factor (By similarity). In adipose tissue and heart, it primarily hydrolyzes stored triglycerides to free fatty acids, while in steroidogenic tissues, it principally converts cholesteryl esters to free cholesterol for steroid hormone production (By similarity). {ECO:0000250|UniProtKB:P15304, ECO:0000250|UniProtKB:P54310, ECO:0000269|PubMed:15716583, ECO:0000269|PubMed:15955102, ECO:0000269|PubMed:19800417, ECO:0000269|PubMed:8812477}. | Alternative splicing;Cell membrane;Cholesterol metabolism;Cytoplasm;Diabetes mellitus;Hydrolase;Lipid degradation;Lipid droplet;Lipid metabolism;Membrane;Obesity;Phosphoprotein;Reference proteome;Steroid metabolism;Sterol metabolism | PATHWAY: Glycerolipid metabolism; triacylglycerol degradation. | The protein encoded by this gene has a long and a short form, generated by use of alternative translational start codons. The long form is expressed in steroidogenic tissues such as testis, where it converts cholesteryl esters to free cholesterol for steroid hormone production. The short form is expressed in adipose tissue, among others, where it hydrolyzes stored triglycerides to free fatty acids. [provided by RefSeq, Jul 2008]. | hsa:3991; | caveola [GO:0005901]; cytosol [GO:0005829]; lipid droplet [GO:0005811]; membrane [GO:0016020]; 1,2-diacylglycerol acylhydrolase activity [GO:0102259]; 1,3-diacylglycerol acylhydrolase activity [GO:0102258]; acylglycerol lipase activity [GO:0047372]; all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity [GO:0047376]; hormone-sensitive lipase activity [GO:0033878]; retinyl-palmitate esterase activity [GO:0050253]; sterol esterase activity [GO:0004771]; triglyceride lipase activity [GO:0004806]; cholesterol metabolic process [GO:0008203]; diacylglycerol catabolic process [GO:0046340]; ether lipid metabolic process [GO:0046485]; lipid catabolic process [GO:0016042]; protein phosphorylation [GO:0006468]; triglyceride catabolic process [GO:0019433] | 11574428_Observational study of gene-disease association. (HuGE Navigator) 11731226_Observational study of gene-disease association. (HuGE Navigator) 11850754_Observational study of gene-disease association. (HuGE Navigator) 11979403_genes of C3, hormone-sensitive lipase, and PPARgamma may exert a modifying effect on lipid and glucose metabolism in familial combined hypersensitivity 12514936_Observational study of gene-disease association. (HuGE Navigator) 12518034_overexpression of HSL, despite increased lipase activity, does not lead to enhanced lipolysis 12534454_HSL i6 A5 HOMOZYGOSITY IS A RISK FACTOR FOR BODY FAT ACCUMULATION 12534454_Observational study of gene-disease association. (HuGE Navigator) 12701046_high concentrations of estradiol significantly increased both hormone-sensitive lipase expression and glycerol release relative to control 12727985_lipolytic catecholamine resistance of sc adipocytes in polycystic ovary syndrome is probably due to a combination of decreased amounts of beta(2)-adrenergic receptors, the regulatory II beta-component of protein kinase A, and hormone-sensitive lipase 12730334_High adrenaline levels can stimulate hormone-sensitive lipase(HSL) activity regardless of metabolic milieu. Large increases in adrenaline during exercise are able to further stimulate contraction-induced increase in HSL activity. 12765952_The presence of a catalytically inactive variant of this enzyme is associated with decreased lipolysis in abdominal subcutaneous adipose tissue of obese subjects 12970365_role of cyclic GMP in natriuretic peptide-mediated phosphorylation in adipocytes 14984467_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15231718_AMPK is a major regulator of skeletal muscle HSL activity that can override beta-adrenergic stimulation 15260473_catalytic serine of hormone-sensitive lipase is highly reactive and similar behavior was also observed with lipases with no lid domain covering their active site, or with a deletion in the lid domain 15308678_5'AMP-activated protein kinase phosphorylates hormone-sensitive lipase on Ser565 in human skeletal muscle during exercise with reduced muscle glycogen. Apparently, HSL Ser565 phosphorylation by AMPK during exercise 15345679_mechanism of infertility in HSL-deficient males is cell autonomous and resides in postmeiotic germ cells, because HSL expression in these cells is in itself sufficient to restore normal fertility 15456755_a pre-lipolysis complex containing at least AFABP and HSL exists 15609025_Basal HSL is decreased in patients with type 2 diabetes mellitus, and this may be a consequence of elevated plasma insulin levels. 15871848_Observational study of gene-disease association. (HuGE Navigator) 16243839_Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase 16534522_Observational study of gene-disease association. (HuGE Navigator) 16534522_the HSL C-60G polymorphism is associated with increased waist circumference in non-obese subjects 16690773_adrenergic stimulation contributes to the increase in HSL activity that occurs in human skeletal muscle in the first minute of exercise at 65% and 90% VO2 peak 16752181_HSL gene expression shows a regulation according to obesity status and is associated with increased adipose tissue lipase activity. 16822962_although HSL expression and Ser(659) phosphorylation in skeletal muscle during exercise is sex specific, total muscle HSL activity measured in vitro was similar between sexes 16940551_maternal type 1 diabetes is associated with TG accumulation and increased EL and HSL gene expression in placenta 17026959_hormone-sensitive lipase 17074755_Adipose triglyceride lipase and hormone sensitive lipase are responsible for more than 95% of the triacylglycerol hydrolase activity present in murine white adipose tissue. 17318300_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17318300_The combined effect of LIPC, LIPE and ApoCIII gene polymorphisms may increase the likelihood of gestational hypertension, but seemingly not of preeclampsia. 17327373_Adipose triglyceride lipase (ATGL) is less important than hormone-sensitive lipase (HSL) in regulating catecholamine-induced lipolysis. Both lipases regulate basal lipolysis in human adipocytes. ATGL expression is not influenced by obesity or PCOS. 17356053_In obese subjects, insulin resistance and hyperinsulinemia are strongly associated with ATGL and HSL mRNA and protein expression, independent of fat mass 17587400_Dydrogesterone and norethisterone increase secretion of HSL from abdominaal adipocytes. 18249203_Observational study of gene-disease association. (HuGE Navigator) 18249203_variation of the HSL gene might be associated with a physiological effect on in vivo beta-adrenoceptor-mediated fat oxidation 18383440_Real-time PCR revealed that large adipocytes expressed higher mRNA levels of hormone sensitive lipase. 18398140_Obesity is accompanied by impaired fasting glycerol release, lower HSL protein expression, and serine phosphorylation. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18820256_Mapping of the hormone-sensitive lipase binding site on the adipocyte fatty acid-binding protein (AFABP). Identification of the charge quartet on the AFABP/aP2 helix-turn-helix domain. 18824087_analysis of human, mouse and ovine Hormone Sensitive Lipase 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19018281_PKA activates human HSL against lipid substrates in vitro primarily through phosphorylation of Ser649 and Ser650 19164092_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19164092_These results suggest that the associations between physical activity and body fat and plasma lipoprotein/lipid concentrations in men are dependent on the LIPE C-60G polymorphism,. 19336475_Observational study of gene-disease association. (HuGE Navigator) 19369647_Observational study of gene-disease association. (HuGE Navigator) 19433586_results suggest that ATGL/CGI-58 acts independently of HSL and precedes its action in the sequential hydrolysis of triglycerides in human hMADS adipocytes 19491387_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19695247_TNFalpha decreased ATGL and HSL protein content and triglycerides (TG)-hydrolase activity but increased basal lipolysis due to a marked reduction in perilipin (PLIN) protein content 19913121_Observational study of gene-disease association. (HuGE Navigator) 20017959_The present study aimed at comparing expression and subcellular distribution of perilipin and hormone-sensitive lipase in two abdominal adipose tissues of lean and obese women. 20495294_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20495294_Those findings indicate improvement and conservation of lifestyle depending on genetic predisposition in ADIPOQ, PLIN and LIPE should be encouraged. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20734064_Observational study of gene-disease association. (HuGE Navigator) 20855565_Observational study of gene-disease association. (HuGE Navigator) 20926921_Data show that hormone-sensitive lipase activity is reduced in adipose tissue of patients with and without diabetes, while lipoprotein lipase activity is reduces only in patients with diabetes. 21081692_It is concluded that ACTH via the PKA pathway stimulates expression of SF-1, which activates transcription of LIPE presumably by interaction with putative binding sequences within promoter A 21241784_Studies indicate tha HSL is regulated by reversible phosphorylation on five critical residues. 21498783_Data indicate that altered ATGL and HSL expression in skeletal muscle could promote DAG accumulation and disrupt insulin signaling and action. 21543206_Resveratrol increased adipose triglyceride lipase gene and protein expressions, an effect that was not observed for hormone-sensitive lipase in human SGBS adipocytes. 21680814_total lipase, ATGL and HSL activities were higher in visceral white adipose tissue of cancer patients compared with individuals without cancer and higher in cancer patients with cachexia compared with cancer patients without cachexia 21826994_LIPE C-60G variation can inhibit the decrease of LDL-C and the increases of HDL-C and apo A-I in young healthy males, and can inhibit the decrease of LDL-C and the increase of insulin in young healthy females induced by a high-carbohydrate diet. 21919688_suggests that genetic variation of HSL may be a risk factor for male infertility 21933124_Enzyme promiscuity in the hormone-sensitive lipase family of proteins. 22553833_M. leprae suppresses lipid degradation through inhibition of HSL expression. 23688034_Serum triglyceride was significantly up-regulated in men with the (CG + GG) genotype of HSL promoter polymorphism. 24848981_These findings indicate the physiological significance of HSL in adipocyte function and the regulation of systemic lipid and glucose homeostasis and underscore the severe metabolic consequences of impaired lipolysis. 25475467_Identification of a homozygous nonsense variant p.Ala507fsTer563 in hormone sensitive lipase as the likely cause of the lipodystrophy phenotype in siblings. 25819461_Despite reductions in intramyocellular lipolysis and HSL expression, overexpression of HSL did not rescue defects in insulin action in skeletal myotubes from obese type 2 diabetic subjects. 27862896_The homozygous null LIPE mutation could result in marked inhibition of lipolysis from some adipose tissue depots and thus may induce an extremely rare phenotype of MSL and partial lipodystrophy in adulthood associated with complications of insulin resistance, such as diabetes, hypertriglyceridemia and hepatic steatosis. 31150775_HSL regulates vitamin A metabolism in quiescent hepatic stellate cells. 33445064_Dietary fat content and adipose triglyceride lipase and hormone-sensitive lipase gene expressions in adults' subcutaneous and visceral fat tissues. | ENSMUSG00000003123 | Lipe | 2.858089e+02 | 1.1569244 | 0.210294582 | 0.3578440 | 3.204579e-01 | 0.5713325768 | 0.89885610 | No | Yes | 3.016473e+02 | 52.758753 | 2.417640e+02 | 43.972570 |
| ENSG00000080371 | 23011 | RAB21 | protein_coding | Q9UL25 | FUNCTION: Small GTPase involved in membrane trafficking control (PubMed:18804435, PubMed:25648148). During the mitosis of adherent cells, controls the endosomal trafficking of integrins which is required for the successful completion of cytokinesis (PubMed:18804435). Regulates integrin internalization and recycling, but does not influence the traffic of endosomally translocated receptors in general (By similarity). As a result, may regulate cell adhesion and migration (By similarity). Involved in neurite growth (By similarity). Following SBF2/MTMT13-mediated activation in response to starvation-induced autophagy, binds to and regulates SNARE protein VAMP8 endolysosomal transport required for SNARE-mediated autophagosome-lysosome fusion (PubMed:25648148). Modulates protein levels of the cargo receptors TMED2 and TMED10, and required for appropriate Golgi localization of TMED10 (PubMed:31455601). {ECO:0000250|UniProtKB:P35282, ECO:0000250|UniProtKB:Q6AXT5, ECO:0000269|PubMed:18804435, ECO:0000269|PubMed:25648148, ECO:0000269|PubMed:31455601}. | 3D-structure;Acetylation;Cell projection;Cytoplasmic vesicle;Direct protein sequencing;Endoplasmic reticulum;Endosome;GTP-binding;Golgi apparatus;Lipoprotein;Membrane;Methylation;Nucleotide-binding;Prenylation;Protein transport;Reference proteome;Transport | This gene belongs to the Rab family of monomeric GTPases, which are involved in the control of cellular membrane traffic. The encoded protein plays a role in the targeted trafficking of integrins via its association with integrin alpha tails. As a consequence, the encoded protein is involved in the regulation of cell adhesion and migration. Expression of this gene is associated with a poor prognosis for glioma patients. This gene is downregulated by the tumor suppressor miR-200b, and miRNA-200b is itself downregulated in glioma tissues. [provided by RefSeq, Nov 2015]. | hsa:23011; | axon cytoplasm [GO:1904115]; cleavage furrow [GO:0032154]; cytoplasmic side of early endosome membrane [GO:0098559]; cytoplasmic side of plasma membrane [GO:0009898]; cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; endomembrane system [GO:0012505]; endoplasmic reticulum membrane [GO:0005789]; endosome [GO:0005768]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; synapse [GO:0045202]; trans-Golgi network [GO:0005802]; vesicle membrane [GO:0012506]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; anterograde axonal transport [GO:0008089]; intracellular protein transport [GO:0006886]; positive regulation of dendrite morphogenesis [GO:0050775]; positive regulation of early endosome to late endosome transport [GO:2000643]; positive regulation of receptor-mediated endocytosis [GO:0048260]; protein stabilization [GO:0050821]; Rab protein signal transduction [GO:0032482]; regulation of axon extension [GO:0030516]; regulation of exocytosis [GO:0017157] | 16754960_Rab21 (and Rab5) associate with the cytoplasmic domains of alpha-integrin chains, and their expression influences the endo/exocytic traffic of integrins. This function of Rab21 is dependent on its GTP/GDP cycle and proper membrane targeting 18804435_Results show that targeted trafficking of integrins to and from the cleavage furrow is required for successful cytokinesis, and that this is regulated by Rab21. 19745841_show that TI-VAMP interacts with the Vps9 domain and ankyrin-repeat-containing protein Varp, a guanine nucleotide exchange factor of the small GTPase Rab21, through a specific domain herein called the interacting domain. 22525675_Taken together, these data suggest that Rab21 plays a negative role in the EGF-mediated MAPK signaling pathway. Overexpression of Rab21 attenuated EGF-mediated mitogen-activated protein kinase (MAPK) signaling by inducing EGFR degradation. 24477653_High RAB21 expression is associated with glioma. 25648148_Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. 28547526_we identified a novel gamma-secretase-associating protein Rab21 and illustrate that Rab21 promotes gamma-secretase internalization and translocation to late endosome/lysosome. Moreover, silencing of Rab21 decreases the gamma-secretase activity in APP processing thus production of Abeta. 29270202_Rab21 silencing can induce apoptosis and inhibit proliferation in human glioma cells. 30471852_Rab21 regulates LPS-induced pro-inflammatory responses by promoting TLR4 endosomal traffic and downstream signaling activation 31383875_Colorectal cancer cells respond differentially to autophagy inhibition in vivo. 35163051_Rab21 Protein Is Degraded by Both the Ubiquitin-Proteasome Pathway and the Autophagy-Lysosome Pathway. 35220907_Circular RNA Circ_0008043 promotes the proliferation and metastasis of hepatocellular carcinoma cells by regulating the microRNA (miR)-326/RAB21 axis. | ENSMUSG00000020132 | Rab21 | 6.508584e+02 | 1.2580347 | 0.331171748 | 0.2947853 | 1.267643e+00 | 0.2602093991 | 0.79684153 | No | Yes | 7.353761e+02 | 115.153758 | 5.330439e+02 | 85.659476 | |
| ENSG00000081154 | 57092 | PCNP | protein_coding | Q8WW12 | FUNCTION: May be involved in cell cycle regulation. | Acetylation;Alternative splicing;Cell cycle;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:57092; | nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cell cycle [GO:0007049]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein ubiquitination [GO:0016567] | 14741369_NIRF has ubiquitination activity, the hallmark of a ubiquitin ligase. PCNP was readily ubiquitinated in 293 and COS-7 cells, and NIRF ubiquitinated PCNP in vitro as well as in vivo. 29716528_PCNP mediates the proliferation, migration, and invasion of human neuroblastoma cells through mitogen-activated protein kinase and PI3K/AKT/mTOR signaling pathways, implying that PCNP is a therapeutic target for patients with neuroblastoma. 32548978_PCNP promotes ovarian cancer progression by accelerating beta-catenin nuclear accumulation and triggering EMT transition. 33629294_TIPE2 and PCNP expression abnormalities in peripheral blood mononuclear cells associated with disease activity in rheumatoid arthritis: a meta-analysis. | ENSMUSG00000071533 | Pcnp | 1.451877e+03 | 1.3321780 | 0.413786813 | 0.3128360 | 1.753737e+00 | 0.1854076751 | 0.77865017 | No | Yes | 1.663568e+03 | 288.254230 | 1.091546e+03 | 194.189079 | ||
| ENSG00000081189 | 4208 | MEF2C | protein_coding | Q06413 | FUNCTION: Transcription activator which binds specifically to the MEF2 element present in the regulatory regions of many muscle-specific genes. Controls cardiac morphogenesis and myogenesis, and is also involved in vascular development. Enhances transcriptional activation mediated by SOX18. Plays an essential role in hippocampal-dependent learning and memory by suppressing the number of excitatory synapses and thus regulating basal and evoked synaptic transmission. Crucial for normal neuronal development, distribution, and electrical activity in the neocortex. Necessary for proper development of megakaryocytes and platelets and for bone marrow B-lymphopoiesis. Required for B-cell survival and proliferation in response to BCR stimulation, efficient IgG1 antibody responses to T-cell-dependent antigens and for normal induction of germinal center B-cells. May also be involved in neurogenesis and in the development of cortical architecture (By similarity). Isoforms that lack the repressor domain are more active than isoform 1. {ECO:0000250|UniProtKB:Q8CFN5, ECO:0000269|PubMed:11904443, ECO:0000269|PubMed:15340086, ECO:0000269|PubMed:15831463, ECO:0000269|PubMed:15834131, ECO:0000269|PubMed:9069290, ECO:0000269|PubMed:9384584}. | Acetylation;Activator;Alternative splicing;Apoptosis;Cytoplasm;DNA-binding;Developmental protein;Differentiation;Disease variant;Epilepsy;Isopeptide bond;Mental retardation;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This locus encodes a member of the MADS box transcription enhancer factor 2 (MEF2) family of proteins, which play a role in myogenesis. The encoded protein, MEF2 polypeptide C, has both trans-activating and DNA binding activities. This protein may play a role in maintaining the differentiated state of muscle cells. Mutations and deletions at this locus have been associated with severe cognitive disability, stereotypic movements, epilepsy, and cerebral malformation. Alternatively spliced transcript variants have been described. [provided by RefSeq, Jul 2010]. | hsa:4208; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; postsynapse [GO:0098794]; protein-containing complex [GO:0032991]; sarcoplasm [GO:0016528]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; histone deacetylase binding [GO:0042826]; minor groove of adenine-thymine-rich DNA binding [GO:0003680]; protein heterodimerization activity [GO:0046982]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; apoptotic process [GO:0006915]; B cell homeostasis [GO:0001782]; B cell proliferation [GO:0042100]; B cell receptor signaling pathway [GO:0050853]; blood vessel development [GO:0001568]; blood vessel remodeling [GO:0001974]; cardiac ventricle formation [GO:0003211]; cell differentiation [GO:0030154]; cell morphogenesis involved in neuron differentiation [GO:0048667]; cellular response to calcium ion [GO:0071277]; cellular response to fluid shear stress [GO:0071498]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to parathyroid hormone stimulus [GO:0071374]; cellular response to transforming growth factor beta stimulus [GO:0071560]; cellular response to trichostatin A [GO:0035984]; cellular response to xenobiotic stimulus [GO:0071466]; chondrocyte differentiation [GO:0002062]; endochondral ossification [GO:0001958]; epithelial cell proliferation involved in renal tubule morphogenesis [GO:2001013]; excitatory postsynaptic potential [GO:0060079]; germinal center formation [GO:0002467]; glomerulus morphogenesis [GO:0072102]; heart development [GO:0007507]; heart looping [GO:0001947]; humoral immune response [GO:0006959]; learning or memory [GO:0007611]; MAPK cascade [GO:0000165]; melanocyte differentiation [GO:0030318]; muscle cell fate determination [GO:0007521]; muscle organ development [GO:0007517]; myotube differentiation [GO:0014902]; negative regulation of blood vessel endothelial cell migration [GO:0043537]; negative regulation of gene expression [GO:0010629]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of ossification [GO:0030279]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of vascular associated smooth muscle cell migration [GO:1904753]; negative regulation of vascular associated smooth muscle cell proliferation [GO:1904706]; negative regulation of vascular endothelial cell proliferation [GO:1905563]; nephron tubule epithelial cell differentiation [GO:0072160]; nervous system development [GO:0007399]; neural crest cell differentiation [GO:0014033]; neuron development [GO:0048666]; neuron differentiation [GO:0030182]; neuron migration [GO:0001764]; osteoblast differentiation [GO:0001649]; outflow tract morphogenesis [GO:0003151]; platelet formation [GO:0030220]; positive regulation of alkaline phosphatase activity [GO:0010694]; positive regulation of B cell proliferation [GO:0030890]; positive regulation of behavioral fear response [GO:2000987]; positive regulation of bone mineralization [GO:0030501]; positive regulation of cardiac muscle cell differentiation [GO:2000727]; positive regulation of cardiac muscle cell proliferation [GO:0060045]; positive regulation of gene expression [GO:0010628]; positive regulation of macrophage apoptotic process [GO:2000111]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of myoblast differentiation [GO:0045663]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of osteoblast differentiation [GO:0045669]; positive regulation of skeletal muscle cell differentiation [GO:2001016]; positive regulation of skeletal muscle tissue development [GO:0048643]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; primary heart field specification [GO:0003138]; regulation of AMPA receptor activity [GO:2000311]; regulation of dendritic spine development [GO:0060998]; regulation of germinal center formation [GO:0002634]; regulation of megakaryocyte differentiation [GO:0045652]; regulation of neuron apoptotic process [GO:0043523]; regulation of neurotransmitter secretion [GO:0046928]; regulation of NMDA receptor activity [GO:2000310]; regulation of synapse assembly [GO:0051963]; regulation of synaptic activity [GO:0060025]; regulation of synaptic plasticity [GO:0048167]; regulation of synaptic transmission, glutamatergic [GO:0051966]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; renal tubule morphogenesis [GO:0061333]; response to ischemia [GO:0002931]; secondary heart field specification [GO:0003139]; sinoatrial valve morphogenesis [GO:0003185]; skeletal muscle tissue development [GO:0007519]; smooth muscle cell differentiation [GO:0051145]; ventricular cardiac muscle cell differentiation [GO:0055012] | 11744164_MEF2C carboxy terminus (aa 387-473) deletion enhances transcriptional activation. AA 312-367 coupled with aa 1-86 activates transcription. Deletion of aa 312-350 decreases transcription. 11792813_The C-terminal region in MEF2C contains signals that are necessary to localize the histone deacetylase 4/MEF2 complex to the nucleus. 12061776_The DNA-binding TEA domain of transcription factor TEF-1 interacts with the MADS/MEF2 domain of MEF2C. 12135755_upregulation of expression by C-terminus of myogenin 12376544_The skeletal muscle-specific transcription cofactor vestigial-like 2 interacts with the C-terminal domain of MEF2C. 15084602_myogenin and myocyte enhancer factor-2 expression are triggered by membrane hyperpolarization during human myoblast differentiation 15831463_MEF2C is acetylated by p300 both in vitro and in vivo, which enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. 15834131_A conserved pattern of alternative splicing in vertebrate MEF2 (myocyte enhancer factor 2) genes generates an acidic activation domain in MEF2 proteins selectively in tissues where MEF2 target genes are highly expressed. (MEF2) 16469744_data show a dosage-dependent cardiomyopathic phenotype and a progressive reduction in ventricular performance associated with MEF2A or MEF2C overexpression 16478538_Phosphorylation of MEF2C at S396 aids sumoylation at K391, which recruits yet unidentified corepressors to inhibit transcription. Sumoylation motifs with a phosphorylated serine or an acidic residue at the +5 position might be more efficiently sumoylated. 16504037_Study demonstrates that human intestinal cell BCMO1 expression is dependent on the functional cooperation between peroxisome proliferator-activated receptor-gamma and myocyte enhancer factor 2 isoforms. 17611778_Overexpression of MEF2C is associated with hepatocellular carcinoma 17903302_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18067759_Results show the altered interaction between the different variants in the PGC-1alpha gene and MEF2C may attribute to the susceptibility to type 2 diabetes in the southern Chinese population. 18079734_In T-ALL cell lines, MEF2C is activated by NKX2-5 at both the RNA and protein levels. MEF2C consistently inhibits expression of NR4A1/NUR77, which regulates apoptosis via BCL2 transformation. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18950845_Observational study of gene-disease association. (HuGE Navigator) 19011954_Thus, ERK5 phosphorylation contributes to MEF2C activation and subsequent HASMC hypertrophy induced by Ang II 19065516_the association of the 482G/A polymorphism of the PGC-1alpha gene with type 2 diabetes and the quantitative and qualitative binding force changes between the PGC-1alpha domain mutant and MEF2C 19093215_MEF2 proteins are an important component in Galpha13-mediated angiogenesis. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19592390_These results strongly suggest that haploinsufficiency of MEF2C is responsible for severe mental retardation with stereotypic movements, seizures and/or cerebral malformations. 19720801_MEF2C is involved in the effect of insulin on skeletal muscle in type 2 diabetes. 19751190_Studies indicate that MAML1 functions as a coactivator for the tumor suppressor p53, MEF2C, beta-catenin and Notch signaling. 19828686_Muscle-growth geene MEF2C gene expression is enhanced by essential amino acid ingestion. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20333642_These deletions further support that haploinsufficiency of MEF2C is responsible for severe mental retardation, seizures, and hypotonia. 20412115_we present two additional patients with severe mental retardation, autism spectrum disorder and epilepsy, carrying a very small deletion encompassing the MEF2C gene. 20513142_Data indicate that MEF2C missense de novo mutations in severe mental retardation showed diminished MECP2 and CDKL5 expression. 20513142_Observational study of gene-disease association. (HuGE Navigator) 20610535_Binding sequences for the Mef2 family of transcription factors were enriched in the AR-bound regions, and data show that several Mef2c-dependent genes are direct targets of AR, suggesting a functional interaction between Mef2c and AR in skeletal muscle. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20709755_RNA-binding protein Muscleblind-like 3 (MBNL3) disrupts myocyte enhancer factor 2 (Mef2) {beta}-exon splicing 21060863_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21481790_NKX2-1, NKX2-2, and MEF2C define oncogenic pathways in T cell acute lymphoblastic leukemia (T-ALL). 21652706_knockdown of bone-specific transcription factors, Runx2 and osterix by shRNAi knockdown of Mef2c, suggests that Mef2c lies upstream of these two important factors in the cascade of gene expression in osteoblasts. 21849497_MEF2C is a transcriptional regulator of homeostasis in rod photoreceptor cells 21901155_Data report that forced expression of constitutively active MEF2C (MEF2CA) generates significantly greater numbers of neurons with dopaminergic properties in vitro. 22073279_suggest that PRL-3 functions downstream of the VEGF/MEF2C pathway in endothelial cells and may play an important role in tumor angiogenesis 22275376_a novel signaling cascade that links RhoA-mediated calcium sensitivity to MEF2-dependent myocardin expression in VSMCs through a mechanism involving p38 MAPK, PP1alpha, and CPI-17. 22363514_The present study investigated the effect of heart failure aetiology on Ca(+2) handling proteins and NFAT1, MEF2C and GATA4 (transcription factors) in the same cardiac tissue. 22449245_Mutations in MEF2C are probably a very rare cause of Rett syndrome. 22718505_15 bp-deletion and C-insertion in the 5'UTR region of MEF2C could affect hypertrophic cardiomyopathy, potentially by affecting expression of MEF2C. 22798246_genetic association study in population of 1,012 Han women in China: Data suggest that an SNP in MEF2C (rs1366594) is associated with bone mineral density of lumbar spine and hip joint in aging women. 22997707_Allele frequencies of three Alu insertions that are located in MEF2C (two of them) and TAX1BP1 genes significantly differ between cohorts of healthy donors and ALL(acute lymphoblastic leukemia) patients. 23001426_A targeted search for MEF2C mutations could be applied to patients with a severe intellectual deficiency associated with absence of language and hypotonia, strabismus, and epilepsy. 23174904_Validated miR-223 targets MEF2C and PTBP2 were significantly upregulated in chronic myeloid leukemia samples. 23226416_SREBP-1 regulate muscle protein synthesis through the downregulation of the expression of MYOD1, MYOG and MEF2C factors. 23468913_Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart. 23572186_variants at MEF2C were associated with forearm bone mineral density (BMD), implicating this gene in the determination of BMD at forearm. [meta-analysis] 23776548_One variant, rs2194025 on chromosome 5q14 near the myocyte enhancer factor 2C MEF2C gene, was associated with retinal arteriolar caliber in meta-analysis. 23888946_We identified MEF2C as a novel transcription factor that regulates Nampt expression through specific interaction sites at the promoter; its regulatory role was highly dependent on epigenetic modulations, especially under hypoxia conditions 24008018_MEF2C alpha- variants are significantly expressed during neuronal cell differentiation, indicating a putative role of these variants in development. 24290359_Results identify redox-mediated protein posttranslational modifications, including S-nitrosylation and sulfonation of a critical cysteine residue in MEF2, as an early event contributing to neuronal damage in Parkinson's disease induced by mitochondrial toxins. 24337390_MEF2C binding in inflammatory pathways is associated with its role in bone density 24412363_MEF2 regulatory network is disrupted in myotonic dystrophy cardiac tissue leading to altered expression of a large number of miRNA and mRNA targets. 24988463_MEF2C/alpha-2-macroglobulin axis functions in endothelial cells as a negative feed-back mechanism that adapts sprouting activity to the oxygen concentration thus diminishing inappropriate and excess angiogenesis. 25328135_The overall effect of MEF2C in hepatocellular carcinoma progression regulation was dictated by its subcellular distribution. 25336633_MEF2 is the key cis-acting factor that regulates expression of a number of transcriptional targets involved in pulmonary vascular homeostasis, including microRNAs 424 and 503, connexins 37, and 40, and Kruppel Like Factors 2 and 4. 25352737_Single nucleotide polymorphisms in ALDOB, MAP3K1, and MEF2C are associated with cataract. 25404735_Alternative splicing of the alpha-exon of MEF2C regulates myogenesis. 25416133_Combinations that resulted in higher protein levels of Mef2c enhanced reprogramming efficiency of cardiac myocytes. 25691421_this is the first report of a Greek-Cypriot patient with a MEF2C-related phenotype highlighting the rich variability in phenotypic expression and the ethnic diversity associated with this condition. 25733682_MEF2C and MEF2D interact with the E3 ligase F-box protein SKP2, which mediates their subsequent degradation through the ubiquitin-proteasome system. 25789873_MEF2C regulates the expression of G2/M checkpoint genes (14-3-3gamma, Gadd45b and p21) and the sub-cellular localization of CYCLIN B1. 26172269_BCL2 inhibitors may be a therapeutic candidate in vivo for patients with ETP-ALL with high expression levels of MEF2C. 26184978_MiR-135b-5p and MiR-499a-3p Promote Cell Proliferation and Migration in Atherosclerosis by Directly Targeting MEF2C 26426104_The finding of a jugular pit in this patient facilitated the diagnosis, and he is, to our knowledge, the third case of jugular pit in association with 5q14.3 deletion incorporating the MEF2C locus. 26487643_Data show that high myocyte enhancer factor 2C (MEF2C) mRNA expression leads to overexpression of MEF2C protein, and these findings provide the rationale for therapeutic targeting of MEF2C transcriptional activation in acute myeloid leukemia. 26864752_We describe the prenatal identification of 5q14.3 duplication, including MEF2C, in a monochorionic twin pregnancy with corpus callosum anomalies, confirmed by autopsy. To the best of our knowledge, this cerebral finding has been observed for the first time in 5q14.3 duplication patients, possibly widening the neurological picture of this scarcely known syndrome. A pathogenetic role of MEF2C overexpression in brain develop 26900922_early B cell factor-1 (EBF1) was identified as a co-regulator of gene expression with MEF2C. 26921792_MEF2C mRNA level is up-regulated in both sporadic and SOD1 + ALS patients. 26923194_Study provides evidence that Mef2c cooperated with Sp1 to activate human AQP1 transcription by binding to its proximal promoter in human umbilical cord vein endothelial cells indicating that AQP1 is a direct target of Mef2c in regulating angiogenesis and vasculogenesis of endothelial cells. 27144530_Key role for miR-214 in modulation of MEF2C-MYOCD-LMOD1 signaling. 27255693_New MEF2C mutation in MEF2C haploinsufficiency syndrome 27268728_Long non-coding RNA uc.167 influences cell proliferation, apoptosis and differentiation of P19 cells by regulating Mef2c. 27276684_MEF2C rs190982 polymorphism has a role in late-onset Alzheimer's disease in Han Chinese 27297623_a MEF2C and CEBPA correlation in CML disease progression 27337099_Data show that microRNA miR-27a was essential for the shift of mesenchymal stem cells (MSCs) from osteogenic differentiation to adipogenic differentiation in osteoporosis by targeting myocyte enhancer factor 2 c (Mef2c). 27479909_Single nucleotide polymorphism in MEF2C gene is associated with major depressive disorder. 27553283_Findings suggest that a single introduction of the three cardiomyogenic transcription factor (GATA4, cand TBX5)genes using polyethyleneimine (PEI)-based transfection is sufficient for transdifferentiation of adipose-derived stem cells (hADSCs) towards the cardiomyogenic lineage. 27616567_we identified novel associations in WLS , ARHGAP1 , and 5' of MEF2C ( P- values < 8x10 - 5 ; false discovery rate (FDR) q-values < 0.01) that were much more strongly associated with BMD compared to the GWAS SNPs. 27664809_Our analysis consistently identified significant sub-networks associated with the interacting transcription factors MEF2C and TWIST1, genes not previously associated with spontaneous preterm births , both of which regulate processes clearly relevant to birth timing. 27907007_The mRNA expressions of PPP3CB and MEF2C were significantly up-regulated, and CAMK1 and PPP3R1 were significantly down-regulated in mitral regurgitation(MR) patients compared to normal subjects. Moreover, MR patients had significantly increased mRNA levels of PPP3CB, MEF2C and PLCE1 compared to aortic valve disease patients 28017720_Mef2c is highly expressed in the retina where it modulates photoreceptor-specific gene expression 28456137_We report heterozygous MEF2C loss-of-function as a possible cause of question mark ear associated with intellectual deficiency. 28473437_Endothelial Mef2c regulates the endothelial actin cytoskeleton and inhibits smooth muscle cell migration into the intima. 28482719_MEF2C expression levels were significantly associated with or may even be predictive of the response to glucocorticoid treatment. 28799067_Overexpression of MEF2C decreased miR-448-induced VSMCs proliferation and migration. 28821601_Combined with automated 2D nano-scale chromatography, Accumulated ion monitoring achieved subattomolar limits of detection of endogenous proteins in complex biological proteomes. This allowed quantitation of absolute abundance of the human transcription factor MEF2C at approximately 100 molecules/cell, and determination of its phosphorylation stoichiometry from as little as 1 mug of extracts isolated from 10,000 human ... 28902616_This study indicates MEF2C as a new gene responsible for human dilated cardiomyopathy (DCM), which provides novel insight into the mechanism underpinning DCM, suggesting potential implications for development of innovative prophylactic and therapeutic strategies for DCM, the most prevalent form of primary myocardial disease. 29104469_the mutation significantly diminished the synergistic activation between MEF2C and GATA4, another cardiac core transcription factor that has been causally linked to Congenital heart disease (CHD). 29112298_MEF2C mRNA expression levels in AD subjects are significantly lower than those in control subjects and are correlated with disease severity. 29413154_At levels of condFDR < 0.01 and conjFDR < 0.05; we identified 5 ADHD-associated loci, 3 of these being shared between ADHD and EA. 29431698_High levels of phosphorylation of S222 of MEF2C are associated with primary chemotherapy resistance in an independent cohort of cytogenetically normal and Acute Myeloid Leukemia. 29468350_This study firstly associates MEF2C loss-of-function mutation with double outlet right ventricle in humans, which provides novel insight into the molecular pathogenesis of congenital heart diseases. 29526696_Targeting of LKB1 or SIK3 diminishes histone acetylation at MEF2C-bound enhancers and deprives leukemia cells of the output of this essential TF. 29678826_HuR upregulates MEF2C mRNA expression by protecting MEF2C mRNA from degradation, and consequently, the elevated MEF2C enhances SCN5A mRNA transcription. 29714661_The regulation mechanism of MIG6 and suggests potential implications for the therapeutic strategies of gefitinib resistance through inhibiting MEF2C in hepatic cancer cells. 29863696_Fourteen patients were studied including four patients with mutations in the MEF2C gene revealed by exome sequencing 29995456_MEF2 family, particularly MEF2C, is a key regulator underlying the transcriptional signature of bed rest and, hence, ultimately also skeletal muscle alterations induced by systemic unloading in humans. 30006604_The previously identified major depressive disorder risk variant rs10514299 in TMEM161B-MEF2C predicts neuronal correlates of reward processing in an alcohol dependence phenotype, possibly explaining part of the shared pathophysiology and comorbidity between the disorders. 30335237_MEF2C SNPs rs1366594, rs12521522 and rs11951031 were examined in women for northern Mexico. No significant association was found with bone mineral density, although some trends were seen. 30376817_Novel MEF2C point mutations in Chinese patients with Rett (-like) syndrome or non-syndromic intellectual disability have been reported. 30445463_our results disentangle a complex regulatory network governing neuronal MEF2C expression that involves multiple distal enhancers. In addition, the characterized neuronal enhancers pose as novel candidates to screen for mutations in neurodevelopmental disorders, such as Rett-like syndrome. 31094920_A novel MEF2C-SS18 gene fusion and unique histologic and immunophenotypic features characterize a heretofore undefined low-grade salivary adenocarcinoma for described as a microsecretory adenocarcinoma. 31140610_The results above showed that MEF2C was involved in the process of promoting the differentiation of stem cells into cardiac myocytes by miR-199a-3p inhibitors. 31254364_homeostasis and physiological function of AQP1 in endothelial health are maintained by the MEF2C and miR-133a-3p.1 regulatory circuit 31900516_A novel MEF2C mutation in lymphoid neoplasm diffuse large B-cell lymphoma promotes tumorigenesis by increasing c-JUN expression. 32003456_Identification of FOXH1 mutations in patients with sporadic conotruncal heart defect. 32017034_High expression of myocyte enhancer factor 2C predicts poor prognosis for adult acute myeloid leukaemia with normal karyotype. 32046534_MEF2C is associated with ADHD risk. 32186750_Methylationassociated silencing of miR638 promotes endometrial carcinoma progression by targeting MEF2C. 32410261_MicroRNA-23 suppresses osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting the MEF2C-mediated MAPK signaling pathway. 32975584_Genes influenced by MEF2C contribute to neurodevelopmental disease via gene expression changes that affect multiple types of cortical excitatory neurons. 33270893_Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression. 33303006_MiR-218 affects hypertrophic differentiation of human mesenchymal stromal cells during chondrogenesis via targeting RUNX2, MEF2C, and COL10A1. 33568691_MEF2C shapes the microtranscriptome during differentiation of skeletal muscles. 33831796_Electroclinical features of MEF2C haploinsufficiency-related epilepsy: A multicenter European study. 33984142_MEF2C silencing downregulates NF2 and E-cadherin and enhances Erastin-induced ferroptosis in meningioma. 33999292_Identification of a novel mutation in MEF2C gene in an atypical patient with frontotemporal lobar degeneration. 34022131_Non-coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss-of-function mechanisms. 34184825_Comprehensive investigation of the phenotype of MEF2C-related disorders in human patients: A systematic review. 34570228_A dominant-negative SOX18 mutant disrupts multiple regulatory layers essential to transcription factor activity. 34731014_MEF2 is a key regulator of cognitive potential and confers resilience to neurodegeneration. 34779502_MicroRNA190b expression predicts a good prognosis and attenuates the malignant progression of pancreatic cancer by targeting MEF2C and TCF4. 34946961_Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women. 34991657_Progress on the roles of MEF2C in neuropsychiatric diseases. 35130621_MEF2C ameliorates learning, memory, and molecular pathological changes in Alzheimer's disease in vivo and in vitro 35357565_MEF2C gene variations are associated with ADHD in the Chinese Han population: a case-control study. 35406681_Activin A Causes Muscle Atrophy through MEF2C-Dependent Impaired Myogenesis. | ENSMUSG00000005583 | Mef2c | 1.054135e+02 | 0.7216929 | -0.470543010 | 0.4747364 | 9.228343e-01 | 0.3367318817 | 0.82704387 | No | Yes | 9.697850e+01 | 30.394159 | 1.130287e+02 | 36.384169 | |
| ENSG00000081791 | 9812 | DELE1 | protein_coding | Q14154 | FUNCTION: [DAP3-binding cell death enhancer 1]: Key activator of the integrated stress response (ISR) following mitochondrial stress (PubMed:32132706, PubMed:32132707). In response to mitochondrial stress, cleaved by the protease OMA1, generating the DAP3-binding cell death enhancer 1 short form (DELE1(S) or S-DELE1), which translocates to the cytosol and activates EIF2AK1/HRI to trigger the ISR (PubMed:32132706, PubMed:32132707). Essential for the induction of death receptor-mediated apoptosis through the regulation of caspase activation (PubMed:20563667). {ECO:0000269|PubMed:20563667, ECO:0000269|PubMed:32132706, ECO:0000269|PubMed:32132707}.; FUNCTION: [DAP3-binding cell death enhancer 1 short form]: Protein kinase activator generated by protein cleavage in response to mitochondrial stress, which accumulates in the cytosol and specifically binds to and activates the protein kinase activity of EIF2AK1/HRI (PubMed:32132706, PubMed:32132707). It thereby activates the integrated stress response (ISR): EIF2AK1/HRI activation promotes eIF-2-alpha (EIF2S1) phosphorylation, leading to a decrease in global protein synthesis and the induction of selected genes, including the transcription factor ATF4, the master transcriptional regulator of the ISR (PubMed:32132706, PubMed:32132707). {ECO:0000269|PubMed:32132706, ECO:0000269|PubMed:32132707}. | Apoptosis;Cytoplasm;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;TPR repeat;Transit peptide | hsa:9812; | cytosol [GO:0005829]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; protein kinase binding [GO:0019901]; protein serine/threonine kinase activator activity [GO:0043539]; extrinsic apoptotic signaling pathway via death domain receptors [GO:0008625]; HRI-mediated signaling [GO:0140468]; integrated stress response signaling [GO:0140467]; regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043281] | 20563667_identifies a novel DAP3-binding protein termed death ligand signal enhancer (DELE); demonstrates the biological significance of DELE for apoptosis signal mediated by death receptors 20877624_Observational study of gene-disease association. (HuGE Navigator) 32132706_the mitochondrial protease OMA1 and the poorly characterized protein DELE1, together with HRI, constitute the missing pathway that is triggered by mitochondrial stress; these findings could be used to inform future strategies to modulate the cellular response to mitochondrial dysfunction in the context of human disease 32132707_mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway; the OMA1-DELE1-HRI pathway represents a potential therapeutic target that could enable fine-tuning of the integrated stress response for beneficial outcomes in diseases that involve mitochondrial dysfunction 35163244_The Mitochondrial PHB2/OMA1/DELE1 Pathway Cooperates with Endoplasmic Reticulum Stress to Facilitate the Response to Chemotherapeutics in Ovarian Cancer. | ENSMUSG00000024442 | Dele1 | 2.732770e+03 | 1.0562529 | 0.078955267 | 0.2596654 | 9.360590e-02 | 0.7596420978 | 0.95123134 | No | Yes | 2.352193e+03 | 195.976025 | 2.323873e+03 | 198.566406 | ||
| ENSG00000082438 | 22837 | COBLL1 | protein_coding | Q53SF7 | 3D-structure;Alternative splicing;Phosphoprotein;Reference proteome;Repeat | hsa:22837; | extracellular exosome [GO:0070062]; actin monomer binding [GO:0003785]; cadherin binding [GO:0045296] | 19401544_The gene ratio test with the COBLL1/ARHGDIA genes for survival of patients with malignant pleural mesothelioma has robust predictive value. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20468071_Observational study of gene-disease association. (HuGE Navigator) 21569526_We provide evidence that ARHGDIA, COBLL1, and TM4SF1 are negative regulators of apoptosis in cultured tumor cells. 23463496_genetic association study in children/adolescents in Sardinia: Data suggest that an SNP in COBLL1 (rs7607980; TC or CC) is associated with lower insulin resistance and lower serum insulin levels in overweight/obese children/adolescents. 26757982_Aggregation of rare variants in COBLL1were nominally associated with higher waist-to-hip ratio in European-Americans. 27185377_The expression of COBLL1, LPL, and ZAP70 corresponded to patient prognosis and to IGHV mutational status, although not absolutely. When we combined all three markers together and performed the ROC analysis, AUC increased compared to the AUC of individual gene expression. 28232743_High-level expression of Cobll1 promotes the transformation of CML and generates drug resistance by increasing IKKgamma stability, which results in activation of the canonical NF-kappaB pathway. 28258026_polymorphism, rs7607980 of COBLL1 didnot show any significant association with T2D 29122990_identified COBLL1 as a novel interaction partner of ROR1 29686105_nuclear COBLL1 interacts with AR to enhance complex formation with CDK1 and facilitates AR phosphorylation for genomic binding in castration-resistant prostate cancer model cells. | ENSMUSG00000034903 | Cobll1 | 1.467811e+02 | 1.0657554 | 0.091876363 | 0.4502929 | 3.968842e-02 | 0.8420910343 | 0.96866180 | No | Yes | 1.730760e+02 | 49.466811 | 1.095281e+02 | 32.381540 | |||
| ENSG00000082482 | 3776 | KCNK2 | protein_coding | O95069 | FUNCTION: Ion channel that contributes to passive transmembrane potassium transport (PubMed:23169818). Reversibly converts between a voltage-insensitive potassium leak channel and a voltage-dependent outward rectifying potassium channel in a phosphorylation-dependent manner (PubMed:11319556). In astrocytes, forms mostly heterodimeric potassium channels with KCNK1, with only a minor proportion of functional channels containing homodimeric KCNK2. In astrocytes, the heterodimer formed by KCNK1 and KCNK2 is required for rapid glutamate release in response to activation of G-protein coupled receptors, such as F2R and CNR1 (By similarity). {ECO:0000250|UniProtKB:P97438, ECO:0000269|PubMed:10784345, ECO:0000269|PubMed:11319556, ECO:0000269|PubMed:23169818}.; FUNCTION: [Isoform 4]: Does not display channel activity but reduces the channel activity of isoform 1 and isoform 2 and reduces cell surface expression of isoform 2. {ECO:0000250}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Ion channel;Ion transport;Membrane;Phosphoprotein;Potassium;Potassium channel;Potassium transport;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes one of the members of the two-pore-domain background potassium channel protein family. This type of potassium channel is formed by two homodimers that create a channel that leaks potassium out of the cell to control resting membrane potential. The channel can be opened, however, by certain anesthetics, membrane stretching, intracellular acidosis, and heat. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:3776; | apical plasma membrane [GO:0016324]; astrocyte projection [GO:0097449]; calyx of Held [GO:0044305]; cell surface [GO:0009986]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of plasma membrane [GO:0005887]; neuronal cell body [GO:0043025]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; voltage-gated potassium channel complex [GO:0008076]; outward rectifier potassium channel activity [GO:0015271]; potassium channel inhibitor activity [GO:0019870]; potassium ion leak channel activity [GO:0022841]; cardiac ventricle development [GO:0003231]; cellular response to hypoxia [GO:0071456]; cochlea development [GO:0090102]; G protein-coupled receptor signaling pathway [GO:0007186]; memory [GO:0007613]; negative regulation of cardiac muscle cell proliferation [GO:0060044]; negative regulation of DNA biosynthetic process [GO:2000279]; positive regulation of cell death [GO:0010942]; positive regulation of cellular response to hypoxia [GO:1900039]; potassium ion transmembrane transport [GO:0071805]; response to axon injury [GO:0048678]; response to mechanical stimulus [GO:0009612]; stabilization of membrane potential [GO:0030322] | 12368289_TREK-1 channels may function as sensors that couple the metabolic state of the cell to membrane potential, perhaps through an associated ATP-binding protein 14522822_During hypoxia, modulation of hTREK1 cannot be accomplished by parameters known to be perturbed in brain ischemia. hTREK1 regulation in brain will be more relevant during alkalosis than during ischemia or acidosis. 15685212_TREK-1 is inhibited by fluoxetine and norfluoxetine. 15883010_hypoxic inhibition: (a) requires the C-terminal domain of the channel; (b) does not involve redox modulation of the C-terminal domain cysteine residues C365 and C399; and (c) is critically dependent on the glutamate residue at position 306 16006563_receptor- and kinase-induced inhibition of TREK-1 background potassium channels is mediated by sequential phosphorylation 16250016_human osteoblasts functionally express TREK-1 and that these channels contribute, at least in part, to the resting membrane potential of human osteoblast cells. We hypothesise a possible role for TREK-1 in mechanotransduction, leading to bone remodelling. 17035301_VOCCs and TREK channels have been implicated in mechanotransduction signaling pathways in numerous connective tissue cell types. 17375039_TREK1, the most thoroughly studied K(2P) channel, has a key role in the cellular mechanisms of neuroprotection, anaesthesia, pain and depression--{REVIEW} 18288090_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18288090_These findings indicate that genetic variation in KCNK2 may identify individuals at risk for treatment resistance. More broadly, they indicate the utility of animal models in identifying genes for pharmacogenetic studies of antidepressant response. 18474599_voltage-dependent C-type gating acceleration by protons represents a novel mechanism for K2P2.1 outward rectification. 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18579071_(Review) KCNK2, the gene encoding K2P2.1, can generate either full-length or K2P2.1D1-56 via alternative translation initiation, a mechanism which increases protein diversity by giving rise to two or more proteins from a single mRNA strand. 18676988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18854423_Both TASK-3 and TREK-1 are functionally operational in the adrenocortical H295R cell line, modulate membrane potential and aldosterone secretion. 19130888_Results highlight the important role of K(2P)2.1 channels as receptors for mediators known to cause nociception. 19621370_Observational study of gene-disease association. (HuGE Navigator) 19621370_These findings indicate that TREK1 genotypes are associated with individual differences in reward-related brain activity. 19741570_KCNK2 is related to the susceptibility to major depressive disorder 19741570_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19837167_Results describe the regulation of neuronal K(2P)2.1 (KCNK2, TREK-1) channel activity by resting membrane potential. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20083228_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20362547_these data suggest that beta-COP plays a critical role in the forward transport of TREK1 channel to the plasma membrane. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20811500_role for TREK-1 in contributing to uterine quiescence during gestation 21069514_The data of this study suggested that TREK-1 is associated with NSC proliferation and probably is a modulator of the effect that fluoxetine attenuates the inhibitory neurogenesis induced by glucocorticoid hormones. 21262820_inhibition of TREK1 current by fluoxetine is found to be accompanied by dissociation of the C-terminal domain from the membrane. 21822218_These results demonstrate that the primary activation mechanisms in TREK-1 reside close to, or within the selectivity filter and do not involve gating at the cytoplasmic bundle crossing. 21886777_Cochlin interacts with TREK-1 and annexin A2. 21964404_Potassium channels, in particular K2P channels, are expressed and functional in the apical membrane of airway epithelial cells 21965685_the important role of selectivity filter gating in the regulation of TREK-1 by the extracellular K(+) and proton. 22563662_TREK-1 is expressed by both nucleus pulposus and annulus fibrosus cells of the human intervertebral disc. 22811574_each of four TREK-1 splice variants interacts with full-length wild-type TREK-1 and in vivo, such interactions may contribute to a preterm labor phenotype. 23275623_we report the expression of Trek-1 in human alveolar epithelial cells and propose that Trek-1 deficiency may alter both IL-6 translation and transcription in AECs without affecting Ca(2+) signaling 23305490_TASK and TREK-1 are involved in regulation of cell proliferation and in control of resting membrane potentials in endometrial epithelial cells. 23479219_High TREK-1 expression is associated with epithelial ovarian cancer. 23804201_In human embryonic kidney (HEK-293) cells stably expressing TREK-1, outward currents at 80 mV increased from 91.0 +/- 23.8 to 247.5 +/- 73.3 pA/pF. 23933981_study analyzed the role of TWIK-related potassium channel-1 (TREK1)in endothelial cells and the blood-brain barrier(BBB); blocking TREK1 increased leukocyte transmigration; TREK1 activation had the opposite effect 24154701_Activation of K(2)P channel-TREK1 mediates the neuroprotection induced by sevoflurane preconditioning. 24196565_results suggest that the TREK-1e splice variant may interfere with the vesicular traffic of full-length TREK-1 channels from the ER to the plasma membrane. 24509840_A number of mutations that affect TREK1 channel gating occlude the action of fenamates but only in the longer form of TREK1. 24586773_TREK-1 deficient alveolar epithelial cells have less F-actin and are more deformable making them more resistant to stretch-induced injury. 24801307_Response of the human detrusor to stretch is regulated by TREK-1 25168769_Modulation of K2P 2.1 and K2P 10.1 K(+) channel sensitivity to carvedilol by alternative mRNA translation initiation 25197053_PLD2, but not PLD1, directly binds to the C terminus of TREK1 and TREK2. 25482670_During conductance simulation experiments, both TASK-3 and TREK-1 channels were able to repolarise the membrane once AP threshold was reached 25529528_Nasal epithelia express Trek1 that can be suppressed by allergic response. 25683610_Trek1 expression facilitates the restoration of intestinal epithelial barrier functions in an allergic environment. 25778785_We conclude that Trek1 is critical to maintain the nasal epithelial barrier function. 25962960_Data suggest that potassium channel protein TREK-1 (TREK-1) might be a biomarker in castration resistance free survival (CRFS) judgment of prostate cancer (PCa), as well as a potential therapeutic target. 26332952_How ion channels sense mechanical force: insights from mechanosensitive K2P channels TRAAK, TREK1, and TREK2. 26400398_presence of TREK-1 variants correlated to reduced TREK-1 activity, suggesting a pathological role for TREK-1 variants in preterm labor 27384506_decreased TREK-1 expression in the aganglionic and ganglionic bowel observed in Hirschsprung's disease may alter intestinal epithelial barrier function leading to the development of enterocolitis 27397543_TREK-1 overexpression suppresses CHO cell proliferation by inhibiting the activity of PKA and p38/MAPK signaling pathways and subsequently inducing G1 phase cell arrest. 27443495_Exposure to a set of different K(+) channel inhibitors, and the TREK-1 (K2P2.1)-specific activator BL1249, TREK-1 was identified as the main component of pH-regulated current. A voltage-sensor dye (VF2.1.Cl) was used to monitor effects of pH and BL1249 on Vm in more physiological conditions and TREK-1-mediated current was found as critical player in setting Vm. 28005193_Atrial TREK-1 expression was reduced in atrial fibrillation patients with concomitant severe heart failure 28242754_The authors identified a heterozygous point mutation in the selectivity filter of the stretch-activated K2P potassium channel TREK-1 (KCNK2 or K2P2.1). This mutation introduces abnormal sodium permeability and stretch-activation hypersensitivity to TREK-1. 28539337_downregulated in overactive detrusor 28676394_The M2-glycine hinge controls the macroscopic currents of TREK1 channels. 28693035_data reveal a druggable K2P site that stabilizes the C-type gate 'leak mode' and provide direct evidence for K2P selectivity filter gating 29103993_Role of TWIK-related potassium channel-1 in chronic rhinosinusitis. 29577872_The results suggest that prolonged stretch enhances the expression/activity of TREK-1 channel, leading to decreased myometrium contractile activity and maintained healthy term pregnancy particularly in multiple pregnancy. 30350878_The frequencies of rs758937019-CT genotype (P = 0.0016) and rs758937019-T allele (P = 0.0022) were significantly higher in the group with overactive lower urinary tract symptoms 30446509_Polymodal TREK-1 channels are required to maintain the membrane potential, Ca2+ homeostasis, and pressure sensitivity in trabecular meshwork cells. 30576235_The PIP2-dependent converse regulation of TREKs by Lys and R3-pCt with Gly implies structural flexibility. 30805940_The activity of TREK-1 has a key role for the correct functioning of the alveolar epithelium. 31782179_Astrocytic AEG-1 regulates expression of TREK-1 under acute hypoxia. 32320829_A regulatory domain in the K2P2.1 (TREK-1) carboxyl-terminal allows for channel activation by monoterpenes. 33025137_Intracellular activation of full-length human TREK-1 channel by hypoxia, high lactate, and low pH denotes polymodal integration by ischemic factors. 34038243_Human adrenal glomerulosa cells express K2P and GIRK potassium channels that are inhibited by ANG II and ACTH. 35177207_Involvement of TREK1 channels in the proliferation of human hepatic stellate LX-2 cells. | ENSMUSG00000037624 | Kcnk2 | 1.626245e+01 | 1.5555400 | 0.637415499 | 0.7334272 | 7.963365e-01 | 0.3721909514 | No | Yes | 1.911672e+01 | 6.914358 | 1.278480e+01 | 4.608675 | ||
| ENSG00000083812 | 25799 | ZNF324 | protein_coding | O75467 | FUNCTION: May be involved in transcriptional regulation. May be involved in regulation of cell proliferation. {ECO:0000305|PubMed:11779640}. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:25799; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; cell population proliferation [GO:0008283]; G1/S transition of mitotic cell cycle [GO:0000082]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000004500 | Zfp324 | 9.226913e+02 | 1.0263269 | 0.037490337 | 0.3365031 | 1.207362e-02 | 0.9125045397 | 0.98338795 | No | Yes | 8.458413e+02 | 130.432399 | 8.629974e+02 | 136.322740 | |||
| ENSG00000083844 | 9422 | ZNF264 | protein_coding | O43296 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a zinc finger protein and belongs to the krueppel C2H2-type zinc-finger protein family. Zinc finger proteins are often localized in the nucleus, bind nucleic acids, and regulate transcription. [provided by RefSeq, Jan 2010]. | hsa:9422; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 3.440519e+02 | 0.7354203 | -0.443359014 | 0.2958189 | 2.159445e+00 | 0.1416958525 | 0.76841160 | No | Yes | 2.828490e+02 | 41.157157 | 3.802361e+02 | 55.614534 | ||||
| ENSG00000084090 | 56910 | STARD7 | protein_coding | Q9NQZ5 | FUNCTION: May play a protective role in mucosal tissues by preventing exaggerated allergic responses. {ECO:0000250|UniProtKB:Q8R1R3}. | Coiled coil;Mitochondrion;Reference proteome;Transit peptide | hsa:56910; | mitochondrial outer membrane [GO:0005741]; lipid binding [GO:0008289] | 17672918_the apparent occurrence of an unusual TG 3' splice site in intron 1 is discussed 19679347_Beta-catenin and TCF4 activate the human StarD7 gene interacting with its promoter region through Wnt/beta-catenin signalling. 20042613_These results suggest that StarD7 facilitates the delivery of phosphatidylcholine to mitochondria in non-vesicular system. 21622533_The SF-1 and beta-catenin pathway convergence on StarD7 expression may have important implications in the phospholipid uptake and transport, contributing to the normal trophoblast development. 22063720_StarD7 behaves as a fusogenic protein in model and cell membrane bilayers. 22952907_StarD7 modulates ABCG2 multidrug transporter level, cell migration, proliferation, and biochemical and morphological differentiation marker expression in a human trophoblast cell model. 27301951_MFSD2B, CCL20 and STAT1, or STARD7 and ZNF512 genes may be risk or protect factors in prognosis of ADC; HTR2B, DPP4, and TGFBRAP1 genes may be risk factors in prognosis of SQC. 27554972_The results indicate that StarD7 contributes to the modulation of cellular redox homeostasis. 28821867_this study demonstrated that StarD7, a non-vesicular carrier of PC, is anchored onto the OMM via its N-terminal TM domain. 29301859_PARL preserves mitochondrial membrane homeostasis via STARD7 processing and is emerging as a critical regulator of protein localization between mitochondria and the cytosol 29343537_The data point at StarD7 as a candidate effector protein by which ceramides may exert part of their mitochondria-mediated cytotoxic effects. 31664034_An ATTTC expansion in the first intron of STARD7 segregated with Familial Adult Myoclonic Epilepsy (FAME2, OMIM: 607876)in 158 typically affected individuals. Expression of STARD7 mRNA and protein abundance was not altered in fibroblast cell lines derived from individuals with FAME2 ATTTC repeat expansions. 31664034_Familial Adult Myoclonic Epilepsy type 2 is caused by an expansion of an ATTTC pentamer within the first intron of STARD7. 32071354_The phosphatidylcholine transfer protein StarD7 is important for myogenic differentiation in mouse myoblast C2C12 cells and human primary skeletal myoblasts. 34416390_Role of the lipid transport protein StarD7 in mitochondrial dynamics. | ENSMUSG00000027367 | Stard7 | 1.471484e+04 | 0.6444715 | -0.633811537 | 0.3132140 | 4.138350e+00 | 0.0419220084 | 0.63602073 | No | Yes | 1.005801e+04 | 1413.043888 | 1.676671e+04 | 2414.824455 | ||
| ENSG00000086232 | 27102 | EIF2AK1 | protein_coding | Q9BQI3 | FUNCTION: Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (EIF2S1/eIF-2-alpha) in response to various stress conditions (PubMed:32132706, PubMed:32132707). Key activator of the integrated stress response (ISR) required for adaptation to various stress, such as heme deficiency, oxidative stress, osmotic shock, mitochondrial dysfunction and heat shock (PubMed:32132706, PubMed:32132707). EIF2S1/eIF-2-alpha phosphorylation in response to stress converts EIF2S1/eIF-2-alpha in a global protein synthesis inhibitor, leading to a global attenuation of cap-dependent translation, while concomitantly initiating the preferential translation of ISR-specific mRNAs, such as the transcriptional activator ATF4, and hence allowing ATF4-mediated reprogramming (PubMed:32132706, PubMed:32132707). Acts as a key sensor of heme-deficiency: in normal conditions, binds hemin via a cysteine thiolate and histidine nitrogenous coordination, leading to inhibit the protein kinase activity (By similarity). This binding occurs with moderate affinity, allowing it to sense the heme concentration within the cell: heme depletion relieves inhibition and stimulates kinase activity, activating the ISR (By similarity). Thanks to this unique heme-sensing capacity, plays a crucial role to shut off protein synthesis during acute heme-deficient conditions (By similarity). In red blood cells (RBCs), controls hemoglobin synthesis ensuring a coordinated regulation of the synthesis of its heme and globin moieties (By similarity). It thereby plays an essential protective role for RBC survival in anemias of iron deficiency (By similarity). Similarly, in hepatocytes, involved in heme-mediated translational control of CYP2B and CYP3A and possibly other hepatic P450 cytochromes (By similarity). May also regulate endoplasmic reticulum (ER) stress during acute heme-deficient conditions (By similarity). Also activates the ISR in response to mitochondrial dysfunction: HRI/EIF2AK1 protein kinase activity is activated upon binding to the processed form of DELE1 (S-DELE1), thereby promoting the ATF4-mediated reprogramming (PubMed:32132706, PubMed:32132707). {ECO:0000250|UniProtKB:Q9Z2R9, ECO:0000269|PubMed:32132706, ECO:0000269|PubMed:32132707, ECO:0000269|PubMed:32197074}. | ATP-binding;Alternative splicing;Disulfide bond;Kinase;Nucleotide-binding;Phosphoprotein;Protein synthesis inhibitor;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene acts at the level of translation initiation to downregulate protein synthesis in response to stress. The encoded protein is a kinase that can be inactivated by hemin. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2008]. | hsa:27102; | cytoplasm [GO:0005737]; ATP binding [GO:0005524]; eukaryotic translation initiation factor 2alpha kinase activity [GO:0004694]; heme binding [GO:0020037]; protein homodimerization activity [GO:0042803]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; acute inflammatory response [GO:0002526]; HRI-mediated signaling [GO:0140468]; integrated stress response signaling [GO:0140467]; iron ion homeostasis [GO:0055072]; macrophage differentiation [GO:0030225]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of hemoglobin biosynthetic process [GO:0046986]; negative regulation of translational initiation by iron [GO:0045993]; phagocytosis [GO:0006909]; protein autophosphorylation [GO:0046777]; protoporphyrinogen IX metabolic process [GO:0046501]; regulation of eIF2 alpha phosphorylation by heme [GO:0010999]; response to iron ion starvation [GO:1990641] | 12767237_autophosphorylation of Thr485 is essential for the hyperphosphorylation and activation of HRI and is required for the acquisition of the eIF2alpha kinase activity 19133234_These data establish for the first time, the possible mechanisms of regulation of hHRI gene expression under normal physiological condition, hemin exposure and stress. 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 23357686_During lead-stress, the regulation of hHRI mRNA translation is mediated through its 5'-untranslated region (UTR) that interacts with specific trans-acting factors. 23392680_Dephosphorylation of eIF2alpha, specifically in the cortex, is both correlated with and necessary for normal memory consolidation. 23874749_Infection-associated functions of HRI (an eIF2alpha kinase) are independent of its activity as a regulator of protein synthesis. 27557499_data suggests that glutamatergic stimulation induces HRI activation by NO to trigger GluN2B expression and this process would be relevant to maintain postsynaptic activity in cortical neurons 27888007_structural and functional stability of HRI 27974558_The antiviral kinase PKR plays a critical role in controlling HCMV replication. 30026227_Taken together, these results suggest HRI as a potential therapeutic target for hemoglobinopathies. 30170366_Activation of HRI is mediated by Hsp90 during stress through modulation of the HRI-Hsp90 complex 31554636_Recently, HRI has been implicated in the regulation of human fetal hemoglobin production. Therefore, HRI-integrated stress response has emerged as a potential therapeutic target for hemoglobinopathies. especially under iron/heme deficiency and beta-thalassemia. [review] 32132707_mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway; the OMA1-DELE1-HRI pathway represents a potential therapeutic target that could enable fine-tuning of the integrated stress response for beneficial outcomes in diseases that involve mitochondrial dysfunction 32197074_Human disorders associated with deleterious variants in EIF2AK1 and EIF2AK2 have not been reported. 32299090_The HRI-regulated transcription factor ATF4 activates BCL11A transcription to silence fetal hemoglobin expression. 32892501_The eIF2alpha kinase HRI in innate immunity, proteostasis, and mitochondrial stress. 33168630_The eIF2alpha kinase HRI triggers the autophagic clearance of cytosolic protein aggregates. | ENSMUSG00000029613 | Eif2ak1 | 6.204284e+03 | 1.1270058 | 0.172494904 | 0.2496884 | 4.766971e-01 | 0.4899221939 | 0.87511746 | No | Yes | 6.659057e+03 | 768.541135 | 5.193963e+03 | 614.909596 | |
| ENSG00000086712 | 55787 | TXLNG | protein_coding | Q9NUQ3 | FUNCTION: May be involved in intracellular vesicle traffic. Inhibits ATF4-mediated transcription, possibly by dimerizing with ATF4 to form inactive dimers that cannot bind DNA. May be involved in regulating bone mass density through an ATF4-dependent pathway. May be involved in cell cycle progression. {ECO:0000269|PubMed:15911876, ECO:0000269|PubMed:18068885}. | Alternative splicing;Cell cycle;Coiled coil;Cytoplasm;Membrane;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene encodes a member of the taxilin family. The encoded protein binds to the C-terminal coiled-coil region of syntaxin family members 1A, 3A and 4A, and may play a role in intracellular vesicle trafficking. This gene is up-regulated by lipopolysaccharide and the gene product may be involved in cell cycle regulation. The related mouse protein was also shown to inhibit activating transcription factor 4-mediated transcription and thus regulate bone mass accrual. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2009]. | hsa:55787; | cytosol [GO:0005829]; nuclear membrane [GO:0031965]; DNA-binding transcription factor binding [GO:0140297]; syntaxin binding [GO:0019905]; cell cycle [GO:0007049]; regulation of bone mineralization [GO:0030500]; regulation of cell cycle [GO:0051726]; regulation of cell cycle process [GO:0010564] | 15184072_identification of isoforms of taxilin; interaction with syntaxin; tissue distribution [beta-taxilin, gamma-taxilin]. 18068885_evaluation of the effects of a novel environmental lipopolysaccharide-responding (Elrg) gene on the regulation of proliferation and cell cycle of the hepatoma-derived cell line, HepG2 22082360_[review] First evidence of a functional interaction between ATF4, FIAT (factor-inhibiting ATF4-mediated transcription) and alphaNAC (nascent polypeptide-associated complex and coactivator alpha). 29225051_Taken together with the result that gamma-taxilin protein expression levels were decreased at the onset of mitosis, we propose that gamma-taxilin participates in Nek2A-mediated centrosome disjunction as a negative regulator through its interaction with Nek2A. 35119360_alpha-/gamma-Taxilin are required for centriolar subdistal appendage assembly and microtubule organization. | ENSMUSG00000038344 | Txlng | 1.053873e+03 | 0.8537188 | -0.228167125 | 0.3312163 | 4.751836e-01 | 0.4906120894 | 0.87511746 | No | Yes | 9.577901e+02 | 173.942502 | 1.037609e+03 | 193.146266 | |
| ENSG00000087116 | 9509 | ADAMTS2 | protein_coding | O95450 | FUNCTION: Cleaves the propeptides of type I and II collagen prior to fibril assembly (By similarity). Does not act on type III collagen (By similarity). Cleaves lysyl oxidase LOX at a site downstream of its propeptide cleavage site to produce a short LOX form with reduced collagen-binding activity (PubMed:31152061). {ECO:0000250|UniProtKB:P79331, ECO:0000269|PubMed:31152061}. | Alternative splicing;Collagen degradation;Disulfide bond;Ehlers-Danlos syndrome;Extracellular matrix;Glycoprotein;Hydrolase;Metal-binding;Metalloprotease;Protease;Reference proteome;Repeat;Secreted;Signal;Zinc;Zymogen | hsa:9509; | collagen-containing extracellular matrix [GO:0062023]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; metalloendopeptidase activity [GO:0004222]; metallopeptidase activity [GO:0008237]; zinc ion binding [GO:0008270]; collagen catabolic process [GO:0030574]; collagen fibril organization [GO:0030199]; extracellular matrix organization [GO:0030198]; lung development [GO:0030324]; protein processing [GO:0016485]; skin development [GO:0043588]; spermatogenesis [GO:0007283] | Mouse_homologues 17929299_The extent of liver fibrosis is reduced in ADAMTS2-deficient mice in comparison with their wild type littermates. 19795413_Data show thatincreased mRNA of Lamb3 and Adamts2 and enhanced activities of metalloproteinases MMP1, 2 and 7 in transgenic mice expressing the HLA-DQ8. 20574651_Data show that ADAMTS-2 is able to reduce proliferation of endothelial cells, and to induce their retraction and detachment from the substrate resulting in apoptosis. 23491141_There is no significant genotype effect of ADAMTS2 on age, height, weight, BMI or sex on risk of Achilles tendon pathology. 26927563_Data show that the high titer and specific rabbit anti-ADAMTS2 antibody has been prepared successfully. 28373586_ADAMTS2 plays a critical role in cardiac hypertrophy induced by pressure overload in mouse model. 29649548_Mice lacking Adamts2 and Adamts14 develop spontaneous atopic dermatitis due to immune dysregulation. 31491533_ADAMTS-2 significantly contributes to the N-t cleavage and inactivation of Reelin in the postnatal cerebral cortex and hippocampus, but much less in the cerebellum. | ENSMUSG00000036545 | Adamts2 | 3.777037e+02 | 1.1991585 | 0.262022377 | 0.3174487 | 6.700663e-01 | 0.4130285044 | 0.84733931 | No | Yes | 3.348829e+02 | 43.524117 | 2.449281e+02 | 32.934697 | ||
| ENSG00000087152 | 56970 | ATXN7L3 | protein_coding | Q14CW9 | FUNCTION: Component of the transcription regulatory histone acetylation (HAT) complex SAGA, a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates both histones H2A and H2B (PubMed:18206972, PubMed:21746879). The SAGA complex is recruited to specific gene promoters by activators such as MYC, where it is required for transcription. Required for nuclear receptor-mediated transactivation. Within the complex, it is required to recruit USP22 and ENY2 into the SAGA complex (PubMed:18206972). Regulates H2B monoubiquitination (H2Bub1) levels. Affects subcellular distribution of ENY2, USP22 and ATXN7L3B (PubMed:27601583). {ECO:0000255|HAMAP-Rule:MF_03047, ECO:0000269|PubMed:18206972, ECO:0000269|PubMed:21746879, ECO:0000269|PubMed:27601583}. | 3D-structure;Activator;Alternative splicing;Chromatin regulator;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | Mouse_homologues mmu:217218; | DUBm complex [GO:0071819]; nucleus [GO:0005634]; SAGA complex [GO:0000124]; nuclear receptor coactivator activity [GO:0030374]; transcription coactivator activity [GO:0003713]; zinc ion binding [GO:0008270]; chromatin organization [GO:0006325]; histone deubiquitination [GO:0016578]; histone H3 acetylation [GO:0043966]; histone monoubiquitination [GO:0010390]; monoubiquitinated histone deubiquitination [GO:0035521]; monoubiquitinated histone H2A deubiquitination [GO:0035522]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357] | 18206972_ATXN7L3, USP22, & ENY2 are required cofactors for full transcriptional activity by nuclear receptors. The deubiquitinase activity of TFTC/STAGA HAT counteracts heterochromatin silencing & is a positive cofactor for in vivo nuclear receptor activation. 20634802_The solution structures of the SCA7 domain of both ATXN7 and ATXN7L3 reveal a new, common zinc-finger motif at the heart of two distinct folds, providing a molecular basis for the observed functional differences. 21746879_Downregulation of ATXN7L3 by short hairpin RNA speci fi cally inactivated the SAGA deubiquitination activity, leading to a strong increase of global H2B ubiquitination and a moderate increase of H2A ubiquitination. 27132940_ATXN7L3 and ENY2 orchestrate activities of multiple deubiquitinating enzymes, including USP27x and USP51, and that imbalances in these activities likely potentiate human diseases including cancer. 31737900_Long non-coding RNA DSCAM-AS1 contributes to the tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3 axis. 33186807_ATXN7L3 positively regulates SMAD7 transcription in hepatocellular carcinoma with growth inhibitory function. | ENSMUSG00000059995 | Atxn7l3 | 4.632175e+03 | 1.1692073 | 0.225530762 | 0.2880481 | 6.105336e-01 | 0.4345869549 | 0.85294698 | No | Yes | 4.609837e+03 | 396.625663 | 3.905541e+03 | 345.070085 | ||
| ENSG00000087266 | 6452 | SH3BP2 | protein_coding | P78314 | FUNCTION: Binds differentially to the SH3 domains of certain proteins of signal transduction pathways. Binds to phosphatidylinositols; linking the hemopoietic tyrosine kinase fes to the cytoplasmic membrane in a phosphorylation dependent mechanism. | 3D-structure;Alternative splicing;Disease variant;Phosphoprotein;Reference proteome;SH2 domain;SH3-binding | The protein encoded by this gene has an N-terminal pleckstrin homology (PH) domain, an SH3-binding proline-rich region, and a C-terminal SH2 domain. The protein binds to the SH3 domains of several proteins including the ABL1 and SYK protein tyrosine kinases , and functions as a cytoplasmic adaptor protein to positively regulate transcriptional activity in T, natural killer (NK), and basophilic cells. Mutations in this gene result in cherubism. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2009]. | hsa:6452; | phosphotyrosine residue binding [GO:0001784]; SH3 domain binding [GO:0017124]; signal transduction [GO:0007165] | 12200378_Regulation of FcepsilonRI-mediated degranulation by an adaptor protein 3BP2 in rat basophilic leukemia RBL-2H3 cells. 15345594_3BP2 may regulate b cell receptor-mediated gene activation through Vav proteins. 15751964_Adaptor protein SH3BP2 regulates transcription factors through its tyrosine phosphorylation and SH2 domain. 16177062_CD244-3BP2 association regulates cytolytic function but not IFN-gamma release 16713042_no mutations...in giant cell granuloma 16802602_How SH3BP2 affects leukocyte signaling and influences cherubism 17147794_a novel A1517G missense mutation at the SH3BP2 gene in a Chinese family with multiple affected individuals with cherubism was identified 17156730_Mutated in a rare human disease involved in cranial-facial development called cherubism, suggesting a role for 3BP2 in regulating osteoclast and hematopoietic cell function. [REVIEW] 17306257_unexpected role of 3BP2 in endocytic and cytoskeletal regulation through its interaction with CIN85 and HIP-55 17321449_A new mutation in a family affected with cherubism 17544554_People with Giant Cell Granuloma of the Jaw do not harbour cherubism-related germline SH3BP2 mutations. 18479751_3BP2 acts downstream of SAP, increases CD244 phosphorylation and links the receptor with PI3K, Vav, PLC gamma, and PKC downstream events in order to achieve maximum natural killer cell killing function. 18596838_Point mutations in the SH3BP2 gene have been revealed in cherubism patients. 19017279_2 novel mutations were found; heterozygous missense mutation c.1442A>T (Q481L) in exon 11 in one sporadic case of CGCL and heterozygous germline and tumor tissue missense mutation c.320C>T (T107M) in exon 4 in one patient with cherubism. 19576004_The SH-3BP-2 mutation may participate in the differentiation and maturation of osteoclast-like cells in the lesion of cherubism. 19833725_3BP2 induces the protein complex with cellular signaling molecules through phosphorylation of Tyr(183) and SH2 domain leading to the activation of NFAT in B cells 20002873_No SH3BP2 gene mutation was found in PGCL. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20872577_over expression of SH3BP2 in RAW 264.7 cells potentiates sRANKL-stimulated phosphorylation of PLCgamma1 and PLCgamma2. 21680150_if a primary genetic defect is the cause for CGCG it is either located in SH3BP2 gene exons not yet related to cherubism or in a different gene. 21794028_P416R mutation of 3BP2 causes the gain of function in B cells by increasing the interaction with specific signaling molecules. 22795151_In the first family, a missense mutation Arg415Gln was found in exon 9 of SH3BP2 in all affected individuals. The unaffected individuals did not have the mutation. In the second family, a missense mutation Pro418Thr was identified in exon 9 of the SH3BP2 22820184_These results demonstrate that PARP1 regulates expression of SH3BP2. 23083484_Authors conclude that a novel p.Asp419Tyr alteration in SH3BP2 to be a cherubism-causing mutation in a Turkish family. 24608212_A c.1244G>A (p.Arg415Gln) mutation in SH3BP2 gene causes cherubism in a Turkish family 25810396_The adaptor 3BP2 is required for KIT receptor expression and human mast cell survival. 28721660_All members featured a heterozygous missense c.1244G>C; p.Arg415Pro SH3BP2 mutation 29885053_The current study shows that SH3BP2 is expressed in primary tumors and cell lines from Gastrointestinal stromal tumors (GISTs) patients and that SH3BP2 silencing leads to a downregulation of oncogenic KIT and PDGFRA expression and an increase in apoptosis in imatinib-sensitive and imatinib-resistant GIST cells. 33941395_SH3BP2-related fibro-osseous disorders of the maxilla and mandible: A systematic review. 35279646_Cherubism in three siblings.', trans 'A cherubismus elofordulasa harom testverben. | ENSMUSG00000054520 | Sh3bp2 | 2.238023e+03 | 1.0905563 | 0.125064213 | 0.2943927 | 1.769582e-01 | 0.6740004520 | 0.93064572 | No | Yes | 1.859354e+03 | 233.329233 | 1.791308e+03 | 230.582697 | |
| ENSG00000088325 | 22974 | TPX2 | protein_coding | Q9ULW0 | FUNCTION: Spindle assembly factor required for normal assembly of mitotic spindles. Required for normal assembly of microtubules during apoptosis. Required for chromatin and/or kinetochore dependent microtubule nucleation. Mediates AURKA localization to spindle microtubules (PubMed:18663142, PubMed:19208764). Activates AURKA by promoting its autophosphorylation at 'Thr-288' and protects this residue against dephosphorylation (PubMed:18663142, PubMed:19208764). TPX2 is inactivated upon binding to importin-alpha (PubMed:26165940). At the onset of mitosis, GOLGA2 interacts with importin-alpha, liberating TPX2 from importin-alpha, allowing TPX2 to activates AURKA kinase and stimulates local microtubule nucleation (PubMed:26165940). {ECO:0000269|PubMed:18663142, ECO:0000269|PubMed:19208764, ECO:0000269|PubMed:26165940}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;Isopeptide bond;Microtubule;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:22974; | axon hillock [GO:0043203]; cytosol [GO:0005829]; intercellular bridge [GO:0045171]; microtubule cytoskeleton [GO:0015630]; mitotic spindle [GO:0072686]; nuclear microtubule [GO:0005880]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spindle [GO:0005819]; spindle pole [GO:0000922]; importin-alpha family protein binding [GO:0061676]; microtubule binding [GO:0008017]; protein kinase activator activity [GO:0030295]; protein kinase binding [GO:0019901]; activation of protein kinase activity [GO:0032147]; apoptotic process [GO:0006915]; cell division [GO:0051301]; microtubule nucleation [GO:0007020]; mitotic cell cycle [GO:0000278]; mitotic spindle assembly [GO:0090307]; negative regulation of microtubule depolymerization [GO:0007026]; regulation of mitotic spindle organization [GO:0060236] | 12177045_TPX2 is required for targeting Aurora-A kinase to the spindle apparatus and Aurora-A might regulate the function of TPX2 during spindle assembly 12389033_spindle formation requires the function of TPX2 to generate a stable bipolar spindle with overlapping antiparallel microtubule arrays 12477396_observations reveal a structural role for hTPX2 in spindles and provide evidence for a balance between microtubule-based motor forces and structural spindle components 12612055_molecular cloning; overexpression provokes accumulation of cells in G(2)-M phase and subsequent polyploidization, suggesting that excess repp86 may interfere with correct nuclear division 16287863_APC/C(Cdh1) controls the stability of TPX2, thereby ensuring accurate regulation of the spindle assembly in the cell cycle 16489064_Aberrant expression of TPX2 may play important role(s) in both malignant transformation of respiratory epithelium and progression of squamous cell lung cancer and could serve as a prognostic predictor for the disease. 17705509_TPX2 binding decreases the size and accessibility of a hydrophobic pocket, adjacent to the ATP site, to inhibitors. 17716627_Data show that Siah2 is an important mediator of repp86 protein degradation. 18663142_Chromosome nucleation is involved in spindle pole separation and setting spindle length. A second Aurora A-independent function of TPX2 is required to bipolarize spindles. 19148505_human TPX2 mRNA is closely linked to increased or abnormal cell proliferation in malignant salivary gland tumors. 19208764_Data provide a molecular explanation for the assembly of the apoptotic microtubule network, and suggest important similarities with the process of RanGTP- and TPX2-mediated mitotic spindle formation. 19424574_Overexpression of TPX2 is associated with oral squamous cell carcinomas. 19773279_Observational study of gene-disease association. (HuGE Navigator) 19801554_Data show that Aurora A(S155R) mutant reduced cellular activity and mislocalization are due to loss of interaction with TPX2. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20599806_TPX2 expression is associated with the progression of malignant astrocytoma. 20708655_association of Aurora-A and TPX2 gives rise to a novel functional unit with oncogenic properties.[review] 21099343_Decreased AurA-TPX2 complex formation in response to irradiation results from reduced cellular levels of TPX2, as a result of protein degradation and decreased translation of TPX2 mRNA. 21147853_TPX2 protects Aurora-A from degradation both in interphase and in mitosis. 21187329_Results demonstrate a role for PP6 as the T-loop phosphatase regulating Aurora A activity bound to its activator TPX2 during mitotic spindle formation. 21347367_two switches determining Aurora A activation 22207630_TPX2 promote 20q amplicon-driven progression of colorectal adenoma to carcinoma. 22307108_TPX2 shows potential to be used as a new marker for cervical cancer diagnosis and therapy. 22560880_The data support the role of TPX2 as a novel co-activator of Aurora kinase B. 22761906_AIM1, ERGIC1, and TPX2 were shown to be highly expressed especially in prostate cancer tissues, and high mRNA expression of ERGIC1 and TMED3 associated with AR and ERG oncogene expression 23045526_the regulation of gamma-H2AX signals by TPX2 is not associated with apoptosis or the mitotic functions of TPX2. 23328114_Data indicate that the sensitivity of cell-lines with amplification of AURKA depends upon the activity of the kinase, which correlates with the expression of the regulatory gene products TPX2 and HMMR/RHAMM. 23333597_GLIPR1 interacts with Hsc70, and GLIPR1 overexpression or Hsc70 knockdown leads to transcriptional suppression of AURKA and TPX2. 23357462_Data indicate that expression-based risk indices of three genes UBE2C, TPX2, and MELK were more strongly associated with poor 5-year survival in adenocarcinoma patients. 23444224_Data show that five genes CKAP5, KPNB1, RAN, TPX2 and KIF11 were shown to be essential for tumor cell survival in both head and neck squamous cell carcinoma (HNSCC)and non-small cell lung cancer (NSCLC), but most particularly in HNSCC. 23725757_The expression of TPX2 protein and mRNA were correlated with invasive depth and lymphatic metastasis of esophageal squamous cell carcinoma. 23873098_Data indicate that TPX2 (target protein for Xklp2) may play a role in the development and progression of bladder carcinoma, and suggest that inhibition of TPX2 level may be a novel strategy for therapy of the patients with bladder carcinoma. 23963785_TPX2 expression is associated with cell proliferation and poor prognosis among patients with resected esophageal squamous cell carcinoma 24341487_TPX2 plays an important role in promoting tumorigenesis and metastasis of human colon cancer, and may represent a novel prognostic biomarker and therapeutic target for the disease 24488334_TPX2 overexpression is associated with medullary thyroid carcinoma. 24556998_This review provides an historic overview of the discovery of TPX2 and summarizes its cytoskeletal and signaling roles with relevance to cancer therapies. [review] 24625450_RAN nucleo-cytoplasmic transport and mitotic spindle assembly partners XPO7 and TPX2 have roles in serous epithelial ovarian cancer 24718984_The results demonstrated that TPX2 is important in the regulation of tumor growth in cervical cancer and therefore may be a potential therapeutic target as a novel treatment strategy. 24867643_the molecular mechanisms of two distinct activation strategies (autophosphorylation and TPX2-mediated activation) in human Aurora A kinase, was elucidated. 24875404_Identify RHAMM as a critical regulator of TPX2 location/ Aurora kinase A signaling and suggest that RHAMM ensures bipolar spindle assembly and mitotic progression through the integration of biochemical and structural pathways. 25239289_TPX2 siRNA transfection significantly reduced tumor growth. 25302620_In vitro studies found that TPX2 knockdown significantly inhibited cell proliferation and viability in both Hep3B and HepG2 cells. 25365214_our study is the first indication of a constitutive control of TPX2 on H4K16ac levels, with potential implications for DNA damage response. 25632068_Propose TPX2 and AURKA as novel co-regulators on the MYC pathway in colorectal neoplasms. 25830658_a causative link between altered function of AURKA-HMMR-TPX2-TUBG1 and breast carcinogenesis in BRCA1/2 mutation carriers 25914189_These results indicated that TPX2 has an impact on tumor angiogenesis in pancreatic cancer. 26018074_Dimeric, but not monomeric, Eg5 was differentially inhibited by full-length and truncated TPX2, demonstrating that dimerization or residues in the neck region are important for the interaction of TPX2 with Eg5. 26240182_Aurora A-dependent TPX2 phosphorylation controls mitotic spindle length through regulating microtubule flux. 26257190_the levels and distribution of TPX2 are likely to be determinants of when and where kinesin-5 acts in neurons. 26279431_TPX2 was a target gene of miR-491. 26414402_Data show that cytoskeleton associated protein 5 (chTOG) only weakly promotes importin-regulated microtubule nucleation, but acts synergistically with TPX2 protein. 26624896_expression of both TPX2 and PD-L1 are associated with persistence/recurrence of cervical intraepithelial neoplasia after cervical conization 27053618_MiR-491 inhibits hepatocellular carcinoma cell proliferation, invasion and migration by downregulating the expression of TPX2. 27314162_detection of TPX2 overexpression could serve as a prognostic marker and therapeutic target for gastric cancer 27775325_This study concludes that the helical region of TPX2 folds upon binding Aurora-A, and that stabilization of this helix does not compromise Aurora-A activation. 28069036_High TPX2 expression is associated with gastric cancer. 28108243_We show TPX2, a regulator of Aurora-A, is associated with high grade and stage of ccRCC, and is an independent predictor of recurrence. 28636807_TPX2 was correlated with cell radioresistance 28799673_Data found that TPX2 was highly expressed in human bladder cancer tissues. Its overexpression promoted bladder cancer growth and correlated with tumor grade and stage, lymph node metastasis, and poor prognosis. These results support that TPX2 acts as a tumor promoter in human bladder tumor development. 28869599_Study suggests AURKA and TPX2 as potential stratification markers for taxane-based radiochemotherapy. lung adenocarcinoma cohort, high expression levels of AURKA and TPX2 were associated with specifically improved overall survival upon taxane-based radiochemotherapy. 29120325_Together, these results suggest a molecular mechanism of how the Ran-GTP gradient can regulate TPX2-dependent microtubule formation. 29276125_Eg5 localization and centrosome separation in prophase depend on the nuclear microtubule-associated protein TPX2, a pool of which localizes to the centrosomes before nuclear envelope breakdown. This localization involves the kinase Nek9, which phosphorylates TPX2 nuclear localization signal preventing its interaction with importin and nuclear import. 29548937_VPS28, an ESCRT-I protein, interacts with Gbetagamma and Eg5-TPX2. VPS28 organizes Aster Microtubules via Gbetagamma. VPS28 recruits Eg5 and TPX2 to regulate mitotic spindle bipolarity. 29865033_TPX2 promotes the proliferation and migration of human OC cells by regulating PLK1 expression. 29925526_These findings unveil a novel function of ADD1 in maintaining spindle pole integrity through its interaction with TPX2. 30123975_The results show that at metaphase both Eg5-EGFP and TPX2-EGFP are enriched toward spindle poles, but only TPX2-EGFP is enriched relative to microtubules. Eg5-EGFP and TPX2-EGFP show distinct localization patterns in anaphase, with Eg5-EGFP relocalizing to the midzone earlier than TPX2-EGFP. 30177840_findings reveal that BRCA2-deficient cancer cells show enhanced sensitivity to inactivation of TPX2 or its partner Aurora-A, which points at an actionable dependency of genomically unstable cancers. 30257313_Inhibiting TPX2 by miR-330-3p suppresses the proliferation of melanoma cell lines; miR-330-3p/TPX2 pathway expressed differently between melanoma patients and healthy people 30257343_TPX2 expression in cholangiocarcinoma tissues was significantly higher than that paracancerous tissue. 30412141_A PRISMA-compliant systematic review and meta-analysis found that overexpression of TPX2 is related to poor survival rate in most solid tumors, which indicates that the expression level of TPX2 is a significant prognostic parameter and potential therapeutic target in various solid tumors. 30454896_TPX2 silencing negatively regulates the PI3K/AKT and activates p53 signaling pathway by which breast cancer cells proliferation were inhibited whereas cellulars apoptosis were accelerated. 30588191_Study indicated that AURKA, CDC20 and TPX2 are over-expressed in smoking related lung adenocarcinoma tissues and their higher mRNA expression levels have a worse prognosis. 30602538_Mitotic regulators TPX2 and Aurora A protect DNA forks during replication stress by counteracting 53BP1 function. 30633364_We showed that miR-8075 that was downregulated in cervical cancer tissues repressed cervical cancer cell proliferation, migration, and invasion. Furthermore, we validated that upregulation of TPX2 by hsa_circRNA_101996-mediated inhibition of miR-8075 contributed to cervical cancer proliferation, migration, and invasion 31272499_Results suggest activation of CDK5 is associated with HCC tumorigenesis. CDK5-mediated phosphorylation and stabilization of TPX2 promotes hepatocellular proliferation and tumorigenicity. 31494045_TPX2 as a Novel Prognostic Indicator and Promising Therapeutic Target in Triple-negative Breast Cancer. 31638175_TPX2 may suppress the growth of Hepatocellular carcinoma by regulating the PI3K/AKT signaling pathway and thus, TPX2 may be a potential target for the treatment of liver cance 31676293_This review highlights the physiological and pathophysiological properties of AURKA, the structure of the AURKA/TPX2 complex and the main structural features that can be explored for the design of selective AURKA inhibitors. 31799669_MiR-491 suppresses migration and invasion via directly targeting TPX2 in breast cancer. 32041138_Excess TPX2 Interferes with Microtubule Disassembly and Nuclei Reformation at Mitotic Exit. 32193808_Aurora A kinase and its activator TPX2 are potential therapeutic targets in KRAS-induced pancreatic cancer. 32239565_Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. 32356447_miR-485-3p suppresses colorectal cancer via targeting TPX2. 32503626_Epigenome-wide association study in healthy individuals identifies significant associations with DNA methylation and PBMC extract VEGF-A concentration. 32535036_Knockdown of circ_0003340 induces cell apoptosis, inhibits invasion and proliferation through miR-564/TPX2 in esophageal cancer cells. 32705231_Identification of early stage recurrence endometrial cancer biomarkers using bioinformatics tools. 32748897_TPX2 Promotes Metastasis and Serves as a Marker of Poor Prognosis in Non-Small Cell Lung Cancer. 33029085_Comprehensive analysis of TPX2-related ceRNA network as prognostic biomarkers in lung adenocarcinoma. 33093951_Structural bioinformatics predicts that the Retinitis Pigmentosa-28 protein of unknown function FAM161A is a homologue of the microtubule nucleation factor Tpx2. 33275894_The Aurora-A/TPX2 Axis Directs Spindle Orientation in Adherent Human Cells by Regulating NuMA and Microtubule Stability. 33556637_TPX2 mediates prostate cancer epithelial-mesenchymal transition through CDK1 regulated phosphorylation of ERK/GSK3beta/SNAIL pathway. 33622270_High nuclear TPX2 expression correlates with TP53 mutation and poor clinical behavior in a large breast cancer cohort, but is not an independent predictor of chromosomal instability. 33907841_RHPN1AS1 promotes ovarian carcinogenesis by sponging miR4855p and releasing TPX2 mRNA. 34287649_Aurora A kinase activation: Different means to different ends. 34302807_Interaction of spindle assembly factor TPX2 with importins-alpha/beta inhibits protein phase separation. 34597712_Secretory autophagy-induced bladder tumour-derived extracellular vesicle secretion promotes angiogenesis by activating the TPX2-mediated phosphorylation of the AURKA-PI3K-AKT axis. 34982022_Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis. 35273149_Downregulation of TPX2 impairs the antitumor activity of CD8+ T cells in hepatocellular carcinoma. 35356748_TPX2 Serves as a Cancer Susceptibility Gene and Is Closely Associated with the Poor Prognosis of Endometrial Cancer. | ENSMUSG00000027469 | Tpx2 | 6.720516e+03 | 1.1414474 | 0.190864428 | 0.2654993 | 5.170038e-01 | 0.4721225625 | 0.86976192 | No | Yes | 7.672026e+03 | 1163.984231 | 5.394124e+03 | 839.536816 | ||
| ENSG00000089006 | 27131 | SNX5 | protein_coding | Q9Y5X3 | FUNCTION: Involved in several stages of intracellular trafficking. Interacts with membranes containing phosphatidylinositol 3-phosphate (PtdIns(3P)) or phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2) (PubMed:15561769). Acts in part as component of the retromer membrane-deforming SNX-BAR subcomplex. The SNX-BAR retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) and is involved in endosome-to-plasma membrane transport for cargo protein recycling. The SNX-BAR subcomplex functions to deform the donor membrane into a tubular profile called endosome-to-TGN transport carrier (ETC) (Probable). Does not have in vitro vesicle-to-membrane remodeling activity (PubMed:23085988). Involved in retrograde transport of lysosomal enzyme receptor IGF2R (PubMed:17148574, PubMed:18596235). May function as link between endosomal transport vesicles and dynactin (Probable). Plays a role in the internalization of EGFR after EGF stimulation (Probable). Involved in EGFR endosomal sorting and degradation; the function involves PIP5K1C isoform 3 and is retromer-independent (PubMed:23602387). Together with PIP5K1C isoform 3 facilitates HGS interaction with ubiquitinated EGFR, which initiates EGFR sorting to intraluminal vesicles (ILVs) of the multivesicular body for subsequent lysosomal degradation (Probable). Involved in E-cadherin sorting and degradation; inhibits PIP5K1C isoform 3-mediated E-cadherin degradation (PubMed:24610942). Plays a role in macropinocytosis (PubMed:18854019, PubMed:21048941). {ECO:0000269|PubMed:18854019, ECO:0000269|PubMed:21048941, ECO:0000269|PubMed:24610942, ECO:0000303|PubMed:15561769, ECO:0000303|PubMed:19619496, ECO:0000303|PubMed:23085988}. | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Endocytosis;Endosome;Host-virus interaction;Lipid-binding;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene encodes a member of the sorting nexin family. Members of this family contain a phox (PX) domain, which is a phosphoinositide binding domain, and are involved in intracellular trafficking. This protein functions in endosomal sorting, the phosphoinositide-signaling pathway, and macropinocytosis. This gene may play a role in the tumorigenesis of papillary thyroid carcinoma. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Sep 2013]. | hsa:27131; | brush border [GO:0005903]; cytosol [GO:0005829]; early endosome membrane [GO:0031901]; endosome [GO:0005768]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; extrinsic component of endosome membrane [GO:0031313]; intracellular membrane-bounded organelle [GO:0043231]; macropinocytic cup [GO:0070685]; perinuclear region of cytoplasm [GO:0048471]; phagocytic cup [GO:0001891]; retromer complex [GO:0030904]; retromer, tubulation complex [GO:0030905]; ruffle [GO:0001726]; tubular endosome [GO:0097422]; cadherin binding [GO:0045296]; D1 dopamine receptor binding [GO:0031748]; dynactin binding [GO:0034452]; phosphatidylinositol binding [GO:0035091]; phosphatidylinositol-3,5-bisphosphate binding [GO:0080025]; phosphatidylinositol-4-phosphate binding [GO:0070273]; phosphatidylinositol-5-phosphate binding [GO:0010314]; epidermal growth factor catabolic process [GO:0007174]; intracellular protein transport [GO:0006886]; negative regulation of blood pressure [GO:0045776]; pinocytosis [GO:0006907]; positive regulation of insulin receptor signaling pathway [GO:0046628]; positive regulation of renal sodium excretion [GO:0035815]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of macroautophagy [GO:0016241]; retrograde transport, endosome to Golgi [GO:0042147] | 15561769_SNX5 is localized to a subdomain of the early endosomes and is recruited to the cell membrane following EGF stimulation. 17148574_SNX5 and SNX6 may constitute functional equivalents of Vps17p in mammalian retromer 18596235_DOCK180 regulates CI-MPR trafficking via SNX5 and this function is independent of its guanine nucleotide exchange factor activity toward Rac1 19204726_Observational study of gene-disease association. (HuGE Navigator) 22486813_Data indicate induction of caspase-2 by sorting nexin 5 (SNX5) in papillary thyroid carcinoma 23195037_internalization of agonist-activated D1R is regulated by both SNX5 and GRK4, and that SNX5 is critical to the recycling of the receptor to the plasma membrane 25825816_Sorting nexin 5 and dopamine d1 receptor regulate the expression of the insulin receptor in human renal proximal tubule cells 28226239_The Chlamydia trachomatis IncE binding site is unique to human SNX5 and related family members SNX6 and SNX32. 29444945_sorting nexin 5 (SNX5), a component of the retromer and part of the retrograde transport pathway, interacts with UL35 proteins. binding of UL35 proteins to SNX5 was required for efficient viral replication and for transport of the most abundant cytomegalovirus glycoprotein B to the cytoplasmic viral assembly center. 29743363_High SNX5 expression is associated with alphavirus infections. 30419003_Results indicate that SNX5 constitutive phosphorylation that mimics the mutant S226E decreases the active SNX5 in these cells. The phosphorylation of SNX5 regulates the dimerization with SNX1 or SNX2, and this suggests that it controls membrane trafficking and protein sorting. 31576058_The studies identify a sorting motif and provide molecular insight into an evolutionary conserved coat complex, the 'Endosomal SNX-BAR sorting complex for promoting exit 1' (ESCPE-1), which couples sorting motif recognition to the BAR-domain-mediated biogenesis of cargo-enriched tubulo-vesicular transport carriers. 31806368_SNX5 inhibits RLR-mediated antiviral signaling by targeting RIG-I-VISA signalosome. 31819169_Upregulation of SNX5 predicts poor prognosis and promotes hepatocellular carcinoma progression by modulating the EGFR-ERK1/2 signaling pathway. 33328639_Sorting nexin 5 mediates virus-induced autophagy and immunity. | ENSMUSG00000027423 | Snx5 | 3.250050e+03 | 0.7208029 | -0.472323254 | 0.2811223 | 2.713601e+00 | 0.0994961605 | 0.75783482 | No | Yes | 2.381260e+03 | 303.287598 | 3.350480e+03 | 436.918477 | |
| ENSG00000089234 | 8315 | BRAP | protein_coding | Q7Z569 | FUNCTION: Negatively regulates MAP kinase activation by limiting the formation of Raf/MEK complexes probably by inactivation of the KSR1 scaffold protein. Also acts as a Ras responsive E3 ubiquitin ligase that, on activation of Ras, is modified by auto-polyubiquitination resulting in the release of inhibition of Raf/MEK complex formation. May also act as a cytoplasmic retention protein with a role in regulating nuclear transport. {ECO:0000269|PubMed:14724641, ECO:0000303|PubMed:10777491}. | Alternative splicing;Coiled coil;Cytoplasm;Metal-binding;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene was identified by its ability to bind to the nuclear localization signal of BRCA1 and other proteins. It is a cytoplasmic protein which may regulate nuclear targeting by retaining proteins with a nuclear localization signal in the cytoplasm. [provided by RefSeq, Jul 2008]. | hsa:8315; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; ubiquitin ligase complex [GO:0000151]; identical protein binding [GO:0042802]; nuclear localization sequence binding [GO:0008139]; nucleic acid binding [GO:0003676]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; MAPK cascade [GO:0000165]; negative regulation of signal transduction [GO:0009968]; protein ubiquitination [GO:0016567]; Ras protein signal transduction [GO:0007265] | 14724641_modulates sensitivity of the MAP kinase cascade to stimulus-dependent activation by limiting functional assembly of the core enzymatic components through the inactivation of KSR 15340083_Monocytic differentiation of the promyelomonocytic cell lines U937 and HL60 is associated with the upregulation of Brap2 expression concomitantly with the upregulation and cytoplasmic relocalization of p21. 18332145_the capacity of IMP to inhibit signal propagation through Raf to MEK is a consequence of disrupting KSR1 homooligomerization and B-Raf/c-Raf hetero-oligomerization 18380995_The most current evidence suggests that it may be more beneficial in those with BRCA mutations because tumors associated with these mutations are likely to be estrogen-receptor positive. 18577512_p21 Ras/Imp regulate cytokine production and migration in CD4 T cells [Imp] 19009954_Carriers of mutations in the genes BRCA1/2 may present a specific high risk group for PABC especially at younger ages. 19152073_The authors found that Brap2, which has intrinsic RING domain dependent E3 ligase activity, facilitates HsCdc14A Lys-63 linked ubiquitin modification, indicating that Brap2 may be the ubiquitin E3 Ligase of HsCdc14A. 19198608_Observational study of gene-disease association. (HuGE Navigator) 19198608_SNPs in BRAP associated with risk of myocardial infarction in Asian populations.BRAP knockdown in cultured coronary endothelial cells suppressed activation of NF-kappaB. 19713974_Data indicate that one SNP in BRAP showed(rs11066001) a significant association in allele frequency distribution with CAD in both the Japanese and Korean populations. 20040518_All results are consistent with BRAP2 being a novel, phosphorylation-regulated negative regulator of nuclear import, with potential as an antiviral agent. 21301165_Polymorphism of 270 A > G in BRAP is Associated with Lower Ankle-Brachial Index and peripheral artery disease. 21670849_Data show that BRAP conferred a risk for carotid plaque and intima-medial thickness. 22022282_our approach, which is applicable to any set of interval scale traits that is heritable and exhibits evidence of phenotypic clustering, identified three new loci in or near APOC1, BRAP, and PLCG1, which were associated with multiple phenotype domains. 22078348_Several BRCA1 mutations were observed in Pakistani breast cancer patients with moderate family history. 22085839_BRAP gene is associated with the extent of coronary atherosclerosis and has a synergistic effect with diabetes on the occurrence of significant CAD in the Chinese population 22122968_ANRIL on 9p21 and BRAP were both associated with ankle brachial index in a Taiwanese population. 22965072_genetic association studies in population of Chinese Han young adults in Beijing: Data suggest that 2 SNPs in BRAP (rs11066001; rs3782886) are associated with decreased risk of metabolic syndrome in this population. 23105109_The dominant effect of prolonged USP15 depletion upon signal amplitude is due to a decrease in CRAF levels while allowing for the possibility that USP15 may also function to dampen MAPK signaling through direct stabilization of BRAP. 23356535_The BRAP polymorphism may not play an important role in ischemic stroke in the Taiwanese population. 23526420_BAP1 expression closely correlates with age, clinical stage, pathologic differentiation and histological type in colorectal cancer (CRC). BAP1 may serve as a novel prognostic biomarker for CRC. 23554956_Brap2 regulates temporal control of NF-kappaB localization mediated by inflammatory response. 23707952_ectopic expression of BRAP2 inhibits nuclear localization of HMG20A and NuMA1, and prevents nuclear envelope accumulation of SYNE2. 24454952_BRAP gene may confer vulnerability for schizophrenia in Han Chinese population. 25820252_Together the results indicate for the first time that BRAP2 may play an important NRNI role in germ cells of the testis, with an additional, scaffold/structural role in mature spermatozoa. 27245511_Average dietary energy in lunch, BRAP SNP rs3782886, and GHRL SNP rs696217 were associated with obesity, and BRAP rs3782886 was associated with other metabolic abnormalities 27742670_IMPACT data were consistent with increased risks of onset among BRCA1 and BRCA2 mutation carriers 28562329_Six polymorphisms (rs12229654 at 12q24.1, rs671 of ALDH2, rs11066015 of ACAD10, rs2074356 and rs11066280 of HECTD4, and rs3782886 of BRAP) were found to be associated with both systolic and diastolic blood pressure, with those at 12q24.1 or in ACAD10 or BRAP being novel determinants of blood pressure in Japanese. 28780075_Expression of BRAP is increased in esophageal squamous cell carcinoma samples compared with non-tumor esophageal tissues; increased expression correlates with reduced patient survival time and promotes metastasis of xenograft tumors in mice. 30703135_Mutation in BRAP gene is associated with childhood pulmonary arterial hypertension. 31776938_BRCA1-associated protein inhibits glioma cell proliferation and migration and glioma stem cell self-renewal via the TGF-beta/PI3K/AKT/mTOR signalling pathway. 32843694_Potential mechanisms underlying the association between single nucleotide polymorphism (BRAP and ALDH2) and hypertension among elderly Japanese population. 33093602_CUX2, BRAP and ALDH2 are associated with metabolic traits in people with excessive alcohol consumption. | ENSMUSG00000029458 | Brap | 6.457188e+02 | 1.2885432 | 0.365740920 | 0.3100114 | 1.408782e+00 | 0.2352586274 | 0.78818582 | No | Yes | 7.308756e+02 | 119.594144 | 4.826789e+02 | 81.163976 |
| ENSG00000089597 | 23193 | GANAB | protein_coding | Q14697 | FUNCTION: Catalytic subunit of glucosidase II that cleaves sequentially the 2 innermost alpha-1,3-linked glucose residues from the Glc(2)Man(9)GlcNAc(2) oligosaccharide precursor of immature glycoproteins (PubMed:10929008). Required for PKD1/Polycystin-1 and PKD2/Polycystin-2 maturation and localization to the cell surface and cilia (PubMed:27259053). {ECO:0000269|PubMed:10929008, ECO:0000269|PubMed:27259053}. | Alternative splicing;Direct protein sequencing;Disease variant;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Glycosidase;Golgi apparatus;Hydrolase;Phosphoprotein;Reference proteome;Signal | PATHWAY: Glycan metabolism; N-glycan metabolism. {ECO:0000269|PubMed:10929008}. | This gene encodes the alpha subunit of glucosidase II and a member of the glycosyl hydrolase 31 family of proteins. The heterodimeric enzyme glucosidase II plays a role in protein folding and quality control by cleaving glucose residues from immature glycoproteins in the endoplasmic reticulum. Expression of the encoded protein is elevated in lung tumor tissue and in response to UV irradiation. Mutations in this gene cause autosomal-dominant polycystic kidney and liver disease. [provided by RefSeq, Jul 2016]. | hsa:23193; | endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; glucosidase II complex [GO:0017177]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; melanosome [GO:0042470]; membrane [GO:0016020]; alpha-glucosidase activity [GO:0090599]; carbohydrate binding [GO:0030246]; glucan 1,3-alpha-glucosidase activity [GO:0033919]; hydrolase activity, hydrolyzing O-glycosyl compounds [GO:0004553]; RNA binding [GO:0003723]; carbohydrate metabolic process [GO:0005975]; N-glycan processing [GO:0006491] | 17672918_the apparent occurrence of an unusual TG 3' splice site in intron 18 is discussed 19737279_In males being treated for infertility, neutral alpha-glucosidase activity correlated with the percentage of sperm DNA fragmentation. 21642380_Loss of the tumor suppression function of Hsp60 or GANAB contribute to aggressive cancers. 24008518_The high frequency of glucosidase II overexpression, which to the best of our knowledge has not been previously described, indicates its crucial roles in lung tumorigenesis and is thus a valuable biomarker 27259053_Mutations in GANAB gene is associated with Autosomal-Dominant Polycystic Kidney and Liver Disease. 28375157_used whole exome sequencing in a discovery cohort of 102 unrelated patients who were excluded for mutations in the 2 most common polycystic liver disease genes, PRKCSH and SEC63, to identify heterozygous loss-of-function mutations in 3 additional genes, ALG8, GANAB, and SEC61B. Similarly to PRKCSH and SEC63, these genes encode proteins that are integral to the protein biogenesis pathway in the endoplasmic reticulum. 28784653_Case Report: GANAB mutation leading to autosomal dominant polycystic kidney disease. 29243290_a novel intronic nine base pair deletion in the vicinity of the GANAB exon 24 splice donor, is reported. 33097077_Novel GANAB variants associated with polycystic liver disease. | ENSMUSG00000071650 | Ganab | 4.402037e+04 | 1.0458527 | 0.064679729 | 0.3009921 | 4.594373e-02 | 0.8302781149 | 0.96751127 | No | Yes | 4.059474e+04 | 3720.439571 | 3.837808e+04 | 3606.687465 |
| ENSG00000089639 | 51291 | GMIP | protein_coding | Q9P107 | FUNCTION: Stimulates, in vitro and in vivo, the GTPase activity of RhoA. {ECO:0000269|PubMed:12093360}. | 3D-structure;Alternative splicing;Coiled coil;GTPase activation;Metal-binding;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | This gene encodes a member of the ARHGAP family of Rho/Rac/Cdc42-like GTPase activating proteins. The encoded protein interacts with the Ras-related protein Gem through its N-terminal domain. Separately, it interacts with RhoA through a RhoGAP domain, and stimulates RhoA-dependent GTPase activity. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]. | hsa:51291; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; GTPase activator activity [GO:0005096]; metal ion binding [GO:0046872]; activation of GTPase activity [GO:0090630]; intracellular signal transduction [GO:0035556]; negative regulation of GTPase activity [GO:0034260]; regulation of small GTPase mediated signal transduction [GO:0051056] | 12093360_signalling pathways controlled by two proteins of the Ras superfamily, RhoA and Gem, are linked via the action of the RhoGAP protein Gmip 16086184_Observational study of gene-disease association. (HuGE Navigator) 16086184_Variations in the GMIP gene can confer susceptibility to major depressive disorder. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20351714_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22438581_during exocytosis, actin depolymerization commences near the secretory organelle, not the plasma membrane, and secretory granules use a JFC1- and GMIP-dependent molecular mechanism to traverse cortical actin. 25173885_These findings demonstrate a new role of Gem/Gmip/RhoA signaling in cortical actin regulation during early mitotic stages | ENSMUSG00000036246 | Gmip | 3.495043e+02 | 1.5247390 | 0.608562337 | 0.3060312 | 3.949931e+00 | 0.0468732955 | 0.65752408 | No | Yes | 3.350539e+02 | 50.427561 | 2.137473e+02 | 33.329243 | |
| ENSG00000089693 | 8079 | MLF2 | protein_coding | Q15773 | Cytoplasm;Nucleus;Phosphoprotein;Reference proteome | hsa:8079; | cytoplasm [GO:0005737]; membrane [GO:0016020]; nucleus [GO:0005634]; regulation of transcription, DNA-templated [GO:0006355] | 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 24876273_RPL39 and MLF2 have roles in tumor initiation and metastasis in breast cancer that involve nitric oxide synthase signaling 27840155_Mutation in HTT causes Huntington's disease (HD); aggregates of mutated HTT cause apoptosis in neurons of HD patients. Data suggest that both MLF1 and MLF2 preferentially interact with mutated N-terminal HTT; MLF1/MLF2 reduce number of neurons (Neuro2A cell line) containing mutant HTT aggregates and subsequent apoptosis. (HTT = Huntingtin protein; MLF = myeloid leukemia factor) 31831854_The role of phosphorylation of MLF2 at serine 24 in BCR-ABL leukemogenesis. 32342107_Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. 32703400_CRL4(DCAF8) and USP11 oppositely regulate the stability of myeloid leukemia factors (MLFs). | ENSMUSG00000030120 | Mlf2 | 1.380862e+04 | 1.2845863 | 0.361303825 | 0.3110783 | 1.347964e+00 | 0.2456342868 | 0.78892886 | No | Yes | 1.528825e+04 | 1907.691035 | 1.007088e+04 | 1289.252478 | |||
| ENSG00000090060 | 10914 | PAPOLA | protein_coding | P51003 | FUNCTION: Polymerase that creates the 3'-poly(A) tail of mRNA's. Also required for the endoribonucleolytic cleavage reaction at some polyadenylation sites. May acquire specificity through interaction with a cleavage and polyadenylation specificity factor (CPSF) at its C-terminus. {ECO:0000269|PubMed:19224921}. | ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Isopeptide bond;Magnesium;Manganese;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transferase;Ubl conjugation;mRNA processing | The protein encoded by this gene belongs to the poly(A) polymerase family. It is required for the addition of adenosine residues for the creation of the 3'-poly(A) tail of mRNAs. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2011]. | hsa:10914; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; manganese ion binding [GO:0030145]; polynucleotide adenylyltransferase activity [GO:0004652]; RNA binding [GO:0003723]; mRNA 3'-end processing [GO:0031124]; mRNA polyadenylation [GO:0006378]; regulation of mRNA 3'-end processing [GO:0031440]; RNA polyadenylation [GO:0043631] | 20174964_Observations imply that translational regulation is among the conserved mechanisms regulating PAPOLA expression. 25896913_The ability of PABPN1 to promote splicing requires its RNA binding and, to a lesser extent, poly(A)polymerase (PAP) - stimulatory functions. 26496945_Lack of CstF-64 binding excludes PAPalpha from Star-PAP target pre-mRNA UTRs.CstF-64 and 3'-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding poly A polymerase. 28096519_Whereas PABPN1 strongly increases the activity of its cognate poly(A) polymerase in vitro, Pab2 was unable to stimulate Pla1 to any significant extent. 33712453_PAPOLA contributes to cyclin D1 mRNA alternative polyadenylation and promotes breast cancer cell proliferation. | ENSMUSG00000021111 | Papola | 1.703979e+03 | 0.8273339 | -0.273458461 | 0.3316987 | 6.702610e-01 | 0.4129606092 | 0.84733931 | No | Yes | 1.702788e+03 | 312.336413 | 1.851754e+03 | 348.261401 | |
| ENSG00000090989 | 55763 | EXOC1 | protein_coding | Q9NV70 | FUNCTION: Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane.; FUNCTION: (Microbial infection) Has an antiviral effect against flaviviruses by affecting viral RNA transcription and translation through the sequestration of elongation factor 1-alpha (EEF1A1). This results in decreased viral RNA synthesis and decreased viral protein translation. {ECO:0000269|PubMed:19889084}. | Alternative splicing;Antiviral defense;Coiled coil;Cytoplasm;Direct protein sequencing;Exocytosis;Host-virus interaction;Phosphoprotein;Protein transport;Reference proteome;Transport | The protein encoded by this gene is a component of the exocyst complex, a multiple protein complex essential for targeting exocytic vesicles to specific docking sites on the plasma membrane. Though best characterized in yeast, the component proteins and functions of the exocyst complex have been demonstrated to be highly conserved in higher eukaryotes. At least eight components of the exocyst complex, including this protein, are found to interact with the actin cytoskeletal remodeling and vesicle transport machinery. Alternatively spliced transcript variants encoding distinct isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:55763; | cytoplasm [GO:0005737]; cytoplasmic side of apical plasma membrane [GO:0098592]; cytosol [GO:0005829]; exocyst [GO:0000145]; Flemming body [GO:0090543]; membrane [GO:0016020]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; defense response to virus [GO:0051607]; exocytosis [GO:0006887]; Golgi to plasma membrane transport [GO:0006893]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of protein secretion [GO:0050714]; protein transport [GO:0015031]; regulation of macroautophagy [GO:0016241] | 18541705_The exocyst subunits Sec3 and Sec8 interact with the polarity protein IQGAP1 and that this interaction is triggered by active Cdc42 and RhoA, which are essential for matrix degradation. 19889084_This study identified human Sec3 exocyst protein (hSec3p) as a novel interacting partner of West Nile virus and Dengue virus C protein. 19889837_Sec3 associates with a subset of Exocyst complexes that are enriched at desmosomes. 23522008_This study portrayed the non-structural function of C protein that helped the flavivirus to nullify the antiviral activity of hSec3p by accelerating its degradation and facilitating efficient binding of elongation factor 1alpha with flaviviral RNA genome. 25591774_Ezrin/Sec3 complexes are essential for MUC5AC secretion in NE-stimulated airway epithelial cells and that PIP2 is of critical importance in the formation of these complexes. 28694212_rs13117307 is associated with cervical squamous cell carcinoma. rs13117307 SNP upregulated EXCO1 transcript levels in the non-cancerous cervical and in the cervical SCC tissue. 31495494_study provides evidence that Sec3 is involved in TGF-beta induced cell migration and epithelial-mesenchymal transition processes, presumably through the regulation of PI3K/Akt signaling activation in A549 cancer cells | ENSMUSG00000036435 | Exoc1 | 2.604777e+02 | 0.8165587 | -0.292371572 | 0.4209948 | 4.819401e-01 | 0.4875448271 | 0.87511746 | No | Yes | 2.393517e+02 | 55.426307 | 2.816480e+02 | 66.838958 | |
| ENSG00000091127 | 54517 | PUS7 | protein_coding | Q96PZ0 | FUNCTION: Pseudouridylate synthase that catalyzes pseudouridylation of RNAs (PubMed:28073919, PubMed:29628141, PubMed:30778726). Acts as a regulator of protein synthesis in embryonic stem cells by mediating pseudouridylation of RNA fragments derived from tRNAs (tRFs): pseudouridylated tRFs inhibit translation by targeting the translation initiation complex (PubMed:29628141). Also catalyzes pseudouridylation of mRNAs: mediates pseudouridylation of mRNAs with the consensus sequence 5'-UGUAG-3' (PubMed:28073919). In addition to mRNAs and tRNAs, binds other types of RNAs, such as snRNAs, Y RNAs and vault RNAs, suggesting that it can catalyze pseudouridylation of many RNA types (PubMed:29628141). {ECO:0000269|PubMed:28073919, ECO:0000269|PubMed:29628141, ECO:0000269|PubMed:30778726}. | 3D-structure;Acetylation;Alternative splicing;Disease variant;Dwarfism;Isomerase;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;tRNA processing | hsa:54517; | nucleus [GO:0005634]; enzyme binding [GO:0019899]; pseudouridine synthase activity [GO:0009982]; RNA binding [GO:0003723]; mRNA pseudouridine synthesis [GO:1990481]; negative regulation of translation [GO:0017148]; pseudouridine synthesis [GO:0001522]; regulation of hematopoietic stem cell differentiation [GO:1902036]; regulation of mesoderm development [GO:2000380]; tRNA pseudouridine synthesis [GO:0031119] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 30778726_Our results confirm that PUS7 is a bona fide Mendelian disease gene and expand the list of human diseases caused by impaired pseudouridylation. 31451225_Pseudouridine synthase 7 is a nuclear protein involved in stem cell development and intellectual disabilities, and is a novel interactor of SIRT1. The binding regions are predicted and analyzed based on molecular docking studies. The direct interaction occurs between SIRT1 and PUS7, as evidenced by pull-down studies and surface plasmon resonance (SPR) assay. 33811565_Pseudouridylate Synthase 7 Promotes Cell Proliferation and Invasion in Colon Cancer Through Activating PI3K/AKT/mTOR Signaling Pathway. 33990203_HSP90-dependent PUS7 overexpression facilitates the metastasis of colorectal cancer cells by regulating LASP1 abundance. 34718722_The human pseudouridine synthase PUS7 recognizes RNA with an extended multi-domain binding surface. 35051350_Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. 35144859_PUS7 deficiency in human patients causes profound neurodevelopmental phenotype by dysregulating protein translation. | ENSMUSG00000057541 | Pus7 | 6.561706e+02 | 1.1390375 | 0.187815261 | 0.3274501 | 3.233185e-01 | 0.5696201588 | 0.89885610 | No | Yes | 7.214219e+02 | 113.257622 | 6.152614e+02 | 99.153899 | ||
| ENSG00000091136 | 3912 | LAMB1 | protein_coding | P07942 | FUNCTION: Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components. Involved in the organization of the laminar architecture of cerebral cortex. It is probably required for the integrity of the basement membrane/glia limitans that serves as an anchor point for the endfeet of radial glial cells and as a physical barrier to migrating neurons. Radial glial cells play a central role in cerebral cortical development, where they act both as the proliferative unit of the cerebral cortex and a scaffold for neurons migrating toward the pial surface. {ECO:0000269|PubMed:23472759}. | 3D-structure;Basement membrane;Cell adhesion;Coiled coil;Disulfide bond;Extracellular matrix;Glycoprotein;Laminin EGF-like domain;Lissencephaly;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal | Laminins, a family of extracellular matrix glycoproteins, are the major noncollagenous constituent of basement membranes. They have been implicated in a wide variety of biological processes including cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis. Laminins are composed of 3 non identical chains: laminin alpha, beta and gamma (formerly A, B1, and B2, respectively) and they form a cruciform structure consisting of 3 short arms, each formed by a different chain, and a long arm composed of all 3 chains. Each laminin chain is a multidomain protein encoded by a distinct gene. Several isoforms of each chain have been described. Different alpha, beta and gamma chain isomers combine to give rise to different heterotrimeric laminin isoforms which are designated by Arabic numerals in the order of their discovery, i.e. alpha1beta1gamma1 heterotrimer is laminin 1. The biological functions of the different chains and trimer molecules are largely unknown, but some of the chains have been shown to differ with respect to their tissue distribution, presumably reflecting diverse functions in vivo. This gene encodes the beta chain isoform laminin, beta 1. The beta 1 chain has 7 structurally distinct domains which it shares with other beta chain isomers. The C-terminal helical region containing domains I and II are separated by domain alpha, domains III and V contain several EGF-like repeats, and domains IV and VI have a globular conformation. Laminin, beta 1 is expressed in most tissues that produce basement membranes, and is one of the 3 chains constituting laminin 1, the first laminin isolated from Engelbreth-Holm-Swarm (EHS) tumor. A sequence in the beta 1 chain that is involved in cell attachment, chemotaxis, and binding to the laminin receptor was identified and shown to have the capacity to inhibit metastasis. [provided by RefSeq, Aug 2011]. | hsa:3912; | basement membrane [GO:0005604]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; laminin complex [GO:0043256]; laminin-1 complex [GO:0005606]; laminin-10 complex [GO:0043259]; laminin-2 complex [GO:0005607]; laminin-8 complex [GO:0043257]; perinuclear region of cytoplasm [GO:0048471]; extracellular matrix structural constituent [GO:0005201]; integrin binding [GO:0005178]; structural molecule activity [GO:0005198]; animal organ morphogenesis [GO:0009887]; basement membrane assembly [GO:0070831]; cell adhesion [GO:0007155]; cell migration [GO:0016477]; endodermal cell differentiation [GO:0035987]; neuron projection development [GO:0031175]; neuronal-glial interaction involved in cerebral cortex radial glia guided migration [GO:0021812]; odontogenesis [GO:0042476]; positive regulation of cell migration [GO:0030335]; positive regulation of epithelial cell proliferation [GO:0050679]; substrate adhesion-dependent cell spreading [GO:0034446]; tissue development [GO:0009888] | 11891225_Laminin-10/11 and fibronectin differentially prevent apoptosis induced by serum removal via phosphatidylinositol 3-kinase/Akt- and MEK1/ERK-dependent pathways (Laminin 10; separate entry for Laminin 11). 12615822_mRNA encoding laminin-alpha1, -beta1, and -gamma1 chains was expressed in 90% of endometriotic lesions. 12743034_Data suggest that laminin-10 (alpha5beta1gamma1) is required for hair follicle development and report the first use of exogenous protein to correct a cutaneous developmental defect. 15706419_alteration of LAMB1 expression is associated with the genesis and development of uterine leiomyoma 15967115_prostate cancer cells expressing high levels of MT1-MMP have increased invasive potential through their ability to degrade and invade Ln-10 barriers 15987446_laminin beta1 chain (a constituent of laminin-8) was typically found in vessel walls of carcinomas and their metastases but not in those of normal breast 16146715_laminin isoform changes are associated with brain tumor invasion and angiogenesis [review] 18162078_Real-time PCR showed that ETOH significantly altered the expression of genes involved in cell adhesion. There was an increase in the expression of alpha and beta Laminins 1, beta Integrins 3 and 5, Secreted phosphoprotein1 and Sarcoglycan epsilon. 18450753_laminin supports platelet adhesion depending on the interaction of VWF and GPIb-IX-V under pathophysiological high shear flow 18474427_Meningiomas increase in size through increased production of extracellular matrix; furthermore, the proliferation of cells typically associated with neoplasia requires considerable interaction with the extracellular matrix. 18691630_Elevated expression of laminin beta1 mRNA and protein in the hippocampus suggests that laminin beta1 may play a role in the development of epileptic seizures in patients with intractable epilepsy. 19058789_Observational study of gene-disease association. (HuGE Navigator) 19147489_beta2 chain-containing laminins (beta2-laminins) bound more avidly to alpha3beta1 and alpha7X2beta1 integrins than beta1 chain-containing laminins (beta1-laminins). 19416897_laminin-111 (alpha(1), beta(1), gamma(1)), which is expressed during embryonic development but absent in normal or dystrophic skeletal muscle, increased alpha(7)-integrin expression in mouse and DMD patient myoblasts 19526105_The presence of nuclear beta-catenin and high content of mmp-9 in the tumor were associated with abnormal accumulation of laminin in the cytoplasm. These changes were characteristic of colorectal cancer with high invasive metastatic potential. 19834535_Observational study of gene-disease association. (HuGE Navigator) 19915572_LAMB1 new susceptibility loci for ulcerative colitis 19915572_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21276136_results suggest that Lm3B11/3B21 may be required for normal mature vessels and interfere with tumor angiogenesis 21370991_Laminin beta1 and integrin alpha2 expression is elevated in the anterior temporal neocortex tissue from patients with intractable epilepsy. 21744425_We did not find an association of rs6949033 (LAMB1) with ulcerative colitis. 23472759_Homozygous deleterious mutations in LAMB1. 23679161_The phenotype due to LAMB2 mutations appears to be similar between different ethnic groups. 24497408_Upregulation of LAMB1 expression was highly correlated with the downregulation of miR-124-5p. LAMB1 protein expression was suppressed by miR-124-5p. 24804215_Extracellular matrix proteins expression profiling in chemoresistant variants of the A2780 ovarian cancer cell line. 24951930_Laminins 411 and 421 differentially promote tumor cell migration via alpha6beta1 integrin and MCAM (CD146). 25664710_LAMB1 single nucleotide polymorphism rs886774 was associated with early onset Ulcerative colitis and, thus, suggests a fundamentally different mechanism of early disease pathogenesis in Ulcerative colitis versus Crohn's disease. 25774865_An association was found between LAMB1 rs2158836 polymorphisms and symptom severity in Korean autism spectrum disorder patients. 26057585_These results suggest that MUC5B production can be regulated by ECM components and that MUC5B is upregulated by fibronectin and laminin via the integrin, ERK, and NF-kappaB dependent pathway. 26359947_LAMB1 is a potential serological biomarker that could be used in conjunction with carcinoembryonic antigen for diagnosis of colorectal cancer. 28214208_Our results suggest that an impairment in adhesion exists in vitiligo skin, which is supported by the diminished immunohistochemical expression of laminin, beta1 integrin, ICAM-1 and VCAM-1. 28444932_LAMB1 may affect the initiation and progression of pneumoconiosis, or serve as a potential biomarker of pneumoconiosis for diagnosis and genetic susceptibility. 28783171_In vitro activation of PDGFR-alpha leads to translational activation of LAMB1, which in turn induces an invasive and metastatic phenotype of hepatocellular carcinoma cells exhibiting K19 expression. 29108990_Treatment of Neuroscreen-1 (NS-1) cells with laminin-1 or YIGSR peptide, which corresponds to a sequence in laminin-1 beta1 chain that binds to 67LR, induced a decrease in the cell-surface expression of 67LR and caused its internalization. 29479990_High cytoplasmic laminin expression is associated with Oral Squamous Cell Carcinomas. 29599141_Age-related modulation of laminin beta1 versus beta2 chain expression changes the functional properties and phenotype of endothelial cells. The dysregulation of the extracellular matrix during vascular aging may contribute to age-associated impairment of organ function and fibrosis. 30920297_LAMB1 identified as Liver Fibrosis-specific gene, was significantly associated with Liver Fibrosis progression and could be combined to discriminate Liver Fibrosis stages regardless of any etiology. 31180068_Phosphorylation mapping of LAMB1 has been reported. 32085798_CRISPR-Cas9-mediated labelling of the C-terminus of human laminin beta1 leads to secretion inhibition. 32356431_Alterations in the immunoreactivity of laminin, type IV collagen and alpha3beta1 integrin in diabetic rat ovarian follicles. 33435161_Upregulation of LAMB1 via ERK/c-Jun Axis Promotes Gastric Cancer Growth and Motility. 33784890_LAMB1 Is Related to the T Stage and Indicates Poor Prognosis in Gastric Cancer. 34606115_End-Truncated LAMB1 Causes a Hippocampal Memory Defect and a Leukoencephalopathy. 35436411_Lamin B1 is a potential therapeutic target and prognostic biomarker for hepatocellular carcinoma. | ENSMUSG00000002900 | Lamb1 | 1.473199e+03 | 1.0743614 | 0.103479396 | 0.2889581 | 1.284361e-01 | 0.7200590649 | 0.94084174 | No | Yes | 1.556397e+03 | 267.533805 | 1.237874e+03 | 218.549356 | |
| ENSG00000092020 | 55012 | PPP2R3C | protein_coding | Q969Q6 | FUNCTION: May regulate MCM3AP phosphorylation through phosphatase recruitment (By similarity). May act as a negative regulator of ABCB1 expression and function through the dephosphorylation of ABCB1 by TFPI2/PPP2R3C complex (PubMed:24333728). May play a role in the activation-induced cell death of B-cells (By similarity). {ECO:0000250|UniProtKB:Q9JK24, ECO:0000269|PubMed:24333728}. | Alternative splicing;Calcium;Cone-rod dystrophy;Cytoplasm;Deafness;Disease variant;Metal-binding;Nucleus;Reference proteome;Repeat | hsa:55012; | centrosome [GO:0005813]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; metal ion binding [GO:0046872]; activation of protein kinase activity [GO:0032147]; B cell homeostasis [GO:0001782]; cortical cytoskeleton organization [GO:0030865]; microtubule cytoskeleton organization [GO:0000226]; positive regulation of B cell differentiation [GO:0045579]; regulation of antimicrobial humoral response [GO:0002759]; regulation of dephosphorylation [GO:0035303]; regulation of mitochondrial depolarization [GO:0051900]; spleen development [GO:0048536]; T cell homeostasis [GO:0043029] | Mouse_homologues 16129705_G5PR is required for the B cell receptor-mediated proliferation associated with the prevention of activation-induced cell death in mature B cells. 18022237_G5PR regulates the thymocyte unique apoptotic signal involved in JNK-mediated Caspase-3 activation but not in Bim activation. 25601926_findings suggest that G5PR is involved in both the survival of B cells and the differentiation of peritoneal B1a cells into autoantibody-producing plasma cells. | ENSMUSG00000021022 | Ppp2r3c | 1.905691e+02 | 0.9530049 | -0.069444503 | 0.3970353 | 3.028204e-02 | 0.8618518255 | 0.97337236 | No | Yes | 1.677424e+02 | 36.118782 | 1.801391e+02 | 39.650600 | ||
| ENSG00000092199 | 3183 | HNRNPC | protein_coding | P07910 | FUNCTION: Binds pre-mRNA and nucleates the assembly of 40S hnRNP particles (PubMed:8264621). Interacts with poly-U tracts in the 3'-UTR or 5'-UTR of mRNA and modulates the stability and the level of translation of bound mRNA molecules (PubMed:12509468, PubMed:16010978, PubMed:7567451, PubMed:8264621). Single HNRNPC tetramers bind 230-240 nucleotides. Trimers of HNRNPC tetramers bind 700 nucleotides (PubMed:8264621). May play a role in the early steps of spliceosome assembly and pre-mRNA splicing. N6-methyladenosine (m6A) has been shown to alter the local structure in mRNAs and long non-coding RNAs (lncRNAs) via a mechanism named 'm(6)A-switch', facilitating binding of HNRNPC, leading to regulation of mRNA splicing (PubMed:25719671). {ECO:0000269|PubMed:12509468, ECO:0000269|PubMed:16010978, ECO:0000269|PubMed:25719671, ECO:0000269|PubMed:7567451, ECO:0000269|PubMed:8264621}. | 3D-structure;Acetylation;Alternative splicing;Coiled coil;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribonucleoprotein;Spliceosome;Ubl conjugation;mRNA processing;mRNA splicing | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene can act as a tetramer and is involved in the assembly of 40S hnRNP particles. Multiple transcript variants encoding at least two different isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:3183; | actin cytoskeleton [GO:0015629]; catalytic step 2 spliceosome [GO:0071013]; chromatin [GO:0000785]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; spliceosomal complex [GO:0005681]; telomerase holoenzyme complex [GO:0005697]; identical protein binding [GO:0042802]; mRNA 3'-UTR binding [GO:0003730]; N6-methyladenosine-containing RNA binding [GO:1990247]; nucleosomal DNA binding [GO:0031492]; poly(U) RNA binding [GO:0008266]; RNA binding [GO:0003723]; telomerase RNA binding [GO:0070034]; 3'-UTR-mediated mRNA stabilization [GO:0070935]; chromatin remodeling [GO:0006338]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of telomere maintenance via telomerase [GO:0032211]; osteoblast differentiation [GO:0001649]; RNA splicing [GO:0008380] | 11877401_phosphorylation of heterogeneous nuclear ribonucleoprotein C1/C2 in response to physiologic levels of hydrogen peroxide in endothelial cells 12482981_nuclear ribonucleoproteins C1 and C2 are part of the RNP complex that forms on XIAP IRES, the cellular levels of hnRNPC1 and -C2 parallel the activity of XIAP IRES and the overexpression of hnRNPC1-C2 specifically enhanced translation of XIAP IRES 12509468_hnRNP C, via internal ribosomal entry site binding, modulates translation of c-myc mRNA in a cell cycle phase-dependent manner 12564933_the acidic C-terminal domain of hnRNP-C1/C2 could be a regulatory domain and may play an important role in the regulation of mRNA binding by hnRNP-C1/C2. 15494373_a novel nuclear export is activated by the ROCK signaling pathway to exclude hnRNP C1/C2 from nucleus, by which the compartmentalization of specific hnRNP components is disturbed in apoptotic cells 15687492_CK1alpha-mediated phosphorylation modulates the mRNA binding ability of hnRNP-C 15731220_findings suggest that hnRNP C1 has a role in positive-strand RNA synthesis in poliovirus-infected cells, possibly at the level of initiation 15767428_upstream element in human papillomavirus type 16 interacted specifically with CstF-64, hnRNP C1/C2 & polypyrimidine tract binding protein, suggesting these factors were enhancing or regulating polyadenylation at the HPV-16 early polyadenylation signal 16010978_Direct in vivo interaction of hnRNPC with the urokinase receptor mRNA 3'UTR was demonstrated. 16217013_Results describe the purification of a locus control region-associated remodeling complex consisting of heterogeneous nuclear ribonucleoprotein C1/C2, SWI/SNF, and MeCP1 as a single homogeneous complex. 16960656_These results demonstrate that hnRNP C1/C2 is involved in maintenance of cellular homeostasis besides cellular differentiation and proliferation. 17071612_in addition to its RNA-processing functions, hnRNP C1/C2 may be a key determinant of the temporal patterns of VDRE occupancy 17159903_regulation of IRES-mediated translation by hnRNP C1/C2 and Unr might be important in mitosis 18296503_A novel cis-element in the 5' coding region of p53 mRNA and its interaction with heterogeneous nuclear ribonucleoprotein (hnRNP)C1/C2, is described. 18471994_Intracellular NS1 protein of dengue virus interacts with hnRNP C1/C2. 18494499_The RNA binding domain of heterogeneous nuclear ribonuclear protein C binds and stabilizes urokinase-type plasminogen activator receptor (uPAR) mRNA, attributing the key role of this domain in hnRNPC-mediated uPAR regulation in lung epithelial cells. 18508286_The H(2)O(2) responsive pre-mRNA binding protein hnRNP-C is up-regulated in atherosclerosis. 19405953_This protein has been found differentially expressed in the Wernicke's Area from patients with schizophrenia. 19628962_anti-hnRNP C1/C2 antibody hampered splicing of SMN1 exon 7, but did not affect splicing of SMN2 exon 7 19740742_endogenous hnRNP C and PTEN interact and co-localize within the nucleus 20164237_The authors report here that the 5' end of poliovirus negative-strand RNA is capable of interacting with endogenous hnRNP C, as well as with poliovirus nonstructural proteins. 20189623_The authors propose that hnRNP C interacts with poliovirus RNA and replication proteins to increase the efficiency of viral genomic RNA synthesis. 20473314_FMRP represses translation by recruiting APP mRNA to processing bodies, whereas hnRNP C promotes APP translation by displacing FMRP, thereby relieving the translational block. 20601959_HNRNP C recognizes uridine tracts with a defined long-range spacing consistent with HNRNP particle organization. 20873769_Studies indicated that DDX21, HNRNPC, and RCC2 were isolated from Ku86 multicomponent complex in response to DNA damage. 21774814_One of the identified proteins, hnRNPC, was found to interact with small hepatitis delta virus antigen in vitro and in vivo in human liver cells. 22404445_p27kip1 upregulated by hnRNPC1/2 antagonizes CagA-mediated pathogenesis. 22461616_study shows mechanism by which Pol II transcripts are classified according to length; heterotetramer of hnRNP C1/C2 measures the length of the transcripts like a molecular ruler, by binding to unstructured RNA regions longer than 200 to 300 nucleotides 22907752_The data indicated that hnRNPC controls the aggressiveness of glioblastoma cells through the regulation of PDCD4. Silencing of hnRNPC lowered miR-21 levels, in turn increasing the expression of PDCD4, suppressing Akt and p70S6K activation. 23374342_By preventing U2AF65 binding to Alu elements, hnRNP C plays a critical role as a genome-wide sentinel protecting the transcriptome. 23585894_Heterogeneous nuclear ribonucleoprotein C is a key regulator of BRCA gene expression and homologous recombination-based DNA repair. 23973260_down-regulation of MALAT-1 expression compromised the cytoplasmic translocation of hnRNP C in the G2/M phase and resulted in G2/M arrest 24743263_PTBP1 and hnRNP C repress exon 3 inclusion, and that downregulation of PTBP1 inhibited BIM-mediated apoptosis. 25354590_HnRNP C, YB-1 and hnRNP L coordinately enhance skipping of human MUSK exon 10 to generate a Wnt-insensitive MuSK isoform. 25719671_m(6)A-switch-regulated HNRNPC-binding activities affect the abundance as well as alternative splicing of target mRNAs, demonstrating the regulatory role of m(6)A-switches on gene expression and RNA maturation 25878250_Our results suggested that hnRNP C1 controls HPV16 late gene expression. 25890165_These findings suggest that hnRNP C1/C2 is involved in dengue virus replication at the stage of viral RNA synthesis. 26048669_APP levels then decrease progressively as a function of age in close relationship with the gradual normalization of FMRP and hnRNP C levels. 27672039_These data indicate that hnRNPC1/C2 binds to both DNA and RNA and influences both gene expression and RNA splicing, but these actions do not appear to be linked through 1,25(OH)2D-mediated induction of transcription. 28031331_These data suggest an interplay between CELF2 and hnRNP C as the mechanistic basis for activation-dependent alternative splicing of TRAF3 exon 8. 28102850_Endoplasmic reticulum resident chaperone GRP78, mitochondrial protein Prohibitin and heterogeneous nuclear ribonucleoprotein hnRNPC (C1/C2) have been shown to interact with viral RNA. Hence it is proposed that these are the principle candidates governing endoplasmic reticulum stress-induced apoptosis in JEV infection. 28264987_Our results indicate that a balance between lncRNA SNHG1 and hnRNPC regulates p53 activity and p53-dependent apoptosis upon doxorubicin treatment, and further indicate that a change in lncRNA subcellular localization under specific circumstances is biologically significant. 29899543_Data show that miR-744-5p expression directly downregulated mRNA and protein expression of nuclear factor I X (NFIX) and heterogeneous nuclear ribonucleoprotein C (HNRNPC). 30158112_essentialness of HNRNPC in the breast cancer cells was attributed to its function in controlling the endogenous dsRNA and the down-stream interferon response. This is a novel extension from the previous understandings about HNRNPC in binding with introns and regulating RNA splicing. 30824187_LncRNA LBX2-AS1 promoted cell migration and epithelial-mesenchymal transition process in esophageal squamous cell carcinoma. LBX2-AS1 interacting with HNRNPC to promote ZEB1/2 mRNA stability. 31147722_Overexpression of heterogeneous nuclear ribonucleoprotein C (hnRNPC) has a critical role in the establishment of alternative cleavage and polyadenylation (APA) profiles characteristic for metastatic colon cancer cells by regulating poly(A) site. 31234696_hnRNP C1/C2 for positive and negative-strand RNAs guides the final +/- RNA ratio, providing first insight in the regulation of the positive to negative-strand RNA ratio in enteroviruses. 32526707_M6A-related bioinformatics analysis reveals that HNRNPC facilitates progression of OSCC via EMT. 33177247_Prognostic risk signature based on the expression of three m6A RNA methylation regulatory genes in kidney renal papillary cell carcinoma. 33474615_Identification of genetic variants in m(6)A modification genes associated with pancreatic cancer risk in the Chinese population. 34244793_RBFOX2 alters splicing outcome in distinct binding modes with multiple protein partners. 34271104_HNRNPC impedes m(6)A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. 34297039_ADAR and hnRNPC deficiency synergize in activating endogenous dsRNA-induced type I IFN responses. 34965173_Identification of HnRNPC as a novel Tau exon 10 splicing factor using RNA antisense purification mass spectrometry. 35277915_HNRNPC regulates RhoA to induce DNA damage repair and cancer-associated fibroblast activation causing radiation resistance in pancreatic cancer. 35344508_A systematic pan-cancer study demonstrates the oncogenic function of heterogeneous nuclear ribonucleoprotein C. | ENSMUSG00000060373 | Hnrnpc | 2.742523e+04 | 0.4952333 | -1.013819629 | 0.3477553 | 8.563946e+00 | 0.0034288537 | 0.21643274 | No | Yes | 1.559870e+04 | 3088.109419 | 3.371955e+04 | 6845.571417 | |
| ENSG00000092203 | 9878 | TOX4 | protein_coding | O94842 | FUNCTION: Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell cycle progression during the transition from mitosis into interphase. {ECO:0000269|PubMed:20516061}. | Alternative splicing;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome | hsa:9878; | chromatin [GO:0000785]; nucleus [GO:0005634]; PTW/PP1 phosphatase complex [GO:0072357]; chromatin DNA binding [GO:0031490]; regulation of transcription by RNA polymerase II [GO:0006357] | 19293638_the coordinated spatial and temporal regulation of LCP1 and PNUTS may be a novel mechanism to control the expression of genes that are critical for certain physiological and pathological processes. 20516061_mammalian Wdr82 functions in a variety of cellular processes; PTW/PP1 phosphatase complex (PNUTS, Tox4, Wdr82, PP1) has a role in the regulation of chromatin structure during the transition from mitosis into interphase 21184731_interaction between TOX4 and platinated DNA 22496870_Compared with TOX4, expression of TOX1, TOX2 and TOX3 in normal lung was 25, 44, and 88%lower, respectively, supporting the premise that reduced promoter activity confers increased susceptibility to methylation during lung carcinogenesis. 23127401_The formation of vasculogenic mimicry is inhibited by reducing the expression of Mig-7 in gastric cancer cells. 24312278_a regulation of LEDGF interaction with chromatin by cellular partners of its PWWP domain could be involved in several processes linked to LEDGF tethering properties, such as lentiviral integration, DNA repair or transcriptional regulation 34914893_TOX4, an insulin receptor-independent regulator of hepatic glucose production, is activated in diabetic liver. 35365735_TOX4 facilitates promoter-proximal pausing and C-terminal domain dephosphorylation of RNA polymerase II in human cells. | ENSMUSG00000016831 | Tox4 | 3.330846e+03 | 1.0821734 | 0.113931692 | 0.3035343 | 1.421801e-01 | 0.7061232500 | 0.93718130 | No | Yes | 3.487913e+03 | 353.131052 | 2.952155e+03 | 306.631697 | ||
| ENSG00000092208 | 8487 | GEMIN2 | protein_coding | O14893 | FUNCTION: The SMN complex catalyzes the assembly of small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome, and thereby plays an important role in the splicing of cellular pre-mRNAs (PubMed:18984161, PubMed:9323129). Most spliceosomal snRNPs contain a common set of Sm proteins SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP (Sm core) (PubMed:18984161). In the cytosol, the Sm proteins SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG (5Sm) are trapped in an inactive 6S pICln-Sm complex by the chaperone CLNS1A that controls the assembly of the core snRNP (PubMed:18984161). To assemble core snRNPs, the SMN complex accepts the trapped 5Sm proteins from CLNS1A (PubMed:18984161, PubMed:9323129). Binding of snRNA inside 5Sm ultimately triggers eviction of the SMN complex, thereby allowing binding of SNRPD3 and SNRPB to complete assembly of the core snRNP (PubMed:31799625). Within the SMN complex, GEMIN2 constrains the conformation of 5Sm, thereby promoting 5Sm binding to snRNA containing the snRNP code (a nonameric Sm site and a 3'-adjacent stem-loop), thus preventing progression of assembly until a cognate substrate is bound (PubMed:31799625, PubMed:21816274, PubMed:16314521). {ECO:0000269|PubMed:16314521, ECO:0000269|PubMed:18984161, ECO:0000269|PubMed:21816274, ECO:0000269|PubMed:31799625, ECO:0000269|PubMed:9323129}. | 3D-structure;Alternative initiation;Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome;mRNA processing;mRNA splicing | This gene encodes one of the proteins found in the SMN complex, which consists of several gemin proteins and the protein known as the survival of motor neuron protein. The SMN complex is localized to a subnuclear compartment called gems (gemini of coiled bodies) and is required for assembly of spliceosomal snRNPs and for pre-mRNA splicing. This protein interacts directly with the survival of motor neuron protein and it is required for formation of the SMN complex. A knockout mouse targeting the mouse homolog of this gene exhibited disrupted snRNP assembly and motor neuron degeneration. [provided by RefSeq, Aug 2011]. | hsa:8487; | cytosol [GO:0005829]; Gemini of coiled bodies [GO:0097504]; nuclear body [GO:0016604]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; SMN complex [GO:0032797]; SMN-Sm protein complex [GO:0034719]; spliceosomal complex [GO:0005681]; mRNA processing [GO:0006397]; negative regulation of RNA binding [GO:1905215]; RNA splicing [GO:0008380]; RNA splicing, via transesterification reactions [GO:0000375]; spliceosomal complex assembly [GO:0000245]; spliceosomal snRNP assembly [GO:0000387] | 11943600_Spinal muscular atrophy and amyotropic lateral sclerosis patients have decreased SIP 1-alpha and increased SIP 1-beta expression levels compared to normal tissues. 15964810_SIP1 determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. 16731905_binds to HIV-1 integrase and facilitates viral cDNA synthesis and subsequent steps that precede integration in vivo 17308308_Gemin2 plays an important role in small nuclear ribonucleoprotein assembly through the stabilization of the survival of motor neuron oligomer/complex via novel self-interaction 18247312_Observational study of gene-disease association. (HuGE Navigator) 18247312_relationship between the feature of gene mutations and single nucleotide polymorphisms of SIP1 gene and HSCR 19915660_SIP1 appears to stabilize functional multimer forms of IN, thereby promoting the assembly of IN and RT on viral RNA to allow efficient reverse transcription, which is a prerequisite for efficient HIV-1 infection 21732698_purified SMN-GEMIN2 fusion protein enhanced the RAD51-mediated homologous pairing much more efficiently than GEMIN2 alone 21816274_Study identified Gemin2 as the protein that binds a pentamer of Sm proteins comprised of SmD1/D2 and SmF/E/G; the crystal structure of this complex bound to SMN's Gemin2 binding domain to 2.5 A was determined. 21913013_Overexpression of SIP1 and downregulation of E-cadherin is associated with delayed neck metastasis in stage I/II oral tongue squamous cell carcinoma after partial glossectomy 22607171_several conserved SMN residues, including the sites of two SMA patient mutations, are not required for binding to Gemin2. Instead, they form a conserved SMN/Gemin2 surface that may be functionally important for snRNP assembly. 23939045_Two monoclonal antibodies against SMN (survival-of-motor-neurons) protein bind to its site of interaction with gemin2. 26092730_investigate the oligomeric nature of the SMN.Gemin2 complexes from humans and fission yeast (hSMN.Gemin2 and ySMN.Gemin2) 26828962_The authors propose that Gemin2 is a versatile hub for ribonucleoprotein exchange that functions broadly in RNA metabolism. 27974620_Specific mutations in the yeast Sm protein ring expose a requirement for assembly factor Brr1, a homolog of human Gemin2. 31799625_Here we show, using crystallographic and biochemical approaches, that Gemin2 of the SMN complex enhances RNA specificity of SmD1/D2/F/E/G via a negative cooperativity between Gemin2 and RNA in binding SmD1/D2/F/E/G. | ENSMUSG00000060121 | Gemin2 | 2.233612e+02 | 0.8150640 | -0.295014789 | 0.3828884 | 5.808130e-01 | 0.4459938164 | 0.85808635 | No | Yes | 2.050929e+02 | 41.945331 | 2.338068e+02 | 48.863506 | |
| ENSG00000092531 | 8773 | SNAP23 | protein_coding | O00161 | FUNCTION: Essential component of the high affinity receptor for the general membrane fusion machinery and an important regulator of transport vesicle docking and fusion. | 3D-structure;Acetylation;Alternative splicing;Cell junction;Cell membrane;Coiled coil;Direct protein sequencing;Lipoprotein;Membrane;Palmitate;Phosphoprotein;Protein transport;Reference proteome;Repeat;Synapse;Synaptosome;Transport | Specificity of vesicular transport is regulated, in part, by the interaction of a vesicle-associated membrane protein termed synaptobrevin/VAMP with a target compartment membrane protein termed syntaxin. These proteins, together with SNAP25 (synaptosome-associated protein of 25 kDa), form a complex which serves as a binding site for the general membrane fusion machinery. Synaptobrevin/VAMP and syntaxin are believed to be involved in vesicular transport in most, if not all cells, while SNAP25 is present almost exclusively in the brain, suggesting that a ubiquitously expressed homolog of SNAP25 exists to facilitate transport vesicle/target membrane fusion in other tissues. The protein encoded by this gene is structurally and functionally similar to SNAP25 and binds tightly to multiple syntaxins and synaptobrevins/VAMPs. It is an essential component of the high affinity receptor for the general membrane fusion machinery and is an important regulator of transport vesicle docking and fusion. Two alternative transcript variants encoding different protein isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:8773; | adherens junction [GO:0005912]; azurophil granule [GO:0042582]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; mitochondrion [GO:0005739]; neuron projection [GO:0043005]; nucleoplasm [GO:0005654]; phagocytic vesicle membrane [GO:0030670]; plasma membrane [GO:0005886]; presynapse [GO:0098793]; SNARE complex [GO:0031201]; specific granule [GO:0042581]; specific granule membrane [GO:0035579]; tertiary granule membrane [GO:0070821]; SNAP receptor activity [GO:0005484]; syntaxin binding [GO:0019905]; exocytosis [GO:0006887]; histamine secretion by mast cell [GO:0002553]; membrane fusion [GO:0061025]; post-Golgi vesicle-mediated transport [GO:0006892]; protein transport [GO:0015031]; synaptic vesicle fusion to presynaptic active zone membrane [GO:0031629]; synaptic vesicle priming [GO:0016082]; vesicle fusion [GO:0006906]; vesicle targeting [GO:0006903] | 11842301_SNAP-23 and syntaxin-4 are expressed in human eosinophils and are likely candidates for association with VAMP-2 during docking, which is followed by exocytosis. 12121992_cleaved by calpain in activated platelets 12209004_CFTR-mediated chloride currents are inhibited by introducing excess SNAP-23 into HT29-Cl.19A epithelial cells. 12475239_A synaptosome-associated protein of 23 kDa(SNAP23), the ubiqitously expressed homologue of SNAP25, has been shown to interact directly with ubiquitous kinesin heavy chain (uKHC). 12551899_Palmitoylated peptides from the cysteine-rich domain of this protein cause membrane fusion depending on peptide length, position of cysteines, and extent of palmitoylation. 12556468_an examination of the homotetrameric structure of the N-terminal domain of this protein 12930825_studies show that synaptosomal-associated protein-23 is phosphorylated in platelets during cell activation through a protein kinase C-related mechanism at two or more sites 16272324_Because SNAP-23 plays a central role in SNAREs complex formation, it was chosen to examine possible functional implications of the SNARE system in plasma cell Ig secretion. 17485553_expression of SNAP-23 and syntaxin-2 on the extracellular surface of the platelet plasma membrane. 17825825_SNAP-23 is not essential for constitutive exocytosis of secreted alkaline phosphatase 18457912_The expression of the SNARE protein SNAP-25 and its cellular homologue SNAP-23, as well as syntaxin1 and VAMP (vesicle-associated membrane protein) in samples of normal parathyroid tissue, chief cell adenoma, and parathyroid carcinoma, was examined. 18665925_Overexpression of a dominant negative SNAP-23 mutant suppressed expression of P-selectin, CD40L, CD41, CD61, release from dense granules and platelet aggregation 19086053_Observational study of gene-disease association. (HuGE Navigator) 19910495_the importance of SNAP23 in the trafficking of matrix metalloproteinases during degradation of extracellular matrix substrates and subsequent cellular invasion 20416077_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20519516_SNAP23 and SNAP25 palmitoylation is regulated by DHHC palmitoyl transferases 21642540_Introduction of the SNARE domain of SNAP-23 into neutrophils as an HIV transactivator of transcription (TAT) fusion protein significantly inhibits exocytosis of neutrophil granule subsets without altering signal transduction pathway activation. 21768361_Data show that knockdown of SNAP-23 inhibited the production of virus. 21981832_STX-3 and SNAP-23 are crucial for the release of all chemokines in mature human mast cells 23087210_Results suggest that phagosomal SNAP-23 is one of the key players regulating the phagosomal environment in macrophages. 24373201_Data suggest SNAP23 and VAMP3 (vesicle-associated membrane protein 3) participate in interleukin-1beta-, interleukin-1 receptor-, calcium signaling-dependent secretion/exocytosis of interleukin-6 and tumor necrosis factor alpha from synoviocytes. 24496451_The association of Src, EGFR and beta1 integrin is dependent upon membrane traffic that is mediated by syntaxin13 (officially known as STX12) and SNAP23. 24807903_Increased level of SNAP23-Syntaxin4-VAMP7 interaction correlates with decreased Syntaxin4 phosphorylation and trafficking of MT1-MMP to invadopodia during cellular invasion. 26266817_these data suggest that SNAP23 is a key component of the endothelial SNARE machinery that mediates endothelial exocytosis. 26733245_Localization of SNAP23 was found in plasma membrane, lipid droplets and mitochondria of skeletal muscle. 27550144_Study showed that SNAP23 was recruited to the close sites of lipid droplet (LD) in HCV-infected cells, implying that SNAP23 was required for HCV-induced LD enlargement and, HCV production. 27795355_A novel regulatory mechanism for SNAP23-dependent mast cell activation of T. vaginalis-secreted LTB4 involving surface trafficking of BLT1. 27855700_Study identified SNAP23 as a novel oncogene in Ovarian Cancer (OC). It is over-expressed in OC and could promote the proliferation, migration and invasion of OC in vitro. 28067230_PKM2 promotes tumor cell exosome release via phosphorylating protein SNAP23. 28716920_Knockdown of VAMP3 and SNAP23 reduces endothelial secretion of miR-126-3p and miR-200a-3p, as well as the proliferation, migration, and suppression of contractile markers in smooth muscle cells caused by vascular endothelial cell-coculture. 29127297_Rab5 is essential for FcepsilonRI-triggered association of the SNARE protein SNAP23 with the secretory granules. 29352103_Data suggest that acylation of SNAP23 (synaptosome associated protein 23) and STX11 (syntaxin-11) regulates exocytosis in platelets; maintaining acylation states of SNAP23 and STX11 is important for platelet function. 29908998_SNAP23 suppressed progression of cervical cancer and induced cell cycle G2/M arrest via upregulating p21(cip1) and downregulating CyclinB1 30943982_Results show that SNAP23 is phosphorylated by HOTAIR promoting the release of exosome from hepatocellular carcinoma cells. 31229437_issue and biofluid enrichment analyses show broad representation of EVs from across the body without bias towards kidney or urine proteins. Among the proteins linked to neurological diseases, SNAP23 and calbindin were the most elevated in Parkinson's disease with 86% prediction success for disease diagnosis in the discovery cohort and 76% prediction success in the replication cohort. 31757226_Reduction in SNAP-23 Alters Microfilament Organization in Myofibrobastic Hepatic Stellate Cells. 32051343_SNAP23 depletion enables more SNAP25/calcium channel excitosome formation to increase insulin exocytosis in type 2 diabetes. 32949647_IKKbeta activation promotes amphisome formation and extracellular vesicle secretion in tumor cells. 33213278_Pancreas-specific SNAP23 depletion prevents pancreatitis by attenuating pathological basolateral exocytosis and formation of trypsin-activating autolysosomes. 33446221_Let-7a regulates EV secretion and mitochondrial oxidative phosphorylation by targeting SNAP23 in colorectal cancer. 34088337_LINC00511 drives invasive behavior in hepatocellular carcinoma by regulating exosome secretion and invadopodia formation. | ENSMUSG00000027287 | Snap23 | 4.673816e+02 | 0.9712312 | -0.042113279 | 0.3495636 | 1.399616e-02 | 0.9058256916 | 0.98325866 | No | Yes | 4.913692e+02 | 93.032558 | 4.362048e+02 | 84.757341 | |
| ENSG00000092847 | 26523 | AGO1 | protein_coding | Q9UL18 | FUNCTION: Required for RNA-mediated gene silencing (RNAi). Binds to short RNAs such as microRNAs (miRNAs) or short interfering RNAs (siRNAs), and represses the translation of mRNAs which are complementary to them. Lacks endonuclease activity and does not appear to cleave target mRNAs. Also required for transcriptional gene silencing (TGS) of promoter regions which are complementary to bound short antigene RNAs (agRNAs). {ECO:0000269|PubMed:16289642, ECO:0000269|PubMed:16936728, ECO:0000269|PubMed:18771919}. | 3D-structure;Cytoplasm;Disease variant;RNA-binding;RNA-mediated gene silencing;Reference proteome;Repressor;Ribonucleoprotein;Transcription;Transcription regulation;Translation regulation;Ubl conjugation | This gene encodes a member of the argonaute family of proteins, which associate with small RNAs and have important roles in RNA interference (RNAi) and RNA silencing. This protein binds to microRNAs (miRNAs) or small interfering RNAs (siRNAs) and represses translation of mRNAs that are complementary to them. It is also involved in transcriptional gene silencing (TGS) of promoter regions that are complementary to bound short antigene RNAs (agRNAs), as well as in the degradation of miRNA-bound mRNA targets. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. A recent study showed this gene to be an authentic stop codon readthrough target, and that its mRNA could give rise to an additional C-terminally extended isoform by use of an alternative in-frame translation termination codon. [provided by RefSeq, Nov 2015]. | hsa:26523; | cytoplasm [GO:0005737]; cytoplasmic ribonucleoprotein granule [GO:0036464]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; P-body [GO:0000932]; polysome [GO:0005844]; ribonucleoprotein complex [GO:1990904]; RISC complex [GO:0016442]; RISC-loading complex [GO:0070578]; core promoter sequence-specific DNA binding [GO:0001046]; double-stranded RNA binding [GO:0003725]; miRNA binding [GO:0035198]; RNA binding [GO:0003723]; RNA polymerase II complex binding [GO:0000993]; single-stranded RNA binding [GO:0003727]; miRNA loading onto RISC involved in gene silencing by miRNA [GO:0035280]; miRNA mediated inhibition of translation [GO:0035278]; negative regulation of angiogenesis [GO:0016525]; nuclear-transcribed mRNA catabolic process [GO:0000956]; positive regulation of transcription by RNA polymerase II [GO:0045944]; pre-miRNA processing [GO:0031054]; production of miRNAs involved in gene silencing by miRNA [GO:0035196]; RNA secondary structure unwinding [GO:0010501] | 10512872_describes cloning rat sequence and used RNA interference to show that the GERp95 orthologue in C. elegans is important for maturation of germ-line stem cells in the gonad. 15152257_2.6 A crystal structure of the PAZ domain from human Argonaute eIF2c1 bound to both ends of a 9-mer siRNA-like duplex 16936726_These results establish a connection between RNA interference components AGO1 and TRBP2, RNAPII transcription and Polycomb-regulated control of gene expression. 16936728_AGO1 and AGO2 associate with promoter DNA in cells treated with antigene RNAs (agRNAs), and inhibiting expression of AGO1 or AGO2 reverses transcriptional and post-transcriptional silencing. 17482383_a unique amino acid sequence in human DICER protein is essential for binding to Argonaute family proteins 17891150_Results describe a repetitive motif within Tas3, termed the 'Argonaute hook', that is conserved from yeast to humans and binds Ago1 and 2 through their PIWI domains in vitro and in vivo. 17932509_Data show that Argonaute1 and 2 reside in three complexes with distinct Dicer and RNA-induced silencing complex activities, and that the putative RNA-binding protein RBM4 is required for microRNA-guided gene regulation. 18345015_findings show that miRNA function is effected by AGO1-GW182 complexes and the role of GW182 in silencing goes beyond promoting deadenylation 18771919_The specificity of RNA interference depends on the concentration of Ago1, Ago3, and Ago4 relative to Ago2. 19047128_Observational study of gene-disease association. (HuGE Navigator) 19138993_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19393748_EIF2C1 protein expressed during the differentiation of N-type and S-type cells decreased from one of I-type cells to the central period of differentiation. 19723326_Data show that Ago1 and Ago2 (which encode argonautes, the key proteins forming the RNA-induced silencing complex (RISC)) had significantly higher expression levels in ER- than in ER+ breast cancer. 19767416_Study analyzed the association of Argonaute/EIF2C-miRNA complexes with target mRNAs and the degradation of these messages. 19851984_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19946268_The authors demonstrate that AGO1 uses only miRNA duplexes when assembling translational repression-competent complexes, whereas AGO2 can use both miRNA and siRNA duplexes. 20211803_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20721975_Observational study of gene-disease association. (HuGE Navigator) 20721975_These data suggest that Axl acts as a tumor lymphatic metastasis-associated gene, and may function partly through the regulation of Cyr61. 20732906_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21846468_Data show that Ago1, but not Ago2, overexpression in neuroblastoma cells causes slowing of the cell cycle, decreased cellular motility, and a stronger apoptotic response to UV. 22025453_DGCR8, AGO1, AGO2, PACT, and TARBP1 expression levels were significantly higher in the epithelial skin cancer groups than the healthy controls (P > 0.05). 22231398_Mammalian PUM2-Ago-eEF1A inhibited translation of nonadenylated and polyadenylated reporter mRNAs in vitro. 22474261_Ago3 is able to load microRNAs efficiently in the absence of Ago1 and Ago2, despite a significant loss of global microRNA expression 22647351_Drosha, Dicer, Argonaute 1, and Argonaute 2 are differentially expressed at different metastatic sites in ovarian carcinoma compared with primary carcinomas. 22858679_Study observed a dramatic difference in AGO1 and AGO2 associated miRNA profiles in blood plasma. The lack of correlation between AGO1 and AGO2 miRNA content in the plasma can be explained by the fact that many tissues contribute to the extracellular miRNA content. 22961379_AGO1 and AGO2 proteins couple RNA polymerase II elongation to chromatin modification 23019594_Ago2 and Ago1 can slice, and thus functionally bind, preannealed siRNA-mRNA duplexes. 23426184_Tumor xenografts and human cancer specimens indicate that AGO1-mediated translational desuppression of VEGF may be associated with tumor angiogenesis and poor prognosis. 23665583_PIWI-domain mutations in Ago1 may misarrange the catalytic center. Replacing Ago1 cluster 2 by the sequence found in Ago2 fully activated the Ago1 PIWI domain only when the Ago1 PIWI domain was placed into the Ago2 backbone. 23696926_Aberrant expression of argonaute-1/-2 in human renal cell carcinoma is possibly involved with tumorigenesis and prognosis. 23746446_Completion of the tetrad, combined with a mutation on a loop adjacent to the active site of hAgo1, results in slicer activity that is substantially enhanced by swapping in the N domain of hAgo2. 23748378_The Ago1 N domain performed best when juxtaposed with cognate PAZ and MID. This exemplifies the importance of proper intermolecular-domain interactions. 23809764_Evolutionary amino acid changes to hAGO1 were readily reversible, suggesting that loading of guide RNA and pairing of seed-based miRNA and target RNA constrain its sequence drift. 24086155_nuclear Ago1 directly interacts with RNA Polymerase II and is widely associated with chromosomal loci throughout the genome with preferential enrichment in promoters of transcriptionally active genes 25313066_Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing 25656609_EIF2C2, Dicer, and Drosha are more highly expressed in bladder carcinoma, promote the development of bladder cancer, and suggested a poor prognosis 26242502_Blocking AGO1, AGO2, or TRBP expression changes expression levels and nuclear distribution of RNAi factors Dicer, TNRC6A (GW182), and TRBP. 27518285_Low AGO1 expression is associated with melanoma. 27669275_We selected five single nucleotide polymorphisms (SNPs) (rs7813, rs2740349, rs2291778, rs910924, rs595961) in two key microRNA biosynthesis genes (GEMIN4 and AGO1) and systematically evaluated the association between these SNPs, the gene-environment interaction and lung cancer risk. This is the first study showing that rs7813 and rs595961 could be meaningful as genetic markers for lung cancer risk. 28555645_E-cadherin silencing relies on the formation of a complex between the paRNA and microRNA-guided Argonaute 1 that, together, recruit SUV39H1 and induce repressive chromatin modifications in the gene promoter 28781232_In miRNA-mediated gene silencing, the physical interaction between human Argonaute (hAgo) and GW182 (hGW182) is essential for facilitating the downstream silencing of the targeted mRNA. hGW182 can recruit up to three copies of hAgo via its three GW motifs. This may explain the observed cooperativity in miRNA-mediated gene silencing. 29256262_the polymorphisms of the AGO1 and AGO2 genes, the expression levels of which correlated with the proportion of Th17 cells, were associated with the development and prognosis of Graves' disease. 29487329_AGO1 may promote hepatocellular carcinoma metastasis through TGF-beta pathway. 30213762_AGO1 missense mutation was identified in a patient with syndromic form of intellectual disability/autism spectrum disorder. 30770397_Effects of the PIWI/MID domain of Argonaute protein on the association of miRNAi's seed base with the target have been reported. 30858133_Long A-T repeats up-regulated many genes in cancer that can be targeted by AGO1 to change the expression of many genes and limited cancer growth. 31330067_Study reports the regulation of AGO1 at translational level and demonstrates a programmed translational readthrough (PTR) of the AGO1 transcript, which results in an isoform termed as Ago1x. This novel isoform does not cause post-transcriptional gene silencing due to its inability to interact with GW182. Interestingly, this process is positively regulated by let-7a miRNA. 31629687_Nuclear AGO1 Regulates Gene Expression by Affecting Chromatin Architecture in Human Cells. 31666609_Association study of AGO1 and AGO2 genes polymorphisms with recurrent pregnancy loss. 31724726_The association of AGO1 (rs595961G>A, rs636832A>G) and AGO2 (rs11996715C>A, rs2292779C>G, rs4961280C>A) polymorphisms and risk of recurrent implantation failure. 31910330_Epidemiological evidence for associations between variants in microRNA or biosynthesis genes and lung cancer risk. 32143906_Association between genetic variants in genes encoding Argonaute proteins and cancer risk: A meta-analysis. 32393053_Suppression of Endothelial AGO1 Promotes Adipose Tissue Browning and Improves Metabolic Dysfunction. 32400052_Differential expression of microRNA in serum fractions and association of Argonaute 1 microRNAs with heart failure. 32673398_Nuclear role for human Argonaute-1 as an estrogen-dependent transcription coactivator. 32752954_Molecular crowding and conserved interface interactions of human argonaute protein-miRNA-target mRNA complex. 32812257_Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation. 33233493_The Importance of AGO 1 and 4 in Post-Transcriptional Gene Regulatory Function of tRF5-GluCTC, an Respiratory Syncytial Virus-Induced tRNA-Derived RNA Fragment. 33327713_Mixed-lineage leukemia protein modulates the loading of let-7a onto AGO1 by recruiting RAN. 33600493_RanBP2/Nup358 enhances miRNA activity by sumoylating Argonautes. 33832481_POU2F2 promotes the proliferation and motility of lung cancer cells by activating AGO1. 34288373_LncRNA PVT1 promotes the progression of ovarian cancer by activating TGF-beta pathway via miR-148a-3p/AGO1 axis. 35060114_De novo variants in AGO1 recapitulate a heterogeneous neurodevelopmental disorder phenotype. | ENSMUSG00000041530 | Ago1 | 4.003034e+03 | 1.0523763 | 0.073650609 | 0.2785433 | 7.042579e-02 | 0.7907178416 | 0.95850794 | No | Yes | 3.574703e+03 | 320.090296 | 3.543404e+03 | 325.451913 | |
| ENSG00000092871 | 117584 | RFFL | protein_coding | Q8WZ73 | FUNCTION: E3 ubiquitin-protein ligase that regulates several biological processes through the ubiquitin-mediated proteasomal degradation of various target proteins. Mediates 'Lys-48'-linked polyubiquitination of PRR5L and its subsequent proteasomal degradation thereby indirectly regulating cell migration through the mTORC2 complex. Ubiquitinates the caspases CASP8 and CASP10, promoting their proteasomal degradation, to negatively regulate cell death downstream of death domain receptors in the extrinsic pathway of apoptosis. Negatively regulates the tumor necrosis factor-mediated signaling pathway through targeting of RIPK1 to ubiquitin-mediated proteasomal degradation. Negatively regulates p53/TP53 through its direct ubiquitination and targeting to proteasomal degradation. Indirectly, may also negatively regulate p53/TP53 through ubiquitination and degradation of SFN. May also play a role in endocytic recycling. {ECO:0000269|PubMed:15069192, ECO:0000269|PubMed:17121812, ECO:0000269|PubMed:18382127, ECO:0000269|PubMed:18450452, ECO:0000269|PubMed:22609986}. | 3D-structure;Alternative splicing;Apoptosis;Cell membrane;Cytoplasm;Endosome;Lipoprotein;Membrane;Metal-binding;Palmitate;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:15069192}. | hsa:117584; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endosome membrane [GO:0010008]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; recycling endosome membrane [GO:0055038]; metal ion binding [GO:0046872]; p53 binding [GO:0002039]; protease binding [GO:0002020]; protein kinase binding [GO:0019901]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; apoptotic process [GO:0006915]; negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis [GO:2001271]; negative regulation of extrinsic apoptotic signaling pathway via death domain receptors [GO:1902042]; negative regulation of signal transduction by p53 class mediator [GO:1901797]; negative regulation of tumor necrosis factor-mediated signaling pathway [GO:0010804]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein K48-linked ubiquitination [GO:0070936]; regulation of fibroblast migration [GO:0010762]; regulation of signal transduction by p53 class mediator [GO:1901796]; regulation of TOR signaling [GO:0032006]; ubiquitin-dependent protein catabolic process [GO:0006511] | 18382127_CARPs together with MDM2 enhance p53 degradation, thereby inhibiting p53-mediated cell death. 18450452_acts at the level of endocytic vesicles to limit the intensity of TNF-induced NF-kappaB activation by the regulated elimination of a necessary signaling component within the receptor complex 19305408_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 21308745_data indicate AE induces caspase-8-mediated activation of mitochondrial death pathways by decreasing the stability of CARP mRNAs in a p53-independent manner. 24114843_EGF, which signals through CRAF, and an activated BRAF mutant also activate PKC and stimulate cell migration through up-regulating RFFL expression. 28827789_it is interesting to note that a human lncRNA does exist within the 5'-UTR intronic region of the human RFFL gene . Given the rat data, our study may serve as a translational foundation for considering this human lncRNA as a candidate regulator for cardiovascular diseases. 30401747_RFFL is an important regulator of voltage-gated hERG potassium channel activity and therefore cardiac repolarization and that this ubiquitination-mediated regulation requires parts of the ERAD pathway. 30659120_The integral function of the endocytic recycling compartment is regulated by RFFL-mediated ubiquitylation of Rab11 effectors. | ENSMUSG00000020696 | Rffl | 2.716214e+02 | 1.0214896 | 0.030674489 | 0.3155245 | 9.287046e-03 | 0.9232272589 | 0.98588240 | No | Yes | 2.114994e+02 | 31.367577 | 1.932195e+02 | 29.512629 | |
| ENSG00000093167 | 9209 | LRRFIP2 | protein_coding | Q9Y608 | FUNCTION: May function as activator of the canonical Wnt signaling pathway, in association with DVL3, upstream of CTNNB1/beta-catenin. Positively regulates Toll-like receptor (TLR) signaling in response to agonist probably by competing with the negative FLII regulator for MYD88-binding. {ECO:0000269|PubMed:15677333, ECO:0000269|PubMed:19265123}. | Alternative splicing;Coiled coil;Direct protein sequencing;Phosphoprotein;Reference proteome;Wnt signaling pathway | The protein encoded by this gene, along with MYD88, binds to the cytosolic tail of toll-like receptor 4 (TLR4), which results in activation of nuclear factor kappa B signaling. The ubiquitin-like protein FAT10 prevents the interaction of the encoded protein and TLR4, thereby inactivating the nuclear factor kappa B signaling pathway. In addition, this protein can downregulate the NLRP3 inflammasome by recruiting the caspase-1 inhibitor Flightless-I to the inflammasome complex. [provided by RefSeq, Jan 2017]. | hsa:9209; | LRR domain binding [GO:0030275]; regulation of transcription, DNA-templated [GO:0006355]; Wnt signaling pathway [GO:0016055] | 15677333_These data suggest that LRRFIP2 plays an important role in transducing Wnt signals. 19265123_LRR-binding MyD88 interactor LRRFIP2 is a positive regulator of NF-kappa B activity and is also a positive regulator of cytokine production in lipopolysaccharide-stimulated macrophages, suggesting a functional role in TLR4-mediated inflammatory response. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21220426_Unambiguous characterization of site-specific phosphorylation of leucine-rich repeat Fli-I-interacting protein 2 (LRRFIP2) in Toll-like receptor 4 (TLR4)-mediated signaling. 21785361_A novel exonic rearrangement affecting MLH1 and the contiguous LRRFIP2 is a founder mutation in Portuguese Lynch syndrome families 23036196_FATylation of LRRFIP2 occurs on two distinct sites, each being modified by a single FAT10 moiety. 27431477_Data show that the ability of Ca(2+) to accentuate the activity of NLRP3 inflammasome is abrogated in Flightless-I (FliI) and leucine-rich repeat FliI-interaction protein 2 (LRRFIP2)-knockdown macrophages. | ENSMUSG00000032497 | Lrrfip2 | 1.251277e+03 | 1.1479257 | 0.199029283 | 0.2793961 | 5.031175e-01 | 0.4781335164 | 0.87261335 | No | Yes | 1.220623e+03 | 156.576526 | 9.588957e+02 | 126.238041 | |
| ENSG00000093183 | 9117 | SEC22C | protein_coding | Q9BRL7 | FUNCTION: May be involved in vesicle transport between the ER and the Golgi complex. {ECO:0000269|PubMed:9501016}. | Alternative splicing;ER-Golgi transport;Endoplasmic reticulum;Membrane;Protein transport;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the SEC22 family of vesicle trafficking proteins. The encoded protein is localized to the endoplasmic reticulum and may play a role in the early stages of ER-Golgi protein trafficking. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2011]. | hsa:9117; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; protein transport [GO:0015031] | 28414125_These results indicate that the splicing-dependent changes in the number of TMDs allow Sec22c to regulate the subcellular localization in cooperation with ARF4, implying that Sec22c will function at the Golgi as well as the ER. | ENSMUSG00000061536 | Sec22c | 2.393748e+03 | 0.6664003 | -0.585539012 | 0.2707517 | 4.629220e+00 | 0.0314318498 | 0.58378765 | No | Yes | 1.755963e+03 | 174.011929 | 2.754801e+03 | 278.787227 | |
| ENSG00000095066 | 29911 | HOOK2 | protein_coding | Q96ED9 | FUNCTION: Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). Contributes to the establishment and maintenance of centrosome function. May function in the positioning or formation of aggresomes, which are pericentriolar accumulations of misfolded proteins, proteasomes and chaperones. FHF complex promotes the distribution of AP-4 complex to the perinuclear area of the cell (PubMed:32073997). {ECO:0000269|PubMed:17140400, ECO:0000269|PubMed:17540036, ECO:0000269|PubMed:18799622, ECO:0000269|PubMed:32073997}. | Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Golgi apparatus;Microtubule;Phosphoprotein;Protein transport;Reference proteome;Transport | Hook proteins are cytosolic coiled-coil proteins that contain conserved N-terminal domains, which attach to microtubules, and more divergent C-terminal domains, which mediate binding to organelles. The Drosophila Hook protein is a component of the endocytic compartment.[supplied by OMIM, Apr 2004]. | hsa:29911; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; FHF complex [GO:0070695]; intracellular membrane-bounded organelle [GO:0043231]; microtubule [GO:0005874]; trans-Golgi network [GO:0005802]; dynein light intermediate chain binding [GO:0051959]; identical protein binding [GO:0042802]; microtubule binding [GO:0008017]; cytoplasmic microtubule organization [GO:0031122]; cytoskeleton-dependent intracellular transport [GO:0030705]; early endosome to late endosome transport [GO:0045022]; endocytosis [GO:0006897]; endosome organization [GO:0007032]; endosome to lysosome transport [GO:0008333]; lysosome organization [GO:0007040]; protein localization to perinuclear region of cytoplasm [GO:1905719]; protein transport [GO:0015031] | 17540036_Hook2 contributes to the establishment and maintenance of the pericentrosomal localization of aggresomes by promoting the microtubule-based delivery of protein aggregates to pericentriolar aggresomes. 19793914_Results identify Hook2, a linker protein that is essential for regulation of the microtubule network at the centrosome, as a binding partner of CENP-F. 21998199_Hook2 localizes at the Golgi apparatus and centrosome/basal body, a strategic partitioning for ciliogenesis. Hook2 interacts with PCM1. 27624926_We first demonstrate that Hook2 is essential for the polarized Golgi re-orientation towards the migration front. Depletion of Hook2 results in a decrease of PAR6alpha at the centrosome during cell migration, while overexpression of Hook2 in cells induced the formation of aggresomes with the recruitment of PAR6alpha, aPKC and PAR3 29228058_differential methylation profile of the HOOK2 gene in individuals with T2D and obesity might be related to the attendant T2D, but further studies are required to identify the potential role of HOOK2 gene in T2D disease 32397755_Coding Variants in HOOK2 and GTPBP3 May Contribute to Risk of Primary Angle Closure Glaucoma | ENSMUSG00000052566 | Hook2 | 9.950514e+02 | 0.9248627 | -0.112688896 | 0.2632046 | 1.845932e-01 | 0.6674552933 | 0.92913032 | No | Yes | 8.554141e+02 | 112.277090 | 1.016358e+03 | 137.172455 | |
| ENSG00000099290 | 387680 | WASHC2A | protein_coding | Q641Q2 | FUNCTION: Acts at least in part as component of the WASH core complex whose assembly at the surface of endosomes inhibits WASH nucleation-promoting factor (NPF) activity in recruiting and activating the Arp2/3 complex to induce actin polymerization and is involved in the fission of tubules that serve as transport intermediates during endosome sorting. Mediates the recruitment of the WASH core complex to endosome membranes via binding to phospholipids and VPS35 of the retromer CSC. Mediates the recruitment of the F-actin-capping protein dimer to the WASH core complex probably promoting localized F-actin polymerization needed for vesicle scission. Via its C-terminus binds various phospholipids, most strongly phosphatidylinositol 4-phosphate (PtdIns-(4)P), phosphatidylinositol 5-phosphate (PtdIns-(5)P) and phosphatidylinositol 3,5-bisphosphate (PtdIns-(3,5)P2). Involved in the endosome-to-plasma membrane trafficking and recycling of SNX27-retromer-dependent cargo proteins, such as GLUT1. Required for the association of DNAJC13, ENTR1, ANKRD50 with retromer CSC subunit VPS35. Required for the endosomal recruitment of CCC complex subunits COMMD1 and CCDC93 as well as the retriever complex subunit VPS35L. {ECO:0000269|PubMed:25355947, ECO:0000269|PubMed:28892079}. | Alternative splicing;Cell membrane;Endosome;Lipid-binding;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:387680; | cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; intracellular membrane-bounded organelle [GO:0043231]; nucleolus [GO:0005730]; plasma membrane [GO:0005886]; WASH complex [GO:0071203]; phosphatidylinositol phosphate binding [GO:1901981]; retromer complex binding [GO:1905394]; protein localization to endosome [GO:0036010]; protein transport [GO:0015031]; retrograde transport, endosome to Golgi [GO:0042147] | 26956659_SNX27 interaction with FAM21 is required for the precise localization of SNX27 at an endosomal subdomain. 29735545_Study reports that nuclear IGF1R associates with advanced tumor stage and is recruited selectively to regulatory regions of chromatin including JUN and FAM21A promoters. JUN and FAM21A identified as mediators of cell survival and IGF-induced migration, properties that tumors require to attain advanced stage. | ENSMUSG00000024104 | Washc2 | 1.713829e+03 | 0.8631558 | -0.212307133 | 0.2906837 | 5.262786e-01 | 0.4681756425 | 0.86803566 | No | Yes | 1.510135e+03 | 222.603933 | 1.619364e+03 | 244.718346 | ||
| ENSG00000099622 | 1153 | CIRBP | protein_coding | Q14011 | FUNCTION: Cold-inducible mRNA binding protein that plays a protective role in the genotoxic stress response by stabilizing transcripts of genes involved in cell survival. Acts as a translational activator. Seems to play an essential role in cold-induced suppression of cell proliferation. Binds specifically to the 3'-untranslated regions (3'-UTRs) of stress-responsive transcripts RPA2 and TXN. Acts as a translational repressor (By similarity). Promotes assembly of stress granules (SGs), when overexpressed. {ECO:0000250, ECO:0000269|PubMed:11574538, ECO:0000269|PubMed:16513844}. | 3D-structure;Activator;Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repressor;Stress response | hsa:1153; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spliceosomal complex [GO:0005681]; mRNA 3'-UTR binding [GO:0003730]; RNA binding [GO:0003723]; small ribosomal subunit rRNA binding [GO:0070181]; translation repressor activity [GO:0030371]; mRNA stabilization [GO:0048255]; positive regulation of mRNA splicing, via spliceosome [GO:0048026]; positive regulation of translation [GO:0045727]; response to cold [GO:0009409]; response to UV [GO:0009411]; stress granule assembly [GO:0034063] | 12819390_results suggest that cold inducible RNA binding protein may participate in the cell cycle regulation of normal endometrium and the loss of its expression may be involved in endometrial carcinogenesis 15075239_RBM3 and CIRP are adaptatively expressed in response to hypoxia by a mechanism that involves neither HIF-1 nor mitochondria 19158277_CIRP enhanced extracellular signal-regulated kinase 1 and 2 (ERK1/2) phosphorylation, and treatment with an MEK inhibitor decreased the proliferation caused by CIRP. 19277990_pharmacological modulation of RBM3 and CIRBP may represent novel therapeutic approaches for prostate cancer. 23437386_CIRBP contributes to ultraviolet light- and lipopolysaccharide-induced expression of IL1beta by regulating NFkappaB activity. 23437386_CIRBP regulates the expression of interleukin-1beta in an NF-kappaB-dependent fashion. 24097189_report increased levels of cold-inducible RNA-binding protein (CIRP) in the blood of individuals admitted to the surgical intensive care unit with hemorrhagic shock 25027624_High levels of CIRP protein expression was associated with a short survival rate in oral squamous cell carcinoma patients. 25187386_Cirp promotes the development of intestinal inflammation and colorectal tumors through regulating apoptosis and production of TNFalpha and IL23 in inflammatory cells. 25934796_The expression of CIRP in pituitary adenoma is closely related with tumor proliferation and invasion, and its significantly elevated expression level indicates post-op recurrence. 26188505_CIRP inhibits DNA damage-induced apoptosis by regulating p53 protein.CIRP suppresses p53 upregulation during apoptosis.CIRP regulates the expression of genes involved in apoptosis. 26361390_Study shows that CIRP elevated plasma concentration is significantly associated with poor prognosis among patients with sepsis. Therefore, CIRP is a potential predictor of sepsis prognosis. 26673712_CIRP protein regulates telomerase activity in a temperature-dependent manner by regulating the level of TERT mRNAs. 26824322_Clinically, CIRP overexpression is significantly correlated with Cushing's disease recurrence. CIRP appears to play a critical tumorigenesis function in Cushing's disease and its expression might be a useful biomarker for tumor recurrence. 26824423_hnRNP A18 can promote tumor growth in in vivo models by coordinating the translation of pro-survival transcripts to support the demands of proliferating cells and increase survival under cellular stress. 26936526_In human abdominal aortic aneurysm (AAA) tissue, Cold-inducible RNA-binding protein (CIRP) exhibited a 5.6-fold and 93% increase in mRNA and protein expression, respectively. In a rat AAA model, CIRP was upregulated significantly in a time-dependent manner in the serum and AAA tissue. 27184164_CIRP was expressed in the bronchi of human COPD patients and was involved in inflammatory factors and MUC5AC expression after cold stimulation through the ERK and NF-kappaB pathways 27315340_The serum and synovial concentrations of CIRP in the rheumatoid arthritis patients were increased, suggesting that CIRP mediates inflammation and is a potential marker for synovial inflammation. 27395339_We found out that the link between CIRP and Snail is mediated by ERK and p38 pathways. EMT is a critical component of carcinoma metastasis and invasion. As demonstrated in this study, the biological role of CIRP in EMT may explain why CIRP overexpression has been associated with a bad prognosis in cancer patients. 27423012_this study shows that CIRP expression in bronchial airway epithelial cells of patients with chronic obstructive pulmonary disease is higher than that in healthy person 27477308_cold temperature can induce an airway inflammatory response and excess mucus production via a CIRP-mediated increase in mRNA stability and protein translation 27837912_We generated a transgenic mouse model overexpressing human CIRP in the mammary epithelium to ask if it plays a role in mammary gland development. Effects of CIRP overexpression on mammary gland morphology, cell proliferation, and apoptosis were studied from puberty through pregnancy, lactation and weaning 27840101_Increased synovial fluid CIRP concentrations were closely associated with the severity of knee osteoarthritis 27864909_CIRP Expression Is Induced in Skin Cancer Cells and in Keratinocytes Exposed to Lower-Dose But Not Higher-Dose UVB Radiation. 28368279_Crystal structure of the hnRNP A18 RNA recognition motif has been reported. 28373441_Low CIRP expression is associated with colon Cancer. 28536481_Cold induction of serine and arginine rich splicing factor 5 (SRSF5) is independent of cold-inducible RNA-binding protein (CIRP) and RNA-binding motif protein 3 (RBM3). 29432179_This study reports that cold-inducible RNA-binding protein (CIRBP) is a newly identified key regulator in DNA double-strand break (DSB) repair. 29981150_CIRP mediates the activation of STAT3-Bag-1 signaling cascade via activating the Janus kinase family proteins and NF-kappaB signaling pathways upon UV irradiation. 30315244_CIRBP could be a novel oncogene in human bladder cancer inducing transcription of HIF-1alpha. 31054221_Compared with pre-operation levels, CIRP levels significantly increased 6 h after cardiovascular surgery with CPB. Multiple linear regression analysis showed that the length of CPB time contributed to CIRP production (P=0.013). 31405566_downregulation of CIRP may render cardiac cells prone to apoptosis in heart failure. 32027618_Extracellular CIRP as an endogenous TREM-1 ligand to fuel inflammation in sepsis. 32689999_Cold-inducible RNA-binding protein might determine the severity and the presences of major/minor criteria for severe community-acquired pneumonia and best predicted mortality. 33172965_Cold-inducible RNA binding protein promotes breast cancer cell malignancy by regulating Cystatin C levels. 33429432_CIRP Sensitizes Cancer Cell Responses to Ionizing Radiation. 33638934_Human antigen R (HuR) and Cold inducible RNA-binding protein (CIRP) influence intestinal mucosal barrier function in ulcerative colitis by competitive regulation on Claudin1. 33902703_Cold-inducible RNA-binding protein (CIRP) potentiates uric acid-induced IL-1beta production. 33991007_ATP regulates RNA-driven cold inducible RNA binding protein phase separation. 34233365_CIRP Secretion during Cardiopulmonary Bypass Is Associated with Increased Risk of Postoperative Acute Kidney Injury. 34351954_Clinical relevance for circulating cold-inducible RNA-binding protein (CIRP) in patients with adult-onset Still's disease. 34465343_CIRP promotes the progression of non-small cell lung cancer through activation of Wnt/beta-catenin signaling via CTNNB1. 34953031_The role of Cold-Inducible RNA-binding protein in respiratory diseases. | ENSMUSG00000045193 | Cirbp | 1.245217e+04 | 1.1388164 | 0.187535148 | 0.2733991 | 4.708882e-01 | 0.4925788495 | 0.87511746 | No | Yes | 1.052595e+04 | 658.545196 | 8.979659e+03 | 576.220134 | ||
| ENSG00000099849 | 8045 | RASSF7 | protein_coding | Q02833 | FUNCTION: Negatively regulates stress-induced JNK activation and apoptosis by promoting MAP2K7 phosphorylation and inhibiting its ability to activate JNK. Following prolonged stress, anti-apoptotic effect stops because of degradation of RASSF7 protein via the ubiquitin-proteasome pathway. Required for the activation of AURKB and chromosomal congression during mitosis where it stimulates microtubule polymerization. {ECO:0000269|PubMed:20629633, ECO:0000269|PubMed:21278800}. | Alternative splicing;Apoptosis;Coiled coil;Cytoplasm;Cytoskeleton;Reference proteome;Ubl conjugation | hsa:8045; | centriolar satellite [GO:0034451]; cytoplasm [GO:0005737]; apoptotic process [GO:0006915]; regulation of microtubule cytoskeleton organization [GO:0070507]; signal transduction [GO:0007165] | 15098441_rare alleles of HRas1 minisatellite are associated with increased risk of papillary thyroid cancer formation in children and adolescents after Chernobyl accident 19367319_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20629633_RASSF7 regulates the microtubule cytoskeleton and is required for spindle formation, Aurora B activation and chromosomal congression during mitosis. 21278800_RASSF7 acts in concert with N-Ras to constitute a stress-sensitive temporary mechanism of apoptotic regulation. 22695170_The RASSF gene family members RASSF5, RASSF6 and RASSF7 show frequent DNA methylation in neuroblastoma. 26569555_truncated RASSF7 could act as an oncogene in a small subset of tumours where it is mutated in this way. 26884887_Suggest that loss of RASSF7 expression results in apoptosis in nucleus pulposus cells in human intervertebral disc degeneration. 29729697_RASSF7 promotes cell proliferation through activating MEK1/MEK2-ERK1/ERK2 signaling pathway in hepatocellular carcinoma. 30139745_RASSF7 competed with MAX in the formation of a heterodimeric complex with c-Myc and attenuated its occupancy on target gene promoters to regulate transcription. | ENSMUSG00000038618 | Rassf7 | 1.328011e+03 | 1.0641234 | 0.089665480 | 0.3201478 | 7.806741e-02 | 0.7799336896 | 0.95670131 | No | Yes | 1.400907e+03 | 194.210978 | 1.241640e+03 | 177.205696 | ||
| ENSG00000099917 | 51586 | MED15 | protein_coding | Q96RN5 | FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. Required for cholesterol-dependent gene regulation. Positively regulates the Nodal signaling pathway. {ECO:0000269|PubMed:12167862, ECO:0000269|PubMed:16630888, ECO:0000269|PubMed:16799563}. | 3D-structure;Activator;Alternative splicing;Cytoplasm;Direct protein sequencing;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Triplet repeat expansion;Ubl conjugation | The protein encoded by this gene is a subunit of the multiprotein complexes PC2 and ARC/DRIP and may function as a transcriptional coactivator in RNA polymerase II transcription. This gene contains stretches of trinucleotide repeats and is located in the chromosome 22 region which is deleted in DiGeorge syndrome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jun 2014]. | hsa:51586; | core mediator complex [GO:0070847]; cytoplasm [GO:0005737]; mediator complex [GO:0016592]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription coregulator activity [GO:0003712]; regulation of transcription by RNA polymerase II [GO:0006357]; stem cell population maintenance [GO:0019827] | 10235267_The protein encoded by this gene was identified as ARC105, a 105kDa component of the Activator-Recruited Cofactor (ARC) that is involved in mediating gene activation by several classes of transcriptional regulators. 12497610_Observational study of gene-disease association. (HuGE Navigator) 14691453_Methylation of TIG1 in prostatic neoplasms correlates with methylation of the retinoic acid receptor beta gene. 15318033_Limited role of exon 7 PCQAP polymorphisms in the pathogenesis of schizophrenia. 15318033_Observational study of gene-disease association. (HuGE Navigator) 15455391_TIG1 gene Silencing by promoter hypermethylation is associated with nasopharyngeal carcinoma 16799563_SREBPs use the evolutionarily conserved ARC105 (also called MED15) subunit to activate target genes 16904669_results suggest that TRIM11, with the ubiquitin-proteasome pathway, regulates ARC105 function in TGFbeta signaling 20025940_The data identify PC2 as a novel PLAGL2-binding protein and important mediator of PLAGL2 transactivation. 22916034_MED15 and PUM1 proteins with coiled-coil domains are potent enhancers of polyQ-mediated ataxin-1 protein misfolding and proteotoxicity in vitro and in vivo. 24374838_these findings implicate MED15 in CRPC, and as MED15 is evolutionary conserved, it is likely to emerge as a lethal phenotype in other therapeutic-resistant diseases, and not restricted to our disease model. 25791637_MED15 overexpression is a clonal event during head and neck squamous cell carcinoma progression. 26377566_MED15 is differentially expressed in tumor-free testis and testicular germ cell tumors 26457685_MED15 as a potential biomarker for head and neck squamous cell carcinoma 27974704_MED15 overexpression arises during androgen deprivation therapy via hyper-activation of PI3K/Akt/mTOR signaling pathway in prostate cancer cells. 29400661_MED15 does seem to play a tumour promoting role in the progression and metastatic spread of renal cell carcinoma 31828108_In this article, we described MED15-TFE3 renal cell carcinoma (RCC), a rare gene subtype of Xp11 translocation RCCs, that was confirmed by FISH and RNA sequencing. The tumor demonstrated different morphological features and immunophenotypic characteristics with the cases reported in literatures, expanding our understanding on heterogeneity of MED15-TFE3 RCC. 32439976_MED15, transforming growth factor beta 1 (TGF-beta1), FcgammaRIII (CD16), and HNK-1 (CD57) are prognostic biomarkers of oral squamous cell carcinoma. 33772081_MED15 prion-like domain forms a coiled-coil responsible for its amyloid conversion and propagation. 33904398_Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to Mediator. | ENSMUSG00000012114 | Med15 | 4.907816e+03 | 1.3240353 | 0.404941593 | 0.2935686 | 1.910364e+00 | 0.1669228064 | 0.77611497 | No | Yes | 5.033161e+03 | 469.864204 | 3.549943e+03 | 340.434291 | |
| ENSG00000099940 | 9342 | SNAP29 | protein_coding | O95721 | FUNCTION: SNAREs, soluble N-ethylmaleimide-sensitive factor-attachment protein receptors, are essential proteins for fusion of cellular membranes. SNAREs localized on opposing membranes assemble to form a trans-SNARE complex, an extended, parallel four alpha-helical bundle that drives membrane fusion. SNAP29 is a SNARE involved in autophagy through the direct control of autophagosome membrane fusion with the lysososome membrane. Plays also a role in ciliogenesis by regulating membrane fusions. {ECO:0000269|PubMed:23217709, ECO:0000269|PubMed:25686250, ECO:0000269|PubMed:25686604}. | 3D-structure;Autophagy;Cell membrane;Cell projection;Cilium;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Golgi apparatus;Ichthyosis;Membrane;Neuropathy;Palmoplantar keratoderma;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene, a member of the SNAP25 gene family, encodes a protein involved in multiple membrane trafficking steps. Two other members of this gene family, SNAP23 and SNAP25, encode proteins that bind a syntaxin protein and mediate synaptic vesicle membrane docking and fusion to the plasma membrane. The protein encoded by this gene binds tightly to multiple syntaxins and is localized to intracellular membrane structures rather than to the plasma membrane. While the protein is mostly membrane-bound, a significant fraction of it is found free in the cytoplasm. Use of multiple polyadenylation sites has been noted for this gene. [provided by RefSeq, Jul 2008]. | hsa:9342; | autophagosome [GO:0005776]; autophagosome membrane [GO:0000421]; azurophil granule membrane [GO:0035577]; centrosome [GO:0005813]; ciliary pocket membrane [GO:0020018]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi membrane [GO:0000139]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; presynapse [GO:0098793]; SNARE complex [GO:0031201]; SNAP receptor activity [GO:0005484]; syntaxin binding [GO:0019905]; autophagosome maturation [GO:0097352]; autophagosome membrane docking [GO:0016240]; cilium assembly [GO:0060271]; exocytosis [GO:0006887]; membrane fusion [GO:0061025]; protein transport [GO:0015031]; synaptic vesicle fusion to presynaptic active zone membrane [GO:0031629]; synaptic vesicle priming [GO:0016082]; vesicle fusion [GO:0006906]; vesicle targeting [GO:0006903] | 11317222_Observational study of gene-disease association. (HuGE Navigator) 15890653_SNAP-29 acts as a negative modulator for neurotransmitter release, probably by slowing recycling of the SNARE-based fusion machinery and synaptic vesicle turnover 15968592_a SNAP29 mutation codes for a SNARE protein involved in intracellular trafficking and causes a novel neurocutaneous syndrome characterized by cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma 19086053_Observational study of gene-disease association. (HuGE Navigator) 19350501_Observational study of gene-disease association. (HuGE Navigator) 20305790_SNAP29 mediated membrane fusion has an important role in endocytic recycling and consequently, in cell motility 21073448_a causal relationship between defective function of SNAP29 and the pleiotropic manifestations of CEDNIK syndrome 23231787_This work implicates SNAP29 as a major modifier of variable expressivity in 22q11.2 DS patients. 25419848_In mammalian cells, mutating the O-GlcNAc sites in SNAP-29, promotes the formation of a SNAP-29-containing SNARE complex, increases fusion between autophagosomes and endosomes/lysosomes, and promotes autophagic flux. 25551675_support a role of Snap29 at key steps of membrane trafficking, and predict that signaling defects may contribute to the pathogenesis of cerebral dysgenesis 25958742_phenotypic variability in Arab families with c.223delG mutation affected by cerebral dysgenesis, neuropathy, ichthyosis and keratoderma syndrome 27647876_Here, we identify a novel role for Snap29, an unconventional SNARE, in promoting kinetochore assembly during mitosis in Drosophila and human cells. Snap29 localizes to the outer kinetochore and prevents chromosome mis-segregation and the formation of cells with fragmented nuclei. 29454964_NEK3 kinase phosphorylates SNAP29 on its serine 105. NEK3-mediated phosphorylation determines membrane localization of SNAP29. Membrane-associated SNAP29 regulates Golgi and focal adhesion structures. 30555541_These findings suggest that down-regulation of OGT enhances cisplatin-induced autophagy via SNAP-29, resulting in cisplatin-resistant ovarian cancer. 30742775_autophagic degradation critically determines the production of HBV virions and HBsAg; this is controlled by the SNAP29-VAMP8 interaction 31811899_Silencing DAPK3 blocks the autophagosome-lysosome fusion by mediating SNAP29 in trophoblast cells under high glucose treatment. 32817423_Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion. 34069872_Generation and Characterization of a CRISPR/Cas9-Mediated SNAP29 Knockout in Human Fibroblasts. 34535638_Alpha-Synuclein defects autophagy by impairing SNAP29-mediated autophagosome-lysosome fusion. | ENSMUSG00000022765 | Snap29 | 1.398504e+03 | 0.9282439 | -0.107424148 | 0.2596207 | 1.721535e-01 | 0.6782049777 | 0.93237187 | No | Yes | 1.410273e+03 | 164.166480 | 1.351919e+03 | 161.458897 | |
| ENSG00000100139 | 85377 | MICALL1 | protein_coding | Q8N3F8 | FUNCTION: Probable lipid-binding protein with higher affinity for phosphatidic acid, a lipid enriched in recycling endosome membranes. On endosome membranes, may act as a downstream effector of Rab proteins recruiting cytosolic proteins to regulate membrane tubulation. May be involved in a late step of receptor-mediated endocytosis regulating for instance endocytosed-EGF receptor trafficking. Alternatively, may regulate slow endocytic recycling of endocytosed proteins back to the plasma membrane. May indirectly play a role in neurite outgrowth. {ECO:0000269|PubMed:19864458, ECO:0000269|PubMed:20801876, ECO:0000269|PubMed:21795389, ECO:0000269|PubMed:23596323}. | 3D-structure;Coiled coil;Endocytosis;Endosome;LIM domain;Membrane;Metal-binding;Phosphoprotein;Protein transport;Reference proteome;Transport;Zinc | hsa:85377; | extrinsic component of membrane [GO:0019898]; late endosome [GO:0005770]; late endosome membrane [GO:0031902]; recycling endosome membrane [GO:0055038]; cadherin binding [GO:0045296]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; phosphatidic acid binding [GO:0070300]; small GTPase binding [GO:0031267]; endocytic recycling [GO:0032456]; endocytosis [GO:0006897]; neuron projection development [GO:0031175]; plasma membrane tubulation [GO:0097320]; protein localization to endosome [GO:0036010]; protein targeting to membrane [GO:0006612]; receptor-mediated endocytosis [GO:0006898]; slow endocytic recycling [GO:0032458] | 19864458_These data implicate MICAL-L1 as an unusual type of Rab effector that regulates endocytic recycling by recruiting and linking EHD1 and Rab8a on membrane tubules. 21795389_novel insights into the MICAL-L1/Rab protein complex that can regulate EGFR trafficking at late endocytic pathways. 21951725_Rab35 is a critical upstream regulator of MICAL-L1 and Arf6, while both MICAL-L1 and Arf6 regulate Rab8a function. 23596323_Cooperation of MICAL-L1, pacsin 2 (syndapin2), and phosphatidic acid in tubular recycling endosome biogenesis. 24481818_MICAL-L1-mediated recruitment of EHD1 to Src-containing recycling endosomes is required for the release of Src from the perinuclear endocytic recycling compartment in response to growth factor stimulation. 25248744_DGKalpha generates phosphatidic acid to drive its own recruitment to tubular recycling endosomes via its interaction with MICAL-L1 25287187_suggesting an EHD1-independent function for MICAL-L1 earlier in mitosis 28714518_Findings found that ectopic expression of MICALL1 significantly inhibited the proliferation of HCT116 colorectal cancer cells. Also, MICALL1 expression was found to be suppressed in colorectal cancer with p53 mutations. 31615969_Data support the notion that a pool of centriolar gamma-tubulin and/or alpha-tubulin-beta-tubulin heterodimers anchor MICAL-L1 to the centriole, where it might recruit EHD1 to promote ciliogenesis. 33334886_Defining the protein and lipid constituents of tubular recycling endosomes. 33972531_A junctional PACSIN2/EHD4/MICAL-L1 complex coordinates VE-cadherin trafficking for endothelial migration and angiogenesis. 34100897_MICAL-L1 is required for cargo protein delivery to the cell surface. | ENSMUSG00000033039 | Micall1 | 4.161540e+03 | 1.6431281 | 0.716444957 | 0.3225189 | 4.922167e+00 | 0.0265142391 | 0.55176060 | No | Yes | 5.044253e+03 | 696.148656 | 2.554303e+03 | 362.129922 | ||
| ENSG00000100167 | 55964 | SEPTIN3 | protein_coding | Q9UH03 | FUNCTION: Filament-forming cytoskeletal GTPase (By similarity). May play a role in cytokinesis (Potential). {ECO:0000250, ECO:0000305}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Cell junction;Cytoplasm;Cytoskeleton;GTP-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Synapse | This gene belongs to the septin family of GTPases. Members of this family are required for cytokinesis. Expression is upregulated by retinoic acid in a human teratocarcinoma cell line. The specific function of this gene has not been determined. Alternative splicing of this gene results in several transcript variants encoding different isoforms. [provided by RefSeq, May 2018]. | hsa:55964; | cell division site [GO:0032153]; microtubule cytoskeleton [GO:0015630]; neuron projection [GO:0043005]; presynapse [GO:0098793]; presynaptic cytoskeleton [GO:0099569]; septin complex [GO:0031105]; septin ring [GO:0005940]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; identical protein binding [GO:0042802]; molecular adaptor activity [GO:0060090]; cellular protein localization [GO:0034613]; cytoskeleton-dependent cytokinesis [GO:0061640] | 15200238_Observational study of gene-disease association. (HuGE Navigator) 21082023_Data show that Septins of the SEPT6 group preferentially interacted with septins of the SEPT2 group, SEPT3 group and SEPT7 group. 23163726_septins from the SEPT3 subgroup may be important determinants of polymerization by occupying the terminal position in octameric units which themselves form the building blocks of at least some heterofilaments 24787956_Data indicate that forchlorfenuron (FCF) exhibits differential binding preference for septins SEPT2 and SEPT3. 29051266_SUMOylation of human septins is critical for septin filament bundling and cytokinesis. 30254212_argeted RNA sequencing was used to screen 60 melanoma patient-derived xenograft (PDX) models for BRAF fusions. We identified three unique BRAF fusions, including a novel SEPT3-BRAF fusion, occurring in four tumors (4/60, 6.7%), all of which were 'pan-negative' (lacking other common mutations) (4/18, 22.2%). 32910969_Molecular Recognition at Septin Interfaces: The Switches Hold the Key. | ENSMUSG00000022456 | Septin3 | 1.108483e+03 | 1.0498809 | 0.070225654 | 0.3020612 | 5.452633e-02 | 0.8153663782 | 0.96373941 | No | Yes | 1.044108e+03 | 77.643014 | 9.482919e+02 | 72.700446 | |
| ENSG00000100239 | 9701 | PPP6R2 | protein_coding | O75170 | FUNCTION: Regulatory subunit of protein phosphatase 6 (PP6). May function as a scaffolding PP6 subunit. Involved in the PP6-mediated dephosphorylation of NFKBIE opposing its degradation in response to TNF-alpha. {ECO:0000269|PubMed:16769727}. | Alternative splicing;Cytoplasm;Phosphoprotein;Reference proteome | The protein encoded by this gene is a regulatory protein for the protein phosphatase-6 catalytic subunit. Together, these proteins act as a significant T-loop phosphatase for Aurora A, an essential mitotic kinase. Loss of function of either the regulatory or catalytic subunit of protein phosphatase-6 interferes with spindle formation and chromosome alignment. [provided by RefSeq, May 2017]. | hsa:9701; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; protein phosphatase binding [GO:0019903]; protein phosphatase regulator activity [GO:0019888]; regulation of phosphoprotein phosphatase activity [GO:0043666] | ENSMUSG00000036561 | Ppp6r2 | 4.929916e+03 | 1.0597571 | 0.083733593 | 0.2707110 | 9.535379e-02 | 0.7574781619 | 0.95013975 | No | Yes | 4.443468e+03 | 392.053941 | 4.340471e+03 | 393.003751 | ||
| ENSG00000100307 | 23492 | CBX7 | protein_coding | O95931 | FUNCTION: Component of a Polycomb group (PcG) multiprotein PRC1-like complex, a complex class required to maintain the transcriptionally repressive state of many genes, including Hox genes, throughout development. PcG PRC1 complex acts via chromatin remodeling and modification of histones; it mediates monoubiquitination of histone H2A 'Lys-119', rendering chromatin heritably changed in its expressibility. Promotes histone H3 trimethylation at 'Lys-9' (H3K9me3). Binds to trimethylated lysine residues in histones, and possibly also other proteins. Regulator of cellular lifespan by maintaining the repression of CDKN2A, but not by inducing telomerase activity. {ECO:0000269|PubMed:19636380, ECO:0000269|PubMed:21047797, ECO:0000269|PubMed:21060834, ECO:0000269|PubMed:21282530}. | 3D-structure;Chromatin regulator;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation | This gene encodes a protein that contains the CHROMO (CHRomatin Organization MOdifier) domain. The encoded protein is a component of the Polycomb repressive complex 1 (PRC1), and is thought to control the lifespan of several normal human cells. [provided by RefSeq, Oct 2016]. | hsa:23492; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PcG protein complex [GO:0031519]; PRC1 complex [GO:0035102]; chromatin organization [GO:0006325]; developmental process involved in reproduction [GO:0003006]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of transcription elongation from RNA polymerase II promoter [GO:0032968]; response to xenobiotic stimulus [GO:0009410] | 14647293_controls cellular lifespan through regulation of both the p16(Ink4a)/Rb and the Arf/p53 pathways 15897876_CBX7 represses melanoma, p16, inhibits CDK4 and p14Arf expression in normal and tumor-derived prostate cells, affecting their growth 17374722_CBX7 is a chromobox protein causally linked to cancer development 18686603_In human glioma, CBX7 is down-regulated by the inhibition of miR-9 at posttranscriptional level. 18701502_Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer 18984978_Downregulation of CBX7 is associated with urothelial tumor progression. 19706751_the ability of CBX7 to positively regulate E-cadherin expression by interacting with HDAC2 and inhibiting its activity on the E-cadherin promoter would account for the correlation between the loss of CBX7 expression and a highly malignant phenotype 20541999_Data show that chromobox 7 (CBX7) within the polycomb repressive complex 1 binds to ANRIL, and both CBX7 and ANRIL are found at elevated levels in prostate cancer tissues. 20542683_data reported indicate that the evaluation of CBX7 expression may represent a valid tool in the prognosis of colon cancer since a reduced survival of colorectal cancer patients is associated with the loss of CBX7 expression. 20723236_CBX7 acts as an oncogene in the carcinogenesis and progression of gastric cancer, and it may regulate tumorigenesis, cell migration and cancer metastasis partially via p16(INK4a) regulatory pathway. 21060834_found that expression of CBX7 in gastric carcinoma tissues with p16 methylation was significantly lower than that in their corresponding normal tissues, which showed a negative correlation with transcription of p16 in gastric mucosa 22041561_cbx7 expression was significantly downregulated in multiple human cancer tissues. 22214847_These data suggest that CBX7 is a tumor suppressor and that its loss plays a key role in the pathogenesis of cancer. 22226354_MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. 24375438_we showed for the first time that CBX7 was associated with a decreased prognosis for clear cell adenocarcinoma of the ovary 24865347_Results suggest that the loss of CBX7 expression might play a critical role in advanced stages of carcinogenesis by deregulating the expression of specific effector genes. 25351982_CBX7-mediated epigenetic induction of DKK-1 is crucial for the inhibition of breast tumorigenicity, suggesting that CBX7 could be a potential tumor suppressor in breast cancer. 25595895_CBX7/HMGA1b/NF-kappaB could take part in the same transcriptional mechanism that finally leads to the regulation of SPP1 gene expression in papillary thyroid carcinoma. 25596753_Aberrantly expressed miR-9 contributes to T24 cells invasion, partly through directly down-regulating CBX7 protein expression in bladder transitional cell carcinoma 25881303_Cbx7 is downregulated in CCs, and Cbx7 expression-low tumors correlated with lymph metastasis and poor overall survival of CC patients. 26216446_these results suggest that the retention of CBX7 expression may play a role in the modulation of chemosensitivity of lung cancer patients to the treatment with irinotecan and etoposide 26343356_Data show that polycomb-group proteins BMI1, PHC3, CBX6 and CBX7 expression was significantly increased during imatinib treatment. 26416703_the miR-9 family of microRNAs (miRNAS) downregulates the expression of CBX7. In turn, CBX7 represses miR-9-1 and miR-9-2 as part of a regulatory negative feedback loop. 27291091_Study found that Cbx7 was downregulated in glioma cell lines and tumors and identifies it as an inhibitor of glioma cell migration through its inhibitory effect on YAP/TAZ-CTGF-hippo signaling axis and underscores the importance of epigenetic inactivation of Cbx7 in gliomagenesis. 27449098_Data suggest that miR-375 leads to the activation of oncogenic signatures and tumor progression by targeting chromobox homolog 7 protein (CBX7). 28030829_Suggest CBX7 is an important tumor suppressor that negatively modulates PTEN/Akt signaling during pancreatic tumorigenesis. 28388562_CBX7 inhibits epithelial-to-mesenchymal transformation and invasion in glioma 28460453_our results validate the assumption that CBX7 is a tumor suppressor of gliomas. Moreover, CBX7 is a potential and novel prognostic biomarker in glioma patients. We also clarified that CBX7 silences CCNE1 via the combination of CCNE1 promoter and the recruitment of HDAC2. 29422082_CBX7 positively regulates stem cell-like characteristics of gastric cancer cells by inhibiting p16 and activating AKT-NF-kappaB-miR-21 pathway. 29717132_Potential role of CBX7 in regulating pluripotency of adult human pluripotent-like olfactory stem cells in stroke model has been demonstrated. 30759399_Mass spectrometry analysis revealed several non-histone protein interactions between CBX7 and the H3K9 methyltransferases SETDB1, EHMT1, and EHMT2. These CBX7-binding proteins possess a trimethylated lysine peptide motif highly similar to the canonical CBX7 target H3K27me3. 30826432_CBX7 and PRMT1 contribute to regulate E-cadherin expression through several mechanisms. 30990338_Decreased CBX7 expression levels were correlated with liver cirrhosis in HCC patients. Furthermore, the survival times of HCC patients who were CBX7-expression-negative were shorter than HCC patients who were CBX7-expression-positive. Results show that downregulation of CBX7 is related to HCC progression and a poor prognosis in HCC patients. 31211140_findings suggest that CBX4 rs2289728 and CBX7 rs139394 are protective SNPs against HCC. The two SNPs may reduce the risk of HCC while suppressing the expression of CBX4 and CBX7. 32205869_CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. 32495862_MicroRNA-18a suppresses ovarian carcinoma progression by targeting CBX7 and regulating ERK/MAPK signaling pathway and epithelial-to-mesenchymal transition. 33245100_Regulation of circGOLPH3 and its binding protein CBX7 on the proliferation and apoptosis of prostate cancer cells. 33400401_Multiomics integrative analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of breast cancer. 34035231_CBX7 suppresses urinary bladder cancer progression via modulating AKR1B10-ERK signaling. 34353139_Neuroprotection of chromobox 7 knockout in the mouse after cerebral ischemia-reperfusion injury via nuclear factor E2-related factor 2/hemeoxygenase-1 signaling pathway. | ENSMUSG00000053411 | Cbx7 | 6.076722e+02 | 1.0641800 | 0.089742133 | 0.2917027 | 9.437168e-02 | 0.7586913462 | 0.95067621 | No | Yes | 4.939466e+02 | 63.455551 | 5.005858e+02 | 66.069824 | |
| ENSG00000100320 | 23543 | RBFOX2 | protein_coding | O43251 | FUNCTION: RNA-binding protein that regulates alternative splicing events by binding to 5'-UGCAUGU-3' elements. Prevents binding of U2AF2 to the 3'-splice site. Regulates alternative splicing of tissue-specific exons and of differentially spliced exons during erythropoiesis (By similarity). RNA-binding protein that seems to act as a coregulatory factor of ER-alpha. {ECO:0000250, ECO:0000269|PubMed:11875103}. | 3D-structure;Alternative splicing;Cytoplasm;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;mRNA processing;mRNA splicing | This gene is one of several human genes similar to the C. elegans gene Fox-1. This gene encodes an RNA binding protein that is thought to be a key regulator of alternative exon splicing in the nervous system and other cell types. The protein binds to a conserved UGCAUG element found downstream of many alternatively spliced exons and promotes inclusion of the alternative exon in mature transcripts. The protein also interacts with the estrogen receptor 1 transcription factor and regulates estrogen receptor 1 transcriptional activity. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:23543; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor binding [GO:0140297]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; transcription corepressor activity [GO:0003714]; dendrite morphogenesis [GO:0048813]; intracellular estrogen receptor signaling pathway [GO:0030520]; mRNA processing [GO:0006397]; negative regulation of transcription, DNA-templated [GO:0045892]; nervous system development [GO:0007399]; neuromuscular process controlling balance [GO:0050885]; radial glia guided migration of Purkinje cell [GO:0021942]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; regulation of cell population proliferation [GO:0042127]; regulation of definitive erythrocyte differentiation [GO:0010724]; RNA metabolic process [GO:0016070]; RNA splicing [GO:0008380] | 16260614_Fox-1 and Fox-2 isoforms specifically activate splicing of neuronally regulated exons, which requires UGCAUG enhancer elements 16537540_Fox-1 and Fox-2 splicing factors have roles in alternative splicing of protein 4.1R 18573872_Fox-1/Fox-2 proteins block prespliceosome complex formation at two distinct steps through binding to two functionally important UGCAUG elements. 18573884_These results establish hnRNP H and hnRNP F as being repressors of exon inclusion and suggest that Fox proteins enhance their ability to antagonize ASF/SF2. 18794351_predict thousands of Fox-1/2 targets with conserved binding sites, at a false discovery rate of approximately 24%, including many validated experimentally, suggesting a surprisingly extensive splicing regulatory networks 19136955_These findings suggest that FOX2 functions as a critical regulator of a splicing network, and that FOX2 is important for the survival of human embryonic stem cells. 20131247_Fox-2 plays an integral role in the regulation of LH2 splicing and knockdown of Fox-2 may suggest a novel approach to strategies directed against scleroderma. 21747913_the negative regulation of Rbfox2 by Rbfox3 through a novel mechanism 21876675_functional significance of EMT-associated alternative splicing depletion of RBFOX2 in mesenchymal cells 22083953_This study characterizes the mechanism by which RBFOX2 regulates protein 4.1R exon 16 splicing through the downstream intronic element UGCAUG. 22666429_FOX-2 is involved in splicing of ataxin-2 transcripts and that this splicing event is altered by overexpression of ataxin-1 23143756_RBFOX2 polymorphism is associated with breast cancer. 23149937_RBFOX2 drives mesenchymal tissue-specific splicing in both normal and cancer tissues. 24048253_MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. 25065397_RBFOX2 SNPs showed evidence for effects across multiple reading and language traits. 25087874_Results show that the conserved Rbfox2 RNA binding protein regulates 30% of the splicing transitions observed during myogenesis and is required for the specific step of myoblast fusion. 25524026_RBFOX proteins can facilitate the splicing of micro-exons. We also found that PTBP1 likely regulates the inclusion of micro-exons, possibly by repressing the inclusion of micro-exons that are enhanced by RBFOX proteins and other splicing factors.[RBFOX] 25921069_CPSF2 and SYMPK, are RBFOX2 cofactors for both inclusion and exclusion of internal exons. 27001519_Some of the widespread cellular functions of Rbfox2 protein are attributable to regulation of miRNA biogenesis, and might include the mis-regulation of miR-20b and miR-107 in cancer and neurodegeneration. 27146458_Data show that while RBFOX1 and RBFOX2 do not mediate neuron-specific processing of UBE3A-ATS, these proteins play important roles in developing neurons and are not completely functionally redundant. 27211866_RBFox2 interactis with chromatin in a nascent RNA-dependent manner. RBFox2 inactivation eradicates PRC2 targeting on the majority of bivalent gene promoters and leads to transcriptional de-repression. 27239029_RBFOX2 dysregulation by dominant-negative RBFOX2 is an early pathogenic event in diabetic hearts. 27485310_Rbfox2 nonsense mutation is associated with hypoplastic left heart syndrome. 27911856_Results showed that the expression patterns of these genes were indicative of the onset of EMT in in-vitro models, but not in tissue samples. However, the ratio between ESRP1 or ESRP2 and RBFOX2 significantly decreased during EMT and positively correlated with the EMT-specific phenotype in cell models. Low ESRP1/RBFOX2 ratio value was associated with a higher risk of metastasis in early breast cancer patients. 28894257_The authors report that a subset of cell cycle-related genes including retinoblastoma 1 is the target of Rbfox2 in cytoplasmic stress granules, and Rbfox2 regulates the retinoblastoma 1 mRNA and protein expression levels during and following stress exposure. 28993448_Rbfox2 modulates the functions of vascular CaV1.2 calcium channel by dynamically regulating the expressions of alternative exons 9* and 33, which in turn affects the vascular myogenic tone. 30294913_Results provide evidence that RBFOX2 expression in epithelial ovarian cancer is regulated by MALAT1 which its alternative splicing. 31128090_Data show that exon 10, responsible for nuclear localization, of RBFOX2. was absent in calcific tendons. 32109384_Aberrant Expression of a Non-muscle RBFOX2 Isoform Triggers Cardiac Conduction Defects in Myotonic Dystrophy. 32762877_Trophoblast lineage specific expression of the alternative splicing factor RBFOX2 suggests a role in placental development. 32807990_Concentration-dependent splicing is enabled by Rbfox motifs of intermediate affinity. 34009296_ERG transcription factors have a splicing regulatory function involving RBFOX2 that is altered in the EWS-FLI1 oncogenic fusion. 34180133_RBFOX2/GOLIM4 Splicing Axis Activates Vesicular Transport Pathway to Promote Nasopharyngeal Carcinogenesis. 34244793_RBFOX2 alters splicing outcome in distinct binding modes with multiple protein partners. 34346508_Alternative microexon splicing by RBFOX2 and PTBP1 is associated with metastasis in colorectal cancer. 34548489_SON drives oncogenic RNA splicing in glioblastoma by regulating PTBP1/PTBP2 switching and RBFOX2 activity. | ENSMUSG00000033565 | Rbfox2 | 5.537330e+03 | 1.0429835 | 0.060716376 | 0.2565198 | 5.639747e-02 | 0.8122832571 | 0.96274656 | No | Yes | 5.025798e+03 | 380.876025 | 4.743302e+03 | 368.775383 | |
| ENSG00000100325 | 84164 | ASCC2 | protein_coding | Q9H1I8 | FUNCTION: Plays a role in DNA damage repair as component of the ASCC complex. Recruits ASCC3 and ALKBH3 to sites of DNA damage by binding to polyubiquitinated proteins that have 'Lys-63'-linked polyubiquitin chains (PubMed:29144457). Part of the ASC-1 complex that enhances NF-kappa-B, SRF and AP1 transactivation (PubMed:12077347). Involved in activation of the ribosome quality control (RQC) pathway, a pathway that degrades nascent peptide chains during problematic translation (PubMed:32099016). As part of the ribosome quality control trigger (RQT) complex, recognizes ZNF598-dependent ubiquitination of stalled ribosomes (By similarity). {ECO:0000250|UniProtKB:P53137, ECO:0000269|PubMed:12077347, ECO:0000269|PubMed:29144457, ECO:0000269|PubMed:32099016}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | hsa:84164; | activating signal cointegrator 1 complex [GO:0099053]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ubiquitin binding [GO:0043130]; DNA repair [GO:0006281]; regulation of transcription, DNA-templated [GO:0006355]; rescue of stalled ribosome [GO:0072344]; ribosome-associated ubiquitin-dependent protein catabolic process [GO:1990116] | 33139697_The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations. 33686958_Identification of key genes in coronary artery disease: an integrative approach based on weighted gene co-expression network analysis and their correlation with immune infiltration. | ENSMUSG00000020412 | Ascc2 | 5.254457e+03 | 1.0759001 | 0.105544121 | 0.2907638 | 1.317133e-01 | 0.7166622524 | 0.94050500 | No | Yes | 5.035245e+03 | 368.365987 | 4.450258e+03 | 334.314948 | ||
| ENSG00000100376 | 55007 | FAM118A | protein_coding | Q9NWS6 | Acetylation;Alternative splicing;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:55007; | integral component of membrane [GO:0016021]; identical protein binding [GO:0042802] | 19680542_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000022434 | Fam118a | 1.066555e+03 | 0.9292005 | -0.105938187 | 0.2684022 | 1.578173e-01 | 0.6911740424 | 0.93376105 | No | Yes | 9.853452e+02 | 69.088957 | 1.068359e+03 | 76.701802 | |||
| ENSG00000100379 | 79734 | KCTD17 | protein_coding | Q8N5Z5 | FUNCTION: Is a positive regulator of ciliogenesis, playing a crucial role in the initial steps of axoneme extension. It acts as a substrate-adapter for CUL3-RING ubiquitin ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of TCHP, a protein involved in ciliogenesis down-regulation (PubMed:25270598). May be involved in endoplasmic reticulum calcium ion homeostasis (PubMed:25983243). {ECO:0000269|PubMed:25270598, ECO:0000269|PubMed:25983243}. | 3D-structure;Alternative splicing;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Disease variant;Dystonia;Reference proteome;Ubl conjugation pathway | This gene encodes a protein that belongs to a conserved family of potassium channel tetramerization domain (KCTD)-containing proteins. The encoded protein functions in ciliogenesis by acting as a substrate adaptor for the cullin3-based ubiquitin-conjugating enzyme E3 ligase, and targets trichoplein, a keratin-binding protein, for degradation via polyubiquitinylation. A mutation in this gene is associated with autosomal dominant myoclonic dystonia 26. [provided by RefSeq, Nov 2016]. | hsa:79734; | Cul3-RING ubiquitin ligase complex [GO:0031463]; cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; cullin family protein binding [GO:0097602]; identical protein binding [GO:0042802]; cell projection organization [GO:0030030]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; positive regulation of cilium assembly [GO:0045724]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein homooligomerization [GO:0051260] | 25983243_A missense mutation in KCTD17 causes autosomal dominant myoclonus-dystonia. | ENSMUSG00000033287 | Kctd17 | 2.617954e+03 | 1.5357483 | 0.618941783 | 0.3189162 | 3.778407e+00 | 0.0519180573 | 0.67565452 | No | Yes | 2.948921e+03 | 380.759865 | 1.732823e+03 | 230.148949 | |
| ENSG00000100413 | 171568 | POLR3H | protein_coding | Q9Y535 | FUNCTION: DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts, such as Epstein-Barr virus-encoded RNAs (EBERs) induce type I interferon and NF- Kappa-B through the RIG-I pathway (By similarity). {ECO:0000250, ECO:0000269|PubMed:19609254, ECO:0000269|PubMed:19631370}. | 3D-structure;Alternative splicing;Antiviral defense;DNA-directed RNA polymerase;Immunity;Innate immunity;Nucleus;Reference proteome;Transcription | hsa:171568; | centrosome [GO:0005813]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; RNA polymerase III complex [GO:0005666]; DNA binding [GO:0003677]; DNA-directed 5'-3' RNA polymerase activity [GO:0003899]; defense response to virus [GO:0051607]; innate immune response [GO:0045087]; nucleobase-containing compound metabolic process [GO:0006139]; transcription by RNA polymerase III [GO:0006383]; transcription initiation from RNA polymerase III promoter [GO:0006384] | 19246067_The authors demonstrate the interaction of both RNA polymerase I and III with hepatitis delta virus RNA, both in vitro and in human cells. 20843307_Findings reveal that RNA polymerase III-dependent EBER expression through induction of cellular transcription factors and add to the repertoire of EBNA1's transcription-regulatory properties. 30830215_A pathogenic homozygous missense mutation (c.149A>G; p.Asp50Gly) in the POLR3H gene in two unrelated families with primary ovarian insufficiency was identified. | ENSMUSG00000022476 | Polr3h | 7.090340e+03 | 1.2063323 | 0.270627351 | 0.3099077 | 7.599835e-01 | 0.3833336713 | 0.84293011 | No | Yes | 8.181158e+03 | 935.909072 | 5.746045e+03 | 674.761799 | ||
| ENSG00000100441 | 23351 | KHNYN | protein_coding | O15037 | 3D-structure;Phosphoprotein;Reference proteome | The protein encoded by this gene contains a ribonuclease NYN domain and belongs to the N4BP1 family. The protein is a cofactor for the zinc finger antiviral protein (ZAP protein) which targets viral RNA for degradation and restricts SARS-CoV-2 infection. [provided by RefSeq, Sep 2021]. | hsa:23351; | cytoplasmic ribonucleoprotein granule [GO:0036464]; nucleus [GO:0005634]; endoribonuclease activity [GO:0004521]; mRNA binding [GO:0003729]; RNA phosphodiester bond hydrolysis, endonucleolytic [GO:0090502] | 20800603_Observational study of gene-disease association. (HuGE Navigator) 31284899_KHNYN as a novel cofactor for ZAP to target CpG-containing retroviral RNA for degradation. | ENSMUSG00000047153 | Khnyn | 3.051607e+03 | 1.0740146 | 0.103013539 | 0.2807800 | 1.349932e-01 | 0.7133100837 | 0.93974050 | No | Yes | 2.654377e+03 | 158.380636 | 2.462867e+03 | 151.007764 | ||
| ENSG00000100473 | 1690 | COCH | protein_coding | O43405 | FUNCTION: Plays a role in the control of cell shape and motility in the trabecular meshwork. {ECO:0000269|PubMed:21886777}. | 3D-structure;Alternative splicing;Deafness;Disease variant;Disulfide bond;Extracellular matrix;Glycoprotein;Hearing;Non-syndromic deafness;Reference proteome;Repeat;Secreted;Signal | The protein encoded by this gene is highly conserved in human, mouse, and chicken, showing 94% and 79% amino acid identity of human to mouse and chicken sequences, respectively. Hybridization to this gene was detected in spindle-shaped cells located along nerve fibers between the auditory ganglion and sensory epithelium. These cells accompany neurites at the habenula perforata, the opening through which neurites extend to innervate hair cells. This and the pattern of expression of this gene in chicken inner ear paralleled the histologic findings of acidophilic deposits, consistent with mucopolysaccharide ground substance, in temporal bones from DFNA9 (autosomal dominant nonsyndromic sensorineural deafness 9) patients. Mutations that cause DFNA9 have been reported in this gene. Alternative splicing results in multiple transcript variants encoding the same protein. Additional splice variants encoding distinct isoforms have been described but their biological validities have not been demonstrated. [provided by RefSeq, Oct 2008]. | hsa:1690; | collagen-containing extracellular matrix [GO:0062023]; extracellular region [GO:0005576]; collagen binding [GO:0005518]; defense response to bacterium [GO:0042742]; positive regulation of innate immune response [GO:0045089]; regulation of cell shape [GO:0008360]; sensory perception of sound [GO:0007605] | 11709536_Areas that express COCH mRNA as determined by in situ hybridization, and to the regions of the inner ear which show histological abnormalities in autosomal dominant sensorineural deafness and vestibular disorder, DFNA9. 12928864_findings suggest that COCH mutations are unlikely to cause abnormalities in secretion and suggest that extracellular events might cause autosomal dominant sensorineural deafness (DFNA9) pathology 14501450_A multigeneration Belgian family with late-onset progressive sensorineural hearing loss--Linkage to DFNA9 was confirmed and mutation analysis revealed a P51S mutation in the COCH gene. 15579465_Cochlin, a protein associated with deafness disorder DFNA9, is present in glaucomatous but absent in normal trabecular meshwork 16835921_A new COCH mutation is identified which causes autosomal dominant hearing impairment. 16951386_Cochlin-specific interferon-gamma-producing T cells are implicated in the etiopathogenesis of autoimmune sensorineural hearing loss. 17138532_Haplotype analysis placed the late onset autosomal dominant hereditary non-syndromic hearing loss locus within a 7.6 cM genetic interval defined by marker D14S1021 and D14S70, overlapping with the DFNA9 locus 17264471_the phenotype associated with the novel COCH (G87W) mutation is largely similar to that associated with the P51S and G88E mutation carriers 17368553_Data analysis demonstrated a significant association between vertical corneal striae and the Pro51Ser and Gly88Glu mutations in the COCH gene in DFNA9 families 1, 2, and 3 with cochleovestibular dysfunction. 17561763_This is a report of the audiological and vestibular characteristics of a Dutch DFNA9 family with a novel mutation, I109T, in the LCCL domain of COCH 17926100_A prominent but previously unreported ribbon-like pattern of cochlin in the basilar membrane was demonstrated, suggesting an important role for cochlin in the structure of the basilar membrane. 17944208_All affected family members with a COCH mutation in the vWFA2 domain shared sensorineural hearing loss with full penetrance starting between the second and fifth decade of life. 18312449_novel mutations in the vWFA2 domain of the COCH gene were identified in Chinese families with autosomal dominant sensorineural non-syndromic hearing loss (HL) 9 19013156_The second von Willebrand type A domain of cochlin has affinity for type II collagen, as well as type I and type IV collagens whereas the LCCL-domain of cochlin has no affinity for these proteins. 19098315_These results support the finding that the observed increased cochlin expression in glaucomatous TM is due to relative elevated abundance of transcription factors. 19161137_causative mutation in the COCH gene in American families associated with superior semicircular canal dehiscence.(280-5) 19657184_By RT-PCR, we found that full-length cochlin was expressed in all organs examined, with a splice variant in the heart. By Western blot, we detected short isoforms (11-17 kDa) in the perilymph. 19933177_Cochlin expression was effective in decreasing outflow facility and increasing pressure in cultured anterior segment, suggesting possible involvement of cochlin in IOP elevation in vivo. 20105107_present in the perilymph, not in cerebrospinal fluid 20228067_study suggests a possible molecular mechanism underlying DFNA9 hearing loss and provides an in vitro model that may be used to explore protein-misfolding diseases in genera 20237496_Observational study of gene-disease association. (HuGE Navigator) 20447147_The causative gene of autosomal dominant non-syndromic hearing loss in the Korean family and a recurrent mutation in the COCH gene, were identified. 21046548_The onset of the hearing loss, in the 2nd or 3rd decade of life, is earlier than in most DFNA9 families. The progression of hearing loss and vestibular dysfunction in the American family is typical of other DFNA9 families with mutations in this domain. 21774451_The phenotype associated with the I109N COCH mutation is largely similar to that associated with the I109T, P51S, G87W, and G88E mutation carriers. However, subtle differences seem to exist in terms of age of onset and rate of progression. 21886777_Cochlin interacts with TREK-1 and annexin A2. 22139968_The data cannot confirm the association described previously between superior semicircular canal dehiscence and the presence of mutations in COCH gene. 22610276_the instability of mutant cochlin is the major driving force for cochlin aggregation in the inner ear in DFNA9 patients carrying the COCH p.F527C mutation 22931125_Identification of a novel missense mutation in COCH in a Chinese family with autosomal dominant non-syndromic progressive sensorineural hearing loss. 23660400_COCH and SLC26A5 mRNA are expressed in specific structures and cells of the inner ear in archival human temporal bone 23993205_Chinese DFNA9 family associated with novel COCH mutation with genotype-phenotype correlation. 24063017_This study suggests lack of association of both COCH and TNFA with primary open-angle glaucoma pathogenesis. 24275721_new variants in genes such as COCH is associated with nonsyndromic deafness and vestibular dysfunction. 24662630_A new phenotypic and characteristic radiologic feature of DFNA9 has been discovered. 25049087_prominent in the incudomalleal joint, incudostapedial joint, and the pars tensa of the tympanic membrane 25230692_This is the first report showing failure of mutant cochlin transport through the secretory pathway, abolishment of cochlin secretion, and formation and retention of dimers and large multimeric intracellular aggregates 25780252_Targeted exon resequencing of selected genes using next-generation sequencing identified 3 COCH (one known, two novel) mutations in a cohort of hearing loss patients in Japan. 26256111_the impaired post-translational cleavage of cochlin mutants may be associated with pathological mechanisms underlying DFNA9-related sensorineural hearing loss. 26631968_This family is the first case of a truncating COCH variant and supports the hypothesis that COCH haploinsufficiency is not the cause of hearing loss in humans. 26758463_Distinct vestibular phenotypes depending on the location of COCH mutations were demonstrated, and this study correlates a genotype of p.G38D in COCH to the phenotype of bilateral total vestibular loss, therefore expanding the vestibular phenotypic spectrum of DFNA9 to range from bilateral vestibular loss without episodic vertigo to MD-like features with devastating episodic vertigo 27083884_This study showed that Mendelian sensorineural hearing loss exhibits vestibular dysfunction, including DFNA9, DFNA11, DFNA15 and DFNA28. 28005267_COCH expression is significantly downregulated in human masticatory mucosa during wound healing 28099493_c.889G>A (p.C162Y) Mutation in COCH leads to vestibular dysfunction and autosomal dominant nonsyndromic deafness 9.The p.C162Y mutation causes either disruption of LCCL domain fragment cleavage or aggregation of mutant cochlin. 28116169_A missense mutation in the LCCL domain of COCH was associated with autosomal dominant nonsyndromic sensorineural hearing loss in a Chinese family. 29305555_Recessive dystrophic epidermolysis bullosa patients displayed lower levels of systemic cochlin LCCL domain with subsequently impaired macrophage response in infected wounds. 29449721_A homozygous nonsense c.292C>T(p.Arg98*) COCH variant was identified in two brothers with prelingual hearing impairment. 30806805_The aim of this study was to carry out a systematic review of all reported hearing and vestibular function data in P51S COCH mutation carriers and its correlation with age.[review] 30904974_we selectively filtered out several reports describing DFNA9 patients with MD-like symptoms caused by COCH mutation 31126177_In 3 families with hearing impairment, whole exome sequencing revealed 3 novel variants in KCNQ4, LHFPL5 and COCH genes. Another variant in a homozygous state (c.116T>A, p.L39X) was identified in the COCH gene which encodes a secretory protein. 32562050_Novel loss-of-function mutations in COCH cause autosomal recessive nonsyndromic hearing loss. 32635986_Cochlin-cleaved LCCL is a dual-armed regulator of the innate immune response in the cochlea during inflammation. 32939038_Homozygote loss-of-function variants in the human COCH gene underlie hearing loss. 33421658_On the pathophysiology of DFNA9: Effect of pathogenic variants in the COCH gene on inner ear functioning in human and transgenic mice. 33710989_A Novel COCH Mutation Affects the vWFA2 Domain and Leads to a Relatively Mild DFNA9 Phenotype. 34369416_Genotype-phenotype Correlation Study in a Large Series of Patients Carrying the p.Pro51Ser (p.P51S) Variant in COCH (DFNA9): Part I-A Cross-sectional Study of Hearing Function in 111 Carriers. 35020687_Does Vestibulo-Ocular Reflex (VOR) Gain Correlate With Radiological Findings in the Semi-Circular Canals in Patients Carrying the p.Pro51Ser (P51S) COCH Variant Causing DFNA9? Relationship Between the Three-Dimensional Video Head Impulse Test (vHIT) and MR/CT Imaging. | ENSMUSG00000020953 | Coch | 7.373318e+02 | 0.9502337 | -0.073645718 | 0.3315077 | 4.938158e-02 | 0.8241428831 | 0.96654947 | No | Yes | 6.633556e+02 | 110.905818 | 6.731509e+02 | 115.377650 | |
| ENSG00000100504 | 5836 | PYGL | protein_coding | P06737 | FUNCTION: Allosteric enzyme that catalyzes the rate-limiting step in glycogen catabolism, the phosphorolytic cleavage of glycogen to produce glucose-1-phosphate, and plays a central role in maintaining cellular and organismal glucose homeostasis. {ECO:0000269|PubMed:22225877}. | 3D-structure;Acetylation;Allosteric enzyme;Alternative splicing;Carbohydrate metabolism;Cytoplasm;Disease variant;Glycogen metabolism;Glycogen storage disease;Glycosyltransferase;Nucleotide-binding;Phosphoprotein;Pyridoxal phosphate;Reference proteome;Transferase | This gene encodes a homodimeric protein that catalyses the cleavage of alpha-1,4-glucosidic bonds to release glucose-1-phosphate from liver glycogen stores. This protein switches from inactive phosphorylase B to active phosphorylase A by phosphorylation of serine residue 15. Activity of this enzyme is further regulated by multiple allosteric effectors and hormonal controls. Humans have three glycogen phosphorylase genes that encode distinct isozymes that are primarily expressed in liver, brain and muscle, respectively. The liver isozyme serves the glycemic demands of the body in general while the brain and muscle isozymes supply just those tissues. In glycogen storage disease type VI, also known as Hers disease, mutations in liver glycogen phosphorylase inhibit the conversion of glycogen to glucose and results in moderate hypoglycemia, mild ketosis, growth retardation and hepatomegaly. Alternative splicing results in multiple transcript variants encoding different isoforms.[provided by RefSeq, Feb 2011]. | hsa:5836; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; secretory granule lumen [GO:0034774]; AMP binding [GO:0016208]; ATP binding [GO:0005524]; bile acid binding [GO:0032052]; glucose binding [GO:0005536]; glycogen phosphorylase activity [GO:0008184]; identical protein binding [GO:0042802]; linear malto-oligosaccharide phosphorylase activity [GO:0102250]; purine nucleobase binding [GO:0002060]; pyridoxal phosphate binding [GO:0030170]; SHG alpha-glucan phosphorylase activity [GO:0102499]; vitamin binding [GO:0019842]; 5-phosphoribose 1-diphosphate biosynthetic process [GO:0006015]; glucose homeostasis [GO:0042593]; glycogen catabolic process [GO:0005980]; glycogen metabolic process [GO:0005977]; necroptotic process [GO:0070266]; response to bacterium [GO:0009617] | 15223230_Observational study of genotype prevalence. (HuGE Navigator) 15223230_Susceptibility to excessive liver glycogen storage in patients with type 1 diabetes. 17705025_Deficiency of liver glycogen phosphorylase is predominantly the result of missense mutations affecting enzyme activity. There are no common mutations and the severity of clinical symptoms varies significantly. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 32126244_The vitamin B6-regulated enzymes PYGL and G6PD fuel NADPH oxidases to promote skin inflammation. 32268899_Description of two GSD VI patients expanding the spectrum of PYGL mutations. 32892177_Glycogen storage disease type VI can progress to cirrhosis: ten Chinese patients with GSD VI and a literature review. 33879691_Glycogen storage disease type VI with a novel PYGL mutation: Two case reports and literature review. 34516362_Long noncoding RNA KCNMB2-AS1 promotes the development of esophageal cancer by modulating the miR-3194-3p/PYGL axis. 34675331_Human hair follicles operate an internal Cori cycle and modulate their growth via glycogen phosphorylase. 35037470_Identification of PYGL as a key prognostic gene of glioma by integrated bioinformatics analysis. | ENSMUSG00000021069 | Pygl | 4.747157e+03 | 1.0292561 | 0.041601985 | 0.2748742 | 2.331605e-02 | 0.8786380510 | 0.97727938 | No | Yes | 5.051366e+03 | 718.584283 | 4.245986e+03 | 619.658845 | |
| ENSG00000100519 | 5706 | PSMC6 | protein_coding | P62333 | FUNCTION: Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair. PSMC6 belongs to the heterohexameric ring of AAA (ATPases associated with diverse cellular activities) proteins that unfolds ubiquitinated target proteins that are concurrently translocated into a proteolytic chamber and degraded into peptides. {ECO:0000269|PubMed:1317798}. | 3D-structure;ATP-binding;Acetylation;Cytoplasm;Direct protein sequencing;Nucleotide-binding;Nucleus;Phosphoprotein;Proteasome;Reference proteome | The 26S proteasome is a multicatalytic proteinase complex with a highly ordered structure composed of 2 complexes, a 20S core and a 19S regulator. The 20S core is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. The 19S regulator is composed of a base, which contains 6 ATPase subunits and 2 non-ATPase subunits, and a lid, which contains up to 10 non-ATPase subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides. This gene encodes one of the ATPase subunits, a member of the triple-A family of ATPases which have a chaperone-like activity. Pseudogenes have been identified on chromosomes 8 and 12. [provided by RefSeq, Jul 2008]. | hsa:5706; | cytosol [GO:0005829]; cytosolic proteasome complex [GO:0031597]; extracellular exosome [GO:0070062]; inclusion body [GO:0016234]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; proteasome accessory complex [GO:0022624]; proteasome complex [GO:0000502]; proteasome regulatory particle, base subcomplex [GO:0008540]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; identical protein binding [GO:0042802]; proteasome-activating activity [GO:0036402]; protein-macromolecule adaptor activity [GO:0030674]; positive regulation of inclusion body assembly [GO:0090261]; positive regulation of proteasomal protein catabolic process [GO:1901800]; positive regulation of RNA polymerase II transcription preinitiation complex assembly [GO:0045899]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; ubiquitin-dependent ERAD pathway [GO:0030433] | 20478047_N protein of SARS Coronavirus interacts with the host cell proteasome subunit p42. 24875235_Evidence of a sex-specific association of PSMC6 genetic variants with subtypes of juvenile idiopathic arthritis. 25801217_Advanced oxidation protein products down-regulate the expression of calcium transport channels through p44/42 MAPK signaling mechanisms in the small intestinal epithelium. 26661414_Our results provide evidence on new T1DM-susceptible loci in the PSMA3, PSMA6 and PSMC6 proteasome genes and give a new insight into the T1DM pathogenesis 30559147_PSMC6 was differently expressed in melanosis coli tissues.PSMC6 expression was related to cell apoptosis. 32017075_PSMC6 promotes osteoblast apoptosis through inhibiting PI3K/AKT signaling pathway activation in ovariectomy-induced osteoporosis mouse model. 34239933_The Silence of PSMC6 Inhibits Cell Growth and Metastasis in Lung Adenocarcinoma. | ENSMUSG00000021832 | Psmc6 | 8.239836e+02 | 0.8932293 | -0.162897592 | 0.3771271 | 1.839435e-01 | 0.6680059500 | 0.92913032 | No | Yes | 6.541715e+02 | 131.295693 | 8.153288e+02 | 167.505552 | |
| ENSG00000100523 | 80821 | DDHD1 | protein_coding | Q8NEL9 | FUNCTION: Phospholipase that hydrolyzes phosphatidic acid, including 1,2-dioleoyl-sn-phosphatidic acid. The different isoforms may change the substrate specificity (PubMed:22922100). Required for the organization of the endoplasmic reticulum exit sites (ERES), also known as transitional endoplasmic reticulum (tER) (PubMed:17428803). {ECO:0000269|PubMed:17428803, ECO:0000269|PubMed:22922100}. | Alternative splicing;Cytoplasm;Hereditary spastic paraplegia;Hydrolase;Lipid degradation;Lipid metabolism;Neurodegeneration;Phosphoprotein;Reference proteome | This gene is a member of the intracellular phospholipase A1 gene family. The protein encoded by this gene preferentially hydrolyzes phosphatidic acid. It is a cytosolic protein with some mitochondrial localization, and is thought to be involved in the regulation of mitochondrial dynamics. Overexpression of this gene causes fragmentation of the tubular structures in mitochondria, while depletion of the gene results in mitochondrial tubule elongation. Deletion of this gene in male mice caused fertility defects, resulting from disruption in the organization of the mitochondria during spermiogenesis. In humans, mutations in this gene have been associated with hereditary spastic paraplegia (HSP), also known as Strumpell-Lorrain disease, or, familial spastic paraparesis (FSP). This inherited disorder is characterized by progressive weakness and spasticity of the legs. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Aug 2015]. | hsa:80821; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; metal ion binding [GO:0046872]; phospholipase activity [GO:0004620]; lipid catabolic process [GO:0016042]; positive regulation of mitochondrial fission [GO:0090141] | 20359546_DDHD1-LPI-GPR55 axis to be involved in functions in the brain. 24599962_a possible mechanism of PA regulation of the mitochondrial membrane and demonstrate an in vivo function of PA-PLA1 in the organization of mitochondria during spermiogenesis. 24989667_Two novel heterozygous mutations in DDHD1 were found in the affected members of one family, with clinical features overlapping the SPG28 subtype. 27216551_the novel mutation in DDHD1 is the causative variant for the SPG28 patient that is the first record of the disease in Japanese population. 28818478_A novel homozygous mutation in DDHD1 was identified in a patient with hereditary spastic paraplegia, retinal dystrophy and a pattern of neurodegeneration with brain iron accumulation. 33952459_Expression of Lysophosphatidylinositol Signaling-relevant Molecules in Colorectal Cancer. 34089703_Phosphorylation of human phospholipase A1 DDHD1 at newly identified phosphosites affects its subcellular localization. | ENSMUSG00000037697 | Ddhd1 | 5.246274e+02 | 1.0590358 | 0.082751397 | 0.3694454 | 4.913846e-02 | 0.8245692520 | 0.96682921 | No | Yes | 3.189452e+02 | 60.980108 | 3.093395e+02 | 60.476521 | |
| ENSG00000100554 | 51382 | ATP6V1D | protein_coding | Q9Y5K8 | FUNCTION: Subunit of the V1 complex of vacuolar(H+)-ATPase (V-ATPase), a multisubunit enzyme composed of a peripheral complex (V1) that hydrolyzes ATP and a membrane integral complex (V0) that translocates protons (PubMed:33065002). V-ATPase is responsible for acidifying and maintaining the pH of intracellular compartments and in some cell types, is targeted to the plasma membrane, where it is responsible for acidifying the extracellular environment (By similarity). May play a role in cilium biogenesis through regulation of the transport and the localization of proteins to the cilium (PubMed:21844891). {ECO:0000250|UniProtKB:P39942, ECO:0000269|PubMed:21844891, ECO:0000269|PubMed:33065002}. | 3D-structure;Cell projection;Cilium biogenesis/degradation;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Hydrogen ion transport;Ion transport;Membrane;Reference proteome;Transport | This gene encodes a component of vacuolar ATPase (V-ATPase), a multisubunit enzyme that mediates acidification of eukaryotic intracellular organelles. V-ATPase dependent organelle acidification is necessary for such intracellular processes as protein sorting, zymogen activation, receptor-mediated endocytosis, and synaptic vesicle proton gradient generation. V-ATPase is composed of a cytosolic V1 domain and a transmembrane V0 domain. The V1 domain consists of three A and three B subunits, two G subunits plus the C, D, E, F, and H subunits. The V1 domain contains the ATP catalytic site. The V0 domain consists of five different subunits: a, c, c', c', and d. Additional isoforms of many of the V1 and V0 subunit proteins are encoded by multiple genes or alternatively spliced transcript variants. This gene encodes the V1 domain D subunit protein. [provided by RefSeq, Jul 2008]. | hsa:51382; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; plasma membrane [GO:0005886]; proton-transporting V-type ATPase complex [GO:0033176]; specific granule membrane [GO:0035579]; vacuolar proton-transporting V-type ATPase, V1 domain [GO:0000221]; proton-transporting ATPase activity, rotational mechanism [GO:0046961]; cilium assembly [GO:0060271]; protein localization to cilium [GO:0061512]; regulation of macroautophagy [GO:0016241] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25006744_Top single-nucleotide polymorphism rs9590614 in VMA8 is located within genes that function in cell-cell signaling and cell migration. | ENSMUSG00000021114 | Atp6v1d | 4.891082e+02 | 0.8427468 | -0.246828822 | 0.3189889 | 6.017532e-01 | 0.4379098957 | 0.85439760 | No | Yes | 4.155252e+02 | 65.953215 | 4.850216e+02 | 78.624618 | |
| ENSG00000100575 | 26520 | TIMM9 | protein_coding | Q9Y5J7 | FUNCTION: Mitochondrial intermembrane chaperone that participates in the import and insertion of multi-pass transmembrane proteins into the mitochondrial inner membrane. May also be required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space. {ECO:0000269|PubMed:14726512}. | 3D-structure;Acetylation;Chaperone;Disulfide bond;Membrane;Metal-binding;Mitochondrion;Mitochondrion inner membrane;Protein transport;Reference proteome;Translocation;Transport;Zinc | TIMM9 belongs to a family of evolutionarily conserved proteins that are organized in heterooligomeric complexes in the mitochondrial intermembrane space. These proteins mediate the import and insertion of hydrophobic membrane proteins into the mitochondrial inner membrane.[supplied by OMIM, Apr 2004]. | hsa:26520; | mitochondrial inner membrane [GO:0005743]; mitochondrial intermembrane space [GO:0005758]; mitochondrial intermembrane space protein transporter complex [GO:0042719]; mitochondrion [GO:0005739]; TIM22 mitochondrial import inner membrane insertion complex [GO:0042721]; chaperone binding [GO:0051087]; membrane insertase activity [GO:0032977]; protein homodimerization activity [GO:0042803]; zinc ion binding [GO:0008270]; protein insertion into mitochondrial inner membrane [GO:0045039]; protein targeting to mitochondrion [GO:0006626]; sensory perception of sound [GO:0007605] | 14726512_in contrast to yeast, only a small fraction of Tim9-Tim10a-Tim10b complex is in a stable association with Tim22 16387659_The crystal structure of TIM9.10 described here reveals a previously undescribed alpha-propeller topology in which helical 'blades' radiate from a narrow central pore lined with polar residues. 20877624_Observational study of gene-disease association. (HuGE Navigator) 27720672_in gastric cancer patients, a borderline association was found between overexpression of TIMM9 and vascular invasion; patients with high expression levels of TIMM9 achieved a significantly lower disease-free survival rate compared with those with low expression levels | ENSMUSG00000021079 | Timm9 | 6.519778e+02 | 1.2459986 | 0.317302470 | 0.3115221 | 1.036284e+00 | 0.3086871842 | 0.81631081 | No | Yes | 8.244296e+02 | 149.330343 | 5.108495e+02 | 95.251769 | |
| ENSG00000100578 | 9786 | KIAA0586 | protein_coding | Q9BVV6 | FUNCTION: Required for ciliogenesis and sonic hedgehog/SHH signaling. Required for the centrosomal recruitment of RAB8A and for the targeting of centriole satellite proteins to centrosomes such as of PCM1. May play a role in early ciliogenesis in the disappearance of centriolar satellites that preceeds ciliary vesicle formation (PubMed:24421332). Involved in regulation of cell intracellular organization. Involved in regulation of cell polarity (By similarity). Required for asymmetrical localization of CEP120 to daughter centrioles (By similarity). {ECO:0000250|UniProtKB:E9PV87, ECO:0000250|UniProtKB:Q1G7G9, ECO:0000269|PubMed:24421332}. | Alternative splicing;Cell projection;Ciliopathy;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Joubert syndrome;Phosphoprotein;Reference proteome | This gene encodes a conserved centrosomal protein that functions in ciliogenesis and responds to hedgehog signaling. Mutations in this gene causes Joubert syndrome 23. Alternative splicing results in multiple transcript variants and protein isoforms. [provided by RefSeq, Aug 2016]. | hsa:9786; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; photoreceptor inner segment [GO:0001917]; cilium assembly [GO:0060271]; regulation of establishment of protein localization [GO:0070201]; smoothened signaling pathway [GO:0007224] | 16702409_The chicken ortholog functions in regulation of the Gli repressor and activator proteins in the Hedgehog signaling pathway. 24421332_Talpid3 and Cep290 play overlapping and distinct roles in ciliary vesicle formation through regulation of centriolar satellite accretion and Rab8a 26026149_Intersection of this data with whole exome results from 145 individuals with unexplained Joubert syndrome identified six families with predominantly compound heterozygous mutations in KIAA0586. 26096313_we show that biallelic KIAA0586 mutations are associated with relatively mild JS in square2.5% of families with JS. 26166481_Mutations in KIAA0586 cause lethal ciliopathies ranging from a hydrolethalus phenotype to short-rib polydactyly syndrome. 26386044_biallelic deleterious mutations in KIAA0586 lead to Joubert syndrome with or without Jeune asphyxiating thoracic dystrophy. 26386247_The authors demonstrate KIAA0586 protein localization at the basal body in human and mouse photoreceptors, as is common for Joubert syndrome proteins, and also in pericentriolar locations. 27146717_In the absence of PCM1, Mib1 destabilizes Talpid3 through poly-ubiquitylation and suppresses cilium assembly. 30120217_Considering this and the high allele frequency of 0.003117 in the gnomAD database, we conclude that c.428delG represents a JBTS disease-causing variant only if present in compound heterozygous state with a more severe KIAA0586 variant, but not in a homozygous situation. 30258116_Talpid3, C2CD3, and OFD1 differentially regulate the assembly of centriole sub-distal appendages, the CEP350/FOP/CEP19 module, centriolar satellites, and actin networks. 30988386_CEP120 interacts with C2CD3 and Talpid3 and is required for centriole appendage assembly and ciliogenesis. 31326647_Loss of KIAA0586 protein (TALPID3) function can cause both severe lethal and mild cilia-related developmental disorders [Review] 32381069_This lead to the identification of the most common variant detected in patients with JBTS23 (OMIM# 616490), rs534542684, in compound heterozygosity with a 8.3 kb deletion in KIAA0586, not previously reported. 33326788_Talpid3-Mediated Centrosome Integrity Restrains Neural Progenitor Delamination to Sustain Neurogenesis by Stabilizing Adherens Junctions. | ENSMUSG00000034601 | 2700049A03Rik | 4.969956e+02 | 1.2052296 | 0.269308024 | 0.3489652 | 5.815399e-01 | 0.4457093625 | 0.85808635 | No | Yes | 5.146624e+02 | 123.458076 | 3.052881e+02 | 75.290058 | |
| ENSG00000100592 | 23002 | DAAM1 | protein_coding | Q9Y4D1 | FUNCTION: Binds to disheveled (Dvl) and Rho, and mediates Wnt-induced Dvl-Rho complex formation. May play a role as a scaffolding protein to recruit Rho-GDP and Rho-GEF, thereby enhancing Rho-GTP formation. Can direct nucleation and elongation of new actin filaments. Involved in building functional cilia (PubMed:16630611, PubMed:17482208). Involved in the organization of the subapical actin network in multiciliated epithelial cells (By similarity). Together with DAAM2, required for myocardial maturation and sarcomere assembly (By similarity). {ECO:0000250|UniProtKB:B0DOB5, ECO:0000250|UniProtKB:Q8BPM0, ECO:0000269|PubMed:16630611, ECO:0000269|PubMed:17482208}. | 3D-structure;Actin-binding;Alternative splicing;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome;Wnt signaling pathway | Cell motility, adhesion, cytokinesis, and other functions of the cell cortex are mediated by reorganization of the actin cytoskeleton and several formin homology (FH) proteins have been associated with these processes. The protein encoded by this gene contains two FH domains and belongs to a novel FH protein subfamily implicated in cell polarity. A key regulator of cytoskeletal architecture, the small GTPase Rho, is activated during development by Wnt/Fz signaling to control cell polarity and movement. The protein encoded by this gene is thought to function as a scaffolding protein for the Wnt-induced assembly of a disheveled (Dvl)-Rho complex. This protein also promotes the nucleation and elongation of new actin filaments and regulates cell growth through the stabilization of microtubules. Alternative splicing results in multiple transcript variants encoding distinct proteins. [provided by RefSeq, Jul 2012]. | hsa:23002; | ciliary basal body [GO:0036064]; cytosol [GO:0005829]; membrane [GO:0016020]; motile cilium [GO:0031514]; plasma membrane [GO:0005886]; stress fiber [GO:0001725]; actin binding [GO:0003779]; identical protein binding [GO:0042802]; small GTPase binding [GO:0031267]; actin cytoskeleton organization [GO:0030036]; Wnt signaling pathway, planar cell polarity pathway [GO:0060071] | 16630611_DAAM1 was shown to bind to the SH3 domain of CIP4 in vivo. 17021034_Results report that Profilin1 is an effector downstream of Daam1 required for cytoskeletal changes during gastrulation. 17482208_The DAAM1 FH2 domain structure, determined at 2.25 A resolution, and DAAM1 actin binding activity 17986009_Crystal structure of human DAAM1 formin homology 2 domain 18162551_a carboxyl-terminal binding partner, Dvl, has a role in activation of Daam1 18218625_mDia1 and Daam1 are platelet actin assembly factors having distinct efficiencies, and they are directly regulated by Rho GTPases 20223827_Fli-I promotes the GTP-bound active Rho-mediated relief of the autoinhibition of Daam1 and mDia1. Thus, Fli-I is a novel positive regulator of Rho-induced linear actin assembly mediated by DRFs. 20351293_DAAM1 regulates endothelial cell growth through MT stabilization in a cell type-selective manner 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20927366_DAAM1 is a formin required for centrosome re-orientation during cell migration. 22655072_Wnt5a promotes breast cancer cell migration via Dvl2/Daam1/RhoA. 22869791_A developmentally restricted function of miR-490 and its putative DAAM1 target are associated with exaggerated megakaryocytopoiesis and/or proplatelet formation. 24696301_Daam1 plays a dual role in the phagocytic uptake of borreliae. 26792835_Kif26b, together with Dvl3/Daam1, initiates cell polarity through the control of planar cell polarity-signaling pathway-dependent activation in endothelial cells. 27760153_DAAM1 organizes actin filaments into a nodal complex 28284839_Kank1 plays a crucial role in regulating the activity of RhoA through retrieving excess Daam1 and balancing the activities of RhoA and its effectors. 28288669_High Daam-1 expression may upregulate the Wnt/PCP pathway and cause idiopathic pulmonary arterial hypertension 29207169_Results provide evidence that Daam1 activates RhoA to regulate Wnt5ainduced glioblastoma cell invasion. 29339084_miR-613 is involved in cell migration and invasion of triple-negative breast cancer cells via targeting Daam1/RhoA signaling pathway. 29746581_coiling phagocytosis is a mechanism for borrelial internalization by neuroglial cells mediated by Daam1 29752407_Integrin alphavbeta3-associated DAAM1 is essential for collagen-induced invadopodia extension and cell haptotaxis in breast cancer cells 31104928_Our data suggest that miR-208-5p/DAAM1 axis participates in ovarian cancer migration and invasion. 31406243_findings reveal a novel mechanism by which reversible tyrosine phosphorylation of DAAM1 by Src and PTPN3 regulates actin dynamics and lung cancer invasivenes 31757662_Overexpressed DAAM1 correlates with metastasis and predicts poor prognosis in breast cancer. 31894282_Wnt5a/ROR1 activates DAAM1 and promotes the migration in osteosarcoma cells. 31972124_DAAM1 and PREP are involved in human spermatogenesis. 32772041_Dishevelled Associated Activator Of Morphogenesis (DAAM) Facilitates Invasion of Hepatocellular Carcinoma by Upregulating Hypoxia-Inducible Factor 1alpha (HIF-1alpha) Expression. 33742497_SNHG15 facilitated malignant behaviors of oral squamous cell carcinoma through targeting miR-188-5p/DAAM1. 33974241_Disheveled-associated activator of morphogenesis 2 promotes invasion of colorectal cancer by activating PAK1 and promoting MMP7 expression. 34360857_Preliminary Investigation on the Involvement of Cytoskeleton-Related Proteins, DAAM1 and PREP, in Human Testicular Disorders. | ENSMUSG00000034574 | Daam1 | 2.759461e+02 | 0.8191466 | -0.287806377 | 0.3420442 | 7.201814e-01 | 0.3960843999 | 0.84483554 | No | Yes | 2.693519e+02 | 61.648198 | 2.669848e+02 | 62.669338 | |
| ENSG00000100596 | 9517 | SPTLC2 | protein_coding | O15270 | FUNCTION: Serine palmitoyltransferase (SPT). The heterodimer formed with LCB1/SPTLC1 constitutes the catalytic core. The composition of the serine palmitoyltransferase (SPT) complex determines the substrate preference. The SPTLC1-SPTLC2-SPTSSA complex shows a strong preference for C16-CoA substrate, while the SPTLC1-SPTLC2-SPTSSB complex displays a preference for C18-CoA substrate. Plays an important role in de novo sphyngolipid biosynthesis which is crucial for adipogenesis (By similarity). {ECO:0000250|UniProtKB:P97363, ECO:0000269|PubMed:19416851, ECO:0000269|PubMed:19648650, ECO:0000269|PubMed:20920666}. | 3D-structure;Acyltransferase;Disease variant;Endoplasmic reticulum;Lipid metabolism;Membrane;Neurodegeneration;Neuropathy;Pyridoxal phosphate;Reference proteome;Sphingolipid metabolism;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; sphingolipid metabolism. | This gene encodes a long chain base subunit of serine palmitoyltransferase. Serine palmitoyltransferase, which consists of two different subunits, is the key enzyme in sphingolipid biosynthesis. It catalyzes the pyridoxal-5-prime-phosphate-dependent condensation of L-serine and palmitoyl-CoA to 3-oxosphinganine. Mutations in this gene were identified in patients with hereditary sensory neuropathy type I. [provided by RefSeq, Mar 2011]. | hsa:9517; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; serine C-palmitoyltransferase complex [GO:0017059]; pyridoxal phosphate binding [GO:0030170]; serine C-palmitoyltransferase activity [GO:0004758]; adipose tissue development [GO:0060612]; ceramide biosynthetic process [GO:0046513]; positive regulation of lipophagy [GO:1904504]; sphinganine biosynthetic process [GO:0046511]; sphingolipid biosynthetic process [GO:0030148]; sphingomyelin biosynthetic process [GO:0006686]; sphingosine biosynthetic process [GO:0046512] | 12207934_results suggest that SPTLC2 mutations are not a common cause for genetic sensory neuropathies. 12445191_an increase in transepidermal water loss is an obligatory trigger for the upregulation of serine palmitoyltransferase mRNA expression in human epidermis 17331073_Results suggest that functional serine palmitoyltransferase is not a dimer, but a higher organized complex, composed of three distinct subunits (SPTLC1, SPTLC2 and SPTLC3) with a molecular mass of 480 kDa. 19416851_discovery of 2 proteins, ssSPTa and ssSPTb, which each interacts with both hLCB1 and hLCB2, suggesting that there are 4 distinct human SPT isozymes. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20920666_Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. 23658386_Mutations in SPTLC2 are associated with increased deoxySL formation causing hereditary sensory and autonomic neuropathy type 1 (HSANI) in a familial study. 24175284_The activities of the hLCB2a mutants were measured in the presence of ssSPTa and ssSPTb and was found that all decrease enzyme activity. 26573920_2 families had late-onset autosomal dominant HSAN1C caused by a new variant in SPTLC2, c.547C>T, p.(Arg183Trp). The variant changed a conserved amino acid. 30952607_HSAN-I-associated mutations in serine palmitoyltransferase subunit SPTLC2 dampened human T cell responses. SPTLC2 underpins protective immunity by translating extracellular stimuli into intracellular anabolic signals and antagonizes endoplasmic reticulum stress to promote T cell metabolic fitness. 30955194_A Novel Variant (Asn177Asp) in SPTLC2 Causing Hereditary Sensory Autonomic Neuropathy Type 1C. 31509666_Elevated levels of atypical deoxysphingolipids, caused by variant SPTLC1 or SPTLC2 or by low serine levels, were risk factors for macular telangiectasia type 2, as well as for peripheral neuropathy. 33031402_Increased expression of serine palmitoyl transferase and ORMDL3 polymorphism are associated with eosinophilic inflammation and airflow limitation in aspirin-exacerbated respiratory disease. 33558762_Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex. 34090020_Rare mutations in ATL3, SPTLC2 and SCN9A explaining hereditary sensory neuropathy and congenital insensitivity to pain in a Brazilian cohort. | ENSMUSG00000021036 | Sptlc2 | 1.510384e+03 | 0.7129549 | -0.488117377 | 0.2698759 | 3.212014e+00 | 0.0730994327 | 0.71859192 | No | Yes | 1.201184e+03 | 183.433413 | 1.529604e+03 | 238.968062 |
| ENSG00000100650 | 6430 | SRSF5 | protein_coding | Q13243 | FUNCTION: Plays a role in constitutive splicing and can modulate the selection of alternative splice sites. | Acetylation;Alternative splicing;Direct protein sequencing;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;mRNA processing;mRNA splicing | The protein encoded by this gene is a member of the serine/arginine (SR)-rich family of pre-mRNA splicing factors, which constitute part of the spliceosome. Each of these factors contains an RNA recognition motif (RRM) for binding RNA and an RS domain for binding other proteins. The RS domain is rich in serine and arginine residues and facilitates interaction between different SR splicing factors. In addition to being critical for mRNA splicing, the SR proteins have also been shown to be involved in mRNA export from the nucleus and in translation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:6430; | cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; mRNA binding [GO:0003729]; protein kinase B binding [GO:0043422]; RNA binding [GO:0003723]; cellular response to insulin stimulus [GO:0032869]; liver regeneration [GO:0097421]; mRNA processing [GO:0006397]; mRNA splice site selection [GO:0006376]; mRNA splicing, via spliceosome [GO:0000398]; positive regulation of RNA splicing [GO:0033120]; regulation of cell cycle [GO:0051726] | 12665590_role in c-H-ras alternative splicing regulation 15123677_SR proteins 9G8, SC35, ASF/SF2, and SRp40 have effects on the utilization of the A1 to A5 splicing sites of HIV-1 RNA 15163745_activates the ESE (exon splicing enhancer) which regulates HIV-1 rev, env, vpu, and nef gene expression 16103121_SRp40 regulates the switch in splicing from production of CREMtau(2)alpha to CREMalpha 16990281_SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 interact competitively at the HIV-1 Tat exon 2 splicing site 19843576_SRp40 antagonizes ASF/SF2 and SRp55 by competing for binding to certain sites in exon 5, thereby promoting TF exon 5 exclusion, an event unique to asTF biosynthesis. 20376328_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20427542_Here, the authors report that specific SR proteins, particularly SRp40 and SRp55, promote human immunodeficiency virus type 1 (HIV-1) Gag translation from unspliced (intron-containing) viral RNA. 22100336_We show that changes in alternative splicing of hnRNP A/B, affected by up regulation of SRSF5 (SRp40) or by treatment with C6-ceramide, occur within supraspliceosomes. 22205602_Relative levels of SRp20, SRp30c, and SRp40 in TM cells control differential expression of the two alternatively spliced isoforms of the GR and thereby regulate GC responsiveness. 25665148_Suggest that SRp40 might be associated with GRalpha transcripts in systemic lupus erythematosus patients. 26670336_Enhanced SRSF5 Protein Expression Reinforces Lamin A mRNA Production in HeLa Cells and Fibroblasts of Progeria Patients 27565915_the up-regulated expression of SRSF 5-7 proteins in LC with much more profound up-regulation in SCLC than in NSCLC and suggest that up-regulation of the SRSFs is related to SCLC proliferation. Moreover, we identified SRSF5 as a novel detection marker for SCLC and pleural metastatic cancer cells. 28536481_Cold induction of serine and arginine rich splicing factor 5 (SRSF5) is independent of cold-inducible RNA-binding protein (CIRP) and RNA-binding motif protein 3 (RBM3). 28592444_Posttranslational modification of SR proteins underlies the regulation of their mRNA export activities and distinguishes pluripotent from differentiated cells. 28854257_this study demonstrates that HRS acts as a key component of TLR7 signaling to orchestrate immune and inflammatory responses during EV71 infection 28867611_we show that ErbB3 interacts with the ESCRT-0 subunit Hrs both in the presence and absence of heregulin. This indicates an ESCRT-mediated sorting of ErbB3 to late endosomes and lysosomes, and in line with this we show that impaired ESCRT function leads to an endosomal accumulation of ErbB3. 29857020_Study found that SRSF5 is a novel target of SRSF3. SRSF5 is overexpressed in oral squamous cell carcinoma (OSCC) and functions as an oncogene. Its downregulation in OSCC cell lines slows cell growth, cycle progression, and tumor growth. The expression of SRSF5 seems to controlled by an autoregulation mechanism. 29891722_In this study, we uncover an alternative role for the ESCRT-0 component hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) in promoting the constitutive recycling of transmembrane proteins. We find that endosomal localization of the actin nucleating factor Wiscott-Aldrich syndrome protein and SCAR homologue (WASH) requires HRS, which occupies adjacent endosomal subdomains 29942010_upon glucose intake, the splicing factor SRSF5 is specifically induced through Tip60-mediated acetylation on K125, which antagonizes Smurf1-mediated ubiquitylation. SRSF5 promotes the alternative splicing of CCAR1 to produce CCAR1S proteins, which promote tumor growth by enhancing glucose consumption and acetyl-CoA production. 31106485_Antitumor activity of SR splicing-factor 5 knockdown by downregulating pyruvate kinase M2 in non-small cell lung cancer cells. 32400287_Genes involved in glucocorticoid receptor signalling affect susceptibility to mood disorders. | ENSMUSG00000021134 | Srsf5 | 5.948637e+03 | 0.8270412 | -0.273968870 | 0.3250021 | 7.098853e-01 | 0.3994819014 | 0.84485965 | No | Yes | 3.707394e+03 | 704.557748 | 5.342822e+03 | 1040.842806 | |
| ENSG00000100796 | 55671 | PPP4R3A | protein_coding | Q6IN85 | FUNCTION: Regulatory subunit of serine/threonine-protein phosphatase 4. May regulate the activity of PPP4C at centrosomal microtubule organizing centers. The PPP4C-PPP4R2-PPP4R3A PP4 complex specifically dephosphorylates H2AX phosphorylated on 'Ser-140' (gamma-H2AX) generated during DNA replication and required for DNA DSB repair. {ECO:0000269|PubMed:18614045}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Cytoskeleton;Nucleus;Phosphoprotein;Reference proteome | hsa:55671; | cytosol [GO:0005829]; microtubule organizing center [GO:0005815]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; protein phosphatase 4 complex [GO:0030289] | 22349239_The N-terminal of BLU was observed to interact with the C-terminal of SMEK1, a regulatory subunit of protein phosphatase 4. Furthermore, we determined the binding domains that are required for interaction between BLU and sMEK1. The N-terminal of BLU was observed to interact with the C-terminal of sMEK1. 26378810_Loss of sMEK1 is associated with Ovarian Neoplasms. 27878292_sMEK1 significantly inhibited BMI-1-stimulated oncogenesis. 29130521_A variant in PPP4R3A was associated with reduced metabolic decline in Alzheimer's disease patients. | ENSMUSG00000041846 | Ppp4r3a | 1.530293e+03 | 1.0211337 | 0.030171754 | 0.3158646 | 8.984602e-03 | 0.9244839006 | 0.98588240 | No | Yes | 1.739079e+03 | 314.791669 | 1.413841e+03 | 262.738282 | ||
| ENSG00000100889 | 5106 | PCK2 | protein_coding | Q16822 | FUNCTION: Catalyzes the conversion of oxaloacetate (OAA) to phosphoenolpyruvate (PEP), the rate-limiting step in the metabolic pathway that produces glucose from lactate and other precursors derived from the citric acid cycle. {ECO:0000250}. | Acetylation;Alternative splicing;Decarboxylase;GTP-binding;Gluconeogenesis;Lyase;Manganese;Metal-binding;Mitochondrion;Nucleotide-binding;Phosphoprotein;Reference proteome;Transit peptide | PATHWAY: Carbohydrate biosynthesis; gluconeogenesis. | This gene encodes a mitochondrial enzyme that catalyzes the conversion of oxaloacetate to phosphoenolpyruvate in the presence of guanosine triphosphate (GTP). A cytosolic form of this protein is encoded by a different gene and is the key enzyme of gluconeogenesis in the liver. Alternatively spliced transcript variants have been described. [provided by RefSeq, Apr 2014]. | hsa:5106; | cytosol [GO:0005829]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; GTP binding [GO:0005525]; manganese ion binding [GO:0030145]; phosphoenolpyruvate carboxykinase (GTP) activity [GO:0004613]; phosphoenolpyruvate carboxykinase activity [GO:0004611]; cellular response to dexamethasone stimulus [GO:0071549]; cellular response to glucose stimulus [GO:0071333]; cellular response to insulin stimulus [GO:0032869]; cellular response to tumor necrosis factor [GO:0071356]; gluconeogenesis [GO:0006094]; glycerol biosynthetic process from pyruvate [GO:0046327]; hepatocyte differentiation [GO:0070365]; NADH oxidation [GO:0006116]; oxaloacetate metabolic process [GO:0006107]; positive regulation of insulin secretion [GO:0032024]; propionate catabolic process [GO:0019543]; response to lipid [GO:0033993]; response to lipopolysaccharide [GO:0032496]; response to starvation [GO:0042594] | 12916001_Observational study of gene-disease association. (HuGE Navigator) 15315819_Results suggest that pepck2 gene expression is regulated by its 5' flanking region up to 822 bp, and 317 bp upstream of transcriptional start point. 16132948_Observational study of gene-disease association. (HuGE Navigator) 17097062_Wild-type AREBP, but not Ser(470) to Ala(470) substituted non-phosphorylating mutant, represses gene expression of the phosphoenolpyruvate carboxykinase (PEPCK), a key enzyme of gluconeogenesis. 18660489_Observational study of gene-disease association. (HuGE Navigator) 19521512_skeletal muscle PEPCK has a role in determining physical activity levels 19587243_Increased transcriptional expression of PEPCK1 and G6Pc does not account for increased gluconeogenesis and fasting hyperglycemia in patients with type 2 diabetes mellitus. 20682687_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20797423_Endoplasmic reticulum stress triggers suppression of AMPK while increasing C/EBPbeta and pCREB expression which activates PEPCK gene transcription. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21733854_results reveal a novel link between glucose metabolism and the DNA damage signaling pathway and suggest a possible role for PEPCK and G6P in the DNA damage response 22955269_expression of HCV nonstructural component NS5A in Huh7 or primary hepatocytes stimulated PEPCK gene expression and glucose output in HepG2 cells. 24602180_Expression of phosphoenolpyruvate carboxykinase linked to chemoradiation susceptibility of human colon cancer cells. 24632615_PEPCK activity was elevated threefold in lung cancer samples over normal lungs and its activation mediates an adaptive response to glucose depletion in lung cancer. 24973213_Amino acid limitation and ER stress inducers, conditions that activate the amino acid response (AAR) and the unfolded protein response (UPR), stimulate PCK2 gene transcription in tumor cell lines. 26036577_When autophagy was blocked, the level of glucose-6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK) was reduced in HepG2 cells and not in Hep3B cells. 26474064_Mitochondrial PCK2 regulates metabolic adaptation and enables glucose-independent tumor growth in various neoplasms. 26481663_Results indicate that PEPCK promotes tumor growth by increasing glucose and glutamine metabolism, increases anabolic metabolism and promotes mTORC1 activity. 26556724_ApoA-IV colocalizes with NR4A1, which suppresses G6Pase and PEPCK gene expression at the transcriptional level, reducing hepatic glucose output and lowering blood glucose. 28166201_Downregulation of PCK2 remodels tricarboxylic acid cycle in tumor-repopulating cells of melanoma 29731064_Our study indicates that the T allele of the rs4982856 single-nucleotide polymorphisms in the PCK2 gene may be a risk factor for glucose intolerance after kidney transplantation 31574189_This study unveiled a novel role for PCK2 in integrating autophagy and bone formation, providing a potential target for stem cell-based bone tissue engineering that may lead to improved therapies for metabolic bone diseases. 32036699_The relationship between PEPCK metabolism in the urinary tract and insulin resistance and diabetes is reported. 32045650_Hypoxia increases the rate of renal gluconeogenesis via hypoxia-inducible factor-1-dependent activation of phosphoenolpyruvate carboxykinase expression. 33052225_Restoring the epigenetically silenced PCK2 suppresses renal cell carcinoma progression and increases sensitivity to sunitinib by promoting endoplasmic reticulum stress. 33142842_PURalpha Promotes the Transcriptional Activation of PCK2 in Oesophageal Squamous Cell Carcinoma Cells. 33893083_Proteomic Analysis of Hepatocellular Carcinoma Tissues With Encapsulation Shows Up-regulation of Leucine Aminopeptidase 3 and Phosphoenolpyruvate Carboxykinase 2. 34020084_Phosphoenolpyruvate carboxykinase in cell metabolism: Roles and mechanisms beyond gluconeogenesis. 34520823_PCK2 opposes mitochondrial respiration and maintains the redox balance in starved lung cancer cells. 34650006_A novel mutation in PCK2 gene causes primary angle-closure glaucoma. | ENSMUSG00000040618 | Pck2 | 1.408955e+03 | 1.3484276 | 0.431278078 | 0.3091129 | 1.936633e+00 | 0.1640346018 | 0.77593452 | No | Yes | 1.612684e+03 | 209.377184 | 1.035878e+03 | 138.538557 |
| ENSG00000100968 | 4776 | NFATC4 | protein_coding | Q14934 | FUNCTION: Ca(2+)-regulated transcription factor that is involved in several processes, including the development and function of the immune, cardiovascular, musculoskeletal, and nervous systems (PubMed:7749981, PubMed:11514544, PubMed:11997522, PubMed:17875713, PubMed:17213202, PubMed:18668201, PubMed:25663301). Involved in T-cell activation, stimulating the transcription of cytokine genes, including that of IL2 and IL4 (PubMed:7749981, PubMed:18668201, PubMed:18347059). Along with NFATC3, involved in embryonic heart development. Involved in mitochondrial energy metabolism required for cardiac morphogenesis and function (By similarity). Transactivates many genes involved in the cardiovascular system, including AGTR2, NPPB/BNP (in synergy with GATA4), NPPA/ANP/ANF and MYH7/beta-MHC (By similarity). Involved in the regulation of adult hippocampal neurogenesis. Involved in BDNF-driven pro-survival signaling in hippocampal adult-born neurons. Involved in the formation of long-term spatial memory and long-term potentiation (By similarity). In cochlear nucleus neurons, may play a role in deafferentation-induced apoptosis during the developmental critical period, when auditory neurons depend on afferent input for survival (By similarity). Binds to and activates the BACE1/Beta-secretase 1 promoter, hence may regulate the proteolytic processing of the amyloid precursor protein (APP) (PubMed:25663301). Plays a role in adipocyte differentiation (PubMed:11997522). May be involved in myoblast differentiation into myotubes (PubMed:17213202). Binds the consensus DNA sequence 5'-GGAAAAT-3' (Probable). In the presence of CREBBP, activates TNF transcription (PubMed:11514544). Binds to PPARG gene promoter and regulates its activity (PubMed:11997522). Binds to PPARG and REG3G gene promoters (By similarity). {ECO:0000250|UniProtKB:D3Z9H7, ECO:0000250|UniProtKB:Q8K120, ECO:0000269|PubMed:11514544, ECO:0000269|PubMed:11997522, ECO:0000269|PubMed:17213202, ECO:0000269|PubMed:17875713, ECO:0000269|PubMed:18347059, ECO:0000269|PubMed:18668201, ECO:0000269|PubMed:25663301, ECO:0000269|PubMed:7749981, ECO:0000305}. | 3D-structure;Activator;Alternative splicing;Cytoplasm;DNA-binding;Developmental protein;Differentiation;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a member of the nuclear factor of activated T cells (NFAT) protein family. The encoded protein is part of a DNA-binding transcription complex. This complex consists of at least two components: a preexisting cytosolic component that translocates to the nucleus upon T cell receptor stimulation and an inducible nuclear component. NFAT proteins are activated by the calmodulin-dependent phosphatase, calcineurin. The encoded protein plays a role in the inducible expression of cytokine genes in T cells, especially in the induction of interleukin-2 and interleukin-4. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]. | hsa:4776; | chromatin [GO:0000785]; cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; peroxisome proliferator activated receptor binding [GO:0042975]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; brain-derived neurotrophic factor receptor signaling pathway [GO:0031547]; branching involved in blood vessel morphogenesis [GO:0001569]; calcineurin-NFAT signaling cascade [GO:0033173]; cellular respiration [GO:0045333]; cellular response to lithium ion [GO:0071285]; cellular response to UV [GO:0034644]; heart development [GO:0007507]; inflammatory response [GO:0006954]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; long-term memory [GO:0007616]; long-term synaptic potentiation [GO:0060291]; muscle cell development [GO:0055001]; negative regulation of chromatin binding [GO:0035562]; negative regulation of dendrite morphogenesis [GO:0050774]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of pri-miRNA transcription by RNA polymerase II [GO:1902894]; negative regulation of protein binding [GO:0032091]; negative regulation of synapse maturation [GO:2000297]; negative regulation of Wnt signaling pathway [GO:0030178]; positive regulation of apoptotic signaling pathway [GO:2001235]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of tumor necrosis factor production [GO:0032760]; regulation of transcription by RNA polymerase II [GO:0006357]; smooth muscle cell differentiation [GO:0051145] | 12939651_Observational study of gene-disease association. (HuGE Navigator) 12939651_Polymorphism of the NFATC4 gene plays a role in the development of human cardiac hypertrophy. 16219765_NFAT3 may play a role in estrogen receptor signaling in breast cancer cells 16489119_This study demonstrates that silica was able to activate NFAT3 in an oxygen radical-dependent manner, which was required for TNF-alpha induction. 16645724_ionizing radiation is able to enhance cyclin D1 transcription induced by B[a]PDE, and NFAT3 is involved in the regulation of cyclin D1 transcription by B[a]PDE or B[a]PDE plus ionizing radiation 17044076_Neuronal nuclear translocation of NFAT requires a functional cytoskeleton. 17194453_overexpression of NFAT3 in cell lines originated from kidney decreased dose-dependently both ERalpha and ERbeta transcriptional activities in a ligand-independent manner. 17213202_RSK2 is an important kinase for NFAT3 in mediating myotube differentiation 17875713_Results provided direct evidence for the anti-oncogenic potential of the NFAT3 transcription factor. 18034994_Data show that the calcineurin pathway is activated in hypertrophic myocardium as demonstrated by increased calcineurin activity and expression of calcineurin A-beta and B, and GATA-4, and a shift of cytoplasmic NFAT-3 into the nucleus. 18668201_estrogen receptor may play a critical role in regulation of NFAT3 transcriptional activity 19653005_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19784808_Data show that inhibition of NFAT3 activation by shNFAT3 significantly downregulated tumor necrosis factor (TNF)-alpha induction, its receptor TNFR1, caspase 10, caspase 3, and poly (ADP-ribose) polymerase. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20101218_an earlier unknown NFAT3/LCN2 axis that critically controls motility in breast cancer 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21047202_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21081043_The expression of COX-2 was significantly associated with the expressions of transcription factors NFAT3 and c-Fos in nonsmall cell lung cancer. 22164265_Syndecan-4 is essential for development of concentric myocardial hypertrophy via stretch-induced activation of the calcineurin-NFAT pathway 22234350_Polymorphisms in the NFATc4 gene may confer certain protection or predisposition for new-onset diabetes after transplantation (NODAT). 22378890_Dendritic spine loss and dendritic branching simplification induced by amyloid-beta peptide exposure are mimicked by constitutively active NFATC4, and abolished when NFATC4 activation is blocked by the genetically encoded inhibitor VIVIT. 22977251_Data indicate that NFATc3 undergoes rapid dephosphorylation and nuclear translocation that are essentially complete within 20 min, although NFATc4 remains phosphorylated and localized to the cytosol. 23888774_Expression level of PPP3R1 and GATA4, and NFATC4 genes for transcription factors did not differ in studied subgroups of patients. 24257415_This is the first study to provide evidence of new and differential roles for NFAT3 and SMAD3 in the osteoarthritis process in the regulation of miR-140 transcription 25422138_Suggest nuclear NF-AT3 and NF-AT4 participates in atrial structural remodeling, and that PICP and TGF-beta1 levels may be sensitive serum biomarkers to estimate atrial structural remodeling with atrial fibrillation. 25514788_NFAT3 expression plays a role in regulating CXCR4 expression. 26527057_Data indicate that RNA interference of NFAT isoforms NFATc1, NFATc2, NFATc3 and NFATc4 regulate gene expression differentially in human retinal microvascular endothelial cells (HRMEC). 27307045_Ca(2+)/calcineurin (CaN)/nuclear factor of activated T-cells (NFAT) c4 axis is required for neuritin-induced Kv4.2 transcriptional expression and potentiation of IA densities in cerebellum granule neurons. 27893713_These results provided evidence supporting the oncogenic potential of NFAT3 and suggested that CDK3-mediated phosphorylation of NFAT3 has an important role in skin tumorigenesis. 28760926_NFATc1 knockdown strongly reduced the number and the surface area of myotubes, NFATc4 knockdown increased the surface area of myotubes and reduced the pool of reserve cells. 29180489_TBX5 deficiency-mediated downregulation of NFAT3 is crucial for the high cytokine-producing activity of T cells 32182216_NFATC4 promotes quiescence and chemotherapy resistance in ovarian cancer. 32805187_NULP1 Alleviates Cardiac Hypertrophy by Suppressing NFAT3 Transcriptional Activity. | ENSMUSG00000023411 | Nfatc4 | 3.872916e+02 | 1.0532609 | 0.074862864 | 0.3011029 | 6.242956e-02 | 0.8026963224 | 0.96141221 | No | Yes | 2.985339e+02 | 45.913174 | 3.136151e+02 | 49.886225 | |
| ENSG00000100997 | 26090 | ABHD12 | protein_coding | Q8N2K0 | FUNCTION: Lysophosphatidylserine (LPS) lipase that mediates the hydrolysis of lysophosphatidylserine, a class of signaling lipids that regulates immunological and neurological processes (PubMed:25290914, PubMed:30237167, PubMed:30420694, PubMed:30720278, PubMed:30643283). Represents a major lysophosphatidylserine lipase in the brain, thereby playing a key role in the central nervous system (By similarity). Also able to hydrolyze oxidized phosphatidylserine; oxidized phosphatidylserine is produced in response to severe inflammatory stress and constitutes a proapoptotic 'eat me' signal (PubMed:30643283). Also has monoacylglycerol (MAG) lipase activity: hydrolyzes 2-arachidonoylglycerol (2-AG), thereby acting as a regulator of endocannabinoid signaling pathways (PubMed:22969151, PubMed:24027063). Has a strong preference for very-long-chain lipid substrates; substrate specificity is likely due to improved catalysis and not improved substrate binding (PubMed:30237167). {ECO:0000250|UniProtKB:Q99LR1, ECO:0000269|PubMed:22969151, ECO:0000269|PubMed:24027063, ECO:0000269|PubMed:25290914, ECO:0000269|PubMed:30237167, ECO:0000269|PubMed:30420694, ECO:0000269|PubMed:30643283, ECO:0000269|PubMed:30720278}. | Alternative splicing;Cataract;Deafness;Disease variant;Endoplasmic reticulum;Glycoprotein;Hydrolase;Lipid metabolism;Membrane;Neuropathy;Reference proteome;Retinitis pigmentosa;Transmembrane;Transmembrane helix | This gene encodes an enzyme that catalyzes the hydrolysis of 2-arachidonoyl glycerol (2-AG), the main endocannabinoid lipid transmitter that acts on cannabinoid receptors, CB1 and CB2. The endocannabinoid system is involved in a wide range of physiological processes, including neurotransmission, mood, appetite, pain appreciation, addiction behavior, and inflammation. Mutations in this gene are associated with the neurodegenerative disease, PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract), resulting from an inborn error of endocannabinoid metabolism. Alternatively spliced transcript variants encoding different isoforms have been noted for this gene.[provided by RefSeq, Jan 2011]. | hsa:26090; | AMPA glutamate receptor complex [GO:0032281]; dendrite cytoplasm [GO:0032839]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; acylglycerol lipase activity [GO:0047372]; lysophospholipase activity [GO:0004622]; palmitoyl-(protein) hydrolase activity [GO:0008474]; phospholipase activity [GO:0004620]; acylglycerol catabolic process [GO:0046464]; adult walking behavior [GO:0007628]; arachidonic acid metabolic process [GO:0019369]; glycerophospholipid catabolic process [GO:0046475]; monoacylglycerol catabolic process [GO:0052651]; phosphatidylserine catabolic process [GO:0006660]; phospholipid catabolic process [GO:0009395]; protein depalmitoylation [GO:0002084]; regulation of inflammatory response [GO:0050727]; response to auditory stimulus [GO:0010996] | 20797687_Mutations in the ABHD12 gene cause polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract disease. 22938382_After the identification of the ABHD12 mutation in this family, one patient underwent neurological examination which revealed ataxia, but no polyneuropathy 22969151_Data show that the three hydrolases are genuine MAG lipases; medium-chain saturated MAGs were the best substrates for hABHD6 and hMAGL, whereas hABHD12 preferred the 1 (3)- and 2-isomers of arachidonoylglycerol. 23490117_ABHD12 mutations are not a frequent cause of ataxia at least in Southern Italy 24027063_This is the first report of compound heterozygosity in PHARC and the first study to describe how a mutation might affect ABHD12 expression and function. 24697911_Null mutations in the ABHD12 gene lead to PHARC syndrome, a neurodegenerative disease including polyneuropathy, hearing loss, cerebellar ataxia, RP, and early-onset cataract. Our study allowed us to report 5 new mutations in ABHD12. 25743180_ABHD12 mutation in 2 PHARC (Polyneuropathy, Hearing Loss, Ataxia, Retinitis Pigmentosa, And Cataract)patients who suffered from deaf-blindness and dysfunctional central and peripheral nervous systems. 27890673_Study identified a new missense mutation, p.T253R, in ABHD12, which is functionally linked to the neurodegenerative disease PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and early-onset cataract), as demonstrated by its deactivation of monoacylglycerol lipase activity and inability to rescue zebrafish abhd12 knockdown phenotypes. 28448692_This study presented the various mutation of ABHD12 responsible for PHARC syndrome. 30237167_ABHD12 is enriched on the endoplasmic reticulum membrane, where most of the very-long-chain fatty acids are biosynthesized in cells. 30974196_Findings in Usher Syndrome predicted the potential function of this gene in the development of hearing and vision loss, particularly with regard to impaired signal transmission, and identified a novel nonsense variant to expand the variant spectrum in ABHD12. 32366405_ABHD12 plays a crucial role in cell proliferation, migration, and invasion of breast cancer cells. 32462874_Mapping the Neuroanatomy of ABHD16A, ABHD12, and Lysophosphatidylserines Provides New Insights into the Pathophysiology of the Human Neurological Disorder PHARC. 34223797_Expanding the clinical phenotype in patients with disease causing variants associated with atypical Usher syndrome. 34573385_The Phenotypic Spectrum of Patients with PHARC Syndrome Due to Variants in ABHD12: An Ophthalmic Perspective. | ENSMUSG00000032046 | Abhd12 | 3.031873e+03 | 1.1091910 | 0.149507781 | 0.3047582 | 2.379761e-01 | 0.6256716931 | 0.91638726 | No | Yes | 2.801411e+03 | 263.969404 | 2.373286e+03 | 229.840761 | |
| ENSG00000101266 | 1457 | CSNK2A1 | protein_coding | P68400 | FUNCTION: Catalytic subunit of a constitutively active serine/threonine-protein kinase complex that phosphorylates a large number of substrates containing acidic residues C-terminal to the phosphorylated serine or threonine (PubMed:11239457, PubMed:11704824, PubMed:16193064, PubMed:19188443, PubMed:20625391, PubMed:22406621, PubMed:24962073). Regulates numerous cellular processes, such as cell cycle progression, apoptosis and transcription, as well as viral infection (PubMed:12631575, PubMed:19387552, PubMed:19387551). May act as a regulatory node which integrates and coordinates numerous signals leading to an appropriate cellular response (PubMed:12631575, PubMed:19387552, PubMed:19387551). During mitosis, functions as a component of the p53/TP53-dependent spindle assembly checkpoint (SAC) that maintains cyclin-B-CDK1 activity and G2 arrest in response to spindle damage (PubMed:11704824, PubMed:19188443). Also required for p53/TP53-mediated apoptosis, phosphorylating 'Ser-392' of p53/TP53 following UV irradiation. Can also negatively regulate apoptosis (PubMed:11239457). Phosphorylates the caspases CASP9 and CASP2 and the apoptotic regulator NOL3 (PubMed:16193064). Phosphorylation protects CASP9 from cleavage and activation by CASP8, and inhibits the dimerization of CASP2 and activation of CASP8 (PubMed:16193064). Regulates transcription by direct phosphorylation of RNA polymerases I, II, III and IV. Also phosphorylates and regulates numerous transcription factors including NF-kappa-B, STAT1, CREB1, IRF1, IRF2, ATF1, ATF4, SRF, MAX, JUN, FOS, MYC and MYB (PubMed:19387550, PubMed:12631575, PubMed:19387552, PubMed:19387551, PubMed:23123191). Phosphorylates Hsp90 and its co-chaperones FKBP4 and CDC37, which is essential for chaperone function (PubMed:19387550). Mediates sequential phosphorylation of FNIP1, promoting its gradual interaction with Hsp90, leading to activate both kinase and non-kinase client proteins of Hsp90 (PubMed:30699359). Regulates Wnt signaling by phosphorylating CTNNB1 and the transcription factor LEF1 (PubMed:19387549). Acts as an ectokinase that phosphorylates several extracellular proteins (PubMed:19387550, PubMed:12631575, PubMed:19387552, PubMed:19387551). During viral infection, phosphorylates various proteins involved in the viral life cycles of EBV, HSV, HBV, HCV, HIV, CMV and HPV (PubMed:19387550, PubMed:12631575, PubMed:19387552, PubMed:19387551). Phosphorylates PML at 'Ser-565' and primes it for ubiquitin-mediated degradation (PubMed:20625391, PubMed:22406621). Plays an important role in the circadian clock function by phosphorylating ARNTL/BMAL1 at 'Ser-90' which is pivotal for its interaction with CLOCK and which controls CLOCK nuclear entry (By similarity). Phosphorylates CCAR2 at 'Thr-454' in gastric carcinoma tissue (PubMed:24962073). {ECO:0000250|UniProtKB:P19139, ECO:0000269|PubMed:11239457, ECO:0000269|PubMed:11704824, ECO:0000269|PubMed:16193064, ECO:0000269|PubMed:19188443, ECO:0000269|PubMed:20625391, ECO:0000269|PubMed:22406621, ECO:0000269|PubMed:23123191, ECO:0000269|PubMed:24962073, ECO:0000269|PubMed:30699359, ECO:0000303|PubMed:12631575, ECO:0000303|PubMed:19387549, ECO:0000303|PubMed:19387550, ECO:0000303|PubMed:19387551, ECO:0000303|PubMed:19387552}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Biological rhythms;Cell cycle;Disease variant;Kinase;Mental retardation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase;Wnt signaling pathway | Casein kinase II is a serine/threonine protein kinase that phosphorylates acidic proteins such as casein. It is involved in various cellular processes, including cell cycle control, apoptosis, and circadian rhythm. The kinase exists as a tetramer and is composed of an alpha, an alpha-prime, and two beta subunits. The alpha subunits contain the catalytic activity while the beta subunits undergo autophosphorylation. The protein encoded by this gene represents the alpha subunit. Multiple transcript variants encoding different protein isoforms have been found for this gene. [provided by RefSeq, Apr 2018]. | hsa:1457; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; NuRD complex [GO:0016581]; plasma membrane [GO:0005886]; protein kinase CK2 complex [GO:0005956]; Sin3 complex [GO:0016580]; ATP binding [GO:0005524]; Hsp90 protein binding [GO:0051879]; identical protein binding [GO:0042802]; kinase activity [GO:0016301]; protein N-terminus binding [GO:0047485]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; apoptotic process [GO:0006915]; cell cycle [GO:0007049]; chaperone-mediated protein folding [GO:0061077]; negative regulation of apoptotic signaling pathway [GO:2001234]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of ubiquitin-dependent protein catabolic process [GO:2000059]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of cell growth [GO:0030307]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of protein catabolic process [GO:0045732]; positive regulation of Wnt signaling pathway [GO:0030177]; protein phosphorylation [GO:0006468]; regulation of cell cycle [GO:0051726]; regulation of chromosome separation [GO:1905818]; rhythmic process [GO:0048511]; signal transduction [GO:0007165]; Wnt signaling pathway [GO:0016055] | 11756447_uPA-dependent VSMC adhesion is a function of selective Vn phosphorylation by the ectoprotein kinase CK2 11827164_Generation of mutants of CK2alpha which are dependent on the beta-subunit for catalytic activity 11827166_HIV-1 Rev transactivator: a beta-subunit directed substrate and effector of protein kinase CK2 11827167_Protein kinase CK2: signaling and tumorigenesis in the mammary gland. 11827168_Response of cancer cells to molecular interruption of the CK2 signal. 11827170_Functional specialization of CK2 isoforms and characterization of isoform-specific binding partners 11827171_Characterization of CK2 holoenzyme variants with regard to crystallization 11827174_Transcriptional coordination of the genes encoding catalytic (CK2alpha) and regulatory (CK2beta) subunits of human protein kinase CK2. 11827176_Consequences of CK2 signaling to the nuclear matrix. 11827177_Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus 11940573_Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. 11956194_Unique activation mechanism of protein kinase CK2. The N-terminal segment is essential for constitutive activity of the catalytic subunit but not of the holoenzyme 12037680_Protein kinase CK2 dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with beta-TRCp and enhances beta-catenin degradation [UBC3B] 12145206_FGF-1 binds to both the catalytic alpha-subunit and to the regulatory beta-subunit of CK2. FGF-1 & CK2 alpha are shown to interact in vivo. A correlation between the mitogenic potential of FGF-1 mutants & their ability to bind to CK2 alpha was observed. 12568341_Data point to a particular role of the catalytic alpha and alpha' subunits of protein kinase CK2, which may be different from their roles in the holoenzyme. 12628923_This protein is associated with the COP9 signalosome. 12659875_HSF1 activation by heat is correlated with the thermal activation of nuclear CK2 and overexpression of CK2 activates HSF1 12748192_results indicate that protein kinase CKII may control IkappaBalpha and p27Kip1 degradation and thereby G1/S phase transition through the phosphorylation of threonine 10 within CKBBP1 protein 12860116_Crystal structure of a C-terminal deletion mutant of human protein kinase CK2 catalytic subunit 14962846_Evaluation of different pathways involved in death signaling suggest that the regulation of a critical proapoptotic step in HuH-7 cells by CK2alpha' is mediated by a JNK signaling cascade. 15108354_structure activity relationship: the PA 382-384 mutant exhibits an increased thermal and proteolytic stability 15218032_might play an important role in vivo in regulating the function and transport activity of ABCA1 and possibly of other members of the ABCA subfamily 15287743_Utilizing a kinase-driven assay biochemical purification, the authors identified casein kinase II (CKII) from HeLa cell nuclear extract as a cellular phosphoprotein pp32 kinase. 15342635_CK2 acts as an inhibitor of Cdk5 in the brain 15557471_Involvement of ubiquitous protein kinase CK2 in angiogenesis. Naturally derived CK2 inhibitors may be useful for treatment of proliferative retinopathies. 15659405_CK2 regulates the DNA-binding ability of SSRP1 and that this regulation may be responsive to specific cell stresses. 15740749_Data demonstrate that CK2alpha possesses sophisticated structural adaptations in favour of dual-cosubstrate specificity. 15818404_constitutive phosphorylation by CK2 may be required for maximal activation of Akt/PKB 15955816_CK2 may have the capacity to differentially regulate U1 and U6 transcription even though SNAP(C) is universally utilized for human snRNA gene transcription 16107342_multiple kinases, including CK2 and GSK3beta, participate in PTEN phosphorylation and GSK3beta may provide feedback regulation of PTEN 16133877_Protein kinase CK2 is characterized by an extremely high stability that might be due to its association with other intracellular proteins, enhanced half-life or lower vulnerability towards proteolytic degradation. 16157582_protein kinase CK2 is inactive in CCVs because of the fact that it is bound to the clathrin-coated vesicle (CCV) membrane via an interaction between phosphatidylinositol 4,5-bisphosphate in the CCV membrane and the active site in CK2 16225457_In vitro phosphorylation of eIF2beta also pointed to Ser2 as a preferred site for CK2 phosphorylation 16227438_CK2 may be involved in the regulation of cell cycle progression by associating with and phosphorylating a key molecule for translation initiation. 16308564_casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia 16342410_CK2 is potentially a highly plausible target for cancer therapy. 16426576_phosphorylation of the (T300) residue is dependent on CK2 and is a necessary and functional prerequisite for TSPY's transport into the nucleus 16540521_Dynamics of nucleolar reformation is ATP/GTP-dependent, sensitive to temperature, and creatin kinase-2 driven. 16581776_CK2 regulates NKX3-1 in prostate cells. 16651637_Based on its retinal localization, CK2 may be considered a new immunohistochemical astrocytic marker, and combination of CK2 inhibitors and octreotide may be a promising future treatment for proliferative retinopathies. 16751801_role of CK2beta for cell survival and might allow the design of novel proapoptotic agents targeting this protein 16806200_CK2 activity in Chinese Hamster Ovary (CHO) cells is entirely accounted for by the holoenzyme. 16837460_v-Src-dependent down-regulation of the Ste20-like kinase SLK through casein kinase II. 16874460_These studies suggested that CK2 might regulate SAG-SCF E3 ligase activity through modulating SAG's stability, rather than its enzymatic activity directly. 16880508_CK2 has the potential to regulate Pol I transcription at multiple levels, in preinitiation complex (PIC) formation, activation, and reinitiation of transcription. 17009010_CK2 plays an active role in NF-kappaB signaling in intestinal epithelial cell lines and may represent a possible target for intervention. 17106255_CK2-site phosphorylation of p53 is induced in DeltaNp63 expressing basal stem cells in UVB irradiated human skin 17113388_CK2 is required for the assembly and cycling of Wnt-enhancer complexes in vivo. 17524418_Results describe the 1.6 A resolution crystal structure of a fully active C-terminal deletion mutant of human CK2alpha liganded by two sulfate ions, and compare this structure systematically with representative structures of related CMGC kinases. 17545155_extracellular phosphorylation of collagen XVII by ecto-CK2 inhibits its shedding by TACE and represents novel mechanism to regulate adhesion and motility of epithelial cells 17629615_Suggest a novel role of CK2 in breast cancer resistance to antiestrogens. 17636126_cytotoxic adaptor function of NS1 was confirmed with fusion peptides, where the tropomyosin-binding domain of NS1 and CKIIalpha are physically linked 17670747_CK2 phosphorylation triggers an allosteric inhibition of the SNAP190 Myb DNA binding domain 17699575_EB2-mediated production of infectious EBV virions is regulated by CK2 phosphorylation at one or more of the serine residues Ser-55, -56, and -57 17894550_ABC50 N-terminal region interacts with eukaryotic initiation factor eIF2 and is a target for regulatory phosphorylation by CK2 17923166_The number of predicted CK2 sites correlated with the degree of in vitro and in vivo phosphorylation of NS5A by CK2. CK2-dependent phosphorylation of NS5A is heterogeneous among different HCV genotypes and clinical isolates. 17935135_CK2 may be a key mediator for HDAC1 and HDAC2 activation under hypoxia in tumor cells 18026141_splicing activity is significantly influenced by the CK2-hPrp3p interaction 18062282_There was a positive correlation between the expressions of CK2alpha and CK2beta in laryngeal squamous cell carcinoma. 18089732_Human herpesvirus 6Binduces p53 Ser392 phosphorylation by an atypical pathway independent of CK2 protein and p38 kinases. 18242640_The complex structure between a C-terminal deletion mutant of human CK2alpha and the ATP-competitive inhibitor emodin is studies in comparison with with a previously published complex structure of emodin and maize CK2alpha. 18347021_CK2-mediated phosphorylation of TRF1 plays an important role in modulating telomere length homeostasis by determining the levels of TRF1 at telomeres 18363813_Protease activated receptor 1 activation of platelet is associated with an increase in protein kinase CK2 activity 18548200_CK2 is tightly associated with TAFII250 and is the principal activity responsible for TAFII250-mediated phosphorylation of Mdm2. 18553055_Studies in yeast and in human cells demonstrate that the different forms of CK2 interact with a large number of cellular proteins. 18553059_Transcript amounts of the subunits CK2alpha and CK2beta and holoenzyme CK2 activity in 34 muscle biopsies of human patients with different muscle pathologies, were determined. 18560764_CK2 involvement in the phenomenon of the drug resistance was investigated. 18563533_Casein kinase II activity is increased by the binding of haptoglobin 1-1-hemoglobin to CD163. 18566753_CK2 activity was investigated both in vitro and in cultured cells by using anti-[pSer13]-Cdc37 antibody. 18566754_These data identify a key post-translational mechanism that controls PML protein levels in cancer cells and suggest that CK2 inhibitors may be beneficial anti-cancer drugs. 18574673_These results suggest that one of the modes of CK2-mediated modulation of apoptotic activity is via its impact on cellular inhibitors of apoptosis proteins. 18607692_A crystal of a C-terminally truncated variant of human CK2alpha to which two molecules of the inhibitor 5,6-dichloro-1-beta-D-ribo-furanosyl-benzimidazole (DRB) are bound, is analyzsed. 18614797_CK2 binds to the NHE3 C terminus and stimulates basal NHE3 activity by phosphorylating a separate single site on the NHE3 C terminus 18649047_CK2 may be involved in the regulation of cell cycle progression through the phosphorylation of a key molecule for translation initiation and of nuclear substrates upon activation of CK2 by itself. 18662771_CK2 catalyzes the phosphorylation of tyrosine residues in mammalian cells. 18682247_KIF5C, a member of the kinesin 1 heavy chain family, is a substrate for protein kinase CK2. 18710613_Mitoxantrone is a strong inhibitor of recombinant human protein kinase CK2 in vitro. Apoptosis induced by mitoxantrone in HL-60 cells has no correlation to intracellular CK2 activity. 18790693_InsP(6) specifically binds to CK2alpha and disrupts the interaction between CK2alpha and Nopp140 with an IC(50) value of 25 microM, thereby attenuating the Nopp140-mediated repression of CK2 activity. 18824508_study presents unbound three-dimensional structure of a CK2beta construct that is fully capable of CK2alpha recruitment and quantify its affinity to CK2alpha thermodynamically 19027835_DNA methylation-dependent down-regulation of transcription factors Sp1, Ets1 and NF-kappaB might be involved in silencing of the CKII alpha and CKII alpha' genes during cellular senescence. 19027835_DNA methylation-dependent down-regulation of transcription factors Sp1, Ets1 and NF-kappaB might be involved in silencing of the CKII alpha and CKII alpha' genes during cellular senescence.[CKII alpha'] 19035320_PrP protein can bind to protein kinase CK2 both in native and recombinant forms in vitro. 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19282287_Ikaros is controlled by the CK2 and PP1 pathways and that a balance between these two signal transduction pathways is essential for normal cellular function and for the prevention of malignant transformation. 19361447_CK2beta dimer stabilizes the glycine-rich loop in the extended active conformation known from the majority of CK2alpha structures. 19398558_protein kinase CK2 phosphorylates Ser(361) on Sgt1, and this phosphorylation inhibits Sgt1 dimerization 19486891_Extracellular protein kinase CK2 binds to the extracellular domain of NRP1 which is also phosphorylated by CK2 both in vitro and in vivo. 19542537_CK2-dependent phosphorylation controls the nuclear localization, aggregation and stability of ataxin-3. 19556345_These studies reveal multisite phosphorylation of IGFBP-3 that both positively and negatively regulate its apoptotic potential. 19596613_The constitutive phosphorylation of XRCC1 in the chromatin and its DNA damage-induced recruitment to the nuclear matrix are critical for foci formation. 19616514_PDCD5 is phosphorylated in vitro by both CK2alpha subunit and by the CK2. 19723109_Data show that decreasing CK2 activity increases intracellular hydrogen peroxide, creating an intracellular environment conducive for death execution. 19726289_Protein kinase CK2 plays an important role in the radiosensitivity of the nasopharyngeal carcinoma cells. 19839995_Observational study of gene-disease association. (HuGE Navigator) 19855935_This study shows that the activation of the p53-p21(Cip1/WAF1) pathway acts as a major mediator of cellular senescence induced by CKII inhibition. 19904978_CK2 is the kinase responsible for phosphorylating Pax3 and oncogenic fusion protein Pax3-FOXO1 at serine205 in proliferating mouse primary myoblasts and in a variety of translocation-containing alveolar rhabdomyosarcoma cell lines. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19923321_These results highlight serines 11 and 92 as new players in Snail1 regulation and suggest the participation of CK2 and PKA in the modulation of Snail1 functionality. 19933278_MSK1 and MSK2 are differentially regulated by CK2 during the UV response and that MSK2 is the major protein kinase responsible for the UV-induced phosphorylation of p65 at Ser(276) that positively regulates NF-kappaB activity in MDA-MB-231 cells 19941816_The EGFR-ERK-CK2-mediated phosphorylation of alpha-catenin promotes beta-catenin transactivation and tumor cell invasion. 20021963_Protein kinase CK2alpha plays an important role in the proliferation and invasion of nasopharyngeal carcinoma. 20026644_CK-II-dependent PPARgamma phosphorylation at Ser16 and Ser21 is necessary for CRM1/Ran/RanBP3-mediated nucleocytoplasmic translocation of PPARgamma. 20157113_CK2-mediated pathways reversibly regulate purinosome assembly 20356928_The interface between apoptosis initiation and execution by determining caspase-8 activation, Bid cleavage and mitochondrial engagement (onset of mitochondrial depolarisation) in individual HeLa, was analysed. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20526659_These data show that Tax-1 is phosphorylated in vitro by the pleiotropic human serine/threonine kinase CK2 at three residues, Ser-336, Ser-344 and Thr-351, close to and within its C-terminal PDZ-binding motif. 20545769_CK2 plays a crucial role in the ATR-dependent checkpoint pathway through its ability to phosphorylate Ser-341 and Ser-387 of the Rad9 subunit of the 9-1-1 complex. 20610541_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20625391_Data show that the CSNK2A1P gene is a functional proto-oncogene in human cancers and its functional polymorphism appears to degrade PML differentially in cancer cells. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20664522_Herein, the authors have identified polo-like kinase 1 (Plk1) and casein kinase 2 (CK2) as two kinases of CLIP-170 and mapped S195 and S1318 as their respective phosphorylation sites. 20671611_The prognostic significance of the genes casein kinase 2 alpha subunit (CSNK2A1), anti-apoptosis clone-11 (AAC-11), and tumor metastasis suppressor NME1 in completely resected non-small cell lung cancer (NSCLC) patients, was analysed. 20711232_These results indicate that CK2 has an important role in the modulation of DNA-PKcs activity and its phosphorylation status providing important insights into the mechanisms by which DNA-PKcs is regulated in vivo. 20718998_Reduced Casein Kinase II enhances chemosensitivity to gemcitabine in pancreatic cancer. 20719947_an interaction between EBNA1 and the host CK2 kinase is crucial for EBNA1 to disrupt PML bodies and degrade PML proteins. 20807566_Protein kinase CK2 inhibition results in activation of the receptor mediated apoptosis pathway via the ER stress response highlight the importance of CK2 inhibitors. 20864032_The CK2 phospho-dependent interaction between TEL2 and the R2TP complex affects phosphatidylinositol 3-kinase-related kinase functions and is essential for mTOR and SMG1 stability in vivo. 21093442_Data presented the crystal structures of CK2alpha in complex with CX-4945 and adenylyl phosphoramidate at 2.7 and 1.3 A, respectively. 21162802_Protein kinase CK2alpha plays an important role in the apoptosis of human laryngeal carcinoma cells possibly by decreasing bcl-2/Bax. 21165564_Sox4 stimulates ss-catenin activity through induction of CK2. 21182307_CK2 is a central regulator of topo I hyperphosphorylation. 21270425_The effect of a mutation in the follicle-stimulating hormone receptor in a protein kinase-CK2 consensus site that alters degradation of follicle-stimulating hormone (FSH) is studied. 21359197_Overexpression of nuclear CK2alpha can be a useful marker for predicting the outcome of patients with colorectal cancer. 21486957_Findings identify CK2 as a crucial protein involved in the formation and clearance of aggresomes, and hence in cell viability in response to misfolded protein stress. 21518957_Data suggest that phospho-Ser81 progesterone receptors (PR) provides a platform for casein kinase 2 recruitment and regulation of selected PR-B target genes. 21527517_we report that the kinase CK2 is a novel interaction partner of JAKs and is essential for JAK-STAT activation 21559372_Twist expression can be regulated at the post-translational level through phosphorylation by CK2, which increases Twist stability in response to IL-6 stimulation 21559479_CK2 down regulation facilitates TNF-alpha-mediated chondrocyte death through apoptosis and autophagy 21576649_A novel mechanism in the development of cardiac hypertrophy is demonstrated when transgenic CK2alpha1 activates histone deacetylase 2 (HDAC2) via phosphorylating HDAC2 serine394. 21697493_Human papillomavirus type 38 E7 protein promotes keratinocyte proliferation by negatively regulating actin cytoskeleton fiber formation through the CK2-MEK-ERK-Rho pathway and by inhibiting eEF1A and its effects on actin cytoskeleton remodeling. 21702981_CK2alpha plays an essential role in the development of colorectal cancer. 21711111_CK2 inhibitors enhance the radiosensitivity of human non-small cell lung cancer cells through inhibition of stat3 activation 21735091_The Arabidopsis co-chaperone protein p23 is a CK2 target. 21735095_Persistence of high CK2 level in R (resistant)-CEM, as opposed to S (sensitive)-CEM, is accompanied by the presence of an immunospecific form of Cdc37. 21739153_In hsCK2alpha' an open conformation of the interdomain hinge/helix alphaD region that is critical for ATP-binding is found corresponding to an incomplete catalytic spine. In contrast hsCK2alpha often adopts the canonical, PKA-like version of the catalytic spine. 21739154_The nuclear localization signal sequence-dependent Grp94 kinase in the cell lysate is identical with CK2. 21750986_CK2 appears to play an important role in the biology of B-cell chronic lymphocytic leukemia. 21750987_Inhibition of CK2 activity by three different inhibitors led to a down-regulation of the level of cdc25C. 21750988_An interaction of the adiponectin receptor 1 with the tetrameric complex and protein kinase CK2 as a key player in adiponectin signaling, is reported. 21777522_These data suggest a novel cellular function of CK-2 as a transcriptional co-activator of AP-2alpha. 21871133_The primary mechanisms by which apigenin kill multiple myeloma cells is by targeting the trinity of CK2-Cdc37-Hsp90. 21936567_our results demonstrate that EEF1D is a bona fide physiological CK2 substrate for CK2 phosphorylation 21968188_CKII downregulation induces p53 stabilization by negatively regulating SIRT1 deacetylase activity during senescence 22027148_the pleotropic Serine/Threonine kinase CK2 is implicated in the regulation of IL-6 expression in a model of inflammatory breast cancer. 22184066_identify serine 118 in the transactivation domain of YY1 as the site of CK2alpha phosphorylation, proximal to a caspase 7 cleavage site 22186626_The unique flexibility of the CK2alpha catalytic subunit in the hinge region, the p-loop and the beta4 beta5 loop, show that there is no clear-cut correlation between the conformations of these flexible zones. 22190719_The human adenovirus type 5 E1B protein is phosphorylated by protein kinase CK2. 22209233_CK2 regulates vaccinia virus dissemination and actin tail formation. 22267120_The the CK2 catalytic subunit CK2alpha is modified by O-linked beta-N-acetyl-glucosamine (O-GlcNAc) on Ser347, proximal to a cyclin-dependent kinase phosphorylation site (Thr344). 22317921_The connection with proliferation and migration, as well as the activation of ZIP7 by CK2, a kinase that is antiapoptotic and promotes cell division 22351691_Results suggest CK2 as a novel regulator of the endoplasmic reticulum (ER) stress/unfolded protein response (UPR) cascades and HSP90 function in myeloma cells and offer the groundwork to design novel combination treatments for this disease. 22404984_Protein kinase CK2 co-localizes with gamma-H2AX to sites of genetic lesions and modulation of its expression levels affects the cellular DNA damage response. 22506723_A new crystal structure of the CK2 holoenzyme was determined that plays an essential role in the formation of inactive polymeric assemblies. 22562247_Unbalanced expression of CK2 subunits may drive epithelial-to-mesenchymal transition, thereby contributing to tumour progression. 22609407_ATF4 is involved in ER stress signalling through the AARE, which further supports our finding that CK2 inhibition provokes an amino acid induced response pathway. 22675025_CK2alpha-NCoR cascade selectively represses the transcription of IP-10 and promotes oncogenic signaling in human esophageal cancer cells. 22718630_Phosphorylation of S284 by protein kinase CK2 significantly decreases nuclear targeting of IPMK in HEK-293 cells. 22767590_that high intracellular PKCK2 activity confers anoikis resistance on esophageal cancer cells by inducing E- to N-cadherin switching. 22768056_The inhibition of CK2alpha down-regulates Hh/Gli signaling and subsequently reduces stem-like side population in human lung cancer cells. 22810236_Recovery of RNA polymerase III transcription from the glycerol-repressed state: revisiting the role of protein kinase CK2 in Maf1 phosphoregulation. 22814265_CK2 and human malignant tumor 22868753_analysis of crystal packing of human protein kinase CK2 catalytic subunit in complex with resorufin or other ligands 22904299_A CD5-CK2 signaling pathway represents a major signaling cascade initiated by CD5 that regulates the threshold of T cell activation and T helper (Th) cell differentiation. 23007634_our findings identify CK2 as a novel component of the autophagic machinery and underline the potential of its downregulation to kill glioblastoma cells by overcoming the resistance to multiple anticancer agents. 23017496_Functional proteomics strategy for validation of protein kinase inhibitors reveals new targets for a TBB-derived inhibitor of protein kinase CK2. 23137536_the present results show that miR-186, miR-216b, miR-337-3p, and miR-760 cooperatively promote cellular senescence through the p53-p21(Cip1/WAF1) pathway by CKII downregulation-mediated ROS production in HCT116 cells. 23153582_Casein kinase 2alpha inhibition decreases the total protein level of beta-catenin and cell growth. 23185622_CK2-mediated hyperphosphorylation of topoisomerase I targets serine 506, enhances topoisomerase I-DNA binding, and increases cellular camptothecin sensitivity. 23287549_Phosphorylation of Sec63 by CK2 enhanced its binding to Sec62 23317504_Brd4 association with p53 is modulated by casein kinase II (CK2)-mediated phosphorylation of a conserved acidic region in Brd4 23349870_These results suggest that Sec31 phosphorylation by CK2 controls the duration of COPII vesicle formation, which regulates ER-to-Golgi trafficking. 23360763_Data indicate that by altering the scaffold of CK2 inhibitors to obtain selective inhibitors of DYRK1A and PIM1. 23416530_Studies indicate systematic platforms for studying CK2 inhibitors. 23474121_crystal structure of CK2alpha with a 13-meric cyclic peptide derived from the C-terminal CK2beta segment 23492774_Activation of protein kinase CK2 attenuates FOXO3a functioning in a PML-dependent manner. 23523798_PI3K is involved in CKII inhibition-mediated cellular senescence in HCT116 cells. 23580615_Data indicate that protein S phosphorylation by casein kinase 1 (CK1) and casein kinase 2 (CK2) increased activated proten C cofactor activity. 23599180_CK2alpha' exhibits a striking preference for caspase-3 phosphorylation in cells as compared to CK2alpha and that CK2beta exhibits the capacity to abolish caspase-3 phosphorylation 23612983_Enzyme inhibition studies and mutational analyses demonstrated that protein kinase CK2-catalyzed phosphorylation of HDAC1 and -2 is crucial for the dissociation of these two enzymes. 23651443_inhibition of CK2alpha down-regulates Notch1 signalling and subsequently reduces a cancer stem-like cell population in human lung cancer cells 23732914_These results identify casein kinase 2 as a new target of PD-1 and reveal an unexpected mechanism by which PD-1 decreases PTEN protein expression while increasing PTEN activity, thereby inhibiting the PI3K/Akt signaling axis. 23816881_The study identifies casein kinase 2 as an NRF1-binding protein and finds that the knockdown of casein kinase 2 enhances the Nrf1-dependent expression of the proteasome subunit genes. 23885116_These findings identify CK2 as an upstream activating kinase of PAK1, providing a novel mechanism for PAK1 activation. 23979991_HspA1A also interacts with CK2 and enhances the kinase activities of CK2 during DNA repair. 24012109_knockdown of CK2alpha expression by siRNA restores the sensitivity of resistant LAMA84 cells to low imatinib concentrations. 24021586_upregulation of VEGF in primary RPE cells was blocked by a chemical inhibitor of protein kinase CK2 known to suppress induction of ATF4 and VEGF by OxPLs 24036851_The CSNK2A1 gene has gene dosage gains in glioblastoma, and is significantly associated with the classical glioblastoma subtype. 24098452_miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-alpha, CCNJ, and MEGF9. 24121351_crystal structure of CK2alpha with 21 loci of alternative conformations, including a niacin, 19 ethylene glycols and 346 waters, was determined at 1.06 A resolution to an Rwork of 14.0% 24175891_Asymmetric alpha2beta2 tetramers of casein kinase 2 are organized in trimeric rings that correspond to inactive forms of the enzyme. The new crystal structures presented here reveal the symmetric architecture of the isolated active tetramers. 24178769_The CFTR Ser511 can be phosphorylated by the combined action of tyrosine kinases and CK2 and disclose a new mechanism of hierarchical phosphorylation where the role of the priming kinase is that of removing negative determinants. 24218616_CK2 activity is inhibited by Nopp140 and reactivated by inositol hexakisphosphate by competitive binding at the substrate recognition site of CK2. 24268649_Phosphorylation of the CK2 site on SMRT significantly increased affinity for SHARP. 24297901_These data support a model in which activated Akt enhances rRNA synthesis both by preventing TIF-IA degradation and phosphorylating CK2alpha, which in turn phosphorylates TIF-IA. 24440309_Protein kinase CK2 alpha is involved in the phosphorylation of the ESCRT-III subunits CHMP3 and CHMP2B, as well as of VPS4B/SKD1, an ATPase that mediates ESCRT-III disassembly. 24457960_Phosphorylation by CK2 impairs Par-4 proapoptotic functions. 24486797_LSD1 is a novel substrate of protein kinase CK2alpha. 24616922_Data suggest that overexpression of CK2a (casein kinase 2, alpha) in liver promotes oncogenesis by mediating EGF/EGFR- (epidermal growth factor/EGF receptor)-induced HDAC2 (histone deacetylase 2) expression in hepatocarcinogenesis. 24686080_the inhibition of protein kinase CK2 transiently inhibits cell proliferation through the induction of G1 cell cycle arrest and attenuation of protein synthesis by phosphorylating eIF2 alpha. 24718935_HCV core up-regulates HAMP gene transcription via a complex signaling network that requires both SMAD/BMP and STAT3 pathways and CK2 involvement 24726840_Overexpression of CK2 (alpha, alpha' or beta subunits) by transient transfection resulted in decreased Stat3 Ser-727 phosphorylation. 24742922_we characterize CK2 as an essential component of the Jak/STAT pathway 24831064_Genetic suppression of CK2alpha enhances the apoptosis induced by cisplatin in laryngeal carcinoma cells. 24840952_Study revealved that RPL22 binds CK2alpha in lung cancer cells. 24897506_protein kinase CK2 has a role in bone marrow stromal cell-fueled multiple myeloma growth and osteoclastogenesis 24962073_CK2 alpha is an independent prognostic indicator for gastric carcinoma patients and is involved in tumorigenesis by regulating the phosphorylation of DBC1. 25052887_CagA(+) H. pylori upregulates cellular invasiveness and motility through casein kinase 2. 25120778_These findings highlight the potential role of dysregulated miRNA expression regulated by CK2 in breast cancer. 25241897_Results show that CK2alpha may have an important role in brain tumor-initiating cell maintenance through the regulation of beta-catenin in glioblastoma. 25379016_CK2 phosphorylates and inhibits TAp73 tumor suppressor function to promote expression of cancer stem cell genes and phenotype in head and neck cancer. 25422081_Over-expressed CK2alpha positively regulate Hh/Gli1 signaling in human mesothelioma. 25476899_JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. 25788269_CK2 is widely expressed in follicular, Burkitt and diffuse large B-cell lymphomas and may have role in malignant B-cell growth. 25805179_our findings provide new insights on the potential relevance of the CK2-mediated phosphorylation of B23/NPM in cancer cells, revealing at the same time the potentialities of its pharmacological manipulation for cancer therapy 25837326_CDK11 and CK2 expression are individually essential for breast cancer cell survival, including TNBC. 25872870_Data show that casein kinase 2 (CK2)-mediated phosphorylation of deubiquitylating enzyme OTUB1 at Ser16 causes nuclear accumulation of OTUB1. 25882195_These phosphopeptides include altogether 69 phosphoresidues, a large proportion of which (almost 50%) are generated by CK2, while the others do not conform to the CK2 consensus 25887626_CK2-phosphorylation of eIF3j at Ser127 promotes the assembly of the eIF3 complex, a crucial step in the activation of the translation initiation machinery. 25891901_Overall results, confirm that a balance of hydrophobic and electrostatic interactions contribute predominantly relative to possible intermolecular halogen/hydrogen bonding ,in binding of halogenated benzotriazoles to the ATP-binding site of hCK2alpha 25995454_Casein kinase 2-mediated phosphorylation of Hsp90beta and stabilization of PXR is a key mechanism in the regulation of MDR1 expression. 25998125_Phosphorylation of KCNQ2 and KCNQ3 anchor domains by protein kinase CK2 augments binding to AnkG. 26083323_ATG16L1 as a bona fide physiological CSNK2 and PPP1 substrate, which reveals a novel molecular link from CSNK2 to activation of the autophagy-specific ATG12-ATG5-ATG16L1 complex and autophagy induction 26160174_There is a major role of the CK2alpha-interacting protein CKIP-1 in activation of PAK1 for neoplastic prostate cells transformation. 26165401_The combination treatment of TRAIL and the CK2 inhibitor decreased p65 nuclear translocation... the treatment of a sub-dose of TRAIL, downregulation of CK2, | ENSMUSG00000074698 | Csnk2a1 | 4.809946e+03 | 1.0438963 | 0.061978462 | 0.2695018 | 5.338821e-02 | 0.8172690496 | 0.96412647 | No | Yes | 4.573219e+03 | 573.400357 | 4.220672e+03 | 542.549611 | |
| ENSG00000101413 | 58490 | RPRD1B | protein_coding | Q9NQG5 | FUNCTION: Interacts with phosphorylated C-terminal heptapeptide repeat domain (CTD) of the largest RNA polymerase II subunit POLR2A, and participates in dephosphorylation of the CTD by RPAP2. Transcriptional regulator which enhances expression of CCND1. Promotes binding of RNA polymerase II to the CCDN1 promoter and to the termination region before the poly-A site but decreases its binding after the poly-A site. Prevents RNA polymerase II from reading through the 3' end termination site and may allow it to be recruited back to the promoter through promotion of the formation of a chromatin loop. Also enhances the transcription of a number of other cell cycle-related genes including CDK2, CDK4, CDK6 and cyclin-E but not CDKN1A, CDKN1B or cyclin-A. Promotes cell proliferation. {ECO:0000269|PubMed:22231121, ECO:0000269|PubMed:22264791, ECO:0000269|PubMed:24399136, ECO:0000269|PubMed:24997600}. | 3D-structure;Acetylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | hsa:58490; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA polymerase II, holoenzyme [GO:0016591]; identical protein binding [GO:0042802]; RNA polymerase II complex binding [GO:0000993]; dephosphorylation of RNA polymerase II C-terminal domain [GO:0070940]; mRNA 3'-end processing [GO:0031124]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of cell cycle process [GO:0010564] | 22264791_CREPT increases cyclin D1 transcription during tumorigenesis. 24452636_RPRD1B overexpression promotes tumor growth and accelerates cell cycle progression. 24589584_This study reports that K-H functions in RNAPII regulation, and aids in stabilizing interactions between transcription termination factors, localizing Xrn2 to the 3'-end of genes and ultimately suppressing R-loop formation. 24982424_study suggests that CREPT acts as an activator to promote transcriptional activity of the beta-catenin.TCF4 complex in response to Wnt signaling. 24997600_RPRD1A and RPRD1B associate directly with RPAP2 phosphatase and coordinate the dephosphorylation of RNAPII phospho-S5 by RPAP2. 25400738_CREPT displays unique immunostaining for retroperitoneal leiomyosarcoma tissue and can be used to supplement other currently available markers. 26819409_a detailed description of proteins associating with K-H/RPRD1B in higher-order protein complexes is required to further elucidate its role in various cellular processes 27773816_In addition, CREPT overexpression significantly promoted tumor growth in vivo. Mechanism study showed that CREPT may regulate cell proliferation and cell cycle through the regulation on cyclin D3, CDK4 and CDK6. 28369091_Consistently, samples from oral squamous cell carcinoma (OSCC) patients exhibited a noticeably stronger CREPT expression than noncancerous samples. In contrast, knocking down of CREPT in OSCC cell lines significantly reduced proliferation, colony formation and migration as well as the expression of cyclin D1 and c-Myc, but promoted apoptosis. 28611204_Ccisplatin-induced peripheral neuropathy(isIPN) is associated with age, modifiable risk factors, and genetically determined expression level of RPRD1B Further study of implicated genes could elucidate the pathophysiologic underpinnings of CisIPN. 28893536_Mechanistically, CREPT regulated beta-catenin/TCF4/cyclin D1 pathway in BC. In conclusion, the data suggested that miR-138/CREPT involved BC progression, providing potential therapeutic targets for BC. 29397041_CREPT is closely relevant to the proliferation of NSCLC cells. 29398868_Aberrant overexpression of CREPT contributes to tumorigenesis of colorectal cancer by promoting cell proliferation and accelerating the cell cycle, and confers sensitivity to 5-fluorouracil. 29402413_Altogether, our results provided a novel insight into CREPT in regulating gastric cancer progression through apoptosis regulated by ROS/p53 pathways. 29563608_CREPT plays a pivotal oncogenic role in colorectal carcinogenesis through promoting Wnt/beta-catenin pathway via cooperating with p300. CREPT may serve as a prognostic biomarker of patients with colorectal cancer. 29938829_Data show that miR-383 targeted the 3' untranslated regions (3'-UTR) of CREPT mRNA directly. 30518842_The study provides a mechanism by which gastric tumor cells maintain their high proliferation rate via coordination of AURKB and CREPT on the expression of CCNB1. 30995540_these results demonstrate that CREPT exerts an oncogenic role in glioma and its expression is regulated by miR-596. 31054975_Crosstalk between RNA Pol II C-terminal domain acetylation and phosphorylation via RPRD1B protein and its isoforms has been reported. 31939329_Overexpression of cell-cycle related and expression-elevated protein in tumor (CREPT) in malignant cervical cancer. 32327260_MiR-139-5p inhibits the proliferation of gastric cancer cells by targeting Regulation of Nuclear Pre-mRNA Domain Containing 1B. 32866332_RPRD1B is a potentially molecular target for diagnosis and prevention of human papillomavirus E6/E7 infection-induced cervical cancer: A case-control study. 33125631_CREPT serves as a biomarker of poor survival in pancreatic ductal adenocarcinoma. 33431892_CREPT is required for murine stem cell maintenance during intestinal regeneration. 33531691_CREPT/RPRD1B promotes tumorigenesis through STAT3-driven gene transcription in a p300-dependent manner. 33838964_Immunohistochemical expression levels of cyclin D1 and CREPT reflect the course and prognosis in oral precancerous lesions and squamous cell carcinoma. | ENSMUSG00000027651 | Rprd1b | 1.515937e+03 | 0.8014408 | -0.319332141 | 0.2923439 | 1.216151e+00 | 0.2701173878 | 0.79962285 | No | Yes | 1.264706e+03 | 174.614901 | 1.622882e+03 | 229.389125 | ||
| ENSG00000101773 | 5932 | RBBP8 | protein_coding | Q99708 | FUNCTION: Endonuclease that cooperates with the MRE11-RAD50-NBN (MRN) complex in DNA-end resection, the first step of double-strand break (DSB) repair through the homologous recombination (HR) pathway (PubMed:17965729, PubMed:19202191, PubMed:19759395, PubMed:20064462, PubMed:26721387). HR is restricted to S and G2 phases of the cell cycle and preferentially repairs DSBs resulting from replication fork collapse (PubMed:17965729, PubMed:19202191). Key determinant of DSB repair pathway choice, as it commits cells to HR by preventing classical non-homologous end-joining (NHEJ) (PubMed:19202191). Functions downstream of the MRN complex and ATM, promotes ATR activation and its recruitment to DSBs in the S/G2 phase facilitating the generation of ssDNA (PubMed:16581787, PubMed:17965729, PubMed:19759395, PubMed:20064462). Component of the BRCA1-RBBP8 complex that regulates CHEK1 activation and controls cell cycle G2/M checkpoints on DNA damage (PubMed:15485915, PubMed:16818604). During immunoglobulin heavy chain class-switch recombination, promotes microhomology-mediated alternative end joining (A-NHEJ) and plays an essential role in chromosomal translocations (By similarity). {ECO:0000250|UniProtKB:Q80YR6, ECO:0000269|PubMed:15485915, ECO:0000269|PubMed:16581787, ECO:0000269|PubMed:16818604, ECO:0000269|PubMed:17965729, ECO:0000269|PubMed:19202191, ECO:0000269|PubMed:19759395, ECO:0000269|PubMed:20064462, ECO:0000269|PubMed:26721387}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Chromosome;Coiled coil;DNA damage;DNA repair;DNA-binding;Disease variant;Dwarfism;Endonuclease;Hydrolase;Isopeptide bond;Meiosis;Mental retardation;Mitosis;Nuclease;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene is a ubiquitously expressed nuclear protein. It is found among several proteins that bind directly to retinoblastoma protein, which regulates cell proliferation. This protein complexes with transcriptional co-repressor CTBP. It is also associated with BRCA1 and is thought to modulate the functions of BRCA1 in transcriptional regulation, DNA repair, and/or cell cycle checkpoint control. It is suggested that this gene may itself be a tumor suppressor acting in the same pathway as BRCA1. Three transcript variants encoding two different isoforms have been found for this gene. More transcript variants exist, but their full-length natures have not been determined. [provided by RefSeq, Jul 2008]. | hsa:5932; | BRCA1-C complex [GO:0070533]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; site of double-strand break [GO:0035861]; transcription repressor complex [GO:0017053]; damaged DNA binding [GO:0003684]; double-strand/single-strand DNA junction binding [GO:0000406]; double-stranded DNA binding [GO:0003690]; flap-structured DNA binding [GO:0070336]; identical protein binding [GO:0042802]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; single-stranded DNA binding [GO:0003697]; single-stranded DNA endodeoxyribonuclease activity [GO:0000014]; transcription corepressor activity [GO:0003714]; Y-form DNA binding [GO:0000403]; blastocyst hatching [GO:0001835]; cell division [GO:0051301]; DNA double-strand break processing involved in repair via single-strand annealing [GO:0010792]; DNA repair [GO:0006281]; double-strand break repair via homologous recombination [GO:0000724]; G1/S transition of mitotic cell cycle [GO:0000082]; meiotic cell cycle [GO:0051321]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; regulation of transcription by RNA polymerase II [GO:0006357] | 11751867_These findings reveal a novel complex between BRCA1, LMO4, and CtIP and indicate a role for LMO4 as a repressor of BRCA1 activity in breast tissue. 16287852_a corepressor complex containing CtIP/CtBP facilitates RBP-Jkappa/SHARP-mediated repression of Notch target genes 16581787_CTIP activates its own and cyclin D promoters via the E2F/RB pathway during G1/S progression. 17112672_Since CtIP plays important roles in cell cycle checkpoint control and it has been implicated in tumorigenesis, our data suggest that TRB3 may be involved in these biological processes through interacting with CtIP. 17546052_Data show that CtIP expression is induced by AdE1A during viral infection and that reduction of CtIP expression with RNA interference can retard virus replication. 17965729_findings establish evolutionarily conserved roles for CtIP-like proteins in controlling DSB resection, checkpoint signalling and homologous recombination 18007598_Results idientify COM1, a novel plant gene essential for meiosis that is related to the human CtIP and the yeast COM1/SAE2 gene. 18095152_the BRCA1 interacting protein CTIP has a role in breast cancer 18171670_cell cycle-dependent complex formation of BRCA1, CtIP, and MRN contributes to the activation of HR-mediated DSB repair in the S and G(2) phases of the cell cycle. 18171986_CtIP silencing might be a novel mechanism for the development of tamoxifen resistance in breast cancer, suggesting that CtIP is likely associated with estrogen receptor function 18676825_Stresses and stimuli that reduce CtBP-mediated repression are associated with increased p16 expression; therefore, CtBP may provide a common final target for regulating the balance among tumor suppression, regenerative capacity, and senescence 18950845_Observational study of gene-disease association. (HuGE Navigator) 19202191_unlike cells expressing wild-type CtIP, cells expressing the Thr-to-Glu mutant resect DSBs even after cyclin-dependent kinase inhibition 19270026_Observational study of gene-disease association. (HuGE Navigator) 19270026_Single nucleotide polymorphisms in CASP5 and RBBP8 gene are associated with survival in ovarian cancer patients. 19342888_CtIP is recruited to S-phase DNA replication foci through a novel motif functioning as replication foci targeting sequence. 19357644_CtIP is required not only for repair of double strand breaks by homologous recombination in S/G2 phase but also for microhomology-mediated end-joining in G1 19584272_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19633668_Data show that silencing of Rad50 or CtIP decreases end-joining efficiency in the same pathway as Mre11. 19690177_Observational study of gene-disease association. (HuGE Navigator) 20007691_Data found that HMGA2, along with a dozen of other genes, was co-repressed by ZBRK1, BRCA1, and CtIP. 20064462_CtIP facilitates the transition from DSB sensing to processing, by binding to the DNA at double-strand breaks (DSBs) after DSB sensing and ATM activation and then promoting DNA resection, leading to checkpoint activation and homologous recombination. 20546612_Observational study of gene-disease association. (HuGE Navigator) 20635389_Observational study of gene-disease association. (HuGE Navigator) 20974687_BCR-ABL promotes mutagenic DSB repair with the DSB end-processing protein CtIP acting as the key mediator downstream of BCR-ABL 20975951_Studies indicate that codon-based models of gene evolution yielded statistical support for the recurrent positive selection of five NHEJ genes during primate evolution: XRCC4, NBS1, Artemis, POLlambda, and CtIP. 21052091_This study identifies new functions of CtIP and EXO1 in DNA end resection and provides new information on the regulation of DNA double-strand breaks repair pathways, which is a key factor in the maintenance of genome integrity. 21998596_the SCKL2 mutation creates an alternative splicing site leading to both the normal and aberrant CtIP proteins coexisting in the cells of patients and carriers 22231403_The authors show that, in human and mouse, Mre11 controls these events through a direct interaction with CDK2 that is required for CtIP phosphorylation and BRCA1 interaction in normally dividing cells. 22544744_The severe repair defects of CtIP dimerization mutants are likely due to the failure in localization to chromosomal DSBs upon DNA damage. 22733999_show that CHK1 was rapidly and robustly activated before detectable end resection 22832221_CtIP binds to ATM protein proximal promoter, but after DNA damage Ctip is released. 23333306_RIF1 accumulation at DSB sites is strongly antagonized by BRCA1 and its interacting partner CtIP 23349304_Microsatellite instability dependent mutations were detected in CtIP in myeloid malignancies conferring hypersensitivity to PARP inhibitors. 23353824_Data suggest that overexpression of LMO4 may disrupt some of the normal tumour suppressor activities of CtIP, thereby contributing to breast cancer progression. 23468639_These studies reveal one important mechanism to regulate cell-cycle-dependent activation of HR upon DNA damage by coupling CDK- and ATM-mediated phosphorylation of CtIP through modulating the interaction of CtIP with Nbs1 23696749_Taken together, these findings strongly suggest that an ATM-dependent CREB-miR-335-CtIP axis influences the selection of HRR for repair of certain DSB lesions 23994874_BRCA1/CtIP-mediated processing of the second end of the break controls the annealing step that normally terminates synthesis-dependent strand annealing, thereby suppressing the error-prone long-tract gene conversion outcome. 24196441_our data demonstrate that CtIP is required for DNA damage-induced P21 induction 24403251_Low or no expression of RBBP8 correlates with high-grade breast cancer, poor prognosis and with nodal metastasis. 24440292_study identified a homozygous mutation in RBBP8, which co-segregates with microcephaly-associated intellectual disability syndrome in a Pakistani family; also identified a heterozygous deletion encompassing the NRXN1 in this family, which is present in 2 affected sibs with complex phenotype and the mother with mild phenotype 24556218_Study identified CtIP as a novel interaction partner of FANCD2. CtIP binds and stabilizes FANCD2 in a DNA damage- and FA core complex-independent manner, suggesting that FANCD2 monoubiquitination is dispensable for its interaction with CtIP. 24794434_Data indicate that FANCD2 primes CtIP-dependent resection during HR after ICL induction but that CtIP helps prevent illegitimate recombination in FA cells. 24837675_These studies suggest that an end resection-independent CtIP function is important for processing double-strand break ends with secondary structures to promote homologous recombination. 24837676_human CtIP is a 5' flap endonuclease and this activity is required in some contexts for the efficient function of CtIP. 25053769_Possible association of SOX-17 and RBBP8 with brain arteriovenous malformations, genes involved in cell cycle progression, deserves further investigation 25267294_Plk3 binds to CtIP phosphorylated at S327 via its Polo box domains, which is necessary for robust damage-induced CtIP phosphorylation at S327 and subsequent CtIP phosphorylation at T847. 25308476_The CtIP 3'UTR is directly targeted by miR-19a and miR-19b. 25349192_CtIP interacts with Cdh1 through a conserved KEN box, mutation of which impedes ubiquitylation and downregulation of CtIP both during G1 and after DNA damage in G2. 25771978_findings provide strong evidence that CtIP is continuously recruited to DSBs downstream of both the initiation and extension step of resection 25909997_BRCA1 and CtIP contribute to DSB resection by recruiting Dna2 to damage sites, thus ensuring the robust DSB resection necessary for efficient homologous recombination. 25957490_CtIP is a DNA damage response protein at the intersection of DNA metabolism. (Review) 26333564_Homozygous RBBP8 mutation is associated with microcephaly, intellectual disability, short stature and brachydactyly. 26387952_USP4 cooperates with CtIP in DNA double-strand break end resection. 26502057_Data show that ubiquitin E2 enzymes UBE2D1/2/3 and E3 ligase RNF138 accumulate at DNA-damage sites and act at early resection stages by promoting CtIP protein ubiquitylation and accrual. 26713604_Low level of CtIP expression is associated with breast cancer. 27494840_53BP1/RIF1 has a role in limiting BRCA1/CtIP-mediated end resection to control double strand break repair pathway choice 27561354_Study identifies the KLHL15 as a new interaction partner of CtIP and show that KLHL15 promotes CtIP protein turnover via the ubiquitin-proteasome pathway. 27641979_ATM-dependent phosphorylation of CtIP and the epistatic and coordinated actions of MRE11 and CtIP nuclease activities are required to limit the stable loading of Ku on single-ended DNA double-strand breaks. 27798638_his shows that 53BP1 protects both close and distant DSEs from degradation and that the association of unprotection with distance between DSEs favors ECS capture. Reciprocally, silencing CtIP lessens ECS capture both in control and 53BP1-depleted cells. We propose that close ends are immediately/rapidly tethered and ligated, whereas distant ends first require synapsis of the distant DSEs prior to ligation 27814491_The results illuminate the important role of Nbs1 and CtIP in determining the substrates and consequences of human Mre11/Rad50 nuclease activities on protein-DNA lesions. 27940552_And-1 interacts with CtIP and that these interactions are required for DNA damage checkpoint maintenance, thereby linking DNA processing with prolonged cell cycle arrest to allow sufficient time for DNA repair. 28065643_we reveal that reprogramming is associated with high levels of DNA end resection, a critical step in homologous recombination. Moreover, the resection factor CtIP is essential for cell reprogramming and establishment of iPSCs, probably to repair reprogramming-induced DNA damage. 28481869_Data delineates the regulatory mechanisms of GATA3 in DNA double-strand breaks repair and strongly suggests that it might act as a tumor suppressor by promoting CtIP expression and homologous recombination to stabilize genomes. 28623092_CtIP/Ctp1/Sae2/Com1 role in removal of DNA double strand breaks through DSB repair by homologous recombination is reviewed. 28740167_Data show that SUMO E3 ligase CBX4 sumoylates subpopulation of CtIP to regulate recruitment to breaks and resection. 29445424_Data demonstrate that RBBP8 is almost exclusively methylated in 45% primary bladder cancer, suggesting the development of methylation in a distinct molecular context. A close association between RBBP8 methylation and its gene expression in primary tumors as well as after demethylation treatment in vitro indicate that RBBP8 methylation could be responsible for its gene inactivation. 29855336_HGF/c-Met regulates expression of SAE2 and cirRNA CCDC66 to increase epithelial-to-mesenchymal transition and drug resistance of lung adenocarcinoma cells. 30202980_Data sufgest that polo like kinase 1 (PLK1) may target retinoblastoma binding protein 8 (RBBP8; CtIP) to promote error-prone microhomology-mediated end joining (MMEJ) and inactivate the G2/M checkpoint. 30569153_Bioinformatics predicted the putative binding site of miR18a5p in the 3' untranslated region of CtIP/RBBP8. Study using nasopharyngeal carcinoma cell line further confirmed that miR18a5p could directly bind with RBBP8 and inhibit RBBP8 expression. 30601117_Data show that retinoblastoma binding protein 8 (CtIP) is a tetrameric protein adopting a dumbbell architecture in which DNA binding domains are connected by long coiled-coils. 30622325_Prognosis value of RBBP8 expression in plasma cell myeloma. 30657944_We propose that BRCA1-CTIP and MRE11 prepare nascent DNA ends, blocked from synthesis by CTNAs, for further repair. 30915754_CtIP contributes to non-homologous end joining formation through interacting with ligase IV and promotion of TMZ resistance in glioma cells. 31340001_The N-terminus of CTCF is able to bind to only MRE11 and its C-terminus is incapable of binding to MRE11 and CtIP, thereby resulting in compromised CtIP recruitment, DSB resection and HR. Overall, this suggests an important function of CTCF in DNA end resection through the recruitment of CtIP at DSBs. 31378812_CtIP is required for faithful replication through telomeres via its roles at stalled replication tracts. 31501894_CSB interacts with BRCA1 to promote DNA end resection 31636387_RBBP8/CtIP suppresses P21 expression by interacting with CtBP and BRCA1 in gastric cancer. 31934630_DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. 32241893_Study revealed that phosphorylated CtIP (pCtIP) dramatically stimulates the motor activity of DNA2. This accelerates degradation of RPA-coated ssDNA by DNA2, showing the need for the motor activity of DNA2 to facilitate resection. CtIP is thus a cofactor not only of MRE11, but also of DNA2, and domains of CtIP required for the stimulation of MRE11-RAD50-NBS1 (MRN) complex and DNA2 are physically separate. 32347940_The internal region of CtIP negatively regulates DNA end resection. 32379725_Germline RBBP8 variants associated with early-onset breast cancer compromise replication fork stability. 32392243_CtlP expression is regulated by both ACL1 and eIF4A1 proteins which cooperates to help stabilizing the most abundant splicing form of CtIP mRNA. In fact, the G4 5'-UTR sequence of CtIP is necessary for both eIF4A1 and ALC1 functions. 32683422_SCF(SKP2) regulates APC/C(CDH1)-mediated degradation of CTIP to adjust DNA-end resection in G2-phase. 32817418_Phosphorylated CtIP bridges DNA to promote annealing of broken ends. 33406258_SUMOylation mediates CtIP's functions in DNA end resection and replication fork protection. 33901493_CtIP suppresses primary microRNA maturation and promotes metastasis of colon cancer cells in a xenograft mouse model. 34270086_A 24-generation-old founder mutation impairs splicing of RBBP8 in Pakistani families affected with Jawad syndrome. 35203293_RNAi Screening Uncovers a Synthetic Sick Interaction between CtIP and the BARD1 Tumor Suppressor. 35414773_IKZF1 selectively enhances homologous recombination repair by interacting with CtIP and USP7 in multiple myeloma. | ENSMUSG00000041238 | Rbbp8 | 6.232415e+02 | 0.9041477 | -0.145369622 | 0.3621794 | 1.564753e-01 | 0.6924225905 | 0.93376105 | No | Yes | 7.039996e+02 | 139.514701 | 7.049212e+02 | 143.383199 | |
| ENSG00000101986 | 215 | ABCD1 | protein_coding | P33897 | FUNCTION: ATP-dependent transporter of the ATP-binding cassette (ABC) family involved in the transport of very long chain fatty acid (VLCFA)-CoA from the cytosol to the peroxisome lumen (PubMed:11248239, PubMed:15682271, PubMed:16946495, PubMed:18757502, PubMed:21145416, PubMed:23671276, PubMed:29397936, PubMed:33500543). Coupled to the ATP-dependent transporter activity has also a fatty acyl-CoA thioesterase activity (ACOT) and hydrolyzes VLCFA-CoA into VLCFA prior their ATP-dependent transport into peroxisomes, the ACOT activity is essential during this transport process (PubMed:33500543, PubMed:29397936). Thus, plays a role in regulation of VLCFAs and energy metabolism namely, in the degradation and biosynthesis of fatty acids by beta-oxidation, mitochondrial function and microsomal fatty acid elongation (PubMed:23671276, PubMed:21145416). Involved in several processes; namely, controls the active myelination phase by negatively regulating the microsomal fatty acid elongation activity and may also play a role in axon and myelin maintenance. Controls also the cellular response to oxidative stress by regulating mitochondrial functions such as mitochondrial oxidative phosphorylation and depolarization. And finally controls the inflammatory response by positively regulating peroxisomal beta-oxidation of VLCFAs (By similarity). {ECO:0000250|UniProtKB:P48410, ECO:0000269|PubMed:11248239, ECO:0000269|PubMed:15682271, ECO:0000269|PubMed:16946495, ECO:0000269|PubMed:18757502, ECO:0000269|PubMed:21145416, ECO:0000269|PubMed:23671276, ECO:0000269|PubMed:29397936, ECO:0000269|PubMed:33500543}. | ATP-binding;Disease variant;Endoplasmic reticulum;Glycoprotein;Hydrolase;Lysosome;Membrane;Mitochondrion;Nucleotide-binding;Peroxisome;Phosphoprotein;Reference proteome;Translocase;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. ABC proteins transport various molecules across extra- and intra-cellular membranes. ABC genes are divided into seven distinct subfamilies (ABC1, MDR/TAP, MRP, ALD, OABP, GCN20, White). This protein is a member of the ALD subfamily, which is involved in peroxisomal import of fatty acids and/or fatty acyl-CoAs in the organelle. All known peroxisomal ABC transporters are half transporters which require a partner half transporter molecule to form a functional homodimeric or heterodimeric transporter. This peroxisomal membrane protein is likely involved in the peroxisomal transport or catabolism of very long chain fatty acids. Defects in this gene have been identified as the underlying cause of adrenoleukodystrophy, an X-chromosome recessively inherited demyelinating disorder of the nervous system. [provided by RefSeq, Jul 2008]. | hsa:215; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; integral component of peroxisomal membrane [GO:0005779]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; mitochondrial membrane [GO:0031966]; perinuclear region of cytoplasm [GO:0048471]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; ABC-type fatty-acyl-CoA transporter activity [GO:0015607]; acyl-CoA hydrolase activity [GO:0047617]; ADP binding [GO:0043531]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; ATPase-coupled transmembrane transporter activity [GO:0042626]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; long-chain fatty acid transporter activity [GO:0005324]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; alpha-linolenic acid metabolic process [GO:0036109]; fatty acid beta-oxidation [GO:0006635]; fatty acid elongation [GO:0030497]; fatty acid homeostasis [GO:0055089]; linoleic acid metabolic process [GO:0043651]; long-chain fatty acid catabolic process [GO:0042758]; long-chain fatty acid import into peroxisome [GO:0015910]; myelin maintenance [GO:0043217]; negative regulation of cytokine production involved in inflammatory response [GO:1900016]; negative regulation of reactive oxygen species biosynthetic process [GO:1903427]; neuron projection maintenance [GO:1990535]; peroxisomal membrane transport [GO:0015919]; peroxisome organization [GO:0007031]; positive regulation of fatty acid beta-oxidation [GO:0032000]; positive regulation of unsaturated fatty acid biosynthetic process [GO:2001280]; regulation of cellular response to oxidative stress [GO:1900407]; regulation of fatty acid beta-oxidation [GO:0031998]; regulation of mitochondrial depolarization [GO:0051900]; regulation of oxidative phosphorylation [GO:0002082]; sterol homeostasis [GO:0055092]; very long-chain fatty acid catabolic process [GO:0042760]; very long-chain fatty acid metabolic process [GO:0000038]; very long-chain fatty-acyl-CoA catabolic process [GO:0036113] | 10551832_ALDP and ALDRP interact via their carboxy termini. ALDP mutations P484R and R591Q abolish this interaction. This interaction was demonstrated using human ALDP and murine ALDRP. 10551832_ALDP homodimerizes via the C-terminal cytosolic domain [361-745]. Residues Pro-484 and Arg-591 are important for the interaction. 10551832_ALDP interacts with PMP70. This interaction occurs via the C-terminus of ALDP [361-745] and the C-terminus of PMP70 [338-659]. ALDP mutations P484R and R591Q abolish the interaction. 10737980_Fifteen new mutations are described in Adrenoleukodystrophy patients 11438993_mutational analysis in patients with X-linked adrenoleukodystrophy 11992258_Contiguous deletion of the X-linked adrenoleukodystrophy gene (ABCD1) and DXS1357E: a novel neonatal phenotype similar to peroxisomal biogenesis disorders. 12175782_Eight novel mutations are described. 12509471_ALDP facilitates the interaction between peroxisomes and mitochondria, resulting, when ALDP is deficient in X-ALD, in increased VLCFA accumulation 12530690_Mutations are heterogeneously distributed over functional domains of ALDP and alter peroxisomal transport function. 12579499_The splice mutation in 5' end of intron 5 leading to abnormal splice in exon 5 and exon 6 appears to be one of the causes of X-linked recessive adrenoleukodystrophy. 12624723_Six different missense mutations in ALD were identified in seven Japanese families. 14556192_For the first time, mutations in ABCD1 are identified in Chinese adrenoleukodystrophy patients in the mainland of China. 14767898_There were no hot spot mutations in ABCD1 gene in China, mutations in gene were found over 70% of patients with ALD and the ABCD1 gene mutations identified revealed no obvious correlation between the type of mutation and phenotype. 15001567_ABCA1-independent but cytoskeleton-dependent cholesterol removal pathway may help to prevent early atherosclerosis in Tangier disease. 15772093_Accumulation of very long-chain fatty acids does not affect mitochondrial function in ABCD1 protein deficiency. 15781447_analysis of the PEX19-binding site of human adrenoleukodystrophy protein 16018167_Adrenomyeloneuropathy must be considered in the differential diagnosis of spastic paraparesis in men or women. We report an ABCD1 gene mutation in the French-Canadian population, which should lead to the recognition of other cases in the future. 16087056_over half of the mutations (19/34) were located in exon 1 and exon 6, suggesting possible hot exons 16331554_Data show that fetus 1 had R617G mutation on his ABCD1 gene and he was an adrenoleukodystrophy hemizygote. Fetus 2 had no P534R mutation on his ABCD1 gene and he was a normal hemizygote. 17285533_ABCD1 gene mutations were found in 4 cases of X-linked adrenoleukodystrophy with high VLCFAs levels of amniocytes, no mutation was found in other 4 cases with normal VLCFAs levels of amniocytes. 17504626_Observational study of genotype prevalence. (HuGE Navigator) 17542813_mutant ALDPs, which have a mutation in COOH-terminal half of ALDP, including S606L, R617H, & H667D, were degraded by proteasomes after dimerization. region between transmembrane domain 2 and 3 is important for the targeting of ALDP to the peroxisome. 17662307_This study examined a patient with Adult onset cerebral form of X-linked adrenoleukodystrophy with dementia of frontal lobe type with new L160P mutation in ABCD1 gene. 17761426_ALDP-encoding mRNA is most abundant in tissues with high energy requirements such as heart, muscle, liver, and the renal and endocrine systems. ALDP selectively occurs in specific cell types of the brain. 17828604_ALDP deficiency enhances metabolic distress in oligodendrocytes that are compromised a priori by destabilised myelin. The age at which this occurs precedes the onset of axonal degeneration in Abcd1-deficient mice. 18306728_ABCD1 mutation in the ethiopathogenesis of X-linked adrenoleukodystrophy. Its defect causes accumulation of the very long chain fatty acids in the tissues of the central and peripheral nervous system, adrenal glands and in the body fluids. 18481121_A family with combined point mutations of the hemophilia A (F8)and X-linked adrenoleukodystrophy (ABCD1) genes. 18973459_This study concluded that de novo mutations occurred in this gene resulting in the disease. 19204726_Observational study of gene-disease association. (HuGE Navigator) 19343046_Observational study of gene-disease association. (HuGE Navigator) 19406751_study reports 3 novel ABCD1 gene variants (c.67_83del17, c.395G>A, c.1938_1939dupGG) in 3 unrelated Indian families with X-linked adrenoleukodystrophy 19787628_ABCD1 downregulation may be involved in human renal tumorigenesis. 20042197_A family harbors a novel deletion of 1 base pair in exon 8 at nucleotide position 2245 (2245delA) in the ABCD1 gene. 20376793_Three female patients heterozygous for ABCD1 gene mutation were first reported in China, and a novel mutation, p.H283R, was identified in this X-linked adrenoleukodystrophy family. 20661612_Observational study of gene-disease association. (HuGE Navigator) 21145416_HsABCD1 and HsABCD2 have distinct substrate specificities 21273699_Novel mutation in ATP-binding domain of ABCD1 gene in adrenoleukodystrophy is reported. 21700483_Amongst 489 X-linked adrenoleukodystrophy families, 20 cases in which the ABCD1 mutation was de novo in the index case, indicating that the mutation arose in the maternal germ line. 21889498_standardized conformation sensitive gel electrophoresis (CSGE) method to detect mutations in ABCD1 gene in twenty Indian patients with X-ALD. The results were confirmed by sequencing. Genotype-phenotype correlation was also attempted 21966424_Single germ line mutation was identified in each index case in ABCD1 gene. Results detected 4 novel mutations (2 missense and 2 deletion/insertion) and 3 novel SNPS. Data observed a variable protein expression in different patients. 22280810_These results indicate that preferential X chromosome inactivation leads to the favored expression of the mutant ABCD1 allele. 22994209_Array comparative genomic hybridization analysis suggested that the deletion was a genomic rearrangement in the 90-kb span starting in exon 4 and included ABCD1 23123468_Very long chain fatty acid (VLCFA) is beta-oxidized in ABCD1-dependent pathway, but the ABCD1-independent peroxisomal and mitochondrial beta-oxidation pathways significantly contribute to VLCFA beta-oxidation in astrocytes 23300730_Identification of novel mutations in ABCD1 in unrelated Argentinean X-linked adrenoleukodystrophy patients 23469258_Adrenoleukodystrophy and skewed x chromosome inactivation in favor of the mutatnt ABCD1 allele is associated with symptoms manifestation in heterozygotes from a Chinese pedigree. 23566833_Identified 8 mutationsof ABCD1 , including one novel deletion (c.1477_1488+11del23) and 7 known mutations. 23671276_in contrast to yeast cells, very long-chain acyl-CoA esters are transported into peroxisomes by ABCD1 independently of additional synthetase activity 23835273_This study unveil unequivocally that cryptic splicing-induced aberrant messenger-RNA carrying an internal frameshift deletion results from an intronic mutation in the ABCD1 gene. 24154795_We describe four unrelated women with a late-onset progressive spastic paraparesis and heterozygous mutations in the ABCD1 gene 24480483_X-inactivation pattern of the ABCD1 gene is associated with symptomatic status in female X-linked adrenoleukodystrophy carriers. 24597975_both BCAP31 and ABCD1 were associated with hepatic cholestasis and death before 1 year. Remarkably, a patient with an isolated deletion at the 3'-end of SLC6A8 had a similar severe phenotype as seen in BCAP31 deficiency 25044748_Distal Xq28 microdeletions: clarification of the spectrum of contiguous gene deletions involving ABCD1, BCAP31, and SLC6A8 with a new case and review of the literature. 25234129_Exome sequencing in two brothers with distinct phenotype including congenital language disorder, growth retardation, intellectual disability and urinary and fecal incontinence, identifies missense mutations in ABCD1 and DACH2. 25275259_We detected the same mutation of the ABCD1 gene in two unrelated patients with X-linked adrenoleukodystrophy. 25393703_As a result of loss of ABCD1, there is pathogenic accumulation of very long chain fatty acids which leads to mitochondrial dysfunction. 25835712_The current study demonstrates that a single splicing mutation affects the ABCD1 transcripts and the ALDP protein function. 26454440_CCALD is the most common phenotype (64%) in our Chinese patients with X-ALD. Eight novel mutations in the ABCD1 gene identified are disease-causing mutations. 26686776_Phenotypic variability in a Tunisian family with X-linked adrenoleukodystrophy caused by the p.Gln316Pro novel mutation 27084228_This study showed that the mutations of were detected in SPG11, ATL1, NIPA1, and ABCD1 in patient with hereditary spastic paraplegia. 27337030_To assist in the evaluation process, the New York Newborn Screening Program also routinely performs Sanger sequencing to determine if there are mutations in the ABCD1 gene. 27766264_ABCD1 and ABCD2 are involved in the transport of long and very long chain fatty acids (VLCFA) or their CoA-derivatives into peroxisomes with different substrate specificities, while ABCD3 is involved in the transport of branched chain acyl-CoA into peroxisomes.ABCD4 is deduced to take part in the transport of vitamin B12 from lysosomes into the cytosol. 28601575_a novel heterozygous mutation IVS4+2T>A (c.1393+2T>A) of the ABCD1 gene are associated with different clinical phenotypes in a family with adrenoleukodystrophy. 28911205_Expression of human ABCD1 in oligodendrocytes rescued apoptosis in the abcd1 mutant. 29136088_In X-linked adrenoleukodystrophy, lack of ABCD1 function causes increased capillary flow heterogeneity in asymptomatic hemizygotes predominantly in the white matter regions and developmental stages with the highest probability for conversion to cerebral disease. 29966135_Description of a new clinical variant of adult adrenomyeloneuropathy that is possibly caused by a novel ABCD1 gene mutation. 31074578_A state-wide Minnesota newborn screening for X-linked adrenoleukodystrophy revealed ABCD1 mutations in affected newborns and their relatives. 31665121_ABCD1 gene mutation is associated with X-linked adrenoleukodystrophy. 32075856_Hexacosenoyl-CoA is the most abundant very long-chain acyl-CoA in ATP binding cassette transporter D1-deficient cells. 32416190_Proteasome-dependent protein quality control of the peroxisomal membrane protein Pxa1p. 33127985_Adrenoleukodystrophy siblings with a novel ABCD1 missense variant presenting with phenotypic differences: a case report and literature review. 33373044_Evolution of adrenoleukodystrophy model systems. 33500543_Acyl-CoA thioesterase activity of peroxisomal ABC protein ABCD1 is required for the transport of very long-chain acyl-CoA into peroxisomes. 34056752_Drug discovery for X-linked adrenoleukodystrophy: An unbiased screen for compounds that lower very long-chain fatty acids. 34069712_A Large Family with p.Arg554His Mutation in ABCD1: Clinical Features and Genotype/Phenotype Correlation in Female Carriers. 34198763_Peroxisomal ABC Transporters: An Update. 34347682_Diverse clinical manifestations of X-linked adrenoleukodystrophy in a Chinese family with identical multisite variants of ABCD1 gene. 34649108_Investigating ABCD1 mutations in a Taiwanese cohort with hereditary spastic paraplegia phenotype. 34754073_Structural basis of acyl-CoA transport across the peroxisomal membrane by human ABCD1. 34943935_Molecular Biomarkers for Adrenoleukodystrophy: An Unmet Need. 34946879_Biochemical Studies in Fibroblasts to Interpret Variants of Unknown Significance in the ABCD1 Gene. 35013584_Structures of the human peroxisomal fatty acid transporter ABCD1 in a lipid environment. 35053399_Structure and Function of the ABCD1 Variant Database: 20 Years, 940 Pathogenic Variants, and 3400 Cases of Adrenoleukodystrophy. | ENSMUSG00000031378 | Abcd1 | 8.305202e+02 | 1.1411332 | 0.190467155 | 0.3334755 | 3.295950e-01 | 0.5658976724 | 0.89758870 | No | Yes | 8.071694e+02 | 127.480994 | 6.363428e+02 | 103.598808 | |
| ENSG00000102119 | 2010 | EMD | protein_coding | P50402 | FUNCTION: Stabilizes and promotes the formation of a nuclear actin cortical network. Stimulates actin polymerization in vitro by binding and stabilizing the pointed end of growing filaments. Inhibits beta-catenin activity by preventing its accumulation in the nucleus. Acts by influencing the nuclear accumulation of beta-catenin through a CRM1-dependent export pathway. Links centrosomes to the nuclear envelope via a microtubule association. EMD and BAF are cooperative cofactors of HIV-1 infection. Association of EMD with the viral DNA requires the presence of BAF and viral integrase. The association of viral DNA with chromatin requires the presence of BAF and EMD. Required for proper localization of non-farnesylated prelamin-A/C. {ECO:0000269|PubMed:15328537, ECO:0000269|PubMed:16680152, ECO:0000269|PubMed:16858403, ECO:0000269|PubMed:17785515, ECO:0000269|PubMed:19323649}. | 3D-structure;Acetylation;Actin-binding;Cardiomyopathy;Direct protein sequencing;Disease variant;Emery-Dreifuss muscular dystrophy;Membrane;Microtubule;Nucleus;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | Emerin is a serine-rich nuclear membrane protein and a member of the nuclear lamina-associated protein family. It mediates membrane anchorage to the cytoskeleton. Dreifuss-Emery muscular dystrophy is an X-linked inherited degenerative myopathy resulting from mutation in the emerin gene. [provided by RefSeq, Jul 2008]. | hsa:2010; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; microtubule [GO:0005874]; nuclear envelope [GO:0005635]; nuclear inner membrane [GO:0005637]; nuclear membrane [GO:0031965]; nuclear outer membrane [GO:0005640]; nucleoplasm [GO:0005654]; spindle [GO:0005819]; actin binding [GO:0003779]; beta-tubulin binding [GO:0048487]; cadherin binding [GO:0045296]; cellular response to growth factor stimulus [GO:0071363]; muscle contraction [GO:0006936]; muscle organ development [GO:0007517]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of fibroblast proliferation [GO:0048147]; nuclear membrane organization [GO:0071763]; positive regulation of protein export from nucleus [GO:0046827]; regulation of canonical Wnt signaling pathway [GO:0060828]; skeletal muscle cell differentiation [GO:0035914] | 11792821_Disease-linked emerin proteins all remain active for binding directly to barrier-to-autointegration factor (BAF) both in vitro and in vivo, suggesting that disease results from loss of interactions between emerin and lamin A or putative novel partner(s). 11973618_genes known to be responsible for Emery-Dreifuss muscular dystrophy 12493765_emerin binds directly to a transcriptional repressor, GCL, and the emerin-repressor complexes might be regulated by barrier to autointegration factor 12755701_emerin has a regulatory role, as well as a structural role in the cell nucleus 12783988_the lamin a-emerin complex might have a role in muscular dystrophy and cardiomyopathy 15009215_These results suggest that Btf localization is regulated by apoptotic signals, and that loss of emerin binding to Btf may be relevant to muscle wasting in Emery-Dreifuss muscular dystrophy. 15109603_Data describe the mobility of barrier-to-autointegration factor to its partners emerin, LAP2 beta, and MAN1 in the nuclear membrane of living HeLa cells. 15328537_emerin may disrupt transcription factors and nuclear envelope architecture by weakening a nuclear actin network 15681850_Data describe the direct binding of the nuclear membrane protein MAN1 to emerin in vitro. 16204256_phosphorylation at Ser175 regulates the dissociation of emerin from BAF 16246140_Data show that chromosome positioning is largely unaffected in lymphoblastoid cell lines containing emerin or A-type lamin mutations. 16283426_We show that epitope masking in the nucleus is often responsible for failure to detect emerin in human and rat tissues. Pig spleen had fewer emerin-positive nuclei than lung (5% vs. 32%), although their emerin content was similar by Western blotting. 16371512_Ser-4 phosphorylation inhibits BAF binding to emerin and lamin A, and thereby weakens emerin-lamin interactions during both mitosis and interphase. 16481476_Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation, with deficient cells displaying delayed differentiation kinetics that may underlie dystrophic phenotypes. 16680152_emerin, an integral inner-nuclear-envelope protein, is necessary for HIV-1 infection 16761279_Review summarizes growing evidence that emerin has both architectural and gene-regulatory roles in the nucleus which may contribute to the pathology and mechanism of Emery-Dreifuss muscular dystrophy. 16823856_We found that in inclusion-body myositis (IBM) muscle vacuoles were immunoreactive for the inner nuclear membrane proteins emerin and lamin A/C. 16858403_Emerin regulates the flux of beta-catenin, an important transcription coactivator, into the nucleus. 16997877_Altered nuclear envelope elasticity caused by loss of emerin could contribute to increased nuclear fragility in Emery-Dreifuss muscular dystrophy patients with mutations in the emerin gene. 17067998_Lmo7 positively regulates many EDMD-relevant genes (including emerin), and is feedback-regulated by binding to emerin. 17097067_Mislocalization of emerin to the endoplasmic reticulum in human cells lacking A-type lamins leads to its degradation and provides the first evidence that its degradation is mediated by the proteasome. 17117676_Neither emerin nor LMNA mutations in a subset of families with EDMD-like phenotypes that may imply an existence of other genes causing similar disorders. 17462627_The characterisation of the residues both in emerin and in nesprin-1alpha and -2beta which are involved in their interaction is reported. 17536044_findings highlight the crucial role of lamin A/C-emerin interactions, with evidence for synergistic effects of these mutations that lead to Emery-Dreifuss muscular dystrophy as the worsened result of digenic mechanism in this family 17620497_Mutations in EMD may indicate a limb-girdle muscular dystrophy phenotype. Identification of emerin deficiency among patients with limb-girdle muscular dystrophy is essential to prevent cardiac catastrophe. 17652388_The results suggest a model in which pUS3 and PKCdelta that has been recruited by pUL34 hyperphosphorylate emerin, leading to disruption of its connections with lamin proteins and contributing to the disruption of the nuclear lamina. 17785515_significant fraction of emerin is located at the outer nuclear membrane and peripheral ER, where it interacts directly with the centrosome. 18266676_mutation of EMD can underlie X-linked familial atrial fibrillation; Lys37del is associated with epithelial cell emerin deficiency, as in Emery-Dreifuss muscular dystrophy, yet it causes electrical atriomyopathy in the absence of skeletal muscle disease 18400857_emerin is dispensable for HIV-1 infections. 18646565_Mutations in the genes for nuclear envelope proteins of emerin (EMD) and lamin A/C (LMNA) are known to cause Emery-Dreifuss muscular dystrophy (EDMD) and limb girdle muscular dystrophy (LGMD). 19022376_Knockdown of A-type lamins and emerin in HeLa and C2C12 stimulated phosphorylation and nuclear translocation of ERK as well as activation of genes encoding downstream transcription factors. 19126678_process of adipogenesis is affected by a dynamic link between complexes of emerin and lamins A/C at the nuclear envelope and nucleocytoplasmic distribution of beta-catenin, to influence cellular plasticity and differentiation. 19302538_Decoration of the NM by emerin represents an original approach to identify papillary thyroid carcinomas nuclear shape 19323649_Non-farnesylated and farnesylated carboxymethylated lamin A precursors in human fibroblasts modifies emerin localization. 19727227_An association of Mel18 with emerin was observed in Hutchinson-Gilford progeria syndrome, but not in WT cells. 19759913_BAF and emerin have dynamic roles in genome integrity and might help couple DNA damage responses to the nuclear lamina network 19789182_These findings suggest roles for emerin as a downstream effector and signal integrator for tyrosine kinase signaling pathway(s) at the nuclear envelope. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20054742_Certain patients presenting a typical EDMD phenotype in which no mutations in the EMD or LMNA genes can be confirmed. This may indicate that an Emery-Dreifuss-like dystrophy could also be associated with mutations in other genes. 20474083_Observational study of genetic testing. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21486941_Data provide evidence that 4.1R has functional interactions with emerin and A-type lamin that impact upon nuclear architecture, centrosome-nuclear envelope association and the regulation of beta-catenin transcriptional co-activator activity. 21697856_Data augment the number of EMD mutations by 13.8%, equating to an increase of 5.2% in the total known EMD mutations and to an increase of 6.0% in the number of different mutations. 22480903_Genetic testing identified the mutation in the EMD gene, confirming X-linked recessive (XR) EDMD. The patient's asymptomatic mother was confirmed as a carrier. 23623980_Immunofluorescence assay and biochemical analysis of infected or transfected cells showed that Kaposi's sarcoma-associated herpesvirus p29 expression resulted in delocalization and hyperphosphorylation of emerin. 23873439_Emerin, a conserved LEM-domain protein, is among the few nuclear membrane proteins for which extensive basic knowledge--biochemistry, partners, functions, localizations, posttranslational regulation, roles in development and links to human disease 24014020_Emerin and BAF associated only in histone- and lamin-B-containing fractions. The S173D mutation specifically and selectively reduced GFP-emerin association with BAF by 58% 24375709_Results highlight the interactions at the nuclear envelope where mutations in the EMD and TMPO gene in combination with mutations in SUN1 have an impact on several components of the network. 24819607_These data suggest a new role of EMD as an enhancer of autophagosome formation in the C16-ceramide autophagy pathway in colon cancer cells. 24950247_the nucleoplasmic domains of Samp1 and Emerin can bind directly to each other. 24997722_Findings show a novel EMD deletion causing rare clinical presentations which broaden the heterogeneous spectrum of phenotypes attributed to EMD mutations and provide new insight of genotype-phenotype correlations between EMD mutations and EDMD symptoms. 25052089_Association of emerin with nuclear BAF in cells required the LEM domain (residues 1-47). 25502304_X-linked Emery-Dreifuss muscular dystrophy may occur along with dilative cardiomyopathy. 27865926_results indicate that emerin negatively regulates Notch signaling by promoting the retention of the NICD at the nuclear membrane 27960036_It has been concluded that the LEM domain, responsible for binding to the chromatin protein BAF, undergoes a conformational change during self-assembly of emerin N-terminal region. 28238874_Epstein-Barr virus early lytic protein BFRF1 alters emerin distribution and post-translational modifications. 28290476_Emerin plays a crucial role in nuclear invagination and in the nuclear calcium transient in Emery-Dreifuss muscular dystrophy. 29510091_the mobility of YFP-Emerin was higher in Samp1 knock out cells and lower in cells overexpressing Samp1, suggesting that Samp1 significantly attenuates the mobility of Emerin in the nuclear envelope. The mobility of Emerin depends on RanGTP. The affinity between Samp1 and Emerin is decreased in the presence of Ran, suggesting that Ran attenuates the interaction between Samp1 and Emerin. 30137533_Dimerization of barrier to autointegration factor 1 protein (BAF) is essential for both lamin A/C and emerin binding. 30154147_emerin is a mediator of nuclear shape stability in cancer; destabilization of emerin can promote metastasis 30188764_evaluation of the diagnostic value of emerin and CD56 in papillary thyroid carcinoma, using immunohistochemistry 30449594_Hop stabilises emerin and that loss of Hop alters nuclear structure via emerin degradation. 30542735_Study data revealed that low expression of emerin contributes not only to the oval shape of nuclei, but also to the enlargement in nuclear size in lung adenocarcinoma pathological specimens. 30871242_EmeryDreifuss muscular dystrophy type 1 phenotype may be strengthened by the toxicity of truncated emerin expressed in patients with certain nonsense mutations in EMD. 31117946_Taken together, these studies underscore a remarkable interplay between Lamin A/C and Emerin in modulating cytoskeletal organization of actin and NM1 that impinges on chromatin dynamics and function in the interphase nucleus. 32517247_Emerin Phosphorylation during the Early Phase of the Oxidative Stress Response Influences Emerin-BAF Interaction and BAF Nuclear Localization. 32824881_The Molecular Basis and Biologic Significance of the beta-Dystroglycan-Emerin Interaction. 33771882_Defects in Emerin-Nucleoskeleton Binding Disrupt Nuclear Structure and Promote Breast Cancer Cell Motility and Metastasis. 33822520_Immunohistochemical Expression of Emerin in Renal Cell Carcinomas: Does It Contribute to Histopathological Features and WHO/ISUP Grading in Predicting Prognosis? 34734530_Emerin knockdown induces the migration and invasion of hepatocellular carcinoma cells by up-regulating the cytoplasmic p21. | ENSMUSG00000001964 | Emd | 1.101296e+04 | 1.5446881 | 0.627315576 | 0.3165340 | 3.938535e+00 | 0.0471918616 | 0.65905990 | No | Yes | 1.389601e+04 | 1974.345574 | 7.212566e+03 | 1051.607152 | |
| ENSG00000102743 | 10166 | SLC25A15 | protein_coding | Q9Y619 | FUNCTION: Ornithine-citrulline antiporter. Connects the cytosolic and the intramitochondrial reactions of the urea cycle by exchanging cytosolic ornithine with matrix citrulline (PubMed:12807890). The stoichiometry is close to 1:1 (By similarity). {ECO:0000250|UniProtKB:A0A0G2K309, ECO:0000269|PubMed:12807890}. | Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene is a member of the mitochondrial carrier family. The encoded protein transports ornithine across the inner mitochondrial membrane from the cytosol to the mitochondrial matrix. The protein is an essential component of the urea cycle, and functions in ammonium detoxification and biosynthesis of the amino acid arginine. Mutations in this gene result in hyperornithinemia-hyperammonemia-homocitrullinuria (HHH) syndrome. There is a pseudogene of this locus on the Y chromosome.[provided by RefSeq, May 2009]. | hsa:10166; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; L-ornithine transmembrane transporter activity [GO:0000064]; mitochondrial L-ornithine transmembrane transport [GO:1990575]; urea cycle [GO:0000050] | 11668643_novel mutations in patients with hyperornithinemia, hyperammonemia, and homocitrullinuria syndrome 12807890_expression, reconstitution, functional characterization, and tissue distribution of two human isoforms 14759633_Hyperornithinemia, hyperammonemia, and homocitrullinuria syndrome with evidence of mitochondrial dysfunction due to a novel SLC25A15 (ORNT1) gene mutation in a Palestinian family. 16256388_The DeltaF 188 mutant was not incorporated into the membrane to the same extent as wild type, but retained significant residual activity and lost stereospecificity. 16376511_A novel R275X mutation of the SLC25A15 gene in a Japanese patient with the HHH syndrome 17825324_The three patients were homozygous for a novel mutation in ORNT1 with a Gly220Arg change. We suggest including HHH syndrome in the differential diagnosis of patients found to have stroke-like lesions on brain MRI. 18978333_Clinical presentations and outcomes varied significantly in HHH syndrome patients homozygous for delF188 mutations in SLC25A15. 19242930_16 additional Hyperornithinemia-hyperammonemia-homocitrullinuria cases were collected and the spectrum of SLC25A15/ORC1 mutations, was expanded. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22262851_characterized mutations of the proposed substrate binding site in ORC1 and ORC2; demonstrated that the residue at position 179 in the 2 soforms is largely responsible for the difference in their substrate specificity;concluded that Arg-179 is a key residue in the opening of the carrier to the matrix side 22292090_Useful insights for in-depth understanding of the molecular mechanism of the HHH syndrome and developing effective drugs against the disease. 22465082_Mutation analysis revealed two novel mutations in the ORNT1 gene. 23266187_Compares and contrasts all the known human SLC25A* genes and includes functional information. 30121301_Structure/function relationships of the human mitochondrial ornithine/citrulline carrier by Cys site-directed mutagenesis. Relevance to mercury toxicity. 30403179_Overexpression of SLC25A15 is involved in the progression of melanoma and may predict the prognosis of melanoma. | ENSMUSG00000031482 | Slc25a15 | 1.638940e+03 | 0.5692123 | -0.812961242 | 0.3382346 | 5.829868e+00 | 0.0157563084 | 0.44888036 | No | Yes | 1.041547e+03 | 193.569819 | 1.952609e+03 | 371.487156 | |
| ENSG00000102904 | 55815 | TSNAXIP1 | protein_coding | Q2TAA8 | FUNCTION: Possible role in spermatogenesis. {ECO:0000250|UniProtKB:Q99P25}. | Alternative splicing;Coiled coil;Cytoplasm;Developmental protein;Differentiation;Reference proteome;Spermatogenesis | hsa:55815; | cytoplasm [GO:0005737]; perinuclear region of cytoplasm [GO:0048471]; cell differentiation [GO:0030154]; spermatogenesis [GO:0007283] | 21683752_rs1630250 was associated in males w/ methamphetamine dependence in allele analysis. results not significant after Bonferroni correction. findings suggest TSNAX does not play a role in methamphetamine dependence in Japanese population | ENSMUSG00000031893 | Tsnaxip1 | 1.437010e+02 | 0.8835668 | -0.178588819 | 0.3650963 | 2.474025e-01 | 0.6189099902 | 0.91466270 | No | Yes | 8.015164e+01 | 18.899230 | 1.054108e+02 | 25.425204 | ||
| ENSG00000102931 | 23568 | ARL2BP | protein_coding | Q9Y2Y0 | FUNCTION: Together with ARL2, plays a role in the nuclear translocation, retention and transcriptional activity of STAT3. May play a role as an effector of ARL2. {ECO:0000269|PubMed:18234692}. | 3D-structure;Alternative splicing;Cell projection;Ciliopathy;Cilium;Cytoplasm;Cytoskeleton;Disease variant;Mitochondrion;Nucleus;Reference proteome | ADP-ribosylation factor (ARF)-like proteins (ARLs) comprise a functionally distinct group of the ARF family of RAS-related GTPases. The protein encoded by this gene binds to ARL2.GTP with high affinity but does not interact with ARL2.GDP, activated ARF, or RHO proteins. The lack of detectable membrane association of this protein or ARL2 upon activation of ARL2 is suggestive of actions distinct from those of the ARFs. This protein is considered to be the first ARL2-specific effector identified, due to its interaction with ARL2.GTP but lack of ARL2 GTPase-activating protein activity. [provided by RefSeq, Jul 2008]. | hsa:23568; | centrosome [GO:0005813]; cilium [GO:0005929]; cytosol [GO:0005829]; midbody [GO:0030496]; mitochondrial intermembrane space [GO:0005758]; mitochondrial matrix [GO:0005759]; nucleus [GO:0005634]; spindle [GO:0005819]; GTPase regulator activity [GO:0030695]; transcription coactivator activity [GO:0003713]; maintenance of protein location in nucleus [GO:0051457]; positive regulation of tyrosine phosphorylation of STAT protein [GO:0042531]; signal transduction [GO:0007165] | 19368893_Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector. 21665939_overexpression of the amino (N)-terminal region of G3BP, including the binding region for BART mRNA, dominant-negatively inhibits formation of the complex between endogenous G3BP and BART mRNA, and increases the expression of BART. 21833473_Our results imply that BART increases active RhoA by inhibiting ARL2 function, which in turn inhibits invasiveness of cancer cells. 21901094_We identify a subset of BART miRNAs that are restricted to Latency III in normal infection but are up regulated in tumors that express Latency I and II. 22532868_These results imply that BART contributes to regulating PKCalpha activity through binding to ANX7, thereby affecting the invasiveness of pancreatic cancer cells. 22745590_Our results imply that BART regulates actin-cytoskeleton rearrangements at membrane ruffles through modulation of the activity of Rac1, which, in turn, inhibits pancreatic cancer cell invasion. 23685147_EBV-miR-BART1 could influence the expression of metabolism-associated genes and might be involved in cancer metabolism in nasopharyngeal carcinoma 23849777_Mutations in ARL2BP cause autosomal-recessive retinitis pigmentosa. 24899173_EBV also downregulates two immediate early genes by miR-BART20-5p. 26135619_Alteration of EBV encoded miR-BART1 expression results in an increase in migration and invasion of nasopharyngeal carcinoma in vitro and causes metastasis in vivo. EBV-miR-BART1 directly targets the cellular tumour suppressor PTEN. 27790702_Subsequent analysis of 844 index cases did not reveal further pathogenic chances in ARL2BP indicating that mutations in ARL2B are a rare cause of arRCD (about 0.1%) in a large cohort of French patients. 30210231_This study identified two homozygous variants in ARL2BP as a rare cause of autosomal recessive retinitis pigmentosa. Further studies are required to define the underlying disease mechanism causing retinal degeneration as a result of mutations in ARL2BP and any phenotype-genotype correlation associated with residual levels of the wild-type transcript. 31425546_Study identified multiple ciliopathy phenotypes associated with mutations in ARL2BP in human patients and in a mouse knockout model. Spermiogenesis is impaired, resulting in abnormally shaped heads, shortened and mis-assembled sperm tails, and loss of axonemal doublets. ARL2BP is required for the structural maintenance of cilia as well as of the sperm flagellum, and that its deficiency leads to syndromic ciliopathy. 33438581_ARL3 activation requires the co-GEF BART and effector-mediated turnover. 34681596_Epstein-Barr Virus miR-BART1-3p Regulates the miR-17-92 Cluster by Targeting E2F3. | ENSMUSG00000031776 | Arl2bp | 2.361062e+03 | 0.9045830 | -0.144675187 | 0.2684230 | 2.968089e-01 | 0.5858898994 | 0.90467248 | No | Yes | 2.363035e+03 | 290.806604 | 2.235896e+03 | 282.231441 | |
| ENSG00000102977 | 65057 | ACD | protein_coding | Q96AP0 | FUNCTION: Component of the shelterin complex (telosome) that is involved in the regulation of telomere length and protection. Shelterin associates with arrays of double-stranded TTAGGG repeats added by telomerase and protects chromosome ends. Without its protective activity, telomeres are no longer hidden from the DNA damage surveillance and chromosome ends are inappropriately processed by DNA repair pathways. Promotes binding of POT1 to single-stranded telomeric DNA. Modulates the inhibitory effects of POT1 on telomere elongation. The ACD-POT1 heterodimer enhances telomere elongation by recruiting telomerase to telomeres and increasing its processivity. May play a role in organogenesis. {ECO:0000269|PubMed:15181449, ECO:0000269|PubMed:16166375, ECO:0000269|PubMed:16880378, ECO:0000269|PubMed:17237768, ECO:0000269|PubMed:20231318, ECO:0000269|PubMed:25205116, ECO:0000269|PubMed:25233904}. | 3D-structure;Alternative splicing;Chromosome;DNA-binding;Disease variant;Dyskeratosis congenita;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Telomere;Ubl conjugation | This gene encodes a protein that is involved in telomere function. This protein is one of six core proteins in the telosome/shelterin telomeric complex, which functions to maintain telomere length and to protect telomere ends. Through its interaction with other components, this protein plays a key role in the assembly and stabilization of this complex, and it mediates the access of telomerase to the telomere. Multiple transcript variants encoding different isoforms have been found for this gene. This gene, which is also referred to as TPP1, is distinct from the unrelated TPP1 gene on chromosome 11, which encodes tripeptidyl-peptidase I. [provided by RefSeq, Jul 2008]. | hsa:65057; | chromosome, telomeric region [GO:0000781]; nuclear body [GO:0016604]; nuclear telomere cap complex [GO:0000783]; nucleoplasm [GO:0005654]; shelterin complex [GO:0070187]; telomerase holoenzyme complex [GO:0005697]; DNA polymerase binding [GO:0070182]; protein-containing complex binding [GO:0044877]; telomeric DNA binding [GO:0042162]; embryonic limb morphogenesis [GO:0030326]; establishment of protein localization to telomere [GO:0070200]; intracellular protein transport [GO:0006886]; negative regulation of telomere maintenance via telomerase [GO:0032211]; positive regulation of single-stranded telomeric DNA binding [GO:0060381]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomere maintenance via telomerase [GO:0032212]; protection from non-homologous end joining at telomere [GO:0031848]; protein localization to chromosome, telomeric region [GO:0070198]; segmentation [GO:0035282]; skeletal system development [GO:0001501]; telomere assembly [GO:0032202]; telomere capping [GO:0016233]; telomere maintenance [GO:0000723]; telomere maintenance via telomerase [GO:0007004]; urogenital system development [GO:0001655] | 15181449_a paper that firstly reported cloning of human TTP1 (PTOP) and its biological function at telomere. TPP1 interacts with both POT1 and TIN2, heterodimerizes with POT1 and regulates POT1 telomeric recruitment and telomere length. 15380063_TINT1 localized to telomeres via TIN2, where it functions as a negative regulator of telomerase-mediated telomere elongation. 16504561_Sequencing of ACD in 15 patients with clinical features of IMAGe syndrome, adrenal hypoplasia congenita, or congenital adrenal insufficiency revealed no coding mutations, but three novel SNPs were identified 16880378_coordinated interactions among TPP1, TIN2, TRF1, and TRF2 may ensure robust assembly of the telosome, telomere targeting of its subunits, and, ultimately, regulated telomere maintenance 17237767_findings highlight the critical role of TPP1 in telomere maintenance, and support a yin-yang model in which TPP1 and POT1 function as a unit to protect human telomeres, by both positively and negatively regulating telomerase access to telomere DNA 17237768_crystal structure of human TPP1 reveals an oligonucleotide/oligosaccharide-binding fold that is structurally similar to the beta-subunit of the telomere end-binding protein of a ciliated protozoan; TPP1 is the missing beta-subunit of human POT1 17373762_REVIEW:TPP1 as a critical mediator of control of telomerase activity 17466001_No mutations were identified in ACD in this collection of patients with ACTH resistance phenotypes. However, the newly identified SNPs in ACD should be more closely examined for possible links to disease. 17466001_Observational study of genotype prevalence. (HuGE Navigator) 17632522_Tpp1 is required for the protective function of Pot1 proteins. 19424630_Increased expression of TPP1 correlates with resistance to radiation in human laryngeal cancer cell lines. 19648609_Studies indicate that TPP1 and POT1can form heterodimers that bind to the telomeric single-stranded DNA, an activity that is central for telomere end capping. 19766477_Observational study of gene-disease association. (HuGE Navigator) 20094033_Results support a model in which POT1-TPP1 enhances telomerase processivity in a manner markedly different from the sliding clamps used by DNA polymerases. 20404094_TIN2-anchored TPP1 plays a major role in the recruitment of telomerase to telomeres in human cells. 20937264_Observational study of gene-disease association. (HuGE Navigator) 21209389_Mouse gene deletion experiments revealed DNA-damage-response pathways that threaten chromosome ends and how the components of the telomeric shelterin complex prevent activation of these pathways.[Shelterin] 21355086_The presence of dysfunctional telomeres in chronic lymphocytic leukemia did not correlate with telomere shortening or chromatin marks deregulation but with a down-regulation of 2 shelterin genes: ACD and TINF2. 21461822_the human POT1-TPP1 complex is a processivity factor for telomerase 21829167_UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication 21833529_Altered expression of TPP1 might contribute to persistent proliferation of fibroblast-like synovial cells in rheumatoid arthritis. 22863003_Study shows that the OB-fold domain of the telomere-binding protein TPP1 recruits telomerase to telomeres through an association with the telomerase reverse transcriptase TERT; data define a potential interface for telomerase-TPP1 interaction required for telomere maintenance and implicate defective telomerase recruitment in telomerase-related disease. 23103865_seven separation-of-function mutations map to a patch of amino acids on the surface of TPP1, the TEL patch, that both recruits telomerase to telomeres and promotes high-processivity DNA synthesis 23509301_Blocking TPP1 S111 phosphorylation by mutating residue S111 led to reduced telomerase association and telomere shortening. 23616058_POT1-TPP1 binds telomeric DNA in a coordinated manner to facilitate assembly of the nucleoprotein complexes into a state that is more accessible to enzymatic activity. 23862686_Telomere damage and reduced TPP1 dimerization as a result of Akt inhibition was also accompanied by diminished recruitment of TPP1 and POT1 to the telomeres. 24260532_Elevated expression of TPP1 in human colorectal cancer cells could protect telomere from DNA damage and confer radioresistance. 24513288_Down-regulation of TPP1 induced cell apoptosis in telomerase-negative osteosarcoma cell line. 24516170_G-quadruplex formation of telomeres significantly enhances the ability of POT1/TPP1 to block RPA's access to telomeres. 24721976_Shelterin protein TPP 1 interacts with hTERT and recruits hTERT onto the telomeres, suggesting that TPP 1 might be involved in regulation of telomere shortening. 24780581_TPP1 has recently emerged as a primary contributor in protecting telomere DNA and in recruiting telomerase to the telomere ends. (Review) 25128433_TPP1 provides an essential step of telomerase activation as well as feedback regulation of telomerase by telomere length, which is necessary to determine the appropriate telomere length set point in human embryonic stem cells 25172512_TPP1 is a binding partner and substrate for the deubiquitinating enzyme USP7. 25205116_The data support a causal relationship between a TPP1 mutation and bone marrow disorders in a family. 25263700_these data provide a molecular basis by which POT1-TPP1 increases the processivity of telomerase15. Further, we show that this increased processivity may arise from the dynamic sliding of POT1-TPP1 that induces fast translocation of telomerase. 25271372_Mutations have been identified in the TEN-domain of TERT that disrupt the interaction of telomerase with TPP1 in vivo and in vitro but have very little effect on the catalytic activity of telomerase. 25505254_Clustering of novel mutations in the POT1 binding domain of ACD was statistically higher (P = .005) in melanoma probands compared with population control individuals (n = 6785). 26345285_a novel ACD mutation(p.G223V)is detected; ACD is a novel gene involved in childhood pre-B acute lymphoblastic leukemia and may play a functional role in enhancing leukemia cell survival 26365187_The conservation between fission yeast Tpz1-Pot1 and human TPP1-POT1 interactions resulted in mapping a human melanoma-associated POT1 mutation (A532P) to the TPP1-POT1 interface. 26503784_that the insertion in fingers domain can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner 27173378_Binding of POT1-TPP1 unfolds telomere secondary structure to assist loading of additional heterodimers. 27396482_We found that NEK6-mediated phosphorylation of TPP1 Ser255 in G2/M phase regulates the association between telomerase activity and TPP1. Furthermore, we found evidence that POT1 negatively regulates TPP1 phosphorylation because the level of Ser255 phosphorylation was elevated when telomeres were elongated by a POT1 mutant lacking its OB-fold domains 27655633_Together, these functional data combined with biophysical analyses and homology modeling provide a molecular understanding of the diverse contributions of TPP1 in telomere maintenance. 27807141_We report the crystal structure of a mutant TPP1 implicated in dyskeratosis congenita (DC) to reveal how the mutation disrupts a region of the protein essential for telomerase function. Furthermore, we demonstrated that this mutation, when introduced into a human cell line, is sufficient to cause the cellular underpinnings of DC. 28393830_A defective POT1-TPP1 complex leads to longer and fragile telomeres, which in turn promotes genomic instability and cancer. 28393832_several missense mutations in human cancers that disrupt the POT1C-TPP1 interaction, resulting in POT1 instability, were identified. 29386102_show that TPP1 NOB is critical for telomerase function and demonstrate that the telomerase interaction surface on TPP1 is more elaborate than previously appreciated 29891727_Missense variant rs149418249 in the 11th exon of TPP1 (ACD) is significantly associated with colorectal cancer risk. 30064976_This study reports the first instance of homozygous TPP1 OB-fold variants in patients from 2 unrelated families who have overlapping phenotypes with dyskeratosis congenital. 30080989_is involved in glioblastoma radioresistance, likely through the modulation of telomerase activity, proliferation, and apoptosis. 30611803_shorter telomere length and downregulation of the major shelterin components TRF2 and TPP1 (ACD) leading to 'telomere uncapping', might play a critical role in recurrent pregnancy loss. 31158366_Authors found that TPP1(ACD) upon binding to TIN2 induces changes that expand TIN2 binding capacity, such that TIN2 can accommodate both TRF1 and TRF2 simultaneously. Authors suggest a molecular model that explains why ACD is essential for the stable formation of TRF1-TIN2-TRF2 core complex. 31383750_All of the TIN2 isoforms stimulated telomerase to similar extents. Mutations in the TPP1 TEL patch abrogated this stimulation, suggesting that TIN2 functions with TPP1/POT1 to stimulate telomerase processivity. 31515401_Pulmonary fibrosis linked to variants in the ACD gene, encoding the telomere protein TPP1. 31685617_The assembly of telomere ssDNA substrates of differing lengths bound by POT1-TPP1 heterodimers was affected by a POT1 missense mutation. There was a telomerase inhibitory role when several native POT1-TPP1 proteins coat physiologically relevant lengths of telomere ssDNA. 32171974_Shelterin and the replisome: at the intersection of telomere repair and replication. 32903138_Telomere length set point regulation in human pluripotent stem cells critically depends on the shelterin protein TPP1. 32976206_Constitutional variants in POT1, TERF2IP, and ACD genes in patients with melanoma in the Polish population. 33446513_Identification and characterization of novel ACD variants: modulation of TPP1 protein level offsets the impact of germline loss-of-function variants on telomere length. 33549587_Active and Passive Destabilization of G-Quadruplex DNA by the Telomere POT1-TPP1 Complex. 33673424_A Role for Human DNA Polymerase lambda in Alternative Lengthening of Telomeres. 33822766_TPP1 mutagenesis screens unravel shelterin interfaces and functions in hematopoiesis. 34534331_Multiple hPOT1-TPP1 cooperatively unfold contiguous telomeric G-quadruplexes proceeding from 3' toward 5', a feature due to a 3'-end binding preference and to structuring of telomeric DNA. 35201900_Structural basis of human telomerase recruitment by TPP1-POT1. | ENSMUSG00000038000 | Acd | 1.802720e+03 | 1.0789996 | 0.109694330 | 0.3184190 | 1.191787e-01 | 0.7299269647 | 0.94303490 | No | Yes | 1.649119e+03 | 194.022537 | 1.416097e+03 | 171.238665 | |
| ENSG00000103037 | 79918 | SETD6 | protein_coding | Q8TBK2 | FUNCTION: Protein-lysine N-methyltransferase. Monomethylates 'Lys-310' of the RELA subunit of NF-kappa-B complex, leading to down-regulation of NF-kappa-B transcription factor activity (PubMed:21131967, PubMed:30189201, PubMed:21515635). Monomethylates 'Lys-8' of H2AZ (H2AZK8me1) (PubMed:23324626). Required for the maintenance of embryonic stem cell self-renewal (By similarity). Methylates PAK4. {ECO:0000250|UniProtKB:Q9CWY3, ECO:0000269|PubMed:21131967, ECO:0000269|PubMed:21515635, ECO:0000269|PubMed:23324626, ECO:0000269|PubMed:30189201}. | 3D-structure;Alternative splicing;Methylation;Methyltransferase;Nucleus;Reference proteome;S-adenosyl-L-methionine;Transferase | This gene encodes a methyltransferase that adds a methyl group to the histone H2AZ, which is involved in nuclear receptor-dependent transcription. The protein also interacts with several endogenous proteins which are involved in nuclear hormone receptor signaling. A related pseudogene is located on chromosome 2. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:79918; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; NF-kappaB binding [GO:0051059]; protein-lysine N-methyltransferase activity [GO:0016279]; S-adenosyl-L-methionine binding [GO:1904047]; histone lysine methylation [GO:0034968]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; peptidyl-lysine monomethylation [GO:0018026]; regulation of inflammatory response [GO:0050727]; stem cell differentiation [GO:0048863]; stem cell population maintenance [GO:0019827] | 21131967_SETD6 mono-methylate Rela on K310 and negatively regulates RelA transcription activity 21131967_methylation of the NF-kappaB results in repression of NF-kappaB signaling 23324626_SETD6 monomethylates H2AZ on lysine 7. 24751716_Several chromatin proteins that associate with SETD6 are identified and SETD6 is described as an essential factor for nuclear receptor signaling and cellular proliferation. 26780326_these findings demonstrate that SETD6 negatively regulates the Nrf2-mediated oxidative stress response through a physical and catalytically independent interaction with DJ1 at chromatin. 26841865_PAK4 methylation by SETD6 promotes the activation of the Wnt/beta-catenin pathway. 28122346_High SETD6 expression is associated with bladder cancer. 28973356_Findings suggest that the identified mutation impairs the normal function of SETD6, which may result in the deregulation of the different pathways in which it is involved, contributing to the increased susceptibility to cancer in this FCCTX family. 30189201_Study demonstrates that SETD6 monomeric, dimeric and trimeric forms are stabilized by the methyl donor, S-adenosyl-l-methionine. SETD6 has auto-methylation activity at K39 and K179, which serves as the major auto-methylation sites with a moderate auto-methylation activity toward K372. Data support a model by which SETD6 auto-methylation and self-interaction positively regulate its enzymatic activity in vitro. 30226578_WDR5 was identified as a substrate of SETD6 and it was revealed that the methylation of specific lysines (K207/K325) of WDR5 was critical for maintaining global histone H3K4me3 levels and promoting breast cancer cell proliferation and migration 30622182_During mitosis SETD6 binds and methylates PLK1 31081088_MiR-411 inhibits gastric cancer proliferation and migration through targeting SETD6. 31370726_Lysine methyltransferase SETD6 modifies histones on a glycine-lysine motif. 33710605_Silencing of SETD6 inhibits the tumorigenesis of oral squamous cell carcinoma by inhibiting methylation of PAK4 and RelA. 34039605_BRD4 methylation by the methyltransferase SETD6 regulates selective transcription to control mRNA translation. | ENSMUSG00000031671 | Setd6 | 1.484784e+03 | 0.8611042 | -0.215740271 | 0.2687257 | 6.478999e-01 | 0.4208645154 | 0.84954815 | No | Yes | 1.235277e+03 | 171.970534 | 1.646701e+03 | 234.726196 | |
| ENSG00000103197 | 7249 | TSC2 | protein_coding | P49815 | FUNCTION: In complex with TSC1, this tumor suppressor inhibits the nutrient-mediated or growth factor-stimulated phosphorylation of S6K1 and EIF4EBP1 by negatively regulating mTORC1 signaling (PubMed:12271141, PubMed:28215400). Acts as a GTPase-activating protein (GAP) for the small GTPase RHEB, a direct activator of the protein kinase activity of mTORC1 (PubMed:15340059). May also play a role in microtubule-mediated protein transport (By similarity). Also stimulates the intrinsic GTPase activity of the Ras-related proteins RAP1A and RAB5 (By similarity). {ECO:0000250|UniProtKB:P49816, ECO:0000269|PubMed:12271141, ECO:0000269|PubMed:15340059, ECO:0000269|PubMed:28215400}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Epilepsy;GTPase activation;Host-virus interaction;Membrane;Phosphoprotein;Reference proteome;Tumor suppressor;Ubl conjugation | Mutations in this gene lead to tuberous sclerosis complex. Its gene product is believed to be a tumor suppressor and is able to stimulate specific GTPases. The protein associates with hamartin in a cytosolic complex, possibly acting as a chaperone for hamartin. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:7249; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; lysosome [GO:0005764]; membrane [GO:0016020]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; postsynaptic density [GO:0014069]; TSC1-TSC2 complex [GO:0033596]; GTPase activator activity [GO:0005096]; Hsp90 protein binding [GO:0051879]; phosphatase binding [GO:0019902]; protein homodimerization activity [GO:0042803]; small GTPase binding [GO:0031267]; anoikis [GO:0043276]; endocytosis [GO:0006897]; heart development [GO:0007507]; insulin-like growth factor receptor signaling pathway [GO:0048009]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of insulin receptor signaling pathway [GO:0046627]; negative regulation of mitophagy [GO:1901525]; negative regulation of phosphatidylinositol 3-kinase signaling [GO:0014067]; negative regulation of protein kinase activity [GO:0006469]; negative regulation of protein kinase B signaling [GO:0051898]; negative regulation of TOR signaling [GO:0032007]; negative regulation of Wnt signaling pathway [GO:0030178]; neural tube closure [GO:0001843]; positive chemotaxis [GO:0050918]; positive regulation of GTPase activity [GO:0043547]; positive regulation of macroautophagy [GO:0016239]; protein import into nucleus [GO:0006606]; protein kinase B signaling [GO:0043491]; protein localization [GO:0008104]; regulation of cell cycle [GO:0051726]; regulation of endocytosis [GO:0030100]; regulation of insulin receptor signaling pathway [GO:0046626]; regulation of small GTPase mediated signal transduction [GO:0051056]; vesicle-mediated transport [GO:0016192] | 11112665_Observational study of gene-disease association. (HuGE Navigator) 11403047_missense mutation in the GTPase activating protein homology region in families with tuberous sclerosis complex 11686512_negative regulators of cell division; control of transition from G0/G1 to S phase 11781698_Detected two sequence changes involving the TSC2 stop codon. 11811958_calmodulin signaling in the propagation of this TSC2 activity 11836366_Fluorescence in situ hybridization analysis in a patient with an acrofacial dysostosis-like phenotype, tuberous sclerosis, and polycystic kidney disease shows a microdeletion of approximately 280 kb including the TSC2 gene on chromosome 16p13.3. 12015165_Novel TSC1 and TSC2 mutations in Japanese patients with tuberous sclerosis complex. 12062115_TSC1 and TSC2 mutations in tuberous sclerosis - used DHPLC analysis to facilitate the detection of a mosaic mutation, in the presence of a coexisting constitutional polymorphism. 12127687_effect on EHEN-induced renal and hepatocarcinogenesis in the suppressor gene transgenic rats 12172553_TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. 12176984_tuberin binds with 14-3-3 zeta to regulate phosphorylation of ribosomal protein S6 12271141_hamartin and tuberin function together to inhibit mammalian target of rapamycin (mTOR)-mediated signaling to eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) and ribosomal protein S6 kinase 1 (S6K1) 12364343_associates with 14-3-3 in vivo 12468542_TSC2 expression is regulated by 14-3-3 beta in human cells 12547695_A link exists between this protein and kidney diseases 12581886_Estrogen action induces tyrosine phosphatase activity that regulates stability of tuberin, which may play a crucial role in cellular specific functions such as endocytosis. 12582162_MK2 phosphorylates TSC2, which creates a 14-3-3 binding site and thus regulates the cellular function of the TSC2 tumor suppressor protein 12711473_Mutation in TSC2 and activation of mammalian target of rapamycin signalling pathway in renal angiomyolipoma. 12766909_We conclude that the hamartin/tuberin complex exerted a direct effect on the morphology and adhesive properties of 293 cells through regulation of the level and/or activity of cellular E-cadherin/beta-catenin 12773159_mutated in suberous sclerosis (REVIEW). 12773162_Mutated in tuberous sclerosis. 12773163_mutated in sporadic tumors (REVIEW) 12820960_TSC2 is a GAP for rheb and insulin-mediated rheb activation is PI3K dependent. 12842888_TSC2 binds to rheb and has a role in S6 kinase activation 12869586_Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. 12894220_Human TSC2 triggers mammalian cell size reduction and a dominant-negative TSC2 mutant induces increased size. 14551205_data support a model in which phosphorylation of hamartin regulates the function of the hamartin-tuberin complex during the G2/M phase of the cell cycle 14559897_the TSC1.TSC2 complex is regulated by pam and its ortholog highwire 14680818_Western blot analyses confirmed the deregulation of 14-3-3 proteins upon ectopic overexpression of TSC1 and TSC2. 14756965_Observational study of gene-disease association. (HuGE Navigator) 14756965_TSC2 gene, which is responsible for tuberous sclerosis was identified, and all the exons of TSC2 were analyzed by using polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) from peripheral blood of 28 patients . 14871804_A novel mechanism of post-translational inactivation of the TSC2 protein, tuberin, by physiologically inappropriate phosphorylation is demonstated. 14985384_people with TSC2 mutations were significantly more likely than those with TSC1 mutations to have autistic disorder, a low IQ, and a history of infantile spasms; low IQ was independently associated with both TSC2 mutations and a history of infantile spasm 15059224_Down regulation or loss of tuberin and/or hamartin expression may be permissive to fibrocyte proliferation or promote collagen production leading to fibroepithelial polyp formation. 15066998_Tuberin (TSC2) interact with smad2/smad3 during TGF-beta1 growth regulation. 15072102_tsc2 gene expression is reduced in the majority of subependymal giant cell astrocytomas 15150271_inhibition of B-Raf kinase via Rheb is an mTOR-independent function of tuberin 15175323_Binding with HPV16 E6 causes the proteasome-mediated degradation of Tuberin 15231735_Peutz-Jeghers syndrome and other benign tumor syndromes could be caused by dysregulation of the TSC2 pathway 15340059_To investigate the function of TSC2 and Rheb in mTOR signaling, we analyzed the TSC2-stimulated Rheb GTPase activity. 15355997_Tuberin has a role in binding p27 and negatively regulating its interaction with Skp2 15477556_Cortical tuber giant cells in a case of epileptogenic tuberous sclerosis showed predominantly nuclear hamartin, cytosolic tuberin, and hyperphosphorylation of S6. 15579767_data suggest that PGE2 signaling may promote endometrial tumorigenesis by inactivation of tuberin after its phosphorylation via the Akt signaling pathway 15595939_A total of 12 mutations were detected in 24 Indian TSC families in TSC genes. 15624760_Five of 6 subependymal giant cell astrocytomas(SEGAs) also showed evidence of biallelic mutation of TSC1 or TSC2, suggesting that SEGAs develop due to complete loss of a functional tuberin-hamartin complex. 15647351_monitored 14 previously uncharacterized and six known phosphorylation events after phorbol ester stimulation in the ERK/p90 ribosomal S6 kinase-signaling targets, TSC1 and TSC2, and a protein kinase C-dependent pathway to TSC2 phosphorylation 15798777_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15851026_Erk may modulate mTOR signaling and contribute to disease progression through phosphorylation and inactivation of TSC2. 15874888_Observational study of gene-disease association. (HuGE Navigator) 15963462_identified three sites of TSC2 phosphorylation and a novel site of TSC1 phosphorylation, and investigated the roles of these sites in regulating the activity of the TSC1-TSC2 complex 16053916_investigation of the ability of different opioid receptors to regulate the phosphorylation and degradation of tuberin 16192644_Growth of smooth muscle cells derived from TSC2-renal angiomyolipoma demonstrates that epidermal growth factor is required. 16211238_study provides new insights into cellular roles of TSC proteins and promotes discussion on whether separable functions of these proteins might be associated with clinical differences of TSC1- and TSC2-associated disease 16213898_Reduced expression of tuberin might be involved in the progression of pancreatic cancer. 16237225_patient should be considered as having Subependymal giant cell astrocytoma that developed from two somatic hit mutations in TSC2 16258273_Tuberous sclerosis tumor suppressors TSC1 and TSC2 form a protein complex that integrates and transmits cellular growth factor and stress signals to negatively regulate checkpoint kinase TOR activity, as described in this review. 16341938_Data show that tuberin protein levels are decreased in the frontal cortex of patients with Alzheimer's disease. 16388022_TSC2 may play a critical role in modulating cell migration and invasiveness, which contributes to the pathobiology of LAM. 16537497_Overexpression of TSC2 rescues the migration phenotype of myr-Akt1-expressing tumor cells, and high levels of TSC2 in breast cancer patients correlate with increased metastasis and reduced survival. 16554133_Denaturing high performance liquid chromatography and DNA sequencing analysis of TSC1 and TSC2 revealed 13 types of mutations (30%). Nine novel mutations of TSC2 were identified. 16624901_The genome of Schizosaccharomyces pombe contains tsc1(+) and tsc2(+), homologs of human Tsc1 and Tsc2, respectively. Deletion of either tsc1(+) or tsc2(+) affects gene induction upon nitrogen starvation. 16835931_mutation in the TSC2 gene has a role in acrochordons and pancreatic islet-cell tumors in tuberous sclerosis [case report] 16897363_According to Knudson's two-hit model of tumorigenesis, second-hit mutation and resulting loss of heterozygosity of a tumor suppressor gene (tsc1 and tsc2) is necessary for tumor formation 16905638_Fractionation of synchronized airway smooth cells showed that tuberin enters the nucleus in late G(1), and passage through the cell cycle is necessary for nuclear entry. 16940165_documents the incidence, natural history, and outcome of cardiac tumors in patients with TSC in the largest series yet reported and provides a comparison of these features with TSC1 versus TSC2 mutation 17018601_The TSC/Rheb/mTOR pathway plays a critical role in the regulation of E(2)-induced proliferation. 17114181_mTOR-dependent pathways have roles in IFN signaling and 4E-BP1 and TSC1-TSC2 are key components in the generation of IFN-dependent biological responses 17114346_These findings suggest a link between tuberin nuclear localization and a variety of intracellular signaling events that have direct implications with respect to the role of tuberin in the pathology of tuberous sclerosis and lymphangioleiomyomatosis. 17234746_These functional data indicate that the Crumbs complex is a potential regulator of the mTORC1 pathway, cell metabolism and survival through a direct interaction with TSC1/2. 17273797_During conditions of cell stress, GADD34 forms a stable complex with tuberous sclerosis complex (TSC) 1/2, causes TSC2 dephosphorylation, and inhibits signaling by mammalian target of the rapamycin (mTOR). 17287951_We conclude that large deletions in TSC1 and TSC2 account for about 0.5 and 6% of mutations seen in TSC patients, respectively, and MLPA is a highly sensitive and accurate detection method, including for mosaicism. 17304050_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17373211_implications for the development of cystic kidney disease [REVIEW] 17458623_study presents the cytoplasmic/nuclear distribution of tuberin in cell lines 17470459_p27 localization during the mammalian cell cycle is under the control of the tumor suppressor tuberin 17592551_Angiomyolipoma-derived smooth muscle TSC2-/- cells express survivin when exposed to IGF-1. Survivn expression is also triggered whenever culture conditions perturb normal TSC2-/- cell function. 17632432_Pulmonary lymphangioleiomyomatosis can appear sporadically or be associated with tuberous sclerosis with abnormalities of the TSC2 suppressor gene. 17671177_Erk-mediated TSC2 phosphorylation occurred at a high incidence and positively correlated with mitogen activated protein kinase and mammalian target of rapamycin activation in Tuberous Sclerosis and cancers 17888633_novel tandem-duplication mutation od TSC2 in Chinese tuberous sclerosis patient 17922028_This is the first description of a functional interaction between the tumor suppressor tuberin and the oncogene Ras in regulating apoptosis. 17975002_CD44v6-positive sorted lymphangioleiomyomatosis cells showed loss of heterozygosity at the TSC2 locus; binding of CD44v6 antibody resulted in loss of cell viability. 17989114_Tuberin regulates a specific DNA repair enzyme, OGG1. This regulation may be important in the pathogenesis of kidney tumors in patients with tuberous sclerosis complex. 18032745_Patients with a TSC1 mutation are more likely to have a less severe neurologic and cognitive phenotype than those with a TSC2 mutation. 18060739_Analysis of 15 tuberous sclerosis patient samples in which deletions in TSC2 extended into PKD1 showed no evidence of clustering of breakpoints near the polypyrimidine tract 18085521_Our observations of frequent deletion of TSC2 and the mTOR signalling pathway provide evidence that the oncogenetic lineage of PEComa, as a distinct TSC2-linked neoplasm, is similar to that of angiomyolipoma. 18094073_IFNbeta augments TSC2-dependent inhibition of TSC2-null ELT3 and human lymphangioleiomyomatosis-derived cell proliferation. 18302728_Functional characterisation of TSC2 variants can help identify pathogenic changes in individuals with tuberous sclerosis complex, and assist in the diagnosis and genetic counselling of the index cases and/or other family members 18320306_Results suggest that tuberin dysfunction may represent a mechanism for neuronal damage in Alzheimer's disease (AD), Parkinson's disease with dementia (PD/DLB), and a mouse model of PD. 18342602_Data show that loss of TSC1 or TSC2 in cell lines and mouse or human tumors causes endoplasmic reticulum (ER) stress and activates the unfolded protein response (UPR). 18368626_loss of tuberin in balloon cells of both cortical dysplasia type IIB in tuberous sclerosis complex (TSC)-related and sporadic patients suggests that Focal cortical dysplasia type IIB may represent the focal form of TSC. 18381890_Results indicate that FBW5-DDB1-CUL4-ROC1 is an E3 ubiquitin ligase regulating TSC2 protein stability and TSC complex turnover. 18411301_the TSC1-TSC2 complex inhibits mTORC1 and activates mTORC2, which through different mechanisms promotes Akt activation 18451215_Inactivation of TSC2 via loss of expression or phosphorylation occurred frequently in endometrial carcinoma to activate mTOR signaling 18538015_Involvement of TSC genes and other members of the mTOR signaling pathway in the pathogenesis of oral squamous cell carcinoma. LOH and promoter methylation are two important mechanisms for downregulation of TSC genes. 18794342_in addition to the kinase LKB1, the Tsc1-Tsc2 complex, acting through TORC1, also modulates SAD to regulate axon formation 18794346_results reveal key roles of TSC1/TSC2 in neuronal polarity, suggest a common pathway regulating polarization/growth in neurons and cell size in other tissues, and have implications for the understanding of the pathogenesis of TSC 18807177_This study suggests a pivotal role of PI3 K, MAPK and mTOR pathways, via tuberin, in post-transcriptional control of CXCR4 18848473_These data identify the TSC2-mTOR pathway as a key regulator of innate immune homeostasis with broad clinical implications for infectious and autoimmune diseases, vaccination, cancer, and transplantation. 18926585_Dysregulation of the TSC-mTOR pathway may cause not only tumor development but also metabolic disorders such as diabetes and its comp 18958173_Anti-EGFR antibody efficiently and specifically inhibits human TSC2-/- smooth muscle cell proliferation. 18974095_a novel interaction between DAPK and TSC2 proteins that has revealed a positive link between growth factor stimulation of DAPK and mTORC1 signaling that may ultimately affect autophagy, cell survival, or apoptosis. 19005330_TSC2 which codes for tuberin plays a central role in regulating cell survival and proliferation signaling pathways 19058789_Observational study of gene-disease association. (HuGE Navigator) 19086053_Observational study of gene-disease association. (HuGE Navigator) 19175396_Observational study of gene-disease association. (HuGE Navigator) 19250671_Hypermethylation and downregulation of TSC2 gene is associated with acute myeloid leukemia. 19259131_Data suggest that the three mutations were most likely de novo, as parents of affected patients did not present any features of TSC. 19265534_The data suggest that tuberin and OGG1 are important proteins in the pathogenesis of angiomyolipoma in tuberous sclerosis complex patients. 19332694_Cyst-like cortical tubers in the brain are strongly associated with TSC2 gene mutation in tuberous sclerosis complex. 19332694_Observational study of gene-disease association. (HuGE Navigator) 19357198_A hypomorphic allele of Tsc2 highlights the role of TSC1/TSC2 in signaling to AKT and models mild human TSC2 alleles 19357198_AKT activity is downregulated when Tsc1/Tsc2 function is reduced. 19395678_found increased MMP-2 expression in cells lacking TSC1/TSC2 compared with their respective controls 19419980_Conducted a retrospective review of the chest computed tomography (CT) of 45 female and 20 male patients with tuberous sclerosis complex (TSC) and found cysts consistent with Lymphangioleiomyomatosis (LAM) in 22 (49%) women and two (10%) men. 19422538_mineralized focal cortical dysplasias with balloon cells revealed an increased frequency of TSC2 allelic variants but not TSC1 19443708_that methylation of the TSC2 promoter might cause a complete loss of tuberin in TSC2 cells. 20042714_Tuberin-null cells become nonadherent and invasive and these nonadherent cells express cleaved forms of beta-catenin. 20145209_ARD1 functions as an inhibitor of the mTOR pathway and that dysregulation of the ARD1-TSC2-mTOR axis may contribute to cancer development 20146692_In the brain, TSC2 has been implicated in cell body size, dendritic arborization, axonal outgrowth and targeting, neuronal migration, cortical lamination, and spine formation [REVIEW]. 20160076_results identify a cytoplasmic pathway for ROS-induced ATM activation of TSC2 to regulate mTORC1 signaling and autophagy, identifying an integration node for the cellular damage response 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20219685_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20237422_These findings link TACC3 to novel structural and cell division functions of TSC2. 20304964_p22(phox)-based Nox oxidases maintain HIF-2alpha protein expression through inactivation of tuberin and downstream activation of ribosomal protein S6 kinase 1/4E-BP1 pathway 20354165_Observational study of gene-disease association. (HuGE Navigator) 20363874_AMPK functions to inhibit IGF-I-stimulated PI3K pathway activation through stimulation of IRS-1 serine 794 phosphorylation. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20530489_TSC2 has a role in controlling cell polarity and migration by regulating CDC42 and RAC1 activation 20605525_The rare occurrence of complete loss of TSC1/TSC2 function in human tumors suggests that retaining growth suppressor activity might be beneficial during tumour evolution, perhaps by promoting survival when cells grow in a nutrient-limited environment. 20622004_Observational study of gene-disease association. (HuGE Navigator) 20639436_Lymphangioleiomyomatosis (LAM), occurring sporadically (S-LAM) or in patients with tuberous sclerosis complex (TSC), results from abnormal proliferation of LAM cells exhibiting mutations or loss of heterozygosity (LOH) of the TSC genes, TSC1 or TSC2. 20658316_Observational study of gene-disease association. (HuGE Navigator) 20658316_analysis of polymorphic variants in TSC1 and TSC2 and their association with breast cancer phenotypes 20671064_One of the targets of HtrA1 activity during fetal development is the TSC2-TSC1 pathway. 20818424_Rheb controls proliferation of TSC2-deficient cells by a mechanism that involves regulation of AMPK and p27, and that Rheb is a potential target for TSC/LAM therapy. 20882401_These findings establish a mouse model for TSC-related anxiety phenotypes and suggest that anxiety disorders in TSC have a biological foundation. 21036916_Lymphangioleiomyomatosis is characterized by cystic lung destruction, resulting from proliferation of smooth-muscle-like cells, which have mutations in the tumor suppressor gene TSC2. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21134130_TSC2 binds to the death domain of DAPK. This interaction is required for TSC2 to reduce DAPK protein levels and half-life. DAPK is regulated by the lysosome pathway. Lysosome inhibition blocks TSC2-mediated degradation of DAPK. 21145542_Increased expression of tuberin in human uterine leiomyoma. 21243421_Rattus norvegicus and Mus musculus TSC1 and TSC2 showed very high similarity to the human sequences, this was not the case for Danio rerio, Drosophila melanogaster, Strongylocentrotus purpuratus, Schizosaccharomyces pombe or Disctyostelium discoideum. 21252315_study identified six TSC2 mutations, one indel, one nonsense, and four missense in pancreatic neuroendocrine tumors 21309039_Functional assessment of variants in the TSC2 genes identified in individuals with Tuberous Sclerosis Complex 21329690_REVIEW: TSC signaling in the CNS 21332470_TSC2 R1200W variant, and four other TSC2 missense variants associated with a mild TSC phenotype, confirmed that the changes disrupted the TSC1-TSC2 function. The TSC1-TSC2 interaction was not affected by the amino acid substitution. 21345208_results support the possibility that allele-specific variation in TSC mRNA expression contributes to the variable severity of symptoms in tuberous sclerosis complex patients 21407201_Human TSC2-null fibroblast-like cells induce hair follicle neogenesis and hamartoma morphogenesis. 21412983_TSC1, TSC2 and Rheb function independently of TORC1 in tuberous sclerosis complex.[Review] 21419848_Tuberous sclerosis complex protein TSC2 plays a critical role in Purkinje cell survival by regulating endoplasmic reticulum and oxidative stress. 21449900_In a cohort of tuberous sclerosis patients, there was a trend towards greater severity for patients with TSC2 mutations compared with their TSC1 counterparts, particularly for autistic spectrum disorder 21533174_presence of TSC2 mutations, in addition to TSC1 mutations, underlines the involvement of mTOR signaling in urothelial carcinoma 21555252_This study suggested that allelic imbalances of TSC2 in nonlesional focal epileptic tissue. 21784859_Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. 21795849_ULK1 negatively regulates the kinase activity of mTORC1 and cell proliferation in a manner independent of Atg5 and TSC2 21846442_Eighty percent of cardiac rhabdomyomas are associated with tuberous sclerosis. In this case, molecular testing for tuberous sclerosis identified two variants in the TSC2 gene. 21900748_Tuberin regulates the cellular localization of cyclin B1. 21910228_Two females cases with typical manifestations of Tuberous sclerosis complex, horseshoe kidney, and an identical variant c.5138G>A in exon 39 (p.Arg1713His) of TSC2 gene. 21949787_Mutations in TSC2 is associated with angiomyolipoma. 22090422_study found tuberin and PRAS40 to be potent anti-apoptotic gatekeepers in early mammalian stem-cell differentiation; data allow new insights into the regulation of early stem-cell maintenance and differentiation and identify a new role of the tumor suppressor tuberin and the oncogenic protein PRAS40 22189265_TSC2 protein-truncating mutations and small in-frame mutations are associated with distinctly different intelligence profiles, providing further evidence that different types and locations of TSC germline mutations 22251200_Patients with mutations in TSC2 tended to have a higher frequency of hepatic angiomyolipomas than those with mutations in TSC1 or those with no mutations detected. 22287548_The results defined the TSC2-mTOR pathway as a key determinant in the differentiation of monocytes into M2 phenotype tumor-associated macrophages that promote angiogenesis. 22456611_Perivascular epithelioid cell tumors with TFE3 gene fusions demonstrated intact, robust tuberin protein labeling and no TSC2 loss of heterozygosity. 22490766_This study presented that the mutation rate of the TSC1 and TSC2 genes in Korean patients with tuberous sclerosis complex was 100%. 22608477_TSC2 mutations are more frequent in patients with retinal findings than in those without retinal findings. 22707510_Patient with TSC2 1801A>G mutation was found to have five facial features of TSC, including a rash of facial angiofibromas, a shagreen patch, a forehead plaque, gingival fibromas, and dental pitting. 22795129_The TSC1-TSC2-TBC1D7 (TSC-TBC) complex is the functional complex that senses specific cellular growth conditions and possesses Rheb-GAP activity. 22867869_Missense mutations located in the central region of TSC2 (exons 23-33) are associated with significantly reduced incidence of infantile spasms. 22903760_Data suggest that different, nonterminating TSC2 mutations can have distinct effects on TSC1-TSC2 function, and therefore, on Tuberous sclerosis complex (TSC) pathology. 23217510_Genetic investigation of the coding exons of TSC1 and TSC2 revealed a 4 bp deletion at nucleotides 3693-3696 in exon 30 of the TSC2 gene. 23254740_Two novel TSC2 mutations in Chinese patients with tuberous sclerosis complex. 23348097_Our results suggest that XLID CUL4B mutants are defective in promoting TSC2 degradation and positively regulating mTOR signaling in neocortical neurons. 23389244_No differences emerged in mutation distributions and types in precedent studies, excepting low frequency of the TSC2 nonsense mutation 23689538_Our results suggest that tuberin and p27 are aberrantly expressed in malignant breast tissue 23818547_Pim2 directly phosphorylates TSC2 on Ser-1798 and relieves the suppression of TSC2 on mTOR-C1, leading to multiple myeloma cells proliferation. 23846400_An increased frequency of C>G/G>C and C>T/G>A mutations in the coding strand was found in TSC2. 23867796_OPG stimulated proliferation of cells cultured from explanted LAM lungs, and selectively induced migration of lymphangioleiomyomatosis cells identified by the loss of heterozygosity for TSC2. 23878245_Data indicate a nitrosative-stress signaling pathway that engages ATM, LKB1, AMPK and TSC2 tumor suppressors to repress mTORC1 and regulate autophagy. 23878397_Data indicate that sunitinib inhibited TORC1 in endothelial cells in a Tsc1/Tsc2-dependent manner 23947572_Data show growth-inhibitory and proapoptotic effects of simvastatin on TSC2-null lymphangioleiomyomatosis cells compared with atorvastatin. 24075384_Prenatal diagnosis of an intrathoracic lesion with a family history of parental epilepsy should raise a suspicion of fetal cardiac rhabdomyoma and tuberous sclerosis. 24077282_Alpha B-crystallin has an essential role in TSC1/2 complex deficiency-mediated tumorigenesis. 24271014_TSC2 somatic second-hit mutations are associated with angiofibroma development in tuberous sclerosis. 24318044_This work indicates a novel role for this TSC2 gene, which encodes an activator of cell proliferation in response to androgen stimulation. 24398473_Covalent modification of TSC2 by iNOS-derived NO is associated with impaired TSC2/TSC1 dimerization, mTOR pathway activation, and proliferation of human melanoma. 24444419_oxidative stress induces Tnfaip8 l1/Oxi-beta, which results in increased autophagy by its exclusive binding with FBXW5 to stabilize TSC2 24529380_These data suggest that regulation of TSC2 subcellular localization may be a general mechanism to control its activity and place TSC2 in the amino-acid-sensing pathway to TORC1. 24599401_Findings indicate that neuronal Tsc1/2 complex activity is required for the coordinated regulation of autophagy by AMPK. 24606538_The features of alpha-smooth muscle cells of a patient affected by lymphangioleiomyomatosis associated with Tuberous sclerosis complex, named LAM/TSC cells, bearing a TSC2 mutation and an epigenetic defect causing the absence of tuberin, were investigated. 24683199_Two novel gross deletions of TSC2 gene in Malay patients with tuberous sclerosis complex and TSC2/PKD1 contiguous gene deletion syndrome, respectively. 24698169_In children with tuberous sclerosis complex, nonsense mutations in the TSC2 gene had a correlation with autistic behavior. 24748662_TSC2 also functions as a transcription factor. 24794161_study describes 2 cases of genetically proven TCS2, sharing the same genotype; detected a novel, small and in frame deletion/insertion TSC2 mutation on exon 30 (c.3664_3665delinsTT-p.Asp1222Phe) 24917535_TSC2 mutations are associated with a more severe, earlier presenting tuberous sclerosis complex phenotype. 25114899_Studied conditions that increase the sensitivity of cancer cells to MK-2206. and found reduction by salinomycin of Akt and downregulation of pAkt, pGSk3beta, pTSC2, and p4EBP1 by cotreatment with MK-2206. 25185584_This study demonstrates that TSC2-deficient tumor cells are hypersensitive to oxidative stress-dependent cell death, and provide critical proof of concept that TSC2-deficient cells can be therapeutically targeted 25281918_This is the first mutation and multiplex ligation-dependent probe amplification (MLPA) analyses of TSC2 in Korean Angiomyolipomas that focus on tuberous sclerosis complex. 25355409_A short segment of chromosome 16 encodes the tumor suppressor gene tuberin as well as the protein polycystin 1, which are responsible for tuberous sclerosis complex type 2 and autosomal-dominant polycystic kidney disease type 1, respectively. 25432535_Multiple mutations in TSC2 during kidney development lead to severe phenotype of multifocal renal cell carcinoma. 25476905_In TSC2-deficient angiomyolipoma patient cells, IRF7 is a pivotal factor in the Rheb/mTOR pathway. 25498131_Compared to patients with TSC1 mutations, individuals with TSC2 mutations had a significantly higher frequency of epilepsy and tended to have a higher frequency of infantile spasms. 25563326_a novel frame shift Tuberous Sclerosis Complex-2 Mutation in three patients with Tuberous sclerosis complex but with different severity of symptoms 25565629_PLK1 protein levels are increased in hamartin and tuberin deficient cells and Lymphangioleiomyomatosis patient-derived specimens, and that this increase is rapamycin-sensitive. 25654764_TSC2/mTORC1 signaling contributes to the maintenance of intestinal epithelium homeostasis by regulating Notch activity. 25724664_Data shows frequent loss of TSC2 in hepatocellular carcinoma cells (HCC) and that TSC2-null cell lines were more sensitive to mTOR inhibition by everolimus suggesting that TSC2 loss is a predictive biomarker for the response to everolimus in HCC patients. 25780943_these results demonstrate that TSC2-deficient cells have enhanced choline phospholipid metabolism and reveal a novel function of the TSC proteins in choline lysoglycerophospholipid metabolism 25782670_Results confirm strong association between TSC2 mutation and angiomyolipoma burden, and they indicate that everolimus response occurs regardless of mutation type or location or when no mutation in TSC1 or TSC2 has been identified. 25927202_previously unidentified TSC1 and TSC2 mutations in tuberous sclerosis complex 25972538_pUL38 can activate mTORC1 in both TSC2-dependent and -independent manners. 26252095_A novel frame-shifting mutation c.4258-4261delTCAG in the TSC2 gene is associated with tuberous sclerosis in a Chinese family. 26318033_AKT3 has a role in prostate cancer proliferation through regulation of Akt, B-Raf, and TSC1/TSC2 26393489_Tuberous sclerosis is a syndrome caused by dominant mutations in Tuberin (TSC2),causing Autism spectrum disorder - like behaviors, seizures, intellectual disability and characteristic brain and skin lesions. 26408672_IQ/DQ correlates inversely with predicted levels and/or deleterious biochemical effects of mutant TSC1 or TSC2 protein in tuberous sclerosis complex. 26412398_PAK2 is a direct effector of TSC1-TSC2-RHEB signaling and a new target for rational drug therapy in TSC. 26540169_TSC-related tumors can increase the mutation detection rate, indicate that it is not likely that a third TSC gene exists, and enable provision of genetic counseling to the substantial population of TSC individuals who are currently NMI 26563443_results confirm the consistent finding of TSC2 mutations in LAM samples, and highlight the benefit of laser capture microdissection and in-depth allele analyses for detection, such as NGS 26703369_Our evidence suggests that variants in TSC2 exons 25 or 31 are very unlikely to cause classical TSC, although a role for these exons in tissue/stage specific development cannot be excluded. 26728384_Data show that 10 pathogenic mutations were quickly identified, 7 were located in tuberous sclerosis 1 protein (TSC1) and 3 were observed in tuberous sclerosis 2 protein (TSC2). 26742086_By interfering with TSC-Rheb complex, arginine relieves allosteric inhibition of Rheb by TSC. Arginine cooperates with growth factor signaling which further promotes dissociation of TSC2 from lysosomes and activation of mTORC1. 26831717_Mutations in MTOR, TSC1, or TSC2 were more common in patients who experienced clinical benefit from rapalogs than in those who progressed. 26868506_Lysosomal recruitment of TSC2 is a universal response to stimuli that inactivate | ENSMUSG00000002496 | Tsc2 | 8.604492e+03 | 1.1012679 | 0.139165484 | 0.2744572 | 2.547230e-01 | 0.6137689194 | 0.91292972 | No | Yes | 8.177330e+03 | 1028.509926 | 7.540045e+03 | 972.802918 | |
| ENSG00000103199 | 26048 | ZNF500 | protein_coding | O60304 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:26048; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 1.402735e+03 | 1.2382393 | 0.308290142 | 0.2967308 | 1.085746e+00 | 0.2974157062 | 0.81383297 | No | Yes | 1.283509e+03 | 141.849133 | 9.938069e+02 | 113.025323 | |||||
| ENSG00000103264 | 79791 | FBXO31 | protein_coding | Q5XUX0 | FUNCTION: Component of some SCF (SKP1-cullin-F-box) protein ligase complex that plays a central role in G1 arrest following DNA damage. Specifically recognizes phosphorylated cyclin-D1 (CCND1), promoting its ubiquitination and degradation by the proteasome, resulting in G1 arrest. May act as a tumor suppressor. {ECO:0000269|PubMed:16357137, ECO:0000269|PubMed:19412162}. | 3D-structure;Alternative splicing;Cell cycle;DNA damage;Mental retardation;Phosphoprotein;Reference proteome;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene is a member of the F-box family. Members are classified into three classes according to the substrate interaction domain, FBW for WD40 repeats, FBL for leucing-rich repeats, and FBXO for other domains. This protein, classified into the last category because of the lack of a recognizable substrate binding domain, has been proposed to be a component of the SCF ubiquitination complex. It is thought to bind and recruit substrate for ubiquitination and degradation. This protein may have a role in regulating the cell cycle as well as dendrite growth and neuronal migration. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:79791; | centrosome [GO:0005813]; cytosol [GO:0005829]; neuronal cell body [GO:0043025]; SCF ubiquitin ligase complex [GO:0019005]; cyclin binding [GO:0030332]; anaphase-promoting complex-dependent catabolic process [GO:0031145]; cellular response to DNA damage stimulus [GO:0006974]; mitotic G1 DNA damage checkpoint signaling [GO:0031571]; positive regulation of dendrite morphogenesis [GO:0050775]; positive regulation of neuron migration [GO:2001224]; protein ubiquitination [GO:0016567]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146] | 16357137_Loss of FBXO31 is associated with breast cancer 19412162_ectopic expression of FBXO31 acts through a proteasome-directed pathway to mediate the degradation of cyclin D1, an important regulator of progression from G1 to S phase, resulting in arrest in G1 21537837_CGH array indicated that cases having cyclin D1 with increased copy number were significantly associated with elevated FBXO31 expression levels (P<0.05). FBXO31 could be a novel and robust prognostic marker for ESCC. 22632973_The Skp2 SCF complex, not TRAF6, is a critical E3 ligase for ErbB-receptor-mediated Akt ubiquitination and membrane recruitment in response to EGF. 23469015_ascribe a role for FBXO31 in dendrite growth and neuronal migration in the developing cerebellar cortex. Taken together, we uncovered the centrosomal E3 ligase FBXO31-SCF as a novel regulator of neuronal development 24623383_Truncation of the E3 ubiquitin ligase component FBXO31 causes non-syndromic autosomal recessive intellectual disability. 24828503_FBXO31 interacts with Cdt1 and regulates the degradation of Cdt1 in G2 phase. 24936062_uncover a new mechanism of deactivation of MKK6-p38 and substantiate a novel regulatory role of FBXO31 in stress response 25115392_Expression of FBXO31 inhibited xenograft tumor growth in mice. miR-20a and miR-17 mimics inhibited, whereas the inhibitor of miR-20a and miR-17 increased, the expression of FBXO31, respectively. miR-20a and miR-17 directly bind to the 3'-UTR of FBXO31. 26124108_FBXO31-mediated loss of MDM2 leads to elevated levels of p53, resulting in growth arrest. In cells depleted of FBXO31, MDM2 is not degraded and p53 levels do not increase following genotoxic stress 28500896_FBXO31 targets and ubiquitylates Slug for proteasomal degradation. However, this mechanism is repressed in breast tumors where miR-93 and miR-106a are overexpressed. Our study further unravels an interesting mechanism whereby Slug drives the expression of miR-93 and miR-106a, thus establishing a positive feedback loop to maintain an invasive phenotype 28905993_in addition to revealing that FBXO31 is an independent prognostic marker for esophageal squamous cell carcinoma , our findings substantiate a novel regulatory role of FBXO31 in tumorigenesis and drug resistance of esophageal squamous cell carcinoma 29117943_Results identified FBXO31 as a novel E3 ubiquitin ligase of Snail1, and its low expression contributes to an aberrant accumulation of Snail1 in gastric cancer. Furthermore, F-box domain of FBXO31 and the phosphorylation of Snail1 are essential for the interaction and degradation. These findings reveal the important function of FBXO31 on the regulation of Snail1 protein level in gastric cancer. 29279382_Findings indicate the substrate specificity of the F-box protein FBXO31 and the mechanism of FBXO31-regulated cyclin D1 protein turnover. 29343641_results reveal how alterations in FBXO31 phosphorylation, mediated by AKT and ATM, underlie physiological regulation of FBXO31 levels in unstressed and genotoxically stressed cells 30341246_Higher mRNA expression levels of FBXO1, FBXO31, SKP2, and FBXO5 were significantly associated with worse prognosis for breast cancer patients. (Review) 31413110_FBXO31 plays a pivotal role in preserving genomic integrity by maintaining low cyclin A levels during the G1 phase for faithful genome duplication and segregation 32800832_F-box protein FBXO31 modulates apoptosis and epithelial-mesenchymal transition of cervical cancer via inactivation of the PI3K/AKT-mediated MDM2/p53 axis. 32989326_Mutations disrupting neuritogenesis genes confer risk for cerebral palsy. 33538099_MicroRNA-210 targets FBXO31 to inhibit tumor progression and regulates the Wnt/beta-catenin signaling pathway and EMT in esophageal squamous cell carcinoma. 33675180_Variant recurrence confirms the existence of a FBXO31-related spastic-dystonic cerebral palsy syndrome. 34686346_Loss of FBXO31-mediated degradation of DUSP6 dysregulates ERK and PI3K-AKT signaling and promotes prostate tumorigenesis. 34839191_Effects and mechanisms of FBXO31 on Taxol chemoresistance in esophageal squamous cell carcinoma. 35019165_Novel variants underlying autosomal recessive neurodevelopmental disorders with intellectual disability in Iranian consanguineous families. | ENSMUSG00000052934 | Fbxo31 | 2.389031e+03 | 1.2285686 | 0.296978440 | 0.3158077 | 8.722226e-01 | 0.3503407339 | 0.82969965 | No | Yes | 2.310778e+03 | 297.645251 | 1.675462e+03 | 221.764000 |
| ENSG00000103510 | 84148 | KAT8 | protein_coding | Q9H7Z6 | FUNCTION: Histone acetyltransferase which may be involved in transcriptional activation (PubMed:12397079, PubMed:22020126). May influence the function of ATM (PubMed:15923642). As part of the MSL complex it is involved in acetylation of nucleosomal histone H4 producing specifically H4K16ac (PubMed:16227571, PubMed:16543150, PubMed:21217699, PubMed:22547026, PubMed:22020126). As part of the NSL complex it may be involved in acetylation of nucleosomal histone H4 on several lysine residues (PubMed:20018852, PubMed:22547026). That activity is less specific than the one of the MSL complex (PubMed:20018852, PubMed:22547026). Can also acetylate TP53/p53 at 'Lys-120'. {ECO:0000269|PubMed:12397079, ECO:0000269|PubMed:15923642, ECO:0000269|PubMed:16227571, ECO:0000269|PubMed:16543150, ECO:0000269|PubMed:20018852, ECO:0000269|PubMed:21217699, ECO:0000269|PubMed:22020126, ECO:0000269|PubMed:22547026, ECO:0000269|PubMed:31794431}. | 3D-structure;Acetylation;Activator;Acyltransferase;Alternative splicing;Chromatin regulator;Chromosome;Disease variant;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Transferase;Zinc;Zinc-finger | This gene encodes a member of the MYST histone acetylase protein family. The encoded protein has a characteristic MYST domain containing an acetyl-CoA-binding site, a chromodomain typical of proteins which bind histones, and a C2HC-type zinc finger. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2012]. | hsa:84148; | histone acetyltransferase complex [GO:0000123]; kinetochore [GO:0000776]; MLL1 complex [GO:0071339]; MSL complex [GO:0072487]; NSL complex [GO:0044545]; nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; enzyme binding [GO:0019899]; H3 histone acetyltransferase activity [GO:0010484]; H4 histone acetyltransferase activity [GO:0010485]; histone acetyltransferase activity (H4-K16 specific) [GO:0046972]; histone acetyltransferase activity (H4-K5 specific) [GO:0043995]; histone acetyltransferase activity (H4-K8 specific) [GO:0043996]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; methylated histone binding [GO:0035064]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; chromatin organization [GO:0006325]; histone acetylation [GO:0016573]; histone H4-K16 acetylation [GO:0043984]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; myeloid cell differentiation [GO:0030099]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of autophagy [GO:0010506]; regulation of dosage compensation by inactivation of X chromosome [GO:1900095] | 15923642_These results suggest that hMOF influences the function of ATM. 16024812_hMOF has a role in DNA damage response during cell cycle progression. 16227571_A multisubunit human histone acetylase complex that contains homologs of the Drosophila MSL proteins MOF, MSL1 (hampin A), MSL2, and MSL3 was described. This complex is responsible for histone H4 lysine-16 acetylation of all cellular chromosomes. 17694080_hMOF is an important component of many cellular processes and plays role the in cell malignant transformation. 17967868_MOF is an essential factor for embryogenesis and oncogenesis 18058815_downregulation of hMOF protein expression was associated with lower survival rates identifying hMOF as an independent prognostic marker for clinical outcome in univariate as well as multivariate analyses 19578370_Data show that MOF acetylates TIP5, the largest subunit of NoRC, at a single lysine residue, K633, adjacent to the TIP5 RNA-binding domain, and that SIRT1 (removes the acetyl group from K633. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20018852_Although MSL-associated MOF acetylates nucleosomal histone H4 almost exclusively on lysine 16, NSL-associated MOF exhibits a relaxed specificity and also acetylates nucleosomal histone H4 on lysines 5 and 8. 20479123_MOF activity was associated with general chromatin upon DNA damage and colocalized with the synaptonemal complex in male meiocytes. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21321083_data indicate that H4K20me3 invokes gene repression by antagonizing hMOF-mediated H4K16Ac 21502975_hMOF is autoacetylated in vitro and in vivo, and SIRT1, the deacetylase for H4K16Ac, is responsible for deacetylating acetylated hMOF. 21691301_hMOF employs a novel regulatory mechanism of acetyltransferase activities 22020126_MYST protein acetyltransferase activity requires active site lysine autoacetylation. 22152480_RNAi-mediated silencing of MOF reduced both gene activation and tumor suppression by FOXP3, while both somatic mutations in clinical cancer samples and targeted mutation of FOXP3 in mouse prostate epithelial cells disrupted nuclear localization of MOF. 22586264_uncover novel pathways in which SIRT1 dynamically interacts with and regulates hMOF and TIP60 through deacetylation and provide additional mechanistic insights by which SIRT1 regulates DNA damage response 22918831_new insights into the mechanism and function of MYST HAT autoacetylation. 23628702_The hMOF mediated S phase entry by regulating H4K16ac in the Skp2 promoter region in NSCLC cells. 23638218_Our results demonstrate an important role of KAT8 in cancer 23863932_induction of autophagy is coupled to reduction of histone H4 lysine 16 acetylation through downregulation of the histone acetyltransferase hMOF; and this histone modification regulates the outcome of autophagy 24126058_MOF acetylation of DBC1 inhibits binding to SirT1 and serves as a mechanism that connects DNA damage signaling to SirT1 and cell fate determination. 24452485_low expression of hMOF was strongly correlated with tumor differentiation and survival of patients with gastric cancer. While in patients with renal cell carcinoma, downregulation of hMOF was connected to ccRCC and tissues with T1 tumor status. 24452550_review of regulation and function 24571482_hMOF was overexpressed in human non-small cell lung cancer and was a predictor of poor survival. 24702180_Functional interactions of MYST1 with androgen receptor and NF-KB are critical for prostate cancer progression. 24802406_MOF expression was down-regulated in failing hearts at protein and mRNA levels. 24898892_MOF mediates Notch signaling by manipulating Histone H4 acetylation. 24953651_Mutant MOF-T392A expression abrogates DSB repair in S/G2 phase cells. MOF-T392A has delayed 53BP1 dissociation and decreased DNA association. 25181338_The histone acetyltransferase hMOF suppresses hepatocellular carcinoma growth by targeting the expression of SIRT6. 25483274_Results show the expression of hMOF mRNA and protein was significantly downregulated in ovarian epithelial cancer tissues, and patients with high hMOF levels showed improved survival as compared to those with low hMOF levels. 25873202_Data found downregulation of hMOF in gastric cancer cells and tissues. Declined hMOF expression, but not high level of HDAC4, may account for global histone H4K16ac suggesting that loss of hMOF expression may be involved in gastric cancer progression. 26032517_EZH2 (enhancer of zeste homolog 2) was up-regulated in human oral tongue squamous cell carcinoma tissues and its level positively correlated with level of hMOF. 26091365_MOF is highly enriched in induced pluripotent stem cells (iPSCs), and MOF expression is upregulated during the reprogramming process. The ectopic expression of MOF promotes reprogramming. MOF affects Wdr5 and endogenous Oct4 expression. 26387537_This work identifies MOF as a key regulator of cellular stress response in glomerular podocytes. 26960573_Along with the PHF20/MOF complex, G9a links the crosstalk between ERalpha methylation and histone acetylation that governs the epigenetic regulation of hormonal gene expression. 27038808_recent results indicate MOF is an upstream regulator of the ATM (ataxia-telangiectasia mutated) protein, the loss of which is responsible for ataxia telangiectasia (AT). ATM is a key regulatory kinase that interacts with and phosphorylates multiple substrates that influence critical, cell-cycle control and DNA damage repair pathways in addition to other pathways. 27268279_Data describe a trans-histone modification pathway involving PKN1/histone H3 threonine 11 phosphorylation followed by WDR5/MLL histone methyltransferase and KAT8/histone acetyltransferase recruitment to effect androgen-dependent gene activation and prostate cancer cell proliferation. 27292636_These findings provide insight into the regulation of LSD1 and Epithelial-to-Mesenchymal Transition (EMT) and identify MOF as a critical suppressor of EMT and tumor progression. 27382063_these studies point to the critical and specific role of hMOF Lys-274 autoacetylation in hMOF stability and cognate substrate acetylation and argues that binding of Ac-CoA to hMOF likely drives Lys-274 autoacetylation for subsequent cognate substrate acetylation. 27733505_our findings reveal that TET1 forms a complex with hMOF to modulate its function and the level of H4K16Ac ultimately affect gene expression and DNA repair. 27768893_MOF is a dual-transcriptional regulator of nuclear and mitochondrial genomes connecting epigenetics and metabolism. 28202522_Histone acetyltransferase activity of MOF is required to sustain MLL-AF9 leukemia and may be important for multiple AML subtypes. 28991411_Novel importin alpha1-specific nuclear localization signals were identified in the N-terminal of MOF. 29321206_a novel function for the MYST KATs as lysine propionyltransferases 30598260_MOF promotes endometrial cancer cell growth and proliferation in vitro and in vivo. Clinical evidence indicates that expression MOF is decreased and positively correlated with that of ER-alpha in endometrial cancer tissues. 30842237_KAT8 is down-regulating virus-induced IFN-I production by acetylating the transcription factor IRF3 at lysine 359, inhibiting the recruitment of IRF3 to IFN-I promoters, and decreasing its transcriptional activity. 31794431_Lysine acetyltransferase 8 is involved in cerebral development and syndromic intellectual disability. 31943753_The histone acetyltransferase hMOF promotes vascular invasion in hepatocellular carcinoma. 31990345_Human MOF was highly expressed in endometrial carcinoma and associated with proliferation. High expression of human MOF was associated with late-stage cancer, lymph node metastasis and short survival time, and it was also an independent prognostic risk factor for endometrial carcinoma. 32661120_Histone Acetyltransferase MOF Orchestrates Outcomes at the Crossroad of Oncogenesis, DNA Damage Response, Proliferation, and Stem Cell Development. 33376138_Dysregulation of intercellular signaling by MOF deletion leads to liver injury. 33467728_Substrate Scope for Human Histone Lysine Acetyltransferase KAT8. 33544437_MOF upregulates the estrogen receptor alpha signaling pathway by its acetylase activity in hepatocellular carcinoma. 33657400_Complex-dependent histone acetyltransferase activity of KAT8 determines its role in transcription and cellular homeostasis. 34285225_TGFbeta promotes fibrosis by MYST1-dependent epigenetic regulation of autophagy. 35191804_Ropivacaine represses the proliferation, invasion, and migration of glioblastoma via modulating the microRNA-21-5p/KAT8 regulatory NSL complex subunit 2 axis. | ENSMUSG00000030801 | Kat8 | 1.969749e+03 | 1.0313449 | 0.044526840 | 0.2678354 | 2.769674e-02 | 0.8678237841 | 0.97473316 | No | Yes | 1.855924e+03 | 140.861473 | 1.649786e+03 | 128.666706 | |
| ENSG00000103707 | 123263 | MTFMT | protein_coding | Q96DP5 | FUNCTION: Methionyl-tRNA formyltransferase that formylates methionyl-tRNA in mitochondria and is crucial for translation initiation. {ECO:0000269|PubMed:21907147, ECO:0000269|PubMed:25288793}. | Alternative splicing;Disease variant;Mitochondrion;Primary mitochondrial disease;Protein biosynthesis;Reference proteome;Transferase;Transit peptide | The protein encoded by this nuclear gene localizes to the mitochondrion, where it catalyzes the formylation of methionyl-tRNA. [provided by RefSeq, Jun 2011]. | hsa:123263; | mitochondrion [GO:0005739]; methionyl-tRNA formyltransferase activity [GO:0004479]; conversion of methionyl-tRNA to N-formyl-methionyl-tRNA [GO:0071951] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 21907147_Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. 24461907_We provide detailed clinical descriptions on eleven MTFMT patients and review five previously reported cases 25288793_Data indicate that methionyl-tRNA formyltransferase (MTF) mutation initiated poor formylation of mitochondrial methionyl-tRNA and thereby reduced mitochondrial translation efficiency, causing Leigh syndrome. 25288793_Recessive mutations in MTFMT underlie defects of the mitochondrial respiratory chain, leading to multi-system disease that includes Leigh syndrome. This paper reports on the biochemical activity of such mutant alleles. 32133637_First report of childhood progressive cerebellar atrophy due to compound heterozygous MTFMT variants. 32636430_MTFMT deficiency correlates with reduced mitochondrial integrity and enhanced host susceptibility to intracellular infection. | ENSMUSG00000059183 | Mtfmt | 2.981224e+02 | 1.0710670 | 0.099048693 | 0.3032408 | 1.038024e-01 | 0.7473135008 | 0.94694596 | No | Yes | 3.624316e+02 | 54.200323 | 2.628883e+02 | 40.549059 | |
| ENSG00000104081 | 90427 | BMF | protein_coding | Q96LC9 | FUNCTION: May play a role in apoptosis. Isoform 1 seems to be the main initiator. | 3D-structure;Alternative splicing;Apoptosis;Reference proteome | The protein encoded by this gene belongs to the BCL2 protein family. BCL2 family members form hetero- or homodimers and act as anti- or pro-apoptotic regulators that are involved in a wide variety of cellular activities. This protein contains a single BCL2 homology domain 3 (BH3), and has been shown to bind BCL2 proteins and function as an apoptotic activator. This protein is found to be sequestered to myosin V motors by its association with dynein light chain 2, which may be important for sensing intracellular damage and triggering apoptosis. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:90427; | acrosomal vesicle [GO:0001669]; cytosol [GO:0005829]; mitochondrial outer membrane [GO:0005741]; myosin complex [GO:0016459]; plasma membrane [GO:0005886]; anoikis [GO:0043276]; cellular response to UV [GO:0034644]; negative regulation of autophagy [GO:0010507]; positive regulation of apoptotic process [GO:0043065]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of release of cytochrome c from mitochondria [GO:0090200] | 14574334_Up or downregulation of Bmf isoforms may have a role in regulating growth and survival in B cells and leukemic B-CLL cells 16484005_There was no somatic mutation of BH3 domains of Bad, Bmf and Bcl-G genes in transitional cell carcinoma samples. The data presented here indicate that BH3 domain mutation of these genes is rare in TCCs and may not contribute to the pathogenesis of TCCs. 16830229_histone hyperacetylation may enhance ionizing radiation-induced death via activation of Bmf transcription, thereby implying Bmf as a key molecule for HDAC inhibitors (FK228 and CBHA)-mediated enhancing effect on IR-induced cell death 17360431_Bmf is a central mediator of anoikis in mammary cells and a target of oncogenes that contribute to the progression of glandular epithelial tumors 17557568_data presented here indicate that BH3 domain mutation of the proapoptotic genes Bad, Bmf and Bcl-G is rare in laryngeal squamous cell carcinoma and may not contribute to the apoptosis-resistance mechanisms of laryngeal squamous cell carcinoma 18354037_arsenic trioxide upregulated expression of Bmf, Noxa, and Bim. Silencing of Bmf, Noxa, and Bim significantly protected MM cells from ATO-induced apoptosis 19267218_Bmf is upregulated by PS-341 and has a crucial role in PS-341-mediated glioma cell death through JNK phosphorylation. 19300516_Our data indicate a synergistic role for both bim and Bmf in an apoptotic pathway leading to the clearance of Neisseria gonorrhoeae -infected cells. 19641506_Bmf supports Bim in regulating cell death processes in response to many stimuli. Review. 19671867_MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. 20706276_characterization of the bmf gene locus; molecular basis of the generation of the 2 major isoforms of Bmf; provide evidence that Bmf can act as a sensor for stress that associates with the repression of the conventional CAP-dependent translation machinery 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20861305_Data show that hypoxic conditions inhibit anoikis and block expression of proapoptotic BH3-only family members Bim and Bmf in epithelial cells. 21673109_BMF is induced in human IEC by the loss of cell attachment and is likely to play an important role in the regulation of IEC survival 23192964_Diva binds peptides derived from the BH3 domain of several other proapoptotic Bcl-2 proteins, including mouse Harakiri, Bid, Bak and Bmf. 24901046_Overexpression of ApoL2 did not induce cell death on its own. ApoL2 did not sensitize or protect cells from overexpression of the BH3-only proteins Bmf or Noxa. 25321483_On the corresponding BMF gene promoter, loss of HDAC8 was associated with signal transducer and activator of transcription 3 (STAT3)/specificity protein 3 (Sp3) transcription factor exchange and recruitment of p300. 26181206_these findings suggest that p53-R273H can specifically drive AKT signaling and suppress BMF expression, resulting in enhanced cell survivability and anoikis resistance. 27524613_The findings are consistent with rs539846 influencing chronic lymphocytic leukemia (CLL) susceptibility through differential RELA binding, with direct modulation of BMF expression impacting on anti-apoptotic BCL2, a hallmark of oncogenic dependency in CLL. 27539959_Reciprocal regulation of BMF and BIRC5 is linked to Eomes overexpression in colorectal cancer. 27899442_Early generated B1 B cells with restricted BCRs become chronic lymphocytic leukemia with continued c-Myc and low Bmf expression 32620849_Characterization of an alternative BAK-binding site for BH3 peptides. 33149265_The transcription factor IRF4 represses proapoptotic BMF and BIM to licence multiple myeloma survival. 33658012_FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. | ENSMUSG00000040093 | Bmf | 2.517114e+02 | 0.8892437 | -0.169349240 | 0.3405386 | 2.475171e-01 | 0.6188287825 | 0.91466270 | No | Yes | 2.089557e+02 | 40.270534 | 2.435693e+02 | 48.147339 | |
| ENSG00000104213 | 5157 | PDGFRL | protein_coding | Q15198 | Disease variant;Disulfide bond;Glycoprotein;Immunoglobulin domain;Reference proteome;Repeat;Secreted;Signal | This gene encodes a protein with significant sequence similarity to the ligand binding domain of platelet-derived growth factor receptor beta. Mutations in this gene, or deletion of a chromosomal segment containing this gene, are associated with sporadic hepatocellular carcinomas, colorectal cancers, and non-small cell lung cancers. This suggests this gene product may function as a tumor suppressor. [provided by RefSeq, Jul 2008]. | hsa:5157; | extracellular region [GO:0005576]; platelet activating factor receptor activity [GO:0004992]; platelet-derived growth factor beta-receptor activity [GO:0005019] | 20237496_Observational study of gene-disease association. (HuGE Navigator) 20333786_Results indicate that PDGFRL functions as a tumor suppressor, inhibiting the growth of colorectal cancer cells. 22926996_Data indicate the association between SNP rs17633132:C/T in PDGFRL with Behcet disease and suggest that the PDGFRL gene may be involved in Behcet disease by modulating its transcription. 23944365_review elucidates the role of tumor stroma interactions, the roles of PDGF receptor signaling in cancer-associated fibroblasts via alteration of stromal matrix composition and the mitogenic effects of cancer-derived PDGFs. | ENSMUSG00000031595 | Pdgfrl | 9.432853e+01 | 0.7565946 | -0.402407634 | 0.3787303 | 1.129673e+00 | 0.2878450386 | 0.81106118 | No | Yes | 7.918544e+01 | 11.942166 | 9.730346e+01 | 14.944175 | ||
| ENSG00000104343 | 55284 | UBE2W | protein_coding | Q96B02 | FUNCTION: Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins (PubMed:20061386, PubMed:21229326). Specifically monoubiquitinates the N-terminus of various substrates, including ATXN3, MAPT/TAU, POLR2H/RPB8 and STUB1/CHIP, by recognizing backbone atoms of disordered N-termini (PubMed:23560854, PubMed:23696636, PubMed:25436519). Involved in degradation of misfolded chaperone substrates by mediating monoubiquitination of STUB1/CHIP, leading to recruitment of ATXN3 to monoubiquitinated STUB1/CHIP, and restriction of the length of ubiquitin chain attached to STUB1/CHIP substrates by ATXN3. After UV irradiation, but not after mitomycin-C (MMC) treatment, acts as a specific E2 ubiquitin-conjugating enzyme for the Fanconi anemia complex by associating with E3 ubiquitin-protein ligase FANCL and catalyzing monoubiquitination of FANCD2, a key step in the DNA damage pathway (PubMed:19111657, PubMed:21229326). In vitro catalyzes 'Lys-11'-linked polyubiquitination. UBE2W-catalyzed ubiquitination occurs also in the presence of inactive RING/U-box type E3s, i.e. lacking the active site cysteine residues to form thioester bonds with ubiquitin, or even in the absence of E3, albeit at a slower rate (PubMed:25436519). {ECO:0000269|PubMed:19111657, ECO:0000269|PubMed:20061386, ECO:0000269|PubMed:21229326, ECO:0000269|PubMed:23560854, ECO:0000269|PubMed:23696636, ECO:0000269|PubMed:25436519}. | 3D-structure;ATP-binding;Alternative splicing;DNA damage;DNA repair;Nucleotide-binding;Nucleus;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000255|PROSITE-ProRule:PRU00388}. | This gene encodes a nuclear-localized ubiquitin-conjugating enzyme (E2) that, along with ubiquitin-activating (E1) and ligating (E3) enzymes, coordinates the addition of a ubiquitin moiety to existing proteins. The encoded protein promotes the ubiquitination of Fanconi anemia complementation group proteins and may be important in the repair of DNA damage. There is a pseudogene for this gene on chromosome 1. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2012]. | hsa:55284; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ubiquitin conjugating enzyme activity [GO:0061631]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; cellular response to misfolded protein [GO:0071218]; DNA repair [GO:0006281]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein K11-linked ubiquitination [GO:0070979]; protein monoubiquitination [GO:0006513]; protein polyubiquitination [GO:0000209]; protein quality control for misfolded or incompletely synthesized proteins [GO:0006515] | 16368532_human Ubiquitin-conjugating enzyme (E2) is homologous to the Arabidopsis thaliana UBC-16 gene product and contains 2 nuclear localization signals 21229326_UBE2W regulates FANCD2 monoubiquitination by mechanisms different from UBE2T and HRR6. 23696636_Ube2w has novel enzymatic properties that direct ubiquitination of the N terminus of substrates 25436519_We show the ubequitin-conjugating enzyme (E2) Ube2w uses a unique mechanism to facilitate the specific ubiquitination of the alpha-amino group 26101372_TRIM5alpha requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. 27185577_Thus, although Rnf4 and Ube2w functionally interact in vitro, these genetic experiments indicate that in response to DNA damage Ube2w and Rnf4 function in distinct pathways. 31518613_we show in vitro that UBE2W can modify the N-terminus of both a-synuclein and a tau tetra-repeat domain with a single ubiquitin. We demonstrate that an engineered N-terminal ubiquitin modification changes the aggregation process of both proteins, resulting in the formation of structurally distinct aggregates 33931024_Prognostic value of ubiquitin E2 UBE2W and its correlation with tumor-infiltrating immune cells in breast cancer. | ENSMUSG00000025939 | Ube2w | 1.404823e+02 | 1.0095081 | 0.013652554 | 0.3846070 | 1.243721e-03 | 0.9718672924 | 0.99430550 | No | Yes | 1.243776e+02 | 26.932867 | 1.089350e+02 | 24.028953 |
| ENSG00000104419 | 10397 | NDRG1 | protein_coding | Q92597 | FUNCTION: Stress-responsive protein involved in hormone responses, cell growth, and differentiation. Acts as a tumor suppressor in many cell types. Necessary but not sufficient for p53/TP53-mediated caspase activation and apoptosis. Has a role in cell trafficking, notably of the Schwann cell, and is necessary for the maintenance and development of the peripheral nerve myelin sheath. Required for vesicular recycling of CDH1 and TF. May also function in lipid trafficking. Protects cells from spindle disruption damage. Functions in p53/TP53-dependent mitotic spindle checkpoint. Regulates microtubule dynamics and maintains euploidy. {ECO:0000269|PubMed:15247272, ECO:0000269|PubMed:15377670, ECO:0000269|PubMed:17786215, ECO:0000269|PubMed:9766676}. | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Charcot-Marie-Tooth disease;Cytoplasm;Cytoskeleton;Direct protein sequencing;Membrane;Microtubule;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Reference proteome;Repeat | This gene is a member of the N-myc downregulated gene family which belongs to the alpha/beta hydrolase superfamily. The protein encoded by this gene is a cytoplasmic protein involved in stress responses, hormone responses, cell growth, and differentiation. The encoded protein is necessary for p53-mediated caspase activation and apoptosis. Mutations in this gene are a cause of Charcot-Marie-Tooth disease type 4D, and expression of this gene may be a prognostic indicator for several types of cancer. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, May 2012]. | hsa:10397; | adherens junction [GO:0005912]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; recycling endosome membrane [GO:0055038]; cadherin binding [GO:0045296]; gamma-tubulin binding [GO:0043015]; microtubule binding [GO:0008017]; small GTPase binding [GO:0031267]; cellular response to hypoxia [GO:0071456]; DNA damage response, signal transduction by p53 class mediator [GO:0030330]; mast cell activation [GO:0045576]; negative regulation of cell population proliferation [GO:0008285]; peripheral nervous system myelin maintenance [GO:0032287]; response to metal ion [GO:0010038]; signal transduction [GO:0007165] | 9251681_NDRG1 up-regulation during colon epithelial cell differentiation. First publication on tissue and subcellular expression of NDRG1. 11835375_Observational study of genotype prevalence. (HuGE Navigator) 12046693_examination as diagnostic aid due to overexpression in cancer cells 12429530_Cap43 overexpression in cancer cells involves a state of hypoxia characteristic of cancer cells where the Cap43 protein becomes a signature for this hypoxic state 12432451_Expression of this protein, a differentiation-related gene, has wide normal human tissue distribution and is in the SW480 colon neoplasm tumor cell line. 12767066_Downregulation of Cap43 gene by von Hippel-Lindau tumor suppressor protein in renal cancer cells. 12872253_Observational study of genotype prevalence. (HuGE Navigator) 12872253_Two disease causing mutations in NDRG1 in Charcot-Marie-Tooth Disease patients. 12962147_Ndrg1 gene is a Myc negative target in human neuroblastomas and other cell types with overexpressed N- or c-myc 14966915_NDRG1 gene may play an important role in colorectal carcinogenesis 15247272_Rit42 plays a role in the regulation of microtubule dynamics and the maintenance of euploidy 15251988_Increased Ndrg1 expression following Fe chelation was related to the permeability and antiproliferative activity of chelators. Ndrg1 up-regulation after iron chelation occurred at the transcriptional level. 15377670_NDRG1 is necessary but not sufficient for p53-mediated caspase activation and apoptosis 15461589_results identify NDRG1 and NDRG2 as physiological substrates for SGK1, and demonstrate that phosphorylation of NDRG1 by SGK1 primes it for phosphorylation by GSK3 15867226_Drg1 expression may be associated with a less aggressive, indolent colorectal cancer. 15922294_NDRG1 interacts with APO A-I and A-II and may have a role in the general mechanisms of HDL-mediated cholesterol transport 16288478_Collectively these findings establish the importance of intracellular ascorbate levels for the regulation of expression of CA IX and NDRG1/Cap43. 16314423_NDRG1 interacts with SIRT1/p53 signaling to attenuate hypoxic injury in human trophoblasts 16622835_NDRG1 was up-regulated in differentiated leukemic cells after hypoxia or CoCl2 treatment via HIF-1a, which was reported involved in cell differentiation. The results indicated NDRG1 might play important role in the regulation of cell differentiation. 16707596_E2-induced down-regulation of Cap43 seems to be mediated through ER-alpha-dependent pathways in breast cancer cells both in culture and in patients 16778198_Analysis of specimens from 65 patients with pancreatic ductal adenocarcinoma showed a significant association between Cap43 expression and tumor microvascular density, depth of invasion, and histopathologic grading. 16832411_NDRG1 will not serve as a reliable marker of tumour cells in the pancreas, but may serve as a marker of differentiation. 16920733_The discovery that iron chelators also increase Ndrg-1 expression further augments their antitumor activity and provides a novel strategy for the treatment of cancer and its metastasis. [REVIEW] 17069588_Decreased expression of NDRG! mRNA expression was accompanied by the local progression of esophageal squamous cell carcinoma. 17220478_identified 58 proteins that interact with NDRG1 in prostate cancer cells 17316623_hypoxia is a potent inducer of NDRG1 in HCCs, albeit requiring additional stimuli within the tumour microenvironment for its recruitment to the membrane 17488873_NDRG1 mRNA remains associated with polysomes during hypoxia. 17569115_NDRG1 expression was correlated with various clinicopathological features and clinical outcomes in colorectal cancer depending on the race/ethnicity of the patients. 17601350_Observational study of gene-disease association. (HuGE Navigator) 17786215_NDRG1 knockdown cells show a delay in recycling transferrin, conversely NDRG1 overexpressing cells reveal an increase in rate of transferrin recycling 17786215_The N-myc down regulated Gene1 (NDRG1) is a Rab4a effector involved in vesicular recycling of E-cadherin. 17909017_Egr-1 regulates NDRG1 transcription through an overlapping Egr-1/Sp1 binding site that acts as a major site of positive regulation of the NDRG1 promoter by hypoxia signaling. 18377423_Development of endometrial carcinoma is associated with an overexpression of NDRG1 and the loss of PTEN expression. 18455888_our results demonstrate that, in common with NDRG2, human NDRG1 can be indirectly transcriptionally down-regulated by Myc via interaction with the NDRG1 core promoter. 18582504_Ndrg-1 reduced the protein levels of cathepsin C which plays a role in invasion, indicating a potential mechanism of its anti-metastatic role in pancreatic cancer cells 18602353_Localization of N-myc downstream-regulated gene 1 and its significant correlation with p53 expression may play an important role in gastric cancer progression. 18653908_Data suggest that alteration of PTEN may upregulate NDRG1, which may be an important gene in facilitating endometrium carcinogenesis. 18977241_Observational study of gene-disease association. (HuGE Navigator) 19020710_NDRG1/Cap43 may have a role in portal vein invasion and intrahepatic metastasis in human hepatocellular carcinoma 19046768_the role of NDRG1 protein in myeloid leukemic cell differentiation. 19082468_NDRG1 represents an additional diagnostic marker for brain tumor detection 19259744_the present study provides for the first time that the NDRG1 mRNA level is statistically significantly correlated with lymph node metastases, metastases to distant organs, and the degree of tumor cells differentiation. 19337694_Decreased expression of NDRG1 in glioma is related to tumor progression. 19414333_Overexpression of Cap43 is associated with malignant status of esophageal cancer 19491262_study suggests a novel mechanism by which NDRG1/Cap43 modulates tumor angiogenesis/growth and infiltration of macrophages/neutrophils through attenuation of NF-kappaB signaling 19649210_This study demonstrates that most ERG-overexpressing prostate cancers harbor hormonally regulated TMPRSS2-ERG, SLC45A3-ERG, or NDRG1-ERG fusions 19682504_NDRG1 is not a classic client protein but interacts with Hsp90 and is still dually regulated by Hsp90 at a transcriptional and post-translational level. We suggest for the first time GSK3beta as a new client protein of Hsp90. 19760510_Silencing of NDRG1 decreased endothelial cell migration. 19775754_we found significantly higher NDRG1 mRNA levels in granulocytes of healthy donors than in primary acute myeloid leukemia (AML) cells 19800102_NDRG1/Cap43 could be a novel marker for malignant progression and poor prognosis in prostate cancer, plausibly in its close association with the down-regulation of E-cadherin expression. 20173668_These findings indicate that NDRG1 gene reactivation in PaCa cell lines by pharmacologic reversal of DNA methylation and histone deacetylation occurs via an indirect mechanism. 20348948_Data identify a novel mechanism for TBX2-driven oncogenesis and highlight the importance of NDRG1 as a growth control gene in breast tissue. 20369286_NDRG1 over-expression is associated with breast cancer. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20388062_NDRG1 plays a significant role in carcinogenesis and preventing the metastasis and invasion of gastric cancer cells 20403915_Observational study of gene-disease association. (HuGE Navigator) 20416281_Phosphorylation of NDRG1/Cap43 at both serine330 and threonine346 is required for its suppressive action on the NF-kappaB signaling pathway and CXC chemokine expression in pancreatic cancer cells. 20573444_The selective interruption of NDRG1 signaling may prove to be therapeutically useful in HCCs, particularly in the advanced infiltrative type of tumors exposed to hypoxic environments. 20664979_NDRG1/Cap43 promotes differentiation and invasion in osteosarcoma cells. 20853080_Increased NDRG1 expression is associated with advanced T stages and poor vascularization in non-small cell lung cancer. 21052891_Downregulation of NDRG1 promotes invasion of human gastric cancer through MMP-2. 21181370_Up-regulated NDRG1 may play a role in the suppression of malignant cell growth, invasion and metastasis of human cervical cancer. 21184144_NDRG1 plays vital roles in tumor metastasis suppression and is frequently silenced in metastatic colon cancers. 21221878_Low NDRG1 expression is associated with breast cancer. 21236457_NDRG1 completely inhibits growth in anchorage-independent assays and in vivo supports a biological function as a tumor suppressor in pancreatic cancer. 21454613_KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFkappaB complex in human prostate cancer. 21463610_These results suggest that regulation of invasion and metastasis by NDRG1 is a highly complicated process. 21575456_As2O3 and ATRA down-regulated the expression of NDRG-1 in HeLa cells. 21706236_NDRG1 was upregulated in osteosarcoma 21708134_this work demonstrates that NDRG1 phosphorylation, by the protein kinase SGK1, is temporally and spatially controlled during the cell cycle, suggesting a role for NDRG1 in successful mitosis. 21735144_our findings suggest that NDRG1 expression could be used as a novel prognostic factor for patient survival and might be a potential therapeutic target in primary gallbladder carcinoma 21763068_Results suggested that NDRG1 was an important molecule in controlling HCC metastasis and thus suggested as a novel biomarker for predicting HCC recurrence after LT. 21775055_Data suggest that NDRG1/Cap43 might play a key role in the cell cycle control of G0/G1 in HCC cells. 21784644_Tissue samples from four patients with central neurocytoma (CN) have nuclear and cytoplasmic/membranous expression of NDRG1 protein in highly differentiated CN tumor cells. 21909787_Data suggest that NDRG1 expression might participate in the carcinogenesis and progression of invasive breast cancer, and may serve as an important biomarker for invasive breast cancer. 21912630_Upon reoxygenation, overexpression of NDRG1 significantly inhibited cell migration 21976667_a link between (*)NO, chelatable iron, and regulation of NDRG1 expression and signaling in tumor cells. 22187533_Results suggest that N-myc downstream regulated gene-1 expression is enhanced by 3,3'-diindolylmethane in poorly differentiated cells and followed by induction of apoptosis. 22453918_chelators inhibit the TGF-beta-induced EMT via a process consistent with NDRG1 up-regulation and elucidates the mechanism of their activity. 22462691_The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. 22481237_Loss of NDRG1 is associated with tumor angiogenesis in lung cancer. 22678098_Overexpression of NDRG1 in Ishikawa cells greatly inhibited cell proliferation and migration. 22722710_Data indicate that NDRG1/Cap43 overexpression may be of predictive value in determining the prognosis of lung cancer patients. 22844455_NDRG1 plays its pivotal role in the malignant progression of gastric cancer through epithelial mesenchymal transition 22972152_NDRG1 plays a metastasis suppressor role in oral and oropharyngeal squamous cell carcinomas. 22978647_Data indicate that the most frequent form is SH3TC2 gene (CMT4C; 57.14%), followed by HK1 gene causative of CMT4G (CMT4G/HMSN-Russe 25%) and NDRG1 p.R148X in CMT type 4D (CMT4D/HMSN-Lom; 17.86%). 23099645_Loss of NDRG1 expression may contribute to gastric cancer development or progression. 23188716_NDRG1 inhibited an important regulatory pathway mediated by ROCK1/pMLC2 pathway that modulates stress fiber assembly. 23192272_NDRG1 may play positive but dispensable roles in the progression of esophageal squamous cell carcinoma. 23296375_Low NDRG1 expression is a significant and independent prognostic indicator in neuroblastoma by multivariate analysis. 23377825_Our results suggest that CTAB and irinotecan could be further explored for their potential clinical benefit in patients with NDRG1 deficient PCa. 23437357_NDRG1 is regulated by eukaryotic initiation factor 3a (eIF3a) during cellular stress caused by iron depletion. 23526365_HIF-1alpha up-regulates NDRG1 expression through binding to NDRG1 promoter, leading to proliferation of lung cancer A549 cells. 23634903_Sequence analysis predicted a pseudotrypsin protease cleavage site between Cys49-Gly50. Such cleavage of NDRG1 in cancer cells may result in loss of NDRG1 tumour suppressive activity. 23671130_this review summarizes the underlying molecular mechanisms of the antimetastatic effects of NDRG1 in cancer cells. 23813961_NDRG1 is a novel regulator of multivesicular body formation and endosomal LDL receptor trafficking. The deficiency of functional NDRG1 in Charcot-Marie-Tooth disease type 4D might impair lipid processing and differentiation of myelinating cells. 23846687_activation of JNK/AP-1 thus seems to promote tumor angiogenesis in relationship to NDRG1-induced inflammatory stimuli by gastric cancer cells. 23874544_NDRG1 is a novel favorable predictor for the prognosis in CRC patients. 23874837_The results suggest that miR-182 plays an important role in the proliferation of human prostate cancer cells by directly suppressing the tumor supressor gene NDRG1. 23899187_These findings suggest that the DNA methylation status of NDRG1 gene may play an important role in the pathogenesis and/or development of breast cancer 23973486_Nuclear NDRG1 expression in colorectal neoplasms was significantly higher than in the normal colorectal mucosa, and yet the normal colorectal mucosa showed no nuclear expression. 23996628_We detected mutation in the NDRG1 gene in two families and mutation in the HK1 gene in the other two families. These mutations cause Charcot-Marie-Tooth Disease, Type 4D and Charcot-Marie-Tooth disease, type 4g, respectively. 23999030_These findings suggest that nuclear NDRG1 has tumor suppressive effects, and the NDRG1 expression may have clinical values in ccRCC. 24136616_a ~6.25 kb homozygous intragenic duplication in NDRG1, intragenic copy-number variation resulted in a homozygous duplication of exons 6-8 that caused decreased mRNA expression of NDRG1 24302615_study demonstrated that NDRG1 is a potential therapeutic target for HCC because its suppression triggers senescence of HCC cells through activating glycogen synthase kinase-3beta-p53 pathway, thereby inhibiting tumor progression 24367102_NDRG1 has a role in MGMT-dependent resistance to alkylating chemotherapy in glioma and glioblastoma 24386364_BTF3, HINT1, NDRG1 and ODC1 protein over-expression in human prostate cancer tissue 24498060_Results suggest that N-myc downstream-regulated gene 1 (NDRG1) may have roles connected to vesicle transport. 24532803_the role of NDRG1 as an autophagic inhibitor that is important for understanding its mechanism of action. 24626771_NDRG1 acts as a tumor suppressor in ovarian carcinogenesis and may be a potential therapeutic target in this disease. 24747552_NDR1 has been activated by Thr444 phosphorylation, the activities of hMOB1 and MST1 are not required anymore to maintain elevated NDR1 kinase activity and for NDR1 to support centrosome overduplication. 24829151_NDRG1 is shown to modulate the WNT-beta-catenin pathway by inhibiting the nuclear translocation of beta-catenin. 24970329_NDRG1 silencing in human pancreatic cancer cells leads to a phenotype with more aggressive tumor growth and metastasis. 24985974_Down-regulation of NDRG1 in gastric cancer metastatic progression was correlated to E-cadherin and MMP-9. NDRG1 acts as a tumor suppressor gene. 25081069_miR-769-3p can functionally regulate NDRG1 during changes in oxygen concentration. 25110805_no significant difference in serum levels between pre-eclampsia and normotensive 25152373_A functional link between SET-mediated NDRG1 regulation. 25162997_NDRG1 as a molecular target to inhibit the epithelial-mesenchymal transition: the case for developing inhibitors of metastasis--{review} 25218595_Knockdown of NDRG1 enhanced F-actin expression. 25335733_Variants of NDRG1 mRNA were transcriptionally regulated after HepG2 and MCF-7 cells were treated by iron chelators, resulting in domination of NDRG1 mRNA Variant 1 (V1) in the HepG2 calls and domination of NDRG1 mRNA Variant 2 (V2) in the MCF-7 cells 25520514_Microarray analyses of cellular gene expression identified N-myc downstream regulated gene 1 (NDRG1) as a putative target of Epstein-Barr virus BamHI A rightward transcripts (BART) locus miRNAs. 25712528_NDRG1 is SUMOylated at Lysine 14.SUMO modification destabilizes the protein stability of NDRG1. 25777142_Overexpression of NDRG1 inhibits human glioma proliferation and invasion via phosphoinositide 3-kinase/AKT pathways. 25860930_NDRG1 decreases phosphorylation of c-Src at Tyr416 by down-regulating EGFR expression and activation. It also affected Rac1, p130Cas, CrkII, and c-Abl. NDRG1 decreases cell migration via c-src inhibition. 26028238_pomegranate juice-mediated decrease in cell death in hypoxia is partially mediated by NDRG1 in BeWo cells but not in primary trophoblasts. 26202882_Results show that aberrant methylation of NDRG1 promoter is an important mechanism for gene silencing, playing a major role in tumor occurrence and progression of protate cancer (Pca). Reversing this process may be used for PCa treatment. 26297987_NDRG1 overexpression leads to reduced tumor growth and angiogenesis in experimental glioma via upregulation of TNFSF15. In NDRG1 overexpressing glioma antiangiogenic treatment does not yield a therapeutic response. 26324937_TP53 loss leads to abnormal centrosome numbers and genomic instability mediated by NDRG1 26349604_GLI1 expression in both H441 and PW cells was associated with increased expression of NDRG1, a gene known to be downregulated by the MYC family of proteins, indicating that upregulation of NDRG1 by GLI1 is not cell-type specific. 26359353_Data indicate that N-myc downstream regulated gene 1 (NDRG1) competitively bind to glycogen synthase kinase 3beta (GSK-3beta) and orphan nuclear receptor (Nur77) to prevent beta-catenin degradation. 26418878_NDRG1 inhibits stemness of colorectal cancer via down-regulation of nuclear beta-catenin and CD44 26534963_Data suggest that NDRG1 down-regulates expression and activation of HER1/EGFR, HER2/ERBB2, and HER3/ERBB3 in response to epidermal growth factor (EGF) ligand in pancreatic/colonic neoplasm cells. 26653549_NDRG1 could increase the resistance of neuroblastoma cells to chemotherapeutic drugs, with its positive regulation on drug resistant proteins. 26852918_Also, overexpression of AHR facilitated cell proliferation and migration via up-regulation of NDRG1. 26895766_Data suggest that, in colonic/prostatic neoplasm cells, increased expression of NDRG1 decreases activating phosphorylation of FAK and paxillin; silencing/inhibition of NDRG1 results in opposite effect and inhibits neoplasm cell migration/adhesion. 27154576_Cell proliferation and invasion effect were remarkably enhanced when NDRG1 was silencing. 27338835_NDRG1 appears to prevent epithelial-mesenchymal transition (EMT)-induced metastasis by attenuating NF-kappaB signaling in metastasis of colorectal cancer (CRC). 27414086_Study elucidates a mechanism of NDRG1-regulated Wnt pathway activation and EMT via affecting TLE2 and beta-catenin expression in esophageal cancer cells. 27655496_The positive rates of NDRG1expression was 63. 83.33% (40/48)and 27.78% (5/18) in the controls, respectively. High expressions of NDRG1 and VEGF, NDRG1 and VEGF influenced both the occurrence and development of CA 27698340_Authors confirm that the decrease of GOLPH3 that promotes the apoptosis of glioma cells may be regulated by the activation of NDRG1 and cleaved capcase 3. There was a inverse association between GOLPH3 and NDRG1 in glioma samples. 27716814_The mean nerve NDRG1 expression score was 5.4. 27894074_Data show that LSD1 affects motility and invasiveness of neuroblastoma cells by modulating the transcription of the metastasis suppressor NDRG1. Mechanistically, results found that LSD1 co-localizes with MYCN at the promoter region of the NDRG1 gene and inhibits its expression. 28003645_Hereditary motor and sensory neuropathy-type Lom in 12 Czech patients, carrying NDRG1 mutation and one unusual case due to uniparental isodisomy of chromosome 8 has been reported. 28075464_NDRG1 role in proliferation, invasion and migration of pancreatic cancer 28191699_strong NDRG1 expression in ciliated epithelial cells in nasal tissues sampled from patients with chronic rhinosinusitis. NDRG1 gene knockdown decreased the transepithelial electrical resistance and increased the dextran permeability. NDRG1 knockdown disrupted tight junctions of airway epithelial cells. NDRG1 knockdown significantly decreased only claudin-9 expression, but did not decrease other claudin family molecules. 28219902_NDR1 interacts with TRAF3 and interferes with the association of TRAF3 and IL-17R, resulting in increased formation of the activation complex IL-17R-Act1, which is required for the downstream signaling and production of pro-inflammatory factors 28292472_Compared with the normal term pregnancies, the expression of both NDRG1 mRNA and protein were significantly high in placentas from preeclampsia, and the expression of NDRG1 in early-onset preeclampsia was higher than that in late-onset preeclampsia. 28346422_we demonstrated that the novel molecule of cell migration and invasion, caveolin-1, has direct interaction with NDRG1 in human colorectal cancer cells 28350132_The current study is the first to elucidate a unique facet of the potent tumor/metastasis suppressor NDRG1 in the regulation of pancreatic ductal adenocarcinoma (PDAC)glycolysis, leading to important insights into the mechanism by which NDRG1 exert inhibitory function in PDAC. 28371345_The research findings provide novel insights suggesting that loss of N-myc downregulated gene 1 (NDRG1) leads to a decrease in actin-mediated cellular motility but an increase in cellular invasion, resulting in increased tumor dissemination which positively impacts metastatic outcome. 28456659_NDRG1 prevented the degradation of c-Myc through Skp2-mediated ubiquitination in tumor cells. NDRG1 directly interacted with Skp2, and decreased phosphorylation of Skp2 through inactivation of CDK2. 28498432_Our experiments revealed that long-term (24 h), but not short-term hypoxia led to the induction of NDRG1 expression in human glioma cell lines. NDRG1 expression was found to correlate with the protein expression of HIF-1alpha, SP1, CEBPalpha, YB-1 and Smad7 28537875_NDRG1 over-expression promoted apoptosis in colorectal cancer cells whereas depletion of NDRG1 resulted in resistance to oxaliplatin treatment. 28545025_these data show that NDRG1 is regulated by the oncogenic MAP kinase-interacting kinase pathway, a target for cancer therapy 28615452_Data suggest that NDRG1 attenuates oncogenic signaling by inhibiting formation of EGFR/HER2 and HER2/HER3 heterodimers and by down-regulating EGFR via a mechanism involving its degradation. (NDRG1 = N-myc downstream regulated gene 1 protein; EGFR = epidermal growth factor receptor; HER = human epidermal growth factor receptor) [REVIEW] 28776325_our study described two homozygous missense mutations in NDRG1 in CMT patients and mentioned the function of NDRG1 in protein recycling, which plays an important role in myelination of the peripheral nerve system. 29118118_HCV coopts the MYC pathway responsible for NDRG1 expression and phosphorylation that regulates lipid droplet formation and metabolism. NDRG1 appears to restrict HCV by suppressing the lipid droplet formation that is necessary for HCV assembly. 29248714_Findings indicate a positive feedback loop between cancer associated fibroblast (CAFs) and box Q1 (FOXQ1)/N-myc downstream-regulated gene 1 (NDRG1) axis in neoplastic cells to drive hepatocellular carcinoma (HCC) initiation, suggesting new potential therapeutic targets for HCC. 29431240_These findings indicate that NDRG1 expression is associated with both prognosis and c-Myc expression in lung adenocarcinoma 29467385_Up-regulation of NDRG1 by DpdtC is a promising therapeutic approach in HER2-overexpressed cancers. 29524631_present study shows that HER4 and/or NDRG1 might play a critical role for the cell survival and chemo-resistance of Osteosarcoma (OS), and could be used as potential therapeutic targets in OS. 29569762_Our findings suggest that NDRG1 positively regulates haemangioma proliferation. 29679718_These results indicate the N-terminus region and phosphorylation at Ser330 could be crucial for NDRG1 nuclear localization and function. PTEN silencing indicated that p-NDRG1 (Thr346) could be regulated differentially in different tumor cell-types, indicating PTEN may be involved in the mechanism(s) underlying the pleiotropic activity of NDRG1. 29712969_We propose a nucleolar role of TARG1 in ribosome assembly or quality control that is stalled when TARG1 is re-located to sites of DNA damage. 29730307_NDRG1 plays a positive role in highly pathogenic avian influenza A/H5N1 virus replication by suppressing the canonical NF-kappaB signaling pathway. 29738439_In summary, CAPE attenuates Nasopharyngeal carcinoma (NPC)cell proliferation and invasion by upregulating NDRG1 expression via MAPK pathway and by inhibiting phosphorylation of STAT3. Considering the poor prognosis of NPC patients with metastasis, CAPE could be a promising agent against NPC. 29768183_NDRG1 disruption alleviates cisplatin/sodium glycididazole-induced DNA damage response and apoptosis in ERCC1-defective lung cancer cells 29801473_Study suggests that the higher NDRG1 expression may be associated with the radio-resistance of human rectal cancer cells, and it can be a potential therapeutic target for re-sensitizing radio-resistant rectal cancer cells to ionizing radiation. 29898756_our findings show that NDRG1 is a critical regulator of lipid fate in breast cancer cells. The association between NDRG1 and poor prognosis in breast cancer suggests it should play a more prominent role in patient risk assessment. The function of NDRG1 in breast cancer lipid metabolism may represent a promising therapeutic approach in the future. 30313035_Meta-analysis showed that low NDRG1 expression was significantly associated with worse survival in colorectal cancer and pancreatic cancer but higher survival of patients with liver cancer and gallbladder cancer. No significant association was observed between low NDRG1 expression and survival in gastric cancer or esophageal cancer. 30337371_Up-regulation of the kinase gene SGK1 by progesterone activates the AP-1-NDRG1 axis in both PR-positive and -negative breast cancer cells 30407715_Findings indicate the regulatory mechanism of microrchidia family CW-type zinc finger 2 (MORC2) in downregulating N-myc downstream-regulated gene 1 protein (NDRG1), and suggest MORC2 as a potential therapeutic target for colorectal cancer (CRC). 30497328_this is the first paper to reveal that different fragments of the same lncRNA, NDRG1-OT1, could produce opposite effects within a single target gene, NDRG1. 30538287_The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. 30561520_therapies targeting NDRG1 could be a new strategy to inhibit metastasis, and as such, we examined novel anticancer agents, namely di-2-pyridylketone thiosemicarbazones, which upregulate NDRG1 30679310_The NDRG1 promotes EGFR down-regulation through the EGFR inhibitor MIG6, which leads to late endosomal/lysosomal processing of EGFR. 30763642_The studies elucidated the complex and intricate regulatory pathways involving both proteasomal and autophagic processing of the metastasis suppressor protein, NDRG1. 30811506_Data demonstrated that NDRG1 is critical for mediating LANA to recruit PCNA onto terminal repeat (TR) of KSHV genome, and facilitates viral DNA replication and episome persistence. Findings suggest that NDRG1 plays an important role in KSHV viral genome replication, and provide new clues for understanding of KSHV persistence. 30914736_Upregulation of NDRG1 predicts poor outcome and facilitates disease progression by influencing the EMT process in bladder cancer. 30922920_CCAT2 acts as an oncogene by up-regulating NDRG1. 30981813_NDRG1 enhances the anti-proliferative and anti-migratory activity of Dp44mT. This increased efficacy could be related to the following effects in the presence of Dp44mT and NDRG1, namely: markedly increased activation of the PERK target, eukaryotic translation initiation factor 2alpha (eIF2alpha); the maintenance of activating transcription factor 4 (ATF4) expression. 31242951_annexin A2 (ANXA2), N-myc downstream regulated gene 1 (NDRG1) and signal transducer and activator of transcription 1 (STAT1) as biomarkers of LACC patients' responsiveness to CRT. 31694959_Merkel Cell Polyomavirus Downregulates N-myc Downstream-Regulated Gene 1, Leading to Cellular Proliferation and Migration. 31723002_High level expression of NDRG1 in tumors significantly correlated with better prognosis of glioblastoma (GBM) patients. Loss of NDRG1 in GBM cells upregulated GSK3beta levels and promoted cell proliferation. GSK3beta phosphorylated serine and threonine sites in the C-terminal domain of NDRG1 and limited the protein stability of NDRG1. Study identifies the NDRG1/GSK3beta signaling pathway as a key growth regulatory prog... 31831018_NDRG1 was down-regulated in colorectal cancer (CRC) tissues and correlated with tumor size and patient survival. NDRG1 inhibited CRC tumor proliferation through increasing p21 expression via suppressing p21 ubiquitylation. 32106831_GR, Sgk1, and NDRG1 in esophageal squamous cell carcinoma: their correlation with therapeutic outcome of neoadjuvant chemotherapy. 32112078_NDRG1 deficiency is associated with regional metastasis in oral cancer by inducing epithelial-mesenchymal transition. 32165297_we provide the first direct evidence that NDRG1-driven change in mitochondrial dynamics and aerobic glycolysis maintain cells survival in HCC during hypoxia 32335136_NDRG1 suppresses basal and hypoxia-induced autophagy at both the initiation and degradation stages and sensitizes pancreatic cancer cells to lysosomal membrane permeabilization. 32470876_Increased NDRG1 expression suppresses angiogenesis via PI3K/AKT pathway in human placental cells. 32537867_NDRG1 suppressed the expression of series oncogenes as well as the proliferation, metastasis and invasion of VHL-deficient ccRCC cells in vitro and vivo. 32557341_LINC01419 promotes cell proliferation and metastasis in hepatocellular carcinoma by enhancing NDRG1 promoter activity. 32757096_MicroRNA-1469-5p promotes the invasion and proliferation of pancreatic cancer cells via direct regulating the NDRG1/NF-kappaB/E-cadherin axis. 33305529_Crystal and solution structure of NDRG1, a membrane-binding protein linked to myelination and tumour suppression. 33398037_Identification of PIM1 substrates reveals a role for NDRG1 phosphorylation in prostate cancer cellular migration and invasion. 33484481_Breaking the cycle: Targeting of NDRG1 to inhibit bi-directional oncogenic cross-talk between pancreatic cancer and stroma. 34077484_NDRG1 facilitates lytic replication of Kaposi's sarcoma-associated herpesvirus by maintaining the stability of the KSHV helicase. 34298089_FOXP1 and NDRG1 act differentially as downstream effectors of RAD9-mediated prostate cancer cell functions. 34355586_MiR-188-3p and miR-133b Suppress Cell Proliferation in Human Hepatocellular Carcinoma via Post-Transcriptional Suppression of NDRG1. 34385595_NDRG1 enhances the sensitivity of cetuximab by modulating EGFR trafficking in colorectal cancer. 34512147_The role of N-cadherin/c-Jun/NDRG1 axis in the progression of prostate cancer. 34785213_The metastasis suppressor NDRG1 directly regulates androgen receptor signaling in prostate cancer. 34893874_NDRG1 in Aggressive Breast Cancer Progression and Brain Metastasis. 34913078_Microrchidia family CWtype zinc finger 2 promotes the proliferation, invasion, migration and epithelialmesenchymal transition of glioma by regulating PTEN/PI3K/AKT signaling via binding to Nmyc downstream regulated gene 1 promoter. 34956089_NDRG1 Activity in Fat Depots Is Associated With Type 2 Diabetes and Impaired Incretin Profile in Patients With Morbid Obesity. | ENSMUSG00000005125 | Ndrg1 | 1.786657e+03 | 1.0585135 | 0.082039663 | 0.3071670 | 7.231398e-02 | 0.7879969503 | 0.95754084 | No | Yes | 1.630943e+03 | 184.835438 | 1.535506e+03 | 178.801036 | |
| ENSG00000104983 | 729440 | CCDC61 | protein_coding | Q9Y6R9 | FUNCTION: Microtubule-binding centrosomal protein required for centriole cohesion, independently of the centrosome-associated protein/CEP250 and rootletin/CROCC linker (PubMed:31789463). In interphase, required for anchoring microtubule at the mother centriole subdistal appendages and for centrosome positioning (PubMed:31789463). During mitosis, may be involved in spindle assembly and chromatin alignment by regulating the organization of spindle microtubules into a symmetrical structure (PubMed:30354798). Has been proposed to play a role in CEP170 recruitment to centrosomes (PubMed:30354798). However, this function could not be confirmed (PubMed:31789463). Plays a non-essential role in ciliogenesis (PubMed:31789463, PubMed:32375023). {ECO:0000269|PubMed:30354798, ECO:0000269|PubMed:31789463, ECO:0000269|PubMed:32375023}. | 3D-structure;Acetylation;Alternative splicing;Cell projection;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | hsa:729440; | centriolar satellite [GO:0034451]; centriolar subdistal appendage [GO:0120103]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; microtubule organizing center [GO:0005815]; identical protein binding [GO:0042802]; microtubule binding [GO:0008017]; cell projection organization [GO:0030030]; centriole assembly [GO:0098534]; mitotic spindle assembly [GO:0090307] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 30354798_Ccdc61 controls centrosomal localization of Cep170 and is required for spindle assembly and symmetry. 31789463_hVFL3/CCDC61 is the human orthologue of proteins required for anchoring distinct sets of cytoskeletal fibers to centrioles in unicellular eukaryotes. 32375023_CCDC61/VFL3 Is a Paralog of SAS6 and Promotes Ciliary Functions. | ENSMUSG00000074358 | Ccdc61 | 1.755452e+02 | 1.2898242 | 0.367174431 | 0.3356478 | 1.173589e+00 | 0.2786649968 | 0.80653959 | No | Yes | 1.780359e+02 | 30.750922 | 1.498254e+02 | 26.992050 | ||
| ENSG00000105136 | 79744 | ZNF419 | protein_coding | Q96HQ0 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA | hsa:79744; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 27826023_previously unreported alternative splicing patterns in the genes ZNF419 and DKKL1 | ENSMUSG00000000103+ENSMUSG00000053211 | Zfy2+Zfy1 | 2.880920e+02 | 0.7825141 | -0.353811369 | 0.3110453 | 1.271714e+00 | 0.2594453679 | 0.79684153 | No | Yes | 1.841382e+02 | 32.296184 | 2.748099e+02 | 48.790661 | |
| ENSG00000105146 | 6795 | AURKC | protein_coding | Q9UQB9 | FUNCTION: Serine/threonine-protein kinase component of the chromosomal passenger complex (CPC), a complex that acts as a key regulator of mitosis. The CPC complex has essential functions at the centromere in ensuring correct chromosome alignment and segregation and is required for chromatin-induced microtubule stabilization and spindle assembly. Plays also a role in meiosis and more particularly in spermatogenesis. Has redundant cellular functions with AURKB and can rescue an AURKB knockdown. Like AURKB, AURKC phosphorylates histone H3 at 'Ser-10' and 'Ser-28'. AURKC phosphorylates the CPC complex subunits BIRC5/survivin and INCENP leading to increased AURKC activity. Phosphorylates TACC1, another protein involved in cell division, at 'Ser-228'. {ECO:0000269|PubMed:15316025, ECO:0000269|PubMed:15499654, ECO:0000269|PubMed:15670791, ECO:0000269|PubMed:15938719, ECO:0000269|PubMed:21493633, ECO:0000269|PubMed:21531210, ECO:0000269|PubMed:27332895}. | 3D-structure;ATP-binding;Alternative splicing;Cell cycle;Cell division;Centromere;Chromosome;Cytoplasm;Cytoskeleton;Kinase;Meiosis;Mitosis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the Aurora subfamily of serine/threonine protein kinases. The encoded protein is a chromosomal passenger protein that forms complexes with Aurora-B and inner centromere proteins and may play a role in organizing microtubules in relation to centrosome/spindle function during mitosis. This gene is overexpressed in several cancer cell lines, suggesting an involvement in oncogenic signal transduction. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2008]. | hsa:6795; | chromosome passenger complex [GO:0032133]; chromosome, centromeric region [GO:0000775]; condensed chromosome [GO:0000793]; cytoplasm [GO:0005737]; midbody [GO:0030496]; nucleus [GO:0005634]; spindle [GO:0005819]; spindle microtubule [GO:0005876]; spindle midzone [GO:0051233]; spindle pole centrosome [GO:0031616]; ATP binding [GO:0005524]; histone serine kinase activity [GO:0035174]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; attachment of spindle microtubules to kinetochore [GO:0008608]; cell division [GO:0051301]; histone modification [GO:0016570]; meiotic cell cycle [GO:0051321]; mitotic spindle midzone assembly [GO:0051256]; mitotic spindle organization [GO:0007052]; oocyte development [GO:0048599]; positive regulation of cytokinesis [GO:0032467]; protein phosphorylation [GO:0006468]; regulation of cytokinesis [GO:0032465]; spermatogenesis [GO:0007283] | 15316025_association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C 15499654_Aurora-C interacts with the inner centromere protein (INCENP) at the carboxyl terminal end spanning the conserved IN box domain. 15670791_Therefore, we speculated that Aurora C-SV might also contribute to the regulation of chromosome segregation and cytokinesis. 15917996_Aurora-C is a chromosomal passenger protein that disrupts the association of INCENP with Aurora-B and may serve as a key regulator in cell division 15938719_Aurora-c is directly associated with surwvivin and is required for cytokinesis. 16258285_A role of Aurora-C in the development and progression of cancer especially in the presence of mutated p53. 17435757_Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. 17637569_Results demonstrate that human Auroras a regulator of cell morphology and cell growth, and that the non-conserved T191 in the activation loop of Auroras of major importance for its regulation. 18309533_Aurora C phosphorylates TRF2 19147683_A functional AURKC protein is necessary for male meiotic cytokinesis while its absence does not impair oogenesis. 19147683_Observational study of gene-disease association. (HuGE Navigator) 19649632_Data show that SNS-314 is a novel, potent, and selective inhibitor of Aurora kinases A, B, and C. 19713763_Aurora-C can perform the same essential functions as Aurora-B in mitosis: regulation of kinetochore-microtubule attachments, the spindle assembly checkpoint, and cytokinesis 20508983_Observational study of gene-disease association. (HuGE Navigator) 20629650_Aurora-C mRNA was reduced to 0.20 +/- 0.32 fold (P < 0.01) in 13 seminomas and up-regulated in one case 21353399_We confirm in this study the research interest of the recurrent mutation c.144delC in the gene AURKC in male infertility with high rates of large-headed spermatozoa 21493633_Aurora C mRNA is up-regulated with Aurora B after activation of the embryonic genome and both proteins were detected in early Day 4 embryos. 21531210_the study demonstrated that TACC1 localizes at the midbody during cytokinesis and interacts with and is a substrate of Aurora-C, which warrant further investigation in order to elucidate the functional significance of this interaction. 21710690_we show that elevated AURKC increases the proliferation, transformation and migration of cancer cells 21733974_Molecular analysis of the AURKC gene was carried out in two brothers presenting with a typical large-headed spermatozoa phenotype. Both affected brothers were heterozygous for the c.144delC mutation in the AURKC gene. 22046298_Overexpressed Aurora-C (AURKC) is an oncogene. Overexpression of Aurora-C induces abnormal cell division resulting in centrosome amplification and multinucleation. Cells overexpressing active Aurora-C induced tumour formation in nude mice. 22888167_Investigators identified a new non-sense mutation in aurora kinase C, p.Y248*, in 10 unrelated individuals of European and North African origin; this mutation is associated with macrozoospermia and male infertility. 23075505_The immunoreactivity profile of AURKA, AURKB and AURKC in this study showed a significantly reduced expression in PCa cases compared to BPH cases. 23292864_expression of AURKC, OIP5, PIWIL2 and TAF7L differed between patients with Acute myeloid leukemia, myelodysplastic syndrome and healthy controls in a gender-dependent manner 23581231_Here we provide evidence for the first time of Aurora-C overexpression and gene amplification. 24484996_Our data indicate that the AURKC c.144delC mutation has a relatively high carrier frequency in the Moroccan population. 24603334_Aberrantly expressed Aurora-C in somatic cancer cells may impair spindle checkpoint assembly by displacing the centromeric localization of chromosomal passenger complexes. 24632603_This review will describe the functions of each Aurora kinase which include Aurora A (AURKA), Aurora B (AURKB) and Aurora C (AURKC summarize their involvement in leukemia and discuss inhibitor development and efficacy in leukemia clinical trials 25219909_AURKC mutations are more frequent than Klinefelter syndrome and constitute the leading genetic cause of infertility in North African men. 25369539_Data indicate that the selective Aurora A, B, and C inhibitor SAR156497 showed antineoplastic activity in HCT116 cell xenograft model. 25990457_Low AURKC expression is associated with cancer. 25994570_Overexpression of AURKC is associated with breast tumors. 26118178_may play an important role in the development of colorectal cancer 26341096_Homozygous c.144delC mutation in AURKC gene in infertile men with macrozoospermia in the Tunisian population 27332895_Aurora-C interactions with members of the Chromosome Passenger Complex (CPC), Survivin and Inner Centromere Protein (INCENP) in reference to known Aurora-B interactions to understand the functional significance of Aurora-C overexpression in human cancer cells, is reported. 27779748_Patients presenting with a monomorphic teratozoospermia such as globozoospermia or macrospermia with more than 85% of the spermatozoa presenting this specific abnormality have been analyzed permitting to identify several key genes for spermatogenesis such as AURKC and DPY19L2. 28369513_Identification and characterization of AURKB and AURKC variants associated with maternal aneuploidy has been reported. 28930610_Two novel DMRs, located in RPS6KA4/MIR1237 and the AURKC promoter, were found to be hypermethylated in WT 29283376_The data suggest that AKA is the vertebrate ancestral gene, and that AKB and AKC resulted from gene duplication in placental mammals. 29738175_Teratozoospermia is not correlated with c.144delC mutation in the AURKC gene in the men of the Sichuan area. Therefore, large-scale genotyping of the AURKC gene may not be necessary clinically among Chinese patients with idiopathic teratozoospermia. 30077875_The epigenetic targets AURKB, AURKC and DNMT3B, and the global DNA methylation profile are regulated during HIV-1 replication in CD4+ T cells, and this regulation can be influenced by the activation state of the cell at the time of infection. 30545223_This study showed the role of Lactobacilli in down-regulation of TSGA10, AURKC, OIP5 and AKAP4 genes. Such expression change might be involved in the anticancer effects of these Lactobacilli. The underlying mechanisms of these observations are not clear but epigenetic modulatory mechanisms may participate in this process. 30594972_These results further support the important role of AURKC in male infertility and guide the practitioner in optimal decision making for patients with macrozoospermia. 31521144_AURKA (rs1047972 and rs911160), AURKB (rs2241909 and rs2289590) and AURKC (rs11084490) are associated with a higher risk of GC susceptibility in Bosnian population. 32399982_Mutational analysis of Aurora kinase C gene in Egyptian patients with macrozoospermia. 32452402_Aurora kinase mRNA expression is reduced with increasing gestational age and in severe early onset fetal growth restriction. 33118205_Two frequent loss-of-function mutations in Aurora Kinase C gene in Algerian infertile men with macrozoospermia. 33172986_Aurora B and C kinases regulate chromosome desynapsis and segregation during mouse and human spermatogenesis. 33725274_Purging human ovarian cortex of contaminating leukaemic cells by targeting the mitotic catastrophe signalling pathway. 34030642_Co-relation with novel phosphorylation sites of IkappaBalpha and necroptosis in breast cancer cells. 34461108_The oncogenic role of meiosis-specific Aurora kinase C in mitotic cells. 34465813_Genetic variations in AURORA cell cycle kinases are associated with glioblastoma multiforme. 35218477_Aurora kinase genetic polymorphisms: an association study in Down syndrome and spontaneous abortion. | ENSMUSG00000070837 | Aurkc | 1.525671e+01 | 0.9522499 | -0.070587790 | 0.7327345 | 9.256852e-03 | 0.9233517767 | No | Yes | 1.084016e+01 | 4.218840 | 1.172351e+01 | 4.641157 | ||
| ENSG00000105369 | 973 | CD79A | protein_coding | P11912 | FUNCTION: Required in cooperation with CD79B for initiation of the signal transduction cascade activated by binding of antigen to the B-cell antigen receptor complex (BCR) which leads to internalization of the complex, trafficking to late endosomes and antigen presentation. Also required for BCR surface expression and for efficient differentiation of pro- and pre-B-cells. Stimulates SYK autophosphorylation and activation. Binds to BLNK, bringing BLNK into proximity with SYK and allowing SYK to phosphorylate BLNK. Also interacts with and increases activity of some Src-family tyrosine kinases. Represses BCR signaling during development of immature B-cells. {ECO:0000269|PubMed:8617796, ECO:0000269|PubMed:9057631}. | 3D-structure;Adaptive immunity;Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunity;Immunoglobulin domain;Membrane;Methylation;Phosphoprotein;Reference proteome;Signal;Transmembrane;Transmembrane helix | The B lymphocyte antigen receptor is a multimeric complex that includes the antigen-specific component, surface immunoglobulin (Ig). Surface Ig non-covalently associates with two other proteins, Ig-alpha and Ig-beta, which are necessary for expression and function of the B-cell antigen receptor. This gene encodes the Ig-alpha protein of the B-cell antigen component. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:973; | B cell receptor complex [GO:0019815]; external side of plasma membrane [GO:0009897]; integral component of membrane [GO:0016021]; membrane raft [GO:0045121]; multivesicular body [GO:0005771]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; transmembrane signaling receptor activity [GO:0004888]; adaptive immune response [GO:0002250]; B cell activation [GO:0042113]; B cell differentiation [GO:0030183]; B cell proliferation [GO:0042100]; B cell receptor signaling pathway [GO:0050853] | 11907094_Unlike the B29 octamer motif, the mb-1 octamer motif does not have the essential sequence required for OBF-1/Bob1 interaction, therefore its promoter is not transactivated by OBF-1/Bob1. 12403343_B cell-restricted mb-1 gene: expression, function, and lineage infidelity. Review. 12651942_There is a somatic hypermutation of this gene in B-cell lymphoma and multiple myeloma. 15492262_Results indicate that PAX5 is a more specific marker than CD79a for B-cell ALL diagnosis. 15591116_lower levels of B-cell receptor surface expression observed in chronic lymphocytic leukemia are accounted for by an impaired glycosylation and folding of the mu and CD79a chains. 16271957_FISH findings indicate that CD79a, despite its specificity for B-cell differentiation, represented the aberrant presence of a B-cell antigen in leukemias of distinct myeloid linage. 17350472_Acute leukemia with t(8;21) coexpresses CytCD79a represent biphenotypic acute leukemia. 17374736_Anti-CD79b antibodies downregulated surface B-cell receptor and were trafficked to the lysosomal-like major histocompatibility complex class II-positive compartment of a mouse xenograft model of non-Hodgkin lymphoma. 17701175_Ig-alpha was phosphorylated in all myeloma IgG BCR isolates, the 31-kD variant being phosphorylated most frequently. It was not phosphorylated in normal control B cells. 18160827_Median survival time of cytoplasmic CD79 alpha positive group was shorter than that of cytCD79a negative group in acute myeloid leukemia. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19883431_we determined the significance of loss or downregulation of CD79a, overexpression of cyclinD1, and c-maf expression in bone marrow specimens with plasma cell myeloma 19913121_Observational study of gene-disease association. (HuGE Navigator) 20102401_CD79a may be helpful marker in the differential diagnosis of classical Hodgkin's lymphoma and primary mediastinal B-cell lymphoma 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21324920_STN produced significant antitumor effects in a mouse xenograft model of CD79A/B-mutated DLBCL. 24146823_CD79a plays a functional role in the tumor promoting effects of myeloid cells, and may represent a novel target for cancer therapy. 24769851_Phosphorylation of CD79a causes a decrease in helical propensity in the C-terminal region, for CD79b, the opposite was observed and phosphorylation resulted in an increase of helical propensity in the C-terminal part. 24974109_High intensity of caries is associated with increased levels of some salivary components - sIgA, histatin-5 and lactoperoxidase. 26540570_Data show that Bruton tyrosine kinase (BTK)inhibitor Ibrutinib augments MALT lymphoma associated translocation protein (MALT1) inhibition by S-Mepazine in CD79 antigen mutant activated B cell-subtype (ABC) of diffuse large B cell lymphoma (DLBCL). 27915469_The incidence of MYD88 and CD79B mutations in patients with CD5(+) DLBCL is lower than that in patients with DLBCL-SS, suggesting that CD5(+) DLBCL is not the same disease as DLBCL-SS in terms of gene mutation status. 30396184_Aberrant CD79a and/or PAX5 expression can be found in Acute Myelogenous Leukemia cases with RUNX1 mutations even without the translocation t(8; 21). 32641598_Clinicopathological significance of CD79a expression in classic Hodgkin lymphoma. 33452446_CD79a promotes CNS-infiltration and leukemia engraftment in pediatric B-cell precursor acute lymphoblastic leukemia. | ENSMUSG00000003379 | Cd79a | 1.333264e+01 | 1.8255070 | 0.868297195 | 0.7708584 | 1.296901e+00 | 0.2547800234 | No | Yes | 1.738301e+01 | 6.567750 | 9.120041e+00 | 3.772680 | ||
| ENSG00000105516 | 1628 | DBP | protein_coding | Q10586 | FUNCTION: This transcriptional activator recognizes and binds to the sequence 5'-RTTAYGTAAY-3' found in the promoter of genes such as albumin, CYP2A4 and CYP2A5. It is not essential for circadian rhythm generation, but modulates important clock output genes. May be a direct target for regulation by the circadian pacemaker component clock. May affect circadian period and sleep regulation. | Activator;Biological rhythms;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | The protein encoded by this gene is a member of the PAR bZIP transcription factor family and binds to specific sequences in the promoters of several genes, such as albumin, CYP2A4, and CYP2A5. The encoded protein can bind DNA as a homo- or heterodimer and is involved in the regulation of some circadian rhythm genes. [provided by RefSeq, Jul 2014]. | hsa:1628; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; circadian rhythm [GO:0007623]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 20174623_Observational study of gene-disease association. (HuGE Navigator) 21411511_Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBalpha, RIP140, and PGC1alpha) genes. 25503293_the expression of DBP and MAPK in epilepsy patients was upregulated, suggesting a possible pathogenetic role in IE. 31659740_Vitamin D binding protein and risk of renal cell carcinoma in the prostate, lung, colorectal and ovarian cancer screening trial. 32255717_The Association Between VDR and GC Polymorphisms and Lung Cancer Risk: A Systematic Review and Meta-Analysis. 32816832_Expression of clock gene Dbp in omental and mesenteric adipose tissue in patients with type 2 diabetes. | ENSMUSG00000059824 | Dbp | 4.978859e+02 | 1.4596708 | 0.545643029 | 0.3533942 | 2.413395e+00 | 0.1203012270 | 0.75783482 | No | Yes | 5.660745e+02 | 82.782945 | 3.416466e+02 | 51.748658 | |
| ENSG00000105778 | 23080 | AVL9 | protein_coding | Q8NBF6 | FUNCTION: Functions in cell migration. {ECO:0000269|PubMed:22595670}. | Alternative splicing;Endosome;Membrane;Methylation;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:23080; | cytoplasm [GO:0005737]; integral component of membrane [GO:0016021]; recycling endosome [GO:0055037]; cell migration [GO:0016477] | 32678693_Clinical significance and functions of miR-203a-3p/AVL9 axis in human non-small-cell lung cancer. 33880595_hsa_circ_0058357 acts as a ceRNA to promote nonsmall cell lung cancer progression via the hsamiR243p/AVL9 axis. | ENSMUSG00000029787 | Avl9 | 7.876149e+02 | 0.6766766 | -0.563461529 | 0.2875425 | 3.770890e+00 | 0.0521518619 | 0.67565452 | No | Yes | 6.369790e+02 | 92.008022 | 8.600649e+02 | 126.876169 | ||
| ENSG00000105849 | 221830 | POLR1F | protein_coding | Q3B726 | FUNCTION: DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase I which synthesizes ribosomal RNA precursors. Through its association with RRN3/TIF-IA may be involved in recruitment of Pol I to rDNA promoters. | DNA-directed RNA polymerase;Nucleus;Phosphoprotein;Reference proteome;Transcription | hsa:221830; | nucleoplasm [GO:0005654]; RNA polymerase I complex [GO:0005736]; DNA-directed 5'-3' RNA polymerase activity [GO:0003899]; cellular response to leukemia inhibitory factor [GO:1990830]; DNA-templated transcription, initiation [GO:0006352] | 12438708_describe the localisation and characterisation of the TWIST NEIGHBOR (TWISTNB) gene | ENSMUSG00000020561 | Polr1f | 3.298054e+02 | 0.7936564 | -0.333413497 | 0.3577184 | 8.248415e-01 | 0.3637684196 | 0.83355620 | No | Yes | 3.062487e+02 | 58.561011 | 3.238714e+02 | 63.485006 | ||
| ENSG00000105875 | 29062 | WDR91 | protein_coding | A4D1P6 | FUNCTION: Functions as a negative regulator of the PI3 kinase/PI3K activity associated with endosomal membranes via BECN1, a core subunit of the PI3K complex. By modifying the phosphatidylinositol 3-phosphate/PtdInsP3 content of endosomal membranes may regulate endosome fusion, recycling, sorting and early to late endosome transport (PubMed:26783301). It is for instance, required for the delivery of cargos like BST2/tetherin from early to late endosome and thereby participates indirectly to their degradation by the lysosome (PubMed:27126989). May play a role in meiosis (By similarity). {ECO:0000250|UniProtKB:Q7TMQ7, ECO:0000269|PubMed:26783301, ECO:0000269|PubMed:27126989}. | 3D-structure;Alternative splicing;Coiled coil;Endosome;Membrane;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:29062; | cytosol [GO:0005829]; early endosome membrane [GO:0031901]; extrinsic component of endosome membrane [GO:0031313]; late endosome membrane [GO:0031902]; phosphatidylinositol 3-kinase regulator activity [GO:0035014]; early endosome to late endosome transport [GO:0045022]; regulation of cellular protein catabolic process [GO:1903362]; regulation of phosphatidylinositol 3-kinase activity [GO:0043551] | 27126989_suggest a role for the WDR81-WDR91 complex in the fusion of endolysosomal compartments and the absence of WDR81 leads to impaired receptor trafficking and degradation 28860274_WDR91 serves as a Rab7 effector that is essential for neuronal development by facilitating endosome conversion in the endosome-lysosome pathway. | ENSMUSG00000058486 | Wdr91 | 1.919327e+03 | 0.8655609 | -0.208292799 | 0.2804625 | 5.580921e-01 | 0.4550299823 | 0.86227340 | No | Yes | 1.410528e+03 | 122.486606 | 1.722272e+03 | 152.915543 | ||
| ENSG00000106089 | 6804 | STX1A | protein_coding | Q16623 | FUNCTION: Plays an essential role in hormone and neurotransmitter calcium-dependent exocytosis and endocytosis (PubMed:26635000). Part of the SNARE (Soluble NSF Attachment Receptor) complex composed of SNAP25, STX1A and VAMP2 which mediates the fusion of synaptic vesicles with the presynaptic plasma membrane. STX1A and SNAP25 are localized on the plasma membrane while VAMP2 resides in synaptic vesicles. The pairing of the three SNAREs from the N-terminal SNARE motifs to the C-terminal anchors leads to the formation of the SNARE complex, which brings membranes into close proximity and results in final fusion. Participates in the calcium-dependent regulation of acrosomal exocytosis in sperm (PubMed:23091057). Plays also an important role in the exocytosis of hormones such as insulin or glucagon-like peptide 1 (GLP-1) (By similarity). {ECO:0000250|UniProtKB:O35526, ECO:0000269|PubMed:23091057, ECO:0000269|PubMed:26635000}. | Alternative splicing;Cell junction;Cell membrane;Coiled coil;Cytoplasmic vesicle;Exocytosis;Isopeptide bond;Membrane;Neurotransmitter transport;Phosphoprotein;Reference proteome;Secreted;Synapse;Synaptosome;Transmembrane;Transmembrane helix;Transport;Ubl conjugation;Williams-Beuren syndrome | This gene encodes a member of the syntaxin superfamily. Syntaxins are nervous system-specific proteins implicated in the docking of synaptic vesicles with the presynaptic plasma membrane. Syntaxins possess a single C-terminal transmembrane domain, a SNARE [Soluble NSF (N-ethylmaleimide-sensitive fusion protein)-Attachment protein REceptor] domain (known as H3), and an N-terminal regulatory domain (Habc). Syntaxins bind synaptotagmin in a calcium-dependent fashion and interact with voltage dependent calcium and potassium channels via the C-terminal H3 domain. This gene product is a key molecule in ion channel regulation and synaptic exocytosis. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Sep 2009]. | hsa:6804; | acrosomal vesicle [GO:0001669]; actomyosin [GO:0042641]; axon [GO:0030424]; cytosol [GO:0005829]; endomembrane system [GO:0012505]; extracellular region [GO:0005576]; glutamatergic synapse [GO:0098978]; integral component of membrane [GO:0016021]; integral component of presynaptic membrane [GO:0099056]; integral component of synaptic vesicle membrane [GO:0030285]; neuron projection [GO:0043005]; nuclear membrane [GO:0031965]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; presynaptic active zone membrane [GO:0048787]; SNARE complex [GO:0031201]; synaptic vesicle [GO:0008021]; synaptobrevin 2-SNAP-25-syntaxin-1a complex [GO:0070044]; synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex [GO:0070032]; synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex [GO:0070033]; voltage-gated potassium channel complex [GO:0008076]; ATP-dependent protein binding [GO:0043008]; calcium channel inhibitor activity [GO:0019855]; calcium-dependent protein binding [GO:0048306]; chloride channel inhibitor activity [GO:0019869]; identical protein binding [GO:0042802]; kinase binding [GO:0019900]; myosin head/neck binding [GO:0032028]; protein domain specific binding [GO:0019904]; protein N-terminus binding [GO:0047485]; protein-containing complex binding [GO:0044877]; SNAP receptor activity [GO:0005484]; SNARE binding [GO:0000149]; transmembrane transporter binding [GO:0044325]; calcium-ion regulated exocytosis [GO:0017156]; exocytosis [GO:0006887]; insulin secretion [GO:0030073]; intracellular protein transport [GO:0006886]; positive regulation of calcium ion-dependent exocytosis [GO:0045956]; positive regulation of catecholamine secretion [GO:0033605]; positive regulation of excitatory postsynaptic potential [GO:2000463]; positive regulation of neurotransmitter secretion [GO:0001956]; positive regulation of norepinephrine secretion [GO:0010701]; protein localization to membrane [GO:0072657]; protein sumoylation [GO:0016925]; regulation of insulin secretion [GO:0050796]; regulation of synaptic vesicle priming [GO:0010807]; response to gravity [GO:0009629]; secretion by cell [GO:0032940]; SNARE complex assembly [GO:0035493]; synaptic vesicle docking [GO:0016081]; synaptic vesicle endocytosis [GO:0048488]; synaptic vesicle exocytosis [GO:0016079]; synaptic vesicle fusion to presynaptic active zone membrane [GO:0031629]; vesicle docking [GO:0048278]; vesicle fusion [GO:0006906] | 11719842_Observational study of gene-disease association. (HuGE Navigator) 12114505_SNAP-25 traffics to the plasma membrane by a syntaxin-independent mechanism. 12198139_Munc18b binds to syntaxins 1A, 2, and 3 and regulates vesicle transport to the apical plasma membrane 12209004_CFTR channels are coordinately regulated by two cognate t-SNAREs, SNAP-23 (synaptosome-associated protein of 23 kDa) and syntaxin 1A. 12446681_interacts with CFTR protein differently from interactions with SNARE. 15175344_Syntaxin/Munc18 interactions in the late events during vesicle fusion and release in exocytosis 15202772_interacts with dopamine transporter 15219469_A significant genetic association was found between schizophrenia and an intron 7 single nucleotide polymorphism (SNP) tested. Haplotype analysis supported the association with several significant values that appear to be driven by the intron 7 SNP. 15316009_cleavage of APP but not syntaxin 1 is independent of cell surface regulation by extracellular ligands 15339904_Syn-1A binds both NBFs of SUR1 and SUR2A but appears to exhibit distinct interactions with NBF2 of these SUR proteins in modulating the KATP channels in islet beta cells and cardiac myocytes 16672225_Syntaxin-1A actions on sulfonylurea receptor 2A blocks acidic pH-induced cardiac K(ATP) channel activation 17032905_Norepinephrine transpossrter/syntaxin 1A complex rapidly redistributes, upon amphetamine treatment, when mechanisms supported by the transporter's NH2 terminus are eliminated. 17264080_analysis of the spatially distinct modes of munc18-syntaxin 1 interaction 17506992_the mechanisms involved in Syn1A-K(v) interactions vary significantly between K(v) channels, thus providing a wide scope for Syn1A modulation of exocytosis and membrane excitability 17543282_the H(abc) domain has a role in membrane trafficking and targeting of syntaxin 1A 17912268_A lower frequency of the PRM -352T allele of the STX1A gene in overweight/obese subjects impaires glucose regulation, particularly among individuals with combined glucose intolerance and overt diabetes. 17912268_Observational study of gene-disease association. (HuGE Navigator) 18065728_We have identified a new three-gene classifier that is independent of and improves on stage to stratify early-stage NSCLC patients with significantly different prognoses. 18457912_The expression of the SNARE protein SNAP-25 and its cellular homologue SNAP-23, as well as syntaxin1 and VAMP (vesicle-associated membrane protein) in samples of normal parathyroid tissue, chief cell adenoma, and parathyroid carcinoma, was examined. 18512733_Observational study of gene-disease association. (HuGE Navigator) 18512733_SNARE complex-related genes STX1A, VAMP2 and SNAP25 do not play a major role in susceptibility to schizophrenia in the Japanese population 18593506_Observational study of gene-disease association. (HuGE Navigator) 18593506_STX1A might influence the serotonergic system during neurodevelopment 19004828_Direct interaction of otoferlin with syntaxin 1A, SNAP-25, and the L-type voltage-gated calcium channel Cav1.3. 19129200_Homology with vesicle fusion mediator syntaxin-1a predicts determinants of epimorphin/syntaxin-2 function in mammary epithelial morphogenesis.( 19368856_Observational study of gene-disease association. (HuGE Navigator) 19368856_This analysis revealed significant differences in both allele frequencies and genotype distributions of the STX1A gene in migraine. 19557857_Observational study of gene-disease association. (HuGE Navigator) 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20385907_Observational study of gene-disease association. (HuGE Navigator) 20385907_We confirmed the involvement of syntaxin 1A in migraine susceptibility regarding rs941298. In addition, we found rs6951030 to also be associated in Portuguese migraine patients. 20422020_syntaxin 1A, a neuronal regulator of presynaptic vesicle release, may play a role in WS and be a component of the cellular pathway determining human intelligence. 20471030_This protein has been found differentially expressed in thalami from patients with schizophrenia. 20564318_Results suggest that variants in RNASEL contribute to susceptibility to early onset and familial forms of prostate cancer. 21118708_association of STX1A with autism in a trio association study; in the anterior cingulate gyrus region, STX1A expression in the autism group was found to be significantly lower than controls; suggests a possible role of STX1A in pathogenesis of autism 21173146_ATP regulates pancreatic beta-cell K(ATP) channel activity, not only by its direct actions on Kir6.2 pore subunit, but also via ATP modulation of Syn-1A binding to SUR1. 21669024_Exocytotic dysfunctions in schizophrenia are probably related to an imbalance of the interaction between munc18-1a and SNARE (mainly syntaxin-1A) complex. 21789195_Recombinant alpha-SNAP-M105I has greater affinity for the cytosolic portion of immunoprecipitated syntaxin than the wild type protein. 21916482_Forming an acceptor SNARE complex between syntaxin-1A and SNAP-25 weakens but does not abrogate cholesterol-controlled cluster formation and indicates that the reconstitution process results in equal incorporation of protein at either lipid composition. 21976501_Syntaxin 1A effects may be additive but can be blocked at different concentration ranges of calmodulin, suggesting selective presynaptic targeting to directly regulate exocytosis. 22130660_A Ca(v)3.2/syntaxin-1A signaling complex controls T-type channel activity and low-threshold exocytosis. 22250207_Our results provide support for the hypothesis that STX1A represents a susceptibility gene for migraine 22264512_DrrA activation of the Rab1 GTPase on plasma membrane-derived organelles stimulated the tethering of endoplasmic reticulum-derived vesicles, resulting in vesicle fusion through the pairing of Sec22b with the plasma membrane syntaxin proteins. 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 22404429_Nesca directly binds KIF5B, kinesin light-chain and syntaxin-1 22411134_Direct interaction between syntaxin 1A and the Kv2.1 C-terminus is required for efficient insulin exocytosis and glucose-stimulated insulin secretion. 22571925_The histone modification marks were significantly increased in bipolar disorder and major depression and this effect was correlated with significant increases in Syn1a gene expression. 22791290_Platelets deficient in Munc18b from a Familial Hemophagocytic Lymphohistiocytosis type 5 had secretion defect. 23242284_In dementia with Lewy bodies patients there were lower levels of syntaxin in visual cortex compared to controls. 23403573_syntaxin 1 and SNAP-25 cooperate as SNARE proteins to support neuron survival. 23572023_The clinical relevance of STX1A variants in CF 23801330_the preferential binding of CAPS1 to open syntaxin-1 can contribute to the stabilization of the open state of syntaxin-1 during its transition from 'closed' state to the SNARE complex formation. 23858467_N-peptide and LE mutation have no effect on the global conformation of the Munc18a-Syx1a complex. 24164654_no associaton with idiopathic generalized epilepsy was found regarding Intron 7 rs1569061 of Syntaxin 1A gene, MnlI rs3746544 and DdeI rs1051312 polymorphisms of SNAP-25 gene compared with healthy subjects 24218570_Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. 24375629_We described clinical, genetic, and functional data from 17 families with a diagnosis of benign familial neonatal epilepsy caused by KCNQ2 or KCNQ3 mutations and we showed that some mutations lead to a reduction of Q2 channel regulation by syntaxin-1A. 24429282_PIP2 affects islet beta-cell KATP channels not only by its actions on Kir6.2 but also by sequestering Syn-1A to modulate Syn-1A availability and its interactions with SUR1 on PM. 25445064_SNARE complex genes and their interactions may play a significant role in susceptibility and working memory of ADHD. 25803850_Blockade of the SNARE protein syntaxin 1 inhibits glioblastoma tumor growth. 26918652_Mislocalization of syntaxin-1 was found in pluripotent stem cells from epileptic encephalopathy patient. 27072493_Our results suggest that, as in the CNS, CADM1 interactions drive exocytic site assembly and promote actin network formation. These results support the broader hypothesis that the effects of cell-cell contact on beta-cell maturation and function are mediated by the same extracellular protein interactions that drive the formation of the presynaptic exocytic machinery. These interactions may be therapeutic targets for re... 27458546_A significant interactive two-locus model of STX1A_rs4363087|VAMP2_rs2278637 (presynaptic genes) was observed among SVC variants in all epilepsy cases. 28235601_Some Autism spectrum disorder patients had haploidy of STX1A gene and lower STX1A gene expression. 28722652_Analysing protein mobility, cluster size and accessibility to myc-epitopes the authors show that forces acting on the transmembrane segment produce loose clusters, while cytoplasmic protein interactions mediate a tightly packed state. 30916996_The N-terminal of the SNAP25 loop region binds with membrane, and this interaction induced a disorder-to-order conformational change of the loop, resulting in enhanced interaction between the C-terminal of the SNAP25 loop and syx-1. SNARE-complex assembly efficiency decreased when the electrostatic interaction between C-terminal of the SNAP25 loop and syx-1 was disrupted. 30976917_Using a case-control study to explore the association between STX1A gene and children with ADHD in Chinese Han population, our results suggest STX1A genetic variants might contribute to the susceptibility of children ADHD. 31192914_Results suggest a role of Stx-1A rs4717806 SNP in ischemic heart disease, possibly due to its influence in Stx-1A expression and, at cascade, to insulin secretion and to glucose dependent metabolism. 32059362_Syntaxin 1: A Novel Robust Immunophenotypic Marker of Neuroendocrine Tumors. 32815282_De novo STXBP1 mutation in a child with developmental delay and spasticity reveals a major structural alteration in the interface with syntaxin 1A. 33217562_Oligomeric alpha-Syn and SNARE complex proteins in peripheral extracellular vesicles of neural origin are biomarkers for Parkinson's disease. 34764822_Syntaxin-1 and Insulinoma-Associated Protein 1 Expression in Breast Neoplasms with Neuroendocrine Features. | ENSMUSG00000007207 | Stx1a | 6.326893e+02 | 1.0017115 | 0.002466998 | 0.2978931 | 6.918102e-05 | 0.9933636615 | 0.99877177 | No | Yes | 4.933838e+02 | 64.926501 | 5.310976e+02 | 71.723794 | |
| ENSG00000106266 | 29886 | SNX8 | protein_coding | Q9Y5X2 | FUNCTION: May be involved in several stages of intracellular trafficking. May play a role in intracellular protein transport from early endosomes to the trans-Golgi network. {ECO:0000269|PubMed:19782049}. | Endosome;Lipid-binding;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:29886; | cytosol [GO:0005829]; early endosome membrane [GO:0031901]; intracellular membrane-bounded organelle [GO:0043231]; identical protein binding [GO:0042802]; phosphatidylinositol binding [GO:0035091]; early endosome to Golgi transport [GO:0034498]; intracellular protein transport [GO:0006886] | 21973056_SNX4, but not SNX1 and SNX8, is associated with the Rab11-recycling endosomes and that a high frequency of SNX4-mediated tubule formation is observed as endosomes undergo Rab4-to-Rab11 transition. 24311514_SNX8: A candidate gene for 7p22 cardiac malformations including tetralogy of fallot. 29180417_SNX8 mediates IFNG-triggered non-canonical signaling pathway and host defense against Listeria monocytogenes. 34231239_Fatty Acid Synthase-Suppressor Screening Identifies Sorting Nexin 8 as a Therapeutic Target for NAFLD. 34524084_A PX-BAR protein Mvp1/SNX8 and a dynamin-like GTPase Vps1 drive endosomal recycling. | ENSMUSG00000029560 | Snx8 | 1.930948e+03 | 1.2889581 | 0.366205413 | 0.3240941 | 1.257940e+00 | 0.2620414454 | 0.79788700 | No | Yes | 2.012808e+03 | 278.191831 | 1.401181e+03 | 198.902869 | ||
| ENSG00000106477 | 95681 | CEP41 | protein_coding | Q9BYV8 | FUNCTION: Required during ciliogenesis for tubulin glutamylation in cilium. Probably acts by participating in the transport of TTLL6, a tubulin polyglutamylase, between the basal body and the cilium. {ECO:0000269|PubMed:22246503}. | Alternative splicing;Autism;Autism spectrum disorder;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Joubert syndrome;Methylation;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene encodes a centrosomal and microtubule-binding protein which is predicted to have two coiled-coil domains and a rhodanese domain. In human retinal pigment epithelial cells the protein localized to centrioles and cilia. Mutations in this gene have been associated with Joubert Syndrome 15; an autosomal recessive ciliopathy and neurological disorder. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2012]. | hsa:95681; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cilium [GO:0005929]; cytosol [GO:0005829]; membrane [GO:0016020]; cilium assembly [GO:0060271]; protein polyglutamylation [GO:0018095]; protein transport [GO:0015031] | 21438139_Three rare potentially pathogenic variants were identified in the TSGA14 gene, which encodes a centrosomal protein. 22246503_The data identified CEP41 mutations as a cause of Joubert syndrome and implicated tubulin post-translational modification in the pathogenesis of human ciliary dysfunction. 22456293_In cortices, the MEST promoter was hemimethylated, as expected for a differentially methylated imprinting control region, whereas the COPG2 and TSGA14 promoters were completely demethylated, typical for transcriptionally active non-imprinted genes. 30664616_Missense variants in one gene, CEP41, associated significantly with Autism Spectrum Disorder (ASD). Homozygous gene-disrupting variants in CEP41 were initially found to be responsible for recessive Joubert syndrome. 31885126_CEP41-mediated ciliary tubulin glutamylation drives angiogenesis through AURKA-dependent deciliation. | ENSMUSG00000029790 | Cep41 | 4.277175e+02 | 1.1178418 | 0.160715984 | 0.3081268 | 2.740116e-01 | 0.6006533849 | 0.90910497 | No | Yes | 4.126163e+02 | 57.600228 | 3.607540e+02 | 51.647157 | |
| ENSG00000106683 | 3984 | LIMK1 | protein_coding | P53667 | FUNCTION: Serine/threonine-protein kinase that plays an essential role in the regulation of actin filament dynamics. Acts downstream of several Rho family GTPase signal transduction pathways (PubMed:10436159, PubMed:11832213, PubMed:12807904, PubMed:15660133, PubMed:16230460, PubMed:18028908, PubMed:22328514, PubMed:23633677). Activated by upstream kinases including ROCK1, PAK1 and PAK4, which phosphorylate LIMK1 on a threonine residue located in its activation loop (PubMed:10436159). LIMK1 subsequently phosphorylates and inactivates the actin binding/depolymerizing factors cofilin-1/CFL1, cofilin-2/CFL2 and destrin/DSTN, thereby preventing the cleavage of filamentous actin (F-actin), and stabilizing the actin cytoskeleton (PubMed:11832213, PubMed:15660133, PubMed:16230460, PubMed:23633677). In this way LIMK1 regulates several actin-dependent biological processes including cell motility, cell cycle progression, and differentiation (PubMed:11832213, PubMed:15660133, PubMed:16230460, PubMed:23633677). Phosphorylates TPPP on serine residues, thereby promoting microtubule disassembly (PubMed:18028908). Stimulates axonal outgrowth and may be involved in brain development (PubMed:18028908). {ECO:0000269|PubMed:10436159, ECO:0000269|PubMed:11832213, ECO:0000269|PubMed:12807904, ECO:0000269|PubMed:15660133, ECO:0000269|PubMed:16230460, ECO:0000269|PubMed:18028908, ECO:0000269|PubMed:22328514, ECO:0000269|PubMed:23633677}.; FUNCTION: [Isoform 3]: Has a dominant negative effect on actin cytoskeletal changes. Required for atypical chemokine receptor ACKR2-induced phosphorylation of cofilin (CFL1). {ECO:0000269|PubMed:10196227}. | 3D-structure;ATP-binding;Alternative splicing;Cell projection;Cytoplasm;Cytoskeleton;Kinase;LIM domain;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase;Ubl conjugation;Williams-Beuren syndrome;Zinc | There are approximately 40 known eukaryotic LIM proteins, so named for the LIM domains they contain. LIM domains are highly conserved cysteine-rich structures containing 2 zinc fingers. Although zinc fingers usually function by binding to DNA or RNA, the LIM motif probably mediates protein-protein interactions. LIM kinase-1 and LIM kinase-2 belong to a small subfamily with a unique combination of 2 N-terminal LIM motifs and a C-terminal protein kinase domain. LIMK1 is a serine/threonine kinase that regulates actin polymerization via phosphorylation and inactivation of the actin binding factor cofilin. This protein is ubiquitously expressed during development and plays a role in many cellular processes associated with cytoskeletal structure. This protein also stimulates axon growth and may play a role in brain development. LIMK1 hemizygosity is implicated in the impaired visuospatial constructive cognition of Williams syndrome. Alternative splicing results in multiple transcript variants encoding distinct isoforms.[provided by RefSeq, Feb 2011]. | hsa:3984; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; lamellipodium [GO:0030027]; membrane [GO:0016020]; neuron projection [GO:0043005]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; ATP binding [GO:0005524]; heat shock protein binding [GO:0031072]; metal ion binding [GO:0046872]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; actin cytoskeleton organization [GO:0030036]; Fc-gamma receptor signaling pathway involved in phagocytosis [GO:0038096]; negative regulation of ubiquitin-protein transferase activity [GO:0051444]; nervous system development [GO:0007399]; positive regulation of actin filament bundle assembly [GO:0032233]; positive regulation of axon extension [GO:0045773]; positive regulation of stress fiber assembly [GO:0051496]; protein phosphorylation [GO:0006468]; Rho protein signal transduction [GO:0007266]; signal transduction [GO:0007165] | 11925442_Mitosis-specific activation of LIM motif-containing protein kinase and roles of cofilin phosphorylation and dephosphorylation in mitosis. 12323073_identification as interaction partner of 14-3-3 zeta 12777619_LIM kinase 1 (LIMK1), a critical regulator of actin dynamics, plays a regulatory role in tumor cell invasion. 12821664_LIMK1 has a role in regulating cell division and invasive property of prostate cancer cells; the effect is not mediated by phosphorylation of cofilin 14530263_LIM-kinase 1 is translocated to the nucleus after being bound by p57kip2 15023529_LIMK1 is enriched in both axonal and dendritic growth cones of E18 hippocampal pyramidal neurons where it is found in punctae that extend far out into filopodia, as well as in a perinuclear region identified as Golgi. 15189451_LIMK1 is cleaved and activated during antiFas antibody-induced apoptosis. Expression of an N-terminally truncated LMK1 fragment induced membrane blebbing. 15220930_LATS1 is a novel cytoskeleton regulator that affects cytokinesis by regulating actin polymerization through negative modulation of LIMK1. 15660133_a multi-protein complex consisting of SSH-1L, LIMK1, actin, and the scaffolding protein, 14-3-3zeta, is involved, along with the kinase, PAK4, in the regulation of ADF/cofilin activity 15897190_LIMK1 activity is required for thrombin-induced modulation of microtubule destabilization and actin polymerization 16204183_Rnf6 controls cellular LIMK1 concentrations and indicate a new function for the ubiquitin/proteasome system in regulating local growth cone actin dynamics 16381000_results suggested an important role for LIMK1 signaling in breast cancer tumor growth, angiogenesis and invasion and a regulatory connection between LIMK1 and the uPA system 16455074_LIMK1 may play a role different from that of LIMK2 in regulating mitotic spindle organization, chromosome segregation, and cytokinesis during the cell division cycle. 16611674_Elastin and LIMK1 SNPs effect in the at-risk haplotype possibly by weakening the vascular wall and promoting the development of IA 16611674_Observational study of gene-disease association. (HuGE Navigator) 16763828_Taken together, our novel findings presented in this paper implicate that LIMK1 signaling indeed plays a pivotal role in the regulation of EGFR trafficking through the endocytic pathway in invasive tumor cells. 17287949_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17294230_the structure, regulation and function; possible contributions to human disease (Review) 17512523_These findings demonstrate a cell-type dependent functional interaction between parkin and LIMK1 and provide new evidence that links parkin and LIMK1 in the pathogenesis of familial PD. 18000399_gamma-tubulin associates with phosphorylated LIMK1 and LIMK2 but not with dephosphorylated LIMK1 or LIMK2. 18028908_LIMK1 phosphorylation of p25 blocks p25 activity, thus promoting microtubule disassembly. 18079118_LIMK1-mediated cofilin phosphorylation is required for accurate spindle orientation by stabilizing cortical actin networks during mitosis. 18096821_PDGF participates in actin dynamics by dual regulation of cofilin activity via LIMK and SSH1L in aortic smooth muscle cells 18424072_These results support a model whereby hepatocyte growth factor-stimulated cell migration also requires a cofilin phosphorylation step that is mediated by PAK4. 18566305_Observational study of gene-disease association. (HuGE Navigator) 18978953_LIMK1 may mediate TGF-beta-dependent signaling during ocular inflammation. 19056482_Observational study of gene-disease association. (HuGE Navigator) 19662944_findings indicate that spatial deficits associated with Williams Syndrome also affect large-scale spatial processing and suggest that hemizygous deletion of LIMK1 is not sufficient to account for any of the spatial deficits associated with this syndrome. 19734939_the p57(Kip2) control of LIMK-1 ultimately affects cell mobility negatively. 19778628_DGCR6L, a novel PAK4 interacting protein, regulated PAK4-mediated migration of human gastric cancer cells via LIMK1. 20047470_Both LIMK1 and LIMK2 single knock down led to a reduction of invasion and metastatic behavior in the zebrafish xenograft metastasis assay. 20061392_phosphorylation of annexin 1 regulates the angiogenic effect that is associated with the activation of the p38/LIM kinase 1 axis by VEGF 20100465_LIMK1 mediates both the mesenchymal and amoeboid modes of invasion of HT1080 cells. 20347121_FXa induced myosin light chain phosphorylation and LIMK1 activation resulting in cofilin inactivation 20622363_High LIMK1 is associated with cervical cancer dysplasia. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20694560_Observational study of gene-disease association. (HuGE Navigator) 20873970_Changes in the expression of cytoskeletal regulatory proteins such as LIMK and cofilin may play a role in weakening thoracic aortic medial tissue, as a precondition to thoracic aortic dissection. 21219645_LIM kinase1 modulates function of membrane type matrix metalloproteinase 1 in prostate cancer cells 21228795_evealed a SNP, rs6460071 located on LIMK1 gene (P = 0.00069) to be significantly associated with increased risk of intracranial aneurysm 21321123_LIM kinase 1 modulates cortical actin and CXCR4 cycling and is activated by HIV-1 to initiate viral infection. 21682918_LIMK1 activity in both the cytoplasmic and nuclear compartments promotes breast cancer progression. 21868383_LIMK1 has a dual role in regulating lamellipodium extension by decelerating actin retrograde flow and polymerization. 21923909_Data describe the role of LIMK1-mediated CFL1 pathway and actin dynamics in retinoid receptor mediated function and show that LIMK1-mediated phosphocycling of CFL1 plays a role in maintaining actin homeostasis and receptor activity. 22214762_Aur-A physically associates with LIMK1 and activates it through phosphorylation, which is important for its centrosomal and spindle pole localization. 22715593_demonstrated that LIMK1, a key regulator of actin cytoskeleton, was overexpressed at both mRNA and protein levels in MG63/VCR cells and the higher LIMK1 protein level was correlated with higher level of phosphorylated cofilin 22883572_A novel activity of Rho in promoting a complex between fascin-1 and LIMK1/2 that modulates the interaction of fascin-1 with actin, was identified. 22906279_LIMK1 is associated with the development of ovarian cancer and with the level of tumour differentiation in patients with ovarian carcinoma. 23086941_BDNF-induced TrkB dimerization led to LIMK1 dimerization and transphosphorylation independent of TrkB kinase activity, which could further enhance the activation and stabilization of LIMK1 23167938_The authors believe that the Williams-Beuren syndrome chromosome region can be a possible target for further investigation as the genetic basis of keratoconus. 23239465_Overexpression of LIMK1 is associated with breast cancer growth and invasiveness. 23338717_Overexpression of LIMK1 is associated with the development and progression of glioblastoma multiforme. 23620575_LIMK1 plays a novel role in selectively mediating GPIb-IX-dependent TXA2 synthesis and thrombosis. 23700283_the conserved region in the LIMK1 3' UTR is involved in regulating LIMK1 expression at the post-transcriptional level 23702034_DAPK functions as a scaffold for the LIMK/cofilin complex and triggers a closer interaction of both proteins under TNF stimulation. 24039253_Thus this study provides evidence for a novel role of LIMK1 and SSH-1L in selectively regulating endothelial cell inflammation associated with intravascular coagulation. 24063279_These results indicate that the overexpression of LIMK1 is tightly associated with an aggressive phenotype of lung cancer cells, knockdown of LIMK1 suppressed cell migration and invasion, enhanced chemosensitivity, suggesting a potential therapeutic target for lung cancer. 24390089_Ectopic expression of LIMK1 dramatically dampened mir-27b action of cancer inhibition. 24696479_ROCK1- and LIMK1-regulated phosphorylation of cofilin played a role in retrovirus release from host cells and in cell-cell transmission events. 24858712_miR-20a plays a role as a tumor suppressor in thyroid cancer cells and targets LIMK1 25019203_miR-20a is involved in the tumor inhibition of cutaneous squamous cell carcinoma by directly targeting LIMK1 gene. 25107909_the functional relevance of the LRAP25-MRCK complex in LIMK1-cofilin signaling and the importance of LRAP adaptors as key determinants of MRCK cellular localization and downstream specificities. 25190487_Data show that the downregulation of miR-138 induced the upregulation of Limk1 in ovarian cancer (OC) cells suggesting that these 2 genes may play a key role in the migration and invasion of OC cells. 25307528_LIMK1 has a role in regulating human trophoblast invasion/differentiation and is down-regulated in preeclampsia 25344584_Results show that in addition to a potential role in promoting metastasis, changes in LIMK1 and LIMK2 expression and/or activity might contribute to AR function in prostate cancer via regulation of microtubule cytoskeletal dynamics. 25359780_CXCL12/CXCR4 signaling has a role in docetaxel-induced microtubule stabilization via p21-activated kinase 4-dependent activation of LIMK1 25529997_LIMK1 is overexpressed in endometrial stromal cells. 25661344_LIMK1 is overexpressed in gastric cancer. 25884247_Gene knockdown/rescue experiments reveal that LIMK1 palmitoylation is essential for normal spine actin polymerization, for spine-specific structural plasticity and for long-term spine stability. 25981171_This study showed that LIMK1 messenger RNA levels was significantly upregulated in subjects with schizophrenia in laminar and cellular samples. 27116935_In turn, LIMK1 and LIMK2 are required for MT1-MMP-dependent matrix degradation and cell invasion in a three-dimensional type I collagen environment. 27153537_Report structural basis for noncanonical substrate recognition of cofilin-1/LIMK1 to regulation actin cytoskeleton dynamics. 27178114_LIMK1 expression is negatively regulated in granular layers-negative psoriatic epidermis or IL-22/IL-24-treated hyperproliferative reconstituted epidermis. These findings suggest a novel regulatory mechanism and a potent role of LIMK1 in psoriatic epidermis. 27273096_increased BMPR2 signal transduction is linked to fragile X syndrome (FXS) and that the BMPR2-LIMK1 pathway is a putative therapeutic target in patients with FXS and possibly other forms of autism 28358024_Results indicated that LIM kinase 1 was a potential target molecule for the inhibitory effect of diallyl disulfide on colon cancer cell migration and invasion. 28440035_The present study demonstrates that miR-145 plays an important role in inhibiting cell migration by directly targeting PAK4, and identifies miR-145-PAK4-LIMK1-cofilin as a novel regulatory pathway that contributed to colorectal cancer metastasis. 29058761_Once activated, c-Abl kinase regulated the activity of Vav1, which further affected Rac1/PAK1/LIMK1/cofilin signaling pathway. 29188531_findings suggest that miR-519d-3p regulates the LIMK1/CFL1 pathway in breast cancer 29925632_Results show that silencing LIMK1 leads to defective spindle organization and centrosome integrity. Dynein light intermediate chains were identified as the downstream mediators of LIMK1 in the maintenance of centrosome integrity. 30837367_Our findings revealed that increased LIMK1 protein levels may contribute to atrial fibrosis, and suggested that LIMK1 might be involved in AF development by promoting fibrogenesis associated with TGF-beta 30861189_LIMK1 depletion enhances fasudil-dependent inhibition of urethral fibroblast proliferation and migration. 31026254_ROCK1 and ROCK2 regulated cell proliferation in PC-3 and DU145 prostate cancer cells. Protein levels of phosphorylated LIM kinase 1 (p-LIMK1) and matrix metalloproteinase-2 (MMP-2) were reduced in ROCK1 and ROCK2 siRNA transfected cells. CONCLUSIONS In PC-3 and DU145 human prostate cancer cells, ROCK promoted cell proliferation and migration by targeting LIMK1 and MMP-2. 31043701_ROCK1 but not LIMK1 or PAK2 is a key regulator of apoptotic membrane blebbing and cell disassembly. 31277940_It has been found that LIMK1/LIMK2 is critical for GPER activation-induced breast cancer cell migration. 31641031_LIMK1and LIMK2 expression status is associated with glioblastoma (GBM) grade and poor prognosis. Simultaneous knockdown of both isoforms strongly reduced the invasive motility of continuous culture models and human GBM tumor-initiating cells. LIMK1/2 functionally regulated cell invasiveness, in part, by disrupting polarized cell motility under confinement and cell chemotaxis. 31652302_LIMK1 robustly phosphorylates both serine and tyrosine residues, establishing LIMK1 as a dual-specificity kinase. 32141553_Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. 32168432_Up-regulation of LIMK1 expression in prostate cancer is correlated with poor pathological features, lymph node metastases and biochemical recurrence. 32196612_MiR-429-5p attenuates the migration and invasion of malignant melanoma by targeting LIMK1. 32463132_Effects of PAK4/LIMK1/Cofilin-1 signaling pathway on proliferation, invasion, and migration of human osteosarcoma cells. 32641387_Human NK Cell Cytoskeletal Dynamics and Cytotoxicity Are Regulated by LIM Kinase. 32860848_Knockdown of RhoGDI2 represses human gastric cancer cell proliferation, invasion and drug resistance via the Rac1/Pak1/LIMK1 pathway. 33649781_Alantolactone suppresses the metastatic phenotype and induces the apoptosis of glioblastoma cells by targeting LIMK kinase activity and activating the cofilin/Gactin signaling cascade. 33686242_LIMK1 nuclear translocation promotes hepatocellular carcinoma progression by increasing p-ERK nuclear shuttling and by activating c-Myc signalling upon EGF stimulation. 33760158_FOXD3AS1/miR1283p/LIMK1 axis regulates cervical cancer progression. 33883692_LIMK1 promotes peritoneal metastasis of gastric cancer and is a therapeutic target. 34079084_Long non-coding RNA Lnc-408 promotes invasion and metastasis of breast cancer cell by regulating LIMK1. 34107579_[Regulatory effects of LIM kinase 1 on the proliferation and metastasis of hepatocellular carcinoma cells]. 34381117_Cleavage and activation of LIM kinase 1 as a novel mechanism for calpain 2-mediated regulation of nuclear dynamics. 34686503_The Diagnostic and Prognostic Value of LIMK1/2: A Pan-Cancer Analysis. | ENSMUSG00000029674 | Limk1 | 2.106911e+03 | 1.3286327 | 0.409942332 | 0.3210125 | 1.624058e+00 | 0.2025269074 | 0.78445471 | No | Yes | 2.298605e+03 | 293.445567 | 1.466648e+03 | 192.562762 | |
| ENSG00000106829 | 7091 | TLE4 | protein_coding | Q04727 | FUNCTION: Transcriptional corepressor that binds to a number of transcription factors. Inhibits the transcriptional activation mediated by PAX5, and by CTNNB1 and TCF family members in Wnt signaling. The effects of full-length TLE family members may be modulated by association with dominant-negative AES. Essential for the transcriptional repressor activity of SIX3 during retina and lens development and for SIX3 transcriptional auto-repression (By similarity). {ECO:0000250}. | Acetylation;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;WD repeat;Wnt signaling pathway | hsa:7091; | beta-catenin-TCF complex [GO:1990907]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; chromatin binding [GO:0003682]; DNA-binding transcription factor binding [GO:0140297]; transcription corepressor activity [GO:0003714]; cellular response to leukemia inhibitory factor [GO:1990830]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of transcription by RNA polymerase II [GO:0000122]; Wnt signaling pathway [GO:0016055] | 17060451_Gbx2 and Otx2 interact with the WD40 domain of Groucho/Tle corepressors 18258796_Knockdown of TLE1 or TLE4 levels increased the rate of cell division of the AML1-ETO-expressing Kasumi-1 cell line, whereas forced expression of either TLE1 or TLE4 caused apoptosis and cell death. 19714205_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19893608_germ cell-specific RBMY and hnRNP G-T proteins were more efficient in stimulating TLE4-T incorporation than somatically expressed hnRNP G protein. 20816195_Observational study of gene-disease association. (HuGE Navigator) 20889853_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 22169276_Grg4 recruits the arginine methyltransferase PRMT5 to chromatin resulting in symmetric H4R3 dimethylation. 22927467_Displacing coactivators CREB-binding protein/p300 while promoting the recruitment of a corepressor, Grg4. 24099773_Groucho related gene Grg4 robustly activates the expression of a BMP reporter gene, as well as enhancing and sustaining the upregulation of the endogenous Id1 gene induced by BMP7. 25631048_PPM1B interacts with Groucho 4 and is localized to DNA in a Groucho-dependent manner, and phosphatase activity is required for transcriptional silencing. 26701208_the results suggested that TLE4, a potential prognostic biomarker for colorectal cancer, plays an important role in the development and progression of human colorectal cancer. 27486062_the loss of TLE4 confers proliferative advantage to leukemic cells, simultaneous with an upregulation of a pro- inflammatory signature mediated through aberrant increases in Wnt signaling activity 28262390_Genes TGFB1, TLE4 and MUC22 are associated with the risk of childhood asthma in Chinese population. 35099008_TLE4 regulates muscle stem cell quiescence and skeletal muscle differentiation. | ENSMUSG00000024642 | Tle4 | 2.812657e+03 | 1.2583159 | 0.331494196 | 0.2493178 | 1.789672e+00 | 0.1809661707 | 0.77664849 | No | Yes | 2.772648e+03 | 200.011458 | 2.244231e+03 | 166.213535 | ||
| ENSG00000107077 | 23081 | KDM4C | protein_coding | Q9H3R0 | FUNCTION: Histone demethylase that specifically demethylates 'Lys-9' and 'Lys-36' residues of histone H3, thereby playing a central role in histone code. Does not demethylate histone H3 'Lys-4', H3 'Lys-27' nor H4 'Lys-20'. Demethylates trimethylated H3 'Lys-9' and H3 'Lys-36' residue, while it has no activity on mono- and dimethylated residues. Demethylation of Lys residue generates formaldehyde and succinate. {ECO:0000269|PubMed:16603238, ECO:0000269|PubMed:28262558}. | 3D-structure;Alternative splicing;Chromatin regulator;Dioxygenase;Iron;Metal-binding;Nucleus;Oxidoreductase;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene is a member of the Jumonji domain 2 (JMJD2) family. The encoded protein is a trimethylation-specific demethylase, and converts specific trimethylated histone residues to the dimethylated form. This enzymatic action regulates gene expression and chromosome segregation. Chromosomal aberrations and changes in expression of this gene may be found in tumor cells. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2015]. | hsa:23081; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; pericentric heterochromatin [GO:0005721]; androgen receptor binding [GO:0050681]; enzyme binding [GO:0019899]; histone demethylase activity [GO:0032452]; histone H3-methyl-lysine-36 demethylase activity [GO:0051864]; histone H3-methyl-lysine-9 demethylase activity [GO:0032454]; histone H3-tri/dimethyl-lysine-9 demethylase activity [GO:0140684]; zinc ion binding [GO:0008270]; blastocyst formation [GO:0001825]; chromatin remodeling [GO:0006338]; histone H3-K36 demethylation [GO:0070544]; histone H3-K9 demethylation [GO:0033169]; negative regulation of histone H3-K9 trimethylation [GO:1900113]; positive regulation of cell population proliferation [GO:0008284]; regulation of androgen receptor signaling pathway [GO:0060765]; regulation of stem cell differentiation [GO:2000736]; regulation of transcription by RNA polymerase II [GO:0006357]; stem cell population maintenance [GO:0019827] | 16732293_GASC1 interacts with H3K9me3; three members of this subfamily of proteins demethylate H3K9me3/me2 in vitro through a hydroxylation reaction requiring iron and alpha-ketoglutarate as cofactors 17277772_JMJD2C and LSD1 interact and both demethylases cooperatively stimulate androgen receptor-dependent gene transcription. 17611647_JMJD2C is the histone demethylase implicated in the epigenetic reprogramming during the early embryogenesis. 18649358_Observational study of gene-disease association. (HuGE Navigator) 18713068_Results show that many genes regulated by hypoxia and HIF-1alpha show patterns of induction with JMJD (Jumonji-domain containing)1A and JMJD2B demonstrating robust, and JMJD2C more modest, up-regulation by hypoxia. 18927281_Gene rearrangements of JMJD2C is associated with mucosa-associated lymphoid tissue lymphoma. 19339270_Observational study of gene-disease association. (HuGE Navigator) 19784073_GASC1 is a driving oncogene in the 9p23-24 amplicon in human breast cancer. 20127736_identification of genetic alterations & expression changes of LSD1, JHDM2A & GASC1 in prostate cancer (PC); as no genetic alterations & only modest expression changes were found, it is unlikely they play a major role in progression of PC 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20410850_Observational study of gene-disease association. (HuGE Navigator) 20410850_The strongest association in obsessive-compulsive disorder has been found at rs301443 (P=0.000067) residing between SLC1A1 and JMJD2C at 9p24. 21575637_inhibition measurements showed significant selectivity between KDM4C and KDM6A 22072270_This study presented that KDM4C showed strong associations with alcohol withdrawal symptoms (p < 5 x 10-5). 23067339_JMJD2C displays an optimal activity in vitro at alphaKG concentrations similar to those found in cancer cells, with implications for the regulation of histone demethylation activity in cancer versus normal cells. 23129632_JMJD2C decreases trimethylation of histone H3 at lysine 9, and enhances HIF-1 binding to hypoxia response elements, thereby activating transcription of proteins that are required for metabolic reprogramming and for lung metastasis. 23148692_Patients with GASC1 positive tumors have better breast cancer specific survival and respond better to radiotherapy and hormonal treatment 23692052_analysis of the nickel-induced inhibition of truncated constructs of JMJD2A and JMJD2C histone demethylases using X-ray absorption spectroscopy 23698634_KDM4C maintains the sphere-forming capacity in CRCs by mediating the beta-catenin-dependent transcription of JAG1 in a feed-forward manner. 24285722_KDM4C promotes transcriptional activation by removing the repressive histone mark, H3K9me3, from its target genes. Variation in its expression leads to differences in the growth of normal and some cancer cells. 24371038_GASC1 expression is higher in adenocarcinoma than squamous cell carcinoma. Smoking decreases GASC1 expression in tumor cells, indicating that tobacco smoke may influence the methylation of histone 3 lysine residues in lung cancer. 24952432_Data suggest that D396N polymorphism of JmjC domain-containing histone demethylase JMJD2C affects the prognosis of breast cancer by altering caspase-3 cleavage and the ability of double strand DNA break repair which may contribute to therapy resistance. 25636512_JMJD2B and JMJD2C play an important role in the pathology of osteosarcoma via the up-regulation of FGF2. 26747609_KDM4C recognition of H3K4me3 stimulates demethylation of H3K9me3 in cis on peptide and mononucleosome substrates. 26766589_Pharmacological inhibition of KDM4C/PRMT1 suppresses transcription and transformation ability of MLL fusions 28236704_JMJD2C regulated the activities of lung cancer cells by directly controlling the expression of CUL4A in JMJD2C over-expression cell line. 28396876_histone H3 lysine 9 methylation reduction, which may be due to the downregulation of methyltransferase SUV39H2 and the upregulation of demethylase KDM4C, was found in CD4(+) T lymphocytes of Latent autoimmune diabetes in adult patients 29374629_we have identified KDM4C as a new risk gene shared between systemic vasculitides, consistent with the increasing evidences of the crucial role that the epigenetic mechanisms have in the development of complex immune-mediated conditions. 29438700_LSD1 and JMJD2C disable oncogenic Ras- or Braf-induced senescence by enabling the expression of E2F target genes 30537731_CircZMYM2 repressed the expression of its target miR-335-5p. MiR-335-5p attenuated pancreatic cancer development via inhibition of JMJD2C. 31406252_data indicate that circKDM4C might have considerable potential as a prognostic biomarker in breast cancer, and support the notion that therapeutic targeting of circKDM4C/miR-548p/PBLD axis may be a promising treatment approach for breast cancer patients 31665047_Data found that JMJD2C was overexpressed in colorectal cancer (CRC) tumor and metastatic foci, and CRC patients with lower JMJD2C expression had better prognosis. JMJD2C could enhance the metastatic abilities of CRC cells in vitro and in vivo by regulating the histone methylation level of MALAT1 promoter, thereby up-regulating the expression of MALAT1 and enhancing the activity of beta-catenin signaling pathway. 31888886_Wnt-Induced Stabilization of KDM4C Is Required for Wnt/beta-Catenin Target Gene Expression and Glioblastoma Tumorigenesis. 32483441_Harnessing stemness and PD-L1 expression by AT-rich interaction domain-containing protein 3B in colorectal cancer. 32972441_microRNA-216b enhances cisplatin-induced apoptosis in osteosarcoma MG63 and SaOS-2 cells by binding to JMJD2C and regulating the HIF1alpha/HES1 signaling axis. 33279929_Rare genetic variants in the gene encoding histone lysine demethylase 4C (KDM4C) and their contributions to susceptibility to schizophrenia and autism spectrum disorder. 33462212_Histone demethylase KDM4C controls tumorigenesis of glioblastoma by epigenetically regulating p53 and c-Myc. 33469678_JMJD2C triggers the growth of multiple myeloma cells via activation of betacatenin. 33558705_USP9X-mediated KDM4C deubiquitination promotes lung cancer radioresistance by epigenetically inducing TGF-beta2 transcription. 33649841_GASC1 promotes glioma progression by enhancing NOTCH1 signaling. 33692332_GASC1 promotes hepatocellular carcinoma progression by inhibiting the degradation of ROCK2. 34265399_UHRF1 promotes androgen receptor-regulated CDC6 transcription and anti-androgen receptor drug resistance in prostate cancer through KDM4C-Mediated chromatin modifications. 34548470_Disruption of KDM4C-ALDH1A3 feed-forward loop inhibits stemness, tumorigenesis and chemoresistance of gastric cancer stem cells. 35046387_JMJD2C-mediated long non-coding RNA MALAT1/microRNA-503-5p/SEPT2 axis worsens non-small cell lung cancer. 35343073_Inhibition of KDM4C/c-Myc/LDHA signalling axis suppresses prostate cancer metastasis via interference of glycolytic metabolism. 35414503_Histone Demethylase GASC1 Inhibitor Targeted GASC1 Gene to Inhibit the Malignant Transformation of Esophageal Cancer through the NOTCH-MAPK Signaling Pathway. | ENSMUSG00000028397 | Kdm4c | 2.494767e+02 | 0.9028456 | -0.147448857 | 0.3655974 | 1.629099e-01 | 0.6864914429 | 0.93348335 | No | Yes | 2.458461e+02 | 49.482437 | 2.579264e+02 | 53.090634 | |
| ENSG00000107099 | 81704 | DOCK8 | protein_coding | Q8NF50 | FUNCTION: Guanine nucleotide exchange factor (GEF) which specifically activates small GTPase CDC42 by exchanging bound GDP for free GTP (PubMed:28028151, PubMed:22461490). During immune responses, required for interstitial dendritic cell (DC) migration by locally activating CDC42 at the leading edge membrane of DC (By similarity). Required for CD4(+) T-cell migration in response to chemokine stimulation by promoting CDC42 activation at T cell leading edge membrane (PubMed:28028151). Is involved in NK cell cytotoxicity by controlling polarization of microtubule-organizing center (MTOC), and possibly regulating CCDC88B-mediated lytic granule transport to MTOC during cell killing (PubMed:25762780). {ECO:0000250|UniProtKB:Q8C147, ECO:0000269|PubMed:22461490, ECO:0000269|PubMed:25762780, ECO:0000269|PubMed:28028151}. | Alternative splicing;Cell membrane;Cell projection;Chromosomal rearrangement;Cytoplasm;Disease variant;Guanine-nucleotide releasing factor;Membrane;Mental retardation;Phosphoprotein;Reference proteome | This gene encodes a member of the DOCK180 family of guanine nucleotide exchange factors. Guanine nucleotide exchange factors interact with Rho GTPases and are components of intracellular signaling networks. Mutations in this gene result in the autosomal recessive form of the hyper-IgE syndrome. Alternatively spliced transcript variants encoding different isoforms have been described.[provided by RefSeq, Jun 2010]. | hsa:81704; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; lamellipodium membrane [GO:0031258]; leading edge membrane [GO:0031256]; membrane [GO:0016020]; guanyl-nucleotide exchange factor activity [GO:0005085]; cellular response to chemokine [GO:1990869]; dendritic cell migration [GO:0036336]; immunological synapse formation [GO:0001771]; memory T cell proliferation [GO:0061485]; negative regulation of T cell apoptotic process [GO:0070233]; positive regulation of establishment of T cell polarity [GO:1903905]; positive regulation of GTPase activity [GO:0043547]; positive regulation of T cell migration [GO:2000406]; regulation of small GTPase mediated signal transduction [GO:0051056]; small GTPase mediated signal transduction [GO:0007264] | 15304341_involvement of DOCK8 in processes that affect the organisation of filamentous actin. 18060736_rare mutations in the DOCK8 gene may contribute to some cases of autosomal dominant mental retardation 19640199_Under-expression of DOCK8 is associated with hepatocellular carcinoma. 19776401_Autosomal recessive DOCK8 deficiency is associated with a novel variant of combined immunodeficiency. 20004785_Autosomal-recessive mutations in DOCK8 are responsible for many, although not all, cases of autosomal-recessive hyper-IgE syndrome. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20596660_Chromosome 9p loss is the hallmark of squamous cell carcinoma, and DMRT1, DMRT3 and DOCK8 genes at 9p24.3 might be of interest for the study of the pathophysiology of SCC as potential targets for therapeutic measures. 21058221_Several AR-HIES patients have recently been shown to harbour mutations in the gene for dedicator of cytokinesis 8 (DOCK8). Here, we present the long-term outcome of a girl having received a hematopoietic stem cell graft. 21178271_DOCK8 deficiency and clinical manifestations of hyper IgE syndromes (Review) 21178272_Mutations in DOCK8 lead to DOCK8 immunodeficiency syndrome characterized by recurrent viral infections, severe atopy, and early onset malignancy. (Review) 21763205_A 2-bp deletion at codon 510 in exon 14 causing a frameshift mutation was found in 3 homozygous siblings with Job syndrome and their heterozygous first-cousin parents. 21931011_Rates of malignancy and overall mortality in patients with DOCK8 deficiency were higher than in those with Job's syndrome 21969276_Findings help to explain why DOCK8-deficient patients are susceptible to recurrent infections and provide new insights into how T-cell memory is sustained. 22006977_These findings highlight a key role for DOCK8 in the survival and function of human and mouse CD8 T cells. 22534316_DOCK8 deficiency (a newly described combined primary immunodeficiency disease) accounted for 15% of combined immune deficiency cases in the National Primary Immunodeficiency Disorders Registry in Kuwait, a country with high prevalence of consanguinity. 22581261_DOCK8 mediates an MyD88 signaling pathway essential for TLR9-driven B-cell proliferation aand immunoglobulin production. 22876580_DOCK8 encodes dedicator of cytokinesis 8. 22942019_We used this new approach to analyse exome data from 24 patients with primary immunodeficiencies. Our analysis identified two novel causative deletions in the genes GATA2 and DOCK8 22968740_Three novel DOCK8 mutations and two large deletions are found in thirteen patients with autosomal recessive hyper-IgE syndrome in a single center experience. 23374272_nonsense mutation in the CLEC7A gene, p.Tyr238X, rs16910526 and a novel homozygous frameshift variant in exon 27 of the DOCK8 gene, c.3193delA were identified in brothers with an immunodeficiency syndrome characterized by severe eczema, milk and egg allergies, infections, diarrhea, failure to thrive and, in two of the three, lymphoma lymphoma. 23380217_DOCK8 deficiency results in severely impaired natural killer cell function because of an inability to form a mature lytic immunologic synapse through targeted synaptic F-actin accumulation 23455509_Dedicator of cytokinesis 8 interacts with talin and Wiskott-Aldrich syndrome protein to regulate NK cell cytotoxicity. 23859592_Two novel large deletions, del1-14 exons and del8-18 exons, of DOCK8 have been identified in two siblings with the adaptive immune deficiencies. 23891736_DOCK8 deficiency results in defective antibody responses and undirected plasma cell expansion in the lymph nodes, as part of a combined immunodeficiency cured by hematopoietic stem cell transplantation. 23911989_Clinical features of immunodeficiency syndrome are associated with DOCK8 mutations. (Review) 23929855_DOCK8 is required for the development and survival of mature NKT cells. 24698323_Biallelic mutations in the DOCK8 gene cause autosomal-recessive hyper-IgE syndrome. 24898675_Hyper-IgE syndromes and atopic dermatitis patients showed different sensitization pattern of serum IgE corresponding to the allergic disease manifestations and Th-cell subset data, suggesting a key role of DOCK8 in the development of food allergy 25218284_Dedicator of cytokinesis 8-deficient patients have a breakdown in peripheral B-cell tolerance and defective regulatory T cells 25220305_Mutations of DOCK8 in three children, two of whom developed sclerosing cholangitis, are reported. 25332498_This is a case of systemic lupus erythematosus with hyper-immunoglobulin E syndrome documented as DOCK8 deficiency. 25422492_DOCK8-regulated shape integrity of lymphocytes prevents cytothripsis and promotes antiviral immunity in the skin. 25428919_CD147 has a role in promoting Src-dependent activation of Rac1 signaling through STAT3/DOCK8 during the motility of hepatocellular carcinoma cells 25724123_DOCK8 deficiency is likely in patients with severe viral infections, allergies, and/or low IgM levels. 26235511_Letter/Case Report: DOCK8 homozygous mutation leading to primary immune deficiency. 26592211_comparative study provides a long-term observation of DOCK8- and STAT3-hyper-IgE syndrome patients with regard to clinical and laboratory findings, and assesses the activation and cytokine secretion of lymphocytes after in vitro stimulation 26659092_mutations in Chinese patients with hyper-IgE syndrome 26799599_Our results suggest that decreased expression of DOCK8 in response to CRH might disturb the immunosuppressive function of Tregs and contribute to stress-induced aggravation of AD symptoms. 26883462_A novel DOCK8 sequence insertion caused primary immunodeficiency in two siblings from a consanguineous family. 27350570_Our results showed that DOCK8-deficient patients have a profound defect in TH17 differentiation related to decreased STAT3 phosphorylation, translocation to the nucleus, and transcriptional activity 27554822_The CD4+ T-cell compartment is greatly altered in the absence of DOCK8. Specifically, DOCK8-deficient patients have increased TH2 cells and defects in TH1 and TH17 cell differentiation 27890707_Sequence analysis identified two copies of missense mutation, c.4346C > T, in the coding region of the DOCK8 gene in a family with 3 patients with autosomal recessive Hyper-IgE syndrome. 28065530_In severe atopic eczema the dermatologist should initially suspect and document a mutation of DOCK8. 28067314_EPAS1 links DOCK8 deficiency to atopic skin inflammation via IL-31 induction in CD4thorn T cells. 28366940_Recent advances in the understanding of DOCK8 function are summarized, paying particular attention to an emerging role as a signaling intermediate to promote immune responses to diverse external stimuli. [Review] 29058101_DOCK8 deficiency may present severe immune dysregulation with features that may overlap with those of immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) and other IPEX-like disorders 29419892_In our patients, a novel homozygous c.646-647delCT was found in the DOCK8 coding region, leading to a frame shift and resulting in a premature stop codon at position 226. 29472447_a novel mechanism for Dock8 regulation of BCR signaling by regulating cd19 transcription 29930340_CNVs disrupting the DOCK8 gene have clinical relevance, regardless of whether they are deletions or duplications. 30120291_our finding of significantly decreased DOCK8 expression and altered DOCK8 interaction network in ERU might explain changes in immune response and shows the contribution of DOCK8 in pathomechanisms of spontaneous autoimmune diseases. 30252138_This comprehensive cytokine profile in HIES patients reveals distinctive biomarkers that differentiate between the DOCK8-deficient and AD patients. 30397357_Expansion of the skin virome in DOCK8-deficient patients underscores the importance of immune surveillance in controlling eukaryotic viral colonization and infection. 30689480_We conclude that DOCK8 protein is expressed in resting human neutrophils and DOCK8 expression is increased after stimulation with either phorbol 12-myristate 13-acetate or N-Formylmethionyl-leucyl-phenylalanine. Most patients with a disease-causing mutation in DOCK8 have normal neutrophil functions, while a minority showed a mild to moderate chemotactic defect. 31242861_Dedicator of cytokinesis 8 (DOCK8) deficiency (MIM #243700) is a rare disease, leads to a combined primary immunodeficiency (PID), and accounts for the autosomal recessive-hyper immunoglobulin E syndrome (AR-HIES). DOCK8 deficiency status characterizes by recurrent infections. Whole-exome sequencing supported the diagnosis of PID by identifying a homozygous DOCK8 mutation. 31596517_ILC3 deficiency and generalized ILC abnormalities in DOCK8-deficient patients. 31630188_The OCK8 is a Cdc42-specific GEF that regulates interstitial migration of dendritic cells. 32372632_Exome-first Approach Identified Novel Homozygous Dedicator of Cytokinesis 8 (DOCK8) Mutations in Three Unrelated Iranian Pedigrees Suspected with Hyper-IgE Syndrome. 33290277_Somatic reversion of pathogenic DOCK8 variants alters lymphocyte differentiation and function to effectively cure DOCK8 deficiency. 33545980_Prenatal detection of terminal 9p24.3 microduplication encompassing DOCK8 gene: A variant of likely benign. 33634762_Novel Variants of DOCK8 Deficiency in a Case Series of Iranian Patients. 33948880_DOCK8-related Immunodeficiency Syndrome (DIDS): Report of Novel Mutations in Iranian Patients. 35278713_Lack of DOCK8 impairs the primary biologic functions of human NK cells and abrogates CCR7 surface expression in a WASP-independent manner. | ENSMUSG00000052085 | Dock8 | 6.729552e+01 | 0.9436244 | -0.083715415 | 0.4249331 | 3.805054e-02 | 0.8453416909 | 0.96957465 | No | Yes | 7.912014e+01 | 22.073179 | 6.978611e+01 | 20.277528 | |
| ENSG00000107282 | 320 | APBA1 | protein_coding | Q02410 | FUNCTION: Putative function in synaptic vesicle exocytosis by binding to Munc18-1, an essential component of the synaptic vesicle exocytotic machinery. May modulate processing of the amyloid-beta precursor protein (APP) and hence formation of APP-beta. Component of the LIN-10-LIN-2-LIN-7 complex, which associates with the motor protein KIF17 to transport vesicles containing N-methyl-D-aspartate (NMDA) receptor subunit NR2B along microtubules (By similarity). {ECO:0000250|UniProtKB:B2RUJ5}. | 3D-structure;Alternative splicing;Cytoplasm;Golgi apparatus;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | The protein encoded by this gene is a member of the X11 protein family. It is a neuronal adapter protein that interacts with the Alzheimer's disease amyloid precursor protein (APP). It stabilizes APP and inhibits production of proteolytic APP fragments including the A beta peptide that is deposited in the brains of Alzheimer's disease patients. This gene product is believed to be involved in signal transduction processes. It is also regarded as a putative vesicular trafficking protein in the brain that can form a complex with the potential to couple synaptic vesicle exocytosis to neuronal cell adhesion. [provided by RefSeq, Jul 2008]. | hsa:320; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendritic spine [GO:0043197]; glutamatergic synapse [GO:0098978]; Golgi apparatus [GO:0005794]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; presynaptic active zone membrane [GO:0048787]; Schaffer collateral - CA1 synapse [GO:0098685]; synaptic vesicle [GO:0008021]; amyloid-beta binding [GO:0001540]; axo-dendritic transport [GO:0008088]; cell adhesion [GO:0007155]; chemical synaptic transmission [GO:0007268]; gamma-aminobutyric acid secretion [GO:0014051]; glutamate secretion [GO:0014047]; in utero embryonic development [GO:0001701]; intracellular protein transport [GO:0006886]; locomotory behavior [GO:0007626]; multicellular organism growth [GO:0035264]; nervous system development [GO:0007399]; protein-containing complex assembly [GO:0065003]; regulation of gene expression [GO:0010468]; regulation of synaptic vesicle exocytosis [GO:2000300] | 12016213_Munc18a acts through direct and indirect interactions with X11 proteins and powerfully regulates APP metabolism and Abeta secretion. 12849748_Coexpression of X11alpha with amyloid protein precursor retarded its maturation, prolonged its half-life, and inhibited amyloid protein precursor, Abeta40, and Abeta42 secretion. 12970358_human X11alpha inhibits Abeta production and deposition in vivo in the brain in transgenic mice harboring a familial Alzheimer's disease mutant APP that produces increased levels of Abeta. 14970211_Involvement of X11L in the phosphorylation of APP family proteins in cellular stress and suggest that X11L protein may be . 15699037_X11alpha and X11beta have roles in beta-amyloid precursor protein processing 16413130_Mediates exocytosis and decreases beta-amyloid peptide formation in Alzheimer disease. 18836734_Data show that neuronal munc18-1-interacting protein 1 is an inclusion body component in neuronal intranuclear inclusion disease identified by anti-SUMO-1-immunocapture. 19720620_ApoEr2 regulates cell movement, and both X11alpha and Reelin enhance this effect. 20016085_A novel consensus sequence for interaction with the PDZ-1 and PDZ-2 domains of amyloid precursor protein (APP)-interacting proteins Mint1, Mint2, and Mint3 is reported, with multiple novel interactors for these proteins. 20160714_Methylation of MINT1 was significantly more prevalent in UC-CRC cases compared with controls. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20468060_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20531236_Our findings show a new function for X11alpha that may impact on Alzheimer's disease pathogenesis. 21178287_Transcriptional co-activators Taz and Yap mediate signaling via the amyloid beta protein precursor paralogues APLP1 and APLP2 through interactions with Mint1 and Mint3. 21763699_Study identified the conserved binding site for the peptide on the CASK calmodulin kinase domain. A related EPIWVMRQ peptide from Mint1 was also discovered to be sufficient for binding. 22355143_an autoinhibitory mechanism in Mint1 is important for regulating APP processing and may provide novel therapies for Alzheimer's disease 23737971_Mint1 826 bridges APP to the small GTPase 24742670_Mints are necessary for activity-induced APP and PS1 trafficking and provide insight into the cellular fate of APP in endocytic pathways essential for Abeta production. 26865271_Mint1 causes amyloid precursor protein accumulation in the trans-Golgi network. 32793962_Status of Gene Methylation and Polymorphism in Different Courses of Ulcerative Colitis and Their Comparison with Sporadic Colorectal Cancer. 34403115_RNA sequencing and functional studies of patient-derived cells reveal that neurexin-1 and regulators of this pathway are associated with poor outcomes in Ewing sarcoma. | ENSMUSG00000024897 | Apba1 | 9.998387e+01 | 2.0596432 | 1.042394421 | 0.3924961 | 7.186487e+00 | 0.0073454638 | 0.31685983 | No | Yes | 1.089307e+02 | 29.865333 | 5.212889e+01 | 14.842482 | |
| ENSG00000107371 | 51010 | EXOSC3 | protein_coding | Q9NQT5 | FUNCTION: Non-catalytic component of the RNA exosome complex which has 3'->5' exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. In the nucleus, the RNA exosome complex is involved in proper maturation of stable RNA species such as rRNA, snRNA and snoRNA, in the elimination of RNA processing by-products and non-coding 'pervasive' transcripts, such as antisense RNA species and promoter-upstream transcripts (PROMPTs), and of mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm. The RNA exosome may be involved in Ig class switch recombination (CSR) and/or Ig variable region somatic hypermutation (SHM) by targeting AICDA deamination activity to transcribed dsDNA substrates. In the cytoplasm, the RNA exosome complex is involved in general mRNA turnover and specifically degrades inherently unstable mRNAs containing AU-rich elements (AREs) within their 3' untranslated regions, and in RNA surveillance pathways, preventing translation of aberrant mRNAs. It seems to be involved in degradation of histone mRNA. The catalytic inactive RNA exosome core complex of 9 subunits (Exo-9) is proposed to play a pivotal role in the binding and presentation of RNA for ribonucleolysis, and to serve as a scaffold for the association with catalytic subunits and accessory proteins or complexes. EXOSC3 as peripheral part of the Exo-9 complex stabilizes the hexameric ring of RNase PH-domain subunits through contacts with EXOSC9 and EXOSC5. {ECO:0000269|PubMed:11782436, ECO:0000269|PubMed:17545563, ECO:0000269|PubMed:19056938, ECO:0000269|PubMed:21255825}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Disease variant;Exosome;Isopeptide bond;Neurodegeneration;Nucleus;RNA-binding;Reference proteome;Ubl conjugation;rRNA processing | This gene encodes a non-catalytic component of the human exosome, a complex with 3'-5' exoribonuclease activity that plays a role in numerous RNA processing and degradation activities. Related pseudogenes of this gene are found on chromosome 19 and 21. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jun 2012]. | hsa:51010; | cytoplasm [GO:0005737]; cytoplasmic exosome (RNase complex) [GO:0000177]; cytosol [GO:0005829]; exosome (RNase complex) [GO:0000178]; nuclear exosome (RNase complex) [GO:0000176]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcriptionally active chromatin [GO:0035327]; 3'-5'-exoribonuclease activity [GO:0000175]; RNA binding [GO:0003723]; CUT catabolic process [GO:0071034]; DNA deamination [GO:0045006]; exonucleolytic catabolism of deadenylated mRNA [GO:0043928]; exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [GO:0000467]; isotype switching [GO:0045190]; nuclear polyadenylation-dependent rRNA catabolic process [GO:0071035]; nuclear polyadenylation-dependent tRNA catabolic process [GO:0071038]; nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription [GO:0071049]; nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5' [GO:0034427]; polyadenylation-dependent snoRNA 3'-end processing [GO:0071051]; positive regulation of isotype switching [GO:0045830]; RNA catabolic process [GO:0006401]; RNA processing [GO:0006396]; rRNA processing [GO:0006364]; U4 snRNA 3'-end processing [GO:0034475] | 12419256_Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. 23564332_We identified a homozygous mutation [c.395A > C/p.D132A] in EXOSC3 in four patients with muscle hypotonia, developmental delay, spinal anterior horn involvement, and prolonged survival, consistent with the 'mild PCH1 phenotype'. 23883322_The same mutation c.92G-->C, p.G31A in EXOSC3 was found in three unrelated Czech Roma patients with Pontocerebellar hypoplasia type 1 23975261_The present study indicates that EXOSC3 mutations can underlie clinical phenotype not classifiable as pontocerebellar hypoplasia type 1. 24524299_study identified new nonsense and missense mutations in the EXOSC3 gene and showed mutations in this gene are exclusively found in pontocerebellar hypoplasia type 1 patients; there are evident genotype-phenotype correlations in EXOSC3-mediated PCH reflected in clinical outcome, age of death and pons hypoplasia 25149867_EXOSC3 mutations were linked to complicated hereditary spastic paraplegia. 27193168_Knock-down of rbm7, exosc8 and exosc3 in zebrafish showed a common pattern of defects in motor neurons and cerebellum. Our data indicate that impaired RNA metabolism may underlie the clinical phenotype by fine tuning gene expression which is essential for correct neuronal differentiation 28053271_Mutations of EXOSC3/Rrp40p associated with pontocerebellar hypoplasia with progressive cerebral atrophy impact ribosomal RNA processing functions of the exosome in S. cerevisiae. 28687512_This is the first case of mitochondrial dysfunction associated with an EXOSC3 mutation, which expands the phenotypic spectrum of pontocerebellar hypoplasia type 1b. 32645003_A Drosophila model of Pontocerebellar Hypoplasia reveals a critical role for the RNA exosome in neurons. 33462000_Two siblings with a novel variant of EXOSC3 extended phenotypic spectrum of pontocerebellar hypoplasia 1B to an exceptionally mild form. 34626022_Aberrant expression of MYD88 via RNA-controlling CNOT4 and EXOSC3 in colonic mucosa impacts generation of colonic cancer. | ENSMUSG00000028322 | Exosc3 | 1.170421e+03 | 0.8163827 | -0.292682445 | 0.2725497 | 1.169430e+00 | 0.2795183958 | 0.80666974 | No | Yes | 9.899290e+02 | 106.891798 | 1.209568e+03 | 133.667314 | |
| ENSG00000107485 | 2625 | GATA3 | protein_coding | P23771 | FUNCTION: Transcriptional activator which binds to the enhancer of the T-cell receptor alpha and delta genes. Binds to the consensus sequence 5'-AGATAG-3'. Required for the T-helper 2 (Th2) differentiation process following immune and inflammatory responses. Positively regulates ASB2 expression (By similarity). {ECO:0000250|UniProtKB:P23772, ECO:0000269|PubMed:23824597}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Deafness;Disease variant;Immunity;Innate immunity;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a protein which belongs to the GATA family of transcription factors. The protein contains two GATA-type zinc fingers and is an important regulator of T-cell development and plays an important role in endothelial cell biology. Defects in this gene are the cause of hypoparathyroidism with sensorineural deafness and renal dysplasia. [provided by RefSeq, Nov 2009]. | hsa:2625; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; E-box binding [GO:0070888]; HMG box domain binding [GO:0071837]; identical protein binding [GO:0042802]; interleukin-2 receptor binding [GO:0005134]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; transcription coactivator binding [GO:0001223]; zinc ion binding [GO:0008270]; anatomical structure formation involved in morphogenesis [GO:0048646]; anatomical structure morphogenesis [GO:0009653]; aortic valve morphogenesis [GO:0003180]; axon guidance [GO:0007411]; canonical Wnt signaling pathway involved in metanephric kidney development [GO:0061290]; cardiac right ventricle morphogenesis [GO:0003215]; cell fate commitment [GO:0045165]; cell fate determination [GO:0001709]; cell maturation [GO:0048469]; cellular response to BMP stimulus [GO:0071773]; cellular response to interferon-alpha [GO:0035457]; cellular response to interleukin-4 [GO:0071353]; cellular response to tumor necrosis factor [GO:0071356]; chromatin remodeling [GO:0006338]; cochlea development [GO:0090102]; defense response [GO:0006952]; developmental growth [GO:0048589]; ear development [GO:0043583]; embryonic hemopoiesis [GO:0035162]; embryonic organ development [GO:0048568]; erythrocyte differentiation [GO:0030218]; humoral immune response [GO:0006959]; immune system development [GO:0002520]; in utero embryonic development [GO:0001701]; innate immune response [GO:0045087]; inner ear morphogenesis [GO:0042472]; kidney development [GO:0001822]; lens development in camera-type eye [GO:0002088]; lymphocyte migration [GO:0072676]; male gonad development [GO:0008584]; mast cell differentiation [GO:0060374]; mesenchymal to epithelial transition [GO:0060231]; mesonephros development [GO:0001823]; negative regulation of cell cycle [GO:0045786]; negative regulation of cell motility [GO:2000146]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of cell proliferation involved in mesonephros development [GO:2000607]; negative regulation of DNA demethylation [GO:1901536]; negative regulation of endothelial cell apoptotic process [GO:2000352]; negative regulation of epithelial to mesenchymal transition [GO:0010719]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation [GO:2000703]; negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation [GO:2000734]; negative regulation of inflammatory response [GO:0050728]; negative regulation of interferon-gamma production [GO:0032689]; negative regulation of interleukin-2 production [GO:0032703]; negative regulation of mammary gland epithelial cell proliferation [GO:0033600]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; nephric duct formation [GO:0072179]; nephric duct morphogenesis [GO:0072178]; neuron migration [GO:0001764]; norepinephrine biosynthetic process [GO:0042421]; otic vesicle development [GO:0071599]; parathyroid gland development [GO:0060017]; parathyroid hormone secretion [GO:0035898]; pharyngeal system development [GO:0060037]; phosphatidylinositol 3-kinase signaling [GO:0014065]; positive regulation of endothelial cell migration [GO:0010595]; positive regulation of histone H3-K14 acetylation [GO:0071442]; positive regulation of histone H3-K9 acetylation [GO:2000617]; positive regulation of interleukin-13 production [GO:0032736]; positive regulation of interleukin-4 production [GO:0032753]; positive regulation of interleukin-5 production [GO:0032754]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of signal transduction [GO:0009967]; positive regulation of T cell differentiation [GO:0045582]; positive regulation of T-helper 2 cell cytokine production [GO:2000553]; positive regulation of thyroid hormone generation [GO:2000611]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription regulatory region DNA binding [GO:2000679]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of ureteric bud formation [GO:0072107]; post-embryonic development [GO:0009791]; pro-T cell differentiation [GO:0002572]; regulation of CD4-positive, alpha-beta T cell differentiation [GO:0043370]; regulation of cellular response to X-ray [GO:2000683]; regulation of cytokine production [GO:0001817]; regulation of epithelial cell differentiation [GO:0030856]; regulation of establishment of cell polarity [GO:2000114]; regulation of histone H3-K27 methylation [GO:0061085]; regulation of histone H3-K4 methylation [GO:0051569]; regulation of nephron tubule epithelial cell differentiation [GO:0072182]; regulation of neuron apoptotic process [GO:0043523]; regulation of neuron projection development [GO:0010975]; response to estrogen [GO:0043627]; response to ethanol [GO:0045471]; response to gamma radiation [GO:0010332]; response to virus [GO:0009615]; response to xenobiotic stimulus [GO:0009410]; signal transduction [GO:0007165]; sympathetic nervous system development [GO:0048485]; T cell differentiation [GO:0030217]; T cell receptor signaling pathway [GO:0050852]; T-helper 2 cell differentiation [GO:0045064]; thymic T cell selection [GO:0045061]; thymus development [GO:0048538]; TOR signaling [GO:0031929]; type IV hypersensitivity [GO:0001806]; ureter maturation [GO:0035799]; ureter morphogenesis [GO:0072197]; ureteric bud formation [GO:0060676]; uterus development [GO:0060065]; ventricular septum development [GO:0003281] | 11937547_Strong GATA-3 binding to an oligonucleotide containing two GATA-3 motifs has been mapped at a DNase hypersensitivity site in cluster IV of the CD8 gene complex. 11970965_Identification of an alternative GATA-3 promoter directing tissue-specific gene expression 12057898_A marked reduction in both GATA-3 mRNA and protein expression in HPV-immortalized cell lines was confirmed in cervical carcinoma cell lines. 12087127_GATA-3 transcriptional imprinting in Th2 lymphocytes: a mathematical model. GATA-3 autoactivation creates a bistable system that can memorize a transient inductive signal. 12960249_VZV virus and mite antigens induce expression of this transcription factor in cord blood mononuclear cells, differenetially affecting Th1 and Th2 cells 14757746_GATA-3 is an important transcription factor in regulating human Th2 cell differentiation in vivo. 14985365_GATA3 mutations may have a role in the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome 15087456_GATA3 plays important roles in the maintenance of the Th2 phenotype and continuous chromatin remodeling of the specific Th2 cytokine gene locus through cell division. 15251440_GATA-3 suppresses IFN-gamma promoter activity independently of binding to cis-regulatory elements 15328158_Erythropoietin (Epo) gene expression is under the control of hypoxia-inducible factor 1 (HIF-1), and is negatively regulated by GATA3. 15361840_GATA3 is involved in growth control and the maintenance of the differentiated state in epithelial cells and may contribute to tumorigenesis in breast cancer. 15563083_GATA-3 contributed to the production of IL-4, IL-5 in patients with allergic rhinitis. 15632006_Expressed in Hodgkin disesase Reed Sternberg cells. 15637551_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15699146_GATA-3 binds and transactivates the NKG2A promoter; coexpression of human GATA-3 with an NKG2A promoter construct in K562 cells leads to enhanced promoter activity. 15826950_interleukin-5 transcription repression by the glucocorticoid receptor targets GATA3 signaling and involves histone deacetylase recruitment 15947486_GATA3 is crucially involved in IL-5 gene transcription in human peripheral CD4-positive t cells. 16087702_strong specific upregulation of Gata-3 impaired nuclear translocation and its cooperative action with the TGF-beta pathway, suggesting that Gata-3 plays a central role in human pancreatic cancer 16317090_DL1-induced activation of the Notch1 pathway controls the lineage commitment of early thymic precursors by altering the levels between Spi-B and GATA-3. 16357129_Low expression of GATA3 is associated with invasive breast carcinomas 16498264_Segmental allergen challenge in asthmatics leads to increased GATA-3, c-maf and T-bet expression in BAL cells but not in bronchial biopsies 16509533_Direct sequencing after PCR amplification of genomic DNA revealed a novel insertional mutation (405insC) in the GATA3 gene of two patients with HDR syndrome (Hypoparathyroidism, sensorineural Deafness and Renal anomaly syndrome). 16912130_Three novel mutations in the GATA3 gene responsible for familial hypoparathyroidism and deafness. 17074191_Imbalance of transcription factors T-bet and GATA-3 may be one of the key factors in immune dysregulation of recurrent aphthous ulcerations. 17075044_the GATA-3/T-bet transcription factor complex regulates the cell-lineage-specific expression of the lymphocyte homing receptors 17078870_GATA3 is probably a mediator for the transcriptional upregulation of MUC1 expression in some breast cancers. 17111354_Overexpression of GATA3 rescues T-helper type 2 (Th2) cytokine expression in protein kinase C theta-deficient transgenic Th2 cells. 17114920_A novel heterozygous deletion frameshift mutation of GATA3 in a Japanese kindred with the hypoparathyroidism, deafness and renal dysplasia syndrome. 17117487_There is an association between expression of Th1/Th2 transcription factors and cytokines (T-bet, GATA-3, IFN-gamma, IL-4,IL-18) in systemic lupus erythematosus. 17210674_No mutations were identified in patients with isolated hypoparathyroidism, thereby indicating that GATA3 abnormalities are more likely to result in two or more of the phenotypic features of the HDR syndrome. 17234745_The human GATA-3 gene is not regulated in response to polarizing signals that are sufficient to direct Th2-specific expression in mouse cells. 17272506_E2A proteins prevent lymphoma cell expansion, at least in part through regulation of Gfi1b and modulation of Gata3 expression. 17277157_GATA-3 nuclear translocation is dependent on its phosphorylation on serine residues by p38 MAPK 17357106_GATA3 establishes a positive feedback loop that increases transgenic T-cell receptor surface expression in developing CD4 lineage cells. 17381824_Gata-3 has a role in specifying and maintaining mammary cell fate 17390031_WNT8B expression in hepatocyte progenitors derived from human ES cells is due to POU5F1 (OCT3/OCT4) and GATA3, and WNT8B expression in diffuse-type gastric cancer is due to POU5F1 and GATA6 17445472_In Dermatophagoides farinae-stimulated PBMCs from patients with asthma, expression of GATA-3 and T-bet and FOPX3 expression is decreased. 17445473_In patients with acute asthma, rosiglitazone could regulate the balance of IFN-gamma and IL-4 by affecting expression of T-bet mRNA and GATA-3. 17616709_GATA-3 is a critical component of the master cell-type-specific transcriptional network including ER alpha and FoxA1 that dictates the phenotype of hormone-dependent breast cancer 17628972_Activation of GATA-3 might be one of the mechanisms for induction of IL-5 expression in chronic rhinosinusitis. 17654061_GATA-3 expression in bone marrow stromal cells from chronic aplastic anemia was significantly higher than controls. Expression levels of GATA genes may influence hematopoiesis in BM microenvironment & relate to the pathogenesis/development of AA. 17658279_Gata3 is a critical element determining inductive T helper (Th) type 2 cell differentiation and limiting Th1 differentiation by Notch. 17703412_Observational study of gene-disease association. (HuGE Navigator) 17845581_Transactivation by GATA-3 increased luciferase expression 100-fold, in the absence of stimulation. Synergistic interactions were demonstrated between Ets1, GATA-3 and AP-1. 18006915_Observational study of gene-disease association. (HuGE Navigator) 18037162_Gene-gene interaction between GATA3 and IL13 polymorphisms can influence the risk of childhood rhinitis. 18037162_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18079734_Direct interactions between NKX2-5 and GATA3 as indicated by co-immunoprecipitation data may contribute to MEF2C regulation. 18154704_Role in the proliferaative and regenerative capacity of mammary stem and progenitor cells. 18212358_The expression patterns of T-Bet and GATA-3 oppose progesterone receptor, suggesting antagonistic function and/or regulation between PR and T-Bet/GATA-3 18260379_The expression of GATA-3 was negatively correlated to the expression of IL-12 in patients with allergic rhinitis. 18268121_GATA-3 is a breast cancer marker almost exclusively expresses among ER-positive tumors. 18410415_A SNP in GATA3 is associated with atopic eczema. This finding highlights the importance of GATA3 as an immune-modulating gene in atopic eczema. 18410415_Observational study of gene-disease association. (HuGE Navigator) 18533032_GATA3 is hypothesized to be integral to the ERalpha pathway 18607915_Observational study of gene-disease association. (HuGE Navigator) 18607915_We aimed to evaluate the association between the SNPs of GATA-3 and AR. 18619618_GATA-3 mRNA expression level was significantly higher preoperatively, and remained higher by postoperative d 7 in patients with postoperative infection after aggressive hepatobiliary pancreatic surgery. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18701459_the T cell receptor-mediated induction of Gfi1 controls Th2 cell differentiation through the regulation of GATA3 protein stability 18772129_cyclic AMP signals enhance histone H3 acetylation at the IL-5 promoter and the concerted binding of GATA-3 and NFATc to the promoter. 18826115_Observational study of gene-disease association. (HuGE Navigator) 18826115_The SNP at rs1269486 of GATA3 is associated with allergic rhinitis in Chinese. 18849568_GlcNAc6ST-1 transcription is coordinated with the NF-kappaB/GATA-3 axis, which is known to figure heavily in Th2 cell differentiation 19043799_Data demonstrate that zinc influences the proliferation and differentiation, and increases GATA-3 expression, of CD34(+) progenitors both in young and old ages. 19057839_A novel mutation in the GATA3 gene in a patient with PTH-deficient hypoparathyroidism. 19059610_reviews the literature from the past 10 y on GATA-3 in normal and pathological states of the mammary gland [review] 19082709_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19082709_there is no evidence that either GATA3 rs570613, or any variant in strong linkage disequilibrium with it, is associated with breast cancer risk in women. 19084267_no statistically significant differences in recurrence or survival rates between GATA3-positive and GATA3-negative breast carcinomas 19092634_expression of S100P, GATA3, and p63 by a majority of ovarian Brenner tumors underscores the similarity between these neoplasms and urothelial proliferations of bladder origin. 19094228_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19189213_It is possible that GATA3 mutations occur earlier in the evolution of breast tumors, compared to BRCA1, BRCA2 or sporadic tumors, and are therefore easier to detect by direct sequencing in the presence of some stromal contamination. 19189213_Observational study of gene-disease association. (HuGE Navigator) 19232384_GATA3 was found to bind the site located at -954/-855 and to be a key regulator of abundant KLK1 expression in keratinocyte. 19247692_Observational study of gene-disease association. (HuGE Navigator) 19248112_GATA-3 expression protects against severe joint inflammation and bone erosion in mice, accompanied by reduced differentiation of Th17 cells, but not Th1 cells, during mBSA-induced arthritis. 19253381_heterozygous nonsense mutation p.R367X of the GATA3 gene in a Chinese family with the HDR syndrome 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19261198_REVIEW: The transcription factor network composed of the estrogen receptor alpha, FOXA1 and GATA3 may control the gene expression pattern in luminal subtype A breast cancers 19336263_decreased levels in kidney and PBMC of patients with mesangial proliferative nephritis 19342088_Observational study of gene-disease association. (HuGE Navigator) 19342088_it might be speculated that genetic variations in GATA3 have no major influence on the origin of asthma. 19346497_acquisition of IL-4Ralpha expression after stimulation was inhibited by IFN-lambda1, as was GATA3 expression 19411068_p18(INK4C) is a downstream target of GATA3, constrains luminal progenitor cell expansion, and suppresses luminal tumorigenesis in the mammary gland 19452711_The overexpression of GATA-3 in patients with allergic rhinitis was related to the increase of ICAM-1. 19473628_GATA-3 is upregulated in peripheral blood mononuclear cells from patients with minimal change nephrotic syndrome. 19483726_GATA3 inhibits primary breast tumor outgrowth and reduces lung metastatic burden by regulating key genes involved in metastatic breast tumor progression. 19549328_FOXA1 and GATA-3 have roles in breast cancer, but FOXA1 is expressed in patients with good outcome, while GATA-3 is a luminal marker 19559773_GATA3 binds to regulatory elements and controls target gene expression through physical interaction with core promoter regions. 19615257_T-bet and GATA-3 play a part role in the regulation of Th1/Th2 balance in esophageal cancer. 19625176_Observational study of gene-disease association. (HuGE Navigator) 19639723_upregulated expression in nasal polyps tissue in Chinese patients 19674970_GATA3 is indispensable for Ang-1-Tie2-mediated signaling in large vessel endothelial cells 19692168_Observational study of gene-disease association. (HuGE Navigator) 19719829_From the described expression pattern, it is highly probable that GATA-3 plays a role in follicular and epidermal morphogenesis. 19723756_A novel missense HDR-associated GATA3 mutation, Thr272Ile, has been identified and shown to result in reduced DNA binding, a partial loss of FOG2 interaction, and a decrease in gene transcription. 19728080_HuR binds to the GATA3 3'UTR. Inhibition of HuR decreased GATA3 mRNA, mRNA stability & protein expression. 19735555_GATA3 binds to the promoter regions of genes responding to low ionizing radiation doses, and that silencing this protein in irradiated human keratinocytes leads to generalised transcriptional deregulation after X-irradiation. 19798694_This review highlights recent understanding of GATA3 in both normal mammary development and tumor differentiation. 19805038_GATA-3 occupies genes in both Th1 and Th2 cells and, unexpectedly, shares a large proportion of targets with T-bet. 19861286_Compared with the normal controls, T-bet mRNA expression decreased whereas GATA-3 mRNA expression increased significantly in patients with systemic lupus erythematosus. 19946260_This study evaluates the association of Forkhead-box protein A1 (FOXA1) and GATA-binding protein 3 (GATA3) expressions with Oncotype DX recurrences scores in 77 cases of patients with ER-positive node-negative breast carcinomas 20130088_I3C is a novel anticancer agent in human cancers that coexpress ERalpha, GATA3, and AhR, a combination found in a large percentage of breast cancers but not in other critical ERalpha target tissues essential to patient health 20154722_GATA3 as a gene involved in transcriptional regulation of Cyclin D1. 20189993_GATA3 drives invasive breast cancer cells to undergo the reversal of epithelial-mesenchymal transition. 20237292_Overexpression of GATA-3 restores interleukin (IL)-4 expression in Src family kinase Lck-deficient T helper (Th) type 2 cells; this indicates that decreased interleukin (IL)-4 expression is due in part to reduced amounts of GATA-3. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20333526_lower expression in PBMC of psoriatic patients 20368097_GATA3-NFAT1 complex may play an important role in the regulation of IL-13 transcription in human T cells. 20503287_Observational study of gene-disease association. (HuGE Navigator) 20660789_Down-regulation of GATA3 in Hodgkin lymphoma cell lines demonstrated its role in the regulation of IL-5, IL-13, STAT4, and other genes. 20696860_Translational increase via T cell receptor (TCR) signaling is specific for GATA-3 and responsible for the TCR-dependent increase in GATA-3 associated with enhanced T helper type (Th)2 cell differentiation in T cells from TCR transgenic mice. 20716621_Observational study of gene-disease association. (HuGE Navigator) 21037568_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21037568_Three new susceptibility loci at 2p16.1 (rs1432295, REL, 8q24.21 (rs2019960, PVT1 and 10p14 (rs501764, GATA3). 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21114556_The T-bet deficiency and the correlation of GATA-3 expression with the increase of IgE are characteristics of long-lasting asthma. 21120445_The Cys321Ser-mutated GATA3 lacked transactivation activity. Moreover, mutated GATA3 exerted a dominant-negative effect over the transactivation activity of wild-type GATA3. 21157112_report clinical phenotype and molecular analysis of GATA3 in 5 Japanese HDR patients, including 2 familial cases; sequence analysis of GATA3 demonstrated three novel (R262G, c1063delC and C318) and two reported mutations (c.432insG and c.1051-1G>T) 21242646_GATA3 abnormalities in patients with HDR syndrome; results consistent with previous finding that GATA3 mutations are usually identified in patients with 2 or 3 of the HDR triad features 21289214_predominance of intraglomerular T-bet expression relative to GATA3 expression associated with poor response to treatment with bolus steroid 21329183_GATA3 may play a crucial role in oestrogen receptor positive breast cancer patients 21334719_Plasmacytoid carcinoma of the bladder is a tumor entity, characterized by specific immunohistochemical markers including positivity for GATA-3. 21399899_Case Report: Report hypomagnesemia in association with HDR syndrome in a patient with GATA3 mutation. 21468546_The ratio of T-bet/GATA-3 can be used as an objective indicator of immune imbalance in patients with allergic asthma. 21611195_The prototypic Th2 cytokine IL-4 was the only cytokine capable of inducing GATA3 in skin explants from healthy donors. 21638273_CD8 T cells express significantly higher levels of GATA-3 and that this increase is associated with augmented production of IL-13 and is more pronounced in patients with diffuse cutaneous systemic sclerosis. 21682737_Allergic inflammatory responses are orchestrated by several transcription factors, particularly NF-kappaB and GATA3. (Review) 21743959_CDKN2A, GATA3, CREBBP, ITGA2, NBL1 and TGM4 were down-regulated in the prostate carcinoma glands compared to the corresponding normal glands 21761347_These results demonstrate a direct positive role of estrogen in regulating Aurora-A expression through activation of the ERalpha-GATA-3 signaling cascade 21878914_All the enhanceosome components comprising estrogen receptor alpha (ERalpha), FOXA1, and GATA3 are necessary for the full repertoire of cancer-associated effects of the ERalpha. 21892208_Expression of GATA3 reprograms BTNBCs to a less aggressive phenotype and inhibits a major mechanism of metastasis through inhibition of LOX. 21930782_A strong association was evident between GATA-3 and caspase-14 expression in preinvasive ductal carcinoma in situ samples, where GATA-3 also displayed prognostic significance 22019771_Compared with transplant recipients not exhibiting operational tolerance operationally tolerant individuals had a predominant regulatory gene expression profile with higher expression of GATA3, TGFB1, TGFBR1, and FOXP3 which was stable over time. 22039304_c-Myb critically regulates IL-13 expression via direct binding to the conserved GATA-3 response element 22120723_the BRCA1-GATA3 interaction is important for the repression of genes associated with triple-negative and basal-like breast cancer (BLBCs) including FOXC1 22336257_Leukocyte T-box expressed in T cells (TBET)/GATA3 mRNA ratio is elevated in patients with acute coronary syndrome, and highest in patients with acute myocardial infarction. 22391183_Th1/Th2 imbalance exists in the acute phase of childhood Hench-Schonlein purpura, which correlates with the abnormal expression of transcription factor T-bet and GATA-3. 22607700_GATA binding protein 3 is down-regulated in bladder cancer yet strong expression is an independent predictor of poor prognosis in invasive tumor. 22706858_GATA3 is a sensitive and specific marker for the diagnosis of breast and urothelial carcinomas. 22909160_Dysregulation of Th1 transcription may contribute to heightened expression of STAT6 and GATA3 leading to exaggerated Th2-driven manifestations of allergic disease. 22951069_GATA3 tethers TCF7L2 to the genome at a subset of sites in MCF7 cells. 22951729_this study demostrtrates a new T-cell regulatory pathway that directly links decreased proteasome degradation of GATA3, CTLA-4 upregulation, and inhibition of T-cell responses. 22982890_GATA3 IHC is a sensitive marker for uro-thelial carcinoma and positive staining is typically nonfocal and moderate or strong in intensity. 22985730_GATA3 enhance induction of POU4F3 and myosin VII 23063330_report that the transcription factor GATA3 was highly expressed by human type 2 innate lymphoid cells 23172872_GATA3 is pivotal in mediating enhancer accessibility at regulatory regions involved in ESR1-mediated transcription 23203342_This report adds to the body of data on genotype-phenotype analysis in HDR syndrome. 23266442_There appears to be no link between GATA-3 positivity and survival in men with breast cancer, whereas in women, GATA-3-positive tumors are typically lower grade with a better prognosis. 23291697_Brain calcification caused by HDR syndrome with GATA3 mutation, presenting as a familial brain calcinosis. 23308012_Gene expression of T-bet and GATA-3 by endometrial stromal cells is controlled by hormonal conditions mimicking decidualization, and cyclic AMP. 23354167_GATA3/miR-29b axis regulates the breast tumor tumour microenvironment and inhibits metastasis. 23375642_GATA3 may be a useful marker for metastatic breast carcinoma, especially triple-negative and metaplastic carcinomas its labeling may help distinguish metaplastic carcinoma from malignant phyllodes tumors. 23395819_Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3 23413906_Data indicate that T-bet mRNA and the ratio of T-bet:GATA-3 mRNA in peripheral blood lymphocytes was lower in the chronic cardiac insufficiency (CCI) intervention group than in the CCI control group. 23428429_Majority of human prostate tumors show loss of active GATA3 as they progress to the aggressive castrate-resistant stage. 23430443_Iranian women who carry the GATA3 gene's 17-CT dinucleotide repeat are at lower risk of developing breast cancer. 23435732_Thus, approximately 20 % ER-positive breast cancers have somatic GATA3 mutations that lead to a loss of GATA3 transactivation activity and altered cell invasiveness 23446338_Increase in the expression of GATA-3 is associated with endoscopic inflammation in patients with ulcerative colitis. 23453625_GATA3 lacks sensitivity for distinguishing sarcomatoid renal cell carcinoma from atypical epithelioid epithelioid angiomyolipomas 23549873_Data established a seven-gene (AR, ESR2, GATA3, GBX2, KRT16, MMP28 and WNT11) prognostic signature to define a subset of triple-negative breast cancer (TNBC). 23577196_GATA3 overexpression results in a reduction of TGFss1 response in MDA-MB-231 (MB-231) cell line 23599157_we evaluated GATA3 expression in urinary bladder paragangliomas, which may closely mimic urothelial carcinomas. GATA3 was positive in 83% of urinary bladder paragangliomas. this will prevent misdiagnosis of urothelial tumor based on GATA3 expression 23604756_GATA3 immunohistochemistry is not helpful in resolving the differential diagnosis between a primary salivary gland neoplasm and metastatic breast cancer 23715162_Report utility of gata3 as an immunohistochemical marker for tumors of the autonomic nervous system. 23906664_our present study clarifies that the aberrant expression profile of GATA-3 in human clear cell renal carcinoma is possibly involved with tumorigenesis, and the complicated mechanism is worthy of further investigation. 23939152_Review: owing to its restricted expression in urothelial and breast carcinomas, GATA3 has proved to be a useful immunohistochemical marker for assisting in distinguishing these 2 groups of neoplasms. 23958551_The expression of GATA3 and TTF1 distinguishes metastatic mammary carcinoma from primary lung carcinoma. 23996088_Variation at 10p12.2 (PIP4K2A) and 10p14 (GATA3) influences risk of acute lymphoblastic anemia and tumor subtype. (Meta-analysis) 24061521_GATA-3 is rarely positive in bladder adenocarcinomas displaying extracellular mucin production. 24121175_findings demonstrate high sensitivity and specificity of the GATA3 diagnostic marker, in regional metastases of urothelial carcinoma 24124001_TGFBR3 co-downregulated with GATA3 is associated with methylation of the GATA3 gene in bladder urothelial carcinoma. 24134931_our study suggests that GATA3 and human papillomavirus detection are useful markers for distinguishing urothelial carcinomas from squamous cell carcinomas of the penile urethra. 24141364_Genotype at the GATA3 SNP was also associated with early treatment response and risk of acute lymphoblastic leukemia relapse. 24145643_GATA3 is a useful marker in the characterization of not only mammary and urothelial but also renal and germ cell tumors, mesotheliomas, and paragangliomas. 24235142_GATA3 abrogated the canonical TGFbeta-Smad signaling by abolishing interactions between Smad4 and its DNA binding elements 24235972_intraglomerular predominance of T-bet over GATA3 might be used as diagnosis maker of ABMR in addition to C4d, especially in C4d-negative cases. 24315206_GATA3 is expressed in a minority of oncocytomas. We found an overall lack of GATA3 expression in primary and metastatic renal cell carcinomas. 24327562_Mafb is responsible for executing one branch of the SGN differentiation program orchestrated by the Gata3 transcriptional network 24338248_In the presence of 100 ng/ml IL-11, GATA-3 transcript abundance rose up to ~85-fold of that measured in untreated cells, whereas T-bet transcripts were lowered merely to ~41% 24346062_inhibits proliferation and induces differentiation of primary keratinocytes 24363163_Inhibition of GATA3 activity or blockade of GATA3 expression may attenuate the interleukin-13 mediated asthma phenotypes. 24415069_GATA-3 expression is associated with a less aggressive phenotype and a better prognosis in patients with hormone receptor-positive/HER2-negative breast cancer. 24421220_GATA3 may be a useful addition to immunostaining panels for serous effusion specimens when metastatic breast carcinoma is a consideration. 24434941_Dutch family with a new mutation (c523_528dup) in GATA3 causing HDR syndrome. Their hearing impairment was congenital, bilateral and symmetric. Audiograms showed mild-to-moderate hearing impairment with a flat audiogram configuration. 24448324_GATA3 plays an important role in the prevention of bladder cancer progression and metastasis by inhibiting cell migration and invasion as well as epithelial-to-mesenchymal transition. 24477928_GATA3 mutations mainly occur in patients with luminal-like breast cancer and have identifiable clinicopathologic and genetic characteristics, highlighting a subgroup of patients with breast cancer in whom limited therapy may be appropriate. 24504018_GATA3 expression independently predicts an unfavorable prognosis in primary gastric adenocarcinoma patients. 24512009_GATA3 immunohistochemistry is a valuable addition in the discrimination of cystic or oncocytic parathyroid lesions 24549642_High expression of FOXA1 and GATA3 was significantly associated with improved disease-free survival. 24614117_Significant association with alleles of two STAT4 markers and nominal association of Autoimmune Addison's disease with alleles at GATA3, is reported. 24622013_Patients with GATA3 mutation present with early-onset sensorineural hearing loss 24660543_Abnormal expression of the transcription factors T-bet and GATA-3 contributes to the imbalance of Thl/Th2 lymphocytes associated with immune dysfunction, leading to the development and progression of aplastic anemia. 24711443_High GATA3 expression is associated with metastatic urothelial carcinoma and distinguishes between metastatic urothelial carcinoma and squamous cell carcinomas. 24713735_GATA-3 is, so far, the best breast-specific immunomarker, especially when encountering ER-negative metastatic breast carcinomas 24745616_GATA-3 expression differed among urothelial carcinoma variants. GATA-3 is a useful marker for confirming the urothelial origin of micropapillary and plasmacytoid urothelial carcinoma variants but not that of sarcomatoid or small cell carcinoma variants. 24758297_with a similar distribution across the genome, comparing to T47D cells. CONCLUSIONS: We propose that this specific, cancer-derived mutation in GATA3 deregulates physiologic protein turnover, stabilizes GATA3 binding across the genome 24766459_Study shows the capacity of human CD4(+) and CD8(+) T cells for stable co-expression of GATA-3 and T-bet in humanized mice and reveals a critical role for IL-12 in regulating this phenotype. 24813204_IFN-alpha/beta disrupts the GATA3-autoactivation loop and promotes epigenetic silencing of a Th2-specific regulatory region within the GATA3 gene. 24820417_Data propose that control of GATA3 levels by Fbw7 contributes to the fine-tuning of T-cell development. 24824028_GATA3 is expressed in most cases of sarcomatoid urothelial carcinoma. 24894987_The effect of GATA3 on SPINK5 expression was indirect and GATA3 alone was insufficient for final differentiation of keratinocytes where full SPINK5 expression was observed. 24925221_Our novel finding of GATA3 positivity in one-third of bladder small cell carcinoma is of potential value in differentiating small cell carcinomas of prostate origin from those of bladder origin. 24926087_Strong GATA-3 expression is a reliable marker of primary breast carcinoma in the appropriate clinical context. GATA-3 reactivity in around 70% of triple-negative breast carcinomas is also clinically useful. 25124981_Identified a novel nonsense mutation which will expand the spectrum of HDR-associated GATA3 mutations. 25150746_GATA3 is a diagnostically useful marker for triple-negative breast cancer and is more sensitive than MGB and GCDFP15 combined. 25188865_This study expands the spectrum of neoplasms known to express GATA-3. 25258321_ASH2L enhances ERalpha expression as a coactivator of GATA3 in breast cancers 25292313_Data show that the expression levels of transcription factors GATA-3 and FOXP3 were upregulated with 1.0 mug/ml galectin-1, while transcription factors TBX21 and RORC expression levels were reduced with both 1.0 and 2.0 mug/ml concentrations of galectin-1. 25351211_Highlevel GATA3 expression is associated with increased selfrenewal and proliferation of neuroblastoma cells. 25410484_In breast cancer, some GATA3 effects shift from tumor suppressing to tumor promoting during tumorigenesis, with deregulation of three genes, BCL2, DACH1, THSD4, representing major GATA3-controlled processes in cancer progression. 25425335_The commonly used breast carcinoma biomarkers vary in their prognostic implications. GCDFP-15 independently indicated a favourable prognosis. GATA-3 and MGB were not associated with outcome. 25552913_The mRNA level of Th2-specific transcription factor GATA-3 was increased in patients with liver cancer. 25575062_Expressions of GATA-3 in both Th2 polarized gammadeltaT cells and CD4T cells were up-regulated. 25611245_UPII, GATA3, and p40, when used in combination, are highly sensitive in the differential diagnosis of invasive urothelial carcinoma. 25651379_overexpression of S100A7 in A431 skin squamous carcinoma cells significantly promoted cell proliferation in vitro and tumor growth in vivo, whereas it suppressed the expression of GATA-3 and caspase-14 25753145_Western blotting demonstrated GATA3 in choriocarcinoma, whereas the placenta, and cervical and endometrial cancer cell lines, were negative 25771973_Identification of a novel GATA3 mutation in a deaf Taiwanese family by massively parallel sequencing 25851711_results support that GATA binding protein 3 immunostain can be a useful tool in differentiating mesonephric lesions from endocervical and endometrial adenocarcinomas 25855136_rs73635312[A] is associated with susceptibility to cutaneous basal cell carcinoma. 25873156_The tested 25 SNPs in TBX21, GATA3, Rorc and Foxp3 did not associate with BD and VKH syndrome. 25901741_GATA-3 augmentation down-regulates Connexin43 in Helicobacter pylori associated gastric carcinogenesis 25906123_novel marker GATA3 stains a significantly higher proportion of both primary and metastatic breast carcinomas than GCDFP15 or mammaglobin with stronger and more diffuse staining, helpful in cases with small tissue samples 25917456_Gata3 interacted with Gcm2 and MafB, two known transcriptional regulators of parathyroid development, and synergistically stimulated the PTH promoter. 26028330_GATA3/G9A/NuRD(MTA3) complex inhibits the invasive potential of breast cancer cells in vitro and suppresses breast cancer metastasis in vivo. 26135559_GATA3 is a highly sensitive and specific marker for mesonephric lesions in the lower ge | ENSMUSG00000015619 | Gata3 | 2.341795e+02 | 1.3237025 | 0.404578919 | 0.3216079 | 1.594996e+00 | 0.2066138014 | 0.78763590 | No | Yes | 2.430321e+02 | 30.225684 | 1.743121e+02 | 22.614398 | |
| ENSG00000107779 | 657 | BMPR1A | protein_coding | P36894 | FUNCTION: On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. Receptor for BMP2, BMP4, GDF5 and GDF6. Positively regulates chondrocyte differentiation through GDF5 interaction. Mediates induction of adipogenesis by GDF6. {ECO:0000250|UniProtKB:P36895}. | 3D-structure;ATP-binding;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Kinase;Magnesium;Manganese;Membrane;Metal-binding;Nucleotide-binding;Receptor;Reference proteome;Serine/threonine-protein kinase;Signal;Transferase;Transmembrane;Transmembrane helix | The bone morphogenetic protein (BMP) receptors are a family of transmembrane serine/threonine kinases that include the type I receptors BMPR1A and BMPR1B and the type II receptor BMPR2. These receptors are also closely related to the activin receptors, ACVR1 and ACVR2. The ligands of these receptors are members of the TGF-beta superfamily. TGF-betas and activins transduce their signals through the formation of heteromeric complexes with 2 different types of serine (threonine) kinase receptors: type I receptors of about 50-55 kD and type II receptors of about 70-80 kD. Type II receptors bind ligands in the absence of type I receptors, but they require their respective type I receptors for signaling, whereas type I receptors require their respective type II receptors for ligand binding. [provided by RefSeq, Jul 2008]. | hsa:657; | dendrite [GO:0030425]; external side of plasma membrane [GO:0009897]; HFE-transferrin receptor complex [GO:1990712]; integral component of membrane [GO:0016021]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; BMP binding [GO:0036122]; BMP receptor activity [GO:0098821]; metal ion binding [GO:0046872]; phosphatase activator activity [GO:0019211]; protein homodimerization activity [GO:0042803]; protein serine/threonine kinase activator activity [GO:0043539]; protein serine/threonine kinase activity [GO:0004674]; SMAD binding [GO:0046332]; transforming growth factor beta receptor activity, type I [GO:0005025]; transmembrane receptor protein serine/threonine kinase activity [GO:0004675]; atrioventricular node cell development [GO:0060928]; atrioventricular valve development [GO:0003171]; BMP signaling pathway [GO:0030509]; BMP signaling pathway involved in heart development [GO:0061312]; cardiac conduction system development [GO:0003161]; cardiac right ventricle morphogenesis [GO:0003215]; cellular response to BMP stimulus [GO:0071773]; cellular response to growth factor stimulus [GO:0071363]; chondrocyte differentiation [GO:0002062]; developmental growth [GO:0048589]; dorsal aorta morphogenesis [GO:0035912]; dorsal/ventral axis specification [GO:0009950]; dorsal/ventral pattern formation [GO:0009953]; ectoderm development [GO:0007398]; embryonic digit morphogenesis [GO:0042733]; embryonic organ development [GO:0048568]; endocardial cushion formation [GO:0003272]; endocardial cushion morphogenesis [GO:0003203]; fibrous ring of heart morphogenesis [GO:1905285]; heart formation [GO:0060914]; hindlimb morphogenesis [GO:0035137]; immune response [GO:0006955]; in utero embryonic development [GO:0001701]; lateral mesoderm development [GO:0048368]; lung development [GO:0030324]; mesendoderm development [GO:0048382]; mesoderm formation [GO:0001707]; mitral valve morphogenesis [GO:0003183]; Mullerian duct regression [GO:0001880]; negative regulation of gene expression [GO:0010629]; negative regulation of muscle cell differentiation [GO:0051148]; negative regulation of neurogenesis [GO:0050768]; negative regulation of smooth muscle cell migration [GO:0014912]; neural crest cell development [GO:0014032]; neural plate mediolateral regionalization [GO:0021998]; odontogenesis of dentin-containing tooth [GO:0042475]; osteoblast differentiation [GO:0001649]; outflow tract morphogenesis [GO:0003151]; outflow tract septum morphogenesis [GO:0003148]; paraxial mesoderm structural organization [GO:0048352]; pharyngeal arch artery morphogenesis [GO:0061626]; pituitary gland development [GO:0021983]; positive regulation of bone mineralization [GO:0030501]; positive regulation of cardiac muscle cell proliferation [GO:0060045]; positive regulation of cardiac ventricle development [GO:1904414]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of mesenchymal cell proliferation [GO:0002053]; positive regulation of osteoblast differentiation [GO:0045669]; positive regulation of pathway-restricted SMAD protein phosphorylation [GO:0010862]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of SMAD protein signal transduction [GO:0060391]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transforming growth factor beta2 production [GO:0032915]; positive regulation of vascular associated smooth muscle cell proliferation [GO:1904707]; protein phosphorylation [GO:0006468]; regulation of cardiac muscle cell proliferation [GO:0060043]; regulation of cellular senescence [GO:2000772]; regulation of lateral mesodermal cell fate specification [GO:0048378]; roof of mouth development [GO:0060021]; somitogenesis [GO:0001756]; stem cell population maintenance [GO:0019827]; transforming growth factor beta receptor signaling pathway [GO:0007179]; tricuspid valve morphogenesis [GO:0003186]; ventricular compact myocardium morphogenesis [GO:0003223]; ventricular septum morphogenesis [GO:0060412]; ventricular trabecula myocardium morphogenesis [GO:0003222] | 11536076_Germline mutations in BMPR1A cause a subset of juvenile polyposis syndrome and Cowden syndrome cases. 12136244_Observational study of gene-disease association. (HuGE Navigator) 12417513_Observational study of gene-disease association. (HuGE Navigator) 12620973_BMPR1A can act as a minor susceptibility gene for PTEN mutation negative Cowden syndrome 15235019_Observational study of genotype prevalence. (HuGE Navigator) 15351706_BMPR-IA may interact with and modulate the activity of a developmentally relevant splicing factor 15940369_A defect in BMPRIA internalization and increased activation of downstream signaling, suggesting that altered BMP receptor trafficking underlies ectopic bone formation in fibrodysplasia ossificans progressiva. 16226113_BMPR1A is a promising marker for evaluating ganglion cells in the enteric nervous system. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16436528_Human granulosa-like tumor cell line KGN expressed BMP type I (BMPR1A and BMPR1B) and type II receptors (BMPR2) and the BMP signaling molecules SMADs (SMAD1 and SMAD5). 16436638_Observational study of genotype prevalence. (HuGE Navigator) 16525031_BMPR1A mutation accounts for hereditary mixed polyposis syndrome and inactivating this gene can initiate colorectal tumourigenesis 16672363_structure of the ternary complex representing the signaling competent complex of BMP-2 bound to the entire extracellular domains of both its type I receptor, BMPR-Ia, & its type II receptor, ActRII, at a resolution of 2.2 angstroms 16886151_Observational study of gene-disease association. (HuGE Navigator) 17101085_Cooperation between this gene and PTEN gene is deleted on chromoome 10 in juvenile polyposis coli. 17513295_SF3b4, known to be localized in the nucleus and involved in RNA splicing, binds BMPR-IA and specifically inhibits BMP-mediated osteochondral cell differentiation 17573831_Linkage analysis suggested a cryptic BMPR1A mutation or the presence of another gene in close proximity to the BMPR1A locus. 17624341_Expression of BMP-4 and BMP-7 and their receptors in human ovaries from fetuses as well as adults. 17873119_5 nonsense, 2 frameshift, 4 missense and 2 splice site mutations were associated with juvenile polyposis syndrome. A 65-BP deletion in intron 4 included -2 of the splice acceptor side of exon 5. 17873119_Observational study of gene-disease association. (HuGE Navigator) 18160401_inactivating BMPR-IA and causing a loss of the BMP-2 tumor suppressor function in colon epithelial cells. 18178612_Large genomic deletions of SMAD4, BMPR1A and PTEN are a common cause of JPS. 18262054_Germline mutation of BMPR1A in a family with juvenile polyposis and colon cancer 18510548_patients with 10q23 microdeletions involving the PTEN and BMPR1A genes have variable clinical phenotypes, which cannot be explained merely by the deletion sizes, and are not restricted to severe infantile juvenile polyposis 18667463_Loss of Bmpr1a, by decreasing MMP2 and/or MMP9 activity, can account for vascular dilatation and persistence of brain microvessels, leading to the impaired organogenesis documented in the brain. 18823382_Observational study of gene-disease association. (HuGE Navigator) 18823382_The overall prevalence of SMAD4 and BMPR1A point mutations and deletions in JPS was 45% in the largest series of patients to date 18937504_The solution structure of BMPR-IA reveals a local disorder-to-order transition upon BMP-2 binding. 19244313_5-HT transactivates the serine kinase receptor, BMPR 1A, to activate Smads 1/5/8 via Rho and Rho kinase in in bovine and human pulmonary artery smooth muscle cells 19438883_we describe the significance of a bone morphogenetic protein receptor type 1A gene mutation in an Irish family with hereditary mixed polyposis syndrome. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19463221_The prevalence of germline mutations of BMPR1A and SMAD4 are about 20% each in the patient with JPS. 19502417_Observational study of gene-disease association. (HuGE Navigator) 19502417_genetic variation in the BMPR1A may play a role in the pathophysiology of human obesity, possibly mediated through effects on mRNA expression 19773747_Germline BMPR1A defect is the disease-causing mutation in 50% of the HMPS families. 20346360_Observational study of gene-disease association. (HuGE Navigator) 20587070_Low BMP2 is associated with epithelial ovarian cancer. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20673479_Data show that blocking both endogenous BMPR1A and BMPR1B almost offset the effect of BMP7 on the proliferation of NCI-H460 cell completely. 20693682_Crystals BMP receptor type IA bound to the antibody Fab fragment belonged to the monoclinic space group P2(1), with unit-cell parameters a=89.32, b=129.25, c=100.24 A, beta=92.27 degrees 20734064_Observational study of gene-disease association. (HuGE Navigator) 20843829_identified the promoter for BMPR1A, in which mutations may be responsible for as many as 10% of juvenile polyposis cases with unknown mutations 20927405_The crystal structure of the complex of the BMPR-IA bound to the Fab AbD1556 revealed that the contact surface of BMPR-IA overlaps extensively with the contact surface for BMP-2 interaction. 21152263_BMPR1A were detected in the human retina and retinoblastoma cell lines. 21258932_Disruption of BMPR1A-mediated BMP1 signalling during the narrow window of early embryogenesis may interfere with normal VBW formation, causing omphalocele phenotype in the Cd chick model. 21412070_Juvenile polyps with a SMAD4 germline mutation were predominantly type B, whereas type A was more common among juvenile polyps with a BMPR1A germline mutation. 21543859_Crystals of GDF5 and BMP receptor IA complex belonged to a monoclinic space group: either I2, with unit-cell parameters a = 63.81, b = 62.85, c = 124.99 A, beta = 95.9 degrees , or C2, with unit-cell parameters a = 132.17, b = 62.78, c = 63.53 A, beta = 112.8 degrees 21640116_Letter: Report the phenotypic spectrum of BMPR1A mutations in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. 21872883_Sp1 was found to be a candidate factor that likely plays a role in the transcriptional regulation of BMPR1A. 22210872_Results suggest that BMPRIA expression identifies thymic NK cell precursors and that BMP signaling is relevant for NK cell differentiation in the thymus. 22293501_generation of TGF-beta and BMP receptor homo- and hetero-oligomers and its roles as a mechanism capable of fast regulation of signaling by these crucial cytokines [review] 22519633_These data support the role of BMPR-1A as an indicator ofosteoarthritis progression in human knees with circumscribed cartilage lesions. 22710174_analysis of promiscuity and specificity in BMP receptor activation [review] 22886282_Results suggest that rs7922846 BMPR1A polymorphism may account for subtle variation in kidney size at birth, reflecting congenital nephron endowment. 22971142_lack of associations between LVM, values of blood pressure, and the BMP4, BMPR1A, BMPR1B, and ACVR1 genotypes 23399955_Seventy-seven patients (13%) were found to have colorectal polyposis-associated mutations, including 20 in BMPR1A (3.3%) 23433720_Bone morphogenetic protein receptor type 1A missense mutations occurring in patients with juvenile polyposis affected cellular localization in an in vitro model. 23531103_Decreased expression of BMPR1A was associated malignant gallbladder lesions. 23593444_BMP receptor antagonists and silencing of BMP type I receptors with siRNA induced cell death, inhibited cell growth, and caused a significant decrease in the expression of inhibitor of differentiation (Id1, Id2, and Id3) family members. 23969729_findings show that a reduction in the expression of BMPRIA is associated with a poorer prognosis in pancreatic cancer 24140593_BMP15 down-regulates StAR expression and decreases progesterone production in human granulosa cells, likely via ALK3-mediated SMAD1/5/8 signaling. 24398041_BMPR1a and BMPR2 are downregulated in cardiac remodeling and heart failure 24480809_High BMPR1A expression is associated with glioma tumorigenesis. 24850914_Data show that USP15 enhances BMP-induced phosphorylation of SMAD1 by interacting with and deubiquitylating ALK3. 24882581_Results support a novel role for miR-885-3p in tumor angiogenesis by targeting BMPR1A, which regulates a proangiogenic factor. 24904118_results provide evidence that HFE induces hepcidin expression via the BMP pathway: HFE interacts with ALK3 to stabilize ALK3 protein and increase ALK3 expression at the cell surface. 24966941_The mRNA/protein expressions of BMPR1alpha was higher in the stenotic colon segment tissue than in the normal colon segment tissue of Hirschsrung disease patients. 25129392_This is the first case report to document coding exon 3 duplication in the BMPR1A gene in a patient with juvenile polyposis syndrome. 26171675_About half of BMPR1A-related polyps displayed loss of heterozygosity, predominantly in the epithelial compartment, compatible with BMPR1A acting as a tumour suppressor gene. 26274893_Authors analyzed human databases from TCGA and survival data from microarrays to confirm BMPR1a tumor promoting functions, and found that high BMPR1a gene expression correlates with decreased survival regardless of molecular breast cancer subtype. 26383923_Duplication of 10q22.3-q23.3 encompassing BMPR1A gene is associated with congenital heart disease, microcephaly, and mild intellectual disability 26585335_BMPR1A(+) ASCs show an enhanced ability for adipogenesis in vitro, as shown by gene expression and histological staining. 26701726_Data show that protein kinase LKB1 physically interacts with BMP type I receptors and requires Smad7 protein to promote downregulation of the receptor. 27146957_Several germline variants in Hamartomatous Polyposis Syndrome genes were detected, among them three in ENG, two in BMPR1A, one in PTEN, and one in SMAD4. Although some of the detected variants have been reported previously none could be definitely pathogenic or likely pathogenic. 27660075_The present study suggests that HNF-4alpha has a suppressive effect on hepcidin expression by inactivating the BMP pathway, specifically via BMPR1A, in HepG2 cells. 27780042_In solid ameloblastoma, positive correlations were observed between the stromal and parenchymal expression of BMP-2 and between the stromal expression of BMP-2 and BMP-4, as well as between the stromal expression of BMPR-II and BMP-4 and the stromal and parenchymal expression of BMPR-II. 28357470_BMPR1A and the ubiquitous isoform of BMPR1B differed in mode of translocation into the endoplasmic reticulum; and (ii) BMPR1A was N-glycosylated while BMPR1B was not, resulting in greater efficiency of processing and plasma membrane expression of BMPR1A. 28660566_Study findings highlight that BMPR1a mutations are not a major contributor of Familial colorectal cancer type X incidence in Newfoundland. 28733457_both bone morphogenetic protein 2 (BMP2) and BMP6 are proangiogenic in vitro and ex vivo and that the BMP type I receptors, activin receptor-like kinase 3 (ALK3) and ALK2, play crucial and distinct roles in this process. 28869862_The binding free energies indicate that ALK-3 preferably binds to BMP-2 instead of BMP-9. The structural analysis shows that ALK-3 binding with BMP-2 occurs in a perfectly symmetry pathway, whereas this symmetry is lost for possible ALK-3 interactions with BMP-9 29396550_IL-6 may be responsible for coformation of new bone and excessive adipose tissue in rhBMP-2-induced bone voids. 29458345_Single nucleotide polymorphisms of the BMPR-1A gene were significantly associated with the development of ossification of the posterior longitudinal ligament of the cervical spine. 29495003_Knockdown of BMPR1a of breast cancer cells suppresses their production of RANKL via p38 pathway and inhibits cancer-induced osteoclastogenesis. 29522511_BMPR1A mutation in superior coloboma 30262802_This high expression of BMPR1A is further increased upon BMP4 exposure. 31062505_dentified an interstitial 10q23.1q23.3 deletion in a buccal mucosa sample of Patient 1 that encompassed PTEN, BMPR1A, and KLLN, among others. In contrast, neither sequencing nor array-CGH analysis identified a pathogenic variant in PTEN or BMPR1A in a blood sample of Patient 2 31259752_Variable Features of Juvenile Polyposis Syndrome With Gastric Involvement Among Patients With a Large Genomic Deletion of BMPR1A. 31769494_BMPR1A and BMPR1B Missense Mutations Cause Primary Ovarian Insufficiency. 32398773_Disease expression in juvenile polyposis syndrome: a retrospective survey on a cohort of 221 European patients and comparison with a literature-derived cohort of 473 SMAD4/BMPR1A pathogenic variant carriers. 32764110_BMPR1A is necessary for chondrogenesis and osteogenesis, whereas BMPR1B prevents hypertrophic differentiation. 33097490_Phenotypic Differences in Juvenile Polyposis Syndrome With or Without a Disease-causing SMAD4/BMPR1A Variant. 33250156_Bone morphogenetic protein signaling regulates skin inflammation via modulating dendritic cell function. 34013359_Anti-Mullerian hormone concentration regulates activin receptor-like kinase-2/3 expression levels with opposing effects on ovarian cancer cell survival. 34743989_Expression of GALNT8 and O-glycosylation of BMP receptor 1A suppress breast cancer cell proliferation by upregulating ERalpha levels. | ENSMUSG00000021796 | Bmpr1a | 3.089039e+02 | 0.8069411 | -0.309464679 | 0.3613067 | 7.167079e-01 | 0.3972259120 | 0.84483554 | No | Yes | 2.637402e+02 | 51.931085 | 2.891735e+02 | 58.170191 | |
| ENSG00000107829 | 6468 | FBXW4 | protein_coding | P57775 | FUNCTION: Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation. Likely to be involved in key signaling pathways crucial for normal limb development. May participate in Wnt signaling. | Developmental protein;Reference proteome;Repeat;Ubl conjugation pathway;WD repeat;Wnt signaling pathway | This gene is a member of the F-box/WD-40 gene family, which recruit specific target proteins through their WD-40 protein-protein binding domains for ubiquitin mediated degradation. In mouse, a highly similar protein is thought to be responsible for maintaining the apical ectodermal ridge of developing limb buds; disruption of the mouse gene results in the absence of central digits, underdeveloped or absent metacarpal/metatarsal bones and syndactyly. This phenotype is remarkably similar to split hand-split foot malformation in humans, a clinically heterogeneous condition with a variety of modes of transmission. An autosomal recessive form has been mapped to the chromosomal region where this gene is located, and complex rearrangements involving duplications of this gene and others have been associated with the condition. A pseudogene of this locus has been mapped to one of the introns of the BCR gene on chromosome 22. [provided by RefSeq, Jul 2008]. | hsa:6468; | cytosol [GO:0005829]; SCF ubiquitin ligase complex [GO:0019005]; ubiquitin ligase complex [GO:0000151]; embryonic limb morphogenesis [GO:0030326]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146]; ubiquitin-dependent protein catabolic process [GO:0006511]; Wnt signaling pathway [GO:0016055] | 12913067_a complex rearrangement associated with a approximately 0.5 Mb tandem duplication ion containing a disrupted extra copy of the DACTYLIN gene and the entire LBX1 and beta-TRCP genes. 16235095_results indicate that genomic rearrangement of SHFM3 is rare among non-syndromic SHFM patients and emphasize the importance of screening for genomic rearrangements even in sporadic cases of SHFM 16761290_Genomic rearrangements involving the SHFM3 locus at chromosome 10q24 is associated with syndromic and non-syndromic split-hand/foot malformation 23658844_biochemical characterization of the novel F-box and WD40 containing protein, FBXW4 | ENSMUSG00000040913 | Fbxw4 | 1.349263e+03 | 1.4506599 | 0.536709364 | 0.2695013 | 3.990970e+00 | 0.0457447107 | 0.65167849 | No | Yes | 1.531972e+03 | 233.993871 | 9.324173e+02 | 146.641264 | |
| ENSG00000108239 | 23232 | TBC1D12 | protein_coding | O60347 | FUNCTION: RAB11A-binding protein that plays a role in neurite outgrowth. {ECO:0000250|UniProtKB:M0R7T9}. | Acetylation;Coiled coil;Endosome;GTPase activation;Phosphoprotein;Reference proteome | hsa:23232; | autophagosome [GO:0005776]; recycling endosome [GO:0055037]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; intracellular protein transport [GO:0006886]; regulation of autophagosome assembly [GO:2000785] | ENSMUSG00000048720 | Tbc1d12 | 1.960524e+02 | 0.7265180 | -0.460929522 | 0.3514382 | 1.697028e+00 | 0.1926770748 | 0.78025017 | No | Yes | 1.640223e+02 | 28.211411 | 2.156315e+02 | 37.522643 | |||
| ENSG00000108344 | 5709 | PSMD3 | protein_coding | O43242 | FUNCTION: Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair. {ECO:0000269|PubMed:1317798}. | 3D-structure;Alternative splicing;Isopeptide bond;Phosphoprotein;Proteasome;Reference proteome;Ubl conjugation | The 26S proteasome is a multicatalytic proteinase complex with a highly ordered structure composed of 2 complexes, a 20S core and a 19S regulator. The 20S core is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. The 19S regulator is composed of a base, which contains 6 ATPase subunits and 2 non-ATPase subunits, and a lid, which contains up to 10 non-ATPase subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. This gene encodes a member of the proteasome subunit S3 family that functions as one of the non-ATPase subunits of the 19S regulator lid. Single nucleotide polymorphisms in this gene are associated with neutrophil count. [provided by RefSeq, Jul 2012]. | hsa:5709; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; proteasome accessory complex [GO:0022624]; proteasome complex [GO:0000502]; proteasome regulatory particle, lid subcomplex [GO:0008541]; secretory granule lumen [GO:0034774]; enzyme regulator activity [GO:0030234]; regulation of protein catabolic process [GO:0042176]; ubiquitin-dependent protein catabolic process [GO:0006511] | 20172861_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20172861_The significant contribution of PSMD3-CSF3 and PLCB4 loci to the regulation of neutrophil count, is demonstrated. 22037903_Study identified significantly white blood cell count (WBC) level associated SNPs of three separate genes GSDMA, MED24, and PSMD3 in European continent (EA) subjects. 22788528_HNF1A gene was associated with C-reactive protein, and the region including PSMD3 and CSF3 genes was associated with white blood cell count. 23303871_Our study suggests that PSMD3 variants are associated with insulin resistance in populations of different ancestries and that these relationships may also be modified by dietary factors. 25515861_A strong association between genetic variant rs2305482 in PSMD3 on chromosome 17 and IFN-induced neutropenia. 27163155_this study shows that haplotypes consisting of single nucleotide polymorphisms harboring PSMD3, CSF3 and MED24 genes are associated with asthma in Slovenian patients 28390009_In thyroid hemiagenesis, genomic examinations revealed the presence of four recurrent defects (three deletions and one duplication) affecting highly conservative proteasome genes PSMA1, PSMA3, and PSMD3. 33712704_Proteasome 26S subunit, non-ATPases 1 (PSMD1) and 3 (PSMD3), play an oncogenic role in chronic myeloid leukemia by stabilizing nuclear factor-kappa B. 34572038_26S Proteasome Non-ATPase Regulatory Subunits 1 (PSMD1) and 3 (PSMD3) as Putative Targets for Cancer Prognosis and Therapy. | ENSMUSG00000017221 | Psmd3 | 1.113859e+04 | 1.1617720 | 0.216326957 | 0.3116820 | 4.829492e-01 | 0.4870894601 | 0.87475557 | No | Yes | 1.119114e+04 | 1140.276586 | 8.618340e+03 | 900.870905 | |
| ENSG00000108424 | 3837 | KPNB1 | protein_coding | Q14974 | FUNCTION: Functions in nuclear protein import, either in association with an adapter protein, like an importin-alpha subunit, which binds to nuclear localization signals (NLS) in cargo substrates, or by acting as autonomous nuclear transport receptor. Acting autonomously, serves itself as NLS receptor. Docking of the importin/substrate complex to the nuclear pore complex (NPC) is mediated by KPNB1 through binding to nucleoporin FxFG repeats and the complex is subsequently translocated through the pore by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to importin-beta and the three components separate and importin-alpha and -beta are re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran from importin. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus. Mediates autonomously the nuclear import of ribosomal proteins RPL23A, RPS7 and RPL5. Binds to a beta-like import receptor binding (BIB) domain of RPL23A. In association with IPO7 mediates the nuclear import of H1 histone. In vitro, mediates nuclear import of H2A, H2B, H3 and H4 histones. In case of HIV-1 infection, binds and mediates the nuclear import of HIV-1 Rev. Imports SNAI1 and PRKCI into the nucleus. {ECO:0000269|PubMed:10228156, ECO:0000269|PubMed:11891849, ECO:0000269|PubMed:19386897, ECO:0000269|PubMed:24699649, ECO:0000269|PubMed:9687515}. | 3D-structure;ADP-ribosylation;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Host-virus interaction;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | Nucleocytoplasmic transport, a signal- and energy-dependent process, takes place through nuclear pore complexes embedded in the nuclear envelope. The import of proteins containing a nuclear localization signal (NLS) requires the NLS import receptor, a heterodimer of importin alpha and beta subunits also known as karyopherins. Importin alpha binds the NLS-containing cargo in the cytoplasm and importin beta docks the complex at the cytoplasmic side of the nuclear pore complex. In the presence of nucleoside triphosphates and the small GTP binding protein Ran, the complex moves into the nuclear pore complex and the importin subunits dissociate. Importin alpha enters the nucleoplasm with its passenger protein and importin beta remains at the pore. Interactions between importin beta and the FG repeats of nucleoporins are essential in translocation through the pore complex. The protein encoded by this gene is a member of the importin beta family. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2013]. | hsa:3837; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; endoplasmic reticulum tubular network [GO:0071782]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; membrane [GO:0016020]; NLS-dependent protein nuclear import complex [GO:0042564]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; nuclear pore [GO:0005643]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; specific granule lumen [GO:0035580]; enzyme binding [GO:0019899]; Hsp90 protein binding [GO:0051879]; importin-alpha family protein binding [GO:0061676]; nuclear import signal receptor activity [GO:0061608]; nuclear localization sequence binding [GO:0008139]; protein domain specific binding [GO:0019904]; RNA binding [GO:0003723]; small GTPase binding [GO:0031267]; zinc ion binding [GO:0008270]; astral microtubule organization [GO:0030953]; establishment of mitotic spindle localization [GO:0040001]; establishment of protein localization [GO:0045184]; mitotic chromosome movement towards spindle pole [GO:0007079]; mitotic metaphase plate congression [GO:0007080]; mitotic spindle assembly [GO:0090307]; NLS-bearing protein import into nucleus [GO:0006607]; protein import into nucleus [GO:0006606]; Ran protein signal transduction [GO:0031291]; ribosomal protein import into nucleus [GO:0006610]; RNA import into nucleus [GO:0006404] | 12551970_nuclear import of the HPV16 E6 oncoprotein in digitonin-permeabilized HeLa cells could be mediated by Kap beta1 13679510_XCTK2 binding to microtubules inhibited in the presence of importin alpha and beta 15194443_Importin alpha/beta-mediated nuclear import machinery is regulated in a cell cycle-dependent manner through the modulation of interaction modes between importins alpha and beta. 15572412_This study shows a direct role of importin beta in control of mitotic spindle pole organization in human cells 15795315_Inhibition by Impbeta results from depletion nuclear RanGTP, and, in support of this mechanism, expression of mRFP-Ran reversed the inhibition. 16552725_These data suggest the importance of receptor endocytosis, endosomal sorting machinery, interaction with importins alpha1/beta1, and exportin CRM1 in EGFR nuclear-cytoplasmic trafficking. 16982803_These results show that maximum nucleocytoplasmic transport velocities can be modulated by at least approximately 10-fold by the importin beta concentration and hence suggest a potential mechanism for regulating the speed of cargo traffic across the NE. 17537211_dengue virus nonstructural protein 5 nuclear localization through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection 17596301_SARS-COV ORF6 protein is localized to the endoplasmic reticulum (ER)/Golgi membrane in infected cells, where it binds to and disrupts nuclear import complex formation by tethering karyopherin alpha 2 and karyopherin beta 1 to the membrane. 17646395_Data show that a novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta1 interactions. 17916694_study found binding interactions with karyopherin-beta1 caused FG domains of nucleoporin Nup153 to collapse into compact molecular conformations; reversible collapse of the FG domains may play an important role in regulating nucleocytoplasmic transport 18028944_crystal structure of Imp beta (127-876) in complex SPN1 (1-65) at 2.8-A resolution reveals that Imp beta adopts an open conformation, which is unique for a functional Imp beta/cargo complex, and resembles the conformation of the Imp beta/RanGTP complex 18238777_importins alpha5 and beta1 associate with Nrf2, an interaction that was blocked by the nuclear import inhibitor SN50 18398821_Observational study of gene-disease association. (HuGE Navigator) 19117056_Crm1, Kpnbeta1 and Kpnalpha2 are overexpressed in cervical cancer and inhibiting the expression of Crm1 and Kpnbeta1, not Kpnalpha2, induces cancer cell death 19884259_replacement of the cap-binding complex by eukaryotic translation initiation factor 4E is promoted by importin beta (IMPbeta) 19925438_[review] Recognition and shuttling of calcineurin into the nucleus by importin-beta requires the nuclear localization sequence of the region spanning amino acids 172-183 of calcineurin A beta. 20711181_Results demonstrate that PP2A-B55alpha and importin-beta1 cooperate in the regulation of postmitotic assembly mechanisms in human cells. 21209321_The mammalian E1 subunits can be imported separately, identify nuclear localization signals (NLSs) in Aos1 and in Uba2, and demonstrate that their import is mediated by importin alpha/beta in vitro and in intact cells. 21278340_Importin beta interacts with the NFAT-tubulin alpha complex rather than NFAT or tubulin alpha alone, resulting in cotranslocation of NFAT and tubulin alpha into the nucleus. 21625522_study defines importin-alpha/importin-beta1/Ran as the molecular mechanism by which STAT3 traffics to the nucleus 21630149_[review] The cellular functions of Ran are mediated by RanGTP interactions with nuclear transport receptors related to importin beta and depend on the existence of chromosome-centered RanGTP gradient. 22020938_Findings suggest that the importin beta1-mediated nuclear localization of DR5 limits the DR5/TRAIL-induced cell death of human tumor cells and thus can be a novel target to improve cancer therapy with recombinant TRAIL and anti-DR5 antibodies. 22125623_Findings suggest that the deregulated activity of E2F in cancer cells causes increased activation of the Kpnbeta1 and Kpnalpha2 promoters, leading to elevated levels of these proteins, and ultimately impacting the cancer phenotype. 22321063_LRRC59 facilitates transport of cytosolic FGF1 through nuclear pores by interaction with Kpns and movement of LRRC59 along the ER and NE membranes 22415091_Nuclear translocation of Wilms' tumour protein involves importins alpha and beta, and a nuclear localisation signal in the third zinc finger 23012356_Identification of a karyopherin beta1/beta2 proline-tyrosine nuclear localization signal in huntingtin protein. 23444224_Data show that five genes CKAP5, KPNB1, RAN, TPX2 and KIF11 were shown to be essential for tumor cell survival in both head and neck squamous cell carcinoma (HNSCC)and non-small cell lung cancer (NSCLC), but most particularly in HNSCC. 23557333_The role of Kpnbeta1 in cancer is only now being elucidated, and recent work points to its potential usefulness as an anti-cancer target. 23906023_Knockdown of KPNB1 reduced the amount of nuclear p65 following TNF stimulation. KPNB1 binding to p65 is NLS dependent. 24132643_we report that the EZH2-miR-30d-KPNB1 signalling pathway is critical for malignant peripheral nerve sheath tumour cell survival in vitro and tumourigenicity in vivo 24398670_the inhibition of endogenous Kpnbeta1 in cervical cancer cells results in a significant increase in mitotic abnormalities and a prolonged mitotic arrest. 24699649_Although there are many kinds of C2H2-type ZFs which have the same fold as Snail, nuclear import by direct recognition of importin beta is observed in a limited number of C2H2-type zinc-finger proteins such as Snail 24712655_This work extends published observations on SAMHD1 nuclear localization to a natural cell type for HIV-1 infection and identifies KPNA2/KPNB1 as cellular proteins important for SAMHD1 nuclear import. 24739174_These data reveal an emergent Kap-centric barrier mechanism that may underlie mechanistic and kinetic control in the nuclear pore complex. 24821838_Ei24 can bind specifically to IMPbeta1 and IMPalpha2 to impede their normal role in nuclear import. 24854174_Data suggest nuclear entry of GLI1 (glioma-associated oncogene homolog, a zinc finger protein) is regulated by unique mechanism via mutually exclusive binding by its nuclear import factor IMB1 (importin B1) and SuFu (suppressor of fused protein). 24953690_The results here establish for the first time that intracellular calcium modulates conventional nuclear import through direct effects on the nuclear transport machinery. 25037261_ARTD15 plays role in nucleocytoplasmic shuttling, through karyopherin-beta1 mono-ADP-ribosylation. [review] 25499977_ARHI competes with RanGTPase and interacts with importin beta via basic-acidic patch interaction, which leads to inhibition of STAT3 translocation. 25643631_Importin beta1 mediates the translocation of NF-kappaB into the nuclei of myeloma cells, thereby regulating proliferation and blocking apoptosis, which provides new insights for targeted myeloma therapies. 25696812_Data reveal a novel role for miR-9 in regulation of the NFAT pathway by targeting KPNB1 and DYRK1B. 25748139_Data show that importin-beta (impbeta) alters the nuclear pore's permeability in a Ran-dependent manner, suggesting that impbeta is a functional component of the nuclear pore complex (NPC). 25794490_High expression of KPNB1 protein is associated with hepatocellular carcinoma. 25890085_DZNep suppressed EZH2/miR-30a,d/KPNB1 signaling. 25960398_This hypersensitivity of malignant cell types to Impbeta1 knockdown raises the exciting possibility of anti-cancer therapies targeted at Impbeta1. 26107903_Importin-beta1 mediates the non-classical nucleocytoplasmic transport of MARVELD1. 26112410_The study revealed a regulatory role of the p97-Npl4-Ufd1 complex in regulating a partial degradation of the NF-kappaB subunit p100. 26216267_Humanin Peptide Binds to Insulin-Like Growth Factor-Binding Protein 3 (IGFBP3) and Regulates Its Interaction with Importin-beta. 26242264_KPNbeta1 was significantly highly expressed in gastric cancer and was correlated with tumor grade as well as poor prognosis. 26319354_Collectively, these data show that KPNB1 is required for timely nuclear import of PER/CRY in the negative feedback regulation of the circadian clock. 26414402_Data show that cytoskeleton associated protein 5 (chTOG) only weakly promotes importin-regulated microtubule nucleation, but acts synergistically with microtubule- associated protein TPX2. 26491019_RBBP4 functions as a novel regulatory factor to increase the efficiency of importin alpha/beta-mediated nuclear import 26498772_Patients with tumors highly expressing Kpnbeta1 have poorer overall survivals. Kpnbeta1 interacts with p65 and enhances cell adhesion-mediated drug resistance. 27528606_Importins, Impbeta, Kapbeta2, Imp4, Imp5, Imp7, Imp9, and Impalpha, show the H3 tail binding more tightly than the H4 tail. The H3 tail binds Kapbeta2 and Imp5 with KD values of 77 and 57 nm, respectively, and binds the other five Importins more weakly. 27568288_KPNB1 might enhance human glioma proliferation via Wnt/beta-Catenin Pathway. 27808165_Results show that importin beta1 is an Epac1 binding partner that regulates Epac1 subcellular localization. 28427184_Results found KPNB1 to be required for the migration and invasion of cervical cancer cells through mediating nuclear import. Its inhibition of KPNB1 interferes with NFkB subcellular localization. 28698143_Therefore, we show for the first time that the nuclear localization of Cat L and its substrate Cux1can be positively regulated by Snail NLS and importin beta1, suggesting that Snail, Cat L and Cux1 all utilize importin beta1 for nuclear import. 28835592_Data suggest that IGFBP5 nuclear import is mediated by KPNA5/KPNB1 complex; nuclear localization sequence of IGFBP5 is critical domain in this nuclear translocation. (IGFBP5 = insulin-like growth factor binding protein-5; KPNA5 = karyopherin subunit alpha-5; KPNB1 = karyopherin subunit beta-1/importin-beta) 28864541_Karyoppherins constitute integral constituents of the nuclear pore complex whose barrier, transport, and cargo release functionalities establish a continuum under a mechanism of Kap-centric control. 29017749_These results showed that BLM enters the nucleus via the importin beta1, RanGDP and NTF2 dependent pathway, demonstrating for the first time the nuclear trafficking mechanism of a DNA helicase. 29251332_Study identified that high expression of Kpnbeta1 in breast cancer (BC) often leads to poor prognosis and that Kpnbeta1-knockdown markedly reduced BC cell proliferation by abrogating nuclear transport of Her2. 29520102_Findings demonstrate that KPNB1 is required for proteostasis maintenance and its inhibition induces apoptosis in glioblastoma cells through UPR-mediated deregulation of Bcl-2 family members. 29622086_our findings suggest that Kpnbeta1 plays a significant role in non-small cell lung cancer progression and chemoresistance 29746765_KPNB1 and Ran protein jointly mediated the nuclear import of NDV M protein, showing that KPNB1 protein interacted with NDV M protein to form binary complex and then entered into the nucleus with the assistance of Ran protein. 29957337_Taken together, these results showed that full-length Pom121 enables efficient HIV-1 pre-integration complex nuclear import, and suggested that this process may rely on KPNB1 dependent classical cargo nuclear transportation way. 30153455_NCAM140 is translocated into the nucleus by an importin-beta1-dependent mechanism. 30445944_A tight balance of Kpnbeta1 expression is required for cellular function. 30742095_Inhibition of KPNB1 could be a new therapeutic target for Prostate cancer treatment. 30887716_KPNB1 is a direct target of miR-101. 31629952_KPNB1 expression was significantly increased in colorectal cancer, contributing to CRC progression and hepatic metastasis by interacting with MET to subsequently control the expression of EMT-related signals. 31642884_KPNA7, an isoform with expression mostly limited to early development, can bind Importin-beta (Imp-beta) in the absence of NLS cargo. 31653923_Thioredoxin-related transmembrane protein 2 (TMX2) regulates the Ran protein gradient and importin-beta-dependent nuclear cargo transport. 33439253_Antivirals that target the host IMPalpha/beta1-virus interface. 33801927_Inhibition of NUPR1-Karyopherin beta1 Binding Increases Anticancer Drug Sensitivity. | ENSMUSG00000001440 | Kpnb1 | 1.407672e+04 | 0.9635750 | -0.053531078 | 0.2481135 | 4.634684e-02 | 0.8295465480 | 0.96751127 | No | Yes | 1.309844e+04 | 1933.079283 | 1.123391e+04 | 1700.207717 | |
| ENSG00000108509 | 23125 | CAMTA2 | protein_coding | O94983 | FUNCTION: Transcription activator. May act as tumor suppressor. {ECO:0000269|PubMed:11925432}. | ANK repeat;Activator;Alternative splicing;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a member of the calmodulin-binding transcription activator protein family. Members of this family share a common domain structure that consists of a transcription activation domain, a DNA-binding domain, and a calmodulin-binding domain. The encoded protein may be a transcriptional coactivator of genes involved in cardiac growth. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Jan 2010]. | hsa:23125; | chromatin [GO:0000785]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; double-stranded DNA binding [GO:0003690]; histone deacetylase binding [GO:0042826]; sequence-specific DNA binding [GO:0043565]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; cardiac muscle hypertrophy in response to stress [GO:0014898]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 29110692_Mutation of CAMTA2 resulting in post-transcriptional inhibition of its own gene activity likely underlies a novel syndromic tremulous dystonia. | ENSMUSG00000040712 | Camta2 | 1.643400e+03 | 1.0937702 | 0.129309664 | 0.2845306 | 2.057779e-01 | 0.6500969712 | 0.92370776 | No | Yes | 1.318383e+03 | 226.488859 | 1.259166e+03 | 222.077570 | |
| ENSG00000108515 | 2027 | ENO3 | protein_coding | P13929 | FUNCTION: Glycolytic enzyme that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate. Appears to have a function in striated muscle development and regeneration. {ECO:0000250|UniProtKB:P15429}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Disease variant;Glycogen storage disease;Glycolysis;Lyase;Magnesium;Metal-binding;Phosphoprotein;Reference proteome | PATHWAY: Carbohydrate degradation; glycolysis; pyruvate from D-glyceraldehyde 3-phosphate: step 4/5. {ECO:0000250|UniProtKB:P15429}. | This gene encodes one of the three enolase isoenzymes found in mammals. This isoenzyme is found in skeletal muscle cells in the adult where it may play a role in muscle development and regeneration. A switch from alpha enolase to beta enolase occurs in muscle tissue during development in rodents. Mutations in this gene have be associated glycogen storage disease. Alternatively spliced transcript variants encoding different isoforms have been described.[provided by RefSeq, Jul 2010]. | hsa:2027; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; membrane [GO:0016020]; phosphopyruvate hydratase complex [GO:0000015]; plasma membrane [GO:0005886]; magnesium ion binding [GO:0000287]; phosphopyruvate hydratase activity [GO:0004634]; glycolytic process [GO:0006096] | 19472918_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 21347838_Pyridoxamine and carnosine protected enolase against the total loss of catalytic activity. 25267339_Molecular genetic analysis of ENO3 gene revealed two novel homozygous missense mutations, (p.Asn151Ser and p.Glu187Lys)in patients presenting with recurrent rhabdomyolysis. 25918939_tRK1 forms a complex with human enolases and interacts with tRK1 and human pre-lysyl-tRNA synthetase (preKARS2) 31697874_Enolase 3 (ENO3) overexpression under serine/threonine kinase 11 (STK11) loss-of-function mutations implies that ENO3 might be a selective anticancer target in STK11-mutant cancer. | ENSMUSG00000060600 | Eno3 | 1.403468e+02 | 0.8168973 | -0.291773373 | 0.3385458 | 7.490060e-01 | 0.3867911210 | 0.84317192 | No | Yes | 6.878453e+01 | 11.050540 | 9.305270e+01 | 15.015688 |
| ENSG00000108559 | 4927 | NUP88 | protein_coding | Q99567 | FUNCTION: Component of nuclear pore complex. {ECO:0000269|PubMed:30543681}. | Acetylation;Coiled coil;Disease variant;Nuclear pore complex;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Translocation;Transport;mRNA transport | The nuclear pore complex is a massive structure that extends across the nuclear envelope, forming a gateway that regulates the flow of macromolecules between the nucleus and the cytoplasm. Nucleoporins, a family of 50 to 100 proteins, are the main components of the nuclear pore complex in eukaryotic cells. The protein encoded by this gene belongs to the nucleoporin family and is associated with the oncogenic nucleoporin CAN/Nup214 in a dynamic subcomplex. This protein is also overexpressed in a large number of malignant neoplasms and precancerous dysplasias. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Mar 2016]. | hsa:4927; | cytosol [GO:0005829]; nuclear inner membrane [GO:0005637]; nuclear outer membrane [GO:0005640]; nuclear pore [GO:0005643]; nucleoplasm [GO:0005654]; structural constituent of nuclear pore [GO:0017056]; transporter activity [GO:0005215]; mitotic cell cycle [GO:0000278]; mRNA export from nucleus [GO:0006406]; protein import into nucleus [GO:0006606]; ribosomal large subunit export from nucleus [GO:0000055]; ribosomal small subunit export from nucleus [GO:0000056] | 14999780_High Nup88 mRNA expression levels correlated with ductal and tubular histology, nuclear grade 3, absence of hormone receptors, expression of c-erb-B2 oncogene and mutant p53 protein, high proliferation, DNA aneuploidy, and axillary node invasion. 15300515_A monoclonal antibody for Nup88 in an experimental model for studying the role played by Nup88 during cell development and differentiation. 16675447_the Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export 17007055_Our data strongly indicate a dichotomous role for Nup88 in non-neoplastic and neoplastic conditions of the liver. 17650673_The antitumor activity of deguelin was related to up-regulating the expression of Nup98 and down-regulating Nup88 protein. 17828493_Gambogic acid exerts its anti-leukemia effects by regulating the expression and distribution of nucleoporin Nup88. 18068677_The formation of high molecular mass complexes containing importin-alpha, Nup153 and Nup88 is increased upon oxidant treatment. 18606815_Nup88 is up-regulated in response to hypertonic stress and acts to retain TonEBP in the nucleus, activating transcription of critical osmoprotective genes. 19751906_Nup88 is significantly overexpressed in high-grade CIN lesions and ISCC compared with normal ectocervical squamous epithelia and CIN1. 20497554_proper expression of Nup88 is critical for preventing aneuploidy formation and tumorigenesis. 20973273_association found between myometrial invasion and Nup88 expression in endometrial carcinoma 21289091_Data suggest that a pool of Nup88 on the nuclear side of the nuclear pore complex provides a novel, unexpected binding site for nuclear lamin A. 21863385_Serum Nup88 might be a candidate for a new biomarker implicated in the development and aggressiveness of colorectal cancer 21896994_Nup88 mRNA was overexpressed in colorectal cancers and the overexpression was associated with cancer development and aggressiveness. 23777819_Nup62 and Nup88 protein levels were significantly decreased upon knockdown of O-GlcNAc transferase. 26731471_These findings demonstrate that the NUP88-NUP98-RAE1-APC/CCDH1 axis contributes to aneuploidy 26839161_Nup88 may be related to the occurrence of endometrial cancers and premalignant lesions 27636375_Overexpression of the NUP88 is associated with cancer. 29724197_Study in HeLa cells revealed that Nup88 can affect the phosphorylation status of vimentin, which may contribute to the Nup88-dependent multinucleated phenotype through changing the organization of vimentin. 30543681_Here we report biallelic mutations in the nucleoporin NUP88 as a novel cause of lethal fetal akinesia deformation sequence (FADS) in two families. We demonstrate that NUP88 depletion affects rapsyn, a key regulator of the muscle nicotinic acetylcholine receptor at the neuromuscular junction. 31186352_Loss of Nup88 inhibited nuclear export of recombination signal-binding protein for immunoglobulin kappaJ region (RBP-J), the DNA-binding component of the Notch pathway. 34331103_Overexpression of the nucleoporin Nup88 stimulates migration and invasion of HeLa cells. 34729665_In focus in HCB. | ENSMUSG00000040667 | Nup88 | 7.716338e+02 | 0.8175366 | -0.290644764 | 0.3100166 | 8.893373e-01 | 0.3456569475 | 0.82969965 | No | Yes | 7.345834e+02 | 126.722351 | 8.642051e+02 | 152.763230 | |
| ENSG00000108588 | 57003 | CCDC47 | protein_coding | Q96A33 | FUNCTION: Component of the PAT complex, an endoplasmic reticulum (ER)-resident membrane multiprotein complex that facilitates multi-pass membrane proteins insertion into membranes (PubMed:32814900). The PAT complex acts as an intramembrane chaperone by directly interacting with nascent transmembrane domains (TMDs), releasing its substrates upon correct folding, and is needed for optimal biogenesis of multi-pass membrane proteins (PubMed:32814900). WDR83OS/Asterix is the substrate-interacting subunit of the PAT complex, whereas CCDC47 is required to maintain the stability of WDR83OS/Asterix (PubMed:32814900). The PAT complex favors the binding to TMDs with exposed hydrophilic amino acids within the lipid bilayer and provides a membrane-embedded partially hydrophilic environment in which the first transmembrane domain binds (PubMed:32814900). Component of a ribosome-associated ER translocon complex involved in multi-pass membrane protein transport into the ER membrane and biogenesis (PubMed:32820719). Involved in the regulation of calcium ion homeostasis in the ER (PubMed:30401460). Required for proper protein degradation via the ERAD (ER-associated degradation) pathway (PubMed:25009997). Has an essential role in the maintenance of ER organization during embryogenesis (By similarity). {ECO:0000250|UniProtKB:Q9D024, ECO:0000269|PubMed:25009997, ECO:0000269|PubMed:30401460, ECO:0000269|PubMed:32814900, ECO:0000269|PubMed:32820719}. | 3D-structure;Alternative splicing;Chaperone;Coiled coil;Disease variant;Endoplasmic reticulum;Glycoprotein;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:57003; | endoplasmic reticulum [GO:0005783]; integral component of endoplasmic reticulum membrane [GO:0030176]; membrane [GO:0016020]; rough endoplasmic reticulum membrane [GO:0030867]; calcium ion binding [GO:0005509]; protein folding chaperone [GO:0044183]; ribosome binding [GO:0043022]; RNA binding [GO:0003723]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; ER overload response [GO:0006983]; ERAD pathway [GO:0036503]; osteoblast differentiation [GO:0001649]; post-embryonic development [GO:0009791]; protein insertion into ER membrane [GO:0045048]; ubiquitin-dependent ERAD pathway [GO:0030433] | 25009997_these findings suggested that calumin serves to maintain the yolk sac integrity through participation in the ERAD activity, contributing to embryonic development. | ENSMUSG00000078622 | Ccdc47 | 1.939545e+03 | 1.1890643 | 0.249826791 | 0.3154491 | 6.209463e-01 | 0.4306958610 | 0.85086128 | No | Yes | 2.170692e+03 | 379.221011 | 1.541608e+03 | 276.333226 | ||
| ENSG00000108602 | 218 | ALDH3A1 | protein_coding | P30838 | FUNCTION: ALDHs play a major role in the detoxification of alcohol-derived acetaldehyde (Probable). They are involved in the metabolism of corticosteroids, biogenic amines, neurotransmitters, and lipid peroxidation (Probable). Oxidizes medium and long chain aldehydes into non-toxic fatty acids (PubMed:1737758). Preferentially oxidizes aromatic aldehyde substrates (PubMed:1737758). Comprises about 50 percent of corneal epithelial soluble proteins (By similarity). May play a role in preventing corneal damage caused by ultraviolet light (By similarity). {ECO:0000250|UniProtKB:P47739, ECO:0000269|PubMed:1737758, ECO:0000305}. | 3D-structure;Acetylation;Cytoplasm;Direct protein sequencing;Lipid metabolism;NAD;NADP;Oxidoreductase;Reference proteome | Aldehyde dehydrogenases oxidize various aldehydes to the corresponding acids. They are involved in the detoxification of alcohol-derived acetaldehyde and in the metabolism of corticosteroids, biogenic amines, neurotransmitters, and lipid peroxidation. The enzyme encoded by this gene forms a cytoplasmic homodimer that preferentially oxidizes aromatic and medium-chain (6 carbons or more) saturated and unsaturated aldehyde substrates. It is thought to promote resistance to UV and 4-hydroxy-2-nonenal-induced oxidative damage in the cornea. The gene is located within the Smith-Magenis syndrome region on chromosome 17. Multiple alternatively spliced variants, encoding the same protein, have been identified. [provided by RefSeq, Sep 2008]. | hsa:218; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; 3-chloroallyl aldehyde dehydrogenase activity [GO:0004028]; alcohol dehydrogenase (NADP+) activity [GO:0008106]; aldehyde dehydrogenase (NAD+) activity [GO:0004029]; aldehyde dehydrogenase [NAD(P)+] activity [GO:0004030]; benzaldehyde dehydrogenase (NAD+) activity [GO:0018479]; cellular aldehyde metabolic process [GO:0006081]; lipid metabolic process [GO:0006629]; xenobiotic metabolic process [GO:0006805] | 9855707_These findings suggest that a high level of expression of ALDH3 in cancerous liver tissues resulted from the expression or activation of at least two nuclear proteins reacting to the ALDH3 promoter region. 12367788_ADH3*1 allele associated with high ADH activity and acetyaldehyde-DNA adducts in lung; may be a risk factor for lung cancer 12706498_ALDH3A1 is a regulatory element of the cellular defense system that protects corneal epithelium against UV-induced oxidative damage 12943535_Enzyme activity and localization of ALDH3A1 in the cornea. 14745454_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15905174_ALDH3A1 may protect corneal epithelial cells against oxidative damage not only through its metabolic function but also by prolonging the cell cycle 18496131_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18536178_In most cases, ALDH3A1 activity in tumor and bordering tissues was higher than in a control group 18621017_These kinetic properties ensure that ALDHs, and particularly ALDH2, can complete the ADH-mediated detoxification. 18854779_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19025616_specific down regulation of ALDH1A1 and ALDH3A1 in Lenti 1+3 cells and in comparison to 12 other ALDH genes detected 19058789_Observational study of gene-disease association. (HuGE Navigator) 19343046_Observational study of gene-disease association. (HuGE Navigator) 19360691_Observational study of gene-disease association. (HuGE Navigator) 19696793_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19894643_the mechanism of salivary ALDH3A1 inactivation must be related to free radical activity, presumably in the salivary gland, but it is not simply correlated with antioxidatn capacity of saliva 20056567_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20102361_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21203538_the UV-induced inactivation of ALDH3A1 is a result of non-native aggregation and associated structural changes rather than specific damage to the active site Cys 21229876_Antihypertensives, nonopioid analgesics and hormonal contraceptives significantly affect the salivary ALDH3A1 activity. 21251908_through modulation of PPARgamma or ALDH3A1, it may be possible to reduce cell proliferation in tumor cells or stimulate cell proliferation in normal cells during tissue regeneration. 22406320_ALDH3A1 provides exceptional protection from the adverse effects of pathophysiological concentrations of 4-HNE such as may occur during periods of oxidative stress 24276407_No correlation between ALDH3A1 expression and patient survival or tumour recurrence was observed.In conclusion, ALDH3A1 is a marker of activation of the Wnt/ss-catenin pathway in hepatocellular carcinoma 24316006_SiRNA-mediated suppression of ALDH3A1 blocked ALDH enzymatic activity and augmented cytotoxicity in CSE-exposed cells. 24762960_While ALDH3A1 was not found in prostate glands, it was present in prostatic intraepithelial neoplasia, further increased in carcinomas, and upregulated in lymphatic metastases. ALDH3A1 increased in DU145-cell-derived lung metastasis vs. local xenografts. 25221425_Reported increased expression of ALDH3A1, PDIA3, and PRDX2 in pterygia using a proteomic approach. These proteins are presumed to have a protective role against oxidative stress-induced apoptosis. 26124079_Down-regulation of ALDH3 activity in trophoblast stem cells initiates trophoblast differentiation during placental development. 26751691_ALDH3A1 has a role in the maintenance of corneal epithelial homeostasis by simultaneously modulating proliferation and differentiation through both enzymatic and non-enzymatic mechanisms 27276244_ALDH3A1 confers a multi-modality resistance phenotype in MCF-7 cells 27279633_ALDH-3A1 expression correlates well with gastric cancer dysplasia and grades, differentiation, lymph node metastasis and cancer stage. 27997560_Molecular docking analysis depicted that Sulforaphane fits into the active site of ALDH3A1, and facilitates the catalytic mechanism of the enzyme. 28526614_ALDH3A1 appears to play an essential role in protecting cellular proteins against aggregation under stress conditions 31138432_higher expression in fetal growth restriction-associated placentas but localized specifically to extravillous trophoblasts 31705020_Gastric cancer depends on aldehyde dehydrogenase 3A1 for fatty acid oxidation. 32006654_Aldehyde dehydrogenase 3A1 confers oxidative stress resistance accompanied by altered DNA damage response in human corneal epithelial cells. 32297178_Exosomes carrying ALDOA and ALDH3A1 from irradiated lung cancer cells enhance migration and invasion of recipients by accelerating glycolysis. 33596621_Association with Corneal Remodeling Related Genes, ALDH3A1, LOX, and SPARC Genes Variations in Korean Keratoconus Patients. | ENSMUSG00000019102 | Aldh3a1 | 4.201479e+01 | 1.5284392 | 0.612059133 | 0.4817742 | 1.627400e+00 | 0.2020630011 | 0.78445471 | No | Yes | 4.722112e+01 | 9.873532 | 2.970650e+01 | 6.736166 | |
| ENSG00000109180 | 54940 | OCIAD1 | protein_coding | Q9NX40 | FUNCTION: Maintains stem cell potency (By similarity). Increases STAT3 phosphorylation and controls ERK phosphorylation (By similarity). May act as a scaffold, increasing STAT3 recruitment onto endosomes (By similarity). Involved in integrin-mediated cancer cell adhesion and colony formation in ovarian cancer (PubMed:20515946). {ECO:0000250|UniProtKB:Q9CRD0, ECO:0000269|PubMed:20515946}. | Alternative splicing;Endosome;Phosphoprotein;Reference proteome | hsa:54940; | endosome [GO:0005768]; Golgi apparatus [GO:0005794]; lysosome [GO:0005764]; membrane [GO:0016020]; endocytosis [GO:0006897]; hematopoietic stem cell homeostasis [GO:0061484]; positive regulation of receptor signaling pathway via JAK-STAT [GO:0046427]; regulation of stem cell differentiation [GO:2000736] | 18328549_Over-expressed in metastatic ovarian cancer. Effect of OCIAD1 on cell adhesion may be related to its function in ovarian cancer. Possibility of OCIAD1's role in tumor metastasis. 20515946_This is the first study to indicate that OCIAD1 is a key player in generating ovarian cancer recurrence. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22081784_OCIAD1 is a potential biomarker of thyroid carcinoma but had no significant additive effect on the risk of distant metastasis. 22726067_Like OCIAD1, OCIAD2 is a cancer-related protein and its expression level increases during the course of malignant progression and is thought to be a very useful marker for evaluating the malignancy of ovarian mucinous tumors. 31221523_high OCIAD1 levels in pancreatic ductal adenocarcinoma promoted tumor cells migration. OCIAD1 exerted its effects by regulating ATM 31931285_OCIAD1 contributes to neurodegeneration in Alzheimer's disease by inducing mitochondria dysfunction, neuronal vulnerability and synaptic damages. 32697788_OCIAD1 is a host mitochondrial substrate of the hepatitis C virus NS3-4A protease. 34034859_Genome-wide CRISPRi screening identifies OCIAD1 as a prohibitin client and regulatory determinant of mitochondrial Complex III assembly in human cells. | ENSMUSG00000029152 | Ociad1 | 1.113504e+03 | 1.2005536 | 0.263699821 | 0.3370990 | 6.164789e-01 | 0.4323587733 | 0.85164585 | No | Yes | 1.121447e+03 | 190.266935 | 9.350218e+02 | 162.689681 | ||
| ENSG00000109184 | 23142 | DCUN1D4 | protein_coding | Q92564 | FUNCTION: Contributes to the neddylation of all cullins by transferring NEDD8 from N-terminally acetylated NEDD8-conjugating E2s enzyme to different cullin C-terminal domain-RBX complexes which are necessary for the activation of cullin-RING E3 ubiquitin ligases (CRLs). {ECO:0000269|PubMed:23201271, ECO:0000269|PubMed:26906416}. | 3D-structure;Alternative splicing;Isopeptide bond;Nucleus;Reference proteome;Ubl conjugation | hsa:23142; | nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; cullin family protein binding [GO:0097602]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin-like protein binding [GO:0032182]; positive regulation of protein neddylation [GO:2000436]; positive regulation of ubiquitin-protein transferase activity [GO:0051443]; protein neddylation [GO:0045116] | ENSMUSG00000051674 | Dcun1d4 | 3.586648e+02 | 0.5512165 | -0.859309015 | 0.3315267 | 6.486240e+00 | 0.0108712660 | 0.37978104 | No | Yes | 2.474077e+02 | 39.343770 | 4.579604e+02 | 73.733416 | |||
| ENSG00000109255 | 10874 | NMU | protein_coding | P48645 | FUNCTION: [Neuromedin-U-25]: Ligand for receptors NMUR1 and NMUR2 (By similarity). Stimulates muscle contractions of specific regions of the gastrointestinal tract. In humans, NmU stimulates contractions of the ileum and urinary bladder. {ECO:0000250|UniProtKB:P12760}.; FUNCTION: [Neuromedin precursor-related peptide 33]: Does not function as a ligand for either NMUR1 or NMUR2. Indirectly induces prolactin release although its potency is much lower than that of neuromedin precursor-related peptide 36. {ECO:0000250|UniProtKB:P12760}.; FUNCTION: [Neuromedin precursor-related peptide 36]: Does not function as a ligand for either NMUR1 or NMUR2. Indirectly induces prolactin release from lactotroph cells in the pituitary gland, probably via the hypothalamic dopaminergic system. {ECO:0000250|UniProtKB:P12760}. | Amidation;Cleavage on pair of basic residues;Neuropeptide;Oxidation;Reference proteome;Secreted;Signal | This gene encodes a member of the neuromedin family of neuropeptides. The encoded protein is a precursor that is proteolytically processed to generate a biologically active neuropeptide that plays a role in pain, stress, immune-mediated inflammatory diseases and feeding regulation. Increased expression of this gene was observed in renal, pancreatic and lung cancers. Alternative splicing results in multiple transcript variants encoding different isoforms. Some of these isoforms may undergo similar processing to generate the mature peptide. [provided by RefSeq, Jul 2015]. | hsa:10874; | extracellular region [GO:0005576]; terminal bouton [GO:0043195]; neuromedin U receptor binding [GO:0042922]; signaling receptor binding [GO:0005102]; type 1 neuromedin U receptor binding [GO:0031839]; type 2 neuromedin U receptor binding [GO:0031840]; energy homeostasis [GO:0097009]; G protein-coupled receptor signaling pathway [GO:0007186]; neuropeptide signaling pathway [GO:0007218]; positive regulation of smooth muscle contraction [GO:0045987]; positive regulation of synaptic transmission [GO:0050806]; regulation of feeding behavior [GO:0060259]; regulation of grooming behavior [GO:2000821]; temperature homeostasis [GO:0001659] | 14623274_These results suggest that NMU plays a role in feeding behavior and catabolic functions via corticotropin-releasing hormone. 15187020_NmU expression is related to Myb and that the NmU/NMU1R axis constitutes a previously unknown growth-promoting autocrine loop in myeloid leukemia cells 15331768_irreversible binding of NmU to its receptors 16878152_Overexpression of neuromedin U is associated with bladder tumor formation, lung metastasis and cancer cachexia 16984985_Amino acid variants in NMU associate with overweight and obesity, suggesting that NMU is involved in energy regulation in humans. 16984985_Observational study of gene-disease association. (HuGE Navigator) 17018595_NMU & its cancer-specific receptors, as well as its target genes, are frequently overexpressed in clinical samples of lung cancer and in cell lines, and that those gene products play indispensable roles in the growth and progression of lung cancer cells. 19118941_NmU may be involved in the HGF-c-Met paracrine loop regulating cell migration, invasiveness and dissemination of pancreatic ductal adenocarcinoma. 19519756_[review] Taken together with its vascular actions, NMU may be a functional link between energy balance and the cardiovascular system and may provide a future target for therapies directed against the disorders that comprise metabolic syndrome. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21791076_Inactivation of the von Hippel-Lindau tumour suppressor gene induces Neuromedin U expression in renal cancer cells. 23936460_a role for NMU gene through interaction with ADRB2 gene in bone strength regulation 24876102_our results defined NmU as a candidate drug response biomarker for HER2-overexpressing cancers and as a candidate therapeutic target to limit metastatic progression and improve the efficacy of HER-targeted drugs. 25871004_polymorphisms in the gene may be useful in identifying women at risk for osteoporosis. 27279246_Overexpression of Nmu may be involved in the process of regional metastasis of HNSCC, and may serve as a novel and valuable biomarker for predicting regional metastasis in patients with HNSCC. 28235053_This study shows an association between a NMU haplotype and anthropometric indices, mainly linked to fat mass, which appears to be age- and sex-specific in children. 28340506_Results indicate a mechanism of action for neuromedin U (NmU) in HER2-overexpressing breast cancer that enhances resistance to HER2-targeted drugs through conferring cancer stem cell (CSC) characteristics and expansion of the CSC phenotype. 28423716_NMU might contribute to progression of NMUR2-positive breast cancer 29107108_The anti-tumorigenic effect of HAND2-AS1 in endometrioid endometrial carcinoma was mediated by down-regulating neuromedin U expression. 30096454_Results identified NMU as highly expressed in non-small cell lung cancer (NSCLC) tissues and cell lines. Furthermore, integrative bioinformatics analyses demonstrate that NMU may confer the alectinib resistance in NSCLC via multiple mechanisms. 31326439_Association between variants of neuromedin U gene and taste thresholds and food preferences in European children: Results from the IDEFICS study. 32013887_The prognostic value of neuromedin U in patients with hepatocellular carcinoma. | ENSMUSG00000029236 | Nmu | 1.112608e+02 | 1.0129796 | 0.018605171 | 0.3799093 | 2.473947e-03 | 0.9603305475 | 0.99184539 | No | Yes | 9.881514e+01 | 15.942669 | 9.951619e+01 | 16.379336 | |
| ENSG00000109475 | 6164 | RPL34 | protein_coding | P49207 | FUNCTION: Component of the large ribosomal subunit. {ECO:0000269|PubMed:23636399, ECO:0000269|PubMed:25901680, ECO:0000269|PubMed:25957688, ECO:0000305|PubMed:12962325}. | 3D-structure;Acetylation;Cytoplasm;Direct protein sequencing;Endoplasmic reticulum;Isopeptide bond;Phosphoprotein;Reference proteome;Ribonucleoprotein;Ribosomal protein;Ubl conjugation | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 60S subunit. The protein belongs to the L34E family of ribosomal proteins. It is located in the cytoplasm. This gene originally was thought to be located at 17q21, but it has been mapped to 4q. Overexpression of this gene has been observed in some cancer cells. Alternative splicing results in multiple transcript variants, all encoding the same isoform. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome. [provided by RefSeq, Feb 2016]. | hsa:6164; | cytosol [GO:0005829]; cytosolic large ribosomal subunit [GO:0022625]; cytosolic ribosome [GO:0022626]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; cadherin binding [GO:0045296]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; cytoplasmic translation [GO:0002181]; translation [GO:0006412] | 20332099_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 26323242_RPL34 plays a critical role in cell proliferation, cell cycle distribution and apoptosis of human malignant gastric cells. 26526135_over-expressed RPL34 may promote malignant proliferation of NSCLC cells, thus playing an important role in development and progress of NSCLC 27845896_this study shows that ribosomal protein L34 promotes the proliferation, invasion and metastasis of pancreatic cancer cells 27883047_RPL34 plays an important role in the proliferation of osteosarcoma cells. 28109079_RPL34 functions as an oncogene that modulates the proliferation and metastasis of esophageal cancer cells in part through the inactivation of the PI3K/Akt signaling pathway 28697409_The RPL34 gene was highly expressed in OSCC, while silencing RPL34 could block cell proliferation and metastasis, but promote cell apoptosis, suggesting the RPL34 gene to be a new promising clinical target for OSCC therapy. 30103068_The present study combines an individual-based phenotypic profiling with a transdiagnostic approach and shows that several ribosomal genes including RPL34 is involved in stress vulnerability across nonclinical and clinical conditions. 30216512_Findings indicated that knockdown of RPL34 inhibits the proliferation and migration of glioma cells through the inactivation of JAK/STAT3 signaling pathway. 33675124_RPL34-AS1-induced RPL34 inhibits cervical cancer cell tumorigenesis via the MDM2-P53 pathway. | ENSMUSG00000062006 | Rpl34 | 5.169993e+03 | 0.5283619 | -0.920401547 | 0.2922737 | 9.896269e+00 | 0.0016561421 | 0.15688645 | No | Yes | 3.883713e+03 | 685.490017 | 6.563803e+03 | 1187.404403 | |
| ENSG00000109586 | 51809 | GALNT7 | protein_coding | Q86SF2 | FUNCTION: Glycopeptide transferase involved in O-linked oligosaccharide biosynthesis, which catalyzes the transfer of an N-acetyl-D-galactosamine residue to an already glycosylated peptide. In contrast to other proteins of the family, it does not act as a peptide transferase that transfers GalNAc onto serine or threonine residue on the protein receptor, but instead requires the prior addition of a GalNAc on a peptide before adding additional GalNAc moieties. Some peptide transferase activity is however not excluded, considering that its appropriate peptide substrate may remain unidentified. {ECO:0000269|PubMed:10544240, ECO:0000269|PubMed:11925450}. | 3D-structure;Disulfide bond;Glycosyltransferase;Golgi apparatus;Lectin;Manganese;Membrane;Metal-binding;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000305|PubMed:10544240, ECO:0000305|PubMed:11925450}. | This gene encodes GalNAc transferase 7, a member of the GalNAc-transferase family. The enzyme encoded by this gene controls the initiation step of mucin-type O-linked protein glycosylation and transfer of N-acetylgalactosamine to serine and threonine amino acid residues. This enzyme is a type II transmembrane protein and shares common sequence motifs with other family members. Unlike other family members, this enzyme shows exclusive specificity for partially GalNAc-glycosylated acceptor substrates and shows no activity with non-glycosylated peptides. This protein may function as a follow-up enzyme in the initiation step of O-glycosylation. [provided by RefSeq, Jul 2008]. | hsa:51809; | extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; carbohydrate binding [GO:0030246]; metal ion binding [GO:0046872]; polypeptide N-acetylgalactosaminyltransferase activity [GO:0004653]; carbohydrate metabolic process [GO:0005975]; O-glycan processing [GO:0016266]; protein O-linked glycosylation [GO:0006493] | 19844573_MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7 21741600_Ectopic expression of miR-30b/30d promoted the metastatic behavior of melanoma cells by directly targeting the GalNAc transferase GALNT7. 22399294_miR-214 is a new regulator of GALNT7, and both miR-214 and GALNT7 play important roles in the pathogenesis of cervical cancer 24482044_The study of miR-34a, miR-34c and its novel target, GALNT7, may serve as novel potential makers for laryngeal squamous cell carcinoma therapy. 25809707_GALNT7 and CDK16 were confirmed to be the direct targets of miR-494. These results suggested that miR-494 play an inhibitory role in the tumorigenesis of NPC 26503214_miR-494 and GALNT7 play oncogenic roles in nasopharyngeal carcinoma. 27619677_High expression of GALNT7 promotes migration and invasion and lymph node metastasis in esophageal squamous cell carcinoma. 29970122_Results indicated that SNHG7 facilitated the proliferation and metastasis as a competing endogenous RNA to regulate GALNT7 expression by sponging miR-34a in CRC cell lines. 30685086_crystallographic analysis of GalNAc-T7, a GalNAc-T capable of glycosylating consecutive sites, and of its complex with the donor substrate UDP-GalNAc 32416102_Long non-coding RNA TP73-AS1 contributes to glioma tumorigenesis by sponging the miR-103a/GALNT7 pathway. | ENSMUSG00000031608 | Galnt7 | 2.780432e+02 | 1.1351547 | 0.182888897 | 0.3689064 | 2.445114e-01 | 0.6209665428 | 0.91517038 | No | Yes | 2.985083e+02 | 58.610982 | 2.271285e+02 | 45.856543 |
| ENSG00000109756 | 9693 | RAPGEF2 | protein_coding | Q9Y4G8 | FUNCTION: Functions as a guanine nucleotide exchange factor (GEF), which activates Rap and Ras family of small GTPases by exchanging bound GDP for free GTP in a cAMP-dependent manner. Serves as a link between cell surface receptors and Rap/Ras GTPases in intracellular signaling cascades. Acts also as an effector for Rap1 by direct association with Rap1-GTP thereby leading to the amplification of Rap1-mediated signaling. Shows weak activity on HRAS. It is controversial whether RAPGEF2 binds cAMP and cGMP (PubMed:23800469, PubMed:10801446) or not (PubMed:10608844, PubMed:10548487, PubMed:11359771). Its binding to ligand-activated beta-1 adrenergic receptor ADRB1 leads to the Ras activation through the G(s)-alpha signaling pathway. Involved in the cAMP-induced Ras and Erk1/2 signaling pathway that leads to sustained inhibition of long term melanogenesis by reducing dendrite extension and melanin synthesis. Provides also inhibitory signals for cell proliferation of melanoma cells and promotes their apoptosis in a cAMP-independent nanner. Regulates cAMP-induced neuritogenesis by mediating the Rap1/B-Raf/ERK signaling through a pathway that is independent on both PKA and RAPGEF3/RAPGEF4. Involved in neuron migration and in the formation of the major forebrain fiber connections forming the corpus callosum, the anterior commissure and the hippocampal commissure during brain development. Involved in neuronal growth factor (NGF)-induced sustained activation of Rap1 at late endosomes and in brain-derived neurotrophic factor (BDNF)-induced axon outgrowth of hippocampal neurons. Plays a role in the regulation of embryonic blood vessel formation and in the establishment of basal junction integrity and endothelial barrier function. May be involved in the regulation of the vascular endothelial growth factor receptor KDR and cadherin CDH5 expression at allantois endothelial cell-cell junctions. {ECO:0000269|PubMed:10548487, ECO:0000269|PubMed:10608844, ECO:0000269|PubMed:10608883, ECO:0000269|PubMed:10801446, ECO:0000269|PubMed:10934204, ECO:0000269|PubMed:11359771, ECO:0000269|PubMed:12391161, ECO:0000269|PubMed:16272156, ECO:0000269|PubMed:17724123, ECO:0000269|PubMed:21840392, ECO:0000269|PubMed:23800469}. | 3D-structure;Cell junction;Cell membrane;Cytoplasm;Developmental protein;Differentiation;Endosome;Epilepsy;GTPase activation;Guanine-nucleotide releasing factor;Membrane;Neurogenesis;Phosphoprotein;Reference proteome;Ubl conjugation | Members of the RAS (see HRAS; MIM 190020) subfamily of GTPases function in signal transduction as GTP/GDP-regulated switches that cycle between inactive GDP- and active GTP-bound states. Guanine nucleotide exchange factors (GEFs), such as RAPGEF2, serve as RAS activators by promoting acquisition of GTP to maintain the active GTP-bound state and are the key link between cell surface receptors and RAS activation (Rebhun et al., 2000 [PubMed 10934204]).[supplied by OMIM, Mar 2008]. | hsa:9693; | apical plasma membrane [GO:0016324]; bicellular tight junction [GO:0005923]; cell-cell junction [GO:0005911]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endocytic vesicle [GO:0030139]; integral component of plasma membrane [GO:0005887]; late endosome [GO:0005770]; membrane [GO:0016020]; neuron projection [GO:0043005]; neuronal cell body [GO:0043025]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; synapse [GO:0045202]; beta-1 adrenergic receptor binding [GO:0031697]; calcium ion binding [GO:0005509]; cAMP binding [GO:0030552]; diacylglycerol binding [GO:0019992]; GTPase activator activity [GO:0005096]; guanyl-nucleotide exchange factor activity [GO:0005085]; PDZ domain binding [GO:0030165]; phosphatidic acid binding [GO:0070300]; WW domain binding [GO:0050699]; adenylate cyclase-activating adrenergic receptor signaling pathway [GO:0071880]; blood vessel development [GO:0001568]; brain-derived neurotrophic factor receptor signaling pathway [GO:0031547]; cAMP-mediated signaling [GO:0019933]; cellular response to cAMP [GO:0071320]; cellular response to cGMP [GO:0071321]; cellular response to nerve growth factor stimulus [GO:1990090]; establishment of endothelial barrier [GO:0061028]; establishment of endothelial intestinal barrier [GO:0090557]; forebrain neuron development [GO:0021884]; G protein-coupled receptor signaling pathway [GO:0007186]; intracellular signal transduction [GO:0035556]; MAPK cascade [GO:0000165]; microvillus assembly [GO:0030033]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of dendrite morphogenesis [GO:0050774]; negative regulation of melanin biosynthetic process [GO:0048022]; nerve growth factor signaling pathway [GO:0038180]; neuron migration [GO:0001764]; neuron projection development [GO:0031175]; neuropeptide signaling pathway [GO:0007218]; positive regulation of cAMP-dependent protein kinase activity [GO:2000481]; positive regulation of cAMP-mediated signaling [GO:0043950]; positive regulation of dendritic cell apoptotic process [GO:2000670]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of GTPase activity [GO:0043547]; positive regulation of neuron migration [GO:2001224]; positive regulation of neuron projection development [GO:0010976]; positive regulation of protein binding [GO:0032092]; positive regulation of protein kinase activity [GO:0045860]; positive regulation of vasculogenesis [GO:2001214]; protein localization to plasma membrane [GO:0072659]; Rap protein signal transduction [GO:0032486]; regulation of cell junction assembly [GO:1901888]; regulation of synaptic plasticity [GO:0048167]; small GTPase mediated signal transduction [GO:0007264]; ventricular system development [GO:0021591] | 11799111_Regulation of G protein-linked guanine nucleotide exchange factors for Rho, PDZ-RhoGEF, and LARG by tyrosine phosphorylation: evidence of a role for focal adhesion kinase 12196628_PSD-95/Dlg/ZO-1 homology (PDZ)-RhoGEF was isolated from mouse brain as a plexin-B1-specific interacting protein 17724123_Data show that the interaction of PDZ-GEF1 with an internalized neurotrophin receptor transported to late endosomes induces sustained activation of both Rap1 and ERK and neurite outgrowth. 21840392_PDZ-GEF activates Rap1 under resting conditions to stabilize cell-cell junctions and maintain basal integrity 22422452_DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. 23800469_Data indicate that expression of Rapgef2 in embryonic kidney 293T cells enables cAMP-Rap1-ERK signaling. 23885123_JAM-A regulates epithelial permeability via association with ZO-2, afadin, and PDZ-GEF1 to activate Rap2c and control contraction of the apical cytoskeleton. 24290981_We report that in response to factors that promote cell motility, the Rap guanine exchange factor RAPGEF2 is rapidly phosphorylated by I-kappa-B-kinase-beta and casein kinase-1alpha and consequently degraded by the proteasome. 24340059_NEDD4-1 regulates cell migration and invasion through ubiquitination of CNrasGEF in vitro. 24858808_local accumulation of PDZGEF at the apical membrane during establishment of epithelial polarity is mediated by electrostatic interactions between positively charged side chains in the PDZ domain and negatively charged phosphatidic acid. 25189171_Cdk5-mediated phosphorylation of RapGEF2 controls neuronal migration in the developing cerebral cortex. 25332235_SCF(FBXW11) bound, polyubiquitylated, and destabilized RAPGEF2, a guanine nucleotide exchange factor that activates the small GTPase RAP1 31171376_Disruption of MAGI2-RapGEF2-Rap1 signaling contributes to podocyte dysfunction in congenital nephrotic syndrome caused by mutations in MAGI2. 33345400_RAPGEF2 mediates oligomeric Abeta-induced synaptic loss and cognitive dysfunction in the 3xTg-AD mouse model of Alzheimer's disease. | ENSMUSG00000062232 | Rapgef2 | 6.180309e+02 | 0.8757594 | -0.191393508 | 0.3122127 | 3.786326e-01 | 0.5383359717 | 0.88950730 | No | Yes | 6.284389e+02 | 107.185794 | 6.468795e+02 | 113.206071 | |
| ENSG00000110080 | 6484 | ST3GAL4 | protein_coding | Q11206 | FUNCTION: A beta-galactoside alpha2-3 sialyltransferase involved in terminal sialylation of glycoproteins and glycolipids (PubMed:8288606, PubMed:8611500). Catalyzes the transfer of sialic acid (N-acetyl-neuraminic acid; Neu5Ac) from the nucleotide sugar donor CMP-Neu5Ac onto acceptor Galbeta-(1->3)-GalNAc- and Galbeta-(1->4)-GlcNAc-terminated glycoconjugates through an alpha2-3 linkage (PubMed:8288606, PubMed:8611500). Plays a major role in hemostasis. Responsible for sialylation of plasma VWF/von Willebrand factor, preventing its recognition by asialoglycoprotein receptors (ASGPR) and subsequent clearance. Regulates ASGPR-mediated clearance of platelets (By similarity). Participates in the biosynthesis of the sialyl Lewis X epitopes, both on O- and N-glycans, which are recognized by SELE/E-selectin, SELP/P-selectin and SELL/L-selectin. Essential for selectin-mediated rolling and adhesion of leukocytes during extravasation (PubMed:25498912). Contributes to adhesion and transendothelial migration of neutrophils likely through terminal sialylation of CXCR2 (By similarity). In glycosphingolipid biosynthesis, sialylates GM1 and GA1 gangliosides to form GD1a and GM1b, respectively (PubMed:8288606). Metabolizes brain c-series ganglioside GT1c forming GQ1c (By similarity). Synthesizes ganglioside LM1 (IV3Neu5Ac-nLc4Cer), a major structural component of peripheral nerve myelin (PubMed:8611500). {ECO:0000250|UniProtKB:P61131, ECO:0000250|UniProtKB:Q91Y74, ECO:0000269|PubMed:25498912, ECO:0000269|PubMed:8288606, ECO:0000269|PubMed:8611500}. | Alternative splicing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Lipid metabolism;Membrane;Reference proteome;Secreted;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000305|PubMed:8288606}.; PATHWAY: Glycolipid biosynthesis. {ECO:0000305|PubMed:8288606}. | This gene encodes a member of the glycosyltransferase 29 family, a group of enzymes involved in protein glycosylation. The encoded protein is targeted to Golgi membranes but may be proteolytically processed and secreted. The gene product may also be involved in the increased expression of sialyl Lewis X antigen seen in inflammatory responses. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2011]. | hsa:6484; | extracellular region [GO:0005576]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; beta-galactoside (CMP) alpha-2,3-sialyltransferase activity [GO:0003836]; monosialoganglioside sialyltransferase activity [GO:0047288]; N-acetyllactosaminide alpha-2,3-sialyltransferase activity [GO:0008118]; neolactotetraosylceramide alpha-2,3-sialyltransferase activity [GO:0004513]; sialyltransferase activity [GO:0008373]; cognition [GO:0050890]; glycolipid biosynthetic process [GO:0009247]; glycoprotein biosynthetic process [GO:0009101]; keratan sulfate biosynthetic process [GO:0018146]; lipid glycosylation [GO:0030259]; O-glycan processing [GO:0016266]; oligosaccharide biosynthetic process [GO:0009312]; positive regulation of blood coagulation [GO:0030194]; positive regulation of leukocyte tethering or rolling [GO:1903238]; protein glycosylation [GO:0006486]; protein sialylation [GO:1990743]; sialylation [GO:0097503]; viral protein processing [GO:0019082] | 12375029_down-regulation of ST3Gal IV mRNA may be one of the factors associated with the malignant progression of human renal cell carcinoma. 12565846_Transcriptional regulation of human Galbeta1,3GalNAc/Galbeta1, 4GlcNAc alpha2,3-sialyltransferase (hST3Gal IV) gene in testis and ovary cell lines. 17054948_IL-1 beta-induced sLeX expression on HuH-7 cells was suppressed by transfection of gene-specific small interference RNAs against FUT VI and ST3Gal IV but not against FUT IV and ST3Gal III. 19520807_Thrombocytopenia in mice deficient in the St3gal4 sialyltransferase gene (St3Gal-IV(-/-) mice) is caused by the recognition of terminal galactose residues exposed on the platelet surface in the absence of sialylation. 19781661_Expression of ST3Gal IV in several gastrointestinal cell lines is correlated with the expression of sialyl Lewis x at the cell surface. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20383541_These data suggest that the ST3GAL4 gene is responsible for biosynthesis of the viral receptor and may play a crucial role in infection of H5N1 avian influenza virus in humans. 21140242_These results indicated that gastric cancer tissues expressed high levels of alpha 2,3-linked sialic acid residues, ST3Gal IV, and ST6Gal I. 22691873_TNF increases the expression of alpha2,3-sialyltransferase gene ST3GAL4 23726834_the role of ST3Gal IV in the acquisition of adhesive, migratory and metastatic capabilities and, secondly, in analyzing the expression of ST3Gal III and ST3Gal IV in pancreatic adenocarcinoma tissues, was investigated. 23799130_The expression of ST3GAL4 leads to SLe(x) antigen expression in gastric cancer cells which in turn induces an increased invasive phenotype through the activation of c-Met 24606438_The polymorphism analysis showed that at SNP rs10893506, genotypes CC and CT of the ST3GAL4 B3 promoter were associated with the presence of premalignant lesions and cervical cancer. 25086711_There may be a racial/ethnic-specific association, and/or sex-specific association between the ST3GAL4 rs11220462 SNP and serum lipid parameters in some ethnic groups. 25498912_ST3Gal-4 is the primary sialyltransferase regulating the synthesis of E-, P-, and L-selectin ligands on human myeloid leukocytes 26258617_Expression levels of sialyltransferases ST3GAL1 and ST3GAL4 were upregulated in the HRMECs after high-glucose stimulation. 27584569_study illustrates the power of next-generation sequencing in the discovery of new genetic variants and a significant ethnic diversity in the ST3GAL4 gene 27821620_TNF-responsive element in an intronic region of the ST3GAL4 gene, whose TNF-dependent activity is repressed by ERK/p38 and MSK1/2 inhibitors. This TNF-responsive element contains potential binding sites for ETS1 and ATF2 transcription factors related to TNF signaling. 28512058_ST3GAL IV affects apoptotic signal, cell proliferation and the effectiveness of imatinib treatment in chronic myeloid leukemia cells. 29667779_MiR-193a-3p and miR-224 promoted the cell proliferation and decreased apoptosis by targeting alpha-2,3-sialyltransferase IV in renal cell carcinoma. 29749491_The V1 transcript of the ST3GAL4 demonstrated significant decreased expression in premalignant and malignant cervical tissues. 31914669_a cross-restoration of each of the three genes in ST3GAL6 KO cells showed that overexpression of ST3GAL6 sufficiently rescued the total alpha2,3-sialylation levels, cell morphology, and alpha2,3-sialylation of EGFR, whereas the alpha2,3-sialylation levels of beta1 were greatly enhanced by an overexpression of ST3GAL4 32295993_Identification and characterization of the V3 promoter of the ST3GAL4 gene. 33080315_MiR-193b modulates osteoarthritis progression through targeting ST3GAL4 via sialylation of CD44 and NF-small ka, CyrillicB pathway. 33524390_Rab11-mediated post-Golgi transport of the sialyltransferase ST3GAL4 suggests a new mechanism for regulating glycosylation. | ENSMUSG00000032038 | St3gal4 | 1.232289e+03 | 1.3431499 | 0.425620272 | 0.3150898 | 1.822604e+00 | 0.1770035926 | 0.77663851 | No | Yes | 1.328817e+03 | 140.823582 | 8.950200e+02 | 97.930552 |
| ENSG00000110169 | 3263 | HPX | protein_coding | P02790 | FUNCTION: Binds heme and transports it to the liver for breakdown and iron recovery, after which the free hemopexin returns to the circulation. | Direct protein sequencing;Disulfide bond;Glycoprotein;Heme;Host-virus interaction;Iron;Metal-binding;Reference proteome;Repeat;Secreted;Signal;Transport | This gene encodes a plasma glycoprotein that binds heme with high affinity. The encoded protein is an acute phase protein that transports heme from the plasma to the liver and may be involved in protecting cells from oxidative stress. [provided by RefSeq, Apr 2009]. | hsa:3263; | blood microparticle [GO:0072562]; collagen-containing extracellular matrix [GO:0062023]; endocytic vesicle lumen [GO:0071682]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; heme transmembrane transporter activity [GO:0015232]; metal ion binding [GO:0046872]; cellular iron ion homeostasis [GO:0006879]; heme metabolic process [GO:0042168]; heme transport [GO:0015886]; hemoglobin metabolic process [GO:0020027]; positive regulation of humoral immune response mediated by circulating immunoglobulin [GO:0002925]; positive regulation of immunoglobulin production [GO:0002639]; positive regulation of interferon-gamma-mediated signaling pathway [GO:0060335]; positive regulation of tyrosine phosphorylation of STAT protein [GO:0042531] | 12675843_mesangial hemopexin is able to affect glomerular anionic sites, it is conceivable that stimulated mesangium may contribute to enhanced glomerular permeability in corticosteroid responsive nephrotic syndrome through local hemopexin release. 15558018_The conserved binding ability of the Hemopexin domains suggests that CD44 may act as a core molecule assembling multiple Membrane-type 1 matrix metalloproteinases on the cell surface. 15697212_The ability of hemopexin to bind metal ions raises the possibility that this protein may participate in the transport of metal ions in blood or in the exchange of metal ions between proteins. 15697213_Thermal denaturation of the human hemopexin-heme complex is investigated under a variety of solution conditions to identify factors that influence heme release, including the potential presence of transition metal ions or heme iron reduction. 16014037_hemopexin molecule as such can potentially act as a toxic protease, leading to proteinuria and glomerular alterations 16079987_Hemopexin in minimal change nephropathy relapse subjects may exist in an altered isoform, showing enhanced protease activity as compared with subjects in remission, subjects with other forms of primary glomerulopathy, or healthy controls. 17185359_Hemopexin and hemopexin domains of human proteins fulfill functions in activation of MMPs, inhibition of MMPs, dimerization, binding of substrates or ligands, cleavage of substrates, and endocytosis by LRP-1 and LRP-2. 17229156_HPX-heme(II)-NO appears to act as an efficient scavenger of peroxynitrite and of strong oxidants and nitrating species following the reaction of peroxynitrite with CO2 17636883_The structural consequences of metal ion binding to the form of hemopexin that dominates in plasma have been evaluated. 18044975_these observations result from the binding of heme in form beta with an orientation that differs from the crystallographically observed binding orientation for rabbit hemopexin 19863188_Results indicate that for the discrimination of AD from ND control subjects, measurement of a set of markers including Abeta1-42, ApoA1 and HPX improved diagnostic performance over that obtained by measurement of Abeta1-42 alone. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19966041_In a family with a hypomaturation-type enamel defect, mutational and haplotype analyses revealed an amelogenesis imperfecta-causing point mutation in exon 6 of MMP20 that results in a single amino acid substitution in the hemopexin domain. 20149159_elevated Hx predicts late graft failure in kidney transplantation 20610401_hemopexin directly interacts with FLVCR 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21193411_mutations of amino acids involved in the interaction weakened the dimer interaction of Hpx domains in solution, and incorporation of these mutations into the full-length enzyme significantly inhibited dimer-dependent functions on the cell surface 21404362_Studies indicate that hemopexin lacks the catalytic triad that is characteristic of many serine proteases but possesses two highly exposed Arg-Gly-Glu sequences that may promote interaction with cell surfaces. 22579751_In conclusion, the current work has identified potential T1DM biomarkers and one of these, hemopexin, can be modulated by glucose through a ROS-dependent mechanism. 22772444_The findings suggest that hemopexin can modulate the role of hemoglobin in sterile and infectious inflammation 22993068_In sum, these data indicated that AKI-associated hepatic stress generates Hpx, which gains renal tubule access. 23254305_activated hemopexin might be considered as a factor mediating ang II effects upon blood pressure by modulating AT1-R availability 23350672_hemopexin will be neuroprotective after traumatic brain injury, with heme release in the CNS, and during the ensuing inflammation. 23620477_Hemopexin is overexpressed in the RPE of diabetic patients with DME and induces the breakdown of RPE cells in vitro. 24613679_The Bach1-dependent repression of the HO-1 expression is under the control of the Hemopexin-dependent uptake of extracellular he 24969553_Data show that HPX, POTEE and ApoA1 are deregulated in breast tumors suggesting un important role in breast tumorigenesis. 25888135_Plasma hemopexin levels were decreased or markedly decreased in patients with burns or sepsis and in premature infants. 26339767_Heme scavenging is a major mechanism by which Hx defends against oxidative stress and related inflammatory disorders. [Review] 26908325_HPX is upregulated in non-small cell lung cancer patients compared to those with benign lung disease or no lung disease. 27155336_There were significantly higher serum concentrations of fetal hemoglobin and alpha1-microglobulin and significantly lower first trimester serum concentrations of hemopexin in patients who later developed preeclampsia. 28596380_Data suggest that apo-hemopexin isolated from plasma exchibits low endogenous levels of tyrosine nitration in the peptide YYCFQGNQFLR in the heme-binding site of human hemopexin, which was similarly nitrated in rabbit and rat hemopexins; heme binding by hemopexin declined as tyrosine nitration proceeded in vitro. 30226858_Study found that hemopexin was significantly higher in acute T cell-mediated rejection (TCMR) patients and could be detected in urine of kidney transplant recipients which makes it s potential diagnostic protein for TCMR. 32915914_Hemopexin and alpha1-microglobulin heme scavengers with differential involvement in preeclampsia and fetal growth restriction. 32966797_Scavenging of Labile Heme by Hemopexin Is a Key Checkpoint in Cancer Growth and Metastases. 33813281_Widespread severe cerebral elevations of haptoglobin and haemopexin in sporadic Alzheimer's disease: Evidence for a pervasive microvasculopathy. 34564750_Evaluation of the relationship between intravascular hemolysis and clinical manifestations in sickle cell disease: decreased hemopexin during vaso-occlusive crises and increased inflammation in acute chest syndrome. | ENSMUSG00000030895 | Hpx | 3.607936e+01 | 0.8663149 | -0.207036608 | 0.5559614 | 1.297377e-01 | 0.7187041524 | 0.94084174 | No | Yes | 3.430798e+01 | 15.354321 | 3.752900e+01 | 17.018205 | |
| ENSG00000110442 | 29099 | COMMD9 | protein_coding | Q9P000 | FUNCTION: May modulate activity of cullin-RING E3 ubiquitin ligase (CRL) complexes (PubMed:21778237). May down-regulate activation of NF-kappa-B (PubMed:15799966). Modulates Na(+) transport in epithelial cells by regulation of apical cell surface expression of amiloride-sensitive sodium channel (ENaC) subunits (PubMed:23637203). {ECO:0000269|PubMed:15799966, ECO:0000269|PubMed:23637203, ECO:0000305|PubMed:21778237}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasmic vesicle;Ion transport;Nucleus;Reference proteome;Sodium;Sodium transport;Transcription;Transcription regulation;Transport;Ubl conjugation pathway | hsa:29099; | cytosol [GO:0005829]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; secretory granule lumen [GO:0034774]; cholesterol homeostasis [GO:0042632]; sodium ion transport [GO:0006814] | 27871936_COMMD9 participates in TFDP1/E2F1 activation and plays a critical role in non-small cell lung cancer. | ENSMUSG00000027163 | Commd9 | 2.572494e+03 | 0.9710804 | -0.042337418 | 0.3025124 | 1.949765e-02 | 0.8889491994 | 0.97918210 | No | Yes | 2.490932e+03 | 214.412173 | 2.299358e+03 | 203.263393 | ||
| ENSG00000110497 | 55626 | AMBRA1 | protein_coding | Q9C0C7 | FUNCTION: Substrate-recognition component of a DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complex involved in cell cycle control and autophagy (PubMed:20921139, PubMed:23524951, PubMed:24587252, PubMed:33854232, PubMed:33854235, PubMed:33854239, PubMed:32333458). The DCX(AMBRA1) complex specifically mediates the polyubiquitination of target proteins such as BECN1, CCND1, CCND2, CCND3, ELOC and ULK1 (PubMed:23524951, PubMed:33854232, PubMed:33854235, PubMed:33854239). Acts as an upstream master regulator of the transition from G1 to S cell phase: AMBRA1 specifically recognizes and binds phosphorylated cyclin-D (CCND1, CCND2 and CCND3), leading to cyclin-D ubiquitination by the DCX(AMBRA1) complex and subsequent degradation (PubMed:33854232, PubMed:33854235, PubMed:33854239). By controlling the transition from G1 to S phase and cyclin-D degradation, AMBRA1 acts as a tumor suppressor that promotes genomic integrity during DNA replication and counteracts developmental abnormalities and tumor growth (PubMed:33854232, PubMed:33854235, PubMed:33854239). AMBRA1 also regulates the cell cycle by promoting MYC dephosphorylation and degradation independently of the DCX(AMBRA1) complex: acts via interaction with the catalytic subunit of protein phosphatase 2A (PPP2CA), which enhances interaction between PPP2CA and MYC, leading to MYC dephosphorylation and degradation (PubMed:25803737, PubMed:25438055). Acts as a regulator of Cul5-RING (CRL5) E3 ubiquitin-protein ligase complexes by mediating ubiquitination and degradation of Elongin-C (ELOC) component of CRL5 complexes (PubMed:25499913, PubMed:30166453). Acts as a key regulator of autophagy by modulating the BECN1-PIK3C3 complex: controls protein turnover during neuronal development, and regulates normal cell survival and proliferation (PubMed:21358617). In normal conditions, AMBRA1 is tethered to the cytoskeleton via interaction with dyneins DYNLL1 and DYNLL2 (PubMed:20921139). Upon autophagy induction, AMBRA1 is released from the cytoskeletal docking site to induce autophagosome nucleation by mediating ubiquitination of proteins involved in autophagy (PubMed:20921139). The DCX(AMBRA1) complex mediates 'Lys-63'-linked ubiquitination of BECN1, increasing the association between BECN1 and PIK3C3 to promote PIK3C3 activity (By similarity). In collaboration with TRAF6, AMBRA1 mediates 'Lys-63'-linked ubiquitination of ULK1 following autophagy induction, promoting ULK1 stability and kinase activity (PubMed:23524951). Also activates ULK1 via interaction with TRIM32: TRIM32 stimulates ULK1 through unanchored 'Lys-63'-linked polyubiquitin chains (PubMed:31123703). Also acts as an activator of mitophagy via interaction with PRKN and LC3 proteins (MAP1LC3A, MAP1LC3B or MAP1LC3C); possibly by bringing damaged mitochondria onto autophagosomes (PubMed:21753002, PubMed:25215947). Also activates mitophagy by acting as a cofactor for HUWE1; acts by promoting HUWE1-mediated ubiquitination of MFN2 (PubMed:30217973). AMBRA1 is also involved in regulatory T-cells (Treg) differentiation by promoting FOXO3 dephosphorylation independently of the DCX(AMBRA1) complex: acts via interaction with PPP2CA, which enhances interaction between PPP2CA and FOXO3, leading to FOXO3 dephosphorylation and stabilization (PubMed:30513302). May act as a regulator of intracellular trafficking, regulating the localization of active PTK2/FAK and SRC (By similarity). Also involved in transcription regulation by acting as a scaffold for protein complexes at chromatin (By similarity). {ECO:0000250|UniProtKB:A2AH22, ECO:0000269|PubMed:20921139, ECO:0000269|PubMed:21358617, ECO:0000269|PubMed:21753002, ECO:0000269|PubMed:23524951, ECO:0000269|PubMed:24587252, ECO:0000269|PubMed:25215947, ECO:0000269|PubMed:25438055, ECO:0000269|PubMed:25499913, ECO:0000269|PubMed:25803737, ECO:0000269|PubMed:30166453, ECO:0000269|PubMed:30217973, ECO:0000269|PubMed:30513302, ECO:0000269|PubMed:31123703, ECO:0000269|PubMed:32333458, ECO:0000269|PubMed:33854232, ECO:0000269|PubMed:33854235, ECO:0000269|PubMed:33854239}. | Alternative splicing;Autophagy;Cell cycle;Cell junction;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Developmental protein;Differentiation;Endoplasmic reticulum;Isopeptide bond;Methylation;Mitochondrion;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Tumor suppressor;Ubl conjugation;Ubl conjugation pathway;WD repeat | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:23524951, ECO:0000269|PubMed:30166453, ECO:0000269|PubMed:33854232, ECO:0000269|PubMed:33854235, ECO:0000269|PubMed:33854239}. | hsa:55626; | autophagosome [GO:0005776]; axoneme [GO:0005930]; Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; focal adhesion [GO:0005925]; intracellular membrane-bounded organelle [GO:0043231]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; GTPase binding [GO:0051020]; protein phosphatase binding [GO:0019903]; ubiquitin ligase-substrate adaptor activity [GO:1990756]; ubiquitin protein ligase binding [GO:0031625]; autophagosome assembly [GO:0000045]; autophagy [GO:0006914]; autophagy of mitochondrion [GO:0000422]; cell cycle [GO:0007049]; cell differentiation [GO:0030154]; mitophagy [GO:0000423]; nervous system development [GO:0007399]; positive regulation of autophagy [GO:0010508]; positive regulation of free ubiquitin chain polymerization [GO:1904544]; positive regulation of mitophagy [GO:1901526]; positive regulation of phosphatidylinositol 3-kinase activity [GO:0043552]; positive regulation of protein dephosphorylation [GO:0035307]; positive regulation of regulatory T cell differentiation [GO:0045591]; protein polyubiquitination [GO:0000209]; regulation of G1/S transition of mitotic cell cycle [GO:2000045]; response to mitochondrial depolarisation [GO:0098780] | 20921139_When autophagy is induced, ULK1 phosphorylates AMBRA1, releasing the autophagy core complex from dynein. 21358617_In this study, the authors show that AMBRA1 binds preferentially the mitochondrial pool of the antiapoptotic factor BCL-2, and that this interaction is disrupted following autophagy induction. 22441670_Ambra1 is an important target of apoptotic proteases resulting in the dismantling of the autophagic machinery and the accomplishment of the cell death program 23069654_role that Ambra1 in the switch between autophagy and apoptosis 23429496_Data suggest that activated autophagy is associated with the progression of pancreatic ductal adenocarcinoma and that the overexpression of autophagy-related proteins Atg5, Ambra1, beclin-1, LC3B and Bif-1 is significantly correlated with poor outcome. 23524951_findings show under non-autophagic conditions, mTOR inhibits AMBRA1 by phosphorylation, whereas on autophagy induction, AMBRA1 is dephosphorylated; in this condition, AMBRA1, interacting with TRAF6, supports ULK1 ubiquitylation by LYS-63-linked chains, and its stabilization, self-association and function 23551272_A schizophrenia-related risk variant in AMBRA1 (rs11819869) is involved in various aspects of impulsivity, and this involvement occurs on a behavioral as well as an imaging genetics level. 24587252_Ambra1 is a crucial regulator of autophagy and apoptosis in colorectal cancer cells 25438055_a de-regulation of c-Myc correlates with increased tumorigenesis in AMBRA1-defective systems, thus supporting a role for AMBRA1 as a haploinsufficient tumour suppressor gene. 25499913_AMBRA1 interacts with cullin E3 ubiquitin ligases to regulate autophagy dynamics 25803737_Both AMBRA1 and BECLIN 1 affect c-Myc regulation, but through two different pathways. 26086269_Ambra1 mRNA translocation to P-bodies and translational suppression correlated with increased cell death. 26423274_An increased expression of AMBRA1 and SQSTM1. 26763392_Ambra1 is a crucial regulator of autophagy and apoptosis in ovarian cancer cells subject to cisplatin to maintain the balance between autophagy and apoptosis. Ambra1-targeting inhibition might sensitize ovarian cancer cells to chemotherapy. 27875637_Results showed that AMBRA1 is a novel hub binding protein of alpha-synuclein and plays a central role in the pathogenesis of multiple system atrophy through the degradative dynamics of alpha-synuclein. 28059583_These data suggest a new and interesting role of MIR7-3HG as an anti-autophagic MIRNA that may affect oncogenesis through the regulation of the tumor suppressor AMBRA1. 28224423_Results show that the expression of AMBRA1 and Beclin-1 is increased in human gastric adenocarcinoma (GC) tissues. High protein expression of AMBRA1 and Beclin-1 is correlated with tumor invasion and is an independent poor prognostic marker in GC patients. 28994820_Data support a role of AMBRA1/Ambra1 partial loss-of-function genotypes for female autistic traits. Moreover, they suggest Ambra1 heterozygous mice as a novel multifaceted and construct-valid genetic mouse model for female autism. 29101240_Ambra1 induces autophagy and desensitizes human prostate cancer cells to cisplatin. 30027574_Ambra1 plays an important role in regulating the sensitivity of breast cancer cells to epirubicin. Regulatory effect of Ambra1 on epirubicin sensitivity is achieved through the regulation of autophagy by targeting ATG12. 30166453_CRL4(AMBRA)(1) modulates interleukin-6/STAT3 signaling and HIV-1 infectivity that are regulated by CRL5(SOCS)(3) and CRL5(VIF), respectively. Thus, by discovering a substrate of CRL4(AMBRA)(1), ELOC, the shared adapter of CRL5 ubiquitin ligases, we uncovered a novel CRL cross-regulation pathway. 30217973_AMBRA1 regulates mitophagy through a novel pathway, in which HUWE1 and IKKalpha are key factors, shedding new lights on the regulation of mitochondrial quality control and homoeostasis in mammalian cells. 30513302_AMBRA1 is a key modulator of T cell differentiation. Through its ability to interact with the phosphatase PP2A, AMBRA1 promotes the FOXO3 stability. 31056744_Epidermal autophagy and beclin 1 regulator 1 and loricrin: a paradigm shift in the prognostication and stratification of the American Joint Committee on Cancer stage I melanomas. 31434979_HUWE1 controls MCL1 stability to unleash AMBRA1-induced mitophagy. 32333458_Rare mutations in the autophagy-regulating gene AMBRA1 contribute to human neural tube defects. 32616651_The autophagy protein Ambra1 regulates gene expression by supporting novel transcriptional complexes. 33172332_HPV sensitizes OPSCC cells to cisplatin-induced apoptosis by inhibiting autophagy through E7-mediated degradation of AMBRA1. 33562550_The Role of Cardiolipin as a Scaffold Mitochondrial Phospholipid in Autophagosome Formation: In Vitro Evidence. 33854232_AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. 33854235_CRL4(AMBRA1) is a master regulator of D-type cyclins. 33854239_The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. 33953176_Loss of Ambra1 promotes melanoma growth and invasion. 34302498_Targeting cancer stem cells in medulloblastoma by inhibiting AMBRA1 dual function in autophagy and STAT3 signalling. 34362797_AMBRA1 Promotes TGFbeta Signaling via Nonproteolytic Polyubiquitylation of Smad4. 34495753_The Long-Lost Ligase: CRL4(AMBRA1) Regulates the Stability of D-Type Cyclins. 34769507_AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination. | ENSMUSG00000040506 | Ambra1 | 3.708367e+03 | 1.2438902 | 0.314859192 | 0.3010442 | 1.088275e+00 | 0.2968538299 | 0.81383297 | No | Yes | 3.952618e+03 | 416.073728 | 2.862783e+03 | 309.546236 | |
| ENSG00000110583 | 79829 | NAA40 | protein_coding | Q86UY6 | FUNCTION: N-alpha-acetyltransferase that specifically mediates the acetylation of the N-terminal residues of histones H4 and H2A (PubMed:21935442, PubMed:25619998). In contrast to other N-alpha-acetyltransferase, has a very specific selectivity for histones H4 and H2A N-terminus and specifically recognizes the 'Ser-Gly-Arg-Gly sequence' (PubMed:21935442, PubMed:25619998). Acts as a negative regulator of apoptosis (PubMed:26666750). May play a role in hepatic lipid metabolism (By similarity). {ECO:0000250|UniProtKB:Q8VE10, ECO:0000269|PubMed:21935442, ECO:0000269|PubMed:25619998, ECO:0000269|PubMed:26666750}. | 3D-structure;Acyltransferase;Alternative splicing;Cytoplasm;Lipoprotein;Myristate;Nucleus;Reference proteome;Transferase | hsa:79829; | centriolar satellite [GO:0034451]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; H2A histone acetyltransferase activity [GO:0043998]; H4 histone acetyltransferase activity [GO:0010485]; peptide-serine-N-acetyltransferase activity [GO:1990189]; histone H2A acetylation [GO:0043968]; histone H4 acetylation [GO:0043967]; lipid metabolic process [GO:0006629]; N-terminal protein amino acid acetylation [GO:0006474] | 19695338_a novel protein acetyltransferase Patt1 might be involved in the development of hepatocellular carcinoma 21935442_Results strongly suggest that human Naa40p/NatD is conserved from yeast. Thus, the NATs of all classes of N-terminally acetylated proteins in humans now appear to be accounted for. 24248912_The knowledge of detailed molecular architecture of Patt1 is not only the key to understanding its mechanistic functional properties but it also opens the possibility of rational drug and protein design experiments 26666750_findings reveal an anti-apoptotic role for Naa40 and exhibit its potential as a therapeutic target in colorectal cancers. 29030587_NatD-mediated acetylation of histone H4 serine 1 competes with phosphorylation by CK2alpha at the same residue, leading to the upregulation of Slug and lung cancer progression. 30858358_The findings strengthen the importance of NAA40 to maintain colorectal cancer cell growth. We show that NAA40 oncogenic properties stimulate the global levels of H4R3me2s by transcriptionally activating PRMT5 methyltransferase which in turn modulates the expression of key downstream cancer-related genes. | ENSMUSG00000024764 | Naa40 | 2.786768e+03 | 1.0835655 | 0.115786404 | 0.2866296 | 1.670633e-01 | 0.6827350472 | 0.93332240 | No | Yes | 2.335012e+03 | 206.875451 | 2.283745e+03 | 207.479859 | ||
| ENSG00000110851 | 11108 | PRDM4 | protein_coding | Q9UKN5 | FUNCTION: May function as a transcription factor involved in cell differentiation. | 3D-structure;DNA-binding;Metal-binding;Methyltransferase;Nucleus;Reference proteome;Repeat;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Zinc;Zinc-finger | The protein encoded by this gene is a transcription factor of the PR-domain protein family. It contains a PR-domain and multiple zinc finger motifs. Transcription factors of the PR-domain family are known to be involved in cell differentiation and tumorigenesis. An elevated expression level of this gene has been observed in PC12 cells treated with nerve growth factor, beta polypeptide (NGF). This gene is located in a chromosomal region that is thought to contain tumor suppressor genes. [provided by RefSeq, Jul 2008]. | hsa:11108; | cytoplasm [GO:0005737]; histone methyltransferase complex [GO:0035097]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; histone methyltransferase binding [GO:1990226]; metal ion binding [GO:0046872]; methyltransferase activity [GO:0008168]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; histone H4-R3 methylation [GO:0043985]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of gene expression [GO:0010468]; transcription by RNA polymerase II [GO:0006366] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21604305_PRDM4 zinc knuckle by NMR spectroscopy. 28228349_Both Prdm 4 and Prdm 5 are expressed in human corneal endothelium, primary hCECs and in HCECs-12 cells, characterised by expression of the Na(+)/K(+)-ATPase. 29669796_results demonstrate that YAP promotes cell invasion by inducing leukocyte-specific integrin expression, and identify PRDM4 as a novel transcription factor for YAP targets. | ENSMUSG00000035529 | Prdm4 | 1.718142e+03 | 1.1172606 | 0.159965785 | 0.2644908 | 3.681001e-01 | 0.5440416778 | 0.89063840 | No | Yes | 1.804952e+03 | 215.296311 | 1.459031e+03 | 178.615829 | |
| ENSG00000110987 | 605 | BCL7A | protein_coding | Q4VC05 | Alternative splicing;Chromosomal rearrangement;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | This gene is directly involved, with Myc and IgH, in a three-way gene translocation in a Burkitt lymphoma cell line. As a result of the gene translocation, the N-terminal region of the gene product is disrupted, which is thought to be related to the pathogenesis of a subset of high-grade B cell non-Hodgkin lymphoma. The N-terminal segment involved in the translocation includes the region that shares a strong sequence similarity with those of BCL7B and BCL7C. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:605; | SWI/SNF complex [GO:0016514]; negative regulation of transcription, DNA-templated [GO:0045892] | 15897551_Promoter hypermethylation of BCL7a is associated with cutaneous T-cell lymphoma 18663754_deletion of genes BCL7A in early-stage mycosis fungoides. 19336552_Observational study of gene-disease association. (HuGE Navigator) 19336552_Variants in BCL7A were strongly related to diffuse large B-cell lymphoma. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 22856870_Data suggest that BCL7A may play an important role in cutaneous T-cell lymphoma (CTCL) carcinogenesis. 23043359_Report describes BCL7A protein expression in normal lymphoid tissues and lymphomas using immunohistochemistry. 29166413_BCL7A, BRWD3, and AUTS2 demonstrate significantly higher mutation frequencies among AA cases. These genes are all involved in translocations in B-cell malignancies. Moreover, we detected a significant difference in mutation frequency of TP53 and IRF4 with frequencies higher among CA cases. Our study provides rationale for interrogating diverse tumor cohorts to best understand tumor genomics across populations. 31077237_Survival analysis showed that, compared with those who had higher levels of BCL7A expression, patients with ovarian cancer and low levels of BCL7A generally had shorter overall/relapse-free survival times. 32576963_Frequent mutations in the amino-terminal domain of BCL7A impair its tumor suppressor role in DLBCL. 33415690_MiR-501-3p promotes osteosarcoma cell proliferation, migration and invasion by targeting BCL7A. 34362400_BCL7A as a novel prognostic biomarker for glioma patients. | ENSMUSG00000029438 | Bcl7a | 2.484580e+03 | 1.1055247 | 0.144731266 | 0.2892162 | 2.547092e-01 | 0.6137785558 | 0.91292972 | No | Yes | 2.362162e+03 | 150.447289 | 2.130981e+03 | 139.409130 | ||
| ENSG00000111011 | 65117 | RSRC2 | protein_coding | Q7L4I2 | Acetylation;Alternative splicing;Coiled coil;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:65117; | RNA binding [GO:0003723] | 17203224_RSRC2 might play a role in cell proliferation and progression of esophageal cancer 28416606_our findings suggest that paclitaxel targets the TRA2A-RSRC2 splicing pathway, and deregulated TRA2A and RSRC2 expression may confer paclitaxel resistance. In addition, our study demonstrates that expression of TRA2A in conjunction with RSRC2 may provide valuable molecular biomarker evidence for triple-negative breast cancer (TNBC)clinical treatment decisions and patient outcome | ENSMUSG00000029422 | Rsrc2 | 1.668264e+03 | 1.1109411 | 0.151782376 | 0.3268569 | 2.138914e-01 | 0.6437343400 | 0.92210426 | No | Yes | 1.674911e+03 | 278.548146 | 1.478186e+03 | 252.124724 | |||
| ENSG00000111186 | 81029 | WNT5B | protein_coding | Q9H1J7 | FUNCTION: Ligand for members of the frizzled family of seven transmembrane receptors. Probable developmental protein. May be a signaling molecule which affects the development of discrete regions of tissues. Is likely to signal over only few cell diameters (By similarity). {ECO:0000250}. | Developmental protein;Disulfide bond;Extracellular matrix;Glycoprotein;Lipoprotein;Reference proteome;Secreted;Signal;Wnt signaling pathway | The WNT gene family consists of structurally related genes which encode secreted signaling proteins. These proteins have been implicated in oncogenesis and in several developmental processes, including regulation of cell fate and patterning during embryogenesis. This gene is a member of the WNT gene family. It encodes a protein which shows 94% and 80% amino acid identity to the mouse Wnt5b protein and the human WNT5A protein, respectively. Alternative splicing of this gene generates 2 transcript variants. [provided by RefSeq, Jul 2008]. | hsa:81029; | endocytic vesicle membrane [GO:0030666]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; Golgi lumen [GO:0005796]; plasma membrane [GO:0005886]; cytokine activity [GO:0005125]; frizzled binding [GO:0005109]; signaling receptor binding [GO:0005102]; canonical Wnt signaling pathway [GO:0060070]; cell fate commitment [GO:0045165]; cellular response to retinoic acid [GO:0071300]; chondrocyte differentiation [GO:0002062]; fat cell differentiation [GO:0045444]; lens fiber cell development [GO:0070307]; muscle cell differentiation [GO:0042692]; neuron differentiation [GO:0030182]; positive regulation of cell migration [GO:0030335]; positive regulation of convergent extension involved in gastrulation [GO:1904105]; positive regulation of non-canonical Wnt signaling pathway [GO:2000052]; wound healing [GO:0042060] | 12165812_WNT5b is upregulated by beta-estradiol in tumor cell lines 15386214_WNT5B gene may contribute to conferring susceptibility to type 2 diabetes and may be involved in the pathogenesis of this disease through the regulation of adipocyte function. 15972578_In leiomyoma cells, the regulation of Wnt5b expression by retinoids appears to be attenuated. 18555673_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18555673_Variation in WNT5B predisposes to type 2 diabetes in the absence of obesity. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19863181_Results demonstrated significant up-regulation of WNT-3, WNT-4, WNT-5B, WNT-7B, WNT-9A, WNT-10A, and WNT-16B in patients with CLL compared to normal subjects. 21766608_This is significantly related to fat cell differentiation, suggesting its possible involvement in the onset of diabetes mellitus type 2 and metabolic syndrome. 21980498_Differentiation paralleled the activation of Wnt5/Calmodulin signalling by autocrine/paracrine intense secretion of Wnt5a and Wnt5b 23817222_This study shows that the WNT5B germline variant rs2010851 was significantly identified as a stage-dependent prognostic marker for CC patients after 5-fluorouracil-based adjuvant therapy. 24040439_TGF-beta/Smad, integrin/integrin-linked kinase (ILK) and wnt/beta-catenin signaling pathways are involved in epithelial to mesenchymal transition in human renal fibrogenesis. 24220306_These results indicate that Wnt5b is involved in the migration ability of OSCC cells through active Cdc42 and RhoA. 24344732_WNT3 and WNT5B are critical factors, secreted from mesenchymal cancer cells, for instigating the epithelial cancer cell invasion. 24399733_Ursolic acid suppressed the expression of Wnt5alpha/beta and beta-catenin, inducing apoptosis in prostate cancer cells. 24564888_WNT5B is elevated in both in the tumor and the patients' serum from triple negative breast cancer. 24841200_Knocking down Wnt5b expression reduced phospho-PKCA levels and cell migration. 24944491_Report increased Wnt5b levels in adriamycin treated liver cancer stem cells within 4h of treatment. 25246426_Results demonstrate that the Wnt5B pathway may play an important role in atypical teratoid rhabdoid tumor biology. 25425204_The interaction between Wnt isoforms and their LRP6 cognate receptor depends on the jutting loop present in Wnt3a but absent in Wnt5b. 26733379_This study identified WNT5B and CTNNBL1 for peak bone mineral density and body composition in males from the Han Chinese ethnic group. 26861993_these results suggest that TGF-beta1 stimulates HSC-4 cell invasion through the Slug/Wnt-5b/MMP-10 signalling axis. 27036869_WNT-5B induces IL-6 and CXCL8 secretion in pulmonary fibroblasts. 27121420_overexpression of WNT5B significantly increased cell proliferation, migration and invasion capacities of the COLO 205 cells in vitro. WNT5B may play an important role in the tumorigenesis of colorectal cancer. 27126693_exaggerated WNT-5B expression upon cigarette smoke exposure in the bronchial epithelium of COPD patients leads to TGF-beta/Smad3-dependent expression of genes related to airway remodelling 27593938_the data suggest that WNT5B induces tube formation by regulating the expression of Snail and Slug proteins through activation of canonical and non-canonical WNT signalling pathways. 27762090_results suggest that Wnt5b-associated exosomes promote cancer cell migration and proliferation in a paracrine manner 27932350_Noncanonical Wnt signaling via Wnt5a/5b/11 may have role in the pathogenesis of aortic valve calcification. 28078366_were not able to replicate or further verify the genetic association of polymorphisms in WNT4 and WNT5B with bone mineral density 29847788_IMP3 also facilitates the transcription of SLUG, which is necessary for TAZ nuclear localization and activation, by a mechanism that is also mediated by WNT5B. 31666682_WNT5B exerts oncogenic effects and is negatively regulated by miR-5587-3p in lung adenocarcinoma progression. 32092330_Up-regulated miR-374a-3p relieves lipopolysaccharides induced injury in CHON-001 cells via regulating Wingless-type MMTV integration site family member 5B. 32561739_The lncRNA RP11-142A22.4 promotes adipogenesis by sponging miR-587 to modulate Wnt5beta expression. 34536574_Exosome-mediated circ_0001846 participates in IL-1beta-induced chondrocyte cell damage by miR-149-5p-dependent regulation of WNT5B. 34906330_Estrogen receptor alpha and NFATc1 bind to a bone mineral density-associated SNP to repress WNT5B in osteoblasts. | ENSMUSG00000030170 | Wnt5b | 2.638372e+02 | 1.1371731 | 0.185451926 | 0.3478287 | 2.805035e-01 | 0.5963713791 | 0.90859245 | No | Yes | 2.688965e+02 | 33.945244 | 2.374350e+02 | 31.157564 | |
| ENSG00000111237 | 51699 | VPS29 | protein_coding | Q9UBQ0 | FUNCTION: Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. The recruitment of the CSC to the endosomal membrane involves RAB7A and SNX3. The SNX-BAR retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) and is involved in endosome-to-plasma membrane transport for cargo protein recycling. The SNX3-retromer mediates the retrograde endosome-to-TGN transport of WLS distinct from the SNX-BAR retromer pathway. The SNX27-retromer is believed to be involved in endosome-to-plasma membrane trafficking and recycling of a broad spectrum of cargo proteins. The CSC seems to act as recruitment hub for other proteins, such as the WASH complex and TBC1D5. Required to regulate transcytosis of the polymeric immunoglobulin receptor (pIgR-pIgA) (PubMed:15247922, PubMed:21725319, PubMed:23563491). Acts also as component of the retriever complex. The retriever complex is a heterotrimeric complex related to retromer cargo-selective complex (CSC) and essential for retromer-independent retrieval and recycling of numerous cargos such as integrin alpha-5/beta-1 (ITGA5:ITGB1) (PubMed:28892079). In the endosomes, retriever complex drives the retrieval and recycling of NxxY-motif-containing cargo proteins by coupling to SNX17, a cargo essential for the homeostatic maintenance of numerous cell surface proteins associated with processes that include cell migration, cell adhesion, nutrient supply and cell signaling (PubMed:28892079). The recruitment of the retriever complex to the endosomal membrane involves CCC and WASH complexes (PubMed:28892079). Involved in GLUT1 endosome-to-plasma membrane trafficking; the function is dependent of association with ANKRD27 (PubMed:24856514). {ECO:0000269|PubMed:24856514, ECO:0000269|PubMed:28892079, ECO:0000303|PubMed:15247922, ECO:0000303|PubMed:21725319, ECO:0000303|PubMed:23563491}.; FUNCTION: (Microbial infection) The heterotrimeric retromer cargo-selective complex (CSC) mediates the exit of human papillomavirus from the early endosome and the delivery to the Golgi apparatus. {ECO:0000269|PubMed:25693203}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Endosome;Host-virus interaction;Membrane;Metal-binding;Protein transport;Reference proteome;Transport;Zinc | This gene belongs to a group of vacuolar protein sorting (VPS) genes that, when functionally impaired, disrupt the efficient delivery of vacuolar hydrolases. The protein encoded by this gene is a component of a large multimeric complex, termed the retromer complex, which is involved in retrograde transport of proteins from endosomes to the trans-Golgi network. This VPS protein may be involved in the formation of the inner shell of the retromer coat for retrograde vesicles leaving the prevacuolar compartment. Alternative splice variants encoding different isoforms and representing non-protein coding transcripts have been found for this gene. [provided by RefSeq, Aug 2013]. | hsa:51699; | cytosol [GO:0005829]; early endosome [GO:0005769]; endosome [GO:0005768]; endosome membrane [GO:0010008]; intracellular membrane-bounded organelle [GO:0043231]; late endosome [GO:0005770]; retromer complex [GO:0030904]; retromer, cargo-selective complex [GO:0030906]; metal ion binding [GO:0046872]; endocytic recycling [GO:0032456]; intracellular protein transport [GO:0006886]; retrograde transport, endosome to Golgi [GO:0042147] | 15788412_analysis of the phosphodiesterase/nuclease-like fold and two protein-protein interaction sites in human VPS29 16737443_It was demonstrated that recombinant human Vps29 displays in vitro phosphatase activity towards a serine-phosphorylated peptide, containing the acidic-cluster dileucine motif of the cytoplasmatic tail of the CI-M6PR. 17101778_These observations indicate that the mammalian retromer complex assembles by sequential association of SNX1/2 and Vps26-Vps29-Vps35 subcomplexes on endosomal membranes and that SNX1 and SNX2 play interchangeable but essential roles. 17891154_crystal structure of a VPS29-VPS35 subcomplex showing how the metallophosphoesterase-fold subunit VPS29 acts as a scaffold for the carboxy-terminal half of VPS35 19531583_Membrane recruitment of the cargo-selective retromer subcomplex VPS35/29/26 is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. 21629666_Conclusion is that VPS29 is a metal ion-independent, rigid scaffolding domain, which is essential but not sufficient for incorporation of retromer into functional endosomal transport assemblies. 24684791_Mutations in the gene composing the retromer cargo recognition subunit are not a common cause of Parkinson's disease. 25475142_This study demonstrated that Genetic variability of VPS29 in parkinsonism. 25937119_Data indicate that vesicular transport proteins VPS35 and VPS29 influence the levels of the other subunit of retromer. 26965691_The retromer complex is a highly conserved membrane trafficking assembly composed of three proteins - Vps26, Vps29 and Vps35, which are impaired in neurodegenerative diseases. (Review) 28892079_Heterotrimer composed of DSCR3, C16orf62 and VPS29 orchestrates endosomal cargo retrieval and recycling. 29146912_RidL is critical for binding of the L. pneumophila effector to the Vps29 retromer subunit and displacement of the regulator TBC1D5. 32737286_Retromer stabilization results in neuroprotection in a model of Amyotrophic Lateral Sclerosis. 33024112_Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29. | ENSMUSG00000029462 | Vps29 | 6.661893e+02 | 0.6142083 | -0.703200015 | 0.3025033 | 5.333353e+00 | 0.0209210958 | 0.51358130 | No | Yes | 4.520557e+02 | 63.978340 | 7.822679e+02 | 112.613098 | |
| ENSG00000111364 | 57696 | DDX55 | protein_coding | Q8NHQ9 | FUNCTION: Probable ATP-binding RNA helicase. | ATP-binding;Alternative splicing;Helicase;Hydrolase;Nucleotide-binding;Phosphoprotein;RNA-binding;Reference proteome | This gene encodes a member of protein family containing a characteristic Asp-Glu-Ala-Asp (DEAD) motif. These proteins are putative RNA helicases, and may be involved in a range of nuclear processes including translational initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Multiple alternatively spliced transcript variants have been found for this gene. Pseudogenes have been identified on chromosomes 1 and 12. [provided by RefSeq, Feb 2016]. | hsa:57696; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724] | 33048000_The DExD box ATPase DDX55 is recruited to domain IV of the 28S ribosomal RNA by its C-terminal region. | ENSMUSG00000029389 | Ddx55 | 1.457246e+03 | 0.8195032 | -0.287178571 | 0.2685829 | 1.121703e+00 | 0.2895520513 | 0.81232121 | No | Yes | 1.086622e+03 | 134.337111 | 1.476084e+03 | 186.574738 | |
| ENSG00000111639 | 51258 | MRPL51 | protein_coding | Q4U2R6 | 3D-structure;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transit peptide | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 39S subunit protein. Pseudogenes corresponding to this gene are found on chromosomes 4p and 21q. [provided by RefSeq, Jul 2008]. | hsa:51258; | mitochondrial inner membrane [GO:0005743]; mitochondrial large ribosomal subunit [GO:0005762]; mitochondrial ribosome [GO:0005761]; mitochondrion [GO:0005739]; structural constituent of ribosome [GO:0003735]; mitochondrial translation [GO:0032543]; translation [GO:0006412] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000030335 | Mrpl51 | 4.715053e+03 | 0.6806474 | -0.555020484 | 0.2839016 | 3.844511e+00 | 0.0499090843 | 0.66791998 | No | Yes | 3.697967e+03 | 395.496342 | 5.114430e+03 | 560.372426 | ||
| ENSG00000111641 | 4839 | NOP2 | protein_coding | P46087 | FUNCTION: Involved in ribosomal large subunit assembly (PubMed:24120868). S-adenosyl-L-methionine-dependent methyltransferase that specifically methylates the C(5) position of cytosine 4447 in 28S rRNA (Probable). May play a role in the regulation of the cell cycle and the increased nucleolar activity that is associated with the cell proliferation (Probable). {ECO:0000269|PubMed:24120868, ECO:0000305, ECO:0000305|PubMed:23913415}. | Acetylation;Alternative splicing;Citrullination;Isopeptide bond;Methyltransferase;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribosome biogenesis;S-adenosyl-L-methionine;Transferase;Ubl conjugation;rRNA processing | hsa:4839; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; rRNA (cytosine-C5-)-methyltransferase activity [GO:0009383]; maturation of LSU-rRNA [GO:0000470]; positive regulation of cell population proliferation [GO:0008284]; regulation of signal transduction by p53 class mediator [GO:1901796]; ribosomal large subunit assembly [GO:0000027]; RNA methylation [GO:0001510]; rRNA base methylation [GO:0070475] | 25481415_Findings suggest a newly discovered role of Nop2 in both mature neurons and in cells possibly involved in the regeneration of nervous tissue 26196125_p120 has an rRNA:5-Methylcytosine-Methyltransferase activity. 26906424_NOL1 represents a new route by which telomerase activates transcription of cyclin D1 gene, thus maintaining cell proliferation capacity 32176734_Mechanistically, NOP2 associates with HIV-1 5' LTR, interacts with HIV-1 TAR RNA by competing with HIV-1 Tat protein, as well as contributes to TAR m5C methylation. RNA MTase catalytic domain (MTD) of NOP2 mediates its competition with Tat and binding with TAR. Overall, these findings verified that NOP2 suppresses HIV-1 transcription and promotes viral latency. 32554858_Long noncoding RNA LINC00963 induces NOP2 expression by sponging tumor suppressor miR-542-3p to promote metastasis in prostate cancer. | ENSMUSG00000038279 | Nop2 | 5.619680e+03 | 0.9606421 | -0.057929038 | 0.2609592 | 4.930919e-02 | 0.8242697325 | 0.96655419 | No | Yes | 4.557167e+03 | 284.806146 | 4.959874e+03 | 318.131051 | ||
| ENSG00000111652 | 50813 | COPS7A | protein_coding | Q9UBW8 | FUNCTION: Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of SCF-type E3 ligase complexes, leading to decrease the Ubl ligase activity of SCF-type complexes such as SCF, CSA or DDB2. The complex is also involved in phosphorylation of p53/TP53, JUN, I-kappa-B-alpha/NFKBIA, ITPK1 and IRF8/ICSBP, possibly via its association with CK2 and PKD kinases. CSN-dependent phosphorylation of TP53 and JUN promotes and protects degradation by the Ubl system, respectively. {ECO:0000269|PubMed:11285227, ECO:0000269|PubMed:11337588, ECO:0000269|PubMed:12628923, ECO:0000269|PubMed:12732143, ECO:0000269|PubMed:9535219}. | 3D-structure;Acetylation;Coiled coil;Cytoplasm;Direct protein sequencing;Host-virus interaction;Nucleus;Phosphoprotein;Reference proteome;Signalosome | This gene encodes a component of the COP9 signalosome, an evolutionarily conserved multi-subunit protease that regulates the activity of the ubiquitin conjugation pathway. Alternatively spliced transcript variants that encode the same protein have been described. [provided by RefSeq, Mar 2014]. | hsa:50813; | COP9 signalosome [GO:0008180]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; COP9 signalosome assembly [GO:0010387]; protein deneddylation [GO:0000338] | Mouse_homologues 17077498_This protein was purified to homogeneity and screened for crystallization. | ENSMUSG00000030127 | Cops7a | 3.388398e+03 | 1.3190943 | 0.399547666 | 0.2970143 | 1.821333e+00 | 0.1771547454 | 0.77663851 | No | Yes | 3.709885e+03 | 337.648650 | 2.528881e+03 | 236.628608 | |
| ENSG00000111679 | 5777 | PTPN6 | protein_coding | P29350 | FUNCTION: Modulates signaling by tyrosine phosphorylated cell surface receptors such as KIT and the EGF receptor/EGFR. The SH2 regions may interact with other cellular components to modulate its own phosphatase activity against interacting substrates. Together with MTUS1, induces UBE2V2 expression upon angiotensin II stimulation. Plays a key role in hematopoiesis. {ECO:0000269|PubMed:11266449}. | 3D-structure;Alternative splicing;Cytoplasm;Hydrolase;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome;Repeat;SH2 domain | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. N-terminal part of this PTP contains two tandem Src homolog (SH2) domains, which act as protein phospho-tyrosine binding domains, and mediate the interaction of this PTP with its substrates. This PTP is expressed primarily in hematopoietic cells, and functions as an important regulator of multiple signaling pathways in hematopoietic cells. This PTP has been shown to interact with, and dephosphorylate a wide spectrum of phospho-proteins involved in hematopoietic cell signaling. Multiple alternatively spliced variants of this gene, which encode distinct isoforms, have been reported. [provided by RefSeq, Jul 2008]. | hsa:5777; | alpha-beta T cell receptor complex [GO:0042105]; cell-cell junction [GO:0005911]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; specific granule lumen [GO:0035580]; tertiary granule lumen [GO:1904724]; cell adhesion molecule binding [GO:0050839]; phosphorylation-dependent protein binding [GO:0140031]; phosphotyrosine residue binding [GO:0001784]; protein kinase binding [GO:0019901]; protein tyrosine phosphatase activity [GO:0004725]; SH2 domain binding [GO:0042169]; SH3 domain binding [GO:0017124]; transmembrane receptor protein tyrosine phosphatase activity [GO:0005001]; abortive mitotic cell cycle [GO:0033277]; B cell receptor signaling pathway [GO:0050853]; cell differentiation [GO:0030154]; cytokine-mediated signaling pathway [GO:0019221]; epididymis development [GO:1905867]; G protein-coupled receptor signaling pathway [GO:0007186]; hematopoietic progenitor cell differentiation [GO:0002244]; intracellular signal transduction [GO:0035556]; megakaryocyte development [GO:0035855]; natural killer cell mediated cytotoxicity [GO:0042267]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of humoral immune response mediated by circulating immunoglobulin [GO:0002924]; negative regulation of interleukin-6 production [GO:0032715]; negative regulation of MAP kinase activity [GO:0043407]; negative regulation of mast cell activation involved in immune response [GO:0033007]; negative regulation of peptidyl-tyrosine phosphorylation [GO:0050732]; negative regulation of T cell proliferation [GO:0042130]; negative regulation of T cell receptor signaling pathway [GO:0050860]; negative regulation of tumor necrosis factor production [GO:0032720]; peptidyl-tyrosine dephosphorylation [GO:0035335]; peptidyl-tyrosine phosphorylation [GO:0018108]; platelet aggregation [GO:0070527]; platelet formation [GO:0030220]; positive regulation of cell adhesion mediated by integrin [GO:0033630]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; protein dephosphorylation [GO:0006470]; regulation of apoptotic process [GO:0042981]; regulation of B cell differentiation [GO:0045577]; regulation of ERK1 and ERK2 cascade [GO:0070372]; regulation of G1/S transition of mitotic cell cycle [GO:2000045]; regulation of release of sequestered calcium ion into cytosol [GO:0051279]; regulation of type I interferon-mediated signaling pathway [GO:0060338]; T cell costimulation [GO:0031295] | 11826756_The Lyn/CD22/SHP-1 pathway is important in autoimmunity. Naive and tolerant B-cells differ in their calcium signaling in response to antigenic stimulation. 11858824_peripheral blood monocytes treated for 5--8 days with GM-CSF (i.e. mature Mphi) acquired the capacity to assemble tyrosine phosphatase SHP-1 with CD9, alpha3-beta1 integrin and their CD46. 11907066_SHP-1 is recruited to the NK cell immune synapse within 1 min in both cytolytic and noncytolytic conjugates, allowing clear distinction between noncytolytic and cytolytic interactions at this early time. 11986327_interacts with siglec-11 11987243_evidence of cytokine-regulated SHP-1 nuclear localization mediated by a bipartite nuclear localization signal, suggesting that SHP-1 regulates nuclear signaling in cell growth control. 12130517_ILT2 triggering on cutaneous T-cell lymphoma (CTCL) cells leads to the recruitment of SHP-1 and to the specific inhibition of CTCL malignant cell proliferation in Sezary syndrome induced by CD3/T-cell receptor. (TCR) stimulation. 12145687_In the T cell line HUT-78, JAK3 was found to be highly phosphorylated. These results suggest that SHP-1 might be involved in maintaining the IL-2R/JAK3 signaling pathway under control and point towards a role of SHP-1 in the pathogenesis of the disease. 12438221_Functional loss of SHP1 is associated with the pathogenesis of leukemias/lymphomas. 12459556_participation in regulation of Stat6 dephosphorylation 12482860_crystal structure of the C-terminal truncated human SHP-1 in the inactive conformation at 2.8-A resolution and refined the structure to a crystallographic R-factor of 24.0% 12571228_SHP-1 may be involved in the regulation of beta-catenin transcriptional function and in the negative control of intestinal epithelial cell proliferation 12591278_show the existence of multiple transcripts of SHP-1 in human transformed T cells and normal PBMCs and give evidence supporting the concept that SHP-1 can negatively regulate growth of malignant T cells 12646642_SHP-1 plays a pivotal role in reorganization of cytoskeletal architecture by inducing actin dephosphorylation. 12734331_A blockade in early signaling events observed with TCR antagonists occurs via a negative intracellular signal that is mediated by SHP-1. 12832410_Associates with phosphorylated Fc gamma RIIa to modulate signaling events in myeloid cells 12917349_Vav1 trapping was independent of actin polymerization, suggesting that inhibition of cellular cytotoxicity occurs through an early dephosphorylation of Vav1 by SHP-1, which blocks actin-dependent activation signals. 14551136_SHP-1 inhibits PRL receptor and EPO receptor signaling by recruitment and targeting of SOCS-1 to Jak2, highlighting a new mechanism of SHP-1 regulation of cytokine-receptor signaling. 14630083_Down-regulation of SHP1 is associated with adult T-cell leukemia 14691303_SHP1 gene silencing is one of the events critical to the onset of malignant lymphomas/leukemias in humans. 14976049_hypermethylated in multiple myeloma 15070900_SHP-1 can dephosphorylate alpha-actinin in vitro and in vivo. 15197735_CNS myelination was significantly reduced in SHP-1-deficient mice relative to their normal littermates at multiple times during the active period of myelination. 15339845_Mice lacking SHIP-1 have defects in the erythroid and myeloid compartments similar to those in mice lacking Lyn or SHP-1. 15456853_inactivation of Ros by SHP-1-mediated dephosphorylation plays a role in the regulation of complex stability 15557341_TFG is a novel protein able to modulate SHP-1 activity. 15574429_transformation of TF-1 cells with FLT3/ITD mutations suppressed the activity of SHP-1 by approximately 3-fold 15579525_SHP-1 may be a novel target molecule to sensitize erythropoietin action in hemodialysis patients with hyporesponsive anemia 15701718_SHP-1 acts at multiple stages of hematopoietic differentiation to alter lineage balance. Expression of WT SHP-1 reduced myeloid colony numbers while increasing the numbers of secondary embryoid bodies and mixed hematopoietic colonies obtained 15746253_SRIF inhibitory effects on thyroid medullary carcinoma cell proliferation are mediated, at least in part, by SHP-1, which acts through a MAPK-dependent mechanism 15870198_STAT3 may transform cells by inducing epigenetic silencing of SHP-1 in cooperation with DNMT1 and histone deacetylase 1 16326706_Shp-1 mediates the antiproliferative activity of tissue inhibitor of metalloproteinase-2 16501054_Our results suggest an alteration of the SHP-1 modulation by GM-CSF in lipid rafts of PMN with aging. These alterations could contribute to the decreased GM-CSF effects on PMN. 17046078_SHP1 activation, association with Src and dephosphorylation of specific proteins were dependent on extracellular calcium and maintenance of a higher cytosolic calcium plateau. 17079228_Protein phosphatase 6 specifically down-regulates transforming growth factor beta-activated kinase 1 (TAK1) through dephosphorylation of threonine-187 in the activation loop, suppressing inflammatory responses via TAK1 signaling pathways. 17142110_increased activity of inhibitory molecules, such as Src homology domain-containing protein tyrosine phosphatase-1 (SHP-1) and suppressors of cytokine signalling (SOCS), is responsible for age-related failure of GM-CSF to stimulate neutrophil functions 17218319_findings provide an insight into the structure of the hematopoietic cell-specific P2 promoter of the SHP-1 gene and identify PU.1 as the transcriptional activator of the P2 promoter 17227821_These data suggest a mechanism of IC-mediated inhibition of IFN-gamma signaling, which requires the ITAM-containing FcgammaRI, as well as the ITIM-dependent phosphatase SHP-1, ultimately resulting in the suppression of STAT1 phosphorylation. 17239936_SHP-1 loss correlates with, and may contribute to, progression of cutaneous T-cell lymphoma 17272397_novel mechanism regulating GH signaling in which estrogen receptor modulators enhance GH activation of the JAK2/STAT5 pathway in a cell-type-dependent manner by attenuating protein tyrosine phosphatase activities 17404032_In human erythrocytes SHP-1 is present in membranes from resting cells, but in 5% of the protein amount. SHP-1 ensures band 3 dephosphorylation. 17561098_SHP-1 and SHP-2, two structurally related cytoplasmic protein-tyrosine phosphatases with different cellular functions and cell-specific expression patterns, were compared for their intrinsic susceptibility to oxidation by H(2)O(2). 17579069_A novel 6-mer motif SKHKED within the carboxyl terminus of SHP-1 is necessary and sufficient for lipid raft localization and for the function of SHP-1 as a negative regulator of T-cell receptor signaling. 18086677_Thus the phosphorylation and dephosphorylation of caspase-8, mediated by Lyn and SHP-1, respectively, represents a novel, dynamic post-translational mechanism for the regulation of neutrophil apoptosis 18174230_Observational study of gene-disease association. (HuGE Navigator) 18209728_Mononuclear leukocytes of multiple sclerosis patients have a deficiency in SHP-1 and increased inflammatory gene expression. 18441283_SHP-1 plays an important role in regulating oxidative stress. 18502033_TIMP1 functions as a differentiating and survival factor of GC B cells by modulating CD44 and SHP1 in the late centrocyte/post-GC stage, regardless of EBV infection. 18543080_Our results demonstrate putative roles of SHP-1 and SHP-2 in the progression of both Condyloma acuminatum and cervical cancer after HPV infection. 18948549_in HTLV-1-transformed CD4+ T cells, TIPS causes dissociation of transcription factors from the core SHP-1 P2 promoter, which in turn leads to subsequent DNA methylation, an important early step for leukemogenesis. 18952289_IL-10 suppresses the co-stimulatory effect of CD2 in T cells that have intact SHP-1, demonstrating an essential role of SHP-1 in IL-10-mediated direct T cell suppression. 19096001_Downregulation of platelet responsiveness upon contact with LDL by the protein-tyrosine phosphatases SHP-1 and SHP-2. 19104650_Leishmania can exploit a host protein tyrosine phosphatase, namely SHP-1, to directly inactivate IRAK-1. 19166311_The kinetic and mechanistic details of reversible oxidation of SHP-1 and SHP-2, were investigated. 19204726_Observational study of gene-disease association. (HuGE Navigator) 19234487_Lyn is downstream of Jak2, and Jak2 maintains activated Lyn kinase in CML through the SET-PP2A-Shp1 pathway. 19379557_5-aza-2'-deoxycytidine can re-express the silent SHP-1 gene, inhibit cell proliferation and induce apoptosis in K562 cells. 19398961_macrophages of multiple sclerosis patients display a deficiency of SHP-1 expression, heightened activation of STAT6, STAT1, and NF-kappaB and a corresponding inflammatory profile 19542910_Data suggest that age-specific interactions between Siglec-9 and SHP-1 may influence the altered inflammatory responsiveness and longevity of neonatal polymorphonuclear neutrophils. 19543515_increased frequency of promoter hypermethylation in SHP1 gene in acute myeloid leukemia compared to chronic myeloid leukemia. 19551406_Demethylation in HT-29 cells recovered the constitutive expression of SOCS-1 and SHP-1. 19591923_The SHP-1:PDK1 complex between the cytoplasm and the nucleus is dependent on a specific sequence of events; initial recruitment of PDK1 to perinuclear PIPs, followed by SHP-1 mediated entry via the nuclear pore complex and PDK-1 mediated nuclear export. 19619438_As(2)O(3) could cause demethylation and re-expression of SHP-1 in T2 lymphoma cells, and may inhibit proliferation and induce apoptosis. 19749791_p53 repression of SHP-1 expression leads to trkA-Y674/Y675 phosphorylation and trkA-dependent suppression of breast-cancer cell proliferation. 19789387_SHP-1 is phosphorylated in a dynamic fashion after endorepellin stimulation. It is an essential mediator of endorepellin activity. A novel functional interaction between the integrin alpha2 subunit & SHP-1 is described. 19838216_Data show that SHP-1 knockdown increases p27stability, decreases the CDK6 levels, inducing retinoblastoma protein hypophosphorylation, downregulation of cyclin E and thereby a decrease in the CDK2 activity. 19874234_SHP-1 plays a minor role in determining autoimmune hemolytic anemia and its expression is upregulated in monocytes from these patients. 19950550_As2O3 can effectively cause demethylation and inhibit the growth of tumor by reactivating the SHP-1 gene transcription. 20068065_Plumbagin inhibits STAT3 activation pathway through the induction of SHP-1 and this may mediate the sensitization of STAT3 overexpressing cancers to chemotherapeutic agents. 20117097_SHP-1 plays an important role in airway mucin production through regulating oxidative stress. 20130595_Forced expression of SOCS3 and SHP-1 inhibits IFN-alpha signaling in psoriatic T cells. 20196786_5-aza-dc can induce SHP1 expression and inhibit JAK2/STAT3/STAT5 signalling 20351292_TonEBP/OREBP is extensively regulated by phosphatases, including SHP-1, whose inhibition by high NaCl increases phosphorylation of TonEBP/OREBP at Y143, contributing to the nuclear localization and activation of TonEBP/OREBP 20398180_These results suggest that calpain-dependent cleavage of SHP-1 and SHP-2 may contribute to protein tyrosine dephosphorylation in Jurkat T cell death induced by Entamoeba histolytica. 20424160_Results support the notion that the tumor suppressor effects of SHP1 on NPM-ALK are dependent on its ability to bind to this oncogenic protein. 20687222_SHP-1 may be a therapeutic target in prostate cancer, even when there is an IL-6-related growth advantage. 20696858_SHP-1 has disparate effects on subpopulations of responding CD8 T lymphocytes, reducing the number of short-lived effector cells generated without affecting the size of the memory precursor effector cell pool that leads to long-term memory formation. 20840866_SHP-1 negatively controls tyrosine phosphorylation of beta-catenin, thus inhibiting its association with TATA-binding protein and promotes beta-catenin degradation into the proteasome through a GSK3beta-dependent mechanism. 21291405_REVIEW: role of the protein tyrosine phosphatase SHP-1 on cell-cycle regulation and its possible roles in tumour onset and progression 21357539_Transgenic SHP-1 inhibits signaling through multiple receptors and regulates NF-kappaB and transcription factor activator protein (AP)-1 signaling in dendritic cells. 21406173_an alteration in PTPN6/SHP1 may contribute to neutrophilic dermatoses 21465528_Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation 21505184_Immunoreceptor-tyrosine-based-activation-motifs suppressive signals subvert the activating function of rafts to promote incorporation of receptors into supramolecular domains where signaling molecules are deactivated by SHP-1. 21505186_Inhibitory ITAM signaling traps activating receptors with the phosphatase SHP-1 to form polarized 'inhibisome' clusters. 21536801_Data indicate that CD300F is an active regulator of TLR-mediated macrophage activation through association with SHP-1 involving MyD88 and/or TRIF. 21604205_SHP-1 is expressed significantly higher in multisystem than single-system Langerhans cell histiocytosis 21719561_in serum- or IGF-1-stimulated breast cancer MCF-7 cells, JNK induces SHP1 expression through the binding of AP-4 and RFX-1 transcription factors to the epithelial tissue-specific SHP1 promoter. 21799016_SHP-1 antagonizes the action of receptor and non-receptor-tyrosine kinases on GIV and down-regulates the phospho-GIV-PI3K-Akt axis of signaling. 21806449_impaired signaling is associated with enhanced phagocytosis in neonatal monocyte-derived macrophages 21818116_data demonstrate that constitutive activation of STAT3 in Sezary syndrome is not due to the loss of SHP-1, but is mediated by constitutive aberrant activation of JAK family members 21821701_SHP-1 could be a new biologic indicator at baseline of IMA sensitivity in patients with chronic myelogenous leukemia. 21900501_These findings showed a novel role for SHP-1 in the regulation of Akt activity through the modulation of the ghrelin/GHSR1a system signaling 21964525_Methylation at the SHP-1 promoter in plasma is associated with non-small cell lung cancer. 22043923_regulation of the TLR3/TRIF-mediated pathway required the combined action of SHP-1 and SHP-2, which could be accomplished by CD300f but not CD300a. 22180308_Data show that silencing of SHP-1 by RNA interference abolished the effects of dovitinib on p-STAT3, indicating that SHP-1, a protein tyrosine phosphatase, mediates the effects of dovitinib. 22210881_Data show that that SPL/RGS/SHP1 complexes are present in resting platelets where constitutive phosphorylation of SPL(Y398) creates an atypical binding site for SHP-1. 22258937_it is indicated that the SHP-1 methylation plays important role in the pathogenesis of myelodysplastic syndrome via activating the JAK/STAT pathway probably 22371396_SHP-1 plays a critical role in abrogating M. pneumoniae-induced IL-8 production in nonasthmatic airway epithelial cells through inhibition of PI3K/Akt and NF-kappaB activity, but it is defective in asthma. 22458809_Studies indicate that CD45, nonreceptor SH2 domain-containing PTP-1 (SHP-1) and PEST domain-enriched tyrosine phosphatase (PEP) are expressed at elevated levels in immune cells and play essential roles in T cell homeostasis. 22458980_Increased SHP-1 promoter methylation may relate in part to decreased SHP-1 expression and increased leukocyte-mediated inflammation in multiple sclerosis. 22488585_PTPN6 epigenetic silencing was recurrent in primary effusion lymphoma & diffuse large B-cell lymphoma, with a higher prevalence in the non-germinal centre subtype. It was associated with the activation of the Jak-Stat pathway. 22539788_Mice lacking protein tyrosine phosphatase Shp1 specifically in dendritic cells (DCs) develop splenomegaly associated with more CD11c+ DCs. 22589543_LST1 is expressed specifically in leukocytes of the myeloid lineage, where it localizes to the tetraspanin-enriched microdomains and it binds SHP-1 and SHP-2 phosphatases. 22730659_Abnormal methylation of SHP-1 gene promoter might have tentative role in the pathogenesis and progression of myelodysplastic syndrome. 22738830_IFN-beta and IFN-gamma/TNF-alpha decrease erythrophagocytosis by human monocytes in vitro, and this effect does not apparently require an increase in SIRP-alpha or SHP-1 expression. 22797910_Data suggest that protein tyrosine phosphatase non-receptor type 6 (SHP-1) may interact with EGF receptor (EGFR) to inhibit proliferation. 22960265_This study is the first to identify and define an interaction between SHP-1 and an SH3 domain-containing protein. Our findings provide an alternative way that SHP-1 can be linked to potential substrates. 23074279_Data show that TIMP-2-mediated inhibition of vascular endothelial cell permeability involves an integrin alpha3beta1-Shp-1-cAMP/protein kinase A-dependent vascular endothelial cadherin cytoskeletal association. 23391724_The global role of SHP-1 in regulating CD8 positive T cell activation. 23406209_These results suggest the involvement of the ZAP70 and PTPN6 genes in the genetic component conferring a general susceptibility to Crohn's disease and ulcerative colitis, respectively. 23729600_Data suggest growth retardation in idiopathic short stature is related to delayed activation of JAK2 (Janus kinase 2) by PTP1B (protein tyrosine phosphatase, non-receptor type 1) and SHP1 in response to GH/GHR (growth hormone/GH receptor) signaling. 23766558_The endothelial tyrosine phosphatase SHP-1 plays an important role for vascular hemostasis in vivo, which is crucial in TNF alpha -induced endothelial inflammation. 23824557_A high level of SHP1P2 methylation of hilar lymph nodes from stage I NSCLC patients is associated with early relapse of disease. 23842094_SHP1 decreases the radiosensitivity of non-small cell lung cancer cells through affecting cell cycle distribution 23948750_results strongly suggest that expression of NTRK1-pY674/pY675 together with wild-type TP53 and low levels of PTPN6 expression are predictors of improved disease-free survival in breast cancer 23979523_studies have uncovered novel epigenetic mechanisms of PTPN6 suppression and suggest that PTPN6 may be a potential target of epigenetic therapy in DLBCL. 24100145_study identified novel SHP-1 transcripts in epithelial cancer cells; each product of ORF SHP-1 transcripts has phosphatase activity and isoforms with an N-SH2 deletion have increased phosphatase activity and novel protein-protein interactions; study documents utilization of promoter 2 by SHP-1 transcripts and a noncoding transcript in epithelial cells 24510345_PTPN6 expression may be a factor contributing to poor survival for anaplastic glioma patients. 24587194_hERG and hEAG channels are regulated by Src and by SHP-1 tyrosine phosphatase via an ITIM region in the cyclic nucleotide binding domain. 24647617_decreased expression levels of SHP-1 caused by aberrant promoter hypermethylation may play a key role in the progression of CML by dysregulating BCR-ABL1, AKT, MAPK, MYC and JAK2/STAT5 signaling. 24793756_SHP-1 expression ws lower in PBMCs from unmedicated schizophrenics. The promoter region was hypermethylated. Silencing SHP-1 induced IKK/NF-kB and pro-inflammatory cytokines. SHP-1 expression may explain 30% of the clinical negative symptom variance. 24824657_We demonstrated that the association and oxidation of c-Src and SHP-1 by ROS are key steps in enhancing OC survival, which are responsible for increased bone loss when ovarian function ceases. 24886428_A PKC-SHP1 signaling axis desensitizes Fcgamma receptor signaling by reducing the tyrosine phosphorylation of CBL and regulates FcgammaR mediated phagocytosis. 24952874_transcription factor RFX-1 regulates SC-2001-mediated SHP-1 Phosphatase transcription in hepatocellular carcinoma cells. 25619838_SHP-1 is a potent suppressor of epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. 25635370_SHP-1(I) presented concordance between unmethylated promoter region and tumor for breast or prostate. 25785436_Results show that dissociation of SHP-1 from spinophilin is followed by an increase in the binding of spinophilin to PP1. 25799543_Data show that SHP-1 promotes HIF-1alpha degradation under hypoxic conditions leading to a reduction in VEGF synthesis and secretion and impairing epithelial cells proliferation. 25824741_Inactivation of SHP1 is associated with myeloproliferative neoplasm. 25897665_Soluble egg antigens glycans are essential for induction of enhanced SOCS1 and SHP1 levels in dendritic cells via the mannose receptor. 25962492_Phosphorylation of ATR and CHK1 did not show significant differences in the three cell groups. Overexpression of SHP-1 in the CNE-2 cells led to radioresistance and the radioresistance was related to enhanced DNA DSB repair. 26081980_miR-4649-3p is downregulated in nasopharyngeal carcinoma cell lines accompanied with SHP-1 upregulation. 26215037_SHP-1 has a critical role in radioresistance, cell cycle progression, and senescence of nasopharyngeal carcinoma cells 26334669_cAMP signalling of Bordetella adenylate cyclase toxin through the SHP-1 phosphatase activates the BimEL-Bax pro-apoptotic cascade in phagocytes. 26373709_This study also found no correlation of SHP-1 expression at diagnosis with response to treatment, although a trend for lower SHP-1 expression was noted in the very small non-responders' group of the 3-month therapeutic milestone. 26473472_MiR-378g enhanced radiosensitivity partially by targeting SHP-1 in NPC cells. 26508024_Overexpression of SHP1 downregulates the JAK2/STAT3 pathway to modulate various target genes and inhibit cell proliferation, migration, and invasion in gastric cancer cells. 26565811_N225K and A550V PTPN6 mutations cause loss-of-function leading to JAK3 mediated deregulation of STAT3 pathway and uncover a mechanism that tumor cells can use to control PTPN6 substrate specificity. 26597461_Data demonstrate for the first time that SHP1 methylation has high specificity for diagnosis of endometrial carcinoma, while CDH13 promoter methylation plays a role in the earlier stage. 26679051_A combination of sorafenib and SC-43 is a synergistic SHP-1 agonist duo which reduces tumor size and prolonged survival time. 26742467_SYK, LYN and PTPN6 were markedly elevated in atherosclerotic plaques of carotid atherosclerosis patients. 26781335_Quinalizarin enhances radiosensitivity of nasopharyngeal carcinoma cells partially by suppressing SHP-1 expression 26878112_In addition to their role in NK cell activation by hematopoietic cells, the SLAM-SAP-SHP1 pathways influence responsiveness toward nonhematopoietic targets by a process akin to NK cell 'education'. 26959741_we demonstrated that SHP-1 dephosphorylates PKM2Y105 to inhibit the Warburg effect and nucleus-dependent cell proliferation, and the dephosphorylation of PKM2Y105 by SHP-1 determines the efficacy of targeted drugs for hepatocellular carcinoma treatment 27216862_this review focalizes upon the implication of SHP-1 in the pathogenesis of autoimmune disorders, and addresses developing therapeutic strategies targeting SHP-1 27364975_Data suggest that SHP-1/p-STAT3/VEGF-A axis is a potential therapeutic target for metastatic triple-negative breast cancer (TNBC). 27572445_The results reveal that SHP1 is the long-sought phosphatase that can antagonize Helicobacter pylori CagA. Augmented Helicobacter pylori CagA activity, via SHP1 inhibition, might also contribute to the development of Epstein-Barr virus-positive gastric cancer. 27585521_Hyperglycemia induces SHP-1 promoter epigenetic modifications, causing its persistent expression and activity and leading to insulin resistance, podocyte dysfunction, and DN. 27644671_study evaluated SHP1-P2 methylation levels in the lymph nodes of colorectal cancer (CRC) patients; hypothesized that SHP1-P2 methylation levels would be higher in metastatic lymph nodes 27814644_Analyzed gene expression profiles of monocytes from symptomatic congestive heart failure patients; there is a down-regulation of the phosphatase SHP-1 which induces a significant activation of TAK-1/IKK/NF-kB signaling. 27959415_Low SHP1 expression is associated with primary central nervous system lymphoma. 28182003_luteolin inhibited STAT3 activation through disrupting the binding of HSP-90 to STAT3, which promoted its interaction to SHP-1. 28187032_the absence of SHP-1 in immunohistochemical analysis might serve as a marker of poor prognosis for a subset of high-grade endometrial cancer. 28210822_SHP1 DNA methylation in in patients with B cell non-Hodgkin lymphoma 28250424_we found that THEMIS directly regulated the catalytic activity of the tyrosine phosphatase SHP-1. 28295507_crocin induced the expression of SHP-1, a tyrosine protein phosphatase, and pervanadate treatment reversed the crocin-induced downregulation of STAT3, suggesting the involvement of a protein tyrosine phosphatase. 28369102_Results provide evidence that repression of SHP-1, SHP-2 and SOCS-1 gene expression in patient plasma cells supports the constitutive activation of the JAK/STAT3 pathway and probably leads to higher degrees of bone marrow invasion. 28376405_these results indicate that DNMT1 mediates aberrant methylation and silencing of SHP-1 gene in chronic myelogenous leukemia cells 28378014_Biophysical assay for tethered signaling reactions reveals tether-controlled activity for the phosphatase SHP-1. 28416389_The Shp1 functions as a positive regulator and acts in a novel mechanism through promoting EGFR protein expression in human epithelial cells. 28465325_These findings show a novel role for Shp-1 in the regulation of IEC growth and secretory lineage allocation, possibly via modulation of PI3K/Akt-dependent signaling pathways. 28480959_PTPN6 is associated with progression of chronic myeloid leukaemia. Low expression level of PTPN6 was associated with DNA methylation and regulated by histone acetylation 28533521_this study provided strong in vivo and cellular evidences that hepatocyte SHP-1 plays a cardinal role in the production of inflammatory mediators that contribute to endotoxemia. 28606940_the role of Shp1 in myeloid cells and how its dysregulation affects immune function, which can impact human disease. 28692056_These findings have provided first lines of evidences that PDZK1 expression is negatively correlated with SHP-1 activation and poor clinical outcomes in clear cell renal cell carcinoma (ccRCC) . PDZK1 was identified as a novel tumor suppressor in ccRCC by negating SHP-1 activity 28790195_these data highlight a signaling pathway in which SHP-1 acts through CrkII to reshape the pattern of Rap1 activation in the immunological synapse. 28978467_M. tuberculosis-initiated human mannose receptor signaling regulates macrophage recognition and vesicle trafficking by gamma Fc receptors, Grb2, and SHP-1. 29058147_observations suggest that Chikungunya virus (CHIKV) has the ability to induce host PTPN6 expression, and induction of PTPN6 may favour the attenuation of the pro-inflammatory immune response of the host, which is otherwise detrimental for the survival of CHIKV and establishment of an infection 29288235_Data suggest that targeting the ring finger 6 protein (RNF6)/protein-tyrosine phosphatase SHP-1 (SHP-1)/signal transducer and activator of transcription 3 (STAT3) axis provides a potential therapeutic option for RNF6-amplified tumors. 29298416_FcgammaRIIB expressed in neurons functions as a receptor for alpha-syn fibrils and mediates cell-to-cell transmission of alpha-syn. SHP-1 and 2 are activated downstream by alpha-syn fibrils through FcgammaRIIB and play an important role in cell-to-cell transmission of alpha-syn. 29449322_actomyosin retrograde flow controls the immune response of primary human Natural killer cells through a novel interaction between beta-actin and the SH2-domain-containing protein tyrosine phosphatase-1 (SHP-1), converting its conformation state, and thereby regulating Natural killer cell cytotoxicity. 29466992_Our data revealed that HNF1A-AS1 is a direct transactivation target of HNF1alpha in HCC cells and involved in the anti-HCC effect of HNF1alpha. HNF1A-AS1 functions as phosphatase activator through the direct interaction with SHP-1. 29776962_The nonreceptor protein tyrosine phosphatase SHP-1 acts as a tumor suppressor in hepatocellular carcinoma 29931651_PTPN6 is required for the cell-surface expression of TRPM4 channels in HEK293 cells. 29961065_This study demonstrates that VB inhibits glioblastoma cell proliferation, migration, and invasion while promoting apoptosis via SHP-1 activation and inhibition of STAT3 phosphorylation. 30112836_Data show that E3 ubiquitin-protein ligase RNF38 (RNF38) interacts with the nonreceptor tyrosine phosphatase SH2-containing protein tyrosine phosphatase 1 (SHP-1) and induces the polyubiquitination of SHP-1, which leads to destabilization of SHP-1. 30420593_These findings support the notion that control of pSTAT3 in TCL tumors is complex and involves epigenetic silencing of the tyrosine phosphatase PTPN6. 30470842_Study demonstrates that miR-152 can inhibit lymphoma growth via suppressing DNMT1-mediated silencing of SOCS3 and SHP-1. These data demonstrate a new mechanism for the development of NHL. 30674631_HoxA10 interacts with p38 MAPK to repress the activation of p38 MAPK and STAT3 and recruits and facilitates SHP-1 to catalyze the dephosphorylation of p38 MAPK and STAT3. 30764849_SHP1 and SHP2 are relevant mediators of differentiation in acute myeloid leukemia cells 30816454_PTPN6 may serve as tumor suppressor gene in ESCC and inhibit esophageal cancer cell proliferation and invasion. The P2 is frequently methylated in esophageal cancer cells and ESCC tissues, and this may be one of the epigenetic mechanisms implicated in PTPN6 silencing in ESCC. 31000475_A rare in-silico-predicted damaging variant (Ala455Thr) in the protein tyrosine phosphatase non-receptor type 6 (PTPN6) gene is shown to cause early-onset emphysema. 31427082_The Ang1 induced the dissociation of the tyrosine phosphatase SHP-1 from the Tie2 receptor and increased the binding of SHP-1 to JAK1, JAK2, and STAT3, which are IL-6 downstream signaling proteins. 31550807_revealed that the SHP-1 methylation rate was positively correlated with the positive rate of STAT3 phosphorylation 31836852_SHP1 regulates a STAT6-ITGB3 axis in FLT3ITD-positive AML cells. 32004441_synapse formed by 4-1BB-encoding CAR recruits the THEMIS-SHP1 phosphatase complex that attenuates CAR-CD3zeta phosphorylationk 32343611_Decrease in SHP-1 enhances myometrium remodeling via FAK activation leading to labor. 32390601_Immunosuppressive receptor LILRB1 acts as a potential regulator in hepatocellular carcinoma by integrating with SHP1. 32437509_PD-1 and BTLA regulate T cell signaling differentially and only partially through SHP1 and SHP2. 32454905_The Analysis of PTPN6 for Bladder Cancer: An Exploratory Study Based on TCGA. 32674045_Correlation between SHP-1 and carotid plaque vulnerability in humans. 32779804_SHP-1 suppresses the antiviral innate immune response by targeting TRAF3. 32917126_Vitamin D3-VDR-PTPN6 axis mediated autophagy contributes to the inhibition of macrophage foam cell formation. 33133093_Shp1 Loss Enhances Macrophage Effector Function and Promotes Anti-Tumor Immunity. 33264612_SHP-1 Regulates Antigen Cross-Presentation and Is Exploited by Leishmania to Evade Immunity. 33615510_PTPN6 promotes chemosensitivity of colorectal cancer cells via inhibiting the SP1/MAPK signalling pathway. 33639070_SHP1 Decreases Level of P-STAT3 (Ser727) and Inhibits Proliferation and Migration of Pancreatic Cancer Cells. 33736531_The role of Src homology region 2 domain-containing phosphatase-1 hypermethylation in the classification of patients with myelodysplastic syndromes and its association with signal transducer and activator of transcription 3 phosphorylation in skm-1 cells. 33990399_The Protein Tyrosine Phosphatase SHP-1 (PTPN6) but Not CD45 (PTPRC) Is Essential for the Ligand-Mediated Regulation of CD22 in BCR-Ligated B Cells. 34088320_The phosphatase Shp1 interacts with and dephosphorylates cortactin to inhibit invadopodia function. 34535085_Circular RNA-0007059 protects cell viability and reduces inflammation in a nephritis cell model by inhibiting microRNA-1278/SHP-1/STAT3 signaling. 34910686_METTL14 benefits the mesenchymal stem cells in patients with | ENSMUSG00000004266 | Ptpn6 | 1.847991e+01 | 1.1372961 | 0.185607877 | 0.7250925 | 6.050023e-02 | 0.8057070540 | 0.96141221 | No | Yes | 1.337418e+01 | 5.848429 | 1.211166e+01 | 5.512165 | |
| ENSG00000111785 | 55188 | RIC8B | protein_coding | Q9NVN3 | FUNCTION: Guanine nucleotide exchange factor (GEF), which can activate some, but not all, G-alpha proteins by exchanging bound GDP for free GTP. Able to potentiate G(olf)-alpha-dependent cAMP accumulation suggesting that it may be an important component for odorant signal transduction. {ECO:0000250}. | Alternative splicing;Cytoplasm;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome | hsa:55188; | cell cortex [GO:0005938]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; G-protein alpha-subunit binding [GO:0001965]; guanyl-nucleotide exchange factor activity [GO:0005085]; G protein-coupled receptor signaling pathway [GO:0007186]; regulation of G protein-coupled receptor signaling pathway [GO:0008277] | 12652642_We identified a protein member of the synembryn family as one of the interacting proteins in human brain. Gqalpha also interacts with synembryn. Synembryn translocates to the plasma membrane in response to carbachol and isoproterenol. 20133939_Ric-8B plays a critical and specific role in the control of G alpha(s) protein levels by modulating G alpha(s) ubiquitination and positively regulates G(s) signaling 26729411_these data show the existence of functional CREB and C/EBP binding sites in the human RIC8B gene promoter, a particular distribution of these sites and demonstrate a relevant role of CREB in stimulating transcriptional activity of this gene. 26966186_overexpression of ARMS blocked NGF-mediated secretion, without affecting basal secretion, a decrease in ARMS resulted in potentiation. Similar effects were observed with synembryn-B, a protein that interacts directly with ARMS. Downstream of ARMS and synembryn-B are Galphaq and Trio proteins, which modulate the activity of Rac1 in response to NGF | ENSMUSG00000035620 | Ric8b | 3.413635e+02 | 0.8593449 | -0.218690869 | 0.3410192 | 4.168660e-01 | 0.5185050276 | 0.88281356 | No | Yes | 2.904629e+02 | 57.963785 | 3.085205e+02 | 62.889011 | ||
| ENSG00000112062 | 1432 | MAPK14 | protein_coding | Q16539 | FUNCTION: Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK14 is one of the four p38 MAPKs which play an important role in the cascades of cellular responses evoked by extracellular stimuli such as proinflammatory cytokines or physical stress leading to direct activation of transcription factors. Accordingly, p38 MAPKs phosphorylate a broad range of proteins and it has been estimated that they may have approximately 200 to 300 substrates each. Some of the targets are downstream kinases which are activated through phosphorylation and further phosphorylate additional targets. RPS6KA5/MSK1 and RPS6KA4/MSK2 can directly phosphorylate and activate transcription factors such as CREB1, ATF1, the NF-kappa-B isoform RELA/NFKB3, STAT1 and STAT3, but can also phosphorylate histone H3 and the nucleosomal protein HMGN1. RPS6KA5/MSK1 and RPS6KA4/MSK2 play important roles in the rapid induction of immediate-early genes in response to stress or mitogenic stimuli, either by inducing chromatin remodeling or by recruiting the transcription machinery. On the other hand, two other kinase targets, MAPKAPK2/MK2 and MAPKAPK3/MK3, participate in the control of gene expression mostly at the post-transcriptional level, by phosphorylating ZFP36 (tristetraprolin) and ELAVL1, and by regulating EEF2K, which is important for the elongation of mRNA during translation. MKNK1/MNK1 and MKNK2/MNK2, two other kinases activated by p38 MAPKs, regulate protein synthesis by phosphorylating the initiation factor EIF4E2. MAPK14 interacts also with casein kinase II, leading to its activation through autophosphorylation and further phosphorylation of TP53/p53. In the cytoplasm, the p38 MAPK pathway is an important regulator of protein turnover. For example, CFLAR is an inhibitor of TNF-induced apoptosis whose proteasome-mediated degradation is regulated by p38 MAPK phosphorylation. In a similar way, MAPK14 phosphorylates the ubiquitin ligase SIAH2, regulating its activity towards EGLN3. MAPK14 may also inhibit the lysosomal degradation pathway of autophagy by interfering with the intracellular trafficking of the transmembrane protein ATG9. Another function of MAPK14 is to regulate the endocytosis of membrane receptors by different mechanisms that impinge on the small GTPase RAB5A. In addition, clathrin-mediated EGFR internalization induced by inflammatory cytokines and UV irradiation depends on MAPK14-mediated phosphorylation of EGFR itself as well as of RAB5A effectors. Ectodomain shedding of transmembrane proteins is regulated by p38 MAPKs as well. In response to inflammatory stimuli, p38 MAPKs phosphorylate the membrane-associated metalloprotease ADAM17. Such phosphorylation is required for ADAM17-mediated ectodomain shedding of TGF-alpha family ligands, which results in the activation of EGFR signaling and cell proliferation. Another p38 MAPK substrate is FGFR1. FGFR1 can be translocated from the extracellular space into the cytosol and nucleus of target cells, and regulates processes such as rRNA synthesis and cell growth. FGFR1 translocation requires p38 MAPK activation. In the nucleus, many transcription factors are phosphorylated and activated by p38 MAPKs in response to different stimuli. Classical examples include ATF1, ATF2, ATF6, ELK1, PTPRH, DDIT3, TP53/p53 and MEF2C and MEF2A. The p38 MAPKs are emerging as important modulators of gene expression by regulating chromatin modifiers and remodelers. The promoters of several genes involved in the inflammatory response, such as IL6, IL8 and IL12B, display a p38 MAPK-dependent enrichment of histone H3 phosphorylation on 'Ser-10' (H3S10ph) in LPS-stimulated myeloid cells. This phosphorylation enhances the accessibility of the cryptic NF-kappa-B-binding sites marking promoters for increased NF-kappa-B recruitment. Phosphorylates CDC25B and CDC25C which is required for binding to 14-3-3 proteins and leads to initiation of a G2 delay after ultraviolet radiation. Phosphorylates TIAR following DNA damage, releasing TIAR from GADD45A mRNA and preventing mRNA degradation. The p38 MAPKs may also have kinase-independent roles, which are thought to be due to the binding to targets in the absence of phosphorylation. Protein O-Glc-N-acylation catalyzed by the OGT is regulated by MAPK14, and, although OGT does not seem to be phosphorylated by MAPK14, their interaction increases upon MAPK14 activation induced by glucose deprivation. This interaction may regulate OGT activity by recruiting it to specific targets such as neurofilament H, stimulating its O-Glc-N-acylation. Required in mid-fetal development for the growth of embryo-derived blood vessels in the labyrinth layer of the placenta. Also plays an essential role in developmental and stress-induced erythropoiesis, through regulation of EPO gene expression. Isoform MXI2 activation is stimulated by mitogens and oxidative stress and only poorly phosphorylates ELK1 and ATF2. Isoform EXIP may play a role in the early onset of apoptosis. Phosphorylates S100A9 at 'Thr-113'. {ECO:0000269|PubMed:10330143, ECO:0000269|PubMed:10747897, ECO:0000269|PubMed:10943842, ECO:0000269|PubMed:11154262, ECO:0000269|PubMed:11333986, ECO:0000269|PubMed:15905572, ECO:0000269|PubMed:16932740, ECO:0000269|PubMed:17003045, ECO:0000269|PubMed:17724032, ECO:0000269|PubMed:19893488, ECO:0000269|PubMed:20188673, ECO:0000269|PubMed:20932473, ECO:0000269|PubMed:9430721, ECO:0000269|PubMed:9687510, ECO:0000269|PubMed:9792677, ECO:0000269|PubMed:9858528}.; FUNCTION: (Microbial infection) Activated by phosphorylation by M.tuberculosis EsxA in T-cells leading to inhibition of IFN-gamma production; phosphorylation is apparent within 15 minute and is inhibited by kinase-specific inhibitors SB203580 and siRNA (PubMed:21586573). {ECO:0000269|PubMed:21586573}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Apoptosis;Cytoplasm;Direct protein sequencing;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Stress response;Transcription;Transcription regulation;Transferase;Ubl conjugation | The protein encoded by this gene is a member of the MAP kinase family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. This kinase is activated by various environmental stresses and proinflammatory cytokines. The activation requires its phosphorylation by MAP kinase kinases (MKKs), or its autophosphorylation triggered by the interaction of MAP3K7IP1/TAB1 protein with this kinase. The substrates of this kinase include transcription regulator ATF2, MEF2C, and MAX, cell cycle regulator CDC25B, and tumor suppressor p53, which suggest the roles of this kinase in stress related transcription and cell cycle regulation, as well as in genotoxic stress response. Four alternatively spliced transcript variants of this gene encoding distinct isoforms have been reported. [provided by RefSeq, Jul 2008]. | hsa:1432; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; glutamatergic synapse [GO:0098978]; mitochondrion [GO:0005739]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; secretory granule lumen [GO:0034774]; spindle pole [GO:0000922]; ATP binding [GO:0005524]; enzyme binding [GO:0019899]; MAP kinase activity [GO:0004707]; MAP kinase kinase activity [GO:0004708]; mitogen-activated protein kinase p38 binding [GO:0048273]; NFAT protein binding [GO:0051525]; protein phosphatase binding [GO:0019903]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; 3'-UTR-mediated mRNA stabilization [GO:0070935]; angiogenesis [GO:0001525]; apoptotic process [GO:0006915]; bone development [GO:0060348]; cartilage condensation [GO:0001502]; cell morphogenesis [GO:0000902]; cell surface receptor signaling pathway [GO:0007166]; cellular response to ionizing radiation [GO:0071479]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to lipoteichoic acid [GO:0071223]; cellular response to tumor necrosis factor [GO:0071356]; cellular response to vascular endothelial growth factor stimulus [GO:0035924]; cellular response to virus [GO:0098586]; cellular senescence [GO:0090398]; chemotaxis [GO:0006935]; chondrocyte differentiation [GO:0002062]; DNA damage checkpoint signaling [GO:0000077]; fatty acid oxidation [GO:0019395]; glucose metabolic process [GO:0006006]; intracellular signal transduction [GO:0035556]; lipopolysaccharide-mediated signaling pathway [GO:0031663]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of hippo signaling [GO:0035331]; negative regulation of inflammatory response to antigenic stimulus [GO:0002862]; osteoblast differentiation [GO:0001649]; osteoclast differentiation [GO:0030316]; p38MAPK cascade [GO:0038066]; peptidyl-serine phosphorylation [GO:0018105]; placenta development [GO:0001890]; platelet activation [GO:0030168]; positive regulation of brown fat cell differentiation [GO:0090336]; positive regulation of cardiac muscle cell proliferation [GO:0060045]; positive regulation of cyclase activity [GO:0031281]; positive regulation of erythrocyte differentiation [GO:0045648]; positive regulation of gene expression [GO:0010628]; positive regulation of glucose import [GO:0046326]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of muscle cell differentiation [GO:0051149]; positive regulation of myoblast differentiation [GO:0045663]; positive regulation of myoblast fusion [GO:1901741]; positive regulation of myotube differentiation [GO:0010831]; positive regulation of protein import into nucleus [GO:0042307]; positive regulation of reactive oxygen species metabolic process [GO:2000379]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of cytokine production involved in inflammatory response [GO:1900015]; regulation of ossification [GO:0030278]; regulation of synaptic membrane adhesion [GO:0099179]; regulation of transcription by RNA polymerase II [GO:0006357]; response to dietary excess [GO:0002021]; response to insulin [GO:0032868]; response to muramyl dipeptide [GO:0032495]; response to muscle stretch [GO:0035994]; signal transduction [GO:0007165]; signal transduction in response to DNA damage [GO:0042770]; skeletal muscle tissue development [GO:0007519]; stress-induced premature senescence [GO:0090400]; striated muscle cell differentiation [GO:0051146]; transmembrane receptor protein serine/threonine kinase signaling pathway [GO:0007178]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11756422_PI3K promotes assembly of adherens junctions, which, in turn, control p38 MAPK activation and enterocyte differentiation. 11773065_p38 Map kinase pathway is a common effector for type I IFN and transforming growth factor beta signaling in human hematopoietic progenitors and plays a critical role in the induction of the suppressive effects of these cytokines on normal hematopoiesis 11788789_endotoxemia induced a phosphorylation and activation of p38 MAPK 11848686_mediates the cell death induced by PrP106-126 11852102_TRAIL/Apo2L-induced apoptosis is mediated by ROS-activated p38 MAP kinase followed by caspase activation in HeLa cells. 11896401_allosteric binding site for a diaryl urea class of highly potent and selective inhibitors against human p38 MAP kinase 11994493_p38alpha MAP kinase is significantly activated in the inflamed colonic mucosa of Crohn's disease patients and is linked to the induction of TNF-alpha secretion and signaling in this system. 12009331_involvement of P38 MAPK, AP-1 and NF-kappa B transcription factors in the IL-1 induction of MMPs in chondrocytes 12051955_Specificity of arachidonic acid-induced inhibition of growth and activation of c-jun kinases and p38 mitogen-activated protein kinase in hematopoietic cells 12067303_Activation of p38 mitogen-activated protein kinase and nuclear factor-kappaB in tumour necrosis factor-induced eotaxin release of human eosinophils. 12071847_UVB-mediated activation of p38 mitogen-activated protein kinase enhances resistance of normal human keratinocytes to apoptosis by stabilizing cytoplasmic p53. 12085235_Distinct roles for phosphoinositide 3-kinase, mitogen-activated protein kinase and p38 MAPK in mediating cell cycle progression of breast cancer cells. 12126643_P38 MAPK has a role in inducing IL-8 gene transcription via AP-1 activation in hVSMCs 12164931_activation of this stress-associated pathway has been reported for many stimuli, a unique translocation of p38 from the cytoplasm to the detergent-resistant fraction is a specific event during hapten-induced activation of antigen-presenting cells. 12170268_parthenolide inhibits LPS- but not TNF-alpha-induced maturation of human monocyte-derived dendritic cells by inhibition of the p38 mitogen-activated protein kinase pathway 12183063_both Ca(2+)-dependent and p38 kinase-mediated phosphorylation events, causes reassembly of the actin cytoskeleton and subsequent appearance of membrane blebs at the plasma membrane. 12200131_ITGB1 activated by ERK1/2, p38 MAPK after hypoxia 12232043_p38 MAPK has a role in STAT1 dependent trans activation independent of serine phosphorylation 12296996_As(2)O(3) is able to induce the apoptotic activity in K562 cells, and its apoptotic mechanism may be associated with the activation of p38. 12324467_there is a mechanically coupled transcriptional circuit that activates p38, tethers p38 to actin filaments, and promotes binding of p38 to Sp1 in the nucleus 12370831_p38 play a critical role in the UVA-induced expression of COX-2 in keratinocytes and may serve as a potential drug target in the chemoprevention of skin cancer. 12384504_p38 MAP Kinase suppresses the function of Mirk as a transcriptional activator only when cells are proliferating 12429732_p38 alpha is regulated by TauAlphaBeta1beta in physiological conditions 12473116_p38MAPK and caspase are required for zinc-induced apoptosis in human leukemia HL-60 cells 12478662_JNK-1 and p38 play a role in apoptosis induced by capsaicin in H-ras-transformed tumor cells 12505871_data suggest that thrombin induces p38 mitogen-activated protein kinase activation, which leads to NF-kappa B mobilization to the nucleus and causes the upregulation of E-selectin and subsequent leukocyte recruitment 12509443_p38 MAP kinase is involved in a positive feedback loop regulating macrophage signaling with MKK6 12519482_p38 signaling is down-regulated in postinjury circulating neutrophils 12581156_p38 mitogen-activated protein kinase defines the common senescence-signaling pathway. 12595530_Lipopolysaccharide stimulates p38-dependent induction of antiviral genes , such as myxovirus resistance-1 (MX1) in neutrophils independently of paracrine factors. 12606502_upregulation of the p38 mitogen-activated protein kinase pathway might contribute to the loss of GLUT4 protein expression observed in adipose tissue from type 2 diabetic patients 12631076_activated p38 MAPK pathway may play a role in the pathogenesis of peritoneal fibrosis. 12654092_p38 MAPK activation is central to IL-10 production of cells from patients with schistosomiasis mansoni and the levels of phosphorylated p38 MAPK influences the amount of Il-10 produced in cells from intestinal and hepatosplenic patients 12663671_p38 and ERK1/2 have roles in coordinating cellular migration and proliferation in epithelial wound healing 12698197_This additivism, where doxorubicin acts via p53 expression and vinorelbine through p38 MAP KINASE activation, may contribute to the high clinical response rate when the two drugs are used together in the treatment of breast cancer. 12702497_results indicate that epithelial TLR5 mediates p38 activation and subsequently regulates flagellin-induced IL-8 expression independently of NF-kappaB, probably by influencing IL-8 mRNA translation 12784337_These findings suggested that reduction of the p38 MAPK cascade may account, in part, for the resistance to apoptosis, leading to the unrestricted cell growth of human HCC. 12810082_Results suggest that tissue or plasma fibronectin may modulate the intestinal epithelial response to repetitive deformation through inhibted activation of p38 and jun kinases. 12855667_p38 has a role in apoptosis in human melanoma cells by modulating depolarization of mitochondrial membrane potential 12882963_p38 MAPK pathway predominantly regulates deadenylation, rather than decay of the mRNA body 12933809_transforming growth factor-beta promotes osteoclastogenesis in monocytes by stimulation of the p38 mitogen activated protein kinase but continuous exposure abrogates osteoclastogenesis by down-regulation of receptor activator of NF-KB(RANK) expression 13679851_p38 is selectively activated by the new subfamily of Ste20-like kinases. 14552880_p38 MAPK is activated at early stages of neurofibrillary degeneration in AD hippocampus. The p38 activation may also be linked to neurodegeneration through mechanisms other than neurofibrillary tangle formation 14578350_p38alpha regulates activation of downstream effectors that participate in the induction of IFN-dependent gene transcription to mediate IFN-responses 14592977_TAB1 participates in a SAPK2a/p38alpha-mediated feedback control of TAK1 14648593_p38 MAPK modulates STAT1 phosphorylation in IFN-gamma signaling in fetal brain astrocytes. 14714552_epidermal p44/42 and p38 mitogen-activated protein kinases are activated by acute cutaneous barrier disruption 14715645_Pressure distension of intact human saphenous vein induces activation of p38, and this is associated with apoptosis. Inhibition of p38 kinase activity in saphenous vein smooth muscle cells and intact vein reduces apoptosis. 14970175_p38-MAPK can directly phosphorylate and inhibit the activities of caspase-8 and caspase-3 and thereby hinder neutrophil apoptosis, and, in so doing, regulate the inflammatory response. 14989597_activation of p38MAPK might contribute significantly to the pathology of motor neurons in ALS. 15132952_dual signaling pathways involving TGF-beta1, an antiapoptotic pathway mediated by the Smad pathway involving p21, and an apoptosis-permissive pathway mediated in part by p38 MAPK. 15133024_p38alpha and Smad signaling pathways have roles in MSK1-mediated transforming growth factor-beta-mediated responses 15170913_MIF activates rheumatoid arthritis COX-2 and IL-6 expression via p38 MAP kinase activation and induces IL-8 via p38 and ERK MAP kinase-independent pathways. 15232685_Increased p38 MAPK signalling is a feature of human and experimental diabetic nephropathy 15245437_dual effect of UVB, to stimulate maturation or to induce apoptosis in monocyte derived dendritic cells, depending on the dosage, via p38 MAPK pathway. 15246972_cAMP concentration in endothelial cells prevents thrombin-induced ICAM-1 expression by inhibiting p38 MAPK activation, which in turn prevents phosphorylation of RelA/p65 and transcriptional activity of the bound NF-kappaB. 15256218_PMA-induced activations of ERK and p38 kinase are parallel events that are both required for inhibition of NO-induced death of Molt4 cells 15257094_endotoxin increases sTRAIL where the p38 MAP kinase signaling pathway is involved 15284239_active p38 mutants emulate the conformational changes imposed naturally by dual phosphorylation 15292176_ERK suppresses stress-induced apoptosis downstream of mitochondrial alterations by maintaining XIAP levels and oxidants block this effect through activation of p38 and protein phosphatases 15292225_Suppression of glucocorticoid receptor function by activated p38 may be important as a mechanism of resistance to glucocorticoids seen in many patients with chronic inflammatory conditions. 15304097_cholesterol depletion alters involucrin gene expression through activation of p38alpha/beta. 15304486_RANKL-induced cathepsin K gene expression is cooperatively regulated by the combination of the transcription factors and p38 MAP kinase in a gradual manner. 15310753_Data show that sustained activation of p38(MAPK) is essential for the death cascade following exposure of Ewing's sarcoma tumor cells to bFGF and provide evidence that activation of p38(MAPK) results in an up-regulation of the death receptor p75(NTR). 15315972_p38 is a central component of NK cell ability to directly respond to dsRNA pathogen-associated molecular pattern (PAMP). 15319438_NF-kappaB and pp38 mitogen-activated protein kinase have roles in endoplasmic reticulum stress-stimulated expression of cyclooxygenase 2 15322009_Apoptosis of neutrophils from patients with active TB is induced by the interaction with the whole M. tuberculosis via Toll-like receptor 2 (TLR2), and involves the p38 mitogen-activated protein kinase (MAPK) pathway 15345584_the synergistic induction of IFN-gamma by interleukin-12 and IL-18 in natural killer cells is mediated at least in part by p38-dependent and 3' UTR-mediated stabilization of interferon-gamma mRNA 15375157_a specific requirement for p150(Glued)/dynein/functional microtubules in activation of MKK3/6 and p38 MAPKs in vivo. 15388348_Data show that Exip, but not p38alpha, binds to Toll interacting protein, which is involved in interleukin-1 (IL-1) signaling pathway, and impaired NF-kappaB activity. 15489375_These findings indicate that signaling via PKCdelta-p38 kinase-linked cascade specifically induces expression of VCAM-1 in lung epithelium in response to TNF-alpha and that this effect is both functionally and clinically significant. 15514847_low-density shock waves enhance IL-2 expression through a mechanism that involves p38 MAPK activation 15516490_MAP kinase kinase3- and 6-dependent activation of the alpha-isoform of p38 MAP kinase is required for the cytoskeletal changes induced by neutrophil adherence and influences subsequent neutrophil migration toward endothelial cell junctions 15527495_important for the control of interleukin-6 and IL-8 release by human brain microviascular endoethelial cells during interaction with Neisseria meningitidis 15541600_To gain insight into cellular mechanisms that regulate apoptosis, survival and proliferation, we studied the modulation of p38 mitogen-activated protein kinase (MAPK) and the activation state of cdc2 kinase, which regulates progression into mitosis. 15542843_activation of p38 MAPK and c-Jun N-terminal kinase pathways by hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and TNFR1/TNFa expression 15550393_basal PAK6 kinase activity was repressed by a p38 mitogen-activated protein (MAP) kinase antagonist 15567067_p38 MAPK plays a critical role in GDF-8's function as a negative regulator of muscle growth 15569672_the early and temporary activation of PP2A in neutrophils impaired not only the p38 MAPK-mediated inhibition of caspase 3 but also restored the activity to caspase 3 that had already been phosphorylated and thereby inactivated 15578697_LMP1-mediated p38/SAPK2 activation induces expression of the chemokine IP-10 and regulates transcript stability 15592496_The expression of tenascin-W is dependent on p38MAPK and JNK signaling pathways in mammary tumors. 15625302_Changes in extracellular pressure during infection or inflammation regulate macrophage phagocytosis by a FAK-dependent inverse effect on p38 MAPKalpha that might subsequently downregulate ERK. 15632082_Data suggest that Rit is involved in a novel pathway of neuronal development and regeneration by coupling specific trophic factor signals to sustained activation of the B-Raf/ERK and p38 MAP kinase cascades. 15634764_We demonstrate that one of the mitogen-activated protein kinases (MAPKs), p38 MAPK, regulates the serotonin transporter (SERT). 15650392_NF-kappaB appears crucial in ICAM-1 induction, but p38 activation itself is not sufficient to confer HO-1 expression and may not be involved in HO-1 and ICAM-1 induction in human tumor cells 15665518_ANP recruits a signal pathway associated with p38 MAPK, NHE-1 and PLD responsible for ROS production, suggesting a possible role for ANP as novel modulator of ROS generation in HepG2 cells 15668322_These results indicate that B lymphocyte stimulator (BLyS)-activated p38 mitogen-activated protein kinase plays an essential role in BLyS-induced activation-induced cytidine deaminase expression and Ig class switch recombination in human B cells. 15677464_H-Ras-specific activation of Rac-MKK3/6-p38 pathway has a role in invasion and migration of breast epithelial cells 15708845_p38alpha and -beta mediate UV-induced, AP-1-mediated, c-Fos phosphorylation 15737629_Finally, we conclude that the up-regulation of CCR5 and CXCR4 can at least partially contribute to the down-regulation of transfactor YY1 which is p38 MAPK pathway-dependent and this up-regulation has little relationship with MTB and HIV co-infection. 15737736_Pharmacological inhibition of oxidative stress diminished the elevated p38, JNK activity and PARP cleavage, and enhanced PD cybrid viability. 15753227_p38 MAP kinase plays a pivotal role in the signal transduction pathways regulating the number of endothelial progenitor cells ex vivo. 15755725_IL-1b decreases expression of the ENaC alpha-subunit in alveolar epithelial cells via a p38 MAPK-dependent signaling pathway 15784503_both p38 and ERK1 are spatially regulated in different uterine regions during pregnancy/labor 15790570_MEK6E activates p38 and results in phosphorylation of its downstream substrate, heat shock protein 27 15791651_Effect of hibuscus polyphenol extracs in human gastric carcinoma cells is mediated via p53 signaling and p38 MAPK/FasL cascade pathway. 15797859_p38 mediates EGF receptor activation after oxidant injury; Src activates MMK3, which, in turn, activates p38; and the EGF receptor signaling pathway plays a critical role in renal epithelial cell dedifferentiation 15817653_p38 MAPK has a role in the pathway of GC-induced apoptosis of lymphoid cells 15833736_transcriptional induction of HO-1 gene expression by AEBSF is mediated via activation of a PKB, p38 MAPK signaling pathway 15843433_a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module is regulated by Protein Kinase A gating and then regulates invasion in breast cancer cell lines 15845648_Together, our data suggest that inhibition of p38 MAPK may be an attractive target to limit inflammatory responses that are mediated by TNF-alpha. 15849811_FACL4 is involved in the hepatocellular carcinoma tumorigenesis and both cAMP and p38 MAPK pathways are associated with the regulation of FACL4 15851480_ERK2 and p38 have complementary effects in the control of platelet adhesion to collagen in a shear stress-dependent manner 15862947_p38 MAPK pathway was found to be involved in liganded estrogen receptor beta repression activity by ErbB2 signaling. 15894558_STAT3 signaling pathway is involved in IL-6-induced neuroendocrine(NE) differentiation and that p38 MAPK is associated with IL-6-stimulated growth regulation in NSCLC-NE cells. 15901830_the PI3-kinase/p38(MAPK)/CREB pathway contributes to the EGF activation of NF-IL6beta gene expression 15917247_Pellino3 is a novel upstream regulator of p38 MAPK and activates CREB in a p38-dependent manner 15919053_mast cell chymase activates ERK and p38 probably through G-protein-coupled receptor, and the ERK but not p38 cascade may have a crucial role in chymase-induced migration of eosinophils 15946948_Reduction-oxidation regulation ofgamma-glutamylcysteine synthetase expression is mediated by p38 mitogen-activated protein kinase signaling. 15950779_IL-1beta-induced cPLA2 and COX-2 gene expression is modulated through the p38 MAPK pathway in both neuroglioma and neuroblastoma cells. 15964514_activation of PKC, p38(MAPK), JNK, ERK1/2, and Nrf2 by oxidized LDL in human smooth muscle cells leads to HO-1 induction, constituting an adaptive response against oxidative injury that can be ameliorated by vitamin C 16027723_study shows a new role for p38alpha in contact inhibition and provides a mechanistic basis for the recently proposed tumour suppressive function of this MAPK pathway 16098034_contribution of p38 MAPK signaling to the TGF-beta-mediated regulation of the human alpha2(I) collagen gene in normal dermal fibroblasts and constitutive upregulated expression of type I collagen and fibronectin in systemic sclerosis 16099944_T-cad overexpression in vascular endothelial cells protects against stress-induced apoptosis through activation of the PI3K/Akt/mTOR survival signal pathway and concomitant suppression of the p38 MAPK proapoptotic pathway 16123320_alpha-lipoic acid induces HO-1 expression in THP-1 monocytic cells via Nrf2 and p38 16133687_biological responses of OPG and sRANKL synthesis in osteoblasts to the application of cyclic tensile strain are regulated via the p38 MAPK pathway 16139274_Data support a role for p38 MAPK in cannabinoid receptor 2-induced apoptosis of human leukaemia cells. 16153249_determination of whether Rickettsia rickettsii infection of endothelial cells activates p38 and/or c-jun N-terminal 16164755_These studies suggest that HCV core protein can lead to enhanced p38- and gC1qR-dependent IL-8 expression. 16197369_an independent role for p38 MAPK and JNK in LPA-induced IL-8 expression and secretion via NF-kappaB and AP-1 transcription respectively in human bronchial epithelial cells. 16214108_Importantly, inhibition of p38alpha MAPK blocks the release of vascular endothelial growth factor and suppresses tumor-promoted endothelial cell migration, a key step in angiogenesis. 16242072_A3 receptor stimulation activates p44/p42 and p38 mitogen-activated protein kinases, which are required for A3-induced increase of HIF-1alpha and Ang-2 16251188_the capacity of LDLs to induce fibroblast spreading and accelerate wound closure relies on their ability to stimulate IL-8 secretion in a p38 MAPK-dependent manner 16256071_Taken together, these results indicate a novel role for CIN85 in the regulation of cellular stress response via the MAPK pathways. 16256948_Collectively, our results demonstrate that PGE(2) at physiologically relevant concentrations induces COX-2 expression in human NPE cells via activation of EP(2)- and EP(4) receptors and phosphorylation of p38 and p42/44 MAPKs. 16257181_The results suggest that oncogenic K-Ras enhances the malignant phenotype and identify the mitogen-activated protein kinase p38 as a target to inhibit oncogenic K-Ras-induced pancreatic tumor cell migration. 16278378_in pancreatic cancer cells, the p38-mediated phosphorylation of HIF-1alpha contributed to the inhibition of HIF-1alpha and von Hippel-Lindau tumor suppressor protein interaction during ischemia 16308312_p38 MAPK and Nrf2 have roles in phenolic acid-induced P-form phenol sulfotransferase expression in human hepatoma HepG2 cells 16338464_p38 regulates PPARgamma expression and activity in term human trophoblasts. 16342939_selectivity pocket compounds prevent MKK6-dependent activation of p38alpha in addition to inhibiting catalysis by activated p38alpha 16344056_The signaling pathway for colonocyte apoptosis following toxin A exposure involves p38-dependent activation of p53 and subsequent induction of p21(WAF1/CIP1), resulting in cytochrome c release and caspase-3 activation through Bak induction. 16380078_activation of p38MAPK is required for the inhibitory effect of LDL on Na+/H+ antiport and thereby for LDL-dependent sensitization in human platelets 16410245_activation of the c-Abl-PKCdelta-Rac1-p38 MAPK pathway in response to ionizing radiation signals conformational changes of Bak and Bax, resulting in mitochondrial activation-mediated apoptotic cell death in human non-small cell lung cancer cells 16410316_p160 coactivator glucocorticoid receptor-interacting protein 1 (GRIP1) is phosphorylated and potentiated by the p38 MAPK signaling cascade in vitro and in vivo. 16446363_HCV core expression leads to deregulation of the mitotic checkpoint via a p38/PKR-dependent pathway 16467496_In conclusion, increased TGF-beta2 transcription in response to serum stimulation in KFs appears to be mediated by the p38 MAPK pathway. 16469115_p38 MAPK is a potential therapeutic target, inhibition of which may maintain the cellularity of articular chondrocytes by inhibiting cell death and its amplification signal and by increasing cell proliferation. 16478922_These results suggest that the p38 kinase, through its ability to control Erk activation levels, acts as a gatekeeper, which prevents inappropriate IL-2 production. 16516911_Asymmetric dimethylarginine induces apoptosis of endothelial cell via elevation of intracellular oxidant production, which involves p38 MAPK/caspase-3-dependent signaling pathway. 16532349_the inhibition of p38 MAP kinase might enhance anti-inflammatory tendencies in the MH7A cells 16538384_Data suggest that 2-methoxyestradiol 2 inhibits estradiol-stimulated proliferation and induces apoptosis in ovarian carcinoma cells, and that activation of p38 and phosphorylation of Bcl-2 plays a critical role in the mechanism. 16554354_p38 MAPK may play an important role in Stat-6 phosphorylation and the induction of IL-4, Bax, Bcl-2 & Bcl-xL and a putative role in protecting cells from undergoing apoptosis. 16613612_S100A8/A9 heterodimer, secreted extracellularly from activated tissue macrophages, may amplify proinflammatory cytokine responses through activation of NF-kappaB and p38 MAPK pathways in rheumatoid arthritis. 16627995_p38 has emerged as an important regulator of both embryonic development and cancer progression [review] 16645187_Our data indicate that TGF-beta1 induces endothelial barrier dysfunction involving Smad2-dependent p38 activation, resulting in RhoA activation by possible transcriptional regulation. 16648477_Determinants that control the specific interactions between TAB1 and MAPK14 are identified. 16648633_involvement of p38 MAP kinase in the hyaluronan oligosaccharide induction of MMP-13 16683917_role for p38 MAP kinase in the regulation of EGFR endocytosis 16697418_p38 kinase plays an important role in iron chelator-mediated cell death. 16699462_p38 plays a key role during small intestinal ischemia reperfusion injury through a proapoptotic action on the tip of villi. 16699726_ERK1/2, JNK1/2 and p38 mapk pathways are all required for B[a]P-induced G1/S transition 16709866_Platelet activating factor signaling requires beta-arrestin-1 recruitment of a p38 MAP kinase signalosome for transduction before neutrophil endosomal scission. 16720051_two inhibitors of p38 MAPK and protein tyrosine kinase pathways could be used as combined targets for inhibiting matrix metalloproteinase-9 expression in inflamed tissues 16769768_Halofuginone inhibits NF-kappaB and p38 MAPK in activated T cells. 16790501_Acidic pH exposure protects HEMEC through induction of Hsps and activation of MAPK and PI3 kinase pathway. 16814421_Results show that activation of c-Jun N-terminal protein kinase, ERKs 1 and 2, and p38 MAP kinase is critical for Hs683 glioma cell migration induced by GDNF. 16820791_TGF-beta1 regulates COX-2 expression in human mesangial cells through the activation of ERK1/2, p38 MAPK, and PI3K. 16843435_doxorubicin-stimulated phosphorylation of BCL2-antagonist of cell death in cells expressing dominant negative p38 MAPK was impeded by the inhibition of PI3-kinase 16861915_MAPKAP kinase-2 phosphorylates CDC25B on multiple sites including S169, S323, S353 and S375, while p38SAPK phosphorylates CDC25B on S249. 16864945_High glucose accelerates the onset of endothelial progenitor cell senescence leading to the impairment of proliferative activity, which might be related to the phosphorylation of p38 MAPK. 16869889_These studies establish p38 MAP kinase-mediated activation of ATF-2 as a significant mechanism in amylin-evoked beta-cell death, which may serve as a target for pharmaceutical intervention and effective suppression of beta-cell failure in type-2 diabetes. 16899715_Our results indicate that CD23 expression in these human intestinal epithelial cells is mediated through the p38 MAPK pathway. 16912047_E2F and Wip1 are modulators of E2F1-induced apoptosis involving p38 MAP kinase 16924234_DUSP26 effectively dephosphorylates p38 and has a little effect on extracellular signal-regulated kinase in anaplastic thyroid cancer. 16924420_These results suggest that p38 affects the phosphorylation of Smad2 and Smad3 differentially during TGF-beta signaling in human dental pulp cells and ERK1/2 might be involved in the process. 16927023_This study is the first to demonstrate that H2O2 induces a Rac1/JNK1/p38 signaling cascade, and that JNK and p38 activation is important for H2O2-induced apoptosis as well as apoptosis-inducing factor/Bax translocation of retinal pigment epithelial cells. 16935849_All trans-retinoic acidinduces apoptosis in Barrett's epithelial cells and sensitizes them to apoptosis induced by UV-B irradiation via activation of p38. 16951426_Icreased nitration of phospho-p38 mitogen-activated protein kinase occurs in the pre-eclamptic placenta. Its catalytic activity & the concentration of phospho-p38 MAPK was also reduced. 16964394_p38 MAPK and JNK pathways play an important role in VEGF secretion from malignant glioma cells under normoxic conditions. 16973972_p38 MAPK and NF-kappaB were crucial for the hydrolase-modified LDL-induced IL-8 secretion from the macrophages. 17000004_These data suggest that M. bovis BCG induces human beta defensin-2 mRNA expression in A549 lung epithelial cells at least in part mediated through intracellular calcium flux as well as activation of p38MAPK 17012370_These results indicate that 2ME-induced barrier disruption is governed by microtubule depolymerization and p38- and ROCK-dependent mechanisms. 17028194_p38alpha plays an important role in extravasation of tumor cells, possibly through regulating the formation of tumor-platelet aggregates and their interaction with the endothelium involved in a step of hematogenous metastasis 17030506_results suggest that p38 MAP kinase is involved not only in activating NADPH oxidase stimulated by fMLP but also in determining the extent of its activity. 17030510_accumulation of C(16)-ceramide in mitochondria formed from the protein kinase C-dependent salvage pathway results at least in part from the action of longevity-assurance homologue 5, and the generated ceramide modulates the p38 cascade via PP1 17035228_hSef isoforms can control signal specificity and subsequent cell fate by utilizing different mechanisms to modulate RTK signaling 17050539_SOX9 mRNA regulation in human articular chondrocytes involves p38 MAPK activation and mRNA stabilization 17054722_Sin1 binds directly to both ATF-2 and p38. 17055984_Phosphorylation of p38 by GRK2 at the docking groove unveils a novel mechanism for inactivating p38MAPK. 17059827_high-resolution crystal structures of the intrinsically active p38alpha mutants reveal that local alterations in the L16 loop region promote kinase activation 17085432_p38 MAPK as an upstream regulator of nSMase2 17088247_hyperactive variants of p38alpha induce, whereas hyperactive variants of p38gamma suppress, activating protein 1-mediated transcription 17107667_epidermal growth factor (EGF)-mediated induction of transforming growth factor-beta receptor type II expression may participate in a synergistic interplay between EGF and transforming growth factor-beta signaling pathway 17126905_These findings are consistent with the hypothesis that cytokine-induced autophosphorylation of P38 MAPK in primary granulocytes depends on the interaction with TAB1. 17141198_Further characterization of enzyme kinetics showed the binding mode of MT4 was competitive with the ATP substrate-binding site of p38alpha MAPK. 17142767_role for p38 MAPK and actin but not protein kinase C zeta in generating the neutrophil respiratory burst to beta-glucan 17188240_Our results confirm that ultrasound induces apoptosis via a pathway that involves Bak, Bcl-2, and caspases, but not ROS. 17196171_possibility that the IL-6 mediated androgen-independent proliferation of PC-3 and DU145 prostate cancer cells is regulated at least partly via SAPKs signaling pathway especially through p38MAPK activation.gh p38MAPK activation 17202865_Study demonstrates that BMP-2 promotes chondrogenesis by activating p38 mitogen-activated protein kinase, which downregulates Wnt-7a/b-catenin signaling responsible for proteasomal degradation of Sox9. 17207971_p38 MAPK/NF-kappaB/cyclin D1 signaling pathway might participate in the oncogenesis of extramammary Paget's disease 17219054_Results indicate that stimulation of heme oxygenase-1 expression by hypericin-PDT is a cytoprotective mechanism governed by the p38(MAPK) and PI3K pathways. 17222367_The expression of p-p38 and uPA was negatively correlated to prognosis of breast cancer. 17229385_Data show that HCMV could activate p38MAPK pathway and trigger apoptosis and interfere cell cycle in U251 cells. 17234706_p38 MAPkinase mediates palmitate-induced apoptosis and may participate in coronary endothelial injury in diabetes. 17241234_a new activating mutation site in p38alpha is identified. 17244678_a cross-talk of cell-cycle regulation and p38-dependent signal transduction is required for the suppressor function of iTregs. 17255097_Binding of MAP-kinase-activated kinase 2 (MK2): the C-terminal regulatory domain of MK2 binds in the docking groove of p38alpha, and the ATP-binding sites of both kinases are at the heterodimer interface. 17255949_Mxi2 is a key spatial regulator for ERK1/2 and we disclose an unprecedented stimulus-independent mechanism for ERK nuclear import. 17277157_GATA-3 nuclear translocation is dependent on its phosphorylation on serine residues by p38 MAPK 17284589_Epac1 and Epac2 induce a potent activation of the Ca2+-sensitive big K+ channel, slight membrane hyperpolarization, and increased after-hyperpolarization in cultured cerebellar granule cells. These effects involve activation of Rap and p38 MAPK. 17303142_Ad-PGIS gene transfer reduced PPAR delta expression and inhibited neointimal formation after balloon injury in accordance with the reduction in the phosphorylation of p38 MAPK 17303382_These results strongly suggest that hypoxia-induced apoptosis is regulated through signal transduction in which inactivation of ERK1/2 leads to activation of p38, which then triggers caspase cascade as an execution mechanism of apoptosis. 17303384_These results suggest that in human tracheal smooth muscle cells, activation of p42/p44 MAPK, p38, and JNK pathways, at least in part, mediated through NF-kappaB, is essential for lipopolysaccharide-induced VCAM-1 gene expression. 17307333_These lines of evidence suggested that a Gq-coupled receptor activates specifically p38 MAPK through lipid rafts and Src kinase activation, in which flotillins positively modulate the Gq signaling. 17316568_Intermittent hypoxia/reoxygenation activates NF-kappaB in an IKK-dependent manner signaled at least in part via activation of p38 MAPK. 17332506_NHERF1 overexpression enhances the invasive phenotype in breast cancer cells, both alone and in synergy with exposure to the tumor microenvironment, via the coordination of PKA-gated RhoA/p38 signaling. 17334397_results indicate that p38a | ENSMUSG00000053436 | Mapk14 | 1.939361e+03 | 0.6002609 | -0.736338342 | 0.2677224 | 7.433939e+00 | 0.0064005077 | 0.29566279 | No | Yes | 1.363294e+03 | 194.439830 | 2.180293e+03 | 318.223183 | |
| ENSG00000112130 | 9025 | RNF8 | protein_coding | O76064 | FUNCTION: E3 ubiquitin-protein ligase that plays a key role in DNA damage signaling via 2 distinct roles: by mediating the 'Lys-63'-linked ubiquitination of histones H2A and H2AX and promoting the recruitment of DNA repair proteins at double-strand breaks (DSBs) sites, and by catalyzing 'Lys-48'-linked ubiquitination to remove target proteins from DNA damage sites. Following DNA DSBs, it is recruited to the sites of damage by ATM-phosphorylated MDC1 and catalyzes the 'Lys-63'-linked ubiquitination of histones H2A and H2AX, thereby promoting the formation of TP53BP1 and BRCA1 ionizing radiation-induced foci (IRIF). Also controls the recruitment of UIMC1-BRCC3 (RAP80-BRCC36) and PAXIP1/PTIP to DNA damage sites. Also recruited at DNA interstrand cross-links (ICLs) sites and catalyzes 'Lys-63'-linked ubiquitination of histones H2A and H2AX, leading to recruitment of FAAP20/C1orf86 and Fanconi anemia (FA) complex, followed by interstrand cross-link repair. H2A ubiquitination also mediates the ATM-dependent transcriptional silencing at regions flanking DSBs in cis, a mechanism to avoid collision between transcription and repair intermediates. Promotes the formation of 'Lys-63'-linked polyubiquitin chains via interactions with the specific ubiquitin-conjugating UBE2N/UBC13 and ubiquitinates non-histone substrates such as PCNA. Substrates that are polyubiquitinated at 'Lys-63' are usually not targeted for degradation. Also catalyzes the formation of 'Lys-48'-linked polyubiquitin chains via interaction with the ubiquitin-conjugating UBE2L6/UBCH8, leading to degradation of substrate proteins such as CHEK2, JMJD2A/KDM4A and KU80/XRCC5: it is still unclear how the preference toward 'Lys-48'- versus 'Lys-63'-linked ubiquitination is regulated but it could be due to RNF8 ability to interact with specific E2 specific ligases. For instance, interaction with phosphorylated HERC2 promotes the association between RNF8 and UBE2N/UBC13 and favors the specific formation of 'Lys-63'-linked ubiquitin chains. Promotes non-homologous end joining (NHEJ) by promoting the 'Lys-48'-linked ubiquitination and degradation the of KU80/XRCC5. Following DNA damage, mediates the ubiquitination and degradation of JMJD2A/KDM4A in collaboration with RNF168, leading to unmask H4K20me2 mark and promote the recruitment of TP53BP1 at DNA damage sites (PubMed:11322894, PubMed:14981089, PubMed:17724460, PubMed:18001824, PubMed:18001825, PubMed:18006705, PubMed:18077395, PubMed:18337245, PubMed:18948756, PubMed:19015238, PubMed:19124460, PubMed:19202061, PubMed:19203578, PubMed:19203579, PubMed:20550933, PubMed:21558560, PubMed:21857671, PubMed:21911360, PubMed:22266820, PubMed:22373579, PubMed:22531782, PubMed:22705371, PubMed:22865450, PubMed:22980979). Following DNA damage, mediates the ubiquitination and degradation of POLD4/p12, a subunit of DNA polymerase delta. In the absence of POLD4, DNA polymerase delta complex exhibits higher proofreading activity (PubMed:23233665). In addition to its function in damage signaling, also plays a role in higher-order chromatin structure by mediating extensive chromatin decondensation. Involved in the activation of ATM by promoting histone H2B ubiquitination, which indirectly triggers histone H4 'Lys-16' acetylation (H4K16ac), establishing a chromatin environment that promotes efficient activation of ATM kinase. Required in the testis, where it plays a role in the replacement of histones during spermatogenesis. At uncapped telomeres, promotes the joining of deprotected chromosome ends by inducing H2A ubiquitination and TP53BP1 recruitment, suggesting that it may enhance cancer development by aggravating telomere-induced genome instability in case of telomeric crisis. Promotes the assembly of RAD51 at DNA DSBs in the absence of BRCA1 and TP53BP1 Also involved in class switch recombination in immune system, via its role in regulation of DSBs repair. May be required for proper exit from mitosis after spindle checkpoint activation and may regulate cytokinesis. May play a role in the regulation of RXRA-mediated transcriptional activity. Not involved in RXRA ubiquitination by UBE2E2 (PubMed:11322894, PubMed:14981089, PubMed:17724460, PubMed:18001824, PubMed:18001825, PubMed:18006705, PubMed:18077395, PubMed:18337245, PubMed:18948756, PubMed:19015238, PubMed:19124460, PubMed:19202061, PubMed:19203578, PubMed:19203579, PubMed:20550933, PubMed:21558560, PubMed:21857671, PubMed:21911360, PubMed:22266820, PubMed:22373579, PubMed:22531782, PubMed:22705371, PubMed:22865450, PubMed:22980979). {ECO:0000269|PubMed:11322894, ECO:0000269|PubMed:14981089, ECO:0000269|PubMed:17724460, ECO:0000269|PubMed:18001824, ECO:0000269|PubMed:18001825, ECO:0000269|PubMed:18006705, ECO:0000269|PubMed:18077395, ECO:0000269|PubMed:18337245, ECO:0000269|PubMed:18948756, ECO:0000269|PubMed:19015238, ECO:0000269|PubMed:19124460, ECO:0000269|PubMed:19202061, ECO:0000269|PubMed:19203578, ECO:0000269|PubMed:19203579, ECO:0000269|PubMed:20550933, ECO:0000269|PubMed:21558560, ECO:0000269|PubMed:21857671, ECO:0000269|PubMed:21911360, ECO:0000269|PubMed:22266820, ECO:0000269|PubMed:22373579, ECO:0000269|PubMed:22531782, ECO:0000269|PubMed:22705371, ECO:0000269|PubMed:22865450, ECO:0000269|PubMed:22980979, ECO:0000269|PubMed:23233665}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Chromatin regulator;Chromosome;Cytoplasm;DNA damage;DNA repair;Direct protein sequencing;Host-virus interaction;Metal-binding;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Telomere;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000255|HAMAP-Rule:MF_03067}. | The protein encoded by this gene contains a RING finger motif and an FHA domain. This protein has been shown to interact with several class II ubiquitin-conjugating enzymes (E2), including UBE2E1/UBCH6, UBE2E2, and UBE2E3, and may act as an ubiquitin ligase (E3) in the ubiquitination of certain nuclear proteins. This protein is also known to play a role in the DNA damage response and depletion of this protein causes cell growth inhibition and cell cycle arrest. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2012]. | hsa:9025; | chromosome, telomeric region [GO:0000781]; cytosol [GO:0005829]; midbody [GO:0030496]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; site of double-strand break [GO:0035861]; ubiquitin ligase complex [GO:0000151]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; identical protein binding [GO:0042802]; protein homodimerization activity [GO:0042803]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; cell cycle [GO:0007049]; cell division [GO:0051301]; cellular response to DNA damage stimulus [GO:0006974]; double-strand break repair [GO:0006302]; double-strand break repair via nonhomologous end joining [GO:0006303]; histone exchange [GO:0043486]; histone H2A K63-linked ubiquitination [GO:0070535]; histone H2A ubiquitination [GO:0033522]; histone H2B ubiquitination [GO:0033523]; interstrand cross-link repair [GO:0036297]; isotype switching [GO:0045190]; negative regulation of transcription elongation from RNA polymerase II promoter [GO:0034244]; positive regulation of DNA repair [GO:0045739]; protein autoubiquitination [GO:0051865]; protein K48-linked ubiquitination [GO:0070936]; protein K63-linked ubiquitination [GO:0070534]; protein ubiquitination [GO:0016567]; response to ionizing radiation [GO:0010212]; spermatid development [GO:0007286]; spermatogenesis, exchange of chromosomal proteins [GO:0035093]; ubiquitin-dependent protein catabolic process [GO:0006511] | 14981089_results suggest a novel function of RNF8 as a regulator of RXR alpha-mediated transcriptional activity through interaction between their respective N-terminal regions 17724460_Regulates the rate of exit from mitosis and cytokinesis. 18001824_RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. 18001825_RNF8 is a novel DNA-damage-responsive protein that integrates protein phosphorylation and ubiquitylation signaling and plays a critical role in the cellular response to genotoxic stress. 18006705_results demonstrate how the DNA-damage response is orchestrated by ATM-dependent phosphorylation of MDC1 and RNF8-mediated ubiquitination 18077395_the human Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage 18171988_RNF8 is the human orthologue of the yeast protein Dma1p 18337245_RNF8 ubiquitylation pathways are essential for 53BP1 regulation in response to ionizing radiation, whereas RNF8-independent pathways contribute to 53BP1 targeting and phosphorylation in response to UV light and other forms of DNA replication stress 18550271_Depletion of RAP80 or RNF8 impairs the translocation of BRCA1 to DNA damage sites and results in defective cell cycle checkpoint control and DSB repair 18948756_RNF8 readily mono-ubiquitinates PCNA in the presence of UbcH5c, and polyubiquitinates PCNA in the added presence of Ubc13/Uev1a. 19124460_subset of PTIP.PA1 complex is recruited to DNA damage sites via the RNF8-dependent pathway and is required for cell survival in response to DNA damage. 19797077_Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20075863_Data identify RNF8 and RNF168, cellular histone ubiquitin ligases responsible for anchoring repair factors at sites of damage, as new targets for ICP0-mediated degradation. 20080757_Ubiquitin ligase does not protect cells from Nutlin-3-mediated apoptosis, indicating that RNF8 does not regulate 53BP1 protein. 20081839_Data show that the ATM signalling mediator proteins MDC1, RNF8, RNF168 and 53BP1 are also required for heterochromatic DSB repair. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20713529_Phosphorylated NPM1 may interact with RNF8-dependent ubiquitin conjugates at sites of DNA damage. 21412056_The E3 Ubiquitin ligases, RNF8 and RNF168, are recruited to DNA damage foci in late mitosis, presumably to prime sites for the DNA damage response, 53BP1, recruitment in early G1. 21558560_the differential requirement for the ubiquitin ligase RNF8 in facilitating repair of replication stress-associated DNA damage 21635870_These results suggest that RNF8 is downregulated in many cancer cells and inversely correlated with Plk1. 21664912_Studies indicate that Non-proteolytic ubiquitylation of chromatin surrounding DSBs, mediated by the RNF8/RNF168 ubiquitin ligase cascade, has emerged as a key mechanism for restoration of genome integrity. 21698222_The viral protein ICP0 targets RNF8 and RNF168 for degradation, thereby preventing the deposition of repressive ubiquitin marks and counteracting this repair protein recruitment. 21774837_entire coding region and splice junctions of RNF8, UBC13 and MMS2 genes were screened for mutations in affected index cases from 123 Northern Finnish breast cancer families 21857671_Data show that depletion of RNF8, as well as of the E3 ligase RNF168, reduces telomere-induced genome instability. 22266820_The authors find that RNF8 regulates the abundance of the nonhomologous end-joining (NHEJ) repair protein KU80 at sites of DNA damage. 22373579_The authors propose that the RNF8-dependent degradation of JMJD2A regulates DNA repair by controlling the recruitment of 53BP1 at DNA damage sites. 22405594_By mimicking a cellular phosphosite, ICP0 binds RNF8 via the RNF8 forkhead associated (FHA) domain. Phosphorylation of ICP0 T67 by CK1 recruits RNF8 for degradation and thereby promotes viral transcription 22508508_In response to double-strand breaks, both HERC2 and RNF168 were specifically modified with SUMO1 at double-strand break sites in a manner dependent on the SUMO E3 ligase PIAS4. 22531782_A new mechanism of chromatin remodelling-assisted ubiquitylation was shown, which involves cooperation between CHD4 and RNF8 to create a local chromatin environment permissive to the assembly of checkpoint and repair machineries at DNA lesions. 22589545_Data show RING finger (RNF) E3 ubiquitin ligase RNF8 dimerizes and binds to E2 ubiquitin-conjugating complex Ubc13/Mms2 with formation of Lys-63 ubiquitin chains, whereas the RNF168 RING domain is a monomer and does not catalyze Lys-6 ubiquitylation. 22705371_Data indicate that RNF8 and FAAP20 (C1orf86) are needed for efficient Fanconi anemia group D2 protein FANCD2 monoubiquitination. 22814251_RNF8 may play a role in protein synthesis, possibly linking the nucleolar exit of this factor to the attenuation of protein synthesis in response to DNA damage. 22865450_Data indicate that RNF8-dependent chromatin ubiquitination is required for RAD51 assembly in BRCA1/53BP1-depleted cells. 23038782_PALB2 localization depends on the presence of MDC1, RNF8, RAP80 and Abraxas upstream of BRCA1. 23115235_Nbs1 is one important target of RNF8 to regulate DNA DSB repair. 23230272_The PARP1-dependent localization of BAL1-BBAP functionally limits both early and delayed DNA damage and enhances cellular viability independent of ATM, MDC1, and RNF8. 23233665_The identification of RNF8 allows new insights into the integration of the control of p12 degradation by different DNA damage signaling pathways. 23442799_RNF8 maintains genome stability through independent, yet analogous, nuclear and cytoplasmic ubiquitylation activities 23615962_findings implicate USP44 in negative regulation of the RNF8/RNF168 pathway 25304081_Finding that RNF8 is less abundant than RNF168 identifies RNF8 as a rate-limiting determinant of focal repair complex assembly. 25337968_Depletion of RNF8 or RNF168 blocks the degradation of diffusely localized nuclear 53BP1. 25374327_considered potential associations of 14 single nucleotide polymorphisms (SNPs) in RNF8 and BRDT genes in Chinese patients with non-obstructive azoospermia 25417706_Significantly restored tolerance of RAD18-/- and RNF8-/- cells to camptothecin and olaparib without affecting Rad51 focus formation. 25483088_our results indicate that down-regulation of RNF8 mediated by miR-214 impedes DNA damage response to induce chromosomal instability in ovarian cancers, which may facilitate the understanding of mechanisms underlying chromosomal instability. 25955491_Results indicate that RNF8 recruits and ubiquitinates many factors to repair DNA damage thereby conferring radioresistance to nasopharyngeal cancer cells. 26381412_define a novel function for ATDC in the RNF8-mediated DNA damage response and implicate RNF8 binding as a key determinant of the radioprotective function of ATDC 26507658_These findings elucidate deeply and extensively the mechanism of RNF8/RNF168 and USP11 to maintain the proper status of ubiquitylation gammaH2AX to repair double strand breaks. 26734725_The interaction of MDC1 with RNF8, but not with ATM requires WRAP53beta, suggesting that WRAP53beta facilitates the former interaction without altering phosphorylation of MDC1 by ATM. 26788910_High RNF8 expression is associated with bladder cancer. 27259701_Data report that RNF8 is overexpressed in highly metastatic breast cell lines and its overexpression can induce EMT in breast cancer cells. Furthermore, RNF8 is aberrantly expressed in invasive breast cancer and positively correlates with lymph node metastasis. 27310875_The present findings indicate that WRAP53beta and RNF8 are rate-limiting factors in the repair of DNA double-strand breaks and raise the possibility that upregulation of WRAP53beta may contribute to genomic stability in and survival of cancer cells. 27550454_The attenuated DNA damage localization of RNF8 resulting from INT6 depletion could be attributed to the defective retention of ATM. 27618486_RNF8-promoted Twist ubiquitination is required for Twist localization to the nucleus for subsequent epithelial-mesenchymal transition and cancer stem cells functions, thereby conferring chemoresistance. 27993934_In late S/G2 phase, the DNA damage-responsive E3 ligase RNF8 conjugates K63-linked ubiquitin chains to tankyrase 1, while in G1 phase such ubiquitin chains are removed by BRISC, an ABRO1/BRCC36-containing deubiquitinase complex. 28194753_Findings indicate direct interaction of dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2) with ring finger protein (C3HC4 type) 8 (RNF8) in regulating response to DNA damage. 28216286_study describes RNF8 as a co-activator of ERalpha increases ERalpha stability via post-transcriptional pathway, and provides a new insight into mechanisms for RNF8 to promote cell growth of ERalpha-positive breast cancer. 28499869_The study identifies a previously unrecognized role for RNF8 in the negative regulation of NF-kappaB activation by targeting and deactivating the IKK complex. 28507061_the role of RNF8-mediated histone H3 polyubiquitylation in the regulation of histone H3 stability and chromatin modification, is reported. 28525740_RNF8- and Ube2S-dependent Lys11-linkage ubiquitin conjugation plays an important role in regulating DNA damage-induced transcriptional silencing, distinct from the role of Lys63-linkage ubiquitin in the recruitment of DNA damage repair proteins 53BP1 and BRCA1. 28983621_the present study described the noncovalent interaction between the E3 ubiquitin ligase RNF8 and SUMO2/3 and indicated that this interaction promoted DSB repair. 29127375_data are in line with a model in which HUWE1 primes histone H1 with ubiquitin to allow ubiquitin chain elongation by RNF8, thereby stimulating the RNF8-RNF168 mediated DDR. 29581593_L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage, regulating the DNA damage response pathway. 30445466_UV-induced NONO degradation by RNF8 is required for timely termination of intra-S-phase checkpoint signaling and continued cell cycle progression. 31294443_the miR-214-RNF8 axis has a role in epithelial-mesenchymal transition in breast cancer; miR-214 acts as a tumor suppressor 32031738_These findings suggested that RNF8 promotes efficient DNA double-strand break repair by inhibiting the pro-apoptotic activity of p53 through regulating the function of Tip60. 32427332_RNF8 has both KU-dependent and independent roles in chromosomal break repair. 32453758_during HTLV-1 infection, Tax activates RNF8 to assemble nuclear K63-pUbs that sequester DDR factors in Tax speckles, disrupting DDR signaling and DSB repair. 32502756_RNF8 promotes high linear energy transfer carbon-ion-induced DNA double-stranded break repair in serum-starved human cells. 32549753_RNF8 induces beta-catenin-mediated c-Myc expression and promotes colon cancer proliferation. 32753472_RNF8 Promotes Epithelial-Mesenchymal Transition in Lung Cancer Cells via Stabilization of Slug. 33205415_The ubiquitin ligase RNF8 regulates Rho GTPases and promotes cytoskeletal changes and motility in triple-negative breast cancer cells. 33710666_RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage. 35428760_RNF8 up-regulates AR/ARV7 action to contribute to advanced prostate cancer progression. | ENSMUSG00000098374+ENSMUSG00000090083 | Gm28043+Rnf8 | 1.031938e+03 | 0.9539106 | -0.068073999 | 0.2734225 | 6.175355e-02 | 0.8037455418 | 0.96141221 | No | Yes | 1.009361e+03 | 139.931624 | 9.154281e+02 | 130.200705 |
| ENSG00000112146 | 26268 | FBXO9 | protein_coding | Q9UK97 | FUNCTION: Substrate recognition component of a SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of TTI1 and TELO2 in a CK2-dependent manner, thereby directly regulating mTOR signaling. SCF(FBXO9) recognizes and binds mTORC1-bound TTI1 and TELO2 when they are phosphorylated by CK2 following growth factor deprivation, leading to their degradation. In contrast, the SCF(FBXO9) does not mediate ubiquitination of TTI1 and TELO2 when they are part of the mTORC2 complex. As a consequence, mTORC1 is inactivated to restrain cell growth and protein translation, while mTORC2 is activated due to the relief of feedback inhibition by mTORC1. {ECO:0000269|PubMed:23263282}. | Alternative splicing;Cytoplasm;Phosphoprotein;Reference proteome;TPR repeat;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of the ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into 3 classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbxs class. Alternative splicing of this gene generates at least 3 transcript variants diverging at the 5' terminus. [provided by RefSeq, Jul 2008]. | hsa:26268; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; SCF ubiquitin ligase complex [GO:0019005]; ubiquitin ligase complex [GO:0000151]; ubiquitin-protein transferase activity [GO:0004842]; fat cell differentiation [GO:0045444]; innate immune response [GO:0045087]; protein ubiquitination [GO:0016567]; regulation of TOR signaling [GO:0032006]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146] | 20459822_ICK and FBX9 are divergently transcribed from a bidirectional promoter that is GC-rich and contains a CpG island. 35025031_F-box only protein 9 and its role in cancer. | ENSMUSG00000001366 | Fbxo9 | 2.146550e+03 | 1.0326573 | 0.046361585 | 0.2672192 | 3.043371e-02 | 0.8615097798 | 0.97337236 | No | Yes | 1.948689e+03 | 229.547313 | 1.751200e+03 | 211.444361 |
| ENSG00000112739 | 8899 | PRPF4B | protein_coding | Q13523 | FUNCTION: Has a role in pre-mRNA splicing. Phosphorylates SF2/ASF. | 3D-structure;ATP-binding;Acetylation;Isopeptide bond;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Spliceosome;Transferase;Ubl conjugation;mRNA processing;mRNA splicing | Pre-mRNA splicing occurs in two sequential transesterification steps, and the protein encoded by this gene is thought to be involved in pre-mRNA splicing and in signal transduction. This protein belongs to a kinase family that includes serine/arginine-rich protein-specific kinases and cyclin-dependent kinases (CDKs). This protein is regarded as a CDK-like kinase (Clk) with homology to mitogen-activated protein kinases (MAPKs). [provided by RefSeq, Jul 2008]. | hsa:8899; | catalytic step 2 spliceosome [GO:0071013]; chromosome [GO:0005694]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; ATP binding [GO:0005524]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; RNA binding [GO:0003723]; mRNA cis splicing, via spliceosome [GO:0045292]; mRNA splicing, via spliceosome [GO:0000398]; protein phosphorylation [GO:0006468]; RNA splicing [GO:0008380] | 12077342_Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. 15452250_interacts specifically with HIV-2 gag 17513757_PRP4, a serine/threonine protein kinase, is identified as a Kruppel-like factor 13 (KLF13)-binding protein; PRP4 phosphorylates KLF13 19902070_Short-term exercise resulted in a significant increase of mRNA expression of genes encoding proteins involved in the formation of precatalytic splisosome: PRPF4B. 20118938_The authors provide evidence that PRP6 and PRP31 are directly phosphorylated by human PRP4 kinase (PRP4K) concomitant with their incorporation into B complexes. 23466643_data indicate that miR-371-5p, which is highly expressed in hepatocellular carcinoma, promotes the growth of HCC cells in vitro and in vivo by activating cell cycle progression at the G1/S checkpoint by directly targeting PRPF4B 23686430_expression and activity are closely associated with the survival and regulation of apoptotic events in colon cancer cells 25602630_PRP4K functions as a HER2-regulated modifier of taxane sensitivity and is a prognostic biomarker for better survival in taxane-treated ovarian cancer patients. PRP4K knock-down results in 'mitotic slippage' in response to taxanes, and reduced PRP4K expression is associated with both intrinsic and acquired resistance of ovarian and breast cancers to taxanes. 25602630_PRP4K functions as a HER2-regulated modifier of taxane sensitivity that may have prognostic value as a marker of better overall survival in taxane-treated ovarian cancer patients. 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that PRPF4B is a putative expression variability marker for the 3-day, 8-cell embryo stage. 26712520_PRP4K is novel estrogen regulated kinase, and its levels can be reduced by 4-OHT in ER+ breast cancer cells altering their response to taxanes 28892043_Low PRP4K expression correlates with significantly worse overall survival in high-grade serous ovarian cancer, and its depletion increased metastasis in the mouse ID8 ovarian carcinoma model and promoted both sustained growth factor signalling in detached conditions and anoikis resistance in human cervical, breast and ovarian cancer cells. 28892043_Study identifies a novel role for PRP4K in regulating the endosomal trafficking of EGFR leading to altered anoikis sensitivity in epithelial cancer cells. 29695716_PRP4K inhibits proliferation and invasiveness of cultured breast cancer cells and its high expression correlates with good prognosis in breast cancer patients. 30104601_a SNP in PRPF4B was associated with anhedonia 31278301_PRPF4B is essential for triple-negative breast cancer metastasis formation in vivo, making PRPF4B a candidate for further drug development. 32576716_PRP4 Kinase Domain Loss Nullifies Drug Resistance and Epithelial-Mesenchymal Transition in Human Colorectal Carcinoma Cells. 34674320_Haploinsufficient tumor suppressor PRP4K is negatively regulated during epithelial-to-mesenchymal transition. 35328513_PRP4 Induces Epithelial-Mesenchymal Transition and Drug Resistance in Colon Cancer Cells via Activation of p53. | ENSMUSG00000021413 | Prpf4b | 1.355356e+03 | 0.9659560 | -0.049970693 | 0.3374188 | 2.240922e-02 | 0.8810036011 | 0.97741608 | No | Yes | 1.121414e+03 | 189.848196 | 1.252480e+03 | 217.212963 | |
| ENSG00000113272 | 54974 | THG1L | protein_coding | Q9NWX6 | FUNCTION: Adds a GMP to the 5'-end of tRNA(His) after transcription and RNase P cleavage. This step is essential for proper recognition of the tRNA and for the fidelity of protein synthesis (Probable). Also functions as a guanyl-nucleotide exchange factor/GEF for the MFN1 and MFN2 mitofusins thereby regulating mitochondrial fusion (PubMed:25008184, PubMed:27307223). By regulating both mitochondrial dynamics and bioenergetic function, it contributes to cell survival following oxidative stress (PubMed:25008184, PubMed:27307223). {ECO:0000269|PubMed:25008184, ECO:0000269|PubMed:27307223, ECO:0000305|PubMed:21059936}. | 3D-structure;Cytoplasm;Disease variant;GTP-binding;Magnesium;Membrane;Metal-binding;Mitochondrion;Mitochondrion outer membrane;Neurodegeneration;Nucleotide-binding;Nucleotidyltransferase;Reference proteome;Transferase;tRNA processing | The protein encoded by this gene is a mitochondrial protein that is induced by high levels of glucose and is associated with diabetic nephropathy. The encoded protein appears to increase mitochondrial biogenesis, which could lead to renal fibrosis. Another function of this protein is that of a guanyltransferase, adding GMP to the 5' end of tRNA(His). Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2015]. | hsa:54974; | cytosol [GO:0005829]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; transferase complex [GO:1990234]; ATP binding [GO:0005524]; GTP binding [GO:0005525]; guanyl-nucleotide exchange factor activity [GO:0005085]; identical protein binding [GO:0042802]; magnesium ion binding [GO:0000287]; nucleotidyltransferase activity [GO:0016779]; tRNA binding [GO:0000049]; tRNA guanylyltransferase activity [GO:0008193]; mitochondrial fusion [GO:0008053]; protein homotetramerization [GO:0051289]; response to oxidative stress [GO:0006979]; stress-induced mitochondrial fusion [GO:1990046]; tRNA modification [GO:0006400]; tRNA processing [GO:0008033] | 15459185_ICF45 is a highly conserved novel protein, which is expressed in a cell cycle-dependent manner and seemed to be involved in cell cycle progression and cell proliferation 18508967_induced in high glucose-1 (IHG-1)which increases in diabetic nephropathy, may enhance the actions of TGF-beta1 and contribute to the development of tubulointerstitial fibrosis 20877624_Observational study of gene-disease association. (HuGE Navigator) 21078997_The catalytic domain of Thg1 shares both a common architecture and a two-metal ion-dependent mechanism with canonical 5'-3' DNA polymerases. 21784897_IHG-1 increases mitochondrial biogenesis by promoting PGC-1alpha-dependent processes, potentially contributing to the pathogenesis of renal fibrosis 22136300_The high-resolution crystal structure of human Thg1 reveals remarkable structural similarity between canonical DNA/RNA polymerases and eukaryotic Thg1. 25008184_IHG-1 is a novel regulator of both mitochondrial dynamics and bioenergetic function and contributes to cell survival following oxidant stress. Increased IHG-1 expression may contribute to pathogenesis of diabetic kidney disease. 27307223_Study proposes that homozygosity for the p.Val55Ala mutation in tRNA-histidine guanylyltransferase 1 like (THG1L) is the cause of the abnormal mitochondrial network in the patient fibroblasts, likely by interfering with THG1L activity towards MFN2. 33682303_Severe epileptic encephalopathy associated with compound heterozygosity of THG1L variants in the Ashkenazi Jewish population. 33758037_Analysis of GTP addition in the reverse (3'-5') direction by human tRNA(His) guanylyltransferase. | ENSMUSG00000011254 | Thg1l | 3.047639e+02 | 1.1728843 | 0.230060686 | 0.2992475 | 6.052461e-01 | 0.4365833794 | 0.85332708 | No | Yes | 3.448918e+02 | 42.261939 | 2.514542e+02 | 31.775811 | |
| ENSG00000113318 | 4437 | MSH3 | protein_coding | P20585 | FUNCTION: Component of the post-replicative DNA mismatch repair system (MMR). Heterodimerizes with MSH2 to form MutS beta which binds to DNA mismatches thereby initiating DNA repair. When bound, the MutS beta heterodimer bends the DNA helix and shields approximately 20 base pairs. MutS beta recognizes large insertion-deletion loops (IDL) up to 13 nucleotides long. After mismatch binding, forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. | 3D-structure;ATP-binding;DNA damage;DNA repair;DNA-binding;Nucleotide-binding;Phosphoprotein;Reference proteome | The protein encoded by this gene forms a heterodimer with MSH2 to form MutS beta, part of the post-replicative DNA mismatch repair system. MutS beta initiates mismatch repair by binding to a mismatch and then forming a complex with MutL alpha heterodimer. This gene contains a polymorphic 9 bp tandem repeat sequence in the first exon. The repeat is present 6 times in the reference genome sequence and 3-7 repeats have been reported. Defects in this gene are a cause of susceptibility to endometrial cancer. [provided by RefSeq, Mar 2011]. | hsa:4437; | membrane [GO:0016020]; mismatch repair complex [GO:0032300]; MutSbeta complex [GO:0032302]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP-dependent activity, acting on DNA [GO:0008094]; DNA insertion or deletion binding [GO:0032135]; enzyme binding [GO:0019899]; DNA repair [GO:0006281]; maintenance of DNA repeat elements [GO:0043570]; mismatch repair [GO:0006298]; mitotic recombination [GO:0006312]; negative regulation of DNA recombination [GO:0045910]; positive regulation of helicase activity [GO:0051096]; reciprocal meiotic recombination [GO:0007131]; somatic recombination of immunoglobulin gene segments [GO:0016447] | 11756455_hMSH2-hMSH3 did not appear to bind any of the 8-oxo-G containing DNA substrates nor was there enhanced ATPase or ADP --> ATP exchange activities. 11895912_Observational study of gene-disease association. (HuGE Navigator) 12014680_Frequent LOH at hMLH1, a highly variable SNP in hMSH3, and negligible coding instability occur in ovarian cancer. 14510941_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14625810_Observational study of genotype prevalence. (HuGE Navigator) 14871813_MSH3 abrogation may be a predictor of metastatic disease or even favor tumor cell spread in MLH1-deficient colorectal cancers. 15541380_This is the first report suggesting that genetic and epigenetic alterations in the human MSH3 gene might play a significant role in the progression of bladder tumors. 16552576_Plays a key role in the formation of CTG repeat expansions over successive generations in DM1 transgenic mice. 16774946_Observational study of gene-disease association. (HuGE Navigator) 17205513_Observational study of gene-disease association. (HuGE Navigator) 17205513_Polymorphisms in the mismatch repair gene, MSH3 is associated with colorectal cancer 17494052_Observational study of gene-disease association. (HuGE Navigator) 17676485_Alterations in TGF-betaRII, BAX, IGFIIR, caspase-5, hMSH3 and hMSH6 genes of microsatellite instability are rare in urinary bladder carcinoma and they are not associated with microsatellite instability or the presence of p53 mutations. 17950544_Mutations at the mononucleotide repeats within the hMSH3 gene occurred in certain basal cell carcinomas, not always in association with microsatellite instability . 18355840_Mismatch repair gene MSH3 polymorphism is associated with the risk of sporadic prostate cancer 18355840_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18364438_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18683134_Observational study of gene-disease association. (HuGE Navigator) 18701435_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18723338_Observational study of gene-disease association. (HuGE Navigator) 18922920_Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19115210_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19377479_The human DNA mismatch complex MSH2-MSH3 recognizes small loops by a mechanism different from that of MSH2-MSH6 for single-base mismatches. 19525234_MutSbeta displays identical biochemical and biophysical activities when interacting with a (CAG)(n) hairpin and a mismatch.[MutSbeta] 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19730683_Observational study of gene-disease association. (HuGE Navigator) 19894224_methylation of hMLH1 and hMSH3 is age related and thus may play an important role in gastric carcinogenesis in the elderly. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20154325_the MutSbeta-MutLalpha interaction is mediated in part by residues ((L/I)SRFF) embedded within the MSH3 PCNA-binding motif 20160730_Strong role of MutSbeta in insertion-deletions repair indicates MSH3 deficiency in tumours with low dinucleotide and no mononucleotide repeat instability. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20632816_Endoscopic biopsy provides equal accuracy and easier interpretation of MMRP expression immunostaining compared to surgical resection specimens. 20708618_Loss of hMSH3 corresponds with multiple tetranucleotide frameshifts. The association between EMAST and ulcerated tumors might result from increased inflammation. 20845481_Nondysplastic epithelium from hamartomatous polyposis syndrome polyps harbors hMSH3 defects, which may prime neoplastic transformation. 20869096_No association of tumor necrosis with expression of p53, bcl-2, and mismatch repair protein status was observed in colorectal cancers. 21128252_We hypothesise a model in which variants of the MSH3 gene behave as low-risk alleles that contribute to the risk of colon cancer in Lynch families, mostly with other low-risk alleles of MMR genes. 21285347_Results provide novel evidence that MSH3 deficiency contributes to the cytotoxicity of platinum drugs through deficient DSB repair. 21344488_Stress treatment of mouse cells with ethanol or hydrogen peroxide caused the re-distribution of MSH3 into nuclear bodies containing the proliferating cell nuclear antigen (PCNA), a known binding partner of MutSbeta. 21960445_Complex MSH2/MSH3 discriminates between a repair-competent and a repair-resistant loop by sensing the conformational dynamics of the junctions. MSH2/MSH3 binds, bends and dissociates from repair-competent loops to signal downstream repair. 21974800_polymorphisms in MSH3 do not contribute to cancer risk in a population of Lynch syndrome patients with colorectal cancer 22110587_two novel HLA-A0201-restricted cytotoxic T cell epitopes derived from a (-1) frameshift mutation of a coding A(8) tract within the MSH3 gene 22179786_Studies indicate thar eukaryotes MutSbeta, a heterodimer of Msh2 and Msh3, recognizes insertion-deletion loops (IDLs) of 1-15 nucleotides, as well as DNA with a 3' single-stranded overhang. 22249440_The high frequency of loss of heterozygosity as well as the aberrant protein expression in some tumors indicates an involvement of MSH3 impairment in colorectal cancer with low-level microsatellite instability. 22343000_The binding of HIF-1alpha complexes to hypoxia response element sites is necessary for down-regulation of hMSH3 in both wt-p53 and mut-p53 cells. 22941650_siRNA knockdown of the MutSbeta subunits MSH2 or MSH3 impeded expansion of threshold-length CTG*CAG repeats. 23226332_Oxidative stress, which causes a shift of hMSH3's subcellular location, may contribute to an hMSH3 loss-of-function phenotype by sequestering it to the cytosol 23339595_Single nucleotide polymorphisms in MSH3 are associated with myelodysplastic syndromes. 23724141_MSH3 status can regulate the DNA damage response and extent of apoptosis induced by chemotherapy. 24934723_Methylation of MSH3 together with exposure to tobacco smoke is involved in esophageal carcinogenesis. 25461668_IL6 signaling disrupts the nuclear localization of hMSH3 and DNA repair, leading to elevated microsatellite alterations at selected tetranucleotide repeats in cancer cell lines. 25598504_Our data present, for the first time, evidence that inherited MLH1 c.-93G>A, MSH2 c.211 + 9C>G, MSH3 c.3133G>A, and EXO1 c.1765G>A abnormalities of DNA MMR pathway are important determinants of head and neck squamous cell carcinoma 25966119_Data show that single nucleotide polymorphisms in MutS homolog 3 (MSH3) had an impact on the chemotherapy response and prognosis of advanced non-small cell lung cancer (NCSLC) patients who were treated with platinum-based chemotherapy. 26212458_The mismatch-binding protein MutS beta, a heterodimer of MSH2 and MSH3, activates ATR in response to DNA double-strand breaks. 26617824_Our meta-analysis results demonstrated that MSH3 rs26279 G > A polymorphism is associated with an increased risk of overall cancer, especially for the colorectal cancer and breast cancer. 26994442_Three polymorphisms in MSH3 were associated with variation in somatic instability in myotonic dystrophy type 1. 27476653_data suggest that MSH3 mutations represent an additional recessive subtype of colorectal adenomatous polyposis 27546332_MSH2-MSH3 not only stimulates pol beta to copy through the repeats but also enhances formation of the flap precursor for expansion. 28093084_Findings indicate that carriers of the MSH5 rs707939 T allele, the MSH2 rs6544991 C allele, the MSH3 rs6151627 and rs6151670 G alleles, and the MSH3 rs7709909 T allele have poor toxicity tolerance to platinum-based chemotherapy in non-small cell lung cancer patients. 28528517_The role of MSH3 in 11 Lynch Syndrome patients with truncating MSH6 germline variants and an unexplained MSH2 protein loss.Heterozygous MSH3 defects alone do not seem to induce a Lynch Syndrome phenotype 28642124_MSH3 is probably a modifier of disease progression in Huntington's disease. 28656302_MSH3 was frequently inactivated by promoter methylation and its mRNA and protein expression correlated with the primary tumor stage in nasopharyngeal carcinoma. 28973443_Msh3-/- cells are severely defective for CTG*CAG repeat expansions but show full activity on contractions. Msh3 overexpression led to high expansion activity and elevated levels of MutSbeta complex, indicating that MutSbeta abundance drives expansions. Expression of 2 Msh3 polymorphic variants at normal levels showed no detectable change in expansions. These polymorphisms primarily affect Msh3 protein stability, not ac... 30590005_Study suggests the SNPs rs12513549, rs33013, and rs6151627 in MSH3 are associated with the risk of acute adverse events (AEs). Therefore, these polymorphisms may be potential independent biomarkers for predicting AEs and prognosis in rectal cancer patients receiving postoperative CRT. 31342644_The genotype frequency distribution of rs26279 (A>G) in MSH3 was found to be significantly different between idiopathic male infertility and control (p < 0.05), as well as azoospermia. 31549400_EMAST status as a beneficial predictor of fluorouracil-based adjuvant chemotherapy for Stage II/III colorectal cancer. 31789935_Inflammation-Associated Microsatellite Alterations Caused by MSH3 Dysfunction Are Prevalent in Ulcerative Colitis and Increase With Neoplastic Advancement. 31953835_Genetic Polymorphism of Mismatch Repair Genes and Susceptibility to Prostate Cancer. 32284349_The Human DNA Mismatch Repair Protein MSH3 Contains Nuclear Localization and Export Signals That Enable Nuclear-Cytosolic Shuttling in Response to Inflammation. 33596761_Coordinated roles of SLX4 and MutSbeta in DNA repair and the maintenance of genome stability. 34250384_Prevalence and Characterization of Biallelic and Monoallelic NTHL1 and MSH3 Variant Carriers From a Pan-Cancer Patient Population. | ENSMUSG00000014850 | Msh3 | 3.479468e+02 | 0.9003476 | -0.151446030 | 0.3428037 | 1.880159e-01 | 0.6645731503 | 0.92913032 | No | Yes | 3.430756e+02 | 70.113302 | 3.175459e+02 | 66.658595 | |
| ENSG00000113396 | 28965 | SLC27A6 | protein_coding | Q9Y2P4 | FUNCTION: Involved in translocation of long-chain fatty acids (LFCA) across the plasma membrane. Thought to function as the predominant fatty acid protein transporter in heart (PubMed:12556534). Has acyl-CoA ligase activity for long-chain and very-long-chain fatty acids (VLCFAs) (By similarity). {ECO:0000250|UniProtKB:E9Q9W4, ECO:0000269|PubMed:12556534}. | Cell membrane;Fatty acid metabolism;Ligase;Lipid metabolism;Lipid transport;Membrane;Nucleotide-binding;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the fatty acid transport protein family (FATP). FATPs are involved in the uptake of long-chain fatty acids and have unique expression patterns. Alternatively spliced transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:28965; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; sarcolemma [GO:0042383]; arachidonate-CoA ligase activity [GO:0047676]; fatty acid transmembrane transporter activity [GO:0015245]; long-chain fatty acid transporter activity [GO:0005324]; long-chain fatty acid-CoA ligase activity [GO:0004467]; nucleotide binding [GO:0000166]; oleate transmembrane transporter activity [GO:1901480]; very long-chain fatty acid-CoA ligase activity [GO:0031957]; long-chain fatty acid transport [GO:0015909]; very long-chain fatty acid metabolic process [GO:0000038] | 12556534_FATP6 is involved in heart LCFA uptake, in which it may play a role in the pathogenesis of lipid-related cardiac disorders. 20965718_Observational study of gene-disease association. (HuGE Navigator) 21920065_The FATP6-7T>A polymorphism may protect from traits of the metabolic syndrome and CVD. 30911270_Inverse correlation between SLC27A6 expression and breast tumoral tissues.SLC27A6role in the cell growth and cell cycle regulation in non-tumorigenic breast cells. | ENSMUSG00000024600 | Slc27a6 | 2.351281e+01 | 1.0093616 | 0.013443037 | 0.6317870 | 4.462904e-04 | 0.9831454744 | 0.99700442 | No | Yes | 1.984627e+01 | 4.584153 | 2.008123e+01 | 4.752591 | |
| ENSG00000113441 | 4012 | LNPEP | protein_coding | Q9UIQ6 | FUNCTION: Release of an N-terminal amino acid, cleaves before cysteine, leucine as well as other amino acids. Degrades peptide hormones such as oxytocin, vasopressin and angiotensin III, and plays a role in maintaining homeostasis during pregnancy. May be involved in the inactivation of neuronal peptides in the brain. Cleaves Met-enkephalin and dynorphin. Binds angiotensin IV and may be the angiotensin IV receptor in the brain. {ECO:0000269|PubMed:11389728, ECO:0000269|PubMed:11707427, ECO:0000269|PubMed:1731608}. | 3D-structure;Acetylation;Alternative splicing;Aminopeptidase;Cell membrane;Direct protein sequencing;Glycoprotein;Hydrolase;Membrane;Metal-binding;Metalloprotease;Phosphoprotein;Protease;Reference proteome;Secreted;Signal-anchor;Transmembrane;Transmembrane helix;Zinc | This gene encodes a zinc-dependent aminopeptidase that cleaves vasopressin, oxytocin, lys-bradykinin, met-enkephalin, dynorphin A and other peptide hormones. The protein can be secreted in maternal serum, reside in intracellular vesicles with the insulin-responsive glucose transporter GLUT4, or form a type II integral membrane glycoprotein. The protein catalyzes the final step in the conversion of angiotensinogen to angiotensin IV (AT4) and is also a receptor for AT4. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:4012; | cytoplasm [GO:0005737]; cytoplasmic vesicle membrane [GO:0030659]; cytosol [GO:0005829]; early endosome lumen [GO:0031905]; extracellular region [GO:0005576]; integral component of plasma membrane [GO:0005887]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; aminopeptidase activity [GO:0004177]; metalloaminopeptidase activity [GO:0070006]; metallopeptidase activity [GO:0008237]; peptide binding [GO:0042277]; zinc ion binding [GO:0008270]; antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent [GO:0002480]; cell-cell signaling [GO:0007267]; female pregnancy [GO:0007565]; negative regulation of cold-induced thermogenesis [GO:0120163]; peptide catabolic process [GO:0043171]; protein catabolic process [GO:0030163]; protein polyubiquitination [GO:0000209]; proteolysis [GO:0006508]; regulation of blood pressure [GO:0008217]; signal transduction [GO:0007165]; SMAD protein signal transduction [GO:0060395] | 12080061_Identification of a tankyrase-binding motif in this protein 12569180_Interleukin-1beta stimulates expression in BeWo choriocarcinoma cells 12700100_The expression of IRAP/P-LAP is not limited to fat and muscle cells, and the subcellular distribution of IRAP/P-LAP is regulated by different peptide hormones and exercise. 12871575_Of a range of peptides tested, only vasopressin, oxytocin, and met-enkephalin were rapidly cleaved by IRAP. We propose that the physiological effects of AT4 ligands result from inhibition of IRAP cleavage of neuropeptides involved in memory processing 14527672_Placental leucine aminopeptidase/oxytocinase gene regulation by activator protein-2 in BeWo cell model of human trophoblast differentiation. 15187412_This review characterizes insulin-regulated IRAP in muscle and fat cells and discusses how impaired IRAP action may play a role in the development of complications in type 2 diabetes. 15894523_P-LAP/OTase is type II integral membrane protein, which is converted to soluble form existing in maternal serum by metalloproteases. It may be involved in maintaining pregnancy homeostasis via metabolizing peptides such as OT & vasopressin[review] 16054015_oxytocinase subfamily of M1 aminopeptidases play important roles in the maintenance of homeostasis including maintenance of normal pregnancy, memory retention, blood pressure regulation and antigen presentation [review] 16113565_RCAS1 and CAP may play a role in the downregulation of the maternal immune response during pregnancy and may participate in the initiation of the labor 16136012_no significant increase of oxytocinase plasma level in first trimester spontaneous abortions 17059388_insulin-stimulated IRAP translocation remained intact despite substantial GLUT4 knockdown 17373876_In addition to its capacity to degrade a range of peptides, placental-leucine aminopeptidase (P-LAP) has novel functions that impact on normal cells and neoplastic cells. 17391061_Substrate degradation studies using vasopressin & Leu-enkephalin showed that replacement of G428 by either D, E or Q selectively abolished the catalysis of Leu-enkephalin, while [A429G]IRAP & [N432A]IRAP mutants were incapable of cleaving both substrates 17692401_fluorimetrically analysis of membrane-bound and soluble Cystinyl aminopeptidase activity in the three main renal cancers: clear cell (CCRCC), papillary (PRCC), and chromophobe (ChRCC) renal cell carcinomas 17999179_Data show that placental leucine aminopeptidase (P-LAP) plays important roles in the regulation of blood pressure under both the physiological and pathological conditions. 18398343_Insulin-regulated aminopeptidase (IRAP)/AT4 receptors are involved in neither the regulation of RBF or CBF nor in the handling of renal sodium. 18502721_study identifies smooth muscle cell alpha actin positive ACE2 and AT4R in blood vessels as well as in angiogenic vessels, indicating a possible role for these enzymes in pathological disease 18996364_Triton-slowed APN as well as PLAP is present in the serum of pregnant women. 19071192_distinct biological effects of Angiotensin II 3-8 fragment, denoted as Angiotensin IV, and high affinity Ang IV binding to the AT(4) receptor. [review] 19498108_study found a role for peptide trimming by IRAP in cross-presentation; in dendritic cells, IRAP was localized to a Rab14+ endosomal storage compartment in which it interacted with MHC class I molecules 19578876_Observational study of gene-disease association. (HuGE Navigator) 20150869_The probable involvement of P-LAP in trophoblast invasion and development of preeclampsia. 20304486_Expression of AT4R was increased in term placentae, with a significant reduction in pre-eclampsia placentae. 20592285_Although PLAP has the necessary enzymatic properties to participate in generating or destroying major histocompatibility class I-presented peptides, its trimming behavior during antigen processing is distinct from that of ERAP1. 21314638_S1 specificity pocket of the aminopeptidases that generate antigenic peptides. 21330387_The genetic variation in LNPEP (vasopressinase) is associated with 28-day mortality in septic shock and is associated with biologic effects on vasopressin clearance and serum sodium regulation 21348480_investigation of domain structure of IRAP; catalytic domains; inhibitor-binding domains; zinc binding sites 23500679_Activities of aminopeptidases N and B and insulin-regulated aminopeptidase could be useful non-invasive biomarkers of Alzheimer's disease from the earliest stages. 23889750_An association between maternal common polymorphisms in LNPEP and susceptibility to preterm birth was observed. 23897274_We identified the missense variant rs2303138 (p.Ala763Thr) within the LNPEP gene associated with psoriasis. 25408552_Determined is the crystal structure of human IRAP revealing a closed, four domain arrangement with a large, mostly buried cavity abutting the active site. 26311161_The substrate Angiotensin II, the enzymes aminopeptidases-A, B, M as well as IRAP were detected in the jejunal mucosa. 26366890_Vasopressinase might be a potential early biomarker for acute kidney injury after cardiopulmonary bypass. 27501164_Structural and biological characterization of three low molecular weight aryl sulfonamides: binding modes to human IRAP were explored by docking calculations combined with molecular dynamics simulations and binding affinity estimations using the linear interaction energy method. Two of these drug-like IRAP inhibitors can alter dendritic spine morphology and increase spine density in primary cultures of hippocampal neurons 27834335_The PCR-RFLP is a simple and reliable method that allows a quick genotyping for the rs4869317 SNP of LNPEP gene. 28035472_This study shown that the schizophrenia patients, the numerical density of IRAP-expressing neurons in the paraventricular and the suprachiasmatic nuclei is significantly reduced, which might be associated with the reduction in neurophysin-containing neurons in these nuclei in schizophrenia. 28328206_This study describes a crystal structure of insulin-regulated aminopeptidase in complex with a recently developed bioactive and selective inhibitor at 2.53 A resolution. 30581499_LNPEP expression in the left atrium in mitral regurgitation patients significantly differed from those in aortic valve disease patients and normal controls. 32619880_The role of LNPEP and ANPEP gene polymorphisms in the pathogenesis of pre-eclampsia. | ENSMUSG00000023845 | Lnpep | 4.099759e+02 | 0.6231023 | -0.682459111 | 0.3357626 | 4.012435e+00 | 0.0451658853 | 0.64935422 | No | Yes | 3.155203e+02 | 52.998292 | 5.078033e+02 | 86.573561 | |
| ENSG00000113575 | 5515 | PPP2CA | protein_coding | P67775 | FUNCTION: PP2A is the major phosphatase for microtubule-associated proteins (MAPs) (PubMed:22613722). PP2A can modulate the activity of phosphorylase B kinase casein kinase 2, mitogen-stimulated S6 kinase, and MAP-2 kinase (PubMed:22613722). Cooperates with SGO2 to protect centromeric cohesin from separase-mediated cleavage in oocytes specifically during meiosis I (By similarity). Can dephosphorylate SV40 large T antigen and p53/TP53 (PubMed:17245430). Activates RAF1 by dephosphorylating it at 'Ser-259' (PubMed:10801873). Mediates dephosphorylation of WEE1, preventing its ubiquitin-mediated proteolysis, increasing WEE1 protein levels, and promoting the G2/M checkpoint (PubMed:33108758). Mediates dephosphorylation of MYC; promoting its ubiquitin-mediated proteolysis: interaction with AMBRA1 enhances interaction between PPP2CA and MYC (PubMed:25438055). Mediates dephosphorylation of FOXO3; promoting its stabilization: interaction with AMBRA1 enhances interaction between PPP2CA and FOXO3 (PubMed:30513302). {ECO:0000250|UniProtKB:P63330, ECO:0000269|PubMed:10801873, ECO:0000269|PubMed:17245430, ECO:0000269|PubMed:22613722, ECO:0000269|PubMed:25438055, ECO:0000269|PubMed:30513302, ECO:0000269|PubMed:33108758, ECO:0000269|PubMed:9920888}. | 3D-structure;Alternative splicing;Centromere;Chromosome;Cytoplasm;Cytoskeleton;Direct protein sequencing;Disease variant;Hydrolase;Manganese;Meiosis;Mental retardation;Metal-binding;Methylation;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome;Ubl conjugation | This gene encodes the phosphatase 2A catalytic subunit. Protein phosphatase 2A is one of the four major Ser/Thr phosphatases, and it is implicated in the negative control of cell growth and division. It consists of a common heteromeric core enzyme, which is composed of a catalytic subunit and a constant regulatory subunit, that associates with a variety of regulatory subunits. This gene encodes an alpha isoform of the catalytic subunit. [provided by RefSeq, Jul 2008]. | hsa:5515; | chromosome, centromeric region [GO:0000775]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; membrane [GO:0016020]; membrane raft [GO:0045121]; microtubule cytoskeleton [GO:0015630]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein phosphatase type 2A complex [GO:0000159]; spindle pole [GO:0000922]; synapse [GO:0045202]; GABA receptor binding [GO:0050811]; metal ion binding [GO:0046872]; phosphoprotein phosphatase activity [GO:0004721]; protein C-terminus binding [GO:0008022]; protein heterodimerization activity [GO:0046982]; protein serine phosphatase activity [GO:0106306]; protein serine/threonine phosphatase activity [GO:0004722]; protein threonine phosphatase activity [GO:0106307]; protein tyrosine phosphatase activity [GO:0004725]; tau protein binding [GO:0048156]; apoptotic process [GO:0006915]; ceramide metabolic process [GO:0006672]; meiotic cell cycle [GO:0051321]; mesoderm development [GO:0007498]; negative regulation of cell growth [GO:0030308]; negative regulation of epithelial to mesenchymal transition [GO:0010719]; negative regulation of protein kinase B signaling [GO:0051898]; negative regulation of tyrosine phosphorylation of STAT protein [GO:0042532]; peptidyl-serine dephosphorylation [GO:0070262]; peptidyl-threonine dephosphorylation [GO:0035970]; positive regulation of microtubule binding [GO:1904528]; positive regulation of protein serine/threonine kinase activity [GO:0071902]; protein dephosphorylation [GO:0006470]; regulation of cell adhesion [GO:0030155]; regulation of cell differentiation [GO:0045595]; regulation of DNA replication [GO:0006275]; regulation of G1/S transition of mitotic cell cycle [GO:2000045]; regulation of growth [GO:0040008]; regulation of microtubule binding [GO:1904526]; regulation of protein phosphorylation [GO:0001932]; regulation of transcription, DNA-templated [GO:0006355]; regulation of Wnt signaling pathway [GO:0030111]; response to lead ion [GO:0010288]; response to organic substance [GO:0010033]; RNA splicing [GO:0008380]; second-messenger-mediated signaling [GO:0019932] | 11929874_A functional role for the B56 alpha-subunit of protein phosphatase 2A in ceramide-mediated regulation of Bcl2 phosphorylation status and function 12054646_regulated by hydrogen peroxide and glutathionylation 12186863_role in p38 mitogen-activated protein kinase-mediated regulation of the c-Jun NH(2)-terminal kinase pathway in human neutrophils 12370081_Results describe the cell cycle expression, subcellular distribution, and metabolic stability of protein phosphatase 2Abeta in comparison with 2Aalpha. 15252037_the A subunit and alpha-4 (mTap42) require charged residues in separate but overlapping surface regions to associate with the back side of PP2Ac and modulate phosphatase activity 17693927_Studies suggest that the target of regulation by PP2A includes upstream kinases in the JNK MAPK pathway. 17803990_We propose that stabilization of this inactive, nuclear PP2A pool is a major in vivo function of PME-1. 17852432_down-regulation of expression of PP2A catalytic subunit in CAPE treated cells 17941990_PP2Calpha may possess tumor-suppressing properties, and it thereby sets the stage for more elaborate analyses on its involvement in the development and progression of cancer. 18299321_PP2A functions as a negative regulator in TGF-beta1-induced TAK1 activation 18430997_study found that the level of the catalytic subunit of PP2A(PP2Ac) was dramatically decreased in adult Down syndrome brain, and this decrease correlated negatively with tau leveland phosphorylation at several abnormal hyperphosphorylation sites 18477699_Aurora-B is an EB1-interacting protein; EB1 stimulates Aurora-B activity through antagonizing its dephosphorylation/inactivation by PP2A 18583989_These data shed new light on the significance of negative feedback regulation of NF-kappaB and identifies PP2A as the key regulator of this process. 18586681_1) PAK plays a required role in hyperosmotic signaling through the PI3K/pTEN/Cdc42/PP2Calpha/p38 pathway, and 2) PAK and PP2Calpha modulate the effects of this pathway on focal adhesion dynamics. 18852266_PP2A appears to function as an endogenous regulator of SK1 phosphorylation 18928587_The activity of PP2A decreases with increasing concentration of arsenic trioxide during the apoptosis of NB4 and MR2 cells. 18949047_Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A. 18977201_The catalytic subunit of protein phosphatase 2A (PP2A-C) is the protein interacting with GPR54. 19126401_The role of PP2A in the phosphorylation of P301L mutant human tau and in neurofibrillary tangle pathology in vivo was determined. 19155497_The PP2Ac alpha promoter defines a cyclic AMP response element (CRE) site flanked by CpG motifs in human T cells; methylation of the PP2Ac alpha promoter affects CREB binding to the CRE motif and suppresses its activity. 19356149_study shows that hydrogen peroxide increases the association of PP2A with occludin by a Src kinase-dependent mechanism, and that PP2A activity is involved in hydrogen peroxide-induced disruption of tight junctions in Caco-2 cell monolayers 19360341_Expression of tyrosine-phosphorylated PP2A (pY307-PP2A) was highly increased in the HER-2/neu positive breast tumours, and significantly correlated to tumour progression, thus enhancing its potential prognostic value. 19553685_a new role for mTOR and alpha4/PP2Ac in the control of STAT1 nuclear content, and the expression of interferon-gamma-sensitive genes involved in immunity and apoptosis. 20100830_VCP/p97-mediated inducible nitric-oxide synthase-dependent Tyr nitration of PP2A increases the levels of phosphatases PP2A and DUSP1 to contribute to the refractory response of conditioned cells 20106966_cAMP-stimulated protein phosphatase 2A activity associated with muscle A kinase-anchoring protein (mAKAP) signaling complexes inhibits the phosphorylation and activity of the cAMP-specific phosphodiesterase PDE4D3 20133759_Data show that HUNK reconstitution in basal breast cancer cell lines prevented protein phosphatase 2-A (PP2A from binding to CFL-1. 20363734_Neuroprotectin D1 induces dephosphorylation of Bcl-xL in a PP2A-dependent manner during oxidative stress and promotes retinal pigment epithelial cell survival 20558158_Silencing of the PP2A/C subunit causes the HER-2/neu positive breast cancer cells to undergo apoptosis via p38 MAPK-related pathways. Phosphorylated Y307-PP2A plays an essential role in cell survival in these cells. 20615878_Cross-talk between serine/threonine protein phosphatase 2A and protein tyrosine phosphatase 1B regulates Src activation and adhesion of integrin alphaIIbbeta3 to fibrinogen. 20663872_identification of a PP2A trimeric holoenzyme containing B55alpha, which plays a major role in restricting the phosphorylation state of p107 and inducing its activation in human cells. 20677014_Observational study of gene-disease association. (HuGE Navigator) 20711181_Results demonstrate that PP2A-B55alpha and importin-beta1 cooperate in the regulation of postmitotic assembly mechanisms in human cells. 21072166_Data show that the I84P mutation or the IIQ/VTR(83-85) and T89A substitutions in the Vpr(77-92) sequence prevent PP2A(1) binding. 21151953_protein serine/threonine phosphatase PP2Calpha activation efficiently prevents liver fibrosis 21172653_The recruitment of PP2A to UNC5H2/B allows the activation of DAPk via a PP2A-mediated dephosphorylation and that this mechanism is involved in angiogenesis regulation. 21346232_PP2Acalpha represents a new methylation-sensitive gene that contributes to the pathogenesis of systemic lupus erythematosus. 21355954_Protein phosphatase 2A was not associated with carcinoma progression of lung neoplasms. 21393425_modulation of PP2A activity may represent an alternative therapeutic approach for the treatment of advanced androgen-independent prostate cancer. 21454489_both the C-terminal Mid1-binding domain and the PP2Ac-binding determinants are required for Alpha4-mediated protection of PP2Ac from polyubiquitination and degradation. 21590681_evidence of an association between PPP2CA polymorphisms and elevated PP2Ac transcript levels in T cells, which implicates a new molecular pathway for systemic lupus erythematosus susceptibility in European Americans, Hispanic Americans, and Asians. 21791414_Hyperphosphorylation of autoantigenic targets of paraproteins is due to inactivation of PP2A. 21835928_dephosphorylation of pS776-ATXN1 by PP2A regulates the interaction of ATXN1 with the splicing factors RBM17 and U2AF65 21959857_The PP2A activity was unchanged in failing hearts despite an increase of both total PP and PP1 activity. 21996587_Deletion of residue Cys269 alters the conformation and activity of PP2A(D) and influences the interaction between PP2A and various metal ions, notably Mn(2+). 22031698_analysis of resistance of Akt kinase and PP2A to dephosphorylation through ATP-dependent conformational plasticity 22135143_The results show that a low-dose treatment of MCLR in FL cells for 6 h induced an increase in PP2A activity, and a high-dose treatment of MCLR for 24 h decreased the activity of PP2A, as expected. 22167190_Identification and characterization of an alternatively spliced isoform of the human protein phosphatase 2Aalpha catalytic subunit. 22192750_A biological strategy, used by various viruses based on the targeting of PP2A enzymes by viral proteins, in order to specifically deregulate cellular pathways of their hosts. 22299668_Studies indicate that the Ser/Thr phosphatases PP1 and PP2A are responsible for the dephosphorylation and activation of Rb proteins. 22314363_EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex 22443683_Data indicate that PP2A holoenzyme biogenesis and activity are controlled by five PP2A modulators, consisting of alpha4, PTPA, LCMT1, PME-1 and TIPRL1, which serve to prevent promiscuous phosphatase activity until the holoenzyme is completely assembled. 22479341_Data show that knockdown of the catalytic subunit of PP1 (PP1Calpha), but not PP2A (PP2ACalpha), increased pS137-PFN1 levels. 22685328_Semaphorin 3A impinged upon the basal activity of the serine phosphatase PP2A and disrupted PP2A interaction with VE-cadherin, leading to cell-cell junction disorganization and increased permeability. 22732552_GSK-3beta can inhibit PP2A by increasing the inhibitory L309-demethylation involving upregulation of PME-1 and inhibition of PPMT1 22904676_disruption of PP2Ac methylation may contribute to cancer, and modulation of this methylation may serve as an anticancer target. 22961627_Together, these observations suggest that PP2A regulates hTERT subcellular localization, in addition to its inhibitory effects on telomerase activity 23064762_These results demonstrate that acetaldehyde-induced disruption of tight junctions is mediated by PP2A translocation to tight junctions and dephosphorylation of occludin on threonine residues. 23183242_Data suggest that inhibition of PPP2CA activity impairs osteoclastogenesis by regulating RANKL (receptor activator of nuclear factor-kappa B ligand) and OPG (osteoprotegerin) expression in osteoblasts. 23184344_Inhibition of PP2A by okadaic acid curtailed the free fatty acids induced upregulation of SREBP1 expression, fatty acid synthase promoter transcriptional activity and lipid accumulation in HepG2 cells 23185379_activation of PP2A or inactivation of the p38MAPK-MAPKAPK2-Hsp27 has a role in survival of cancer stem cells under hypoxia and serum depletion via decrease in PP2A activity 23290789_Results suggest that Tgase-2/PP2A/JNK might be a novel axis that affects N-cadherin switching in the epithelial-mesenchymal-transition (EMT) of A549 lung cancer cells. 23434594_SPRY2, PTEN, and PP2A status is an important determinant of prostate cancer progression. 23651848_Our data reveal a protective role of TWEAK in glucose homeostasis and identify PP2A as a new driver in the modulation of TNF-alpha signaling by sTWEAK. 23677989_Results reveal that nilotinib induces the autophagy-inducing activity is associated with PP2A-regulated AMPK phosphorylation. 23720779_Data indicate that XIAP E3 ligase mediates cyclin D1 transcription via PP2A-regulated c-Jun/AP-1 activation. 23775084_Data suggest that enhanced PP2Ac in systemic lupus erythematosus (SLE) T-cells may dephosphorylate and activate the signaling pathway upstream of DNMT1, thus disturbing the tight control of methylation-sensitive genes involved in SLE pathogenesis. 23892082_Unlike yeast TIP41, TIPRL has a positive effect on mTORC1 signaling through the association with PP2Ac. 23901063_PP2Ac is upregulated in tumorous liver biopsies and negatively correlated with p53 proapoptotic gene expression. 24054836_Reduced expression of PP2A/PR65 in ameloblastoma compared with normal oral mucosa indicates that PP2A/PR65 is involved in the occurrence and development of ameloblastoma. 24064353_PP2A holoenzyme and PP6 were found stably associated with U1 snRNP; findings indicate that these phosphatases regulate splicing catalysis involving U1 snRNP and suggest an important evolutionary conserved role of PP2A family phosphatases in pre-mRNA splicing 24100030_a novel mechanism by which the interaction between PP2A and Src in the context of caspase-8 activation modulates TRAIL sensitivity in cancer cells. 24145130_the alpha4 N-terminus binding to endogenous PP2Ac and PABP, and the C-terminus to EDD, is reported. 24269630_Activation of BAFF-mediated PP2A-Erk1/2 pathway and B-cell proliferation is dependent upon the intracellular divalent Ca2+-ion. 24382322_Data indicate that TCDD or omeprazole caused protein phosphatase 2A (PP2A)-mediated dephosphorylation of Sp1 transcription factor at Ser-59 and induced CYP1A1 transcription. 24462681_PP2A constitutively dephosphorylates dCK in cells and negatively regulates its activity. 24550388_This result combined with a number of biophysical analyses provide evidence that the coiled coil domain of striatin 3 and the PP2A A subunit form a stable core complex with a 2:2 stoichiometry 24621013_Findings suggest the possible contribution of SET protein to the tumor progression and the utility of protein phosphatase 2A (PP2A) activator, FTY720 for treatment of alveolar soft part sarcoma (ASPS). 24642616_Our findings suggest that PPP2CA downregulation serves as a molecular link between gain of castration-resistance and aggressive PCa phenotype, and its restoration could be an effective preventive/therapeutic approach against the advanced disease. 24696731_These results suggest that the aberrant expression of PP2A in human clear cell renal cell carcinoma 24770487_PPP2AC activity is required for SREBP-2 DNA binding. 24841198_this study suggests that the tightly linked regulatory loop comprised of the SIK2-PP2A and CaMKI and PME-1 networks may function in fine-tuning cell proliferation and stress response. 24958351_decreased eEF2 phosphorylation, mediated by increased PP2A activity, contributes to resistance to HER2 inhibition and may provide novel targets for therapeutic intervention in HER2 positive breast cancer which is resistant to HER2 targeted therapies. 25003662_Phosphorylated PP2A is an alteration that determines poor outcome in metastatic colorectal cancer. 25015035_Studies indicate that oncoprotein CIP2A (KIAA1524) controls oncogenic cellular signals by suppressing protein phosphatase 2A (PP2A). 25152517_Data suggest that expression of PP2A (protein phosphatase 2 catalytic subunit alpha) and PTEN (phosphatase and tensin homolog) is down-regulated in adenomyosis as compared to normal endometrium; expression of survivin appears to be up-regulated. 25207814_MID1 catalyzes the ubiquitination of protein phosphatase 2A and mutations within its Bbox1 domain disrupt polyubiquitination of alpha4 but not of PP2Ac in X-linked Opitz syndrome. 25219467_GSK-3b and PP2A regulate each other and control tau phosphorylation both directly and indirectly through each other. 25253569_possible role of an miR-155-PP2Ac loop in regulating IL-2 release 25567480_Increased PP2A activity and reduced phosphorylation of PP2A were observed in alpha-synuclein overexpression primary cortex neurons. 25612003_Idiopathic pulmonary fibrosis fibroblast interaction with polymerized type I collagen results in an aberrant PP2A/HDAC4 axis, which suppresses miR-29, causing a pathologic increase in type I collagen expression. 25645927_HSP105 depletion disrupts the integration of protein phosphatase 2A into the beta-catenin degradation complex, favoring the hyperphosphorylation and degradation of beta-catenin. 25708299_Mutations in the structurally buried D38 residue of PP2Calpha (PPM1A) redefined the water-mediated hydrogen network in the active site and selectively disrupted M2 metal ion binding. 25939762_Data indicate that LB100 attenuates protein phosphatase 2A (PP2A) activity alone and following radiation. 25945834_Data show that the biogical effect of inhibitor-2 of protein phosphatase-2A (SET) on proliferation and invasion was mediated by the inhibition of the protein phosphatase 2A (PP2A). 25956027_Binding of calmodulin changes the calcineurin regulatory region to a less dynamic conformation. 25979969_Data suggest that activation of TAZ (tafazzin) inhibits adipogenesis in mesenchymal stem cells; interaction of TAZ and protein phosphatases (PP1A, PP2A) up-regulates dephosphorylation and transport of TAZ to cell nucleus. 26053095_Data show that the protein phosphatase 2A (PP2A)/c-jun N-Terminal Kinase (JNK)/Sp1 transcription factor/CDK1 kinase pathway and the autophagy/cyclin kinase inhibitor p21 pathway participated in G2/M cell cycle arrest triggered by PP2A inhibitors. 26234767_Alterations affecting PP2A subunits together with the deregulation of endogenous PP2A inhibitors such as CIP2A and SET have been described as contributing mechanisms to inactivate PP2A in prostate cancer. 26310906_Concurrent mTORC1 inactivation and PP2A-B55alpha stimulation fuel ULK1-dependent autophagy. 26378614_that loss of glucocerebrosidase function may contribute to SNCA accumulation through inhibition of autophagy via PPP2A inactivation 26563471_Data suggest a critical role for the I2PP2A protein (SET)-protein phosphatase-2A (PP2A) signaling axis in Pten protein (Pten) deficient castration resistant prostate cancer (CRPC) progression. 26575017_Data show that downregulating proto-oncogene protein Akt (p-Akt) by inhibiting PP2A inhibitor SET-mediated protein phosphatase 2A (PP2A) inactivation determined the pro-apoptotic effects of EMQA and paclitaxel combination treatment. 26592247_Both PP-1 and PP-2A are directly involved in regulating eye development, and are aberrantly expressed in cataract and glaucoma patients. (Review) 26618405_PP2Ac upregulation has a poor prognostic impact on the overall survival of hepatocellular carcinoma (HCC) patients and contributes to the aggressiveness of HCC. PP2Ac may represent a potential therapeutic target for HCC. 26876307_Data show that loss of epithelial membrane protein 2 (EMP2) is involved in sphingosylphosphorylcholine (SPC)-induced phosphorylation of keratin 8 (K8) via ubiquitination of protein phosphatase 2 (PP2A) through alpha4 phosphoprotein by caveolin-1 (cav-1). 27169767_Knockdown of Alpha4 preferentially impacts the expression of PP4c and PP6c compared to expression levels of PP2Ac. 27306323_Suspension survival mediated by PP2A-STAT3-Col XVII determines tumor initiation and metastasis in cancer stem cells. 27334924_these data support a role for the novel PP2Ac-CIN85 complex in supporting integrin-dependent platelet function by dampening the phosphatase activity. 27517624_Results show that miR-199b is a tumor suppressor emerges as a potential contributing mechanism to inhibit PP2A via PP2A inhibitor SET (SET) overexpression in metastatic colorectal cancer (mCRC). 27531894_B55alpha-PP2A mutations in acute myeloid leukemia have roles in leukemogenesis by promoting AKT T308 phosphorylation and sensitivity to AKT inhibitor-induced growth arrest 27600565_This work has significantly advanced our understanding of the RACK1/PP2A complex and suggests a pro-carcinogenic role for the RACK1/PP2A interaction. This work suggests that approaches to target the RACK1/PP2A complex are a viable option to regulate PP2A activity and identifies a novel potential therapeutic target in the treatment of breast cancer. 27611305_RAB9 competes with the catalytic subunit PPP2CA in binding to PPP2R1A. This competitive association has an important role in controlling the PP2A catalytic activity. 27612201_protein phosphatase 2A (PP2A)-mediated Raf-MEK-ERK signaling was involved in glutaminolysis in endothelial cells. 27696687_Binding of PP2A and Akt increased in response to cAMP or phosphatidic acid (PA), suggesting that their binding is directly responsible for the inactivation of Akt during decidualization. 27913678_Studies indicate that protein phosphatase methylesterase-1 (PME-1) negatively regulates protein phosphatase 2A (PP2A) activity by highly complex mechanisms. 28144936_The results showed SNPs near PPP2CA were associated with decreased expression of PPP2CA mRNA in PBMCs from patients with SLE and suggested that the mRNA expression of the gene may be correlated with the pathogenesis of SLE. 28904398_These findings support our hypothesis that genetic variants in PPP2CA may be implicated in gastric cancer susceptibility in Chinese population. 29066346_Moreover, PP2Acalpha2-overexpressed cells demonstrated increased expression of IGBP1, activated mTORC1 signaling to reduce basal autophagy and increased anchorage-independent growth. Our study provides new insights into the complex mechanisms of PP2A regulation. 29450633_Data show that protein phosphatase-2A (PP2A) was upregulated in lung adenocarcinoma cell lines that were transfected with midline 1 E3 ubiquitin-protein ligase (MID1)-siRNA, suggesting MID1 negatively regulates PP2A in lung adenocarcinoma. 29979448_This study demonstrated an association of PPP2CA (rs10491322 and rs7704116) with systemic lupus erythematosus susceptibility in a Chinese Han population. Furthermore, the minor allele of PPP2CA rs10491322 as a risk factor was correlated with immunologic disorders for systemic lupus erythematosus. 29996119_alpha-Syn bound to PP2A Calpha by the hydrophobic interaction and upregulated its activity. Blocking the hydrophobic domain of alpha-Syn or hydrophilic mutation on the residue I123 in PP2A Calpha all reduced PP2A activity upregulation by alpha-Syn. 30144452_PP2A overexpression is associated with lung metastasis in colorectal cancer . 30275201_High PP2A expression is associated with Colorectal Cancer Cell Invasiveness. 30296597_PPP2CA may act as an oncogene in the progression of colorectal cancer 30453012_the phosphorylation levels of several substrates of PP2A, namely Akt, S6 kinase, and GSK3beta, were decreased in CLN8 disease patient fibroblasts. 30595372_pathogenic PPP2CA variants impair PP2A-B56(delta) functionality, suggesting that PP2A-related neurodevelopmental disorders constitute functionally converging intellectual disability syndromes 30679389_These data demonstrate that LB-100 is a catalytic inhibitor of both PP2AC and PPP5C and suggest that the observed antitumor activity might be due to an additive effect achieved by suppressing both PP2A and PPP5C. 30684253_P2ACalpha negatively regulates LPS-induced cytokine secretion of Alveolar macrophages by NF-kappaB and MAPK pathways. Together, these findings provide the evidence to guide the development of novel therapeutic options targeting PP2ACalpha for Acute respiratory distress syndrome/acute lung injury. 30927143_These results suggest that miR-650/PPP2CA axis could be modulated to interfere with motile ability of thyroid carcinoma cells 30943283_High PP2A expression is associated with diffuse intrinsic pontine gliomas. 31164634_Hydrogen peroxide (H2O2) inhibits protein phosphatase 2 A activity during mitosis. 31461344_Ca2+-activated Cl- channels encoded by the Tmem16a gene are regulated by calmodulin-dependent protein kinase II (CaMKII) and protein phosphatases 1 (PP1) and 2A (PP2A). 31605257_PP2A is involved in apoptotic induction of hepatocytes after brain death by specific suppression of Akt. 31992581_Antibodies recognizing the C terminus of PP2A catalytic subunit are unsuitable for evaluating PP2A activity and holoenzyme composition. 32123010_AKT Regulates Mitotic Progression of Mammalian Cells by Phosphorylating MASTL, Leading to Protein Phosphatase 2A Inactivation. 32681005_Dissecting the sequence determinants for dephosphorylation by the catalytic subunits of phosphatases PP1 and PP2A. 32900880_Quantitative kinase and phosphatase profiling reveal that CDK1 phosphorylates PP2Ac to promote mitotic entry. 32966759_Integrator Recruits Protein Phosphatase 2A to Prevent Pause Release and Facilitate Transcription Termination. 33128264_SIK2 represses AKT/GSK3beta/beta-catenin signaling and suppresses gastric cancer by inhibiting autophagic degradation of protein phosphatases. 33184242_High-glucose-induced apoptosis, ROS production and pro-inflammatory response in cardiomyocytes is attenuated by metformin treatment via PP2A activation. 34100859_Evolutionary crossroads of cell signaling: PP1 and PP2A substrate sites in intrinsically disordered regions. 34425033_PP-1beta and PP-2Aalpha modulate cAMP response element-binding protein (CREB) functions in aging control and stress response through de-regulation of alphaB-crystallin gene and p300-p53 signaling axis. 34485508_Inhibiting PP2Acalpha Promotes the Malignant Phenotype of Gastric Cancer Cells through the ATM/METTL3 Axis. 34502355_Mechanisms of Systolic Cardiac Dysfunction in PP2A, PP5 and PP2AxPP5 Double Transgenic Mice. 34521817_Programmed cell death 10 promotes metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma via PP2Ac-mediated YAP activation. 34780974_PP2A Catalytic Subunit alpha promotes fibroblast activation and kidney fibrosis via ERK pathway. 34902541_Protein phosphatase 2A in the healthy and failing heart: New insights and therapeutic opportunities. | ENSMUSG00000020349 | Ppp2ca | 1.618608e+03 | 1.1310870 | 0.177709907 | 0.2923867 | 3.690940e-01 | 0.5434985307 | 0.89063840 | No | Yes | 1.713506e+03 | 254.048960 | 1.414254e+03 | 215.036295 | |
| ENSG00000113649 | 10915 | TCERG1 | protein_coding | O14776 | FUNCTION: Transcription factor that binds RNA polymerase II and inhibits the elongation of transcripts from target promoters. Regulates transcription elongation in a TATA box-dependent manner. Necessary for TAT-dependent activation of the human immunodeficiency virus type 1 (HIV-1) promoter. {ECO:0000269|PubMed:11604498, ECO:0000269|PubMed:9315662}. | 3D-structure;Activator;Alternative splicing;Coiled coil;Direct protein sequencing;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a nuclear protein that regulates transcriptional elongation and pre-mRNA splicing. The encoded protein interacts with the hyperphosphorylated C-terminal domain of RNA polymerase II via multiple FF domains, and with the pre-mRNA splicing factor SF1 via a WW domain. Alternative splicing results in multiple transcripts variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:10915; | nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; RNA binding [GO:0003723]; RNA polymerase binding [GO:0070063]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; transcription corepressor activity [GO:0003714]; transcription elongation regulator activity [GO:0003711]; ubiquitin-like protein conjugating enzyme binding [GO:0044390]; mRNA processing [GO:0006397]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription elongation from RNA polymerase II promoter [GO:0034244]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription elongation from RNA polymerase II promoter [GO:0032968]; RNA splicing [GO:0008380] | 12821179_Specific alleles in GluR6 and CA150 locus were only observed in HD patients. 15456888_Results suggest an essential role of WW/FF domain-containing factors such as FBP11 and CA150 in pre-mRNA splicing that likely occurs in concert with transcription in vivo. 16644732_CA150 is a co-repressor of C/EBP proteins and provides a possible mechanism for how C/EBPalpha can repress transcription of specific genes 16782886_Sequences located at both the amino and carboxyl regions of CA150 are required to assemble transcription/splicing complexes, which may be involved in the coupling of those processes. 17018562_GRIN2A and TCERG1 may show true association with residual age of onset for Huntington's disease in genetic association tests in 443 affected people from a large set of kindreds from Venezuela. 18187414_The interferon consensus sequence-binding protein (ICSBP/IRF8) represses PTPN13 gene transcription in differentiating myeloid cells 19660470_Data provide the first crystal structure of an FF domain and insights into the tandem nature of the FF domains and suggest that, in addition to protein binding, FF domains might be involved in DNA binding. 19715701_Data gave a model for FF domain organization within tandem arrays suggests a general mechanism by which individual FF domains can manoeuvre to achieve optimal recognition of flexible binding partners, such as the intrinsically-disordered phosphoCTD 19845895_Observational study of gene-disease association. (HuGE Navigator) 20215116_mutation of the SUMO acceptor lysine residues enhanced TCERG1 transcriptional activity, indicating that SUMO modification negatively regulates TCERG1 transcriptional activity 21503969_TCERG1 can inhibit C/EBPalpha activity regardless of the latter's location in the nucleus 22158966_We propose that TCERG1 modulates the elongation rate of RNAPII to relieve pausing, thereby activating the proapoptotic Bcl-x(S) 5' splice site. 22453921_The FF4 and FF5 domains of transcription elongation regulator 1 (TCERG1) target proteins to the periphery of speckles. 23436654_Specific interaction of the transcription elongation regulator TCERG1 with RNA polymerase II requires simultaneous phosphorylation at Ser2, Ser5, and Ser7 within the carboxyl-terminal domain repeat. 24165037_This study reveals that TCERG1 regulates HIV-1 transcriptional elongation by increasing the elongation rate of RNAPII and phosphorylation of Ser 2 within the carboxyl-terminal domain. 26264132_The QA repeat domain of TCERG1 is required for relocalization of CEBPalpha. 26462236_TCERG1 sensitizes a cell to apoptotic agents, thus promoting apoptosis by regulating the alternative splicing of both the Bcl-x and Fas/CD95 genes. 26873599_TCERG1 binds independently to elongation and splicing complexes, thus performing their coupling by transient interactions rather than by stable association with one or the other complexes. 27844289_TCERG1 affects expression of multiple mRNAs involved in neuron projection development. 31636114_Results of this study suggest that TCERG1 plays an important role in Cajal body formation and snRNP biogenesis. | ENSMUSG00000024498 | Tcerg1 | 2.506129e+03 | 1.1783326 | 0.236746788 | 0.3148864 | 5.651186e-01 | 0.4522052166 | 0.86203713 | No | Yes | 2.738729e+03 | 520.839674 | 1.834505e+03 | 358.159813 | |
| ENSG00000113658 | 4090 | SMAD5 | protein_coding | Q99717 | FUNCTION: Transcriptional modulator activated by BMP (bone morphogenetic proteins) type 1 receptor kinase. SMAD5 is a receptor-regulated SMAD (R-SMAD). | 3D-structure;Acetylation;Cytoplasm;DNA-binding;Direct protein sequencing;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Zinc | The protein encoded by this gene is involved in the transforming growth factor beta signaling pathway that results in an inhibition of the proliferation of hematopoietic progenitor cells. The encoded protein is activated by bone morphogenetic proteins type 1 receptor kinase, and may be involved in cancer. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2014]. | hsa:4090; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; heteromeric SMAD protein complex [GO:0071144]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; SMAD protein complex [GO:0071141]; DEAD/H-box RNA helicase binding [GO:0017151]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; I-SMAD binding [GO:0070411]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; ubiquitin protein ligase binding [GO:0031625]; anatomical structure morphogenesis [GO:0009653]; BMP signaling pathway [GO:0030509]; bone development [GO:0060348]; cardiac conduction system development [GO:0003161]; cardiac muscle contraction [GO:0060048]; cartilage development [GO:0051216]; cell differentiation [GO:0030154]; cellular response to organic cyclic compound [GO:0071407]; embryonic pattern specification [GO:0009880]; erythrocyte differentiation [GO:0030218]; germ cell development [GO:0007281]; Mullerian duct regression [GO:0001880]; osteoblast fate commitment [GO:0002051]; positive regulation of osteoblast differentiation [GO:0045669]; positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus [GO:1901522]; positive regulation of transcription, DNA-templated [GO:0045893]; protein phosphorylation [GO:0006468]; signal transduction [GO:0007165]; SMAD protein signal transduction [GO:0060395]; transforming growth factor beta receptor signaling pathway [GO:0007179]; ureteric bud development [GO:0001657] | 12060751_regulatory signals are active at transcriptionally subnuclear sites 12064918_Smad5 activation by BMP4 in human hematopoietic cells results in significantly increased proliferation of erythroid progenitors and formation of glycophorin-A+ cells. 12473652_up-regulated Smad5 mediates apoptosis of gastric epithelial cells induced by Helicobacter pylori infection 12849988_Results demonstrate a mitochondrial distribution of Smad5 in non-stimulated chondroprogenitor cells. 14670176_SMAD5 undergoes copy number gain and increased expression, rather than loss of expression, and therefore does not act as a tumor-suppressor gene in hepatocellular carcinoma 15877825_In mature human B cells, BMP-6 inhibited cell growth, and rapidly induced phosphorylation of Smad5. 16243555_We used homology-modeling techniques to generate a reliable molecular model of the Smad5 MH1 domain based on the crystal structure of Smad3 MH1 domain 16247476_activation by BMP-2 in lung cancer cells 16436528_Human granulosa-like tumor cell line KGN expressed BMP type I (BMPR1A and BMPR1B) and type II receptors (BMPR2) and the BMP signaling molecules SMADs (SMAD1 and SMAD5). 17359969_These data suggest autocrine TGF-beta1 antagonizes BMP signaling through modulation of inducible Smad6 and the activity of BMP specific Smad1/5. 17847004_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18298822_SMAD5 gene was found to be associated with schizophrenia. 18310319_The shear-induced G(2)/M arrest and corresponding changes in G(2)/M regulatory protein expression and activity were mediated by alpha(v)beta(3) and beta(1) integrins through bone morphogenetic protein receptor type IA-specific Smad1 and Smad5. 18393632_SMAD 2/3 signaling directly supports NANOG expression, while SMAD 1/5/8 activation moderately represses SOX2. 18684712_ALK2(R206H) with Smad1 or Smad5 induced osteoblastic differentiation that could be inhibited by Smad7 or dorsomorphin. 18775991_Endoglin promotes transforming growth factor beta-mediated Smad 1/5/8 signaling and inhibits endothelial cell migration through its association with GIPC 19211612_Observational study of gene-disease association. (HuGE Navigator) 19244313_5-HT transactivates the serine kinase receptor, BMPR 1A, to activate Smads 1/5/8 via Rho and Rho kinase in in bovine and human pulmonary artery smooth muscle cells 19247629_Observational study of gene-disease association. (HuGE Navigator) 19269967_increases of p16(INK4a) and p21(WAF1/cip1) expression in response to BMP4 were mediated by the Smad1/5/8 signaling pathway. 19819941_Data suggest that the SMAD family, possibly through disruption of SMAD1/5 or activation of SMAD2/3 may contribute to the pathogenesis of JGCT in humans. 20148926_Studies indicate SMAD5beta isoform plays role in hematopoietic stem cell homeostasis. 20522807_Prostacyclin analogues inhibited smooth muscle cell proliferation and prevented progression of pulmonary hypertension while enhancing Smad1/5 phosphorylation and Id1 gene expression. 20734064_Observational study of gene-disease association. (HuGE Navigator) 21454478_CHIP inhibits the signaling activities of Smad1/5 by recruiting Smad1/5 from the functional R-/Co-Smad complex and further promoting the ubiquitination/degradation of Smad1/5 in a chaperone-independent manner. 21945631_Results show that BMP4-induced changes in OvCa cell morphology and motility are Smad-dependent with shRNA targeting Smads 1, 4, and 5. 22013049_found that restoration of SMAD5, in addition to the TGF-beta type II receptor, which was epigenetically silenced by the latent viral protein latency-associated nuclear antigen, sensitized BC3 cells to the cytostatic effect of TGF-beta signaling 22452883_Data suggest that Smads 1, 5 and 8 as potential prognostic markers and therapeutic targets for mTOR inhibition therapy of prostate cancer. 22897816_TNF activated NF-kappaB pathway and inhibited the phosphorylation of Smad 1/5/8 and BMP-2-induced osteoblastic differentiation in BMMSCs 22964636_our studies establish that loss of SMAD1/5 leads to upregulation of PDGFA in ovarian granulosa cells 22966907_CD44 signaling regulates phosphorylation of RUNX2. Localization of RUNX2 in the nucleus requires phosphorylation of Smad-5 by integrin alphavbeta3 signaling. 23384547_ATP production by NaF promotes hypertrophy-like changes via activation of phospho-Smad5 23387849_Oscillatory shear stress induces synergistic interactions between specific BMPRs and integrin to activate Smad1/5 through the Shc/FAK/ERK pathway 23804438_a detailed computational model for TGF-beta signalling that incorporates elements of previous models together with crosstalking between Smad1/5/8 and Smad2/3 channels through a negative feedback loop dependent on Smad7. 23918166_Results indicate that BMP/Smad signaling pathway was altered during the period of osteogenesis, and that the activities of p-Smad1/5 were required for Saos-2 cells viability and differentiation induced by fluoride. 24021264_the shear-induced apoptosis and autophagy are mediated by bone morphogenetic protein receptor type (BMPR)-IB, BMPR-specific Smad1 and Smad5, and p38 mitogen-activated protein kinase. 24163009_among the 15 SNPs, rs3206634 was significantly associated with KD in a recessive model (odds ratio = 2.31, p = 0.019), whereas there was no association between any of the 15 SNPs and CALs. 24339730_Specific gene siRNAs knock-down further confirmed the osteogenic effects of Genistein on BMP2, SMAD5 and RUNX2 protein expression 24828823_Inhibiting Smurf1 mediated ubiquitination of Smad1/5. 24928394_Our results indicated that KGN promoted the type-I collagen synthesis of dermal fibroblasts in vitro and in the dermis of mice through activation of the smad4/smad5 pathway. 25505291_balance between Smad1/5- and Smad2/3-dependent signaling defines the outcome of the effect of TGF-beta on atherosclerosis where Smad1/5 is responsible for proatherogenic effects 25754204_The polycomb group protein L3MBTL1 represses a SMAD5-mediated hematopoietic transcriptional program in human pluripotent stem cells. 25880873_adult human Sertoli cells assumed similar morphological features, stable global gene expression profiles and numerous proteins, and activation of AKT and SMAD1/5 during long-period culture. 26339396_Overexpression of the BMP4/SMAD4/SMAD5 signaling pathway could predict poor clinical outcome in skull base chordomas, suggesting activation of this pathway is involved in chordoma pathogenesis. 26400397_miR-23a and miR-27a target SMAD5 and regulate apoptosis in human granulosa cells via the FasL-Fas pathway 26809090_Our findings suggest that suppression of miR-222-3p activity promoted osteogenic differentiation hBMSCs through regulating Smad5-RUNX2 signaling axis. 27025722_the BMP-2/Smad1/5/RUNX2 signaling pathway participates in the silicon-mediated induction of COL-1 and osteocalcin synth 27727244_Here the involvement of the pathway in adult brain function is suggested. This exploratory study establishes a strategy to better identify neuronal molecular signatures that are potentially associated with mental illness and cognitive deficits. We propose that the SMAD pathway may be a novel target in addressing cognitive deficit of SZ in future studies. 27848974_Differential expression of TGF-beta superfamily members and role of Smad1/5/9-signalling in chondral versus endochondral chondrocyte differentiation. 28675158_Smad5 acts as a intracellular pH messenger and maintains the bioenergetic homeostasis of cells by regulating cytoplasmic metabolic machinery. 29029022_Data suggest that differences in expression levels in granulosa-like tumor cells and granulosa cells (GCs) from patients with polycystic ovary syndrome (PCOS) are due to increased expression of microRNA-27a-3p in GCs caused by insulin resistance in PCOS; microRNA-27a-3 expression is up-regulated in GCs in PCOS; overexpression of miR-27a-3p inhibits SMAD5 expression and promotes apoptosis. (SMAD5 = SMAD family member 5) 29192241_these results suggested that miR-21 be mechanistically implicated in the regulation of osteogenic differentiation of hPDLSCs by targeting Smad5. 29376829_Here, the authors show that TGF-beta-induced SMAD1/5 phosphorylation requires members of two classes of type I receptor, TGFBR1 and ACVR1, and establish a new paradigm for receptor activation where TGFBR1 phosphorylates and activates ACVR1, which phosphorylates SMAD1/5. 29852786_We demonstrated high expression of miR-145 associated with late stage and unfavorable prognosis of esophageal cancer. We identified SMAD5 as direct target of miR-145, the suppressed expression of which consequently led to increased cell proliferation and migration/invasion. 30367512_Erbin is mainly localized in endothelial cells in human umbilical veins and plays a critical role in endothelial cell migration and tubular formation via the Smad1/5 pathway. 30683863_Memory T cells targeting oncogenic mutations in KRAS, SMAD5, and MUC4 detected in peripheral blood of colon cancer patients have been isolated. 30953749_Cyclic AMP appears to promote SMAD1/5/8 pathway activity intracellularly and has the ability to activate canonical SMAD1/5/8 downstream targets. 31945413_LncRNA AFAP1-AS1 promotes osteoblast differentiation of human aortic valve interstitial cells through regulating miR-155/SMAD5 axis. 31951299_LINC01410 promotes cell proliferation and migration of cholangiocarcinoma through modulating miR-124-3p/SMAD5 axis. 32271402_KCNQ1OT1 regulates osteogenic differentiation of hBMSC by miR-320a/Smad5 axis. 32883538_Long non-coding RNA LINC01116 accelerates the progression of keloid formation by regulating miR-203/SMAD5 axis. 33371778_Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. 34974806_Long non-coding RNA H19 aggravates keloid progression by upregulating SMAD family member 5 expression via miR-196b-5p. 34978464_Role of lncRNA LINC01194 in hepatocellular carcinoma via the miR-655-3p/SMAD family member 5 axis. | ENSMUSG00000021540 | Smad5 | 6.284145e+02 | 0.8540137 | -0.227668884 | 0.3513310 | 4.110537e-01 | 0.5214351599 | 0.88429638 | No | Yes | 4.819978e+02 | 95.992175 | 5.730676e+02 | 116.782200 | |
| ENSG00000113719 | 57222 | ERGIC1 | protein_coding | Q969X5 | FUNCTION: Possible role in transport between endoplasmic reticulum and Golgi. {ECO:0000303|PubMed:15308636}. | Alternative splicing;Direct protein sequencing;Disease variant;ER-Golgi transport;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a cycling membrane protein which is an endoplasmic reticulum-golgi intermediate compartment (ERGIC) protein which interacts with other members of this protein family to increase their turnover. [provided by RefSeq, Jul 2008]. | hsa:57222; | COPII-coated ER to Golgi transport vesicle [GO:0030134]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of Golgi membrane [GO:0030173]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890] | 15308636_ERGIC-32 functions as a modulator of the hErv41-hErv46 complex by stabilizing hErv46 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22761906_AIM1, ERGIC1, and TPX2 were shown to be highly expressed especially in prostate cancer tissues, and high mRNA expression of ERGIC1 and TMED3 associated with AR and ERG oncogene expression 28317099_Homozygous pathogenic variant in the endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 protein (ERGIC1) gene was identified in arthrogryposis multiplex congenita (AMC) neuropathic type. 28970727_Data indicate that abnormal ERGIC1 and DNA-PKcs expression may play an important role in gastric cancer initiation. 29549924_ERGIC1 might play an inhibitory role in the initiation and progression of gastric cancer. 34037256_Bi-allelic loss of ERGIC1 causes relatively mild arthrogryposis. | ENSMUSG00000001576 | Ergic1 | 7.890983e+03 | 1.1946227 | 0.256554988 | 0.3052391 | 7.106671e-01 | 0.3992224460 | 0.84485965 | No | Yes | 7.604923e+03 | 632.437384 | 6.138730e+03 | 523.947107 | |
| ENSG00000114383 | 11334 | TUSC2 | protein_coding | O75896 | FUNCTION: May function as a tumor suppressor, inhibiting colony formation, causing G1 arrest and ultimately inducing apoptosis in homozygous 3p21.3 120-kb region-deficient cells. | Cell cycle;Lipoprotein;Myristate;Phosphoprotein;Reference proteome;Tumor suppressor | This gene is a highly conserved lung cancer candidate gene. No other information about this gene is currently available. [provided by RefSeq, Jul 2008]. | hsa:11334; | mitochondrion [GO:0005739]; cell cycle [GO:0007049]; cell maturation [GO:0048469]; inflammatory response [GO:0006954]; natural killer cell differentiation [GO:0001779]; negative regulation of interleukin-17 production [GO:0032700]; neutrophil-mediated killing of gram-negative bacterium [GO:0070945]; phagocytosis [GO:0006909]; positive regulation of interleukin-10 production [GO:0032733]; regulation of mitochondrial membrane potential [GO:0051881]; regulation of reactive oxygen species metabolic process [GO:2000377] | 11593436_Overexpression of candidate tumor suppressor geneFUS1 isolated from the 3p21.3 homozygous deletionregion leads to G1 arrest and growth inhibition oflung cancer cells. 15126327_Myristoylation is required for Fus1-mediated tumor-suppressing activity and suggest a novel mechanism for the inactivation of tumor suppressors in lung cancer. 17545076_Data show that the recombinant FUS1 plasmid has been successfully cloned, which allows highly efficient FUS1 expression in E.coli strain Rosetta (DE3)2 plys. 18077375_These results suggest that miR-378 enhances cell survival, tumor growth, and angiogenesis through repression of the expression of two tumor suppressors, Sufu and Fus-1. 18172250_FUS1 gene and Fus1 protein abnormalities could be used to develop new strategies for molecular cancer therapy for a significant subset of lung tumors. 19671678_These results suggest that the three miRNAs are negative regulators of Fus1 expression in lung cancers. 19852844_The goal of this study was to analyze possible involvement of TUSC2 in malignant pleural mesothelioma. 21273575_Absence of tumor suppressor FUS1 protein expression is associated with bone and soft tissue sarcomas. 21645495_RNA sequence elements in FUS1 UTRs that regulate FUS1 protein express were identified. 24146957_The tumor suppressor gene TUSC2 (FUS1) sensitizes NSCLC to the AKT inhibitor MK2206 in LKB1-dependent manner. 24394498_TUSC2P and TUSC2 3'-UTR expression inhibits cell proliferation, survival, migration, invasion and colony formation, and increases tumor cell death. TUSC2P and TUSC2 3'-UTR bind to miRNAs and arrest their functions, leading to increased TUSC2 translation. 26053020_The therapeutic activity of TUSC2 could extend the use of erlotinib to lung cancer patients with wildtype EGFR. 26367773_FOXP1, TP53INP1, TNFAIP3, and TUSC2 were identified as miR-19a targets. 27661106_Data show that TUSC2 is a direct target of miR-584, which is transcriptionally regulated by TWIST1. 27845352_these findings show that the combination of TUSC2-erlotinib induces additional novel vulnerabilities that can be targeted with Auranofin. 30219035_TUSC2P can suppresses the tumor function of esophageal squamous cell carcinoma by regulating TUSC2 expression and may also serve as a prognostic factor for esophageal squamous cell carcinoma patients. 30896880_Low TUSC2 expression is associated with bladder cancer. 31481748_MicroRNA-138 is a Prognostic Biomarker for Triple-Negative Breast Cancer and Promotes Tumorigenesis via TUSC2 repression. 31973107_The TUSC2 Tumour Suppressor Inhibits the Malignant Phenotype of Human Thyroid Cancer Cells via SMAC/DIABLO Protein. 35167936_NEDD4 degrades TUSC2 to promote glioblastoma progression. | ENSMUSG00000010054 | Tusc2 | 3.740026e+03 | 1.2330631 | 0.302246602 | 0.3170135 | 9.056362e-01 | 0.3412749199 | 0.82750349 | No | Yes | 4.015350e+03 | 505.916354 | 2.720688e+03 | 351.983826 | |
| ENSG00000114541 | 23150 | FRMD4B | protein_coding | Q9Y2L6 | FUNCTION: Member of GRP1 signaling complexes that are acutely recruited to plasma membrane ruffles in response to insulin receptor signaling. May function as a scaffolding protein that regulates epithelial cell polarity by connecting ARF6 activation with the PAR3 complex. Plays a redundant role with FRMD4A in epithelial polarization. {ECO:0000250|UniProtKB:Q920B0}. | Alternative splicing;Cell junction;Coiled coil;Cytoplasm;Cytoskeleton;Isopeptide bond;Phosphoprotein;Reference proteome;Tight junction;Ubl conjugation | hsa:23150; | adherens junction [GO:0005912]; bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; extracellular space [GO:0005615]; ruffle [GO:0001726]; establishment of epithelial cell polarity [GO:0090162] | Mouse_homologues 29947801_his study reveals a critical role of FRMD4B in maintaining ELM integrity and in rescuing morphological abnormalities of the outer nuclear layer in photoreceptor dysplasia. | ENSMUSG00000030064 | Frmd4b | 3.302406e+01 | 0.8652868 | -0.208749782 | 0.5426300 | 1.427042e-01 | 0.7056073078 | 0.93718130 | No | Yes | 3.450811e+01 | 8.907184 | 3.681224e+01 | 10.003436 | ||
| ENSG00000114738 | 7867 | MAPKAPK3 | protein_coding | Q16644 | FUNCTION: Stress-activated serine/threonine-protein kinase involved in cytokines production, endocytosis, cell migration, chromatin remodeling and transcriptional regulation. Following stress, it is phosphorylated and activated by MAP kinase p38-alpha/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. MAPKAPK2 and MAPKAPK3, share the same function and substrate specificity, but MAPKAPK3 kinase activity and level in protein expression are lower compared to MAPKAPK2. Phosphorylates HSP27/HSPB1, KRT18, KRT20, RCSD1, RPS6KA3, TAB3 and TTP/ZFP36. Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to dissociate HSP27/HSPB1 from large small heat-shock protein (sHsps) oligomers and impair their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating tumor necrosis factor (TNF) and IL6 production post-transcriptionally: acts by phosphorylating AU-rich elements (AREs)-binding proteins, such as TTP/ZFP36, leading to regulate the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major post-transcriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity leading to inhibition of dependent degradation of ARE-containing transcript. Involved in toll-like receptor signaling pathway (TLR) in dendritic cells: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. Also acts as a modulator of Polycomb-mediated repression. {ECO:0000269|PubMed:10383393, ECO:0000269|PubMed:15563468, ECO:0000269|PubMed:18021073, ECO:0000269|PubMed:20599781, ECO:0000269|PubMed:8626550, ECO:0000269|PubMed:8774846}. | 3D-structure;ATP-binding;Acetylation;Cytoplasm;Disease variant;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the Ser/Thr protein kinase family. This kinase functions as a mitogen-activated protein kinase (MAP kinase)- activated protein kinase. MAP kinases are also known as extracellular signal-regulated kinases (ERKs), act as an integration point for multiple biochemical signals. This kinase was shown to be activated by growth inducers and stress stimulation of cells. In vitro studies demonstrated that ERK, p38 MAP kinase and Jun N-terminal kinase were all able to phosphorylate and activate this kinase, which suggested the role of this kinase as an integrative element of signaling in both mitogen and stress responses. This kinase was reported to interact with, phosphorylate and repress the activity of E47, which is a basic helix-loop-helix transcription factor known to be involved in the regulation of tissue-specific gene expression and cell differentiation. Alternate splicing results in multiple transcript variants that encode the same protein. [provided by RefSeq, Sep 2011]. | hsa:7867; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; calcium-dependent protein serine/threonine kinase activity [GO:0009931]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; MAP kinase kinase activity [GO:0004708]; mitogen-activated protein kinase binding [GO:0051019]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; intracellular signal transduction [GO:0035556]; macropinocytosis [GO:0044351]; peptidyl-serine phosphorylation [GO:0018105]; protein autophosphorylation [GO:0046777]; response to cytokine [GO:0034097]; response to lipopolysaccharide [GO:0032496]; signal transduction [GO:0007165]; toll-like receptor signaling pathway [GO:0002224]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11975836_In the preconditioned heart, genes for MAPKAP kinase 3 were up-regulated. 15302577_3pK is transported to the cytoplasm upon both stress and mitogenic stimulation. While kinetics of nuclear export are similar in both situations, the activation pattern differs substantially. 19208361_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19937655_A high-resolution (1.9 A) crystal structure of the highly homologous MK3 in complex with a pharmaceutical lead compound is presented. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23127979_Identified CREB activators MAPKAPK3 and FHL5 as mediators of intimal hyperplasia in vein graft samples. 23487458_Hepatitis C virus core protein interacted with MAPKAPK3 through amino acid residues 41 to 75 of core and the N-terminal half of kinase domain of MAPKAPK3. 25693418_findings reveal MK2/MK3 as crucial stress-responsive kinases that promote autophagy through Beclin 1 S90 phosphorylation 25853770_MK3 modulation affects BMI1-dependent and independent cell cycle check-points 26744326_A dominant mutation in MAPKAPK3 causes a new retinal dystrophy involving Bruch's membrane and retinal pigment epithelium. 27474146_The natural history of MCRPE is in relation to the role of MAPKAPK3 in BM modeling, vascular endothelial growth factor activity, retinal pigment epithelial responses | ENSMUSG00000032577 | Mapkapk3 | 5.661791e+03 | 1.3060899 | 0.385254174 | 0.3149492 | 1.486855e+00 | 0.2227049997 | 0.78763590 | No | Yes | 6.026649e+03 | 742.560755 | 3.911550e+03 | 494.791370 | |
| ENSG00000114853 | 92999 | ZBTB47 | protein_coding | Q9UFB7 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:92999; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000013419 | Zfp651 | 1.845261e+03 | 1.1657878 | 0.221305201 | 0.2873703 | 5.851113e-01 | 0.4443157904 | 0.85798841 | No | Yes | 1.902888e+03 | 182.075561 | 1.600716e+03 | 157.758943 | |||
| ENSG00000114861 | 27086 | FOXP1 | protein_coding | Q9H334 | FUNCTION: Transcriptional repressor (PubMed:18347093, PubMed:26647308). Can act with CTBP1 to synergistically repress transcription but CTPBP1 is not essential (By similarity). Plays an important role in the specification and differentiation of lung epithelium. Acts cooperatively with FOXP4 to regulate lung secretory epithelial cell fate and regeneration by restricting the goblet cell lineage program; the function may involve regulation of AGR2. Essential transcriptional regulator of B-cell development. Involved in regulation of cardiac muscle cell proliferation. Involved in the columnar organization of spinal motor neurons. Promotes the formation of the lateral motor neuron column (LMC) and the preganglionic motor column (PGC) and is required for respective appropriate motor axon projections. The segment-appropriate generation of spinal cord motor columns requires cooperation with other Hox proteins. Can regulate PITX3 promoter activity; may promote midbrain identity in embryonic stem cell-derived dopamine neurons by regulating PITX3. Negatively regulates the differentiation of T follicular helper cells T(FH)s. Involved in maintenance of hair follicle stem cell quiescence; the function probably involves regulation of FGF18 (By similarity). Represses transcription of various pro-apoptotic genes and cooperates with NF-kappa B-signaling in promoting B-cell expansion by inhibition of caspase-dependent apoptosis (PubMed:25267198). Binds to CSF1R promoter elements and is involved in regulation of monocyte differentiation and macrophage functions; repression of CSF1R in monocytes seems to involve NCOR2 as corepressor (PubMed:15286807, PubMed:18799727, PubMed:18347093). Involved in endothelial cell proliferation, tube formation and migration indicative for a role in angiogenesis; the role in neovascularization seems to implicate suppression of SEMA5B (PubMed:24023716). Can negatively regulate androgen receptor signaling (PubMed:18640093). Acts as a transcriptional activator of the FBXL7 promoter; this activity is regulated by AURKA (PubMed:28218735). {ECO:0000250|UniProtKB:P58462, ECO:0000269|PubMed:15286807, ECO:0000269|PubMed:18640093, ECO:0000269|PubMed:18799727, ECO:0000269|PubMed:24023716, ECO:0000269|PubMed:25267198, ECO:0000269|PubMed:26647308, ECO:0000269|PubMed:28218735, ECO:0000305|PubMed:18347093, ECO:0000305|PubMed:24023716}.; FUNCTION: [Isoform 8]: Involved in transcriptional regulation in embryonic stem cells (ESCs). Stimulates expression of transcription factors that are required for pluripotency and decreases expression of differentiation-associated genes. Has distinct DNA-binding specifities as compared to the canonical form and preferentially binds DNA with the sequence 5'-CGATACAA-3' (or closely related sequences) (PubMed:21924763). Promotes ESC self-renewal and pluripotency (By similarity). {ECO:0000250|UniProtKB:P58462, ECO:0000269|PubMed:21924763}. | 3D-structure;Alternative splicing;Chromosomal rearrangement;DNA-binding;Disease variant;Isopeptide bond;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene belongs to subfamily P of the forkhead box (FOX) transcription factor family. Forkhead box transcription factors play important roles in the regulation of tissue- and cell type-specific gene transcription during both development and adulthood. Forkhead box P1 protein contains both DNA-binding- and protein-protein binding-domains. This gene may act as a tumor suppressor as it is lost in several tumor types and maps to a chromosomal region (3p14.1) reported to contain a tumor suppressor gene(s). Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:27086; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; androgen receptor binding [GO:0050681]; core promoter sequence-specific DNA binding [GO:0001046]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; protein self-association [GO:0043621]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; cellular response to DNA damage stimulus [GO:0006974]; endothelial cell activation [GO:0042118]; macrophage activation [GO:0042116]; monocyte activation [GO:0042117]; negative regulation of androgen receptor signaling pathway [GO:0060766]; negative regulation of B cell apoptotic process [GO:0002903]; negative regulation of gene expression [GO:0010629]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; osteoclast development [GO:0036035]; osteoclast differentiation [GO:0030316]; positive regulation of B cell receptor signaling pathway [GO:0050861]; positive regulation of endothelial cell migration [GO:0010595]; positive regulation of interleukin-21 production [GO:0032745]; positive regulation of smooth muscle cell proliferation [GO:0048661]; regulation of chemokine (C-X-C motif) ligand 2 production [GO:2000341]; regulation of defense response to bacterium [GO:1900424]; regulation of endothelial tube morphogenesis [GO:1901509]; regulation of gene expression [GO:0010468]; regulation of inflammatory response [GO:0050727]; regulation of interleukin-1 beta production [GO:0032651]; regulation of interleukin-12 production [GO:0032655]; regulation of macrophage colony-stimulating factor production [GO:1901256]; regulation of monocyte differentiation [GO:0045655]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of tumor necrosis factor production [GO:0032680]; response to lipopolysaccharide [GO:0032496]; T follicular helper cell differentiation [GO:0061470] | 11751404_The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. 12692134_Foxp1, although broadly expressed, is further regulated by tissue-specific alternative splicing of its functionally important sequence domains 14701752_complex regulatory mechanism underlying Foxp1, Foxp2, and Foxp4 activity, demonstrating that Foxp1, Foxp2, and Foxp4 are the first Fox proteins reported whose activity is regulated by homo- and heterodimerization 15056695_FOXP1 and FOXP2 expression patterns in human fetal brain are strikingly similar to those in the songbird, including localization to subcortical structures that function in sensorimotor integration and the control of skilled, coordinated movement 15161711_FOXP1 is a potential ER coregulator in human breast carcinoma and may also independently regulate additional important pathways that control the progression of breast cancer 15286807_Integrin engagement regulates monocyte differentiation through FOXP1. 15703784_This study identifies FOXP1 as a new translocation partner of IGH in a site-dependent subset of MALT lymphomas. 15709173_Results suggest that FOXP1 expression may be important in diffuse large B-cell lymphoma (DLBCL) pathogenesis. 16200457_The heterogeneity of FOXP1 expression in germinal centre-derived lymphomas, may have more to do with the transforming events underlying these distinct types of lymphoma than with their common origin. 16258506_Tumors with exclusively cytoplasmic expression of FOXP1 were linked with deep myometrial invasion. 16636337_FOXP1 expression is an independent prognostic factor in MALT lymphomas. A subgroup of nongerminal center DLBCLs (Diffuse Large B-Cell Lymphomas, those marked by FOXP1 expression and trisomy 3 and 18) might represent a large-cell variant of MALT lymphomas. 16673020_rearrangement of FOXP1 is detected in a subset of large B-cell lymphomas with extranodal presentation 16952980_these data implicate specific members of the FOX family of TFs (FOXC1, C2, P1, P4, and O1A) not previously suggested in heart failure pathogenesis. 17199743_Our results suggest that MALT1-specific translocations and FOXP1 rearrangements are not commonly involved in pathogenesis. 17477366_There may be a hormonal and hypoxia independent regulatory mechanism coordinating the expression of HIFs, the AR, and FOXP1 in prostate tumors. 18026833_Expression of the forkhead transcription factor FOXP1 is associated with that of estrogen receptor-beta in primary invasive breast carcinomas. 18077790_The expression of potentially oncogenic smaller FOXP1 isoforms may resolve the previously contradictory findings that FOXP1 represents a favorable prognostic marker in breast cancer and an adverse risk factor in B-cell lymphomas. 18332236_FOXP1 protein is present in human endometrium with evidence of cycle stage-dependent changes in expression. FOXP1 expression was found in endometriotic lesions but not in endometrial adenocarcinoma 18487996_Mechanisms other than translocation and copy number changes are responsible for FOXP1 overexpression in lymphoma. 18640093_FOXP1 directly interacts with androgen receptor (AR) and negatively regulates AR signaling ligand-dependently. 18925695_There was an association between FOXP1 and diffuse large B-cell lymphoma: significant relationship between BCL2 expression and FOXP1 genetic abnormalities. 19141121_expression of Bcl-2 and FOXP1 is associated with the non-germinal center phenotype, but only Bcl-2 expression continues to be of prognostic significance in DLBCL patients treated with immunochemotherapy 19352412_FOXP1 mutations have a role in developmental verbal dyspraxia 19352412_Observational study of gene-disease association. (HuGE Navigator) 19365123_FOXP1 expression is correlated with morphic histology and may be a biomarker for assessment of malignant transformation. 19417623_present in patients with higher clinical stages of chronic lymphocytic leukemia 19432679_FOXP1 might play an important role in plasma cell neoplasm. 19438754_Atypical FOXP1 expression in malignant plasma cells that show several simultaneous translocations 19448593_The expression of CD10, Bcl-6, MUM1/IRF4, Bcl-2, and FOXP1 was determined immunohistochemically from 88 samples of diffuse large B-cell lymphoma patients treated uniformly with R-CHOP 19487025_Our results provide strong evidence that relative FOXP1 isoform abundance is associated with NFkappaB activity in follicular lymphoma, and could potentially be used as a marker for this gene signature 19622517_Results suggest that FOXP1 demonstrates different expression patterns in familial breast cancers than sporadic tumours, even in tumours showing similar phenotypes, and also suggest a different role of FOXP1 as a tumour suppressor in familial tumours. 19680146_truncated FOXP1 isoforms are preferentially overexpressed in primary central nervous system lymphomas 19760589_FOXP1 is not rearranged in Splenic marginal zone lymphoma 19913121_Observational study of gene-disease association. (HuGE Navigator) 19925052_Studies illustrated tools for cut-off level determination with prognostic tumor-related biomarkers Bcl-2, Bcl-6, CD10, FOXP1, MUM1, and Cyclin E in DLBCL. 20190138_These results demonstrate a role of PKCdelta in alpha(M)beta(2)-mediated Foxp1 regulation in monocytes. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20526340_Data show a replicated association of generalized vitiligo with variants at 3p13 encompassing FOXP1. 20526340_Observational study of gene-disease association. (HuGE Navigator) 20579129_FOXP1 overexpression was associated with poor disease-specific survival in all nodal diffuse large B-cell lymphomas. There was no correlation between FOXP1 gene aberrations and either FOXP1 protein expression or survival. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20848658_Found three heterozygous overlapping deletions solely affecting the FOXP1 gene. All three patients had moderate mental retardation and significant language and speech deficits. 20950788_both FOXP1 and FOXP2 are associated with language impairment, but decrease of the former has a more global impact on brain development than that of the latter. 21120478_FOXP1 might play a role in the pathogenesis of nodal diffuse large B-cell lymphoma . 21154235_Overexpression of FOXP1 is present in a considerable proportion of primary cutaneous large B cell lymphoma, leg type and might indicate an unfavourable prognosis. 21210727_FOXP1 expression negatively correlated with Ki67 expression in clear cell renal cell carcinomas (p = .036). 21416545_Data show that FOXP1 C61S and C61Y mutants existed as a mixture by NMR analysis. 21460242_Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. 21655267_The results indicate that in cancer cells E2A, FOXO1 and FOXP1 regulate RAG1 and RAG2 expression, which initiates Ig gene rearrangement much in the way similar to B lymphocytes. 21660567_patients with Non-Hodgkin's lymphoma of mucosa-associated lymphoid tissue that are positive for both PIK3CA and FOXP1 had a worse overall and progression-free survival 21683954_FoxP1 is expressed by different cell types in atherosclerotic lesions and associated with more stable plaque characteristics and intraplaque TGFbeta signaling 21901488_FOXP1 plays an important role in proliferation of breast cancer cells by modulating estrogen signaling. 21924763_Study shows that tembryonic stem cell (ESC)-specific isoform of FOXP1 stimulates expression of transcription factor genes required for pluripotency, OCT4, NANOG, NR5A2, and GDF3, while concomitantly repressing genes required for ESC differentiation. 21927029_A reverse correlation between CTGF and miR-504, miR-504 and FOXP1, and a positive correlation between CTGF and FOXP1 were shown. 21962897_The PAX5-FOXP1 translocation is associated with B-cell acute lymphoblastic leukemia. 22368156_FOXP1 gene expression carries a poor prognosis in thyroid diffuse large B-cell lymphomas. 22401769_forkhead box P1 overexpression was associated with an improved overall survival of the patients with peripheral T-cell lymphoma, not otherwise specified 22422806_These results support a role for FOXP1 as an oncogene in hepatocellular carcinoma. 22476979_Double-positive immunoreactivities of FOXA1 and FOXP1 were significantly associated with a favorable prognosis for the relapse-free and overall survival in with tamoxifen-treated breast cancer. 22492998_Foxp1 is a transcriptional repressor of immune signaling in the central nervous system. 22736042_Our data demonstrated the close relationship between FOXP1 nuclear expression and the occurrence of large tumor cells in MALT lymphoma 22809882_FOXP1 and p65 expression are adverse risk factors in diffuse large B-cell lymphoma 22904134_Expression of FoxP1 messenger RNA (mRNA) and protein was significantly higher in NonSmall Cell Lung Carcinoma tissue than in corresponding peritumoral tissue (P = .013 and P < .001, respectively). 23580662_FOXP1 is physiologically downregulated in germinalo center B cells and that aberrant expression of FOXP1 impairs mechanisms triggered by B-cell activation, potentially contributing to B-cell lymphomagenesis. 23766104_Results suggest that haploinsufficiency of FOXP1 is associated with human congenital heart defects. 23810008_Data suggest that FOXP1 and FOXA1 (forkhead box A1) are primary estrogen receptors alpha (ERalpha) target genes and critical transcription factors that regulate ERalpha activity in breast cancer. [REVIEW] 24023716_data indicate that FoxP1 is essential for the angiogenic function of endothelial cells and functions as a suppressor of the inhibitory guidance cue semaphorin 5B, suggesting A function of FoxP1 in the regulation of neovascularization. 24211718_High Foxp1 expression is associated with hepatocarcinoma. 24214399_Autistic traits and other behavioral problems are likely to be associated with haploinsufficiency of FOXP1 24232982_Decreased miR-34a expression and increased FOXP1, p53, and BCL2 coexpression predict poor overall survival for MALT lymphoma and diffuse large B-cell lymphoma patients 24416450_Non-immunoglobulin heavy chain rearrangements of FOXP1 are usually acquired during clinical course of various lymphoma subtypes. 24787006_miR-150 can influence the relative expression of GAB1 and FOXP1 and the signaling potential of the B-cell receptor 24893616_our results may suggest a possible broader role of FOXP1 in the pathogenesis and progression of myelodysplasia and acute myeloid leukemia. 24908370_The significant expression of FOXP1 may be helpful to some extent in the pathologic diagnosis of cervical mucinous minimal deviation adenocarcinoma 25266127_This study demonstrated that Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behavior and mental disorders. 25267198_Through direct repression of proapoptotic genes, (aberrant) expression of FOXP1 complements (constitutive) NF-kappaB activity to promote B-cell survival and can thereby contribute to B-cell homeostasis and lymphomagenesis. 25329375_results suggest a novel mechanism in which AR-induced FOXP1 functions as a direct modulator of the AR and FOXA1 centric global transcriptional network. 25406647_Our results suggest that down-regulation of FOXP1 expression is a common event in high-risk neuroblastoma pathogenesis and may contribute to tumor progression and unfavorable patient outcome. 25500588_Data indicate that the mRNA level for forkhead box P1 (FOXP1) and estrogen receptor beta (ERbeta) in ovarian carcinoma tissues decreased, while the expression level of estrogen receptor alpha (ERalpha) mRNA increased. 25650440_FOXP1 has a role in potentiating Wnt/beta-catenin signaling in diffuse large B cell lymphoma 25778315_Abnormal expression of FOXP1 in renal cell carcinoma may create progression of tumor from low grade to high grade by regulating the HIF-1-VEGF pathway. 25868900_Foxp1 mediates programming of limb-innervating motor neurons from mouse and human embryonic stem cells. 25895457_FOXP1 expression is closely related to the degree of malignancy of epithelial ovarian cancer and may be a reliable index of the chemoresistance and prognosis of ovarian cancer 26054682_Over-expression of FOXP1 and SPINK1 may participate in the carcinogenesis of hepatitis B virus related cirrhosis. 26289642_FOXP1 overexpression specifically inhibits formation of IgG- but not IgM-secreting Plasma Cells. 26367773_FOXP1, TP53INP1, TNFAIP3, and TUSC2 were identified as miR-19a targets. 26383589_we provide supportive evidence that genetic variants at FOXP1, BARX1, and FOXF1 confer risk for the development of EAC. 26460480_FOXP1 - novel candidate genes validated in a large case-control sample of schizophrenia. 26489674_Studied the expression of FOXP1 in colorectal cancer and its potential associations with outcome in colorectal cancer. 26494785_integral role for FoxP1 in regulating signaling pathways vulnerable in autism and the specific regulation of striatal pathways important for vocal communication 26500140_FOXP1 represents a novel regulator of genes targeted by the class II MHC transactivator CIITA and CD74. 26647308_The findings highlight that de novo FOXP1 variants are a cause of sporadic intellectual disability and emphasize the importance of this transcription factor in neurodevelopment. 26654944_FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. 26729899_S1PR2 is repressed by FOXP1 in activated B-cell and germinal center B-cell DLBCL cell lines with aberrantly high FOXP1 levels; S1PR2 expression is further inversely correlated with FOXP1 expression in 3 DLBCL patient cohorts. 26787899_Cell-line derived FOXP1 target genes that were highly correlated with FOXP1 expression in primary DLBCL accurately segregated the corresponding clinical subtypes of a large cohort of primary DLBCL isolates 26854715_Authors identified forkhead box protein P1 (FOXP1) as a direct target of miR-504 using microarray analysis and a luciferase assay. 26898077_FOXP1 is present in normal cells of erythroid and myeloid linages and may have a possible role in development of all hematopoetic cells as well as possible involvement in neoplasm development of myeloid disorders. 27056922_the activity of multiple alternate FOXP1 promoters to produce multiple protein isoforms is likely to regulate B-cell maturation. 27167345_EBV-miR-BART11 plays a crucial role in the promotion of inflammation-induced nasopharyngeal carcinoma (NPC) and gastric cancer (GC) carcinogenesis by directly targeting and inhibiting FOXP1 tumor-suppressive effects. 27224915_Increased frequency of FOXP2 expression significantly correlated with FOXP1-positivity, and FOXP1 co-immunoprecipitated FOXP2 from activated B-cell-diffuse large B-cell lymphoma (ABC-DLBCL) cells. 27276253_Results suggest that domain swapping in FoxP1 is at least partially linked to monomer folding stability and follows an unusual three-state folding mechanism, which might proceed via transient structural changes rather than requiring complete protein unfolding as do most domain-swapping proteins 27457567_Prognostic value of decreased FOXP1 protein expression in various tumors, is reported. 27588474_High FOXP1 expression is associated with acute lymphoblastic leukemia. 27618020_FOXP1 knockdown significantly suppressed growth of HCC cells and induced G1/S phase arrest. 27806315_Data indicate that forkhead box P1 protein (FOXP1) as a target of microRNA miR-92a in primary mediastinal large B-cell lymphoma (PMBL). 27859969_FOXP1 has protein-protein interaction with NFAT1 on DNA and enhances breast cancer cell migration by repressing NFAT1 transcriptional activity. 27861791_these data identify FOXP1 as an essential transcriptional regulator for primary human CD4(+) T cells and suggest its potential important role in the development of PTCL. 27909217_these novel insights into the function of FOXP1 isoforms in controlling the transcriptional program, survival and differentiation of B cells advance our understanding of the role of FOXP1 in lymphomagenesis and further enhance the value of FOXP1 for diagnostics, prognostics, and treatment of DLBCL patients. 27924626_Blimp1, Foxp1 and pStat3 are expressed in extranodal diffuse large B-cell lymphomas 28232582_results reveal a novel regulatory pathway, underscoring a previously unknown and interconnected key role of PUM1/2 and FOXP1 in regulating normal hematopoietic stem/progenitor cell and leukemic cell growth. 28344757_Study identified USP15 as having recurrent de novo loss of function mutations and discovered evidence supporting two other known genes with recurrent de novo variants (FOXP1 and KDM5B). 28466777_Kaplan-Meier survival analysis showed that pancreatic ductal adenocarcinoma patients with negative forkhead box P1 and forkhead box O3a expression survived significantly shorter than patients with positive forkhead box P1 and forkhead box O3a expression (p = 0.000). 28550168_Although the mutant huntingtin gene is expressed widely, neurons of the striatum and cortex are selectively affected in Huntington's disease (HD). Our results suggest that this selectivity is attributable to the reduced expression of Foxp1, a protein expressed selectively in striatal and cortical neurons that plays a neuroprotective role in these cells. 28735298_FOXP1-related intellectual disability syndrome (ID) is a recognisable entity with wide clinical spectrum and frequent systemic involvement; more ID and neuromotor delay, sensorineural hearing loss and feeding difficulties are more common in patients with interstitial 3p deletions versus patients with monogenic FOXP1 defects; Mutations result in impaired transcriptional repression and/or reduced protein stability 28741757_we have identified a novel de novo missense variant in FOXP1 that is identical to the most well-studied etiological variant in FOXP2. Functional characterization revealed clear similarities between these equivalent mutations in terms of their impact on protein function. 28884888_Two rare novel FOXP1 variants associated with a phenotype similar to Mental Retardation with Language Impairment and with or without Autistic Features (MIM 613670). 28890397_The tumour suppressors FOXP1 and NKX3.1, strongly implicated in PCa development, were identified as key transcription factors regulating TPbeta expression through Prm3 in both PCa cell lines. 29090079_The majority of pathogenic missense and in-frame mutations lie in the DNA-binding domain. The mutations perturb amino acids necessary for binding to the DNA or interfere with the domain swapping that mediates FOXP1 dimerization. 29262329_FOXP1 expression is epigenetically regulated by PRMT5. 29365100_etiological mutations of FOXP1 and FOXP2, known to cause neurodevelopmental disorders, severely disrupted the interactions with FOXP-interacting transcription factors. 29666340_the varients of FOXP1 and FOXF1 genes are functionally associated with oesophageal adenocarcinoma in Chinese population. 29713984_Having a SNP in the FOXP1 gene in the absence of Reflux symptoms had an odds ratio of developing Barrett's esophagus of 1.5. 29739037_Upregulated circulating miR-139 level may be a potential biomarker for patients with metabolic disorders via suppressing the expression of forkhead box O1(FoxO1) and forkhead box P1 (FoxP1). 29772443_MiR-92a may act as a tumor inducer in OSCC by suppressing FOXP1 expression. 29934982_It was shown that LINC01614 promoted the expression of FOXP1 by inhibiting miR-217, which ultimately stimulated the development of lung adenocarcinoma. 29969624_The findings, in addition to expanding the molecular spectrum of FOXP1 mutations, emphasize the emerging role of WGS in identifying small balanced chromosomal rearrangements responsible for neurodevelopmental disorders and not detected by conventional cytogenetics. 30031607_The FOXP1 can suppressed the transcription activity of miR-181d-5p by acting as a transcription inhibitor. The miR-181d-5p-FOXP1 feedback loop modulates cell proliferation and metastasis in osteosarcoma. 30068241_miR-29b recedes the progression of multiple myeloma (MM) via downregulating FOXP1, which may provide a potential biological target for MM treatment. 30098431_Results show that FOXP1 acts as the functional protein of SNHG12. Its expression is regulated by SNHG12 and miR-101-3p in glioma cells. 30111844_High FOXP1 expression is associated with chronic b cell lymphocytic leukaemia. 30134017_In summary, STAT3 gene was a transcriptional regulator of FOXP1. Depleted STAT3 restrained cell proliferation and invasion, promoted cell apoptosis in glioma cells. This molecular mechanism between STAT3 and FOXP1 can serve as a therapeutic target for glioma treatment. 30181650_Co-occurrence of mutations in FOXP1 and PTCH1 in a girl with extreme megalencephaly, callosal dysgenesis and profound intellectual disability. 30213873_Low miR-150 levels in follicular lymphoma transformation lead to upregulation of its target, namely FOXP1 protein, which is a known positive regulator of cell survival, as well as B-cell receptor and NF-kappaB signaling in malignant B cells. 30274777_miR-374b-5p-FOXP1 feedback loop regulates tumor progression in ovarian cancer. 30385778_Here we present three patients with the same FOXP1 mutation, c.1574G>A (p.R525Q), that results in the characteristic loss of transcription repression activity. This mutation, however, represents the first reported FOXP1 mutation that does not result in cytoplasmic or nuclear aggregation of the protein but maintains normal nuclear localization. 30409062_Low FOXP1 expression is associated with the development of lung adenocarcinoma. 30579865_Data identify FOXP1 as an important negative regulator of immune responses in breast cancer (BC) via its regulation of cytokines expression. 30831269_identification of FOXP1 as a multi-promoter gene regulated by Mac-1 through a complex signaling network involving IRAK-1, HDAC4, CaMKIIdelta and a novel cloned FOXP1-IT1 long non-coding RNA, whose gene is embedded within FOXP1 itself. 30931977_The protonation state of an evolutionarily conserved histidine modulates domainswapping stability of FoxP1. 30935821_the current study not only establishes a novel regulatory axis of miR-3687/FOXP1 regarding regulation of cyclin E2 expression in bladder cancer (BC) cells, but also provides strong suggestive evidence that miR-3687 and FOXP1 may be promising targets in therapeutic strategies for human BC. 30937446_FOXP1 circular RNA has a role in sustaining mesenchymal stem cell identity via microRNA inhibition 31103631_miR-449b-5p interacted with FOXP1 to regulate cell proliferation, migration, invasion and radiosensitivity in CC 31235696_This study revealed that FOXP1-induced CLRN1-AS1 regulated cellular functions in pituitary prolactinoma by sponging miR-217 to release the DKK1/Wnt/beta-catenin signaling pathway. 31271729_Down-regulation of forkhead box protein P1 (FOXP1) increases prostanoid TP receptor beta (TPbeta) expression in THP-1 monocytes. 31494106_FOXP1 expression in the mesangial cells under high glucose (diabetic nephropathy) conditions.FOXP1 role in the mesangial cells proliferation, extracellular matrix and oxidative stress response. 31623628_These results demonstrated that circFOXP1 serve as a prognostic biomarker and critical regulator in GBC progression and Warburg effect, suggesting a potential target for GBC treatment. 31686194_FOXP-1 can represent a novel useful diagnostic marker in the differential diagnosis between pediatric-type follicular lymphoma and follicular hyperplasi 31696487_MicroRNA-92a regulates the development of cutaneous malignant melanoma by mediating FOXP1. 31698267_Circ-FOXP1 was significantly upregulated in HCC tissues, serum and cell lines., which was attributed to the upregulation of oncogenic transcription factor SOX9. 31699980_In pheochromocytoma patients,there was enrichment of FOXP1 expression in beige adipocytes in the vicinity of the vasculature within omental white adipose tissue. 31704154_FOXP1 plays an essential role in esophageal squamous cell carcinoma progression and prognosis and may be a useful biomarker for predicting survival. 31885310_miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. 31895100_Expression of FOXP1 and SOX10 by ameloblastomas differentiates them from basaloid salivary gland neoplasms. 32049024_Foxp1 Regulates Neural Stem Cell Self-Renewal and Bias Toward Deep Layer Cortical Fates. 32150333_Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. 32530809_Epstein-Barr virus-encoded miR-BART11 promotes tumor-associated macrophage-induced epithelial-mesenchymal transition via targeting FOXP1 in gastric cancer. 32633368_Circle RNA FOXP1 promotes cell proliferation in lung cancer by regulating miR-185-5p/Wnt1 signaling pathway. 32648149_Targeting the forkhead box protein P1 pathway as a novel therapeutic approach for cardiovascular diseases. 32735805_Intrinsically Disordered Regions of the DNA-Binding Domain of Human FoxP1 Facilitate Domain Swapping. 32808501_Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. 32891193_Mutations of the Transcriptional Corepressor ZMYM2 Cause Syndromic Urinary Tract Malformations. 32951006_A novel epigenetic regulation of circFoxp1 on Foxp1 in colon cancer cells. 32996692_CircFOXP1/FOXP1 promotes osteogenic differentiation in adipose-derived mesenchymal stem cells and bone regeneration in osteoporosis via miR-33a-5p. 33139493_Elevated MicroRNA 183 Impairs Trophoblast Migration and Invasiveness by Downregulating FOXP1 Expression and Elevating GNG7 Expression during Preeclampsia. 33372387_LINC01116 promotes proliferation and migration of endometrial stromal cells by targeting FOXP1 via sponging miR-9-5p in endometriosis. 33427368_Novel FOXP1 pathogenic variants in two Indian subjects with syndromic intellectual disability. 33716296_FOXP1 drives osteosarcoma development by repressing P21 and RB transcription downstream of P53. 33830472_CircRNA_OTUD7A upregulates FOXP1 expression to facilitate the progression of diffuse large B-cell lymphoma via acting as a sponge of miR-431-5p. 33887587_The circACC1/miR-29c-3p/FOXP1 network plays a key role in gastric cancer by regulating cell proliferation. 34298089_FOXP1 and NDRG1 act differentially as downstream effectors of RAD9-mediated prostate cancer cell functions. 34514002_Circular RNA FOXP1 Induced by ZNF263 Upregulates U2AF2 Expression to Accelerate Renal Cell Carcinoma Tumorigenesis and Warburg Effect through Sponging miR-423-5p. 34783112_Impact of FOXP1 rs2687201 genetic variant on the susceptibility to HCV-related hepatocellular carcinoma in Egyptians. 34839505_[Clinical features and genetic analysis of three children with mental retardation, language impairment and autistic features due to de novo variants of FOXP1 gene]. 35098439_LncRNA PVT1 facilitates DLBCL development via miR-34b-5p/Foxp1 pathway. 35142784_The Role of Survivin and Transcription Factor FOXP1 in Scarring After Glaucoma Surgery. | ENSMUSG00000030067 | Foxp1 | 6.565021e+02 | 1.2856655 | 0.362515314 | 0.2681574 | 1.863936e+00 | 0.1721711663 | 0.77611497 | No | Yes | 6.976047e+02 | 79.266074 | 5.060685e+02 | 59.274887 | |
| ENSG00000114942 | 1933 | EEF1B2 | protein_coding | P24534 | FUNCTION: EF-1-beta and EF-1-delta stimulate the exchange of GDP bound to EF-1-alpha to GTP. | 3D-structure;Acetylation;Direct protein sequencing;Elongation factor;Isopeptide bond;Phosphoprotein;Protein biosynthesis;Reference proteome;Ubl conjugation | This gene encodes a translation elongation factor. The protein is a guanine nucleotide exchange factor involved in the transfer of aminoacylated tRNAs to the ribosome. Alternative splicing results in three transcript variants which differ only in the 5' UTR. [provided by RefSeq, Jul 2008]. | hsa:1933; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; eukaryotic translation elongation factor 1 complex [GO:0005853]; guanyl-nucleotide exchange factor activity [GO:0005085]; translation elongation factor activity [GO:0003746]; translational elongation [GO:0006414] | 14623968_TCTP preferentially stabilized the GDP form of eEF1A, and, furthermore, impaired the GDP exchange reaction promoted by eEF1Bbeta. 15341733_Results indicate an evolutionary lineage of translation initiation factor eIF2alpha/gamma from the functionally related elongation factor eEF1Balpha/eEF1A complex. 19487283_Cathepsin D and eEF1 are promising markers for the detection of cellular senescence induced by a variety of treatments. 25436608_Both eEF1A1 and eEF1A2 colocalise with all eEF1B subunits, in such close proximity that they are highly likely to be in a complex. 29572982_It has been postulated that the N-terminus region of EF1beta may be responsible for its dimerization and the C-terminus region of this protein modulates the formation of an ordered EF1beta-gamma oligomer, a structure that may be essential in the elongation step of eukaryotic protein biosynthesis. 30572058_high eEF1Balpha expression is associated with poor overall survival and may serve as an independent prognostic factor of gastric cancer. 30590147_the protein-binding domain of eEF1Bbeta shows flexible spatial organization which may be needed for interaction with eEF1Bgamma or other protein partners. 31845318_New evidence that biallelic loss of function in EEF1B2 gene leads to intellectual disability. 34420305_Proteogenomics Integrating Novel Junction Peptide Identification Strategy Discovers Three Novel Protein Isoforms of Human NHSL1 and EEF1B2. | ENSMUSG00000025967 | Eef1b2 | 8.316477e+03 | 0.5935207 | -0.752629743 | 0.3085834 | 6.032110e+00 | 0.0140479315 | 0.42783848 | No | Yes | 5.929783e+03 | 1158.957925 | 9.808337e+03 | 1965.493246 | |
| ENSG00000115392 | 55120 | FANCL | protein_coding | Q9NW38 | FUNCTION: Ubiquitin ligase protein that mediates monoubiquitination of FANCD2 in the presence of UBE2T, a key step in the DNA damage pathway (PubMed:12973351, PubMed:16916645, PubMed:17938197, PubMed:19111657, PubMed:24389026). Also mediates monoubiquitination of FANCI (PubMed:19589784). May stimulate the ubiquitin release from UBE2W. May be required for proper primordial germ cell proliferation in the embryonic stage, whereas it is probably not needed for spermatogonial proliferation after birth. {ECO:0000269|PubMed:12973351, ECO:0000269|PubMed:16916645, ECO:0000269|PubMed:17938197, ECO:0000269|PubMed:19111657, ECO:0000269|PubMed:19589784, ECO:0000269|PubMed:24389026}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;DNA damage;DNA repair;Fanconi anemia;Metal-binding;Nucleus;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a ubiquitin ligase that is a member of the Fanconi anemia complementation group (FANC). Members of this group are related by their assembly into a common nuclear protein complex rather than by sequence similarity. This gene encodes the protein for complementation group L that mediates monoubiquitination of FANCD2 as well as FANCI. Fanconi anemia is a genetically heterogeneous recessive disorder characterized by cytogenetic instability, hypersensitivity to DNA crosslinking agents, increased chromosomal breakage, and defective DNA repair. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2018]. | hsa:55120; | cytoplasm [GO:0005737]; Fanconi anaemia nuclear complex [GO:0043240]; intracellular membrane-bounded organelle [GO:0043231]; nuclear body [GO:0016604]; nuclear envelope [GO:0005635]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; gamete generation [GO:0007276]; interstrand cross-link repair [GO:0036297]; protein monoubiquitination [GO:0006513]; regulation of cell population proliferation [GO:0042127] | 12417526_Deficiency of Fancl (also called Pog) is the cause of gcd mouse, which has a reduced number of primordial germ cells during the embryonic stage. 12606378_FANCL is necessary for primordial germ cell proliferation during the embryonic stage but not necessary for spermatogonia proliferation in adulthood. Thus, mouse FancL-/- males are infertile at 7 to 12 weeks but gain fertility thereafter. 12973351_data suggest that PHF9 has a crucial role in the Fanconi anemia pathway as the likely catalytic subunit required for monoubiquitination of FANCD2 16474167_FANCL, via its WD40 region, binds the FA complex and, via its PHD, recruits an as-yet-unidentified E2 for mono-ubiquitination of FANCD2 17106252_Abnormal FANCL expression is the cause leading to a defective Ranconi anemia-BRCA pathway, conferring sensitivity of a lung cancer cell line to mitomycin C> 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18607065_the first report to describe hypermethylation of FANCC in leukemia 19589784_Upon the occurrence of DNA damage, FANCI becomes monoubiquitinated on Lys-523 by the UBE2T-FANCL pair. 19690177_Observational study of gene-disease association. (HuGE Navigator) 19737859_Observational study of gene-disease association. (HuGE Navigator) 19737859_results rule out a major role of FANCL in familial breast cancer susceptibility 20407210_expression of a novel splice variant of FA complementation group L (FANCL), named FAVL, can impair the FA pathway in non-FA tumor cells and act as a tumor promoting factor 21297076_FANCL is associated with acute lung injury in mice 21543111_genetic diversity in FANCA, FANCC and FANCL does not support an association of these genes with cervical cancer susceptibility in the Swedish population. 21697891_FA DNA repair genes, FANCD2, FANCL, and FANCC, are transcriptionally upregulated differently in melanoma compared with non-melanoma skin cancer 22653977_Suppression of FANCL expression in normal CD34(+) stem and progenitor cells results in fewer beta-catenin active cells and inhibits expansion of multilineage progenitors. 22828653_FAVL elevation can increase the tumorigenic potential of bladder cancer cells, including the invasive potential that confers the development of advanced bladder cancer. 23783032_a signal transduction pathway involved in self-renewal and survival of hematopoietic stem cells also functions to stabilize FANCL and suggesting that FANCL participates directly in support of stem cell function. 25754594_Loss-of-Function FANCL Mutations Associate with Severe Fanconi Anemia Overlapping the VACTERL Association 26385482_Using small interfering RNA (siRNA), knockdown of FANCF, FANCL, or FANCD2 inhibited function of the FA/BRCA pathway in A549, A549/DDP and SK-MES-1 cells, and potentiated sensitivity of the three cells to cisplatin. 28419882_A novel homozygous mutation c.822_823insCTTTCAGG (p.Asp275LeufsX13) in the FANCL gene identified in a Chinese patient with Fanconi anemia. 31513304_A founder variant in the South Asian population leads to a high prevalence of FANCL Fanconi anemia cases in India. 32048394_FANCL gene mutations in premature ovarian insufficiency. 32420600_Characterization of FANCL variants observed in patient cancer cells. 33394227_Severe telomere shortening in Fanconi anemia complementation group L. | ENSMUSG00000004018 | Fancl | 1.090043e+03 | 0.8772538 | -0.188933854 | 0.3275943 | 3.238552e-01 | 0.5692999832 | 0.89878854 | No | Yes | 7.162890e+02 | 122.468086 | 7.944441e+02 | 138.935453 |
| ENSG00000115414 | 2335 | FN1 | protein_coding | P02751 | FUNCTION: Fibronectins bind cell surfaces and various compounds including collagen, fibrin, heparin, DNA, and actin (PubMed:3024962, PubMed:3900070, PubMed:3593230, PubMed:7989369). Fibronectins are involved in cell adhesion, cell motility, opsonization, wound healing, and maintenance of cell shape (PubMed:3024962, PubMed:3900070, PubMed:3593230, PubMed:7989369). Involved in osteoblast compaction through the fibronectin fibrillogenesis cell-mediated matrix assembly process, essential for osteoblast mineralization (By similarity). Participates in the regulation of type I collagen deposition by osteoblasts (By similarity). Acts as a ligand for the LILRB4 receptor, inhibiting FCGR1A/CD64-mediated monocyte activation (PubMed:34089617). {ECO:0000250|UniProtKB:P11276, ECO:0000269|PubMed:3024962, ECO:0000269|PubMed:34089617, ECO:0000269|PubMed:3593230, ECO:0000269|PubMed:3900070, ECO:0000269|PubMed:7989369}.; FUNCTION: [Anastellin]: Binds fibronectin and induces fibril formation. This fibronectin polymer, named superfibronectin, exhibits enhanced adhesive properties. Both anastellin and superfibronectin inhibit tumor growth, angiogenesis and metastasis. Anastellin activates p38 MAPK and inhibits lysophospholipid signaling. {ECO:0000269|PubMed:11209058, ECO:0000269|PubMed:15665290, ECO:0000269|PubMed:19379667, ECO:0000269|PubMed:8114919}. | 3D-structure;Acute phase;Alternative splicing;Angiogenesis;Cell adhesion;Cell shape;Direct protein sequencing;Disease variant;Disulfide bond;Dwarfism;Extracellular matrix;Glycoprotein;Heparin-binding;Isopeptide bond;Oxidation;Phosphoprotein;Pyrrolidone carboxylic acid;Reference proteome;Repeat;Secreted;Signal;Sulfation | This gene encodes fibronectin, a glycoprotein present in a soluble dimeric form in plasma, and in a dimeric or multimeric form at the cell surface and in extracellular matrix. The encoded preproprotein is proteolytically processed to generate the mature protein. Fibronectin is involved in cell adhesion and migration processes including embryogenesis, wound healing, blood coagulation, host defense, and metastasis. The gene has three regions subject to alternative splicing, with the potential to produce 20 different transcript variants, at least one of which encodes an isoform that undergoes proteolytic processing. The full-length nature of some variants has not been determined. [provided by RefSeq, Jan 2016]. | hsa:2335; | apical plasma membrane [GO:0016324]; basement membrane [GO:0005604]; blood microparticle [GO:0072562]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; extracellular exosome [GO:0070062]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; fibrinogen complex [GO:0005577]; plasma membrane [GO:0005886]; platelet alpha granule lumen [GO:0031093]; chaperone binding [GO:0051087]; collagen binding [GO:0005518]; disordered domain specific binding [GO:0097718]; enzyme binding [GO:0019899]; extracellular matrix structural constituent [GO:0005201]; heparin binding [GO:0008201]; identical protein binding [GO:0042802]; integrin binding [GO:0005178]; peptidase activator activity [GO:0016504]; protease binding [GO:0002020]; protein C-terminus binding [GO:0008022]; proteoglycan binding [GO:0043394]; signaling receptor binding [GO:0005102]; acute-phase response [GO:0006953]; angiogenesis [GO:0001525]; biological process involved in interaction with symbiont [GO:0051702]; blood coagulation, fibrin clot formation [GO:0072378]; calcium-independent cell-matrix adhesion [GO:0007161]; cell adhesion [GO:0007155]; cell-matrix adhesion [GO:0007160]; cell-substrate junction assembly [GO:0007044]; endodermal cell differentiation [GO:0035987]; heart development [GO:0007507]; integrin activation [GO:0033622]; integrin-mediated signaling pathway [GO:0007229]; negative regulation of monocyte activation [GO:0150102]; negative regulation of transforming growth factor beta production [GO:0071635]; nervous system development [GO:0007399]; neural crest cell migration involved in autonomic nervous system development [GO:1901166]; peptide cross-linking [GO:0018149]; platelet aggregation [GO:0070527]; positive regulation of axon extension [GO:0045773]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of gene expression [GO:0010628]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of substrate-dependent cell migration, cell attachment to substrate [GO:1904237]; regulation of cell shape [GO:0008360]; regulation of ERK1 and ERK2 cascade [GO:0070372]; regulation of protein phosphorylation [GO:0001932]; response to wounding [GO:0009611]; substrate adhesion-dependent cell spreading [GO:0034446] | 11500098_Binding of fibronectin by Trichomonas vaginalis is influenced by iron and calcium 11605051_Observational study of gene-disease association. (HuGE Navigator) 11751853_We show a novel alternative pathway of apoptosis that is regulated by the alternatively spliced V region and high-affinity heparin-binding domain of fibronectin 11768240_Potential mechanism of fibronectin deposits in acute renal failure induced by mercuric chloride 11832485_Alternative splicing of the IIICS domain in fibronectin governs the role of the heparin II domain in fibrillogenesis and cell spreading. 11864705_Increased plasma fibronectin levels in patients with acute myocardial infarction complicated with left ventricular thrombus; multivariate analysis showed that plasma fibronectin levels were not an independent predictor of LV thrombus formation. 11867643_Low density lipoprotein receptor-related protein (LRP) functions as a catabolic receptor for Fn 11891225_Laminin-10/11 and fibronectin differentially prevent apoptosis induced by serum removal via phosphatidylinositol 3-kinase/Akt- and MEK1/ERK-dependent pathways (Laminin 10; separate entry for Laminin 11). 11928812_differential role of u-PA and t-PA as inducers of fibronectin mRNA 11937267_investigation of the role of direct contact with stroma and the ECM protein fibronectin in the preservation of normal but not chronic myelogenous leukemia primitive hematopoietic progenitors 12091360_Adhesion of hairy cells to fibronectin inhibits interferon-alpha-induced apoptosis. 12105189_fragment from fibronectin's III1 module localizes to lipid rafts and stimulates cell growth and contractility 12225805_Human fibronectin and MMP-2 collagen binding domains compete for collagen binding sites and modify cellular activation of MMP-2. 12270980_proximal tubular cell fibronectin production induced by oleate-complexed recombinant human serum albumin was mediated via protein kinase C activation 12388756_These data indicate that fibronectin polymerization is a critical regulator of extracellular matrix organization and stability. 12401883_familial combined hyperlipidemia plasma stimulated secretion of several distinct hepatic proteins, among which fibronectin was identified 12468382_FN may be partly involved in the increase in SPARC (secreted protein, acidic and rich in cysteine) glycoprotein expression by TGF-beta 1 in human periodontal ligament (HPL) cells. 12486316_REVIEW: Substance p-fibronectin-cytokine interactions in myeloproliferative disorders with bone marrow fibrosis. 12497612_Observational study of gene-disease association. (HuGE Navigator) 12535082_Streptococcus pyogenes isolates were able to recruit collagen type IV via surface-bound fibronectin; fibronectin-mediated collagen recruitment represents a novel aggregation, colonization and immune evasion mechanism of S. pyogenes 12538453_FN may have a role in the pathogenesis of papillary thyroid carcinoma 12538576_all six modules of the gelatin-binding domain contribute to the interaction with gelatin either directly by contacting the ligand or indirectly through module-module interactions 12621118_differences in fibronectin gene expression between the SWS port-wine-derived fibroblasts and the Sturge-Weber syndrome (sws) normal skin-derived fibroblasts are consistent with the presence of a hypothesized somatic mutation underlying SWS. 12631068_Fibronectin secretion by renal fibroblasts was increased upon exposure to high glucose, but with delayed kinetics compared to TGF-beta1-induced fibronectin. 12736686_Pathogenic bacteria, Staphylococcus aureus and Streptococcus pyogenes, attach to human fibronectin through a tandem beta-zipper 12761244_AngRem104 is a novel human gene potentially involved in regulation of fibronectin induced by AngII in human mesangial cells. 12802498_An increased concentration of wild-type GFAT in mesangial cells is enhanced both TGF-beta1 and fibronectin. 12810082_Results suggest that tissue or plasma fibronectin may modulate the intestinal epithelial response to repetitive deformation through inhibted activation of p38 and jun kinases. 14558920_an additional marker of malignant potential of thyroid nodular lesions (oncofetal fibronectin) 14602715_levels of fibronectin in the extracellular matrix are regulated by uPAR and integrin receptors 14645245_FN and IGFBP-5 bind to each other, and this binding negatively regulates the ligand-dependent action of IGFBP-5 by triggering IGFBP-5 proteolysis. 14662883_Fibronectin plus TNF-alpha stabilizes granulocyte macrophage-colony-stimulating factor (GM-CSF) mRNA, increases GM-CSF secretion, and prolongs in vitro eosinophil survival. 14737076_Abundant expression of fibronectin is a major feature of leukemic dendritic cells differentiated from patients with acute myeloid leukemia 15161923_the aggrecanase activity of ADAMTS4 is inhibited by fibronectin through interaction with their C-terminal domains 15213410_crystal structure of the 2F1(3)F1 module pair 15247227_human fibronectin binding to streptococcus requires specific recognition of sequential F1 modules 15265957_Adding 110-120 kDa fibronectin fragments to normal blood replicated changes in monocyte CD49e, CD62L, CD11b, & CD86 seen in vivo in HIV-1 patients. FNf caused monocytes to release proteinase-3. Monocyte behavior in HIV-1 patients may be affected by FNf. 15331608_Data show that squamous cell carcinoma cells escape suspension-induced, p53-mediated anoikis by forming multicellular aggregates that use fibronectin survival signals mediated by integrin alpha(v) and focal adhesion kinase. 15384859_fibronectin concentration significantly elevated in ovarian cancer patients with recurrent disease compared with ovarian cancer patients without recurrence 15456495_demonstrate that integrins alpha(5)beta(1) and alpha(v)beta(3) support fibronectin-dependent platelet adhesion and the generation of filopodia but that, in contrast to the integrin alpha(IIb)beta(3), are unable to promote formation of lamellipodia 15456743_Site-directed mutagenesis of amino acids coordinating Ca2+ drastically decreased the binding of C-reactive protein to fibronectin (Fn), indicating that the Ca2+ -binding site indeed formed the Fn-binding site. 15467744_Fibronectin and type IV collagen activate ERalpha AF-1 by the c-Src pathway in breast cancer 15485890_integrin-dependent function of the ninth and tenth FIII domains of human fibronectin depends on interdomain tilt angle 15645125_adhesion of uveal melanoma cells to the extracellular matrix proteins fibronectin, type IV collagen and laminin might confer resistance to the chemotherapeutic agent cisplatin 15652337_The heparin II domain of fibronectin mediated the incorporation of paxillin into focal adhesions. 15699160_Fibronectin enhances IL-1 beta-dependent airway smooth muscle cell secretory responses through a beta 1 integrin-dependent mechanism. 15721303_SGK1 activation has a role in oncofetal fibronectin expression 15835821_Fibronectin and ezrin were upregulated in primary laryngeal carcinoma. 15855153_Fibronectin 1 and RAB10 show elevated expression in uninvolved psoriatic epidermis. 15890670_FN modulates thyroid cancer cell adhesiveness and, at least in part, mediates vitamin D actions on neoplastic cell growth. 15912204_The relative amounts of the 60, 90 and 100 kDa FN fragments were 2-3 times higher in seminal plasmas with abnormal semen characteristics than in the normozoospermic group, suggesting that seminal plasma FN fragments may contribute to fertilization. 15961545_FN has a role in controlling translation through beta1 integrin and eukaryotic initiation factors 4 and 2 coordinated pathways 16083879_Chemical shift differences between the fibronectin glycoforms have revealed an intimate interaction between one N-linked sugar and the polypeptide that is critical for gelatin binding. 16091757_Over-expression of oncofetal fibronectin is associated with thyroid carcinoma 16150826_binding with C terminus of apolipoprotein [a] 16159961_These results provide evidence for an additional level of control of fibronectin deposition through cell interactions with the local microenvironment. 16277979_the role of the extra domain A (EDA) and type III connecting segment (IIICS) of fibronectin in fiber assembly, topographical distribution and proteolytic cleavage 16336961_Results show fibronectin plays a role in the regulation of hematopoiesis by binding to FLRG and follistatin. 16375583_Observational study of gene-disease association. (HuGE Navigator) 16378831_platelet fibronectin may have a role in progression of coronary artery disease 16463680_The expression of FN protein was higher in cortex cataract than in nuclear cataract. 16495290_Elevated glucose concentrations increased TSP-1 synthesis, which was associated with reduced cell proliferation, increased TGF-beta1 bioactivity, and stimulation of FN synthesis 16621928_Results indicate that the adhesion mechanism by embryonal fibronectin may be involved in human blastocyst implantation. 16757476_constitutive FN degradation, as well as UV-induced degradation, is ubiquitination dependent and controlled by beta-TrCP 16839338_review goes over roles of fibronectin in platelet functions with a focus on fibronectin assembly within developing platelet thrombi. 16928957_Flow induced alpha2beta1 activation in cells on collagen, but not on fibronectin or fibrinogen. Conversely, alpha5beta1 and alphavbeta3 are activated on fibronectin and fibrinogen, but not collagen. 16978691_IL-1, FN, and FN-derivatives have interrelated roles in determining biomaterial-modulated macrophage function [review] 16982604_The role of the matricryptic site of the fibronectin terminal repeat III1 in cell proliferation and growth is reported. 16984808_Review highlights the role of fibronectin as a key orchestrator for the assembly of multiple extracellular matrix proteins. 16986166_Data suggest that natriuretic peptide receptor A (NPR-A) interacts with fibronectin to enhance brain natriuretic peptide (BNP) activation of cyclic GMP and that a small NPR-A-specific RGD peptide augments this action of BNP. 17003032_the FN1 alpha-helix is involved in an Ig5-FN1 interaction that is critical for the correct positioning of Ig5 N-glycans for polysialylation 17015452_analysis of L-isoaspartate formation and gain of function in fibronectin 17027088_These findings suggest that interaction with fibronectin through integrin alpha5 plays an important role in human extravillous trophoblast invasion. 17136547_Downregulated expression of migration-inducing FN-isoforms in contrast to unchanged FN receptor expression may contribute to the observed alterations of Crohn's disease colonic lamina propria fibroblasts migration. 17187346_Our results indicate that Src maintains fibronectin matrix at the cell surface through its effect on integrin activity and paxillin phosphorylation. 17266699_fibronectin does not have an obvious role in development or progression of coronary artery disease 17273763_epithelial fibronectin suppression is not only involved with malignancy, but can also occur with benign processes that involve leukocyte migration 17416372_local presence of endogenous Tlr4 ligand Fibronectin EDA is not associated with in an unstable plaque phenotype in humans 17425334_Interaction of fibronectin carboxy-terminal regions with the fibrin alpha C chain occurs through two types of binding sites, one with high affinity and another one with much lower affinity, both of which play a role during hemostasis and wound healing. 17459242_Smad 7 transfection reduces the expression of COL1A2 mRNA and the synthesis of type I collagen, but has no effect on fibronectin mRNA expression. 17468934_ED-B fibronectin expression is closely associated with inflammation in atherosclerosis. 17490871_The IIICS region of FN is a novel zinc-binding module. 17512904_Twist regulates cell motility and invasion in gastric cancer cell lines, probably through the N-cadherin and fibronectin production 17526550_MspI and HaeIIIb FN gene polymorphisms may be clinically relevant to define the risk of lymphoma development in MCsn. 17526550_Observational study of gene-disease association. (HuGE Navigator) 17554369_TNF-alpha suppresses TGF-beta1-induced myofibroblast (fibroproliferative) phenotypic genes, for example, alpha-SMA, collagen type 1A, and fibronectin at the mRNA level. 17596138_Fibrinogen has a role in controlling human platelet fibronectin internalization and cell-surface retention 17636193_study concludes that increased platelet fibronectin may partially contribute to platelet aggregation in patients with coronary artery disease 17704536_Expression of onfFN mRNA characterizes not only papillary thyroid carcinoma but also ATC. onfFN mRNA or protein may be a useful marker to identify anaplastic carcinoma cells and may be considered as an optimistic target in molecular-based therapy of ATC. 17879163_These findings provide evidence that mutations in fibronectin, induce anoikis in human squamous cell carcinoma cells by modulating integrin alpha v-mediated phosphorylation of FAK and ERK. 17929131_high molecular weight hyaluronan suppresses HBFN-f-activated NF-kappaB in rheumatoid arthritis chondrocytes 17949711_Results show that neutrophil gelatinase-associated lipocalin (NGAL or lipocalin-2)is co-expressed with migration stimulating factor (MSF or FN1) by keratinocytes and acts as a functional inhibitor of MSF. 18006840_expression of the alternatively spliced extra-domain A (EDA) of fibronectin in the neovasculature of metastases and primary tumors of human cancer patients was shown 18042364_TSG-6 is a high affinity ligand that can mediate fibronectin interactions with other matrix components and modulate some interactions of fibronectin with cells 18064631_These data provide the first evidence that IL-18 and FN stimulate each other's expression. 18083324_These results suggest that fibrinogen and fibronectin facilitate the adherence of Paracoccidioides brasiliensis conidia to A549 cells probably through the interaction with adhesin-type molecules or a sialic acid based recognition system. 18268355_dominant mutations in FN1 accounted for 40% of cases of Glomerulopathy with fibronectin (FN) deposits in our study group 18313373_These results are consistent with the hypothesis that ScpB binds to a motif created by the juxtaposition of multiple Fn molecules. 18332212_The TP0136 protein was exposed on the surface of the bacterial outer membrane and bound to the host extracellular matrix glycoproteins fibronectin and laminin. 18334936_Filtering bleb scarring is associated with an abundant expression of TGF-beta receptors in activated fibroblasts and the deposition of the fibrogenic ED-A fibronectin splice-variant. 18340378_MMP-2 enhanced peritoneal adhesion of ovarian cancer cells through cleavage of ECM proteins fibronectin (FN) and vitronectin (Vn) into small fragments 18343220_KLK7 is able to cleave fibronectin in a time-dependent manner, but not laminin. 18348696_Increased production of a fibronectin isoform containing the oncofetal domain and its release in the circulation in patients with primary biliary cirrhosis is at least partially responsible for the decrease in bone formation. 18451110_Results advance a new model wherein Fn polymerization serves as a structural scaffolding that displays adhesive ligands on a mechanically ideal substratum for promoting neovessel development. 18451144_A link between fibronectin expression and epithelial cell growth during development and oncogenesis in the mammary gland. 18474427_Meningiomas increase in size through increased production of extracellular matrix; furthermore, the proliferation of cells typically associated with neoplasia requires considerable interaction with the extracellular matrix. 18499669_TG-FN binding to syndecan-4 activates PKCalpha leading to its association with beta(1) integrin, reinforcement of actin-stress fiber organization, and MAPK pathway activation 18577581_Caveolin-1-dependent beta1 integrin and fibronectin endocytosis plays a critical role in fibronectin matrix turnover. 18633626_Inflammation-induced modulation of FN-isoform production is involved in the alterations of migratory potential of CLPF isolated from CD mucosa. 18690341_elevated plasma fibronectin levels are associated with venous thromboembolism (VTE) especially in males, and extend the potential association between biomarkers and risk factors for arterial atherothrombosis and VTE 18713862_four crystal structures that together comprise the structures of two complete high-affinity binding repeats for Fn from FnBPA and FnBPB, each in complex with four of the N-terminal modules of Fn 18716775_Plasma fibronectin level is increased in patients with severe coronary artery disease, namely, those who have at least one critically stenosed coronary vessel, compared to patients with normal coronary arteries. 18776591_analysis of fibronectin expression in benign hyperplasia and cancer of the prostate 18801363_These data suggest that SOCS-1 inhibits high glucose-induced overexpression of TGF-beta1 and synthesis of fibronectin in human mesangial cells, which may be via JAK/STAT pathway. 18810851_Genotyping for HaeIIIb fibronectin gene polymorphisms may be clinically relevant to define the predisposition to the major clinical manifestations in mixed cryoglobulinemic syndrome. 18810851_Observational study of gene-disease association. (HuGE Navigator) 18824166_a new mechanism of fibroblast activation, which is initiated by interaction of fibronectin with its integrin receptors. 18829569_MAGE A3 is a functional integrator of diverse signals, including FGFR2 and FN, to modulate cancer progression. 18957516_Caveolin-1 plays a dual role in the fibronectin assembly regulated by uPAR signaling. 18958156_The simultaneous positivity for thyroid specific factor TTF-1 and onfFN suggest they might represent putative thyroid cancer stem-like cells. 19020673_analysis of the unfolding of the tenth type-III repeat of human fibronectin 19031824_Dynamic mechanical forces could regulate the expression of collagen type I and fibronectin mRNA in periodontal ligament fibroblasts. 19037100_Mapping studies revealed that the major binding interaction between fibrillins and fibronectin involves the collagen/gelatin-binding region between domains FNI(6) and FNI(9). 19094228_Observational study of gene-disease association. (HuGE Navigator) 19160003_enhanced capacity of fibronectin-adherent HL60 cells to differentiate into macrophages in response to PMA. 19161998_The 320-kDa and 280-kDa FN forms as well as FN fragments appeared in the pleural effusion and plasma of patients suffering from lung inflammation or cancer in significantly higher relative amounts in both lung cancer groups than in the inflammation. 19212764_relative amount of EDA-FN in synovial fluid, but not in plasma samples, was significantly lower in the early RA group than in established and late RA. In contrast, its level did not correlate with the CRP concentration 19224684_Observational study of gene-disease association. (HuGE Navigator) 19224684_The risk of pneumoconiosis increases in workers with the FN (MspICC or HaeIIIb AA) genotype after exposure to dust. Workers both carrying FN (HaeIIIb AA) and (MspICC) genotypes are more susceptible to pneumoconiosis. 19251642_role for fibronectin in collagen proteolysis and tissue remodeling 19342448_Regulation of estrogen-mediated fibronectin matrix assembly and epidermal growth factor receptor transactivation by GPR30. 19345424_These results established a novel in vitro dynamic BBB model. We also demonstrated the dependence of leukocyte-endothelial interactions in this model on alpha4 integrins and FN-CS1. 19391127_Human umbilical vein endothelial cells showed dose-dependent upregulation of fibronectin mRNA exposed to various D-glucose levels. 19404402_Borrelia burgdorferi virulence factor BBK32 induces the formation of Fn aggregates that are indistinguishable from those formed by anastellin. 19417080_isoforms of a group B streptococcus-secreted component named Fib displayed differential binding capacities for fibronectin, fibrinogen, and C4BP 19460753_Erythroid differentiation stimulated by hemin was greatly enhanced when K562 cells were induced to adhere to Fibronectin by activating VLA-5 with TNIIIA2, a peptide derived from tenascin-C. 19541353_fibronectin might be a critical molecule produced by T(H)1 cells involved in pathogen eradication. 19553700_the angioinhibitory action of NK4 involves impaired extracellular assembly of fibronectin mediated by perlecan-NK4 association 19564406_Results describe the measurement of the force-dependent lifetimes of single bonds between a fibronectin fragment and an integrin alpha(5)beta(1)-Fc fusion protein or membrane alpha(5)beta(1). 19581300_LigB was shown to bind to the N-terminal domain of fibronectin. 19616291_HindIII polymorphism of the FN1 gene is highly associated with calcium oxalate stone disease 19616291_Observational study of gene-disease association. (HuGE Navigator) 19617627_FN binds BMP1-like proteinases in vivo and that FN is an important determinant of the in vivo activity levels of BMP1-like proteinases. 19701604_Serum fibronectin concentration in patients with hemorrhagic fever with renal syndrome (HFRS) was increased with disease phases and severity, suggesting the value of detection of fibronectin levels for evaluating HFRS disease progression and severity. 19701759_higher expression in the fetal skin than in adult skin 19730683_Observational study of gene-disease association. (HuGE Navigator) 19760097_AR could mediate the TGF-beta1-induced FN production, which may associate with AP-1 activation. 19821053_Exposure to high glucose and overexpression AR increase the expression of fibronectin. 19834535_Observational study of gene-disease association. (HuGE Navigator) 19847669_AR is a potent regulator of TGF-beta1 induced expression of FN in human mesangial cells. 19880379_the third FN type III repeat (FnIII3) and several fragments containing the repeat promote cell spreading and migration of human dermal fibroblasts (HDFs), whereas the fourth repeat (FnIII4) did not. 19893244_-beta1 shRNAs could obviously inhibit the expression levels of TGF-beta1, CTGF, FN and cellular proliferation stimulated by HSA in HK2 cells. 19893454_Data show that the expression of collagen types I, III and fibronectin was significantly higher in pancreatic cancer, and the expression of collagen type IV, laminin and vitronectin was significantly lower in pancreatic cancer. 19903114_vaginal fetal fibronectin not changed by vaginal ultrasound 19913121_Observational study of gene-disease association. (HuGE Navigator) 19914350_Upregulation of fibronectin-1 protects Dicer knockdown HEK293T cells against apoptosis. 19935699_findings consistently indicate that PPAR-beta may facilitate differentiation and inhibit the cell-fibronectin adhesion of colon cancer. 20007695_Data show that potentiated and sustained adhesion to fibronectin via VLA-4 causally induces apoptosis also in various types of hematopoietic tumor cells. 20012564_Increase in urinary fibronectin is associated with bladder tumor. 20043904_The identification of a fibronectin-binding domain in ameloblastin might permit interesting applications for dental implantology. 20097172_these data suggest that hypoxia promotes oral squamous cell carcinoma cell invasion that is elicited by HIF-1alpha-dependent alpha5 integrin and fibronectin induction. 20097751_iso-DGR sequences spontaneously converted from NGR are cryptic and do not mediate the interaction of the 70K region of FN with the cell surface during FN assembly 20145126_Mda-9/syntenin acts as a molecular adaptor linking PKCalpha and FAK activation in a pathway of FN adhesion by human cancer cells. 20161770_Data demonstrated that the inclusion of the alternatively spliced angiogenesis-associated ED-B leads to the unmasking of the FNIII 8 B-C loop that is cryptic in FN molecules lacking ED-B. 20201928_Exposure to ethanol drastically enhances the adhesion of cultured breast cancer cells to fibronectin and increases the expression of focal adhesions. 20210243_Fibronectin promotes paxillin (tyr118) phosphorylation and invasiveness of AGS cells. 20232238_An increased expression of fibronectin and tenascin-C mRNA in association to histological damage and in valvular heart disease compared to coronary artery disease could be shown. 20357207_Fibronectin may modulate the directional migration of corneal epithelial cells 20359090_The expression of Col I, Col IV and Fn was closely related to tumor invasion, the regional lymph node metastasis and other pathological features in laryngeal squamous cell carcinoma. 20364278_our findings suggest that both fibronectin and ASGPR mediate HBsAg binding to the cell surface, which provides further evidence for the potential roles of these two proteins in mediating HBV binding to liver cells. 20403349_Steric hindrance determines the motogenic activity of MSF and Fn, and that both molecules contain cryptic bioactive fragments. 20419094_in the presence of laminin and fibronectin, DC maintain a 'more immature' phenotype and express higher levels of key endocytic receptors, and become better endocytic cells, but possess superior ability to induce antigen-specific T cell differentiation. 20452482_Observational study of gene-disease association. (HuGE Navigator) 20463177_Observational study of gene-disease association. (HuGE Navigator) 20470202_Oncofetal fibronectin transcripts are highly abundant in the peripheral blood of patients with papillary thyroid carcinoma. 20484935_Elevated urinary fibronectin was identified as an independent factor predicting kidney function deterioration. 20489157_Data show that metastatic outgrowth is associated with activation of the transforming growth factor-beta signaling pathway and concerted up-regulation of POSTN, FN1, COL-I, and VCAN genes. 20495339_plays role in bronchial smooth muscle cells migration in the process of airway remodeling 20505078_Both IFN-gamma and mGBP-2 also inhibit cell spreading initiated by platelet-derived growth factor treatment, which is also accompanied by inhibition of Rac activation by mGBP-2. 20506163_Data show that occupancy of EPCR by protein C switches signaling specificity so that activation of PAR-1 by thrombin inhibits TNF-alpha-mediated synthesis of IL-6 and IL-8 and down-regulates TGF-beta-mediated expression of collagen type 4 and fibronectin. 20530259_MSF is a novel marker associated with the M2 stage of macrophage polarization expressed by tumor-conditioned macrophages; MSF may contribute to macrophage-mediated promotion of cancer cell invasion and metastasis. 20552237_high molecular weight of HA suppressed the FN-f-activated p38 via ICAM-1 and the CD44 in RA chondrocytes 20600851_Migration Stimulating Factor, a truncated form of fibronectin that promotes the migration of many cell types, inhibits AKT activity in human fibroblasts 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20628340_data indicate that higher levels of EDB isoform and FN-f are associated with intervertebral disc degeneration; this shift in alternative splicing may reflect an attempt of tissue repair and remodeling 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20638438_FN induces MMP-9 in HEp-2 cells mainly by involving integrin receptor alpha5beta1 and involves activation of multiple signaling pathways which independently or in 'cross-talk' to each other finally leads to the transactivation of the MMP-9 gene. 20643357_in migrating cells, a fraction of the endocytosed fibronectin receptor, alpha 5 beta 1 integrin, is sorted into multivesicular endosomes together with fibronectin and degraded in lysosomes 20670094_Fetal fibronectin as a predictor of spontaneous preterm birth. 20671107_Data show that FN III12-14 is a highly promiscuous ligand for GFs and also holds great potential in clinical healing applications. 20672308_FN upregulates osteoclast activity despite inhibiting osteoclast formation and that these effects involve NO and IL-1beta signaling. 20672350_Observational study of gene-disease association. (HuGE Navigator) 20673868_Observational study of gene-disease association. (HuGE Navigator) 20686611_The interplay between alpha5-integrin and sFn contributes significantly to tissue cohesion and, depending on their level of expression, can mediate a shift from liquid to elastic behavior. 20708078_Tenascin-C contains cryptic sites which can control tissue levels of fibrillar fibronectin either by preventing de novo fibril assembly or reducing levels of deposited fibronectin. 20725648_In the plasma group, the mean N-terminal FN domain immunoreactivity was about one fourth that of the cell-binding and C-terminal domains, whereas in cerebrospinal fluid they were nearly equal. 20811396_The findings that fibroblast survival is dependent on FN-growth factor binding domains underscore the critical importance of pericellular matrix for cell survival 20814177_CTGF cooperates with FN in enhancing the attachment and migration of corneal epithelial cells. 20819642_Glioma cells may induce vascular epithelial cells to express fibronectin, and in turn fibronectin could promote glioma cell invasion. 20851879_IGFBP-3 can either inhibit or enhance EGF-mediated growth of breast epithelial cells dependent upon the presence of fibronectin 20860816_High FN1 mRNA expression is associated with renal cell cancer. 20939933_rhFNHN29 and rhFNHC36 inhibit adhesion and invasion of liver cancer cells 21036168_syndecan-2 and syndecan-4 regulate the compensatory effect of TG-FN on osteoblast cell adhesion and actin cytoskeletal formation in the presence of RGD peptides 21036738_study suggests that FN and tissue transglutaminase (TG2) facilitate the metastatic activity of A431 tumor cells, and this may be partly attributed to TG2 enhancement of the association of FN and beta integrin 21098633_These results highlight a crucial role for caveolin-1 in negative regulation of membrane microdomain mobility, thereby affecting endocytosis of bacteria-engaged integrins. 21131589_Megakaryocyte adhesion to type I collagen promotes inhibits pro-platelet formation through the release of cFN to the plasma membrane. This regulatory mechanism is dependent on the engagement of FN receptors and on transglutaminase FXIII-A activity. 21138686_autocrine soluble fibronectin of osteosarcoma MG-63 cells may be involved in cell shape maintaining 21148425_Impaired splicing of fibronectin is associated with thoracic aortic aneurysm formation in patients with bicuspid aortic valve. 21160032_FN upregulation of cGMP-dependent protein kinase (PKG-I) protein content is due to increased mRNA expression via augmented transcriptional activity of the PKG-I promoter region. 21178109_Cav1.2 calcium channel association with alpha5beta1-integrin depends on adhesion to fibronectin; interactions between alpha5beta1-integrin and fibronectin modulate L-type calcium channel entry. 21190414_Fetal fibronectin may be used as a primary screening tool for prediction of preterm birth in asymptomatic high-risk women. 21224775_Data show that a molecular complex of fibronectin and aggrecan predicts response to lumbar ESI for radiculopathic back pain with HNP. 21311138_Danshensu can protect from oxidative stress through inhibiting the expression of FN and Col-I induced by high glucose. 21325030_Kindlin-2 regulates podocyte adhesion and fibronectin deposition through interactions with phosphoinositides and integrin b1 and b3. 21330459_Data show that ITP-79 modulated binding of TG2 to fibronectin. 21347378_LigB could facilitate leptospirosis transmission by binding fibronectin and fibrinogen 21429937_Mesenchymal stem cell migration is regulated by fibronectin through alpha5beta1-integrin-mediated activation of PDGFR-beta and potentiation of PDGF-BB signals. 21472136_GALNT6-fibronectin pathway should be a critical component for breast cancer development and progression. 21508391_Increase expression of sialic fibronectin correlates with lymph node metastasis of thyroid malignant neoplasmas. 21573010_This work provides a broad overview of matrix remodeling at the cell-material interface, establishing correlations between surface chemistry, fibronectin adsorption, cell adhesion and signaling, matrix reorganization and degradation. 21630266_We conclude that TGF-beta1 may both switch on and switch off migration stimulating factor (MSF) expression in a manner criti | ENSMUSG00000026193 | Fn1 | 1.422562e+03 | 1.0611639 | 0.085647558 | 0.2568486 | 1.113890e-01 | 0.7385682617 | 0.94515436 | No | Yes | 1.482953e+03 | 200.712737 | 1.151888e+03 | 160.114184 | |
| ENSG00000115484 | 10575 | CCT4 | protein_coding | P50991 | FUNCTION: Component of the chaperonin-containing T-complex (TRiC), a molecular chaperone complex that assists the folding of proteins upon ATP hydrolysis (PubMed:25467444). The TRiC complex mediates the folding of WRAP53/TCAB1, thereby regulating telomere maintenance (PubMed:25467444). As part of the TRiC complex may play a role in the assembly of BBSome, a complex involved in ciliogenesis regulating transports vesicles to the cilia (PubMed:20080638). The TRiC complex plays a role in the folding of actin and tubulin (Probable). {ECO:0000269|PubMed:20080638, ECO:0000269|PubMed:25467444, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cell projection;Chaperone;Cytoplasm;Cytoskeleton;Direct protein sequencing;Methylation;Nucleotide-binding;Phosphoprotein;Reference proteome | The chaperonin containing TCP1 (MIM 186980) complex (CCT), also called the TCP1 ring complex, consists of 2 back-to-back rings, each containing 8 unique but homologous subunits, such as CCT4. CCT assists the folding of newly translated polypeptide substrates through multiple rounds of ATP-driven release and rebinding of partially folded intermediate forms. Substrates of CCT include the cytoskeletal proteins actin (see MIM 102560) and tubulin (see MIM 191130), as well as alpha-transducin (MIM 139330) (Won et al., 1998 [PubMed 9819444]).[supplied by OMIM, Mar 2008]. | hsa:10575; | cell body [GO:0044297]; cell projection [GO:0042995]; centrosome [GO:0005813]; chaperonin-containing T-complex [GO:0005832]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; melanosome [GO:0042470]; microtubule [GO:0005874]; nucleoplasm [GO:0005654]; zona pellucida receptor complex [GO:0002199]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; protein folding chaperone [GO:0044183]; RNA binding [GO:0003723]; unfolded protein binding [GO:0051082]; binding of sperm to zona pellucida [GO:0007339]; chaperone-mediated protein folding [GO:0061077]; positive regulation of establishment of protein localization to telomere [GO:1904851]; positive regulation of protein localization to Cajal body [GO:1904871]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomerase RNA localization to Cajal body [GO:1904874]; positive regulation of telomere maintenance via telomerase [GO:0032212]; protein folding [GO:0006457]; protein stabilization [GO:0050821]; scaRNA localization to Cajal body [GO:0090666]; toxin transport [GO:1901998] | 23612981_Both CCT4 and CCT5 homo-oligomers have the property of forming 8-fold double rings absent the other subunits, and these complexes carry out chaperonin reactions without other partner subunits. 24320561_Chaperonin CCT is required for the correct folding of eIF3h and eIF3i subunits. 34732665_CCT4 suppression inhibits tumor growth in hepatocellular carcinoma by interacting with Cdc20. | ENSMUSG00000007739 | Cct4 | 7.684811e+03 | 0.6745329 | -0.568039284 | 0.2724589 | 4.372708e+00 | 0.0365189171 | 0.61802832 | No | Yes | 6.735576e+03 | 975.880647 | 9.248173e+03 | 1373.262646 | |
| ENSG00000115561 | 51652 | CHMP3 | protein_coding | Q9Y3E7 | FUNCTION: Probable core component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and mostly are delivered to lysosomes enabling degradation of membrane proteins, such as stimulated growth factor receptors, lysosomal enzymes and lipids. The MVB pathway appears to require the sequential function of ESCRT-O, -I,-II and -III complexes. ESCRT-III proteins mostly dissociate from the invaginating membrane before the ILV is released. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis and the budding of enveloped viruses (HIV-1 and other lentiviruses). ESCRT-III proteins are believed to mediate the necessary vesicle extrusion and/or membrane fission activities, possibly in conjunction with the AAA ATPase VPS4. Selectively binds to phosphatidylinositol 3,5-bisphosphate PtdIns(3,5)P2 and PtdIns(3,4)P2 in preference to other phosphoinositides tested. Involved in late stages of cytokinesis. Plays a role in endosomal sorting/trafficking of EGF receptor. Isoform 2 prevents stress-mediated cell death and accumulation of reactive oxygen species when expressed in yeast cells. {ECO:0000269|PubMed:14505570, ECO:0000269|PubMed:15707591, ECO:0000269|PubMed:16740483, ECO:0000269|PubMed:17331679, ECO:0000269|PubMed:18076377}. | 3D-structure;Alternative splicing;Apoptosis;Cell cycle;Cell division;Coiled coil;Cytoplasm;Endosome;Isopeptide bond;Lipoprotein;Membrane;Myristate;Phosphoprotein;Protein transport;Reference proteome;Transport;Ubl conjugation | This gene encodes a protein that sorts transmembrane proteins into lysosomes/vacuoles via the multivesicular body (MVB) pathway. This protein, along with other soluble coiled-coil containing proteins, forms part of the ESCRT-III protein complex that binds to the endosomal membrane and recruits additional cofactors for protein sorting into the MVB. This protein may also co-immunoprecipitate with a member of the IFG-binding protein superfamily. Alternative splicing results in multiple transcript variants. Read-through transcription also exists between this gene and the upstream ring finger protein 103 (RNF103) gene. [provided by RefSeq, Nov 2010]. | hsa:100526767;hsa:51652; | amphisome membrane [GO:1904930]; autophagosome membrane [GO:0000421]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; ESCRT III complex [GO:0000815]; extracellular exosome [GO:0070062]; kinetochore [GO:0000776]; kinetochore microtubule [GO:0005828]; late endosome [GO:0005770]; lysosomal membrane [GO:0005765]; midbody [GO:0030496]; multivesicular body [GO:0005771]; multivesicular body membrane [GO:0032585]; nuclear pore [GO:0005643]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; phosphatidylcholine binding [GO:0031210]; ubiquitin-specific protease binding [GO:1990381]; apoptotic process [GO:0006915]; autophagosome maturation [GO:0097352]; autophagy [GO:0006914]; endosome transport via multivesicular body sorting pathway [GO:0032509]; late endosome to lysosome transport [GO:1902774]; late endosome to vacuole transport [GO:0045324]; macroautophagy [GO:0016236]; midbody abscission [GO:0061952]; mitotic metaphase plate congression [GO:0007080]; multivesicular body assembly [GO:0036258]; multivesicular body sorting pathway [GO:0071985]; multivesicular body-lysosome fusion [GO:0061763]; negative regulation of cell death [GO:0060548]; nuclear membrane reassembly [GO:0031468]; nucleus organization [GO:0006997]; plasma membrane repair [GO:0001778]; protein polymerization [GO:0051258]; protein transport [GO:0015031]; regulation of centrosome duplication [GO:0010824]; regulation of early endosome to late endosome transport [GO:2000641]; regulation of mitotic spindle assembly [GO:1901673]; suppression of viral release by host [GO:0044790]; ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway [GO:0043162]; viral budding from plasma membrane [GO:0046761]; viral budding via host ESCRT complex [GO:0039702]; viral release from host cell [GO:0019076] | 12878588_In rodents, this protein selectively binds phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 3,4-bisphosphate and may participate in endosomal trafficking. 16302002_data suggest that mac25/IGFBP-rP1 and 25.1 (NEDF) may play a functional role in the NE differentiation of NSCLC cell lines and may provide a novel therapeutic target for treating lung cancers, in particular NSCLC with NE differentiation 16554368_ESCRT subunit is important for degradation of the epidermal growth factor receptor (EGFR) and for transport of the receptor from endosomes to lysosomes. 17146056_CHMP proteins are regulated through an autoinhibitory switch mechanism that allows tight control of endosomal sorting complex ESCRT-III assembly 17261583_A dominant-negative version of CHMP3, which specifically prevents targeting of AMSH (STAMBP) to endosomes, inhibits degradation but not internalization of epidermal growth factor receptor. 17331679_Data demonstrate that the VPS24 gene gives rise to two functionally distinct proteins, one of which is involved in the ESCRT pathway and another novel protein that serves an anti-apoptotic role. 17686835_expression of dominant negative forms of Vps4 and Vps24, two components of the MVB pathway, rresult in an impairment in infectious herpes simplex virus assembly/egress 17711858_UBPY MIT domain and another ubiquitin isopeptidase, AMSH, reveals common interactions with CHMP1A and CHMP1B but a distinct selectivity of AMSH for CHMP3/VPS24, a core subunit of the ESCRT-III complex, and UBPY for CHMP7. 18687924_study found the ESCRT-III proteins CHMP2A & CHMP3 could assemble in vitro into helical tubular structures that expose their membrane interaction sites on the outside of the tubule; VPS4 could bind on the inside of the tubule & disassemble the tubes 19525971_Data show that the N-terminal core domains of increased sodium tolerance-1 (IST1) and charged multivesicular body protein-3 (CHMP3) form equivalent four-helix bundles, revealing that IST1 is a previously unrecognized ESCRT-III family member. 21827950_tight coupling of ESCRT-III CHMP3 and AMSH functions and provide insight into the regulation of ESCRT-III 24440309_Protein kinase CK2 alpha is involved in the phosphorylation of the ESCRT-III subunits CHMP3 and CHMP2B, as well as of VPS4B/SKD1, an ATPase that mediates ESCRT-III disassembly. 31541468_miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charged multivesicular body protein 3 through mitogen-activated protein kinase signaling. 34597582_RetroCHMP3 blocks budding of enveloped viruses without blocking cytokinesis. | ENSMUSG00000053119 | Chmp3 | 1.462914e+03 | 0.9055229 | -0.143177048 | 0.2555939 | 3.144762e-01 | 0.5749463415 | 0.89968088 | No | Yes | 1.398177e+03 | 139.271993 | 1.554804e+03 | 158.334132 | |
| ENSG00000115694 | 10494 | STK25 | protein_coding | O00506 | FUNCTION: Oxidant stress-activated serine/threonine kinase that may play a role in the response to environmental stress. Targets to the Golgi apparatus where it appears to regulate protein transport events, cell adhesion, and polarity complexes important for cell migration. {ECO:0000269|PubMed:15037601}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Golgi apparatus;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the germinal centre kinase III (GCK III) subfamily of the sterile 20 superfamily of kinases. The encoded enzyme plays a role in serine-threonine liver kinase B1 (LKB1) signaling pathway to regulate neuronal polarization and morphology of the Golgi apparatus. The protein is translocated from the Golgi apparatus to the nucleus in response to chemical anoxia and plays a role in regulation of cell death. A pseudogene associated with this gene is located on chromosome 18. Multiple alternatively spliced transcript variants have been observed for this gene. [provided by RefSeq, Dec 2012]. | hsa:10494; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein homodimerization activity [GO:0042803]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; establishment of Golgi localization [GO:0051683]; establishment or maintenance of cell polarity [GO:0007163]; Golgi localization [GO:0051645]; Golgi reassembly [GO:0090168]; intrinsic apoptotic signaling pathway in response to hydrogen peroxide [GO:0036481]; positive regulation of axonogenesis [GO:0050772]; positive regulation of stress-activated MAPK cascade [GO:0032874]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; response to hydrogen peroxide [GO:0042542]; response to oxidative stress [GO:0006979]; signal transduction [GO:0007165] | 15037601_14-3-3zeta is a specific substrate for YSK1 that localizes to the Golgi apparatus, and potentially links YSK1 signaling at the Golgi apparatus with protein transport. 17253963_Ste20 family member (MAP4K3) that is required for maximal S6K (S6 kinase)/4E-BP1 [eIF4E (eukaryotic initiation factor 4E) that is required for maximal S6K (S6 kinase). 18364353_SOK1 entry into the nucleus is important for the cell death response 20592472_In cultured human endothelial cells, CCM3 and STK25 regulated barrier function in a manner similar to CCM2, and STKs negatively regulated Rho by directly activating moesin. 21396913_The results of the study results indicate that attenuation of SOK1 enhanced cell migration and lead to changes in the expression of migration-associated proteins such as FAK and GM130. 22391949_Significantly higher STK25 levels were observed in the skeletal muscle of type 2 diabetic patients, compared with individuals with normal glucose tolerance. 22652780_PDCD10 might be a regulatory adaptor required for STK25 functions, which differ distinctly depending on the redox status of the cells that may be potentially related to tumor progression. 22782892_Down-modulation of STK25, but not STK24, rescued medulloblastoma cells from NGF-induced TrkA-dependent cell death, suggesting that STK25 is part of the death-signaling pathway initiated by TrkA and CCM2. 23889253_This review notes that all 3 human germinal center kinase III genes consist of 12 exons and are ubiquitously expressed. In SOK1 and MST4, exon 1 encodes a 5' untranslated region, but this is not the case for MST3. 25550858_Our results suggest that MST4, STK25 and PDCD10 are upregulated in prostate cancer and may play roles in prostate tumorigenesis. 26553096_STK25 regulates lipid partitioning in human liver cells by controlling TAG synthesis as well as lipolytic activity and thereby NEFA release from lipid droplets for beta-oxidation and TAG secretion. 29996891_Our results demonstrated that STK25 negatively regulates the proliferation and glycolysis via GOLPH3-dependent mTOR signaling. Accordingly, STK25 could be a potential therapeutic target for the treatment of CRC. 30646538_hYSK1 blocks the p21(WAF1/Cip1) functions by direct interaction and inhibits the p16(INK4a) expression and induces MMP-2 expression by its regulations of SP-1 transcriptional activity under the hypoxia conditions. 30948712_Loss of STK25 promotes YAP/TAZ activation and enhanced cellular proliferation, even under normally growth-suppressive conditions both in vitro and in vivo. 32292165_STK25 suppresses Hippo signaling by regulating SAV1-STRIPAK antagonism. 33170807_Depletion of protein kinase STK25 ameliorates renal lipotoxicity and protects against diabetic kidney disease. 34624527_Antagonizing STK25 Signaling Suppresses the Development of Hepatocellular Carcinoma Through Targeting Metabolic, Inflammatory, and Pro-Oncogenic Pathways. 35217361_Loss of serine/threonine protein kinase 25 in retinal ganglion cells ameliorates high glucose-elicited damage through regulation of the AKT-GSK-3beta/Nrf2 pathway. | ENSMUSG00000026277 | Stk25 | 1.326778e+04 | 1.1548172 | 0.207664455 | 0.2896823 | 5.146956e-01 | 0.4731132097 | 0.87032151 | No | Yes | 1.314153e+04 | 1355.126647 | 1.049202e+04 | 1110.014908 | |
| ENSG00000115904 | 6654 | SOS1 | protein_coding | Q07889 | FUNCTION: Promotes the exchange of Ras-bound GDP by GTP (PubMed:8493579). Probably by promoting Ras activation, regulates phosphorylation of MAP kinase MAPK3 in response to EGF (PubMed:17339331). Catalytic component of a trimeric complex that participates in transduction of signals from Ras to Rac by promoting the Rac-specific guanine nucleotide exchange factor (GEF) activity (By similarity). {ECO:0000250|UniProtKB:Q62245, ECO:0000269|PubMed:17339331, ECO:0000269|PubMed:8493579}. | 3D-structure;Alternative splicing;Disease variant;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome | This gene encodes a protein that is a guanine nucleotide exchange factor for RAS proteins, membrane proteins that bind guanine nucleotides and participate in signal transduction pathways. GTP binding activates and GTP hydrolysis inactivates RAS proteins. The product of this gene may regulate RAS proteins by facilitating the exchange of GTP for GDP. Mutations in this gene are associated with gingival fibromatosis 1 and Noonan syndrome type 4. [provided by RefSeq, Jul 2008]. | hsa:6654; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; GTPase complex [GO:1905360]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; DNA binding [GO:0003677]; GTPase activator activity [GO:0005096]; guanyl-nucleotide exchange factor activity [GO:0005085]; protein heterodimerization activity [GO:0046982]; SH3 domain binding [GO:0017124]; axon guidance [GO:0007411]; B cell homeostasis [GO:0001782]; blood vessel morphogenesis [GO:0048514]; cardiac atrium morphogenesis [GO:0003209]; cytokine-mediated signaling pathway [GO:0019221]; epidermal growth factor receptor signaling pathway [GO:0007173]; eyelid development in camera-type eye [GO:0061029]; Fc-epsilon receptor signaling pathway [GO:0038095]; hair follicle development [GO:0001942]; heart trabecula morphogenesis [GO:0061384]; leukocyte migration [GO:0050900]; midbrain morphogenesis [GO:1904693]; multicellular organism growth [GO:0035264]; neurotrophin TRK receptor signaling pathway [GO:0048011]; pericardium morphogenesis [GO:0003344]; positive regulation of epidermal growth factor receptor signaling pathway [GO:0045742]; positive regulation of small GTPase mediated signal transduction [GO:0051057]; Ras protein signal transduction [GO:0007265]; regulation of cell population proliferation [GO:0042127]; regulation of MAP kinase activity [GO:0043405]; regulation of pro-B cell differentiation [GO:2000973]; regulation of T cell differentiation in thymus [GO:0033081]; regulation of T cell proliferation [GO:0042129]; regulation of transcription by RNA polymerase II [GO:0006357]; roof of mouth development [GO:0060021]; signal transduction [GO:0007165]; vitellogenesis [GO:0007296] | 11868160_mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1 14551916_Observational study of gene-disease association. (HuGE Navigator) 14656442_study of tandem histone folds in the structure of the N-terminal segment of the ras activator Son of Sevenless 15010862_p21Ras, hSOS1, and p120GAP are not involved in polycystic ovary disease 16760435_The full-length Grb2 proteins mediate negative regulation of the intrinsic Ras guanine-nucleotide exchange activity of hSos1. 16906159_In T cells, the SH2 domain of GRB2 binds phosphorylated tyrosines on the adaptor protein LAT and the GRB2 SH3 domains associate with the proline-rich regions of SOS1 and CBL. 17062749_increased fibroblast numbers and collagen matrix changes are associated with mutation [in hereditary gingival fibromatosis] 17075039_crystal structure of the Rem and Cdc25 domains of Sos1 determined at 2.0-A resolution in the absence of Ras 17143282_Missense mutations in SOS1 associated with Noonan syndrome. 17143285_SOS1 mutants as a major cause of Noonan syndrome. 17283063_The unusual RasGRP-SOS interplay results in sensitive and robust Ras activation that cannot be achieved with either activator alone. 17315019_it was concluded that SOS1 does not act as a proto-oncogene in juvenile myelomonocytic leukemia 17437339_The pleckstrin homology domain of human SOS1 seems to function as a sensor domain in detecting the prenylation status of Ras bound to the distal site of SOS1. 17486115_Our findings establish a crucial role for PLD2 in the coupling of extracellular signals to Sos-mediated Ras activation, and provide new insights into the spatial coordination of this activation event. 17510059_study reports that mutant SOS1 contributes an increased and sustained activation of ERK signaling in Hereditary gingival fibromatosis 1 fibroblasts under serum-starved conditions 17586837_SOS1 is the second most common Noonan gene but plays no major role in cardio-facio-cutaneous syndrome 18064648_findings suggest that SOS1 is not a significant human oncogene in most cancers. Furthermore, NS patients with SOS1 mutations may not be at increased risk of developing cancer 18454158_Study shows that SOS responds to the membrane density of Ras molecules, to their GTP loading and to the concentration of phosphatidylinositol-4,5-bisphosphate, and that the integration of these signals potentiates the release of autoinhibition. 18651097_Patients with SOS1 mutations range from Noonan syndrome to cardio-facio-cutaneous syndrome. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18778683_Structural basis of the differential binding of the SH3 domains of GRB2 and SOS1 are reported. 18972187_Mutational analysis of SOS1 gene in acute myeloid leukemia. 18972187_Observational study of gene-disease association. (HuGE Navigator) 19020799_Mutations in the SOS1 are associated with Noonan syndrome. 19020799_Observational study of gene-disease association. (HuGE Navigator) 19077116_Observational study of gene-disease association. (HuGE Navigator) 19133693_Observational study of gene-disease association. (HuGE Navigator) 19206169_Observational study of gene-disease association. (HuGE Navigator) 19289468_Loss of MADD expression resulted in reduced Grb2 and Sos1/2 recruitment to the TNFR1 complex and decreased Ras and MEKK1/2 activation 19352411_analysis of SOS1 and PTPN11 mutations in five cases of Noonan syndrome with multiple giant cell lesions 19438935_the involvement of SOS1 gene in a family with the Noonan-like/multiple giant cell lesion phenotype 19464300_The tandem SH3A and SH3B domains of Tks5 constitute a versatile module for the implementation of isoform-specific protein-protein interactions. 19724911_Enhanced expression of SOS1 is detected in prostate cancer epithelial cells from African-American men. 19911011_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19953625_First report describing different tumor types in Noonan syndrome with germline SOS1 mutations. 20005866_The binding of Grb2 adaptor to its downstream partners Sos1 and Gab1 docker is under tight allosteric regulation. 20030748_A case mimicking CS with SOS1 T158A substitution, which has not been reported previously in CS, revealed the complex relationship between the genotype and phenotype of overlapping syndromes of the RAS/RAF/MEK/ERK pathway. 20133692_a new crystal structure of SOS at 3.2 A resolution was presented that contains the histone domain and the DH-PH unit in addition to the catalytic segment (SOSHDPC, residues 1-1049). 20133694_Sos-histone domain plays a critical role in governing the catalytic output of Sos through the coupling of membrane recruitment to the release of autoinhibition. 20186801_Observational study of gene-disease association. (HuGE Navigator) 20302979_Observational study of gene-disease association. (HuGE Navigator) 20305546_Noonan-like/multiple giant cell lesion syndrome with mutations in the SOS1 gene 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20389169_Based on our results, it is possible that a subtle dysfunction (expression) of the SOS1 gene is involved in the development of the most common male reproductive tract disorder - unilateral or bilateral cryptorchidism 20543023_Observational study of gene-disease association. (HuGE Navigator) 20607846_Noonan syndrome is due to a SOS1 missense mutation and rhabdomyosarcoma. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20673819_two unrelated patients with Noonan syndrome carrying the same heterozygous SOS1 missense mutation (c.1867T > A/p.F623I) 20683980_mutation analysis performed on RAF1, SOS1, and GRB2, in 24 Noonan syndrome patients previously found to be negative for PTPN11 and KRAS mutations; SOS1 may have a role of modifier gene that might contribute the variable expressivity of the disease 20724475_Multiple decisive phosphorylation sites for the negative feedback regulation of SOS1 via ERK. 20950586_This study investigated the regulation of the previously uncharacterized SOS1 gene promoter by the aryl hydrocarbon receptor and its ligands in HepG2 cells. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21387466_this report expanded the available information about the molecular diversity of SOS1 mutations underlying Noonan syndrome, and have provided a more comprehensive assessment of the clinical features associated with those molecular lesions. 21674833_The researchers found evidence that there were significant differences between the D2S441 locus in the Maghreb population and other populations. 21746917_Comparison of RasGEF expression at different developmental stages showed that relative to Sos2 and RasGRP1, Sos1 is most abundant in DN thymocytes, but least abundant in DP thymocytes. 21851854_We present a stochastic mathematical model describing intra-molecular regulation of hSos1 activity. 22042618_CIIA functions as a molecular switch for the GEF activity of SOS1, directing this activity toward Rac1. 22217993_SOS1 over-expression may play a role in the regulation of the RAS/mitogen-activated protein kinase pathway in the skin, in the hair follicle proliferation and cell cycle, suggesting new perspectives in understanding the pathogenesis of hirsutism. 22344067_T-cell antigen receptor (TCR)-mediated Erk activation requires RasGRP1, but not Grb2/Sos. 22360309_multiple binding sites within Sos1 provide a physical route for Grb2 to hop in a flip-flop manner from one site to the next through facilitated diffusion, and such rapid exchange forms the basis of cooperativity driving bivalent binding of Grb2 to Sos1 22396725_Modeling and simulation of aggregation of membrane protein LAT with molecular variability in the number of binding sites for cytosolic Grb2-SOS1-Grb2 23132018_Oncogenic K-Ras promotes the activation of wild-type H- and N-Ras and this activation is mediated by oncogenic K-Ras-dependent allosteric stimulation of Sos. 23334917_Although Sos1 and Gab1 recognize two non-overlapping sites within the Grb2 adaptor, allostery promotes the formation of two distinct pools of Grb2-Sos1 and Grb2-Gab1 binary signaling complexes in lieu of a composite Sos1-Grb2-Gab1 ternary complex. 23528009_The SOS1 T158A mutation altered the phosphorylation of gene products involved in both RAS/MAPK and PI3K/AKT pathways. 23528987_The PR domain displays a highly dynamic conformational basin in agreement with the knowledge that the intrinsically unstructured proteins rapidly interconvert between an ensemble of conformations. 23589333_study established that the presence of SOS1, but not its enzymatic activity, is critical for p38 activation 23817964_MiR-124 inhibits the growth of glioblastoma through the downregulation of SOS1. 24497027_sustained Erk signaling and T-cell activation depend on both Sos1 and RasGRP1. 24522193_CIIA functions as a negative modulator of the SOS1-Ras signaling events initiated by peptide growth factors including EGF. 25062969_HGF-related mutation g.126,142-126,143insC in exon 21 was not found in any of the 6 affected individuals from three families. 25071181_Increased expression of SOS1 increases NFkappaB activation in several types of cancer cells, and ablation of SOS1 inhibits EGF-induced NFkappaB activation in these cells, indicating that SOS1 is a component of the pathway connecting EGFR to NFkappaB activation. 25394671_SOS1 and Ras regulate epithelial tight junction formation in the human airway through EMP1. 25469565_These data demonstrated the negative regulation between miR-146a and SOS1 and between miR-370 and GADD45beta and that these regulations are influenced by enterovirus 71 to induce apoptosis. 25624485_Stabilized alpha helices of son of sevenless 1 directly inhibit wild-type and mutant forms of KRAS. 25712082_The present study provides a first evidence of allelic imbalance of SOS1 and pinpoints this condition as a possible mechanism underlying a different penetrance of some SOS1-mutated alleles in unrelated carriers 25825487_Combined rational design and a high throughput screening platform for identifying chemical inhibitors of a Ras-activating enzyme. 25980493_In non-apoptotic cells, nuclear EGFR induced SOS1 expression by directly binding to the SOS1 promoter. 26077951_rs963731 is associated with corticobasal degeneration. 26447228_findings suggest that targeting the Src/Abl/Sos1/Rac pathway may represent a double-edged sword to control both cancer-invasive capacities and cancer-related inflammation. 26565026_Ras.GDP weakly binds to the catalytic but not to the allosteric site of Sos. 26708403_Data show that a heterozygous son of sevenless homolog 1 (SOS1) gene frameshift mutation (c.3266dup or c.3248dup) was identified in each patient. 27304678_a computational methodology that overlays any variant database onto the somatic mutations in all cancer exomes identified activating SOS1 mutations associated with Noonan syndrome as significantly altered in melanoma 27412770_Data indicate dynamic of H-Ras functional cycle as controlled by son of sevenless homolog 1 (Sos). 27501536_Grb2-independent interactions are sufficient to retain human SOS on the membrane for many minutes, during which a single SOS molecule could processively activate thousands of Ras molecules. 28425619_In summary, patients from two families with history of non-syndromic autosomal-dominant HGF from Malopolska and Mazovia provinces in Poland had not been affected by HGF type 1, caused by a single-cytosine insertion in exon 21 of the SOS-1 gene. 28433619_Mutation scanning of the entire coding sequence of SOS1 gene identified seven intronic variants and one new exonic substitution (c.138G > A). The seven common intronic variants were not considered to be of pathogenic importance. The exonic substitution c.138G > A was found to be absent in 100 ethnically matched normal control chromosomes, not expected to have functional significance based on prediction bioinformatics tool 28452363_a distinct role of the C-terminal proline-rich (PR) domain of the full-length son of sevenless homolog 1 (SOS1) protein. 28456002_Findings identified p.Ser548Arg missense mutation in Son of Sevenless Homolog 1 (SOS1) in the boy, confirmed in his mother. 28819285_SOS1 is required for full transformation and critically contribute to the leukemogenic potential of BCR-ABL. 28884940_We report on a novel pathogenic mutation in the SOS1 gene and a large clinical spectrum in a Noonan syndrome family with ten genetically confirmed affected individuals. 29074966_Noonan Syndrome mutations in SOS1 protein affects SOS1 translocation to the plasma membrane. 29233017_SOS1 was statistically significantly associated with gestational diabetes mellitus risk after adjusting for multiple comparisons 29408703_In the present study, we analyze the 14-3-3/SOS1 protein-protein interaction (PPI) by different biochemical assays and report the high resolution crystal structure of a 13-mer motif of SOS1 bound to 14-3-3 zeta 29696744_Disease-causing variants were identified in 0.6% (four of 692) of individuals with HCM, including three missense variants in the PTPN11, SOS1, and BRAF genes. Overall, 36 variants of uncertain significance (VUSs) were identified, averaging approximately 3VUSs/100 cases. 29855605_There was an additive effect of the alleles in the four genes associated with good growth response. For rs3110697 within IGFBP3, rs1045992 in CYP19A1 and rs2888586 in SOS1, the variant associated with better growth response showed higher transcriptional activity with r-hGH treatment 29907801_The overall VUS frequency was twice as high in prenatal specimens (58.1 vs. 29.3%). PTPN11 and SOS1 had a 1.5-fold higher VUS frequency in the prenatal cohort (10.7 vs. 7.4% and 95 vs. 61.1%, respectively). The diagnostic yield was 3.7% for prenatal samples, with a higher yield of 12.3% in fetuses with cystic hygroma as a sole finding, and 21.3% for postnatal 30635434_Transcriptional profiling reveals that the expression of mutant SOS1 leads to the upregulation of MYC target genes and genes associated with Ras transformation. 30714452_Enhanced expression of SOS1 is associated with uveal malignant melanoma. 31368652_six patients with SOS1 variants and Congenital heart defects and cardiomyopathy (male to female ratio 2:1) including two novel variants, were identified. 31882608_Molecular Dynamics model of peptide-protein conjugation: case study of covalent complex between Sos1 peptide and N-terminal SH3 domain from Grb2. 32603605_SOS1 Gain-of-Function Variants in Dilated Cardiomyopathy. 32752665_SOS1 interacts with Grb2 through regions that induce closed nSH3 conformations. 33446575_Epigenetic and Posttranscriptional Modulation of SOS1 Can Promote Breast Cancer Metastasis through Obesity-Activated c-Met Signaling in African-American Women. 33549576_Noonan syndrome patient-specific induced cardiomyocyte model carrying SOS1 gene variant c.1654A>G. 33760128_lncRNA DUXAP8 inhibits papillary thyroid carcinoma cell apoptosis via sponging the miR20b5p/SOS1 axis. 34232285_The intramolecular allostery of GRB2 governing its interaction with SOS1 is modulated by phosphotyrosine ligands. 35041140_Knockdown of circRNA-Memo1 Reduces Hypoxia/Reoxygenation Injury in Human Brain Endothelial Cells Through miRNA-17-5p/SOS1 Axis. 35152853_miR-148b-3p, as a tumor suppressor, targets son of sevenless homolog 1 to regulate the malignant progression in human osteosarcoma. | ENSMUSG00000024241 | Sos1 | 8.527106e+02 | 1.4951980 | 0.580336532 | 0.3626962 | 2.578482e+00 | 0.1083255025 | 0.75783482 | No | Yes | 1.171796e+03 | 261.479959 | 6.622049e+02 | 151.857141 | |
| ENSG00000116001 | 7072 | TIA1 | protein_coding | P31483 | FUNCTION: RNA-binding protein involved in the regulation of alternative pre-RNA splicing and mRNA translation by binding to uridine-rich (U-rich) RNA sequences (PubMed:8576255, PubMed:11106748, PubMed:12486009, PubMed:17488725). Binds to U-rich sequences immediately downstream from a 5' splice sites in a uridine-rich small nuclear ribonucleoprotein (U snRNP)-dependent fashion, thereby modulating alternative pre-RNA splicing (PubMed:11106748, PubMed:8576255). Preferably binds to the U-rich IAS1 sequence in a U1 snRNP-dependent manner; this binding is optimal if a 5' splice site is adjacent to IAS1 (By similarity). Activates the use of heterologous 5' splice sites; the activation depends on the intron sequence downstream from the 5' splice site, with a preference for a downstream U-rich sequence (PubMed:11106748). By interacting with SNRPC/U1-C, promotes recruitment and binding of spliceosomal U1 snRNP to 5' splice sites followed by U-rich sequences, thereby facilitating atypical 5' splice site recognition by U1 snRNP (PubMed:11106748, PubMed:12486009, PubMed:17488725). Activates splicing of alternative exons with weak 5' splice sites followed by a U-rich stretch on its own pre-mRNA and on TIAR mRNA (By similarity). Acts as a modulator of alternative splicing for the apoptotic FAS receptor, thereby promoting apoptosis (PubMed:11106748, PubMed:1934064, PubMed:17488725). Binds to the 5' splice site region of FAS intron 5 to promote accumulation of transcripts that include exon 6 at the expense of transcripts in which exon 6 is skipped, thereby leading to the transcription of a membrane-bound apoptotic FAS receptor, which promotes apoptosis (PubMed:11106748, PubMed:1934064, PubMed:17488725). Binds to a conserved AU-rich cis element in COL2A1 intron 2 and modulates alternative splicing of COL2A1 exon 2 (PubMed:17580305). Also binds to the equivalent AT-rich element in COL2A1 genomic DNA, and may thereby be involved in the regulation of transcription (PubMed:17580305). Binds specifically to a polypyrimidine-rich controlling element (PCE) located between the weak 5' splice site and the intronic splicing silencer of CFTR mRNA to promote exon 9 inclusion, thereby antagonizing PTB1 and its role in exon skipping of CFTR exon 9 (PubMed:14966131). Involved in the repression of mRNA translation by binding to AU-rich elements (AREs) located in mRNA 3' untranslated regions (3' UTRs), including target ARE-bearing mRNAs encoding TNF and PTGS2 (By similarity). Also participates in the cellular response to environmental stress, by acting downstream of the stress-induced phosphorylation of EIF2S1/EIF2A to promote the recruitment of untranslated mRNAs to cytoplasmic stress granules (SGs), leading to stress-induced translational arrest (PubMed:10613902). Formation and recruitment to SGs is regulated by Zn(2+) (By similarity). Possesses nucleolytic activity against cytotoxic lymphocyte target cells (PubMed:1934064). {ECO:0000250|UniProtKB:P52912, ECO:0000269|PubMed:10613902, ECO:0000269|PubMed:11106748, ECO:0000269|PubMed:12486009, ECO:0000269|PubMed:14966131, ECO:0000269|PubMed:17488725, ECO:0000269|PubMed:17580305, ECO:0000269|PubMed:1934064, ECO:0000269|PubMed:8576255}.; FUNCTION: [Isoform Short]: Displays enhanced splicing regulatory activity compared with TIA isoform Long. {ECO:0000269|PubMed:17488725}. | 3D-structure;Acetylation;Alternative splicing;Amyotrophic lateral sclerosis;Apoptosis;Cytoplasm;Disease variant;Neurodegeneration;Nucleus;RNA-binding;Reference proteome;Repeat;mRNA processing;mRNA splicing | The product encoded by this gene is a member of a RNA-binding protein family and possesses nucleolytic activity against cytotoxic lymphocyte (CTL) target cells. It has been suggested that this protein may be involved in the induction of apoptosis as it preferentially recognizes poly(A) homopolymers and induces DNA fragmentation in CTL targets. The major granule-associated species is a 15-kDa protein that is thought to be derived from the carboxyl terminus of the 40-kDa product by proteolytic processing. Alternative splicing resulting in different isoforms has been found for this gene. [provided by RefSeq, May 2017]. | hsa:7072; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; nuclear stress granule [GO:0097165]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; mRNA 3'-UTR AU-rich region binding [GO:0035925]; mRNA 3'-UTR binding [GO:0003730]; poly(A) binding [GO:0008143]; RNA binding [GO:0003723]; apoptotic process [GO:0006915]; negative regulation of cytokine production [GO:0001818]; negative regulation of translation [GO:0017148]; protein localization to cytoplasmic stress granule [GO:1903608]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; regulation of mRNA splicing, via spliceosome [GO:0048024]; stress granule assembly [GO:0034063] | 11106748_apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing 12486009_binding of TIA-1 in the vicinity of a 5' ss helps to stabilize U1 snRNP recruitment, at least in part, via a direct interaction with U1-C 12885872_Data suggest that TIA-1 functions as a translational silencer of cyclooxygenase-2 (COX-2) expression and support the hypothesis that dysregulated RNA-binding of TIA-1 promotes COX-2 expression in neoplasia. 12949814_Increased TIA-1 gene expression is associated with sensitize endothelial cells to proapoptotic stimuli present in the tumor microenvironment and enhance NK cell cytotoxic activity against cancer cells in advanced soft tissue sarcoma 14966131_data indicate that, in CFTR exon 9, TIA-1 binding to the polypyrimidine-rich controlling element recruits U1 small nuclear ribonucleoprotein to the weak 5'-ss and induces exon inclusion 15280467_activated during HSV-1 infection and accumulated in cytoplasm of cells 6 hr after infection 16227602_TIA-1 represses the translation of target transcripts. 16820934_Results describe the gene expression of tristetraprolin, T-cell intracellular antigen and Hu antigen R in synovial tissues from rheumatoid arthritis and osteoarthritis patients. 17135269_FAST K synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing 17488725_TIAR regulates the relative expression of TIA-1 isoforms. 17493234_Cytotoxic molecule (CM) expression, specifically TIA1 and granzyme B, is predictive of prognosis in Hodgkin's-like anaplastic large cell lymphoma. 17580305_dual role for TIA-1 in shuttling between DNA and RNA ligands to co-regulate COL2A1 expression at the level of transcription and pre-mRNA alternative splicing. 17599736_Positive correlations between TIA1 protein gene expression in patients with rheumatoid and healthy persons. 17711853_TIA-1-induced polysome disassembly is required for enhanced mRNA decay 18456862_Simultaneous knockdown of TIA1 and TIAL1 resulted in increased skipping of alternatively spliced exons associated with U-rich motifs, but did not affect alternatively spliced exons that are not associated with U-rich motifs. 18642007_Basophilic inclusions from patients with adult-onset atypical motor neuron disease were distinctly labeled with the antibodies against poly(A)-binding protein 1, T cell intracellular antigen 1, and ribosomal protein S6. 18753794_These data demonstrated that TIA-1 inhibits HBsAg expression by interacting with the posttranscriptional regulatory element (PRE) of hepatitis B virus. 18775331_Reults describe codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. 19110540_TIA1 and TIAL1 regulate the alternate splicing of lysyl hydroxylase 2 19386911_The carboxyl-terminal domain of TIA-1 is responsible for its recruitment into inclusions containing mutant huntingtin protein. 19615357_Sam68 is recruited into stress granules through complexing with TIA-1 in response to oxidative stress 19641607_Host immune responses to EOC vary widely according to histological subtype and the extent of residual disease. TIA-1, FoxP3 and CD20 emerge as new positive prognostic factors in high-grade serous EOC from optimally debulked patients 20599318_Down-regulation of the IGFBP-3 transcripts correlated with the up-regulation of the TIA-1 transcripts in primary HCC biopsies. 20675271_Data show that apoptotic (TIAR and TIA-1) marker expression in thyroid tissues from adolescents with immune thyroid diseases is higher than in non-immune thyroid diseases. 20980400_Severe hypoxia caused co-aggregation of TIAR/TIA-1 and these proteins suppressed HIF-1alpha expression. 21048981_Data show that TIA1 and TIAL1 bind at the same positions on human RNAs, and are consistent with a model where TIA proteins shorten the time available for definition of an alternative exon by enhancing recognition of the preceding 5' splice site. 21179245_TIA-1 cytotoxic granule-associated RNA binding protein has a role in preventing progression of mismatch repair-proficient colorectal cancer 21189287_TIA1 and TIAR proteins are intron-associated positive regulators of SMN2 exon 7 splicing. 21257637_TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. 21284605_A role for TIA proteins as growth/tumour-suppressor genes. 21846467_Results characterise the C-terminal RRM2 and RRM3 domains of T-cell intracellular antigen-1 protein. 22154808_Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. 23348830_the TIA1 mutation causes perturbed RNA splicing and cellular stress resulting in WDM. 23401021_Welander distal myopathy is caused by mutated TIA1 through a dominant pathomechanism probably involving altered stress granule dynamics. 23830997_Work described here examined the punctate pattern of SRp20 localization in the cytoplasm of poliovirus-infected cells, demonstrating the partial co-localization of SRp20 with the stress granule marker protein TIA-1. 23902765_RNA binding mediated by either isolated RRM3 or the RRM23 construct is controlled by slight environmental pH changes due to the protonation/deprotonation of TIA-1 RRM3 histidine residues. 24433873_TIA1 gene expression do not predict prognosis in patients diagnosed with cutaneous T-cell lymphoma. 24566137_TIA1 inhibition of the exon 8 exclusion led to a decrease in SIRT1-Exon8 mRNA levels. 24682828_Structural insights into the role of binding avidity and the contributions of the TIA-1 RNA recognition motifs for recognition of pyrimidine-rich RNAs. 24766723_TIA proteins can function as long-term regulators of the ACTB mRNA metabolism in mouse and human cells. 24927121_TIA1-knockdown HeLa cells show an increase in ribosomes and translational machinery components. 25224594_Alternative splicing of TIA-1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. 25405991_TIA proteins play a role in the regulation and/or modulation of cellular homeostasis related to focal/cell adhesion, extracellular matrix and membrane and cytoskeleton dynamics. 25673011_SERPINE1 mRNA dissociates from the translational repressor proteins Ago2 and TIA-1 upon platelet activation 26363455_results suggest that TIA-1 and TIAR are two new host factors that interact with 5-UTR of EV71 genome and positively regulate viral replication 26681690_AT1R mRNA is regulated by TIA-1 in a ER stress-dependent manner. 26738979_This study showed that reactive oxygen species such as H2O2 oxidize the cytoplasmic stress granules (SG)-nucleating protein TIA1, thereby inhibiting SG assembly. 26958940_findings uncover a novel oncogenic function of TIA1 in esophageal tumorigenesis 27612012_Downregulation of TIA-1-enhanced mitochondrial elongation, whereas ectopic expression of TIA-1 resulted in mitochondria fragmentation. In addition, TIA-1 increased mitochondrial activity, including the rate of ATP synthesis and oxygen consumption. 28174264_YAP (Yes-associated protein) expression negatively regulates TIA1 (Rox8 ortholog) expression and cell invasion in human cancer cells. 28184449_Here we designed UC-rich and CU-rich 10-nt sequences for engagement of both RRM2 and RRM3 and demonstrated that the TIA-1 RRM23 construct preferentially binds the UC-rich RNA ligand. Together our data support a specific mode of TIA-1 RRM23 interaction with target oligonucleotides consistent with the role of TIA-1 in binding RNA to regulate gene expression 28193846_The results provide a mechanism for exon 16 3' splice site activation in which a coordinated effort among TIA1, Pcbp1, and RBM39 stabilizes or increases U2 snRNP recruitment, enhances spliceosome A complex formation, and facilitates exon definition through RBM39-mediated splicing regulation. 28257633_miR-19a could promote cell proliferation and migration in CRC cells and accelerated tumor growth in xenograft mice by targeting TIA1. 28298474_Data suggest that TPD52 (tumor protein D52) and a TPD52 fragment (residues 78-280) along with TIA-1 (T-cell intracellular antigen-1) and TIAR (TIA-1-related protein) contribute to mRNA stability as cis-acting and trans-acting factors; 3prime-untranslated regions of TPD52, TPD53, and TPD54 regulate expression of their respective genes in a post-transcriptional manner by altering mRNA stability. 28817800_studied a novel Amyotrophic Lateral Sclerosis/Frontotemporal Dementia (ALS/FTD) family and identified the P362L mutation in the low-complexity domain (LCD) of TIA1; genetic association analyses showed an increased burden of TIA1 LCD mutations in ALS patients compared to controls; TIA1 mutations significantly increased the propensity of TIA1 protein to undergo phase transition 29216908_The purpose of this report is to provide a detailed description of the clinical and neuropathological features associated with the recently identified TIA1 mutations that cause ALS +/- FTD. 29298433_These findings suggest that Zn(2+) is a physiological ligand of TIA-1, acting as a stress-inducible second messenger to promote multimerization of TIA-1 and subsequent localization into stress granules. 29370934_TIA1 is a novel causative gene of amyotrophic lateral sclerosis. 29457785_identification of a TIA1 variant that drives myodegeneration in multisystem proteinopathy with SQSTM1 mutations 29496454_expressions of TIA-1 and MFF were augmented in the cancerous liver tissues compared to the corresponding non-tumor tissues at mRNA and protein level, while the levels of miR-200a-3p and miR-27a/b were relatively lower in the cancerous liver tissues 29555582_TIA-1 knockdown by siRNA mimicked the effect of SAHA on COX-2 expression. These findings suggest SAHA can prevent TGF-beta1-induced COX-2 repression in lung fibroblasts post-transcriptionally through a novel TIA-1-dependent mechanism and provide new insights into the mechanisms underlying its potential antifibrotic activity 29699721_TIA1 LCD mutations are not common in Chinese ALS/ALS-FTD 29773329_TIA1 mutation is an uncommon genetic cause for ALS in the Chinese population. 29886022_TIA1 variants are not associated with frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) in European patient cohort. 30144499_The repression of TIA-1 by miR-487a promoted cell proliferation and suppressed cell apoptosis in vitro, and the knockdown of miR-487a had the opposite effects. 31541970_Single-nucleotide polymorphism in the human TIA1 gene interacts with stressful life events to predict the development of pathological anxiety symptoms in a Swedish population. 31941779_Typical Stress Granule Proteins Interact with the 3' Untranslated Region of Enterovirus D68 To Inhibit Viral Replication. 33621982_Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1. 34310938_Disease-associated mutations affect TIA1 phase separation and aggregation in a proline-dependent manner. 34410578_Identification of TIA1 mRNA targets during human neuronal development. 35163320_The Multifunctional Faces of T-Cell Intracellular Antigen 1 in Health and Disease. | ENSMUSG00000071337 | Tia1 | 4.509996e+02 | 0.6882898 | -0.538911896 | 0.3561953 | 2.295135e+00 | 0.1297798973 | 0.76070573 | No | Yes | 3.683296e+02 | 66.927678 | 5.863636e+02 | 108.584447 | |
| ENSG00000116171 | 6342 | SCP2 | protein_coding | P22307 | FUNCTION: [Isoform SCPx]: Plays a crucial role in the peroxisomal oxidation of branched-chain fatty acids (PubMed:10706581). Catalyzes the last step of the peroxisomal beta-oxidation of branched chain fatty acids and the side chain of the bile acid intermediates di- and trihydroxycoprostanic acids (DHCA and THCA) (PubMed:10706581). Also active with medium and long straight chain 3-oxoacyl-CoAs. Stimulates the microsomal conversion of 7-dehydrocholesterol to cholesterol and transfers phosphatidylcholine and 7-dehydrocholesterol between membrances, in vitro (By similarity). Isoforms SCP2 and SCPx cooperate in peroxisomal oxidation of certain naturally occurring tetramethyl-branched fatty acyl-CoAs (By similarity). {ECO:0000250|UniProtKB:P11915, ECO:0000250|UniProtKB:P32020, ECO:0000269|PubMed:10706581}.; FUNCTION: [Isoform SCP2]: Mediates the transfer of all common phospholipids, cholesterol and gangliosides from the endoplasmic reticulum to the plasma membrane. May play a role in regulating steroidogenesis (PubMed:17157249, PubMed:8300590, PubMed:7642518). Stimulates the microsomal conversion of 7-dehydrocholesterol to cholesterol (By similarity). Also binds fatty acids and fatty acyl Coenzyme A (CoA) such as phytanoyl-CoA. Involved in the regulation phospholipid synthesis in endoplasmic reticulum enhancing the incorporation of exogenous fatty acid into glycerides. Seems to stimulate the rate-limiting step in phosphatidic acid formation mediated by GPAT3. Isoforms SCP2 and SCPx cooperate in peroxisomal oxidation of certain naturally occurring tetramethyl-branched fatty acyl-CoAs (By similarity). {ECO:0000250|UniProtKB:P11915, ECO:0000250|UniProtKB:P32020, ECO:0000269|PubMed:17157249, ECO:0000269|PubMed:7642518, ECO:0000269|PubMed:8300590}. | 3D-structure;Acetylation;Acyltransferase;Alternative promoter usage;Alternative splicing;Cytoplasm;Direct protein sequencing;Endoplasmic reticulum;Lipid metabolism;Lipid transport;Lipid-binding;Mitochondrion;Peroxisome;Phosphoprotein;Reference proteome;Transferase;Transport | This gene encodes two proteins: sterol carrier protein X (SCPx) and sterol carrier protein 2 (SCP2), as a result of transcription initiation from 2 independently regulated promoters. The transcript initiated from the proximal promoter encodes the longer SCPx protein, and the transcript initiated from the distal promoter encodes the shorter SCP2 protein, with the 2 proteins sharing a common C-terminus. Evidence suggests that the SCPx protein is a peroxisome-associated thiolase that is involved in the oxidation of branched chain fatty acids, while the SCP2 protein is thought to be an intracellular lipid transfer protein. This gene is highly expressed in organs involved in lipid metabolism, and may play a role in Zellweger syndrome, in which cells are deficient in peroxisomes and have impaired bile acid synthesis. Alternative splicing of this gene produces multiple transcript variants, some encoding different isoforms.[provided by RefSeq, Aug 2010]. | hsa:6342; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; peroxisomal matrix [GO:0005782]; peroxisome [GO:0005777]; protein-containing complex [GO:0032991]; acetyl-CoA C-acyltransferase activity [GO:0003988]; acetyl-CoA C-myristoyltransferase activity [GO:0050633]; cholesterol binding [GO:0015485]; cholesterol transfer activity [GO:0120020]; fatty-acyl-CoA binding [GO:0000062]; long-chain fatty acyl-CoA binding [GO:0036042]; oleic acid binding [GO:0070538]; phosphatidylcholine transfer activity [GO:0120019]; phosphatidylinositol transfer activity [GO:0008526]; propanoyl-CoA C-acyltransferase activity [GO:0033814]; propionyl-CoA C2-trimethyltridecanoyltransferase activity [GO:0050632]; signaling receptor binding [GO:0005102]; sterol binding [GO:0032934]; alpha-linolenic acid metabolic process [GO:0036109]; bile acid biosynthetic process [GO:0006699]; bile acid metabolic process [GO:0008206]; fatty acid beta-oxidation [GO:0006635]; fatty acid beta-oxidation using acyl-CoA oxidase [GO:0033540]; inositol trisphosphate biosynthetic process [GO:0032959]; intracellular cholesterol transport [GO:0032367]; lipid hydroperoxide transport [GO:1901373]; peroxisome organization [GO:0007031]; phospholipid transport [GO:0015914]; positive regulation of intracellular cholesterol transport [GO:0032385]; positive regulation of steroid metabolic process [GO:0045940]; progesterone biosynthetic process [GO:0006701]; protein localization to plasma membrane [GO:0072659]; regulation of phospholipid biosynthetic process [GO:0071071]; steroid biosynthetic process [GO:0006694] | 12356316_data for the first time showed that while the N-terminal membrane binding domain of SCP(2) was itself inactive in mediating intermembrane sterol transfer, it nevertheless potentiated the ability of SCP(2) to enhance sterol transfer 12641450_plays a hitherto unrecognized role in intracellular phosphatidylinositol transfer, distribution, and signaling 14563822_SCP2 in the cellular defense against oxidative damage and found that a fluorescent fatty acid analog bound to SCP2 is protected against H2O2/Cu2+-induced oxidative damage 14661971_Overexpression of human SCP-2 in murine fibroblasts significantly alters the sterol dynamics of caveolae/lipid rafts, but not nonlipid raft domains, to facilitate retention of cholesterol within the cell. 15449949_By trafficking cholesterol hydroperoxides and phospholipid hydroperoxides in addition to parent lipids, SCP2 may exacerbate cell injury under oxidative stress conditions 17418802_Long chain fatty acyl-coenzyme A (CoA)s are confirmed to be high affinity ligands for SCP2, while long chain fatty acyl-carnitines are demonstrated for the first time not to interact with SCP2. 18465878_the importance of the N-terminal presequence in regulating SCP-2 structure, cholesterol localization within the ligand binding site, membrane association, and, potentially, intracellular targeting 19020914_Results describe the dynamical effect of sterol carrier protein-2 (SCP-2) interacting between aqueous dispersions of dehydroergosterol monohydrate microcrystal donors and acceptors. 19598235_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20656919_cellular SCP-2 not only binds and translocates cholesterol but also cholesterol hydroperoxides, thus expanding their redox toxicity and signaling ranges under oxidative stress conditions 20677014_Observational study of gene-disease association. (HuGE Navigator) 20677014_Statistical analysis indicated that six genes, NFATC2, SCP2, CACNA1C, TCRA, POLE, and FAM3D, were associated with narcolepsy. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21375735_The Peroxisomal targeting signal 1 in Scp2 is autonomous and is essential for binding to pex5. 24510313_We conclude that SCP-2 is a low affinity binding protein for arachidonylethanolamine that can facilitate its cellular uptake but does not contribute significantly to intracellular sequestration of AEA. 27097688_Mice harboring a deletion of the Scp2 locus present a modulated diurnal accumulation of lipids in the liver and a perturbed activation of several signaling pathways including PPARalpha, SREBP, LRH-1, TORC1 and its upstream regulators. 27311714_imported protein sterol carrier protein 2 (SCP2) occupies only a subregion of larger peroxisomes, highlighting the heterogeneous distribution of proteins even within the peroxisome. 28284963_We (1) analyzed the structural basis of the fold and the classification of SCP2 domains; (2) identified structure-determined sequence features; (3) compared the lipid binding cavity of SCP2 and other lipid binding proteins; (4) surveyed proposed mechanisms of SCP2 mediated lipid transfer between membranes; and (5) uncovered a possible new function of the SCP2 domain as a protein-protein recognition device. 31407919_Methylation dependent microRNA 1285-5p and sterol carrier proteins 2 in type 2 diabetes mellitus 34541823_LINC00261 elevation inhibits angiogenesis and cell cycle progression of pancreatic cancer cells by upregulating SCP2 via targeting FOXP3. | ENSMUSG00000028603 | Scp2 | 6.009614e+02 | 0.9954697 | -0.006550650 | 0.3383340 | 3.720909e-04 | 0.9846100334 | 0.99716006 | No | Yes | 5.818567e+02 | 103.415040 | 5.653881e+02 | 103.013122 | |
| ENSG00000116288 | 11315 | PARK7 | protein_coding | Q99497 | FUNCTION: Multifunctional protein with controversial molecular function which plays an important role in cell protection against oxidative stress and cell death acting as oxidative stress sensor and redox-sensitive chaperone and protease (PubMed:17015834, PubMed:20304780, PubMed:18711745, PubMed:12796482, PubMed:19229105, PubMed:25416785, PubMed:26995087, PubMed:28993701). It is involved in neuroprotective mechanisms like the stabilization of NFE2L2 and PINK1 proteins, male fertility as a positive regulator of androgen signaling pathway as well as cell growth and transformation through, for instance, the modulation of NF-kappa-B signaling pathway (PubMed:12612053, PubMed:15502874, PubMed:14749723, PubMed:17015834, PubMed:21097510, PubMed:18711745). Has been described as a protein and nucleotide deglycase that catalyzes the deglycation of the Maillard adducts formed between amino groups of proteins or nucleotides and reactive carbonyl groups of glyoxals (PubMed:25416785, PubMed:28596309). But this function is rebuted by other works (PubMed:27903648, PubMed:31653696). As a protein deglycase, repairs methylglyoxal- and glyoxal-glycated proteins, and releases repaired proteins and lactate or glycolate, respectively. Deglycates cysteine, arginine and lysine residues in proteins, and thus reactivates these proteins by reversing glycation by glyoxals. Acts on early glycation intermediates (hemithioacetals and aminocarbinols), preventing the formation of advanced glycation endproducts (AGE) that cause irreversible damage (PubMed:25416785, PubMed:28013050, PubMed:26995087). Also functions as a nucleotide deglycase able to repair glycated guanine in the free nucleotide pool (GTP, GDP, GMP, dGTP) and in DNA and RNA. Is thus involved in a major nucleotide repair system named guanine glycation repair (GG repair), dedicated to reversing methylglyoxal and glyoxal damage via nucleotide sanitization and direct nucleic acid repair (PubMed:28596309). Protects histones from adduction by methylglyoxal, controls the levels of methylglyoxal-derived argininine modifications on chromatin (PubMed:30150385). Able to remove the glycations and restore histone 3, histone glycation disrupts both local and global chromatin architecture by altering histone-DNA interactions as well as histone acetylation and ubiquitination levels (PubMed:30150385, PubMed:30894531). Displays a very low glyoxalase activity that may reflect its deglycase activity (PubMed:22523093, PubMed:31653696, PubMed:28993701). Eliminates hydrogen peroxide and protects cells against hydrogen peroxide-induced cell death (PubMed:16390825). Required for correct mitochondrial morphology and function as well as for autophagy of dysfunctional mitochondria (PubMed:19229105, PubMed:16632486). Plays a role in regulating expression or stability of the mitochondrial uncoupling proteins SLC25A14 and SLC25A27 in dopaminergic neurons of the substantia nigra pars compacta and attenuates the oxidative stress induced by calcium entry into the neurons via L-type channels during pacemaking (PubMed:18711745). Regulates astrocyte inflammatory responses, may modulate lipid rafts-dependent endocytosis in astrocytes and neuronal cells (PubMed:23847046). In pancreatic islets, involved in the maintenance of mitochondrial reactive oxygen species (ROS) levels and glucose homeostasis in an age- and diet dependent manner. Protects pancreatic beta cells from cell death induced by inflammatory and cytotoxic setting (By similarity). Binds to a number of mRNAs containing multiple copies of GG or CC motifs and partially inhibits their translation but dissociates following oxidative stress (PubMed:18626009). Metal-binding protein able to bind copper as well as toxic mercury ions, enhances the cell protection mechanism against induced metal toxicity (PubMed:23792957). In macrophages, interacts with the NADPH oxidase subunit NCF1 to direct NADPH oxidase-dependent ROS production, and protects against sepsis (By similarity). {ECO:0000250|UniProtKB:Q99LX0, ECO:0000269|PubMed:11477070, ECO:0000269|PubMed:12612053, ECO:0000269|PubMed:12855764, ECO:0000269|PubMed:12939276, ECO:0000269|PubMed:14749723, ECO:0000269|PubMed:15181200, ECO:0000269|PubMed:15502874, ECO:0000269|PubMed:15976810, ECO:0000269|PubMed:16390825, ECO:0000269|PubMed:17015834, ECO:0000269|PubMed:18626009, ECO:0000269|PubMed:18711745, ECO:0000269|PubMed:19229105, ECO:0000269|PubMed:20186336, ECO:0000269|PubMed:20304780, ECO:0000269|PubMed:21097510, ECO:0000269|PubMed:22523093, ECO:0000269|PubMed:23792957, ECO:0000269|PubMed:23847046, ECO:0000269|PubMed:25416785, ECO:0000269|PubMed:26995087, ECO:0000269|PubMed:28013050, ECO:0000269|PubMed:28596309, ECO:0000269|PubMed:28993701, ECO:0000269|PubMed:30150385, ECO:0000269|PubMed:30894531, ECO:0000269|PubMed:9070310}. | 3D-structure;Acetylation;Autophagy;Cell membrane;Chaperone;Copper;Cytoplasm;DNA damage;DNA repair;Direct protein sequencing;Disease variant;Endoplasmic reticulum;Fertilization;Hydrolase;Inflammatory response;Isopeptide bond;Lipoprotein;Membrane;Mitochondrion;Neurodegeneration;Nucleus;Oxidation;Palmitate;Parkinson disease;Parkinsonism;Phosphoprotein;Protease;RNA-binding;Reference proteome;Stress response;Tumor suppressor;Ubl conjugation;Zymogen | The product of this gene belongs to the peptidase C56 family of proteins. It acts as a positive regulator of androgen receptor-dependent transcription. It may also function as a redox-sensitive chaperone, as a sensor for oxidative stress, and it apparently protects neurons against oxidative stress and cell death. Defects in this gene are the cause of autosomal recessive early-onset Parkinson disease 7. Two transcript variants encoding the same protein have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:11315; | adherens junction [GO:0005912]; axon [GO:0030424]; cell body [GO:0044297]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; membrane raft [GO:0045121]; mitochondrial intermembrane space [GO:0005758]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; PML body [GO:0016605]; presynapse [GO:0098793]; androgen receptor binding [GO:0050681]; cadherin binding [GO:0045296]; copper ion binding [GO:0005507]; cupric ion binding [GO:1903135]; cuprous ion binding [GO:1903136]; cytokine binding [GO:0019955]; DNA-binding transcription factor binding [GO:0140297]; enzyme binding [GO:0019899]; glyoxalase (glycolic acid-forming) activity [GO:1990422]; identical protein binding [GO:0042802]; kinase binding [GO:0019900]; L-dopa decarboxylase activator activity [GO:0036478]; mercury ion binding [GO:0045340]; mRNA binding [GO:0003729]; oxidoreductase activity, acting on peroxide as acceptor [GO:0016684]; oxygen sensor activity [GO:0019826]; peptidase activity [GO:0008233]; peroxiredoxin activity [GO:0051920]; protein deglycase activity [GO:0036524]; protein homodimerization activity [GO:0042803]; protein-containing complex binding [GO:0044877]; scaffold protein binding [GO:0097110]; signaling receptor binding [GO:0005102]; small protein activating enzyme binding [GO:0044388]; superoxide dismutase copper chaperone activity [GO:0016532]; transcription coactivator activity [GO:0003713]; tyrosine 3-monooxygenase activator activity [GO:0036470]; ubiquitin-like protein conjugating enzyme binding [GO:0044390]; ubiquitin-specific protease binding [GO:1990381]; activation of protein kinase B activity [GO:0032148]; adult locomotory behavior [GO:0008344]; autophagy [GO:0006914]; cellular detoxification of aldehyde [GO:0110095]; cellular detoxification of methylglyoxal [GO:0140041]; cellular response to glyoxal [GO:0036471]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to oxidative stress [GO:0034599]; detection of oxidative stress [GO:0070994]; detoxification of copper ion [GO:0010273]; detoxification of mercury ion [GO:0050787]; DNA repair [GO:0006281]; dopamine uptake involved in synaptic transmission [GO:0051583]; glucose homeostasis [GO:0042593]; glutathione deglycation [GO:0036531]; glycolate biosynthetic process [GO:0046295]; glyoxal metabolic process [GO:1903189]; guanine deglycation [GO:0106044]; guanine deglycation, glyoxal removal [GO:0106046]; guanine deglycation, methylglyoxal removal [GO:0106045]; histone modification [GO:0016570]; hydrogen peroxide metabolic process [GO:0042743]; inflammatory response [GO:0006954]; insulin secretion [GO:0030073]; lactate biosynthetic process [GO:0019249]; membrane depolarization [GO:0051899]; membrane hyperpolarization [GO:0060081]; methylglyoxal catabolic process to lactate [GO:0061727]; methylglyoxal metabolic process [GO:0009438]; mitochondrion organization [GO:0007005]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell death [GO:0060548]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway [GO:2001268]; negative regulation of death-inducing signaling complex assembly [GO:1903073]; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902236]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of gene expression [GO:0010629]; negative regulation of hydrogen peroxide-induced cell death [GO:1903206]; negative regulation of hydrogen peroxide-induced neuron death [GO:1903208]; negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway [GO:1903384]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of neuron death [GO:1901215]; negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway [GO:1905259]; negative regulation of oxidative stress-induced cell death [GO:1903202]; negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway [GO:1903377]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; negative regulation of protein acetylation [GO:1901984]; negative regulation of protein binding [GO:0032091]; negative regulation of protein export from nucleus [GO:0046826]; negative regulation of protein K48-linked deubiquitination [GO:1903094]; negative regulation of protein kinase activity [GO:0006469]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of protein sumoylation [GO:0033234]; negative regulation of protein ubiquitination [GO:0031397]; negative regulation of reactive oxygen species biosynthetic process [GO:1903427]; negative regulation of TRAIL-activated apoptotic signaling pathway [GO:1903122]; negative regulation of ubiquitin-protein transferase activity [GO:0051444]; negative regulation of ubiquitin-specific protease activity [GO:2000157]; peptidyl-arginine deglycation [GO:0036527]; peptidyl-cysteine deglycation [GO:0036526]; peptidyl-lysine deglycation [GO:0036528]; positive regulation of acute inflammatory response to antigenic stimulus [GO:0002866]; positive regulation of androgen receptor activity [GO:2000825]; positive regulation of autophagy of mitochondrion [GO:1903599]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of dopamine biosynthetic process [GO:1903181]; positive regulation of gene expression [GO:0010628]; positive regulation of interleukin-8 production [GO:0032757]; positive regulation of L-dopa biosynthetic process [GO:1903197]; positive regulation of L-dopa decarboxylase activity [GO:1903200]; positive regulation of mitochondrial electron transport, NADH to ubiquinone [GO:1902958]; positive regulation of NAD(P)H oxidase activity [GO:0033864]; positive regulation of oxidative phosphorylation uncoupler activity [GO:2000277]; positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway [GO:1902177]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of protein localization to nucleus [GO:1900182]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of pyrroline-5-carboxylate reductase activity [GO:1903168]; positive regulation of reactive oxygen species biosynthetic process [GO:1903428]; positive regulation of superoxide dismutase activity [GO:1901671]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription regulatory region DNA binding [GO:2000679]; positive regulation of tyrosine 3-monooxygenase activity [GO:1903178]; protein deglycation, glyoxal removal [GO:0036529]; protein deglycation, methylglyoxal removal [GO:0036530]; protein deglycosylation [GO:0006517]; protein stabilization [GO:0050821]; Ras protein signal transduction [GO:0007265]; regulation of androgen receptor signaling pathway [GO:0060765]; regulation of histone acetylation [GO:0035065]; regulation of histone ubiquitination [GO:0033182]; regulation of inflammatory response [GO:0050727]; regulation of mitochondrial membrane potential [GO:0051881]; regulation of neuron apoptotic process [GO:0043523]; regulation of supramolecular fiber organization [GO:1902903]; single fertilization [GO:0007338] | 12446870_show that DJ-1 mutations are associated with PARK7, a monogenic form of human parkinsonism 12548343_Autosomal recessive early onset parkinsonism is linked to three loci: PARK2, PARK6, and PARK7. 12612053_androgen receptor is positively regulated by DJ-1, which antagonizes the function of negative regulators, including DJBP, a novel DJ-1-binding protein 12761214_disease-causing L166P mutation has disrupted the C-terminal region and the dimerization of the protein. 12796482_DJ-1 contains an additional alpha-helix at the C-terminal region, which blocks the putative catalytic site of DJ-1 and appears to regulate the enzymatic activity. 12815653_We describe clinical and neuroimaging features of the 4 patients in the original PARK7-linked kindred. 12851414_mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system 12914946_crystal structure of DJ-1 at 1.6 A resolution locates Leu166 in the middle of a helix and thus predicts that the L166P mutation will bend the helix and impact the dimerization of DJ-1 12939276_structural comparisons between DJ-1 protein, Hsp31, and an Archaea protease lead to the identification of the chaperone activity of DJ-1 14557580_Observational study of gene-disease association. (HuGE Navigator) 14557580_Whether a polymorphism (g.168_185del) within exon 1 of DJ-1 contributes to the risk of sporadic Parkinson disease in a Finnish case-control series was analyzed. This gene does not play a major role in genetic predisposition to PD in this population 14579415_localization in testis, sperm, and semen; DJ-1 may play an important and as yet uncharacterized role in spermatogenesis and fertilization in humans 14598065_By positional cloning within the refined PARK7 critical region we recently identified mutations in the DJ-1 gene in the two PARK7-linked families. 14607841_DJ-1 mutants are rich in beta-strand and alpha-helix conformation and have different suceptibilies to thermal denaturation 14625045_Observational study of gene-disease association. (HuGE Navigator) 14638971_The PARK7 gene is not a common locus for early onset autosomal recessive parkinsonism. 14652021_These results suggest that loss of protective activity of DJ-1 from neuro-toxicity induced by these stresses contributes to neuronal cell death in AR-EOP with mutant DJ-1. 14705119_different neurodegenerative diseases may have similar pathological mechanisms, and that these processes likely include DJ-1 14712351_DJ-1 has a role in neurodegeneration offers and progression of Parkinson's disease (review) 14713311_L166P mutation may contribute to the loss of normal DJ-1 function and are likely to be the underlying cause of early onset PD in affected members of the Italian kindred. 14872018_Early-onset Parkinson disease is associated with single heterozygous loss-of-function DJ-1 mutations, including a heterozygous deletion of exons 5 to 7 and an 11-base pair deletion, removing the invariant donor splice site in intron 5. 14872018_Observational study of gene-disease association. (HuGE Navigator) 14985393_DNA mutational analysis in patients with recessively-inherited early onset parkinsonism 15018843_DJ-1(A NEWLY DISCOVERED MUTANT GENE) IS LINKED TO A GENETICALLY ISOLATED POPULATION IN THE SOUTHWEST OF THE NETHERLANDS AND PRESENTS WITH EARLY-ONSET AUTOSOMAL-RECESSIVE PARKINSONISM. (SEE PAGE 80 UNDER PARK7(DJ-1) 15108293_Observational study of gene-disease association. (HuGE Navigator) 15219840_analyzed other mutants of DJ-1 found in Parkinson's disease patients, including M26I, R98Q, and D149A, as well as L166P 15254937_Mutations in the DJ-1 gene are rare in early-onset Parkinson's disease in both sporadic and familial cases. 15304593_Observational study of gene-disease association. (HuGE Navigator) 15304593_SNP1 (position 4,345 bp) & SNP3 (position 16,491 bp) were significantly associated with Parkinson disease in women. Because the DJ1 protein regulates the androgen receptor, these gender-specific findings may be genuine. 15308309_Observational study of gene-disease association. (HuGE Navigator) 15365989_A new mutation, c.192G>C (p.E64D), associated with early-onset Parkinson disease was found in a Turkish patient. It does not alter the structure, but affects expression & nuclear localization of the protein. 15372597_Observational study of gene-disease association. (HuGE Navigator) 15502874_DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation 15503154_DJ-1 is the third of four genes known to be definitively causal in familial Parkinson disease 15542239_Observational study of gene-disease association. (HuGE Navigator) 15703819_DJ-1 positively regulates p53 through Topors-mediated sumoylation 15717024_linkage to six chromosomal regions and have identified three causative genes: PARK1 (alpha-synuclein), PARK2 (parkin), and PARK7 (DJ-1) in Parkinson disease 15766664_Data show that DJ-1 is a key negative regulator of PTEN that may be a useful prognostic marker for cancer. 15790595_Parkinson disease-associated PARK7 is a transcriptional co-activator that protects against neuronal apoptosis. 15935068_We confirmed the pathological co-localization of DJ-1 with other neurodegenerative disease-associated proteins, as well as the decrease in DJ-1 solubility in disease tissue. In addition, we report the presence of DJ-1 in a large molecular complex. 15944198_DJ-1 is an integral mitochondrial protein that may have important functions in regulating mitochondrial physiology. Our findings of DJ-1's mitochondrial localization may have important implications for understanding the pathogenesis of Parkinson's disease 15970950_Observational study of genotype prevalence. (HuGE Navigator) 15976810_DJ-1 was sumoylated on a lysine residue at K130 by PIASxalpha or PIASy. The K130 mutation abrogated all of the functions of DJ-1, including ras-dependent transformation, cell growth promotion and anti-UV-induced apoptosis activities. 15983381_DJ-1 sequesters Daxx in the nucleus, prevents it from gaining access to the cytoplasm, from binding to and activating its effector kinase apoptosis signal-regulating kinase 1, and therefore, from triggering the ensuing death pathway. 16157901_Observational study of gene-disease association. (HuGE Navigator) 16227205_DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity 16240358_A novel homozygous mutation in exon 7 (E163K) and a new homozygous mutation (g.168_185dup) in the promoter region of the gene of parkinsonism-dementia-amyotrophic lateral sclerosis complex. 16316629_distinct cysteine residues of DJ-1 harbor differential roles in relation to its structure and function 16331561_Data show that DJ1 mutations were rare in Chinese patients with autosomal recessive early-onset Parkinsonism. 16403519_Results suggest that DJ-1 may act as an oxidative-stress-induced chaperone to prevent alpha-synuclein fibrillation. 16517609_DJ-1 oxidative damage has a role in sporadic Parkinson and Alzheimer diseases 16570276_We also demonstrate the expression and secretion of DJ-1 oncoprotein by uveal melanoma cells. 16632486_PINK1 and DJ-1 may have a role in early-onset Parkinson's disease and physically associate and collaborate to protect cells against stress via complex formation 16707095_DJ-1 may have a protective role against oxidative stress during the early stage of Parkinson's Disease 16731528_DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor 16781058_Elevated levels of oxidation of DJ-1 were observed in brain tissues of patients with Parkinson's and Alzheimer's diseases. 17015834_DJ-1's effect on Nrf2 and subsequent effects on antioxidant responses may explain how DJ-1 affects the etiology of both cancer and Parkinson's disease. 17017532_mutations in DJ-1 cause autosomal recessive forms of Parkinson disease-REVIEW 17034344_The Upregulation of DJ-1 were observed in human glioma cells exposed to Reactive Oxygen Species(ROS). 17085780_Review. Loss of DJ-1 causes autosomal recessive PD. DJ-1 inclusions are seen in other alpha-synucleopathies & tauopathies. Its function is still unknown, but it is associated with many cell processes. 17120294_Together, these findings indicate neuronal and synaptic expression of DJ-1 in primate subcortical brain regions and suggest a physiological role for DJ-1 in the survival and/or function of nigral-striatal neurons. 17451229_These results suggest that (i) the M26I substitution and over-oxidation destabilize dimeric DJ-1, and (ii) the oxidation of cysteine 106 contributes to DJ-1 destabilization. 17487420_Oxidative stress to cells abrogates secretion of DJ-1; secreted DJ-1 degrades aggregated transthyretin to protect against the onset of familial amyloidotic polyneuropathy. 17504761_analysis of the DJ-1 protein and its mutants that are associated with Parkinson Disease 17599367_Comparing DJ-1 expression in dopamine neurons on hemi-sections from controls and patients, this study could not detect any difference between controls, idiopathic Parkinson and schizophrenia cases. 17671684_DJ-1 and PA28alpha may have roles in the onset of hepatocarcinogenesis 17846173_Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. 17882163_exerts a neuroprotective effect by reducing ROS-mediated neuronal injury 17949781_These findings suggest that overexpression of EIF-5A2 in colorectal carcinomas may be important in the acquisition of a metastatic phenotype and plays an important role in colorectal carcinoma development and progression. 18045143_Review describes the main advances in our understanding of the function of DJ-1, a ubiquitous protein first described as an oncogene, now implicated in the pathophysiology of Parkinson's disease (PD), and its interaction with other PD-associated proteins. 18162323_DJ-1 was secreted into the serum of both controls and PD patients. There was no difference between the levels of secreted DJ-1 and correlations of levels of DJ-1 with age, clinical severity of PD and level of oxidative stress were not found. 18181649_our results suggest that the protective function of DJ-1 can be compromised by diverse perturbations in its structural integrity, particularly near the junctions of secondary structural elements. 18331584_DJ-1 protein acts as a versatile pro-survival factor in dopaminergic neurons, activating different protective mechanisms in a cellular model of Parkinson's disease. 18430896_DJ-1 is specifically expressed in thyroid carcinomas and not in the normal thyroid; the protein modulates the response to TRAIL-mediated apoptosis in human neoplastic thyroid cells, at least partially through its antioxidant property 18436956_The extent of structural perturbations associated with five different Parkinson's disease linked DJ-1mutations: L166P, E64D, M26I, A104T, and D149A was evaluated. 18486522_Observational study of gene-disease association. (HuGE Navigator) 18570440_Search of the Protein Data Bank (PDB) produces several candidate hydrogen-bonded aspartic/glutamic acid-cysteine interactions, which are particularly common in the DJ-1 superfamily. 18586035_These findings suggest that DJ-1 is secreted through microdomains and that oxidation of DJ-1 at C106 facilitates the secretion. 18626009_DJ-1 associates with RNA targets in cells and the brain, including mitochondrial genes, genes involved in glutathione metabolism, and members of the PTEN/PI3K cascade. 18632777_DJ-1 levels are significantly increased in the cerebrospinal fluid of patients with multiple sclerosis and may be associated wigh disease progression. 18647263_Our data support the conclusion that the major cellular DJ-1 response to stroke in the human brain is astrocytic, and that there is a temporal correlation between DJ-1 expression in these cells and advanced infarct age. 18689799_inactivation of DJ-1 causes decreased expression of the human MnSOD 18704525_Observational study of gene-disease association. (HuGE Navigator) 18706098_MMP-9, DJ-1 and A1BG proteins are overexpressed in pancreatic juice from pancreatic ductal adenocarcinoma 18707128_Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson's disease 18717316_siRNA can effectively inhibit DJ-1 expression, resulting in the reduced proliferation and the enhanced apoptosis in Hep-2 cells. 18722352_DJ-1 might be involved in leukemogesis through regulating cell growth, proliferation, and apoptosis. 18722801_Observational study of gene-disease association. (HuGE Navigator) 18722801_Thie results of this study support the growing importance of mutations in non-coding portion of human genome, and confirm that alterations in DJ-1 are a cause, even if rare, of early-onset Parkinson's disease. 18785233_Mutations of DJ-1 gene are found in Chinese families with autosomal recessive early-onset Parkinsonism 18817430_Proteomic analysis of oxidative stress-responsive proteins in pneumonocytes: insight into the regulation of DJ-1 expression is reported. 18822273_These findings suggest that DJ-1 influences several neuroprotective pathways and that the E163K mutation impairs the mechanism that is more specific to oxidative stress. 18841573_Observational study of gene-disease association. (HuGE Navigator) 18841573_the frequencies of three polymorphisms in DJ-1 (g.168-185del; SNP405, refSNP ID:rs3766606 and 293 G/A) and their association with sporadic Parkinson's disease 18922803_analysis of crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner 18973254_3 new DJ-1 mutations were found: a homozygous 3-base deletion in exon 7 (c.555-557delGCC;p.P158del), a heterozygous duplication involving the first 5 exons, & a heterozygous nonsynonymous substitution in exon 7 (c.535G>A;p.A179T). 18973254_Observational study of gene-disease association. (HuGE Navigator) 18974921_DJ-1 may serve as a novel neuroprotective modality. 19006224_No DJ-1 miutations were found in a Taiwanese cohort of 58 sporadic and 10 familial Parkinson disease patients. 19014922_altered expression of proteins that underlie pathogenesis of SCA17 19023331_These results suggest an active role of DJ-1 missense mutants in the control of cell death and position TTRAP as a new player in the arena of neurodegeneration. 19064576_DJ-1 plays a very important role in transformation and progression of ESCC and may be used as a prognostic marker. 19124468_formation of Cys106-sulfinic acid is a key modification that regulates the protective function of DJ-1. 19144925_Loss of DJ-1 in human cell lines and transformed mouse fibroblasts decreases the transcription of a variety of HIF1-responsive genes during hypoxia. 19224617_Observational study of gene-disease association. (HuGE Navigator) 19224617_This study found that no associations were seen for DJ1 in Australia patient with Parkinson's disease 19229105_Functional ubiquitin E3 ligase complex consisting of parkinson disease (PD)-associated Parkin, PINK1, and DJ-1 to promote degradation of un-/misfolded proteins. 19335984_DJ-1 gene expression may play an important role in the invasion and metastasis of hepatocellular carcinoma. 19371728_DJ-1 does affect the ERK1/2 signaling pathway and change the susceptibility of cells to oxidative stress. 19384955_High DJ-1 expression is associated with clear cell renal cell carcinoma. 19405094_Observational study of gene-disease association. (HuGE Navigator) 19425177_findings suggest an active role for DJ-1 in the autophagic response produced by paraquat, providing evidence for the role of Parkinson's disease-related proteins in the autophagic degradation pathway 19429112_None had causative mutation in DJ-1, suggesting DJ-1 mutation is very rare among patients with familial and sporadic parkinsonism from Asian countries and those with other ethnic background. 19429112_Observational study of gene-disease association. (HuGE Navigator) 19680261_PARK7 protein loss of function could be due to impaired caspase-6 proteolysis and we document the fact that various PARK7 protein mutations could lead to PD pathology through distinct molecular mechanisms. 19686841_REVIEW: DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders 19692353_LRRK2 with other Parkinson's disease causing genes, Parkin, DJ-1 and PINK-1. 19703902_DJ-1-dependent activation of dopamine synthesis occurs through interaction of tyrosine hydroxylase and 4-dihydroxy-L-phenylalanine (L-DOPA) decarboxylase with DJ-1 19716892_Region-specific decreases are observed in DJ-1 mRNA levels in putamen, frontal cortex, parietal cortex and cerebellum in Parkinson's patients, while an up-regulation is observed in the amygdala and entorhinal cortex. 19733211_These results suggest the oxidative modification of DJ-1 in Parkinson's disease patients and the potential application of the antibody for diagnosis of Parkinson's disease at early-stage. 19822128_These findings suggest that DJ-1 is an integral mitochondrial protein and that DJ-1 plays a role in maintenance of mitochondrial complex I activity. 19825160_novel sequence variant in the highly conserved DJ-1 gene significantly reduced DJ-1 promoter activity, which indicates the possible importance of the 16 bp sequence in transcriptional regulation of DJ-1 in Parkinson disease 19968671_Observational study of gene-disease association. (HuGE Navigator) 19968671_polymorphisms located in a region spanning 3535 bp from the promoter to the intron 2 of the DJ-1 gene confer risk to sporadic Parkinson's Disease. 20014998_Loss of DJ-1 occurring through its oxidative modification and subsequent proteolysis mediated through dysregulation of thiol disulfide oxidoreductase may contribute to pathogenesis of sporadic Parkinson's disease. 20061558_DJ-1/p53 interactions contribute to apoptosis resistance in clonal myeloid cells and may serve as a prognostic marker in patients with myelodysplastic syndromes. 20127688_Our data suggest that the increased DJ-1/NuRD interaction is a general anti-stress response regulated by okadaic acid-induced modifications of DJ-1. 20146068_Mutations of DJ-1 and PTEN-induced putative kinase 1 (PINK1) gene are also found in Chinese patients diagnosed with sporadic early onset parkinsonism and mutations of the Parkin gene. 20146068_Observational study of gene-disease association. (HuGE Navigator) 20150646_The present study demonstrated that the survival of patients with astrocytomas was correlated with the nuclear DJ-1 status of the tumor. The DJ-1 molecule might therefore play an important role as a tumor suppressor in astrocytomas. 20156966_Wild-type DJ-1 possesses chaperone activity, which is abolished by the L166P mutation which is reversed by the expression of the cochaperone BAG1. 20186336_Study provides evidence for a critical role of DJ-1 in mitochondrial homeostasis by connecting basal autophagy and mitochondrial integrity in Parkinson's disease. 20213747_DJ-1, by directly inhibiting ASK1, may act as a negative regulator in ASK1 signaling cascades. 20300974_the DJ-1 gene may have an important role in the carcinogenesis of laryngeal carcinoma 20395301_Parkinson disease-associated DJ-1 is required for the expression of the glial cell line-derived neurotrophic factor receptor RET in human neuroblastoma cells 20497343_This is the first study to show that DJ-1 staining intensity in glioblastomas varies directly with p53 nuclear immunoreactivity and inversely with EGFR amplification. 20506312_Studies indicate that molecular genetic analyses have identified five disease genes associated with familial Parkinson disease; SNCA, PARK2, PINK1, PARK7 and LRRK2. 20510502_DJ-1 regulates autophagy through activating JNK-mediated beclin 1 transcription, p62 degradation, thus may play a role in tumorigenesis. 20540987_Total DJ-1 or alpha-synuclein in plasma alone is not useful as biomarkers for Parkinson's disease diagnosis or progression/severity. 20543466_Exon rearrangement mutation in the DJ-1 gene is rare in Chinese patients with autosomal recessive early-onset parkinsonism. 20634198_TRAF6 stimulates the accumulation of insoluble and polyubiquitinated mutant DJ-1 into cytoplasmic aggregates. In human post-mortem brains of Parkinson's disease patients, TRAF6 protein colocalizes with aSYN in Lewy bodies. 20639397_The DJ-1-dependent mitochondrial defects contribute to the increased sensitivity to oxidative stress-induced cell death that has been previously reported. 20673799_upregulated in inflammatory multiple sclerosis lesions 20730377_Studies suggest results suggest an active role for DJ-1 in the autophagic response produced by PQ, opening the door to new strategies for PD therapy. 20736773_Our findings showed, for the first time, overexpression of DJ-1 at both transcriptional and protein levels in gestational trophoblastic disease 20806408_Both L10P and P158DEL DJ-1 mutations alter protein structure in a manner that prevents self-interaction, but both protein variants maintain sufficient protein folding to interact with DJ-1 protein. 20842103_Data show that mitochondrial fragmentation induced by expression of alpha-synuclein is rescued by coexpression of PINK1, parkin or DJ-1 but not the PD-associated mutations PINK1 G309D and parkin Delta1-79 or by DJ-1 C106A. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20938049_Human DJ-1-specific transcriptional activation of tyrosine hydroxylase gene. 20940149_DJ-1 works in parallel to the PINK1/parkin pathway to maintain mitochondrial function in the presence of an oxidative environment 21055329_The expression of DJ-1 protein in laryngeal squamous cell carcinoma is higher than that in control laryngeal mucous tissues. Overexpression of DJ-1 is associated with poor overall survival in LSCC patients. 21097510_DJ-1 enhances cell survival through the binding of Cezanne, a negative regulator of NF-kappaB. 21160133_plays roles in transcriptional regulation and the anti-oxidative stress reaction. 21317550_DJ-1 may act to maintain mitochondrial function during oxidative stress and thereby alter mitochondrial dynamics and autophagy indirectly. 21322020_Two novel DJ-1 mutations, c.91-2A>G affecting splicing and c.319G>C causing Ala107Pro, were observed among patients with AAO of <31 years, suggesting that PD in a high fraction (>12%) of this group of Iranian patients. 21410067_DJ-1 expression is significantly upregulated in hepatocellular carcinoma (HCC), and its expression level correlates with clinicopathological variables and prognosis of HCC patients. 21445310_Abnormally high levels of DJ-1 expression may be involved in endometriosis, possibly by stimulating endometrial cell survival, proliferation, migration, and invasion 21535932_Pathogenic DJ-1 mutations are restricted in certain populations and compared with the parkin gene are unlikely to be of clinical importance in Parkinson's disease patients living in eastern India. 21556124_a positive correlation between DJ-1 and survivin gene expression in laryngeal carcinoma was found 21575935_DJ-1 protein levels are reduced in moderate asthenozoospermia patients; DJ-1 concentration is positively correlated with sperm motility and sperm superoxide dismutase activity indicated by partial correlation analysis. 21645620_DJ-1 associates with membranous organelles including synaptic membranes to exhibit its normal function. 21670963_DJ-1 expression was found to be significantly correlated with lymphatic metastasis of non-small cell lung cancer. 21785459_DJ-1 protects against TRAIL-induced apoptosis through the regulation of death-inducing signaling complex (DISC) formation. 21796667_DJ-1 regulates SOD1 expression through Erk1/2-Elk1 pathway in its protective response to oxidative insult. 21819105_DJ-1 has a role as an oxidative stress sensor and partner of a molecular machinery notorious for its involvement in cell fate decisions 21852238_DJ-1 protects cells against UVB-induced cell death dependent on its oxidation and its association with mitochondrial Bcl-X(L) 21893204_the crystal structure of recombinant DJ-1 at 1.56A resolution was solved, allowing to capture Cys106 in the reduced state for the first time. 21943684_Nuclear factor erythroid-derived 2-like 2 and its stabilizing protein DJ1 affect the prognosis of patients with lung cancer by inducing an elevated stress response to oxidative damage. 22024154_Data suggest that DJ-1 upregulates tyrosine hydroxylase expression by activating its transcription factor Nurr1 via the ERK1/2 pathway. 22066264_Dj-1 protein is a perspective biomarker candidate for PCa. 22132160_Electrophilic p-quinone formed from 6-hydroxydopamine induces DJ-1 oxidation by decreasing intracellular glutathione. 22157000_oxidative stress-dependent chaperone function of YajL and DJ-1 22173095_Data suggest that direct proteasomal endoproteolytic cleavage of PARK7 L166P is the mechanism of degradation contributing to the loss-of-function of the mutant protein. 22223849_data represent the first identification of an important function of DJ-1, which is to regulate the invasion and metastasis properties of pancreatic ductal adenocarcinoma through the ERK/uPA cascade 22233331_Pathogenic DJ1 mutations do not seem to play a major role in Turkey. 22272336_Data show that ENOA, PARK7 and Beta-actin are proper reference standards in obesity studies based on omental fat. 22321799_YajL and possibly DJ-1 function as covalent chaperones involved in the detection of sulfenylated proteins by forming mixed disulfides with them and that these disulfides are subsequently reduced by low-molecular-weight thiols. 22404125_data indicate that breast cancer cells secrete DJ-1 protein in vivo, and that its level is a potential indicator of breast cancer in patients with nipple discharge 22428580_these studies suggest that DJ-1 is involved in the regulation of mitochondrial dynamics through modulation of DLP1 expression 22445250_Pathogenic mutations were not detected in SNCA, PINK1, or DJ-1 in this study. 22492997_wild-type DJ-1 induces the expression of thioredoxin 1, a protein disulfide oxidoreductase 22508282_[review] Targeted markers alpha-synuclein and DJ-1 are two proteins intimately associated with premotor Parkinson's disease pathogenesis. 22511790_This evidence not only provides strong evidence for the role of endogenous Pink1 in neuronal survival, but also supports a role of DJ-1 and Parkin acting parallel or downstream of endogenous Pink1 to mediate survival in a mammalian in vivo context. 22515803_the oxidation of conserved cysteine residue to its sulfinate form (Cys-SO(2)(-)) results in considerable thermal stabilization of both Drosophila DJ-1beta and human DJ-1 22523093_Human DJ-1 is a novel glyoxalases converting glyoxal or methylglyoxal to glycolic or lactic acid. 22526154_Results suggest that the serum level of DJ-1 may be a potential biomarker for pancreatic cancer, and that DJ-1 plays critical roles in the pancreatic tumor chemoresistance. 22532838_L166P mutation of PARK7 gene alters rRNA biogenesis inhibiting TTRAP localization to the nucleolus and enhancing its recruitment into cytoplasmic aggregates with a mechanism that depends in part on TRAF6 activity. 22554508_These results indicate that wild-type DJ-1, but not Parkinson's disease-derived mutant DJ-1, stimulates VMAT2 activity and that C106 is necessary for the stimulating activity of DJ-1 toward VMAT2. 22555455_The generation by caspase 6 of a new N-terminal fragment of DJ-1 with a nuclear localization co-operate to trigger oxidation of DJ-1, loss of mitochondrial integrity, ROS production and finally apoptosis. 22571429_Identification of potential BAG1-DJ1 mutant interaction interfaces. 22664331_DJ-1 protein protected dopaminergic neurons in two PD model rats by increasing antioxidant capacity and inhibiting alpha-synuclein expression. 22666465_DJ-1 participates in metabolism of fatty acid synthesis through transcriptional regulation of the LDLR gene 22836768_Decreased levels of DJ-1 in Fuchs endothelial corneal dystrophy at baseline and under oxidative stress correlate with impaired Nrf2 nuclear translocation and heightened cell susceptibility to apoptosis. 22892098_Our findings suggest that mutant DJ-1(L166P) impairs cells by differentially regulating mitochondrial Bax/Bcl-XL functions. 22956510_This systematic review included information from >5800 unique cases.The weighted mean proportion of cases with PARK2 (parkin), PINK1, and PARK7 (DJ-1) mutations was 8.6%, 3.7%, and 0.4%, respectively. PINK1 mutations were more common in Asian subjects. 22960331_DJ-1 variants were found in 3.9% of Parkinson's patients in India. 22968650_In areas of high, but not low incidence of gastric cancer, both MIF and DJ-1 have elevated serum concentrations in gastric cancer, compared with controls. This suggests there are differt mechanisms of disease pathogenesis in high- and low-incidence areas. 22985211_cell proliferation, migration, and invasion properties of CL1-0, CL1-5, and A549 cells were significantly diminished when the expression of their PARK7 proteins was reduced 23015639_Report changes in DJ-1 cell signaling associated with ischemic/idiopathic cardiomyopathies. 23076395_Cytoplasmic DJ-1 associated with worse survival in ependymoma patients. 23149933_DJ-1 downregulates DUSP1 expression under conditions of oxidative stress. 23151319_Our data suggested that DJ-1 over-expression was linked to nodal status, and might be an independent prognostic marker for patients with SSCC 23182168_A deficiency or constitutive activation of DJ-1 can have implications in mast cell-driven allergic diseases, such as asthma and anaphylaxis. 23183826_Parkinson's disease-associated mutations in DJ-1 modulate its dimerization in living cells. 23241025_the P158Delta point mutation may contribute to neurodegeneration by protein destabilization and hence loss of DJ-1 function. 23250426_promotes angiogenesis and osteogenesis via activation of FGF receptor-1 signaling 23347603_Overexpression of the DJ-1 protein in the placentas of severe preeclampsia patients is thought to be a causative or compensatory mechanism in response to hypoxia. 23418303_The Parkinson disease-related protein DJ-1 counteracts mitochondrial impairment induced by the tumour suppressor protein p53 by enhancing endoplasmic reticulum-mitochondria tethering. 23447676_a parkin-dependent ER-stress-associated modulation of DJ-1 and identifies p53 and XBP-1 as two major actors acting downstream of parkin in this signaling cascade in cells and in vivo. 23530187_DJ-1 is a unique and nonredundant antioxidant that functions independent of other antioxidative pathways in the cellular defense against ROS 23592371_low DJ-1 protein expression is a significant predictor of pathological complete remission after neoadjuvant chemotherapy in breast cancer patients. 23593018_DJ-1beta protects flies against oxidative stress- and UV-induced apoptosis by regulating the subcellular localization and gene expression of DLP, thus implying that Daxx-induced apoptosis is involved in the pathogenesis of DJ-1-associated PD. 23714193_DJ-1 and beta-catenin may contribute to the development and recurrence of glioma 23727824_High DJ-1 expression is associated with endometrial cancer. 23743200_Data suggest that DJ-1 and PYCR1 are on the same pathway of anti-oxidative stress protection of the cells. 23792957_results show that expression of DJ-1 enhances the cells' protective mechanisms against induced metal toxicity and that this protection is lost for DJ-1 PD mutations A104T and D149A 23831022_analysis of the recognition sequence for DJ-1 protease and its interactions with KIF1B and ABL1 23844142_Results show that higher levels of DJ-1 and oxDJ-1 are present in retinal pigment epithelial (RPE) cells from age-related macular degeneration (AMD) donors. 23847046_Results show DJ-1 protein associated with lipid rafts. This novel function of DJ-1 in lipid rafts, may contribute the pathogenesis of Parkinson's Disease. 23902708_Study emphasized the effectiveness of high DJ-1 expression in predicting poor survival of astrocytoma patients, when compared to MIB-1. 23986421_The frequency of the PARK2, PINK1, PARK7 mutations among Polish EO-PD patients seems to be low. 24052110_In the present study, we screened two indel polymorphisms in the 5'UTR of the gene and found that the frequency of both is low in South African PD patients and controls. 24070869_DJ-1 expression and its oxidation state ar | ENSMUSG00000028964 | Park7 | 6.614006e+03 | 0.4998409 | -1.000459158 | 0.2900025 | 1.167820e+01 | 0.0006323662 | 0.09090595 | No | Yes | 4.468454e+03 | 706.215306 | 8.241161e+03 | 1334.854323 | |
| ENSG00000116337 | 271 | AMPD2 | protein_coding | Q01433 | FUNCTION: AMP deaminase plays a critical role in energy metabolism. Catalyzes the deamination of AMP to IMP and plays an important role in the purine nucleotide cycle. {ECO:0000269|PubMed:23911318}. | 3D-structure;Alternative splicing;Disease variant;Hereditary spastic paraplegia;Hydrolase;Metal-binding;Methylation;Neurodegeneration;Nucleotide metabolism;Phosphoprotein;Reference proteome;Zinc | PATHWAY: Purine metabolism; IMP biosynthesis via salvage pathway; IMP from AMP: step 1/1. {ECO:0000269|PubMed:23911318}. | The protein encoded by this gene is important in purine metabolism by converting AMP to IMP. The encoded protein, which acts as a homotetramer, is one of three AMP deaminases found in mammals. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2012]. | hsa:271; | cytosol [GO:0005829]; AMP deaminase activity [GO:0003876]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; AMP metabolic process [GO:0046033]; cyclic purine nucleotide metabolic process [GO:0052652]; energy homeostasis [GO:0097009]; IMP biosynthetic process [GO:0006188]; IMP salvage [GO:0032264] | 12745092_N-terminal extensions of the AMPD2 polypeptide influence ATP regulation of isoform L. 18493842_This is a first report evidencing the pattern of AMPD genes expression in neoplastic human liver. 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 23911318_Study concluded that AMPD2 as necessary for guanine nucleotide biosynthesis and protein translation and provide evidence that AMP deaminase activity is critical during neurogenesis. Patients with mutations in AMPD2 have characteristic brain imaging features of pontocerebellar hypoplasia due to loss of brainstem and cerebellar parenchyma. 24755741_In human HepG2 cells, AMPD2 activation counterregulates AMPK and increases intracellular glucose production, in association with up-regulation of PEPCK and G6Pc. 25496463_tofacitinib increases the cellular levels of adenosine, which is known to have anti-inflammatory activity, through the downregulation of AMPD2. This would be a novel functional aspect of tofacitinib. 28168832_Here we report the clinical and genetic analysis of an individual with PCH9 secondary to a novel missense variant with strong evidence of pathogenicity, located outside the catalytic domain of AMPD2 29463858_The existence of various AMPD2 isoforms with different functions possibly explains the variability in phenotypes associated with AMPD2 variants: variants leaving some of the isoforms intact may cause spastic paraplegia type 63 , while those affecting all isoforms may result in the severe and early-onset Pontocerebellar hypoplasia type 9. 30267407_Data suggest that adenosine monophosphate deaminase 2 (AMPD2) may serve as a biomarker for outcome prediction in undifferentiated pleomorphic sarcoma (UPS). 30577810_These data demonstrate a novel mechanism in Systemic lupus erythematosus development that involves the targeting of AMPD2 expression by NovelmiRNA-25. 31833174_Homozygous variants in AMPD2 and COL11A1 lead to a complex phenotype of pontocerebellar hypoplasia type 9 and Stickler syndrome type 2. | ENSMUSG00000027889 | Ampd2 | 5.007370e+03 | 1.0689723 | 0.096224497 | 0.2704149 | 1.265302e-01 | 0.7220571278 | 0.94084174 | No | Yes | 4.753829e+03 | 488.957554 | 4.349157e+03 | 459.019064 |
| ENSG00000116560 | 6421 | SFPQ | protein_coding | P23246 | FUNCTION: DNA- and RNA binding protein, involved in several nuclear processes. Essential pre-mRNA splicing factor required early in spliceosome formation and for splicing catalytic step II, probably as a heteromer with NONO. Binds to pre-mRNA in spliceosome C complex, and specifically binds to intronic polypyrimidine tracts. Involved in regulation of signal-induced alternative splicing. During splicing of PTPRC/CD45, a phosphorylated form is sequestered by THRAP3 from the pre-mRNA in resting T-cells; T-cell activation and subsequent reduced phosphorylation is proposed to lead to release from THRAP3 allowing binding to pre-mRNA splicing regulatotry elements which represses exon inclusion. Interacts with U5 snRNA, probably by binding to a purine-rich sequence located on the 3' side of U5 snRNA stem 1b. May be involved in a pre-mRNA coupled splicing and polyadenylation process as component of a snRNP-free complex with SNRPA/U1A. The SFPQ-NONO heteromer associated with MATR3 may play a role in nuclear retention of defective RNAs. SFPQ may be involved in homologous DNA pairing; in vitro, promotes the invasion of ssDNA between a duplex DNA and produces a D-loop formation. The SFPQ-NONO heteromer may be involved in DNA unwinding by modulating the function of topoisomerase I/TOP1; in vitro, stimulates dissociation of TOP1 from DNA after cleavage and enhances its jumping between separate DNA helices. The SFPQ-NONO heteromer binds DNA (PubMed:25765647). The SFPQ-NONO heteromer may be involved in DNA non-homologous end joining (NHEJ) required for double-strand break repair and V(D)J recombination and may stabilize paired DNA ends; in vitro, the complex strongly stimulates DNA end joining, binds directly to the DNA substrates and cooperates with the Ku70/G22P1-Ku80/XRCC5 (Ku) dimer to establish a functional preligation complex. SFPQ is involved in transcriptional regulation. Functions as transcriptional activator (PubMed:25765647). Transcriptional repression is mediated by an interaction of SFPQ with SIN3A and subsequent recruitment of histone deacetylases (HDACs). The SFPQ-NONO-NR5A1 complex binds to the CYP17 promoter and regulates basal and cAMP-dependent transcriptional activity. SFPQ isoform Long binds to the DNA binding domains (DBD) of nuclear hormone receptors, like RXRA and probably THRA, and acts as transcriptional corepressor in absence of hormone ligands. Binds the DNA sequence 5'-CTGAGTC-3' in the insulin-like growth factor response element (IGFRE) and inhibits IGF-I-stimulated transcriptional activity. Regulates the circadian clock by repressing the transcriptional activator activity of the CLOCK-ARNTL/BMAL1 heterodimer. Required for the transcriptional repression of circadian target genes, such as PER1, mediated by the large PER complex through histone deacetylation (By similarity). Required for the assembly of nuclear speckles (PubMed:25765647). Plays a role in the regulation of DNA virus-mediated innate immune response by assembling into the HDP-RNP complex, a complex that serves as a platform for IRF3 phosphorylation and subsequent innate immune response activation through the cGAS-STING pathway (PubMed:28712728). {ECO:0000250|UniProtKB:Q8VIJ6, ECO:0000269|PubMed:10847580, ECO:0000269|PubMed:10858305, ECO:0000269|PubMed:10931916, ECO:0000269|PubMed:11259580, ECO:0000269|PubMed:11525732, ECO:0000269|PubMed:11897684, ECO:0000269|PubMed:15590677, ECO:0000269|PubMed:20932480, ECO:0000269|PubMed:25765647, ECO:0000269|PubMed:28712728, ECO:0000269|PubMed:8045264, ECO:0000269|PubMed:8449401}. | 3D-structure;Acetylation;Activator;Alternative splicing;Biological rhythms;Chromosomal rearrangement;Coiled coil;Cytoplasm;DNA damage;DNA recombination;DNA repair;DNA-binding;Direct protein sequencing;Immunity;Innate immunity;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;mRNA processing;mRNA splicing | hsa:6421; | chromatin [GO:0000785]; cytosol [GO:0005829]; dendrite cytoplasm [GO:0032839]; nuclear matrix [GO:0016363]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; paraspeckles [GO:0042382]; RNA polymerase II transcription regulator complex [GO:0090575]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone deacetylase binding [GO:0042826]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; transcription cis-regulatory region binding [GO:0000976]; activation of innate immune response [GO:0002218]; alternative mRNA splicing, via spliceosome [GO:0000380]; chromosome organization [GO:0051276]; dendritic transport of messenger ribonucleoprotein complex [GO:0098963]; double-strand break repair via homologous recombination [GO:0000724]; histone H3 deacetylation [GO:0070932]; innate immune response [GO:0045087]; mRNA processing [GO:0006397]; negative regulation of circadian rhythm [GO:0042754]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway [GO:1902177]; positive regulation of sister chromatid cohesion [GO:0045876]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of circadian rhythm [GO:0042752]; regulation of transcription, DNA-templated [GO:0006355]; rhythmic process [GO:0048511]; RNA splicing [GO:0008380] | 12403470_Here we show that PSF interacts with p54nrb.PSF bind U5 snRNA with both the sequence and structure of stem 1b contributing to binding specificity. 12417296_Review article of PSF as a multi-functional nuclear protein. 12944487_p54nrb and PSF have properties of key factors mediating INS function and likely define a novel mRNA regulatory pathway that is hijacked by HIV-1. 15590677_The PSF.p54(nrb) complex cooperates with Ku protein to form a functional preligation complex with substrate DNA 15668243_Data suggest that polypyrimidine tract-binding protein-associated splicing factor acts as a progesterone receptor corepressor and contributes to the functional withdrawal of progesterone and the initiation of human labor. 16024807_recruitment of PSF to activated promoters and the carboxyl-terminal domain of RNA polymerase II provides a mechanism by which transcription and pre-mRNA processing are coordinated within the cell. 16731528_DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor 16832349_Expression of PSF-TFE3 in renal epithelial cells plays an important role in the initiation and maintenance of oncogenic phenotype in papillary renal cell carcinomas 16938326_Mass spectrometric analysis of the complex revealed the polypyrimidine tract-binding protein-associated splicing factor (PSF) as a novel Hepatitis Delta virus RNA-interacting protein. 17318576_These results suggest that IbeA interacts with polypyrimidine tract-binding protein (PTB)-associated splicing factor (PSF) for the E. coli K1 invasion of HBMEC.[IbeA] 17452459_These data demonstrated that PSF and p54nrb complex with AR and play a key role in modulating AR-mediated gene transcription. 17507659_Identification of PSF, p54(nrb), PTB, and U1A as proteins specifically bound to the COX-2 polyadenylation signal upstream sequence elements . 17537995_identify PSF as a novel nucleophosmin 1/anaplastic lymphoma kinase-binding protein and substrate 17639083_XRN2 associates with p54nrb/NonO(p54)-protein-associated splicing factor (PSF), multifunctional proteins involved in several nuclear processes. 17965020_Findings suggest that the PSF.p54nrb complex is a novel MAP kinase signal-integrating kinases substrate that binds the mRNA for tumor necrosis factor alpha. 18445785_TFII-I, PARP1, and SFPQ proteins, each previously implicated in gene regulation, form a complex controlling transcription of DYX1C1. Allelic differences in the promoter or 5'UTR of DYX1C1 may affect factor binding and thus regulation of the gene. 18655028_Co-expression of PSF relocates oncogenic RING finger protein 43 (RNF43) from the nuclear periphery to the nucleoplasm. 19339282_These findings are suggestive of a role for myometrial PSF as a nuclear co-regulator in the regulation of specific hormone receptor genes and their target hormone response genes. 19447914_results suggest that PSF may have dual functions in homologous recombination and RNA processing through its N-terminal and central regions, respectively. 19809274_Melanotic Xp11 translocation renal cancer: a case with PSF-TFE3 gene fusion and up-regulation of melanogenetic transcripts. 19812236_Disruption of the growth hormone receptor polypyrimidine tract causes aberrant mRNA splicing resulting in growth hormone insensitivity. 19874820_PSF (polypyrimidine tract-binding protein-associated splicing factor) and p54(nrb), two highly related proteins involved in transcription and RNA processing, are identified as new binding partners of hnRNP M. 20421735_SFPQ/NONO heterodimer is involved in the early stage of the DSB response. 20605917_In response to hyperosmotic stress, p38 also regulates formation of complexes between hDlg and PSF. 20932480_GSK3/TRAP150, complex regulates CD45 alternative splicing and demonstrate a paradigm for signal transduction from the cell surface to the RNA processing machinery through the multifunctional protein PSF. 21106524_these results identify PSF as a repressor of STAT6-mediated transcription that functions through recruitment of HDAC to the STAT6 transcription complex, and delineates a novel regulatory mechanism of IL-4 signaling 21144806_PSF influences repair via direct, local, interaction with the DNA substrate 21566083_RVxF motifs play an important role in controlling the multifunctional properties of p54nrb and PSF in the regulation of gene transcription 21881826_Data reveal a new player in tau exon 10 alternative splicing regulation and uncover a previously unknown mechanism of PSF in regulating tau pre-mRNA splicing. 22114566_results indicate that SFPQ/PSF is a host factor essential for influenza virus transcription that increases the efficiency of viral mRNA polyadenylation 22156371_The results suggested that PSF may function as an activator for the meiosis-specific recombinase DMC1. 22213094_Localisation of TopBP1 at DNA damage sites was noticed as early as 5 s following damage induction, whereas p54(nrb) and PSF localised there after 20 s. 22558197_In Alzheimer's disease, Pick's disease, and Frontotemporal Lobar Degeneration Tau-mediates nuclear depletion and cytoplasmic accumulation of SFPQ. 23131771_Partial knockdown of Annexin A2 and PSF showed decrease in p53 IRES activity and reduced levels of both the p53 isoforms. 23158102_PSF and MATR3 are cellular host factors that bind viral RNA and promote Rev activity. 23516550_the effects of PSF on cell proliferation, tumor growth, and cell signaling associated with PPARgamma 23618401_Results suggest that Llme23 can function as an oncogenic RNA and directly associate the polypyrimidine tract-binding (PTB) protein-associated splicing factor (PSF)-binding lncRNA with melanoma. 24288667_The results of this study identify a new physiological role for the PSF-LC3B axis as a potential endogenous modulator of colon cancer treatment. 24507715_NEAT1 plays an important role in the innate immune response through the transcriptional regulation of antiviral genes by the stimulus-responsive cooperative action of NEAT1 and SFPQ. 25025966_MALAT1 binds to SFPQ releasing PTBP2 from the SFPQ/PTBP2 complex, the increased SFPQ-detached PTBP2 promotes cell proliferation and migration in colorectal cancer. 25605962_The ability of the SFPQ/NONO complex to form varying protein assemblies, in conjunction with the effect of post-translational modifications of SFPQ modulating mRNA binding, suggests key roles affecting mRNP dynamics within the cell. 25765647_coiled-coil interaction motif thus provides a molecular explanation for the functional aggregation of SFPQ that directs its role in regulating many aspects of cellular nucleic acid metabolism 26261210_The data suggest a model in which TRAP150 interacts with dimeric PSF to block access of RNA to RRM2, thereby regulating the activity of PSF toward a broad set of splicing events in T cells. 26274027_PSF/SFPQ is a very common gene fusion partner in TFE3 rearrangement-associated perivascular epithelioid cell tumors (PEComas) and melanotic Xp11 translocation renal cancers. 27259250_GAPLINC has a role in promoting colorectal cancer invasion via binding to PSF/NONO and partly by stimulating the expression of SNAI2 27924002_The SFPQ*NONO complex contains an RNA binding domain, and prior work has demonstrated diverse roles in RNA metabolism. It is thus plausible that the additional repair function of NONO, revealed in cell-based assays, could involve RNA interaction. 28315422_Describe the unusual morphology and expanded the morphologic spectrum of SFPQ/PSF-TFE3 renal cell carcinomas. 28633417_PSF was found to be an important internal ribosome entry site trans acting factor (ITAF) for efficient translation of coxsackievirus B3 RNA. 28846091_Mechanistic dissection reveals that NEAT1 broadly interacts with the NONO-PSF heterodimer as well as many other RNA-binding proteins and that multiple RNA segments in NEAT1, including a 'pseudo pri-miRNA' near its 3' end, help attract the Drosha-DGCR8 Microprocessor. 29346433_Vault RNAs partially induces drug resistance of human tumor cells MCF-7 by binding to the RNA/DNA-binding protein PSF and inducing oncogene GAGE6 29376859_SFPQ dislocation was found enhanced with tau co-transfection and tau co-transfection further resulted in extended DNA disorganization in N2a cells. Overall, our results indicate that dysregulation and dislocation of SFPQ and subsequent DNA disorganization might be a novel pathway in the progression of Alzheimer's disease and frontotemporal dementia. 29530979_Analytical ultracentrifugation to estimate the dimerization equilibrium of the SFPQ-containing dimers revealed that the SFPQ-containing dimers dissociate at low micromolar concentrations and that the heterodimers have higher affinities than the homodimer. Moreover, we observed that the apparent dissociation constant for the SFPQ/PSPC1 heterodimer was over 6-fold lower than that of the SFPQ/NONO heterodimer. 29789581_SFPQ intron retention and nuclear loss are molecular hallmarks of familial and sporadic of amyotrophic lateral sclerosis. 30725116_Findings suggest a PARP polymerase (PARP)-dependent role for non-POU domain containing, octamer-binding protein NONO (NONO) and splicing factor proline-glutamine rich protein (SFPQ) in insulin-like growth factor binding protein 3 (IGFBP-3)-dependent DNA double-strand break (DSB) repair. 30931476_Dido3 is an adaptor that controls SFPQ utilization in RNA splicing 31036027_The protein binding study identified the association of host protein Polypyrimidine tract binding protein and associated splicing factor (PSF) with HIV-1 integrase. Thus our study emphasizes the negative influence of PSF on HIV-1 replication. 31981477_SFPQ Is an FTO-Binding Protein that Facilitates the Demethylation Substrate Preference. 32034402_we propose that dysregulation of zinc availability and/or localization in neuronal cells may represent a mechanism for the imbalance in the nucleocytoplasmic distribution of SFPQ, which is an emerging hallmark of neurodegenerative diseases including Alzheimer's disease (AD) and amyotrophic lateral sclerosis (ALS) 32332923_Splicing factor proline- and glutamine-rich (SFPQ) protein regulates platinum response in ovarian cancer-modulating SRSF2 activity. 32661324_AR-induced long non-coding RNA LINC01503 facilitates proliferation and metastasis via the SFPQ-FOSL1 axis in nasopharyngeal carcinoma. 32966782_SFPQ Depletion Is Synthetically Lethal with BRAF(V600E) in Colorectal Cancer Cells. 32998269_The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration. 33284322_Binding and transport of SFPQ-RNA granules by KIF5A/KLC1 motors promotes axon survival. 33476259_The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. 33495278_SFPQ regulates the accumulation of RNA foci and dipeptide repeat proteins from the expanded repeat mutation in C9orf72. 33729478_ALS/FTD-causing mutation in cyclin F causes the dysregulation of SFPQ. | ENSMUSG00000028820 | Sfpq | 1.614111e+04 | 0.9637202 | -0.053313757 | 0.2629479 | 4.193247e-02 | 0.8377485524 | 0.96856922 | No | Yes | 1.622785e+04 | 1906.805844 | 1.538163e+04 | 1853.353981 | ||
| ENSG00000116754 | 9295 | SRSF11 | protein_coding | Q05519 | FUNCTION: May function in pre-mRNA splicing. | Acetylation;Alternative splicing;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes 54-kD nuclear protein that contains an arginine/serine-rich region similar to segments found in pre-mRNA splicing factors. Although the function of this protein is not yet known, structure and immunolocalization data suggest that it may play a role in pre-mRNA processing. Alternative splicing results in multiple transcript variants encoding different proteins. In addition, a pseudogene of this gene has been found on chromosome 12.[provided by RefSeq, Sep 2010]. | hsa:9295; | nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; RNA binding [GO:0003723]; mRNA processing [GO:0006397]; RNA splicing [GO:0008380] | 16943417_SRp54 interacts with a purine-rich element tau in exon 10 and antagonizes Tra2beta, an SR-domain-containing protein that enhances exon 10 inclusion. 18832053_The G2/M arrest induced by MIAs is required for p54(nrb) phosphorylation. CDK activity is required for MIA-induced phosphorylation of p54(nrb). 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24657192_Regulation of gene expression programmes by serine-arginine rich splicing factors. | ENSMUSG00000055436 | Srsf11 | 4.336141e+03 | 0.8440852 | -0.244539477 | 0.3271098 | 5.713655e-01 | 0.4497168220 | 0.86048048 | No | Yes | 3.183182e+03 | 535.105116 | 4.238455e+03 | 730.282578 | |
| ENSG00000116786 | 23207 | PLEKHM2 | protein_coding | Q8IWE5 | FUNCTION: Plays a role in lysosomes movement and localization at the cell periphery acting as an effector of ARL8B. Required for ARL8B to exert its effects on lysosome location, recruits kinesin-1 to lysosomes and hence direct their movement toward microtubule plus ends. Binding to ARL8B provides a link from lysosomal membranes to plus-end-directed motility (PubMed:28325809, PubMed:22172677, PubMed:25898167, PubMed:24088571). Critical factor involved in NK cell-mediated cytotoxicity. Drives the polarization of cytolytic granules and microtubule-organizing centers (MTOCs) toward the immune synapse between effector NK lymphocytes and target cells (PubMed:24088571). Required for maintenance of the Golgi apparatus organization (PubMed:22172677). May play a role in membrane tubulation (PubMed:15905402). {ECO:0000269|PubMed:15905402, ECO:0000269|PubMed:22172677, ECO:0000269|PubMed:24088571, ECO:0000269|PubMed:25898167, ECO:0000269|PubMed:28325809}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Lysosome;Membrane;Phosphoprotein;Reference proteome | This gene encodes a protein that binds the plus-end directed microtubule motor protein kinesin, together with the lysosomal GTPase Arl8, and is required for lysosomes to distribute away from the microtubule-organizing center. The encoded protein belongs to the multisubunit BLOC-one-related complex that regulates lysosome positioning. It binds a Salmonella effector protein called Salmonella induced filament A and is a critical host determinant in Salmonella pathogenesis. It has a domain architecture consisting of an N-terminal RPIP8, UNC-14, and NESCA (RUN) domain that binds kinesin-1 as well as the lysosomal GTPase Arl8, and a C-terminal pleckstrin homology domain that binds the Salmonella induced filament A effector protein. Naturally occurring mutations in this gene lead to abnormal localization of lysosomes, impaired autophagy flux and are associated with recessive dilated cardiomyopathy and left ventricular noncompaction. [provided by RefSeq, Feb 2017]. | hsa:23207; | endosome membrane [GO:0010008]; lysosomal membrane [GO:0005765]; kinesin binding [GO:0019894]; Golgi organization [GO:0007030]; lysosome localization [GO:0032418]; natural killer cell mediated cytotoxicity [GO:0042267]; positive regulation of membrane tubulation [GO:1903527]; regulation of protein localization [GO:0032880] | 15905402_A dynamic process of kinesin recruitment in Salmonella-infected cells is down-regulated by the SifA-mediated recruitment of SKIP on membranes [SKIP] 26464484_The association of PLEKHM2 mutation with dilated cardiomyopathy and left ventricular noncompaction supports the importance of autophagy for normal cardiac function. | ENSMUSG00000028917 | Plekhm2 | 5.264089e+03 | 1.3984025 | 0.483779679 | 0.3236115 | 2.227145e+00 | 0.1356042423 | 0.76582991 | No | Yes | 6.088111e+03 | 823.524927 | 3.771008e+03 | 523.686626 | |
| ENSG00000116885 | 127700 | OSCP1 | protein_coding | Q8WVF1 | FUNCTION: May be involved in drug clearance in the placenta. {ECO:0000269|PubMed:16006562}. | Alternative splicing;Cell membrane;Membrane;Reference proteome;Transport | hsa:127700; | basal plasma membrane [GO:0009925]; cytoplasm [GO:0005737]; plasma membrane [GO:0005886]; transmembrane transporter activity [GO:0022857]; xenobiotic detoxification by transmembrane export across the plasma membrane [GO:1990961] | 12819961_We cloned a novel gene NOR(1), and the Glu58Gly polymorphism of NOR(1) may be involved in the development and/or progression of NPC suggesting that NOR(1) could be a candidate tumor repressor gene related with NPC. 16006562_hOSCP1 is a novel polyspecific organic solute carrier protein responsible for drug clearance from the human placenta 18237537_NOR(1) gene may have some effects on liver cancer by up-regulating the expression of these proteins. 19727524_NOR1 protein is predominantly expressed in human nasopharynx and tracheal tissues and is frequently down-expressed in NPC. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21803736_Frequent epigenetic inactivation of the NOR1 gene in nasopharyngeal carcinoma (NPC) suggests that it may be a critical tumor suppressor involved in the development of nasopharyngeal carcinoma. 23831407_Data revealed novel aspects of the interplay between autophagy and apoptosis in nasopharyngeal carcinoma cells, which underlies the tumor suppression function of NOR1. 26781989_Results identified NOR1 as a downstream target gene of miR-199a which is critical for regulating cell proliferation and migration by targeting and upregulating NOR1 in human HepG2 cells. 29227538_Abberant expression of NOR1 protein in tumor associated macrophages contributes to the development of DEN-induced hepatocellular carcinoma. | ENSMUSG00000042616 | Oscp1 | 2.681782e+02 | 0.7848011 | -0.349601076 | 0.3165970 | 1.234004e+00 | 0.2666297310 | 0.79861760 | No | Yes | 2.210410e+02 | 24.531504 | 2.811960e+02 | 31.546987 | ||
| ENSG00000116984 | 4548 | MTR | protein_coding | Q99707 | FUNCTION: Catalyzes the transfer of a methyl group from methylcob(III)alamin (MeCbl) to homocysteine, yielding enzyme-bound cob(I)alamin and methionine in the cytosol (PubMed:27771510). MeCbl is an active form of cobalamin (vitamin B12) used as a cofactor for methionine biosynthesis. Cob(I)alamin form is regenerated to MeCbl by a transfer of a methyl group from 5-methyltetrahydrofolate (PubMed:27771510). The processing of cobalamin in the cytosol occurs in a multiprotein complex composed of at least MMACHC, MMADHC, MTRR (methionine synthase reductase) and MTR which may contribute to shuttle safely and efficiently cobalamin towards MTR in order to produce methionine (PubMed:27771510). {ECO:0000269|PubMed:27771510, ECO:0000303|PubMed:27771510}. | 3D-structure;Alternative splicing;Amino-acid biosynthesis;Cobalamin;Cobalt;Cytoplasm;Disease variant;Metal-binding;Methionine biosynthesis;Methyltransferase;Phosphoprotein;Reference proteome;Repeat;S-adenosyl-L-methionine;Transferase;Zinc | PATHWAY: Amino-acid biosynthesis; L-methionine biosynthesis via de novo pathway; L-methionine from L-homocysteine (MetH route): step 1/1. | This gene encodes the 5-methyltetrahydrofolate-homocysteine methyltransferase. This enzyme, also known as cobalamin-dependent methionine synthase, catalyzes the final step in methionine biosynthesis. Mutations in MTR have been identified as the underlying cause of methylcobalamin deficiency complementation group G. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, May 2014]. | hsa:4548; | cytosol [GO:0005829]; cobalamin binding [GO:0031419]; methionine synthase activity [GO:0008705]; zinc ion binding [GO:0008270]; axon regeneration [GO:0031103]; cellular response to nitric oxide [GO:0071732]; cobalamin metabolic process [GO:0009235]; methionine biosynthetic process [GO:0009086]; methylation [GO:0032259]; nervous system development [GO:0007399]; pteridine-containing compound metabolic process [GO:0042558]; response to axon injury [GO:0048678] | 10791559_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, gene-environment interaction, and healthcare-related. (HuGE Navigator) 11074217_Observational study of gene-disease association. (HuGE Navigator) 11074524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11186937_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11204591_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 11257268_Observational study of gene-disease association. (HuGE Navigator) 11342450_Observational study of gene-disease association. (HuGE Navigator) 11584084_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11592436_Observational study of gene-disease association. (HuGE Navigator) 11672761_Observational study of genotype prevalence, gene-disease association, and gene-gene interaction. (HuGE Navigator) 11684547_Observational study of gene-gene interaction. (HuGE Navigator) 11920232_polymorphisms are not risk factors for venous thromboembolism [methionine synthase and methylenetrahydrofolate reductase] 12015064_Observational study of gene-disease association. (HuGE Navigator) 12020105_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12068375_Hyperhomocysteinemia due to methionine synthase deficiency, cblG: structure of the MTR gene, genotype diversity, and recognition of a common mutation, P1173L. 12091127_Observational study of gene-disease association. (HuGE Navigator) 12111380_Observational study of gene-disease association. (HuGE Navigator) 12111380_polymorphisms in methionine synthase is associated with increased risk for neural tube defects 12154064_Observational study of gene-disease association. (HuGE Navigator) 12164325_Observational study of genotype prevalence. (HuGE Navigator) 12372672_Observational study of gene-disease association. (HuGE Navigator) 12375236_Variants influence the risk of spina bifida via the maternal rather than the embryonic genotype. 12482550_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12642343_Observational study of gene-disease association. (HuGE Navigator) 12648076_Observational study of gene-disease association. (HuGE Navigator) 12649067_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12649067_results indicate that MTRR and MTR genes may interact to increase the infants' Neural tube defects risks 12670934_B12 supplementation does not alter mRNA or protein turnover rates but induces translational up-regulation of MTR by shifting the mRNA from the ribonucleoprotein to the polysome pool. 12716294_Observational study of gene-disease association. (HuGE Navigator) 12810988_Results of screening mutations 2756A-->G and 66A-->G in MTR and MTRR genes respectively show that are might have an effect on NTDs incidence (neural tube defects). 12855226_Observational study of gene-disease association. (HuGE Navigator) 12876480_Observational study of gene-disease association. (HuGE Navigator) 12893022_Despite absence of association with plasma homocysteine, the GG genotype represented a four-fold increased risk of coronary disease when compared to AA genotype. 12893022_Observational study of gene-disease association. (HuGE Navigator) 12923861_Observational study of gene-disease association. (HuGE Navigator) 12923861_polymorphisms in methionine synthase is associated with increased risk for Down syndrome 12964809_Observational study of genotype prevalence. (HuGE Navigator) 14744749_Observational study of gene-disease association. (HuGE Navigator) 14977639_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 15044114_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15098482_Observational study of gene-disease association. (HuGE Navigator) 15111988_Observational study of genotype prevalence. (HuGE Navigator) 15117811_Polymorphisms of methionine Synthase is associated wtih bladder cancer 15138479_Observational study of gene-disease association. (HuGE Navigator) 15148588_D919G polymorphism of methionine synthase gene has a role in blood pressure response to antihypertensive agents 15148588_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15159311_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15198953_Observational study of gene-disease association. (HuGE Navigator) 15201366_Observational study of gene-disease association. (HuGE Navigator) 15202865_Methionine synthase gene variation in parents is associated with occurrence of congenital heart defects in offspring 15205694_Observational study of gene-disease association. (HuGE Navigator) 15354395_Observational study of gene-disease association. (HuGE Navigator) 15514969_Observational study of gene-environment interaction. (HuGE Navigator) 15551285_Observational study of gene-disease association. (HuGE Navigator) 15598777_Observational study of gene-disease association. (HuGE Navigator) 15612980_Observational study of gene-disease association. (HuGE Navigator) 15719048_Observational study of gene-disease association. (HuGE Navigator) 15790587_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15797993_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15820491_Observational study of gene-disease association. (HuGE Navigator) 15820491_The effect of C677T methylenetetrahydrofolate reductase and A2756G methionine synthase polymorphisms could affect the relative risk for MI is reported. 15866085_Observational study of gene-disease association. (HuGE Navigator) 15889417_Observational study of gene-disease association. (HuGE Navigator) 15894670_Finds that MTR variant AG and AG/GG genotypes associated with a significantly increased squamous cell head and neck cancer risk. 15894670_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15979034_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16006998_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16013960_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16051610_analysis of the B12-responsive internal ribosome entry site (IRES) element in human methionine synthase 16051637_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16115349_Observational study of gene-disease association. (HuGE Navigator) 16142417_Observational study of gene-disease association. (HuGE Navigator) 16142417_The methionine synthase G-allele was significantly higher in the older than in the younger individuals and the influence of the genotype on male subjects became relevant at a younger age as opposed to female subjects suggesting a gender-dependent effect. 16268464_Observational study of gene-disease association. (HuGE Navigator) 16284371_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16328059_Observational study of gene-disease association. (HuGE Navigator) 16333305_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16365025_Observational study of gene-disease association. (HuGE Navigator) 16407418_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16470748_Observational study of gene-disease association. (HuGE Navigator) 16485733_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16580699_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16709328_Observational study of gene-disease association. (HuGE Navigator) 16712703_Mothers with c.2756AG or GG genotype displayed a 2.195-fold increased risk of having a child with non-syndromic cleft lip with or without cleft palate. 16712703_Observational study of gene-disease association. (HuGE Navigator) 16778415_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16799656_Observational study of gene-disease association. (HuGE Navigator) 16861746_Observational study of gene-disease association. (HuGE Navigator) 16894458_Observational study of gene-disease association. (HuGE Navigator) 16947783_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16985020_Clinical trial of gene-disease association. (HuGE Navigator) 17009228_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17024475_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17035141_Observational study of gene-disease association. (HuGE Navigator) 17074544_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17119065_Observational study of gene-disease association. (HuGE Navigator) 17119065_the methionine synthase polymorphism c.2756A>G alters susceptibility to glioblastoma multiforme 17119116_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17151928_Observational study of gene-disease association. (HuGE Navigator) 17152488_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17160942_Observational study of gene-disease association. (HuGE Navigator) 17220339_Observational study of gene-disease association. (HuGE Navigator) 17287836_a significant association of the MTR polymorphism c.2756A>G (D919G) with diastolic blood pressure was found which might explain the beneficial effects of its G-allele on vascular disease and disease-free longevity 17311259_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17311260_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17339646_MTR genotype is not associated with cognitive functioning, cognitive decline, or survival among nonagenarians 17339646_Observational study of gene-disease association. (HuGE Navigator) 17412799_Observational study of gene-disease association. (HuGE Navigator) 17417062_Observational study of gene-disease association. (HuGE Navigator) 17436311_Observational study of gene-disease association. (HuGE Navigator) 17449906_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17450230_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17454638_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17461517_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17522601_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17522601_Serum folate and MTHFR C677T and MTR A2576G gene polymorphisms were the determinants for tHcy levels. 17533396_Observational study of gene-disease association. (HuGE Navigator) 17546637_Observational study of gene-disease association. (HuGE Navigator) 17551576_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17551576_study of whether C677T & A1298C MTHFR, A2756G MTR & -786 C/T eNOS polymorphisms are associated with NonValvular Atrial Fibrillation; data show the polymorphisms were not associated with an increased risk of NVAF per se or in combination 17553479_Observational study of gene-disease association. (HuGE Navigator) 17574963_Observational study of gene-disease association. (HuGE Navigator) 17581676_Observational study of gene-disease association. (HuGE Navigator) 17581676_Results suggest that the alterations of folate metabolism related to MTHFR, MTR and MTRR polymorphisms are not involved in clefting in South Brazil. 17590289_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17595805_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17596206_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17611986_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17617021_Observational study of gene-disease association. (HuGE Navigator) 17636160_Observational study of gene-disease association. (HuGE Navigator) 17655928_Observational study of gene-disease association. (HuGE Navigator) 17655928_These results suggest, for the first time, a role for the MTR A2756G polymorphism in MM risk in our country. 17664238_study found that MTR 2756AG (919DG) or GG (919GG) genotype exhibited 2.005-fold increased risk of systemic lupus erythematosus (95% CI = 1.177-3.416, P = 0.0146) 17691219_Observational study of gene-disease association. (HuGE Navigator) 17691219_The results indicate that of the enzymes studied the polymorphisms of MTR no significant differences in intensity of turnover of circulating thiols between AD and PD patients. 17702010_Observational study of gene-disease association. (HuGE Navigator) 17726616_Observational study of gene-disease association. (HuGE Navigator) 17726616_no association between methionine synthase A2756G polymorphism insertion polymorphism and cancer of the upper gastrointestinal tract. 17822659_Observational study of gene-disease association. (HuGE Navigator) 17853476_Observational study of gene-disease association. (HuGE Navigator) 17853476_The risk of having a child with congenital malformation or FACS was three to four times higher for mothers who were MTHFR 677TT homozygotes compared with MTHFR 677CC homozygotes 17891500_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17904392_Observational study of gene-disease association. (HuGE Navigator) 17925002_Observational study of gene-disease association. (HuGE Navigator) 17967524_Homozygosis for the MTR 2756CC genotype was not related to prostatic carginogenesis 17967524_Observational study of gene-disease association. (HuGE Navigator) 17993766_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17993766_These findings suggest that in obese subjects, homocysteine cycle efficiency is impaired by MTHFR, MTR, and MTRR inability to supply methyl-group donors, providing evidence that MTHFR, MTR, and MTRR gene polymorphisms are genetic risk factors for obesity. 18004208_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18023275_Observational study of gene-disease association. (HuGE Navigator) 18034637_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18053808_Observational study of gene-disease association. (HuGE Navigator) 18060320_Hcy concentrations were significantly increased in the MTR 2756AG heterozygous genotype compared to the MTR 2756AA wild-type genotype. 18060320_Observational study of gene-disease association. (HuGE Navigator) 18174236_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18199722_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18204969_Observational study of gene-disease association. (HuGE Navigator) 18222012_A 2756 A - G polymorhpism in the MTR gene seems to be associated with homocysteine. 18249021_Observational study of gene-disease association. (HuGE Navigator) 18273817_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18273817_The presence of the genetic polymorphism of MTR is a maternal risk factor for Down syndrome in Brazil. 18281142_Observational study of gene-disease association. (HuGE Navigator) 18281142_The SNP 2756 A>G in the methionine synthase gene does not appear to influence the plasma Hcy levels. 18322994_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18329006_Our data did not reveal significant association between the methionine synthase polymorphism and Alzheimer disease development 18351371_Methylenetetrahydrofolate reductase and Methionine synthase polymorphisms might have protective effect on the risk of cervical cancer in the North Indian women. 18351371_Observational study of gene-disease association. (HuGE Navigator) 18378576_Observational study of gene-disease association, gene-gene interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18386810_Observational study of gene-disease association. (HuGE Navigator) 18406541_Observational study of gene-disease association. (HuGE Navigator) 18427977_Observational study of genetic testing. (HuGE Navigator) 18435414_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18447718_Observational study of gene-disease association. (HuGE Navigator) 18447718_The results of this study suggest that genetic variants of methionine metabolism are associated with meningioma formation. 18483342_Observational study of gene-disease association. (HuGE Navigator) 18483342_The MTR A2756G genotype showed a non-significant association between homozygosity and decreased risk of glioblastoma. 18485163_Observational study of gene-disease association. (HuGE Navigator) 18498768_Observational study of gene-disease association. (HuGE Navigator) 18510611_Observational study of gene-disease association. (HuGE Navigator) 18607581_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18620331_Observational study of gene-disease association. (HuGE Navigator) 18635682_Observational study of gene-disease association. (HuGE Navigator) 18636124_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18700049_Methionine synthase is associated with risk and extent of ulcerative colitis, at least in Central China 18700049_Observational study of gene-disease association. (HuGE Navigator) 18771981_Observational study of gene-disease association. (HuGE Navigator) 18771981_This study suggests the maternal and fetal MTR2756G allele is an important risk factor in the development of uteroplacental insufficiency. 18785313_Observational study of gene-disease association. (HuGE Navigator) 18799873_Observational study of gene-disease association. (HuGE Navigator) 18801628_Data did not show differences in the distribution of MTR 2756A>G and MTHFD1 1958G>A polymorphic variants postmenopausal women with and without depression. The MTR GG genotype exhibited a 5.750-fold increased risk of depression in postmenopausal women. 18801628_Observational study of gene-disease association. (HuGE Navigator) 18815869_Observational study and meta-analysis of gene-disease association and gene-gene interaction. (HuGE Navigator) 18815869_combined genotypes of MTHFR and methionine synthase polymorphisms were associated with significantly increased risk of bladder cancer. 18830263_Observational study of gene-disease association. (HuGE Navigator) 18836720_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18842806_Observational study of gene-disease association. (HuGE Navigator) 18842997_Observational study of gene-disease association. (HuGE Navigator) 18842997_The presence of MTR A2756G mutant allele and MTHFR C677T mutant allele in BRCA carriers was associated with increased breast cancer risk. 18843018_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18843018_Risk of pancreatic cancer was increased with alcohol consumption in subjects with the MTR 2756 AA genotype. 18844488_Observational study of gene-disease association. (HuGE Navigator) 18844488_significant but not independent association between the 2756A>G polymorphism of the MTR (presence of G allele) and MI in the Tunisian population 18983896_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18988749_Observational study of genetic testing. (HuGE Navigator) 18992148_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18996879_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19019492_Observational study of gene-disease association. (HuGE Navigator) 19020309_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19034339_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19048631_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19064578_Observational study of gene-disease association. (HuGE Navigator) 19074750_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19112534_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19124508_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19124508_The MTR A2756G interacted with alcohol and folate intake to influence the risk of colorectal adenoma. 19159907_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19177501_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19240236_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19240236_Overall, we found no evidence that common polymorphisms of the MTHFR and MTR genes are associated with promoter methylation of E-cadherin, p16, and RAR-beta2 genes in breast cancer. 19263808_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19283448_A2756G genotype not associated with plasma homocysteine, folate, or methylmalonic acid 19283448_Observational study of gene-disease association. (HuGE Navigator) 19328558_Observational study of gene-disease association. (HuGE Navigator) 19332728_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19336370_Observational study of gene-disease association. (HuGE Navigator) 19349296_MTHFR 677C>T/1298A>G and MTR 2756A>G polymorphism distribution in both recipients (R) and donors (D) showed no significant difference between matches with loss of graft function and those with long-term graft survival 19349296_Observational study of gene-disease association. (HuGE Navigator) 19353223_Observational study of gene-disease association. (HuGE Navigator) 19389261_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19394322_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19420105_Observational study of gene-disease association. (HuGE Navigator) 19427504_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19493349_Observational study of gene-disease association. (HuGE Navigator) 19504454_Observational study of gene-disease association. (HuGE Navigator) 19533788_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19588544_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19588544_The strongest result obtained by this study was for an additive effect between smoking status, slow NAT2 variants, MTR 2756*G and MTHFR 677*T alleles, in affecting bladder cancer risk 19619240_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19649727_Observational study of gene-disease association. (HuGE Navigator) 19649727_The polymorphism distribution of genes encoding MTR, 5,10-methylenetetrahydrofolate dehydrogenase, 5,10-methenyltetrahydrofolate cyclohydrolase and MTHFD1 and MTHFR in patients with larynx cancer, was examined. 19683694_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19706843_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19706844_Meta-analysis of gene-disease association. (HuGE Navigator) 19729796_Maternal folate-related polymorphisms studied here (CBS, MTR, RFC-1, and TC) have no influence on trisomy 21 susceptibility in subjects of Brazilian population. 19729796_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19737740_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19751277_No asssociation between MTR single nucleotide polymorphism 2756A > G and both, follicular lymphoma and diffuse, large B-Cell lymphoma was observed in a cohort of swedish patients. 19760026_Observational study of gene-disease association. (HuGE Navigator) 19760026_there was no significant association between most SNPs in methionine synthase , and the risk of cervical intraepithelial neoplasia and cervical cancer in Korean women. 19774638_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19776626_Polymorphisms in methionine synthase, methionine synthase reductase and serine hydroxymethyltransferase, folate and alcohol intake, and colon cancer risk. 19776634_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19777601_Observational study of gene-disease association. (HuGE Navigator) 19826453_MTR A2756G polymorphism is a candidate gene polymorphism for cancer susceptibility regardless of environmental factors. 19826453_Meta-analysis of gene-disease association. (HuGE Navigator) 19834535_Observational study of gene-disease association. (HuGE Navigator) 19837268_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19838916_A case-control study in Japan failed to find significant associations between polymorphisms of MTR or MTHFR (5,10-ethylenetetrahydrofolate reductase) genes and breast cancer risk. 19838916_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19843671_MTR polymorphisms are associated with colorectal cancer. 19843671_Observational study of gene-disease association. (HuGE Navigator) 19858780_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19936946_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20031554_Observational study of gene-disease association. (HuGE Navigator) 20056620_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20082058_Observational study of gene-disease association. (HuGE Navigator) 20085490_Meta-analysis of gene-disease association. (HuGE Navigator) 20101025_Genetic variation in the methionine synthase is associated with childhood leukemia. 20101025_Observational study of gene-disease association. (HuGE Navigator) 20111902_Meta-analysis of gene-disease association. (HuGE Navigator) 20111902_the MTR A2756G polymorphism may contribute to susceptibility to breast cancer among Europeans. 20180013_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20209990_Observational study of gene-disease association. (HuGE Navigator) 20235210_Uncategorized study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20237949_Observational study of gene-disease association. (HuGE Navigator) 20310006_Observational study of gene-disease association. (HuGE Navigator) 20310006_This study presents evidence for an association between the MTR A2756G polymorphism and retinoblastoma susceptibility in a Northeast population from Brazil 20378615_Observational study of gene-disease association. (HuGE Navigator) 20437058_Observational study of genetic testing. (HuGE Navigator) 20445573_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20445573_Our results suggest that the effects of methylenetetrahydrofolate reductase C677T and methionine synthase A2756G gene polymorphisms may have pivotal roles in the aetiology of essential hypertension and blood pressure response to benazepril. 20447924_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20458436_Observational study of gene-disease association. (HuGE Navigator) 20466634_Data show that MTHFR 677C>T and MTRR 66A>G polymorphisms are two independent risk factors for DS pregnancies in young women, but RFC-1 80G>A and MTR 2756A>G polymorphism are not independent risk factor. 20466634_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20490431_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20490431_data provide evidence that supports an association between MTR polymorphism and the risk of head and neck squamous cell cancer 20532609_Environmental factors played a more important role in urothelial carcinoma carcinogenesis than MTHFR or methionine synthase (MS) gene polymorphism. 20532609_Observational study of gene-disease association. (HuGE Navigator) 20544798_Observational study of gene-disease association. (HuGE Navigator) 20549016_Germ line polymorphisms in the folate- and methyl-associated genes MTHFR, MTR and MTRR, were analyzed in colorectal cancer patient cohort to find a possible link between these genetic variants and p16 hypermethylation. 20549016_Observational study of gene-disease association. (HuGE Navigator) 20615890_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20647221_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20670920_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20723587_Observational study of gene-disease association. (HuGE Navigator) 20737570_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20852008_Observational study of gene-disease association. (HuGE Navigator) 20883119_Observational study of gene-disease association. (HuGE Navigator) 20890936_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20955826_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20960050_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21041608_Observational study | ENSMUSG00000021311 | Mtr | 3.899825e+03 | 0.8911732 | -0.166222310 | 0.2622980 | 3.945771e-01 | 0.5299032239 | 0.88754250 | No | Yes | 3.730333e+03 | 619.277342 | 3.321454e+03 | 565.567740 |
| ENSG00000117114 | 23266 | ADGRL2 | protein_coding | O95490 | FUNCTION: Calcium-independent receptor of low affinity for alpha-latrotoxin, an excitatory neurotoxin present in black widow spider venom which triggers massive exocytosis from neurons and neuroendocrine cells. Receptor probably implicated in the regulation of exocytosis. {ECO:0000250|UniProtKB:O88923}. | Alternative splicing;Disulfide bond;G-protein coupled receptor;Glycoprotein;Lectin;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Transducer;Transmembrane;Transmembrane helix | This gene encodes a member of the latrophilin subfamily of G-protein coupled receptors. The encoded protein participates in the regulation of exocytosis. The proprotein is thought to be further cleaved within a cysteine-rich G-protein-coupled receptor proteolysis site into two chains that are non-covalently bound at the cell membrane. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2014]. | hsa:23266; | integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; neuron projection [GO:0043005]; carbohydrate binding [GO:0030246]; G protein-coupled receptor activity [GO:0004930]; latrotoxin receptor activity [GO:0016524]; adenylate cyclase-activating G protein-coupled receptor signaling pathway [GO:0007189]; brain development [GO:0007420]; cell surface receptor signaling pathway [GO:0007166]; G protein-coupled receptor signaling pathway [GO:0007186] | 30340542_The results of this study suggest that this ADGRL2 variation impedes the proper development of the cerebellum resulting in RES which is thought to occur when the cerebellar primordium develops and probably results from abnormal function of genes expressed during initial patterning of the mesencephalon-rhombencephalon 33798451_Adhesion GPCR Latrophilin-2 Specifies Cardiac Lineage Commitment through CDK5, Src, and P38MAPK. 34581723_LPHN2 inhibits vascular permeability by differential control of endothelial cell adhesion. | ENSMUSG00000028184 | Adgrl2 | 7.467065e+02 | 0.7923774 | -0.335740272 | 0.3300241 | 1.006655e+00 | 0.3157055252 | 0.81949717 | No | Yes | 7.359741e+02 | 162.251031 | 6.592645e+02 | 149.081337 | |
| ENSG00000117360 | 9129 | PRPF3 | protein_coding | O43395 | FUNCTION: Plays role in pre-mRNA splicing as component of the U4/U6-U5 tri-snRNP complex that is involved in spliceosome assembly, and as component of the precatalytic spliceosome (spliceosome B complex). {ECO:0000269|PubMed:26912367, ECO:0000269|PubMed:28781166, ECO:0000305|PubMed:20595234}. | 3D-structure;Alternative splicing;Direct protein sequencing;Disease variant;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Retinitis pigmentosa;Spliceosome;Ubl conjugation;mRNA processing;mRNA splicing | The removal of introns from nuclear pre-mRNAs occurs on complexes called spliceosomes, which are made up of 4 small nuclear ribonucleoprotein (snRNP) particles and an undefined number of transiently associated splicing factors. This gene product is one of several proteins that associate with U4 and U6 snRNPs. Mutations in this gene are associated with retinitis pigmentosa-18. [provided by RefSeq, Jul 2008]. | hsa:9129; | Cajal body [GO:0015030]; cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spliceosomal complex [GO:0005681]; U2-type precatalytic spliceosome [GO:0071005]; U4/U6 x U5 tri-snRNP complex [GO:0046540]; identical protein binding [GO:0042802]; RNA binding [GO:0003723]; mRNA processing [GO:0006397]; mRNA splicing, via spliceosome [GO:0000398]; RNA splicing [GO:0008380]; RNA splicing, via transesterification reactions [GO:0000375]; spliceosomal tri-snRNP complex assembly [GO:0000244] | 11773002_Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. 11773002_The human HPRP3 gene, the orthologue of the yeast pre-mRNA splicing factor (PRP3) 11971898_role in the recruitment of Hprp4p for the U4/U6 snRNP assembly 12714658_Nine mutations, six of which are novel, in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31, causing adRP have been identified in the Spanish population. 12875835_Free and complexed cyclophilin H have virtually identical conformations suggesting that the U4/U6-60K binding site is pre-shaped and the peptidyl-prolyl-cis/trans isomerase activity is unaffected by complex formation 15085354_We conclude that the Thr494Met mutation in the HPRP3 gene causes ADRP in Japanese patients. This mutation was found in 1% of patients with ADRP in Japan 15541726_PAP-1 interacted with Prp3p but not Prp31p in human cells and yeast, and the basic region of PAP-1 and the C-terminal region of Prp3p, regions beside spots found in retinitis pigmentosa mutations, were needed for binding 17517693_splicing factor PRPF3 mutations cause retinal degeneration and form detrimental aggregates in photoreceptor cells 17932117_Findings suggest that the loss of Hprp3p phosphorylation at Thr494 is a key step for initiating Thr494Met aberrant interactions within U4/U6 snRNP complex and these are likely linked to the retinitis pigmentosa type 18 phenotype. 18026141_splicing activity is significantly influenced by the CK2-hPrp3p interaction 18211889_U2AF35 and hPrp3 interactions with SPF30 can occur simultaneously, thereby potentially linking 3' splice site recognition with tri-small nuclear ribonucleoprotein addition 18395097_TASP1, EPS15R, and PRPF3 expression were significantly induced in HCCs of transgenic EGF2B mice as was P2 promoter-driven HNF4alpha 18553058_These data support the notion about individual roles for CK2alpha and CK2alpha' in the splicing process. 20309403_A mutation in the PRPF3 gene is rare compared to other genes causing autosomal dominant retinitis pigmentosa. 20801516_Observational study of genetic testing. (HuGE Navigator) 21378395_RP-PRPF defects affect the stoichiometry of spliceosomal small nuclear RNAs. Mutant PRPF3 proteins stably associated with tri-snRNPs. 27886254_Results from whole-exome sequencing identified 2 variants c.1345C > G (p.R449G) and c.1532A > C (p.H511P) in PRPF3 which co-segregate with retinitis pigmentosa in two families respectively. 28379520_SUMO conjugation plays a role during mRNA splicing processes including a role for Prp3 SUMOylation in U4/U6*U5 tri-snRNP formation and/or recruitment. | ENSMUSG00000015748 | Prpf3 | 2.212266e+03 | 0.9777893 | -0.032404476 | 0.2871572 | 1.267755e-02 | 0.9103519289 | 0.98338795 | No | Yes | 2.141011e+03 | 273.084816 | 2.270750e+03 | 297.041243 | |
| ENSG00000117411 | 8704 | B4GALT2 | protein_coding | O60909 | FUNCTION: Responsible for the synthesis of complex-type N-linked oligosaccharides in many glycoproteins as well as the carbohydrate moieties of glycolipids (PubMed:9405390). Can produce lactose (PubMed:9405390). {ECO:0000269|PubMed:9405390}. | Alternative splicing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Manganese;Membrane;Metal-binding;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | This gene is one of seven beta-1,4-galactosyltransferase (beta4GalT) genes. They encode type II membrane-bound glycoproteins that appear to have exclusive specificity for the donor substrate UDP-galactose; all transfer galactose in a beta1,4 linkage to similar acceptor sugars: GlcNAc, Glc, and Xyl. Each beta4GalT has a distinct function in the biosynthesis of different glycoconjugates and saccharide structures. As type II membrane proteins, they have an N-terminal hydrophobic signal sequence that directs the protein to the Golgi apparatus and which then remains uncleaved to function as a transmembrane anchor. By sequence similarity, the beta4GalTs form four groups: beta4GalT1 and beta4GalT2, beta4GalT3 and beta4GalT4, beta4GalT5 and beta4GalT6, and beta4GalT7. The enzyme encoded by this gene synthesizes N-acetyllactosamine in glycolipids and glycoproteins. Its substrate specificity is affected by alpha-lactalbumin but it is not expressed in lactating mammary tissue. Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2011]. | hsa:8704; | Golgi apparatus [GO:0005794]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity [GO:0003831]; galactosyltransferase activity [GO:0008378]; lactose synthase activity [GO:0004461]; metal ion binding [GO:0046872]; N-acetyllactosamine synthase activity [GO:0003945]; carbohydrate metabolic process [GO:0005975]; cerebellar Purkinje cell layer development [GO:0021680]; glycosylation [GO:0070085]; locomotory behavior [GO:0007626]; memory [GO:0007613]; protein glycosylation [GO:0006486]; visual learning [GO:0008542] | 15504978_Significance of core 2 GlcNAc-T in the pathogenesis of capillary occlusion in diabetic retinopathy. 15939404_Thus, these results suggest that among human beta4GalTs, beta4GalT-II is a major regulator of the synthesis of glycans involved in neuronal development. 17470362_beta4GalT-II increases HeLa cell apoptosis induced by cisplatin depending on its Golgi localization, which indicates that beta4GalT-II might contribute to the therapeutic efficiency of cisplatin for cervix cancer. 18211920_results suggested that beta1,4GalT II might serve as a target gene of p53 transcription factor during adriamycin-induced HeLa cell apoptosis, which elucidated a new mechanism of p53-mediated cell apoptosis 19524017_Data show that Beta4-Gal-transferase (beta4GalT) extends core 2 and forms the backbone structure for biologically important epitopes. 27004849_Study provides evidence that mutations in B3GNT2, B4GALT2, and ST6GALNAC2 underlie aberrant glycosylation, and contribute to the pathogenesis of molecular subsets of colon and other gastrointestinal malignancies. 30020015_In individuals under dual antiplatelet therapy, B4GALT2 c.909C>T is an independent genetic predictor of on-treatment platelet reactivity. | ENSMUSG00000028541 | B4galt2 | 4.550319e+03 | 1.4277234 | 0.513716474 | 0.3277501 | 2.459139e+00 | 0.1168428426 | 0.75783482 | No | Yes | 5.273350e+03 | 750.888995 | 3.075363e+03 | 449.642910 |
| ENSG00000117448 | 10327 | AKR1A1 | protein_coding | P14550 | FUNCTION: Catalyzes the NADPH-dependent reduction of a wide variety of carbonyl-containing compounds to their corresponding alcohols. Displays enzymatic activity towards endogenous metabolites such as aromatic and aliphatic aldehydes, ketones, monosaccharides and bile acids, with a preference for negatively charged substrates, such as glucuronate and succinic semialdehyde (PubMed:10510318). Functions as a detoxifiying enzyme by reducing a range of toxic aldehydes. Reduces methylglyoxal and 3-deoxyglucosone, which are present at elevated levels under hyperglycemic conditions and are cytotoxic. Involved also in the detoxification of lipid-derived aldehydes like acrolein (By similarity). Plays a role in the activation of procarcinogens, such as polycyclic aromatic hydrocarbon trans-dihydrodiols, and in the metabolism of various xenobiotics and drugs, including the anthracyclines doxorubicin (DOX) and daunorubicin (DAUN) (PubMed:18276838, PubMed:11306097). Displays no reductase activity towards retinoids (By similarity). {ECO:0000250|UniProtKB:P50578, ECO:0000250|UniProtKB:P51635, ECO:0000269|PubMed:10510318, ECO:0000269|PubMed:11306097, ECO:0000269|PubMed:18276838}. | 3D-structure;Acetylation;Cell membrane;Cytoplasm;Direct protein sequencing;Lipid metabolism;Membrane;NADP;Oxidoreductase;Phosphoprotein;Reference proteome | This gene encodes a member of the aldo/keto reductase superfamily, which consists of more than 40 known enzymes and proteins. This member, also known as aldehyde reductase, is involved in the reduction of biogenic and xenobiotic aldehydes and is present in virtually every tissue. Multiple alternatively spliced transcript variants of this gene exist, all encoding the same protein. [provided by RefSeq, Jan 2011]. | hsa:10327; | apical plasma membrane [GO:0016324]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; synapse [GO:0045202]; alditol:NADP+ 1-oxidoreductase activity [GO:0004032]; aldo-keto reductase (NADP) activity [GO:0004033]; allyl-alcohol dehydrogenase activity [GO:0047655]; D-threo-aldose 1-dehydrogenase activity [GO:0047834]; glucuronolactone reductase activity [GO:0047941]; glycerol dehydrogenase [NADP+] activity [GO:0047956]; L-glucuronate reductase activity [GO:0047939]; methylglyoxal reductase (NADPH-dependent, acetol producing) [GO:1990002]; aldehyde catabolic process [GO:0046185]; cellular detoxification of aldehyde [GO:0110095]; D-glucuronate catabolic process [GO:0042840]; daunorubicin metabolic process [GO:0044597]; doxorubicin metabolic process [GO:0044598]; glucuronate catabolic process to xylulose 5-phosphate [GO:0019640]; glutathione derivative biosynthetic process [GO:1901687]; L-ascorbic acid biosynthetic process [GO:0019853]; lipid metabolic process [GO:0006629] | 15769597_structure of Apo R268A human aldose reductase reveals hinges and latches that control the kinetic mechanism 16543247_Observational study of gene-disease association. (HuGE Navigator) 17149600_Observational study of gene-disease association. (HuGE Navigator) 18495158_the binding site residues deviating between ALR1 and ALR2 influence ligand affinity in a complex interplay, presumably involving changes of dynamic properties and differences of the solvation/desolvation balance upon ligand binding 18676680_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19423521_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 21276435_AKR1A1 has little effect on the production of gamma-hydroxybutyrate 23643085_AKR1A1 could be involved in the metabolism of 4-hydroxynonenal and play a role in the resistance to oxidative stress 23747692_prostaglandin F synthase activity of human and bovine aldo-keto reductases 29763686_Data (including data from studies in knockout and transgenic mice) suggest that AKR1A1 in liver is involved in bioactivation of xenobiotic/carcinogen thioacetamide (TAA); Akr1a-/- knockout mice are resistant to TAA-induced liver injury/hepatotoxicity. 33158254_(5-Hydroxy-4-oxo-2-styryl-4H-pyridin-1-yl)-acetic Acid Derivatives as Multifunctional Aldose Reductase Inhibitors. | ENSMUSG00000028692 | Akr1a1 | 3.320777e+03 | 1.1636889 | 0.218705414 | 0.2974751 | 5.436220e-01 | 0.4609355172 | 0.86547338 | No | Yes | 3.425385e+03 | 405.821021 | 2.562073e+03 | 311.797406 | |
| ENSG00000117592 | 9588 | PRDX6 | protein_coding | P30041 | FUNCTION: Thiol-specific peroxidase that catalyzes the reduction of hydrogen peroxide and organic hydroperoxides to water and alcohols, respectively (PubMed:9497358, PubMed:10893423). Can reduce H(2)O(2) and short chain organic, fatty acid, and phospholipid hydroperoxides (PubMed:10893423). Also has phospholipase activity, can therefore either reduce the oxidized sn-2 fatty acyl group of phospholipids (peroxidase activity) or hydrolyze the sn-2 ester bond of phospholipids (phospholipase activity) (PubMed:10893423, PubMed:26830860). These activities are dependent on binding to phospholipids at acidic pH and to oxidized phospholipds at cytosolic pH (PubMed:10893423). Plays a role in cell protection against oxidative stress by detoxifying peroxides and in phospholipid homeostasis (PubMed:10893423). Exhibits acyl-CoA-dependent lysophospholipid acyltransferase which mediates the conversion of lysophosphatidylcholine (1-acyl-sn-glycero-3-phosphocholine or LPC) into phosphatidylcholine (1,2-diacyl-sn-glycero-3-phosphocholine or PC) (PubMed:26830860). Shows a clear preference for LPC as the lysophospholipid and for palmitoyl CoA as the fatty acyl substrate (PubMed:26830860). {ECO:0000269|PubMed:10893423, ECO:0000269|PubMed:26830860, ECO:0000269|PubMed:9497358}. | 3D-structure;Acetylation;Antioxidant;Cytoplasm;Direct protein sequencing;Hydrolase;Lipid degradation;Lipid metabolism;Lysosome;Multifunctional enzyme;Oxidoreductase;Peroxidase;Phosphoprotein;Redox-active center;Reference proteome;Transferase | Mouse_homologues NA; + ;NA | The protein encoded by this gene is a member of the thiol-specific antioxidant protein family. This protein is a bifunctional enzyme with two distinct active sites. It is involved in redox regulation of the cell; it can reduce H(2)O(2) and short chain organic, fatty acid, and phospholipid hydroperoxides. It may play a role in the regulation of phospholipid turnover as well as in protection against oxidative injury. [provided by RefSeq, Jul 2008]. | hsa:9588; | azurophil granule lumen [GO:0035578]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; membrane [GO:0016020]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; 1-acylglycerophosphocholine O-acyltransferase activity [GO:0047184]; cadherin binding [GO:0045296]; calcium-independent phospholipase A2 activity [GO:0047499]; glutathione peroxidase activity [GO:0004602]; identical protein binding [GO:0042802]; phospholipase A2 activity [GO:0004623]; thioredoxin peroxidase activity [GO:0008379]; ubiquitin protein ligase binding [GO:0031625]; cell redox homeostasis [GO:0045454]; cellular oxidant detoxification [GO:0098869]; glycerophospholipid catabolic process [GO:0046475]; positive regulation of mRNA splicing, via spliceosome [GO:0048026]; response to oxidative stress [GO:0006979] | 12121978_Data show that p29 is a novel protein that associates with p67, has peroxiredoxin activity, and has a potential role in protecting the NADPH oxidase by inactivating H(2)O(2) or altering signaling pathways affected by H(2)O(2) [neutrophil protein p29 ] 12193653_overexpression protects cells against phospholipid peroxidation-mediated membrane damage [1-cys peroxiredoxin] 12650976_The significant increase in peroxiredoxin 6 level in frontal cortex of patients with Pick's disease is useful in discriminating it from Down syndrome/Alzheimer's disease. 14751239_AOP2 protects hyperglycemia-induced lens epithelial cell apoptosis; this molecule may have the potential to prevent hyperglycemia-mediated complications in diabetes 15890616_Prdx6 is an important antioxidant enzyme and has a major role in lung phospholipid metabolism [review] 15941719_Protects HeLa cells from H(2)O(2)-induced cell death. 16186110_Saitohin interacts with peroxiredoxin 6, a unique member of that family that is bifunctional and the levels of which increase in Pick disease. 16330552_SP-A and Prdx6 directly interact, which provides a mechanism for regulation of the PLA(2) activity of Prdx6 by SP-A 17653765_overexpression of peroxiredoxin 6 is associated with oligodendroglioma 17980029_Overexpression of peroxiredoxin 6 leads to a more invasive phenotype and metastatic potential in human breast cancer, at least in part, through regulation of the levels of uPAR, Ets-1, MMP-9, RhoC and TIMP-2 expression. 18025307_PRDX6 is required for blood vessel integrity in wounded skin. 18386021_In brain tissue of patients with Alzheimer's disease , many blood vessels exhibited peroxiredoxin 6 staining that appeared to be due to the astrocytic foot processes. 18619034_two potential interaction partners of Prx6: the calcium-activated cysteine endopeptidase calpain and the p50RhoGAP protein of the family of Sec14-like proteins. 18826942_H2O2-mediated hyperoxidation of Prdx6 induces cell cycle arrest at the G2/M transition through up-regulation of iPLA2 activity. 18973804_expression is upregulated during response to oxidative stress 19140803_The MAPKs can mediate phosphorylation of Prdx6 at Thr-177 with a consequent marked increase in its aiPLA(2) activity. 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19236840_Prdx6 binds to reduced phospholipid at acidic pH but at cytosolic pH binds only phospholipid that is oxidized compatible with a role for Prdx6 in the repair of peroxidized cell membranes 19405953_This protein has been found differentially expressed in the Wernicke's Area from patients with schizophrenia. 19566940_Overexpression of peroxiredoxin I and is associated with breast carcinoma. 19572226_PRDX6 attenuates oxidative stress- and TGFbeta-induced abnormalities of human trabecular meshwork cells. 19700648_The localization of Prdx6 in acidic organelles and consequent PLA2 activity depend on a novel 10-aa peptide located at positions 31-40 of the protein. 19889963_Extrinsic PRDX6 prevents UV-triggered cell death and abnormal protein expression in PRDX6-deficient lens epitheial cells. 19937138_promotes lung cancer cell invasion by inducing urokinase-type plasminogen activator via p38 kinase, phosphoinositide 3-kinase, and Ak 20237496_Observational study of gene-disease association. (HuGE Navigator) 20354123_This study is the first to define the functions of the enzymatic activities of PRDX6 in metastasis and to show the involvement of arachidonic acid in PRDX6 action in intact cells. 20485444_Observational study of gene-disease association. (HuGE Navigator) 20532202_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20796224_It could be detected that PRDX6 is associated with tumorigenesis in tonge squamous cell carcinoma. 20829884_Prx6 modulates TRAIL signaling as a negative regulator of caspase-8 and caspase-10 in death-inducing signaling complex formation of TRAIL-resistant metastatic cancer cells. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21166495_The antioxidant Prdx 6 protects ovarian cancer cells against cisplatin-induced apoptosis via inactivation of the caspase signaling pathway. 21346153_Results suggest that ERK and p38 MAPK regulate subcellular localization of Prdx6 by activation of 14-3-3epsilon as a chaperone protein, resulting in its translocation to acidic organelles. 21415860_Results provide new insights into the distinct roles of bifunctional Prdx6 with peroxidase and PLA(2) activities in oxidative stress-induced and TNF-induced apoptosis, respectively. 21627785_Study revealed novel SNPs within the PRDX6 gene and its 5' and 3' flanking regions via direct sequencing. 22178385_Data suggest that Prdx6 serves an additional biochemical or structural role in supporting optimal NADPH oxidase activity. 22215146_Data suggest that SNPs of peroxiredoxin 1, 2 and 6 are not associated with esophageal cancer. 22236188_expression and function of Prdx1 and Prdx6 in MCF-7 and noncancerous MCF-10A cell lines; found elevated Prdx1 expression in MCF-7 cells and comparable expression of Prdx6; data suggest synergistic role for Prdx1 and Prdx6 in MCF-10A cells 22492841_decreased levels, combined with higher thiol oxidation (and probable inhibition) of PRDXs (particularly PRDX6), are noted when human sperm function is altered. 22663767_a change in the conformation of Prdx6 upon its phosphorylation is the basis for enhancement of PLA(2) enzymatic activity 22678913_(2) activity of Prdx6-PLA(2) in intact cells mediates its ability to enhance phox activity in response to fMLF 22985558_The protein and mRNA expressions of Prx1 and Prx6 increased significantly in the order of normal brain tissue, grade II astrocytoma, grade III astrocytoma and grade IV astrocytoma. 23113308_Drug resistance formation was accompanied by a significant increase in the expression of PRDX1, PRDX2, PRDX3, PRDX6 genes in all cancer cell strains, which confirms the importance of redox-dependent mechanisms into development of cisplatin resistance. 23158669_The level of Prx6 protein is lower in gastric cancer tissues than in normal para-cancer tissue. Prx6 expression is significantly correlated with the differentiation degree of GC. 23164639_Oxidation of the catalytic cysteine in Prdx6 is required for its interaction with PiGST, the interaction plays an important role in regenerating the peroxidase activity of Prdx6. 23643677_overexpression of PRDX6 promotes lung tumor growth via increased glutathione peroxidase and iPLA2 activities through the upregulation of the AP-1 and JNK pathways. 23815338_KGF can activate an antioxidant response element in a promoter without reactive oxygen species involvement and that KGF and Dex can synergistically activate the PRDX6 promoter and protect cells from oxidative stress 24316730_Hypoexpression of PRDX6 is associated with papillary thyroid carcinomas. 24391750_Apoptosis related proteins ERp29, PRDX6 and MPO were differentially expressed in placentas of pregnant women with intrahepatic cholestasis and in healthy pregnant women. 24512906_our findings indicate that PRDX6 promotes lung tumor growth via increased glutathione peroxidase and iPLA2 activities. 24910119_Reactive oxygen species-evoked aberrant sumoylation signaling affects Prdx6 activity by reducing Prdx6 abundance. 25748205_Several regions of reduced PRDX6 are in a substantially different conformation from that of the crystal structure of the peroxidase catalytic intermediate. The differences between the two structures likely reflect catalysis-related conformational changes. 25935550_High intensity of cytoplasmic Prx VI expression in pretreatment diffuse large B-cell lymphoma samples predicts worse outcome. 25938937_PRDX6 may serve as a biomarker for traumatic brain injury and that autoimmune profiling is a viable strategy for the discovery of novel biomarkers 26279427_PRDX6 protects ARPE-19 cells from hydrogen peroxide-induced oxidative stress and apoptosis. 26285655_AA (arachidonic acid) as a crucial effector of PRDX6-dependent proliferation and inducer of Src family kinase activation. 26293541_PRDX6 inhibited the carcinogenesis of hepatocellular carcinoma, and the calcium-independent phospholipase A2 activity of PRDX6 promoted cancer cell death induced by TNF-alpha. 26398495_PRDX6-anion exchanger 1 interaction contributes to the maintenance of anion exchanger 1 during cellular stress such as during metabolic acidosis. 26447207_Delivery of a protein transduction domain-mediated Prdx6 protein protected against oxidative stress-evoked neuronal cell death. 26560306_Our study provides new insight into the initial regulatory mechanisms of mitophagy and reveals the protective role of PRDX6 in the clearance of damaged mitochondria. 26647763_Prdx6 and its PLA2 activity have a protective role in donors after brain death livers. 26830860_Prdx6 is a complete enzyme comprising both PLA2and LPCAT activities 26891882_Peroxidase activity was markedly reduced by mutation at either of the Leu sites and was essentially abolished by the double mutation, while PLA2 activity was unaffected. Decreased peroxidase activity following mutation of the interfacial leucines presumably is mediated via impaired heterodimerization of Prdx6 with piGST that is required for reduction and re-activation of the oxidized enzyme. 26921317_Thr can substitute for Ser for the enzymatic activities of Prdx6 but not for its targeting to LB. These results confirm an important role for LB Prdx6 in the degradation and remodeling of lung surfactant phosphatidylcholine. 27094494_the transition-state substrate analogue inhibitor of Prdx6 phospholipase A2 activity (MJ-33) was shown to suppress Nox1 activity, suggesting Nox1 activity is regulated by the phospholipase activity of Prdx6. Finally, wild type Prdx6, but not lipase or peroxidase mutant forms, supports Nox1-mediated cell migration in the HCT-116 colon epithelial cell model of wound closure 27353378_Thus far only the crystal structure of Prx in the oxidized state has been reported. In this study, we present the crystal structures of human Prx6 in the reduced (SH) and the sulfinic acid (SO2H) forms. 27484502_These findings implicate miR-371-3p as a suppressor of PRDX6 and suggest that co-targeting of peroxiredoxin 6 or modulating miR-371-3p expression together with targeted cancer therapies may delay or prevent acquired drug resistance. 27554973_The constitutive elevations of Prdx6 and NF-kappaB during Clonorchis sinensis infection may be associated with more severe persistent hepatobiliary abnormalities mediated by clonorchiasis. 27932289_Peroxiredoxin 6 has a role in the repair of peroxidized cell membranes and cell signaling [review] 27934969_The increased HDL and plasma levels of PRDX6 in AAA patients support the potential of PRDX6 as a new biomarker of AAA. 28055018_Mutational analysis of functional sites showed that both peroxidase and PLA2 active sites were necessary for mutant Prdx6 function, and that Prdx6 phosphorylation (at T177 residue) was essential for optimum PLA2 activity.Mutant Prdx6 at its Sumo1 sites escapes and abates this adverse process by maintaining its integrity and gaining function 28293090_PRX6 protein is associated with fetal esophageal development and cancer differentiation. 28393051_the data demonstrate that Prdx6 interrupts the formation of TRAF6-ECSIT complex induced by TLR4 stimulation, leading to suppression of bactericidal activity because of inhibited mROS production in mitochondria and the inhibition of NF-kappaB activation in the cytoplasm. 28513872_These observations demonstrated that the expression and localization of NPM affected the homeostatic balance of oxidative stress in tumor cells via PRDX6 protein. The regulation axis of NPM/PRDX/ROS may provide a novel therapeutic target for cancer treatment. 29109765_The present data suggest that PS2 mutations suppress lung tumor development by inhibiting the iPLA2 activity of PRDX6 via a gamma-secretase cleavage mechanism and may explain the inverse relationship between lung cancer and Alzheimer's disease incidence. 29792351_These findings illuminate the effects of PRDX6 during osteogenic differentiation of hDPSCs, and also suggest that regulating PRDX6 may improve the clinical utility of stem cell-based regenerative medicine for the treatment of bone diseases. 29912414_The Ca2+-iPLA2 activity of PRDX6 is the most important and first line of defense against oxidative stress in human spermatozoa, maintaining cell viability and DNA integrity. 30104402_These results suggest that there are distinct prognostic values of PRDX family members in patients with ovarian cancer, and that the expression of PRDX3, PRDX5, and PRDX6 mRNAs are a useful prognostic indicator in the effect of chemotherapy in ovarian cancer patients. 30120980_These findings highlight the importance of Prdx6 as an essential regulator of oxidative stress in the eye. 30413111_we examined the therapeutic efficacy of the Ginkgolic acid (GA), a Sumoylation antagonist, to disrupt aberrant Sumoylation signaling in human and mouse lens epithelial cells (LECs) ...our study revealed an unprecedented role for GA in LECs and provided new mechanistic insights into the use of GA in rescuing LECs from aging/oxidative stress-evoked dysregulation of Sp1/Prdx6 protective molecules 30562740_These findings demonstrated that PRDX6 expression plays a characteristic growth-promoting role in Colorectal Cancer metastasis. 31036877_The findings reveal an essential role of PRDX6 in protecting cells against ferroptosis and provide a potential target to improve the antitumor activity of ferroptosis-based chemotherapy. 31519597_Patients with low Prx6 levels might be more prone to treatment-related adverse effects through elevated levels of oxidative stress. 31651026_PRDX6 contributes to the regulation of reactive oxygen species production and the PI3K/AKT pathway for the maintenance of sperm survival. 32018008_The anti-oxidant enzyme, Prdx6 might have cis-acting regulatory sequence(s). 32051828_PRDX6 Promotes the Differentiation of Human Mesenchymal Stem (Stromal) Cells to Insulin-Producing Cells. 32643149_Peroxiredoxins are involved in the pathogenesis of multiple sclerosis and neuromyelitis optica spectrum disorder. 32784474_Clock Protein Bmal1 and Nrf2 Cooperatively Control Aging or Oxidative Response and Redox Homeostasis by Regulating Rhythmic Expression of Prdx6. 32798391_[Expression of Peroxiredoxin-6 Gene in Patients with Acute Myeloid Leukemia and Its Clinical Significance]. 33035814_Knockout of PRDX6 induces mitochondrial dysfunction and cell cycle arrest at G2/M in HepG2 hepatocarcinoma cells. 33060708_Structural basis of peroxidase catalytic cycle of human Prdx6. 33547545_KLF9 regulates PRDX6 expression in hyperglycemia-aggravated bupivacaine neurotoxicity. 33561454_Peroxiredoxin 6 translocates to the plasma membrane of human sperm under oxidative stress during cryopreservation. 33631924_Identification of peroxiredoxin 6 as a potential lung-adenocarcinoma biomarker for predicting chemotherapy response by proteomic analysis. 33958651_pH induced conformational alteration in human peroxiredoxin 6 might be responsible for its resistance against lysosomal pH or high temperature. 34937537_Role of Innate Immunity and Oxidative Stress in the Development of Type 1 Diabetes Mellitus. Peroxiredoxin 6 as a New Anti-Diabetic Agent. | ENSMUSG00000026701+ENSMUSG00000050114 | Prdx6+Prdx6b | 1.808634e+04 | 1.0124780 | 0.017890547 | 0.2777425 | 4.178449e-03 | 0.9484599027 | 0.99022718 | No | Yes | 1.859731e+04 | 1548.131863 | 1.595908e+04 | 1362.548162 |
| ENSG00000118058 | 4297 | KMT2A | protein_coding | Q03164 | FUNCTION: Histone methyltransferase that plays an essential role in early development and hematopoiesis (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac) (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:24235145, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place (PubMed:25561738, PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity (PubMed:19187761, PubMed:26886794). Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9' (PubMed:19187761). Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state (PubMed:20010842). Required for transcriptional activation of HOXA9 (PubMed:12453419, PubMed:20677832, PubMed:20010842). Promotes PPP1R15A-induced apoptosis (PubMed:10490642). Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin (By similarity). Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate (PubMed:24235145). {ECO:0000250|UniProtKB:P55200, ECO:0000269|PubMed:10490642, ECO:0000269|PubMed:12453419, ECO:0000269|PubMed:15960975, ECO:0000269|PubMed:19187761, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:20010842, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:24235145, ECO:0000269|PubMed:26886794, ECO:0000305|PubMed:20677832}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Biological rhythms;Bromodomain;Chromatin regulator;Chromosomal rearrangement;DNA-binding;Direct protein sequencing;Isopeptide bond;Metal-binding;Methylation;Methyltransferase;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repeat;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a transcriptional coactivator that plays an essential role in regulating gene expression during early development and hematopoiesis. The encoded protein contains multiple conserved functional domains. One of these domains, the SET domain, is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity which mediates chromatin modifications associated with epigenetic transcriptional activation. This protein is processed by the enzyme Taspase 1 into two fragments, MLL-C and MLL-N. These fragments reassociate and further assemble into different multiprotein complexes that regulate the transcription of specific target genes, including many of the HOX genes. Multiple chromosomal translocations involving this gene are the cause of certain acute lymphoid leukemias and acute myeloid leukemias. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Oct 2010]. | hsa:4297; | cytosol [GO:0005829]; histone methyltransferase complex [GO:0035097]; MLL1 complex [GO:0071339]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; histone methyltransferase activity (H3-K4 specific) [GO:0042800]; identical protein binding [GO:0042802]; lysine-acetylated histone binding [GO:0070577]; minor groove of adenine-thymine-rich DNA binding [GO:0003680]; protein homodimerization activity [GO:0042803]; protein-cysteine methyltransferase activity [GO:0106363]; unmethylated CpG binding [GO:0045322]; zinc ion binding [GO:0008270]; apoptotic process [GO:0006915]; chromatin organization [GO:0006325]; circadian regulation of gene expression [GO:0032922]; embryonic hemopoiesis [GO:0035162]; histone H3-K4 dimethylation [GO:0044648]; histone H3-K4 methylation [GO:0051568]; histone H3-K4 monomethylation [GO:0097692]; histone H3-K4 trimethylation [GO:0080182]; histone H4-K16 acetylation [GO:0043984]; negative regulation of DNA methylation [GO:1905642]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of transporter activity [GO:0032411]; protein-containing complex assembly [GO:0065003]; regulation of histone H3-K14 acetylation [GO:0071440]; regulation of histone H3-K9 acetylation [GO:2000615] | 11733351_Nine (22.5%) of 40 patients exhibited MLL rearrangements. Three (33.3%) of these nine patients had granulocytic sarcoma and were younger than 12 months of age. 11809673_SEPTIN6, a human homologue to mouse Septin6, is fused to MLL in infant acute myeloid leukemia with complex chromosomal abnormalities involving 11q23 and Xq24. 11921290_An MLL rearrangement evolving as a secondary abnormality within a preexisting leukemic clone has been reported in a case of adult acute myeloid leukemia. 11943339_MLL-AF4 fusion gene rearrangements occurs between chromosome 4 and 11 in pre B-cell lymphoblastic leukemia 12034519_MLL amplification in myeloid malignancies: clinical, molecular, and cytogenetic findings 12095151_Fusion of MLL and MSF in adult de novo acute myelomonocytic leukemia (M4) with t(11;17)(q23;q25). 12096348_MLL-SEPTIN6 fusion recurs in novel translocation of chromosomes 3, X, and 11 in infant acute myelomonocytic leukaemia and in t(X;11) in infant acute myeloid leukaemia 12127405_FISH analysis in infant AML-M5 showed a complex rearrangement between chromosomes 10 and 11, disrupting the MLL gene, a paracentric inversion of the 11q13-q23 fragment translocated to 10p12. AF10 was the fusion partner gene of MLL in this rearrangement. 12138900_The correlation between infant leukemia and in utero exposure to topoisomerase II (topo-II) inhibitor has been clarified. in vitro effect of etoposide on cleavage of the MLL gene in cord and peripheral blood mononuclear cells (MNCs). 12192052_the MLL-AFX fusion protein transforms myeloid progenitors and interferes with forkhead protein function 12203795_Fusion of MLL to one of the AF4 family members (e.g., LAF4) may determine a proB-cell phenotype in infant leukemia. 12393701_MLL is proteolytically processed into 2 fragments with opposite transcriptional properties 12461747_A novel chromosomal inversion at 11q23 in acute myeloid leukemia fuses the MLL gene to the CALM gene. 12482972_This paper demonstrates that full length precursor MLL undergoes site-specific proteolysis to generate a heterodimeric complex consisting of N-terminal 320 kD and C-terminal 180kD of MLL. 12482972_we report that MLL is normally cleaved at two conserved sites (D/GADD and D/GVDD). MLL cleavage generates N-terminal p320 (N320) and C-terminal p180 (C180) fragments, which form a stable complex that localizes to a subnuclear compartment. 12547160_t(10;11)(p11.2;q23) involving this and ABI-1 genes is associated with congenital acute monocytic leukemia 12637319_myeloid gene dysregulation is dispensable in leukemic transformation mediated by MLL fusion proteins; dysregulation of HOX gene family members is implicated as a dominant mechanism of leukemic transformation induced by chimeric MLL oncogenes 12646957_demonstrated that TET1 is fused to MLL in a case of pediatric acute myeloid leukemia containing the t(10;11)(q22;q23) 12660026_Tandem duplication of the MLL gene may occur in AML with a partial 11q trisomy. Thus, systematic screening of this molecular defect should be performed in patients with unbalanced translocations involving 11q22 approximately q23-->qter 12665591_SWI/SNF complexes show considerable heterogeneity, and one or more may be involved in the etiology of leukemia by cooperating with MLL fusion proteins. 12665971_polymorphism within the MLL BCR has only a suggestive association with t-AML development. 12696071_MLL-CBL fusion was the result of an interstitial deletion in patients with de novo acute myeloid leukemia (FAB-M1). 12732503_loss of heterozygosity in the MLL gene in childhood ALL 12743608_There is fusion of an AF4-related gene, LAF4, to this gene in childhood acute lymphoblastic leukemia with t(2;11)(q11;q23). 12829790_Histone deacetylase 1 interacts with the MLL repression domain, partially mediating its activity; binding of Cyp33 to the adjacent MLLPolycomb group proteins HPC2 & BMI-1 & the corepressor C-terminal-binding protein also bind the MLL repression domain. 12946992_findings confirm the MLL gene as a prominent target of 11q23 amplification and provide further evidence for an etiologic role for MLL gain of function in myeloid malignancies 14504097_FLT3-D835/I836 mutations are one of the second genetic events in infant ALL with MLL rearrangements or pediatric ALL with hyperdiploidy 14551139_new mechanism for MLL-EEN-mediated leukemogenesis in which MLL-EEN interferes with the Ras-suppressing activities of EBP through direct interaction. 14562113_lack of HOX gene expression in acute lymphoblastic leukemia B-cells was not due to a nonfunctional MLL/AF4 14580777_Involvement of the MLL gene located at chromosome region 11q23 is a frequent occurrence in both acute myelocytic leukemia and acute lymphoblastic leukemia. 14636557_This paper identifies the conserved protease, Taspase1, responsible for MLL proteolysis. 14704031_Review. The normal MLL gene plays a key role in developmental regulation of gene expression (including HOX genes), & in leukemia this function is subverted by breakage, recombination, and chimeric fusion with one of 40 or more alternative partner genes. 14751928_GPHN as an MLL-GPHN chimera is able to transform hematopoietic progenitors; a tubulin-binding domain of GPHN is necessary and sufficient for this activity and also confers oligomerization capacity on MLL protein, which may contribute to leukemogenesis 14990976_the cell cycle control exerted by MLL-AF4 may be responsible of resistance to cell-death promoting stimuli in leukemia carrying the t(4;11) translocation. 15132992_Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Review. 15160920_suggest that MLL aberrations may regulate MEIS1 and HOXA9 gene expression in ALL-derived cell lines, while AML-derived cell lines express these genes independently of the MLL status 15199122_MLL associates with a cohort of proteins shared with the yeast and human SET1 histone methyltransferase complexes; studies link menin with the MLL histone methyltransferase machinery, with implications for Hox gene expression in leukemia pathogenesis 15221006_role of interaction with SIAH1 and SIAH2 proteins in t(4,11) pathobiology 15542854_the CXXC DNA binding domain has a critical role in MLL-associated oncogenesis, most likely via epigenetic recognition of CpG DNA sites within the regulatory elements of target genes 15592432_Some leukemic patients are inherently more susceptible to the genotoxic effect of topo 2 inhibitors. The degree and persistence of MLL cleavage may identify patients at risk. 15618957_NQO1 gene is an MLL-independent risk factor, in the leukemogenic process of this subtype of infant lymphoblastic leukemia. 15618964_Different mechanisms might be included in leukemogenesis by MLL rearrangements. (review) 15621793_case of congenital acute myeloblastic leukemia with granulocytic sarcomata arising in a patient with proven MLL rearrangement 15640349_that regulation of cyclin-dependent kinase inhibitor transcription by cooperative interaction between menin and MLL plays a central role in menin's activity as a tumor suppressor 15670842_TAF-Ibeta plays an important role in MLL-mediated transcription and possibly chromatin maintenance 15710585_MLL and FLT3 gene duplications could have roles in myeloid malignancies 15774615_Silencing of MLL WT in MLL(PTD/WT) blasts was reversed by DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors, and MLL WT induction was associated with selective sensitivity to cell death 15815718_the identified molecular expression pattern of MLL fusion gene samples and biological networks revealed new insights into the aberrant transcriptional program in 11q23/MLL leukemias 15968309_Observational study of genetic testing. (HuGE Navigator) 15996925_MLL with a partial tandem duplication is expressed in minimal residual disease in acute myeloid leukemi 16019540_translocation t(4;11)(q21;q23) involving the MLL gene may result from chemotherapy for Burkitt's leukemia [case report] 16029447_Observational study of gene-disease association. (HuGE Navigator) 16076867_Observational study of genotype prevalence. (HuGE Navigator) 16086288_In the Acute Myeloid Leukemia patients, detected MLL rearrangements consisting of MLL/AF6, MLL/AF9, MLL/AF17 , MLL/ELL and MLL partial tandem duplication. MLL rearrangement include chromosome translocation and partial tandem duplication. 16138343_Translocation 11q23 and MLL gene rearrangements observed in acute myeloid leukemia in a 15-yeaar-old girl. 16138344_A child with chronic granulomatous disease (CGD) who developed acute lymphoblastic leukemia who was found to have a mutation om the MLL gene. 16169901_MLL-AF4 inhibits or activates CDKN1B expression. 16193084_analysis of sequence motifs surrounding the cleaved region of MLL showed the presence of both a predicted nuclear matrix attachment sequence and a potential strong binding site for topoisomerase II, flanking the site of cleavage 16215946_The patients with immunophenotype of Pre-B-acute lymphoblastic leukemia were found to carry: MLL/AF4, MLL/AF9, MLL/AF10, MLL/AFX-MLL/AF6-MLL/ELL, MLL/AF6-MLL/ELL, and dupMLL. 16253272_Results describe the solution structure of the ternary complex formed by cooperative binding of activation domains from the c-Myb and mixed lineage leukemia transcription factors to the KIX domain of CREB binding protein. 16328057_Down-regulation of MLL is not necessary to abolish malignant phenotypes by induction of terminal monocyte-macrophage differentiation in leukaemic cells carrying t(9;11)(p22;q23). 16357944_Topoisomerase II inhibitors are cell cycle-specific DNA-damaging agents which correlate with chromosomal translocations involving the mixed-lineage leukemia/myeloid lymphoid leukemia(MLL) gene on chromosome 11. 16386788_The TCR gene rearrangements in childhood B-lineage acute lymphoblastic leukemia was associated with expression of MLL chimeric oncogene. 16433901_determination of the relative positions of MLL, AF4 and ENL genes, in two lymphoblastic and two myeloid human cell lines 16604156_analysis of menin's role as a tumor suppressor and binding sites of Rbbp5 and MLL1 16843108_Molecular analysis using rtPCR followed by sequencing confirmed the expression of an MLL-SEPT6 fusion transcript with a novel sequence. 16878130_WDR5 mediates interactions of the MLL1 catalytic unit both with the common structural platform and with the histone substrate. 16970032_constitutional duplication 11q23 de novo involves the MLL gene. 16990798_The structure of the human MLL CXXC domain was determined, by multidimensional Nuclear magnetic resonance spectroscopy. 17047146_MLL regulates endothelial-cell migration via HoxA9 and EphB4, whereas sprout formation requires MLL-dependent signals beyond HoxA9 and HoxD3. 17205059_Acute myeloid leukemia cells harbor an MLL translocation with a recurrent partner gene of MLL. 17252016_Transcriptional read-through and subsequent splicing - or trans-splicing - to other transcription units is a novel property of disrupted MLL genes in order to create functional MLL fusions - at least at the mRNA level. 17268512_Immunobiological subgroups in 133 infant acute lymphoblastic leukemia (ALL) cases as assessed by MLL rearrangements 17280715_secondary hematopoietic malignancies involving MLL confirms secondary chronic myelomonocytic leukemia is associated with t(9;11) 17341662_The clinical impact of MLL partial tandem duplication (MLL-PTD) was evaluated in 238 adults aged 18 to 59 years with cytogenetically normal de novo acute myeloid leukemia. 92% of patients with MLL-PTD achieved complete remission compared with 83% without. 17410185_t(4;11) patients exhibit reciprocal MLL alleles, but due to the individual recombination events, provide different pathological disease mechanisms 17550846_Observational study of genotype prevalence. (HuGE Navigator) 17578910_HCF-1 was required for recruitment of the histone methyltransferases Set1 and MLL1, leading to histone H3K4 trimethylation and transcriptional activation. 17581865_identify specific miRNAs up-regulated in leukemias triggered by All1 fusions. All1 fusion protein-mediated recruitment of Drosha to target genes encoding miRNAs is demonstrated 17597811_mutated NPM1 leads to dysregulated HOX expression via a different mechanism than MLL rearrangement 17612494_During the G1-to-S phase transition, HCF-1 recruits the mixed-lineage leukemia (MLL) and Set-1 histone H3 lysine 4 methyltransferases to E2F-responsive promoters and induces histone methylation and transcriptional activation. 17698583_Data demonstrate that the Drosophila homologs of mixed-lineage leukemia protein and host cell factor 1, called Trithorax and dHCF, are both cleaved by Drosophila taspase 1. 17763464_AML patients with FLT3-ITD, but not D835Mt, showed a poor prognosis. AML patients with MLL-PTD were also correlated with poor prognosis in this study. 17851550_NG2 antigen diagnosis of MLL rearrangements in pediatric patients 17854671_Thus, t(4;11)(p12;q23) with MLL and FRYL involvement represents a new recurring 11q23 translocation, to date seen only in therapy-related acute leukemias. 17889710_analysis of a rare MLL-AF4 (MLL-AFF1) fusion rearrangement in infant leukemia 17905612_review considers recent advances in understanding of the role of MLL gene rearrangements in pediatric leukemia [review] 17908926_An essential post-translational regulation of MLL by the cell cycle ubiquitin/proteasome system (UPS) assures the temporal necessity of MLL in coordinating cell cycle progression. 17942719_Chromatin remodeling mechanisms at GABAergic gene promoters, including MLL1-mediated histone methylation, operate throughout an extended period of normal prefrontal cortex development and play a role in the neurobiology of schizophrenia. 17957188_Review. MLL methylates histone H3K4 & upregulates gene expression including multiple Hox genes. Leukaemogenic translocations encode MLL fusion proteins missing H3K4 methyltransferase activity & transform haematopoietic cells into leukaemia stem cells. 18007578_mature 'sIg+' B-ALL with non FAB-L3 morphology displaying 11q23-MLL rearrangement, and especially t(9;11), occurring in young children with a poor prognosis and requiring treatment intensification. 18024407_screened the entire FLT3 coding sequence in MLL rearranged and hyperdiploid ALL cases for yet unidentified additional genetic alterations using denaturing D-HPLC; a small minority of samples, 7% and 10% respectively, carried genetic alterations. 18082152_CGBP interacts with MLL1, MLL2 as well as Set1 H3-Lysine 4 (H3K4) specific methyl-transferases and plays critical roles in regulations of MLL target genes. 18450602_Observational study of gene-disease association. (HuGE Navigator) 18538732_Expression of MLL-AF9 in human CD34+ cells induces acute myeloid, lymphoid, or mixed-lineage leukemia in immunodeficient mice. 18559874_Observational study of gene-disease association. (HuGE Navigator) 18566324_In acute myeloid leukemia with mixed lineage leukemia partial tandem duplication, the SLC5A8 tumor suppressor gene promoter was more frequently hypermethylated. 18598942_Lens epithelium-derived growth factor critically associates with MLL and menin at the nexus of transcriptional pathways that are recurrently targeted in diverse diseases. 18617057_The most common abnormality was the p16 deletion (22.8%), followed by MLL and ETV6/RUNX1 rearrangements (8.7%) in this mexican population. 18642054_monocytic differentiation and a poor prognosis may also be associated with acute myeloid leukemia with the variant MLL/SEPT9 fusion transcript 18656690_MLL and a t(6;14)(q25;q32) in two adults is reported. 18656699_A novel MLL-SEPT2 fusion variant in therapy-related myelodysplastic syndrome is reported. 18676843_data support the hypothesis that the binding of Cyp33 to the MLL third PHD finger switches the MLL function from transactivation to repression 18776924_ectopic expression of C/EBPepsilon, as well as C/EBPalpha, can induce the monocytic differentiation of myelomonocytic leukemic cells with MLL-fusion gene through the downregulation of Myc 18806775_GSK3 paradoxically supports MLL leukaemia cell proliferation and transformation by a mechanism that ultimately involves destabilization of the cyclin-dependent kinase inhibitor p27(Kip1). 18829457_WDR5's recognition of arginine 3765 of MLL1 is essential for the assembly and enzymatic activity of the MLL1 core complex in vitro 18829459_WDR5 recognizes Arg-3765 of MLL1, which is essential for the assembly and enzymatic activity of the MLL1 core complex 19028982_These results indicate an increase in acute lymphoblastic leukaemia aberrations in childhood ALL survivors years after completion of therapy. 19029444_Even genetically complex leukemias can be reversed on inactivation of the initiating MLL fusion. 19059939_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19109563_results show that MEIS1 expression is important for MLL-rearranged leukemias and suggest that MEIS1 promotes cell-cycle entry 19144982_Characteristic features of MLL(+) patients were significantly lower CD10 expression, expression of the NG2 antigen, a higher white blood count at diagnosis, and female sex 19155294_The identification and validation of consistent changes of gene expression in human and murine MLL rearrangement leukemias provide important insights into the genetic base for MLL-associated leukemogenesis. 19187761_the crystal structure of the MLL1 SET domain in complex with cofactor product AdoHcy and a histone H3 peptide 19208841_DDB1-CUL4 and MLL1 complexes constitute a novel pathway that mediates p16 activation during oncogenic checkpoint response. 19219072_A molecular mechanism by which the recruitment of a H3K4 histone methyltransferase complex on the promoter of a NF-kappaB-dependent gene induces its expression. 19220463_studies demonstrate that MLL1 and H3K4 methylation have distinct dynamics during the cell cycle and play critical roles in the differential expression of Hox genes associated with cell cycle regulation 19229974_MLL gene rearrangement is associated with recurrent and refractory acute lymphoblastic leukemia 19262598_760 MLL-rearranged biopsy samples obtained from 384 pediatric & 376 adult leukemia patients were characterized at the molecular level for breakpoint distribution & fusion translocation partner genes. 19264358_estradiol and 4-hydroxyestradiol, which may occur during pregnancy, may cause aberrations of the MLL gene and could thus effect leukemogenesis occurring in utero. 19269851_controls the maintenance of established memory Th2 cells via histone methyltransferase activity 19309322_Observational study of gene-disease association. (HuGE Navigator) 19438726_Results demonstrated that MLL1 gene expression is sensitive to toxic stress and Sp1 plays crucial roles in the stress-induced upregulation of MLL1. 19454493_NRIP3 is a novel translocation partner of MLL in acute myeloid leukemia 19528237_Results identify a previously unrecognized role of MLL in modifying telomeric chromatin and provide evidence for the functional interaction between MLL, p53, and the shelterin complex in the regulation of telomeric transcription and stability. 19528532_MLL gene rearrangment ism associated with acute myeloid leukemia. 19556245_the mechanism of multiple lysine methylation by the MLL1 core complex involves the sequential addition of two methyl groups at two distinct active sites within the complex. 19631984_NOTCH1 and MLL abnormalities are primary leukemogenic hits in early T-ALL. 19722759_We conclude that MLL may play an important role in the lymphomagenesis of IVLBCL at least in a subset of cases. 19729989_Stduies indicate that leukemogenic MLL translocations fuse the common MLL N-terminus (approximately 1,400 aa) in frame with more than 60 translocation partner genes. 19763085_Data show here that the DHQY HCF-1-binding sequence permits E2F1 to stimulate both DNA damage and apoptosis, and that HCF-1 and the MLL family of H3K4 methyltransferases have important functions in these processes. 19835597_These results suggest that the conspicuous expression of the tumor suppressor genes BEX2, IGSF4 and TIMP3 in MLLmu acute myeloid leukemias cell lines is the consequence of altered epigenetic properties of MLL fusion proteins. 19855078_Data show that inhibition of aberrant DNA methylation may be an important novel therapeutic strategy for MLL-rearranged ALL in infants. 19940862_Haploinsufficiency of the MLL and TOB2 genes in lymphoid malignancy. 19956635_Uncategorized study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19965632_Data show that down-regulation of MCL-1 in prednisolone-resistant MLL-rearranged leukemia cells by RNA interference, to some extent, led to prednisolone sensitization. 20064463_Data suggest that MLL perpetuates active transcription in dividing cells by associating with gene promoters packaged within condensed mitotic chromosomes. 20093773_Mixed-lineage leukemia (MLL) is a proto-oncogene frequently involved in chromosomal translocations associated with acute leukemia. 20113834_Three-way translocation involving MLL, MLLT1, and a novel third partner, NRXN1, in a patient with acute lymphoblastic leukemia and t(2;19;11) (p12;p13.3;q23). 20113838_Coexistence of alternative MLL-SEPT9 fusion transcripts in an acute myeloid leukemia with t(11;17)(q23;q25). 20139053_MLL gene rearrangement is associated with maternal acute lymphoctic leukemia during pregnancy. 20206559_In acute leukemia patients, fluorescent in situ hybridization with a panel of probes coupled with long distance inverse-PCR was used to identify chromosomal rearrangements involving the MLL gene. 7 unusual chromosomal rearrangements were identified. 20215641_Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. 20299091_MLL-MLLT10 gene rearrangment is associated with mixed phenotype acute leukemia. 20332322_In B-ALL, Gal-1 is a highly sensitive and specific biomarker of MLL rearrangement that is likely induced by a MLL-dependent epigenetic modification. 20362031_findings suggest that MLL-AF4 family fusion oncoproteins can activate Elk-1 through Ras/MEK/extracellular signal-regulated kinase (ERK) pathway and strongly support the role of Ras signaling in the pathogenesis of MLL-rearranged leukemia 20395514_Ras activation seems to complement the MLLT3-MLL oncogene in transformation. 20454944_Regardless of the precise mechanism, a consistent effect of MLL fusion oncogenes in human leukemia is to induce high level expression of HOXA and MEIS1 genes. 20493169_high levels of histone acetylation--the hallmarks of active chromosome regions in vivo--can increase the affinity of reconstituted nucleosomes to the SET domain of ALL-1 histone methyltransferase in a defined system in vitro. 20520634_Observational study of gene-disease association. (HuGE Navigator) 20541251_Results highlight the role of PHD3-Bromo cassette as a regulatory platform, orchestrating MLL1 binding of H3K4me3/2 marks and cyclophilin-mediated repression through Histone deacetylase recruitment. 20541477_MLL interacts directly with the polymerase associated factor complex (PAFc) via Paf1 and CTR9. PAFc augments MLL and MLL-AF9 mediated transcriptional activation of Hoxa9 and their interaction is essential for leukemogenesis. 20558618_characterized a novel recurrent chromosomal aberration, inv(11)(p15q23) in 2 patients with acute myeloid leukemia. This aberration is predicted to result in the expression of a NUP98-MLL fusion protein that is unable to interact with menin. 20581860_The role of the interaction between MOZ and MLL in CD34+ cells in which both proteins have a critical role in hematopoietic cell-fate decision. 20658337_From the overall results obtained, nsSNP with id (rs1784246) at the amino acid position Q1198P could be considered as deleterious mutation in the acute leukemia caused by MLL gene. 20677832_mechanism for the MLL PHD3 domain to act as a switch between activation and repression 20682395_Case represents an additional MLL-SEPT9-positive AML that was considered to be related to therapy. RT-PCR and sequencing analyses demonstrated MLL-SEPT9 fusion transcripts with the breakpoint of MLL exon 8/SEPT9 exon 2 and MLL exon 9/SEPT9 exon 2. 20691474_Lack cryptic MLL rearrangement is associated with myelodysplastic syndromes with deletions of chromosome 11q. 20818375_results identify MLL as a key constituent of the mammalian DNA damage response pathway and show that deregulation of the S-phase checkpoint incurred by MLL translocations probably contributes to the pathogenesis of human MLL leukaemias 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20844554_studies provide evidence for the importance of the regulation of Pol II elongation in disease pathogenesis and suggest that MLL chimaeras function by licensing Pol II transcription elongation without the appropriate checkpoints 20854876_The findings suggest that MLL fusion proteins deregulate genes critical for leukemia by excessive recruitment and impaired dissociation of EAP from target loci 20855524_Data report that ANCCA directly interacts with E2F1 to E2F3 in its complex with MLL and that its N terminus interacts with both the N and C termini of E2F1. 20858288_Data show that high cell density and LMP1 expression induced apoptosis in NPC cells and subsequently resulted in MLL bcr cleavage at the MAR/SAR region. 20861917_overexpression of the BRE gene is predominantly found in MLL-rearranged AML with t(9;11)(p22;q23). 20869771_Overexpression of MLL-AF4 fusion protein and not AF4-MLL is associated with t(4;11) leukemias. 20920256_CT45A2 is a novel spliced MLL fusion partner in de novo biphenotypic acute leukemia [case report] 20961854_Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias 20969867_nature of interactions between CREB binding protein and mixed-lineage leukemia protein 20979663_The data suggests that reduced expression of wild-type MLL can contribute to GC resistance in ALL patients both with and without MLL-translocations. 20980053_A variant form of t(11;22)(q23;q13)/MLL-EP300 fusion transcript is associated with acute myeloid leukemia with myelodysplasia-related changes 21030982_findings suggest that the AF4-MLL protein disturbs the fine-tuned activation cycle of promoter-arrested RNA Pol II and causes altered histone methylation signatures 21106533_WRAD is a new H3K4 methyltransferase with functions that include regulating the substrate and product specificities of the MLL1 core complex 21116279_provide additional evidence that they should be tested for their efficacy in MLL-rearranged infant acute lymphoblastic leukemia in in vivo models 21124902_MLL1 SET domain make direct contacts with the substrates and contribute to the formation of a joint catalytic center. 21135039_results provide the structural basis to control WDR5-RbBP5-Ash2L-MLL1 activity and a tool to manipulate stem cell differentiation programs 21135858_MLL-AF4 leukemogenesis has been particularly difficult to model and bona fide MLL-AF4 disease models do not exist so far.[review] 21331072_MLL partner genes drive distinct gene expression profiles and genomic alterations in pediatric acute myeloid leukemia. 21389315_MLL-AF4 expression was insufficient to initiate leukemogenesis on its own, indicating that either additional hits (or reciprocal AF4-MLL product) may be required to initiate ALL 21436736_High frequency of MLL-ENL Fusion/t(11;19) is associated with infant leukemia. 21464370_genetic association studies: investigation of association of MLL gene rearrangement and chromosome aberrations with prognosis/treatment outcome in pediatric patients with acute myeloid leukemia in Italy 21493871_Single Nucleotide Polymorphisms in MLL is associated with childhood acute lymphoblastic leukemia. 21518926_MLL fusion-bound gene regions exhibit an aberrant distribution pattern of MLL in human leukemic cells. 21565404_MLL gene rearrangements in amniocytes from fetuses of mothers is associated with smoking. 21706045_miR-143 functions as a tumor suppressor in MLL-AF4 B-cell acute lymphoblastic leukemia. 21741597_These data point to DOT1L as a potential therapeutic target in MLL-rearranged leukemia. 21757704_data provide a structural basis for understanding the role of menin as a tumor suppressor protein and as an oncogenic co-factor of mixed lineage leukemia fusion proteins 21836617_Targeting MLL1 by RNA interference inhibited the expression of HIF2alpha and target genes, including vascular endothelial growth factor (VEGF). 21900057_The study reports here a series of 27 patients with acute leukemia in whom the MLL fusion partner gene was identified by LDI-PCR and the fusion gene confirmed by dual color FISH using BAC clones. 21937695_Within the group of MLL-AF9-positive patients, high BRE expression predicted superior survival, while normal BRE expression predicted extremely poor survival. 21953510_Data from follow-up study in Rome suggest a particularly favorable prognosis for patients with acute lymphoblastic leukemia expressing the MLL/ENL fusion gene. 21959947_Data describe immunocytotoxicity against MLL-AF9-negative U937 cells but not positive MLL-AF9 THP-1 cells, suggesting that the toxocity ting was not antibody-mediated by antibodies against MLL-AF9 protein. 21988239_alterations of CBL gene and MLL gene rearrangement (MLL-R) may cooperatively play a pathogenic role in the development of infant acute lymphoblastic leukaemia with MLL-R. 21998658_MLL translocations relieve oncogenic MLL-PG fusions from the repressive MLL 3'UTR, contributing to higher activity of these genes and leukaemia development 22012064_Studies identified a physical and functional interaction between RUNX1 (AML1) and MLL and show that both are required to maintain the histone lysine 4 trimethyl mark (H3K4me3) at 2 critical regulatory regions of the AML1 target gene PU.1. 22039304_the c-Myb/GATA-3 complex recruits MLL to the GATA-3 response element for histone modification of the IL-13 locus during the differentiation of memory Th2 cells. 22046413_that hPaf1/PD2 in association with MLL1 regulates methylation of H3K4 residues, as well as interacts and regulates nuclear shuttling of chromatin remodeling protein CHD1, facilitating its function in pancreatic cancer cells 22052166_MLL duplication is associated with B-cell lymphoblastic lymphoma. 22137486_Five years after the initial diagnosis, investigators identified a complex rearrangement of the MLL gene without progression to acute leukemia. 22174154_A model whereby ASB2 contributes to hematopoietic differentiation, in part, through MLL degradation;deletion of the PHD/Bromo region renders MLL fusion proteins resistant to ASB2-mediated degradation and may contribute to leukemogenesis. 22212479_MLL-AF4 enhances the specification of hemogenic precursors from hESCs but strongly impairs further hematopoietic commitment in favor of an endothelial cell fate. 22304832_Abl-interactor 2 (ABI2) is a novel MLL translocation partner in acute myeloid leukemia. 22327296_crystal structures of human menin in its free form and in complexes with MLL1 or with JUND, or with an MLL1-LEDGF heterodimer 22382894_MLL partial tandem duplication in adults aged 60 years and older is not associated with cytogenetically normal acute myeloid leukemia. 22427200_Identification of genes that define transcription factor networks and important genetic pathways acting during progression of leukemia induced by MLL fusion oncogenes. 22583166_The GST-tagged SET-domain polypeptide of ALL-1 was assayed for binding to assembled mononucleosomes and nucleosome-dimer particles, either intact or remodeled with purified yeast Isw2. 22674806_Here we demonstrate, for the first time, that Mll partial tandem duplication contributes to leukemogenesis as a gain-of-function mutation and describe a novel murine model closely recapitulating human acute myeloid leukemia. 22705992_findings indicate that high FLT3 expression identifies MLL-AF4+ ALL patients at very high risk of treatment failure and poor survival, emphasizing the value of ongoing/future clinical trials for FLT3 inhibitors 22740449_The underlying pathogenic role of MLL partial tandem duplication in the clonal evolution of human leukemia. 22795537_De novo mutations in MLL cause Wiedemann-Steiner syndrome. 22814291_the MLL gene is etiopathogenetically relevant fo | ENSMUSG00000002028 | Kmt2a | 5.185968e+03 | 1.1702332 | 0.226796090 | 0.2619158 | 7.396126e-01 | 0.3897849952 | 0.84317192 | No | Yes | 5.494305e+03 | 802.846752 | 4.634327e+03 | 694.568626 | |
| ENSG00000118197 | 83479 | DDX59 | protein_coding | Q5T1V6 | 3D-structure;ATP-binding;Alternative splicing;Ciliopathy;Cytoplasm;Disease variant;Helicase;Hydrolase;Isopeptide bond;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ubl conjugation;Zinc;Zinc-finger | hsa:83479; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724] | 23972372_These data strongly implicate DDX59 RNA helicase family member in the pathogenesis of orofaciodigital syndrome. 28711741_The novel DDX59 mutation is associated with oral-facial-digital syndrome. 29127725_Analysis of human brain gene expression provides evidence that DDX59 is enriched in oligodendrocytes and might act within pathways of leukoencephalopathies-associated genes. 29133145_Study presents evidence that DDX59 protein can be upregulated by MEK/ERK/MAP kinase pathway downstream of EGFR and Ras signaling. Increased expression of DDX59 mediates the tumorigenecity of EGFR and Ras in lung cancer. 31111544_MicroRNA-628-5p inhibits cell proliferation in glioma by targeting DDX59. | ENSMUSG00000026404 | Ddx59 | 2.565264e+02 | 0.7845668 | -0.350031875 | 0.3398178 | 1.045432e+00 | 0.3065615901 | 0.81492114 | No | Yes | 2.301571e+02 | 36.286282 | 3.129236e+02 | 50.325936 | |||
| ENSG00000118482 | 23469 | PHF3 | protein_coding | Q92576 | 3D-structure;Alternative splicing;Isopeptide bond;Metal-binding;Methylation;Phosphoprotein;Reference proteome;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of a PHD finger-containing gene family. This gene may function as a transcription factor and may be involved in glioblastomas development. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2014]. | hsa:23469; | nucleus [GO:0005634]; metal ion binding [GO:0046872]; transcription, DNA-templated [GO:0006351] | 11856869_novel member of a large class of regulatory proteins containing a LAP motif, and loss of its expression in glioblastoma may contribute to glioma development 15906353_Survival of patients with GLEA2 antibodies was increased to 17.4 months and for patients with PHF3 antibodies to 14.7 months, as compared to 7.2 months for patients without GLEA2 or PHF3 antibodies in glioblastoma. 22096494_Data conclude that the PHF3-PTP4A1 region appears to harbor a causal locus for alcohol dependence, and proteins encoded by PHF3 and/or PTP4A1 might play a functional role in the disorder. 23324950_PTP4A1-PHF3-EYS variants were associated with alcohol dependence. 24961364_We confirmed with our previous findings that PTP4A1-PHF3-EYS variants were significantly associated with alcohol dependence. 34667177_PHF3 regulates neuronal gene expression through the Pol II CTD reader domain SPOC. | ENSMUSG00000048874 | Phf3 | 5.660496e+02 | 1.3854872 | 0.470393426 | 0.3895166 | 1.456510e+00 | 0.2274862265 | 0.78792184 | No | Yes | 5.115885e+02 | 121.965808 | 3.455249e+02 | 84.571896 | ||
| ENSG00000118495 | 5325 | PLAGL1 | protein_coding | Q9UM63 | FUNCTION: Acts as a transcriptional activator (PubMed:9722527). Involved in the transcriptional regulation of type 1 receptor for pituitary adenylate cyclase-activating polypeptide. {ECO:0000269|PubMed:18299245, ECO:0000269|PubMed:9722527}. | Activator;Alternative splicing;DNA-binding;Diabetes mellitus;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a C2H2 zinc finger protein that functions as a suppressor of cell growth. This gene is often deleted or methylated and silenced in cancer cells. In addition, overexpression of this gene during fetal development is thought to be the causal factor for transient neonatal diabetes mellitus (TNDM). Alternative splicing and the use of alternative promoters results in multiple transcript variants encoding two different protein isoforms. The P1 downstream promoter of this gene is imprinted, with preferential expression from the paternal allele in many tissues. [provided by RefSeq, Nov 2015]. | hsa:5325; | Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; apoptotic process [GO:0006915]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of cell cycle [GO:0051726]; regulation of transcription by RNA polymerase II [GO:0006357] | 10655556_PLAGL1 gene is imprinted, with preferential expression from the paternal allele in many tissues. 11935319_genetically imprinted in neonatal diabetes 12473647_This candidate tumor suppressor gene, located at chromosome 6q24-25, is epigenetically regulated in cancer 12529403_Zac contains transactivation activities that are differentially controlled by DNA binding. 15286800_ZAC expression was studied in transgenic mice. 15581945_The altered expression and LOH of the LOT1 locus support the gene's potential role in the pathogenesis of ovarian cancer and possibly in other types of cancer. 15592663_Mutated in multiple congenital abnormalities including neonatasl diabstes, hypoplastic pasnscreas, intestinal atresia and gallbladdre dysplasia. 15888726_Mutations in ZAC may contribute to Beckwith-Wiedemann syndrome. 16112421_Down-regulation of PLAGL1 is associated with extraskeletal myxoid chondrosarcoma tumors 16179495_Loss of ZAC expression is associated with basal cell carcinomas of skin. 16733217_Using the ZAC1-specific marker located at 6q24-25, LOH or allelic imbalance was observed. 16809786_coordinated binding of Zac zinc fingers and C terminus to p300 regulates HAT function by increasing histone and acetyl coenzyme A affinities and catalytic activity 16873690_Observational study of gene-disease association. (HuGE Navigator) 16928428_a transgene carrying the human HYMAI/PLAGL1 differentially methylated region was methylated in the correct parent-origin-specific manner in mice and this was sufficient to confer imprinted expression from the transgene 17178896_a functional relationship between Zac and PPARgamma that could be relevant to the understanding of tumorigenesis and diabetes as well. 17341487_Tissue-specific imprinting of PLAGL1 gene results from variable use of monoallelic and biallelic promoters. 17805016_Absence of unmethylated ZAC1 sequence was highly concurrent with ZAC1 region LOH or 6q loss and with lack of ZAC1 expression, suggesting preferential loss of nonimprinted, expressed ZAC1 allele in capillary hemangioblastoma. 18644983_Zac1 might be involved in regulating the p21(WAF1/Cip1) gene and protein expression through its protein-protein interaction with p53 and HDAC1 in HeLa cells. 18663001_Here it is shown that Zac1, together with the coactivators p300 and PCAF, recruit to the p21(Cip1) promoter during the differentiation of embryonic stem cells into neurons. 19155788_No evidence was found that polymorphisms in ZAC1 might influence anthropometric, biochemical or clinical parameters in French individuals. 19155788_Observational study of gene-disease association. (HuGE Navigator) 19423711_The zinc-finger protein ZAC1 is up-regulated under hypertonic stress and negatively regulates expression of sorbitol dehydrogenase, allowing for accumulation of sorbitol as a compatible organic osmolyte. 19694203_Our results do not indicate a relevant role of mutations in LOT1(ZAC1/PLAGL1) in the etiology of Silver-Russell Syndrome. 20175198_Data indicates that downregulation of ZAC occurs in diffuse large B-cell lymphoma. As ZAC expression in normal B lymphocytes is derived from promoter P2, in DLBCL we analyzed both promoter P1 and P2 for gene expression, LOH and abnormal methylation. 20363751_ZAC1 is a novel and previously unknown regulator of cardiomyocyte Glut4 expression and glucose uptake; MEF2 is a regulator of ZAC1 expression in response to induction of hypertrophy 20487506_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20518900_ZAC1 may have a role in tumorigenesis of von Hippel-Lindau associated central nervous system hemangioblastoma. 21305342_The specific expression of ZAC in the human fetal beta-cells supports it as the gene involved in transient neonatal diabetes mellitus 21872827_Our study suggests the involvement of the imprinted ZAC gene in the pathogenesis of pheochromocytoma. 22001409_Zac1 is able to interact directly with the Sp1-responsive element in the p21(WAF1/Cip1) gene promoter. 22227369_This work indicates that Zac1 functions are regulated, at least in part, via non-covalent interactions with SUMO-1 for the induction of p21, which is important for the modulation of apoptosis. 22784302_detected a synonymous variation in the protein-coding exon-2 of PLAGL1 in isolated VSD patients. It is possible that the etiology of isolated VSD might not be directly linked with this mutation 23153226_Based on these findings we conclude that the imprinted gene expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 are conserved between human and bovine 23295672_The imprinted PLAGL1 domain is flanked by CTCF-binding sites conserved between species in both expressing and non-expressing cells. 23415968_Paternal methylation aberrations at imprinting control regions of DLK1-GTL2, MEST (PEG1), and ZAC (PLAGL1) and global methylation levels are not associated with idiopathic recurrent spontaneous miscarriages. 24098585_ZAC1 and SSTR2 are down-regulated in non-functioning pituitary adenomas but not in somatotropinomas. 24260468_research identified a specific CpG site where determination of the methylation status was associated with both metastasis-free and overall survival in undifferentiated sarcoma. 24316753_There is positive correlations between the ZAC1 DMR methylation index (MI) and estimated fetal weight (EFW) at 32 weeks of gestation, weight at birth and weight at one year of age (respectively, r = 0.15, 0.09, 0.14; P values = 0.01, 0.15, 0.03). 24993786_Fetal growth can be influenced by altered expression of the PLAGL1 gene network in human placenta. 25091631_DNA deletion and promoter hyper-methylation both contribute to the down-regulation of PLAGL1 in gastric adenocarcinoma 26134521_Results suggest that dysregulation of PLAGL1 expression may be involved in the progression of colorectal cancer (CRC) but the patient survival data do not confirm applicability of the PLAGL1 expression level as a prognostic factor in CRC. 26851016_High PLAGL1 mRNA and protein levels were associated with Clear Cell Renal Cell Carcinoma. 27178226_PLAGL1 methylation/expression may be altered after assisted reproductive technologies. 28260048_These results suggest that OCT attenuates SGC-7901 cell proliferation by enhancing P300-HAT activity through the interaction of ZAC and P300, causing a reduction in pS10-H3 and an increase in acK14-H3. These findings provide insight for future research on OCT and further demonstrate the potential of OCT to be used as a therapeutic agent for gastric cancer 28957425_report here a novel PLAGL1 promoter (P5) derived from the insertion of a primate-specific, MIR3 SINE retrotransposon. P5 is highly utilized in lymphocytes, particularly in T cells, and like P2, directs biallelic transcription 28985358_Plagl1 is a transcription factor that coordinated the regulation of a subset of imprinted gene network genes and controlled extracellular matrix composition. 30070601_Results suggest that PLAGL1 methylation is lower in very preterm infants compared to their full term counterparts, even when controlling for perinatal confounders 31296310_Focusing on ISL1 and PLAGL1 the authors found no statistically significant differences in methylation status at these loci between patients and control samples. However, 20 CpGs in PLAGL1 and 19 CpGs in ISL1 showed at least 25% increase or decrease of DNA methylation. 32678267_Gene expression profiling identifies the role of Zac1 in cervical cancer metastasis. 32693431_Imprinting aberrations of SNRPN, ZAC1 and INPP5F genes involved in the pathogenesis of congenital heart disease with extracardiac malformations. 33171905_Transcription Factor PLAGL1 Is Associated with Angiogenic Gene Expression in the Placenta. 34355256_Recurrent fusions in PLAGL1 define a distinct subset of pediatric-type supratentorial neuroepithelial tumors. | ENSMUSG00000019817 | Plagl1 | 1.543885e+03 | 1.1943944 | 0.256279292 | 0.2863345 | 8.041499e-01 | 0.3698555007 | 0.83664500 | No | Yes | 1.436629e+03 | 206.407904 | 1.322145e+03 | 194.770834 | |
| ENSG00000118600 | 10329 | RXYLT1 | protein_coding | Q9Y2B1 | FUNCTION: Acts as a UDP-D-xylose:ribitol-5-phosphate beta1,4-xylosyltransferase, which catalyzes the transfer of UDP-D-xylose to ribitol 5-phosphate (Rbo5P) to form the Xylbeta1-4Rbo5P linkage on O-mannosyl glycan (PubMed:27733679, PubMed:29477842) (Probable). Participates in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1), which is required for binding laminin G-like domain-containing extracellular proteins with high affinity (PubMed:25279699, PubMed:27601598, PubMed:27733679) (Probable). {ECO:0000269|PubMed:25279699, ECO:0000269|PubMed:27601598, ECO:0000269|PubMed:27733679, ECO:0000269|PubMed:29477842, ECO:0000305|PubMed:27130732}. | Congenital muscular dystrophy;Disease variant;Dystroglycanopathy;Golgi apparatus;Lissencephaly;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:25279699, ECO:0000269|PubMed:27130732, ECO:0000269|PubMed:27733679, ECO:0000269|PubMed:29477842}. | This gene encodes a type II transmembrane protein that is thought to have glycosyltransferase function. Mutations in this gene result in cobblestone lissencephaly. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, May 2013]. | hsa:10329; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of plasma membrane [GO:0005887]; nucleoplasm [GO:0005654]; ribitol beta-1,4-xylosyltransferase activity [GO:0120053]; protein O-linked mannosylation [GO:0035269] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23217329_TMEM5 mutations were frequently associated with gonadal dysgenesis and neural tube defects, and ISPD mutations were frequently associated with brain vascular anomalies. 27130732_TMEM5 is a UDP-xylosyl transferase that elaborates the O-mannose glycan structure on alpha-dystroglycan. The authors demonstrate in a zebrafish model as well as in a human patient that defects in TMEM5 result in muscular dystrophy in combination with abnormal brain development. 27733679_TMEM5 acts as a UDP-d-xylose:ribitol-5-phosphate beta1,4-xylosyltransferase in the biosynthetic pathway of O-mannosyl glycan. 29477842_Fukutin, FKRP, and TMEM5 form a complex while maintaining each of their enzyme activities. Data showed that endogenous fukutin and FKRP enzyme activities coexist with TMEM5 enzyme activity, and suggest the possibility that formation of this enzyme complex may contribute to specific and prompt biosynthesis of glycans that are required for dystroglycan function. | ENSMUSG00000034620 | Rxylt1 | 3.280944e+02 | 0.8729088 | -0.196097171 | 0.3359573 | 3.343352e-01 | 0.5631174376 | 0.89601175 | No | Yes | 2.780663e+02 | 43.938676 | 3.040422e+02 | 49.164486 |
| ENSG00000118707 | 60436 | TGIF2 | protein_coding | Q9GZN2 | FUNCTION: Transcriptional repressor, which probably repress transcription by binding directly the 5'-CTGTCAA-3' DNA sequence or by interacting with TGF-beta activated SMAD proteins. Probably represses transcription via the recruitment of histone deacetylase proteins. {ECO:0000269|PubMed:11427533}. | Alternative splicing;DNA-binding;Homeobox;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | The protein encoded by this gene is a DNA-binding homeobox protein and a transcriptional repressor, which appears to repress transcription by recruiting histone deacetylases to TGF beta-responsive genes. This gene is amplified and over-expressed in some ovarian cancers. Alternative splicing results in multiple transcript variants. A related pseudogene has been identified on chromosome 1. Read-through transcription also exists between this gene and the neighboring downstream C20orf24 (chromosome 20 open reading frame 24) gene. [provided by RefSeq, Dec 2010]. | hsa:60436; | centrosome [GO:0005813]; chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; negative regulation of transcription by RNA polymerase II [GO:0000122]; nodal signaling pathway [GO:0038092]; positive regulation of neuron differentiation [GO:0045666]; regulation of gastrulation [GO:0010470]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; retina development in camera-type eye [GO:0060041] | 20040491_Shows that the homologous mouse Tgif2 gene is necessary for gastrulation. 26330748_Corneal fibroblasts demonstrate the expression of TGIF1 and TGIF2 transcription factors. These transcriptional repressors are critical, at least partially, in mediating the antifibrotic effect of vorinostat in the cornea. 26464633_miR-34a could inhibit tumor invasion and metastasis in gastric cancer by targeting Tgif2 26638007_expression of miR-148a was regulated by DNA methylation and targeted by TGIF2. Its methylation may be a potential prognostic indicator in skin cancer 27448300_Data show that TGFB-induced factor 2 protein (TGIF2) is involved in microRNA miR-541-3p-regulated cell proliferation, invasion, migration, as well as cell cycle of non-small lung cancer (NSCLC) cells. 27712592_Authors suggest that miR-148a inhibits OC cell proliferation and invasion partly through inhibition of TGFI2. Therefore, this study highlights the importance of the miR-148a/TGFI2 axis in the malignant progression of OC. 27956704_Tgifs regulate ciliogenesis and suggests that Evi5l mediates at least part of this effect. 29431269_MiR-129-5p could inhibit glioma cell progression by targeting TGIF2, shining light for the development of target treatment for glioma 29895719_miR-34 inhibits growth and promotes apoptosis of osteosarcoma cells through targeted regulation of TGIF2 expression. 30632703_our study identified three mRNAs (TBX21, TGIF2, and CYCS) that were significantly altered between high- and low-risk patients with breast cancer. The three-mRNA model was independent and predicted the prognosis of patients robustly. Furthermore, this model could predict survival probability precisely in patients with or without metastasis. 30825648_Overexpression of TGIF1 lowers levels of cholesterol, triglycerides, and apoB. Overexpression of TGIF2 has more limited effects on lipid metabolism. 31113203_miRNA-34a could induce osteoarthritis synovial cell apoptosis via regulation of TGIF2 in vitro 32572908_TGIF2 promotes cervical cancer metastasis by negatively regulating FCMR. 32709240_miR-541 serves as a prognostic biomarker of osteosarcoma and its regulatory effect on tumor cell proliferation, migration and invasion by targeting TGIF2. 33714261_Circ-RNF13, as an oncogene, regulates malignant progression of HBV-associated hepatocellular carcinoma cells and HBV infection through ceRNA pathway of circ-RNF13/miR-424-5p/TGIF2. 34471941_Circular RNA circMMP1 Contributes to the Progression of Glioma Through Regulating TGIF2 Expression by Sponging miR-195-5p. 34741437_Circular RNA circCPA4 promotes tumorigenesis by regulating miR-214-3p/TGIF2 in lung cancer. | ENSMUSG00000062175 | Tgif2 | 3.185417e+03 | 1.1560602 | 0.209216479 | 0.3011032 | 4.881404e-01 | 0.4847580083 | 0.87421307 | No | Yes | 3.323023e+03 | 270.402693 | 2.611790e+03 | 218.499072 | |
| ENSG00000118939 | 7347 | UCHL3 | protein_coding | P15374 | FUNCTION: Deubiquitinating enzyme (DUB) that controls levels of cellular ubiquitin through processing of ubiquitin precursors and ubiquitinated proteins. Thiol protease that recognizes and hydrolyzes a peptide bond at the C-terminal glycine of either ubiquitin or NEDD8. Has a 10-fold preference for Arg and Lys at position P3'', and exhibits a preference towards 'Lys-48'-linked ubiquitin chains. Deubiquitinates ENAC in apical compartments, thereby regulating apical membrane recycling. Indirectly increases the phosphorylation of IGFIR, AKT and FOXO1 and promotes insulin-signaling and insulin-induced adipogenesis. Required for stress-response retinal, skeletal muscle and germ cell maintenance. May be involved in working memory. Can hydrolyze UBB(+1), a mutated form of ubiquitin which is not effectively degraded by the proteasome and is associated with neurogenerative disorders. {ECO:0000269|PubMed:19154770, ECO:0000269|PubMed:21762696, ECO:0000269|PubMed:22689415, ECO:0000269|PubMed:2530630, ECO:0000269|PubMed:9790970}. | 3D-structure;Cytoplasm;Hydrolase;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway | Mouse_homologues NA; + ;NA | The protein encoded by this gene is a member of the deubiquitinating enzyme family. Members of this family are proteases that catalyze the removal of ubiquitin from polypeptides and are divided into five classes, depending on the mechanism of catalysis. This protein may hydrolyze the ubiquitinyl-N-epsilon amide bond of ubiquitinated proteins to regenerate ubiquitin for another catalytic cycle. Alternative splicing results in multiple transcript variants that encode different protein isoforms. [provided by RefSeq, Aug 2012]. | hsa:7347; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; NEDD8-specific protease activity [GO:0019784]; peptidase activity [GO:0008233]; thiol-dependent deubiquitinase [GO:0004843]; ubiquitin binding [GO:0043130]; post-translational protein modification [GO:0043687]; protein catabolic process [GO:0030163]; protein deubiquitination [GO:0016579]; protein ubiquitination [GO:0016567]; ubiquitin-dependent protein catabolic process [GO:0006511] | 15531586_1.45 A resolution crystal structure of human UCH-L3 in complex with the inhibitor ubiquitin vinylmethylester, an inhibitor that forms a covalent adduct with the active site cysteine of ubiquitin-specific proteases 16402389_Upregulation of UCH-L3 is associated with Uterine Cervical Neoplasms 19047059_Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. 19154770_These results indicate that mono-Ub and Ub dimers may regulate the enzymatic functions of UCH-L1 and UCH-L3, respectively, in vivo. 19343046_Observational study of gene-disease association. (HuGE Navigator) 19476499_Untangling the folding mechanism of the 5(2)-knotted protein UCH-L3 19636824_identification and the sequence-specific assignments for 186 out of the 218 UCH-L3 (230 less 12 prolines) backbone 15N and amide proton resonances 21762696_impaired UCH-L3 function may contribute to the accumulation of full length UBB(+1) in various pathologie 22679485_Data identified of UCHL3 as an essential deubiquitinase in adipogenesis. 25194810_UCH-L3 is a novel regulator of epithelial-mesenchymal transition and cell migration in prostate cancer cells. 25369561_Using a series of engineered protein substrates, which are similar in size yet differ in secondary structure, we demonstrate that thermal stability is a key factor that significantly affects UCH-L3 hydrolysis. 27780264_Study demonstrated positive correlations between the level and activity of UCHL3 and sperm characteristics and function suggesting that UCHL3 may play a role in male infertility. 28583475_The authors find that UCHL3 regulates COPS5-dependent deneddylation of Cullin1, which is an essential component of SCF(beta-TrCP) complex and associated with SCF(beta-TrCP) activities. The authors further demonstrate that UCHL3 upregulates the levels of SCF(beta-TrCP) substrates including IFN-I receptor IFNAR1, which enhances IFN-I mediated signaling pathway and antiviral activity. 29898404_Depletion of UCHL3 increases TDP1 ubiquitylation and turnover rate and sensitizes cells to TOP1 poisons. Overexpression of UCHL3, but not a catalytically inactive mutant, suppresses TDP1 ubiquitylation and turnover rate. 30559450_UCHL3 facilitates cellular viability after DSB induction by antagonizing Ku80 ubiquitylation to enhance Ku80 retention at sites of damage. 31477831_Our results indicate that highly expressed UCHL3 enhances inflammation by stabilizing TRAF2, which in turn facilitates tumourigenesis in ovarian cancer, and that UCHL3 is a potential target for ovarian cancer patients with increased inflammation. 31642235_UCH-L3 might play a role in epithelial ovarian cancer pathogenesis and progression 32180254_UCH-L3 promotes non-small cell lung cancer proliferation via accelerating cell cycle and inhibiting cell apoptosis. 32486284_Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration. 32546741_The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. 32738097_Deubiquitylase UCHL3 regulates bi-orientation and segregation of chromosomes during mitosis. 33238157_K27-Linked Diubiquitin Inhibits UCHL3 via an Unusual Kinetic Trap. 33616859_UCHL3 promotes aerobic glycolysis of pancreatic cancer through upregulating LDHA expression. 34016790_Silencing UCHL3 enhances radio-sensitivity of non-small cell lung cancer cells by inhibiting DNA repair. 35053210_Rational Development and Characterization of a Ubiquitin Variant with Selectivity for Ubiquitin C-Terminal Hydrolase L3. 35088238_Knockdown of UCHL3 inhibits esophageal squamous cell carcinoma progression by reducing CRY2 methylation. | ENSMUSG00000022111+ENSMUSG00000035337 | Uchl3+Uchl4 | 6.954104e+02 | 1.1668797 | 0.222655876 | 0.3254996 | 4.655881e-01 | 0.4950238590 | 0.87543103 | No | Yes | 7.464703e+02 | 132.167749 | 5.894160e+02 | 107.165981 |
| ENSG00000118971 | 894 | CCND2 | protein_coding | P30279 | FUNCTION: Regulatory component of the cyclin D2-CDK4 (DC) complex that phosphorylates and inhibits members of the retinoblastoma (RB) protein family including RB1 and regulates the cell-cycle during G(1)/S transition (PubMed:8114739, PubMed:18827403). Phosphorylation of RB1 allows dissociation of the transcription factor E2F from the RB/E2F complex and the subsequent transcription of E2F target genes which are responsible for the progression through the G(1) phase (PubMed:8114739, PubMed:18827403). Hypophosphorylates RB1 in early G(1) phase (PubMed:8114739, PubMed:18827403). Cyclin D-CDK4 complexes are major integrators of various mitogenenic and antimitogenic signals (PubMed:8114739, PubMed:18827403). {ECO:0000269|PubMed:18827403, ECO:0000269|PubMed:8114739}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Cyclin;Cytoplasm;Disease variant;Membrane;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene belongs to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance through the cell cycle. Cyclins function as regulators of CDK kinases. Different cyclins exhibit distinct expression and degradation patterns which contribute to the temporal coordination of each mitotic event. This cyclin forms a complex with CDK4 or CDK6 and functions as a regulatory subunit of the complex, whose activity is required for cell cycle G1/S transition. This protein has been shown to interact with and be involved in the phosphorylation of tumor suppressor protein Rb. Knockout studies of the homologous gene in mouse suggest the essential roles of this gene in ovarian granulosa and germ cell proliferation. High level expression of this gene was observed in ovarian and testicular tumors. Mutations in this gene are associated with megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 3 (MPPH3). [provided by RefSeq, Sep 2014]. | hsa:894; | chromatin [GO:0000785]; cyclin D2-CDK4 complex [GO:0097129]; cyclin-dependent protein kinase holoenzyme complex [GO:0000307]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cyclin-dependent protein serine/threonine kinase regulator activity [GO:0016538]; protein kinase binding [GO:0019901]; cell division [GO:0051301]; mitotic cell cycle phase transition [GO:0044772]; negative regulation of apoptotic process [GO:0043066]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0045737]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; positive regulation of protein phosphorylation [GO:0001934]; regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0000079] | 11992406_Activation of cyclin D1 and D2 promoters by human T-cell leukemia virus type I tax protein is associated with IL-2-independent growth of T cells 12112307_Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. 12771922_cyclin d2 hypermethylation associated with loss of cyclin d2 expression in subset of gastric cancer 12777997_results show that cyclin D2 is complexed with p27, leading to a model for testicular germ cell tumors whereby the overexpression of cyclin D2 leads to the functional sequestration of p27 in the presence of CCNE and CCND2, favoring cell proliferation 14581343_methylation of Cyclin D2 in prostate cancers correlates with clinicopathological features of poor prognosis 14601057_hypermethylated in invasive and in in situ lobular breast cancer 14612939_DNA hypomethylation is a mechanism underlying the increased expression of cyclin D2 in cancer cells and demethylation of cyclin D2 may be involved in development and progression of gastric carcinoma 15169570_p21/cyclin D2/cdk4 complex is not an inhibitory complex for the cyclin D2/cdk4 complex in HTLV-1 infected cells. 15245432_E2 may stimulate the growth of keratinocytes by inducing cyclin D2 expression via CREB phosphorylation by protein kinase A, dependent on cAMP. 15509806_conditional activation of FoxO3a leads to accumulation of BCL6 and down-regulation of cyclin D2 at protein and mRNA levels 15755896_Cyclin D1, D2, or D3 expression appears to be increased and/or dysregulated in virtually all MM tumors despite their low proliferative capacity 15797629_This suggests that dysregulation of CCND2 and CDK4 plays a specific role in WT tumorigenesis. 16548914_the recurrent T-ALL-associated t(12;14) results in overexpression of cyclin D2 17016690_Cyclin D2 promoter methylation and gene silencing may play an important functional role in prostate carcinogenesis. 17409409_Observational study of gene-disease association. (HuGE Navigator) 17486076_cyclin D2 expression in normal and malignant hematopoietic cells is regulated by ubiquitin/proteasome-dependent degradation triggered by Thr280 phosphorylation by GSK3beta or p38, which is induced by inhibition of the PI3K pathway 17873913_Responsible for neoplastic cell transformation when coexpressed with an activated Ras protein. 17882269_role for D-type cyclins in the excessive basal-cell proliferation and perturbed keratinocyte differentiation in the psoriatic epidermis 18174243_Observational study of gene-disease association. (HuGE Navigator) 18281541_Observational study of gene-disease association. (HuGE Navigator) 18391076_cyclin E expression in 2 t(11;14)-negative mantle cell lymphomas characterized by a cryptic t(2;14)(p24;q32) and identification of MYCN as a new lymphoma oncogene associated with a blastoid mantle cell lymphoma 18431519_Data demonstrate that RNA interference of genes encoding cyclin D1 and cyclin D2 (CCND1 and CCND2, respectively) inhibits proliferation and is progressively cytotoxic in human myeloma cells. 18483325_Forty one percent of breast cancers were associated with methylation of cyclin d2. 18504428_Tax-induced cell-cycle progression in T cells is mediated, at least in part, through cell-type-specific activation of the cyclin D2 and cdk6 genes through NF-kappaB and may be important for the cell-type-specific oncogenesis. 18507837_Observational study of gene-disease association. (HuGE Navigator) 18635970_All three D-type cyclins promoted robust hepatocyte proliferation and marked liver growth, although cyclin D3 stimulated less DNA synthesis than D1 or D2. 18714005_in response to cellular activation in T cells and B cells, a PTB-containing stability complex forms that contains binding sites for Rab8A and cyclin D(2) transcripts and increases their mRNA half-lifes 18827403_a pattern of translocalization suggests a spatial separation of the cyclin D-Cdk complex from pRb and DNA in the nucleus to regulate the G1-S transition 18930031_These results suggest that miR-302b plays an important role in maintaining the pluripotency of embryonal carcinoma cells and probably embryonic stem cells , by post-transcriptional regulation of Cyclin D2 expression. 18950845_Observational study of gene-disease association. (HuGE Navigator) 19124506_Observational study of gene-disease association. (HuGE Navigator) 19252133_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19258477_Observational study of gene-disease association. (HuGE Navigator) 19379567_Valproic acid could down-regulate mRNA expression of cyclin D2 in the kasumi-1 leukemic cell line. 19464003_study suggests that downstream signaling components of the PI3K/Akt pathway, GSK3 & cyclin D2 as well as the significant interaction between PTEN-PDK and between pAkt-pGSK3beta, are involved in the survival and proliferation of leiomyomas. 19508551_In colorectal adenocarcinoma expression of cyclin D2 at the margin was associated with vascular invasion, lymph node metastasis and liver metastasis 19577536_restoration of CCND2 expression potentially prevents the carcinogenesis of prostate cancer, which is mostly androgen receptor -dependent in the initial settings. 19608671_As a result of the mantle cell lymphoma translocation, cycD2 mRNA was highly over-expressed when compared with normal lymphoid tissue and other B-cell non-Hodgkin's lymphomas 19618401_Data show that methylation of CCND-2, p16, RAR-beta and RASSF-1a was significantly more prevalent in tumor than in normal tissue specimens. 19641124_data presented here indicate that cD2 is not necessary for cortical intermediate progenitor cells (IPCs) to emerge but that cD2 is used in progenitors as they transition into and expand their IPC numbers 19679881_the knockdown of cyclin D1 is compensated by the upregulation of cyclin D2, a more powerful strategy would be to inhibit both cyclin D1 and cyclin D2. 19738611_Observational study of gene-disease association. (HuGE Navigator) 19890848_Cyclin D2 and the CDK substrate p220(NPAT) are required for self-renewal of human embryonic stem cells. 19945765_hypermethylation in tumors from non-small cell lung cancer patients is associated with gender and histologic type 20066902_cyclin D2+ cells represent a pool of leukemic cells with the potential to enter the dividing compartment. 20077038_High cyclin D2 is associated with higher grade (III and IV) of astrocytoma. 20414251_Observational study of gene-disease association. (HuGE Navigator) 20414251_One common polymorphism in the 5'-untranslated region (that is, rs1049606) and the most common haplotype (CCND-ht1 [T-C-T-A-T]), however, were significantly associated with hepatatis B virus clearance. 20418948_the 3'UTR of E2F2 and CCND2 were directly bound to let-7a and let-7a down-regulated the expression of E2F2 and CCND2, suppressing prostate cancer cell proliferation in culture 20473882_High level of CCND2 expression is associated with nasopharyngeal carcinoma. 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 20939487_Zinc can accelerate the proliferation and DNA reproduction of cord blood-derived mesenchymal stem cells and increase the contents of cyclin D2, CDK4, and cellular total protein. 21039844_Our result supported the finding that ubiquitin-specific protease 22 can interact with cyclin D2 and active cyclin D2 activity. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21059263_sonic hedgehog-GLI1 downstream target genes PTCH1, Cyclin D2, Plakoglobin, PAX6 and NKX2.2 are differently regulated in medulloblastoma and astrocytoma 21479697_High expression of cyclin D2 is associated with mantle cell lymphoma. 21504716_Authors present a CD5-positive, CCND1-negative B-cell lymphoma with a novel translocation involving CCND2 and the immunoglobulin lambda (IGL) gene. 21521747_Major regulator of CCND2 expression in pancreatic beta cells is glucose acting via glycolysis and calcium channels; glucose up-regulates nuclear CCND2 in quiescent cells and down-regulates CCND2 in replicating cells. 21745168_Data show that combination of siRNA-mediated inhibition of the cyclin D1 and cyclin D2 along with chemotherapeutic agents could potentially be used to lower the effective doses of the chemotherapeutic agents and reduce drug-related toxicities. 21790895_Cyclin D2 is overexpressed in the proliferation centers of CLL/SLL and this is due, in part, to the upregulation of NF-kappaB-related pathways. 21878650_ethanol-induced neuronal apoptosis follows both the mitochondria-mediated (miR-497- and BCL2-mediated) and non-mitochondria-mediated (miR-302b- and CCND2-mediated) pathway. 22004425_Single nucleotide polymorphisms of CCND2, RAD23B, GRP78, CEP164, MDM2, and ALDH2 genes were significantly associated with development and recurrence of hepatocellular carcinoma in Japanese patients with hepatitis C virus. 22048254_CCND2, was the most 'dietary sensitive' genes, as methylation of their promoters was associated with intakes of at least two out of the eight dietary methyl factors examined. 22323446_FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. 22330340_Downregulation of miR-1, -206 and -29 stabilizes the expression of PAX3 and CCND2 in both embryonal and alveolar rhabdomyosarcoma. 22395070_The authors demonstrate that Cyclin D2 is also expressed in the developing human cortex within similar domains, thus indicating that its role as a fate determinant is ancient and conserved. 22534667_The present study suggests that reduced expression of cyclin D2 in stage III NSCLC may be associated with poor RFS. And, cyclin D2 may have a distinct role from cyclin D1 in NSCLC. 22848417_miR-182, a p53 dependent miRNA, suppressed the expression of MITF, BCL2, cyclin D2 and functioned as a potent tumor suppressor in uveal melanoma cells. 22853316_Transgenic K562 cells have distinct gene expression profiles both in steady state and during terminal erythroid differentiation, with GATA1s expression characterised by lack of repression of MYB, CCND2 and SKI. 22954617_Experimental verification of the ability of this small RNA molecule to regulate the expression of CCND2, a gene with documented oncogenic activity, confirms its functional role as a miRNA. 22964630_our results demonstrate that cyclin D2 has a critical role in cell cycle progression and the tumorigenicity of glioblastoma stem cells 23255553_Chromosomal rearrangements of the CCND2 locus were detected in 55% of the cases, with an IG gene as partner in 18 of 22, in particular with light chains (10 IGK@ and 5 IGL@)for mantle cell lymphoma. 23266556_CCND2 gene polymorphism is associated in the pathogenesis of colorectal cancers. 23348698_miR-206 could suppress gastric carcinoma cell proliferation at least partially through targeting cyclinD2 expression. 23428540_miR-154 plays a prominent role in prostate cancer proliferation by suppressing CCND2 23466356_Cyclin D2 is a direct target of miR-206 in breast cancer cells 23591848_SUMO modification of ATF3 influences CCND1/2 activity and cellular proliferation of prostate cancer PC3 and DU145 cells and explains at least in part how ATF3 functions to regulate cancer development. 23989931_Study reveals molecular insight into how the Ets family transcription factor Pea3 favors EMT and contributes to tumorigenesis via a negative regulatory loop with Cyclin D2, a new Pea3 target gene. 24249158_Frequent aberrations of CCND2 and RB1 is associated with intracranial germ cell tumors 24464100_A low-frequency (1.47%) variant in intron 1 of CCND2, rs76895963[G], reduces risk of type 2 diabetes by half (odds ratio (OR) = 0.53, P = 5.0 x 10(-21)) and is correlated with increased CCND2 expression. 24705253_De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. 24743557_results provide evidence that CCND2 polymorphism rs3217927 may be involved in the etiology of childhood ALL, and the GG genotype of rs3217927 may modulate the genetic susceptibility to childhood ALL in the Chinese population. 24992041_Together, this study has uncovered a positive role of cyclin D2 in hepatitis B virus replication. 25193387_Stepwise Cox regression modelling suggested that the methylation of genes HSPB1, CCND2 and DPYS contributed objective prognostic information to Gleason score and PSA with respect to prostate cancer-related death. 25572712_miR206 inhibits glioma progression via the regulation of cyclinD2 and that miR206 may be a novel biomarker with potential for use as a therapeutic target in gliomas. 25605810_Study establishes that a low-frequency allele in CCND2 halves the risk of type 2 diabetes primarily through enhanced insulin secretion. 25605810_study establishes that a low frequency allele in CCND2 halves the risk of type 2 diabetes primarily through enhanced insulin secretion 25673160_OY-TES-1 downregulation in liver cancer cells promotes cell proliferation by upregulating CCND2 and CDCA3. 25738424_Methylation changes were enriched in MSX1, CCND2, and DAXX at specific loci within the hippocampus of patients with schizophrenia and bipolar disorder. 25817794_that re-introducing CDK6, cyclin-D1 or cyclin-D2 expression partially, but significantly, rescues cells from the suppression of cell proliferation and cell cycle arrest mediated by miR-340 25824739_Cyclin D2 hypermethylation is associated with breast cancer. 25960238_the dysregulation of miR-206-CCND2 axis may contribute to the aggressive progression and poor prognosis of human gastric cancer. 25961636_Treatment of rSCC-61 and SCC-61 with the DNA hypomethylating agent 5-aza-2'deoxycitidine increased CCND2 levels only in rSCC-61 cells, while treatment with the control reagent cytosine arabinoside did not influence the expression of this gene 25980436_CCND2 was identified as a putative target gene for SMYD3 transcriptional regulation, through trimethylation of H4K20.Our results support a proto-oncogenic role for SMYD3 in prostate carcinogenesis, mainly due to its methyltransferase enzymatic activity. 26221902_Up-regulation of clyclinD2 regulates laryngeal squamous cell carcinoma cell growth. 26225959_miR-198 inhibited HaCaT cell proliferation by directly targeting CCND2. 26290144_The results do not support our hypothesis that common germline genetic variants in the CCND2 genes is associated with the risk of developing medulloblastoma. 26341922_these results highlight the impact of CCND2 3'UTR shortening on miRNA-dependent regulation of CCND2 in multiple myeloma. 26707908_this study shows that miR-124-3p may negatively regulate the transcription of the STAT3 by interfering with its 3'UTR, and the degradation of STAT3 affects its downstream expression of such as p-STAT3, CCND2 and MMP-2 26782072_Bioinformatics analysis further revealed cyclin D2 (CCND2) and AKT3, putative tumor promoters, as potential targets of miR610. Data from reporter assays showed that miR610 directly binds to the 3'untranslated 26986233_miR-155 overexpression plays a promoting role in the proliferative, migratory and invasive behavior of OSCC cells. Its effects on OSCC are possibly associated with its regulation of the BCL6/cyclin D2 axis. 27146121_CCND1 is downregulated, whereas CCND2 is not, following ionizing radiation (IR) . Both CCND1- and CCND2-expressing MM cells arrested in S/G2/M, and did not differ in other cell-cycle proteins or sensitivity to IR.Differential expression of D-cyclin does not appear to affect cell-cycle response to IR, and is unlikely to underlie differential sensitivity to DNA damage. 27494902_these results demonstrated that miR-146a-5p could suppress the proliferation and cell cycle progression in non-small cell lung cancer cells by inhibiting the expression of CCND1 and CCND2 27541004_cyclin D2 acts as regulator of cell cycle proteins affecting SAMHD1-mediated HIV-1 restriction in non-proliferating macrophages. 27572135_data indicate that linc00598 plays an important role in cell cycle regulation and proliferation through its ability to regulate the transcription of CCND2. 27583477_data suggested that loss of CCND2 expression is closely associated with the promoter aberrant methylation 27633099_the phosphorylation levels of ErbB2, ErbB3 and Akt and the protein levels of cyclin D1 were decreased by lapatinib treatment of HSC3, HSC4 and Ca9-22 cells 27843138_Mutation in the CCND2 gene is associated with acute myeloid leukemia. 27855694_The surface immune molecule CD274 plays a critical role in the proliferation of leukemia-initiating cells, LICs. The CD274/JNK/Cyclin D2 pathway promotes the cell cycle entry of LIC. 27918592_MiR-497 significantly suppressed cell proliferation by arresting the cell cycle through the CCND2 protein. 27923660_CCND2-AS1 promotes glioma cells proliferation and growth in a process that involves Wnt and beta-catenin 28000032_focal gain of CCND2 and adjacent regions was seen in 8 of 9 (89%) gemistocytic IDH mutant astrocytomas 28630439_present study provides novel insight into the genetics of myeloid malignancies, namely Philadelphia-negative neutrophilic leukemias. Our work suggests that in addition to the commonly recurring classes of genes that are frequently mutated in these malignancies, recurrent mutations in cyclin D2, and perhaps other cell cycle regulators, have biochemical and therapeutic consequences and may play important roles in the pathog 28643014_NAV2 and CCND2 are novel candidate prognostic markers in uterine leiomyosarcoma and uterine low-grade endometrial stromal sarcoma, respectively. 28674394_the lower expression levels of CCND2 markedly correlated with prostate tumor progression 28933597_High CCND2 expression is associated with Metastasis of Colorectal Cancer. 29018036_CCND2 overexpression activates cell cycle progression in induced pluripotent stem cell-derived cardiomyocytes and results in enhanced cardiac repair. 29471894_MiR-615 as a tumor suppressor by targeting cyclin D2 in prostate cancer. 29662889_miR-29b suppressed cellular proliferation and promoted apoptosis of pulmonary artery smooth muscle cells, possibly through the inhibition of Mcl-1 and CCND2. 29764627_Our findings showed that by promoting adherence to HDP and increasing intake of some vegetables and fruits, the risk of MetS or its components reduced in G allele carriers; these associations were not observed in the AA genotype group. 29945347_GACAT3 promotes breast cancer malignancy by sponging miR-497, leading to the enhancement of its endogenous target CCND2. These results suggest that GACAT3/miR-497/CCND2 is a potential therapeutic target and biomarker for breast cancer 29997218_Our results provide novel insights into the function of EBNA3C on cell progression by regulating the cyclin D2 protein and raise the possibility of the development of new anticancer therapies against EBV-associated cancers. 30068546_PRC1-targeting element appears to act in concert with an adjacent cytosine-phosphate-guanine (CpG) island to arrange for the robust binding of PRC1 and PRC2 to repressed CCND2 30193733_Acute genome duplication in haploids restored cyclin D2 expression to diploid level, indicating that the regulation of cyclin D2 expression is directly linked to ploidy. 30227870_Our results indicate that miR-4317 can reduce Non-small cell lung cancer (NSCLC) cell growth and metastasis by targeting FGF9 and CCND2. These findings provide new evidence of miR-4317 as a potential non-invasive biomarker and therapeutic target for NSCLC. 30300583_that RUNX1/ETO maintains leukemia by promoting cell cycle progression and identifies G1 CCND-CDK complexes as promising therapeutic targets for treatment of RUNX1/ETO-driven acute myeloid leukemia 30429236_LncRNA GAS8-AS1 suppresses papillary thyroid carcinoma cell growth through the miR-135b-5p/CCND2 axis. 30520141_Overexpression of CCND2 suppressed FOXD2-AS1 knockdown-induced inhibition of glioma malignancy. Taken together, our findings highlight the FOXD2-AS1/miR-185-5p/CCND2 axis in the glioma development 30538135_CCND2/CCND3 rearrangements with immunoglobulin genes are associated with cyclin D1(-) mantle cell lymphoma. 30664171_the present study demonstrated that miR6713p exerted its tumorsuppressive roles via directly targeting CCND2 in nonsmall cell lung cancer 30864742_the results indicated that miR145 serves a critical role in suppressing the biological behavior of EOC cells by targeting CCND2 and E2F3. 30950241_CCND2 played an oncogenic role in gastric cancer, whereas it could also be a tumor suppressor in NSCLC. 30988120_Mechanism for IL-15-Driven B Cell Chronic Lymphocytic Leukemia Cycling: Roles for AKT and STAT5 in Modulating Cyclin D2 and DNA Damage Response Proteins. 30999112_The role and regulation of UCA1/miR-204/CCND2 regulatory axis in pCGs. 31253987_Long non-coding RNA JPX correlates with poor prognosis and tumor progression in non-small-cell lung cancer by interacting with miR-145-5p and CCND2. 31392505_the results of rescue experiments suggested that the oncogenic role of KCNQ1OT1 was performed through sponging miR-4458 and upregulating CCND2 during osteosarcoma development, providing a novel perspective of intervention in osteosarcoma management. 31432138_the findings suggested that upregulation of TP73AS1 promoted cervical cancer progression by promoting CCND2 via the suppression of miR607 expression. 31432162_findings demonstrated that cisplatin could effectively inhibit lung adenocarcinoma cell proliferation by decreasing cyclin D2 expression via miR93. 31511084_we first identified JAK2/STAT3/CCND2 signaling as a resistance mechanism for the persistent growth of CSCs after RT, suggesting potential biomarkers and regimens for improving outcomes among CRC patients. 31527584_PICOT binding to chromatin-associated EED negatively regulates cyclin D2 expression by increasing H3K27me3 at the CCND2 gene promoter. 31568682_Prognostic impact of glioblastoma stem cell markers OLIG2 and CCND2. 32113162_BAP18 is involved in upregulation of CCND1/2 transcription to promote cell growth in oral squamous cell carcinoma. 32414847_CCND2 mutations are infrequent events in BCR-ABL1 negative myeloproliferative neoplasm patients. 32820798_CircTHBS1 targeting miR-211/CCND2 pathway to promote cell proliferation and migration potential in primary cystitis glandularis cells. 32828867_miR-451a suppresses the development of breast cancer via targeted inhibition of CCND2. 33320844_Measurement of Cyclin D2 (CCND2) Gene Promoter Methylation in Plasma and Peripheral Blood Mononuclear Cells and Alpha-Fetoprotein Levels in Patients with Hepatitis B Virus-Associated Hepatocellular Carcinoma. 33560216_CCND2 amplification is an independent adverse prognostic factor in IDH-mutant lower-grade astrocytoma. 33634562_Medulloblastoma in the setting of megalencephaly polymicrogyria polydactyly hydrocephalus. 33676174_Identification of CXCL13/CXCR5 axis's crucial and complex effect in human lung adenocarcinoma. 34077003_Decreased ERbeta expression and high cyclin D1 expression may predict early CRC recurrence in high-risk Duke's B and Duke's C stage. 34087052_Proximal variants in CCND2 associated with microcephaly, short stature, and developmental delay: A case series and review of inverse brain growth phenotypes. 34102226_Downregulation of miR-892b inhibits the progression of osteoarthritis via targeting cyclin D1 and cyclin D2. 34223793_Hsa_circ_0008360 sponges miR-186-5p to target CCND2 to modulate high glucose-induced vascular endothelial dysfunction. 34255617_Long noncoding RNA FAM225B facilitates proliferation and metastasis of nasopharyngeal carcinoma cells by regulating miR-613/CCND2 axis. 34334537_Hypermethylation of Cyclin D2 Predicts Poor Prognosis of Hepatitis B Virus-Associated Hepatocellular Carcinoma after Hepatectomy. 34521827_Chromatin-based, in cis and in trans regulatory rewiring underpins distinct oncogenic transcriptomes in multiple myeloma. 34664684_Circular RNA hsa_circ_105039 promotes cardiomyocyte differentiation by sponging miR17 to regulate cyclinD2 expression. 34801563_Reduced expression of microRNA-432-5p by DNA methyltransferase 3B leads to development of colorectal cancer through upregulation of CCND2. 34995730_Expression of obesity- and type-2 diabetes-associated genes in omental adipose tissue of individuals with obesity. | ENSMUSG00000000184 | Ccnd2 | 3.354766e+03 | 0.9550031 | -0.066422612 | 0.2910761 | 5.356128e-02 | 0.8169783525 | 0.96393083 | No | Yes | 3.052603e+03 | 269.466820 | 3.069683e+03 | 277.988494 | |
| ENSG00000119471 | 84263 | HSDL2 | protein_coding | Q6YN16 | FUNCTION: Has apparently no steroid dehydrogenase activity. {ECO:0000269|PubMed:19703561}. | 3D-structure;Acetylation;Alternative splicing;Hydroxylation;NADP;Oxidoreductase;Peroxisome;Reference proteome | hsa:84263; | membrane [GO:0016020]; mitochondrion [GO:0005739]; peroxisome [GO:0005777]; oxidoreductase activity [GO:0016491] | 20583170_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 27658777_High expression of HSDL2 is associated with glioma. 29894468_Study shows that HSDL2 upregulation is associated with ovarian cancer progression. HSDL2 knockdown inhibited cell proliferation, colony formation, motility, and tumorigenesis. 31101684_Hydroxysteroid dehydrogenase like 2 (HSDL2) expression was increased in papillary thyroid carcinoma (PTC) tissues and cells, which could promote tumor progression in vitro and in vivo. 31217732_Study reported that HSDL2 was upregulated in two human bladder cancer cell lines 5637 and T24 compared to normal human urothelial cells. Lentiviral-mediated HSDL2 knockdown inhibited the proliferation and colony formation while promoted the apoptosis of human bladder cancer T24 cells in vitro and in nude mice T24 derived xenografts in vivo. Results suggest that HSDL2 plays an oncogenic role in bladder cancer. 32211805_Knockdown of HSDL2 inhibits lung adenocarcinoma progression via down-regulating AKT2 expression. 33738911_Lipid metabolism regulator human hydroxysteroid dehydrogenase-like 2 (HSDL2) modulates cervical cancer cell proliferation and metastasis. | ENSMUSG00000028383 | Hsdl2 | 3.456505e+02 | 0.9765516 | -0.034231889 | 0.3737967 | 8.442703e-03 | 0.9267900608 | 0.98598977 | No | Yes | 3.141488e+02 | 59.544404 | 3.184489e+02 | 61.803853 | ||
| ENSG00000119682 | 9870 | AREL1 | protein_coding | O15033 | FUNCTION: E3 ubiquitin-protein ligase that catalyzes 'Lys-11'- or 'Lys-33'-linked polyubiquitin chains, with some preference for 'Lys-33' linkages (PubMed:25752577). E3 ubiquitin-protein ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates (PubMed:23479728, PubMed:31578312). Ubiquitinates SEPTIN4, DIABLO/SMAC and HTRA2 in vitro (PubMed:23479728). Modulates pulmonary inflammation by targeting SOCS2 for ubiquitination and subsequent degradation by the proteasome (PubMed:31578312). {ECO:0000269|PubMed:23479728, ECO:0000269|PubMed:25752577, ECO:0000269|PubMed:31578312}. | 3D-structure;Alternative splicing;Apoptosis;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:23479728, ECO:0000269|PubMed:25752577}. | hsa:9870; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; apoptotic process [GO:0006915]; negative regulation of apoptotic process [GO:0043066]; positive regulation of protein catabolic process [GO:0045732]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein K11-linked ubiquitination [GO:0070979]; protein K33-linked ubiquitination [GO:1990390]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; regulation of inflammatory response [GO:0050727]; ubiquitin-dependent protein catabolic process [GO:0006511] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 27162139_new protein isoform encoded by KIAA0317, termed fibrosis-inducing E3 ligase 1 (FIEL1), which potently stimulates the TGFbeta signaling pathway through the site-specific ubiquitination of PIAS4. 31732561_The AREL1 HECT domain assembled Lys(33)-, Lys(48)-, and Lys(63)-linked polyubiquitin chains. E701A substitution in the AREL1 HECT domain substantially increased its autopolyubiquitination and SMAC ubiquitination activity, whereas deletion of the last three amino acids at the C terminus completely abrogated AREL1 autoubiquitination and reduced SMAC ubiquitination. 32151281_Epigenetic modulation of AREL1 and increased HLA expression in brains of multiple system atrophy patients. | ENSMUSG00000042350 | Arel1 | 3.203075e+03 | 0.8435295 | -0.245489612 | 0.2856489 | 7.422689e-01 | 0.3889350448 | 0.84317192 | No | Yes | 2.401457e+03 | 261.808308 | 3.067721e+03 | 342.601357 | |
| ENSG00000119685 | 23093 | TTLL5 | protein_coding | Q6EMB2 | FUNCTION: Polyglutamylase which modifies tubulin, generating polyglutamate side chains on the gamma-carboxyl group of specific glutamate residues within the C-terminal tail of tubulin. Preferentially mediates ATP-dependent initiation step of the polyglutamylation reaction over the elongation step. Preferentially modifies the alpha-tubulin tail over a beta-tail (By similarity). Required for CCSAP localization to both polyglutamylated spindle and cilia microtubules (PubMed:22493317). Increases the effects of transcriptional coactivator NCOA2/TIF2 in glucocorticoid receptor-mediated repression and induction and in androgen receptor-mediated induction (PubMed:17116691). {ECO:0000250|UniProtKB:Q8CHB8, ECO:0000269|PubMed:17116691, ECO:0000269|PubMed:22493317}. | ATP-binding;Alternative splicing;Cell projection;Cilium;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Disease variant;Ligase;Magnesium;Metal-binding;Microtubule;Nucleotide-binding;Nucleus;Reference proteome;Transcription | This gene encodes a member of the tubulin tyrosine ligase like protein family. This protein interacts with two glucocorticoid receptor coactivators, transcriptional intermediary factor 2 and steroid receptor coactivator 1. This protein may function as a coregulator of glucocorticoid receptor mediated gene induction and repression. This protein may also function as an alpha tubulin polyglutamylase.[provided by RefSeq, Feb 2010]. | hsa:23093; | centrosome [GO:0005813]; cilium [GO:0005929]; cytosol [GO:0005829]; microtubule [GO:0005874]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; tubulin binding [GO:0015631]; tubulin-glutamic acid ligase activity [GO:0070740]; microtubule cytoskeleton organization [GO:0000226]; protein polyglutamylation [GO:0018095]; retina development in camera-type eye [GO:0060041] | 17116691_STAMP is an important new, downstream component of GR action in both gene activation and gene repression. 20374646_This study indicates that a physiological function of STAMP in several settings is to modify cell growth rates in a manner that can be independent of steroid hormones. 24791901_this study has performed exome sequencing in 28 individuals with a similar disease phenotype and subsequently used a casecontrol approach to identify mutations in TTLL5 as a cause of recessive retinal dystrophy. 28173158_5 homozygous variants [p.(Asp594fs), p.(Gln117*), p.(Met712fs), p.(Ile756Phe) and p.(Glu543Lys)] in TTLL5, in 8 patients from 6 families were identified. 2 male patients carrying truncating TTLL5 variants also displayed a reduction in sperm motility and infertility, whereas those carrying missense changes were fertile. TTLL has multiple viable isoforms, being highly expressed in retina, testis and spermatozoon flagellum. 28356705_in a study of 3 family members from 2 generations, identified in a previously misdiagnosed incomplete congenital stationary night blindness (icCSNB) case a splice-site mutation in intron 3 of TTLL5 (c.182-3_182-1delinsAA); reinvestigation of the clinical data corrected the diagnosis to cone dystrophy 30517872_Binding of CSAP to TTLL5 promotes relocalization of TTLL5 toward microtubules. 34203883_Novel TTLL5 Variants Associated with Cone-Rod Dystrophy and Early-Onset Severe Retinal Dystrophy. 35365235_Expanding the phenotype of TTLL5-associated retinal dystrophy: a case series. | 8.590802e+02 | 0.9458432 | -0.080327084 | 0.2866288 | 8.099959e-02 | 0.7759473407 | 0.95540959 | No | Yes | 8.186973e+02 | 128.744264 | 7.992742e+02 | 128.928777 | |||
| ENSG00000119801 | 51646 | YPEL5 | protein_coding | P62699 | FUNCTION: Component of the CTLH E3 ubiquitin-protein ligase complex that selectively accepts ubiquitin from UBE2H and mediates ubiquitination and subsequent proteasomal degradation of the transcription factor HBP1 (PubMed:29911972). Required for normal cell proliferation (By similarity). {ECO:0000250|UniProtKB:Q65Z55, ECO:0000269|PubMed:29911972}. | Cytoplasm;Cytoskeleton;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Zinc | hsa:51646; | extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; microtubule organizing center [GO:0005815]; midbody [GO:0030496]; mitotic spindle pole [GO:0097431]; nucleus [GO:0005634]; tertiary granule lumen [GO:1904724]; ubiquitin ligase complex [GO:0000151]; metal ion binding [GO:0046872]; cell population proliferation [GO:0008283] | 20508983_Observational study of gene-disease association. (HuGE Navigator) 20580816_YPEL5 protein of the YPEL gene family is involved in the cell cycle progression by interacting with two distinct proteins RanBPM and RanBP10. 23382248_results indicate that SCN-iPS cells provide a useful disease model for SCN, and the activation of the Wnt3a/beta-catenin pathway may offer a novel therapy for SCN with ELANE mutation 27705791_YPEL5 silencing enhanced the induction of IFNB1 by pattern recognition receptors and phosphorylation of TBK1/IKBKE kinases, whereas co-immunoprecipitation experiments revealed that YPEL5 interacted physically with IKBKE. 28173693_results demonstrate the functional conservation between yeast Moh1 and human YPEL5, and their involvement in mitochondria-dependent apoptosis induced by DNA damage 33411363_METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. | ENSMUSG00000039770 | Ypel5 | 2.522123e+02 | 0.8219037 | -0.282958798 | 0.3403848 | 6.911688e-01 | 0.4057671108 | 0.84651196 | No | Yes | 2.453657e+02 | 43.801745 | 2.472327e+02 | 45.224171 | ||
| ENSG00000119844 | 54812 | AFTPH | protein_coding | Q6ULP2 | FUNCTION: Component of clathrin-coated vesicles (PubMed:15758025). Component of the aftiphilin/p200/gamma-synergin complex, which plays roles in AP1G1/AP-1-mediated protein trafficking including the trafficking of transferrin from early to recycling endosomes, and the membrane trafficking of furin and the lysosomal enzyme cathepsin D between the trans-Golgi network (TGN) and endosomes (PubMed:15758025). {ECO:0000269|PubMed:15758025}. | Alternative splicing;Cytoplasm;Cytoplasmic vesicle;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | hsa:54812; | AP-1 adaptor complex [GO:0030121]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; trans-Golgi network membrane [GO:0032588]; clathrin binding [GO:0030276]; intracellular transport [GO:0046907]; protein transport [GO:0015031] | 15758025_the aftiphilin/p200/gamma-synergin complex facilitates AP-1 function 18815278_Data show that by recruiting aftiphilin/gamma-synergin in addition to clathrin, AP-1 coordinates formation of Weibel-Palade bodies with their acquisition of a regulated secretory phenotype. 33090728_Expression profiles and prognostic significance of AFTPH in different tumors. | ENSMUSG00000049659 | Aftph | 2.673872e+02 | 0.7208559 | -0.472217115 | 0.3499985 | 1.763980e+00 | 0.1841289030 | 0.77834741 | No | Yes | 2.404684e+02 | 42.153812 | 3.134100e+02 | 56.108505 | ||
| ENSG00000120451 | 399979 | SNX19 | protein_coding | Q92543 | FUNCTION: Plays a role in intracellular vesicle trafficking and exocytosis (PubMed:24843546). May play a role in maintaining insulin-containing dense core vesicles in pancreatic beta-cells and in preventing their degradation. May play a role in insulin secretion (PubMed:24843546). Interacts with membranes containing phosphatidylinositol 3-phosphate (PtdIns(3P)) (By similarity). {ECO:0000250|UniProtKB:Q6P4T1, ECO:0000269|PubMed:24843546}. | Alternative splicing;Cytoplasmic vesicle;Endosome;Exocytosis;Lipid-binding;Membrane;Protein transport;Reference proteome;Transport | Islet antigen-2 (IA-2) is an autoantigen in type 1 diabetes and plays a role in insulin secretion. IA-2 is found in dense-core secretory vesicles and interacts with the product of this gene, a sorting nexin. In mouse pancreatic beta-cells, the encoded protein influenced insulin secretion by stabilizing the number of dense-core secretory vesicles. [provided by RefSeq, Dec 2016]. | hsa:399979; | cytoplasm [GO:0005737]; early endosome membrane [GO:0031901]; intracellular membrane-bounded organelle [GO:0043231]; phosphatidylinositol binding [GO:0035091]; chondrocyte differentiation [GO:0002062]; dense core granule maturation [GO:1990502]; exocytosis [GO:0006887]; insulin secretion [GO:0030073] | 17975119_Observational study of gene-disease association. (HuGE Navigator) 18073581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19023099_Observational study of gene-disease association. (HuGE Navigator) 19730683_Observational study of gene-disease association. (HuGE Navigator) 19752551_Observational study of gene-disease association. (HuGE Navigator) 30635639_Schizophrenia risk variants influence multiple classes of transcripts of sorting nexin 19 (SNX19). 33649454_Single molecule in situ hybridization reveals distinct localizations of schizophrenia risk-related transcripts SNX19 and AS3MT in human brain. 34315878_SNX19 restricts endolysosome motility through contacts with the endoplasmic reticulum. | ENSMUSG00000031993 | Snx19 | 2.499362e+02 | 0.8843188 | -0.177361561 | 0.3007077 | 3.392036e-01 | 0.5602893044 | 0.89534708 | No | Yes | 2.314658e+02 | 60.275534 | 2.687723e+02 | 72.035181 | |
| ENSG00000120526 | 84955 | NUDCD1 | protein_coding | Q96RS6 | Alternative splicing;Cytoplasm;Immunity;Nucleus;Phosphoprotein;Reference proteome;Tumor antigen | hsa:84955; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; immune system process [GO:0002376] | 14688378_An alternative promoter (CML66-S short isoform) has been identified in combination with alternative splicing as a novel mechanism for regulation of the epitope generation of a self-tumor antigen. 18534745_CML66 may play an oncogenic role in ways of favoring tumor cells proliferation, invasion and metastasis-associated with multiple pathways. 18754882_study identified a novel gene, OVA66, which was expressed significantly higher in cancer patients than normal controls; IgG level against OVA66 was significantly elevated in the serum of cancer patients from different histological types of cancer [OVA66] 29021621_the different NudCD1 isoforms have unique interacting partners, with the first isoform binding to a putative RNA helicase named DHX15 involved in mRNA splicing. 29461594_NudCD1 significantly increased in renal cell carcinoma and was positively correlated with cell proliferation, migration, and invasion 30833190_OVA66 overexpression in the cancer cell lines promoted VEGF secretion, tumour growth and angiogenesis in vitro and in vivo 32634806_NudCD1 Promotes the Proliferation and Metastasis of Non-Small Cell Lung Cancer Cells through the Activation of IGF1R-ERK1/2. 33336728_Analysis of NudCD1 and NF-kappaBeta in the early detection and course evaluation of renal cancer. | ENSMUSG00000038736 | Nudcd1 | 1.331399e+02 | 0.9183623 | -0.122864706 | 0.4329410 | 7.916811e-02 | 0.7784279468 | 0.95606951 | No | Yes | 1.257441e+02 | 26.093445 | 1.302592e+02 | 27.554324 | |||
| ENSG00000120616 | 80314 | EPC1 | protein_coding | Q9H2F5 | FUNCTION: Component of the NuA4 histone acetyltransferase (HAT) complex, a multiprotein complex involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A (PubMed:14966270). The NuA4 complex plays a direct role in repair of DNA double-strand breaks (DSBs) by promoting homologous recombination (HR) (PubMed:27153538). The NuA4 complex is also required for spermatid development by promoting acetylation of histones: histone acetylation is required for histone replacement during the transition from round to elongating spermatids (By similarity). In the NuA4 complex, EPC1 is required to recruit MBTD1 into the complex (PubMed:32209463). {ECO:0000250|UniProtKB:Q8C9X6, ECO:0000269|PubMed:14966270, ECO:0000269|PubMed:27153538, ECO:0000269|PubMed:32209463}. | 3D-structure;Activator;Alternative splicing;Chromatin regulator;Cytoplasm;Differentiation;Direct protein sequencing;Growth regulation;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a member of the polycomb group (PcG) family. The encoded protein is a component of the NuA4 histone acetyltransferase complex and can act as both a transcriptional activator and repressor. The encoded protein has been linked to apoptosis, DNA repair, skeletal muscle differentiation, gene silencing, and adult T-cell leukemia/lymphoma. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2012]. | hsa:80314; | intracellular membrane-bounded organelle [GO:0043231]; NuA4 histone acetyltransferase complex [GO:0035267]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; Piccolo NuA4 histone acetyltransferase complex [GO:0032777]; histone acetylation [GO:0016573]; histone H2A acetylation [GO:0043968]; histone H4 acetylation [GO:0043967]; negative regulation of gene expression, epigenetic [GO:0045814]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of cell cycle [GO:0051726]; regulation of growth [GO:0040008]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription, DNA-templated [GO:0006351] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 17192267_Epc1 plays a role in the initiation of skeletal muscle differentiation, and its interaction with Hop is required for the full activity. 18984674_Observational study of gene-disease association. (HuGE Navigator) 19484761_SNP array CGH analysis of the breakpoint region in 3 ATLL-related cell lines and 4 patient samples revealed the chromosomal breakpoints are localized within the EPC1 gene locus in an ATLL-derived cell line (SO4) and in one patient with acute-type ATLL. 24166297_EPC1 and EPC2 are components of a complex that directly or indirectly serves to prevent MYC accumulation and AML cell apoptosis, thus sustaining oncogenic potential 25755723_Silencing EPC1 by short hairpin RNA technology had the inhibition effects on cell proliferation and tumor growth in lung cancer 26350215_Epigenetic factor EPC1 is a master regulator of DNA damage response by interacting with E2F1 to silence death and activate metastasis-related gene signatures 32209463_Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex. | ENSMUSG00000024240 | Epc1 | 2.995151e+02 | 0.7010600 | -0.512390112 | 0.3191538 | 2.526482e+00 | 0.1119495453 | 0.75783482 | No | Yes | 2.571290e+02 | 39.939698 | 3.616509e+02 | 57.332076 | |
| ENSG00000121578 | 8702 | B4GALT4 | protein_coding | O60513 | FUNCTION: Galactose (Gal) transferase involved in the synthesis of terminal N-acetyllactosamine (LacNac) unit present on glycan chains of glycoproteins and glycosphingolipids (PubMed:9792633, PubMed:17690104, PubMed:12511560, PubMed:32827291). Catalyzes the transfer of Gal residue via a beta1->4 linkage from UDP-Gal to the non-reducing terminal N-acetyl glucosamine 6-O-sulfate (6-O-sulfoGlcNAc) in the linearly growing chain of both N- and O-linked keratan sulfate proteoglycans. Cooperates with B3GNT7 N-acetyl glucosamine transferase and CHST6 and CHST1 sulfotransferases to construct and elongate mono- and disulfated disaccharide units [->3Galbeta1->4(6-sulfoGlcNAcbeta)1->] and [->3(6-sulfoGalbeta)1->4(6-sulfoGlcNAcbeta)1->] within keratan sulfate polymer (PubMed:17690104). Transfers Gal residue via a beta1->4 linkage to terminal 6-O-sulfoGlcNAc within the LacNac unit of core 2 O-glycans forming 6-sulfo-sialyl-Lewis X (sLex). May contribute to the generation of sLex epitope on mucin-type glycoproteins that serve as ligands for SELL/L-selectin, a major regulator of leukocyte migration (PubMed:12511560). In the biosynthesis pathway of neolacto-series glycosphingolipids, transfers Gal residue via a beta1->4 linkage to terminal GlcNAc of a lactotriaosylceramide (Lc3Cer) acceptor to form a neolactotetraosylceramide (PubMed:9792633). {ECO:0000269|PubMed:12511560, ECO:0000269|PubMed:17690104, ECO:0000269|PubMed:9792633}. | Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Lipid metabolism;Manganese;Membrane;Metal-binding;Reference proteome;Secreted;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000305|PubMed:12511560}.; PATHWAY: Glycolipid biosynthesis. {ECO:0000305|PubMed:9792633}. | This gene is one of seven beta-1,4-galactosyltransferase (beta4GalT) genes. They encode type II membrane-bound glycoproteins that appear to have exclusive specificity for the donor substrate UDP-galactose; all transfer galactose in a beta1,4 linkage to similar acceptor sugars: GlcNAc, Glc, and Xyl. Each beta4GalT has a distinct function in the biosynthesis of different glycoconjugates and saccharide structures. As type II membrane proteins, they have an N-terminal hydrophobic signal sequence that directs the protein to the Golgi apparatus and which then remains uncleaved to function as a transmembrane anchor. By sequence similarity, the beta4GalTs form four groups: beta4GalT1 and beta4GalT2, beta4GalT3 and beta4GalT4, beta4GalT5 and beta4GalT6, and beta4GalT7. The enzyme encoded by this gene appears to mainly play a role in glycolipid biosynthesis. Two alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:8702; | Golgi apparatus [GO:0005794]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; galactosyltransferase activity [GO:0008378]; metal ion binding [GO:0046872]; N-acetyllactosamine synthase activity [GO:0003945]; UDP-galactosyltransferase activity [GO:0035250]; carbohydrate metabolic process [GO:0005975]; glycosylation [GO:0070085]; keratan sulfate biosynthetic process [GO:0018146]; lactosylceramide biosynthetic process [GO:0001572]; membrane lipid metabolic process [GO:0006643]; protein glycosylation [GO:0006486] | 28228616_expression of the beta4GalT4 gene is controlled by Sp1, and Sp1 plays a key role in the activation of the beta4GalT4 gene in colon cancer cells. 32827291_N-glycosylation of the human beta1,4-galactosyltransferase 4 is crucial for its activity and Golgi localization. 34270095_The critical role of B4GALT4 in promoting microtubule spindle assembly in HCC through the regulation of PLK1 and RHAMM expression. | ENSMUSG00000022793 | B4galt4 | 3.379882e+02 | 0.8942424 | -0.161262063 | 0.3347383 | 2.297392e-01 | 0.6317172508 | 0.91702041 | No | Yes | 2.660784e+02 | 43.004525 | 3.118421e+02 | 51.355231 |
| ENSG00000121749 | 64786 | TBC1D15 | protein_coding | Q8TC07 | FUNCTION: Acts as a GTPase activating protein for RAB7A. Does not act on RAB4, RAB5 or RAB6 (By similarity). {ECO:0000250}. | Acetylation;Alternative splicing;Cytoplasm;GTPase activation;Phosphoprotein;Reference proteome | This gene encodes a member of the Ras-like proteins in brain-GTPase activating protein superfamily that share a conserved Tre-2/Bub2/Cdc16 domain. The encoded protein interacts with Ras-like protein in brain 5A and may function as a regulator of intracellular trafficking. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Apr 2009]. | hsa:64786; | cytoplasm [GO:0005737]; extracellular region [GO:0005576]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; intracellular protein transport [GO:0006886]; regulation of GTPase activity [GO:0043087] | 16055087_This paper primarily describes mouse TBC1D15 but also reports the primary structure of human TBC1D15. 20363736_although only TBC1D15/Rab7-GAP altered Rab7-GTP levels, both Rab7-GAP and mVps39 regulate lysosomal morphology and play a role in maintaining growth factor dependence 23077178_Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. 24337944_TBC1D15 is required for the accumulation of RhoA. 24569479_Demonstrate that TBC1D15 and TBC1D17 mediate proper autophagic encapsulation of mitochondria by regulating Rab7 activity at the interface between mitochondria and isolation membranes. 30316925_Report provides evidence for a new function of TBC1D15 in the control of GLUT4 vesicles translocation through regulating Rab7 activity. Glucose uptake was greatly reduced when TBC1D15 was knocked out. 31664461_Here, we demonstrate that this cellular cholesterol imbalance is due to AnxA6 promoting Rab7 inactivation via TBC1D15, a Rab7-GAP. 31958036_IFNB/interferon-beta regulates autophagy via a MIR1-TBC1D15-RAB7 pathway. 32555153_p53 destabilizing protein skews asymmetric division and enhances NOTCH activation to direct self-renewal of TICs. | ENSMUSG00000020130 | Tbc1d15 | 2.535629e+02 | 0.8788753 | -0.186269534 | 0.4050337 | 2.042911e-01 | 0.6512792838 | 0.92370776 | No | Yes | 2.104408e+02 | 49.399350 | 2.365185e+02 | 56.773128 | |
| ENSG00000121905 | 3208 | HPCA | protein_coding | P84074 | FUNCTION: Calcium-binding protein that may play a role in the regulation of voltage-dependent calcium channels (PubMed:28398555). May also play a role in cyclic-nucleotide-mediated signaling through the regulation of adenylate and guanylate cyclases (By similarity). {ECO:0000250|UniProtKB:P84076, ECO:0000269|PubMed:28398555}. | 3D-structure;Calcium;Cytoplasm;Disease variant;Dystonia;Lipoprotein;Membrane;Metal-binding;Myristate;Reference proteome;Repeat | The protein encoded by this gene is a member of neuron-specific calcium-binding proteins family found in the retina and brain. This protein is associated with the plasma membrane. It has similarities to proteins located in the photoreceptor cells that regulate photosignal transduction in a calcium-sensitive manner. This protein displays recoverin activity and a calcium-dependent inhibition of rhodopsin kinase. It is identical to the rat and mouse hippocalcin proteins and thought to play an important role in neurons of the central nervous system in a number of species. [provided by RefSeq, Jul 2008]. | hsa:3208; | axon [GO:0030424]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendrite cytoplasm [GO:0032839]; dendrite membrane [GO:0032590]; dendritic spine head [GO:0044327]; extrinsic component of membrane [GO:0019898]; glutamatergic synapse [GO:0098978]; neuronal cell body membrane [GO:0032809]; perikaryon [GO:0043204]; actin binding [GO:0003779]; calcium ion binding [GO:0005509]; identical protein binding [GO:0042802]; kinase binding [GO:0019900]; activation of phospholipase D activity [GO:0031584]; brain development [GO:0007420]; calcium-mediated signaling [GO:0019722]; cellular response to calcium ion [GO:0071277]; cellular response to electrical stimulus [GO:0071257]; cellular response to monosodium glutamate [GO:1904009]; inner ear development [GO:0048839]; negative regulation of guanylate cyclase activity [GO:0031283]; positive regulation of adenylate cyclase activity [GO:0045762]; positive regulation of protein targeting to membrane [GO:0090314]; regulation of postsynaptic neurotransmitter receptor internalization [GO:0099149]; regulation of voltage-gated calcium channel activity [GO:1901385]; response to Aroclor 1254 [GO:1904010]; response to ketamine [GO:1901986]; response to L-glutamate [GO:1902065]; retina development in camera-type eye [GO:0060041] | 14638856_Hippocalcin is a sensitive Ca2+ sensor capable of responding to increases in intracellular Ca2+ concentration over the narrow dynamic range of 200-800 nM free Ca2+. 16053445_The structural factors affecting the binding of hippocalcin to phosphatidylinositol 4,5-bisphosphate at the cell surface and the Golgi apparatus are described. 19332348_Hippocalcin and MLK2 were colocalized in the halo of Lewy bodies in Parkinson disease patients, and neither protein was detected in normal pigmented neurons. 19686238_diminished hippocalcin expression does not contribute to Huntington disease-related neurodegeneration 22544757_analysis of a novel acetylation cycle of transcription co-activator Yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents 23782819_Current known dystonia genes include those related to dopamine metabolism, transcription factor, cytoskeleton, transport of glucose and sodium ion, etc. 25799108_Mutations in HPCA cause autosomal-recessive primary isolated dystonia. 27145302_Sequence analysis of the HPCA gene in 505 patients with dystonia did not reveal variants in the coding regions of HPCA. 27771228_None of the patients enrolled was found to carry HPCA mutations, rising suspicion that these probably represent a very rare cause of dystonia in childhood-adolescence. 28398555_hippocalcin forms oligomers upon calcium binding and directly interacts with VGCCs. The dystonia-causing mutations did not affect protein stability or folding. In common for both T71N and A190T mutants was an impaired calcium-dependent oligomerisation and increased intracellular calcium influx after KCl depolarisation. 30145809_HPCA mutations lead to recessively inherited dystonia 31486848_MiR-24-3p is an important miRNA that regulates neuronal differentiation by controlling HPCA expression. | ENSMUSG00000028785 | Hpca | 1.603829e+01 | 0.9550894 | -0.066292344 | 0.7973313 | 7.381611e-03 | 0.9315329515 | No | Yes | 1.493053e+01 | 5.687066 | 1.295078e+01 | 5.350246 | ||
| ENSG00000121988 | 84083 | ZRANB3 | protein_coding | Q5FWF4 | FUNCTION: DNA annealing helicase and endonuclease required to maintain genome stability at stalled or collapsed replication forks by facilitating fork restart and limiting inappropriate recombination that could occur during template switching events (PubMed:21078962, PubMed:22704558, PubMed:22705370, PubMed:22759634, PubMed:26884333). Recruited to the sites of stalled DNA replication by polyubiquitinated PCNA and acts as a structure-specific endonuclease that cleaves the replication fork D-loop intermediate, generating an accessible 3'-OH group in the template of the leading strand, which is amenable to extension by DNA polymerase (PubMed:22759634). In addition to endonuclease activity, also catalyzes the fork regression via annealing helicase activity in order to prevent disintegration of the replication fork and the formation of double-strand breaks (PubMed:22705370, PubMed:22704558). {ECO:0000269|PubMed:21078962, ECO:0000269|PubMed:22704558, ECO:0000269|PubMed:22705370, ECO:0000269|PubMed:22759634, ECO:0000269|PubMed:26884333}. | 3D-structure;ATP-binding;Alternative splicing;Chromosome;DNA damage;DNA repair;Endonuclease;Helicase;Hydrolase;Metal-binding;Methylation;Multifunctional enzyme;Nuclease;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | hsa:84083; | nuclear replication fork [GO:0043596]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; ATP-dependent DNA/DNA annealing activity [GO:0036310]; endodeoxyribonuclease activity [GO:0004520]; helicase activity [GO:0004386]; K63-linked polyubiquitin modification-dependent protein binding [GO:0070530]; metal ion binding [GO:0046872]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; DNA rewinding [GO:0036292]; negative regulation of DNA recombination [GO:0045910]; replication fork processing [GO:0031297]; replication fork protection [GO:0048478]; response to UV [GO:0009411] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21078962_structural and functional differences between AH2 and HARP suggest that different annealing helicases have distinct functions in the cell. 22704558_ZRANB3 translocase, a SNF2 family member related to the SIOD disorder SMARCAL1 protein, is recruited by polyubiquitinated PCNA to promote fork restart following replication arrest. 22705370_AH2 is recruited to stalled replication forks and that cells depleted of AH2 are hypersensitive to replication stresses. 22759634_a role for ZRANB3 in the replication stress response and suggest new insights into how DNA repair is coordinated with DNA replication to maintain genome stability. 26884333_Here is described a substrate recognition domain within ZRANB3 that is needed for it to recognize forked DNA structures, hydrolyze ATP, catalyze fork remodeling, and act as a structure-specific endonuclease. 28621305_PCNA and ATP-dependency serve as a multi-layered regulatory mechanism that modulates ZRANB3 activity at replication forks. 28886337_Damage-induced fork reversal in mammalian cells requires PCNA ubiquitination, UBC13, and K63-linked polyubiquitin chains, previously involved in error-free damage tolerance. Fork reversal in vivo also requires ZRANB3 translocase activity and its interaction with polyubiquitinated PCNA, pinpointing ZRANB3 as a key effector of error-free DNA damage tolerance. 28954549_A large number of SNF2 family, DNA and ATP-dependent motor proteins are needed during transcription, DNA replication, and DNA repair to manipulate protein-DNA interactions and change DNA structure. SMARCAL1, ZRANB3, and HLTF are three related members of this family with specialized functions that maintain genome stability during DNA replication. [review] 29053959_depletion of SMARCAL1, a SNF2-family DNA translocase that remodels stalled forks, restores replication fork stability and reduces the formation of replication stress-induced DNA breaks and chromosomal aberrations in BRCA1/2-deficient cells. In addition to SMARCAL1, other SNF2-family fork remodelers, including ZRANB3 and HLTF, cause nascent DNA degradation and genomic instability 30150681_Nuclear RNR-alpha antagonizes cell proliferation by directly inhibiting ZRANB3 31324766_ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. 32971328_Time for remodeling: SNF2-family DNA translocases in replication fork metabolism and human disease. | ENSMUSG00000036086 | Zranb3 | 1.059971e+02 | 0.6491859 | -0.623296504 | 0.3990846 | 2.296536e+00 | 0.1296628809 | 0.76070573 | No | Yes | 8.571800e+01 | 17.922831 | 1.158448e+02 | 24.561701 | ||
| ENSG00000122034 | 2971 | GTF3A | protein_coding | Q92664 | FUNCTION: Involved in ribosomal large subunit biogenesis. Binds the approximately 50 base pairs internal control region (ICR) of 5S ribosomal RNA genes. It is required for their RNA polymerase III-dependent transcription and may also maintain the transcription of other genes (PubMed:24120868). Also binds the transcribed 5S RNA's (By similarity). {ECO:0000250|UniProtKB:P17842, ECO:0000269|PubMed:24120868}. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;RNA-binding;Reference proteome;Repeat;Ribosome biogenesis;Transcription;Transcription regulation;Zinc;Zinc-finger | The product of this gene is a zinc finger protein with nine Cis[2]-His[2] zinc finger domains. It functions as an RNA polymerase III transcription factor to induce transcription of the 5S rRNA genes. The protein binds to a 50 bp internal promoter in the 5S genes called the internal control region (ICR), and nucleates formation of a stable preinitiation complex. This complex recruits the TFIIIC and TFIIIB transcription factors and RNA polymerase III to form the complete transcription complex. The protein is thought to be translated using a non-AUG translation initiation site in mammals based on sequence analysis, protein homology, and the size of the purified protein. [provided by RefSeq, Jul 2008]. | hsa:2971; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 5S rRNA binding [GO:0008097]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; ribosomal large subunit biogenesis [GO:0042273]; rRNA transcription [GO:0009303]; transcription by RNA polymerase III [GO:0006383] | 33925358_Comprehensive Analysis of Prognostic and Genetic Signatures for General Transcription Factor III (GTF3) in Clinical Colorectal Cancer Patients Using Bioinformatics Approaches. | ENSMUSG00000016503 | Gtf3a | 1.992019e+03 | 0.9597804 | -0.059223693 | 0.2710123 | 4.774089e-02 | 0.8270419745 | 0.96748040 | No | Yes | 1.746699e+03 | 210.485171 | 1.922060e+03 | 237.325281 | |
| ENSG00000123143 | 5585 | PKN1 | protein_coding | Q16512 | FUNCTION: PKC-related serine/threonine-protein kinase involved in various processes such as regulation of the intermediate filaments of the actin cytoskeleton, cell migration, tumor cell invasion and transcription regulation. Part of a signaling cascade that begins with the activation of the adrenergic receptor ADRA1B and leads to the activation of MAPK14. Regulates the cytoskeletal network by phosphorylating proteins such as VIM and neurofilament proteins NEFH, NEFL and NEFM, leading to inhibit their polymerization. Phosphorylates 'Ser-575', 'Ser-637' and 'Ser-669' of MAPT/Tau, lowering its ability to bind to microtubules, resulting in disruption of tubulin assembly. Acts as a key coactivator of androgen receptor (AR)-dependent transcription, by being recruited to AR target genes and specifically mediating phosphorylation of 'Thr-11' of histone H3 (H3T11ph), a specific tag for epigenetic transcriptional activation that promotes demethylation of histone H3 'Lys-9' (H3K9me) by KDM4C/JMJD2C. Phosphorylates HDAC5, HDAC7 and HDAC9, leading to impair their import in the nucleus. Phosphorylates 'Thr-38' of PPP1R14A, 'Ser-159', 'Ser-163' and 'Ser-170' of MARCKS, and GFAP. Able to phosphorylate RPS6 in vitro. {ECO:0000269|PubMed:11104762, ECO:0000269|PubMed:12514133, ECO:0000269|PubMed:17332740, ECO:0000269|PubMed:18066052, ECO:0000269|PubMed:20188095, ECO:0000269|PubMed:21224381, ECO:0000269|PubMed:21754995, ECO:0000269|PubMed:24248594, ECO:0000269|PubMed:8557118, ECO:0000269|PubMed:8621664, ECO:0000269|PubMed:9175763}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cell membrane;Chromatin regulator;Coiled coil;Cytoplasm;Endosome;Kinase;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase;Ubl conjugation | The protein encoded by this gene belongs to the protein kinase C superfamily. This kinase is activated by Rho family of small G proteins and may mediate the Rho-dependent signaling pathway. This kinase can be activated by phospholipids and by limited proteolysis. The 3-phosphoinositide dependent protein kinase-1 (PDPK1/PDK1) is reported to phosphorylate this kinase, which may mediate insulin signals to the actin cytoskeleton. The proteolytic activation of this kinase by caspase-3 or related proteases during apoptosis suggests its role in signal transduction related to apoptosis. Alternatively spliced transcript variants encoding distinct isoforms have been observed. [provided by RefSeq, Jul 2008]. | hsa:5585; | cleavage furrow [GO:0032154]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endosome [GO:0005768]; midbody [GO:0030496]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; androgen receptor binding [GO:0050681]; ATP binding [GO:0005524]; calcium-dependent protein kinase C activity [GO:0004698]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; histone deacetylase binding [GO:0042826]; histone kinase activity (H3-T11 specific) [GO:0035402]; nuclear receptor coactivator activity [GO:0030374]; protein kinase activity [GO:0004672]; protein kinase C activity [GO:0004697]; protein kinase C binding [GO:0005080]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; small GTPase binding [GO:0031267]; B cell apoptotic process [GO:0001783]; B cell homeostasis [GO:0001782]; chromatin organization [GO:0006325]; epithelial cell migration [GO:0010631]; histone H3-T11 phosphorylation [GO:0035407]; hyperosmotic response [GO:0006972]; intracellular signal transduction [GO:0035556]; negative regulation of B cell proliferation [GO:0030889]; negative regulation of protein kinase activity [GO:0006469]; peptidyl-serine phosphorylation [GO:0018105]; protein phosphorylation [GO:0006468]; regulation of androgen receptor signaling pathway [GO:0060765]; regulation of cell motility [GO:2000145]; regulation of germinal center formation [GO:0002634]; regulation of immunoglobulin production [GO:0002637]; regulation of transcription by RNA polymerase II [GO:0006357]; renal system process [GO:0003014]; signal transduction [GO:0007165]; spleen development [GO:0048536] | 12514133_Data show that stimulation of the RhoA effector protein kinase C-related kinase (PRK) signalling cascade results in a ligand-dependent superactivation of androgen receptors both in vivo and in vitro. 12761180_PKNalpha functions as not only an upstream activator of MLTKalpha but also a putative scaffold protein for the p38gamma MAPK signaling pathway 14514689_analysis of the interaction between the small G proteins Rac1 and RhoA and protein kinase C-related kinase 1 15123640_Data suggest that hyaluronan-CD44 interaction with Rac1-protein kinase N gamma plays a pivotal role in phospholipase C gamma1-regulated calcium signaling and cortactin-cytoskeleton function required for keratinocyte cell-cell adhesion and differentiation. 15791647_PKN1 mediates arsenite-induced delay of the G(2)/M transition by binding to and phosphorylating Cdc25C 16331689_Data strengthen the hypothesis that Cyclin T2a plays a role in muscle differentiation, and propose PKNalpha as a novel partner of Cyclin T2a in this process. 17301291_Human pregnancy is characterized by increases in PKN1 expression in the myometrium. 18429822_Study identifies TRAF1 as a substrate of PKN1 kinase activity in vitro and in vivo, and show that this phosphorylation event is required for attenuating downstream kinase activities. 18519042_Deregulation of PKN1 may contribute to the pathogenic process in amyotrophic lateral sclerosis. 18713748_Protein kinase C-related kinase and ROCK are required for thrombin-induced endothelial cell permeability downstream from Galpha12/13 and Galpha11/q 19427017_PRK1 is present in various malignancies, but especially in ovarian serous carcinomas 19913121_Observational study of gene-disease association. (HuGE Navigator) 20188095_Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21224381_A-kinase anchoring protein (AKAP)-Lbc anchors a PKN-based signaling complex involved in alpha1-adrenergic receptor-induced p38 activation. 21351730_Data show that just one contact site as being relevant for binding of RhoA and domain from PRK1, and the HR1b domain was found not to contribute to RhoA binding. 21754995_PKN isoforms are not simply redundant in supporting migration, but appear to be linked through isoform specific regulatory domain properties to selective upstream signals. It 22893700_Protein kinase N1 is a novel substrate of NFATc1-mediated cyclin D1-CDK6 activity and modulates vascular smooth muscle cell division and migration leading to inward blood vessel wall remodeling. 24114839_Protein kinase N1 inhibits Wnt/b-catenin signaling and apoptosis in melanoma cells. 24248594_Data indicate that Salmonella SspH1 catalyzes the ubiquitination and proteasome-dependent degradation of PKN1 in cells. 25504435_Transcriptome and interactome analyses uncover that PRK1 regulates expression of migration-relevant genes by interacting with the scaffold protein sperm-associated antigen 9 (SPAG9/JIP4). 26296974_TXA2-mediated neoplastic responses in prostate adenocarcinoma PC-3 cells occur through a PRK1/PRK2-dependent mechanism. 27919031_Steady-state kinetic analysis revealed that PKN1-3 follows a sequential ordered Bi-Bi kinetic mechanism, where peptide substrate binding is preceded by ATP binding. This kinetic mechanism was confirmed by additional kinetic studies for product inhibition and affinity of small molecule inhibitors. 28222172_PKN1 activity was up-regulated by the active RhoA mutant (G14V) and suppressed by RhoA T19N. PKN1 siRNA interrupted the ability of RhoA to promote ESC proliferation and DNA synthesis. The effect of RhoA on ESC proliferation is mediated by activation of the PKN1-cyclin D1 pathway in vitro. 28875501_Pkn1 is not required for tumorigenesis initiated by loss of Pten. Triple knockout of Pten, Pkn1, and Pkn2 in mouse prostate results in squamous cell carcinoma, an uncommon but therapy-resistant form of prostate cancer. 30742064_PKN1 transduces androgen-responsiveness to serum response factor in prostate neoplasms 31045890_Secretory carcinoma of the skin associated with the presence of novel NFIX-PKN1 translocation. 31125460_Characterization of the novel cardiolipin binding regions identified on the protease and lipid activated PKC-related kinase 1 31981797_Cyclin-dependent kinase 1-mediated phosphorylation of protein kinase N1 promotes anchorage-independent growth and migration. 32127582_Novel roles of PRK1 and PRK2 in cilia and cancer biology. | ENSMUSG00000057672 | Pkn1 | 9.395811e+03 | 1.4637601 | 0.549679154 | 0.3417857 | 2.575446e+00 | 0.1085335224 | 0.75783482 | No | Yes | 1.122423e+04 | 1798.581894 | 6.052995e+03 | 995.305744 | |
| ENSG00000123213 | 57486 | NLN | protein_coding | Q9BYT8 | FUNCTION: Hydrolyzes oligopeptides such as neurotensin, bradykinin and dynorphin A. {ECO:0000250}. | 3D-structure;Acetylation;Cytoplasm;Hydrolase;Metal-binding;Metalloprotease;Mitochondrion;Protease;Reference proteome;Transit peptide;Zinc | This gene encodes a member of the metallopeptidase M3 protein family that cleaves neurotensin at the Pro10-Tyr11 bond, leading to the formation of neurotensin(1-10) and neurotensin(11-13). The encoded protein is likely involved in the termination of the neurotensinergic signal in the central nervous system and in the gastrointestinal tract.[provided by RefSeq, Jun 2010]. | hsa:57486; | extracellular region [GO:0005576]; mitochondrial intermembrane space [GO:0005758]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; metalloendopeptidase activity [GO:0004222]; peptide binding [GO:0042277]; G protein-coupled receptor signaling pathway [GO:0007186]; peptide metabolic process [GO:0006518]; proteolysis [GO:0006508]; regulation of gluconeogenesis [GO:0006111]; regulation of skeletal muscle fiber differentiation [GO:1902809] | 17251185_Mutations at only two residues (Arg-470 and Arg-498) are required to swap specificity with thimet oligopeptidase, a result that is confirmed by testing the two-mutant constructs. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 29183787_The crystal structure of human neurolysin(E475Q) in complex with the products of neurotensin cleavage at 2.7A revealed a closed conformation with an internal cavity that restricts substrate length and highlighted the mechanism of enzyme opening/closing that is necessary for substrate binding and catalytic activity. 32269163_The mitochondrial peptidase, neurolysin, regulates respiratory chain supercomplex formation and is necessary for AML viability. | ENSMUSG00000021710 | Nln | 5.633961e+02 | 1.0264457 | 0.037657365 | 0.3548263 | 1.103400e-02 | 0.9163418266 | 0.98426458 | No | Yes | 4.937476e+02 | 101.172214 | 3.964696e+02 | 83.265350 | |
| ENSG00000123352 | 65244 | SPATS2 | protein_coding | Q86XZ4 | Cytoplasm;Phosphoprotein;Reference proteome | hsa:65244; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; RNA binding [GO:0003723] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 32118724_Gene Set Enrichment Analysis (GSEA) was used to identify signaling pathways related to SPATS2 expression. SPATS2 expression could be a novel diagnostic and prognostic biomarker in liver cancer. 33037180_SPATS2, negatively regulated by miR-145-5p, promotes hepatocellular carcinoma progression through regulating cell cycle. 35077478_SPATS2 is positively activated by long noncoding RNA SNHG5 via regulating DNMT3a expression to promote hepatocellular carcinoma progression. | ENSMUSG00000051934 | Spats2 | 5.685310e+02 | 0.9115519 | -0.133603249 | 0.2751614 | 2.351821e-01 | 0.6277077509 | 0.91646120 | No | Yes | 5.112875e+02 | 61.973663 | 5.347467e+02 | 66.351118 | |||
| ENSG00000123374 | 1017 | CDK2 | protein_coding | P24941 | FUNCTION: Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (By similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability (By similarity). Phosphorylates CDK2AP2 (PubMed:12944431). Phosphorylates ERCC6 which is essential for its chromatin remodeling activity at DNA double-strand breaks (PubMed:29203878). {ECO:0000250|UniProtKB:P97377, ECO:0000269|PubMed:10499802, ECO:0000269|PubMed:10884347, ECO:0000269|PubMed:10995386, ECO:0000269|PubMed:10995387, ECO:0000269|PubMed:11051553, ECO:0000269|PubMed:11113184, ECO:0000269|PubMed:12944431, ECO:0000269|PubMed:15800615, ECO:0000269|PubMed:17495531, ECO:0000269|PubMed:18372919, ECO:0000269|PubMed:19966300, ECO:0000269|PubMed:20079829, ECO:0000269|PubMed:20147522, ECO:0000269|PubMed:20195506, ECO:0000269|PubMed:20935635, ECO:0000269|PubMed:21262353, ECO:0000269|PubMed:21319273, ECO:0000269|PubMed:21596315, ECO:0000269|PubMed:28666995, ECO:0000269|PubMed:29203878}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;DNA damage;DNA repair;Endosome;Kinase;Magnesium;Meiosis;Metal-binding;Mitosis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;S-nitrosylation;Serine/threonine-protein kinase;Transferase | This gene encodes a member of a family of serine/threonine protein kinases that participate in cell cycle regulation. The encoded protein is the catalytic subunit of the cyclin-dependent protein kinase complex, which regulates progression through the cell cycle. Activity of this protein is especially critical during the G1 to S phase transition. This protein associates with and regulated by other subunits of the complex including cyclin A or E, CDK inhibitor p21Cip1 (CDKN1A), and p27Kip1 (CDKN1B). Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2014]. | hsa:1017; | Cajal body [GO:0015030]; centrosome [GO:0005813]; chromosome, telomeric region [GO:0000781]; condensed chromosome [GO:0000793]; cyclin A1-CDK2 complex [GO:0097123]; cyclin A2-CDK2 complex [GO:0097124]; cyclin E1-CDK2 complex [GO:0097134]; cyclin E2-CDK2 complex [GO:0097135]; cyclin-dependent protein kinase holoenzyme complex [GO:0000307]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endosome [GO:0005768]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; X chromosome [GO:0000805]; Y chromosome [GO:0000806]; ATP binding [GO:0005524]; cyclin binding [GO:0030332]; cyclin-dependent protein kinase activity [GO:0097472]; cyclin-dependent protein serine/threonine kinase activity [GO:0004693]; magnesium ion binding [GO:0000287]; protein domain specific binding [GO:0019904]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cell division [GO:0051301]; cellular response to nitric oxide [GO:0071732]; cellular senescence [GO:0090398]; centriole replication [GO:0007099]; centrosome duplication [GO:0051298]; DNA repair [GO:0006281]; DNA replication [GO:0006260]; G1/S transition of mitotic cell cycle [GO:0000082]; G2/M transition of mitotic cell cycle [GO:0000086]; histone phosphorylation [GO:0016572]; meiotic cell cycle [GO:0051321]; mitotic G1 DNA damage checkpoint signaling [GO:0031571]; negative regulation of transcription by RNA polymerase II [GO:0000122]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of DNA-dependent DNA replication initiation [GO:0032298]; positive regulation of transcription, DNA-templated [GO:0045893]; potassium ion transport [GO:0006813]; protein phosphorylation [GO:0006468]; Ras protein signal transduction [GO:0007265]; regulation of anaphase-promoting complex-dependent catabolic process [GO:1905784]; regulation of G2/M transition of mitotic cell cycle [GO:0010389]; regulation of gene expression [GO:0010468]; regulation of gene silencing [GO:0060968]; signal transduction [GO:0007165] | 11907280_Cyclin A/Cdk2 and cyclin E/cdk2 continuously shuttle between the nucleus and the cytoplasm 12049628_results argue that TTK-associated CDK2 may function to maintain target-specific phosphorylation of RNA Pol II that is essential for Tat transactivation of HIV-1 promoter 12081504_Activation mechanism role of cyclin binding versus phosphorylation 12114499_CDK2/cyclin E is required for Tat-dependent transcription in vitro. 12149264_CDK2 binding to cyclin E is required to drive cells from G(1) into S phase 12531694_Interferon gamma reduces the activity of Cdk4 and Cdk2, inhibiting he G1 cell cycle in human hepatocellular carcinoma cells. 12676582_CDK2 is not required for sustained cell division. 12729791_Data suggest that the interaction between PKCeta and cyclin E is carefully regulated, and is correlated with the inactivated form of the cyclin E/Cdk2 complex. 12732645_IRF1 represses CDK2 gene expression by interfering with SP1-dependent transcriptional activation. 12801928_role in regulating Cdc25A half life 12810668_TGF-beta 1 inhibition requires early G(1) induction and stabilization of p21 protein, which binds to & inhibits cyclin E-CDK2 and cyclin A-CDK2 kinase activity rather than direct modulation of cyclin or CDK protein levels as seen in other systems. 12857729_Cdk2 has a role in phosphorylation of the NF-Y transcription factor 12912980_CDK2 has a role in the G2 DNA damage checkpoint 12915577_Kaposi's sarcoma-associated herpesvirus K-bZIP physically associates with cyclin-CDK2 and downmodulates its kinase activity. 12947099_it is evident that B-Myb protein may promote cell proliferation by a non-transcriptional mechanism that involves release of active cyclin/cyclin dependent kinase 2 from cyclin-dependent inhibitor 1C p57(KIP2) 12954644_Inhibition of Cdk2 by 1,25-(OH)2D3 may thus involve two mechanisms: 1) reduced nuclear Cdk2 available for cyclin binding and activation and 2) impairment of cyclin E-Cdk2-dependent p27 degradation through cytoplasmic mislocalization of Cdk2. 14506259_kinetic insight into the basis for selecting suboptimal specificity determinants for the phosphorylation of cellular substrates 14536078_multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation 14550307_CDK2 binds to SU9516 at Leu83 and Glu81 14551212_CDK2 activation process through phosphorylation is examined using 2D PAGE 14562046_Epstein-Barr virus can inhibit genotoxin-induced G1 arrest downstream of p53 by preventing the inactivation of CDK2 14612403_p220 is an essential downstream component of the cyclin E/Cdk2 signaling pathway and functions to coordinate multiple elements of the G1/S transition. 14645251_CDK2-cyclin E, without prior CDK4-cyclin D activity, can phosphorylate and inactivate pRb, activate E2F, and induce DNA synthesis. 14646596_significant difference in their biochemical properties between CDK4/cyclin D1 and CDK2/cyclin A affecting regulation of cellular RB function 14694185_cyclin-dependent kinase (CDK)2, -4, and -6 were down-regulated from the myelocytes/metamyelocytes stages and onward 14701826_CDK2 complexes have roles in G(1)/S deregulation and tumor progression 14985333_CDK2 regulates beta-catenin phosphorylation/ degradation 15004027_Cdk2 and Cdk4 phosphorylate human Cdt1 and induce its degradation 15024385_Binding to Cdk2-cyclin A is accompanied by p27 folding, and kinetic data suggest a sequential mechanism that is initiated by binding to cyclin A 15063782_We also found that cyclin A/CDK2 phosphorylates Axin, thereby enhancing its association with beta-catenin. 15159402_study provides evidence that the cyclin A1-cyclin dependent kinase 2 complex plays a role in several signaling pathways important for cell cycle control and meiosis 15178429_interacts with dephosphorylated NIRF 15199159_cyclin A-cdk2 plays an ancillary noncatalytic role in the ubiquitination of p27(KIP1) by the SCF(skp2) complex 15226429_Results identify an important role for CDK2 in the maintenance of genomic stability, acting via an ATM- and ATR-dependent pathway. 15309028_after CDK4/6 inactivation, the fate of pancreatic tumor cells depends on the ability to modulate CDK2 activity 15355984_Data suggest that cyclin D1-Cdk2 complexes mediate some of the transforming effects of cyclin D1 and demonstrate that the cyclin D1-Cdk2 fusion protein is a useful model to investigate the biological functions of cyclin D1-Cdk2 complexes. 15456866_These findings establish a novel function for cyclin A1 and CDK2 in DNA double strand break repair following radiation damage. 15572662_Phosphborylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of CDK2. 15601848_cyclin A/Cdk2 has a role as a progesterone receptor coactivator 15607961_CDK2 depletion suppressed growth and cell cycle progression in melanoma and may be a suitable drug target in melanoma. 15611077_Inhibition of CDK2 kinase by indole-3-carbinol is accompanied by selective alterations in cyclin E composition. 15632290_molecular dynamics study on the complex CDK2 with the peptide substrate HHASPRK 15649889_Results demonstrate that a peptide derived from the alpha5 helix of cyclin A significantly inhibits kinase activity of complexes harboring CDK2, and forms stable complexes with CDK2-cyclin A. 15660127_crystal structure of phospho-CDK2 in complex with a truncated cyclin E1 (residues 81-363) at 2.25 A resolution 15665273_CDK2-BRCA1-Nucleophosmin pathway coordinately functions in cell growth and tumor progression pathways. 15671017_HTm4 binding to KAP.Cdk2.cyclin A complex enhances the phosphatase activity of KAP, dissociates cyclin A, and facilitates KAP dephosphorylation of Cdk2 15695825_Results present a comprehensive description of the dynamic behavior of cyclin-dependent kinase 2 in complex with cyclin A. 15707957_Puralpha has been shown to colocalize with cyclin A/Cdk2 and to coimmunoprecipitate with cyclin A during S-phase and we show that this interaction is mediated by a specific affinity of Puralpha for Cdk2. 15890360_Rapid binding of p27 domain 1 to cyclin A tethers the inhibitor to the binary Cdk2/cyclin A complex 15922732_CDK2 translational down-regulation may be a key regulatory event in replicative senescence of endothelial cells. 15944161_origin recognition complex 2 has an unexpected role in CDK2 activation, a linkage that could be important for maintaining genomic stability 15964852_Cdk2 destabilizes p21 via the cy2 cyclin-binding motif and p21 phosphorylation 16036217_Our results demonstrate that differential regulation of Cdc2 and Cdk2 activity by different doses of doxorubicin may contribute to the induction of two modes of cell death in hepatoma cells, either apoptosis or cell death through mitotic catastrophe. 16082200_CINP is part of the Cdc7-dependent mechanism of origin firing and a functional and physical link between Cdk2 and Cdc7 complexes at the origins 16082227_CDK2 inhibition modifies the dynamics of chromatin-bound minichromosome maintenance complex and replication protein A 16085226_results indicate that CDK2 participates in Tat-mediated HIV-1 transcription and may serve as a potential therapeutic target 16150942_Cdk2 inhibition decreases the efficiency of chemical induction of KSHV lytic transcripts ORF 50 and 26. Importantly, Cdk2 activity is also essential for replication in other human herpesviruses 16258277_A new concept indicates in this review that both Cdk2 and/or Cdc2 can drive cells through G1/S phase in parallel. 16262700_Cdk2 dependent phosphorylation(s) cannot be a critical trigger of replicon initiation in response to reoxygenation after several hours of hypoxia, at least in the T24 cells studied 16343435_We propose that during TNFalpha-induced apoptosis, PKCdelta-mediated phosphorylation of p21(WAF1/CIP1) at (146)Ser attenuates the Cdk2 binding of p21(WAF1/CIP1) and thereby upregulates Cdk2 activity. 16407256_molecular analysis of the CDK5/p25 and CDK2/cyclin A systems 16504183_Cyclin-dependent kinases regulate the transcriptional activity of FOXM1c; a combination of three phosphorylation sites mediates the Cyclin E and Cyclin A/CDK2 effects. 16540140_Here, we show that human papillomavirus type 16 16E1--E4 is also able to associate with cyclin A and Cdk2 during the G2 phase of the cell cycle. 16575928_The interaction between roscovitine and cyclin-dependent kinase 2 (cdk2) was investigated by performing correlated ab initio quantum-chemical calculations. 16707497_the phospho-CDK2/cyclin A recruitment site has a role in substrate recognition 16762841_Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha 16765349_suggest a novel retinoic acid (RA)-signaling, by which RA-induced p21 induction and complex formation with cyclin E/CDK2 diverts CDK2 function from normally driving proliferation to alternatively promoting apoptosis 16824683_Membrane depolarization may stimulate cellular proliferation by augmenting the expression of cyclin E leading to increases in Cdk2 activity and RB phosphorylation in a neuroblastoma cell line. 16912045_the Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism 16912201_Breast cancer cells lacking cancer predisposition genes BRCA1 are more sensitive to CDK2 inhibitors. 17001081_analysis of the NBI1-binding site on cyclin A which inhibits the catalytic activity of the complex cyclin-dependent kinase 2-cyclin A 17013093_progression of melanoma is associated with changes in CDK-2 expression level 17038621_functional interaction between CDK2 and FOXO1 provides a mechanism that regulates apoptotic cell death after DNA strand breakage 17095507_Kinetic and crystallographic analyses of CDK2-cyclin A complexes reveal that this inhibitory mechanism operates through steric blockade of peptide substrate binding. 17207508_Review highlights an alternative role for CDK2 in the regulation of progesterone receptor signaling. 17293600_TopBP1 necessary for the G(1)/S transition: one for activating cyclin E/CDK2 kinase and the other for loading replication components onto chromatin to initiate DNA synthesis. 17361108_Our results demonstrate that CDK2 is capable of autophosphorylation at Thr160. 17371838_results argue that Mdm2 is needed for full inhibition of Cdk2 activity by p21, thereby positively contributing to p53-dependent cell cycle arrest 17386261_Both Cdk1 and -2 require cyclin binding and T loop phosphorylation for full activity. 17409409_Observational study of gene-disease association. (HuGE Navigator) 17495531_The structure of phospho-CDK2/cyclin B is reported. pCDK2/cyclin B is less discriminatory in substrate recognition than CDK2/cyclin A & has properties of both an S-phase & an M-phase kinase. CDK2/cyclin B is effective against S phase substrates. 17638878_ATRIP is a CDK2 substrate, and CDK2-dependent phosphorylation of S224 regulates the ability of ATR-ATRIP to promote cell cycle arrest in response to DNA damage 17713927_Phosphorylation on a conserved Thr14 can inhibit activities of both the kinases, but phosphorylating another conserved Tyr15, however, can lead to totally opposite inhibition and stimulation consequences in CDK2 and CDK5. 18042686_The conserved rigid regions are important for nucleotide binding, catalysis, and substrate recognition; most flexible regions correlate with those where large conformational changes occur during CDK2 regulation processes. 18156799_cdk2 activity is necessary for the survival of human DLBCL. 18174243_Observational study of gene-disease association. (HuGE Navigator) 18199752_major Cdk2-dependent multiple gene regulatory events are present in pemphigus vulgar 18202766_serum starvation induces G1 arrest through suppression of Skp2-dependent CDK2 activity and Skp2-independent CDK4 activity in human SK-OV-3 ovarian cancer cells 18208561_growth arrest by SmE directly correlates with the reduction of cyclin E, CDK2, CDC25C and CDC2 expression, and up-regulation of p27Kip 18236071_Findings strongly demonstrate that retinoblastoma (RB) and cyclin-dependent kinase 2 (CDK2) on one side and cytokeratin 8 (CK8) and epidermal growth factor receptor 2 (HER2) on the other may affect the clinical course of the disease in 56% of patients. 18276582_Cyclin E and SV40 small T antigen cooperate to bypass quiescence and contribute to transformation by activating CDK2 in human fibroblasts 18281541_Observational study of gene-disease association. (HuGE Navigator) 18345036_Bim-mediated apoptosis following actin damage due to deregulation of Cdk2 and the cell cycle by the absence of functional p53. 18372919_G2 phase cyclin A/cdk2 controls the timing of entry into mitosis by controlling the subsequent activation of cyclin B/cdk1, but also has an unexpected role in coordinating the activation of cyclin B/cdk1 at the centrosome and in the nucleus 18400748_disruption of the spindle-assembly checkpoint does not directly influence p53 activation, but the shortening of the mitotic arrest allows cyclin E-CDK2 to be activated before the accumulation of p21(CIP1/WAF1). 18408738_Results suggest that GSK-3 regulates nuclear p27 Kip1 expression through downregulation of Skp2 expression and regulates p27 Kip1 assembly with CDK2, playing a critical role in the G0/G1 arrest associated with intestinal cell differentiation. 18470542_The structures of fully active cyclin-dependent kinase-2 (CDK2)complexed with ATP and peptide substrate, CDK2 after the catalytic reaction, and CDK2 inhibited by phosphorylation at Thr14/Tyr15 were studied using molecular dynamics simulations. 18507837_Observational study of gene-disease association. (HuGE Navigator) 18617527_Cdk2-associated complexes, by targeting SHP-1 for proteolysis, counteract the ability of SHP-1 to block cell cycle progression of intestinal epithelial cells 18635963_Cyclin A-CDK activity during G(1) would result in an inhibition of progression into the S phase. 18667424_the cyclin A-CDK2 complex may be a potential effector of NFATs, specifically NFATc1, in mediating SMC multiplication leading to neointima formation. 18784074_Cdk2 negatively regulates the activity of hPXR, and suggest an important role for Cdk2 in regulating hPXR activity and CYP3A4 expression in hepatocytes passing through the cell cycle 18806832_This suggests an important role for CDK2 in cell cycle regulation in hESCs that are likely to bear significant impacts on the maintenance of their pluripotent phenotype. 18941885_Observational study of gene-disease association. (HuGE Navigator) 19061641_Cyclin A assembles with Cdk1 only after complex formation with Cdk2 reaches a plateau during late S and G2 phases. 19066288_These findings establish phosphorylation events by CDKs 1 and 2 as key regulators of Discs Large 1 localisation and function. 19091404_Notch-1 may be mediated through regulating the expression of cell cycle regulatory proteins cyclin D1, CDK2 and p21 and the activity of Akt signaling 19101503_These results demonstrate that double phosphorylation of CDK2 peptides increases the stoichiometry of metal ion binding, and hence may contribute to the previously observed regulation of CDK2 activity by metal ions. 19103742_the pathway of apoptin-induced apoptosis and show that it essentially depends on abnormal phosphatidylinositol 3-kinase (PI3-kinase)/Akt activation, resulting in the activation of the cyclin-dependent kinase CDK2 19124506_Observational study of gene-disease association. (HuGE Navigator) 19166026_Overexpression of CDK2 was strongly correlated with abnormal proliferation in laryngeal squamous cell carcinoma. 19197163_Results show that human Cdk2 is a functional homolog for most of Ime2 functions. 19201832_disruption of Smad2 function by CDK2 phosphorylation acts as a mechanism for TGF-beta resistance in multiple myeloma. 19258477_Observational study of gene-disease association. (HuGE Navigator) 19258477_Strengthened signals in imputation-based analysis at CDK2 SNPs rs2069391, rs2069414 and rs17528736 lend evidence to the role of cell cycle genes in ovarian cancer etiology. 19321444_The combination of st and deregulated cyclin E result in cooperative and coordinated activation of both an essential origin licensing factor, CDC6, and an activity required for origin firing, CDK2, resulting in progression from quiescence to S phase. 19440053_Co-depletion of Cdc6 and p53 in normal cells restored Cdk2 activation and Rb phosphorylation, permitting them to enter S phase with a reduced rate of replication. 19594747_Observational study of gene-disease association. (HuGE Navigator) 19596857_resistance of oral squamous carcinoma to IFNgamma is not due to deficiency in STAT1-dependent signaling but from a defect in the signaling component that mediates IFNgamma-induced down-regulation of CcnA2 and Cdk2 expression 19609547_Four genes previously not examined in that respect in laryngeal carcinoma, occurred to be good markers of the neoplasm. They are: metal-proteinase ADAM12, cyclin-dependent kinase 2-CDK2, kinesin 14-KIF14, suppressor 1 of checkpoint-CHES1. 19631451_Data demonstrate that the novel anticancer mechanism of hinokitiol involves accumulation of p27, down-regulation of pRb, Skp2, and impairment of Cdk2 function. 19703905_cyclin A/cdk2-dependent phosphorylation of APC affects astral microtubule attachment to the cortical surface in mitosis 19706521_Results suggest that simple but robust rules encoded in the CDK2 structure play a dominant role in predefining the mechanisms of ligand binding, which may be advantageously exploited in designing inhibitors. 19723060_Studies indicate that roscovitine arrests the cell cycle is direct inhibition of CDK1, a mitotic regulator, and CDK2, involved in G1/S transition. 19724860_Overexpression of Notch1 in laryngeal carcinoma cell line was coupled with the downregulation of cdk2 19738611_Observational study of gene-disease association. (HuGE Navigator) 19797611_results show that the expression of UGT1A1 and CYP2B6 is negatively regulated through a CDK2 signaling pathway linked to cell cycle progression in HepG2 and SW480 cells 19822658_Results underscore the crucial role of cyclin A2-CDK2 in regulating the PLK1-SCF(beta-TrCP1)-EMI1-APC/C axis and CDC6 to trigger genome reduplication after the activity of CDK1 is suppressed. 19829063_Since CAC1 interacts with CDK2 and promotes the kinase activity of CDK2 protein, we propose that CAC1 is a novel cell cycle associated protein capable of promoting cell proliferation. 19838212_Chk1 signalling causes centrosome amplification after ionizing radiation by upregulating Cdk2 activity through activating phosphorylation. 19838216_Data show that SHP-1 knockdown increases p27stability, decreases the CDK6 levels, inducing retinoblastoma protein hypophosphorylation, downregulation of cyclin E and thereby a decrease in the CDK2 activity. 19854217_expression upregulation is critical for TLR9-stimulated proliferation of kung cancer cells 19858290_Export was also reduced by Cdk inhibition or cyclin A RNA interference, suggesting that cyclin A/Cdk complexes contribute to Wee1 export. 19885547_aberrant regulation of S100P in HCC might activate cyclin D1 and CDK expression and contribute to the mitogenic potential of tumor cells during Hepatocellular carcinoma carcinogenesis. 19960406_Cellular production of IGFBP-3 leads to G1 cell cycle arrest with inhibition of CDK2 and CDK4. 19966300_Data show that Myc repressed Ras-induced senescence, and that Cdk2 interacted with Myc at promoters, where it affected Myc-dependent regulation of genes, including those of proteins known to control senescence. 20017906_FUS-DDIT3 and the normal DDIT3 bind CDK2. 20062077_Results directly show that the inhibition of Cdk1 activity and the persistence of Cdk2 activity in G2 cells induces endoreplication without mitosis. 20068231_Results show that most of the up-regulated sites phosphorylated by cyclin-dependent CDK1 or CDK2 were almost fully phosphorylated in mitotic cells. 20079829_the nitric oxide-mediated biphasic effect was dependent on Cdk2 nitrosylation/activation and the loss of mitochondrial potential 20147522_central roles for CDK2 nuclear-cytoplasmic trafficking and cyclin E in the mechanism of 1,25-(OH)(2)D(3)-mediated growth inhibition in prostate cancer cells 20195506_These findings demonstrate that Cdk2 maintains a balance of S-phase regulatory proteins and thereby coordinates subsequent p53-independent G(2)/M checkpoint activation. 20399812_Data describe the properties of a mutant form of Cdk2 identified during large-scale sequencing of protein kinases from cancerous tissue. 20422243_Triticum aestivum-5B2 (( Ta ) 5B2) is suggested to be a wheat analogue of human CDK2 enzyme. 20444741_Conclude that cisplatin likely activates both caspase-dependent and -independent cell death, and Cdk2 is required for both pathways. 20465575_In addition to having a pivotal role in the up-regulation of IL-2 and IL-2RA gene expression, IKK controls the expression of cyclin D3, cyclin E and CDK2, and the stability SKP2 and its co-factor CKS1B, through mechanisms independent of IL-2. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20512928_Hr and VDR interact via multiple protein-protein interfaces, catalyzing histone demethylation to effect chromatin remodeling and repress the transcription of VDR target genes that control the hair cycle. 20694007_protein phosphatase 1 competition with Cdk-cyclins for retinoblastoma protein(Rb) binding is sufficient to retain Rb activity and block cell-cycle advancement. 20711190_cyclin-dependent kinases (Cdks), especially Cdk1 and Cdk2, promote interphase nuclear pore complex formation in human dividing cells. 20844047_Nuclear export of HPV31 E1 is inhibited by Cdk2 phosphorylation at two serines residues, S92 and S106. 20935635_The results demonstrate that CDK2-mediated phosphorylation is a key mechanism governing EZH2 function and that there is a link between the cell-cycle machinery and epigenetic gene silencing. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21062975_Data show that miR-302 simultaneously suppressed both the cyclin E-CDK2 and cyclin D-CDK4/6 pathways to block>70% of the G1-S cell cycle transition. 21099355_Overexpression of human Cdk2 resulted in a defect in the G1 to S transition and a reduction in viability. 21233845_MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. 21262353_Cdk2 functions via a Cdk2/SHP-1/beta-catenin/CEACAM1 axis, and show that Cdk2 has the capacity to regulate insulin internalization. 21264535_XPD may play an important role in cell apoptosis of hepatoma by inducing an over-expression of p53, but suppressing expressions of c-myc and cdk2 21319273_CDK2 downregulation causes high apoptosis at the early time points 21325496_Conclude that in cisplatin induced-kidney injury phosphorylation of p21 by Cdk2 limits the effectiveness of p21 to inhibit Cdk2. 21454540_the ability of Emi1 to inhibit APC/C is negatively regulated by CDKs 21515670_cyclin E and CDK2 genes are key physiological effectors of the c-ETS1 proto-oncogene. Furthermore, c-ETS1 is indispensable for the hepatotropic action of HBx in cell cycle deregulation. 21565702_Transient binding of a second catalytic magnesium activates the structure and dynamics of CDK2 kinase for catalysis. 21596315_The deubiquitinase USP37 binds CDH1 and removes degradative polyubiquitin from cyclin A. USP37 was induced by E2F factors in G1, peaked at G1/S, and was degraded in late mitosis. Phosphorylation of USP37 by CDK2 stimulated its full activity. 21646351_anti-oncogenic role of miR-372 may be through control of cell growth and cell cycle progression by down-regulating the cell cycle genes CDK2 and cyclin A1 21658603_Cdk2 is required for cell proliferation. 21769424_RT-PCR and Western blotting results revealed that both mRNA and protein levels of CDK2 were significantly higher in tumor tissues. 21871181_NF-Y binds to CCAAT sequences in the Cyclin A promoter, as well as to those in the promoters of cell cycle G2 regulators such as CDC2, Cyclin B and CDC25C. 21918011_Epstein-Barr virus Rta-mediated transactivation of p21 and 14-3-3sigma arrests cells at the G1/S transition by reducing cyclin E/CDK2 activity. 21941773_The expression level of CDK2 protein did not change significantly in silica-induced human embryo lung fibroblasts. 21965652_excess of MCM3 up-regulates the phosphorylation of CHK1 Ser-345 and CDK2 Thr-14. 22084169_The S-phase-specific cyclin-dependent kinase 2 was required for robust activation of ATR in response to diverse chemotherapeutic agents. 22231403_The authors show that, in human and mouse, Mre11 controls these events through a direct interaction with CDK2 that is required for CtIP phosphorylation and BRCA1 interaction in normally dividing cells. 22467868_Lin-28 homologue A (LIN28A) promotes cell cycle progression via regulation of cyclin-dependent kinase 2 (CDK2), cyclin D1 (CCND1), and cell division cycle 25 homolog A (CDC25A) expression in cancer. 22474407_CDK2 inhibition drastically diminishes anchorage-independent growth of human cancer cells and cells transformed with various oncogenes 22479189_low molecular weight cyclin E (LMW-E) requires CDK2-associated kinase activity to induce mammary tumor formation by disrupting acinar development 22673765_The activation of p21(Waf1/Cip1) was significantly up-regulated over time, but there was no change in the level of CDK2 expression by treatment of HEK293 cells with various concentrations of veterinary antibiotics. 22718829_Human cytomegalovirus IE1/2 expression was downregulated by cyclin A2, CDK1 and CDK2. 22819841_exposure of cancer cells (such as HeLa and MCF7 cells) to H2O2 increased CDK2 activity with no accompanying change in the PCNA level, leading to cell proliferation. 22927831_By a chemical-genetic approach study identified Nbs1 as a target of Cdk2, and mapped the phosphorylation to a conserved CDK consensus recognition site. 22951823_cellular CDK2 phosphorylates the functionally critical S/T-P sites of the hepadnavirus core CTD and is incorporated into viral capsids 23028682_cyclin A-Cdk2 regulates apoptosis through a mechanism that involves Rad9phosphorylation 23065011_human papillomavirus E4 proteins can interact with cyclin A and cdk2, which may contribute to viral manipulation of the host cell cycle. 23082202_Cdk2 also binds the N-terminal domain of Fbw7-gamma as well as SLP-1. 23140174_CDK2 phosphorylates CDK9 on Ser 90 and thereby contributes to HIV-1 transcription. 23184662_EEF2 phosphorylation by cyclin A-cyclin-dependent kinase 2 (CDK2) on a novel site, serine 595 (S595), directly regulates T56 phosphorylation by eEF2K. 23185313_This study aimed to explore the effects of single nucleotide polymorphisms in CDK2 and CCNE1 on breast cancer risk, progression and survival in a Chinese Han population. 23230143_Findings revealed a novel function of simultaneous p27 and CDK2 cytoplasmic mislocalization in mediating growth-factor-regulated cell proliferation, migration and invasion. 23300027_possible relationship between the CDK2 deleterious variants and the drug-binding ability 23321641_Constitutive Cdk2 activity promotes aneuploidy while altering the spindle assembly and tetraploidy checkpoints. 23390492_Constitutive CCND1/CDK2 expression contributes to neoplastic mammary epithelial cell transformation. 23390529_The prolyl isomerase Pin1 acts synergistically with CDK2 to regulate the basal activity of estrogen receptor alpha in breast cancer. 23446853_Aurora-A kinase-induced centrosome amplification was mediated by Cdk2 kinase. 23479742_the up-regulation of CDK2 by CUL4B is achieved via the repression of miR-372 and miR-373, which target CDK2. 23532886_Data indicate that TG02 blocked signaling by CDKs 1, 2, 7, and 9 and ERK5, leading to potent and highly consistent antimyeloma activity. 23643165_The expression of CDK2 mRNA significantly decreased in P(CDK2-siRNA). 23671119_A specific and essential roles for Cdk2 inhibitory phosphorylation in the successful execution of the replication stress checkpoint response and in maintaining genome integrity. 23720738_MCM7 is a substrate of cyclin E/Cdk2 and can be phosphorylated on Ser-121. 23727278_Data indicate that different binding sites of cyclin-dependent kinase (CDK2) contributing towards the binding of inhibitors. 23737759_CDK7 involved in phosphorylation/activation of CDK4 and CDK6; existence of CDK4-activating kinase(s) other than CDK7; and novel CDK7-dependent positive feedbacks mediated by p21 phosphorylation by CDK4 and CDK2 to sustain CDK4 activation. 23776131_FBXO28 activity and stability are regulated during the cell cycle by CDK1/2-mediated phosphorylation of FBXO28, which is required for its efficient ubiquitylation of MYC. 23781148_antitumor effects of DOC-1R may be mediated by negatively regulating G1 phase progression and G1/S transition through inhibiting CDK2 expression and activation 23787073_This study indicates that genetic polymorphisms of AURKA, BRCA1 and CCNE1 may affect ovarian cancer susceptibility in Chinese Han women. 24075009_Cells decide at the end of mitosis to either start the next cell cycle by immediately building up CDK2 activity or to enter a transient G0-like state by suppressing CDK2 activity. 24204256_PKC activation then triggered activation of cdk-2, which became further activated by caspase-3. 24216307_Two nuclear export signals of Cdc6 work cooperatively and distinctly for the cytoplasmic translocation of Cdc6 phosphorylated by cyclin A/Cdk2. 24240190_CDK2 knockdown alters the profile of Rb phosphorylation in coronary artery smooth muscle cells, as well as the proliferative response of these cells to mitogenic stimulation. 24386425_Of the total, the deregulation of several genes (CDK1, CDK2, CDK4, MCM2, MCM3, MCM4, EIF3a and RPN2) were potentially associated with disease development and progression. 24444383_MYC-dependent breast cancer cells possess high MYC expression and high level of MYC phosphorylation, but are not sensitive to inhibition of CDK2. 24520316_CRIF1 may play a regulatory role in the BM microenvironment-induced leukemia cell cycle arrest possibly through interacting with CDK2 and acting as a cyclin-dependent kinase inhibitor. 24623419_Authors identified and validated two additional host proteins interacting with human SAMHD1, namely, cyclin-dependent kinase 2 (CDK2) and S-phase kinase-associated protein 2 (SKP2). 24671051_Expression of Notch1, -2, and -3, CDK2, and CCNE1 was significantly decreased by upregulation of ALDH1A1 in A549 cells, but increased by its interruption in A549s cells. 24700371_In the subsequent molecular experiments, western blot analysis and kinase activity detection demonstrated that TAMs can significantly boost the expression levels and activities of CDK2 and CDK4 in SKOV3 cells. 24820417_Results show that CDK2 phosphorylates Thr-156 in GATA3. 24911186_Report structure-based discovery of allosteric inhibitors of CDK2. 24922574_CDK2 Supports HIV-1 Reverse Transcription in CD4+ T Cells.HIV-1 reverse transcriptase Is a Substrate for CDK2-Dependent Phosphorylation 24935000_It is concluded that non-response to everolimus is characterized by increased cdk2/cyclin A, driving RCC cells into the G2/M-phase. VPA hinders everolimus non-response by diminishing cdk2/cyclin A. 24947816_More effective packing and interactions between CDK2 and LMW cyclin E isoforms, however, produce more efficient protein-protein complexes that accelerate the cell division processes in cancer cells, where these cyclin E isoforms are overexpressed. 25015816_CDK2 was strongly linked to cell cycle progression and coordinated SAMHD1 phosphorylation and inactivation. 25071185_Cdk1 activity blocks lysosomal degradation of HIF-1alpha and increases HIF-1alpha protein stability and transcriptional activity. By contrast, Cdk2 activity promotes lysosomal degradation of HIF-1alpha at the G1/S phase transition. 25136960_A positive correlation between cdk2/cyclin A expression level and tumor growth. Amygdalin, therefore, may block tumor growth. 25149358_for both oncogene- and DNA damage-induced cellular senescence, CDK2 transcript and protein are decreased in a p53- and RB-dependent manner, and this repression is necessary for cell-cycle exit during senescence 25154617_Which is mutated at the CDK2 phosphorylation site. 25218592_The Cell Cycle Profiling - Risk Score (C2P-RS) based on CDK1 and CDK2 specific activities was significantly associated with relapse in breast cancers. 25218637_Data indicate that tumour suppressor RASSF1A | ENSMUSG00000025358 | Cdk2 | 5.818831e+03 | 0.8785267 | -0.186841881 | 0.3033813 | 3.857290e-01 | 0.5345529905 | 0.88844615 | No | Yes | 4.668930e+03 | 409.102078 | 5.595744e+03 | 502.429819 | |
| ENSG00000123427 | 25895 | EEF1AKMT3 | protein_coding | Q96AZ1 | FUNCTION: Protein-lysine methyltransferase that selectively mono-, di- and trimethylates 'Lys-165' of the translation elongation factors EEF1A1 and EEF1A2 in an aminoacyl-tRNA and GTP-dependent manner. EEF1A1 methylation by EEF1AKMT3 is dynamic as well as inducible by stress conditions, such as ER-stress, and plays a regulatory role on mRNA translation. {ECO:0000269|PubMed:28108655, ECO:0000269|PubMed:28663172}. | 3D-structure;Alternative splicing;Cytoplasm;Cytoskeleton;Methyltransferase;Reference proteome;S-adenosyl-L-methionine;Transferase | hsa:25895; | centrosome [GO:0005813]; chromosome [GO:0005694]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; protein-containing complex [GO:0032991]; heat shock protein binding [GO:0031072]; methyltransferase activity [GO:0008168]; protein-lysine N-methyltransferase activity [GO:0016279]; peptidyl-lysine methylation [GO:0018022] | 20190274_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22948820_The METTL21B protein is here suggested to be a protein methyltransferase, as it is shown to belong to a family of 10 human methyltransferases, some of which are shown to have lysine specific protein methyltransferase activity. 28108655_The present study identifies METTL21B as the enzyme responsible for methylation of eEF1A on Lys-165 and shows that this modification is dynamic, inducible and likely of regulatory importance. 28663172_Results have shown that METTL21B is a protein methyltransferase that methylates both isoforms of translation elongation factor eEF1A at lysine 165. Also, further data suggests that METTL21B has an evolutionarily important role for the methylation of lysine 165 in eEF1A. 34446611_METTL21B is a prognostic biomarker and potential therapeutic target in low-grade gliomas. | ENSMUSG00000080115 | Eef1akmt3 | 3.387917e+02 | 0.9036410 | -0.146178285 | 0.3081519 | 2.257792e-01 | 0.6346713282 | 0.91844201 | No | Yes | 2.884331e+02 | 26.275870 | 3.181533e+02 | 29.786459 | ||
| ENSG00000123815 | 79934 | COQ8B | protein_coding | Q96D53 | FUNCTION: Atypical kinase involved in the biosynthesis of coenzyme Q, also named ubiquinone, an essential lipid-soluble electron transporter for aerobic cellular respiration (PubMed:24270420). Its substrate specificity is unclear: does not show any protein kinase activity. Probably acts as a small molecule kinase, possibly a lipid kinase that phosphorylates a prenyl lipid in the ubiquinone biosynthesis pathway. Required for podocyte migration (PubMed:24270420). {ECO:0000250|UniProtKB:Q8NI60, ECO:0000269|PubMed:24270420}. | ATP-binding;Alternative splicing;Cell membrane;Cytoplasm;Disease variant;Kinase;Membrane;Mitochondrion;Nucleotide-binding;Reference proteome;Transferase;Transmembrane;Transmembrane helix | This gene encodes a protein with two copies of a domain found in protein kinases. The encoded protein has a complete protein kinase catalytic domain, and a truncated domain that contains only the active and binding sites of the protein kinase domain, however, it is not known whether the protein has any kinase activity. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2011]. | hsa:79934; | cytosol [GO:0005829]; extrinsic component of mitochondrial inner membrane [GO:0031314]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; kinase activity [GO:0016301]; lipid binding [GO:0008289]; cerebellar Purkinje cell layer morphogenesis [GO:0021692]; ubiquinone biosynthetic process [GO:0006744] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 24270420_Mutations in ADCK4 resulted in reduced CoQ10 levels and reduced mitochondrial respiratory enzyme activity in cells isolated from individuals with steroid-resistant nephrotic syndrome & transformed lymphoblasts. 28405841_Study in Korean cohort revealed that ADCK4 mutations should be considered in older children presenting with steroid resistant focal segmental glomerulosclerosis. The association with medullary nephrocalcinosis may be an additional diagnostic indicator. 29194833_a COQ8B polymorphism, present in 50% of the European population (NM_024876.3:c.521A > G, p.His174Arg), affects stability of the protein and could represent a risk factor for secondary Coenzyme Q deficiencies or for other complex traits. 30352687_a novel ABC1 domain-localized pathogenic mutation responsible for ADCK4-glomerulopathy was identified, further supporting the importance of the C-terminal portion of ADCK4. 32489187_Absence of Long Noncoding RNA H19 Promotes Childhood Nephrotic Syndrome through Inhibiting ADCK4 Signal. 33033902_Transcription factor Kruppel-like factor 5 positively regulates the expression of AarF domain containing kinase 4. 33084234_The role of novel COQ8B mutations in glomerulopathy and related kidney defects. 34172776_Whole-exome sequencing reveals a novel homozygous mutation in the COQ8B gene associated with nephrotic syndrome. | ENSMUSG00000003762 | Coq8b | 1.763314e+03 | 1.3341745 | 0.415947386 | 0.3086922 | 1.822256e+00 | 0.1770449282 | 0.77663851 | No | Yes | 1.769779e+03 | 233.329792 | 1.225708e+03 | 166.158511 | |
| ENSG00000123992 | 23549 | DNPEP | protein_coding | Q9ULA0 | FUNCTION: Aminopeptidase with specificity towards an acidic amino acid at the N-terminus. Likely to play an important role in intracellular protein and peptide metabolism. {ECO:0000269|PubMed:9632644}. | 3D-structure;Acetylation;Alternative initiation;Aminopeptidase;Cytoplasm;Hydrolase;Metal-binding;Metalloprotease;Phosphoprotein;Protease;Reference proteome;Zinc | The protein encoded by this gene is an aminopeptidase which prefers acidic amino acids, and specifically favors aspartic acid over glutamic acid. It is thought to be a cytosolic protein involved in general metabolism of intracellular proteins. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2016]. | hsa:23549; | blood microparticle [GO:0072562]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; aminopeptidase activity [GO:0004177]; identical protein binding [GO:0042802]; metallopeptidase activity [GO:0008237]; zinc ion binding [GO:0008270]; peptide metabolic process [GO:0006518] | 12413488_Identification of histidine residues important in the catalysis and structure of aspartyl aminopeptidase 22720794_The crystal structure of human DNPEP complexed with zinc and a substrate analogue aspartate-beta-hydroxamate reveals a dodecameric machinery built by domain-swapped dimers. 23948443_aspartyl aminopeptidase was upregulated in colorectal cancer tissues, and greater activity correlated significantly with the absence of lymph node metastases and with better overall survival 31228326_Aspartyl Aminopeptidase Suppresses Proliferation, Invasion, and Stemness of Breast Cancer Cells via Targeting CD44. | ENSMUSG00000026209 | Dnpep | 2.499670e+03 | 1.1397550 | 0.188723734 | 0.2971273 | 4.041284e-01 | 0.5249648606 | 0.88560482 | No | Yes | 2.535153e+03 | 253.357740 | 2.085637e+03 | 214.116606 | |
| ENSG00000124074 | 84080 | ENKD1 | protein_coding | Q9H0I2 | Alternative splicing;Phosphoprotein;Reference proteome | hsa:84080; | ciliary base [GO:0097546]; cytoplasmic microtubule [GO:0005881]; microtubule cytoskeleton [GO:0015630] | 33724673_Enkurin domain containing 1 (ENKD1) regulates the proliferation, migration and invasion of non-small cell lung cancer cells. | ENSMUSG00000013155 | Enkd1 | 9.295314e+02 | 1.0890217 | 0.123032685 | 0.3162840 | 1.517870e-01 | 0.6968334275 | 0.93520380 | No | Yes | 8.756589e+02 | 105.086660 | 7.595437e+02 | 93.961088 | |||
| ENSG00000124459 | 7596 | ZNF45 | protein_coding | Q02386 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7596; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; multicellular organism development [GO:0007275]; regulation of transcription by RNA polymerase II [GO:0006357] | 19195718_Observational study of gene-disease association. (HuGE Navigator) 32363570_our study discovered 3 novel population-specific functional genetic variants (rs6913677, rs2078267, rs8100011) in 2 novel (SLC22A11 and ZNF45) and 1 earlier reported gene (BAI3) for BMI in Indians. Our study decodes key genomic loci underlying obesity phenotype in Indians that may serve as prospective drug targets in future | ENSMUSG00000074282 | Zfp94 | 2.633630e+02 | 0.7840975 | -0.350895118 | 0.3354752 | 1.087462e+00 | 0.2970341705 | 0.81383297 | No | Yes | 2.552299e+02 | 40.977811 | 2.863412e+02 | 46.883374 | ||
| ENSG00000124784 | 83732 | RIOK1 | protein_coding | Q9BRS2 | FUNCTION: Involved in the final steps of cytoplasmic maturation of the 40S ribosomal subunit. Involved in processing of 18S-E pre-rRNA to the mature 18S rRNA. Required for the recycling of NOB1 and PNO1 from the late 40S precursor (PubMed:22072790). The association with the very late 40S subunit intermediate may involve a translation-like checkpoint point cycle preceeding the binding to the 60S ribosomal subunit (By similarity). Despite the protein kinase domain is proposed to act predominantly as an ATPase (By similarity). The catalytic activity regulates its dynamic association with the 40S subunit (By similarity). In addition to its role in ribosomal biogenesis acts as an adapter protein by recruiting NCL/nucleolin the to PRMT5 complex for its symmetrical methylation (PubMed:21081503). {ECO:0000250|UniProtKB:G0S3J5, ECO:0000250|UniProtKB:Q12196, ECO:0000269|PubMed:21081503, ECO:0000269|PubMed:22072790}. | 3D-structure;ATP-binding;Cytoplasm;Direct protein sequencing;Hydrolase;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Ribosome biogenesis;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene competes with pICln for inclusion in the protein arginine methyltransferase 5 complex. This complex targets substrates for dimethylation. The encoded protein is essential for the last steps in the maturation of 40S subunits. [provided by RefSeq, Jan 2017]. | hsa:83732; | cytosol [GO:0005829]; methyltransferase complex [GO:0034708]; nucleoplasm [GO:0005654]; preribosome, small subunit precursor [GO:0030688]; ATP binding [GO:0005524]; hydrolase activity [GO:0016787]; metal ion binding [GO:0046872]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; maturation of SSU-rRNA [GO:0030490]; positive regulation of rRNA processing [GO:2000234]; ribosomal small subunit biogenesis [GO:0042274] | 21081503_RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. 22072790_These data reveal a role of hRio1 in the final stages of cytoplasmic pre-40S maturation 23459592_These results imply that, in glioblastoma cells, constitutive Akt signaling drives RIO kinase overexpression, which creates a feedforward loop that promotes and maintains oncogenic Akt activity through stimulation of mTor signaling. 24948609_The RIO kinase fold generates a versatile ATPase enzyme, which in the case of Rio1 is activated following the Rio2 step to regulate one of the final 40S maturation events, at which time the 60S subunit is recruited for final quality control check. 27068473_methionine adenosyltransferase II alpha (MAT2A), and the arginine methyltransferase, PRMT5, as vulnerable enzymes in cells with MTAP deletion. 27909846_Docking studies revealed that TIBI can occupy the ATP-binding site of Rio1 in a manner similar to toyocamycin, and enhances the thermostability of the enzyme. 28499923_we demonstrate that RIOK1 promotes lung colonization in vivo and that RIOK1 is overexpressed in different subtypes of human lung- and breast cancer. 29384474_Together, this study highlights the importance of a RIOK1 methylation-phosphorylation switch in determining colorectal and gastric cancer development. 32599985_Elevated Expression of RIOK1 Is Correlated with Breast Cancer Hormone Receptor Status and Promotes Cancer Progression. 33908345_The final step of 40S ribosomal subunit maturation is controlled by a dual key lock. | ENSMUSG00000021428 | Riok1 | 1.208482e+03 | 0.9628539 | -0.054611148 | 0.3099823 | 3.073699e-02 | 0.8608284429 | 0.97337236 | No | Yes | 1.204483e+03 | 183.228381 | 1.187657e+03 | 185.303667 | |
| ENSG00000124920 | 745 | MYRF | protein_coding | Q9Y2G1 | FUNCTION: [Myelin regulatory factor]: Constitutes a precursor of the transcription factor. Mediates the autocatalytic cleavage that releases the Myelin regulatory factor, N-terminal component that specifically activates transcription of central nervous system (CNS) myelin genes (PubMed:23966832). {ECO:0000269|PubMed:23966832}.; FUNCTION: [Myelin regulatory factor, C-terminal]: Membrane-bound part that has no transcription factor activity and remains attached to the endoplasmic reticulum membrane following cleavage. {ECO:0000269|PubMed:23966832}.; FUNCTION: [Myelin regulatory factor, N-terminal]: Transcription factor that specifically activates expression of myelin genes such as MBP, MOG, MAG, DUSP15 and PLP1 during oligodendrocyte (OL) maturation, thereby playing a central role in oligodendrocyte maturation and CNS myelination. Specifically recognizes and binds DNA sequence 5'-CTGGYAC-3' in the regulatory regions of myelin-specific genes and directly activates their expression. Not only required during oligodendrocyte differentiation but is also required on an ongoing basis for the maintenance of expression of myelin genes and for the maintenance of a mature, viable oligodendrocyte phenotype (PubMed:23966832). {ECO:0000269|PubMed:23966832}. | 3D-structure;Acetylation;Activator;Alternative splicing;Autocatalytic cleavage;Coiled coil;Cytoplasm;DNA-binding;Differentiation;Disease variant;Endoplasmic reticulum;Glycoprotein;Hydrolase;Membrane;Nucleus;Protease;Reference proteome;Transcription;Transcription regulation;Transmembrane;Transmembrane helix | This gene encodes a transcription factor that is required for central nervous system myelination and may regulate oligodendrocyte differentiation. It is thought to act by increasing the expression of genes that effect myelin production but may also directly promote myelin gene expression. Loss of a similar gene in mouse models results in severe demyelination. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2014]. | hsa:745; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; peptidase activity [GO:0008233]; sequence-specific DNA binding [GO:0043565]; central nervous system myelin maintenance [GO:0032286]; central nervous system myelination [GO:0022010]; oligodendrocyte development [GO:0014003]; oligodendrocyte differentiation [GO:0048709]; positive regulation of myelination [GO:0031643]; positive regulation of transcription, DNA-templated [GO:0045893]; protein autoprocessing [GO:0016540] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23966832_A Bacteriophage tailspike domain promotes self-cleavage of a human membrane-bound transcription factor, the myelin regulatory factor MYRF. 28160598_molecular mechanisms underlying the homo-trimerization of Myrf N-terminal fragments and the homo-trimeric DNA binding of Myrf N-terminal fragments 28631093_The relative scarcity of oligodendrocyte lineage cells expressing MYRF in demyelinated MS lesions demonstrates, for the first time, that chronic lesions lack oligodendrocytes that express this necessary transcription factor for remyelination and supports the notion that a failure to fully differentiate underlies remyelination failure. 29265453_Functional defects of MYRF are likely to be causally associated with encephalopathy with extensive myelin vacuolization 29446546_Heterozygous variants in MYRF should be considered in patients with variants of Scimitar syndrome and urogenital anomalies. 30532227_We identified four unrelated individuals with congenital diaphragmatic hernia with damaging de novo variants in MYRF (P = 5.3x10(-8)), including one likely gene-disrupting (LGD) and three deleterious missense (D-mis) variants. Eight additional individuals with de novo LGD or missense variants were identified from our other genetic studies or from the literature. 30985895_The results suggest that MYRF is a novel causative gene of 46,XY and 46,XX disorders of sex development and MYRF is a transcription factor regulating coelomic epithelium and/or coelomic epithelium derived cells proliferation and migration, which is essential for development of multiple organs. 31048900_Identified a canonical splice site alteration upstream of the last exon of the gene encoding myelin regulatory factor (MYRF c.3376-1G>A), which produced a stable RNA transcript, leading to a frameshift mutation p.Gly1126Valfs*31 in the C-terminus of the protein. 31069960_results underline the importance of including SLC18A3 sequencing in the differential diagnostics of fetuses with arthrogryposis, FADS, or LMPS of unknown etiology 31172260_These evidence all support that truncation mutations in the C-terminal region of MYRF are responsible for autosomal dominant high hyperopia in these families. 31266062_This is the first trio-based whole-genome sequencing (WGS), study for nanophthalmos, revealing the potential role of de novo mutations (DNMs) in MYRF and rare inherited genetic variants in PRSS56 and MFRP. 31700225_C-terminal variants in MYRF, which are expected to escape nonsense-mediated decay, represent a rare cause of autosomal dominant nanophthalmos with or without dextrocardia or congenital diaphragmatic hernia. 31964908_Functional mechanism and pathogenic potential of MYRF ICA domain mutations implicated in birth defects. 32052405_The genetic and clinical landscape of nanophthalmos and posterior microphthalmos in an Australian cohort. 32997974_Pancreatic Cancer Cells Require the Transcription Factor MYRF to Maintain ER Homeostasis. 33670313_Common Myelin Regulatory Factor Gene Variants Predisposing to Excellence in Sports. 33798553_Functional mechanisms of MYRF DNA-binding domain mutations implicated in birth defects. 33864620_MYRF: A Mysterious Membrane-Bound Transcription Factor Involved in Myelin Development and Human Diseases. 35136345_Evaluation of MYRF as a candidate gene for primary angle closure glaucoma. | ENSMUSG00000036098 | Myrf | 2.656356e+02 | 1.3172668 | 0.397547624 | 0.3016115 | 1.759695e+00 | 0.1846625133 | 0.77834741 | No | Yes | 2.178571e+02 | 40.238593 | 1.807659e+02 | 34.414926 | |
| ENSG00000125046 | 51066 | SSUH2 | protein_coding | Q9Y2M2 | FUNCTION: Plays a role in odontogenesis. {ECO:0000269|PubMed:27680507}. | Alternative splicing;Cytoplasm;Disease variant;Nucleus;Reference proteome | hsa:51066; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; odontogenesis [GO:0042476] | 20205943_Expression and synthesis of fls485 are found in surface lining epithelia of normal human intestinal mucosa and deriving epithelial cell lines. 27680507_our observations demonstrate that SSUH2 disrupts dental formation and that this novel gene, together with other odontogenesis genes, is involved in tooth development. | ENSMUSG00000034387 | Ssu2 | 3.037749e+01 | 1.0449978 | 0.063499873 | 0.6043752 | 1.111668e-02 | 0.9160301059 | 0.98426458 | No | Yes | 2.474857e+01 | 10.185172 | 2.434210e+01 | 10.155725 | ||
| ENSG00000125122 | 26231 | LRRC29 | transcribed_unitary_pseudogene | Q8WV35 | FUNCTION: Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation. | Leucine-rich repeat;Reference proteome;Repeat;Ubl conjugation pathway | This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into 3 classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbls class and, in addition to an F-box, contains 9 tandem leucine-rich repeats. Two transcript variants encoding the same protein have been found for this gene. Other variants may occur, but their full-length natures have not been characterized. [provided by RefSeq, Jul 2008]. | 1.190758e+02 | 1.3996962 | 0.485113764 | 0.4067147 | 1.401552e+00 | 0.2364638366 | 0.78818582 | No | Yes | 1.087624e+02 | 20.118681 | 7.184383e+01 | 14.055298 | ||||||
| ENSG00000125319 | 78995 | HROB | protein_coding | Q8N3J3 | FUNCTION: DNA-binding protein involved in homologous recombination that acts by recruiting the MCM8-MCM9 helicase complex to sites of DNA damage to promote DNA repair synthesis. {ECO:0000269|PubMed:31467087}. | Alternative splicing;Chromosome;DNA damage;DNA recombination;DNA repair;DNA synthesis;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome | hsa:78995; | nucleus [GO:0005634]; site of DNA damage [GO:0090734]; single-stranded DNA binding [GO:0003697]; cellular response to DNA damage stimulus [GO:0006974]; DNA synthesis involved in DNA repair [GO:0000731]; female gamete generation [GO:0007292]; interstrand cross-link repair [GO:0036297]; male gamete generation [GO:0048232]; recombinational repair [GO:0000725] | 19079262_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 30370607_The C17orf53 SNP rs227584 is associated with human bone density and bone fractures risk. 31467087_C17orf53/HROB is an OB-fold-containing factor involved in HR that acts by recruiting the MCM8-MCM9 helicase to sites of DNA damage to promote DNA synthesis 32528060_MCM8IP activates the MCM8-9 helicase to promote DNA synthesis and homologous recombination upon DNA damage. 32853826_C17orf53 is identified as a novel gene involved in inter-strand crosslink repair. 34707299_Meiotic genes in premature ovarian insufficiency: variants in HROB and REC8 as likely genetic causes. | ENSMUSG00000034773 | Hrob | 1.019956e+03 | 1.1377554 | 0.186190425 | 0.3017065 | 3.827990e-01 | 0.5361090369 | 0.88864621 | No | Yes | 1.038687e+03 | 108.076560 | 8.284917e+02 | 88.857581 | ||
| ENSG00000125354 | 23157 | SEPTIN6 | protein_coding | Q14141 | FUNCTION: Filament-forming cytoskeletal GTPase. Required for normal organization of the actin cytoskeleton. Involved in cytokinesis. May play a role in HCV RNA replication. Forms a filamentous structure with SEPTIN12, SEPTIN6, SEPTIN2 and probably SEPTIN4 at the sperm annulus which is required for the structural integrity and motility of the sperm tail during postmeiotic differentiation (PubMed:25588830). {ECO:0000269|PubMed:17229681, ECO:0000269|PubMed:17803907, ECO:0000305|PubMed:25588830}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Cell projection;Centromere;Chromosome;Cilium;Coiled coil;Cytoplasm;Cytoskeleton;Differentiation;Direct protein sequencing;Flagellum;GTP-binding;Host-virus interaction;Kinetochore;Nucleotide-binding;Phosphoprotein;Reference proteome;Spermatogenesis | This gene is a member of the septin family of GTPases. Members of this family are required for cytokinesis. One version of pediatric acute myeloid leukemia is the result of a reciprocal translocation between chromosomes 11 and X, with the breakpoint associated with the genes encoding the mixed-lineage leukemia and septin 2 proteins. This gene encodes four transcript variants encoding three distinct isoforms. An additional transcript variant has been identified, but its biological validity has not been determined. [provided by RefSeq, Jul 2008]. | hsa:23157; | axon terminus [GO:0043679]; cell division site [GO:0032153]; cleavage furrow [GO:0032154]; kinetochore [GO:0000776]; microtubule cytoskeleton [GO:0015630]; midbody [GO:0030496]; septin collar [GO:0032173]; septin complex [GO:0031105]; septin ring [GO:0005940]; sperm annulus [GO:0097227]; spindle [GO:0005819]; synaptic vesicle [GO:0008021]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; molecular adaptor activity [GO:0060090]; cell differentiation [GO:0030154]; cellular protein localization [GO:0034613]; cilium assembly [GO:0060271]; cytoskeleton-dependent cytokinesis [GO:0061640]; mitotic cytokinesis [GO:0000281]; spermatogenesis [GO:0007283] | 12096348_MLL-SEPTIN6 fusion recurs in novel translocation of chromosomes 3, X, and 11 in infant acute myelomonocytic leukaemia and in t(X;11) in infant acute myeloid leukaemia 16843108_Molecular analysis using rtPCR followed by sequencing confirmed the expression of an MLL-SEPT6 fusion transcript with a novel sequence. 16914550_septin 2, 6, and 7 complexes make up polymerized filaments 17229681_These results indicate that the host proteins hnRNP A1 and septin 6 play important roles in the replication of HCV through RNA-protein and protein-protein interactions. 17637674_crystal structures of the human SEPT2 G domain and the heterotrimeric human SEPT2-SEPT6-SEPT7 complex 17803907_Demonstrate connection between septins/SOCS7/NCK signaling and the DNA damage response. 18047794_co-expression of SEPT12 altered the filamentous structure of SEPT6 in Hela cells 21082023_Data show that Septins of the SEPT6 group preferentially interacted with septins of the SEPT2 group, SEPT3 group and SEPT7 group. 21788367_The SEPT6 provides the directional guidance cues necessary for polarizing the epithelial microtubule network. 25380047_Septin6 and Septin7 GTP binding proteins regulate AP-3- and ESCRT-dependent multivesicular body biogenesis 25519054_MiR-223-3p might target gene SEPT6 and promoted the biological behavior of prostate cancer. 29051266_SUMOylation of human septins is critical for septin filament bundling and cytokinesis. 29943706_Results demonstrate that in stored platelets, septine-2 and septin-6 mRNAs have miR- 223 target sites, septin-2 and septin-6 are in complex with Ago-2. The results demonstrate that like in nucleated cells, enucleated platelets also have microRNA-based mechanisms for the regulation of their septins. 30315255_regulates various biological behaviors in hepatic stellate cells through TGF-beta1/Smad, mitogen-activated protein kinases and phosphatidylinositol-3-kinase/protein kinase B signaling pathways, thus promoting liver fibrosis 31865373_Septin2 mediates podosome maturation and endothelial cell invasion associated with angiogenesis. 32910969_Molecular Recognition at Septin Interfaces: The Switches Hold the Key. 33639214_Orientational Ambiguity in Septin Coiled Coils and its Structural Basis. 33664458_LSD1-mediated stabilization of SEPT6 protein activates the TGF-beta1 pathway and regulates non-small-cell lung cancer metastasis. 33846777_SEPT6 drives hepatocellular carcinoma cell proliferation, migration and invasion via the Hippo/YAP signaling pathway. 34116125_An atomic model for the human septin hexamer by cryo-EM. | ENSMUSG00000050379 | Septin6 | 1.807769e+03 | 1.5332932 | 0.616633600 | 0.2788074 | 4.864800e+00 | 0.0274099782 | 0.55921720 | No | Yes | 2.443577e+03 | 278.787308 | 1.383385e+03 | 162.500783 | |
| ENSG00000125388 | 2868 | GRK4 | protein_coding | P32298 | FUNCTION: Specifically phosphorylates the activated forms of G protein-coupled receptors. GRK4-alpha can phosphorylate rhodopsin and its activity is inhibited by calmodulin; the other three isoforms do not phosphorylate rhodopsin and do not interact with calmodulin. GRK4-alpha and GRK4-gamma phosphorylate DRD3. Phosphorylates ADRB2. {ECO:0000269|PubMed:19520868, ECO:0000269|PubMed:8626439}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Kinase;Lipoprotein;Nucleotide-binding;Palmitate;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the guanine nucleotide-binding protein (G protein)-coupled receptor kinase subfamily of the Ser/Thr protein kinase family. The protein phosphorylates the activated forms of G protein-coupled receptors thus initiating its deactivation. This gene has been linked to both genetic and acquired hypertension. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2013]. | hsa:2868; | cell cortex [GO:0005938]; cytoplasm [GO:0005737]; photoreceptor disc membrane [GO:0097381]; ATP binding [GO:0005524]; G protein-coupled receptor kinase activity [GO:0004703]; protein kinase activity [GO:0004672]; rhodopsin kinase activity [GO:0050254]; protein phosphorylation [GO:0006468]; receptor internalization [GO:0031623]; regulation of G protein-coupled receptor signaling pathway [GO:0008277]; regulation of rhodopsin mediated signaling pathway [GO:0022400]; regulation of signal transduction [GO:0009966]; signal transduction [GO:0007165] | 12164861_critical role of GRK4, relative to GRK2, in the homologous desensitization of D1 receptors in renal proximal tubule cells. 12446468_Observational study of gene-disease association. (HuGE Navigator) 12881416_role in phosphorylation-independent desensitization of GABA(B) receptor 15097232_Genetic variation in GRK4gamma was associated with hypertension 15097232_Observational study of gene-disease association. (HuGE Navigator) 16338988_GRK4 constitutively phosphorylates the D1 receptor in the absence of agonist activation. 16461192_Observational study of gene-disease association. (HuGE Navigator) 16461192_data indicate that the R65L polymorphism of the GRK4 gene plays a role in blood pressure regulation in adolescents and young adults. 16636198_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16940246_The 65L allele of GRK4 gene is associated with stress-induced urinary sodium excretion. 17044852_Genetic variation in Gprotein-coupled receptor kinase 4 was associated with hypertension in northern Han Chinese. 17044852_Observational study of gene-disease association. (HuGE Navigator) 17259440_We conclude that the elevated blood pressure of hGRK4gamma A142V transgenic mice is due mainly to the effect of hGRK4gamma A142V transgene acting via D(1)R and increased ROS production is not a contributor. 18190783_This evidence suggests the kinase domain of GRK4gamma is responsible for GPCR regulation and GRK4 binding to inactive Galpha(s) and Gbeta may explain the constitutive activity of GRK4gamma towards Galpha(s)-coupled receptors. 18413491_Observational study of gene-disease association. (HuGE Navigator) 18413491_The GRK4 Ala142Val polymorphism did not contribute to the phenotypes in renal handling of sodium 18711006_The potential implication of GRK4 promoter variation in blood pressure control in humans is unmistakable. 18711008_The 4 identified genetic variants within this region exert allele-specific impact on both cell type- and stimulation-dependent transcription and may affect the expression balance of renal G protein-coupled receptor kinase 4. 19119263_hypertensive African American men with a GRK4 A142 variant were less responsive to metoprolol if they had a GRK4 L65 variant 19520868_GRK4, specifically the GRK4-gamma and GRK4-alpha isoforms, phosphorylates the D(3) receptor and is crucial for its signaling in human proximal tubule cells 19842931_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20153824_Dopamine and G protein-coupled receptor kinase 4 in the kidney: role in blood pressure regulation. 20308035_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20424473_Observational study of gene-disease association. (HuGE Navigator) 20448640_Observational study of gene-disease association. (HuGE Navigator) 20448640_The G-protein-coupled receptor kinase 4 (T-rs1024323-C and T-rs1801058-C) polymorphisms were not associated with a risk of preeclampsia in Northern Han Chinese. 20537417_Observational study of gene-disease association. (HuGE Navigator) 20577119_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21544086_GRK-4 polymorphisms predict blood pressure response to dietary modification in Black subjects with mild-to-moderate hypertension. 22639547_The significant association was also found for rs2960306 polymorphism in the GRK4 gene among Europeans. 23195037_internalization of agonist-activated D1R is regulated by both SNX5 and GRK4, and that SNX5 is critical to the recycling of the receptor to the plasma membrane 23839509_Buffalo colostrum beta-lactoglobulin inhibits VEGF-induced angiogenesis by interacting with G protein-coupled receptor kinase 24218433_This study provides a mechanism by which GRK4, via regulation of arterial AT(1)R expression and function, participates in the pathogenesis of conduit vessel abnormalities in hypertension. 25023003_Mutations in GRK4 gene is associated with stress fracture. 25732908_the association between GRK4(142V) and a larger decrease in blood pressure with angiotensin receptor blockers in hypertensive patients. 25768006_A high sodium intake markedly increased the obesity risk in variants of GRK4 A486V regardless of sex amongst Korean children. 25870190_GRK4 genetic polymorphism plays an important role in the regulation of hypertension. 26134571_GRK4alpha is more similar to GRK2 than GRK6. A fully ordered kinase C-tail reveals interactions linking the C-tail with important determinants of kinase activity, including the alphaB helix, alphaD helix, and the P-loop 26250109_The C-terminal region of FMRP interacts with a domain of GRK4 mRNA that is folded in four stem loops. 26667412_hGRK4gamma(142V) phosphorylates histone deacetylase type 1 and promotes its nuclear export to the cytoplasm, resulting in increased AT1R expression and greater pressor response to angiotensin II. 26730182_Subgroup analysis for ethnicity showed that rs1024323 locus of GRK4 gene was associated with hypertension in Caucasians (OR =1.826, 95% CI =1.215-2.745, P=0.004) but not in East Asians and Africans. [meta-analysis] 26820533_Thus, we provided new insight into the function of GRKs in agonist-unstimulated GPCR trafficking using a recombinant AM1 receptor and further determined the region of the CLR C-tail responsible for this GRK function. 27045027_GRK4 gene variants in humans (GRK4 A>65L, A>142V, and A>486V) are associated with Hypertension in several but not all ethnic groups. GRK4 gene variants impair the function of D1R and D3R and abet the function of AT1R. These lead to enhancement of renal sodium reabsorption and impaired ability to excrete a sodium load that eventually leads to hypertension. Acknowledgments 28189851_On normal salt diet, renal CuZnSOD and ECSOD proteins were similar but renal MnSOD was lower in hGRK4g486V than Non-T mice and remained low on high salt diet. hGRK4gammawild-type mice were normotensive and hGRK4g142V mice were hypertensive but both were salt-resistant and in normal redox balance. Chronic tempol treatment partially prevented the salt-sensitivity of hGRK4g486V mice. 28912086_A novel function of GRK4 on triggering a p53-independent cellular senescence, which involves an intricate signaling network. 31349021_Proteomics analysis of G protein-coupled receptor kinase 4-inhibited cellular growth of HEK293 cells. 32349533_Renal Dopamine Receptors and Oxidative Stress: Role in Hypertension. 33127268_Associations of SUCNR1, GRK4, CAMK1D gene polymorphisms and the susceptibility of type 2 diabetes mellitus and essential hypertension in a northern Chinese Han population. 33280021_The role of G protein-coupled receptor kinase 4 in cardiomyocyte injury after myocardial infarction. 33899625_Genetic variants of GRK4 influence circadian rhythm of blood pressure and response to candesartan in hypertensive patients. 34297769_Targeted capture sequencing identifies genetic variations of GRK4 and RDH8 in Han Chinese with essential hypertension in Xinjiang. | ENSMUSG00000052783 | Grk4 | 1.995616e+02 | 0.9116327 | -0.133475385 | 0.3108675 | 1.825694e-01 | 0.6691744136 | 0.92913032 | No | Yes | 1.987612e+02 | 23.424189 | 2.247014e+02 | 27.313721 | |
| ENSG00000125458 | 30833 | NT5C | protein_coding | Q8TCD5 | FUNCTION: Dephosphorylates the 5' and 2'(3')-phosphates of deoxyribonucleotides, with a preference for dUMP and dTMP, intermediate activity towards dGMP, and low activity towards dCMP and dAMP. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Hydrolase;Magnesium;Metal-binding;Nucleotide metabolism;Nucleotide-binding;Phosphoprotein;Reference proteome | This gene encodes a nucleotidase that catalyzes the dephosphorylation of the 5' deoxyribonucleotides (dNTP) and 2'(3')-dNTP and ribonucleotides, but not 5' ribonucleotides. Of the different forms of nucleotidases characterized, this enzyme is unique in its preference for 5'-dNTP. It may be one of the enzymes involved in regulating the size of dNTP pools in cells. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Nov 2011]. | hsa:30833; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; 5'-nucleotidase activity [GO:0008253]; identical protein binding [GO:0042802]; IMP 5'-nucleotidase activity [GO:0050483]; metal ion binding [GO:0046872]; nucleotidase activity [GO:0008252]; pyrimidine nucleotide binding [GO:0019103]; allantoin metabolic process [GO:0000255]; dCMP catabolic process [GO:0006249]; dephosphorylation [GO:0016311]; dGMP catabolic process [GO:0046055]; dTMP catabolic process [GO:0046074]; dUMP catabolic process [GO:0046079]; IMP catabolic process [GO:0006204]; pyrimidine deoxyribonucleotide catabolic process [GO:0009223]; UMP catabolic process [GO:0046050] | 12418222_Data show that PN-I shows a higher affinity for oxynucleosides rather than deoxynucleosides, while the opposite is true for PN-II. 12714505_Mutation analysis in the P5N-I gene led to the identification of 3 novel mutations 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000020736 | Nt5c | 2.031476e+03 | 1.2761857 | 0.351838225 | 0.3075507 | 1.317393e+00 | 0.2510605355 | 0.79219522 | No | Yes | 2.233847e+03 | 310.895061 | 1.467539e+03 | 210.005142 | |
| ENSG00000125863 | 8195 | MKKS | protein_coding | Q9NPJ1 | FUNCTION: Probable molecular chaperone that assists the folding of proteins upon ATP hydrolysis (PubMed:20080638). Plays a role in the assembly of BBSome, a complex involved in ciliogenesis regulating transports vesicles to the cilia (PubMed:20080638). May play a role in protein processing in limb, cardiac and reproductive system development. May play a role in cytokinesis (PubMed:28753627). {ECO:0000269|PubMed:20080638, ECO:0000269|PubMed:28753627}. | ATP-binding;Bardet-Biedl syndrome;Chaperone;Ciliopathy;Cytoplasm;Cytoskeleton;Disease variant;Mental retardation;Nucleotide-binding;Nucleus;Obesity;Reference proteome;Sensory transduction;Vision | This gene encodes a protein which shares sequence similarity with other members of the type II chaperonin family. The encoded protein is a centrosome-shuttling protein and plays an important role in cytokinesis. This protein also interacts with other type II chaperonin members to form a complex known as the BBSome, which involves ciliary membrane biogenesis. This protein is encoded by a downstream open reading frame (dORF). Several upstream open reading frames (uORFs) have been identified, which repress the translation of the dORF, and two of which can encode small mitochondrial membrane proteins. Mutations in this gene have been observed in patients with Bardet-Biedl syndrome type 6, also known as McKusick-Kaufman syndrome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2013]. | hsa:8195; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; kinociliary basal body [GO:1902636]; motile cilium [GO:0031514]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; unfolded protein binding [GO:0051082]; artery smooth muscle contraction [GO:0014824]; brain morphogenesis [GO:0048854]; cartilage development [GO:0051216]; cerebral cortex development [GO:0021987]; chaperone-mediated protein complex assembly [GO:0051131]; cilium assembly [GO:0060271]; convergent extension involved in gastrulation [GO:0060027]; detection of mechanical stimulus involved in sensory perception of sound [GO:0050910]; determination of left/right symmetry [GO:0007368]; developmental process [GO:0032502]; face development [GO:0060324]; fat cell differentiation [GO:0045444]; gonad development [GO:0008406]; heart development [GO:0007507]; heart looping [GO:0001947]; hippocampus development [GO:0021766]; intracellular transport [GO:0046907]; melanosome transport [GO:0032402]; negative regulation of actin filament polymerization [GO:0030837]; negative regulation of appetite by leptin-mediated signaling pathway [GO:0038108]; negative regulation of blood pressure [GO:0045776]; negative regulation of gene expression [GO:0010629]; negative regulation of GTPase activity [GO:0034260]; non-motile cilium assembly [GO:1905515]; photoreceptor cell maintenance [GO:0045494]; pigment granule aggregation in cell center [GO:0051877]; positive regulation of multicellular organism growth [GO:0040018]; protein folding [GO:0006457]; regulation of cilium beat frequency involved in ciliary motility [GO:0060296]; regulation of stress fiber assembly [GO:0051492]; sensory perception of smell [GO:0007608]; social behavior [GO:0035176]; spermatid development [GO:0007286]; striatum development [GO:0021756]; vasodilation [GO:0042311]; visual perception [GO:0007601] | 11567139_Unaffected individuals in 2 pedigrees had 2 but not all 3 mutations that affecteds had which suggests that Bardet-Biedle syndrome might not be a single-gene recessive disease but a complex trait requiring three mutant alleles to manifest the phenotype. 12837689_The presence of three mutant alleles in the BBS family correlates with a more severe Bardet-Biedl phenotype. 15483080_Observational study of gene-disease association. (HuGE Navigator) 15731008_MKKS/BBS6 is a novel centrosomal component required for cytokinesis 17003356_Observational study of gene-disease association. (HuGE Navigator) 18094050_These results indicate that the MKKS mutants have an abnormal conformation and that chaperone-dependent degradation mediated by CHIP is a key feature of McKusick-Kaufman syndrome/Bardet-Biedl syndrome diseases. 18813213_Observational study of gene-disease association. (HuGE Navigator) 18813213_results suggest that genetic variation in the MKKS gene may play a role in the development of obesity and the metabolic syndrome. 19077438_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19247371_genetic variations at MKKS gene influence the risk of metabolic syndrome 19876004_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20472660_Using sequence analysis, the role of BBS6, 10 and 12 was assessed in a Bardet-Biedl syndrome patient population comprising 93 cases from 74 families. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20801516_Observational study of genetic testing. (HuGE Navigator) 23432027_Novel mutation (c.1272+1G>A) in BBS6 found in Tunisian families with Bardet-Biedl syndrome. 23671934_Three uORFs (uMKKS0, uMKKS1 and uMKKS2) are reported, and they can repress the translation of the downstream MKKS ORF. uMKKS1 and uMKKS2 are highly conserved in mammals and they encode two different mitochondrial membrane proteins respectively. 23716571_Findings indicate that Bbs proteins play a central role in the regulation of the actin cytoskeleton and control the cilia length through alteration of RhoA levels. 24400638_we report here, for the first time, in Indian population, a novel, different profile of mutations in BBS genes (BBS3, BBS9, BBS10 and BBS2) compared to worldwide (BBS1 and 10) reports. 26900326_We identified a novel H395R substitution in MKKS/BBS6 that results in a unique phenotype of only retinitis pigmentosa and polydactyly. 28624958_found compound heterozygous variants (c.1192C>T, p.Q398* and c.1175C>T, p.T392M) in MKKS in both the siblings, and these were likely to be pathogenic variants 28761321_Two novel mutations and three previously reported variants, identified in the present study, further extend the body of evidence implicating BBS6, BBS7, BBS8, and BBS10 in causing Bardet-Biedl Syndrome. 29232001_Novel sequence variants in the MKKS gene cause Bardet-Biedl syndrome in two consanguineous families with intra- and inter-familial variable phenotypes. 31989739_Novel mutation in MKKS/BBS6 linked with arRP and polydactyly in a family of North Indian origin. 33741323_Apparent but unconfirmed digenism in an Iranian consanguineous family with syndromic Retinal Disease. | ENSMUSG00000027274 | Mkks | 5.539173e+02 | 0.7459933 | -0.422765331 | 0.3507071 | 1.473904e+00 | 0.2247307957 | 0.78763590 | No | Yes | 4.580248e+02 | 74.935464 | 6.303121e+02 | 105.528611 | |
| ENSG00000125864 | 631 | BFSP1 | protein_coding | Q12934 | FUNCTION: Required for the correct formation of lens intermediate filaments as part of a complex composed of BFSP1, BFSP2 and CRYAA (PubMed:28935373). Involved in altering the calcium regulation of MIP water permeability (PubMed:30790544). {ECO:0000269|PubMed:28935373, ECO:0000269|PubMed:30790544}. | Acetylation;Alternative splicing;Cataract;Cell membrane;Coiled coil;Cytoplasm;Cytoskeleton;Eye lens protein;Intermediate filament;Lipoprotein;Membrane;Myristate;Phosphoprotein;Reference proteome;Repeat | This gene encodes a lens-specific intermediate filament-like protein named filensin. The encoded protein is expressed in lens fiber cells after differentiation has begun. This protein functions as a component of the beaded filament which is a cytoskeletal structure found in lens fiber cells. Mutations in this gene are the cause of autosomal recessive cortical juvenile-onset cataract. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:631; | cell cortex [GO:0005938]; cytoplasm [GO:0005737]; intermediate filament [GO:0005882]; plasma membrane [GO:0005886]; structural constituent of cytoskeleton [GO:0005200]; structural constituent of eye lens [GO:0005212]; cell maturation [GO:0048469]; intermediate filament organization [GO:0045109]; lens fiber cell development [GO:0070307] | 17225135_This is the first report of a mutation in the BFSP1 gene associated with human inherited cataracts. 24319337_The crystallin beta cluster on chromosome 22, GJA3, and BFSP1 play a major role in maintaining lens transparency. 24379646_A novel mutation (c.1042G>A) at exon 7 of BFSP1, which creates a substitution of an aspartate to an asparagine (p.D348N) was identified to be associated with autosomal dominant congenital cataract in a Chinese family. 28935373_The results suggest that the N-terminal domain of CRYAA is required during in vitro complex formation with filensin and phakinin. | ENSMUSG00000027420 | Bfsp1 | 1.463621e+02 | 0.8071130 | -0.309157393 | 0.3511626 | 7.823507e-01 | 0.3764231672 | 0.83999158 | No | Yes | 1.402876e+02 | 18.714901 | 1.789112e+02 | 24.577284 | |
| ENSG00000125871 | 92667 | MGME1 | protein_coding | Q9BQP7 | FUNCTION: Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance. Has preference for 5'-3' exonuclease activity but is also capable of endoduclease activity on linear substrates. Necessary for maintenance of proper 7S DNA levels. Probably involved in mitochondrial DNA (mtDNA) repair, possibly via the processing of displaced DNA containing Okazaki fragments during RNA-primed DNA synthesis on the lagging strand or via processing of DNA flaps during long-patch base excision repair. Specifically binds 5-hydroxymethylcytosine (5hmC)-containing DNA in stem cells. {ECO:0000255|HAMAP-Rule:MF_03030, ECO:0000269|PubMed:23313956, ECO:0000269|PubMed:23358826}. | 3D-structure;DNA damage;DNA repair;Disease variant;Exonuclease;Hydrolase;Mitochondrion;Nuclease;Phosphoprotein;Primary mitochondrial disease;Progressive external ophthalmoplegia;Reference proteome | The protein encoded by this gene is a nuclear-encoded mitochondrial protein necessary for the maintenance of mitochondrial genome synthesis. The encoded protein is a RecB-type exonuclease and primarily cleaves single-stranded DNA. Defects in this gene have been associated with mitochondrial DNA depletion syndrome-11. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2015]. | hsa:92667; | mitochondrion [GO:0005739]; single-stranded DNA exodeoxyribonuclease activity [GO:0008297]; mitochondrial DNA repair [GO:0043504]; mitochondrial DNA replication [GO:0006264]; mitochondrial genome maintenance [GO:0000002] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 23313956_Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. 23358826_Overexperssion of Ddk1 decreases the levels of 7S DNA, suggesting an important role of the protein in 7S DNA regulation. 24986917_MGME1-mediated mtDNA processing is essential for faithful mitochondrial genome replication and might be required for intramolecular recombination of mtDNA. 27220468_MGME1 processes flaps into ligatable nicks in concert with DNA polymerase gamma during mtDNA replication. 28711739_A novel frameshift deletion in MGME1 causes early onset cerebellar ataxia. 29572490_MGME1 is part of a termination complex acting at the end of the D-loop region where it modulates mtDNA replication and H-strand transcription termination 30247721_Besides the conserved two-cation-assisted catalytic mechanism, structural analysis of HsMGME1 and comparison with homologous proteins also clarified substrate binding and cleavage directionalities of the DNA double-strand break repair complexes RecBCD and AddAB. | ENSMUSG00000027424 | Mgme1 | 9.737199e+02 | 0.9277184 | -0.108241193 | 0.3222106 | 1.119485e-01 | 0.7379365793 | 0.94494829 | No | Yes | 9.951559e+02 | 172.077912 | 9.554502e+02 | 169.453570 | |
| ENSG00000126012 | 8242 | KDM5C | protein_coding | P41229 | FUNCTION: Histone demethylase that specifically demethylates 'Lys-4' of histone H3, thereby playing a central role in histone code (PubMed:28262558). Does not demethylate histone H3 'Lys-9', H3 'Lys-27', H3 'Lys-36', H3 'Lys-79' or H4 'Lys-20'. Demethylates trimethylated and dimethylated but not monomethylated H3 'Lys-4'. Participates in transcriptional repression of neuronal genes by recruiting histone deacetylases and REST at neuron-restrictive silencer elements. Represses the CLOCK-ARNTL/BMAL1 heterodimer-mediated transcriptional activation of the core clock component PER2 (By similarity). {ECO:0000250|UniProtKB:P41230, ECO:0000269|PubMed:17320160, ECO:0000269|PubMed:17320161, ECO:0000269|PubMed:17468742, ECO:0000269|PubMed:26645689, ECO:0000269|PubMed:28262558}. | 3D-structure;Alternative splicing;Biological rhythms;Chromatin regulator;Dioxygenase;Disease variant;Iron;Isopeptide bond;Mental retardation;Metal-binding;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene is a member of the SMCY homolog family and encodes a protein with one ARID domain, one JmjC domain, one JmjN domain and two PHD-type zinc fingers. The DNA-binding motifs suggest this protein is involved in the regulation of transcription and chromatin remodeling. Mutations in this gene have been associated with X-linked cognitive disability. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2009]. | hsa:8242; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; histone demethylase activity [GO:0032452]; histone H3-methyl-lysine-4 demethylase activity [GO:0032453]; histone H3-tri/di/monomethyl-lysine-4 demethylase activity [GO:0034647]; zinc ion binding [GO:0008270]; chromatin remodeling [GO:0006338]; histone H3-K4 demethylation [GO:0034720]; negative regulation of transcription, DNA-templated [GO:0045892]; response to toxic substance [GO:0009636]; rhythmic process [GO:0048511] | 16538222_JARID1C may have a role in X-Linked Mental Retardation 16541399_JARID1C appears to be one of the more frequently mutated genes in X-linked mental retardation. 17320160_We show that the X-linked mental retardation gene SMCX, which encodes a JmjC-domain protein, reversed histone H3 lysine 4 to di- and mono- but not unmethylated products. 17468742_has H3K4 tri-demethylase activity; functions as transcriptional repressor; loss of JARID1C/SMCX activity impairs REST-mediated neuronal gene regulation, thereby contributing to SMCX-associated X-linked mental retardation 18078810_Thus, SMCX is a novel Smad3 corepressor that may antagonize the tumor suppressing activity of the TGF-beta/Smad3 signaling pathway and thereby contribute to tumorigenesis. 18697827_male patients with mental retardation, short stature and hyperreflexia should be considered candidates for mutations in the JARID1C gene. 19346402_human NOT4 can polyubiquitinate human JARID1C/SMCX, a homolog of Jhd2, suggesting that this is likely a conserved mechanism 19826449_The two novel changes impair JARID1C protein function and are disease-causing mutations in the families reported. 20054297_identification of inactivating mutations in two genes--SETD2, a histone H3 lysine 36 methyltransferase, and JARID1C, a histone H3 lysine 4 demethylase--as well as mutations in the histone H3 lysine 27 demethylase, UTX in clear cell renal cell carcinoma 20538609_the JmjN domain of Jhd2 is important for its protein stability, and the plant homeodomain (PHD) finger mediates its chromatin association independent of H3K4 methylation 21575681_we describe clinical and genetic findings of a Brazilian family co-segregating a novel nonsense mutation (c.2172C>A) in exon 15 of KDM5C gene 21725364_Data show that von Hippel-Lindau (VHL) inactivation decreases H3K4Me3 levels through JARID1C, which alters gene expression and suppresses tumor growth. 22326837_Herein we present a large family with X Linked Intellectual Disability caused by a novel mutation c.2T > C in the start codon of the KDM5C gene 22611640_The KDM5C mutation carriers had higher mean scores on the abstract/visual and quantitative sections of the Stanford-Binet Intelligence Scale: Fourth Edition and lower mean short term memory scores. 23246292_We established that ARX polyA alterations damage the regulation of KDM5C expression. 23356856_DNA methylation at these three genes in blood correlated with dosage of KDM5C. 24029645_Mutation frequencies among CT images of clear cell RCCs were as follows: KDM5C, 6.9% (16 of 233). 24583395_Results indicate a KDM5C pathogenic mutational frequency of 0.7% among males with probable X-linked intellectual disability (XLID). 25016185_KDM5C is functionally involved in proliferation control of prostate cancer cells 25222147_These findings suggest that E2 recruits histone-modifying cellular proteins to the HPV LCR, resulting in transcriptional repression of E6 and E7. 25666439_Mutations in KDM5C gene in patients are described and their effect on gene expression, stability and catalytic activity is examined. Patient fibroblasts do not show global changes in histone methylation but several up-regulated genes were identified. 25712104_Data indicate the role for histone demethylase KDM5C/JARID1C in a specific phase of DNA replication in mammalian cells, through its demethylase activity on histone H3K4me3. 26059843_the extent of the duplicated regions in each case encompassing a minimum of three known disease genes TSPYL2, KDM5C and IQSEC2, is reported. 26182878_BRMS1 expression in human breast cancer is negatively correlated with JARID1C expression. Our results, for the first time, portray a pivotal role of JARID1C in regulating metastatic behaviors of breast cancer cells 26503415_Expression of KDM5C in hepatocellular carcinoma tumor cells promoted cell migration and tumor invasion. 26551685_these data suggest that inactivation of JARID1C in renal cancer leads to heterochromatin disruption, genomic rearrangement, and aggressive ccRCCs. 26580603_study of non-synonymous mutations in the KDM5C ARID domain and evaluates effects of 2 syndromic Claes-Jensen-type disease-associated missense mutations (A77T and D87G) and 3 non-classified missense mutations (R108W, N142S, and R179H); analysis indicates, among the non-classified mutations, R108W is possibly a disease-associated mutation, and N142S and R179H are probably harmless 26621457_KDM5C is overexpressed in breast cancer cells and miR-138 regulates its expression. 26645689_Characterization of a Linked Jumonji Domain of the KDM5/JARID1 Family of Histone H3 Lysine 4 Demethylases. 26858085_KDM5C expression was generally lower in cancer lesions compared with matched nontumor tissues. KDM5C has a vital function in inhibiting cell mobility, which is at least partially controlled by the p53. 26919706_The two mutations are present on the same maternal haplotype, suggesting that a postzygotic somatic mutation or a reversion error occurred at an early embryonic stage in the mother, leading to switched KDM5C mutations in the affected siblings. 27058665_Findings reveal a RACK7/KDM5C-regulated, dynamic interchange between histone H3K4me1 and H3K4me3 at active enhancers, representing an additional layer of regulation of enhancer activity. Authors propose that RACK7/KDM5C functions as an enhancer 'brake' to ensure appropriate enhancer activity, which, when compromised, could contribute to tumorigenesis. 27211531_Results suggest that KDM5C mutations predispose to X-linked intellectual disability which accounts for 1-4% of cases in male patient. 27696497_The predicted structure of KDM5C was used to investigate the effects of disease-causing mutations, and it was shown that the mutations alter domain stability and inter-domain interactions. 27869828_Mutation in KDM5C gene is associated with cancer more frequently in males. 27896428_In vitro models indicated that KDM5C suppressed miR-320a transcription by directly binding to the promoter of miR-320a to prevent histone methylation. KITLG, an essential gene for ovarian development and primordial germ cell survival, was a direct target of miR-320a and that it was downregulated in 45,X fetal gonadal tissues. 28408295_SETD2 and KDM5C mutations were associated with prolonged overall survival in patients with metastatic clear cell renal cell carcinoma 29339538_HPV16 E6 protein interacts with histone demethylase KDM5C in ovarian cancer.HPV16 E6 protein destabilizes the KDM5C.KDM5C regulates cervical cancer cell EGFR and c-MET expression by modulating their super-enhancer H3K4 methylation dynamics. 30257334_In colon cancer cells, KDM5c might downregulate ABCC1 expression by demethylating the ABCC1 H3K4me3 in the TSS region, which can promote multidrug resistance 30921702_The BRD4-KDM5C-PTEN may be a new oncogenic pathway in CRPC development. 31221981_This article reviewed the most recent findings regarding cancer-specific metabolic reprogramming and the tumor-suppressive roles of IDH1/2, JARID1C/KDM5C and UTX/KDM6A. [review] 31691806_Studies indicate that KDM5C is a conserved and druggable effector molecule across a number of NDDs for whom the use of SAHA may be considered a potential therapeutic strategy. 32279304_Further delineation of the female phenotype with KDM5C disease causing variants: 19 new individuals and review of the literature. 32392963_[Two cases of X-linked mental retardation, Claes-Jensen syndrome caused by variation of KDM5C gene]. 32732223_X- and Y-Linked Chromatin-Modifying Genes as Regulators of Sex-Specific Cancer Incidence and Prognosis. 33306820_[Clinical features and gene variant of a pedigree affected with X-linked recessive mental retardation Claes-Jensen type]. 33400184_Relationship between visceral adipose tissue and genetic mutations (VHL and KDM5C) in clear cell renal cell carcinoma. 33953726_Predictive Value of KDM5C Alterations for Immune Checkpoint Inhibitors Treatment Outcomes in Patients With Cancer. 33977073_The Dual Function of KDM5C in Both Gene Transcriptional Activation and Repression Promotes Breast Cancer Cell Growth and Tumorigenesis. 34089235_Caregiver-reported characteristics of children diagnosed with pathogenic variants in KDM5C. 34356104_Analysis of a Set of KDM5C Regulatory Genes Mutated in Neurodevelopmental Disorders Identifies Temporal Coexpression Brain Signatures. 34522206_Deficiency of the X-inactivation escaping gene KDM5C in clear cell renal cell carcinoma promotes tumorigenicity by reprogramming glycogen metabolism and inhibiting ferroptosis. 34928233_HOXD3 Up-regulating KDM5C Promotes Malignant Progression of Diffuse Large B-Cell Lymphoma by Decreasing p53 Expression 35080113_Lysine demethylase 5C epigenetically reduces transcription of ITIH1 that results in augmented progression of liver hepatocellular carcinoma. 35331081_Lysine (K)-specific demethylase 5C regulates the incidence of severe preeclampsia by adjusting the expression of bone morphogenetic protein-7. | ENSMUSG00000025332 | Kdm5c | 1.375840e+04 | 1.2493277 | 0.321151995 | 0.2807482 | 1.307594e+00 | 0.2528307679 | 0.79332654 | No | Yes | 1.431027e+04 | 1374.542953 | 1.082413e+04 | 1066.581951 | |
| ENSG00000126246 | 79713 | IGFLR1 | protein_coding | Q9H665 | FUNCTION: Probable cell membrane receptor for the IGF-like family proteins. Binds IGFL1 and IGFL3 with a higher affinity. May also bind IGFL2. {ECO:0000269|PubMed:21454693}. | Alternative splicing;Cell membrane;Membrane;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:79713; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886] | 21454693_Murine insulin growth factor-like (IGFL) and human IGFL1 proteins are induced in inflammatory skin conditions and bind to a novel tumor necrosis factor receptor family member, IGFLR1. | ENSMUSG00000036826 | Igflr1 | 3.763929e+02 | 1.7297519 | 0.790565092 | 0.3633691 | 4.639173e+00 | 0.0312500618 | 0.58378765 | No | Yes | 4.096653e+02 | 62.111947 | 2.296074e+02 | 36.301465 | ||
| ENSG00000126705 | 27245 | AHDC1 | protein_coding | Q5TGY3 | Acetylation;DNA-binding;Isopeptide bond;Mental retardation;Methylation;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | This gene encodes a protein containing two AT-hooks, which likely function in DNA binding. Mutations in this gene were found in individuals with Xia-Gibbs syndrome. [provided by RefSeq, Jun 2014]. | hsa:27245; | DNA binding [GO:0003677] | 24791903_this study hasidentified AHDC1 de novo truncating mutations that most likely cause syndromic expressive language delay, hypotonia, and sleep apnea. 30615951_Microdeletion and microduplication of 1p36.11p35.3 involving AHDC1 contribute to neurodevelopmental disorder. 30729726_De novo heterozygous variants in AHDC1 gene were identified in two patients with partial growth hormone deficiency. 31390932_we unveiled that LINC01133 may function as a ceRNA for miR-4784 to advance AHDC1 expression, intensifying CC cell malignant phenotypes and EMT process, which may demonstrate the implied value of LINC01133 as a therapeutic target for CC patients. 31737670_Three rare mutations of AHDC1 in patients with OSA in Chinese Hanindividuals. 33644933_Phenotypic and protein localization heterogeneity associated with AHDC1 pathogenic protein-truncating alleles in Xia-Gibbs syndrome. 34229113_Phenotypic heterogeneity and mosaicism in Xia-Gibbs syndrome: Five Danish patients with novel variants in AHDC1. | ENSMUSG00000037692 | Ahdc1 | 6.862872e+02 | 1.4544230 | 0.540446904 | 0.2894347 | 3.471404e+00 | 0.0624382828 | 0.69812420 | No | Yes | 7.192701e+02 | 104.540416 | 5.200911e+02 | 78.012004 | ||
| ENSG00000126883 | 8021 | NUP214 | protein_coding | P35658 | FUNCTION: Part of the nuclear pore complex (PubMed:9049309). Has a critical role in nucleocytoplasmic transport (PubMed:31178128). May serve as a docking site in the receptor-mediated import of substrates across the nuclear pore complex (PubMed:31178128, PubMed:8108440). {ECO:0000269|PubMed:31178128, ECO:0000269|PubMed:9049309, ECO:0000303|PubMed:8108440}.; FUNCTION: (Microbial infection) Required for capsid disassembly of the human adenovirus 5 (HadV-5) leading to release of the viral genome to the nucleus (in vitro). {ECO:0000269|PubMed:25410864}. | 3D-structure;Acetylation;Alternative splicing;Chromosomal rearrangement;Coiled coil;Disease variant;Glycoprotein;Host-virus interaction;Isopeptide bond;Nuclear pore complex;Nucleus;Phosphoprotein;Protein transport;Proto-oncogene;Reference proteome;Repeat;Translocation;Transport;Ubl conjugation;mRNA transport | The nuclear pore complex is a massive structure that extends across the nuclear envelope, forming a gateway that regulates the flow of macromolecules between the nucleus and the cytoplasm. Nucleoporins are the main components of the nuclear pore complex in eukaryotic cells. This gene is a member of the FG-repeat-containing nucleoporins. The protein encoded by this gene is localized to the cytoplasmic face of the nuclear pore complex where it is required for proper cell cycle progression and nucleocytoplasmic transport. The 3' portion of this gene forms a fusion gene with the DEK gene on chromosome 6 in a t(6,9) translocation associated with acute myeloid leukemia and myelodysplastic syndrome. Alternative splicing of this gene results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Dec 2015]. | hsa:8021; | cytoplasmic side of nuclear pore [GO:1990876]; cytosol [GO:0005829]; nuclear inner membrane [GO:0005637]; nuclear outer membrane [GO:0005640]; nuclear pore [GO:0005643]; nucleoplasm [GO:0005654]; nuclear export signal receptor activity [GO:0005049]; nuclear localization sequence binding [GO:0008139]; structural constituent of nuclear pore [GO:0017056]; mitotic cell cycle [GO:0000278]; mRNA export from nucleus [GO:0006406]; protein export from nucleus [GO:0006611]; protein import into nucleus [GO:0006606]; regulation of cell cycle [GO:0051726]; regulation of nucleocytoplasmic transport [GO:0046822]; RNA export from nucleus [GO:0006405] | 12191473_Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. 12917407_A surface hydrophobic corridor within the MH2 domain of Smad3 is critical for association with CAN/Nup214 and nuclear import, whereas Smad4 interaction with CAN/Nup214, and nuclear import requires structural elements present only in the full-length Smad4 14671643_leukemia SET-CAN fusion protein inhibited proliferation of U937 cells, most likely by interfering with hCRM1, but it also partially blocks differentiation 14766228_Nup214 has a role in regulating TTP localization 15361874_NUP214-ABL1 expression defines a new subgroup of individuals with T-ALL 16045929_Nup214 domain-specific localization by immunoelectron microscopy. Transport studies show that the spatial distribution of the FG-repeat domains of both Nup153 and Nup214 shifts in a transport-dependent manner. 16675447_the Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export 16943420_The Nup214/Nup88 complex is required for efficient CRM1-mediated transport, supporting a model involving a high-affinity binding site for CRM1 at Nup214 in the terminal steps of export. 17264208_A mechanism is proposed by which the C-terminal peptide extension is involved in nuclear pore complex assembly. 17296573_TAF-Ialpha/CAN and TAF-Ibeta/CAN fusion transcripts are generated by chromosome 9q34 deletion in acute myeloid leukemia 17620317_Impairment of erythroid and megakaryocytic differentiation by a leukemia-associated and t(9;9)-derived product, SET/TAF-IBeta-CAN/Nup214. 18096310_Set/TAF-Ibeta acts as a ligand-activated GR-responsive transcriptional repressor, while Set-Can does not retain physiologic responsiveness to ligand-bound glucocorticoid receptor. 18299449_SET-NUP214 binds in the promoter regions of specific HOXA genes, where it interacts with CRM1 and DOT1L, which may transcriptionally activate specific members of the HOXA cluster. 18614052_NUP214-ABL1 fusion is unique because of its requisite localization to the nuclear pore complex for its transforming potential. 18784740_a thorough biochemical comparative analysis of NUP214-ABL1 and BCR-ABL1 show that, despite their common tyrosine kinase domain, the two fusion proteins differ in many critical catalytic properties 18923437_FISH revealed a heterogeneous pattern of NUP214-ABL1 amplification. NUP214-ABL1 gene requires amplification for oncogenicity; it is part of a multistep process of leukemogenesis; and it can be a late event present only in subpopulations 19166587_Cell lines LOUCY and MEGAL express the recently described SET-NUP214 fusion gene. 19219046_Using in vitro assays, the authors demonstrate that NUP214 decreases both the RNA binding and ATPase activities of DBP5. 19229862_These results indicate that CAN/Nup214 interacts with VDR and modulates its function as a transcription factor. 19386703_CAN/Nup214 is a nuclear receptor for the herpesvirus capsid. 19828735_Oxidative stress up-regulated the binding of Crm1 to Ran and affected multiple repeat-containing nucleoporins by changing their localization, phosphorylation, O-glycosylation, or interaction with other transport components. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20970119_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21489623_NUP214-ABL1 positive T-cell acute lymphoblastic leukemia patient shows an initial favorable response to imatinib therapy post relapse 21691751_Used MALDI-TOF MS 40-plex assay for testing 40 loci within 21 high-ranking breast cancer CAN-genes. No mutation could be found in 6 cell lines. A single breast cancer tissue sample showed heterozygosity at locus c.5834G>A within the ZFYVE26 gene. 21720744_Data describe the relatively high incidence of SET-NUP214 rearrangement in adult T-ALLs, and demonstrate comprehensive clinical, phenotypic, and genetic characteristics of this entity. 22558357_Nucleoporin p62 (NUP62) and nucleoporin 214 (NUP214) are differentially distributed between nuclear pore complexes. 23264634_Several phenylalanine-glycine motives in the nucleoporin Nup214 are essential for binding of the nuclear export receptor CRM1. 23831628_The expression of endogenous Nup214 is significantly down-regulated by the reverse inserted lentiviral promoter 23872305_NUP214-ABL1-mediated cell proliferation in T-cell acute lymphoblastic leukemia is dependent on the LCK kinase and various interacting proteins. 24073922_the expression of the fusion gene DEK-NUP214 leads to increased cellular proliferation. We show that this is dependent on upregulation of the signal transduction protein mTOR with subsequent effects on protein synthesis and glucose metabolism. 24441146_t(6;9)/DEK-NUP214 represents a unique subtype of acute myeloid leukemia with a high risk of relapse. 24449214_When compared with SET-NUP214-negative patients, SET-NUP214-positive patients showed a significantly higher rate of corticosteroid resistance (91% vs 44%; P = .003) and chemotherapy resistance (100% vs 44%; P = .0001). 25410864_Both in vitro hexon binding and in vivo nuclear import of the adenovirus genome were strongly reduced in Nup214-depleted cells suggesting that Nup214 is a major binding site of adenovirus during infection. 25743594_We have identified NUP214, a member of the massive nuclear pore complex, as a novel miR-133b target. 27613868_SQSTM1-Nup214, although mostly cytoplasmic, also forms nuclear bodies and inhibits nuclear protein but not poly(A)(+) RNA export. 29109093_RNA-sequencing proved to be a valuable tool for the detection of a fusion of genes DEK and NUP214 in a leukemia that showed cryptic cytogenetic rearrangement of chromosome band 9q34. 29483668_19 patients with neurodevelopmental disorders harboring a rare deletion inherited from a healthy parent were investigated by whole-exome sequencing to search for SNV on the contralateral segment. This strategy allowed us to identify a candidate variant in two patients in the NUP214 and NCOR1 genes. 30107177_oncogenic kinase NUP214-ABL1, through its downstream effector STAT5, directly cooperates with TLX1 at the transcriptional level. 30758658_we propose that while NUP214 complete deficiency may be lethal in humans, partial deficiency results in a novel autosomal recessive disorder characterized by severe encephalopathy and early death. 31178128_Pathogenic Variants in NUP214 Cause 'Plugged' Nuclear Pore Channels and Acute Febrile Encephalopathy. 31186352_Loss of Nup214 inhibited nuclear export of recombination signal-binding protein for immunoglobulin kappaJ region (RBP-J), the DNA-binding component of the Notch pathway. NUP214 fusion proteins, causative for certain cases of T-cell acute lymphatic leukemia, potentially contribute to tumorigenesis via a Notch-dependent mechanism. 31755865_these findings indicate that CRM1 acts as a key molecule that connects leukemogenic proteins to aberrant HOX gene regulation either via nucleoporin-CRM1 interaction (for SET-Nup214) or NES-CRM1 interaction (for NPM1c). 34320268_SET-NUP214 and MLL cooperatively regulate the promoter activity of the HoxA10 gene. 34551474_[Prognostic significance of DEK-NUP214 fusion gene in patients with acute myeloid leukemia after allogeneic hematopoietic stem cell transplantation]. 34753234_[SET-NUP214-positive pediatric acute myeloid leukemia: a report of two cases]. | ENSMUSG00000001855 | Nup214 | 5.759028e+03 | 1.0952895 | 0.131312236 | 0.2779609 | 2.213211e-01 | 0.6380351292 | 0.91967810 | No | Yes | 5.648262e+03 | 446.223265 | 4.925001e+03 | 399.446939 | |
| ENSG00000126953 | 1678 | TIMM8A | protein_coding | O60220 | FUNCTION: Mitochondrial intermembrane chaperone that participates in the import and insertion of some multi-pass transmembrane proteins into the mitochondrial inner membrane. Also required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space. The TIMM8-TIMM13 complex mediates the import of proteins such as TIMM23, SLC25A12/ARALAR1 and SLC25A13/ARALAR2, while the predominant TIMM9-TIMM10 70 kDa complex mediates the import of much more proteins. Probably necessary for normal neurologic development. {ECO:0000269|PubMed:11489896, ECO:0000269|PubMed:15254020}. | Chaperone;Deafness;Disease variant;Disulfide bond;Dystonia;Membrane;Metal-binding;Mitochondrion;Mitochondrion inner membrane;Phosphoprotein;Protein transport;Reference proteome;Translocation;Transport;Zinc | This translocase is involved in the import and insertion of hydrophobic membrane proteins from the cytoplasm into the mitochondrial inner membrane. The gene is mutated in Mohr-Tranebjaerg syndrome/Deafness Dystonia Syndrome (MTS/DDS) and it is postulated that MTS/DDS is a mitochondrial disease caused by a defective mitochondrial protein import system. Defects in this gene also cause Jensen syndrome; an X-linked disease with opticoacoustic nerve atrophy and muscle weakness. This protein, along with TIMM13, forms a 70 kDa heterohexamer. Alternative splicing results in multiple transcript variants encoding distinct isoforms.[provided by RefSeq, Mar 2009]. | hsa:1678; | mitochondrial inner membrane [GO:0005743]; mitochondrial intermembrane space [GO:0005758]; mitochondrial intermembrane space protein transporter complex [GO:0042719]; mitochondrion [GO:0005739]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; nervous system development [GO:0007399]; protein insertion into mitochondrial inner membrane [GO:0045039] | 12745081_Interaction of TIMM8a with the signal transduction adaptor molecule STAM1. 15710860_Intronic mutations in the DDP1 gene can also cause X-linked dystonia-deafness syndrome. 16332536_Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. 16411215_Mutation in TIMM8a is associated with deafness-dystonia (Mohr-Tranebjaerg) syndrome 17534980_A sporadic 42-year-old man with MTS presenting with postlingual deafness, adult-onset progressive dystonia with marked arm tremor, mild spasticity of the legs, and visual disturbance due to a novel mutation in the DDP1 gene. 17999202_mRNA expression demonstrate increased TIMM8A mRNA levels in cultured fibroblasts from a patient with Mohr-Tranebjaerg Syndrome. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21984432_knockdown of the TIMM8A gene by RNA interference did not show an influence on the oxygen respiration rate and the mitochondrial membrane potentia 23418071_The results of this study demonistrated that the syndrome of deafness-dystonia is cause by mutation of Timm8a. 31682224_The authors show that human Tim8a is required for the assembly of Complex IV in neurons, which is mediated through a transient interaction with Complex IV assembly factors, in particular the copper chaperone COX17. 32820032_Frameshift mutation of Timm8a1 gene in mouse leads to an abnormal mitochondrial structure in the brain, correlating with hearing and memory impairment. | ENSMUSG00000048007 | Timm8a1 | 9.329738e+02 | 0.7348374 | -0.444502965 | 0.2798174 | 2.488288e+00 | 0.1146964338 | 0.75783482 | No | Yes | 9.095470e+02 | 97.123120 | 1.057949e+03 | 115.648523 | |
| ENSG00000127419 | 84286 | TMEM175 | protein_coding | Q9BSA9 | FUNCTION: Organelle-specific potassium channel specifically responsible for potassium conductance in endosomes and lysosomes (PubMed:26317472, PubMed:28723891, PubMed:32267231, PubMed:33505021, PubMed:32228865). Forms a potassium-permeable leak-like channel, which regulates lumenal pH stability and is required for autophagosome-lysosome fusion (PubMed:26317472, PubMed:28723891, PubMed:32267231, PubMed:33505021, PubMed:32228865). Constitutes the major lysosomal potassium channel (PubMed:26317472, PubMed:28723891). Constitutes the pore-forming subunit of the lysoK(GF) complex, a complex activated by extracellular growth factors (PubMed:33505021). The lysoK(GF) complex is composed of TMEM175 and AKT (AKT1, AKT2 or AKT3), a major target of growth factor receptors: in the complex, TMEM175 channel is opened by conformational changes by AKT, leading to its activation (PubMed:33505021). The lysoK(GF) complex is required to protect neurons against stress-induced damage (PubMed:33505021). {ECO:0000269|PubMed:26317472, ECO:0000269|PubMed:28723891, ECO:0000269|PubMed:32228865, ECO:0000269|PubMed:32267231, ECO:0000269|PubMed:33505021}. | 3D-structure;Alternative splicing;Endosome;Ion channel;Ion transport;Lysosome;Membrane;Neurodegeneration;Parkinson disease;Parkinsonism;Phosphoprotein;Potassium;Potassium channel;Potassium transport;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:84286; | endosome [GO:0005768]; integral component of endosome membrane [GO:0031303]; integral component of lysosomal membrane [GO:1905103]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; potassium channel activity [GO:0005267]; potassium ion leak channel activity [GO:0022841]; neuron cellular homeostasis [GO:0070050]; phagosome-lysosome fusion [GO:0090385]; potassium ion transmembrane transport [GO:0071805]; regulation of lysosomal lumen pH [GO:0035751] | 25914293_Data indicated that GBA and TMEM175/GAK significantly alter age at onset in PD. 28193887_TMEM175 deficiency impairs lysosomal and mitochondrial function and increases alpha-synuclein aggregation. 28723891_structure of TMEM175 represents a novel architecture of a tetrameric cation channel whose ion selectivity mechanism appears to be distinct from that of the classical K(+) channel family 31261387_Data suggest that the TMEM175 p.M393T variant is responsible for the main signal in the chromosome 4p16.3 locus therefore conferring risk for Parkinson disease through its phosphorylation of alpha-synuclein. 31658403_Coding variants in TMEM175 are likely to be responsible for the association in the TMEM175/GAK/DGKQ locus, which could be mediated by affecting glucosylceramidase activity. 32228865_Gating and selectivity mechanisms for the lysosomal K(+) channel TMEM175. 33505021_A growth-factor-activated lysosomal K(+) channel regulates Parkinson's pathology. | ENSMUSG00000013495 | Tmem175 | 1.454151e+03 | 1.0772446 | 0.107345899 | 0.2728283 | 1.553124e-01 | 0.6935094536 | 0.93437240 | No | Yes | 1.181771e+03 | 128.151761 | 1.162509e+03 | 129.489999 | ||
| ENSG00000127483 | 50809 | HP1BP3 | protein_coding | Q5SSJ5 | FUNCTION: Component of heterochromatin that maintains heterochromatin integrity during G1/S progression and regulates the duration of G1 phase to critically influence cell proliferative capacity (PubMed:24830416). Mediates chromatin condensation during hypoxia, leading to increased tumor cell viability, radio-resistance, chemo-resistance and self-renewal(PubMed:25100860). {ECO:0000269|PubMed:24830416, ECO:0000269|PubMed:25100860}. | 3D-structure;Acetylation;Alternative splicing;Chromosome;DNA-binding;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | hsa:50809; | chromosome [GO:0005694]; nuclear speck [GO:0016607]; nucleosome [GO:0000786]; nucleus [GO:0005634]; DNA binding [GO:0003677]; nucleosome binding [GO:0031491]; cellular response to hypoxia [GO:0071456]; heterochromatin organization [GO:0070828]; nucleosome assembly [GO:0006334]; regulation of cell population proliferation [GO:0042127]; regulation of nucleus size [GO:0097298]; regulation of transcription, DNA-templated [GO:0006355] | 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 20042602_HP1-BP74 directly binds to HP1, and its middle region associates with linker DNA at the entry/exit site of nucleosomal DNA in vitro 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 24317310_found two biomarker loci at HP1BP3 and TTC9B, which predicted postpartum depression 24830416_HP1BP3 protein maintains heterochromatin integrity during G1-S progression and regulates the duration of G1 phase to critically influence cell proliferative capacity. 25100860_Quantitative profiling of chromatome dynamics reveals an important role for HP1BP3 in hypoxia-induced oncogenesis. 25662603_HP1BP3 is a component of heterochromatin, and its recruitment is dependent on a functional interaction with HP1 proteins. 26402843_HP1BP3 regulates hepatic expression of IGF1 and its binding proteins in mice, thus modulating the endocrine IGF1 pathway. 26503311_DNA methylation at early antenatal time points associated with changes in estradiol and allopregnanolone and postpartum depression 27425409_HP1BP3 promotes co-transcriptional miRNA processing via chromatin retention of nascent pri-miRNA transcripts. 31819167_A fail-safe system to prevent oncogenesis by senescence is targeted by SV40 small T antigen. | ENSMUSG00000028759 | Hp1bp3 | 4.279588e+03 | 0.9442728 | -0.082724433 | 0.3126232 | 6.965052e-02 | 0.7918463042 | 0.95937374 | No | Yes | 3.146366e+03 | 489.973346 | 3.620824e+03 | 577.969886 | ||
| ENSG00000128000 | 163131 | ZNF780B | protein_coding | Q9Y6R6 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000250}. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:163131; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 1.969238e+02 | 0.4659967 | -1.101608315 | 0.3468866 | 9.801039e+00 | 0.0017441329 | 0.16179612 | No | Yes | 6.517108e+01 | 28.802547 | 1.695529e+02 | 76.574217 | |||||
| ENSG00000128245 | 7533 | YWHAH | protein_coding | Q04917 | FUNCTION: Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. Negatively regulates the kinase activity of PDPK1. {ECO:0000269|PubMed:12177059}. | 3D-structure;Acetylation;Direct protein sequencing;Phosphoprotein;Reference proteome | This gene product belongs to the 14-3-3 family of proteins which mediate signal transduction by binding to phosphoserine-containing proteins. This highly conserved protein family is found in both plants and mammals, and this protein is 99% identical to the mouse, rat and bovine orthologs. This gene contains a 7 bp repeat sequence in its 5' UTR, and changes in the number of this repeat have been associated with early-onset schizophrenia and psychotic bipolar disorder. [provided by RefSeq, Jun 2009]. | hsa:7533; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; intercalated disc [GO:0014704]; plasma membrane [GO:0005886]; actin binding [GO:0003779]; enzyme binding [GO:0019899]; glucocorticoid receptor binding [GO:0035259]; identical protein binding [GO:0042802]; insulin-like growth factor receptor binding [GO:0005159]; protein domain specific binding [GO:0019904]; protein heterodimerization activity [GO:0046982]; sodium channel regulator activity [GO:0017080]; transmembrane transporter binding [GO:0044325]; cellular protein localization [GO:0034613]; glucocorticoid catabolic process [GO:0006713]; glucocorticoid receptor signaling pathway [GO:0042921]; intracellular protein transport [GO:0006886]; membrane depolarization during action potential [GO:0086010]; negative regulation of dendrite morphogenesis [GO:0050774]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of neuron differentiation [GO:0045664]; regulation of sodium ion transmembrane transporter activity [GO:2000649]; regulation of sodium ion transport [GO:0002028]; regulation of synaptic plasticity [GO:0048167]; signal transduction [GO:0007165]; substantia nigra development [GO:0021762] | 11121172_Observational study of gene-disease association. (HuGE Navigator) 11996670_Results show that three 14-3-3 isoforms, beta, gamma and eta, are DAL-1/Protein 4.1B-binding proteins. 12176995_phosphorylation of 14-3-3 binding site of myeloid leukemia factor 1 by MADM 12480176_Data demonstrate that 14-3-3 proteins are colocalized with Lewy bodies in Parkinson disease, but there was no specific staining for the 14-3-3 eta subunit. 16459651_Observational study of gene-disease association. (HuGE Navigator) 16545136_gremlin 1 is overexpressed in human cancers and interacts with YWHAH protein 17611984_Detection of only 2 (14-3-3 eta and gamma) out of 7 different isoforms in synovial fluid suggests they are specific to the site of joint inflammation 18950845_Observational study of gene-disease association. (HuGE Navigator) 19034380_This protein has been found differentially expressed in the temporal lobe from patients with schizophrenia. 19160447_Observational study of gene-disease association. (HuGE Navigator) 19160447_The association of YWHAH with bipolar disorder in a large sample, consisting of 1211 subjects from 318 nuclear families including 554 affected offspring, was investigated. 19558434_14-3-3beta, 14-3-3gamma, 14-3-3epsilon, 14-3-3eta and 14-3-3theta isoforms interact with the GPIb-IX complex in platelets 20388496_The 14-3-3 eta, beta, gamma and sigma isoforms were negatively expressed in meningioma 21416292_Data indicate that gene analysis revealed an up-regulation of all four 14-3-3 isoforms beta, eta, gamma, and sigma. 21432775_relocation of p33ING1b from the nucleus to the cytoplasm, where the protein is tethered by 14-3-3eta, participates in tumorigenesis and progression in HNSCC 21739144_This is the first genetic association study of YWHAH with sporadic Creutzfeldt-Jakob disease populations. 22562251_14-3-3eta may be required for mitotic progression and may be considered as a novel anti-cancer strategy in combination with microtubule inhibitors. 24507884_This study identified YWHAH significantly associated loci with a biologically plausible role in schizophrenia. 24751211_Extracellular 14-3-3eta activates key signalling cascades and induces factors associated with the pathogenesis of rheumatoid arthritis 27256400_14-3-3 eta isoform colocalizes TDP-43 on the coarse granules in the anterior horn cells of patients with sporadic amyotrophic lateral sclerosis. 27717743_The impact of AKT1 on glucocorticoid receptor (GR)-induced transcriptional activity in cooperation with phospho-serine/threonine-binding protein 14-3-3, was examined. 29512774_Results show that YWHAZ amplification indicates a better survival trend in bladder cancer. Its knockdown promotes both in vitro and in vivo tumorigenesis in bladder cancer and may be a novel biomarker for bladder cancer. 29704241_14-3-3eta inhibits LDHA by direct interaction in the setting of complex I dysfunction, highlighting the role of 14-3-3eta overexpression and inefficient oxidative phosphorylation in oncocytoma mitochondrial biogenesis. 30022679_14-3-3eta protein: a promising biomarker for rheumatoid arthritis. 30143532_findings reveal that Akt activity determines the phosphorylation status of TBC1D7 at the phospho-switch Ser-124, which governs binding to either 14-3-3 or beta-TrCP2, resulting in increased or decreased stability of TBC1D7, respectively. 30358169_Study found that 14-3- 3zeta bond to HBx in HBV-positive hepatocellular carcinoma (HCC) cell lines. 14-3- 3zeta- silenced cancer cells displayed weakened migration and invasion, which was accompanied by decreased HBx expression. 30742689_Results uncover a novel function of 14-3-3eta to promote the MDA5-dependent IFNbeta induction pathway by reducing the immunostimulatory potential of viral dsRNA within MDA5 activation signaling pathway. 31333006_Serum 14-3-3eta protein levels in patients with rheumatoid arthritis were significantly higher as compared to healthy individuals 31478661_Proteomic and Interactome Approaches Reveal PAK4, PHB-2, and 14-3-3eta as Targets of Overactivated Cdc42 in Cellular Responses to Genomic Instability. 32080735_14-3-3eta Protein in Rheumatoid Arthritis: Promising Diagnostic Marker and Independent Risk Factor for Osteoporosis. 32676916_NACHO and 14-3-3 promote expression of distinct subunit stoichiometries of the alpha4beta2 acetylcholine receptor. 32807493_MiR-660-5p promotes the progression of hepatocellular carcinoma by interaction with YWHAH via PI3K/Akt signaling pathway. 33373444_Genomics Links Inflammation With Neurocognitive Impairment in Children Living With Human Immunodeficiency Virus Type-1. 33400046_The significance of serum 14-3-3eta level in rheumatoid arthritis patients. | ENSMUSG00000018965 | Ywhah | 3.807456e+03 | 0.8045687 | -0.313712488 | 0.2777866 | 1.306996e+00 | 0.2529392958 | 0.79350582 | No | Yes | 3.464165e+03 | 383.927104 | 3.874175e+03 | 440.105355 | |
| ENSG00000128607 | 23008 | KLHDC10 | protein_coding | Q6PID8 | FUNCTION: Substrate-recognition component of a Cul2-RING (CRL2) E3 ubiquitin-protein ligase complex of the DesCEND (destruction via C-end degrons) pathway, which recognizes a C-degron located at the extreme C terminus of target proteins, leading to their ubiquitination and degradation (PubMed:29779948). The C-degron recognized by the DesCEND pathway is usually a motif of less than ten residues and can be present in full-length proteins, truncated proteins or proteolytically cleaved forms (PubMed:29779948). The CRL2(KLHDC10) complex specifically recognizes proteins with a proline-glycine (Pro-Gly) at the C-terminus, leading to their ubiquitination and degradation (PubMed:29779948). Participates in the oxidative stress-induced cell death through MAP3K5 activation (PubMed:23102700). Inhibits PPP5C phosphatase activity on MAP3K5 (PubMed:23102700). Acts as a regulator of necroptosis (By similarity). {ECO:0000250|UniProtKB:Q6PAR0, ECO:0000269|PubMed:23102700, ECO:0000269|PubMed:29779948}. | Alternative splicing;Cytoplasm;Kelch repeat;Methylation;Nucleus;Reference proteome;Repeat;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:29779948}. | hsa:23008; | Cul2-RING ubiquitin ligase complex [GO:0031462]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; ubiquitin ligase-substrate adaptor activity [GO:1990756]; positive regulation of stress-activated MAPK cascade [GO:0032874]; ubiquitin-dependent protein catabolic process via the C-end degron rule pathway [GO:0140627] | 23102700_Suggest that Slim/KLHDC10 is an activator of ASK1, contributing to oxidative stress-induced cell death through the suppression of PP5. | ENSMUSG00000029775 | Klhdc10 | 1.686560e+03 | 0.6579338 | -0.603985565 | 0.2544857 | 5.406448e+00 | 0.0200624961 | 0.50208266 | No | Yes | 1.389745e+03 | 201.742718 | 1.864328e+03 | 277.008740 | |
| ENSG00000128641 | 4430 | MYO1B | protein_coding | O43795 | FUNCTION: Motor protein that may participate in process critical to neuronal development and function such as cell migration, neurite outgrowth and vesicular transport. {ECO:0000250}. | ATP-binding;Actin-binding;Alternative splicing;Calmodulin-binding;Isopeptide bond;Motor protein;Myosin;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | hsa:4430; | actin cytoskeleton [GO:0015629]; actin filament [GO:0005884]; apical part of cell [GO:0045177]; brush border [GO:0005903]; cell periphery [GO:0071944]; cytoplasm [GO:0005737]; early endosome [GO:0005769]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; filopodium [GO:0030175]; microvillus [GO:0005902]; myosin complex [GO:0016459]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; vesicle [GO:0031982]; actin filament binding [GO:0051015]; ATP binding [GO:0005524]; cadherin binding [GO:0045296]; calmodulin binding [GO:0005516]; microfilament motor activity [GO:0000146]; phosphatidylinositol-3,4,5-trisphosphate binding [GO:0005547]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; actin filament bundle assembly [GO:0051017]; actin filament organization [GO:0007015]; actin filament-based movement [GO:0030048]; post-Golgi vesicle-mediated transport [GO:0006892]; vesicle transport along actin filament [GO:0030050] | 12486594_At apical surfaces of supporting cells that surround hair cells in auditory epithelia of postnatal rats. In vestibular epithelia, present in ring within apical pole of hair cell. Expression was prominent only immediately after birth. 16219689_observations suggest that myosin 1b controls the traffic of protein cargo in multivesicular endosomes most probably through its ability to modulate with actin the morphology of these sorting endosomes 18599791_Sensing molecular tension is crucial for a wide array of cellular processes. Myosin I dramatically alters its motile properties in response to tension. 21666684_Actin and myosin 1 regulate organelle shape and uncover an important function for myosin 1b in the initiation of post-Golgi carrier formation by regulating actin assembly and remodelling trans-Golgi network membranes. 24709651_Myosin 1b catch-bond properties facilitate tube extraction under conditions of increasing membrane tension by reducing the density of myo1b required to pull tubes. 25421751_Results showed that aberrant overexpression of Myo1b in human head and neck squamous cell carcinoma increased cancer cell motility via enhanced large protrusion formation of cell membrane and promotes lymph node metastasis. 26195670_Myosin 1 functions as an effector of EphB2/ephrinB signaling, controls cell morphology, and thereby cell repulsion. 26545583_the overexpression of miR-363 reduces cellular migration in head and neck cancer and reveal the biological relationship between miR-363, myosin 1b, and HPV-positive SCCHN. 28088345_Identify MTO1b as a novel pericyte marker. 29115582_Data demonstrated that the passenger strand of miR145 acted as an antitumor miRNA through targeting MYO1B in HNSCC cells. The involvement of dual strands of premiR145 (miR1455p and miR1453p) in the regulation of HNSCC pathogenesis is a novel concept in present RNA research. 29395313_Data suggested a potential role of Myo1b in cervical carcinogenesis and tumor progression and provided novel insights into the mechanism of how this factor promotes cell proliferation, migration, and invasion in CC cells. 30481162_identify SRSF1 as an important oncodriver that integrates AS control of MYO1B into promotion of gliomagenesis and represents a potential prognostic biomarker and target for glioma therapy. 31040242_The authors report here the identification of a novel Toxoplasma effector protein that is exported from the parasitophorous vacuole in a MYR1-dependent manner and localizes to the host's nucleus. Parasites lacking this inducer of host cyclin E (HCE1) are unable to modulate E2F transcription factor target genes, including cyclin E, and exhibit a substantial growth defect. 31729365_F-actin depolymerizes when sliding on immobilized Myo1b. | ENSMUSG00000018417 | Myo1b | 3.227496e+02 | 0.9060133 | -0.142395861 | 0.3805614 | 1.400823e-01 | 0.7081992441 | 0.93786306 | No | Yes | 3.409993e+02 | 69.370127 | 3.436753e+02 | 71.623828 | ||
| ENSG00000128656 | 1123 | CHN1 | protein_coding | P15882 | FUNCTION: GTPase-activating protein for p21-rac and a phorbol ester receptor. Involved in the assembly of neuronal locomotor circuits as a direct effector of EPHA4 in axon guidance. | 3D-structure;Acetylation;Alternative splicing;Disease variant;GTPase activation;Metal-binding;Neurogenesis;Phosphoprotein;Reference proteome;SH2 domain;Zinc;Zinc-finger | This gene encodes GTPase-activating protein for ras-related p21-rac and a phorbol ester receptor. It is predominantly expressed in neurons, and plays an important role in neuronal signal-transduction mechanisms. Mutations in this gene are associated with Duane's retraction syndrome 2 (DURS2). Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Apr 2011]. | hsa:1123; | cytosol [GO:0005829]; ephrin receptor binding [GO:0046875]; GTPase activator activity [GO:0005096]; metal ion binding [GO:0046872]; ephrin receptor signaling pathway [GO:0048013]; intracellular signal transduction [GO:0035556]; motor neuron axon guidance [GO:0008045]; positive regulation of GTPase activity [GO:0043547]; regulation of axonogenesis [GO:0050770]; regulation of GTPase activity [GO:0043087]; regulation of small GTPase mediated signal transduction [GO:0051056] | 15013773_we show that alpha-chimaerin is a Cdk5p35-binding protein; in transfected HeLa cells, Cdk5p35 and alpha-chimaerin displayed an overlapping distribution pattern 17197532_These two pedigrees double the published pedigrees known to map to the DURS2 locus and can thus contribute toward the search for the DURS2 gene 17197533_DRS (Duane retraction syndrome) linked to the DURS2 locus is associated with bilateral abnormalities of many orbital motor nerves, and structural abnormalities of all EOMs except those innervated by the inferior division of CN3. 18249095_Identify chimaerins as candidates for the downmodulation of Rac1 in T-lymphocytes and, in addition, uncover a novel regulatory mechanism that mediates their activation in T-cells. 18653847_studying families with a variant form of Duane's retraction syndrome (DURS2-DRS), causative heterozygous missense mutations in CHN1 were identified; these are gain-of-function mutations that increase alpha2-chimaerin RacGAP activity 18826946_analysis of an autoinhibitory mechanism that restricts C1 domain-mediated activation of the Rac-GAP alpha2-chimaerin 19541263_Considerable intrafamilial clinical variability was observed in this Duane syndrome pedigree that carried a alpha2-chimaerin mutation 20034095_study concludes that CHN1 mutations are not a major cause of Duane's retraction syndrome among individuals with sporadic disease 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20468071_Observational study of gene-disease association. (HuGE Navigator) 20535495_We found no evidence for a causative involvement of CHN1 mutations in congenital ocular motor anomalies different from autosomal dominant Duane's retraction syndrome 21555619_Members of families segregating Duane retraction syndrome (DRS) as an autosomal dominant trait should be screened for mutations in the CHN1 gene, enhancing genetic counseling and permitting earlier diagnosis. 21715346_Analysis of the current pedigree expands the phenotypic spectrum of hyperactivating CHN1 mutations to include vertical strabismus and supraduction deficits in the absence of Duane retraction syndrome. 25331612_The patients most similar belong to families with a small or absent cranial nerve VI and septo-optic hypoplasia who carry CHN1 mutations 25676811_reduced alpha1-chimaerin expression in the brain of Alzheimer's disease cases, suggesting a role in the upregulation of Rac1 activity during the disease process. 27072986_CHN1 and TNFAIP3 are candidate biomarkers for esophageal squamous cell carcinomas 28160556_this study shows that serum chemerin is an independent risk factor of prognosis of non-small cell lung cancer patients 28346224_These findings reveal a role for ephrin bidirectional signaling upstream of mutant alpha2-chimaerin in Duane retraction syndrome, which may contribute to the selective vulnerability of abducens motor neurons in this disorder 29031989_CHN1 mutations were identified in 2 bilateral cases and in 1 parent of 1 affected case. One mutation is novel and occurred with additional vertical gaze abnormalities. 29872820_The results suggest that chemerin plays an important role in inhibiting the cell proliferation of hepatocellular carcinomas by interfering with cellular iron homeostasis in this type of tumors. 33004823_Identification of a novel CHN1 p.(Phe213Val) variant in a large Han Chinese family with congenital Duane retraction syndrome. 33109127_Upregulation of miR-205 induces CHN1 expression, which is associated with the aggressive behaviour of cervical cancer cells and correlated with lymph node metastasis. 33667650_CHN1 and duane retraction syndrome: Expanding the phenotype to cranial nerves development disease. 34152250_Bioinformatics-based analysis of the association between the A1-chimaerin (CHN1) gene and gastric cancer. 34238315_CHN1 promotes epithelial-mesenchymal transition via the Akt/GSK-3beta/Snail pathway in cervical carcinoma. | ENSMUSG00000056486 | Chn1 | 2.923660e+02 | 1.1450451 | 0.195404462 | 0.3314224 | 3.443020e-01 | 0.5573564937 | 0.89423300 | No | Yes | 3.577995e+02 | 67.982031 | 2.332581e+02 | 45.784743 | |
| ENSG00000128699 | 94101 | ORMDL1 | protein_coding | Q9P0S3 | FUNCTION: Negative regulator of sphingolipid synthesis. {ECO:0000269|PubMed:20182505}. | Endoplasmic reticulum;Membrane;Reference proteome;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA | hsa:94101; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; SPOTS complex [GO:0035339]; cellular sphingolipid homeostasis [GO:0090156]; ceramide metabolic process [GO:0006672]; negative regulation of ceramide biosynthetic process [GO:1900060] | 17928364_Adoplin-1/ORMDL-1 displays reduced expression in association with presenilin mutations; Adoplin-1 & two highly homologous genes (adoplin-2, -3) are a gene family that encodes transmembrane proteins that may play roles in gamma-secretase complex maturation 23066021_ORMDL1, ORMDL2 and ORMDL3 mediate the feedback response in ceramide biosynthesis. 25395622_Expression of the ORMDLS, modulators of serine palmitoyltransferase, is regulated by sphingolipids in mammalian cells. 27313060_ORMDL1 downregulation modestly increased levels of sphingosine, S1P, and ceramide in HepG2 cells. 33145351_Expression Patterns and Prognostic Values of ORMDL1 in Different Cancers. | ENSMUSG00000100679+ENSMUSG00000026097 | Gm28778+Ormdl1 | 8.655989e+02 | 0.6996718 | -0.515249673 | 0.3240872 | 2.501026e+00 | 0.1137721250 | 0.75783482 | No | Yes | 6.630845e+02 | 105.685508 | 1.022298e+03 | 166.563870 | |
| ENSG00000129173 | 79733 | E2F8 | protein_coding | A0AVK6 | FUNCTION: Atypical E2F transcription factor that participates in various processes such as angiogenesis and polyploidization of specialized cells. Mainly acts as a transcription repressor that binds DNA independently of DP proteins and specifically recognizes the E2 recognition site 5'-TTTC[CG]CGC-3'. Directly represses transcription of classical E2F transcription factors such as E2F1: component of a feedback loop in S phase by repressing the expression of E2F1, thereby preventing p53/TP53-dependent apoptosis. Plays a key role in polyploidization of cells in placenta and liver by regulating the endocycle, probably by repressing genes promoting cytokinesis and antagonizing action of classical E2F proteins (E2F1, E2F2 and/or E2F3). Required for placental development by promoting polyploidization of trophoblast giant cells. Acts as a promoter of sprouting angiogenesis, possibly by acting as a transcription activator: associates with HIF1A, recognizes and binds the VEGFA promoter, which is different from canonical E2 recognition site, and activates expression of the VEGFA gene. {ECO:0000269|PubMed:15897886, ECO:0000269|PubMed:16179649, ECO:0000269|PubMed:18202719, ECO:0000269|PubMed:22903062}. | 3D-structure;Activator;Cell cycle;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation | This gene encodes a member of a family of transcription factors which regulate the expression of genes required for progression through the cell cycle. The encoded protein regulates progression from G1 to S phase by ensuring the nucleus divides at the proper time. Multiple alternatively spliced variants, encoding the same protein, have been identified. [provided by RefSeq, Jan 2012]. | hsa:79733; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; RNA polymerase II transcription regulator complex [GO:0090575]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity [GO:0001217]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; identical protein binding [GO:0042802]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; cell cycle comprising mitosis without cytokinesis [GO:0033301]; cell population proliferation [GO:0008283]; chorionic trophoblast cell differentiation [GO:0060718]; hepatocyte differentiation [GO:0070365]; negative regulation of cytokinesis [GO:0032466]; negative regulation of transcription by RNA polymerase II [GO:0000122]; placenta development [GO:0001890]; positive regulation of DNA endoreduplication [GO:0032877]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; sprouting angiogenesis [GO:0002040]; trophoblast giant cell differentiation [GO:0060707] | 15897886_The similarities between E2F-7 and E2F-8 define a new subgroup of the E2F family, and further imply that E2F-7 and E2F-8 may act through overlapping mechanisms in mediating cell cycle control. 16179649_E2F8 may have an important role in repressing the expression of E2F-target genes in the S-phase of the cell cycle. 18202719_E2F7 and E2F8 act upstream of E2F1, and influence the ability of cells to undergo a DNA-damage response. 20068156_Analyses indicated that E2F8 could bind to regulatory elements of cyclin D1, regulating its transcription and promoting accumulation of S-phase cells. Findings suggest that E2F8 contributes to the oncogenic potential of HCC 23064264_The results identify E2F8 as a repressor and E2F1 as an activator of a transcriptional network controlling polyploidization in mammalian cells. 25892397_transcription factor E2F8 is involved in the polyploidization during mouse and human decidualization 26089541_E2F8 is overexpressed in Lung Cancer and is required for the growth of LC cells. E2F8 knockdown reduced the expression of UHRF1. These findings implicate E2F8 as a novel therapeutic target for LC treatment. 26992224_High E2F8 expression is associated with breast cancer. 27454291_E2F8-mediated transcriptional repression is a critical tumor suppressor mechanism during postnatal liver development 27683099_Results showed that E2F8 is up-regulated in metastatic prostate cancer and associated with poor prognosis. These results indicate that E2F8 is a crucial transcription regulator controlling cell cycle and survival in prostate cancer cells. 28270228_Study provides evidence that E2F8 functions as a proliferation-related oncogene in papillary thyroid cancer progression. Moreover, miR-144 appears to be a tumor suppressor through direct inhibition of E2F8. 28605876_E2F8 can shorten cisplatin induced G2/M arrest by promoting MASTL mediated mitotic progression in ER+ breast cancer cells, conferring drug resistance. 29615147_miR-223-5p suppressed NSCLC progression through targeting E2F8. 30144184_MiR-1258 may function as a suppressive factor by negatively controlling E2F8 30951518_Authors detected the expression of hsa_circ_0002468, miR-561, and E2F8 by using quantitative real-time polymerase chain reaction (qRT-PCR) analyses. 31471336_These results demonstrate that E2F8 modulates the growth of human colon cancer cells through promoting the NF-kappaB pathway. 31733123_NONO post-transcriptionally regulates the expression of cell proliferation-related genes by binding to their mRNAs, as exemplified by S-phase-associated kinase 2 and E2F transcription factor 8. 31929759_E2F8 regulates the proliferation and invasion through epithelial-mesenchymal transition in cervical cancer. 31990034_Carcinogenesis effects of E2F transcription factor 8 (E2F8) in hepatocellular carcinoma outcomes: an integrated bioinformatic report. 31995441_Cell cycle oscillators underlying orderly proteolysis of E2F8. 32703494_E2F transcription factor 8 promotes proliferation and radioresistance in glioblastoma. 32823614_E2F8 Induces Cell Proliferation and Invasion through the Epithelial-Mesenchymal Transition and Notch Signaling Pathways in Ovarian Cancer. | ENSMUSG00000046179 | E2f8 | 2.892982e+02 | 1.2052132 | 0.269288423 | 0.3506789 | 5.918172e-01 | 0.4417172646 | 0.85687843 | No | Yes | 3.403490e+02 | 65.083642 | 2.410775e+02 | 47.564103 | |
| ENSG00000129245 | 9513 | FXR2 | protein_coding | P51116 | FUNCTION: RNA-binding protein. | 3D-structure;Cytoplasm;Phosphoprotein;RNA-binding;Reference proteome;Repeat | The protein encoded by this gene is a RNA binding protein containing two KH domains and one RCG box, which is similar to FMRP and FXR1. It associates with polyribosomes, predominantly with 60S large ribosomal subunits. This encoded protein may self-associate or interact with FMRP and FXR1. It may have a role in the development of fragile X cognitive disability syndrome. [provided by RefSeq, Jul 2008]. | hsa:9513; | axon [GO:0030424]; cytoplasm [GO:0005737]; cytoplasmic ribonucleoprotein granule [GO:0036464]; cytosol [GO:0005829]; cytosolic large ribosomal subunit [GO:0022625]; dendritic filopodium [GO:1902737]; dendritic spine [GO:0043197]; dendritic spine neck [GO:0044326]; growth cone [GO:0030426]; membrane [GO:0016020]; neuronal cell body [GO:0043025]; nucleus [GO:0005634]; polysome [GO:0005844]; postsynaptic density [GO:0014069]; presynapse [GO:0098793]; identical protein binding [GO:0042802]; mRNA 3'-UTR binding [GO:0003730]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; translation regulator activity [GO:0045182]; negative regulation of translation [GO:0017148]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of response to DNA damage stimulus [GO:2001022]; positive regulation of translation [GO:0045727]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; regulation of filopodium assembly [GO:0051489]; regulation of mRNA stability [GO:0043488] | 19487368_FXR1P and FXR2P KH2 domains bind G-quadruplex and kissing complex RNA with the same affinity as the FMRP KH2 domain. 20442204_These results indicate that FMR1 gene function is evolutionarily conserved in neural mechanisms and cannot be compensated by either FXR1 or FXR2, but that all three proteins can substitute for each other in non-neuronal requirements. 21072162_Data show that the nuclear localization signals of the FXR1 and FXR2 comprise tandem Tudor domain architectures. 26612855_An accumulation of 8 SNPs in the fragile gene family (FMR1, FXR1 and FXR2)were found associated with autistic traits in a sample of male patients. 28767039_Inhibition of the remaining family member FXR1 selectively blocks cell proliferation in human cancer cells containing homozygous deletion of both TP53 and FXR2 in a collateral lethality manner. 32706158_Spatial control of nucleoporin condensation by fragile X-related proteins. 32878499_Fragile X-related protein family: a double-edged sword in neurodevelopmental disorders and cancer. | ENSMUSG00000018765 | Fxr2 | 5.332722e+03 | 1.1883979 | 0.249018011 | 0.2827676 | 7.800741e-01 | 0.3771184737 | 0.84010627 | No | Yes | 5.687878e+03 | 534.382070 | 4.116309e+03 | 397.271836 | |
| ENSG00000129925 | 58986 | PGAP6 | protein_coding | Q9HCN3 | FUNCTION: Involved in the lipid remodeling steps of GPI-anchor maturation. Lipid remodeling steps consist in the generation of 2 saturated fatty chains at the sn-2 position of GPI-anchor proteins (GPI-AP). Has phospholipase A2 activity that removes an acyl-chain at the sn-2 position of GPI-anchors during the remodeling of GPI. Required for the shedding of the GPI-AP TDGF1, but not CFC1, at the cell surface. Shedding of TDGF1 modulates Nodal signaling by allowing soluble TDGF1 to act as a Nodal coreceptor on other cells (PubMed:27881714). Also indirectly involved in the translocation of RAC1 from the cytosol to the plasma membrane by maintaining the steady state amount of CAV1-enriched plasma membrane subdomains, stabilizing RAC1 at the plasma membrane (PubMed:27835684). In contrast to myomaker (TMEM8C), has no fusogenic activity (PubMed:26858401). {ECO:0000269|PubMed:26858401, ECO:0000269|PubMed:27835684, ECO:0000269|PubMed:27881714}. | Cell membrane;Disulfide bond;EGF-like domain;Glycoprotein;Hydrolase;Lipid metabolism;Lysosome;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:58986; | extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; lysosomal membrane [GO:0005765]; plasma membrane [GO:0005886]; phospholipase A2 activity [GO:0004623]; lipid metabolic process [GO:0006629] | Mouse_homologues 27881714_PGAP6 plays a critical role in Nodal signaling modulation through CRIPTO shedding. | ENSMUSG00000024180 | Pgap6 | 2.889986e+03 | 1.1949539 | 0.256954970 | 0.3118970 | 6.812015e-01 | 0.4091732991 | 0.84661257 | No | Yes | 2.723733e+03 | 313.987830 | 2.038997e+03 | 241.520283 | ||
| ENSG00000130305 | 55695 | NSUN5 | protein_coding | Q96P11 | FUNCTION: S-adenosyl-L-methionine-dependent methyltransferase that specifically methylates the C(5) position of cytosine 3782 (m5C3782) in 28S rRNA (PubMed:23913415, PubMed:31428936, PubMed:31722427). m5C3782 promotes protein translation without affecting ribosome biogenesis and fidelity (PubMed:31428936, PubMed:31722427). Required for corpus callosum and cerebral cortex development (By similarity). {ECO:0000250|UniProtKB:Q8K4F6, ECO:0000269|PubMed:23913415, ECO:0000269|PubMed:31428936, ECO:0000269|PubMed:31722427}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Methyltransferase;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;S-adenosyl-L-methionine;Transferase;Williams-Beuren syndrome;rRNA processing | This gene encodes a member of an evolutionarily conserved family of proteins that may function as methyltransferases. This gene is located in a larger region of chromosome 7 that is deleted in Williams-Beuren syndrome, a multisystem developmental disorder. There are two pseudogenes for this gene located in the same region of chromosome 7. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2013]. | hsa:55695; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; rRNA (cytosine-C5-)-methyltransferase activity [GO:0009383]; cerebral cortex development [GO:0021987]; cognition [GO:0050890]; corpus callosum development [GO:0022038]; oligodendrocyte development [GO:0014003]; positive regulation of translation [GO:0045727]; regulation of myelination [GO:0031641]; RNA methylation [GO:0001510]; rRNA base methylation [GO:0070475] | 11978965_Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome 31428936_Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. 31722427_Phenotypic consequences of NSUN5 deficiency in mammalian cells include decreased proliferation and size, which can be attributed to a reduction in total protein synthesis by altered ribosomes. 33177158_NSUN5 Facilitates Viral RNA Recognition by RIG-I Receptor. | ENSMUSG00000000916 | Nsun5 | 3.448385e+03 | 1.3020509 | 0.380785796 | 0.3097017 | 1.522122e+00 | 0.2172985994 | 0.78763590 | No | Yes | 3.651046e+03 | 465.242109 | 2.450073e+03 | 320.641495 | |
| ENSG00000130558 | 10439 | OLFM1 | protein_coding | Q99784 | FUNCTION: Contributes to the regulation of axonal growth in the embryonic and adult central nervous system by inhibiting interactions between RTN4R and LINGO1. Inhibits RTN4R-mediated axon growth cone collapse (By similarity). May play an important role in regulating the production of neural crest cells by the neural tube (By similarity). May be required for normal responses to olfactory stimuli (By similarity). {ECO:0000250|UniProtKB:O88998, ECO:0000250|UniProtKB:Q9IAK4}. | 3D-structure;Alternative splicing;Cell junction;Cell projection;Coiled coil;Developmental protein;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Reference proteome;Secreted;Signal;Synapse | This gene product shares extensive sequence similarity with the rat neuronal olfactomedin-related ER localized protein. While the exact function of the encoded protein is not known, its abundant expression in brain suggests that it may have an essential role in nerve tissue. Several alternatively spliced transcripts encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:10439; | axon [GO:0030424]; axonal growth cone [GO:0044295]; endoplasmic reticulum [GO:0005783]; extracellular space [GO:0005615]; neuronal cell body [GO:0043025]; perikaryon [GO:0043204]; synapse [GO:0045202]; atrioventricular valve formation [GO:0003190]; cardiac epithelial to mesenchymal transition [GO:0060317]; negative regulation of gene expression [GO:0010629]; nervous system development [GO:0007399]; neuronal signal transduction [GO:0023041]; positive regulation of apoptotic process [GO:0043065]; positive regulation of epithelial to mesenchymal transition [GO:0010718]; positive regulation of gene expression [GO:0010628]; regulation of axon extension [GO:0030516] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21968811_Wnt activation suppresses Olfm-1 expression, and this may predispose a favorable microenvironment of the retained embryo in the Fallopian tube, leading to ectopic pregnancy in humans 25999259_Down-regulation of OLFM1 in fallopian tube epithelial cells enhances spheroid attachment to fallopian tube. 26121352_OLF domains from H. sapiens olfactomedin-1 and M. musculus gliomedin were biophysically, biochemically, and structurally characterized. 26377223_human chorionic gonadotropin stimulated miR-212, which down-regulated OLFM1 and CTBP1 expression in fallopian and endometrial epithelial cells to favor spheroid attachment 27555280_OLFM1 is a negative regulator of non-canonical NF-kappaB signalling by interacting with and inhibiting NIK. Thus, OLFM1 may serve as a valuable biomarker and therapeutic target for colorectal cancer (CRC) patients. | ENSMUSG00000026833 | Olfm1 | 1.240431e+02 | 0.9407334 | -0.088142158 | 0.3899011 | 5.006545e-02 | 0.8229494224 | 0.96628825 | No | Yes | 9.194986e+01 | 14.553135 | 9.887787e+01 | 16.085800 | |
| ENSG00000130638 | 25814 | ATXN10 | protein_coding | Q9UBB4 | FUNCTION: Necessary for the survival of cerebellar neurons. Induces neuritogenesis by activating the Ras-MAP kinase pathway. May play a role in the maintenance of a critical intracellular glycosylation level and homeostasis. {ECO:0000250}. | Alternative splicing;Ciliopathy;Cytoplasm;Methylation;Neurodegeneration;Phosphoprotein;Reference proteome;Spinocerebellar ataxia | This gene encodes a protein that may function in neuron survival, neuron differentiation, and neuritogenesis. These roles may be carried out via activation of the mitogen-activated protein kinase cascade. Expansion of an ATTCT repeat from 9-32 copies to 800-4500 copies in an intronic region of this locus has been associated with spinocerebellar ataxia, type 10. Alternatively spliced transcript variants have been described.[provided by RefSeq, Jul 2016]. | hsa:25814; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendrite [GO:0030425]; extracellular space [GO:0005615]; membrane [GO:0016020]; neuronal cell body [GO:0043025]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; cilium assembly [GO:0060271]; nervous system development [GO:0007399]; neuron projection development [GO:0031175] | 11891842_frequency of SCA10 in spinocerebellar ataxia in France 15148151_Observational study of genotype prevalence. (HuGE Navigator) 16385455_Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10 suggest that the purity of the expanded repeat element may be a disease modifier. 17846122_These results implicate replication origin activity as one molecular mechanism associated with the instability of ataxin 10(ATTCT)n tracts that are longer than normal length. 18386626_cause Spinocerebellar ataxia type 10 by expansion of the ATTCT pentanucleotide repeat in intron 9 of the gene 19147916_The normal reference standard for ATXN10 gene's ATTCT pentanucleotide repeat of 9-32 is verified in the Chinese Han group. 19171184_Nucleosome formation on pure and interrupted ATTCT pentanucleotides associated with spinocerebellar ataxia type 10 (SCA10), is reported. 19234597_evidence for an ancestral common origin for SCA10 in Latin America, which might have arisen in an ancestral Amerindian population and later have been spread into the mixed populations of Mexico and Brazil. 19259763_Observational study of gene-disease association. (HuGE Navigator) 19835171_Ataxin 10 and eukaryotic initiation factors interacts with M2 protein of influenza A virus. 19936807_Data suggest that SCA10 could be related to chromatin structure abnormalities caused by the expansion and not to an abnormal or abnormally expressed ATXN10. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20548952_suggesting that the loss of function of hnRNP K plays a key role in cell death of SCA10. 21565611_Network building strategy led to the proposal of candidates for new ciliopathy disease genes, leading to the identification of the first human mutations in the Nephronophthisis gene Ataxin10 (ATXN10) and Joubert syndrome gene Tectonic2 (TCTN2). 21827910_The expansion of the attct repeat in intron 9 of atxn10 is may caused Spinocerebellar ataxia type 10. 21857149_Plk1 phosphorylates Ataxin-10 on Ser 77 and Thr 82. 23026538_The SCA10 pentanucleotide repeat expansion was not found among a group of Cypriot ataxia patients. All had 10-19 ATTCT repeats. Controls had 11-20 repeats, with 14 being the most common number. 24318420_the presence of repeat interruptions in SCA10 repeat expansion may have a role in epilepsy phenotype 25630585_This is the first description of a family with two SCA mutations with affected subjects having a combined SCA2 and SCA10 phenotype. 25666058_Inhibition of Aurora B or expression of the S12A mutant renders reduced interaction between Ataxin-10 and polo-like kinase 1 (Plk1), a kinase previously identified to regulate Ataxin-10 in cytokinesis. 26039897_Data suggest precursor mRNA for SCA10 (crystalized using two model AUUCU SCA10 repeats) exhibits the following conformation: the two asymmetric RNA molecules are antiparallel to each other and the interaction is stabilized by multiple hydrogen bonds. 26295943_Single molecule real time sequencing of long tandem nucleotide repeats in spinocerebellar ataxia ATXN10 reveals unique insight of repeat expansion structure in three unrelated patients. 28423040_Inheritance patterns of ATCCT repeat interruptions in spinocerebellar ataxia type 10 expansions of ataxin-10 has been reported. 28542277_Polymorphism in ATXN10 gene is associated with spinocerebellar ataxia type 10. 28905220_The 19-CGGC-14 shared haplotype was found in 47% of Brazilian and in 63% of Peruvian families. Frequencies from both are statistically different from Brazilian controls but not Quechua controls. The most frequent haplotype in Quechuas, 19-15-CGGC-14-10, is found in 50% of Brazilian and in 65% of Peruvian patients. The ATTCT expansion may have arisen in a Native American chromosome. 29169923_Ataxin-10 is involved in Golgi membrane dynamics 31342269_ATXN10 allele distribution in a healthy Amerindian population from Peru does not differ from that of other populations. 31377949_The most common clinical presentation of SCA10 in Brazilian families was pure cerebellar ataxia. | ENSMUSG00000016541 | Atxn10 | 2.775933e+03 | 1.1149008 | 0.156915332 | 0.2757175 | 3.288694e-01 | 0.5663255719 | 0.89778071 | No | Yes | 3.159923e+03 | 493.288800 | 2.441161e+03 | 391.051019 | |
| ENSG00000130649 | 1571 | CYP2E1 | protein_coding | P05181 | FUNCTION: A cytochrome P450 monooxygenase involved in the metabolism of fatty acids (PubMed:10553002, PubMed:18577768). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase) (PubMed:10553002, PubMed:18577768). Catalyzes the hydroxylation of carbon-hydrogen bonds. Hydroxylates fatty acids specifically at the omega-1 position displaying the highest catalytic activity for saturated fatty acids (PubMed:10553002, PubMed:18577768). May be involved in the oxidative metabolism of xenobiotics (Probable). {ECO:0000269|PubMed:10553002, ECO:0000269|PubMed:18577768, ECO:0000305|PubMed:9348445}. | 3D-structure;Direct protein sequencing;Endoplasmic reticulum;Fatty acid metabolism;Heme;Iron;Lipid metabolism;Membrane;Metal-binding;Microsome;Mitochondrion;Mitochondrion inner membrane;Monooxygenase;NADP;Oxidoreductase;Reference proteome | PATHWAY: Lipid metabolism; fatty acid metabolism. {ECO:0000269|PubMed:10553002, ECO:0000269|PubMed:18577768}. | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This protein localizes to the endoplasmic reticulum and is induced by ethanol, the diabetic state, and starvation. The enzyme metabolizes both endogenous substrates, such as ethanol, acetone, and acetal, as well as exogenous substrates including benzene, carbon tetrachloride, ethylene glycol, and nitrosamines which are premutagens found in cigarette smoke. Due to its many substrates, this enzyme may be involved in such varied processes as gluconeogenesis, hepatic cirrhosis, diabetes, and cancer. [provided by RefSeq, Jul 2008]. | hsa:1571; | cytoplasm [GO:0005737]; endoplasmic reticulum membrane [GO:0005789]; Golgi membrane [GO:0000139]; intracellular membrane-bounded organelle [GO:0043231]; intrinsic component of endoplasmic reticulum membrane [GO:0031227]; mitochondrial inner membrane [GO:0005743]; 4-nitrophenol 2-monooxygenase activity [GO:0018601]; arachidonic acid epoxygenase activity [GO:0008392]; aromatase activity [GO:0070330]; enzyme binding [GO:0019899]; heme binding [GO:0020037]; Hsp70 protein binding [GO:0030544]; Hsp90 protein binding [GO:0051879]; iron ion binding [GO:0005506]; long-chain fatty acid omega-1 hydroxylase activity [GO:0120319]; monooxygenase activity [GO:0004497]; oxidoreductase activity [GO:0016491]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen [GO:0016709]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen [GO:0016712]; oxygen binding [GO:0019825]; steroid hydroxylase activity [GO:0008395]; 4-nitrophenol metabolic process [GO:0018960]; benzene metabolic process [GO:0018910]; carbon tetrachloride metabolic process [GO:0018885]; epoxygenase P450 pathway [GO:0019373]; halogenated hydrocarbon metabolic process [GO:0042197]; heterocycle metabolic process [GO:0046483]; lipid hydroxylation [GO:0002933]; long-chain fatty acid biosynthetic process [GO:0042759]; long-chain fatty acid metabolic process [GO:0001676]; monoterpenoid metabolic process [GO:0016098]; organic acid metabolic process [GO:0006082]; response to bacterium [GO:0009617]; response to ethanol [GO:0045471]; response to organonitrogen compound [GO:0010243]; response to ozone [GO:0010193]; steroid metabolic process [GO:0008202]; triglyceride metabolic process [GO:0006641]; xenobiotic metabolic process [GO:0006805] | 11051375_Observational study of gene-disease association. (HuGE Navigator) 11104220_Observational study of gene-disease association. (HuGE Navigator) 11191882_Observational study of genotype prevalence. (HuGE Navigator) 11198676_Observational study of genotype prevalence. (HuGE Navigator) 11207032_Observational study of genotype prevalence. (HuGE Navigator) 11236836_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11259352_Observational study of genotype prevalence and gene-environment interaction. (HuGE Navigator) 11263781_Observational study of gene-environment interaction. (HuGE Navigator) 11275366_Observational study of gene-disease association. (HuGE Navigator) 11305777_Observational study of gene-environment interaction. (HuGE Navigator) 11331106_Observational study of gene-disease association. (HuGE Navigator) 11377232_Observational study of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 11389775_Observational study of gene-disease association. (HuGE Navigator) 11389775_There is a correlation between the RsaI polymorphism homozygous uncut genotype in the CYP2E1 gene and a higher relative risk of nasopharyngeal carcinoma development in the Thai or Chinese populations in Thailand. 11406608_Observational study of gene-disease association. (HuGE Navigator) 11410713_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11422615_Observational study of gene-disease association. (HuGE Navigator) 11503278_Observational study of gene-disease association. (HuGE Navigator) 11520401_Observational study of gene-disease association. (HuGE Navigator) 11535247_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11535253_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11641039_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11675150_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11696658_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11697456_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11700262_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11719088_Observational study of gene-disease association. (HuGE Navigator) 11746208_Observational study of genotype prevalence. (HuGE Navigator) 11748356_Observational study of gene-disease association. (HuGE Navigator) 11751440_Observational study of genotype prevalence. (HuGE Navigator) 11766168_Observational study of gene-disease association. (HuGE Navigator) 11774269_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11776598_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11782477_increased translation of collagen mRNA by CYP2E1-derived reactive oxygen species is responsible for the increase in collagen protein 11798822_Observational study of gene-disease association. (HuGE Navigator) 11802217_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11815398_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11854903_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11869835_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11895912_Observational study of gene-disease association. (HuGE Navigator) 11907164_Damage to mitochondria may be a critical step for cellular toxicity by CYP2E1-derived reactive oxygen species 11911601_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11960914_CYP2A6/2A7 and CYP2E1 expression in human oesophageal mucosa: regional and inter-individual variation in expression and relevance to nitrosamine metabolism 11964928_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11966948_Observational study of gene-disease association. (HuGE Navigator) 12010862_Individuals possessing more susceptible CYP2E1 c2c2 genotypes were more likely to reveal p53 overexpression. Susceptible CYP2E1 genotypes may modulate the mutation of the p53 gene among VCM-exposed workers. 12010862_Observational study of gene-disease association. (HuGE Navigator) 12047484_Observational study of genotype prevalence. (HuGE Navigator) 12055050_Observational study of gene-environment interaction. (HuGE Navigator) 12063626_Observational study of gene-disease association. (HuGE Navigator) 12080432_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12115524_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12172927_Observational study of gene-disease association. (HuGE Navigator) 12198369_In the CYP2E1 gene 5'-flanking region polymorphism, patients with esophageal cancer showed significantly higher frequency of the A4/A4 genotype compared with the control subjects (p = 0.02), but no difference was found in patients with lung cancer. 12198369_Observational study of gene-disease association. (HuGE Navigator) 12211622_Observational study of gene-disease association. (HuGE Navigator) 12355548_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12365037_Observational study of genotype prevalence. (HuGE Navigator) 12376502_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12376511_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12397416_Observational study of gene-disease association. (HuGE Navigator) 12452057_expression of human cytochrome P450 2E1 gene in embryonic nasopharynx, nasopharyngeal cancer cell lines and tissue 12454736_Observational study of gene-disease association. (HuGE Navigator) 12460800_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12469218_Overexpression of this enzyme overexpression up-regulates both non-specific delta-aminolevulinate synthase and heme oxygenase-1 in a human hepatoma cell line. 12490624_substitution of residue 363 in cyp2e1 resulted in significant alterations of the metabolite profile for the side chain hydroxylation of 7-butoxycoumarin 12540498_Observational study of gene-disease association. (HuGE Navigator) 12548461_Observational study of gene-disease association. (HuGE Navigator) 12552594_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12552594_The CYP2E1 variant genotype did not significantly increase the risk for neoplasia. 12554615_Polymorphisms of alcohol-metabolizing enzymes: analyses of mutations on the CYP2E1, ADH2, ADH3 and ALDH2 genes in a Mexican-American population living in the Los Angeles area. 12561466_Observational study of gene-disease association. (HuGE Navigator) 12563175_CYP2E1 genetic polymorphism may be associated with susceptibility to breast cancer in alcohol-consuming women. 12563175_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12569554_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12579334_Observational study of gene-disease association. (HuGE Navigator) 12601351_hepatic cytochrome P450 2E1 activity and lymphocyte cytochrome P450 2E1 expression are enhanced in nondiabetic nonalcoholic steatohepatitis 12668988_CYP 2E1 genetic polymorphism may be associated with susceptibility to antituberculosis drug-induced hepatitis. 12668988_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12705718_Observational study of gene-disease association. (HuGE Navigator) 12707490_Observational study of gene-disease association. (HuGE Navigator) 12707490_This study shown that the genotypes of CYP2E1 are associated with clinical features of alcoholics. 12710951_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12718576_Observational study of genotype prevalence. (HuGE Navigator) 12718671_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12732844_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12738724_Down-regulation of cytochrome P450 CYP2E1 is associated with breast cancer 12739102_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12759747_Observational study of gene-disease association. (HuGE Navigator) 12760253_Observational study of gene-disease association. (HuGE Navigator) 12767509_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12771559_Observational study of gene-disease association. (HuGE Navigator) 12774019_Proteasome activity plays an important role in modulating CYP2E1-mediated toxicity in HepG2 cells by regulating CYP2E1 levels and by removal of oxidized proteins. 12777398_HO-1 induction was observed in the livers of chronic alcohol-fed mice or pyrazole-treated rats, conditions known to elevate CYP2E1 levels. 12777398_P450 2E1 has a role in inducing heme oxygenase-1 through ERK MAPK pathway 12777962_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 12777962_association of CYP2E1*1D with alcohol and nicotine dependence suggests that CYP2E1 may contribute to the development of these dependencies 12777965_CYP2E1 and NQO1 genotypes may play an important role in development of smoking related bladder cancer among Korean men 12777965_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12813050_CYP2E1 has a role in oxidative stress causing mitochondrial damage along with phospholipase A2 12824748_Observational study of gene-disease association. (HuGE Navigator) 12824892_Observational study of gene-disease association. (HuGE Navigator) 12851035_Arg76 is closely associated with the function of CYP2E1, and that the genetic polymorphism of cytochrome p450 2E1 is one cause of interindividual differences in the toxicity of xenobiotics 12860273_These results show that CYP2E1 protein is expressed in both tumour and normal breast tissue with an increased expression in breast tumours. 12883487_hepatic CYP2E1 activity is up-regulated in morbidly obese subjects 12883749_Observational study of gene-disease association. (HuGE Navigator) 12915519_Observational study of gene-disease association. (HuGE Navigator) 12939804_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12940444_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12960506_Transcriptional activity of the mutant allele of the tandem repeat polymorphism in the 5'-flanking region of the CYP2E1 gene is greater than that of the wild type. 14500779_CYP2E1 was clearly expressed in human fetal liver. 14510941_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14527082_Levels of CYP2E1 are elevated under a variety of physiological and pathophysiological conditions, and after acute and chronic alcohol treatment. 14527082_biochemical and toxicological properties of CYP2E1(review) 14535982_Observational study of gene-disease association. (HuGE Navigator) 14578150_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14606109_The genetic polymorphism of CYPIIE1 on the position of Pst I and Rsa I is related to the susceptibility of fatty liver. 14634838_Observational study of genotype prevalence. (HuGE Navigator) 14646291_Observational study of gene-disease association. (HuGE Navigator) 14669323_The cDNA of human CYP2E1 can be successfully cloned, and a cell line, HepG2-CYP2E1 can efficiently express mRNA and has CYP2E1 activity. 14681495_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14691069_Observational study of gene-disease association. (HuGE Navigator) 14695651_Observational study of genotype prevalence. (HuGE Navigator) 14695664_enhanced injury in hepatocytes over expressing both Hepatitis C virus core protein and CYP2E1 is mediated by increases in oxidative stress. 14696128_CYP2E1 polymorphisms are associated with incomplete intestinal metaplasia in a high-risk area of stomach cancer 14696128_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14751678_Observational study of gene-disease association. (HuGE Navigator) 14757192_Observational study of gene-disease association. (HuGE Navigator) 14991750_Observational study of gene-disease association. (HuGE Navigator) 15036355_The objective of this study was to analyze the effect of the green tea flavanol epigallocatechin-3-gallate (EGCG), which has been shown to prevent alcohol-induced liver damage, on CYP2E1-mediated toxicity in HepG2 cells overexpressing CYP2E1 (E47 cells). 15061915_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15064998_Observational study of gene-disease association. (HuGE Navigator) 15066574_Observational study of gene-disease association. (HuGE Navigator) 15112335_Observational study of gene-disease association. (HuGE Navigator) 15125228_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15138035_Observational study of gene-disease association. (HuGE Navigator) 15162526_CYP2E1 is a unique gene expressing in liver but did not express in hepatocellular carcinoma 15177663_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15182482_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15215328_Observational study of gene-disease association. (HuGE Navigator) 15215328_polymorphism and susceptibility to cirrhosis or pancreatitis in alcoholics 15220553_Observational study of gene-disease association. (HuGE Navigator) 15226677_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15289170_Observational study of gene-disease association. (HuGE Navigator) 15318112_Observational study of gene-disease association. (HuGE Navigator) 15327835_Observational study of gene-disease association. (HuGE Navigator) 15349722_marked impairment of CYP enzyme activity during allograft rejection which is presumably secondary to an increased intragraft production of proinflammatory cytokines and NO. 15355699_Observational study of gene-disease association. (HuGE Navigator) 15370874_Observational study of gene-disease association. (HuGE Navigator) 15491310_Observational study of gene-disease association. (HuGE Navigator) 15519646_Observational study of gene-disease association. (HuGE Navigator) 15532721_overexpression of human CYP2E1 activates acetaminophen to reactive metabolites which damage mitochondria, form protein adducts, and result in toxicity to HepG2 cells 15536330_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15632182_increased hepatocyte CYP2E1 expression and the presence of steatohepatitis result in the down-regulation of insulin signaling 15633127_The aim of this study was to determine whether hepatitis C virus core protein and alcohol-inducible cytochrome P450 2E1 contribute to reactive oxygen species production and cytotoxicity in human hepatoma cells. 15640066_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15646021_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15660387_Nicotinamide adenine dinucleotide phosphate oxidase-derived oxidants are critical for development of ethanol-induced liver injury. CYP2E1 is required for induction of oxidative stress to DNA and may play role in ethanol-associated hepatocarcinogenesis. 15712341_Observational study of gene-disease association. (HuGE Navigator) 15714076_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, gene-gene interaction, gene-environment interaction, pharmacogenomic / toxicogenomic, and healthcare-related. (HuGE Navigator) 15734972_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15753073_To address this question, we determined whether DLPC protects against alcohol-induced cytotoxicity in HepG2 cells expressing CYP2E1. 15763499_a decrease of Hsp60 in the cytoplasmic fraction of dilated cardiomyopathy-affected left ventricles was observed; at the same time an increase in P450 2E1 expression in dilated hearts' cytoplasmic fractions was observed 15769360_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15774926_Observational study of gene-disease association. (HuGE Navigator) 15780023_Observational study of gene-disease association. (HuGE Navigator) 15793883_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15793883_the CYP2E1 genotype may influence individual susceptibility to development of gastric cardia cancer, and that the risk increases significantly in smokers. 15849806_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 15899651_Observational study of gene-disease association. (HuGE Navigator) 15902904_Polymorphisms of the alcohol metabolism-related CYP2E1 gene are significantly different in Korean patients with alcoholism and Korean control subjects without alcoholism. 15914211_Observational study of gene-disease association. (HuGE Navigator) 15914277_Observational study of gene-disease association. (HuGE Navigator) 15928955_Observational study of gene-disease association. (HuGE Navigator) 15938845_Observational study of gene-disease association. (HuGE Navigator) 15952134_The CYP2E1 genotype frequencies are 66.7% for type of c1/c1, 30% for type of c1/c2, 3.3% for type of c2/c2 with the allele frequencies 0.818 for C1 and 0.182 for C2, and the gene distribution matched the equilibrium law of Hardy-Weinberg. 15961886_CYP2E1 priming could explain the sensitization of Kupffer cells to LPS activation by ethanol, a critical early step in alcoholic liver disease 15968714_CYP2E1 wild type (c1/c1) increased the susceptibility to esophageal squamous cell cancer risk in Kazakh individuals with GSTM1 presence genotype 15968714_Observational study of gene-disease association. (HuGE Navigator) 15991278_Observational study of gene-disease association. (HuGE Navigator) 16006997_Observational study of gene-disease association. (HuGE Navigator) 16019049_Low concentrations of iron and arachidonic acid synergistically interacted with CYP2E1 to produce cell toxicity, suggesting these nutrients may act as priming or sensitizing agents to alcohol-induced liver injury. 16039674_Observational study of gene-disease association. (HuGE Navigator) 16043197_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16052683_A positive relationship between ethanol-induced oxidative damage in human primary cultured hepatocytes and CYP2E1 activity was exhibited 16125881_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16126235_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16137184_A PCR method for identifying polymorphism. 16137184_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 16142352_Observational study of gene-disease association. (HuGE Navigator) 16142352_the A4/A4 genotype of the 5'-flanking region of CYP2E1 has a role in development of non-small cell lung carcinoma in Japanese patients 16172237_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16235983_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16235992_Observational study of genotype prevalence. (HuGE Navigator) 16253141_Protein concentration of CYP2E1 is significantly lower by ~81% in the sigmoid colon compared to the level in the descending colon. 16311924_We report here for the first time three novel alternative spliced mRNA transcripts which are more frequently present in lung carcinoma cell lines as in hepatocyte cell lines. 16324524_Observational study of gene-disease association. (HuGE Navigator) 16337880_Potentiation of serum deprivation-induced cell death by CYP2E1 may contribute to the sensitivity of the liver to alcohol-induced ischemia and growth factor deprivation. 16365683_Observational study of gene-disease association. (HuGE Navigator) 16372174_Observational study of gene-disease association. (HuGE Navigator) 16380384_CYP2E1 has a role in sensitizing cells to the toxicity caused by depletion of glutathione 16385451_Observational study of gene-disease association. (HuGE Navigator) 16393248_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16424825_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16425414_Observational study of gene-disease association. (HuGE Navigator) 16425414_frequencies of this high-activity polymorphism in alcohol related patient groups 16440362_Meta-analysis of gene-disease association. (HuGE Navigator) 16459354_Observational study of gene-disease association. (HuGE Navigator) 16470306_Observational study of genotype prevalence. (HuGE Navigator) 16471212_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16484137_Observational study of gene-disease association. (HuGE Navigator) 16488179_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16497268_These results suggest that CYP2E1 mediated oxidative stress downregulates the expression of GRP proteins in HepG2 cells and oxidative stress is an important mechanism in causing ER dysfunction in these cells. 16535827_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16551616_Nrf2, through up-regulation of glutamate-cysteine ligase and increase of GSH levels, protects against CYP2E1-dependent AA toxicity 16600530_Observational study of gene-disease association. (HuGE Navigator) 16634857_Observational study of genotype prevalence. (HuGE Navigator) 16634857_studies suggest that the frequency of the c2 alleles in the Polish population is low; it seems, however, that they pose the risk of alcoholic cirrhosis 16679316_CYP2E1-b(5) complex model was constructed, leading to improved insights into the protein interaction. 16720291_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16721740_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16758119_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16758119_findings indicated a gene-gene interaction between CYP2E1 and GSTM1 was accessible to developing breast cancer in Taiwanese women without the habits of cigarette smoking and alcohol consumption 16770646_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16834659_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16837478_Meta-analysis of gene-disease association and gene-environment interaction. (HuGE Navigator) 16841220_Observational study of gene-disease association. (HuGE Navigator) 16962935_increased toxic interactions by TGF-beta1 plus CYP2E1 can occur by a mechanism involving increased production of intracellular ROS and depletion of GSH, resulting in mitochondrial membrane damage and loss of membrane potential, followed by apoptosis 16985026_Observational study of gene-disease association. (HuGE Navigator) 16985032_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17007050_Expression of Thy1 decreased in differentiated ADSC and BMSC. Expression of albumin, CYP2E1, and CYP3A4 increased in differentiated BMSC and ADSC. Hepatic gene activation may involve increased C/EBPbeta and HNF4alpha. 17016589_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17022435_Observational study of gene-disease association. (HuGE Navigator) 17034788_Ethanol induced CYP2E1 generates oxidative stress that is responsible for the decrease in proteasome activity. 17059334_Observational study of genotype prevalence. (HuGE Navigator) 17078101_Observational study of gene-disease association. (HuGE Navigator) 17118447_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17119198_Observational study of gene-disease association. (HuGE Navigator) 17119944_This report is the first demonstration that thalassemia major is associated with an alteration of CYP2E1 and CYP3A4 activities; this could modify the sensitivity of thalassemia patients to the toxic or therapeutic effects of drugs. 17134659_Observational study of gene-disease association. (HuGE Navigator) 17134659_This documents the presence of a polymorphism of CYP2E1 that is overexpressed in alcoholic Otomies, in which the variant allele (A1 of CYP2E1/TaqI) is associated with increased susceptibility to alcoholism. 17146594_The expression of CYP2E1 in Nasopharyngeal carcinoma (NPC) cells, embryonic nasopharyngeal epithelial tissue (ENET) specimens, and NPC biopsies was analysed. 17156750_comparison of substrate dynamics in CYP2E1 and CYP2A6 17176083_The unusually rapid carbon monoxide binding kinetics of P450 2E1 indicate that it is more dynamically mobile than other P450s and thus able to more readily interconvert among alternate conformations. 17178637_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17219769_Observational study of genetic testing. (HuGE Navigator) 17264406_Observational study of genotype prevalence. (HuGE Navigator) 17264406_The frequency of the CYP2E1 allele distribution was found to be markedly different between Chinese and South African populations. 17284772_Induction of DNA strand breaks and oxidative stress in HeLa cells by ethanol is dependent on CYP2E1 expression. 17292341_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17331164_The aim of this study was to determine the role of these polymorphisms for the transcriptional regulation of the human CYP2E1 gene in general transcription and during enzyme induction 17361553_Observational study of gene-disease association. (HuGE Navigator) 17367411_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17373732_Differences exist in protein levels of certain CYPs in non-malignant esophageal tissue (e.g. CYP2C8, CYP3A4, CYP3A5, and CYP2E1) between SCC patients and healthy subjects and may contribute to the development of squamous-cell carcinoma in the esophagus. 17380320_Observational study of genotype prevalence. (HuGE Navigator) 17380320_The genotype frequencies of CYP2E1*7B in Turkish population were found to be similar to those of other Caucasian populations 17384900_Observational study of gene-disease association. (HuGE Navigator) 17427487_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17427487_Our studies suggest that the frequency of allele c2 in Polish population is low, but the presence of c2 allele may be a risk factor for the alcohol liver cirrhosis. 17440116_the level of CYP2E1 is induced by hypertonic condition via TonEBP transactivation. The present study suggests that osmotic status may influence individual responses to the substrate of CYP2E1. 17442289_Observational study of gene-disease association. (HuGE Navigator) 17442289_The CYP2E1 and NAT2 variants associated with COPD 17486761_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17498780_Observational study of gene-disease association. (HuGE Navigator) 17559142_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17559142_examined genetic polymorphisms in the alcohol dehydrogenase 3, aldehyde dehydrogenase 2, and cytochrome P450 2E1 genes in 505 patients with histologically confirmed lung cancer and 256 noncancer controls to determine association with alcohol drinking 17564586_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17577619_Gastric cancer in patients younger than 40 years is closely associated with H. pylori infection, but not with genetic characteristics. 17577619_Observational study of gene-disease association. (HuGE Navigator) 17584020_Chlorzoxazone metabolism in vivo remains the only available method for CYP2E1 phenotyping. 17603900_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17611777_CYP2E1 polymorphism is not associated with the risk of head and neck cancer 17611777_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17617119_CYP2E1 activity cannot be used to distinguish nonalcoholic steatotis patients from steatohepatitis patients. 17627011_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17659824_Genetic polymorphism in CYP2E1 may be responsible for individual differences in susceptibility to liver fibrosis with regard to chronic vinyl chloride monomer exposure. 17659824_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17695473_Observational study of gene-disease association. (HuGE Navigator) 17885617_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17916905_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17935737_Findings suggest that human and cynomolgus monkey CYP2E1 enzymes have high homology in their amino acid sequences, and that their enzymatic properties are considerably similar. 17950035_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17963298_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17963298_The CYP2E1 c2/c2 genotype increases susceptibility to rectal cancer and the gene-environmental interactions between the CYP2E1 polymorphism and smoking or alcohol drinking exist for colorectal neoplasia in general. 17996038_Individuals carrying the *5A or *6 alleles of CYP2E1 are associated with gastric cancer in drinkers 17996038_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18025800_Individuals carrying combinations of CYP2E1*5B, *6 and *7B variants together are likely associated with the risk of developing childhood ALL. 18025800_Observational study of gene-disease association. (HuGE Navigator) 18028774_Observational study of gene-disease association. (HuGE Navigator) 18028774_carrying c2 allele and DD genotype of CYP2E1 conferreded an elevated risk for esophageal squamous cell carcinoma. 18034693_Observational study of gene-disease association. (HuGE Navigator) 18034693_The H6, H7, and H9 haplotypes may play certain roles in different clinical phenotypes in Mexican American alcoholics. In addition, our data suggest that the H1, H2, and H3 haplotypes are associated with alcohol drinking and smoking. 18056994_biochemical analysis of CYP2E1 substrate inhibition 18211048_QSAR analysis for substrate specificity of six CYP isoforms, revealing that CYP2C9 substrates are anionic compounds, while CYP2D6 substrates are cationic, and CYP2E1 substrates are smaller compou | ENSMUSG00000025479 | Cyp2e1 | 8.836447e+01 | 0.5399040 | -0.889225083 | 0.5302796 | 2.691718e+00 | 0.1008710077 | 0.75783482 | No | Yes | 3.009776e+01 | 12.690289 | 6.627195e+01 | 28.308840 |
| ENSG00000130733 | 78992 | YIPF2 | protein_coding | Q9BWQ6 | Acetylation;Endosome;Golgi apparatus;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:78992; | Golgi apparatus [GO:0005794]; Golgi medial cisterna [GO:0005797]; Golgi trans cisterna [GO:0000138]; integral component of membrane [GO:0016021]; late endosome membrane [GO:0031902]; trans-Golgi network [GO:0005802]; transport vesicle [GO:0030133]; small GTPase binding [GO:0031267]; vesicle-mediated transport [GO:0016192] | 31189879_YIPF2 is a new trafficking determinant essential for CD147 glycosylation and transport. YIPF2-controlled ER-Golgi trafficking signature promotes CD147-medated malignant phenotypes in hepatocellular carcinoma. 32303681_YIPF2 promotes chemotherapeutic agent-mediated apoptosis via enhancing TNFRSF10B recycling to plasma membrane in non-small cell lung cancer cells. | ENSMUSG00000032182 | Yipf2 | 2.923003e+03 | 1.3780120 | 0.462588483 | 0.3043007 | 2.320548e+00 | 0.1276750412 | 0.76070573 | No | Yes | 2.934809e+03 | 366.612465 | 2.006960e+03 | 257.538758 | |||
| ENSG00000130734 | 84971 | ATG4D | protein_coding | Q86TL0 | FUNCTION: [Cysteine protease ATG4D]: Cysteine protease that plays a key role in autophagy by mediating both proteolytic activation and delipidation of ATG8 family proteins (PubMed:21177865, PubMed:29458288, PubMed:30661429). The protease activity is required for proteolytic activation of ATG8 family proteins: cleaves the C-terminal amino acid of ATG8 proteins MAP1LC3 and GABARAPL2, to reveal a C-terminal glycine (PubMed:21177865). Exposure of the glycine at the C-terminus is essential for ATG8 proteins conjugation to phosphatidylethanolamine (PE) and insertion to membranes, which is necessary for autophagy (By similarity). In addition to the protease activity, also mediates delipidation of ATG8 family proteins (PubMed:29458288, PubMed:33909989). Catalyzes delipidation of PE-conjugated forms of ATG8 proteins during macroautophagy (PubMed:29458288, PubMed:33909989). Also involved in non-canonical autophagy, a parallel pathway involving conjugation of ATG8 proteins to single membranes at endolysosomal compartments, by catalyzing delipidation of ATG8 proteins conjugated to phosphatidylserine (PS) (PubMed:33909989). ATG4D plays a role in the autophagy-mediated neuronal homeostasis in the central nervous system (By similarity). Compared to other members of the family (ATG4A, ATG4B or ATG4C), constitutes the major protein for the delipidation activity, while it promotes weak proteolytic activation of ATG8 proteins (By similarity). Involved in phagophore growth during mitophagy independently of its protease activity and of ATG8 proteins: acts by regulating ATG9A trafficking to mitochondria and promoting phagophore-endoplasmic reticulum contacts during the lipid transfer phase of mitophagy (PubMed:33773106). {ECO:0000250|UniProtKB:Q8BGV9, ECO:0000250|UniProtKB:Q9Y4P1, ECO:0000269|PubMed:21177865, ECO:0000269|PubMed:29458288, ECO:0000269|PubMed:30661429, ECO:0000269|PubMed:33773106, ECO:0000269|PubMed:33909989}.; FUNCTION: [Cysteine protease ATG4D, mitochondrial]: Plays a role as an autophagy regulator that links mitochondrial dysfunction with apoptosis. The mitochondrial import of ATG4D during cellular stress and differentiation may play important roles in the regulation of mitochondrial physiology, ROS, mitophagy and cell viability. {ECO:0000269|PubMed:19549685, ECO:0000269|PubMed:22441018}. | Alternative splicing;Apoptosis;Autophagy;Cytoplasm;Hydrolase;Mitochondrion;Phosphoprotein;Protease;Protein transport;Reference proteome;Thiol protease;Transport;Ubl conjugation pathway | Autophagy is the process by which endogenous proteins and damaged organelles are destroyed intracellularly. Autophagy is postulated to be essential for cell homeostasis and cell remodeling during differentiation, metamorphosis, non-apoptotic cell death, and aging. Reduced levels of autophagy have been described in some malignant tumors, and a role for autophagy in controlling the unregulated cell growth linked to cancer has been proposed. This gene belongs to the autophagy-related protein 4 (Atg4) family of C54 endopeptidases. Members of this family encode proteins that play a role in the biogenesis of autophagosomes, which sequester the cytosol and organelles for degradation by lysosomes. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:84971; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; cysteine-type peptidase activity [GO:0008234]; apoptotic process [GO:0006915]; autophagy [GO:0006914]; mitophagy [GO:0000423]; protein delipidation [GO:0051697]; protein localization to phagophore assembly site [GO:0034497]; protein transport [GO:0015031]; proteolysis [GO:0006508] | 19549685_Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. 22441018_the import of Atg4D during cellular stress and differentiation may play important roles in the regulation of mitochondrial physiology, reactive oxygen species, mitophagy and cell viability. 23508006_The actions of ATG4 family members (particularly ATG4B) are required for the control of autophagosome fusion with late, degradative compartments in differentiating human erythroblasts. 27265029_This study highlights the transcriptional inactivation mechanisms of ATG2B, ATG4D, ATG9A and ATG9B promoter methylation status and the possible origin of autophagy signal pathway repression in invasive ductal carcinomas. 28834690_study elucidated a novel Malat1-miR-101-STMN1/RAB5A/ATG4D regulatory network that Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma cells 30374741_High ATG4D expression is associated with Colorectal Cancer. 30661429_Human HAP1 and HeLa cells lacking ATG4B exhibit a severe but incomplete defect in LC3/GABARAP processing and autophagy. By further genetic depletion of ATG4 isoforms using CRISPR-Cas9 and siRNA we uncover that ATG4A, ATG4C and ATGD all contribute to residual priming activity, which is sufficient to enable lipidation of endogenous GABARAPL1 on autophagic structures. 33773106_ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. 33988247_Pathogenic variants of ATG4D in infertile men with non-obstructive azoospermia identified using whole-exome sequencing. 34313895_Silencing of ATG4D suppressed proliferation and enhanced cisplatin-induced apoptosis in hepatocellular carcinoma through Akt/Caspase-3 pathway. | ENSMUSG00000002820 | Atg4d | 2.615817e+03 | 1.3306338 | 0.412113639 | 0.3147163 | 1.712447e+00 | 0.1906681947 | 0.78025017 | No | Yes | 3.032788e+03 | 438.348445 | 1.884044e+03 | 279.906303 | |
| ENSG00000130816 | 1786 | DNMT1 | protein_coding | P26358 | FUNCTION: Methylates CpG residues. Preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand, that is essential for epigenetic inheritance. Associates with chromatin during G2 and M phases to maintain DNA methylation independently of replication. It is responsible for maintaining methylation patterns established in development. DNA methylation is coordinated with methylation of histones. Mediates transcriptional repression by direct binding to HDAC2. In association with DNMT3B and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells (PubMed:24623306). Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs) (PubMed:24623306). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing (PubMed:24623306). Promotes tumor growth (PubMed:24623306). {ECO:0000269|PubMed:16357870, ECO:0000269|PubMed:18413740, ECO:0000269|PubMed:18754681, ECO:0000269|PubMed:24623306}. | 3D-structure;Acetylation;Activator;Alternative splicing;Chromatin regulator;DNA-binding;Deafness;Disease variant;Isopeptide bond;Metal-binding;Methylation;Methyltransferase;Neuropathy;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene encodes an enzyme that transfers methyl groups to cytosine nucleotides of genomic DNA. This protein is the major enzyme responsible for maintaining methylation patterns following DNA replication and shows a preference for hemi-methylated DNA. Methylation of DNA is an important component of mammalian epigenetic gene regulation. Aberrant methylation patterns are found in human tumors and associated with developmental abnormalities. Variation in this gene has been associated with cerebellar ataxia, deafness, and narcolepsy, and neuropathy, hereditary sensory, type IE. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:1786; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; pericentric heterochromatin [GO:0005721]; replication fork [GO:0005657]; DNA (cytosine-5-)-methyltransferase activity [GO:0003886]; DNA binding [GO:0003677]; DNA-methyltransferase activity [GO:0009008]; methyl-CpG binding [GO:0008327]; promoter-specific chromatin binding [GO:1990841]; RNA binding [GO:0003723]; zinc ion binding [GO:0008270]; cellular response to amino acid stimulus [GO:0071230]; cellular response to bisphenol A [GO:1903926]; DNA methylation [GO:0006306]; DNA methylation involved in embryo development [GO:0043045]; DNA methylation-dependent heterochromatin assembly [GO:0006346]; maintenance of DNA methylation [GO:0010216]; negative regulation of gene expression [GO:0010629]; negative regulation of histone H3-K9 methylation [GO:0051573]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of vascular associated smooth muscle cell apoptotic process [GO:1905460]; negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching [GO:1905931]; positive regulation of DNA methylation-dependent heterochromatin assembly [GO:0090309]; positive regulation of gene expression [GO:0010628]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of vascular associated smooth muscle cell proliferation [GO:1904707]; Ras protein signal transduction [GO:0007265] | 11834837_recruited to the RARbeta2 promoter by the PML-RAR fusion protein 11932749_DNMT1 and DNMT3b cooperate to silence genes in human cancer cells 12145218_the human de novo enzymes hDNMT3a and hDNMT3b form complexes with the major maintenance enzyme hDNMT1. 12496760_DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. 12530095_DNMT1 gene copy number does not influence susceptibility to development of malignant lymphoproliferative disease. 12538344_Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway 12548018_DNA methylation is tightly coupled to replication through physical interaction of DNMT1 and core components of the replication machinery. 12576480_reduction in DNMT1 triggers intra-S-phase arrest of DNA replication proposed to protect the genome from extensive DNA demethylation. 12594811_Decrease of DNA methyltransferase 1 expression relative to cell proliferation in transitional cell carcinoma. 12637155_mutational inactivation of the DNMT1 gene that potentially causes a genome-wide alteration of DNA methylation status may be a rare event during human carcinogenesis 12738984_Over-expressed in squamous cell carcinoma of the mouth. 12789259_We investigated occupancy of ER-alpha promoter by pRb2/p130-E2F4/5-HDAC1-SUV39 H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 complexes, and provided a link between pRb2/p130 and chromatin-modifying enzymes in the regulation of ER-alpha transcription 12804601_structural homology model for human DNA methyltransferase 1 14559786_DNMT1 plays a key role in methylation maintenance, DNMT3b may act as an accessory to support the function in ovarian cancer cells. 14577574_gene expression as a likely mechansism in underlying causes of changes in DNA methylation in aging and tumorigenesis 14583449_Activation of p53 reduces binding and relieves transcriptional repression of the Dnmt1gene, whereas loss of p53, a frequent, early event in tumorigenesis, may significantly contribute to aberrant genomic methylation. 14684836_hypothesis that the increase of DNA-methyltransferase 1 expression in telencephalic GABAergic interneurons of schizophrenia patients causes a promoter hypermethylation of reelin and GAD(67) and perhaps of other genes expressed in these interneurons 15087453_DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation 15220328_DNMT1 activity contributes to the preservation of the correct organization of large heterochromatic regions 15289832_inhibition of DNA methylation by DNMT1 by an antisense oligodeoxynucleotide influences cell morphology and adhesion 15340041_Expression levels of DNMT1 in tumor cells may affect the effectiveness of doxorubicin in chemotherapy. 15375672_Observational study of gene-disease association. (HuGE Navigator) 15526354_The average mRNA level of Dnmt1 gene from cancerous tissue was higher and that of mbd2 gene from cancerous tissue was lower than that from non-cancerous tissue 15657147_DNMT1- and p53-mediated methylation of the survivin promoter, suggesting cooperation between p53 and DNMT1 in gene silencing. 15735013_Results suggest a novel mechanism for gene silencing mediated by RUNX1/MTG8 and support the combination of HDAC and DNMT inhibitors as a novel therapeutic approach for t(8;21) AML. 15755728_DNMT1 protein levels are elevated in breast cancer tissues and in MCF-7 breast cancer cells relative to normal human mammary epithelial cells without a concomitant increase in DNMT1 mRNA or proliferative fraction. 15762053_Increased DNMT1 gene expression appears to be in lymphomas and maybe associated with oncogenesis in this group of tumors. 15799776_Unlike Dnmt1, pre-existing cytosine methylation at CpG sites or non-CpG sites does not stimulate Dnmt3a activity in vitro and in vivo. 15870198_STAT3 may transform cells by inducing epigenetic silencing of SHP-1 in cooperation with DNMT1 and histone deacetylase 1 15956212_Direct role of Dnmt1 in the restoration of epigenetic information during DNA repair. 16053511_These data suggest that increased DNMT1 protein expression participates in multistage pancreatic carcinogenesis from the precancerous stage to malignant progression of ductal carcinomas and may be a biological predictor of poor prognosis. 16227093_necessary for both class switching and somatic hypermutation in B cells 16314526_DNMT1 is necessary for proper PcG body assembly independent of DNMT-associated histone deacetylase activity. 16322686_Breast cancer cells have a prominent loss of DNA methylation accompanied by altered expression of maintenance DNA methyltransferase DNMT1. 16423997_data suggests a potential role for DNMT1 in the initiation of promoter CpG island hypermethylation in human cancer cells 16424002_human cancer cells may differ in their reliance on DNMT1 for maintaining DNA methylation 16497664_down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells 16537562_DNMT1 protein overexpression may be responsible for aberrant DNA methylation during multistage carcinogenesis of the pancreas 16769694_In human cells maintenance of XIST methylation is controlled differently than global genomic methylation and in the absence of both DNMT1 and DNMT3B. 16801630_Inhibitors of DNMT1 may have clinical relevance for immune modulation by augmentation of cytokine effects and/or expression of tumor-associated antigens. 16861352_STAT3 binds in vitro to 2 STAT3 SIE/GAS-binding sites identified in promoter 1 and enhancer 1 of the DNMT1 gene 16897079_suppression of DNMT-1 might be related to the pathogenesis of atopic dermatitis, especially in whom serum IgE level is high 16960727_study demonstrated that expression of DNMT1 is clearly regulated in both impaired spermatogenesis and development of embryonal carcinoma, while HDAC1 expression is not regulated during aberrant germ cell differentiation 16963560_Data suggest that DNMT1 might be essential for maintenance of DNA methylation, proliferation, and survival of cancer cells. 16998846_Sex-specific time windows for concomitant upregulation of DNMT1 are associated with prenatal remethylation of the human male and female germ line. 17015478_depletion of DNMT1 with either antisense or small interfering RNA (siRNA) specific to DNMT1 activates a cascade of genotoxic stress checkpoint proteins 17030625_AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability 17053888_The increased DNMT1 expression was more frequent in adenocarcinoma vs. squamous cell carcinoma (42% vs 19%). Smokers with ~65 packyears showed 4.17 times higher risk of increased DNMT1 expression compared to those who smoked 45 packyears in adenocarcinoma 17067458_DNMT1 expression was correlated to the methylation of RASSF1A, tumor grade and stage, which implied that DNMT1 may contribute to the carcinogenesis and development of adenoid cystic carcinoma. 17081533_The genes DNMT1, DNMT3A, and DNMT3B were over-expressed in the ectopic endometrium as compared with normal control subjects or the eutopic endometrium of women with endometriosis. 17085482_Direct cooperation between DNMT1 and G9a provides a mechanism of coordinated DNA and H3K9 methylation during cell division. 17093909_These findings suggested that DNMT1 was associated with the malignant phenotype, and dysregulation of DNMT1 expression was present in tumor cells of colorectal cancer. 17178861_LMP1 induces formation of a transcriptional repression complex, composed of DNMT1 and histone deacetylase, which locates on E-cadherin gene promoter. 17312023_DNMT1 is essential for the maintenance of DNA methylation patterns in human cells. 17322882_DNMT1 is required for faithfully maintaining DNA methylation patterns in human cancer cells and is essential for their proliferation and survival. 17470536_Direct interactions between HP1 and DNMT1 mediate silencing of euchromatic genes. 17492476_LAT, ZAP-70, and DNMT1 protein levels in CD4(+) T cells can be associated with systemic lupus erythematosus. 17532557_DNMT1 plays a key role in maintenance of methylation, and DNMT3B may act as an accessory DNA methyltransferase to epigenetically silence CXCL12 expression in MCF-7 and AsPC1 cells 17538945_Progressive up-regulation of the gene encoding DNMT1 was found in the colorectal adenoma-carcinoma sequence. 17657744_SUV39H1 is significantly associated with DNMT1, but not with euchromatic promoter methylation in colorectal cancer 17673620_data suggest that UHRF1 may help recruit DNMT1 to hemimethylated DNA to facilitate faithful maintenance of DNA methylation 17698033_mahanine can reverse an epigenetically silenced gene, RASSF1A in prostate cancer cells by inhibiting DNMT activity that in turn down-regulates a key cell cycle regulator, cyclin D1 17716861_These results suggested that PKB enhanced DNMT1 stability and maintained DNA methylation and chromatin structure, which might contribute to cancer cell growth. 17893234_DNA methyltransferase (DNMT) 1 over-expression is not a secondary result of increased cell proliferative activity but is significantly correlated with the CpG island methylator phenotype. 17931718_establish a link between HESX1 and DNMT1 and suggest a novel mechanism for the repressing properties of HESX1 17934516_The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. 17965595_DNMT1 expression is increased in schizophrenia (SZ) and bipolar (BP) disorder brains. [REVIEW] 17991895_reveals novel functions for DNMT1 as a component of the cellular response to DNA damage, which may help optimize patient responses to this agent in the future 18038118_mutant p53 loses its ability to suppress DNMT1 expression, and thus enhances methylation levels of the p16 ( ink4A ) promoter and subsequently down-regulates p16(ink4A )protein. 18049164_In hepatocellular carcinoma, DNMT1 is necessary to maintain the methylation of CpG islands in certain tumor-related genes. 18198215_DNMT-1 has a direct suppressive role in 15-LOX-1 transcriptional silencing that is independent of 15-LOX-1 promoter DNA methylation 18202356_Parental Dnmt1 is a modifier of transmission of alleles at an unlinked chromosomal region and perhaps has a role in the genesis of transmission ratio distortion. 18204201_The subcellular localization of DNMT1 can be altered by the addition of IL-6, and this process is greatly enhanced by phosphorylation of the DNMT1 nuclear localization signal (NLS) by PKB/AKT. 18252747_Study shows that the maintenance DNA methyltransferase Dnmt1, which preserves the patterns of CpG methylation, plays a key role in CAG repeat instability in human cells and in the male and female mouse germlines. 18253830_DNMT1 was over expressed in gastric neoplasms. 18404674_DNMT1 protein overexpression might contribute to aberrant DNA hypermethylation of specific tumor suppressor genes in endometrial cancers. 18413740_Chromatin immunoprecipitation analysis of the BAG-1 promoter in DNMT1-overexpressing or DNMT3B-overexpressing colon cells show a permissive chromatin status assoscsiated with DNA binding of BORIS. 18414412_DNMT1 may play an important role in modulating NSCLC patient survival and thus be useful for identifying NSCLC patients who would benefit most from aggressive therapy. 18499700_Observational study of gene-disease association. (HuGE Navigator) 18499700_The association between sequence variants of DNMT1 and 3B and mutagen sensitivity induced by BPDE supports the involvement of these DNMTs in protecting the cell from DNA damage. 18505931_histone deacetylase inhibition promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells 18563322_DNMT1 and DNMT3b will increase their biological effects and have a synergistic effect on suppressing the growth of cholangiocarcinoma 18567946_The HIV-1 responsive element resides in the 5' most 420 bp of the -1634 to +71 DNMT1 promoter; positioning of this truncated promoter proximal to a hybrid SV40-DNMT1 reporter results in HIV-1-dependent regulation 18680430_study shows endogenous DNMT1 & CFP1 interact in vivo; CFP1 interaction with Setd1A or Setd1B not required for its interaction with DNMT1; result indicates CFP1 intersects cytosine methylation machinery independently of its association with Setd1 complexes 18754681_The CXXC domain in the amino terminus region of DNMT1 cooperates with the catalytic domain for DNA methyltransferase activity. 18829110_elevated DNMT1 expression may in particular contribute to ineffective erythropoiesis in MDS. 18931722_DNMT1 and HLA-DRalpha are prognostic factors for hepatocelluar carcinoma. 19016755_Interaction between DNMT1 and DNA replication reactions in the SV40 in vitro replication system is reported. 19019634_The ratio of MBD-2/DNMT-1 might be valuable in explanation of hypomethylation and evaluation of clinical activity of systemic luopus erythematosus. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19124506_Observational study of gene-disease association. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19173286_mapped the dimerization domain to the targeting sequence TS that is located in the center of the N-terminal domain (amino acids 310-629) 19211935_miR-29b down-regulates DNMT1 indirectly by targeting Sp1. 19236003_Incorporation of 6-thioguanine into the CpG site affects the methylation of the cytosine residue by both DNMT1 and HpaII. The effect on cytosine methylation is dependent on the position of 6-thioguanine with respect to the cytosine to be methylated. 19246518_Observational study of gene-disease association. (HuGE Navigator) 19275888_p21(WAF1)-p300-DNMT1 pathway may play a pivotal role to ensure regulated DNMT1 expression and DNA methylation in mammalian cell division. 19279403_Panobinostat treatment depletes DNMT1 levels and enhances decitabine mediated repression of JunB and loss of survival of human acute leukemia cells. 19282482_signaling through SET7 represents a means of DNMT1 enzyme turnover 19292979_DNMT1 levels in tumor cells may determine the effectiveness of doxorubicin in chemotherapy. 19339266_EBV latent membrane protein 2A (LMP2A) up-regulated DNMT1, leading to an increase in methylation of the PTEN promoter in stomach cancer. 19386473_In patient with schizophrenia the DNA-methyltransferase 1 was up regulation in telencephalic GABAergic neurons. 19394279_Data showed that the CD70, perforin and KIR2DL4 promoters are demethylated in CD4(+)CD28(-) T cells, and that DNA methyltransferase 1 (Dnmt1) and Dnmt3a levels are decreased in this subset. 19399408_Silencing DNMT3b and DNMT1 could inhibit the cell growth and promote apoptosis of bladder carcinoma cells. 19399937_DNA methyltransferase 1 (DNMT1) expression was increased in hepatocellular carcinoma compared to non-neoplastic liver tissues and the incidence of DNMT1 immunoreactivity in HCCs correlated significantly with poor tumor differentiation. 19424621_Higher expression of DNMT1 and HDAC1 correlated with advanced stages of the disease and reflect the malignancy of pancreatic carcinoma. 19450230_SUMOylation significantly enhances the methylase activity of DNMT1 both in vitro and in chromatin. 19465937_the role of promoter region DNA hypermethylation as a mediator of gene silencing in glioblastoma multiforme cell line using RNAi directed against DNMT1 and DNMT3b was investigated. 19468253_Our results were not able to demonstrate a clear correlation between DNMT1 and DNMT3a immunoexpression and salivary gland neoplasms development. 19531770_Increase of DNMT1 expression is associated with multiple myeloma and plasma cell leukemias. 19539327_genetic and phenotypic consequences of silencing DNMT1 in PC3 cells are markedly different from those in colon and gastric cancers, indicating that DNMT1 preferentially targets certain gene promoters 19763880_The expression of DNMT1, DNMT2, DNMT3A and DNMT3B in pediatric acute lymphoblastic leukemia patients, was investigated. 19798569_Our data suggest there is no apparent association of common DNMT-1 and DNMT-3B polymorphisms with the risk of breast cancer in Chinese women. 19896490_Observational study of gene-disease association. (HuGE Navigator) 19932585_Our results indicate that DNMT1 plays the main role in maintenance of methylation of CXCR4 promoter, while DNMT3B may function as an accessory DNA methyltransferase to modulate CXCR4 expression in AsPC1 cells. 19936946_Observational study of gene-disease association. (HuGE Navigator) 19957555_Downregulation of DNMT1 can inhibit cervical cancer cell proliferation and induce cell apoptosis. 20044957_The DNMT 1 is the key enzyme responsible for DNA methylation, which often occurs in CpG islands located near the regulatory regions of genes and affects transcription of specific genes. Two intron polymorphisms of DNMT1 might affect HBV clearance. 20071334_DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta 20071580_Data suggest that KSHV miRNA targets multiple pathways to maintain KHSV latency, including repression of the viral protein Rta and a cellular factor, Rbl2, and increased cellular DNMT, in regulating global epigenetic reprogramming. 20081831_DNMT1 is essential for epidermal progenitor cell function 20093774_Carcinogne-induced DNMT1 accumulation and subsequent hypermethylation of the promoter of tumor suppressor genes may lead to tumorigenesis and poor prognosis. 20147412_Hepatitis B virus-induced overexpression of DNMTs leads to viral DNA methylation and decreased viral gene expression and also leads to methylation of host CpG islands. 20192566_No association between DNMT1 and colorectal cancer in Iranian patients. 20192608_No association between DNMT1 polymorphisms and gastric cancer. 20192608_Observational study of gene-disease association. (HuGE Navigator) 20228804_Dnmt1 and Dnmt3a transgenes are required for synaptic plasticity, learning and memory through their overlapping roles in maintaining DNA methylation and modulating neuronal gene expression in adult central nervous system neurons. 20335008_Our results suggest that one potential mechanism by which varenicline may decrease cigarette smoking in schizophrenia is by decreasing DNMT1 mRNA. 20354000_Our results indicate that DNMT-mediated gene silencing may play a role in inflammation-associated colon tumorigenesis. 20381114_expression of DNA methyltransferase 1, 3a, and 3b showed significantly higher levels in stage IV tumors than in stage I or II tumors 20398055_DNMT1 and DNMT3b silencing sensitizes human hepatoma cells to TRAIL-mediated apoptosis via up-regulation of TRAIL-R2/DR5 and caspase-8. 20428781_thymine DNA glycosylase was not up-regulated in DNMT1 nor 3B knockdown cancer cells 20570896_Data provided compelling evidence that deregulation of DNMT1 is associated with gain of transcriptional activation of Sp1 and/or loss of repression of p53. 20588031_Increased expression of DNMT1 in non-neoplastic epithelium may precede or be a relatively early event in ulcerative colitis-associated tumorigenesis. 20592467_interaction between hNaa10p and DNMT1 was required for E-cadherin silencing through promoter CpG methylation, and E-cadherin repression contributed to the oncogenic effects of hNaa10p. 20593030_Observational study of gene-disease association. (HuGE Navigator) 20613874_results reveal that the disruption of Dnmt1/PCNA/UHRF1 interactions acts as an oncogenic event and that one of its signatures (i.e. the low level of mMTase activity) is a molecular biomarker associated with a poor prognosis in GBM patients 20620135_Regulation of DNMT1 and DNMT3A by HBx promoted hypermethylation of p16(INK4A) promoter region 20840813_The expression of DNMT1 was significantly higher in lung cancer tissues than in corresponding normal lung tissues. 20875141_In MCF7 cells, ANT2 knockdown enhanced the expression and activity of DNA methyltransferase 1 (DNMT1). 20920981_Observational study of gene-disease association. (HuGE Navigator) 20920981_The current work shows that polymorphisms of the DNMT1 gene in exons may affect the individual intraductal carcinoma risk in women of the northeast of China. 20937307_The anti-TNFalpha biological agents do not seem to affect DNA methylation and mRNA expressions of DNMT1 and MBD2 in RA 20940144_our studies provide compelling additional evidence for DNMT1 acting as a regulator of genome integrity and as an early responder to DNA DSBs. 20951977_lower levels of DNMT1 may be related with smoking habit. significantly higher mean percentage of DNMT1 immunoreactivity in non-smokers 20970125_The mRNA of DNMT1 (DNA-methyltransferase 1) is expressed in the endometrium across the menstrual cycle. 20980350_Depletion of DNMT1 did not induce cellular invasion in MCF-7 and ZR-75-1 non-invasive breast cancer cell lines. 21042757_HPV-16 E6 may act through p53/ DNMT1 to regulate the development of cervical cancer. 21045206_a previously unknown mode of regulation of DNMT1 protein stability through the coordinated action of an array of DNMT1-associated proteins. 21078759_This study demonstrated that patients with SLE had a significantly lower level of DNA methylation than the controls, and that expression of both DNMT1 and MBD2 mRNA was significantly increased in the SLE patients compared with controls. 21151116_Modifications to DNMT1 mediated by AKT1 and SET7, affect cellular DNMT1 levels. 21163962_structure of DNMT1 composed of CXXC, tandem bromo-adjacent homology (BAH1/2), and methyltransferase domains bound to DNA-containing unmethylated CpG sites; studies identify an autoinhibitory mechanism of DNA methylation 21229291_we report significant overexpression of DNMT1 and DNMT3B in glioma 21266713_Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. 21268065_Findings suggest that, by balancing Dnmt1 ubiquitination, Usp7 and Uhrf1 fine tune Dnmt1 stability. 21296890_association of Dnmt1 and alpha-synuclein might mediate aberrant subcellular localization of Dnmt1 21311766_Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. 21316665_Expression level of DNMT1 is significantly higher in secretory-phase endometrium compared with proliferative endometrium and menstrual endometrium. 21321201_Mitochondrial DNMT1 appears to be responsible for mtDNA cytosine methylation. 21347439_DNA methylation recovery was mediated by the major human DNA methyltransferase, DNMT1 21353307_Results implied that IL-6 expression was regulated by promoter demethylation induced by down-regulation of DNMT activity. 21389349_the naked DNA- and polynucleosome-binding activities of Dnmt1 are inhibited by the RFTS domain, which functions by virtue of binding the catalytic domain to the exclusion of DNA 21395176_Increased expression of DNMT1 may initiate the oncogenesis of laryngeal squamous cell carcinoma. Smoking may induce expression. 21458988_Low DNMT1 expression defines a subgroup of GC patients with better outcomes following platinum/5FU-based neoadjuvant chemotherapy. In vitro data support a functional relationship between DNMT1 and cisplatin sensitivity. 21459093_Study provides evidence for nuclear receptor mediated regulation of Dnmt1 expression through ERRgamma and SHP crosstalk. 21478913_These findings show an important role for p53 in the progression of serous borderline ovarian tumors to an invasive carcinoma, and suggest that downregulation of E-cadherin by DNMT1-mediated promoter methylation contributes to this process. 21523767_DNMT1 could be a significant clinical predictor for stage and treatment response of bladder cancer. 21532572_Here we show that mutations in DNMT1 cause both central and peripheral neurodegeneration in one form of hereditary sensory and autonomic neuropathy with dementia and hearing loss. 21539677_The proportion of cells that express DNMT1 at the mRNA and protein levels in esophageal squamous cell carcinoma clinical samples is generally higher than in paired non-cancerous tissues. 21565170_these results suggest that dysregulation of cell cycle via CDKs could induce abnormal phosphorylation of DNMT1 and lead to DNA hypermethylation often observed in cancer cells. 21565830_findings describe a new mechanism for the regulation of DNMT1 and aberrant DNA hypermethylation in colorectal cancer 21573703_DNA methylation of the BNIP3 promoter was mediated by DNMT1 via the MEK pathway 21592522_Data show that the levels of DNMT1 mRNA were significantly decreased in a depressive but not in a remissive state of MDD and BPD. 21619587_Results indicate that phosphorylation of human DNMT1 by protein kinase C is isoform-specific and provides the first evidence of cooperation between PKCzeta and DNMT1 in the control of the DNA methylation patterns of the genome. 21636528_These results offer an explanation for how and why unmethylated microsatellite repeats can be destabilized in cells with decreased DNMT1 levels and uncover a novel and important role for PARP in this process. 21735817_DNMT1 as a key enzyme in maintaing of proper methylation pattern is a attractive target in anti-tumor therapy. 21756783_PARP1 could regulate DNA methylation by inhibiting the enzyme activity of DNMT1 in bronchial epithelial cells exprosed to B(a)P. 21757290_Data show that Core inhibited p16 expression by inducing promoter hypermethylation via up-regulation of DNMT1 and DNMT3b. 21791605_Data show that trichostatin A treatment reduces global DNA methylation and the DNMT1 protein level and alters DNMT1 nuclear dynamics and interactions with chromatin. 21826395_Dnmt1 was principally expressed in the nucleus & cytoplasm of NeuN-positive neurons, but not in GFAP-positive astrocytes. Levels were significantly increased in patients with temporal lobe epilepsy. 21887463_Significantly higher levels of CXCR4, DNMT3A, DNMT3B and DNMT1 transcript (p=0.0058, 0.0163, 0.0003 and <0.0001, respectively) levels in cancer tissue as compared to normal samples. 21947282_SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities 21962230_MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma 21966451_Results suggest that DNMT1 operates either as a functional intermediary or in cooperation with E2F1 inhibiting AR gene expression in a methylation independent manner. 21999220_DNMT1 silencing is associated with malignant phenotype and methylated tumor suppressor gene expression in cervical cancer 22048249_analysis of DNMT1, expression showed peaks of mRNA transcripts in primary spermatocytes and in mature ejaculated spermatozoa, with DNMT1 transcript level being the most abundant in all cell stages. I 22072770_Elevated expression of DNMT1 or DNMT3B alone is not required to maintain restricted latency. 22110720_DNMT1 and DNMT3a are regulated by GLI1 in pancreatic cancer, and DNMT1 is its direct target gene 22167392_DNMT1 was over-expressed in primary tumors and cell lines, while knockdown of DNMT1 using siRNA could decrease methylation level of miR-148a promoter and restore its expression 22219193_Regulation of human RNA polymerase III transcription by DNMT1 and DNMT3a DNA methyltransferases. 22236544_Increased expression of DNMT1 was observed in oral lichen planus compared to the control group 22264301_DNMT1 hypermethylation is a late part of glioma progression. 22295098_reduced microRNA-152 can lose an inhibitory effect on DNA methyltransferase, which leads to hypermethylation of the ERalpha gene and a decrease of ERalpha level 22301400_Increased DNMT1 gene expression is associated with carcinogenesis in pancreatic ductal adenocarcinoma. 22317856_supported the idea that NNK-induced DNMT1 expression may result from protein stabilization. Increased DNMT1 protein expression may play a critical role in the malignant progression of larynx. 22328086_Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. 22349384_Decreased expression levels of DNMT1, DNMT3a, and DNMT3b in the ectopic endometrium and eutopic endometrium may play a role in patients with abnormal epigenetics which may lead to endometriosis. 22362755_In human hepatocellular carcinoma, increased DNMT1 expression is negatively correlated with SHP levels. 22403725_Wnt7a is lost by methylation in a subset of tumors and that this methylation is maintained by DNMT1 22455563_MeCP2, DNMT1 and H4Ac expression levels did not correlate with any of tested clinicopathological parameters in breast invasive ductal carcinoma tissues 22520950_data suggest that overexpression of DNMT1, DNMT3a, and DNMT3b might represent a critical event responsible for the epigenetic inactivation of multiple tumor suppressor genes, leading to the development of aggressive forms of sporadic breast cancer 22547080_Pregnancy and the DNMT1a genotype independently influence the arsenic methylation phenotype 22739025_induction of DNMT1 expression in the chronically inflamed colon may release IL-6 signaling towards signal transducer and activator of transcription 3 from inhibition through SOCS3 increasing the propensity to malignant transformation. 22768205_The data from this study provided the first evidence for differential expression of DNMTs proteins in ovarian cancer tissues and their associations with clinicopathological and survival data in sporadic ovarian cancer patients. 22795133_Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells 22879518_MT-ND6 transcriptional regulation dependent on the enhanced expression of the mitochondrial-targeted isoform of the DNA (cytosine-5) methyl transferase 1 (DNMT1). 22894906_A review of the various regulatory mechanisms and interactions of DNMT1. 22898998_increased Dnmt1 expression in DDX20-deficient cells hypermethylated the promoters of metallothionein genes, resulting in decreased metallothionein expression leading to enhanced NF-kappaB activity 22942708_DNMT1, DNMT2 and DNMT3A may play important roles in gastric cancer carcinogenesis. 22975348_these data demonstrated for the first time that ERalpha could upregulate DNMT1 expression by directly binding to the DNMT1 promoter region in MFC-7(ER )/PTX cells. 23039890_The DNMT1+32204GG genotype correlates with DNA hypomethylation. 23041765_Ciprofloxacin has antifibrotic actions in Systemic sclerosis dermal and lung fibroblasts via the downregulation of Dnmt1, the upregulation of Fli1. 23049933_Results suggested that SNPs of DNMT1 could be used as genotypic markers for predicting genetic susceptibilities to H.pylori infection and risks in gastric atrophy. 23064049_results indicate DNMT1 is essential for maintenance of colon cancer stem-like cells/cancer-initiating cells and short-term suppression of DNMT1 might be sufficient to disrupt cancer stem-like cells/cancer-initiating cells 23072722_Data suggest that LSD1 (lysine-specific demethylase 1) is critical in the regulation of cell proliferation, but not an absolute requirement for the stabilization of either p53 or DNMT1(DNA methyltransferase 1). 23079992_Two SNPs, rs16999593 in DNMT1 and rs2424908 in DNMT3B, are significantly associated with breast cancer risk. 23125218_Upregulation of DNMT1 is associated with neoplastic transformation. 23127209_Abnormal expression levels of DNMT1 and methyl-CpG binding domain protein 2 demethylase mRNA may be important causes of the global hypomethylation observed in CD4+T cells in systemic lupus erythematosus. 23242655_mutant p53 protein binds to the promoter of ESR1 through direct interaction with HDAC1 and indirect interaction with DNMT1, MeCP2 proteins in the ER-negative MDA-MB-468 cells. 23249948_HBP1 represses the DNMT1 gene through binding a high-affinity site in the DNMT1 promoter. 23254386_Higher levels of expression of DNMT1 in prostate cancer cells is associated with a more aggressive phenotype. 23318422_were involved in ovarian cancer cisplatin resistance in vitro and in vivo by targeting DNMT1 directly. 23364257_Both expression level and activity of DNMT1 were inversely correlated with the expression level of BNIP3 in colon carcinoma cells after treatment with chemotherapeutic agents and radiation 23365052_DNMT1 mutations in patients presenting with familial frontotemporal dementia or primary memory decline may also have sensory neuropathy and hearing loss, or phenotype for narcolepsy. 23393137_mammalian DNMT1, DN | ENSMUSG00000004099 | Dnmt1 | 1.337774e+04 | 1.1590770 | 0.212976456 | 0.2517335 | 7.252423e-01 | 0.3944297112 | 0.84317192 | No | Yes | 1.396529e+04 | 1045.061122 | 1.157424e+04 | 888.627355 | |
| ENSG00000130827 | 55558 | PLXNA3 | protein_coding | P51805 | FUNCTION: Coreceptor for SEMA3A and SEMA3F. Necessary for signaling by class 3 semaphorins and subsequent remodeling of the cytoskeleton. Plays a role in axon guidance in the developing nervous system. Regulates the migration of sympathetic neurons, but not of neural crest precursors. Required for normal dendrite spine morphology in pyramidal neurons. May play a role in regulating semaphorin-mediated programmed cell death in the developing nervous system. Class 3 semaphorins bind to a complex composed of a neuropilin and a plexin. The plexin modulates the affinity of the complex for specific semaphorins, and its cytoplasmic domain is required for the activation of down-stream signaling events in the cytoplasm. | Cell membrane;Disulfide bond;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of the plexin class of proteins. The encoded protein is a class 3 semaphorin receptor, and may be involved in cytoskeletal remodeling and as well as apoptosis. Studies of a similar gene in zebrafish suggest that it is important for axon pathfinding in the developing nervous system. This gene may be associated with tumor progression. [provided by RefSeq, Aug 2013]. | hsa:55558; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; semaphorin receptor complex [GO:0002116]; semaphorin receptor activity [GO:0017154]; axon guidance [GO:0007411]; branchiomotor neuron axon guidance [GO:0021785]; facial nerve structural organization [GO:0021612]; hippocampus development [GO:0021766]; negative chemotaxis [GO:0050919]; negative regulation of axon extension involved in axon guidance [GO:0048843]; negative regulation of cell adhesion [GO:0007162]; neuron projection extension [GO:1990138]; neuron projection guidance [GO:0097485]; positive regulation of axonogenesis [GO:0050772]; positive regulation of cytoskeleton organization [GO:0051495]; pyramidal neuron development [GO:0021860]; regulation of cell migration [GO:0030334]; regulation of cell shape [GO:0008360]; regulation of GTPase activity [GO:0043087]; semaphorin-plexin signaling pathway [GO:0071526]; semaphorin-plexin signaling pathway involved in axon guidance [GO:1902287]; trigeminal nerve structural organization [GO:0021637] | 19480842_Data show that the expression of Sema3A receptors (neuropilin-1 (NRP-1), NRP-2, plexin A1, plexin A2, and plexin A3) significantly increased during M-CSF-mediated differentiation of monocytes into macrophages. 21925246_in vitro analysis on PLXNA3 also suggest that this gene may have some form of growth suppressive role in breast cancer, in addition to a similar role for the gene previously reported in ovarian cancer. 25518740_Data indicate that plexin A1-4 (PLXNA1-4) mediation of neuroanatomical traits can be detected using in vivo neuroimaging techniques. 28536997_Exome analysis of Kuwaiti multiple sclerosis patients revealed a missense variant (rs5945430) in Plexin A3 (PLXNA3) gene (Xq28) associated with male-specific MS severity and disability. 34740135_Semaphorin-Plexin Signaling: From Axonal Guidance to a New X-Linked Intellectual Disability Syndrome. | ENSMUSG00000031398 | Plxna3 | 2.683110e+03 | 0.9488335 | -0.075773144 | 0.2625386 | 8.384505e-02 | 0.7721528019 | 0.95442153 | No | Yes | 1.819553e+03 | 354.986016 | 2.221739e+03 | 444.570709 | |
| ENSG00000130985 | 7317 | UBA1 | protein_coding | P22314 | FUNCTION: Catalyzes the first step in ubiquitin conjugation to mark cellular proteins for degradation through the ubiquitin-proteasome system (PubMed:1606621, PubMed:1447181, PubMed:33108101). Activates ubiquitin by first adenylating its C-terminal glycine residue with ATP, and thereafter linking this residue to the side chain of a cysteine residue in E1, yielding a ubiquitin-E1 thioester and free AMP (PubMed:1447181). Essential for the formation of radiation-induced foci, timely DNA repair and for response to replication stress. Promotes the recruitment of TP53BP1 and BRCA1 at DNA damage sites (PubMed:22456334). {ECO:0000269|PubMed:1447181, ECO:0000269|PubMed:1606621, ECO:0000269|PubMed:22456334, ECO:0000269|PubMed:33108101}. | 3D-structure;ATP-binding;Acetylation;Alternative initiation;Cytoplasm;Direct protein sequencing;Disease variant;Ligase;Mitochondrion;Neurodegeneration;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:1447181, ECO:0000269|PubMed:1606621}. | The protein encoded by this gene catalyzes the first step in ubiquitin conjugation to mark cellular proteins for degradation. This gene complements an X-linked mouse temperature-sensitive defect in DNA synthesis, and thus may function in DNA repair. It is part of a gene cluster on chromosome Xp11.23. Alternatively spliced transcript variants that encode the same protein have been described. [provided by RefSeq, Jul 2008]. | hsa:7317; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; RNA binding [GO:0003723]; ubiquitin activating enzyme activity [GO:0004839]; cellular response to DNA damage stimulus [GO:0006974]; protein modification by small protein conjugation [GO:0032446]; protein ubiquitination [GO:0016567]; ubiquitin-dependent protein catabolic process [GO:0006511] | 11447495_Observational study of gene-disease association. (HuGE Navigator) 15202508_In conclusion, nUBE1L is a novel human E1 like gene highly expressed in adult testis, which plays key role in ubiquitin system, and accordingly influences spermatogenesis and male fertility. 16595681_analysis of ATP.Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme 17060614_unusual type of regulation at the level of the E1 enzyme is likely to affect numerous cellular processes and may represent a strategy to coordinate multiple phenotypic changes upon differentiation by using E1 as a 'master switch.' 17597759_Human Uba6 and Ube1 have distinct preferences for E2 charging in vitro, and their specificity depends in part on their C-terminal ubiquitin-fold domains, which recruit E2s 18179898_Large-scale mutation analysis in genes resulted in detection of 3 rare novel variants in exon 15 of UBE1 that segregate with X-linked infantile spinal muscular atrophy. 18661401_high-level expression of UBE1 was seen inextranodal NK/T cell lymphoma, nasal type; there was no significant difference between UBE1 expression and patients' outcome 19723899_Observational study of gene-disease association. (HuGE Navigator) 20653130_Ubiquitin and UBE1 are upregulated in tic epilepsy. 22279528_Data show that largazole and ester/keto analogs inhibit ubiquitin E1 activation. 22370482_Decrease in free ubiquitin levels under stress allows NEDD8 to be conjugated through Ube1. 22456334_UBA1 knockdown impairs formation of ubiquitin conjugates at DNA double-strand breaks. 22999844_Diminished ubiquitylation phenotype observed in enteropathogenic Escherichia coli infected cells corresponds to a strong reduction in the abundance of both Ube1 and Uba6. 25138535_Ubiquitin-activating enzyme is necessary for 17beta-estradiol-induced breast cancer cell proliferation and migration. 25207809_A ubiquitin shuttle DC-UbP reconciles protein ubiquitination and deubiquitination via linking UbE1 and USP5 enzymes. 25209502_We expressed and purified the N-terminal domains of human E1 and determined their crystal structures, which contain inactive adenylation domain (IAD) and the first catalytic cysteine half-domain (FCCH) 25768649_Conjugation of the ubiquitin activating enzyme UBE1 with the ubiquitin-like modifier FAT10 targets it for proteasomal degradation 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that UBA1 is a putative expression variability marker for the 3-day, 8-cell embryo stage. 26432019_recent findings put UBA1 at the center of cellular homeostasis and neurodegeneration [review] 28134249_Polyubiquitination and proteasomal degradation of ezrin and CUGBP1 require Uba6, but not Uba1, and that Uba6 is involved in the control of ezrin localization and epithelial morphogenesis. These data suggest that distinctive substrate pools exist for Uba1 and Uba6 that reflect non-redundant biological roles for Uba6. 30279270_hUBA1 shares a conserved overall structure and mechanism with previously characterized yeast orthologs, but displays subtle structural differences, particularly within the active site. 31700050_Differential Inhibition of Human and Trypanosome Ubiquitin E1S by TAK-243 Offers Possibilities for Parasite Selective Inhibitors. 32315024_Ubiquitination-activating enzymes UBE1 and UBA6 regulate ubiquitination and expression of cardiac sodium channel Nav1.5. 33108101_Somatic Mutations in UBA1 and Severe Adult-Onset Autoinflammatory Disease. 33355669_Herpes simplex virus 1 infection induces ubiquitination of UBE1a. 33779074_Somatic Mutations in UBA1 Define a Distinct Subset of Relapsing Polychondritis Patients With VEXAS. 34048852_Adult-onset autoinflammation caused by somatic mutations in UBA1: A Dutch case series of patients with VEXAS. 34074684_Somatic Mutation in UBA1 and ANCA-associated Vasculitis. 34213531_Atypical splice-site mutations causing VEXAS syndrome. 34301924_GRP78 determines glioblastoma sensitivity to UBA1 inhibition-induced UPR signaling and cell death. 34344988_Prevalence of UBA1 mutations in MDS/CMML patients with systemic inflammatory and auto-immune disease. 34391501_Vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic (VEXAS) syndrome: fevers, myalgia, arthralgia, auricular chondritis, and erythema nodosum. 34474632_Looking for somatic mutations in UBA1 in patients with chronic myelomonocytic leukemia associated with systemic inflammation and autoimmune diseases. 35094194_UBA1 gene mutation in giant cell arteritis. | ENSMUSG00000001924 | Uba1 | 5.026891e+04 | 1.2653253 | 0.339508387 | 0.3004094 | 1.285068e+00 | 0.2569588215 | 0.79605964 | No | Yes | 5.244828e+04 | 5111.651479 | 3.799271e+04 | 3797.329529 |
| ENSG00000131018 | 23345 | SYNE1 | protein_coding | Q8NF91 | FUNCTION: Multi-isomeric modular protein which forms a linking network between organelles and the actin cytoskeleton to maintain the subcellular spatial organization. As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex involved in the connection between the nuclear lamina and the cytoskeleton. The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning. May be involved in nucleus-centrosome attachment and nuclear migration in neural progenitors implicating LINC complex association with SUN1/2 and probably association with cytoplasmic dynein-dynactin motor complexes; SYNE1 and SYNE2 may act redundantly. Required for centrosome migration to the apical cell surface during early ciliogenesis. May be involved in nuclear remodeling during sperm head formation in spermatogenenis; a probable SUN3:SYNE1/KASH1 LINC complex may tether spermatid nuclei to posterior cytoskeletal structures such as the manchette. {ECO:0000250|UniProtKB:Q6ZWR6, ECO:0000269|PubMed:11792814, ECO:0000269|PubMed:18396275}. | 3D-structure;Actin-binding;Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Differentiation;Disease variant;Disulfide bond;Emery-Dreifuss muscular dystrophy;Golgi apparatus;Membrane;Neurodegeneration;Nucleus;Phosphoprotein;Reference proteome;Repeat;Spermatogenesis;Transmembrane;Transmembrane helix | This gene encodes a spectrin repeat containing protein expressed in skeletal and smooth muscle, and peripheral blood lymphocytes, that localizes to the nuclear membrane. Mutations in this gene have been associated with autosomal recessive spinocerebellar ataxia 8, also referred to as autosomal recessive cerebellar ataxia type 1 or recessive ataxia of Beauce. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:23345; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; meiotic nuclear membrane microtubule tethering complex [GO:0034993]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; nuclear outer membrane [GO:0005640]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; P-body [GO:0000932]; postsynaptic membrane [GO:0045211]; sarcomere [GO:0030017]; actin binding [GO:0003779]; actin filament binding [GO:0051015]; cytoskeleton-nuclear membrane anchor activity [GO:0140444]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; lamin binding [GO:0005521]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; Golgi organization [GO:0007030]; muscle cell differentiation [GO:0042692]; nuclear matrix anchoring at nuclear membrane [GO:0090292]; nucleus organization [GO:0006997]; spermatogenesis [GO:0007283] | 12408964_Vertebrate Nesprin-1 and nesprin-2 proteins are orthologous to Drosophila melanogaster muscle protein MSP-300 14709720_The role of Syne-1 in cytokinesis involves an interaction with kinesin II. 15093733_Enaptin is an element of the nuclear membrane and the actin cytoskeleton. 16079285_The Nesprin-1 is essential for the interaction with a C-terminal region in SUN1 protein. 16875688_These findings indicate that GSRP-56 is a small Golgi-localized splice-isoform of the spectrin-repeat-containing Syne1 gene. 17159980_SYNE1 is the first identified gene responsible for a recessively inherited pure cerebellar ataxia. 17462627_The characterisation of the residues both in emerin and in nesprin-1alpha and -2beta which are involved in their interaction is reported. 17503513_This study identified a cluster of French-Canadian families with a new recessive ataxia of relatively pure cerebellar type caused by mutations in SYNE1. 17761684_Screening for DNA variations in the genes encoding nesprin-1 (SYNE1) and nesprin-2 (SYNE2) in 190 probands with Emery Dreifuss muscular dystrophy identified four heterozygous missense mutations. 18649358_Observational study of gene-disease association. (HuGE Navigator) 18709643_Loss of Drop1 expression at early stages in carcinomas of the uterus, cervix, kidney, lung, thyroid and pancreas. 18827015_association with human nesprin-3 appeared to be stronger for torsinADeltaE than for torsinA. TorsinA also associated with the KASH domains of nesprin-1 and -2 19542096_splice site mutation of SYNE-1 gene found in the family is responsible for arthrogryposis multiplex congenita. 19944109_analysis of nsprin-1 mutations in human and murine cardiomyopathy 20056644_Meta-analysis of gene-disease association. (HuGE Navigator) 20056644_a single nucleotide polymorphism downstream of ESR1, a gene called spectrin repeat containing, nuclear envelope 1, was associated with invasive ovarian cancer risk 20108321_Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development 20201924_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20351715_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20655839_Nesprin-1 depletion increased the number of focal adhesions and substrate traction while decreasing the speed of cell migration 22565781_The findings of this study add to the evidence that association of SYNE1 mutation influences susceptibility to bipolar and unipolar mood disorders. 22632968_Study presents crystal structures of the human SUN2-KASH1/2 complex, i.e. SUN2 complexed with the C-terminal 29 residues of human Nesprin-1 or -2 (the core of the LINC complex). 22728879_These results indicate that nesprin-1 knockdown releases the nucleus from the tension of F-actin bound to the nucleus, thereby increasing allowance for deformation before stretching, and that F-actin bound to the nucleus through nesprin-1 causes sustainable force transmission to the nucleus. 22768332_Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. 23325900_we report 4 novel homozygous SYNE1 mutations in 3 Japanese patients with cerebellar ataxia 23959263_Two novel truncating mutations were found among the French-Canadian participants, and 2 other novel mutations were found in a patient from France and a patient from Brazil. 24280874_FOXE1 and SYNE1 hypermethylation markers demonstrated significantly increased expression in neoplastic tissue. 24387768_role of CSMD1 and SYNE1 in the etiology of bipolar disorder 24718612_The significance of these shorter isoforms of nesprin, were evaluated. 24781983_a role for Nesprin-1 in the DNA damage response pathway 24862572_These data identify p50(Nesp1) as a multi-functional P-body component and microtubule scaffold necessary for RNA granule dynamics and provides evidence for P-body and stress granule micro-heterogeneity. 24931616_nesprin-1 and nesprin-2 both regulate nuclear and cytoplasmic architecture. 25091525_A report of a family with a SYNE1 gene mutation that seems to cause an 'Emery-Dreifuss muscular dystrophy-like' phenotype. 25315199_ur results show that SNPs rs9371601 and rs3093664 in the SYNE1 and TNF genes respectively, are associated with menstrual migraine. 25538088_our data suggest that SYNE1 and FOXE1 are promising markers for colorectal cancer detection. 26008743_Drosophila melanogaster larval muscles, exhibiting both elastic features contributed by the stretching capacity of MSP300 (nesprin) and rigidity provided by a perinuclear network of microtubules stabilized by Shot (spectraplakin) and EB1. 26330482_SNP rs79575945 in ESR1 gene is associated with cancers of endometrioid subtype and resulted in the expression of SYNE1. 26704904_A full length transcript homologous to rat CPG2 exists within human SYNE1. A full length transcript homologous to rat CPG2 exists within human SYNE1. 26791211_Mechanostimulation mediates nuclear changes in oligodendrocyte progenitor cells via the SYNE1 complex. 27086870_findings revise the view that SYNE1 ataxia causes mainly a relatively pure cerebellar recessive ataxia and that it is largely limited to Quebec. Instead, complex phenotypes with a wide range of extra-cerebellar neurological and nonneurological dysfunctions are frequent, including in particular motor neuron and brainstem dysfunction. 27178001_This study demonstrate four novel truncating mutations in SYNE1 in pedigrees from British, Sri Lankan and Turkish origin in patient with cerebellar ataxia. 27350129_The results show that nesprin-1-alpha2 is dynamically controlled and may be involved in some process occurring during early myofibre formation, such as re-positioning of nuclei. 27620326_The frequency of CACNA1C rs10848683 in genetic high-risk individuals was double that in controls. For SYNE1 rs214950, higher frequencies were found in the genetic high-risk group than in controls. Polymorphisms in CACNA1C and SYNE1 could confer a greater risk of developing Schizophrenia and Bipolar Disorder in individuals who are already at high risk because of their family history. 27782104_Nonsense mutation in the ultimate exon of full-length SYNE1 causes congenital onset of muscular weakness with distal arthrogryposis. 28178086_Authors screened 937 BPD samples for genetic variation in SYNE1 exons 14-33, which covers the CPG2 region, using high-resolution melt analysis. Nine patients are compound heterozygotes for variants in SYNE1/CPG2, suggesting that rare coding variants may contribute significantly towards the complex genetic architecture underlying BPD. 28398466_three novel rare variants (R8272Q, S8381C and N8406K) in the C-terminus of the SYNE1 gene (nesprin-1) were identified in seven dilated cardiomyopathy patients by mutation screening. Expression of these mutants caused nuclear morphology defects and reduced lamin A/C and SUN2 staining at the Nuclear envelope. 28455503_The functional integrity of lamin and nesprin-1 is thus required to modulate the FHOD1 activity and the inside-out mechanical coupling that tunes the cell internal stiffness to match that of its soft, physiological-like environment. 28583108_We reported a novel mutation in exon 46 on codon 2304 (G2304R) of the SYNE1 gene in a Chinese family with Emery-Dreifuss muscular dystrophy-like features, and 100 healthy individuals did not show such mutation. 30573412_Three novel mutations are described in SYNE1-ataxia patients. 30610203_Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function. 31049853_SYNE1 gene mutations were identified as a cause of late-onset pure cerebellar syndrome 31236099_Our explorative analyses of the biological impact of rs9371601 suggested that this SNP was significantly associated with the methylation of a CpG site (cg01844274, p = 5.0510(- 6)) within SYNE1 in human dorsal lateral prefrontal cortex (DLPFC) tissues. CONCLUSIONS: Our data confirms the association between rs9371601 and BPD 31578382_Nesprin-1-alpha2 associates with kinesin at myotube outer nuclear membranes, but is restricted to neuromuscular junction nuclei in adult muscle. 32281752_SYNE1-QK1 SNPs, G x G and G x E interactions on the risk of hyperlipidaemia. 32889669_Autosomal Recessive Cerebellar Ataxia Type 1: Phenotypic and Genetic Correlation in a Cohort of Chinese Patients with SYNE1 Variants. 33472039_Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. | ENSMUSG00000096054 | Syne1 | 3.149712e+02 | 0.6449929 | -0.632644773 | 0.3101485 | 3.881373e+00 | 0.0488246232 | 0.66791998 | No | Yes | 2.994337e+02 | 67.159018 | 3.338793e+02 | 76.793112 | |
| ENSG00000131089 | 23229 | ARHGEF9 | protein_coding | O43307 | FUNCTION: Acts as guanine nucleotide exchange factor (GEF) for CDC42. Promotes formation of GPHN clusters (By similarity). {ECO:0000250|UniProtKB:Q9QX73, ECO:0000269|PubMed:10559246}. | 3D-structure;Alternative splicing;Cell junction;Cytoplasm;Disease variant;Epilepsy;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome;SH3 domain;Synapse | The protein encoded by this gene is a Rho-like GTPase that switches between the active (GTP-bound) state and inactive (GDP-bound) state to regulate CDC42 and other genes. This brain-specific protein also acts as an adaptor protein for the recruitment of gephyrin and together these proteins facilitate receceptor recruitement in GABAnergic and glycinergic synapses. Defects in this gene are the cause of startle disease with epilepsy (STHEE), also known as hyperekplexia with epilepsy, as well as several other types of cognitive disability. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2017]. | hsa:23229; | cytosol [GO:0005829]; postsynaptic density [GO:0014069]; guanyl-nucleotide exchange factor activity [GO:0005085]; regulation of small GTPase mediated signal transduction [GO:0051056] | 11727829_Here we identified residues critical for interaction with gephyrin in the linker region between the SH3 and the DH domains of collybistin. 15215304_translocates gephyrin to submembrane microaggregates; collybistin mutation (G55A)is found in exon 2 of the ARHGEF9 gene in a patient with clinical symptoms of both hyperekplexia and epilepsy 18208356_Results show that hPEM-2 is a target protein of Smurf1. 19911011_Observational study of gene-disease association. (HuGE Navigator) 20622020_Study propose that the collybistin-gephyrin complex has an intimate role in the clustering of GABA(A)Rs containing the alpha2 subunit. 21633362_Data indicate that ARHGEF9 is likely to be responsible for syndromic X-linked mental retardation associated with epilepsy. 21807943_major regulator of GABAergic postsynaptic gephyrin clustering 22033413_These results reveal that G(s) and G(q) signalings regulate hPEM-2 functions through PKA and c-Src in Neuro-2a neuroblastoma cells, respectively. 22778260_Phosphorylation of gephyrin in hippocampal neurons by cyclin-dependent kinase CDK5 at Ser-270 is dependent on collybistin. 24297911_The enhancement of Cb-induced gephyrin clustering by GTP-TC10 does not depend on the guanine nucleotide exchange activity of Cb but involves an interaction that resembles reported interactions of other small GTPases with their effectors 25678704_Impairment of the membrane lipid binding activity of Collybistin R290H and a consequent defect in inhibitory synapse maturation represent a likely molecular pathomechanism of epilepsy and mental retardation in humans. 25898924_Collybistin forms a complex with mTOR and eIF3 and by sequestering these proteins downregulates mTORC1 signaling and protein synthesis potentially contributing to intellectual disability and autism. 27238888_Autism spectrum disorder patient with the smallest inactivating deletion in the collybistin gene. 29130122_Study identified a novel mutation in ARHGEF9, c.868C > T/p.R290C, which co-segregated with epileptic encephalopathy, and validated its association with epileptic encephalopathy. Further analysis revealed that all ARHGEF9 mutations were associated with intellectual disability. 31283007_Identification of TAF1, SAT1, and ARHGEF9 as DNA methylation biomarkers for hepatocellular carcinoma. 31942680_Clinical and Molecular Characterization of Three Novel ARHGEF9 Mutations in Patients with Developmental Delay and Epilepsy. 32939676_De novo ARHGEF9 missense variants associated with neurodevelopmental disorder in females: expanding the genotypic and phenotypic spectrum of ARHGEF9 disease in females. | ENSMUSG00000025656 | Arhgef9 | 8.066551e+02 | 0.9468107 | -0.078852129 | 0.2737675 | 8.422062e-02 | 0.7716572000 | 0.95442153 | No | Yes | 7.429247e+02 | 86.403991 | 8.258947e+02 | 98.333617 | |
| ENSG00000131115 | 7770 | ZNF227 | protein_coding | Q86WZ6 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7770; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 2.243738e+02 | 0.8263448 | -0.275184247 | 0.3515367 | 5.930189e-01 | 0.4412540933 | 0.85687843 | No | Yes | 1.943414e+02 | 34.295192 | 2.137218e+02 | 38.558853 | |||||
| ENSG00000131149 | 23199 | GSE1 | protein_coding | Q14687 | Acetylation;Alternative splicing;Coiled coil;Methylation;Phosphoprotein;Reference proteome | This gene encodes a proline-rich protein with coiled coil domains that may be a subunit of a BRAF35-HDAC (BHC) histone deacetylase complex. This gene may function as an oncogene in breast cancer and enhanced expression of the encoded protein has been observed in breast cancer patients. [provided by RefSeq, May 2017]. | hsa:23199; | 26828271_In this study, we reported that GSE1 is overexpression in breast cancer and silencing of GSE1 significantly suppressed breast cancer cells proliferation, migration and invasion. 29367342_High GSE1 expression is associated with gastric cancer growth and metastasis. 33623790_Overexpression of GSE1 Related to Trastuzumab Resistance in Gastric Cancer Cells. | ENSMUSG00000031822 | Gse1 | 2.898525e+03 | 1.2150038 | 0.280960816 | 0.2601419 | 1.167496e+00 | 0.2799163106 | 0.80713123 | No | Yes | 2.871130e+03 | 268.779623 | 2.234395e+03 | 214.979290 | |||
| ENSG00000131375 | 23473 | CAPN7 | protein_coding | Q9Y6W3 | FUNCTION: Calcium-regulated non-lysosomal thiol-protease. {ECO:0000250}. | 3D-structure;Acetylation;Hydrolase;Nucleus;Phosphoprotein;Protease;Reference proteome;Repeat;Thiol protease | Calpains are ubiquitous, well-conserved family of calcium-dependent, cysteine proteases. The calpain proteins are heterodimers consisting of an invariant small subunit and variable large subunits. The large subunit possesses a cysteine protease domain, and both subunits possess calcium-binding domains. Calpains have been implicated in neurodegenerative processes, as their activation can be triggered by calcium influx and oxidative stress. The function of the protein encoded by this gene is not known. An orthologue has been found in mouse but it seems to diverge from other family members. The mouse orthologue is thought to be calcium independent with protease activity. [provided by RefSeq, Jul 2008]. | hsa:23473; | centrosome [GO:0005813]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; calcium-dependent cysteine-type endopeptidase activity [GO:0004198]; endopeptidase activity [GO:0004175]; MIT domain binding [GO:0090541]; positive regulation of epithelial cell migration [GO:0010634]; self proteolysis [GO:0097264] | 20849418_Results demonstrate that human calpain 7 is proteolytically active, and imply that calpain 7 is activated by ESCRT-III-related protein IST1. 21616915_The detected enhancement of autolysis of mGFP-fused calpain-7 by coexpression with CHMP1B and observed further activation by additional coexpression of IST1 in HEK293T cells. 23497113_Mutational analysis of calpain-7 reveals the importance of not only the N-terminal microtubule-interacting and trafficking domains but also the C-terminal C2 domain-like domains for proteolytic activity. 23855590_Data indicate that CAPN 7 promotes endometrial stromal cell (hESC) migration and invasion by increasing the activity of MMP-2 via an increased ratio of MMP-2 to TIMP-2. 24953135_The proteolytic activity of CAPN7 is important for the acceleration of EGFR degradation via the endosomal sorting pathway. | ENSMUSG00000021893 | Capn7 | 3.649893e+02 | 1.0104200 | 0.014955151 | 0.4022992 | 1.411648e-03 | 0.9700290055 | 0.99408977 | No | Yes | 3.770517e+02 | 87.796690 | 3.652449e+02 | 87.119678 | |
| ENSG00000131480 | 314 | AOC2 | protein_coding | O75106 | FUNCTION: Has a monoamine oxidase activity with substrate specificity for 2-phenylethylamine and tryptamine. May play a role in adipogenesis. May be a critical modulator of signal transmission in retina. {ECO:0000269|PubMed:17400359, ECO:0000269|PubMed:19588076}. | Alternative splicing;Calcium;Catecholamine metabolism;Cell membrane;Copper;Cytoplasm;Disulfide bond;Glycoprotein;Membrane;Metal-binding;Oxidoreductase;Reference proteome;Signal;TPQ | Copper amine oxidases catalyze the oxidative conversion of amines to aldehydes and ammonia in the presence of copper and quinone cofactor. This gene shows high sequence similarity to copper amine oxidases from various species ranging from bacteria to mammals. The protein contains several conserved motifs including the active site of amine oxidases and the histidine residues that likely bind copper. It may be a critical modulator of signal transmission in retina, possibly by degrading the biogenic amines dopamine, histamine, and putrescine. This gene may be a candidate gene for hereditary ocular diseases. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jul 2008]. | hsa:314; | cytoplasm [GO:0005737]; plasma membrane [GO:0005886]; aliphatic-amine oxidase activity [GO:0052595]; aminoacetone:oxygen oxidoreductase(deaminating) activity [GO:0052594]; copper ion binding [GO:0005507]; electron transfer activity [GO:0009055]; phenethylamine:oxygen oxidoreductase (deaminating) activity [GO:0052596]; primary amine oxidase activity [GO:0008131]; quinone binding [GO:0048038]; tryptamine:oxygen oxidoreductase (deaminating) activity [GO:0052593]; amine metabolic process [GO:0009308]; catecholamine metabolic process [GO:0006584]; visual perception [GO:0007601]; xenobiotic metabolic process [GO:0006805] | 19588076_AOC2 mRNA is expressed in many tissues, however, the only tissues with detectable AOC2-like enzyme activity is found in the eye. 27766585_Adipose tissue SSAO activity did not vary according to anatomical location and/or metabolic status in severely obese women. | ENSMUSG00000078651 | Aoc2 | 1.631073e+02 | 0.6466777 | -0.628881195 | 0.3325202 | 3.522623e+00 | 0.0605365544 | 0.69358774 | No | Yes | 1.066517e+02 | 15.310176 | 1.839685e+02 | 26.579123 | |
| ENSG00000131652 | 79228 | THOC6 | protein_coding | Q86W42 | FUNCTION: Acts as component of the THO subcomplex of the TREX complex which is thought to couple mRNA transcription, processing and nuclear export, and which specifically associates with spliced mRNA and not with unspliced pre-mRNA. TREX is recruited to spliced mRNAs by a transcription-independent mechanism, binds to mRNA upstream of the exon-junction complex (EJC) and is recruited in a splicing- and cap-dependent manner to a region near the 5' end of the mRNA where it functions in mRNA export to the cytoplasm via the TAP/NFX1 pathway. The TREX complex is essential for the export of Kaposi's sarcoma-associated herpesvirus (KSHV) intronless mRNAs and infectious virus production. Plays a role in apoptosis negative control involved in brain development. {ECO:0000269|PubMed:15833825, ECO:0000269|PubMed:15998806, ECO:0000269|PubMed:17190602, ECO:0000269|PubMed:18974867, ECO:0000269|PubMed:23621916}. | 3D-structure;Alternative splicing;Apoptosis;Developmental protein;Disease variant;Mental retardation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Transport;WD repeat;mRNA processing;mRNA splicing;mRNA transport | This gene encodes a subunit of the multi-protein THO complex, which is involved in coordination between transcription and mRNA processing. The THO complex is a component of the TREX (transcription/export) complex, which is involved in transcription and export of mRNAs. A missense mutation in this gene is associated with a neurodevelopmental disorder called Beaulieu-Boycott-Innes syndrome. [provided by RefSeq, Dec 2016]. | hsa:79228; | nuclear body [GO:0016604]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; THO complex [GO:0000347]; THO complex part of transcription export complex [GO:0000445]; transcription export complex [GO:0000346]; RNA binding [GO:0003723]; apoptotic process [GO:0006915]; central nervous system development [GO:0007417]; mRNA export from nucleus [GO:0006406]; mRNA processing [GO:0006397]; negative regulation of apoptotic process [GO:0043066]; RNA splicing [GO:0008380]; viral mRNA export from host cell nucleus [GO:0046784] | 19773279_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 26739162_In addition to confirming the morbid nature of THOC6 by providing an independent homozygous apparently loss of function allele in a patient with a compatible phenotype, our data also expand THOC6-related phenotype to include previously unreported imperforate anus and undescended testicles. 27102954_Results indicate three unrelated patients with bi-allelic mutations in THOC6 associated with intellectual disability and additional clinical features. | ENSMUSG00000041319 | Thoc6 | 3.545112e+03 | 1.2238463 | 0.291422378 | 0.3170943 | 8.439831e-01 | 0.3582600933 | 0.83211403 | No | Yes | 3.736058e+03 | 467.271966 | 2.702752e+03 | 347.184979 | |
| ENSG00000131941 | 85415 | RHPN2 | protein_coding | Q8IUC4 | FUNCTION: Binds specifically to GTP-Rho. May function in a Rho pathway to limit stress fiber formation and/or increase the turnover of F-actin structures in the absence of high levels of RhoA activity. {ECO:0000269|PubMed:12221077}. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;Phosphoprotein;Reference proteome | This gene encodes a member of the rhophilin family of Ras-homologous (Rho)-GTPase binding proteins. The encoded protein binds both GTP- and GDP-bound RhoA and GTP-bound RhoB and may be involved in the organization of the actin cytoskeleton. [provided by RefSeq, Apr 2009]. | hsa:85415; | cytosol [GO:0005829]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; glomerular filtration [GO:0003094]; negative regulation of stress fiber assembly [GO:0051497]; signal transduction [GO:0007165] | 12221077_Rhophilin-2 may function normally in a Rho pathway to limit stress fiber formation and/or increase the turnover of F-actin structures in the absence of high levels of RhoA activity 12473120_may play a key role between RhoB and potential downstream elements needed under stimulation of the thyrotropin/cAMP pathway in thyrocytes 19011631_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20437058_Observational study of genetic testing. (HuGE Navigator) 20501757_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20530476_Observational study of gene-disease association. (HuGE Navigator) 20638935_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20648012_Observational study of gene-disease association. (HuGE Navigator) 20659471_Observational study of gene-disease association. (HuGE Navigator) 21071539_Observational study of gene-disease association. (HuGE Navigator) 23774217_These effects were implemented through RHPN2-mediated activation of RhoA. 25799222_Single nucleotide polymorphism in RHPN2 gene is associated with colorectal cancer. 26349980_We found significant association between RHPN2 gene rs10411210 and colorectal cancer in European and Asian populations, but not American population 29949196_It may contribute to the pathogenesis of nonsyndromic orofacial clefts in Chinese populations. 31847864_The hBMSCs-derived exosomal miR-205 retards prostate cancer progression by inhibiting RHPN2, suggesting that miR-205 may present a predictor and potential therapeutic target for prostate cancer. 34382920_P76RBE silencing inhibits ovarian cancer cell proliferation, migration, and invasion via suppressing the integrin beta1/NF-kappaB pathway. | ENSMUSG00000030494 | Rhpn2 | 4.550623e+02 | 0.9554639 | -0.065726674 | 0.2851859 | 5.405046e-02 | 0.8161593506 | 0.96373941 | No | Yes | 4.727272e+02 | 57.727773 | 4.414150e+02 | 55.462786 | |
| ENSG00000132122 | 54558 | SPATA6 | protein_coding | Q9NWH7 | FUNCTION: Required for formation of the sperm connecting piece during spermiogenesis. Sperm connecting piece is essential for linking the developing flagellum to the head during late spermiogenesis. May be involved in myosin-based microfilament transport through interaction with myosin subunits. {ECO:0000250|UniProtKB:Q3U6K5}. | Alternative splicing;Cell projection;Cilium;Developmental protein;Differentiation;Flagellum;Glycoprotein;Isopeptide bond;Phosphoprotein;Reference proteome;Secreted;Signal;Spermatogenesis;Ubl conjugation | hsa:54558; | extracellular region [GO:0005576]; sperm connecting piece [GO:0097224]; myosin light chain binding [GO:0032027]; cell differentiation [GO:0030154]; motile cilium assembly [GO:0044458]; spermatogenesis [GO:0007283] | 26464655_SPATA6 may play an important role in testicular germ cell tumors, and down-regulation of SPATA6 could lead to apoptosis of testicular germ cell tumors. 31838782_Increased DNA methylation in the spermatogenesis-associated (SPATA) genes correlates with infertility. | ENSMUSG00000034401 | Spata6 | 6.827826e+01 | 1.2692687 | 0.343997501 | 0.4364061 | 5.933655e-01 | 0.4411206484 | 0.85677282 | No | Yes | 7.616375e+01 | 13.480141 | 5.559569e+01 | 10.190860 | ||
| ENSG00000132153 | 22907 | DHX30 | protein_coding | Q7L2E3 | FUNCTION: RNA-dependent helicase (PubMed:29100085). Plays an important role in the assembly of the mitochondrial large ribosomal subunit (PubMed:25683715, PubMed:29100085). Required for optimal function of the zinc-finger antiviral protein ZC3HAV1 (By similarity). Associates with mitochondrial DNA (PubMed:18063578). Involved in nervous system development and differentiation through its involvement in the up-regulation of a number of genes which are required for neurogenesis, including GSC, NCAM1, neurogenin, and NEUROD (By similarity). {ECO:0000250|UniProtKB:Q5BJS0, ECO:0000250|UniProtKB:Q99PU8, ECO:0000269|PubMed:18063578, ECO:0000269|PubMed:25683715, ECO:0000269|PubMed:29100085}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Disease variant;Helicase;Hydrolase;Mental retardation;Mitochondrion;Mitochondrion nucleoid;Nucleotide-binding;Phosphoprotein;RNA-binding;Reference proteome;Ribosome biogenesis | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this DEAD box protein family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. The family member encoded by this gene is a mitochondrial nucleoid protein that associates with mitochondrial DNA. It has also been identified as a component of a transcriptional repressor complex that functions in retinal development, and it is required to optimize the function of the zinc-finger antiviral protein. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Feb 2013]. | hsa:22907; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrial nucleoid [GO:0042645]; mitochondrion [GO:0005739]; ribonucleoprotein granule [GO:0035770]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; chromatin binding [GO:0003682]; double-stranded RNA binding [GO:0003725]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; central nervous system development [GO:0007417]; mitochondrial large ribosomal subunit assembly [GO:1902775] | 18022663_Here, we provide evidence that overexpression of an RNA helicase named DHX30 enhances HIV-1 gene expression, but leads to the generation of viruses that package significantly low levels of viral RNA and exhibit severely decreased infectivity. 18063578_Identifies DHX30 as a nucleoid protein. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 29100085_Provide molecular insight into how DHX30 dysfunction might cause a neurodevelopmental disorder. 31754723_A double-stranded RNA is required for the interaction between a host restriction factor DHX30 and the NS1 protein of influenza A virus. 32234473_Nutlin-Induced Apoptosis Is Specified by a Translation Program Regulated by PCBP2 and DHX30. 34020708_Genotype-phenotype correlations and novel molecular insights into the DHX30-associated neurodevelopmental disorders. 34180050_De novo pathogenic DHX30 variants in two cases. | ENSMUSG00000032480 | Dhx30 | 2.031150e+04 | 1.2696504 | 0.344431265 | 0.2968523 | 1.349013e+00 | 0.2454506710 | 0.78892886 | No | Yes | 2.106917e+04 | 2210.165697 | 1.524011e+04 | 1639.829312 | |
| ENSG00000132170 | 5468 | PPARG | protein_coding | P37231 | FUNCTION: Nuclear receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the nuclear receptor binds to DNA specific PPAR response elements (PPRE) and modulates the transcription of its target genes, such as acyl-CoA oxidase. It therefore controls the peroxisomal beta-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. ARF6 acts as a key regulator of the tissue-specific adipocyte P2 (aP2) enhancer. Acts as a critical regulator of gut homeostasis by suppressing NF-kappa-B-mediated proinflammatory responses. Plays a role in the regulation of cardiovascular circadian rhythms by regulating the transcription of ARNTL/BMAL1 in the blood vessels (By similarity). {ECO:0000250|UniProtKB:P37238, ECO:0000269|PubMed:16150867, ECO:0000269|PubMed:20829347, ECO:0000269|PubMed:23525231, ECO:0000269|PubMed:9065481}.; FUNCTION: (Microbial infection) Upon treatment with M.tuberculosis or its lipoprotein LpqH, phosphorylation of MAPK p38 and IL-6 production are modulated, probably via this protein. {ECO:0000269|PubMed:25504154}. | 3D-structure;Activator;Alternative splicing;Biological rhythms;Cytoplasm;DNA-binding;Diabetes mellitus;Disease variant;Glycoprotein;Metal-binding;Nucleus;Obesity;Phosphoprotein;Receptor;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a member of the peroxisome proliferator-activated receptor (PPAR) subfamily of nuclear receptors. PPARs form heterodimers with retinoid X receptors (RXRs) and these heterodimers regulate transcription of various genes. Three subtypes of PPARs are known: PPAR-alpha, PPAR-delta, and PPAR-gamma. The protein encoded by this gene is PPAR-gamma and is a regulator of adipocyte differentiation. Additionally, PPAR-gamma has been implicated in the pathology of numerous diseases including obesity, diabetes, atherosclerosis and cancer. Alternatively spliced transcript variants that encode different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:5468; | chromatin [GO:0000785]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; receptor complex [GO:0043235]; RNA polymerase II transcription regulator complex [GO:0090575]; alpha-actinin binding [GO:0051393]; arachidonic acid binding [GO:0050544]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; DNA binding domain binding [GO:0050692]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; double-stranded DNA binding [GO:0003690]; E-box binding [GO:0070888]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; LBD domain binding [GO:0050693]; nuclear receptor activity [GO:0004879]; peptide binding [GO:0042277]; prostaglandin receptor activity [GO:0004955]; protein C-terminus binding [GO:0008022]; protein self-association [GO:0043621]; R-SMAD binding [GO:0070412]; retinoid X receptor binding [GO:0046965]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; STAT family protein binding [GO:0097677]; transcription cis-regulatory region binding [GO:0000976]; transcription coregulator binding [GO:0001221]; zinc ion binding [GO:0008270]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; BMP signaling pathway [GO:0030509]; cell differentiation [GO:0030154]; cell fate commitment [GO:0045165]; cell maturation [GO:0048469]; cellular response to hypoxia [GO:0071456]; cellular response to insulin stimulus [GO:0032869]; cellular response to low-density lipoprotein particle stimulus [GO:0071404]; epithelial cell differentiation [GO:0030855]; fatty acid metabolic process [GO:0006631]; glucose homeostasis [GO:0042593]; hormone-mediated signaling pathway [GO:0009755]; innate immune response [GO:0045087]; lipid homeostasis [GO:0055088]; lipid metabolic process [GO:0006629]; lipoprotein transport [GO:0042953]; long-chain fatty acid transport [GO:0015909]; macrophage derived foam cell differentiation [GO:0010742]; monocyte differentiation [GO:0030224]; mRNA transcription by RNA polymerase II [GO:0042789]; negative regulation of angiogenesis [GO:0016525]; negative regulation of blood vessel endothelial cell migration [GO:0043537]; negative regulation of BMP signaling pathway [GO:0030514]; negative regulation of cardiac muscle hypertrophy in response to stress [GO:1903243]; negative regulation of cellular response to transforming growth factor beta stimulus [GO:1903845]; negative regulation of cholesterol storage [GO:0010887]; negative regulation of connective tissue replacement involved in inflammatory response wound healing [GO:1904597]; negative regulation of gene expression [GO:0010629]; negative regulation of gene silencing by miRNA [GO:0060965]; negative regulation of inflammatory response [GO:0050728]; negative regulation of interferon-gamma-mediated signaling pathway [GO:0060336]; negative regulation of lipid storage [GO:0010888]; negative regulation of macrophage derived foam cell differentiation [GO:0010745]; negative regulation of MAP kinase activity [GO:0043407]; negative regulation of mitochondrial fission [GO:0090258]; negative regulation of osteoblast differentiation [GO:0045668]; negative regulation of pathway-restricted SMAD protein phosphorylation [GO:0060394]; negative regulation of pri-miRNA transcription by RNA polymerase II [GO:1902894]; negative regulation of receptor signaling pathway via STAT [GO:1904893]; negative regulation of sequestering of triglyceride [GO:0010891]; negative regulation of signaling receptor activity [GO:2000272]; negative regulation of SMAD protein signal transduction [GO:0060392]; negative regulation of smooth muscle cell proliferation [GO:0048662]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of transforming growth factor beta receptor signaling pathway [GO:0030512]; negative regulation of vascular associated smooth muscle cell proliferation [GO:1904706]; negative regulation of vascular endothelial cell proliferation [GO:1905563]; peroxisome proliferator activated receptor signaling pathway [GO:0035357]; placenta development [GO:0001890]; positive regulation of adiponectin secretion [GO:0070165]; positive regulation of cholesterol efflux [GO:0010875]; positive regulation of DNA binding [GO:0043388]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of fat cell differentiation [GO:0045600]; positive regulation of fatty acid metabolic process [GO:0045923]; positive regulation of gene expression [GO:0010628]; positive regulation of low-density lipoprotein receptor activity [GO:1905599]; positive regulation of pathway-restricted SMAD protein phosphorylation [GO:0010862]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of SMAD protein signal transduction [GO:0060391]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of vascular associated smooth muscle cell apoptotic process [GO:1905461]; regulation of blood pressure [GO:0008217]; regulation of cholesterol transporter activity [GO:0060694]; regulation of circadian rhythm [GO:0042752]; regulation of lipid metabolic process [GO:0019216]; regulation of transcription by RNA polymerase II [GO:0006357]; response to lipid [GO:0033993]; response to nutrient [GO:0007584]; retinoic acid receptor signaling pathway [GO:0048384]; rhythmic process [GO:0048511]; signal transduction [GO:0007165]; white fat cell differentiation [GO:0050872] | 11069206_Observational study of gene-disease association. (HuGE Navigator) 11079814_Observational study of gene-disease association. (HuGE Navigator) 11242476_Observational study of gene-disease association. (HuGE Navigator) 11246892_Observational study of gene-environment interaction. (HuGE Navigator) 11248748_Observational study of gene-disease association. (HuGE Navigator) 11289055_Observational study of gene-disease association. (HuGE Navigator) 11315829_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11334419_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11402923_Observational study of gene-disease association. (HuGE Navigator) 11409297_Observational study of gene-disease association. (HuGE Navigator) 11444435_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11466580_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11508283_Observational study of gene-disease association. (HuGE Navigator) 11511919_Observational study of gene-disease association. (HuGE Navigator) 11522688_Observational study of gene-disease association. (HuGE Navigator) 11554739_The expression of the human PPARgamma gene is controlled by RORalpha1. 11554760_PPAR-gamma induces pancreatic cancer cell apoptosis 11596673_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11793024_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11809750_role in inducing cyclooxygenase-2 in monocytes 11836319_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11836575_Expression of peroxisome proliferator-activated receptor gamma and the growth inhibitory effect of its synthetic ligands in human salivary gland cancer cell lines. 11840500_Observational study of gene-disease association. (HuGE Navigator) 11845236_Reduced lipolysis as possible cause for greater weight gain in subjects with the Pro12Ala polymorphism in PPARgamma2 11847231_Peroxisome proliferator-activated receptor gamma agonists inhibit HIV-1 replication in macrophages by transcriptional and post-transcriptional effects. 11862322_Observational study of gene-disease association. (HuGE Navigator) 11872365_Observational study of gene-disease association. (HuGE Navigator) 11872365_the role of peroxisome proliferator-activated receptor gamma (PPARgamma) in high-fat diet induced-obesity and insulin resistance by gene targeting and case-control study using the common PPARgamma2 polymorphism 11872366_Pharmacologic activation of PPARgamma in vascular cells may provide a novel therapeutic approach to retard diabetes-associated vascular disease. 11872377_the roles of PPARgamma and the actions of PPARgamma ligands in the cardiovascular system 11872694_Observational study of gene-disease association. (HuGE Navigator) 11877444_T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities 11884452_Activation of PPAR gamma by oxidized lipoproteins (oxLDL) promotes macrophage desensitization by reducing oxLDL-stimulated oxygen radical formation, an important determinant of the activation/deactivation balance in macrophages. 11888683_The presence of PPAR gamma in bladder cancers having characteristics of low malignancy is associated with its role in modulating expression of those angiogenic factors (e.g., bFGF and PDECGF) that may be responsible for malignancy in bladder tumor cells. 11897617_cPLA(2) plays a critical role in PPAR-mediated gene transcription in HepG2 cells 11897821_Observational study of gene-disease association. (HuGE Navigator) 11903058_Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor delta-mediated transactivation 11914642_PPARgamma augments TNF-family induced apoptosis 11916624_Lack of association between peroxisome proliferator-activated receptor-gamma-2 gene variants and the occurrence of coronary heart disease in patients with diabetes mellitus. 11916624_Observational study of gene-disease association. (HuGE Navigator) 11923467_Retinoid X receptor alpha is implicated in the nuclear reorganization of PPAR gamma and suggest that PPAR gamma colocalizes with RXR alpha at specific locations within the nucleus independent of added ligand. 11924722_Observational study of gene-disease association. (HuGE Navigator) 11924722_PPARgamma2 pro12Ala polymorphism and insulin resistance in Japanese hypertensive patients. 11928067_Pro12Ala polymorphism of PPAR(gamma2) gene is not associated with diabetic retinopathy but is associated with dyslipidemia in male type 2 diabetic patients 11934839_activation of PPARgamma in human CD4-positive T cells limits the expression of proinflammatory cytokines, such as IFNgamma 11948400_Activation of peroxisome proliferator-activated receptor-gamma stimulates the growth arrest and DNA-damage inducible 153 gene in non-small cell lung carcinoma cells. 11948965_PPAR-gamma is induced in prostate cancer; PPAR-gamma ligands may mediate its antiproliferative effect against prostate cancer cells through differentiation 11953889_Inhibition of human chondrosarcoma cell growth via apoptosis by peroxisome proliferator-activated receptor-gamma. 11956653_Frequent polymorphism of peroxisome proliferator activated receptor gamma gene in colorectal cancer containing wild-type K-ras gene. 11972302_Observational study of gene-disease association. (HuGE Navigator) 11972302_Pro12Ala polymorphism of the gene is associated with reduced type 2 diabetes risk and increased insulin sensitivity 11979403_genes of C3, HSL, and PPARgamma may exert a modifying effect on lipid and glucose metabolism in familial combined hyperlipidemia 11980626_Observational study of gene-disease association. (HuGE Navigator) 11980898_differential regulation of vascular endothelial growth factor expression in bladder cancer cells (peroxisome proliferative activated receptor, beta) 12015306_PPRE in intron 1 of the ACBP gene is a bona fide PPARgamma-response element. 12029076_findings suggest an involvement of PPARgamma in trophoblast differentiation during normal placental development; down-regulation of PPARgamma may contribute to trophoblastic diseases such as hydatidufirm mole and choriocarcinoma 12048686_Observational study of gene-disease association. (HuGE Navigator) 12054675_PPAR activators inhibit endothelial cell migration by targeting Akt. 12055328_adipose tissue peroxisome proliferator-activated receptor gamma1 mRNA concentration is positively regulated by eicosapentaenoic acid 12056809_Activation of peroxisome proliferator-activated receptor gamma inhibits osteoprotegerin gene expression in human aortic smooth muscle cells 12062858_Differential effect of variants in the development of type 2 diabetes between native Japanese and Japanese Americans. 12062858_Observational study of gene-disease association. (HuGE Navigator) 12065695_PPARgamma is a target for induction of apoptosis in rheumatoid synovial cells 12075579_Observational study of gene-disease association. (HuGE Navigator) 12077117_85kD PLA2 mediates PPARG activation in lung epithelial cells 12079854_Observational study of gene-disease association. (HuGE Navigator) 12080321_reduces growth of esophageal cancer 12080444_obesity is associated with an inverse relationship between PPARgamma and retinoic acid receptor alpha expressions in subcutaneous adipose tissue 12082592_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12086968_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12086968_effect of polymorphism on insulin sensitivity and metabolism; interactions with birth size 12107164_Activation of endothelial PPAR-gamma has a potent anti-inflammatory role. 12118251_we describe a family in which five individuals with severe insulin resistance, but no unaffected family members, were compound heterozygous with respect to frameshift/premature stop mutations in two unlinked genes, PPARG and PPP1R3A 12145143_the Pro12Ala polymorphism in PPAR-gamma2 represents the first genetic variant with a broad impact on the risk of common type 2 diabetes 12145174_Association of the Pro12Ala polymorphism with 3-year incidence of type 2 diabetes and body weight change in the Finnish Diabetes Prevention Study 12145174_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12145184_Observational study of gene-disease association. (HuGE Navigator) 12145184_polymorphism Pro12Ala is associated with nephropathy in type 2 diabetes 12148087_Observational study of gene-disease association. (HuGE Navigator) 12148087_The Pro115Gln polymorphism has no epidemiological impact on morbid obesity 12161538_Expression of PAX8-PPAR gamma 1 rearrangements in both follicular thyroid carcinomas and adenomas. 12161548_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 12165097_negative regulatory function for PPARgamma on cytokine and MMP production together with inhibition of cytokine-mediated inflammatory responses in rheumatoid synovial cells 12189136_role of 13-(S)-HODE in upregulating the MAP kinase signaling pathway and subsequently downregulating PPARgamma 12193733_Nitric oxide-activated anti-inflammatory properties of the peroxisome proliferator-activated receptor (PPAR)gamma in monocytes/macrophages down-regulate p47phox and attenuate the respiratory burst. 12213872_Differential effects of adiposity on messenger ribonucleic acid expression in human adipocytes 12218380_Observational study of gene-disease association. (HuGE Navigator) 12218380_analysis of peroxisome proliferator-activated receptor-gamma2 genotype showed that leptin levels were determined by the Pro12Ala mutation in type-2 diabetic women but not in men 12230498_melanoma cell growth may be modulated through peroxisome proliferator-activated receptor gamma. 12324185_Observational study of gene-disease association. (HuGE Navigator) 12324185_There is no association between PPAR-gamma polymorphism and the occurence or severity of preeclampsia. 12324573_In esophageal cancer tissues, the expression of PPAR gamma was decreased compared with normal esophageal epithelium. 12354130_Observational study of gene-disease association. (HuGE Navigator) 12370112_Observational study of gene-disease association. (HuGE Navigator) 12370112_the Pro12Ala polymorphism of the PPARgamma-2 gene promotes peripheral deposition of adipose tissue and increased insulin sensitivity for a given BMI. 12395215_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12397176_mediates the proliferative action of mast-cell tryptase: possible relevance to human fibrotic disorders 12397391_mRNA levels of peroxisome proliferator-activated receptor gamma are not altered during fasting. 12401439_Binding of PPARG ligands (troglitazone or BRL49653) to monocyte PPARG inhibited TXA(2) production and enhanced PGE(2) production of PBMC 12404189_Observational study of gene-disease association. (HuGE Navigator) 12406034_Observational study of gene-disease association. (HuGE Navigator) 12429071_Observational study of gene-disease association. (HuGE Navigator) 12439219_Observational study of gene-disease association. (HuGE Navigator) 12439219_lack of association of codon 12 polymorphism with breast cancer and body mass 12453902_Observational study of gene-disease association. (HuGE Navigator) 12453919_Candidate gene sequencing showed that all four affected subjects were heterozygous for a novel T-->A mutation at PPARG nucleotide 1164 in exon 5 that predicted substitution of phenylalanine at codon 388 by leucine (F388L). 12457461_Data demonstrate that Egr-1, AP1 and Smad are part components of the transforming growth factor beta signal transduction pathway that regulates PPAR gamma expression. 12468551_PPARgamma has a role in inhibiting intestinal epithelial cell growth by regulating levels of TSC-22 12485829_With RXR-alpha, forms a heterodimer involved in the regulation of human trophoblast invasion 12502512_Observational study of gene-disease association. (HuGE Navigator) 12502512_We investigated the phenotypic appearance of the two polymorphisms (Pro12Ala and exon 6 C-->T) in PPARgamma among elderly twins and plasma glucose and insulin profiles among MZ and DZ twins. 12502716_role of intact AF-2 domain for ligand-dependent and ligand-independent interaction and coactivation 12519876_Detection of the PAX8-PPAR gamma fusion oncogene in both follicular thyroid carcinomas and adenomas. 12535639_Results suggest that activation of peroxisome proliferative activated receptor gamma (PPARgamma) may represent a novel approach for the treatment of pancreatic cancer by increasing PTEN levels and inhibiting PI3K activity. 12536206_natural mutations in human PPARgamma, associated with severe insulin resistance and diabetes mellitus, exhibit perturbations in the dynamic behavior of helix 12 12538445_activity of PPARgamma and PPARgamma ligands in cancer [review] 12544508_Observational study of gene-environment interaction. (HuGE Navigator) 12551936_that fatty acids and rosiglitazone directly stimulate transcription of the LRP gene through activation of PPARgamma and increase functional LRP expression. 12565902_Data describe the expression of peroxisome proliferator-activated receptor gamma (PPAR gamma) co-activator 1, PPAR gamma, insulin receptor substrate-1, glucose transporter isoform-4, and mitochondrial uncoupling protein-1 in adipose tissue. 12568179_Observational study of gene-disease association. (HuGE Navigator) 12588773_Functional polymorphism in a STAT5B site of PPAR gamma 3 promoter region affects height and lipid homeostasis via the STAT5B pathway. 12588773_Observational study of gene-disease association. (HuGE Navigator) 12591919_codon 422 may be a part of a co-factor(s) interaction domain necessary for PPARgamma to induce terminal differentiation in epithelial, but not adipocyte, cell lineages 12594295_A novel anti-inflammatory pathway is revealed in human airway smooth muscle, where PPAR gamma activation by natural and synthetic ligands inhibits serum-induced cell growth and induces apoptosis. 12601634_Observational study of gene-disease association. (HuGE Navigator) 12601634_PPARgamma gene polymorphism is associated with exercise-mediated changes of insulin resistance in healthy men. 12601635_Observational study of gene-disease association. (HuGE Navigator) 12601635_conclude that the Pro12Ala polymorphism of the PPAR-gamma2 gene was associated with increased ox-LDL autoantibodies in type 2 diabetic subjects 12609711_Observational study of gene-disease association. (HuGE Navigator) 12609711_PPAR-gamma may play a role in prostate carcinogenesis; however, the Pro12Ala polymorphism does not appear to play a significant role in prostate cancer risk in this large cohort of male Finnish smokers. 12610044_Clinical trial of gene-environment interaction. (HuGE Navigator) 12615657_The activation of PPARgamma-dependent pathways induces the differentiated phenotype in proliferative VSMCs, and this induction is mediated, in part, through a GATA-6-dependent transcriptional mechanism. 12615696_PPARgamma and/or RXR activation inhibit foam cell formation through enhanced cholesterol efflux despite increased oxLDL uptake 12615821_Observational study of gene-disease association. (HuGE Navigator) 12615821_Role for PPARgamma gene polymorphism in pathogenesis of polycystic ovary syndrome (PCOS). Presence of Ala isoform being protective against development of PCOS. 12620489_PPAR-gamma may play a role in the pathogenesis of endometriosis related to the production of IL-6 and some other cytokines. 12630956_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12630956_PPAR-gamma P12A polymorphism can modulate the association between dietary fat intake and components of the metabolic syndrome 12663371_A common A for P substitution at codon 12 in the PPARG2 was associated with reduced incidence of myocardial infarction. 12663371_Observational study of gene-disease association. (HuGE Navigator) 12673785_Observational study of gene-disease association. (HuGE Navigator) 12679463_common Pro12Ala polymorphism of the peoxisome proliferator-activated receptor gamma2 gene has minor influence on mRNA expression of peroxisome proliferator-activated receptor gamma target genes in adipose tissue of obese subjects 12709137_Observational study of gene-disease association. (HuGE Navigator) 12714563_PPARgamma expression is reduced or lacking in all of the angiogenic plexiform lesions of pulmonary hypertensive lungs and in the vascular lesions of a rat model of severe PH (pulmonary hypertension). 12714582_Expression of PPARgamma in activated T-cells may regulate apoptosis & their immunological response in inflammation, depending on agonists released by near-by cells. 12716762_Observational study of gene-disease association. (HuGE Navigator) 12716762_The polymorphism (Pro12Ala) of this receptor beneficially influences insulin resistance and its tracking from childhood to adulthood. 12727991_results suggest that conventional follicular thyroid carcinomas develop through at least two distinct molecular pathways initiated by either RAS point mutation or PAX8-peroxisome proliferator-activated receptor gamma rearrangement 12730300_PPARG has a role in regulating glucose and lipid uptake and oxidation and preadipocyte differentiation 12730867_commensal intestinal flora affects the expression of PPAR gamma and that PPAR gamma expression is considerably impaired in patients with ulcerative colitis 12732844_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12739018_Amino acid substistution is not associated with insulin resistance phenotype. 12746759_Observational study of gene-disease association. (HuGE Navigator) 12746759_The Pro115 Gln variant, but not the Pro12Ala mutation in the PPAR-gamma 2 gene, could be a rare cause of severe insulin resistance. 12763030_Transcriptional activation of hepatic scavenger receptor class B, type I is stimulated by peroxisome proliferator-activated receptor gamma. 12765846_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 12765972_Observational study of gene-disease association. (HuGE Navigator) 12765972_Pro12Ala of this gene is associated with lower serum insulin levels in nonobese African Americans 12767049_activity in prostate cancer cells 12776192_functional downregulation of PPARgamma is a key event in multiple types of thyroid neoplasia 12779083_Treatment of human breast cancer cell line MDA-MB-231 with the synthetic PPARgamma ligands pioglitazone, rosiglitazone, GW7845 or its natural ligand 15-deoxy-delta 12, 14-prostaglandin J2(15d-PGJ2) led to inhibition of the invasiveness of this cell line. 12800106_Because PPARgamma(2) is mainly expressed in adipose tissue, one of the main regulatory effects of the polymorphism may well be the more efficient suppression of (possibly intra-abdominal) lipolysis. 12800106_Observational study of gene-disease association. (HuGE Navigator) 12800228_Troglitazone, peroxisome proliferator-activated receptor gamma ligand, inhibits growth of HepG2 cells and induces apoptosis. COX-2 mRNA and protein and Bcl-2 was down-regulated. Bax and Bak was up-regulated. Activity of caspase-3 was elevated. 12801525_Receptor 'C(k)'-dependent signalling pathway regulates PPAR-gamma gene transcription; a proposed molecular cross-talk pathway between Receptor-C(k) and PPAR-gamma may play a role in control of leukemogenesis. 12801610_Expression of PPARgamma was increased to greatest degree by highly-oxLDL. DNA binding activity of PPARgamma was increased only by mildly- and moderately-oxLDL. 12805087_Results suggest that granulocyte macrophage colony-stimulating factor regulates lung homeostasis via peroxisome proliferator-activated receptor-gamma-dependent pathways. 12813462_PPARgamma participates in the regulation of caveolin 1 and 2 gene expression in human carcinoma cells. 12821652_Rev-Erbalpha as a target gene of PPARgamma in adipose tissue and demonstrate a role for this nuclear receptor as a promoter of adipocyte differentiation. 12827242_Adiponectin is a genetic factor associated with insulin sensitivity. Interactions with PPARgamma2 genotypes modified this association. 12829631_Expression plays a role in energy homeostasis in obese women. 12829658_A functional variant in this protein's promoter is associated with predictors of obesity and NIDDM in Pima Indians. 12829658_Observational study of gene-disease association. (HuGE Navigator) 12829715_Peroxisome proliferator-activated receptor gamma and ligands inhibit surfactant protein B gene expression in the lung 12829999_a novel p53-dependent mechanism in the PPARgamma ligand-mediated inhibition of cholangiocarcinoma growth and suggest a potential therapeutic role of PPARgamma ligands in the treatment of human cholangiocarcinoma. 12839938_Ligands of this receptor affect the expression of RAR beta in cancer cells 12839942_Common polymorphisms in this protein are associated with increased risk of colorectal cancer. 12839942_Observational study of gene-disease association. (HuGE Navigator) 12861348_although activation of PPARgamma appears to have beneficial effects on atherosclerosis and heart failure, it is still largely uncertain whether PPARgamma ligands prevent the development of cardiovascular diseases-REVIEW 12866036_Down-regulation of peroxisome-proliferator activated receptor-gamma is associated with local recurrence and metastasis in breast cancer 12915690_reduced expression of PPARgamma might be an early event in colonic tumorigenesis 12923071_PPARgamma activation enhances cell surface ENaCalpha via up-regulation of SGK1 in collecting duct cells. 12923396_Observational study of gene-disease association. (HuGE Navigator) 12923396_significant contribution of the PPARgamma2(Pro12Ala) and eNOS(4a/b) gene polymorphisms to hypertensive risk in our population (odds ratio, 1.9 and 1.6, respectively), confirmed by multiple logistic regression analysis. 12923397_Observational study of gene-disease association. (HuGE Navigator) 12923397_The common Pro12Ala polymorphism in PPARgamma is associated with lower diastolic blood pressure in male subjects with type 2 diabetes. 12949056_plays an important role in the differentiation of intestinal cells 12970322_lack of the PAX8/PPAR gamma rearrangement in the anaplastic thyroid carcinoma group suggests that the tumorigenic pathway in these tumors is likely to be independent of this fusion 12974743_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14506127_Observational study of gene-disease association. (HuGE Navigator) 14506127_PPARG genotype is an important factor in physiological responses to dietary fat in humans. 14506281_PPARgamma and PPARdelta have roles in transcriptional activation of Spermidine/sperm-ine N1-acetyltransferase, which leads to reduced tissue polyamine contents in human colon cancer cells 14514601_Observational study of gene-disease association. (HuGE Navigator) 14514601_inverse association between the PPARgamma variant 12Ala allele and risk of type 2 diabetes. 14515149_15d-PGJ(2) differently upregulates MMP-1 expression in HMEC-1 through induction of oxidative stress, and inhibits uPA synthesis partly by activation of peroxisome proliferative activated receptor, gamma (PPARgamma). 14550563_non-hydrolysed, oxidised phospholipids in oxLDL activate PPARgamma as a cellular defence mechanism against macrophage death 14566836_15-hydroxy-eicosatetraenoic acid (15S-HETE), an endogenous ligand for PPARgamma, was significantly decreased in the serum of patients with colorectal cancer 14574455_Observational study of genotype prevalence, gene-disease association, and gene-gene interaction. (HuGE Navigator) 14581147_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 14581147_Pro12Ala amino acid variant of the PPARgamma2 isoform is not associated with susceptibility to type 2 diabetes. 14581151_PPARgamma can reduce plasminogen activator inhibitor type 1 production in vascular endothelial cells directly by transcriptional repression. 14581158_Japanese with the Ala12 allele had a significantly lower value of carotid artery intima-media thickness than those without it. 14581158_Observational study of gene-disease association. (HuGE Navigator) 14587029_phosphorylation of PPARgamma was mediated through the ciglitazone-induced activation of Erk1/2 14604894_Observational study of gene-disease association. (HuGE Navigator) 14616762_Observational study of gene-disease association. (HuGE Navigator) 14616762_We report that the PPARG C161T polymorphism is associated with the survival of IgAN patients without hypertension 14627349_Treatment of gastric cancer patients with Vioxx resulted in a significant decrease in plasma and tumor contents of both progastrin and gastrin, and this was accompanied by the increment in tumor expression of COX-2, PPARy, Bax, and caspase-3. 14633865_Observational study of gene-disease association. (HuGE Navigator) 14633865_The presence of the Ala allele may confer protection from diabetic nephropathy in patients with type 2 diabetes. 14657011_Transcriptional silencing via recruitment of corepressor adds to dominant-negative inhibition of wild type by P467L and V290M mutants. Introduction of artificial mutation (L318A) disrupting corepressor interaction abrogated dominant-negative activity. 14659862_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14668325_study shows that peroxisome proliferator-activated receptor gamma (PPARgamma) agonists strongly regulate myeloperoxidase gene expression through the Alu element repeats 14671186_Higher frequency of C-->T substitution in exon 6 of PPAR-gamma gene in polycystic ovary syndrome (PCOS). May play role in complex pathogenesis of obesity in PCOS. Pro(12)Ala polymorphism does not seem to affect body mass index in PCOS women. 14671186_Observational study of gene-disease association. (HuGE Navigator) 14671211_Expression of adipophilin is enhanced during trophoblast differentiation and is up-regulated by ligand-activated PPARgamma/retinoid X receptor. May contribute to fatty acid uptake by placenta. 14671555_Observational study of gene-disease association. (HuGE Navigator) 14677049_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14680975_Observational study of gene-disease association. (HuGE Navigator) 14680975_PPARgamma is a quantitative trait locus for free fatty acid and glycerol, against a background of insulin resistance for adipose tissue lipid metabolism, and therefore as a modifier gene in familial combined hyperlipidemia. 14681322_The high-level expression of PPARgamma may provide a potential molecular target for the treatment of salivary duct carcinoma using agonist ligands. 14681835_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14681835_study examined the modification by peroxisome proliferative activated receptor gamma genotype of the association between energy expenditure, dietary polyunsaturated to saturated fatty acid ratio, and fasting insulin level, a measure of insulin resistance 14686729_High-levels of PPARgamma expression is associated with glioblastoma 14702344_restoration of PPARgamma reverses the activated hepatic stellate cells to the quiescent phenotype and suppresses AP-1 activity via a physical interaction between PPARgamma and JunD 14729856_Observational study of gene-disease association. (HuGE Navigator) 14729856_The Pro12Ala polymorphism was more associated with plasma lipids and fasting glucose concentrations, whereas the C1431T polymorphism was related to the risk of diabetes. 14730381_Ala12 variant of PPARgamma affords protection from Type 2 diabetes, and this protection is modulated by additional common variation at the PPARG locus. 14730381_Observational study of gene-disease association. (HuGE Navigator) 14747257_polymorphic in metabolic syndrome phenotype in Brazil 14974928_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14981210_Observational study of gene-disease association. (HuGE Navigator) 14988278_Association of this gene's single nucleotide polymorphism with type 2 diabetes. 15015141_Observational study of gene-disease association. (HuGE Navigator) 15015141_fatty acid binding protein 4 and PPARgamma work together to influence a biologic pathway affecting insulin sensitivity and body composition 15023995_mPGES-1 and Egr-1 are novel targets of PPARgamma and inhibition of mPGES-1 gene transcription may be one of the mechanisms by which PPARgamma regulates inflammatory responses 15041706_PPARgamma ligands inhibit human lung carcinoma cell growth and induce apoptosis by stimulating the cyclin-dependent kinase inhibitor p21. 15054105_PPARgamma and EGFR signalling have roles in urothelial terminal differentiation 15072437_x ray crystallography to determine binding sites 15077315_By abrogating of TGF beta-induced stimulation of collagen gene expression, myofibroblast transdifferentiation, and Smad-dependent promoter activity in normal fibroblasts, PPAR gamma may play a physiologic role in the regulation of the profibrotic response 15077315_Peroxisome Proliferator Activated Receptor-gamma(PPAR-g) disrupts Transforming Growth Factor-beta (TGF-b) signaling and profibrotic responses in skin fibroblasts. 15083308_Observational study of gene-disease association. (HuGE Navigator) 15083308_Our findings argue against a significant contribution of PPARgamma variations to the genetic basis of psoriasis. 15111325_signaling through PPAR-gamma can mediate transitional differentiation of urothelial cells, modulated by growth regulatory programs 15113752_high glucose upregulates PPAR-gamma and when significantly induced demonstrates anti-proliferative and anti-inflammatory effects. 15128052_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15136115_Observational study of gene-disease association. (HuGE Navigator) 15144586_Observational study of gene-disease association. (HuGE Navigator) 15144586_Peroxisome proliferative activated receptor gamma C161-T may play a important role in carotid artery arteriosclerosis. 15156314_polymorphisms of the PPARg gene are not associated with the incidence of restenosis and adverse clinical events at 30 days or 1 year after coronary artery stenting in patients with diabetes mellitus. 15161789_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15161789_Pro12Ala variant of the PPAR-gamma receptor gene did not explain the failure of patients with NIDDM to increase insulin sensitivity. 15165749_the net result of the pleiotropic effects of PPAR-gamma ligands is improvement of insulin sensitivity, although undesired side-effects limit the utility of this therapy [review] 15187150_15d-PGJ(2)-induced changes were related to functional alteration of the glucocorticoid receptor and did not require the engagement of PPARgamma 15193259_there is an in vivo crosstalk between the HER-kinase axis and PPARgamma pathways, ultimately leading to negative regulation of PPARgamma activity and tumor growth inhibition 15201543_Hypertensive subjects with low birth weight or short length at birth and the Pro12Pro variant had raised systolic blood pressure 15201543_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15202783_In Polish PPAR-gamma2 12Pro allele carriers, obesity correlated with free fatty acid tolerance in males, and insulin resistance in females. 15211802_Observational study of gene-disease association. (HuGE Navigator) 15217350_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15220243_Upregulated by pioglitazone in type 2 diabetes. 15236769_Observational study of gene-disease association. (HuGE Navigator) 15254719_both PPARgamma and its coactivator PGC-1 play important roles in the development and | ENSMUSG00000000440 | Pparg | 2.675005e+02 | 0.9799328 | -0.029245304 | 0.2977698 | 9.641690e-03 | 0.9217797482 | 0.98588240 | No | Yes | 2.568078e+02 | 34.795112 | 2.160106e+02 | 30.119634 | |
| ENSG00000132716 | 50717 | DCAF8 | protein_coding | Q5TAQ9 | FUNCTION: May function as a substrate receptor for CUL4-DDB1 E3 ubiquitin-protein ligase complex. {ECO:0000269|PubMed:16949367, ECO:0000269|PubMed:16964240}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Methylation;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation pathway;WD repeat | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a WD repeat-containing protein that interacts with the Cul4-Ddb1 E3 ligase macromolecular complex. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jul 2009]. | hsa:50717; | Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein ubiquitination [GO:0016567] | 21424732_while RNF10 and WDR42A or VP22 alone showed distinct subcellular localization patterns, RNF10 and WDR42A were relocated when co-expressed with VP22 or its homologues; these potential host cell factors of VP22 might expand the list of host targets of VP22 22500989_WDR42A is a nucleocytoplasmic shuttling protein. 24500646_DCAF8 p.R317C mutation is responsible for specific variety of HMSN2 with infrequent giant axons and mild cardiomyopathy. 32703400_CRL4(DCAF8) and USP11 oppositely regulate the stability of myeloid leukemia factors (MLFs). 35280675_CRL4-DCAF8L1 Regulates BRCA1 and BARD1 Protein Stability. | ENSMUSG00000026554 | Dcaf8 | 6.292748e+03 | 0.7897035 | -0.340617090 | 0.2717857 | 1.562853e+00 | 0.2112479028 | 0.78763590 | No | Yes | 4.250434e+03 | 571.379566 | 6.203071e+03 | 854.753190 |
| ENSG00000132768 | 1802 | DPH2 | protein_coding | Q9BQC3 | FUNCTION: Required for the first step in the synthesis of diphthamide, a post-translational modification of histidine which occurs in translation elongation factor 2 (EEF2). {ECO:0000250|UniProtKB:Q9CR25}. | Acetylation;Alternative splicing;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine;Transferase | PATHWAY: Protein modification; peptidyl-diphthamide biosynthesis. | This gene is one of two human genes similar to the yeast gene dph2. The yeast gene was identified by its ability to complement a diphthamide mutant strain, and thus probably functions in diphthamide biosynthesis. Diphthamide is a post-translationally modified histidine residue present in elongation factor 2 (EF2) that is the target of diphtheria toxin ADP-ribosylation. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2016]. | hsa:1802; | cytosol [GO:0005829]; 2-(3-amino-3-carboxypropyl)histidine synthase activity [GO:0090560]; peptidyl-diphthamide biosynthetic process from peptidyl-histidine [GO:0017183] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 34302311_Spin-Regulated Electron Transfer and Exchange-Enhanced Reactivity in Fe4 S4 -Mediated Redox Reaction of the Dph2 Enzyme During the Biosynthesis of Diphthamide. | ENSMUSG00000028540 | Dph2 | 5.029695e+03 | 1.1980472 | 0.260684704 | 0.2893246 | 8.084724e-01 | 0.3685722665 | 0.83653490 | No | Yes | 5.198884e+03 | 479.305253 | 4.013159e+03 | 380.078557 |
| ENSG00000132780 | 4678 | NASP | protein_coding | P49321 | FUNCTION: Required for DNA replication, normal cell cycle progression and cell proliferation. Forms a cytoplasmic complex with HSP90 and H1 linker histones and stimulates HSP90 ATPase activity. NASP and H1 histone are subsequently released from the complex and translocate to the nucleus where the histone is released for binding to DNA. {ECO:0000250|UniProtKB:Q99MD9}. | Acetylation;Alternative splicing;Cell cycle;Coiled coil;Cytoplasm;DNA replication;Isopeptide bond;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;TPR repeat;Transport;Ubl conjugation | This gene encodes a H1 histone binding protein that is involved in transporting histones into the nucleus of dividing cells. Multiple isoforms are encoded by transcript variants of this gene. The somatic form is expressed in all mitotic cells, is localized to the nucleus, and is coupled to the cell cycle. The testicular form is expressed in embryonic tissues, tumor cells, and the testis. In male germ cells, this protein is localized to the cytoplasm of primary spermatocytes, the nucleus of spermatids, and the periacrosomal region of mature spermatozoa. [provided by RefSeq, Jul 2008]. | hsa:4678; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; histone binding [GO:0042393]; protein-containing complex binding [GO:0044877]; blastocyst development [GO:0001824]; CENP-A containing chromatin assembly [GO:0034080]; DNA replication [GO:0006260]; DNA replication-dependent chromatin assembly [GO:0006335]; DNA replication-independent chromatin assembly [GO:0006336]; G1/S transition of mitotic cell cycle [GO:0000082]; histone exchange [GO:0043486]; male gonad development [GO:0008584]; nucleosome assembly [GO:0006334]; protein transport [GO:0015031]; response to testosterone [GO:0033574] | 16728391_NASP and the linker histones are key players in the assembly of chromatin after DNA replication 18782834_NASP forms distinct, high specificity complexes with histones H3 and H4. 19439102_NASP belongs to a network of genes and gene functions that are critical for cell survival 20164540_NASP and RCAS1 proteins were more frequently expressed in ovarian cancer tissues than with normal ovarian tissue and serous cystadenomas and MRE11 was less frequently expressed 20167597_A deletion analysis of sNASP revealed that the central region, amino acid residues 26-325, of sNASP is responsible for nucleosome assembly in vitro. 21496299_tNASP is critical for the survival of prostate cancer cells; targeting tNASP expression can lead to a new approach for prostate cancer treatment. 21965532_sNASP interacts with linker and core histones through distinct structural domains. 22194605_an increase of miR-29a, and hence decrease of Nasp, may contribute to inhibit cell proliferation during postnatal organ development. 22195965_The insights into NASP function and the existence of a tunable reservoir in mammalian cells demonstrate that contingency is integrated into the histone supply chain to respond to unexpected changes in demand. 22968912_Studies indicate that histone chalerones nucleoplasmin (NPM2/NPM3) preferentially associated with histones H2A-H2B in the egg and the nuclear autoantigenic sperm protein (NASP) families. 24951090_NASP family Proteins are highly conserved throughout the eukaryotes and posses four TPR motifs. It was likely present in the last common ancestor of eukaryotes possibly representing an important innovation regarding H3/H4 transport mechanism. Different TPR motifs in NASP have evolved at different rates with TPR1/4 evolving faster than TPR2/3. 25615412_These results provide evidence that tNASP is ubiquitously produced in various types of human tissues and promotes in vitro nucleosome assembly with H3 variant specificity. 25669170_Our research demonstrates that knockdown of tNASP effectively inhibits the proliferation and causes G1 phase arrest through ERK/MAPK signal pathway. 26673727_findings reveal a new mode of interaction between a TPR repeat domain of histone chaperone sNASP and an evolutionarily conserved peptide motif found in canonical H3 and in all histone H3 variants 28173777_NASP is targeted by miR-29c in gastric cancer cells. miR-29c can decrease NASP expression and the effects observed following miR-29c overexpression are partially due to NASP depletion. 30076957_Results found that NASP expression levels were upregulated in liver tumors. Its down-regulation inhibited liver cells from forming tumors. Also, NASP depletion led to a global decrease of histone H3K9me1 modification associated with newly H3 processing, which occurred directly at the promoters of up-regulated anti-tumor genes BACH2 and RunX1T1. 31797734_miR-381-3p targeted and suppressed NASP gene, reduced the viability, migration, invasion, EMT of HNSCC cells, demonstrating that miR-381-3p has the potential to be a therapeutic target in inhibiting the progression of HNSCC 34633056_Knockdown of long noncoding RNA colorectal neoplasia differentially expressed inhibits hepatocellular carcinoma progression by mediating the expression of nuclear autoantigenic sperm protein. 34786730_UBR7 acts as a histone chaperone for post-nucleosomal histone H3. | ENSMUSG00000028693 | Nasp | 1.140353e+04 | 0.9015076 | -0.149588431 | 0.2516848 | 3.500360e-01 | 0.5540927732 | 0.89296216 | No | Yes | 8.921997e+03 | 835.710968 | 9.755770e+03 | 936.802669 | |
| ENSG00000132950 | 9205 | ZMYM5 | protein_coding | Q9UJ78 | FUNCTION: Functions as a transcriptional regulator. {ECO:0000269|PubMed:17126306}. | 3D-structure;Alternative splicing;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:9205; | nucleus [GO:0005634]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122] | ENSMUSG00000040123 | Zmym5 | 6.176569e+01 | 0.6437045 | -0.635529511 | 0.4984049 | 1.547819e+00 | 0.2134576993 | 0.78763590 | No | Yes | 3.985003e+01 | 9.897293 | 6.878344e+01 | 17.153674 | |||
| ENSG00000133069 | 9911 | TMCC2 | protein_coding | O75069 | FUNCTION: May be involved in the regulation of the proteolytic processing of the amyloid precursor protein (APP) possibly also implicating APOE. {ECO:0000269|PubMed:21593558}. | Alternative splicing;Coiled coil;Endoplasmic reticulum;Membrane;Methylation;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:9911; | endomembrane system [GO:0012505]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; amyloid precursor protein metabolic process [GO:0042982] | ENSMUSG00000042066 | Tmcc2 | 1.032667e+03 | 1.3992100 | 0.484612477 | 0.3391680 | 2.037196e+00 | 0.1534923157 | 0.77404149 | No | Yes | 1.155506e+03 | 172.843091 | 6.777187e+02 | 104.564227 | |||
| ENSG00000133835 | 3295 | HSD17B4 | protein_coding | P51659 | FUNCTION: Bifunctional enzyme acting on the peroxisomal beta-oxidation pathway for fatty acids. Catalyzes the formation of 3-ketoacyl-CoA intermediates from straight-chain, 2-methyl-branched-chain fatty acids bile acid intermediates. With EHHADH, catalyzes the hydration of trans-2-enoyl-CoA and the dehydrogenation of 3-hydroxyacyl-CoA, but with opposite chiral specificity (PubMed:10671535). {ECO:0000269|PubMed:10671535, ECO:0000269|PubMed:15060085, ECO:0000269|PubMed:8902629, ECO:0000269|PubMed:9089413}. | 3D-structure;Acetylation;Alternative splicing;Deafness;Direct protein sequencing;Disease variant;Fatty acid metabolism;Isomerase;Lipid metabolism;Lyase;NAD;Oxidoreductase;Peroxisome;Phosphoprotein;Reference proteome | PATHWAY: Lipid metabolism; fatty acid beta-oxidation. {ECO:0000269|PubMed:10706581, ECO:0000269|PubMed:15060085, ECO:0000269|PubMed:9089413, ECO:0000269|PubMed:9482850}. | The protein encoded by this gene is a bifunctional enzyme that is involved in the peroxisomal beta-oxidation pathway for fatty acids. It also acts as a catalyst for the formation of 3-ketoacyl-CoA intermediates from both straight-chain and 2-methyl-branched-chain fatty acids. Defects in this gene that affect the peroxisomal fatty acid beta-oxidation activity are a cause of D-bifunctional protein deficiency (DBPD). An apparent pseudogene of this gene is present on chromosome 8. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, May 2014]. | hsa:3295; | cytosol [GO:0005829]; membrane [GO:0016020]; peroxisomal matrix [GO:0005782]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; (3R)-hydroxyacyl-CoA dehydrogenase (NAD) activity [GO:0106386]; 17-beta-hydroxysteroid dehydrogenase (NAD+) activity [GO:0044594]; 3-hydroxyacyl-CoA dehydrogenase activity [GO:0003857]; 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity [GO:0033989]; enoyl-CoA hydratase activity [GO:0004300]; isomerase activity [GO:0016853]; long-chain-enoyl-CoA hydratase activity [GO:0016508]; protein homodimerization activity [GO:0042803]; androgen metabolic process [GO:0008209]; estrogen metabolic process [GO:0008210]; fatty acid beta-oxidation [GO:0006635]; medium-chain fatty-acyl-CoA metabolic process [GO:0036112]; osteoblast differentiation [GO:0001649]; Sertoli cell development [GO:0060009]; very long-chain fatty acid metabolic process [GO:0000038]; very long-chain fatty-acyl-CoA metabolic process [GO:0036111] | 15644212_crystal structure of 2-enoyl-CoA hydratase 2 16766224_Deficiency of this enzyme in man causes a severe developmental syndrome with abnormalities in several organs but in particular in the brain, leading to death within the first year of life. 18086758_Observational study of gene-disease association. (HuGE Navigator) 18281655_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18281655_Observational study of gene-disease association. (HuGE Navigator) 18415690_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18794456_HSD17B4 mRNA is expressed in human skin, at similar levels in men and women. HSD17B4 levels are not altered by topical 17-beta-estradiol treatment. 19100308_HSD17B4 is not only associated with the presence of prostate cancer, but is also a significant independent predictor of poor patient outcome. 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19453261_Observational study of gene-disease association. (HuGE Navigator) 19574343_Observational study of gene-disease association. (HuGE Navigator) 19598235_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19627379_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19776291_Observational study of gene-disease association. (HuGE Navigator) 19776291_rs11205 in HSD17B4 was associated with testicular germ cell tumor. Risk doubled per copy of the minor A allele. Homozygosity of this allele quadrupled the risk vs. homozygous major G allele. The risk was increased both for seminoma & nonseminoma. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20214802_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20566640_MFE2 anchors its substrate around the region from Trp(249) to Arg(251) and positions the substrate along the hydrophobic cavity in the proper direction toward the catalytic center 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634197_Meta-analysis of gene-disease association. (HuGE Navigator) 20673864_Perrault syndrome and DBP deficiency overlap clinically and Perrault syndrome is genetically heterogeneous. 20734064_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 20949532_The diagnosis of a type III DBPD with a missense mutation (T15A) in the HSD17B4 gene, coding for D-bifunctional protein (DBP), could be established. 22265031_Epistasis between the HSD17B4 and thyroglobulin polymorphisms is associated with premature ovarian failure. A haplotype in the HSD17B4 gene was identified that was significantly associated with resistance to POF 23181892_Specific combination of compound heterozygous mutations in 17beta-hydroxysteroid dehydrogenase type 4 (HSD17B4) defines a new subtype of D-bifunctional protein deficiency. 23308274_Structural MFE-2 instability is the molecular basis of D-bifunctional protein deficiency type III. 23313254_Molecular models of domain structure of MFE-2 from human, C. elegans, and Drosophila melanogaster lend support to possible structural role of MFE-2 domains including SCP-2L (sterol carrier protein 2-like) domain in human and C. elegans proteins. 25448063_Results show that HSD17B4 is highly expressed in hepatocellular carcinoma (HCC) cells and activated NF-kappaB co-localized with the NF-kappaB-responsive element of HSD17B4 suggesting that HSD17B4 plays an important role in aggravated HCC progression. 26970254_ere we present eight families affected by Perrault syndrome. In five families we identified novel or previously reported variants in HSD17B4, LARS2, CLPP and C10orf2 28186977_we identified that methylation of the promoter CpG island of HSD17B4 was associated with the pathological complete response of HER2-positive breast cancer to trastuzumab and chemotherapy with a specificity of 79% 28296597_This study reveals a crosstalk between acetylation and chaperone-mediated autophagy degradation in HSD17B4 regulation. 28830375_Our findings supported that HSD17B4 was one of the genes contributing to Perrault syndrome in this consanguineous Chinese Han family. 30540494_In HEK293 cells, the D-bifunctional protein (HSD17B4) and the peroxisomal ABC transporter ABCD3 are essential in peroxisomal oxidation of lauric and palmitic acid. 30747222_HSD17B4 overexpression increased STAT3 activation. 32678070_Acetylation-mediated degradation of HSD17B4 regulates the progression of prostate cancer. 33115767_Rapid whole-genome sequencing identifies a homozygous novel variant, His540Arg, in HSD17B4 resulting in D-bifunctional protein deficiency disorder diagnosis. 33935042_The Peroxisomal Localization of Hsd17b4 Is Regulated by Its Interaction with Phosphatidylserine. | ENSMUSG00000024507 | Hsd17b4 | 1.768371e+03 | 0.9702203 | -0.043615667 | 0.3232694 | 1.827398e-02 | 0.8924685423 | 0.98019346 | No | Yes | 1.438768e+03 | 270.390493 | 1.447080e+03 | 278.866772 |
| ENSG00000133983 | 51241 | COX16 | protein_coding | Q9P0S2 | FUNCTION: Required for the assembly of the mitochondrial respiratory chain complex IV (CIV), also known as cytochrome c oxidase (PubMed:29355485, PubMed:29381136, PubMed:33169484). Promotes the insertion of copper into the active site of cytochrome c oxidase subunit II (MT-CO2/COX2) (PubMed:29355485, PubMed:29381136). Interacts specifically with newly synthesized MT-CO2/COX and its copper center-forming metallochaperones SCO1, SCO2 and COA6 (PubMed:29381136). Probably facilitates MT-CO2/COX2 association with the MITRAC assembly intermediate containing MT-CO1/COX1, thereby participating in merging the MT-CO1/COX1 and MT-CO2/COX2 assembly lines (PubMed:29381136). {ECO:0000269|PubMed:29355485, ECO:0000269|PubMed:29381136, ECO:0000269|PubMed:33169484}. | Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Primary mitochondrial disease;Reference proteome;Transmembrane;Transmembrane helix | hsa:51241; | integral component of mitochondrial inner membrane [GO:0031305]; mitochondrial cytochrome c oxidase assembly [GO:0033617] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 29355485_COX16 is required for assembly of cytochrome C oxidase in human cells and is involved in copper delivery to COX2. 29381136_Here, the authors find that COX16, a protein required for cytochrome c oxidase assembly, interacts specifically with newly synthesized COX2 and its copper center-forming metallochaperones SCO1, SCO2, and COA6. The recruitment of SCO1 to the COX2-module is COX16- dependent and patient-mimicking mutations in SCO1 affect interaction with COX16. | ENSMUSG00000091803 | Cox16 | 8.034561e+02 | 0.6228795 | -0.682974997 | 0.2937841 | 5.427958e+00 | 0.0198168364 | 0.49835793 | No | Yes | 6.052943e+02 | 87.833964 | 9.201631e+02 | 136.425066 | ||
| ENSG00000134030 | 9811 | CTIF | protein_coding | O43310 | FUNCTION: Specifically required for the pioneer round of mRNA translation mediated by the cap-binding complex (CBC), that takes place during or right after mRNA export via the nuclear pore complex (NPC). Acts via its interaction with the NCBP1/CBP80 component of the CBC complex and recruits the 40S small subunit of the ribosome via eIF3. In contrast, it is not involved in steady state translation, that takes place when the CBC complex is replaced by cytoplasmic cap-binding protein eIF4E. Also required for nonsense-mediated mRNA decay (NMD), the pioneer round of mRNA translation mediated by the cap-binding complex playing a central role in nonsense-mediated mRNA decay (NMD). {ECO:0000269|PubMed:19648179}. | Acetylation;Alternative splicing;Cytoplasm;Nonsense-mediated mRNA decay;Phosphoprotein;Reference proteome;Translation regulation | CTIF is a component of the CBP80 (NCBP1; MIM 600469)/CBP20 (NCBP2; MIM 605133) translation initiation complex that binds cotranscriptionally to the cap end of nascent mRNA. The CBP80/CBP20 complex is involved in a simultaneous editing and translation step that recognizes premature termination codons (PTCs) in mRNAs and directs PTC-containing mRNAs toward nonsense-mediated decay (NMD). On mRNAs without PTCs, the CBP80/CBP20 complex is replaced with cytoplasmic mRNA cap-binding proteins, including EIF4G (MIM 600495), and steady-state translation of the mRNAs resumes in the cytoplasm (Kim et al., 2009 [PubMed 19648179]).[supplied by OMIM, Dec 2009]. | hsa:9811; | cytosol [GO:0005829]; perinuclear region of cytoplasm [GO:0048471]; RNA binding [GO:0003723]; translation activator activity [GO:0008494]; nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184]; regulation of translational initiation [GO:0006446] | 19648179_a new MIF4G domain-containing protein, CTIF that interacts directly with CBP80 and is part of the CBP80/20-dependent translation initiation complex [CBP80/20-dependent translation initiation factor, CTIF] 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21840310_Study reveals that Ago2, but not Ago2F2V2, inhibits the binding of CBP80/20 to cap structure. 26264041_association of SNPs in LCP1 and CTIF with hearing 29789535_CTIF was identified as a novel PARylation target of the TNKS2 in the centrosomal region of cells, which plays a role in the distribution of the centrosomal satellites. 33103564_CBP80/20-dependent translation initiation factor (CTIF) inhibits HIV-1 Gag synthesis by targeting the function of the viral protein Rev. 34232997_Translation mediated by the nuclear cap-binding complex is confined to the perinuclear region via a CTIF-DDX19B interaction. | ENSMUSG00000052928 | Ctif | 1.293029e+03 | 1.2864423 | 0.363386704 | 0.2843071 | 1.622912e+00 | 0.2026862927 | 0.78445471 | No | Yes | 1.291734e+03 | 129.410054 | 9.614093e+02 | 99.333151 | |
| ENSG00000134058 | 1022 | CDK7 | protein_coding | P50613 | FUNCTION: Serine/threonine kinase involved in cell cycle control and in RNA polymerase II-mediated RNA transcription. Cyclin-dependent kinases (CDKs) are activated by the binding to a cyclin and mediate the progression through the cell cycle. Each different complex controls a specific transition between 2 subsequent phases in the cell cycle. Required for both activation and complex formation of CDK1/cyclin-B during G2-M transition, and for activation of CDK2/cyclins during G1-S transition (but not complex formation). CDK7 is the catalytic subunit of the CDK-activating kinase (CAK) complex. Phosphorylates SPT5/SUPT5H, SF1/NR5A1, POLR2A, p53/TP53, CDK1, CDK2, CDK4, CDK6 and CDK11B/CDK11. CAK activates the cyclin-associated kinases CDK1, CDK2, CDK4 and CDK6 by threonine phosphorylation, thus regulating cell cycle progression. CAK complexed to the core-TFIIH basal transcription factor activates RNA polymerase II by serine phosphorylation of the repetitive C-terminal domain (CTD) of its large subunit (POLR2A), allowing its escape from the promoter and elongation of the transcripts (PubMed:9852112). Phosphorylation of POLR2A in complex with DNA promotes transcription initiation by triggering dissociation from DNA. Its expression and activity are constant throughout the cell cycle. Upon DNA damage, triggers p53/TP53 activation by phosphorylation, but is inactivated in turn by p53/TP53; this feedback loop may lead to an arrest of the cell cycle and of the transcription, helping in cell recovery, or to apoptosis. Required for DNA-bound peptides-mediated transcription and cellular growth inhibition. {ECO:0000269|PubMed:10024882, ECO:0000269|PubMed:11113184, ECO:0000269|PubMed:16327805, ECO:0000269|PubMed:17373709, ECO:0000269|PubMed:17386261, ECO:0000269|PubMed:17901130, ECO:0000269|PubMed:19015234, ECO:0000269|PubMed:19071173, ECO:0000269|PubMed:19136461, ECO:0000269|PubMed:19450536, ECO:0000269|PubMed:19667075, ECO:0000269|PubMed:20360007, ECO:0000269|PubMed:9372954, ECO:0000269|PubMed:9840937, ECO:0000269|PubMed:9852112}. | 3D-structure;ATP-binding;Acetylation;Cell cycle;Cell division;Cytoplasm;DNA damage;DNA repair;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase | The protein encoded by this gene is a member of the cyclin-dependent protein kinase (CDK) family. CDK family members are highly similar to the gene products of Saccharomyces cerevisiae cdc28, and Schizosaccharomyces pombe cdc2, and are known to be important regulators of cell cycle progression. This protein forms a trimeric complex with cyclin H and MAT1, which functions as a Cdk-activating kinase (CAK). It is an essential component of the transcription factor TFIIH, that is involved in transcription initiation and DNA repair. This protein is thought to serve as a direct link between the regulation of transcription and the cell cycle. [provided by RefSeq, Jul 2008]. | hsa:1022; | CAK-ERCC2 complex [GO:0070516]; cyclin-dependent protein kinase activating kinase holoenzyme complex [GO:0019907]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; fibrillar center [GO:0001650]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; transcription factor TFIIH core complex [GO:0000439]; transcription factor TFIIH holo complex [GO:0005675]; transcription factor TFIIK complex [GO:0070985]; ATP binding [GO:0005524]; ATP-dependent activity, acting on DNA [GO:0008094]; cyclin-dependent protein serine/threonine kinase activity [GO:0004693]; protein C-terminus binding [GO:0008022]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; RNA polymerase II CTD heptapeptide repeat kinase activity [GO:0008353]; cell cycle [GO:0007049]; cell division [GO:0051301]; DNA repair [GO:0006281]; phosphorylation of RNA polymerase II C-terminal domain [GO:0070816]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein phosphorylation [GO:0006468]; protein stabilization [GO:0050821]; regulation of cell cycle [GO:0051726]; regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0000079]; regulation of G1/S transition of mitotic cell cycle [GO:2000045]; snRNA transcription by RNA polymerase II [GO:0042795]; transcription by RNA polymerase II [GO:0006366]; transcription initiation from RNA polymerase II promoter [GO:0006367] | 11062157_Observational study of genotype prevalence. (HuGE Navigator) 12140753_The cyclin H/cdk7/Mat1 kinase activity is regulated by CK2 phosphorylation of cyclin H. 15530371_determined the crystal structure of human CDK7 in complex with ATP at 3 A resolution 15695176_Confocal microscopy revealed the co-localization of PKC-iota with CAK/cdk7 in both the cytoplasm and nucleus of U-373 MG glioma cells, supporting its role in cell signaling 16327805_Cdk7 accomplishes dual functions in cell-cycle control and transcription not through promiscuity but through distinct, stringent modes of substrate recognition. 17012222_The reduced CDK7 kinase activity is responsible for the inactivation of CDC2/p34 kinase and the irreversible G(2)/M phase cell-cycle arrest of a human gastric carcinoma cell line. 17386261_Different modes of CDK activation by Cdk7 at two distinct execution points in the cell cycle. 17628022_Retinoic-acid-induced RAR-CAK signaling events appear to proceed intrinsically during granulocytic development of normal primitive hematopoietic cells. ALDH-governed RA availability may mediate this process by initiating RAR-CAK signaling. 17707548_These results suggested that genetic variants in CAK genes, Cdk7, cyclin H, MAT1, might modulate the risk of lung cancer in a gene-gene interaction mode, which consist to the biochemical interaction of corresponding proteins. 17901130_Data revealed that SF1 and CDK7 reside in the same complex and CDK inhibitor roscovitine blocked phosphorylation of SF1, and an inactive form of CDK7 repressed the phosphorylation level and the transactivation capacity of SF1. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18950027_The expression of cyclin H and CDK7 protein in proliferating hemangiomas was significantly higher than that in involuting hemangiomas and normal skin tissues. 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19487459_Results suggest that CDK4 might not be phosphorylated by CDK7 in intact cells but is more likely phosphorylated by another, presumably proline-directed kinase(s). 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19667075_A role of Cdk7 in regulating elongation is further suggested by enhanced histone H4 acetylation and diminished histone H4 trimethylation on lysine 36-two marks of elongation-within genes when the kinase was inhibited. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19738611_Observational study of gene-disease association. (HuGE Navigator) 19941161_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19941161_Our findings suggest that the CDK7 TT genotype and the combined genetic polymorphisms of CDK7 and ESR1P325P are associated with breast cancer in Korean women. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20622004_Observational study of gene-disease association. (HuGE Navigator) 22021906_glioma cells may be proliferating through a novel PI (3)-kinase-/PKC-iota/Cdk7/cdk2-mediated pathway. 22447844_Data show that the cyclin-dependent kinase 7/9 inhibitor (CDK7/9 inhibitor) potently inhibits FIP1L1-PDGFRalpha-positive Bcr-Abl-positive chronic myeloid leukemia (CML) cells. 23027873_three CTD kinases, CDK7, CDK9, and BRD4, engage in cross-talk, modulating their subsequent C-terminal domain phosphorylation, and also phosphorylate TAF7 23064645_Cdk7 thus acts through TFIIE and DSIF to establish, and through P-TEFb to relieve, barriers to elongation: incoherent feedforward that might create a window to recruit RNA-processing machinery. 23393140_Interaction with cyclin H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal binding protein 2 (CtBP2) and promotes cancer cell migration 23456497_CCNH and CDK7 may play an important role in the tumorigenesis and development of esophageal squamous cell carcinoma. 23532886_Data indicate that TG02 blocked signaling by CDKs 1, 2, 7, and 9 and ERK5, leading to potent and highly consistent antimyeloma activity. 23622515_Activating phosphorylation of Cdk7 rises concurrently with that of Cdk4 as cells exit quiescence and accelerates Cdk4 activation in vitro. 23737759_CDK7 involved in phosphorylation/activation of CDK4 and CDK6; existence of CDK4-activating kinase(s) other than CDK7; and novel CDK7-dependent positive feedbacks mediated by p21 phosphorylation by CDK4 and CDK2 to sustain CDK4 activation. 25047832_Data suggest a quantitative contribution of CDK7 to mRNA synthesis, which is critical for cellular homeostasis. 25820824_Our results indicated that CCNH/CDK7-CtBP2 axis may augment ESCC cell migration, and targeting the interaction of both may provide a novel therapeutic target of esophageal squamous cell carcinoma . 26257281_Cdk7 broadly influences transcription and capping. 26406377_Study shows that triple-negative but not hormone receptor-positive breast cancer cells are exceptionally dependent on CDK7, a transcriptional cyclin-dependent kinase. 27155449_Data indicate that cyclin dependent kinase 7 (CDK7) is overexpressed in gastric cancer cell lines and tissues. 27301701_Expressions of components of the CAK complex, CDK7, MAT1, and Cyclin H are elevated in breast cancer. 27562173_High expression of MMP14 and CDK7 was independent prognostic factors for overall survival in patients with gastric cancer. 27899423_MYC promotes mRNA cap methylation and protein production of Wnt/beta-catenin signaling transcripts through recruitment of cyclin-dependent kinase 7 (CDK7) and consequently RNMT to gene promoters. 28134252_Study demonstrate that CDK7 activity is necessary to maintain the transcriptional program induced by STAT proteins that are activated both aberrantly by mutation and by extracellular cues in T-cell lymphomas. 28400279_Taken together, these findings elucidated a novel mechanism of prostate cancer progression. Thus, SNHG1 might serve as a potential target for prostate cancer therapies. 28423697_Our studies have demonstrated the essential role of endogenous PRL and CDK7 in the upregulation of PRLR by E2 and provide insights for therapeutic approaches that will mitigate the transcription/expression of PRLR and its participation in breast cancer progression fueled by E2 and PRL via their cognate receptors. 28768201_these results implicate a CDK7-dependent 'CTD code' that regulates chromatin marks in addition to RNA processing and pol II pausing. 29507396_In this study Hep3B and Huh7 cells were infected with two CDK7 shRNAs, both of which reduced CDK7 levels. Inhibition of CDK7 impaired proliferation of Hep3B and Huh7 cells in both short-term IncuCyte(R) cell proliferation assays and long-term colony formation assays. 29743242_CDK7-mediated Ser-175 phosphorylation is a downstream nuclear event essential for facilitating CDK9 T-loop interactions with Tat 29981747_the oncogenic roles of CDK7 in hepatocellular carcinoma and suggested that CDK7 might be a promising therapeutic target 30451989_Targeting CDK7 increases the stability of Snail to promote the dissemination of colorectal cancer. 30473182_Our findings indicate that aberrant CDK7 overexpression associates with T-stage and reduced survival in oral squamous cell carcinoma. 31399555_combining THZ1 with a BCL2/BCL-XL inhibitor ABT-263 synergized in impairing cell growth and driving apoptosis. Our results demonstrate CDK7 as a potential target in treating CCA. Combinations of CDK7 inhibition and BCL2/BCL-XL inhibition may offer a novel therapeutic strategy for CCA. 31462705_Dual HER2 and CDK7 inhibition induced tumor regression in two HER2iR BC xenograft models in vivo. Our data support the utilization of CDK7 inhibition as an additional therapeutic avenue that blocks the activation of genes engaged by multiple HER2iR kinases. 31755214_Inhibition of cyclin-dependent kinase 7 down-regulates yes-associated protein expression in mesothelioma cells. 31765738_our findings reveal a novel therapeutic strategy of pharmacological inhibitions of BRD4 and CDK7 against head and neck squamous cell carcinoma. 32397800_An integrative multi-omics approach uncovers the regulatory role of CDK7 and CDK4 in autophagy activation induced by silica nanoparticles. 33287368_Efficacy of the CDK7 Inhibitor on EMT-Associated Resistance to 3rd Generation EGFR-TKIs in Non-Small Cell Lung Cancer Cell Lines. 33503275_Pharmacological inhibition of CDK7 by THZ1 impairs tumor growth in p53-mutated HNSCC. 34004188_Cyclin-Dependent Kinase 7 Promotes Th17/Th1 Cell Differentiation in Psoriasis by Modulating Glycolytic Metabolism. 34296805_CDK7 and MITF repress a transcription program involved in survival and drug tolerance in melanoma. 35367753_Inhibition of CDK7-dependent transcriptional addiction is a potential therapeutic target in synovial sarcoma. | ENSMUSG00000069089 | Cdk7 | 2.625278e+02 | 1.3505500 | 0.433547079 | 0.3926642 | 1.222474e+00 | 0.2688756135 | 0.79897038 | No | Yes | 2.890527e+02 | 60.096808 | 1.897744e+02 | 40.588933 | |
| ENSG00000134108 | 55207 | ARL8B | protein_coding | Q9NVJ2 | FUNCTION: Small GTPase which cycles between active GTP-bound and inactive GDP-bound states (PubMed:15331635, PubMed:16537643). In its active state, binds to a variety of effector proteins playing a key role in the regulation of lysosomal positioning which is important for nutrient sensing, natural killer cell-mediated cytotoxicity and antigen presentation. Along with its effectors, orchestrates lysosomal transport and fusion (PubMed:16650381, PubMed:16537643, PubMed:28325809, PubMed:25898167). Localizes specifically to lysosomal membranes and mediates anterograde lysosomal motility by recruiting PLEKHM2, which in turn recruits the motor protein kinesin-1 on lysosomes. Required for lysosomal and cytolytic granule exocytosis (PubMed:22172677, PubMed:29592961, PubMed:24088571). Critical factor involved in NK cell-mediated cytotoxicity. Drives the polarization of cytolytic granules and microtubule-organizing centers (MTOCs) toward the immune synapse between effector NK lymphocytes and target cells (PubMed:24088571). In neurons, mediates the anterograde axonal long-range transport of presynaptic lysosome-related vesicles required for presynaptic biogenesis and synaptic function (By similarity). Also acts as a regulator of endosome to lysosome trafficking pathways of special significance for host defense (PubMed:21802320). Regulates cargo trafficking to lysosomes by binding to PLEKHM1 and recruiting the HOPS subunit VPS41, resulting in functional assembly of the HOPS complex on lysosomal membranes (PubMed:16537643, PubMed:25908847). Plays an important role in cargo delivery to lysosomes for antigen presentation and microbial killing. Directs the intersection of CD1d with lipid antigens in lysosomes, and plays a role in intersecting phagosomes with lysosomes to generate phagolysosomes that kill microbes (PubMed:25908847, PubMed:21802320). Involved in the process of MHC II presentation. Regulates the delivery of antigens to lysosomes and the formation of MHC II-peptide complexes through the recruitment of the HOPS complex to lysosomes allowing the fusion of late endosomes to lysosomes (By similarity). May play a role in chromosome segregation (PubMed:15331635). {ECO:0000250|UniProtKB:Q9CQW2, ECO:0000269|PubMed:15331635, ECO:0000269|PubMed:16537643, ECO:0000269|PubMed:16650381, ECO:0000269|PubMed:21802320, ECO:0000269|PubMed:22172677, ECO:0000269|PubMed:24088571, ECO:0000269|PubMed:25898167, ECO:0000269|PubMed:25908847, ECO:0000269|PubMed:28325809, ECO:0000269|PubMed:29592961}.; FUNCTION: (Microbial infection) During Mycobacterium tuberculosis (Mtb) infection, is required for plasma membrane repair by controlling the exocytosis of lysosomes in macrophages. ARL8B secretion pathway is crucial to control the type of cell death of the M. tuberculosis-infected macrophages, distinguishing avirulent from virulent Mtb induced necrotic cell death. {ECO:0000269|PubMed:29592961}.; FUNCTION: (Microbial infection) During infection, coronaviruses such as SARS-CoV-2 and the chaperone HSPA5/GRP78 are probably co-released through ARL8B-dependent lysosomal exocytic pathway for unconventional egress. {ECO:0000269|PubMed:33157038}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Cell junction;Cell projection;Cytoplasm;Cytoskeleton;Direct protein sequencing;Endosome;GTP-binding;Hydrolase;Lysosome;Membrane;Nucleotide-binding;Protein transport;Reference proteome;Synapse;Transport | hsa:55207; | axon cytoplasm [GO:1904115]; cytolytic granule membrane [GO:0101004]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; late endosome membrane [GO:0031902]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; membrane [GO:0016020]; midbody [GO:0030496]; spindle midzone [GO:0051233]; synapse [GO:0045202]; vacuolar membrane [GO:0005774]; alpha-tubulin binding [GO:0043014]; beta-tubulin binding [GO:0048487]; G protein activity [GO:0003925]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; anterograde axonal transport [GO:0008089]; antigen processing and presentation following phagocytosis [GO:0002747]; antigen processing and presentation of polysaccharide antigen via MHC class II [GO:0002505]; autophagosome-lysosome fusion [GO:0061909]; calcium ion regulated lysosome exocytosis [GO:1990927]; cell cycle [GO:0007049]; cell division [GO:0051301]; chromosome segregation [GO:0007059]; endosomal transport [GO:0016197]; endosome to lysosome transport of low-density lipoprotein particle [GO:0090117]; late endosome to lysosome transport [GO:1902774]; lysosome localization [GO:0032418]; natural killer cell mediated cytotoxicity [GO:0042267]; phagosome-lysosome fusion [GO:0090385]; plasma membrane repair [GO:0001778]; protein transport [GO:0015031]; viral exocytosis [GO:0046754] | 15331635_Results suggest that the novel GTPases Gie1 and Gie2 associate with microtubules, and might be involved in chromosome segregation. 16650381_Arl8b is involved in trafficking processes for lysosomes. 19398576_Data show that with MAK3 knockdown, p53 is stabilized and phosphorylated and there is a significant transcriptional activation of proapoptotic genes downstream of p53, and that localization of Arl8b is altered, suggesting that Arl8b is a Mak3 substrate. 21802320_a critical regulator of cargo delivery to lysosomes 21824248_The authors find that Arl8B is required for kinesin-1 recruitment to Salmonella-containing vacuoles, migration of the vacuoles to the cell periphery 24 h after infection and for cell-to-cell transfer of bacteria to neighbouring cells. 24088571_a small GTP-binding protein, ADP-ribosylation factor-like 8b (Arl8b), is identified as a critical factor required for NK cell-mediated cytotoxicity. 25908847_Arl8b regulates the association of the human HOPS complex with lysosomal membranes. 27105540_Authors present evidence that Arl8b facilitates lipid hydrolysis to maintain efficient metabolism for a proliferative capacity in low nutrient environments, suggesting a likely explanation for the complete inability of Arl8b-depleted tumor cells to grow in vivo. 27808481_Data indicate that E3 ubiquitin-protein ligase RNF167 ubiquitinates Arl8B at the lysine residue K141 and reduces the level of the ADP-ribosylation factor-like 8B Arl8B protein. 28993467_LAMTOR1 is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. 29592961_These findings suggest that membrane repair mediated by Arl8b may be an important mechanism distinguishing avirulent from virulent M. tuberculosis-induced necrotic cell death 30212607_Arl8b contributes to the recruitment of LAMP1 to Salmonella-induced tubules. Salmonella SifA protein interacts with Arl8b. | ENSMUSG00000030105 | Arl8b | 6.560259e+02 | 0.7209304 | -0.472068196 | 0.2963168 | 2.451846e+00 | 0.1173867298 | 0.75783482 | No | Yes | 5.869082e+02 | 107.255041 | 6.965065e+02 | 130.339590 | ||
| ENSG00000134186 | 55119 | PRPF38B | protein_coding | Q5VTL8 | FUNCTION: May be required for pre-mRNA splicing. {ECO:0000305}. | Acetylation;Alternative splicing;Coiled coil;Nucleus;Phosphoprotein;Reference proteome;Spliceosome;mRNA processing;mRNA splicing | hsa:55119; | precatalytic spliceosome [GO:0071011]; RNA binding [GO:0003723]; mRNA processing [GO:0006397]; RNA splicing [GO:0008380] | ENSMUSG00000027881 | Prpf38b | 1.872968e+03 | 0.9209048 | -0.118876057 | 0.2944287 | 1.644307e-01 | 0.6851095822 | 0.93348335 | No | Yes | 1.450421e+03 | 206.059186 | 1.752534e+03 | 254.948972 | |||
| ENSG00000134255 | 10390 | CEPT1 | protein_coding | Q9Y6K0 | FUNCTION: Catalyzes both phosphatidylcholine and phosphatidylethanolamine biosynthesis from CDP-choline and CDP-ethanolamine, respectively. Involved in protein-dependent process of phospholipid transport to distribute phosphatidyl choline to the lumenal surface. Has a higher cholinephosphotransferase activity than ethanolaminephosphotransferase activity. {ECO:0000269|PubMed:10191259, ECO:0000269|PubMed:10893425, ECO:0000269|PubMed:12216837}. | Endoplasmic reticulum;Glycoprotein;Lipid biosynthesis;Lipid metabolism;Magnesium;Manganese;Membrane;Metal-binding;Nucleus;Phospholipid biosynthesis;Phospholipid metabolism;Phosphoprotein;Reference proteome;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Phospholipid metabolism; phosphatidylethanolamine biosynthesis; phosphatidylethanolamine from ethanolamine: step 3/3. {ECO:0000269|PubMed:10191259, ECO:0000269|PubMed:10893425}.; PATHWAY: Phospholipid metabolism; phosphatidylcholine biosynthesis; phosphatidylcholine from phosphocholine: step 2/2. {ECO:0000269|PubMed:10191259, ECO:0000269|PubMed:10893425}. | This gene codes for a choline/ethanolaminephosphotransferase, which functions in the synthesis of choline- or ethanolamine- containing phospholipids. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2016]. | hsa:10390; | endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; nuclear membrane [GO:0031965]; 1-alkenyl-2-acylglycerol choline phosphotransferase activity [GO:0047359]; diacylglycerol cholinephosphotransferase activity [GO:0004142]; ethanolaminephosphotransferase activity [GO:0004307]; metal ion binding [GO:0046872]; lipid metabolic process [GO:0006629]; phosphatidylethanolamine biosynthetic process [GO:0006646] | 12216837_A yeast expression system devoid of endogenous cholinephosphotransferase and ethanolaminephosphotransferase activities to assess the diradylglycerol specificity of CEPT1 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 32551884_Association of the -629C>A (rs1800775) CETP Polymorphism with the Development of Essential Hypertension in Mexican Population. 32576654_Locations and contributions of the phosphotransferases EPT1 and CEPT1 to the biosynthesis of ethanolamine phospholipids. 33214136_CEPT1-Mediated Phospholipogenesis Regulates Endothelial Cell Function and Ischemia-Induced Angiogenesis Through PPARalpha. 34331935_Differential contributions of choline phosphotransferases CPT1 and CEPT1 to the biosynthesis of choline phospholipids. | ENSMUSG00000040774 | Cept1 | 4.846678e+02 | 0.4527814 | -1.143113241 | 0.3631851 | 9.443301e+00 | 0.0021192167 | 0.17592917 | No | Yes | 2.857489e+02 | 63.690023 | 6.012025e+02 | 136.601454 |
| ENSG00000134262 | 10717 | AP4B1 | protein_coding | Q9Y6B7 | FUNCTION: Component of the adaptor protein complex 4 (AP-4). Adaptor protein complexes are vesicle coat components involved both in vesicle formation and cargo selection. They control the vesicular transport of proteins in different trafficking pathways (PubMed:10066790, PubMed:10436028). AP-4 forms a non clathrin-associated coat on vesicles departing the trans-Golgi network (TGN) and may be involved in the targeting of proteins from the trans-Golgi network (TGN) to the endosomal-lysosomal system. It is also involved in protein sorting to the basolateral membrane in epithelial cells and the proper asymmetric localization of somatodendritic proteins in neurons. AP-4 is involved in the recognition and binding of tyrosine-based sorting signals found in the cytoplasmic part of cargos, but may also recognize other types of sorting signal (Probable). {ECO:0000269|PubMed:10066790, ECO:0000269|PubMed:10436028, ECO:0000305|PubMed:10066790, ECO:0000305|PubMed:10436028}. | 3D-structure;Alternative splicing;Golgi apparatus;Hereditary spastic paraplegia;Membrane;Neurodegeneration;Protein transport;Reference proteome;Transport | Mouse_homologues NA; + ;NA | This gene encodes a subunit of a heterotetrameric adapter-like complex 4 that is involved in targeting proteins from the trans-Golgi network to the endosomal-lysosomal system. Mutations in this gene are associated with cerebral palsy spastic quadriplegic type 5 (CPSQ5) disorder. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2011]. | hsa:10717; | AP-4 adaptor complex [GO:0030124]; clathrin adaptor complex [GO:0030131]; cytosol [GO:0005829]; endosome lumen [GO:0031904]; extrinsic component of membrane [GO:0019898]; trans-Golgi network [GO:0005802]; trans-Golgi network membrane [GO:0032588]; clathrin binding [GO:0030276]; protein localization [GO:0008104]; protein targeting [GO:0006605]; vesicle-mediated transport [GO:0016192] | 22290197_Data suggest that AP4B1 mutations cause spastic paraplegia type 47 and should be considered in early onset spastic paraplegia with intellectual disability. 24781758_Novel homozygous 2-bp deletion in AP4B1 was found in siblings with intellectual disability and spastic tetraplegia. 29193663_AP4B1-associated HSP and other AP-4-deficiency syndromes should be suspected in infants and children with hypotonia progressing to spastic paraplegia, delayed motor and speech development, and suggestive findings on brain imaging particularly thinning of the posterior aspect of the corpus callosum. 29430868_we report a novel AP4B1 homozygous mutation in two siblings and review the phenotype of AP-4 deficiency, speculating on a possible role of AP-4 complex in eye development. 30337681_AP4B1 mutation is associated with hereditary spastic paraplegia. 32166732_AP4B1-associated hereditary spastic paraplegia: expansion of phenotypic spectrum related to homozygous p.Thr387fs variant. | ENSMUSG00000032952+ENSMUSG00000105053 | Ap4b1+Gm43064 | 1.088088e+03 | 0.6428319 | -0.637486603 | 0.2673040 | 5.615635e+00 | 0.0178009313 | 0.47969316 | No | Yes | 7.445344e+02 | 72.311603 | 1.217743e+03 | 120.453348 |
| ENSG00000134285 | 51303 | FKBP11 | protein_coding | Q9NYL4 | FUNCTION: PPIases accelerate the folding of proteins during protein synthesis. | Alternative splicing;Isomerase;Membrane;Reference proteome;Rotamase;Signal;Transmembrane;Transmembrane helix | FKBP11 belongs to the FKBP family of peptidyl-prolyl cis/trans isomerases, which catalyze the folding of proline-containing polypeptides. The peptidyl-prolyl isomerase activity of FKBP proteins is inhibited by the immunosuppressant compounds FK506 and rapamycin (Rulten et al., 2006 [PubMed 16596453]).[supplied by OMIM, Mar 2008]. | hsa:51303; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; peptidyl-prolyl cis-trans isomerase activity [GO:0003755]; chaperone-mediated protein folding [GO:0061077] | 23749938_results suggest a progressively elevated expression of FKBP11 during the development of hepatocellular carcinoma 28385890_observed changes in activity of six rER-resident PPIases, cyclophilin B (encoded by the PPIB gene), FKBP13 (FKBP2), FKBP19 (FKBP11), FKBP22 (FKBP14), FKBP23 (FKBP7), and FKBP65 (FKBP10), due to posttranslational modifications of proline residues in the substrate. 33945996_FKBP11 promotes cell proliferation and tumorigenesis via p53-related pathways in oral squamous cell carcinoma. 35456020_FK506-Binding Protein 11 Is a Novel Plasma Cell-Specific Antibody Folding Catalyst with Increased Expression in Idiopathic Pulmonary Fibrosis. | ENSMUSG00000003355 | Fkbp11 | 6.047629e+02 | 0.6513960 | -0.618393200 | 0.3180229 | 3.746587e+00 | 0.0529154644 | 0.67693166 | No | Yes | 3.637495e+02 | 92.292321 | 6.726522e+02 | 174.539163 | |
| ENSG00000134287 | 377 | ARF3 | protein_coding | P61204 | FUNCTION: GTP-binding protein that functions as an allosteric activator of the cholera toxin catalytic subunit, an ADP-ribosyltransferase. Involved in protein trafficking; may modulate vesicle budding and uncoating within the Golgi apparatus. | 3D-structure;Alternative splicing;Cytoplasm;ER-Golgi transport;GTP-binding;Golgi apparatus;Lipoprotein;Myristate;Nucleotide-binding;Protein transport;Reference proteome;Transport | ADP-ribosylation factor 3 (ARF3) is a member of the human ARF gene family. These genes encode small guanine nucleotide-binding proteins that stimulate the ADP-ribosyltransferase activity of cholera toxin and play a role in vesicular trafficking and as activators of phospholipase D. The gene products include 6 ARF proteins and 11 ARF-like proteins and constitute 1 family of the RAS superfamily. The ARF proteins are categorized as class I (ARF1, ARF2,and ARF3), class II (ARF4 and ARF5) and class III (ARF6) and members of each class share a common gene organization. The ARF3 gene contains five exons and four introns. [provided by RefSeq, Jul 2008]. | hsa:377; | extracellular exosome [GO:0070062]; Golgi membrane [GO:0000139]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; intracellular protein transport [GO:0006886]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890]; vesicle-mediated transport [GO:0016192] | 12135740_expression and subcellular redistribution by heregulin 19913121_Observational study of gene-disease association. (HuGE Navigator) 20357002_These results suggest that Arf3 plays a unique function at the trans-Golgi network that likely involves recruitment by a specific receptor. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22971977_ARF1 and ARF3 are redundantly required for the integrity of recycling endosomes 23783033_ARF1+ARF3 are required for integrity of recycling endosomes but are involved in distinct transport pathways: they are required for the transferrin recycling pathway from endosomes to the plasma membrane. 26067373_activated ARF3 is associated with Unc93B1 and TLR9, suggesting that ARF3 conducts TLR9 trafficking by forming the TLR9-Unc93B1-ARF3 complex. 26330566_observations indicate that Arf1 and Arf3 as well as Arf6 play important roles in cytokinesis. 28098897_Upregulation of ARF3 is associated with ovarian cancer. | ENSMUSG00000051853 | Arf3 | 1.514073e+04 | 1.3438030 | 0.426321635 | 0.3015940 | 1.993851e+00 | 0.1579387920 | 0.77593452 | No | Yes | 1.497112e+04 | 1571.449544 | 1.007450e+04 | 1084.795908 | |
| ENSG00000134444 | 57614 | RELCH | protein_coding | Q9P260 | FUNCTION: Regulates intracellular cholesterol distribution from recycling endosomes to the trans-Golgi network through interactions with RAB11 and OSBP (PubMed:29514919). Functions in membrane tethering and promotes OSBP-mediated cholesterol transfer between RAB11-bound recycling endosomes and OSBP-bound Golgi-like membranes (PubMed:29514919). {ECO:0000269|PubMed:29514919}. | Acetylation;Alternative splicing;Coiled coil;Endosome;Golgi apparatus;Lipid transport;Phosphoprotein;Reference proteome;Repeat;Transport | hsa:57614; | recycling endosome [GO:0055037]; trans-Golgi network [GO:0005802]; intracellular cholesterol transport [GO:0032367] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 29514919_These data suggest that RELCH promotes nonvesicular cholesterol transport from recycling endosomes to the trans-Golgi network through membrane tethering. | ENSMUSG00000026319 | Relch | 3.231547e+02 | 0.5488372 | -0.865549927 | 0.3888400 | 4.948747e+00 | 0.0261095832 | 0.55078242 | No | Yes | 2.210587e+02 | 51.662382 | 4.057263e+02 | 97.041873 | ||
| ENSG00000134602 | 51765 | STK26 | protein_coding | Q9P289 | FUNCTION: Serine/threonine-protein kinase that acts as a mediator of cell growth (PubMed:11641781, PubMed:17360971). Modulates apoptosis (PubMed:11641781, PubMed:17360971). In association with STK24 negatively regulates Golgi reorientation in polarized cell migration upon RHO activation (PubMed:27807006). Phosphorylates ATG4B at 'Ser-383', thereby increasing autophagic flux (PubMed:29232556). {ECO:0000269|PubMed:11641781, ECO:0000269|PubMed:17360971, ECO:0000269|PubMed:27807006, ECO:0000269|PubMed:29232556}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Apoptosis;Cytoplasm;Golgi apparatus;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The product of this gene is a member of the GCK group III family of kinases, which are a subset of the Ste20-like kinases. The encoded protein contains an amino-terminal kinase domain, and a carboxy-terminal regulatory domain that mediates homodimerization. The protein kinase localizes to the Golgi apparatus and is specifically activated by binding to the Golgi matrix protein GM130. It is also cleaved by caspase-3 in vitro, and may function in the apoptotic pathway. Several alternatively spliced transcript variants of this gene have been described, but the full-length nature of some of these variants has not been determined. [provided by RefSeq, Jul 2008]. | hsa:51765; | apical plasma membrane [GO:0016324]; cell periphery [GO:0071944]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; Golgi-associated vesicle [GO:0005798]; membrane [GO:0016020]; perinuclear region of cytoplasm [GO:0048471]; vesicle membrane [GO:0012506]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; magnesium ion binding [GO:0000287]; protein homodimerization activity [GO:0042803]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; apoptotic process [GO:0006915]; cellular response to starvation [GO:0009267]; microvillus assembly [GO:0030033]; negative regulation of cell migration [GO:0030336]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of apoptotic process [GO:0042981]; regulation of hydrogen peroxide-induced cell death [GO:1903205]; response to hydrogen peroxide [GO:0042542] | 11741893_cloning of a germinal center iii kinase that induces apoptosis 12810671_MST4 expression in prostate carcinoma tumor samples & cell lines correlated with tumorigenicity & AR status. Overexpression induced anchorage-independent growth & tumorigenesis.It may have a role in the signal transduction in prostate cancer progression. 17360971_Results show that PDCD10 modulation of ERK signaling is mediated by MST4, and that PDCD10 may be a regulatory adaptor necessary for MST4 function, suggesting a link between cerebral cavernous malformation and the ERK-MAPK cascade via PDCD10/MST4. 19386264_These data define a brush border induction pathway downstream of the Lkb1/Strad/Mo25 polarization complex, yet separate from other polarity events. 22750858_crystal of the CCM3-MST4 C-terminal domain complex belonged to space group P4(1)2(1)2 or P4(3)2(1)2, with unit-cell parameters a = 69.10, b = 69.10, c = 117.57 A 23541896_CCM3 forms a stable complex with MST4 in vivo to promote cell proliferation and migration synergistically in a manner dependent on MST4 kinase activity. 23889253_In this review, the germinal center kinase III subfamily of mammalian Ste20 (sterile 20)-like group of serine/threonine protein kinases comprises Ste20-like/oxidant-stress-response kinase 1, MST3 mammalian Ste20-like kinase 3 and MST4. 24374310_Mutation of this residue was sufficient to switch the phosphorylation site preference for multiple kinases, including the serine-specific kinase PAK4 and the threonine-specific kinase MST4. 24859810_This study describes the key role of MST4 in facilitating the epithelial-mesenchymal transition process in hepatocellular carcinoma 25550858_Our results suggest that MST4, STK25 and PDCD10 are upregulated in prostate cancer and may play roles in prostate tumorigenesis. MST4 may be a helpful marker for identifying prostate cancer. 25642822_Findings reveal a mechanism by which TRAF6 is regulated and highlight a role for MST4 in limiting inflammatory responses. 25650755_these data identify the MST4 kinase as a novel candidate to mediate human pituitary tumorigenesis in a hypoxic environment and position it as a potential therapeutic target. 28219358_Low MST-4 expression is associated with Graves' disease. 29232556_Our work describes an MST4-ATG4B signaling axis that influences glioblastoma autophagy and malignancy 30072378_Because of divergent evolution of key interface residues, MST4 and MOB4 could disrupt assembly of the MST1-MOB1 complex through alternative pairing and thereby increased YAP activity. Collectively, these findings identify the MST4-MOB4 complex as a noncanonical regulator of the Hippo-YAP pathway with an oncogenic role in PC 31464346_A network-based variable selection approach for identification of modules and biomarker genes associated with end-stage kidney disease. 31539108_MicroRNA-486-5p inhibits ovarian granulosa cell proliferation and participates in the development of PCOS via targeting MST4. 31557882_The expression of three genes (STK26, KCNT2, CASP12) was correlated with the prognosis of skin cutaneous melanoma (SCM). STK26 and KCNT2 were significantly different between normal skin and SCM. These three hub genes have potential value as predictors for accurate diagnosis and prognosis of SCM in the future. 32271880_MST4 kinase suppresses gastric tumorigenesis by limiting YAP activation via a non-canonical pathway. 32340525_Tripartite motif containing 59 (TRIM59) promotes esophageal cancer progression via promoting MST4 expression and ERK pathway. 32539131_Downregulation of MST4 Underlies a Novel Inhibitory Role of MicroRNA Let-7a in the Progression of Retinoblastoma. 34240584_An MST4-pbeta-Catenin(Thr40) Signaling Axis Controls Intestinal Stem Cell and Tumorigenesis. | ENSMUSG00000031112 | Stk26 | 4.299691e+02 | 1.0940404 | 0.129666020 | 0.3716755 | 1.236861e-01 | 0.7250705511 | 0.94131543 | No | Yes | 4.381751e+02 | 87.425244 | 3.736008e+02 | 76.507008 | |
| ENSG00000134644 | 9698 | PUM1 | protein_coding | Q14671 | FUNCTION: Sequence-specific RNA-binding protein that acts as a post-transcriptional repressor by binding the 3'-UTR of mRNA targets. Binds to an RNA consensus sequence, the Pumilio Response Element (PRE), 5'-UGUANAUA-3', that is related to the Nanos Response Element (NRE) (PubMed:21572425, PubMed:18328718, PubMed:21653694, PubMed:21397187). Mediates post-transcriptional repression of transcripts via different mechanisms: acts via direct recruitment of the CCR4-POP2-NOT deadenylase leading to translational inhibition and mRNA degradation (PubMed:22955276). Also mediates deadenylation-independent repression by promoting accessibility of miRNAs (PubMed:18776931, PubMed:20818387, PubMed:20860814, PubMed:22345517). Following growth factor stimulation, phosphorylated and binds to the 3'-UTR of CDKN1B/p27 mRNA, inducing a local conformational change that exposes miRNA-binding sites, promoting association of miR-221 and miR-222, efficient suppression of CDKN1B/p27 expression, and rapid entry to the cell cycle (PubMed:20818387). Acts as a post-transcriptional repressor of E2F3 mRNAs by binding to its 3'-UTR and facilitating miRNA regulation (PubMed:22345517, PubMed:29474920). Represses a program of genes necessary to maintain genomic stability such as key mitotic, DNA repair and DNA replication factors. Its ability to repress those target mRNAs is regulated by the lncRNA NORAD (non-coding RNA activated by DNA damage) which, due to its high abundance and multitude of PUMILIO binding sites, is able to sequester a significant fraction of PUM1 and PUM2 in the cytoplasm (PubMed:26724866). Involved in neuronal functions by regulating ATXN1 mRNA levels: acts by binding to the 3'-UTR of ATXN1 transcripts, leading to their down-regulation independently of the miRNA machinery (PubMed:25768905, PubMed:29474920). Plays a role in cytoplasmic sensing of viral infection (PubMed:25340845). In testis, acts as a post-transcriptional regulator of spermatogenesis by binding to the 3'-UTR of mRNAs coding for regulators of p53/TP53. Involved in embryonic stem cell renewal by facilitating the exit from the ground state: acts by targeting mRNAs coding for naive pluripotency transcription factors and accelerates their down-regulation at the onset of differentiation (By similarity). Binds specifically to miRNA MIR199A precursor, with PUM2, regulates miRNA MIR199A expression at a postranscriptional level (PubMed:28431233). {ECO:0000250|UniProtKB:Q80U78, ECO:0000269|PubMed:18328718, ECO:0000269|PubMed:18776931, ECO:0000269|PubMed:20818387, ECO:0000269|PubMed:20860814, ECO:0000269|PubMed:21397187, ECO:0000269|PubMed:21572425, ECO:0000269|PubMed:21653694, ECO:0000269|PubMed:22345517, ECO:0000269|PubMed:22955276, ECO:0000269|PubMed:25340845, ECO:0000269|PubMed:25768905, ECO:0000269|PubMed:26724866, ECO:0000269|PubMed:28431233, ECO:0000269|PubMed:29474920}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Differentiation;Disease variant;Methylation;Neurodegeneration;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Spermatogenesis;Spinocerebellar ataxia;Translation regulation | This gene encodes a member of the PUF family, evolutionarily conserved RNA-binding proteins related to the Pumilio proteins of Drosophila and the fem-3 mRNA binding factor proteins of C. elegans. The encoded protein contains a sequence-specific RNA binding domain comprised of eight repeats and N- and C-terminal flanking regions, and serves as a translational regulator of specific mRNAs by binding to their 3' untranslated regions. The evolutionarily conserved function of the encoded protein in invertebrates and lower vertebrates suggests that the human protein may be involved in translational regulation of embryogenesis, and cell development and differentiation. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:9698; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; P-body [GO:0000932]; miRNA binding [GO:0035198]; mRNA 3'-UTR binding [GO:0003730]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; adult locomotory behavior [GO:0008344]; mRNA destabilization [GO:0061157]; positive regulation of gene silencing by miRNA [GO:2000637]; positive regulation of RIG-I signaling pathway [GO:1900246]; posttranscriptional gene silencing [GO:0016441]; posttranscriptional regulation of gene expression [GO:0010608]; production of miRNAs involved in gene silencing by miRNA [GO:0035196]; regulation of cell cycle [GO:0051726]; regulation of chromosome segregation [GO:0051983]; regulation of gene silencing by miRNA [GO:0060964]; regulation of mRNA stability [GO:0043488]; spermatogenesis [GO:0007283]; stem cell differentiation [GO:0048863] | 18211679_The reference genes of choice when performing RT-qPCR on normal and malignant breast specimens should be either the collected group of 3 genes (TBP, RPLP0 and PUM1) employed as an average, or PUM1 as a single gene. 18328718_Description of the structures of hPum Puf domain complexed to two noncognate RNAs, CycB(reverse) and Puf5. 18411299_ribonomic analysis of mRNAs associated with ribonucleoproteins containing an endogenous human PUF protein, Pum1 19319195_Puf-A transcripts were uniformly distributed in early embryos, but became restricted primarily to eyes and ovaries at a later stage of development 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20818387_Pumilio-1 (PUM1) is a ubiquitously expressed RBP that was shown to interact with p27-3' UTR. 21397187_Three distinct modes of target RNA binding by PUM1 around the fifth mRNA base were observed. 22345517_human Pumilio homologs Pum 1 and Pum 2 repress the translation of E2F3 by binding to the E2F3 3' untranslated region (UTR) and also enhance the activity of multiple E2F3 targeting microRNAs (miRNAs) 22916034_MED15 and PUM1 proteins with coiled-coil domains are potent enhancers of polyQ-mediated ataxin-1 protein misfolding and proteotoxicity in vitro and in vivo. 23625657_The promoting effect of hPuf-A in tumorigenesis might be correlated with the regulation of its associated mRNAs. 25512524_PUM repeats have now been identified in proteins that function in pre-rRNA processing, including human Puf-A and yeast Puf6. 26517885_Pumilios (including PUM1 and PUM2) are RNA-binding proteins with Puf domains made up of 8 poorly conserved Puf repeats, 3 helix bundles arranged in rainbow architecture, where each repeat recognizes a single base of the RNA-binding sequence. [REVIEW] 26724866_Identification of NORAD-interacting proteins revealed that this lncRNA functions as a multivalent binding platform for PUM proteins, with the capacity to sequester a significant fraction of the total cellular pool of PUM1 and PUM2.Studies reveal unanticipated roles for a lncRNA and PUMILIO proteins in the maintenance of genomic stability. 27157388_A recent paper by the Mendell group identifies NORAD, a novel lncRNA that is regulated in response to DNA damage and plays a key role in maintaining genome integrity by modulating the activity the RNA binding proteins PUM2 and PUM1. 28232582_results reveal a novel regulatory pathway, underscoring a previously unknown and interconnected key role of PUM1/2 and FOXP1 in regulating normal hematopoietic stem/progenitor cell and leukemic cell growth. 28760986_Results indicate that pumilio RNA binding family member 1 (PUM1) is a negative regulator of RNA helicase LGP2 (LGP2), a master regulator of innate immunity genes expressed in a cascade fashion. 29145611_PUM1 SNP is associated with Osteoporosis and Obesity. 29165587_Data indicate that RNAs-including mRNAs and non-coding RNAs-that are functionally regulated by Pumilio proteins, PUM1 and PUM2. 29297114_Variants in PUM1 may not contribute to primary ovarian insufficiency in Han Chinese women 29385536_the extent of Pumilio binding to the endogenous RGC-32 mRNA in EBV-infected cell lines also correlated with RGC-32 protein expression. Our data demonstrate the importance of RGC-32 for the survival of EBV-immortalised B cells and identify Pumilio as a key regulator of RGC-32 translation. 29428722_Thus, PUM1 promotes the development and progression of ovarian cancer, which may occur via the above-mentioned molecules. 29474920_Studies in patient-derived cells revealed that the missense mutations reduced PUM1 protein levels by approximately 25% in the adult-onset cases and by approximately 50% in the infantile-onset cases; levels of known PUM1 targets increased accordingly. 30269240_Using the novel PUM1 and PUM2 mRNA target SIAH1 as a model, this study shows mechanistic differences between PUM1 and PUM2 and between NANOS1, 2, and 3 paralogues in the regulation of SIAH1. 30333515_A transcriptome-wide approach was used to determine the binding profiles and inter-dependencies of Argonaute2 (AGO2), Pumilio (PUM1 and PUM2) sites on mRNA 3' untranslated regions (3'UTRs). 31078383_We have used the RNA-MaP platform to directly measure equilibrium binding for thousands of designed RNAs and to construct a predictive model for RNA recognition by the human Pumilio proteins PUM1 and PUM2. 31343408_PUM binding is required for maintenance of genomic stability by NORAD whereas binding of RBMX is dispensable for this function. 31395860_our results revealed that PUM1 knockdown suppressed cell growth, invasion, and metastasis, and promoted apoptosis by activating the PERK/eIF2/ATF4 signaling pathway in Pancreatic ductal adenocarcinoma (PDAC) cells. PUM1 could be a potential target to develop pharmaceuticals and novel therapeutic strategies for the treatment of PDAC. 31422002_Investigating PUM1 mutations in a Taiwanese cohort with cerebellar ataxia. 32316190_Characterization of RNP Networks of PUM1 and PUM2 Post-Transcriptional Regulators in TCam-2 Cells, a Human Male Germ Cell Model. 32375027_Systematic Analysis of Targets of Pumilio-Mediated mRNA Decay Reveals that PUM1 Repression by DNA Damage Activates Translesion Synthesis. 32437472_The results suggest that PUM1-2 affects the expression of pluripotency genes as well as the efficiency of the cardiac differentiation process. 32753408_Principles of mRNA control by human PUM proteins elucidated from multimodal experiments and integrative data analysis. 33397688_Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome. 33508364_RNA binding protein PUM1 promotes colon cancer cell proliferation and migration. 33522650_PUM1 and RNase P genes as potential cell-free DNA markers in breast cancer. 34108682_NORAD-induced Pumilio phase separation is required for genome stability. 34681026_Identification of Novel Endogenous Controls for qPCR Normalization in SK-BR-3 Breast Cancer Cell Line. 34705895_Novel regulators of PrPC biosynthesis revealed by genome-wide RNA interference. 34734756_PUM1 modulates trophoblast cell proliferation and migration through LRP6. | ENSMUSG00000028580 | Pum1 | 2.854236e+03 | 1.3209502 | 0.401576094 | 0.3070044 | 1.709845e+00 | 0.1910054303 | 0.78025017 | No | Yes | 3.095178e+03 | 498.230517 | 2.065750e+03 | 341.280849 | |
| ENSG00000134697 | 29889 | GNL2 | protein_coding | Q13823 | FUNCTION: GTPase that associates with pre-60S ribosomal subunits in the nucleolus and is required for their nuclear export and maturation (By similarity). May promote cell proliferation possibly by increasing p53/TP53 protein levels, and consequently those of its downstream product CDKN1A/p21, and decreasing RPL23A protein levels (PubMed:26203195). {ECO:0000250, ECO:0000269|PubMed:26203195}. | 3D-structure;Acetylation;GTP-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Ribosome biogenesis | hsa:29889; | membrane [GO:0016020]; nucleolus [GO:0005730]; nucleus [GO:0005634]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; RNA binding [GO:0003723]; ribosome biogenesis [GO:0042254] | 25452588_GNL2 and SEC13 are required for nonsense-mediated mRNA decay pathway in human cells. 26203195_NGP-1 promotes cell cycle progression through the activation of the p53/p21(Cip-1/Waf1) pathway. 34337013_Study of the G Protein Nucleolar 2 Value in Liver Hepatocellular Carcinoma Treatment and Prognosis. 34965383_Functional analysis of the 1p34.3 risk locus implicates GNL2 in high-grade serous ovarian cancer. | ENSMUSG00000028869 | Gnl2 | 1.575918e+03 | 1.0661774 | 0.092447503 | 0.3327173 | 7.580462e-02 | 0.7830654877 | 0.95712716 | No | Yes | 1.553000e+03 | 261.824395 | 1.425932e+03 | 246.618427 | ||
| ENSG00000134852 | 9575 | CLOCK | protein_coding | O15516 | FUNCTION: Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. Regulates the circadian expression of ICAM1, VCAM1, CCL2, THPO and MPL and also acts as an enhancer of the transactivation potential of NF-kappaB. Plays an important role in the homeostatic regulation of sleep. The CLOCK-ARNTL/BMAL1 heterodimer regulates the circadian expression of SERPINE1/PAI1, VWF, B3, CCRN4L/NOC, NAMPT, DBP, MYOD1, PPARGC1A, PPARGC1B, SIRT1, GYS2, F7, NGFR, GNRHR, BHLHE40/DEC1, ATF4, MTA1, KLF10 and also genes implicated in glucose and lipid metabolism. Promotes rhythmic chromatin opening, regulating the DNA accessibility of other transcription factors. The CLOCK-ARNTL2/BMAL2 heterodimer activates the transcription of SERPINE1/PAI1 and BHLHE40/DEC1. The preferred binding motif for the CLOCK-ARNTL/BMAL1 heterodimer is 5'-CACGTGA-3', which contains a flanking Ala residue in addition to the canonical 6-nucleotide E-box sequence (PubMed:23229515). CLOCK specifically binds to the half-site 5'-CAC-3', while ARNTL binds to the half-site 5'-GTGA-3' (PubMed:23229515). The CLOCK-ARNTL/BMAL1 heterodimer also recognizes the non-canonical E-box motifs 5'-AACGTGA-3' and 5'-CATGTGA-3' (PubMed:23229515). CLOCK has an intrinsic acetyltransferase activity, which enables circadian chromatin remodeling by acetylating histones and nonhistone proteins, including its own partner ARNTL/BMAL1. Represses glucocorticoid receptor NR3C1/GR-induced transcriptional activity by reducing the association of NR3C1/GR to glucocorticoid response elements (GREs) via the acetylation of multiple lysine residues located in its hinge region (PubMed:21980503). The acetyltransferase activity of CLOCK is as important as its transcription activity in circadian control. Acetylates metabolic enzymes IMPDH2 and NDUFA9 in a circadian manner. Facilitated by BMAL1, rhythmically interacts and acetylates argininosuccinate synthase 1 (ASS1) leading to enzymatic inhibition of ASS1 as well as the circadian oscillation of arginine biosynthesis and subsequent ureagenesis (PubMed:28985504). Drives the circadian rhythm of blood pressure through transcriptional activation of ATP1B1 (By similarity). {ECO:0000250|UniProtKB:O08785, ECO:0000269|PubMed:14645221, ECO:0000269|PubMed:18587630, ECO:0000269|PubMed:21659603, ECO:0000269|PubMed:21980503, ECO:0000269|PubMed:22284746, ECO:0000269|PubMed:23229515, ECO:0000269|PubMed:23785138, ECO:0000269|PubMed:24005054, ECO:0000269|PubMed:28985504}. | 3D-structure;Activator;Acyltransferase;Biological rhythms;Cytoplasm;DNA damage;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Transferase;Ubl conjugation | The protein encoded by this gene plays a central role in the regulation of circadian rhythms. The protein encodes a transcription factor of the basic helix-loop-helix (bHLH) family and contains DNA binding histone acetyltransferase activity. The encoded protein forms a heterodimer with ARNTL (BMAL1) that binds E-box enhancer elements upstream of Period (PER1, PER2, PER3) and Cryptochrome (CRY1, CRY2) genes and activates transcription of these genes. PER and CRY proteins heterodimerize and repress their own transcription by interacting in a feedback loop with CLOCK/ARNTL complexes. Polymorphisms in this gene may be associated with behavioral changes in certain populations and with obesity and metabolic syndrome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]. | hsa:9575; | chromatin [GO:0000785]; chromatoid body [GO:0033391]; chromosome [GO:0005694]; CLOCK-BMAL transcription complex [GO:1990513]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; chromatin DNA binding [GO:0031490]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; E-box binding [GO:0070888]; histone acetyltransferase activity [GO:0004402]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; cellular response to ionizing radiation [GO:0071479]; circadian regulation of gene expression [GO:0032922]; circadian rhythm [GO:0007623]; DNA damage checkpoint signaling [GO:0000077]; negative regulation of glucocorticoid receptor signaling pathway [GO:2000323]; negative regulation of transcription, DNA-templated [GO:0045892]; photoperiodism [GO:0009648]; positive regulation of circadian rhythm [GO:0042753]; positive regulation of inflammatory response [GO:0050729]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein acetylation [GO:0006473]; regulation of circadian rhythm [GO:0042752]; regulation of hair cycle [GO:0042634]; regulation of insulin secretion [GO:0050796]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; regulation of type B pancreatic cell development [GO:2000074]; response to redox state [GO:0051775]; signal transduction [GO:0007165]; spermatogenesis [GO:0007283] | 10198158_essential regulator of circadian rhythms 11927173_Observational study of gene-disease association. (HuGE Navigator) 12464098_Observational study of gene-disease association. (HuGE Navigator) 12841365_Observational study of gene-disease association. (HuGE Navigator) 12897057_BMAL1 and CLOCK have roles in circadian system control 12898572_Observational study of gene-disease association. (HuGE Navigator) 14582141_Observational study of gene-disease association. (HuGE Navigator) 14582141_a role for the CLOCK gene polymorphism in the regulation of long-term illness recurrence in bipolar disorder 14750904_CLOCK was cloned & sequenced and its circadian expression studied. 15331141_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15475734_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15572273_Observational study of gene-disease association. (HuGE Navigator) 15578592_Findings suggest the significance of the association of the 3111C/C allele of hClock with evening preference in a Japanese population sample. 15578592_Observational study of gene-disease association. (HuGE Navigator) 15905809_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15917222_RNA interference against beta-TRCP greatly decreases Clock-dependent gene expression in tissue culture cells, indicating that beta-TRCP controls endogenous Per1 activity and the circadian clock by directly targeting Per1 for degradation 15952199_Findings may suggest that CLOCK genotype influences the time course of insomnia during antidepressant treatment. 15952199_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16024980_Single Nucleotide Polymorphism in the 3'-untranslated region of clock gene is associated with Sleep Disorders 16125818_Single nucleotide polymorphism is not associated with narcolepsy. 16232160_Observational study of gene-disease association. (HuGE Navigator) 16474406_a molecular genetic screen in mammalian cells to identify mutants of the circadian transcriptional activators CLOCK and BMAL1. 16528748_Observational study of gene-disease association. (HuGE Navigator) 17106427_Observational study of gene-disease association. (HuGE Navigator) 17116390_Observational study of gene-disease association. (HuGE Navigator) 17116390_The results of this study suggests that the T3111C polymorphism of the CLOCK gene is associated with schizophrenia. 17221848_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17221848_compared to T/T homozygotes, bipolar depressed carriers of the C allele had a similar degree of severity of depression, but showed higher activity levels in the evening, a delayed sleep onset and a reduced amount of sleep during the night 17239050_Observational study of gene-disease association. (HuGE Navigator) 17264841_Observational study of gene-disease association. (HuGE Navigator) 17364575_Observational study of gene-disease association. (HuGE Navigator) 17364575_These results show that there is no association between either polymorphism T3111C or T257G in the Clock gene with diurnal preference or delayed sleep phase syndrome (DSPS). 17428266_CLOCK 3111 T/C SNP was associated with activity levels in the second part of the day, neuropsychological performance and BOLD fMRI correlates (interaction of genotype and moral valence of the stimuli). 17428266_Observational study of gene-disease association. (HuGE Navigator) 17469042_The difference between the mean value for the lowest expression individual (DeltaCT 8.8) and the highest expression individual (DeltaCT 3.7) revealed an approximately 34-fold difference in relative clock gene expression levels. 17516548_Observational study of gene-disease association. (HuGE Navigator) 17516548_polymorphism of the CLOCK gene confers a predisposition to a lifetime lower body weight in patients with anorexia nervosa and bulimia nervosa 17551301_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17696255_Observational study of gene-disease association. (HuGE Navigator) 17696255_Our study suggests a potential role of the CLOCK polymorphisms and their haplotypes in susceptibility to nonalcoholic fatty liver disease and disease severity. 17701674_Allelic variants interaction of CLOCK gene and GNB3 subunit gene with diurnal preference. 17701674_Observational study of gene-disease association. (HuGE Navigator) 17848551_peripheral clock in vascular endothelial cells regulates TM gene expression and that the oscillation of TM expression may contribute to the circadian variation of cardiovascular events 17948273_Observational study of gene-disease association. (HuGE Navigator) 17948273_This is the first study suggesting that a polymorphism of a gene within the circadian 'clock' mechanism is a direct or linked contributing factor in adult ADHD 17994337_CLOCK/BMAL1-mediated activation of PER1 by AP1 and E-Box elements is distinct from peripheral transcriptional modulation via cAMP-induced CREB and C/EBP. 18071340_Observational study of gene-disease association. (HuGE Navigator) 18071340_the Clock gene CGC haplotype may be protective for the development of obesity and support the hypothesis that genetic variation in the Clock gene may play a role in the development of the metabolic syndrome, type 2 diabetes and cardiovascular disease. 18228528_A multi-locus interaction between rs6442925 in the 5' upstream of BHLHB2, rs1534891 in CSNK1E, and rs534654 near the 3' end of the CLOCK gene, however, is significantly associated with bipolar disorder 18228528_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18283403_study does not support the hypothesis that the T3111C CLOCK polymorphism is associated with cluster headache 18314271_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18314271_Present findings show for the first time that the 3111T/C SNP of the CLOCK gene is not associated to human obesity and/or BED, but it seems to predispose obese individuals to a higher BMI. 18379422_No relationship between hCLOCK T3111C polymorphism and endometriosis, nor any effect of the polymorphism on the relationship of shift work to endometriosis. 18379422_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18411297_DEC1, along with DEC2, plays a role in the finer regulation and robustness of the molecular clock CLOCK/BMAL1 18419323_Evidence linking circadian rhythms, the Clock gene, and bipolar disorder is discussed, along with the possible biology that underlies this connection. Review. 18430226_The regulators of clock-controlled transcription PER2, CRY1 and CRY2 differ in their capacity to interact with each single component of the BMAL1-CLOCK heterodimer and, in the case of BMAL1, also in their interaction sites. 18458078_HAT gene expression is required for cisplatin resistance and Clock and Tip60 regulate not only transcription, but also DNA repair, through periodic histone acetylation 18541547_Observational study of gene-disease association. (HuGE Navigator) 18541547_Putative role of the CLOCK polymorphism and related haplotypes in susceptibility to obesity. 18587630_The transcription of human nocturnin gene displayed circadian oscillations in Huh7 cells (a human hepatoma cell line) and was regulated by CLOCK/BMAL1 heterodimer via the E-box of nocturnin promoter. 18957941_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18974966_Impairment of the circadian clock protein appears to be closely associated with the pathophysiology of type 2 diabetes in humans. 19224106_Data show that there are not significant association between CLOCK gene and pathophysiology of Japanese schizophrenia, bipolar disorder and major depressive disorder. 19224106_Observational study of gene-disease association. (HuGE Navigator) 19296127_expression of CLOCK in breast cancer is associated significantly with 3-year survival 19328558_Observational study of gene-disease association. (HuGE Navigator) 19347611_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19347611_Results indicate that CLOCK genotype may be a predictor of fluvoxamine treatment response in Japanese major depressive disorder. 19470168_Observational study of gene-disease association. (HuGE Navigator) 19605937_PER2 is inhibited by BMAL2-CLOCK, which reemphasizes its negative role and a positive role of BMAL2 in circadian transcription 19693801_Observational study of gene-disease association. (HuGE Navigator) 19714310_Evidence suggests metabolic processes feed back into the circadian clock, affecting clock and per2 gene expression and timing of behaviour. 19839995_Observational study of gene-disease association. (HuGE Navigator) 19846548_CLOCK polymorphisms interact with fatty acids to modulate metabolic syndrome X traits. 19846548_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19874574_Observational study of gene-disease association. (HuGE Navigator) 19888304_A novel association of genetic variation at CLOCK with total energy intake, which was particularly relevant for single nucleotide polymorphism rs3749474. 19888304_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19934327_Observational study of gene-disease association. (HuGE Navigator) 19946213_This investigation begins the characterization of a complex phospho-regulatory site that controls the activity and degradation of CLOCK, a core transcription factor that is essential for circadian behavior. 19967263_The frequencies of the shorter allele (4 repeats) in the PER3 gene and the T allele in the CLOCK gene among Asians (0.86 and 0.84, respectively) were significantly higher than among Caucasians (0.69 and 0.71, respectively). 20065968_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20065968_The genetic variation in the rs1801260 CLOCK was associated with obesity at baseline and also affected weight loss and patients with the variant allele (G) lost significantly less weight i(P=0.008) compared with wild type. 20072116_Observational study of gene-disease association. (HuGE Navigator) 20081528_We show that endotoxin causes profound suppression of circadian clock gene expression, clearly manifested in human peripheral blood leukocytes, neutrophils, and monocytes. 20124474_Circadian genes influence tumorigenesis, and identify a set of circadian gene variants as candidate breast cancer susceptibility biomarkers. 20149345_Observational study of gene-disease association. (HuGE Navigator) 20149345_We report an association between variants of the human CLOCK gene and sleep duration in two independent populations. This adds another putative function for CLOCK besides its possible involvement in circadian timing, depression, obesity, and personality. 20174623_Observational study of gene-disease association. (HuGE Navigator) 20180986_Observational study of gene-disease association. (HuGE Navigator) 20364331_Observational study of gene-disease association. (HuGE Navigator) 20364331_The T3111C (RS1801260) polymorphism of hClock gene is associated with schizophrenia, but it seems that the length polymorphism of 18 exon of hPer3 may not be associated with schizophrenia. 20368993_Observational study of gene-disease association. (HuGE Navigator) 20370469_Observational study of gene-disease association. (HuGE Navigator) 20370469_Results do not support the hypothesis of an effect of the T3111C CLOCK variant on sleep disturbances in major depressive disorder. 20396431_Observational study of gene-disease association. (HuGE Navigator) 20551151_Clock genes have been implicated in cancer-related functions; in this work, we investigated CLOCK as a possible target of somatic mutations in microsatellite unstable colorectal cancers. 20554694_Observational study of gene-disease association. (HuGE Navigator) 20560707_Meta-analysis of gene-disease association. (HuGE Navigator) 20560707_meta-analysis shows no association between rs1801260 and mood disorders or depression severity and points out necessity of further research to better understand underlying biological machinery of circadian dysfunction in subjects with mood disorders 20600471_Observational study of gene-disease association. (HuGE Navigator) 20600471_This study suggested that CLOCK 3111T/C is significant allelic and genotypic associations with bipolar disorder 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20653450_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20704703_Observational study of gene-disease association. (HuGE Navigator) 20704703_This study provides evidence for the possible involvement of CLOCK in susceptibility to attention deficit hyperactivity disorder 20861012_ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression 20876123_Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. 20886252_Clock T3111C SNPs do not influence circadian rhythmicity in healthy Italian population. 20886252_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20961464_Observational study of gene-disease association. (HuGE Navigator) 20961464_the Clock 3111 T/C SNP might be associated with the existence of sdLDL small dense low-density lipoprotein 20978934_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21050724_Variations in genes implicated in circadian regulation or its related downstream pathways may be important in the regulation of antropomorphic parameters in patients with schizophrenia during long-term treatment with SGA. 21123577_Disruption of transgenic Clock gene function promotes alterations in nucleus accumbens microcircuits. 21164265_Circadian CLOCK-mediated regulation of target-tissue sensitivity to glucocorticoids: implications for cardiometabolic diseases. 21386998_Influence of the CLOCK gene may extend to a broad range of variables linked with human behaviors. 21463335_SiRNA specific inhibition of either Clock or Per2 protein significantly inhibited alcohol-induced monolayer hyperpermeability in intestinal cells. 21479263_REV-ERBalpha plays a dual role in regulation of the activity of the BMAL1/CLOCK heterodimer by regulation of expression of both the BMAL1 and CLOCK genes. 21628555_did not find a significant association between the CLOCK 3111C/T polymorphism and morningness/eveningness or any circadian marker 21714069_Failure to replicate previous research in relation to PERIOD3 and CLOCK concurs with previous research suggesting that the effects of these genes are small and may be related to population composition. 21734043_The authors report that CLOCK is a component of the transcriptional complex that includes TFIID, ICP4, ICP27, and ICP22. 21773969_CLOCK gene polymorphisms may serve as an independent prognostic marker for colorectal cancer patients. 21898035_We conclude that menopause transition induces several changes in the genotype of the adipose tissue chronobiological machinery related to an increased risk of developing MetS. 21980503_CLOCK-mediated circadian acetylation of the human GR may function as a target-tissue, gene-specific counter regulatory mechanism to the actions of diurnally fluctuating cortisol 22003994_Our study did not provide evidence for the association between the CLOCK gene 3111C/T polymorphism and personality traits in the Japanese population. 22080789_association between the three tagging SNPs (rs3736544, rs1801260, and rs3749474) in CLOCK and risk of bipolar disorder (n=867) and major depressive disorder (n=139) compared to controls (n=889) in the Japanese population 22193177_results suggest that altered promoter methylation may contribute to the abnormal expression of clock genes in Parkinson's disease 22232930_The genotype frequency distributions of the polymorphism of the CLOCK gene examined in the EAH and CAD patients were statistically significantly different from that in the individuals without clinical signs of these diseases. 22284746_Findings establish that Clock regulates Thpo and Mpl expression in vivo, and demonstrate an important link between the body's circadian timing mechanisms and megakaryopoiesis. 22537959_There is an association between CLOCK gene T3111C polymorphism and both attention deficit hyperactivity disorder and related sleep disturbances in children. 22578325_RAI1 is a positive transcriptional regulator of CLOCK, pinpointing a novel and important role for this gene in the circadian oscillator 22709985_No significant differences were found in breakfast frequency, energy intake, or HRV between CLOCK 3111T/C minor C allele (T/C or C/C) and T/T subjects. 22806005_Borderline personality disorder may be associated with circadian typology, and perhaps to CLOCK genotypes. 22823872_The combination of a sensitive genotype (3111C/C) and environmental stress increases vulnerability to circadian rhythm sleep pattern disruption in females. 22905804_disturbances in clock expression may result in the disruption of the control of normal circadian rhythm, thus benefiting the survival of glioma cells and promoting carcinogenesis. 22999556_Genetic variation in the CLOCK gene is associated with semen quality in idiopathic male infertility. 23003921_DNA methylation levels at different CpG sites of CLOCK, BMAL1 and PER2 genes were analyzed in sixty normal-weight, overweight and obese women following a 16-week weight reduction program. 23033538_Data indicate that Clock-Bmal1 regulates the response to glucocorticoids in peripheral tissues through acetylation of the glucocorticoid receptor (GR), possibly antagonizing the biologic actions of diurnally fluctuating circulating cortisol. 23129285_Expression of the CLOCK, BMAL1, and PER1 circadian genes in human oral mucosa cells as dependent on CLOCK gene polymorphic variants. 23131019_A significant interaction was observed between the 3111T/C SNP of CLOCK gene and sex for overweight/obesity risk 23160374_enhancement of ERalpha transcriptional activity exerted by wild-type but not mutant (2K/2R) CLOCK in response to estrogen indicated that sumoylation of CLOCK may have an important role in estrogen-dependent signaling 23183326_C genetic variants in CLOCK 3111T/C display a less robust circadian rhythm than TT and a delayed acrophase that characterizes 'evening-type' subjects. 23221812_findings suggest that the CLOCK C3111T polymorphism may affect personality traits in healthy Japanese subjects. 23229515_CLOCK His84 and BMAL1 Leu125 are the key residues for the mutual recognition between CLOCK and BMAL1 bHLH domains. 23242607_Loss of Clock expression is associated with skin tumors. 23357097_among APOEepsilon4 noncarriers, C carriers in CLOCK gene rs 4580704 were associated with a high susceptibility of Alzheimer's disease 23395176_O-GlcNAc transferase (OGT) promotes expression of BMAL1/CLOCK target genes and affects circadian oscillation of clock genes in vitro and in vivo. 23496259_TNF -alpha modulates expression of the clock gene in rheumatoid arthritis. 23527142_Genetic variability in the CLOCK gene might be associated with male infertility. 23546644_Studies indicate that in the cytoplasm, PER3 protein heterodimerizes with PER1, PER2, CRY1, and CRY2 proteins and enters into the nucleus, resulting in repression of CLOCK-BMAL1-mediated transcription. 23584858_Data suggest that clock genes Per1, Cry1, Clock, and Bmal1 and their protein products may be directly involved in the daytime-dependent regulation and adaptation of hormone synthesis. 23768840_Selected genetic polymorphisms in the AHR-signaling pathway (i.e., AHRR) and CLOCK may play a role in decreasing the risk for experiencing insomnia during the menopausal transition. 23781009_the CLOCK rs1554483 G allele was associated with increased susceptibility to Alzheimer's disease. 23792158_we suggest that as a target of glioma suppressor miR-124, CLOCK positively regulates glioma proliferation and migration by reinforcing NF-kappaB activity 23871470_Common variants in CLOCK are not associated with measures of sleep duration in people of european ancestry . 23912676_Among APOE epsilon4 noncarriers, C carriers in CLOCK gene 3111T/C were associated with a high susceptibility of Alzheimer's disease in a Chinese population. 23970287_CLOCK may interact with HIF-1alpha/ARNT and activate VEGF to stimulate tumor angiogenesis and metastasis. 24037774_The 311T>C polymorphism in the CLOCK1 gene significantly increases the risk for CRC development while it does not affect the outcome of CRC patients. 24068320_This study failed to show a mediating role of evening preference, ongoing efforts are needed to identify the mechanisms by which the CLOCK gene determines ADHD-related traits. 24235147_The Cdk5-dependent regulation of CLOCK function is mediated by alterations of its stability and subcellular distribution. 24328727_data support the notion that a chronic consumption of a healthy diet may play a contributing role in triggering glucose metabolism by interacting with the rs1801260 SNP at CLOCK gene locus in metabolic syndrome patients 24332565_Sleep disruptions may contribute to increases in depressive symptoms via their impact on cognitive control and variation in the CLOCK gene may be associated with sleep quality. 24510388_A population specific variation of MTHFR and hCLOCK genes also highlights ethnicity specific risk management. 24597447_In an arousal model, prolonged wakefulness has a negative effect on subjective sleepiness and mood in three clock gene PERIOD3 polymorphisms analyzed. 24636202_No association with SNPs or haplotypes of the CLOCK gene was observed in prophylactic lithium response. 24789043_E2 promoted the binding of ERalpha to the EREs (estrogen-response elements) of CLOCK promoter, thereby up-regulating the transcription of CLOCK 24818524_The CLOCK 3111T/C polymorphism was not significantly associated with overweight or sleep duration. 24821610_SNP (rs6855837) in the CLOCK gene is significant correlation with genotype and allele frequency in lung cancer. 24824748_CLOCK polymorphisms are associated with increased susceptibility of schizophrenic patients to restless legs syndrome 24892753_The results of this study present no evidence for an association of CLOCK polymorphisms with juvenile myoclonic epilepsy. 24905098_CLOCK 3111 T/C SNP interacts with emotional eating behavior to modulate total weight loss. 24919398_Two single nucleotide polymorphisms in RORA were associated with breast cancer in the whole sample and among postmenopausal women, and we also reported an association with CLOCK, RORA, and NPAS2 in the analyses at the gene level 25070164_There is not a significant difference in the expression of CLOCK, BMAL1, and PER1 in buccal epithelial cells of patients with essential arterial hypertension regardless of patient genotype. 25309987_The observed interaction effects provide converging evidence that the clock gene and OXT/AVP systems are intertwined and contribute to human prosociality. 25527757_Data suggest that influence of obesity-associated CLOCK SNPs (rs12649507, rs6858749) on sleep duration and macronutrient intake can be ameliorated via habitual longer sleep duration and favorable dietary profile (lower protein; higher PUFA). 25625359_the circadian gene hClock promoted CRC progression and inhibit tumor cell apoptosis in vitro and in vivo, while silencing hClock was able to reverse this effect. 25775462_minor polymorphisms of CLOCK may be associated with poor morning gastric motility, and may have a combinatorial effect with PER3. 25936801_PASD1 is a circadian bHLH-PAS paralog repressor of CLOCK protein expression. 25976934_Our results suggest that the circadian gene human Clock may play an important role in carcinogenesis by inhibiting apoptotic cell death via attenuating proapoptotic signaling 26134245_CLOCK, ARNTL, and NPAS2 gene polymorphisms may have a role in seasonal variations in mood and behavior 26164627_these findings suggest that sumoylation plays a critical role in the spatiotemporal co-activation of CLOCK-BMAL1 by CBP for immediate-early Per induction and the resetting of the circadian clock. 26181468_Variants of the CLOCK gene are associated with idiopathic male infertility and therefore may be applied as a risk factor of male infertility. 26204460_CLOCK rs1801260 modulated the relationship between early stress, adult history of attempted suicide and current suicide ideation 26247999_possible circadian rhythm in full-term placental expression 26299356_Upregulation of CLOCK is associated with the expression of HIF-1alpha and VEGF in varicose veins. 26370682_found that overexpression of both Clock and Bmal1 suppressed cell growth 26374515_Clock circadian regulator (CLOCK) gene single nucleotide polymorphism rs1801260 minor allele C showed a significantly higher association with the prevalence of diabetes in the Japanese population independent of body mass index (BMI). 26390085_Findings indicate that CLOCK protein plays an important role in fertility and its knockdown leads to reduction in reproduction and increased miscarriage risk. 26453284_The CLOCK 3111T > C polymorphism could be an independent risk factor for irregular menstrual cycles, irrespective of psychological distress and endocrine or metabolic conditions in Korean adolescents. 26553137_Clock gene variants associated with obesity and sleep duration. [review] 26690565_The possession of CLOCK rs3749474 may influence the effect of reducing the percentage intake of dietary fat on obesity-associated variables and carrying this SNP might benefit more than others from weight loss treatment involving dietary fat restriction. 26739996_CLOCK gene variation is associated with incidence of type-2 diabetes and cardiovascular diseases in type-2 diabetic subjects. Suggest diet-gene interaction. 26850841_when overexpressed, c-MYC is able to repress Per1 transactivation by BMAL1/CLOCK via targeting selective E-box sequences. Importantly, upon serum stimulation, MYC was detected in BMAL1 protein complexes 26923944_Our findings suggest that CLOCK and CRY1 polymorphisms might be involved in individual susceptibility to abdominal obesity in Chinese Han population. 27335043_This study showed that Lack of Association between Genetic Polymorphism of clock gene with Late Onset Depression and Alzheimer's Disease in a Sample of a Brazilian Population This study showed that Lack of Association between Genetic Polymorphism of PER2 gene with Late Onset Depression and Alzheimer's Disease in a Sample of a Brazilian Population 27339606_Evening chronotype is associated with higher obesity in severely obese subjects and with lower weight loss effectiveness after bariatric surgery. In addition, circadian preferences interact with CLOCK 3111T/C for obesity. The circadian and genetic assessment could provide tailored weight loss recommendations in subjects who underwent bariatric surgery. 27373683_TFEB regulates PER3 expression via glucose-dependent effects on CLOCK/BMAL1 27492458_Low Clock gene expression is associated with colorectal liver metastases. 27614897_Immunostaining of CLOCK and PER2 protein was detected in the granulosa cells of dominant antral follicles but was absent in the primordial, primary, or preantral follicles of human ovaries.Oscillating expression of the circadian gene PER2 can be induced by testosterone in human granulosa cells in vitro. Expression of STAR also displayed an oscillating pattern after testosterone stimulation 27996307_CLOCK rs1801260*C and PER3(4/4) influence myelination processes by regulating sleep quality and quantity. 28058089_a novel mechanism of action of CLOCK in human umbilical vein endothelial cells, is reported. 28290876_the CLOCK gene polymorphism is one of factors determining elastic properties of vascular wall 28384165_CLOCK gene expression mean was significantly higher in morning than in evening during Shabaan 28466652_We found a significant association with CLOCK rs12649507 and cluster headache 28498393_These findings suggest that upregulation of the circadian gene hClock plays an important role in metastasis of colorectal cancer. 28514207_Results indicated that inhibiting the circadian gene Clock expression can reverse the cisplatin resistance of ovarian cancer SKOV3/DDP cell lines by affecting the protein expression of drug resistance genes during which autophagy plays an important role. 28645331_CLOCK SNP rs2070062 is associated with shorter sleep duration. 28780642_The present study confirmed that circadian dysfunction is associated with the risk of PD in a Chinese population at the genetic level and provided a potential genetic locus related to PD and CLOCK rs1801260 polymorphism is associated with susceptibility to Parkinson's Disease. 28853078_No association between 3111T/C Clock gene polymorphism and complaints of patients with insomnia was detected. 28899534_Study found three gene variants (CLOCK-rs4864548, PEMT-rs936108, and GHRELIN-rs696217) that exhibited uncorrected gene-by-sleep duration interactions in relation to BMI z-scores in a cohort of New Zealand European children. However, no interactions were identified in percentage body fat differences. Notably, these interactions are evident without detectable effects on sleep duration. 28985504_ASS1 acetylation by CLOCK exhibits circadian oscillation in human cells and mouse liver, possibly caused by rhythmic interaction between CLOCK and ASS1, leading to the circadian regulation of ASS1 and ureagenesis. 29064730_The CLOCK SNP may affect daytime gastric motility less than food stimulation. 29180440_As a novel CLOCK-dependent diurnal gene, TIMP3 inhibits the expression of inflammatory cytokines that are up-regulated by UV irradiation in human keratinocytes. 29196536_CLOCK regulates the expression of genes involved in neuronal migration, and a functional assay showed that CLOCK knockdown increased neuronal migratory distance. 29271044_Low expression of CLOCK protein is associated with kidney tumor. 29316898_Rhythmic luciferase activity from clock gene luciferase reporter cells lines was used to test the effect of p38 MAPK inhibition on clock properties as determined using the damped sine fit and Levenberg-Marquardt algorithm.Glioma treatment with p38 MAPK inhibitors may be more effective and less toxic if administered at the appropriate time of the day. 29318394_Association between HCRTR2, ADH4,CLOCK gene polymorphisms and cluster headache was not significant in the present study. 29324865_results of this study suggest that genetic variability in the ARNTL and CLOCK genes might be associated with risk for multiple sclerosis 29621412_The frequency of the TT-genotype and 3111T-allele of the Clock 3111T/C gene polymorphic marker in the Caucasian women with insomnia is higher as compared to control. 29658882_Basal and insulin-stimulated glucose uptake were significantly reduced upon CLOCK depletion. The findings suggest an essential role for the circadian coordination of skeletal muscle glucose homeostasis and lipid metabolism in humans. 29673458_The proliferation of cells significantly decreased after the expression of CLOCK was knocked down in HepG2 cells. 29768442_data suggest that SNP variability in the CLOCK gene for rs6850524 and rs11932595 is associated with idiopathic recurrent spontaneous abortion in pregnancies from Slovenia and Serbia 29860109_Loss of protective endothelial CLOCK expression aggravates TGF-beta/ROCK1-modulated endothelial to mesenchymal transition progression, which contributes to the vulnerability of human carotid plaque. 29943823_the messenger RNA expression of brain and muscle ARNT-like 1 (BMAL1) and circadian locomotor output cycles kaput (CLOCK) genes is altered by low doses (5 mJ/cm(2) ) of ultraviolet B in the immortalized HaCat human keratinocyte cell line. 30003419_In patients wit | ENSMUSG00000029238 | Clock | 1.888132e+02 | 0.8846402 | -0.176837343 | 0.4080411 | 1.874753e-01 | 0.6650262362 | 0.92913032 | No | Yes | 1.613728e+02 | 37.297291 | 1.720815e+02 | 40.573575 | |
| ENSG00000134899 | 2073 | ERCC5 | protein_coding | P28715 | FUNCTION: Single-stranded structure-specific DNA endonuclease involved in DNA excision repair (PubMed:8206890, PubMed:8090225, PubMed:8078765, PubMed:7651464, PubMed:32821917, PubMed:32522879). Makes the 3'incision in DNA nucleotide excision repair (NER) (PubMed:8090225, PubMed:8078765, PubMed:32821917, PubMed:32522879). Binds and bends DNA repair bubble substrate and breaks base stacking at the single-strand/double-strand DNA junction of the DNA bubble (PubMed:32522879). Plays a role in base excision repair (BER) by promoting the binding of DNA glycosylase NTHL1 to its substrate and increasing NTHL1 catalytic activity that removes oxidized pyrimidines from DNA (PubMed:9927729). Involved in transcription-coupled nucleotide excision repair (TCR) which allows RNA polymerase II-blocking lesions to be rapidly removed from the transcribed strand of active genes (PubMed:16246722). Functions during the initial step of TCR in cooperation with ERCC6/CSB to recognized stalled RNA polymerase II (PubMed:16246722). Also, stimulates ERCC6/CSB binding to the DNA repair bubble and ERCC6/CSB ATPase activity (PubMed:16246722). Required for DNA replication fork maintenance and preservation of genomic stability (PubMed:26833090, PubMed:32522879). Involved in homologous recombination repair (HRR) induced by DNA replication stress by recruiting RAD51, BRCA2, and PALB2 to the damaged DNA site (PubMed:26833090). During HRR, binds to the replication fork with high specificity and stabilizes it (PubMed:32522879). Also, acts upstream of HRR, to promote the release of BRCA1 from DNA (PubMed:26833090). {ECO:0000269|PubMed:16246722, ECO:0000269|PubMed:26833090, ECO:0000269|PubMed:32522879, ECO:0000269|PubMed:32821917, ECO:0000269|PubMed:7651464, ECO:0000269|PubMed:8078765, ECO:0000269|PubMed:8090225, ECO:0000269|PubMed:8206890, ECO:0000269|PubMed:9927729}. | 3D-structure;Acetylation;Alternative splicing;Chromosome;Cockayne syndrome;DNA damage;DNA repair;DNA-binding;Deafness;Disease variant;Dwarfism;Endonuclease;Hydrolase;Magnesium;Metal-binding;Nuclease;Nucleus;Phosphoprotein;Reference proteome;Xeroderma pigmentosum | Mouse_homologues NA; + ;NA | This gene encodes a single-strand specific DNA endonuclease that makes the 3' incision in DNA excision repair following UV-induced damage. The protein may also function in other cellular processes, including RNA polymerase II transcription, and transcription-coupled DNA repair. Mutations in this gene cause xeroderma pigmentosum complementation group G (XP-G), which is also referred to as xeroderma pigmentosum VII (XP7), a skin disorder characterized by hypersensitivity to UV light and increased susceptibility for skin cancer development following UV exposure. Some patients also develop Cockayne syndrome, which is characterized by severe growth defects, cognitive disability, and cachexia. Read-through transcription exists between this gene and the neighboring upstream BIVM (basic, immunoglobulin-like variable motif containing) gene. [provided by RefSeq, Feb 2011]. | hsa:2073; | chromosome [GO:0005694]; nucleoplasm [GO:0005654]; nucleotide-excision repair complex [GO:0000109]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; bubble DNA binding [GO:0000405]; damaged DNA binding [GO:0003684]; double-stranded DNA binding [GO:0003690]; endodeoxyribonuclease activity [GO:0004520]; endonuclease activity [GO:0004519]; enzyme activator activity [GO:0008047]; metal ion binding [GO:0046872]; protein homodimerization activity [GO:0042803]; protein N-terminus binding [GO:0047485]; protein-containing complex binding [GO:0044877]; RNA polymerase II complex binding [GO:0000993]; single-stranded DNA binding [GO:0003697]; base-excision repair, AP site formation [GO:0006285]; double-strand break repair via homologous recombination [GO:0000724]; negative regulation of apoptotic process [GO:0043066]; nucleotide-excision repair [GO:0006289]; nucleotide-excision repair, DNA incision, 3'-to lesion [GO:0006295]; nucleotide-excision repair, DNA incision, 5'-to lesion [GO:0006296]; regulation of catalytic activity [GO:0050790]; response to UV [GO:0009411]; response to UV-C [GO:0010225]; transcription-coupled nucleotide-excision repair [GO:0006283]; UV protection [GO:0009650] | 12494477_Observational study of gene-disease association. (HuGE Navigator) 12644470_results show that XPG endonuclease has distinct requirements for binding and cleaving DNA substrates 12865926_Observational study of gene-disease association. (HuGE Navigator) 12869423_Observational study of gene-disease association. (HuGE Navigator) 14688016_Observational study of gene-disease association. (HuGE Navigator) 14729591_Observational study of gene-disease association. (HuGE Navigator) 15082767_Results suggest that the Cockayne syndrome phenotype results from C-terminal truncations in the XPG (xeroderma pigmentosum) gene in mice and humans. 15328203_XPG expression in solid tumors may be a useful marker to predict their sensitivity to irofulven. 15494739_Observational study of gene-disease association. (HuGE Navigator) 15572672_A short region of XPGC has been defined as necessary for TFIIH interaction and stable recruitment to sites of UV damage. 15590680_the N-terminal portion of the spacer region is particularly important for nucleotide excision repair progression by mediating the XPG-TFIIH interaction and XPG substrate specificity 15746040_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15849729_Observational study of gene-disease association. (HuGE Navigator) 15992842_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16043197_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16094634_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16167068_XPG suppresses UV-induced apoptosis via its endonuclease function. 16195237_Observational study of gene-disease association. (HuGE Navigator) 16246722_The XPG binds transcription-sized DNA bubbles through two domains not required for incision and functionally interacts with Cockayne Syndrome Group B on these bubbles to stimulate its ATPase activity. 16258177_Observational study of gene-disease association. (HuGE Navigator) 16284370_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16284373_Observational study of gene-disease association. (HuGE Navigator) 16343742_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16399771_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16492920_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16507781_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16536785_Observational study of gene-disease association. (HuGE Navigator) 16537713_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16609022_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16646069_Observational study of genotype prevalence. (HuGE Navigator) 16646069_potential implication of the XPG Asp1104His polymorphism in the occurrence of chromosomal translocations associated with specific subtypes of sarcomas 16738949_Observational study of gene-disease association. (HuGE Navigator) 16823510_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16979838_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16985021_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17078101_Observational study of gene-disease association. (HuGE Navigator) 17121236_Observational study of gene-disease association. (HuGE Navigator) 17164380_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17299578_Meta-analysis of gene-disease association. (HuGE Navigator) 17313739_Observational study of gene-disease association. (HuGE Navigator) 17374967_Observational study of gene-disease association. (HuGE Navigator) 17438655_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17466625_XPG forms a stable complex with TFIIH, which is active in transcription and nucleotide excision repair 17476281_Observational study of gene-disease association. (HuGE Navigator) 17494052_Observational study of gene-disease association. (HuGE Navigator) 17575242_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17685459_Observational study of gene-disease association. (HuGE Navigator) 17712032_Observational study of gene-disease association. (HuGE Navigator) 17825393_Observational study of gene-disease association. (HuGE Navigator) 17855454_Observational study of gene-disease association. (HuGE Navigator) 17855454_SNPs associated with prognosis of lung cancer was mapped to ERCC5. 17893230_CEBPG regulates ERCC5 expression and this regulation is modified by E2F1/YY1 interactions. 17932351_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18026184_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18077223_molecular basis of disease caused by TFIIH and XPG mutations [review] 18079701_During nucleotide excision repair of DNA, the recruitment of Pol delta is associated with release of XPG and replication protein A (RPA). 18204222_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18204222_patients treated with first-line oxaliplatin/fluoropyrimidine harboring both XPG C/C and XPA A/G or A/A profiles have a longer survival and TTP. 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18320070_Observational study of gene-disease association. (HuGE Navigator) 18478970_Observational study of genotype prevalence. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18701435_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18709642_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18767034_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18825991_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18830263_Observational study of gene-disease association. (HuGE Navigator) 18854777_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18990748_Meta-analysis of gene-disease association. (HuGE Navigator) 19029193_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19096231_Observational study of gene-disease association. (HuGE Navigator) 19096231_These findings offer evidence of the association between polymorphisms [ XPG Asp1104Asp (GG) and XPD Asn312Asn (AA)] and decreased risk for cervical carcinoma or cervical squamous cell carcinoma. 19116388_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19124519_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19157633_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19157633_the polymorphic status of XPG His46His was associated with susceptibility of chemotherapy in advanced non-small cell lung carcinoma 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19177501_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19270000_Observational study of gene-disease association. (HuGE Navigator) 19280628_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19289372_the combination of high BRCA1 and low XPG expression increases the risk of shorter survival in early non-small-cell lung cancer 19350405_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19404856_the ERCC5 mutation may contribute to development of gastric and colorectal carcinomas with MSI 19414392_Observational study of gene-disease association. (HuGE Navigator) 19430706_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19430706_Results showed that polymorphism in XPG His46His was associated with a decreased treatment response, but was not statistically significant. 19434073_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19444904_Genetic polymorphisms in ERCC5 is associated with Laryngeal cancer risk associated with smoking and alcohol consumption. 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19573080_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19661089_Observational study of gene-disease association. (HuGE Navigator) 19661089_PTGS2 and ERCC5 were associated with stomach cancer risk in a Chinese population. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19693700_Observational study of gene-disease association. (HuGE Navigator) 19693700_There was significant difference in the frequency of the His/His variant genotype between cases and controls indicating a probable role of XPG in host viral interactions. 19740755_BRG1 stimulates the recruitment of XPG and PCNA to successfully culminate the nucleotide excision repair. 19773279_Observational study of gene-disease association. (HuGE Navigator) 19789190_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19878615_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19954624_Observational study of gene-disease association. (HuGE Navigator) 20150366_Observational study of gene-disease association. (HuGE Navigator) 20183911_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20183911_Polymorphisms in the ERCC5 gene is associated with with breast cancer and this association was more pronounced in women with lengthy estrogen exposure. 20199546_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20232390_Observational study of gene-disease association. (HuGE Navigator) 20233728_rs751402 A allele and rs2296147 T allele are associated with higher Allele-specific expression of ERCC5 T allele transcript at rs1047768 in normal human bronchial epithelial cells. 20391347_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20460046_Observational study of gene-disease association. (HuGE Navigator) 20460046_XPC and XPG polymorphisms do not independently affect the susceptibility to hepatocellular carcinoma, but the joint effect of C allele of XPC Lys939Gln and female sex may modify the risk. 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20530453_Observational study of gene-disease association. (HuGE Navigator) 20601096_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20731661_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20813000_Meta-analysis of gene-disease association. (HuGE Navigator) 20868484_XPG gene expression can be influenced by an epigenetic mechanism. Restoration of NER activity through XPG gene transfer or treatment with demethylating agents restored sensitivity to nemorubicin. 21390047_ERCC5 codon 1104 and ERCC2 codon 751 polymorphisms are independent prognostic factors in patients with cutaneous melanoma. 21424776_This meta-analysis suggests that XPG Asp1104His polymorphism is not associated with increased breast cancer risk. 21426550_The XPC, ERCC2 and ERCC5 variants don't affect the tumors stage and grade. 21558802_novel function for XPG in S phase that is, at least in part, performed coordinately with WRN, and which may contribute to the severity of the phenotypes that occur upon loss of XPG. 21670956_Statistically significant increased risk of prostate cancer was observed on individuals that possess the His/His genotype of Asp 1104His of XPG. 22108238_These findings suggest that genetic variation in ERCC5 may not affect the risk of SCCHN, although rs4150351 C variant genotypes were associated with an increased expression of ERCC5 mRNA and nonsignificantly decreased risk of SCCHN 22213216_XPG Asp1104His polymorphism might contribute to the identification of patients with increased risk for colorectal carcinoma 22371296_study provided statistical evidence that the XPG rs873601 SNP, which has an effect on the gene expression in a recessive manner, was associated with risk for gastric cancer among an Eastern Chinese population. 22659345_Single nucleotide polymorphisms (SNPs) of OGG1, XRCC1, ERCC5, and XRCC4 were significantly associated with the overall survival in patients with hepatitis B virus-associated hepatocellular carcinoma. 22771116_XPG endonuclease promotes DNA breaks and DNA demethylation at promoters allowing the recruitment of CTCF and gene looping, which is further stabilized by XPF. 22781116_Down-regulating XPG in epithelial ovarian cancer cells resulted in reduced cell growth and increased susceptibility to cisplatin. 22815677_it is unlikely that the ERCC5 Asp1104His polymorphism may contribute to individual susceptibility to cancer risk 22848513_These findings suggested that ERCC5 polymorphisms may contribute to risk of ESCC in Eastern Chinese populations 22981091_ERCC rs751402 CC genotype is significantly associated with decreased risk of OSCC, whereas the T allotype is correlated with higher risk of OSCC 22982416_investigated effects of two putatively functional polymorphisms in ERCC5 promoter region, rs751402 (+25A>G) and rs2296147 (+202C>T), and their potential interaction with environment factors on the risk of developing gastric cancer 23246108_The XPG gene polymorphism was also related to bladder cancer yet it was prevalent in female non-smokers. 23255472_This work adds novel allelic pathological variants to the panel of known XPG gene mutants and indicates that the mutated XPG proteins are unable to protect the cells from some genotoxic stresses. 23330005_The highest staining was detected in adrenal gland, breast, colon, heart, kidney, thyroid and tongue; in tumors, positive staining was observed in 9 of 10 breast cancer samples and in all 5 ovarian cancer and 5 sarcomas samples. 23370536_Three XPG missense mutations in the I-region of XPG impair repair and transcription and result in severe xeroderma pigmentosum/Cockayne syndrome. 23424205_The results demonstrated that 50 microg/mL beta-glucan significantly repressed the expression of the ERCC5 gene, no change in CASP9 expression, and induction of the CYP1A1 gene. 23464443_Three SNPs of XPG, rs2296147T>C, rs2094258C>T and rs873601G>A, were genotyped. 23534771_The ERCC5 Asp1558His G/G genotype was associated with elevated susceptibility to gliomas and meningiomas. 23621222_polymorphisms in rs1047768 C/T and rs2296147 C/T are associated with response to platinum-based chemotherapy in advanced non-small-cell lung cancer, and XPG polymorphisms could be predictive of prognosis. 23626689_Associations of the MTHFR Glu429Ala and ERCC5 His46His polymorphisms with survival were identified in two colorectal cancer patient cohorts. 23679317_Single nucleotide polymorphisms in the ERCC5 gene is associated with bone malignant tumors. 23818366_ERCC5 SNPs do not appear to play a major role in lymphoma susceptibility in a Spanish population. 23886164_XPG polymorphisms are associated with response to chemotherapy in osteosarcoma. 24061640_XPG Asp1104His polymorphism was not associated with bladder cancer risk. 24192772_Homozygoty for the wild-type Asp1104 SNP of the ERCC5 gene was found in 2 cases of relapsed osteosarcoma, who responded to trabectedin. 24354460_A novel mutation in ERCC5, causative of a pellagra-like condition linked to xeroderma pigmentosum/Cockayne syndrome complex. 24563277_genetic variations in ERCC1 rs11615 and ERCC5 rs17655 are associated with laryngeal cancer risk in a Chinese population, especially in ever smokers and drinkers. 24596032_ERCC5 rs17655 polymorphism may not contribute to genetic susceptibility for lung cancer 24615090_Results found that polymorphisms in XPG rs2296147 and CSB rs2228526 were significantly associated with prostate cancer susceptibility in the Chinese population analyzed. 24615519_study provided statistical evidence that XPG rs2296147T>C and rs873601G>A polymorphisms may be used as surrogate markers toward individualizing non-small cell lung cancer treatment strategies 24700531_Our data broaden the reported clinical spectrum of ERCC5 mutations and provide further evidence of genotype-phenotype correlation with truncating mutations being associated with severe phenotypes. 24737519_XRCC1 Arg399Gln and XPG His46His might significantly affect the clinical outcomes of platinum-based chemotherapy. 24782167_our study indicated that XRCC1 Arg399Gln and ERCC5 His46His might significantly influence the response to chemotherapy 24802942_Meta-analysis suggests that XPF Arg415Gln polymorphism may be associated with decreased lung cancer risk and XPG Asp1104His may be a low-penetrant risk factor in some cancers development. 24990617_The XPC rs2228000 TT genotype is associated with shorter overall survival in gastric cancer. 25023406_the ERCC2 Lys751Gln and ERCC5 His46His polymorphisms might influence osteosarcoma prognosis. 25231183_This meta-analysis suggested that the XPG Asp1104His polymorphism was a risk factor for melanoma susceptibility. 25311495_Data show increased risk of leukemia with XPCC protein (XPC) 939Gln/Gln genotype, ERCC2 protein (XPD) 751Gln allele may be protective against chronic myeloid leukemia and acute myeloid leukemia, and no significant risk for the ERCC5 protein (XPG) gene. 25332048_conclusion, our findings suggest that XPG Asp1104His polymorphism may increase the susceptibility of CRC, especially in Asian populations. 25483071_CDT2 mediated XPG elimination from DNA damage sites clears the chromatin space needed for repair. 25644244_ERCC5 single nucleotide polymorphism rs751402 is associated with breast cancer characteristics and risk in the Han population of northwest China. 25729984_the rs2296147 and rs2094258 polymorphisms of XPG, could be used as surrogate markers, leading to individualization of non-small cell lung cancer treatment strategies. 25835182_It has been found that the genotype 751Gln/Gln and allele Gln of ERCC2 gene and allele Asp of 312Asn/Asp polymorphism of ERCC2 gene may be associated with an increased risk of colorectal cancer. 25987016_XPG Asp1104His polymorphism is a risk factor for head and neck cancer susceptibility. 26045839_our study indicated that ERCC5 rs2094258 polymorphism may contribute to the risk of breast cancer. 26149386_The results suggest that the XPG-TFIIH complex is involved in transcription elongation and that defects in this association may partly account for Cockayne syndrome in xeroderma pigmentosum group G/Cockayne Syndrome patients. 26225711_ERCC5 rs17655 polymorphism might contribute to genetic susceptibility to colorectal cancer. 26264164_The purpose of this study was to assess the previously reported inconsistent association of polymorphisms in ERCC1 (rs11615, rs3212986), ERCC2 (rs13181, rs1799793, rs238406), and ERCC5 (rs17655) with the development of brain tumors. 26338418_Individuals that harbor uORF1 of ERCC5 have a marked resistance to platinum-based agents 26411687_Helicobacter Pylori introduces double-stranded DNA breaks by the nucleotide excision repair endonucleases XPF and XPG, which, together with RelA, are recruited to chromatin in a highly coordinated, type IV secretion system-dependent manner. 26820236_The results indicate that XPG rs873601G>A polymorphism may be associated with the risk of stomach cancer. 26833090_Results show that XPG partners with BRCA1 and BRCA2 to maintain genomic stability through homologous recombination, and its loss causes DNA breaks, chromosome aberrations, and replication fork stalling. 26887052_This study showed that XPG rs2296147 CT/TT variants conferred significant survival disadvantage in CRC patients in term of PFS. 27019310_the XPG gene rs2094258 C>T polymorphism may contribute to neuroblastoma susceptibility. 27051028_GG genotype of rs17655 was correlated with an increased risk of gastric cancer compared to the CC genotype. rs1047768 and rs751402 were not significantly correlated with gastric cancer risk. 27137888_Xpg thus helps to adequately induce DNA damage responses after IR, thereby keeping the expansion of damaged cells under control. This represents a new function of Xpg in the response to IR, in addition to its well-characterized role in nucleotide excision repair. 27175691_The ERCC5 promoter polymorphisms at -763 and +25 may be important functional variants and predictors of clinical outcome of advanced colorectal cancer patients who received oxaliplatin chemotherapy. 27228234_The rs751402 C/T SNP T allele and the T/T genotype were associated with an increased risk of GCA in younger individuals (>61 years) (odds ratio [OR] = 1.33 and 1.77, 95% confidence interval [CI] = 1.00-1.76 and 1.12-3.30, respectively). The rs873601 G/A SNP was not associated with susceptibility to GCA. 27235448_In NBEC, T allele at SNP rs2296147 upregulates ERCC5. 27266804_XPG mRNA expression was not predictive of trabectedin efficacy as single agent in hormone-positive, HER-2-negative advanced breast cancer. 27323134_we found that XPG rs2094258, rs751402, and rs17655 do not influence the development of breast cancer in a Chinese population 27323149_The results from this meta-analysis indicate that the XPG gene Asp1104His polymorphism is associated with lung cancer risk, especially in Asians. 27323158_). In conclusion, we suggest that the rs2094258 and rs751402 polymorphisms of ERCC5 are not connected to the development of this disease under codominant, dominant, and recessive models. 27323165_our study suggests that the rs17655 polymorphism in XPG is associated with an increased risk of gastric cancer. The results of our findings should be further validated by further large sample size studies 27323183_we suggest that the ERCC5 rs751402 polymorphism is associated with development of gastric cancer 27706622_The ERCC5 rs751402 gene polymorphism may influence the susceptibility to gastric cancer in the Chinese population. 27929383_These results indicated that none of the selected XPG polymorphism could significantly alter gastric cancer susceptibility alone. 28314991_No strong evidence was found to support the use of XPG polymorphisms as tumor response and prognostic factors of patients with NSCLC receiving a platinum-based treatment regimen--(REVIEW) 28351583_Relevant SNPs in DNA repair (ERCC1 and ERCC5) and apoptosis (MDM2 and TP53) genes might influence the severity of radiation-related side-effects in HNSCC patients. Prospective clinical SNP-based validation studies are needed on these bases 28416771_this meta-analysis shows that XPG gene polymorphisms are associated with lung cancer and gastric cancer 28514298_Meta-analysis indicated that the ERCC1 rs3212986 polymorphism and 2 polymorphisms in ERCC2 gene (rs13181 and rs1799793) contributed to the susceptibility of glioma. However, no association was observed between glioma risk and ERCC1 rs11615, ERCC2 rs238406, and ERCC5 rs17655 polymorphisms. 28704715_Overexpression of human XPG and FEN1 increases genome instability in U2OS cells 28796034_meta-analysis suggested that the rs873601 polymorphism was significantly associated with overall cancer risk. The moderate effects of rs751402 and rs2296147 polymorphism on cancer susceptibility might be highly dependent on cancer type and ethnicity, respectively 28832189_Nine case-control studies involving 3540 cases and 3953 controls were included in the meta-analysis, which revealed that the XPG rs751402 polymorphism is positively associated with GC risk and could be viewed as a risk factor of GC in three genetic models. The XPG gene rs751402 polymorphism is associated with an increased risk of GC in Chinese Han populations. This fi nding should be veri fi ed by larger studies. 28952217_The polymorphic locus on ERCC5, rs2296147, could reduce the risk of esophageal cancer. 29049208_XPG gene polymorphism rs751402 was associated with increased susceptibility to gastric cancer in Chinese populations (Meta-Analysis) 29148016_This meta-analysis indicates that the XPG rs751402 polymorphism may be a risk factor for gastric cancer in the Chinese population. 29434449_There is a multifarious interaction between the DNA repair gene ERCC5 SNPs (rs2094258 and rs873601) and the metabolic gene GSTP1 rs1695, which may form the basis for various inter-individual susceptibilities to atrophic gastritis in Chinese population. 29506519_Genome maintenance genes comprised by chronic obstructive pulmonary disease (COPD)-associated bronchial epithelial cell expression patterns were enriched for SNPs with cis-regulatory function, including a putative cis-rSNP in ERCC5 that was associated with COPD risk. 29732643_Xeroderma pigmentosum group G gene rs2094258 polymorphism may be associated with an increased risk of gastric cancer in Southern China (Meta-Analysis) 29779017_This meta-analysis indicated that the XPG gene rs17655 G>C polymorphism was associated with increased overall cancer risk 30139812_the present study reported on the association between XPG gene polymorphisms and myoma risk. The observed data indicated that SNP rs873601 G>A contributes to uterine leiomyoma susceptibility in a Southern Chinese population. 30255276_Both rs2296147 and rs1047768 SNPs were found to be associated (P < 0.05) with the risk of breast cancer. XPG rs1047768 was significantly associated with decreased PFS (HR 1.72; 95% CI 1.0-2.8) in breast cancer cases (P = 0.013) which was demonstrated by median time of 26 months for T > C variant. No association was found between XPG rs2296147 polymorphism and survival analysis among breast cancer cases. 30517302_The Asp1104His polymorphism of ERCC5 was associated with the risk and 5-year survival rate of colorectal as well as treatment sensitivity to oxaliplatin. 30522358_XPG 2228959 C/A polymorphism contributes to protective effect in North Indian lung cancer patients. 2228959 C/A polymorphism might be associated with favorable prognosis in lung cancer risk. 30527102_For XPG rs17655, our results showed that the SNP was not associated with the risk of preeclampsia. 30539843_ERCC5 rs17655 polymorphism may be not associated with overall head and neck cancer (HNC) risk. In a subgroup meta-analysis, the results suggest that the ERCC5 rs17655 polymorphism is probably associated with HNC risk in European, but the results should be interpreted with caution for the low number of studies. [meta-analysis] 30672443_The aim of the study is to investigate the relation between XPG and XPD gene variants in the DNA repair system and oral squamous cell cancers. 31450912_Polymorphisms in XPG is associated with Oral Pre Cancer and Cancer. 31470908_Using a graphical representation, from 708 identified polymorphisms, a reduced list of 115 candidates was obtained. Then, by analyzing each gene and the distribution of variant alleles, several candidates were highlighted such as UGT1A9, PTPN22, and ERCC5. These genes were already associated with the transport, the metabolism, and even the sensitivity to imatinib in previous studies. 31558863_Gastric cancer (GC) patients with the rs2094258 CT + CC genotype of XPG showed worse survival than those with the TT genotype, and CC genotype has unfavorable prognosis compared with the TT + CT genotype. The increase in C alleles of rs2094258 were associated with the long-term survival of GC cases. The XPG rs2094258 polymorphism may be associated with overall survival in GC patients. 32052936_Prenatal diagnosis of cerebro-oculo-facio-skeletal syndrome: Report of three fetuses and review of the literature. 32557569_Early-onset nucleotide excision repair disorders with neurological impairment: Clues for early diagnosis and prognostic counseling. 32683874_Association of XPG rs17655G>C and XPF rs1799801T>C Polymorphisms with Susceptibility to Cutaneous Malignant Melanoma: Evidence from a Case-Control Study, Systematic Review and Meta-Analysis', trans 'Vztah mezi polymorfismem XPG rs17655G>C a XPF rs1799801T>C a nachylnosti k malignimu melanomu kůže: důkazy ze studie připadů a kontrol, systematicky přehled a metaanalyza. 32812509_Association of ERCC5 Genetic Polymorphisms With Cirrhosis and Liver Cancer. 32821917_The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair. 33219753_COFS type 3 in an Indian family with antenatally detected arthrogryposis. 33393424_XPG gene polymorphisms and glioma susceptibility: a two-centre case-control study. 33469680_Role of ERCC5 polymorphisms in nonsmall cell lung cancer risk and responsiveness/toxicity to cisplatinbased chemotherapy in the Chinese population. 34205418_Transcriptional Stress Induces Chromatin Relocation of the Nucleotide Excision Repair Factor XPG. 34909875_Relationship of ERCC5 genetic polymorphisms with metastasis and recurrence of gastric cancer. | ENSMUSG00000026048+ENSMUSG00000041684 | Ercc5+Bivm | 5.521276e+02 | 1.1219632 | 0.166025346 | 0.3530998 | 2.268708e-01 | 0.6338538859 | 0.91812318 | No | Yes | 4.815300e+02 | 87.026988 | 4.670826e+02 | 86.472503 |
| ENSG00000134900 | 7174 | TPP2 | protein_coding | P29144 | FUNCTION: Cytosolic tripeptidyl-peptidase that releases N-terminal tripeptides from polypeptides and is a component of the proteolytic cascade acting downstream of the 26S proteasome in the ubiquitin-proteasome pathway (PubMed:25525876, PubMed:30533531). It plays an important role in intracellular amino acid homeostasis (PubMed:25525876). Stimulates adipogenesis (By similarity). {ECO:0000250|UniProtKB:Q64514, ECO:0000269|PubMed:25525876, ECO:0000269|PubMed:30533531}. | Acetylation;Aminopeptidase;Cytoplasm;Direct protein sequencing;Disease variant;Hydrolase;Nucleus;Phosphoprotein;Protease;Reference proteome;Serine protease | This gene encodes a mammalian peptidase that, at neutral pH, removes tripeptides from the N terminus of longer peptides. The protein has a specialized function that is essential for some MHC class I antigen presentation. The protein is a high molecular mass serine exopeptidase; the amino acid sequence surrounding the serine residue at the active site is similar to the peptidases of the subtilisin class rather than the trypsin class. [provided by RefSeq, Jul 2008]. | hsa:7174; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; aminopeptidase activity [GO:0004177]; endopeptidase activity [GO:0004175]; identical protein binding [GO:0042802]; serine-type endopeptidase activity [GO:0004252]; tripeptidyl-peptidase activity [GO:0008240]; amino acid homeostasis [GO:0080144]; protein polyubiquitination [GO:0000209]; proteolysis [GO:0006508] | 15716107_the promoter could be localized to a 215 bp fragment upstream of the initiation codon. 16762321_TPPII appears to promote malignant cell growth by allowing exit from mitosis and the survival of cells with severe mitotic spindle damage. 16849449_TPP2 plays a specialized role in antigen processing and one that is not essential for the generation of most presented peptides. 17343995_This investigation reveals that TPP II expression could be regulated through both positive and negative regulatory elements. 17901116_Expression of mRNA for MuRF-1 increased approximately 3-fold at 10 days without changes in MAFbx or tripeptidyl peptidase II mRNA, but all decreased between 10 and 21 days of muscle disuse. 18286573_Results indicate that TPPII is dispensable for the generation of proteasome-dependent HLA class I ligands and, the enzyme is not involved significantly in generating the proteasome-independent HLA-B27-bound peptide repertoire. 19155470_Cross-presentation of NY-ESO-1/ISCOMATRIX cancer vaccine was proteasome independent and requires the cytosolic protease tripeptidyl peptidase II. 19587004_MHC class I-restricted LMP1 epitopes studied in this work are two of very few epitopes known to date to be processed proteasome independently by tripeptidyl peptidase II. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21134372_Results suggest an important function of TPPII in the maintenance of viral growth and may have implications for anti-viral therapy. 21771670_Current knowledge about TPPII with a focus on structural aspects. 22266401_Previously unknown differences between TPP II orthologues and subtilisin as well as features that might be conserved within the entire family of subtilisin-like serine peptidases. 22483107_obtained a 3D structure of the human TPPII 22986808_Study showed that overexpression of Tripeptidyl peptidase II (TPP2) occurs frequently during oral carcinogenesis and might be associated with the progression of Oral Squamous Cell Carcinoma (OSCC) via Spindle Assembly Checkpoint(SAC) activation. 25303791_TPPII, MYBBP1A and CDK2 form a protein-protein interaction network. 25414442_Early-onset Evans syndrome, immunodeficiency, and premature immunosenescence associated with TPP2 deficiency have been described in two consanguineous siblings. 25525876_Study found that autosomal recessive TPP2 mutations cause recurrent infections, autoimmunity, and neurodevelopmental delay in humans. 26041847_TPP2 mediates many important cellular functions by controlling ERK1 and ERK2 phosphorylation. 26169984_Novel interactions of TPPII, p53, and SIRT7 presented in this study might contribute to the knowledge of the regulatory effects of these proteins on apoptotic pathways and to the understanding mechanisms of aging and lifespan regulation. 33586135_Immune deficiency, autoimmune disease and intellectual disability: A pleiotropic disorder caused by biallelic variants in the TPP2 gene. | ENSMUSG00000041763 | Tpp2 | 3.497396e+02 | 0.8969138 | -0.156958777 | 0.3882960 | 1.603920e-01 | 0.6887959114 | 0.93352105 | No | Yes | 3.257556e+02 | 68.183899 | 3.813145e+02 | 81.842883 | |
| ENSG00000135164 | 9988 | DMTF1 | protein_coding | Q9Y222 | FUNCTION: Transcriptional activator which activates the CDKN2A/ARF locus in response to Ras-Raf signaling, thereby promoting p53/TP53-dependent growth arrest (By similarity). Binds to the consensus sequence 5'-CCCG[GT]ATGT-3' (By similarity). Isoform 1 may cooperate with MYB to activate transcription of the ANPEP gene. Isoform 2 may antagonize transcriptional activation by isoform 1. {ECO:0000250, ECO:0000269|PubMed:12917399}. | 3D-structure;Activator;Alternative splicing;Cell cycle;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Tumor suppressor | Mouse_homologues NA; + ;NA | This gene encodes a transcription factor that contains a cyclin D-binding domain, three central Myb-like repeats, and two flanking acidic transactivation domains at the N- and C-termini. The encoded protein is induced by the oncogenic Ras signaling pathway and functions as a tumor suppressor by activating the transcription of ARF and thus the ARF-p53 pathway to arrest cell growth or induce apoptosis. It also activates the transcription of aminopeptidase N and may play a role in hematopoietic cell differentiation. The transcriptional activity of this protein is regulated by binding of D-cyclins. This gene is hemizygously deleted in approximately 40% of human non-small-cell lung cancer and is a potential prognostic and gene-therapy target for non-small-cell lung cancer. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2008]. | hsa:9988; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; transcription cis-regulatory region binding [GO:0000976]; cell cycle [GO:0007049]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 12917399_hDMP1beta antagonizes hDMP1alpha activity and cellular functions of hDMP1 may be regulated by cellular hDMP1 isoform levels 15010895_Observational study of gene-disease association. (HuGE Navigator) 17936562_Loss of heterozygosity (LOH) of the hDMP1 gene was detectable in approximately 35% of human lung carcinomas, which was found in mutually exclusive fashion with LOH of INK4a/ARF or that of P53. DMP1 is a pivotal tumor suppressor for human lung cancers. 17972942_WT1 downregulation during myeloid differentiation of NB4 and HL60 leukemic cell lines is associated with increased tumor repressor hDMP1 mRNA levels 20351715_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 22331460_new mechanism of p53 activation mediated by direct physical interaction between Dmp1 and p53. 23045280_LOH for hDMP1 was associated with luminal A category and longer relapse-free survival 23938323_This study reveals a pivotal role of combined Dmp1 loss and cyclin D1 overexpression in breast cancer. 25965824_Low DMTF1 expression is associated with in bladder cancer. 26187004_findings imply that DMP1alpha- and beta-ratios are tightly regulated in hematopoietic cells and DMP1beta antagonizes DMP1alpha transcriptional regulation of ARF resulting in the alteration of cellular control with a gain in proliferation 28257090_Study demonstrated that an alternative cyclin D-binding myb-like transcription factor 1 (DMTF1) pre-mRNA splicing isoform, DMTF1 beta, is increasingly expressed in breast cancer and promotes mammary tumorigenesis in a transgenic mouse model. [review]. 30100063_Cisplatin sensitivity in breast cancer cells is associated with a DMTF1beta splice variant expression. 30592263_the results of the study demonstrated that miR6753p directly regulated the expression of DMTF1, which contributed to the further regulation of Colorectal cancer cell proliferation. 30599775_DMTF1 is hemizygously deleted in 35-42% of human cancers and is associated with longer survival. [review] 33120969_Survival of Lung Cancer Patients Dependent on the LOH Status for DMP1, ARF, and p53. 33367929_Mechanisms regulating DMTF1beta/gamma expression and their functional interplay with DMTF1alpha. | ENSMUSG00000042508+ENSMUSG00000058670 | Dmtf1+Dmtf1l | 9.273199e+02 | 0.5460106 | -0.872999096 | 0.3768882 | 5.151363e+00 | 0.0232281339 | 0.53536256 | No | Yes | 5.215026e+02 | 114.042606 | 1.089523e+03 | 243.753332 |
| ENSG00000135249 | 60561 | RINT1 | protein_coding | Q6NUQ1 | FUNCTION: Involved in regulation of membrane traffic between the Golgi and the endoplasmic reticulum (ER); the function is proposed to depend on its association in the NRZ complex which is believed to play a role in SNARE assembly at the ER. May play a role in cell cycle checkpoint control (PubMed:11096100). Essential for telomere length control (PubMed:16600870). {ECO:0000269|PubMed:11096100, ECO:0000269|PubMed:16600870, ECO:0000305}. | Cell cycle;Coiled coil;Cytoplasm;Direct protein sequencing;Disease variant;ER-Golgi transport;Endoplasmic reticulum;Membrane;Protein transport;Reference proteome;Transport | This gene encodes a protein first identified for its ability to interact with the RAD50 double strand break repair protein, with the resulting interaction implicated in the regulation of cell cycle progression and telomere length. The encoded protein may also play a role in trafficking of cellular cargo from the endosome to the trans-Golgi network. Mutations in this gene may be associated with breast cancer in human patients. [provided by RefSeq, Oct 2016]. | hsa:60561; | cytosol [GO:0005829]; Dsl1/NZR complex [GO:0070939]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; mitotic G2 DNA damage checkpoint signaling [GO:0007095]; protein transport [GO:0015031]; regulation of ER to Golgi vesicle-mediated transport [GO:0060628]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890] | 16571679_RINT-1 coordinates the localization and function of ZW10 by serving as a link between ZW10 and the SNARE complex comprising syntaxin 18. 17470549_These findings suggest that RINT-1 serves as a novel tumor suppressor essential for maintaining the dynamic integrity of the Golgi apparatus and the centrosome, a prerequisite to their proper coordination during cell division. 17699596_Rab6 regulates distinct Golgi trafficking pathways involving two separate protein complexes: ZW10/RINT-1 and COG. 19369418_Results together suggest that NAG links between p31 and ZW10-RINT-1 and is involved in Golgi-to-ER transport. 23074196_RINT1 was validated as a novel glioblastoma oncogene 23885118_RINT-1 is also required for endosome-to-trans-Golgi network trafficking. 25050558_Results identified RINT1 as a putative breast cancer predisposition gene that also seems to be associated with risk for a spectrum of other cancers similar to those that have previously been described for Lynch syndrome. 25304616_Results suggest that high RAD50 interactor 1 (RINT1) expression may represent a risk factor for low-grade gliomas (LGGs)-related seizures. 27530925_RINT-1 interacts with MSP58 and UBF within nucleoli and plays a role in ribosomal gene transcription. 27544226_Study presents evidence that in the Caucasian population, RINT1 does not represent a moderate- penetrance breast cancer susceptibility gene, and find no evidence to support an association of RINT1 mutation with Lynch syndrome-related cancer incidence in breast cancer families. 28031358_HPV E2 protein targets Rad50-interacting protein 1 (Rint1) to facilitate virus genome replication. 28264000_these data are more consistent with the hypothesis that RINT1 functions as an oncogene rather than a tumor suppressor gene in the context of colorectal cancer. 31204009_RINT1 Bi-allelic Variations Cause Infantile-Onset Recurrent Acute Liver Failure and Skeletal Abnormalities. 33531371_RINT1 Regulates SUMOylation and the DNA Damage Response to Preserve Cellular Homeostasis in Pancreatic Cancer. | ENSMUSG00000028999 | Rint1 | 3.636637e+02 | 0.6686771 | -0.580618443 | 0.3277793 | 2.988267e+00 | 0.0838698765 | 0.74082963 | No | Yes | 2.696065e+02 | 44.898631 | 4.524525e+02 | 76.634937 | |
| ENSG00000135476 | 9700 | ESPL1 | protein_coding | Q14674 | FUNCTION: Caspase-like protease, which plays a central role in the chromosome segregation by cleaving the SCC1/RAD21 subunit of the cohesin complex at the onset of anaphase. During most of the cell cycle, it is inactivated by different mechanisms. {ECO:0000269|PubMed:10411507, ECO:0000269|PubMed:11509732}. | 3D-structure;Alternative splicing;Autocatalytic cleavage;Chromosome partition;Cytoplasm;Direct protein sequencing;Hydrolase;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease | Stable cohesion between sister chromatids before anaphase and their timely separation during anaphase are critical for chromosome inheritance. In vertebrates, sister chromatid cohesion is released in 2 steps via distinct mechanisms. The first step involves phosphorylation of STAG1 (MIM 604358) or STAG2 (MIM 300826) in the cohesin complex. The second step involves cleavage of the cohesin subunit SCC1 (RAD21; MIM 606462) by ESPL1, or separase, which initiates the final separation of sister chromatids (Sun et al., 2009 [PubMed 19345191]).[supplied by OMIM, Nov 2010]. | hsa:9700; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitotic spindle [GO:0072686]; nucleus [GO:0005634]; catalytic activity [GO:0003824]; cysteine-type endopeptidase activity [GO:0004197]; cysteine-type peptidase activity [GO:0008234]; apoptotic process [GO:0006915]; establishment of mitotic spindle localization [GO:0040001]; homologous chromosome segregation [GO:0045143]; meiotic chromosome separation [GO:0051307]; meiotic spindle organization [GO:0000212]; mitotic cytokinesis [GO:0000281]; mitotic sister chromatid segregation [GO:0000070]; negative regulation of sister chromatid cohesion [GO:0045875]; positive regulation of mitotic metaphase/anaphase transition [GO:0045842] | 12194817_Data suggest that separase is required for sister chromatid separation during mitosis in human cells, and that securin inhibits separase by blocking the access of substrates to the active site of separase. 12297314_Anaphase specific auto-cleavage of separase. 12672959_Processing, localization, and requirement of separase for normal anaphase progression. 15880121_density map at a resolution of 25 A of negatively stained separase-securin complex 16177575_Separase function is not restricted to anaphase initiation; its role in promoting loss of sister chromatid cohesion might be preferentially at arms but not centromeres. 17102637_nuclear exclusion is important to prevent cohesin cleavage during interphase in the absence of securin and the phosphorylation inhibition 17604273_Protein phosphatase 2A and separase form a complex regulated by separase autocleavage 17974570_Phosphorylation promotes complex formation indirectly, possibly by inducing a conformational change in full-length separase. 18003702_The complete removal of cohesin from chromosome arms depends on separase. 18616699_Separase has targets involved in regulation of G(2) to M progression after DNA damage in lung cancer cells. 18728194_These results collectively suggest that Separase is an oncogene, whose overexpression alone in mammary epithelial cells is sufficient to induce aneuploidy and tumorigenesis in a p53 mutant background. 19008095_Observational study of gene-disease association. (HuGE Navigator) 19342897_Aurora B kinase activity helps coordinate the association of separase with chromosome and the initiation of sister-chromatid separation. 19345191_Study reports that cohesin cleavage by human separase requires DNA in a sequence-nonspecific manner. 19351757_Separase might be an oncogene, whose overexpression induces tumorigenesis. Separase overexpression and aberrant nuclear localization are common in many tumor types and may predict outcome in some human cancers. 19758559_Plk1 and separase act at different times during M phase to license centrosome duplication, reminiscent of their roles in removing cohesin from chromosomes 20508983_Observational study of gene-disease association. (HuGE Navigator) 21041660_Plk1-mediated phosphorylation of Cdc6 promotes the interaction of Cdc6 and Cdk1, leading to the attenuation of Cdk1 activity, release of separase, and subsequent anaphase progression. 21126432_SMC3 and separase are upregulated and securin is downregulated in malignant transformation of BEAS-2B cells induced by coal tar pitch smoke extracts. 21272169_Results identify a new functional role of securin and separase in the modulation of membrane traffic and protein secretion that implicates regulation of V-ATPase assembly and function. 22542101_Kendrin is a novel and crucial substrate for separase (ESPL1) at the centrosome, protecting the engaged centrioles from premature disengagement and thereby blocking reduplication until the cell passes through mitosis. 22814604_By consecutively acting as a protease and a cdk1 inhibitor, separase coordinates two key processes to achieve simultaneous and abrupt separation of sister chromatids. 23798554_Mutation of the homologous position in PTTG1 (H(134)) switched PTTG1 from an inhibitor into an activator of ESP1. 24792645_Separase is an oncogene whose overexpression induces tumorigenesis, and indicate that Separase overexpression and aberrant nuclear localization are common in many tumor types and may predict outcome in some human malignancies. 25086634_High ESPL1 mRNA expression was associated with luminal B breast cancers. 25299182_Recruitment and activation of separase at centrosomes are two distinct steps that do not require microtubules. 25921067_Data indicate that separase is subject to native-state cis/trans isomerization by peptidyl-prolyl-isomerase Pin1. 26087013_Separase protein levels decrease and Separase proteolytic activity increases exclusively in b3a2 p210BCR-ABL-positive cell lines under Imatinib treatment. 26267133_The assay was used to quantify Separase proteolytic activity in leukemic cell lines and peripheral blood samples from leukemia patients. 27495871_Proximity mapping of human separase has been presented. 27966791_Studies identified and characterized the role of separase in mitosis, meiosis, non-canonical roles, its regulation, as a regulator of centriole disengagement, non proteolytic roles, diverse substrates, structural insights, and association of separase with cancer. [review] 28859055_High ESP expression is associated with breast cancer. 29370237_Separase activity measurement may therefore be useful as a novel additional molecular marker for disease monitoring 30305303_Thus, tethering of separase to DSBs and confined cohesin cleavage promote DSB repair in G2 cells. Importantly, this conserved interphase function of separase protects mammalian cells from oncogenic transformation. 31729382_LPE motif on the Scc1 substrate is required for rapid and specific cleavage by separase. 32253454_data suggests an association between high separase activity, residual BCR-ABL1 gene expression, and enhanced proliferative capacity in hematopoietic cells within the leukemic niche of TKI-treated chronic phase CML. 32322059_human cells that enter mitosis with already active separase rapidly undergo death in mitosis owing to direct cleavage of anti-apoptotic MCL1 and BCL-XL by separase; separase-triggered apoptosis enforces minimal length of mitosis 32322060_results identify an unexpected function of SGO2 in mitotically dividing cells and a mechanism of separase regulation that is independent of securin but still supervised by the spindle assembly checkpoint 32574725_Identification and genomic analysis of pedigrees with exceptional longevity identifies candidate rare variants. 33282948_TTK, CDC25A, and ESPL1 as Prognostic Biomarkers for Endometrial Cancer. 33308056_Serum ESPL1 Can Be Used as a Biomarker for Patients With Hepatitis B Virus-Related Liver Cancer: A Chinese Case-Control Study. 34180834_[Separase, a key-player of mitosis: A new target for cancer therapy?]', trans 'La separase, proteine-cle de la mitose - Une nouvelle cible therapeutique anti-cancereuse ? 34290405_Structural basis of human separase regulation by securin and CDK1-cyclin B1. | ENSMUSG00000058290 | Espl1 | 7.272820e+03 | 1.2222520 | 0.289541802 | 0.2964475 | 9.501533e-01 | 0.3296802705 | 0.82505234 | No | Yes | 7.057078e+03 | 731.015513 | 5.649330e+03 | 600.483641 | |
| ENSG00000135605 | 7006 | TEC | protein_coding | P42680 | FUNCTION: Non-receptor tyrosine kinase that contributes to signaling from many receptors and participates as a signal transducer in multiple downstream pathways, including regulation of the actin cytoskeleton. Plays a redundant role to ITK in regulation of the adaptive immune response. Regulates the development, function and differentiation of conventional T-cells and nonconventional NKT-cells. Required for TCR-dependent IL2 gene induction. Phosphorylates DOK1, one CD28-specific substrate, and contributes to CD28-signaling. Mediates signals that negatively regulate IL2RA expression induced by TCR cross-linking. Plays a redundant role to BTK in BCR-signaling for B-cell development and activation, especially by phosphorylating STAP1, a BCR-signaling protein. Required in mast cells for efficient cytokine production. Involved in both growth and differentiation mechanisms of myeloid cells through activation by the granulocyte colony-stimulating factor CSF3, a critical cytokine to promoting the growth, differentiation, and functional activation of myeloid cells. Participates in platelet signaling downstream of integrin activation. Cooperates with JAK2 through reciprocal phosphorylation to mediate cytokine-driven activation of FOS transcription. GRB10, a negative modifier of the FOS activation pathway, is another substrate of TEC. TEC is involved in G protein-coupled receptor- and integrin-mediated signalings in blood platelets. Plays a role in hepatocyte proliferation and liver regeneration and is involved in HGF-induced ERK signaling pathway. TEC regulates also FGF2 unconventional secretion (endoplasmic reticulum (ER)/Golgi-independent mechanism) under various physiological conditions through phosphorylation of FGF2 'Tyr-215'. May also be involved in the regulation of osteoclast differentiation. {ECO:0000269|PubMed:10518561, ECO:0000269|PubMed:19883687, ECO:0000269|PubMed:20230531, ECO:0000269|PubMed:9753425}. | 3D-structure;ATP-binding;Adaptive immunity;Cell membrane;Cytoplasm;Cytoskeleton;Direct protein sequencing;Immunity;Kinase;Lipid-binding;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;SH2 domain;SH3 domain;Transferase;Tyrosine-protein kinase;Zinc;Zinc-finger | The protein encoded by this gene belongs to the Tec family of non-receptor protein-tyrosine kinases containing a pleckstrin homology domain. Tec family kinases are involved in the intracellular signaling mechanisms of cytokine receptors, lymphocyte surface antigens, heterotrimeric G-protein coupled receptors, and integrin molecules. They are also key players in the regulation of the immune functions. Tec kinase is an integral component of T cell signaling and has a distinct role in T cell activation. This gene may be associated with myelodysplastic syndrome. [provided by RefSeq, Jul 2008]. | hsa:7006; | cytoskeleton [GO:0005856]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; phospholipid binding [GO:0005543]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; adaptive immune response [GO:0002250]; B cell receptor signaling pathway [GO:0050853]; integrin-mediated signaling pathway [GO:0007229]; intracellular signal transduction [GO:0035556]; peptidyl-tyrosine autophosphorylation [GO:0038083]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; protein phosphorylation [GO:0006468]; regulation of platelet activation [GO:0010543]; T cell receptor signaling pathway [GO:0050852]; tissue regeneration [GO:0042246] | 11940595_Chemotactic factor-induced recruitment and activation of Tec family kinases in human neutrophils 12049818_The AF2 domain of the orphan nuclear receptor is essential for the transcriptional activity of the oncogenic fusion protein EWS 12573241_specificity in tyrosine phosphorylation of SH3 domains in vitro is high and, therefore, SH3 domain phosphorylation is required for a distinct function in signaling 14993283_Tec overexpression in lymphocyte cell lines is sufficient to induce phospholipase Cgamma (PLC-gamma) phosphorylation and NFAT (nuclear factor of activated T cells) activation. 15184383_Btk/Tec kinases control sustained calcium signaling via site-specific phosphorylation of key residues within the phospholipase C gamma2 SH2-SH3 linker 15492005_SHIP1 and SHIP2 interact preferentially with Tec and inactivate it by de-phosphorylation of local PtdIns 3,4,5-P(3) and inhibition of Tec membrane localization 18171525_Overexpression of Tec is associated with the tumorigenesis and development of liver cancer. 18512796_Tec is the principal kinase of the Tec family that plays a major role in the responses of human neutrophils to monosodium urate crystals, which are likely to be involved in the initiation and perpetuation of gout 18564921_Observational study of gene-disease association. (HuGE Navigator) 19393603_LPS-induced actin polymerization as well as MCP-1, tumor necrosis factor-alpha, interleukin-6, and interleukin-1beta expression are dependent on Tec kinase activity. 19883687_inhibits CD25 expression in human T-lymphocytes 19913121_Observational study of gene-disease association. (HuGE Navigator) 20230531_Tec kinase has a crucial role in regulating FGF2 secretion under various physiological conditions 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22581839_results suggest that the oncogenic effect of the t(3;9) translocation may be due to the TFG-TEC chimeric protein and that fusion of the TFG (NTD) to the TEC protein produces a gain-of-function chimeric product 24722985_These results provide the first evidence that Nef interacts with cytoplasmic tyrosine kinases of the Tec family and suggest that Nef provides a mechanistic link between HIV-1 and Itk signaling in the viral life cycle. 27382052_using RNA interference, the identified compounds corroborate the role of Tec kinase in unconventional secretion of FGF2. 28631381_TEC is yet another regulator of FGF2-mediated Human pluripotent stem cells pluripotency and differentiation. 28935395_High TEC expression is associated with the development of Mallory Denk Bodies in balloon cells in alcoholic hepatitis. 31474037_Tec may mediate the production and release of pro-inflammatory cytokine IL-8 from human alveolar epithelial cells A549 induced by LPS via the p38 MAPK and ERK MAPK signal pathways. 34637843_TEC kinase stabilizes PLK4 to promote liver cancer metastasis. | ENSMUSG00000029217 | Tec | 3.741264e+01 | 0.4572543 | -1.128931232 | 0.5235142 | 4.637849e+00 | 0.0312741759 | 0.58378765 | No | Yes | 2.705098e+01 | 7.502868 | 5.981698e+01 | 16.775448 | |
| ENSG00000135632 | 10322 | SMYD5 | protein_coding | Q6GMV2 | FUNCTION: Histone methyltransferase that specifically trimethylates 'Lys-20' of histone H4 to form trimethylated histone H4 lysine 20 (H4K20me3) which represents a specific tag for epigenetic transcriptional repression (By similarity). In association with the NCoR corepressor complex, is involved in the repression of toll-like receptor 4 (TLR4)-target inflammatory genes in macrophages by catalyzing the formation of H4K20me3 at the gene promoters (By similarity). Plays an important role in embryonic stem (ES) cell self-renewal and differentiation (By similarity). Promotes ES cell maintenance by silencing differentiation genes through deposition of H4K20me3 marks (By similarity). Maintains genome stability of ES cells during differentiation through regulation of heterochromatin formation and repression of endogenous repetitive DNA elements by depositing H4K20me3 marks (PubMed:28951459). {ECO:0000250|UniProtKB:Q3TYX3, ECO:0000269|PubMed:28951459}. | Metal-binding;Methyltransferase;Reference proteome;S-adenosyl-L-methionine;Transferase;Zinc;Zinc-finger | hsa:10322; | histone methyltransferase activity (H4-K20 specific) [GO:0042799]; metal ion binding [GO:0046872]; histone H4-K20 trimethylation [GO:0034773]; negative regulation of gene expression, epigenetic [GO:0045814]; negative regulation of transposition [GO:0010529]; regulation of stem cell differentiation [GO:2000736]; regulation of stem cell division [GO:2000035] | 28951459_depletion of SMYD5 in human colon and lung cancer cells results in increased tumor growth and upregulation of genes overexpressed in colon and lung cancers, respectively. The findings implicate an important role for SMYD5 in maintaining chromosome integrity by regulating heterochromatin and repressing endogenous repetitive DNA elements during differentiation. 33676231_Sperm chromatin-condensing protamine enhances SMYD5 thermal stability. | ENSMUSG00000033706 | Smyd5 | 3.119191e+03 | 1.2552673 | 0.327994570 | 0.2956559 | 1.230870e+00 | 0.2672378202 | 0.79861760 | No | Yes | 3.253163e+03 | 343.495339 | 2.327329e+03 | 252.623351 | ||
| ENSG00000135677 | 2799 | GNS | protein_coding | P15586 | Alternative splicing;Calcium;Direct protein sequencing;Disease variant;Glycoprotein;Hydrolase;Lysosome;Metal-binding;Mucopolysaccharidosis;Phosphoprotein;Reference proteome;Signal | The product of this gene is a lysosomal enzyme found in all cells. It is involved in the catabolism of heparin, heparan sulphate, and keratan sulphate. Deficiency of this enzyme results in the accumulation of undegraded substrate and the lysosomal storage disorder mucopolysaccharidosis type IIID (Sanfilippo D syndrome). Mucopolysaccharidosis type IIID is the least common of the four subtypes of Sanfilippo syndrome. [provided by RefSeq, Jul 2008]. | hsa:2799; | azurophil granule lumen [GO:0035578]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; lysosomal lumen [GO:0043202]; glycosaminoglycan binding [GO:0005539]; metal ion binding [GO:0046872]; N-acetylglucosamine-6-sulfatase activity [GO:0008449]; sulfate binding [GO:0043199]; sulfuric ester hydrolase activity [GO:0008484]; glycosaminoglycan catabolic process [GO:0006027]; keratan sulfate catabolic process [GO:0042340] | 12624138_The Sanfilippo syndrome type D patient was found to be homozygous for a single base pair deletion (c1169delA), which will cause a frameshift and premature termination of N-acetylglucosamine-6-sulphatase. 16990043_A large intragenic deletion of 8723 bp encompassing exons 2 and 3 has been identified, the first large intragenic deletion to be reported in any of the four Sanfilippo subtypes. Q272X has also been found. 17998446_Sanfilippo syndrome type D has 3 novel mutations in the GNS Gene. 19650410_We identified the novel homozygous single base pair insertion, c.1226GinsG, which leads to a frame-shift and a premature truncation of the GNS protein (p.R409Rfs21X). 20232353_12 new patients and 15 novel mutations were identified in Mucopolysaccharidosis type IIID. 28334745_Mice deficient in GNS showed lysosomal storage pathology and a phenotype that closely resembled human MPSIIID. Moreover, treatment of the GNS-deficient animals with GNS-encoding adeno-associated viral (AAV) vectors of serotype 9 delivered to the cerebrospinal fluid completely corrected pathological storage, improved lysosomal functionality in the CNS and somatic tissues, resolved neuroinflammation | ENSMUSG00000034707 | Gns | 3.160732e+03 | 0.7265803 | -0.460805766 | 0.2542622 | 3.198233e+00 | 0.0737178683 | 0.71893069 | No | Yes | 2.652268e+03 | 308.592459 | 3.229966e+03 | 384.987692 | ||
| ENSG00000135763 | 9816 | URB2 | protein_coding | Q14146 | FUNCTION: Essential for hematopietic stem cell development through the regulation of p53/TP53 pathway. {ECO:0000250|UniProtKB:B0V0U5}. | Nucleus;Reference proteome | hsa:9816; | aggresome [GO:0016235]; midbody [GO:0030496]; nucleolus [GO:0005730]; regulation of signal transduction by p53 class mediator [GO:1901796]; ribosome biogenesis [GO:0042254] | ENSMUSG00000031976 | Urb2 | 4.340296e+03 | 0.9262460 | -0.110532628 | 0.2777636 | 1.595195e-01 | 0.6895992783 | 0.93352105 | No | Yes | 3.751802e+03 | 244.538032 | 3.974656e+03 | 265.641961 | |||
| ENSG00000135953 | 84804 | MFSD9 | protein_coding | Q8NBP5 | Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:84804; | integral component of membrane [GO:0016021]; transmembrane transporter activity [GO:0022857] | 29049335_propose MFSD4A and MFSD9 to be novel transporters, belonging to disparate SLC families. Both proteins were located to neurons in mouse brain, and their mRNA expression levels were affected by the diet | ENSMUSG00000041945 | Mfsd9 | 6.361473e+02 | 1.1886208 | 0.249288488 | 0.2733901 | 8.297223e-01 | 0.3623528475 | 0.83355620 | No | Yes | 7.136623e+02 | 78.037048 | 5.826792e+02 | 65.181189 | |||
| ENSG00000135974 | 79074 | C2orf49 | protein_coding | Q9BVC5 | Alternative splicing;Nucleus;Phosphoprotein;Reference proteome | hsa:79074; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; tRNA-splicing ligase complex [GO:0072669]; embryonic morphogenesis [GO:0048598] | ENSMUSG00000010290 | AI597479 | 3.425850e+02 | 1.1411989 | 0.190550251 | 0.3564613 | 2.809100e-01 | 0.5961053725 | 0.90859245 | No | Yes | 3.393096e+02 | 64.405019 | 2.708446e+02 | 52.525657 | ||||
| ENSG00000136169 | 83852 | SETDB2 | protein_coding | Q96T68 | FUNCTION: Histone methyltransferase involved in left-right axis specification in early development and mitosis. Specifically trimethylates 'Lys-9' of histone H3 (H3K9me3). H3K9me3 is a specific tag for epigenetic transcriptional repression that recruits HP1 (CBX1, CBX3 and/or CBX5) proteins to methylated histones. Contributes to H3K9me3 in both the interspersed repetitive elements and centromere-associated repeats. Plays a role in chromosome condensation and segregation during mitosis. {ECO:0000269|PubMed:20404330}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Chromatin regulator;Chromosome;Developmental protein;Metal-binding;Methyltransferase;Mitosis;Nucleus;Reference proteome;S-adenosyl-L-methionine;Transferase;Zinc | This gene encodes a member of a family of proteins that contain a methyl-CpG-binding domain (MBD) and a SET domain and function as histone methyltransferases. This protein is recruited to heterochromatin and plays a role in the regulation of chromosome segregation. This region is commonly deleted in chronic lymphocytic leukemia. Naturally-occuring readthrough transcription occurs from this gene to the downstream PHF11 (PHD finger protein 11) gene. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2016]. | hsa:83852; | chromosome [GO:0005694]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; histone methyltransferase activity (H3-K9 specific) [GO:0046974]; histone-lysine N-methyltransferase activity [GO:0018024]; zinc ion binding [GO:0008270]; cell division [GO:0051301]; chromosome segregation [GO:0007059]; heart looping [GO:0001947]; heterochromatin organization [GO:0070828]; histone H3-K9 methylation [GO:0051567]; left/right axis specification [GO:0070986]; mitotic cell cycle [GO:0000278]; negative regulation of gene expression [GO:0010629]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of DNA methylation-dependent heterochromatin assembly [GO:0090309] | 20085599_Observational study of gene-disease association. (HuGE Navigator) 20404330_Results provide evidence that CLLD8/KMT1F is recruited to heterochromatin regions and contributes in vivo to the deposition of trimethyl marks in concert with SUV39H1/KMT1A.SUV39H1/KMT1A. 26378653_Results indicate that the IgE-associated AT/G polymorphism (rs386770867) regulates transcription of SETDB2. 26572639_the relation of genetic variation in SETDB2-and its paralogue SETDB1-with different handedness phenotypes in 950 healthy adult participants, was investigated. 26709698_these findings identify Setdb2 as a novel regulator of the immune system in acute respiratory viral infection. 27572307_our data indicated that SETDB2 is often over-expressed in gastric cancer tissues and cell lines and SETDB2 overexpression significantly accelerated cell proliferation, migration and invasion of gastric cancer cells 29099276_SETDB2 transcripts were overexpressed in renal cell tumor subtypes and associated with prognosis. 29234167_Findings implicate SET domain bifurcated 2 (SETDB2) locus in the longstanding links of handedness with asthma and other atopic diseases. 29694893_an oncogenic role for the protein lysine methyltransferase SETDB2 in leukemia pathogenesis. It is overexpressed in pre-BCR(+) ALL and required for their maintenance in vitro and in vivo. SETDB2 expression is maintained as a direct target gene of the chimeric transcription factor E2A-PBX1 in a subset of ALL and suppresses expression of the cell-cycle inhibitor CDKN2C through histone H3K9 tri-methylation 30850015_A number of converging phenotypes outline a stress-responsive mechanism for SETDB1 and SETDB2 activation and subsequent increased tumor survival, providing novel insights into epigenetic biology. [review] 31350176_Setdb2 regulates macrophage plasticity during normal and pathologic wound repair 34479991_Coronavirus induces diabetic macrophage-mediated inflammation via SETDB2. | ENSMUSG00000071350 | Setdb2 | 1.637187e+02 | 0.6304537 | -0.665537567 | 0.4149933 | 2.479406e+00 | 0.1153458232 | 0.75783482 | No | Yes | 1.132154e+02 | 24.625283 | 1.791942e+02 | 39.382952 | |
| ENSG00000136295 | 80727 | TTYH3 | protein_coding | Q9C0H2 | FUNCTION: Probable large-conductance Ca(2+)-activated chloride channel. May play a role in Ca(2+) signal transduction. {ECO:0000269|PubMed:15010458}. | 3D-structure;Alternative splicing;Calcium;Cell membrane;Chloride;Chloride channel;Glycoprotein;Ion channel;Ion transport;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the tweety family of proteins. Members of this family function as chloride anion channels. The encoded protein functions as a calcium(2+)-activated large conductance chloride(-) channel. [provided by RefSeq, Jul 2008]. | hsa:80727; | chloride channel complex [GO:0034707]; extracellular exosome [GO:0070062]; plasma membrane [GO:0005886]; chloride channel activity [GO:0005254]; intracellular calcium activated chloride channel activity [GO:0005229]; volume-sensitive chloride channel activity [GO:0072320]; chloride transport [GO:0006821]; ion transmembrane transport [GO:0034220] | 18577513_Nedd4-2 differentially interacts with and regulates TTYH1-3 31558800_Identification and characterization of a BRAF fusion oncoprotein with retained autoinhibitory domains. 34385445_Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions. 34796468_Upregulation of TTYH3 promotes epithelial-to-mesenchymal transition through Wnt/beta-catenin signaling and inhibits apoptosis in cholangiocarcinoma. | ENSMUSG00000036565 | Ttyh3 | 5.570309e+03 | 1.2783927 | 0.354331088 | 0.3063721 | 1.323511e+00 | 0.2499629169 | 0.79138062 | No | Yes | 5.580852e+03 | 632.472286 | 4.188175e+03 | 487.236943 | |
| ENSG00000136319 | 91875 | TTC5 | protein_coding | Q8N0Z6 | FUNCTION: Cofactor involved in the regulation of various cellular mechanisms such as actin regulation, autophagy, chromatin regulation and DNA repair (PubMed:18451878, PubMed:31727855). In non-stress conditions, interacts with cofactor JMY in the cytoplasm which prevents JMY's actin nucleation activity and ability to activate the Arp2/3 complex. Acts as a negative regulator of nutrient stress-induced autophagy by preventing JMY's interaction with MAP1LC3B, thereby preventing autophagosome formation (By similarity). Involves in tubulin autoregulation by promoting its degradation in response to excess soluble tubulin (PubMed:31727855). To do so, associates with the active ribosome near the ribosome exit tunnel and with nascent tubulin polypeptides early during their translation, triggering tubulin mRNA-targeted degradation (PubMed:31727855). Following DNA damage, phosphorylated by DNA damage responsive protein kinases ATM and CHEK2, leading to its nuclear accumulation and stability. Nuclear TTC5/STRAP promotes the assembly of a stress-responsive p53/TP53 coactivator complex, which includes the coactivators JMY and p300, thereby increasing p53/TP53-dependent transcription and apoptosis. Also recruits arginine methyltransferase PRMT5 to p53/TP53 when DNA is damaged, allowing PRMT5 to methylate p53/TP53. In DNA stress conditions, also prevents p53/TP53 degradation by E3 ubiquitin ligase MDM2 (By similarity). Upon heat-shock stress, forms a chromatin-associated complex with heat-shock factor 1 HSF1 and p300/EP300 to stimulate heat-shock-responsive transcription, thereby increasing cell survival (PubMed:18451878). Mitochondrial TTC5/STRAP interacts with ATP synthase subunit beta ATP5F1B which decreased ATP synthase activity and lowers mitochondrial ATP production, thereby regulating cellular respiration and mitochondrial-dependent apoptosis. Mitochondrial TTC5/STRAP also regulates p53/TP53-mediated apoptosis (By similarity). {ECO:0000250|UniProtKB:Q99LG4, ECO:0000269|PubMed:18451878, ECO:0000269|PubMed:31727855}. | 3D-structure;Cytoplasm;Cytoplasmic vesicle;DNA damage;DNA repair;Disease variant;Mental retardation;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat | hsa:91875; | cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; ribosome binding [GO:0043022]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to starvation [GO:0009267]; DNA repair [GO:0006281]; positive regulation of mRNA catabolic process [GO:0061014]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 21147850_TTC5 is a novel cofactor regulating GR function in a stress-dependent manner. 23559008_TTC5 and EP300 cooperate to prevent excessive accumulation of MYC in AML cells and their sensitization to cell death. 24091941_Human TTC5, a novel tetratricopeptide repeat domain containing gene, activates p53 and inhibits AP-1 pathway. 31648166_TRIP13 interference inhibited the proliferation and metastasis of thyroid cancer cells through regulating TTC5/p53 pathway and epithelial-mesenchymal transition related genes expression. 31727855_this study identified tetratricopeptide protein 5 (TTC5) as a tubulin-specific ribosome-associating factor that triggers cotranslational degradation of tubulin mRNAs in response to excess soluble tubulin. 32439809_Bi-allelic TTC5 variants cause delayed developmental milestones and intellectual disability. 34947995_Evaluation of the Role of p53 Tumour Suppressor Posttranslational Modifications and TTC5 Cofactor in Lung Cancer. | ENSMUSG00000006288 | Ttc5 | 1.074639e+03 | 0.8580727 | -0.220828130 | 0.2904770 | 5.783395e-01 | 0.4469639022 | 0.85840418 | No | Yes | 9.743914e+02 | 99.419893 | 1.078911e+03 | 112.552220 | ||
| ENSG00000136527 | 6434 | TRA2B | protein_coding | P62995 | FUNCTION: Sequence-specific RNA-binding protein which participates in the control of pre-mRNA splicing. Can either activate or suppress exon inclusion. Acts additively with RBMX to promote exon 7 inclusion of the survival motor neuron SMN2. Activates the splicing of MAPT/Tau exon 10. Alters pre-mRNA splicing patterns by antagonizing the effects of splicing regulators, like RBMX. Binds to the AG-rich SE2 domain in the SMN exon 7 RNA. Binds to pre-mRNA. {ECO:0000269|PubMed:12165565, ECO:0000269|PubMed:12761049, ECO:0000269|PubMed:15009664, ECO:0000269|PubMed:9546399}. | 3D-structure;Acetylation;Activator;Alternative splicing;Direct protein sequencing;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repressor;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes a nuclear protein which functions as sequence-specific serine/arginine splicing factor which plays a role in mRNA processing, splicing patterns, and gene expression. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2011]. | hsa:6434; | nuclear inner membrane [GO:0005637]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; spliceosomal complex [GO:0005681]; identical protein binding [GO:0042802]; mRNA binding [GO:0003729]; pre-mRNA binding [GO:0036002]; protein domain specific binding [GO:0019904]; RNA binding [GO:0003723]; cellular response to glucose stimulus [GO:0071333]; cerebral cortex regionalization [GO:0021796]; embryonic brain development [GO:1990403]; mRNA splicing, via spliceosome [GO:0000398]; positive regulation of mRNA splicing, via spliceosome [GO:0048026]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; regulation of RNA splicing [GO:0043484]; RNA splicing, via transesterification reactions [GO:0000375] | 12531473_interacts with calcitonin/CGRP exon 4 exonic splice enhancer; required for calcitonin splicing in vitro 12649279_results implicate the human tau gene as a target gene for the alternative splicing regulator Tra2 beta protein, suggesting that Tra2 beta may play a role in aberrant tau exon 10 alternative splicing 12798777_Tra2beta1 transcript levels are developmentally regulated in a tissue- and temporal-specific pattern, although expression is ubiquitous. 14520560_Reduced SMN protein levels cause a reduction in the amount of its interacting proteins and of Htra2-beta1 in both discordant and non-discordant spinal muscular atrophy phenotypes. 19037821_hTra2-beta1 expression in cervical cancer. The observed shuttle process of this splicing factor with higher concentrations in the nucleus effects on the cellular function and tumor biology, leading to the worse patient outcome. 20074680_DARPP-32 changes the usage of tra2-beta1 dependent alternative exons in a concentration-dependent manner, suggesting that the DARPP-32:tra2-beta1 interaction is a molecular link between signaling pathways and pre-mRNA processing. 20607830_nuclear hnRNP G level as well as hTra2-beta1 level were independent prognostic factors for endometrial cancer progression-free survival 20926394_Results indicates that the Human Transformer2-beta RNA recognition motif recognizes two types of RNA sequences in different RNA binding modes. 21803291_reduced expression of SFRS10, as observed in tissues from obese humans, alters LPIN1 splicing, induces lipogenesis, and therefore contributes to metabolic phenotypes associated with obesity. 23255807_findings show the inclusion of both HIV-1 exon 3 and vpr mRNA processing is promoted by an exonic splicing enhancer (ESEvpr) localized between exonic splicing silencer ESSV and 5'ss D3; the ESEvpr sequence was found to be bound by members of the Tra2 protein family 23361474_Oxidative stress-responsive Transformer 2beta may play an important role in colon cancer growth. 23396973_analysis of mechanisms by which the subcellular and subnuclear localization of Tra2beta proteins are regulated 23748175_Specific induction of hTra2beta1 due to Alternative splicing is associated with epithelial ovarian cancer. 24098751_SFRS10 is not expressed in normal human retinae but is upregulated in Age-related macular degeneration retinae. 24865968_Facilitating Tra2beta-dependent inclusion of exons in target pre-mRNAs. 24952301_Results indicate that transformer 2beta (Tra2beta) was involved in the tumorigenesis of NSCLC and might be a potential therapeutic target of non-small cell lung cancer (NSCLC). 25208576_Simultaneous depletion of Tra2alpha and Tra2beta induces substantial shifts in splicing of endogenous Tra2beta target exons, and both constitutive and alternative target exons are under dual Tra2alpha-Tra2beta control. 25342468_findings suggest that Tra2beta regulates apoptosis by modulating Bcl-2 expression through its competition with miR-204. This novel function may have a crucial role in tumor growth. 25884434_Results showed that HNRNPG and HTRA2-BETA1 were specific antagonistic regulators of ERa exon7 splicing and increased HNRNPG levels were associated with improved clinical outcome of endometrial cancer through up-regulation of ERaD7 expression. 25970345_Overexpression of either Tra2alpha or Tra2beta results in a marked reduction in HIV-1 Gag/ Env expression. 26013829_SRPK1 is a regulator of Tra2beta1 splicing function and individual domains engage in considerable cross-talk, assuming novel functions with regard to RNA binding, splicing, and catalysis. 26261585_Tra2beta was significantly upregulated in prostate carcinoma, and multivariate analysis confirmed Tra2beta as an independent prognostic factor. 26298634_that TRA2beta promotes glioma cell growth and migration, and could be a candidate for molecular targeting during gene therapy treatments of glioma 28467182_It seems that a new signaling axis, SIRT1-SFRS10-LPIN1 axis, acting in the pathogenesis of alcoholic fatty liver disease exists. 29161765_Findings demonstrate that Tra2b is upregulated in non-small cell lung cancer (NSCLC) and is associated with poor prognosis. Furthermore, Tra2b was identified as a direct target of miR-335. These results indicate that the interaction between miR-335 and Tra2b may play a vital role in the pathogenesis of NSCLC. 31311954_HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells. 32289378_MicroRNA-330-3p represses the proliferation and invasion of laryngeal squamous cell carcinoma through downregulation of Tra2beta-mediated Akt signaling. 32682951_Bone marrow mesenchymal stem cell-derived exosomal miR-206 inhibits osteosarcoma progression by targeting TRA2B. 32964954_Tra2beta protects against the degeneration of chondrocytes by inhibiting chondrocyte apoptosis via activating the PI3K/Akt signaling pathway. | ENSMUSG00000022858 | Tra2b | 4.143665e+03 | 0.5628880 | -0.829080253 | 0.2867923 | 8.343300e+00 | 0.0038711227 | 0.22411432 | No | Yes | 2.896738e+03 | 457.431927 | 5.082493e+03 | 822.319798 | |
| ENSG00000136574 | 2626 | GATA4 | protein_coding | P43694 | FUNCTION: Transcriptional activator that binds to the consensus sequence 5'-AGATAG-3' and plays a key role in cardiac development and function (PubMed:24000169, PubMed:27984724). In cooperation with TBX5, it binds to cardiac super-enhancers and promotes cardiomyocyte gene expression, while it down-regulates endocardial and endothelial gene expression (PubMed:27984724). Involved in bone morphogenetic protein (BMP)-mediated induction of cardiac-specific gene expression. Binds to BMP response element (BMPRE) DNA sequences within cardiac activating regions (By similarity). Acts as a transcriptional activator of ANF in cooperation with NKX2-5 (By similarity). Promotes cardiac myocyte enlargement (PubMed:20081228). Required during testicular development (PubMed:21220346). May play a role in sphingolipid signaling by regulating the expression of sphingosine-1-phosphate degrading enzyme, sphingosine-1-phosphate lyase (PubMed:15734735). {ECO:0000250|UniProtKB:P46152, ECO:0000250|UniProtKB:Q08369, ECO:0000269|PubMed:15734735, ECO:0000269|PubMed:20081228, ECO:0000269|PubMed:21220346, ECO:0000269|PubMed:24000169, ECO:0000269|PubMed:27984724}. | 3D-structure;Activator;Alternative splicing;Atrial septal defect;Cardiomyopathy;DNA-binding;Disease variant;Metal-binding;Methylation;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a member of the GATA family of zinc-finger transcription factors. Members of this family recognize the GATA motif which is present in the promoters of many genes. This protein is thought to regulate genes involved in embryogenesis and in myocardial differentiation and function, and is necessary for normal testicular development. Mutations in this gene have been associated with cardiac septal defects. Additionally, alterations in gene expression have been associated with several cancer types. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2015]. | hsa:2626; | chromatin [GO:0000785]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA polymerase II transcription regulator complex [GO:0090575]; co-SMAD binding [GO:0070410]; DNA binding [GO:0003677]; DNA-binding transcription activator activity [GO:0001216]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; NFAT protein binding [GO:0051525]; protein kinase binding [GO:0019901]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; zinc ion binding [GO:0008270]; aortic valve morphogenesis [GO:0003180]; atrial septum morphogenesis [GO:0060413]; atrial septum primum morphogenesis [GO:0003289]; atrial septum secundum morphogenesis [GO:0003290]; atrioventricular canal development [GO:0036302]; atrioventricular node development [GO:0003162]; atrioventricular valve formation [GO:0003190]; cardiac muscle tissue regeneration [GO:0061026]; cardiac right ventricle morphogenesis [GO:0003215]; cardiac ventricle morphogenesis [GO:0003208]; cell fate commitment [GO:0045165]; cell growth involved in cardiac muscle cell development [GO:0061049]; cell-cell signaling [GO:0007267]; cellular response to glucose stimulus [GO:0071333]; embryonic foregut morphogenesis [GO:0048617]; embryonic heart tube anterior/posterior pattern specification [GO:0035054]; endocardial cushion development [GO:0003197]; endoderm development [GO:0007492]; heart looping [GO:0001947]; intestinal epithelial cell differentiation [GO:0060575]; male gonad development [GO:0008584]; negative regulation of apoptotic signaling pathway [GO:2001234]; negative regulation of autophagy [GO:0010507]; negative regulation of cardiac muscle cell apoptotic process [GO:0010667]; negative regulation of oxidative stress-induced cell death [GO:1903202]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of angiogenesis [GO:0045766]; positive regulation of BMP signaling pathway [GO:0030513]; positive regulation of cardioblast differentiation [GO:0051891]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of vascular endothelial growth factor production [GO:0010575]; regulation of cardiac muscle cell contraction [GO:0086004]; regulation of protein kinase B signaling [GO:0051896]; regulation of transcription, DNA-templated [GO:0006355]; response to mechanical stimulus [GO:0009612]; response to vitamin A [GO:0033189]; response to xenobiotic stimulus [GO:0009410]; transdifferentiation [GO:0060290]; ventricular septum development [GO:0003281]; wound healing [GO:0042060] | 12530677_GATA-4 and GATA-6 mRNAs are readily detectable from gestational week 19 in human adrenal cortex. 12606287_role in trans-activation of bone morphogenetic protein 4 and regulation of mammalian organogenesis 12606418_In postnatal ovary, granulosa cells of growing follicles express FOG-2, partially overlapping with expression of mullerian-inhibiting substance. Important role for FOG-2 and GATA transcription factors in developing ovary. 12775767_Nkx-2.5 and GATA-4 play prime roles in Dio2 gene regulation in the human heart and suggests that it is their synergistic action in humans that causes the differential expression of the cardiac Dio2 gene between humans and rats. 12845333_results implicate GATA4 as a genetic cause of human cardiac septal defects, perhaps through its interaction with TBX5 12907682_Wild-type SF1 but not SF1 G35E is a potent transactvator in GAGA4-expressing cells. Results strengthen the importance of a GATA-4/SF-1 cooperation for MIS transcription; disruption of this synergism might lead to abnormal sex differentiation. 14583613_GATA-4 is critical for transcription of the erythropoietin (Epo) gene in hepatocytes and may contribute to the switch in the site of Epo gene expression from the fetal liver to the adult kidney 14612389_in colorectal cancer and gastric cancer promoter hypermethylation and transcriptional silencing are frequent for GATA-4 and -5 14984931_plays a transactivator role in the regulation of the transcription of the microsomal epoxide hydrolase gene (EPHX1) 15153544_transcription factors GATA4 and YY1 are involved in the regulation of FcgammaRIIb expression, and that the expression variants of FcgammaRIIb lead to altered cell signaling, which may contribute to autoimmune pathogenesis in humans. 15235040_A novel atrial septal defect causative GATA4 mutation in a Japanese family. 15337742_GATA4 is a SUMO-1-targeted transcription factor and together with PIAS1 is a potent regulator of cardiac gene activity 15389642_optimal claudin-2 expression in the gut relies on the presence of GATA-4, suggesting a role for this factor in intestinal regionalization 15585625_Hypermethylation of the GATA4 is associated with lung cancer 15653675_NKX2.5 inhibits myocyte differentiation and myotube formation, and up-regulates Gata4 and Tbx5 expression 15666845_Steroidogenically active human adrenocortical cells were weakly positive for GATA-4, whereas steroidogenically inactive cells were totally GATA-4 negative. In contrast, both cell lines expressed GATA-6. 16110260_GATA-4 detected in ovarian GCTs has retained normal function 16137232_expression of GATA-4 and GATA-6 is up-regulated prior to the transcriptional activation of Nkx 2.5 during cardiogenesis 16159935_Aggressive granulosa cell tumors(GCTs) retain high GATA-4 expression, whereas larger tumors lose proliferation-suppressing anti-Mullerian hormone expression. High GATA-4 expression in GCTs may serve as marker of poor prognosis. 16337738_Methylation of GATA-4 was associated with ovarian carcinogenesis 16470721_a GATA4 mutation may have a role in Tetralogy of Fallot 16604480_two novel GATA4 mutations associated with congenital heart defects in Chinese patients. This suggests that the transcription factor GATA4 may play an important role in cardiogenesis 16607277_Altered histone modification of the promoter loci is one mechanism responsible for the silencing of GATA transcription factors. 16823849_Frequent silencing of GATA-4 and GATA-5 in human esophageal neoplasia is associated with gene promoter hypermethylation. 17211834_Data show that Oct-4, Rex-1, and Gata-4 expression in human mesenchymal stem cells increases cell differentiation efficiency but not hTERT expression. 17253934_GATA4 mutations are relatively rare among congenital heart disease patients. 17253934_Observational study of genotype prevalence. (HuGE Navigator) 17290010_TGF-beta signaling regulates gut epithelial gene expression by targeting GATA4. 17352393_GATA4 mutations that result in deficits in transactivation ability are consistently associated with congenital heart disease suggesting that normal transactivation properties of GATA4 are required for proper cardiac development. 17352393_Observational study of genotype prevalence. (HuGE Navigator) 17403900_Cooperative interaction between HNFA4 and GATA4 and GATA6 regulates ABCG5 and ABCG8. 17548362_analysis of the nuclear shuttling pathways of GATA-4 that represent an additional mechanism of gene regulation 17584735_Tbx18 interacts with Gata4 and Nkx2-5 and competes Tbx5-mediated activation of the cardiac Natriuretic peptide precursor type a-promoter. Tbx18 down-regulates Tbx6-activated Delta-like 1 expression in the somitic mesoderm in vivo 17592645_somatic GATA4 mutations in the 3'-UTR may provide an additional molecular rationale for congenital heart disease 17643447_Observational study of gene-disease association. (HuGE Navigator) 17643447_These data establish the phenotypic spectrum of heterozygous Gata4 mutation in mice, and suggest that heterozygous GATA4 mutation leads to partially overlapping phenotypes in humans. 17805225_Observational study of gene-disease association. (HuGE Navigator) 17912029_While hypermethylation of GATA-5 seems to be a universal feature among human tumors, infrequent methylation of GATA-4, and its corresponding overexpression, appears unique to pancreatic cancer from other tumor types reported thus far. 18055909_4 missense sequence variants (Gly93Ala, Gln316Glu, Ala411Val, Asp425Asn) occurred in patients with cardiac septal defects. 2 led to polarity changes. Non-synonymous GATA4 sequence variants sometimes occur in septal defects & rarely in conotruncal defects. 18055909_Observational study of gene-disease association. (HuGE Navigator) 18076106_identification of a novel GATA4 mutation in a patients with ASDII 18227727_the majority of the ovarian surface epithelial carcinomas retained GATA-4 expression, whereas approximately two-thirds of the carcinomas had mislocalization or loss of GATA-6 expression 18405344_The results suggest differential but overlapping functions for GATA-4 and GATA-6 in the normal gastrointestinal mucosa. Furthermore, GATA-4, GATA-6 and Ihh expression is altered in premalignant dysplastic lesions and reduced in overt cancer. 18426912_HNF4alpha regulates thyroid hormone homeostasis through transcriptional regulation of the Dio1 gene with GATA4 and KLF9 18653721_GATA-4 influences granulosa cell fate by transactivating Bcl-2. 18672102_Observational study of gene-disease association. (HuGE Navigator) 18672102_findings are useful in understanding the prevalence of GATA4 mutations and the correlation between the GATA4 genotype and the congenital heart disease phenotype in Chinese patients 18983250_Translocation of the transcription factors NKX2.5 and GATA4 to the nucleus was observed in all the cultures of mesenchymal stem cells during the differentiation process. 19008335_GATA4 mutations would have a lesser impact on gonadal gene transcription and function. 19018306_Observational study of gene-disease association. (HuGE Navigator) 19276186_It is proposed that the negative feedback loop linking ERBB2 and GATA4 plays a role in the transcriptional dysregulation of ERBB2 gene expression in breast cancer. 19302747_Observational study of gene-disease association. (HuGE Navigator) 19302747_The results provided the primary data on congenital heart disease phenotype associated with GATA4 mutation in the Chinese Uygur population. 19353247_TGF-beta1 drives GATA-4 expression during intestinal inflammation, these two components cooperating to promote epithelial healing. 19353638_elevated levels of GATA4 protein, due to a GATA4 gene duplication are not sufficient to cause congenital heart disease. 19479054_This study suggests that NKX2-5 modulates the beta-catenin and GATA4 transcriptional activities in developing human cardiac myocytes. 19509152_Methylation of GATA4/5 is a common and specific event in colorectal carcinomas, and GATA4/5 exhibit tumor suppressive effects in colorectal cancer cells in vitro. GATA4 methylation in fecal DNA may be of interest for colorectal cancer detection. 19543315_GATA4 is expressed in the embryonic and adult CNS and acts as a negative regulator of astrocyte proliferation and growth. 19649254_loss of GATA4 expression through changes in chromatin conformation suggests a potential non-phenotypic initiating event, leading to subsequent loss of GATA6, morphological transformation, and ultimate tumorigenesis 19678963_Analysis of the genetic background of the parents of the patient showed for the first time that a new mutation of GATA4 can cause sporadic atrial septal defects 19678963_Observational study of gene-disease association. (HuGE Navigator) 19781215_Mutations in the transcription factor GATA-4 may be related to congenital cardiac septal defects in Han Chinese patients. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19915893_Mutations in GATA4 might be related to congenital cardiac septal defects in Han ancestry patients 19915893_Observational study of gene-disease association. (HuGE Navigator) 19918328_These findings support previous studies implicating GATA4 in pancreatic cancer and offer new avenues for investigation into this aggressive tumor type. 19995889_Data suggest GATA-4 may be involved in up-regulation of expression of CYP2C9 in hepatocytes. 20041118_The 5' region of the mouse, rat, and human GATA4 genes, was characterized. 20081228_Cdk9 forms a functional complex with the p300/GATA4 and is required for p300/GATA4- transcriptional pathway during cardiomyocyte hypertrophy 20133952_In cooperation with Cdx2, HNF-1alpha acts as a key factor on human intestinal cells to trigger the onset of their functional differentiation program whereas GATA-4 appears to promote morphological changes. 20201924_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20206639_These data indicate that GATA-4 plays an important role in the transcriptional regulation of CYP2C19 expression 20222162_Epigenetic inactivation of GATA-4 by methylation of CpG islands is an early frequent event during gastric carcinogenesis and is significantly correlated with H. pylori infection. 20347099_Identified a novel M310V mutation in GATA4 gene that leads to hereditary atrial septal defect in a Chinese family. 20363377_Mutations in the cardiac transcription factor GATA4 is associated with lone atrial fibrillation. 20363377_Observational study of gene-disease association. (HuGE Navigator) 20450724_Observational study of gene-disease association. (HuGE Navigator) 20450724_The mutated GATA4 gene may be responsible for congenital heart disease in a subset of patients. 20554787_Overexpressing GATA4 protects granulosa cell tumors from TRAIL-induced apoptosis. 20585342_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20585342_genetic variations in GATA4 might influence relapse and treatment response to acamprosate in alcohol-dependent patients via modulation of atrial natriuretic peptide plasma levels. 20592452_GATA4 copy number variations was not associated with congenital heart disease. 20592452_Observational study of gene-disease association. (HuGE Navigator) 20595226_Studies identify a novel link between wood smoke exposure and gene promotor methylation of the p16 and GATA4 genes that synergistically increases the risk for reduced lung function in cigarette cmokers. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20654103_Observational study of gene-disease association. (HuGE Navigator) 20654103_Three novel heterozygous missense GATA4 mutations were identified in patients with congenital atrial septal defect. 20659440_T280M mutation of GATA4 is suggested to be associated with atrial septal defect in Chinese family 20807224_Observational study of gene-disease association. (HuGE Navigator) 20874241_these data do not support GATA4 mutations as a common cause of congenital heart malformation. 20931527_A novel missense GATA4 mutation leads to congenital ventricular septal defect. 21055141_Observational study of gene-disease association. (HuGE Navigator) 21055141_Two novel mutations of GATA4 gene are identified in two unrelated congenital atrial septal defect patients. 21110066_GATA4 missense mutations may be associated with congenital heart defects in pediatric Chinese patients. 21220346_Data demonstrate the key role of GATA4 in human testicular development. 21276881_Study identified NKX2.5 and GATA4 constitutional variants in CHD cohort. 21330551_show that Gata4 and Smad4 cooperatively activated the Id2 promoter, that human GATA4 mutations abrogated this activity, and that Id2 deficiency in mice could cause atrioventricular septal defects 21373748_The findings underscore the pathogenic link between compromised GATA4 function and congenital Atrial septal defect , providing new insight into the molecular mechanism involved in this common form of congenital cardiovascular anomalies. 21464220_Re-expression of GATA4 in human glioblastoma Multiforme cell lines, primary cultures, and brain tumor-initiating cells suppressed tumor growth in vitro 21519287_Our study shows no evidence of somatic NKX2-5, GATA4 and HAND1 mutations playing a role in the pathogenesis of Tetralogy of Fallot . Findings suggest that the GATA4 and HAND1 germline mutations are associated with non-syndromic congenital heart disease. 21609320_The findings of the present study (i) indicate that GATA-4 may participate in the control of hepcidin expression, and (ii) suggest that alteration of its expression could contribute to the development of iron-related disorders. 21631294_The involvement of specific SNPs of GATA4 in the manifestation of congenital heart defects, reported for the first time in an Indian scenario. 21637914_The findings expand the spectrum of mutations in GATA4 linked to ventricular septal defects and provide more insight into the molecular mechanism involved in VSD. 21673957_the common GATA4 variant S377G is likely to be relatively benign in terms of its participation in congenital heart defect and Patent foramen ovale/stroke 21708142_Two novel heterozygous GATA4 mutations, p.S70T and p.S160T in patients with familial atrial fibrillation 21815254_In the 28 of 32 nonsyndromic patients who underwent molecular testing, no mutation in GATA4 and NKX2.5 genes were detected. 21834050_This case suggests that the gene for GATA4, a transcription factor previously implicated in early cardiac development, is a candidate gene for the Left ventricular noncompaction phenotype. 21874226_Two novel heterozygous GATA4 mutations of p.G16C and p.H28D, were identified in 2 unrelated Chinese families with atrial fibrillation. 21933911_Cardiac defects are infrequent findings in individuals with 8p23.1 genomic duplications containing GATA4. 21945496_The combination of different sources of data has revealed an association of GATA4 with triglyceride levels in humans. 22011241_NKX2-5 and GATA4 were the first congenital heart defects-causing genes identified by linkage analysis in large affected families. 22018271_Heterogeneous methylation in the promoter region of GATA4 was associated with cancer progression in non-small cell lung cancer. 22043484_Somatic mutations in NKX2-5, GATA4, and HAND1 are not a common cause of tetralogy of Fallot or hypoplastic left heart. 22101736_The p.R43W variant was predicted to be a pathogenic mutation, and the functional analysis demonstrated that the GATA4 R43W mutant protein resulted in significantly decreased transcriptional activity compared with its wild-type counterpart. 22472323_The results indicated that histone deacetylation is a silencing mechanism for GATA4 expression in AFP-producing gastric cancer cells. 22500510_data suggest that these sequence variants within the promoter region of the GATA4 gene may contribute to the ventricular septal defect etiology by altering its gene expression 22552926_The findings expand the mutation spectrum of GATA4 linked to Atrial fibrillation (AF), and further support the notion that compromised GATA4 confers genetic susceptibility to AF. 22648249_These findings expand the mutation spectrum of GATA4 linked to ventricular septal defect. 22773876_a novel role for GATA-4 and TAL1 to affect skeletal myogenic differentiation and EPO response via cross-talk with Sirt1. 22862823_Our findings provide evidence of association between alcohol dependence and variation in GATA4 at the gene level. 22959235_genomic GATA4 and TFAP2B missense mutations may be associated with nonfamilial congenital heart disease with diverse clinical phenotypes in patients with congenital heart disease from southern China 23029311_Sex Cord Stromal Tumors in childhood exhibited an embryonal gonadal phenotype, expressing a FOG-2/GATA-4 pattern in keeping with embryonal gonads. 23138528_Identified a novel GATA binding protein 4 (GATA4) variant in a familial case of congenital diaphragmatic hernia (CDH), using whole exome sequencing (WES), and identified a de novo GATA4 variant in sporadic CDH. 23215884_we found a relationship between genetic variation in GATA-4 and the acenocoumarol maintenance dose 23239811_Results thus identify a novel epigenetic mechanism by which MYC activates GATA4 leading to metastasis in lung adenocarcinoma. 23320456_GATA4 and DcR1 promoter hypermethylation is tumor specific event in glioblastoma but they promoter methylation cannot be considered as a prognostic marker of glioblastoma survival. 23626780_Functional mutations in GATA4 in patients with congenital heart disease included effects on transcriptional activity, subcellular localization and DNA binding affinity of some of the mutant GATA4 proteins. 23745586_C-terminus of GATA4 is critical to maintain DNA binding. 23746174_Significantly higher methylation was observed in genes NTKR1, GATA4 and WIF1 in the ovarian cancer group compared with the control group. 23888774_Expression level of PPP3R1 and GATA4, and NFATC4 genes for transcription factors did not differ in studied subgroups of patients. 24000169_GATA4 loss-of-function mutations underlie familial tetralogy of fallot. 24041700_the GATA4 mutant was associated with significantly decreased transcriptional activity and remarkably reduced synergistic activation 24083978_These findings demonstrate a novel ERRgamma/GATA4 signal cascade in the development of cardiac hypertrophy. 24089524_nicotine induced the binding of GATA4 or GATA6 to Sp1 on the alpha7-nAChR promoter, thereby inducing its transcription and increasing its levels in human SCC-L. 24145767_These results confirm that methylation in the GATA4 promoter region could play an important role in ovarian carcinogenesis, and show new loci which are highly methylated only in ovarian cancer samples 24182332_DNA methylation status of NKX2-5, GATA4 and HAND1 in patients with tetralogy of fallot 24284823_Variants in the human GATA4 locus are associated with an increased risk for type-1 diabetes mellitus. 24314346_Lower alcohol-cue-induced activations in G-allele carriers as compared with AA-homozygotes in the bilateral amygdala. A stronger alcohol-specific amygdala response predicted a lowered risk for relapse to heavy drinking in the AA-homozygotes. 24330461_Our study identified the GATA4 transcription factor as an independent risk factor for congenital heart disease and coronary artery disease, myocardial infarction and a metabolic risk trait for cardiovascular diseases. 24365599_Observations have uncovered novel functions of miR-200c and GATA4 in regulating hESC renewal and differentiation. 24366163_The findings expand the mutational spectrum of GATA4 linked to Dilated cardiomyopathy and provide novel insight into the molecular etiology involved in DCM. 24415412_Mesenchymal GATA4 activity regulates hepatic stellate cell activation and inhibits the liver fibrogenic process. 24416423_The molecular interactions of FOXL2, GATA4, and SMAD3 and their roles in the regulation of CCND2 using co-immunoprecipitation, promoter transactivation, and cell viability assays in human granulosa cell tumor cells, were investigated. 24498650_A novel GATA4 mutation, c.955ANG (p.K319E), was identified. 24505034_We found that both of these factors significantly suppressed gelsolin-induced cardiac hypertrophy through p38/GATA4 signaling pathway. 24651021_Study showed that there is significantly higher methylation in GATA4 gene in the endometrial cancer group compared with samples of non-neoplastic endometrium. 24681789_Early cardiac marker gene GATA4 levels in peripheral blood mononuclear cells reflect severity in stable coronary artery disease. 24687970_In conclusion HER2 and GATA4 are new molecular prognostic markers of GCT recurrence, which could be utilized to optimize the management and follow-up of patients with early-stage GCTs 24696446_The results show that GATA4 mutations/deletions are a cause of neonatal or childhood-onset diabetes with or without exocrine insufficiency. 24743694_Art27 interacts with GATA4, FOG2 and NKX2.5 and is a novel co-repressor of cardiac genes. 24862985_GATA4 plays important roles in the progression of breast carcinoma from an early stage and immunohistochemical GATA4 status is considered a potent prognostic factor in human breast cancer patients. 24866383_NEXN as a novel gene for ASD and its function to inhibit GATA4 established a critical regulation of an F-actin binding protein on a transcription factor in cardiac development 24973415_The present study is the first to suggest that GATA-4 gene methylation status may independently predict health status in individuals with COPD. 25017055_3 novel heterozygous GATA4 mutations, p.V39L, p.P226Q and p.T279S, were identified in 3 unrelated patients with sporadic dilated cardiomyopathy. The altered amino acids after missense mutations were completely conserved across species. 25025186_No copy number variations of the gene were detected. GST pull-down assays demonstrated that all potentially deleterious variants, including those previously reported, did not impair the interaction with GATA4 25053715_KLF5/GATA4/GATA6 may promote gastric cancer development by engaging in mutual crosstalk, collaborating to maintain a pro-oncogenic transcriptional regulatory network in gastric cancer cells. 25099673_we identified a mutation in the GATA4 Kozak sequence that likely contributes to the pathogenesis of Atrial septal defect. 25203927_There was no evidence of a role for NKX2-5 and GATA4 CNV in fetal CHD; therefore, these CNV may not be common in fetal CHD in China 25272520_We identified 17 mutations, including five non-synonymous sequence changes, one synonymous variant and one variation in the 5' UTR, three intronic changes and seven deletions. We found no evidence of gene GATA4 somatic sequence variants 25416133_Lower levels of Gata4 expression enhanced cardiac myocyte reprogramming efficiency. 25524324_Germline mutations in the NKX2-5, GATA4, and CRELD1 genes do not appear to be associated with CHD in Mexican DS patients. 25873328_this study confirms that GATA4 M310V mutation may lead to the development of the congenital heart defect, ASD. 25928801_GATA4 genetic variations are associated with congenital heart disease 26071180_Data show that the combination of GATA binding protein 4 (Gata4), T-box transcription factor 5 (Tbx5) and BRG1-associated factor 60C protein (Baf60c) is sufficient for inducing adipose tissue-derived mesenchymal stem cells (ADMSCs) to form cardiomyocytes. 26376067_Whole exome sequencing results on four-generation Chinese family with atrial septal defect (ASD) identified a novel mutation in GATA4 gene at the methylation position associated with ASD. 26381449_This study showed that GATA4 gene involved in neuronal growth and cerebellum development and associated with neurological and psychological disorders. 26404840_GATA4 accumulates in multiple tissues, including the aging brain, and could contribute to aging and its associated inflammation. 26490736_Kaplan-Meier survival analysis revealed significantly shorter overall survival in pediatric Acute myeloid leukemia with GATA4 promoter methylation but multivariate analysis shows that it is not an independent factor. 26861571_The definitive endoderm and foregut endoderm differentiation capabilities of Wnt pathway-modulated cells were determined based on the expression levels of the endodermal transcription factors SOX17 and FOXA2 and those of the transcription activator GATA4 and the alpha-fetoprotein (AFP) gene, respectively. 26946174_results demonstrate that cGMP-PKG signaling mediates transcriptional activity of GATA4 and links defective GATA4 and PKG-1alpha mutations to the development of human heart disease. 27064867_Mutations of GATA4 appear to be responsible for some cardiac septal defects. The aim of this work was to screen for mutations in the GATA4 gene in sample of Egyptian patients affected by isolated and non-isolated cardiac septal defects. Identified are two coding variants and four non-coding ones of GATA4 gene, but further confirmation study for familial segregation detection was recommended. 27118528_common variants in 3'UTR of the GATA4 gene jointly interact, affecting the congenital heart disease susceptibility, probably by altering microRNA posttranscriptional regulation 27154817_NKX2.5 and GATA4 gene mutations might participate in the development of congenital heart disease and can promote bone marrow derived stroma cell differentiate into cardiomyocytes. 27208796_findings demonstrate that RACK1 is involved in p300/GATA4-dependent hypertrophic responses in cardiomyocytes and is a promising therapeutic target for heart failure 27374936_The role of GATA4 was elucidated in alcohol dependence susceptibility by identifying rare genetic variants. 27391137_study identified a novel mutation in GATA4 that likely contributed to the Congenital Heart Disease in this family. This finding expanded the spectrum of GATA4 mutations and underscored the pathogenic correlation between GATA4 mutations and Congenital Heart Disease. 27397865_Study identified effects of GATA4 variant [(SNP) rs13273672] on regional gray matter (GM) volume in alcohol dependence: higher GM volume in the hypothalamus and caudate in the AA genotype group compared to the AG/GG group. GM volume specific to GATA4 variant predicted heavy relapse risk within 60 d following discharge for both caudate and amygdala and within 90 d for the amygdala only. 27426723_Hence, the variant distribution of NKX2-5, GATA4 and TBX5 are tightly associated with particular Congenital heart disease subtypes. Further structure-modelling analysis revealed that these mutated amino acid residuals maintain their DNA-binding ability and structural stability 27553283_Findings suggest that a single introduction of the three cardiomyogenic transcription factor (GATA4, cand TBX5)genes using polyethyleneimine (PEI)-based transfection is sufficient for transdifferentiation of adipose-derived stem cells (hADSCs) towards the cardiomyogenic lineage. 27598297_miR-126 inhibits the migration and invasion of glioma cells, which may be linked to GATA4 as a target gene. 27599506_Study demonstrated downregulation of expression of pancreatic master genes SOX9, FOXA2, and GATA4 (2-, 5-, and 4-fold, respectively) and in PANC1 pancreatic cancer cell line stimulated with TGFbeta1 27788486_This report demonstrates that GATA4 promotes oncogenesis by inhibiting miR125b-dependent suppression of DKK3 expression. This GATA4/miR125b/DKK3 axis may be a major regulator of growth, migration, invasion, and survival in hepatoma cells. 27894866_Our studies suggest that GATA5 but especially GATA4 are main contributors to SCN5A gene expression, thus providing a new paradigm of SCN5A expression regulation that may shed new light into the understanding of cardiac disease. 27984724_GATA4-G296S mutation led to failure of GATA4 and TBX5-mediated repression at non-cardiac genes and enhanced open chromatin states at endothelial/endocardial promoters. These results reveal how disease-causing missense mutations can disrupt transcriptional cooperativity, leading to aberrant chromatin states and cellular dysfunction, including those related to morphogenetic defects. 28161810_Subsequent functional analyses revealed that the transcriptional activity and Western blot of A167D mutant GATA4 protein were not altered in a Chinese Han population. These variants may be involved in other mechanisms underlying Conotruncal heart defect (CTD) or may be unrelated to CTD occurrence. 28196600_study found that the formation of pancreatic progenitors cells is highly sensitive to the GATA6 and GATA4 gene dosage 28241454_We confirmed the significance of the HNF1B and GATA4 hypermethylation with emphasis on the need of selecting the most relevant sites for analysis. We suggest selected CpGs to be further examined as a potential positive prognostic factor. 28263493_Studied prevalence of GATA binding protein 4 (GATA4) exon 1 mutation in Egyptian patients with isolated congenital heart defects. 28349834_our results indicate that since high endogenous levels of transcription factor GATA4 likely protect hepatoblastoma cells from doxorubicin-induced apoptosis, these cells can be rendered more sensitive to the drug by downregulation of GATA4. 28381408_although relatively infrequent in patients with hypertrophic cardiomyopathy, GATA2, 4 and 6 transcription factors may represent a novel insight into the molecular mechanisms related to the pathogenesis of hypertrophic cardiomyopathy 28393293_GATA4 is a regulator of osteoblastic differentiation via the p38 signaling pathways. 28471988_Meta-analysis suggested that GATA4 99 G>T and 487 C>T mutations may not be related to the incidence of congenital heart disease (CHD). However, GATA4 354 A>C mutation was significantly associated with CHD risk. 28484278_direct binding of GATA4 to the GNAI3 promoter, both in vitro and in vivo, is reported. 28541271_Report a genome-wide association scan of 466 bicuspid aortic valve cases and 4,660 age, sex and ethnicity-matched controls with replication in up to 1,326 cases and 8,103 controls. We identify association with a noncoding variant 151 kb from the gene encoding the cardiac-specific transcription factor, GATA4, and near-significance for p.Ser377Gly in GATA4. 28669928_GATA4 induces autocrine BMP2 signaling in endothelial cells 28758902_disruption of GATA4-mediated transactivation in hepatocellular carcinoma suppresses hepatocyte epithelial differentiation to sustain replicative precursor phenotype 28843068_This study attempts to correlate the pattern of intronic variants of GATA4 gene which might provide new insights to unravel the possible molecular etiology of congenital heart disease. 28849107_GATA4 was a transcription factor that activated mouse double minute 2 homolog (MDM2) and B cell lymphoma 2 (BCL2) expression in ALL cells. 29018978_when ZFPM2R698Q was co-transfected with GATA4, BNP promoter activity increased significantly, whereas co-transfection with ZFPM2R736L and GATA4 did not significantly increase BNP promoter activity. This suggests that the R698Q mutation may affect the ability of ZFPM2 to bind GATA4. 29104469_the mutation significantly diminished the synergistic activation between MEF2C and GATA4, another cardiac core transcription factor that has been causally linked to Congenital heart disease (CHD). 29138836_GATA4 may inhibit diabetesinduced endothelial dysfunction by acting as a transcription factor for NOX4 expression. 29377543_we found two nucleotide deletions which one of them was novel and one new indel mutation resulting in frame shift mutation, and 4 synonymous variations or polymorphism in 6 of patients and 3 of normal individuals 29415147_GATA4 variants were not associated with Alcohol Use Disorder (AUD) in either the European ancestry or African ancestry groups after correcting for multiple comparisons. Rs10112596 demonstrated a significant relationship with an anxiety measure among the African ancestry group with AUD. 29717129_GATA4 regulates angiogenesis and persistence of inflammation in rheumatoid arthritis. 29760459_abnormal lamin. proteins trigger paracrine senescence through GATA4-dependent pathway in human mesenchymal stem cells. 29972125_The single nucleotide polymorphisms (SNPs) of NKX2.5, GATA4, and TBX5 are highly associated with congenital heart diseases in the Chinese population, but not significant in the SNPs | ENSMUSG00000021944 | Gata4 | 1.516086e+03 | 1.3674364 | 0.451473739 | 0.3203757 | 1.994727e+00 | 0.1578475168 | 0.77593452 | No | Yes | 1.634694e+03 | 189.865512 | 1.030461e+03 | 123.284693 | |
| ENSG00000136895 | 84253 | GARNL3 | protein_coding | Q5VVW2 | Alternative splicing;GTPase activation;Phosphoprotein;Reference proteome | hsa:84253; | cytoplasm [GO:0005737]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; regulation of small GTPase mediated signal transduction [GO:0051056] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000038860 | Garnl3 | 2.728375e+02 | 0.5844766 | -0.774782862 | 0.3415954 | 4.945034e+00 | 0.0261657250 | 0.55119367 | No | Yes | 1.417729e+02 | 33.303704 | 2.649326e+02 | 63.356702 | |||
| ENSG00000136937 | 4686 | NCBP1 | protein_coding | Q09161 | FUNCTION: Component of the cap-binding complex (CBC), which binds cotranscriptionally to the 5'-cap of pre-mRNAs and is involved in various processes such as pre-mRNA splicing, translation regulation, nonsense-mediated mRNA decay, RNA-mediated gene silencing (RNAi) by microRNAs (miRNAs) and mRNA export. The CBC complex is involved in mRNA export from the nucleus via its interaction with ALYREF/THOC4/ALY, leading to the recruitment of the mRNA export machinery to the 5'-end of mRNA and to mRNA export in a 5' to 3' direction through the nuclear pore. The CBC complex is also involved in mediating U snRNA and intronless mRNAs export from the nucleus. The CBC complex is essential for a pioneer round of mRNA translation, before steady state translation when the CBC complex is replaced by cytoplasmic cap-binding protein eIF4E. The pioneer round of mRNA translation mediated by the CBC complex plays a central role in nonsense-mediated mRNA decay (NMD), NMD only taking place in mRNAs bound to the CBC complex, but not on eIF4E-bound mRNAs. The CBC complex enhances NMD in mRNAs containing at least one exon-junction complex (EJC) via its interaction with UPF1, promoting the interaction between UPF1 and UPF2. The CBC complex is also involved in 'failsafe' NMD, which is independent of the EJC complex, while it does not participate in Staufen-mediated mRNA decay (SMD). During cell proliferation, the CBC complex is also involved in microRNAs (miRNAs) biogenesis via its interaction with SRRT/ARS2 and is required for miRNA-mediated RNA interference. The CBC complex also acts as a negative regulator of PARN, thereby acting as an inhibitor of mRNA deadenylation. In the CBC complex, NCBP1/CBP80 does not bind directly capped RNAs (m7GpppG-capped RNA) but is required to stabilize the movement of the N-terminal loop of NCBP2/CBP20 and lock the CBC into a high affinity cap-binding state with the cap structure. Associates with NCBP3 to form an alternative cap-binding complex (CBC) which plays a key role in mRNA export and is particularly important in cellular stress situations such as virus infections. The conventional CBC with NCBP2 binds both small nuclear RNA (snRNA) and messenger (mRNA) and is involved in their export from the nucleus whereas the alternative CBC with NCBP3 does not bind snRNA and associates only with mRNA thereby playing a role only in mRNA export. NCBP1/CBP80 is required for cell growth and viability (PubMed:26382858). {ECO:0000269|PubMed:11551508, ECO:0000269|PubMed:12093754, ECO:0000269|PubMed:15059963, ECO:0000269|PubMed:15361857, ECO:0000269|PubMed:16186820, ECO:0000269|PubMed:16317009, ECO:0000269|PubMed:17190602, ECO:0000269|PubMed:17873884, ECO:0000269|PubMed:18369367, ECO:0000269|PubMed:19632182, ECO:0000269|PubMed:19648179, ECO:0000269|PubMed:26382858, ECO:0000269|PubMed:7651522, ECO:0000269|PubMed:8069914}. | 3D-structure;Acetylation;Coiled coil;Cytoplasm;Direct protein sequencing;Isopeptide bond;Nonsense-mediated mRNA decay;Nucleus;Phosphoprotein;RNA-mediated gene silencing;Reference proteome;Translation regulation;Transport;Ubl conjugation;mRNA capping;mRNA processing;mRNA splicing;mRNA transport | The product of this gene is a component of the nuclear cap-binding protein complex (CBC), which binds to the monomethylated 5' cap of nascent pre-mRNA in the nucleoplasm. The encoded protein promotes high-affinity mRNA-cap binding and associates with the CTD of RNA polymerase II. The CBC promotes pre-mRNA splicing, 3'-end processing, RNA nuclear export, and nonsense-mediated mRNA decay. [provided by RefSeq, Jul 2008]. | hsa:4686; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; mRNA cap binding complex [GO:0005845]; nuclear cap binding complex [GO:0005846]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; RNA cap binding complex [GO:0034518]; mRNA binding [GO:0003729]; RNA 7-methylguanosine cap binding [GO:0000340]; RNA binding [GO:0003723]; RNA cap binding [GO:0000339]; 7-methylguanosine mRNA capping [GO:0006370]; defense response to virus [GO:0051607]; gene silencing by RNA [GO:0031047]; histone mRNA metabolic process [GO:0008334]; mRNA cis splicing, via spliceosome [GO:0045292]; mRNA export from nucleus [GO:0006406]; mRNA transcription by RNA polymerase II [GO:0042789]; nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184]; positive regulation of cell growth [GO:0030307]; positive regulation of mRNA 3'-end processing [GO:0031442]; positive regulation of mRNA splicing, via spliceosome [GO:0048026]; positive regulation of RNA binding [GO:1905216]; pre-mRNA cleavage required for polyadenylation [GO:0098789]; regulation of mRNA processing [GO:0050684]; regulation of translational initiation [GO:0006446]; RNA catabolic process [GO:0006401]; RNA splicing [GO:0008380]; snRNA export from nucleus [GO:0006408]; spliceosomal complex assembly [GO:0000245] | 12374755_crystal structure at 2.1 A resolution of CBP20 and CBP80 bound to an m(7)GpppG cap analogue 12434151_Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. 16156639_The organization of the CBP80-CBP20 complex suggests how the activity of eIF4G in translation initiation could be regulated through a dynamic network of overlapping intra- and intermolecular interactions. 16186820_During nonsense-mediated mRNA decay, CBP80 interacts with Upf1 and promotes the interaction of Upf1 with Upf2 but not with Stau1. 16317009_Association of CBC with PARN might have importance in the regulated recruitment of PARN to the nonsense-mediated decay pathway during the pioneer round of translation. 17499042_We show that NELF interacts with the nuclear cap binding complex (CBC), a heterodimeric, multifunctional factor that plays important roles in several mRNA processing steps, and the two factors together participate in the 3' end processing of histone mRNAs 19026660_The authors could distinguish two tyrosines, Y43 and Y20, in stabilization of the cap inside the cap-binding complex binding pocket. 19648179_a new MIF4G domain-containing protein, CTIF (CBP80/20-dependent translation initiation factor) that interacts directly with CBP80 and is part of the CBP80/20-dependent translation initiation complex 19668212_resolution X-ray structure for a cap-binding complex (CBC)-importin-alpha complex that provides a detailed picture for how importin-alpha binds to the CBP80 subunit of the CBC. 20691628_UPF1 binds PTC-containing mRNA more efficiently than the corresponding PTC-free mRNA in a way that is promoted by the UPF1-CBP80 interaction. 21402597_Viral mRNA associates with the influenza virus A NS1 and the cellular NCBP1 of the nuclear cap-binding complex. 21447822_Cellular processes by which CBP80-CBP20 -bound messenger ribonucleoproteins (mRNPs) are remodeled to eIF4E-bound mRNPs, are overviewed. 22493286_down-regulation of CTIF using a small interfering RNA causes a redistribution of CBP80 from polysome fractions to subpolysome fractions, without significant consequence to eIF4E distribution 26382858_Study proposes the existence of an alternative cap-binding complex involving NCBP1 and NCBP3 that plays a key role in mRNA biogenesis. 30239828_Results suggest that rev protein, HIV-1 (Rev) regulates the association of nuclear cap-binding protein NCBP 80 kDa subunit (CBP80) and eukaryotic translation initiation factor 4A1 (eIF4AI) with the unspliced mRNA in the cytoplasm and the nucleus, respectively, during HIV-1 replication. 31448526_NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. 32960270_Affinity proteomic dissection of the human nuclear cap-binding complex interactome. 34232997_Translation mediated by the nuclear cap-binding complex is confined to the perinuclear region via a CTIF-DDX19B interaction. | ENSMUSG00000028330 | Ncbp1 | 1.285307e+03 | 0.7918173 | -0.336760544 | 0.2898116 | 1.331450e+00 | 0.2485474840 | 0.79042389 | No | Yes | 1.060614e+03 | 155.888261 | 1.260072e+03 | 189.564646 | |
| ENSG00000136986 | 79139 | DERL1 | protein_coding | Q9BUN8 | FUNCTION: Functional component of endoplasmic reticulum-associated degradation (ERAD) for misfolded lumenal proteins (PubMed:15215856, PubMed:33658201). Forms homotetramers which encircle a large channel traversing the endoplasmic reticulum (ER) membrane (PubMed:33658201). This allows the retrotranslocation of misfolded proteins from the ER into the cytosol where they are ubiquitinated and degraded by the proteasome (PubMed:33658201). The channel has a lateral gate within the membrane which provides direct access to membrane proteins with no need to reenter the ER lumen first (PubMed:33658201). May mediate the interaction between VCP and the misfolded protein (PubMed:15215856). Also involved in endoplasmic reticulum stress-induced pre-emptive quality control, a mechanism that selectively attenuates the translocation of newly synthesized proteins into the endoplasmic reticulum and reroutes them to the cytosol for proteasomal degradation (PubMed:26565908). By controlling the steady-state expression of the IGF1R receptor, indirectly regulates the insulin-like growth factor receptor signaling pathway (PubMed:26692333). {ECO:0000269|PubMed:15215856, ECO:0000269|PubMed:26565908, ECO:0000269|PubMed:26692333, ECO:0000269|PubMed:33658201}.; FUNCTION: (Microbial infection) In case of infection by cytomegaloviruses, it plays a central role in the export from the ER and subsequent degradation of MHC class I heavy chains via its interaction with US11 viral protein, which recognizes and associates with MHC class I heavy chains. Also participates in the degradation process of misfolded cytomegalovirus US2 protein. {ECO:0000269|PubMed:15215855, ECO:0000269|PubMed:15215856}. | 3D-structure;Acetylation;Alternative splicing;Endoplasmic reticulum;Host-virus interaction;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transmembrane;Transmembrane helix;Transport;Unfolded protein response | The protein encoded by this gene is a member of the derlin family. Members of this family participate in the ER-associated degradation response and retrotranslocate misfolded or unfolded proteins from the ER lumen to the cytosol for proteasomal degradation. This protein recognizes substrate in the ER and works in a complex to retrotranslocate it across the ER membrane into the cytosol. This protein may select cystic fibrosis transmembrane conductance regulator protein (CFTR) for degradation as well as unfolded proteins in Alzheimer's disease. Alternative splicing results in multiple transcript variants that encode different protein isoforms. [provided by RefSeq, Aug 2012]. | hsa:79139; | Derlin-1 retrotranslocation complex [GO:0036513]; Derlin-1-VIMP complex [GO:0036502]; early endosome [GO:0005769]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum quality control compartment [GO:0044322]; Hrd1p ubiquitin ligase ERAD-L complex [GO:0000839]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of membrane [GO:0016021]; late endosome [GO:0005770]; membrane [GO:0016020]; ATPase binding [GO:0051117]; identical protein binding [GO:0042802]; MHC class I protein binding [GO:0042288]; misfolded protein binding [GO:0051787]; protease binding [GO:0002020]; protein-containing complex binding [GO:0044877]; signal recognition particle binding [GO:0005047]; signaling receptor activity [GO:0038023]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-specific protease binding [GO:1990381]; endoplasmic reticulum unfolded protein response [GO:0030968]; ER-associated misfolded protein catabolic process [GO:0071712]; ERAD pathway [GO:0036503]; establishment of protein localization [GO:0045184]; positive regulation of protein binding [GO:0032092]; positive regulation of protein ubiquitination [GO:0031398]; protein destabilization [GO:0031648]; response to unfolded protein [GO:0006986]; retrograde protein transport, ER to cytosol [GO:0030970]; ubiquitin-dependent ERAD pathway [GO:0030433] | 15215855_Derlin-1 is an important factor for the extraction of certain aberrantly folded proteins from the mammalian ER 15215856_Derlin-1 interacts with US11, a virally encoded ER protein that specifically targets MHC class I heavy chains for export from the ER, as well as with VIMP, a novel membrane protein that recruits the p97 ATPase and its cofactor 16055502_Derlin-1 interacts with the N-terminal domain of PNGase via its cytosolic C-terminus. PNGase distributed in two populations; ER-associated and free in the cytosol, which suggests the deglycosylation process can proceed at either site 16186510_Derlin-1 is part of a retrotranslocation channel that is associated with both the polyubiquitination and p97-ATPase machineries at the endoplasmic reticulum membrane. 16954204_Derlin-1 recognizes misfolded, nonubiquitylated CFTR to initiate its dislocation and degradation early in the course of CFTR biogenesis, perhaps by detecting structural instability within the first transmembrane domain 17453418_The pool of active Derlin-1 in the ER membrane can be modulated in response to ER stress. 17872946_SVIP is an endogenous inhibitor of ERAD that acts through regulating the assembly of the gp78-p97/VCP-Derlin1 complex. 18048502_DERL1-containing protein mediates the endoplasmic reticulum-associated degradation of V2 vasopressin receptors. 18094046_These findings indicate that Derlin-1 facilitates the retro-translocation of Cholera toxin. 18205950_derlin-1 overexpression in breast cancer, together with its function in relieving ER stress-induced apoptosis, suggests that regulation of the ER stress response pathway may be critical in the development and progression of breast cancer. 18227067_KCa3.1 and KCa2.3 are translocated out of the endoplasmic reticulum associated with Derlin-1. 18927294_Overexpression of DERL1 is associated with neoplasms 19114714_Derlin-1 did not modify KCNQ1 expression level, and no interaction between endogenous KCNQ1 and Derlin-1 could be detected. 21184741_Derlin-1 is a negative regulator for both glycosylated and non-glycosylated BCRP expression and provide a novel posttranslational regulatory mechanism of BCRP by Derlin-1. 22238364_These results indicate that ApoB after lipidation is dislocated from the ER lumen to the LD surface for proteasomal degradation and that Derlin-1 and UBXD8 are engaged in the predislocation and postdislocation steps, respectively. 22311976_Derlin-1 expression levels may affect glucose-stimulated insulin secretion by altering surface expression of K(ATP) channels. 22627700_Upregulation of derlin-1 may be associated with endoplasmic reticulum stress in neuronal cells in Alzheimer's disease. 23306155_Derlin-1 is overexpressed in non-small cell lung cancer and promotes invasion by EGFR-ERK-mediated up-regulation of MMP-2 and MMP-9. 24089527_Cav-1 may be a cofactor in the interaction of Derlin-1 and N-glycosylated COX-2 and may facilitate Derlin-1- and p97 complex-mediated COX-2 ubiquitination, retrotranslocation, and degradation. 25030448_TMEM129 contains an unusual cysteine-only RING with intrinsic E3 ligase activity and is recruited to US11 via Derlin-1. 26173415_Results showed that Derlin-1 is overexpressed in colon cancer and promotes proliferation of colon cancer cells. 27572270_data suggest that miR-181d is a tumor suppressor in ESCC inversely regulating its downstream target gene of DERL1. 27714797_insights into the interactions between other SHP-containing proteins and p97N 27977784_Derlin-1 was overexpressed in bladder cancer and was associated with the malignancy of bladder cancer. 28137758_The derlin-1 pathway therefore may represent a significant early checkpoint in the recognition and degradation of ENaC in mammalian cells. 28178653_Derlin-1 is overexpressed in bladder cancer and promotes malignant phenotype through ERK/MMP-2/9 and PI3K/AKT signaling pathway. 28219405_ChIP assays were used to ascertain the correlations between HNF1beta and Derlin-1 in the miR-217/HNF1beta/Derlin-1 pathway in glioma cells 29530993_Overexpression of Derlin-1 is Associated with Non-small Cell Lung Cancer. 29743537_association of the degradation factor HRD1 with the translocon and the rerouting factor Derlin-1 may be necessary for the smooth and effective clearance of ERpQC substrates 29768262_These findings for the first time revealed that miR-598, as a tumor suppressor, negatively regulate DERL1 and Epithelial-Mesenchymal Transition to suppress the invasion and migration in Non-Small Cell Lung Cancer (NSCLC), thereby putatively serving as a novel therapeutic target for NSCLC clinical treatment. 31670413_Derlin-1 exhibits oncogenic activities and indicates an unfavorable prognosis in breast cancer. 31721721_Derlin-1 functions as a growth promoter in breast cancer. 33658201_The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1. 35046954_Derlin-1, as a Potential Early Predictive Biomarker for Nonresponse to Infliximab Treatment in Rheumatoid Arthritis, Is Related to Autophagy. | ENSMUSG00000022365 | Derl1 | 2.908927e+03 | 0.6670650 | -0.584100659 | 0.3039157 | 3.774308e+00 | 0.0520454321 | 0.67565452 | No | Yes | 2.123470e+03 | 299.196327 | 3.404254e+03 | 490.925923 | |
| ENSG00000137070 | 3590 | IL11RA | protein_coding | Q14626 | FUNCTION: Receptor for interleukin-11 (IL11). The receptor systems for IL6, LIF, OSM, CNTF, IL11 and CT1 can utilize IL6ST for initiating signal transmission. The IL11/IL11RA/IL6ST complex may be involved in the control of proliferation and/or differentiation of skeletogenic progenitor or other mesenchymal cells (Probable). Essential for the normal development of craniofacial bones and teeth. Restricts suture fusion and tooth number. {ECO:0000269|PubMed:21741611, ECO:0000305}.; FUNCTION: [Soluble interleukin-11 receptor subunit alpha]: Soluble form of IL11 receptor (sIL11RA) that acts as an agonist of IL11 activity (PubMed:30279168, PubMed:26876177). The IL11:sIL11RA complex binds to IL6ST/gp130 on cell surfaces and induces signaling also on cells that do not express membrane-bound IL11RA in a process called IL11 trans-signaling (PubMed:30279168, PubMed:26876177). {ECO:0000269|PubMed:26876177, ECO:0000269|PubMed:30279168}.; FUNCTION: [Isoform HCR2]: Soluble form of IL11 receptor (sIL11RA) that acts as an agonist of IL11 activity (PubMed:30279168, PubMed:26876177). The IL11:sIL11RA complex binds to IL6ST/gp130 on cell surfaces and induces signaling also on cells that do not express membrane-bound IL11RA in a process called IL11 trans-signaling (PubMed:30279168, PubMed:26876177). {ECO:0000269|PubMed:26876177, ECO:0000269|PubMed:30279168}. | 3D-structure;Alternative splicing;Craniosynostosis;Disease variant;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA; + ;NA | Interleukin 11 is a stromal cell-derived cytokine that belongs to a family of pleiotropic and redundant cytokines that use the gp130 transducing subunit in their high affinity receptors. This gene encodes the IL-11 receptor, which is a member of the hematopoietic cytokine receptor family. This particular receptor is very similar to ciliary neurotrophic factor, since both contain an extracellular region with a 2-domain structure composed of an immunoglobulin-like domain and a cytokine receptor-like domain. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jun 2012]. | hsa:3590; | external side of plasma membrane [GO:0009897]; extracellular region [GO:0005576]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; cytokine binding [GO:0019955]; cytokine receptor activity [GO:0004896]; interleukin-11 binding [GO:0019970]; interleukin-11 receptor activity [GO:0004921]; transmembrane signaling receptor activity [GO:0004888]; cytokine-mediated signaling pathway [GO:0019221]; developmental process [GO:0032502]; head development [GO:0060322]; positive regulation of cell population proliferation [GO:0008284] | 11241561_Observational study of gene-disease association. (HuGE Navigator) 11315919_Observational study of gene-disease association. (HuGE Navigator) 11498264_Observational study of gene-disease association. (HuGE Navigator) 12200462_IL-11Ralpha was expressed in both epithelial and stromal cells, with epithelial staining being more intense 12569176_expression and function in human endometrium 14701802_the interleukin-11 receptor alpha-chain has evidence of antiapoptotic effects in human colonic epithelial cells 14744752_IL-11Ralpha is a candidate target for translational clinical trials against advanced and metastatic prostate cancer. 15512823_interleukin-11 receptor is expressed in CD38-positive cells from patients with multiple myeloma 16291580_High expression of interleukin-11 receptor alpha is associated with Hodgkin's lymphoma 16614887_High interleukin 11 receptor is associated with breast cancer 16964382_IL-11/IL-11R pathway plays an important role in the progression of colorectal adenocarcinoma. 17332920_The objectives of this study were to clarify the role of IL-11 and IL-11Ralpha in human gastric carcinoma. 18941632_binding site of il-11 to IL-11R alpha is characterized 18987331_IL11 inhibits human extravillous trophoblast invasion via STAT3, indicating a likely role for IL11 in the decidual restraint of EVT invasion during normal pregnancy. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20553623_IL11 as well as IL11RA are likely to play a role in the progression of endometrial carcinoma. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21982075_IL11RA promoter polymorphism--rs1061758--may be associated with the risk of papillary thyroid cancer in the Korean population. 22075555_data suggest that IL-11Ralpha-CAR T cells may represent a new therapy for patients with osteosarcoma pulmonary metastases 22433466_Constructed a designer cytokine Hyper IL-11 (H11), which is exclusively composed of naturally existing components. It contains the full length sIL-11Ralpha connected with the mature IL-11 protein, and acts as an agonist on cells expressing the gp130 molec 25524575_These results indicate that IL-11Ralpha is a potential target for the development of molecular targeted therapy and noninvasive tumor imaging in human osteosarcoma. 26551279_Data suggest domains D1-D3, which contain cytokine binding module, determine which cytokine can activate interleukin-6 or interleukin-11 alpha-receptor subunits; stalk, transmembrane, or intracellular regions do not participate in ligand selectivity. 26876177_Proteolysis of the IL-11R represents a molecular switch that controls the IL-11 trans-signaling pathway which is the target in intestinal tumorigenesis, lung carcinomas, and asthma. 27920471_Report IL11RA and MELK amplification in gastric cancer cell lines and primary gastric adenocarcinomas. 27922075_Cancer-associated fibroblasts treated with cisplatin facilitate chemoresistance of lung adenocarcinoma through IL-11/IL-11R/STAT3 signaling pathway. 28186993_High IL11RA expression is associated with endometrioid tumours. 29237553_we show by molecular replacement that Arg-112 does not participate in binding of IL-11 to its receptors IL-11R and glycoprotein 130 (gp130). Recombinant IL-11 R112H expressed in E. coli displays a correct four-helix-bundle folding topology, and binds with similar affinity to IL-11R and the IL-11/IL-11R/gp130 complex. 29523682_The results presented here reveal an additional function in classic IL-11 signaling, highlighting the importance of the IL-11R stalk in IL-11 signaling 29533934_The D2 domain of the IL-11R contains two N-linked glycans, which are dispensable for its biological activity, but differentially control maturation and intracellular trafficking of the receptor. 29901200_The results indicate that miR-23b regulates IL-11 and IL-11Ralpha expression, and it might act as an anti-oncogenic agent in the progression of Hepatocellular Carcinoma by directly downregulating IL-11 expression. 29926465_identified six missense mutations in IL11RA, a gene encoding the alpha subunit of interleukin 11 receptor, 4 of them being novel, including 2 in the Ig-like C2-type domain 30811827_IL11RA mutation is associated with craniosynostosis with dental anomalies syndrome. 32277509_Multiple craniosynostosis and facial dysmorphisms with homozygous IL11RA variant caused by maternal uniparental isodisomy of chromosome 9. 32332100_The structure of the extracellular domains of human interleukin 11alpha receptor reveals mechanisms of cytokine engagement. 33566379_Interleukin-11 (IL-11) receptor cleavage by the rhomboid protease RHBDL2 induces IL-11 trans-signaling. 33590875_IL11 is elevated in systemic sclerosis and IL11-dependent ERK signalling underlies TGFbeta-mediated activation of dermal fibroblasts. 34530329_Interleukin-11 receptor expression on monocytes is dispensable for their recruitment and pathogen uptake during Leishmania major infection. 35331937_The outcome of targeted NGS screening in patients with syndromic forms of sagittal and pansynostosis - IL11RA is an emerging core-gene for pansynostosis. | ENSMUSG00000078735+ENSMUSG00000073889+ENSMUSG00000073876 | Il11ra2+Il11ra1+Gm13305 | 5.551887e+02 | 0.8585888 | -0.219960774 | 0.2782926 | 6.196559e-01 | 0.4311751748 | 0.85105150 | No | Yes | 4.245330e+02 | 62.522183 | 5.603670e+02 | 84.349583 |
| ENSG00000137434 | 347744 | C6orf52 | protein_coding | Q5T4I8 | Alternative splicing;Reference proteome | hsa:347744; | 4.797612e+01 | 0.7286657 | -0.456671067 | 0.4764982 | 9.281703e-01 | 0.3353388242 | 0.82570033 | No | Yes | 3.031451e+01 | 6.228992 | 4.336402e+01 | 8.847707 | |||||||
| ENSG00000137494 | 338699 | ANKRD42 | protein_coding | Q8N9B4 | ANK repeat;Alternative splicing;Reference proteome;Repeat | hsa:338699; | Mouse_homologues nucleus [GO:0005634]; NF-kappaB binding [GO:0051059]; positive regulation of cytokine production involved in inflammatory response [GO:1900017]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of NF-kappaB transcription factor activity [GO:0051092] | 17123353_the cloning and characterization of a new protein, termed SARP (several ankyrin repeat protein), which is shown to interact with all isoforms of PP1 by a variety of techniques. | ENSMUSG00000041343 | Ankrd42 | 3.578516e+02 | 0.7203184 | -0.473293294 | 0.2995874 | 2.483723e+00 | 0.1150296606 | 0.75783482 | No | Yes | 2.546235e+02 | 32.893425 | 3.628236e+02 | 47.385712 | |||
| ENSG00000137497 | 4926 | NUMA1 | protein_coding | Q14980 | FUNCTION: Microtubule (MT)-binding protein that plays a role in the formation and maintenance of the spindle poles and the alignement and the segregation of chromosomes during mitotic cell division (PubMed:7769006, PubMed:17172455, PubMed:19255246, PubMed:24996901, PubMed:26195665, PubMed:27462074). Functions to tether the minus ends of MTs at the spindle poles, which is critical for the establishment and maintenance of the spindle poles (PubMed:12445386, PubMed:11956313). Plays a role in the establishment of the mitotic spindle orientation during metaphase and elongation during anaphase in a dynein-dynactin-dependent manner (PubMed:23870127, PubMed:24109598, PubMed:24996901, PubMed:26765568). In metaphase, part of a ternary complex composed of GPSM2 and G(i) alpha proteins, that regulates the recruitment and anchorage of the dynein-dynactin complex in the mitotic cell cortex regions situated above the two spindle poles, and hence regulates the correct oritentation of the mitotic spindle (PubMed:23027904, PubMed:22327364, PubMed:23921553). During anaphase, mediates the recruitment and accumulation of the dynein-dynactin complex at the cell membrane of the polar cortical region through direct association with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), and hence participates in the regulation of the spindle elongation and chromosome segregation (PubMed:22327364, PubMed:23921553, PubMed:24996901, PubMed:24371089). Binds also to other polyanionic phosphoinositides, such as phosphatidylinositol 3-phosphate (PIP), lysophosphatidic acid (LPA) and phosphatidylinositol triphosphate (PIP3), in vitro (PubMed:24996901, PubMed:24371089). Also required for proper orientation of the mitotic spindle during asymmetric cell divisions (PubMed:21816348). Plays a role in mitotic MT aster assembly (PubMed:11163243, PubMed:11229403, PubMed:12445386). Involved in anastral spindle assembly (PubMed:25657325). Positively regulates TNKS protein localization to spindle poles in mitosis (PubMed:16076287). Highly abundant component of the nuclear matrix where it may serve a non-mitotic structural role, occupies the majority of the nuclear volume (PubMed:10075938). Required for epidermal differentiation and hair follicle morphogenesis (By similarity). {ECO:0000250|UniProtKB:E9Q7G0, ECO:0000269|PubMed:11163243, ECO:0000269|PubMed:11229403, ECO:0000269|PubMed:11956313, ECO:0000269|PubMed:12445386, ECO:0000269|PubMed:16076287, ECO:0000269|PubMed:17172455, ECO:0000269|PubMed:19255246, ECO:0000269|PubMed:22327364, ECO:0000269|PubMed:23027904, ECO:0000269|PubMed:23870127, ECO:0000269|PubMed:23921553, ECO:0000269|PubMed:24109598, ECO:0000269|PubMed:24371089, ECO:0000269|PubMed:24996901, ECO:0000269|PubMed:25657325, ECO:0000269|PubMed:26195665, ECO:0000269|PubMed:26765568, ECO:0000269|PubMed:27462074, ECO:0000269|PubMed:7769006, ECO:0000305|PubMed:10075938, ECO:0000305|PubMed:21816348}. | 3D-structure;ADP-ribosylation;Acetylation;Alternative splicing;Cell cycle;Cell division;Cell membrane;Chromosome;Chromosome partition;Coiled coil;Cytoplasm;Cytoskeleton;Glycoprotein;Isopeptide bond;Lipid-binding;Lipoprotein;Membrane;Microtubule;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes a large protein that forms a structural component of the nuclear matrix. The encoded protein interacts with microtubules and plays a role in the formation and organization of the mitotic spindle during cell division. Chromosomal translocation of this gene with the RARA (retinoic acid receptor, alpha) gene on chromosome 17 have been detected in patients with acute promyelocytic leukemia. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2013]. | hsa:4926; | cell cortex [GO:0005938]; cell cortex region [GO:0099738]; centrosome [GO:0005813]; chromosome [GO:0005694]; cortical microtubule [GO:0055028]; cytoplasmic microtubule bundle [GO:1905720]; cytosol [GO:0005829]; dendrite [GO:0030425]; extracellular exosome [GO:0070062]; extrinsic component of plasma membrane [GO:0019897]; Golgi membrane [GO:0000139]; lateral cell cortex [GO:0097575]; lateral plasma membrane [GO:0016328]; microtubule bundle [GO:0097427]; microtubule minus-end [GO:0036449]; microtubule plus-end [GO:0035371]; mitotic spindle [GO:0072686]; mitotic spindle astral microtubule [GO:0061673]; mitotic spindle midzone [GO:1990023]; mitotic spindle pole [GO:0097431]; neuronal cell body [GO:0043025]; nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; spindle [GO:0005819]; spindle microtubule [GO:0005876]; spindle pole [GO:0000922]; spindle pole centrosome [GO:0031616]; disordered domain specific binding [GO:0097718]; dynein complex binding [GO:0070840]; microtubule binding [GO:0008017]; microtubule minus-end binding [GO:0051011]; microtubule plus-end binding [GO:0051010]; phosphatidylinositol binding [GO:0035091]; protein C-terminus binding [GO:0008022]; protein domain specific binding [GO:0019904]; protein-containing complex binding [GO:0044877]; structural molecule activity [GO:0005198]; tubulin binding [GO:0015631]; anastral spindle assembly [GO:0055048]; astral microtubule organization [GO:0030953]; cell division [GO:0051301]; chromosome segregation [GO:0007059]; establishment of mitotic spindle orientation [GO:0000132]; meiotic cell cycle [GO:0051321]; microtubule bundle formation [GO:0001578]; nucleus organization [GO:0006997]; positive regulation of BMP signaling pathway [GO:0030513]; positive regulation of chromosome segregation [GO:0051984]; positive regulation of chromosome separation [GO:1905820]; positive regulation of hair follicle development [GO:0051798]; positive regulation of intracellular transport [GO:0032388]; positive regulation of keratinocyte differentiation [GO:0045618]; positive regulation of microtubule polymerization [GO:0031116]; positive regulation of mitotic spindle elongation [GO:1902846]; positive regulation of protein localization to cell cortex [GO:1904778]; positive regulation of protein localization to spindle pole body [GO:1902365]; positive regulation of spindle assembly [GO:1905832]; regulation of metaphase plate congression [GO:0090235]; regulation of mitotic spindle organization [GO:0060236] | 11956313_A domain within the C-terminal tail of NuMA interacts with tubulin and induces bundling and stabilisation of microtubules and leads to formation of abnormal mitotic spindles. 12508117_NuMA is cleaved differently in Jurkat T and HeLa cells, suggesting that different sets of caspases are activated in these cell lines. The normal diffuse intranuclear distribution of NuMA changed during apoptosis. 14737102_role in development of myelodi leukemia with promyelocytic features 15388855_Proteins and open reading frames with a NuMA C terminus distal portion like region were found in a diverse set of vertebrate species including mammals, birds, amphibia, and early teleost fish. 15561764_Multiple mechanisms regulate NUMA1 dynamics at spindle poles. 15684076_concluded that variations in the NuMA gene are likely responsible for the observed increased breast cancer risk 16146802_NuMA plays diverse important roles in vertebrate cells [review] 17108325_NuMA has a role in mammary epithelial differentiation by influencing the organization of chromatin. 17172455_point to the Rae1-NuMA interaction as a critical element for normal spindle formation in mitosis 17293864_Observational study of gene-disease association. (HuGE Navigator) 17401638_NuMA is a structural element in maintaining nuclear integrity. 17609108_critical spindle pole-associated mechanism, called the END (Emi1/NuMA/dynein-dynactin) network, that spatially restricts APC/C activity in early mitosis 18331640_Observational study of gene-disease association. (HuGE Navigator) 18331640_The entire coding region and exon-intron boundaries of NuMa were screened in 92 familial breast cancer patients & the results do not support the role of NuMA variants as breast cancer susceptibility alleles. 18417561_Kaposi's sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation. 19124506_Observational study of gene-disease association. (HuGE Navigator) 19522705_Inhibiting Crm1 in early metaphase causes the formation of excess acentriolar spindle poles containing NuMA and B23, but does not affect centrosome numbers. 19615282_The levels of urinary NMP22 and CK18 in the patients with transitional cell carcinoma of the bladder were significantly higher than those in the non-transitional cell carcinoma of the bladder. 19759176_Data suggest that pADPr provides a dynamic cross-linking function at spindle poles by extending from covalent modification sites on PARP-5a and NuMA and binding noncovalently to NuMA and that this function helps promote assembly of exactly two poles. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20467816_These results suggest that NuMA may provide structural support in the interphase nucleus by contributing to the organization of chromatin. 20479129_Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. 20508983_Observational study of gene-disease association. (HuGE Navigator) 21255834_Phenotype onset is correlated with NuMA-RARalpha copy number; mice with higher copy number developing disease later than those with lower copy number. 21297155_Accurate distribution of NuMA is important for oocyte maturation, zygote and embryo development in humans. Proper assembly of NuMA is likely necessary for bipolar spindle organization and human oocyte developmental competence. 21406448_NuMA is expressed in interphase nuclei of fibroblasts and oocytes. 21865670_nuclear matrix protein 22 (nuclear mitotic apparatus protein, NuMA) has a role in upper tract urothelial tumors. 22023725_During apoptotic rearrangement of interchromatin granule clusters, the nuclear matrix (NuMa rearrangement) and chromatin are closely associated. This process occurs in defined stages and depends on the activity of protein phosphatases, caspases and CAD. 22552228_Without functional NuMA, microtubules lose connection to meiosis I spindle poles, resulting in highly disorganized early spindle assembly. 22619067_Low NUMA1 is associtated with glioblastoma. 22719996_NuMA expression was upregulated in tumours, with a significant association with disease stage in mucinous EOC subtypes, lymph node involvement and patient age 22977735_Studies indicate that the Inscuteable (Insc)and NuMA are mutually exclusive interactors of LGN. 23097092_Phosphorylation of NuMA by aurora-A is important for cell survival. 23368718_Numa regulates spindle assemby in conjunction with Eg5. 23389635_Data indicate that dynein- and astral microtubule-mediated transport of Galphai/LGN/nuclear mitotic apparatus (NuMA) complex from cell cortex to spindle poles. 23589328_NuMA is required for the recruitment of cyclin-dependent kinase 8, a component of the Mediator complex and a promoter of p53-mediated p21 gene function. 23707952_ectopic expression of BRAP2 inhibits nuclear localization of HMG20A and NuMA1, and prevents nuclear envelope accumulation of SYNE2. 23828576_ectopic expression of NuMA can manipulate endogenous p53 and p21 transcriptional expression during interphase. 23921553_NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. 24165937_Hepatocyte Par1b defines lumen position in concert with the position of the astral microtubule anchoring complex LGN-NuMA to yield the distinct epithelial division phenotypes. 24309115_The mitosis-dependent dynamic SUMO-1 modification of NuMA might contribute to NuMA-mediated formation and maintenance of mitotic spindle poles during mitosis. 24350565_Retinoblastoma protein (pRB) have a novel function in regulating the mitotic function of NuMA and spindle organization, which are required for proper cell cycle progression. 24725408_Study finds that frictional forces increase nonlinearly with microtubule-associated proteins (MAP) velocity across microtubules and depend on filament polarity, with NuMA's friction being lower when moving toward minus ends, EB1's lower toward plus ends, and PRC1's exhibiting no directional preference. 24976592_Letter: risk factors for false positive results when using urinary NMP22 as biomarker for early detection of bladder cancer. 24996901_NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane.During anaphase correct NuMA localization is mediated by direct membrane phospholipid binding. 25451259_Seven NuMA isoforms generated by alternative splicing were categorized into 3 groups: long, middle and short. Both exons 15 and 16 in long NuMA were 'hotspot' for alternative splicing. Lower expression of short NuMA was observed in cancer cells compared with nonneoplastic controls. 25488052_Results show that at low grade of disease, NMP22 test provided a significantly higher sensitivity for the detection of recurrent urothelial carcinoma of the bladder compared to voided urine cytology specimens. 26766442_Suppressor APC domain containing 2 negatively regulates the localization of LGN at the cell cortex, likely by competing with NuMA for its binding 26832443_Aurora-A governs the dynamic exchange between the cytoplasmic and the spindle pole-localized pools of NuMA. Aurora-A phosphorylates directly the C terminus of NuMA on three Ser residues, of which Ser1969 determines the dynamic behavior and the spindle orientation functions of NuMA. 27462074_findings reveal a direct physical link between two important regulators of mitotic progression and demonstrate the critical role of the NuMA-Astrin interaction for accurate cell division. 27829174_Chimeric proteins constructed by fusion of LANA of Kaposi's sarcoma-associated herpesvirus with the NuMA could bind with ori-P and enhance replication of an ori-P-containing plasmid. 28045117_The results show how E-cadherin instructs the assembly of the LGN/NuMA complex at cell-cell contacts, and define a mechanism that couples cell division orientation to intercellular adhesion. 28209915_Low post translational modifications of NuMA protein is associated with neoplasms. 28469279_It has been reported that the Galectin-3/NuMA interaction is functionally important for the spindle pole organization; spindle pole cohesion requires glycosylation-mediated localization of NuMA. 28748856_Short isoform of NuMA might be functioned as a putative role of tumor suppressor. Further studies should be made to illuminate the relationship between ACTN4, MYBL2, and tumor progression. 28824318_All urine samples were analyzed by voided urine and bladder washing cytology, NMP22 and UBC rapid test (qualitatively and quantitatively). The best cutoff (highest Youden index; >/=6.7 ng/ml) for the quantitative UBC was determined by receiver operating characteristic curves. 28939615_Importin-alpha/-beta regulates the NuMA functioning required for assembly of higher-order microtubule structures including the mitotic spindle. 28981686_The function of nuclear mitotic apparatus protein (NuMA) in rDNA transcription and p53-independent nucleolar stress response suggests a central role for NuMA in cellular homeostasis. 29185983_Here, the authors use quantitative imaging and laser ablation to show that NuMA targets dynactin to spindle microtubule minus-ends, localizing dynein activity there. 29222185_The p37 negatively regulates this function of PP1, resulting in lower cortical NuMA levels and correct spindle orientation. 29669740_MISP directly interacts with ezrin and that SLK/LOK-activated ezrin ensures appropriate cortical MISP levels in mitosis by competing with MISP for actin-binding sites at the cell cortex. 30572664_Knockdown of the NS1-interacting nuclear mitotic apparatus protein 1 (NUMA1) significantly reduced infectious virus yield, suggesting NUMA1 plays important roles in Influenza A virus maturation. 30812030_A mechanism involving the sequestration of 53BP1 by NuMA in the absence of DNA damage. Such a mechanism may have evolved to disable repair functions and may be a decisive factor for tumor responses to genotoxic treatments. 31101817_Study reports the crystal structure of NuMA:LGN hetero-hexamers, and unveil their role in promoting the assembly of active cortical dynein/dynactin motors that are required in orchestrating oriented divisions in polarized cells. 31290065_The authors show an interaction of Human papillomaviruses 5 and 8 oncoproteins E6 and E7 with the nuclear mitotic apparatus protein 1 (NuMA). 31312011_IFT88 controls NuMA enrichment at k-fibers minus-ends to facilitate their re-anchoring into mitotic spindles. 31727776_NuMA1 promotes axon initial segment assembly through inhibition of endocytosis. 31732560_LGN has a role in cell cortex recruitment of NuMA 31782546_NuMA promoted the initial step of spindle bipolarization in early mitosis 32343689_Study identified new genes that rescue cell death induced by BMI1 inhibitors and selected NUMA1 for follow-up studies. Results unveiled a novel mitotic mechanism underlying BMI1-associated lethality using CRISPR-Cas9 derived knockouts of NUMA1 in both HAP1 cells and non-small lung cancer cell lines and highlighted a mechanism relying on mitotic arrest upon loss of BMI1 and a new genetic resistance mechanism. 32845810_NuMA interaction with chromatin is vital for proper chromosome decondensation at the mitotic exit. 33044554_The mitotic protein NuMA plays a spindle-independent role in nuclear formation and mechanics. 33186548_Ran-GTP Is Non-essential to Activate NuMA for Mitotic Spindle-Pole Focusing but Dynamically Polarizes HURP Near Chromosomes. 33397061_Tissue Nuclear Matrix Protein Expression 22 in Various Grades and Stages of Bladder Cancer. 34165167_Nuclear IGF1R interacts with NuMA and regulates 53BP1dependent DNA doublestrand break repair in colorectal cancer. 34887424_NuMA regulates mitotic spindle assembly, structural dynamics and function via phase separation. 35143306_Mechanism of spindle pole organization and instability in human oocytes. | ENSMUSG00000066306 | Numa1 | 1.045958e+04 | 1.1852316 | 0.245169042 | 0.2802863 | 7.692617e-01 | 0.3804455462 | 0.84161313 | No | Yes | 1.099042e+04 | 842.378374 | 8.767659e+03 | 689.713496 | |
| ENSG00000137992 | 1629 | DBT | protein_coding | P11182 | FUNCTION: The branched-chain alpha-keto dehydrogenase complex catalyzes the overall conversion of alpha-keto acids to acyl-CoA and CO(2). It contains multiple copies of three enzymatic components: branched-chain alpha-keto acid decarboxylase (E1), lipoamide acyltransferase (E2) and lipoamide dehydrogenase (E3). Within this complex, the catalytic function of this enzyme is to accept, and to transfer to coenzyme A, acyl groups that are generated by the branched-chain alpha-keto acid decarboxylase component. | 3D-structure;Acetylation;Acyltransferase;Direct protein sequencing;Disease variant;Lipoyl;Maple syrup urine disease;Mitochondrion;Phosphoprotein;Reference proteome;Transferase;Transit peptide | The branched-chain alpha-keto acid dehydrogenase complex (BCKD) is an inner-mitochondrial enzyme complex involved in the breakdown of the branched-chain amino acids isoleucine, leucine, and valine. The BCKD complex is thought to be composed of a core of 24 transacylase (E2) subunits, and associated decarboxylase (E1), dehydrogenase (E3), and regulatory subunits. This gene encodes the transacylase (E2) subunit. Mutations in this gene result in maple syrup urine disease, type 2. Alternatively spliced transcript variants have been described, but their biological validity has not been determined. [provided by RefSeq, Jul 2008]. | hsa:1629; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; microtubule cytoskeleton [GO:0015630]; mitochondrial alpha-ketoglutarate dehydrogenase complex [GO:0005947]; mitochondrial matrix [GO:0005759]; mitochondrial nucleoid [GO:0042645]; mitochondrion [GO:0005739]; acetyltransferase activity [GO:0016407]; dihydrolipoyllysine-residue (2-methylpropanoyl)transferase activity [GO:0043754]; lipoic acid binding [GO:0031405]; ubiquitin protein ligase binding [GO:0031625]; branched-chain amino acid catabolic process [GO:0009083] | 11448970_two novel type IB MSUD mutations in Israeli patients, which affect the E1beta subunit in the decarboxylase (E1) component of the branched-chain alpha-ketoacid dehydrogenase complex 11509994_Mutation in DBT causes a subset of maple syrup urine disease in Ashkenazi Jewish population. 14768949_a distinct subset of antimitochondrial antibodies recognize sequences on branched-chain acyltransferase which located outside of the lipoyl binding domain, in primary biliary cirrhosis and overlap syndrome with autoimmune hepatitis 16861235_presence of the interdomain linker restricts the motional freedom of the hbSBD more significantly than hbLBD, and that the linker region likely exists as a soft rod rather than a flexible string in solution. 17922217_in our cohort more severe enzyme & clinical phenotypes of variant maple syrup urine disease were mainly associated with specific genotypes in BCKDHA gene; milder enzyme & clinical phenotypes were associated with specific genotypes in BCKDHB & DBT genes 18378174_30 Maple syrup urine disease Portuguese patients studied; 17 putative mutations have been identified (6 in BCKDHA, 5 in BCKDHB and 6 in DBT); 7 of are described for the first time. 18533943_Examination of the deletion mutation in the E2 (DBT) gene facilitated early MSUD diagnosis and was beneficial for the determination of the proper course of treatment. 19480318_In 37% (12 patients) of a total of 64 alleles, the supposed maple syrup urine disease-causing mutations in Turkish patients were located in the BCKDHA gene, in 44% (14 patients) in the BCKDHB gene and in 19% (6 patients) in the DBT gene. 20570198_4 novel mutations in DBT gene resulting in intermittent maple syrup urine disease in 7 Norwegian patients; pathogenic effect of the mutations is depletion of cellular protein; intermittent form of MSUD appears to be due to residual R301C mutant protein 20877624_Observational study of gene-disease association. (HuGE Navigator) 23313820_Deletion in DBT gene is associated with maple syrup urine disease. 24268812_The novel DBT mutation c.650-651insT was more prevalent than the deleted 4.7-kb heterozygote in the Amis population. The reported 4.7-kb deletion indicating a possible founder mutation may be preserved. 34883003_Three novel mutations of the BCKDHA, BCKDHB and DBT genes in Chinese children with maple syrup urine disease. | ENSMUSG00000000340 | Dbt | 3.507282e+02 | 0.8971810 | -0.156529070 | 0.3516339 | 1.887562e-01 | 0.6639538309 | 0.92913032 | No | Yes | 2.227773e+02 | 40.364056 | 2.729416e+02 | 50.300030 | |
| ENSG00000138138 | 84896 | ATAD1 | protein_coding | Q8NBU5 | FUNCTION: Outer mitochondrial translocase required to remove mislocalized tail-anchored transmembrane proteins on mitochondria (PubMed:24843043). Specifically recognizes and binds tail-anchored transmembrane proteins: acts as a dislocase that mediates the ATP-dependent extraction of mistargeted tail-anchored transmembrane proteins from the mitochondrion outer membrane (By similarity). Also plays a critical role in regulating the surface expression of AMPA receptors (AMPAR), thereby regulating synaptic plasticity and learning and memory (By similarity). Required for NMDA-stimulated AMPAR internalization and inhibition of GRIA1 and GRIA2 recycling back to the plasma membrane; these activities are ATPase-dependent (By similarity). {ECO:0000250|UniProtKB:P28737, ECO:0000250|UniProtKB:Q9D5T0, ECO:0000269|PubMed:24843043}. | ATP-binding;Alternative splicing;Cell junction;Cell membrane;Disease variant;Membrane;Mitochondrion;Mitochondrion outer membrane;Nucleotide-binding;Peroxisome;Phosphoprotein;Postsynaptic cell membrane;Reference proteome;Synapse;Translocase;Transmembrane;Transmembrane helix | hsa:84896; | cytosol [GO:0005829]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; mitochondrial outer membrane [GO:0005741]; peroxisomal membrane [GO:0005778]; postsynaptic membrane [GO:0045211]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; transmembrane protein dislocase activity [GO:0140567]; extraction of mislocalized protein from mitochondrial outer membrane [GO:0140570]; learning [GO:0007612]; memory [GO:0007613]; negative regulation of synaptic transmission, glutamatergic [GO:0051967]; positive regulation of receptor internalization [GO:0002092] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 29390050_This study showed that the molecular and functional analyses identify an activating ATAD1 mutation as a new cause of severe encephalopathy and congenital stiffness. 31394253_Sorting out how Msp1 maintains mitochondrial membrane proteostasis. 34254279_CircRNA circ-ATAD1 Is Upregulated in Cervical Squamous Cell Carcinoma and Regulates Cell Proliferation and Apoptosis by Suppressing the Maturation of miR-218. 34857012_Circ-ATAD1 is overexpressed in osteosarcoma (OS) and suppresses the maturation of miR-154-5p to increase cell invasion and migration. | ENSMUSG00000013662 | Atad1 | 6.609156e+02 | 0.8597593 | -0.217995235 | 0.3928264 | 3.006598e-01 | 0.5834690743 | 0.90371250 | No | Yes | 5.401765e+02 | 115.022482 | 6.289902e+02 | 137.153575 | ||
| ENSG00000138162 | 10579 | TACC2 | protein_coding | O95359 | FUNCTION: Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors (By similarity). May play a role in organizing centrosomal microtubules. May act as a tumor suppressor protein. May represent a tumor progression marker. {ECO:0000250, ECO:0000269|PubMed:10749935}. | Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Nucleus;Phosphoprotein;Reference proteome | Transforming acidic coiled-coil proteins are a conserved family of centrosome- and microtubule-interacting proteins that are implicated in cancer. This gene encodes a protein that concentrates at centrosomes throughout the cell cycle. This gene lies within a chromosomal region associated with tumorigenesis. Expression of this gene is induced by erythropoietin and is thought to affect the progression of breast tumors. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:10579; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; microtubule organizing center [GO:0005815]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; nuclear receptor binding [GO:0016922]; cell population proliferation [GO:0008283]; cerebral cortex development [GO:0021987]; microtubule cytoskeleton organization [GO:0000226]; mitotic spindle organization [GO:0007052] | 12620397_Defects in TACC2 expression may affect gene regulation, thus contributing to the pathogenesis of some tumors. 14767476_GCN5L2 is a TACC2-binding protein. 15304323_Centrosomal TACC2 is required for mitotic spindle maintenance. 16385451_Observational study of gene-disease association. (HuGE Navigator) 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19671663_Simian virus 40 large T antigen interacting with TACC2 is involved in stabilizing microtubules in mitosis. 20335520_TACC2 may mediate an oncogenic effect on breast cancer cells. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20508983_Observational study of gene-disease association. (HuGE Navigator) 22210879_Results revealed 3 genes highly predictive of event-free survival (EFS), beyond age and MLL status: FLT3, IRX2, and TACC2. 22456197_Data suggest that TACC2 is highly expressed in prostate cancer cells and contributes to cell cycle progression and cell proliferation in androgen-nonresponsive prostate cancer cells; overexpression of TACC2 leads to recurrence of prostate cancer. 25884766_Differential expression of the transcripts TACC2 connects ubiquitin-proteasome system with infection-inflammation in preterm births and preterm premature rupture of membranes. 27333920_These results suggest that TACC2 plays an important role in the cell proliferation of breast carcinoma and therefore immunohistochemical TACC2 status is a candidate of worse prognostic factor in breast cancer cases. 29074988_TACC2 protein is expressed mainly in the nucleus of the endometrial cancer cells. 29843208_This finding suggests that TACC2 may be a useful tool as a candidate biomarker to predict the recurrence and prognosis of hepatocellular carcinoma. 31996486_Cigarette smoke exposure enhances transforming acidic coiled-coil-containing protein 2 turnover and thereby promotes emphysema. 33931666_Nanopore sequencing reveals TACC2 locus complexity and diversity of isoforms transcribed from an intronic promoter. | ENSMUSG00000030852 | Tacc2 | 2.564534e+02 | 1.0673494 | 0.094032510 | 0.3190083 | 8.641839e-02 | 0.7687807900 | 0.95414161 | No | Yes | 2.154812e+02 | 30.553368 | 1.984356e+02 | 29.252339 | |
| ENSG00000138246 | 23317 | DNAJC13 | protein_coding | O75165 | FUNCTION: Involved in membrane trafficking through early endosomes, such as the early endosome to recycling endosome transport implicated in the recycling of transferrin and the early endosome to late endosome transport implicated in degradation of EGF and EGFR (PubMed:18256511, PubMed:18307993). Involved in the regulation of endosomal membrane tubulation and regulates th dynamics of SNX1 on the endosomal membrane; via association with WASHC2 may link the WASH complex to the retromer SNX-BAR subcomplex (PubMed:24643499). {ECO:0000269|PubMed:18256511, ECO:0000269|PubMed:18307993, ECO:0000269|PubMed:24643499}. | Acetylation;Chaperone;Disease variant;Endosome;Membrane;Neurodegeneration;Parkinson disease;Parkinsonism;Protein transport;Reference proteome;Transport | This gene encodes a member of the Dnaj protein family whose members act as co-chaperones of a partner heat-shock protein by binding to the latter and stimulating ATP hydrolysis. The encoded protein associates with the heat-shock protein Hsc70 and plays a role in clathrin-mediated endocytosis. It may also be involved in post-endocytic transport mechanisms via its associations with other proteins, including the sorting nexin SNX1. Mutations in this gene are associated with Parkinson's disease. [provided by RefSeq, Jun 2016]. | hsa:23317; | azurophil granule membrane [GO:0035577]; cytosol [GO:0005829]; early endosome membrane [GO:0031901]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; plasma membrane [GO:0005886]; secretory granule membrane [GO:0030667]; endosome organization [GO:0007032]; osteoblast differentiation [GO:0001649]; protein transport [GO:0015031]; receptor-mediated endocytosis [GO:0006898]; regulation of early endosome to late endosome transport [GO:2000641]; regulation of early endosome to recycling endosome transport [GO:1902954] | 16179350_RME-8 functions in intracellular trafficking and provides the first evidence of a functional role for a DnaJ domain-bearing co-chaperone on endosomes 18256511_hRME is primarily involved in membrane trafficking through early endosomes, but not through degradative organelles, such as multivesicular bodies and late endosomes. 18307993_These data implicate RME-8 in sorting decisions influencing EGFR at the level of endosomes and point to RME-8 as a potential regulatory target in ErbB2-positive breast cancers. 22507240_Found DNAJC13 A2057S variant is probably a rare cause of Tourette syndrome/chronic tic phenotype in Chinese Han patients. 24126164_Missense mutations in DNAJC13 does not play a major role in PD in the Chinese population. 24218364_a pathogenic mutation in DNAJC13, a component of the endosomal recycling system, helps to emphasize the role of endosomal recycling rather than endolysosomal protein degradation in the biology of late-onset Parkinson disease. 24643499_Data propose that the interaction between RME-8 and the WASH complex provides a means to coordinate the activity of the WASH complex with the membrane-tubulating function of the sorting. 25118025_DNAJC13 c.2564A>G (p.(N855S)) was identified in two patients with essential tremor. 25186792_PD associated with a DNAJC13 p.N855S parkinsonism mutation presents as late-onset, often slowly progressive, usually dopamine-responsive typical Parkinsonism. 25393719_Although the contribution of rare genetic variation in DNAJC13 to parkinsonisms remains to be further elucidated, this study suggests that, in addition to p.N855S, other rare variants might affect disease susceptibility 25550792_Re-expression of miR-193b in breast cancer cell lines decreased DNAJC13 (HPS40) and RAB22A expression, providing a mechanism by which mir193-b acts as a tumor suppressor. 26134565_These results further highlight the critical role for phosphatidylinositol 3-phosphate in the RME-8-mediated organizational control of various endosomal activities, including retrograde transport. 26278106_Mutations in exon 24 of DNAJC13 are not a common cause of Parkinson or Lewy body disease among Caucasian populations. 27236598_There is a possibility that specific DNAJC13 variants may play a minor role in PD susceptibility. 29309590_Parkinson's disease-linked DNAJC13 mutation perturbs multi-directional endosomal trafficking, resulting in the aberrant endosomal retention of alpha-synuclein, which might predispose to the neurodegenerative process that leads to Parkinson's disease. 29887357_This study showed the DNAJC13 mutation screening in patients with Parkinson's disease. 32322926_Receptor-mediated endocytosis 8 (RME-8)/DNAJC13 is a novel positive modulator of autophagy and stabilizes cellular protein homeostasis. 33239198_Association study of DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1 with Parkinson's disease. | ENSMUSG00000032560 | Dnajc13 | 6.206744e+02 | 0.8123244 | -0.299872160 | 0.3432051 | 7.660516e-01 | 0.3814412988 | 0.84188609 | No | Yes | 6.093277e+02 | 119.588293 | 6.137993e+02 | 123.581830 | |
| ENSG00000138303 | 51008 | ASCC1 | protein_coding | Q8N9N2 | FUNCTION: Plays a role in DNA damage repair as component of the ASCC complex (PubMed:29997253). Part of the ASC-1 complex that enhances NF-kappa-B, SRF and AP1 transactivation (PubMed:12077347). In cells responding to gastrin-activated paracrine signals, it is involved in the induction of SERPINB2 expression by gastrin. May also play a role in the development of neuromuscular junction. {ECO:0000269|PubMed:12077347, ECO:0000269|PubMed:19074642, ECO:0000269|PubMed:26924529, ECO:0000269|PubMed:29997253}. | Alternative splicing;DNA damage;DNA repair;Direct protein sequencing;Neurodegeneration;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene encodes a subunit of the activating signal cointegrator 1 (ASC-1) complex. The ASC-1 complex is a transcriptional coactivator that plays an important role in gene transactivation by multiple transcription factors including activating protein 1 (AP-1), nuclear factor kappa-B (NF-kB) and serum response factor (SRF). The encoded protein contains an N-terminal KH-type RNA-binding motif which is required for AP-1 transactivation by the ASC-1 complex. Mutations in this gene are associated with Barrett esophagus and esophageal adenocarcinoma. Alternatively spliced transcripts encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Dec 2011]. | hsa:51008; | neuromuscular junction [GO:0031594]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; RNA binding [GO:0003723]; DNA repair [GO:0006281]; regulation of transcription, DNA-templated [GO:0006355] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 19074642_Gastrin activates paracrine networks leading to induction of PAI-2 via MAZ and ASC-1. 19680556_Observational study of gene-disease association. (HuGE Navigator) 21791690_Three major genes, MSR1, ASCC1, and CTHRC1 were associated with Barrett esophagus/esophageal adenocarcinoma 26503956_ASCC1 inhibits NF-kappaB activation and a truncated and inactive variant of ASCC1 is associated with a more severe disease, which could have clinical value for assessing the progression and prognosis of Rheumatoid Arthritis. 28218388_Our patient was also found to have a homozygous frameshift variant (c.157dupG, p.Glu53Glyfs*19) in ASCC1 , thereby representing the second known case. This confirms ASCC1 involvement in a severe neuromuscular disease lying within the spinal muscular atrophy or primary muscle disease spectra 29997253_ASCC1 knockout through a CRISPR/Cas9 approach results in alkylation damage sensitivity in a manner epistatic with ASCC3. 30327447_this work expands the ASCC1 mutation spectrum, sheds light on the muscle histology of the disorder and emphasises the physiological importance of the ASC-1 complex in fetal muscle and bone development. 31880396_A new case of SMABF2 diagnosed in stillbirth expands the prenatal presentation and mutational spectrum of ASCC1. 32653958_Association between non-Caucasian-specific ASCC1 gene polymorphism and osteoporosis and obesity in Korean postmenopausal women. 33931933_Biallelic ASCC1 variants including a novel intronic variant result in expanded phenotypic spectrum of spinal muscular atrophy with congenital bone fractures 2 (SMABF2). 34204919_Inherited Defects of the ASC-1 Complex in Congenital Neuromuscular Diseases. | ENSMUSG00000044475 | Ascc1 | 6.467411e+02 | 0.7091757 | -0.495785046 | 0.2999006 | 2.764807e+00 | 0.0963582956 | 0.75783482 | No | Yes | 5.181546e+02 | 90.370442 | 6.148284e+02 | 109.828932 | |
| ENSG00000138363 | 471 | ATIC | protein_coding | P31939 | FUNCTION: Bifunctional enzyme that catalyzes the last two steps of purine biosynthesis (PubMed:11948179, PubMed:14756554). Acts as a transformylase that incorporates a formyl group to the AMP analog AICAR (5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamide) to produce the intermediate formyl-AICAR (FAICAR) (PubMed:9378707, PubMed:11948179, PubMed:10985775). Can use both 10-formyldihydrofolate and 10-formyltetrahydrofolate as the formyl donor in this reaction (PubMed:10985775). Also catalyzes the cyclization of FAICAR to IMP (PubMed:11948179, PubMed:14756554). Is able to convert thio-AICAR to 6-mercaptopurine ribonucleotide, an inhibitor of purine biosynthesis used in the treatment of human leukemias (PubMed:10985775). Promotes insulin receptor/INSR autophosphorylation and is involved in INSR internalization (PubMed:25687571). {ECO:0000269|PubMed:10985775, ECO:0000269|PubMed:11948179, ECO:0000269|PubMed:14756554, ECO:0000269|PubMed:25687571, ECO:0000269|PubMed:9378707}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Epilepsy;Hydrolase;Mental retardation;Multifunctional enzyme;Purine biosynthesis;Reference proteome;Transferase | PATHWAY: Purine metabolism; IMP biosynthesis via de novo pathway; 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide from 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide (10-formyl THF route): step 1/1. {ECO:0000269|PubMed:11948179}.; PATHWAY: Purine metabolism; IMP biosynthesis via de novo pathway; IMP from 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide: step 1/1. {ECO:0000269|PubMed:11948179}. | This gene encodes a bifunctional protein that catalyzes the last two steps of the de novo purine biosynthetic pathway. The N-terminal domain has phosphoribosylaminoimidazolecarboxamide formyltransferase activity, and the C-terminal domain has IMP cyclohydrolase activity. A mutation in this gene results in AICA-ribosiduria. [provided by RefSeq, Sep 2009]. | hsa:471; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; membrane [GO:0016020]; plasma membrane [GO:0005886]; cadherin binding [GO:0045296]; IMP cyclohydrolase activity [GO:0003937]; phosphoribosylaminoimidazolecarboxamide formyltransferase activity [GO:0004643]; protein homodimerization activity [GO:0042803]; 'de novo' AMP biosynthetic process [GO:0044208]; 'de novo' IMP biosynthetic process [GO:0006189]; 'de novo' XMP biosynthetic process [GO:0097294]; animal organ regeneration [GO:0031100]; brainstem development [GO:0003360]; cellular response to interleukin-7 [GO:0098761]; cerebellum development [GO:0021549]; cerebral cortex development [GO:0021987]; dihydrofolate metabolic process [GO:0046452]; GMP biosynthetic process [GO:0006177]; nucleobase-containing compound metabolic process [GO:0006139]; nucleoside metabolic process [GO:0009116]; response to inorganic substance [GO:0010035]; tetrahydrofolate biosynthetic process [GO:0046654] | 11948179_The kinetic mechanism of the human bifunctional enzyme ATIC 14966129_crystal structure of ATIC 15114530_Deficiency in AICAR transformylase is associated with severe neurological defects and congenital blindness 15457444_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15457444_Polymorphisms of reduced folate carrier,aminoimidazole carboxamide ribonucleotide transformylase,and thymidylate synthase genes contribute to the therapeutic response in rheumatoid arthritis patients to methotrexate. 15677700_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16447238_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16947783_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17009228_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17181924_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17410198_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17439323_Observational study of gene-disease association. (HuGE Navigator) 17530705_Observational study of gene-disease association, pharmacogenomic / toxicogenomic, and genetic testing. (HuGE Navigator) 17617058_AS160 is a common target of insulin, IGF-1, EGF, PMA and AICAR, these stimuli induce distinctive patterns of phosphorylation and 14-3-3 binding, mediated by at least four protein kinases. 18845790_ATIC associated with nucleophosmin-ALK, and its phosphorylation required ALK activity. ALK-mediated ATIC phosphorylation enhanced its enzymatic activity. 19016697_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19193698_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19858780_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19902562_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19936946_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 22180458_Results proved in cultured skin fibroblasts from patients with AICA-ribosiduria that various mutations of ATIC destabilize to various degrees purinosome assembly. 24967362_study suggests that MTHFR C677T and ATIC T675C genotyping combined with clinicopathological data may help to identify patients whom will not benefit from MTX treatment and, therefore, assist clinicians in personalizing RA treatment 25084201_MTHFR, DHFR and ATIC genetic variants can be considered as pharmacogenetic markers of outcome in RA patients under MTX monotherapy. 25240429_genotyping of ATIC rs2372536 and ITPA rs1127354 variants or measuring ITPA activity could be useful to predict methotrexate response in children with juvenile idiopathic arthritis. 25425682_Single nucleotide polymorphisms in ATIC gene is associated with acute graft-versus-host disease. 25687571_Our data show that ATIC is in complex with insulin receptor (IR)and siRNA-mediated partial knockdown of ATIC in HEK293 cells, decreases IR tyrosine phosphorylation and regulates IR endocytosis. Insulin stimulation and ATIC knockdown readily increase level of AMPK-Thr172 phosphorylation in IR complexes.ATIC depletion delayed insulin response of Glut2 translocation in HEK293 cells and decreased AKT-Ser473 phosphorylation. 25687571_PTPLAD1 and AMPK are rapidly compartmentalized within the plasma membrane (PM) and Golgi/endosome fractions after insulin stimulation and that ATIC later accumulates in the Golgi/endosome fraction. 25823786_This study shows that polymorphisms on genes related to the metabolic pathway of pemetrexed, especially, ATIC and GGH genes, would have a therapeutic implication in pemetrexed-treated patients with lung adenocarcinoma 26799664_ATIC 347C>G gene polymorphism may be associated with the development of MTX induced gastrointestinal adverse events. 27379764_The ATIC 347 C/G polymorphism may be associated with non-responsiveness to and or toxicity of methotrexate in Caucasian rheumatoid arthritis patients. 28267080_Pediatric Osteosarcoma patients with ATIC 347C>G exhibited a good histologic response to chemotherapy 29042184_The AICARFT site is capable of independently binding both nucleotide and folate substrates with high affinity however no evidence for positive cooperativity in binding could be detected using the model ligands employed in this study. 29246230_ATIC acts as an oncogenic gene that promotes survival, proliferation and migration by targeting AMPK-mTOR-S6 K1 signaling 32557644_AICA-ribosiduria due to ATIC deficiency: Delineation of the phenotype with three novel cases, and long-term update on the first case. 33226193_Upregulation of ATIC in multiple myeloma tissues based on tissue microarray and gene microarrays. 33778066_Identification and Validation of a Prognostic Model Based on Three Autophagy-Related Genes in Hepatocellular Carcinoma. 33780152_Polymorphism of genes involved in methotrexate pathway: Predictors of response to methotrexate therapy in Indian rheumatoid arthritis patients. 35205374_Impact of Variants in the ATIC and ARID5B Genes on Therapeutic Failure with Imatinib in Patients with Chronic Myeloid Leukemia. | ENSMUSG00000026192 | Atic | 6.821111e+03 | 0.7838512 | -0.351348367 | 0.2648579 | 1.777101e+00 | 0.1825056785 | 0.77830220 | No | Yes | 6.739917e+03 | 930.821393 | 6.943380e+03 | 983.440288 |
| ENSG00000138386 | 4664 | NAB1 | protein_coding | Q13506 | FUNCTION: Acts as a transcriptional repressor for zinc finger transcription factors EGR1 and EGR2. {ECO:0000250}. | 3D-structure;Alternative splicing;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | hsa:4664; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription coregulator activity [GO:0003712]; endochondral ossification [GO:0001958]; myelination [GO:0042552]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of epidermis development [GO:0045682]; regulation of transcription, DNA-templated [GO:0006355]; Schwann cell differentiation [GO:0014037] | 12030330_mutations in the human NABI gene are most likely not involved in the pathogenesis of peripheral neuropathies. 17853270_Methylation pattern of the corresponding NGFI-A binding site in the human glucocorticoid receptor exon 1-F specific promoter in post mortem hippocampal tissue. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21239760_Data show that the suppressive effect of valproic acid on chondrocytes is not due to reduced expression or recruitment of Egr-1 to the mPGES-1 promoter and involves upregulation of NAB1. 23935197_These data suggest that type I IFN stimulation induces a rapid recruitment of a repressive Egr3/Nab1 complex that silences transcription from the ifngr1 promoter. | ENSMUSG00000002881 | Nab1 | 3.034323e+02 | 0.6961136 | -0.522605245 | 0.3787900 | 1.904616e+00 | 0.1675625120 | 0.77611497 | No | Yes | 1.936183e+02 | 40.120389 | 2.897994e+02 | 61.157991 | ||
| ENSG00000138434 | 6744 | ITPRID2 | protein_coding | P28290 | Actin-binding;Alternative splicing;Coiled coil;Cytoplasm;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:6744; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; actin filament binding [GO:0051015]; signaling receptor binding [GO:0005102] | 14673706_localized as a membrane-bound form with extracellular regions; results suggested KRAP might be involved in the regulation of filamentous actin and signals from the outside of the cells 17934691_results suggested that KRAP might be a cytoskeleton-associated protein involving the structural integrity and/or signal transductions in human cancers. 21457704_KRAP is involved in the proper regulation of IP3R-mediated Ca2+ release. 21501587_the critical region of KRAP protein for the regulation of IP(3)R was determined. 21873152_the predicted coiled-coil region & the region adjacent to the coiled-coil region of the carboxyl-terminus of KRAP may be crucial for its interaction with the cytoskeleton or directional targeting toward the apical pole in polarized epithelial cells 26947549_phosphorylation at Ser92 of the sperm-specific antigen 2 (SSFA2)[phospho-SSFA2 (pS92)], was related to poor prognosis. 30712887_These result reveals us that SSFA2 may act as oncogene to promote the progression of glioma 34301929_KRAP tethers IP3 receptors to actin and licenses them to evoke cytosolic Ca(2+) signals. 34787049_Circular RNA UBR1 promotes the proliferation, migration, and invasion but represses apoptosis of lung cancer cells via modulating microRNA-545-5p/SSFA2 axis. | ENSMUSG00000027007 | Itprid2 | 3.934532e+02 | 0.9098380 | -0.136318399 | 0.3784042 | 1.293483e-01 | 0.7191087466 | 0.94084174 | No | Yes | 3.491153e+02 | 70.801810 | 3.615513e+02 | 75.093748 | |||
| ENSG00000138604 | 26035 | GLCE | protein_coding | O94923 | FUNCTION: Converts D-glucuronic acid residues adjacent to N-sulfate sugar residues to L-iduronic acid residues, both in maturing heparan sulfate (HS) and heparin chains. This is important for further modifications that determine the specificity of interactions between these glycosaminoglycans and proteins. {ECO:0000269|PubMed:20118238, ECO:0000269|PubMed:22528493, ECO:0000269|PubMed:30872481}. | 3D-structure;Calcium;Glycoprotein;Golgi apparatus;Isomerase;Membrane;Metal-binding;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | PATHWAY: Glycan metabolism; heparan sulfate biosynthesis. {ECO:0000305|PubMed:22528493, ECO:0000305|PubMed:30872481}.; PATHWAY: Glycan metabolism; heparin biosynthesis. {ECO:0000305}. | hsa:26035; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; calcium ion binding [GO:0005509]; heparosan-N-sulfate-glucuronate 5-epimerase activity [GO:0047464]; protein homodimerization activity [GO:0042803]; racemase and epimerase activity, acting on carbohydrates and derivatives [GO:0016857]; heparan sulfate proteoglycan biosynthetic process [GO:0015012]; heparin biosynthetic process [GO:0030210] | 15853773_The regulation of GLCE expression by 2 cis-acting elements of the beta-catenin-TCF4 complex located in the enhancer region of the promoter are reported. 17985344_in 82-84% of human breast tumors there is either downregulation or loss of D-glucuronyl C5-epimerase mRNA expression and significant decrease of the protein content 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21488854_SNPs in GLCE are associated with triglyceride and HDL-C levels in Turks, and mouse studies support a role for glce in lipid metabolism. 21654676_Loss of D-glucuronyl C-5 epimerase is associated with small-cell lung cancer. 22528493_The biphasic mode of C(5)-epi offers a novel mechanism to regulate the biosynthesis of HS with the desired biological functions. 22805760_A correlation was observed between D-glucuronyl C5-epimerase (GLCE), TCF4 and beta-catenin expression in breast cancer cells and primary tumors, suggesting an important role for TCF4/beta-catenin in regulating GLCE expression both in vitro and in vivo. 22830596_Chondroitin-glucuronate C5-epimerase is a potential candidate for tumour antigen with immunogenicity and the peptides derived from this antigen could be useful in hepatocellular carcinoma immunotherapy. 22968430_positive correlation between miRNA-218 and GLCE mRNA, and negative correlation between miRNA-218 and GLCE protein levels in breast tissues and primary tumors in vivo, supporting a direct involvement of miRNA-218 in posttranscriptional regulation of GLCE 24264315_activation of angiogenesis as a main molecular mechanism of pro-oncogenic effect of GLCE in prostate cancer. 24403231_GLCE may be used as a potential model to study the functional role of intratumor cell heterogeneity in prostate cancer progression. 25594747_C5-epimerase and 2-O-sulfotransferase in association generate extended domains of consecutive GlcNS-IdoA2S Sequence. 27511124_Results show that overexpression of Hsepi alone resulted in an unexpected increase in heparan sulfate (HS) chain length. A Hsepi point-mutant (Y168A), devoid of catalytic activity, failed to affect chain length. Moreover, the effect of Hsepi overexpression on HS chain length was abolished by simultaneous overexpression of 2OST. 27699767_The GLCE gene polymorphism rs3865014 appears to have biological relevance in human pathophysiology. 28734894_The obtained data suggest an involvement of GLCE rs3865014 in breast cancer development. Heterozygous AG genotype might be a risk factor for breast cancer susceptibility in Siberian women and is associated with aggressive ER-negative and triple-negative cancer subtypes. 30872481_Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase. 31834878_Results found the expression of GLCE is significantly decreased in the brains of frontotemporal lobar degeneration cases relative to normal controls, demonstrating the potential disease relevance of the candidate gene identified. Furthermore, knockdown of GLCE in cultured human cells protects against oxidative stress induced pTDP accumulation. 32304324_Elucidating the unusual reaction kinetics of D-glucuronyl C5-epimerase. 34233293_Glycolysis- and immune-related novel prognostic biomarkers of Ewing's sarcoma: glucuronic acid epimerase and triosephosphate isomerase 1. | ENSMUSG00000032252 | Glce | 3.274389e+02 | 0.6953895 | -0.524106794 | 0.3585584 | 2.078523e+00 | 0.1493846535 | 0.77404149 | No | Yes | 2.930286e+02 | 58.712316 | 3.502649e+02 | 71.669141 | |
| ENSG00000138640 | 10144 | FAM13A | protein_coding | O94988 | Alternative splicing;Coiled coil;GTPase activation;Phosphoprotein;Reference proteome | hsa:10144; | cytosol [GO:0005829]; GTPase activator activity [GO:0005096]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165] | 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20010835_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20173748_A new susceptibility locus at 4q22.1 in FAM13A and replicated this association in one case-control group (n = 1,006) and two family-based cohorts. 20173748_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20656943_Observational study of gene-disease association. (HuGE Navigator) 22027142_these results support that FAM13A rs2869967 and XRCC5 rs3821104 are associated with COPD in Chinese Han population. 23169000_FAM13A1 is one of the top hub genes associated with human Treg suppressor function. See Table 1 of the publication. 23891779_the FAM13A locus might be a contributor to chronic obstructive pulmonary disease susceptibility in Chinese Han population. 24587150_Haplotypes carrying major alleles of rs7671167 (C) of FAM13A had a protective effect on lung function amongst South Indian male smokers with COPD. 24651745_CT quantitative analysis of lung parenchyma is very well correlated with COPD candidate gene FAM13A. 25163686_This review gives a brief summary of the current knowledge of FAM13A, and demonstrates the necessity to resolve its biological function besides its well accepted genetic contribution. 25608829_Data indicate that the FAM13A protein CTGA haplotypes interacted with heavy smoking to affect the risk of reduced pulmonary function. 25609086_Fam13a may contribute to human lung diseases. 26310313_This study confirms that the IREB2 variants contribute to an increased risk of lung cancer, whereas FAM13A predisposes to increased susceptibility to chronic obstructive pulmonary disease. 26527870_FAM13A - candidate gene for Chronic Obstructive Pulmonary Disease identified by Genome-wide association studies. 27612410_this study shows that FAM13A confers a risk for airway obstruction in general that is not driven exclusively by cigarette smoking, which is the main risk factor for chronic obstructive pulmonary disease 28137485_FAM13A gene polymorphism showed a significant association with the susceptibility to idiopathic pulmonary fibrosis, with severity of lung function impairment and with poor prognosis. 28569593_noncoding FAM13A polymorphisms were correlated with gene expression levels, and thus were probably in linkage disequilibrium with genetic loci that regulated the expression of FAM13A 29239766_Data show that FAM13A is a modifier gene of Cystic Fibrosis lung phenotype regulating RhoA activity, actin cytoskeleton dynamics and epithelial-mesenchymal transition. 29487953_Gene expression and adipocyte functional studies support the notion that FAM13A and POM121C control adipocyte lipolysis and adipogenesis, respectively, and might thereby be involved in genetic control of systemic insulin sensitivity 29621588_The rs2609255 in FAM13A gene may modify silicosis susceptibility in the Chinese population. 29872291_rs17014601 in FAM13A was significantly associated with COPD in the additive (odds ratio [OR]=1.36, 95% confidence interval [CI]: 1.11-1.67, P=0.003), heterozygote (OR=1.76, 95% CI: 1.33-2.32, P=0.0001), and dominant (OR=1.67, 95% CI: 1.28-2.18, P=0.0001) models. Stratified analyses indicated that the risk was higher in never smokers. 30079747_rs2013701 as a functional variant likely to contribute to the development of COPD 30301961_Our results suggest that FAM13A is dispensable for adipose development and insulin sensitivity. Yet the expression of FAM13A needs to be tightly controlled in adipose precursor cells for their proper survival and downstream adipogenesis. These data provide novel insights into the link between FAM13A and obesity. 30604588_High expression of FAM13A may be associated with an increased risk of liver cirrhosis. Risk of liver cirrhosis was significantly associated with G/A-G/G genotype of rs3017895. 31215377_Role of Polymorphisms of FAM13A, PHLDB1, and CYP24A1 in Breast Cancer Risk. 31539274_Variant genotypes of rs9224 in the FAM13A 3'UTR may modify Lung squamous carcinoma (LUSQ) susceptibility by affecting the binding of miRNA-22-5p and predict a poor prognosis of patients with LUSQ. 32029695_Loss of Family with Sequence Similarity 13, Member A Exacerbates Pulmonary Fibrosis Potentially by Promoting Epithelial to Mesenchymal Transition. 32193374_GWAS-associated variants within the FAM13A locus alter adipose FAM13A expression, which in turn, regulates adipocyte differentiation and contribute to changes in body fat distribution. 33097673_Respiratory traits and coal workers' pneumoconiosis: Mendelian randomisation and association analysis. 34105356_FAM13A as potential therapeutic target in modulating TGF-beta-induced airway tissue remodeling in COPD. 34166600_Connecting COPD GWAS Genes: FAM13A Controls TGFbeta2 Secretion by Modulating AP-3 Transport. 34210319_Family with sequence similarity 13 member A mediates TGF-beta1-induced EMT in small airway epithelium of patients with chronic obstructive pulmonary disease. 34821370_miR30a5p induces the adipogenic differentiation of bone marrow mesenchymal stem cells by targeting FAM13A/Wnt/betacatenin signaling in aplastic anemia. | ENSMUSG00000037709 | Fam13a | 3.109437e+02 | 0.8403680 | -0.250906870 | 0.3628510 | 4.787102e-01 | 0.4890071161 | 0.87511746 | No | Yes | 2.416518e+02 | 45.921110 | 2.667694e+02 | 51.880481 | |||
| ENSG00000138674 | 22872 | SEC31A | protein_coding | O94979 | FUNCTION: Component of the coat protein complex II (COPII) which promotes the formation of transport vesicles from the endoplasmic reticulum (ER) (PubMed:10788476). The coat has two main functions, the physical deformation of the endoplasmic reticulum membrane into vesicles and the selection of cargo molecules (By similarity). {ECO:0000250|UniProtKB:Q9Z2Q1, ECO:0000269|PubMed:10788476}. | 3D-structure;Alternative splicing;Chromosomal rearrangement;Cytoplasm;Cytoplasmic vesicle;ER-Golgi transport;Endoplasmic reticulum;Isopeptide bond;Membrane;Phosphoprotein;Protein transport;Proto-oncogene;Reference proteome;Repeat;Transport;Ubl conjugation;WD repeat | The protein encoded by this gene shares similarity with the yeast Sec31 protein, and is a component of the outer layer of the coat protein complex II (COPII). The encoded protein is involved in vesicle budding from the endoplasmic reticulum (ER) and contains multiple WD repeats near the N-terminus and a proline-rich region in the C-terminal half. It associates with the protein encoded by the SEC13 homolog, nuclear pore and COPII coat complex component (SEC13), and is required for ER-Golgi transport. Monoubiquitylation of this protein by CUL3-KLHL12 was found to regulate the size of COPII coats to accommodate unusually shaped cargo. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Dec 2015]. | hsa:22872; | COPII vesicle coat [GO:0030127]; COPII-coated ER to Golgi transport vesicle [GO:0030134]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum exit site [GO:0070971]; endoplasmic reticulum membrane [GO:0005789]; ER to Golgi transport vesicle membrane [GO:0012507]; intracellular membrane-bounded organelle [GO:0043231]; perinuclear region of cytoplasm [GO:0048471]; vesicle coat [GO:0030120]; calcium-dependent protein binding [GO:0048306]; structural molecule activity [GO:0005198]; COPII-coated vesicle cargo loading [GO:0090110]; endoplasmic reticulum organization [GO:0007029]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; intracellular protein transport [GO:0006886]; response to calcium ion [GO:0051592] | 16161041_Genomic PCR and subsequent sequencing showed that the breakpoints were located in intron 23 of SEC31L1 and intron 20 of anaplastic lymphoma kinase 16407955_three-dimensional reconstruction of Sec13/31 cages at 30 A resolution using cryo-electron microscopy and single particle analysis 16957052_ALG-2 is recruited to endoplasmic reticulum exit sites via Ca(2+)-dependent interaction with Sec31A and in turn stabilizes the localization of Sec31A at these sites. 17196169_In vitro GST pull down analysis demonstrated that ALG-2 and its alternatively spliced isoform interact with the COPII component Sec31A in a Ca2+-dependent manner, and a biotin-labeled ALG-2 overlay assay revealed direct binding of ALG-2 to Sec31A. 17981133_The Sec31 active fragment is accommodated in a binding groove supported in part by Sec23 residue Phe380. 18713835_Data show that coupling of the Sec23/24 and Sec13/31 layers of the COPII coat is required to drive export of collagen from the endoplasmic reticulum, and that efficient COPII assembly is essential for normal craniofacial development during embryogenesis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20834162_the alg2 binding site is one of the key determinants of the retention kinetics of Sec31A at endoplasmic reticulum exit sites 21109691_SEC31A-ALK fusions are recurrent in ALK-positive large B-cell lymphomas. 21325169_t(4;9)(q21;p24) leads to a novel SEC31A-JAK2 fusion in Hodgkin lymphoma 22331354_efficient COPII-dependent secretion, notably assembly of Sec13-Sec31, is required to drive epithelial morphogenesis in both two- and three-dimensional cultures 23349870_These results suggest that Sec31 phosphorylation by CK2 controls the duration of COPII vesicle formation, which regulates ER-to-Golgi trafficking. 24069399_ALG-2 attenuates COPII budding in vitro and stabilizes the Sec23/Sec31A complex. 25006245_ALG-2/Sec31A interactions were not required for the localization of Sec31A to ER exit sites per se but appeared to acutely regulate the stability and trafficking of the cargo receptor p24 and the distribution of the vesicle tether protein p115 25540196_findings suggest that AnxA11 maintains architectural and functional features of the ERES by coordinating with ALG-2 to stabilize Sec31A at the ERES. 25667979_Results show the crystal structure of the complex between ALG-2 and a peptide of Sec31A and found that the peptide binds to the third hydrophobic pocket (Pocket 3) and that ALG-2 recognizing 2 types of motifs at different hydrophobic surfaces of Sec31A. 29604273_USP8 deubiquitinates Sec31A and inhibits the formation of large COPII carriers, thereby suppressing collagen IV secretion. 30464055_We demonstrate through human and Drosophila genetic and in vitro molecular studies, that a severe neurological syndrome is caused by a null mutation in SEC31A, reducing cell viability through enhanced ER-stress response, in line with SEC31A's role in the COP-II complex. | ENSMUSG00000035325 | Sec31a | 3.374091e+03 | 0.8847878 | -0.176596567 | 0.2730819 | 4.083108e-01 | 0.5228281222 | 0.88477544 | No | Yes | 3.672498e+03 | 535.159180 | 3.538251e+03 | 528.932421 | |
| ENSG00000138764 | 901 | CCNG2 | protein_coding | Q16589 | FUNCTION: May play a role in growth regulation and in negative regulation of cell cycle progression. | Alternative splicing;Cell cycle;Cell division;Cyclin;Cytoplasm;Mitosis;Reference proteome | The eukaryotic cell cycle is governed by cyclin-dependent protein kinases (CDKs) whose activities are regulated by cyclins and CDK inhibitors. The 8 species of cyclins reported in mammals, cyclins A through H, share a conserved amino acid sequence of about 90 residues called the cyclin box. The amino acid sequence of cyclin G is well conserved among mammals. The nucleotide sequence of cyclin G1 and cyclin G2 are 53% identical. Unlike cyclin G1, cyclin G2 contains a C-terminal PEST protein destabilization motif, suggesting that cyclin G2 expression is tightly regulated through the cell cycle. [provided by RefSeq, Jul 2008]. | hsa:901; | cyclin-dependent protein kinase holoenzyme complex [GO:0000307]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; cyclin-dependent protein serine/threonine kinase regulator activity [GO:0016538]; cell division [GO:0051301]; mitotic cell cycle phase transition [GO:0044772]; regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0000079] | 11956189_cyclin G2 also associates with various PP2A B' regulatory subunits, as previously shown for cyclin G1 12452053_ectopic expression of cyclin G2 inhibits cell proliferation 16364039_Involved in progression and regulation of telomerase 16608856_Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter 17123511_Cyclin G2 may modulate the cell cycle and cellular division processes through modulation of PP2A and centrosomal associated activities. 18025271_Cyclin G2 expression is modulated by HER2 signaling through multiple pathways including phosphoinositide 3-kinase, c-jun NH(2)-terminal kinase, and mTOR signaling. 18754885_our results suggest that cotylenin A and rapamycin induce inhibition of cancer cell growth through the induction of cyclin G2. 18784254_The antiproliferative effect of Nodal/ALK7 on ovarian cancer cells is in part mediated by cyclin G2. 19559447_Cyclin G2 appears to be a negative cell-cycle regulator in gastric cancer, and its expression seems to be inversely related to gastric cancer progression. 19738611_Observational study of gene-disease association. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 21532621_These findings showed that Nodal signaling promotes cyclin G2 transcription by upregulating FoxO3a expression, inhibiting FoxO3a phosphorylation and enhancing its synergistic interaction with Smads. 21688120_Expression of cyclin G1 and G2 is strongly associated with nasopharyngeal carcinoma cell differentiation. 22589537_Our results suggest that CycG2 contributes to DNA damage-induced G(2)/M checkpoint by enforcing checkpoint inhibition of CycB1-Cdc2 complexes. 22596188_These findings show that FOXA1, but not ER- alpha, is essential for AHR(aryl hydrocarbon receptor) -dependent regulation of CCNG2(cyclin G2 ), assigning a role for FOXA1 in AHR action. 24059861_Regulation of cyclin G2 is a key mechanism whereby insulin, insulin analogues and IGF-I stimulate cell proliferation. 24248541_CCNG2 expression decreased in gastric cancer and correlated significantly T stages, lymph node metastasis, clinical stage, histological grade, and poor overall survival 24272084_CCNG2 may play important roles as a negative regulator to kidney cancer ACHN cell by promoting degradation of CDK2. 24289643_Low CCNG2 expression is associated with thyroid cancer. 24293374_CCNG2 expression decreased in prostate cancer and correlated significantly with lymph node metastasis, clinic stage, and Gleason score. 24297335_CCNG2 may play important roles as a negative regulator to esophageal cancer cell by promoting degradation of CDK2. 24307622_The level of CCNG2 was correlated with T stages, lymph node metastasis, clinic stage, and histological grade (P < 0.05) in colorectal carcinoma. 24339739_Cyclin G2-involved invasion in vitro and in vivo. 24708911_Data shows the human CCNG2 and CDK4 expression of visceral adipose tissue are inversely associated with glucose and insulin resistance. 25117811_miR-1246 expression was associated with chemoresistance and CSC-like properties via CCNG2, and could predict worse prognosis in pancreatic cancer patients 25122062_Data (from studies using BeWo cells, a choriocarcinoma cell line, as model of placentation) suggest that miR-378a-5p (microRNA 378a) inhibits cell differentiation in syncytiotrophoblasts, in part, by down-regulating CCNG2 (cyclin G2) expression. 25309979_miR-93-CCNG2 axis may be involved in Laryngeal squamous cell carcinoma proliferation and progression. 26149128_Neisseria meningitidis caused changes in the abundance of several cell cycle regulatory mRNAs, including the cell cycle inhibitors p21(WAF1/CIP1) and cyclin G2 in human brain microvascular endothelial cells. 26573378_Upregulation of miR1246 mediated the malignant progression of colorectal cancer and is partly attributed to the downregulation of the expression of CycG2. 26876206_Taken together, our novel findings demonstrate that cyclin G2 has potent tumor-suppressive effects in Epithelial ovarian cancer (EOC) by inhibiting EMT through attenuating Wnt/beta-catenin signaling. 27374211_CCNG2 knockdown eradicated the effects of miR-340 silencing. 27753529_CycG2 contributes to signaling networks that limit breast cancer. 28640887_EGF-induced, calpain-mediated proteolysis contributes to the rapid destruction of cyclin G2 and that the PEST domain is critical for EGF/calpain actions 30106441_Low CCNG2 expression is associated with colorectal cancer. 30547803_This study demonstrates that cyclin G2 suppresses Wnt/beta-catenin signaling and inhibits gastric cancer cell growth and migration through Dapper1. 31013711_these results suggest that miR-590-3p promotes ovarian cancer development, in part by directly targeting CCNG2 and FOXO3. 31841213_MiR-1290 targets CCNG2 to promote the metastasis of oral squamous cell carcinoma. 31978940_Cyclin G2 regulates canonical Wnt signalling via interaction with Dapper1 to attenuate tubulointerstitial fibrosis in diabetic nephropathy. 32941407_Cyclin G2 Is Involved in the Proliferation of Placental Trophoblast Cells and Their Interactions with Endothelial Cells. 33205477_Cyclin G2 upregulation impairs migration, invasion, and network formation through RNF123/Dvl2/JNK signaling in the trophoblast cell line HTR8/SVneo, a possible role in preeclampsia. 34452627_Cyclin G2 reverses immunosuppressive tumor microenvironment and potentiates PD-1 blockade in glioma. 34598688_miR-17-5p drives G2/M-phase accumulation by directly targeting CCNG2 and is related to recurrence of head and neck squamous cell carcinoma. | ENSMUSG00000029385 | Ccng2 | 2.667318e+02 | 0.9515744 | -0.071611667 | 0.4045696 | 3.116714e-02 | 0.8598680133 | 0.97321292 | No | Yes | 1.898226e+02 | 46.518158 | 2.237181e+02 | 56.011520 | |
| ENSG00000139044 | 283358 | B4GALNT3 | protein_coding | Q6L9W6 | FUNCTION: Transfers N-acetylgalactosamine (GalNAc) from UDP-GalNAc to N-acetylglucosamine-beta-benzyl with a beta-1,4-linkage to form N,N'-diacetyllactosediamine, GalNAc-beta-1,4-GlcNAc structures in N-linked glycans and probably O-linked glycans. Mediates the N,N'-diacetyllactosediamine formation on gastric mucosa. {ECO:0000269|PubMed:16728562}. | Glycosyltransferase;Golgi apparatus;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | B4GALNT3 transfers N-acetylgalactosamine (GalNAc) onto glucosyl residues to form N,N-prime-diacetyllactosediamine (LacdiNAc, or LDN), a unique terminal structure of cell surface N-glycans (Ikehara et al., 2006 [PubMed 16728562]).[supplied by OMIM, Aug 2008]. | hsa:283358; | Golgi apparatus [GO:0005794]; Golgi cisterna membrane [GO:0032580]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; acetylgalactosaminyltransferase activity [GO:0008376]; N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity [GO:0033842] | 16728562_supra-nuclear expression of beta4GalNAc-T3 is essential for the formation of LacdiNAc on the surface mucous cells and that LacdiNAc and beta4GalNAc-T3 are novel differentiation markers of surface mucous cells in the gastric mucosa. 17579116_up-regulation of beta4GalNAc-T3 may play a critical role in promoting tumor malignanc 18048353_betaGT3 and betaGT4, that are able to transfer GalNAc to GlcNAc in beta1,4-linkage display the necessary glycoprotein specificity in vivo. 20360844_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22722937_Molecular basis for protein-specific transfer of N-acetylgalactosamine to N-linked glycans by the glycosyltransferases beta1,4-N-acetylgalactosaminyl transferase 3 (beta4GalNAc-T3) and beta4GalNAc-T4. 22722940_Peptide-specific transfer of N-acetylgalactosamine to O-linked glycans by the glycosyltransferases beta1,4-N-acetylgalactosaminyl transferase 3 (beta4GalNAc-T3) and beta4GalNAc-T4. 25003232_Data suggest B4GALNT3 regulates cancer stemness and the invasive properties of colon cancer cells through modifying EGFR glycosylation and signaling. 25803323_Identify a new chimeric transcript generated by the fusion of WNK1 and B4GALNT3 genes, correlated with B4GALNT3 overexpression papillary thyroid carcinoma. | ENSMUSG00000041372 | B4galnt3 | 2.198050e+03 | 1.1014511 | 0.139405402 | 0.2737140 | 2.597128e-01 | 0.6103175125 | 0.91246250 | No | Yes | 1.884931e+03 | 187.864157 | 1.780239e+03 | 182.174074 | |
| ENSG00000139182 | 9746 | CLSTN3 | protein_coding | Q9BQT9 | FUNCTION: May modulate calcium-mediated postsynaptic signals. Complex formation with APBA2 and APP, stabilizes APP metabolism and enhances APBA2-mediated suppression of beta-APP40 secretion, due to the retardation of intracellular APP maturation. {ECO:0000250}. | Alternative splicing;Calcium;Cell adhesion;Cell junction;Cell membrane;Cell projection;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Repeat;Signal;Synapse;Transmembrane;Transmembrane helix | hsa:9746; | cell surface [GO:0009986]; dendrite [GO:0030425]; endoplasmic reticulum membrane [GO:0005789]; GABA-ergic synapse [GO:0098982]; glutamatergic synapse [GO:0098978]; Golgi membrane [GO:0000139]; integral component of postsynaptic density membrane [GO:0099061]; postsynaptic membrane [GO:0045211]; protein-containing complex [GO:0032991]; calcium ion binding [GO:0005509]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; positive regulation of protein localization to synapse [GO:1902474]; positive regulation of synapse assembly [GO:0051965]; positive regulation of synaptic transmission [GO:0050806]; regulation of cell growth [GO:0001558]; regulation of presynapse assembly [GO:1905606]; synapse assembly [GO:0007416]; synaptic transmission, GABAergic [GO:0051932]; synaptic transmission, glutamatergic [GO:0035249] | 15037614_Alcadein and amyloid beta-protein precursor regulates FE65-dependent gene transactivation [alcalpha1, alcbeta, alcgamma] 23499467_The C-terminal fragment but not full-length Cst-3 accumulated in dystrophic neurites surrounding amyloidbeta plaques in Tg2576 mouse and Alzheimer disease brains. 25352602_structure of Calsyntenin 3 and its interaction with neurexin 1alpha 26213366_ApoE expression attenuated intracellular trafficking of APP and Alcbeta | ENSMUSG00000008153 | Clstn3 | 2.367144e+03 | 1.0311659 | 0.044276483 | 0.3082664 | 2.101224e-02 | 0.8847456107 | 0.97808588 | No | Yes | 2.054540e+03 | 227.977502 | 2.039559e+03 | 232.337893 | ||
| ENSG00000139219 | 1280 | COL2A1 | protein_coding | P02458 | FUNCTION: Type II collagen is specific for cartilaginous tissues. It is essential for the normal embryonic development of the skeleton, for linear growth and for the ability of cartilage to resist compressive forces. | 3D-structure;Alternative splicing;Calcium;Cataract;Collagen;Deafness;Direct protein sequencing;Disease variant;Disulfide bond;Dwarfism;Extracellular matrix;Glycoprotein;Hydroxylation;Metal-binding;Reference proteome;Repeat;Secreted;Signal;Stickler syndrome | This gene encodes the alpha-1 chain of type II collagen, a fibrillar collagen found in cartilage and the vitreous humor of the eye. Mutations in this gene are associated with achondrogenesis, chondrodysplasia, early onset familial osteoarthritis, SED congenita, Langer-Saldino achondrogenesis, Kniest dysplasia, Stickler syndrome type I, and spondyloepimetaphyseal dysplasia Strudwick type. In addition, defects in processing chondrocalcin, a calcium binding protein that is the C-propeptide of this collagen molecule, are also associated with chondrodysplasia. There are two transcripts identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:1280; | basement membrane [GO:0005604]; collagen trimer [GO:0005581]; collagen type II trimer [GO:0005585]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; extracellular matrix structural constituent [GO:0005201]; extracellular matrix structural constituent conferring tensile strength [GO:0030020]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; MHC class II protein binding [GO:0042289]; platelet-derived growth factor binding [GO:0048407]; proteoglycan binding [GO:0043394]; anterior head development [GO:0097065]; cartilage condensation [GO:0001502]; cartilage development [GO:0051216]; cartilage development involved in endochondral bone morphogenesis [GO:0060351]; cellular response to BMP stimulus [GO:0071773]; central nervous system development [GO:0007417]; chondrocyte differentiation [GO:0002062]; collagen fibril organization [GO:0030199]; embryonic skeletal joint morphogenesis [GO:0060272]; endochondral ossification [GO:0001958]; extracellular matrix organization [GO:0030198]; heart morphogenesis [GO:0003007]; inner ear morphogenesis [GO:0042472]; limb bud formation [GO:0060174]; negative regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001240]; notochord development [GO:0030903]; otic vesicle development [GO:0071599]; proteoglycan metabolic process [GO:0006029]; regulation of gene expression [GO:0010468]; roof of mouth development [GO:0060021]; sensory perception of sound [GO:0007605]; skeletal system development [GO:0001501]; tissue homeostasis [GO:0001894]; visual perception [GO:0007601] | 10729292_The first paper to show that mutations in exon 2 result in a predominantly ocular form of Stickler syndrome 11447232_Sp3 represses the Sp1-mediated transactivation of the human COL2A1 gene in primary and de-differentiated chondrocytes 11708863_This form of premature osteoarthritis may present in childhood and should be considered in the differential diagnosis of childhood arthropathy presenting in the context of a positive family history. 11716775_COL2A1 gene expression in differentiating chondrocytes can be modulated by culture conditions so that its transcriptional activity is repressed in monolayer cultures and rescued to some extent when the cells are switched to polyHEMA substrata. 12096843_Observational study of gene-disease association. (HuGE Navigator) 12186868_TGF-beta1 inhibition of COL2A1 gene transcription in articular chondrocytes is mediated by an increase of the Sp3/Sp1 ratio and by the repression of Sp1 transactivating effects on that gene 12200454_Upstream elements present in the 3'-untranslated region of the gene influence the processing efficiency of overlapping polyadenylation signals. 12223098_identification of TATA-containing core promoter as target of interferon-gamma-mediated inhibition in human chondrocytes 12360016_Observational study of gene-disease association. (HuGE Navigator) 12429249_Linkage of stop codon mutation in exon 2 of the collagen 2A1 gene in a large stickler syndrome family. 12429250_A variant of Stickler syndrome, caused by mutations in exon 2 of COL2A1, may present in families 12511349_Posterior chorioretinal atrophy and vitreous phenotype in a family with Stickler syndrome from a mutation in the COL2A1 gene. 12544472_Mutations of Col2a1 result in Stickler syndrome. 12637574_Egr-1 represses COL2A1 by preventing interactions between Sp1 and the general transcriptional machinery 12713737_SOX9 exerts a bifunctional effect on COL2A1 gene expression in chondrocytes depending on the differentiation state. 12732631_In promoter assays, CBP/p300 enhances Col2a1, which encodes cartilage-specific type II collagen gene promoter activity via Sox9. 12935820_SOX9 is not the key regulator of COL2A1 promoter activity in human adult articular chondrocytes 14644246_Kniest dysplasia with retinal detachment associated with a novel type II collagen gene (COL2A1) mutation. 14729840_A missense mutation in a lethal type case and a 4-base pair deletion in a non-lethal case in COL2A1 of platyspondylic skeletal dysplasia, Torrance type patients. 15082485_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15102076_BMP2 or 4 in pilomatricoma is responsible for induction of proalpha(1)(II) collagen mRNA in overlying epidermal cells resulting in deposition of type II collagen in dermo-epidermal junction 15466413_a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA may be evolutionarily conserved 15522781_single amino acid substitution positions in the collagen triple helix determine their effect on structure of collagen fibrils 15671297_Mutations outside the alternatively spliced exon 2 region of COL2A1 can also result in an ocular only phenotype. There was no evidence that missplicing modifies the phenotype of these mutations 15731776_The presence of type II collagen in the extracellular tumor matrix significantly facilitates the diagnosis of mesenchymal chondrosarcomas in the absence of histologically visible chondroid matrix formation. 15895462_identification of COL2A1 mutations in 56 families that were suspected of having type II collagenopathies, and 38 mutations in 41 families were found 15922184_Observational study of gene-disease association. (HuGE Navigator) 15930420_In families with avascular necrosis of the femoral head, haplotype and sequence analysis of the COL2A1 gene can be used to identify carriers of the mutant allele before the onset of clinical symptoms 16001263_Data demonstrate a significant reduction of collagens I, II and aggrecan mRNA after the initiation of culture compared with mRNA levels in fresh tissue. 16076844_cis elements in the COL2A1 gene modulate the cell type-specific alternative splicing switch of exon 2 during cartilage development 16133074_Observational study of gene-disease association. (HuGE Navigator) 16192646_Trypsin degrades COL2A1 and is expressed and activated in mesenchymally transformed rheumatoid arthritis synovitis tissue. 16329077_When modified by conditions found within the inflamed joint, CII acts as an autoantigen in rheumatoid arthritis 16395149_An 8-year-old boy with type 1 Stickler syndrome showed a novel mutation in intron 11 of the COL2A1 gene 16650379_Mechanical compression increases the level of type II mRNA expression by transcriptional activation possibly through the Sp1 binding sites residing in the proximal region of the COL2A1 gene promoter. 16877351_In comparison with healthy cartilage, Osteoarthritis articular chondrocytes exhibit increased in vivo synthesis of collagen prolyl-4-hydroxylase type II, a pivotal enzyme in collagen triple helix formation. 16978902_Observational study of genotype prevalence. (HuGE Navigator) 17009260_Premature induction of hypertrophy-related molecules (type X collagen and matrix metalloproteinase 13) occurred before production of type II collagen and was followed by up-regulation of alkaline phosphatase activity. 17163530_COL2A1 mutations associated with marked metaphyseal dysplasia with only mild epiphyseal and spondylar changes. 17195216_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17217840_The expression of collagen type II and TGF-beta1, bFGF in adolescent idiopathic scoliosis was similar to congenital scoliosis. 17335825_type II collagen expression was observed very focally within advanced atherosclerotic plaques in crural arteries 17394019_Study found a missense mutation (p.G1170S) in COL2A1 in a Japanese family with an autosomal dominant hip disorder manifesting as Legg-Calve-Perthes disease and showing considerable intra-familial phenotypic variation. 17437277_We present two missense mutations and one apparently silent mutation that each result in Stickler syndrome, but via different molecular mechanisms. 17509551_Familial mutation of G504S of collagen type II alpha (COL2A1) gene results in distinctive spondyloepiphyseal dysplasia congenita. 17568421_Human cells cultured over 5 days increased expression of aggrecan and collagen II in both nucleus and annulus cells under increasing osmolarity. 17580305_dual role for TIA-1 in shuttling between DNA and RNA ligands to co-regulate COL2A1 expression at the level of transcription and pre-mRNA alternative splicing. 17653045_Observational study of gene-disease association. (HuGE Navigator) 17653045_primary findings from the current study suggest involvement in common forms of myopia by COL2A1 17683641_Prostaglandin E2 at lower levels than in inflammation suppress collagenase-mediated COL2A1 cleavage in osteoarthritic cartilage. 17721977_Missense and nonsense mutations in the alternatively-spliced exon 2 of COL2A1 cause the ocular variant of Stickler syndrome 17994563_Molecular analysis of genomic DNA extracted from amniotic cells of the second and third fetuses revealed heterozygosity for a 10370G > T missense mutation (G346V) in the COL2A1 gene in achondrogenesis type II 18023161_COL2A1 mRNA abundance and other aspects of chondrocyte differentiation may be regulated by the use of previously undetermined alternative splice sites 18040638_mRNAs for type II collagen and aggrecan were expressed by MSCs treated with either TGFbeta1 or OP-1; however, substantial matrix production was not induced. 18065760_Interleukin-6 (IL-6) and/or soluble IL-6 receptor down-regulation of human type II collagen gene expression in articular chondrocytes requires a decrease of Sp1.Sp3 ratio and of the binding activity of both factors to the COL2A1 promoter 18177466_Sequence analysis revealed in the three patients a novel COL2A1 mutation (c.1468_1475delinsT) that accounted for a STL syndrome type I phenotype. One patient carries an EYA1 mutation, p.R328X, which was not present in the two other patients. 18276201_One novel DNA variation (c.1266+7G>C) in the COL2A1 gene occurs near a splice site and it was observed to co-segregate with the phenotype in one of the two families with this DNA variation 18288556_Observed no evidence of linkage between COL2A1 locus and developmental dysplasia of the hip. 18383211_Results provide a new insight into the molecular mechanisms of pathological changes caused by mutations in COL2A1 and identify apoptosis as an element of a cellular response to the presence of altered type II collagen mutant molecules. 18512791_in a family study A p.Gly1170Ser mutation of COL2A1 cosegregated with hip osteoarthritis, avascular necrosis of the femoral head, and Legg-Calve-Perthes,and was absent in controls 18523590_Observational study of gene-disease association. (HuGE Navigator) 18523590_associations between clinical outcomes of congenital toxoplasmosis and polymorphisms at ABCA4 and COL2A1 provide novel insight into the molecular pathways that can be affected by congenital infection with this parasite 18553548_Report on a large family with 11 patients with typical Czech dysplasia and sensorineural hearing loss. Mutation analysis documented the COL2A1 c.823C > T (R275C) mutation in all affected individuals. 18636947_Articular chondrocytes maintained a rounded shape and proliferated rapidly showed a reduced, but persistent, production of COL2A1 mRNA. 18655132_collagen II induces first MMPs and pro-inflammatory cytokines and then release of collagen II fragments from mature collagen II fibers 18799084_Type II collagen levels were lower in osteoarthritis than in Rheumatoid arthritis cartilage 18978274_Observational study of gene-disease association. (HuGE Navigator) 19019890_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19019890_results suggest that the studied COL2A1 gene polymorphisms may play a role in the aetiology of hand osteoarthritis and that this effect may be enhanced by repetitive loading work tasks. 19180518_Observational study of gene-disease association. (HuGE Navigator) 19387081_Observational study of gene-disease association. (HuGE Navigator) 19387081_The COL2A1 gene was associated with high-grade myopia in two independent Caucasian family datasets. COL1A1 gene polymorphisms were not associated with myopia 19430638_Observational study of gene-disease association. (HuGE Navigator) 19433093_The molecular mechanism of spondyloepiphyseal dysplasia may be driven not only by structural changes in the architecture of extracellular collagenous matrices, but also by intracellular processes activated by the presence of mutant collagen II molecules. 19473573_declining N-propeptide of collagen IIA (PIIANP) and increasing collagen II C-telopeptide (CTX-II) in early and longstanding rheumatoid arthritis demonstrate unbalanced anabolic and degradative pathways 19756630_An association of a COL2A1 gene polymorphism with advanced stages of osteoarthritis of the knee is evaluated in Mexican Mestizo population. 19756630_Observational study of gene-disease association. (HuGE Navigator) 19764028_Mutation analysis documented the COL2A1 c.823C > T mutation in all Czech dysplasia affected individuals. 19790048_Hypoxia not only induces type II collagen and aggrecan, but it also inhibits type I and type III collagen in the hypoxia-inducible factor 1alpha-dependent redifferentiation of chondrocytes. 19814628_In rheumatoid arthritis at different stages and healthy individuals, procollagen IIA N-peptide (PIIANP) levels in serum exhibited no circadian rhythmicity, and PIIANP in serum was not influenced by physical activity 19913121_Observational study of gene-disease association. (HuGE Navigator) 20131279_mutations in COL2A1 can present as degenerative joint disease in the absence of any other phenotypic clues 20179744_study was to define more precisely phenotype and genotype of Stickler syndrome type 1 by investigating patients with Col2A1 heterozygous mutation; study confirms Stickler syndrome type 1 is predominantly caused by loss-of-function mutations in COL2A1 20204389_Our study demonstrated that the p.Gly1170Ser mutation of COL2A1 caused significant structural alterations in articular cartilage, which are responsible for the new type II collagenopathy. 20529846_Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes 20569194_The effect of parathyroid hormone on expression of COL10 and COL2 in mesenchymal stem cells from osteoarthritic patients and analyzed the potential mechanisms related to its effect, was investigated. 20592578_Data show that an increase in collagen-II, aggrecan, and Sox-9 protein expression in NP and AF regions of the disc was detected in the exercised rats. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20672350_Observational study of gene-disease association. (HuGE Navigator) 20672350_implication of IRF6, COL2A1, and WNT3 in occurrence of nonsyndromic cleft palate (NSCP); likely variation in cartilage collagen II and XI genes, IRF6, and Wnt and FGF signal pathway genes contributes susceptibility to NSCP in Northeast Europe populations 21044884_identification of the GVMGFO discoidin domain receptor binding motif on collagen II 21088911_Observational study of gene-disease association. (HuGE Navigator) 21088911_The G4006A AA homozygous genotype significantly increased in female Chinese knee osteoarthritis patients. 21204228_Association of a p.Pro786Leu variant in COL2A1 with mild spondyloepiphyseal dysplasia congenita in a three-generation family. 21221577_The results suggest that uCTX-I, uCTX-II and sCOMP could identify patients with focal cartilage lesions from an early stage of osteoarthritis of the knee. 21332586_COL2A1 mutations give rise to a spectrum of phenotypes predominantly affecting cartilage and bone from the severe disorders that are perinatally lethal to the milder conditions that are recognised in the post-natal period and childhood. [review] 21442341_children who present with bilateral Perthes-like disease of the hip might have an underlying mutation in the gene encoding type II collagen. 21655647_This study tests the hypothesis that disease severity is characterized by alterations in expression of cartilage-specific genes for aggrecan and collagen type II. 21777803_We present a case in which a Pierre Robin sequence and a positive family history led to the clinical diagnosis of Stickler syndrome, which was confirmed by the identification of a disease-causing novel deletion of 2 nucleotides in the COL2A1 gene. 21843649_COMP and Col2a1 expression are regulated differently during chondrogenesis. COMP is a primary response gene of TGFbeta and its fast induction during chondrogenesis suggests that COMP is suitable for rapidly accessing the chondrogenic potential of stem cells 21853276_nicotine has the same effect on both chondrocytes, obtained either from osteoarthritis patients or from normal human, and the positive effect of smoking in OA may relate to the alteration in metabolism of chondrocytes. 21853455_In inner meniscus cells, mechanical stretch may have an essential role in the epigenetic regulation of COL2A1 expression. 21924244_this study extends the mutation spectrum of spondyloepiphyseal dysplasia congenita (SEDC) in COL2A1 and is helpful in early molecular diagnoses of SEDC. 21940677_A transgenic mouse model represents the first possibility to study B cell tolerance to endogenous matrix protein collagen II antigen. 21993774_The rs1635529 polymorphism was no statistically significant difference in genotype, allele and haplotype frequencies for the other three SNPs between the high myopia group and the control group. 22049074_RB1CC1 protein suppresses type II collagen synthesis in chondrocytes and causes dwarfism 22068351_serum levels of collagen type 2 in Kashin-Beck disease were increased compared with healthy controls and osteoarthritis patients, but the increase was not correlated with disease severity. 22189268_variants detected in either COL2A1 in patients with Stickler syndrome 22241609_Increased expression of collagen II, aggrecan, and cartilage oligomeric matrix protein (COMP), were observed during differentiation of induced pluripotent stem cells from osteoarthritic chondrocytes. 22319617_Neo-antigenic epitopes were generated on (*)OH modified CII which rendered it highly immunogenic and arthritogenic as compared to the unmodified form. 22495950_A novel in-frame deletion c.4458_4460delCTT (p.Phe1486del) in the C-propeptide region of COL2A1 was found in both mother and fetus with spondyloperipheral dysplasia. 22496037_describe five further examples of somatic mosaicism of COL2A1 mutations illustrating the importance of detailed clinical evaluation and molecular testing even in clinically normal parents of affected individuals 22574936_Genetic analysis revealed that all affected family members of one pedigree carried an exon 2 mutation of COL2A1, and in the second pedigree, all affected members carried an FZD4 mutation. 22689318_CTX-II has unique relations with bone markers as compared to other cartilage markers and might reflect bone rather than cartilage metabolism 22711552_study reports on a severe form of skeletal dysplasia in 2 sibs whose phenotype was most consistent with platyspondylic lethal skeletal dysplasia Torrance type; they had an identical heterozygous missense mutation in the triple helical region of COL2A1 c.3545G>A (p.Gly1182Asp)in exon 50 22750747_Intra-articular injection of human mesenchymal stem cells (MSCs) promote rat meniscal regeneration by being activated to express Indian hedgehog that enhances expression of type II collagen. 22791362_A growth chart was constructed for patients with molecularly confirmed congenital spondylo-epiphyseal dysplasia (SEDC) and other COL2A1 related dysplasias. 22863613_Urinary CTX-II concentrations are elevated and associated with knee pain and function in subjects with anterior cruciate ligament reconstruction. 22965811_Systemic or local down-regulation of miR-7 may contribute to the pathogenesis of localized scleroderma via the overexpression of alpha2(I) collagen. 23079993_Extension of the mutation spectrum of spondyloepiphyseal dysplasia (SED) and confirmation of a relationship between mutations in the COL2A1 gene and clinical findings of SED. 23116329_Data indicate T cell specificity antibody to the CII259-273 T cell epitope in B10.DR4.Ncf1*/* mice following immunization with human collagen type II (CII). 23331625_AP-2epsilon indirectly interacts with the core promoter of COL2A1 and subsequently inhibits its transcriptional activity, thus modulating cartilage development. 23370687_Data indicate link protein peptide (LPP) upregulates expression of aggrecan and collagen II at both mRNA and protein levels. 23545312_we identified a novel truncating mutation (p.Lys1444AsnfsX27) in the C-propeptide of type II collagen COL2A1 in an affected Chinese individual with SPD. 23546968_Data indicate that after rhBMP-2 treatment, mRNA expression of type I and II collagens increased significantly more in cervical than in lumbar nucleus pulposus (NP) cells. 23592912_A three-generation Caucasian family variably diagnosed with Stickler and Wagner syndrome was screened for sequence variants in the COL2A1 and VCAN genes. 23618358_NF-kappaB/p65 signaling, as well as Sox9, may contribute to changes in the morphology of uterine carcinosarcomas cells toward the chondrocytic phenotype through modulation of COL2A1 transcription. 23631855_Tensile strain increases expression of CCN2 and COL2A1 by activating TGF-beta-Smad2/3 pathway in chondrocytic cells. 23770606_We identified hypermutability of the major cartilage collagen gene COL2A1, with insertions, deletions and rearrangements identified in 37% of cases. 23851124_Chondrocalcin is internalized by chondrocytes and triggers cartilage destruction via an interleukin-1beta-dependent pathway. 23873758_study indicates that Set7/9 prevents the histone deacetylase activity of SirT1, potentiating euchromatin formation on the promoter site of COL2A1 and resulting in morphology-dependent COL2A1 gene transactivation. 23882137_Trypsin-1 and trypsin-2 appear to have a function in the degradation of vitreous type II collagen. 23918474_A report on a familial apparently balanced reciprocal translocation t(12;15)(q13;q22.2) which disrupts COL2A1 and causes type 1 Stickler syndrome, in a mother and two of her children. 23928235_COL2A1 mutation R275C regularly leads to symmetric spondyloarthropathy,symptoms gradually developing during puberty. 23932928_In this family spondyloepiphyseal dysplasia congenita was caused by one novel missense mutation of c.3257G>T at exon 46 of COL2A1 gene resulting in substitution of glycine (Gly, G) to valine (Val, V) at the 1086 codon (p.Gly1086Val. 24014797_We report 2 generations of 4 male family members with Legg-Calve-Perthes disease-like features and mutation of the COL2A1 gene of the 12q13 chromosome. 24088220_The objective of this study was to determine the concentrations of types I, II and III collagen in six distinct regions of the supraspinatus tendon. 24164106_Genetic analyses showed that both sisters and their mother carried the same mutation in the COL2A1 gene. 24164447_Data from model complexes of MMP-2 (matrix metalloproteinase-2) and triple-helix peptide fragments of COL2A1 suggest that the triple helix is distorted to allow the accommodation of an individual peptide chain within the MMP active site. 24375478_Denaturation of the chimeric collagen increased its affinity for fibronectin, as seen for mammalian collagens. 24386886_association between polymorphism in COL2A1 gene and MP was observed. results suggested COL2A1 gene could be a new susceptibility gene for use in the study of genetic risk factors for MP. 24641900_acetaldehyde up-regulates COL1A2 by modulating the role of Ski and the expression of SMADs 3, 4, and 7. 24728947_urine C2C and trace element level in patients with knee osteoarthritis 24736929_This is the first familial report of G546S mutation in the COL2A1 gene that results in spondyloepiphyseal dysplasia congenita. 24949742_new variants to the repertoire of COL2A1 mutation resulting in related collagenopathies 24971869_In middle-aged women without clinical knee disease, higher uCTX-II levels were associated with early detrimental structural changes at the knee (cartilage defects, tibial bone expansion and bone marrow lesions) at baseline but not over 2 years. 25008205_The study shows that approximately 45% of the collagen IIA synthesis as assessed by the collagen IIA N-terminal propeptide in serum is attributable to genetic effectors while individual and shared environment account for 24% and 31% respectively. 25024164_somatic alterations of the COL2A1 were found in 19.3% of chondrosarcoma and 31.7% of enchondroma cases. 25050885_A mutation in the COL2A1 gene is the causative agent of ONFH in this family. 25124518_Mutations in the gene encoding the type II collagen gene (COL2A1) cause a series of type II collagenopathies that manifest as inheritable skeletal disorders. 25436060_results suggest that COL2A1 is associated with the risk of degenerative lumbar scoliosis in Korean population 25521223_This study demonstrated that Genes overexpressed in Pilomyxoid Astrocytoma vs. Pilocytic Astrocytoma, ranked according to fold-change, included developmental genes H19, DACT2,COL2A1; COL1A1 and IMP3. 25735649_Six new unrelated patients with R989C mutation in COL2A1 gene associated with a severe phenotype of spondyloepiphyseal dysplasia congenita. 25818544_miR-93 contributed to abnormal nucleus pulposus cell type II collagen expression by targeting MMP3, involved in intervertebral disc degeneration. 25863096_A novel missense mutation of c.2224G>A (p.Gly687Ser) in the COL2A1 gene is associated with a Chinese family with spondyloepiphyseal dysplasia congenita. 25900302_A unique case of spondyloepiphyseal dysplasia congenita with mild coxa vara caused by double de novo COL2A1 mutations (p.G504S, p.G612A) located on the same allele. 26030151_a novel mutation, c.620G>A (p.Gly207Glu), in the collagen type II alpha-1 gene; genotype-phenotype relationship between mutations and clinical findings of Spondyloepiphyseal dysplasia congenita 26037341_A novel missense mutation (c.905C>T, p.Ala302Val)found in the coding region of the COL2A1 gene is associated Kniest dysplasia. 26183434_Results identified a novel COL2A1 variant (c.619G>A, p.Gly207Arg) causing a distinct type II collagenopathy with features of progressive pseudorheumatoid dysplasia and spondyloepiphyseal dysplasia, Stanescu type. 26250472_Dysspondyloenchondromatosis is associated with COL2A1 mutation. 26311224_estimated median of 95 months as compared to an estimated median of 16 months for subjects expressing other levels of COL2A1 and SLC6A10P 26345137_Constitutive mutation in COL2A1 gene is associated with Kniest dysplasia and chondrosarcoma. 26443184_study provide an updated list of COL2A1 mutations identified in the literature and in our patients with a genetic diagnosis for bone dysplasia; work confirmed that mutations in this gene are responsible for a wide clinical spectrum ranging from lethal major skeletal abnormalities to isolated arthritis 26545783_Identified is a novel Col2a1 mutant mouse possessing a p.Tyr1391Ser missense mutation. Endoplasmic reticulum stress-mediated apoptosis contributes to a skeletal dysplasia resembling platyspondylic lethal skeletal dysplasia, Torrance type, in this line. 26583473_Overexpression of miR-27b promoted type II collagen expression in nucleus pulposus cells. 26586363_Whole exome sequencing identified a novel COL2A1 mutation that causes mild Spondylo-epiphyseal dysplasia mimicking autosomal dominant brachyolmia 26626311_Half of the Stickler patients (46%) carried a COL2A1 variant, and the molecular spectrum was different across the phenotypes. 26656045_Collagen type II loss is induced by the down-regulation of miR-133a in intervertebral disc degeneration. 26709265_COL2A1 defects in OSTL1 are not confined to mutations in exon 2, c.2678dupC (p.Ala895Serfs*49) and c.3327+ 1G>C. 27059630_We identified three novel heterozygous COL2A1 mutations (Gly537Asp, Gly909Ser, and Gly1149Val) in three unrelated Chinese Spondyloepiphyseal dysplasia congenita families. 27109135_Serum CTX-II levels in human brucellosis were higher than those of healthy controls but serum CTX-II levels in male patients were significantly higher than those of female patients indicating biological changes in cartilage and bone in human brucellosis. 27183340_analysis of the COL2A1 mutation in idiopathic osteonecrosis of the femoral head among patients from 22 Japanese hospitals 27310669_results highlight the contribution of ELF3 to transcriptional regulation of COL2A1 27390512_In this study, three novel and two known mutations in the COL2A1 gene were identified in six of 16 Chinese patients with Stickler syndrome. This is the first study in a cohort of Chinese patients with Stickler syndrome, and the results expand the mutation spectrum of the COL2A1 gene. 27406592_Report examines how a COL2A1 intron 2 de novo variant and polymorphism affect exon 2 inclusion in the COL2A1 transcript, and identifies potential transacting splicing factors that interact with these different pre-mRNA sequences. Also, using a cohort of patients with rhegmatogenous retinal detachment and controls, a significant difference was found in the frequency of the COL2A1 variant rs1635532 between the two groups. 27428952_Decrease in mRNA expression of COL2A1 is associated with Osteoarthritis. 27569273_Unique charge-dependent constraints on collagen recognition by integrin alpha10beta1 have been described. 27881681_These findings suggest a novel mechanism of action of SOX5/6; namely, the SOX9/5/6 combination enhances Col2a1 transcription through a novel enhancer in intron 6 together with the enhancer in intron 1. 27888646_Spondylometaphyseal dysplasia (SMD) corner fracture type is a heterogeneous disorder with a subset of patients showing overlap with type II collagenopathies. Thus, COL2A1 molecular testing should be considered in patients with the finding of corner fracture-like lesions in the settings of SMD. 27991836_The genotype and allele frequencies of the COL2A1 genetic polymorphisms (rs1793953 and rs2276454) and the Aggrecan VNTR polymorphisms differed significantly between the case group and the control group.the genotype and allele frequencies of the COL2A1 genes, rs1793953 and rs2276454, and Aggrecan VNTR significantly differed in terms of Pfirrmann grades III, IV, and V 28059113_Results suggest that COL2A1 was a likely susceptibility gene of Kashin-Beck disease (KBD). COL2A1 may be implicated in the growth and development failure of hand of KBD. 28095098_We report a novel nonsense mutation in exon 2 of COL2A1 that displays incomplete penetrance and/or variable age of onset with extraocular manifestations. 28302318_Femoral and tibial cartilage show a different behaviour concerning expression values for the genes Col1A1, Col2A1 and Agg. 28559201_High type II collagen expression is associated with intervertebral disc degeneration. 28612031_By acting probably as a posttranscriptional regulator with a different efficacy on COL2A1 and COL1A2 expression, miR-29b can contribute to the collagens imbalance associated with an abnormal chondrocyte phenotype. 28728848_Endoplasmic reticulum stress participates in the progress of senescence and apoptosis of osteoarthritic chondrocytes, which manifested in increased expression of ADAMTS5, MMP13, and decreased COL2A1 expression. 28738883_A c.G1636A (p.G546S) mutation in the COL2A1 in 3 members of a family was associated with different metaphyseal changes. Findings revealed a different causative amino acid substitution (glycine to serine) associated with the 'dappling' and 'corner fracture' metaphyseal abnormalities and may provide a useful reference for evaluating the phenotypic spectrum and variability of type II collagenopathies. 28961166_In summary, regarding the association between Type II Collagen Degradation Marker (CTx-II) and VDR polymorphisms in patients with osteochondrosis, this study observed the presence of higher CTx-II circulating levels in patients with bb, Aa ,and TT genotypes, and F and T alleles, in comparison with the healthy controls. 29068597_Strong correlations between the expression of type I, II, IV collagen and osteopontin and the clinical stage of tympanosclerosis indicate the involvement of these proteins in excessive fibrosis and pathological remodeling of the tympanic membrane. 29104872_the novel mutation of COMP may result in intracellular accumulation of the mutant protein. Decreased plasma COMP and increased plasma CTX-II may potentially serve as diagnostic markers of PSACH but may not be applicable in the presymptomatic carrier. 29439465_Retention of misfolded R740C and R789C proteins triggered an ER stress response. R740C and R789C proteins displayed significantly reduced melting temperatures. 29750297_COL2A1 mutation (c.3508G>A) leads to avascular necrosis of the femoral head in a Chinese family. 30015854_The findings of the current study expand the established mutation spectrum of COL2A1, and may facilitate genetic counseling and development of therapeutic strategies for patients with Stickler syndrome. 30130436_Mutation in COL2A1 gene is associated with Retinal detachment and infantile-onset glaucoma in Stickler syndrome. 30181686_In summary, it is not easy to differentiate stickler syndrome (STL) from early-onset high myopia with routine ocular examination in outpatient clinics. Awareness of atypical phenotypes and newly recognized signs may be of help in identifying atypical STL, especially in children at eye clinics. 30187277_Umbilical cord mesenchymal stem cell conditioned medium resumed the collagen II and aggrecan expression in nucleus pulposus mesenchymal stem cells. 30740902_COL2A1 mutation is associated with multiple epiphyseal dysplasia. 31302913_novel mutation in COL2A1 identified in a pedigree affected with spondyloepiphyseal dysplasia congenita 31486735_VEGFA and COL2A1 polymorphisms are significantly associated with the response to inhaled | ENSMUSG00000022483 | Col2a1 | 2.082294e+03 | 1.1165567 | 0.159056458 | 0.3117420 | 2.593781e-01 | 0.6105477058 | 0.91255290 | No | Yes | 1.831960e+03 | 194.061645 | 1.657100e+03 | 180.286715 | |
| ENSG00000139233 | 84298 | LLPH | protein_coding | Q9BRT6 | FUNCTION: In hippocampal neurons, regulates dendritic and spine growth and synaptic transmission. {ECO:0000250|UniProtKB:Q9D945}. | 3D-structure;Chromosome;Isopeptide bond;Nucleus;Reference proteome;Ubl conjugation | hsa:84298; | chromosome [GO:0005694]; nucleolus [GO:0005730]; basal RNA polymerase II transcription machinery binding [GO:0001099]; RNA binding [GO:0003723]; dendrite extension [GO:0097484]; positive regulation of dendritic spine development [GO:0060999] | 33583303_Differential levels of CHMP2B, LLPH, and SLC25A51 proteins in secondary renal amyloidosis. | ENSMUSG00000020224 | Llph | 3.694212e+02 | 1.0195196 | 0.027889501 | 0.3562367 | 5.942362e-03 | 0.9385545610 | 0.98870361 | No | Yes | 3.613178e+02 | 70.666064 | 2.960695e+02 | 59.564278 | ||
| ENSG00000139437 | 84260 | TCHP | protein_coding | Q9BT92 | FUNCTION: Tumor suppressor which has the ability to inhibit cell growth and be pro-apoptotic during cell stress. Inhibits cell growth in bladder and prostate cancer cells by a down-regulation of HSPB1 by inhibiting its phosphorylation. May act as a 'capping' or 'branching' protein for keratin filaments in the cell periphery. May regulate K8/K18 filament and desmosome organization mainly at the apical or peripheral regions of simple epithelial cells (PubMed:15731013, PubMed:18931701). Is a negative regulator of ciliogenesis (PubMed:25270598). {ECO:0000269|PubMed:15731013, ECO:0000269|PubMed:18931701, ECO:0000269|PubMed:25270598}. | Apoptosis;Cell junction;Cell membrane;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Isopeptide bond;Membrane;Mitochondrion;Reference proteome;Tumor suppressor;Ubl conjugation | hsa:84260; | apical cortex [GO:0045179]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; desmosome [GO:0030057]; keratin filament [GO:0045095]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; apoptotic process [GO:0006915]; cell projection organization [GO:0030030]; negative regulation of cell growth [GO:0030308]; negative regulation of cilium assembly [GO:1902018] | 15731013_trichoplein is a keratin 8/18-binding protein that may be involved in the organization of the apical network of keratin filaments and desmosomes in simple epithelial cells 18931701_MITOSTATIN was found within a 3.2-kb transcript for an approximately 62 kDa mitochondrial protein with tumor suppressor activity. It inhibits cell growth, is proapoptotic and downregulates Hsp27. 20930847_Trichoplein/mitostatin is a new regulator of mitochondria-endoplasmic reticulum juxtaposition. 21325031_Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. 24403067_These findings underscore the complexity of PGC-1alpha-mediated mitochondrial homeostasis and establish mitostatin as a key regulator of tumor cell mitophagy and angiostasis. 26880200_Ndel1 acts as a novel upstream regulator of the trichoplein-Aurora A pathway to inhibit primary cilia assembly. 29080840_Soluble matrix-derived cues being transduced downstream of receptor engagement converge upon a newly-discovered nexus of autophagic machinery consisting of Peg3 for endothelial cell autophagy and mitostatin for tumor cell mitophagy. | 7.683169e+02 | 1.0620279 | 0.086821618 | 0.2622279 | 1.102213e-01 | 0.7398923142 | 0.94575143 | No | Yes | 6.997025e+02 | 64.610008 | 6.731179e+02 | 63.874922 | ||||
| ENSG00000139496 | 9818 | NUP58 | protein_coding | Q9BVL2 | FUNCTION: Component of the nuclear pore complex, a complex required for the trafficking across the nuclear membrane. {ECO:0000250|UniProtKB:P70581}. | 3D-structure;Alternative splicing;Coiled coil;Glycoprotein;Membrane;Nuclear pore complex;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Translocation;Transport;mRNA transport | Mouse_homologues NA; + ;NA | This gene encodes a member of the nucleoporin family that shares 87% sequence identity with rat nucleoporin p58. The protein is localized to the nuclear rim and is a component of the nuclear pore complex (NPC). All molecules entering or leaving the nucleus either diffuse through or are actively transported by the NPC. Alternate transcriptional splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jul 2008]. | hsa:9818; | nuclear envelope [GO:0005635]; nuclear inner membrane [GO:0005637]; nuclear outer membrane [GO:0005640]; nuclear pore [GO:0005643]; identical protein binding [GO:0042802]; nuclear localization sequence binding [GO:0008139]; protein-containing complex binding [GO:0044877]; structural constituent of nuclear pore [GO:0017056]; mRNA transport [GO:0051028]; protein transport [GO:0015031]; regulation of protein import into nucleus [GO:0042306] | 7531196_The human gene NUPL1 shares 87% sequence identity with rat nucleoporin p58. 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that NUP58 is a putative expression variability marker for the 3-day, 8-cell embryo stage. 34528284_Non-genetic and genetic rewiring underlie adaptation to hypomorphic alleles of an essential gene. | ENSMUSG00000114797+ENSMUSG00000063895 | Gm49336+Nupl1 | 6.424453e+02 | 0.6278213 | -0.671574055 | 0.3460496 | 3.608835e+00 | 0.0574733681 | 0.68581629 | No | Yes | 4.532843e+02 | 88.297268 | 7.383193e+02 | 146.915411 |
| ENSG00000139631 | 51380 | CSAD | protein_coding | Q9Y600 | FUNCTION: Catalyzes the decarboxylation of L-aspartate, 3-sulfino-L-alanine (cysteine sulfinic acid), and L-cysteate to beta-alanine, hypotaurine and taurine, respectively. The preferred substrate is 3-sulfino-L-alanine. Does not exhibit any decarboxylation activity toward glutamate. {ECO:0000250|UniProtKB:Q9DBE0}. | 3D-structure;Alternative splicing;Decarboxylase;Lyase;Pyridoxal phosphate;Reference proteome | PATHWAY: Organosulfur biosynthesis; taurine biosynthesis; hypotaurine from L-cysteine: step 2/2. | This gene encodes a member of the group 2 decarboxylase family. A similar protein in rodents plays a role in multiple biological processes as the rate-limiting enzyme in taurine biosynthesis, catalyzing the decarboxylation of cysteinesulfinate to hypotaurine. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Sep 2011]. | hsa:51380; | cytoplasm [GO:0005737]; aspartate 1-decarboxylase activity [GO:0004068]; pyridoxal phosphate binding [GO:0030170]; sulfinoalanine decarboxylase activity [GO:0004782]; L-cysteine catabolic process to hypotaurine [GO:0019449]; L-cysteine catabolic process to taurine [GO:0019452]; taurine biosynthetic process [GO:0042412] | 22718265_The presence of cysteine inhibited ADC, CSAD and GDC activity. 26327310_taurine biosynthesis in vertebrates involves two structurally related PLP-dependent decarboxylases (cysteine sulfinic acid decarboxylase and glutamic acid decarboxylase like 1) | ENSMUSG00000023044 | Csad | 3.952574e+02 | 0.7568225 | -0.401973113 | 0.3140425 | 1.632521e+00 | 0.2013546931 | 0.78445471 | No | Yes | 2.419937e+02 | 56.635854 | 3.914228e+02 | 93.561098 |
| ENSG00000139651 | 283337 | ZNF740 | protein_coding | Q8NDX6 | FUNCTION: May be involved in transcriptional regulation. | Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:283337; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000046897 | Zfp740 | 3.291594e+03 | 0.8285650 | -0.271313205 | 0.2928671 | 8.638278e-01 | 0.3526697113 | 0.82969965 | No | Yes | 2.471696e+03 | 253.014977 | 3.173754e+03 | 332.759391 | |||
| ENSG00000139746 | 64062 | RBM26 | protein_coding | Q5T8P6 | Acetylation;Alternative splicing;Coiled coil;Isopeptide bond;Metal-binding;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ubl conjugation;Zinc;Zinc-finger | hsa:64062; | nucleus [GO:0005634]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; mRNA processing [GO:0006397] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 31950173_three additional proteins required for Poly(A) Tail eXosome Targeting (PAXT) function: ZC3H3, RBM26 and RBM27 along with the known PAXT-associated protein, PABPN1, were identified. | ENSMUSG00000022119 | Rbm26 | 2.074658e+03 | 1.3061949 | 0.385370206 | 0.3105042 | 1.549453e+00 | 0.2132161776 | 0.78763590 | No | Yes | 2.729867e+03 | 504.346457 | 1.708078e+03 | 323.900515 | |||
| ENSG00000139926 | 122786 | FRMD6 | protein_coding | Q96NE9 | Alternative splicing;Cell membrane;Cytoplasm;Membrane;Phosphoprotein;Reference proteome | hsa:122786; | apical junction complex [GO:0043296]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; plasma membrane [GO:0005886]; actomyosin structure organization [GO:0031032]; apical constriction [GO:0003383]; cellular protein localization [GO:0034613]; regulation of actin filament-based process [GO:0032970] | 16137681_Results describe a new FERM domain-containing protein called Willin. 21666719_in mammalian cells willin influences Hippo signaling activity by activating the core Hippo pathway kinase cassette 21785462_Human homolog of Drosophila expanded, hEx, functions as a putative tumor suppressor in human cancer cell lines independently of the Hippo pathway 22190428_Genome-wide and gene-based association implicates FRMD6 in Alzheimer disease. 27661120_a novel function of FRMD6 in inhibiting human GBM growth and progression and uncover a novel mechanism by which FRMD6 exerts its anti-GBM activity. 28079891_we report for the first time that CRB3 acts as an upstream regulator of the Hippo pathway to regulate contact inhibition by recruiting other Hippo molecules, such as Kibra and/or FRMD6, in mammary epithelial cells. 31128910_Results indicate that metastasis associated 1 family member 2 (MTA2) represses a cohort of genes including FERM domain containing 6 protein (FRMD6) that are critically involved in the growth and mobility of hepatocellular carcinoma (HCC). 31917993_concluded that lncRNA FRMD6-AS2 repressed uterine corpus endometrial carcinoma, at least in part, by increasing FRMD6 33249427_FRMD6 has tumor suppressor functions in prostate cancer. | ENSMUSG00000048285 | Frmd6 | 9.959661e+01 | 1.0054230 | 0.007802549 | 0.4442294 | 3.130217e-04 | 0.9858842279 | 0.99716006 | No | Yes | 1.211066e+02 | 30.363284 | 8.934080e+01 | 23.172834 | |||
| ENSG00000140043 | 145482 | PTGR2 | protein_coding | Q8N8N7 | FUNCTION: Functions as 15-oxo-prostaglandin 13-reductase and acts on 15-keto-PGE1, 15-keto-PGE2, 15-keto-PGE1-alpha and 15-keto-PGE2-alpha with highest activity towards 15-keto-PGE2 (PubMed:19000823). Overexpression represses transcriptional activity of PPARG and inhibits adipocyte differentiation (By similarity). {ECO:0000250|UniProtKB:Q8VDQ1, ECO:0000269|PubMed:19000823}. | 3D-structure;Alternative splicing;Cytoplasm;Lipid metabolism;NADP;Oxidoreductase;Reference proteome | This gene encodes an enzyme involved in the metabolism of prostaglandins. The encoded protein catalyzes the NADPH-dependent conversion of 15-keto-prostaglandin E2 to 15-keto-13,14-dihydro-prostaglandin E2. This protein may also be involved in regulating activation of the peroxisome proliferator-activated receptor. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2009]. | hsa:145482; | cytoplasm [GO:0005737]; 13-prostaglandin reductase activity [GO:0036132]; 15-oxoprostaglandin 13-oxidase activity [GO:0047522]; prostaglandin metabolic process [GO:0006693] | 15004468_Cloning and tissue distribution of a splicing variant ZADH1b. 19000823_Besides targeting cyclooxygenase, indomethacin inhibits PTGR2 with a binding mode similar to that of 15-keto-PGE(2). 20877624_Observational study of gene-disease association. (HuGE Navigator) 22998775_Functional data and clinical relevance for the role of PTGR2 in gastric cancer, are provided. 26820738_Silencing of PTGR2 expression enhances reactive oxygen species production, suppresses pancreatic cell proliferation, and promotes cell death through increasing 15-keto-PGE2. | ENSMUSG00000072946 | Ptgr2 | 1.145015e+02 | 0.8679290 | -0.204351038 | 0.3774978 | 2.907691e-01 | 0.5897280655 | 0.90598796 | No | Yes | 7.513315e+01 | 14.350390 | 9.757371e+01 | 19.063440 | |
| ENSG00000140044 | 122953 | JDP2 | protein_coding | Q8WYK2 | FUNCTION: Component of the AP-1 transcription factor that represses transactivation mediated by the Jun family of proteins. Involved in a variety of transcriptional responses associated with AP-1 such as UV-induced apoptosis, cell differentiation, tumorigenesis and antitumogeneris. Can also function as a repressor by recruiting histone deacetylase 3/HDAC3 to the promoter region of JUN. May control transcription via direct regulation of the modification of histones and the assembly of chromatin. {ECO:0000269|PubMed:12707301, ECO:0000269|PubMed:12903123, ECO:0000269|PubMed:16026868, ECO:0000269|PubMed:16518400}. | Alternative splicing;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | hsa:122953; | chromatin [GO:0000785]; nucleus [GO:0005634]; cAMP response element binding [GO:0035497]; chromatin binding [GO:0003682]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; leucine zipper domain binding [GO:0043522]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of histone deacetylation [GO:0031065]; regulation of transcription by RNA polymerase II [GO:0006357] | 14627710_c-Jun dimerization protein 2 inhibits cell transformation and has a role as a tumor suppressor gene 16026868_JDP2 is a cellular survival protein whose presence is necessary for normal cellular function 18396163_JDP2 acts as a repressor and could be functionally associated with HDAC3 to inhibit CHOP transcription 18671972_IRF2-BP1 is a JDP2-binding protein enhancing the polyubiquitination of JDP2 and represses ATF2-mediated transcriptional activation from a CRE-containing promoter. 19553667_A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. 20452405_3 SNPs (2 intronic: rs741846 & rs175646; & 1 in the untranslated region: rs8215) & their genotype distribution showed significant association in the Japanese & Korean but not Dutch intracranial aneurysm patients. 20452405_Observational study of gene-disease association. (HuGE Navigator) 20677166_JDP2 expression was downregulated in pancreatic carcinoma & this correlated with metastasis & decreased post-surgery survival. 20950777_The molecular mechanisms that underlie the action of JDP2 in cellular aging and replicative senescence by mediating the dissociation of polycomb repressive complexes from the p16(Ink4a)/Arf locus are discussed. 21525011_JDP2 is crucial to triggering reactivation from latency to lytic replication 22989952_the recruitment of multiple HDAC members to JDP2 and ATF3 is part of their transcription repression mechanism. 24120378_Preeclamptic plasma induces transcription modifications involving the AP-1 transcriptional regulator JDP2 in endothelial cells. 24232097_Results suggest that JDP2 is an integral component of the Nrf2-MafK complex and that it modulates antioxidant and detoxification programs by acting via the ARE. 28315425_In hepatocellular carcinoma, high expression of JDP2 is significantly correlated with smaller tumor size, early stage HCC and better survival. 29941549_These studies establish JDP2 as a novel oncogene in high-risk T cell acute lymphoblastic leukemia. 30721335_JDP2 was downregulated in myelodysplastic syndrome. Its expression was inversely related to disease aggressiveness and AML transformation. JDP2 suppression is a direct result of reduced PU.1. PU.1 and JDP2 expression correlate and are concurrently reduced with the extent of differentiation arrest and aggression/prognosis in MDS/AML. It was upregulated upon azacytidine treatment. 30877624_JDP2 is downregulated in HCC tissues and cells, and overexpressed JDP2 facilitated HCC cell invasion and EMT. 32150333_Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. 33108704_JDP2 is directly regulated by ATF4 and modulates TRAIL sensitivity by suppressing the ATF4-DR5 axis. | ENSMUSG00000034271 | Jdp2 | 1.296947e+02 | 1.8589889 | 0.894518191 | 0.3686674 | 5.929596e+00 | 0.0148886583 | 0.43789671 | No | Yes | 1.329611e+02 | 30.485104 | 6.025424e+01 | 14.394409 | ||
| ENSG00000140105 | 7453 | WARS1 | protein_coding | P23381 | FUNCTION: Isoform 1, isoform 2 and T1-TrpRS have aminoacylation activity while T2-TrpRS lacks it. Isoform 2, T1-TrpRS and T2-TrpRS possess angiostatic activity whereas isoform 1 lacks it. T2-TrpRS inhibits fluid shear stress-activated responses of endothelial cells. Regulates ERK, Akt, and eNOS activation pathways that are associated with angiogenesis, cytoskeletal reorganization and shear stress-responsive gene expression. {ECO:0000269|PubMed:11773625, ECO:0000269|PubMed:11773626, ECO:0000269|PubMed:1373391, ECO:0000269|PubMed:14630953, ECO:0000269|PubMed:28369220}. | 3D-structure;ATP-binding;Alternative splicing;Aminoacyl-tRNA synthetase;Angiogenesis;Cytoplasm;Direct protein sequencing;Disease variant;Ligase;Neurodegeneration;Neuropathy;Nucleotide-binding;Phosphoprotein;Protein biosynthesis;Reference proteome | Aminoacyl-tRNA synthetases catalyze the aminoacylation of tRNA by their cognate amino acid. Because of their central role in linking amino acids with nucleotide triplets contained in tRNAs, aminoacyl-tRNA synthetases are thought to be among the first proteins that appeared in evolution. Two forms of tryptophanyl-tRNA synthetase exist, a cytoplasmic form, named WARS, and a mitochondrial form, named WARS2. Tryptophanyl-tRNA synthetase (WARS) catalyzes the aminoacylation of tRNA(trp) with tryptophan and is induced by interferon. Tryptophanyl-tRNA synthetase belongs to the class I tRNA synthetase family. Four transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:7453; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; ATP binding [GO:0005524]; kinase inhibitor activity [GO:0019210]; protein domain specific binding [GO:0019904]; protein homodimerization activity [GO:0042803]; protein kinase binding [GO:0019901]; tryptophan-tRNA ligase activity [GO:0004830]; angiogenesis [GO:0001525]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of protein kinase activity [GO:0006469]; negative regulation of protein phosphorylation [GO:0001933]; positive regulation of gene expression [GO:0010628]; positive regulation of protein-containing complex assembly [GO:0031334]; regulation of angiogenesis [GO:0045765]; regulation of protein ADP-ribosylation [GO:0010835]; translation [GO:0006412]; tryptophanyl-tRNA aminoacylation [GO:0006436] | 11773625_In this study, we show that a recombinant form of a COOH-terminal fragment of TrpRS is a potent antagonist of vascular endothelial growth factor-induced angiogenesis in a mouse model and of naturally occurring retinal angiogenesis in the neonatal mouse 11773626_Thus, protein synthesis may be linked to the regulation of angiogenesis by a natural fragment of TrpRS 11834741_Recognition by tryptophanyl-tRNA synthetases of discriminator base on tRNATrp from three biological domains 12416978_The recently discovered antiangiogenic and cell-signaling activities of tryptophanyl-tRNA synthetase bioactive fragments are discussed in this review. 14630953_TrpRS may have a role in the maintenance of vascular homeostasis 14660560_results suggest that mammalian and bacterial tryptophanyl-tRNA synthetase might use different mechanisms to recognize the substrate and modeling studies indicate that transfer RNA binds with the dimeric enzyme 14671330_A crystal structure of human tryptophanyl-tRNA synthetase was solved at 2.1 A with a tryptophanyl-adenylate bound at the active site 15628863_May play an important role in the intracellular regulation of protein synthesis under conditions of oxidative stress. 15939065_Observational study of gene-disease association. (HuGE Navigator) 16724112_These crystals captured two conformations of the human tryptophanyl-tRNA synthetase and tRNATrp complex, which are nearly identical with respect to the protein and a bound tryptophan. 16798914_The first crystal structure of human tryptophanyl-tRNA synthetase (hTrpRS) in complex with tRNA(Trp) and Trp which, together with biochemical data, reveals the molecular basis of a novel tRNA binding and recognition mechanism. 17877375_Results provide the first evidence of the involvement of heme in regulation of TrpRS aminoacylation activity. 17999956_the annexin II-S100A10 complex, which regulates exocytosis, forms a ternary complex with TrpRS. 18180246_Analysis of the molecular basis of the mechanisms of the substrate recognition and the activation reaction by tryptophanyl-tRNA synthetase. 19363598_Indoleamine 2,3-dioxygenase (IDO)-expression in antigen-presenting cells (APCs) may control autoimmune responses by depleting the available tryptophan, whereas tryptophanyl-tRNA synthetase (TTS) may counteract this effect 19768679_Tryptophanyl-tRNA synthetase is a multidomain protein exhibiting excellent allosteric communication, and this research has provided valuable structural as well as functional insights into the protein. 19900940_Low tryptophanyl-tRNA synthetase is associated with recurrence in colorectal cancer. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20963594_Mini-tryptophanyl-tRNA synthetase inhibited ischemic angiogenesis in rats. 21442253_Naturally occurring fragments of the two proteins involved in translation, TyrRS and TrpRS, have opposing activities on angiogenesis. 21926542_Tryptophanyl-tRNA synthetase down-regulation by hypoxia may be a factor responsible for low TrpRS in pancreatic tumors with high metastatic ability. 23651343_Indoleamine2,3-dioxygenase and tryptophanyl-tRNA synthetase may play critical roles in the immune pathogenesis of chronic kidney disease. 23670221_Genes within recently identified loci associated with waist-hip ratio (WHR) exhibit fat depot-specific mRNA expression, which correlates with obesity-related traits. Adipose tissue (AT) mRNA expression of 6 genes (TBX15/WARS2, STAB1, PIGC, ZNRF3, GRB14) 24515434_Tryptophanyl-tRNA synthetase expression is up-regulated in patients with rheumatoid arthritis. 26209610_Overexpression of WARS predicts no recurrence and good survival for triple-negative breast cancer patients. 27748732_Based on these results, secretion of full-length tryptophanyl-tRNA synthetase appears to work as a primary defence system against infection, acting before full activation of innate immunity. 28369220_findings establish WARS as a gene whose mutations may cause distal hereditary motor neuropathy and alter canonical and non-canonical functions of tryptophanyl-tRNA synthetase. 29666190_Inhibition of human tryptophanyl-tRNA synthetase (TrpRS) expression by TrpRS-specific siRNAs decreased and overexpression of TrpRS increased Trp uptake into the cells 30153112_Human tryptophanyl-tRNA synthetase is an IFN-gamma-inducible entry factor for Enterovirus. 30355684_Data report a novel noncanonical function of WARS in antiviral defense. WARS is rapidly secreted in response to viral infection and primes the innate immune response by inducing the secretion of proinflammatory cytokines and type I IFNs, resulting in the inhibition of virus replication both in vitro and in vivo. These results suggest that WARS is a member of the antiviral innate immune response. 31033497_Expression of Indoleamine 2, 3-dioxygenase 1 (IDO1) and Tryptophanyl-tRNA Synthetase (WARS) in Gastric Cancer Molecular Subtypes. 31786502_Mini tryptophanyl-tRNA synthetase is required for a synthetic phenotype in vascular smooth muscle cells induced by IFN-gamma-mediated beta2-adrenoceptor signaling. 32899943_Tryptophanyl-tRNA Synthetase 1 Signals Activate TREM-1 via TLR2 and TLR4. 33754909_Identification of Prognostic RBPs in Osteosarcoma. | ENSMUSG00000021266 | Wars | 4.792703e+03 | 0.9104430 | -0.135359411 | 0.2733287 | 2.500416e-01 | 0.6170457546 | 0.91387240 | No | Yes | 4.391741e+03 | 417.342057 | 4.370671e+03 | 425.918156 | |
| ENSG00000140319 | 6727 | SRP14 | protein_coding | P37108 | FUNCTION: Component of the signal recognition particle (SRP) complex, a ribonucleoprotein complex that mediates the cotranslational targeting of secretory and membrane proteins to the endoplasmic reticulum (ER) (PubMed:11089964). SRP9 together with SRP14 and the Alu portion of the SRP RNA, constitutes the elongation arrest domain of SRP (PubMed:11089964). The complex of SRP9 and SRP14 is required for SRP RNA binding (PubMed:11089964). {ECO:0000269|PubMed:11089964}. | 3D-structure;Cytoplasm;Direct protein sequencing;Phosphoprotein;RNA-binding;Reference proteome;Ribonucleoprotein;Signal recognition particle | hsa:6727; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; nucleus [GO:0005634]; secretory granule lumen [GO:0034774]; signal recognition particle, endoplasmic reticulum targeting [GO:0005786]; 7S RNA binding [GO:0008312]; endoplasmic reticulum signal peptide binding [GO:0030942]; RNA binding [GO:0003723]; cotranslational protein targeting to membrane [GO:0006613]; protein targeting to ER [GO:0045047]; SRP-dependent cotranslational protein targeting to membrane [GO:0006614] | 18455985_SRP14 mutant proteins with absence of a peptide elongation delay caused inefficient targeting of preproteins leading to defects in secretion, depletion of proteins in the endogenous membranes, and reduced cell growth. 20348448_mutational study on SRP9/14 that identified and characterized regions and single residues essential for elongation arrest activity 25697503_Dats show that Alu RNA promotes functional binding of Signal Recognition Particle proteins SRP9/14 to 40S ribosomal subunits. | ENSMUSG00000009549 | Srp14 | 3.662138e+03 | 0.5430146 | -0.880937149 | 0.2569542 | 1.136412e+01 | 0.0007487662 | 0.09944063 | No | Yes | 2.558911e+03 | 276.877175 | 4.502062e+03 | 498.422863 | ||
| ENSG00000140374 | 2108 | ETFA | protein_coding | P13804 | FUNCTION: Heterodimeric electron transfer flavoprotein that accepts electrons from several mitochondrial dehydrogenases, including acyl-CoA dehydrogenases, glutaryl-CoA and sarcosine dehydrogenase (PubMed:27499296, PubMed:15159392, PubMed:15975918, PubMed:9334218, PubMed:10356313). It transfers the electrons to the main mitochondrial respiratory chain via ETF-ubiquinone oxidoreductase (ETF dehydrogenase) (PubMed:9334218). Required for normal mitochondrial fatty acid oxidation and normal amino acid metabolism (PubMed:12815589, PubMed:1882842, PubMed:1430199). {ECO:0000269|PubMed:10356313, ECO:0000269|PubMed:12815589, ECO:0000269|PubMed:1430199, ECO:0000269|PubMed:15159392, ECO:0000269|PubMed:15975918, ECO:0000269|PubMed:27499296, ECO:0000269|PubMed:9334218, ECO:0000303|PubMed:17941859, ECO:0000305|PubMed:1882842}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Electron transport;FAD;Flavoprotein;Glutaricaciduria;Mitochondrion;Phosphoprotein;Reference proteome;Transit peptide;Transport | ETFA participates in catalyzing the initial step of the mitochondrial fatty acid beta-oxidation. It shuttles electrons between primary flavoprotein dehydrogenases and the membrane-bound electron transfer flavoprotein ubiquinone oxidoreductase. Defects in electron-transfer-flavoprotein have been implicated in type II glutaricaciduria in which multiple acyl-CoA dehydrogenase deficiencies result in large excretion of glutaric, lactic, ethylmalonic, butyric, isobutyric, 2-methyl-butyric, and isovaleric acids. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:2108; | electron transfer flavoprotein complex [GO:0045251]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; electron transfer activity [GO:0009055]; flavin adenine dinucleotide binding [GO:0050660]; oxidoreductase activity [GO:0016491]; cellular amino acid catabolic process [GO:0009063]; fatty acid beta-oxidation using acyl-CoA dehydrogenase [GO:0033539]; respiratory electron transport chain [GO:0022904] | 11756429_These studies indicate that a series of conformational changes occur during the assembly of the TMADH.ETF electron transfer complex and that the kinetics of assembly observed with mutant TMADH or ETF complexes are much slower 16510302_Tissue samples from 16 unrelated patients with ETF deficiency were analysed and the majority of the patients had mutations in the ETFA gene. 17689999_No mutations in electron-transfer-flavoprotein but maternal riboflavin deficiency led to multiple acyl-CoA dehydrogenation deficiency 19208393_Observational study of gene-disease association. (HuGE Navigator) 20674745_Data established structural hotspots within the ETF fold, and provided a rationale for the prediction of effects of mutations in ETF. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21219902_investigations are compatible with the notion that the ETFalpha-T171 variant displays an altered conformational landscape that results in reduced protein function under thermal stress 21308847_These results are consistent with the electron transfer flavoprotein alpha II domain adopting orientations in solution that deviate from the crystal structure of free ETF towards the active, substrate-bound orientation. 24394546_the mechanism of tert-butyl hydroperoxide-induced an apoptosis cascade and endoplasmic reticulum stress in hepatocyte cells by up-regulation of ETFA, providing a new mechanism for liver injury. 28320150_our results indicate that genetic variants in ETFA may modify individual susceptibility to non-GBM of glioma in the Han Chinese population and support the role of the ETFA genes in the occurrence of glioma. 29301933_Data suggest that ETF (heterodimer of ETFA and ETFB) catalyzes irreversible and pH-dependent oxidation of 8alpha-methyl group of FAD to form to 8-formyl-FAD (8f-FAD). 31418342_Molecular and Clinical Investigations on Portuguese Patients with Multiple acyl-CoA Dehydrogenase Deficiency. 31665600_Molecular Oxygen Binding in the Mitochondrial Electron Transfer Flavoprotein. 31996215_A case report of a mild form of multiple acyl-CoA dehydrogenase deficiency due to compound heterozygous mutations in the ETFA gene. 34405537_NBPF4 mitigates progression in colorectal cancer through the regulation of EZH2-associated ETFA. 34704421_Screening of multiple acyl-CoA dehydrogenase deficiency in newborns and follow-up of patients. | ENSMUSG00000032314 | Etfa | 1.932982e+03 | 0.7427118 | -0.429125627 | 0.3072937 | 1.968996e+00 | 0.1605545191 | 0.77593452 | No | Yes | 1.596118e+03 | 280.436537 | 2.127251e+03 | 382.950306 | |
| ENSG00000140391 | 10099 | TSPAN3 | protein_coding | O60637 | FUNCTION: Regulates the proliferation and migration of oligodendrocytes, a process essential for normal myelination and repair. {ECO:0000250}. | Alternative splicing;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of four hydrophobic domains. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. The use of alternate polyadenylation sites has been found for this gene. Multiple alternative transcripts encoding different isoforms have been described. [provided by RefSeq, Dec 2009]. | hsa:10099; | extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887] | 26212080_Tspan3 is an important regulator of aggressive leukemias. 26930362_Studies indicate that tetraspanin 3 has a critical role in acute myeloid leukemia. 27818272_Tspan3 is a central endocytic membrane component regulating the expression of ADAM10, presenilin and the amyloid precursor protein. 30367526_study concluded that miR-139-5p suppressed the leukemogenesis in acute myeloid leukemia (AML) cells by targeting Tspan3 through inactivation of the PI3K/Akt pathway, providing a better understanding of AML progression. 31837329_High TSPAN3 expression is associated with progression and chemoresistance in acute myeloid leukemia. | ENSMUSG00000032324 | Tspan3 | 1.197951e+04 | 0.6877275 | -0.540091047 | 0.2761322 | 3.874882e+00 | 0.0490137443 | 0.66791998 | No | Yes | 8.012747e+03 | 840.431559 | 1.226794e+04 | 1318.514573 | |
| ENSG00000140455 | 9960 | USP3 | protein_coding | Q9Y6I4 | FUNCTION: Hydrolase that deubiquitinates monoubiquitinated target proteins such as histone H2A and H2B. Required for proper progression through S phase and subsequent mitotic entry. May regulate the DNA damage response (DDR) checkpoint through deubiquitination of H2A at DNA damage sites. Associates with the chromatin. {ECO:0000269|PubMed:17980597}. | Acetylation;Alternative splicing;Cell cycle;Chromatin regulator;DNA damage;Hydrolase;Metal-binding;Nucleus;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway;Zinc;Zinc-finger | hsa:9960; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytoplasmic ribonucleoprotein granule [GO:0036464]; Flemming body [GO:0090543]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; histone binding [GO:0042393]; thiol-dependent deubiquitinase [GO:0004843]; zinc ion binding [GO:0008270]; chromatin organization [GO:0006325]; DNA repair [GO:0006281]; histone deubiquitination [GO:0016578]; mitotic cell cycle [GO:0000278]; regulation of protein stability [GO:0031647]; ubiquitin-dependent protein catabolic process [GO:0006511] | 17980597_Ubiquitin-specific protease 3 (USP3) is a deubiquitinating enzyme for monoubiquitinated histones H2A and H2B; USP3 dynamically associates with chromatin and deubiquitinates H2A/H2B in vivo. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20445134_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 22743239_the role of UCP3 gene in functional status and survival at old age. 24196443_Taken together, the results suggested that USP3 is a negative regulator of ubiquitination signaling, counteracting RNF168- and RNF8-mediated ubiquitination. 28655924_as a direct target of miR-224, USP3 is a potent ceRNA of SMAD4 in CRC. Decreased expression of USP3 mRNA is associated with a poor prognosis and distal metastasis of CRC, which is attributed to further suppression of SMAD4 through the release of miR-224 from the USP3 3'UTR. 28807825_Depletion of USP3 lead to accelerated degradation of p53 in normal cells thereby enhanced cell proliferation and transformation. Reconstitution of wildtype USP3, but not the USP3 C168S mutant, restored the stability of p53 protein and inhibited cell proliferation and transformation. 30168892_Data found that USP3 was overexpressed in gastric cancer (GC) tissues and cells. USP3 overexpression can be a useful biomarker for predicting the outcomes of GC patients. Moreover, USP3 was shown to influence cell proliferation by regulating cell cycle control and metastasis-related proteins. In vivo experiments showed that USP3 promoted GC tumor growth and metastasis. 31056254_study mechanistically links USP3 to TRAF6 in osteoarthritis development; moreover, data support the pursuit of USP3 and TRAF6 as potential targets for osteoarthritis therapies 31234902_SUZ12 is indispensable for USP3-mediated oncogenic activity in gastric cancer . 31624151_USP3 is a new KLF5 deubiquitinase and that USP3 may represent a potential therapeutic target for breast cancer. 31900278_Smoothened Promotes Glioblastoma Radiation Resistance Via Activating USP3-Mediated Claspin Deubiquitination. 32271432_USP3 promotes proliferation of non-small cell lung cancer through regulating RBM4. 32376451_Upregulation of USP3 or downregulation of miR-224-5p restored proliferation and migration by MKN-28 and MGC-803 cells after hsa_circ_0017639 silencing. Upregulation of USP3 restored MKN-28 and MGC-803 cell proliferation and migration after overexpression of miR-224-5p. 32415280_Genome-scale screening of deubiquitinase subfamily identifies USP3 as a stabilizer of Cdc25A regulating cell cycle in cancer. 32918360_LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. 33767157_An ALYREF-MYCN coactivator complex drives neuroblastoma tumorigenesis through effects on USP3 and MYCN stability. 34070420_Ubiquitin-Specific Protease 3 Deubiquitinates and Stabilizes Oct4 Protein in Human Embryonic Stem Cells. 34758762_Protein deubiquitylase USP3 stabilizes Aurora A to promote proliferation and metastasis of esophageal squamous cell carcinoma. 34917199_MicroRNA-146-5p Promotes Pulmonary Artery Endothelial Cell Proliferation under Hypoxic Conditions through Regulating USP3. 34930901_USP3 promotes gastric cancer progression and metastasis by deubiquitination-dependent COL9A3/COL6A5 stabilisation. | ENSMUSG00000032376 | Usp3 | 3.090924e+02 | 0.8273228 | -0.273477703 | 0.3250403 | 7.105163e-01 | 0.3992724720 | 0.84485965 | No | Yes | 2.522079e+02 | 42.849776 | 2.970671e+02 | 51.580560 | ||
| ENSG00000140632 | 84656 | GLYR1 | protein_coding | Q49A26 | FUNCTION: Nucleosome-destabilizing factor that is recruited to genes during transcriptional activation (PubMed:29759984). Facilitates Pol II transcription through nucleosomes (PubMed:29759984). Binds DNA (in vitro) (PubMed:29759984). Recognizes and binds trimethylated 'Lys-36' of histone H3 (H3K36me3) (PubMed:20850016). Promotes KDM1B demethylase activity (PubMed:23260659). Stimulates the acetylation of 'Lys-56' of nucleosomal histone H3 (H3K56ac) by EP300 (PubMed:29759984). Regulates p38 MAP kinase activity by mediating stress activation of p38alpha/MAPK14 and specifically regulating MAPK14 signaling (PubMed:16352664). Indirectly promotes phosphorylation of MAPK14 and activation of ATF2 (PubMed:16352664). The phosphorylation of MAPK14 requires upstream activity of MAP2K4 and MAP2K6 (PubMed:16352664). Putative oxidoreductase (PubMed:23260659). {ECO:0000269|PubMed:16352664, ECO:0000269|PubMed:20850016, ECO:0000269|PubMed:23260659, ECO:0000269|PubMed:29759984}. | 3D-structure;Alternative splicing;Chromosome;DNA-binding;Isopeptide bond;NAD;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:84656; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone binding [GO:0042393]; methylated histone binding [GO:0035064]; NAD binding [GO:0051287]; NADP binding [GO:0050661]; nucleosome binding [GO:0031491]; oxidoreductase activity [GO:0016491]; positive regulation of histone acetylation [GO:0035066]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 30970244_While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. 32370786_Downregulation of GLYR1 contributes to microsatellite instability colorectal cancer by targeting p21 via the p38MAPK and PI3K/AKT pathways. | ENSMUSG00000022536 | Glyr1 | 5.243319e+03 | 1.0028588 | 0.004118481 | 0.2489246 | 2.760318e-04 | 0.9867443923 | 0.99716006 | No | Yes | 4.569569e+03 | 324.483684 | 4.713806e+03 | 343.206994 | ||
| ENSG00000140832 | 91862 | MARVELD3 | protein_coding | Q96A59 | FUNCTION: As a component of tight junctions, plays a role in paracellular ion conductivity. {ECO:0000269|PubMed:20028514}. | Alternative splicing;Cell junction;Membrane;Reference proteome;Tight junction;Transmembrane;Transmembrane helix | hsa:91862; | bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; integral component of membrane [GO:0016021]; mitogen-activated protein kinase kinase kinase binding [GO:0031435]; bicellular tight junction assembly [GO:0070830]; cell-cell junction organization [GO:0045216]; negative regulation of epithelial cell migration [GO:0010633]; negative regulation of epithelial cell proliferation [GO:0050680]; negative regulation of JNK cascade [GO:0046329]; protein localization to cell junction [GO:1902414]; response to osmotic stress [GO:0006970] | 20028514_MarvelD3 co-localises with occludin at tight junctions in intestinal and corneal epithelial cells. 20164257_marvelD3, occludin, and tricellulin define the tight junction-associated MARVEL protein family 21763689_MarvelD3 is transcriptionally downregulated in Snail-induced epithelial-mesenchymal transition during the progression for the pancreatic cancer. 24567356_MarvelD3 couples tight junctions to the MEKK1-JNK pathway to regulate cell behavior and survival. 34338154_Role of tight junction-associated MARVEL protein marvelD3 in migration and epithelial-mesenchymal transition of hepatocellular carcinoma. | ENSMUSG00000001672 | Marveld3 | 2.701305e+01 | 0.8260675 | -0.275668497 | 0.5771209 | 2.206339e-01 | 0.6385573260 | 0.91968733 | No | Yes | 1.690499e+01 | 4.556746 | 1.838338e+01 | 5.072210 | ||
| ENSG00000140983 | 89941 | RHOT2 | protein_coding | Q8IXI1 | FUNCTION: Mitochondrial GTPase involved in mitochondrial trafficking (PubMed:16630562, PubMed:22396657). Probably involved in control of anterograde transport of mitochondria and their subcellular distribution (PubMed:22396657). {ECO:0000269|PubMed:16630562, ECO:0000269|PubMed:22396657}. | 3D-structure;Alternative splicing;Calcium;GTP-binding;Hydrolase;Membrane;Metal-binding;Mitochondrion;Mitochondrion outer membrane;Nucleotide-binding;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Ubl conjugation | Mouse_homologues NA; + ;NA | This gene encodes a member of the Rho family of GTPases. The encoded protein is localized to the outer mitochondrial membrane and plays a role in mitochondrial trafficking and fusion-fission dynamics. [provided by RefSeq, Nov 2011]. | hsa:89941; | integral component of mitochondrial outer membrane [GO:0031307]; membrane [GO:0016020]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; calcium ion binding [GO:0005509]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; cellular homeostasis [GO:0019725]; mitochondrial outer membrane permeabilization [GO:0097345]; mitochondrion organization [GO:0007005]; mitochondrion transport along microtubule [GO:0047497]; regulation of mitochondrion organization [GO:0010821]; small GTPase mediated signal transduction [GO:0007264] | 16630562_Moreover, we show that Miro interacts with the Kinesin-binding proteins, GRIF-1 and OIP106, suggesting that the Miro GTPases form a link between the mitochondria and the trafficking apparatus of the microtubules. 19098100_Miro proteins serve as a [Ca(2+)](c)-sensitive switch and bifunctional regulator for both the motility and fusion-fission dynamics of the mitochondria. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22078885_Study shows that both PINK1 and Parkin halt mitochondrial movement; PINK1 phosphorylates Miro (1 and 2) and thereby initiates the rapid degradation of Miro through a Parkin- and proteasome-dependent pathway. 24256248_The Miro2-mediated mitochondrial transport in neurons and recently highlighted involvement of Miro2 proteins in mitochondrial turnover, emerging as a key process affected in neurodegeneration. 26259702_Miro and Cenp-F promote anterograde mitochondrial movement and proper mitochondrial distribution in daughter cells. 27605430_Show that the C-terminal GTPase of the Parkin primary substrates Miro1 and Miro2 are necessary and sufficient for efficient ubiquitination. We present several new X-ray crystal structures of both Miro1 and Miro2 that reveal substrate recognition and ubiquitin transfer to be specific to particular protein domains and lysine residues. 30111583_Miro1 binds directly to a C-terminal fragment of the Myo19 tail region and that Miro1/2 recruit the Myo19 tail. 30459446_MIRO2 regulate the spatial organization of mitochondria in a microtubule-dependent manner.GBF1 and Arf1 interact with Miro2. 30513825_the data presented here indicate novel catalytic functions of human Miro atypical GTPases through altered catalytic mechanisms. 30787202_This study showed that RAB32 and RHOT2 were associated with aging, and that individuals exhibiting methylation levels of the RAB32 CpG site higher than 10% were observed more prone to disability than people with lower levels. 32948353_The role of RHOT1 and RHOT2 genetic variation on Parkinson disease risk and onset. 33132189_Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase. 34992146_MIRO2 Regulates Prostate Cancer Cell Growth via GCN1-Dependent Stress Signaling. | ENSMUSG00000025733+ENSMUSG00000093593 | Rhot2+Gm20683 | 7.115951e+03 | 1.0717958 | 0.100030056 | 0.2707043 | 1.371894e-01 | 0.7110912248 | 0.93885931 | No | Yes | 6.058898e+03 | 648.931843 | 5.744290e+03 | 631.073225 |
| ENSG00000140987 | 54925 | ZSCAN32 | protein_coding | Q9NX65 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:54925; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 4.917771e+02 | 0.5515973 | -0.858312735 | 0.2971860 | 8.222077e+00 | 0.0041383817 | 0.23179938 | No | Yes | 3.348418e+02 | 47.945921 | 5.907741e+02 | 85.867423 | |||||
| ENSG00000141179 | 58488 | PCTP | protein_coding | Q9UKL6 | FUNCTION: Catalyzes the transfer of phosphatidylcholine between membranes. Binds a single lipid molecule. {ECO:0000269|PubMed:12055623}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Lipid transport;Lipid-binding;Phosphoprotein;Reference proteome;Transport | hsa:58488; | cytosol [GO:0005829]; phosphatidylcholine binding [GO:0031210]; phosphatidylcholine transporter activity [GO:0008525]; lipid transport [GO:0006869]; negative regulation of cold-induced thermogenesis [GO:0120163]; phospholipid transport [GO:0015914] | 12055623_crystal structures of human PC-TP in complex with dilinoleoyl-PtdCho or palmitoyl-linoleoyl-PtdCho reveal that a single well-ordered PtdCho molecule occupies a centrally located tunnel 17266964_phosphatidylcholine transfer protein gene variants are associated with LDL-peak particle size 17704541_Yeast two-hybrid screening using libraries prepared from mouse liver and embryo identified Them2 (thioesterase superfamily member 2) and the homeodomain transcription factor Pax3 (paired box gene 3), respectively, as PC-TP-interacting proteins. 18676680_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 24216752_PC-TP contributes to the racial difference in PAR4-mediated platelet activation. 26585945_Mutations in PCTP gene is associated with prostate cancer 28251237_Identified the variant rs2912553 as contributing to racially differential expression of PCTP. 33770537_PCTP contributes to human platelet activation by enhancing dense granule secretion. | ENSMUSG00000020553 | Pctp | 8.112020e+02 | 0.5466659 | -0.871268697 | 0.3218938 | 7.352606e+00 | 0.0066965601 | 0.30648627 | No | Yes | 5.096615e+02 | 90.245696 | 9.613038e+02 | 173.774764 | ||
| ENSG00000141314 | 162494 | RHBDL3 | protein_coding | P58872 | FUNCTION: May be involved in regulated intramembrane proteolysis and the subsequent release of functional polypeptides from their membrane anchors. {ECO:0000250}. | Alternative splicing;Hydrolase;Membrane;Protease;Reference proteome;Repeat;Serine protease;Transmembrane;Transmembrane helix | hsa:162494; | integral component of membrane [GO:0016021]; calcium ion binding [GO:0005509]; serine-type endopeptidase activity [GO:0004252] | Mouse_homologues 11900977_Cloning and expression of Ventrhoid 27264103_Here the authors show that the mammalian rhomboid protease RHBDL4 (also known as Rhbdd1) promotes trafficking of several membrane proteins, including the EGFR ligand TGFalpha, from the endoplasmic reticulum (ER) to the Golgi apparatus, thereby triggering their secretion by extracellular microvesicles. | ENSMUSG00000017692 | Rhbdl3 | 3.474365e+02 | 1.1633564 | 0.218293097 | 0.3184628 | 4.823888e-01 | 0.4873422935 | 0.87510784 | No | Yes | 3.239810e+02 | 36.044948 | 2.849501e+02 | 32.796929 | ||
| ENSG00000141401 | 3613 | IMPA2 | protein_coding | O14732 | FUNCTION: Can use myo-inositol monophosphates, scylloinositol 1,4-diphosphate, glucose-1-phosphate, beta-glycerophosphate, and 2'-AMP as substrates. Has been implicated as the pharmacological target for lithium Li(+) action in brain. {ECO:0000269|PubMed:17068342}. | 3D-structure;Alternative splicing;Hydrolase;Magnesium;Metal-binding;Reference proteome | PATHWAY: Polyol metabolism; myo-inositol biosynthesis; myo-inositol from D-glucose 6-phosphate: step 2/2. | This locus encodes an inositol monophosphatase. The encoded protein catalyzes the dephosphoylration of inositol monophosphate and plays an important role in phosphatidylinositol signaling. This locus may be associated with susceptibility to bipolar disorder. [provided by RefSeq, Jan 2011]. | hsa:3613; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; inositol monophosphate 1-phosphatase activity [GO:0008934]; inositol monophosphate 3-phosphatase activity [GO:0052832]; inositol monophosphate 4-phosphatase activity [GO:0052833]; metal ion binding [GO:0046872]; protein homodimerization activity [GO:0042803]; inositol biosynthetic process [GO:0006021]; inositol metabolic process [GO:0006020]; inositol phosphate dephosphorylation [GO:0046855]; phosphate-containing compound metabolic process [GO:0006796]; phosphatidylinositol phosphate biosynthetic process [GO:0046854]; signal transduction [GO:0007165] | 11317223_Observational study of gene-disease association. (HuGE Navigator) 14699425_Promoter is polymorphic in bipolar disorder. 15505643_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15557493_IMPA2 may be a febrile seizure susceptibility gene. 6 SNPs were found: -708G/A, -461C/T, IVS1-15G/A, 159T/C, IVS5+13-14insA, & 558C/T. 17068342_IMPA2 has a separate function in vivo from that of IMPA1 17251911_the present study suggests that a promoter haplotype of IMPA2 possibly contributes to risk for bipolar disorder by elevating IMPA2 levels in the brain, albeit the genetic effect varies among populations. 17340635_The crystal structures of human IMPA2 are useful for understanding the effect of nonsynonymous polymorphism reported in IMPA2, and will contribute to further functional analyses of IMPA2. 17388992_Observational study of gene-disease association. (HuGE Navigator) 17388992_data suggest that the genetic variants in the IMPA2 gene are not associated with a risk of febrile seizures in Caucasian patients and patients from various genetic groups are likely to have different genetic causes of febrile seizures 19066393_Single nucleotide polymorphism in IMPA2 gene is associated with acute lymphoblastic leukemia. 19910543_Observational study of gene-disease association. (HuGE Navigator) 20153384_The current study did not support a substantial role of the upregulation of IMPase in bipolar disorder, although the lithium-insensitivity trait seen in IMPA2 transgenic mice might represent some aspect relevant to the inositol depletion hypothesis. 20398908_Observational study of gene-disease association. (HuGE Navigator) 20800640_No association is found between IMPA2 gene polymorphisms and bipolar disorder. 20800640_Observational study of gene-disease association. (HuGE Navigator) 21213002_Authors propose that the human myo-inositol monophosphatase 2 interacts with myo-inositol monophosphates in the three-metal-ion bound form, and proceeds the dephosphorylation through the three-metal-ion theory. 23453640_results suggest that genetic variability at rs669838-IMPA2,rs4853694-INPP1, rs1732170-GSK3b and rs11921360-GSK3b genes is associated with a higher risk of attempting suicide in bipolar patients. 27661109_a correlation between an IMPA2 polymorphism rs589247 and ischemic stroke risk in a northwest Han Chinese 27748550_a promoter polymorphism of IMPA2 possibly contributed to risk for schizophrenia by elevating transcription activity in Han Chinese individuals. 29499505_IMPA2 is a protein-coding gene for a catalytic protein that converts inositol monophosphate into free inositol through dephosphorylation. 31202813_We conclude that miR-25-mediated IMPA2 downregulation constitutes a novel signature for cancer metastasis and poor outcomes in clear cell renal cell carcinoma (ccRCC). We further postulate that the therapeutic targeting of miR-25 can be useful for preventing the metastatic progression of ccRCC associated with IMPA2 downregulation. 32409648_A novel function of IMPA2, plays a tumor-promoting role in cervical cancer. | ENSMUSG00000024525 | Impa2 | 7.283874e+02 | 1.0709421 | 0.098880484 | 0.3173771 | 9.640749e-02 | 0.7561841416 | 0.94946660 | No | Yes | 6.973812e+02 | 79.779678 | 5.796186e+02 | 68.343867 |
| ENSG00000141504 | 112483 | SAT2 | protein_coding | Q96F10 | FUNCTION: Catalyzes the N-acetylation of the amino acid thialysine (S-(2-aminoethyl)-L-cysteine), a L-lysine analog with the 4-methylene group substituted with a sulfur (PubMed:15283699). May also catalyze acetylation of polyamines, such as norspermidine, spermidine or spermine (PubMed:12803540). However, ability to acetylate polyamines is weak, suggesting that it does not act as a diamine acetyltransferase in vivo (PubMed:15283699). {ECO:0000269|PubMed:12803540, ECO:0000269|PubMed:15283699}. | 3D-structure;Acetylation;Acyltransferase;Cytoplasm;Reference proteome;Transferase | hsa:112483; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; diamine N-acetyltransferase activity [GO:0004145]; identical protein binding [GO:0042802]; N-acetyltransferase activity [GO:0008080]; spermidine binding [GO:0019809]; nor-spermidine metabolic process [GO:0046204]; putrescine acetylation [GO:0032920]; spermidine acetylation [GO:0032918]; spermine acetylation [GO:0032919] | 16596569_Crystallization of this protein and its recombinant form and its complexation with coenzyme A. 17119850_The expression of three messengers coding for SAT-1, SAT-2 and GalNAcT-1 in human samples of intestinal cancer and some cell lines (breast cancer and melanomas), was evaluated. 17558023_SSAT2 is an essential component of the ubiquitin ligase complex that regulates hypoxia-inducible factor 1alpha 17875644_SSAT1, which shares 46% amino acid identity with SSAT2, also binds to HIF-1alpha and promotes its ubiquitination/degradation. However, in contrast to SSAT2, SSAT1 acts by stabilizing the interaction of HIF-1alpha with RACK1 18676680_Observational study of gene-disease association. (HuGE Navigator) 18949426_expression of SSAT mediated by Ad-SSAT infection inhibits the growth of colorectal cancer cells 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000069835 | Sat2 | 1.932638e+03 | 1.1370472 | 0.185292190 | 0.3051012 | 3.725406e-01 | 0.5416226767 | 0.89062022 | No | Yes | 2.021830e+03 | 231.844769 | 1.528427e+03 | 180.072348 | ||
| ENSG00000141562 | 26502 | NARF | protein_coding | Q9UHQ1 | Alternative splicing;Nucleus;Phosphoprotein;Reference proteome | Several proteins have been found to be prenylated and methylated at their carboxyl-terminal ends. Prenylation was initially believed to be important only for membrane attachment. However, another role for prenylation appears to be its importance in protein-protein interactions. The only nuclear proteins known to be prenylated in mammalian cells are prelamin A- and B-type lamins. Prelamin A is farnesylated and carboxymethylated on the cysteine residue of a carboxyl-terminal CaaX motif. This post-translationally modified cysteine residue is removed from prelamin A when it is endoproteolytically processed into mature lamin A. The protein encoded by this gene binds to the prenylated prelamin A carboxyl-terminal tail domain. It may be a component of a prelamin A endoprotease complex. The encoded protein is located in the nucleus, where it partially colocalizes with the nuclear lamina. It shares limited sequence similarity with iron-only bacterial hydrogenases. Alternatively spliced transcript variants encoding different isoforms have been identified for this gene, including one with a novel exon that is generated by RNA editing. [provided by RefSeq, Jul 2008]. | hsa:26502; | lamin filament [GO:0005638]; nuclear lamina [GO:0005652]; nuclear lumen [GO:0031981]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; lamin binding [GO:0005521] | 17326827_Creation of a novel Alu-exon and elimination of a premature stop codon by RNA editing. 17326827_RNA editing enables exonization of a nuclear prelamin A recognition factor Alu-exon. 17654502_Prelamin A processing is linked to heterochromatin organization. 18680752_Data suggest that gorilla, chimpanzee, and human nuclear prelamin A recognition factor genes exemplify the versatile interplay of pre- and posttranscriptional modifications leading to novel genetic potential. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20970119_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 31823109_Nuclear prelamin a recognition factor and iron dysregulation in multiple sclerosis. | ENSMUSG00000000056 | Narf | 3.634756e+03 | 0.9170972 | -0.124853525 | 0.2955116 | 1.797156e-01 | 0.6716177412 | 0.92978311 | No | Yes | 3.099573e+03 | 218.923643 | 3.305862e+03 | 239.250066 | ||
| ENSG00000141655 | 8792 | TNFRSF11A | protein_coding | Q9Y6Q6 | FUNCTION: Receptor for TNFSF11/RANKL/TRANCE/OPGL; essential for RANKL-mediated osteoclastogenesis. Involved in the regulation of interactions between T-cells and dendritic cells. {ECO:0000269|PubMed:9878548}. | 3D-structure;Alternative splicing;Cell membrane;Deafness;Disease variant;Disulfide bond;Glycoprotein;Membrane;Metal-binding;Osteopetrosis;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Sodium;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the TNF-receptor superfamily. This receptors can interact with various TRAF family proteins, through which this receptor induces the activation of NF-kappa B and MAPK8/JNK. This receptor and its ligand are important regulators of the interaction between T cells and dendritic cells. This receptor is also an essential mediator for osteoclast and lymph node development. Mutations at this locus have been associated with familial expansile osteolysis, autosomal recessive osteopetrosis, and Paget disease of bone. Alternatively spliced transcript variants have been described for this locus. [provided by RefSeq, Aug 2012]. | hsa:8792; | cytosol [GO:0005829]; external side of plasma membrane [GO:0009897]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; cytokine binding [GO:0019955]; metal ion binding [GO:0046872]; signaling receptor activity [GO:0038023]; transmembrane signaling receptor activity [GO:0004888]; tumor necrosis factor-activated receptor activity [GO:0005031]; adaptive immune response [GO:0002250]; cell-cell signaling [GO:0007267]; circadian temperature homeostasis [GO:0060086]; lymph node development [GO:0048535]; mammary gland alveolus development [GO:0060749]; monocyte chemotaxis [GO:0002548]; multinuclear osteoclast differentiation [GO:0072674]; ossification [GO:0001503]; osteoclast differentiation [GO:0030316]; positive regulation of bone resorption [GO:0045780]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling [GO:0071848]; positive regulation of fever generation by positive regulation of prostaglandin secretion [GO:0071812]; positive regulation of JUN kinase activity [GO:0043507]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; response to cytokine [GO:0034097]; response to interleukin-1 [GO:0070555]; response to lipopolysaccharide [GO:0032496]; response to radiation [GO:0009314]; response to tumor necrosis factor [GO:0034612]; signal transduction [GO:0007165]; TNFSF11-mediated signaling pathway [GO:0071847]; tumor necrosis factor-mediated signaling pathway [GO:0033209] | 11771666_Expansile skeletal hyperphosphatasia is caused by a 15-base pair tandem duplication (84dup15) in TNFRSF11A encoding RANK. 12043011_immunohistochemical localization of this protein and its ligand in deciduous teeth 12200385_MIP-1alpha and MIP-1beta induce expression of RANK ligand by stromal cells, thereby stimulating osteoclast differentiation of preosteoclastic cells. 12296995_TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. 12362049_Analysis of a large Spanish kindred confirms that exon 1 contains an insertional mutation in the RANK gene. 12393586_Long-lived immature dendritic cells mediated by TRANCE-RANK interaction 12568416_a tandem duplication in exon 1 of the TNFRSF11A gene may have a role in familial expansile osteolysis 12709501_associated with external apical root resorption 12929927_the 75dup27 mutation causes a Paget's disease of bone-like phenotype 12933809_transforming growth factor-beta promotes osteoclastogenesis in monocytes by stimulation of the p38 mitogen activated protein kinase but continuous exposure abrogates osteoclastogenesis by down-regulation of receptor activator of NF-KB(RANK) expression 15248232_These data suggest that RANK is expressed by monocytes whose activation by RANKL stimulates directed migration involving phosphatidylinositol 3-kinase, phosphodiesterase, and Src kinases. 15301860_Observational study of genotype prevalence. (HuGE Navigator) 15377473_RANK is expressed in bone marrow stroma cells & endothelial cells but not myeloma cells. It is involed in IL-6 & IL-11 secretion. 15564564_Review. The RANKL/RANK/OPG system may mediate links between the vascular, skeletal, & immune systems and play a central role in regulating the vascular calcification coincident with declines in skeletal mineralization with age, osteoporosis, or disease. 15615497_disruption of the RANKL-RANK axis with OPG inhibited tumor-induced osteoclastogenesis and decreased bone cancer pain. 16052586_Involvement of RANK/RANKL in dendritic cell-T cell interactions during inflammatory process. RANK expression appears to be limited to sites of immune reaction, both in synovium and in lymph nodes. 16083856_PU.1 regulates RANK gene transcription; this may represent one of the key roles of PU.1 in osteoclast differentiation 16151677_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16215261_OPG dimer formation is required for the mechanism of inhibition of the RANK-L/RANK receptor interaction 16240334_We review the etiology of inflammatory bone loss, the RANK/RANK-ligand/OPG pathway, and the clinical development of anti-RANK-ligand therapy. 16249885_Observational study of gene-disease association. (HuGE Navigator) 16270354_By upregulating IL-8, the RANKL/RANK system may contribute to the pathogenesis of B chronic lymphocytic leukemia. 16328004_a functional RANK expressed on osteosarcoma cells 16556708_Expression of RANK-Fc by genetically modified Mesenchymal stem cells may be a feasible option for the prevention of bone loss induced by ovariectomy. 16583245_Observational study of gene-disease association. (HuGE Navigator) 16953816_Observational study of gene-disease association. (HuGE Navigator) 16960694_Observational study of gene-disease association. (HuGE Navigator) 17115234_Observational study of gene-disease association. (HuGE Navigator) 17115234_results suggest that +34863G > A and +35928insdelC polymorphisms in RANK are possible genetic factors for low Bone mineral density in postmenopausal women 17288531_This review describes the most recent knowledge on the OPG-receptor activator of nuclear factor-kappaB (RANK)-RANK ligand (RANKL) triad and its involvement in bone oncology. 17331078_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17447113_Although the mutation in the Iranian and four of the previously described FEO pedigrees was the same, haplotypes based on the intragenic SNPs suggest that the mutations do not share a common descent. 17546619_A major haplotype in block 5 of Tumor Necrosis Factor receptor superfamily member 11a (RANK) was significantly associated with higher stature in Caucasians. 17546619_Observational study of gene-disease association. (HuGE Navigator) 17580719_The expression levels of RANKL and RANK were markedly increased in the perimatrix of cholesteatoma. 17634140_Review highlights the receptor activator of nuclear factor-kappa B ligand (RANKL)/RANK/osteoprotegerin (OPG) system and its role in the regulation of bone resorption. 17876645_Observational study of gene-disease association. (HuGE Navigator) 17876645_RANK contributes to osteoclastic bone resoption in RA patients. 17895323_RANK/RANKL/OPG system mediates the effects of calciotropic hormones and, consequently, alterations in their ratio are key in the development of several clinical conditions--REVIEW 17966895_The OPG/RANKL/RANK system constitutes the important element of controlling the number of active osteoclasts by the osteoblasts. 17982618_cDNA microarray and quantitative RT-PCR analyses demonstrate that RANK-positive osteosarcoma cells are the target of RANKL as well as osteoclasts/osteoclast precursors. 18008334_RANK is expressed on prostate cancer cells and promotes invasion in a RANKL-dependent manner 18061491_differences in the RANK, RANKL, and OPG expression in odontogenic epithelial tumors...could contribute to the differential bone/tooth resorption activity in these lesions 18174230_Observational study of gene-disease association. (HuGE Navigator) 18311801_The relative protection against bone erosions in spondylarthritis cannot be explained by qualitative or quantitative differences in the synovial expression of RANKL, OPG, and RANK. 18367263_Data reveals for the first time that OPG/RANK/RANKL are expressed in the pathological thyroid gland by follicular cells, by malignant parafollicular cells as well as in metastatic lymph node microenvironment. 18606301_Osteoclast-poor osteopetrosis with agammaglobulinemia due to TNFRSF11A (RANK) mutation is reported. 18632421_ligation of RANK on DC cell surfaces is not only a survival stimulus, but also induces a partial and specific mature DC phenotype 18802807_Ineffective modulation of the OPG/RANK/RANKL system in active polyarticular juvenile idiopathic arthritis may account for bone damage in this disease. 18928898_might locally modulate [odontogenic] tumor-associated bone resorption 19008470_Based on their role in atherogenesis, this enhanced expression of RANKL and RANK could contribute to the increased risk of cardiovascular disease in hyperhomocystinemia 19074885_Observational study of gene-disease association. (HuGE Navigator) 19079262_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19085839_RANKL/RANK have roles in bone-associated tumors (review) 19213753_no significant association was detected between the three RANK tag single nucleotide polymorphisms tested and rheumatoid arthrirtis in our white European family sample 19416721_The binding affinity of RANKL for RANK was measured with surface plasmon resonance technology and K(D) value is about 1.09 x 10(-10) M. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19507210_Mutations in TCIRG1, OSTM1, ClCN7, and TNFRSF11A genes were detected in nine, three, one, and one patientswith infantile malignant osteopetrosis, respectively. 19554506_The expression of RANK gene of NF-kappaB pathway in multiple myeloma using bone marrow aspirates obtained at diagnosis is reported. 19578385_A novel 27-bp duplication in exon 1 (78dup27) in TNFRSF11A was found in four affected individuals and one asymptomatic individual for early onset familial Paget's disease of bone. 19672874_High-throughput copy number and gene expression data of 36 microsatellite stable sporadic colon cancers resected from patients of a single institution characterized for mutations in APC, KRAS, TP53 and loss of 18q were analyzed. 19705167_Observational study of gene-disease association. (HuGE Navigator) 19728335_IKKalpha, IKKbeta, RANK, Maspin, c-FLIP, Cip2 and cyclinD1 were found to show significant differences between hepatocellular tumor tissue and its corresponding adjacent tissue. 19890054_Our findings identify a new mechanism of homeostatic regulation of osteoclastogenesis that targets RANK expression and limits bone resorption during infection and inflammation. 19896533_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19896533_Results suggest that gene-gene interactions between RANK and OPG, and RANK and RANKL influence BMD in postmenopausal women. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19940926_data identify an entirely novel and unexpected function for the key osteoclast differentiation factors RANKL/RANK in female thermoregulation and the central fever response in inflammation 20205168_Genetic variation in the RANKL/RANK/OPG signaling pathway influences bone turnover and bone mineral density in European men. 20205168_Observational study of gene-disease association. (HuGE Navigator) 20231205_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20436471_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20436471_These studies provide new insights into the pathogenesis of PDB and identify OPTN, CSF1 and TNFRSF11A as candidate genes for disease susceptibility. 20506523_Results suggest that the RANKL acts through MEK/ERK, which in turn activates IKKalpha/beta and NF-kappaB, resulting in the activation of beta1 integrin and contributing to the migration of human chondrosarcoma cells. 20531232_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20531232_The TNFRSF11A (tumor necrosis factor receptor superfamily member 11a) and TNFSF11(RANKL) genes are associated with the age of onset of menarche and natural menopause in white women 20534768_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20554715_Observational study of gene-disease association. (HuGE Navigator) 20564239_Observational study of gene-disease association. (HuGE Navigator) 20564239_The possible relationship between TNFRSF11A polymorphisms and sporadic Paget's disease of bone, was investigated. 20623281_Our study represents the first preliminary indication about a local increase of RANK concentration during fracture healing immediately after surgery. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20694489_Receptor activator for nuclear factor-kappaB ligand (0.54 +/- 0.26 vs. 0.16 +/- 0.15 pmol/l, P < 0.0005) values were significantly higher in PCOS subjects compared to the control group 20839008_genetic risk for Paget's disease of bone is associated with variants close to CSF1, OPTN, TM7SF4, and TNFRSF11A genes 20857484_These data highlight the central role of RANK/RANKL/OPG pathway as potential therapeutic target not only in bone metastasis management, but also in the adjuvant setting. 20974615_Observational study of gene-disease association. (HuGE Navigator) 21050071_assays demonstrated >300% increases in luciferase activity after RANK motif activation and Z '-factor values over 0.55 21159522_Increasing concentrations of FSH caused a biphasic dose-response, with a maximal (1.5-fold) increase in RANK(high) cells 21270824_RANK might be involved in the development and maintenance of melanoma-initiating cells and possibly in metastatic spreading for melanoma 21326202_RANKL and RANK are involved in mammary/breast cancer metastasis 21352301_no significant association of SNPs with susceptibility to disease in psoriasis and psoriatic arthritis patients 21472776_Constitutive activation of NF-kappaB occurred in HEK293 cells overexpressing wild-type or mutant RANK but not in stably transfected cell lines expressing low levels of each RANK gene. 21479680_results suggested that mechanisms other than the RANKL/RANK signalling pathway might be involved in the osteoclastogenic response mediated by MG63 cells 21514280_hypoxia upregulates RANK and RANKL expression and increases RANKL-induced cell migration via the PI3K/Akt-HIF-1alpha pathway. 21541702_a common genetic variant near the 5'-end of TNFRSF11A, rs7226991, is associated with breast cancer risk in the general population and among carriers of mutations in the breast cancer 2, early onset (BRCA2) gene 21559440_RANK in primary breast tumors has a role in bone metastasis occurrence in breast cancer patients; OPG has a role in survival 21643971_The molecular system of RANK/RANKL/OPG is variably expressed in odontogenic keratocysts, radicular cysts, and ameloblastomas 21742767_Presented is a comprehensive reaction map of the RANKL/RANK-signaling pathway based on an extensive manual curation of the published literature. 21814016_The receptor of RANKL whick stimulates osteoclast differentiation and functionand ists mutation causes osteoclatst abnormalities. (review) 21814020_It is critical to the development and progression of bone metastases in breast neoplasms. (review) 21814022_It is involved in the development of osteoclasts, cooperating with another key molecule, M-CSF.(review) 21814026_the central RANKL/RANK pathway has an important role for thermoregulation.(review) 21964949_There were five RANK SNPs associated with geometric parameters, including rs8083511 associated with distal radius cross-sectional area. 21987421_Two nonsynonymous single nucleotide polymorphisms (SNPs) (C421T, H141Y and T575C, V192A) in the TNFRSF11A gene, associated with Paget's disease of bone, were identified. 22001124_Results describe the relationship between bone metabolism and OPG/RANK/sRANKL concentrations in females with anorexia nervosa. 22001292_AF groups had higher atrial gene expression of OPG/RANK/RANKL axis and RANKL/OPG ratio, particularly in paroxysmal AF. 22023082_The results of the present study suggest that RANK is associated with Age at menarche in Chinese women. 22176920_Radiographic emphysema is correlated with low BMD in current and former smokers with COPD. IL-1beta, IL-6, TNF-alpha, and the osteoporosis-related protein system OPG/RANK/RANKL. 22178057_The atrial expression of RANK (and RANKL/osteoprotegerin ratio) was higher in normal controls compared to persistent atrial fibrillation patients. 22214279_RANK, RANKL and OPG proteins are differentially expressed in periodontal tissues and may play a major role in the bone loss occurring in periodontitis 22271396_Novel mutations in the TNFRSF11A gene in Autosomal recessive osteopetrosis, are reported. 22402034_serum OPG concentration was significantly lower in carotid population compared to femoral population while RANK and RANKL were equally expressed in both arterial beds 22496457_RANK expression in primary breast cancer associates with poor prognosis. 22531921_regulated RANK expression contributes to the fine tuning of PMN migration, for example, on and through inflamed endothelium that is known to express RANKL. 22546829_Genetic variations using representative single nucleotide polymorphisms (SNPs) of OPG (rs2073618), RANK (rs1805034) and RANKL (rs2073618), were analysed. 22705116_summary of recent advances in understanding osteoclastogenic signaling/osteoimmunology: RANKL/RANK/OPG signaling in osteocytes plays role in bone remodeling as seen in bone resorption or arthritis. [REVIEW] 22753650_In hemophilic arthropathy, the synovium highly expressed RANK and RANKL, whereas OPG immunopositivity decreased, suggesting an osteoclastic activation. 22787434_We conclude that RANK/TNFRSF11A is a novel and frequent target for de novo methylation in gliomas, which affects apoptotic activity and focus formation thereby contributing to the molecular pathogenesis of gliomas. 22824341_Novel RANK variants, providing evidence of a complex regulatory pathway for RANK receptor signaling, with possible implications in various systems, tissues and cells. 22848465_the increase in RANK-RANKL expression is a response to podocyte injury, and RANK-RANKL may be a novel receptor-ligand complex for the survival response during podocyte injury. 23116709_One mechanism of RANK inhibition by 1,25(OH)2D3 is down-regulation of the M-CSF receptor c-Fms, which is required for the expression of RANK. 23267146_Our data suggest that RANK, RANKL and OPG may potentially be used as novel prognostic markers for bone metastasis and provide new therapeutic targets in the treatment of breast cancer 23478294_EEIG1 is a novel RANK signaling component controlling RANK-mediated osteoclast formation. 23516466_Differential expression of the RANKL/RANK/OPG system is associated with bone metastasis in human non-small cell lung cancer. 23531404_OPG and RANK but not RANKL genetic polymorphisms influence bone mineral density mainly in the femoral neck in peri- and postmenopausal Chinese women. 23553199_The genotypes, combined genotypes and allele frequencies of C421T and C575T polymorphisms of the RANK gene have not been found to be associated with bone mineral density in Turkish women. 23572233_Data show that cystatin C inhibits osteoclast differentiation and formation by interfering intracellularly with signaling pathways downstream RANK. 23664977_RANK-e5a is produced through splicing out part of exon 5, resulting in a truncation of its extracellular domain. This isoform seems to have lower binding capacity towards RANK ligand and at the same time it is unable to activate NF-kB, neither after RANKL stimuli, nor by its overexpression. Furthermore, RANK-e5a is specifically expressed in human brain raising the possibility of a tissue specific function. 23664977_When TNFRSF11A-e5a is stimulated by RANK ligand, its capability to activate NF-kappaB is reduced compared to the wild type RANK receptor. 23697850_RANKL/RANK signal promotes Bcl-2 and Ki67 and decreases FasL expression, and further as a positive regulator for stimulating the proliferation and growth of DSCs through up-regulating CCL2/CCR2 signal 23702841_The study prospectively evaluated levels of RANK, RANK-L and OPG transcripts in the peripheral blood of 49 consecutive patients with advanced breast, lung or prostate cancer. 23744843_Genome-wide association associated single-nucleotide polymorphisms (SNPs) near candidate genes such as RANK and RANKL suggest that these single nucleotide polymorphisms and/or other variants nearby may be involved in bone phenotype determination. 23766243_RANK signaling interferes with mammary cell commitment, contributing to breast carcinogenesis. 23821519_silencing of miR-503 using a specific antagomir in ovariectomy (OVX) mice increased RANK protein expression, promoted bone resorption 24118270_we could not identify any association between external apical root resorption and two SNPs, rs1805034 from TNFRSF11A (encoding RANK) and rs3102735 from TNFRSF11B (encoding OPG). 24200492_High RANK expression is associated with Avascular Necrosis of Femur Head. 24340030_Mechanistic studies showed that IL-10 downregulated RANK expression in monocytes and thus, inhibited RANKL-induced OC formation 24478054_RANKL, either derived from the prostate tumor or from the host, plays a key role in cancer bone metastasis. 24549600_Changes in bone markers, OPG, sRANKL and/or the OPG/sRANKL ratio exhibited by girls with Anorexia nervosa have been found to be associated with changes in the levels of the selected adipose tissue hormones. 24651623_Single nucleotide polymorphisms of RANK and PTGS1 show genetic associations with osteoproliferative changes in ankylosing spondylitis. 24676805_RANKL and IL-6 mediate direct paracrine-autocrine signaling between cells of the osteoblast lineage and cancer cells. 24726460_These findings demonstrate that RANKL-RANK signal activation is essential to ABC tumor progression. RANKL-targeted therapy may be an effective alternative to surgery in select ABC presentations. 24729980_The effect of rs1054016(RANKL) adds to the evidence that the RANK pathway plays a role in BC pathogenesis and progression with respect to BMFS, emphasizing the connection between BC and bone health. 24737168_Higher RANK expression in the primary breast tumor is associated with a higher sensitivity to chemotherapy, but also a higher risk of relapse and death. 24842377_we identified RANK expression as a negative prognostic factor regarding disease-free survival in osteosarcoma 24891336_The involvement of TNFRSF11A in hereditary recurrent fever highlights the key role of this receptor in innate immunity. 25019155_Functional polymorphisms RANK rs1805034 T>C may be an indicator for individual susceptibility to esophageal squamous cell carcinoma. 25047443_In the present study, we investigated the ability of triptolide, a diterpenoid isolated from Thunder of God Vine, to inhibit signaling by receptor activator of NF-kappaB (RANK) and its ligand (RANKL) and to modulate osteoclastogenesis 25111682_High RANK protein expression is associated with breast cancer. 25138264_Genetic polymorphism in TNFRSF11A influences bone mineral density in post-menopausal women. 25171769_The urinary mRNA of RANK might be used to differentiate histologic subtypes of glomerulonephritis, particularly between minimal change disease and membranous nephropathy. 25268581_RANKL could potentiate migration and invasion ability of RANK-positive hepatocellular carcinoma cells through NF-kappaB pathway mediated epithelial-mesenchymal transition. 25270538_Data show that icariin decreases the expression of osteoprotegerin (OPG), receptor activator of nuclear factor kappa-B ligand (RANKL) and receptor activator of nuclear factor kappa-B (RANK) in interleukin 1 beta-stimulated chondrocytes. 25333856_High levels of RANK, RANKL and OPG expression can help to identify prostate cancer patients at high risk of developing bone metastasis. 25406312_PGRN and PIRO form a new regulatory axis in osteoclastogenesis that is included in RANK signaling in cell fusion and OC resorption of osteoclastogenesis 25464125_The data support an association between SNPs in the RANK/RANKL/OPG signalling pathway and the development of stress fracture injury. 25478860_he top hit rs17069906 (p = 5.6 e-10) is located within the genomic region of RANK, recently demonstrated to be an important player in the adaptive recovery response in podocytes and suggested as a promising therapeutic target in glomerular diseases. 25618600_This review study has focused on the association of RANKL-RANK-OPG pathway in the pathogenesis and progression of giant cell tumor of bone as well as discussed the possible therapeutic strategies by targeting this pathway 25722033_These findings indicate that epidermal leukocytes gradually acquire RANK during gestation - a phenomenon previously observed also for other markers on LCs in prenatal human skin. 25738879_Microcomputed tomography analysis demonstrated that the mice treated with rhRANK exhibited an increased bone volume and structure model index, and decreased trabecular spacing compared with those treated with rhOPG-Fc. 25810067_genetic variation associated with hypertension in Chinese women 25884698_Three single nucleotide polymorphisms of TNFRSF11A (rs4500848, rs6567270 and rs1805034) are associated with Age at menarche and Age at natural menopause in Chinese women. 25893522_Response to sRANKL in normal and tumor cells suggests a role for RANK/ERK-mediated signaling in normal osteoblasts chemotactic migration during bone remodeling that is altered or lost during osteosarcoma tumorigenesis. 25956608_Results demonstrated that the type of inflammatory infiltrate present in periradicular cysts appears to influence the expression of RANK, RANKL, and osteoprotegerin 25973136_this study is the first to identify RANK overexpression as a novel esophageal cancer marker in both Kazakh and Han ethnic esophageal squamous cell carcinoma patients. 26087197_Results suggest that Cbl-b improves the prognosis of RANK-expressing breast cancer patients by inhibiting RANKL-induced breast cancer cell migration and metastasis. 26200837_Src expression showed a significantly positive linear relationship with RANK, suggesting a potential mechanism of the RANKL-RANK axis in regulating breast cancer cell differentiation and antiapoptosis. 26276390_results indicate that the RANK IVVY motif cooperates with the TRAF-binding motifs to promote osteoclastogenesis, which provides novel insights into the molecular mechanism of RANK signaling in osteoclastogenesis. 26398902_high level of mRANKL/RANK expression in cervical cancer lesions plays an important role in the rapid growth of cervical cancer cells possibly through strengthening the dialogue between cervical cancer cells and regulation of IL-8 secretion 26451891_Based on our findings, the functional SNP RANK rs1805034 T>C may be an indicator for individual susceptibility to GCA. 26528707_RANK/OPG ratio of expression in primary ccRCC is associated with BM. 26617755_expression of OPG, RANK and RANKL genes exert a crucial role in the progression of avascular necrosis 26734994_High RANK expression is associated with endometrial metastasis. 26749530_In this review, we will provide a summary of the biological functions of RANK signaling pathway (receptor activator of nuclear factor kappaB ligand RANKL and its receptor RANK ) and downstream pathways in bone remodeling, immunity and epithelial homeostasis, with a particular emphasis on cancer 26762414_The RANK/RANKL system induces chemoresistance. 26914636_Postmenopausal women with Rheumatoid Arthritis, carrying either the RANKL-290T allele or possessing the RANK 575CC genotype were more likely to develop osteoporosis. 26977008_Study showed that endogenous RANK expression changes might influence prostate cancer cell behavior since reduced RANK expression resulted in significantly increased PC-3 cell proliferation and adhesion. 27171030_Twenty-one single-nucleotide polymorphisms (SNPs) of TNFSF11, TNFRSF11A, and TNFRSF11B were genotyped. This study suggests that TNFRSF11B but not TNFSF11 and TNFRSF11A genetic polymorphisms are associated with Type 2 Diabetes Mellitus in Southern Han Chinese women. 27187610_In this study, whole exome sequencing (WES) was successfully used in six patients with malignant infantile osteopetrosis (MIOP) and identified mutations in four MIOP-related genes (CLCN7, TCIRG1, SNX10, and TNFRSF11A). 27191503_RANK SNP rs34945627 has a high allelic frequency in patients with breast cancer and Bone metastases, and is associated with decreased disease-free survival and Overall Survival. 27279652_RANK is frequently expressed by cancer cells in contrast with RANKL which is frequently detected in the tumor microenvironment, and together they participate in every step in cancer development. (Review) 27304650_RANK 575C>T polymorphisms did not show any statistically significant differences between the study groups (Osteoporosis and Osteopenia) and Postmenopausal women. 27322743_In histologically normal tissue of BRCA1-mutation carriers and showed that RANK(+) cells are highly proliferative, have grossly aberrant DNA repair and bear a molecular signature similar to that of basal-like breast cancer. 27507811_Vav3 is a novel TRAF6 interaction partner that functions in the activation of cooperative signaling between T6BSs and the IVVY motif in the RANK signaling complex. 27822475_RANK and CCR6 expressed on monocytes may be novel targets for the regulation of bone resorption in rheumatoid arthritis and osteoporosis. 27862210_Studies showed that the central hypothalamic-pituitary regulatory system, via it's relative hormones, seems to control OPG/RANKL/RANK system function, and the pulsatility and circadian rhythmicity of these hormones may induce an oscillatory fluctuation of the OPG/ RANKL ratio. Also, psycological characteristics may provoke a shift of the OPG/ RANKL ratio towards an unbalanced or a balanced status. [review] 27881737_Studies strongly implicates RANK and RANKL as key molecules involved in the initiation of BRCA1-associated breast cancer. [review] 28244588_OPG rs2073618, RANK rs75404003, and RANKL rs9594782 single nucleotide poymorphisms may predispose LVH in thalassemia patients. 28246602_the OPG/RANKL/RANK system might be directly influenced by genetic variants of NOTCH1 in aortic valve calcification. 28252575_Our results suggest that the polymorphism of the RANKL, RANK, and OPG genes does not make a significant genetic contribution to heel ultrasound measurements in a population of young Caucasian adults. Further studies replicating the results in independent populations are needed to support these initial findings. 28373003_RANK/RANKL signaling is involved in the androgen deprivation therapy-induced acceleration of bone metastasis in castration-insensitive prostate cancer and is inhibited by osteoprotegerin to prevent bone metastasis. 28417335_triple-negative breast cancer (TNBC) patients that expressed both RANK and RANKL proteins had significantly worse RFS and OS than patients with RANK-positive, RANKL-negative tumors. RANKL was an independent, poor prognostic factor for RFS and OS in multivariate analysis in samples that expressed both RANK and RANKL. 28464982_For the RANK gene, the AGTGC haplotype was associated with the lowest risk of presenting chronic joint pain in individuals without TMD (P=0.03). This study supports the hypothesis that changes in the OPG and RANK genes influence the presence of chronic joint pain in individuals with and without TMD. 28483017_KLF5 and TNFRSF11a are related to cervical cancer. KLF5 promotes the proliferation, migration, and invasion of cervical cancer cells partly by upregulating the transcription of TNFRSF11a. 28577080_RANK is increased in hormone receptor negative and basal breast cancer, and correlates with worse recurrence-free survival and risk of bone metastasis. 29025596_EGFR and RANK combinatorial in vitro analyses revealed a significant upregulation of AKT and ERK signaling after EGF stimulation in cell lines and also an increase of breast cancer cell invasiveness. 29074954_The role of the RANK pathway in the skeletal derangement in the chronic lymphocytic leukaemia patients. 29118048_RANK rewires energy homeostasis in human lung cancer cells and promotes expansion of lung cancer stem-like cells. 29122885_findings indicate that C/EBPalpha is a stronger inducer of osteoclast differentiation than c-Fos, partly via C/EBPalpha regulation by the RANK (535)IVVY(538) motif 29146991_KLF5 and TNFRSF11a promote cervical cancer cell proliferation, migration and invasiveness. 29204705_The mRNA expression of RANK was highest in prostate tumour tissue from patients with bone metastases as compared to BPH or locally confined tumours, also shown in clinical subgroups distinguished by Gleason Score or PSA level. 29241686_RANK/RANKL were identified as crucial regulators for BRCA1 mutation-driven breast cancer. Current prevention strategies for BRCA1 mutation carriers are associated with wide-ranging risks; therefore, the search for alternative, non-invasive strategies is of paramount importance 29334613_Reduced miR-144-3p expression in serum and bone mediates osteoporosis pathogenesis by targeting RANK. 29494398_study indicates that the expression of RANK and RANKL in parotid gland neoplasms is associated with the acquisition of a malignant phenotype and this pathway may represent an attractive therapeutic target in patients with parotid gland carcinomas. 29568001_study identified the second disease gene for DOS. TNFRSF11A isoforms may have the different roles in skeletal development and metabolism 29920311_that TNFR2 in esophageal squamous cell carcinoma (ESCC) tissues was positively correlated with progression and poor prognosis of ESCC patients 29920500_Changes in cerebrospinal fluid levels of the RANKL/RANK/OPG axis are associated with multiple sclerosis, particularly at disease onset. 29932437_RANK protein expression increased from normal to malignant endometrium, and the expression level was related with tumor grade but not with stage or the age of subjects in endometrial cancer. 30066839_gammadelta T cells suppressed iDCs osteoclastogenesis by downregulation of the RANK/cFos/ATP6V0D2 signaling pathway. 30093448_Results from current studies also point to a role of RANK/RANKL signaling in patients with multiple myeloma, who have increased se | ENSMUSG00000026321 | Tnfrsf11a | 3.389844e+02 | 0.6413918 | -0.640722241 | 0.3502634 | 3.192094e+00 | 0.0739951837 | 0.71893069 | No | Yes | 2.557391e+02 | 47.758223 | 3.680071e+02 | 70.555974 | |
| ENSG00000141867 | 23476 | BRD4 | protein_coding | O60885 | FUNCTION: Chromatin reader protein that recognizes and binds acetylated histones and plays a key role in transmission of epigenetic memory across cell divisions and transcription regulation. Remains associated with acetylated chromatin throughout the entire cell cycle and provides epigenetic memory for postmitotic G1 gene transcription by preserving acetylated chromatin status and maintaining high-order chromatin structure (PubMed:23589332, PubMed:23317504, PubMed:22334664). During interphase, plays a key role in regulating the transcription of signal-inducible genes by associating with the P-TEFb complex and recruiting it to promoters. Also recruits P-TEFb complex to distal enhancers, so called anti-pause enhancers in collaboration with JMJD6. BRD4 and JMJD6 are required to form the transcriptionally active P-TEFb complex by displacing negative regulators such as HEXIM1 and 7SKsnRNA complex from P-TEFb, thereby transforming it into an active form that can then phosphorylate the C-terminal domain (CTD) of RNA polymerase II (PubMed:23589332, PubMed:19596240, PubMed:16109377, PubMed:16109376, PubMed:24360279). Promotes phosphorylation of 'Ser-2' of the C-terminal domain (CTD) of RNA polymerase II (PubMed:23086925). According to a report, directly acts as an atypical protein kinase and mediates phosphorylation of 'Ser-2' of the C-terminal domain (CTD) of RNA polymerase II; these data however need additional evidences in vivo (PubMed:22509028). In addition to acetylated histones, also recognizes and binds acetylated RELA, leading to further recruitment of the P-TEFb complex and subsequent activation of NF-kappa-B (PubMed:19103749). Also acts as a regulator of p53/TP53-mediated transcription: following phosphorylation by CK2, recruited to p53/TP53 specific target promoters (PubMed:23317504). {ECO:0000269|PubMed:16109376, ECO:0000269|PubMed:16109377, ECO:0000269|PubMed:19103749, ECO:0000269|PubMed:19596240, ECO:0000269|PubMed:22334664, ECO:0000269|PubMed:22509028, ECO:0000269|PubMed:23086925, ECO:0000269|PubMed:23317504, ECO:0000269|PubMed:23589332, ECO:0000269|PubMed:24360279}.; FUNCTION: [Isoform B]: Acts as a chromatin insulator in the DNA damage response pathway. Inhibits DNA damage response signaling by recruiting the condensin-2 complex to acetylated histones, leading to chromatin structure remodeling, insulating the region from DNA damage response by limiting spreading of histone H2AX/H2A.x phosphorylation. {ECO:0000269|PubMed:23728299}. | 3D-structure;Acetylation;Alternative splicing;Bromodomain;Chromatin regulator;Chromosomal rearrangement;Chromosome;DNA damage;Host-virus interaction;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is homologous to the murine protein MCAP, which associates with chromosomes during mitosis, and to the human RING3 protein, a serine/threonine kinase. Each of these proteins contains two bromodomains, a conserved sequence motif which may be involved in chromatin targeting. This gene has been implicated as the chromosome 19 target of translocation t(15;19)(q13;p13.1), which defines an upper respiratory tract carcinoma in young people. Two alternatively spliced transcript variants have been described. [provided by RefSeq, Jul 2008]. | hsa:23476; | chromatin [GO:0000785]; chromosome [GO:0005694]; condensed nuclear chromosome [GO:0000794]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; enzyme binding [GO:0019899]; lysine-acetylated histone binding [GO:0070577]; nucleosomal histone binding [GO:0031493]; P-TEFb complex binding [GO:0106140]; p53 binding [GO:0002039]; RNA polymerase II C-terminal domain binding [GO:0099122]; RNA polymerase II CTD heptapeptide repeat kinase activity [GO:0008353]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; cellular response to DNA damage stimulus [GO:0006974]; chromatin remodeling [GO:0006338]; negative regulation by host of viral transcription [GO:0043922]; negative regulation of DNA damage checkpoint [GO:2000002]; positive regulation of G2/M transition of mitotic cell cycle [GO:0010971]; positive regulation of histone H3-K36 trimethylation [GO:2001255]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription elongation from RNA polymerase II promoter [GO:0032968]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of inflammatory response [GO:0050727]; regulation of phosphorylation of RNA polymerase II C-terminal domain [GO:1901407]; regulation of transcription involved in G1/S transition of mitotic cell cycle [GO:0000083]; regulation of transcription, DNA-templated [GO:0006355] | 12192049_brd4 binds to replication factor C and inhibits progression to S phase 15109495_E2 protein interacts with Brd4 and tethers the viral DNA to the host mitotic chromosomes. 15456879_Data identif a Rap GTPase-activating protein, signal-induced proliferation-associated protein 1 (SPA-1), as a factor that interacts with Brd4. 15795266_Brd4 has recently been proposed to mediate the association between bovine papilomavirus1 E2 and mitotic chromosomes. 15994877_Wild type Brd4 inhibits G(1) to S progression and we also found that the Brd4-NUT fusion augments the inhibition of progression to S phase compared with wild type Brd4. 16611886_establish a broader role for Brd4 in the papillomavirus life cycle than as the chromosome tether for papillomavirus E2 during mitosis 16921027_Brd4 is a component of human papillomavirus-assembled transcriptional silencing complex with a novel function as a cellular cofactor modulating viral gene expression. 16940503_there are distinct functional roles for the two bromodomain proteins RING3/Brd2 and Brd4 in LANA binding 17023018_The major papillomavirus E2-binding protein was identified by mass spectrometry and western blotting as the long isoform of Brd4.[Brd4] 17189190_We show the X-ray crystal structure of the carboxy-terminal domain of Brd4 in complex with HPV-16 E2, and with this information have developed a Brd4-Tat fusion protein that is efficiently taken up by different transformed cells harboring HPV plasmids. 17626100_evidence for a Brd4-independent mechanism of E2-mediated repression and suggests that different cellular factors must be involved in E2-mediated transcriptional activation and repression functions. 17686863_Tax may compete and functionally substitute for Brd4 in P-TEFb regulation.Tax and Brd4 compete for binding to P-TEFb through direct interaction with cyclin T1 17690245_Overexpression of the BRD4 P-TEFb-interacting domain disrupts the interaction between the HIV transactivator Tat and positive transcription elongation factor b and suppresses the ability of Tat to transactivate the HIV promoter 17934517_BRD-NUT fusion proteins contribute to carcinogenesis by associating with chromatin and interfering with epithelial differentiation. 18039861_while the P-TEFb level remains constant, the Brd4-P-TEFb interaction increases dramatically in cells progressing from late mitosis to early G(1). 18500820_The two bromodomains of Brd4 are mainly monomeric in solution; therefore Brd4 should have its own mechanism to reinforce its association with chromatin both in mitotic retention and related cellular processes. 18971272_The authors provide evidence that Brd4 regulates P-TEFb kinase activity by inducing a negative pathway via phosphorylation of CDK9 at threonine 29 (T29) in the HIV transcription initiation complex, inhibiting CDK9 kinase activity. 19038968_coactivator and corepressor function of Brd4 requires at least one intact bromodomain and is mediated by its direct association with E2 proteins encoded by various papillomaviruses 19103749_Brd4 as a novel coactivator of NF-kappaB through specifically binding to acetylated lysine-310 of RelA. 19129460_Bovine papillomavirus type 1 (BPV-1) E2 binds cellular chromatin in complex with Brd4 in both mitotic and interphase cells. 19211738_results indicate that the stability of human papillomavirus E2 is increased upon complex formation with Brd4 19553317_In addition to Brd4's role in mediating E2 transcription and genome tethering activities, these data suggest a potential role for Brd4 in regulating E2 stability and protein levels within papillomavirus-infected cells. 19596781_Brd4 is essential for the maintenance of the cell cycle progression mediated at least in part through the control of transcription of the Aurora B kinase cell cycle regulatory gene. 19846528_Abrogation of the interaction between P-TEFb and Brd4 thus provides a mechanism for E2-mediated repression of the viral oncogenes from the integrated viral genomes in cancer cells. 20036832_Studies indicate that KSHV LANA interacts with Brd2 and Brd4. 20201073_T-loop phosphorylated Cdk9 localizes to nuclear speckle domains which may serve as sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 regulatory complexes. 20676058_Using a patient-derived cell line, the authors show that p300 sequestration into the chromatin-bound BRD4-NUT fusion protein foci is the principal oncogenic mechanism leading to p53 inactivation. 21555454_Functional studies with Brd4 indicate that the ET domain mediates pTEFb-independent transcriptional activation through a subset of these associated factors, including NSD3. 21652721_study provides a novel mechanism by which the BRD4-NUT oncogene perturbs BRD4 functions to block cellular differentiation and to contribute to the oncogenic progression in the highly aggressive NUT midline carcinoma 22035730_The BRDT gene was not expressed in testicular tissue from patients with Sertoli cells only, whereas the other three genes of the BET family retained expression in all the sperm pathologies. 22084242_model where two BRD4 domains, the second bromodomain and the PID, bind P-TEFb and are required for full transcriptional activation of P-TEFb response genes. 22120039_BRD4 is frequently downregulated by aberrant promoter hypermethylation in human colon cancer cell lines and primary tumors. 22334664_novel structural role of Brd4 in supporting the higher chromatin architecture 22645123_Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. 23027873_three CTD kinases, CDK7, CDK9, and BRD4, engage in cross-talk, modulating their subsequent C-terminal domain phosphorylation, and also phosphorylate TAF7 23041316_The inhibition of bromodomain containing 4 increases Tat-dependent transcriptional elongation and Tat-PTEF-b association. 23086925_BRD4-driven Pol II phosphorylation at serine 2 plays an important role in regulating lineage-specific gene transcription in human CD4+ T cells. 23115324_Brd2 and Brd4 proteins mediatE the responses of LFs after growth factor stimulation and drivE the induction of lung fibrosis in mice in response to bleomycin challenge. 23128391_The novel BRD4-NUT fusion in which Exon 15 of BRD4 was fused to Exon 2 of NUT encodes a functional protein that is central to the oncogenic mechanism in these cells. 23144621_Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication 23317504_the ability of Brd4 binding to chromatin and also the recruitment of p53 to regulated promoters 23365439_Recruitment of Brd4 to the human papillomavirus type 16 DNA replication complex is essential for replication of viral DNA. 23722886_The current knowledge pertaining to the role of Brd4 in papillomavirus infections is summarized. [Review] 23728299_data implicate Brd4, previously known for its role in transcriptional control, as an insulator of chromatin that can modulate the signalling response to DNA damage 23818621_BET proteins promote efficient murine leukemia virus integration at transcription start sites. 24049186_BRD2, BRD3, and BRD4 interact with gammaretroviral INs and serve as cofactors for murine leukemia virus integration. 24090311_Data indicate using virtual screening approach to identify BRD4 inhibitors. 24100334_Three crystallographic structures of the N-terminal bromodomain of BRD4 in complex with low-molecular-weight fragments are presented. Similar molecules mimicking acetylated lysine bind the bromodomain with different orientations and interactions. 24146614_A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins. 24189064_Brd4 as a key regulator for Tax-mediated NF-kappaB gene expression 24260471_BRD4 short isoform interacts with RRP1B, SIPA1 and components of the LINC complex at the inner face of the nuclear membrane. 24278023_while Brd4 can enhance replication by concentrating viral processes in specific regions of the host nucleus, this interaction is not absolutely essential for HPV replication. 24297863_BRD4 suppression attenuates expression of MYC in medulloblastoma cells. 24360279_Study reports that a unique cohort of JMJD6 and Brd4 cobound distal enhancers, termed anti-pause enhancers, regulate promoter-proximal pause release of a large subset of transcription units via long-range interactions. 24403256_BRD4 associates with p53 in DNMT3A-mutated leukemia cells. 24445267_BRD4 is epigenetically regulated during hematopoietic differentiation ESCs in the context of a still unknown signaling pathway 24448221_These studies bring new insights for understanding Brd4-mediated stabilization of human papillomavirus 16 E2 protein, and provide an additional mechanism by which the chromatin-associated Brd4 regulates E2 functions. 24497639_the contribution of individual BRD4 amino acids to histone and JQ1 binding 24525235_BRD4-TWIST Interaction increases tumorigenesis in basal-like breast cancer. 24584072_BRD4 binds and may sustain the activity of the regulatory elements and target genes required for the proliferation of T-ALL cells. 24646477_Data indicate that inhibition of BET bromodomain proteins, including BRD4, as a promising bone tumor therapeutic strategy. 24664238_CCHCR1 interacts specifically with the E2 protein of human papillomavirus type 16 on a surface overlapping BRD4 binding 24733848_BET proteins, particularly Brd2 and Brd4, may play a key role in the regulation of Nrf2-dependent antioxidant gene transcription and are hence an important target for augmenting antioxidant responses in oxidative stress-mediated diseases. 24763052_A deregulation of BRD4 diminished the KEAP1/NRF2 axis and led to a disturbed regulation of the inducible heme oxygenase 1 (HMOX1). 24796395_High BRD4 expression is associated with medulloblastoma. 24832099_HPV replication, which hijacks host DNA damage responses, occurs adjacent to highly susceptible fragile sites bound to BRD4, greatly increasing the chances of integration here, as is found in HPV-associated cancers. 24939842_PP1alpha and class I histone deacetylase (HDAC1/2/3) signaling pathways are essential for the stress-induced BRD4 release from chromatin. 25017071_BRD4 occupies transcriptional start sites and estrogen response elements upon estrogen stimulation. 25049379_An unexpected role for the bromodomain and extraterminal domain (BET) proteins BRD2 and BRD4 in maintaining oncogenic IKK activity in activated B-cell-like diffuse large B-cell lymphoma. 25120803_These results suggest that the aberrant expression of BRD4 in human urothelial carcinoma of the bladder is possibly involved in the tumorigenesis and development 25140737_In this study, the authors show that recruitment of positive transcription elongation factor b, a functional interaction partner of Brd4 in transcription activation, is important for E2's transcription activation activity of human papillomavirus 16. 25263550_BRD4, a member of the bromodomain and extraterminal domain (BET) family of epigenetic readers, regulates the self-renewal ability and pluripotency of ESCs. 25284786_BRD4-mediated tumor protection is clinically relevant given that a BRD4 gene signature predicts positive clinical outcome in breast and lung cancer. Results show a protective function for BRD4 and suggest tissue-specific roles for BRD4 in tumorigenesis. 25340539_phosphorylation of serine 243 in the hinge region of HPV-16 E2 is essential for interaction with Brd4 and required for host chromosome binding 25506891_This review describes the recruitment of BRD4 by Twist in the progression of basal-like breast cancer. 25512383_Data support a model in which BRD4-NUT-stimulated histone hyperacetylation recruits additional BRD4 and interacting partners to support transcriptional activation, which underlies the BRD4-NUT oncogenic mechanism in NMC. 25647019_BRD4 was upregulated in pancreatic ductal adenocarcinoma cell lines. Its suppression reduced in vitro viability and proliferation, and decreased in vivo tumor growth synergistically with gemcitabine. BRD4 activated Shh signaling ligand-independently. 25656449_this study reveals a novel function of BRD4 in controlling the germinal center B cell development pathway. 25694599_The results suggest that interactions between TopBP1 and E2 and between Brd4 and E2 are required to correctly initiate human papillomavirus 16 DNA replication but are not required for continuing DNA replication. 25788266_H4K12ac is regulated by estrogen receptor-alpha and is associated with BRD4 function and inducible transcription 25816404_BRD4 expression is a valuable predictor of recurrence and survival in patients with HCC. 25877301_Brd4 is a novel client protein of Hsp90, which protects Brd4 from degradation and contributes to the upregulation of Brd4 in cystic renal epithelial cells and tissues. 25891802_Data suggest that BRD4 is essential for Toll-like receptor-stimulated interferon-beta (IFNB) gene transcription by permitting transcription factors to interact with the IFNB promoter in plasmacytoid dendritic cells. 25892415_Results show that CCNE1 and BRD4 on chromosome 19 were amplified/overexpressed in a substantial cases of epithelial ovarian cancer with no involvement of BRCA genes. 25941994_Here we show that an enhancer proximal to the c-Myc promoter is enriched in H3K27Ac and associated with high occupancy of BRD4, and coincides with a putative c-Myc super-enhancer in MCC cells. 25946208_This study demonstrated that the developed fluorescent probe will also serve as a powerful tool to evaluate BRD4 inhibitors in living cells. 26044852_IL-10 disrupts the Brd4-docking sites to inhibit lipopolysaccharide-induced CXCL8 and TNF-alpha expression in monocytes. 26083714_Essential for MYC expreession increase upon CDK9 inhibition is that the bromodomain protein BRD4 captures P-TEFb from 7SK snRNP to deliver to target genes and also enhances CDK9's activity and resistance to inhibition. 26093279_the BRD4 inhibition can inhibit joint inflammation and FBS-induced migration and invasion of human RA-FLS via decreasing the phosphorylation of c-Jun and activation of NFkappaB. 26111795_Data show that Bromodomain Containing 4 protein (BRD4) binds to the HOTAIR promoter, suggesting that BET proteins can directly regulate long noncoding RNAs (lncRNAs) expression. 26119939_combined administration of lenalidomide and BRD4 inhibitor JQ-1 significantly increased the survival of PEL bearing NOD-SCID mice in an orthotopic xenograft model 26220994_discovered that the translocation oncoprotein BRD4-NUT protein occupies approximately 100-200 extremely broad, cell type-specific hyperacetylated domains in the genome 26224795_BRD4 plays a key role in the pathological phenotype of pulmonary arterial hypertension. 26324948_This study implicates BET Brds as important regulators of IkappaB kinase/NF-kappaB-mediated synovial inflammation of RA and identifies BET proteins as novel therapeutic targets in inflammatory arthritis. 26365679_HPV16 E2-Brd4 interaction is more responsible for the transcriptional activation of host genes rather than repression. 26456956_Data show that knockdown of bromodomain containing 4 (BRD4) reversed the effect of microRNA miR-329 inhibition. 26504077_novel actions of BRD4 and of NELF-E in GR-controlled gene induction have been uncovered. 26575167_High BRD4 expression is associated with hepatocellular carcinoma. 26623725_Ewing sarcoma may be susceptible to treatment with epigenetic inhibitors blocking BRD3/4 activity and the associated pathognomonic EWS-FLT1 transcriptional program. 26626481_Study provides evidence that links the CHD8 chromatin remodeler to the cancer maintenance functions of BRD4 and NSD3. 26707881_In conclusion, our findings revealed that the aberrant expression of BRD4 in thyroid cancer is possibly involved in tumor progression, and JQ1 is potentially an effective chemotherapeutic agent against human thyroid cancer. 26752646_the long noncoding RNA colon cancer-associated transcript 1 (CCAT1) is transcribed from this superenhancer and is exquisitely sensitive to BET inhibition. 26840017_this study suggests that BRD4 is one of the major contributors to the invasion-prone phenotype of non-small cell lung cancer, and a potential therapeutic target of non-small cell lung cancer. 26855180_we found that NF-kappaB, BRD4 and RNA POL II were rapidly distributed at the upstream regions of miR-146a and miR-155, and more importantly mediated the formation of the super enhancers that drive miR-146a and miR-155 transcription. 26858406_findings identify key structural features of the ET domain of Brd4 that allow for interactions with both cellular and viral proteins. 26939702_BRD4 positively regulates EZH2 transcription through upregulation of C-MYC, and is a novel promising target for pharmacologic treatment in transcriptional program intervention against this intractable disease. 27007123_The Brd4 acetyllysine-binding protein of RelA is involved in activation of polyomavirus JC. 27084101_these studies highlight the importance of Brd4 in Helicobacter pylori-induced inflammatory gene expression and suggest that Brd4 could be a potential therapeutic target for the treatment of Helicobacter pylori-triggered inflammatory diseases and cancer 27099234_This study reveals how cells undergoing oncogene-induced senescence acquire a distinctive enhancer landscape that includes formation of super-enhancers adjacent to immune-modulatory genes required for paracrine immune activation. This process links BRD4 and super-enhancers to a tumor-suppressive immune surveillance program that can be disrupted by small molecule inhibitors of the bromo and extra terminal domain family of 27128490_present a method for rapid Sortase A-mediated segmental labelling of the individual bromodomains of BRD4 that provides a powerful strategy that will enable NMR studies of ligand-bromodomain interactions with atomic detail 27159561_Both mouse and human BRD4 have intrinsic histone acetyltransferase activity. 27223260_Findings reveal that PCa-associated ERG can interact and co-occupy with BRD4 in the genome, and suggest this druggable interaction is critical for ERG-mediated cell invasion and PCa progression. 27259267_Diverse gastric cancer cell lines of Asian and Brazilian origins differ in BRD4 and c-MYC expression levels and sensitivity to BET inhibitors. 27294782_DOT1L, via dimethylated histone H3 K79, facilitates histone H4 acetylation, which in turn regulates the binding of BRD4 to chromatin in acute lymphoblastic leukemia. 27425608_A miR-9 mimic represses stimulus-dependent targeting of BRD4. 27440272_Further analysis indicated that JQ1 inhibited the recruitment of BDR4 to the promoter complex of the Myc and Ccnd1 genes in rat thyroid follicular PCCL3 cells, resulting in decreased MYC expression at the mRNA and protein levels to inhibit tumor cell proliferation 27450555_BRD4 binds and stays associated with chromatin during mitosis, bookmarking early G1 genes and reactivating transcription after mitotic silencing. BRD4 acts as a passive scaffold via its recruitment of vital transcription factors and as an active kinase that phosphorylates RNA polymerase II. A model in which BRD4 actively coordinates chromatin structure and transcription is described. Review. 27452461_The response of the kinome to targeted BETi treatment in a panel of BRD4-dependent ovarian carcinoma (OC) cell lines. 27477287_BRD4 phosphorylation regulates HPV E2-mediated viral transcription and cellular MMP-9 expression 27494802_Results suggest structure-based drug design of bromodomain-containing protein 4 (Brd4) inhibitors. 27536004_BRD4 regulates splicing during heat shock by interacting with HSF1 such that under heat stress BRD4 is recruited to nuclear stress bodies, and non-coding SatIII RNA transcripts are up-regulated. 27624132_Thus, Brd4 activates human papillomavirus 16 transcription at this integration site, and strong selection for E6/E7 expression can drive the formation of a super-enhancer-like element to promote oncogenesis. 27651452_BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. 27666594_The studies show that BRD4 inhibition may have therapeutic implications for the treatment of mitochondrial diseases. 27698495_We postulate that BRD4-NUT (B4N) complexes override the preexisting histone code with new posttranslational modifications patterns that reflect aberrant transcription and that epigenetically modulate the nucleosome environment toward the NUT-midline carcinoma (NMC) state. 27716487_The studies show that BRD4 inhibition may have therapeutic implications for the treatment of mitochondrial diseases. 27732564_Data suggest that pharmacological inhibition of BET proteins could be a potential treatment for renal fibrosis. 27764245_this study demonstrates BRD4 has an essential role in HSV infection 27764789_Study identified Brd4 as a novel proline hydroxylation substrate with nearly 60% stoichiometry under normoxia states and its hydroxylation level is enzymatically regulated. 27769352_The identification of BRD4 as the primary target for BET inhibitors in cancer therapy and the discovery of CK2/PP2A-regulated BRD4 phosphorylation as the critical entity linked to gene regulation, drug resistance, and cancer progression highlight the importance of developing new BET inhibitors targeting the phospho region and other domains of BRD4 mediating protein-protein interaction 27793799_These data provide a detailed mechanism for how activated NF-kappaB and BRD4 control epithelial-mesenchymal transition initiation and transcriptional elongation in model airway epithelial cells in vitro and in a murine pulmonary fibrosis model in vivo. 27846392_Results identify BRD4 as essential protein in blastic plasmacytoid dendritic cell neoplasm and TCF4 expression requires BRD4. 27879331_The authors show that, upon infection of primary human keratinocytes with human papillomavirus 18 quasivirus, Brd4 activates viral transcription and replication. 27917784_BRD4 has a role in inhibiting neoplasms characterized by activation of c-MYC 27965149_Brd4 is involved in different steps of the papillomavirus life cycle. (Review) 28012209_peroxisome proliferator activated receptor alpha peroxisome proliferator-activated receptor alpha PPAR-alpha nuclear receptor subfamily 1 group C member 1 peroxisome proliferative activated receptor alpha peroxisome proliferator-activated nuclear receptor alpha variant 3 28063381_Brd4 inhibition attenuates unilateral ureteral obstruction-induced fibrosis by blocking TGF-beta-mediated Nox4 expression. 28073847_Our data strongly support the use of CCR2 and CD180 mRNAs as whole blood pharmacodynamic (PD)biomarkers for BRD4 inhibitors, especially in situations where paired tumor biopsies are unavailable. In addition, they can be used as tumor-based PD biomarkers for hematologic tumors. 28076398_A BRD4 missense mutation was the likely disease-causing mutation in this family with autosomal dominant syndromic congenital cataracts associated with neuro-skeletal anomalies. 28077651_These data validate BRD4 as a major effector of respiratory syncytial virus-induced inflammation and disease. BRD4 is required for coupling NF-kappaB to expression of inflammatory genes and the IRF-RIG-I autoamplification pathway and independently facilitates antiviral interferon-stimulated gene expression. 28100396_any of the molecules or processes in the network could be targeted to curb the oncogenic effects of c-Myc, just as BRD4 can be targeted. And since the targeting of metabolic enzymes has proved effective in mouse tumor models, it might be possible to develop new therapies based on the fact that c-Myc has a role in controlling cellular metabolism 28182006_BRD4 and CDK9 have independent, coordinated roles in promoting the myofibroblast transition 28203693_we also identified a complex genomic rearrangement involving the BRD4-NUT rearrangement underlying the simple t(15;19) karyotype in Nuclear protein in testis (NUT) midline carcinoma 28359301_we have discovered that BRD4-bound super-enhancers provide a powerful tool for enriching and prioritizing PC and BC genetic risk loci 28369619_BRD4 and MYC are essential for the expression of a subgroup of genes induced by class-I HDAC inhibitors. 28391274_research reveals that BRD4 probably play a critical role in Renal Cell Carcinoma progression, and is a new promising target for pharmacological treatment directed against this intractable disease. 28415703_these results highlighted the significant genetic contribution of the ARID1B variant, rs73013281, to susceptibility for HCC, especially in interaction with physical activity. 28446439_Cells harboring the fusion gene are selectively sensitive to small-molecule inhibition of protein targets induced by, or bound to, PAX3-FOXO1-occupied super enhancers. Furthermore, PAX3-FOXO1 recruits and requires the BET bromodomain protein BRD4 to function at super enhancers, resulting in a complete dependence on BRD4 and a significant susceptibility to BRD inhibition 28481868_High BRD4 expression is associated with stomach neoplasms. 28525743_BRD4 represses autophagy and lysosome gene expression. This repression is alleviated during nutrient deprivation through AMPK-SIRT1 signaling, allowing autophagy activation. BRD4 inhibition enhances autophagic flux and lysosomal function and promotes the degradation of protein aggregates. Sakamaki et 28588073_In pluripotent cells, Brd2-Brd4 occupy Nodal gene regulatory elements (NREs), but only Brd4 is required for pluripotency gene expression. Brd4 downregulation facilitates pluripotent exit and drives enhanced Brd2 NRE occupancy, thereby unveiling a specific function for Brd2 in differentiative Nodal-Smad2 signalling 28630312_BRD4 hyperphosphorylation is associated with cellular transformation in NUT midline carcinoma 28673542_Chemical degrader of BET bromodomain proteins, dBET6, led to the unexpected identification of BRD4 as master regulator of global transcription elongation in acute T-cell leukemia. BRD4 loss does not directly affect CDK9 localization. 28681984_Study shows that BRD4 is significantly highly expressed in gastric cancer patients and cell lines and positively regulates the expression of c-MYC through transcription regulation and epigenetic levels. Functionally, these result demonstrate that the knockdown of BRD4 represses the proliferation and induces the apoptosis of gastric cancer cells through repression of c-MYC. 28694331_These findings reveal a novel mechanism by which the HBV genome hijacks the host P-TEFb-containing complexes to promote its own transcription; both the super elongation complex and BRD4 can bind to the HBV genome. 28766687_Our data suggested that downregulation of BRD4 in gallbladder cancer (GBC) cells induced apoptosis by PI3K/AKT pathway. Inhibition of BRD4 expression may be a novel therapeutic strategy for patients with GBC. 28813519_results indicated that in MPNST samples BRD4 mRNA levels were not upregulated and that MPNST cell lines were relatively insensitive to the bromodomain inhibitor JQ1. 28844864_The short isoform of BRD4 cooperates with SWI/SNF nucleosome remodelers to repress HIV transcription during latency, a phenotype reversed by BET inhibitor treatment. 28844955_Genetic and pharmacological inhibition of BRD4 suppressed IL-1beta-induced expression and translocation of HMGB1. Chromatin immunoprecipitation (ChIP) showed the enrichment of BRD4 around the HMGB1 upstream non-promoter region, which diminished with JQ1 treatment. 28854735_Long-term treatment with bromodomain-containing protein 4 (BRD4) inhibitors caused telomere shortening in both mouse and human cells, suggesting BRD4 plays a role in telomere maintenance in vivo. 28981843_Data show that BRD4 controls RUNX2 by binding to the enhancers (ENHs) and each RUNX2 ENH is potentially controlled by a distinct set of TFs and c-JUN as the principal pivot of this regulatory platform. 28991225_Study in melanoma cell and in vivo melanoma models provides evidence for a direct role of BRD4 binding at super-enhancers that drive the expression of PGC-1alphaalpha and SOX10, a transcription factor involved in melanocyte development. 29133261_Together, dual inhibition of BRD4 and PI3K by SF2523 suppresses human prostate cancer cell growth in vitro and in vivo. 29240787_conclude that BRD3/4 and the FLT3-TAK1/NF-kB pathways collectively control a set of targets that are critically important for the survival of human MLL-AF9 cells 29346775_Authors demonstrate a novel role for BRD4 in the formation of oncogenic TMPRSS2-ERG fusions via its involvement in the NHEJ DNA repair pathway. 29379197_BRD4 and NIPBL displayed correlated binding at super-enhancers and appeared to co-regulate developmental gene expression. 29437854_Results provide evidence that both BRD3 and BRD4 are repressors of epithelial-to-mesenchymal transition in breast cancer cells. 29463681_Direct interactions between AIRE, NF-kappaB, and P-TEFb result in efficient transcription of their target genes. 29636547_A new biological function of BRD4. 29716963_the synergistic effect is likely due to two reasons: (i) Plk1 inhibition results in the accumulation of beta-catenin in the nucleus, thus elevation of c-MYC expression, whereas JQ1 treatment directly suppresses c-MYC transcription; (ii) Plk1 and BRD4 dual inhibition acts synergistically in inhibition of AR signaling. 29748248_High BRD4 expression is associated with preeclampsia. 29777702_miR-608 inhibits hepatocellular carcinoma cell proliferation possibly via targeting BET family protein BRD4. 29898995_we describe the dependency of EWS/ETS-driven transcription upon chromatin reader BET bromdomain proteins and investigate the potential of BET inhibitors in treating EWS. EWS/FLI1 and EWS/ERG were found in a transcriptional complex with BRD4, and knockdown of BRD2/3/4 significantly impaired the oncogenic phenotype of EWS cells. RNA-seq analysis following BRD4 knockdown or inhibition with JQ1 revealed an attenuated EWS/ETS 30012592_MS645 blocks BRD4 binding to transcription enhancer/mediator proteins MED1 and YY1 with potency superior to monovalent BET inhibitors, resulting in down-regulation of proinflammatory cytokines and genes for cell-cycle control and DNA damage repair that are largely unaffected by monovalent BrD inhibition. 30015945_miR204 directly binds to UCA1 and the 3'untranslated region of BRD4. Furthermore, UCA1 competed with BRD4 for miR204 binding. miR204 knockdown enhanced BRD4 expression, which can be partially restored by short hairpinUCA1. 30029633_Tumoral BRD4 expression in breast cancer is significantly associated with T-bet+ TILs, clinicopathological features, and a poor disease-free survival in the absence of T-bet+ TILs. 30036377_BRD4 amplification in cancer has oncogenic potential; BRD4-amplified High-grade Serous Ovarian Cancer is a potential patient population that could benefit from BET inhibitors 30057199_DUB3 promotes BET inhibitor resistance and cancer progression by deubiquitinating BRD4. 30076409_results suggest a mechanism by which eRNAs are directly involved in gene regulation by modulating enhancer interactions and transcriptional functions of BRD4 30079805_The discovery of novel BRD4 inhibitors. Communicated by Ramaswamy H. Sarma. 30154425_Ectopic SPZ1 and TWIST1 expression, but not that of TWIST1 alone, enhanced VEGF expression via the recruitment of BRD4, enhancing RNA-Pol II-dependent transcription and inducing metastasis. Acetylation signaling functions in the SPZ1-TWIST1-BRD4 axis in the mediation of EMT and its regulation during tumor initiation and metastasis 30172011_PES1 is transcriptionally regulated by BRD4 and promotes cell proliferation and glycolysis in hepatocellular carcinoma 30185888_Pancreatic stellate cells (PSCs), the predominant fibroblasts in human pancreatic ductal adenocarcinoma tumors, express high levels of PD-L1 and the interplay between IRF1 and BRD4 regulates PD-L1 expression in PSCs. 30206163_we show that BET inhibition across multiple myeloma cell lines resulted in suppressed interleukin (IL)-6 Janus kinase-signal transducers and activators of transcription (JAK-STAT) signaling. INCB054329 displaced binding of BRD4 to the promoter of IL6 receptor (IL6R) leading to reduced levels of IL6R and diminished signaling through STAT3. 30224758_This work sheds light on essential mediators, mechanisms and genome-wide regulatory elements that are responsible for transcriptional addiction in cancer and lays the grou | ENSMUSG00000024002 | Brd4 | 4.744961e+03 | 1.3750319 | 0.459465056 | 0.2945658 | 2.408285e+00 | 0.1206945113 | 0.75783482 | No | Yes | 5.209426e+03 | 529.524470 | 3.497070e+03 | 365.111374 | |
| ENSG00000141971 | 93343 | MVB12A | protein_coding | Q96EY5 | FUNCTION: Component of the ESCRT-I complex, a regulator of vesicular trafficking process. Required for the sorting of endocytic ubiquitinated cargos into multivesicular bodies. May be involved in the ligand-mediated internalization and down-regulation of EGF receptor. {ECO:0000269|PubMed:16895919}. | 3D-structure;Alternative splicing;Cytoplasm;Cytoskeleton;Endosome;Membrane;Nucleus;Phosphoprotein;Protein transport;Reference proteome;SH3-binding;Transport | hsa:93343; | centrosome [GO:0005813]; cytosol [GO:0005829]; endosome membrane [GO:0010008]; ESCRT I complex [GO:0000813]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; late endosome membrane [GO:0031902]; nucleoplasm [GO:0005654]; vesicle [GO:0031982]; lipid binding [GO:0008289]; SH3 domain binding [GO:0017124]; ubiquitin binding [GO:0043130]; endosome to lysosome transport via multivesicular body sorting pathway [GO:0032510]; macroautophagy [GO:0016236]; multivesicular body assembly [GO:0036258]; protein transport [GO:0015031]; receptor catabolic process [GO:0032801]; regulation of epidermal growth factor receptor signaling pathway [GO:0042058]; ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway [GO:0043162]; viral budding [GO:0046755]; viral budding via host ESCRT complex [GO:0039702]; virus maturation [GO:0019075] | 16895919_CFBP is a novel tyrosine-phosphorylated protein that might function as a regulator of CIN85/CD2AP 20654576_These results suggest that the expression of MVB12B may be normally suppressed through the ubiquitin-proteasome pathway that simultaneously regulates the fate of MVB12A and the functions of ESCRT-I. 32424346_A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission. | ENSMUSG00000031813 | Mvb12a | 2.258766e+03 | 1.5947713 | 0.673349516 | 0.3160420 | 4.555531e+00 | 0.0328125890 | 0.58913859 | No | Yes | 2.883567e+03 | 426.036980 | 1.448841e+03 | 220.277713 | ||
| ENSG00000142173 | 1292 | COL6A2 | protein_coding | P12110 | FUNCTION: Collagen VI acts as a cell-binding protein. | Alternative splicing;Cell adhesion;Collagen;Congenital muscular dystrophy;Direct protein sequencing;Disease variant;Extracellular matrix;Glycoprotein;Hydroxylation;Limb-girdle muscular dystrophy;Membrane;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal | This gene encodes one of the three alpha chains of type VI collagen, a beaded filament collagen found in most connective tissues. The product of this gene contains several domains similar to von Willebrand Factor type A domains. These domains have been shown to bind extracellular matrix proteins, an interaction that explains the importance of this collagen in organizing matrix components. Mutations in this gene are associated with Bethlem myopathy and Ullrich scleroatonic muscular dystrophy. Three transcript variants have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:1292; | collagen trimer [GO:0005581]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; extracellular vesicle [GO:1903561]; protein-containing complex [GO:0032991]; sarcolemma [GO:0042383]; collagen binding [GO:0005518]; extracellular matrix structural constituent conferring tensile strength [GO:0030020]; cell adhesion [GO:0007155]; response to glucose [GO:0009749] | 12011280_Haplotype analysis clearly suggested linkage of Ullrich muscular dystrophy to the COL6A1/2 locus in two cases and to the COL6A3 loci in the third case. In the remaining nine patients, primary collagen VI involvement was excluded 12218063_the C-terminal globular domain of COL6A2 is not essential for triple-helix formation but is critical for microfibrillar assembly in Ullrich congenital muscular dystrophy 12297580_A case of Ullrich disease is associated with complete deficiency of collagen VI and compound heterozygous mutations in the collagen VI alpha 2 gene with absence of microfibrils on electron microscopy. 12374585_Bethlem myopathy is an autosomal dominantly inherited myopathy with contractures, caused by mutations in COL6A1 gene, COL6A2 gene or COL6A3 gene. 14981181_In Ullrich syndrome, a heterozygous G-to-A substitution at position +5 in intron 23 & the corresponding heterozygous 6-bp deletion in exon 26 which deleted 1 of the 2 tandem repeats of the sequence CATCGG in nt 2268-2273 & 2274-2279 in COL6A2 ORF. 15563506_dominant mutations are common in Ullrich congenital muscular dystrophy (UCMD). 16075202_diminished COL6A2 mRNA expression found to be primary pathogenic mechanism in UCMD patient 17602442_This study demonstrates a homogenoeous overexpression of the genes encoding for alpha1 and alpha2 chains of collagen type VI in nuchal skin of human trisomy 21 fetuses. 18366090_Study reports 10 unrelated patients with a Ullrich congenital muscular dystrophy clinical phenotype and de novo dominant negative heterozygous splice mutations in COL6A1, COL6A2 and COL6A3. 18852439_Results describe the characteristic features of myosclerosis myopathy with a homozygous collagen type 6A2 mutation responsible for a peculiar pattern of collagen VI defects. 19204726_Observational study of gene-disease association. (HuGE Navigator) 19309692_Four patients affected by Ullrich congenital muscular dystrophy and carrying unusual mutations of COL6 genes affecting RNA splicing, were identified. 19698785_The alpha2(VI) chain modulates matrix-metalloproteinase (MMP) availability by sequestering proMMPs in the extracellular matrix, blocking proteolytic activity. 20106987_the C2A splice variant has a role in recessive COL6A2 C-globular missense mutations in Ullrich congenital muscular dystrophy 20302629_A deletion within intron 1A of the COL6A2 gene, occurring in compound heterozygosity with a small deletion in exon 28, was identified in a BM patient. 23138527_Homozygous COL6A2 mutation, p.Asp215Asn, was identified in both affected siblings. We conclude that the COL6A2 p.Asp215Asn mutation is likely to be responsible for PME (Progressive Myoclonus Epilepsy) in this family. 23452080_COL6A2 is overexpressed in Down syndrome-affected umbilical cords at early and term gestational ages. 24443028_Mutations in each of the three collagen VI genes, COL6A1, COL6A2 and COL6A3, cause four types of muscle disorders: Ullrich congenital muscular dystrophy, Bethlem myopathy, limb-girdle muscular dystrophy, and autosomal recessive myosclerosis. (Review) 24801232_In UCMD, 8 mutations were identified in COL6A2 in Chinese patients. 25204870_Parental mosaicism was confirmed in the four families through quantitative analysis of the ratio of mutant versus wild-type allele (COL6A1, COL6A2, and COL6A3) in genomic DNA from various tissues; consistent with somatic mosaicism, parental samples had lower ratios of mutant versus wild-type allele compared with the fully heterozygote offspring. 25533456_Mutations in COL6A2 gene are associated with aberrant mitochondria in Bethlem myopathy. 26944560_binding of collagen VI to NG2 is essential for the direction of tendon fibroblasts migration in vitro. 27563703_Genetic study showed a missense mutation in COL6A2 (c.820 G>A, p.Gly268Ser) that causes a glycine substitution in the Gly-X-Y collagenous motif, at the beginning of the collagenous triple helical domain. The c.820 G>A mutation segregated in all the affected patients. 32053901_Tendon Extracellular Matrix Remodeling and Defective Cell Polarization in the Presence of Collagen VI Mutations. 32350230_Proteomics Profiling Reveals Insulin-Like Growth Factor 1, Collagen Type VI alpha-2 Chain, and Fermitin Family Homolog 3 as Potential Biomarkers of Plaque Erosion in ST-Segment Elevated Myocardial Infarction. 33537799_Use of RNAsequencing to detect abnormal transcription of the collagen alpha2 (VI) chain gene that can lead to Bethlem myopathy. 33982770_Collagen type VIalpha1 and 2 repress the proliferation, migration and invasion of bladder cancer cells. | ENSMUSG00000020241 | Col6a2 | 2.762815e+03 | 1.0735054 | 0.102329452 | 0.3023812 | 1.135728e-01 | 0.7361126889 | 0.94490947 | No | Yes | 2.400714e+03 | 270.351527 | 2.298778e+03 | 265.662993 | |
| ENSG00000142330 | 11132 | CAPN10 | protein_coding | Q9HC96 | FUNCTION: Calcium-regulated non-lysosomal thiol-protease which catalyzes limited proteolysis of substrates involved in cytoskeletal remodeling and signal transduction. May play a role in insulin-stimulated glucose uptake. {ECO:0000269|PubMed:17572128}. | Alternative splicing;Diabetes mellitus;Hydrolase;Protease;Reference proteome;Repeat;Thiol protease | Calpains represent a ubiquitous, well-conserved family of calcium-dependent cysteine proteases. The calpain proteins are heterodimers consisting of an invariant small subunit and variable large subunits. The large catalytic subunit has four domains: domain I, the N-terminal regulatory domain that is processed upon calpain activation; domain II, the protease domain; domain III, a linker domain of unknown function; and domain IV, the calmodulin-like calcium-binding domain. This gene encodes a large subunit. It is an atypical calpain in that it lacks the calmodulin-like calcium-binding domain and instead has a divergent C-terminal domain. It is similar in organization to calpains 5 and 6. This gene is associated with type 2 or non-insulin-dependent diabetes mellitus (NIDDM), and is located within the NIDDM1 region. Multiple alternative transcript variants have been described for this gene. [provided by RefSeq, Sep 2010]. | hsa:11132; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; calcium-dependent cysteine-type endopeptidase activity [GO:0004198]; cytoskeletal protein binding [GO:0008092]; SNARE binding [GO:0000149]; actin cytoskeleton reorganization [GO:0031532]; cellular component disassembly involved in execution phase of apoptosis [GO:0006921]; cellular response to insulin stimulus [GO:0032869]; positive regulation of glucose import [GO:0046326]; positive regulation of insulin secretion [GO:0032024]; positive regulation of intracellular transport [GO:0032388]; positive regulation of type B pancreatic cell apoptotic process [GO:2000676]; proteolysis [GO:0006508]; type B pancreatic cell apoptotic process [GO:0097050] | 11481585_Observational study of gene-disease association. (HuGE Navigator) 11522685_Observational study of gene-disease association. (HuGE Navigator) 11704924_Observational study of gene-disease association. (HuGE Navigator) 11704924_type 2 diabetes and three calpain-10 gene polymorphisms in Samoans: no evidence of association 11756346_Observational study of gene-disease association. (HuGE Navigator) 11756349_Observational study of gene-disease association. (HuGE Navigator) 11774208_Observational study of gene-disease association. (HuGE Navigator) 11774208_The variation of calpain-10 gene has impact on the variation of clinical metabolic parameter levels related to type 2 diabetes mellitus. 11872216_Observational study of gene-disease association. (HuGE Navigator) 11891023_Observational study of gene-disease association. (HuGE Navigator) 11891023_polymorphism related to increased serum cholesterol, but not to NIDDM in Japan 11891618_Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. 11891618_Observational study of genotype prevalence. (HuGE Navigator) 11932299_Observational study of gene-disease association. (HuGE Navigator) 11935160_Observational study of gene-disease association. (HuGE Navigator) 11935160_polymorphism influences glucose metabolism in human fat cells 11978665_Observational study of gene-disease association. (HuGE Navigator) 11978669_Observational study of gene-disease association. (HuGE Navigator) 11980626_Homozygous combination of calpain 10 gene haplotypes is associated with type 2 diabetes mellitus in a Polish population. 11980626_Observational study of gene-disease association. (HuGE Navigator) 12046551_SNP distribution of CAPN10 gene differs in different Chinese nationalities; studied SNPs in CAPN10 gene may not be the major susceptibility ones of NIDDM in Han people of Northern China 12050223_Observational study of gene-disease association. (HuGE Navigator) 12050223_Variation within the type 2 diabetes susceptibility gene calpain-10 and polycystic ovary syndrome. 12083814_No evidence for involvement of the calpain-10 gene 'high-risk' haplotype combination for non-insulin-dependent diabetes mellitus in early onset obesity. 12107250_Calpain-10 gene polymorphism is associated with reduced beta(3)-adrenoceptor function in human fat cells. a deletion/insertion polymorphism. 12107250_Observational study of gene-disease association. (HuGE Navigator) 12107735_Observational study of gene-disease association. (HuGE Navigator) 12107735_investigation of the influence of polymorphisms in the Calpain-10 gene on microvascular function 12107745_Observational study of gene-disease association. (HuGE Navigator) 12107745_insulin sensitivity of glucose disposal and lipolysis: no influence of common genetic variants 12133483_Observational study of gene-disease association. (HuGE Navigator) 12133483_The UCSNP44 variation of calpain 10 gene on NIDDM1 locus and its impact on plasma glucose levels in type 2 diabetic patients 12137596_Observational study of gene-disease association. (HuGE Navigator) 12137596_detect the association among calpain-10(CAPN-10) gene polymorphism, hypentension and hyperglycemia 12145185_Observational study of gene-disease association. (HuGE Navigator) 12145185_Variation in gene predisposes to insulin resistance and elevated free fatty acid levels. 12161543_CAPN10 alleles are associated with polycystic ovary syndrome 12161543_Observational study of gene-disease association. (HuGE Navigator) 12453914_Gene frequency of the 112/121 at-risk haplotype of CAPN10 is low among Scandinavians and we were unable to demonstrate significant associations between the CAPN10 variants and type 2 diabetes, insulin resistance, or impaired insulin secretion. 12453914_Observational study of gene-disease association. (HuGE Navigator) 12519860_Genetic variations in the 112/121 haplotype combination defined by the UCSNP-43, -19, and -63 alleles in the calpain-10 gene are not a major factor in the occurrence of type 2 diabetes in Japanese. 12519860_Observational study of gene-disease association. (HuGE Navigator) 12905623_Observational study of gene-disease association. (HuGE Navigator) 14500039_Observational study of gene-disease association. (HuGE Navigator) 14574648_confirmation of role in type 2 diabetes susceptibility 14602801_Observational study of gene-disease association. (HuGE Navigator) 14602801_risk alleles and genotypes, within CAPN10 gene, that could be associated with important phenotypic and prognosis differences observed in polycystic ovary patients. 14642065_Observational study of gene-disease association. (HuGE Navigator) 14646187_Calpain 10 gene has potential for use in predicting the incidence of type 2 diabetes by genetic diagnosis p.1766 14730479_Meta-analysis of gene-disease association. (HuGE Navigator) 14741193_Observational study of gene-disease association. (HuGE Navigator) 14741193_The results suggest that variation in CAPN10 affects risk of type 2 diabetes in the mestizo population of central Mexico (Mexico City and Orizaba) and in Mexican Americans (Starr County, Texas). 14749261_variation at CAPN10 in different human populations over a range of phenotypes related to type 2 diabetes 14974344_Observational study of gene-disease association. (HuGE Navigator) 15044459_calpain-10 has a role in beta-cell survival and is suppressed by RyR2 15172858_Observational study of gene-disease association. (HuGE Navigator) 15240652_Observational study of gene-disease association. (HuGE Navigator) 15240652_Reduced CAPN10 expression may be risk factor for features associated with metabolic syndrome in obese subjects, although variation in gene does not seem to contribute to risk for developing obesity per se. 15471947_an isoform of calpain-10 is a Ca2+-sensor that functions to trigger exocytosis in pancreatic beta-cells 15641690_Observational study of gene-disease association. (HuGE Navigator) 15652721_Observational study of gene-disease association. (HuGE Navigator) 15696376_Observational study of gene-disease association. (HuGE Navigator) 15793266_We show that the diabetes gene calpain-10 (CAPN10) plays a role in atherosclerosis, insulin sensitivity and insulin secretion in a population enriched for atherosclerosis and insulin resistance. 15860244_Observational study of gene-disease association. (HuGE Navigator) 15860244_Polymorphism of CAPN 10 might be associated with type 2 diabetes. 15862281_Studies confirm calpain 10 expression in cultured muscle cells and support calpains in insulin-stimulated glucose uptake in human skeletal muscle cells that may be relevant to the pathogenesis of the peripheral insulin resistance in type 2 diabetes. 15926113_Observational study of gene-disease association. (HuGE Navigator) 16028216_Calpain 10 is a molecule of importance to insulin signaling and secretion that may have relevance to the future development of novel therapeutic targets for the treatment of T2D[review] 16186407_study provides evidence that messenger RNA expression of calpain-10 (CAPN10) in skeletal muscle is under genetic control and glucose-tolerant individuals upregulate messenger RNA levels in response to prolonged exposure to fat 16306378_There may be one or more relatively common alleles increasing risk of type 2 diabetes in this local region. 16333311_Observational study of gene-disease association. (HuGE Navigator) 16377260_Observational study of gene-disease association. (HuGE Navigator) 16377260_UCSNP-19 of CAPN10 may be involved in the pathogenesis of diabetes in CF. 16427731_Observational study of gene-disease association. (HuGE Navigator) 16456802_Observational study of genotype prevalence. (HuGE Navigator) 16546286_Observational study of gene-disease association. (HuGE Navigator) 16546286_Results suggest that a novel 111/121 haplotype combination created by the CAPN10 SNP-43, -19, and -63 increases the susceptibility to the metabolic syndrome in patients with type 2 diabetes. 16721485_A novel diplotype in CAPN10 gene is associated with diabetes mellitus, type 2 in the Korean population. 16721485_Observational study of gene-disease association. (HuGE Navigator) 16752174_Observational study of gene-disease association. (HuGE Navigator) 16752174_SNP-43 of CAPN10 may contribute to the risk of diabetes by regulating abdominal obesity in subjects with high risk of type 2 diabetes. 16837224_Meta-analysis of gene-disease association. (HuGE Navigator) 16837224_the linkage disequilibrium and haplotype diversity studies suggest a role for genetic variation in CAPN10 affecting risk of type 2 diabetes in Europeans. 16857402_No consistent evidence of association of the CAPN10 SNP43 or SNP44 with T2D, obesity, or related quantitative traits, was found. 16857402_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 16873988_Calpain-10 (NIDDM1) as a Susceptibility Gene for Common Type 2 Diabetes. 17106059_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17106059_the calpain-10 TGA2AGCA haplotype is associated with an increased risk for PCOS. 17382509_Observational study of gene-disease association. (HuGE Navigator) 17382509_These results indicate that some CAPN10 alleles may be exerting a protective effect on laryngeal cancer risk in the Spanish population. 17454172_CAPN10 gene UCSNP-43 polymorphisms may influence the polycystic ovary syndrome metabolic phenotype 17454172_Observational study of gene-disease association. (HuGE Navigator) 17511963_a defective CAPN10 pre-mRNA processing is responsible for the decreased levels of SNP-43 A-allele transcripts in peripheral white cells of healthy and type 2 diabetes individuals 17559371_Observational study of gene-disease association. (HuGE Navigator) 17559371_These results indicate a recessive model for the effect of CAPN10 variant UCSNP-44 influencing the risk of colorectal cancer and suggest a novel genetic link between type 2 diabetes mellitus and colon carcinoma. 17560157_Fresh hope for the development of novel diabetic treatmentsutilizing either pharmacological activators that specifically target calpain-10, or through targeted calpain-10 gene therapy 17570749_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17572128_Calpain-10 may exert a direct regulatory effect upon the glucose uptake mechanism, raather than by insulin stimulation. 17601994_Observational study of gene-disease association. (HuGE Navigator) 17855447_Observational study of gene-disease association. (HuGE Navigator) 17855447_we did not observe any significant associations of the common SNPs or haplotypes across the CAPN10 gene with diabetes risk in our large and ethnically diverse cohort of postmenopausal women 17964829_human lymphocytes express calpain-10 mRNA and protein, showing a similar expression between diabetic and control subjects, nevertheless in the diabetic group calpain activity was less glucose-sensitive. 18167206_CAPN10 gene variations might play roles in the risk of diabetes and hypertension in northern Han Chinese population. 18167206_Observational study of gene-disease association. (HuGE Navigator) 18241614_genetic variation at the CALPN10 loci may confer higher cardiovascular disease risk in patients with type 2 diabetes mellitus 18248681_Observational study of genotype prevalence. (HuGE Navigator) 18367022_article reviews genetic evidence for association between CAPN10 & type 2 diabetes mellitus; biologic function of calpain 10 is discussed along with results from recent genome-wide association studies that have failed to put CAPN10 among the top signals 18452715_calpain-10 may require a special intracellular localization or interacting partner(s) to acquire proteolytic activity 18487065_In the Tunisian population, study found genetic variants within CAPN10 that are linked with type 2 diabetes and a novel haplotype combination, 121/221, associated with an increased susceptibility. 18487065_Observational study of gene-disease association. (HuGE Navigator) 18554168_Observational study of gene-disease association. (HuGE Navigator) 18554168_We conclude that the calpain 10 SNP-44 gene polymorphism may be accepted as a risk factor in the development of type 2 diabetes mellitus (T2DM) and elevated BMI in T2DM patients in a Turkish population. 18683748_CAPN-10 gene SNP-56 plays a role in glucose and lipid metabolism in Chinese PCOS patients but does not contribute to the genetic susceptibility of PCOS. 18683748_Observational study of gene-disease association. (HuGE Navigator) 18698425_CAPN10 gene is associated with insulin resistance phenotypes in the Spanish Caucasoid population. 18698425_Observational study of gene-disease association. (HuGE Navigator) 18701098_Observational study of gene-disease association. (HuGE Navigator) 18701098_Our data showed that 111 haplotype and 111/121 and 111/111 diplotypes in the CAPN-10 gene were associated with a significantly increased risk of PCOS 18722363_Observational study of gene-disease association. (HuGE Navigator) 18722363_Our data suggest the contribution of CAPN10 gene polymorphism to polycystic ovary syndrome in Chilean women 18936436_Observational study of genotype prevalence. (HuGE Navigator) 19144693_Calpain 10 levels decreased linearly in kidney with age in the absence of changes in calpains 1 or 2. 19193380_CAPN10 may have a role as a common genetic determinant of diabetes and atherosclerosis in Hispanics 19193380_Observational study of gene-disease association. (HuGE Navigator) 19297292_Observational study of gene-disease association. (HuGE Navigator) 19297292_Polymorphism is associated with a high risk of type 2 diabetes only in one Arab sub-group of the Tunisian population. 19336475_Observational study of gene-disease association. (HuGE Navigator) 19337008_Calpain 10 gene SNP 44 allele polymorphism may have a role in PCOS pathogenesis. 19337008_Observational study of gene-disease association. (HuGE Navigator) 19387820_Observational study of gene-disease association. (HuGE Navigator) 19418728_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19418728_SNP43 polymorphism may influence the response to treatment with sulphonylurea and metformin, the expression being dependent on obesity. 19491387_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19565864_Observational study of gene-disease association. (HuGE Navigator) 19570442_CAPN10 SNP-19 is associated with glucose metabolism disorders in pregnant women. 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19643578_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19688040_Observational study of gene-disease association. (HuGE Navigator) 19688040_calpain-10 mRNA was elevated by 64% in pancreatic islets from patients with T2D compared with non-diabetic donors. Moreover, the calpain-10 expression correlated positively with arginine-stimulated insulin release in islets from non-diabetic donors 19752882_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19752882_Patients' higher body mass index and SNP-63 minor T allele carrier status were identified as independent posttransplant diabetes mellitus risk factors. 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20075150_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20119856_Certain three window haplotypes may confer increased risk for T2DM and others may be protective suggesting that genetic variation in CAPN10 gene may be one factor involved in the aetiology of T2DM in Irish adults. 20119856_Observational study of gene-disease association. (HuGE Navigator) 20142250_prostate cancer was positively associated with the CAPN10 rs3792267 G allele 20178008_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20178008_variation in CAPN10 may be associated with increased risk of pancreatic cancer among smokers 20193213_Observational study of gene-disease association. (HuGE Navigator) 20193213_The association with T2DM in different races was evaluated. SNP43-G allele, G/G genotype, 111/221 were risk factors to Mongoloid race. And SNP-C allele, 111/111 haplotype combination were risk factors to Caucasoid race, and SNP44-C allele to Hybrid race. 20368234_Observational study of gene-disease association. (HuGE Navigator) 20368234_the most common haplotype 121 (OR = 0.70 95% CI: 0.50-0.99) was associated with a reduced risk for type 2 diabetes in East Indian population 20406624_CAPN10 gene may play an important role in the pathogenesis of impaired fasting glucose or impaired glucose tolerance in patients with esential hypertension. 20406624_Observational study of gene-disease association. (HuGE Navigator) 20470430_CAPN10 UCSNP-19 variant, and the 111 haplotype contribute to the risk of T2D in Tunisian subjects; no significant associations between CAPN10 diplotypes and T2D were demonstrated for Tunisians. 20470430_Observational study of gene-disease association. (HuGE Navigator) 20570542_Observational study of gene-disease association. (HuGE Navigator) 20570542_calpain 10 UCSNP-19 polymorphism and haplotype 111 contribute to the risk of type 2 diabetes (T2DM) in Tunisian subjects but no significant association between calpain 10 diplotypes and T2DM was demonstrated 20667559_Observational study of gene-disease association. (HuGE Navigator) 20667559_This study raises the possibility that the 2111 haplotype of SNPs -44, -43, -19, and -63 may be associated with type 2 diabetes mellitus, although none of these SNPs may be individually associated with diabetes. 20682687_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20848425_Calpain 10 gene polymorphism is modifying laryngeal cancer risk and mortality in Spanish population. 20848425_Observational study of gene-disease association. (HuGE Navigator) 20881413_Observational study of gene-disease association. (HuGE Navigator) 20881413_SNP-44 polymorphism of the calpain-10 gene has a significant association with T2DM patients in the Gaza strip 20923526_Meta-analysis and uncategorized study of gene-disease association. (HuGE Navigator) 21389182_genetic association studies in a European cohort: CAPN10 SNP (rs2953171) may influence insulin sensitivity by interacting with plasma fatty acid composition in subjects with metabolic syndrome 21906115_Studies indicate UCSNP-63 of CAPN 10 gene was significantly associated with polycystic ovary syndrome (PCOS). 22384174_CAPN10 SNPs and haplotypes are associated with polycystic ovary syndrome among South Indian Women 22796443_analysis of copy number variation of CAPN10 in Thais with type 2 diabetes by multiplex PCR and denaturing high performance liquid chromatography 23021796_Significant association of SNP -43 in CAPN10 with the risk of cardiovascular disease coexisting with T2 Diabetes mellitus. 23262350_We identified reduced Calpain-10 expression in a pediatric population with overweight and obese phenotypes. 23687414_GAEC1 regulates the expression of CAPN10 in esophageal squamous cell carcinoma. Calpain 10 expression is a potential prognostic marker. 23994294_Polymorphisms in the Calpain-10 gene may be risk factors for PCOS, especially among Asian populations.[meta-analysis] 24034724_Used homology modelling technique to study the 3D structure of calpain-10 from Homo sapiens and its interaction with the protease inhibitor SNJ-1715. 24266779_Variations of SNP-43, -63 and Indel-19 of CAPN10 were not associated with an increased risk of developing gestational diabetes mellitus. 24377587_Calpain-10 SNP43 and SNP19 polymorphisms are associated with colorectal cancer. 24429295_results of present meta-analysis indicate an association of T2D with carriers of DD genotype of CAPN10 I/D polymorphism 24612564_We replicated the significant association of rs1801278 and rs3792267 SNPs of the IRS1 and CAPN10 genes with T2DM in the population of Hyderabad. 24779302_Calpain 10 shows association between the single nucleotide polymorphism (SNP)-43, but not SNP-19 nor -63, and type 2 diabetes mellitus in the Kurdish ethnic group of West Iran 24802731_121 haplotype and 122/121 haplotype combination of SNP-19, -44 and -63 in the Calpain-10 gene are associated with the development of type 2 diabetes in Turkish patients. 24993116_This work confirms the association of CAPN10 gene with metabolic components in PCOS and highlights the role of haplotypes as strong and efficient genetic markers. 25238846_Subjects with the GG genotype of the rs2975762 variant of the CAPN10 gene were better responders to dietary intervention, showing increased HDL-C concentrations from the first month of treatment 25327507_The present study provides the first observation of an association between a variant in CAPN10 gene and the response to metformin therapy in patients with type 2 diabetes 25382134_genetic association studies in Asian populations: Data suggest that an SNP in CAPN10 (SNP43 G>A, rs3792267) is associated with type 2 diabetes in Asian populations, especially in Chinese populations. [META-ANALYSIS] 25504243_Genotype I/I of SNP19 in CAPN10 was significantly associated with excess weight in Colombian patients 10-18 years of age even those with physically active lifestyles. 25617558_SNP-19 in CAPN10 may participate in the development of diabetes mellitus type 2 25773692_Different mutations in CAPN10 have already been found in three independent Iranian families 25867367_Data indicate no association between calpain 10 (CAPN10) polymorphisms and type 2 diabetes mellitus. 25982606_SNP-63 and indel-19 variant of the CAPN10 gene do not represent a risk factor for polycystic ovary syndrome in Mexican women of reproductive age. 26376770_due to its association with androgen excess in phenotype A, CAPN 10 gene polymorphism UCSNP-43 could be used as a genetic marker for CVD in young PCOS women. 27324783_association between SNP 63 of CAPN10 and gestational diabetes mellitus is only significant in the heterozygous model 27374856_There were significant differences between the type 2 diabetes mellitus patients and controls in the risk allele distributions of rs3792267 (CAPN10) (P = 0.002), rs1501299 (APM1) (P = 0.017), and rs3760776 (FUT6) (P = 0.031). 28277135_TCF7L2 rs7903146 and 112/112 haplotype of CAPN10 might be associated with gestational diabetes risks. [meta-analysis] 28360393_The study results were suggestive of a positive association between Gly972Arg of IRS1 and PCOS in the south Indian population, while INS, IRS2, PPAR-G and CAPN10 failed to show any association with PCOS in our studied population. 28422847_These findings indicate that the Calpain-10 SNP 43 may be related to obstructive sleep apnea/hypopnea syndrome with ischemic stroke, with SNP 43 GG genotype as a risk factor for obstructive sleep apnea/hypopnea with ischemic stroke 29506634_The 3R/3R genotype of the indel-19 variant of the CAPN-10 gene influenced increased glucose levels in these Mexican women with gestational diabetes mellitus. 30014550_This study provides evidence that SNP43 (G/A) in the CAPN10 gene increases the risk of cognitive impairment in cerebral small vessel disease patients. 30425305_Calpain-10 regulates actin dynamics by proteolysis of microtubule-associated protein 1B 31292430_grade B and grade D diabetes in the CC and TC genotypes of rs2975760 were significantly different from those in the TT genotype.Grade B and grade D diabetes in the AA and AG genotypes of rs3792267 were significantly different compared with those in the GG genotype (P<0.05), and allele A was significantly increased compared with allele G (P<0.05). 31460838_The GSTT1 null genotype frequency is associated with hyperglycemia in type 2 diabetes mellitus, whereas there is no difference in the CAPN10 SNP19 in this condition. 32236617_Clinical significance of sHER2ECD and calpain10 expression in tumor tissues of patients with breast cancer. | ENSMUSG00000026270 | Capn10 | 1.972874e+03 | 1.1366363 | 0.184770669 | 0.2933443 | 3.958577e-01 | 0.5292363013 | 0.88709702 | No | Yes | 1.810804e+03 | 220.166839 | 1.545954e+03 | 193.118409 | |
| ENSG00000142494 | 55244 | SLC47A1 | protein_coding | Q96FL8 | FUNCTION: Solute transporter for tetraethylammonium (TEA), 1-methyl-4-phenylpyridinium (MPP), cimetidine, N-methylnicotinamide (NMN), metformin, creatinine, guanidine, procainamide, topotecan, estrone sulfate, acyclovir, ganciclovir and also the zwitterionic cephalosporin, cephalexin and cephradin. Seems to also play a role in the uptake of oxaliplatin (a new platinum anticancer agent). Able to transport paraquat (PQ or N,N-dimethyl-4-4'-bipiridinium); a widely used herbicid. Responsible for the secretion of cationic drugs across the brush border membranes. {ECO:0000269|PubMed:16330770, ECO:0000269|PubMed:16996621, ECO:0000269|PubMed:17495125, ECO:0000269|PubMed:17509534, ECO:0000269|PubMed:17582384}. | Acetylation;Alternative splicing;Cell membrane;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene is located within the Smith-Magenis syndrome region on chromosome 17. It encodes a protein of unknown function. [provided by RefSeq, Jul 2008]. | hsa:55244; | apical plasma membrane [GO:0016324]; basolateral plasma membrane [GO:0016323]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; vesicle [GO:0031982]; amide transmembrane transporter activity [GO:0042887]; L-amino acid transmembrane transporter activity [GO:0015179]; L-arginine transmembrane transporter activity [GO:0061459]; solute:proton antiporter activity [GO:0015299]; transmembrane transporter activity [GO:0022857]; xenobiotic transmembrane transporter activity [GO:0042910]; amino acid import across plasma membrane [GO:0089718]; L-alpha-amino acid transmembrane transport [GO:1902475]; L-arginine import across plasma membrane [GO:0097638]; organic cation transport [GO:0015695]; transmembrane transport [GO:0055085]; xenobiotic detoxification by transmembrane export across the plasma membrane [GO:1990961]; xenobiotic transport [GO:0042908] | 16330770_MATE1 appears to be the long searched for polyspecific organic cation exporter that directly transports toxic organic cations into urine and bile. 17047166_an oppositely directed H(+) gradient serves as a driving force of tetraethylammonium transport via rMATE1 17495125_The results suggest that hOCT2 and hMATE1 mediate paraquat transport in the kidney. 17509534_hMATE1 and hMATE2-K function together as a detoxication system, by mediating the tubular secretion of intracellular ionic compounds across the brush-border membranes of the kidney. 17855482_Sp1 functions as basal transcriptional regulator of human and rat MATE1 gene through two GC boxes. May be conserved among species. We have identified rSNP of hMATE1 gene (G-32A) (belonging to Sp1-binding site) that affects promoter activity. 18305230_The molecular basis of substrate recognition by MATE1 was investigated via amino acid substitution in the conserved transmembrane regions. 19158817_These findings suggested that the loss of transport activities of the MATE1 G64D and MATE2-K G211V variants were due to the alteration of protein expression in cell surface membranes. 19172157_Genetic variants in multidrug and toxic compound extrusion-1, hMATE1, alter transport function. 19228809_Guanin/adenine single nucleotide polymorphism is associated with a reduction in A1C level suggesting it role in the pharmacokinetics of metformin in diabetics. 19228809_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19536068_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19745787_Observational study of gene-disease association. (HuGE Navigator) 19745787_the rate of transcription of MATE1 is regulated by AP-1 and AP-2rep and that a common promoter variant, g.-66T>C may affect the expression level of MATE1 in human kidney, and ultimately result in variation in drug disposition and response. 19898263_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19898263_interaction between polymorphisms and OCT1 polymorphisms on the glucose lowering effect of metformin 20016398_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20016398_The purpose of this study was to evaluate the effects of heterozygous MATE variants on the disposition of metformin in mice and humans. 20047987_4',6-diamidino-2-phenylindole (DAPI) can be used as a probe substrate for rapid assays of the functionality of the human MATE1 20053795_This study showed that coordinate function of MATE1 with OCT2 likely contributes to the vectorial renal elimination of organic cationic drugs and that altered activity of MATE1 should be considered as a determinant of renal cationic drug elimination. 20067714_characterization of MATE1: kinetic analysis of transport of cimetidine and tetraethylammonium; inhibitors; comparison to hMATE2-K and rMATE1 20682687_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21128598_The results suggest that agmatine disposition may be influenced by hOCT2 and hMATE1, two transporters critical in the renal elimination of xenobiotic compounds. 22242910_Homozygous MATE1 variant could be one of the risk factors for metformin-induced lactic acidosis. 22569819_The 808G>T single-nucleotide polymorphism in OCT2 ameliorated cisplatin-induced nephrotoxicity without alteration of disposition, whereas the rs2289669 G>A single-nucleotide polymorphism in MATE1 had no effect on cisplatin toxicity. 22722930_Twelve transmembrane helices form the functional core of mammalian MATE1 (multidrug and toxin extruder 1) protein. 22735389_Seven polymorphisms in OCT1, OCT2, and MATE1 genes were compared between 53 type 2 diabetes patients with side effects of metformin and 193 metformin users without symptoms of metformin intolerance. 22882994_Twenty percent of patients with diabetes that are homozygous for A-allele of SLC47A1 had twofold reduction in HbA1c in comparison with the patients carrying G-allele. 23267855_The effect of novel promoter variants in MATE1 ... on the pharmacokinetics and pharmacodynamics of metformin 23303678_Decreasing expression of OCT3 and MATE1 in human placenta indicates these transporters may play a role in fetal protection preferentially at earlier stages of gestation. 23630107_The results confirmed that OAT1, OAT3, OCT2, MATE1, and MATE2-K were coexpressed in tubular epithelial cells. 23864433_ADMA and L-arginine are substrates of human CAT2A, CAT2B, OCT2 and MATE1. Transport kinetics of CAT2A, CAT2B, and OCT2 indicate a low affinity, high capacity transport, which may be relevant for renal and hepatic elimination of ADMA or L-arginine 25241911_MATE1 is a membrane transporter for quercetin.MATE1 was highly expressed in peroxisomes and the endoplasmic reticulum as well as in plasma membranes in the liver and intestine. 25753371_Disease progression according to RECIST was also more frequent in carriers of at least one polymorphic MATE1 A-allele (44%) as compared with homozygous carriers of the wild-type G-allele (12.5%) (P=0.07). OCT1 and MATE1 were not associated with PFS. 25862351_MRNA levels of multidrug and toxin extrusion protein 1 (MATE1 or SLC47A1, encoded by 1 of the 11 genes) were significantly lower in patients with FILI. 26004431_SLC47A1 rs2289669 G>A variants improve the glucose-lowering effect of metformin through slowing its excretion in type 2 diabetes populations. 26538438_MATE1 sequesters organic cations within an intracellular compartment that has no influence on secretion in renal proximal tubules. 26784938_MATE1 rs2289669 may be a significant determinant in the renal clearance of metformin in the case of transporter-mediated drug interactions 27025966_MATE1 mRNA levels in peripheral blood cells were significantly higher in patients carrying the minor allele of rs2453579, but not rs2252281, compared to those with other genotypes 27178732_Pazopanib inhibits OCT2, MATE1 and MATE2-K, which are involved in cisplatin secretion into urine, potentiating cisplatin toxicity. 27226103_This study did not identify any of these known SLC47A1 coding SNPs in the Xhosa individuals who participated in this study. 27271370_The impact of assay conditions on IC50 determination is negligible, kinetic characteristics differ among used test substrates, and substrate-dependent inhibition exists for MATE1 and MATE2-K, giving valuable insight into the assessment of clinically relevant MATE-mediated drug interactions in vitro. 27418674_substrate identity exerts comparatively little influence on ligand interaction with MATE1. 27590272_MATE1 polymorphisms were associated with hematological toxicity in non-small cell lung cancer patients. 27635733_MATE1 is the major transporter for the cellular uptake of imatinib and crucial for the therapeutic success in CML patients. We suggest that the detailed analysis of MATE1 expression levels and mutations could be a predictor for the response to imatinib therapy. 28321905_genetic association studies in population in China: Data confirm that an SNP in an intron of SLC47A1 (rs2289669) is associated with hypoglycemic response to metformin in patients with newly diagnosed type 2 diabetes; differential increases in basal GLP1 plasma levels are also related to this SNP. (SLC47A1 = solute carrier family 47 member 1; GLP1 = glucagon-like peptide-1) 28992563_The combination of ENT1, MATE1 and OCT2 SNPs may serve as a predictive and prognostic marker in metastatic colorectal carcinoma patients treated with TAS-102. 29070695_The 5' CpG island of SLC47A1 acts as an enhancer for SLC47A1, and DNA methylation in the CpG island plays a role in interindividual differences in hepatic SLC47A1 expression. 29704007_The present data suggest that MATE1 contributes to renal elimination of trimethylamine-N-oxide. 30433870_The studied SLC47A1 and SLC47A2 SNPs had no influence on the dose requirement or adverse effects of metformin. 30910922_Apical Shear Stress Enhanced Organic Cation Transport in Human OCT2/MATE1-Transfected Madin-Darby Canine Kidney Cells Involves Ciliary Sensing 32708212_Rapid Regulation of Human Multidrug and Extrusion Transporters hMATE1 and hMATE2K. 35163393_Transport Turnover Rates for Human OCT2 and MATE1 Expressed in Chinese Hamster Ovary Cells. | ENSMUSG00000010122 | Slc47a1 | 7.328243e+02 | 0.9782893 | -0.031666987 | 0.2674045 | 1.419052e-02 | 0.9051771520 | 0.98302368 | No | Yes | 6.364375e+02 | 48.837702 | 6.561543e+02 | 51.497113 | |
| ENSG00000142530 | 112703 | FAM71E1 | protein_coding | Q6IPT2 | Alternative splicing;Reference proteome | hsa:112703; | 28930687_The RAB2B-GARIL5 complex promotes cytosolic DNA-induced innate immune responses (Golgi-associated Rab2B interactor-like 5 = GARIL5, also known as FAM71E1) | ENSMUSG00000051113 | Fam71e1 | 4.596321e+02 | 1.3751988 | 0.459640143 | 0.3145146 | 2.154040e+00 | 0.1421952794 | 0.76841160 | No | Yes | 5.052531e+02 | 76.526499 | 3.083632e+02 | 48.601871 | ||||
| ENSG00000142731 | 10733 | PLK4 | protein_coding | O00444 | FUNCTION: Serine/threonine-protein kinase that plays a central role in centriole duplication. Able to trigger procentriole formation on the surface of the parental centriole cylinder, leading to the recruitment of centriole biogenesis proteins such as SASS6, CENPJ/CPAP, CCP110, CEP135 and gamma-tubulin. When overexpressed, it is able to induce centrosome amplification through the simultaneous generation of multiple procentrioles adjoining each parental centriole during S phase. Phosphorylates 'Ser-151' of FBXW5 during the G1/S transition, leading to inhibit FBXW5 ability to ubiquitinate SASS6. Its central role in centriole replication suggests a possible role in tumorigenesis, centrosome aberrations being frequently observed in tumors. Also involved in deuterosome-mediated centriole amplification in multiciliated that can generate more than 100 centrioles. Also involved in trophoblast differentiation by phosphorylating HAND1, leading to disrupt the interaction between HAND1 and MDFIC and activate HAND1. Phosphorylates CDC25C and CHEK2. Required for the recruitment of STIL to the centriole and for STIL-mediated centriole amplification (PubMed:22020124). Phosphorylates CEP131 at 'Ser-78' and PCM1 at 'Ser-372' which is essential for proper organization and integrity of centriolar satellites (PubMed:30804208). {ECO:0000269|PubMed:16244668, ECO:0000269|PubMed:16326102, ECO:0000269|PubMed:17681131, ECO:0000269|PubMed:18239451, ECO:0000269|PubMed:19164942, ECO:0000269|PubMed:21725316, ECO:0000269|PubMed:22020124, ECO:0000269|PubMed:27796307, ECO:0000269|PubMed:30804208}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Cytoskeleton;Dwarfism;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Ubl conjugation | This gene encodes a member of the polo family of serine/threonine protein kinases. The protein localizes to centrioles, complex microtubule-based structures found in centrosomes, and regulates centriole duplication during the cell cycle. Three alternatively spliced transcript variants that encode different protein isoforms have been found for this gene. [provided by RefSeq, Jun 2010]. | hsa:10733; | centriole [GO:0005814]; centrosome [GO:0005813]; cleavage furrow [GO:0032154]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; deuterosome [GO:0098536]; nucleolus [GO:0005730]; nucleus [GO:0005634]; procentriole [GO:0120098]; procentriole replication complex [GO:0120099]; spindle pole [GO:0000922]; XY body [GO:0001741]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; centriole replication [GO:0007099]; cilium assembly [GO:0060271]; de novo centriole assembly involved in multi-ciliated epithelial cell differentiation [GO:0098535]; mitotic cell cycle [GO:0000278]; positive regulation of centriole replication [GO:0046601]; protein phosphorylation [GO:0006468]; regulation of cytokinesis [GO:0032465]; trophoblast giant cell differentiation [GO:0060707] | 15967108_SAK repression by p53 is likely mediated through the recruitment of HDAC repressors, and SAK repression contributes to p53-induced apoptosis 16244668_we identify Plk4 as a key regulator of centriole duplication.These findings provide an attractive explanation for the crucial function of Plk4 in cell proliferation and have implications for the role of Polo kinases in tumorigenesis. 16326102_SAK/PLK4 is required for centriole duplication and flagella development in Drosophila and human cells. 17681131_Overexpression of Polo-like kinase 4 (Plk4) in human cells induces centrosome amplification through the simultaneous generation of multiple procentrioles adjoining each parental centriole. 18239451_Human Plk4 phosphorylates Cdc25C. 19001868_Plk2 mediated centriole duplication is dependent on Plk4 function 19454482_HCT116 cells fail to organize the ninefold symmetry of centrioles due to insufficient Plk4. 19679553_CUL1 may function as a tumor suppressor by regulating PLK4 protein levels and thereby restraining excessive daughter centriole formation at maternal centrioles. 20032307_Data suggest that polo-like kinase 4 activity is restricted to the centrosome to prevent aberrant centriole assembly and sustained kinase activity is required for centriole duplication. 20348415_Plk4 is required for cytokinesis and maintenance of chromosomal stability 20508983_Observational study of gene-disease association. (HuGE Navigator) 20516151_Data suggest that active Plk4 promotes its own degradation by catalyzing betaTrCP binding through trans-autophosphorylation (phosphorylation by the other kinase in the dimer) within homodimers. 20923822_Observational study and genome-wide association study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21059844_Results suggest that Cep152 recruits Plk4 and CPAP to the centrosome to ensure a faithful centrosome duplication process. 21059850_Data show that Cep152 can be phosphorylated by Plk4 in vitro, suggesting that Cep152 acts with Plk4 to initiate centriole formation. 21297952_CDK11(p58), which accumulates only in the vicinity of mitotic centrosomes, directly interacts with the centriole-associated protein kinase Plk4 that regulates centriole number in cells. 21609466_These results highlight the critical role of PLK4 transcriptional deregulation in centriole multiplication in HPV-16 E7-expressing cells. 21725316_The activity of SCF-FBXW5 is negatively regulated by Polo-like kinase 4 (PLK4), which phosphorylates FBXW5 at Ser 151 to suppress its ability to ubiquitylate HsSAS-6. 22349698_STIL cooperates with SAS-6 and PLK4 in the control of centriole number and represents a key centriole duplication factor in human cells. 22456511_Plk4 is a centriole-localized kinase that does not directly regulate cytokinesis. 22829937_Study demonstrated that PLK4 was remarkably downregulated in HCC and could be served as a potential prognostic marker for patients with this deadly disease. 23019411_CAND1 promotes PLK4-mediated centriole overduplication and is frequently disrupted in prostate cancer. 23249732_Preventing Plk4 autoregulation causes centrosome amplification, stabilization of p53, and loss of cell proliferation 23641073_cooperation between Cep192 and Cep152 is crucial for centriole recruitment of Plk4 and centriole duplication during the cell cycle. 23653187_If both p53 and the SAPKK MKK4 are simultaneously inactivated, persistent polo-like kinase 4 activity combined with the lack of SAPK-mediated inhibition of centrosome duplication conspire to induce supernumerary centrosomes under stress. 23821772_Studies indicate that autophosphorylation of Nek7 and Plk4 occurred through an intermolecular mechanism, the kinases Aurora-A and Chk2 followed an intramolecular mechanism. 23974100_PLK4 is a new NFkappaB target gene, providing a direct link between NFkappaB activity and centrosome duplication, with implications for the role of these transcription factors in tumorigenesis. 24277814_Plk4 dynamically localizes to distinct subcentrosomal regions by interacting with two hierarchically regulated scaffolds, Cep192 and Cep152. 24389189_it appears that Nek2 and Plk4 might synergize to promote breast tumorigenesis and may also be involved in tamoxifen and trastuzumab resistance 24498222_p53-Dependent and cell specific epigenetic regulation of the polo-like kinases under oxidative stress. 24588599_HPVs16 and 18 commonly are present in normal oral mucosa and emphasize the importance of distinguishing clinical 24797070_Results demonstrated that Plk4 is under the direct control of the E2F activators in breast cancer cells. 24867403_Studies indicate that overexpression of polo-like kinase 4 (PLK4) is found in several cancer and suggest the PLK4 inhibitors as anticancer therapeutics. 24981932_PLK4 overexpression induces centrosome amplification and chromosome instability and causes the suppression of primary cilia formation. 24997597_Plk4 is intricately regulated in time and space through ordered interactions with two distinct scaffolds, Cep192 and Cep152, and a failure in this process may lead to human cancer. 25043604_Breast cancer cell viability is dependent on PLK4 expression. 25174401_An unexpected activity of Plk4 that promotes cell migration and may underlie an association between increased Plk4 expression, cancer progression and death from metastasis in solid tumor patients. 25320347_Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. 25342035_Negative feedback by centriolar STIL regulates bimodal centriolar distribution of Plk4 and seemingly restricts occurrence of procentriole formation to one site on each parental centriole. 25344692_different levels of impaired PLK4 activity result in growth and cilia phenotypes, providing a mechanism by which microcephaly disorders can occur with or without ciliopathic features 25347426_Decreased PLK4 protein expression due to promoter hypermethylation was negatively correlated with JAK2 overexpression, a common occurrence in hematological malignancies. 25590559_PLK4 functions downstream of ROCK2 to drive centrosome amplification in arrested cells. 25723005_Data suggest polo-like kinase 4 (PLK4) inhibitors as a clinical candidate for cancer therapy. 25795303_these results identify the interaction between Mib1 and Plk4 as a new and important element in the control of centriole homeostasis. 25859044_identified association between aneuploidy of putative mitotic origin and linked genetic variants on chromosome 4 of maternal genomes; associated region contains candidate gene,PLK4, that plays a role in centriole duplication and can alter mitotic fidelity upon minor dysregulation 26101219_Plk4 activity promotes the recruitment of STIL to the centriole and primes the direct binding of STIL to the C terminus of SAS6. 26188084_The authors suggest that the STIL-coiled-coil region/PLK4 interaction mediates PLK4 activation as well as stabilization of centriolar PLK4 and plays a key role in centriole duplication. 26439168_KLF14 transcription is significantly downregulated, whereas Plk4 transcription is upregulated in multiple types of cancers, and there exists an inverse correlation between KLF14 and Plk4 protein expression in human breast and colon cancers. 26452337_Mutations in human PLK4, the protein of which plays critical role in centriole duplication and normal nuclear formation, could be associated with abnormal spermatogenesis leading to Sertoli cell-only syndrome. Aberrant forms of PLK4 might also cause other types of oligozoospermia or sperm fl agellar abnormalities. 26481051_We demonstrate that centrioles promote PLK4 activation through its recruitment and local accumulation. Though centriole removal reduces the proportion of active PLK4, this is rescued by concentrating PLK4 to the peroxisome lumen 26755742_Plk4 directly binds PCM1 and phosphorylates S372. Plk4 depletion leads to the dispersal of centriolar satellites. 27112295_These data show that complementary mechanisms, such as mother-daughter centriole proximity and CDK1-CyclinB interaction with centriolar components, ensure that centriole biogenesis occurs once and only once per cell cycle, raising parallels to the cell-cycle regulation of DNA replication and centromere formation. 27246242_the interaction between Cep78 and the N-terminal catalytic domain of Plk4 is a new and important element in the centrosome overduplication process. 27650967_Heterozygous missense mutation in PLK4 identified in a patient with microcephaly and chorioretinopathy. Aberrant spindle formation was observed in a LCL derived from this patient. Mutant PLK4 proteins demonstrated altered mobility pattern on a western blot suggesting alterations in post-translation modification. 27796307_KAT2A/2B acetylation of PLK4 prevents centrosome amplification 27872092_Our results validate Plk4 as a therapeutic target in cancer patients 27911707_Studies indicate that depletion of any one of the protein kinase polo-like kinase 4 (PLK4) and the two proteins STIL and SAS-6 blocks centriole duplication, and, conversely, overexpression causes centriole amplification. 28238495_Common PLK4 variant rs2305957 is associated with blastocyst formation and early recurrent miscarriage in Chinese women. 28562169_PLK4 specifically phosphorylates CP110 at the S98 position and is an essential step for centriole assembly. 28832566_Homozygous splicing acceptor site transition (c.31-3 A>G) in PLK4 was identified in a family with Seckel syndrome. PLK4 is essential for centriole biogenesis and DNA damage response. 29496738_Plk4 first phosphorylates the extreme N terminus of Ana2, which is critical for subsequent STAN domain modification. Phosphorylation of the central region then breaks the Plk4-Ana2 interaction. This phosphorylation pattern is important for centriole assembly and integrity. 29500190_Plk4 functions as a homeostatic clock, establishing an inverse relationship between growth rate and period to ensure that daughter centrioles grow to the correct size. 29688390_our data support tripolar chromosome segregation as a key mechanism generating complex aneuploidy in cleavage-stage embryos and implicate maternal genotype at a quantitative trait locus spanning PLK4 as a factor influencing its occurrence. 29712910_Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly. 29768346_PLK4 played important roles in regulating cell cycle- and DNA replication-related pathways. E2F could upregulate the expression levels of PLK4 by deregulating the methylations of their promoters to promote the relapse of Acute Lymphoblastic Leukemia. 30197118_blocking of PLK4 or STIL functions leads to centrosome loss followed by both p53-dependent and -independent defects, including prolonged cell divisions, upregulation of p53, chromosome instability, and, importantly, reduction of pluripotency markers and induction of differentiation. 30315225_MiRNA-126/PLK-4 axis is critical for tumorigenesis and progression of HCC. 30529153_Study showed that the mRNA level of PLK4 was significantly associated with glioma grade and inversely correlated with overall survival and temozolomide sensitivity. 30570110_Pololike kinase 4 (PLK4) has been identified as an oncogene, which is overexpressed in various types of human cancer 30604763_Study found that organization of acto-myosin force modulates specifically S-G2 phase length of the cell cycle, PLK4 recruitment at the centrosome and the fidelity of centriole duplication. 30787112_Expression regulation of PLK-4 and SAS-6 has a central role in centriole biogenesis. (Review) 30804208_PLK4 phosphorylates CEP131 at Ser-78 to maintain centriolar satellite integrity. 30816483_Elevated PLK4 expression was observed in high grade glioblastoma (GBM) patients and was associated with poor prognosis. PLK4 expression was markedly elevated by the exogenous overexpression of ATAD2 in GBM cells. Results suggested that the ATAD2dependent transcriptional regulation of PLK4 promoted cell proliferation and tumorigenesis. 30858376_Study shows that Cep63 and Cep152 cooperatively generate a heterotetrameric alpha-helical bundle that functions in conjunction with its neighboring hydrophobic motifs to self-assemble into a higher-order cylindrical architecture capable of recruiting downstream components, including Plk4, a key regulator for centriole duplication. Mutations disrupting the self-assembly abrogate Plk4-mediated centriole duplication. 31000710_Study demonstrated that PLK4 has an ability to phase-separate into condensates via an intrinsically disordered linker and that the condensation properties of PLK4 are regulated by autophosphorylation. Consistently, the dissociation dynamics of centriolar PLK4 are controlled by autophosphorylation. 31097597_The regulatory role of PLK4 in cytokinesis makes it a potential target for therapeutic intervention in appropriately selected cancers. 31115335_Here, the authors show that PLK4 phosphorylates its centriole substrate STIL on a conserved site, S428, to promote STIL binding to CPAP. This phospho-dependent binding interaction is conserved in Drosophila and facilitates the stable incorporation of both STIL and CPAP into the centriole. 31358734_These findings demonstrate that Cep131 is a novel substrate of Plk4, and that phosphorylation or dysregulated Cep131 overexpression promotes Plk4 stabilization and therefore centrosome amplification, establishing a perspective in understanding a relationship between centrosome amplification and cancer development. 31451615_The self-patterning of Plk4 is crucial for the regulation of centriole duplication. 31489978_A cis-eQTL genetic variant in PLK4 confers high risk of hepatocellular carcinoma. 31672968_Plk4 is a unique kinase that utilizes its autophosphorylated noncatalytic cryptic polo-box (CPB) to phase separate and generate a nanoscale spherical condensate. CPB phosphorylation also promotes Plk4's dissociation from the Cep152 tether while binding to downstream STIL, thus allowing Plk4 condensate to serve as an assembling body for centriole biogenesis. 31696485_LncRNA SNHG1 contributes to tumorigenesis and mechanism by targeting miR-338-3p to regulate PLK4 in human neuroblastoma. 31876063_Polo-like kinase 4 correlates with greater tumor size, lymph node metastasis and confers poor survival in non-small cell lung cancer. 31926341_Findings suggest that endoplasmic reticulum stress induces the ATF6 and C/EBPbeta binding, which may increase their DNA-binding affinity and inhibit the transcription activity of the PLK4 gene. 32107292_Direct interaction between CEP85 and STIL mediates PLK4-driven directed cell migration. 32126150_Inhibition of PLK4 might enhance the anti-tumour effect of bortezomib on glioblastoma via PTEN/PI3K/AKT/mTOR signalling pathway. 32200684_Differential expression of AURKA/PLK4 in quiescence and senescence of osteosarcoma U2OS cells. 32519033_YLZ-F5, a novel polo-like kinase 4 inhibitor, inhibits human ovarian cancer cell growth by inducing apoptosis and mitotic defects. 32715931_Clinical Significance of Polo-Like Kinase 4 as a Marker for Advanced Tumor Stage and Dismal Prognosis in Patients With Surgical Gastric Cancer. 32807875_FAM46C/TENT5C functions as a tumor suppressor through inhibition of Plk4 activity. 32908304_TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. 32966175_Growth disadvantage associated with centrosome amplification drives population-level centriole number homeostasis. 33171265_Non-mitotic functions of polo-like kinases in cancer cells. 33176597_CENPE, PRC1, TTK, and PLK4 May Play Crucial Roles in the Osteosarcoma Progression. 33351100_PLK4-phosphorylated NEDD1 facilitates cartwheel assembly and centriole biogenesis initiations. 33402398_Role of Polo-Like Kinase 4 (PLK4) in Epithelial Cancers and Recent Progress in its Small Molecule Targeting for Cancer Management. 33630225_CircKIF2A contributes to cell proliferation, migration, invasion and glycolysis in human neuroblastoma by regulating miR-129-5p/PLK4 axis. 33706192_Polo Like Kinase 4 (PLK4) impairs human bone marrow mesenchymal stem cell (BMSC) viability and osteogenic differentiation. 33756487_Primary Dwarfism, Microcephaly, and Chorioretinopathy due to a PLK4 Mutation in Two Siblings. 34080290_Identification and assessment of PLK1/2/3/4 in lung adenocarcinoma and lung squamous cell carcinoma: Evidence from methylation profile. 34272646_Overexpression of the PLK4 Gene as a Novel Strategy for the Treatment of Autosomal Recessive Microcephaly by Improving Centrosomal Dysfunction. 34342940_Down-regulation of Polo-like kinase 4 (PLK4) induces G1 arrest via activation of the p38/p53/p21 signaling pathway in bladder cancer. 34637843_TEC kinase stabilizes PLK4 to promote liver cancer metastasis. 34933912_Hypoxia Drives Centrosome Amplification in Cancer Cells via HIF1alpha-dependent Induction of Polo-Like Kinase 4. | ENSMUSG00000025758 | Plk4 | 3.263569e+02 | 0.9805409 | -0.028350332 | 0.3636494 | 6.096917e-03 | 0.9377622241 | 0.98859743 | No | Yes | 2.781851e+02 | 50.252080 | 3.012799e+02 | 55.725375 | |
| ENSG00000142751 | 54707 | GPN2 | protein_coding | Q9H9Y4 | FUNCTION: Small GTPase required for proper localization of RNA polymerase II and III (RNAPII and RNAPIII). May act at an RNAP assembly step prior to nuclear import. {ECO:0000250|UniProtKB:Q08726}. | Acetylation;GTP-binding;Hydrolase;Nucleotide-binding;Reference proteome | hsa:54707; | GTP binding [GO:0005525]; GTPase activity [GO:0003924] | ENSMUSG00000028848 | Gpn2 | 2.627905e+03 | 1.2365418 | 0.306311008 | 0.3104353 | 9.840657e-01 | 0.3211970967 | 0.82152963 | No | Yes | 2.777279e+03 | 306.950413 | 1.956614e+03 | 221.874307 | |||
| ENSG00000143036 | 126969 | SLC44A3 | protein_coding | Q8N4M1 | Alternative splicing;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:126969; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; choline transmembrane transporter activity [GO:0015220]; transmembrane transporter activity [GO:0022857]; phosphatidylcholine biosynthetic process [GO:0006656]; transmembrane transport [GO:0055085] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000039865 | Slc44a3 | 5.972300e+01 | 0.8088977 | -0.305970763 | 0.4367097 | 4.812427e-01 | 0.4878599688 | 0.87511746 | No | Yes | 3.367732e+01 | 7.148678 | 4.798984e+01 | 10.178864 | |||
| ENSG00000143110 | 128346 | C1orf162 | protein_coding | Q8NEQ5 | Alternative splicing;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:128346; | integral component of membrane [GO:0016021] | ENSMUSG00000074342 | I830077J02Rik | 2.435858e+01 | 1.2031247 | 0.266786210 | 0.5980457 | 2.068182e-01 | 0.6492727869 | 0.92370776 | No | Yes | 1.603465e+01 | 5.886101 | 1.447328e+01 | 5.347279 | ||||
| ENSG00000143147 | 23432 | GPR161 | protein_coding | Q8N6U8 | FUNCTION: Key negative regulator of Shh signaling, which promotes the processing of GLI3 into GLI3R during neural tube development. Recruited by TULP3 and the IFT-A complex to primary cilia and acts as a regulator of the PKA-dependent basal repression machinery in Shh signaling by increasing cAMP levels, leading to promote the PKA-dependent processing of GLI3 into GLI3R and repress the Shh signaling. In presence of SHH, it is removed from primary cilia and is internalized into recycling endosomes, preventing its activity and allowing activation of the Shh signaling. Its ligand is unknown (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Cell projection;Cilium;Developmental protein;Disulfide bond;G-protein coupled receptor;Glycoprotein;Membrane;Receptor;Reference proteome;Transducer;Transmembrane;Transmembrane helix | The protein encoded by this gene is an orphan G protein-coupled receptor whose ligand is unknown. This gene is overexpressed in triple-negative breast cancer, and disruption of this gene slows the proliferation of basal breast cancer cells. Therefore, this gene is a potential drug target for triple-negative breast cancer. [provided by RefSeq, Mar 2017]. | hsa:23432; | ciliary membrane [GO:0060170]; cilium [GO:0005929]; endocytic vesicle membrane [GO:0030666]; integral component of membrane [GO:0016021]; recycling endosome [GO:0055037]; G protein-coupled receptor activity [GO:0004930]; adenylate cyclase-activating G protein-coupled receptor signaling pathway [GO:0007189]; G protein-coupled receptor signaling pathway [GO:0007186]; negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning [GO:1901621] | 11959142_False positive non-synonymous polymorphisms of G-protein coupled receptor genes. 19064610_Observational study of gene-disease association. (HuGE Navigator) 19536175_Observational study of gene-disease association. (HuGE Navigator) 24599592_G-protein-coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. 27002170_Smoothened determines beta-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. 27357676_Gpr161 is an A-kinase anchoring protein and the cAMP-sensing Gpr161:PKA complex acts as cilium-compartmentalized signalosome 27731925_Gpr161 is a critical factor in the basal suppression machinery of Shh signaling, neural tube morphogenesis and closure. (Review) 29386106_Gpr161 restricts cerebellar granule cell progenitor production by preventing premature and sonic hedgehog-dependent pathway activity, highlighting the importance of basal pathway suppression in tumorigenesis 30256984_rare variants of GPR161 from Spina bifida infants and determination of their functional relevance in the Shh and Wnt signaling pathways 31609649_Authors describe a novel brain tumor predisposition syndrome that is caused by germline GPR161 mutations and characterized by MBSHH in infants. | ENSMUSG00000040836 | Gpr161 | 8.983581e+02 | 1.1689791 | 0.225249160 | 0.3229549 | 4.873602e-01 | 0.4851072386 | 0.87421307 | No | Yes | 8.889020e+02 | 100.691375 | 6.914582e+02 | 80.535378 | |
| ENSG00000143158 | 25874 | MPC2 | protein_coding | O95563 | FUNCTION: Mediates the uptake of pyruvate into mitochondria. {ECO:0000269|PubMed:22628558}. | Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:25874; | integral component of mitochondrial inner membrane [GO:0031305]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; pyruvate transmembrane transporter activity [GO:0050833]; mitochondrial acetyl-CoA biosynthetic process from pyruvate [GO:0061732]; mitochondrial pyruvate transmembrane transport [GO:0006850]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774] | 3022128_characterization of the rat ortholog 19536175_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 23933155_found no significant difference (P > 0.05) in either allele or genotype frequency in the SNPs between patients and controls. 25458841_Tumor cells expressing MPC1 and MPC2 display increased mitochondrial pyruvate oxidation, with no changes in cell growth in adherent culture. 26577410_These results reveal a novel post-translational regulation of MPC1 by Sirt3, which is important for its activity and colon cancer cell growth. 27460766_a significant association between MPC2 variant rs10489202 and Schizophrenia susceptibility in Han Chinese (Meta-Analysis) 27852261_Our study indicates that MPC1 and MPC2 expressions are of prognostic values in PCAs and that positive expression of MPC1 or MPC2 is a predictor of favorable outcome. 28263840_Results indicate mitochondrial pyruvate transporter (MPC) to be the key regulatory junction perturbed by virulent strains of Mycobacterium tuberculosis leading to alteration of mitochondrial metabolic flux and regulation of acetyl-CoA formation. 29472561_In contrast to MPC1, which co-purifies with a host chaperone, we demonstrated that MPC2 homo-oligomers promote efficient pyruvate transport into proteoliposomes. The derived functional requirements and kinetic features of MPC2 resemble those previously demonstrated for MPC in the literature 29845198_Hypoxia induces lactate secretion and glycolytic efflux by downregulating MPC1/MPC2 levels in HUVEC cells. 30087317_MPC2 rs10489202 was genome-wide significantly associated with schizophrenia. The expression quantitative trait loci analysis in lymphoblastoid cell lines from East Asian donors revealed that MPC2 rs10489202 was specifically and significantly associated with the expression of TIPRL gene. 30356033_The data demonstrated that CtBP1 directly bound to the promoters of MPC1 and MPC2 and transcriptionally repressed them, leading to increased levels of free NADH in the cytosol and nucleus, thus positively feeding back CtBP1's functions. 32403431_Characteristic Analysis of Homo- and Heterodimeric Complexes of Human Mitochondrial Pyruvate Carrier Related to Metabolic Diseases. 32664896_Mitochondrial pyruvate carrier: a potential target for diabetic nephropathy. 32708919_The Multifaceted Pyruvate Metabolism: Role of the Mitochondrial Pyruvate Carrier. 33791391_Decreased Expression of MPC2 Contributes to Aerobic Glycolysis and Colorectal Cancer Proliferation by Activating mTOR Pathway. 34664967_Structural Insights into the Human Mitochondrial Pyruvate Carrier Complexes. | ENSMUSG00000026568 | Mpc2 | 9.764457e+02 | 0.5076828 | -0.978000719 | 0.3034599 | 1.025347e+01 | 0.0013642752 | 0.13950843 | No | Yes | 6.242450e+02 | 94.491630 | 1.215539e+03 | 187.725378 | ||
| ENSG00000143162 | 8804 | CREG1 | protein_coding | O75629 | FUNCTION: May contribute to the transcriptional control of cell growth and differentiation. Antagonizes transcriptional activation and cellular transformation by the adenovirus E1A protein. The transcriptional control activity of cell growth requires interaction with IGF2R. {ECO:0000269|PubMed:12934103, ECO:0000269|PubMed:9710587}. | 3D-structure;Direct protein sequencing;Glycoprotein;Growth regulation;Reference proteome;Secreted;Signal | The adenovirus E1A protein both activates and represses gene expression to promote cellular proliferation and inhibit differentiation. The protein encoded by this gene antagonizes transcriptional activation and cellular transformation by E1A. This protein shares limited sequence similarity with E1A and binds both the general transcription factor TBP and the tumor suppressor pRb in vitro. This gene may contribute to the transcriptional control of cell growth and differentiation. [provided by RefSeq, Jul 2008]. | hsa:8804; | azurophil granule lumen [GO:0035578]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; transcription regulator complex [GO:0005667]; transcription corepressor activity [GO:0003714]; regulation of growth [GO:0040008]; regulation of transcription by RNA polymerase II [GO:0006357] | 16344469_putative flavin mononucleotide-binding pocket in CREG is sterically blocked by a loop and several key bulky residues 18267954_CREG promotes a mature smooth muscle cell phenotype and reduces neointimal formation in balloon-injured rat carotid artery. 18472385_We demonstrate that CREG is expressed in the vascular endothelium; data suggests that CREG differentially regulates the growth of the denuded artery wall and smooth vascular muscle cell. 19064610_Observational study of gene-disease association. (HuGE Navigator) 19413895_The expression of CREG improves cardiac functions and inhibits cardiac hypertrophy, inflammation and fibrosis through blocking MEK-ERK1/2-dependent signalling. 19536175_Observational study of gene-disease association. (HuGE Navigator) 19769965_CREG plays a critical role in the inhibition of SMC migration, as well as maintaining SMCs in a mature phenotype 20060003_CREG plays a key role in modulating VSMC apoptosis through the p38 and JNK signal transduction pathways, both in vitro and in s 20951690_Observational study of gene-disease association. (HuGE Navigator) 20951690_no ssociation between common variants of CREG and coronary artery disease in the northern Chinese Han population 21195083_Data suggest that soluble CREG protein can exert its biological function via glycosylation-independent binding to the extracellular domains 11-13 of cell surface M6P/IGF2R, modulating SMC phenotypic switching from contractile to proliferative. 21263217_Cooperation of CREG1 and p16 (INK4a) inhibits the expression of cyclin A and cyclin B by inhibiting promoter activity thereby decreasing mRNA and protein levels; these proteins are required for S-phase entry and G2/M transition. 21872252_CREG plays a critical role in protecting the vascular endothelium from apoptosis, and the protective effort of CREG against ECs apoptosis is through the activation of the VEGF/PI3K/AKT signaling pathway 21939655_Upregulation of CREG expression induced HUVEC migration. 23040447_Suggest that CREG is a novel adventitial fibroblast phenotypic modulator in a p38MAPK-dependent manner. 23518389_miR-31 not only directly binds to its target gene CREG and modulates the vascular smooth muscle cells(VSMC) phenotype through this interaction, but also can be an important biomarker in diseases involving VSMC phenotypic modulation. 23580165_CREG can inhibit NF-kappaB activation, TNF-alpha-induced inflammatory responses and the hyperpermeability of endothelial cells. 24018888_The results suggest a novel role of CREG to promote HUVEC proliferation through the ERK/cyclin E signaling pathway. 24896341_Results indicate that cellular repressor of E1A-stimulated gene 1 protein (CREG1) increases endothelial cell (EC) filopodia formation. 26722374_These results indicate that CREG1 is a down-stream effector of KRAS in a sub-type of non-small cell lung cancer cells and a novel candidate biomarker or therapeutic target for KRAS mutant non-small cell lung cancer. 27784214_Studies demonstrated that CREG may modulate homeostasis of vascular wall cells and inhibit inflammation of vascular tissue cells and macrophages. Mechanistically, CREG behaves like a typical soluble lysosomal protein that regulates the formation and maturation of lysosomes by modulating the small GTPase protein Rab7, to mediate autophagy in vascular vascular tissue cells. [review] 32067910_DNA hypermethylation: A novel mechanism of CREG gene suppression and atherosclerogenic endothelial dysfunction. | ENSMUSG00000040713 | Creg1 | 7.387177e+02 | 0.9072322 | -0.140456173 | 0.3145460 | 1.971239e-01 | 0.6570524308 | 0.92592190 | No | Yes | 6.893143e+02 | 113.761534 | 6.804535e+02 | 115.184029 | |
| ENSG00000143198 | 4259 | MGST3 | protein_coding | O14880 | FUNCTION: Catalyzes oxydation of hydroxy-fatty acids (PubMed:9278457). Also catalyzes the conjugation of a reduced glutathione to leukotriene A4 in vitro (PubMed:9278457). May participate in the lipid metabolism (PubMed:9278457). {ECO:0000269|PubMed:9278457, ECO:0000303|PubMed:9278457}. | Endoplasmic reticulum;Lipid metabolism;Lipoprotein;Lyase;Membrane;Microsome;Oxidoreductase;Palmitate;Reference proteome;Transferase;Transmembrane;Transmembrane helix | This gene encodes a member of the MAPEG (Membrane Associated Proteins in Eicosanoid and Glutathione metabolism) protein family. Members of this family are involved in the production of leukotrienes and prostaglandin E, important mediators of inflammation. This gene encodes an enzyme which catalyzes the conjugation of leukotriene A4 and reduced glutathione to produce leukotriene C4. This enzyme also demonstrates glutathione-dependent peroxidase activity towards lipid hydroperoxides.[provided by RefSeq, May 2011]. | hsa:4259; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nuclear envelope [GO:0005635]; glutathione peroxidase activity [GO:0004602]; identical protein binding [GO:0042802]; leukotriene-C4 synthase activity [GO:0004464]; transferase activity [GO:0016740]; leukotriene biosynthetic process [GO:0019370]; lipid metabolic process [GO:0006629] | 19064610_Observational study of gene-disease association. (HuGE Navigator) 19343046_Observational study of gene-disease association. (HuGE Navigator) 19536175_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25280473_Variants in MGST3 are associated with differences inn hippocampus size in both human brain data sets (ENIGMA MRI) and in the BXD family of mice. MGST3 is also statistically associated to genes linked to neurodegenerative disorders, including Parkinson's, Huntington's, and Alzheimer's diseases. | ENSMUSG00000026688 | Mgst3 | 3.724430e+03 | 1.0251386 | 0.035818954 | 0.2965153 | 1.459594e-02 | 0.9038386370 | 0.98243436 | No | Yes | 3.532251e+03 | 289.797578 | 3.206417e+03 | 270.000409 | |
| ENSG00000143303 | 51093 | METTL25B | protein_coding | Q96FB5 | Alternative splicing;Coiled coil;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:51093; | integral component of membrane [GO:0016021]; rRNA (adenine-N6,N6-)-dimethyltransferase activity [GO:0000179] | 19204726_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000004896 | Rrnad1 | 1.616158e+03 | 0.9640973 | -0.052749328 | 0.2952819 | 3.230315e-02 | 0.8573639409 | 0.97227942 | No | Yes | 1.442536e+03 | 166.748130 | 1.573948e+03 | 186.968019 | |||
| ENSG00000143314 | 79590 | MRPL24 | protein_coding | Q96A35 | 3D-structure;Mitochondrion;Phosphoprotein;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transit peptide | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 39S subunit protein which is more than twice the size of its E.coli counterpart (EcoL24). Sequence analysis identified two transcript variants that encode the same protein. [provided by RefSeq, Jul 2008]. | hsa:79590; | mitochondrial inner membrane [GO:0005743]; mitochondrial large ribosomal subunit [GO:0005762]; mitochondrion [GO:0005739]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; translation [GO:0006412] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000019710 | Mrpl24 | 3.307445e+03 | 1.0843340 | 0.116809188 | 0.2961349 | 1.568840e-01 | 0.6920416782 | 0.93376105 | No | Yes | 3.486393e+03 | 365.780140 | 2.682887e+03 | 289.278592 | ||
| ENSG00000143353 | 127018 | LYPLAL1 | protein_coding | Q5VWZ2 | FUNCTION: Has depalmitoylating activity toward KCNMA1. Does not exhibit phospholipase nor triacylglycerol lipase activity, able to hydrolyze only short chain substrates due to its shallow active site. {ECO:0000269|PubMed:22052940, ECO:0000269|PubMed:22399288}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Hydrolase;Reference proteome | hsa:127018; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; carboxylic ester hydrolase activity [GO:0052689]; lysophospholipase activity [GO:0004622]; palmitoyl-(protein) hydrolase activity [GO:0008474]; protein depalmitoylation [GO:0002084] | 19557161_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 19557161_TFAP2B, LYPLAL1 and MSRA are associated with adiposity and fat distribution. 20703240_Observational study of gene-disease association. (HuGE Navigator) 20724581_Observational study of gene-disease association. (HuGE Navigator) 20935629_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21674055_central obesity-associated variants in LYPLAL1, NRXN3, MSRA, and TFAP2B 21953277_genetic association studies in Danish population: Quantitative metabolic phenotypes in obesity are associated with SNP in various genes: LYPLAL1 (rs4846567) is associated with waist-hip ratio and insulin sensitivity in women. 22052940_LYPLAL1 exhibits neither phospholipase nor triacylglycerol lipase activity, but rather accepts short-chain substrates 22179955_Gene-treatment interactions were observed for short-term weight loss. (LYPLAL1 rs2605100, Plifestyle*SNP = 0.032) 23221025_Our results suggest that LYPLAL1 rs4846567 and NISCH rs6784615 may influence fat distribution in the Japanese population. 26848030_Data provide evidence that variants of MC4R and LYPLAL1 modulate body fat distribution with sexual dimorphism in a Chinese population. 27181159_The LYPLAL1 genotype is associated with differences in eating behavior and loss of extensive body weight following Roux-en-Y gastric bypass (RYGB) surgery. Genotyping and the use of eating behavior-related questionnaires may help to estimate the RYGB-associated therapy success. 27752939_Among Mexicans, the PNPLA3 (rs738409), LYPLAL1 (rs12137855), PPP1R3B (rs4240624), and GCKR (rs780094) polymorphisms may be associated with a greater risk of chronic liver disease among overweight adults. 28645872_Novel LYPLAL1 SNP rs2605100 is associated with childhood hypertension adjusted by obesity. 29648650_Metabolic effects of LYPLAL1 rs12137855-C were similar, but statistically less robust, to the effects of GCKR rs1260326-T. TM6SF2 rs58542926-T displayed opposite metabolic effects when compared with the fatty liver associations. | ENSMUSG00000039246 | Lyplal1 | 1.662393e+02 | 0.9212181 | -0.118385303 | 0.4561872 | 6.673271e-02 | 0.7961547369 | 0.96109962 | No | Yes | 1.616713e+02 | 41.108007 | 1.601576e+02 | 41.750461 | ||
| ENSG00000143379 | 9869 | SETDB1 | protein_coding | Q15047 | FUNCTION: Histone methyltransferase that specifically trimethylates 'Lys-9' of histone H3. H3 'Lys-9' trimethylation represents a specific tag for epigenetic transcriptional repression by recruiting HP1 (CBX1, CBX3 and/or CBX5) proteins to methylated histones. Mainly functions in euchromatin regions, thereby playing a central role in the silencing of euchromatic genes. H3 'Lys-9' trimethylation is coordinated with DNA methylation (PubMed:12869583). Required for HUSH-mediated heterochromatin formation and gene silencing. Forms a complex with MBD1 and ATF7IP that represses transcription and couples DNA methylation and histone 'Lys-9' trimethylation (PubMed:27732843, PubMed:14536086). Its activity is dependent on MBD1 and is heritably maintained through DNA replication by being recruited by CAF-1 (PubMed:14536086,). SETDB1 is targeted to histone H3 by TRIM28/TIF1B, a factor recruited by KRAB zinc-finger proteins. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells (PubMed:24623306). Required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs) (PubMed:24623306). In ESCs, in collaboration with TRIM28, is also required for H3K9me3 and silencing of endogenous and introduced retroviruses in a DNA-methylation independent-pathway (By similarity). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing (PubMed:24623306). The SETDB1-TRIM28-ZNF274 complex may play a role in recruiting ATRX to the 3'-exons of zinc-finger coding genes with atypical chromatin signatures to establish or maintain/protect H3K9me3 at these transcriptionally active regions (PubMed:27029610). {ECO:0000250|UniProtKB:O88974, ECO:0000269|PubMed:12869583, ECO:0000269|PubMed:14536086, ECO:0000269|PubMed:24623306, ECO:0000269|PubMed:27029610, ECO:0000269|PubMed:27732843}. | 3D-structure;Alternative splicing;Chromatin regulator;Chromosome;Coiled coil;Cytoplasm;Isopeptide bond;Metal-binding;Methylation;Methyltransferase;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc | This gene encodes a histone methyltransferase which regulates histone methylation, gene silencing, and transcriptional repression. This gene has been identified as a target for treatment in Huntington Disease, given that gene silencing and transcription dysfunction likely play a role in the disease pathogenesis. Alternatively spliced transcript variants of this gene have been described.[provided by RefSeq, Jun 2011]. | hsa:9869; | chromosome [GO:0005694]; cytoplasm [GO:0005737]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone methyltransferase activity (H3-K9 specific) [GO:0046974]; histone-lysine N-methyltransferase activity [GO:0018024]; promoter-specific chromatin binding [GO:1990841]; zinc ion binding [GO:0008270]; heterochromatin organization [GO:0070828]; histone H3-K9 methylation [GO:0051567]; negative regulation of gene expression [GO:0010629]; negative regulation of single stranded viral RNA replication via double stranded DNA intermediate [GO:0045869]; positive regulation of DNA methylation-dependent heterochromatin assembly [GO:0090309]; Ras protein signal transduction [GO:0007265] | 11959841_Contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins; KAP-1, SETDB1, H3-MeK9, and HP1 are enriched at promoter sequences of a euchromatic gene silenced by the KRAB-KAP-1 repression system 14536086_mAM/hAM facilitates conversion of H3-K9 dimethyl to trimethyl by ESET/SETDB1 15691849_These data suggest that MBD1.MCAF1.SETDB1 complex facilitates the formation of heterochromatic domains, emphasizing the role of MCAF/AM family proteins in epigenetic control, and describe a new family member, MCAF2 (ATF7IP2). 16682412_histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells 17142323_modulation of gene silencing mechanisms, through regulation of the ESET gene is important to neuronal survival and, as such, may be a promising treatment in Huntington's disease patients 17577629_Akt/PKB interacts with the histone H3 methyltransferase SETDB1 and coordinates to silence gene expression. 18498648_SETDB1 can specifically methylate HIV-1 Tat preferentially at lysine 51. 19124506_Observational study of gene-disease association. (HuGE Navigator) 21170338_analysis of a KRAB domain-containing ZNF (ZNF274) that is involved in recruitment of the KAP1 and SETDB1 to specific regions of the human genome 21430779_studies establish SETDB1 as an oncogene in melanoma and underscore the role of chromatin factors in regulating tumorigenesis 23055267_A transgenic Setdb1 model established a link between this gene and behavior. 23770855_SETDB1 is a bona fide oncogene undergoing gene amplification-associated activation in lung cancer. 23943221_SETDB1 expression was upregulated in glioma cell lines and in glioma tissues compared to normal brain, being positively correlated with grade and histological malignancy. 24056368_of ESET plays an essential role in the maintenance of articular cartilage by preventing articular chondrocytes from terminal differentiation and may have implications in joint diseases such as osteoarthritis. 24556744_data suggested that SETDB1 is overexpressed in human PCa. Silencing SETDB1 inhibited PCa cell proliferation, migration and invasion 24658378_overexpression of SETDB1 or LSD1 had no prognostic impact in patients with melanoma 24673285_SETDB1 is associated with frequent methylation of the euchromatic p16(INK) (4A) promoter and several prognostic parameters in melanomas 24760766_This observation suggests that the ZNF274/SETDB1 complex bound to the SNORD116 cluster may protect the Prader-Willi syndrome induced pluripotent cells from DNA demethylation during early development. 25070049_MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the Epithelial-mesenchymal transition of breast cancer stem cells by down regulating the STAT3 pathway 25404354_Report elevated levels of SETDB1 in non-small lung cancers, associated with neoplasm grading and tumor growth. 25477335_Together, our findings defined an essential role for the KMT1E/SMAD2/3 repressor complex in TGFbeta-mediated lung cancer metastasis. 25569264_Authors demonstrate that a KMT1E-containing complex directly interacts with the FcgammaRIIb promoter and that histone H3 at lysine 9 tri-methylation at this promoter is dependent on Setdb1 25715926_Exogenous expression of MyoD reversed transcriptional repression of MyoD promoter-driven lucif-erase reporter by Setdb1 shRNA and rescued myogenic differentiation of C2C12 myoblast cells depleted of endogenous Setdb1. 26143443_Upon HBV infection, cellular mechanisms involving SETDB1-mediated H3K9me3 and HP1 induce silencing of HBV cccDNA transcription through modulation of chromatin structure. 26296461_SETDB1, localized in the nucleus, might undergo degradation by the proteasome and be exported to the cytosol, resulting in its detection mainly in the cytosol. 26471002_regulates cancer cell growth via methylation of p53 26481868_SETDB1 is an oncogene that is frequently up-regulated in human HCCs; the multiplicity of SETDB1 activating mechanisms at the chromosomal, transcriptional, and posttranscriptional levels together facilitates SETDB1 up-regulation in human HCC 26542178_BRCA1 and SETDB1 stand out as the most significant prognostic markers in this group of patients 26813693_results indicate that ATF7IP does not directly modulate SETDB1 catalytic activity, suggesting alternate roles, such as affecting cellular localization or mediating interaction with additional binding partners. 26824986_SETDB1 mutations are associated with malignant pleural mesotheliomas. 26840455_We identified a list of thirty genes repressed by DeltaNp63 in a SETDB1-dependent manner, whose expression is positively correlated to survival of breast cancer patients. These results suggest that p63 and SETDB1 expression, together with the repressed genes, may have diagnostic and prognostic potential 26846621_The SETDB1 protein was closely associated with the prognosis of prostate cancer (PCa). Bioinformatics suggested that SETDB1 might promote PCa bone metastasis through the WNT pathway. In conclusion, SETDB1 might be associated with the development of bone metastases from PCa 26949019_These results suggest that SETDB1- mediated FosB expression is a common molecular phenomenon, and might be a novel pathway responsible for the increase in cell proliferation that frequently occurs during anticancer drug therapy. 27119313_analysis of a 1q21.3 deletion encompassing SETDB1 that provides further support for the role of chromatin modifiers in the etiology of autism spectrum disorder 27164857_Authors observed several complementary mechanisms contributing to the upregulation of SETDB1 in HCC cells. Besides copy number gains at the SETDB1 gene locus at chromosome 1q21 enhanced SETDB1 transcription mediated by the transcription factor SP1 could be detected. 27237050_SETDB1, a major histone H3K9 methyltransferase is monoubiquitinated at the evolutionarily conserved lysine-867 in its SET-Insertion domain. This ubiquitination is directly catalyzed by UBE2E family of E2 enzymes in an E3-independent manner while the conjugated-ubiquitin (Ub) is protected from active deubiquitination. 27732843_these data identify a critical functional role for ATF7IP in heterochromatin formation by regulating SETDB1 abundance in the nucleus. 27798683_These results suggest that the ubiquitination of SETDB1 at lysine 867 controls the expression of its target gene by activating its H3K9 methyltransferase activity. 28887438_SETDB1 triggers silencing of retrotransposons to inhibit the interferon response in acute myeloid leukemia cells. 28913972_SETDB1 protein expression was significantly associated with poor survival and was related to TNM stage. 29233829_Smad3-mediated recruitment of SETDB1 controls Snail1 expression and epithelial-mesenchymal transition in ductal breast carcinoma. 29234025_SETDB1 is enriched at H3K9me3 regions and K9me3/K14ac is enriched at SETDB1 binding sites overlapping with LINE elements, suggesting that recruitment of the SETDB1 complex to K14ac/K9me regions has a role in silencing of active genomic regions. 29739365_Study in hepatocellular carcinoma (HCC) cells proves that SETDB1 promotes the proliferation and migration of cells by forming SETDB1-Tiam1 compounds. SETDB1-Tiam1 compounds were involved in a novel pathway, which regulated epigenetic modification of gene expression in HCC patients samples. 29901162_Increased expression of SETDB1 may predict poor overall survival. 29926931_Identify SETDB1 as a prominent oncogene in breast cancer. The c-MYC-BMI1 axis is essential for SETDB1-mediated breast tumourigenesis. 30054425_SETDB1 expression is the common target of the miR-29 family members in non-small cell lung cancer.TP53 represses the expression of SETDB1 via increasing miR-29s expression in non-small cell lung cancer. 30103804_SETDB1 loss results in decompaction of the Xi chromosome partly through reactivation of an enhancer in the IL1RAPL1 gene. 30105513_Enhancer of Zeste Homolog 2 (EZH2), SET domain, bifurcated 1 protein (SETDB1), lysine-specific histone demethylase 1 (LSD1), histone H3 methylation (H3K9me3 and H3K27me3) expression are altered in colorectal cancer (CRC) and may play a role in colorectal carcinogenesis. 30309377_SETDB1 knockdown might suppress breast cancer progression at least partly by miR-381-3p-related regulation, providing a novel prospect in breast cancer therapy. 30452683_Although these genes are normally expressed at low amounts in hESCs, HTT knockdown (KD) reduces their induction during neural differentiation. Notably, mutant expanded polyglutamine repeats in HTT diminish its interaction with ATF7IP-SETDB1 complex and trigger H3K9me3 in HD-iPSCs. 30483750_Mechanistic investigations in breast cancer cells indicated that SETDB1 acts as an epithelialmesenchymal transition inducer by binding directly to the promoter of the transcription factor Snail. Thus, SETDB1 is involved in breast cancer metastasis and may be a therapeutic target for treating patients with breast cancer. 30545440_Gene network analysis showed that SMAD7 expression is regulated by SETDB1 levels, indicating that up-regulation of SMAD7 by SETDB1 knockdown inhibited BRC metastasis. 30692625_Data suggest that targeting SET domain, bifurcated 1 protein (SETDB1) signalling could be a potential therapeutic strategy for combatting hyperactive AKT serine/threonine kinase 1 (AKT)-driven cancers. 30692626_Findings indicate complicated layers of serine/threonine kinase Akt (Akt) activation regulation coordinated by SET domain, bifurcated 1 protein (SETDB1)-mediated Akt K64 methylation. 30850015_A number of converging phenotypes outline a stress-responsive mechanism for SETDB1 and SETDB2 activation and subsequent increased tumor survival, providing novel insights into epigenetic biology. [review] 31131878_SETDB1 is a major driver of melanoma development. 31276581_CSB reduces H3K9me3 chromatin remodeler SETDB1 and exacerbates cellular aging. 31306481_Histone methyltransferase SETDB1 promotes colorectal cancer proliferation through the STAT1-CCND1/CDK6 axis. 31896605_Network Inference Analysis Identifies SETDB1 as a Key Regulator for Reverting Colorectal Cancer Cells into Differentiated Normal-Like Cells. 32305991_High SET Domain Bifurcated 1 (SETDB1) Expression Predicts Poor Prognosis in Breast Carcinoma. 32393761_SETDB1 promotes the progression of colorectal cancer via epigenetically silencing p21 expression. 32473242_Blocking histone methyltransferase SETDB1 inhibits tumorigenesis and enhances cetuximab sensitivity in colorectal cancer. 32486217_SETDB1-Mediated Silencing of Retroelements. 32503845_SETDB1 is required for intestinal epithelial differentiation and the prevention of intestinal inflammation. 32768498_Evidence that miR-152-3p is a positive regulator of SETDB1-mediated H3K9 histone methylation and serves as a toggle between histone and DNA methylation. 32972752_Knockout of SETDB1 gene using the CRISPR/cas-9 system increases migration and transforming activities via complex regulations of E-cadherin, beta-catenin, STAT3, and Akt. 33044755_SETDB1 promotes gastric carcinogenesis and metastasis via upregulation of CCND1 and MMP9 expression. 33054052_SETDB1 mediated histone H3 lysine 9 methylation suppresses MLL-fusion target expression and leukemic transformation. 33059737_SETDB1 promotes glioblastoma growth via CSF-1-dependent macrophage recruitment by activating the AKT/mTOR signaling pathway. 33064882_Recurrent co-alteration of HDGF and SETDB1 on chromosome 1q drives cutaneous melanoma progression and poor prognosis. 33115801_Histone Methyltransferase SETDB1: A Common Denominator of Tumorigenesis with Therapeutic Potential. 33370431_LINC00476 Suppresses the Progression of Non-Small Cell Lung Cancer by Inducing the Ubiquitination of SETDB1. 33953401_Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. 34324684_Preparation of the ubiquitination-triggered active form of SETDB1 in Escherichia coli for biochemical and structural analyses. 34801472_Histone methyltransferase SETDB1 inhibits TGF-beta-induced epithelial-mesenchymal transition in pulmonary fibrosis by regulating SNAI1 expression and the ferroptosis signaling pathway. 34843122_Expression of SET domain bifurcated histone lysine methyltransferase 1 and its clinical prognostic significance in hepatocellular carcinoma. 35184652_Enhancer of zeste homolog 2 promotes hepatocellular cancer progression and chemoresistance by enhancing protein kinase B activation through microRNA-381-mediated SET domain bifurcated 1. 35372573_Increased Expression of SETDB1 Predicts Poor Prognosis in Multiple Myeloma. | ENSMUSG00000015697 | Setdb1 | 2.722020e+03 | 0.8298010 | -0.269162613 | 0.2606942 | 1.074973e+00 | 0.2998248602 | 0.81383297 | No | Yes | 2.051836e+03 | 175.837969 | 2.370679e+03 | 208.257366 | |
| ENSG00000143384 | 4170 | MCL1 | protein_coding | Q07820 | FUNCTION: Involved in the regulation of apoptosis versus cell survival, and in the maintenance of viability but not of proliferation. Mediates its effects by interactions with a number of other regulators of apoptosis. Isoform 1 inhibits apoptosis. Isoform 2 promotes apoptosis. {ECO:0000269|PubMed:10766760, ECO:0000269|PubMed:16543145}. | 3D-structure;Alternative splicing;Apoptosis;Cytoplasm;Developmental protein;Differentiation;Direct protein sequencing;Isopeptide bond;Membrane;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Ubl conjugation | This gene encodes an anti-apoptotic protein, which is a member of the Bcl-2 family. Alternative splicing results in multiple transcript variants. The longest gene product (isoform 1) enhances cell survival by inhibiting apoptosis while the alternatively spliced shorter gene products (isoform 2 and isoform 3) promote apoptosis and are death-inducing. [provided by RefSeq, Oct 2010]. | hsa:4170; | Bcl-2 family protein complex [GO:0097136]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; BH3 domain binding [GO:0051434]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; protein transmembrane transporter activity [GO:0008320]; cell fate determination [GO:0001709]; cellular homeostasis [GO:0019725]; extrinsic apoptotic signaling pathway in absence of ligand [GO:0097192]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; negative regulation of anoikis [GO:2000811]; negative regulation of apoptotic process [GO:0043066]; negative regulation of autophagy [GO:0010507]; negative regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001240]; negative regulation of intrinsic apoptotic signaling pathway [GO:2001243]; positive regulation of apoptotic process [GO:0043065]; positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway [GO:1903378]; regulation of apoptotic process [GO:0042981]; regulation of response to DNA damage stimulus [GO:2001020]; response to cytokine [GO:0034097] | 11781193_expression in normal, hyperplastic and carcinomatous human prostate 11877256_Myeloid cell factor-1 (Mcl-1)is a critical survival factor for multiple myeloma. 11911971_Expression of apoptotic regulators and their significance in cervical cancer 12057933_Mcl-1 is over epxressed in anaplastic large cell lymphoma cell lines and tumors. 12070027_Antisense strategy shows that Mcl-1 rather than Bcl-2 or Bcl-x(L) is an essential survival protein of human myeloma cells. 12176902_follicular dendritic cells protect CLL B cells against apoptosis, at least in part through a CD44-dependent mechanism involving up-regulation of Mcl-1 12223490_inactivation of Mcl-1 by JNK-dependent phosphorylation may be one of the mechanisms through which oxidative stress induces cellular damage in human cells 12359245_related to ratio of p21(WAF)/cyclin A and Jun kinase phosphorylation to apoptosis in human breast carcinomas 12445202_in basal cell carcinoma cells, the upregulation of the anti-apoptotic Mcl-1 protein by interleukin-6 is mainly through the Janus tyrosine kinase/phosphotidyl inositol 3-kinase/Akt, but not the STAT3 pathway. 12475993_interaction of Mcl-1 with tankyrase 1 leads to the modulation of the apoptosis pathway 12496428_The antiapoptotic effect of the in vitro induction of MCL1 expression in Mycobacterium tuberculosis strain H37Rv-infected macrophages promotes the intracellular survival and proliferation of virulent M. tuberculosis. 12637318_activation of STAT3 was dependent on Ser727 phosphorylation, in the absence of detectable Tyr705 phosphorylation; expression of human STAT3 in murine macrophages rescued inhibition of human Mcl-1 promoter gene activation and cell death induced by NaSal 12660820_mechanisms regulating Mcl-1 levels in MM cells are heterogeneous, and are often independent from IL-6 signaling pathways 12782407_the human EAT gene driven by the EF1 alpha promoter induced hyperplasia of Langerhans islet cells and upregulation of Bax and Bag-1 -- possible heterodimeric partners for EAT in the anti-apoptotic process 12783855_Following UV treatment, Mcl-1 protein synthesis is blocked, the existing pool of Mcl-1 protein is rapidly degraded by the proteasome, and cytosolic Bcl-xL translocates to the mitochondria 12787138_Mcl-1 is an important factor contributing to the chemoresistance of human melanoma in vivo. 12855556_protein geranylgeranylation is critical for regulating myeloma tumor cell survival, possibly through regulating Mcl-1 expression 12901848_data suggest a role for Mcl-1 in protecting endothelial cells against Stx-1-induced apoptosis 12915532_Respiratory syncytial virus mediated the strong induction of antiapoptotic factors of the Bcl-2 family, especially Mcl-1, which might account for the delayed induction of apoptosis in RSV-infected cells. 12960271_fMLP-stimulated neutrophils coordinate the regulation of FOXO transcription factors and the survival factor Mcl-1, a mechanism that may allow neutrophils to alter their survival. 14633975_restoration of MCL-1 expression rescued infected cells from E1A-induced apoptosis 14982947_IL-15 does not increase IL-1alpha or IL-1beta production but induces IL-1Ra release, increases myeloid cell differentiation factor-1 stability, decreases the activity of caspase-3 and caspase-8, resulting in an inhibition of vimentin cleavage 15014070_Mcl-1L degradation by either GrB or caspase-3 interferes with Bim sequestration by Mcl-1L 15077116_These results are consistent with a model in which p53 and Mcl1 have opposing effects on mitochondrial apoptosis by interacting with, and modulating the activity of, the death effector Bak. 15078892_Profound changes in the rate of neutrophil apoptosis following Granulocyte macrophage colony-stimulating factor signaling occur via dynamic changes in the rate of Mcl-1 turnover via the proteasome. 15122313_Mcl-1 function is an effective means of inducing apoptosis in Mcl-1-positive B-cell lymphoma. 15126604_The MCL-1 promoter insertion may identify a high-risk group of CD38-negative CLL patients 15217829_VEGF-induced MM cell proliferation and survival are mediated via Mcl-1; VEGF up-regulates Mcl-1 expression in a time- and dose-dependent manner in 3 human MM cell lines and MM patient cells 15241487_MCL1 is subject to multiple, separate, post-translational phosphorylation events, produced in living versus dying cells at ERK-inducible versus ERK-independent sites 15262975_the fortilin-MCL1 interaction increases cellular resistance to apoptosis by allowing MCL1, an independently antiapoptotic protein, to stabilize another independently antiapoptotic protein, fortilin 15370246_drug sensitivities of CLL leukemic cells correlated inversely with Mcl-1 levels; results suggest that Mcl-1 may contribute to cell survival in CLL; an inverse correlation was found between Mcl-1 expression and Rai stage 15378010_Cleavage of Mcl-1 by caspases modifies its subcellular localization, increases its association with Bim and inhibits its antiapoptotic function. 15550399_Mcl-1 solution structure and analysis of binding by proapoptotic BH3-only ligands 15588513_SDF-1/CXCL12 enhanced cell survival in synergy with other cytokines involves activation of CREB and induction of Mcl-1 and c-Fos 15611089_Overexpression of Mcl-1 protected hepatoma cells against apoptosis induced by tert-butyl hydroperoxide. 15613543_Mcl-1 accumulation is an unwanted molecular consequence of exposure to proteasome inhibitors, which slows down their proapoptotic effects 15626746_MCL-1 is a BCR/ABL-dependent survival factor and interesting target in chronic myeloid leukemia. 15637055_removal of N-terminal domains of Bid by caspase-8 and Mcl-1 by caspase-3 enables the maximal mitochondrial perturbation that potentiates TRAIL-induced apoptosis 15713684_Knockdown results in a significant level of apoptosis in the absence of external apoptotic stimulation 15728130_Mcl-1 is induced by signaling through the B-cell receptor, which promotes survival of chronic lymphocytic leukemia B cells 15753661_Mcl-1 protein is overexpressed in a subset of human NSCLC and enhanced levels of Mcl-1 may protect lung cancer cells from death induced by a variety of pro-apoptotic stimuli. 15842635_Overexpression of anti-apoptotic Mcl-1 may function to enhance the viability of testicular germ cells, thereby leading to tumorigenesis. 15901672_role in sequestering proapoptotic Bak 15902294_The Mcl-1, which has been shown to be essential for the survival of human myeloma cells in vitro, is overexpressed in vivo in MM in relation with relapse and shorter survival. Mcl-1 represents a potential therapeutical target in multiple myeloma. 15940637_autocrine IL-6/Akt signaling pathway enhances Mcl-1 expression in cholangiocarcinoma 15989957_Mule (Mcl-1 ubiquitin ligase E3)is both required and sufficient for the polyubiquitination of Mcl-1 16007132_The interaction of PUMA with MCL1 is not sufficient to prevent the rapid degradation of MCL1. 16027162_The anti-apoptotic poein MCL1 inhibits mitochondrial Ca2+ signals. 16091744_Melphalan-induced apoptosis in multiple myeloma is associated with a cleavage of MCL1 and BIM and a decrease in the MCL1/BIM complex. 16109713_Mcl-1 is downregulated via inhibition of translation after administration of BAY 43-9006 in human leukemia cells 16213503_The unliganded form of Mcl-1 is sensitive to LASU1-mediated degradation of Mcl-1. 16229017_Knockdown of HIF-1alpha induced parallel knockdown of Mcl-1 mRNA and protein expression, whereas Mcl-1 knockdown had no noticeable effect on HIF-1alpha expression. Both of these proteins were shown to be anti-apoptotic. 16289418_Mcl-1 is overexpressed in half of HCC-tissues. ASO targeting Mcl-1 revealed a prominent single agent and chemosensitizing activity against HCC in vitro. 16327976_Mcl-1 is an important factor for the apoptosis resistance of human HCC, and constitutes an interesting target for HCC therapy 16339575_Mcl-1 was critical for the survival of RA synovial fibroblasts, because the forced reduction of Mcl-1 using a Mcl-1 antisense-expressing adenoviral vector induced apoptotic cell death, which was mediated through Bax, Bak, and Bim 16380381_Data demonstrate a novel regulation of tBid by Mcl-1 through protein-protein interaction in apoptotic signaling from death receptors to mitochondria. 16456709_REVIEW: Mcl-1 plays an apical role in many cell death and survival regulatory programs. 16478725_Mcl-1 may serve as a direct substrate for TRAIL-activated caspases implying the existence of a novel TRAIL/caspase-8/Mcl-1/Bim communication mechanism between the extrinsic and the intrinsic apoptotic pathways 16538501_Analysis of B-cell chronic leukemia cells for ZAP 70 expression and the expression of cyclin E, bcl-2, bax, and mcl-1. 16543145_The results demonstrate that the control of MCL-1 stability by GSK-3 is an important mechanism for the regulation of apoptosis by growth factors, PI3K, and AKT. 16725198_This signalling cascade results in viability in a group of patients in which observe an increase of Mcl-1 expression levels after CD5 stimulation in B-CLL patients. 16761109_MCL1 was upregulated by PMA in THP-1 & U937 myeloid leukemia cells, but by microtubule disrupting agents only in THP-1 cells. 16782027_Noxa/Mcl-1 axis is an apoptosis rheostat in dividing cells, in a selective pathway that functions to restrain lymphocyte expansion and can be triggered by glucose deprivation 16822835_The internal EELD domain facilitates mitochondrial targeting of Mcl-1 via a Tom70-dependent pathway. 16901898_Overexpression of Mcl-1 protects HaCaT cells from both ultraviolet and protein kinase C delta-catalytic fragment-induced apoptosis and blocks release of cytochrome c from the mitochondria. 16969094_Mcl-1 is a promising molecular target for antisense oligonucleotide-based treatment strategies for gastric cancer in the future. 16978419_Mycobacterium leprae inhibits apoptosis in THP-1 cells by upregulation of Mcl-1 gene expression and downregulation of Bad and Bak . 17009247_The expression of Mcl-1 was suppressed with small interfering RNA (siRNA) or chemical inhibitors of the phosphatidylinositol 3-kinase (PI 3-kinase)/Akt-1 and signal transducer and activator of transcription 3 (STAT-3) pathways. 17072336_Specific knockdown of Mcl-1 gene expression by small interfering RNA yielded an increase in apoptosis of LNCaP-IL-6+ cells. 17145774_These results demonstrate that anoxia-induced cell death requires the loss of Mcl-1 protein and inhibition of the electron transport chain to negate Bcl-X(L)/Bcl-2 proteins. 17200126_Because of its rapid turnover, Mcl-1 may serve as a convergence point for signals that affect global translation, coupling translation to cell survival and the apoptotic machinery 17227835_GX15-070 induced apoptosis in vitro in MCL cell lines and primary cells from patients with MCL by releasing Bak from Mcl-1 and Bcl-X(L). 17384650_This study is the first to show a clear dissociation between changes in Bcl-2 expression (downregulation) and Bcl-XL, Mcl-1 expression (upregulation) during progression of melanoma. 17387146_Results indicate that the turnover of Mcl-1 by beta-TrCP is an essential mechanism for GSK-3beta-induced apoptosis and contributes to GSK-3beta-mediated tumor suppression and chemosensitization. 17463001_the antiapoptotic function of Mcl-1 is enhanced by serine 64 phosphorylation 17495975_EXEL-0862 induced apoptotic death in EOL-1 cells and imatinib-resistant T674I FIP1L1-PDGFR-alpha-expressing cells, and resulted in significant downregulation of the antiapoptotic protein Mcl-1 through a caspase-3 dependent mechanism. 17498302_Both ionizing radiation and daunorubicin treatment resulted in concerted protein modulations of Mcl-1, Hdm2 and Flt3. 17525735_preformed Bim(EL)/Mcl-1 and Bim(EL)/Bcl-x(L) complexes can be rapidly dissociated following activation of ERK1/2 by survival factors 17545167_tribbles homolog 2-Mcl-1 axis plays an important role in survival factor withdrawal-induced apoptosis of TF-1 erythroleukemia cells. 17553788_stress-induced phosphorylation of eIF2 alpha is directly coupled to mitochondrial apoptosis regulation via translational repression of MCL-1 17561513_The degradation versus stabilized expression of antiapoptotic MCL1 is thus controlled by N-terminal truncation as well as by ERK- and GSK3 (but not G2/M)-induced phosphorylation 17599053_BCR/ABL induces SPK1 expression and increases its cellular activity, leading to upregulation of Mcl-1 in CML cells. 17698840_Mcl-1 confers TRAIL resistance by serving as a buffer for Bak, Bim, and Puma, and sorafenib is a potential modulator of TRAIL sensitivity 17805325_Mcl-1(139) is an HLA-A2-restricted epitope from Mcl-1 recognized spontaneously by cytotoxic T cells in cancer patients. 17823113_These results suggest that the N terminus of MCL-1 plays a major regulatory role, regulating coordinately the mitochondrial (anti-apoptotic) and nuclear (anti-proliferative) functions of MCL-1. 17893147_a caspase-9 signaling cascade induces feedback disruption of the mitochondrion through cleavage of anti-apoptotic Bcl-2, Bcl-xL, and Mcl-1 17928528_Akt and Mcl-1 are major components of a survival pathway that can be activated in CLL B cells by antigen stimulation. 17942758_While TNFalpha had no effect on MCL-1 transcription, it induced expression of another antiapoptotic molecule, BFL-1. 18006817_Mcl-1 degradation primes the cell for Bim and Bax activation and anoikis, which can be blocked by oncogenic signaling in metastatic cells 18025305_MCL1 determines the Bax dependency of Nbk/Bik-induced apoptosis. 18032706_findings not only illustrate MCL1 as an aberrantly expressed reprogramming oncoprotein in follicular lymphomas but also highlight MCL1 as key therapeutic target 18046444_High MCL-1 expression is associated with B-cell chronic lymphocytic leukemia 18088462_STI571 induces the apoptosis of K562 cells by down-regulating the expressions of Mcl-1 and Bcl-xl. 18089567_the pro-survival activity of MCL-1 proceeds via inhibition of BAX function at mitochondria, downstream of its activation and translocation to this organelle. 18178565_distinctions in the behaviors of Bcl-B and Mcl-1 relative to the other anti-apoptotic Bcl-2 family members, where Bcl-B and Mcl-1 display reciprocal abilities to bind and neutralize Bax and Bak. 18208354_Transcription regulation assays with MCL-1 promoter deletion mutants showed that most of the p53 inhibitory effect was mediated by the -41 to +16 bp promoter binding sites only for TATA-binding protein and other basal transcription factors. 18234961_This study identifies mitogen-activated protein kinase/ERK/Mcl-1 as an important survival signaling pathway in the resistance of melanoma cells to Fas-mediated apoptosis. 18292181_Demonstrate that CUGBP2 inhibits Mcl-1 expression by inhibiting Mcl-1 mRNA translation, resulting in driving the cells to apoptosis during the G(2) phase of the cell cycle. 18415656_Anti-myeloma effect of homoharringtonine with concomitant targeting of the myeloma-promoting molecules, Mcl-1, XIAP, and beta-catenin 18452656_Observational study of gene-disease association. (HuGE Navigator) 18452656_Polymorphisms in MCL1 might be one of genetic factors for the risk of clinical tuberculosis development. 18483275_deregulated PKB/AKT stabilizes Mcl-1 expression in a mammalian target of rapamycin (mTOR)-dependent pathway 18495871_Mcl-1, perhaps acting as an adaptor protein, in controlling the ATR-mediated regulation of Chk1 phosphorylation 18543107_Inhibition of the proteasome by cobalt chloride leads to the accumulation of Mcl-1 which acts to limit cobalt chloride induced apoptosis. 18552129_MCL-1 expression is activated by Triiodothyronine, which increases its promoter activity by a non-genomic mechanism using the PI3-K signal transduction pathway. 18599795_The close correlation between Mcl-1 expression and V(H) gene mutation status, CD38 expression, and ZAP-70 expression offers a biologic explanation for their association with adverse prognosis in CLL. 18609706_Bcl-x(L) and, to a lower extent, Mcl-1, are important anti-apoptotic factors in colorectal carcinoma. 18647593_These findings suggest that calpain inhibition delays neutrophil apoptosis via cyclic AMP-independent activation of PKA and PKA-mediated stabilization of Mcl-1 and XIAP. 18676738_HPV 16/18 up-regulates the expression of interleukin-6 and antiapoptotic Mcl-1 in non-small cell lung cancer 18676833_Erk could phosphorylate Mcl-1 at two consensus residues, Thr 92 and 163, which is required for the association of Mcl-1 and Pin1, resulting in stabilization of Mcl-1 18757878_Mutations also dramatically decrease the levels of MLC1 in cells from megalencephalic leukoencephalopathy with subcortical cysts patients. 18768389_IL-3 up-regulates the expression of the antiapoptotic proteins cIAP2, Mcl-1, and Bcl-X(L) and induces a rapid and sustained de novo expression of the serine/threonine kinase Pim1 that closely correlates with cytokine-enhanced survival. 18769617_Mcl-1 up-regulation is primarily required to maintain apoptosis resistance in C. trachomatis-infected cells 19008456_Mcl-1 expression may therefore be useful in predicting poor response to chemoimmunotherapy. 19012245_multiple copy number alterations in chromosome regions implicated in malignancy progression and indicated a strong expression of MAP2K4 and MCL1 genes in rhabdomyosarcoma 19037233_Mcl-1 acts as a major survival protein by inhibiting premature apoptosis in the spinous and granular layers to promote conification, and promotes the robust induction of keratinocyte differentiation markers 19051025_Increased Mcl-1 expression plays a role in hepatoprotection upon cholestatic liver injury. 19077158_MCL1 mRNA expression in myeloma cells was upregulated;but lacked association with overall and event-free survival. 19092849_inhibition of syk prevented the increase in leukemic cell viability induced by sustained BCR engagement and inhibited BCR-induced Akt activation and Mcl-1 upregulation 19099185_review summarizes the current knowledge on the regulation of Mcl-1 expression and discusses the alternative approaches targeting Mcl-1 in human cancer cells 19148187_The results demonstrate that loss of Mcl-1 is a critical heat-sensitive step leading to Bax activation that is controlled by Hsp70. 19285955_This study demonstrated the physical association of the MCL-1 and IEX-1 proteins, the modulatory role of MCL-1 in IEX-1-induced apoptosis, and the role of BIM as an essential downstream molecule for IEX-1-induced cell death. 19288493_Role of RAF/MEK/ERK pathway, p-STAT-3 and Mcl-1 in sorafenib activity in human pancreatic cancer cell lines. 19369967_Modification of alternative splicing of Mcl-1 pre-mRNA using antisense morpholino oligonucleotides induces apoptosis in basal cell carcinoma cells. 19372583_Enhanced Mcl-1 expression is associated with melanoma. 19401626_Down-regulation of Mcl-1L in clear-cell renal cell carcinomas is associated with shortened recurrence-free and disease-specific survival. 19481066_Bz-423 superoxide signals B cell apoptosis via MCL1, BAK, and BAX. 19484260_Late tumour stages of Kaposi sarcoma in tissues from HIV-positive patients are associated with high levels of LANA-1, HIF-1alpha and of the anti-apoptotic proteins, Bcl-2 and Mcl-1. 19503096_Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. (microRNA-512-5p) 19523441_EG-VEGF protects pancreatic cancer cells from apoptosis through upregulation of myeloid cell leukemia-1, an anti-apoptotic protein of the bcl-2 family. 19549371_As(2)O(3) can markedly decrease mitochondrial respiratory function and membrane potential of Raji cells, and down-regulate expression of mcl-1 gene. 19571464_These results imply a potentially important and novel role of the inhibition of Mcl-1 function 19581935_functional and biochemical evidence that Syk regulated chronic lymphocytic leukemia B cell survival through a novel pathway involving PKCdelta and a proteasome-dependent regulation of the anti-apoptotic protein Mcl-1 19582795_Mcl-1 is highly upregulated in gastric cancer and high Mcl-1 expression is correlated with a poor prognosis in gastric cancer patients. 19587033_Inhibition of host gene expression by the VSV M protein resulted in the degradation of Mcl-1 but not Bcl-X(L); inactivation of both Mcl-1 and Bcl-X(L) was required for cells to undergo apoptosis 19605477_hepatitis C virus core protein contains a BH3 domain that regulates apoptosis through specific interaction with human Mcl-1 19616548_Data show that eosinophils treated with R-roscovitine lose mitochondrial membrane potential and the key survival protein Mcl-1 is down-regulated. 19634140_MCL-1 targeted strategies could constitute an efficient therapeutic tool for the treatment of chemoresistant ovarian carcinoma, in association with conventional chemotherapy. 19654003_over-expression of miR-133B increased apoptosis in response to gemcitabine and reduced MCL-1 and BCL2L2 expression. 19676043_Microrna can inhibit MCL1 expression and may induce apoptosis in a glioblbastoma cell line. 19683529_Data show that MCL-1ES interacts with MCL-1L and induces mitochondrial cell death, suggesting that alternative splicing of MCL-1 may control the fate of cells. 19684859_findings suggest that the combination of ABT-737 and Mcl-1 knockdown represents a promising, new treatment strategy for malignant melanoma 19734538_PKC-dependent destabilization of Mcl-1 is a mechanism contributing to hepatocyte lipoapoptosis 19737931_Interference with Puma induction or expression of MCL1 or BCL2 significantly reduced both glucocorticoid- and FAS-induced cell death in p16INK4A-reconstituted leukemia cells. 19748896_Mcl-1 binds with high affinity to peptides resembling natural BH3 domain sequences. 19763916_Increased expression of Mcl-1 protein was observed in 26% of NSCLC cases with EGFR overexpression. 19773546_Bim, and Mcl-1, but not Bad, integrate death signaling triggered by concomitant disruption of the PI3K/Akt and MEK1/2/ERK1/2 pathways in human leukemia cells. 19782681_immediate impact of Mcl-1 protein level manipulations on the course of early acute apoptotic response of colon adenocarcinoma cells to TRAIL. 19808698_the acquisition of FLT3-internal tandem duplications ensures leukemic stem cells survival by up-regulating MCL-1 via constitutive STAT5 activation that is independent of wild-type FLT3 signaling. 19838163_Correlation of LDH-5 expression with clinicopathological factors and with the expression of Bcl-2, Bcl-XL, Mcl-1 and GRP78 was examined in pigmented lesions, including nevi and melanoma at different stages of progression 19840460_2-methoxyestradiol can regulate the apoptosis of myelodysplastic syndrome cells through down-regulating the expression of mcl-1 mRNA and activating caspase-3. 19841168_these data demonstrate for the first time that the intrinsic apoptosis pathway in neutrophils from severely injured patients is blocked at least in part by elevated Mcl-1 levels induced by GM-CSF. 19887550_In the HA14-1 nonresponsive cell lines (SKOV3 and OAW42), small interfering RNA-mediated Mcl-1 downregulation allowed HA14-1-induced massive apoptosis in the absence of chemotherapy. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19933775_The expression of Mcl-1 was significantly reduced when COX-2 was suppressed and knockdown of Mcl-1 substantially sensitized tumor cells to apoptosis. 19942343_Triptolide induced abundant apoptosis with a prominent decline of Bcl-2, Bcl-X(L), survivin and Mcl-1 in myeloproliferative disorder cells. 19949310_CDODO-Me-12 and CDODO-Me-11 downregulated the levels of anti-apoptosis protein Mcl-1 in HL-60, U937 and K562 leukemic cell lines. 19955184_Data suggest that F1L replaces the antiapoptotic activity of Mcl-1 during vaccinia virus infection by interacting with Bak using highly divergent BH domains. 19965632_Data show that down-regulation of MCL-1 in prednisolone-resistant MLL-rearranged leukemia cells by RNA interference, to some extent, led to prednisolone sensitization. 19966861_Data show a novel mechanism for the sensitization to DR-induced apoptosis linking glucose metabolism to Mcl-1 downexpression, and provide a rationale for the combined use of DR ligands with AMPK activators or mTOR inhibitors in the treatment of cancers. 19968497_Observational study of gene-disease association. (HuGE Navigator) 19968719_STAT3 activation is associated with Mcl-1 expression in nasal NK-cell lymphoma 19968986_Mcl-1(128-350) exerts a pro-apoptotic function governed by its capacity to interact with Bax. 20004446_Mcl-1 downregulation could significantly enhance radiosensitivity of pancreatic carcinoma cells in vitro and in vivo. 20023629_the deubiquitinase USP9X stabilizes MCL1 and thereby promotes cell survival; deubiquitinases may stabilize labile oncoproteins in human malignancies 20038816_Sorafenib inhibits ERK1/2 and MCL-1(L) phosphorylation levels resulting in caspase 3-independent cell death in malignant pleural mesothelioma 20051518_Data suggest that MCL1 steady-state expression levels do not affect sensitivity to proteasome-inhibitor treatment in neuronal tumor cells, and that both the repression of Bcl-xL and the activation of Noxa are necessary for bortezomib-induced cell death. 20066663_the crystal structure of human Mcl-1 bound to a BH3 peptide derived from human Bim and the structures for three complexes that accommodate large physicochemical changes at conserved Bim sites was reported. 20066738_HPPCn is a novel hepatic growth factor that can be secreted to culture medium and suppresses apoptosis of hepatocellular carcinoma cells by up-regulating Mcl-1 expression. 20085644_Activation of VEGF165-NRP1-c-MET signaling could confer prostate cancer (PCa) cells survival advantages by up-regulating Mcl-1, contributing to PCa progression. 20156337_Glucocorticoid Response Elements (GREs) within the promoter regulatory regions of the Bcl-2 family members NOXA and Mcl-1 were identified, indicating that they are direct GR transcriptional targets. 20173022_Human cytomegalovirus engages the epidermal growth factor receptor to facilitate upregulation of Mcl-1 and acquisition of the apoptotic resistant phenotype in infected monocytes. 20189983_Apoptosis protection by Mcl-1 and Bcl-2 modulation of inositol 1,4,5-trisphosphate receptor-dependent Ca2+ signaling. 20197552_there was a dynamic increase MCL-1 transcript BFL-1 protein and transcript levels after ABT-737 treatment in resistant cells 20308427_The Mcl-1 rather than Bcl-xL is a major target for sensitization of Bax-deficient tumors for death receptor-induced apoptosis via the Bak pathway. 20392693_analysis of selectivity of MCL-1 in binding BCL-2 homology 3 (BH3) ligands of interest for mammalian biology 20501829_Overexpression of MCL1 is associated with prostate carcinogenesis. 20503275_BH3 mimetic S1 potently induces Bax/Bak-dependent apoptosis by targeting both Bcl-2 and Mcl-1 20526282_Data suggest that phosphorylation of Mcl-1 by CDK1-cyclin B1 and its APC/C(Cdc20)-mediated destruction initiates apoptosis if a cell fails to resolve mitosis. 20571074_Data show that loss of the antiapoptotic protein myeloid cell leukemia 1 (Mcl-1) coincided with mitotic cell death. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20647761_The role for MCL-1 is in coordinating DNA damage mediated checkpoint response, and have broad implications for the importance of MCL-1 in maintenance of genome integrity. 20677014_Observational study of gene-disease association. (HuGE Navigator) 20693279_Lyn-dependent regulation of miR181 is a novel mechanism of regulating Mcl-1 expression and cell survival. 20802294_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21036904_Heterodimerization of BAK and MCL-1 activated by detergent micelles. 21132008_results indicate that targeting Mcl-1 may improve therapy for a subset of NSCLC patients 21138867_Mcl-1 downregulation and Bax activation were also observed in xenografts. 21139567_MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. 21148306_The BH3 alpha-helical mimic BH3-M6 disrupts Bcl-X(L), Bcl-2, and MCL-1 protein-protein interactions with Bax, Bak, Bad, or Bim and induces apoptosis in a Bax- and Bim-dependent manner. 21151390_We report the correlation of Mcl-1 protein expression with higher grade and stage in colorectal cancer. 21216463_Data indicate that P276-00 rapidly and significantly down regulated both Cdk9 and Mcl-1 protein expression levels in a dose and time-dependent manner. 21220745_the mechanism involved in c-Abl regulation of Mcl-1 expression in CLL cells 21228225_The authors demonstrate that HBx enhances cisplatin-induced hepatotoxicity by a mechanism involving degradation of Mcl-1, an antiapoptotic member of the Bcl-2 family. 21247487_that Bim and Mcl-1 have key opposing roles in regulating JAK2V617F cell survival and propose that inactivation of aberrant JAK2 signaling leads to changes in Bim complexes that trigger cell death. 21253591_Mcl-1 is both an upstream regulator and a downstream target of caspase activity in human neutrophils. 21258408_These results indicate that the EGF induced activation of Elk-1 is an important mediator of Mcl-1 expression and cell survival 21292770_Escape from p21-mediated oncogene-induced senescence leads to cell dedifferentiation and dependence on anti-apoptotic Bcl-xL and MCL1 proteins. 21368833_the E3 ubiquitin ligase SCF(FBW7) governs cellular apoptosis by targeting MCL1, a pro-survival BCL2 family member, for ubiquitylation and destruction in a manner that depends on phosphorylation by glycogen synthase kinase 3 21393866_hypoxic tumor cell are sensitized to BH-3 mimetic-induced apoptosis via downregulation of the Bcl-2 protein Mcl-1 21406400_Data suggest that the joint overexpression of MCL1 and MYC may be a useful biomarker for both prognosis and treatment in NSCLC. 21412051_MCL-1 regulates the balance between autophagy and apoptosis. 21423203_Mutations in the Ets-1 binding site or knockdown of Ets-1 inhibited the increase in Mcl-1, indicating that Ets-1 has a critical role in transcriptional upregulation of Mcl-1 21471522_MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. 21504623_Findings indicate that HBx exerts pro-apoptotic effect upon exposure to oxidative stress probably through accelerating the loss of Mcl-1 protein via caspase-3 cascade 21516346_Noxa/Mcl-1 balance is regulated by glucose deprivation as well as by general metabolic stress. 21596068_genetic knockdown of Mcl-1 resulted in marked induction of apoptosis in pancreatic cancer cell lines 21613222_Mitochondrion-dependent N-terminal processing of outer membrane Mcl-1 protein removes an essential Mule/Lasu1 protein-binding site. 21628457_Multiple BH3 mimetics antagonize antiapoptotic MCL1 protein by inducing the endoplasmic reticulum stress response and up-regulating BH3-only protein NOXA. 21659544_Distribution of Bim determines Mcl-1 dependence or codependence with Bcl-xL/Bcl-2 in Mcl-1-expressing myeloma cells. 21670080_In the presence of glucose, activated Akt prevented Loss of Mcl-1 expression & protected cells from growth factor deprivation-induced apoptosis. Mcl-1 associated with & inhibited the proapoptotic protein Bim, contributing to cell survival. 21674276_Our results suggest that Mcl-1 may play an important role in cervical cancer 21730980_findings suggest that reduced Mule/Mcl-1 complex has a significant role in increasing the stability of Mcl-1 in breast cancer cells and increased resistance to apoptosis 21734342_Mcl-1 serves as a predictive co-marker in tumors of the parotid gland. 21750559_Quercetin downregulates Mcl-1 acting directly or indirectly on its mRNA stability and protein degradation. 21824245_Taken together, these findings identify HIF-1alpha as responsible for upregulation of Mcl-1 and the maintenance of apoptosis resistance during Chlamydia infection. 21841196_Mcl-1 is a determinant of cell fate, and ATF6 mediates apoptosis via specific suppression of Mcl-1 through up-regulation of WBP1 21846680_High Mcl-1 is associated with glioblastoma. 21863213_PUMA enhances sensitivity of SKOV3 cells to cisplatin by lowering the threshold set simultaneously by Bcl-x(L) and Mcl-1. 21887682_common variants in MCL1 promoter co | ENSMUSG00000038612 | Mcl1 | 3.212894e+03 | 0.7746841 | -0.368319943 | 0.3040981 | 1.476343e+00 | 0.2243475226 | 0.78763590 | No | Yes | 2.889128e+03 | 539.572735 | 3.451224e+03 | 660.826824 | |
| ENSG00000143387 | 1513 | CTSK | protein_coding | P43235 | FUNCTION: Thiol protease involved in osteoclastic bone resorption and may participate partially in the disorder of bone remodeling. Displays potent endoprotease activity against fibrinogen at acid pH. May play an important role in extracellular matrix degradation. Involved in the release of thyroid hormone thyroxine (T4) by limited proteolysis of TG/thyroglobulin in the thyroid follicle lumen (PubMed:11082042). {ECO:0000269|PubMed:11082042}. | 3D-structure;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Hydrolase;Lysosome;Membrane;Protease;Reference proteome;Secreted;Signal;Thiol protease;Zymogen | The protein encoded by this gene is a lysosomal cysteine proteinase involved in bone remodeling and resorption. This protein, which is a member of the peptidase C1 protein family, is predominantly expressed in osteoclasts. However, the encoded protein is also expressed in a significant fraction of human breast cancers, where it could contribute to tumor invasiveness. Mutations in this gene are the cause of pycnodysostosis, an autosomal recessive disease characterized by osteosclerosis and short stature. [provided by RefSeq, Apr 2013]. | hsa:1513; | apical plasma membrane [GO:0016324]; endolysosome lumen [GO:0036021]; external side of plasma membrane [GO:0009897]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; nucleoplasm [GO:0005654]; collagen binding [GO:0005518]; cysteine-type endopeptidase activity [GO:0004197]; cysteine-type peptidase activity [GO:0008234]; fibronectin binding [GO:0001968]; proteoglycan binding [GO:0043394]; serine-type endopeptidase activity [GO:0004252]; autophagy of mitochondrion [GO:0000422]; bone resorption [GO:0045453]; collagen catabolic process [GO:0030574]; extracellular matrix disassembly [GO:0022617]; immune response [GO:0006955]; intramembranous ossification [GO:0001957]; negative regulation of cartilage development [GO:0061037]; proteolysis [GO:0006508]; proteolysis involved in cellular protein catabolic process [GO:0051603]; thyroid hormone generation [GO:0006590] | 11733367_This study demonstrates for the first time a critical role of Cathepsin K in cartilage degradation by synovial fibroblasts (SFs) in rheumatoid arthritis (RA) that is comparable to its well-known activity in osteoclasts. 12039963_cathepsin K binds with chondroitin sulfate for collagenase activity 12081494_Selective inhibition of the collagenolytic activity by altering its S2 subsite specificity 12125807_review discusses the human disease pycnodysostosis caused by cathepsin K deficiency and cathepsin K activity and regulation 12492488_data suggest that extracellular cysteine proteases may participate in the regulation of kinin levels at inflammatory sites, and clearly support that cathepsin K may act as a potent kininase 12568399_cathepsin K may have a role in contributing to the invasive potential of prostate cancer 12652657_In db/db mice, cathepsin k(ctsk) increased, as did Mitf and TFE3, two transcription factors involved in ctsk induction in osteoclasts. Ctsk was increased in other obese models including A(y), fat, and tubby. 12887056_cathepsin K is capable to degrade aggrecan complexes at specific cleavage sites 14645229_cathepsin K has a role in lysosomal collagenolytic activity 14753734_Observational study of gene-disease association. (HuGE Navigator) 15161653_Cathepsin K plays a pivotal role in lung matrix homeostasis under physiological and pathological conditions. 15304486_RANKL-induced cathepsin K gene expression is cooperatively regulated by the combination of the transcription factors and p38 MAP kinase in a gradual manner. 15737607_These findings suggest that a specific function of human cathepsin X is unlikely to result from sequence specificity, but rather from a combination of its unique positional specificity and the co-localization of enzyme and substrate. 15797245_heparan sulfate proteoglycans can regulate the cellular trafficking and the enzymatic activity of cathepsin X 15826870_results support the role of cathepsin K as a major proteinase in osteoclastic bone resorption 15837295_Several novel ketoamide-based inhibitors of cathepsin K have been identified. 15878337_Cathepsin X is not involved in degradation of extracellular matrix, a proteolytic event leading to tumor cell invasion and metastasis, and its expression, restricted to immune cells suggests a role in phagocytosis and the regulation of immune response. 15929988_Cathepsin K co-localized with TRAP in osteoclast-resorptive compartments, supporting a role for cathepsin K in the extracellular processing of monomeric TRAP in the resorption lacuna. 16337236_Osteoblastic cathepsin K may thus contribute to collagenous matrix maintenance and recycling of improperly processed collagen I 16354158_The presence of active cathepsins L, K and S suggests that they contribute to the extracellular breakdown of the extracellular matrix. 16774752_Active cathepsin X mediates the function of beta(2) integrin receptors during cell adhesion and that it could also be involved in other processes associated with beta(2) integrin receptors such as phagocytosis and T cell activation. 16831915_Cathepsin K is essential for normal bone resorption (review) 16912123_CTSK may be involved in the pathogenesis of obesity by promoting adipocyte differentiation. 16946716_Possiable role in homeostasis of dermal extracellular matrix and dynamic equilibrium between matrix synthesis and proteolytic degradation, by counteracting deposition of matrix proteins during scar formation with its matrix-degrading activity. 17227755_analysis of human cathepsins K, L, and S iunteractions with elastins 17230547_Cathepsin K is constitutively expressed in normal human brain, and is alterated in its expression inscizophrenia. 17397052_molecular characterization of 12 unrelated patients with Pycnodysostosis; mutational profile consisted of 12 different mutations, including nine previously unreported ones 17683065_Cathepsin K expression is of predictive prognostic value for patients with high-grade osteosaromas and metastasis at diagnosis. 17728092_In breast cancer patients, cathepsin K serum levels were significantly lower than in sex matched control group or in patients with primary osteoporosis 17882010_in peri-and postmenopausal women, moderate negative correlation of serum cathepsin K levels with change in femoral neck BMD, but none with change in spinal BMD was found 17991740_Induction of cathepsin K is associated with the activation of protein p38 MAP kinase. 18053985_Myeloma-osteoclast interactions stimulated the production of TRAP, cathepsin K, MMP-1, -9, and uPA 18163891_did not detect any plasminogen degradation by cathepsins B, K and L. 18368130_catK may play an important role in melanoma invasion and metastasis by mediating intracellular degradation of matrix proteins after phagocytosis 18511517_cathepsin K is involved in the cleavage of type II collagen in human articular cartilage in certain OA patients and that it may play a role in both OA pathophysiology and the aging process. 18664495_Cathepsin X causes cytoskeletal rearrangements and stimulates migration of T lymphocytes. 18664521_Comparison of the S2 site between rat and human cathepsin K sequences indicated that two S2 residues at Ser134 and Val160 in rat are varied to Ala and Leu, respectively, in the human enzyme. 18692071_crystal structure of a 1:n complex of cathepsin K:chondroitin 4-sulfate 18765527_CTSK may function as a paracrine factor in breast tumorigenesis. 18949742_cathepsin B, cathepsin H, cathepsin X and cystatin C may have roles in inflammatory breast cancer 18979635_incomplete inactivation may partially explain why active cysteine cathepsins are still found during acute lung inflammation 19060845_Rerpot cathepsin-k expression in pulmonary lymphangioleiomyomatosis. 19194656_Cathepsin X-upregulated T cells exhibit increased homotypic aggregation and polarized, migration-associated morphology in 2D and 3D models. Extended uropods are frequently formed, which subsequently elongate to nanotubes connecting T lymphocytes 19338743_In this review, some of the known features of cathepsin K such as structure, function in bone resorption, gene regulation and its roles in physiological or pathophysiological processes are highlighted. 19371798_Observational study of gene-disease association. (HuGE Navigator) 19396149_Cathepsin-K immunolabeling in both TFE3 and TFEB translocation renal cell carcinomas distinguishes these neoplasms from the more common adult renal cell carcinomas 19428782_cathepsin K might significantly contribute to altered opioid levels in brains of schizophrenics 19433310_Isozymes alpha- and gamma-enolases were identified as targets for cathepsin X. 19446361_Cathepsin X prevents an effective immune response against Helicobacter pylori infection. Cathepsin X mediated activation of Mac-1 suppresses the stimulatory signal in the form of cytokines. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19469903_Skin fibroblasts were found to strongly express catK in lysosomes. 19674475_Our study strengthens the role of a missense mutation in the CTSK gene in the pathogenesis of pycnodysostosis and suggests its prevalence in Pakistani patients 19700761_role of bone marrow cathepsin K in regulation of biological activity of SPARC in bone metastasis 19750481_Gradual cleavage of LFA-1 by cathepsin X enables the transition between intermediate and high affinity LFA-1 19800993_cleaves kinins and converts angiotensin I to angiotensin II thuys generating hormone peptide receptor agonists 19886543_Cathepsin-k appears to be a reliable tool for the precise and rapid identification of small epithelioid granulomas in Crohn's disease. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20305575_A novel missense mutation was identified in CTSK gene in a Pakistani family with 5 individuals affected with autosomal recessive pycnodysostosis. 20430722_Results suggest a role for syndecan-1 and cathepsins D and K in growth and invasiveness of esophageal squamous cell carcinoma. 20450492_identify and characterize sulfated glycosaminoglycans as natural allosteric modifiers of cathepsin K that exploit the conformational flexibility of the enzyme to regulate its activity and stability against autoproteolysis 20494937_The exo-peptidase cathepsin X has been identified as a new member of the group of CXCL-12-degrading enzymes secreted by non-hematopoietic bone marrow cells. Cathepsin X can influence hematopoietic stem and progenitor cell trafficking in the bone marrow. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20727023_a constant and strong expression of cathepsin K in dermatofibroma, but its complete absence in dermatofibrosarcoma protuberans 20837372_serum levels and catios of catK and catL to cysC were not significantly different between patients with coronary artery ectasia and controls 21086906_Cath-K represents a sensitive and specific marker to detect and quantitate granulomatous reactions in interstitial lung diseases, and is particularly useful in chronic hypersensitivity pneumonitis cases. 21099701_The novel homozygous mutation c.908G>A within exon 8 of the CTSK gene, was identified. 21602817_Demonstrate here the differential expression of cathepsin K among neoplasms harboring TFE3 gene fusions. 21715684_Cathepsin K- and L-cleaved chemerin trigger robust migration of human blood-derived plasmacytoid dendritic cells ex vivo 21756348_Cathepsins K, L, and S activity are profiled in 10 mug human breast, lung, and cervical tumors. 21874011_Cathepsin K was found to be constantly and strongly expressed in renal PEComas. 21880134_Cathepsin K gene expression was found to be significantly greater in more degenerated compared to healthier discs 22562303_inflammatory cues and monocyte-endothelial cell interactions upregulate cathepsin K and V activity via a JNK signaling axis 22569264_Data suggest that clusterin increases stability of cathepsin K in dilute/concentrated solutions; clusterin prevents substrate inhibition of cathepsin K; kinetic measurements show that clusterin binds cathepsin K with high affinity. 22569910_the collagenolytic activity of cathepsin K may be increased toward more matured bone collagen. 22614014_show for the first time that bone marrow macrophage-supplied CTSK may be involved in CCL2- and COX-2-driven pathways that contribute to tumor progression in bone 22730330_Cathepsin S cannibalism of cathepsin K as a mechanism to reduce type I collagen degradation. 22974233_Coronin 3 promotes gastric cancer metastasis via the up-regulation of MMP-9 and cathepsin K. 23152410_Cathepsin X deficiency leads to a reduced phosphorylation of the IGF-I receptor in response to IGF-I stimulation. 23326535_Our results demonstrate that increased adhesion, migration and invasiveness of tumor cells depend on the inactivation of the tumor suppressive function of profilin 1 by cathepsin X. 23355199_Cathepsin K immunohistochemistry can be helpful in distinguishing alveolar soft part sarcomas and translocation renal cell carcinomas from some but not all of the lesions in their differential diagnosis. 23369704_These data indicated that high levels of CatK are closely linked with the presence of coronary artery disease. 23483898_Quantification of immunohistochemistry showed that there is no difference in the global expression of CTSD, CTSH and CTSK between asthmatics and non-asthmatics. 23529168_Cardiac mammalian target of rapamycin and extracellular signal-regulated kinases (ERK) signaling cascades were upregulated by pressure overload, the effects of which were attenuated by cathepsin K knockout. 23689398_Identification of a novel insertional frameshift mutation in exon 4 of the CTSK gene in two families with pycnodysostosis. 23871919_Expression of cathepsins K, S and V within keratinocytes is reduced in photoprotected skin of aged women. 23951042_Data indicate that cathepsin K is expressed in oral tongue squamous cell carcinoma (OTSCC) tissue in both carcinoma and tumor microenvironment (TME) cells. 24088021_The basic amino acid clusters in cathepsin K play roles in the formation of collagenolytically active protease complexes. 24134756_Skipping of the 121-bp exon 2 results in elimination of the normal start codon and an expected absence of the wild-type CTSK protein. 24269275_The use of exome sequencing in the molecular diagnosis of 2 siblings initially thought to be affected by 'intermediate osteopetrosis', which identified a homozygous mutation in the CTSK gene, is described. 24342995_Increased plasma CatK levels are linked with the presence of atrial fibrillation. 24583396_catalase expression (or activity) was higher, while intracellular and extracellular Cat S, Cat L, and Cat K activities were lower in the non-invasive CL1-0 cells compared to the highly invasive CL1-5 cells. 24696729_Cathepsin K appears to be consistently and strongly expressed in melanocytic lesions and valuable in distinguishing malignant melanomas from the majority of human cancers. 24719048_synergism between HIV proteins and pro-atherogenic shear stress to increase endothelial cell expression of the powerful protease cathepsin K 24767306_identified five CTSK missense mutations (M1I, I249T, L7P, D80Y and D169N), one nonsense mutation (R312X) and one 301 bp insertion in intron 7, which is revealed as Alu sequence; among them, only L7P and I249 were described previously 24835450_Studies indicate cathepsin X as a target for improving diagnosis and treating cancer patients. 24954318_The data indicated that elevated levels of cathepsin K are closely associated with the presence of CAD and that circulating cathepsin K serves a useful biomarker for CAD. 24958728_results demonstrate that CS plays an important role in contributing to the enhanced efficiency of CatK collagenase activity in vivo 25184245_Two compounds have sizable effects on enzyme activity using interstitial collagen as a natural substrate of cathepsin K and four compounds show a significantly stabilizing effect on cathepsin K. 25279554_The identification of CatK exosites opens up the prospect of designing highly potent inhibitors that selectively inhibit the degradation of therapeutically relevant substrates by this multifunctional protease. 25304337_This mutation (c.480_481insT), (p.L160fsX173) is a novel frameshift mutation. The index case extends the phenotypic spectrum and the list of previously reported mutations in the CTSK gene. 25356585_The presence of high levels of inactive proforms of cathepsin K in GBM tissues and cells indicate that in GBM the proteolytic/collagenolytic role is not its primary function but it plays rather a different yet unknown role. 25422423_one cathepsin K molecule binds to collagen-bound glycosaminoglycans at the gap region and recruits a second protease molecule that provides an unfolding and cleavage mechanism for triple helical collagen 25558848_Cathepsins in Rotator Cuff Tendinopathy: Identification in Human Chronic Tears and Temporal Induction in a Rat Model. 25626674_The study determined almost identical substrate specificities for cysteine cathepsins K, L and S. 25662720_There is an increase in levels of CATK in type 1 Gaucher patients compared to the control group. In the patient group, CATK showed higher levels in patients with bone damage compared to those without it. 25725806_Sequence analysis of the patient's CTSK gene revealed homozygosity for a missense mutation (c.746T>C) in exon 6, which leads to amino change (p.Ile249Thr) in the mature CTSK protein 25809793_SDF-1alpha is involved in homing of CXCR4+ GSLCs and leukocytes and that cathepsin K and osteopontin are involved in the migration of GSLCs out of the niches 26219353_Matrix-metalloproteinase-9 is cleaved and activated by cathepsin K. 26241216_increased expression and activation in osteoarthritis cartilage 26302400_elevated levels of CatK are closely associated with the presence of chronic heart failure and that the measurement of circulating CatK provides a noninvasive method of documenting and monitoring the extent of chronic heart failure 26458004_The above studies not only demonstrated that CTSK was widely expressed in tooth-related cells (including odonto- clasts, periodontal ligament cells, pulp cells and odonto- blasts), but also indicated that CTSK was closely related to the development of tissues in oral and maxillofacial region as well as occurrence and development of many oral diseases. 26988144_Cathepsin K and osteocalcin plasma levels may be suggested as the significant markers of osteopoenia/osteoporosis. In addition, cathepsin K plasma level can be also a valuable marker of severe Coronary Atherosclerosis and Coronary Artery Calcification . 27558267_Six mutations in CTSK were identified in 33 families with pycnodysostosis. The high frequency of pycnodysostosis in Ceara State is the consequence of the high inbreeding in that region. 27709599_positive expression of cathepsin K in melanoma of the skin is associated with other unfavorable prognostic factors 27859061_The allosteric site of cathepsin K has been modified by site-directed mutagenesis, and it was shown that it is involved in specific regulation of the collagenolytic activity of cathepsin K. 28040478_Cathepsin K cleavage of SDF-1alpha inhibits its chemotactic activity towards glioblastoma stem-like cells. 28085175_Patients affected by castration-resistant prostate carcinoma may be tested for cathepsin K, and a positive strong expression (2+) could be a useful predictive biomarker of response to targeted agents, aiding in the selection of patients eligible for these treatments. 28087412_Cathepsin K expression within the subchondral bone of the medial tibia plateau was not associated with osteoclast density or symptomatic knee osteoarthritis. 28117540_The enhanced invasion of carcinomas resulting from cathepsin K overexpression is probably due to the increased cell migration and adhesion. Thus, cathepsin K is implicated not only in protein degradation but also in invasion, migration and adhesion of oral squamous cell carcinomas. 28216213_Increased cathepsin K expression in skull base chordoma was associated with tumor invasion and reduced progression free survival. 28623674_Immunohistochemistry confirmed cathepsin K protein was expressed in lymphangioleiomyomatosis but not control lungs. Cathepsin K gene expression and protein and protease activity were detected in lymphangioleiomyomatosis -associated fibroblasts but not the LAM cell line 621-101. 28766739_measurement of a periostin fragment resulting from in vivo cathepsin K digestion may help to identify subjects at high risk of fracture. 28887075_cathepsin K in the development of chronic subdural hematoma 29029096_results highlight the possibility that the interaction between GAGs and collagen under acidic conditions has a regulatory impact on cathepsin K-mediated bone degradation. 29088253_Of 160 top compounds tested in enzymatic assays, 28 compounds showed blocking of the collagenase activity of Cathepsin K (CatK) at 100 muM. 29303207_our data showed that elevated Cathepsin K in epithelial ovarian cancer can potentiate the metastasis of epithelial ovarian cancer cells 29618339_Upregulation of CTSK seems to be associated with high incidence of lymphatic spread and poor survival in OSCC. CTSK could therefore serve as a predictive biomarker for OSCC. 29796728_Genetic study of eight Egyptian patients with pycnodysostosis: identification of novel CTSK mutations and founder effect. 29859187_Sclerostin is degraded by cathepsin K in vitro. Cathepsin K degradation of sclerostin is affected by hypoxia. 30046941_expression of cathepsin B and X was detected in stromal cells and cancer cells throughout the glioblastoma (GBM) sections, whereas cathepsin K expression was more restricted to arteriole-rich regions in the GBM sections. Metabolic mapping showed that cathepsin B, but not cathepsin K is active in GSC niches. 30072716_data provides an insight into the possible mechanism of oxLDL on osteoclastogenesis suggesting that it does not perturb the packaging of CatK and v-ATPase (V-a3) in the secretory lysosome, but inhibits the fusion of these lysosomes to the ruffled border. The relevance of our findings suggests a distinct link between oxLDL, autophagy and osteoclastogenesis. 30450725_These actions of miR-185-5p agonists were mirrored by in vivo knockdown of CatK in mice with MI. 30931961_Sequential, but not Concurrent, Incubation of Cathepsin K and L with Type I Collagen Results in Extended Proteolysis. 32007579_Expression of elastolytic cathepsins in human skin and their involvement in age-dependent elastin degradation. 32117071_Associations of Serum Cathepsin K and Polymorphisms in CTSK Gene With Bone Mineral Density and Bone Metabolism Markers in Postmenopausal Chinese Women. 32368981_Obesity and Cathepsin K: A Complex Pathophysiological Relationship in Breast Cancer Metastases. 32819572_Cathepsin K is a potent disaggregase of alpha-synuclein fibrils. 33227497_Human cathepsin X/Z is a biologically active homodimer. 33429075_Clinical and genetic evaluation of Danish patients with pycnodysostosis. 33445732_Elevated Expression of Cathepsin K in Periodontal Ligament Fibroblast by Inflammatory Cytokines Accelerates Osteoclastogenesis via Paracrine Mechanism in Periodontal Disease. 33764165_Cathepsin K (Clone EPR19992) Demonstrates Uniformly Positive Immunoreactivity in Renal Oncocytoma, Chromophobe Renal Cell Carcinoma, and Distal Tubules. 33945887_Phenotypic and genotypic spectrum of CTSK variants in a cohort of twenty-five Indian patients with pycnodysostosis. 34103584_Regulatory properties of vitronectin and its glycosylation in collagen fibril formation and collagen-degrading enzyme cathepsin K activity. 34183355_The CD200-CD200R Axis Promotes Squamous Cell Carcinoma Metastasis via Regulation of Cathepsin K. 34433182_Cathepsin K is Superior to HMB45 for the Diagnosis of Pulmonary Lymphangioleiomyomatosis. 34680947_Genetic and Molecular Evaluation: Reporting Three Novel Mutations and Creating Awareness of Pycnodysostosis Disease. 34755658_[Activation of mir-30a-wnt/beta-catenin signaling pathway upregulates cathepsin K expression to promote cementogenic differentiation of periodontal ligament stem cells]. 34831087_Cathepsin K Regulates Intraocular Pressure by Modulating Extracellular Matrix Remodeling and Actin-Bundling in the Trabecular Meshwork Outflow Pathway. 35315254_Clinical and genetic characterization of three Russian patients with pycnodysostosis due to pathogenic variants in the CTSK gene. | ENSMUSG00000028111 | Ctsk | 1.407105e+02 | 0.5052580 | -0.984907752 | 0.3419376 | 7.854097e+00 | 0.0050705679 | 0.25256335 | No | Yes | 8.463007e+01 | 17.371706 | 1.852394e+02 | 37.717838 | |
| ENSG00000143398 | 8394 | PIP5K1A | protein_coding | Q99755 | FUNCTION: Catalyzes the phosphorylation of phosphatidylinositol 4-phosphate (PtdIns(4)P/PI4P) to form phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2/PIP2), a lipid second messenger that regulates several cellular processes such as signal transduction, vesicle trafficking, actin cytoskeleton dynamics, cell adhesion, and cell motility (PubMed:8955136, PubMed:21477596, PubMed:22942276). PtdIns(4,5)P2 can directly act as a second messenger or can be utilized as a precursor to generate other second messengers: inositol 1,4,5-trisphosphate (IP3), diacylglycerol (DAG) or phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3/PIP3) (PubMed:19158393, PubMed:20660631). PIP5K1A-mediated phosphorylation of PtdIns(4)P is the predominant pathway for PtdIns(4,5)P2 synthesis (By similarity). Can also use phosphatidylinositol (PtdIns) as substrate in vitro (PubMed:22942276). Together with PIP5K1C, is required for phagocytosis, both enzymes regulating different types of actin remodeling at sequential steps (By similarity). Promotes particle ingestion by activating the WAS GTPase-binding protein that induces Arp2/3 dependent actin polymerization at the nascent phagocytic cup (By similarity). Together with PIP5K1B, is required, after stimulation by G-protein coupled receptors, for the synthesis of IP3 that will induce stable platelet adhesion (By similarity). Recruited to the plasma membrane by the E-cadherin/beta-catenin complex where it provides the substrate PtdIns(4,5)P2 for the production of PtdIns(3,4,5)P3, IP3 and DAG, that will mobilize internal calcium and drive keratinocyte differentiation (PubMed:19158393). Positively regulates insulin-induced translocation of SLC2A4 to the cell membrane in adipocytes (By similarity). Together with PIP5K1C has a role during embryogenesis (By similarity). Independently of its catalytic activity, is required for membrane ruffling formation, actin organization and focal adhesion formation during directional cell migration by controlling integrin-induced translocation of the small GTPase RAC1 to the plasma membrane (PubMed:20660631). Also functions in the nucleus where it acts as an activator of TUT1 adenylyltransferase activity in nuclear speckles, thereby regulating mRNA polyadenylation of a select set of mRNAs (PubMed:18288197). {ECO:0000250|UniProtKB:P70182, ECO:0000269|PubMed:18288197, ECO:0000269|PubMed:19158393, ECO:0000269|PubMed:20660631, ECO:0000269|PubMed:21477596, ECO:0000269|PubMed:22942276, ECO:0000269|PubMed:8955136}. | ATP-binding;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Isopeptide bond;Kinase;Lipid metabolism;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation | hsa:8394; | cytosol [GO:0005829]; focal adhesion [GO:0005925]; lamellipodium [GO:0030027]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; 1-phosphatidylinositol-3-phosphate 5-kinase activity [GO:0000285]; 1-phosphatidylinositol-4-phosphate 5-kinase activity [GO:0016308]; 1-phosphatidylinositol-5-kinase activity [GO:0052810]; ATP binding [GO:0005524]; kinase binding [GO:0019900]; phosphatidylinositol-3,4-bisphosphate 5-kinase activity [GO:0052812]; actin cytoskeleton reorganization [GO:0031532]; activation of GTPase activity [GO:0090630]; cell chemotaxis [GO:0060326]; cell migration [GO:0016477]; fibroblast migration [GO:0010761]; focal adhesion assembly [GO:0048041]; glycerophospholipid metabolic process [GO:0006650]; keratinocyte differentiation [GO:0030216]; phagocytosis [GO:0006909]; phosphatidylinositol biosynthetic process [GO:0006661]; phospholipid biosynthetic process [GO:0008654]; protein localization to plasma membrane [GO:0072659]; regulation of phosphatidylinositol 3-kinase signaling [GO:0014066]; ruffle assembly [GO:0097178]; signal transduction [GO:0007165] | 12682053_Membrane ruffling requires coordination between this enzyme and Rac signaling. 15870270_in addition to its effects upon Rac activity, Ajuba can also influence cell migration through regulation of PI(4,5)P2 synthesis through direct activation of PIPKIalpha enzyme activity 16979564_identified apoptotic stimuli that initiate a signaling pathway during cell death that leads to caspase-independent downregulation of PIP5Kalpha. 18073347_Type I phosphatidylinositol-4-phosphate-5-kinase (PI5KI) alpha and gamma isoforms were identified as the enzymes responsible for PIP2 synthesis in natural killer cells. 18534983_beta-arrestins direct the localization of PIP5K Ialpha and PIP(2) production to agonist-activated 7TMRs, thereby regulating receptor internalization 18981107_PI4P5-K Ialpha-mediated PIP(2) production is crucial for HIV-1 entry and the early steps of infection in permissive lymphocytes. 19158393_Extracellular calcium (Cao) stimulated PIP5K1alpha recruitment to the E-cadherin-catenin complex in the plasma membrane. 19331818_The data suggest that activation of FcgammaRIIA leads to membrane rafts coalescing into signaling platforms containing PIP5-kinase Ialpha and PI(4,5)P(2). 20426790_Localized production of Phosphatidylinositol 4,5-bisphosphate by PIP5K1A is required for invadopodia formation by breast cancer cells. 20660631_Results define the role of PIPKI-alpha in cell migration and describes a new mechanism for the spatial regulation of Rac1 activity that is critical for cell migration. 23589613_CD28 regulates phosphatidylinositol 4,5-biphosphate turnover by recruiting and activating phosphatidylinositol 4-phosphate 5-kinases alpha in human primary CD4(+) T lymphocytes. 23994136_PIP5K1A is modified by polySUMO-2 only during apoptosis. 26157143_PIP5K1A modulates ribosomal RNA gene silencing through its interaction with histone H3 lysine 9 trimethylation and heterochromatin protein HP1-alpha. 27340655_These results indicate that PP1 is recruited to the extracellular calcium-dependent E-cadherin-catenin-PIP5K1a complex in the plasma membrane to activate PIP5K1a, which is required for PLC-g1 activation leading to keratinocyte differentiation. 27870828_Blockade of IQGAP1 interaction with PIPKIalpha or PI(3)K inhibited PtdIns(3,4,5)P3 generation and signalling, and selectively diminished cancer cell survival. 29851245_The results suggest that PIP5KA is a novel degradative substrate of NEDD4 and that the PIP5KA-dependent phosphatidylinositol 4,5-diphosphate pool contributing to breast cancer cell proliferation through PI3K/Akt activation is negatively controlled by NEDD4. 30104711_PIP5K1alpha was associated with poor patient outcome in triple-negative BC. It increased expression of pSer-473 AKT and invasiveness of triple-negative MDA-MB-231 cells. Inhibition reduced expression of pSer-473 AKT and its downstream effectors in xenograft tumors. In ER(+) cancer cells, PIP5K1alpha acted on pSer-473 AKT, and was in complexes with VEGFR2, serving as co-factor of ER-alpha to regulate activities of targe... 30194290_KRAS-specific interactor that mediates oncogenic KRAS signaling and proliferation 32376619_Type I Phosphatidylinositol-4-Phosphate 5-Kinases alpha and gamma Play a Key Role in Targeting HIV-1 Pr55(Gag) to the Plasma Membrane. 32686865_Phosphatidylinositol-4-phosphate 5-kinase type 1alpha attenuates Abeta production by promoting non-amyloidogenic processing of amyloid precursor protein. 33079727_CLIC1 recruits PIP5K1A/C to induce cell-matrix adhesions for tumor metastasis. 33591981_Cross-species identification of PIP5K1-, splicing- and ubiquitin-related pathways as potential targets for RB1-deficient cells. 34537072_Circ_PIP5K1A regulates cisplatin resistance and malignant progression in non-small cell lung cancer cells and xenograft murine model via depending on miR-493-5p/ROCK1 axis. | ENSMUSG00000028126 | Pip5k1a | 7.141749e+03 | 0.8097122 | -0.304518927 | 0.2845664 | 1.166090e+00 | 0.2802060859 | 0.80760832 | No | Yes | 5.658890e+03 | 603.915823 | 7.344198e+03 | 803.310324 | ||
| ENSG00000143434 | 10500 | SEMA6C | protein_coding | Q9H3T2 | FUNCTION: Shows growth cone collapsing activity on dorsal root ganglion (DRG) neurons in vitro. May be a stop signal for the DRG neurons in their target areas, and possibly also for other neurons. May also be involved in the maintenance and remodeling of neuronal connections. {ECO:0000269|PubMed:12110693}. | Alternative splicing;Cell membrane;Developmental protein;Differentiation;Disulfide bond;Glycoprotein;Membrane;Neurogenesis;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of the semaphorin family. Semaphorins represent important molecular signals controlling multiple aspects of the cellular response that follows CNS injury, and thus may play an important role in neural regeneration. [provided by RefSeq, May 2010]. | hsa:10500; | cell surface [GO:0009986]; cytoplasm [GO:0005737]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; chemorepellent activity [GO:0045499]; semaphorin receptor binding [GO:0030215]; axon guidance [GO:0007411]; negative chemotaxis [GO:0050919]; negative regulation of axon extension [GO:0030517]; negative regulation of axon extension involved in axon guidance [GO:0048843]; neural crest cell migration [GO:0001755]; positive regulation of cell migration [GO:0030335]; semaphorin-plexin signaling pathway [GO:0071526] | 12110693_identification, characterization, and functional study of the two novel human members of the semaphorin gene family 19054571_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000038777 | Sema6c | 6.220539e+02 | 0.7520094 | -0.411177357 | 0.2915889 | 1.925406e+00 | 0.1652619152 | 0.77593452 | No | Yes | 3.942889e+02 | 84.748215 | 5.649350e+02 | 124.441959 | |
| ENSG00000143493 | 25896 | INTS7 | protein_coding | Q9NVH2 | FUNCTION: Component of the Integrator (INT) complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Plays a role in DNA damage response (DDR) signaling during the S phase (PubMed:21659603). May be not involved in the recruitment of cytoplasmic dynein to the nuclear envelope by different components of the INT complex (PubMed:23904267). {ECO:0000269|PubMed:21659603, ECO:0000269|PubMed:23904267, ECO:0000305|PubMed:16239144}. | 3D-structure;Alternative splicing;Chromosome;Cytoplasm;DNA damage;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a subunit of the integrator complex. The integrator complex associates with the C-terminal domain of RNA polymerase II and mediates 3'-end processing of the small nuclear RNAs U1 and U2. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Dec 2010]. | hsa:25896; | chromosome [GO:0005694]; cytoplasm [GO:0005737]; integrator complex [GO:0032039]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cellular response to ionizing radiation [GO:0071479]; DNA damage checkpoint signaling [GO:0000077]; snRNA 3'-end processing [GO:0034472]; snRNA processing [GO:0016180] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000037461 | Ints7 | 1.019950e+03 | 0.5657055 | -0.821876809 | 0.3032479 | 7.017239e+00 | 0.0080728617 | 0.33086870 | No | Yes | 9.356001e+02 | 189.471992 | 1.320569e+03 | 273.973487 | |
| ENSG00000143494 | 79805 | VASH2 | protein_coding | Q86V25 | FUNCTION: Tyrosine carboxypeptidase that removes the C-terminal tyrosine residue of alpha-tubulin, thereby regulating microtubule dynamics and function (PubMed:29146869). Critical for spindle function and accurate chromosome segregation during mitosis since microtuble detyronisation regulates mitotic spindle length and postioning (PubMed:31171830). Acts as an activator of angiogenesis: expressed in infiltrating mononuclear cells in the sprouting front to promote angiogenesis (PubMed:19204325). Plays a role in axon formation (PubMed:31235911). {ECO:0000269|PubMed:19204325, ECO:0000269|PubMed:29146869, ECO:0000269|PubMed:31235911}. | 3D-structure;Alternative splicing;Carboxypeptidase;Cytoplasm;Cytoskeleton;Hydrolase;Phosphoprotein;Protease;Reference proteome;Secreted | hsa:79805; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; extracellular region [GO:0005576]; actin binding [GO:0003779]; metallocarboxypeptidase activity [GO:0004181]; microtubule binding [GO:0008017]; axon development [GO:0061564]; cell-cell fusion [GO:0140253]; labyrinthine layer blood vessel development [GO:0060716]; positive regulation of angiogenesis [GO:0045766]; positive regulation of endothelial cell proliferation [GO:0001938]; proteolysis [GO:0006508]; regulation of angiogenesis [GO:0045765]; syncytium formation by plasma membrane fusion [GO:0000768] | 22438034_vasohibin-1 and vasohibin-2 mRNA are expressed in gastric cancer cells and in tumor-associated macrophages (TAMs), and their expressions are altered by hypoxia. 22614011_VASH2 contributes to the angiogenesis in hepatocellular carcinoma via an Small vasohibin binding protein-mediated paracrine mechanism. 22826464_VASH2 expressed in serous ovarian carcinoma cells promoted tumor growth and peritoneal dissemination by promoting angiogenesis. 23100270_Data suggest that VASH1 is expressed in vascular endothelium to terminate angiogenesis; VASH2 appears to be expressed in other cells (primarily mononuclear leukocytes) to promote angiogenesis. [REVIEW] 23615928_This is the first study to report differences in the intracellular localization of the VASH2 protein and, hence, a new research direction on the study of VASH2 24595063_VASH2 overexpression downregulated wild-type p53. 25184477_VASH1 and VASH2 showed distinctive localization and opposing function on the fetoplacental vascularization. 25269476_MiR200-upregulated Vasohibin 2 promotes the malignant transformation of tumors by inducing epithelial-mesenchymal transition in hepatocellular carcinoma 25916042_Suggest that overexpression of VASH2 in pancreatic ductal adenocarcinoma accelerated the pace of tumor development toward a more serious malignant phenotype and was associated with a poor clinical outcome. 26177649_to the best of our knowledge, these results are the first clinical data indicating that nuclear VASH2, but not cytoplasmic VASH2, promotes cell proliferation by driving the cell cycle from the G0/G1 to S phase. 27702660_VASH2 expression is positively correlated with FGF2 expression and promotes angiogenesis in human luminal breast cancer by transcriptional activation of fibroblast growth factor 2 through non-paracrine mechanisms. 27879017_identified a novel UPS regulatory system in which essential domain architecture (VASH-PS) of VASHs, comprising regions VASH191-180 and VASH280-169 , regulate the cytosolic punctate structure formation in the absence of SVBP. 28064471_The results indicate that VASH2 played a significant role in the epithelial-mesenchymal transition by modulating the TGF-beta signaling. 28327155_These data suggest that VASH2 reduces the chemosensitivity to gemcitabine in pancreatic cancer cells via JUN-dependent transactivation of RRM2. 28882646_The present study suggests a protective effect of miR-200b/c on high glucose induced human retinal microvascular endothelial cell line dysfunction by inhibiting VASH2. 28960674_These results suggest that VASH2 plays an important role in gastric tumor progression via the accumulation of cancer-associated fibroblasts 29039601_VASH2 may promote drug resistance of breast cancer cells through regulating ABCG2 via the AKT signaling pathway. 29042694_These results suggest that VASH-2 could play an important role in the pathogenesis of renal diseases, and that VASH-2 is closely associated with hypertension and impaired glucose tolerance. 29146869_VASH1 and VASH2 but not SVBP alone, increased detyrosination of -tubulin, and purified vasohibins removed the C-terminal tyrosine of -tubulin. 30318866_Study demonstrates that VASH2 promotes malignant behaviors of pancreatic cancer cells by inducing EMT via activation of the Hedgehog signaling pathway. 30940294_VASH2 Promotes Cell Proliferation and Resistance to Doxorubicin in Non-Small Cell Lung Cancer via AKT Signaling. 31074083_VASH2 promotes tumor angiogenesis as a result of paracrine activity and evasion of tumor immunity as a result of altered gene expression in pancreatic ductal adenocarcinoma cells. 31235911_This study examined the crystal structures of human vasohibin 1 and 2 in complex with small vasohibin-binding protein (SVBP) in the absence and presence of different inhibitors and a C-terminal alpha-tubulin peptide. 32494966_Prognostic significance of vasohibin-1 and vasohibin-2 immunohistochemical expression in gastric cancer. 35026646_Vasohibin-1 and -2 in pulmonary lymphangioleiomyomatosis (LAM) cells associated with angiogenic and prognostic factors. | ENSMUSG00000037568 | Vash2 | 1.202561e+02 | 1.3160298 | 0.396192153 | 0.3892017 | 1.013750e+00 | 0.3140061614 | 0.81928722 | No | Yes | 1.380982e+02 | 26.689754 | 1.022032e+02 | 20.288261 | ||
| ENSG00000143502 | 55061 | SUSD4 | protein_coding | Q5VX71 | FUNCTION: Acts as complement inhibitor by disrupting the formation of the classical C3 convertase. Isoform 3 inhibits the classical complement pathway, while membrane-bound isoform 1 inhibits deposition of C3b via both the classical and alternative complement pathways. {ECO:0000269|PubMed:23482636}. | Alternative splicing;Complement pathway;Disulfide bond;Glycoprotein;Immunity;Innate immunity;Membrane;Reference proteome;Repeat;Secreted;Signal;Sushi;Transmembrane;Transmembrane helix | hsa:55061; | extracellular region [GO:0005576]; integral component of membrane [GO:0016021]; complement activation, classical pathway [GO:0006958]; innate immune response [GO:0045087]; negative regulation of complement activation, alternative pathway [GO:0045957]; negative regulation of complement activation, classical pathway [GO:0045959]; regulation of complement activation [GO:0030449] | 20237496_Observational study of gene-disease association. (HuGE Navigator) 20348246_analysis of cloning, expression, refolding, and tissue distribution of Sushi domain-containing protein 4 23482636_Membrane-bound isoform SUSD4a inhibits the classical and alternative complement pathways when expressed on the surface of Chinese hamster cells but not when expressed as a soluble, truncated protein. 26480818_SUSD4 expression in both breast cancer cells and T cells infiltrating the tumor-associated stroma is useful to predict better prognosis of breast cancer patients. | ENSMUSG00000038576 | Susd4 | 8.212111e+01 | 1.4346884 | 0.520737417 | 0.4353238 | 1.393338e+00 | 0.2378421886 | 0.78818582 | No | Yes | 8.662804e+01 | 22.389412 | 6.918954e+01 | 18.302493 | ||
| ENSG00000143515 | 57198 | ATP8B2 | protein_coding | P98198 | FUNCTION: Catalytic component of P4-ATPase flippase complex, which catalyzes the hydrolysis of ATP coupled to the transport of phosphatidylcholine (PC) from the outer to the inner leaflet of the plasma membrane. May contribute to the maintenance of membrane lipid asymmetry. {ECO:0000269|PubMed:25315773}. | ATP-binding;Alternative splicing;Cell membrane;Endoplasmic reticulum;Lipid transport;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Translocase;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene belongs to the family of P-type cation transport ATPases, and to the subfamily of aminophospholipid-transporting ATPases. The aminophospholipid translocases transport phosphatidylserine and phosphatidylethanolamine from one side of a bilayer to another. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:57198; | endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; integral component of plasma membrane [GO:0005887]; phospholipid-translocating ATPase complex [GO:1990531]; plasma membrane [GO:0005886]; trans-Golgi network [GO:0005802]; ATP binding [GO:0005524]; ATPase-coupled intramembrane lipid transporter activity [GO:0140326]; magnesium ion binding [GO:0000287]; phosphatidylcholine flippase activity [GO:0140345]; phosphatidylcholine floppase activity [GO:0090554]; Golgi organization [GO:0007030]; ion transmembrane transport [GO:0034220]; phospholipid translocation [GO:0045332] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 26240149_the predominant P4 ATPases in pure pancreatic beta cells and human and rat pancreatic islets were ATP8B1, ATP8B2, and ATP9A. ATP8B1 and CDC50A were highly concentrated in ISG | ENSMUSG00000060671 | Atp8b2 | 5.774855e+03 | 0.9540407 | -0.067877217 | 0.2736191 | 6.171261e-02 | 0.8038092837 | 0.96141221 | No | Yes | 4.655221e+03 | 338.002429 | 5.203809e+03 | 387.315563 | |
| ENSG00000143756 | 23219 | FBXO28 | protein_coding | Q9NVF7 | FUNCTION: Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation. {ECO:0000250}. | Alternative splicing;Centromere;Chromosome;Kinetochore;Phosphoprotein;Reference proteome;Ubl conjugation pathway | Members of the F-box protein family, such as FBXO28, are characterized by an approximately 40-amino acid F-box motif. SCF complexes, formed by SKP1 (MIM 601434), cullin (see CUL1; MIM 603134), and F-box proteins, act as protein-ubiquitin ligases. F-box proteins interact with SKP1 through the F box, and they interact with ubiquitination targets through other protein interaction domains (Jin et al., 2004 [PubMed 15520277]).[supplied by OMIM, Mar 2008]. | hsa:23219; | kinetochore [GO:0000776]; identical protein binding [GO:0042802]; protein polyubiquitination [GO:0000209] | 23776131_Study identified F-box protein, FBXO28 that controls MYC-dependent transcription by non-proteolytic ubiquitylation. Depletion of FBXO28 or overexpression of an F-box mutant unable to support MYC ubiquitylation results in an impairment of MYC-driven transcription, transformation and tumourigenesis. 24357076_Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. 27754753_Fbxo28 regulates topoisomerase IIalpha decatenation activity and plays an important role in maintaining genomic stability. 28179588_Expression levels of TP53BP2, FBXO28, and FAM53A genes were associated with patient survival specifically in ER-positive, TP53-mutated tumors. 29587369_It was shown on human primary islets that FBXO28 improves pancreatic beta-cell survival under diabetogenic conditions without affecting insulin secretion. 30160831_FBXO28 is a monogenic disease gene and contributes to the complex neurodevelopmental phenotype of the 1q41-q42 gene deletion syndrome. 31678254_SCF(FBXO28)-mediated self-ubiquitination of FBXO28 promotes its degradation. 33280099_FBXO28 causes developmental and epileptic encephalopathy with profound intellectual disability. | ENSMUSG00000047539 | Fbxo28 | 4.836260e+02 | 0.9099077 | -0.136207963 | 0.3452918 | 1.549625e-01 | 0.6938374375 | 0.93453802 | No | Yes | 4.842369e+02 | 92.062174 | 4.707559e+02 | 91.640531 | |
| ENSG00000143786 | 149111 | CNIH3 | protein_coding | Q8TBE1 | FUNCTION: Regulates the trafficking and gating properties of AMPA-selective glutamate receptors (AMPARs). Promotes their targeting to the cell membrane and synapses and modulates their gating properties by regulating their rates of activation, deactivation and desensitization. {ECO:0000269|PubMed:20805473}. | Cell junction;Cell membrane;Membrane;Postsynaptic cell membrane;Reference proteome;Synapse;Transmembrane;Transmembrane helix | hsa:149111; | AMPA glutamate receptor complex [GO:0032281]; dendrite [GO:0030425]; dendritic shaft [GO:0043198]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; ER to Golgi transport vesicle membrane [GO:0012507]; postsynaptic membrane [GO:0045211]; synapse [GO:0045202]; regulation of AMPA receptor activity [GO:2000311]; vesicle-mediated transport [GO:0016192] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23103966_Significant upregulation of CNIH-3 mRNA expression was found in schizophrenia. 26239289_Study's convergent findings in humans and mice support CNIH3 involvement in the pathophysiology of opioid dependence, complementing prior studies implicating the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate system | ENSMUSG00000026514 | Cnih3 | 2.046978e+02 | 1.2512227 | 0.323338617 | 0.3510888 | 8.618370e-01 | 0.3532251391 | 0.82969965 | No | Yes | 1.984800e+02 | 24.807414 | 1.464832e+02 | 19.092908 | ||
| ENSG00000143847 | 8497 | PPFIA4 | protein_coding | O75335 | FUNCTION: May regulate the disassembly of focal adhesions. May localize receptor-like tyrosine phosphatases type 2A at specific sites on the plasma membrane, possibly regulating their interaction with the extracellular environment and their association with substrates (By similarity). {ECO:0000250}. | Alternative splicing;Coiled coil;Cytoplasm;Phosphoprotein;Reference proteome;Repeat | PPFIA4, or liprin-alpha-4, belongs to the liprin-alpha gene family. See liprin-alpha-1 (LIP1, or PPFIA1; MIM 611054) for background on liprins.[supplied by OMIM, Mar 2008]. | hsa:8497; | cell surface [GO:0009986]; cytosol [GO:0005829]; presynaptic active zone [GO:0048786]; synapse [GO:0045202]; synapse organization [GO:0050808] | 19536175_Observational study of gene-disease association. (HuGE Navigator) 20599943_liprin-alpha4 is a hypoxia-induced gene potentially involved in cell-cell adhesion 21829649_presence of Liprin-alpha4 and nickel increased tyrosine phosphatase activity that reduced the global levels of tyrosine phosphorylation in the cell 28460022_The results identify PP1IFA4 loci associated with early onset atrial fibrillation in a Korean population. 29187440_Data show that liprin-alpha4 plays a pivotal role in inducing malignant phenotypes such as increased proliferation and invasion in pancreatic cancer, and that liprin-alpha4 could be a new effective therapeutic target for pancreatic cancer. 30842147_Hypoxia appears to up-regulate the expression of liprin-alpha4, which induces the expression of HIF1alpha. HIF1alpha contributes to increased proliferation and lower chemosensitivity. Therefore, inhibition of liprin-alpha4 or HIF1alpha led to reduced proliferation and increased chemosensitivity of SBC-5 cells. | ENSMUSG00000026458 | Ppfia4 | 2.367398e+02 | 0.7412058 | -0.432053923 | 0.3166365 | 1.878271e+00 | 0.1705308309 | 0.77611497 | No | Yes | 1.060826e+02 | 25.933593 | 1.659683e+02 | 41.536664 | |
| ENSG00000143947 | 6233 | RPS27A | protein_coding | P62979 | FUNCTION: [Ubiquitin]: Exists either covalently attached to another protein, or free (unanchored). When covalently bound, it is conjugated to target proteins via an isopeptide bond either as a monomer (monoubiquitin), a polymer linked via different Lys residues of the ubiquitin (polyubiquitin chains) or a linear polymer linked via the initiator Met of the ubiquitin (linear polyubiquitin chains). Polyubiquitin chains, when attached to a target protein, have different functions depending on the Lys residue of the ubiquitin that is linked: Lys-6-linked may be involved in DNA repair; Lys-11-linked is involved in ERAD (endoplasmic reticulum-associated degradation) and in cell-cycle regulation; Lys-29-linked is involved in proteotoxic stress response and cell cycle; Lys-33-linked is involved in kinase modification; Lys-48-linked is involved in protein degradation via the proteasome; Lys-63-linked is involved in endocytosis, DNA-damage responses as well as in signaling processes leading to activation of the transcription factor NF-kappa-B. Linear polymer chains formed via attachment by the initiator Met lead to cell signaling. Ubiquitin is usually conjugated to Lys residues of target proteins, however, in rare cases, conjugation to Cys or Ser residues has been observed. When polyubiquitin is free (unanchored-polyubiquitin), it also has distinct roles, such as in activation of protein kinases, and in signaling. {ECO:0000269|PubMed:16543144, ECO:0000269|PubMed:34239127, ECO:0000303|PubMed:19754430}.; FUNCTION: [40S ribosomal protein S27a]: Component of the 40S subunit of the ribosome. | 3D-structure;ADP-ribosylation;Acetylation;Cytoplasm;Direct protein sequencing;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Ribonucleoprotein;Ribosomal protein;Ubl conjugation;Zinc;Zinc-finger | Ubiquitin, a highly conserved protein that has a major role in targeting cellular proteins for degradation by the 26S proteosome, is synthesized as a precursor protein consisting of either polyubiquitin chains or a single ubiquitin fused to an unrelated protein. This gene encodes a fusion protein consisting of ubiquitin at the N terminus and ribosomal protein S27a at the C terminus. When expressed in yeast, the protein is post-translationally processed, generating free ubiquitin monomer and ribosomal protein S27a. Ribosomal protein S27a is a component of the 40S subunit of the ribosome and belongs to the S27AE family of ribosomal proteins. It contains C4-type zinc finger domains and is located in the cytoplasm. Pseudogenes derived from this gene are present in the genome. As with ribosomal protein S27a, ribosomal protein L40 is also synthesized as a fusion protein with ubiquitin; similarly, ribosomal protein S30 is synthesized as a fusion protein with the ubiquitin-like protein fubi. Multiple alternatively spliced transcript variants that encode the same proteins have been identified.[provided by RefSeq, Sep 2008]. | hsa:6233; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; cytosolic ribosome [GO:0022626]; cytosolic small ribosomal subunit [GO:0022627]; endocytic vesicle membrane [GO:0030666]; endoplasmic reticulum membrane [GO:0005789]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; membrane [GO:0016020]; mitochondrial outer membrane [GO:0005741]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; small ribosomal subunit [GO:0015935]; vesicle [GO:0031982]; metal ion binding [GO:0046872]; protein tag [GO:0031386]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; ubiquitin protein ligase binding [GO:0031625]; cytoplasmic translation [GO:0002181]; modification-dependent protein catabolic process [GO:0019941]; protein ubiquitination [GO:0016567]; translation [GO:0006412] | 15647830_ubiquitin carboxyl extension protein 1 is overexpressed in prostate cancer 21561866_S27a plays a non-redundant role in mediating p53 activation in response to ribosomal stress via interplaying with MDM2. 22716248_Gene expression profiles showed that RPS27A was down-regulated in epidermolysis bullosa subtypes. 24680683_Knockdown of RPS27a inhibits the proliferation, induces cell cycle arrest and potentiates the effect of imatinib on apoptosis of K562 cells. 24833360_RPS27A expression was found to have a weak inverse correlation with overexpression of multifunctional protein YB-1 in HCC tissues. 25592822_RPS27a appears to be a novel stress sensor in the cell which amplifies p53 response to arrest cell cycle. 26942564_Imatinib-resistant K562/G01 cells expressed significantly higher levels of STAT3 and RPS27a compared with those of K562 cells. 28498418_the full-length cDNA sequence of MMSA-8 was cloned in MM and it was hypothesized that MMSA-8 is MM-associated RPS27A transcript variant 1 28735865_RPS27a enhances viral LMP1-mediated proliferation and invasion, suggesting that RPS27a interacts with LMP1 and stabilizes it by suppressing proteasome-mediated ubiquitination. 28913776_this study shows that interactions of antisera to different Chlamydia and Chlamydophila species with the ribosomal protein RPS27a correlate with impaired protein synthesis in a human choroid plexus papilloma cell line 30085276_MTG1 establishes a quality control checkpoint in mitoribosome assembly. In conclusion, MTG1 controls mitochondrial translation by coupling mtLSU assembly with intersubunit bridge formation using the intrinsic GEF activity acquired by the mtSSU through mS27, a unique occurrence in translational systems. 32129764_USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit. 32751694_The Ubiquitin Gene Expression Pattern and Sensitivity to UBB and UBC Knockdown Differentiate Primary 23132/87 and Metastatic MKN45 Gastric Cancer Cells. 32941527_Rps27a might act as a controller of microglia activation in triggering neurodegenerative diseases. 34166715_PICT1 is critical for regulating the Rps27a-Mdm2-p53 pathway by microtubule polymerization inhibitor against cervical cancer. | ENSMUSG00000020460 | Rps27a | 2.404103e+04 | 0.5858412 | -0.771418392 | 0.2861796 | 7.225663e+00 | 0.0071868616 | 0.31685983 | No | Yes | 2.165445e+04 | 4365.404228 | 2.723294e+04 | 5630.508791 | |
| ENSG00000144034 | 51002 | TPRKB | protein_coding | Q9Y3C4 | FUNCTION: Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine (PubMed:22912744, PubMed:28805828). The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37 (PubMed:22912744, PubMed:28805828). TPRKB acts as an allosteric effector that regulates the t(6)A activity of the complex. TPRKB is not required for tRNA modification (PubMed:22912744, PubMed:28805828). {ECO:0000269|PubMed:28805828, ECO:0000305|PubMed:22912744}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Epilepsy;Mental retardation;Nucleus;Reference proteome;tRNA processing | hsa:51002; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; EKC/KEOPS complex [GO:0000408]; nucleus [GO:0005634]; protein kinase binding [GO:0019901]; telomere maintenance via recombination [GO:0000722]; tRNA threonylcarbamoyladenosine modification [GO:0002949] | 33547416_Crystal structure of the human PRPK-TPRKB complex. | ENSMUSG00000054226 | Tprkb | 2.856896e+02 | 0.9989833 | -0.001467548 | 0.3659787 | 1.600828e-05 | 0.9968076449 | 0.99926282 | No | Yes | 2.509952e+02 | 47.210433 | 2.680997e+02 | 51.600405 | ||
| ENSG00000144161 | 84524 | ZC3H8 | protein_coding | Q8N5P1 | FUNCTION: Acts as a transcriptional repressor of the GATA3 promoter. Sequence-specific DNA-binding factor that binds to the 5'-AGGTCTC-3' sequence within the negative cis-acting element intronic regulatory region (IRR) of the GATA3 gene (By similarity). Component of the little elongation complex (LEC), a complex required to regulate small nuclear RNA (snRNA) gene transcription by RNA polymerase II and III (PubMed:23932780). Induces thymocyte apoptosis when overexpressed, which may indicate a role in regulation of thymocyte homeostasis. {ECO:0000250, ECO:0000269|PubMed:12077251, ECO:0000269|PubMed:12153508, ECO:0000269|PubMed:23932780}. | Apoptosis;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:84524; | Cajal body [GO:0015030]; chromatin [GO:0000785]; histone locus body [GO:0035363]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription elongation factor complex [GO:0008023]; transcriptionally active chromatin [GO:0035327]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding [GO:0001162]; apoptotic process [GO:0006915]; negative regulation of T cell differentiation in thymus [GO:0033085]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of thymocyte apoptotic process [GO:0070245]; positive regulation of transcription by RNA polymerase III [GO:0045945]; response to antibiotic [GO:0046677]; snRNA transcription by RNA polymerase II [GO:0042795]; snRNA transcription by RNA polymerase III [GO:0042796]; T cell homeostasis [GO:0043029] | Mouse_homologues 12077251_Fliz1 binds specifically to a negative cis-acting element in the intronic regulatory region of GATA-3 and, when overexpressed, can function as a transcriptional repressor of the GATA-3 promoter in vitro and in vivo. 30041613_Using RNA silencing to decrease expression of ZC3H8 in a mouse tumor cell line in vitro, then studied phenotype and cell behavior of the tumor cell line, and further studied ability of those cells to form tumors in mice in vivo. | ENSMUSG00000027387 | Zc3h8 | 4.136564e+02 | 0.9438980 | -0.083297066 | 0.3753823 | 4.691396e-02 | 0.8285229734 | 0.96748040 | No | Yes | 3.358366e+02 | 69.550127 | 3.801392e+02 | 80.523843 | ||
| ENSG00000144231 | 5433 | POLR2D | protein_coding | O15514 | FUNCTION: DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB4 is part of a subcomplex with RPB7 that binds to a pocket formed by RPB1, RPB2 and RPB6 at the base of the clamp element. The RBP4-RPB7 subcomplex seems to lock the clamp via RPB7 in the closed conformation thus preventing double-stranded DNA to enter the active site cleft. The RPB4-RPB7 subcomplex binds single-stranded DNA and RNA (By similarity). {ECO:0000250, ECO:0000269|PubMed:9852112}. | 3D-structure;DNA-directed RNA polymerase;Nucleus;Reference proteome;Transcription | This gene encodes the fourth largest subunit of RNA polymerase II, the polymerase responsible for synthesizing messenger RNA in eukaryotes. In yeast, this polymerase subunit is associated with the polymerase under suboptimal growth conditions and may have a stress protective role. A sequence for a ribosomal pseudogene is contained within the 3' untranslated region of the transcript from this gene. [provided by RefSeq, Jul 2008]. | hsa:5433; | cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; P-body [GO:0000932]; RNA polymerase II, core complex [GO:0005665]; DNA-directed 5'-3' RNA polymerase activity [GO:0003899]; nucleotide binding [GO:0000166]; translation initiation factor binding [GO:0031369]; mRNA export from nucleus in response to heat stress [GO:0031990]; nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:0000288]; positive regulation of translational initiation [GO:0045948]; recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex [GO:0034402]; transcription by RNA polymerase II [GO:0006366]; transcription initiation from RNA polymerase II promoter [GO:0006367] | 16282592_Identification of the RNA binding region of Rpb4. 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000024258 | Polr2d | 3.408161e+03 | 0.8609527 | -0.215994085 | 0.2598042 | 6.942164e-01 | 0.4047339053 | 0.84651196 | No | Yes | 3.055276e+03 | 245.638932 | 3.407138e+03 | 280.122877 | |
| ENSG00000144306 | 79634 | SCRN3 | protein_coding | Q0VDG4 | Alternative splicing;Reference proteome | hsa:79634; | cysteine-type exopeptidase activity [GO:0070004]; dipeptidase activity [GO:0016805] | ENSMUSG00000008226 | Scrn3 | 2.357031e+02 | 1.3012000 | 0.379842718 | 0.3454620 | 1.212983e+00 | 0.2707420640 | 0.79972971 | No | Yes | 2.349016e+02 | 42.384132 | 1.885418e+02 | 34.897509 | ||||
| ENSG00000144468 | 84236 | RHBDD1 | protein_coding | Q8TEB9 | FUNCTION: Intramembrane-cleaving serine protease that cleaves single transmembrane or multi-pass membrane proteins in the hydrophobic plane of the membrane, luminal loops and juxtamembrane regions. Involved in regulated intramembrane proteolysis and the subsequent release of functional polypeptides from their membrane anchors. Functional component of endoplasmic reticulum-associated degradation (ERAD) for misfolded membrane proteins. Required for the degradation process of some specific misfolded endoplasmic reticulum (ER) luminal proteins. Participates in the transfer of misfolded proteins from the ER to the cytosol, where they are destroyed by the proteasome in a ubiquitin-dependent manner. Functions in BIK, MPZ, PKD1, PTCRA, RHO, STEAP3 and TRAC processing. Involved in the regulation of exosomal secretion; inhibits the TSAP6-mediated secretion pathway. Involved in the regulation of apoptosis; modulates BIK-mediated apoptotic activity. Also plays a role in the regulation of spermatogenesis; inhibits apoptotic activity in spermatogonia. {ECO:0000269|PubMed:18953687, ECO:0000269|PubMed:22624035}. | 3D-structure;Alternative splicing;Apoptosis;Differentiation;Endoplasmic reticulum;Hydrolase;Membrane;Mitochondrion;Protease;Reference proteome;Serine protease;Spermatogenesis;Transmembrane;Transmembrane helix | hsa:84236; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum quality control compartment [GO:0044322]; integral component of endoplasmic reticulum membrane [GO:0030176]; mitochondrial membrane [GO:0031966]; endopeptidase activity [GO:0004175]; serine-type endopeptidase activity [GO:0004252]; apoptotic process [GO:0006915]; cellular response to unfolded protein [GO:0034620]; cellular response to UV [GO:0034644]; ERAD pathway [GO:0036503]; membrane protein intracellular domain proteolysis [GO:0031293]; membrane protein proteolysis [GO:0033619]; membrane protein proteolysis involved in retrograde protein transport, ER to cytosol [GO:1904211]; negative regulation of apoptotic process [GO:0043066]; positive regulation of protein catabolic process [GO:0045732]; positive regulation of protein processing [GO:0010954]; positive regulation of secretion [GO:0051047]; post-translational protein modification [GO:0043687]; spermatid differentiation [GO:0048515] | 18953687_RHBDD1, a serine protease, modulates BIK-mediated apoptotic activity. 22624035_RHBDD1 is involved in the regulation of a nonclassical exosomal secretion pathway through the restriction of TSAP6. 23534782_Our data show that in vitro RHBDD1 silencing regulates HepG2 cell proliferation and apoptosis. 23669365_RHBDD1 could inhibit cell apoptosis by activating and upregulating c-Jun and its downstream target, Bcl-3 23883433_this study shows that RHBDD1 gene engineering could be used as an effective tool in malignant brain tumor therapy. 27563067_RHBDL4-mediated APP processing provides insight into APP and rhomboid physiology and qualifies for further investigations to elaborate its impact on Alzheimer disease pathology. 28445956_Tissue microarray assays demonstrated a correlation between RHBDD1 and EGFR in colorectal cancer patients. Therefore, our findings indicate that RHBDD1 stimulates EGFR expression by promoting the AP-1 pathway. 29426364_we proved that RHBDD1 can promote CRC metastasis through the Wnt signaling pathway and ZEB1. As a membrane protein, RHBDD1 has the potential and advantage to become a new therapeutic target or clinical biomarker for metastatic CRC. 30143535_The activity of RHBDL4 is regulated by cholesterol likely through a direct binding of cholesterol to the enzyme. 30286765_RHBDD1 promotes breast cancer progression by regulating p-Akt and CDK2 protein levels 31177093_Bioinformatics analyses revealed that BioID hits of RHBDL4 overlap with factors related to protein stress at the ER, including proteins that interact with p97/VCP. PTP1B (protein-tyrosine phosphatase nonreceptor type 1, also called PTPN1) was also identified as a potential proximity factor and interactor of RHBDL4 31243644_RHBDD1 is a direct target of miR-138-5p. MiR-138-5p directly binds to the 3'-UTR of the RHBDD1 transcript to inhibit the expression of RHBDD1 in breast cancer cells. RHBDD1 and miR-138-5p appear to play an opposite role in breast cancer cell migration and invasion. 31693935_miR-145-5p was inversely related with RHBDD1 expression in CRC tissues. miR-145-5p was found to directly bind to RHBDD1 and restrained its expression in CRC cells. miR-145-5p overexpression repressed CRC cell proliferation, invasion, migration and induced apoptosis, and these effects were reversed by RHBDD1 upregulation. 32528541_Silibinin suppresses epithelial-mesenchymal transition in human non-small cell lung cancer cells by restraining RHBDD1. 34581421_RHBDD1 promotes proliferation, migration, invasion and EMT in renal cell carcinoma via the EGFR/AKT signaling pathway. | ENSMUSG00000026142 | Rhbdd1 | 7.104011e+02 | 1.0156082 | 0.022343977 | 0.2810521 | 6.181594e-03 | 0.9373324015 | 0.98859743 | No | Yes | 6.135934e+02 | 77.503577 | 5.718500e+02 | 74.130799 | ||
| ENSG00000144566 | 5868 | RAB5A | protein_coding | P20339 | FUNCTION: Small GTPase which cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular responses such as of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. Active GTP-bound form is able to recruit to membranes different sets of downstream effectors directly responsible for vesicle formation, movement, tethering and fusion. RAB5A is required for the fusion of plasma membranes and early endosomes (PubMed:10818110, PubMed:14617813, PubMed:16410077, PubMed:15378032). Contributes to the regulation of filopodia extension (PubMed:14978216). Required for the exosomal release of SDCBP, CD63, PDCD6IP and syndecan (PubMed:22660413). Regulates maturation of apoptotic cell-containing phagosomes, probably downstream of DYN2 and PIK3C3 (By similarity). {ECO:0000250|UniProtKB:Q9CQD1, ECO:0000269|PubMed:10818110, ECO:0000269|PubMed:14617813, ECO:0000269|PubMed:14978216, ECO:0000269|PubMed:15378032, ECO:0000269|PubMed:16410077, ECO:0000269|PubMed:22660413}. | 3D-structure;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Endocytosis;Endosome;GTP-binding;Glycoprotein;Hydrolase;Lipoprotein;Membrane;Nucleotide-binding;Phagocytosis;Phosphoprotein;Prenylation;Protein transport;Reference proteome;Transport | hsa:5868; | actin cytoskeleton [GO:0015629]; anchored component of synaptic vesicle membrane [GO:0098993]; axon [GO:0030424]; axon terminus [GO:0043679]; clathrin-coated endocytic vesicle membrane [GO:0030669]; cytoplasm [GO:0005737]; cytoplasmic side of early endosome membrane [GO:0098559]; cytosol [GO:0005829]; dendrite [GO:0030425]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; early phagosome [GO:0032009]; endocytic vesicle [GO:0030139]; endomembrane system [GO:0012505]; endosome [GO:0005768]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; intracellular membrane-bounded organelle [GO:0043231]; melanosome [GO:0042470]; membrane raft [GO:0045121]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; phagocytic vesicle [GO:0045335]; phagocytic vesicle membrane [GO:0030670]; plasma membrane [GO:0005886]; postsynaptic early endosome [GO:0098842]; ruffle [GO:0001726]; somatodendritic compartment [GO:0036477]; synaptic vesicle [GO:0008021]; terminal bouton [GO:0043195]; G protein activity [GO:0003925]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; amyloid-beta clearance by transcytosis [GO:0150093]; early endosome to late endosome transport [GO:0045022]; endocytosis [GO:0006897]; intracellular protein transport [GO:0006886]; phagocytosis [GO:0006909]; positive regulation of exocytosis [GO:0045921]; receptor internalization involved in canonical Wnt signaling pathway [GO:2000286]; regulation of autophagosome assembly [GO:2000785]; regulation of endocytosis [GO:0030100]; regulation of endosome size [GO:0051036]; regulation of filopodium assembly [GO:0051489]; regulation of long-term neuronal synaptic plasticity [GO:0048169]; regulation of synaptic vesicle exocytosis [GO:2000300]; synaptic vesicle recycling [GO:0036465]; viral RNA genome replication [GO:0039694] | 11792815_Expression of a GTPase-hydrolysis-defective rab5a affects lysosome biogenesis by alteration of traffic between lysosomes and endosomes. 11884531_Rab5a regulates fusion between pathogen-containing phagosomes and cytoplasmic organelles in human neutrophils 12034881_an endocytotic catalyst, a tandem regulator of thyroid hormone production 12432064_effect of SARA on rab5-mediated endocytosis 12433916_Rab5a has a P-loop backbone amide group, which is required for catalysis 12668728_dynamin2 and Rab5 have roles in endocytosis of lysophosphatidic acid-coupled LPA1/EDG-2 receptors 12761223_In Rab5 overexpressing cells, the levels of beta-cleaved amyloid precursor protein (APP) carboxyl-terminal fragments (betaCTF), the rate-limiting proteolytic intermediate in Abeta generation, were increased 14644159_increase in the concentration of copper in the medium (189 microM) rapidly induces a redistribution of the MNK protein from early sorting endosomes, positive for Rab5-myc protein, to late endosomes, containing the Rab7-myc protein 14669515_Cell cycle was lengthened by blocking or reducing expression of RAB5A G81R mutation. 15016378_identification of a pathway directly linking the small GTPase Rab5, a key regulator of endocytosis, to signal transduction and mitogenesis via APPL1 and APPL2, two Rab5 effectors 15023538_Guanine nucleotide binding state of rab5 has no bearing on the rate of EGFR endocytosis. However, expression of dominant negative rab5 affects downstream endocytic trafficking by slowing the ligand-induced disappearance of total cellular EGFR. 15328530_Rab5 regulates and coordinates different endocytic mechanisms through its effector Rabankyrin-5 15388334_These results suggest that amyotrophic lateral sclerosis 2 C-terminal like (ALS2CL), a novel ALS2 homologue, modulates Rab5-mediated endosome dynamics in HeLa cells. 16554017_Taken together, these results demonstrate that Rab5 is required for insulin receptor membrane trafficking and signaling. 17173037_We show that EGF relocates to the cell centre in a dynein-dependent fashion, concomitant with the sorting away of transferrin receptor, although it remains in Rab5-positive early endosomes. 17301141_Rab5 participates in the hepatitis C virus RNA replication machinery. 17301152_Rab 5 is required for the cellular entry of dengue and West Nile viruses. 17473071_TSH controls Rab5a activity by promoting its GTP-bound state 17524504_Results suggest that Rab5 and RalA regulate P-gp trafficking between the plasma membrane and an intracellular compartment. 17581628_The crystal structures of human APPL1 N-terminal BAR-PH domain motif, is reported. 17611268_Rab5 activation via amyloid precursor protein signal pathway mediates neuronal apoptosis. 18005733_B coxsackievirus entry depends on occludin and require the activity of Rab34, Ras, and Rab5, GTPases known to regulate macropinocytosis. 18725540_SopB mediates PI(3)P production on the SCV indirectly through recruitment of Rab5 and its effector Vps34. 18957427_Rab5 is a critical regulator of syndecan-1 shedding that serves as an on-off molecular switch through its alternation between the GDP-bound and GTP-bound forms. 18974049_caspase 8 has a role as a modulator of p85alpha Rab5-GAP activity and endosomal trafficking 18982021_AdipoR1 is internalized through a clathrin- and Rab5-dependent pathway and that endocytosis may play a role in the regulation of adiponectin signaling. 19032933_Specific residues of RIN1 are required for its interaction with Rab5, binding to the endosomal membranes and subsequent regulation of the fusion reaction. 19118546_Mutations in the Vps9 domain of Rin1 lead to a loss-of-function phenotype, indicating a specific structure-function relationship between Rab5 and Rin1. 19126785_oxytocin receptors localize in vesicles containing the Rab5 and Rab4 small GTPases 19372461_Rab GTPase regulation of VEGFR2 trafficking and signaling in endothelial cells; Endothelial cell migration was increased by Rab5a depletion but decreased by Rab7a depletion 19656886_Borna Disease Virus cell entry was Rab5 dependent and exhibited rapid fusion kinetics. 19723633_suppression of Rab5A or 5B hampered the degradation of EGFR 19759177_Data report the identification of Rab22 as a binding site on early endosomes for direct recruitment of Rabex-5 and activation of Rab5, establishing a Rab22-Rab5 signaling relay to promote early endosome fusion. 19819222_The objective of the current study was to perform a detailed examination of the structural flexibility of the phosphate-binding loop (P-loop) as well as the switch I and II regions of human Rab5a upon its binding with GTP. 19830447_The Rab5(Q79L) not only induces formation of enlarged early endosomes but also causes enlargement of later endocytic profiles. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19923319_These studies identify Rab5 as a key integrator of caspase-8-mediated signal transduction downstream of integrins, regulating cell survival and migration in vivo and in vitro. 20376209_Rab5(Q79L) interacts with the carboxyl terminus of RUFY3 20412119_Rab5a promoted proliferation of ovarian cancer cells, which may be associated with the APPL1-related epidermal growth factor signaling pathway. 20457610_Data demonstrate a key role of Rab and Arf family small GTPases and intracellular trafficking in mTORC1 activation. 20472552_Rab5 and Rab7, were associated with the pathway of autophagosome formation and the fate of intracellular group A streptococcus. 20558162_Increased expression of early endosomal marker Rab5 correlates well with intracellular protein deposition in sporadic motor neuron disease. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20847427_Results indicate selective upregulation of Both rab5 and rab7 levels within basal forebrain, frontal cortex, and hippocampus in mild cognitive impairment and Alzheimer's disease. 20926780_These data suggest that proteasomes regulate claudin-1 localization at the plasma membrane, which changes upon proteasomal inhibition to a Rab5a-mediated endosomal localization. 21104291_Rab5a plays a role in LHR trafficking by facilitating internalization and fusion to early endosomes, increasing the degradation of internalized receptor resulting in a reduction in LHR recycling. 21203429_upon ligand activation, CysLT(1)R is tyrosine-phosphorylated and released from heterodimers with CysLT(2)R and, subsequently, internalizes from the plasma membrane to the nuclear membrane in a clathrin-, arrestin-3-, and Rab-5-dependent manner 21423773_Dmkndelta activates Rab5 function and thus is involved in the early endosomal trafficking. 21500550_RABEX-5 and RAB5 may be involved in the development of breast cancer metastasis. 21640764_results suggest that Rab5 is involved in CB2 endocytosis and that internalized receptors are recycled via a Rab11 associated pathway rather than the rapid Rab4 associated pathway. 21655223_SUN2 was redistributed to endosomes upon overexpression of Rab5, but remained on the nuclear envelope when the SUN domain was deleted. 21669283_Upregulated expression of rab4, rab5, rab7, and rab27 correlates with antemortem measures of cognitive decline in individuals with mild cognitive impairment and Alzheimer's disease. 21835792_Coimmunoprecipitation assay indicated that hepatitis C virus NS4B formed a complex with human Rab5 and Vps34, supporting the notion that Rab5 and Vps34 are involved in NS4B-induced autophagy. 21849022_Knockdown of Rab5a expression decreases cancer cell motility and invasion through integrin-mediated signaling pathway. 21895870_Rab5A is associated with axillary lymph node metastasis in breast cancer patients. 21987812_Rab5 forms a complex with a subset of CENP-F in mitotic cells and regulates the kinetics of release of CENP-F from the nuclear envelope and its accumulation on kinetochores. 22178872_Rab5A also binds the II-III loop of the Ca(v)2.3 calcium channel and exerts an inhibitory effect on APLP1 mediated channel internalization. 22341461_Recruitment of the inositol 5-phosphatases OCRL and Inpp5b to the Yersinia-filled prevacuoles and subsequent phosphatidylinositol 4,5-diphosphate hydrolysis required the association of the GTPase Rab5 with prevacuoles. 22547071_regulation of PI(3)P synthesis by Rab5 and Vps21 is essential for TORC1 function in both contexts. 22942286_TGF-beta1 can induce epithelial-to-mesenchymal transition through reduction in SARA expression, SARA is also basally regulated by its interaction with PI3K. 23048039_Interaction of Rabex-5 with Rab5 depends on interaction of the MIU domain with the ubiquitinated L1 to drive its internalization. 23086000_These findings highlight Rab5 GTPase as a key regulator of P2X4 receptor cell surface expression and internalisation 23182941_Endoplasmic membrane protein complex subunit 6 (EMC6) interacted with both RAB5A and BECN1/Beclin 1 and colocalized with the omegasome marker ZFYVE1/DFCP1. 23291133_Rab5a and APPL1 are overexpressed in breast cancer, and are positively correlated with the HER-2 expression. 23434372_In mammalian cells, p110beta acts as a molecular sensor for growth factor availability and induces autophagy by activating a Rab5-mediated signaling cascade 23454239_Dominant negative (GDP-locked)-Rab5 and -Rab11 reproduced the effects of inhibitors of tubulin and actin, respectively, on the cycling of bradykinin B receptor. 23536683_HBV infection strongly depends on host Rab5 and Rab7 expression. 23606746_Rab5a, Rab8a and Rab14 are major regulators of MT1-MMP trafficking and invasive migration of primary human macrophages. 23687301_In silico screening for palmitoyl substrates reveals a role for DHHC1/3/10 (zDHHC1/3/11)-mediated neurochondrin palmitoylation in its targeting to Rab5-positive endosomes. 23733193_Data indicate that Rab5, Rab7 and Rab11 are involved in RGS4 traffics through plasma membrane recycling or endosome. 23813952_Rab5 activation is required to enhance cancer cell migration and invasion by promoting focal adhesion disassembly. 23815289_Rab1a and Rab5a preferentially bind to binary lipid compositions with higher stored curvature elastic energy. 23940042_clathrin interacts with Rab5 and plays a fundamental role in the entry and intracellular survival of B. abortus via interaction with lipid rafts and actin rearrangement 24466349_vinculin binds to Rab5 and is required for Staphylococcus aureus uptake in cells. 24576301_The predominant pathway mediated by Australian bat lyssavirus G envelope for internalization into HEK293T cells is clathrin-and actin-dependent also requiring Rab5. 24587345_Rab5 isoforms selectively oversee the multiple signaling and trafficking events associated with the endocytic network. 24659799_activation, which is required for cell migration is promoted by Caveolin-1 24727246_Recent studies have shown that the activation of Rab5 is a critical event for maintaining the dynamics of focal adhesions, which is fundamental in regulating not only cell migration but also tumor cell invasion 24788845_Hepatitis B virus can downregulate miR-101-3p expression by inhibiting its promoter activity and that downregulation of miR-101-3p promotes hepatocellular carcinoma cell proliferation and migration by targeting Rab5a. 24848261_FGF21 has a role in promoting endothelial cell angiogenesis through a dynamin-2 and Rab5 dependent pathway 24872409_a novel mechanism by which a PKC-Rab5a-Rac1 axis regulates cytoskeleton remodeling and T-cell migration, both of which are central for the adaptive immune response. 25049275_Overexpression of the GTPase RAB5A, a master regulator of endocytosis, is predictive of aggressive behavior and metastatic ability in human breast cancers. 25152371_This process is inhibited in the presence of a Rab5 dominant-negative mutant. 25179218_TNF-alpha augments invasion of Porphyromonas gingivalis in human gingival epithelial cells through increment of ICAM-1 and activation of Rab5. 25483964_vacuolin-1 activates RAB5A to block autophagosome-lysosome fusion 25566515_Bacteria controled the localization or function of host Rab5 and Rab7, and therefore modify the maturation from early to late phagosomes.[review] 25711083_In serum-deprivated HeLa cells with low endocytic activity there two types of EEA1-vesicles: the first one carries the both EEA1 and Rab5 at high levels; the second consists of weakly decorated EEA1-vesicles that can be both Rab5-positive and -negative. 25763873_Rab5 activation as a tumor cell migration switch 26112597_Rab5a is a key mediator of LPS-induced vascular hyperpermeability. 26168723_findings suggest that Rab5 expression is required to maintain characteristics associated with cell transformation 26181205_Prelamin A/C was translocated to the nuclear membrane and formed a proper nuclear envelope. Rab5a was translocated to the early endosomes. The specific localizations of the prenylated proteins were dependent on intracellular oxygen concentration 26194181_Results indicate that persistent rab5 overactivation through beta-cleaved carboxy-terminal fragment of APP-APPL1 interactions constitutes a novel APP-dependent pathogenic pathway in Alzheimer's disease 26195760_Membrane attack complexes activate noncanonical NF-kappaB by forming a novel Akt(+)NIK(+) signalosome on Rab5(+) endosomes. 26261586_Higher expression of Rab5A was observed in colorectal cancer tissues and Rab5A may be identified as a useful predictor of metastasis and prognosis for CRC. 26344766_Data show that Rab22a- and Rab5a-driven phagosomal uptake is a crucial step in the vesicular cascade that leads to elimination of spirochetes by macrophages. 26443539_Data show that rab5 GTP-Binding Protein (Rab5a) is overexpressed in human hepatocellular carcinoma (HCC) and contributes to cancer cell proliferation and invasion through regulation of FAK and AKT signaling. 26473288_Low expression of RAB5A is associated with metastasis in prostate cancer. 26528697_Findings indicate the role of CMTM7 protein in the regulation of epidermal growth factor receptor (EGFR)-AKT proto-oncogene protein signaling in tumor cells, and as a molecule related to Rab5 GTP-binding protein activation. 26582392_Results demonstrate that DRG2 is an endosomal protein and a key regulator of Rab5 deactivation and Tfn recycling. 26680696_PLD1 recovers the decrease in epidermal growth factor receptor (EGFR) endocytosis induced by HIF-1alpha independent of lipase activity via the Rab5-mediated endosome fusion pathway. 27023526_Rab5a is involved in critical events not only at the beginning of the autophagy process with endosomal formation (initiation), but also later on, being important for autophagosome sealing and fusion with lysosomes through an interplay with Beclin 1. 27556945_results suggest a new mechanism in which Rab5 induces a change in flexibility of EEA1, generating an entropic collapse force that pulls the captured vesicle towards the target membrane to initiate docking and membrane fusion 27607061_TTBK2 down-regulates GluK2 activity by decreasing the receptor protein abundance in the cell membrane via RAB5-dependent endocytosis. 27666726_downregulated Rab5a led to slowed cell growth, decreased numbers of migrated cells, decreased numbers of cells at the G0G1 phase and a higher apoptosis rate. However, PDGF significantly rescued these phenomena caused by siRNA against Rab5a 27867015_CMTM3 decreases EGFR expression, facilitates EGFR degradation, and inhibits the EGF-mediated tumorigenicity of gastric cancer cells by enhancing Rab5 activity. 28090783_structural differences may provide an opportunity to selectively target one Rab5 state and lead to new approaches in the development of Rab5-specific therapies 28103577_High RAS-associated protein RAB5 expression correlated with the presence of lymphatic invasion and venous invasion and low E-cadherin expression. 28243729_our results show that Rab5a is overexpressed in pancreatic cancer and promotes aggressive biological behavior through regulation of the Wnt/beta-catenin signaling pathway. 28455099_Membrane localization and dynamics of geranylgeranylated Rab5 hypervariable region has been reported. 28575494_Low RAB5 expression is associated with glioma progression. 28634871_The results were indicative that rabies virus N proficiently colocalized with Rab5/EEA1 and Rab7/LAMP1 in both cell lines at 24 and 48 h post-infection, while N titers significantly decreased in early infection of rabies virus. 28650718_Ehrlichia obtains host-derived nutrients by inducing RAB5-regulated autophagy using Ehrlichia translocated factor-1 deployed by its type IV secretion system. This manipulation of RAB5 by a bacterial molecule offers a simple strategy for Ehrlichia to avoid destruction in lysosomes and obtain nutrients. 28650977_siRNA knockdown of Rab5a or overexpression of miR-494 in human macrophages significantly inhibits the survival of the parasites 28742203_The expression level of EMC-6 is significantly elevated in cervical cancer, without significant correlation with Beclin1 and Rab5a. 28834690_study elucidated a novel Malat1-miR-101-STMN1/RAB5A/ATG4D regulatory network that Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma cells 28849149_Rab5a is overexpressed in oral cancer tissue samples and promotes the malignant phenotype through EMT and the ERK/MMP2 signaling pathway. 28867190_In conclusion, mutant KRAS promotes endosomal degradation in PDAC cell lines, which is impaired by KRAS silencing. Moreover, KRAS silencing activates RAB5A upregulation and drives PDAC subtype-dependent modulation of endosome trafficking. 28899395_Rab5 was found abundantly localized in macrophage rich areas of human atherosclerotic lesions.Rab5 plays an important role in modulating the intracellular cholesterol of macrophages and consequently mediating the formation of foam cells. 28968219_Here the authors show that Rab5 is monoubiquitinated on K116, K140, and K165. Structural analysis combined with biochemical data revealed that interactions with downstream effectors were impeded in Rab5 monoubiquitinated at K140, whereas GDP release and GTP loading activities were altered in Rab5 monoubiquitinated at K165. 29065764_These results allow to devise a detailed structural model for the process of extraction of GG-Rab5(GDP) by GDI from the membrane and the dissociation from targeting factors and effector proteins prior to GDI binding. 29127297_Rab5 is essential for FcepsilonRI-triggered association of the SNARE protein SNAP23 with the secretory granules. 29360040_The authors demonstrate that RABGEF1, the upstream factor of the endosomal Rab GTPase cascade, is recruited to damaged mitochondria via ubiquitin binding downstream of Parkin. RABGEF1 directs the downstream Rab proteins, RAB5 and RAB7A, to damaged mitochondria, whose associations are further regulated by mitochondrial Rab-GAPs. 29361527_we demonstrate a novel role for the interaction between APPL1 and Rab5 in governing crosstalk between signaling and trafficking pathways on endosomes to affect cancer cell migration 29469808_These findings define a novel pathway whereby Alsin catalyzes the assembly of the Rab5 endocytic machinery on mitochondria. Defects in stress-sensing by endosomes could be crucial for mitochondrial quality control during the onset of amyotrophic lateral sclerosis. 29626103_Low RAB5A expression is associated with polycystic ovary syndrome. 29740032_several p85a mutations found in human cancers may deregulate PTEN and/or Rab5 regulated pathways to contribute to oncogenesis. We also engineered several experimental mutations within the p85a BH domain and identified L191 and V263 as important for both binding and regulation of Rab5 activity 29743547_Protease Activated Receptor2 Promotes Rab5a Mediated Generation of Pro-metastatic Microvesicles 29868450_This study showed that the RAB5A predictors of regional brain atrophy in Parkinson disease. 30333257_Overexpression of Rab5B largely rescued the miR-575-mediated impairment of angiogenesis. 30659094_results define a single, discrete Rab5-binding site in the p110beta helical domain, which may be useful for generating inhibitors to better define the physiological role of Rab5-PI3Kbeta coupling in vivo 30765602_findings show that endosomal/lysosomal RAB5 and RAB7, which regulate mitophagy, are essential for the survival of colon cancer stem cells 31221728_The study reveals an evolutionarily conserved role for the early endocytic marker Rab5 in cytokinetic abscission. 31337623_sorting nexin SNX3 is transported with Rab5a vesicles and that its PX domain enables vesicle-phagosome contact by binding to PI(3)P in the phagosomal coat. Moreover, the C-terminal region of SNX3 recruits galectin-9, a lectin implicated in protein and membrane recycling, which we identify as a further regulator of phagosome compaction. 31358736_The findings suggest that SPIN90, as an adaptor protein, simultaneously binds inactive Rab5 and Gapex5, thereby altering their spatial proximity and facilitating Rab5 activation. 31558725_Data suggest a mechanism whereby flotillins mediate T cell receptor (TCR) sorting into rab5 GTP-binding protein (Rab5) and ras-related protein Rab-11A (Rab11) endosomes. 31705388_EGFR signaling augments TLR4 cell surface expression and function in macrophages via regulation of Rab5a activation. 31811856_Study data indicate that GRK2 and RAB5 play key roles in alpha1B-adrenergic receptor phosphorylation, internalization, and desensitization. The possibility that RAB5 might form part of a signaling complex is suggested, as well as that GDP-Rab5 might interfere with the ability of GRK2 to catalyze alpha1B-adrenergic receptor phosphorylation. 32304339_Protein diaphanous homolog 1 (Diaph1) promotes myofibroblastic activation of hepatic stellate cells by regulating Rab5a activity and TGFbeta receptor endocytosis. 32560826_Rac1-dependent endocytosis and Rab5-dependent intracellular trafficking are required by Enterovirus A71 and Coxsackievirus A10 to establish infections. 33092247_Rab GTPase Mediating Regulation of NALP3 in Colorectal Cancer. 33137306_The G-Protein Rab5A Activates VPS34 Complex II, a Class III PI3K, by a Dual Regulatory Mechanism. 33341673_RAB5A promotes the formation of filopodia in pancreatic cancer cells via the activation of cdc42 and beta1-integrin. 33416169_Long noncoding RNA CASC19 promotes glioma progression by modulating the miR4543p/RAB5A axis and is associated with unfavorable MRI features. 33692360_Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. 33831402_RAB5A effect on metastasis of hepatocellular carcinoma cell line via altering the pro-invasive content of exosomes. 34129971_Propofol Represses Cell Growth and Metastasis by Modulating the Circular RNA Non-SMC Condensin I Complex Subunit G/MicroRNA-200a-3p/RAB5A Axis in Glioma. 34281589_CMTM7 as a novel molecule of ATG14L-Beclin1-VPS34 complex enhances autophagy by Rab5 to regulate tumorigenicity. 34383013_Who's in control? Principles of Rab GTPase activation in endolysosomal membrane trafficking and beyond. 34826799_Nectin stabilization at adherens junctions is counteracted by Rab5a-dependent endocytosis. 35017116_lncRNA HITT inhibits metastasis by attenuating Rab5-mediated endocytosis in lung adenocarcinoma. | ENSMUSG00000017831 | Rab5a | 5.247706e+02 | 1.0944324 | 0.130182894 | 0.3166399 | 1.664434e-01 | 0.6832922132 | 0.93336121 | No | Yes | 5.312835e+02 | 84.719345 | 4.316009e+02 | 70.793797 | ||
| ENSG00000144647 | 84892 | POMGNT2 | protein_coding | Q8NAT1 | FUNCTION: O-linked mannose beta-1,4-N-acetylglucosaminyltransferase that transfers UDP-N-acetyl-D-glucosamine to the 4-position of the mannose to generate N-acetyl-D-glucosamine-beta-1,4-O-D-mannosylprotein. Involved in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1), which is required for binding laminin G-like domain-containing extracellular proteins with high affinity. {ECO:0000269|PubMed:23929950, ECO:0000269|PubMed:27066570}. | 3D-structure;Congenital muscular dystrophy;Disease variant;Dystroglycanopathy;Endoplasmic reticulum;Glycoprotein;Glycosyltransferase;Limb-girdle muscular dystrophy;Lissencephaly;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:23929950}. | This gene encodes a protein with glycosyltransferase activity although its function is not currently known. [provided by RefSeq, Sep 2012]. | hsa:84892; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; acetylglucosaminyltransferase activity [GO:0008375]; glycosyltransferase activity [GO:0016757]; protein O-GlcNAc transferase activity [GO:0097363]; neuron migration [GO:0001764]; protein O-linked glycosylation [GO:0006493]; protein O-linked mannosylation [GO:0035269] | 22958903_Using WES in consanguineous WWS-affected families, we found multiple deleterious mutations in GTDC2 (also known as AGO61). . 24041696_GTDC2 generates CTD110.6 antibody-reactive N-acetylglucosamine epitopes on the O-mannosylated alpha-dystroglycan. 27932460_POMGNT2 when two of the conserved amino acids are replaced. These findings begin to define the selectivity of POMGNT2 and suggest that this enzyme functions as a gatekeeper enzyme to prevent the vast majority of O-mannosylated sites on proteins from becoming modified with glycan structures functional for binding laminin globular domain-containing proteins 33893702_The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan. | ENSMUSG00000066235 | Pomgnt2 | 4.669929e+03 | 1.2312327 | 0.300103427 | 0.3181107 | 8.888306e-01 | 0.3457943878 | 0.82969965 | No | Yes | 4.717367e+03 | 515.068104 | 3.621408e+03 | 405.919675 |
| ENSG00000144659 | 54977 | SLC25A38 | protein_coding | Q96DW6 | FUNCTION: Mitochondrial glycine transporter that imports glycine into the mitochondrial matrix. Plays an important role in providing glycine for the first enzymatic step in heme biosynthesis, the condensation of glycine with succinyl-CoA to produce 5-aminolevulinate (ALA) in the mitochondrial matrix. Required during erythropoiesis. {ECO:0000255|HAMAP-Rule:MF_03064, ECO:0000269|PubMed:19412178, ECO:0000269|PubMed:27476175}. | Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene is a member of the mitochondrial carrier family. The encoded protein is required during erythropoiesis and is important for the biosynthesis of heme. Mutations in this gene are the cause of autosomal congenital sideroblastic anemia (anemia, sideroblastic, 2, pyridoxine-refractory). A related pseudogene is found on chromosome 1. [provided by RefSeq, Aug 2017]. | hsa:54977; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; glycine transmembrane transporter activity [GO:0015187]; erythrocyte differentiation [GO:0030218]; glycine import into mitochondrion [GO:1904983]; heme biosynthetic process [GO:0006783] | 19731322_Twelve CSA probands had biallelic mutations in SLC25A38 21393332_Mutations in the SLC25A38 gene cause severe, non-syndromic, microcytic/hypochromic sideroblastic anemia in many populations. 23115192_Our study identifies appoptosin as a crucial player in apoptosis and a novel pro-apoptotic protein involved in neuronal cell death. 23266187_Compares and contrasts all the known human SLC25A* genes and includes functional information. 24323989_Several missense mutations are found in SLC25A38 in a Chinese population with congenital sideroblastic anemia. 25512395_Letter/Case Report: novel frameshift mutation in SLC25A38 causing congenital sideroblastic anaemia. 26335643_This study findings reveal a novel role for appoptosin in neurological disorders with tau neuropathology, linking caspase-3-mediated tau cleavage to synaptic dysfunction and behavioral/motor defects. 26813789_Appoptosin can interact with mitochondrial outer-membrane fusion proteins and regulates mitochondrial morphology. 26821380_Given the tolerability of glycine and folate in humans, this study points to a potential novel treatment for SLC25A38 congenital sideroblastic anemia 27476175_the biochemical and molecular characterization of yeast Hem25p and human SLC25A38, providing evidence that they are mitochondrial carriers for glycine. In particular, the hem25Delta mutant manifests a defect in the biosynthesis of delta-aminolevulinic acid and displays reduced levels of downstream heme and mitochondrial cytochromes. 28772256_report confirms the considerable variability in manifestations among patients with ALAS2 or SLC25A38 mutations and draws attention to differences in the assessment and the monitoring of iron overload and its complications 29499877_These findings suggest that sideroblastic anemia must be considered a possible etiology in cases with unexplained hemolytic anemia. Furthermore, mutations in SLC25A38 gene could be a prevalent cause of congenital sideroblastic anemia (CSA) in the Iranian population. 31155012_Dentate gyrus volume deficit in schizophrenia. 32605921_Novel frameshift variant (c.409dupG) in SLC25A38 is a common cause of congenital sideroblastic anaemia in the Indian subcontinent. 32790119_Clinical characterization and hematopoietic stem cell transplant outcomes for congenital sideroblastic anemia caused by a novel pathogenic variant in SLC25A38. 34298585_SLC25A38 congenital sideroblastic anemia: Phenotypes and genotypes of 31 individuals from 24 families, including 11 novel mutations, and a review of the literature. 35411037_SLC25A38 as a novel biomarker for metastasis and clinical outcome in uveal melanoma. | ENSMUSG00000032519 | Slc25a38 | 3.956699e+03 | 0.9859385 | -0.020430446 | 0.2994158 | 4.699074e-03 | 0.9453479844 | 0.98943389 | No | Yes | 3.679019e+03 | 266.888921 | 3.490057e+03 | 259.983599 | |
| ENSG00000144724 | 5793 | PTPRG | protein_coding | P23470 | FUNCTION: Possesses tyrosine phosphatase activity. {ECO:0000269|PubMed:19167335}. | 3D-structure;Alternative splicing;Disulfide bond;Glycoprotein;Hydrolase;Membrane;Phosphoprotein;Protein phosphatase;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP possesses an extracellular region, a single transmembrane region, and two tandem intracytoplasmic catalytic domains, and thus represents a receptor-type PTP. The extracellular region of this PTP contains a carbonic anhydrase-like (CAH) domain, which is also found in the extracellular region of PTPRBETA/ZETA. This gene is located in a chromosomal region that is frequently deleted in renal cell carcinoma and lung carcinoma, thus is thought to be a candidate tumor suppressor gene. [provided by RefSeq, Jul 2008]. | hsa:5793; | extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; identical protein binding [GO:0042802]; protein tyrosine phosphatase activity [GO:0004725]; transmembrane receptor protein tyrosine phosphatase activity [GO:0005001]; brain development [GO:0007420]; negative regulation of epithelial cell migration [GO:0010633]; negative regulation of neuron projection development [GO:0010977]; protein dephosphorylation [GO:0006470]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11859871_both estradiol-17beta and zeranol regulate PTPgamma expression in human breast; epithelial-stromal cell interaction is important in regulation of PTPgamma expression by estrogenically active agents 12553013_Estrogenic down-regulation of protein tyrosine phosphatase gamma (PTP gamma) in human breast is associated with estrogen receptor alpha. 14676845_PTPgamma may function as an important modulator in regulating the process of tumorigenesis in human breast. 15897551_Promoter hypermethylation of PTPRG is associated with cutaneous T-cell lymphoma 17963294_Significant difference exists in methylation of gene PTPRG between primary tumor and metastatic lymph nodes of gastric cancer. 18646686_There were significant differences of PTPRG gene methylation and PTPRG mRNA expression between gastric primary cancer and lymph node metastases. 18829573_PTPRG may contribute to tumorigenesis. 19567891_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20133774_findings implicate PTPRG, PTPRZ and CNTNs as a group of receptors and ligands involved in the manifold recognition events that underlie the construction of neural networks 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20651337_PTPRG inhibited breast tumor formation in vivo; PTPRG may up-regulate p21(cip) and p27(kip) proteins through the ERK1/2 pathway. 20959494_Downregulation of Protein tyrosine phosphatase receptor type {gamma} is associated with chronic myeloid leukemia. 21150880_Tumor-specific methylation of the first intron of the receptor protein-tyrosine phosphatase gamma gene was found in both sporadic and Lynch syndrome colorectal cancers. 21795790_found that the RPTP-gamma, RPTP-gamma(V948I, S970T) and RPTP-gamma(C858S, S970T) proteins could be stored at 193 K in their respective crystallization buffers without affecting crystallization or crystal quality 23758498_Our results showed that there was no significant association between any of five reported SNPs of TCF4 and PTPRG genes and the occurrence of Fuchs' endothelial dystrophy; only rs7640737 in PTPRG showed an increased risk for corneal dystrophy. 24496747_PTPRG expression induces dephosphorylation of ERK, a downstream RAS target that may be critical for mutant RAS-induced cell growth. 25158255_Mutations in PDGFRB cause infantile myofibromatosis while a mutation in PTPRG may act as an aggravating factor. 25196286_Germline polymorphisms in PTPRG gene is associated with lung adenocarcinoma. 25299301_SNPs in PTPRG were not associated with FCD in Caucasians. 25624455_PTPRG is a JAK2 phosphatase that negatively regulates leukocyte integrin beta2 activation. 25775014_Results demonstrate that PTPRG plasmatic form (sPTPRG) represent a novel candidate protein biomarker in plasma whose increased expression is associated to hepatocyte damage. 26830138_family-based GWAS of imputed SNPs revealed novel genomic variants in (or near) PTPRG, OSBPL6, and PDCL3 that influence risk for Alzheimer's Disease. rs7609954 in the gene PTPRG, rs1347297 in the gene OSBPL6, and rs1513625 near PDCL3. In addition, rs72953347 in OSBPL6 and two SNPs in the gene CDKAL1 showed marginally significant association with LOAD (rs10456232, P-value=4.76 x 10-7; rs62400067, P-value=3.54 x 10-7). 27602768_miR-19b inhibits PTPRG expression to promote tumorigenesis in human breast cancer 28637510_Co-localization experiments performed with both anti-PTPRG antibodies identified the presence of isoforms and confirmed protein downregulation at diagnosis in the Philadelphia-positive myeloid lineage (including CD34+/CD38bright/dim cells). 29371290_PTPRG and FGFR1 interact and colocalize at the plasma membrane where PTPRG directly dephosphorylates activated FGFR1. 29907679_OPCML and PTPRG, coordinate to repress AXL-dependent oncogenic signalling. 31012177_cMras was a sponge of miRNA-567 and released its direct target, PTPRG. cMras overexpression decreased miR-567 expression and subsequently increased PTPRG expression, while increased miRNA-567 expression blocked the effects induced by cMras. Moreover, PTPRG was downregulated in lung adenocarcinoma and patients with low PTPRG expression exhibited significantly poor prognosis. 32225105_Regulative Loop between beta-catenin and Protein Tyrosine Receptor Type gamma in Chronic Myeloid Leukemia. 32700424_Aberrant DNA methylation of PTPRG as one possible mechanism of its under-expression in CML patients in the State of Qatar. 32955439_PTPRG is an ischemia risk locus essential for HCO3(-)-dependent regulation of endothelial function and tissue perfusion. 33893334_Predictive value of tyrosine phosphatase receptor gamma for the response to treatment tyrosine kinase inhibitors in chronic myeloid leukemia patients. 34162728_Activation of Protein Tyrosine Phosphatase Receptor Type gamma Suppresses Mechanisms of Adhesion and Survival in Chronic Lymphocytic Leukemia Cells. 34906644_Description of PTPRG genetic variants identified in a cohort of Chronic Myeloid Leukemia patients and their ability to influence response to Tyrosine kinase Inhibitors. 35053232_A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease. | 3.612454e+02 | 1.1785646 | 0.237030838 | 0.2911373 | 6.740409e-01 | 0.4116463052 | 0.84733931 | No | Yes | 3.742001e+02 | 44.809566 | 2.845210e+02 | 35.125867 | |||
| ENSG00000145014 | 93109 | TMEM44 | protein_coding | Q2T9K0 | Alternative splicing;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:93109; | integral component of membrane [GO:0016021] | ENSMUSG00000022537 | Tmem44 | 2.527367e+02 | 1.4012244 | 0.486688044 | 0.3624751 | 1.763212e+00 | 0.1842244006 | 0.77834741 | No | Yes | 2.248929e+02 | 60.509937 | 1.892510e+02 | 52.239836 | ||||
| ENSG00000145216 | 81608 | FIP1L1 | protein_coding | Q6UN15 | FUNCTION: Component of the cleavage and polyadenylation specificity factor (CPSF) complex that plays a key role in pre-mRNA 3'-end formation, recognizing the AAUAAA signal sequence and interacting with poly(A) polymerase and other factors to bring about cleavage and poly(A) addition. FIP1L1 contributes to poly(A) site recognition and stimulates poly(A) addition. Binds to U-rich RNA sequence elements surrounding the poly(A) site. May act to tether poly(A) polymerase to the CPSF complex. {ECO:0000269|PubMed:14749727}. | 3D-structure;Alternative splicing;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;mRNA processing | This gene encodes a subunit of the CPSF (cleavage and polyadenylation specificity factor) complex that polyadenylates the 3' end of mRNA precursors. This gene, the homolog of yeast Fip1 (factor interacting with PAP), binds to U-rich sequences of pre-mRNA and stimulates poly(A) polymerase activity. Its N-terminus contains a PAP-binding site and its C-terminus an RNA-binding domain. An interstitial chromosomal deletion on 4q12 creates an in-frame fusion of human genes FIP1L1 and PDGFRA (platelet-derived growth factor receptor, alpha). The FIP1L1-PDGFRA fusion gene encodes a constitutively activated tyrosine kinase that joins the first 233 amino acids of FIP1L1 to the last 523 amino acids of PDGFRA. This gene fusion and chromosomal deletion is the cause of some forms of idiopathic hypereosinophilic syndrome (HES). This syndrome, recently reclassified as chronic eosinophilic leukemia (CEL), is responsive to treatment with tyrosine kinase inhibitors. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Oct 2008]. | hsa:81608; | cytosol [GO:0005829]; mRNA cleavage and polyadenylation specificity factor complex [GO:0005847]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; mRNA polyadenylation [GO:0006378]; pre-mRNA cleavage required for polyadenylation [GO:0098789] | 12660384_Describes the fusion gene Fip1-like-1-PDGFRalpha in patients with idiopathic hypereosinophilic syndrome, mostly responsive to imatinib therapy. 12660384_The hypereosinophilic syndrome may result from a novel fusion tyrosine kinase - FIP1L1-PDGFRalpha - that is a consequence of an interstitial chromosomal deletion. 12842979_observations suggest that the FIP1L1-PDGFRA rearrangement occurs in an early hematopoietic progenitor and suggests that the molecular pathogenesis for a subset of SMCD patients is similar to that of HES 14630792_results indicate that the fusion of FIP1L1 to PDGFRA occurs rarely in leukemia cell lines 15284118_FIP1L1-PDGFRA is a relatively infrequent but treatment-relevant mutation in primary eosinophilia that is indicative of an underlying systemic mastocytosis. 15767428_the newly identified hFip1 protein, which has been shown to enhance polyadenylation through U-rich upstream elements, interacted specifically with the HPV-16 upstream element. 16502585_Fusion protein is detected in hypereosinophilic syndrome and chronic eosinophilic leukemia. 16690743_FIP1L1-PDGFRalpha activation requires disruption of the juxtamembrane domain of PDGFRalpha and is FIP1L1-independent 17261495_data and previous reports suggest that FIP1L1-PDGFRA - positive HES is a distinct clinical entity with myeloproliferative features and showing a poor response to corticosteroid treatment 17299092_FIP1L1 and PDGFRalpha have roles in response to imatinib mesylate in chronic eosinophilic leukemia 17377585_FIP1L1-PDGFRA-positive patients who presented with acute myeloid leukemia (AML, n=5) or lymphoblastic T-cell non-Hodgkin-lymphoma (n=2) in conjunction with AML or Eos-MPD. 17591942_Fluorescence in situ hybridization and reverse transcriptase-polymerase chain reaction protocols were developed for an accurate del(4)(q12q12) and FIP1L1-PDGFRA fusion gene detection. 18706197_FIP1L1-PDGFRalpha emerges as a relatively homogeneous clinicobiological entity that co-exists with other abnormalities of tyrosine kinase family genes. It reflects the disease progression and there is a good response to imatinib. 18987651_characterized FIP1L1-PDGFRA junction sequences from 113 patients at the mRNA (n=113) and genomic DNA (n=85) levels 19118897_studied a new FISH method to detect CHIC2 deletion, FIP1L1/PDGFRA fusion and PDGFRA translocation in patients with myeloid neoplasms associated with eosinophilia 21399396_FIP1L1/PDGFRA fusion gene-positive myeloproliferative disorders with eosinophilia are discussed. 21818111_Treatment with imatinib is associated with an excellent prognosis in FIP1L1-PDGFRA-positive chronic eosinophilic leukemia in first chronic phase 22447844_Data show that the cyclin-dependent kinase 7/9 inhibitor (CDK7/9 inhibitor) potently inhibits FIP1L1-PDGFRalpha-positive Bcr-Abl-positive chronic myeloid leukemia (CML) cells. 22523564_results strongly suggest that JAK2 is activated by Fip1-like1 (FIP1L1)-platelet-derived growth factor receptor alpha (F/P) and is required for F/P stimulation of cellular proliferation and infiltration in chronic eosinophilic leukemia 22944561_description of polycythemia vera concurrent with FIP1L1-PDGFRA-positive myeloproliferative neoplasm with eosinophilia [case report] 23621172_Oncostatin M is a FIP1L1/PDGFRA-dependent mediator of cytokine production in chronic eosinophilic leukemia. 24458279_FP fusion gene favors secondary KIT mutations in MCs via growth and proliferation signals or that a yet unknown mechanism causes genomic instability with independent evolution of FP and KIT D816V 24763514_FIP1L1 differentially contributes to the pathogenesis of distinct types of leukemia. 25761934_F604S exchange in FIP1L1-PDGFRA enhances FIP1L1-PDGFRA protein stability via SHP-2 and SRC and is associated with kinase inhibitor resistance in hypereosinophilic syndrome and chronic eosinophilic leukemia. 27120808_FIP1L1/ PDGFRA associated chronic eosinophilic leukemia has an excellent long-term prognosis following imatinib therapy. 28374041_Case Report: concurrent development of myeloproliferative hypereosinophilic syndrome and lymphomatoid papulosis associated with FIP1L1-PDGFRA gene fusion. 29274231_The authors define the molecular architecture of the core human CPSF complex comprising CPSF160, WDR33, CPSF30 and Fip1 and identify specific domains involved in inter-subunit interactions. Together, these results shed light on the function of CPSF in mediating polyA signal-dependent RNA cleavage and polyadenylation. 29310833_Herein, we report a case of a 53-year-old man with eosinophilia and a well-differentiated extramedullary myeloid tumor with evidence of FIP1L1/PDGFRA rearrangement by fluorescent in situ hybridization in the extramedullary tissue. 30157244_Two identified variants revealed novel candidate genes for hip and knee osteoarthritis. OLIG3 and FIP1L1 have specific roles in transcription and may effect expression of other genes. Identified variants in these genes may thus have a role in the regulatory events leading to osteoarthritis. 32125294_Exquisite response to imatinib mesylate in FIP1L1-PDGFRA-mutated hypereosinophilic syndrome: a 12-year experience of the Polish Hypereosinophilic Syndrome Study Group. 34304603_FIP1L1-PDGFRA-Associated Hypereosinophilic Syndrome as a Treatable Cause of Watershed Infarction. | 1.021224e+03 | 1.3065217 | 0.385731055 | 0.3098275 | 1.533182e+00 | 0.2156354981 | 0.78763590 | No | Yes | 1.110081e+03 | 169.554461 | 7.908935e+02 | 124.143129 | |||
| ENSG00000145348 | 93627 | TBCK | protein_coding | Q8TEA7 | FUNCTION: Involved in the modulation of mTOR signaling and expression of mTOR complex components (PubMed:27040691, PubMed:23977024). Involved in the regulation of cell proliferation and growth (PubMed:23977024, PubMed:24576458). Involved in the control of actin-cytoskeleton organization (PubMed:23977024). {ECO:0000269|PubMed:23977024, ECO:0000269|PubMed:24576458, ECO:0000269|PubMed:27040691}. | Alternative splicing;Cytoplasm;Cytoskeleton;Disease variant;Reference proteome | This gene encodes a protein that contains a protein kinase domain, a Rhodanase-like domain and the Tre-2/Bub2/Cdc16 (TBC) domain. The encoded protein is thought to play a role in actin organization, cell growth and cell proliferation by regulating the mammalian target of the rapamycin (mTOR) signaling pathway. This protein may also be involved in the transcriptional regulation of the components of the mTOR complex. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2014]. | hsa:93627; | cytoplasm [GO:0005737]; midbody [GO:0030496]; mitotic spindle [GO:0072686]; ATP binding [GO:0005524]; GTPase activator activity [GO:0005096]; protein kinase activity [GO:0004672]; actin cytoskeleton organization [GO:0030036]; activation of GTPase activity [GO:0090630]; cell population proliferation [GO:0008283]; intracellular protein transport [GO:0006886]; regulation of TOR signaling [GO:0032006] | 20332099_Observational study of gene-disease association. (HuGE Navigator) 23977024_TBCK may play an important role in cell proliferation, cell growth and actin organization possibly by modulating mTOR pathway. 24576458_localization and function of TBCK 27040691_We have reported a series of 13 individuals from nine unrelated families that harbor biallelic mutation in TBCK and display overlapping features of intellectual disability and hypotonia. This condition is called TBCK-related intellectual disability syndrome. 27040692_We have established that biallelic mutations in TBCK cause a severe neurodevelopmental disorder whose major features include profound developmental delay or cognitive deficit, brain atrophy without microcephaly. 27633981_RNAsequencing showed that the t(4;5)(q24;q31) resulted in recombination of the genes TBCK on 4q24 and P4HA2 on 5q31.1 with generation of an inframe TBCKP4HA2 and the reciprocal but outofframe P4HA2TBCK fusion transcripts. 27748029_We conclude that the c.1854delT variant in the TBCK gene is the mutation causing the congenital brain abnormality in an Arab-Moslem family from northern Israel. 29283439_TBCK-encephaloneuronopathy is a clinically distinguishable syndrome with progressive central and peripheral nervous system dysfunction, consistently observed in patients with the TBCK mutation 30103036_A novel TBCK mutation was identified in two siblings with infantile hypotonia, psychomotor retardation and characteristic facies type 3. 30591081_Evidence that TBC1 domain-containing kinase (TBCK) deficiency disorder associated with homozygous TBCK mutations is a novel type of lysosomal storage disease. 31331056_These findings suggest that miR-1208 acts as a tumor suppressor and targets TBCK directly, thus possessing great potential for use in renal cancer therapy. | ENSMUSG00000028030 | Tbck | 2.227173e+02 | 0.8273186 | -0.273485121 | 0.4119479 | 4.259113e-01 | 0.5140020578 | 0.88019521 | No | Yes | 1.726782e+02 | 38.850422 | 2.305248e+02 | 52.876537 | |
| ENSG00000145685 | 10184 | LHFPL2 | protein_coding | Q6ZUX7 | FUNCTION: Plays a role in female and male fertility. Involved in distal reproductive tract development. {ECO:0000250|UniProtKB:Q8BGA2}. | Fertilization;Membrane;Reference proteome;Transmembrane;Transmembrane helix | This gene is a member of the lipoma HMGIC fusion partner (LHFP) gene family, which is a subset of the superfamily of tetraspan transmembrane protein encoding genes. Mutations in one LHFP-like gene result in deafness in humans and mice, and a second LHFP-like gene is fused to a high-mobility group gene in a translocation-associated lipoma. Alternatively spliced transcript variants have been found, but their biological validity has not been determined. [provided by RefSeq, Jul 2008]. | hsa:10184; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; platelet alpha granule membrane [GO:0031092]; development of primary female sexual characteristics [GO:0046545]; development of primary male sexual characteristics [GO:0046546]; positive regulation of fertilization [GO:1905516]; single fertilization [GO:0007338] | 15905332_The authors present an overview of the LHFP gene family in mouse and humans 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 27402877_Results report evidence for the existence of variants in LHFPL2 and TPM1 with low allele frequencies and large effects on age-at-onset of familial Parkinson's disease. | ENSMUSG00000045312 | Lhfpl2 | 2.761949e+02 | 0.6845530 | -0.546765755 | 0.3039341 | 3.131622e+00 | 0.0767873157 | 0.72306475 | No | Yes | 2.851460e+02 | 37.696749 | 3.892228e+02 | 52.472434 | |
| ENSG00000145725 | 23262 | PPIP5K2 | protein_coding | O43314 | FUNCTION: Bifunctional inositol kinase that acts in concert with the IP6K kinases IP6K1, IP6K2 and IP6K3 to synthesize the diphosphate group-containing inositol pyrophosphates diphosphoinositol pentakisphosphate, PP-InsP5, and bis-diphosphoinositol tetrakisphosphate, (PP)2-InsP4 (PubMed:17690096, PubMed:17702752, PubMed:21222653, PubMed:29590114). PP-InsP5 and (PP)2-InsP4, also respectively called InsP7 and InsP8, regulate a variety of cellular processes, including apoptosis, vesicle trafficking, cytoskeletal dynamics, exocytosis, insulin signaling and neutrophil activation (PubMed:17690096, PubMed:17702752, PubMed:21222653, PubMed:29590114). Phosphorylates inositol hexakisphosphate (InsP6) at positions 1 or 3 to produce PP-InsP5 which is in turn phosphorylated by IP6Ks to produce (PP)2-InsP4 (PubMed:17690096, PubMed:17702752). Alternatively, phosphorylates at position 1 or 3 PP-InsP5, produced by IP6Ks from InsP6, to produce (PP)2-InsP4 (PubMed:17690096, PubMed:17702752). Required for normal hearing (PubMed:29590114). {ECO:0000269|PubMed:17690096, ECO:0000269|PubMed:17702752, ECO:0000269|PubMed:21222653, ECO:0000269|PubMed:29590114}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Deafness;Disease variant;Hearing;Kinase;Non-syndromic deafness;Nucleotide-binding;Phosphoprotein;Reference proteome;Transferase | This gene encodes a member of the histidine acid phosphatase family of proteins. Despite containing a histidine acid phosphatase domain, the encoded protein functions as an inositol pyrophosphate kinase, and is thought to lack phosphatase activity. This kinase activity is the mechanism by which the encoded protein synthesizes high-energy inositol pyrophosphates, which act as signaling molecules that regulate cellular homeostasis and other processes. This gene may be associated with autism spectrum disorder in human patients. [provided by RefSeq, Sep 2016]. | hsa:23262; | cytosol [GO:0005829]; 5-diphosphoinositol pentakisphosphate 3-kinase activity [GO:0102092]; ATP binding [GO:0005524]; diphosphoinositol-pentakisphosphate kinase activity [GO:0033857]; inositol heptakisphosphate kinase activity [GO:0000829]; inositol hexakisphosphate 1-kinase activity [GO:0052723]; inositol hexakisphosphate 3-kinase activity [GO:0052724]; inositol hexakisphosphate 5-kinase activity [GO:0000832]; inositol hexakisphosphate kinase activity [GO:0000828]; inositol-1,3,4,5,6-pentakisphosphate kinase activity [GO:0000827]; inositol metabolic process [GO:0006020]; inositol phosphate biosynthetic process [GO:0032958]; inositol phosphate metabolic process [GO:0043647]; sensory perception of sound [GO:0007605] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22119861_describe the PPIP5K2's conformational dynamics, its unprecedented topological presentation of nucleotide and inositol phosphate, and the charge balance that facilitates partly associative in-line phosphoryl transfer 23240582_the specificity constants for PPIP5K2 revise upwards by one-to-two orders of magnitude the inherent catalytic activities of this enzyme, and we show its equilibrium point favours 80-90% depletion of InsP/-InsP. 26084399_The degree of nuclear localization of hPPIP5K2 was increased when S1006 was rendered non-phosphorylatable by its mutation to Ala. 26204995_SEZ6L, HISPPD1, FEZF1, SAMD11 gene variants may be associated with autism spectrum disorder. 28126903_This study characterized kinetic properties of the bifunctional inositol pyrophosphate 5-diphosphoinositol 1,2,3,4,6-pentakisphosphatekinase/inositol pyrophosphate, 1,5-bisdiphosphoinositol 2,3,4,6-tetrakisphosphate phosphatase activities of full-length diphosphoinositol pentakisphosphate kinase 1 and 2. 29590114_demonstration that PPIP5K2 has a role in hearing in humans indicates that PP-IP signaling is important to hair cell maintenance and function within inner ear 30956131_Data suggest that the enzyme-substrate forces are predictive of the various diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) catalytic activities. 31852976_PPIP5K2 and PCSK1 are Candidate Genetic Contributors to Familial Keratoconus. 34645979_PPIP5K2 promotes colorectal carcinoma pathogenesis through facilitating DNA homologous recombination repair. | ENSMUSG00000040648 | Ppip5k2 | 2.170400e+02 | 0.7177833 | -0.478379667 | 0.4087095 | 1.333265e+00 | 0.2482251174 | 0.79042389 | No | Yes | 1.586368e+02 | 40.776749 | 2.014416e+02 | 52.885949 | |
| ENSG00000145730 | 5066 | PAM | protein_coding | P19021 | FUNCTION: Bifunctional enzyme that catalyzes the post-translational modification of inactive peptidylglycine precursors to the corresponding bioactive alpha-amidated peptides, a terminal modification in biosynthesis of many neural and endocrine peptides (PubMed:12699694). Alpha-amidation involves two sequential reactions, both of which are catalyzed by separate catalytic domains of the enzyme. The first step, catalyzed by peptidyl alpha-hydroxylating monooxygenase (PHM) domain, is the copper-, ascorbate-, and O2- dependent stereospecific hydroxylation (with S stereochemistry) at the alpha-carbon (C-alpha) of the C-terminal glycine of the peptidylglycine substrate (PubMed:12699694). The second step, catalyzed by the peptidylglycine amidoglycolate lyase (PAL) domain, is the zinc-dependent cleavage of the N-C-alpha bond, producing the alpha-amidated peptide and glyoxylate (PubMed:12699694). Similarly, catalyzes the two-step conversion of an N-fatty acylglycine to a primary fatty acid amide and glyoxylate (By similarity). {ECO:0000250|UniProtKB:P14925, ECO:0000269|PubMed:12699694}. | Alternative splicing;Calcium;Cleavage on pair of basic residues;Copper;Cytoplasmic vesicle;Disulfide bond;Glycoprotein;Lipid metabolism;Lyase;Membrane;Metal-binding;Monooxygenase;Multifunctional enzyme;Oxidoreductase;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal;Sulfation;Transmembrane;Transmembrane helix;Vitamin C;Zinc | This gene encodes a multifunctional protein. The encoded preproprotein is proteolytically processed to generate the mature enzyme. This enzyme includes two domains with distinct catalytic activities, a peptidylglycine alpha-hydroxylating monooxygenase (PHM) domain and a peptidyl-alpha-hydroxyglycine alpha-amidating lyase (PAL) domain. These catalytic domains work sequentially to catalyze the conversion of neuroendocrine peptides to active alpha-amidated products. Alternative splicing results in multiple transcript variants, at least one of which encodes an isoform that is proteolytically processed. [provided by RefSeq, Jan 2016]. | hsa:5066; | cell surface [GO:0009986]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; neuron projection [GO:0043005]; perikaryon [GO:0043204]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; secretory granule membrane [GO:0030667]; trans-Golgi network [GO:0005802]; transport vesicle membrane [GO:0030658]; calcium ion binding [GO:0005509]; copper ion binding [GO:0005507]; identical protein binding [GO:0042802]; L-ascorbic acid binding [GO:0031418]; peptidylamidoglycolate lyase activity [GO:0004598]; peptidylglycine monooxygenase activity [GO:0004504]; protein kinase binding [GO:0019901]; zinc ion binding [GO:0008270]; central nervous system development [GO:0007417]; fatty acid primary amide biosynthetic process [GO:0062112]; heart development [GO:0007507]; lactation [GO:0007595]; limb development [GO:0060173]; long-chain fatty acid metabolic process [GO:0001676]; maternal process involved in female pregnancy [GO:0060135]; odontogenesis [GO:0042476]; ovulation cycle process [GO:0022602]; peptide amidation [GO:0001519]; protein amidation [GO:0018032]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of protein secretion [GO:0050708]; regulation of transcription by RNA polymerase II [GO:0006357]; response to copper ion [GO:0046688]; response to estradiol [GO:0032355]; response to glucocorticoid [GO:0051384]; response to hypoxia [GO:0001666]; response to pH [GO:0009268]; response to xenobiotic stimulus [GO:0009410]; response to zinc ion [GO:0010043]; toxin metabolic process [GO:0009404] | 16107699_nuclear retention of PAM mRNA is lost upon expressing the La proteins that lack a conserved nuclear retention element, suggesting a direct association between PAM mRNA and La protein in vivo 20351714_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22080626_Data indicate that catalytic inactivation of PHM caused by pH changes is accompanied by structural change between two states of the protein involving strong Cu-S interaction that does not involve M314. 22554821_Detail the production of the catalytic core of human peptidylglycine alpha-hydroxylating monooxygenase (hPHMcc) in Escherichia coli possessing a N-terminal fusion to thioredoxin (Trx). 24464100_Two missense variants in PAM, encoding p.Asp563Gly (frequency of 4.98%) and p.Ser539Trp (frequency of 0.65%), confer moderately higher risk of type 2 diabetes (OR = 1.23, P = 3.9 x 10(-10) and OR = 1.47, P = 1.7 x 10(-5), respectively) 26296884_Oxygen Sensitivity of the Peptidylglycine alpha-Amidating Monooxygenase (PAM) in Neuroendocrine Cells 26879543_PAM expression is increased in the secretory pathway of differentiated neurons. 26982589_Data suggest that His108 and a substrate molecule are involved in the reductive pathway while His172 and Tyr79 are important in the catalytic pathway in the copper-centered electron transfer catalyzed by peptidylglycine monooxygenase. 28377049_The ancient ability of PAM to localize to ciliary membranes, which release bioactive ectosomes, may be related to its ability to accumulate in intralumenal vesicles and exosomes 29162152_PAM single nucleotide polymorphism rs13175330 is associated with hypertension and insulin resistance in a Korean population. 29997255_PAM polyubiquitinates NMNAT2 and regulates NMNAT2 protein stability and degradation by the proteasome. 30054598_A role for PAM in beta-cell function. 30054598_The T2D risk-associated rs35658696 (p.Asp563Gly) allele of PAM confers decreased PAM expression in human islets. 30054598_The T2D risk-associated rs35658696 (p.Asp563Gly) allele of PAM confers reduced amidating activity. 30054598_siRNA-mediated knockdown of PAM in EndoC-BetaH1 cells caused reductions in insulin secretion and content. These effects were also observed in primary islets from human donors heterozygous for the T2D risk-associated rs35658696 variant. 31984442_PAM haploinsufficiency does not accelerate the development of diet- and human IAPP-induced diabetes in mice. 33197464_Mitochondrial Oxidative Stress Induces Rapid Intermembrane Space/Matrix Translocation of Apurinic/Apyrimidinic Endonuclease 1 Protein through TIM23 Complex. | ENSMUSG00000026335 | Pam | 1.087133e+03 | 0.8603042 | -0.217081163 | 0.2865053 | 5.807959e-01 | 0.4460005054 | 0.85808635 | No | Yes | 1.125779e+03 | 194.146206 | 1.006546e+03 | 178.064096 | |
| ENSG00000145833 | 9879 | DDX46 | protein_coding | Q7L014 | FUNCTION: Plays an essential role in splicing, either prior to, or during splicing A complex formation. {ECO:0000269|PubMed:12234937}. | 3D-structure;ATP-binding;Acetylation;Coiled coil;Helicase;Hydrolase;Isopeptide bond;Lipoprotein;Membrane;Myristate;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes a member of the DEAD box protein family. DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure, such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. The protein encoded by this gene is a component of the 17S U2 snRNP complex; it plays an important role in pre-mRNA splicing. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jul 2014]. | hsa:9879; | Cajal body [GO:0015030]; fibrillar center [GO:0001650]; membrane [GO:0016020]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; mRNA splicing, via spliceosome [GO:0000398] | 12234937_characterization of protein associated with the human 17S U2 snRNP and one of its stable subunits, SF3b; results indicate that Prp5 plays an important role in pre-mRNA splicing, acting during or prior to prespliceosome assembly 14713954_Prp5 forms a bridge between U1 and U2 Small Nuclear Ribonucleoproteins at the time of pre-spliceosome formation 19902070_Short-term exercise resulted in a significant increase of mRNA expression of genes encoding proteins involved in the formation of precatalytic splisosome: DDX46. 25680556_DDX46 is critical for colorectal cancer (CRC) cell proliferation and is a potential therapeutic target for CRC treatment. 27176873_DDX46 is critical for ESCC cells proliferation. 27697093_Knockdown of DDX46 inhibited osteosarcoma cell proliferation. 33000271_DDX46 silencing inhibits cell proliferation by activating apoptosis and autophagy in cutaneous squamous cell carcinoma. 33347858_Knockdown of DDX46 suppresses the proliferation and invasion of gastric cancer through inactivating Akt/GSK-3beta/beta-catenin pathway. | ENSMUSG00000021500 | Ddx46 | 8.824676e+02 | 1.0520744 | 0.073236708 | 0.3158684 | 5.349559e-02 | 0.8170886317 | 0.96398728 | No | Yes | 8.334980e+02 | 144.878845 | 8.522361e+02 | 151.967233 | |
| ENSG00000145860 | 153830 | RNF145 | protein_coding | Q96MT1 | FUNCTION: E3 ubiquitin ligase that catalyzes the direct transfer of ubiquitin from E2 ubiquitin-conjugating enzyme to a specific substrate. In response to bacterial infection, negatively regulates the phagocyte oxidative burst by controlling the turnover of the NADPH oxidase complex subunits. Promotes monoubiquitination of CYBA and 'Lys-48'-linked polyubiquitination and degradation of CYBB NADPH oxidase catalytic subunits, both essential for the generation of antimicrobial reactive oxygen species. Involved in the maintenance of cholesterol homeostasis. In response to high sterol concentrations ubiquitinates HMGCR, a rate-limiting enzyme in cholesterol biosynthesis, and targets it for degradation. The interaction with INSIG1 is required for this function. In addition, triggers ubiquitination of SCAP, likely inhibiting its transport to the Golgi apparatus and the subsequent processing/maturation of SREBPF2, ultimately down-regulating cholesterol biosynthesis. {ECO:0000250|UniProtKB:Q5SWK7}. | Alternative splicing;Endoplasmic reticulum;Membrane;Metal-binding;Reference proteome;Transferase;Transmembrane;Transmembrane helix;Ubl conjugation pathway;Zinc;Zinc-finger | hsa:153830; | endomembrane system [GO:0012505]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270] | 33727652_Genetic variation of RNF145 gene and blood lipid levels in Xinjiang population, China. | ENSMUSG00000019189 | Rnf145 | 1.749670e+03 | 0.7815538 | -0.355582818 | 0.2812774 | 1.613252e+00 | 0.2040352300 | 0.78566520 | No | Yes | 1.373997e+03 | 161.344513 | 1.821384e+03 | 218.926290 | ||
| ENSG00000145996 | 54901 | CDKAL1 | protein_coding | Q5VV42 | FUNCTION: Catalyzes the methylthiolation of N6-threonylcarbamoyladenosine (t(6)A), leading to the formation of 2-methylthio-N6-threonylcarbamoyladenosine (ms(2)t(6)A) at position 37 in tRNAs that read codons beginning with adenine. {ECO:0000250|UniProtKB:Q91WE6}. | 4Fe-4S;Alternative splicing;Diabetes mellitus;Endoplasmic reticulum;Iron;Iron-sulfur;Membrane;Metal-binding;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine;Transferase;Transmembrane;Transmembrane helix;tRNA processing | The protein encoded by this gene is a member of the methylthiotransferase family. The function of this gene is not known. Genome-wide association studies have linked single nucleotide polymorphisms in an intron of this gene with susceptibilty to type 2 diabetes. [provided by RefSeq, May 2010]. | hsa:54901; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; rough endoplasmic reticulum [GO:0005791]; 4 iron, 4 sulfur cluster binding [GO:0051539]; metal ion binding [GO:0046872]; N6-threonylcarbomyladenosine methylthiotransferase activity [GO:0035598]; tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase [GO:0061712]; maintenance of translational fidelity [GO:1990145]; tRNA methylthiolation [GO:0035600] | 17460697_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17460697_Single Nucleotide polymorphism in CDK5 regulatory subunit associated protein 1-like 1 is associated with type 2 diabetes 17463246_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17463248_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17463249_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 17804762_CDKAL1 and HHEX/IDE diabetes-associated alleles are associated with decreased pancreatic beta-cell function, including decreased beta-cell glucose sensitivity that relates insulin secretion to plasma glucose concentration. 17804762_Observational study of gene-disease association. (HuGE Navigator) 17928989_Observational study of gene-disease association. (HuGE Navigator) 17993580_Observational study of gene-disease association. (HuGE Navigator) 18162508_Observational study of gene-disease association. (HuGE Navigator) 18162508_The association of 6 loci with type 2 diabetes risk in Japanese patients is reported. 18210030_Observational study of gene-disease association. (HuGE Navigator) 18252897_Observational study of gene-disease association. (HuGE Navigator) 18264689_Diabetes-associated variants in TCF7L2 and CDKAL1 impair insulin secretion and conversion of proinsulin to insulin. 18264689_Observational study of gene-disease association. (HuGE Navigator) 18285412_CDKAL1 is likely to increase the risk of type 2 diabetes by impairing insulin secretion. 18285412_Observational study of gene-disease association. (HuGE Navigator) 18426861_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18437351_Observational study of gene-disease association. (HuGE Navigator) 18461161_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18469204_Data confirmed the associations of single nucleotide polymorphisms in CDKAL1 with risk for type 2 diabetes in Asians. 18469204_Observational study of gene-disease association. (HuGE Navigator) 18477659_Observational study of gene-disease association. (HuGE Navigator) 18516622_Observational study of gene-disease association. (HuGE Navigator) 18516622_One SNP, rs7754840 in the CDKAL1 gene, presented a significantly stronger effect in the Ashkenazi Jewish population as compared to the general Caucasian population for type 2 diabetes susceptibility. 18544707_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18587394_Genome-wide association study of gene-disease association. (HuGE Navigator) 18591388_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18597214_Gene variants of CDKAL1, PPARG, IGF2BP2, HHEX, TCF7L2, and FTO predispose to type 2 diabetes in the German KORA 500 K study population. 18618095_Observational study of gene-disease association. (HuGE Navigator) 18618095_Variants of CDKAL1 and IGF2BP2 attenuate the first phase of glucose-stimulated insulin secretion but show no effect on the second phase of insulin secretion in hyperglycmia and type 2 diabetes. 18633108_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18633108_The results indicate that in Chinese Hans, common variants in CDKAL1 loci independently or additively contribute to type 2 diabetes risk, likely mediated through beta-cell dysfunction. 18694974_Study show that polymorphisms in CDKAL1 were associated with type 2 diabetes risk in the studied population. 18719881_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18753662_The underlying mechanisms linking CDKAL1, glutamate decarboxylase, and insulin secretion are unclear. A recent case-control study found no association of variation in CDKAL1 with type 1 diabete.s 18766326_Positive association between single nucleotide polymorphisms in this gene with type 2 diabetes in Han Chinese. 18923449_ADAM33, CDKAL1, and PTPN22 may be true psoriasis-risk genes 18923449_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18984664_Observational study of gene-disease association. (HuGE Navigator) 18991055_Observational study of gene-disease association. (HuGE Navigator) 18991055_Single nucleotide polymorphism in CDKAL1 is associated with type 2 diabetes. 19002430_Observational study of gene-disease association. (HuGE Navigator) 19008344_Data show that SNPs in CDKAL1 did not confer a significant risk for type 2 diabetes in Pima Indians. 19008344_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19020323_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19020324_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19033397_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19033397_Type 2 diabetes susceptibility of CDKAL1 was confirmed in Japanese. 19068216_Observational study of gene-disease association. (HuGE Navigator) 19082521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19139842_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19172244_Observational study of gene-disease association. (HuGE Navigator) 19174780_Observational study of gene-disease association. (HuGE Navigator) 19225753_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19228808_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19247372_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19258404_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19258437_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19279076_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19324937_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19380854_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19401414_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19422935_Observational study of gene-disease association. (HuGE Navigator) 19422935_The presence of a C-allele at the CDKAL1 single nucleotide polymorphism rs6908425 and the absence of NOD2 variants were independently associated with development of perianal fistula 19502414_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19587699_CDKAL1 alleles may confer susceptibility to clinically distinct disorders through differential effects on disease-specific cell types. 19587699_Observational study of gene-disease association. (HuGE Navigator) 19592620_Association between lower birth weight and type 2 diabetes risk-conferring alleles at the CDKAL1 locus. 19592620_Observational study of gene-disease association. (HuGE Navigator) 19602701_Meta-analysis and HuGE review of gene-disease association. (HuGE Navigator) 19622614_Observational study of gene-disease association. (HuGE Navigator) 19718565_Observational study of gene-disease association. (HuGE Navigator) 19718565_Single nucleotide polymorphisms in CDKAL1 have no association with polycystic ovary syndrome or related clinical features in Chinese women. 19720844_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19734900_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19741166_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19741467_Observational study of gene-disease association. (HuGE Navigator) 19760754_Observational study of gene-disease association. (HuGE Navigator) 19794065_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19808892_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19862325_Observational study of gene-disease association. (HuGE Navigator) 19862325_there is an association between PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 and type 2 diabetes in the Chinese population 19892838_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19933996_Observational study of gene-disease association. (HuGE Navigator) 20014019_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20043145_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20075150_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20161779_Observational study of gene-disease association. (HuGE Navigator) 20161779_Studies identified significant association between variants in CDKN2A/B, CDKAL1 and TCF7L2, and type 2 diabetes in a Han Chinese cohort, indicating these genes as strong candidates conferring susceptibility to type 2 diabetes across different ethnicities. 20203524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20215779_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20384434_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20424228_Observational study of gene-disease association. (HuGE Navigator) 20460429_Observational study of gene-disease association. (HuGE Navigator) 20460429_Type 2 diabetes susceptibility alleles at CDKAL1 are associated with low body mass index at 8 years in children who were born large for gestational age. 20490451_Data report a novel association between the fetal ADCY5 type 2 diabetes risk allele and decreased birthweight, and confirm in meta-analyses associations between decreased birthweight and the type 2 diabetes risk alleles of HHEX-IDE and CDKAL1. 20490451_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20503258_Observational study of gene-disease association. (HuGE Navigator) 20509872_Observational study of gene-disease association. (HuGE Navigator) 20550665_Observational study of gene-disease association. (HuGE Navigator) 20568056_Meta-analysis of gene-disease association. (HuGE Navigator) 20568056_there are significant associations between CDKAL1 polymorphisms and type 2 diabetes [meta-analysis] 20580033_Observational study of gene-disease association. (HuGE Navigator) 20587610_Observational study of gene-disease association. (HuGE Navigator) 20616309_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20712903_Observational study of gene-disease association. (HuGE Navigator) 20802253_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20816152_Observational study of gene-disease association. (HuGE Navigator) 20847106_Interaction between the CDKAL1 polymorphism and dietary energy intake influences the dysglycemic phenotype leading to MetS, possibly through impaired insulin secretion. The CDKAL1 polymorphism may be a marker for MetS in the Japanese population. 20847106_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20879858_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20886065_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20889853_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20929593_Observational study of gene-disease association. (HuGE Navigator) 21072187_Observational study of gene-disease association. (HuGE Navigator) 21368910_Single nucleotide polymorphism (SNP) analysis revealed that the sequence variant (rs5015480) near HHEX and two SNPs (rs7756992 and rs9465871) in CDKAL1 were associated with the susceptibility of type 2 diabetes mellitus in females, but not in males. 21416855_no relationship of CDKAL1 and KCNQ1 polymorphisms to the earlier onset of type 2 diabetes was observed 21611789_CDKAL1 is involved in the pathogenesis of T2 diabetes through impaired beta-cell function. 21643948_A significant association between Type 2 Diabetes Mellitus, an increased Fasting Plasma Glucose and rs7754840 at CDKAL1 in lean Han Chinese. 21908934_Variants in cdkal1 gene have been reproducibly associated with decreased first-phase insulin secretion and development of type 2 diabetes. 22096510_Six SNP(rs7754840 in CDKAL1, rs391300 in SRR, rs2383208 in CDKN2A/2B, rs4402960 in IGF2BP2, rs10830963 in MTNR1B, rs4607517 in GCK)risk alleles of type 2 diabetes were associated with GDM in pregnant Chinese women. 22119613_CDKAL1 rs7754840 and rs7756992, but not CDKN2A/2B rs10811661, are associated with T2DM in Lebanese. 22290723_CDKAL1 might influence the level of glycosylated hemoglobin 22344221_Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. 22437209_Our findings indicated that genetic variants of CDKAL1 and VEGFA on chromosome 6 may contribute to T2D risk in Chinese population. 22443257_None of the 12 SNPs in the six genes (KCNJ11, TCF7L2, SLC30A8, HHEX, FTO and CDKAL1) uncovered in the genome-wide association studies were associated with polycystic ovary syndrome. 22487833_The associations between SNPs of TCF7L2, CDKAL1, SLC30A8 and HHEX and the development of DR and DN. 22923468_rs7754840 (CDKAL1) was associated in the nonobese type 2 diabetic subgroup, and for rs7903146 (TCF7L2), association was observed for early-onset type 2 diabetes. 23048041_CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) is a tail-anchored protein in the endoplasmic reticulum (ER) of insulinoma cells. 23173044_CDKAL1 may affect such compensatory mechanisms regulating glucose homeostasis through interaction with diet 23670970_TCF7L2 was replicated in this study (P = 0.004; combined analysis P = 3.8 x 10(-6)), and type 2 diabetes SNPs at or near CDKAL1, CDKN2A/B, and IGF2BP2 were associated with CFRD 23840313_Substantiation of the roles of CAMK1D and CDKAL1 in gluconeogenic and glyconeogenic pathways in primary human hepatocytes. 24012816_Evidence for a significant contribution of CDKN2A/B gene rs10811661 and CDKAL1 gene rs7756992 and rs10946398 to type 2 diabetes.[meta-analysis] 24013783_Five of these 14 loci had single-nucleotide polymorphisms in European studies while the other nine were different. Further stepwise conditional analysis identified seven secondary signals and an independent novel locus at the 3' end of CDKAL1. 24112421_Data indicate the potential importance of CDKAL1 protein and homeobox protein HHEX in glucose homeostasis in this Alaska Native population with a low prevalence of type 2 diabetes (T2D). 24185407_CDKAL1 gene rs7756992 A/G polymorphism was significantly associated with T2DM. The person with G allele of CDKAL1 gene rs7756992 A/G polymorphism might be predisposed to T2DM. 24636221_rs7756992 of CDKAL1 gene have a protective effect against diabetic nephropathy. 24695378_This study reports association of CDKAL1-related SNPs with insulin resistance, a clinical marker related to type 2 diabetes in a cross-sectional cohort of Greek children and adolescents of European descent. 24760768_CDKAL1-v1-mediated suppression of CDKAL1 might underlie the pathogenesis of type 2 diabetes in individuals carrying the risk Single-nucleotide polymorphisms. 24898818_Meta-analysis indicated significant association between the IGF2BP2 rs4402960 and CDKAL1 rs7756992 polymorphisms and increased risk of diabetes in Arab populations.[meta-analysis] 25222615_Variants in CDKAL1 are associated with glucose-induced GIP and insulin response. 25370040_We observed novel selection signals in CDKAL1 and NEGR1, well-known diabetes and obesity susceptibility genes 25483131_The results support a central role of CDKAL1 and TCF7L2 in T2DM susceptibility in Southwest Asian populations and provide a plausible component for understanding molecular mechanisms involved in the disease. 25634229_Provide evidence against a role for dysregulated expression of CDKAL1-v1 in mediating the association between intronic SNPs in CDKAL1 and susceptibility to type 2 diabetes. 25723968_The rs7754840 and rs7756992 SNPs of the CDKAL1 gene were found to be associated with Gestational Diabetes Mellitus in this south Indian population. 25785549_study found SNP rs7754840 in CDKAL1, rs864745 in JAZF1, and rs35767 in IGF1 might serve as potential susceptibility loci for type 2 diabetes in the Uyghur population 26119585_rs10946398 associated with markers of impaired insulin secretion 26168825_study identified the association between type 2 diabetes risk variants in CDKAL1 and birthweight in Chinese Han individuals, and the carrier of risk allele within SRR had the trend of reduced birthweight. 26563541_our data suggested that CDKAL1 gene variants have a significant effect on the response to anti-TNF therapies among Psoriasis patients 26830138_family-based GWAS of imputed SNPs revealed novel genomic variants in (or near) PTPRG, OSBPL6, and PDCL3 that influence risk for Alzheimer's Disease. rs7609954 in the gene PTPRG, rs1347297 in the gene OSBPL6, and rs1513625 near PDCL3. In addition, rs72953347 in OSBPL6 and two SNPs in the gene CDKAL1 showed marginally significant association with LOAD (rs10456232, P-value=4.76 x 10-7; rs62400067, P-value=3.54 x 10-7). 26873362_We investigated the association between 8 single-nucleotide polymorphisms (SNPs) at 3 genetic loci (CDKAL1, CDKN2A/2B and FTO) with type 2 diabetes (T2D) in a Uyghur population 27049325_Risk alleles for 6 loci increased glucose levels from birth to 5 years of age (ADCY5, ADRA2A, CDKAL1, CDKN2A/B, GRB10, and TCF7L2 27377502_The multivariate logistic regression analysis with reference to both alleles and genotypes of CDKAL1 SNPs showed significant association, suggesting an important role for this gene in the T2DM pathophysiology. INTERPRETATION & CONCLUSIONS: A significant association was seen of all the three SNPs of CDKAL1 and CDKN2A/B genes with T2DM but none of the two SNPs of HHEX. 27936930_Our results suggest that rs6908425 in CDKAL1 is associated with the risk of developing SAPHO in Han Chinese populations. People who carry the risk allele T of rs6908425 might be more prone to developing SAPHO syndrome. 28406950_Study provides evidence that SNPs of JMJD1C and KCNQ1 are prospectively associated with the risk of type 2 diabetes (T2D) in Korean population. Additionally, CDKAL1 may not be associated with T2D onset over the age of 40. 28502787_More CDKAL1 variants are required to validate the association between CDKAL1 and gestational glycemic traits. 28538172_Forced MT1E expression rescues both hypersensitivity of CDKAL1 mutant cells to glycolipotoxicity and pancreatic beta-cell dysfunction in vitro and in vivo. 28821857_we identified CDKAL1 rs7756992 as a susceptibility locus for DR in a Chinese population with type 2 diabetes. 29372795_CDKAL1 gene is associated with development of type 2 diabetes. For the HHEX/IDE locus, such an association is absent. 29403086_chromosomal region 6p22.3 is a novel susceptibility locus for nsCL/P. The location of the risk variants within the CDKAL1 intronic sequence containing enhancer elements predicted to regulate the SOX4 transcription may suggest that SOX4, rather than CDKAL1, is a potential candidate gene for this craniofacial anomaly. 29544538_Non-significant association was seen in the two single nucleotide polymorphisms (SNPs) of CDKAL1 (rs7754840) and CDKN2A/2B (rs10811661) with gestational diabetes mellitus (GDM). 29674289_We identified and replicated genetic variants associated with cholesterol efflux capacity using a genome-wide association study-based approach. CDKAL1 variants showed correlations with cholesterol efflux capacity independent of HDL-cholesterol levels and other clinical characteristics. 29933462_The CDKAL1 rs9356744 T allele associated with a predisposition to obesity shows a protective effect on HbA1c, 2hPG, and prediabetes in Chinese; BMI is mediator of the association between the genetic variant and HbA1c, 2hPG, and prediabetes. 30185902_There is a significant association between CDKAL1 rs10946398 and type 2 diabetes among Taiwanese men and women. The CC genotype is a risk factor for type 2 diabetes in women with BMI >/= 24 kg/m2, as well as in men regardless of their BMI. The CA genotype appears to be a risk factor for T2D mainly in obese individuals. 30207601_Genetic risk scores were calculated by summing the risk alleles of 4 selected single nucleotide polymorphisms, CDKAL1 rs7754840 and rs9460546, HHEX rs5015480, and OAS3 30350806_significant risk association between rs9465871 polymorphism and obesity and development of T2DM in Egyptian children 31098383_there is an association between the CDKAL1 gene and risk of gestational diabetes mellitus in the Chinese population 31189758_Dysregulation of CDKAL1 is involved in the pathogenesis of ( Growth Hormone-producing pituitary adenomas GHPAs), and modulation of the proteostatic stress response might control CDKAL1 activity and facilitate treatment of GHPAs. 31639799_CDKAL1 rs35612982 (C/T) polymorphism, as a new polymorphism, was associated with the increased risk of T2D in the Han Chinese population. Moreover, the contribution of CDKAL1 polymorphisms to T2D risk seems to be associated with age, gender, BMI, smoking and drinking. 32228543_Loci near TMEM18 (rs6548238), CDKAL1 (rs7754840), and FAIM2 (rs7138803) may be associated with obesity-related indicators, and loci near TMEM18 (rs6548238) and FAIM2 (rs7138803) may increase susceptibility of concurrent type 2 diabetes associated with obesity. 32764395_Dietary Protein and Fat Intake Affects Diabetes Risk with CDKAL1 Genetic Variants in Korean Adults. 32791750_Association of the CDKAL1 polymorphism rs10946398 with type 2 diabetes mellitus in adults: A meta-analysis. 33558148_Interaction between the rs9356744 polymorphism and metabolic risk factors in relation to type 2 diabetes mellitus: The Cardiometabolic Risk in Chinese (CRC) Study. 33856697_The type 2 diabetes mellitus susceptibility gene CDKAL1 polymorphism is associated with depressive symptom in first-episode drug-naive schizophrenic patients. 34169461_Association of diabetes-related variants in ADCY5 and CDKAL1 with neonatal insulin, C-peptide, and birth weight. 34192303_Serum concentrations of SFAs and CDKAL1 single-nucleotide polymorphism rs7747752 are related to an increased risk of gestational diabetes mellitus. 34721291_CDK5 Regulatory Subunit-Associated Protein 1-Like 1 Gene Polymorphisms and Gestational Diabetes Mellitus Risk: A Trial Sequential Meta-Analysis of 13,306 Subjects. 34872638_The risk variant of CDKAL1 (rs7756992) impairs fasting glucose levels and insulin resistance improvements after a partial meal-replacement hypocaloric diet. 35360068_The CDKAL1 rs7747752-Bile Acids Interaction Increased Risk of Gestational Diabetes Mellitus: A Nested Case-Control Study. | ENSMUSG00000006191 | Cdkal1 | 4.281984e+02 | 0.8289863 | -0.270579798 | 0.2815330 | 9.064219e-01 | 0.3410655688 | 0.82750349 | No | Yes | 3.990960e+02 | 65.679797 | 3.578013e+02 | 60.540266 | |
| ENSG00000146350 | 221322 | TBC1D32 | protein_coding | Q96NH3 | FUNCTION: Required for high-level Shh responses in the developing neural tube. Together with CDK20, controls the structure of the primary cilium by coordinating assembly of the ciliary membrane and axoneme, allowing GLI2 to be properly activated in response to Shh signaling (By similarity). {ECO:0000250}. | Alternative splicing;Cell projection;Cilium;Cytoplasm;Developmental protein;Reference proteome | This gene encodes a TBC-domain containing protein. Studies of a similar protein in mouse and zebrafish suggest that the encoded protein is involved in sonic hedgehog signaling, and that it interacts with and stabilizes cell cycle-related kinase. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2014]. | hsa:221322; | cilium [GO:0005929]; cytoplasm [GO:0005737]; determination of left/right symmetry [GO:0007368]; embryonic digit morphogenesis [GO:0042733]; heart development [GO:0007507]; kidney development [GO:0001822]; lens development in camera-type eye [GO:0002088]; non-motile cilium assembly [GO:1905515]; protein localization to cilium [GO:0061512]; retinal pigment epithelium development [GO:0003406]; roof of mouth development [GO:0060021]; smoothened signaling pathway involved in dorsal/ventral neural tube patterning [GO:0060831] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20159594_Studies in mouse and zebrafish implicate that TBC1D32 is involved in sonic hedgehog signaling. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 24285566_study identified 2 cases with a severe ciliopathy phenotype consistent with oro-facio-digital syndrome type IX; the autozygome of each index harbored a single truncating variant and the affected genes (SCLT1 and TBC1D32/C6orf170) have roles in centrosomal biology and ciliogenesis; findings suggest a role of SCLT1 and TBC1D32 in ciliopathy pathogenesis 32060556_Loss-of-Function Variants in TBC1D32 Underlie Syndromic Hypopituitarism. 32573025_Confirming TBC1D32-related ciliopathy in humans. | ENSMUSG00000038122 | Tbc1d32 | 1.037621e+02 | 0.8351513 | -0.259890486 | 0.4975370 | 2.695880e-01 | 0.6036082935 | 0.90922991 | No | Yes | 7.330847e+01 | 22.341280 | 1.059784e+02 | 32.802646 | |
| ENSG00000146555 | 221935 | SDK1 | protein_coding | Q7Z5N4 | FUNCTION: Adhesion molecule that promotes lamina-specific synaptic connections in the retina. Expressed in specific subsets of interneurons and retinal ganglion cells (RGCs) and promotes synaptic connectivity via homophilic interactions. {ECO:0000250|UniProtKB:Q8AV58}. | Alternative splicing;Cell adhesion;Cell junction;Cell membrane;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Reference proteome;Repeat;Signal;Synapse;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the immunoglobulin superfamily. The protein contains six immunoglobulin-like domains and thirteen fibronectin type III domains. Fibronectin type III domains are present in both extracellular and intracellular proteins and tandem repeats are known to contain binding sites for DNA, heparin and the cell surface. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2016]. | hsa:221935; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; synapse [GO:0045202]; identical protein binding [GO:0042802]; behavioral response to cocaine [GO:0048148]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; regulation of dendritic spine development [GO:0060998]; retina layer formation [GO:0010842]; synapse assembly [GO:0007416] | 15213259_dysregulation of sdk-1 protein may play an important role in HIV-associated nephropathy pathogenesis 19851296_Observational study of gene-disease association. (HuGE Navigator) 19851296_SDK1 may be a susceptibility gene for hypertension in Japanese individuals, although the functional relevance of the identified polymorphism was not determined. 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20398908_Observational study of gene-disease association. (HuGE Navigator) 20562105_Data conclude that the up-regulation of sdk-1 in podocytes is an important pathogenic factor in glomerulosclerosis and that the mechanism involves disruption of the actin cytoskeleton possibly via alterations in MAGI-1 function. 23827383_Case-control studies reveal malignant mesothelioma risk associated with variants in the SDK1, CRTAM and RASGRF2 genes. 26463840_SDK1 could possibly serve as a candidate gene for alterations associated with asbestos exposure. | ENSMUSG00000039683 | Sdk1 | 9.869674e+02 | 1.2950307 | 0.372986339 | 0.3535153 | 1.083312e+00 | 0.2979578935 | 0.81383297 | No | Yes | 8.946783e+02 | 121.492228 | 6.275814e+02 | 87.683269 | |
| ENSG00000146670 | 113130 | CDCA5 | protein_coding | Q96FF9 | FUNCTION: Regulator of sister chromatid cohesion in mitosis stabilizing cohesin complex association with chromatin. May antagonize the action of WAPL which stimulates cohesin dissociation from chromatin. Cohesion ensures that chromosome partitioning is accurate in both meiotic and mitotic cells and plays an important role in DNA repair. Required for efficient DNA double-stranded break repair. {ECO:0000269|PubMed:15837422, ECO:0000269|PubMed:17349791, ECO:0000269|PubMed:21111234}. | Cell cycle;Cell division;Chromosome;Cytoplasm;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:113130; | chromatin [GO:0000785]; chromosome [GO:0005694]; chromosome, centromeric region [GO:0000775]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; protein-containing complex binding [GO:0044877]; cell division [GO:0051301]; double-strand break repair [GO:0006302]; mitotic cell cycle [GO:0000278]; mitotic chromosome condensation [GO:0007076]; mitotic metaphase plate congression [GO:0007080]; mitotic sister chromatid cohesion [GO:0007064]; positive regulation of exit from mitosis [GO:0031536]; regulation of cohesin loading [GO:0071922] | 17349791_data indicate that sororin interacts with chromatin-bound cohesin and functions during the establishment or maintenance of cohesion in S or G2 phase, respectively 17361102_Sororin protects centromeric cohesion in response to the spindle checkpoint, but prevents the removal of cohesion by a mechanism independent of the anaphase-promoting complex. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20551060_Transactivation of CDCA5 and its phosphorylation at Ser209 by ERK play an important role in lung cancer proliferation. 21115494_the interaction of the highly conserved motif at the C terminus of sororin with the cohesin complex is critical to its ability to mediate sister chromatid cohesion. 21987589_Sororin is phosphorylated by Cdk1/cyclin B at prophase and acts as a docking protein to bring Plk1 into proximity with SA2, resulting in the phosphorylation of SA2 and the removal of cohesin complexes from chromosomal arms. 22833568_Data show that the MEK-ERK pathway regulates cell cycle-related neuronal apoptosis (CRNA) by elevating the levels of cyclin D1, and the increase in cyclin D1 attenuates the activation of cyclin-dependent kinase 5 (cdk5) by its neuronal activator p35. 24098701_Targeting p35/Cdk5 signalling via CIP-peptide promotes angiogenesis in hypoxia. 25092791_We propose that the Prp19 complex and the splicing machinery contribute to the establishment of cohesion by promoting Sororin accumulation during S phase, and are, therefore, essential to the maintenance of genome stability. 25257309_A transcriptome-wide analysis revealed that SNW1 or PRPF8 depletion affects the splicing of specific introns in a subset of pre-mRNAs, including pre-mRNAs encoding the cohesion protein sororin and the APC/C subunit APC2. 25608232_the C-terminus of Sororin functions as an anchor binding to SA2, which facilitates other conserved motifs on Sororin to interact with other proteins to regulate sister chromatid cohesion and separation. 26177583_phosphorylation plays unexpected roles in regulating the subcellular localization of Sororin. 26497678_The results suggest that CDCA5 functions as a critical gene supporting oral squamous cell carcinoma progression and that targeting CDCA5 may be a useful therapeutic strategy for oral squamous cell carcinoma. 26951638_Sororin participates in the regulation of centromeric cohesion during meiosis in collaboration with SGO2-PP2A. 29326043_Silencing of CDCA5 suppresses proliferation of gastric cancer cells by inducing G1-phase arrest via downregulating CCNE1. 29383807_These results suggest that satellite I RNA plays a role in stabilizing RBMX and Sororin in the ncRNP complex to maintain proper sister chromatid cohesion. 29452217_high CDCA5 expression was also an independent factor of disease-free survival for patients with acral melanoma 29724914_Rec8-Stag3 cohesin is shown to be susceptible to Wapl-dependent ring opening and sororin-mediated protection. 30015982_High CDCA5 expression is associated with hepatocellular carcinoma. 30497429_These findings suggest that CDCA5 expression is associated with poor prognosis in patients with hepatocellular carcinoma. 30657957_We define a molecular pathway through which SLU7 keeps in check the generation of truncated forms of the splicing factor SRSF3 (SRp20) (SRSF3-TR). Behaving as dominant negative, or by gain-of-function, SRSF3-TR impair the correct splicing and expression of the splicing regulator SRSF1 (ASF/SF2) and the crucial SCC protein sororin. 31068217_LMTK2 binds to KLC1 to direct axonal transport of p35 and its loss may contribute to Alzheimer's disease. 32694239_Higher expression of cell division cycle-associated protein 5 predicts poorer survival outcomes in hepatocellular carcinoma. 32759885_Cyclin-Dependent Kinase 1 (CDK1) is Co-Expressed with CDCA5: Their Functions in Gastric Cancer Cell Line MGC-803. 33639053_Downregulation of CDCA5 Can Inhibit Cell Proliferation, Migration, and Invasion, and Induce Apoptosis of Prostate Cancer Cells. 33650660_CDCA5 promotes the progression of prostate cancer by affecting the ERK signalling pathway. 33770322_LINC01515 promotes nasopharyngeal carcinoma progression by serving as a sponge for miR-325 to up-regulate CDCA5. 34077004_CDCA5 is negatively regulated by miR-326 and boosts ovarian cancer progression. | ENSMUSG00000024791 | Cdca5 | 8.811137e+03 | 1.1330939 | 0.180267400 | 0.2987081 | 3.675186e-01 | 0.5443599759 | 0.89063840 | No | Yes | 9.082143e+03 | 736.545613 | 7.392497e+03 | 615.272078 | ||
| ENSG00000146776 | 222255 | ATXN7L1 | protein_coding | Q9ULK2 | Alternative splicing;Reference proteome | hsa:222255; | ENSMUSG00000020564 | Atxn7l1 | 2.150576e+02 | 1.0611819 | 0.085671953 | 0.3129203 | 7.404831e-02 | 0.7855311411 | 0.95754084 | No | Yes | 1.974469e+02 | 28.098155 | 1.849092e+02 | 27.077944 | |||||
| ENSG00000146909 | 64434 | NOM1 | protein_coding | Q5C9Z4 | FUNCTION: Plays a role in targeting PPP1CA to the nucleolus. {ECO:0000269|PubMed:17965019}. | Nucleus;Phosphoprotein;Reference proteome | Proteins that contain MIF4G (middle of eIF4G (MIM 600495)) and/or MA3 domains, such as NOM1, function in protein translation. These domains include binding sites for members of the EIF4A family of ATP-dependent DEAD box RNA helicases (see EIF4A1; MIM 602641) (Simmons et al., 2005 [PubMed 15715967]).[supplied by OMIM, Mar 2008]. | hsa:64434; | nucleolus [GO:0005730]; RNA binding [GO:0003723]; hair follicle maturation [GO:0048820]; ribosomal small subunit biogenesis [GO:0042274] | 17965019_Data demonstrate that NOM1 can target PP1 to the nucleolus and show NOM1 nucleolar localization sequence are required for this targeting activity and conclude that NOM1 is a PP1 nucleolar targeting subunit, the first identified in eukaryotic cells. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21576267_demonstrate direct physical interactions between yeast Sgd1p and Fal1p, and between their human orthologs (NOM1 and eIF4AIII) in vitro and in vivo, identifying human NOM1 as a missing eIF4G-like interacting partner of eIF4AIII 30639046_Results are consistent with a physiologically-relevant interplay between the nuclear IGF1 signaling pathway and nucleolar protein NOM1. | ENSMUSG00000001569 | Nom1 | 2.005993e+03 | 0.9220737 | -0.117046044 | 0.2945522 | 1.573614e-01 | 0.6915975284 | 0.93376105 | No | Yes | 1.828015e+03 | 252.775983 | 2.145524e+03 | 304.023594 | |
| ENSG00000147050 | 7403 | KDM6A | protein_coding | O15550 | FUNCTION: Histone demethylase that specifically demethylates 'Lys-27' of histone H3, thereby playing a central role in histone code (PubMed:17851529, PubMed:17713478, PubMed:17761849). Demethylates trimethylated and dimethylated but not monomethylated H3 'Lys-27' (PubMed:17851529, PubMed:17713478, PubMed:17761849). Plays a central role in regulation of posterior development, by regulating HOX gene expression (PubMed:17851529). Demethylation of 'Lys-27' of histone H3 is concomitant with methylation of 'Lys-4' of histone H3, and regulates the recruitment of the PRC1 complex and monoubiquitination of histone H2A (PubMed:17761849). Plays a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression (By similarity). {ECO:0000250|UniProtKB:O70546, ECO:0000269|PubMed:17713478, ECO:0000269|PubMed:17761849, ECO:0000269|PubMed:17851529, ECO:0000269|PubMed:18003914}. | 3D-structure;Chromatin regulator;Dioxygenase;Iron;Mental retardation;Metal-binding;Methylation;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Zinc | This gene is located on the X chromosome and is the corresponding locus to a Y-linked gene which encodes a tetratricopeptide repeat (TPR) protein. The encoded protein of this gene contains a JmjC-domain and catalyzes the demethylation of tri/dimethylated histone H3. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Apr 2014]. | hsa:7403; | histone methyltransferase complex [GO:0035097]; MLL3/4 complex [GO:0044666]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin DNA binding [GO:0031490]; histone demethylase activity [GO:0032452]; histone H3-tri/di-methyl-lysine-27 demethylase activity [GO:0071558]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; chromatin remodeling [GO:0006338]; heart development [GO:0007507]; histone H3-K27 demethylation [GO:0071557]; histone H3-K4 methylation [GO:0051568]; regulation of gene expression [GO:0010468] | 17500065_UTX associates with MLL3- and MLL4-containing histone H3 K4 methyltransferase complex(es) that also include ASH2L, RBBP5, WDR5, hDPY-30, PTIP, PA1, NCOA6. 17713478_the human JmjC-domain-containing proteins UTX and JMJD3 demethylate tri-methylated Lys 27 on histone H3 17761849_study shows that UTX is a di- and trimethyl H3K27 demethylase; results suggest a concerted mechanism for transcriptional activation in which cycles of H3K4 methylation by MLL2/3 are linked with the demethylation of H3K27 through UTX 17851529_critical role for UTX in regulating H3K27 methylation at the HOX gene loci and in animal posterior development 18003914_The JmjC domain-containing protein UTX specifically demethylates mono-, di- and trimethylated K27 of histone H3 in vitro. 18003914_UTX and JMJD3 may function as H3K27 demethylases in vivo 19330029_Here, we describe inactivating somatic mutations in the histone lysine demethylase gene UTX, pointing to histone H3 lysine methylation deregulation in multiple tumor types 20054297_identification of inactivating mutations in two genes--SETD2, a histone H3 lysine 36 methyltransferase, and JARID1C, a histone H3 lysine 4 demethylase--as well as mutations in the histone H3 lysine 27 demethylase, UTX in clear cell renal cell carcinoma 20123895_UTX removes H3K27me3 and maintains expression of several RB-binding proteins, enabling cell cycle arrest 20442750_Observational study of gene-disease association. (HuGE Navigator) 21209387_The role of the H3K27 demethylases Jmjd3 and UTX in gene expression, is discussed. 21245294_KDM6A- and KDM6B-responsive Homeobox genes are expressed at significantly higher levels, suggesting that HPV16 E7 results in reprogramming of host epithelial cells 21515470_UXT is a potential interactor of HBV Pol. 21575637_inhibition measurements showed significant selectivity between KDM4C and KDM6A 21828135_Novel UTX, DNMT3A, and EZH2 mutations were found in 8%, 10%, and 5.5% of patients with chronic myelomonocytic leukemia. 21841772_H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERalpha ligand dependency 21865393_Correlating with the loss of H3K27me3, human papillomavirus 16 E6/E7-expressing cells exhibited derepression of specific EZH2-, KMD6A-, and BMI1-targeted HOX genes. 22002947_clarified how UTX discriminates H3K27me3/2 from the other methyllysines with distinct roles 22197486_This study identifies KDM6A mutations as another cause of Kabuki syndrome and highlights the growing role of histone methylases and histone demethylases in multiple-congenital-anomaly and intellectual-disability syndromes. 22306297_demonstrate that UTX directly associates with the promoters of the Mll1, Runx1, and Scl genes and modulate their transcription by controlling H3K27me3 marks on respective promoter regions. 22589717_PAN RNA interacts with demethylases JMJD3 and UTX, and the histone methyltransferase MLL2 22801502_identification of Utx as a novel mediator with distinct functions during the re-establishment of pluripotency and germ cell development 22840376_Microdeletions and microduplications have not been identified in the MLL2 and KDM6A genes of a large cohort of patients with Kabuki syndrome. 22907667_KDM6A contributes to the activation of WNT3 and DKK1 at different differentiation stages when WNT3 and DKK1 are required for mesendoderm and definitive endoderm differentiation. 23184418_This study demonistrated that KDM6A mutations were most commonly identified in subgroups in medulloblastoma. 23266085_KDM6A is overexpressed in breast cancer patients with an unfavorable prognosis (mortality at 1 year, p=8.65E-7). 23365460_UTX regulates stem cell migration and hematopoiesis. 23527641_UTX histone demethylase plays important functional role in epigenetic alteration of HOX clusters during retinoic acid-induced neural differentiation. 23644518_PBRM1, KDM6A, SETD2 and BAP1 were unmethylated in all tumor and normal specimens. 23913813_The identification of novel KDM6A mutations in patients with Kabuki syndrome. 24123378_Both Ezh2 and Kdm6a were shown to affect expression of master regulatory genes involved in adipogenesis and osteogenesis. 24465480_results demonstrate that UTX is implicated in IL-4 mediated transcriptional activation of the ALOX15 gene 24491801_High levels of UTX or MLL4 are associated with poor prognosis in patients with breast cancer. 24527667_A report of novel KDM6A mutations in patients with Kabuki syndrome. 24739679_One girl had a novel splice-site mutation in KDM6A. 25071154_Results show that UTX interacts with the retinoic acid receptor alpha (RARalpha) and this interaction is essential for proper differentiation of leukemic U937 cells in response to retinoic acid. 25225064_This study is the first to identify frequent BAP1 and BRCA pathway alterations in bladder cancer, show TERT promoter alterations are independent of other bladder cancer gene alterations, and show KDM6A loss is a driver of the bladder cancer phenotype. 25281733_Mutations in KMT2D gene were identified in 10/16 (62%) of the patients, whereas none of the patients had KDM6A mutations. 25320243_H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia 25972376_Our results provide further support for the similar roles of KMT2D and KDM6A in the etiology of KS by using a vertebrate model organism to provide direct evidence of their roles in the development of organs and tissues affected in KS patients. 26049589_Kabuki syndrome may be caused by mutations in one of two histone methyltransferase genes: KMT2D and KDM6A. 26138514_The KDM6A gene is a histone demethylase specific for histone H3 Lysin 27 and regulates gene transcription [35]. In approximately 24% of urothelial carcinoma, KDM6A is altered. 26303947_UTX is a prominent tumour suppressor that functions as a negative regulator of EMT-induced Cancer Stem Cell-like properties by epigenetically repressing epithelial-mesenchymal transition -TFs. 26431949_Turner Syndrome subjects, who are predisposed to chronic ear infections, had reduced UTX expression in immune cells and decreased circulating CD4(+) CXCR5(+) T cell frequency. 26762983_The results define UTX as a bivalency-resolving histone modifier necessary for stem cell differentiation 26819089_UTX positively regulates E-cadherin expression in colon cancer cells. 26841933_Pathogenic variants in KMT2D resulting in protein truncation in 43% (6/14; of which 3 are novel) of all cases were detected, while analysis of KDM6A was negative. MLPA analysis was negative in all instances. 26898171_Mutations of the epigenetic genes KMT2D and KDM6A cause dysregulation of certain developmental genes and account for the multiple congenital anomalies of the syndrome 27028180_we identified a novel de novo deletion of KDM6A in a Chinese girl with KS. We consider her allergic skin manifestations to be part of the phenotypic spectrum of KS 27151432_Here, we discuss the roles of lysine 27 demethylases, JMJD3 and UTX, in cancer and potential therapeutic avenues targeting these enzymes. Despite a high degree of sequence similarity in the catalytic domain between JMJD3 and UTX, numerous studies revealed surprisingly contrasting roles in cellular reprogramming and cancer, particularly leukemia 27302555_Study presents a mutation screening of patients with Kabuki syndrome type 1 which identified 208 mutations in KMT2D. Two of the KDM6A mutations were maternally inherited and nine were shown to be de novo. 27533081_both UTX and UTY function as dose-dependent suppressors of urothelial bladder cancer development 27869828_Mutation in KDM6A gene is associated with cancer more frequently in males. 27983522_UTX gene expression in renal cell carcinoma and bladder cancer. 28197626_Kdm6a and Kdm6b were found to be significantly overexpressed in Malignant pleural mesothelioma (MPM) at the mRNA level. However, tests examining if targeting therapeutically Kdm6a/b using a specific small molecule inhibitor was potentially useful for treating MPM, revealed that members of the Kdm6 family may not be suitable candidates for therapy 28228601_inactivating mutations of KDM6A, which are common in urothelial bladder carcinoma, are potentially targetable by inhibiting EZH2. 28442529_Two novel missense mutations: p.G325A in the KDM6A gene responsible for Kabuki syndrome and p.G1877V in the SCN1A gene responsible for generalized epilepsy with febrile seizures plus were identified using the TruSight One sequencing panel. 28534508_Study identified a feed-forward loop between UTX and ER in the regulation of hormonally responsive breast carcinogenesis. 28968467_KDM6A and p21CIP1 expression are essential to curb E7 induced replication stress to levels that do not markedly interfere with cell viability 29045832_Rebalance of Histone h3 lysine 27 methylation 3 levels at specific genes through EZH2 inhibitors may be a therapeutic strategy in multiple myeloma cases harboring UTX mutations. 29136510_Data show that more mutations in the histone lysine demethylase KDM6A were present in non-invasive tumors from females than males. 29171124_Depletion of KDM6A inhibits the expression of SOX9, Col2a1, ACAN and results in increased H3K27me3 and decreased H3K4me3 levels. 29351209_High UTX expression is independently associated with a better prognosis in patients with esophageal squamous cell carcinoma (ESCC) and downregulation of UTX increases ESCC cell growth and decreases E-cadherin expression. Our results suggest that UTX may be a novel therapeutic target for patients with ESCC. 29846646_Study in Finnish Caucasian obesity patients demonstrated a different genome-wide DNA methylation pattern in autosomes and X-chromosome between males and females in human liver. Sex-specific differences in liver expression and methylation of KDM6A may contribute to higher HDL-cholesterol levels in females. 29902804_data suggest that haploinsufficiency for KDM6A due to mosaic X chromosome monosomy may be responsible for hyperinsulinism in Turner syndrome. 29907798_we retrospectively evaluated 100 infants with HI lacking a genetic diagnosis, for causative variants in KS genes. Molecular diagnoses of KS were established by identification of pathogenic variants in KMT2D (n = 5) and KDM6A (n = 4). 29973620_ut of twenty-seven genes culminating into leading hubs in the network, we identified two key regulators (KRs) i.e. KDM6A and BDNF 30006524_Lymphomas with low UTX expression express high levels of Efnb1, and cause significantly poor survival. 30107592_The mutation pattern of KMT2D and KDM6A in a cohort of 505 patients clinically diagnosed as Kabuki syndrome was reported. 30166694_The histone demethylase UTX/KDM6A is mutated in up to 10% of cases of multiple myeloma, activating genes by removing the H3K27me3 repressive histone mark, counteracting EZH2 activity. 30556125_KDM6A exhibited essential roles in human PDAC as a tumor suppressor and KDM6A deficiency could be a promising biomarker for unfavorable outcome in PDAC patients and a potential surrogate marker for response to HDAC inhibitors. 30556359_Three of the 27 children had a KDM6A mutation and had thesame hypermobility as the KMT2D subjects. Only one of the Kabuki syndrome chil-dren had a patellar luxation in here previous history and was not con-sidered hypermobile using both scores. 30718900_Study shows that UTX and 53BP1 directly interact and co-occupy promoters in human embryonic stem cells and differentiating neural progenitor cells. Data suggests that the 53BP1-UTX interaction supports the activation of key genes required for human neurodevelopment. 30753822_In clear cell renal cell carcinoma, KDM6A(UTX) is one of the most frequently mutated genes; In 101 cases of patient samples, 12 samples contained KDM6A mutations. Other studies provided additional evidence that KDM6A was highly mutated in different human solid tumors and leukemias 30872525_the H3K27 histone demethylase KDM6A/UTX, but not its paralog KDM6B, is oxygen sensitive. 30948420_targeting the KDM6A-KLF10 feedback loop may be beneficial to attenuate diabetes-induced kidney injury. 31097364_UTY is co-regulated with KDM6A. UTY compensates for KDM6A in eutherian males and is responsible for the association between the loss of the Y chromosome and poor prognosis in a range of cancers. [review] 31201358_our findings highlight KDM6A as a novel mediator of drug resistance in acute myeloid leukemia 31221981_This article reviewed the most recent findings regarding cancer-specific metabolic reprogramming and the tumor-suppressive roles of IDH1/2, JARID1C/KDM5C and UTX/KDM6A. [review] 31285428_EZH2 methyltransferase and JMJD3/UTX demethylases were deregulated during hepatic differentiation of human HepaRG cells. 31335488_Expression of UTX Indicates Poor Prognosis in Patients With Luminal Breast Cancer and is Associated With MMP-11 Expression. 31403472_the X escapee Kdm6a regulates multiple immune response genes, providing a mechanism for sex differences in autoimmune disease susceptibility 31654559_Prenatal and perinatal history in Kabuki Syndrome. 31685800_GATA3 recruits UTX for gene transcriptional activation to suppress metastasis of breast cancer. 31883305_The phenotypic spectrum of Kabuki syndrome in patients of Chinese descent: A case series. 31935506_This study expands the number of naturally occurring KMT2D and KDM6A variants. The discovery of novel pathogenic variants will add to the knowledge on disease-causing variants and the relevance of missense variants in Kabuki syndrome. 31959746_Knock-out of KDM6s (JMJD3 and/or UTX) in human embryonic stem cells (hESCs) showed that KDM6s (JMJD3 and/or UTX)-deficient human ESCs exit pluripotency and commit to neural progenitor cells (NPC) differentiation normally, but the resulting NPCs fail to transit into neurons and glia due to a lack of accessibility at loci essential for neurogenesis. 32071397_Cancer-derived UTX TPR mutations G137V and D336G impair interaction with MLL3/4 complexes and affect UTX subcellular localization. 32125007_Coordinated demethylation of H3K9 and H3K27 is required for rapid inflammatory responses of endothelial cells. 32154941_HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. 32269126_KDM6A-Mediated Expression of the Long Noncoding RNA DINO Causes TP53 Tumor Suppressor Stabilization in Human Papillomavirus 16 E7-Expressing Cells. 32346926_Histone demethylase KDM6A promotes somatic cell reprogramming by epigenetically regulating the PTEN and IL-6 signal pathways. 32427586_Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. 32679064_UTX Regulates Human Neural Differentiation and Dendritic Morphology by Resolving Bivalent Promoters. 32732223_X- and Y-Linked Chromatin-Modifying Genes as Regulators of Sex-Specific Cancer Incidence and Prognosis. 32803813_Update of the genotype and phenotype of KMT2D and KDM6A by genetic screening of 100 patients with clinically suspected Kabuki syndrome. 32867456_[KDM6A mutation and expression in gastric cancer are associated with prognosis]. 32879445_Histone 3 lysine-27 demethylase KDM6A coordinates with KMT2B to play an oncogenic role in NSCLC by regulating H3K4me3. 32929331_Targeted inhibition of KDM6 histone demethylases eradicates tumor-initiating cells via enhancer reprogramming in colorectal cancer. 32977832_UTX/KDM6A suppresses AP-1 and a gliogenesis program during neural differentiation of human pluripotent stem cells. 32989154_Loss of UTX/KDM6A and the activation of FGFR3 converge to regulate differentiation gene-expression programs in bladder cancer. 33174323_Combination of lysine-specific demethylase 6A (KDM6A) and mismatch repair (MMR) status is a potential prognostic factor in colorectal cancer. 33253789_Histone demethylase UTX/KDM6A enhances tumor immune cell recruitment, promotes differentiation and suppresses medulloblastoma. 33314698_Clinical and molecular characterization study of Chinese Kabuki syndrome in Hong Kong. 33456567_KDM6A promotes imatinib resistance through YY1-mediated transcriptional upregulation of TRKA independently of its demethylase activity in chronic myelogenous leukemia. 33546721_Molecular mechanics and dynamic simulations of well-known Kabuki syndrome-associated KDM6A variants reveal putative mechanisms of dysfunction. 34051747_Significance of KDM6A mutation in bladder cancer immune escape. 34257820_hsa-miR-199b-3p Prevents the Epithelial-Mesenchymal Transition and Dysfunction of the Renal Tubule by Regulating E-cadherin through Targeting KDM6A in Diabetic Nephropathy. 34262032_SMARCA4 deficient tumours are vulnerable to KDM6A/UTX and KDM6B/JMJD3 blockade. 34465286_Overexpression of UTX promotes tumor progression in Oral tongue squamous cell carcinoma patients receiving surgical resection: a case control study. 34526716_UTX condensation underlies its tumour-suppressive activity. 34583087_KDM6A Regulates Cell Plasticity and Pancreatic Cancer Progression by Noncanonical Activin Pathway. 34661759_EGFR transcriptionally upregulates UTX via STAT3 in non-small cell lung cancer. 34667079_PROSER1 mediates TET2 O-GlcNAcylation to regulate DNA demethylation on UTX-dependent enhancers and CpG islands. 35022315_KDM6A Depletion in Breast Epithelial Cells Leads to Reduced Sensitivity to Anticancer Agents and Increased TGFbeta Activity. 35073341_Mutually exclusive mutation profiles define functionally related genes in muscle invasive bladder cancer. 35185915_Inhibiting KDM6A Demethylase Represses Long Non-Coding RNA Hotairm1 Transcription in MDSC During Sepsis. | ENSMUSG00000037369 | Kdm6a | 9.481534e+02 | 0.8474366 | -0.238822696 | 0.3246733 | 5.408249e-01 | 0.4620910507 | 0.86581093 | No | Yes | 7.716685e+02 | 126.859171 | 9.323354e+02 | 156.859366 | |
| ENSG00000147123 | 54539 | NDUFB11 | protein_coding | Q9NX14 | FUNCTION: Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone. {ECO:0000269|PubMed:27626371}. | 3D-structure;Alternative splicing;Cardiomyopathy;Disease variant;Electron transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Primary mitochondrial disease;Reference proteome;Respiratory chain;Transit peptide;Transmembrane;Transmembrane helix;Transport | Mouse_homologues NA; + ;NA | The protein encoded by this gene is a subunit of the multisubunit NADH:ubiquinone oxidoreductase (complex I). Mammalian complex I is located at the mitochondrial inner membrane. This protein has NADH dehydrogenase activity and oxidoreductase activity. It transfers electrons from NADH to ubiquinone. Mutations in the human gene are associated with linear skin defects with multiple congenital anomalies 3 and mitochondrial complex I deficiency. [provided by RefSeq, Dec 2016]. | hsa:54539; | mitochondrial inner membrane [GO:0005743]; mitochondrial respiratory chain complex I [GO:0005747]; mitochondrion [GO:0005739]; mitochondrial respiratory chain complex I assembly [GO:0032981] | 17292333_NDUFB11 did not seem to influence risk and age at onset of visual loss in a total of 65 individuals from 35 Italian Leber hereditary optic neuropathy patients. 23246602_the post-transcriptional regulation of the Ndufb11 gene can be involved in the programmed cell death process 25772934_Mutations in NDUFB11, encoding a complex I component of the mitochondrial respiratory chain, cause microphthalmia with linear skin defects syndrome. 25921236_The novel NDUFB11 mutation may cause a complex 1 deficiency in synergy with additional unknown mtDNA variants. 27102574_This is the third report that describes a mutation in NDUFB11, but all are associated with a different phenotype. Our results further expand the molecular spectrum and associated clinical phenotype of NDUFB11 defects. 27488349_recurring mutation, c.276_278del, p.F93del, in NDUFB11, a mitochondrial respiratory complex I-associated protein encoded on the X chromosome, in 5 males with a variably syndromic, normocytic congenital sideroblastic anemia. 30423443_Our findings together with a review of the thirteen previously described patients demonstrate a wide spectrum of clinical features associated with NDUFB11-related complex I deficiency. However, histiocytoid cardiomyopathy and/or congenital sideroblastic anemia could be indicative for mutation in the NDUFB11 gene, while the clinical manifestation of the same mutation can be highly variable | ENSMUSG00000031059+ENSMUSG00000061633 | Ndufb11+Ndufb11b | 9.695818e+03 | 1.2310881 | 0.299933957 | 0.3272852 | 8.324529e-01 | 0.3615641960 | 0.83352869 | No | Yes | 1.026805e+04 | 1351.806550 | 7.136032e+03 | 963.942043 |
| ENSG00000147274 | 27316 | RBMX | protein_coding | P38159 | FUNCTION: RNA-binding protein that plays several role in the regulation of pre- and post-transcriptional processes. Implicated in tissue-specific regulation of gene transcription and alternative splicing of several pre-mRNAs. Binds to and stimulates transcription from the tumor suppressor TXNIP gene promoter; may thus be involved in tumor suppression. When associated with SAFB, binds to and stimulates transcription from the SREBF1 promoter. Associates with nascent mRNAs transcribed by RNA polymerase II. Component of the supraspliceosome complex that regulates pre-mRNA alternative splice site selection. Can either activate or suppress exon inclusion; acts additively with TRA2B to promote exon 7 inclusion of the survival motor neuron SMN2. Represses the splicing of MAPT/Tau exon 10. Binds preferentially to single-stranded 5'-CC[A/C]-rich RNA sequence motifs localized in a single-stranded conformation; probably binds RNA as a homodimer. Binds non-specifically to pre-mRNAs. Plays also a role in the cytoplasmic TNFR1 trafficking pathways; promotes both the IL-1-beta-mediated inducible proteolytic cleavage of TNFR1 ectodomains and the release of TNFR1 exosome-like vesicles to the extracellular compartment. {ECO:0000269|PubMed:12165565, ECO:0000269|PubMed:12761049, ECO:0000269|PubMed:16707624, ECO:0000269|PubMed:18445477, ECO:0000269|PubMed:18541147, ECO:0000269|PubMed:19282290, ECO:0000269|PubMed:21327109}. | 3D-structure;Acetylation;Activator;Alternative splicing;Direct protein sequencing;Glycoprotein;Isopeptide bond;Mental retardation;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repressor;Ribonucleoprotein;Spliceosome;Transcription;Tumor suppressor;Ubl conjugation;mRNA processing;mRNA splicing | This gene belongs to the RBMY gene family which includes candidate Y chromosome spermatogenesis genes. This gene, an active X chromosome homolog of the Y chromosome RBMY gene, is widely expressed whereas the RBMY gene evolved a male-specific function in spermatogenesis. Pseudogenes of this gene, found on chromosomes 1, 4, 9, 11, and 6, were likely derived by retrotransposition from the original gene. Alternatively spliced transcript variants encoding different isoforms have been identified. A snoRNA gene (SNORD61) is found in one of its introns. [provided by RefSeq, Sep 2009]. | hsa:27316; | catalytic step 2 spliceosome [GO:0071013]; euchromatin [GO:0000791]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; ribonucleoprotein complex [GO:1990904]; spliceosomal complex [GO:0005681]; supraspliceosomal complex [GO:0044530]; chromatin binding [GO:0003682]; identical protein binding [GO:0042802]; mRNA binding [GO:0003729]; protein domain specific binding [GO:0019904]; RNA binding [GO:0003723]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; cellular response to interleukin-1 [GO:0071347]; membrane protein ectodomain proteolysis [GO:0006509]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of mRNA splicing, via spliceosome [GO:0048025]; osteoblast differentiation [GO:0001649]; positive regulation of mRNA splicing, via spliceosome [GO:0048026]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein homooligomerization [GO:0051260]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; transcription by RNA polymerase II [GO:0006366] | 16491274_Observational study of gene-disease association. (HuGE Navigator) 16491274_evidence provided that deletions in or around RBMX may be involved in non-obstructive azoospermia(NOA) 16552754_From these results, it seems that the X-chromosome, through its RBM genes, plays a formerly unknown role in the regulation of programmed cell death (apoptosis) in breast cancer. 17387044_p53 modulates DNA DSB repair by, in part, inducing hnRNP G 18445477_This identifies RBMX as an ARTS-1-associated protein that regulates both the constitutive release of TNFR1 exosome-like vesicles and the inducible proteolytic cleavage of TNFR1 ectodomains. 18541147_These studies indicate that hnRNP G promotes the expression of Txnip and mediates its tumor-suppressive effect. 21840245_studied the genetic and expression states of hnRNP G in normal, premalignant and malignant human oral tissues to further understand the relationship between the hnRNP G alterations and the development of human oral cancer 22344029_Data show that RBMX accumulated at DNA lesions through multiple domains in a poly(ADP-ribose) polymerase 1-dependent manner and promoted HR by facilitating proper BRCA2 expression. 22832223_RBMX is a cohesion regulator that maintains the proper cohesion of sister chromatids. 25256757_A sequence deletion within RBMX is identified as associated with with Shashi X-linked intellectual disability syndrome. 25884434_Results showed that HNRNPG and HTRA2-BETA1 were specific antagonistic regulators of ERa exon7 splicing and increased HNRNPG levels were associated with improved clinical outcome of endometrial cancer through up-regulation of ERaD7 expression. 26333388_Host RBMX is required for the maintenance of Borna disease virus nuclear viral factories. 28334903_HNRNPG binds m6A-methylated RNAs through its C-terminal low-complexity region, which self-assembles into large particles in vitro. The Arg-Gly-Gly repeats within the low-complexity region are required for binding to the RNA motif exposed by m6A methylation. 29383807_These results suggest that satellite I RNA plays a role in stabilizing RBMX and Sororin in the ncRNP complex to maintain proper sister chromatid cohesion. 30502052_Study demonstrates a novel role for the splice factor hnRNPG in the pregnant myometrium. hnRNPG is required for the exclusion of exon 7 in the derivation of the dominant negative myometrial ERDelta7 isoform and is negatively regulated in an E2 dependent manner. 31343408_PUM binding is required for maintenance of genomic stability by NORAD whereas binding of RBMX is dispensable for this function. 31445886_hnRNPG associates co-transcriptionally with RNAPII and regulates alternative splicing transcriptome-wide. m(6)A near splice sites in nascent pre-mRNA modulates hnRNPG binding, which influences RNAPII occupancy patterns and promotes exon inclusion. 32494026_RBMX is required for activation of ATR on repetitive DNAs to maintain genome stability. 33564070_RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. 34260915_Deletion of RBMX RGG/RG motif in Shashi-XLID syndrome leads to aberrant p53 activation and neuronal differentiation defects. 34458856_Transcriptional control of CBX5 by the RNA binding proteins RBMX and RBMXL1 maintains chromatin state in myeloid leukemia. | 7.907061e+03 | 1.0743784 | 0.103502247 | 0.2729234 | 1.447404e-01 | 0.7036131381 | 0.93624959 | No | Yes | 8.535439e+03 | 1341.878796 | 6.842536e+03 | 1103.262195 | |||
| ENSG00000147403 | 6134 | RPL10 | protein_coding | P27635 | FUNCTION: Component of the large ribosomal subunit (PubMed:26290468). Plays a role in the formation of actively translating ribosomes (PubMed:26290468). May play a role in the embryonic brain development (PubMed:25316788). {ECO:0000269|PubMed:25316788, ECO:0000269|PubMed:26290468, ECO:0000305|PubMed:12962325}. | 3D-structure;Autism;Autism spectrum disorder;Citrullination;Developmental protein;Direct protein sequencing;Disease variant;Isopeptide bond;Mental retardation;Reference proteome;Ribonucleoprotein;Ribosomal protein;Translation regulation;Ubl conjugation | This gene encodes a ribosomal protein that is a component of the 60S ribosome subunit. The related protein in chicken can bind to c-Jun and can repress c-Jun-mediated transcriptional activation. Some studies have detected an association between variation in this gene and autism spectrum disorders, though others do not detect this relationship. There are multiple pseudogenes of this gene dispersed throughout the genome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2015]. | hsa:6134; | cytosol [GO:0005829]; cytosolic large ribosomal subunit [GO:0022625]; cytosolic ribosome [GO:0022626]; endoplasmic reticulum [GO:0005783]; membrane [GO:0016020]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; translation regulator activity [GO:0045182]; cytoplasmic translation [GO:0002181]; embryonic brain development [GO:1990403]; negative regulation of apoptotic process [GO:0043066]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of translation [GO:0006417]; ribosomal large subunit assembly [GO:0000027]; translation [GO:0006412] | 12138090_QM binds to c-yes at the SH3 domain in tumor cell lines 16331298_Reduction of QM protein expression correlates with tumor grade in prostatic adenocarcinoma 16940977_Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism 17566674_Xq28 (QM gene) may be involved in ovary failure. 18007048_A hexagonal crystal of L10CD was obtained by the sitting-drop vapour-diffusion method. The L10CD crystal diffracted to 2.5 A resolution and belongs to space group P3(1)21 or P3(2)21. 18258260_Characteristic interactions among Arg90-Trp171-Arg139 guide the C-terminal part outside of the central fold 19166581_Observational study of gene-disease association. (HuGE Navigator) 19166581_Our results suggest that RPL10 has no major effect on the susceptibility to autism spectrum disorders 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21567917_mutation analysis of RPL10 in German patients with autism spectrum disorder 23263491_Mutations affect the ribosomal proteins RPL5 and RPL10 in 12 of 122 (9.8%) pediatric T-cell acute lymphoblastic leukemias, with recurrent alterations of Arg98 in RPL10. 25316788_A mutation within the conserved N-terminal end of RPL10, a protein in close proximity to the peptidyl transferase active site of the 60S ribosomal subunit, causes severe defects in brain formation and function. 25846674_report confirms the implication of RPL10 mutations in neurodevelopmental disorders and extends the associated clinical spectrum from autism to syndromic intellectual disability 26290468_Our results expand the mutational and clinical spectrum of RPL10 identifying a new genetic cause of SED and highlight the emerging role of ribosomal proteins in the pathogenesis of neurodevelopmental disorders. 27726420_Mitochondrial Ribosomal Protein L10 regulates cyclin B1/Cdk1 (cyclin-dependent kinase 1) activity and mitochondrial protein synthesis in mammalian cells 28428269_there are 7 mutations in RPL10 in 344 patients, or a mutation frequency of 2% 28744013_Mutation R98S in the RPL10 gene in cells from patients with T-Cell acute lymphocytic leukemia results in elevated expression of JAK-STAT signaling cascade proteins, hyper-reactivity to cytokine stimulation, and sensitization to JAK-STAT inhibitors. 29066376_A rare de novo mutation K78E is associated with severe syndromic intellectual disability and epilepsy. 29930300_RPL10 R98S mutation is associated with T-cell acute lymphoblastic leukemia. 30172100_The regulation of reactive oxygen species level by mitochondrial RPL10 is one of the major extra-ribosomal functions in pancreatic cancer cells, which could be used as an indicator for the tumorigenesis of pancreatic cancer. | 1.521112e+05 | 1.1680278 | 0.224074552 | 0.3255956 | 4.741498e-01 | 0.4910842229 | 0.87511746 | No | Yes | 1.721433e+05 | 25906.520561 | 1.140583e+05 | 17603.172420 | |||
| ENSG00000147437 | 2796 | GNRH1 | protein_coding | P01148 | FUNCTION: Stimulates the secretion of gonadotropins; it stimulates the secretion of both luteinizing and follicle-stimulating hormones. | 3D-structure;Amidation;Cleavage on pair of basic residues;Direct protein sequencing;Disease variant;Hormone;Hypogonadotropic hypogonadism;Kallmann syndrome;Pharmaceutical;Pyrrolidone carboxylic acid;Reference proteome;Secreted;Signal | This gene encodes a preproprotein that is proteolytically processed to generate a peptide that is a member of the gonadotropin-releasing hormone (GnRH) family of peptides. Alternative splicing results in multiple transcript variants, at least one of which is secreted and then cleaved to generate gonadoliberin-1 and GnRH-associated peptide 1. Gonadoliberin-1 stimulates the release of luteinizing and follicle stimulating hormones, which are important for reproduction. Mutations in this gene are associated with hypogonadotropic hypogonadism. [provided by RefSeq, Nov 2015]. | hsa:2796; | extracellular region [GO:0005576]; extracellular space [GO:0005615]; gonadotropin hormone-releasing hormone activity [GO:0005183]; gonadotropin-releasing hormone receptor binding [GO:0031530]; hormone activity [GO:0005179]; cell-cell signaling [GO:0007267]; negative regulation of neuron migration [GO:2001223]; regulation of gene expression [GO:0010468]; regulation of ovarian follicle development [GO:2000354]; reproduction [GO:0000003]; response to ethanol [GO:0045471]; response to steroid hormone [GO:0048545]; signal transduction [GO:0007165] | 11875100_data indicate that the promoter region between -992 and -795 contains elements both essential and sufficient for targeting gene expression to GnRH neurons 12040003_Coupling of GnRH concentration and the GnRH receptor-activated gene program 12054733_JunD activated by LHRH acts as a modulator of cell proliferation and cooperates with the anti-apoptotic and anti-mitogenic functions of LHRH. 12447356_GnRH-II and GnRH-I interact directly with T cells and trigger gene transcription, adhesion, chemotaxis and homing to specific organs, which may be of clinical relevance. 12633791_Observational study of gene-disease association. (HuGE Navigator) 12770744_regulation of GnRH-I and GnRH-II gene expression in the ovary 12788881_Genetic analysis has excluded sequence variations in GNRH1 and GNRHR in four families with recessive IHH, suggesting the existence of a novel, as-yet-undiscovered gene for this condition. 12969578_gonadotropin releasing hormone-II is more effective than gonadotropin releasing hormone-I in stimulating leptin secretion 14565958_Neurons respond to GnRH with time- and dose-dependent increases in GnRH gene expression and protein release. 14594454_Review. The proliferation of human ovarian cancer cell lines is time- and dose-dependently reduced by GnRH and its superagonistic analogs. 14726258_Experimental evidence indicates that GnRH-I is expressed, together with its receptors, in tumors of the reproductive tract. Activation of type I GnRH receptors consistently decreases cell proliferation, mainly by interfering with growth factors. 15001648_GnRH I and GnRH II have both common and discrete cellular distributions in the placenta and decidua and suggest that these two hormones are capable of eliciting their biological actions in an autocrine and/or paracrine manner 15062568_In placental cytotrophoblasts, the upstream transcription start site of GnRH gene was the major one and gave rise to an mRNA level three times higher than the downstream start site. Estradiol downregulated GnRH gene expression in a dose-dependent fashion. 15138251_expression of the GnRH gene is regulated in neurons by TALE homeodomain proteins and Oct-1 15229199_It is suggested that during the early to midfollicular phase the ovaries produce a gonadotrophin surge attenuating factor that antagonizes the pituitary-sensitizing effect of E2 to GnRH. (Gonadotrophin surge attenuating factor) 15283968_These galanin-LHRH and LHRH-galanin contacts may be functional synapses, and they may be the morphological substrate of the galanin-controlled gonadal functions in humans. 15490304_Observational study of gene-disease association. (HuGE Navigator) 15546906_Genetic variation in GNRH1 is not likely to be a substantial modulator of pubertal timing in the general population. 15546906_Observational study of gene-disease association. (HuGE Navigator) 15562029_Progesterone receptor isoform p4 is a potent regulator of GnRHRI at the transcriptional level as well as GnRH I mRNA. 15578334_GnRH-I induced greater proliferation in normal B-cells than IL-2 treatment alone. 15809743_GnRH may protect ovarian cancer cells from stimulated apoptotic cell death. 16061872_LHRH receptors are expressed in human RCC specimens 16157590_analysis of gonadotropin-releasing hormone ligand conformation and receptor selectivity 16359986_GnRH1 gene is not responsible for idiopathic hypogonadotropic hypogonadism, at least as mutations are concerned 17202595_we observed that LHRH-(1-5), the specific processed peptide of LHRH-I, upregulates LHRH-II mRNA expression in Ishikawa cells, an endometrial cell line but does not exert any influence on LHRH-I mRNA levels 17220347_Observational study of gene-disease association. (HuGE Navigator) 17456575_estrogens may exert direct actions upon GnRH neurons exclusively through ER-beta 17605472_Our data indicate a bridging interaction between Arg500 of the N-domain and Arg8 of GnRH that involves a chloride ion may account for its role in the specificity of the N-domain for endoproteolytic cleavage of the substrate at the NH2-terminus in vitro 17680884_Data show that GnRH neurones morphologically interact with astrocytes and tanycytes in the human brain and suggest that glial cells may contribute to the process by which the neuroendocrine brain controls the function of GnRH neurones in humans. 17692113_Observational study of gene-disease association. (HuGE Navigator) 18463157_12% of Kallman syndrome males have KAL1 deletions, but intragenic deletions of the FGFR1, GNRH1, GNRHR, GPR54 and NELF genes are uncommon in Idiopathic hypogonadotropic hypogonadism/Kallman syndrome. 18467526_analysis of how GNRH I and GNRH II inhibit cell growth 18477660_GnRH-I and -II induce apoptosis in human granulosa cells through GnRH-I receptors, which mediate the proteolytic caspase cascade involving caspase-8 (the initiator) and caspase-3 and -7 (the effectors). 18959738_REVIEW: role of GnRH in the control of tumor growth, progression, and dissemination 18959739_summarize the current understanding of the antiproliferative actions of GnRH analogs, as well as the recent observations of GnRH effects on ovarian cancer cell apoptosis and motogenesis,and the molecular mechanisms that mediate GnRH actions 18980792_Data show that over half of meningiomas may be regulated by GnRH-GnRH-R expression in an autocrine fashion. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19086053_Observational study of gene-disease association. (HuGE Navigator) 19190109_the antitumor effect of gonadotropin-releasing hormone type i is mediated by the activation of type I (but not of type II) GnRH-R 19403562_Observational study of gene-disease association. (HuGE Navigator) 19453261_Observational study of gene-disease association. (HuGE Navigator) 19489874_Observational study of gene-disease association. (HuGE Navigator) 19535795_identified a homozygous GNRH1 frameshift mutation in the sequence encoding the N-terminal region of the signal peptide-containing prepro-GnRH in a teenage brother & sister who had normosmic idiopathic hypogonadotropic hypogonadism 19567835_GNRH1 mutations are a genetic cause of idiopathic hypogonadotropic hypogonadism. 19640273_Common variants of the GNRH1 and GNRHR genes are not associated with risk of invasive breast cancer in Caucasians 19640273_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19849976_GNRH1 is mutated in people with familial hypogonadotropic hypogonadism. 20118984_cadherin switching and p120(ctn) signaling as important targets of GnRH function and as novel mediators of invasiveness and tumor progression in ovarian cancer. 20138117_Metabolic signals are integrated at the levels of first-order neurons equipped with the proper receptors, ant that these neurons send their signals towards hypothalamic GnRH neurons which constitute the integrative element of this network. 20188792_The major genes associated with GnRH-dependent pubertal disorders, are reviewed. 20389089_GNRH1 mutations might cause congenital idiopathic (or isolated) hypogonadotropic hypogonadism (Review) 20400076_The role of GnRH-GnRHR signaling at the maternal-fetal interface therefore appears to be limited to the regulation of trophoblast hCG production. 20424473_Observational study of gene-disease association. (HuGE Navigator) 20484732_Review: Using human GnRH deficiency as a paradigm and presenting original data from the screening of numerous candidate genes, we discuss the emerging model of patient-focused clinical genetic research. 20529119_Data describe the distribution and sex dimorphism of kisspeptin-immunoreactive elements in hypothalami, reveal contacts between kisspeptin-immunoreactive fibers and GnRH cells, and colocalize kisspeptins and neurokinin B in the infundibular nucleus. 20634197_Meta-analysis of gene-disease association. (HuGE Navigator) 20727862_Lactation is an important physiological model of the integration of energy balance and reproduction, as it involves activation of potent appetitive neuropeptide systems coupled to a profound inhibition of pulsatile GnRH/LH secretion--REVIEW 20727865_A large literature dealing with various stressors that regulate gonadotrophin-releasing hormone secretion in a variety of species provides evidence that stress modulates GnRH secretion by activating the corticotrophin-releasing factor system--REVIEW 20734064_Observational study of gene-disease association. (HuGE Navigator) 20807514_The sources of glutamatergic inputs to the GnRH neurons are only just beginning to be examined and include the anteroventral periventricular nucleus as well as the possibility that GnRH neurons--REVIEW 20887715_homozygous inactivating mutations in the GNRH1 gene causing hypothalamic hypogonadotropic hypogonadism in patients were reported, validating definitively the pivotal role of GnRH in human pubertal development and reproduction--REVIEW 20951683_We then discuss possible neurobiological mechanisms underlying GnRH pulse generation, and conclude by proposing that kisspeptin neurons in the arcuate nucleus are key players--REVIEW 21368045_Deletion of the cell-specific enhancer region of GnRH implicates the GnRH promoter in mediating pubertal development and periodic reproductive cycling. 21646369_Sleep is associated with a significant decline in GnRH pulse frequency in both older and younger postmenopausal women 21714833_Data show that higher levels of GnRH IgM antibodies were detected in patients with IBS and dysmotility, but not organic GI diseases, compared with healthy controls. 21722705_Mutations in GNRH1 have been associated with both mild and severe forms of GnRH deficiency, and may work in combination with other gene mutations to produce GnRH-deficient phenotypes. 21827674_Assessment of CGB and GNRH1 expression level in cancer patients' blood may be useful for indicating metastatic spread of tumor cells. 21855365_[review] Existing evidence suggests that neural pathways originating in hindbrain and hypothalamic feeding nuclei transmit information concerning glucose availability to GnRH neurons, which integrate numerous physiological signals and metabolic cues. 22024993_molecular mechanisms involved in GnRH/GnRHR signaling at the maternal-fetal interface. [Review] 22074952_data demonstrate that GnRH transcription is repressed by AR via multiple sequences in GnRH-P, including three Oct-1 binding sites, and that this repression requires the complex interaction of several transcription factors 22161498_The role of several class 1 cytokines in regulating GnRH neuronal development, GnRH secretion, and GnRH expression in this REVIEW. 23077052_Spontaneous GnRH release is detected in the median eminence of transgenic mice that produce diurnal variations in GnRH. 23287110_GnRHI, GnRHII and GnRH receptor expression correlated significantly with poor prognosis in breast cancer patients 23404564_Olfactory ensheathing cells form a microenvironment for migrating GnRH-1 neurons during development. 23550003_Kisspeptin resets the GnRH pulse generator in men, but does not appear to do so in women 23550012_Kisspeptin and GnRH pulse generation. 23648337_AXL and MET crosstalk to promote gonadotropin releasing hormone (GnRH) neuronal cell migration and survival. 23735672_This review summarizes the mechanisms used by glial cells to control GnRH neuronal ativity and secretion. 23936060_R31C GNRH1 is the only missense mutation that was identified in a CpG islet in nine congenital hypogonadotropic hypogonadism subjects from four unrelated families, giving evidence for a putative ''hot spot''. 24002956_No abnormalities were found in the patient group for the PROKR2 and GNRH1genes. In addition, no genomic rearrangements were identified in the healthy control individuals for the described genes 24010162_TRH, LH-RH and substance P are not affected in Alzheimer disease and Down's syndrome 24056171_Current research efforts aim to discover the mechanisms responsible for the decoding of the GnRH pulse signal by the gonadotrope. 24095645_This review will summarize the current understanding of the mechanisms by which PACAP modulates gonadotrope function, with a focus on interactions with GnRH. 24264576_GPR101 is a critical requirement for GnRH-(1-5) transactivation of EGFR in Ishikawa cells. 24472523_GnRH, through heterotopic expression of its receptor, may be a potential regulator of CYP11B2 expression levels in some cases of aldosterone-producing adenoma. 24603682_Data indicate that the gene expression pattern is profoundly different between human chorionic gonadotropin (hCG) and gonadotropin-releasing hormone (GnRH) agonist. 24722580_GnRH agonists fail to increase Bax expression and do not potentiate the cytotoxic activity of docetaxel 25248098_Haploinsufficiency of Dmxl2, encoding a synaptic protein, causes infertility associated with a loss of GnRH neurons in humans and mice. 25916694_Melatonin affects the secretion of GnRH, LH and testosterone, improves sperm quality thereby regulating the testicular development and male reproduction. (Review) 25955300_our results showed that GnRH participates in the self-renewal capacity and stemness maintenance of LCSLCs by upregulating the JNK signaling pathway, and GnRH may be useful as an alternative lung cancer stem-like cells therapy. 26308290_Data suggest differences in regulation of expression of PEDF (up-regulation) vs. VEGF (down-regulation) in granulosa cells explain reduced risk of ovarian hyperstimulation syndrome due to ovulation induction using GnRH/GNRHR agonists rather than hCG. 26595427_mutations in the region encoding the decapeptide associated with complete Idiopathic hypogonadotropic hypogonadism 26644469_analysis of information transfer via gonadotropin-releasing hormone receptors to extracellular signal-regulated kinase or nuclear factor of activated T-cells 26660506_GnRH regulates trophoblast invasion via RUNX2-mediated MMP2/MMP9 expression. 26920257_GNRH (and GNRHR) are expressed in trophoblast cell populations and fallopian tube epithelium at tubal ectopic pregnancy sites. 27639272_High LHRH expression is associated with sarcomas of bone and soft tissue. 27793127_High expression of LHRH is associated with ovarian cancer. 28385888_Modeling and high-throughput experimental data uncover the mechanisms underlying Fshb gene sensitivity to gonadotropin-releasing hormone pulse frequency 33652606_Expression of Luteinizing Hormone-Releasing Hormone (LHRH) and Type-I LHRH Receptor in Transitional Cell Carcinoma Type of Human Bladder Cancer. 33964320_The KiNG of reproduction: Kisspeptin/ nNOS interactions shaping hypothalamic GnRH release. 34104122_Investigation of Specific Targeting of Triptorelin-Conjugated Dextran-Coated Magnetite Nanoparticles as a Targeted Probe in GnRH(+) Cancer Cells in MRI. | ENSMUSG00000015812 | Gnrh1 | 1.723776e+01 | 1.2595076 | 0.332859840 | 0.7323487 | 2.056922e-01 | 0.6501649862 | No | Yes | 1.912676e+01 | 8.148789 | 1.665560e+01 | 6.790938 | ||
| ENSG00000147548 | 54904 | NSD3 | protein_coding | Q9BZ95 | FUNCTION: Histone methyltransferase. Preferentially dimethylates 'Lys-4' and 'Lys-27' of histone H3 forming H3K2me2 and H3K27me2. H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation, while 'Lys-27' is a mark for transcriptional repression. {ECO:0000269|PubMed:16682010}. | 3D-structure;Acetylation;Alternative splicing;Chromatin regulator;Chromosomal rearrangement;Chromosome;Coiled coil;Isopeptide bond;Metal-binding;Methyltransferase;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repeat;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene is related to the Wolf-Hirschhorn syndrome candidate-1 gene and encodes a protein with PWWP (proline-tryptophan-tryptophan-proline) domains. This protein methylates histone H3 at lysine residues 4 and 27, which represses gene transcription. Two alternatively spliced variants have been described. [provided by RefSeq, May 2015]. | hsa:54904; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; histone methyltransferase activity (H3-K36 specific) [GO:0046975]; histone-lysine N-methyltransferase activity [GO:0018024]; metal ion binding [GO:0046872]; transcription regulator activator activity [GO:0140537]; chromatin organization [GO:0006325]; histone methylation [GO:0016571]; positive regulation of histone H3-K36 trimethylation [GO:2001255]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription, DNA-templated [GO:0006355] | 20599755_NSD3L depletion increased the invasiveness of MDA-MB-231 breast cancer cells indicating that NSD3L normally restrain cellular metastatic potential. Together the presented data indicates that NSD3L is a candidate tumor suppressor. 20940404_Overexpression of WHSC1L1 gene is associated with breast cancer. 21555454_Functional studies with Brd4 indicate that the ET domain mediates pTEFb-independent transcriptional activation through a subset of these associated factors, including NSD3. 23011637_Data indicate that siRNA attenuated the expression levels of CCNG1 and NEK7, implying that WHSC1L1 appears to activate the expression of CCNG1 and NEK7 in cancer cells. 23269674_methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition 24051013_PPAPDC1B and WHSC1L1 played a major role in regulating the survival of breast cancer, pancreatic adenocarcinoma and small-cell lung cancer-derived cell lines. 24875858_The involvement of the NSD3 methyltransferase as a component of the NUT fusion protein oncogenic complex identifies a new potential therapeutic target. 25494638_The results describe the binding of NSD1, 2 and 3 catalytic domains (CD) on histone tails through recognition of histone-lysine and methylation properties. 25942451_Studies indicate that the NSD methyltransferases NSD1, NSD2/WHSC1/MMSET and NSD3/WHSC1L1 were overexpressed, amplified or somatically mutated in multiple types of cancer, suggesting their critical role in cancer. 26626481_Results demonstrate that the AML maintenance function of BRD4 requires its interaction with the short isoform of NSD3 lacking the methyltransferase domain. This protein is an adaptor that sustains leukemia by linking BRD4 to the CHD8 chromatin remodeler. 27005559_This study demonstrates that over-expression of WHSC1L1 is linked to over-expression of ER-alpha in SUM-44 breast cancer cells and in primary human breast cancers. 27285764_WHSC1L1 and H3K36me2 are enriched in the gene bodies of the cell cycle-related genes CDC6 and CDK2, implying that WHSC1L1 directly regulates the transcription of these gene 28484924_Results extend the in vitro results and show that targeted expression of NSD3 to the mammary gland of FVB mice is oncogenic, consistent with the hypothesis that NSD3 is an important driver oncogene in human breast cancer. 28901481_Studies showed that depletion of NSD3 in osteosarcoma cell lines inhibited cell proliferation and survival, and induced cell apoptosis. RNA sequence analysis of tumor cell lines with NSD3 deletion revealed that NSD3 functions as either a transcriptional activator or a repressor. These results suggest that NSD3 may function as an oncogenic driver in osteosarcoma. 30320908_The observation that certain metabolic pathways are differentially regulated by NSD3s and Pdp3 suggests that, despite the structural similarity between their PWWP domains, the two proteins act by unique mechanisms and may recruit different downstream signaling complexes. 31638140_NSD3S stabilizes MYC through hindering its interaction with FBXW7. 32967925_NSD3-Induced Methylation of H3K36 Activates NOTCH Signaling to Drive Breast Tumor Initiation and Metastatic Progression. 33361816_Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. 33536620_Elevated NSD3 histone methylation activity drives squamous cell lung cancer. 33592170_A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins. 34083510_Targeting H3K36 methyltransferases NSDs: a promising strategy for tumor targeted therapy. 34271259_Histone and DNA binding ability studies of the NSD subfamily of PWWP domains. 34615858_Elevated expression of nuclear receptor-binding SET domain 3 promotes pancreatic cancer cell growth. | ENSMUSG00000054823 | Nsd3 | 9.617805e+02 | 0.7378394 | -0.438621240 | 0.3210320 | 1.857198e+00 | 0.1729485635 | 0.77611497 | No | Yes | 7.810437e+02 | 126.719290 | 9.980371e+02 | 165.639189 | |
| ENSG00000147586 | 28957 | MRPS28 | protein_coding | Q9Y2Q9 | 3D-structure;Acetylation;Disease variant;Mitochondrion;Phosphoprotein;Primary mitochondrial disease;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transit peptide | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein that has been called mitochondrial ribosomal protein S35 in the literature. [provided by RefSeq, Jul 2008]. | hsa:28957; | mitochondrial inner membrane [GO:0005743]; mitochondrial small ribosomal subunit [GO:0005763]; mitochondrion [GO:0005739]; RNA binding [GO:0003723]; mitochondrial translation [GO:0032543] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 30566640_MRPS28 joins the ranks of genes encoding mitoribosomal proteins linked to impaired mitoribosomal biogenesis and function in mitochondrial disease. | ENSMUSG00000040269 | Mrps28 | 4.779572e+02 | 0.4680833 | -1.095162941 | 0.3209143 | 1.150253e+01 | 0.0006950164 | 0.09756134 | No | Yes | 2.915950e+02 | 52.838153 | 6.253083e+02 | 115.200511 | ||
| ENSG00000147649 | 92140 | MTDH | protein_coding | Q86UE4 | FUNCTION: Down-regulates SLC1A2/EAAT2 promoter activity when expressed ectopically. Activates the nuclear factor kappa-B (NF-kappa-B) transcription factor. Promotes anchorage-independent growth of immortalized melanocytes and astrocytes which is a key component in tumor cell expansion. Promotes lung metastasis and also has an effect on bone and brain metastasis, possibly by enhancing the seeding of tumor cells to the target organ endothelium. Induces chemoresistance. {ECO:0000269|PubMed:15927426, ECO:0000269|PubMed:16452207, ECO:0000269|PubMed:18316612, ECO:0000269|PubMed:19111877}. | 3D-structure;Acetylation;Cell junction;Cytoplasm;Endoplasmic reticulum;Membrane;Nucleus;Phosphoprotein;Reference proteome;Tight junction;Transmembrane;Transmembrane helix | hsa:92140; | apical plasma membrane [GO:0016324]; bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; fibrillar center [GO:0001650]; integral component of membrane [GO:0016021]; intercellular canaliculus [GO:0046581]; nuclear body [GO:0016604]; nuclear membrane [GO:0031965]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; double-stranded RNA binding [GO:0003725]; NF-kappaB binding [GO:0051059]; RNA binding [GO:0003723]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; lipopolysaccharide-mediated signaling pathway [GO:0031663]; negative regulation of apoptotic process [GO:0043066]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of angiogenesis [GO:0045766]; positive regulation of autophagy [GO:0010508]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of protein kinase B signaling [GO:0051897]; regulation of transcription by RNA polymerase II [GO:0006357] | 14980505_3D3/lyric is localized to the membrane of the endoplasmic reticulum, nuclear envelope, and the nucleolus. 15093543_overexpression of metadherin is associated with metastatic breast cancer 15383321_a novel protein LYRIC is recruited during the maturation of the tight junction complex. 17397930_findings indicate that AEG-1 might play a pivotal role in the pathogenesis, progression and metastasis of diverse cancers 17563745_AEG-1 is overexpressed in clinical prostate cancer (PC) tissue samples and cultured PC cells compared to benign prostatic hyperplasia tissue samples and normal prostate epithelial cells. 17704808_AEG-1 is an oncogene cooperating with Ha-ras as well as functioning as a downstream target gene of Ha-ras; may perform a central role in Ha-ras-mediated carcinogenesis. 18316612_AEG-1 might function as a coactivator for NF-kappaB, consequently augmenting expression of genes necessary for invasion of glioma cells 18440304_LYRIC/AEG-1 is a negative regulator of BCCIPalpha, promoting proteasomal degradation either through direct interaction, or potentially through an indirect mechanism involving downstream effects of the NF-kappaB signaling pathway. 18519759_AEG-1 protein is a valuable marker of breast cancer progression. 19111877_establish MTDH as an important therapeutic target for simultaneously enhancing chemotherapy efficacy and reducing metastasis risk 19221438_AEG1 plays a central role in regulating diverse aspects of hepatocellular carcinoma pathogenesis and inhibition may lead to an effective therapeutic strategy for this disease. 19304953_Upregulation of AEG-1 plays an important role in the development and pathogenesis of human esophageal squamous cell carcinoma progression and pathogenesis. 19383828_Increased cytoplasmic distribution of LYRIC is associated with prostate cancer. 19448665_AEG-1 may play a crucial role in the pathogenesis of neuroblastoma and could represent a potential target for therapeutic intervention. 19633686_Studies were further confirmed in clinical primary breast cancer specimens, in which high-level expression of AEG-1 was inversely correlated with the expression of FOXO1. 19648967_one mechanism for cells with altered LYRIC/AEG-1 expression to evade apoptosis and increase cell growth during tumourigenesis through the regulation of PLZF repression 19740331_AEG-1 was suggested to be a LPS-responsive gene and involved in LPS-induced inflammatory response. 19940250_findings demonstrate that aberrant AEG-1 expression plays a dominant positive role in regulating oncogenic transformation and angiogenesis 20053777_AEG-1 may play a crucial role in the pathogenesis of glioma. 20236756_AEG-1 gene is clinically significant in human oligodendroglioma. 20300973_AEG-1 may play a role in Wnt/beta-catenin-mediated cancer progression 20388776_Findings show that AEG-1 contributes to glioma progression by enhancing MMP-9 transcription and tumor cell invasiveness, and underscore the importance of AEG-1 in glioma development and progression. 20388796_although AEG-1 does not affect MDR1 gene transcription, it facilitates association of MDR1 mRNA to polysomes, resulting in increased translation 20565850_MTDH overexpression could be identified in proliferative breast lesions and may contribute to breast cancer progression. 20625905_AEG-1 expression in colorectal carcinoma may be associated with tumor progression, and high AEG-1 expression correlates with poor overall survival 20802479_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20845990_AEG-1 protein is overexpressed in RCC and plays an important role in tumor differentiation and progression. 20952513_miR-26a functionally antagonizes human breast carcinogenesis by targeting MTDH and EZH2. 21084864_Observations link AEG-1 overexpression in glioblastoma with hypoxia and glucose deprivation. 21127263_AEG-1-mediated chemoresistance is because of protective autophagy and inhibition of AEG-1 results in a decrease in protective autophagy and chemosensitization of cancer cells. 21150314_Overexpression of AEG-1 increases cell migration and invasion of glioma cells. 21256156_historical perspective of AEG-1; evaluation of AEG-1 as a novel diagnostic/prognostic biomarker for a variety of cancers; understanding of the molecular and biochemical bases of oncogenic properties of AEG-1 [review] 21259255_AEG-1 expression may be related with tumor angiogenesis and progression and is a valuable prognostic factor in patients with triple-negative breast cancer. 21293286_high AEG-1 expression was associated with progression and prognosis of ovarian carcinoma. 21371176_results suggest that MTDH could promote epithelial-mesenchymal transition in breast cancer cells in driving the progression of their aggressive behavior 21408129_All variants of the MTDH gene were investigated and the association of the variants with breast cancer development, was explored. 21412050_Astrocyte elevated gene-1 activates AMPK in response to cellular metabolic stress and promotes protective autophagy. 21448238_Study revealed a modest gene-based significant association between migraine and the metadherin (MTDH) gene, previously identified in the first clinic-based GWA study (GWAS) for migraine (Bonferroni-corrected gene-based P-value=0.026). 21478147_SND1 as a novel MTDH-interacting protein and shown that it is a functionally and clinically significant mediator of metastasis. 21495225_AEG-1 overexpression is associated with carcinogenesis and tumor progression in endometrial cancer. 21543927_AEG-1 is overexpressed in a great portion of epithelial ovarian cancer (EOC) patients with peritoneal dissemination and/or lymph node metastasis and may be clinically useful for predicting metastasis in EOC. 21609571_The expression of EphA7 and/or MTDH might be closely related to the carcinogenesis, progression, clinical biological behaviors and prognosis of gallbladder adenocarcinoma. 21750404_AEG-1 plays a crucial role in osteosarcoma progression through MMP-2, and AEG-1 could be a useful biomarker for the prediction of osteosarcoma progression and prognosis 21750868_Our results suggest that the AEG-1 protein is a valuable marker of GBC progression and could be a potential therapeutic target. 21753766_Data show that AEG-1/MTDH was directly regulated by miR-375. 21852380_AEG-1 contributes to glioma-induced neurodegeneration (a hallmark of this fatal tumo) through regulation of EAAT2 expression. 21957284_Gag interacts with endogenous Lyric via its matrix (MA) and nucleocapsid (NC) domains. 21957284_HIV-1 Gag interacts with endogenous MTDH (Lyric/Aeg-1) via its matrix (MA) and nucleocapsid (NC) domains. Lyric is incorporated into HIV-1 virions and is cleaved by the HIV protease. Lyric influences HIV protein expression and infectivity. 21957284_Via Affinity purifications, MTDH (aka Lyric) interacts with HIV-1 Gag, is incorporated in virions and cleaved by HIV PR. Lyric interaction is conserved for retroviral Gag proteins. Cotransfections suggest a role for Lyric in regulating HIV infectivity. 21964981_MTDH and EphA7 are markers for metastasis and poor prognosis of gallbladder adenocarcinoma. 21976539_MTDH may promote hepatocellular carcinoma metastasis through the induction of epithelial-mesenchymal transition process 22031094_Data show that MiR-375 expression was significantly reduced, and conversely, metadherin (MTDH) was significantly increased in nasopharyngeal carcinoma (NPC) samples. 22056881_miR-375 targets AEG-1 in HCC and suppresses liver cancer cell growth in vitro and in vivo 22133054_AEG-1 expression is associated with salivary gland carcinomas progression. 22195048_MTDH is involved in inflammation-induced tumor progression. 22199357_implicate cytoplasmic MTDH in cell survival and broad drug resistance via association with RNA and RNA-binding proteins. 22204714_astrocyte-elevated gene-1 plays a crucial role in the carcinogenesis and aggressiveness of non-small cell lung cancer, promoting its metastasis by modulating matrix metalloproteinase-9 expression and leading to a poor clinical prognosis. 22246354_Overexpression of EphA7 and/or MTDH might indicate poor prognosis in squamous cell cancer of the tongue. 22340175_AEG-1 might facilitate the proliferation and invasion of breast cancer cells by upregulating HER2/neu expression. 22351252_AEG-1 was highly expressed in colorectal cancer and may be a potential biomarker for liver metastatic tumors. 22367022_MTDH affects the radiosensitivity of cervical cancer cells and that MTDH may be a novel target to improve cervical cancer radiation response. 22372608_Tumour AEG-1 overexpression is associated with poor prognosis and cisplatin resistance in advanced serous ovarian cancer. 22431469_AEG-1 interacted with beta-catenin in CRC cells and AEG-1 expression was closely associated with progression of CRC. AEG-1 might be a potential therapeutic target in CRC. 22470125_study detected MTDH expression in normal liver, chronic hepatitis B and HBV-related hepatocellular carcinoma(HCC)tissues; data showed that MTDH expression levels were elevated in the hepatitis B tissues and especially in the HBV-related HCC tissues compared to normal liver tissues 22643064_AEG-1 is up-regulated, at the mRNA and the protein level, during colorectal cancer development and aggressiveness. 22684557_These results suggested that the expression of MTDH/AEG-1 gene in HCC cell lines of different metastatic potentials was closely positively related to the abilities of orientation chemotaxis and adhesion of HCC cells. 22689379_AEG-1 provides strong protection from senescence, AEG-1 induces marked up-regulation of FXII protein, and is associated with induction of steatosis. 22768080_MTDH contributes to the pathogenesis of diffuse large-B-cell lymphoma mediated by activation of Wnt/beta-catenin pathway 22898150_MTDH protein may be a valuable marker of bladder cancer progression. MTDH expression is associated with poor overall survival in patients with bladder cancer. 22903204_MTDH overexpression contributes to an aggressive phenotype, thus leading to a poor prognosis for primary invasive breast cancer. 22938468_results demonstrated that expression of MTDH at the transcriptional level may be increased in gastric cancer tissue samples but with considerable heterogeneity 22967897_miR-136 might play a tumor-suppressive role in human glioma 23011158_Evidence that elevated expression of AEG-1 protein is correlated with poor prognosis and reduced survival of patients with neuroblastoma (P= 0.031). 23023948_findings demonstrate that AEG-1 plays a crucial role in T-cell non-Hodgkin's lymphoma growth and metastasis 23065261_The increased expression of MTDH and/or SND1 is closely related to carcinogenesis, progression, and prognosis of colon cancer. 23142337_High AEG-1 expression was associated with tongue carcinoma. 23240043_These data suggest that MTDH (-470G>A) could be a useful molecular marker for assessing ovarian cancer risk and for predicting ovarian cancer patient prognosis. 23307243_our results indicated that AEG-1 played a crucial role in the carcinogenesis of non-small cell lung cancer 23364922_MTDH overexpression was tightly associated with more aggressive tumour behaviour and a poor prognosis, indicating that MTDH is a valuable molecular biomarker for laryngeal squamous cell carcinoma progression. 23408429_oncogene metadherin modulates the apoptotic pathway based on the tumor necrosis factor superfamily member TRAIL (Tumor Necrosis Factor-related Apoptosis-inducing Ligand) in breast cancer 23499911_AEG-1 could potentially serve as an alternative target to AURKA for drug design and therapeutic treatment, to achieve more specific killing of AML cells while sparing normal cells. 23595222_MTDH expression is high in cervical cancer, and it contributes to chemoresistance of cervical cancer. 23640911_AEG-1 plays an important role in TGF-beta1-induced epithelial-mesenchymal transition through activation of p38 MAPK in proximal tubular epithelial cells. 23710918_AEG-1 protein might play a critical role in the initiation and progression of laryngeal squamous cell carcinoma 23835593_Overexpression of astrocyte-elevated gene-1 is associated with cervical carcinoma progression and angiogenesis. 23851509_High metadherin is associated with breast tumor growth and metastasis. 23889986_Initial cloning, structure, expression profile, and regulation of expression of AEG-1 protein. [Review] 23889987_A comprehensive analysis of the existing literature to emphasize the common and conflicting findings relative to the clinical significance of AEG-1/MTDH/LYRIC in cancer. [Review] 23889988_AEG-1 function relative to signaling changes, interacting partners, and angiogenesis and new perspectives on its potential as a significant target for the clinical treatment of cancers and other diseases. [review] 23889989_Discovery of agents that can block AEG-1 and its regulated pathways will be beneficial to cancer patients with aberrant expression of AEG-1.[review] 23889990_The multiple functions of AEG-1/MTDH/LYRIC in drug resistance highlight that it is a viable target as an anticancer agent for a wide variety of cancers. [review] 23889991_As an oncogene that promotes aberrant cellular processes within the CNS, AEG-1/MTDH/LYRIC represents an important therapeutic target for the treatment of neurological disease. [review] 23889992_Combination of AEG-1 inhibition and chemotherapy has documented significant efficacy in abrogating human hepatocellular carcinoma(HCC) xenografts.[review] 23910058_LY294002 treatment down-regulated AEG-1 expression in hepatocellular carcinoma 24063540_AEG-1 increases phosphorylation of the p65 subunit of NF-kappaB, and regulates the expression of MMP1 in head and neck squamous cell carcinoma cells. 24119914_AEG-1 gene expression associated with poor prognosis in children with Wilms tumor. 24136747_These findings suggest that AEG-1 may be an epithelial-mesenchymal transition-associated biomarker in human hepatocellular carcinoma and play important roles in the progression of hepatocellular carcinoma 24256614_High expression of astrocyte elevated gene-1 is associated with progression of cervical intraepithelial neoplasia. 24462870_Targeted inhibition of AEG-1 can lead to modification of key elemental characteristics, such as miRNAs, which may become a potential effective therapeutic strategy for CRC 24495449_MTDH/AEG-1 overexpression contributes to the neoplastic phenotype of bladder cancer cells by promoting survival, clonogenicity, and migration 24529480_Specific sites of LYRIC/AEG-1 ubiquitination are essential for regulating LYRIC/AEG-1 localisation and functionally interacting proteins. 24656097_Expression of AEG-1 may be correlated with tumor angiogenesis and metastasis and is a valuable prognostic factor in patients with oral squamous cell carcinoma 24659263_AEG-1 promoter variant -483 A>C may be associated with the susceptibility to HCC in Iranian population 24674449_An association between MTDH rs1835740 SNP and migraine in a Swedish case-control study. 24675891_Knockdown of astrocyte elevated gene-1 (AEG-1) in cervical cancer cells decreases their invasiveness, epithelial to mesenchymal transition, and chemoresistance. 24705862_AEG-1 appears to be a novel therapeutic target for preventing the metastasis of CRC. 24817955_Metadherin contribute to BCR signaling in chronic lymphocytic leukemia. 24829140_AEG-1 was overexpressed in bladder cancer tissues, compared with normal tissues.AEG-1 is associated with tumor progression in nonmuscle-invasive bladder cancer. 24829520_Astrocyte elevated gene-1 mediates glycolysis and tumorigenesis in colorectal carcinoma cells via AMPK signaling 24855648_IL-1beta- or TNF-alpha-induced AEG-1 interaction with NF-kappaB p65 subunit. 24941119_AEG-1 promotes anoikis resistance and orientation chemotaxis in hepatocellular carcinoma cells. 24946951_Determination of MTDH expression in primary CRC may be useful in the earlier detection of lung metastases in patients with high expression and increased risk 24963474_Strong p50, p65, and metadherin expression was associated with a high probability to distinguish ovarian carcinomas over borderline and benign ovarian tumours 24981741_MTDH supports the survival of mammary epithelial cells under oncogenic/stress conditions by interacting with and stabilizing SND1. 24989027_Study suggests that AEG-1 protein was highly expressed in pancreatic ductal adenocarcinoma and associated with poor prognosis of the patients. 25074613_Findings establish a pivotal role for MTDH in prostate cancer progression and metastasis. 25092897_results demonstrate that AEG-1 can promote gastric cancer progression by a positive feedback TLR4/NF-kappaB signaling-related mechanism 25125681_Study establishes AEG-1 as a novel homeostatic regulator of RXR and RXR/RAR that might contribute to hepatocarcinogenesis. 25174891_MTDH mediated metastasis of Osteosarcoma through regulating epithelial-mesenchymal transition. 25182604_Results demonstrate an increased expression of AEG-1 in malignant meningiomas correlated with grade; elevation of AEG-1 promotes malignant tumor progression in meningioma by an increase in cell proliferation, tumorigenicity, and a decrease in apoptosis 25193383_our findings offer in vivo proofs that AEG-1 is essential for NF-kappaB activation and hepatocarcinogenesis, and they reveal new roles for AEG-1 in shaping the tumor microenvironment for HCC development 25197376_our data suggest that AEG-1 may represent a novel prognostic marker for astrocytomas. 25204501_Taken together, our findings suggest that AEG-1 plays a crucial role in the aggressiveness of osteosarcoma via the JNK/c-Jun/MMP-2 pathway. 25266957_MiR-136 plays a key role in temozolomide (TMZ) resistance by targeting AEG-1 in glioma cell line, suggesting that miR-136 can be used to predict a patient's response to TMZ therapy as well as serve as a novel potential maker for glioma therapy. 25304263_Our study uncovers a novel molecular mechanism by which AEG-1 augments glioma progression and offers a rationale to block AEG-1-Akt2 signaling function as a novel GBM treatment. 25323629_microRNA-22 acts as a metastasis suppressor by targeting metadherin in gastric cancer. 25333261_MTDH up-regulation was associated with high-grade serous ovarian carcinoma. 25346496_Data provide evidence that MTDH has a functional role in the progression and metastatic spread of ovarian cancer. 25407490_AEG-1 may be a novel predictor for metastasis and prognosis of the patients with GC. 25417825_MTDH mediates trastuzumab resistance, at least in part, by PTEN inhibition through an NFkappaB-dependent pathway, which may be utilized as a promising therapeutic target for HER2 positive breast cancer. 25428378_we have reported the altered expression of miR-145 in NSCLC cell lines and that miR-145 could modulate cell proliferation and invasion by targeting AEG-1/MTDH 25431427_Data suggest that AEG-1 plays an important role in renal cancer formation and development. 25432696_data suggest that miR-542-3p might function as a tumor suppressor in gastric cancer, potentially by targeting the oncogene AEG-1 25483832_AEG1 is a valuable biomarker for the prediction of ovarian cancer prognosis, and AEG1 inhibition may be a potential therapeutic strategy for ovarian cancer treatment. 25484183_A competitive endogenous RNAs regulatory network among AEG-1, Snail and Vimentin mediated via competitive binding to miR-30a was proved. 25575438_The results demonstrate that MTDH is specifically expressed in B cell of chronic lymphocytic leukemia and exert a preservative role through activation of Wnt signaling pathway 25652471_our results suggest that the evodiamine suppress the proliferation of lung cancer cells, at least, in part, via inhibition of MTDH expression and activation of apoptosis. 25684730_Repression of metadherin inhibits biological behavior of prostate cancer cells and enhances their sensitivity to cisplatin. 25695541_AEG-1 was associated with the progression of cervical squamous cell carcinoma by promoting epithelial-mesenchymal transition via Wnt signaling pathway. 25787750_Real-time quantitative RT-PCR revealed a dose-dependent decrease in RNA-binding factor 1 (AUF-1) and astrocyte elevated gene-1 (AEG-1) messenger RNA (mRNA) levels in TP-1-treated SMMC-7721 cells 25794773_Results indicated that MTDH was a direct downstream target of miR-153 and was involved in the miR-153-induced suppression of the migration and invasion in breast cancer cell. 25824750_Overexpression of astrocyte elevated gene-1 is associated with angiogenesis in cervical cancer. 25880337_These results reveal the critical role of AEG-1 in EMT and suggest that AEG-1 may be a prognostic biomarker and its targeted inhibition may be utilized as a novel therapy for NSCLC. 25902416_Study provides evidence that the knockdown of MTDH signi fi cantly inhibited angiogenesis suggesting that MTDH is a potential therapeutic target for anti-angiogenesis in breast cancer. 25913216_In patients with non-small cell lung cancer, AEG-1 expression was an independent prognostic factor for both overall and disease-free survival. 25944909_AEG-1 thus might play a role in NTIS associated with HCC and other cancers. 25987128_EFEMP1 might indirectly enhance the expression of MMP-2, providing a potential explanation for the role of AEG-1 in metastasis. NF-kappaB pathways might be one of the effective ways which EFEMP1 was induced by AEG-1. 26016795_ectopic miR-217 expression decreased AEG-1 expression and repressed luciferase reporter activity associated with the AEG-1 3'-untranslated region (UTR). 26035424_Data established that AEG-1 is frequently upregulated and functions as an oncogene by regulating several major signaling pathways in hepatocellular carcinoma (HCC) suggesting potential role as a biomarker or target for HCC therapy. [review] 26051629_MTDH might be used as a potential therapeutic target in the lymph node metastasis of ESCC 26096243_The present work aims to investigate the relationship between the expression of AEG-1(astrocyte elevated gene-1), b-FGF(basic-fibroblast growth factor), beta-catenin, Ki-67, TNF-alpha (tumor necrosis factor-alfa) other prognostic parameters in DC (Ductal Carcinomas) and ductal intraepithelial neoplasm. We found a relationship between these factors. 26122237_mRNA and protein expression level of metadherin in breast cancer cells both improved after transfection. The inhibition effect of 1 mg/L doxorubicin and 8 mg/L taxol on breast cancer cells decreased after metadherin transfection. 26134542_AEG-1 mediates CCL3/CCR5-induced EMT development via both Erk1/2 and Akt signaling pathway in CM patients, which indicates CCL3/CCR5-AEG-1-EMT pathway could be suggested as a useful target to affect the progression of CM. 26141861_our work revealed that MTDH promotes CSC accumulation and breast tumorigenicity by regulating TWIST1, deepening the understanding of MTDH function in cancer. 26156805_AEG-1 mediates CCL20/CCR6-induced EMT development via both Erk1/2 and Akt signaling pathway in cervical cancer, which indicates that CCL20/CCR6-AEG-1-EMT pathway could be suggested as a useful target to affect the progression of cervical cancer. 26176806_miR-302c-3p play a pivotal role in the progression of glioma by targeting MTDH and is a potential inhibitor in glioma treatment. 26236947_Data suggest that activation of AMPK (AMP-activated protein kinase) in triple negative breast cancer cells down-regulates expression of MTDH (metadherin) via up-regulation of GSK3B (glycogen synthase kinase 3 beta) and SIRT1 (sirtuin 1) activities. 26287185_Knockdown of MTDH inhibits phosphorylation of AKT and increased apoptosis related protein expression. 26318406_We identified two powerful genes in the liver cancer metastasis process, AEG-1 and AKR1C2. 26336827_Results indicate that microRNA miR-497 directly inhibited the 3'-untranslated regions (3' UTRs) of vascular endothelial growth factor A (VEGFA) and astrocyte elevated gene-1 (AEG-1). 26351209_Identify two novel factors, AKR1C2 (positive factor) and NF1 (negative factor), as the AEG-1 downstream players in the process of metastasis in liver cancer. 26418251_Data showed that AEG1 and MMP7 levels were both significantly increased and strongly correlated in non-small cell lung cancer (NSCLC) tissues. AEG-1 promotes NSCLC cell invasiveness through MAPK-p42/p44-dependent activation of MMP7. 26589417_Our results suggest that underexpression of miR-30a-5p might function as a tumor suppressing miRNA by directly targeting MTDH in HCC and is therefore a potential candidate biomarker for HCC targeting therapy. 26595523_Data indicate metadherin (MTDH) as a direct target gene of microRNA miR-630. 26683226_High MTDH expression is associated with multiple myeloma. 26689985_AEG-1 overexpression is closely associated with epithelial-mesenchymal transition and promote metastasis in tongue squamous cell carcinoma. 26710214_High AEG-1 staining index might be associated with tumor progression and poor survival status in patients with gastrointestinal cancer. Meta-analysis. 26823698_Suggest that miR-377 plays an important role in the development of NSCLC by regulating AEG-1 expression. 26884832_MiR-30a-5p may play an essential role in the cell growth and apoptosis of hepatocellular carcinoma cells, partially via targeting AEG-1. 27059438_rs1835740 associated with the need for neonatal resuscitation and Apgar scores 27090750_In-depth review of the role of AEG-1 in a diverse spectrum of diseases, from cancer to HIV-1 and aging, highlighting mechanistic processes that synergize with known AEG-1 functions and promoting the notion that AEG-1 occupies a unique niche in inflammation, mitochondrial, ER and nucleolar stress, culminating in excitotoxicity and other neurodegenerative outcomes. 27229534_Findings clearly demonstrate that miR-320a suppresses breast cancer metastasis by directly inhibiting MTDH expression. The present study provides a new insight into anti-oncogenic roles of miR-320a. 27339400_Knocking down TSPAN8 in AEG-1-overexpressing human hepatocellular carcinoma (HCC) cells markedly inhibited invasion and migration without affecting proliferation. TSPAN8 knockdown profoundly abrogated AEG-1-induced primary tumor and intrahepatic metastasis in an orthopic xenograft model in athymic nude mice 27434586_Study highlights the importance of interactions among lncRNA HCP5, microRNA-139, and transcription factor RUNX1 in regulating the malignant behavior of glioma cells. HCP5 down-regulated miR-139 to up-regulate RUNX1. RUNX1 promoted AEG-1 expression, which was involved in a series of oncogenic effects in glioma cells. RUNX1 also up-regulated HCP5 expression, which formed a positive feedback loop. 27565732_IL-8 was displayed to be capable of directly interacting with metadherin (MTDH), which in turn can up-regulate IL-8 expression. 27571703_these findings suggested that miR-124 was able to suppress cell proliferation, invasion and migration, as well as the EMT process in cervical carcinomas through directly targeting AEG-1. miR-124 and AEG-1 may be potential 27667169_our findings suggest that AEG-1 promotes mesenchymal transition in glioblastoma through the regulation of the Rho signaling pathway, resulting in tumor invasion, a primary characteristic of malignant brain tumors 27746178_AEG-1 is involved in miR-1297-regulated prostate cancer cell proliferation and invasion.MiR-1297 targets and inhibits AEG-1 in prostate cancer. 27765924_Gene expression data and in silico database analysis showed that the metadherin gene (MTDH) was a direct target of both miR-145-5p and miR-145-3p regulation and high expression of MTDH predicted poorer survival of lung squamous cell carcinoma patients. 27793010_Study shows that expression of AEG-1 was significantly higher in macrophages and associated with elevated expression levels of MMP-9 in hypopharyngeal cancer. These results demonstrate that macrophage AEG-1 promotes tumor invasion through up-regulation of MMP-9 in both macrophages and cancer cells. 27835571_AEG-1 knockdown inhibits migration and invasion, as well as radiation-enhanced invasion both in vitro and in vivo I colonic cancer cells. 27903708_Expression of AEG-1 was strongly associated with stem cell markers CD133 and SOX2. AEG-1 facilitated beta-catenin translocation into the nucleus by forming a complex with LEF1 and beta-catenin, subsequently activating Wnt signaling downstream genes. 27917902_The results indicated that high MTDH expression is significantly correlated with higher mortality in breast, ovarian and cervical cancer. (Meta-analysis) 27923917_High MTDH expression is associated with glioblastomas. 27956703_c-Jun and p300 are novel interacting partners of AEG-1 in gliomas. 28107197_MTDH induces Epithelial-mesenchymal transition-like change and invasion of glioma via the regulation of miR-130b-competing endogenous RNAs, providing the first direct link between MTDH and miRNAs in cancer cells. 28112756_Long noncoding RNA FTX regulated astrocyte-elevated gene-1 (AEG-1) expression through microRNA miR-342-3p. 28174206_miR-26a is capable of suppressing the proliferation and migration of ESCC cells via negative regulation of metadherin 28184926_High MTDH expression is associated with hepatocellular carcinoma. 28276315_Results show that miR-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells through targeting AEG-1. 28323000_Study showed that AEG-1's knock down inhibited cell growth, triggered apoptosis, arrested cell cycle, retarded cell migration and invasion in ovarian cancer cells. These results provide evidence that AEG-1 plays an oncogenic role in ovarian cancer cells. 28401704_Results demonstrated that AEG-1 significantly enhanced invasion capabilities of OVCAR3 ovarian cancer cells and suggested that HIF-1alpha binds to AEG-1 promoter to upregulate its expression, which was correlated with metastasis in ovarian cancer. 28429540_oncogenic role of AEG-1 in melanoma 28534938_High MTDH expression is associated with gastric cancer metastasis. 28627585_Through the analysis of the Cancer Genome Atlas (TCGA) datasets for estrogen receptor (ER)-positive endometrial and breast cancers, this study found that over 25% of all gene expression correlated with MTDH. 28661037_The findings suggest that AEG-1 promotes gastric cancer metastasis through upregulation of eIF4E-mediated MMP-9 and Twist. 28685276_MTDH expression does not correlate with prognosis in the esophageal adenocarcinoma patients. 28714009_Upregulation of MTDH reversed the suppression of glioma cell growth and metastasis by miR-202. 28731152_We found that overexpression of AEG-1 in PTC was positively correlated with lymph node metastasis and MMP2/9 expression. Knockdown of AEG-1 reduced the capacity of migration and invasion through downregulation of MMP2/9 in thyroid cancer cells. Furthermore, we firstly found that AEG-1 interacted with MMP9 in thyroid cancer cells. 28849015_miR448 was downregulated in osteosarcoma and overexpression of miR448 inhibited cell proliferation and invasion by targeting AEG1, providing novel insights into the understanding of osteosarcoma pathogenesis. 28877780_High AEG-1 expression is associated with renal cell carcinoma. 28901527_Study confirmed that hypoxia increased MTDH expression via HIF-1alpha expression. Knockdown of MTDH expression in head and neck squamous cell carcinoma (HNSCC) cell lines interrupted hypoxiainduced metastasis and glycolysis. Furthermore, reduced MTDH expression decreased HIF-1alpha expression. These results indicated the presence of a positive feedback loop between MTDH and HIF-1alpha in HNSCC. 28923415_we have identified a novel mechanism for DIM- and ring-DIM-induced protective autophagy, via induction of AEG-1 and subsequent activation of AMPK. Our findings could facilitate the development of novel drug therapies for prostate cancer that include selective autophagy inhibitors as adjuvants. 28927747_Our findings provided evidence that AEG-1 contributed to the production of inflammatory cytokines, migration and invasion of rheumatoid arthritis fibroblast-like synoviocytes, and underscored the importance of AEG-1 in the inflammation process of rheumatoid arthritis 28938524_miR-384 and AEG-1 may serve as potential targets for the diagnosis and treatment of non-small-cell lung cancer. 28941723_AEG-1 may play important roles at the transcription level in malignant transformation and tumor angiogenesis in non-small cell lung cancer. 28952213_AEG-1 and CXCR4 activate and regulat | ENSMUSG00000022255 | Mtdh | 1.448265e+03 | 0.9625298 | -0.055096817 | 0.2971154 | 3.419487e-02 | 0.8532929317 | 0.97087206 | No | Yes | 1.189333e+03 | 179.888466 | 1.110628e+03 | 172.172535 | ||
| ENSG00000148143 | 58499 | ZNF462 | protein_coding | Q96JM2 | FUNCTION: Zinc finger nuclear factor involved in transcription by regulating chromatin structure and organization (PubMed:20219459, PubMed:21570965). Involved in the pluripotency and differentiation of embryonic stem cells by regulating SOX2, POU5F1/OCT4, and NANOG (PubMed:21570965). By binding PBX1, prevents the heterodimerization of PBX1 and HOXA9 and their binding to DNA (By similarity). Regulates neuronal development and neural cell differentiation (PubMed:21570965). {ECO:0000250|UniProtKB:B1AWL2, ECO:0000269|PubMed:20219459, ECO:0000269|PubMed:21570965}. | 3D-structure;Alternative splicing;DNA-binding;Disease variant;Glycoprotein;Isopeptide bond;Metal-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene belongs to C2H2-type zinc finger family of proteins. It contains multiple C2H2-type zinc fingers and may be involved in transcriptional regulation. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Dec 2016]. | hsa:58499; | nucleus [GO:0005634]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; negative regulation of DNA binding [GO:0043392]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of gene expression [GO:0010468] | 18978678_Observational study of gene-disease association. (HuGE Navigator) 19266077_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20080650_Observational study of gene-disease association. (HuGE Navigator) 20546612_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22826440_Zinc finger protein 462 (ZNF462) is the first human O-GlcNAc-6-phosphate modified protein. 28513610_Loss of function variants in ZNF462 were identified in patients with ptosis, metopic ridging, craniosynostosis, dysgenesis of the corpus callosum, and developmental delay. 29427787_In the current study we describe a patient with a syndromic form of autism spectrum disorder and intellectual disability characterized by metopic craniosynostosis, ptosis and corpus callosum dysgenesis most likely caused by ZNF462 haploinsufficiency. The possible contribution of the disruption of KLF12 by one of the translocation breakpoints remains unclear. 31361404_a multiple congenital anomaly syndrome associated with haploinsufficiency of ZNF462 that has distinct clinical characteristics and facial features, is reported. | ENSMUSG00000060206 | Zfp462 | 2.003944e+02 | 0.8567289 | -0.223089321 | 0.3299178 | 4.414501e-01 | 0.5064233932 | 0.87821327 | No | Yes | 2.124383e+02 | 46.677436 | 2.124474e+02 | 48.214904 | |
| ENSG00000148356 | 90678 | LRSAM1 | protein_coding | Q6UWE0 | FUNCTION: E3 ubiquitin-protein ligase that mediates monoubiquitination of TSG101 at multiple sites, leading to inactivate the ability of TSG101 to sort endocytic (EGF receptors) and exocytic (HIV-1 viral proteins) cargos (PubMed:15256501). Bacterial recognition protein that defends the cytoplasm from invasive pathogens (PubMed:23245322). Localizes to several intracellular bacterial pathogens and generates the bacteria-associated ubiquitin signal leading to autophagy-mediated intracellular bacteria degradation (xenophagy) (PubMed:23245322, PubMed:25484098). {ECO:0000269|PubMed:15256501, ECO:0000269|PubMed:23245322, ECO:0000269|PubMed:25484098}. | Alternative splicing;Autophagy;Charcot-Marie-Tooth disease;Coiled coil;Cytoplasm;Disease variant;Leucine-rich repeat;Metal-binding;Neurodegeneration;Neuropathy;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transferase;Transport;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:15256501, ECO:0000269|PubMed:23245322}. | This gene encodes a ring finger protein involved in a variety of functions, including regulation of signaling pathways and cell adhesion, mediation of self-ubiquitylation, and involvement in cargo sorting during receptor endocytosis. Mutations in this gene have been associated with Charcot-Marie-Tooth disease. Multiple transcript variants encoding different isoforms have been identified for this gene. [provided by RefSeq, Jan 2012]. | hsa:90678; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; autophagy [GO:0006914]; negative regulation of endocytosis [GO:0045806]; positive regulation of autophagosome assembly [GO:2000786]; positive regulation of xenophagy [GO:1904417]; protein autoubiquitination [GO:0051865]; protein catabolic process [GO:0030163]; protein polyubiquitination [GO:0000209]; ubiquitin-dependent endocytosis [GO:0070086]; viral budding [GO:0046755] | 14635195_Results suggest that RIFLE represents a novel signaling protein that mediates components of the Wnt/wingless signaling pathway and cell adhesion in PC12 cells [RIFLE protein]. 15256501_Tal regulates a Tsg101-associated complex responsible for the sorting of cargo into cytoplasm-containing vesicles that bud at the multivesicular body and at the plasma membrane 18077552_Tal polyubiquitinates lysine residues in the C-terminus of uncomplexed Tsg101, resulting in proteasomal degradation. 20616063_LRSAM1 as a component of the antibacterial autophagic response. 20865121_LRSAM1 is a strong candidate for the causal gene for the Charcot-Marie-Tooth disease. 22012984_homozygous mutation in LRSAM1 was proposed as a strong candidate for the disease in a family with recessive axonal polyneuropathy 22781092_Our data further confirms that LRSAM1 mutations are associated with CMT2 of AD inheritance. 23245322_Authors identify LRSAM1 as the E3 ligase responsible for anti-Salmonella autophagy-associated ubiquitination. 24894446_disruption of the C-terminal RING domain confers dominant negative properties to LRSAM1 25484098_Plant homeodomain finger protein 23 negatively regulates cell autophagy by promoting ubiquitination and degradation of E3 ligase LRSAM1 27615052_findings suggest that the mutant LRSAM1 may aberrantly affect the formation of transcription machinery. 27686364_findings demonstrate that the isolated genetic entity Charcot-Marie-Tooth type 2G is caused by a missense mutation in LRSAM1. 28189685_LRSAM1 exhibited self-association in vitro and in vivo. The study found the self-association of LRSAM1 promotes intermolecular ubiquitination and proved a potential N-terminal ubiquitination. 28335037_We identified a novel LRSAM1 missense mutation (c.2120C > T, p.Pro707Leu) mapping to the RING domain. The identified missense mutation, as well as of another recently reported pathogenic missense mutation (c.2081G > A, p.Cys694Tyr), revealed that in vitro ubiquitylation activity was largely abrogated. 29253842_Our study shows the potential function of mir-939 through regulating LRSAM1 in Hirschsprung's disease 29341362_We report a novel LRSAM1 mutation c.2021-2024del (p.E674VfsX11) in 4 members of a Chinese autosomal dominant Charcot-Marie-Tooth disease type 2 family 30826859_The article systematically represents the molecular functions, nature and detailed characterization of LRSAM1 E3 ubiquitin ligase, which are linked to molecular mechanisms of neurodegeneration. (Review) 30996334_Our results confirm the localization of variants in its catalytic C-terminal RING domain and broaden the phenotypic spectrum of LRSAM1-related neuropathies, including painful and predominantly sensory ataxic forms. 31852984_Identification of novel pathogenic copy number variations in Charcot-Marie-Tooth disease. 31982566_Ubiquitin ligase LRSAM1 suppresses neurodegenerative diseases linked aberrant proteins induced cell death. 33207262_LRSAM1 E3 ubiquitin ligase promotes proteasomal clearance of E6-AP protein. 33414056_Location matters - Genotype-phenotype correlation in LRSAM1 mutations associated with rare Charcot-Marie-Tooth neuropathy CMT2P. | ENSMUSG00000026792 | Lrsam1 | 1.034652e+03 | 1.2992498 | 0.377678849 | 0.3043123 | 1.566873e+00 | 0.2106617126 | 0.78763590 | No | Yes | 1.063054e+03 | 98.792840 | 7.571651e+02 | 72.731289 |
| ENSG00000148396 | 9919 | SEC16A | protein_coding | O15027 | FUNCTION: Acts as a molecular scaffold that plays a key role in the organization of the endoplasmic reticulum exit sites (ERES), also known as transitional endoplasmic reticulum (tER). SAR1A-GTP-dependent assembly of SEC16A on the ER membrane forms an organized scaffold defining an ERES. Required for secretory cargo traffic from the endoplasmic reticulum to the Golgi apparatus (PubMed:17192411, PubMed:17005010, PubMed:17428803, PubMed:21768384, PubMed:22355596). Mediates the recruitment of MIA3/TANGO to ERES (PubMed:28442536). Regulates both conventional (ER/Golgi-dependent) and GORASP2-mediated unconventional (ER/Golgi-independent) trafficking of CFTR to cell membrane (PubMed:28067262). Positively regulates the protein stability of E3 ubiquitin-protein ligases RNF152 and RNF183 and the ER localization of RNF183 (PubMed:29300766). Acts as a RAB10 effector in the regulation of insulin-induced SLC2A4/GLUT4 glucose transporter-enriched vesicles delivery to the cell membrane in adipocytes (By similarity). {ECO:0000250|UniProtKB:E9QAT4, ECO:0000269|PubMed:17005010, ECO:0000269|PubMed:17192411, ECO:0000269|PubMed:17428803, ECO:0000269|PubMed:21768384, ECO:0000269|PubMed:22355596, ECO:0000269|PubMed:28067262, ECO:0000269|PubMed:28442536, ECO:0000269|PubMed:29300766}. | Alternative splicing;Cytoplasm;ER-Golgi transport;Endoplasmic reticulum;Golgi apparatus;Membrane;Microsome;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene encodes a protein that forms part of the Sec16 complex. This protein has a role in protein transport from the endoplasmic reticulum (ER) to the Golgi and mediates COPII vesicle formation at the transitional ER. Alternative splicing results in multiple transcript variants that encode different protein isoforms. [provided by RefSeq, Feb 2013]. | hsa:9919; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum exit site [GO:0070971]; endoplasmic reticulum membrane [GO:0005789]; ER to Golgi transport vesicle membrane [GO:0012507]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; organelle membrane [GO:0031090]; perinuclear region of cytoplasm [GO:0048471]; autophagy [GO:0006914]; COPII vesicle coating [GO:0048208]; endoplasmic reticulum organization [GO:0007029]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; Golgi organization [GO:0007030]; Golgi to plasma membrane CFTR protein transport [GO:0043000]; protein exit from endoplasmic reticulum [GO:0032527]; protein localization to endoplasmic reticulum exit site [GO:0070973]; protein localization to plasma membrane [GO:0072659]; protein stabilization [GO:0050821]; response to endoplasmic reticulum stress [GO:0034976]; substantia nigra development [GO:0021762] | 17005010_Sar1-GTP-dependent assembly of Sec16 on the endoplasmic reticulum(ER) membrane forms an organized scaffold defining ER exit sites 17192411_Mammalian cells contain two distinct Sec16 homologues: a large protein Sec16L of 2154 aa and a smaller protein Sec16S of 1060 aa. 17428803_Results suggest that KIAA0310p, a mammalian homologue of yeast Sec16, builds up endoplasmic reticulum (ER) exit sites in cooperation with p125 and plays a role in membrane traffic from the ER. 19638414_These data are consistent with a model where Sec16 acts as a platform for COPII assembly at endoplasmic reticulum exit sites. 21045114_Sec16A remains associated with endoplasmic reticulum exit sites throughout mitosis. 21768384_Data show that knockdown of Sec16B but not Sec16A by RNAi affected the morphology of peroxisomes, inhibited the transport of Pex16 from the ER to peroxisomes, and suppressed expression of Pex3. 25201882_LRRK2 regulates the anterograde endoplasmic reticulum (ER)-Golgi transport through anchoring Sec16A at the endoplasmic reticulum exit sites (ERES). 25526736_growth factors modulate Sec16 protein levels and dynamics. Sec16 acts as part of a coherent feed-forward loop, which integrates secretion and growth factor signaling. 25956157_Results suggest that it is the presence of rare syntenic SEC16A and MAMDC4 deletions that increases susceptibility to axial spondyloarthritis in family members who carry the HLA-B*27 allele. 28067262_these findings highlight a novel function of Sec16A as an essential mediator of endoplasmic reticulum stress-associated unconventional secretion. 28442536_Mammalian endoplasmic reticulum exit sites are organized by TANGO1 acting as a scaffold, in cooperation with Sec16 for efficient secretion. 29187380_Nbeal2 interacts with Dock7, Sec16a, and Vac14. 29300766_Sec16A also stabilized the interacting ubiquitin ligase RNF152, which localizes to the lysosome and has structural similarity with RNF183 | ENSMUSG00000026924 | Sec16a | 8.080000e+03 | 1.1621117 | 0.216748727 | 0.2889839 | 5.602949e-01 | 0.4541414445 | 0.86227340 | No | Yes | 9.142577e+03 | 828.687996 | 7.446569e+03 | 692.697435 | |
| ENSG00000148399 | 92715 | DPH7 | protein_coding | Q9BTV6 | FUNCTION: Catalyzes the demethylation of diphthine methyl ester to form diphthine, an intermediate diphthamide biosynthesis, a post-translational modification of histidine which occurs in translation elongation factor 2 (EEF2) which can be ADP-ribosylated by diphtheria toxin and by Pseudomonas exotoxin A (Eta). {ECO:0000250|UniProtKB:P38332, ECO:0000269|PubMed:19965467, ECO:0000269|PubMed:23486472}. | Hydrolase;Phosphoprotein;Reference proteome;Repeat;WD repeat | PATHWAY: Protein modification; peptidyl-diphthamide biosynthesis. | Diphthamide is a post-translationally modified histidine residue present in elongation factor 2, and is the target of diphtheria toxin. This gene encodes a protein that contains a WD-40 domain, and is thought to be involved in diphthamide biosynthesis. A similar protein in yeast functions as a methylesterase, converting methylated diphthine to diphthine, which can then undergo amidation to produce diphthamide. [provided by RefSeq, Oct 2016]. | hsa:92715; | diphthine methylesterase activity [GO:0061685]; peptidyl-diphthamide biosynthetic process from peptidyl-histidine [GO:0017183] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000026975 | Dph7 | 3.166464e+03 | 1.1316518 | 0.178430178 | 0.2842678 | 3.982108e-01 | 0.5280147096 | 0.88668779 | No | Yes | 2.912401e+03 | 258.920552 | 2.538563e+03 | 231.834169 |
| ENSG00000148459 | 23590 | PDSS1 | protein_coding | Q5T2R2 | FUNCTION: Heterotetrameric enzyme that catalyzes the condensation of farnesyl diphosphate (FPP), which acts as a primer, and isopentenyl diphosphate (IPP) to produce prenyl diphosphates of varying chain lengths and participates in the determination of the side chain of ubiquinone (PubMed:16262699). Supplies nona and decaprenyl diphosphate, the precursors for the side chain of the isoprenoid quinones ubiquinone-9 (Q9)and ubiquinone-10 (Q10) respectively (PubMed:16262699). The enzyme adds isopentenyl diphosphate molecules sequentially to farnesyl diphosphate with trans stereochemistry (PubMed:16262699). {ECO:0000269|PubMed:16262699}. | Alternative splicing;Disease variant;Isoprene biosynthesis;Lipid metabolism;Magnesium;Metal-binding;Mitochondrion;Primary mitochondrial disease;Reference proteome;Transferase;Ubiquinone biosynthesis | PATHWAY: Cofactor biosynthesis; ubiquinone biosynthesis. | The protein encoded by this gene is an enzyme that elongates the prenyl side-chain of coenzyme Q, or ubiquinone, one of the key elements in the respiratory chain. The gene product catalyzes the formation of all trans-polyprenyl pyrophosphates from isopentyl diphosphate in the assembly of polyisoprenoid side chains, the first step in coenzyme Q biosynthesis. The protein may be peripherally associated with the inner mitochondrial membrane, though no transit peptide has been definitively identified to date. Defects in this gene are a cause of coenzyme Q10 deficiency. [provided by RefSeq, Jul 2008]. | hsa:23590; | mitochondrial matrix [GO:0005759]; transferase complex [GO:1990234]; all-trans-decaprenyl-diphosphate synthase activity [GO:0097269]; metal ion binding [GO:0046872]; prenyltransferase activity [GO:0004659]; protein heterodimerization activity [GO:0046982]; trans-hexaprenyltranstransferase activity [GO:0000010]; trans-octaprenyltranstransferase activity [GO:0050347]; isoprenoid biosynthetic process [GO:0008299]; ubiquinone biosynthetic process [GO:0006744] | 16262699_murine and human solanesyl and decaprenyl diphosphate synthases are heterotetramers composed of newly characterized hDPS1 (mSPS1) and hDLP1 (mDLP1) 17332895_A deletirious mutation D308E that causes severe ubiquinone deficiency was identified in PDSS1. 18636124_Observational study of gene-disease association. (HuGE Navigator) 20010493_low CoQ10 is another factor explaining the risk to cardiovascular disorder in depression 20010505_lowered levels of CoQ10 play a role in the pathophysiology of myalgic encephalomyelitis/ chronic fatigue and that symptoms, such as fatigue, and autonomic and neurocognitive symptoms may be caused by CoQ10 depletion 20051244_Data show that expression of either dlp1 or dps1 recovered the thermo-sensitive growth of an E. coli ispB(R321A) mutant and restored IspB activity and production of Coenzyme Q-8. 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 31868205_CircRNA circPDSS1 promotes bladder cancer by down-regulating miR-16. 32232919_Genetic variants in PDSS1 and SLC16A6 of the ketone body metabolic pathway predict cutaneous melanoma-specific survival. 33285023_Missense PDSS1 mutations in CoenzymeQ10 synthesis cause optic atrophy and sensorineural deafness. 34408002_PDSS1-Mediated Activation of CAMK2A-STAT3 Signaling Promotes Metastasis in Triple-Negative Breast Cancer. | ENSMUSG00000026784 | Pdss1 | 3.242744e+02 | 0.8166459 | -0.292217391 | 0.3193641 | 8.036839e-01 | 0.3699942281 | 0.83676049 | No | Yes | 2.798007e+02 | 41.812675 | 3.545507e+02 | 54.060742 |
| ENSG00000148481 | 80013 | MINDY3 | protein_coding | Q9H8M7 | FUNCTION: Hydrolase that can remove 'Lys-48'-linked conjugated ubiquitin from proteins. {ECO:0000269|PubMed:27292798}. | Alternative splicing;Apoptosis;Hydrolase;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway | The protein encoded by this gene contains a caspase-associated recruitment domain and may function in apoptosis. It has been identified as a tumor suppressor in lung and gastric cancers, and a polymorphism in the gene may be associated with gastric cancer risk. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Dec 2015]. | hsa:80013; | nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; Lys48-specific deubiquitinase activity [GO:1990380]; thiol-dependent deubiquitinase [GO:0004843]; apoptotic process [GO:0006915] | 12054670_CARP is a novel caspase recruitment domain containing pro-apoptotic protein. (CARP, CARD containing Protein) 21499297_results indicated that C10ORF97 functions as a novel tumor suppressor by modulating several key G(1)/S-regulatory proteins by interacting with JAB1 24870804_CARP is a potential tumor suppressor of gastric carcinoma and the rs2297882 C>T phenotype of CARP may serve as a predictor of gastric carcinoma. | ENSMUSG00000026767 | Mindy3 | 3.473672e+02 | 1.0147939 | 0.021186685 | 0.3789217 | 3.082330e-03 | 0.9557252043 | 0.99122996 | No | Yes | 3.811268e+02 | 82.072370 | 3.043700e+02 | 67.406767 | |
| ENSG00000148700 | 120 | ADD3 | protein_coding | Q9UEY8 | FUNCTION: Membrane-cytoskeleton-associated protein that promotes the assembly of the spectrin-actin network. Plays a role in actin filament capping (PubMed:23836506). Binds to calmodulin. {ECO:0000269|PubMed:23836506}. | Acetylation;Actin-binding;Alternative splicing;Calmodulin-binding;Cell membrane;Cytoplasm;Cytoskeleton;Disease variant;Isopeptide bond;Membrane;Phosphoprotein;Reference proteome;Ubl conjugation | Adducins are heteromeric proteins composed of different subunits referred to as adducin alpha, beta and gamma. The three subunits are encoded by distinct genes and belong to a family of membrane skeletal proteins involved in the assembly of spectrin-actin network in erythrocytes and at sites of cell-cell contact in epithelial tissues. While adducins alpha and gamma are ubiquitously expressed, the expression of adducin beta is restricted to brain and hematopoietic tissues. Adducin, originally purified from human erythrocytes, was found to be a heterodimer of adducins alpha and beta. Polymorphisms resulting in amino acid substitutions in these two subunits have been associated with the regulation of blood pressure in an animal model of hypertension. Heterodimers consisting of alpha and gamma subunits have also been described. Structurally, each subunit is comprised of two distinct domains. The amino-terminal region is protease resistant and globular in shape, while the carboxy-terminal region is protease sensitive. The latter contains multiple phosphorylation sites for protein kinase C, the binding site for calmodulin, and is required for association with spectrin and actin. Alternatively spliced adducin gamma transcripts encoding different isoforms have been described. The functions of the different isoforms are not known. [provided by RefSeq, Jul 2008]. | hsa:120; | brush border [GO:0005903]; cell cortex [GO:0005938]; cell-cell junction [GO:0005911]; condensed nuclear chromosome [GO:0000794]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; membrane [GO:0016020]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; actin filament binding [GO:0051015]; calmodulin binding [GO:0005516]; structural constituent of cytoskeleton [GO:0005200]; barbed-end actin filament capping [GO:0051016] | 15716695_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15716695_The interaction of ADD1 and ADD3 gene variants in humans is statistically associated with variation in blood pressure, suggesting the presence of epistatic effects among these loci. 15834281_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15963851_The early expression of ADD3 suggests that it may have a role in erythroblasts but is replaced by ADD2 in later stages of erythropoiesis. 16385451_Observational study of gene-disease association. (HuGE Navigator) 18475162_Left ventricular diastolic relaxation is modulated by genetic variation in ADD3. 18475162_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18667944_Observational study of gene-disease association. (HuGE Navigator) 18787518_In ADD1 GlyGly homozygotes, the properties of the brachial artery are related to the ADD3 (A386G) polymorphism, but the underlying mechanism needs further clarification. 18787518_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20810786_These findings suggest novel roles for adducins in stabilization of epithelial junctions and regulation of junctional remodeling. 21164023_gamma-adducin may influence blood pressure homeostasis by modulating renal NaCl transport. 23814265_High expression of ADD3 is associated with glioma. 23836506_Homozygous p.G367D mutation in ADD3 causes spastic diplegic/quadriplegic cerebral palsy and intellectual disability. 23872602_Common genetic variants in 10q24.2 can alter biliary atresia risk by regulating ADD3 expression levels in the liver, and may exert an effect on disease epidemiology and on the general population. 24104524_ADD3 gene may be functionally relevant for the development of biliary atresia 25285724_ADD3 gene plays an important role in biliary atresia pathogenesis. 28033648_ADD3 gene deletion is associated with acute lymphoblastic leukemia. 28902846_MiR-145-5p was confirmed to target ADD3 by luciferase reporter assay. The downregulation of miR-145 may contribute to liver fibrosis in Biliary atresia by upregulating the expression of ADD3. 29508064_ADD3 and ADD3-AS1 variants increased susceptibility to BA, suggesting that these genes may play an additive role in the pathogenesis of the disease. 29685956_The intragenic epistatic association of ADD3 with biliary atresia in Southern Han Chinese population has been reported. 29768408_Data indicate lysine acetyltransferase 2B (KAT2B) as a susceptibility gene for kidney and heart disease in adducin 3 (gamma) protein (ADD3)-associated disorders. 31958485_Loss of cytoskeleton protein ADD3 promotes tumor growth and angiogenesis in glioblastoma multiforme. 32237935_Increased expression of phosphorylated adducin in tumor cells. 32315284_Association of common variation in ADD3 and GPC1 with biliary atresia susceptibility. 33172155_Identification of New Genetic Clusters in Glioblastoma Multiforme: EGFR Status and ADD3 Losses Influence Prognosis. 33196842_QKI-5 regulates the alternative splicing of cytoskeletal gene ADD3 in lung cancer. | ENSMUSG00000025026 | Add3 | 3.149270e+02 | 0.9168884 | -0.125181926 | 0.3783885 | 1.076749e-01 | 0.7428069516 | 0.94662178 | No | Yes | 2.778888e+02 | 51.187313 | 3.152805e+02 | 59.301197 | |
| ENSG00000148840 | 23082 | PPRC1 | protein_coding | Q5VV67 | FUNCTION: Acts as a coactivator during transcriptional activation of nuclear genes related to mitochondrial biogenesis and cell growth. Involved in the transcription coactivation of CREB and NRF1 target genes. {ECO:0000269|PubMed:11340167, ECO:0000269|PubMed:16908542}. | Activator;Alternative splicing;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transcription;Transcription regulation | The protein encoded by this gene is similar to PPAR-gamma coactivator 1 (PPARGC1/PGC-1), a protein that can activate mitochondrial biogenesis in part through a direct interaction with nuclear respiratory factor 1 (NRF1). This protein has been shown to interact with NRF1. It is thought to be a functional relative of PPAR-gamma coactivator 1 that activates mitochondrial biogenesis through NRF1 in response to proliferative signals. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2013]. | hsa:23082; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; nuclear receptor coactivator activity [GO:0030374]; RNA binding [GO:0003723]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 11340167_PRC is a functional relative of PGC-1 that operates through NRF-1 and possibly other activators in response to proliferative signals 14550271_overexpression of the PRC pathway is responsible for mitochondrial proliferation in the context of thyroid oncocytoma 14722127_PGC-1 has a role in regulating gluconeogenic genes along with HNF-4alpha and SREBP-1 15684387_Coordinate regulation of nucleus-encoded mitochondrial transcription factors by NRFs and PGC-1 family coactivators is essential to the control of mitochondrial biogenesis 16385451_Observational study of gene-disease association. (HuGE Navigator) 16511594_This review focuses on the biologic and physiologic functions of the PGC-1 coactivators, with particular emphasis on striated muscle, liver, and other organ systems relevant to common diseases such as diabetes and heart failure. 17341490_Upregulation of PGC-1alpha and PGC-1beta in the colorectal tumor cells can be part of an adaptation mechanism to help overcome the severe consequences of mtDNA mutations on oxidative phosphorylation. 17390079_These results revealed that bile acid inhibits the promoter activity of PGC-1 in an SHP-dependent manner. 17937892_Activation of the SIRT1/PGC-1 pathway, in a metabolic context promotes mitochondrial function and this pathway plays a role in neurodegerative disease. Review. 18343819_The results are consistent with a pathway whereby PRC regulates NRF-2-dependent genes through a multiprotein complex involving HCF-1. 18660489_Observational study of gene-disease association. (HuGE Navigator) 19036724_support a role for PRC in the integration of pathways directing mitochondrial respiratory function and cell growth 19209188_Meta-analysis of gene-disease association. (HuGE Navigator) 19956726_PRC has a role in the rapid modulation of metabolic functions in response to the status of the cell cycle 23364789_A general role for PRC in the adaptive response to cellular dysfunction. 23954632_PRC is a negative regulator of endothelial inflammation; it negatively regulates endothelial adhesion of monocytes via inhibition of NF kappaB activity 27789709_PRC and c-MYC can act in concert through Akt-GSK-3 signaling to reprogram gene expression in response to mitochondrial stress. 33818872_A novel PPRC1 point mutation in a Chinese family with premature ovarian failure: A case study. | ENSMUSG00000055491 | Pprc1 | 7.227094e+03 | 1.1590365 | 0.212926028 | 0.2885754 | 5.432509e-01 | 0.4610885495 | 0.86547338 | No | Yes | 7.540636e+03 | 643.878453 | 6.092975e+03 | 534.259565 | |
| ENSG00000149084 | 51144 | HSD17B12 | protein_coding | Q53GQ0 | FUNCTION: Catalyzes the second of the four reactions of the long-chain fatty acids elongation cycle. This endoplasmic reticulum-bound enzymatic process, allows the addition of two carbons to the chain of long- and very long-chain fatty acids/VLCFAs per cycle. This enzyme has a 3-ketoacyl-CoA reductase activity, reducing 3-ketoacyl-CoA to 3-hydroxyacyl-CoA, within each cycle of fatty acid elongation. Thereby, it may participate in the production of VLCFAs of different chain lengths that are involved in multiple biological processes as precursors of membrane lipids and lipid mediators. May also catalyze the transformation of estrone (E1) into estradiol (E2) and play a role in estrogen formation. {ECO:0000269|PubMed:12482854, ECO:0000269|PubMed:16166196}. | Alternative splicing;Direct protein sequencing;Endoplasmic reticulum;Lipid biosynthesis;Lipid metabolism;Membrane;NADP;Oxidoreductase;Reference proteome;Steroid biosynthesis;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; fatty acid biosynthesis. {ECO:0000269|PubMed:12482854}.; PATHWAY: Steroid biosynthesis; estrogen biosynthesis. {ECO:0000269|PubMed:16166196}. | This gene encodes a very important 17beta-hydroxysteroid dehydrogenase (17beta-HSD) that converts estrone into estradiol in ovarian tissue. This enzyme is also involved in fatty acid elongation. [provided by RefSeq, Oct 2011]. | hsa:51144; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; extracellular matrix [GO:0031012]; fatty acid elongase complex [GO:0009923]; integral component of membrane [GO:0016021]; 17-beta-hydroxysteroid dehydrogenase (NAD+) activity [GO:0044594]; 17-beta-hydroxysteroid dehydrogenase (NADP+) activity [GO:0072582]; 3-oxo-arachidoyl-CoA reductase activity [GO:0102339]; 3-oxo-behenoyl-CoA reductase activity [GO:0102340]; 3-oxo-cerotoyl-CoA reductase activity [GO:0102342]; 3-oxo-lignoceroyl-CoA reductase activity [GO:0102341]; collagen binding [GO:0005518]; fibronectin binding [GO:0001968]; heparin binding [GO:0008201]; long-chain-3-hydroxyacyl-CoA dehydrogenase activity [GO:0016509]; long-chain-fatty-acyl-CoA reductase activity [GO:0050062]; oxidoreductase activity [GO:0016491]; estrogen biosynthetic process [GO:0006703]; extracellular matrix organization [GO:0030198]; fatty acid biosynthetic process [GO:0006633]; long-chain fatty-acyl-CoA biosynthetic process [GO:0035338]; positive regulation of cell-substrate adhesion [GO:0010811] | 16166196_17beta-HSD12 is the major estrogenic 17beta-HSD responsible for the conversion of E1 to E2 in women. 16621523_Location of 17beta-hydroxysteroid dehydrogenase type 12 through the human body. 19190350_17beta-HSD12 is not elated to intratumoral estradiol biosynthesis in human breast carcinoma, but is correlated with production of very long chain fatty acids and tumor progresssion. 19429442_determined the activity and expression levels of known estrogenic 17beta-HSDs, namely types 1, 7 and 12 17beta-HSD in preadipocytes before and after differentiation into mature adipocytes 19460435_There is no difference in catalytic properties between variants of 17beta-HSD types 7 and 12 and wild-type enzymes, while variants p.Glu77Gly and p.Lys183Arg in 17beta-HSD type 5 showed a slightly decreased activity. 19533843_SREBP-1 represents one of the transcriptional regulators of human 17beta-HSD12. 20677014_Observational study of gene-disease association. (HuGE Navigator) 21409596_High HSD17B12 expression is associated with neoplasms. 22903146_HSD17B12 overexpression is shown to be a marker of poor survival in patients with ovarian cancer; expression in the tumor and function of this enzyme facilitates ovarian cancer progression. 23435447_These evidences were suggestive of a 46,XY DSD due to 17betaHSD3 deficiency. An homozygous mutation (IVS3 -1 G>C or c.326-1G>C) of the 17betaHSD3 gene was discovered. 24058506_Significant associations have been found between CpG sites and patient sex, including DNA methylation in CASP6, a gene that may respond to estradiol treatment, and in HSD17B12, which encodes a sex steroid hormone. 29324448_The expression of HSD17B12 increased along with the severity of ovarian cancer, and the expression mimicked COX-2 expression and intensity. This further suggests the involvement of HSD17B12 in AA production, and its coexpression with COX-2 indicates a role for the enzyme in the increased prostaglandin production during ovarian cancer progression. 29788210_Among lung-expressing enzymes only HSD17beta12 exhibited activity against tobacco-specific carcinogen nitrosamine NNK. siRNA knock-down of HSD17beta12 resulted in significant decreases in (R)-NNAL-formation activity in HEK293 cells. These data suggest that both cytosolic and microsomal enzymes are active against NNK and that HSD17beta12 is the major active microsomal reductase that contributes to (R)-NNAL formation in ... 30734280_Study identified two independent SNPs of ELOVL2 rs3734398 T>C and HSD17B12 rs11037684 A>G that predicted cutaneous melanoma disease-specific survival. The ELOVL2 rs3734398 variant CC genotype was found to be associated with a significantly increased mRNA expression level. 31302749_Silencing of hydroxysteroid 17-beta dehydrogenase 12 essentially catalyzes the 3-ketoacyl-CoA reduction step in long-chain fatty acids production, modulates proliferation and migration of breast cancer cells in a cell line-dependent manner. 32132633_Very-long-chain fatty acid metabolic capacity of 17-beta-hydroxysteroid dehydrogenase type 12 (HSD17B12) promotes replication of hepatitis C virus and related flaviviruses. 32340285_An Integrative Phenotype-Genotype Approach Using Phenotypic Characteristics from the UAE National Diabetes Study Identifies HSD17B12 as a Candidate Gene for Obesity and Type 2 Diabetes. 33118286_Functional genetic variant of HSD17B12 in the fatty acid biosynthesis pathway predicts the outcome of colorectal cancer. 34762930_Association of genetic polymorphisms with local steroid metabolism in human benign breasts. | ENSMUSG00000027195 | Hsd17b12 | 5.633095e+02 | 0.8902202 | -0.167765796 | 0.3382768 | 2.440388e-01 | 0.6213042006 | 0.91532851 | No | Yes | 5.644856e+02 | 114.325157 | 5.154426e+02 | 107.057949 |
| ENSG00000149136 | 6749 | SSRP1 | protein_coding | Q08945 | FUNCTION: Component of the FACT complex, a general chromatin factor that acts to reorganize nucleosomes. The FACT complex is involved in multiple processes that require DNA as a template such as mRNA elongation, DNA replication and DNA repair. During transcription elongation the FACT complex acts as a histone chaperone that both destabilizes and restores nucleosomal structure. It facilitates the passage of RNA polymerase II and transcription by promoting the dissociation of one histone H2A-H2B dimer from the nucleosome, then subsequently promotes the reestablishment of the nucleosome following the passage of RNA polymerase II. The FACT complex is probably also involved in phosphorylation of 'Ser-392' of p53/TP53 via its association with CK2 (casein kinase II). Binds specifically to double-stranded DNA and at low levels to DNA modified by the antitumor agent cisplatin. May potentiate cisplatin-induced cell death by blocking replication and repair of modified DNA. Also acts as a transcriptional coactivator for p63/TP63. {ECO:0000269|PubMed:10912001, ECO:0000269|PubMed:11239457, ECO:0000269|PubMed:12374749, ECO:0000269|PubMed:12934006, ECO:0000269|PubMed:16713563, ECO:0000269|PubMed:9489704, ECO:0000269|PubMed:9566881, ECO:0000269|PubMed:9836642}. | 3D-structure;Acetylation;Chromosome;DNA damage;DNA repair;DNA replication;DNA-binding;Direct protein sequencing;Host-virus interaction;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a subunit of a heterodimer that, along with SUPT16H, forms chromatin transcriptional elongation factor FACT. FACT interacts specifically with histones H2A/H2B to effect nucleosome disassembly and transcription elongation. FACT and cisplatin-damaged DNA may be crucial to the anticancer mechanism of cisplatin. This encoded protein contains a high mobility group box which most likely constitutes the structure recognition element for cisplatin-modified DNA. This protein also functions as a co-activator of the transcriptional activator p63. An alternatively spliced transcript variant of this gene has been described, but its full-length nature is not known. [provided by RefSeq, Jul 2008]. | hsa:6749; | FACT complex [GO:0035101]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; histone binding [GO:0042393]; nucleosome binding [GO:0031491]; RNA binding [GO:0003723]; DNA repair [GO:0006281]; DNA replication [GO:0006260]; nucleosome disassembly [GO:0006337]; regulation of chromatin organization [GO:1902275] | 12374749_SSRP1 stimulates p63 activity by associating with this activator at the promoter 15659405_CK2 regulates the DNA-binding ability of SSRP1 and that this regulation may be responsive to specific cell stresses. 16498457_These results demonstrate that SSRP1 degradation during apoptosis is a two-step process coupling caspase cleavage and ubiquitin-dependent proteolysis. 17209051_SSRP1 has Spt16-dependent and -independent roles in regulating gene transcription in human cells. 17672918_occurrence of an unusual TG 3' splice site in intron 1 19625449_CENP-H-containing complex facilitates deposition of newly synthesized CENP-A into centromeric chromatin in cooperation with FACT and CHD1. 19639603_SSRP1 may play a role in the DNA damage response mediated by homologous recombination; SSRP1 physically interacts with a key recombination repair protein, Rad54 19710137_These results indicate that SSRP1 is a novel cellular protein involved in LANA-dependent DNA replication. 19995907_results demonstrate that SSRP1 is crucial for microtubule growth and spindle assembly during mitosis. 21232560_Studies suggest that that even though FACT has rapid chromatin-binding activity, the binding pattern of FACT on chromatin changes after origin licensing, which may contribute to the establishment of its functional link to the DNA replication machinery. 21454601_The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. 21679440_DNA-PK and FACT both play roles in DNA repair. Therefore both are putative targets for therapeutic inhibition. 21969370_specific FACT subunits synchronize interactions with various target sites on individual nucleosomes to generate a high affinity binding event and promote reorganization. 22940245_The HSF1-RPA complex leads to preloading of RNA polymerase II and opens the chromatin structure by recruiting a histone chaperone, FACT 23325844_SETD2 activity modulates FACT recruitment and nucleosome dynamics, thereby repressing cryptic transcription initiation. 23610384_hFACT-H2A/H2B interactions play a key role in overcoming the nucleosomal barrier by Pol II and promoting nucleosome survival during transcription 23839038_In the absence of FACT complex, SSRP1 and SPT16 mRNAs are unstable and inefficiently translated, making reactivation of FACT function unlikely in normal cells. 24357716_A primary role for FACT in RNF20 recruitment chromatin remodeling for initiation of homologous recombination repair. 25000480_EEF1A1, SSRP1, and XRCC6 are novel interacting partners of the mineralocorticoid receptor 25028470_FACT, NF-kappaB, and cell-cycle progression are inhibited by quinacrine, which overcomes resistance to erlotinib in non-small cell lung cancer 26687053_Our studies facilitate the understanding of SSRP1 and provide insights into the molecular mechanisms of interaction with DNA and histones of the FACT complex. 26755331_Results show that SSRP1 upregulation contributed to hepatocellular carcinoma development and the tumor-suppressive miR-497 served as its negative regulator. 26842758_FACT is required for TOP1 binding to H3K4me3 at non-B DNA containing chromatin for the site-specific cleavage. 26966247_AID accesses the H2B N-terminal basic region exposed by partial unwrapping of the nucleosomal DNA, thereby triggering the invasion of FACT into the nucleosome 27146025_These data uncover a previously unknown role for SSRP1 in promoting the activation of the Wnt signaling pathway activity during cellular differentiation. 27216501_The association of LEDGF proteins with the FACT complex and give further support to a role of SSRP1 in HIV-1 infection. 27284163_FACT chaperone stabilizes the soluble CENP-T/-W complex in the cell and promotes dynamics of exchange, enabling CENP-T/-W deposition at centromeres. 27370399_High FACT expression is associated with glioblastoma. 27467129_This discloses a novel property of FACT wherein it has a co-remodeling activity and strongly enhances the remodeling capacity of the chromatin remodelers. Altogether, our data suggest that FACT may acts in concert with RSC to facilitate excision of DNA lesions during the initial step of BER. 27525970_SSRP1/Ets-1/Pim-3 signalling is tightly associated with the proliferation, apoptosis, autophagy, invasion and clonogenicity of nasopharyngeal carcinoma cells, and blockage of this signalling facilitates chemosensitivity of the cells to docetaxel. 28423528_we propose that FACT is both a marker and a target of aggressive breast cancer cells, whose inhibition results in their death or conversion a less aggressive subtype 29048646_Data revealed that SSRP1 is highly overexpressed in glioma tissues at both the mRNA and protein levels and correlated with tumor grade. Further analysis demonstrated that that SSRP1 regulates the proliferation and metastasis of glioma cells via the MAPK signaling pathway. 29514976_Intrinsic DNA-bending activity therefore favors nucleosome assembly by FACT over nucleosome reorganization, but excessive activity impairs FACT release, suggesting a quality control checkpoint during nucleosome assembly 29764934_regulation of SSRP1 by homodimerization 30029006_Findings provide mechanistic insights by which the two subunits of FACT coordinate with each other to fulfill its functions and suggest that FACT may play essential roles in preserving the original histones with epigenetic identity during transcription or DNA replication. 30344095_It has been demonstrated that FACT potentiates H2A.X-dependent signaling of DNA damage. 30548853_Authors confirm that inhibition of FACT at release from quiescence suppressed the p27 degradation capacity resulting in an increased mVenus-p27K(-) signal. FACT plays an important role in promoting the transition from G0 to the proliferative state. 30616889_Long noncoding RNA LOC101927746 interacts with miR-584-3p which targets SSRP1. LOC101927746/miR-584-3p/SSRP1 pathway modulates colorectal cancer progression. 30715484_FACT subunit Spt16 controls UVSSA recruitment to lesion-stalled RNA Pol II and stimulates transcription-coupled nucleotide excision repair. 30762286_we explored the molecular mechanisms accounting for the dysregulation of SSRP1 in colorectal cancer (CRC) and identified microRNA-28-5p (miR-28-5p) as a direct upstream regulator of SSRP1. We concluded that SSRP1 promotes colorectal cancer (CRC) progression and is negatively regulated by miR-28-5p. 31439637_FACT complex is essential for the expeditious hepatocellular carcinoma oxidative stress response and is a potential therapeutic target for HCC treatment. 31575655_Targeting Histone Chaperone FACT Complex Overcomes 5-Fluorouracil Resistance in Colon Cancer. 31775157_suggest a compelling mechanism for how FACT maintains chromatin integrity during polymerase passage, by facilitating removal of the H2A-H2B dimer, stabilizing intermediate subnucleosomal states and promoting nucleosome reassembly 31839745_his study demonstrated that SSRP1 silencing influenced the proliferation and apoptosis of colorectal cancer cells via the AKT signaling pathway. 31960591_CPF impedes cell cycle re-entry of quiescent lung cancer cells through transcriptional suppression of FACT and c-MYC. 32533099_Histone chaperone FACT is essential to overcome replication stress in mammalian cells. 33104782_The role of FACT in managing chromatin: disruption, assembly, or repair? 33688504_SSRP1 Is a Prognostic Biomarker Correlated with CD8(+) T Cell Infiltration in Hepatocellular Carcinoma (HCC). 33852836_Dual targeting of the epigenome via FACT complex and histone deacetylase is a potent treatment strategy for DIPG. 34271103_Histone chaperone FACT complex inhibitor CBL0137 interferes with DNA damage repair and enhances sensitivity of medulloblastoma to chemotherapy and radiation. 34756889_Structural insights into multifunctionality of human FACT complex subunit hSSRP1. 35013515_Electron microscopy analysis of ATP-independent nucleosome unfolding by FACT. | ENSMUSG00000027067 | Ssrp1 | 3.188321e+04 | 1.0687319 | 0.095899982 | 0.2816884 | 1.171290e-01 | 0.7321693940 | 0.94382538 | No | Yes | 3.253975e+04 | 2370.617613 | 2.722931e+04 | 2034.481863 | |
| ENSG00000149273 | 6188 | RPS3 | protein_coding | P23396 | FUNCTION: Involved in translation as a component of the 40S small ribosomal subunit (PubMed:8706699). Has endonuclease activity and plays a role in repair of damaged DNA (PubMed:7775413). Cleaves phosphodiester bonds of DNAs containing altered bases with broad specificity and cleaves supercoiled DNA more efficiently than relaxed DNA (PubMed:15707971). Displays high binding affinity for 7,8-dihydro-8-oxoguanine (8-oxoG), a common DNA lesion caused by reactive oxygen species (ROS) (PubMed:14706345). Has also been shown to bind with similar affinity to intact and damaged DNA (PubMed:18610840). Stimulates the N-glycosylase activity of the base excision protein OGG1 (PubMed:15518571). Enhances the uracil excision activity of UNG1 (PubMed:18973764). Also stimulates the cleavage of the phosphodiester backbone by APEX1 (PubMed:18973764). When located in the mitochondrion, reduces cellular ROS levels and mitochondrial DNA damage (PubMed:23911537). Has also been shown to negatively regulate DNA repair in cells exposed to hydrogen peroxide (PubMed:17049931). Plays a role in regulating transcription as part of the NF-kappa-B p65-p50 complex where it binds to the RELA/p65 subunit, enhances binding of the complex to DNA and promotes transcription of target genes (PubMed:18045535). Represses its own translation by binding to its cognate mRNA (PubMed:20217897). Binds to and protects TP53/p53 from MDM2-mediated ubiquitination (PubMed:19656744). Involved in spindle formation and chromosome movement during mitosis by regulating microtubule polymerization (PubMed:23131551). Involved in induction of apoptosis through its role in activation of CASP8 (PubMed:14988002). Induces neuronal apoptosis by interacting with the E2F1 transcription factor and acting synergistically with it to up-regulate pro-apoptotic proteins BCL2L11/BIM and HRK/Dp5 (PubMed:20605787). Interacts with TRADD following exposure to UV radiation and induces apoptosis by caspase-dependent JNK activation (PubMed:22510408). {ECO:0000269|PubMed:14706345, ECO:0000269|PubMed:14988002, ECO:0000269|PubMed:15518571, ECO:0000269|PubMed:15707971, ECO:0000269|PubMed:17049931, ECO:0000269|PubMed:18045535, ECO:0000269|PubMed:18610840, ECO:0000269|PubMed:18973764, ECO:0000269|PubMed:19656744, ECO:0000269|PubMed:20217897, ECO:0000269|PubMed:20605787, ECO:0000269|PubMed:22510408, ECO:0000269|PubMed:23131551, ECO:0000269|PubMed:23911537, ECO:0000269|PubMed:7775413, ECO:0000269|PubMed:8706699}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;DNA damage;DNA repair;DNA-binding;Direct protein sequencing;Isopeptide bond;Lyase;Membrane;Methylation;Mitochondrion;Mitochondrion inner membrane;Mitosis;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transcription;Transcription regulation;Translation regulation;Ubl conjugation | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 40S subunit, where it forms part of the domain where translation is initiated. The protein belongs to the S3P family of ribosomal proteins. Studies of the mouse and rat proteins have demonstrated that the protein has an extraribosomal role as an endonuclease involved in the repair of UV-induced DNA damage. The protein appears to be located in both the cytoplasm and nucleus but not in the nucleolus. Higher levels of expression of this gene in colon adenocarcinomas and adenomatous polyps compared to adjacent normal colonic mucosa have been observed. This gene is co-transcribed with the small nucleolar RNA genes U15A and U15B, which are located in its first and fifth introns, respectively. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2012]. | hsa:6188; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; cytosolic ribosome [GO:0022626]; cytosolic small ribosomal subunit [GO:0022627]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; membrane [GO:0016020]; mitochondrial inner membrane [GO:0005743]; mitochondrial matrix [GO:0005759]; mitotic spindle [GO:0072686]; NF-kappaB complex [GO:0071159]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; polysome [GO:0005844]; postsynaptic density [GO:0014069]; ribonucleoprotein complex [GO:1990904]; ribosome [GO:0005840]; ruffle membrane [GO:0032587]; class I DNA-(apurinic or apyrimidinic site) endonuclease activity [GO:0140078]; damaged DNA binding [GO:0003684]; DNA binding [GO:0003677]; DNA-(apurinic or apyrimidinic site) endonuclease activity [GO:0003906]; DNA-binding transcription factor binding [GO:0140297]; endodeoxyribonuclease activity [GO:0004520]; enzyme binding [GO:0019899]; Hsp70 protein binding [GO:0030544]; Hsp90 protein binding [GO:0051879]; iron-sulfur cluster binding [GO:0051536]; kinase binding [GO:0019900]; microtubule binding [GO:0008017]; mRNA binding [GO:0003729]; oxidized purine DNA binding [GO:0032357]; oxidized pyrimidine DNA binding [GO:0032358]; protein kinase A binding [GO:0051018]; protein kinase binding [GO:0019901]; protein-containing complex binding [GO:0044877]; RNA binding [GO:0003723]; small ribosomal subunit rRNA binding [GO:0070181]; structural constituent of ribosome [GO:0003735]; supercoiled DNA binding [GO:0097100]; tubulin binding [GO:0015631]; ubiquitin-like protein conjugating enzyme binding [GO:0044390]; apoptotic process [GO:0006915]; cell division [GO:0051301]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to reactive oxygen species [GO:0034614]; cellular response to tumor necrosis factor [GO:0071356]; chromosome segregation [GO:0007059]; cytoplasmic translation [GO:0002181]; DNA repair [GO:0006281]; negative regulation of DNA repair [GO:0045738]; negative regulation of protein ubiquitination [GO:0031397]; negative regulation of translation [GO:0017148]; positive regulation of activated T cell proliferation [GO:0042104]; positive regulation of apoptotic signaling pathway [GO:2001235]; positive regulation of base-excision repair [GO:1905053]; positive regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis [GO:2001272]; positive regulation of DNA N-glycosylase activity [GO:1902546]; positive regulation of DNA repair [GO:0045739]; positive regulation of endodeoxyribonuclease activity [GO:0032079]; positive regulation of gene expression [GO:0010628]; positive regulation of interleukin-2 production [GO:0032743]; positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage [GO:1902231]; positive regulation of JUN kinase activity [GO:0043507]; positive regulation of microtubule polymerization [GO:0031116]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of T cell receptor signaling pathway [GO:0050862]; regulation of apoptotic process [GO:0042981]; response to TNF agonist [GO:0061481]; spindle assembly [GO:0051225]; translation [GO:0006412]; translational initiation [GO:0006413] | 11911468_Electron paramagnetic resonance study reveals a putative iron-sulfur cluster in human rpS3 protein. 14988002_RPS3 is involved in apoptosis. 15518571_Using surface plasmon resonance technology, the authors show that ribosomal protein S3 positively interacts with the human base excision repair enzymes N-glycosylase/apurinic-apyrimidinic lyase OGG1 and APE/Ref-1. 16737853_The S3-K132A mutant retained the ability to cleave abasic DNA, but its capacity to bind 8-oxoG was abrogated completely. 17140567_PEP-1-rpS3 fusion protein can be used in protein therapy for various disorders related to UV, including skin aging and cancer 17179743_S3 is a key protein at the mRNA binding site neighboring mRNA downstream of the codon at the decoding site in the human ribosome. 18045535_RPS3 is an essential but previously unknown subunit of NF-kappaB involved in the regulation of key genes in rapid cellular activation responses. 18973764_hRpS3 may be involved in the uracil-excision pathway, probably by participating in the DNA repair mechanism to remove uracil generated by the deamination of cytosine in DNA, and by preventing C/G-->T/A transition mutations. 19059439_The destiny of rpS3 molecules between translation and DNA repair is regulated by PKCdelta-dependent phosphorylation. 19088750_Protein S3 fragments neighboring mRNA during elongation and translation termination on the human ribosome 19458393_oxidative stress regulates the phosphorylation status of nonribosomal rpS3 by both activating PKCdelta and blocking the PP2A interaction with rpS3 19460357_these results clearly show that arginine methylation of rpS3 plays a critical role in its import into the nucleolus, as well as in small subunit assembly of the ribosome. 19656744_DNA pull-down assays using a 7,8-dihydro-8-oxoguanine duplex oligonucleotide as a substrate found that RPS3 acted as a scaffold for the additional binding of MDM2 and p53. 20217897_when Flag-tagged rpS3 was transiently transfected into 293T cells, the level of endogenous rpS3 gradually decreased regardless of transcription 20709134_PEP-1-rpS3 inhibits inflammatory response cytokines and enzymes by blocking NF-kappaB and MAP kinase, prompting the suggestion that PEP-1-rpS3 can be used as a therapeutic agent against skin inflammation. 21399639_Data show that the IKKbeta-dependent modification of a specific amino acid in RPS3 promoted specific NF-kappaB functions that underlie the molecular pathogenetic mechanisms of E. coli O157:H7. 21871177_The phosphorylation of rpS3 by Cdk1 occurs at Thr221 during G2/M phase. 21968017_rpS3 is covalently modified by SUMO-1 and this post-translational modification regulates rpS3 function by increasing rpS3 protein stability. 22510408_rpS3 is recruited to the DISC and plays a critical role in both genotoxic stress and cytokine induced apoptosis. 23115242_An N-terminal fragment of p65 (amino acids 21-186) can selectively modulate NF-kappaB gene transcription by competing for RPS3 binding to p65. 23131551_rpS3 acts as a microtubule associated protein and regulates spindle dynamics during mitosis. 23188828_A novel radioresistance mechanism through functional orchestration of rpS3, TRAF2, and NF-kappaB in non-small cell lung cancer cells, is reported. 23911537_rpS3 accumulates in the mitochondria to repair damaged DNA due to the decreased interaction between rpS3 and HSP90 in the cytosol. 24211576_These findings suggest that the secreted rpS3 protein is an indicator of malignant tumors. 24239944_RPS3, a component of basic translation machinery operates at initiation and most probably elongation of protein synthesis, is also implicated in various events of the cell life as an extraribosomal player. [Review] 24457201_IkappaBalpha sequesters not only p65 but also RPS3 in the cytoplasm. 25449781_Increased RPS3 expression is associated with osteosarcoma invasion. 26336993_Data show that ribosomal protein S3 (RPS3) knockdown decreased mitochondrial calcium uptake 1 protein (MICU1) expression. 26526615_a novel cell fate determination mechanism to ensure cells undergo programed cell death through interfering with RPS3/NF-kappaB-conferred anti-apoptotic transcription by the fragment from partial p65 cleavage by activated Caspase-3 27384988_Asn 165 residue of rpS3 is a critical site for N-linked glycosylation and passage through the ER-Golgi secretion pathway. 28334742_findings suggest that uS3 residing in the 40S ribosome might perform extra-ribosomal functions related to control of DNA quality 28584194_Short 5'UTR mRNAs are enriched with TISU (translation initiator of short 5'UTR), a 12-nucleotide element directing efficient scanning-independent translation. This study demonstrate that TISU is particularly dependent on eukaryotic initiation factor 1A (eIF1A) which interacts with both RPS3 and RPS10e. 29018126_Results found that BfrB subverts the host innate immune system by binding the NF-kappaB subunit RPS3 and promotes the survival of mycobacteria in macrophages by inhibiting cytokine production in host cells. 29048653_These results reveal that RPS3 upregulates XIAP independently of the NF-kappaB pathway in human breast cancer cells 29371697_RpS3 is a crucial substrate of ubiquitination by RNF138 in glioblastoma.RpS3 role in the radioresistance of glioblastoma. 29875444_These results suggest that Tat inhibits cell proliferation via an interaction with RPS3 and thereby disrupts mitotic spindle formation during HIV-1 infection. These results might provide insight into the mechanism underlying lymphocyte pathogenesis during HIV-1 infection. 30594661_abasic sites, which can occur in mRNAs due to oxidative stress and ageing, are able to interact directly with the uS3 fragment exposed on the 40S subunit surface near the mRNA entry channel during translation. 30972734_Quantitative model is presented for a tandem arrangement of two helicase active sites on the ribosome are tested. One of the proteins involved in helicase activity is RPS3. 31356988_our results revealed the previously unknown crucial role of the uS3 tetrapeptide (60)GEKG(63) in translation initiation related to maintaining the proper structure of the 48S complex, most likely via the prevention of premature mRNA loading into the ribosomal channel. 32748020_Ribosomal protein S3 selectively affects colon cancer growth by modulating the levels of p53 and lactate dehydrogenase. 32828277_Ribosomal protein S3-derived repair domain peptides regulate UV-induced matrix metalloproteinase-1. 34070332_Fucosylated Proteome Profiling Identifies a Fucosylated, Non-Ribosomal, Stress-Responsive Species of Ribosomal Protein S3. 34412652_A novel NF-kappaB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. | ENSMUSG00000030744 | Rps3 | 1.070968e+05 | 1.0111167 | 0.015949544 | 0.3090481 | 2.643888e-03 | 0.9589918445 | 0.99162977 | No | Yes | 1.163952e+05 | 14547.716525 | 9.247413e+04 | 11852.208494 | |
| ENSG00000149294 | 4684 | NCAM1 | protein_coding | P13591 | FUNCTION: This protein is a cell adhesion molecule involved in neuron-neuron adhesion, neurite fasciculation, outgrowth of neurites, etc.; FUNCTION: (Microbial infection) Acts as a receptor for rabies virus. {ECO:0000269|PubMed:9696812}.; FUNCTION: (Microbial infection) Acts as a receptor for Zika virus. {ECO:0000269|PubMed:32753727}. | 3D-structure;Alternative splicing;Cell adhesion;Cell membrane;Disulfide bond;GPI-anchor;Glycoprotein;Host cell receptor for virus entry;Host-virus interaction;Immunoglobulin domain;Lipoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | This gene encodes a cell adhesion protein which is a member of the immunoglobulin superfamily. The encoded protein is involved in cell-to-cell interactions as well as cell-matrix interactions during development and differentiation. The encoded protein plays a role in the development of the nervous system by regulating neurogenesis, neurite outgrowth, and cell migration. This protein is also involved in the expansion of T lymphocytes, B lymphocytes and natural killer (NK) cells which play an important role in immune surveillance. This protein plays a role in signal transduction by interacting with fibroblast growth factor receptors, N-cadherin and other components of the extracellular matrix and by triggering signalling cascades involving FYN-focal adhesion kinase (FAK), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K). One prominent isoform of this gene, cell surface molecule CD56, plays a role in several myeloproliferative disorders such as acute myeloid leukemia and differential expression of this gene is associated with differential disease progression. For example, increased expression of CD56 is correlated with lower survival in acute myeloid leukemia patients whereas increased severity of COVID-19 is correlated with decreased abundance of CD56-expressing NK cells in peripheral blood. Alternative splicing results in multiple transcript variants encoding distinct protein isoforms. [provided by RefSeq, Aug 2020]. | hsa:4684; | anchored component of membrane [GO:0031225]; cell surface [GO:0009986]; collagen-containing extracellular matrix [GO:0062023]; cytosol [GO:0005829]; external side of plasma membrane [GO:0009897]; extracellular region [GO:0005576]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; virus receptor activity [GO:0001618]; cell adhesion [GO:0007155]; commissural neuron axon guidance [GO:0071679]; regulation of semaphorin-plexin signaling pathway [GO:2001260] | 11681838_NCAM 105-115 kDa is a protease- and neuraminidase-susceptible fragment and was correlated with ventricular enlargement in chronic schizophrenia (p = 0.01). Release of NCAM fragments in schizophrenia may be part of the pathogenic mechanism. 11820619_Significance of cell adhesion molecules, CD56/NCAM in particular, in human tumor growth and spreading. 11915324_exists in lipid microdomains and transducts cell signalings via regulating the activation of signal transduction molecules 12003861_PSA-NCAM was found to be expressed in the somata, dendrites and axonal processes of some neurons, which were identified as chandelier cell axon terminals (chandelier terminals), in the adult human entorhinal cortex and neocortex. 12008081_CD56 expression predicts occurrence of CNS disease in acute lymphoblastic leukemia but not CR or survival. CD56 may enable targeting of leukemic cells to tissues that express it. 12121226_CD56 molecules on NK cells interact with fibroblast growth factor receptor 1 on Jurkat T cells to trigger IL-2 production. 12594840_Preferential apoptosis of CD56dim natural killer cell subset in patients with cancer. 12727026_a useful immunohistochemical marker of Merkel cell carcinoma 12791681_which the polysialyltransferases bind to the first fibronectin type III repeat (FN1) of NCAM to polymerize polysialic acid chains on appropriately presented glycans in adjacent regions. 12937148_Strong overexpression of NCAM(CD56) and RUNX1(AML1) is a constant and characteristic feature of cardiomyocytes within or adjacent to scars in ICM. 14688313_Natural killer (NK) cells expressing high CD56 levels are terminally differentiated cells identical to mature NK cells recently activated in the presence of IL-12, and not a functionally distinct subset or progenitors to mature CD56+low NK cells. 14726964_there are two types of CD56+ epithelial cells in the pancreatic duct system: CD56+ endocrine cells are numerous during the early stage of gestation while CD56+ luminal cells may represent developmental and regenerative changes of pancreatic ducts. 14959847_CD56 is expressed in bone marrow of acute myelogenous leukemias but not acute lymphoblastic leukemia 15006709_Alzheimer patients presented values of low molecular weight-NCAM and high molecular weight-NCAM significantly higher than healthy controls of similar age (higher than 130 kDa) 15050861_Observational study of gene-disease association. (HuGE Navigator) 15050861_genetic variations in neural cell adhesion molecule 1 or nearby genes could confer risks associated with bipolar affective disorder in Japanese individuals. 15061198_REVIEW: Prognostic significance of CD56 expressed in multiple myeloma 15223636_CD56 expression was associated with the leukemogenetic mutation at the primitive hematopoietic progenitor cell level 15231874_Unlike wild-type FGFR4, pituitary tumor derived-FGFR4 does not associate with neural cell-adhesion molecule (NCAM). 15246157_Cells expressing this antigen repond to a Wilms tumor cell line feeder cells and are precursors of NK cells. 15356097_Pre-activated, adherent-natural killer cells express low levels of CD56 and CD161 15459479_There is a close relationship between PSA-NCAM expression and neuronal migration. 15528382_CD56bright natural killer cells accumulate in inflammatory lesions and, in the appropriate cytokine environment, can engage with CD14+ monocytes in a reciprocal activatory fashion, thereby amplifying the inflammatory response. 15626024_first pediatric case describing coexpression of CD56 on B-lineage acute lymphoblastic leukemia 15782066_CD56 seems to be the most sensitive marker for the diagnosis of SCC of the uterine cervix 15950781_A significant increase was observed in PSA-NCAM, NCAM-180, NCAM-140, and HSP70 expression as seen by Western blotting and immunocytofluorescent studies in NMDA-treated cultures. 16027151_polysialyltransferase ST8Sia IV/PST recognizes specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule 16172115_analysis of polymerization of polysialic acid on neural cell adhesion molecules 16211277_NCAM is associated not only with a cell-to-cell adhesion mechanism, but also with tumorigenesis, including growth, development and perineural invasion in human salivary gland tumors 16316416_CD56(bright) and CD56(dim) human natural killer-cell subsets exert different functional and cytotoxic activities in response to a live bacterial pathogen. 16406048_The new role of Neural Cell Adhesion Molecules in tumor neo-angiogenesis relevant for endothelial cell organization into capillary-like structures. 16534119_NCAM is hyposialylated in hereditary inclusion body myopathy skeletal) muscle. 16572491_Tests whether activation and expansion of human NK cells with lipopolysaccharide (LPS) reveals differences between identical twins with regaard to C56 antigen. 16627685_The gene expression profile of hepatic stem cells throughout life consists of high levels of expression of neuronal cell adhesion molecule (NCAM). 16690409_identified co-induction of NKG2A and CD56 on activation of TH2 cells 16892559_Isolation of 3 novel isoforms of AML 1 (RUNX1) with different transactivating function, that might be a regulatory element of the NCAM (CD56) overexpression in chronic myocardial ischemia. 17003032_the FN1 alpha-helix is involved in an Ig5-FN1 interaction that is critical for the correct positioning of Ig5 N-glycans for polysialylation 17005551_Co-immunoprecipitation and co-clustering paradigms were used to show that both NCAM and N-cadherin can interact with the 3Ig IIIC isoform of the FGFR1 in a number of cell types. 17043020_NCAM1 along with RUNX1 is overexpressed during stress hematopoiesis in Down syndrome children and may contribute to the development of overt leukemia. 17085484_Observational study of gene-disease association. (HuGE Navigator) 17161382_In the 'Europe only' stratum, there were nominally significant associations with five contiguous SNPs. 17181871_Neuroblastoma cells resistant to anticancer drugs have increased invasive capacity caused by down-regulation of NCAM adhesion receptor. 17208489_GPI-anchored NCAM-120 suppressed rabies virus replication via induction of IFN-ss even though NCAM-120 was able to promote virus penetration into the cells. 17216340_The pattern of serum NCAM bands could be useful to detect brain tumor pathology. NCAM immunostaining of tumors was inversely correlated with the histological grade of malignancy. Loss of NCAM staining was significantly associated with poor prognosis. 17337466_Renal NCAM-expressing interstitial cells can participate in the initial phase of interstitial fibrosis. 17413444_Observational study of gene-disease association. (HuGE Navigator) 17413444_Single nucleotide polymorphisms within NCAM1 contribute differential risk for both bipolar disorder and schizophrenia possibly by alternative splicing of the gene. 17431094_HIV-1 Nef protein up-regulates the ability of dendritic cellss to stimulate the immunoregulatory naturak killer cells. 17467233_PSA-NCAM is expressed in the human PFC neuropil following a laminated pattern and in a subpopulation of mature neurons, which lack doublecortin expression. Most of these cells have been identified as interneurons expressing calbindin 17635242_Although CD56 expression level varies among the cases, this molecule might play some roles in the manner of growth and expansion of CD56-positive B-cell lymphomas 17683591_PSA-NCAM is not involved in masking Siglec-7 17761687_Observational study of gene-disease association. (HuGE Navigator) 17761687_association studies of alcohol dependence and 43 SNPs mapped to the gene cluster of NCAM1, TTC12, ANKK1 and DRD2 17878347_may explain the preferential accumulation of CD56(bright) NK cells often seen in environments rich in reactive oxygen species, such as at sites of chronic inflammation and in tumors 17891186_lack of CD56 expression on MM cells is not a prognostic marker in patients treated with high-dose chemotherapy, but is associated with t(11;14). 17900814_fetal forebrain axonal PSA-NCAM expression is inversely related to primary myelination 17940597_that expansion of the CD56(bright) NK cell subtype in peripheral blood is not a hallmark of TAP deficiency, but can be found in other diseases as well 17971410_Ubiquitylation represents an endocytosis signal for NCAM. 17982624_NCAM expression may be used as a predictor of perineural invasion in adenoid cystic carcinoma 18209097_Monitoring of T cells driven to senescence showed de novo induction of CD56, the prototypic receptor of NK cells. 18213713_Ganglioneuromas and ganglioneuroblastomas express the adhesive 120 kDa NCAM isoform, while neuroblastomas preferentially express the 180 kDa isoform classically involved in cell motility 18231917_determined the clinical characteristics of 204 multiple myeloma (MM) patients and 26 plasma cell leukemia (PCL) patients with regard to CD56 expression. 18261743_NCAM-fibroblast growth factor receptor 1 interaction at the cell surface is likely to depend upon avidity effects due to receptor clustering 18289872_Neural cell adhesion molecule-extracellular domain overexpression disrupts aborization of basket cells during the major period of axon/dendrite growth in transgenic mice. 18323797_identified molecular characteristics of an aggressive subset of pediatric patients with AML through a prospective evaluation of CD56+ neural cell adhesion molecule (NCAM) and CD94 expression 18333845_relative frequency of CD19 and/or CD56 expression in acute myeloid leukemia (AML) with t(8;21) was significantly higher than those without this translocation and co-expression of these two antigens may serve as the surrogate markers for AML with t(8;21) 18353777_direct receptor-receptor interactions are not required for high affinity GDNF binding to NCAM but play an important role in the regulation of NCAM-mediated cell adhesion by GFRalpha1 18368482_results provide evidence that the BCL motif is one of the multiple FGFR binding sites in NCAM 18384787_Enzymatic removal of PSA from NCAM or reduction of polysialyltransferase expression led to reduced association between NCAM and E-cadherin and subsequently increased E-cadherin-mediated cell-cell aggregation and reduced cell migration. 18425046_CD56 is often expressed by a wide variety of spindle cell sarcomas, thus, it has no value in differentiating GYN from non-GYN spindle cell tumors. 18432248_there was increased placental expression of NCAM (neural cell adhesion molecule) in small for gestational age cases 18462256_extramedullary relapse of MM is characterized by loss of CD56 expression 18594005_CD56-expressing gammadelta T lymphocytes are resistant to Fas ligand and chemically induced apoptosis 18601968_No genetic association between NCAM1 gene polymorphisms and schizophrenia in the Chinese population. 18601968_Observational study of gene-disease association. (HuGE Navigator) 18628406_Immunological events associated with the luteinizing hormone surge induce alterations in all subsets of CD56(+) cells in the fertile menstrual cycle 18641363_CD56(bright) NK cells postallogeneic hematopoietic stem cell transplantation exhibit peculiar phenotypic and functional properties 18755075_To identify the gene regulated by PKD2 and c-Myc, we performed gene expression profiling in PKD2 and c-Myc overexpressing cells. NCAM is an important molecule in the cystogenesis induced by PKD2 overexpression. 18828801_NCAM1 exon 12 markers are significant for specific haplotypes in a family sample of comorbid alcohol and drug dependence. 18828801_Observational study of gene-disease association. (HuGE Navigator) 18972120_NCAM represents a marker for neuroblastomas irrespectively of their stages 18979395_N-CAM increased in cytotrophoblasts and decreased in extravillous trophoblasts and decidual cells of preeclamptic subjects. 18990213_The results obtained show that, in man, the expression of PSA-NCAM in selective populations of central and peripheral neurons occurs not only during prenatal life, but also in adulthood. 19086053_Observational study of gene-disease association. (HuGE Navigator) 19153015_CD56 is extremely useful in the diagnosis of papillary thyroid carcinoma 19222860_NCAM-positive neuroblastoma patients had more often metastases at diagnosis, and NCAM expression associated with advanced disease 19235015_propose four potential activators of the CD56 promoter and for CD56 to be involved in proliferation and anti-apoptosis, leading to disease progression in multiple myeloma 19328310_Subcutaneous panniculitis-like T-cell lymphoma with a phenotype showing a deficiency of cd56. 19328558_Observational study of gene-disease association. (HuGE Navigator) 19393299_study implicates the major NCAM isoforms, PSA-NCAM and proteolytically cleaved NCAM in pre- and postnatal development of the human prefrontal cortex 19411161_This data suggests NCAM-180 mRNA expression is altered in a regionally-specific manner in schizophrenia and these changes are associated with the early period following diagnosis. 19507465_The role of NCAM in cell differentiation, survival and plasticity suggests an involvement in the development process of the carotid body and in cellular/molecular changes due to chronic hypoxia. 19587433_Structural change in NCAM1 caused by point mutation may be the reason for astrocytoma tumorigenesis. 19652998_Additional immunohistochemical detection of neuroendocrine differentiation (chromogranin-A, synaptophysin, and neural-cell adhesion molecule) in non-small cell lung cancer is presently not of prognostic importance. 19724850_Class III beta-tubulin and NCAM were expressed in 50 and 68% of basal cell carcinoma 19725832_polysialylated NCAM persistence, up-regulated polysialyltransferase-1 mRNA and previously uncovered defective myelin-associated glycoprotein may be early pathogenetic events in adult-onset autosomal-dominant leukodystrophy 19772585_Expression defines a committed T-lymphocyte lineage to the cytotoxic phenotype 19788570_results suggest a positive role of NCAM in neurogenesis in the periventricular region; expression of NCAM in stem cells might be one of many factors useful for therapeutic approaches in the future 19788615_Mucosal remodeling with alterations of NCAM+ or alpha-SMA+ subepithelial and interstitial cells may play a critical role in UC-associated tumorigenesis. 19864234_CD56 immunohistochemical analysis is useful for detecting residual plasma cell myeloma particularly in morphologically equivocal cases in which light chain restriction cannot be demonstrated, and may serve as a potential response criterion. 19897577_human CD56(bright) NK cells progress through a continuum of differentiation that ends with a CD94(low)CD56(dim) phenotype. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20015889_Neural cell adhesion molecules expressed on mesenchymal stem cells play a crucial role in the human hematopoiesis-supporting ability of the cell line. 20027291_The IL-23 induced cytokines allow for the subsequent production of IL-12 and amplify the IFN-gamma production in the type-1 cytokine pathway. 20029409_Observational study of gene-disease association. (HuGE Navigator) 20049565_induction of SOX4 gene expression might be responsible for the CD56 expression in human myeloma cells 20059553_NCAM in the amygdala mediates consolidation of auditory fear conditioning; increased NCAM transgene expression in the amygdala is among the mechanisms whereby stress facilitates fear conditioning processes in floxed mice. 20083228_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20164549_This study suggested a potential involvement of NCAM expressing neurons in the cognitive deficits in Alzheimer's disease. 20187302_we suggest NCAM as a marker for the WT progenitor cell population. These findings provide novel insights into the cellular hierarchy of WT, having possible implications for future therapeutic options 20231901_SOCS-3 and PIAS-3 upregulation impairs IL-12-mediated interferon-gamma response in CD56 T cells in HCV-infected heroin users 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20414008_analysis of the effect of human colorectal carcinogenesis on the neural cell adhesion molecule expression and polysialylation 20483466_Observational study of gene-disease association. (HuGE Navigator) 20483466_This study suggested that SNP variants in NCAM1 may impact on related traits, particularly by mediating inhibition of aggressiveness. 20524836_we confirmed the expression of NCAM in Wilms tumors, nephrogenic rests, and metanephric mesenchyme and its early derivatives in fetal renal cortex 20538416_CD56 cannot be used as a supplementary tool in the differential diagnosis of non-neoplastic biliary diseases in needle biopsies. 20557674_The predominance of extranodal involvement in our series may be associated with the adhesion-related function of CD56. A high frequency of bcl-6 expression may be associated with a more favorable clinical course and prognosis. 20610389_a decisive role for the neuronal K(+) channel in regulating NCAM-dependent neurite outgrowth and attribute a physiologically meaningful role to the functional interplay of Kir3.3, NCAM, and TrkB in ontogeny 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20664990_Assessment of natural killer-like T CD3+/CD16+ CD56+ cells may be helpful in determining a worsening of clinical course in chronic lymphocytic leukemia. 20684989_Results confirm the favorable impact of NPM1 mutations and identify the adverse prognostic relevance of CD56 expression in this subgroup of AML. 20696944_CD56(dim) NK cells continue to differentiate. During this process, they lose expression of NKG2A, sequentially acquire inhibitory killer cell inhibitory immunoglobulin-like receptors and CD57. 20733159_assessed the transcriptional, phenotypic, and functional differences between CD57(+) and CD57(-) NK cells within the CD56(dim) mature NK subset 20805222_Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera. 20875069_CD56 expression in odontogenic epithelium is highly suggestive of ameloblastoma and can help in differentiating this from odontogenic keratocyst 20932956_PSA-NCAM may modulate the functional interaction between BDNF and its high and low affinity receptors;possible clinical significance of neuronal trophism in cerebellar neurodegenerative disorders 21115007_Data suggest that the intra-cellular part of NCAM inhibits cis-dimerization, an effect mainly dependent on the palmitoylation sites. 21148082_CD56 expression is associated with coexpression of immaturity-associated and T-cell antigens and is an independent adverse prognostic factor for relapse in acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline-based regimens 21178331_CD6 expression on peripheral NK cells marks a novel CD56(dim) subpopulation associated with distinct patterns of cytokine and chemokine secretion. 21212386_Genetic variation in NCAM1 contributes to left ventricular wall thickness in hypertensive families. 21239711_CD56(bright)CD11c(positive) cells play a key role in the interleukin (IL)-18-mediated proliferation of gammadelta T cells. 21457956_Maturation of follicles is accompanied by a decrease in CD56+CD16+ natural killer cells that may have deleterious effects on follicular maturation. 21464126_STX(G421A) shows a dramatic decrease in polySia synthetic activity on NCAM, whereas STX(C621G) does not; polySia shows a dopamine (DA) binding activity 21467162_Excellent prognosis in a subset of patients with ewing sarcoma identified at diagnosis by CD56 using flow cytometry. 21515372_Abnormal synaptic innervation in transgenic mice expressing the NCAM extracellular proteolytic cleavage fragment impairs prefrontal cortex plasticity and alters working memory. 21577011_This study did not support the ncam1 is direct modulation of these genes on temperament 21666061_Natural killer (NK)-92 cell line retains the functional characteristics of the CD56(bright) NK cell subset; consequently, gene silencing of CD56 profoundly inhibits the ability of NK-92 cells to kill activated syngeneic T cells. 21669487_Soluble NCAM status was a significant independent factor predictive of long-term survival in patients with hepatocellular carcinoma 21691800_NCAM-140 interacts with APP, potentially playing a role in neurite outgrowth and neural development. 21717310_CD56 expression is associated with neuroectodermal differentiation in ameloblastomas. 21739604_These results point to NCAM-mediated stimulation of FGFR as a novel mechanism underlying epithelial ovarian carcinoma malignancy and indicate that this interplay may represent a valuable therapeutic target. 21940794_Together, these data indicate that polysialic acid regulates cell motility through NCAM-induced but FGF-receptor-independent signalling to focal adhesions 22001684_Multiple sclerosis is characterized by a dysregulation of CD8+CD56-perforin+ T cells that may play a role in the development of disability. 22081445_The CD56 antigen is an independent prognostic risk factor in patients with acute myeloid leukemia 22085395_Serum NCAM is hyposialylated in hereditary inclusion body myopathy. 22099865_This study demonistrated that the depressed patients showed decreases in PSA-NCAM expression in the basolateral and basomedial amygdala and in the lateral nucleus of bipolar patients 22211388_Prominent CD56 expression by damaged and regenerating muscle fibers in the skin. 22219127_Cerebrospinal fluid NCAM-1 is a potential biomarker for drug-effective epilepsy and drug-refractory epilepsy. 22228741_Dehydroepiandrosterone increased PSA-NCAM expression and inhibited monocyte binding in an estrogen- and androgen receptor-dependent manner. 22276608_CD56 expression in B cell lymphoma is a rare occurrence. 22281821_NCAM and polySia are expressed and developmentally regulated in chick corneas. Both membrane-associated and soluble NCAM isoforms are expressed in chick corneas. 22319021_results suggest that during differentiation CD56(bright) NK cells, similarly to mature activated NK cells, become highly cytotoxic and are relatively resistant to apoptosis induced by TNF family members. 22384114_Cytotoxicity of CD56(bright) NK cells towards autologous activated CD4+ T cells is mediated through NKG2D, LFA-1 and TRAIL and dampened via CD94/NKG2A 22384181_NCAM180 regulates Ric8A membrane localization and potentiates beta-adrenergic response 22423624_assessment of CD56 and CD117 expression by flow cytometry is a sensitive method for diagnostic evaluation of plasma cell neoplasms. 22449227_NCAM1, SYPT and CGA expressions are differently regulated by neuroendocrine phenotype-specific transcription factors in lung cancer cells. 22591692_CD56-positive T cells have a critical role in innate defense against HIV-1 infection. 22732936_TGF-beta1 contributes to HCC-induced vascular alterations by affecting the interaction between HCC-StCs and Ld-MECs through a down-modulation of NCAM expression 22792160_Intra-hepatic accumulation of the functionally impaired CXCR3(+)CD56Bright NK cell subset might be involved in HCV-induced liver fibrosis. 23015367_Bioinformatic analysis of NCAM-associated expression profiles predicted a highly interactive protein network, which further implies potential molecular mechanisms underlying the metastatic processes of thyroid cancer 23022470_In schizophrenia, abnormal PSA-NCAM and GAD67 expression may underlie the alterations observed in inhibitory neurotransmission. 23061666_Data indicate that UCHL1 is a novel interaction partner of both NCAM isoforms that regulates their ubiquitination and intracellular trafficking. 23258197_An unknown factor with a molecular weight >100 kDa plays a critical role in the impairment of CD56(dim) natural killer cells in malignant pleural effusion, which might lead to tumor progression. 23260340_Immunoblots revealed that depressed subjects displayed increased expression of PSA-NCAM. 23292839_the author showed that in cutaneous basal cell carcinoma, the expression of NCAM and c-KIT was high, PDGFRA was intermediate, and chromogranin A and synaptophysin was relatively low 23303482_SNPs in NCAM1 were not significantly associated with heroin dependence. 23365458_GATA2 is required for the maturation of human natural killer cells and the maintenance of the CD56(bright) pool in the periphery. 23418554_Data suggest that multi-antibody assay of TTF1, Vimentin, p63 CD56, chromogranin and synaptophysin may be of special value, especially in diagnosing small biopsies. 23462508_CD56(140kD) up-regulation plays a pivotal role in the pathogenesis of ischemic cardiomyopathy. 23470050_We recommend immunohistochemical analyses for CD123, CD56 and CD4 inblastic plasmacytoid dendritic cell neoplasm patients, particularly in cases where the initial bone marrow study indicates normal morphology. 23480226_Women with age above 35 years and greater than 13% CD56 positive CD16 positive natural killer cells showed the highest risk of further pregnancy loss 23495921_Depletion of NCAM is one of the factors associated with or possibly responsible for disease progression in multiple sclerosis. 23557873_Results suggest that CD3+CD56+NKT cells do not play a role as a mediator in mental symptom such as depression in fibromyalgia syndrome patients. 23635388_CD56 and quantitative Ki-67 along with cytomorphology is a robust immunohistochemical panel to differentiate small cell lung carcinoma from other neuroendocrine neoplasms. 23671285_translational modification of the neural cell adhesion molecule NCAM and the polysialyltransferase ST8SiaII in mammalian semen involves polysialic acid 23716295_NCAM polysialylation in small cell lung cancer progression regulates substrate adherence potential. 23810283_High CD56 expression is associated with relapse in acute myeloid leukemia with t(8;21). 23960070_NCAM and dynein have roles in tethering dynamic microtubules and maintaining synaptic density in cortical neurons 24055371_Depletion of NCAM1(+) cells from human kidney epithelial cells abrogated stemness traits in vitro. 24206578_Positive CD56 expression was found in 23 APL patients. 24240977_Decreased proportion of NK cells expressing CD56dim/CD16 antigens in Franconi anemia patients correlates with the impairment in the differentiation process of the NK cells and of the immune surveillance. 24286519_Data indicate that the CD14bright/CD56+ monocyte subset is expanded in aging individuals as well as in patients with rheumatoid arthritis. 24294395_Case Report: incidentally diagnosed CD56 positive diffuse large B-cell lymphoma. 24349544_The described scFv has potential application in delivery of therapeutics to NCAM1-expressing cells in degenerated IVD. 24365773_Expression of NCAM was associated with worsening hemodynamic parameters and major metabolic genes. 24369228_CD56 can be expressed in normal immature granulocytes at a variety of expression levels in regenerative bone marrow. Attention should be paid when evaluating aberrant antigen expression of CD56 in granulocytes. 24526449_NCAM expression in myelinating Schwann cells is indicative of early Schwann cell abnormalities. 24715165_HBME-1, Galectin-3, Cytokeratin-19 and CD56 were used alone or in panels in a series of papillary thyroid carcinoma (PTC) and thyroid tumors of uncertain malignant potential. 24726913_NCAM140 interacts with ufc1 and its trafficking and endocytosis is upregulated in the presense of Ufm1. 24782118_CD56 positivity did not have any influence on the prognosis of these patients. 24807109_Women, w/ or w/o endometriosis, having larger populations of cytotoxic CD16(+) uterine NK cells and/or higher populations of NKp46(+)CD56(+) cells may be at greater risk of infertility resulting from inflammatory environment during implantation or later 24909369_Increased frequency of ILT2-expressing CD56(dim)CD16(+) NK cells correlates with disease severity of pulmonary tuberculosis. 25137309_results showed that NCAM1 might play an important role in the pathogenesis of autism 25188863_Report differential diagnosis of Primary cutaneous NK/T-cell lymphoma, nasal type and CD56-positive peripheral T-cell lymphoma. 25201755_Homophilic interaction between CD56 molecules may occur in tumor-cell recognition, leading to CIK-mediated cell death. 25326085_Report ontogenic development of nerve fibers in human fetal livers using immunohistochemical detection of NCAM1/neurone-specific enolase expression. 25363560_analysis of CD8+ T cell induction via Vgamma9gammadeltaT cell expansion by CD56(high+) Interferon-alpha-induced dendritic cells 25445624_Results provide direct evidence for NCAM1 as a susceptibility gene for schizophrenia, which offers support to a neurodevelopmental model and neuronal connectivity hypothesis in the onset of schizophrenia 25596273_CD56(low)CD16(low) natural killer cells are multifunctional cells, and that the presence of hematologic malignancies affects their frequency and functional ability at both tumor site and in the periphery. 25619885_improvement in behavioral performance (open-field and grip-strength tests), as well as increased life-span was observed in rodents treated with NCAM-VEGF or NCAM-GDNF co-transfected cells 25628040_Low NCAM1 in the sera is associated with active forms of inflammatory bowel disease. 25723856_CD56 is generally expressed in 70-80%11 of patients with MM, as observed in 69% of the t(14;16)-negative cases in this study. In contrast, none of the t(14;16)-positive cases showed CD56 positivity. 25769453_Determining the P-glycoprotein expression and function of the peripheral blood CD56+ cells may help predict the MDR of NHL, thus has profound guiding significance for NHL treatment 25779340_Human BDCA2+CD123+CD56+ dendritic cells (DCs) related to blastic plasmacytoid dendritic cell neoplasm represent a unique myeloid DC subset. 25889612_Aberrant NCAM expression plays a role in the pathogenesis of keratin producing odontogenic tumors. 25921109_Up-regulated level of serum sNCAM is associated with hepatic encephalopathy in hepatocelular carcinoma patients. 25924702_CD56 overexpression was associated with shorter OS. 25935537_Loss of CD56 expression is associated with extranodal NK/T cell lymphoma. 26013700_Case Report: chronic lymphocytic leukemia with aberrant CD56 and CD57 expression. 26039898_Results show significant increases in proportions of CD56+ T cells in relation to CMV infection in renal transplant patients and suggest that these cells have a cytotoxic function against CMV-infected cells. 26045862_CD56 could potentially become an adjunct diagnostic marker for ectomesenchymal chondromyxoid tumor instead of previously used CD57. 26087825_Renal graft CD56+ cell infiltrates were significantly associated with antibody mediated rejection. 26097548_for differential diagnosis of papillary thyroid cancer it was found that the only marker with both sensitivity and specificity above 90% was CD56 negativity 26147745_this study shows that CD56 expression defines a poor prognosis subset in the cytogenetically intermediate prognosis pediatric AML. 26183877_CD56 expression remains to be a potentially unfavorable prognostic factor in acute promyelocytic leukemia patients. 26186733_CD56 was negative in 96% of the primary malignant thyroid tumors while being expressed in the cytoplasm of 68.5% of the benign thyroid nodules. 26255203_Our findings demonstrate a novel Wnt/beta-catenin-miR-30a-5p-NCAM regulatory axis which plays important roles in controlling glioma cell invasion and tumorigenesis. 26339384_CD56 marker is a useful alternative that is comparable to SSTR2A for the diagnosis of phosphaturic mesenchymal tumors. 26344352_the expression of CD56 in adenomyosis is positively associated with the severity of dysmenorrhea. 26391771_Letter/Case Report: CD56 positive diffuse large B cell lymphoma of the urinary bladder. 26437631_A FOXP3(+)CD3(+)CD56(+)-expressed T-cell population with immunosuppressive function and reduced patient survival has been identified in cancer tissues of human hepatocellular carcinoma. 26460482_The neural cell adhesion molecule (NCAM) is a glycoprotein implicated in cell-cell adhesion, neurite outgrowth and synaptic plasticity. 26463893_NCAM1 deletion is associated with neuroblastoma and ganglioneuroma. 26478212_L1 | ENSMUSG00000039542 | Ncam1 | 1.405924e+03 | 0.9611448 | -0.057174315 | 0.2709330 | 4.432142e-02 | 0.8332567547 | 0.96751127 | No | Yes | 1.260980e+03 | 132.497875 | 1.264289e+03 | 136.235831 | |
| ENSG00000149311 | 472 | ATM | protein_coding | Q13315 | FUNCTION: Serine/threonine protein kinase which activates checkpoint signaling upon double strand breaks (DSBs), apoptosis and genotoxic stresses such as ionizing ultraviolet A light (UVA), thereby acting as a DNA damage sensor. Recognizes the substrate consensus sequence [ST]-Q. Phosphorylates 'Ser-139' of histone variant H2AX at double strand breaks (DSBs), thereby regulating DNA damage response mechanism. Also plays a role in pre-B cell allelic exclusion, a process leading to expression of a single immunoglobulin heavy chain allele to enforce clonality and monospecific recognition by the B-cell antigen receptor (BCR) expressed on individual B-lymphocytes. After the introduction of DNA breaks by the RAG complex on one immunoglobulin allele, acts by mediating a repositioning of the second allele to pericentromeric heterochromatin, preventing accessibility to the RAG complex and recombination of the second allele. Also involved in signal transduction and cell cycle control. May function as a tumor suppressor. Necessary for activation of ABL1 and SAPK. Phosphorylates DYRK2, CHEK2, p53/TP53, FBXW7, FANCD2, NFKBIA, BRCA1, CTIP, nibrin (NBN), TERF1, UFL1, RAD9, UBQLN4 and DCLRE1C (PubMed:9843217, PubMed:9733515, PubMed:10550055, PubMed:10766245, PubMed:10839545, PubMed:10910365, PubMed:10802669, PubMed:10973490, PubMed:11375976, PubMed:12086603, PubMed:15456891, PubMed:19965871, PubMed:30612738, PubMed:30886146, PubMed:26774286). May play a role in vesicle and/or protein transport. Could play a role in T-cell development, gonad and neurological function. Plays a role in replication-dependent histone mRNA degradation. Binds DNA ends. Phosphorylation of DYRK2 in nucleus in response to genotoxic stress prevents its MDM2-mediated ubiquitination and subsequent proteasome degradation. Phosphorylates ATF2 which stimulates its function in DNA damage response. Phosphorylates ERCC6 which is essential for its chromatin remodeling activity at DNA double-strand breaks (PubMed:29203878). Phosphorylates TTC5/STRAP at 'Ser-203' in the cytoplasm in response to DNA damage, which promotes TTC5/STRAP nuclear localization (PubMed:15448695). {ECO:0000269|PubMed:10550055, ECO:0000269|PubMed:10766245, ECO:0000269|PubMed:10802669, ECO:0000269|PubMed:10839545, ECO:0000269|PubMed:10910365, ECO:0000269|PubMed:10973490, ECO:0000269|PubMed:11375976, ECO:0000269|PubMed:12086603, ECO:0000269|PubMed:12556884, ECO:0000269|PubMed:14871926, ECO:0000269|PubMed:15448695, ECO:0000269|PubMed:15456891, ECO:0000269|PubMed:15916964, ECO:0000269|PubMed:16086026, ECO:0000269|PubMed:16858402, ECO:0000269|PubMed:17923702, ECO:0000269|PubMed:19431188, ECO:0000269|PubMed:19965871, ECO:0000269|PubMed:26774286, ECO:0000269|PubMed:29203878, ECO:0000269|PubMed:30612738, ECO:0000269|PubMed:30886146, ECO:0000269|PubMed:9733514, ECO:0000269|PubMed:9733515, ECO:0000269|PubMed:9843217}. | 3D-structure;ATP-binding;Acetylation;Cell cycle;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;DNA damage;DNA-binding;Disease variant;Kinase;Neurodegeneration;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Tumor suppressor | The protein encoded by this gene belongs to the PI3/PI4-kinase family. This protein is an important cell cycle checkpoint kinase that phosphorylates; thus, it functions as a regulator of a wide variety of downstream proteins, including tumor suppressor proteins p53 and BRCA1, checkpoint kinase CHK2, checkpoint proteins RAD17 and RAD9, and DNA repair protein NBS1. This protein and the closely related kinase ATR are thought to be master controllers of cell cycle checkpoint signaling pathways that are required for cell response to DNA damage and for genome stability. Mutations in this gene are associated with ataxia telangiectasia, an autosomal recessive disorder. [provided by RefSeq, Aug 2010]. | hsa:472; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; DNA repair complex [GO:1990391]; intracellular membrane-bounded organelle [GO:0043231]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; peroxisomal matrix [GO:0005782]; spindle [GO:0005819]; 1-phosphatidylinositol-3-kinase activity [GO:0016303]; ATP binding [GO:0005524]; DNA binding [GO:0003677]; DNA-dependent protein kinase activity [GO:0004677]; identical protein binding [GO:0042802]; protein N-terminus binding [GO:0047485]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein-containing complex binding [GO:0044877]; brain development [GO:0007420]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to gamma radiation [GO:0071480]; cellular response to nitrosative stress [GO:0071500]; cellular response to retinoic acid [GO:0071300]; cellular response to X-ray [GO:0071481]; cellular senescence [GO:0090398]; determination of adult lifespan [GO:0008340]; DNA damage checkpoint signaling [GO:0000077]; DNA damage induced protein phosphorylation [GO:0006975]; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest [GO:0006977]; double-strand break repair [GO:0006302]; double-strand break repair via homologous recombination [GO:0000724]; double-strand break repair via nonhomologous end joining [GO:0006303]; establishment of protein-containing complex localization to telomere [GO:0097695]; establishment of RNA localization to telomere [GO:0097694]; female meiotic nuclear division [GO:0007143]; heart development [GO:0007507]; histone mRNA catabolic process [GO:0071044]; histone phosphorylation [GO:0016572]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; lipoprotein catabolic process [GO:0042159]; male meiotic nuclear division [GO:0007140]; meiotic telomere clustering [GO:0045141]; mitotic G2 DNA damage checkpoint signaling [GO:0007095]; mitotic spindle assembly checkpoint signaling [GO:0007094]; multicellular organism growth [GO:0035264]; negative regulation of B cell proliferation [GO:0030889]; negative regulation of telomere capping [GO:1904354]; negative regulation of TORC1 signaling [GO:1904262]; neuron apoptotic process [GO:0051402]; oocyte development [GO:0048599]; ovarian follicle development [GO:0001541]; peptidyl-serine autophosphorylation [GO:0036289]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of apoptotic process [GO:0043065]; positive regulation of cell adhesion [GO:0045785]; positive regulation of cell migration [GO:0030335]; positive regulation of DNA catabolic process [GO:1903626]; positive regulation of DNA damage response, signal transduction by p53 class mediator [GO:0043517]; positive regulation of gene expression [GO:0010628]; positive regulation of histone phosphorylation [GO:0033129]; positive regulation of neuron apoptotic process [GO:0043525]; positive regulation of telomerase catalytic core complex assembly [GO:1904884]; positive regulation of telomere maintenance via telomerase [GO:0032212]; positive regulation of telomere maintenance via telomere lengthening [GO:1904358]; positive regulation of transcription by RNA polymerase II [GO:0045944]; post-embryonic development [GO:0009791]; pre-B cell allelic exclusion [GO:0002331]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; reciprocal meiotic recombination [GO:0007131]; regulation of apoptotic process [GO:0042981]; regulation of autophagy [GO:0010506]; regulation of cell cycle [GO:0051726]; regulation of cellular response to gamma radiation [GO:1905843]; regulation of cellular response to heat [GO:1900034]; regulation of microglial cell activation [GO:1903978]; regulation of signal transduction by p53 class mediator [GO:1901796]; regulation of telomere maintenance via telomerase [GO:0032210]; replicative senescence [GO:0090399]; response to hypoxia [GO:0001666]; response to ionizing radiation [GO:0010212]; signal transduction [GO:0007165]; somitogenesis [GO:0001756]; telomere maintenance [GO:0000723]; thymus development [GO:0048538]; V(D)J recombination [GO:0033151] | 11165203_Observational study of gene-disease association. (HuGE Navigator) 11606401_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11723136_In ataxia telangiectasia-mutated (ATM)-deficient cells the amount of nuclear PP2A heterotrimer relative to heterodimer was not We conclude a novel ATM-dependent mechanism is regulating association of B55 subunits with nuclear PP2A in response to IR 11805335_Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. 11830600_role in breast cancer 11830610_at least two ATM mutations are associated with a sufficiently high risk of breast cancer to be found in multiple-case breast cancer families; genetic susceptibility for breast cancer 11849780_Observational study of gene-disease association. (HuGE Navigator) 11859564_ATM: from phenotype to functional genomics--and back 11875057_mediates phosphorylation at multiple p53 sites in response to ionizing radiation 11889466_A new type of mutation causes a splicing defect in ATM 11897822_ATM mutations in Finnish breast cancer patients. 11927575_lack of role in cellular response to DNA strand-scission enediyne C-1027 11992555_risk of developing BC is 3.6-fold higher among ATM heterozygous women 11996792_Observational study of gene-disease association. (HuGE Navigator) 11996792_The data are compatible with certain missense mutations in ATM predisposing to breast cancer. 12032824_Aberrant methylation of the ATM promoter correlates with increased radiosensitivity in a human colorectal tumor cell line. 12034743_ATM and BLM function together in recognizing abnormal DNA structures by direct interaction and that these phosphorylation sites in BLM are important for radiosensitivity status but not for SCE frequency. 12036913_Observational study of gene-disease association. (HuGE Navigator) 12065055_Roles of DNA-dependent protein kinase and ATM in cell-cycle-dependent radiation sensitivity in human cells. 12072552_dominant negative mutations in breast cancer families 12091354_ATM mutations are rare in familial chronic lymphocytic leukemia. 12149228_ATM mutations contribute to the development of diffuse large B-cell lymphoma 12151394_These findings indicate that ATM activation is not limited to the ionizing radiation-induced response and potentially plays an important role in response to DNA alkylation. 12234250_These observations provide the first link between ATM and LKB1 and suggest that ATM could regulate LKB1 12362033_Subtle constitutional alterations of ATM may impart an increased risk of developing breast cancer and therefore act as a low penetrance, high prevalence gene in the general population. 12376469_deficiency in the repair of UV-induced DNA damage in human skin fibroblasts compromised for the gene 12409306_Data suggest that a novel AMPK family member, ARK5, is the tumor cell survival factor activated by Akt and acts as an ATM kinase under the conditions of nutrient starvation. 12420214_ATM kinase has a role in orchestrating the coordinated induction and transcriptional cooperation of IRF-1 and p53 to regulate p21 expression. 12429935_ATM kinase is not required for signaling when chromatid decatenation is blocked 12473176_Observational study of gene-disease association. (HuGE Navigator) 12473594_Observational study of gene-disease association. (HuGE Navigator) 12511424_A dominant-negative germline missense ATM mutation (8921C>T; Pro2974Leu), located in the PI-3-kinase domain was found in a childhood ALL patient with MLL rearrangement. Altered ATM function plays some pathogenic role in the development of MLL+ leukemia. 12513844_ATM protein plays a critical role in the signal transduction of cell cycle checkpoint, the repair of damaged DNA and the apoptosis. 12519769_ATM has a critical role in the response of hypoxia and reperfusion in solid tumors. 12526805_ATM does not have a critical role in regulating chromosomal fragile site stability 12545170_our data suggest that ATM may mediate cell response to mitogenic factors by tightly regulating the set point of the CaR and thereby modulating the crosstalk between this metabotropic receptor and growth factor receptors 12556884_Cellular irradiation induces rapid intermolecular autophosphorylation of serine 1981 that causes dimer dissociation and initiates cellular ATM kinase activity 12612651_Review. DNA double-strand breaks activate ATM. ATM modulates many signalling pathways. ATM mutations cause the cancer-prone disorder ataxia-telangiectasia. Understanding ATM's action clarifies the relation of defective responses to DNA damage & cancer. 12628935_ATM functions upstream of protein kinase c-delta. 12637545_ATM is a major signal initiator for genotoxin-induced apoptosis but, paradoxically, also contributes to maintenance of cell survival by facilitating recovery/escape from terminal growth arrest 12645530_ATM is activated by ATP in a mechanism involving autophosphorylation 12646636_ATM seems dispensable in the somatic hypermutation and Ig heavy chain variable-diversity- joining recombination processes but is clearly involved in the end joining-repair machinery in class switch recombination. 12650908_In conclusion, effective and prompt IR-induced Rad51 focus formation is cell cycle-regulated and requires both ATM and c-Abl. 12660173_targets tousled like kinasese via DNA damage checkpoint 12673126_ataxia telangiectasia mutated (ATM) gene mutation/deletion is Associated with rhabdomyosarcoma 12676583_ATM is the transducer of the S phase checkpoint and presumably propagates the signal through downstream effector kinases. 12773400_BRCA1 facilitates the ability of ATM and ATR to phosphorylate downstream substrates that directly influence cell cycle checkpoint arrest and apoptosis 12782595_ATM is involved in histone acetylation-mediated gene regulation. 12810666_There is a significant prevalence of ATM mutations in breast and ovarian cancer families. This adds to a growing body of evidence that ATM mutations confer increased susceptibility to breast cancer. 12813460_ATM is composed of two main domains comprising a head and an arm. DNA binding to ATM induces a large conformational movement of the arm-like domain which clamps around the double helix. 12815592_Twenty eight novel mutations in the ATM gene have been identified and their associated haplotypes defined. 12833146_ATM is involved in a signaling pathway that induces ATF3 after ionizing radiation 12860021_analyses of lymphocytes and DNA from 37 familial CLL cases do not support the hypothesis that the ATM gene is a CLL susceptibility gene 12861053_multiple functional domains of NBS1 are required for ATM-dependent activation of CHK2, nuclear focus formation, S phase checkpoint control, and cell survival after exposure to ionizing radiation 12875964_Data suggest that ataxia telangiectasia mutated (ATM) expression is differentially regulated in lymphoid tumors and is likely to reflect their cellular origin. 12882767_ATM missense variants could confer an AT-like phenotype and influence the formation of retinal and choroidal vascular abnormalities. 12915485_Polyglutamine-expanded proteins strongly activated ATM. 12917204_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 12926986_ATM is involved in regulation of transcription factors such as SP1, AP1 and MTF1 12935922_Observational study of gene-disease association. (HuGE Navigator) 12935922_Total missense mutations were significantly elevated in cases of breast neoplasms. 12955071_we propose that ATM activation is not linked solely to DSBs and that ATM participates in initiating signaling pathways in response to replication block and UV-induced DNA damage 12958068_the greater severity of TP53-mutant B-CLLs compared with ATM-mutant B-CLLs is consistent with the additive effect of defective apoptotic and elevated survival responses after DNA damage in these tumors 12969974_rare polymorphic variants of the ATM gene identified in children with Hodgkin disease encode functionally abnormal proteins 14532133_ATM is activated and phosphorylates key players in various branches of the DNA damage response network. 14553952_study the null GSTM1 genotype is an independent risk factor for the development of lung cancer for Turkish population. 14562025_Observational study of gene-disease association. (HuGE Navigator) 14570874_ATM-dependent signaling pathway triggered by DNA damage is dispensable for activation of p38 MAPK and SIPS in response to IR or oxidative stress. 14628072_REVIEWS the ATM gene in sporadic lymphoid malignancies and the apparent paradox between the predominance of nonsense mutations observed in ataxia-telangiectasia patients and the high proportion of missense alterations found in sporadic lymphoid tumours 14643952_Observational study of genotype prevalence. (HuGE Navigator) 14657032_Infection with an adenovirus lacking the E4 region also induces a cellular DNA damage response, with activation of ATM. 14695167_In cells with mutant Nbs1, suppression of 53BP1 led to decreased ATM activation and phosphorylation of ATM substrates. 14695186_Study suggests a general pattern of increased breast cencer risk associated with carrying any one of the ATM variants studied, with a significant association being observed in individuals carrying variants on both ATM alleles. 14695534_Analysis of splicing mutations in the ATM gene. 14735203_Observational study of gene-disease association. (HuGE Navigator) 14744762_ATM-deficient cells repair the majority of DSBs with normal kinetics but fail to repair a subset of breaks. 14744854_ATM plays a critical role in regulating homologous recombination repair but not nonhomologous end joining throughout the cell cycle 14745549_ATM and NBS regulate several genes in common, both of these proteins also have distinct patterns of gene regulation. 14754616_ATM was found to have a minor contribution to the pathogenesis of some B-cell chronic lymphocytic leukemia patients. 14966265_Data describe a novel pathway of ataxia telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK) signaling that results in nuclear factor kappaB (NF-kappaB) activation and chemoresistance in response to DNA damage. 14983937_The presence of the ATM protein at the same or a higher level than that in normal prostate cells might have an important role in the maintenance of the shortened telomeres commonly found in prostate cancer cells. 15024084_Results suggest that E2F1 plays a central role in signaling disturbances in the retinoblastoma growth control pathway and, by upregulation of Chk2 by Atm and Nbs1, may sensitize cells to undergo apoptosis. 15039971_Five haplotypes account for fifty-five percent of ATM mutations in Brazilian patients with ataxia telangiectasia 15042666_Observational study of gene-disease association. (HuGE Navigator) 15048089_Mre11-Rad50-Nbs1 complex serves also as a modulator/amplifier of ATM activity. 15064416_demonstrated that MRN (Mre11, Rad50, and Nbs1 proteins) stimulates the kinase activity of ATM in vitro toward its substrates p53, Chk2, and histone H2AX 15073328_ATM-mediated phosphorylation of CREB in response to DNA damage modulates CREB-dependent gene expression and that dysregulation of the ATM-CREB pathway may contribute to neurodegeneration in ataxia telangiectasia 15084244_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15101044_Alterations in the ATM gene are associated with increased chromosome radiosensitivity. 15164409_Observational study of genotype prevalence. (HuGE Navigator) 15173573_signaling via ATM is necessary for full activation of TonEBP/OREBP 15174027_Adult-onset ataxia telangiectasia due to ATM 5762ins137 mutation homozygosity. 15210935_ATM/ATR-dependent (ataxia-telangiectasia-mutated/ATM- and Rad3-related) checkpoint pathways are directly linked to three members of the MCM complex(MCM2,MCM3,MCM7). 15217510_Observational study of gene-disease association. (HuGE Navigator) 15226443_Data show that DNA damage induces the accumulation of hPMS1, hPMS2, and hMLH1 through ataxia-telangiectasia-mutated (ATM)-mediated protein stabilization. 15234984_Atm phosphorylation events are regulated by nibrin and Mre11-Rad50 15258567_p14ARF-induced inhibition of MCF7 cell proliferation was significantly attenuated by downregulation of ATM by RNAi. 15279774_DNA damage induceds activation of ATM [review] 15279777_hSMG-1 teams with ATM and ATR to insure the overall quality of the transcriptome in human cells [review] 15279780_53BP1 is an activator of ATM in response to DNA damage [review] 15279808_analysis of the domain structure of the ATM molecule, sites of interaction with other proteins and the consequences of specific amino acid changes on function [review] 15280449_ATM is not required for dephosphorylation and inhibition of Polo-like kinase 1 activity following mitotic DNA damage. 15280931_Observational study of gene-disease association. (HuGE Navigator) 15284180_Hexavalent chromium-induced activation of ATM involves the formation of S phase-dependent double stranded breaks. 15314656_TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response 15345673_The present data suggests that the IGF-IR gene is a novel downstream target in an ATM-mediated DNA damage response pathway. 15361830_Chk2 activity is triggered by a greater number of double strand breaks, implying that, below a certain threshold level of lesions, DNA repair can occur through ATM, without enforcing Chk2-dependent checkpoints. 15389585_DNA-PK, ATM and possibly other kinases implicated in H2AX phosphorylation. 15390180_The risk of breast cancer is associated with the alteration of binding domains rather than with the length of the predicted ATM protein. 15450731_Observational study of gene-environment interaction. (HuGE Navigator) 15456759_PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation 15456891_Data show that Artemis interacts with cell cycle checkpoint proteins and is a phosphorylation target of the checkpoint kinases ATM or ATR after exposure of cells to IR or UV irradiation, respectively. 15459181_results implicate ATM in the HRR-mediated rescue of replication forks impaired by thymidine treatment 15485651_IGF-1 induces AMPK-alpha subunit phosphorylation via an ATM-dependent and LKB1-independent pathway 15489221_hydroxyl radicals contribute to the doxorubicin-induced activation of ATM-dependent pathways 15510216_Autophosphorylation of ATM is regulated by protein phosphatase 2A. 15516988_epigenetic silencing of ATM expression occurs in locally advanced breast tumors, and establish a link at the molecular level between reduced ATM function and sporadic breast malignancy 15533933_data reveal activated ATM and ATR exhibit selective substrate specificity in response to DNA damage 15539948_Thus, optimal repair of damaged replication fork lesions likely requires both ATR and ATM. BLM recruits 53BP1 to these lesions independent of its helicase activity, and optimal activation of ATM requires both p53 and BLM helicase activities. 15546858_ATM is specifically activated by IR-induced DSBs, with little or no contribution from SSBs and other types of DNA damage 15546863_PKB/Akt activation in response to insulin or ionizing radiation is mediated through ATM 15574327_ATM, Artemis, and proteins locating to gamma-H2AX foci have roles in double-strand break rejoining 15611093_ATM checkpoint signal transduction is elicited by Epstein-Barr virus lytic replication 15629612_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15713674_a protein interaction domain in the N terminus of ATM is required for DNA damage-induced association of ATM with its target proteins 15788536_sites important for Hdm2-mediated ubiquitination of Hdmx after double-strand break induction; one of these sites, S403, is a direct ATM target 15790808_findings show that the Mre11-Rad50-Nbs1 (MRN) complex acts as a double-strand break sensor for ATM and recruits ATM to broken DNA molecules 15824150_Observational study of gene-disease association. (HuGE Navigator) 15837784_Down-regulation of ATM protein sensitizes human prostate cancer cells to radiation-induced apoptosis 15840767_activation of ATM contributes to high NaCl-induced nuclear translocation of TonEBP/OREBP 15846060_Our results strongly support the physiological relevance of the recently proposed model of ATM autoactivation, and provide further evidence for constitutive activation of the DNA damage machinery during cancer development. 15878096_Observational study of gene-disease association. (HuGE Navigator) 15880680_Observational study of gene-disease association. (HuGE Navigator) 15880680_We identified three new breast cancer families with c.1066-6T>G, and seven families with c.4258C>T. 15880721_Ataxia telangiectasia (A-T) patients from 16 Russian families were assessed for immunological status and ATM haplotype analysis, and screened for ATM mutations. 15916964_Data demonstrate that the protein kinase ATM phosphorylates ATF2 on serines 490 and 498 following ionizing radiation (IR). 15923642_These results suggest that hMOF influences the function of ATM. 15928302_These results confirm a moderate risk of breast cancer in A-T heterozygotes and give some evidence of an excess risk of other cancers but provide no support for large mutation-specific differences in risk. 15929992_the amino terminus of ATM is crucial not only for nuclear localization but also for chromatin association, thereby facilitating the kinase activity of ATM in vivo 15964794_ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. 15964848_host cells activate ATM checkpoint signaling in response to HSV infection 15987456_Observational study of gene-disease association. (HuGE Navigator) 16012708_The pattern of ATM expression was changed from normal mucosa to tumour and less expression of ATM may be related to males. 16014569_Observational study of gene-disease association. (HuGE Navigator) 16049814_Observational study of gene-disease association. (HuGE Navigator) 16082221_ATM directly activates p53 while activating a safe-lock mechanism to inactivate the negative regulators of p53, Mdm2, and Mdmx [review] 16141325_DNA damage induces the rapid acetylation of ATM, dependent on the Tip60 histone acetyltransferase; activation of Tip60 by DNA damage and the recruitment of the ATM-Tip60 complex to sites of DNA damage is independent of ATM's kinase activity. 16167060_Observational study of gene-disease association. (HuGE Navigator) 16184611_This report demonstrates activation of ATM and ATM-mediated phoshorylation of H2AX, both in relation to cell-cycle phase and onset of apoptosis within the same cells. 16221684_ATM controls SV40 viral replication in vivo 16266405_ATM gene founder haplotypes and associated mutations are found in ataxia-telangiectasia 16293623_ATM is required for CREB phosphorylation in UV irradiation-damaged cells. 16319535_ATM is activated by default at each mitotic onset and phosphorylates p53 at Ser15 so as to keep it inactive at centrosomes when the spindle is correctly in place 16325375_Data provide evidence that ATM, a predominantly nuclear kinase, could be relocalized to the plasma membrane by CKIP-1. 16326028_hypothesis that one molecular mechanism by which 11q23 deletions confer a poor prognosis in CLL is via increased TfR expression secondary to ATM loss, resulting in the increased cellular iron import, and hence increased capacity for malignant growth 16327781_we show that efficient ATM-dependent ATR activation in response to DSBs is restricted to the S and G2 cell cycle phases and requires CDK kinase activity. Thus, in response to DSBs, ATR activation is regulated by ATM in a cell-cycle dependent manner 16329039_Reduced ATM expression in breast carcinomas correlated with tumor differentiation and increased microvascular parameters, supporting its role in neoangiogenesis and tumor progression in breast carcinogenesis. 16338099_Observational study of gene-disease association. (HuGE Navigator) 16380133_Two novel mutations in the ATM gene were identified in Chinese ataxia telangiectasia patient. The first one is a novel, homozygous, Gly449Ala mutation. The second one is a compound heterozygous mutation, which consists of a novel,(Gly204Stop)mutation. 16426422_Frequency of activated ATM and phosphorylated H2AX molecules, per apoptotic cell, is comparable. 16426903_ATM is phosphorylated on serine-1981 in lymphoblastoid cell lines (LCLs) derived from Immunodeficiency, Centromeric instability, Facial anomalies syndrome (ICF) patients but not from the other syndromes. 16431910_ATM and Mre11 may stimulate the ATR signaling pathway by converting DNA damage generated by ionizing radiation into structures that recruit and activate ATR 16432227_Myc overexpression causes DNA damage in vivo and the ATM-dependent response to this damage is critical for p53 activation, apoptosis, and the suppression of tumor development 16461339_ATM signals DNA double-strand breaks arising from ionizing radiation through a checkpoint kinase (Chk)2-dependent pathway. 16474133_results indicate that KSHV vIRF1 comprehensively compromises an ATM/p53-mediated DNA damage response checkpoint by targeting both upstream ATM kinase and downstream p53 tumor suppressor 16474151_results indicate that ATM can positively influence HIV-1 Rev function 16474843_ATM and MRN protein complex play role in recruiting ATR to sites of ionizing radiation-induced DNA damage 16478990_nibrin plays an active role in Atm activation and that this function requires nibrin-Atm interaction. 16481012_Plk3 is in a pathway linking ATM, Plk3, Chk2, Cdc25C and Cdc2 in cellular response to DNA damage. 16497724_Observational study of gene-disease association. (HuGE Navigator) 16497931_results show that ATM phosphorylates Ser85 of NEMO in response to genotoxic stress & that this is required for ubiquitination of NEMO; this modification is essential for nuclear export of NEMO & ATM & their interaction with IKK in the cytoplasm 16520463_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16533773_Low expression of ATM was associated with colorectal cancer 16574953_Observational study of gene-disease association. (HuGE Navigator) 16574953_Preeminent associations were identified in SNPs mapping to genes pivotal in the DNA damage-response and cell-cycle pathways, including ATM F858L and P1054R, CHEK2 I157T, BRCA2 N372H, and BUB1B Q349R. 16582589_Myc and E2F1 engage the ATM signaling pathway to activate p53 and induce apoptosis [review] 16603769_FATC domain of ATM mediates the interaction between ATM and Tip60 16622469_Observational study of gene-disease association. (HuGE Navigator) 16627474_nuclear ATM mediates the DSB response in NLCs similarly to in proliferating cells 16628006_2-deoxy-D-glucose reduces the level of constitutive activation of ATM and phosphorylation of histone H2AX 16631465_These observations document a high prevalence of ataxia telangiectasia mutated (ATM) gene and protein expression alterations, suggesting that ATM is involved in childhood non-Hodgkin lymphoma (NHL). 16631604_In conclusion, methylation of the ATM promoter may account for the variable radiosensitivity and heterogeneous ATM expression in a fraction of glioma cells. 16636671_activation of ATM suppresses Bcl-2-induced tumorigenesis, and that attenuation of ATM function may be an important event in breast cancer progression 16638864_Observational study of gene-disease association. (HuGE Navigator) 16651613_SUMOylation and activation of ataxia-telangiectasia-mutated protein, PKCdelta, caspase-3, and nuclear factor kappaB signaling pathways modulate salivary adaptive responses to stress in cells exposed to either 1% O(2) or DFO. 16652348_Heterozygous mutations within the ATM gene are associated with the risk of breast cancer. 16652348_Observational study of gene-disease association. (HuGE Navigator) 16705183_The activation of ATM/ATR/CHK signaling pathways contributes to this G2 checkpoint and highlight the interrelated roles of p14ARF and the Tip60 protein in the initiation of this DNA damage-signaling cascade. 16728507_PI3K-IA activity is necessary for both high NaCl- and ionizing radiation-induced activation of ATM and (ii) high NaCl activates PI3K-IA, which, in turn, contributes to full activation of TonEBP/OREBP via ATM. 16741947_ATR is one of the kinases that is likely involved in phosphorylation of Chk2 in response to ionizing radiation when ATM is deficient. 16765197_Observational study of gene-disease association. (HuGE Navigator) 16824197_ATM (ataxia-telangiectasia mutated) is activated by a variety of noxious agent, including oxidative stress, and ATM deficiency results in an anomalous cellular response to oxidative stress. 16832357_ATM mutations that cause ataxia-telangiectasia in biallelic carriers are breast cancer susceptibility alleles in monoallelic carriers, with an estimated relative risk of 2.37 16832357_Observational study of gene-disease association. (HuGE Navigator) 16849332_ataxia-telangiectasia mutated protein activation has a role in nucleotide excision repair-facilitated cell survival with cisplatin treatment 16858402_There are at least three functionally important radiation-induced autophosphorylation events in ATM. 16905549_Mre11 stabilizes Nbs1 and Rad50 and MRN activates Chk2 downstream from ATM in response to replication-mediated DNA double strand breaks 16914028_Observational study of gene-disease association. (HuGE Navigator) 16914028_Results do not support association of the 5557G>A or ivs38-8T>C variant with increased breast cancer risk or with bilateral breast cancer. 16931761_in response to DNA damage, ATM phosphorylated COP1 on Ser(387) and stimulated a rapid autodegradation mechanism; ionizing radiation triggered an ATM-dependent movement of COP1 from the nucleus to the cytoplasm 16941484_ATM mutations in Italian families with ataxia telangiectasia include two distinct large genomic deletions. 16943424_These results demonstrate a sophisticated control by ATM of a target protein, Hdmx, which itself is one of several ATM targets in the ATM-p53 axis of the DNA damage response. 16949371_Wip1 phosphatase is an integral component of an ATM-dependent signaling pathway. 16951182_ATR/ATM-independent checkpoint response to DNA synthesis inhibition exists in HeLa cells. 16958054_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16966185_Observational study of gene-disease association. (HuGE Navigator) 16997395_PINs are generally regarded as precursors of prostatic carcinoma. Our results suggest that ATM and Chk2 activation at earlier stages of prostate tumorigenesis suppresses tumor progression, with attenuation of ATM activation leading to cancer progression. 16998505_Epidemiological and molecular studies have provided conclusive evidence that ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. [REVIEW] 17001622_Study reports the screening of 782 multiple-case breast cancer families that identified two additional index cases with ATM 7271T>G and phylogenetic sequence analysis showed that Valine2424 is a highly conserved residue. 17008050_ATM regulates ionizing radiation-induced disruption of HDAC1:PP1:Rb complexes. 17019709_Increased ATM expression is associated with esophageal squamous cell carcinoma and its premalignant lesions 17030982_The response of promyelocytic leukemia nuclear bodies to DNA double-strand breaks is regulated by NBS1, ATM, Chk2, and ATR. 17106266_ATM is activated during oxidative burst in phorbol ester-treated human leukocytes. 17121863_a novel pathway, which is connected between ataxia telangiectasia-mutated kinase (ATM) and protein phosphatase-Plk1 in DNA damage response in mitosis 17124492_ATM phosphorylation at Ser1981, a characterised autophosphorylation site, is ATR-dependent and ATM-independent following replication fork stalling or ultraviolet rays treatment. 17132159_Observational study of gene-disease association. (HuGE Navigator) 17151932_Observational study of gene-disease association. (HuGE Navigator) 17151932_study found associations of risk haplotypes and protective haplotypes in p53 for glioblastoma and in ATM for meningioma; study provides new data that could add to our understanding of brain tumour susceptibility 17164260_Common variants in the ATM and CHEK2 genes, in interaction with oestrogen-related exposures, are involved in endometrial cancer aetiology. 17164260_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17166884_current results support the association of two A-T-related ATM mutations, 6903insA and 7570G>C, in addition to 8734A>G, with breast cancer susceptibility 17172861_Smad7 plays a crucial role upstream of ATM and p53 to protect the genome from insults evoked by extracellular stress. 17178844_Function for ATM in the control of mitogenic pathways affecting cell signaling and emphasize the key role of ATM in coordinating the cellular response to DNA damage. 17189255_ATM mediates DNA-PKcs phosphorylation at Thr-2609 as well as at the adjacent (S/T)Q motifs within the Thr-2609 cluster. DNA-PKcs- and ATM-mediated DNA-PKcs phosphorylations are cooperative and required for the full activation of DNA-PKcs 17203191_Observational study of gene-disease association. (HuGE Navigator) 17227291_The levels of CAA and CHP in lymphocytes were increased many-fold during their stimulation. 17242184_ATM regulates G(2)/M checkpoint recovery through inhibitory phosphorylations of Artemis that occur soon after DNA damage, thus setting a molecular switch that, hours later upon completion of DNA repair, allows activation of the Cdk1-cyclin B complex. 17293864_Observational study of gene-disease association. (HuGE Navigator) 17333338_ATM does not appear to represent a breast cancer susceptibility gene in the general African-American population 17333338_Observational study of gene-disease association. (HuGE Navigator) 17341484_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17341604_ATM may be essential for Ku activation to repair DNA damage from oxidative stress and prevent cell death caused by oxidative stress. 17350468_Deletion-specific probes detected a homozygous loss of two anonymous loci in chromosomal band 13q14 in parallel with a heterozygous loss of the ATM gene located in chromosomal band 11q22.3. 17351744_Observational study of gene-disease association. (HuGE Navigator) 17351744_analysis of allelic variants in the ATM gene in Chilean women with breast cancer 17384674_-terminal domains of NBS1 are the major regulatory domains for recombination pathways, very likely through the recruitment and retention of double strand breakage sites in an ATM-independent fashion. 17384681_Alterations in the cellular levels of TIP influence the phosphorylation state of a specific protein substrate of ataxia-telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR) kinases. 17389389_Antisense oligonucleotides corrected prototypic ATM splicing mutations and aberrant ATM function. 17393301_ATM missense variants do not contribute to the risk of contralateral breast cancer, but the combination of radiation therapy and (certain) ATM missense variants seems to accelerate tumor development 17393301_Observational study of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 17409144_EBNA3C can directly regulate the G2/M component of the host cell cycle machinery through ATM/ATR and Chk2, allowing for the release of the checkpoint block 17409195_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17426037_a novel mechanism for activation of the activity of ATM kinase by retinoic acid , and implicate ATM in the regulation of CREB function during RA-induced differentiation. 17428320_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17428325_Observational study of gene-disease association. (HuGE Navigator) 17428325_common polymorphisms in the ATM, BRCA1, BRCA2, CHEK2 and TP53 cancer susceptibility genes are not shown to increase breast cancer risk 17428792_physical interaction between the major DSB signaling kinase, ATM and poly(ADP- | ENSMUSG00000034218 | Atm | 8.069025e+02 | 0.4932849 | -1.019507111 | 0.3342382 | 8.982513e+00 | 0.0027257552 | 0.19044519 | No | Yes | 4.141018e+02 | 90.892594 | 9.888509e+02 | 221.449099 | |
| ENSG00000149503 | 3619 | INCENP | protein_coding | Q9NQS7 | FUNCTION: Component of the chromosomal passenger complex (CPC), a complex that acts as a key regulator of mitosis. The CPC complex has essential functions at the centromere in ensuring correct chromosome alignment and segregation and is required for chromatin-induced microtubule stabilization and spindle assembly. Acts as a scaffold regulating CPC localization and activity. The C-terminus associates with AURKB or AURKC, the N-terminus associated with BIRC5/survivin and CDCA8/borealin tethers the CPC to the inner centromere, and the microtubule binding activity within the central SAH domain directs AURKB/C toward substrates near microtubules (PubMed:15316025, PubMed:12925766, PubMed:27332895). The flexibility of the SAH domain is proposed to allow AURKB/C to follow substrates on dynamic microtubules while ensuring CPC docking to static chromatin (By similarity). Activates AURKB and AURKC (PubMed:27332895). Required for localization of CBX5 to mitotic centromeres (PubMed:21346195). Controls the kinetochore localization of BUB1 (PubMed:16760428). {ECO:0000250|UniProtKB:P53352, ECO:0000269|PubMed:12925766, ECO:0000269|PubMed:15316025, ECO:0000269|PubMed:16760428, ECO:0000269|PubMed:21346195, ECO:0000269|PubMed:27332895}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Centromere;Chromosome;Chromosome partition;Cytoplasm;Cytoskeleton;Kinetochore;Microtubule;Mitosis;Nucleus;Phosphoprotein;Reference proteome | In mammalian cells, 2 broad groups of centromere-interacting proteins have been described: constitutively binding centromere proteins and 'passenger,' or transiently interacting, proteins (reviewed by Choo, 1997). The constitutive proteins include CENPA (centromere protein A; MIM 117139), CENPB (MIM 117140), CENPC1 (MIM 117141), and CENPD (MIM 117142). The term 'passenger proteins' encompasses a broad collection of proteins that localize to the centromere during specific stages of the cell cycle (Earnshaw and Mackay, 1994 [PubMed 8088460]). These include CENPE (MIM 117143); MCAK (MIM 604538); KID (MIM 603213); cytoplasmic dynein (e.g., MIM 600112); CliPs (e.g., MIM 179838); and CENPF/mitosin (MIM 600236). The inner centromere proteins (INCENPs) (Earnshaw and Cooke, 1991 [PubMed 1860899]), the initial members of the passenger protein group, display a broad localization along chromosomes in the early stages of mitosis but gradually become concentrated at centromeres as the cell cycle progresses into mid-metaphase. During telophase, the proteins are located within the midbody in the intercellular bridge, where they are discarded after cytokinesis (Cutts et al., 1999 [PubMed 10369859]).[supplied by OMIM, Mar 2008]. | hsa:3619; | central element [GO:0000801]; chromocenter [GO:0010369]; chromosome passenger complex [GO:0032133]; chromosome, centromeric region [GO:0000775]; cytosol [GO:0005829]; kinetochore [GO:0000776]; lateral element [GO:0000800]; meiotic spindle midzone [GO:1990385]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; midbody [GO:0030496]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; pericentric heterochromatin [GO:0005721]; protein-containing complex [GO:0032991]; spindle [GO:0005819]; protein serine/threonine kinase activator activity [GO:0043539]; chromosome segregation [GO:0007059]; histone phosphorylation [GO:0016572]; meiotic spindle midzone assembly [GO:0051257]; metaphase plate congression [GO:0051310]; mitotic cytokinesis [GO:0000281]; mitotic sister chromatid segregation [GO:0000070]; positive regulation of protein serine/threonine kinase activity [GO:0071902]; regulation of mitotic cytokinesis [GO:1902412] | 12925766_Aurora B kinase activity is stimulated by INCENP and C-terminal region of INCENP is sufficient for activation 15316025_association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C 15796717_Recruitment of MKLP1 to the midzone/midbody by INCENP is a crucial step for the midbody formation and completion of cytokinesis in mammalian cells. 15917996_Aurora-C is a chromosomal passenger protein that disrupts the association of INCENP with Aurora-B and may serve as a key regulator in cell division 16239925_Data show that INCENP has an important role in stabilizing the chromosomal passenger complex, and that Borealin acts to promote binding of Survivin to INCENP. 16378098_INCENP phosphorylation by Cdk1 is necessary for the recruitment of Plk1 to the kinetochore. 16571674_A functional module within the chromosomal passenger complex involving the inner centromere protein INCENP, Survivin, and Borealin. 17623812_protein truncation and in vitro mutagenesis,have identified the nucleolar localization sequences on INCENP 17956729_Borealin and INCENP associate with the helical domain of Survivin to form a tight three-helical bundle. 18752045_High INCENP expression is associated with high grade non-Hodgkin B-cell lymphomas. 19494039_Data show that binding to INCENP is alone critical to the distinct function of Aurora B, and although G198 of Aurora A is required for TPX2 binding, N142G Aurora B retains INCENP binding and Aurora B function. 20372054_Results indicate that INCENP-Aurora B localized at centromeres/inner kinetochores is sufficient to mediate SAC activity upon spindle disruption. 20619651_Dephosphorylation of INCENP at anaphase and the concomitant relocation of the chromosomal passenger protein complex prevents kinetochore recruitment of mitotic checkpoint proteins. 21346195_HP1alpha binding by INCENP or Shugoshin 1 (Sgo1) is dispensable for centromeric cohesion protection during mitosis of human cells, but might regulate yet unknown interphase functions of the chromosome passenger complex (CPC) or Sgo1 at the centromeres. 23562843_v-Src induces the failure of cytokinesis and the delocalization of Mklp1, Aurora B, and INCENP from the spindle midzone. 23848594_Our results provide the structural basis and energetics of the human Aurora-A(G198N) - INCENP complex 25586992_The data suggest that INCENP in the chromosomal passenger complex pathway contributes to estrogen receptor-negative breast cancer susceptibility in the European population. 26460953_Data unveils a novel mechanism of PRMT1-mediated CPC regulation through methylation of INCENP. 27332895_Aurora-C interactions with members of the Chromosome Passenger Complex (CPC), Survivin and Inner Centromere Protein (INCENP) in reference to known Aurora-B interactions to understand the functional significance of Aurora-C overexpression in human cancer cells, is reported. 31306061_Switching of INCENP paralogs controls transitions in mitotic chromosomal passenger complex functions. 31416840_Dysregulation of INCENP contributes to neuroblastoma tumorigenesis 31943174_lncRNA MIAT promotes esophageal squamous cell carcinoma progression by regulating miR-1301-3p/INCENP axis and interacting with SOX2. 31991041_In this study, we designed and synthesized 7-19 residues of inner centromere protein (INCENP)-derived small peptides (INC peptides) as novel survivin-targeting agents. 33355621_An ATM-Chk2-INCENP pathway activates the abscission checkpoint. | ENSMUSG00000024660 | Incenp | 4.009387e+03 | 1.3481935 | 0.431027535 | 0.2629403 | 2.715518e+00 | 0.0993767270 | 0.75783482 | No | Yes | 3.791673e+03 | 220.593959 | 2.792433e+03 | 167.057556 | |
| ENSG00000149657 | 149986 | LSM14B | protein_coding | Q9BX40 | FUNCTION: Required for oocyte meiotic maturation. May be involved in the storage of translationally inactive mRNAs and protect them from degradation (By similarity). Plays a role in control of mRNA translation (By similarity). {ECO:0000250|UniProtKB:Q68FI1, ECO:0000250|UniProtKB:Q8CGC4}. | Acetylation;Alternative splicing;Developmental protein;Isopeptide bond;Methylation;Phosphoprotein;Reference proteome;Ribonucleoprotein;Translation regulation;Ubl conjugation | hsa:149986; | mRNA binding [GO:0003729]; RNA binding [GO:0003723]; regulation of translation [GO:0006417] | ENSMUSG00000039108 | Lsm14b | 2.927619e+03 | 1.0127249 | 0.018242330 | 0.2799493 | 4.274807e-03 | 0.9478698492 | 0.99014376 | No | Yes | 2.502048e+03 | 146.890121 | 2.554279e+03 | 153.800733 | |||
| ENSG00000149743 | 83707 | TRPT1 | protein_coding | Q86TN4 | FUNCTION: Catalyzes the last step of tRNA splicing, the transfer of the splice junction 2'-phosphate from ligated tRNA to NAD to produce ADP-ribose 1''-2'' cyclic phosphate. {ECO:0000305|PubMed:14504659}. | Acetylation;Alternative splicing;NAD;Phosphoprotein;Reference proteome;Transferase;tRNA processing | hsa:83707; | tRNA 2'-phosphotransferase activity [GO:0000215]; regulation of protein kinase activity [GO:0045859]; tRNA processing [GO:0008033]; tRNA splicing, via endonucleolytic cleavage and ligation [GO:0006388] | Mouse_homologues 18094117_Inactivation of the Trpt1 gene, encoding the only known mammalian homolog of Tpt1p, eliminates all detectable 2'-phosphotransferase activity from cultured mouse cells but has no measurable effect on spliced Xbp-1 translation. | ENSMUSG00000047656 | Trpt1 | 8.525536e+02 | 1.2134922 | 0.279164805 | 0.3175445 | 7.728820e-01 | 0.3793269509 | 0.84118691 | No | Yes | 8.218054e+02 | 94.467204 | 6.215000e+02 | 73.743740 | ||
| ENSG00000149809 | 7108 | TM7SF2 | protein_coding | O76062 | FUNCTION: Catalyzes the reduction of the C14-unsaturated bond of lanosterol, as part of the metabolic pathway leading to cholesterol biosynthesis. {ECO:0000269|PubMed:16784888}. | Alternative splicing;Cholesterol biosynthesis;Cholesterol metabolism;Endoplasmic reticulum;Lipid biosynthesis;Lipid metabolism;Membrane;Microsome;NADP;Oxidoreductase;Reference proteome;Steroid biosynthesis;Steroid metabolism;Sterol biosynthesis;Sterol metabolism;Transmembrane;Transmembrane helix | PATHWAY: Steroid biosynthesis; cholesterol biosynthesis. | hsa:7108; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; nuclear inner membrane [GO:0005637]; receptor complex [GO:0043235]; delta14-sterol reductase activity [GO:0050613]; NADP binding [GO:0050661]; oxidoreductase activity, acting on the CH-CH group of donors [GO:0016627]; cholesterol biosynthetic process [GO:0006695]; sterol biosynthetic process [GO:0016126] | 11937680_Results show that the TM7SF2 gene product SR-1 and six chimeras containing SR-1 and Neurospora erg-3 sequences fail to complement sterol c-14 reductase mutants in Neurospora and yeast. 16784888_a primary role in human cholesterol biosynthesis 18660489_Observational study of gene-disease association. (HuGE Navigator) 19940018_LBR mutant variants and sterol reductases can severely interfere with the regular organization of the nuclear envelope and the endoplasmic reticulum. 20138239_Data suggest that, besides the SRE motif, both the inverted CCAAT-box and GC-box2 are essential for full TM7SF2 promoter activation by SREBP-2. 31911440_Twin enzymes, divergent control: The cholesterogenic enzymes DHCR14 and LBR are differentially regulated transcriptionally and post-translationally. | ENSMUSG00000024799 | Tm7sf2 | 1.657275e+03 | 1.3557864 | 0.439129925 | 0.3131476 | 1.966732e+00 | 0.1607951800 | 0.77593452 | No | Yes | 1.683365e+03 | 195.430633 | 1.188036e+03 | 142.011765 | |
| ENSG00000149930 | 9344 | TAOK2 | protein_coding | Q9UL54 | FUNCTION: Serine/threonine-protein kinase involved in different processes such as membrane blebbing and apoptotic bodies formation DNA damage response and MAPK14/p38 MAPK stress-activated MAPK cascade. Phosphorylates itself, MBP, activated MAPK8, MAP2K3, MAP2K6 and tubulins. Activates the MAPK14/p38 MAPK signaling pathway through the specific activation and phosphorylation of the upstream MAP2K3 and MAP2K6 kinases. In response to DNA damage, involved in the G2/M transition DNA damage checkpoint by activating the p38/MAPK14 stress-activated MAPK cascade, probably by mediating phosphorylation of upstream MAP2K3 and MAP2K6 kinases. Isoform 1, but not isoform 2, plays a role in apoptotic morphological changes, including cell contraction, membrane blebbing and apoptotic bodies formation. This function, which requires the activation of MAPK8/JNK and nuclear localization of C-terminally truncated isoform 1, may be linked to the mitochondrial CASP9-associated death pathway. Isoform 1 binds to microtubules and affects their organization and stability independently of its kinase activity. Prevents MAP3K7-mediated activation of CHUK, and thus NF-kappa-B activation, but not that of MAPK8/JNK. May play a role in the osmotic stress-MAPK8 pathway. Isoform 2, but not isoform 1, is required for PCDH8 endocytosis. Following homophilic interactions between PCDH8 extracellular domains, isoform 2 phosphorylates and activates MAPK14/p38 MAPK which in turn phosphorylates isoform 2. This process leads to PCDH8 endocytosis and CDH2 cointernalization. Both isoforms are involved in MAPK14 phosphorylation. {ECO:0000269|PubMed:10660600, ECO:0000269|PubMed:11279118, ECO:0000269|PubMed:12639963, ECO:0000269|PubMed:12665513, ECO:0000269|PubMed:13679851, ECO:0000269|PubMed:16893890, ECO:0000269|PubMed:17158878, ECO:0000269|PubMed:17396146}. | ATP-binding;Alternative splicing;Cell projection;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Kinase;Magnesium;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Transmembrane;Transmembrane helix | This gene encodes a serine/threonine protein kinase that is involved in many different processes, including, cell signaling, microtubule organization and stability, and apoptosis. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Oct 2011]. | hsa:9344; | actin cytoskeleton [GO:0015629]; axon [GO:0030424]; axonal growth cone [GO:0044295]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytoplasmic vesicle membrane [GO:0030659]; cytosol [GO:0005829]; dendritic growth cone [GO:0044294]; integral component of membrane [GO:0016021]; neuron projection [GO:0043005]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; MAP kinase kinase kinase activity [GO:0004709]; mitogen-activated protein kinase kinase binding [GO:0031434]; neuropilin binding [GO:0038191]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activator activity [GO:0043539]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; tau protein binding [GO:0048156]; tau-protein kinase activity [GO:0050321]; actin cytoskeleton organization [GO:0030036]; activation of protein kinase activity [GO:0032147]; apoptotic process [GO:0006915]; axonogenesis [GO:0007409]; basal dendrite arborization [GO:0150020]; basal dendrite morphogenesis [GO:0150019]; cell migration [GO:0016477]; cellular response to DNA damage stimulus [GO:0006974]; focal adhesion assembly [GO:0048041]; intracellular signal transduction [GO:0035556]; MAPK cascade [GO:0000165]; mitotic G2 DNA damage checkpoint signaling [GO:0007095]; neuron projection morphogenesis [GO:0048812]; positive regulation of JNK cascade [GO:0046330]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of protein autophosphorylation [GO:0031954]; positive regulation of stress-activated MAPK cascade [GO:0032874]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; protein targeting to membrane [GO:0006612]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of cell growth [GO:0001558]; regulation of cell shape [GO:0008360]; regulation of MAPK cascade [GO:0043408]; stress-activated MAPK cascade [GO:0051403] | 12639963_PSK interacts with microtubules and affects their organization and stability independently of PSK kinase activity 12665513_mediates signaling from carbachol to p38 mitogen-activated protein kinase and ternary complex factors 16893890_the TAK1-JNK pathway is activated by osmotic stress, while blocking TAK1-mediated NF-kappaB activation; TAO2 regulates TAK1 pathways 17158878_PSK1-alpha is a bifunctional kinase that associates with microtubules, and JNK- and caspase-mediated removal of its C-terminal microtubule-binding domain permits nuclear translocation of the N-terminal region of PSK1-alpha and its induction of apoptosis 19242545_Observational study of gene-disease association. (HuGE Navigator) 22683681_TAOK2 is essential for dendrite morphogenesis; TAOK2 downregulation impairs basal dendrite formation without affecting apical dendrites. 23585562_tau is a good substrate for PSK1 and PSK2 phosphorylation with mass spectrometric analysis of phosphorylated tau revealing more than 40 tau residues as targets of these kinases. 28385331_A mutation in TAOK2 appears to cause a novel form of primary immunodeficiency, characterized by an impaired T cell proliferation upon activation. 28830982_we characterize a compound that inhibits TAOK1 and TAOK2 activity with IC50 values of 11 to 15 nmol/L, is ATP-competitive, and targets these kinases selectively. TAOK inhibition or depletion in centrosome-amplified SKBR3 or BT549 breast cancer cell models increases the mitotic population, the percentages of mitotic cells displaying amplified centrosomes and multipolar spindles, induces cell death, and inhibits cell growt 34879262_TAOK2 is an ER-localized kinase that catalyzes the dynamic tethering of ER to microtubules. 35141166_The circRNA circSIAE Inhibits Replication of Coxsackie Virus B3 by Targeting miR-331-3p and Thousand and One Amino-Acid Kinase 2. | ENSMUSG00000059981 | Taok2 | 1.096647e+04 | 1.0848750 | 0.117528874 | 0.2920167 | 1.611190e-01 | 0.6881283368 | 0.93352105 | No | Yes | 9.771142e+03 | 955.270285 | 9.184596e+03 | 920.995929 | |
| ENSG00000150281 | 1489 | CTF1 | protein_coding | Q16619 | FUNCTION: Induces cardiac myocyte hypertrophy in vitro. Binds to and activates the ILST/gp130 receptor. | Alternative splicing;Cytokine;Reference proteome;Secreted | The protein encoded by this gene is a secreted cytokine that induces cardiac myocyte hypertrophy in vitro. It has been shown to bind and activate the ILST/gp130 receoptor. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2008]. | hsa:1489; | extracellular region [GO:0005576]; extracellular space [GO:0005615]; cytokine activity [GO:0005125]; leukemia inhibitory factor receptor binding [GO:0005146]; cell surface receptor signaling pathway [GO:0007166]; cell-cell signaling [GO:0007267]; leukemia inhibitory factor signaling pathway [GO:0048861]; muscle organ development [GO:0007517]; neuron development [GO:0048666]; neuron differentiation [GO:0030182]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of tyrosine phosphorylation of STAT protein [GO:0042531] | 12234945_upregulation of cardiotrophin-1 in congestive heart failure 12707269_Leukemia inhibitory factor (LIF), cardiotrophin-1, and oncostatin M share structural binding determinants in the immunoglobulin-like domain of LIF receptor 15219667_Our study clearly demonstrates production of CT-1 in the postnatal and adult CNS, specifically by cell types comprising the blood-CSF barrier, and its accumulation in ventricular ependyma. 15339920_Data show that cardiotrophin (CT-1) is a potent regulator of signaling in adipocytes in vitro and in vivo. 17483238_CT-1 and TGF-beta 1 exert differential effects on myofibroblast proliferation and contraction in vitro. A balance of these effects may be important for normal cardiac wound healing. 17940213_adipose tissue can be recognized as a source of CT-1, which could account for the high circulating levels of CT-1 in patients with metabolic syndrome X 17979974_Increased expression of some IL-6 cytokine family members (oncostatin M, gp130, CT-1, LIF) in cutaneous inflammation might contribute to the promotion of hair loss. 18609050_Perioperative plasma brain natriuretic peptide and cardiotrophin-1 measured in off-pump coronary artery bypass. 19155793_Plasma cardiotrophin-1 is associated with progression of heart failure in hypertensive patients. 19273604_Results indicate that transcription factors such as CTF1 can act to delimit chromatin domain boundaries at mammalian telomeres, thereby blocking the propagation of a silent chromatin structure. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20053569_This study aims to analyse cardiotrophin-1 (CT-1) and Tumor Necrosis Factor-alpha (TNF-alpha) plasma levels at rest and during exercise in elite athletes and healthy controls. 20224758_Data show that both protein synthesis and intracellular transport are essential for cardiotrophin-1 induced TNFalpha expression. 20474083_Observational study of genetic testing. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20683337_Observational study of gene-disease association. (HuGE Navigator) 20683337_the 1742(C/G) polymorphism of the human CT-1 gene is associated with LVH in hypertension, and the GG genotype may have a protective role 21572008_CT-1 treatment of myofibroblasts was associated with increased phosphorylation of myosin light chain kinase via myosin light chain kinase to induce cell migration. 21771897_Hypoxia increased cardiotrophin-1 levels in cardiac cells through a direct regulation of CT-1 promoter by HIF-1alpha, protecting cells from apoptosis. 22418690_Cardiotrophin-1 may serve as both a biomarker of left ventricular hypertrophy and dysfunction in hypertensive patients, and is potential target for therapies aimed to prevent and treat hypertensive heart disease beyond blood pressure control. 23856329_A reduction in serum CT-1 levels after a WL program. 23935888_CT-1 induces the proteolytic potential in human aortic endothelial cells by upregulating MMP-1 expression. 24054317_Subjects with impaired glucose tolerance (IGT) and newly diagnosed diabetes (NDD) have significantly higher CT-1 concentrations than those with normal glucose tolerance. IGT and NDD are positively associated with CT-1 concentrations. 24344663_CT-1 is differentially induced in the myocardium of infants with congenital cardiac defects depending on hypoxemia, and may mediate myocardial hypertrophy and dysfunction. 24366078_exaggerated cardiomyocyte production of cardiotrophin-1 in response to increased left ventricular end-diastolic stress may contribute to fibrosis through stimulation of fibroblasts in heart failure of hypertensive origin. 24401751_Study correlates CT-1 levels with ambulatory and central BP, as well as with pulse wave velocity in patients with essential hypertension. 25025664_this study, even though preliminary and awaiting further confirmation by independent replication, provides first evidence that common genetic variation in CTF1 could contribute to insulin sensitivity in humans. 25689900_Data show that cardiotrophin-1 is positively related to brachial-ankle pulse-wave velocity (baPWV) independent of traditional cardiometabolic risk factors for arterial stiffness. 25919795_biliary epithelium-derived CT-1 may exert a profibrogenic potential in PCLD. 26188636_This paper will review many aspects of CT-1 physiological role in several organs and discuss data for consideration in therapeutic approaches. 26334851_CT-1 was found to be associated with Tn-I, which is used to detect myocardial damage after OPCAB surgery. CT-1 may also be used to detect myocardial damage. 26621340_non-diabetic subjects who were overweight or obesity had significantly lower cardiotrophin-1 concentrations than those with normal weight, and both obesity and being overweight were inversely associated with cardiotrophin-1 levels. 26787685_Cardiotrophin-1 levels were not an independent predictor of all-cause mortality in hemodialysis patients. 26867947_a transcriptional factor, Myc-associated zinc finger protein (MAZ), plays an important role in ADAM10 transcription in response to CT-1 in neural stem/progenitor cells. 28749076_Data suggest that, in children with pediatric obesity, lifestyle weight-loss intervention results in down-regulation of serum cardiotrophin-1 (CTF1), interleukin-6 (IL6), and tumor necrosis factor-alpha (TNFA); expression of CTF1, IL6, and TNFA is also down-regulated in peripheral blood mononuclear cells after improvement in adiposity, body mass index, and waist-hip ratio. 28785017_We identify the cytokine cardiotrophin 1 (CT1) as a factor capable of recapitulating the key features of physiologic growth of the heart including transient and reversible hypertrophy of the myocardium, and stimulation of cardiomyocyte-derived angiogenic signals leading to increased vascularity 30408810_study indicates that variations in dietary salt intake affect the serum Cardiotrophin-1 levels in Chinese adults. 30724267_CT-1 may be considered as an additional sensitive and specific prognostic marker complementary to the conventional markers such as NT-proBNP, usable in the stratification of the adverse cardiac events risk in patients with symptomatic, systolic heart failure 33713510_A quantitative detection of Cardiotrophin-1 in chronic heart failure by chemiluminescence immunoassay. | ENSMUSG00000042340 | Ctf1 | 8.842829e+01 | 1.2527761 | 0.325128547 | 0.4090263 | 6.469666e-01 | 0.4211993055 | 0.84954815 | No | Yes | 8.663764e+01 | 14.546336 | 6.894178e+01 | 12.355872 | |
| ENSG00000150401 | 55208 | DCUN1D2 | protein_coding | Q6PH85 | FUNCTION: Contributes to the neddylation of all cullins by transferring NEDD8 from N-terminally acetylated NEDD8-conjugating E2s enzyme to different cullin C-terminal domain-RBX complexes and plays an essential role in the regulation of SCF (SKP1-CUL1-F-box protein)-type complexes activity. {ECO:0000269|PubMed:19617556, ECO:0000269|PubMed:23201271, ECO:0000269|PubMed:26906416}. | 3D-structure;Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation pathway | hsa:55208; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; cullin family protein binding [GO:0097602]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin-like protein binding [GO:0032182]; positive regulation of protein neddylation [GO:2000436]; positive regulation of ubiquitin-protein transferase activity [GO:0051443]; protein neddylation [GO:0045116]; regulation of protein neddylation [GO:2000434] | ENSMUSG00000038506 | Dcun1d2 | 3.115384e+02 | 0.6356042 | -0.653799501 | 0.3243376 | 3.978813e+00 | 0.0460760246 | 0.65303870 | No | Yes | 1.824405e+02 | 33.570878 | 3.318924e+02 | 61.985576 | |||
| ENSG00000150433 | 219854 | TMEM218 | protein_coding | A2RU14 | FUNCTION: May be involved in ciliary biogenesis or function. {ECO:0000250|UniProtKB:Q9CQ44}. | Cell projection;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:219854; | cilium [GO:0005929]; integral component of membrane [GO:0016021] | Mouse_homologues 25161209_tmem218-/- mice develop progressive cystic kidney disease and retinal degeneration, a model for Senior-Loken syndrome | ENSMUSG00000032121 | Tmem218 | 8.897660e+02 | 0.7649694 | -0.386526131 | 0.3006503 | 1.664862e+00 | 0.1969481856 | 0.78383913 | No | Yes | 6.393008e+02 | 65.363732 | 8.737854e+02 | 90.971068 | ||
| ENSG00000150756 | 134145 | ATPSCKMT | protein_coding | Q6P4H8 | FUNCTION: Mitochondrial protein-lysine N-methyltransferase that trimethylates ATP synthase subunit C, ATP5MC1 and ATP5MC2. Trimethylation is required for proper incorporation of the C subunit into the ATP synthase complex and mitochondrial respiration (PubMed:29444090, PubMed:30530489). Promotes chronic pain (PubMed:29444090). Involved in persistent inflammatory and neuropathic pain: methyltransferase activity in the mitochondria of sensory neurons promotes chronic pain via a pathway that depends on the production of reactive oxygen species (ROS) and on the engagement of spinal cord microglia (PubMed:29444090). {ECO:0000269|PubMed:29444090, ECO:0000269|PubMed:30530489}. | Acetylation;Alternative splicing;Membrane;Methyltransferase;Mitochondrion;Reference proteome;S-adenosyl-L-methionine;Transferase;Transmembrane;Transmembrane helix | hsa:134145; | integral component of membrane [GO:0016021]; mitochondrial crista [GO:0030061]; mitochondrion [GO:0005739]; protein-lysine N-methyltransferase activity [GO:0016279]; peptidyl-lysine methylation [GO:0018022]; peptidyl-lysine trimethylation [GO:0018023]; positive regulation of proton-transporting ATP synthase activity, rotational mechanism [GO:1905273]; positive regulation of sensory perception of pain [GO:1904058]; regulation of mitochondrial ATP synthesis coupled proton transport [GO:1905706] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21802380_Genetic alteration of JS-2 was found to be related to location, pathological subtypes and staging of colorectal cancer. 29444090_we uncover a role for methyltransferase activity of FAM173B in the neurobiology of pain. These results also highlight FAM173B methyltransferase activity as a potential therapeutic target to treat debilitating chronic pain conditions 30530489_It has been identified FAM173B as the long-sought KMT responsible for methylation of ATP synthase c-subunit (ATPSc), a key protein in cellular ATP production, and have demonstrated functional significance of ATPSc methylation. 33067627_Expression of mitochondrial TSPO and FAM173B is associated with inflammation and symptoms in patients with painful knee osteoarthritis. | ENSMUSG00000039065 | Atpsckmt | 2.034162e+02 | 0.8542378 | -0.227290306 | 0.3425815 | 4.395790e-01 | 0.5073257451 | 0.87821327 | No | Yes | 1.583395e+02 | 24.316197 | 1.763879e+02 | 27.645320 | ||
| ENSG00000151135 | 90488 | TMEM263 | protein_coding | Q8WUH6 | FUNCTION: May play a role in bone development. {ECO:0000269|PubMed:34238371}. | Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:90488; | integral component of membrane [GO:0016021] | 34238371_TMEM263: a novel candidate gene implicated in human autosomal recessive severe lethal skeletal dysplasia. | ENSMUSG00000060935 | Tmem263 | 3.000351e+02 | 0.8954602 | -0.159298731 | 0.3695963 | 1.822105e-01 | 0.6694804442 | 0.92913032 | No | Yes | 2.911154e+02 | 57.373389 | 2.853402e+02 | 57.661030 | ||
| ENSG00000151332 | 51562 | MBIP | protein_coding | Q9NS73 | FUNCTION: Inhibits the MAP3K12 activity to induce the activation of the JNK/SAPK pathway. Component of the ATAC complex, a complex with histone acetyltransferase activity on histones H3 and H4. {ECO:0000269|PubMed:19103755}. | Acetylation;Alternative splicing;Cytoplasm;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | hsa:51562; | ATAC complex [GO:0140672]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; identical protein binding [GO:0042802]; protein kinase inhibitor activity [GO:0004860]; histone H3 acetylation [GO:0043966]; histone H3-K14 acetylation [GO:0044154]; positive regulation of gene expression [GO:0010628]; positive regulation of JNK cascade [GO:0046330]; regulation of cell cycle [GO:0051726]; regulation of cell division [GO:0051302]; regulation of embryonic development [GO:0045995]; regulation of histone deacetylation [GO:0031063]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; regulation of tubulin deacetylation [GO:0090043] | 32711446_Pathway-Affecting Single Nucleotide Polymorphisms (SNPs) in RPS6KA1 and MBIP Genes are Associated with Breast Cancer Risk. 32963352_MBIP (MAP3K12 binding inhibitory protein) drives NSCLC metastasis by JNK-dependent activation of MMPs. | ENSMUSG00000021028 | Mbip | 1.749477e+02 | 1.0702783 | 0.097985987 | 0.4383421 | 5.045326e-02 | 0.8222764319 | 0.96628438 | No | Yes | 1.652687e+02 | 35.830187 | 1.574405e+02 | 34.962853 | ||
| ENSG00000151422 | 2241 | FER | protein_coding | P16591 | FUNCTION: Tyrosine-protein kinase that acts downstream of cell surface receptors for growth factors and plays a role in the regulation of the actin cytoskeleton, microtubule assembly, lamellipodia formation, cell adhesion, cell migration and chemotaxis. Acts downstream of EGFR, KIT, PDGFRA and PDGFRB. Acts downstream of EGFR to promote activation of NF-kappa-B and cell proliferation. May play a role in the regulation of the mitotic cell cycle. Plays a role in the insulin receptor signaling pathway and in activation of phosphatidylinositol 3-kinase. Acts downstream of the activated FCER1 receptor and plays a role in FCER1 (high affinity immunoglobulin epsilon receptor)-mediated signaling in mast cells. Plays a role in the regulation of mast cell degranulation. Plays a role in leukocyte recruitment and diapedesis in response to bacterial lipopolysaccharide (LPS). Plays a role in synapse organization, trafficking of synaptic vesicles, the generation of excitatory postsynaptic currents and neuron-neuron synaptic transmission. Plays a role in neuronal cell death after brain damage. Phosphorylates CTTN, CTNND1, PTK2/FAK1, GAB1, PECAM1 and PTPN11. May phosphorylate JUP and PTPN1. Can phosphorylate STAT3, but the biological relevance of this depends on cell type and stimulus. {ECO:0000269|PubMed:12972546, ECO:0000269|PubMed:14517306, ECO:0000269|PubMed:19147545, ECO:0000269|PubMed:19339212, ECO:0000269|PubMed:19738202, ECO:0000269|PubMed:20111072, ECO:0000269|PubMed:21518868, ECO:0000269|PubMed:22223638, ECO:0000269|PubMed:7623846, ECO:0000269|PubMed:9722593}. | 3D-structure;ATP-binding;Alternative promoter usage;Alternative splicing;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Kinase;Lipid-binding;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;SH2 domain;Transferase;Tyrosine-protein kinase;Ubl conjugation | The protein encoded by this gene is a member of the FPS/FES family of non-transmembrane receptor tyrosine kinases. It regulates cell-cell adhesion and mediates signaling from the cell surface to the cytoskeleton via growth factor receptors. Alternative splicing results in multiple transcript variants. A related pseudogene has been identified on chromosome X. [provided by RefSeq, Apr 2015]. | hsa:2241; | cell cortex [GO:0005938]; cell junction [GO:0030054]; cell projection [GO:0042995]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; nucleus [GO:0005634]; actin binding [GO:0003779]; ATP binding [GO:0005524]; cadherin binding [GO:0045296]; epidermal growth factor receptor binding [GO:0005154]; gamma-catenin binding [GO:0045295]; lipid binding [GO:0008289]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase binding [GO:0019901]; protein phosphatase 1 binding [GO:0008157]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; signaling receptor binding [GO:0005102]; small GTPase binding [GO:0031267]; actin cytoskeleton reorganization [GO:0031532]; cell adhesion [GO:0007155]; cell differentiation [GO:0030154]; cell population proliferation [GO:0008283]; cell-cell adhesion mediated by cadherin [GO:0044331]; cellular response to insulin stimulus [GO:0032869]; cellular response to macrophage colony-stimulating factor stimulus [GO:0036006]; cellular response to reactive oxygen species [GO:0034614]; chemotaxis [GO:0006935]; cytokine-mediated signaling pathway [GO:0019221]; diapedesis [GO:0050904]; extracellular matrix-cell signaling [GO:0035426]; Fc-epsilon receptor signaling pathway [GO:0038095]; innate immune response [GO:0045087]; insulin receptor signaling pathway via phosphatidylinositol 3-kinase [GO:0038028]; interleukin-6-mediated signaling pathway [GO:0070102]; intracellular signal transduction [GO:0035556]; Kit signaling pathway [GO:0038109]; microtubule cytoskeleton organization [GO:0000226]; negative regulation of mast cell activation involved in immune response [GO:0033007]; peptidyl-tyrosine phosphorylation [GO:0018108]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of actin filament polymerization [GO:0030838]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of epidermal growth factor receptor signaling pathway [GO:0042058]; regulation of fibroblast migration [GO:0010762]; regulation of lamellipodium assembly [GO:0010591]; regulation of mast cell degranulation [GO:0043304]; regulation of protein phosphorylation [GO:0001932]; response to lipopolysaccharide [GO:0032496]; response to platelet-derived growth factor [GO:0036119]; substrate adhesion-dependent cell spreading [GO:0034446]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; tyrosine phosphorylation of STAT protein [GO:0007260] | 11994747_Closing in on the biological functions of Fps/Fes and Fer. A review. 12871378_Fps/Fes and Fer are expressed in human and mouse platelets, and are activated following stimulation with collagen and collagen-related peptide (CRP), suggesting a role in GPVI receptor signaling 16732323_Fer is a regulator of cell-cycle progression in malignant cells and a potential target for cancer intervention. 18985748_FerT coexist in the acroplaxome with phosphorylated cortactin, a regulator of F-actin dynamics[Fer testis ] 19147545_Fer tyrosine kinase level correlates with the development of prostate cancer and aggressiveness of prostate cancer cell lines 19339212_Results suggest that Fer may allow a bypass of focal adhesion kinase-related cell anchorage dependency for intracellular signal transduction in hepatocytes. 19738202_Overexpression of Fer enhanced lamellipodia formation and cell migration in a manner dependent on PLD activity and the PA-FX interaction. 19835603_FER plays a role in the invasion and metastasis of hepatocellular carcinoma cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21122136_tyrosine phosphorylation of RhoGDIalpha by Fer as a mechanism to regulate binding of RhoGDIalpha to Rac 21518868_Overexpression of FER from a cDNA confers quinacrine resistance to several different types of cancer cell lines. 21700879_The A allele of SNP rs10447248 in the FER locus was nominally associated with lower MMW (P = 0.016) but was not associated with LMW (P > 0.05) adiponectin levels 22223638_Transcription of the ferT gene in CC cells was found to be driven by an intronic promoter residing in intron 10 of the fer gene and to be regulated by the Brother of the Regulator of Imprinted Sites (BORIS) transcription factor. 22238358_Fer, a non-receptor-type tyrosine kinase, plays a critical role in synthesis of the laminin-binding glycans on alpha-DG. 23445469_Fer expression correlates with renal cell carcinoma cell proliferation both in vitro and in vivo, and with tumor progression and survival 23699534_data show that Fer kinase is elevated in non-small cell lung cancer tumors and is important for cellular invasion and metastasis 23873028_FER kinase promotes breast cancer metastasis by regulating alpha6- and beta1-integrin-dependent cell adhesion and anoikis resistance. 23906537_Fer contributes to aberrant androgen receptor signaling via pSTAT3 cross-talk during castrate-resistant prostate cancer progression. 23931849_Detection of PJA2-FER fusion mRNA is correlated with poor postoperative survival periods in non-small cell lung cancer. 25533491_FER encodes a cytosolic non-receptor tyrosine kinase that influences neutrophil chemotaxis and endothelial permeability. 25867068_Fer serves as a crucial mediator and amplifier of Src-induced tumor progression. 28245430_Many human tumor types and cancer cell lines express the MAN2A1-FER fusion, which increases proliferation and invasiveness of cancer cell lines and has liver oncogenic activity in mice. 28851893_The FER rs4957796 TT genotype remained a significant covariate for the 90-day mortality risk in the multivariate analysis (hazard ratio, 4.62; 95% CI, 1.58-13.50; p = 0.0050). In conclusion, FER rs4957796 might act as a prognostic variable for survival in patients with severe ARDS due to pneumonia. 29038547_Data show that FER tyrosine kinase (Fer/FerT) activity was disrupted by E260, which selectively evokes metabolic stress in cancer cells by imposing mitochondrial dysfunction and deformation. 29099290_FER mediated HGF-independent regulation of HGFR/MET activates RAC1-PAK1 pathway to potentiate metastasis in ovarian cancer. 29208465_We propose a model for the regulation of Fer based on an intramolecular interaction and the curvature-dependent membrane binding mediated by its intrinsically disordered region. 29540831_FER enhances IGF-1R expression, phosphorylation, and signaling to promote cooperative growth and adhesion signaling that may facilitate cancer progression. 29907877_FER overexpression improves survival through STAT activation enhancing innate immunity and accelerating bacterial clearance in the lung. 29920310_Targeting of both FER and MET may be an effective strategy for therapeutic intervention in ovarian cancer. 31746983_This study supports the critical role of FER and FES tyrosine kinase fusions in the pathogenesis of follicular T-cell lymphoma and provides additional evidence that these can drive follicular T-cell lymphoma in the absence of RHOA mutations. 33411917_Phosphorylation of PKCdelta by FER tips the balance from EGFR degradation to recycling. 33430475_Fer and FerT Govern Mitochondrial Susceptibility to Metformin and Hypoxic Stress in Colon and Lung Carcinoma Cells. 33806191_Loss of Fer Jeopardizes Metabolic Plasticity and Mitochondrial Homeostasis in Lung and Breast Carcinoma Cells. | ENSMUSG00000000127 | Fer | 1.406026e+02 | 0.9113170 | -0.133975176 | 0.4634855 | 8.454433e-02 | 0.7712309971 | 0.95442153 | No | Yes | 1.057228e+02 | 26.720763 | 1.257583e+02 | 32.369868 | |
| ENSG00000151491 | 2059 | EPS8 | protein_coding | Q12929 | FUNCTION: Signaling adapter that controls various cellular protrusions by regulating actin cytoskeleton dynamics and architecture. Depending on its association with other signal transducers, can regulate different processes. Together with SOS1 and ABI1, forms a trimeric complex that participates in transduction of signals from Ras to Rac by activating the Rac-specific guanine nucleotide exchange factor (GEF) activity. Acts as a direct regulator of actin dynamics by binding actin filaments and has both barbed-end actin filament capping and actin bundling activities depending on the context. Displays barbed-end actin capping activity when associated with ABI1, thereby regulating actin-based motility process: capping activity is auto-inhibited and inhibition is relieved upon ABI1 interaction. Also shows actin bundling activity when associated with BAIAP2, enhancing BAIAP2-dependent membrane extensions and promoting filopodial protrusions. Involved in the regulation of processes such as axonal filopodia growth, stereocilia length, dendritic cell migration and cancer cell migration and invasion. Acts as a regulator of axonal filopodia formation in neurons: in the absence of neurotrophic factors, negatively regulates axonal filopodia formation via actin-capping activity. In contrast, it is phosphorylated in the presence of BDNF leading to inhibition of its actin-capping activity and stimulation of filopodia formation. Component of a complex with WHRN and MYO15A that localizes at stereocilia tips and is required for elongation of the stereocilia actin core. Indirectly involved in cell cycle progression; its degradation following ubiquitination being required during G2 phase to promote cell shape changes. {ECO:0000269|PubMed:15558031, ECO:0000269|PubMed:17115031}. | 3D-structure;Actin-binding;Alternative splicing;Cell junction;Cell membrane;Cell projection;Cytoplasm;Deafness;Membrane;Non-syndromic deafness;Phosphoprotein;Reference proteome;SH3 domain;Synapse;Synaptosome;Ubl conjugation | This gene encodes a member of the EPS8 family. This protein contains one PH domain and one SH3 domain. It functions as part of the EGFR pathway, though its exact role has not been determined. Highly similar proteins in other organisms are involved in the transduction of signals from Ras to Rac and growth factor-mediated actin remodeling. Alternate transcriptional splice variants of this gene have been observed but have not been thoroughly characterized. [provided by RefSeq, Jul 2008]. | hsa:2059; | brush border [GO:0005903]; cell cortex [GO:0005938]; extracellular exosome [GO:0070062]; glutamatergic synapse [GO:0098978]; growth cone [GO:0030426]; NMDA selective glutamate receptor complex [GO:0017146]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; ruffle membrane [GO:0032587]; stereocilium [GO:0032420]; stereocilium tip [GO:0032426]; vesicle [GO:0031982]; actin binding [GO:0003779]; signaling adaptor activity [GO:0035591]; small GTPase binding [GO:0031267]; actin crosslink formation [GO:0051764]; actin cytoskeleton reorganization [GO:0031532]; actin filament bundle assembly [GO:0051017]; actin polymerization-dependent cell motility [GO:0070358]; adult locomotory behavior [GO:0008344]; barbed-end actin filament capping [GO:0051016]; behavioral response to ethanol [GO:0048149]; cellular response to leukemia inhibitory factor [GO:1990830]; dendritic cell migration [GO:0036336]; exit from mitosis [GO:0010458]; positive regulation of ruffle assembly [GO:1900029]; Rac protein signal transduction [GO:0016601]; regulation of actin filament length [GO:0030832]; regulation of cell shape [GO:0008360]; regulation of postsynaptic membrane neurotransmitter receptor levels [GO:0099072]; regulation of Rho protein signal transduction [GO:0035023]; Rho protein signal transduction [GO:0007266] | 15273867_Involved in the growth factor-controlled regulation of cell proliferation and differentiation in the seminiferous epithelium. 17115031_These results support a model whereby the synergic bundling activity of the IRSp53-Eps8 complex, regulated by Cdc42, contributes to the generation of actin bundles, thus promoting filopodial protrusions. 17537571_Eps8 is essential for actin dynamics and cell interactions, independent of Eps8-like gene products. 18566210_These findings implicate the involvement of Eps8 in chemoresistance and show its importance in prognosis of cervical cancer patients. 19008210_overexpression of EPS8 in HN4 cells was sufficient to induce growth of non-tumorigenic cells in orthotopic transplantation assays. EPS8 expression in samples of squamous cell carcinoma showed variable expression levels and paralleled expression of MMP-9 19116338_A role of Eps8 in amplifying growth factor receptor signaling in human pituitary tumors to promote proliferation and cell survival. 19448673_Results show that alphavbeta6- and alpha5beta1-integrin-dependent activation of Rac1 was mediated through Eps8. 19528316_the crystal structures of human LanCL1, both free of and complexed with glutathione, revealing glutathione binding to a zinc ion at the putative active site formed by conserved GxxG motifs 20184880_Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells 20351091_Overexpression of EPS8 induced expression of the chemokine ligands CXCL5 and CXCL12 in a FOXM1-dependent manner, which was blocked by LY294002 or a dominant-negative form of AKT 20418908_IRSp53, through its interaction with Eps8, not only affects cell migration but also dictates cellular growth in cancer cells. 20677014_Observational study of gene-disease association. (HuGE Navigator) 21118970_Study implicates that the integrity of SOS1/EPS8/ABI1 tri-complex is a determinant of ovarian cancer metastasis. 21357683_critical role for JNK2 and EPS8 in receptor tyrosine kinase signaling and trafficking to convey distinctly different effects on cell migration. 21814501_Studied generation of filopodia with regards to the dynamic interaction established by Eps8, IRSp53 and VASP with actin filaments. 22449706_The ITSN2 interacts with Eps8 and stimulates the degradation of Eps8 proteins. 22493489_Eps8 is a key regulator of the LPS-stimulated TLR4-MyD88 interaction and contributes to macrophage phagocytosis 22683923_silencing of the protein by siRNA abrogated the migratory and invasive capacity of three different glioblastoma cell lines both in 2-dimensional and 3-dimensional in vitro assays. 22876043_The loss of EPS8 expression in colorectal adenomas and carcinomas suggests that down regulation of this gene contributes to the development of a subset of colorectal cancers. 22897151_Eps8 is frequently expressed in OSCC. The aberrant expression of Eps8 closely correlated with poor survival in patients with OSCC. 23203811_Eps8 functions as a key coordinator in the interplay between FGFR signalling and trafficking. 23229386_We found that Eps8 mediates cell proliferation and survival of glioma cells, at least in part, by affecting phosphorylated ERK and Akt/beta-catenin activities. 23314863_Identify Fbxw5-driven fluctuation of Eps8 levels as an important mechanism that contributes to cell-shape changes during entry into-and exit from-mitosis. 23626693_Novel binding partners and differentially regulated phosphorylation sites clarify Eps8 as a multi-functional adaptor. 24409660_results suggest that Eps8 may serve as a prognostic factor of responsiveness to chemotherapy in AML patients 24741995_EPS8 is an F-actin capping and bundling protein. Mutant mice lacking EPS8 (Eps8-/- mice), which is present in the hair bundle, the sensory antenna of the auditory sensory cells that operate the mechano-electrical transduction 25031323_determined the alpha-synuclein-binding domain of beta-III tubulin and demonstrated that a short fragment containing this domain can suppress alpha-synuclein accumulation in the primary cultured cells 25333707_Eps8 is overexpressed in human breast cancers, possibly by regulating ERK signaling, MMP9, p53 and EMT-like transition to affect breast cancer cell growth, migration and invasion. 25359883_Eps8 is a crucial mediator of Src- and FAK-regulated processes. 25376540_These results indicate that employing the native and modified epitopes identified here in Eps8-based immunotherapy for HLA-A2.1 positive cancer patients may result in efficient anticancer immune responses for diverse tumor types. 25843487_EPS8, as MDR1 and WT1, may be a clinically valuable biomarker for assessing the outcome of ALL patients. 26163656_Erk activity promotes actin bundling by Eps8 to enhance cortex tension and drive the bleb-based migration of cancer cells under non-adhesive confinement. 26976596_Eps8 is required for continuous membrane blebbing. 27573546_Immunohistochemistry revealed that Eps8 was significantly increased in cervical cancer specimens compared with squamous intraepithelial lesion and normal cervical tissues. Additionally, it was revealed that Eps8 expression not only correlated with cervical cancer progression, but also exhibited a close correlation with the epithelialmesenchymal transition (EMT) markers, Ecadherin and vimentin. 28214294_These results indicate that plasma-membrane-associated PTK6 phosphorylates Eps8, which promotes cell proliferation, adhesion, and migration 28608476_Eps8/Abi1/Sos1 tricomplex acts as a key molecular switch altering the balance between Rac1 and Rho activation; its presence or absence in pancreatic ductal adenocarcinoma cells modulates alphavbeta6-dependent functions, resulting in a pro-migratory (Rac1-dependent) or a pro-TGF-beta1 activation (Rho-dependent) functional phenotype 29107665_Eps8 is involved in tumor invasion but not necessarily the development of regional lymph node metastasis 29192326_Data showed that chronic myeloid leukemia (CML) patients expressed a higher level of EPS8 mRNA in bone marrow mononuclear cells. Functional results revealed that EPS8 regulated multiple biological functions such as proliferation, apoptosis, cell cycle, drug sensitivity of CML cells possibly by mediating the regulation of the BCR-ABL/AKT/mTOR signalling pathway. 30203615_LncRNA DSCAM-AS1 acts as a competing endogenous RNA of miR-137 and regulates EPS8 to promote cell reproduction and suppresses cell apoptosis in Tamoxifen-resistant breast cancer 30431134_Comparative analyses revealed that Eps8 protein was abundant in exosomes derived from metastatic pancreatic tumors and ascites and that the amount of exosomal Eps8 was quantitatively correlated with the in vitro cell migratory activity. 30858505_we focused on EPS8 because its expression had the greatest impact on patient prognosis (overall survival, p < 0.0001). Overexpression of EPS8 was detected in PDAC clinical specimens. Knockdown assays with siEPS8 showed that its overexpression enhanced cancer cell proliferation, migration, and invasion 31118055_We identified EE02 as an EGFR/Eps8 complex inhibitor that demonstrated promising antitumor effects in breast cancer and non-small cell lung cancer (NSCLC). Our data suggest that the EGFR/Eps8 complex offers a novel cancer drug target. 32147678_miR-345 inhibits migration and stem-like cell phenotype in gastric cancer via inactivation of Rac1 by targeting EPS8. 33432368_Effects and mechanisms of Eps8 on the biological behaviour of malignant tumours (Review). 33626355_Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex. 34031495_The roles and prognostic significance of ABI1-TSV-11 expression in patients with left-sided colorectal cancer. 34391775_EPS8 supports pancreatic cancer growth by inhibiting BMI1 mediated proteasomal degradation of ALDH7A1. 34637946_Apparent homozygosity for a novel splicing variant in EPS8 causes congenital profound hearing loss. | ENSMUSG00000015766 | Eps8 | 1.743028e+02 | 0.9196263 | -0.120880319 | 0.4387031 | 7.422798e-02 | 0.7852774736 | 0.95754084 | No | Yes | 1.803743e+02 | 41.189635 | 1.831156e+02 | 42.980264 | |
| ENSG00000151502 | 112936 | VPS26B | protein_coding | Q4G0F5 | FUNCTION: Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. The recruitment of the CSC to the endosomal membrane involves RAB7A and SNX3. The SNX-BAR retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) and is involved in endosome-to-plasma membrane transport for cargo protein recycling. The SNX3-retromer mediates the retrograde transport of WLS distinct from the SNX-BAR retromer pathway. The SNX27-retromer is believed to be involved in endosome-to-plasma membrane trafficking and recycling of a broad spectrum of cargo proteins. The CSC seems to act as recruitment hub for other proteins, such as the WASH complex and TBC1D5. May be involved in retrograde transport of SORT1 but not of IGF2R. Acts redundantly with VSP26A in SNX-27 mediated endocytic recycling of SLC2A1/GLUT1 (By similarity). {ECO:0000250|UniProtKB:O75436, ECO:0000250|UniProtKB:Q8C0E2}. | Cytoplasm;Endosome;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:112936; | cytosol [GO:0005829]; early endosome [GO:0005769]; endosome [GO:0005768]; late endosome [GO:0005770]; phagocytic vesicle [GO:0045335]; retromer complex [GO:0030904]; retromer, cargo-selective complex [GO:0030906]; cellular response to interferon-gamma [GO:0071346]; intracellular protein transport [GO:0006886]; regulation of macroautophagy [GO:0016241]; retrograde transport, endosome to Golgi [GO:0042147] | 21920005_Colocalization of Vps26 paralogues with different endosomally located Rab proteins shows prolonged association of Vps26B-retromer with maturing endosomes relative to Vps26A-retromer. 26113136_Authors propose that PAR-2 plasma membrane repopulation is regulated by Vps26B-retromer, describing a potential novel role for this complex. | ENSMUSG00000031988 | Vps26b | 4.105137e+03 | 1.1877656 | 0.248250124 | 0.2795498 | 8.029098e-01 | 0.3702247995 | 0.83676049 | No | Yes | 4.105212e+03 | 245.689072 | 3.265261e+03 | 201.263213 | ||
| ENSG00000151576 | 79691 | QTRT2 | protein_coding | Q9H974 | FUNCTION: Non-catalytic subunit of the queuine tRNA-ribosyltransferase (TGT) that catalyzes the base-exchange of a guanine (G) residue with queuine (Q) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine). {ECO:0000255|HAMAP-Rule:MF_03043}. | 3D-structure;Alternative splicing;Cytoplasm;Membrane;Metal-binding;Mitochondrion;Mitochondrion outer membrane;Reference proteome;Zinc;tRNA processing | This gene encodes a subunit of tRNA-guanine transglycosylase. tRNA-guanine transglycosylase is a heterodimeric enzyme complex that plays a critical role in tRNA modification by synthesizing the 7-deazaguanosine queuosine, which is found in tRNAs that code for asparagine, aspartic acid, histidine, and tyrosine. The encoded protein may play a role in the queuosine 5'-monophosphate salvage pathway. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Feb 2012]. | hsa:79691; | cytoplasm [GO:0005737]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; protein-containing complex [GO:0032991]; transferase complex [GO:1990234]; metal ion binding [GO:0046872]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; queuine tRNA-ribosyltransferase activity [GO:0008479]; tRNA-guanine transglycosylation [GO:0101030] | 20354154_TGT is composed of a catalytic subunit, QTRT1, and QTRTD1, not USP14. QTRTD1 has been implicated as the salvage enzyme that generates free queuine from QMP. 21044367_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000022704 | Qtrt2 | 1.530654e+03 | 0.8896804 | -0.168640945 | 0.3111539 | 2.847278e-01 | 0.5936190338 | 0.90759816 | No | Yes | 1.523969e+03 | 257.358338 | 1.513527e+03 | 262.104874 | |
| ENSG00000151725 | 79682 | CENPU | protein_coding | Q71F23 | FUNCTION: Component of the CENPA-NAC (nucleosome-associated) complex, a complex that plays a central role in assembly of kinetochore proteins, mitotic progression and chromosome segregation. The CENPA-NAC complex recruits the CENPA-CAD (nucleosome distal) complex and may be involved in incorporation of newly synthesized CENPA into centromeres. Plays an important role in the correct PLK1 localization to the mitotic kinetochores. A scaffold protein responsible for the initial recruitment and maintenance of the kinetochore PLK1 population until its degradation. Involved in transcriptional repression. {ECO:0000269|PubMed:12941884, ECO:0000269|PubMed:16716197, ECO:0000269|PubMed:17081991}. | Alternative splicing;Centromere;Chromosome;Coiled coil;Cytoplasm;Host-virus interaction;Isopeptide bond;Kinetochore;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | The centromere is a specialized chromatin domain, present throughout the cell cycle, that acts as a platform on which the transient assembly of the kinetochore occurs during mitosis. All active centromeres are characterized by the presence of long arrays of nucleosomes in which CENPA (MIM 117139) replaces histone H3 (see MIM 601128). MLF1IP, or CENPU, is an additional factor required for centromere assembly (Foltz et al., 2006 [PubMed 16622419]).[supplied by OMIM, Mar 2008]. | hsa:79682; | centriolar satellite [GO:0034451]; cytosol [GO:0005829]; kinetochore [GO:0000776]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chordate embryonic development [GO:0043009] | 15116101_a possible role for MLF1IP deregulation in the genesis of erythroleukemias 15893739_MLF1IP was found in glioblastomas where it was co-localized with MLF1 and nestin. Moreover, it was elevated in the contralateral brain where no tumor cells occurred, suggesting a role in glioma pathogenesis and potentially in other types of malignancies. 17081991_Plk1 self-regulates the Plk1-PBIP1 interaction to timely localize to the kinetochores and promote proper chromosome segregation. 17595757_Deletion mutants of MLF1IP revealed that the N-terminal bipartite nuclear localization signal (NLS) was responsible for nucleolar targeting. 21056971_CENP-U is a novel microtubule binding protein and plays an important role in kinetochore-microtubule attachment through its interaction with Hec1 21454580_Mammalian polo-like kinase 1-dependent regulation of the PBIP1-CENP-Q complex at kinetochores. 23028590_Data propose that CENP-P/O/R/Q/U self-assembles on kinetochores with varying stoichiometry and undergoes a pre-mitotic maturation step that could be important for kinetochores switching into the correct conformation for microtubule-attachment. 25572810_MLF1IP significantly promotes prostate cancer cell proliferation and colony formation and significantly inhibits apoptosis without affecting cell cycle phase arrest in prostate cancer cell lines 25670858_Plk1 regulates the timing of the delocalization and ultimate destruction of the PBIP1.CENP-Q complex. 27378428_This study provides new insights and evidences that MLF1IP over-expression plays important roles in progression of luminal breast cancer. However, the precise cellular mechanisms for MLF1IP in luminal breast cancer need to be further explored. 28173615_MLF1IP promotes normal erythroid proliferation and is involved in the pathogenesis of polycythemia vera. 28677729_In-depth IPA analysis revealed that CENPU was associated with the HMGB1 signaling pathway. qPCR and western blot analysis demonstrated that in the HMGB1 signaling pathway, CENPU knockdown downregulated expression levels of ILB, CXCL8, RAC1 and IL1A 30536323_Centromere protein U promotes cell proliferation, migration and invasion involving Wnt/beta-catenin signaling pathway in non-small cell lung cancer 30849291_CENPU downregulation significantly inhibited LAC cell proliferation, migration and invasion in, which was possibly mediated by PI3K/AKT pathway inactivation 31705927_Centromere protein U (CENPU) enhances angiogenesis in triple-negative breast cancer by inhibiting ubiquitin-proteasomal degradation of COX-2. 33248027_BUB1 and CENP-U, Primed by CDK1, Are the Main PLK1 Kinetochore Receptors in Mitosis. 33596090_Differential requirements for the CENP-O complex reveal parallel PLK1 kinetochore recruitment pathways. 33902008_The lncRNA GATA3-AS1/miR-495-3p/CENPU axis predicts poor prognosis of breast cancer via the PLK1 signaling pathway. 34551298_Bub1 and CENP-U redundantly recruit Plk1 to stabilize kinetochore-microtubule attachments and ensure accurate chromosome segregation. 34872447_Centromere protein U (CENPU) promotes gastric cancer cell proliferation and glycolysis by regulating high mobility group box 2 (HMGB2). 34957303_Abnormal Expression of Centromere Protein U Is Associated with Hepatocellular Cancer Progression. 35122991_Overexpression of MLF1IP promotes colorectal cancer cell proliferation through BRCA1/AKT/p27 signaling pathway. | ENSMUSG00000031629 | Cenpu | 6.407019e+02 | 0.7428523 | -0.428852705 | 0.3314741 | 1.620991e+00 | 0.2029536399 | 0.78445471 | No | Yes | 5.549324e+02 | 90.831091 | 7.722565e+02 | 129.254607 | |
| ENSG00000151929 | 9531 | BAG3 | protein_coding | O95817 | FUNCTION: Co-chaperone for HSP70 and HSC70 chaperone proteins. Acts as a nucleotide-exchange factor (NEF) promoting the release of ADP from the HSP70 and HSC70 proteins thereby triggering client/substrate protein release. Nucleotide release is mediated via its binding to the nucleotide-binding domain (NBD) of HSPA8/HSC70 where as the substrate release is mediated via its binding to the substrate-binding domain (SBD) of HSPA8/HSC70 (PubMed:9873016, PubMed:27474739). Has anti-apoptotic activity (PubMed:10597216). Plays a role in the HSF1 nucleocytoplasmic transport (PubMed:26159920). {ECO:0000269|PubMed:10597216, ECO:0000269|PubMed:24318877, ECO:0000269|PubMed:26159920, ECO:0000269|PubMed:27474739, ECO:0000269|PubMed:9873016}. | Acetylation;Apoptosis;Cardiomyopathy;Chaperone;Cytoplasm;Disease variant;Isopeptide bond;Methylation;Myofibrillar myopathy;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | BAG proteins compete with Hip for binding to the Hsc70/Hsp70 ATPase domain and promote substrate release. All the BAG proteins have an approximately 45-amino acid BAG domain near the C terminus but differ markedly in their N-terminal regions. The protein encoded by this gene contains a WW domain in the N-terminal region and a BAG domain in the C-terminal region. The BAG domains of BAG1, BAG2, and BAG3 interact specifically with the Hsc70 ATPase domain in vitro and in mammalian cells. All 3 proteins bind with high affinity to the ATPase domain of Hsc70 and inhibit its chaperone activity in a Hip-repressible manner. [provided by RefSeq, Jul 2008]. | hsa:9531; | aggresome [GO:0016235]; chaperone complex [GO:0101031]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; neuron projection [GO:0043005]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; stress fiber [GO:0001725]; Z disc [GO:0030018]; adenyl-nucleotide exchange factor activity [GO:0000774]; cadherin binding [GO:0045296]; chaperone binding [GO:0051087]; dynein intermediate chain binding [GO:0045505]; protein carrier chaperone [GO:0140597]; protein-containing complex binding [GO:0044877]; aggresome assembly [GO:0070842]; autophagosome assembly [GO:0000045]; brain development [GO:0007420]; cellular response to heat [GO:0034605]; cellular response to mechanical stimulus [GO:0071260]; cellular response to unfolded protein [GO:0034620]; chaperone-mediated autophagy [GO:0061684]; extrinsic apoptotic signaling pathway in absence of ligand [GO:0097192]; extrinsic apoptotic signaling pathway via death domain receptors [GO:0008625]; muscle cell cellular homeostasis [GO:0046716]; negative regulation of apoptotic process [GO:0043066]; negative regulation of protein targeting to mitochondrion [GO:1903215]; negative regulation of striated muscle cell apoptotic process [GO:0010664]; negative regulation of transcription from RNA polymerase II promoter in response to stress [GO:0097201]; positive regulation of aggrephagy [GO:1905337]; positive regulation of protein export from nucleus [GO:0046827]; positive regulation of protein import into nucleus [GO:0042307]; protein folding [GO:0006457]; protein stabilization [GO:0050821]; protein transport along microtubule [GO:0098840]; spinal cord development [GO:0021510] | 12700638_BAG3 protein has a role in apoptosis in B-chronic lymphocytic leukaemia cells 12706811_heavy metals and temperature regulate BAG3 gene expression. 12750378_findings suggest that heat shock protein-70 is a chaperone driving a multiprotein degradation complex and that the inhibitory co-chaperone CAIR-1 protein functions distal to client ubiquitination 16385451_Observational study of gene-disease association. (HuGE Navigator) 16391524_These results suggest that the Bis induced growth inhibition of HL-60 cells promotes G0/G1 phase arrest via up-regulation of p27, which seems to be a prerequisite for differentiation. 16859681_Results indicate that CAIR-1 may negatively regulate adhesion, focal adhesion assembly, signaling, and migration via its PXXP domain. 17164298_BAG3 downmodulates the apoptotic response to TRAIL in human neoplastic thyroid cells. 17187345_These observations reveal a previously unrecognized cell response, that is, an increase in BAG3, elicited by HIV-1 infection, and may provide a new avenue for the suppression of HIV-1 gene expression. 17507989_The metabolic pathway of BAG3 was studied. 17974966_High levels of BAG3 protein seen in some epithelial cancer cell lines may be relevant to mechanisms of tumor invasion and metastasis. 17996194_Taken together, our results suggest that BAG3 induction might represents as an unwanted molecular consequence of utilizing proteasome inhibitors to combat tumors. 18006506_These results suggested that the HspB8-Bag3 complex might stimulate the degradation of Htt43Q by macroautophagy. 18094623_HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. 18286539_Two of the three putative heat shock-responsive elements (HSEs) in bag3 promoter interact with the heat shock factor (HSF) 1 in vitro and in vivo. 18469860_Activation of BAG3 by Egr-1 in response to FGF-2 in neuroblastoma. 18821563_Downmodulation of BAG3 protein levels allows caspase-3 activation by HIV-1 infection in human primary microglial cells. 19085932_This study conclude mutation in Bag3 defines a novel severe autosomal dominant childhood muscular dystrophy. 19088197_a requirement for BAG3 to augment virus gene expression and demonstrate that the co-chaperone acts independently of promyelocytic leukemia to increase herpes simplex virus replication. 19111544_These results indicate that knocking down BAG3 gene is a promising new approach to enhance the therapeutic potency of Bortezomib in leukemia. 19212330_in a B-cell lymphoma cell line, physical interaction was observed between BTK and BAG3, possibly upon modifications induced by stress 19282432_This study showed that BAG3 protein is downregulated upon JC virus (JCV) infection and that this effect is mediated by JCV T-antigen via repression of the BAG3 promoter. 19352495_Loss of BAG3 under STS injury required sequential caspase cleavage followed by polyubiquitination and proteasomal degradation 19777443_BAG3 gene expression is controlled by its own product. 19845507_Data show that the interaction between HspB6 and Bag3 requires the same regions that are involved in the HspB8-Bag3 association. 19850283_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20018251_identification of BAG3 binding partner (cytosolic chaperonin CCT); BAG3/CTT complex binds to BAG3 promoter & exhibits biological function (i.e., influences correct folding of monomeric actin); study of BAG3 domain participation in binding to CCT 20232307_BAG3 is cleaved during apoptosis of pancreatic cancer cells, disrupting its anti-apoptotic properties. 20368414_BAG3 alters the interaction between HSP70 and IKKgamma, increasing availability of IKKgamma and protecting it from proteasome-dependent degradation; this, in turn, results in increased NF-kappaB activity and survival 20535599_BAG3 intra-cytoplasmic delocalisation is a specific feature of cancer versus non-neoplastic prostate and a candidate new marker for prediction of prostate cancer invasiveness and behaviour. 20605452_These observations suggest that the BAG3 variant of myofibrillar myopathy may result from a spontaneous mutation at an early point of embryonic development and that transmission from a mosaic parent may occur more than once. 20607728_interaction between virus penton base protein and cellular co-chaperone BAG3 positively influences virus life cycle 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20723284_bag3 expression in chronic lymphocytic leukemia patients is markedly higher than that in normal controls, while high bag3 level in CML patients is probably related with drug resistance, but is not related with clinically established prognostic factors. 20800573_BAG3 overexpression increases cell adhesion in Cos7 cells, but not in PDZGEF2 gene knockdown cells indicating that PDZGEF2 is a critical partner for BAG3 in regulating cell adhesion 20800603_Observational study of gene-disease association. (HuGE Navigator) 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20855536_Observational study of gene-disease association. (HuGE Navigator) 21233200_BAG3 expression in glioblastoma cells promotes accumulation of ubiquitinated clients in an Hsp70-dependent manner. 21252941_aggresome-targeting by BAG3 is distinct from previously described mechanisms, as it does not depend on substrate ubiquitination 21302292_BAG3 was identified as a marker of electromagnetic field-induced stress. These results suggest that BAG3 induction by an electromagnetic field may contribute to melanoma cell survival and/or resistance to therapy. 21316839_The incidence of BAG3 positivity was significantly higher at advanced clinical stages of ovarian cancer than at early stages. It is suggected BAG3 binds to MMP2 to positively regulate the process of cell invasion. 21353195_New comprehensive genomic approaches have identified rare variants in BAG3 as causative of dilated cardiomyopathy. 21361913_the first Chinese case of Bag3opathy so far reported. 21423662_These findings indicate a novel function for BAG3 in inhibiting protein aggregation caused by the genetic mutation of CRYAB responsible for human myofibrillar myopathy. 21451574_BAG3 promotes the emergence of castration resistant prostate cancer by modulating IKKalpha nuclear translocation. 21459883_The results show that rare mutations in BAG3 contribute to monogenic forms of the disease, while common variant(s) in the same gene are implicated in sporadic DCM 21472004_Multifaceted interactions underlie BAG3 ability to modulate apoptosis, development, cytoskeleton organization and autophagy. [Review] 21561597_BAG3, although negative in normal brain tissues, was highly expressed in astrocytic tumors and increasingly expressed in more aggressive types of cancer; it was particularly high in glioblastomas. 21683594_This study identified no pathogenic mutations in BAG3, MATR3, PTRF or TCAP in Australian muscular dystrophy. 21696420_we observed upregulation of HSPB8 and BAG3 selectively in astrocytes located within the degenerated areas of patients with protein aggregation diseases 21767525_The complexes formed by Bag3 and HspB8 might have variable stoichiometry and can participate in different processes including clearing of the cell from improperly folded proteins. 21850027_BAG3 is expressed in a proportion of melanoma and in the majority of metastases, so it may be involed in tumor development. 21898660_We report here two heterozygous missense mutations of BAG3 found in familial DCM, which cause abnormal Z-disc assembly and increase the sensitivity to apoptosis in the cardiomyocytes. 21971866_non-small cell lung cancer cells were protected from apoptosis through increasing Bag3 expression and consequently promoted the expression of Bcl-XL and Bcl-2. 22020323_the data support that BAG3 is induced by LPS via JNK and NF-kappaB-dependent signals, and involved in monocytic cell-extracellular matrix interaction, suggesting that BAG3 may have a role in the host response to LPS stimulation 22072743_BAG3 protein sustains anaplastic thyroid tumor growth in vitro and in vivo. The underlying molecular mechanism appears to rely on BAG3 binding to BRAF, thus protecting it from proteasome-dependent degradation. 22302993_findings show that during heat shock recovery NF-kappaB activates selective removal of misfolded or aggregated proteins by controlling expression of BAG3 and HSPB8 and by modulating the level of the BAG3-HspB8 complex 22310281_BAG3 is expressed in endothelial cells. 22734908_This report confirms the association of giant axonal neuropathy with BAG3-associated myofibrillar myopathy 22944597_BAG3 expression level might be a potential marker for prediction of patient outcome. 22984599_Data suggest that by investigating the interplay between large T-antigen (T-Ag) and Bag3 may provid better understanding on the development of JCV-associated diseases. 23108398_BAG3 is a novel substrate of PKCdelta, and PKCdelta-mediated phosphorylation of BAG3 is implicated in EMT and invasiveness of thyroid cancer cells. 23271418_we identified Caspase 3, BAG1 and BAG3 as key targets of ERalpha in neuronal cells that may play a role in ERalpha-mediated neuroprotection. 23341456_Studies indicate BAG3-mediated Mcl-1 stabilization as a potential target for cancer drug discovery. 23395971_Hyperthermia sensitivity in human oral squamous cell carcinoma cells is enhanced upon silencing of BAG3 and inhibition of the JNK pathway. 23434281_Autophagosome formation during chaperone-assisted selective autophagy depends on an interaction of BAG3 with synaptopodin-2. 23434281_BAG3 balances autophagic degradation and YAP/TAZ-mediated transcription of filamin in mechanically strained cells and tissues. BAG3 uses its WW domain to associate with SYNPO2 during autophagosome formation or to bind the YAP/TAZ-inhibitors LATS1/2 and AMOTL1/2 during transcription regulation. This dual function is essential for tissue maintenance and for cell adhesion, migration and proliferation in mammals. 23553442_aberrant Bag-3 expression might be involved in colorectal adenoma-adenocarcinoma sequence and subsequent progression 23575457_Noncanonical autophagy mediated by BAG3 suppresses responsiveness of HepG2 cells to proteasome inhibitors 23582692_Missense mutations in antiapoptotic bag3 protein are found in patients with tako-tsubo cardiomyopathy. 23824909_Functional studies demonstrated that BAG3 interacts with the proteasome and modulates its activity, sustaining cell survival and underlying resistance to therapy through the down-modulation of apoptosis. 23978946_Data suggest that BAG3 expression is increased in chronic lymphocytic leukemia (CLL) cells compared with normal B-cells; knocking down BAG3 expression results in increased apoptosis and decreased cell migration in primary CLL cells. 24080088_these results suggested that through direct interaction BAG3 could prevent the antiapoptotic effect of GRP78 upon genotoxic stress. 24140207_BAG3 stabilized JunD mRNA. 24286317_Bis knockdown resulted in a significant decrease in the migration and invasion of A172 glioma cells. Furthermore, Bis knockdown notably decreased TPA-induced matrix metalloproteinase-9 (MMP-9) activity and mRNA expression. 24318877_we measured the binding of human Hsp72 (HSPA1A) to BAG1, BAG2, BAG3, and the unrelated NEF Hsp105. These studies revealed a clear hierarchy of affinities: BAG3 > BAG1 > Hsp105 >> BAG2. 24365746_BAG3 was overexpressed in hepatocellular carcinoma. BAG3 knockdown resulted in reduction in migration and invasion of cells, which was linked to reversion of epithelial-mesenchymal transition. 24492285_BAG-3 expression correlated with increased HSP70 expression in a subset of systemic T cell lymphoma cases co-expressing the CD30 antigen. 24577090_These results indicate BAG3 as a regulator of ZEB1 expression in epithelial-mesenchymal transition and as a regulator of metastasis in thyroid cancer cells. 24623017_Diminished levels of BAG3 protein may be associated with both familial and non-familial forms of dilated cardiomyopathy. 24722298_These results identify a subgroup of stage III melanoma patients, that is, patients with 2-3 positive lymph nodes, whose clinical behavior is influenced by the expression of the anti-apoptotic BAG3 protein. 24895585_The results suggested that the expression level of BAG3 and HIF-1 alpha is efficient prognostic parameters in patients with HCC after liver transplantation. 24994713_Disrupts the Hsp70-Bag3 interaction. 24997994_Bag3-MVP is an important complex that regulates a potent prosurvival signaling pathway and contributes to chemotherapy resistance in breast cancer 25008357_BAG3 point mutations and large deletions are relatively frequent cause of dilated cardiomyopathy 25046115_BAG3 binding to the ubiquitinated clients occurs through the BAG domain, in competition with BAG1 25149536_A subset of SCLCs over express BAG3 that exerts an anti-apoptotic effect resulting in resistance to chemotherapy. 25175321_Findings reveal a novel role for BAG3 as host protein contributing to HPV18 E6-activated pro-survival strategies. 25204229_BAG3 induced autophagy is Beclin-1 independent 25208129_BAG3 mutations should be considered even in cases with a mild phenotype or an adult onset 25273835_findings showed that the P209L mutation causes BAG3 to aggregate; proposed that the gradual loss of available BAG3(wt) and BAG3(P209L) proteins results in insufficiency leading to myofibrillar disintegration 25347739_Studies indicate that heat shock protein Hsp70 (Hsp70) regulates multiple pathways in cancer cells via interaction with Bcl2-associated athanogene 3 protein (BAG3) co-chaperone. 25361773_Knockdown of BAG3 also significantly suppressed motion and invasion of HK2 cells mediated by FGF-2 25412315_BIS targeting induces cellular senescence through the regulation of 14-3-3 zeta/STAT3/SKP2/p27 in glioblastoma cells. 25448463_BAG3 carries highly penetrant DCM mutations that are associated with a worse prognosis characterized by earlier age of onset. 25483098_HIV-1 Tat protein is able to stimulate autophagy through increasing BAG3 levels in glial cells. 25564440_The Hsp70-Bag3 interaction may be a promising, new target for anticancer therapy. 25738313_To examine the relationships among BAG3, miR-29b and MMP2 in endometrioid adenocarcinoma cells. 25753466_The stress response protein BAG3 may be prognostic for death in patients with acutely decompensated heart failure. 25766323_Upon glucose stimulation BAG3 is phosphorylated by FAK and dissociates from SNAP-25 allowing the formation of the SNARE complex, destabilization of the F-actin network and insulin release. 25766331_by demonstrating a critical role of NIK in mediating NF-kappaB activation and BAG3 induction upon ST80/Bortezomib cotreatment, our study provides novel insights into mechanisms of resistance to proteotoxic stress in RMS 25889438_BAG3 protein loci is involved in the pathophysiology of systolic heart failure. 25968616_Bis expression was higher in squamous cell carcinoma than in adenocarcinoma in Lung Cancer. 26100943_BAG3 is a suitable target for combined therapies aimed at synergistically inducing apoptosis in bladder cancer. 26158518_This newly described ERa-mediated and estrogen response element (ERE)-independent non-canonical autophagy pathway, which involves the function of BAG3 and provides stress resistance in our model systems. 26159920_BAG3 plays a key role in the processing of the nucleocytoplasmic shuttling of HSF1 upon heat stress 26337083_Data show although no any significant differences between patient groups and lean subjects of proteins SYT4, BAG3, APOA1, and VAV3, except for VGF protein, there was a trend between the expression of these four genes and their protein levels. 26496431_our findings suggest the existence of a so-far unrecognized quality control mechanism involving BAG3, HSPB8 and p62/SQSTM1 for accurate remodelling of actin-based mitotic structures that guide spindle orientation. 26522614_BAG3 promotes pancreatic ductal adenocarcinoma growth by activating stromal macrophages. 26526841_modulation of proteostasis is a distinct biological function of sAPPalpha and does not require surface-bound holo-APP. 26541455_MiR-143 enhanced the tumor suppressive effect of shikonin partly through the regulation of BAG3 in glioblastoma stem cells. 26545904_BAG3 is associated with Z-disc maintenance. 26577854_in the present study, we demonstrated that BAG3 overexpression plays a critical role in cell proliferation, migration, and invasion of colorectal cancer. Our data suggests targeted inhibition of BAG3 may be useful for patients with colorectal cancer 26621836_Data indicate a tumor suppressor-like function of Bcl-2 associated athanogene 3 (BAG3) via direct interaction with glucose 6 phosphate dehydrogenase (G6PD) in hepatocellular carcinomas (HCCs) at the cellular level. 26654586_BAG3 maintains the basal amount of LC3B protein by controlling the translation of its mRNA in HeLa and HEK293 cells. 26655271_BAG3-mediated miRNA let-7g and let-7i inhibit proliferation and enhance apoptosis of human esophageal carcinoma cells by targeting the drug transporter ABCC10 and modulates cisplatin resistance. 26707573_The present study, for the first time, examined the expression of central autophagy proteins BAG3 and p62 in testicular cancer 26969713_This study therefore identifies both BAG3 reduction and autophagy promotion as potential therapies for FLNC(W2710X) myofibrillar myopathy, and identifies protein insufficiency due to sequestration, compounded by impaired autophagy, as the cause. 27120977_BAG3 was found to positively regulate Mcl-1 levels by binding to and inhibiting USP9X. Our data show that BAG3 and Mcl-1 are key mediators of resistance to chemotherapy in ovarian cancer 27145367_Our findings suggest that high levels of BIS expression might confer stem-cell-like properties on cancer cells through STAT3 stabilization 27245201_These results provide further insight into the molecular mechanisms involved in the enhancement of hyperthermia sensitivity by the silencing of BAG3 in human oral squamous cell carcinoma cells. 27321750_we report the first mammalian model of a single amino acid mutation of BAG3 (P209L) in exon 3 of Bag3 associated with the development of muscle disease with left ventricular dysfunction and heart failure 27391596_Study report p.H243Tfr*64_BAG3 as a novel pathogenic variation responsible for familial dilated cardiomyopathy. This variation correlates with a more severe phenotype of the disease, mainly in younger individuals. 27414463_These results indicate a possible role for BAG3 protein in the maintenance of cell survival in endometrioid endometrial cancer. 27456361_high expression of BAG3 was detected in a majority of medulloblastoma tissues and predicted poor outcome for medulloblastoma patients. 27474739_These results suggest that Bag1 and Bag3 control the stability of the Hsc70-client complex using at least two distinct protein-protein contacts, providing a previously under-appreciated layer of molecular regulation in the human Hsc70 system. 27570075_It has been demonstrated that HSPB8-BAG3-HSP70 ensures the functionality of stress granules and restores proteostasis by targeting defective ribosomal products for degradation. 27736720_Familial suffering of Dilated cardiomyopathy and carrying a heterozygous large deletion in the BAG3 gene.This gene encodes BCL2-associated athanogene 3 protein. 27756573_The spatial regulation of mTORC1 exerted by BAG3 apparently provides the basis for a simultaneous induction of autophagy and protein synthesis to maintain the proteome under mechanical strain. 27884606_BAG3 bound to Hsp70 at the same time as Hsp22, Hsp27, or alphaB-crystallin, suggesting that it might physically bring the chaperone families together into a complex. 27922674_By showing transcription factor HSF1 activation, we demonstrated that HCA induces the expression of BAG3 through HSF1 activation. More importantly, knockdown of BAG3 expression using siRNA largely inhibited HCA-induced apoptosis, suggesting that BAG3 is actively involved in HCA-induced cancer cell death 28076420_The authors propose that the chaperone-mediated autophagy function of BAG3 represents a specific host defense strategy to counteract the function of VP40 in promoting efficient egress and spread of virus particles. 28181153_HSPB2 competes with HSPB8 for binding to BAG3. In contrast, HSPB3 negatively regulates HSPB2 association with BAG3. 28211974_BAG3 mutations are associated with DCM phenotypes. BAG3 should be added to cardiomyopathy gene panels for screening of DCM patients, and patients previously considered gene elusive should undergo sequencing of the BAG3 gene. 28224639_This case indicates that rigid spine syndrome and sensory-motor axonal neuropathy are key clinical features of BAG3 mutations that should be considered even without cardiac involvement. 28275944_Silencing of HSPB8 markedly decreased the mitotic levels of BAG3 in HeLa cells, supporting its crucial role in BAG3 mitotic functions. The results support a role for the HSPB8-BAG3 chaperone complex in quality control of actin-based structure dynamics that are put under high tension, notably during cell cytokinesis. 28373462_High BAG3 expression Correlates with Sebaceous Gland Carcinoma of the Eyelid. 28624440_These results indicated that at least some oncogenic functions of BAG3 were mediated through posttranscriptional regulation of Skp2 via antagonizing suppressive action of miR-21-5p in ovarian cancer cells. 28669108_variants in TNNT2 and BAG3 are associated with a high propensity to life-threatening cardiomyopathy presenting from childhood and young adulthood. 28696030_higher levels of BAG3 were observed in hypertensive patients compared to healthy controls, and even higher levels in hypertensive diabetic patients compared to healthy subjects. 28703799_BAG3 interacted with CXCR4 mRNA and promoted its expression via its coding and 3'-untranslational regions. 28754666_BAG3 mutation causes adult onset Charcot-Marie-Tooth disease, type 2. 28864347_BAG3 can modulate the levels, localization or activity of its partner proteins, thereby regulating major cell pathways and functions, including apoptosis, autophagy, mechanotransduction, cytoskeleton organisation, motility. 29068469_The HSF1-BAG3-Mcl-1 signal axis is critical for protection of mutant KRAS colon cancer cells from AUY922-induced apoptosis. 29114069_BAG3 directly stabilizes hexokinase 2 mRNA and promotes aerobic glycolysis in pancreatic cancer cells. 29295729_High BAG3 expression is associated with Cervical Cancer Cell Proliferation. 29405094_BAG3 interaction with HSPB8 promotes spatial sequestration of ubiquitinated proteins. BAG3 mutation P209L in the HSPB8-binding motif deregulated association between BAG3 and SQSTM1 and the KEAP1-Nrf2 signaling axis. 29462756_Genetic and pharmacological interference with BAG3 is capable to resensitize TNBC cells to treatment. 29484408_study revealed oncogenic roles of BAG3 in chondrosarcoma and provided mechanisms that the BAG3-modulated the expression of RUNX2 through upregulation of beta-catenin. 29514624_There was no association of SNPs in ADRB1, GRK5 and BAG3 genes with Takotsubo cardiomyopathy. 29962275_HIV-1 Tat induces autophagy by upregulating BAG3 via NF-kappaB signaling. 29986884_BAG3 silencing-mediated correction of F508del-CFTR restores the autophagy pathway, which is defective in F508del-CFTR-expressing cells, likely because of the maladaptive stress response in cystic fibrosis pathophysiology. 29987014_The Hsp70-Bag3 complex therefore functions as an important signaling node that senses proteotoxicity and triggers multiple pathways that control cell physiology, including activation of protein aggregation. 30081850_BAG3 overexpression in colorectal cancer can promote tumor proliferation, migration and invasion. 30106105_These findings provide a molecular basis for understanding of BAG3dependent cell proliferation and survival from the aspect of alteration of gene expression. 30140897_Results show that unique variants in BAG3 found almost exclusively in individuals of African descent were not associated with the onset of dilated cardiomyopathy (DCM) but were associated with a negative influence on the phenotypic response to the development of DCM and worse outcomes of patients with both ischemic and non-ischemic disease. 30145633_this is the first Korean patient with BAG3 mutation. We described a BAG3 mutation patient with atypical phenotype of CMT and myopathy, and those are expected to broaden the clinical spectrum of the disease and help to diagnose it. 30442290_Dilated cardiomyopathy caused by mutations in BAG3 is characterized by high penetrance in carriers > 40years of age and a high risk of progressive heart failure. 30514177_BAG3 may have an important role in HGF-mediated cell proliferation and metastasis in gastric cancer through an ERK and Egr1-dependent pathway. 30559338_BAG3 in particular, and maybe nucleotide exchange factors in general, are a potential Achilles heel of the Hsp70 machinery, where minor malfunctioning results in the entrapment of the whole chaperone complex with disastrous consequences for protein homeostasis. 30631036_These results suggest a novel function for BAG3 in maintaining the level and quality of lamin B, thereby promoting the integrity of the nuclear envelope under stressful conditions. 30771383_The current study revealed that BAG3 did not interact with ISG15, but positively regulated ISG15 expression in pancreatic ductal adenocarcinoma cancer. 30792263_Data show that 56% of Bcl2-associated athanogene 3 protein (BAG3+)/ dilated cardiomyopathy (DCM+) significantly co-expressed mir-154-5p and mir-182-5p, suggesting that co-expression of mir-154-5p and mir-182-5p may potentially show diagnostic value. 30910998_BAG3 promotes autophagic activity via enhancing glutaminolysis and ammonia generation. 31119886_BAG3-positive pancreatic stellate cells promote migration and invasion of pancreatic ductal adenocarcinoma. 31471350_Genetic variations within the BIS gene in the korean patients with dilated cardiomyopathy. 31723063_Investigation of a dilated cardiomyopathy-associated variant in BAG3 using genome-edited iPSC-derived cardiomyocytes. 31801989_BAG3 regulates multiple myeloma cell proliferation through FOXM1/Rb/E2F axis. 31808029_Advances in the role and mechanism of BAG3 in dilated cardiomyopathy. 31823704_Association of GRP78, HIF-1alpha and BAG3 Expression with the Severity of Chronic Lymphocytic Leukemia. 32029272_Downregulation of BIS sensitizes A549 cells for digoxin-mediated inhibition of invasion and migration by the STAT3-dependent pathway. 32360144_The multiple activities of BAG3 protein: Mechanisms. 32472079_BAG3 Pro209 mutants associated with myopathy and neuropathy relocate chaperones of the CASA-complex to aggresomes. 32578139_BAG3 interacts with p53 in endometrial carcinoma. 32696179_BAG3 and BAG6 differentially affect the dynamics of stress granules by targeting distinct subsets of defective polypeptides released from ribosomes. 32956817_Novel BAG3 Variants in African American Patients With Cardiomyopathy: Reduced beta-Adrenergic Responsiveness in Excitation-Contraction. 33030392_Metformin rescues muscle function in BAG3 myofibrillar myopathy models. 33086735_SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA. 33158300_BAG3 Proteomic Signature under Proteostasis Stress. 33989081_BAG3 expression and sarcomere localization in the human heart are linked to HSF-1 and are differentially affected by sex and disease. 34011988_Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. 34099896_An emerging role for BAG3 in gynaecological malignancies. 34111434_BAG3 epigenetically regulates GALNT10 expression via WDR5 and facilitates the stem cell-like properties of platin-resistant ovarian cancer cells. 34117258_Overexpression of human BAG3(P209L) in mice causes restrictive cardiomyopathy. 34180073_BAG3 is a negative regulator of ciliogenesis in glioblastoma and triple-negative breast cancer cells. 34685619_CDK1-Mediated Phosphorylation of BAG3 Promotes Mitotic Cell Shape Remodeling and the Molecular Assembly of Mitotic p62 Bodies. 34741483_BAG3 induces alpha-SMA expression in human fibroblasts and its over-expression correlates with poorer survival in fibrotic cancer patients. 34783111_Lutein induces an inhibitory effect on the malignant progression of pancreatic adenocarcinoma by targeting BAG3/cholesterol homeostasis. 34951719_PKM2 compensates for proteasome dysfunction by mediating the formation of the CHIP-HSP70-BAG3 complex and the aggregation of ubiquitinated proteins. 35008592_Neurodegeneration and Astrogliosis in the Human CA1 Hippocampal Subfield Are Related to hsp90ab1 and bag3 in Alzheimer's Disease. | ENSMUSG00000030847 | Bag3 | 1.997292e+03 | 1.3380460 | 0.420127711 | 0.3034095 | 1.925097e+00 | 0.1652959000 | 0.77593452 | No | Yes | 2.276105e+03 | 234.859394 | 1.476943e+03 | 156.979386 | |
| ENSG00000152127 | 4249 | MGAT5 | protein_coding | Q09328 | FUNCTION: Catalyzes the addition of N-acetylglucosamine (GlcNAc) in beta 1-6 linkage to the alpha-linked mannose of biantennary N-linked oligosaccharides (PubMed:10395745, PubMed:30140003). Catalyzes an important step in the biosynthesis of branched, complex-type N-glycans, such as those found on EGFR, TGFR (TGF-beta receptor) and CDH2 (PubMed:10395745, PubMed:22614033, PubMed:30140003). Via its role in the biosynthesis of complex N-glycans, plays an important role in the activation of cellular signaling pathways, reorganization of the actin cytoskeleton, cell-cell adhesion and cell migration. MGAT5-dependent EGFR N-glycosylation enhances the interaction between EGFR and LGALS3 and thereby prevents rapid EGFR endocytosis and prolongs EGFR signaling. Required for efficient interaction between TGFB1 and its receptor. Enhances activation of intracellular signaling pathways by several types of growth factors, including FGF2, PDGF, IGF, TGFB1 and EGF. MGAT5-dependent CDH2 N-glycosylation inhibits CDH2-mediated homotypic cell-cell adhesion and contributes to the regulation of downstream signaling pathways. Promotes cell migration. Contributes to the regulation of the inflammatory response. MGAT5-dependent TCR N-glycosylation enhances the interaction between TCR and LGALS3, limits agonist-induced TCR clustering, and thereby dampens TCR-mediated responses to antigens. Required for normal leukocyte evasation and accumulation at sites of inflammation (By similarity). Inhibits attachment of monocytes to the vascular endothelium and subsequent monocyte diapedesis (PubMed:22614033). {ECO:0000250|UniProtKB:Q8R4G6, ECO:0000269|PubMed:10395745, ECO:0000269|PubMed:22614033, ECO:0000269|PubMed:30140003}.; FUNCTION: [Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A]: Promotes proliferation of umbilical vein endothelial cells and angiogenesis, at least in part by promoting the release of the growth factor FGF2 from the extracellular matrix. {ECO:0000269|PubMed:11872751}. | 3D-structure;Direct protein sequencing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Membrane;Reference proteome;Secreted;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:10395745, ECO:0000269|PubMed:17142794, ECO:0000269|PubMed:30140003}. | The protein encoded by this gene belongs to the glycosyltransferase family. It catalyzes the addition of beta-1,6-N-acetylglucosamine to the alpha-linked mannose of biantennary N-linked oligosaccharides present on the newly synthesized glycoproteins. It is one of the most important enzymes involved in the regulation of the biosynthesis of glycoprotein oligosaccharides. Alterations of the oligosaccharides on cell surface glycoproteins cause significant changes in the adhesive or migratory behavior of a cell. Increase in the activity of this enzyme has been correlated with the progression of invasive malignancies. [provided by RefSeq, Oct 2011]. | hsa:4249; | extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity [GO:0030144]; manganese ion binding [GO:0030145]; protein phosphatase inhibitor activity [GO:0004864]; negative regulation of protein tyrosine phosphatase activity [GO:1903614]; positive regulation of cell migration [GO:0030335]; positive regulation of receptor signaling pathway via STAT [GO:1904894]; protein N-linked glycosylation [GO:0006487]; protein N-linked glycosylation via asparagine [GO:0018279]; viral protein processing [GO:0019082] | 11864986_Prometastatic effect of N-acetylglucosaminyltransferase V is due to modification and stabilization of active matriptase by adding beta 1-6 GlcNAc branching 11872751_A secreted type of beta 1,6-N-acetylglucosaminyltransferase V (GnT-V) induces tumor angiogenesis without mediation of glycosylation: a novel function of GnT-V distinct from the original glycosyltransferase activity. 15014031_Low GnT-V expression is associated with shorter survival and poor prognosis in pStage I overall Non-small cell lung cancer. 15044007_Data describe the effect of N-acetylglucosaminyltransferase V on the expressions of other glycosyltransferases involved in the synthesis of surface sialyl Lewis X antigen. 15313475_GlcNAc-transferase-V activity is up-regulated in RA and Vit-D(3)-treated hepatoma cell lines. 15809094_GnT-V would contribute to placentation in the early phase of pregnancy, possibly regulating the process of invasion of trophoblast cells 16467879_These results supported the mechanism that blocking of GnT-V expression impaired functions of chaperones and N-glycan-synthesizing enzymes, which caused UPR in vivo. 16638859_GnT-V is closely related to low malignant potential and good prognosis of the patients with bladder cancer. 16924681_Glycosylation caused by GnT-V directs integrin beta1 stability and more delivery to plasma membrane, subsequently promotes Fn-based cell migration and invasion. 17451637_glycosylation change & decrease of transport activity of GLUT1 may be 1 possible mechanism of ER stress induced by down-regulating GnT-V, & GnT-V may contribute to the regulation of glucose uptake by modifying glycosylation of GLUT1 in some tumor cells. 17488527_We investigated mRNA levels of glycosyltransferases, namely GnT-V, and found that it's expression was decreased in HLE-cells resistant to Epirubicin as well as in HLE-cells resistant to Mitoxantrone. 17878270_Tissue inhibitor of metalloproteinase-1 (TIMP-1), was identified as a target protein for GnT-V in human colon cancer cell WiDr. 17971775_Results suggested that high GnT-V expression was correlated with an unfavourable clinical outcome. 18649738_Knock-down of MGAT5 in PC-3 cells attenuated the metastatic ability of prostate cancer cells, as determined by the in vitro invasion assay and the xenograft animal studies. 18845630_GnT-V is expressed in human extravillous trophoblast and is involved in regulating trophoblast invasion through modifications of the oligosaccharide chains of alpha5beta1 integrin. 18931531_GnT-V expression is correlated with a poor prognosis in gastric cancer patients due to metastases. 19225046_decreased GnT-Va activity due to siRNA expression in human carcinoma cells inhibits ligand-induced EGFR internalization, consequently resulting in delayed downstream signal transduction and inhibition of the EGF-induced, invasiveness-related phenotypes. 19236842_overexpression of GnT-V in a hepatoma cell line not only induced the addition of beta1,6 GlcNAc branch to N-glycan of RPTPkappa but also decreased the protein level of RPTPkappa 19403558_MGAT3 and MGAT5 competitively modified E-cadherin N-glycans. 19787216_High expression of N-acetylglucosaminyltransferase V is associated with mucinous tumors of the ovary. 19846580_The GnT-V was fully active without exogenous cation and in the presence of EDTA, and pH optimum for GnT-V was in the range of 6.5-7.0. 19910543_Observational study of gene-disease association. (HuGE Navigator) 19911372_Our results suggest that GnT-V could decrease human hepatoma SMMC-7721 cell adhesion and promote cell proliferation partially through RPTPkappa. 20089585_the modulation of glycosyltransferases MGAT3 and MGAT5 by synthetic N-acetyl-D-glucosamine-calix[4]arene correlated with the improvement of NK cell effector functions and the augmentation of tumor cells sensitivity to NK cell-mediated cytotoxicity. 20117844_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20468071_Observational study of gene-disease association. (HuGE Navigator) 20584650_Inhibiting expression of N-acetylglucosaminyltransferase V inhibits the proliferation of PC-3 cells. 21115203_The rs1257169(G) allele of MGAT5 is associated with lower disease severity in multiple sclerosis 21631992_Findings suggest Mgat5 may play an important role during oncogenesis, identifying a potential therapeutic target for pulmonary adenocarcinoma. 21676538_GnT-V expression is positively correlated with malignancy in nasopharyngeal carcinoma cells 21908519_33 directly measured and 13 derived glycosylation traits in 3533 individuals were identified and three novel gene association (MGAT5, B3GAT1 and SLC9A9) were identified using an additional European cohort. 22537550_These data suggested that GnT-V expression was positively related with malignancy in human hepatocellular carcinoma (HCC) and GnT-V may be both a differentiation marker and a potential target for the treatment of HCC. 22614033_decreased GnT-V activity due to inflammatory cytokine induction in human monocytes resulted in enchancement of integrin alpha5beta1-dependent monocyte-vascular endothelium adhesion and transmigration 22780953_The results suggest that GnT-V may be a potential target for predicting nasopharyngeal carcinoma response to radiotherapy. 23005037_Data show that knockdown of CD147 inhibited MMP-2 activity of GnT-V-overexpressing cells, indicating that aberrant beta1,6-branches on CD147 is crucial for the induction of MMPs (matrix metalloproteinases) in SMMC-7721 cells. 23107376_The combination of intratumoral MGAT5 expression and TNM or Kiel staging systems had a better predictive power for overall survival. 23351704_The results of this study have shown that the MGAT5 intronic variants rs4953911 and rs3814022 correlate with lower N-glycan branching, reduced surface CTLA-4 in human CD4 + T cell blasts, and associate with multiple sclerosis. 23357422_GnT-V directs cancer progression by modulating MMPs in cancer. 23563846_GnT-V plays a significant role in metastasis and invasion in gastric cancer cells. 23671930_GnT-III determines E-cadherin-mediated tumor suppression, and GnT-V regulates E-cadherin-mediated tumor invasion. 23811795_The results identified Mgat5-mediated beta-1-6-GlcNAc branched N-glycosylation and following activation of EGFR as a potential novel upstream molecular event for PAK1-induced anoikis resistance in hepatoma cells. 24334766_Study disclose that a deficiency in branched N-glycosylation on TCR due to a reduced MGAT5 gene expression is a new molecular mechanism underlying UC pathogenesis. 24399258_These results contribute to new insight into the underlying molecular mechanisms of GnT-V regulation in gastric cancer with potential translational clinical applications. 24726881_functional characterization of N-acetylglucosaminyltransferases III and V in human melanoma cells 25395405_Human Mgat5 increases amino acid uptake, intracellular levels of glycolytic and TCA intermediates, as well as HEK293 cell growth. 25876794_High expression of GnT-V was observed in infiltrating cells in skin section samples from systemic and localized patients with scleroderma. 25944901_UDP-galactose (SLC35A2) and UDP-N-acetylglucosamine (SLC35A3) Transporters Form Glycosylation-related Complexes with Mannoside Acetylglucosaminyltransferases (Mgats). 26098720_MGAT5 protein and gene expression in in uveal and cutaneous melanoma cells 26109616_Mgat5 plays an important role in early spontaneous miscarriage in humans. 26293457_Gnt-V caused tumour growth more quickly 26349781_role in the inhibition of trophoblast cell invasion and migration during early pregnancy by direct or indirect regulation of MMP2/9 activity 26526581_Tunicamycin, an inhibitor of N-glycan biosynthesis, was also able to enhance the radiosensitivity of U251 cells. Thus, our results suggest that development of therapeutic approaches targeting N-linked beta1,6-GlcNAc branches which are encoded by N-acetylglucosaminyltransferase V may be a promising strategy in glioblastoma treatment 26531171_the knockdown of GnTV significantly suppressed the proliferation, migration and invasion (P<0.05) of the SMMC7721/R cells. 26583147_the level of TGFBR1 and early osteogenic differentiation were abolished in the DPSCs transfected with siRNA for GnT-V knockdown...GnT-V plays a critical role in the hexosamine-induced activation of TGF-b signaling and osteogenic differentiation 26760037_binding of recombinant Gal-3 to the RPE cell surface and inhibitory effects on RPE attachment and spreading largely dependent on interaction with Mgat5 modified N-glycans 26821880_The best homology model is consistent with available experimental data. The three-dimensional model, the structure of the enzyme catalytic site and binding information obtained for the donor and acceptor can be useful in studies of the catalytic mechanism and design of inhibitors of GnT-V. 27334383_Our data suggest that oxidative stress induces the overexpression of MGAT5 via the regulation of the focal adhesion kinase-extracellular signal-regulated kinase signaling pathway, which, in turn, affects the function of endothelial cells, which then participates in the pathogenesis of preeclampsia. 27965091_PTPalpha is identified as a novel substrate of N-Acetylglucosaminyltransferase V (GnT-V) and could be a factor regulating promotion of migration in breast cancer cells 30140003_The acceptor-GnT-V complex structure suggests a catalytic mechanism, explains the previously observed inhibition of GnT-V by branching enzyme GnT-III, and provides a basis for the rational design of drugs targeting N-glycan branching. 30143259_GnT-V enhances gemcitabine chemosensitivity via modulation of human ENT1 N-glycosylation and transport activity in T24cells, providing new insights into how N-glycosylation drives antitumor drug sensitivity during chemotherapy for patients with cancer. 30274773_Results suggest increased post-translational modification of RPTPmu N-glycans by GnT-V attenuates its tyrosine phosphatase activity and promotes glioma cell migration through PLCgamma-PKC pathways. 32352685_Genetic Variants of the MGAT5 Gene Are Functionally Implicated in the Modulation of T Cells Glycosylation and Plasma IgG Glycome Composition in Ulcerative Colitis. 32890705_Recognition of glycan and protein substrates by N-acetylglucosaminyltransferase-V. 32945502_Nglycosylation and receptor tyrosine kinase signaling affect claudin3 levels in colorectal cancer cells. 33879098_The polymorphisms of FGFR2 and MGAT5 affect the susceptibility to COPD in the Chinese people. 33894774_Glioma stem cells invasive phenotype at optimal stiffness is driven by MGAT5 dependent mechanosensing. 35104505_N-acetylglucosaminyltransferase-V requires a specific noncatalytic luminal domain for its activity toward glycoprotein substrates. | ENSMUSG00000036155 | Mgat5 | 3.666598e+03 | 1.0172395 | 0.024659331 | 0.2956790 | 6.970995e-03 | 0.9334599541 | 0.98773389 | No | Yes | 3.031035e+03 | 261.540756 | 3.089487e+03 | 273.260590 |
| ENSG00000152253 | 57405 | SPC25 | protein_coding | Q9HBM1 | FUNCTION: Acts as a component of the essential kinetochore-associated NDC80 complex, which is required for chromosome segregation and spindle checkpoint activity (PubMed:14699129, PubMed:14738735). Required for kinetochore integrity and the organization of stable microtubule binding sites in the outer plate of the kinetochore (PubMed:14738735, PubMed:14699129). The NDC80 complex synergistically enhances the affinity of the SKA1 complex for microtubules and may allow the NDC80 complex to track depolymerizing microtubules (PubMed:23085020). {ECO:0000269|PubMed:14699129, ECO:0000269|PubMed:14738735, ECO:0000269|PubMed:23085020}. | 3D-structure;Cell cycle;Cell division;Centromere;Chromosome;Coiled coil;Kinetochore;Mitosis;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a protein that may be involved in kinetochore-microtubule interaction and spindle checkpoint activity. [provided by RefSeq, Jul 2008]. | hsa:57405; | cytosol [GO:0005829]; kinetochore [GO:0000776]; Ndc80 complex [GO:0031262]; nucleus [GO:0005634]; attachment of spindle microtubules to kinetochore [GO:0008608]; cell division [GO:0051301]; chromosome segregation [GO:0007059]; mitotic spindle organization [GO:0007052] | 14699129_hSPC25 is an essential kinetochore component that plays a significant role in proper execution of mitotic events 15961401_the Spc24, Spc25, Nuf2, and Ndc80/Hec1 complex is a faithful copy of the endogenous Ndc80 complex 27197203_SPC25 overexpression is associated with Breast Cancer. 29432994_SPC25 overexpression significantly increased the cancer stem cell properties and invasion of A549 cells. 29709477_SPC25 expressed by extracellular matrix stiffening is required for lung cancer cell proliferation. 30408771_These data suggest that SPC25 may be highly expressed in the CSC-like cells in PrC and could be a promising target for effective treatment of PrC. 31400751_These findings suggest that SPC25 levels are higher in more malignant breast cancer subtypes and are associated with poor prognosis in breast cancer patients 32351050_SPC25 may promote proliferation and metastasis of hepatocellular carcinoma via p53. 33408271_SPC25 overexpression promotes tumor proliferation and is prognostic of poor survival in hepatocellular carcinoma. 35293598_SPC25 promotes hepatocellular carcinoma metastasis via activating the FAK/PI3K/AKT signaling pathway through ITGB4. | ENSMUSG00000005233 | Spc25 | 4.069835e+02 | 1.0592895 | 0.083096940 | 0.3461606 | 5.671456e-02 | 0.8117661550 | 0.96274656 | No | Yes | 4.505737e+02 | 84.700387 | 3.638477e+02 | 70.280082 | |
| ENSG00000152404 | 143884 | CWF19L2 | protein_coding | Q2TBE0 | 3D-structure;Alternative splicing;Coiled coil;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:143884; | post-mRNA release spliceosomal complex [GO:0071014]; mRNA splicing, via spliceosome [GO:0000398] | 18398821_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000025898 | Cwf19l2 | 2.171437e+02 | 0.7542014 | -0.406978198 | 0.3645546 | 1.210214e+00 | 0.2712896984 | 0.79972971 | No | Yes | 1.643234e+02 | 28.503597 | 2.326485e+02 | 40.927510 | |||
| ENSG00000152495 | 814 | CAMK4 | protein_coding | Q16566 | FUNCTION: Calcium/calmodulin-dependent protein kinase that operates in the calcium-triggered CaMKK-CaMK4 signaling cascade and regulates, mainly by phosphorylation, the activity of several transcription activators, such as CREB1, MEF2D, JUN and RORA, which play pivotal roles in immune response, inflammation, and memory consolidation. In the thymus, regulates the CD4(+)/CD8(+) double positive thymocytes selection threshold during T-cell ontogeny. In CD4 memory T-cells, is required to link T-cell antigen receptor (TCR) signaling to the production of IL2, IFNG and IL4 (through the regulation of CREB and MEF2). Regulates the differentiation and survival phases of osteoclasts and dendritic cells (DCs). Mediates DCs survival by linking TLR4 and the regulation of temporal expression of BCL2. Phosphorylates the transcription activator CREB1 on 'Ser-133' in hippocampal neuron nuclei and contribute to memory consolidation and long term potentiation (LTP) in the hippocampus. Can activate the MAP kinases MAPK1/ERK2, MAPK8/JNK1 and MAPK14/p38 and stimulate transcription through the phosphorylation of ELK1 and ATF2. Can also phosphorylate in vitro CREBBP, PRM2, MEF2A and STMN1/OP18. {ECO:0000269|PubMed:10617605, ECO:0000269|PubMed:17909078, ECO:0000269|PubMed:18829949, ECO:0000269|PubMed:7961813, ECO:0000269|PubMed:8065343, ECO:0000269|PubMed:8855261, ECO:0000269|PubMed:8980227, ECO:0000269|PubMed:9154845}. | 3D-structure;ATP-binding;Adaptive immunity;Calcium;Calmodulin-binding;Cytoplasm;Glycoprotein;Immunity;Inflammatory response;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The product of this gene belongs to the serine/threonine protein kinase family, and to the Ca(2+)/calmodulin-dependent protein kinase subfamily. This enzyme is a multifunctional serine/threonine protein kinase with limited tissue distribution, that has been implicated in transcriptional regulation in lymphocytes, neurons and male germ cells. [provided by RefSeq, Jul 2008]. | hsa:814; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; fibrillar center [GO:0001650]; glutamatergic synapse [GO:0098978]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; postsynapse [GO:0098794]; ATP binding [GO:0005524]; calcium-dependent protein serine/threonine kinase activity [GO:0009931]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; adaptive immune response [GO:0002250]; inflammatory response [GO:0006954]; intracellular signal transduction [GO:0035556]; long-term memory [GO:0007616]; myeloid dendritic cell differentiation [GO:0043011]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of transcription, DNA-templated [GO:0045893]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of osteoclast differentiation [GO:0045670]; regulation of T cell differentiation in thymus [GO:0033081]; signal transduction [GO:0007165] | 12065094_CaMKIV proteins were found in the nucleus of epithelial ovarian cancer tissue. CaMKIV expression was significantly associated with clinical stage (P<0.01), histological grade (P<0.01), and clinical outcome (P<0.01). 14701808_sequestration of CaMKK may be the molecular mechanism by which catalytically inactive mutants of CaMKIV exert their 'dominant-negative' functions within the cell 15143065_the Ca(2+)/CaM binding-autoinhibitory domain of CaMKIV is required for association of the kinase with PP2A 15591024_calcium/CaMKIV signaling pathway may play an important role in the excitation-mediated regulation of corticotropin releasing hormone synthesis 15665723_the function of CaMK II is essential for PAF-induced macrophage priming, while CaMK IV is not specific for priming by PAF and appears to have a direct link in TLR4-mediated events 15840651_CaMKIV is expressed in human sperm and may have a role in the regulation of human sperm motility 15841182_Results identify calcium/calmodulin-dependent kinase IV as being responsible for the increased expression of CREM and the decreased production of interleukin-2 in systemic lupus erythematosus T cells. 17909078_a novel link between TLR4 and a calcium-dependent signaling cascade comprising CaMKIV-CREB-Bcl-2 that is essential for DC survival. 18053176_Transgenic CaMKIV plays a modulatory role in the nucleus accumbens in anxiety-like behavior of adult CaMKIV variant mice. 18606955_Observational study of gene-disease association. (HuGE Navigator) 18660489_Observational study of gene-disease association. (HuGE Navigator) 18829949_CaMK-4 expression correlates positively with the ability to form long-term memory and implicates the decline of CaMKIV signaling mechanisms in age-related memory deficits. 19001277_CaMKIV plays a critical role in the development and persistence of cocaine-induced behaviors, through mechanisms dissociated from acute effects on gene expression and CREB-dependent transcription. 19017650_hnRNP L is an essential component of CaMKIV-regulated alternative splicing through CA repeats, with its phosphorylation likely playing a critical role. 19386606_a group of RNA elements are responsive to PKA and CaMKIV from in vivo selection 19436069_CaMKIV is a molecular link between Group I mGluRs and fragile X mental retardation protein in anterior cingulate cortex neurons 19506079_analysis of regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification 19538941_these data indicate that the B subunits alpha and delta are essential for the interaction of PP2A with CaMKIV. 19633294_Data show that RA-induced repression of the CaMKIV signaling pathway may represent an early event in retinoid-dependent neuronal differentiation. 20171262_These findings suggest that PLC/CAMK IV-NF-kappaB is involved in RAGE mediated signaling pathway in human endothelial cells. 20378615_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 21514275_The regulation of RORalpha activity by PKA as well as CaMK-IV provides a new link in the signalling network that regulates metabolic processes such as glycogen and lipid metabolism. 21612516_Prolongevity genes are activated by CAMKIV, the levels of which are influenced by rs10491334, a single-nucleotide polymorphism associated with human longevity. 22897820_study suggests that the mutations in CAMK4 may lead to abnormal semen parameters 23049845_CaMK4 regulates beta-cell proliferation and apoptosis in a CREB-dependent manner and CaMK4-induced IRS-2 expression is important in these processes 23103515_Phosphorylated Notch1-IC by CaMKIV increases Notch1-IC stability, which enhances osteoclast differentiation. 24442360_An imbalance of specific isoforms of CYFIP1, an FMRP interaction partner, and CAMK4, a transcriptional regulator of the FMRP gene, modulates risk for autism spectrum disorders. 24667640_CaMK4-dependent activation of AKT/mTOR and CREM-alpha underlies autoimmunity-associated Th17 imbalance. 25446257_Expression of CaMKIV inhibits autophosphorylation and activation of CaMKII, and elicits G0/G1cell cycle arrest,impairing cell proliferation. 26909912_The T-allele of rs10491334 in CAMK4 was associated with hypertension in the Uygur group. 27032767_Within the pH range 5.0-11.5, CAMK4 maintained both its secondary and tertiary structures, along with its function, whereas significant aggregation was observed at acidic pH (2.0-4.5). 27298345_hTau accumulation impairs synapse and memory by CaN-mediated suppression of nuclear CaMKIV/CREB signaling. 27659345_A positive association was not observed between rs10491334 in the CAMK4 gene and longevity in a Chinese population. 28734942_Genotype and allele frequencies of CAMKIV gene SNPs differed significantly between alcohol dependence patients and control subjects. The results of the present study suggest that CAMKIV might be a candidate alcohol dependence gene. 28744811_vanillin binds strongly to the active site cavity of CAMKIV and stabilized by a large number of non-covalent interactions. 29985166_CaMK4 is pivotal in immune and nonimmune podocyte injury and that its targeted cell-specific inhibition preserves podocyte structure and function and should have therapeutic value in lupus nephritis and podocytopathies, including focal segmental glomerulosclerosis. 30113881_Clinical disease severity directly correlates with calmodulin-dependent kinase IV (CaMKIV) activation, as does expression of proinflammatory cytokines and histologic features of colitis. In wild-type mice, CaMKIV activation is associated with increases in expression of 2 cell cycle proarrest signals: p53 and p21 30462889_CaMK4 could be responsible for glycolysis, which contributes to the production of IL-17, and CaMK4 may contribute to aberrant expression of GLUT1 in T cells from patients with active SLE. 31624237_MiR-129-5p inhibits liver cancer growth by targeting calcium calmodulin-dependent protein kinase IV (CAMK4). 31976761_rs2300782 of gene CAMK4 is associated with diabetic retinopathy incidence and severity among Chinese Hui population. 31978801_Comparative transcriptome analysis reveals a potential role for CaMK4 in gammadeltaT17 cells from systemic lupus erythematosus patients with lupus nephritis. 32460794_CAMKK2-CAMK4 signaling regulates transferrin trafficking, turnover, and iron homeostasis. 32572897_MiR-507 inhibits the growth and invasion of trophoblasts by targeting CAMK4. 32738170_CaMK4 promotes abortion-related Th17 cell imbalance by activating AKT/mTOR signaling pathway. 33784256_Aberrantly glycosylated IgG elicits pathogenic signaling in podocytes and signifies lupus nephritis. | ENSMUSG00000038128 | Camk4 | 8.007904e+01 | 0.3877946 | -1.366635361 | 0.3950316 | 1.132098e+01 | 0.0007663645 | 0.09944063 | No | Yes | 3.388735e+01 | 10.800991 | 8.454188e+01 | 27.577933 | |
| ENSG00000152795 | 9987 | HNRNPDL | protein_coding | O14979 | FUNCTION: Acts as a transcriptional regulator. Promotes transcription repression. Promotes transcription activation in differentiated myotubes (By similarity). Binds to double- and single-stranded DNA sequences. Binds to the transcription suppressor CATR sequence of the COX5B promoter (By similarity). Binds with high affinity to RNA molecules that contain AU-rich elements (AREs) found within the 3'-UTR of many proto-oncogenes and cytokine mRNAs. Binds both to nuclear and cytoplasmic poly(A) mRNAs. Binds to poly(G) and poly(A), but not to poly(U) or poly(C) RNA homopolymers. Binds to the 5'-ACUAGC-3' RNA consensus sequence. {ECO:0000250, ECO:0000269|PubMed:9538234}. | Acetylation;Activator;Alternative splicing;Cytoplasm;DNA-binding;Direct protein sequencing;Disease variant;Isopeptide bond;Limb-girdle muscular dystrophy;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has two RRM domains that bind to RNAs. Three alternatively spliced transcript variants have been described for this gene. One of the variants is probably not translated because the transcript is a candidate for nonsense-mediated mRNA decay. The protein isoforms encoded by this gene are similar to its family member HNRPD. [provided by RefSeq, May 2011]. | hsa:9987; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spliceosomal complex [GO:0005681]; DNA binding [GO:0003677]; double-stranded DNA binding [GO:0003690]; poly(A) binding [GO:0008143]; poly(G) binding [GO:0034046]; RNA binding [GO:0003723]; single-stranded DNA binding [GO:0003697]; regulation of gene expression [GO:0010468]; RNA processing [GO:0006396] | 16011250_Study shows that there is interaction of the intracellular domain of beta-amyloid precursor protein with JKTBP2, indicating that JKTBP2 may have an important function in AD formation. 17592041_The data of this study show that JKTBP1 and the 14-nt element act independently to mediate NRF internal ribosome entry segment activity. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18381662_overexpression of JKTBP1 in LNCaP cells leads to abnormal cell proliferation 21300069_The results indicate that JKTBP1 regulates the level of NRF protein expression by binding to both NRF 5' and 3' UTRs. 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 23642268_hnRNP DL and CNBP are novel antigens for SLE patients 24647604_A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G. 29263134_Both hnRNP D and DL are able to control their own expression by alternative splicing of cassette exons in their 3'UTRs. Exon inclusion produces mRNAs degraded by nonsense-mediated decay. Moreover, hnRNP D and DL control the expression of one another by the same mechanism. 30052712_Heterogeneous nuclear ribonucleoprotein D-like (HNRPDL) is aberrantly expressed in specimens from colorectal cancer patients. 30447347_High HNRNPDL expression is associated with cervical cancer. 31488872_HNRPDL transforms hematopoietic cells and a novel HNRPDL/PBX1 axis plays an important role in human CML CD34(+) cells | ENSMUSG00000029328 | Hnrnpdl | 6.504754e+03 | 1.0865184 | 0.119712541 | 0.2982049 | 1.597119e-01 | 0.6894219305 | 0.93352105 | No | Yes | 6.016481e+03 | 853.501463 | 5.469775e+03 | 795.610408 | |
| ENSG00000152969 | 152789 | JAKMIP1 | protein_coding | Q96N16 | FUNCTION: Associates with microtubules and may play a role in the microtubule-dependent transport of the GABA-B receptor. May play a role in JAK1 signaling and regulate microtubule cytoskeleton rearrangements. {ECO:0000269|PubMed:14718537, ECO:0000269|PubMed:15277531, ECO:0000269|PubMed:17532644}. | Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Membrane;Microtubule;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:152789; | cytoplasm [GO:0005737]; extrinsic component of membrane [GO:0019898]; microtubule [GO:0005874]; ribonucleoprotein complex [GO:1990904]; GABA receptor binding [GO:0050811]; kinase binding [GO:0019900]; microtubule binding [GO:0008017]; RNA binding [GO:0003723]; cognition [GO:0050890]; protein transport [GO:0015031] | 14718537_Marlin-1 functions to regulate the cellular levels of GABA(B) R2 subunits, which may have significant effects on the production of functional GABA(B) receptor heterodimers 15277531_Marlin1 has a role in cell polarization, segregation of signaling complexes, and vesicle traffic, some of which may involve Jak tyrosine kinases 17761393_We have identified four new transcripts of 2975 bp, 1743 bp, 2189 bp and 2420 bp respectively, named Jakmip1B, Jakmip1C, Jakmip1D and Jakmip1E 17804789_Observational study of gene-disease association. (HuGE Navigator) 18941173_Jakmip1 is a novel effector memory gene that restrains T cell-mediated cytotoxicity. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23481296_JAKMIP1 associates with and possibly contributes to the Wnt/beta-catenin pathway activity through its influence on downstream Wnt target proteins, including beta-catenin. | ENSMUSG00000063646 | Jakmip1 | 1.390045e+02 | 1.0401358 | 0.056771891 | 0.3488946 | 2.741210e-02 | 0.8684985129 | 0.97498746 | No | Yes | 1.226178e+02 | 16.052714 | 1.161844e+02 | 15.613003 | ||
| ENSG00000153060 | 146279 | TEKT5 | protein_coding | Q96M29 | FUNCTION: May be a structural component of the sperm flagellum. {ECO:0000250|UniProtKB:G5E8A8}. | Cell projection;Cilium;Coiled coil;Flagellum;Reference proteome | hsa:146279; | microtubule cytoskeleton [GO:0015630]; nucleus [GO:0005634]; sperm flagellum [GO:0036126]; cilium assembly [GO:0060271]; cilium movement involved in cell motility [GO:0060294] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000039179 | Tekt5 | 1.347196e+01 | 0.4030083 | -1.311118518 | 0.7801740 | 2.746068e+00 | 0.0974939303 | No | Yes | 5.182920e+00 | 3.541028 | 1.472243e+01 | 10.445064 | |||
| ENSG00000153113 | 831 | CAST | protein_coding | P20810 | FUNCTION: Specific inhibition of calpain (calcium-dependent cysteine protease). Plays a key role in postmortem tenderization of meat and have been proposed to be involved in muscle protein degradation in living tissue. | Acetylation;Alternative splicing;Isopeptide bond;Palmoplantar keratoderma;Phosphoprotein;Protease inhibitor;Reference proteome;Repeat;Thiol protease inhibitor;Ubl conjugation | The protein encoded by this gene is an endogenous calpain (calcium-dependent cysteine protease) inhibitor. It consists of an N-terminal domain L and four repetitive calpain-inhibition domains (domains 1-4), and it is involved in the proteolysis of amyloid precursor protein. The calpain/calpastatin system is involved in numerous membrane fusion events, such as neural vesicle exocytosis and platelet and red-cell aggregation. The encoded protein is also thought to affect the expression levels of genes encoding structural or regulatory proteins. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jun 2010]. | hsa:831; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; membrane [GO:0016020]; cadherin binding [GO:0045296]; calcium-dependent cysteine-type endopeptidase inhibitor activity [GO:0010859]; endopeptidase inhibitor activity [GO:0004866]; RNA binding [GO:0003723]; inhibition of cysteine-type endopeptidase activity [GO:0097340]; negative regulation of type B pancreatic cell apoptotic process [GO:2000675]; presynaptic active zone organization [GO:1990709] | 11849768_Observational study of gene-disease association. (HuGE Navigator) 11849768_involved in the proteolysis of amyloid precursor protein, which is thought to be abnormal in patients with Alzheimer's disease 12482888_Overexpression of calpastatin reduced muscle atrophy during 10 day unloading period. Overexpression completely prevented shift in myofibrillar myosin content from slow to fast isoforms, which normally occurs in muscle unloading. 14559243_insights into how the calpain/calpastatin network is spatially and temporally regulated in cells binding to the extracellular matrix 14612448_calpastatin and calpain-1 represent critical proximal elements in a cascade of pro-apoptotic events leading to Bax, mitochondria, and caspase-3 activation 14992597_Calpastatin amino acid side chains at leucine-11 and isoleucine-18 interact with hydrophobic pockets in calpain, and each of these interactions is indispensable for effective inhibition of calpain. 15014085_Overexpression of calpastatin in transgenic mice is associated with an increase in GLUT4 protein. 15950654_A complete calpain-calpastatin system is expressed in the human oocyte and may play a role in the various calcium-mediated processes occurring during activation of human oocytes. 17137217_Observational study of gene-disease association. (HuGE Navigator) 17950697_CS(L) modulates Ca2+-channel activity through interacting with the calmodulin-binding site on the C-terminal tail of the Cav1.2 channel. 18165173_The activity of calpain in human peripheral blood lymphocytes, was estimated by assessing the levels of limited proteolysis of calpastatin. 18195134_Genome-wide association study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18340456_Results suggest a regulation on the calpain-calpastatin expression response to muscle damaging eccentric exercise, but not concentric exercise. 18498295_genetically determined IL-1alpha levels may modulate transcription of calpain and calpastatin 18519038_NMR data of calapastin tripartite binding mode to capain induced by calcium were presented. 18537264_performed a full NMR assignment of hCSD1 to characterize it in its solution state 18544539_c-Myc regulates calpain activity through calpastatin; apoptosis induced by calpain inhibition is dependent on c-Myc, and calpastatin knockdown promotes transformation in c-Myc-negative cells 18724972_Observational study of gene-disease association. (HuGE Navigator) 18793761_Proof of concept that the calpastatin-based reagents may be useful to selectively detect the active conformation of calpain. 18809371_Role of the calpain-calpastatin system in the density-dependent growth arrest. 19103264_recruitment of calpastatin into aggregates allows translocation and activation of protease to the membranes; on the contrary, the presence of large amounts of calpastatin in the cytosol prevents both processes, protecting the cell from proteolysis 19365859_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19756344_Overexpression of calpain-2 and low expression of calpastatin may involve in the pathological development of stress urinary incontinence. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20127884_A confirmation study reports that a single nucleotide polymorphism and CAST are associated with Parkinson disease. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21424381_Variations in CAST gene is associated with femoral neck Low bone mineral density. 21531560_The data supports the hypothesis that calpastatin may play a role in regulating the initial metastatic dissemination of breast cancer. 21839844_intervertebral discs at early stages of degeneration expressed low levels of calpastatin, and few cells expressed degenerative enzymes. At more advanced stages of degeneration, expression and number of cells immunopositive for calpastatin were higher 21937422_CSL competes with CaM as a partial agonist for the site in the IQ domain in the C-terminal region of the Cav1.2 channel, which may be involved in activation of the channel. 21983488_calpain basal activity becomes measurable at a significant extent in peripheral blood mononuclear cells from cystic fibrosis patients due to a 40-60% decrease in both calpastatin protein and inhibitory activity 22046434_calpastatin-calpain balance varied during Th-1,2, and -17 development; overexpression of a domain of calpastatin suppressed production of IL-6 and IL-17 by Th cells and of IL-6 by fibroblasts; suppression of IL-6 occurred by reducing NF-kappaB signaling 22206846_Neuronal overexpression of calpastatin modulates amyloid precursor protein processing in brains of transgenic beta-amyloid depositing mice. 22288381_Mycoplasma hyorhinis membrane lipoproteins induce calpastatin upregulation in human cells 22572592_Calpastatin overexpression reduces calpain-mediated proteolysis and behavioral impairment after traumatic brain injury. 23140395_The results suggest that calpain-2 and calpastatin expression is important in pancreatic cancers, influencing disease progression 23449483_Linkage analysis and genetic association support involvement of CAST gene in the genetic susceptibility to keratoconus. 23707532_HER2 inhibited calpain-1 activity through upregulating calpastatin, an endogenous calpain inhibitor. 23951044_Calpastatin gene (CAST) is not associated with late onset sporadic Parkinson's disease in the Han Chinese population. 24462690_Calpastatin binds with Cav1.2 amino acid motifs in a calcium dependent manner. 25648699_Loss of CAST in angiogenic ECs facilitates mu-calpain-induced SOCS3 degradation, which amplifies pathological angiogenesis through interleukin-6/STAT3/VEGF-C axis. 25683118_We describe PLACK syndrome, as a clinical entity of defective epidermal adhesion, caused by loss-of-function mutations in calpastatin. 26974350_calpains and calpastatin in patients with idiopathic pulmonary arterial hypertension (PAH) and mice with hypoxic or spontaneous (SM22-5HTT(+) strain) PH, were investigated. 27715410_Kaposi sarcoma-associated herpesvirus reduces CAST (calpastatin) and consequently decreases ATG5 expression in both THP-1 monocytoid cells and primary monocytes. 29428799_single nucleotide polymorphisms in CAST gene confer risk for keratoconus susceptibility in Han Chinese population 29572388_Unexpected role of the L-domain of calpastatin during the autoproteolytic activation of human erythrocyte calpain. 30448882_Low CAST expression is associated with ovarian cancer. 30959065_new human calpastatin skipped of the inhibitory region protects calpain-1 from inactivation and degradation. 31392520_A new recessive form of genetic peeling skin syndrome with peeling skin, leukonychia, acral punctate keratoses, cheilitis and knuckle pads (termed PLACK syndrome) shown associated with the loss of function mutations in the CAST gene encoding calpastatin. 34191270_Variants in Genes of Calpain System as Modifiers of Spinocerebellar Ataxia Type 3 Phenotype. 34440632_Critical Roles of Calpastatin in Ischemia/Reperfusion Injury in Aged Livers. | ENSMUSG00000021585 | Cast | 9.196999e+02 | 1.3544975 | 0.437757731 | 0.3416123 | 1.642189e+00 | 0.2000253246 | 0.78383913 | No | Yes | 1.142794e+03 | 214.330194 | 7.296225e+02 | 140.582417 | |
| ENSG00000153208 | 10461 | MERTK | protein_coding | Q12866 | FUNCTION: Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to several ligands including LGALS3, TUB, TULP1 or GAS6. Regulates many physiological processes including cell survival, migration, differentiation, and phagocytosis of apoptotic cells (efferocytosis). Ligand binding at the cell surface induces autophosphorylation of MERTK on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with GRB2 or PLCG2 and induces phosphorylation of MAPK1, MAPK2, FAK/PTK2 or RAC1. MERTK signaling plays a role in various processes such as macrophage clearance of apoptotic cells, platelet aggregation, cytoskeleton reorganization and engulfment (PubMed:32640697). Functions in the retinal pigment epithelium (RPE) as a regulator of rod outer segments fragments phagocytosis. Plays also an important role in inhibition of Toll-like receptors (TLRs)-mediated innate immune response by activating STAT1, which selectively induces production of suppressors of cytokine signaling SOCS1 and SOCS3. {ECO:0000269|PubMed:17005688, ECO:0000269|PubMed:32640697}. | 3D-structure;ATP-binding;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Immunoglobulin domain;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Proto-oncogene;Receptor;Reference proteome;Repeat;Retinitis pigmentosa;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | This gene is a member of the MER/AXL/TYRO3 receptor kinase family and encodes a transmembrane protein with two fibronectin type-III domains, two Ig-like C2-type (immunoglobulin-like) domains, and one tyrosine kinase domain. Mutations in this gene have been associated with disruption of the retinal pigment epithelium (RPE) phagocytosis pathway and onset of autosomal recessive retinitis pigmentosa (RP). [provided by RefSeq, Jul 2008]. | hsa:10461; | cytoplasm [GO:0005737]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; photoreceptor outer segment [GO:0001750]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; rhabdomere [GO:0016028]; ATP binding [GO:0005524]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; cell migration [GO:0016477]; cell surface receptor signaling pathway [GO:0007166]; cell-cell signaling [GO:0007267]; natural killer cell differentiation [GO:0001779]; negative regulation of cytokine production [GO:0001818]; negative regulation of leukocyte apoptotic process [GO:2000107]; negative regulation of lymphocyte activation [GO:0051250]; nervous system development [GO:0007399]; neutrophil clearance [GO:0097350]; phagocytosis [GO:0006909]; platelet activation [GO:0030168]; positive regulation of kinase activity [GO:0033674]; positive regulation of phagocytosis [GO:0050766]; protein kinase B signaling [GO:0043491]; protein phosphorylation [GO:0006468]; retina development in camera-type eye [GO:0060041]; secretion by cell [GO:0032940]; spermatogenesis [GO:0007283]; substrate adhesion-dependent cell spreading [GO:0034446]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; vagina development [GO:0060068] | 11727200_retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively 12768229_Gas6 receptors were not upregulated in any of the allograft groups, except for the Axl receptor, which increased only in acute tubular necrosis. 15111602_The present study reports the identification of R844C, the first putative pathogenic MERTK missense mutation that results in severe retinal degeneration with childhood onset 15130911_mer, presumably through activation by its ligand Gas6, participates in regulation of platelet function in vitro and platelet-dependent thrombosis in vivo. 15733062_Axl, Sky and Mer are Gas6 receptors that enhance platelet activation and regulate thrombotic responses 16675557_Transforming Mer signals may contribute to T-cell leukemogenesis, and abnormal Mer expression may be a novel therapeutic target in pediatric acute lymhpocytic leukemia therapy. 16710167_Mutations in the MERTK gene are relatively rare in Japanese patients with autosomal recessive retinitis pigmentosa. 16837475_Observational study of gene-disease association. (HuGE Navigator) 17005688_Here we show the involvement of members of the Tyro3 receptor tyrosine kinase family-Axl, Dtk, and Mer-in cell entry of filoviruses. 17047157_cleavage results in production of a soluble Mer protein that prevented Gas6-mediated stimulation of membrane-bound Mer. Mer inhibition led to defective macrophage-mediated engulfment of apoptotic cells and decreased platelet aggregation 17626743_Mer is highly expressed on Jurkat cells, and could inhibit cell apoptosis via Bcl-2 signaling pathway. 18039660_Tyr-867 in Mer receptor tyrosine kinase allows for dissociation of multiple signaling pathways for phagocytosis of apoptotic cells and down-modulation of lipopolysaccharide-inducible NF-kappaB transcriptional activation 18174230_Observational study of gene-disease association. (HuGE Navigator) 18246816_Overexpressed Mer tyrosine kinase receptor can inhibit the migration and angiogenesis of HMEC-1 cells through VEGF-C/VEGFR-2 signal pathway. 18250462_MerTK macrophage receptor is an essential component required for serum-stimulated phagocytosis of apoptotic cells. 18587056_MERTK, a cell surface receptor that recognizes apoptotic cells, is expressed on human alveolar macrophages (AMs), and its expression is up-regulated in AMs of cigarette smokers. 18620092_Possible strategies for targeted inhibition of the TAM family in the treatment of human cancer. 18815424_IVS16+1G>T disrupts the splice donor site causing exon 16 skipping. Absence of exon 16 causes a frameshift and the introduction of a premature termination codon into exon 17 creating an altered mRNA with a seriously affected tyrosine kinase domain. 18922854_human protein S can inhibit the expression and activity of SR-A through Mer RTK in macrophages, suggesting that human protein S is a modulator for macrophage functions in uptaking of modified lipoproteins 19403518_The splice site mutation c.2189+1G>T in MERTK causes rod-cone dystrophy with a distinct macular phenotype. 19541935_there was a negative correlation between Gas6 and soluble Axl and Mer in established multiple sclerosis lesions. In addition, increased levels of soluble Axl and Mer were associated with increased levels of mature ADAM17, mature ADAM10, and Furin 19922767_data suggest that growth arrest-specific-6 (GAS6)/MER receptor tyrosine kinase axis regulates homing and survival of the E2A/PBX1-positive B-cell precursor acute lymphoblastic leukemia in the bone marrow niche 20130272_Homozygosity mapping and mutation analysis in the distant family member affected by RP revealed a homozygous mutation in MERTK, but no CEP290 mutations. 20300561_The phenotype associated with these identified MERTK mutations is of a childhood onset rod-cone dystrophy with early macular atrophy. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20538656_results strongly suggest that the nonsense mutation in MERTK, leading to premature termination of the protein, is responsible for retinitis pigmentosa phenotype in the affected individuals of the Pakistani family. 20546121_Gas6 and Tyro3, Axl and Mer (TAM) receptors have roles in human and murine platelet activation and thrombus stabilization 20591486_Observational study of genetic testing. (HuGE Navigator) 20637106_correlation of decreased protein S levels with lupus disease activity is consistent with a role for the TAM receptors in scavenging apoptotic cells and controlling inflammation 20654219_The lower activity of PKC and the higher expression of MERTK are very important for sustaining the phagocytic process of ROS by human retinal pigment epithelial cells. 20664904_Findings indicate genetic association between MERTK and TYRO3 allelic variants and carotid atherosclerosis. 20664904_Observational study of gene-disease association. (HuGE Navigator) 20801516_Observational study of genetic testing. (HuGE Navigator) 20959443_PMN-Ect interacted with the macrophages by activation of the MerTK pathway responsible for down-modulation of the proinflammatory signals generated by ZymA. 20978472_Tubby and Tulp1 are bridging molecules with their N-terminal region as MERTK-binding domain and C-terminal region as phagocytosis prey-binding domain. 21347448_MERTK gene is a novel risk gene for multiple sclerosis susceptibility 21384080_overexpressed in atherosclerotic carotid plaques 21496228_plasma concentrations of sMer and sTyro3 were significantly increased in patients with active systemic lupus erythematosis and Rheumatoid arthritis 21542987_Gene expression profile is changed by the dysfunction of Mer during retinal pigment epithelium phagocytosis.( 21677792_A novel MERTK deletion is a common founder mutation in the Faroe Islands and is responsible for a high proportion of retinitis pigmentosa cases. 21792939_This study identified galectin-3 as a new MerTK ligand by an advanced dual functional cloning strategy. 22180149_MERTK mutations lead to severe retinitis pigmentosa with discrete dot-like autofluorescent deposits at early stages, which are a hallmark of this MERTK-specific dystrophy. 22363695_The present study explores Mer behavior following prolonged exposure to Gas6. 22469987_Mer receptor tyrosine kinase promotes invasion and survival in glioblastoma multiforme. 22841784_The single nucleotide polymorphisms rs4374383 and rs9380516 were linked to the functionally related genes MERTK and TULP1, which encode factors involved in phagocytosis of apoptotic cells by macrophages. 22890323_These results indicate that Mer and Axl have complementary and overlapping roles in Non-small cell lung cancer 22942426_MerTK expression levels adapt to changing immunologic environment, being suppressed in M1 and M2a macrophages and in dendritic cells. 22997156_Oxidized of polyunsaturated fatty acids contained in the tissue close to necrotic core of human carotid plaques are strong inducers of Adam17, which in turn may cleave the extracellular domain of Mertk giving rise to sMer. 23065156_The tyrosine kinase receptor MER is activated by PROS and mediates its inhibitory effect on VEGF-A-induced EC proliferation. 23390493_MERTK signaling in the retinal pigment epithelium involves a cohort of SH2-domain proteins with the potential to regulate both cytoskeletal rearrangement and membrane movement. 23474756_data suggest a role for Mer in acute myeloid leukemogenesis and indicate that targeted inhibition of Mer may be an effective therapeutic strategy in pediatric and adult AML 23585477_MERTK expression correlates with disease progression in metastatic melanomas, primary melanomas, and nevi 23617806_MERTK has a role in regulating melanoma cell migration and survival and differentially regulates cell behavior relative to AXL 23662598_[review] Receptor tyrosine kinases Tyro-3, Axl and Mer, collectively designated as TAM, are involved in the clearance of apoptotic cells. 23835724_MerTK expression in circulating innate immune cells is increased in patients with septic shock in comparison with healthy volunteers and trauma patients and its persistent overexpression after septic shock is associated with adverse outcome. 24741600_Both mMer and sMer levels significantly increased in SLE and positively correlated with disease activity and severity. The upregulation of MerTK expression may serve as a biomarker of the disease activity and severity of SLE. 24939420_The MER receptor pathway promotes wound repair in macrophages and epithelial cell growth. 25074926_These studies demonstrate that, despite their similarity, TYRO3, AXL, and MER are likely to perform distinct functions in both immunoregulation and the recognition and removal of apoptotic cells 25074939_These data collectively identify MERTK as a significant link between cancer progression and efferocytosis, and a potentially unrealized tumor-promoting event when MERTK is overexpressed in epithelial cells. 25102945_Inhibition of the Gas6 receptor Mer or therapeutic targeting of Gas6 by warfarin is a promising strategy for the treatment of multiple myeloma. 25428221_Mer expression correlates with CNS positivity upon initial diagnosis in t(1;19)-positive pediatric acute lymphoblastic leukemia patients. 25450174_The key role of the MERTK could be demonstrated in HMDM engulfing dying cells using gene silencing as well as blocking antibodies. Similar pathways were found upregulated in living ARPE-19 engulfing anoikic ARPE-19 cells. 25479139_Patients with ACLF have increased numbers of immunoregulatory monocytes and macrophages that express MERTK and suppress the innate immune response to microbes. The number of these cells correlates with disease severity and the inflammatory response. 25624460_MERTK on DCs controls T cell activation and expansion through the competition for PROS1 interaction with MERTK in the T cells. MERTK is a potent suppressor of T cell response. 25695599_results identify Mer as a receptor uniquely capable of both tethering ACs to the macrophage surface and driving their subsequent internalization. 25762638_UNC1666 is a novel potent small molecule tyrosine kinase inhibitor that decreases oncogenic signaling and myeloblast survival by dual Mer/Flt3 inhibition. 25826078_Mer enhances malignant phenotype and pharmacological inhibition of Mer overcomes resistance of non-small cell lung cancer to EGFR-targeted agents. 25878564_Significantly increased levels of sMer, sTyro3 and sAxl may be important factors contributing to the deficit in phagocytosis ability in systemic lupus erythematosus . 25881761_The mRNA expression levels of Tyro-3, Axl were decreased in pSS patients. When considering the plasma level, increased levels of soluble Mer was observed with statistically significant difference. 26263531_Upon differentiation of these iPSC towards RPE, patient-specific RPE cells exhibited defective phagocytosis, a characteristic phenotype of MERTK deficiency observed in human patients and animal models 26316303_Utilizing an ex vivo co-cultivation approach to model key cellular and molecular events found in vivo during infarction, cardiomyocyte phagocytosis was found to be inefficient, in part due to myocyte-induced shedding of macrophage MERTK 26427420_Studies indicate that c-Mer receptor tyrosine kinase MERTK mutations cause retinal degenerations. 26427450_Data indicate that AAV2-VMD2-c-mer proto-oncogene protein (hMERTK) provided up to 6.5 months photoreceptor rescue in the RCS rat, and also had a major protective effect in Mertk-null mice. 26427488_Data show that activated AMP-activated protein kinase (AMPK) limits retinal pigment epithelial cells (RPE) phagocytic activity by abolishing retinal photoreceptor cell outer segment (POS)-induced activation of c-mer proto-oncogene tyrosine kinase (MerTK). 26596542_The rs4374383 AA genotype, associated with lower intrahepatic expression of MERTK, is protective against F2-F4 fibrosis in patients with non-alcoholic fatty liver disease (NAFLD). 26768676_STK 11 testing can confirm those at risk of Peutz-Jeghers syndrome, who require lifelong surveillance, and possibly release those with a simple dermatosis, such as Laugier-Hunziker syndrome, from invasive and thus potentially harmful surveillance. 26819373_Combined Mertk (and Mfge8) deficiency in macrophages blunted VEGFA release from infarcted hearts. 26962228_The current study demonstrates the contribution of the TAM receptor MerTK to the phagocytosis of myelin by human adult microglia and monocyte-derived macrophages. 26990204_One of the associated variants was also found to be linked with increased expression of MERTK in monocytes and higher expression of MERTK was associated with either increased or decreased risk of developing MS, dependent upon HLA-DRB1*15:01 status. 27028863_these data suggest that endogenous GAS6 and Mer receptor signaling contribute to the establishment of prostate cancer stem cells in the bone marrow microenvironment 27081701_MERTK is frequently overexpressed in head and neck squamous cell carcinoma and plays an important role in tumor cell motility. 27122965_We report a novel missense mutation (c.3G>A, p.0?) in the MERTK gene that causes severe vision impairment in a patient. 27486820_Study identified the Gas6/TAM receptor pathway with Tyro3 and Mer as novel targets in colorectal cancer. 27649555_The broad-spectrum activity mediated by UNC2025 in leukemia patient samples and xenograft models, alone or in combination with cytotoxic chemotherapy, supports continued development of MERTK inhibitors for treatment of leukemia 27753136_Knockdown of MERTK by shRNA in prostate cancer cells induced a decreased ratio of P-Erk1/2 to P-p38, increased expression of p27, NR2F1, SOX2, and NANOG, induced higher levels of histone H3K9me3 and H3K27me3, and induced a G1/G0 arrest, all of which are associated with dormancy. 27801848_In this paper, we review the biology of the Gas6/Tyro3, Axl, and MerTK(collectively named TAM system)and the current evidence supporting its potential role in the pathogenesis of multiple sclerosis . 27827458_Study describes a novel cellular pathway involved in diabetic efferocytosis, wherein diabetes-induced decrease in miR-126 expression results in upregulation of ADAM9 expression that in-turn leads proteolytic cleavage of MerTK and formation of inactive soluble Mer. Decrease in MerTK phosphorylation leads to reduced downstream cytoskeletal signaling required for engulfment and thus decreases efferocytosis. 28067670_evidence that proteolytic cleavage of the macrophage efferocytosis receptor c-Mer tyrosine kinase (MerTK) reduces efferocytosis and promotes plaque necrosis and defective resolution. 28127639_The expression of MerTK and AxlTK varied according to the deposition of immunoglobulin and complements on glomeruli. Both MerTK and AxlTK expressions were increased on glomeruli and varied according to pathological classifications. 28184013_Phosphatidylserine mediated hyperactivation of Mertk.MERTK promotes epithelial cell efferocytosis in a tyrosine kinase-dependent manner.MERTK role in AKT-dependent drug resistance. 28251492_Small molecule and antibody inhibitors of AXL and MER have recently been described, and some of these have already entered clinical trials. The optimal design of treatment strategies to maximize the clinical benefit of these AXL and MER targeting agents are discussed in relation to the different cancer types and the types of resistance encountered. 28324114_A 48 bp insertion sequence was buried within the breakpoint; 18 bps shared homology to MIR4435-2HG and LINC00152, and 30 bp mapped to MERTK. The deletion cosegregated with arRP in the family. 28334911_MERTK G > A variant affects liver disease, nutrient oxidation and glucose metabolism in NAFLD. 28462455_Sequence analysis revealed that the proband was a compound heterozygote with two independent mutations in MERTK, a novel nonsense mutation (c.2179C > T) and a previously reported missense variant (c.2530C > T). The proband's affected brother also had both mutations 28668213_Patients with macroalbuminuria diabetes had higher circulating levels of sMer and more urinary soluble Tyro3 and sMer than normoalbuminuric diabetics. Increased clearance of sTyro3 and sMer was associated with loss of tubular Tyro3 and Mer expression in diabetic nephropathy tissue. During in vitro diabetes, human kidney cells had down-regulation of Tyro3 and Mer mRNA and increased shedding of sTyro3 and sMer. 28851810_Monocyte-induced MerTK cleavage on proreparative MHCII(LO) cardiac macrophages is a novel contributor to myocardial ischemic reperfusion injury. 28916522_this study shows that viral infection sensitizes fetal membranes by MERTK Inhibition 29299721_The present study provides a meaningfully negative result demonstrating that rare variants in MERTK are not associated with AMD. The study also demonstrates the role of large sample size genetic studies utilizing whole-genome sequencing as a powerful tool that can resolve clinically relevant questions regarding the genetic basis of ophthalmic disease. 29359540_We observed that the frequency for the wild-type haplotype was higher in the control group, compared to that in the group of patients with COPD, in the subgroup analysis of current smokers, although the difference was not statistically significant 29437494_The targeted NGS strategy employed provides an efficient tool for RP pathogenic gene detection. This study identified a new autosomal recessive mutation in the RP-related gene MERTK, which expands the spectrum of RP disease-causing mutations 29553850_MerTK mediates STAT3-KRAS/SRC-signaling axis for glioma stem cell maintenance 29659094_Mutations in MERTK have been associated with severe autosomal recessive retinal dystrophies in the RCS rat and in humans. We present here a comprehensive review of all reported MERTK disease causing variants with the associated phenotype 30091033_cooperation between CD14 and MerTK may foster the clearance of apoptotic neutrophils by human monocytes/macrophages 30093568_MERTK expression was increased in cell lines and patient-derived xenografts treated with AXL inhibitors. 30254055_activation of MerTK in human macrophages led to ERK-mediated expression of the gene encoding SERCA2, which decreased the cytosolic Ca(2+) concentration and suppressed the activity of calcium/calmodulin-dependent protein kinase II 30416333_Patients with retinitis pigmentosa (RP) in this study were carriers of two novel allelic mutations in the MERTK gene, a missense variant in exon 17 and an approximate 91 kb genomic deletion. Mapping of the deletion breakpoints allowed molecular testing of a cohort of patients with RP with allele-specific PCR. 30541554_Axl and Tyro3, but not Mertk, have an important role in platelet activation and thrombus formation 30619243_data do not support a particular role for TAM receptors or for activated CD11b in the association of platelet-derived EVs with monocytes and granulocytes in the circulation. 30765874_Mertk expression was not significantly higher in non-muscle invasive bladder cancers (MIBCs) and MIBCs, compared to normal urothelium. Loss-of-function experiments in vitro and in a mouse xenograft model showed that Mertk depletion had only a minor impact on cell viability. 30790467_Three novel loss-of-function mutations in MERTK gene were identified in Chinese patients with retinitis pigmentosa. 30851773_MERTK has a role in retinal pigment epithelium as a regulator of rod outer segments' phagocytosis. Due to c.1647T > G substitution, the stop codon (p.Tyr549Ter) appears early in the transcript. 30944303_Authors report that SAV1, a Hippo signaling component, inhibits Akt, a function independent of its role in Hippo signaling. Binding to a proline-tyrosine motif in the Akt-PH domain, SAV1 suppresses Akt activation by blocking Akt's movement to plasma membrane. Authors further identify cancer-associated SAV1 mutations with impaired ability to bind Akt, leading to Akt hyperactivation. 31221805_Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. 31230815_we identify an unexpected role of TAM kinases (Tyro3, Axl, and Mer)as promoters of necroptosis, a pro-inflammatory necrotic cell death. Pharmacologic or genetic targeting of TAM kinases results in a potent inhibition of necroptotic death in various cellular models. We identify phosphorylation of MLKL Tyr376 as a direct point of input from TAM kinases into the necroptosis signaling 31451482_TAM Family Receptor Kinase Inhibition Reverses MDSC-Mediated Suppression and Augments Anti-PD-1 Therapy in Melanoma. 31583026_Gas6/MERTK signaling components are upregulated in liver fibrosis. (Review) 31655933_Increased sMer, but not sAxl, sTyro3, and Gas6 relate with active disease in juvenile systemic lupus erythematosus. 31805065_These results suggest that deletion of MERTK in human pulmonary microvascular endothelial cells in vitro and in all cells in vivo aggravates the inflammatory response. 31839486_Macrophage MerTK Promotes Liver Fibrosis in Nonalcoholic Steatohepatitis. 32051695_New Insights into the Role of Tyro3, Axl, and Mer Receptors in Rheumatoid Arthritis. 32160519_MERTK-Dependent Ensheathment of Photoreceptor Outer Segments by Human Pluripotent Stem Cell-Derived Retinal Pigment Epithelium. 32454903_Role of Gas6 and TAM Receptors in the Identification of Cardiopulmonary Involvement in Systemic Sclerosis and Scleroderma Spectrum Disorders. 32640697_Mertk Interacts with Tim-4 to Enhance Tim-4-Mediated Efferocytosis. 32973744_Potential Oncogenic Effect of the MERTK-Dependent Apoptotic-Cell Clearance Pathway in Starry-Sky B-Cell Lymphoma. 32976546_Pathogenic variants of AIPL1, MERTK, GUCY2D, and FOXE3 in Pakistani families with clinically heterogeneous eye diseases. 33101282_Expression of TAM-R in Human Immune Cells and Unique Regulatory Function of MerTK in IL-10 Production by Tolerogenic DC. 33119085_A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding. 33234241_Recent advancements in role of TAM receptors on efferocytosis, viral infection, autoimmunity, and tissue repair. 33234243_TAM receptors and their ligand-mediated activation: Role in atherosclerosis. 33234244_Post-translational modifications of the ligands: Requirement for TAM receptor activation. 33234245_Immunological role of TAM receptors in the cancer microenvironment. 33353011_Bi-Allelic Pathogenic Variations in MERTK Including Deletions Are Associated with an Early Onset Progressive Form of Retinitis Pigmentosa. 33512463_Mer tyrosine kinase as a possible link between resolution of inflammation and tissue fibrosis in IgG4-related disease. 33532004_Role of the Gas6/TAM System as a Disease Marker and Potential Drug Target. 33571503_MerTK activity is not necessary for the proliferation of glioblastoma stem cells. 33609509_Deterioration of phagocytosis in induced pluripotent stem cell-derived retinal pigment epithelial cells established from patients with retinitis pigmentosa carrying Mer tyrosine kinase mutations. 33761885_Differential polarization and the expression of efferocytosis receptor MerTK on M1 and M2 macrophages isolated from coronary artery disease patients. 33859405_Microglia use TAM receptors to detect and engulf amyloid beta plaques. 34035216_Ligand-dependent kinase activity of MERTK drives efferocytosis in human iPSC-derived macrophages. 34207717_MERTK-Mediated LC3-Associated Phagocytosis (LAP) of Apoptotic Substrates in Blood-Separated Tissues: Retina, Testis, Ovarian Follicles. 34289798_MERTK retinopathy: biomarkers assessing vision loss. 34415994_MERTK on mononuclear phagocytes regulates T cell antigen recognition at autoimmune and tumor sites. 35036076_MerTK-mediated efferocytosis promotes immune tolerance and tumor progression in osteosarcoma through enhancing M2 polarization and PD-L1 expression. 35163068_High Glucose Impairs Expression and Activation of MerTK in ARPE-19 Cells. | ENSMUSG00000014361 | Mertk | 3.066842e+02 | 0.6778571 | -0.560946897 | 0.3431228 | 2.675820e+00 | 0.1018828673 | 0.75783482 | No | Yes | 2.368023e+02 | 46.563459 | 3.227048e+02 | 65.052017 | |
| ENSG00000153310 | 51571 | CYRIB | protein_coding | Q9NUQ9 | FUNCTION: Negatively regulates RAC1 signaling and RAC1-driven cytoskeletal remodeling (PubMed:31285585, PubMed:30250061). Regulates chemotaxis, cell migration and epithelial polarization by controlling the polarity, plasticity, duration and extent of protrusions. Limits Rac1 mediated activation of the Scar/WAVE complex, focuses protrusion signals and regulates pseudopod complexity by inhibiting Scar/WAVE-induced actin polymerization (PubMed:30250061). Protects against Salmonella bacterial infection. Attenuates processes such as macropinocytosis, phagocytosis and cell migration and restrict sopE-mediated bacterial entry (PubMed:31285585). Restricts also infection mediated by Mycobacterium tuberculosis and Listeria monocytogenes (By similarity). Involved in the regulation of mitochondrial dynamics and oxidative stress (PubMed:29059164). {ECO:0000250|UniProtKB:Q921M7, ECO:0000269|PubMed:29059164, ECO:0000269|PubMed:30250061, ECO:0000269|PubMed:31285585}. | 3D-structure;Alternative splicing;Isopeptide bond;Lipoprotein;Membrane;Mitochondrion;Myristate;Reference proteome;Ubl conjugation | hsa:51571; | cilium [GO:0005929]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; membrane [GO:0016020]; mitochondrion [GO:0005739]; platelet alpha granule lumen [GO:0031093]; MHC class Ib protein binding, via antigen binding groove [GO:0023030]; small GTPase binding [GO:0031267]; cellular response to molecule of bacterial origin [GO:0071219]; negative regulation of actin filament polymerization [GO:0030837]; negative regulation of small GTPase mediated signal transduction [GO:0051058]; positive regulation of interferon-gamma production [GO:0032729]; positive regulation of memory T cell activation [GO:2000568]; positive regulation of T cell activation [GO:0050870]; positive regulation of T cell mediated cytotoxicity [GO:0001916]; regulation of cell migration [GO:0030334]; regulation of chemotaxis [GO:0050920]; regulation of establishment of cell polarity [GO:2000114]; regulation of mitochondrial fission [GO:0090140] | 21555518_FAM49B is a target gene of the BACH1 transcription factor according to ChIP-seq analysis in HEK 293 cells. 25284053_Three proteins-matrix metalloproteinase-9, neutrophil elastase, and FAM49B-were significantly lower in abundance in samples from women with endometriosis. 29059164_Low fam49B expression is associated with pancreatic ductal adenocarcinoma metastasis. 29632189_FAM49B inhibits T cell activation by repressing Rac activity and modulating cytoskeleton reorganization. 31285585_CYFIP related Rac1 interactor B (CYRI) binds to the small GTPase RAC1 through a conserved domain present in CYFIP proteins. CYRI negatively regulates RAC1 signalling, thereby attenuating processes such as macropinocytosis, phagocytosis and cell migration. This enables CYRI to counteract Salmonella at various stages of infection. The bacterial effector SopE, a RAC1 activator, selectively targets CYRI following infection. 32071545_TASP1 Promotes Gallbladder Cancer Cell Proliferation and Metastasis by Up-regulating FAM49B via PI3K/AKT Pathway. 33217330_Structural Basis of CYRI-B Direct Competition with Scar/WAVE Complex for Rac1. | ENSMUSG00000022378 | Cyrib | 6.028480e+02 | 0.9140355 | -0.129677828 | 0.3449916 | 1.390906e-01 | 0.7091867522 | 0.93786306 | No | Yes | 5.762648e+02 | 108.734837 | 5.747894e+02 | 111.200836 | ||
| ENSG00000153574 | 22934 | RPIA | protein_coding | P49247 | FUNCTION: Catalyzes the reversible conversion of ribose-5-phosphate to ribulose 5-phosphate and participates in the first step of the non-oxidative branch of the pentose phosphate pathway. {ECO:0000269|PubMed:14988808}. | Disease variant;Isomerase;Methylation;Neuropathy;Phosphoprotein;Reference proteome | PATHWAY: Carbohydrate degradation; pentose phosphate pathway; D-ribose 5-phosphate from D-ribulose 5-phosphate (non-oxidative stage): step 1/1. {ECO:0000269|PubMed:14988808}. | The protein encoded by this gene is an enzyme, which catalyzes the reversible conversion between ribose-5-phosphate and ribulose-5-phosphate in the pentose-phosphate pathway. This gene is highly conserved in most organisms. The enzyme plays an essential role in the carbohydrate metabolism. Mutations in this gene cause ribose 5-phosphate isomerase deficiency. A pseudogene is found on chromosome 18. [provided by RefSeq, Mar 2010]. | hsa:22934; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; identical protein binding [GO:0042802]; monosaccharide binding [GO:0048029]; ribose-5-phosphate isomerase activity [GO:0004751]; D-ribose metabolic process [GO:0006014]; pentose-phosphate shunt [GO:0006098]; pentose-phosphate shunt, non-oxidative branch [GO:0009052]; ribose phosphate metabolic process [GO:0019693] | 14988808_RPI is the second known inborn error in the reversible phase of the pentose-phosphate-pathway, confirming that defects in pentose and polyol metabolism constitute a new area of inborn metabolic disorders 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25429733_Study provides new insight into the molecular mechanisms by which RPIA overexpression can induce oncogenesis in hepatocellular carcinoma. 25528729_In this work, through an in silico comparative analysis between the genomes of Leishmania major and Homo sapiens, the enzyme ribose 5-phosphate isomerase (R5PI) was indicated as a promising molecular target. 26248089_CRC cells that overexpressed miR124 or with knockdown of RPIA or PRPS1 had reduced DNA synthesis and proliferation, whereas cells incubated with an inhibitor of miR124 had significantly increased DNA synthesis and proliferation and formed more colonies. 27328773_The results are consistent with a model in which RPIA suppresses autophagy and LC3 processing by modulation of redox signaling. 30177747_SRC-2 controls the pentose phosphate pathway through RPIA in human endometrial cancer cells. 31247379_We report on a subject with RPIA associated progressive leukoencephalopathy with elevated urine arabitol and ribitol levels and a novel missense variant c.770T>C p.(Ile257Thr) in exon 8 of RPIA. We also compare the phenotypes of all the four subjects. Our report confirms the phenotype and the genetic cause of this condition. | ENSMUSG00000053604 | Rpia | 1.573672e+03 | 0.8804344 | -0.183712630 | 0.2895185 | 4.179013e-01 | 0.5179860985 | 0.88251380 | No | Yes | 1.333146e+03 | 134.018937 | 1.501425e+03 | 154.482385 |
| ENSG00000153827 | 9320 | TRIP12 | protein_coding | Q14669 | FUNCTION: E3 ubiquitin-protein ligase involved in ubiquitin fusion degradation (UFD) pathway and regulation of DNA repair (PubMed:19028681, PubMed:22884692). Part of the ubiquitin fusion degradation (UFD) pathway, a process that mediates ubiquitination of protein at their N-terminus, regardless of the presence of lysine residues in target proteins (PubMed:19028681). Acts as a key regulator of DNA damage response by acting as a suppressor of RNF168, an E3 ubiquitin-protein ligase that promotes accumulation of 'Lys-63'-linked histone H2A and H2AX at DNA damage sites, thereby acting as a guard against excessive spreading of ubiquitinated chromatin at damaged chromosomes (PubMed:22884692). In normal cells, mediates ubiquitination and degradation of isoform p19ARF/ARF of CDKN2A, a lysine-less tumor suppressor required for p53/TP53 activation under oncogenic stress (PubMed:20208519). In cancer cells, however, isoform p19ARF/ARF and TRIP12 are located in different cell compartments, preventing isoform p19ARF/ARF ubiquitination and degradation (PubMed:20208519). Does not mediate ubiquitination of isoform p16-INK4a of CDKN2A (PubMed:20208519). Also catalyzes ubiquitination of NAE1 and SMARCE1, leading to their degradation (PubMed:18627766). Ubiquitination and degradation of target proteins is regulated by interaction with proteins such as MYC, TRADD or SMARCC1, which disrupt the interaction between TRIP12 and target proteins (PubMed:20829358). Mediates ubiquitination of ASXL1: following binding to N(6)-methyladenosine methylated DNA, ASXL1 is ubiquitinated by TRIP12, leading to its degradation and subsequent inactivation of the PR-DUB complex (PubMed:30982744). {ECO:0000269|PubMed:18627766, ECO:0000269|PubMed:19028681, ECO:0000269|PubMed:20208519, ECO:0000269|PubMed:20829358, ECO:0000269|PubMed:22884692, ECO:0000269|PubMed:30982744}. | Acetylation;Alternative splicing;Autism spectrum disorder;DNA damage;DNA repair;Disease variant;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:18627766, ECO:0000269|PubMed:20208519, ECO:0000269|PubMed:30982744}. | The protein encoded by this gene is an E3 ubiquitin-protein ligase involved in the degradation of the p19ARF/ARF isoform of CDKN2A, a tumor suppressor. The encoded protein also plays a role in the DNA damage response by regulating the stability of USP7, which regulates tumor suppressor p53. [provided by RefSeq, Jan 2017]. | hsa:9320; | cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; thyroid hormone receptor binding [GO:0046966]; ubiquitin protein ligase activity [GO:0061630]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; negative regulation of double-strand break repair [GO:2000780]; negative regulation of histone H2A K63-linked ubiquitination [GO:1901315]; protein polyubiquitination [GO:0000209]; regulation of double-strand break repair [GO:2000779]; regulation of embryonic development [GO:0045995]; ubiquitin-dependent protein catabolic process [GO:0006511] | 18627766_TRIP12 promotes degradation of APP-BP1 by catalyzing its ubiquitination, which in turn modulates the neddylation pathway. 19028681_The HECT domain of TRIP12 ubiquitinates substrates of the ubiquitin fusion degradation pathway. 19730683_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20699639_ULF is a bona fide E3 ligase for ARF and also suggest that ULF is an important target for activating the ARF-p53 axis in human acute myeloid leukaemia cells. 20829358_Data show that the mechanism of BAF155-mediated stabilization of BAF57 involves blocking its ubiquitination by preventing interaction with TRIP12. 22124266_we found somatic mutations of HERC2, HERC3, TRIP12, UBE2Q1 and UBE4B genes in gastric carcinoma and colorectal carcinomas with microsatellite instability 22561347_data indicate that TRADD shuttles dynamically from the cytoplasm into the nucleus to modulate the interaction between p19(Arf) and its E3 ubiquitin ligase ULF, thereby promoting p19(Arf) protein stability and tumour suppression 22884692_Study shows that TRIP12 and UBR5, two HECT domain ubiquitin E3 ligases, control accumulation of RNF168, a rate-limiting component of a pathway that ubiquitylates histones after DNA breakage. 23209776_HUWE1 and TRIP12 collaborate in degradation of ubiquitin-fusion proteins and misframed ubiquitin. 24961348_An exon3-skipping event in TRIP12 was detected in acute myeloid leukemia patients at remission 25355311_Data indicate that E3 ubiquitin ligase thyroid hormone receptor-interacting protein 12 (TRIP12) promotes proteasomal degradation of pancreas transcription factor 1a (PTF1a)and regulates PTF1a activities. 27425591_p16 overexpression led to downregulation of TRIP12, which in turn led to increased RNF168 levels, repressed DNA damage repair (DDR), increased 53BP1 foci and enhanced radioresponsiveness. 27800609_modulatory role for Trip12 in the USP7-dependent DNA damage response 27848077_We describe the TRIP12-associated phenotype, showing that TRIP12 is a risk gene for non-syndromic intellectual disability with and without autism spectrum disorder, and that TRIP12 mutation carriers present with a broad phenotypic range within the neurodevelopmental phenotypes. 28251352_nine presented pathogenic variants further document that TRIP12 haploinsufficiency causes a childhood-onset neurodevelopmental disorder 31814248_Clark-Baraitser syndrome is associated with a nonsense alteration in the autosomal gene TRIP12. 31964993_The E3 ubiquitin ligase TRIP12 participates in cell cycle progression and chromosome stability. 32424948_Novel de novo TRIP12 mutation reveals variable phenotypic presentation while emphasizing core features of TRIP12 variations. 32755579_The Ubiquitin Ligase TRIP12 Limits PARP1 Trapping and Constrains PARP Inhibitor Efficiency. 33567268_TRIP12 promotes small-molecule-induced degradation through K29/K48-branched ubiquitin chains. 33824312_Proteasomal degradation of the tumour suppressor FBW7 requires branched ubiquitylation by TRIP12. 33963176_TRIP12 has an inhibitory role in Epithelial-Mesenchymal Transition (EMT) and possibly, metastasis 34644545_TRIP12 ubiquitination of glucocerebrosidase contributes to neurodegeneration in Parkinson's disease. | ENSMUSG00000026219 | Trip12 | 3.189798e+03 | 0.5400992 | -0.888703715 | 0.3125138 | 7.986814e+00 | 0.0047119252 | 0.24765550 | No | Yes | 2.585235e+03 | 518.732080 | 4.003551e+03 | 823.397448 |
| ENSG00000153933 | 8526 | DGKE | protein_coding | P52429 | FUNCTION: Membrane-bound diacylglycerol kinase that converts diacylglycerol/DAG into phosphatidic acid/phosphatidate/PA and regulates the respective levels of these two bioactive lipids (PubMed:15544348, PubMed:19744926, PubMed:22108654, PubMed:21477596, PubMed:23949095). Thereby, acts as a central switch between the signaling pathways activated by these second messengers with different cellular targets and opposite effects in numerous biological processes (PubMed:8626589, PubMed:15544348). Also plays an important role in the biosynthesis of complex lipids (PubMed:8626589). Displays specificity for diacylglycerol substrates with an arachidonoyl acyl chain at the sn-2 position, with the highest activity toward 1-octadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycerol the main diacylglycerol intermediate within the phosphatidylinositol turnover cycle (PubMed:19744926, PubMed:22108654, PubMed:23274426). Can also phosphorylate diacylglycerol substrates with a linoleoyl acyl chain at the sn-2 position but much less efficiently (PubMed:22108654). {ECO:0000269|PubMed:15544348, ECO:0000269|PubMed:19744926, ECO:0000269|PubMed:21477596, ECO:0000269|PubMed:22108654, ECO:0000269|PubMed:23274426, ECO:0000269|PubMed:23949095, ECO:0000303|PubMed:15544348, ECO:0000303|PubMed:8626589}. | ATP-binding;Alternative splicing;Cytoplasm;Disease variant;Hemolytic uremic syndrome;Kinase;Lipid metabolism;Membrane;Metal-binding;Nucleotide-binding;Reference proteome;Repeat;Transferase;Transmembrane;Transmembrane helix;Zinc;Zinc-finger | PATHWAY: Lipid metabolism; glycerolipid metabolism. {ECO:0000305|PubMed:15544348}. | Diacylglycerol kinases are thought to be involved mainly in the regeneration of phosphatidylinositol (PI) from diacylglycerol in the PI-cycle during cell signal transduction. When expressed in mammalian cells, DGK-epsilon shows specificity for arachidonyl-containing diacylglycerol. DGK-epsilon is expressed predominantly in testis. [provided by RefSeq, Jul 2008]. | hsa:8526; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; glutamatergic synapse [GO:0098978]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; diacylglycerol kinase activity [GO:0004143]; kinase activity [GO:0016301]; metal ion binding [GO:0046872]; NAD+ kinase activity [GO:0003951]; diacylglycerol metabolic process [GO:0046339]; intracellular signal transduction [GO:0035556]; lipid phosphorylation [GO:0046834]; modulation of chemical synaptic transmission [GO:0050804]; phosphatidic acid biosynthetic process [GO:0006654]; phosphatidylinositol biosynthetic process [GO:0006661]; platelet activation [GO:0030168]; protein kinase C-activating G protein-coupled receptor signaling pathway [GO:0007205] | 17455907_The hydrophobic domain of diacylglycerol kinase epsilon does not contribute to substrate specificity but plays a role in permanently sequestering the enzyme to a membrane. 18004883_The alpha and zeta isoforms of diacylglycerol kinase are inhibited by 2,3-dioleoylglycerol, but not the more substrate-selective epsilon isoform 19329342_Observational study of gene-disease association. (HuGE Navigator) 19744926_Substrate specificity of diacylglycerol kinase epsilon is determined by selectivity of the sn-1 and sn-2 acyl chains phosphatidic acid or diacylglycerol. 20546612_Observational study of gene-disease association. (HuGE Navigator) 21194521_Data show that Diacylglycerol that 2-arachidonoyl glycerol is a very poor substrate for either the epsilon or the zeta isoforms of diacylglycerol kinases. 21725595_DGK activity is reduced by oxidative stress in human mesangial cells cultured under high glucose conditions. 21833457_A role for diacylglycerol kinase (DGK) and its downstream product phosphatidic acid (PA) in ANCA-induced neutrophil exocytosis, is reported. 22108654_diacylglycerol kinase-epsilon (DGKepsilon) has less preference for the acyl chain at the sn-1 position of diacylglycerol (DAG) than the one at the sn-2 position 22266092_The region responsible for this arachidonoyl specificity is the lipoxygenase (LOX)-like motif found in the accessory domain, adjacent to DGKvarepsilon's catalytic site. 22511757_Inhibition of lipid signaling enzyme diacylglycerol kinase epsilon attenuates mutant huntingtin toxicity. 23261795_Substrate specificity of DGKE is not a consequence of competition with hydrolysis of ATP. 23274426_performed homozygosity mapping and whole exome sequencing in a Turkish consanguineous family and identified DGKE gene variants as the cause of a membranoproliferative-like glomerular microangiopathy 23542698_Recessive mutations in DGKE cause atypical hemolytic-uremic syndrome. 23630338_Fully activating high-density transfected muscarinic receptors (M1Rs) by oxotremorine-M (Oxo-M) leads to similar calcium, DAG, and PKC signals, but PIP2 is depleted. 24119575_DGKzeta localizes to the nucleus and is considered to regulate nuclear diacylglycerol signaling.[review] 24511134_Our study expands the clinical phenotypes associated with mutations in DGKE and challenges the benefits of complement blockade treatment in such patients 25135762_Data suggest that complement dysregulation influences the onset and disease severity in carriers of diacylglycerol kinase-epsilon mutations 25498910_DGKE silencing in resting endothelial cells does not affect complement activation at their surface. 25854283_Report DGKE intronic mutations located beyond the exon-intron boundaries in familial hemolytic uremic syndrome. 26018111_Letter/Case Report: atypical haemolytic uraemic syndrome in a Japanese patient with DGKE genetic mutations. 28526779_mutations can lead to atypical hemolytic uremic syndrome or membranoproliferative glomerulonephritis 32413569_Various phenotypes of disease associated with mutated DGKE gene. 33986189_Loss of diacylglycerol kinase epsilon causes thrombotic microangiopathy by impairing endothelial VEGFA signaling. | ENSMUSG00000000276 | Dgke | 2.933084e+02 | 0.7364046 | -0.441429508 | 0.3531364 | 1.579943e+00 | 0.2087689373 | 0.78763590 | No | Yes | 2.048074e+02 | 40.450502 | 3.148833e+02 | 63.183639 |
| ENSG00000153936 | 9653 | HS2ST1 | protein_coding | Q7LGA3 | FUNCTION: Catalyzes the transfer of sulfate to the C2-position of selected hexuronic acid residues within the maturing heparan sulfate (HS). 2-O-sulfation within HS, particularly of iduronate residues, is essential for HS to participate in a variety of high-affinity ligand-binding interactions and signaling processes. Mediates 2-O-sulfation of both L-iduronyl and D-glucuronyl residues (By similarity). {ECO:0000250}. | Alternative splicing;Disease variant;Disulfide bond;Glycoprotein;Golgi apparatus;Membrane;Mental retardation;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | Heparan sulfate biosynthetic enzymes are key components in generating a myriad of distinct heparan sulfate fine structures that carry out multiple biologic activities. This gene encodes a member of the heparan sulfate biosynthetic enzyme family that transfers sulfate to the 2 position of the iduronic acid residue of heparan sulfate. The disruption of this gene resulted in no kidney formation in knockout embryonic mice, indicating that the absence of this enzyme may interfere with the signaling required for kidney formation. Two alternatively spliced transcript variants that encode different proteins have been found for this gene. [provided by RefSeq, Aug 2008]. | hsa:9653; | Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; heparan sulfate 2-O-sulfotransferase activity [GO:0004394]; sulfotransferase activity [GO:0008146]; glycosaminoglycan biosynthetic process [GO:0006024]; heparan sulfate proteoglycan biosynthetic process, enzymatic modification [GO:0015015]; heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process [GO:0015014] | 17227754_analysis of differences and similarities various residues play in the biological roles of the HS-2OST and CS-2OST enzymes 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 25594747_C5-epimerase and 2-O-sulfotransferase in association generate extended domains of consecutive GlcNS-IdoA2S Sequence. 32573871_HS2ST1-dependent signaling pathways determine breast cancer cell viability, matrix interactions, and invasive behavior. 33159882_Bi-allelic Pathogenic Variants in HS2ST1 Cause a Syndrome Characterized by Developmental Delay and Corpus Callosum, Skeletal, and Renal Abnormalities. | ENSMUSG00000040151 | Hs2st1 | 5.106556e+02 | 1.0554888 | 0.077911203 | 0.3189266 | 5.971255e-02 | 0.8069508496 | 0.96141221 | No | Yes | 5.260150e+02 | 90.688189 | 4.449045e+02 | 78.612225 | |
| ENSG00000153989 | 116150 | NUS1 | protein_coding | Q96E22 | FUNCTION: With DHDDS, forms the dehydrodolichyl diphosphate synthase (DDS) complex, an essential component of the dolichol monophosphate (Dol-P) biosynthetic machinery. Both subunits contribute to enzymatic activity, i.e. condensation of multiple copies of isopentenyl pyrophosphate (IPP) to farnesyl pyrophosphate (FPP) to produce dehydrodolichyl diphosphate (Dedol-PP), a precursor of dolichol phosphate which is utilized as a sugar carrier in protein glycosylation in the endoplasmic reticulum (ER) (PubMed:21572394, PubMed:25066056, PubMed:28842490, PubMed:32817466). Synthesizes long-chain polyprenols, mostly of C95 and C100 chain length (PubMed:32817466). Regulates the glycosylation and stability of nascent NPC2, thereby promoting trafficking of LDL-derived cholesterol. Acts as a specific receptor for the N-terminus of Nogo-B, a neural and cardiovascular regulator (PubMed:16835300). {ECO:0000269|PubMed:16835300, ECO:0000269|PubMed:21572394, ECO:0000269|PubMed:25066056, ECO:0000269|PubMed:28842490, ECO:0000269|PubMed:32817466}. | 3D-structure;Angiogenesis;Congenital disorder of glycosylation;Developmental protein;Differentiation;Disease variant;Endoplasmic reticulum;Glycoprotein;Lipid metabolism;Magnesium;Membrane;Mental retardation;Metal-binding;Receptor;Reference proteome;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:25066056}.; PATHWAY: Lipid metabolism. {ECO:0000269|PubMed:25066056, ECO:0000269|PubMed:28842490}. | This gene encodes a type I single transmembrane domain receptor, which is a subunit of cis-prenyltransferase, and serves as a specific receptor for the neural and cardiovascular regulator Nogo-B. The encoded protein is essential for dolichol synthesis and protein glycosylation. This gene is highly expressed in non-small cell lung carcinomas as well as estrogen receptor-alpha positive breast cancer cells where it promotes epithelial mesenchymal transition. This gene is associated with the poor prognosis of human hepatocellular carcinoma patients. Naturally occurring mutations in this gene cause a congenital disorder of glycosylation and are associated with epilepsy. A knockout of the orthologous gene in mice causes embryonic lethality before day 6.5. Pseudogenes of this gene have been defined on chromosomes 13 and X. [provided by RefSeq, May 2017]. | hsa:116150; | dehydrodolichyl diphosphate synthase complex [GO:1904423]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; dehydrodolichyl diphosphate synthase activity [GO:0045547]; metal ion binding [GO:0046872]; angiogenesis [GO:0001525]; cell differentiation [GO:0030154]; cholesterol homeostasis [GO:0042632]; dolichol biosynthetic process [GO:0019408]; dolichyl diphosphate biosynthetic process [GO:0006489]; positive regulation of blood vessel endothelial cell migration [GO:0043536]; positive regulation of nitric-oxide synthase activity [GO:0051000]; protein glycosylation [GO:0006486]; protein mannosylation [GO:0035268]; regulation of intracellular cholesterol transport [GO:0032383]; vascular endothelial growth factor signaling pathway [GO:0038084] | 16835300_identify a previously uncharacterized Nogo-B receptor specific for the amino terminus of Nogo-B 17764014_In s-IBM muscle the Nogo-B increase may represent an attempt by muscle fiber to decrease A beta production. However, the increase of Nogo-B seems insufficient because A beta continues to accumulate and the disease progresses. 19723497_The Nogo-B receptor localizes primarily to the endoplasmic reticulum and regulates the stability of nascent Niemann-Pick type C2 protein. 21572394_Nogo-B receptor (NgBR) is an essential component of the dolichol monophosophate (Dol-P) biosynthetic machinery. Loss of NgBR results in a robust deficit in cis-isoprenyltransferase (IPTase) activity and Dol-P production. 24223763_NgBR is a new molecular marker for breast cancer. 24568601_Nogo-B receptor mediates pulmonary endothelial cell angiogenesis response through Akt/endothelial nitric oxide synthase pathway. 25066056_Described is a family with a congenital disorder of glycosylation caused by a loss of function mutation in the conserved C terminus of Nogo-B receptor - R290H. 25075030_Significant NgBR mRNA down-regulation was associated with larger primary tumor size (p=0.039), lymph node involvement (p=0.039) and advancement stage (p=0.0054). 25173099_These findings provide new insights for understanding the roles of NgBR in regulating breast epithelial cell transform during the pathogenesis of breast cancer. 25202063_Nogo-B receptor expression correlates negatively with malignancy grade and ki-67 antigen expression in invasive ductal breast carcinoma. 26840457_High NgBR expression is associated with chemoresistance in hepatocellular carcinoma. 28602162_The data suggest that miR-26a plays a key role in VEGF-mediated angiogenesis through the modulation of eNOS activity, which is mediated by its ability to regulate NgBR expression by directly targeting the NgBR 3'-UTR. 28842490_findings show that eukaryotic cis-PT is composed of the NgBR and hCIT subunits. The strong conservation of the RXG motif among NgBR orthologs indicates that this subunit is critical for the synthesis of polyprenol diphosphates and cellular function. 29331415_Data show that Nogo-B receptor (NgBR) knockdown inhibited epithelial-mesenchymal transition (EMT) in non-small cell lung cancer (NSCLC) cells in vitro and metastasis of NSCLC cells in vivo. 29346419_Small Angle X-ray Scattering (SAXS) analysis reveals the radius of gyration (Rg) of our NgBR construct to be 18.2 A with a maximum particle dimension (Dmax) of 61.0 A. Ab initio shape modeling returns a globular molecular envelope with an estimated molecular weight of 23.0 kD closely correlated with the calculated molecular weight 29373839_Data suggest that Nogo-B receptor (NgBR) expression is essential to promoting estrogen receptor alpha (ERalpha) positive breast cancer cell resistance to paclitaxel. 29904947_The Nogo-B receptor was highly expressed in human hepatocellular carcinoma (HCC) cell lines and in the tissue of patients with HCC and promoted human HCC cell growth by increasing the Akt phosphorylation in human HCC cells 30106141_NgBR role in cancer.NgBR is able to promote N-glycosylation to attenuate endoplasmic reticulum stress and the unfolded protein response.[review] 30208932_Results suggest that Nogo-B receptor (NgBR) is a potential therapeutic target for increasing the sensitivity of estrogen receptor alpha (ERalpha)-positive breast cancer to tamoxifen. 30348779_Coding mutations in NUS1 contribute to Parkinson's disease. 31154456_NUS1 was upregulated in IUAs tissues, and the high expression level of NUS1 was positively correlated with the severity of IUAs. NUS1 promoted cell proliferation in vitro. NUS1 overexpression on cell migration and invasion promoted the EMT process in vitro and in vivo. 31656175_we report two unrelated Japanese patients with a novel, recurrent, de novo NUS1 variant, who presented with epileptic seizures with involuntary movement, ataxia, intellectual disability and scoliosis. 32246992_Nogo-B fosters HCC progression by enhancing Yap/Taz-mediated tumor-associated macrophages M2 polarization. 32485575_NUS1 mutation in a family with epilepsy, cerebellar ataxia, and tremor. 32817466_Structural elucidation of the cis-prenyltransferase NgBR/DHDDS complex reveals insights in regulation of protein glycosylation. 32871760_Salivary NUS1 and RCN1 Levels as Biomarkers for Oral Squamous Cell Carcinoma Diagnosis. 33077723_Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase complex. 33184037_Assessment of the association between NUS1 variants and essential tremor. 33309333_Replication assessment of NUS1 variants in Parkinson's disease. 33400686_NOGOB receptor-mediated RAS signaling pathway is a target for suppressing proliferating hemangioma. 33548880_Contribution of coding/non-coding variants in NUS1 to late-onset sporadic Parkinson's disease. 34326813_Common Variants in NUS1 and GP2 Genes Contributed to the Risk of Gestational Diabetes Mellitus. 34635350_Low-frequency and rare coding variants of NUS1 contribute to susceptibility and phenotype of Parkinson's disease. | ENSMUSG00000023068 | Nus1 | 1.350112e+03 | 0.7717209 | -0.373848903 | 0.3140487 | 1.384394e+00 | 0.2393541360 | 0.78818582 | No | Yes | 1.247842e+03 | 225.226822 | 1.361437e+03 | 251.867193 |
| ENSG00000154079 | 135154 | SDHAF4 | protein_coding | Q5VUM1 | FUNCTION: Plays an essential role in the assembly of succinate dehydrogenase (SDH), an enzyme complex (also referred to as respiratory complex II) that is a component of both the tricarboxylic acid (TCA) cycle and the mitochondrial electron transport chain, and which couples the oxidation of succinate to fumarate with the reduction of ubiquinone (coenzyme Q) to ubiquinol (PubMed:24954416). Binds to the flavoprotein subunit SDHA in its FAD-bound form, blocking the generation of excess reactive oxigen species (ROS) and facilitating its assembly with the iron-sulfur protein subunit SDHB into the SDH catalytic dimer (By similarity). {ECO:0000250|UniProtKB:P38345, ECO:0000269|PubMed:24954416}. | Chaperone;Mitochondrion;Reference proteome;Transit peptide | hsa:135154; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; enzyme activator activity [GO:0008047]; cellular respiration [GO:0045333]; innate immune response [GO:0045087]; mitochondrial respiratory chain complex II assembly [GO:0034553]; positive regulation of succinate dehydrogenase activity [GO:1904231] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000026154 | Sdhaf4 | 2.198846e+02 | 0.8358180 | -0.258739325 | 0.3058871 | 7.183797e-01 | 0.3966759264 | 0.84483554 | No | Yes | 1.536531e+02 | 27.304818 | 2.071255e+02 | 37.430961 | ||
| ENSG00000154548 | 135295 | SRSF12 | protein_coding | Q8WXF0 | FUNCTION: Splicing factor that seems to antagonize SR proteins in pre-mRNA splicing regulation. {ECO:0000269|PubMed:11684676}. | Nucleus;RNA-binding;Reference proteome;mRNA processing;mRNA splicing | hsa:135295; | cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; RS domain binding [GO:0050733]; unfolded protein binding [GO:0051082]; mRNA 5'-splice site recognition [GO:0000395]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of mRNA splicing, via spliceosome [GO:0048025]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; spliceosomal tri-snRNP complex assembly [GO:0000244] | ENSMUSG00000054679 | Srsf12 | 1.205640e+02 | 0.9987789 | -0.001762813 | 0.3655185 | 2.294854e-05 | 0.9961777781 | 0.99926282 | No | Yes | 1.220442e+02 | 18.238254 | 1.222050e+02 | 18.875451 | |||
| ENSG00000154803 | 201163 | FLCN | protein_coding | Q8NFG4 | FUNCTION: GTPase-activating protein that plays a key role in the cellular response to amino acid availability through regulation of the mTORC1 signaling cascade controlling the MiT/TFE factors TFEB and TFE3 (PubMed:17028174, PubMed:18663353, PubMed:21209915, PubMed:24081491, PubMed:24095279, PubMed:31704029, PubMed:31672913). Activates mTORC1 by acting as a GTPase-activating protein: specifically stimulates GTP hydrolysis by RRAGC/RagC or RRAGD/RagD, promoting the conversion to the GDP-bound state of RRAGC/RagC or RRAGD/RagD, and thereby activating the kinase activity of mTORC1 (PubMed:24095279, PubMed:31704029, PubMed:31672913). The GTPase-activating activity is inhibited during starvation and activated in presence of nutrients (PubMed:31672913). Acts as a key component for mTORC1-dependent control of the MiT/TFE factors TFEB and TFE3, while it is not involved in mTORC1-dependent phosphorylation of canonical RPS6KB1/S6K1 and EIF4EBP1/4E-BP1 (PubMed:21209915, PubMed:24081491, PubMed:31672913). In low-amino acid conditions, the lysosomal folliculin complex (LFC) is formed on the membrane of lysosomes, which inhibits the GTPase-activating activity of FLCN, inactivates mTORC1 and maximizes nuclear translocation of TFEB and TFE3 (PubMed:31672913). Upon amino acid restimulation, RRAGA/RagA (or RRAGB/RagB) nucleotide exchange promotes disassembly of the LFC complex and liberates the GTPase-activating activity of FLCN, leading to activation of mTORC1 and subsequent cytoplasmic retention of TFEB and TFE3 (PubMed:31672913). Indirectly acts as a positive regulator of Wnt signaling by promoting mTOR-dependent cytoplasmic retention of MiT/TFE factor TFE3 (PubMed:31272105). Required for the exit of hematopoietic stem cell from pluripotency by promoting mTOR-dependent cytoplasmic retention of TFE3, thereby increasing Wnt signaling (PubMed:30733432). Acts as an inhibitor of browning of adipose tissue by regulating mTOR-dependent cytoplasmic retention of TFE3 (By similarity). In response to flow stress, regulates STK11/LKB1 accumulation and mTORC1 activation through primary cilia: may act by recruiting STK11/LKB1 to primary cilia for activation of AMPK resided at basal bodies, causing mTORC1 down-regulation (PubMed:27072130). Together with FNIP1 and/or FNIP2, regulates autophagy: following phosphorylation by ULK1, interacts with GABARAP and promotes autophagy (PubMed:25126726). Required for starvation-induced perinuclear clustering of lysosomes by promoting association of RILP with its effector RAB34 (PubMed:27113757). {ECO:0000250|UniProtKB:Q8QZS3, ECO:0000269|PubMed:17028174, ECO:0000269|PubMed:18663353, ECO:0000269|PubMed:21209915, ECO:0000269|PubMed:24081491, ECO:0000269|PubMed:24095279, ECO:0000269|PubMed:25126726, ECO:0000269|PubMed:27072130, ECO:0000269|PubMed:27113757, ECO:0000269|PubMed:30733432, ECO:0000269|PubMed:31272105, ECO:0000269|PubMed:31672913, ECO:0000269|PubMed:31704029}. | 3D-structure;Alternative splicing;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;GTPase activation;Lysosome;Membrane;Nucleus;Phosphoprotein;Reference proteome;Tumor suppressor | This gene is located within the Smith-Magenis syndrome region on chromosome 17. Mutations in this gene are associated with Birt-Hogg-Dube syndrome, which is characterized by fibrofolliculomas, renal tumors, lung cysts, and pneumothorax. Alternative splicing of this gene results in two transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:201163; | centrosome [GO:0005813]; cilium [GO:0005929]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; mitotic spindle [GO:0072686]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; GTPase activator activity [GO:0005096]; guanyl-nucleotide exchange factor activity [GO:0005085]; protein-containing complex binding [GO:0044877]; cell-cell junction assembly [GO:0007043]; cellular response to amino acid starvation [GO:0034198]; cellular response to starvation [GO:0009267]; energy homeostasis [GO:0097009]; hemopoiesis [GO:0030097]; in utero embryonic development [GO:0001701]; lysosome localization [GO:0032418]; negative regulation of ATP biosynthetic process [GO:2001170]; negative regulation of brown fat cell differentiation [GO:1903444]; negative regulation of cell growth [GO:0030308]; negative regulation of cell migration [GO:0030336]; negative regulation of cell proliferation involved in kidney development [GO:1901723]; negative regulation of cellular respiration [GO:1901856]; negative regulation of cold-induced thermogenesis [GO:0120163]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of gene expression [GO:0010629]; negative regulation of mitochondrial DNA metabolic process [GO:1901859]; negative regulation of mitochondrion organization [GO:0010823]; negative regulation of muscle tissue development [GO:1901862]; negative regulation of post-translational protein modification [GO:1901874]; negative regulation of protein kinase B signaling [GO:0051898]; negative regulation of protein localization to nucleus [GO:1900181]; negative regulation of Rho protein signal transduction [GO:0035024]; negative regulation of TOR signaling [GO:0032007]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of apoptotic process [GO:0043065]; positive regulation of autophagy [GO:0010508]; positive regulation of cell adhesion [GO:0045785]; positive regulation of GTPase activity [GO:0043547]; positive regulation of intrinsic apoptotic signaling pathway [GO:2001244]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of TOR signaling [GO:0032008]; positive regulation of TORC1 signaling [GO:1904263]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transforming growth factor beta receptor signaling pathway [GO:0030511]; regulation of cytokinesis [GO:0032465]; regulation of histone acetylation [GO:0035065]; regulation of pro-B cell differentiation [GO:2000973]; regulation of protein phosphorylation [GO:0001932]; regulation of Ras protein signal transduction [GO:0046578]; regulation of TOR signaling [GO:0032006]; TOR signaling [GO:0031929] | 12204536_Mutations lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome 12471204_Clinical and genetic studies of four sporadic BHD cases and four families with a total of 23 affected subjects 12746401_Mutations in sporadic colorectal carcinomas and colorectal carcinoma cell lines with microsatellite instability 12907635_BHD is involved in the entire spectrum of histological types of renal tumors, suggesting its major role in kidney cancer tumorigenesis. 15579035_potential role of BHD as a tumor suppressor gene (review) 15852235_Germ-line mutation spectrum and phenotype analysis were expanded in a large cohort of families with Birt-Hogg-Dube syndrome. 15956655_These results support a role for BHD as a tumor suppressor gene that predisposes to the development of renal tumors when both copies are inactivated. 16636660_May regulate tumorigenesis through modulating stem cells in human. 16870330_Findings show that the BHD gene is a rare target in microsatellite instability (MSI)-high gastric cancer, and BHD mutation tends to occur downstream in the mutational events of other major MSI-high target genes. 17028174_Results suggest that FLCN, mutated in Birt-Hogg-Dube syndrome, and its interacting partner FNIP1 may be involved in energy and/or nutrient sensing through the AMPK and mTOR signaling pathways. 17496196_Germline mutations of the BHD gene are involved in some patients with multiple lung cysts & pneumothorax. 4 were new: 3 deletions or insertions in exons 6, 12 & 13, & a splice acceptor site mutation in intron 5 resulting in an in-frame deletion of exon 6. 18206534_The UOK 257 cell line: a novel model for studies of the human Birt-Hogg-Dube gene pathway. 18234728_The first germline missense mutation in BHD c.1978A>G (K508R) in a patient who presented with bilateral multifocal renal oncocytomas, is reported. 18505456_FLCN mutation contributes to not only familial but also 'apparently sporadic' patients with isolated primary spontaneous pneumothorax 18505456_Observational study of gene-disease association. (HuGE Navigator) 18573707_Birt-Hogg-Dube (BHD) syndrome, showing the occurrence of two frameshift mutations located respectively in exons 5 (802insA) and 9 (1345delAAAG) of the FLCN gene. A novel homozygous sequence variant in the intron 9 (IVS9 +5C>T) was also found. 18579543_Mutations in the folliculin gene are associated with cystic lung lesions in an otherwise morphological normal lung and predispose to spontaneous pneumothorax [case report] 18709329_study reports an Asian family with Birt-Hogg-Dube syndrom with a BHD germline mutation 19116017_Genetic variation in folliculin does not appear to be a major risk factor for severe COPD 19116017_Observational study of gene-disease association. (HuGE Navigator) 19234517_Folliculin regulates the activity of TORC1, and a new paradigm in which both inappropriately high and inappropriately low levels of TORC1 activity can be associated with renal tumorigenesis. 19327534_In the siblings reported, no FLCN mutations were identified 19483054_A family with lung cysts and spontaneous pneumothorax in 3 generations had a deletion mutation in exon 10 of the FLCN gene. 20227563_We report cases involving a new mutation in three unrelated families of Birt-Hogg-Dube syndrome. 20413710_An Birt-Hogg-Dube syndrome protein germline mutation was found in 23 (63.9%) of the 36 patients. A large genomic deletion was identified in two of the remaining 13 patients. 20522427_Data show that germline FLCN mutations were not detected in 50 patients with familial non-syndromic colorectal cancer. 20618353_Data show that FLCN mutations were found in 9 of 19 (47%) families. 21079084_plays role in tumor suppression and inhibition on rapamycin pathway 21209915_FLCN tumor suppressor gene inactivation induces TFE3 transcriptional activity by increasing its nuclear localization 21258407_Birt-Hogg-Dube (BHD) protein-deficient cells exhibited defects in cell-intrinsic apoptosis that correlated with reduced expression of the BH3-only protein Bim, which was similarly observed in all human and mouse BHD-related tumors examined. 21412933_This report confirms that large intragenic FLCN deletions can cause Birt-Hogg-Dube syndrome and documents the first large intragenic FLCN duplication in a Birt-Hogg-Dube syndrome patient 21496834_Genetic testing for Birt-Hogg-Dube should be considered in the treatment algorithm of patients with bilateral renal masses and known oncocytoma 21538689_FLCN mutations throughout the coding sequence, and suggest that multiple protein domains contribute to folliculin stability and tumor suppressor activity. 22146830_confirmed a high yield of FLCN mutations in clinically defined BHD families, we found a substantially increased lifetime risk of renal cancer of 16% for FLCN mutation carriers 22211584_Germline mutation analysis of the FCLN gene in mother and daughter cases with renal cell neoplasms showed a deletion of 18 bp in exon 5(c.332_349del/p.H111_Q116del), predicting an alteration of the amino acid sequence of 'HPSHPQ' replaced by a single amino acid, 'L'. 22446046_Study reports a novel in-frame deletion mutation p.F143del (c.427_429delTTC) in exon 6 of FLCN gene in a Korean proband and her two sisters. 22709692_The FLCN-FNIP complex deregulated in Birt-Hogg-Dube syndrome is absolutely required for B-cell differentiation. 22965878_Findings suggest that aspects of folliculin tumour suppressor function are linked to interaction with p0071 and the regulation of RhoA signalling. 22977732_Data indicate that folliculin-CT is structurally similar to the DENN domain of DENND1B. 23077212_FLCN functions as a tumor suppressor by negatively regulating rRNA synthesis. 23139756_These data support a model in which dysregulation of the FLCN-p0071 interaction leads to alterations in cell adhesion, cell polarity, and RhoA signaling. 23150719_FLCN deficiency and subsequent increased PPARGC1A expression result in increased mitochondrial function and oxidative metabolism as the source of cellular energy, which may drive hyperplastic transformation. 23155228_These findings identify novel pathways and targets linked to folliculin tumour suppressor activity. 23264078_FLCN germ-line mutation is associated with spontaneous pneumothorax and renal cancer 23414156_Birt-Hogg-Dube syndrome in a patient with melanoma and a novel mutation in the FCLN gene. 23416984_data indicate that: (a) apoptotic cell death in FLCN-null cells can be triggered by SSH2 knockdown through cell cycle arrest; (b) SSH2 represents a potential therapeutic target for the development of agents for the treatment of BHD syndrome related tumors 23784378_FLCN localizes to motile and non-motile cilia, centrosomes and the mitotic spindle. Alteration of FLCN levels can cause changes to the onset of ciliogenesis. In three-dimensional culture, abnormal expression of FLCN disrupts polarized growth of kidney cells and deregulates canonical Wnt signalling. Findings suggest that BHD is a ciliopathy, with symptoms at least partly due to abnormal ciliogenesis. 23784378_Results suggest that Birt-Hogg-Dube (BHD)syndrome-causing FLCN mutants may retain partial functionality. Thus, several BHD symptoms may be due to abnormal levels of FLCN rather than its complete loss. 23874397_Tumor suppression function of FLCN may be linked to its impact on the cell cycle. 24095279_FLCN and its binding partners, FNIP1/2, are Rag-interacting proteins with GAP activity for RagC/D, but not RagA/B. 24346394_A rare mutation of the folliculin gene was detected in the patient and family members with Birt-Hogg-Dube syndrome with pulmonary cysts or pneumothorax, but no skin or renal lesions. 24434776_Findings suggest that folliculin deficient renal cell carcinoma cells are highly sensitive to irradiation due to increased autophagic cell death, unlike other types of renal cell carcinoma. 24762438_loss of FLCN constitutively activates AMPK, resulting in PGC-1alpha-mediated mitochondrial biogenesis and increased ROS production 24996715_Birt-Hogg-Dube (BHD) syndrome is a recently discovered autosomal-dominant disease caused by a mutation in the folliculin gene. 25126726_The FLCN-GABARAP association is modulated by the presence of either folliculin-interacting protein (FNIP)-1 or FNIP2 and further regulated by ULK1. 25583493_Two predominant genes, ephrin type A receptor 6 (EPHA6) and folliculin (FLCN), with mutations exclusive to African American CRCs, are by genetic and biological criteria highly likely African American CRC driver genes. 25594584_FLCN-related renal cell carcinomas showed overexpression of GPNMB and underexpression of FLCN, whereas sporadic tumors showed inverted patterns. 25807935_This report documents the first identification of founder mutations in FLCN, as well as expands mutation spectrum of the gene 25827758_Case Report: FLCN deletion mutation in members of Indian Birt-Hogg-Dube syndrome family. 26342594_We identified a hitherto unreported pathogenic FLCN frameshift deletion c.563delT (p.Phe188Serfs*35) in a family of a 46-year-old woman presented with macrohematuria due to bilateral chromophobe renal carcinomas 26398834_FLCN irregulation in lung cysts of primary spontaneous pneumothorax is not associated with promoter methylation. 26418749_mTOR inhibitor, sirolimus, suppresses the tumor's growth, suggesting that mTOR inhibitors might be effective in control of FLCN-deficient RCC. 26439621_we show that glycogen accumulates in kidneys from mice lacking FLCN and in renal tumors from a BHD patient 27220747_Seventy-six of 156 FLCN mutation carriers (120 probands and 36 sibs, 48.7%) had skin papules; however, cutaneous manifestations were so subtle that only one patient voluntarily consulted dermatologists. Japanese Asian BHD families have three FLCN mutational hotspots. 27258496_For patients whose clinical features are atypical, detection of germline mutation in FLCN gene would help confirm diagnosis. The 2 mutations we reported would expand the mutation spectrum of FLCN gene associated with BHD syndrome 27484154_The molecular insight into the folliculin dynamics in the presence and absence of mutations may provide valuable information regarding the interactions essential for normal functioning of cellular process and the molecular basis of Birt-Hogg-Dube syndrome 27486260_A nonsense mutation of FLCN was found in a spontaneous pneumothorax family. The results expand the mutational spectrum of FLCN in patients with Birt-Hogg-Dube syndrome. 27633572_We report Smith-Magenis syndrome who presents bilateral renal tumors. This is most likely related to haploinsufficiency of FLCN gene, located in the deleted region 27734835_the study describes the FLCN mutation spectrum in Danish Birt-Hogg-Dube (BHD) syndrome patients, and contributes to a better understanding of BHD syndrome and management of BHD patients. 27780965_DNA sequence analyses determined that there was a two base pair deletion in exon 4 of the FLCN gene, confirming the diagnosis of BHD syndrome. 28039480_SCFbeta-TRCP negatively regulates the FLCN complex by promoting FNIP2 degradation in Birt-Hogg-Dube syndrome-associated renal cancer. 28558743_In the folliculin gene, a similar genotype spectrum but different mutant loci was determined in Chinese patients with Birt-Hogg-Dube syndrome compared with European and American patients. 28656962_FLCN functions as a Rab7A GTPase-activating protein.Negative regulation of EGFR signalling by FLCN. 28775225_FLCN gene analysis revealed a heterozygous FLCN{NM_144997.5}:c.1285dupC mutation in all affected members. The clinical features of BHD syndrome are heterogeneous with wide intra-familial and interfamilial variation. It is caused by mutations of the FLCN gene. 28785590_The present identification of two mutations not only further supports the important role of tumor suppressor FLCN in Birt-Hogg-Dube syndrome and primary spontaneous pneumothorax, but also expands the spectrum of FLCN mutations. 28970150_Germline mutations in the FLCN gene are responsible for the autosomal dominant inherited disorder Birt-Hogg-Dube syndrome. 29357828_Two FLCN mutations have been identified in Birt-Hogg-Dube syndrome patients: One is an insertion mutation previously reported in three Asian families; while the other is the first mutation (originally found in Asian populations) that has ever been detected in a French family. 29767721_Germline FLCN mutations in Birt-Hogg-Dube syndrome associated kidney cancer. 30446510_FLCN promoted the loading of proton-coupled amino acid transporter 1 on Rab11A protein 31272105_Data found that canonical Wnt signaling is decreased in FLCN-deficient mesenchymal cells associated with decreased levels of multiple components of the Wnt enhanceosome. This phenotype can be rescued by silencing TFE3. These findings lead to the hypothesis that lung cysts in Birt-Hogg-Dube may arise through defective mesenchymal signaling via Wnt. 31567476_novel nonsense mutation of FLCN gene was found in a large family with primary spontaneous pneumothorax in China 31625278_Results found that a heterozygous mutation in exon 11 of FLCN c. 1273C>T (p.Gln425Ter), which was identified for the first time, might cause isolated familial spontaneous pneumothorax. 31672913_structure of the lysosomal FLCN complex (LFC) reveals how FLCN and FNIP2 assemble into one complex by means of an intricate domain arrangement, as well as how they interact with the G domains of the Rag GTPases in their inactive state 31709497_Intratumoral heterogeneity of FLCN somatic mutations in gastric and colorectal cancers. 31806376_FLCN alteration drives metabolic reprogramming towards nucleotide synthesis and cyst formation in salivary gland. 32279295_New Developments in the Pathogenesis of Pulmonary Cysts in Birt-Hogg-Dube Syndrome. 32934076_Quantitative genetic screening reveals a Ragulator-FLCN feedback loop that regulates the mTORC1 pathway. 33064845_Ciliary localization of folliculin mediated via a kinesin-2-binding motif is required for its functions in mTOR regulation and tumor suppression. 33137092_Folliculin variants linked to Birt-Hogg-Dube syndrome are targeted for proteasomal degradation. 33298956_Blood and lymphatic systems are segregated by the FLCN tumor suppressor. 33459596_Loss of FLCN-FNIP1/2 induces a non-canonical interferon response in human renal tubular epithelial cells. 33609526_FLCN regulates transferrin receptor 1 transport and iron homeostasis. 34031471_Folliculin haploinsufficiency causes cellular dysfunction of pleural mesothelial cells. 34229741_Novel folliculin gene mutations in Polish patients with Birt-Hogg-Dube syndrome. 34381247_The tumor suppressor Folliculin also known as FLCN, functions as an intracellular uncompetitive inhibitor of Lactate Dehydrogenase-A and a regulator of the Warburg effect. 34779410_Folliculin impairs breast tumor growth by repressing TFE3-dependent induction of the Warburg effect and angiogenesis. 35176117_A retrospective two centre study of Birt-Hogg-Dube syndrome reveals a pathogenic founder mutation in FLCN in the Swedish population. | ENSMUSG00000032633 | Flcn | 1.670551e+03 | 1.0325088 | 0.046154018 | 0.2818688 | 2.651185e-02 | 0.8706565426 | 0.97573306 | No | Yes | 1.467606e+03 | 133.823514 | 1.438583e+03 | 134.566318 | |
| ENSG00000154856 | 147495 | APCDD1 | protein_coding | Q8J025 | FUNCTION: Negative regulator of the Wnt signaling pathway. Inhibits Wnt signaling in a cell-autonomous manner and functions upstream of beta-catenin. May act via its interaction with Wnt and LRP proteins. May play a role in colorectal tumorigenesis. {ECO:0000269|PubMed:12384519, ECO:0000269|PubMed:20393562}. | Cell membrane;Disease variant;Glycoprotein;Hypotrichosis;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix;Wnt signaling pathway | This locus encodes an inhibitor of the Wnt signaling pathway. Mutations at this locus have been associated with hereditary hypotrichosis simplex. Increased expression of this gene may also be associated with colorectal carcinogenesis.[provided by RefSeq, Sep 2010]. | hsa:147495; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; Wnt-protein binding [GO:0017147]; astrocyte cell migration [GO:0043615]; hair follicle development [GO:0001942]; negative regulation of Wnt signaling pathway [GO:0030178]; regulation of odontogenesis of dentin-containing tooth [GO:0042487]; Wnt signaling pathway [GO:0016055] | 20200978_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20393562_APCDD1 is a novel inhibitor of the Wnt signalling pathway with an essential role in human hair growth 21152411_Data show that the methylated VAPA-APCDD1 DNA in maternal plasma is predominantly derived from the fetus, and this novel fetal epigenetic marker in maternal plasma is useful for the noninvasive detection of fetal trisomy 18. 22512811_mutation in the APCDD1 gene is responsible for hereditary hypotrichosis simplex in a large Chinese family. 25592970_Unusual role of APCDD1 in dental follicle cells during osteogenic differentiation. APCDD1 sustains the expression and activation of beta-catenin. 25946682_This study demonstrated a critical role for Apcdd1 in OL differentiation after white matter injury that points to a potential therapeutic approach for inhibiting Wnt signaling in these disorders. 28242765_these novel findings suggest that APCDD1 positively regulates adipogenic differentiation and that its down-regulation by miR-130 during diet-induced obesity may contribute to impaired adipogenic differentiation and obesity-related metabolic disease. 28698141_Thus, we have provided the first evidence that APCDD1 expression is epigenetically silenced in OS, which may facilitate invasion and metastasis of OS cells. 30496486_Through integrative analysis, we identify MEIS1 as a super-enhancer-driven oncogene, which co-operates with EWS-FLI1 in transcriptional regulation, and plays a key pro-survival role in Ewing sarcoma. Moreover, APCDD1, another super-enhancer-associated gene, acting as a downstream target of both MEIS1 and EWS-FLI1, is also characterized as a novel tumor-promoting factor in this malignancy 33673279_Whole Exome Sequencing Identifies APCDD1 and HDAC5 Genes as Potentially Cancer Predisposing in Familial Colorectal Cancer. | ENSMUSG00000071847 | Apcdd1 | 7.841884e+01 | 0.5945982 | -0.750012980 | 0.4337641 | 2.980425e+00 | 0.0842771182 | 0.74082963 | No | Yes | 3.765761e+01 | 9.332810 | 6.983761e+01 | 17.613557 | |
| ENSG00000154889 | 65258 | MPPE1 | protein_coding | Q53F39 | FUNCTION: Metallophosphoesterase required for transport of GPI-anchor proteins from the endoplasmic reticulum to the Golgi. Acts in lipid remodeling steps of GPI-anchor maturation by mediating the removal of a side-chain ethanolamine-phosphate (EtNP) from the second Man (Man2) of the GPI intermediate, an essential step for efficient transport of GPI-anchor proteins. {ECO:0000269|PubMed:19837036, ECO:0000269|PubMed:29374258}. | Alternative splicing;ER-Golgi transport;GPI-anchor biosynthesis;Golgi apparatus;Hydrolase;Manganese;Membrane;Metal-binding;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:65258; | cis-Golgi network [GO:0005801]; endoplasmic reticulum exit site [GO:0070971]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; nucleoplasm [GO:0005654]; GPI anchor binding [GO:0034235]; GPI-mannose ethanolamine phosphate phosphodiesterase activity [GO:0062050]; manganese ion binding [GO:0030145]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; GPI anchor biosynthetic process [GO:0006506] | 11978971_cDNA cloning, genomic organization and expression of the novel human metallophosphoesterase gene MPPE1 on chromosome 18p11.2 19328558_Observational study of gene-disease association. (HuGE Navigator) 19837036_Data demonstrate that glycosylphosphatidylinositol(GPI) glycan acts as an endoplasmic reticulum-exit signal and suggest that glycan remodeling mediated by PGAP5 regulates GPI-anchored proteins transport in the early secretory pathway. 19859903_Observational study of gene-disease association. (HuGE Navigator) 19859903_These results provide evidence of an association between a polymorphism in the MPPE1 gene and bipolar disorder. 33054568_Metallophosphoesterase 1, a novel candidate gene in hepatocellular carcinoma malignancy and recurrence. | ENSMUSG00000062526 | Mppe1 | 4.333850e+02 | 0.9988356 | -0.001680869 | 0.2818856 | 3.574576e-05 | 0.9952296557 | 0.99926282 | No | Yes | 3.766544e+02 | 40.956944 | 3.933981e+02 | 43.229559 | ||
| ENSG00000154928 | 2047 | EPHB1 | protein_coding | P54762 | FUNCTION: Receptor tyrosine kinase which binds promiscuously transmembrane ephrin-B family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling. Cognate/functional ephrin ligands for this receptor include EFNB1, EFNB2 and EFNB3. During nervous system development, regulates retinal axon guidance redirecting ipsilaterally ventrotemporal retinal ganglion cells axons at the optic chiasm midline. This probably requires repulsive interaction with EFNB2. In the adult nervous system together with EFNB3, regulates chemotaxis, proliferation and polarity of the hippocampus neural progenitors. In addition to its role in axon guidance plays also an important redundant role with other ephrin-B receptors in development and maturation of dendritic spines and synapse formation. May also regulate angiogenesis. More generally, may play a role in targeted cell migration and adhesion. Upon activation by EFNB1 and probably other ephrin-B ligands activates the MAPK/ERK and the JNK signaling cascades to regulate cell migration and adhesion respectively. Involved in the maintenance of the pool of satellite cells (muscle stem cells) by promoting their self-renewal and reducing their activation and differentiation (By similarity). {ECO:0000250|UniProtKB:Q8CBF3, ECO:0000269|PubMed:12223469, ECO:0000269|PubMed:12925710, ECO:0000269|PubMed:18034775, ECO:0000269|PubMed:9430661, ECO:0000269|PubMed:9499402}. | 3D-structure;ATP-binding;Alternative splicing;Cell adhesion;Cell membrane;Cell projection;Direct protein sequencing;Endosome;Glycoprotein;Kinase;Membrane;Neurogenesis;Nucleotide-binding;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase;Ubl conjugation | Ephrin receptors and their ligands, the ephrins, mediate numerous developmental processes, particularly in the nervous system. Based on their structures and sequence relationships, ephrins are divided into the ephrin-A (EFNA) class, which are anchored to the membrane by a glycosylphosphatidylinositol linkage, and the ephrin-B (EFNB) class, which are transmembrane proteins. The Eph family of receptors are divided into 2 groups based on the similarity of their extracellular domain sequences and their affinities for binding ephrin-A and ephrin-B ligands. Ephrin receptors make up the largest subgroup of the receptor tyrosine kinase (RTK) family. The protein encoded by this gene is a receptor for ephrin-B family members. [provided by RefSeq, Jul 2008]. | hsa:2047; | axon [GO:0030424]; cytosol [GO:0005829]; dendrite [GO:0030425]; early endosome membrane [GO:0031901]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; filopodium tip [GO:0032433]; glutamatergic synapse [GO:0098978]; integral component of plasma membrane [GO:0005887]; membrane raft [GO:0045121]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; axon guidance receptor activity [GO:0008046]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein-containing complex binding [GO:0044877]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; transmembrane-ephrin receptor activity [GO:0005005]; angiogenesis [GO:0001525]; axon guidance [GO:0007411]; camera-type eye morphogenesis [GO:0048593]; cell chemotaxis [GO:0060326]; cell-substrate adhesion [GO:0031589]; central nervous system projection neuron axonogenesis [GO:0021952]; dendritic spine development [GO:0060996]; dendritic spine morphogenesis [GO:0060997]; detection of temperature stimulus involved in sensory perception of pain [GO:0050965]; ephrin receptor signaling pathway [GO:0048013]; establishment of cell polarity [GO:0030010]; hindbrain tangential cell migration [GO:0021934]; immunological synapse formation [GO:0001771]; modulation of chemical synaptic transmission [GO:0050804]; negative regulation of satellite cell differentiation [GO:1902725]; negative regulation of skeletal muscle satellite cell proliferation [GO:1902723]; neural precursor cell proliferation [GO:0061351]; neurogenesis [GO:0022008]; optic nerve morphogenesis [GO:0021631]; positive regulation of kinase activity [GO:0033674]; positive regulation of synapse assembly [GO:0051965]; protein autophosphorylation [GO:0046777]; regulation of ERK1 and ERK2 cascade [GO:0070372]; regulation of JNK cascade [GO:0046328]; regulation of neuron death [GO:1901214]; retinal ganglion cell axon guidance [GO:0031290]; skeletal muscle satellite cell activation [GO:0014719]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11140838_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12084815_Human platelets express EphA4 and EphB1, and the ligand, ephrinB1. Forced clustering of EphA4 or ephrinB1 led to cytoskeletal reorganization, adhesion to fibrinogen, and alpha-granule secretion. 14576067_Eph/ephrin signaling enhances the ability of platelet agonists to cause aggregation provided that those agonists can increase cytosolic Ca(++) and this is accomplished in part by activating Rap1 15722342_analysis of EphB1, EphB2, and EphB4-binding peptides interaction with antagonists with ephrin-like affinity 18034775_The ubiquitin ligase Cbl induces the ubiquitination and lysosomal degradation of activated EphB1, a process requiring EphB1 and Src kinase activity. 18057206_Transgenic EphB1 and ephrin-B3 cooperatively regulate the proliferation and migration of neural progenitors in the hippocampus. 18424888_Loss of expression of EphB1 protein in gastric carcinoma is associated with invasion and metastasis 18628988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18649358_Observational study of gene-disease association. (HuGE Navigator) 18931529_EphB1 may have roles in the pathogenesis and development of colorectal cancer. 19736353_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21041834_No association is found for EPH receptor B1 and susceptibility to schizophrenia. 21041834_Observational study of gene-disease association. (HuGE Navigator) 21085126_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21124932_Data shew that the identification of three novel candidates as EPH receptor genes might indicate a link between perturbed compartmentalization of early neoplastic lesions and breast cancer risk and progression. 21763378_EPHB1 polymorphisms may be associated with susceptibility to hepatocellular carcinoma in the Korean population. 21804545_Data show that EphB receptors interact with E-cadherin and with the metalloproteinase ADAM10 at sites of adhesion. 23118026_EphB1 stimulation triggered approximately 50% serine-threonine PTEN dephosphorylation and PTEN-Cbl complex disruption, a process requiring PTEN protein phosphatase activity. 24121831_Low EphB1 expression is associated with glioma. 24427352_Our data indicate that loss of EphB1 protein is associated with metastasis and poorer survival in patients with serous ovarian cancer 24606480_EphB1 and Ephrin-B could be regarded as independent good prognostic factors and important biological markers for Squamous cell/adenosquamous carcinoma and adenocarcinoma of gallbladder. 24677421_The study presents the first structure of the EphB1 tyrosine kinase domain determined by X-ray crystallography to 2.5A. 24716914_The genes CD248, Ephb1 and P2RY2 were detected as the top overexpressed in GC biopsies. 25120806_Our results indicate that EphB1 may be involved in carcinogenesis of renal cell carcinoma 25879388_In medulloblastoma cell lines, EphB1 downregulation or knockdown reduced cell growth, viability, cell-cycle regulator expression, and migration, but increased radiosensitivity and the percentage of cells in G1 phase of the cell cycle. 25944917_The tumor-suppressor function of EphB1 is clinically relevant across many malignancies, suggesting that EphB1 is an important regulator of common cancer cell transforming pathways. 27028544_Association of EPHB1 rs11918092 with symptoms of schizophrenia in Chinese Zhuang and Han populations. 27541794_investigate NET could modulate one's attention orientation to facial expressions, we categorized individuals according to the genotypes of the -182 T/C (rs2242446) polymorphism. Our results indicated that the -182 T/C polymorphism significantly modulated attention orientation to facial expressions, of which the CC genotype facilitated attention reorientation to the locations where cued faces were previously presented. 28108514_some of the mutations found in EPHB1 may contribute to an increased invasive capacity of cancers. 29550816_SUMOylation of EphB1 repressed activation of its downstream signaling molecule PKC-gamma, and consequently inhibited neuroblastoma tumorigenesis. 30401746_EphB1 and EphA1 phosphorylate the Cx32CT domain residue Tyr(243) Unlike for Cx43, the tyrosine phosphorylation of the Cx32CT increased gap junction intercellular communication. 33356837_Peripheral EphrinB1/EphB1 signalling attenuates muscle hyperalgesia in MPS patients and a rat model of taut band-associated persistent muscle pain. 33627480_Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain. | ENSMUSG00000032537 | Ephb1 | 3.100868e+02 | 0.9979830 | -0.002912879 | 0.2957883 | 9.786473e-05 | 0.9921069278 | 0.99877177 | No | Yes | 2.853447e+02 | 25.101948 | 2.791236e+02 | 25.212398 | |
| ENSG00000154945 | 91369 | ANKRD40 | protein_coding | Q6AI12 | ANK repeat;Acetylation;Direct protein sequencing;Reference proteome;Repeat | hsa:91369; | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000020864 | Ankrd40 | 1.714028e+03 | 1.0044431 | 0.006395855 | 0.2630136 | 5.895558e-04 | 0.9806286524 | 0.99700442 | No | Yes | 1.786136e+03 | 216.325958 | 1.591997e+03 | 197.855545 | ||||
| ENSG00000155229 | 64210 | MMS19 | protein_coding | Q96T76 | FUNCTION: Key component of the cytosolic iron-sulfur protein assembly (CIA) complex, a multiprotein complex that mediates the incorporation of iron-sulfur cluster into apoproteins specifically involved in DNA metabolism and genomic integrity (PubMed:29848660). In the CIA complex, MMS19 acts as an adapter between early-acting CIA components and a subset of cellular target iron-sulfur proteins such as ERCC2/XPD, FANCJ and RTEL1, thereby playing a key role in nucleotide excision repair (NER), homologous recombination-mediated double-strand break DNA repair, DNA replication and RNA polymerase II (POL II) transcription (PubMed:22678362, PubMed:22678361, PubMed:29225034, PubMed:23585563). As part of the mitotic spindle-associated MMXD complex, plays a role in chromosome segregation, probably by facilitating iron-sulfur (Fe-S) cluster assembly into ERCC2/XPD (PubMed:20797633). Together with CIAO2, facilitates the transfer of Fe-S clusters to the motor protein KIF4A, which ensures proper localization of KIF4A to mitotic machinery components to promote the progression of mitosis (PubMed:29848660). Indirectly acts as a transcriptional coactivator of estrogen receptor (ER), via its role in iron-sulfur insertion into some component of the TFIIH-machinery (PubMed:11279242). {ECO:0000269|PubMed:11279242, ECO:0000269|PubMed:20797633, ECO:0000269|PubMed:22678361, ECO:0000269|PubMed:22678362, ECO:0000269|PubMed:23585563, ECO:0000269|PubMed:29225034, ECO:0000269|PubMed:29848660}. | Acetylation;Activator;Alternative splicing;Chromosome partition;Cytoplasm;Cytoskeleton;DNA damage;DNA repair;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | hsa:64210; | CIA complex [GO:0097361]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; microtubule organizing center [GO:0005815]; MMXD complex [GO:0071817]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spindle [GO:0005819]; transcription factor TFIIH holo complex [GO:0005675]; enzyme binding [GO:0019899]; estrogen receptor binding [GO:0030331]; protein-macromolecule adaptor activity [GO:0030674]; signaling receptor complex adaptor activity [GO:0030159]; transcription coactivator activity [GO:0003713]; cellular response to DNA damage stimulus [GO:0006974]; chromosome segregation [GO:0007059]; DNA metabolic process [GO:0006259]; DNA repair [GO:0006281]; iron-sulfur cluster assembly [GO:0016226]; nucleotide-excision repair [GO:0006289]; phosphorelay signal transduction system [GO:0000160]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; positive regulation of transcription, DNA-templated [GO:0045893]; protein maturation by iron-sulfur cluster transfer [GO:0097428]; response to hormone [GO:0009725]; transcription, DNA-templated [GO:0006351] | 11328871_Cloning of the human MMS19 genes and functional complementation in Saccharomyces cerevisiae 16385451_Observational study of gene-disease association. (HuGE Navigator) 16797255_MMS19 HEAT repeat domain is essential for MMS19 function in NER and transcription, while domains A and B, within MMS19 N-terminus, modulate the balance between DNA repair and transcription. 19318433_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19318433_Single nucleotide polymorphisms in MMS19L is associated with pancreatic cancer. 20522537_Observational study of gene-disease association. (HuGE Navigator) 20797633_Results indicate that the MMS19-XPD protein complex is required for proper chromosome segregation, an abnormality of which could contribute to the pathogenesis in some cases of xeroderma pigmentosum. 22678361_study demonstrates MMS19 forms a complex with the cytoplasmic Fe-S assembly (CIA) proteins CIAO1, IOP1 and MIP18; cytoplasmic MMS19 also binds to multiple nuclear Fe-S proteins involved in DNA metabolism; propose that MMS19 functions as a platform to facilitate Fe-S cluster transfer to proteins critical for DNA replication and repair 22678362_identified MMS19 as a member of the cytosolic iron-sulfur protein assembly (CIA) machinery; MMS19 functions as part of the CIA targeting complex that interacts with and facilitates iron-sulfur cluster insertion into apoproteins involved in methionine biosynthesis, DNA replication, DNA repair, and telomere maintenance 23150669_The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. 23585563_MMS19 interacts with target proteins. MIP18 has a role to bridge MMS19 and CIAO1. CIAO1 also binds IOP1. 23632208_Polymorphisms in ERCC1, codon-118 and MMS19 genes are not associated with clinical response to platinum or survival. 23679317_Single nucleotide polymorphisms in the MMS19L gene is associated with bone malignant tumors. 23886164_MMS19L polymorphisms are associated with response to chemotherapy in osteosarcoma. 25892874_Suggest that MMS19 may be a potential new predictor of metastasis and chemoradiotherapy response in esophageal squamous cell carcinoma. 27235625_POLE1 is phosphorylated at serine-1940 after DNA damage and interacts with the iron-sulfur complex chaperones CIAO1 and MMS19. 27802208_We therefore propose the expression level of MMS19 as a candidate predictive marker of ACT benefit in resected NSCLC patients. 29035693_findings suggest that MMS19 plays an essential role in maintaining mitochondrial genome stability 32632277_Structural insights into Fe-S protein biogenesis by the CIA targeting complex. | ENSMUSG00000025159 | Mms19 | 6.019967e+03 | 1.0269222 | 0.038326951 | 0.2813465 | 1.890305e-02 | 0.8906447877 | 0.97949221 | No | Yes | 5.177417e+03 | 398.753629 | 4.990796e+03 | 394.513463 | ||
| ENSG00000155304 | 6782 | HSPA13 | protein_coding | P48723 | FUNCTION: Has peptide-independent ATPase activity. | ATP-binding;Endoplasmic reticulum;Microsome;Nucleotide-binding;Reference proteome;Signal | The protein encoded by this gene is a member of the heat shock protein 70 family and is found associated with microsomes. Members of this protein family play a role in the processing of cytosolic and secretory proteins, as well as in the removal of denatured or incorrectly-folded proteins. The encoded protein contains an ATPase domain and has been shown to associate with a ubiquitin-like protein. [provided by RefSeq, Jul 2008]. | hsa:6782; | cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; intracellular membrane-bounded organelle [GO:0043231]; mitochondrion [GO:0005739]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; heat shock protein binding [GO:0031072]; misfolded protein binding [GO:0051787]; protein folding chaperone [GO:0044183]; unfolded protein binding [GO:0051082]; cellular response to unfolded protein [GO:0034620]; chaperone cofactor-dependent protein refolding [GO:0051085]; protein refolding [GO:0042026] | 11319647_Observational study of gene-disease association. (HuGE Navigator) 16087163_Observational study of gene-disease association. (HuGE Navigator) 16087163_in the Japanese population, STCH might be a new candidate for conferring susceptibility to gastric cancer 18793616_These results suggest that STCH has a role in cell survival via modulation of the TRAIL-mediated cell death pathway. 19793966_An entirely novel path is revealed toward therapeutic intervention of tauopathies by inhibition of the previously untargeted ATPase activity of Hsp70. 23303189_novel role of STCH in the regulation of pHi through site-specific interactions with NBCe1-B and NHE1 and subsequent modulation of membrane transporter expression. 32547538_Hspa13 Promotes Plasma Cell Production and Antibody Secretion. 33672238_Differential Effects of STCH and Stress-Inducible Hsp70 on the Stability and Maturation of NKCC2. | ENSMUSG00000032932 | Hspa13 | 2.025560e+02 | 0.7558013 | -0.403921135 | 0.3931931 | 1.026214e+00 | 0.3110494873 | 0.81737113 | No | Yes | 1.689526e+02 | 34.573269 | 2.047748e+02 | 42.639237 | |
| ENSG00000155329 | 54819 | ZCCHC10 | protein_coding | Q8TBK6 | Alternative splicing;Metal-binding;Reference proteome;Zinc;Zinc-finger | hsa:54819; | nucleic acid binding [GO:0003676]; zinc ion binding [GO:0008270] | 31138778_ZCCHC10 exerts its tumor-suppressive effects by stabilizing the p53 protein and can be used a potential prognostic marker and therapeutic target in lung adenocarcinoma. | ENSMUSG00000018239 | Zcchc10 | 2.029423e+02 | 0.9507347 | -0.072885339 | 0.3260182 | 4.930783e-02 | 0.8242721063 | 0.96655419 | No | Yes | 3.140148e+02 | 36.922395 | 2.851377e+02 | 34.060991 | |||
| ENSG00000155893 | 92370 | PXYLP1 | protein_coding | Q8TE99 | FUNCTION: Responsible for the 2-O-dephosphorylation of xylose in the glycosaminoglycan-protein linkage region of proteoglycans thereby regulating the amount of mature glycosaminoglycan (GAG) chains. Sulfated glycosaminoglycans (GAGs), including heparan sulfate and chondroitin sulfate, are synthesized on the so-called common GAG-protein linkage region (GlcUAbeta1-3Galbeta1-3Galbeta1-4Xylbeta1-O-Ser) of core proteins, which is formed by the stepwise addition of monosaccharide residues by the respective specific glycosyltransferases. Xylose 2-O-dephosphorylation during completion of linkage region formation is a prerequisite for the initiation and efficient elongation of the repeating disaccharide region of GAG chains. {ECO:0000269|PubMed:24425863}. | Alternative splicing;Glycoprotein;Golgi apparatus;Hydrolase;Membrane;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | hsa:92370; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; phosphatase activity [GO:0016791]; chondroitin sulfate proteoglycan biosynthetic process [GO:0050650]; dephosphorylation [GO:0016311]; glycosaminoglycan biosynthetic process [GO:0006024]; positive regulation of heparan sulfate proteoglycan biosynthetic process [GO:0010909] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24425863_this study describes the cloning of a human cDNA encoding a novel protein designated 2-phosphoxylose phosphatase capable of dephosphorylating this Xyl residue. | ENSMUSG00000043587 | Pxylp1 | 3.193618e+02 | 1.1913543 | 0.252602501 | 0.3011293 | 7.022179e-01 | 0.4020394402 | 0.84607954 | No | Yes | 3.476961e+02 | 44.089948 | 2.721674e+02 | 35.470932 | ||
| ENSG00000156011 | 23362 | PSD3 | protein_coding | Q9NYI0 | FUNCTION: Guanine nucleotide exchange factor for ARF6. {ECO:0000250}. | Alternative splicing;Cell junction;Cell membrane;Cell projection;Coiled coil;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome;Synapse | hsa:23362; | postsynaptic density [GO:0014069]; ruffle membrane [GO:0032587]; guanyl-nucleotide exchange factor activity [GO:0005085]; regulation of ARF protein signal transduction [GO:0032012] | 16270321_Down regulated in ovarian cancer or absent in ovarian cancer and impact survival. 18649358_Observational study of gene-disease association. (HuGE Navigator) 20080650_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25296758_EFA6R regulated ARF6 localization and thereby actin stress fiber loss. 28237857_Common variants in PSD3 were associated with obesity, T2D and HDL level 35102341_PSD3 downregulation confers protection against fatty liver disease. | ENSMUSG00000030465 | Psd3 | 2.821905e+02 | 0.6574287 | -0.605093624 | 0.3429543 | 3.085213e+00 | 0.0790069081 | 0.73138229 | No | Yes | 2.351951e+02 | 50.341460 | 3.046751e+02 | 66.354525 | ||
| ENSG00000156026 | 90550 | MCU | protein_coding | Q8NE86 | FUNCTION: Mitochondrial inner membrane calcium uniporter that mediates calcium uptake into mitochondria (PubMed:21685888, PubMed:21685886, PubMed:23101630, PubMed:22904319, PubMed:23178883, PubMed:22829870, PubMed:22822213, PubMed:24332854, PubMed:23755363, PubMed:26341627). Constitutes the pore-forming and calcium-conducting subunit of the uniporter complex (uniplex) (PubMed:23755363). Activity is regulated by MICU1 and MICU2. At low Ca(2+) levels MCU activity is down-regulated by MICU1 and MICU2; at higher Ca(2+) levels MICU1 increases MCU activity (PubMed:24560927, PubMed:26903221). Mitochondrial calcium homeostasis plays key roles in cellular physiology and regulates cell bioenergetics, cytoplasmic calcium signals and activation of cell death pathways. Involved in buffering the amplitude of systolic calcium rises in cardiomyocytes (PubMed:22822213). While dispensable for baseline homeostatic cardiac function, acts as a key regulator of short-term mitochondrial calcium loading underlying a 'fight-or-flight' response during acute stress: acts by mediating a rapid increase of mitochondrial calcium in pacemaker cells (PubMed:25603276). participates in mitochondrial permeability transition during ischemia-reperfusion injury (By similarity). Regulates glucose-dependent insulin secretion in pancreatic beta-cells by regulating mitochondrial calcium uptake (PubMed:22904319, PubMed:22829870). Mitochondrial calcium uptake in skeletal muscle cells is involved in muscle size in adults (By similarity). Regulates synaptic vesicle endocytosis kinetics in central nerve terminal (By similarity). Involved in antigen processing and presentation (By similarity). {ECO:0000250|UniProtKB:Q3UMR5, ECO:0000269|PubMed:21685886, ECO:0000269|PubMed:21685888, ECO:0000269|PubMed:22822213, ECO:0000269|PubMed:22829870, ECO:0000269|PubMed:22904319, ECO:0000269|PubMed:23101630, ECO:0000269|PubMed:23178883, ECO:0000269|PubMed:23755363, ECO:0000269|PubMed:24332854, ECO:0000269|PubMed:24560927, ECO:0000269|PubMed:25603276, ECO:0000269|PubMed:26341627, ECO:0000269|PubMed:26903221}. | 3D-structure;Acetylation;Alternative splicing;Calcium;Calcium channel;Calcium transport;Coiled coil;Ion channel;Ion transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Phosphoprotein;Reference proteome;Transit peptide;Transmembrane;Transmembrane helix;Transport | This gene encodes a calcium transporter that localizes to the mitochondrial inner membrane. The encoded protein interacts with mitochondrial calcium uptake 1. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2012]. | hsa:90550; | calcium channel complex [GO:0034704]; integral component of mitochondrial inner membrane [GO:0031305]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; uniplex complex [GO:1990246]; calcium channel activity [GO:0005262]; identical protein binding [GO:0042802]; uniporter activity [GO:0015292]; actin filament reorganization [GO:0090527]; calcium import into the mitochondrion [GO:0036444]; calcium-mediated signaling [GO:0019722]; glucose homeostasis [GO:0042593]; mitochondrial calcium ion homeostasis [GO:0051560]; mitochondrial calcium ion transmembrane transport [GO:0006851]; positive regulation of insulin secretion [GO:0032024]; positive regulation of mitochondrial calcium ion concentration [GO:0051561]; positive regulation of mitochondrial fission [GO:0090141]; positive regulation of neutrophil chemotaxis [GO:0090023]; protein complex oligomerization [GO:0051259] | 18614015_CCDC109A was identified in this large-scale proteomics analysis as a mitochondrial protein broadly expressed in many mouse tissues. The human protein was also confirmed to localize to mitochondria. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21685886_MCU is an oligomeric protein residing in the mitochondrial inner membrane that interacts with MICU1 and is necessary for mitochondrial calcium uniport. 21685888_Functional characterization of the MCU protein; shows that it is necessary for mitochondrial calcium uptake. 22904319_the crucial role of MICU1 and MCU in mitochondrial Ca(2+) uptake in pancreatic beta-cells and their involvement in the positive feedback required for sustained insulin secretion. 23400777_The mitochondrial calcium uniporter (MCU): molecular identity and physiological roles. 23602897_study demonstrates that MCU overexpression is a feature of some breast cancers and that MCU overexpression may offer a survival advantage against some cell death pathways 23755363_analyses establish that MCU encodes the pore-forming subunit of the uniporter channel 24366263_We review not only the biochemical identities and structures of the proteins required for mitochondrial Ca2+ uptake but also their implications in different physiopathological contexts. [review] 24430870_SLC25A23 augments mitochondrial Ca(2) uptake, interacts with MCU, and induces oxidative stress-mediated cell death. 24815697_ERp57 can regulate the expression of the mitochondrial calcium uniporter (MCU) and modulate mitochondrial calcium uptake 25640838_MCU plays a critical role in breast cancer cell migration by regulating store-operated Ca(2) entry. 25753332_MCU-VDAC1 complex regulates mitochondrial Ca(2+) uptake and oxidative stress-induced apoptosis 25824785_Loss of heterozygosity of MCU gene on chromosome 10q is associated with pancreatic cancer. 25999421_Studies indicate the existence of an inner mitochondrial protein termed the mitochondrial calcium uniporter for calcium transport. 26341627_results suggest that N terminal domain of MCU is essential for the modulation of MCU function, although it does not affect the uniplex formation 26489515_Our experiments provide novel details about how MCU/EMRE is regulated by MICU1 and an original approach to investigate MCU/EMRE activation in intact cells. 26968367_The molecular structure and regulation of the MCU complex in addition to its pathophysiological role are discussed with particular attention to striated muscle tissues. Review. 27001609_Data suggest that MCU regulator (EMRE) might be a structural factor for opening of the mitochondrial calcium uniporter (MCU)-forming pore. 27099988_Here, the authors determine the transmembrane orientation of EMRE, and show that its known MCU-activating function is mediated by the interaction of transmembrane helices from both proteins. 27138568_MCU downregulation hampered cell motility and invasiveness and reduced tumor growth, lymph node infiltration, and lung metastasis in triple-negative breast cancer xenografts. In MCU-silenced cells, production of mitochondrial reactive oxygen species (mROS) is blunted and expression of the hypoxia-inducible factor-1alpha (HIF-1alpha) is reduced, suggesting a signaling role for mROS and HIF-1alpha, downstream of mitochon... 27288019_Study indicates that inhibition of mitochondrial calcium uniporter can inhibit excessive mitophagy and protect the neurocytes from ischemia/reperfusion injury. 27471128_decreased MCU expression in hypertensive with mutation cells contributed to dysregulated Ca(2+) uptake into the mitochondria 27627464_It has been shown that the propagation of the TRPV1-induced cytosolic calcium and sodium fluxes into mitochondria is dependent on coordinated activity of NCLX and MCU. 27642082_Mitochondrial Ca(2+) uptake is controlled by protein arginine methyl transferase 1 that asymmetrically methylates MICU1, resulting in decreased Ca(2+) sensitivity. UCP2/3 normalize Ca(2+) sensitivity of methylated MICU1 and, thus, re-establish mitochondrial Ca(2+) uptake activity. 28039397_The studies findings in aging human skeletal muscle confirm the data obtained in mice and propose mitochondrial calcium uniporter and mitochondria-related proteins as potential pharmacological targets to counteract age-related muscle loss. 28262504_Reveal a distinct functional role for Cys-97 in mitochondrial reactive oxygen species sensing and regulation of MCU activity. 28351840_Regulation of mitochondrial Ca(2+) suggests that MCU may play a pivotal role in the development of fibrosis and could potentially be a therapeutic target for pulmonary fibrosis. 28396416_The results highlight the dynamic nature of uniporter subunit assembly, which must be tightly regulated to ensure proper mitochondrial responses to intracellular Ca(2+) signals. 28650465_High MCU expression is associated with metastasis in hepatocellular carcinoma. 28777009_Mitochondrial calcium uniporter plays an important role in hyperglycaemia-induced endothelial cell dysfunction. 28790027_MCU expression returned to physiological levels in visceral adipose tissue of patients after weight loss by bariatric surgery. Altered mitochondrial calcium flux in fat cells may play a role in obesity and diabetes and may be associated with the differential metabolic profiles of visceral and subcutaneous adipose tissue. 29241542_MICU2 restricts spatial crosstalk between InsP3R and MCU channels by regulating threshold and gain of MICU1-mediated inhibition and activation of MCU. 29594867_VDAC1 allows Ca(2+) access to the MCU, facilitating transport of Ca(2+) to the matrix, and also from the IMS to the cytosol. Intra-mitochondrial Ca(2+) controls energy production and metabolism by modulating critical enzymes in the tricarboxylic acid (TCA) cycle and fatty acid oxidation. 30020827_Wild-type TRPM2 but not Ca(2+)-impermeable mutant E960D reconstituted phosphorylation and expression of Pyk2 and CREB in TRPM2-depleted cells exposed to doxorubicin. Results demonstrate that TRPM2 expression protects the viability of neuroblastoma through Src, Pyk2, CREB, and MCU activation, which play key roles in maintaining mitochondrial function and cellular bioenergetics 30082385_MICU1 imparts the mitochondrial uniporter with the ability to discriminate between Ca(2+) and Mn(2+). 30403999_MICU1 deletion sensitizes human cells to manganese-dependent cell death by disinhibiting MCU-mediated manganese uptake. 30638448_MICU1 confers Ca(2+)-dependent gating of the uniporter by blocking/unblocking MCU. 31473487_These observations suggest that macrophage MCU-mediated metabolic reprogramming contributes to fibrotic repair after lung injury. 31907514_MCU deficiency blocks cell proliferation by disrupting cytoplasmic Ca2+ transients due to altered Drp1 hospitalization. 32317369_Mitochondrial pyruvate and fatty acid flux modulate MICU1-dependent control of MCU activity. 32371956_MCU-induced mitochondrial calcium uptake promotes mitochondrial biogenesis and colorectal cancer growth. 32762847_Structural insights into the Ca(2+)-dependent gating of the human mitochondrial calcium uniporter. 32769116_Evolutionary divergence reveals the molecular basis of EMRE dependence of the human MCU. 32777646_Uncorking MCU to let the calcium flow. 32862359_Structure of intact human MCU supercomplex with the auxiliary MICU subunits. 33067576_MCU-dependent negative sorting of miR-4488 to extracellular vesicles enhances angiogenesis and promotes breast cancer metastatic colonization. 33296646_Mechanisms of EMRE-Dependent MCU Opening in the Mitochondrial Calcium Uniporter Complex. 34189138_MCU That Is Transcriptionally Regulated by Nrf2 Augments Malignant Biological Behaviors in Oral Squamous Cell Carcinoma Cells. 34499925_The Mitochondrial Ca(2+) uniporter is a central regulator of interorganellar Ca(2+) transfer and NFAT activation. 34847839_Dihydroartemisinin represses oral squamous cell carcinoma progression through downregulating mitochondrial calcium uniporter. | ENSMUSG00000009647 | Mcu | 4.985433e+02 | 0.6859845 | -0.543752175 | 0.3284791 | 2.718762e+00 | 0.0991749138 | 0.75783482 | No | Yes | 3.734431e+02 | 62.860332 | 5.826937e+02 | 100.073491 | |
| ENSG00000156170 | 137682 | NDUFAF6 | protein_coding | Q330K2 | FUNCTION: Involved in the assembly of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I) at early stages. May play a role in the biogenesis of complex I subunit MT-ND1. {ECO:0000269|PubMed:18614015, ECO:0000269|PubMed:22019594}. | Alternative splicing;Cytoplasm;Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Nucleus;Primary mitochondrial disease;Reference proteome;Transit peptide | This gene encodes a protein that localizes to mitochondria and contains a predicted phytoene synthase domain. The encoded protein plays an important role in the assembly of complex I (NADH-ubiquinone oxidoreductase) of the mitochondrial respiratory chain through regulation of subunit ND1 biogenesis. Mutations in this gene are associated with complex I enzymatic deficiency. [provided by RefSeq, Nov 2011]. | hsa:137682; | cytoplasm [GO:0005737]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; transferase activity [GO:0016740]; biosynthetic process [GO:0009058]; mitochondrial respiratory chain complex I assembly [GO:0032981] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 22019594_C8orf38 is a crucial factor required for the translation and/or integration of ND1 into an early-stage assembly intermediate 23509070_In a forward genetic screen to identify genes that cause neurodegeneration, we identified sicily, the Drosophila melanogaster homologue of human C8ORF38, the loss of which causes Leigh syndrome. 27466185_Affected kidney and lung showed specific loss of the mitochondria-located NDUFAF6 isoform and ultrastructural characteristics of mitochondrial dysfunction. Accordingly, affected tissues had defects in mitochondrial respiration and complex I biogenesis that were corrected with NDUFAF6 cDNA transfection. Our results demonstrate that the Acadian variant of Fanconi Syndrome results from mitochondrial respiratory chain complex 27623250_This paper confirms NDUFAF6 as a genuine morbid gene and proposes the coupling of exome sequencing with mRNA analysis as a method useful for enhancing the exome sequencing detection rate when the simple application of classical inheritance models fails. 28476317_NDUFAF6 encodes a complex I assembly factor and mutations result in complex I deficiency, Leigh syndrome or Acadian variant Fanconi syndrome. Human NDUFAF6 is a mitochondria-targeted 333-amino acid protein belonging to the family of squalene and phytoene synthases. 30642748_NDUFAF6-related Leigh syndrome is a relevant cause of childhood onset dystonia and isolated bilateral striatal necrosis [review] 35237031_Genetic Effects of NDUFAF6 rs6982393 and APOE on Alzheimer's Disease in Chinese Rural Elderly: A Cross-Sectional Population-Based Study. | ENSMUSG00000050323 | Ndufaf6 | 3.507909e+02 | 0.6989925 | -0.516651051 | 0.3121772 | 2.645759e+00 | 0.1038265593 | 0.75783482 | No | Yes | 2.521494e+02 | 35.160859 | 3.806716e+02 | 53.971718 | |
| ENSG00000156273 | 571 | BACH1 | protein_coding | O14867 | FUNCTION: Transcriptional regulator that acts as repressor or activator, depending on the context. Binds to NF-E2 DNA binding sites. Plays important roles in coordinating transcription activation and repression by MAFK (By similarity). Together with MAF, represses the transcription of genes under the control of the NFE2L2 oxidative stress pathway (PubMed:24035498). {ECO:0000250|UniProtKB:P97302, ECO:0000269|PubMed:24035498}. | 3D-structure;Activator;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a transcription factor that belongs to the cap'n'collar type of basic region leucine zipper factor family (CNC-bZip). The encoded protein contains broad complex, tramtrack, bric-a-brac/poxvirus and zinc finger (BTB/POZ) domains, which is atypical of CNC-bZip family members. These BTB/POZ domains facilitate protein-protein interactions and formation of homo- and/or hetero-oligomers. When this encoded protein forms a heterodimer with MafK, it functions as a repressor of Maf recognition element (MARE) and transcription is repressed. Multiple alternatively spliced transcript variants have been identified for this gene. [provided by RefSeq, May 2009]. | hsa:571; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA polymerase II transcription regulator complex [GO:0090575]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; heme binding [GO:0020037]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; DNA repair [GO:0006281]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription from RNA polymerase II promoter in response to hypoxia [GO:0061418]; regulation of transcription involved in G1/S transition of mitotic cell cycle [GO:0000083]; regulation of transcription involved in G2/M transition of mitotic cell cycle [GO:0000117]; regulation of transcription, DNA-templated [GO:0006355] | 12511571_Bach1 functions as a hypoxia-inducible repressor for the HO-1 gene, thereby contributing to fine-tuning of oxygen homeostasis in human cells 14504288_nuclear export of Bach1 constitutes an important regulatory mechanism to relieve the Bach1-mediated repression of genes such as heme oxygenase 1 14660636_Bach1 binding to the Maf recognition element in the microlocus control region is blocked by heme in the beta-globin regulation pathway 15068237_BACH1 plays a role in the development of Alzheimer's disease (AD)-like neuropathology in Down syndrome and in pathogenesis of AD per se. 15068251_BACH1 protein is significantly overexpressed in fetal Down syndrome cerebral cortex and may contribute to abnormal brain development or at least to defective transcription machinery in Down syndrome. 15464985_These results indicated that heme plays an important role in the induction of alpha-globin gene expression through disrupting the interaction of Bach1 and the NA site in HS-40 enhancer in erythroid cells. 15465821_Bach-1 has a specific and selective ability to repress expression of hepatic heme oxygenase-1 15613547_BACH1 acts as a transcriptional repressor in the regulation of Maf recognition-element-dependent genes in megakaryocytes. 16530877_key role of down-regulation of Bach1 and up-regulation of heme oxygenase 1 in diminishing cytotoxic effects of hepatitis C virus proteins in human hepatocytes. 16724942_Reviewer presents the Bach1-heme oxygenase(HO)-1 system as an important defense mechanism against oxidative stress--a mechanism in which Bach1 is a critical regulator of HO-1. 16771696_Data show that beta-carotene, combined with cigarette smoke condensate (TAR), regulates heme oxygenase-1 (HO-1) via its transcriptional factor Bach1 and modulates cell growth. 17065227_The effect of Bach1 and Nrf2 on heme oxygenase 1 expression via cobalt protoporphyrin in human liver cells is reported. 17881434_BACH-1 protein levels are low in cells expressing either the human miR-155 or miR-K12-11 of Kaposi's sarcoma-associated herpesvirus. 18550526_Loss of BACH1 function in human keratinocytes results almost exclusively in HMOX1 induction 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19591297_Bach1 inhibits expression of oxidative stress responsive genes by competing with Nrf2, the key activator of oxidative stress response. Bach1 inhibits p53-dependent cellular senescence induced by oxidative stress 19618874_regulates oxidative stress response in cellular senescence. (review) 19822148_ERK(1/2) and JNK are involved in cigarette smoking-induced biphasic HO-1 expression by a specific regulation of Nrf2/Keap1-Bach1. 19874887_prevents radiation-induced upregulation of heme oxygenase 1 gene expression in keratinocytes 20127796_miR-196 directly acts on the 3'-UTR of Bach1 messenger RNA and translationally represses the expression of this protein. 20345481_Bach1 plays a critical role in regulating HO-1 gene expression in AML cells and its expression suppresses their survival by downregulating HO-1 expression. 21555518_The BACH1 target genes in HEK 293 cells are involved in heme degradation, redox regulation, cell cycle/apoptosis pathways and subcellular transport processes. 21555518_The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. 21812759_We conclude that BACH1 is a bona fide Nrf2 target gene and that induction of BACH1 by Nrf2 may serve as a feedback-inhibitory mechanism for antioxidant-response-element-mediated gene regulation. 22289179_Bach1 depletion resulted in disordered mitotic chromosome alignment, which was rescued by Bach1 mutants lacking the BTB or DNA binding domains, suggesting its transcription-independent mechanism. 22307849_miR-155 may function as an oncogene by targeting BACH1 22698995_let-7 miRNA directly acts on the 3'-UTR of Bach1 and negatively regulates expression of this protein, and thereby up-regulates HMOX1 gene expression. 23455180_In colorectal cancer, Nrf2 expression is closely correlated with Keap1 in the tumor and with Bach1 in the normal tissue. 23737527_Data indicate that transcription factors MafK and Bach1 regulate expression of heme oxygenase-1 (HO-1). 23738048_The arsenic-induced Nrf2 pathway activation in hepatocytes suggested that the translocation of Bach1 was associated with the regulation of Nrf2 pathway by arsenic. 24035498_Identify a role for SCF(FBXL17) in controlling the threshold for NRF2-dependent gene activation via BACH1 repressor turnover. 24366869_CXCR3-B mediates a growth-inhibitory signal in breast cancer cells through the modulations of nuclear translocation of Bach-1 and Nrf2 and down-regulation of HO-1. 24395801_BACH1 acts in a double-negative (overall positive) feedback loop to inhibit RKIP transcription in breast cancer cells 24613679_The Bach1-dependent repression of the HO-1 expression is under the control of the Hx-dependent uptake of extracellular heme 24752012_Higher HMOX1 expression correlated with higher expression of Bach-1 (Spearman's rho = 0.586, p = 0.000001) and miR-122 (Spearman's rho = 0.270, p = 0.014059). 25391381_This study demonstrated that Bach1 overexpression in Down syndrome correlates with the alteration of the HO-1/BVR-a system. 26123998_Bach1 suppresses angiogenesis after ischemic injury and impairs Wnt/beta-catenin signaling by disrupting the interaction between beta-catenin and TCF4 and by recruiting histone deacetylase 1 to the promoter of TCF4-targeted genes. 26244607_sensitizer-induced up-regulation of both the endogenous HMOX1 and the luciferase constructs under the control of the HMOX1-ARE or the full HMOX1 promoter appear to be under the control of both Nrf2 and Bach1. 26377036_This study indicates that the expression of BACH1 could be elevated as a compensatory mechanism to decrease the globin chain imbalance as well as to reduce the oxidative stress found in hemoglobin E/beta-thalassemia. 26422990_Cyanidin-3-O-glucoside protects HUVECs from palmitic acid-induced injuryby modulating the balance of Nrf2 versus Bach1 inside the nucleus so influencing upregulation of electrophile responsive element mediated gene expression. 26445536_heme oxygenase-1 expression is induced by gold nanoparticles through Nrf2 activation and Bach1 export in human vascular endothelial cells 26698668_Induction of GSH-related genes xCT and GCLM were oxygen and Bach1-insensitive during long-term culture under 5% O2, providing the first evidence that genes related to GSH synthesis mediate protection afforded by Nrf2-Keap1 defense pathway 27057283_Bach1 suppresses cell proliferation and induces cell-cycle arrest and apoptosis. 27108804_A short while ago some studies suggested that BACH1 is involved in cancers, especially breast cancer. This factor is mentioned as a novel master regulator which adjusts several genes involved in bone metastasis of breast cancer, especially CXCR4 and MMP1, two main genes in the enhancement of cancer cell migration and invasion to distant organs. [Review] 27657827_BACH1 down-regulation in HT29 colon cancer cells had no effect on cell growth but did inhibit cell migration by decreasing metastasis-related genes expression. 28000777_BACH1 overexpression impaired the association between p53 and SP1 via competitive binding p53, and antagonized the impact of p53 on MGMT expression. 28349828_Our results suggest that the bach1-specific small interfering RNA effectively decrease CXCR4 receptor, matrix metalloproteinase-9 expression and breast adenocarcinoma cells invasive, also increased the expression of tumor-suppressive microRNA-203 and miR-145 28645578_The electrophilic character of quinones ensure their conjugation with Bach1, which is important for the downregulation of Bach1 and the upregulation of Nrf2 signaling. 28685309_these findings suggested that increased miR-155 expression in activated monocytes leads to enhanced phagocytic activity via BACH-1 regulation in beta-thalassemia/HbE. 28790431_In conclusion, these findings highlight the central role of Bach1 in HO-1-dependent neuronal response to oxidative stress. 28889753_BACH1 is overexpressed in prostate cancer 29125538_In light of a pivotal role of NRF2 and BACH1 in response to oxidative stress and regulation of HO-1, we examined if smoke-induced HO-1 expression is modulated through the NRF2/BACH1 axis. We demonstrated that smoke causes significant nuclear translocation of NRF2, but only a slight decrease in nuclear BACH1. 29459360_BACH1 performs stabilization of mitotic spindle orientation together with HMMR and CRM1 in mitosis, and that the cell cycle-specific phosphorylation switches the transcriptional and mitotic functions of BACH1. 29481800_BACH1 may inhibit the progression of colorectal cancer through BACH1/CXCR4 pathway. 29930735_The authors found substantially lower BACH1 expression in PDAC compared with normal pancreatic tissues and the rs372883T allele had significantly lower BACH1 levels than the rs372883C allele in both tumor and normal tissues. 30250186_Anemia of inflammation and myelodysplastic syndrome might involve reduced activity of Bach 1 and Bach2 30316884_This is the first study to explore the relationship between tagSNPs of BACH1 and ATDH in a Chinese cohort. Based on this cohort, genetic polymorphisms of BACH1 may be associated with susceptibility to ATDH in the Chinese population. 30480817_These results revealed that miR-142-3p could target Bach-1 in breast cancer cells leading to the reduction of EMT-related proteins and reduced cell proliferation, invasion, and migration; the results also demonstrated that miR-142-3p could regulate important tumor suppressor miRNAs in breast cancer cells. 30597358_Nfr2 and Bach1 expression are altered in colorectal carcinoma but in different ways 30654010_Study identified Bach1 as a key regulator that controls multiple factors essential for epithelial ovarian cancer (EOC) metastasis and growth. Bach1 promotes EMT gene expression by recruiting HMGA2 in EOC cells to promote EMT and ovarian cancer metastasis. 30706605_A proposed model indicates a key role for miR-let-7c targeting Bach1 to transactivate HO-1-mediated antiviral actions against hepatitis C virus . 30825204_Combined targeting of HMGA2 and Bach1 may be an effective therapeutic strategy to treat breast cancer. 30842661_BACH1 gene expression inversely correlates with electron transport chain gene expression in tumours from patients with breast cancer and in other tumour types; mitochondrial metabolism can be exploited by targeting BACH1 to sensitize breast cancer and potentially other tumour tissues to mitochondrial inhibitors 30891497_It has been reported that the Usp7 is a direct target of Bach1; that Bach1 interacts with Nanog, Sox2, and Oct4, and that Bach1 facilitates their deubiquitination and stabilization via the recruitment of Usp7, thereby maintaining stem cell identity and self-renewal. 31067541_results suggested that the IL-16 rs859, CYP19A1 rs4646, and BACH1 rs372883 polymorphisms have potential roles in the genetic susceptibility to IgAN in Chinese Han population. 31257027_BACH1 activates transcription of Hexokinase 2 and Gapdh and increases glucose uptake, glycolysis rates, and lactate secretion, thereby stimulating glycolysis-dependent metastasis of mouse and human lung cancer cells. 31591481_Oncogenic HOXB8 is driven by MYC-regulated super-enhancer and potentiates colorectal cancer invasiveness via BACH1. 31919242_BACH1 Promotes Pancreatic Cancer Metastasis by Repressing Epithelial Genes and Enhancing Epithelial-Mesenchymal Transition. 31939443_this study demonstrated that BACH1 and MALAT1 are upregulated synchronously in patients with TNBC who had poor clinical outcomes. Our results indicate that BACH1 and MALAT1 could be important prognostic factors and potential therapeutic targets in TNBC. 32003018_CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. 32132179_BACH family members regulate angiogenesis and lymphangiogenesis by modulating VEGFC expression. 32228546_LncRNA MIR17HG inhibits non-small cell lung cancer by upregulating miR-142-3p to downregulate Bach-1. 32344103_MicroRNA-based regulatory mechanisms underlying the synergistic antioxidant action of quercetin and catechin in H2O2-stimulated HepG2 cells: Roles of BACH1 in Nrf2-dependent pathways. 32390598_Circular RNA circ_0000337 contributes to osteosarcoma via the miR-4458/BACH1 pathway. 32534959_Defective BACH1/HO-1 regulatory circuits in cystic fibrosis bronchial epithelial cells. 32707301_Circ_0123996 promotes cell proliferation and fibrosisin mouse mesangial cells through sponging miR-149-5p and inducing Bach1 expression. 33596919_Chronic intermittent hypoxia promoted lung cancer stem cell-like properties via enhancing Bach1 expression. 33831787_CircRNA circBACH1 (hsa_circ_0061395) serves as a miR-656-3p sponge to facilitate hepatocellular carcinoma progression through increasing SERBP1 expression. 33878446_BACH1 is transcriptionally inhibited by TET1 in hepatocellular carcinoma in a microRNA-34a-dependent manner to regulate autophagy and inflammation. 33932125_BACH1 promotes the progression of esophageal squamous cell carcinoma by inducing the epithelial-mesenchymal transition and angiogenesis. 34160287_BACH1 Binding Links the Genetic Risk for Severe Periodontitis with ST8SIA1. 34339740_The transcription factor BACH1 at the crossroads of cancer biology: From epithelial-mesenchymal transition to ferroptosis. 34390174_Circ_0087862 promotes the progression of colorectal cancer by sponging miR-142-3p and up-regulating BACH1 expression. 34464885_MicroRNA-532-5p upregulation protects neurological deficits after ischemic stroke through inhibition of BTB and CNC homology 1. 34482423_Regulatory mechanisms of heme regulatory protein BACH1: a potential therapeutic target for cancer. 34600339_Exosomes derived from mesenchyml stem cells ameliorate oxygen-glucose deprivation/reoxygenation-induced neuronal injury via transferring MicroRNA-194 and targeting Bach1. 34605540_BACH1, the master regulator of oxidative stress, has a dual effect on CFTR expression. 34737234_Bach1 derepression is neuroprotective in a mouse model of Parkinson's disease. 34863556_A heme-regulatable chemodynamic nanodrug harnessing transcription factor Bach1 against lung cancer metastasis. 34923423_BACH1 as a potential target for immunotherapy in glioblastomas. 34949193_BTB and CNC homology 1 (Bach1) induces lung cancer stem cell phenotypes by stimulating CD44 expression. 35067159_Knockdown of long non-coding RNA SNHG8 suppresses the progression of esophageal cancer by regulating miR-1270/BACH1 axis. 35154476_Overexpression of BACH1 mediated by IGF2 facilitates hepatocellular carcinoma growth and metastasis via IGF1R and PTK2. 35283228_CAPE and its synthetic derivative VP961 restore BACH1/NRF2 axis in Down Syndrome. 35406740_A Heme-Binding Transcription Factor BACH1 Regulates Lactate Catabolism Suggesting a Combined Therapy for Triple-Negative Breast Cancer. | ENSMUSG00000025612 | Bach1 | 1.703307e+02 | 1.1578829 | 0.211489369 | 0.3826914 | 3.005211e-01 | 0.5835559466 | 0.90371250 | No | Yes | 1.985280e+02 | 35.397757 | 1.685832e+02 | 30.818276 | |
| ENSG00000156599 | 25921 | ZDHHC5 | protein_coding | Q9C0B5 | FUNCTION: Palmitoyltransferase that catalyzes the addition of palmitate onto various protein substrates such as CTNND2, CD36, STAT3 and S1PR1 thus plays a role in various biological processes including cell adhesion, fatty acid uptake, bacterial sensing or cardiac functions (PubMed:21820437, PubMed:29185452, PubMed:31402609). Plays an important role in the regulation of synapse efficacy by mediating palmitoylation of delta-catenin/CTNND2, thereby increasing synaptic delivery and surface stabilization of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs). Under basal conditions, remains at the synaptic membrane through FYN-mediated phosphorylation that prevents association with endocytic proteins (PubMed:26334723). Neuronal activity enhances the internalization and trafficking of DHHC5 from spines to dendritic shafts where it palmitoylates delta-catenin/CTNND2 (PubMed:26334723). Regulates cell adhesion at the plasma membrane by palmitoylating GOLGA7B and DSG2 (PubMed:31402609). Plays a role in innate immune response by mediating the palmitoylation of NOD1 and NOD2 and their proper recruitment to the bacterial entry site and phagosomes (PubMed:31649195). Participates also in fatty acid uptake by palmitoylating CD36 and thereby targeting it to the plasma membrane. Upon binding of fatty acids to CD36, gets phosphorylated by LYN leading to inactivation and subsequent CD36 caveolar endocytosis (PubMed:32958780). Controls oligodendrocyte development by catalyzing STAT3 palmitoylation (By similarity). {ECO:0000250|UniProtKB:Q8VDZ4, ECO:0000269|PubMed:21820437, ECO:0000269|PubMed:26334723, ECO:0000269|PubMed:29185452, ECO:0000269|PubMed:31402609, ECO:0000269|PubMed:31649195, ECO:0000269|PubMed:32958780}. | Acyltransferase;Alternative splicing;Cell junction;Cell membrane;Immunity;Innate immunity;Lipid transport;Lipoprotein;Membrane;Methylation;Palmitate;Phosphoprotein;Reference proteome;Synapse;Transferase;Transmembrane;Transmembrane helix;Transport | hsa:25921; | dendrite [GO:0030425]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; phagocytic vesicle [GO:0045335]; plasma membrane [GO:0005886]; palmitoyltransferase activity [GO:0016409]; protein-cysteine S-palmitoyltransferase activity [GO:0019706]; positive regulation of pattern recognition receptor signaling pathway [GO:0062208]; positive regulation of protein localization to phagocytic vesicle [GO:1905171]; positive regulation of protein localization to plasma membrane [GO:1903078]; protein palmitoylation [GO:0018345] | 19801377_Data show that palmitoyl acyltransferases DHHC5, DHHC6, and DHHC8 appear to be S-acylated on three cysteine residues within a novel CCX(7-13)C(S/T) motif downstream of a conserved Asp-His-His-Cys cysteine-rich domain. 21820437_ZDHHC5 and SSTR5 are colocalized at the plasma membrane and coexpression of ZDHHC5 increased palmitoylation of SSTR5 whereas knock-down of endogenous ZDHHC5 by siRNAs decreased it. 25573953_Data indicate that DHHC5 has oncogenic capacity and contributes to tumor formation in non-small cell lung cancer. 28739689_We demonstrated that LXR stimulation decreases mRNA and protein expression of FLOT2 and DHHC5 in MCF-7 cells. LXR stimulation also reduces Akt phosphorylation and its localization at the plasma membrane 28775165_High ZDHHC5 expression is associated with Glioma. 30605677_DHHC4 and DHHC5 function at different subcellular localizations to control the palmitoylation, plasma membrane localization, and fatty acid uptake activity of the scavenger receptor CD36. 31402609_This work uncovers a novel mechanism of DHHC5 regulation by Golga7b and demonstrates a role for the DHHC5/Golga7b complex in the regulation of cell adhesion. 31547976_DHHC5 Mediates beta-Adrenergic Signaling in Cardiomyocytes by Targeting Galpha Proteins. 31649195_ZDHHC5-mediated S-palmitoylation is indispensable for NOD1/2 recruitment to bacteria containing phagosomes. 31885423_Study findings identified six miRNAs related to gastric adenocarcinoma prognosis and suggested that downregulated miR-96-5p might induce cell apoptosis via upregulating ZDHHC5 expression in MGC-803 cells. 32737405_Control of protein palmitoylation by regulating substrate recruitment to a zDHHC-protein acyltransferase. 33415776_Regulation and function of the palmitoyl-acyltransferase ZDHHC5. 34961524_The interactions of ZDHHC5/GOLGA7 with SARS-CoV-2 spike (S) protein and their effects on S protein's subcellular localization, palmitoylation and pseudovirus entry. | ENSMUSG00000034075 | Zdhhc5 | 4.966117e+03 | 0.9543236 | -0.067449568 | 0.3024575 | 5.039116e-02 | 0.8223840227 | 0.96628438 | No | Yes | 4.060659e+03 | 383.138671 | 4.498595e+03 | 435.029771 | ||
| ENSG00000156639 | 60685 | ZFAND3 | protein_coding | Q9H8U3 | 3D-structure;Metal-binding;Reference proteome;Zinc;Zinc-finger | hsa:60685; | DNA binding [GO:0003677]; zinc ion binding [GO:0008270] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 27862917_Replication of lead type 2 diabetes mellitus SNPs in GLIS3, KCNK16, and ZFAND3 was observed in American Indians. Sex-specific T2DM signals in GLIS3 and ZFAND3, which are distinct from the East Asian GWAS signals, were also identified. 28580277_ZFAND3 was identified as a type 2 diabetes susceptibility gene. ZFAND3 plays a role in insulin secretion in vitro. 33311477_AN1-type zinc finger protein 3 (ZFAND3) is a transcriptional regulator that drives Glioblastoma invasion. | ENSMUSG00000044477 | Zfand3 | 1.788671e+03 | 1.1645750 | 0.219803590 | 0.3190612 | 4.911868e-01 | 0.4833983663 | 0.87379319 | No | Yes | 1.811971e+03 | 220.602379 | 1.361012e+03 | 170.405295 | |||
| ENSG00000156983 | 7862 | BRPF1 | protein_coding | P55201 | FUNCTION: Scaffold subunit of various histone acetyltransferase (HAT) complexes, such as the MOZ/MORF and HBO1 complexes, which have a histone H3 acetyltransferase activity (PubMed:16387653, PubMed:24065767, PubMed:27939640). Plays a key role in HBO1 complex by directing KAT7/HBO1 specificity towards histone H3 'Lys-14' acetylation (H3K14ac) (PubMed:24065767). Some HAT complexes preferentially mediate histone H3 'Lys-23' (H3K23ac) acetylation (PubMed:27939640). Positively regulates the transcription of RUNX1 and RUNX2 (PubMed:18794358). {ECO:0000269|PubMed:16387653, ECO:0000269|PubMed:18794358, ECO:0000269|PubMed:24065767, ECO:0000269|PubMed:27939640}. | 3D-structure;Acetylation;Activator;Alternative splicing;Bromodomain;Chromatin regulator;Chromosome;Cytoplasm;DNA-binding;Disease variant;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a bromodomain, PHD finger and chromo/Tudor-related Pro-Trp-Trp-Pro (PWWP) domain containing protein. The encoded protein is a component of the MOZ/MORF histone acetyltransferase complexes which function as a transcriptional regulators. This protein binds to the catalytic MYST domains of the MOZ and MORF proteins and may play a role in stimulating acetyltransferase and transcriptional activity of the complex. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:7862; | chromosome [GO:0005694]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; MOZ/MORF histone acetyltransferase complex [GO:0070776]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; acetyltransferase activator activity [GO:0010698]; DNA binding [GO:0003677]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; histone H3 acetylation [GO:0043966]; histone H3-K14 acetylation [GO:0044154]; histone H3-K23 acetylation [GO:0043972]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription, DNA-templated [GO:0006355] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20400950_Identified is the PWWP domain of bromo and plant homeodomain (PHD) finger-containing protein 1 (BRPF1) as a histone H3 (H3K36me3) binding module; determined is the structure of this domain in complex with an H3K36me3-derived peptide. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 24258712_MOZ-TIF2/BRPF1 complex upregulates HOX genes mediated by MOZ-dependent histone acetylation, leading to the development of leukemia. 24333487_novel interactions of the BRPF1 bromodomain with multiple acetyllysine residues on the N-terminus of histones show that it preferentially selects for H2AK5ac, H4K12ac, and H3K14ac 25281266_critical insights into the molecular mechanism of ligand binding by the BRPF1 bromodomain 27939639_comparison of the clinical symptoms of individuals carrying mutations or small deletions of BRPF1 alone or SETD5 alone with those of individuals with deletions encompassing both BRPF1 and SETD5; leads to conclusion that both genes contribute to the phenotypic severity of 3p25 deletion syndrome but that some specific features, such as ptosis and blepharophimosis, are mostly driven by BRPF1 haploinsufficiency 27939640_data indicate that aberrations in the chromatin regulator gene BRPF1 cause histone H3 acetylation deficiency and a previously unrecognized intellectual disability syndrome 31711755_Molecular Basis for the PZP Domain of BRPF1 Association with Chromatin. 31851932_Truncated BRPF1 Cooperates with Smoothened to Promote Adult Shh Medulloblastoma. 34285329_Bromodomain-containing protein BRPF1 is a therapeutic target for liver cancer. | ENSMUSG00000001632 | Brpf1 | 3.738080e+03 | 1.1579922 | 0.211625539 | 0.2932585 | 5.254265e-01 | 0.4685360174 | 0.86818070 | No | Yes | 3.535324e+03 | 235.917123 | 3.013077e+03 | 206.620354 | |
| ENSG00000157110 | 11030 | RBPMS | protein_coding | Q93062 | FUNCTION: Acts as a coactivator of transcriptional activity. Required to increase TGFB1/Smad-mediated transactivation. Acts through SMAD2, SMAD3 and SMAD4 to increase transcriptional activity. Increases phosphorylation of SMAD2 and SMAD3 on their C-terminal SSXS motif, possibly through recruitment of TGFBR1. Promotes the nuclear accumulation of SMAD2, SMAD3 and SMAD4 proteins (PubMed:26347403). Binds to poly(A) RNA (PubMed:17099224, PubMed:26347403). {ECO:0000269|PubMed:17099224, ECO:0000269|PubMed:26347403}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transcription;Transcription regulation | This gene encodes a member of the RNA recognition motif family of RNA-binding proteins. The RNA recognition motif is between 80-100 amino acids in length and family members contain one to four copies of the motif. The RNA recognition motif consists of two short stretches of conserved sequence, as well as a few highly conserved hydrophobic residues. The encoded protein has a single, putative RNA recognition motif in its N-terminus. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jun 2013]. | hsa:11030; | cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; P-body [GO:0000932]; mRNA binding [GO:0003729]; poly(A) binding [GO:0008143]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; transcription coactivator activity [GO:0003713]; positive regulation of pathway-restricted SMAD protein phosphorylation [GO:0010862]; positive regulation of SMAD protein signal transduction [GO:0060391]; response to oxidative stress [GO:0006979]; RNA processing [GO:0006396] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25281386_RBPMS1 is a critical repressor of AP-1 signaling and RBPMS1 activation may be a useful strategy for cancer treatment. 27166999_Data indicate RNA Binding Protein with Multiple Splicing (RBPMS), Regulator of Chromosome Condensation and POZ Domain Containing Protein 1 (RCBTB1), and Zinc Finger protein 608 (ZNF608) as miR-21-3p target genes. 27273514_ERG is recruited to mRNAs via interaction with the RNA-binding protein RBPMS, and it promotes mRNA decay by binding CNOT2, a component of the CCR4-NOT deadenylation complex. 27592836_Study indicates that the RNA binding increases the stability of RNA-recognition motif (RRM) in RBPMS domain, but residue mutations of RRM domain induce the fluctuation of complex systems through weakening of hydrogen bonds, conformational change or loss of binding affinity. 28003515_Conserved binding of GCAC motifs by MEC-8, couch potato, and the RBPMS protein family has been reported. 28017375_observation of a strong in vivo erythropoietic effect for RBPMS but not for GTF2E2, supporting the statistical fine-mapping at this locus and demonstrating that RBPMS is a regulator of erythropoiesis 29423656_The possible involvement of the GC box 1 at position - 54 in transcriptional regulation of Rbpms was corroborated by EMSA, which showed formation of a DNA-protein complex in the presence of the oligonucleotide corresponding to this Sp1-binding site. 29542167_the key role of miR-21-3p in CRC 29743723_RBPMS silencing confers resistance to MM cells. 35008958_Reduced RBPMS Levels Promote Cell Proliferation and Decrease Cisplatin Sensitivity in Ovarian Cancer Cells. | ENSMUSG00000031586 | Rbpms | 2.267290e+02 | 1.6853216 | 0.753023941 | 0.3580620 | 4.433758e+00 | 0.0352348562 | 0.60993432 | No | Yes | 2.426778e+02 | 30.537738 | 1.423235e+02 | 18.858335 | |
| ENSG00000157152 | 6854 | SYN2 | protein_coding | Q92777 | FUNCTION: Neuronal phosphoprotein that coats synaptic vesicles, binds to the cytoskeleton, and is believed to function in the regulation of neurotransmitter release. May play a role in noradrenaline secretion by sympathetic neurons (By similarity). {ECO:0000250}. | Alternative splicing;Cell junction;Phosphoprotein;Reference proteome;Schizophrenia;Synapse | This gene is a member of the synapsin gene family. Synapsins encode neuronal phosphoproteins which associate with the cytoplasmic surface of synaptic vesicles. Family members are characterized by common protein domains, and they are implicated in synaptogenesis and the modulation of neurotransmitter release, suggesting a potential role in several neuropsychiatric diseases. This member of the synapsin family encodes a neuron-specific phosphoprotein that selectively binds to small synaptic vesicles in the presynaptic nerve terminal. Polymorphisms in this gene are associated with abnormal presynaptic function and related neuronal disorders, including autism, epilepsy, bipolar disorder and schizophrenia. Alternative splicing of this gene results in multiple transcript variants. The tissue inhibitor of metalloproteinase 4 gene is located within an intron of this gene and is transcribed in the opposite direction. [provided by RefSeq, Feb 2014]. | hsa:6854; | glutamatergic synapse [GO:0098978]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; Schaffer collateral - CA1 synapse [GO:0098685]; SNARE complex [GO:0031201]; synapse [GO:0045202]; synaptic vesicle membrane [GO:0030672]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; calcium-ion regulated exocytosis [GO:0017156]; chemical synaptic transmission [GO:0007268]; neurotransmitter secretion [GO:0007269]; synaptic vesicle clustering [GO:0097091] | 15271586_Observational study of gene-disease association. (HuGE Navigator) 15271586_This study suggests a positive association between synapsin II and schizophrenia, implying that synapsin II is involved in the etiology of schizophrenia. 15449241_synapsin II variants are associated with susceptibility to schizophrenia. 16131404_Observational study of gene-disease association. (HuGE Navigator) 17766091_Syn2 is likely to be involved in the etiology or pathogenesis of schizophrenia. 17913586_Meta-analysis of gene-disease association. (HuGE Navigator) 19665806_A case-control study with synapsin II was conducted in 506 bipolar disorder patients and 507 healthy individuals from the Han Chinese population. No association was found in this study. 19665806_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20034013_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20034013_Results from our study indicate the involvement of SYN2 gene polymorphism in conferring risk to epilepsy; however, the genetic variant does not seem to modulate drug-response in epilepsy pharmacotherapy. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21465568_SYN2 rs3773364 A>G polymorphism is not a risk factor for susceptibility to epilepsy in this case-control study and meta-analysis. 22384280_Synapsin II is involved in the molecular pathway of lithium treatment in bipolar disorder 23956174_Results identify SYN2 as a novel predisposing gene for autism spectrum disorders (ASD) and strengthen the hypothesis that a disturbance of synaptic homeostasis underlies ASD. 25088614_Both GABRA6 and Synapsin II polymorphisms are important risk factors for the development of idiopathic generalized epilepsy in a South Indian population. 27515700_These findings contribute to previous work showing dysregulation of Synapsins, particularly SYN2, in mood disorders and improve our understanding of the regulatory mechanisms that precipitate these changes likely leading to the BD or MDD phenotype. 29763751_Our findings support that ASB16-AS1 and SYN2 may represent two novel functional genes underlying bone mineral density variation | ENSMUSG00000009394 | Syn2 | 4.520082e+02 | 1.5686603 | 0.649532945 | 0.3011737 | 4.776516e+00 | 0.0288504349 | 0.57061453 | No | Yes | 4.329429e+02 | 60.063094 | 2.395792e+02 | 34.360915 | |
| ENSG00000157764 | 673 | BRAF | protein_coding | P15056 | FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Allosteric enzyme;Cardiomyopathy;Cell membrane;Chromosomal rearrangement;Cytoplasm;Deafness;Direct protein sequencing;Disease variant;Ectodermal dysplasia;Isopeptide bond;Kinase;Membrane;Mental retardation;Metal-binding;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Serine/threonine-protein kinase;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein belonging to the RAF family of serine/threonine protein kinases. This protein plays a role in regulating the MAP kinase/ERK signaling pathway, which affects cell division, differentiation, and secretion. Mutations in this gene, most commonly the V600E mutation, are the most frequently identified cancer-causing mutations in melanoma, and have been identified in various other cancers as well, including non-Hodgkin lymphoma, colorectal cancer, thyroid carcinoma, non-small cell lung carcinoma, hairy cell leukemia and adenocarcinoma of lung. Mutations in this gene are also associated with cardiofaciocutaneous, Noonan, and Costello syndromes, which exhibit overlapping phenotypes. A pseudogene of this gene has been identified on the X chromosome. [provided by RefSeq, Aug 2017]. | hsa:673; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; identical protein binding [GO:0042802]; MAP kinase kinase activity [GO:0004708]; MAP kinase kinase kinase activity [GO:0004709]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; scaffold protein binding [GO:0097110]; animal organ morphogenesis [GO:0009887]; cellular response to calcium ion [GO:0071277]; epidermal growth factor receptor signaling pathway [GO:0007173]; establishment of protein localization to membrane [GO:0090150]; MAPK cascade [GO:0000165]; negative regulation of apoptotic process [GO:0043066]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of gene expression [GO:0010628]; positive regulation of glucose transmembrane transport [GO:0010828]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; protein phosphorylation [GO:0006468]; trehalose metabolism in response to stress [GO:0070413] | 8621729_MEK1 interacts with B-Raf. 12068308_somatic missense mutations in 66% of malignant melanomas and at lower frequency in a wide range of human cancers 12198537_BRAF mutations in colorectal cancers occur only in tumours that do not carry mutations in a RAS gene known as KRAS, and BRAF mutation is linked to the proficiency of these tumours in repairing mismatched bases in DNA 12447372_High frequency of BRAF mutations in nevi 12619120_The V599E BRAF mutation appears to be a somatic mutation associated with melanoma development and/or progression in a proportion of affected individuals. 12644542_results demonstrate that the mutational status of BRAF and KRAS is distinctly different among histologic types of ovarian serous carcinoma, occurring most frequently in invasive micropapillary serous carcinomas and its precursors, serous borderline tumors 12670889_High prevalence of BRAF mutations in thyroid cancer is genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. 12697856_activating BRAF mutations may be an important event in the development of papillary thyroid cancer 12753285_cAMP activates ERK and increases proliferation of autosomal dominant polycystic kindey epithelial cells through the sequential phosphorylation of PKA, B-Raf and MAPK in a pathway separate from the classical receptor tyrosine kinase cascade 12778069_gene is mutated in skin melanoma, but not in uveal melanomas 12810628_13 germline BRAF variants, 4 of which were silent mutations in coding regions & 9 nucleotide substitutions in introns, were found in melanoma patients and melanoma family, but none appeared statistically likely to be a melanoma susceptibility gene. 12821662_B-raf is involved in adhesion-independent ERK1/2 signaling in melanocytes 12824225_Data suggest that BRAF T1796A activating mutation is not common in primary uveal melanoma. 12855697_B-Raf has a role in extracellular signal-regulated kinase (ERK) signaling in T cells and prevents antigen-presenting cell-induced anergy 12879021_BRAF has a role in in squamous cell carcinoma of the head and neck through uncommon mutations 12881714_The BRAF(V599E) mutation appears to be an alternative event to RET/PTC rearrangement rather than to RAS mutations, which are rare in PTC. BRAF(V599E) may represent an alternative pathway to oncogenic MAPK activation in PTCs without RET/PTC activation. 12893203_Mucinous ovarian cancers without a KRAS mutation have not sustained alternative activation of this signaling pathway through mutation of the BRAF oncogene. 12917419_3 cell lines derived from human choroidal melanoma express B-Raf containing the V599E mutation and showed a 10-fold increase in endogenous B-RafV599E kinase activity and a constitutive activation of the MEK/ERK pathway that is independent of Ras 12931219_Mutations are not detectable in plasma cell leukemia and multiple myeloma. 12970315_mutation of BRAF gene could be a potentially useful marker of prognosis of patients with advanced thyroid cancers 14501284_Our findings of a high frequency of BRAF mutations at codon 599 in benign melanocytic lesions of the skin indicate that this mutation is not sufficient by itself for malignant transformation. 14507635_Both BRAF and FBXW7 mutations functionally activate kinase effectors important in pancreatic cancer and extend potential options for therapeutic targeting of kinases in treatment of phenotypically distinct pancreatic adenocarcinoma subsets. 14513361_BRAF mutations, which are present in a variety of other human cancers, do not seem to be involved in gastric cancer development 14522897_Uceal melanomas arise independent of oncogenic BRAF and NRAS mutations. 14534542_BRAF mutations were seen in stomach neoplasms. 14602780_BRAF mutations are restricted to papillary carcinomas and poorly differentiated and anaplastic carcinomas arising from papillary carcinomas 14612909_BRAF is occasionally mutated in NHL, and BRAF mutation may contribute to tumor development in some NHLs 14618633_None of the cases of gastric cancer showed braf mutations 14639609_Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas 14668801_Missense mutation is marker of colonic but not gastric cancer. 14688025_Mutations were found in exon 15 in colorectal adenocarcinoma. 14691295_Our data indicate that BRAF gene mutations are rare to absent events in uveal melanoma of humans. 14695152_NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression 14695993_BRAF mutations are associated with proximal colon tumors with mismatch repair deficiency and MLH1 hypermethylation. 14719068_New enriched PCR-RFLP assay for detecting mutations of BRAF codon 599 mutation in pleural mesotheliomas. 14722037_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 14724583_RAS or BRAF mutations are detected in about 32% of all Barrett's adenocarcinomas; the disruption of the Raf/MEK/ERK (MAPK) kinase pathway is a frequent but also early event in the development of Barrett's adenocarcinoma 14734469_BRAF mutations are frequently present in sporadic colorectal cancer with methylated hMLH1 14961576_Mutations in BRAF gene is associated with malignant melanomas 14966563_These studies identify isoprenylcysteine carboxyl methyltransferase as a potential target for reducing the growth of K-Ras- and B-Raf-induced malignancies. 15001635_The lack or low prevalence of BRAF mutation in other thyroid neoplasms is consistent with the notion that other previously defined genetic alterations on the same signaling pathway are sufficient to cause tumorigenesis in most thyroid neoplasms. 15009714_possible cooperation between BRAF activation and PTEN loss in melanoma development. 15009715_mutations in the BRAF gene and to some extent in the N-ras gene represent early somatic events that occur in melanocytic nevi 15014028_BRAF mutation may be acquired during development of metastasis but is not a significant factor for primary melanoma development and disease outcome. 15077125_ovarian serous cystadenomas do not contain mutations in either BRAF or KRAS genes 15104286_These results suggest that the BRAF mutation is unlikely to be involved in gastric carcinogenesis. 15126572_BRAF(V599E) is more common genetic alteration found to date in adult sporadic papillary thyroid carcinomas (PTCs). It is unique for this thyroid cancer histotype, and it might drive the development of PTCs of classic papillary subtype. 15140228_The finding of tandem mutations in thin melanomas makes it more likely that they arise as a simultaneous rather than sequential event. 15145515_Radiation-induced tumors have a low prevalence of BRAF point mutations and high prevalence of RET/PTC rearrangements 15150271_B-Raf kinase activity regulation by tuberin and Rheb is mammalian target of rapamycin (mTOR)-independent 15161700_mucosal melanomas of the head and neck do not frequently harbor an activating mutation of BRAF 15179189_in contrast to cutaneous melanoma, BRAF does not appear to be involved in the pathogenesis of uveal melanoma 15186612_BRAF mutations are rather rare in solitary cold adenomas and adenomatous nodules and do not explain the molecular etiology of ras mutation-negative cold thyroid nodules. 15191558_activation of this gene may be one of the early events in the pathogenesis of some melanomas. 15263001_B-Raf and ERK are activated by cyclic AMP after calcium restriction 15273715_mutated in papillary thyroid cancer. 15277467_In this study, this BRAF mutation was demonstrated in some conjunctival melanoma tissue samples, suggesting that some conjunctival melanomas may share biological features in common with cutaneous melanoma. 15313890_Data suggest that SPRY2, an inhibitor of ERK signaling, may be bypassed in melanoma cells either by down-regulation of its expression in WT BRAF cells, or by the presence of the BRAF mutation. 15330192_Mutations within the BRAF gene are useful markers for the differential diagnosis between Spitz nevus and malignant melanoma. 15331929_we found 19 cases (38%) to harbor somatic B-raf exon 15 mutations. 15339934_Data provide evidence that B-Raf is a positive regulator of T cell receptor-mediated sustained ERK activation, which is required for NFAT activation and the full production of IL-2. 15373778_BRAF(V599E) mutation is seven times higher in lesions with structural changes and 13 times higher in growing lesions as compared with lesions without changes 15488754_REVIEW: our understanding of B-RAF as an oncogene and of its role in cancer 15489648_Mutations of BRAF or KRAS oncogenes are early events in the serrated polyp neoplasia pathway. CpG island methylation plays a role in serrated polyp progression to colorectal carcinoma. 15538400_mutated in childhood acute lymphoblastic leukemia. 15577314_BRAF mutations are associated with conjunctival neoplasms 15630448_AKAP9-BRAF fusion was preferentially found in radiation-induced papillary carcinomas developing after a short latency, whereas BRAF point mutations were absent in this group 15632082_Data suggest that Rit is involved in a novel pathway of neuronal development and regeneration by coupling specific trophic factor signals to sustained activation of the B-Raf/ERK and p38 MAP kinase cascades. 15653554_a novel Ras-independent ERK1/2 activation system in which p110gamma/Raf-1/MEK1/2 and PKA/B-Raf/MEK1/2 cooperate to activate ERK1/2. 15702478_We found mutations in p53, K-ras, and BRAF genes in 35%, 30%, and 4% of tumors, respectively, and observed a minimal or no co-presence of these gene alterations. 15705790_KSHV-infected cell lines expressed higher levels of B-Raf and VEGF-A; B-Raf-induced VEGF-A expression was demonstrated to be sufficient to enhance tubule formation in endothelial cells 15710605_autoinhibition was negatively regulated by acidic substitutions at phosphorylation sites within the activation loop 15765445_Mutations in the BRAF protooncogene (V599E)may be an alternative pathway of tumorigenesis of familial colorectal cancer. 15782118_BRAF mutations proved to be absent in tumors from hereditary nonpolyposis colorectal cancer syndrome (HNPCC) families with germline mutations in the MMR genes MLH1 and MSH2. 15791479_The data of this study suggest that activating mutations of B-RAF are not a frequent event in gliomas; nevertheless, when present they are associated with high-grade malignant lesions. 15791648_B-raf mutations surrounding Thr439 found in human cancers are unlikely to contribute to increased oncogenic properties of B-raf 15824163_Observational study of gene-disease association. (HuGE Navigator) 15842051_These results suggest that BRAF mutations do not have a role in tumorigenesis of neuroendocrine gastroenteropancreatic tumors. 15880523_Anaplastic thyroid carcinomas which are derived from papillary carcinomas are due to BRAF and p53 mutations 15904951_Observational study of gene-disease association. (HuGE Navigator) 15935100_B-raf V599E and V599K oncogenic mutations are likely to affect melanocyte-specific pathways controlling proliferation and differentiation 15968271_Observational study of gene-disease association. (HuGE Navigator) 15968271_The increasing frequency of BRAF mutations as a function of age could help account for the well documented but poorly understood observation that age is a relevant prognostic indicator for patients with papillary thyroid carcinoma. 15980887_BRAF mutation occurs later in thyroid tumor progression and is restricted mainly to papillary thyroid carcinoma and anaplastic thyroid carcinoma 15994075_Observational study of gene-disease association. (HuGE Navigator) 15998781_Role of BRAF mutation in facilitating metastasis and progression of papillary thyroid cancer in lymph nodes. 16007166_determination of mutation specific gene expression profiles in papillary thyroid carcinoma 16007203_Single-cell clones with efficient knockdown of (V 600 E)B-RAF could be propagated in the presence of basic fibroblast growth factor but underwent apoptosis or senescence-like growth arrest upon withdrawal of this growth factor 16015629_Observational study of gene-disease association. (HuGE Navigator) 16024606_Observational study of gene-disease association. (HuGE Navigator) 16079850_sustained BRAF(V600E) expression in human melanocytes induces cell cycle arrest, which is accompanied by the induction of both p16(INK4a) and senescence-associated acidic beta-galactosidase (SA-beta-Gal) activity, a commonly used senescence marker 16096377_BRAF mutation in melanoma is most likely to occur prior to the development of metastatic disease 16098042_Although BRAF and NRAS mutations are likely to be important for the initiation and maintenance of some melanomas, other factors might be more significant for proliferation and prognosis in subgroups of aggressive melanoma 16098042_Observational study of gene-disease association. (HuGE Navigator) 16123397_The results showed that conjunctival nevi, similar to skin nevi, have a high frequency of oncogenic BRAF mutations. 16129781_These data suggest that MITF is an anti-proliferation factor that is down-regulated by B-RAF signaling and that this is a crucial event for the progression of melanomas that harbor oncogenic B-RAF. 16143028_Observational study of gene-disease association. (HuGE Navigator) 16144912_Mutations of the BRAF gene are partly involved in the malignant transformation of the endometrium. 16144912_Observational study of gene-disease association. (HuGE Navigator) 16172610_selective reduction in catalytic activity and expression of B-Raf but not Raf-1 suggest that B-Raf may be playing an important role in altered ERK signaling in brain of suicide subjects, and thus in the pathophysiology of suicide 16174717_In patients with papillary thyroid cancer, BRAF mutation is associated with poorer clinicopathological outcomes and independently predicts recurrence. 16174717_Observational study of gene-disease association. (HuGE Navigator) 16179867_As the BRAF oncogene is frequently found to be mutated in human cutaneous melanomas, it may constitute a risk factor for melanoma formation within CMN and DMN. 16179870_The oncogenic B-raf mutations V599E and V599K, as early events in melanocyte transformation, persist throughout metastasis with important prognostic implications. 16181240_Observational study of gene-disease association. (HuGE Navigator) 16181547_Observational study of gene-disease association. (HuGE Navigator) 16199894_copy number gain may represent another mechanism of BRAF activation in thyroid tumors 16268813_Observational study of gene-disease association. (HuGE Navigator) 16354196_Observational study of gene-disease association. (HuGE Navigator) 16354196_The estimated proportion of attributable risk of melanoma due to variants in BRAF is 1.6%, but the burden of disease associated with this variant is greater than that associated with the major melanoma locus (CDKN2A) which has a risk of 0.2%. 16354586_Mutation and elevated expression of BRAF is associated with the development of testicular germ cell tumors 16361694_The authors have developed and run a high-throughput screen to find inhibitors of V600E BRAF using an enzyme cascade assay in which oncogenic BRAF activates MEK1, which in turn activates ERK2, which then phosphorylates the transcription factor ELK1. 16364920_Data suggest that B-RAF activates C-RAF through a mechanism involving 14-3-3 mediated heterooligomerization and C-RAF transphosphorylation. 16371460_V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. 16373964_activating mutations of PDGFR-alpha, c-kit and B-RAF are absent in gliosarcomas 16376942_Observational study of gene-disease association. (HuGE Navigator) 16376942_V599E BRAF mutation was uncommon in Japanese lung cancer. 16382052_aberrant B-Raf activity in angiomyolipomas leads to abnormal cellular differentiation and migration [review] 16397024_Observational study of genotype prevalence. (HuGE Navigator) 16413100_The most frequent B-RAF gene alterations are not involved in prostate carcinogenesis 16417232_BRAF mutation does not seem to be sufficient to produce MAPK activation in melanocytic nevi. 16424035_gain-of-function BRAF signaling is strongly associated with in vivo tumorigenicity 16439621_findings demonstrate that heterogeneous de novo missense mutations in three genes within the mitogen-activated protein kinase pathway, BRAF, MEK1 and MEK2 cause cardio-facio-cutaneous syndrome 16452469_wild-type B-Raf-mediated ERK1/2 activation plays a major role in proliferation and transformation of uveal melanocytes; Raf-1 is not involved in this activation 16452550_Observational study of gene-disease association. (HuGE Navigator) 16462768_NRAS and BRAF activating mutations can coexist in the same melanoma, but are mutually exclusive at the single-cell level 16474404_Cardio-facio-cutaneous (CFC) syndrome involves dysregulation of the RAS-RAF-ERK pathway. 16487015_Observational study of gene-disease association. (HuGE Navigator) 16537381_Merlin and MLK3 can interact in situ and merlin can disrupt the interactions between B-Raf and Raf-1 or those between MLK3 and either B-Raf or Raf-1. 16547495_Melanoma cells require either B-RAF or phosphoinositide-3 kinase activation for protection from anoikis. 16601293_BRAF V600E is associated with a high risk of recurrence and less differentiated papillary thyroid carcinoma due to the impairment of Na+/I- targeting to the membrane 16601293_Observational study of gene-disease association. (HuGE Navigator) 16618717_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16687919_Observational study of genotype prevalence. (HuGE Navigator) 16691193_Observational study of gene-disease association. (HuGE Navigator) 16691193_UV light is not necessarily required for the acquisition of the BRAF(V600E) mutation, and non-mutagenic effects of UV light to melanocytes may be more important in the nevogenesis 16721785_BRAF mutations are as uncommon as KRAS mutations in prostate adenocarcinoma 16728573_B-RAF (V600E) was confirmed to be associated with the papillary growth pattern, but not with poorer differentiated papillary thyroid carcinoma variants. 16728573_Observational study of gene-disease association. (HuGE Navigator) 16773193_among 23 melanomas located at body sites with chronic UV exposure, only a single tumour harboured the B-raf V599E mutation which was a significantly lower frequency in comparison to melanomas from sun-protected body sites 16786134_a BRAFT1799A mutation may have a role in poor differentiation of thyroid carcinoma 16799476_A subset of Spitz nevi, some with atypical histologic features, possess BRAF mutations. The BRAF mutational status does not separate all Spitz nevi from spitzoid melanomas and non-Spitz types of melanocytic proliferations, contrary to previous reports. 16803888_Rheb has a central role in the regulation of the Ras/B-Raf/C-Raf/MEK signaling network 16804544_CpG island methylator phenotype-positive colorectal tumors represent a distinct subset, encompassing almost all cases of tumors with BRAF mutation 16809487_findings show that MC1R variants are strongly associated with BRAF mutations in non-chronic sun-induced damage melanomas; in this subtype, risk for melanoma associated with MC1R is due to increase in risk of developing melanomas with BRAF mutations 16845322_BRAF mutation is associated with melanoma and melanocytic nevi. 16858395_Thus, we propose that the hitherto unidentified function of the B-Raf amino-terminal region is to mediate calcium-dependent activation of B-Raf and the following MEK activation, which may occur in the absence of Ras activation. 16858683_Aberrant methylation and hence silencing of TIMP3, SLC5A8, DAPK and RARbeta2, in association with BRAF mutation, may be an important step in PTC tumorigenesis and progression. 16879389_BRAF mutation was frequent in hyperplastic polyps (67%) and sessile serrated adenomas (81%). 16912199_B-RAF has been identified as the most mutated gene in invasive cells and therefore an attractive therapeutic target in melanoma. 16918136_BRAF mutations are associated with colorectal cancers 16918957_Observational study of gene-disease association. (HuGE Navigator) 16924241_Expression of p27Kip1 in melanoma is regulated by B-RAF at the mRNA level and via B-RAF control of Cks1/Skp2-mediated proteolysis. 16932278_Single nucleotide polymorphism found exclusively in papillary thyroid carcinoma. 16937524_BRAF, K-ras and BAT26 are expressed in colorectal polyps and stool 16937524_BRAF, K-ras and BAT26 are expressed in colorectal polyps and stool [BAT26] 16946010_Braf mutations in thyroid tumorigenesis. 16953233_Concomitant KRAS and BRAF mutations increased along progression of MSS colorectal cancer, suggesting that activation of both genes is likely to harbour a synergistic effect 16959844_BRAFV600E activates not only MAPK but also NF-kappaB signaling pathway in human thyroid cancer cells, leading to an acquisition of apoptotic resistance and promotion of invasion. 16960555_Expression of active mutants of B-Raf induces fibronectin. 16964379_Extracellular signal-regulated kinase-3 (ERK3/MAPK6) is highly expressed in response to BRAF signaling. 16973828_Observational study of gene-disease association. (HuGE Navigator) 16987295_BRAF T1976A mutation is present at high frequency in benign naevi such as Spitz and Reed. 17001349_data support a model in which mutational activation of BRAF in human melanomas contributes to constitutive induction of NF-kappaB activity and to increased survival of melanoma cells 17018604_Normally, BRAF alone is responsible for signaling to MEK. However, when RAS is mutated in melanoma, melanocytes switch their signaling from BRAF to CRAF. 17044028_Activating BRAF mutation is associated with papillary thyroid carcinoma 17060774_BRAF mutation remained a significant prognostic factor for lymph node metastasis (odds ratio = 10.8, 95% confidence interval, 3.5-34.0, P < 0.0001). 17074813_phosphorylation on both S365 and S429 participate in the differential regulation of B-Raf isoforms through distinct mechanisms 17097223_data provide evidence that oncogenic properties of BRAF contribute to the tumorigenesis of intraductal papillary mucinous neoplasm/carcinoma (IPMN/IPMC), but at a lower frequency than KRAS 17119056_BRAF-V600E mutations are mainly involved in colorectal cancer families characterized by an increased risk of other common malignancies 17119447_Association with preexisting nevi and pronounced infiltration of lymphocytes was significantly higher in BRAF mutated melanoma tumours 17148775_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17148775_Previously identified associations between smoking and colon cancer, whether microsatellite unstable or stable, appear to be explained by the association of smoking with BRAF mutation. 17159915_BRAF(T1799A) mutation is associated with a lower rate of tumor proliferation. 17159915_Observational study of gene-disease association. (HuGE Navigator) 17170014_RASSF1A methylation was observed in a high frequency in endometrioid endometrial carcinoma whereas K-ras and B-raf mutations were observed in a low frequency 17179987_The role for BRAF activation in thyroid cancer development and establishing the potential therapeutic efficacy of BRAF-targeted agents in patients with thyroid cancerwill be reviewed. 17186541_BRAF mutation is associated with thyroid carcinogenesis 17186541_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17195912_Observational study of gene-disease association. (HuGE Navigator) 17195912_there is a subgroup of colorectal carcinomas which develop via the microsatellite instability pathway that carry an alteration of the BRAF gene 17199737_Absence of association between BRAF mutation and activation of MAPK pathway in papillary thyroid carcinoma suggests the presence of mechanisms that downregulate MAPK activation. 17227125_Copy gain of PDGFB occurs in a subset of tumors showing no evidence of mutated BRAF or rearranged ret, suggesting that copy gain of PDGFB may underlie the increased expression of platelet-derived growth factor described recently in the literature. 17270239_Observational study of gene-disease association. (HuGE Navigator) 17297294_characterization of the T1799-1801del and A1799-1816ins BRAF mutations in papillary thyroid cancer; the two new mutations resulted in constitutive activation of the BRAF kinase and caused NIH3T3 cell transformation 17302867_Overexpression of B-Raf mRNA and protein may be a feature of nonfunctioning pituitary adenomas, highlighting overactivity of the Ras-B-Raf-MAP kinase pathway in these tumors. 17309670_BRAF gene plays a 'gatekeeper' role but does not act as a predisposition gene in the development of low-grade ovarian serous carcinomas 17309670_Observational study of gene-disease association. (HuGE Navigator) 17312306_Observational study of genetic testing. (HuGE Navigator) 17315191_BRAFV600E represents a detectable marker in the plasma/serum from melanoma patients for monitoring but not diagnostic purposes 17318013_B-RAF mutations are a rare event in pituitary tumorigenesis. 17355635_The aim of this study was to identify the effect that BRAF oncogene has on post-transcriptional regulation in papillary thyroid carcinoma by using microRNA analysis. 17360030_findings show that RASSF1A hypermethylation and KRAS mutations and BRAF mutations are inversely correlated and play an important role in the development of cervical adenocarcinomas 17366577_mutational analysis of KRAS, BRAF, and MAP2K1/2 in 56 patients with CFC syndrome; comparison of the genotype-phenotype correlation of CFC with that of Costello syndrome suggest a significant clinical overlap but not genotype overlap. 17387744_BRAF(V600E) mutation is identified in a subset of cutaneous metastases from papillary thyroid carcinomas 17393356_Observational study of gene-disease association. (HuGE Navigator) 17393356_data suggest that BRAF mutations might be present less frequently than KRAS mutations in Greek patients with colorectal carcinomas 17440063_finding of a strong association between BRAF mutations and serrated histology in hyperplastic aberrant crypt foci supports the idea that these lesions are an early, sentinel, or a potentially initiating step on the serrated pathway to colorectal carcinoma 17453004_BRAF V600E mutation was occasionally observed in anaplastic carcinomas with papillary carcinoma. 17453358_Observational study of gene-disease association. (HuGE Navigator) 17454879_MSI is rare in UC-related neoplasia as well as non-neoplastic lesions, and does not contribute to the development of dysplasia. 17464312_prevalence of BRAF mutation and RET/PTC were determined in diffuse sclerosing variant of papillary thyroid carcinoma; none of the cases showed a BRAF mutation 17483702_Molecular diagnosis and careful observations should be considered in children with Cardio-facio-cutaneous syndrome because they have germline mutations in BRAF and might develop malignancy. 17487277_Observational study of gene-disease association. (HuGE Navigator) 17487504_c-kit expression is not alternative to BRAF and/or KRAS activation. 17488796_BRAF V600E mutation in PTCs is associated with reduced expression of key genes involved in iodine metabolism 17507627_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17507627_data indicate that both early-life UV exposure and nevus propensity contribute to occurrence of BRAF+ melanoma, whereas nevus propensity and later-life sun exposure influence the occurrence of NRAS+ melanoma 17516929_analysis of a BRAF mutation-associated gene expression signature in melanoma 17518771_low rate of RAS-RAF mutations (2/22, 9.1%) observed in Spitz melanocytic nevi suggests that these lesions harbor as yet undetected activating mutations in other components of the RAS-RAF-MEK-ERK-MAPK pathway 17520704_Meta-analysis of gene-disease association. (HuGE Navigator) 17520704_frequency of the BRAF mutation and the associations between BRAF mutation and clinicopathologic parameters in papillary thyroid carcinoma were evaluated by meta-analysis 17525723_T1790A BRAF mutation (L597Q) in childhood acute lymphoblastic leukemia is a functional oncogene 17535994_The heterogeneous distribution of BRAF mutations suggests that discrete tumor foci in multifocal PTC may occur as independent tumors. 17542667_Presence of BRAF V600E in very early stages of papillary thyroid carcinoma. 17548320_influence of B-RAF-specific RNA interference on the proliferation and apoptosis of gastric cancer BGC823 cell line 17566669_Observational study of gene-disease association. (HuGE Navigator) 17566669_We conclude that screening for BRAF 15 exon mutation is an efficient tool in the diagnostic strategy for HNPCC 17635919_In contrast to C-RAF that requires farnesylated H-Ras, cytosolic B-RAF associates effectively and with significantly higher affinity with both farnesylated and nonfarnesylated H-Ras. 17663506_KLF6 and p53 mutations are involved in the development of nonpolypoid colorectal carcinoma, whereas K-ras and B-raf mutations are not 17671688_PPARbeta/delta has a role in growth of RAF-induced lung adenomas 17685465_BRAF V600E mutation in papillary carcinoma of the thyroid may facilitate tumor cell growth and progression once seeded in the lymph nodes. 17693984_Observational study of genotype prevalence, gene-disease association, and genetic testing. (HuGE Navigator) 17693984_There was no coexistence of BRAF (V600E) mutation in papillary thyroid carcinoma. 17696195_data showed differences in gene expression between nevi with and without the V600E BRAF mutation. Moreover, nevi with mutations showed over-expression of genes involved in melanocytic senescence and cell cycle inhibition 17699719_RNA interference and pharmacologic approaches were used to assess the role of B-Raf activation in the growth of human melanomas and additionally determined if a similar role for mutant B-Raf is seen for colorectal carcinoma cell lines. 17704260_5 unreported mutations (T241P, Q262R, G464R, E501V, N581K) were found in cardio-facio-cutaneous syndrome. A hotspot in exon 6 at Q257 was found. 17714762_diffuse expression of wild-type and/or mutant B-Raf may be involved in the tumorigenic process 17717450_BRAF V600E mutation is primarily present in conventional papillary thyroid cancer; it is associated with an aggressive tumor phenotype and higher risk of recurrent and persistent disease in patients with conventional papillary thyroid cancer 17717450_Observational study of gene-disease association. (HuGE Navigator) 17721188_Develompment of malignant strumo ovarii with papillary thyroid carcinoma features is associated with BRAF mutations. 17727338_BRAF(V600E) mutation detected on fine-needle aspiration biopsy specimens, more than RET/PTC rearrangements, is highly specific for papillary thyroid carcinoma. 17785355_BRAF V600E mutation is associated with high-risk papillary thyroid carcinoma 17785355_Observational study of gene-disease association. (HuGE Navigator) 17786355_BRAFV600E mutations were found in 41.2% of the papillary thyroid carcinomas 17854396_Papillary thyroid cancers with no 131I uptake had a high frequency of BRAF mutations. 17878251_MEK inhibition is cytostatic in papillary thyroid cancer and anaplastic thyroid cancer cells bearing a BRAF mutation 17911174_effects of a MEK inhibitor, CI-1040, on thyroid cancer cells, some of which, particularly cell proliferation and tumor growth, seemed to be BRAF mutation or RAS mutation selective 17914558_BRAF mutation is associated as early as the hyperplastic polyp stage followed by microsatellite instability at the carcinoma stage 17924122_Examined associations between BRAF mutations, morphology, and apoptosis in early colorectal cancer. 17940185_BRAF mutation represents a novel indicator of the progression and aggressiveness of papillary thyroid cancer (Review) 17942568_BRAF interacts with PLCepsilon1 in nephrotic syndrome type 3. Both proteins are coexpressed and colocalize in developing and mature glomerular podocytes. 17962436_In this small study, the T1799A BRAF mutation was identified in almost half of the iris melanoma tissues samples examined. This finding suggests that there may be genetic as well as clinical differences between iris and posterior uvea | ENSMUSG00000002413 | Braf | 5.429863e+02 | 1.0614003 | 0.085968832 | 0.3084310 | 7.709076e-02 | 0.7812793372 | 0.95675823 | No | Yes | 5.442745e+02 | 99.886629 | 4.083061e+02 | 76.819422 | |
| ENSG00000157823 | 10239 | AP3S2 | protein_coding | P59780 | FUNCTION: Part of the AP-3 complex, an adaptor-related complex which is not clathrin-associated. The complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and may be directly involved in trafficking to lysosomes. In concert with the BLOC-1 complex, AP-3 is required to target cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals. | Alternative splicing;Cytoplasmic vesicle;Golgi apparatus;Membrane;Protein transport;Reference proteome;Transport | hsa:10239; | AP-3 adaptor complex [GO:0030123]; axon cytoplasm [GO:1904115]; cytoplasmic vesicle membrane [GO:0030659]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; anterograde axonal transport [GO:0008089]; anterograde synaptic vesicle transport [GO:0048490]; Golgi to vacuole transport [GO:0006896]; intracellular protein transport [GO:0006886]; vesicle-mediated transport [GO:0016192] | 20966410_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000063801 | Ap3s2 | 2.519336e+03 | 0.9063345 | -0.141884555 | 0.3192810 | 1.929608e-01 | 0.6604637181 | 0.92746340 | No | Yes | 1.789191e+03 | 290.566746 | 1.838368e+03 | 306.301035 | ||
| ENSG00000158290 | 8450 | CUL4B | protein_coding | Q13620 | FUNCTION: Core component of multiple cullin-RING-based E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins (PubMed:14578910, PubMed:16322693, PubMed:16678110, PubMed:18593899, PubMed:29779948, PubMed:30166453, PubMed:33854232, PubMed:33854239, PubMed:22118460). The functional specificity of the E3 ubiquitin-protein ligase complex depends on the variable substrate recognition subunit (PubMed:14578910, PubMed:16678110, PubMed:18593899, PubMed:29779948, PubMed:22118460). CUL4B may act within the complex as a scaffold protein, contributing to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme (PubMed:14578910, PubMed:16678110, PubMed:18593899, PubMed:22118460). Plays a role as part of the E3 ubiquitin-protein ligase complex in polyubiquitination of CDT1, histone H2A, histone H3 and histone H4 in response to radiation-induced DNA damage (PubMed:14578910, PubMed:16678110, PubMed:18593899). Targeted to UV damaged chromatin by DDB2 and may be important for DNA repair and DNA replication (PubMed:16678110). A number of DCX complexes (containing either TRPC4AP or DCAF12 as substrate-recognition component) are part of the DesCEND (destruction via C-end degrons) pathway, which recognizes a C-degron located at the extreme C terminus of target proteins, leading to their ubiquitination and degradation (PubMed:29779948). The DCX(AMBRA1) complex is a master regulator of the transition from G1 to S cell phase by mediating ubiquitination of phosphorylated cyclin-D (CCND1, CCND2 and CCND3) (PubMed:33854232, PubMed:33854239). The DCX(AMBRA1) complex also acts as a regulator of Cul5-RING (CRL5) E3 ubiquitin-protein ligase complexes by mediating ubiquitination and degradation of Elongin-C (ELOC) component of CRL5 complexes (PubMed:30166453). Required for ubiquitination of cyclin E (CCNE1 or CCNE2), and consequently, normal G1 cell cycle progression (PubMed:16322693, PubMed:19801544). Regulates the mammalian target-of-rapamycin (mTOR) pathway involved in control of cell growth, size and metabolism (PubMed:18235224). Specific CUL4B regulation of the mTORC1-mediated pathway is dependent upon 26S proteasome function and requires interaction between CUL4B and MLST8 (PubMed:18235224). With CUL4A, contributes to ribosome biogenesis (PubMed:26711351). {ECO:0000269|PubMed:14578910, ECO:0000269|PubMed:16322693, ECO:0000269|PubMed:16678110, ECO:0000269|PubMed:18235224, ECO:0000269|PubMed:18593899, ECO:0000269|PubMed:19801544, ECO:0000269|PubMed:22118460, ECO:0000269|PubMed:26711351, ECO:0000269|PubMed:29779948, ECO:0000269|PubMed:30166453, ECO:0000269|PubMed:33854232, ECO:0000269|PubMed:33854239}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;DNA damage;DNA repair;Disease variant;Dwarfism;Isopeptide bond;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:10230407, ECO:0000269|PubMed:16678110, ECO:0000269|PubMed:18593899, ECO:0000269|PubMed:29779948, ECO:0000269|PubMed:30166453, ECO:0000269|PubMed:33854232, ECO:0000269|PubMed:33854239}. | This gene is a member of the cullin family. The encoded protein forms a complex that functions as an E3 ubiquitin ligase and catalyzes the polyubiquitination of specific protein substrates in the cell. The protein interacts with a ring finger protein, and is required for the proteolysis of several regulators of DNA replication including chromatin licensing and DNA replication factor 1 and cyclin E. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:8450; | Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; Cul4B-RING E3 ubiquitin ligase complex [GO:0031465]; cullin-RING ubiquitin ligase complex [GO:0031461]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleoplasm [GO:0005654]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; cellular response to DNA damage stimulus [GO:0006974]; G1/S transition of mitotic cell cycle [GO:0000082]; histone H2A monoubiquitination [GO:0035518]; neuron projection development [GO:0031175]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; positive regulation of protein catabolic process [GO:0045732]; proteasomal protein catabolic process [GO:0010498]; protein ubiquitination [GO:0016567]; ribosome biogenesis [GO:0042254]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146]; UV-damage excision repair [GO:0070914] | 16322693_human CUL4B and cyclin E proteins interact with each other and the CUL4B complexes can polyubiquitinate the CUL4B-associated cyclin E 16407252_Cul4B, PCNA, and DDB1 are involved in the degradation of Cdt1 after ultraviolet radiation 17236139_The relatively high frequency of CUL4B mutations in this series indicates that it is one of the most commonly mutated genes underlying XLMR and suggests that its introduction into clinical diagnostics should be a high priority. 17273978_Mutation in CUL4B causes X-linked mental retardation 17392787_a fat-soluble ligand-dependent ubiquitin ligase complex in human cell lines, in which dioxin receptor (AhR) is integrated as a component of a novel cullin 4B ubiquitin ligase complex, CUL4B(AhR) 18235224_CUL4-DDB1 ubiquitin ligase interacts with Raptor and regulates the mTORC1-mediated signaling pathway through ubiquitin-dependent proteolysis. 18593899_DDB1-CUL4B(DDB2) E3 ligase may have a distinctive function in modifying the chromatin structure at the site of UV lesions to promote efficient NER. 19056892_CUL4A and CUL4B are therefore components of a conserved Wnt-induced proteasome targeting (WIPT) complex that regulates p27(KIP1) levels and cell cycle progression in mammalian cells. 19295130_Cells depleted of Dda1 spontaneously accumulated double-stranded DNA breaks in a similar way to Cul4A-, Cul4B- or Wdr23-depleted cells, indicating that Dda1 interacts physically and functionally with cullin-RING E3 ligases complexes. 19801544_Data show that that RNA interference of CUL4B led to an inhibition of cell proliferation and a prolonged S phase, due to the overaccumulation of cyclin E. 19818632_Studies indicate that CUL4 uses a large beta-propeller protein, DDB1, as a linker to interact with a subset of WD40 proteins. 20002452_The CUL4B gene is associated with X-linked mental retardation syndrome. 20005570_CUL4B is over-expressed in placenta in intra-uterine growth restriction. 20064923_the interplay between CUL4A and CUL4B in pathogenesis of CUL4B-deficiency in humans 20932471_This study identifies CRL4-Cdt2 ubiquitin ligase to promote the ubiquitin-dependent proteolysis of the histone H4 methyltransferase Set8 during S-phase of the cell cycle and after UV-irradiation in a reaction that is dependent on PCNA. 20951943_Increased PRMT5 activity mediates key events associated with cyclin D1-dependent neoplastic growth, including CUL4 repression, CDT1 overexpression, and DNA rereplication. 21352845_the unexpected association of defective CUL4B with syndromal X-linked mental retardation in humans 21795677_Cullin 4B protein ubiquitin ligase targets peroxiredoxin III for degradation. 21816345_CUL4B targets WDR5 for ubiquitylation and degradation in the nucleus. 22992378_The data suggest that unneddylated Cul4B isoforms specifically inhibits beta-catenin degradation during mitosis. 23238014_Cullin4B-Ring E3 ligase complex (CRL4B) is physically associated with PRC2. CRL4B possesses an intrinsic transcription repressive activity by promoting H2AK119 monoubiquitination. CUL4B promotes cancer cell proliferation, invasion, and tumorigenesis in vitro and in vivo. 23348097_Our results suggest that XLID CUL4B mutants are defective in promoting TSC2 degradation and positively regulating mTOR signaling in neocortical neurons 23357576_Studies indicate Jun activation domain-binding protein Jab1 as a substrate for CUL4B E3 ligase. 23479742_the up-regulation of CDK2 by CUL4B is achieved via the repression of miR-372 and miR-373, which target CDK2. 23649548_Investigated CUL4B expression pattern in patients with colon cancer; immunohistochemistry and PCR study showed that high CUL4B expression was significantly associated with colon cancer progression and pathogenesis. 24292684_CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing 24452595_these observations establish an important negative regulatory role of CUL4B on p53 stability. 24719410_HIV-1 Vpr can trigger G2 cell cycle arrest in the absence of either CUL4A or CUL4B. 24898194_The intellectual disability phenotype is caused by aberrant splicing and removal of intron 7 from CUL4B gene primary transcript. 25189186_Results demonstrated that CUL4B promotes cell proliferation and inhibits the apoptosis of osteosarcoma cells. 25385192_Data show that CUL4B variants are associated with a wide range of cerebral malformations and suggest an important role in brain through its interaction with WDR62, a protein in which variants were identified in patients with cerebral malformations. 25430888_CUL4B can up-regulate Wnt/beta-catenin signalling in human HCC through transcriptionally repressing Wnt antagonists and thus contributes to the malignancy of HCC. 25464270_results established a critical role of CUL4B in negatively regulating the p53-ROS positive feedback loop that drives cellular senescence 25542213_Results show that CUL4A- and CUL4B-mediated polyubiquitination of gamma-tubulin for its degradation. 25970626_FBXO44-mediated degradation of RGS2 protein uniquely depends on a Cul4B/DDB1 complex. 26021757_Our data are consistent with the idea that the CUL4A/B-DDB1-CRBN complex catalyses the polyubiquitination and thus controls the degradation of CLC-1 channels. 26617747_these results showed that knockdown of CUL4B inhibit proliferation and promotes apoptosis of colorectal cancer cells through suppressing the Wnt/beta-catenin signaling pathway 27656838_these results suggest that knockdown of CUL4B inhibited the proliferation and invasion through suppressing the Wnt/beta-catenin signaling pathway in NSCLC cells. Therefore, CUL4B may represent a novel therapeutic target for the treatment of NSCLC. 27899484_CUL4B protein levels in human subcutaneous adipose tissue is negatively correlated with body mass index. 27974468_findings revealed that CUL4A and CUL4B are differentially associated with etiologic factors for pulmonary malignancies and are independent prognostic markers for the survival of distinct lung cancer subtypes 28164432_This study found that microRNA-194 (miR-194) and CUL4B protein were inversely correlated in cancer specimens and demonstrated that miR-194 could downregulate CUL4B by directly targeting its 3'-UTR. 28225217_CUL4B regulates protein turnover and homeostasis in response to dopamine stimulation. 28816568_lack of any significant mutations in patients with azoospermia due to Sertoli-cell-only syndrome 28886238_The CUL4B interacts with WD-40 proteins through the adaptor protein DNA damage-binding protein 1 (DDB1) to target substrates for ubiquitylation. 29106389_High CUL4B expression promotes gastric cancer invasion and metastasis. 30229816_In osteosarcoma tissues, expression of CUL4B is higher and expression of miRNA-708 is lower. 30483755_Taken together, the findings of this study suggest that the miR381/miR489mediated expression of CUL4B modulates the proliferation and invasion of GC cells via the Wnt/betacatenin pathway, which indicates that the miR381/miR489CUL4B axis is critical in the control of GC tumourigenesis. 30609075_Authors demonstrate that CUL4B upregulates the expression of C-MYC at post-transcriptional level through epigenetic silencing of miR-33b-5p. CUL4B-induced oncogenic activity in PCa by targeting C-MYC is repressed by miR-33b-5p. 30612524_Enhanced CUL4B expression was discovered in diffuse large B-cell lymphoma tissues and cells. CUL4B promoted the growth of diffuse large B-cell lymphoma cells both in vitro and in vivo, which was by autophagy mediated by JNK signaling. 30883036_Results demonstrate a key contribution of CUL4B overexpression in the malignant behavior of head and neck squamous cell carcinoma (HNSCC) cells, at least in part through the stimulation of angiogenesis and the activation of the Wnt/beta-catenin signaling pathway. 30898011_An autophagy-related (ATG) protein that plays a critical role in autophagosome biogenesis, is a direct substrate of CUL4-RING ubiquitin ligases (CRL4s). 30945295_miR-194-5p, was significantly downregulated in intervertebral disc degeneration samples and could bind to the three prime untranslated regions (3'-UTRs) of both CUL4A and CUL4B, thereby downregulating their expression. 31111526_CUL4B regulates cancer stem-like traits of prostate cancer cells by targeting BMI1 via miR200b/c, which might give novel insight into how CUL4B promotes prostate cancer progression through regulating cancer stem-like traits. 31329620_These results show that ubiquitinated Apaf-1 may activate caspase-9 under conditions of proteasome impairment. 31407591_circZFR exhibited a carcinogenic role by sponging miR-101-3p and regulating CUL4B expression in NSCLC 31448526_NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. 31729179_Alu-mediated Xq24 deletion encompassing CUL4B, LAMP2, ATP1B4, TMEM255A, and ZBTB33 genes causes Danon disease in a female patient. 32275162_Downregulation of lncRNA ZEB1-AS1 Represses Cell Proliferation, Migration, and Invasion Through Mediating PI3K/AKT/mTOR Signaling by miR-342-3p/CUL4B Axis in Prostate Cancer. 32466489_CUL4-DDB1-CRBN E3 Ubiquitin Ligase Regulates Proteostasis of ClC-2 Chloride Channels: Implication for Aldosteronism and Leukodystrophy. 32587774_CUL4 E3 ligase regulates the proliferation and apoptosis of lung squamous cell carcinoma and small cell lung carcinoma. 32622365_Cul4B promotes the progression of ovarian cancer by upregulating the expression of CDK2 and CyclinD1. 33022894_Cullin 4B regulates cell survival and apoptosis in clear cell renal cell carcinoma as a target of microRNA-217. 33227394_CUL4B promotes aggressive phenotypes of renal cell carcinoma via upregulating c-Met expression. 33506897_Effect and mechanism of miR-217 on drug resistance, invasion and metastasis of ovarian cancer cells through a regulatory axis of CUL4B gene silencing/inhibited Wnt/beta-catenin signaling pathway activation. 33638154_CUL4B renders breast cancer cells tamoxifen-resistant via miR-32-5p/ER-alpha36 axis. 34002487_LncRNA SNHG12 regulates the miR-101-3p/CUL4B axis to mediate the proliferation, migration and invasion of non-small cell lung cancer. 34011786_Cul4b Promotes Progression of Malignant Cutaneous Melanoma Patients by Regulating CDKN2A. 34026424_CUL4B Promotes Breast Carcinogenesis by Coordinating with Transcriptional Repressor Complexes in Response to Hypoxia Signaling Pathway. 34119472_CUL4(high) Lung Adenocarcinomas Are Dependent on the CUL4-p21 Ubiquitin Signaling for Proliferation and Survival. 34153655_Cullin-4B promotes cell proliferation and invasion through inactivation of p53 signaling pathway in colorectal cancer. 34338146_Inhibition of MicroRNA miR-101-3p on prostate cancer progression by regulating Cullin 4B (CUL4B) and PI3K/AKT/mTOR signaling pathways. | ENSMUSG00000031095 | Cul4b | 1.145333e+03 | 1.0409535 | 0.057905649 | 0.3125906 | 3.409552e-02 | 0.8535037909 | 0.97089672 | No | Yes | 1.245397e+03 | 228.040227 | 9.705243e+02 | 182.413589 |
| ENSG00000158417 | 9669 | EIF5B | protein_coding | O60841 | FUNCTION: Plays a role in translation initiation. Translational GTPase that catalyzes the joining of the 40S and 60S subunits to form the 80S initiation complex with the initiator methionine-tRNA in the P-site base paired to the start codon. GTP binding and hydrolysis induces conformational changes in the enzyme that renders it active for productive interactions with the ribosome. The release of the enzyme after formation of the initiation complex is a prerequisite to form elongation-competent ribosomes. {ECO:0000250|UniProtKB:P39730}. | Cytoplasm;Direct protein sequencing;GTP-binding;Hydrolase;Initiation factor;Metal-binding;Nucleotide-binding;Phosphoprotein;Protein biosynthesis;Reference proteome | Accurate initiation of translation in eukaryotes is complex and requires many factors, some of which are composed of multiple subunits. The process is simpler in prokaryotes which have only three initiation factors (IF1, IF2, IF3). Two of these factors are conserved in eukaryotes: the homolog of IF1 is eIF1A and the homolog of IF2 is eIF5B. This gene encodes eIF5B. Factors eIF1A and eIF5B interact on the ribosome along with other initiation factors and GTP to position the initiation methionine tRNA on the start codon of the mRNA so that translation initiates accurately. [provided by RefSeq, Jul 2008]. | hsa:9669; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; translation initiation factor activity [GO:0003743]; regulation of translational initiation [GO:0006446] | 12569173_determination of binding site on eukaryotic initiation factor 1A 17161026_Transfected nuclear factor 2 trans activates the IF2 promoter in a cell line. 17568775_binding of eIF5B might induce conformational changes in both subunits, with ribosomal segments wrapping around the factor 18572216_3Cpro-mediated cleavage of eIF5B may thus play an accessory role in the shutoff of translation that occurs in enterovirus-infected cells. 21697471_The authors show that the cleavage of initiation factor eIF5B during enteroviral infection, along with the viral internal ribosome entry site, plays a role in mediating viral translation under conditions that are nonpermissive for host cell translation. 25261552_eIF5B overexpression promotes maturation of G0-like immature oocytes and causes cell death, an alternative to G0, in serum-starved THP1 cells. 27325746_results indicate that the interactions between eIF1A and eIF5B are being continuously rearranged during translation initiation; presentation of a model how the dynamic eIF1A/eIF5B interaction network can promote remodeling of the translation initiation complexes, and the roles in the process played by intrinsically disordered protein segments 27694689_eIF5B promoted hepatocellular carcinoma cell proliferation and migration in vitro and in vivo partly through increasing ASAP1 expression. 27959964_eIF5B silencing provides a negative feedback to deactivate MAPK signaling 29298419_aerobic eukarya retained eIF5B/IF2 to remodel anaerobic pathways during episodes of oxygen deficiency. 30211544_results indicate that in humans, eIF5B displacing eIF2 from Met-tRNAi upon subunit joining may be coupled to eIF1A displacing eIF5 from eIF5B, allowing the eIF5:eIF2-GDP complex to leave the ribosome. 30670698_Data suggest that eIF5B represents a regulatory node, allowing cancer cells to evade apoptosis by promoting the translation of pro-survival proteins from IRES-containing mRNAs. 31671279_Eukaryotic initiation factor 5B (eIF5B) regulates temozolomide-mediated apoptosis in brain tumour stem cells (BTSCs). 32984844_eIF5B drives integrated stress response-dependent translation of PD-L1 in lung cancer. 33123915_Depletion of eukaryotic initiation factor 5B (eIF5B) reprograms the cellular transcriptome and leads to activation of endoplasmic reticulum (ER) stress and c-Jun N-terminal kinase (JNK). 34525951_eIF5B regulates the expression of PD-L1 in prostate cancer cells by interacting with Wig1. 34923394_Dynamic interaction network involving the conserved intrinsically disordered regions in human eIF5. | ENSMUSG00000026083 | Eif5b | 3.642217e+03 | 1.1844771 | 0.244250259 | 0.3172825 | 6.008686e-01 | 0.4382468157 | 0.85439760 | No | Yes | 4.355825e+03 | 768.426955 | 3.188309e+03 | 577.170011 | |
| ENSG00000158805 | 92822 | ZNF276 | protein_coding | Q8N554 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;Centromere;Chromosome;DNA-binding;Kinetochore;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:92822; | chromosome [GO:0005694]; kinetochore [GO:0000776]; nucleus [GO:0005634]; chromatin insulator sequence binding [GO:0043035]; sequence-specific double-stranded DNA binding [GO:1990837]; zinc ion binding [GO:0008270]; regulation of transcription by RNA polymerase II [GO:0006357] | Mouse_homologues 35137157_Transcription factor Zfp276 drives oligodendroglial differentiation and myelination by switching off the progenitor cell program. | ENSMUSG00000001065 | Zfp276 | 6.505816e+03 | 0.9181718 | -0.123163951 | 0.2534507 | 2.326680e-01 | 0.6295525415 | 0.91646120 | No | Yes | 4.718025e+03 | 601.218471 | 5.757715e+03 | 752.019740 | ||
| ENSG00000159023 | 2035 | EPB41 | protein_coding | P11171 | FUNCTION: Protein 4.1 is a major structural element of the erythrocyte membrane skeleton. It plays a key role in regulating membrane physical properties of mechanical stability and deformability by stabilizing spectrin-actin interaction. Recruits DLG1 to membranes. Required for dynein-dynactin complex and NUMA1 recruitment at the mitotic cell cortex during anaphase (PubMed:23870127). {ECO:0000269|PubMed:23870127}. | 3D-structure;Actin-binding;Alternative splicing;Calmodulin-binding;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;Direct protein sequencing;Elliptocytosis;Glycoprotein;Hereditary hemolytic anemia;Mitosis;Nucleus;Phosphoprotein;Pyropoikilocytosis;Reference proteome;Transport | The protein encoded by this gene, together with spectrin and actin, constitute the red cell membrane cytoskeletal network. This complex plays a critical role in erythrocyte shape and deformability. Mutations in this gene are associated with type 1 elliptocytosis (EL1). Alternatively spliced transcript variants encoding different isoforms have been described for this gene.[provided by RefSeq, Oct 2009]. | hsa:2035; | basolateral plasma membrane [GO:0016323]; cell cortex [GO:0005938]; cell junction [GO:0030054]; cortical cytoskeleton [GO:0030863]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; intercellular bridge [GO:0045171]; mitotic spindle [GO:0072686]; nuclear body [GO:0016604]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; spectrin-associated cytoskeleton [GO:0014731]; 1-phosphatidylinositol binding [GO:0005545]; actin binding [GO:0003779]; calmodulin binding [GO:0005516]; phosphoprotein binding [GO:0051219]; protein C-terminus binding [GO:0008022]; protein N-terminus binding [GO:0047485]; spectrin binding [GO:0030507]; structural constituent of cytoskeleton [GO:0005200]; actin cytoskeleton organization [GO:0030036]; actomyosin structure organization [GO:0031032]; cell cycle [GO:0007049]; cell division [GO:0051301]; cortical actin cytoskeleton organization [GO:0030866]; positive regulation of protein binding [GO:0032092]; positive regulation of protein localization to cell cortex [GO:1904778]; regulation of calcium ion transport [GO:0051924]; regulation of cell shape [GO:0008360]; regulation of intestinal absorption [GO:1904478] | 11737230_Reassignment of the EPB4.1 gene to 1p36 and assessment of its involvement in neuroblastomas. 12044158_With deletions or mutations, the ability of the 8 amino acid motif (LKKNFMES) of the spectrin-actin-binding domain of erythrocyte protein 4.1 recombinant peptides to form ternary complexes with spectrin and actin is remarkably diminished. 12130521_falcipain-2-mediated cleavage of protein 4.1 occurs immediately after lysine 437, which lies within a region of the spectrin-actin-binding domain critical for erythrocyte membrane stability. 12239178_A splicing alteration of pre-mRNA generates 2 protein isoforms with distinct assembly to spindle poles in mitotic cells 12427749_A 4.1R isoform expressing the leucine-rich sequence binds to the export receptor CRM1 in a RanGTP-dependent fashion, whereas this does not occur in a mutant whose two conserved hydrophobic residues are substituted 12522012_synthesis of structurally distinct 4.1R protein isoforms in various cell types is regulated by a novel mechanism requiring coordination between upstream transcription initiation events and downstream alternative splicing events 12601556_A novel member of the protein 4.1 family was cloned; it has focal expression in the ovary. 12807908_protein 4.1R has a role in recruiting hDlg to the lateral membrane in epithelial cells 12901833_Protein 4.1R functions as an important tumor suppressor in the molecular pathogenesis of meningioma 12960380_interaction with nuclear actin during nuclear assembly in vitro 15040429_alpha-spectrin ubiquitination at repeats 20 and 21 increases the dissociation of the spectrin-protein-4.1-actin ternary complex thereby regulating protein 4.1's ability to stimulate the spectrin-actin interaction 15184364_135-kDa non-erythroid 4.1R has a role in cell division 15525677_protein 4.1R mitotic regulation involves phosphorylation by cdc2 kinase 15564380_4.1R plays a key role at the centrosome, contributing to the maintenance of a radial microtubule organization 15611095_protein 4.1 phosphorylation modulates erythrocyte membrane mechanical function 15714879_Alternative splicing isoforms are present in muscular dystsrophy skeletal muscle. 15731777_4.1R loss of expression was statistically more common in ependymomas. 15834631_We speculate that over the repetitive cycles of heart muscle contraction and relaxation, 4.1s are likely to locate, support, and coordinate functioning of key membrane-bound macromolecular assemblies. 16060676_4.1R binds to the separate calponin homology CH1 and CH2 domains of beta I spectrin. 16157202_EPB41 gene expression was unchanged in all analyzed meningiomas. This suggests that involvement of the EPB41 gene (4.1R protein) in meningioma pathogenesis should be reconsidered. 16254212_interaction of protein 4.1 with TRPC4 is required for activation of the endothelial ISOC channel. 16368534_protein 4.1R interactions with membrane proteins are regulated by Ca2+ and calmodulin [review] 16537540_Fox-1 and Fox-2 splicing factors have roles in alternative splicing of protein 4.1R 16881872_4.1R60 isoforms are constitutively self-associated, whereas 4.1R80 and 4.1R135 self-association is prevented by intramolecular interactions. 17087826_The interaction of Plasmodium falciparum EBA-181 with the highly conserved 10 kDa domain of 4.1R provides new insight into the molecular mechanisms utilized by P. falciparum during erythrocyte entry 17298666_A decreased expression pattern of the 4.1R protein was observed in the erythrocytes from patients with atypical NA. 17715393_A regulated splicing event in protein 4.1R pre-mRNA-the inclusion of exon 16-encoding peptides for spectrin-actin binding-occurs in late erythroid differentiation 17994571_A deficit in protein 4.1R is recurrent in myeloid malignancies and should be particularly investigated when deletion del (20 q) is present, since this chromosomal abnormality was present in four out of six patients. 18079699_In the 4.1R gene, intrasplicing ultimately determines N-terminal protein structure and function. 18212055_4.1R makes crucial contributions to the structural integrity of centrosomes & mitotic spindles which normally enable mitosis and anaphase to proceed with coordinated precision. 18691159_Findings enable us to offer potential new insights into the differential contribution of 4.1R isoforms, 4.1R(80) and 4.1R(135), to membrane assembly during terminal erythroid differentiation. 18952129_p55 binds to two distinct sites within the FERM domain, and the alternatively spliced exon 5 is necessary for the membrane targeting of protein 4.1R in epithelial cells. 19338061_Study reports the first NMR-derived structure of the 4.1R FERM alpha-lobe domain and propose a new glycophorin C-binding site on the 4.1R FERM alpha-lobe domain, based on our NMR experiments. 19624891_Proteins in the membrane skeleton protein 4.1 family are weakly expressed in non-small cell lung cancer and are related to tumor cell differentiation. 19729518_4.1R gene expression involves transcriptional regulation coupled with a complex splicing regulatory network. 19794081_4.1R plays a role in the phosphatidylserine exposure signaling pathway that is of fundamental importance in red cell turnover. 20007969_Data suggest that one or both of proteins 4.1 and 4.2 cause a portion of band 3 to localize near the spectrin-actin junctions and provide another point of attachment between the membrane skeleton and the lipid bilayer. 20797695_Four EPB41 SNPs showed allelic and genotypic associations with MP in first stage. In the second stage, the allele rs4654388 showed the strongest significant association with MP. rs4654388 G-allele was associated with a significantly increased risk of MP. 20797695_Observational study of gene-disease association. (HuGE Navigator) 20863723_in addition to two known minor shortened and stable spliceoforms, a 4.1R splicing mutation activates an intronic cryptic splice site, which results in a nonsense mRNA major isoform, targeted to degradation in intact cells by Nonsense-mediated mRNA decay. 21486941_Data provide evidence that 4.1R has functional interactions with emerin and A-type lamin that impact upon nuclear architecture, centrosome-nuclear envelope association and the regulation of beta-catenin transcriptional co-activator activity. 21750196_Data show that protein 4.1R is necessary for the localization of IQGAP1 to the leading edge of cells migrating into a wound, whereas IQGAP1 is not required for protein 4.1R localization. 21808284_a novel gene region, EPB41, which may be associated with smoking cessation, along with gene regions in CNR1 that may be targeted to further elucidate the etiology of gender differences in smoking behaviors. 21848512_apo-calmodulin stabilizes the 4.1R N-terminal domain through interaction with its beta-strand-rich C-lobe and provide a novel function for calmodulin, i.e. structural stabilization of 4.1R 22083953_This study characterizes the mechanism by which RBFOX2 regulates protein 4.1R exon 16 splicing through the downstream intronic element UGCAUG. 22197999_Further studies involving siRNA-mediated knockdowns of spectrin, adducin, or p4.1 revealed that those proteins are needed for efficient docking of enterohaemorrhagic Escherichia coli to host cells. 22731252_4.1R regulates NHE1 activity through a direct protein-protein interaction that can be modulated by intracellular pH and Na(+) and Ca(2+) concentrations. 23663475_Plasmodium falciparum PF3D7_0402000 was identified as a new binding partner for the major erythrocyte cytoskeletal protein, 4.1R. 23943871_Results suggest a previously unidentified role for the scaffolding protein 4.1R in locally controlling CLASP2 behavior, CLASP2 cortical platform turnover and GSK3 activity, enabling correct MT organization and dynamics essential for cell polarity. 24081810_Calcium mediates the conformation-based 4.1R FERM domain binding to membrane proteins by calmodulin. 24607279_We conclude that PIP2 may play an important role as a modulator of apo-CaM binding to 4.1R(80) throughout evolution. 24912669_The 4.1R, 4.1N and 4.1B are all expressed at the lateral membrane as well as cytoplasm of epithelial cells, suggesting a potentially redundant role of these proteins. 27453575_identification of EPB41 as a hepatocellular carcinoma susceptibility gene in vitro and in vivo; consistent with this notion, EPB41 expression is significantly decreased in HCC tissue specimens, especially in portal vein metastasis or intrahepatic metastasis, compared to normal tissues 27667160_Using Next-Generation sequencing, we identified the causative genetic mutations in fifteen patients with clinically suspected hereditary elliptocytosis and hereditary pyropoikilocytosis and correlated the identified mutations with the clinical phenotype and ektacytometry profile. 27981895_Our study shows alternative polyadenylation to be an additional mechanism for the generation of 4.1 protein diversity in the already complex EPB41-related genes. Understanding the diversity of EPB41 RNA processing is essential for a full appreciation of the many 4.1 proteins expressed in normal and pathological tissues. 28570402_6 Single Nucleotide Polymorphisms within the EPB41 gene are significantly associated with Mandibular Prognathism (rs2762686, rs2788888, rs4654388, rs502393, rs11581096, and rs488113). The G-allele of SNP, assigned as rs4654388, showed the strongest link with an increased risk of Mandibular Prognathism in the Chinese population. 30227690_the role of lnc-EPB41-1-1 in congenital pouch colon 31776189_Epithelial-specific isoforms of protein 4.1R promote adherens junction assembly in maturing epithelia. 31950797_The c.1215G>A mutation of the EPB41 gene probably accounts for the Elliptocytosis in this family 33242559_EPB41 suppresses the Wnt/beta-catenin signaling in non-small cell lung cancer by sponging ALDOC. 33942936_Clinical and molecular genetic analysis of a Chinese family with hereditary elliptocytosis caused by a novel mutation in the EPB41 gene. | ENSMUSG00000028906 | Epb41 | 5.993182e+03 | 0.9435996 | -0.083753224 | 0.2655171 | 9.713297e-02 | 0.7552976948 | 0.94910089 | No | Yes | 5.229768e+03 | 622.472134 | 5.208686e+03 | 635.648208 | |
| ENSG00000159214 | 149473 | CCDC24 | protein_coding | Q8N4L8 | Alternative splicing;Coiled coil;Reference proteome | hsa:149473; | blastocyst hatching [GO:0001835] | ENSMUSG00000078588 | Ccdc24 | 1.626754e+02 | 1.3827701 | 0.467561338 | 0.3262347 | 2.064074e+00 | 0.1508065124 | 0.77404149 | No | Yes | 1.553307e+02 | 21.825411 | 1.202285e+02 | 17.673558 | ||||
| ENSG00000159335 | 5763 | PTMS | protein_coding | P20962 | FUNCTION: Parathymosin may mediate immune function by blocking the effect of prothymosin alpha which confers resistance to certain opportunistic infections. | Acetylation;Direct protein sequencing;Immunity;Phosphoprotein;Reference proteome | hsa:5763; | nucleus [GO:0005634]; DNA replication [GO:0006260]; immune system process [GO:0002376] | 19204726_Observational study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) | 5.751401e+03 | 1.4654787 | 0.551371965 | 0.3402301 | 2.614993e+00 | 0.1058579591 | 0.75783482 | No | Yes | 6.892547e+03 | 1080.935086 | 3.818787e+03 | 614.799049 | ||||
| ENSG00000159445 | 117145 | THEM4 | protein_coding | Q5T1C6 | FUNCTION: Has acyl-CoA thioesterase activity towards medium and long-chain (C14 to C18) fatty acyl-CoA substrates, and probably plays a role in mitochondrial fatty acid metabolism. Plays a role in the apoptotic process, possibly via its regulation of AKT1 activity. According to PubMed:11598301, inhibits AKT1 phosphorylation and activity. According to PubMed:17615157, enhances AKT1 activity by favoring its phosphorylation and translocation to plasma membrane. {ECO:0000269|PubMed:11598301, ECO:0000269|PubMed:17615157, ECO:0000269|PubMed:19168129, ECO:0000269|PubMed:19421406, ECO:0000269|PubMed:19453107, ECO:0000269|PubMed:22871024}. | 3D-structure;Acetylation;Apoptosis;Cell membrane;Cell projection;Cytoplasm;Fatty acid metabolism;Hydrolase;Lipid metabolism;Membrane;Mitochondrion;Mitochondrion inner membrane;Phosphoprotein;Reference proteome;Transit peptide | Protein kinase B (PKB) is a major downstream target of receptor tyrosine kinases that signal via phosphatidylinositol 3-kinase. Upon cell stimulation, PKB is translocated to the plasma membrane, where it is phosphorylated in the C-terminal regulatory domain. The protein encoded by this gene negatively regulates PKB activity by inhibiting phosphorylation. Transcription of this gene is commonly downregulated in glioblastomas. [provided by RefSeq, Jul 2008]. | hsa:117145; | cytosol [GO:0005829]; mitochondrial inner membrane [GO:0005743]; mitochondrial intermembrane space [GO:0005758]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; myristoyl-CoA hydrolase activity [GO:0102991]; palmitoyl-CoA hydrolase activity [GO:0016290]; fatty acid metabolic process [GO:0006631]; protein kinase B signaling [GO:0043491]; regulation of mitochondrial membrane permeability involved in apoptotic process [GO:1902108] | 17615157_CTMP induces translocation of Akt to the membrane and thereby increases the level of Akt phosphorylation. As a result, CTMP enhances various cellular activities that are principally mediated by the PI3-kinase/Akt pathway. 19168129_Proper maturation of CTMP is essential for its pro-apoptotic function. CTMP delays PKB phosphorylation following cell death induction, suggesting that CTMP regulates apoptosis via inhibition of PKB. 19604401_Phosphorylation on Ser37/Ser38 of CTMP is important for the prevention of mitochondrial localization of CTMP, eventually leading to cell death by binding to heat shock protein 70. 19679565_Data show low or moderate methylation was found in seven selected genes BAD, BBC3, CAV1, CDK2AP1, NPM1, PRKCDBP and THEM4. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22200884_Suggest that CTMP may therefore play a critical role in mitochondrial-mediated apoptosis in lung cancer cells. 27328758_Results find that head and neck squamous cell carcinoma (HNSCC) tumor tissues and cell lines had high levels of CTMP expression. These data suggest that CTMP functions as a positive regulator of Akt and facilitates HNSCC invasion such as LN metastasis by regulating EMT in a Snail-dependent manner indicating the oncogenic activity of CTMP in HNSCC, and its expression is an independent predictor of clinical prognosis. 27447863_CTMP expression may be considered as a prognostic biomarker in HER2-enriched breast cancer and high expression may indicate a utility for AKT-inhibition in these patients. 32654229_Enolase-phosphatase 1 acts as an oncogenic driver in glioma. | ENSMUSG00000028145 | Them4 | 7.737865e+02 | 0.9435152 | -0.083882339 | 0.2788466 | 8.896481e-02 | 0.7654973296 | 0.95299881 | No | Yes | 6.124975e+02 | 78.016310 | 7.359379e+02 | 95.811609 | |
| ENSG00000159640 | 1636 | ACE | protein_coding | P12821 | FUNCTION: Converts angiotensin I to angiotensin II by release of the terminal His-Leu, this results in an increase of the vasoconstrictor activity of angiotensin. Also able to inactivate bradykinin, a potent vasodilator. Has also a glycosidase activity which releases GPI-anchored proteins from the membrane by cleaving the mannose linkage in the GPI moiety. | 3D-structure;Alternative splicing;Carboxypeptidase;Cell membrane;Cytoplasm;Direct protein sequencing;Disulfide bond;Glycoprotein;Hydrolase;Membrane;Metal-binding;Metalloprotease;Phosphoprotein;Protease;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix;Zinc | This gene encodes an enzyme involved in blood pressure regulation and electrolyte balance. It catalyzes the conversion of angiotensin I into a physiologically active peptide angiotensin II. Angiotensin II is a potent vasopressor and aldosterone-stimulating peptide that controls blood pressure and fluid-electrolyte balance. This angiotensin converting enzyme (ACE) also inactivates the vasodilator protein, bradykinin. Accordingly, the encoded enzyme increases blood pressure and is a drug target of ACE inhibitors, which are often prescribed to reduce blood pressure. This enzyme additionally plays a role in fertility through its ability to cleave and release GPI-anchored membrane proteins in spermatozoa. Many studies have associated the presence or absence of a 287 bp Alu repeat element in this gene with the levels of circulating enzyme. This polymorphism, as well as mutations in this gene, have been implicated in a wide variety of diseases including cardiovascular pathophysiologies, psoriasis, renal disease, stroke, and Alzheimer's disease. Regulation of the homologous ACE2 gene may be involved in progression of disease caused by several human coronaviruses, including SARS-CoV and SARS-CoV-2. Alternative splicing results in multiple transcript variants encoding both somatic (sACE) and male-specific testicular (tACE) isoforms. [provided by RefSeq, Sep 2020]. | hsa:1636; | basal plasma membrane [GO:0009925]; brush border membrane [GO:0031526]; endosome [GO:0005768]; external side of plasma membrane [GO:0009897]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; lysosome [GO:0005764]; plasma membrane [GO:0005886]; sperm midpiece [GO:0097225]; actin binding [GO:0003779]; bradykinin receptor binding [GO:0031711]; carboxypeptidase activity [GO:0004180]; chloride ion binding [GO:0031404]; endopeptidase activity [GO:0004175]; exopeptidase activity [GO:0008238]; heterocyclic compound binding [GO:1901363]; metallodipeptidase activity [GO:0070573]; metallopeptidase activity [GO:0008237]; mitogen-activated protein kinase binding [GO:0051019]; mitogen-activated protein kinase kinase binding [GO:0031434]; peptidyl-dipeptidase activity [GO:0008241]; tripeptidyl-peptidase activity [GO:0008240]; zinc ion binding [GO:0008270]; aging [GO:0007568]; amyloid-beta metabolic process [GO:0050435]; angiogenesis involved in coronary vascular morphogenesis [GO:0060978]; angiotensin maturation [GO:0002003]; animal organ regeneration [GO:0031100]; antigen processing and presentation of peptide antigen via MHC class I [GO:0002474]; arachidonic acid secretion [GO:0050482]; blood vessel diameter maintenance [GO:0097746]; blood vessel remodeling [GO:0001974]; bradykinin catabolic process [GO:0010815]; brain development [GO:0007420]; cell proliferation in bone marrow [GO:0071838]; cellular response to aldosterone [GO:1904045]; cellular response to glucose stimulus [GO:0071333]; eating behavior [GO:0042755]; embryo development ending in birth or egg hatching [GO:0009792]; female pregnancy [GO:0007565]; heart contraction [GO:0060047]; hematopoietic stem cell differentiation [GO:0060218]; hormone catabolic process [GO:0042447]; kidney development [GO:0001822]; lung alveolus development [GO:0048286]; male gonad development [GO:0008584]; mononuclear cell proliferation [GO:0032943]; negative regulation of calcium ion import [GO:0090281]; negative regulation of gap junction assembly [GO:1903597]; negative regulation of gene expression [GO:0010629]; negative regulation of glucose import [GO:0046325]; negative regulation of protein binding [GO:0032091]; negative regulation of renal sodium excretion [GO:0035814]; neutrophil mediated immunity [GO:0002446]; peptide catabolic process [GO:0043171]; positive regulation of apoptotic process [GO:0043065]; positive regulation of inflammatory response [GO:0050729]; positive regulation of neurogenesis [GO:0050769]; positive regulation of peptidyl-cysteine S-nitrosylation [GO:2000170]; positive regulation of peptidyl-tyrosine autophosphorylation [GO:1900086]; positive regulation of protein binding [GO:0032092]; positive regulation of protein tyrosine kinase activity [GO:0061098]; positive regulation of systemic arterial blood pressure [GO:0003084]; positive regulation of vasoconstriction [GO:0045907]; posttranscriptional regulation of gene expression [GO:0010608]; proteolysis [GO:0006508]; regulation of angiotensin metabolic process [GO:0060177]; regulation of blood pressure [GO:0008217]; regulation of hematopoietic stem cell proliferation [GO:1902033]; regulation of renal output by angiotensin [GO:0002019]; regulation of smooth muscle cell migration [GO:0014910]; regulation of systemic arterial blood pressure by renin-angiotensin [GO:0003081]; regulation of vasoconstriction [GO:0019229]; response to dexamethasone [GO:0071548]; response to hypoxia [GO:0001666]; response to laminar fluid shear stress [GO:0034616]; response to lipopolysaccharide [GO:0032496]; response to nutrient levels [GO:0031667]; response to thyroid hormone [GO:0097066]; response to xenobiotic stimulus [GO:0009410]; sensory perception of pain [GO:0019233]; spermatogenesis [GO:0007283]; vasoconstriction [GO:0042310] | 11024215_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11027844_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11029323_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11030378_Observational study of gene-disease association. (HuGE Navigator) 11035682_Observational study of gene-disease association. (HuGE Navigator) 11036822_Observational study of gene-disease association. (HuGE Navigator) 11053482_Observational study of gene-disease association. (HuGE Navigator) 11054622_Observational study of gene-disease association. (HuGE Navigator) 11071585_Observational study of gene-disease association. (HuGE Navigator) 11078932_Observational study of gene-disease association. (HuGE Navigator) 11079515_Observational study of gene-disease association. (HuGE Navigator) 11079661_Observational study of gene-disease association. (HuGE Navigator) 11082147_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11084577_Observational study of gene-disease association. (HuGE Navigator) 11085286_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11106322_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11106834_Observational study of gene-disease association. (HuGE Navigator) 11116112_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11116113_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11121171_Observational study of gene-disease association. (HuGE Navigator) 11122103_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11122322_Observational study of gene-disease association. (HuGE Navigator) 11124487_Observational study of gene-disease association. (HuGE Navigator) 11136175_Observational study of gene-disease association. (HuGE Navigator) 11137090_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11142763_Observational study of gene-disease association. (HuGE Navigator) 11155292_Observational study of gene-disease association. (HuGE Navigator) 11155741_Observational study of gene-disease association. (HuGE Navigator) 11168787_Observational study of gene-disease association. (HuGE Navigator) 11181802_Observational study of gene-disease association. (HuGE Navigator) 11200871_Observational study of gene-disease association. (HuGE Navigator) 11204304_Observational study of gene-disease association. (HuGE Navigator) 11208361_Observational study of gene-disease association. (HuGE Navigator) 11208365_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11208681_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11208755_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11210078_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11213892_Observational study of gene-disease association. (HuGE Navigator) 11217909_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11230288_Observational study of gene-disease association. (HuGE Navigator) 11231997_Observational study of gene-disease association. (HuGE Navigator) 11234416_Observational study of gene-disease association. (HuGE Navigator) 11239522_Observational study of gene-disease association. (HuGE Navigator) 11244007_Observational study of gene-disease association. (HuGE Navigator) 11248758_Observational study of gene-disease association. (HuGE Navigator) 11250978_Observational study of gene-disease association. (HuGE Navigator) 11256792_Observational study of gene-disease association. (HuGE Navigator) 11257269_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11265121_Observational study of gene-disease association. (HuGE Navigator) 11273991_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11280044_Observational study of gene-disease association. (HuGE Navigator) 11283791_Observational study of gene-disease association. (HuGE Navigator) 11286523_Observational study of gene-disease association. (HuGE Navigator) 11288810_Observational study of gene-disease association. (HuGE Navigator) 11289708_Observational study of gene-disease association. (HuGE Navigator) 11300226_Observational study of gene-disease association. (HuGE Navigator) 11305733_Observational study of gene-disease association. (HuGE Navigator) 11311153_Observational study of gene-disease association. (HuGE Navigator) 11313676_Observational study of gene-disease association. (HuGE Navigator) 11317203_Observational study of gene-disease association. (HuGE Navigator) 11328206_Observational study of gene-disease association. (HuGE Navigator) 11330506_Observational study of gene-disease association. (HuGE Navigator) 11330874_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11341749_Observational study of genotype prevalence. (HuGE Navigator) 11344227_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11345362_Observational study of gene-disease association. (HuGE Navigator) 11352521_Observational study of gene-disease association. (HuGE Navigator) 11354635_Observational study of gene-disease association. (HuGE Navigator) 11354780_Observational study of gene-disease association. (HuGE Navigator) 11354781_Observational study of gene-disease association. (HuGE Navigator) 11355019_Observational study of gene-disease association. (HuGE Navigator) 11357377_Observational study of genotype prevalence. (HuGE Navigator) 11359065_Observational study of genotype prevalence. (HuGE Navigator) 11361058_Observational study of gene-disease association. (HuGE Navigator) 11377823_Observational study of gene-disease association. (HuGE Navigator) 11381371_Observational study of gene-disease association. (HuGE Navigator) 11387483_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11399938_Observational study of gene-disease association. (HuGE Navigator) 11401071_Observational study of gene-disease association. (HuGE Navigator) 11401115_Observational study of gene-disease association. (HuGE Navigator) 11402126_Observational study of gene-disease association. (HuGE Navigator) 11408790_Observational study of gene-disease association. (HuGE Navigator) 11409168_Observational study of gene-disease association. (HuGE Navigator) 11421129_Observational study of gene-disease association. (HuGE Navigator) 11422735_Observational study of gene-disease association. (HuGE Navigator) 11427204_Observational study of gene-disease association. (HuGE Navigator) 11428725_Observational study of gene-disease association. (HuGE Navigator) 11431175_Observational study of gene-disease association. (HuGE Navigator) 11436125_Observational study of gene-disease association. (HuGE Navigator) 11436566_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11437027_Observational study of genotype prevalence. (HuGE Navigator) 11446493_Observational study of gene-disease association. (HuGE Navigator) 11447495_Observational study of gene-disease association. (HuGE Navigator) 11463587_Observational study of gene-disease association. (HuGE Navigator) 11473656_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 11474225_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11484170_Observational study of gene-disease association. (HuGE Navigator) 11485372_Observational study of gene-disease association. (HuGE Navigator) 11487079_Observational study of gene-disease association. (HuGE Navigator) 11498459_Observational study of gene-disease association. (HuGE Navigator) 11501342_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11505631_Observational study of gene-disease association. (HuGE Navigator) 11507973_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11509536_Observational study of gene-disease association. (HuGE Navigator) 11518842_Observational study of gene-disease association. (HuGE Navigator) 11522714_Observational study of gene-disease association. (HuGE Navigator) 11522715_Observational study of gene-disease association. (HuGE Navigator) 11525534_Observational study of gene-disease association. (HuGE Navigator) 11527624_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11531970_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11532108_Observational study of gene-disease association. (HuGE Navigator) 11544438_Observational study of gene-disease association. (HuGE Navigator) 11545752_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 11568114_Observational study of gene-disease association. (HuGE Navigator) 11568784_Observational study of gene-disease association. (HuGE Navigator) 11569699_Observational study of gene-disease association. (HuGE Navigator) 11577832_Observational study of genotype prevalence. (HuGE Navigator) 11593098_Clinical trial of gene-environment interaction. (HuGE Navigator) 11596779_Observational study of gene-disease association. (HuGE Navigator) 11599244_Observational study of gene-disease association. (HuGE Navigator) 11640993_Observational study of gene-disease association. (HuGE Navigator) 11665795_Observational study of gene-disease association. (HuGE Navigator) 11665795_The DD genotype of ACE polymorphism is a risk factor for coronary artery disease and coronary stent restenosis in Japanese patients. 11668351_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11677359_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11688760_Observational study of gene-disease association. (HuGE Navigator) 11691520_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11692002_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11696022_Observational study of gene-disease association. (HuGE Navigator) 11696658_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11696688_Observational study of gene-disease association. (HuGE Navigator) 11699055_Observational study of gene-disease association. (HuGE Navigator) 11707217_Observational study of gene-disease association. (HuGE Navigator) 11707686_Observational study of gene-disease association. (HuGE Navigator) 11711521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11711524_Observational study of gene-disease association. (HuGE Navigator) 11712321_Observational study of gene-disease association. (HuGE Navigator) 11714857_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11715070_Observational study of gene-disease association. (HuGE Navigator) 11718065_Observational study of gene-disease association. (HuGE Navigator) 11720769_Observational study of gene-disease association. (HuGE Navigator) 11723093_Observational study of gene-disease association. (HuGE Navigator) 11728946_Observational study of gene-disease association. (HuGE Navigator) 11732226_Observational study of gene-disease association. (HuGE Navigator) 11733623_Observational study of gene-disease association. (HuGE Navigator) 11737220_Observational study of gene-disease association. (HuGE Navigator) 11747437_During shedding of the ectodomain of ACE, a minimum requirement is an exposed or unhindered sequence of minimum length between the extracellular domain and the membrane, within or adjacent to otherwise compact, folded domains. 11749850_Observational study of genotype prevalence. (HuGE Navigator) 11756350_Observational study of gene-disease association. (HuGE Navigator) 11768721_Observational study of gene-disease association. (HuGE Navigator) 11768721_association between left ventricular hypertrophy and the C825T allele of the G-protein beta3 subunit gene in Arabs. (G PROTEIN BETA3) 11770799_It appears that the DD genotype (of ACE gene) is associated with progression of Japanese type 2 diabetic nephropathy. 11770799_Observational study of gene-disease association. (HuGE Navigator) 11773214_Observational study of gene-disease association. (HuGE Navigator) 11774217_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11775123_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11776100_Observational study of gene-disease association. (HuGE Navigator) 11776522_Observational study of gene-disease association. (HuGE Navigator) 11781693_Observational study of gene-disease association. (HuGE Navigator) 11787479_Observational study of gene-disease association. (HuGE Navigator) 11791024_Observational study of gene-disease association. (HuGE Navigator) 11791802_Observational study of gene-disease association. (HuGE Navigator) 11793101_Observational study of gene-disease association. (HuGE Navigator) 11796676_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11802972_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11803189_Observational study of gene-disease association. (HuGE Navigator) 11818755_Observational study of gene-disease association. (HuGE Navigator) 11818755_vasomotor properties are influenced by the I/D polymorphism of the ACE gene. 11835945_Observational study of gene-disease association. (HuGE Navigator) 11836444_Observational study of gene-disease association. (HuGE Navigator) 11840772_Meta-analysis of gene-disease association. (HuGE Navigator) 11842043_ACE insertion/deletion polymorphism and submaximal exercise cardiovascular hemodynamics in postmenopausal women. 11842043_Observational study of gene-disease association. (HuGE Navigator) 11851058_Observational study of gene-disease association. (HuGE Navigator) 11851983_Homozygosity for the insertion allele of the I/D polymorphism is associated with adaptation to high altitude. 11851983_Observational study of genotype prevalence. (HuGE Navigator) 11852788_Observational study of genotype prevalence. (HuGE Navigator) 11855311_Observational study of gene-disease association. (HuGE Navigator) 11859923_ACE polymorphism is not a risk factor for the development of cerebral infarction in a Korean population. 11859923_Observational study of gene-disease association. (HuGE Navigator) 11860787_Observational study of gene-disease association. (HuGE Navigator) 11861692_Observational study of gene-disease association. (HuGE Navigator) 11865575_Observational study of gene-disease association. (HuGE Navigator) 11870238_Observational study of gene-disease association. (HuGE Navigator) 11879185_effects of monoclonal antibodies (mAbs) to eight different epitopes on the N-terminal domain of ACE on shedding 11882570_Observational study of gene-disease association. (HuGE Navigator) 11882570_alleles of the ACE gene may be a predisposing factor for developing white matter lesions in essential hypertensive patients. 11888948_Observational study of gene-disease association. (HuGE Navigator) 11893827_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11899582_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11903317_Angiotensin I-converting enzyme and metabolism of the haematological peptide N-acetyl-seryl-aspartyl-lysyl-proline. 11903319_Different aortic reflection wave responses following long-term angiotensin-converting enzyme inhibition and beta-blocker in essential hypertension 11903350_A prospective evaluation of the angiotensin-converting enzyme D/I polymorphism and left ventricular remodeling in the 'Healing and Early Afterload Reducing Therapy' study. 11903350_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 11906289_Toward an optimal joint recognition of the S1' subsites of endothelin converting enzyme-1 (ECE-1), angiotensin converting enzyme (ACE), and neutral endopeptidase (NEP). 11909563_Observational study of gene-disease association. (HuGE Navigator) 11910300_Polymorphisms of genes encoding kininase II as risk factors for orthostatic hypotension. 11910301_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 11912262_Enhanced responses of blood pressure, renal function, and aldosterone to angiotensin I in the DD genotype are blunted by low sodium intake. MAP, renal hemodynamic parameters, and aldosterone concentrations to AngI are enhanced for DD genotype. 11912262_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11917060_Observational study of gene-disease association. (HuGE Navigator) 11918733_Observational study of gene-disease association. (HuGE Navigator) 11918988_Observational study of gene-disease association. (HuGE Navigator) 11920854_Observational study of gene-disease association. (HuGE Navigator) 11923700_Observational study of gene-disease association. (HuGE Navigator) 11924723_Observational study of gene-disease association. (HuGE Navigator) 11926202_Observational study of gene-disease association. (HuGE Navigator) 11927778_Observational study of genotype prevalence. (HuGE Navigator) 11927778_Results suggest ethnic heterogeneity in the angiotensin-converting enzyme gene with a significant gene cline with higher insertion allele frequency. 11938025_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11941276_Observational study of gene-disease association. (HuGE Navigator) 11951494_Observational study of gene-disease association. (HuGE Navigator) 11956052_Observational study of gene-disease association. (HuGE Navigator) 11956670_Observational study of genotype prevalence. (HuGE Navigator) 11963567_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11985486_the ACE gene has been recognised as a top candidate gene in cardiovascular diseases 11990733_Observational study of gene-disease association. (HuGE Navigator) 11990733_may play a role in enhanced endurance performance but this is not mediated by differences in VO2max or the heart rate/VO2 relationship in response to training. 11992568_Observational study of gene-disease association. (HuGE Navigator) 11992568_in a study of 261 patients with Alzheimer disease, no association found between genotype counts or allelic frequencies of DCP1, the gene encoding angiotensin-converting enzyme 11994001_Selective inhibition of the C-domain of angiotensin I converting enzyme by bradykinin potentiating peptides. 11994975_The ACE DD genotype was associated with myocardial infarction in African-American males but not in females. Racial/ethnic and sex differences in genotype distribution of the ACE 4656(CT)(2/3) polymorphism. 12006218_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 12010139_Observational study of gene-disease association. (HuGE Navigator) 12015245_Increased risk of systemic sclerosis in ACE D allele carriers. 12015245_Observational study of gene-disease association. (HuGE Navigator) 12015366_Observational study of gene-disease association. (HuGE Navigator) 12015946_Observational study of gene-disease association. (HuGE Navigator) 12023681_Observational study of gene-disease association. (HuGE Navigator) 12023681_There is little association between ACE I/D polymorphism and hypertension in the general Japanese population. 12031704_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12031990_ACE DD genotype is associated with an increased susceptibility to type 2 diabetes 12031990_Observational study of gene-disease association. (HuGE Navigator) 12032106_Observational study of gene-disease association. (HuGE Navigator) 12032106_gene insertion/deletion polymorphism associated with 1998 World Health Organization definition of metabolic syndrome in Chinese type 2 diabetic patients 12043242_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12043886_Observational study of gene-disease association. (HuGE Navigator) 12043886_insertion/deletion polymorphisms of ACE gene did not affect susceptibility to systemic lupus erythematosus, lupus nephritis and the vascular manifestations, including Raynaud's phenomenon, in Korean SLE patients 12047032_Observational study of gene-disease association. (HuGE Navigator) 12070247_Observational study of gene-disease association. (HuGE Navigator) 12081578_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12082592_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12084438_Observational study of gene-disease association. (HuGE Navigator) 12095411_Observational study of gene-disease association. (HuGE Navigator) 12099691_Observational study of gene-disease association. (HuGE Navigator) 12099691_We have been the first to demonstrate that an Alu element insertion exerts protective effects against age-related macular degeneration (AMD). 12102452_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12113906_Observational study of gene-disease association. (HuGE Navigator) 12114335_Observational study of gene-disease association. (HuGE Navigator) 12119485_Observational study of gene-disease association. (HuGE Navigator) 12122874_The ACE polymorphism does not seem to be an independent risk factor for stroke. 12122980_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12123487_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12123491_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12126783_Observational study of gene-disease association. (HuGE Navigator) 12126783_polymorphic in normoalbuminuric type 1 diabetic patients 12127851_Observational study of gene-disease association. (HuGE Navigator) 12133420_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12133420_investigate the relationship of gene polymorphism of angiotension converting enzyme (ACE), aldosterone synthase (CYP11B2) with essential hypertension (EH) and left ventricular hypertrophy (LVH) in hypertensive patients 12133519_Observational study of gene-disease association. (HuGE Navigator) 12135330_Observational study of gene-disease association. (HuGE Navigator) 12143941_Angiotensin-converting enzyme insertion/deletion (I/D) gene polymorphism plays a role in determining the inter-individual variability of circulating angiotensin-converting enzyme activity and intracellular angiotensin-converting enzyme levels. 12143941_Observational study of gene-disease association. (HuGE Navigator) 12147330_Hypothalamic-pituitary-adrenocortical axis dysregulation in patients with major depression is influenced by the insertion/deletion polymorphism in the angiotensin I-converting enzyme gene. 12147330_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12147333_Observational study of gene-disease association. (HuGE Navigator) 12147333_The angiotensin 1-converting enzyme insertion (I)/deletion (D) polymorphism does not influence the extent of amyloid or tau pathology in patients with sporadic Alzheimer's disease. 12147786_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12147786_in type 1 diabetic patients, the ACE and the PC-1 genes interact in increasing the individual risk of having a faster diabetic nephropathy progression 12152254_Observational study of gene-disease association. (HuGE Navigator) 12153971_Observational study of gene-disease association. (HuGE Navigator) 12160518_angiotensin converting enzyme is downregulated by TNF-alpha in differentiating human macrophages 12164879_Observational study of gene-disease association. (HuGE Navigator) 12165743_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12165743_The presence of the D allele on the angiotensin I-converting enzyme (ACE) gene in middle-aged hypertensive patients determines higher circulating ACE activity but not increased sympathetic activity in response to submaximal exercise. 12165749_Observational study of gene-disease association. (HuGE Navigator) 12165749_Results suggest the additive effect of angiotensin I-converting enzyme (ACE) and heart chymase (CMA) gene polymorphisms on the increase in left ventricular mass in NIDDM patients. 12166538_Observational study of gene-disease association. (HuGE Navigator) 12166654_Observational study of gene-disease association. (HuGE Navigator) 12169209_A candidate gene associated with essential hypertension. 12169829_Observational study of gene-disease association. (HuGE Navigator) 12169829_The incidence of the angiotensin-converting enzyme insertion genotype was higher in retinopathy of prematurity cases compared to non-ROP controls. 12172317_Carotid and femoral intima-media thickness were assessed in subjects genotyped for the presence of the ACE D, aldosterone synthase -344T and alpha-adducin 460Trp alleles. 12172317_Observational study of gene-disease association. (HuGE Navigator) 12172760_Observational study of gene-disease association. (HuGE Navigator) 12172761_Observational study of gene-disease association. (HuGE Navigator) 12173033_high-resolution genetic mapping of the QTL influencing circulating ACE activity 12176567_impact of polymorphism on renal allograft function, blood pressure, and proteinuria under ACE inhibition 12186695_in leukemia patients, the ACE concentration was found to be significantly higher in the bone marrow than in the peripheral blood, with level correlated with blast percentages, indicating presence of a local BM renin angiotensin system 12194796_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12196500_Observational study of gene-disease association. (HuGE Navigator) 12203048_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12204015_Observational study of gene-disease association. (HuGE Navigator) 12204859_Observational study of gene-disease association. (HuGE Navigator) 12205735_Observational study of gene-disease association. (HuGE Navigator) 12208484_ACE I/D polymorphism is not a significant risk factor for peripheral arterial disease. 12208484_Observational study of gene-disease association. (HuGE Navigator) 12217990_Meta-analysis of gene-disease association. (HuGE Navigator) 12220450_Observational study of gene-disease association. (HuGE Navigator) 12222694_Observational study of gene-disease association. (HuGE Navigator) 12297007_Angiotensin 1-converting enzyme gene polymorphism is associated with hypertension 12297007_Observational study of gene-disease association. (HuGE Navigator) 12297438_Observational study of gene-disease association. (HuGE Navigator) 12356632_Observational study of gene-disease association. (HuGE Navigator) 12359135_Observational study of gene-disease association. (HuGE Navigator) 12360157_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12360613_Ace gene polymorphism is associated with acute myocardial infarction, stable angina, postinfarction cardiosclerosis. 12360613_Observational study of gene-disease association. (HuGE Navigator) 12362316_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12362316_relationship between angiotensin I converting enzyme gene insertion/deletion polymorphism and Alzheimer disease (AD) 12371972_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12372063_Observational study of gene-disease association. (HuGE Navigator) 12372418_the boundaries of the ACE ectodomain are Asp40 at the N-terminus and Gly615 at the C-terminus 12388790_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12394950_Observational study of gene-disease association. (HuGE Navigator) 12398019_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12406857_Observational study of gene-disease association. (HuGE Navigator) 12419932_Observational study of gene-disease association. (HuGE Navigator) 12419936_Observational study of genetic testing. (HuGE Navigator) 12422143_Meta-analysis of gene-disease association. (HuGE Navigator) 12425488_Observational study of gene-disease association. (HuGE Navigator) 12426159_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12442545_Observational study of gene-disease association. (HuGE Navigator) 12449516_Observational study of gene-disease association. (HuGE Navigator) 12453999_The ACE insertion/deletion polymorphism is not associated with the metabolic syndrome in Brazilian type 2 diabetic patients. 12454231_Observational study of gene-disease association. (HuGE Navigator) 12458570_Observational study of gene-disease association. (HuGE Navigator) 12459519_Observational study of gene-disease association. (HuGE Navigator) 12468767_Observational study of gene-disease association. (HuGE Navigator) 12469628_Observational study of gene-disease association. (HuGE Navigator) 12469902_A significant, moderate, male, gender-specific independent association between a polymorphism and high blood pressure was found 12469902_Observational study of gene-disease association. (HuGE Navigator) 12471298_Observational study of gene-disease association. (HuGE Navigator) 12471298_The I/D-ACE gene polymorphism has no important effect on susceptibility to acute mountain sickness or high-altitude pulmonary edema. 12476328_Observational study of gene-disease association. (HuGE Navigator) 12476328_combined action of this protein and and GNB3 in major depression: may have a link tocaardiovascular disease 12476891_No statistically significant differences between groups were found in the allele frequency and genotype distribution for ACE and AGT polymorphisms. 12476891_Observational study of gene-disease association. (HuGE Navigator) 12478352_Observational study of gene-disease association. (HuGE Navigator) 12478352_the DD genotype and D allele of angiotensin converting enzyme may be a genetic susceptibility factor contributing to scar formation in vesicoureteric reflux 12480755_Observational study of gene-disease association. (HuGE Navigator) 12484503_Both the ACE and chymase-like enzyme activities in the aneurysmal aortae were significantly higher than those in the control aortae. 12484507_ACE polymorphism does not appear to have significant association with blood pressure, changes in blood pressure or sex in Japanese subjects, who have a more homogeneous genetic background than any other group reported to date. 12484507_Observational study of gene-disease association. (HuGE Navigator) 12484511_Linear model of interaction between ACE genotype and conventional risk factors showed systolic blood pressure-ACE genotype interaction were significantly associated with intima-media thickness. 12484511_Observational study of gene-environment interaction. (HuGE Navigator) 12511523_Angiotensin-converting enzyme gene I alle | ENSMUSG00000020681 | Ace | 1.073378e+02 | 1.6965632 | 0.762615203 | 0.3985854 | 3.563392e+00 | 0.0590669183 | 0.68938192 | No | Yes | 1.133078e+02 | 23.195147 | 7.630150e+01 | 16.367919 | |
| ENSG00000159917 | 9310 | ZNF235 | protein_coding | Q14590 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene product belongs to the zinc finger protein superfamily, members of which are regulatory proteins characterized by nucleic acid-binding zinc finger domains. The encoded protein is a member of the Kruppel family of zinc finger proteins, and contains Kruppel-associated box (KRAB) A and B domains and 15 tandemly arrayed C2H2-type zinc fingers. It is an ortholog of the mouse Zfp93 protein. This gene is located in a cluster of zinc finger genes on 19q13.2. [provided by RefSeq, Jul 2008]. | hsa:9310; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 7.809832e+01 | 0.7761354 | -0.365619739 | 0.4280266 | 7.304962e-01 | 0.3927223931 | 0.84317192 | No | Yes | 5.816110e+01 | 16.363950 | 9.700794e+01 | 27.281230 | ||||
| ENSG00000160145 | 8997 | KALRN | protein_coding | O60229 | FUNCTION: Promotes the exchange of GDP by GTP. Activates specific Rho GTPase family members, thereby inducing various signaling mechanisms that regulate neuronal shape, growth, and plasticity, through their effects on the actin cytoskeleton. Induces lamellipodia independent of its GEF activity. {ECO:0000269|PubMed:10023074}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Cytoskeleton;Disulfide bond;Guanine-nucleotide releasing factor;Immunoglobulin domain;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;SH3 domain;Serine/threonine-protein kinase;Transferase | Huntington's disease (HD), a neurodegenerative disorder characterized by loss of striatal neurons, is caused by an expansion of a polyglutamine tract in the HD protein huntingtin. This gene encodes a protein that interacts with the huntingtin-associated protein 1, which is a huntingtin binding protein that may function in vesicle trafficking. [provided by RefSeq, Apr 2016]. | hsa:8997; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extrinsic component of membrane [GO:0019898]; nucleoplasm [GO:0005654]; postsynaptic density [GO:0014069]; ATP binding [GO:0005524]; guanyl-nucleotide exchange factor activity [GO:0005085]; metal ion binding [GO:0046872]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; axon guidance [GO:0007411]; central nervous system development [GO:0007417]; ephrin receptor signaling pathway [GO:0048013]; intracellular signal transduction [GO:0035556]; nervous system development [GO:0007399]; protein phosphorylation [GO:0006468]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165]; vesicle-mediated transport [GO:0016192] | 14742910_we have identified multiple transcriptional start sites in rats and humans. These multiple transcriptional start sites result in full-length Kalirin transcripts possessing different 5' ends encoding proteins with differing amino termini 15950621_Kalirin GEF1 domain induces lamellipodia through activation of Pak, where Guanine nucleotide exchange factor (GEF) activity is not required. 17357071_Three SNPs from the kalirin (KALRN) gene are associated with early-onset coronary artery disease. 17640372_ARF6 recruits KALRN to the cell membrane facilitating Rac activation. 17851188_Our observation is the first to relate kalirin to Alzheimer's disease. Kalirin was consistently under-expressed in Alzheimer's disease hippocampus. 18199770_Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. 18839057_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18953434_Observational study of gene-disease association. (HuGE Navigator) 19706030_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20080650_Observational study of gene-disease association. (HuGE Navigator) 20107840_Observational study of gene-disease association. (HuGE Navigator) 20107840_Two SNPs in the KALRN gene region (rs17286604 and rs11712619)constitute risk factors for ischemic stroke. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20604901_SNX1 and SNX2 interact with Kalirin-7. Overexpression of SNX1 or SNX2 and Kalirin-7 partially redistributes both SNXs to the plasma membrane, and results in RhoG-dependent lamellipodia formation. 20730383_Studies indicate that Kalirin-7 plays a key role in excitatory synapse formation and function. 21041834_Missense mutations in KALRN may be genetic risk factors for schizophrenia. 21041834_Observational study of gene-disease association. (HuGE Navigator) 21664346_KALRN gene variation is not associated with overall ischemic stroke 22120753_We found Kalirin-9 expression to be paradoxically increased in schizophrenia 22194219_Neuronal guanine nucleotide exchange factor (GEF) kalirin is emerging as a key regulator of structural and functional plasticity at dendritic spines. 22429885_The kalirin expression were reduced in Alzheimer disease with psychosis. 22458949_In both anterior cingulate cortex (ACC) and dorsolateral prefrontal cortex (DLPFC), study found a reduction of Duo expression and PAK1 phosphorylation in schizophrenia. Cdc42 protein expression was decreased in ACC but not in DLPFC 22720673_The age-at-onset of Huntington disease (HD) is not associated with eleven SNPs, including SNP rs10934657 in the kalirin gene in 680 European HD patients. 25224588_A sequence variant in human KALRN impairs protein ability to activate Rac1 and coincides with reduced cortical thickness. 25316661_consider the GG genotype and the G allele of rs9289231 polymorphism of KALRN to be genetic risk factors for CAD in an Iranian population, especially in early-stage atherosclerotic vascular disease 25917671_4 KALRN gene SNPs were studied in Han ischemic stroke patients. rs11712619 seemed associated with lacunar stroke until risk factors were considered. re6438833 was significantly associated with ischemic and lacunar stroke. 27218147_GG genotype and the G allele of the rs9289231 polymorphism of KALRN and the rs224766 polymorphism of ADIPOQ genes may be considered genetic risk factors for Iranian type 2 diabetic patients with coronary artery disease. 27421267_DNA sequencing provided evidence linking KALRN to monogenic intellectual disability in two patients. 28152519_Data suggest protein levels of kalirin and CHD7 in circulating extracellular vesicles (EVs) as endothelial dysfunction markers to monitor vascular condition in hypertensive patients with albuminuria. 28706949_The GG genotype and G allele of SNP rs7620580 were associated with a risk for ischemic stroke with an adjusted OR of 3.195 and an OR of 1.446, respectively. Haplotype analysis revealed that A-T-G,G-T-A, and A-T-A haplotypes were associated with ischemic stroke. Our results provide evidence that kalirin gene variations were associated with ischemic stroke in the Chinese Han population. 29241584_The data of this study reveal a novel mechanism for disease-associated single nucleotide variants of KALARN and provide a platform for modeling morphological changes in mental disorders. 29554915_Combination of polymorphisms in the NOD2, IL17RA, EPHA2 and KALRN genes could play a significant role in the development of sarcoidosis by maintaining a chronic pro-inflammatory status in macrophages 29789657_The interaction of kalirin with the C-terminal region of Htt influences the function of kalirin and modulates the cytotoxicity induced by C-terminal Htt. 30232674_SNPs of the KALRN gene are associated with intracranial atherosclerotic stenosis in the northern Chinese population. 31801062_Synaptic Kalirin-7 and Trio Interactomes Reveal a GEF Protein-Dependent Neuroligin-1 Mechanism of Action. 33037113_KALRN mutations promote antitumor immunity and immunotherapy response in cancer. 33658318_Kalirin-RAC controls nucleokinetic migration in ADRN-type neuroblastoma. | ENSMUSG00000061751 | Kalrn | 1.009943e+02 | 0.7366281 | -0.440991729 | 0.3962007 | 1.250186e+00 | 0.2635168886 | 0.79788700 | No | Yes | 9.030836e+01 | 22.130272 | 1.061312e+02 | 26.713499 | |
| ENSG00000160201 | 7307 | U2AF1 | protein_coding | Q01081 | FUNCTION: Plays a critical role in both constitutive and enhancer-dependent splicing by mediating protein-protein interactions and protein-RNA interactions required for accurate 3'-splice site selection. Recruits U2 snRNP to the branch point. Directly mediates interactions between U2AF2 and proteins bound to the enhancers and thus may function as a bridge between U2AF2 and the enhancer complex to recruit it to the adjacent intron. {ECO:0000269|PubMed:22158538, ECO:0000269|PubMed:25311244, ECO:0000269|PubMed:8647433}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Metal-binding;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Spliceosome;Zinc;Zinc-finger;mRNA processing;mRNA splicing | This gene belongs to the splicing factor SR family of genes. U2 auxiliary factor, comprising a large and a small subunit, is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. This gene encodes the small subunit which plays a critical role in both constitutive and enhancer-dependent RNA splicing by directly mediating interactions between the large subunit and proteins bound to the enhancers. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:102724594;hsa:7307; | Cajal body [GO:0015030]; catalytic step 2 spliceosome [GO:0071013]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; spliceosomal complex [GO:0005681]; U2AF complex [GO:0089701]; metal ion binding [GO:0046872]; pre-mRNA 3'-splice site binding [GO:0030628]; RNA binding [GO:0003723]; mRNA processing [GO:0006397]; mRNA splicing, via spliceosome [GO:0000398]; RNA splicing [GO:0008380] | 11830556_A misspliced form of the cholecystokinin-B/gastrin receptor in pancreatic carcinoma: role of reduced sellular U2AF35 and a suboptimal 3'-splicing site leading to retention of the fourth intron. 12297299_U2AF35 RRM is unstructured in solution but its tertiary structure is induced upon binding to U2AF65. 15096518_U2AF1 gene expression may provide a mechanism by which the relative cellular concentration and availability of U2AF(35) protein isoforms are modulated, thus contributing to the finely tuned control of splicing events in different tissues. 15899895_U2AF35 appears to be completely dispensable for splicing in nuclear extracts prepared from adenovirus late-infected cells 15950937_Binding assays revealed that IpaH9.8 has a specific affinity to U2AF(35), a mammalian splicing factor, which interferes with U2AF(35)-dependent splicing as assayed for IgM pre-mRNA 16043505_identified and spatially localized sites of direct interaction between U2AF35 and U2AF65 in vivo in live cell nuclei. 16809543_DEK enforces 3' splice site discrimination by U2AF; DEK phosphorylated at serines 19 and 32 associates with U2AF35, facilitates the U2AF35-AG interaction and prevents binding of U2AF65 to pyrimidine tracts not followed by AG 16855028_Taken together our results demonstrate that U2AF35a is essential for HeLa cell division and suggest a novel role for both U2AF35 protein isoforms as regulators of alternative splicing of a specific subset of genes. 16940179_Results describe the roles of the two subunits of U2AF, U2AF65 and 35, in the selection between alternative 3' splice sites associated with polypyrimidine tracts of different strengths. 18211889_U2AF35 and hPrp3 interactions with SPF30 can occur simultaneously, thereby potentially linking 3' splice site recognition with tri-small nuclear ribonucleoprotein addition 18285458_SF1 and U2AF form extraspliceosomal complexes before and after taking part in the assembly of catalytic spliceosomes. 20514852_Observational study of gene-disease association. (HuGE Navigator) 22158538_Here we show that a missense mutation affecting the serine at codon 34 (Ser34) in U2AF1 was recurrently present in 13 out of 150 (8.7%) subjects with de novo MDS, and we found suggestive evidence of an increased risk of progression to sAML 22323480_U2AF1 and SRSF2 mutations are frequent in chronic myelomonocytic leukemia and advanced forms of MDS. U2AF1 and SRSF2 mutations are predictive for shorter survival. 22325350_hnRNP A1 forms a ternary complex with the U2AF heterodimer on AG-containing/uridine-rich RNAs, while it displaces U2AF from non-AG-containing/uridine-rich RNAs, an activity that requires the glycine-rich domain of hnRNP A1 22389253_In univariate analysis, mutated SRSF2 predicted shorter overall survival and more frequent acute myeloid leukemia progression compared with wild-type SRSF2, whereas mutated U2AF1, ZRSR2 had no impact on patient outcome. 23029227_U2AF1 mutation is a recurrent event at a low frequency in acute myeloid leukemia and myelodysplastic syndrome and influences overall survival. 23280334_data suggest that SF3B1, U2AF1 and SRSF2 mutations occur not only in myeloid lineage tumors but also in lymphoid lineage tumors; data suggest that the splicing gene mutations play important roles in the pathogenesis of hematologic tumors, but rarely in solid tumors 23335386_SRSF2 is the most frequently mutated spliceosome gene in chronic myelomonocytic leukemia, but neither it nor SF3B1 or U2AF35 mutations are prognostically relevant. 23775717_U2AF1 mutations play a significant role in myeloid leukemogenesis due to selective missplicing of tumor-associated genes 23861105_genetic association studies in a population in Taiwan: Data suggest that U2AF1 mutations are associated with poor prognosis/survival in patients with myelodysplastic syndrome and shorter time-to-leukemia transformation in young patients. 24097336_study describes incidence and phenotypic and prognostic relevance of U2AF1 mutations in primary myelofibrosis(PMF); U2AF1 mutations cluster with JAK2V617F, ASXL1 mutations and normal karyotype; U2AF1 mutations are strongly and inter-independently associated with anemia and thrombocytopenia 24498085_Data indicate somatic mutations in the splicing factor U2AF1 across 12 cancer types. 25231745_Molecular monitoring of patients having undergone AHSCT for PMF should not be restricted to JAK2, MPL or CALR, but all mutations present in primary fibrotic neoplastic myeloproliferation should be included to interpret abnormal blood values after AHSCT 25267526_mutations influence the similarity of splicing programs in leukemias, but do not give rise to widespread splicing failure. 25271374_A mutant U2AF1 (S34F) found in a variety of cancer types results in delayed splicing and disruption of kinetic competition during transcription. 25311244_The S34F mutation alters U2AF1 function in the context of specific RNA sequences, leading to aberrant alternative splicing of target genes, some of which may be relevant for myelodysplastic syndromes pathogenesis. 25326705_U2AF has the capacity to directly define ~88% of functional 3' splice sites in the human genome; numerous U2AF binding events also occur in intronic locations. 25412851_In multivariate analysis, U2AF1 and TP53 mutations retained independent prognostic significance across 93 cases of acute myeloid leukemia 25964599_The mutational status of the SRSF2, U2AF1 and ZRSR2 did not affect the response rate or survival in MDS patients who had received first-line decitabine treatment. 26508027_Mutations in ASXL1, U2AF1, and SF3B1 are common in Chinese patients with myelodysplastic syndromes. 27058230_in primary myelofibrosis, anemia was significantly associated with U2AF1 mutation; study confirms previous observation regarding the association of mutant U2AF1 with anemia supporting its role in hematopoiesis and in the pathogenesis of PMF-associated anemia 27184077_The U2AF35(S34F) mutation alters interaction with CFIm59, leading to increased use of a distal cleavage and polyadenylation site in the ATG7 pre-mRNA, decreasing levels of ATG7 protein and defective autophagy, ultimately leading to transformation. 27435003_data confirm MLL-PTD and, to a lesser extent, FLT3-ITD as common events in +11 AML.6, 7, 8 However, the high mutation frequencies of U2AF1 and genes involved in methylation (DNMT3A, IDH2) have hitherto not been reported in +11 AML 27566151_Alternative splicing of U2AF1 reveals a shared repression mechanism for duplicated exons controlled by SRF3. 27602765_this study characterized novel candidate pediatric T-cell acute lymphoblastic leukemia driver mutation in splicesome factor U2AF1 27639445_The aberrantly spliced target genes and deregulated cellular pathways associated with the commonly mutated splicing factor genes in myelodysplastic syndromes (SF3B1, SRSF2 and U2AF1) are being identified, illuminating the molecular mechanisms underlying the disease. (Review) 27776121_Our results provide mechanistic explanations of the magnitude of splicing changes observed in U2AF1-mutant cells and why tumors harboring U2AF1 mutations always retain an expressed copy of the wild-type allele 27799531_In summary, we provide insight into the underlying molecular mechanism of U2AF binding to 3' splice sites. 28067246_The splicing effects of sudemycin and U2AF1 can be cumulative in cells exposed to both perturbations-drug and mutation, as compared with cells exposed to either alone. 28372848_The frequently mutated SF3B1 residues contact the pre-mRNA splice site. Based on structural homology with other spliceosome subunits, and recent findings of altered RNA binding by mutant U2AF1 proteins, we suggest that affected U2AF1 residues also contact pre-mRNA. 28436936_Expression of U2AF1S34F in human hematopoietic progenitors results in impaired erythroid differentiation as a result of poor hemoglobinization and reduced growth of erythroid progenitors. 28893951_U2AF35 missense mutation is associated with alternative 5' splice site that impacts splicing regulation in cancer. 28938223_infer that U2AF1 S34 mutations characterize a distinct subgroup of myelodysplastic syndrome 29057546_U2AF1 mutations are one of the earliest genetic events in myelodysplastic syndrome patients and different types of U2AF1 mutations have distinct clinical and biological characteristics. 29321554_In univariable analysis, patients carrying mutations in DNMT3A, U2AF1, and EZH2 had worse overall and relapse-free survival. 29516544_Among 52 patients with U2AF1 mutations, 28 (54%) harbored S34 (25 S34F and 3 S34Y), 22 (42%) Q157 (16 Q157P and 6 Q157R), and two R156H 29535431_U2AF1 mutation types in primary myelofibrosis. 29649018_Langerhans Cell Histiocytosis is a clonally heterogenous disease, with multiple driver mutations ranging from BRAF mutations to TP53 and U2AF1 mutations likely playing a role in the pathogenesis. 29991672_Study thus points to an active role of U2AF1 S34F mutant protein in inducing cell cycle dysregulation and mitotic stress. 30152885_lenalidomide response was adversely affected by U2AF1 mutations and high risk karyotype in MDS 30194306_Although 13 of 66 patients with the U2AF1 Q157 mutation had EZH2 mutations or deletions, no genetic alteration of EZH2 was found in the U2AF1 S34-mutated (n=32). 30334576_Enasidenib-induced eosinophilic differentiation in a patient with acute myeloid leukaemia with IDH2 and U2AF1 mutations. 30842218_U2AF1 in association with its binding partner U2AF2, binds mature RNA in the cytoplasm and functions asa translational repressor 30846499_mutations in SF3B1, U2AF1, and SRSF2 enhance NFkappaB activity and LPS-induced inflammatory cytokine production in macrophages, patient-derived cell lines, and mouse and human myeloid cells 31011167_Inhibition of IRAK4-L abrogates leukaemic growth, particularly in acute myeloid leukaemia (AML) cells with higher expression of the IRAK4-L isoform. Collectively, mutations in U2AF1 induce expression of therapeutically targetable 'active' IRAK4 isoforms and provide a genetic link to activation of chronic innate immune signalling in myelodysplastic syndromes and AML. 31124956_This meta-analysis indicates a positive effect of SF3B1 and an adverse prognostic effect of SRSF2, U2AF1, and ZRSR2 mutations in patients with myelodysplastic syndrome. 31144421_Knockdown of spliceosome U2AF1 significantly inhibits the development of human erythroid cells. 31504847_These findings unveil distinct roles of duplicated tandem exon-derived U2AF1 isoforms in the regulation of the transcriptome and suggest U2AF1a-driven 5'-UTR alternative splicing as a molecular mechanism of mTOR-regulated translational control. 31605415_U2AF1 was significantly positively associated with PA28gamma in oral squamous cell carcinoma 31754743_meta-analysis indicates that U2AF1 mutants are independent, detrimental prognostic factors for overall survival (OS) and acute myeloid leukemia transformation in patients with de novo MDS, as well as associating with shorter OS in subgroups of low- or intermediate-1-IPSS, U2AF1(S34) and U2AF1(Q157/R156). 31826693_Prognostic significance of U2AF1 mutations in myelodysplastic syndromes: a meta-analysis. 31836708_Study in lung adenocarcinoma biopsy specimens found that U2AF1 S34F preferentially binds and modulates splicing of introns containing CAG trinucleotides at their 3' splice junctions. U2AF1 S34F induces cell invasiveness and this appears to be in part mediated via preferential splicing of the SLC34A2-ROS1 long isoform, whose expression also increases tumor cell invasion. 31992135_Deoxynivalenol globally affects the selection of 3' splice sites in human cells by suppressing the splicing factors, U2AF1 and SF1. 32023759_prognosis of myelodysplastic syndromes patients with U2AF1 mutation 32027245_acute myeloid leukemia patients with U2AF1 mutation positive have a poor prognosis as compared with the wild type group 32116123_The zinc finger domains in U2AF26 and U2AF35 have diverse functionalities including a role in controlling translation. 32150510_The core spliceosomal factor U2AF1 controls cell-fate determination via the modulation of transcriptional networks. 32358566_Clinical presentation and differential splicing of SRSF2, U2AF1 and SF3B1 mutations in patients with acute myeloid leukemia. 32958768_Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1. 33122737_Differential U2AF1 mutation sites, burden and co-mutation genes can predict prognosis in patients with myelodysplastic syndrome. 33137094_Ribosome biogenesis is a downstream effector of the oncogenic U2AF1-S34F mutation. 33274830_Prevalent intron retention fine-tunes gene expression and contributes to cellular senescence. 33283729_[Correlation between U2AF1 Gene Mutation Characteristics and Clinical Manifestations and Prognosis in Patients with Myelodysplastic Syndrome]. 33314767_U2AF1 expression is a novel and independent prognostic indicator of childhood T-lineage acute lymphoblastic leukemia. 33347855_U2AF - Hypoxia-induced fas alternative splicing regulator. 33477033_Down regulation of U2AF1 promotes ARV7 splicing and prostate cancer progression. 33930289_Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. 34183647_U2AF1 mutation promotes tumorigenicity through facilitating autophagy flux mediated by FOXO3a activation in myelodysplastic syndromes. 34215620_Nonsense-Mediated RNA Decay Is a Unique Vulnerability of Cancer Cells Harboring SF3B1 or U2AF1 Mutations. 34520118_Identification of a small molecule splicing inhibitor targeting UHM domains. 34581783_Bilineal evolution of a U2AF1-mutated clone associated with acquisition of distinct secondary mutations. 34893123_[Effect of U2AF1 Mutation to Inflammatory Cytokine Expression in SKM-1 Cells through FOXO3a-Bim Signaling Pathway]. 34896936_Decreased CD177(pos) neutrophils in myeloid neoplasms is associated with NPM1, RUNX1, TET2, and U2AF1 S34F mutations. 35303483_Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. | ENSMUSG00000061613 | U2af1 | 7.661768e+02 | 0.9294561 | -0.105541405 | 0.2881498 | 1.361148e-01 | 0.7121743727 | 0.93941595 | No | Yes | 5.832986e+02 | 79.840940 | 6.724964e+02 | 93.612601 | |
| ENSG00000160294 | 8888 | MCM3AP | protein_coding | O60318 | FUNCTION: [Isoform GANP]: As a component of the TREX-2 complex, involved in the export of mRNAs to the cytoplasm through the nuclear pores (PubMed:20005110, PubMed:20384790, PubMed:23591820, PubMed:22307388). Through the acetylation of histones, affects the assembly of nucleosomes at immunoglobulin variable region genes and promotes the recruitment and positioning of transcription complex to favor DNA cytosine deaminase AICDA/AID targeting, hence promoting somatic hypermutations (PubMed:23652018). {ECO:0000269|PubMed:20005110, ECO:0000269|PubMed:20384790, ECO:0000269|PubMed:22307388, ECO:0000269|PubMed:23591820, ECO:0000269|PubMed:23652018}.; FUNCTION: [Isoform MCM3AP]: Binds to and acetylates the replication protein MCM3. Plays a role in the initiation of DNA replication and participates in controls that ensure that DNA replication initiates only once per cell cycle (PubMed:11258703, PubMed:12226073). Through the acetylation of histones, affects the assembly of nucleosomes at immunoglobulin variable region genes and promotes the recruitment and positioning of transcription complex to favor DNA cytosine deaminase AICDA/AID targeting, hence promoting somatic hypermutations (PubMed:23652018). {ECO:0000269|PubMed:11258703, ECO:0000269|PubMed:12226073, ECO:0000269|PubMed:23652018}. | 3D-structure;Acetylation;Acyltransferase;Alternative promoter usage;Chromosome;Coiled coil;Cytoplasm;Immunity;Mental retardation;Methylation;Neuropathy;Nuclear pore complex;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Transferase;Translocation;Transport;mRNA transport | The minichromosome maintenance protein 3 (MCM3) is one of the MCM proteins essential for the initiation of DNA replication. The protein encoded by this gene is a MCM3 binding protein. It was reported to have phosphorylation-dependent DNA-primase activity, which was up-regulated in antigen immunization induced germinal center. This protein was demonstrated to be an acetyltransferase that acetylates MCM3 and plays a role in DNA replication. The mutagenesis of a nuclear localization signal of MCM3 affects the binding of this protein with MCM3, suggesting that this protein may also facilitate MCM3 nuclear localization. This gene is expressed in the brain or in neuronal tissue. An allelic variant encoding amino acid Lys at 915, instead of conserved Glu, has been identified in patients with mild intellectual disability. [provided by RefSeq, Jan 2014]. | hsa:8888; | chromosome [GO:0005694]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear membrane [GO:0031965]; nuclear pore nuclear basket [GO:0044615]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription export complex 2 [GO:0070390]; chromatin binding [GO:0003682]; H3 histone acetyltransferase activity [GO:0010484]; histone acetyltransferase activity [GO:0004402]; histone binding [GO:0042393]; nucleic acid binding [GO:0003676]; mRNA export from nucleus [GO:0006406]; nucleosome organization [GO:0034728]; poly(A)+ mRNA export from nucleus [GO:0016973]; protein transport [GO:0015031]; somatic hypermutation of immunoglobulin genes [GO:0016446] | 12226073_inhibits initiation of DNA replication via binding to MCM3 12885157_plays a certain important role in the maturation of immunoglobulin or selection of B cells in germinal centers during the immune response to TD-Ag. A selective function of GANP molecule on B cell proliferation and differentiation might exist. 19578742_abnormal over-expression of GANP together with AID might be associated with rigorous DNA damage, potentially causing the malignant development of Cholangiocarcinoma (CCAs) during long-term inflammation. 19686285_GANP protects cells from cellular senescence caused by DNA damage and that a significant decrease in GANP expression leads to malignancy by generating hyperploidy and chromosomal instability (CIN). 20005110_GANP depletion inhibits mRNA export, with retention of mRNPs and NXF1 in punctate foci within the nucleus. 20384790_Data show that human germinal center-associated nuclear protein (GANP) is critically involved in cell proliferation at the mitotic phase through its selective support of shugoshin-1 mRNA export. 20507984_GANP may serve as an essential link required to transport AID to B-cell nuclei and to target AID to actively transcribed IgV regions 20714864_HBV DNA integration sites into human genome were random, and MCM3AP was a new site. 21195085_MCM3AP and GANP are different proteins, occupying different locations in the cell and transcribed from different promoters 22395445_GANP, a homologue of yeast Sac3 that is involved in mRNA export, is indispensable for ensuring the stability of human genomic DNA and GANP knockdown causes apoptosis and necrosis of p53-insufficient cancer cells. 22942428_GANP transgene may play a critical role in Lyn tyrosine-protein kinase-mediated signaling during selection of high-affinity B cells in peripheral lymphoid organs. 23094019_The cellular protein MCM3AP is required for inhibition of cellular DNA synthesis by the IE86 protein of human cytomegalovirus. 23652018_GANP-mediated chromatin modification promotes transcription complex recruitment and positioning at immunoglobulin variable loci to favour AID targeting. 24123876_A homozygous potentially pathogenic variant (c.2743G>A) was identified, which encodes amino acid Lys, instead of conserved Glu, at the position 915, in patients with mild intellectual disability. 24198285_GANP is induced in activated T4 cells & physically interacts with A3G. GANP is encapsidated in HIV-1 virions & modulates A3G packaging into the cores. Upregulation increases A3G-catalyzed viral G-->A hypermutation, suppressing infectivity. 26615982_MCM3AP and POMP Mutations Cause a DNA-Repair and DNA-Damage-Signaling Defect in an Immunodeficient Child 26749495_These results indicated that the GANP protein is associated with breast cancer resistance. 28633435_The identification of MCM3AP variants in affected individuals from multiple centres establishes it as a disease gene for childhood-onset recessively inherited Charcot-Marie-Tooth neuropathy with intellectual disability. 30782188_Circular RNA MCM3AP-AS1 directly bound to miR-194-5p and acted as competing endogenous RNA while subsequently facilitating miR-194-5p target gene FOXA1 expression in hepatocellular carcinoma cells. 30810967_The G allele of rs2839178 at the GANP locus was significantly associated with reduced breast cancer risk and longer disease-free survival in breast cancer patients, showing a consistent direction in the association between susceptibility and clinical outcome. 31241196_MCM3AP encodes germinal center-associated nuclear protein (GANP), a protein involved in the export of certain messenger RNAs from the nucleus to the cytoplasm 32202298_Distinct effects on mRNA export factor GANP underlie neurological disease phenotypes and alter gene expression depending on intron content. 32319184_Genetic spectrum of MCM3AP and its relationship with phenotype of Charcot-Marie-Tooth disease. 32827560_The critical role of germinal center-associated nuclear protein in cell biology, immunohematology, and hematolymphoid oncogenesis. 32940099_Long noncoding RNA MCM3AP antisense RNA 1 is downregulated in chronic obstructive pulmonary disease and regulates human bronchial smooth muscle cell proliferation. | ENSMUSG00000001150 | Mcm3ap | 7.869734e+03 | 1.1093731 | 0.149744681 | 0.2611235 | 3.349190e-01 | 0.5627768343 | 0.89583618 | No | Yes | 6.985707e+03 | 357.224624 | 6.447958e+03 | 338.294055 | |
| ENSG00000160298 | 54058 | C21orf58 | protein_coding | P58505 | Alternative splicing;Reference proteome | hsa:54058; | ENSMUSG00000009114 | 2610028H24Rik | 1.754163e+03 | 0.7966459 | -0.327989425 | 0.2590601 | 1.580401e+00 | 0.2087028873 | 0.78763590 | No | Yes | 1.183298e+03 | 152.179522 | 1.672909e+03 | 220.256994 | |||||
| ENSG00000160410 | 92799 | SHKBP1 | protein_coding | Q8TBC3 | FUNCTION: Inhibits CBL-SH3KBP1 complex mediated down-regulation of EGFR signaling by sequestration of SH3KBP1. Binds to SH3KBP1 and prevents its interaction with CBL and inhibits translocation of SH3KBP1 to EGFR containing vesicles upon EGF stimulation. {ECO:0000250|UniProtKB:Q6P7W2}. | 3D-structure;Acetylation;Alternative splicing;Lysosome;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:92799; | lysosome [GO:0005764]; identical protein binding [GO:0042802]; positive regulation of epidermal growth factor receptor signaling pathway [GO:0045742]; protein homooligomerization [GO:0051260] | 16733801_hSB1 may function as a regulator of cathepsin B-mediated apoptosis. 21830225_SHKBP1 disturbed the translocation of CIN85 to EGFR-containing vesicles. 25457385_Genetic variant in the BCL11A (rs1427407), but not HBS1-MYB (rs6934903) loci associate with fetal hemoglobin levels in Indian sickle cell disease patients. 31138318_Study suggests that an elevated SHKBP1/miR-499a ratio is a molecular signature that characterizes the erlotinib-resistant overall survival of osteosarcoma cells, which may have clinical value as a predictive biomarker. | ENSMUSG00000089832 | Shkbp1 | 2.868588e+03 | 1.3212442 | 0.401897166 | 0.3063035 | 1.714075e+00 | 0.1904574862 | 0.78025017 | No | Yes | 3.180184e+03 | 384.961125 | 2.212366e+03 | 275.375483 | ||
| ENSG00000160633 | 6294 | SAFB | protein_coding | Q15424 | FUNCTION: Binds to scaffold/matrix attachment region (S/MAR) DNA and forms a molecular assembly point to allow the formation of a 'transcriptosomal' complex (consisting of SR proteins and RNA polymerase II) coupling transcription and RNA processing (PubMed:9671816). Functions as an estrogen receptor corepressor and can also bind to the HSP27 promoter and decrease its transcription (PubMed:12660241). Thereby acts as a negative regulator of cell proliferation (PubMed:12660241). When associated with RBMX, binds to and stimulates transcription from the SREBF1 promoter (By similarity). {ECO:0000250|UniProtKB:D3YXK2, ECO:0000269|PubMed:12660241, ECO:0000269|PubMed:9671816}. | Acetylation;Alternative splicing;DNA-binding;Direct protein sequencing;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a DNA-binding protein which has high specificity for scaffold or matrix attachment region DNA elements (S/MAR DNA). This protein is thought to be involved in attaching the base of chromatin loops to the nuclear matrix but there is conflicting evidence as to whether this protein is a component of chromatin or a nuclear matrix protein. Scaffold attachment factors are a specific subset of nuclear matrix proteins (NMP) that specifically bind to S/MAR. The encoded protein is thought to serve as a molecular base to assemble a 'transcriptosome complex' in the vicinity of actively transcribed genes. It is involved in the regulation of heat shock protein 27 transcription, can act as an estrogen receptor co-repressor and is a candidate for breast tumorigenesis. This gene is arranged head-to-head with a similar gene whose product has the same functions. Multiple transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Jan 2011]. | hsa:6294; | midbody [GO:0030496]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; double-stranded DNA binding [GO:0003690]; RNA binding [GO:0003723]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; chromatin organization [GO:0006325]; intracellular estrogen receptor signaling pathway [GO:0030520]; regulation of mRNA processing [GO:0050684]; regulation of transcription by RNA polymerase II [GO:0006357] | 12837281_HSP27 is a nuclear speckle component in unstressed cells in tissue culture. It is also associated with the nucleolar compartment. 14587024_REVIEW: possibility that SAFB1 and SAFB2 are novel breast tumor suppressor genes, and how they might function in this role, are discussed 14702179_HSP27 expression may have useful diagnostic use for the prognosis of mouth squamous cell carcinoma. 15066997_SAFB1 represses ERalpha activity via indirect association with histone deacetylation and interaction with the basal transcription machinery 16195251_SAFB1 was shown to interact directly with the nuclear receptor corepressor N-CoR. 16326836_SAFB1 interacts in pull-down assays not only with PPARgamma but also with all nuclear receptors tested 16475161_PP2A-mediated dephosphorylation of HSP27 and tau correlated with PP2A-induced preservation of endothelial cell cytoskeleton 17643427_SAFB may direct the reorganization and segregation of nuclear RNA and DNA prior to endonuclease-mediated DNA cleavage. 18154639_Over expression of HSP27 is associated with intrahepatic cholangiocarcinoma 18772145_hXOR is a tumor suppressor-targeted gene and the phosphorylation of SAFB1 is regulated by OSM, which provides a molecular basis for understanding the role of SAFB1-regulated hXOR transcription in cytokine stimulation and tumorigenesis 19077293_SAFB1 is not likely to be causative of the hereditary breast cancer syndrome in west Swedish breast cancer families 19106221_Importance of ERE-BP as an attenuator of normal ERalpha signaling in vivo and is a novel target for modulation by selective estrogen receptor modulators. 19137425_This study shows that low SAFB protein levels predict poor prognosis of breast cancer patients, suggesting critical functions of SAFB1 and SAFB2 in breast cancer cells. 19674106_The enzymatic activity of SR protein kinases 1 and 1a is negatively affected by interaction with scaffold attachment factors B1 and 2. 19901029_Study confirms the primary role of SAFB1/SAFB2 as corepressors and also uncovers a previously unknown role for SAFB1 in the regulation of immune genes and in estrogen-mediated repression of genes. 21130767_Data show that binding of p53 to SAFB1 had a significant functional outcome, since SAFB1 was shown to suppress p53-mediated reporter gene expression. 21527249_transcriptional repressor SAFB1 is modified by both SUMO1 and SUMO2/3, and this modification is necessary for its full repressive activity. 22566185_Results indicate that SAFB1 and SAFB2 are crucial repressors for ERalpha dynamics in association with the nuclear matrix and that their synergistic regulation of ERalpha mobility is sufficient for inhibiting ERalpha function. 23893242_SAFB1 formed a complex with the histone methyltransferase EZH2. 24055346_Data indicate that scaffold attachment factor SAFB1 is transiently recruited to DNA breaks in a poly(ADP-ribose)-polymerase 1- and poly(ADP-ribose)-dependent manner. 25800734_reveals an unexpected role of SUMO-1 and SAFB in the stimulatory coupling of promoter binding, transcription initiation and RNA processing 26273616_Single depletion of either SAFB1 or SAFB2 leads to an increase in expression of the other SAFB protein. 26694817_The expression of coding and non-coding genes with SAFB1 cross-link sites was altered by SAFB1 knockdown. The isoform-specific expression of neural cell adhesion molecule (NCAM1) and ASTN2 was influenced by SAFB1. 27731383_Depletion of SAFB1 reduced FUS's localization to chromatin-bound fraction and splicing activity, suggesting SAFB1 could tether FUS to chromatin compartment thorough N-terminal DNA-binding motif. Moreover, FUS interacts with another nuclear matrix-associated protein, Matrin3. 28627136_Data suggest that ERH interacts directly in nucleus with C-terminal Arg-Gly-rich region of SAFB1/SAFB2 and this multimer co-localizes in insoluble nuclear fraction; binding of ERH reverses inhibition exerted by SAFB1/SAFB2 on SRPK1. (ERH = enhancer of rudimentary homolog protein; SAFB = scaffold attachment factor B; SRPK1 = splicing kinase SR protein kinase-1) 28912140_SAFB regulated the activity of NF-kappaB signaling in CRC by targeting TAK1 This novel mechanism provides a comprehensive understanding of both SAFB and the NF-kappaB signaling pathway in the progression of CRC and indicates that the SAFB-TAK1-NF-kappaB axis is a potential target for early therapeutic intervention in CRC progression 29887524_SAFB1 binds to the HIV-1 LTR and physically interacts with phosphorylated RNA polymerase II, repressing HIV-1 transcription initiation and elongation. 31677973_Depletion of SAFB leads to changes in 3D genome organization. 32580238_Abnormal scaffold attachment factor 1 expression and localization in spinocerebellar ataxias and Huntington's chorea. 33257571_Scaffold association factor B (SAFB) is required for expression of prenyltransferases and RAS membrane association. 34067147_Inhibition of HSF1 and SAFB Granule Formation Enhances Apoptosis Induced by Heat Stress. 34129097_Subcellular dynamics of estrogen-related receptors involved in transrepression through interactions with scaffold attachment factor B1. | ENSMUSG00000071054 | Safb | 8.202443e+03 | 1.2171672 | 0.283527316 | 0.2489615 | 1.297427e+00 | 0.2546837576 | 0.79582240 | No | Yes | 9.218744e+03 | 904.057587 | 6.592127e+03 | 663.506692 | |
| ENSG00000160752 | 2224 | FDPS | protein_coding | P14324 | FUNCTION: Key enzyme in isoprenoid biosynthesis which catalyzes the formation of farnesyl diphosphate (FPP), a precursor for several classes of essential metabolites including sterols, dolichols, carotenoids, and ubiquinones. FPP also serves as substrate for protein farnesylation and geranylgeranylation. Catalyzes the sequential condensation of isopentenyl pyrophosphate with the allylic pyrophosphates, dimethylallyl pyrophosphate, and then with the resultant geranylpyrophosphate to the ultimate product farnesyl pyrophosphate. | 3D-structure;Acetylation;Alternative splicing;Cholesterol biosynthesis;Cholesterol metabolism;Cytoplasm;Disease variant;Host-virus interaction;Hydroxylation;Isoprene biosynthesis;Lipid biosynthesis;Lipid metabolism;Magnesium;Metal-binding;Reference proteome;Steroid biosynthesis;Steroid metabolism;Sterol biosynthesis;Sterol metabolism;Transferase | PATHWAY: Isoprenoid biosynthesis; farnesyl diphosphate biosynthesis; farnesyl diphosphate from geranyl diphosphate and isopentenyl diphosphate: step 1/1.; PATHWAY: Isoprenoid biosynthesis; geranyl diphosphate biosynthesis; geranyl diphosphate from dimethylallyl diphosphate and isopentenyl diphosphate: step 1/1. | This gene encodes an enzyme that catalyzes the production of geranyl pyrophosphate and farnesyl pyrophosphate from isopentenyl pyrophosphate and dimethylallyl pyrophosphate. The resulting product, farnesyl pyrophosphate, is a key intermediate in cholesterol and sterol biosynthesis, a substrate for protein farnesylation and geranylgeranylation, and a ligand or agonist for certain hormone receptors and growth receptors. Drugs that inhibit this enzyme prevent the post-translational modifications of small GTPases and have been used to treat diseases related to bone resorption. Multiple pseudogenes have been found on chromosomes 1, 7, 14, 15, 21 and X. Multiple transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Oct 2008]. | hsa:2224; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; dimethylallyltranstransferase activity [GO:0004161]; geranyltranstransferase activity [GO:0004337]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; cholesterol biosynthetic process [GO:0006695]; farnesyl diphosphate biosynthetic process [GO:0045337]; geranyl diphosphate biosynthetic process [GO:0033384] | 15713990_This study provides the first evidence of the presence of FPPs activity in human CRC. Moreover, FPPs enzyme was found to play a significant role in colon cancer proliferation. 17198737_mitochondrial targeting of FPS may be widespread among eukaryotes 17368768_findings suggest that a single nucleotide polymorphism in the FDPS gene (rs2297480) may be a genetic marker for lower bone mineral density in postmenopausal Caucasian women 17387528_Observational study of gene-disease association. (HuGE Navigator) 18494934_FDPS is involved in the resistance to zoledronic acid of osteosarcoma cells. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18687167_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19056481_characterized functionally the minimal basal promoter of the human FDPS gene by means of deletion mutants and we have identified two cis-acting elements which modulate the FDPS gene expression and are recognized by Pax5 and OCT-1 transcription factors 19494338_FPPS knockdown cells activated Vgamma9Vdelta2 T cells, as measured by increased levels of CD69 and CD107a, killing of FPPS knockdown cells, and induction of IFN-gamma secretion 20191015_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20450493_characterized the sterol-response-element-binding protein 2 and nuclear factor Y-binding site in the farnesyl diphosphate synthase promoter 20877624_Observational study of gene-disease association. (HuGE Navigator) 21151198_Common polymorphisms of the FDPS gene influence the response to bisphosphonates in osteoporotic women. 21196316_The A/C rs2297480 polymorphism of FDPS was highly differently distributed among osteonecrosis-of-the-jaw patients and controls, with a correlation between AA carrier status and occurrence of ONJ after 18-24 months of treatment with bisphosphonates. 22278941_findings reveal a FDPS-dependent mechanism in the internalization and down-regulation of beta2AR, identify FDPS as a potential target for improving the therapeutic efficacy of beta-agonists 22338925_first study on the gene FDPS rs2297480 SNP in postmenopausal Thai women.The effect did not contribute to the baseline of bone mineral density nor bone turnover markers. 22407328_FPPS was more highly expressed in prostate cancer vs. normal prostate tissue. The association of FPPS with established histopathological risk parameters and biochemical recurrence implicates a contribution of the mevalonate pathway to PC progression. 23234314_The crystal structure of human FPPS in complex with a novel bisphosphonate YS0470 and in the absence of a second substrate showed partial ordering of the tail in the closed conformation. 23238007_LRP5 and FDPS loci age-specifically affect skeletal traits in healthy fertile women. 23277274_FPPS might play an important role in Ang II-induced cardiac hypertrophy and fibrosis in vivo, at least in part through RhoA, p-38 MAPK and TGF-beta1. 23847096_The iPA-driven modulation of FDPS can cause an enhancement of post-translational prenylation essential for the biological activity of key proteins in NK signaling and effector functions, such as Ras. 23998921_Data indicate compounds represent a new structural class of farnesyl pyrophosphate synthase (hFPPS) inhibitors and suggest a development of therapeutics. 24311107_Results suggest that polymorphisms of the FDPS gene may influence the bone response to drugs targeting the mevalonate pathway, like statins. 24369118_These observations suggest that an increase in the expression of endogenous FPPS could confer at least partial resistance to the pharmacological effect of N-BP drugs such as ZOL in vivo 24534219_our study indicated that DR patients have higher VEGF levels than diabetic patients without retinopathy, and -2578A/C (rs699947) and +405C/G (rs2010963) may be important factors in determining serum VEGF levels. 24598914_A co-crystal structure of human farnesyl pyrophosphate synthase in complex with a bisphosphonate and two molecules of inorganic phosphate. 24927548_The results identify new classes of FPPS inhibitors, diterpenoids and sesquiterpenoids, that bind to the IPP site and may be of interest as anticancer and antiinfective drug leads. 25630225_These results are consistent with the previously proposed hypothesis that the allosteric pocket of human FPPS, located near the active site, plays a feed-back regulatory role for this enzyme. 28098152_Farnesyl pyrophosphate (FPP) allosterically regulated the activity of farnesyl pyrophosphate synthase. 29036218_Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase 29075041_Deregulated expression and activity of Farnesyl Diphosphate Synthase (FDPS) in Glioblastoma 29337059_FPPS mediates TGF-beta1-induced lung cancer cell invasion and epithelial-to-mesenchymal transition via the RhoA/Rock1 pathway. 30561051_A novel premature termination mutation in FDPS in a Chinese family with disseminated superficial actinic porokeratosis. 30914801_FDPS plays an oncogenic role in PTEN-deficient Prostate cancer through GTPase/AKT axis. Identifying mevalonate pathway proteins could serve as a therapeutic target in PTEN dysregulated tumors. 31774873_FDPS rs2297480 is associated with postmenopausal osteoporosis. 34751146_Novel missense mutations of MVK and FDPS gene in Chinese patients with disseminated superficial actinic porokeratosis. 34885721_Towards an Improvement of Anticancer Activity of Benzyl Adenosine Analogs. | ENSMUSG00000059743 | Fdps | 1.058131e+04 | 0.8373289 | -0.256133711 | 0.2919487 | 7.699404e-01 | 0.3802354845 | 0.84161313 | No | Yes | 8.729526e+03 | 598.281366 | 1.017096e+04 | 714.554266 |
| ENSG00000160767 | 10712 | FAM189B | protein_coding | P81408 | Alternative splicing;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | This gene is located near the gene for the lysosomal enzyme glucosylceramidase; a deficiency in this enzyme is associated with Gaucher disease. The encoded protein has been identified as a potential binding partner of a WW domain-containing protein which is involved in apoptosis and tumor suppression. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2010]. | hsa:10712; | integral component of membrane [GO:0016021]; WW domain binding [GO:0050699] | 23317259_Overexpression of COTE1 promotes cellular invasion of hepatocellular carcinoma. 24899407_our findings suggest that the cytoplasmic protein COTE1 contributes to hepatocellular carcinoma tumorigenesis by regulating cell proliferation through the modulation of WWOX signaling 34124264_High FAM189B Expression and Its Prognostic Value in Patients with Gastric Cancer. | ENSMUSG00000032657 | Fam189b | 6.784752e+03 | 1.4180796 | 0.503938564 | 0.3110020 | 2.626039e+00 | 0.1051237035 | 0.75783482 | No | Yes | 7.666021e+03 | 911.710555 | 4.723321e+03 | 576.669930 | ||
| ENSG00000160789 | 4000 | LMNA | protein_coding | P02545 | FUNCTION: Lamins are components of the nuclear lamina, a fibrous layer on the nucleoplasmic side of the inner nuclear membrane, which is thought to provide a framework for the nuclear envelope and may also interact with chromatin. Lamin A and C are present in equal amounts in the lamina of mammals. Recruited by DNA repair proteins XRCC4 and IFFO1 to the DNA double-strand breaks (DSBs) to prevent chromosome translocation by immobilizing broken DNA ends (PubMed:31548606). Plays an important role in nuclear assembly, chromatin organization, nuclear membrane and telomere dynamics. Required for normal development of peripheral nervous system and skeletal muscle and for muscle satellite cell proliferation (PubMed:10080180, PubMed:22431096, PubMed:10814726, PubMed:11799477, PubMed:18551513). Required for osteoblastogenesis and bone formation (PubMed:12075506, PubMed:15317753, PubMed:18611980). Also prevents fat infiltration of muscle and bone marrow, helping to maintain the volume and strength of skeletal muscle and bone (PubMed:10587585). Required for cardiac homeostasis (PubMed:10580070, PubMed:12927431, PubMed:18611980, PubMed:23666920). {ECO:0000269|PubMed:10080180, ECO:0000269|PubMed:10580070, ECO:0000269|PubMed:10587585, ECO:0000269|PubMed:10814726, ECO:0000269|PubMed:11799477, ECO:0000269|PubMed:12075506, ECO:0000269|PubMed:12927431, ECO:0000269|PubMed:15317753, ECO:0000269|PubMed:18551513, ECO:0000269|PubMed:18611980, ECO:0000269|PubMed:22431096, ECO:0000269|PubMed:23666920, ECO:0000269|PubMed:31548606}.; FUNCTION: Prelamin-A/C can accelerate smooth muscle cell senescence. It acts to disrupt mitosis and induce DNA damage in vascular smooth muscle cells (VSMCs), leading to mitotic failure, genomic instability, and premature senescence. | 3D-structure;Acetylation;Alternative splicing;Cardiomyopathy;Charcot-Marie-Tooth disease;Coiled coil;Congenital muscular dystrophy;Direct protein sequencing;Disease variant;Emery-Dreifuss muscular dystrophy;Intermediate filament;Isopeptide bond;Limb-girdle muscular dystrophy;Lipoprotein;Methylation;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Prenylation;Reference proteome;Ubl conjugation | The nuclear lamina consists of a two-dimensional matrix of proteins located next to the inner nuclear membrane. The lamin family of proteins make up the matrix and are highly conserved in evolution. During mitosis, the lamina matrix is reversibly disassembled as the lamin proteins are phosphorylated. Lamin proteins are thought to be involved in nuclear stability, chromatin structure and gene expression. Vertebrate lamins consist of two types, A and B. Alternative splicing results in multiple transcript variants. Mutations in this gene lead to several diseases: Emery-Dreifuss muscular dystrophy, familial partial lipodystrophy, limb girdle muscular dystrophy, dilated cardiomyopathy, Charcot-Marie-Tooth disease, and Hutchinson-Gilford progeria syndrome. [provided by RefSeq, Apr 2012]. | hsa:4000; | cytosol [GO:0005829]; intermediate filament [GO:0005882]; lamin filament [GO:0005638]; nuclear body [GO:0016604]; nuclear envelope [GO:0005635]; nuclear lamina [GO:0005652]; nuclear matrix [GO:0016363]; nuclear membrane [GO:0031965]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; site of double-strand break [GO:0035861]; identical protein binding [GO:0042802]; structural molecule activity [GO:0005198]; cellular protein localization [GO:0034613]; cellular response to hypoxia [GO:0071456]; DNA double-strand break attachment to nuclear envelope [GO:1990683]; establishment or maintenance of microtubule cytoskeleton polarity [GO:0030951]; muscle organ development [GO:0007517]; negative regulation of cardiac muscle hypertrophy in response to stress [GO:1903243]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of mesenchymal cell proliferation [GO:0072201]; negative regulation of release of cytochrome c from mitochondria [GO:0090201]; nuclear envelope organization [GO:0006998]; positive regulation of cell aging [GO:0090343]; positive regulation of gene expression [GO:0010628]; positive regulation of histone H3-K9 trimethylation [GO:1900114]; protein import into nucleus [GO:0006606]; protein localization to nucleus [GO:0034504]; regulation of cell migration [GO:0030334]; regulation of protein localization to nucleus [GO:1900180]; regulation of protein stability [GO:0031647]; regulation of telomere maintenance [GO:0032204]; ventricular cardiac muscle cell development [GO:0055015] | 11243729_Observational study of gene-disease association. (HuGE Navigator) 11440372_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11792809_Some lamin A mutants causing disease can be aberrantly localized, partially disrupt the endogenous lamina and alter emerin localization, whereas others localize normally in transfected cells. 11792810_Certain dilated cardiomyopathy- and Emery-Dreyfuss muscular dystrophy-associated LMNA mutations result in misassembly of A-type lamins and give rise to a variety of nuclear structure abnormalities which may contribute to disease progression. 11792811_A population of cultured skin fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene present dysmorphic nuclei or a disorganization of the nuclear lamina, or both. 11792821_Lamin A-binding is mediated by the central region of emerin, residues 70-178, outside the LEM-domain. 11799477_homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse 11897440_LMNA gene mutations account for 33% of the dilated cardiomyopathies with atrioventricular block, all familial autosomal dominant. 11973618_genes known to be responsible for Emery-Dreifuss muscular dystrophy 12015247_Novel missense mutations in two families with the Dunnigan variety of familial partial lipodystrophy, cardiac conduction system defects, and cardiomyopathy suggest a multisystem dystrophy syndrome due to LMNA mutations. 12018485_1H, 13C and 15N resonance assignments of the C-terminal domain 12032588_LMNA mutation is associated with autosomal dominant Emery-Dreifuss mucular dystrophy amd limb-girdle muscular dystrophy 12057196_solution structure of the human lamin A/C C-terminal globular domain which contains specific mutations causing four different heritable diseases 12075506_mutation causes mandibuloacral dysplasia 12112001_regions of PKC-alpha that are crucial for binding to lamin A, and vice versa 12138353_Absence of mutations in exon 8 of the gene in combination antiretroviral therapy-associated partial lipodystrophy. 12145775_Observational study of gene-disease association. (HuGE Navigator) 12196663_Mutations in the lamin A/C gene found associated with lipodystrophy, cardiac abnormalities, and muscular dystrophy. 12409453_In this study, we identify a novel interaction between lamin A/C and hsMOK2 by using the yeast two-hybrid system 12467734_The autosomal recessive axonal Charcot-Marie-Tooth type 2 due to mutation (c.892C>T-p.R298C) in a gene encoding Lamin A/C nuclear envelope proteins and the first gene in which a mutation leads to autosomal recessive Charcot-Marie-Tooth type 2. 12524233_LMNA missense mutations in nondiabetic carriers with Dunnigan-type familial partial lipodystrophy (FPLD) are associated with elevated levels of serum C-reactive protein and free fatty acid, particularly in women. 12628721_Mutations in LMNA cause a severe and progressive dilated cardiomyopathy in a relevant proportion of patients. 12673789_French family affected with a new phenotype composed of autosomal dominant severe dilated cardiomyopathy & a specific quadriceps muscle myopathy;identified missense mutation in the lamin A/C gene that cosegregated with the disease 12702809_Hutchinson-Gilford progeria appears to represent a novel laminopathy, caused by a single heterozygous splicing mutation in the LMNA gene, leading to a major loss of Lamin A expression, intimately associated to nuclear alterations 12714972_point mutations in Hutchinson-Gilford progeria syndrome 12718522_The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. 12729796_Results suggest that nuclear aggregate formation is in part due to overexpression of lamin A, but that there are also mutant-specific effects. 12768443_LMNA mutation was studied in Hutchinson-Gilford progeria. It does not occur in Wiedemann-Rautenstrauch progeroid syndrome. 12783988_the lamin a-emerin complex might have a role in muscular dystrophy and cardiomyopathy 12844477_In this study, we excluded mutations within the complete coding region and the promoter of LMNA and the CRABP II gene in HIV-1 infected patients 12920062_A specific phenotype characterized by early atrial fibrillation is associated with LMNA mutation. 13129702_REVIEW: The recent explosion in the number of identified mutations within the LMNA gene, which encodes two protein products lamins A and C, has allowed the identification of an allelic series of disorders, all caused by mutations within this one gene 14597414_Nuclear lamin A/C aggregates and a reduced incorporation of bromouridine were noted in fibroblasts from a familial partial lipodystrophy patient carrying an R482L lamin A/C mutation, demonstrating RNA transcription interference 14644157_data demonstrate that lamin C and lamin A interact in vivo directly with nesprin-1alpha and with emerin and that lamin A or C is sufficient for the correct anchorage of emerin and nesprin-1alpha at the nuclear envelope in human cells 14675861_A new LMNA mutation (1621C>T, R541C) was found in two members of a French family with a history of ventricular rhythm disturbances and an uncommon form of systolic left ventricle dysfunction. 14985400_LMNA represents the first gene implicated in both recessive and dominant forms of Charcot-Marie-Tooth disease 15026149_identified the epitope recognized by a new panel of mAbs against lamin A/C as a sequence of 9 amino acids that contain a complete beta-strand of the Ig-like globular domain; the major site of lipodystrophy missense mutation, R482 is present in the epitope 15080529_Review. Naturally occurring mutations in LMNA have been shown to be responsible for distinct diseases called laminopathies, including dilated cardiomyopathy with or without conduction defect and with or without variable skeletal muscle involvement. 15205219_Observational study of gene-disease association. (HuGE Navigator) 15205219_The H566H polymorphism was associated with metabolic syndrome and also higher mean fasting triglyceride and lower mean HDL-cholesterol concentrations in the Old Order Amish. 15205220_Review. Laminopathies are genetic diseases that encompass a wide spectrum of phenotypes with diverse tissue pathologies and result mainly from mutations in the LMNA gene encoding nuclear lamin A/C. 15219508_Observational study of gene-disease association. (HuGE Navigator) 15284226_the distinctive ensemble of heterotypic lamin interactions in a particular cell type affects the stability of the lamin polymer 15298354_a novel mutation associated with familial partial lipodystrophy 15317753_Heterozygous splicing mutation in the LMNA gene, leading to the complete or partial loss of exon 11 in mRNAs encoding Lamin A in restrictive dermopathy was found. 15342704_partial splice site selection in Hutchinson-Gilford progeria syndrome 15372542_mutations in lamins A and C may lead to a weakening of a structural support network in the nuclear envelope in fibroblasts and that nuclear architecture changes depend upon the location of the mutation 15551023_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 15622532_this is the first report of a patient combining features of these two phenotypes because of a single mutation(S143F) mutation in LMNA. 15636422_Observational study of gene-disease association. (HuGE Navigator) 15639119_This study identified a novel mutation in the 5' region of the LMNA gene -3del15, resulting in the loss of 15 nucleotides from -3 to +12, including the translation ATG initiator codon. 15748902_Our findings suggest a loss of function of A-type lamin mutant proteins in the organization of intranuclear chromatin and predict the loss of gene regulatory function in laminopathies. 15798706_Mutations in LMNA cause a severe and progressive dilated cardiomyopathy in a relevant proportion of patients. 15919811_This study further extends the vast range of diseases linked to LMNA mutations and identifies another genetic cause for the type A insulin resistance syndrome. 15961312_Two patients with 'Dropped head syndrome' due to mutations in LMNA genes. 15982412_results suggest that a mutant truncated lamin A, even when expressed at low levels, causes defective cell stability, which may be responsible for phenotypic abnormalities in Hutchinson-Gilford progeria syndrome 15998779_Two homozygous missense LMNA mutations involving the arginine 527 and alanine 529 residues cause MAD with subtle variations in phenotype. 16084085_Missense mutation in peripheral nerve disease in an individual. 16117820_Observational study of gene-disease association. (HuGE Navigator) 16126733_These results implicate the abnormal farnesylation of progerin in the cellular phenotype in HGPS cells and suggest that FTIs may represent a therapeutic option for patients with HGPS 16129833_blocking farnesylation of authentic progerin in transiently transfected HeLa, HEK 293, and NIH 3T3 cells with farnesyltransferase inhibitors (FTIs) restored normal nuclear architecture 16179429_Our results identify the absence of A-type lamin expression as a novel marker for undifferentiated ES cells and further support a role for nuclear lamins in cell maintenance and differentiation. 16218190_These data identify specific functional roles for the emerin-lamin C- and emerin-lamin A- containing protein complexes and is the first report to suggest that the A-type lamin mutations may be differentially dysfunctional for the same LMNA mutation. 16239243_The presence of the expanded CGG-repeat FMR1 mRNA results in reduced cell viability as well as the disruption of the normal architecture of lamin A/C within the nucleus. 16246140_Data show that chromosome positioning is largely unaffected in lymphoblastoid cell lines containing emerin or A-type lamin mutations. 16248985_Thus, we did not find evidence for uniquely interacting partner proteins using this approach, but did identify four new lamin A/C interactive partners 16262891_Observational study of gene-disease association. (HuGE Navigator) 16266469_The missense mutation E82K in LMNA gene was associated with a malignant phenotype of severe clinical symptoms of familial dilated cardiomyopathy. 16288872_This study reports a case of early onset myopathy due to a heterozygous LMNA mutation in exon 9, characterized by the presence of a marked number of cytoplasmic bodies with extensive myofibrillar abnormalities and Z-disk disruption in skeletal muscle. 16289535_The redistribution into lamin A-/pre-lamin A-containing aggregates of proteins such as pRb and SREBP1a could represent a key aspect underlying the molecular pathogenesis of certain laminopathies. 16344005_lamin A has a role in sensitivity to DNA damaging agents, the DNA damage response, and a senescent phenotype 16357800_Description of the clinical, morphological and biological features that should lead clinicians to consider the diagnosis of laminopathy in a diabetic patient. (review) 16371512_Ser-4 phosphorylation inhibits BAF binding to emerin and lamin A, and thereby weakens emerin-lamin interactions during both mitosis and interphase. 16410549_Results suggest that the C-terminus of nuclear titin binds lamins A and B in vivo and might contribute to nuclear organization during interphase. 16415973_The Charcot-Marie-Tooth diseases resulted from the mutations of LMNA gene are rare. 16461887_The mutant lamin A (progerin) accumulates in the nucleus in a cellular age-dependent manner. 16481476_Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation, with deficient cells displaying delayed differentiation kinetics that may underlie dystrophic phenotypes. 16518869_caspase-6 and its cleavage of lamin A are critical in apoptotic signaling triggered by resveratrol in the colon carcinoma cells, which can be activated in the absence of Bax or p53 16645051_observations implicate lamin A in physiological aging 16697197_Lamin A/C mutations form 'nuclear aggregates' when overexpressed by transfection and in cultured skin fibroblasts from EDMD patients. However, inappropriate lamin A/C assembly may be preventable by manipulation of cell growth conditions. 16738054_The epigenetic changes described most likely represent molecular mechanisms responsible for the rapid progression of premature aging in Hutchinson-Gilford Progeria Syndrome (HGPS) patients. 16772334_A subset of lamin A mutants might hinder the response of components of the DNA repair machinery to DNA damage by altering interactions with chromatin. 16823856_We found that in inclusion-body myositis (IBM) muscle vacuoles were immunoreactive for the inner nuclear membrane proteins emerin and lamin A/C. 16825283_Expression of a lamin A mutant that induces alterations in nuclear morphology can cause tissue and organ damage in mice with a normal complement of wild-type lamins. 16981056_The most frequently encountered mutations associated with Dilated Cardiomiopathy are found in LMNA, coding for lamins A and C, intermediate filament proteins. 17090536_This review summarizes the abnormalities caused by an LMNA gene mutation which targets the nuclear envelope, where it interferes with the integrity of the nuclear envelope and causes misshapen cell nuclei, leading to progeroid syndromes. 17097067_Mislocalization of emerin to the endoplasmic reticulum in human cells lacking A-type lamin leads to its degradation and provides the first evidence that its degradation is mediated by the proteasome. 17117676_Neither emerin nor LMNA mutations in a subset of families with EDMD-like phenotypes that may imply an existence of other genes causing similar disorders. 17136397_If neurogenic atrophy is combined with a cardiac disease in a family, this should prompt LMNA mutation analysis. 17150192_Since it has been reported that progeroid features are associated with increased extracellular matrix in dermal tissues, we compared a subset of these components in fibroblast cultures from LMNA mutants with those of control fibroblasts. 17227891_Results suggest that LAP2alpha and lamin A/C are involved in controlling retinoblastoma protein localization and phosphorylation, and a lack or mislocalization of either protein leads to cell cycle arrest in fibroblasts. 17291448_data suggest that the unprenylated prelamin A is not toxic to the cells 17301031_The major causal mutation associated with HGPS triggers abnormal messenger RNA splicing of the lamin A gene leading to changes in the nuclear architecture. 17327437_Eight tag single nucleotide polymorphisms in the LMNA locus were genotyped in 7,495 Danish whites and related to metabolic and anthropometric traits. 17327437_Observational study of gene-disease association. (HuGE Navigator) 17327460_A meta-analysis and a case-control study evaluating the role of LMNA mutations in the development of type 2 diabetes are reported. 17327460_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17327461_Observational study of gene-disease association. (HuGE Navigator) 17327461_The results of 2 case-control studies of type 2 diabetes mellitus and a separate study of metabolic syndrome and LMNA polymorphisms is reported. 17334235_found three LMNA mutations including a case having a previously described (Glu161Lys) mutation and two having novel mutations (Glu53Val and Glu186Lys) 17352743_a change in the amount of lamin A, rather than appearance of its truncated form, is responsible for growth retardation in affected cells 17360326_These results provide insights into the mechanisms responsible for premature aging and also shed light on the role of lamins in the normal process of human aging. 17360355_Progerin/LADelta50 mislocalizes into insoluble cytoplasmic aggregates and membranes during mitosis and causes abnormal chromosome segregation and binucleation. 17428859_US3 kinase activity regulates HSV-1 capsid nuclear egress at least in part by phosphorylation of lamin A/C 17454124_a lamin A/C mutation has a role in amyotrophic quadricipital syndrome with cardiac involvement 17459035_LMNA p.G608G mutation results in a uniform phenotype through early to mid-childhood, in keeping with that described in classical Hutchinson-Gilford progeria syndrome 17469202_Unusual LMNA mutations associated with severe progeria. 17536044_findings highlight the crucial role of lamin A/C-emerin interactions, with evidence for synergistic effects of these mutations that lead to Emery-Dreifuss muscular dystrophy as the worsened result of digenic mechanism in this family 17605093_Malignant mutation in lamin A/C gene causing progressive conduction system disease and early death in limb-girdle muscular dystrophy. 17612587_LMNA mutations and protease inhibitor treatment result in accumulation of farnesylated prelamin A and oxidative stress that trigger premature cellular senescence. 17701980_The findings from these cases further expand the clinical spectrum associated with mutations in the LMNA gene. 17711925_Mutations in the LMNA gene are responsible for several laminopathies, including lipodystrophies, with complex genotype/phenotype relationships 17718387_Mutations in LMNA gene are the cause of many different diseases, called laminopathies. Among laminopathies are muscle tissue diseases, adipose tissue diseases and also progerias, the premature aging syndromes. 17760566_Results indicate that pathogenic mutations in lamin A/C lead to sequestration of hsMOK2 into nuclear aggregates, which may deregulate MOK2 target genes. 17848622_RNA interference (RNAi) knockdown of XPA in Hutchinson-Gilford progeria syndrome cells partially restored double-strand breaks repair as evidenced by Western blot analysis, immunofluorescence and comet assays 17870066_Our findings raise a hypothesis that changes in lamina organization may cause accelerated telomere attrition, with different kinetics for overexpession of wild-type and mutant lamin A, which leads to rapid replicative senescence and progroid phenotypes. 17881656_The S143F lamin A/C point mutation causes a phenotype combining features of myopathy and progeria. Dermal fibroblast cells have dysmorphic nuclei containing numerous blebs and lobulations, which progressively accumulate as cells age in culture. 17893350_both patients had markedly increased neck fat content, specifically surrounding the trachea.The association with sleep apnea might be related to the repartitioning of adipose tissue in patients with this type of lipodystrophy. 17935239_Homozygous missense mutation in LMNA gene is associated with Mandibuloacral dysplasia and severe progressive skeletal changes 17987279_Nonsense mediated decay is not sufficient to completely prevent the expression of truncated lamin A and that even trace amounts of it may negatively interfere with structural and/or regulatory functions of lamin A/C. 17994215_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18031308_A novel clinical form of familial partial lipodystrophy 2, due to a mutation affecting lamin A only, with cardiac involvement. 18035086_study highlights the role of LMNA mutations in dilated cardiomyopathy and related disorders. A severe phenotype in p.N195K mutation carriers and preferential cardiac conduction disease in p.R225X carriers was encountered. 18041775_present case illustrates that mutations of the LMNA codon cause familial partial lipodystrophy that is inherited in an autosomal dominant pattern with variable expressivity 18077842_Correlation between LMNA adipose expression and cytokine and adipogenic gene markers in HIV-positive patients, regardless of presence or absence of lipodystrophy. 18093584_These results may be used to evaluate downstream effects of FTIs or other prelamin A inhibitors potentially useful for the therapy of laminopathies. 18308323_Systematic comparison of the mechanical behavior of the wild-type protein and a missense mutated protein with the point mutation p.Glu358Lys show that the nanomechanical tensile behavior of the dimer segment does not vary with the mutation. 18311132_Data suggest that Lamin A-dependent misregulation of adult stem cells is associated with accelerated aging. 18337098_patients bearing a LMNA gene mutation associated to an apparently selective cardiac phenotype may present subclinical skeletal muscle involvement 18339564_We present a 6-year-old girl with premature aging associated with mild myopathy, displaying muscle weakness, joint contractures and hyporeflexia. Genetic analysis revealed rare heterozygous point mutation in lamin A/C gene. 18348272_Association of homozygous LMNA mutation R471C with new phenotype: mandibuloacral dysplasia, progeria, and rigid spine muscular dystrophy. 18364375_In these LMNA-linked lipodystrophic patients, the prevalence of PCOS, infertility, and gestational diabetes was higher than in the general population. 18396274_The influence of expression of the Emery-Dreifuss muscular dystrophy R453W mutation and of the Dunnigan-type partial lipodystrophy R482W mutation of lamin A on transcription and epigenetic regulation of the myogenin gene and on chromatin organization. 18442054_bone resorption activity of osteoclasts obtained in the presence of high prelamin A levels is lower with respect to control osteoclasts 18442998_Expression of progerin leads to alterations in nuclear morphology, which may underlie pathology in Hutchinson-Gilford progeria syndrome. 18478590_This report provides further evidence of the extreme phenotypic diversity and low penetrance associated with the R644C mutation. 18497734_Lamin A/C may be involved in the adipocyte gene profile observed in obesity and type 2 diabetes. 18502446_D192G mutation in LMNA gene may lead to the disruption of the nuclear wall in cardiomyocytes, thus supporting the mechanical hypothesis of dilated cardiomyopathy development in humans, which might be mutation-specific. 18524819_lamin A/C, lamin B1, and viral US3 kinase have roles in viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane 18538321_The inability of lamin C mutants to join the nuclear rim in the absence of lamin A is a potential pathophysiological mechanism for laminopathies. 18549403_Founder effect and estimation of the age of the c.892C>T (p.Arg298Cys) mutation in LMNA associated to Charcot-Marie-Tooth subtype CMT2B1 in families from North Western Africa. 18551513_study describes a new entity of congenital muscular dystrophies caused by de novo LMNA mutations 18564364_the first cases of laminopathies from Russia are reported: In 10 unrelated families, 9 different mutations were identified and three phenotypes were observed 18585512_Observational study of gene-disease association. (HuGE Navigator) 18604166_results indicate that accumulation of the lamin A precursor protein determines a defect in DNA damage response after X-ray exposure, supporting a crucial role of lamin A in regulating DNA repair process and cell cycle control 18606848_These results suggest that SUMO modification is important for normal lamin A function and implicate an involvement for altered sumoylation in the E203G/E203K lamin A cardiomyopathies. 18611980_A mutation within the LMNA gene is associated with heart-hand syndrome of Slovenian type. 18612243_The silencing of lamin A/C expression resulted in a decrease in the volume and surface area of chromosome territories, especially in chromosomes with high heterochromatin content. 18643848_In human keratinocytes, we found BMP-4 facilitates trichohyalin (THH) transcription, and lamin C plays a key role in the posttranslational stabilization of THH. 18646565_Mutations in the genes for nuclear envelope proteins of emerin (EMD) and lamin A/C (LMNA) are known to cause Emery-Dreifuss muscular dystrophy (EDMD) and limb girdle muscular dystrophy (LGMD). 18667561_Binding of T. foetus to LMN-1 rendered the parasite toxic to HeLa cell monolayers. 18691775_we present a family with sudden cardiac death in the absence of left ventricular dysfunction, related to a Lamin A/C mutation 18714339_Report links A-type lamin expression to colorectal tumour progression and raises the profile from one implicated in multiple but rare genetic conditions to a gene involved in one of the commonest diseases in the Western World. 18714801_Studiy identified a large French Canadian family with the LGMD 1B phenotype and a cardiac conduction disease phenotype that carried a new new (IVS9-3C > G) LMNA gene mutation. 18767923_lamin A/C is essential for proper RANKL-dependent osteoblastogenesis 18795223_Testing LMNA in families of people with familial dilated cardiomyopathy is recommended because genotype information in an individual could definitely be useful for the clinician. 18805829_prelamin A accumulation in peripheral scAT is associated with a reduced expression of several genes involved in adipogenesis. 18816602_We report the clinical characteristics, genetic analysis, and muscle biopsy findings of a family with Emery-Dreifuss muscular dystrophy and a novel mutation (Leu162Pro) in the LMNA gene. 18830724_Meta-analysis of gene-disease association. (HuGE Navigator) 18843043_Data show that hTERT activity or inactivation of p53 can suppress the cell proliferation defects associated with lamin A mutants that are incorrectly processed. 18848371_Observational study of gene-disease association. (HuGE Navigator) 18848371_The mechanisms underlying the associations with cognitive impairment and LOAD require further elucidation, but both genes are interesting candidates for involvement in age-related cognitive impairment. 18923140_Results suggest that prelamin A is imported directly into the nucleus where it is processed by Zmpste24 and Icmt, which exhibit a dual localization to the inner nuclear membrane as well as the ER membrane. 18926329_Dilated cardiomyopathies caused by LMNA gene defects are highly penetrant, adult onset, malignant diseases characterized by a high rate of heart failure and life-threatening arrhythmias. 18946024_plasma-membrane-anchored growth factor pro-amphiregulin binds A-type lamin and regulates global transcription 18950579_The intranuclear accumulation of lamin A precursors which cannot be fully processed & exert a toxic effect on nuclear homeostasis leads to various genetic syndromes. Review. 18959190_During detailed studies of the cells from this patient the nuclear lamina aberrations were detected. 18982914_Progeria is caused by mutation in the gene of LMNA, encoding a nuclear protein, lamin A, which has been shown to affect RNA polymerase II transcription. 19015316_Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. 19022376_Knockdown of A-type lamins and emerin in HeLa and C2C12 stimulated phosphorylation and nuclear translocation of ERK as well as activation of genes encoding downstream transcription factors. 19084400_We present a consanguineous family in which two children have early onset LMNA-related myopathy likely due to paternal germinal mosaicism. 19124654_Lamin A/C-mediated neuromuscular junction defects contribute to the autosomal dominant Emery-Dreifuss muscular dystrophy disease phenotype 19126678_process of adipogenesis is affected by a dynamic link between complexes of emerin and lamins A/C at the nuclear envelope and nucleocytoplasmic distribution of beta-catenin, to influence cellular plasticity and differentiation. 19141474_Silencing lamin B1 expression dramatically increases the lamina meshwork size and the mobility of nucleoplasmic lamin A 19144202_lamin A/C is involved in the pathogenesis of gastric carcinoma 19172989_lamin A Delta 150 transcript is present in unaffected controls but its expression is >160-fold lower than in HGPS patients. Lamin A Delta 150 transcript increases in late passage cells from HGPS patients and parental controls. 19201734_Transgenic mice express the LMNA mutation that causes familial partial lipodystrophy of the Dunnigan type (FPLD2). The phenotype in FPLD-transgenic mice resembles human FPLD2, including lack of fat accumulation, insulin resistance, and fatty liver. 19204888_Male subjects with familial partial lipodystrophy due to a lamin A/C R482W mutation may develop metabolic abnormalities. including hypoleptinemia 19220582_The R439C mutation causes oligomerization of the C-terminal globular domain of lamins A and C, which increases its binding affinity for DNA. 19247430_Regulation of the D4Z4 array depends on both the number of repeats and the presence of CTCF and A-type Lamins in facio-scapulo-humeral dystrophy. 19270485_Atypical Werner's syndrome with the severe metabolic complications, the extent of the lipodystrophy is associated with A133L mutation in the LMNA gene and these patients present with phenotypically heterogeneous disorders. 19283854_Two unrelated young women experienced premature ovarian failure, and both were found to have the same heterozygous novel missense mutation c.176T>G in exon 1 of the LMNA gene. 19318026_Observational study of genetic testing. (HuGE Navigator) 19323649_Non-farnesylated and farnesylated carboxymethylated lamin A precursors in human fibroblasts modifies emerin localization. 19328042_mutations in LMNA were identified in individuals with isolated cardiac involvement, specifically DCM, AVB, and infrequently atrial arrhythmias, including AF 19351612_inhibition of the prelamin A endoprotease ZMPSTE24 mostly elicits accumulation of full-length prelamin A in its farnesylated form, while loss of the prelamin A cleavage site causes accumulation of carboxymethylated prelamin A in progeria cells 19384091_Lamin A/C deficiency is an important cause of dilated cardiomyopathy. 19401371_Common variation in the lamin a/c gene does not contribute to the etiology of PCOS in women of European ancestry. 19401371_Observational study of gene-disease association. (HuGE Navigator) 19424285_Mutations in the LMNA gene do not cause axonal Charcot-Marie-Tooth. 19424285_Observational study of gene-disease association. (HuGE Navigator) 19427440_LMNA mutations rarely cause lone AF and routine genetic testing of LMNA in these patients does not appear warranted. 19427440_Observational study of gene-disease association. (HuGE Navigator) 19442658_This is the first study of a direct link between LaA mutant expression and reduced nuclear protein import. 19446900_We describe 7 transplanted heart recipients from a single family with limb-girdle muscular dystrophy type 1B linked to a mutation of the LMNA gene in the splice donor site of the exon 9 (IVS 9+1:g>a). 19490114_Data demonstrated that the phosphorylation of hsMOK2 interfered with its ability to bind lamin A/C. 19524666_Results suggest that a functional emerin-lamin A/C complex is required for cell spreading and proliferation, possibly acting through ERK1/2 signalling. 19574635_The crystal structure of the lamin A/C mutant R482W, a variant that causes FPLD, has been determined at 1.5 A resolution. 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19589617_Impaired nuclear functions lead to increased senescence and inefficient cell differentiation in human myoblasts with a dominant p.R545C mutation in the LMNA gene. 19638735_Lamin A/C gene mutations may have a role in familial cardiomyopathy with advanced atrioventricular block and arrhythmia 19644448_These findings reveal the existence of an 80 bp D4Z4 human subtelomeric repeat sequence that is sufficient to position an adjacent telomere to the nuclear periphery in a CTCF and A-type lamins-dependent manner. 19645629_results suggest that LMNA, ZMPSTE24, and LBR sequence variations are not major genetic determinants involved in scleroderma pathogenesis 19672032_Observational study of gene-disease association. (HuGE Navigator) 19680556_Observational study of gene-disease association. (HuGE Navigator) 19768759_the exon 1 c.178 C/G, p.Arg 60 Gly LMNA gene mutation is associated with a novel phenotype featuring cardiac involvement followed by late lipodystrophy, diabetes, and peripheral axonal neuropathy. 19775189_Results show that decline in Hela lamin A/C expression correlates with modified cell signaling and minor alterations in metabolism coupled with changes in expression in structural proteins of the cytoskeleton. 19841875_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19855837_analysis of Drosophila and human A-type lamins 19859838_A clinical picture related to the LMNA | ENSMUSG00000028063 | Lmna | 4.071104e+03 | 1.2135278 | 0.279207199 | 0.3194929 | 7.663496e-01 | 0.3813487226 | 0.84181165 | No | Yes | 4.147504e+03 | 538.411058 | 3.003974e+03 | 400.612784 | |
| ENSG00000160932 | 4061 | LY6E | protein_coding | Q16553 | FUNCTION: GPI-anchored cell surface protein that regulates T-lymphocytes proliferation, differentiation, and activation. Regulates the T-cell receptor (TCR) signaling by interacting with component CD3Z/CD247 at the plasma membrane, leading to CD3Z/CD247 phosphorylation modulation (By similarity). Restricts the entry of human coronaviruses, including SARS-CoV, MERS-CoV and SARS-CoV-2, by interfering with spike protein-mediated membrane fusion (PubMed:32641482). Plays also an essential role in placenta formation by acting as the main receptor for syncytin-A (SynA). Therefore, participates in the normal fusion of syncytiotrophoblast layer I (SynT-I) and in the proper morphogenesis of both fetal and maternal vasculatures within the placenta. May also act as a modulator of nicotinic acetylcholine receptors (nAChRs) activity (By similarity). {ECO:0000250|UniProtKB:Q64253, ECO:0000269|PubMed:32641482}.; FUNCTION: (Microbial infection) Promotes entry, likely through an enhanced virus-cell fusion process, of various viruses including HIV-1, West Nile virus, dengue virus and Zika virus (PubMed:28130445). In contrast, the paramyxovirus PIV5, which enters at the plasma membrane, does not require LY6E (PubMed:28130445, PubMed:29610346). Mechanistically, adopts a microtubule-like organization upon viral infection and enhances viral uncoating after endosomal escape (PubMed:28130445, PubMed:30190477). {ECO:0000269|PubMed:28130445, ECO:0000269|PubMed:29610346, ECO:0000269|PubMed:30190477}. | Cell membrane;Disulfide bond;GPI-anchor;Glycoprotein;Lipoprotein;Membrane;Reference proteome;Signal | This gene belongs to the human Ly6 gene family and encodes a glycosylphosphatidyl-inositol (GPI)-anchored cell surface protein. The protein plays an important role in T cell physiology, oncogenesis and immunological regulation. The protein is also involved in modulation of viral infection by coronaviruses, SARS-CoV, MERS-CoV and SARS-CoV-2. [provided by RefSeq, Aug 2021]. | hsa:4061; | anchored component of membrane [GO:0031225]; extracellular region [GO:0005576]; plasma membrane [GO:0005886]; synapse [GO:0045202]; acetylcholine receptor binding [GO:0033130]; acetylcholine receptor inhibitor activity [GO:0030550]; acetylcholine receptor signaling pathway [GO:0095500]; adrenal gland development [GO:0030325]; cell surface receptor signaling pathway [GO:0007166]; epinephrine secretion [GO:0048242]; in utero embryonic development [GO:0001701]; negative regulation of viral entry into host cell [GO:0046597]; norepinephrine metabolic process [GO:0042415]; organ growth [GO:0035265]; ventricular cardiac muscle tissue morphogenesis [GO:0055010] | 18755862_Increased expression of the type I interferon-inducible gene, lymphocyte antigen 6 complex locus E, in peripheral blood cells is predictive of lupus activity in a large cohort of Chinese lupus patients. 18990604_Two SCA3 and one SCA2 cases have been identified which show autosomal dominant inheritance in Parkinson disease. 19672991_This study suggested that a mutation in SCA2 or SCA3/MJD may be one of the genetic causes of Parkinson's disease in china. 20237496_Observational study of gene-disease association. (HuGE Navigator) 25225669_the LY6E pathway in monocytes represents one of negative feedback mechanisms that counterbalance monocyte activation which may serve as a potential target for immune intervention. 25344775_LY6E level of gene expression may serve as good biomarkers for systemic lupus erythematosus diagnosis. 27197181_LY6E overexpression is associated with Breast Cancer Progression, Immune Escape, and Drug Resistance. 27589564_High LY6E expression is associated with breast cancer. 28130445_LY6E is a positive modulator of HIV-1 infection in primary human peripheral blood mononuclear cells, immortalized CD4+ T lymphoid cells, and macrophages. HIV Long Terminal Repeat-driven HIV-1 gene expression is also enhanced by LY6E, suggesting additional roles of LY6E in HIV-1 replication. 29448250_LY6E knockdown by targeted-siRNA inhibited gastric cancer cell survival and proliferation and induced G1-S cell cycle arrest and apoptosis in gastric cancer cells 30190477_LY6E belongs to a growing class of interferon-inducible factors that broadly enhance viral infectivity in an interferon-independent manner. 30674630_LY6E downregulates the cell surface receptor CD4. 32694157_A Phase I Study of DLYE5953A, an Anti-LY6E Antibody Covalently Linked to Monomethyl Auristatin E, in Patients with Refractory Solid Tumors. 32839948_Protein LY6E as a candidate for mediating transport of adeno-associated virus across the human blood-brain barrier. 35310607_Multidimension Analysis of the Prognostic Value, Immune Regulatory Function, and ceRNA Network of LY6E in Individuals with Colorectal Cancer. | ENSMUSG00000022587 | Ly6e | 8.290892e+03 | 1.2784531 | 0.354399215 | 0.3277702 | 1.160010e+00 | 0.2814635623 | 0.80799359 | No | Yes | 8.528852e+03 | 1127.060675 | 6.066612e+03 | 822.526346 | |
| ENSG00000161082 | 60680 | CELF5 | protein_coding | Q8N6W0 | FUNCTION: RNA-binding protein implicated in the regulation of pre-mRNA alternative splicing. Mediates exon inclusion and/or exclusion in pre-mRNA that are subject to tissue-specific and developmentally regulated alternative splicing. Specifically activates exon 5 inclusion of cardiac isoforms of TNNT2 during heart remodeling at the juvenile to adult transition. Binds to muscle-specific splicing enhancer (MSE) intronic sites flanking the alternative exon 5 of TNNT2 pre-mRNA. {ECO:0000269|PubMed:11158314}. | 3D-structure;Alternative splicing;Cytoplasm;Nucleus;RNA-binding;Reference proteome;Repeat;mRNA processing | This gene encodes a member of the the CELF/BRUNOL protein family, which contain two N-terminal RNA recognition motif (RRM) domains, one C-terminal RRM domain, and a divergent segment of 160-230 aa between the second and third RRM domains. Members of this protein family regulate pre-mRNA alternative splicing and may also be involved in mRNA editing and translation. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jan 2012]. | hsa:60680; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA binding [GO:0003729]; pre-mRNA binding [GO:0036002]; RNA binding [GO:0003723]; mRNA splice site selection [GO:0006376]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381] | ENSMUSG00000034818 | Celf5 | 1.752168e+02 | 0.9702129 | -0.043626773 | 0.3327848 | 1.754742e-02 | 0.8946151857 | 0.98078971 | No | Yes | 1.392948e+02 | 21.829487 | 1.603019e+02 | 25.637533 | ||
| ENSG00000161179 | 150223 | YDJC | protein_coding | A8MPS7 | FUNCTION: Probably catalyzes the deacetylation of acetylated carbohydrates an important step in the degradation of oligosaccharides. {ECO:0000250|UniProtKB:Q53WD3}. | Alternative splicing;Carbohydrate metabolism;Hydrolase;Magnesium;Metal-binding;Reference proteome | hsa:150223; | deacetylase activity [GO:0019213]; magnesium ion binding [GO:0000287]; carbohydrate metabolic process [GO:0005975] | ENSMUSG00000041774 | Ydjc | 6.918462e+03 | 1.5247772 | 0.608598425 | 0.3480911 | 3.018395e+00 | 0.0823249097 | 0.73777071 | No | Yes | 9.556282e+03 | 1534.755779 | 5.347854e+03 | 881.703793 | |||
| ENSG00000161217 | 5130 | PCYT1A | protein_coding | P49585 | FUNCTION: Catalyzes the key rate-limiting step in the CDP-choline pathway for phosphatidylcholine biosynthesis. {ECO:0000269|PubMed:10480912, ECO:0000269|PubMed:7918629}. | Acetylation;Cone-rod dystrophy;Cytoplasm;Disease variant;Dwarfism;Endoplasmic reticulum;Lipid biosynthesis;Lipid metabolism;Membrane;Nucleotidyltransferase;Nucleus;Phospholipid biosynthesis;Phospholipid metabolism;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation | PATHWAY: Phospholipid metabolism; phosphatidylcholine biosynthesis; phosphatidylcholine from phosphocholine: step 1/2. {ECO:0000269|PubMed:10480912, ECO:0000269|PubMed:7918629}. | This gene belongs to the cytidylyltransferase family and is involved in the regulation of phosphatidylcholine biosynthesis. Mutations in this gene are associated with spondylometaphyseal dysplasia with cone-rod dystrophy. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Aug 2015]. | hsa:5130; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; glycogen granule [GO:0042587]; nuclear envelope [GO:0005635]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; calmodulin binding [GO:0005516]; choline-phosphate cytidylyltransferase activity [GO:0004105]; identical protein binding [GO:0042802]; phosphatidylcholine binding [GO:0031210]; protein homodimerization activity [GO:0042803]; CDP-choline pathway [GO:0006657]; phosphatidylcholine biosynthetic process [GO:0006656] | 17184542_Analyses showed genotype effects of PCYT1A genes on spina bifida risk, but did not show evidence of gene-nutrient. interactions. 17184542_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19737740_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20661636_N-Methylaspartate induced nitric oxide synthase activation and nuclear factor-kB subunit p65 nuclear translocation in A549 cells were responsible for decreased CTP:phosphocholine cytidylyltransferase A expression 20662904_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 24387990_We report loss-of-function mutations in PCYT1A as the cause of spondylometaphyseal dysplasia with cone-rod dystrophy. 24387991_Mutations in PCYT1A cause spondylometaphyseal dysplasia with cone-rod dystrophy 24889630_PCYT1A-generated phosphatidylcholine has a role in the normal function of white adipose tissue and insulin action 26165797_CCT contributes to phospholipid compositional homeostasis. [Review] 28272537_PCYT1A mutations were identified in patients with isolated retinal dystrophy without any skeletal involvement from two Italian families. 28509322_There is no correlation between single PCYT1A rs712012 and PCYT1A rs7639752 polymorphisms and the incidence of intrauterine fetal death. 30055775_Our study shows that choline intake in Polish pregnant women is inadequate and that polymorphisms of PEMT rs12325817 and PCYT1A rs7639752 are associated with betaine but not choline concentrations. 30559292_Mutations in PCYT1A gene is associated with impaired enzyme kinetics and folding resulting in lipodystrophy, spondylometaphyseal dysplasia with cone-rod dystrophy, and isolated retinal dystrophy 31488547_CCTA activation by membrane binding is sensitive to mutations in the alphaE and J segments, especially within or proximal to the alphaE hinge. 31488548_The membrane-adsorbed, folded allosteric linker of CCTA may partially cover the active site cleft and pull it close to the membrane surface. 31517566_De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. 32186954_Differential dephosphorylation of CTP:phosphocholine cytidylyltransferase upon translocation to nuclear membranes and lipid droplets. 32703435_PCYT1A suppresses proliferation and migration via inhibiting mTORC1 pathway in lung adenocarcinoma. 33521992_Methylome and transcriptome profiling revealed epigenetic silencing of LPCAT1 and PCYT1A associated with lipidome alterations in polycystic ovary syndrome. | ENSMUSG00000005615 | Pcyt1a | 1.586380e+03 | 0.6885235 | -0.538422185 | 0.2932322 | 3.336933e+00 | 0.0677407547 | 0.71041829 | No | Yes | 1.170593e+03 | 162.751075 | 1.680110e+03 | 239.112329 |
| ENSG00000161860 | 256126 | SYCE2 | protein_coding | Q6PIF2 | FUNCTION: Major component of the transverse central element of synaptonemal complexes (SCS), formed between homologous chromosomes during meiotic prophase. Requires SYCP1 in order to be incorporated into the central element. May have a role in the synaptonemal complex assembly, stabilization and recombination (By similarity). {ECO:0000250|UniProtKB:Q505B8}. | 3D-structure;Cell cycle;Cell division;Chromosome;Coiled coil;Meiosis;Nucleus;Reference proteome | The protein encoded by this gene is part of the synaptonemal complex formed between homologous chromosomes during meiotic prophase. The encoded protein associates with SYCP1 and SYCE1 and is found only where chromosome cores are synapsed. [provided by RefSeq, Dec 2012]. | hsa:256126; | central element [GO:0000801]; chromosome [GO:0005694]; nucleoplasm [GO:0005654]; cell division [GO:0051301]; synaptonemal complex assembly [GO:0007130] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22870393_a model of chromosome synapsis driven by growth of SYCE2-TEX12 higher-order structures within the CE of the SC. 34373646_Structural basis of meiotic chromosome synaptic elongation through hierarchical fibrous assembly of SYCE2-TEX12. | ENSMUSG00000003824 | Syce2 | 2.888855e+01 | 0.7636880 | -0.388944719 | 0.5679298 | 4.574362e-01 | 0.4988246287 | 0.87548575 | No | Yes | 1.740166e+01 | 4.754563 | 1.542976e+01 | 4.348597 | |
| ENSG00000161921 | 58191 | CXCL16 | protein_coding | Q9H2A7 | FUNCTION: Acts as a scavenger receptor on macrophages, which specifically binds to OxLDL (oxidized low density lipoprotein), suggesting that it may be involved in pathophysiology such as atherogenesis (By similarity). Induces a strong chemotactic response. Induces calcium mobilization. Binds to CXCR6/Bonzo. {ECO:0000250}. | Cell membrane;Chemotaxis;Cytokine;Disulfide bond;Glycoprotein;Membrane;Reference proteome;Secreted;Signal;Transmembrane;Transmembrane helix | hsa:58191; | extracellular region [GO:0005576]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; chemokine activity [GO:0008009]; low-density lipoprotein particle receptor activity [GO:0005041]; scavenger receptor activity [GO:0005044]; chemotaxis [GO:0006935]; positive regulation of cell growth [GO:0030307]; positive regulation of cell migration [GO:0030335]; receptor-mediated endocytosis [GO:0006898]; response to cytokine [GO:0034097]; response to interferon-gamma [GO:0034341]; response to tumor necrosis factor [GO:0034612]; T cell chemotaxis [GO:0010818] | 12902461_SR-PSOX/CXCL16 may play an important role in facilitating uptake of various pathogens and chemotaxis of T and NKT cells by antigen presenting cells through its chemokine domain. 14625285_CXCL16 may play an important role in the development and progression of atherosclerotic vascular disease 14634054_SR-PSOX/CXCL16 is a unique molecule that not only attracts T cells and NKT cells toward DCs but also supports their firm adhesion to DCs. 14699018_SR-PSOX/CXCL16 may be involved in CD8+ T cell recruitment through VLA-4 activation and stimulation of IFN-[gamma] production by CD8+ T cells during inflammatory valvular heart disease. 14988089_This chemokine/scavenger receptor could serve as a molecular link between lipid metabolism and immune activity in the atherosclerotic lesion. 15128827_This report demonstrates that CXCL16 is a novel substrate for a disintegrin and metalloproteinase domain (ADAM)10-mediated ectodomain shedding. 15555552_Smooth muscle cells express CXCL16 in atherosclerotic lesions, which may play a role in the attraction of T cells to atherosclerotic lesions and contribute to the cellular internalisation of modified LDL. 15634930_CXCL16 promotes lymphocyte adhesion to epithelial cells and may function to attract and retain effector cells that support biliary and hepatocyte destruction in inflammatory liver disease. 15836657_Observational study of gene-disease association. (HuGE Navigator) 15883016_CXCL16 may act as a novel angiogenic factor for HUVEC and that ERK is involved as an important signaling molecule to mediate its angiogenic effects 15934948_Transmembrane chemokine CXCL16 is expressed in the brain by malignant and inflamed astroglial cells, shed to a soluble form and targets not only activated T cells but also glial cells themselves. 16142401_Findings suggest that retinoid signaling might be a pathway modulating inflammatory response by regulating CXCL16 expression in a cell-specific manner. 16200580_CXCL16 plays an important role in T cell accumulation and stimulation in rheumatoid arthritis synovium 16431903_CXCL16 plays important roles in human extravillous cytotrophoblast invasion and placentation. 16849465_The membrane-bound scavenger receptor CXCL16 expressed on the surface of plasmacytoid dendritic cells contributes to the binding, uptake, and stimulatory activity of D class CpG oligonucleotides. 17300746_evaluate the contribution of soluble CXCL16 to the scavenging of oxLDL and its potential as a marker for cardiovascular disease in patients with rheumatoid arthritis 17363916_CXCL16 is constitutively expressed on the surface of human epidermal keratinocytes, released upon cell activation or photodamage and may then target CXCR6-expressing T cells in the dermis 17703412_Observational study of gene-disease association. (HuGE Navigator) 17803654_Bacterial infection causes the upregulation of CXCL16 in gallbladder epithelia, leading to the chemoattraction of macrophages via CXCL16-CXCR6 interaction and formation of the characteristic histology of xanthogranulomatous cholecystitis. 17855433_The CXCL16 is a novel mediator of the innate immune reactivities of epidermal keratinocytes. 18194461_Hyperhomocysteinemia up-regulates CXCL16 leading to increased recruitment of CXCR6(+) lymphocytes and scavenging of modified lipids via a potential involvement of a PPAR-gamma-dependent mechanism 18195710_Re-expression of CXCL16 reduced growth of an RCC cell line in vitro. 18248772_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18248772_in Crohn's disease (CD), the CXCL16 p.Ala181Val polymorphism is not a disease susceptibility gene but associated with younger age at disease onset and higher frequency of ileal involvement 18250446_Functions as one of the factors in trophoblast-conditioned culture medium that interacts with CXCR6 and recruits and maintains monocytes, T cells, and gammadelta T cells residing in human decidua. 18279707_Increased level of serum-soluble CXCL16 was independently associated with acute coronary syndromes 18293410_CXCL16 appears to be a novel growth factor for schwannomas of different localization. 18339644_Soluble CXCL16 could be linked to atherogenesis not only as a marker of inflammation, but also as a potential inflammatory mediator. 18344492_CXCL16 functions, through CXCR6, as a novel chemotactic factor for prostate cancer cells. 18373975_The regulation and functional role of CXCL16 in human mesangial cells was studied. 18480749_CXCL16 and ADAM10 are involved in the recruitment of T cells to the kidney 18514099_CXCL16 levels in the acute coronary syndrome group were higher than controls and stable angina pectoris group 18565283_sirna decreased the expression of the SRPSOX gene by inhibiting macrophage-derived foam cell formation 18636150_CXCL16 seems to play an important role in the pathobiology of pancreatic cancer 18760678_CXCR6 and CXCR3 act coordinately with respective ligands and are involved in the pathophysiology of Juvenile Idiopathic Arthritis-associated inflammatory processes. 19070478_data demonstrate that downregulation of CXCL16 plays an important role in renal cancer development and progression, and that CXCL16 in renal cell carcinoma is an independent prognostic marker for better patient survival 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19415545_bronchial epithelial cells secreted relatively high basal levels of CXCL16, chemotactic for CXCR6 expressing T cells from blood 19426159_regulation of CXCL16, ADAM10 and oxLDL expression may be an early event in the onset of diabetic nephropathy 19435795_we presume important roles for CXCL16, ADAM10, and ADAM17 in the development of membranous nephropathy 19494317_CXCL16 mediates atheroprotection through its scavenger role in macrophages and not by cell-cell adhesion 19528340_In patients with an acute coronary syndrome, CXCL16 levels obtained within 24 hours of admission are associated with long-term mortality after adjustment for other risk factors 19575365_Myofibroblastic hepatic stellate cells release soluble Hedgehog ligands that stimulate cholangiocytes to produce Cxcl16 and recruit NKT cells. 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19605674_CXCL16 levels were increased in patients with systemic sclerosis, and correlated with the extent of skin sclerosis, suggesting that CXCL16 may have a role in the development of skin fibrosis in systemic sclerosis. 19674076_Our findings suggest a complex interaction between CXCL16 and HIV, promoting both inflammatory and anti-inflammatory effects as well as HIV replication, partly dependent on accompanying HIV replication. 19690611_data suggest that CXCL16 and CXCR6 may mark cancers arising in an inflammatory milieu and mediate pro-tumorigenic effects of inflammation through effects on cancer cell growth and by inducing the migration and proliferation of tumor-associated leukocytes 19729872_sSR-PSOX/CXCL16 is a biomarker for acute coronary syndrome, which would provide additional diagnostic information besides TnT and sLOX-1 19913121_Observational study of gene-disease association. (HuGE Navigator) 19919988_Report increased production of CXCL16 in clinical heart failure: a possible role in extracellular matrix remodeling. 19954776_Observational study of gene-disease association. (HuGE Navigator) 19954776_SNP rs3744700 of CXCL16 gene is independently associated with the development of CAD in Chinese Han population, and GG homozygote which is associated with increased expression of CXCL16 may have a promoting effect on CAD. 20334513_Overall, our data indicate that up-regulation of CXCL16 is a common response of tumor cells to radiation, and they have important implications for the use of local radiotherapy in combination with immunotherapy. 20503287_Observational study of gene-disease association. (HuGE Navigator) 20621591_Observational study of gene-disease association. (HuGE Navigator) 20621591_Serum CXCL16 concentration is significantly associated with atherosclerotic stroke and carotid atherosclerosis 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20646641_No CXCL16 protein was detected in clinical prostate cancer samples. CXCL16 could promote the in vitro migration and invasion of PC3 and LNCap cell lines. 20848509_CXCL16 mRNA and protein expression is up-regulated in intestinal inflammation in vitro and in Crohn's disease patients, suggesting an important role for this chemokine in intestinal inflammation. 20960183_The co-localization of SARS-CoVN and CXCL16 in the cytoplasm of HEK293FT cells was also shown using confocal laser scanning microscopy. 21233446_The CXCL16 A181V mutation selectively inhibits monocyte adhesion to CXCR6 but is not associated with human coronary heart disease. 21303517_CXCL16 and CXCR6 are elevated in Systemic sclerosis (SSc) serum and on SSc dermal Endothelial cells, respectively 21467783_Plasma level of CXCL16 is an independent predictor of the prognosis of the patients with intermediate coronary lesions. 21468586_High CXCL16/CXCR6 expression may be related to aggressive cancer behavior, and high CXCL16 expression to bone metastases. 21471570_SR-PSOX/CXCL16 plays a critical role in colonic inflammation and could be a potential therapeutic target for patients with inflammatory bowel diseases. 21492481_These findings suggest that mutagenesis of the basic amino acids in the chemokine domain of SR-PSOX may contribute to atherogenesis. 21505717_enhanced platelet expression in patients with acute coronary syndrome compared to patients with stable angina 21527066_The expressions of CXCL12/CXCR4 and CXCL16/CXCR6 were significantly higher in epithelial ovarian carcinomas than in normal epithelial ovarian tissues or benign epithelial ovarian tumors. 21612780_Suggest that CXCL16 may promote the adhesion of monocytes to the endothelium during early atherogenesis and that accumulating cytokines enhance CXCL16-mediated adhesion by upregulating CXCL16 expression. 21638128_is associated with Henoch-Schonlein purpura 21773780_CXCL16 and CXCR6 might be involved in the pathophysiology of endometriosis through regulation of the inflammatory response. 21845497_Soluble CXCL16 may play an important role in liver metastases through the induction of epithelial-mesenchymal transition. 22055341_An increase in plasma CXCL16 during the first days after the initial event is associated with an adverse outcome in patients with acute ischemic stroke. 22113484_The results indicated that CXCL16-CXCR6 interactions mediate homing of CD8+ T cells into human skin, and thereby contribute to psoriasis pathogenesis. 22278019_TWEAK upregulates the expression of the chemokine CXCL16 in tubular epithelium and this may contribute to kidney tubulointerstitial inflammation. 22378888_CXCL16 mediates cross talk between astrocytes and neighboring neurons and in pathological conditions counteracts neuronal cell death. 22454615_data suggest that expression levels of a surface receptor, namely, CXCL16, correlate with B cell responses mediated by TLR9 in common variable immunodeficiency 22627199_CXCL16 levels are significantly increased in gout patients with and without chronic kidney disease, and are independently associated with renal function. 22863086_The expression and serum concentration of CXC chemokine ligand 16 could indicate the aggressiveness and prognosis of gastric carcinomas. 23009930_study revealed some conformational motifs of basic amino acid residues, especially R76 with K79 in SR-PSOX, may form a common functional motif for its critical functions; R78 in SR-PSOX has the potential action to stabilize the function of oxLDL uptake and bacterial phagocytosis 23229614_CXCL16 was found to be abundantly expressed in human meningioma samples of different malignant grades. 23398954_Data suggest that plasma level of CXCL16 is up-regulated in subjects with metabolic syndrome and correlates with severity of carotid atherosclerosis (i.e., intima-media thickness and plaque index). 23428418_This data indicates that human P2X7 activation induces the rapid shedding of CXCL16 and that this process involves ADAM10. 23628207_CXCL16 is highly expressed by glial tumor and stroma cells whereas CXCR6 defines a subset of cells with stem cell character. 23633118_These results indicate that CXCL16 and its receptor CXCR6 may be a central ligand/receptor pair that is closely associated with EPC recruitment and blood vessel formation in the rheumatoid arthritis joint. 23743627_CXCL16, inversely correlated with CD99 expression in Hodgkin Reed-Sternberg (H/RS) cells. 24064021_Suggest that expression of transmembrane CXCL16 on surface of plasmacytoid dendritic cell might contribute to high serum IFN-alpha levels seen in patients with Behcet's disease. 24069377_CXCL16 could be a novel biomarker and potential predictor of disease activity in MS. 24284794_Erythrocyte adhesion to immobilized platelets requires phosphatidylserine at the erythrocyte surface and CXCL16 as well as CD36 expression on platelets. 24302814_CXCL16/CXCR6 interaction may play an important role in modifying the response of pDCs to environmental danger signals 24460887_Serum CXCL16 was increased in patients with active Primary nephrotic syndrome and correlated with blood lipids, urine protein and immune and inflammation responses 24489966_Serum CXCL16 may be an indicator of renal injury in subjects with type 2 diabetes mellitus. 24507753_Our results suggest that the CXCL16/CXCR6 axis appears to be important in the progression of Ewing sarcoma family tumor 24513807_CXCL16, iNKT cell-associated cell marker Valpha24, and CD1d were significantly upregulated in esophageal biopsies from EoE patients and correlated with the expression of inflammatory mediators associated with allergy. 24518602_High serum sCXCL16 is a prognostic marker for poor survival of OC patients, possibly reflecting ADAM-10 and ADAM-17 pro-metastatic activity. 24854635_No significant association of the CXCL16 polymorphism was established either with soluble CXCL16 plasma levels or with clinical parameters and course of multiple sclerosis. 24861945_High circulating CXCL16 levels are associated to human kidney and cardiovascular disease and urinary CXCL16 may increase in kidney injury. 24897301_CXCL16 and CXCR6 coexpression is associated with invasiveness of lung cancer. 25015061_Elevated serum sCXCL16 levels were discovered in the systemic lupus erythematosus patients with cutaneous and renal involvement. 25142184_CXCL16 T123V181 haplotype is a moderate genetic risk factor for the development of carotid plaque. 25223819_CXCL16 expression is enhanced in inflammatory cardiomyopathy and is an independent predictor of death in patients with heart failure. 25372401_injured hepatocytes up-regulated CXCL16 expression, indicating that scavenging functions of CXCL16 might be additionally involved in the pathogenesis of NAFLD. 25391425_Silencing CXCL16 could phenocopy the effects of miR-451 on phenotypes of osteosarcoma cells. 25661686_Serum CXCL16 is increased in severe pancreatitis with infected pancreatic necrosis and identifies patients who benefit from surgical necrosectomy 25904061_Endothelial CXCL16's action on platelets is not limited to platelet activation. Immobilised CXCL16 also acts as a potent novel platelet adhesion ligand, inducing platelet adhesion to the human vessel wall. 26021984_we examined the expression of CXCL16 and CXCR6 and their relations to prognosis in 335 unselected patients with NSCLC, and investigated possible relationships with our previously studied immunologic and angiogenic markers 26045830_PPARG rs1152002, AGTR1 rs5186, CXCL16 rs3744700 and LGALS2 rs7291467 polymorphisms may be closely related to the development of coronary heart disease 26058873_These results indicate that MEK inhibitor diminishes Nasopharyngeal carcinoma cell proliferation and NPC-induced osteoclastogenesis via modulating CCL2 and CXCL16 expressions. 26272362_high protein expression of CXCL16 and high protein co-expression of CXCL16/CXCR6 in prostate cancer were independent predictors for a worse clinical outcome 26345917_HMGB-1, CXCL16, miRNA-30a, and urinary TGF-ss1 were highly expressed in PNS patients and may play important roles in the pathogenesis and development of PNS. 26499307_CXCL16 is released into the circulation as a result of cardiac surgery and that high post-operative CXCL16 levels are associated with an increased severity of post-operative organ dysfunctions. 26621504_In conclusion, rheumatoid arthritis synovial fibroblasts were activated by CXCL16 to produce RANKL via pathways involving JAK2/STAT3 and p38/MAPK 26707275_Our data suggest that CXCL16 induces angiogenesis in autocrine manner via ERK, Akt, p38 pathways and HIF-1alpha modulation 26708384_Wnt5a-Ror2 signaling enhances expression and secretion of CXCL16 in mesenchymal stem cells thereby activating CXCR6 expressed on tumor cells to promote proliferation. 26796342_Cell proliferation enhancing and anti-apoptosis activity requires the intracellular domain and apparently the dimerization of the transmembrane chemokine ligand. 26799186_High CXCL16 expression is associated with prostate cancer. 27098626_Serum CXCL16 may be a novel biomarker for the diagnosis of patients with diabetic coronary artery disease. 27355560_Higher levels CXCL16 may be a biomarker for predicting stroke incidence and might contribute to plaque destabilization. 27665581_eGFR and serum albumin had an independent and significant negative correlation with plasma CXCL16 in diabetic kidney disease. 27725631_activation of cancer-associated fibroblasts and expression of CXCL16, shown to be a monocyte chemoattractant. 27784296_Transmembrane-CXCL16 specifically acts as a receptor for soluble-CXCL16 in human meningioma cells. 27826097_Our study demonstrated that CXCL16-CXCR6 mediates CD8(+) T-cell skin trafficking under oxidative stress in patients with vitiligo, and CXCL16 expression in human keratinocytes induced by ROS is, at least in part, caused by unfolded protein response activation 27869573_The expression of CXCL16 and and its receptor, CXCR6, their immunolocalization in disc tissue and their presence following exposure of cultured human annulus fibrosus cells to proinflammatory cytokines are reported. 27877078_Inflammation contributed to foam cell formation in the radial arteries of ESRD patients via activation of the CXCL16/CXCR6 pathway, which may be regulated by P2X7R. 28286356_CXCL16 single nucleotide polymorphisms significantly impacted myocardial infarction risk in a Chinese Han population. 28628472_IFN-gamma, CXCL16 and uPAR are promising as effective biomarkers of disease activity, renal damage, and the activity of pathological lesions in systemic lupus erythematosus. 28633141_Study showed, for the first time, highly significant relationships of circulating CXCL16 level with cardiac injury markers in dialysis patients. 28647282_CXCL16/CXCR6-mediated adhesion of human peripheral blood mononuclear cells to inflamed endothelium. 28698473_Data showed that reverse signaling via CXCL16 promotes migration in CXCL16-expressing melanoma and glioblastoma cells, but does not affect proliferation or protection from chemically-induced apoptosis. 28722105_Serum CXCL16 levels are significantly increased in patients with gallstone, and are independently associated with liver injury in Chinese population, suggesting that CXCL16 may be a biomarker of liver injury in subjects with gallstone or NAFLD. Hepatic CXCL16 mRNA and protein levels were also significantly increased in gallstone patients 28759013_Data suggest that primary cells from papillary renal cell carcinoma secrete the chemokines IL8, CXCL16, and chemerin; these chemokines attract primary human monocytes and induce shift/transdifferentiation in monocytes toward M2 macrophage/foam cell phenotype. (IL8 = interleukin-8; CXCL16 = C-X-C motif chemokine ligand 16) 28816285_This study shows that the expression of CXCL16 and its receptor CXCR6 was highly elevated in rheumatoid arthritis fibroblast-like synoviocytes 28856928_Women with gestational diabetes mellitus and preeclampsia had a dysregulated CXC chemokine ligand 16 during pregnancy, and in gestational diabetes mellitus, the increase in CXC chemokine ligand 16 early in pregnancy and after 5 years was strongly associated with their lipid profile. 28886489_increased plasma sCXCL16 might be implicated in the pathogenesis of immune thrombocytopenia and have a relationship with Th1/Th2 imbalance. 28942364_Study data link innate immune stimulation to CXCL16 up-regulation and neutrophil infiltration into skin. CXCL16 could therefore represent a potent future target for treatment of psoriasis. 29353287_Enhanced CXCL16 expression in lung cancer tissue promoted the proliferation and invasion of lung cancer cells. CXCL16 might promote proliferation and invasion of lung cancer by regulating the NF-kappaB pathway. 29588487_the data shown in the present study provide evidence to support the hypothesis that CXCL16 secreted by trophoblast cells is a key molecule involved in decidual M2 MPhi polarization, which in turn regulates the killing ability of NK cells, thereby contributing to the homeostatic and immune-tolerant milieu required for successful fetal development 29779473_Activation of the CXCL16/CXCR6 Axis by TNF-alpha Contributes to Ectopic Endometrial Stromal Cells Migration and Invasion.. These findings indicate that the CXCL16/CXCR6 axis may contribute to the progression of endometriosis and could be served as a potential target for diagnosis and treatment. 29800106_CXCL16/CXCR6 signaling axis plays a functional role in metabolic syndrome endothelial dysfunction and abdominal aortic aneurysm formation. 29909746_Study suggested that the CXCL16/CXCR6 axis may contribute to maintaining normal pregnancy by reducing the secretion of cytotoxic factor granzyme B of decidual gammadelta T cells and promoting the expression of antiapoptotic marker Bcl-xL of trophoblasts. 29981574_Plasma CXCL16 levels were elevated for 4 weeks after minimally invasive colorectal resection for cancer. Also, WF CXCL16 levels were 3-10 times greater than the corresponding plasma concentrations. 30049511_demonstrate that macrophages can promote the migration and invasion of ovarian carcinoma cells by affecting the CXCL16/CXCR6 pathway 30053055_CXCL16 was highly expressed in patients with nonalcoholic fatty liver disease (NAFLD), suggesting that it may contribute to steatotic and fibrotic progression. In vitro, CXCL16 treatment led to severe steatosis of hepatocytes in the hepatocyte-stellate cell coculture system. CXCL16 may be a potential noninvasive marker of NAFLD and a future potential therapeutic target to treat NAFLD. 30542347_CXCL16/CXCR6 signaling acts directly on mouse glioma cells. 30792527_Higher CXCL16 exodomain is associated with aggressive ovarian cancer and promotes the disease by CXCR6 activation and MMP modulation. 30974114_Auithors demonstrated that CXCR6-CXCL16 axis promotes DTX resistance and acts as a counter-defense mechanism. After CXCR6 activation, cell death in response to DTX was inhibited, and blocking of CXCR6 potentiated DTX cytotoxicity. 31033093_This study suggests that soluble CXCL16 (sCXCL16) enhances TNF-alpha-induced apoptosis of diffuse large B-cell lymphoma cells, which may involve a positive feedback loop consisting of TNF-alpha, ADAM10, sCXCL16 proteins, and members of the NF-kappaB pathway. 31199046_CXCL16/CXCR6 is involved in LPS-induced acute lung injury via P38 signalling. 31270884_In addition, knockdown of Notch1 markedly inhibited the expression of urokinase plasminogen activator (uPA) and its receptor uPAR, and chemokines C-C motif chemokine ligand 2 and C-X-C motif chemokine ligand 16, indicating that these factors are downstream targets of Notch1. 31379980_serum level elevated in pulmonary fibrosis 31527616_CXCL16 positively correlated with M2-macrophage infiltration, enhanced angiogenesis, and poor prognosis in thyroid cancer. 31598726_Urinary chemokine C-X-C motif ligand 16 and endostatin as predictors of tubulointerstitial fibrosis in patients with advanced diabetic kidney disease. 31752131_Results provide evidence that high expression level of CXCL16 mRNA in regional lymph nodes of colon cancer patients is a sign of a poor prognosis. 31753588_CXCL16/CXCR6 axis promotes bleomycin-induced fibrotic process in MRC-5 cells via the PI3K/AKT/FOXO3a pathway. 32012403_These results suggest that CXCL16 produced through Ror2-mediated signaling in mesenchymal stem or stromal cells (MSC) within the tumor microenvironment acts on MKN45 cells in a paracrine manner to activate the CXCR6-STAT3 pathway, which, in turn, induces expression of Ror1 in MKN45 cells, thereby promoting tumor progression 32163231_The clinical significance of CXCL16 in the treatment of advanced non-small cell lung cancer. 32171552_CXCL16 silencing alleviates hepatic ischemia reperfusion injury during liver transplantation by inhibiting p38 phosphorylation. 32346064_Entry and exit of chemotherapeutically-promoted cellular dormancy in glioblastoma cells is differentially affected by the chemokines CXCL12, CXCL16, and CX3CL1. 32365786_Differentially Expressed Genes of Natural Killer Cells Can Distinguish Rheumatoid Arthritis Patients from Healthy Controls. 32535333_The chemokine CXCL16 can rescue the defects in insulin signaling and sensitivity caused by palmitate in C2C12 myotubes. 32974919_Circ_0000524/miR-500a-5p/CXCL16 axis promotes podocyte apoptosis in membranous nephropathy. 33306334_Evaluation of serum CXC chemokine ligand 16 (CXCL16) as a novel inflammatory bio- marker or familial Mediterranean fever disease 33549109_Role of CXCL16 in BLM-induced epithelial-mesenchymal transition in human A549 cells. 33637127_C-X-C motif chemokine 16, modulated by microRNA-545, aggravates myocardial damage and affects the inflammatory responses in myocardial infarction. 33800554_The Role of CXCL16 in the Pathogenesis of Cancer and Other Diseases. 34275795_Application effect of tirofiban on percutaneous coronary intervention in patients with acute coronary syndrome and its postoperative effect on C-X-C motif chemokine ligand 16 level and myocardial perfusion. 34345211_CXCL16 Promotes Gastric Cancer Tumorigenesis via ADAM10-Dependent CXCL16/CXCR6 Axis and Activates Akt and MAPK Signaling Pathways. 34353074_Expression and significance of serum soluble fms-like tyrosine kinase 1 (sFlt-1), CXC chemokine ligand 16 (CXCL16), and lipocalin 2 (LCN-2) in pregnant women with preeclampsia. 34487815_Interleukin-18 accelerates cardiac inflammation and dysfunction during ischemia/reperfusion injury by transcriptional activation of CXCL16. 34882892_P2X7 receptor inhibition attenuates podocyte injury by oxLDL through deregulating CXCL16. 34943917_CXCL16/CXCR6 Axis in Adipocytes Differentiated from Human Adipose Derived Mesenchymal Stem Cells Regulates Macrophage Polarization. 35120907_Clinical significance and role of CXCL16 in anti-neutrophil cytoplasmic autoantibody-associated vasculitis. | ENSMUSG00000018920 | Cxcl16 | 2.254685e+02 | 1.3539661 | 0.437191565 | 0.3809808 | 1.317964e+00 | 0.2509578307 | 0.79219522 | No | Yes | 2.052991e+02 | 42.559607 | 1.169246e+02 | 25.316767 | ||
| ENSG00000161956 | 26168 | SENP3 | protein_coding | Q9H4L4 | FUNCTION: Protease that releases SUMO2 and SUMO3 monomers from sumoylated substrates, but has only weak activity against SUMO1 conjugates (PubMed:16608850, PubMed:32832608). Deconjugates SUMO2 from MEF2D, which increases its transcriptional activation capability (PubMed:15743823). Deconjugates SUMO2 and SUMO3 from CDCA8 (PubMed:18946085). Redox sensor that, when redistributed into nucleoplasm, can act as an effector to enhance HIF1A transcriptional activity by desumoylating EP300 (PubMed:19680224). Required for rRNA processing through deconjugation of SUMO2 and SUMO3 from nucleophosmin, NPM1 (PubMed:19015314). Plays a role in the regulation of sumoylation status of ZNF148 (PubMed:18259216). Functions as a component of the Five Friends of Methylated CHTOP (5FMC) complex; the 5FMC complex is recruited to ZNF148 by methylated CHTOP, leading to desumoylation of ZNF148 and subsequent transactivation of ZNF148 target genes (PubMed:22872859). Deconjugates SUMO2 from KAT5 (PubMed:32832608). {ECO:0000269|PubMed:15743823, ECO:0000269|PubMed:16608850, ECO:0000269|PubMed:18259216, ECO:0000269|PubMed:18946085, ECO:0000269|PubMed:19015314, ECO:0000269|PubMed:19680224, ECO:0000269|PubMed:22872859, ECO:0000269|PubMed:32832608}. | Cytoplasm;Hydrolase;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway | The reversible posttranslational modification of proteins by the addition of small ubiquitin-like SUMO proteins (see SUMO1; MIM 601912) is required for numerous biologic processes. SUMO-specific proteases, such as SENP3, are responsible for the initial processing of SUMO precursors to generate a C-terminal diglycine motif required for the conjugation reaction. They also have isopeptidase activity for the removal of SUMO from high molecular mass SUMO conjugates (Di Bacco et al., 2006 [PubMed 16738315]).[supplied by OMIM, Jun 2009]. | hsa:26168; | cytoplasm [GO:0005737]; MLL1 complex [GO:0071339]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; SUMO-specific isopeptidase activity [GO:0070140]; protein desumoylation [GO:0016926] | 18259216_These results define SENP3 as an essential factor for ribosome biogenesis and suggest that deconjugation of SUMO2 from NPM1 by SENP3 is critically involved in 28S rRNA maturation. 18639523_SMT3IP1 desumoylates nucleophosmin and might controls its physiological functions at both the nucleolus and other subcellular compartments. 18946085_Data describe a mitotic SUMO2/3 conjugation-deconjugation cycle of Borealin and further assign a regulatory function of RanBP2 and SENP3 in the mitotic SUMO pathway. 19015314_Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19680224_SENP3 is a redox sensor that regulates HIF-1 transcriptional activity under oxidative stress through the de-SUMOylation of p300 19773279_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20181954_SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. 20337593_Results demonstrate that SENP1 is the most efficient SUMO protease acting on Elk-1, and that SENP3 has little effect on Elk-1. SENP2 has an intermediate effect, but its ability to activate Elk-1 is independent from its SUMO-deconjugating activity. 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20924358_Hsp90/SENP3 association protects SENP3 from CHIP-mediated ubiquitination and subsequent degradation, but this effect of Hsp90 requires the presence of CHIP. 21316347_These results suggest that SMT3IP1 is a new regulator of the p53-Mdm2 pathway. 22684029_The shift of HIF-1 transactivation by reactive oxidative species is correlated with and dependent on the biphasic redox sensing of SENP3 that leads to the differential SENP3/p300 interaction and the consequent fluctuation in the p300 SUMOylation status. 22872859_Recruitment of the protein complex 5FMC to Zbp-89, a zinc-finger transcription factor, affects its sumoylation status and transactivation potential.[5FMC] 23467634_ROS induced SENP3 redistribution from the nucleoli to the nucleoplasm. 23524851_In a novel adaptive pathway to extreme cell stress, dynamic changes in SENP3 stability and regulation of Drp1 SUMOylation are crucial determinants of cell fate. 24930734_SENP3 is associated with MLL1/MLL2 complexes and catalyzes deSUMOylation of RbBP5. 25216525_SENP3, which is increased in gastric cancer cells, potentiates the transcriptional activity of FOXC2 through de-SUMOylation, in favor of the induction of specific mesenchymal gene expression in gastric cancer metastasis. 25288641_Evidence show that mTOR-mediated phosphorylation of SENP3 facilitates the interaction with NPM1, thereby promoting nucleolar targeting. 26511642_High SENP3 expression is associated with stomach neoplasms. 27016777_In early-onset pre-eclampsia, enhanced deSUMOylation of HIF1A by SENP3 may in part contribute to increased HIF1A activity and stability found in this pathology. 27181202_Our data identified SUMOylation as a previously undescribed post-translational modification of STAT3 and SENP3 as a critical positive modulator of tobacco- or cytokine-induced STAT3 activation. 27814492_The data demonstrate that SUMO2 conjugation and SENP3-driven deSUMOylation of PELP1 is instrumental for ordered progression of ribosome maturation, and they provide molecular insight into the dynamics of ribosome maturation. 27853276_Important role of SENP3 in lipid metabolism during the development of non-alcoholic fatty liver disease. 28262828_Data show that increasing dynamin-related protein 1 (Drp1) SUMOylation by knocking down SUMO1-sentrin-SMT3 specific protease 3 (SENP3) reduces both Drp1 binding to mitochondrial fission factor protein (Mff) and stress-induced cytochrome c release. 28351334_our findings for the first time specifically supported that SUMO-specific protease 3 might play an important role in the regulation of epithelial ovarian cancer progression and could serve as a potential biomarker for prognosis as well as provide a promising therapeutic target against epithelial ovarian cancer 28747609_SENP3 knockdown reduced cadmium-induced caspase 3 cleavage and cell death in PC12 cells, while SENP3 overexpression enhanced cell death. 29438989_SENP3 phosphorylation decreased its interaction with Topo II alpha, resulting in reduced SENP3 deSUMOylation activity on Topo II alpha. Furthermore, we observed mitotic arrest, increased chromosome instability, and promotion of tumorigenesis in cells expressing a nonphosphorylatable SENP3 mutant 29576508_we provide the first direct evidence that SENP3 upregulation pivotally contributes to myocardial ischemia reperfusion (MIR) injury in a Drp1-dependent manner, and suggest that SENP3 suppression may hold therapeutic promise for constraining MIR injury. 30640896_SENP3 expression level decreased in HBV-infected hepatocytes in various models including HepG2-NTCP cell lines and a humanized mouse model. Downregulation of SENP3 reduced HBV replication and boosted host protein translation. 30973885_we performed ischemia and ischemia-reperfusion experiments in the cardiomyocyte H9C2 cell line. Similar to whole hearts, ischemia induced a decrease in cytosolic SENP3. Furthermore, shRNA-mediated knockdown of SENP3 led to an increase in the rate of cell death upon reperfusion. 31914638_SUMO1-catalyzed SUMOylation and SENP3-mediated deSUMOylation of NLRP3 orchestrate the inflammasome activation. 32049023_SENP3 Suppresses Osteoclastogenesis by De-conjugating SUMO2/3 from IRF8 in Bone Marrow-Derived Monocytes. 33030103_High SENP3 Expression Promotes Cell Migration, Invasion, and Proliferation by Modulating DNA Methylation of E-Cadherin in Osteosarcoma. 33558447_Combined identification of ARID1A, CSMD1, and SENP3 as effective prognostic biomarkers for hepatocellular carcinoma. 34313310_SUMO proteases SENP3 and SENP5 spatiotemporally regulate the kinase activity of Aurora A. | ENSMUSG00000005204 | Senp3 | 8.322746e+03 | 1.1894464 | 0.250290248 | 0.3017124 | 6.913629e-01 | 0.4057011876 | 0.84651196 | No | Yes | 8.851509e+03 | 1016.001810 | 6.561395e+03 | 772.877082 | |
| ENSG00000162062 | 80178 | TEDC2 | protein_coding | Q7L2K0 | FUNCTION: Acts as a positive regulator of ciliary hedgehog signaling. Required for centriole stability. {ECO:0000250|UniProtKB:Q6GQV0}. | Alternative splicing;Cell projection;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | hsa:80178; | centriole [GO:0005814]; cilium [GO:0005929]; cytoplasm [GO:0005737]; positive regulation of smoothened signaling pathway [GO:0045880] | ENSMUSG00000024118 | Tedc2 | 2.381311e+03 | 1.1666676 | 0.222393576 | 0.3054943 | 5.312535e-01 | 0.4660803535 | 0.86742514 | No | Yes | 2.255432e+03 | 267.977776 | 1.746849e+03 | 213.267232 | |||
| ENSG00000162065 | 57465 | TBC1D24 | protein_coding | Q9ULP9 | FUNCTION: May act as a GTPase-activating protein for Rab family protein(s) (PubMed:20727515, PubMed:20797691). Involved in neuronal projections development, probably through a negative modulation of ARF6 function (PubMed:20727515). Involved in the regulation of synaptic vesicle trafficking (PubMed:31257402). {ECO:0000269|PubMed:20727515, ECO:0000269|PubMed:20797691, ECO:0000269|PubMed:31257402}. | Alternative splicing;Cell junction;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Deafness;Disease variant;Epilepsy;GTPase activation;Membrane;Mental retardation;Non-syndromic deafness;Phosphoprotein;Reference proteome;Synapse | This gene encodes a protein with a conserved domain, referred to as the TBC domain, characteristic of proteins which interact with GTPases. TBC domain proteins may serve as GTPase-activating proteins for a particular group of GTPases, the Rab (Ras-related proteins in brain) small GTPases which are involved in the regulation of membrane trafficking. Mutations in this gene are associated with familial infantile myoclonic epilepsy. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2011]. | hsa:57465; | cell junction [GO:0030054]; cytoplasm [GO:0005737]; cytoplasmic vesicle membrane [GO:0030659]; neuromuscular junction [GO:0031594]; plasma membrane [GO:0005886]; terminal bouton [GO:0043195]; GTPase activator activity [GO:0005096]; neuron death in response to oxidative stress [GO:0036475]; neuron projection development [GO:0031175] | 20727515_Two compound heterozygous missense mutations (D147H and A509V) in TBC1D24, a gene of unknown function, are responsible for familial infantile myoclonic epilepsy. 20797691_A pathogenic mutation was identified in TBC1D24. 23343562_Findings expand the spectrum of the TBC1D24 mutation phenotype and the transcript isoforms. 23517570_A TBC1D24 mutation associated with focal epilepsy, cognitive impairment and cerebro-cerebellar malformation is found in a family with a homozygous TBC1D24 mutation. 23526554_we describe a familial form of MMPSI due to mutation in TBC1D24, revealing a devastating epileptic phenotype associated with TBC1D24 dysfunction. 24291220_Mutations in TBC1D24 seem to be an important cause of DOORS syndrome and can cause diverse phenotypes. 24315024_Novel variations in TBC1D24 do not allow prediction of functional phenotypes that might explain, at least in part, the symptoms of malignant migrating partial seizures of infancy (MMPSI). 24387994_Recessive alleles of TBC1D24 can cause either epilepsy or nonsyndromic deafness in human. 24729539_TBC1D24 mutation causes autosomal-dominant nonsyndromic hearing loss. 24729547_that the p.Ser178Leu mutation of TBC1D24 is a probable cause for dominant, nonsyndromic hearing impairment. Identification of TBC1D24 as the stereocilia-expressing gene may shed new light on its specific function in the inner ear. 25557349_This report supports previous observations that mutations in TBC1D24 cause diverse phenotypes 26371875_mutations in TBC1D24 gene are a frequent cause (>2%) of NSHL in Morocco 27281533_TBC1D24-related epilepsy syndromes show marked phenotypic pleiotropy, with multisystem involvement and severity spectrum ranging from isolated deafness (not studied here), benign myoclonic epilepsy restricted to childhood with complete seizure control and normal intellect, to early-onset epileptic encephalopathy with severe developmental delay and early death 27502353_TBC1D24-related epilepsy can manifest with hypotonia, developmental delays, and a variety of focal-onset seizures. 27541164_Here, we present a familial case of a lethal early-onset epileptic encephalopathy, associated with two novel compound heterozygous missense variants on the TBC1D24 gene, which were detected by exome sequencing 29176366_We identified a homozygous single base alteration, c.1415 G>A;p.G428R, in TBC1D24 gene. This mutation was found in the proband's parents and elder sister as heterozygous. The c.1415G>A mutation has not been reported previously. The c.1415G>A was considered to be damaging by SIFT software 29893377_Silencing TBC1D24 inhibited MCF-7 cells growth in vitro and in vivo. TBC1D24 promoted breast carcinoma growth through the IGF1R/PI3K/AKT pathway. 30154457_TBC1d24-ephrinB2 interaction regulates contact inhibition of locomotion in neural crest cell migration 30180405_he clinical feature of TBC1D24 gene mutation related epilepsy was focal myoclonus, and tended to develop into myoclonic status epilepticus, and could be aggravated by infections, and terminated by sleep or sedation drugs. 30245510_eight individuals with epilepsy and developmental delay who share overlapping microdeletions at 16p13.3 including TBC1D24, ATP6V0C, and PDPK1 30335140_We then further investigated TBC1D24 haploinsufficiency in vivo and demonstrate that TBC1D24 is also crucial for normal presynaptic function: genetic disruption of Tbc1d24 expression in the mouse leads to an impairment of endocytosis and an enlarged endosomal compartment in neurons with a decrease in spontaneous neurotransmission. 30858606_TBC1D24 regulates axonal outgrowth and membrane trafficking at the growth cone in rodent and human neurons. 31112829_Multifocal myoclonus, epilepsia partialis continua (EPC), and fever-induced seizures were the most prominent features of epilepsy patients with TBC1D24 mutations. The best therapeutic strategy to terminate EPC might be using chloral hydrate to induce sleep and control the infection and temperature. Hearing loss and abnormal brain MRIs might exacerbate the condition during follow-up. 31257402_In a Drosophila model neuronally expressing human TBC1D24, the TBC1D24G501R TLDc mutation causes activity-induced locomotion and synaptic vesicle trafficking defects, but TBC1D24R360H is benign. The phenotypes of the TBC1D24G501R mutation are consistent with oxidative stress sensitivity, rescued by antioxidants. TLDc domain mutations of TBC1D24 cause Rolandic-type focal motor epilepsy and exercise-induced dystonia. 32475639_TBC1D24 regulates recycling of clathrin-independent cargo proteins mediated by tubular recycling endosomes. 32663648_Disrupted oxidative stress resistance: A homozygous mutation in the catalytic (TLDc) domain of TBC1D24 gene associated with epileptic encephalopathy. 32987832_Mouse Models of Human Pathogenic Variants of TBC1D24 Associated with Non-Syndromic Deafness DFNB86 and DFNA65 and Syndromes Involving Deafness. 33063868_Novel variants in TBC1D24 associated with epilepsy and deafness: Report of two cases. 33986365_TBC1D24 emerges as an important contributor to progressive postlingual dominant hearing loss. | ENSMUSG00000036473 | Tbc1d24 | 1.708589e+03 | 1.3881149 | 0.473126966 | 0.2703500 | 3.101681e+00 | 0.0782114804 | 0.72639154 | No | Yes | 1.728238e+03 | 153.291596 | 1.206069e+03 | 110.048055 | |
| ENSG00000162599 | 4774 | NFIA | protein_coding | Q12857 | FUNCTION: Recognizes and binds the palindromic sequence 5'-TTGGCNNNNNGCCAA-3' present in viral and cellular promoters and in the origin of replication of adenovirus type 2. These proteins are individually capable of activating transcription and replication. | Activator;Alternative splicing;DNA replication;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene encodes a member of the NF1 (nuclear factor 1) family of transcription factors. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2011]. | hsa:4774; | cell junction [GO:0030054]; chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; DNA replication [GO:0006260]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; synapse maturation [GO:0060074]; ureter development [GO:0072189]; viral genome replication [GO:0019079] | 10518556_Disruption of the Nfia gene in the mouse causes perinatal lethality, agenesis of the corpus callosum and hydrocephalus. 15458926_Role in the expression mechanism of hNaPi-IIb gene transcription. 15466411_nuclear factor I has a role in the intrinsic control of cerebellar granule neuron gene expression 15598822_Distribution of nuclear factor I binding sites correlate with Z-DNA forming regions in human chromosome 22. 16325577_Data show that human granulocytic differentiation is controlled by a regulatory circuitry involving miR-223 and two transcriptional factors, NFI-A and C/EBPalpha. 17010934_The NF1-A transcription factor plays an important role in the transcriptional activation of the TR2 orphan receptor gene expression via a promoter activating cis-element. 17530927_The mouse Nfia mutant phenotype and the common features among five human cases indicate that NFIA haploinsufficiency contributes to a novel human central nervous system malformation syndrome that can also include ureteral and renal defects. 19540848_NFI family of transcription factors plays a key role in the regulation of both the B-FABP and GFAP genes in malignant glioma cells. 19542302_in early hematopoiesis, the NFI-A expression level acts as a novel factor channeling HPCs into either the E or G lineage 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22807310_NFIA expression in white matter lesions of human newborns with neonatal HIE, as well active MS lesions in adults, revealed that it is similarly expressed in oligodendrocyte progenitors and not oligodendrocytes. 22925353_These data suggest that genetic variants in the NF1A gene region may predispose to seasonal patterned of mania in bipolar disorder. 23946414_These studies represent the first characterization of miR-223/NFIA axis function in glioma 24305710_High nuclear factor IA expression is associated with glioblastomas. 24462883_This report presents the first case of an intragenic deletion within the NFIA gene that is still consistent with classic clinical phenotypes present in previously reported cases of chromosome 1p31.3 related deletion. 24560411_a strong candidate gene for asthma and allergic rhinitis 25220407_TGF-beta-mediated suppression of ANT2 through NF1/Smad4 complexes contributes to oxidative stress and DNA damage during induction of cellular senescence. 25265644_RP5-833A20.1/miR-382-5p/NFIA pathway was essential to the regulation of cholesterol homeostasis and inflammatory reactions. 25544502_NFI-A is involved in the miR-21-induced expression of IL-10 in B cells in nasopharyngeal carcinoma; Il-10 is capable of suppressing CD8+ T-cell activities. 25714559_this family also carried a microdeletion affecting solely the NFIA gene, this study substantiates the importance of this gene in craniofacial development. 26329426_microRNA-136 targeted and degraded NFIA, which induced the release of microRNA-223, promoting CD11b expression. Direct base pairing occurs between miR-136 and the 3' UTR of NFIA mRNA. 27267730_Dihydrocapsaicin can significantly decrease proinflammatory cytokines through enhancing NFIA and inhibiting NF-kappaB expression 27664977_Altogether, these results demonstrated that miR-370 suppressed hepatitis B virus gene expression and replication through repressing NFIA expression, which stimulates hepatitis B virus replication via direct regulation on hepatitis B virus Enhancer I activities. 27779670_These results demonstrated that RP5833A20.1 inhibited tumor cell proliferation, induced apoptosis and inhibited cellcycle progression by suppressing the expression of NFIA in U251 cells. 27994064_Data define a previously unknown nuclear factor I-A-nuclear factor-kappaB feed-forward regulation that may contribute to glioblastoma cell survival. 28075452_miR-191 was upregulated in patients with middle- and late-stage NSCLC, and in NSCLC cell lines, under mild hypoxic conditions. miR-191 promoted the proliferation and migration of NSCLC under chronic hypoxic conditions, and this promotion may be associated with its targeting of NFIA. 28076901_Studies indicate the role of nuclear factor one (NFIs) as epigenetic regulators in cancer. 28323865_We verified that NFIA binds to the IGFBP2 promoter and transcriptionally enhances IGFBP2 expression levels. We identified that NFIA-mediated IGFBP2 signaling pathways are involved in miR-302b-induced glioma cell death. 28941020_The consistent overlap in clinical presentation provides further evidence of the critical role of NFIA haploinsufficiency in the development of the 1p32-p31 microdeletion syndrome phenotype. 29577671_High NFIA expression in tumors of patients with esophageal squamous cell carcinoma (ESCC) is correlated with lymph node metastasis and poor differentiation. NFIA is found to be an independent predictor of poor prognosis in patients with ESCC. 30590051_This study reveals NFIA as a pro-endocrine factor in the pancreas, acting to repress Mib1, inhibit Dll1 endocytosis and thus promote escape from Notch activation. 30804533_Transient expression of NFIA is sufficient to trigger glial competency of human pluripotent stem cell-derived neural stem cells within 5 days and to convert these cells into astrocytes in the presence of glial-promoting factors, as compared to 3-6 months using current protocols. 31178144_NFIA enhances cell radiosensitivity by downregulating p-AKT and p-ERK in non-small cell lung cancer 31498149_NFIA is highly expressed in reactive astrocytes in neurological injury and we identify unique roles across distinct injury states and regions of the CNS. 31502558_our results indicated that miR-212-3p targeting NFIA might serve as a promising target for bladder cancer (BC) 31614149_High Nuclear factor I A expresion is associated with temozolomide resistance in glioblastoma via activation of nuclear factor kappaB pathway. 31760595_Low expression of NFIA is associated with high-grade glioblastoma. 33640202_Genome-wide association study identifies susceptibility loci of brain atrophy to NFIA and ST18 in Alzheimer's disease. 33932136_The miR-223/nuclear factor I-A axis regulates inflammation and cellular functions in intestinal tissues with necrotizing enterocolitis. 33973697_Recurrent NFIA K125E substitution represents a loss-of-function allele: Sensitive in vitro and in vivo assays for nontruncating alleles. 34969759_Nuclear Factor IA Is Down-regulated in Muscle-invasive and High-grade Bladder Cancers. | ENSMUSG00000028565 | Nfia | 5.758907e+02 | 0.7834375 | -0.352109844 | 0.2785122 | 1.547558e+00 | 0.2134963124 | 0.78763590 | No | Yes | 4.163779e+02 | 68.215662 | 4.742148e+02 | 79.602201 | |
| ENSG00000162729 | 93185 | IGSF8 | protein_coding | Q969P0 | FUNCTION: May play a key role in diverse functions ascribed to CD81 and CD9 such as oocytes fertilization or hepatitis C virus function. May regulate proliferation and differentiation of keratinocytes. May be a negative regulator of cell motility: suppresses T-cell mobility coordinately with CD81, associates with CD82 to suppress prostate cancer cell migration, regulates epidermoid cell reaggregation and motility on laminin-5 with CD9 and CD81 as key linkers. May also play a role on integrin-dependent morphology and motility functions. May participate in the regulation of neurite outgrowth and maintenance of the neural network in the adult brain. {ECO:0000269|PubMed:11504738, ECO:0000269|PubMed:12750295, ECO:0000269|PubMed:12752121, ECO:0000269|PubMed:14662754, ECO:0000269|PubMed:15070678}. | Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunoglobulin domain;Lipoprotein;Membrane;Palmitate;Phosphoprotein;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a member the EWI subfamily of the immunoglobulin protein superfamily. Members of this family contain a single transmembrane domain, an EWI (Glu-Trp-Ile)-motif and a variable number of immunoglobulin domains. This protein interacts with the tetraspanins CD81 and CD9 and may regulate their role in certain cellular functions including cell migration and viral infection. The encoded protein may also function as a tumor suppressor by inhibiting the proliferation of certain cancers. Alternate splicing results in multiple transcript variants that encode the same protein. [provided by RefSeq, Sep 2011]. | hsa:93185; | extracellular exosome [GO:0070062]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; plasma membrane [GO:0005886]; cell motility [GO:0048870]; nervous system development [GO:0007399]; single fertilization [GO:0007338]; skeletal muscle tissue development [GO:0007519] | 12750295_EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. 15070678_EWI-2-dependent reorganization of alpha4beta1-CD81 complexes on the cell surface is responsible for EWI-2 effects on integrin-dependent morphology and motility functions 16537545_EWI proteins EWI-2 and EWI-F, alpha3beta1 and alpha6beta4 integrins, and protein palmitoylation have contrasting effects on cell surface CD9 organization 16690612_EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins 17785435_Important functions of recently activated dendritic cells are thus critically modulated by the newly discovered HSPA8-EWI-2 interaction. 18382656_CD81 partner EWI-2wint inhibits hepatitis C virus entry [EWI-2wint] 19107234_EWI-2 causes a substantial molecular reorganization of multiple molecules known to affect proliferation and/or invasion of astrocytes and/or glioblastomas. 21343309_analysis of interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 22689882_The EWI-2/alpha-actinin complex is involved in regulation of the actin cytoskeleton at T cell immune and virological synapses, providing a link between membrane microdomains and the formation of polarized membrane structures involved in T cell recognition. 23351194_Authors demonstrated that EWI-2wint promotes CD81 clustering and confinement in CD81-enriched areas. 25656846_EWI-2 negatively regulates TGF-beta signaling and its downstream events including cytostasis (in vitro and in vivo), EMT-like changes, cell migration, CD271-dependent invasion, and lung metastasis (in vivo). 31757023_EWI-2 Inhibits Cell-Cell Fusion at the HIV-1 Virological Presynapse. 33605506_EWI-2 controls nucleocytoplasmic shuttling of EGFR signaling molecules and miRNA sorting in exosomes to inhibit prostate cancer cell metastasis. | ENSMUSG00000038034 | Igsf8 | 2.581732e+03 | 1.0579821 | 0.081315228 | 0.3260225 | 6.141165e-02 | 0.8042785243 | 0.96141221 | No | Yes | 2.316845e+03 | 296.576608 | 2.123157e+03 | 278.962532 | |
| ENSG00000162735 | 5824 | PEX19 | protein_coding | P40855 | FUNCTION: Necessary for early peroxisomal biogenesis. Acts both as a cytosolic chaperone and as an import receptor for peroxisomal membrane proteins (PMPs). Binds and stabilizes newly synthesized PMPs in the cytoplasm by interacting with their hydrophobic membrane-spanning domains, and targets them to the peroxisome membrane by binding to the integral membrane protein PEX3. Excludes CDKN2A from the nucleus and prevents its interaction with MDM2, which results in active degradation of TP53. {ECO:0000269|PubMed:10051604, ECO:0000269|PubMed:10704444, ECO:0000269|PubMed:11259404, ECO:0000269|PubMed:11883941, ECO:0000269|PubMed:14709540, ECO:0000269|PubMed:15007061}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Lipoprotein;Membrane;Methylation;Peroxisome;Peroxisome biogenesis;Peroxisome biogenesis disorder;Phosphoprotein;Prenylation;Reference proteome;Zellweger syndrome | This gene is necessary for early peroxisomal biogenesis. It acts both as a cytosolic chaperone and as an import receptor for peroxisomal membrane proteins (PMPs). Peroxins (PEXs) are proteins that are essential for the assembly of functional peroxisomes. The peroxisome biogenesis disorders (PBDs) are a group of genetically heterogeneous autosomal recessive, lethal diseases characterized by multiple defects in peroxisome function. These disorders have at least 14 complementation groups, with more than one phenotype being observed for some complementation groups. Although the clinical features of PBD patients vary, cells from all PBD patients exhibit a defect in the import of one or more classes of peroxisomal matrix proteins into the organelle. Defects in this gene are a cause of Zellweger syndrome (ZWS), as well as peroxisome biogenesis disorder complementation group 14 (PBD-CG14), which is also known as PBD-CGJ. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2010]. | hsa:5824; | brush border membrane [GO:0031526]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; integral component of membrane [GO:0016021]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; protein-containing complex [GO:0032991]; ATPase binding [GO:0051117]; peroxisome membrane class-1 targeting sequence binding [GO:0036105]; peroxisome membrane targeting sequence binding [GO:0033328]; protein carrier chaperone [GO:0140597]; protein N-terminus binding [GO:0047485]; chaperone-mediated protein folding [GO:0061077]; establishment of protein localization to peroxisome [GO:0072663]; negative regulation of lipid binding [GO:1900131]; peroxisome fission [GO:0016559]; peroxisome membrane biogenesis [GO:0016557]; peroxisome organization [GO:0007031]; protein import into peroxisome membrane [GO:0045046]; protein stabilization [GO:0050821]; protein targeting to peroxisome [GO:0006625] | 10777694_LDRP (ABCD2) interacts with both farnesylated wild-type and farnesylation-deficient mutant PEX19. This interaction is mediated by amino acids 1-218 of ALDRP. 10777694_MP70 (ABCD3) interacts with both farnesylated wild-type and farnesylation-deficient mutant PEX19. 11883941_ALDRP interacts with PEX19 splice variants PEX19-delta-E2 and PEX19-delta-E8. 11883941_MP70 interacts with PEX19 splice variants PEX19-delta-E2 and PEX19p-delta-E8. 11883941_a considerable functional diversity of the proteins encoded by two PEX19 splice variants and thereby provide first experimental evidence for specific biological functions of the different predicted domains of the PEX19 protein. 14709540_PEX19 binds and stabilizes newly synthesized PMPs in the cytosol, binds to multiple PMP targeting signals (mPTSs), interacts with the hydrophobic domains of PMP targeting signals, and is essential for PMP targeting and import. 14713233_Interaction of PEX3 and PEX19 visualized by fluorescence resonance energy transfer (FRET). 14715663_Pex19p has a role in assembly of PTS-receptor docking complexes 15007061_Results suggest that PEX3 plays a selective, essential, and direct role in class I peroxisomal membrane protein import as a docking factor for PEX19. 15252024_human Pex19p domain architecture and activity 15781447_analysis of the PEX19-binding site of human adrenoleukodystrophy protein 16280322_Pex19p translocates the membrane peroxins from the cytosol to peroxisomes in an ATP- and Pex3p-dependent manner and then shuttles back to the cytosol 16344115_Pex19p binds to PMP70 co-translationally and keeps PMP70 in a proper conformation for the localization to peroxisome. 16791427_Nonfarnesylated and farnesylated human Pex19p display a similar affinity towards a select set of peroxisomal membrane proteins. 16895967_Data suggest that Pex19p probably functions as a chaperone for membrane proteins and transports them to peroxisomes by anchoring to Pex3p using residues 12-73 and 40-131. 18174172_either one or two tryptophan residues of Pex3p (Trp-104 and Trp-224) are directly involved in binding to Pex19p. 18782765_targeting of hFis1 to peroxisomes and mitochondria are independent events and support a direct, Pex19p-dependent targeting of peroxisomal tail-anchored proteins. 19197237_N-terminal domain of Pex14, Pex14(N), adopts a three-helical fold. Pex5 and Pex19 ligand helices bind competitively to the same surface in Pex14(N) albeit with opposite directionality. 20531392_data indicate a divided N-terminal and C-terminal structural arrangement in Pex19p, which is reminiscent of a similar division in the Pex5p receptor, to allow separation of cargo-targeting signal recognition and additional functions. 20554521_The crystal structure of the cytosolic domain of PEX3 in complex with a PEX19-derived peptide. PEX3 adopts a novel fold that is best described as a large helical bundle. 21102411_The Pex19p peptide contains a characteristic motif, consisting of the leucine triad (Leu18, Leu21, Leu22), and Phe29, which are critical for the Pex3p binding and peroxisome biogenesis. 22624858_PEX3-PEX19 interaction is crucial for de novo formation of peroxisomes in peroxisome-deficient cells. 23460677_PEX19 formed a complex with the peroxisomal tail anchored protein PEX26 in the cytosol and translocated it directly to peroxisomes by a TRC40-independent class I pathway. 25062251_Thus within the cell, PEX3 is stabilized by PEX19 preventing PEX3 aggregation. 26018079_suggest a novel regulatory mechanism for peroxisome biogenesis through the interaction between Pex19p and PLA/AT-3 27295553_that newly synthesized UBXD8 is post-translationally inserted into discrete ER subdomains by a mechanism requiring cytosolic PEX19 and membrane-integrated PEX3, proteins hitherto exclusively implicated in peroxisome biogenesis 28281558_The results demonstrate an allosteric mechanism for the modulation of PEX19 function by farnesylation. 29282281_Hnf4-induced lipotoxicity and accumulation of free fatty acids is the cause for mitochondrial damage in consequence of peroxisome loss in Pex19 mutants 29396426_Thus, PEX19 and PEX3 peroxisome biogenesis factors provide an alternative posttranslational route for membrane insertion of the reticulon homology domain-containing proteins, implying that endoplasmic reticulum membrane shaping and peroxisome biogenesis may be coordinated. 29500918_The summarize recent insights into the biogenesis and function of Lipid droplets and peroxisomes with emphasis on the role of PEX19 in these processes. 30776093_studies indicate that Pex3 and Pex19 can also facilitate sorting of certain membrane proteins to other cellular organelles, including the endoplasmic reticulum, lipid droplets, and mitochondria 34108265_Viperin interacts with PEX19 to mediate peroxisomal augmentation of the innate antiviral response. 35215846_Dengue and Zika Virus Capsid Proteins Contain a Common PEX19-Binding Motif. | ENSMUSG00000003464 | Pex19 | 3.418070e+03 | 0.6862699 | -0.543151954 | 0.3391126 | 2.626948e+00 | 0.1050634891 | 0.75783482 | No | Yes | 2.181387e+03 | 366.417730 | 3.394212e+03 | 584.181852 | |
| ENSG00000162923 | 80232 | WDR26 | protein_coding | Q9H7D7 | FUNCTION: G-beta-like protein involved in cell signal transduction (PubMed:15378603, PubMed:19446606, PubMed:22065575, PubMed:23625927, PubMed:27098453, PubMed:26895380). Acts as a negative regulator in MAPK signaling pathway (PubMed:15378603). Functions as a scaffolding protein to promote G beta:gamma-mediated PLCB2 plasma membrane translocation and subsequent activation in leukocytes (PubMed:22065575, PubMed:23625927). Core component of the CTLH E3 ubiquitin-protein ligase complex that selectively accepts ubiquitin from UBE2H and mediates ubiquitination and subsequent proteasomal degradation of the transcription factor HBP1 (PubMed:29911972). Acts as a negative regulator of the canonical Wnt signaling pathway through preventing ubiquitination of beta-catenin CTNNB1 by the beta-catenin destruction complex, thus negatively regulating CTNNB1 degradation (PubMed:27098453). Serves as a scaffold to coordinate PI3K/AKT pathway-driven cell growth and migration (PubMed:26895380). Protects cells from oxidative stress-induced apoptosis via the down-regulation of AP-1 transcriptional activity as well as by inhibiting cytochrome c release from mitochondria (PubMed:19446606). Protects also cells by promoting hypoxia-mediated autophagy and mitophagy (By similarity). {ECO:0000250|UniProtKB:F1LTR1, ECO:0000269|PubMed:15378603, ECO:0000269|PubMed:19446606, ECO:0000269|PubMed:23625927, ECO:0000269|PubMed:26895380, ECO:0000269|PubMed:27098453, ECO:0000269|PubMed:29911972}. | Alternative splicing;Cytoplasm;Disease variant;Mental retardation;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Repeat;WD repeat | This gene encodes a member of the WD repeat protein family. WD repeats are minimally conserved regions of approximately 40 amino acids typically bracketed by gly-his and trp-asp (GH-WD), which may facilitate formation of heterotrimeric or multiprotein complexes. Members of this family are involved in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation. Two transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:80232; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; GID complex [GO:0034657]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161] | 15378603_WDR26 may act as a negative regulator in MAPK signaling pathway and play an important role in cell signal transduction. 19446606_The results of this study indicated that WDR26 was up-regulated by oxidative stress and played a key role in H2O2-induced SH-SY5Y cell death, which may be mediated by the down-regulation of AP-1 transcriptional activity. 20171191_these data suggest that MIP2 may participate in the progression of cell proliferation in H9c2 cells. 22065575_WDR26 is a novel Gbetagamma-binding protein that is required for the efficacy of Gbetagamma signaling and leukocyte migration 23625927_WDR26 functions as a scaffolding protein to promote PLCbeta2 membrane translocation and interaction with Gbetagamma, thereby enhancing PLCbeta2 activation in leukocytes. 25918994_The WDR26 gene was differentially methylated in monozygotic twins discordant for depressive disorder. 26895380_WDR26 serves as a scaffold that fosters assembly of a specific signaling complex consisting of Gbetagamma, PI3Kbeta and AKT2 in breast cancer. 27098453_WDR26 affected beta-catenin levels. WDR26/Axin binding is involved in the ubiquitination of beta-catenin. 27835684_It has been shown that WDR26 promotes Rac1 membrane translocation following a Coro1A-like and Coro1A-dependent mechanism. 28686853_haploinsufficiency of WDR26 contributes to the pathology of 1q41q42 microdeletion syndrome. 32958140_WD40 Repeat Protein 26 Negatively Regulates Formyl Peptide Receptor-1 Mediated Wound Healing in Intestinal Epithelial Cells. 33506510_Skraban-Deardorff syndrome: Six new cases of WDR26-related disease and expansion of the clinical phenotype. 33675273_Expanding the clinical phenotype of the ultra-rare Skraban-Deardorff syndrome: Two novel individuals with WDR26 loss-of-function variants and a literature review. 33905682_GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme. | ENSMUSG00000038733 | Wdr26 | 3.365587e+03 | 1.1696588 | 0.226087767 | 0.2766437 | 6.714801e-01 | 0.4125360498 | 0.84733931 | No | Yes | 3.539767e+03 | 488.964836 | 2.707458e+03 | 383.640141 | |
| ENSG00000163050 | 56997 | COQ8A | protein_coding | Q8NI60 | FUNCTION: Atypical kinase involved in the biosynthesis of coenzyme Q, also named ubiquinone, an essential lipid-soluble electron transporter for aerobic cellular respiration (PubMed:25498144, PubMed:21296186, PubMed:25540914, PubMed:27499294). Its substrate specificity is unclear: does not show any protein kinase activity (PubMed:25498144, PubMed:27499294). Probably acts as a small molecule kinase, possibly a lipid kinase that phosphorylates a prenyl lipid in the ubiquinone biosynthesis pathway, as suggested by its ability to bind coenzyme Q lipid intermediates (PubMed:25498144, PubMed:27499294). Shows an unusual selectivity for binding ADP over ATP (PubMed:25498144). {ECO:0000269|PubMed:25498144, ECO:0000269|PubMed:27499294, ECO:0000305|PubMed:21296186, ECO:0000305|PubMed:25540914}. | 3D-structure;ATP-binding;Alternative splicing;Direct protein sequencing;Disease variant;Kinase;Membrane;Mitochondrion;Neurodegeneration;Nucleotide-binding;Primary mitochondrial disease;Reference proteome;Transferase;Transit peptide;Transmembrane;Transmembrane helix;Ubiquinone biosynthesis | PATHWAY: Cofactor biosynthesis; ubiquinone biosynthesis. {ECO:0000269|PubMed:25498144}. | This gene encodes a mitochondrial protein similar to yeast ABC1, which functions in an electron-transferring membrane protein complex in the respiratory chain. It is not related to the family of ABC transporter proteins. Expression of this gene is induced by the tumor suppressor p53 and in response to DNA damage, and inhibiting its expression partially suppresses p53-induced apoptosis. Alternatively spliced transcript variants have been found; however, their full-length nature has not been determined. [provided by RefSeq, Jul 2008]. | hsa:56997; | extrinsic component of mitochondrial inner membrane [GO:0031314]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; ADP binding [GO:0043531]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; phosphorylation [GO:0016310]; ubiquinone biosynthetic process [GO:0006744] | 11888884_play an important role in mediating p53-inducible apoptosis through the mitochondrial pathway. 18319072_CABC1 gene mutations in four ubiquinone-deficient patients in three distinct families were reported. 18319074_Five additional mutations in ADCK3 were found in three patients with sporadic ataxia, including one known to have CoQ10 deficiency in muscle. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22036850_These observations highlight the importance of screening for a potentially treatable cause, CABC1/ADCK3 mutations 25216398_a structural foundation for investigating the role of transmembrane association in regulating the biological activity of ADCK3 25498144_Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. 25540914_work reveals Mg(2+)-dependent ATPase activity of ADCK3, providing strong support for the theoretical prediction of this protein being a functional atypical kinase. 26866375_ADCK3/COQ8 localises to mitochondrial cristae and is targeted to this organelle via the presence of an N-terminal localisation signal 27499294_Loss of COQ8a results in cerebellar ataxia and coenzyme Q deficiency. 30968303_expand the clinical, molecular and biochemical spectrum of ADCK3 related CoQ10 deficiencies 32337771_Clinico-Genetic, Imaging and Molecular Delineation of COQ8A-Ataxia: A Multicenter Study of 59 Patients. 33622667_Photoparoxysmal response in ADCK3 autosomal recessive ataxia: a case report and literature review. | ENSMUSG00000026489 | Coq8a | 6.718403e+03 | 1.1342352 | 0.181719895 | 0.2986601 | 3.705033e-01 | 0.5427300311 | 0.89062022 | No | Yes | 6.627886e+03 | 630.283456 | 5.399703e+03 | 527.101873 |
| ENSG00000163071 | 132671 | SPATA18 | protein_coding | Q8TC71 | FUNCTION: Key regulator of mitochondrial quality that mediates the repairing or degradation of unhealthy mitochondria in response to mitochondrial damage. Mediator of mitochondrial protein catabolic process (also named MALM) by mediating the degradation of damaged proteins inside mitochondria by promoting the accumulation in the mitochondrial matrix of hydrolases that are characteristic of the lysosomal lumen. Also involved in mitochondrion degradation of damaged mitochondria by promoting the formation of vacuole-like structures (named MIV), which engulf and degrade unhealthy mitochondria by accumulating lysosomes. The physical interaction of SPATA18/MIEAP, BNIP3 and BNIP3L/NIX at the mitochondrial outer membrane regulates the opening of a pore in the mitochondrial double membrane in order to mediate the translocation of lysosomal proteins from the cytoplasm to the mitochondrial matrix. {ECO:0000269|PubMed:21264221, ECO:0000269|PubMed:21264228, ECO:0000269|PubMed:22292033}. | Alternative splicing;Coiled coil;Cytoplasm;Membrane;Mitochondrion;Mitochondrion outer membrane;Phosphoprotein;Reference proteome | This gene encodes a p53-inducible protein that is able to induce lysosome-like organelles within mitochondria that eliminate oxidized mitochondrial proteins, thereby contributing to mitochondrial quality control. Dysregulation of mitochondrial quality control is associated with cancer and degenerative diseases. The encoded protein mediates accumulation of the lysosome-like mitochondrial organelles through interaction with B cell lymphoma 2 interacting protein 3 and B cell lymphoma 2 interacting protein 3 like at the outer mitochondrial membrane, which allows translocation of lysosomal proteins to the mitochondrial matrix from the cytosol. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2016]. | hsa:132671; | cytoplasm [GO:0005737]; intracellular membrane-bounded organelle [GO:0043231]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; identical protein binding [GO:0042802]; cellular response to DNA damage stimulus [GO:0006974]; mitochondrial protein catabolic process [GO:0035694]; mitophagy by induced vacuole formation [GO:0035695] | 16088906_beta-sarcoglycan and SPATA18 may have a role in limb-girdle muscular dystrophy type 2E 21264221_Mieap induces intramitochondrial lysosome-like organella that plays a critical role in mitochondrial quality control by eliminating oxidized mitochondrial proteins 21264228_Mieap induced vacuole-like structures (designated as MIV for Mieap-induced vacuole), which engulfed and degraded the unhealthy mitochondria by accumulating lysosomes 21300779_show that SPATA18 transcription is induced by p53 in a variety of cell types of both human and mouse origin 22292033_The physical interaction of Mieap, BNIP3 and NIX at the mitochondrial outer membrane may play a critical role in the translocation of lysosomal proteins from the cytoplasm to the mitochondrial matrix. 30290054_Results show that Mieap induced caspase-dependent mitochondrial apoptosis in breast cancer (BC) cells. Mieap protein was downregulated and its promoter methylated in BC with more aggressive and malignant phenotypes. 30808977_Mieap-induced accumulation of lysosomes within mitochondria (MALM) regulates gastric cancer cell invasion under hypoxia by suppressing reactive oxygen species accumulation. 32453416_DNA damage invokes mitophagy through a pathway involving Spata18. 32458504_Causative role for defective expression of mitochondria-eating protein in accumulation of mitochondria in thyroid oncocytic cell tumors. 32736677_p53/Mieap-regulated mitochondrial quality control plays an important role as a tumor suppressor in gastric and esophageal cancers. 35269894_SPATA18 Expression Predicts Favorable Clinical Outcome in Colorectal Cancer. | ENSMUSG00000029155 | Spata18 | 7.921494e+01 | 0.8810124 | -0.182765768 | 0.4557679 | 1.627044e-01 | 0.6866787911 | 0.93348335 | No | Yes | 7.493812e+01 | 17.088545 | 8.208607e+01 | 19.171399 | |
| ENSG00000163214 | 90957 | DHX57 | protein_coding | Q6P158 | FUNCTION: Probable ATP-binding RNA helicase. | ATP-binding;Alternative splicing;Coiled coil;Helicase;Hydrolase;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | hsa:90957; | ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724] | ENSMUSG00000035051 | Dhx57 | 1.002667e+03 | 0.6591970 | -0.601218329 | 0.3277954 | 3.211869e+00 | 0.0731059124 | 0.71859192 | No | Yes | 1.075090e+03 | 219.802063 | 1.244787e+03 | 261.158093 | |||
| ENSG00000163608 | 25871 | NEPRO | protein_coding | Q6NW34 | FUNCTION: May play a role in cortex development as part of the Notch signaling pathway. Downstream of Notch may repress the expression of proneural genes and inhibit neuronal differentiation thereby maintaining neural progenitors. May also play a role in preimplentation embryo development. {ECO:0000250|UniProtKB:Q8R2U2}. | Alternative splicing;Developmental protein;Disease variant;Dwarfism;Nucleus;Phosphoprotein;Reference proteome | hsa:25871; | nucleolus [GO:0005730]; nucleus [GO:0005634]; negative regulation of neuron differentiation [GO:0045665]; positive regulation of Notch signaling pathway [GO:0045747] | 31250547_Our report delineates the clinical and radiological characteristics of an emerging ribosomopathy caused by biallelic variants in NEPRO | ENSMUSG00000036208 | Nepro | 6.449807e+02 | 0.8267493 | -0.274478108 | 0.3194120 | 7.271008e-01 | 0.3938245631 | 0.84317192 | No | Yes | 5.730652e+02 | 94.700962 | 6.349693e+02 | 107.448989 | ||
| ENSG00000163659 | 25976 | TIPARP | protein_coding | Q7Z3E1 | FUNCTION: ADP-ribosyltransferase that mediates mono-ADP-ribosylation of glutamate, aspartate and cysteine residues on target proteins (PubMed:23275542, PubMed:25043379, PubMed:30373764). Acts as a negative regulator of AHR by mediating mono-ADP-ribosylation of AHR, leading to inhibit transcription activator activity of AHR (PubMed:23275542, PubMed:30373764). {ECO:0000269|PubMed:23275542, ECO:0000269|PubMed:25043379, ECO:0000269|PubMed:30373764}. | ADP-ribosylation;Glycosyltransferase;Metal-binding;NAD;Nucleus;Reference proteome;Transferase;Zinc;Zinc-finger | This gene encodes a member of the poly(ADP-ribose) polymerase superfamily. Studies of the mouse ortholog have shown that the encoded protein catalyzes histone poly(ADP-ribosyl)ation and may be involved in T-cell function. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:25976; | nucleus [GO:0005634]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; metal ion binding [GO:0046872]; NAD+ ADP-ribosyltransferase activity [GO:0003950]; protein ADP-ribosylase activity [GO:1990404]; androgen metabolic process [GO:0008209]; cellular response to organic cyclic compound [GO:0071407]; estrogen metabolic process [GO:0008210]; face morphogenesis [GO:0060325]; female gonad development [GO:0008585]; hemopoiesis [GO:0030097]; kidney development [GO:0001822]; negative regulation of gene expression [GO:0010629]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of protein catabolic process [GO:0045732]; post-embryonic development [GO:0009791]; protein ADP-ribosylation [GO:0006471]; protein auto-ADP-ribosylation [GO:0070213]; protein mono-ADP-ribosylation [GO:0140289]; roof of mouth development [GO:0060021]; skeletal system morphogenesis [GO:0048705]; smooth muscle tissue development [GO:0048745]; vasculogenesis [GO:0001570] | 12851707_TIPARP (DKFZp434J214) gene is amplified in HNSCC. TIPARP, FLJ22693 and ZAP proteins with TPH, WW and PARP-like domains constitute the TIPARP family. 20852632_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20852632_Single nucleotide polymorphism in TIPARP is associated with ovarian cancer. 24806346_Knockdown of TiPARP, but not AHRR, increased 2,3,7,8-tetrachlorodibenzo-p-dioxin - induced CYP1A1 mRNA and AHR protein levels. 26814197_these data identify a new mechanism of LXR regulation that involves TIPARP, ADP-ribosylation and MACROD1. 28213497_TIPARP is a viral RNA-sensing pattern recognition receptors that mediates antiviral responses triggered by BAX- and BAK1-dependent mitochondrial damage 29274782_Moreover, time course and promoter activity assays suggest that TIPARP and TIPARP-AS1 work in concert to regulate AHR signaling. Collectively, these data show an added level of complexity in the AHR signaling cascade which involves lncRNAs, whose functions remain poorly understood. 29938598_circHECTD1 functions as an endogenous MIR142 (microRNA 142) sponge to inhibit MIR142 activity, resulting in the inhibition of TIPARP (TCDD inducible poly[ADP-ribose] polymerase) expression with subsequent inhibition of astrocyte activation via macroautophagy/autophagy. 30373764_Mutation of cysteine 39 to alanine resulted in a small, but significant, reduction in TCDD inducible poly(ADP-ribose) polymerase protein (TIPARP) autoribosylation activity. 32482854_TiPARP forms nuclear condensates to degrade HIF-1alpha and suppress tumorigenesis. 33475084_Chemical genetics and proteome-wide site mapping reveal cysteine MARylation by PARP-7 on immune-relevant protein targets. 33475085_Identification of PARP-7 substrates reveals a role for MARylation in microtubule control in ovarian cancer cells. 33572475_Post-Transcriptional Regulation of PARP7 Protein Stability Is Controlled by Androgen Signaling. 33799807_PARP7 and Mono-ADP-Ribosylation Negatively Regulate Estrogen Receptor alpha Signaling in Human Breast Cancer Cells. 33976187_Androgen signaling uses a writer and a reader of ADP-ribosylation to regulate protein complex assembly. 34264286_PARP7 mono-ADP-ribosylates the agonist conformation of the androgen receptor in the nucleus. 34375612_PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity. | ENSMUSG00000034640 | Tiparp | 1.784094e+02 | 1.1098162 | 0.150320807 | 0.3913885 | 1.465986e-01 | 0.7018072516 | 0.93624959 | No | Yes | 2.093322e+02 | 45.718956 | 1.408093e+02 | 31.820993 | |
| ENSG00000163681 | 7871 | SLMAP | protein_coding | Q14BN4 | FUNCTION: May play a role during myoblast fusion. {ECO:0000250}. | 3D-structure;Alternative splicing;Cell membrane;Coiled coil;Cytoplasm;Cytoskeleton;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a component of a conserved striatin-interacting phosphatase and kinase complex. Striatin family complexes participate in a variety of cellular processes including signaling, cell cycle control, cell migration, Golgi assembly, and apoptosis. The protein encoded by this gene is a coiled-coil, tail-anchored membrane protein with a single C-terminal transmembrane domain that is posttranslationally inserted into membranes. Mutations in this gene are associated with Brugada syndrome, a cardiac channelopathy. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2015]. | hsa:7871; | integral component of plasma membrane [GO:0005887]; microtubule organizing center [GO:0005815]; sarcolemma [GO:0042383]; smooth endoplasmic reticulum [GO:0005790]; muscle contraction [GO:0006936]; protein localization to plasma membrane [GO:0072659]; regulation of membrane depolarization during cardiac muscle cell action potential [GO:1900825]; regulation of sodium ion transmembrane transport [GO:1902305]; regulation of voltage-gated sodium channel activity [GO:1905150] | 23064965_Mutations in SLMAP may cause Brugada syndrome via modulating the intracellular trafficking of hNav1.5 channel. 25880194_The data suggests the potential role of SLMAP single nucleotide polymorphism as a risk factor for the susceptibility of diabetic retinopathy among type 2 diabetes patients in the Qatari population. 29063833_Here, the authors discover SAV1-mediated inhibition of the PP2A complex STRIPAK(SLMAP) as a key mechanism of MST1/2 activation. | ENSMUSG00000021870 | Slmap | 6.934924e+02 | 1.2002206 | 0.263299602 | 0.3431267 | 5.825035e-01 | 0.4453327158 | 0.85808635 | No | Yes | 8.005986e+02 | 179.688675 | 5.528521e+02 | 127.428878 | |
| ENSG00000163702 | 84818 | IL17RC | protein_coding | Q8NAC3 | FUNCTION: Receptor for IL17A and IL17F, major effector cytokines of innate and adaptive immune system involved in antimicrobial host defense and maintenance of tissue integrity (By similarity). Receptor for IL17A and IL17F, major effector cytokines of innate and adaptive immune system involved in antimicrobial host defense and maintenance of tissue integrity. Receptor for IL17A and IL17F homodimers as part of a heterodimeric complex with IL17RA (PubMed:16785495). Receptor for the heterodimer formed by IL17A and IL17B as part of a heterodimeric complex with IL17RA (PubMed:18684971). Has also been shown to be the cognate receptor for IL17F and to bind IL17A with high affinity without the need for IL17RA (PubMed:17911633). Upon binding of IL17F homodimer triggers downstream activation of TRAF6 and NF-kappa-B signaling pathway (PubMed:16785495, PubMed:32187518). Induces transcriptional activation of IL33, a potent cytokine that stimulates group 2 innate lymphoid cells and adaptive T-helper 2 cells involved in pulmonary allergic response to fungi (By similarity). Promotes sympathetic innervation of peripheral organs by coordinating the communication between gamma-delta T cells and parenchymal cells. Stimulates sympathetic innervation of thermogenic adipose tissue by driving TGFB1 expression (By similarity). Binding of IL17A-IL17F to IL17RA-IL17RC heterodimeric receptor complex triggers homotypic interaction of IL17RA and IL17RC chains with TRAF3IP2 adapter through SEFIR domains. This leads to downstream TRAF6-mediated activation of NF-kappa-B and MAPkinase pathways ultimately resulting in transcriptional activation of cytokines, chemokines, antimicrobial peptides and matrix metalloproteinases, with potential strong immune inflammation (PubMed:18684971, PubMed:17911633). Primarily induces neutrophil activation and recruitment at infection and inflammatory sites (By similarity). Stimulates the production of antimicrobial beta-defensins DEFB1, DEFB103A, and DEFB104A by mucosal epithelial cells, limiting the entry of microbes through the epithelial barriers (By similarity). {ECO:0000250|UniProtKB:Q8K4C2, ECO:0000269|PubMed:16785495, ECO:0000269|PubMed:17911633, ECO:0000269|PubMed:18684971, ECO:0000269|PubMed:32187518}.; FUNCTION: [Isoform 5]: Receptor for both IL17A and IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 6]: Does not bind IL17A or IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 7]: Does not bind IL17A or IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 8]: Receptor for both IL17A and IL17F. {ECO:0000269|PubMed:16785495}. | 3D-structure;Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Inflammatory response;Membrane;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene encodes a single-pass type I membrane protein that shares similarity with the interleukin-17 receptor (IL-17RA). Unlike IL-17RA, which is predominantly expressed in hemopoietic cells, and binds with high affinity to only IL-17A, this protein is expressed in nonhemopoietic tissues, and binds both IL-17A and IL-17F with similar affinities. The proinflammatory cytokines, IL-17A and IL-17F, have been implicated in the progression of inflammatory and autoimmune diseases. Multiple alternatively spliced transcript variants encoding different isoforms have been detected for this gene, and it has been proposed that soluble, secreted proteins lacking transmembrane and intracellular domains may function as extracellular antagonists to cytokine signaling. [provided by RefSeq, Feb 2011]. | hsa:84818; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; interleukin-17 receptor activity [GO:0030368]; inflammatory response [GO:0006954] | 16688746_IL-17RL exists as multiple isoforms due to extensive alternative splicing. Changes in RNA IL-17RL splicing occur in advanced cancers. 16785495_The biologic activity of IL-17 is dependent on a complex composed of receptors IL-17RA and IL-17RC. 17911633_IL-17RC functions as a receptor for both IL-17A and IL-17F; a soluble version of this protein should be an effective antagonist of IL-17A and IL-17F mediated inflammatory diseases. 18097068_IL-17A-induced IL-6, IL-8, and CCL20 secretion was dependent on both IL-17RA and IL-17RC, which are overexpressed in RA patients. 18684971_Interleukin-17F is inhibited by the IL-17RC receptor, a combination of soluble IL-17RA/IL-17RC receptors is required for inhibition of the IL-17F/IL-17A activity. 20173024_IL-17 and its receptor IL-17RC are involved in rheumatoid arthritis synovial fluid-mediated chemotaxis in human lung microvascular endothelial cell culture. 22744455_Overall, our study found a significant association of IL-17RC gene polymorphisms with AIS in a Chinese Han population, indicating IL-17RC gene may be as a susceptibility gene for AIS. 22898922_Data show that IL-17RA, IL-17RC, IL-22R1, ERK1/2 MAPK and NF-kappaB pathways are involved in Th17 cytokine-induced proliferation. 22999050_IL-17RC predisposes to the development of adolescent idiopathic scoliosis in a Chinese Han population. 24885153_results suggested a potential involvement of IL-17RC+CD8+ T cells in pathogenesis of ocular sarcoidosis 25918342_human IL-17RC is essential for mucocutaneous immunity to C. albicans but is otherwise largely redundant. 26731132_methylation of IL17RC could play as a marker in CNV and degeneration of RPE cells in vitro. 27155366_Interleukin 17A (IL17a) and interleukin-23 (IL-23) - dependent interleukin-17 receptor C (IL-17RC) are expressed by sputum and neutrophils in deltaF508-CFTR protein (F508del) cystic fibrosis patients. 29584788_IL-17RC rs708567 polymorphism in A/A genotype, G/G genotype, and G/a genotype did not seem to influence RA susceptibility in Tunisian population. 29695654_Genetic Variants on IL-17 are associated with development of atherosclerotic diseases. 29764467_Five SNPs in the IL17RC (and COL6A1) genes were found to be associated with susceptibility to ossification of the posterior longitudinal ligament in Han Chinese patients. 30024651_Genetic study revealed no association between IL-17RC, and SNPs and acute kidney transplant graft rejection. Nevertheless, a significant improvement of graft survival was found in kidney transplant recipients carrying the IL-17RC*G/G, and *G/A genotypes. 31291973_An increase in IL17RC gene expression levels in peripheral blood samples was found in ossification of the posterior longitudinal ligament patients. 31481525_Interleukin-17 receptor C gene polymorphism reduces treatment effect and promotes poor prognosis of ischemic stroke. 32187518_The crystal structure of the interleukin 17 receptor C (IL-17RC):interleukin 17F (IL-17F) complex provides a structural basis for IL-17F signaling through IL-17RC. 35167487_IL-17 Receptor C Signaling Controls CD4(+) TH17 Immune Responses and Tissue Injury in Immune-Mediated Kidney Diseases. | ENSMUSG00000030281 | Il17rc | 3.121703e+02 | 1.1436326 | 0.193623670 | 0.3398258 | 3.238306e-01 | 0.5693146086 | 0.89878854 | No | Yes | 2.437697e+02 | 37.536464 | 1.994345e+02 | 31.882354 | |
| ENSG00000163811 | 23160 | WDR43 | protein_coding | Q15061 | FUNCTION: Ribosome biogenesis factor that coordinates hyperactive transcription and ribogenesis (PubMed:17699751). Involved in nucleolar processing of pre-18S ribosomal RNA. Required for optimal pre-ribosomal RNA transcription by RNA polymerase I (PubMed:17699751). Essential for stem cell pluripotency and embryonic development. In the nucleoplasm, recruited by promoter-associated/nascent transcripts and transcription to active promoters where it facilitates releases of elongation factor P-TEFb and paused RNA polymerase II to allow transcription elongation and maintain high-level expression of its targets genes (By similarity). {ECO:0000250|UniProtKB:Q6ZQL4, ECO:0000269|PubMed:17699751}. | 3D-structure;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ribosome biogenesis;Transcription;Transcription regulation;Ubl conjugation;WD repeat;rRNA processing | hsa:23160; | chromatin [GO:0000785]; fibrillar center [GO:0001650]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; RNA polymerase II complex binding [GO:0000993]; transcription elongation regulator activity [GO:0003711]; positive regulation of rRNA processing [GO:2000234]; positive regulation of transcription by RNA polymerase I [GO:0045943]; regulation of stem cell population maintenance [GO:2000036]; regulation of transcription elongation from RNA polymerase II promoter [GO:0034243]; rRNA processing [GO:0006364] | 31128943_RNA targets ribogenesis Factor WDR43 to chromatin for transcription and pluripotency control. | ENSMUSG00000041057 | Wdr43 | 9.727700e+02 | 0.8126979 | -0.299208913 | 0.3290072 | 8.145197e-01 | 0.3667873584 | 0.83581079 | No | Yes | 7.608820e+02 | 129.677888 | 9.269294e+02 | 161.825457 | ||
| ENSG00000163923 | 116832 | RPL39L | protein_coding | Q96EH5 | Reference proteome;Ribonucleoprotein;Ribosomal protein | This gene encodes a protein sharing high sequence similarity with ribosomal protein L39. Although the name of this gene has been referred to as 'ribosomal protein L39' in the public databases, its official name is 'ribosomal protein L39-like'. It is not currently known whether the encoded protein is a functional ribosomal protein or whether it has evolved a function that is independent of the ribosome. [provided by RefSeq, Jul 2008]. | hsa:116832; | cytosolic large ribosomal subunit [GO:0022625]; structural constituent of ribosome [GO:0003735]; spermatogenesis [GO:0007283]; translation [GO:0006412] | 12359333_locus, structure and expression of the gene and its relevance to the biological activities of the ribosome 24452241_Results show the RP paralog Rpl39l is highly expressed in Embryonic Stem Cells and its expression strongly correlates with hepatocellular carcinoma tumor (HCC) samples with high tumor grading and alpha-fetoprotein level giving it diagnostic potential. 24815453_The ribosomal protein L39-L gene may have effects on the drug resistance mechanism of lung cancer A549 cells. 24998577_High RPL39L expression is associated with drug-resistant lacrimal gland adenoid cystic carcinoma. | 7.653191e+02 | 0.7498165 | -0.415390583 | 0.3312935 | 1.573756e+00 | 0.2096624529 | 0.78763590 | No | Yes | 6.060030e+02 | 71.084182 | 7.706678e+02 | 92.596049 | ||||
| ENSG00000163935 | 51460 | SFMBT1 | protein_coding | Q9UHJ3 | FUNCTION: Histone-binding protein, which is part of various corepressor complexes. Mediates the recruitment of corepressor complexes to target genes, followed by chromatin compaction and repression of transcription. Plays a role during myogenesis: required for the maintenance of undifferentiated states of myogenic progenitor cells via interaction with MYOD1. Interaction with MYOD1 leads to the recruitment of associated corepressors and silencing of MYOD1 target genes. Part of the SLC complex in germ cells, where it may play a role during spermatogenesis. {ECO:0000269|PubMed:17599839, ECO:0000269|PubMed:23349461, ECO:0000269|PubMed:23592795}. | Alternative splicing;Chromatin regulator;Differentiation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Spermatogenesis;Transcription;Transcription regulation | This gene shares high similarity with the Drosophila Scm (sex comb on midleg) gene. It encodes a protein which contains four malignant brain tumor repeat (mbt) domains and may be involved in antigen recognition. [provided by RefSeq, Jun 2012]. | hsa:51460; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; transcription corepressor activity [GO:0003714]; cell differentiation [GO:0030154]; chromatin organization [GO:0006325]; negative regulation of muscle organ development [GO:0048635]; negative regulation of transcription, DNA-templated [GO:0045892]; spermatogenesis [GO:0007283] | 17599839_The tandem MBT repeats form a functional structure required for biological activity of hSFMBT and predict similar properties for other MBT domain-containing proteins. 21325761_A segmental copy number loss of the SFMBT1 gene may be involved in the pathological process in some individuals with ventriculomegaly/idiopathic normal pressure hydrocephalus. 22479346_Data identified IGF1, SLC4A4, WWOX, and SFMBT1 as hypertension susceptibility genes by gene based association scan and gene expression analysis. 23349461_novel mechanisms accounting for SFMBT1-mediated transcription repression and revealed an essential role of Sfmbt1 in regulating MyoD-mediated transcriptional silencing 23592795_When bound to its gene targets, SFMBT1 recruits its associated proteins and causes chromatin compaction and transcriptional repression 23928305_The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition. 27861535_The present study demonstrates that a copy number loss within intron 2 of the SFMBT1 gene may be a genetic risk factor for shunt-responsive definite iNPH. 28789618_copy number variation analysis identified deletions in SFMBT1 associated with fasting plasma glucose in a Han Chinese population 29886071_SFMBT1 knockdown was demonstrated to inhibit cell growth and induced apoptosis, which was consistent with the function of miR-20a- 3p upregulation in HaCaT cells. 32023483_High SFMBT1 expression is associated with clear cell renal cell carcinomas. 33481017_Multi-omics analysis to identify susceptibility genes for colorectal cancer. 34978167_Regulatory Variant rs2535629 in ITIH3 Intron Confers Schizophrenia Risk By Regulating CTCF Binding and SFMBT1 Expression. | ENSMUSG00000006527 | Sfmbt1 | 9.074842e+02 | 0.8880345 | -0.171312308 | 0.2688276 | 4.091524e-01 | 0.5224000438 | 0.88476243 | No | Yes | 8.332356e+02 | 82.739652 | 8.415630e+02 | 85.541641 | |
| ENSG00000163950 | 7884 | SLBP | protein_coding | Q14493 | FUNCTION: RNA-binding protein involved in the histone pre-mRNA processing (PubMed:8957003, PubMed:9049306, PubMed:12588979, PubMed:19155325). Binds the stem-loop structure of replication-dependent histone pre-mRNAs and contributes to efficient 3'-end processing by stabilizing the complex between histone pre-mRNA and U7 small nuclear ribonucleoprotein (snRNP), via the histone downstream element (HDE) (PubMed:8957003, PubMed:9049306, PubMed:12588979, PubMed:19155325). Plays an important role in targeting mature histone mRNA from the nucleus to the cytoplasm and to the translation machinery (PubMed:8957003, PubMed:9049306, PubMed:12588979, PubMed:19155325). Stabilizes mature histone mRNA and could be involved in cell-cycle regulation of histone gene expression (PubMed:8957003, PubMed:9049306, PubMed:12588979, PubMed:19155325). Involved in the mechanism by which growing oocytes accumulate histone proteins that support early embryogenesis (By similarity). Binds to the 5' side of the stem-loop structure of histone pre-mRNAs (By similarity). {ECO:0000250|UniProtKB:P97440, ECO:0000269|PubMed:12588979, ECO:0000269|PubMed:19155325, ECO:0000269|PubMed:8957003, ECO:0000269|PubMed:9049306}. | 3D-structure;Alternative splicing;Cytoplasm;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribonucleoprotein;Ubl conjugation;mRNA processing | This gene encodes a protein that binds to the stem-loop structure in replication-dependent histone mRNAs. Histone mRNAs do not contain introns or polyadenylation signals, and are processed by endonucleolytic cleavage. The stem-loop structure is essential for efficient processing but this structure also controls the transport, translation and stability of histone mRNAs. Expression of the protein is regulated during the cell cycle, increasing more than 10-fold during the latter part of G1. [provided by RefSeq, Jul 2008]. | hsa:7884; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; histone mRNA stem-loop binding complex [GO:0062073]; histone pre-mRNA 3'end processing complex [GO:0071204]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; histone pre-mRNA DCP binding [GO:0071208]; histone pre-mRNA stem-loop binding [GO:0071207]; identical protein binding [GO:0042802]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; cap-dependent translational initiation [GO:0002191]; mRNA 3'-end processing by stem-loop binding and cleavage [GO:0006398]; mRNA transport [GO:0051028] | 12588979_SLBP is the only cell cycle-regulated factor required for histone pre-mRNA processing 15546920_human HBP/SLBP is essential for the coordinate synthesis of DNA and histone proteins and is required for progression through the cell division cycle 15829567_SLBP is imported into the cell nucleus during the cell cycle. 15916543_SLBP is required for efficient DNA replication probably because a decreased ability to assemble chromatin results in a decrease in the rate of DNA replication 16492733_Removal of the phosphoryl group from T230 by either dephosphorylation or mutation results in a 7-fold reduction in the affinity of SLBP for the stem-loop RNA 16931877_replication-dependent histone mRNAs are likely to be the sole target of SLBP 16982637_SLBP is required for efficient histone 3'-UTR end processing. 17499042_Here we show that NELF interacts with the cap binding complex (CBC), a factor that plays important roles in mRNA processing steps, and the two factors together participate in the 3' end processing of histone mRNAs, through association with the SLBP. 18025107_identified five conserved residues in a 15-amino-acid region in the amino-terminal portion of SLBP, each of which is required for translation. 18490441_Study concludes that the increase in cyclin A/Cdk1 activity at the end of S phase triggers degradation of SLBP at S/G(2). 19155325_These results suggest a previously undescribed role for SLBP in histone mRNA export. 22328085_haploinsufficiency of SLBP and/or WHSC2 (NELF-A) contributes to several novel cellular phenotypes of WHS. 22439849_The nuclear magnetic resonance and kinetic studies presented here provide a framework for understanding how SLBP recognizes histone mRNA and highlight possible structural roles of phosphorylation and proline isomerization in RNA binding proteins 22907757_This paper shows that SLBP is a substrate for the prolyl isomerase Pin1. Pin1, along with PP2A, facilitates dissociation of the SLBP-histone mRNA complex at the end of S-phase, thereby promoting histone mRNA decay and SLBP ubiquitination. 23234701_Using yeast two-hybrid screening, the authors identify CT initiation factor-interacting protein (CTIF) as a protein that binds directly to SLBP. SLBP preferentially associates with the CT complex of histone mRNAs consisting of CBP80/CBP20, but not with the eIF4E/eIF4G (ET) complex, as has been proposed. Rapid degradation of histone mRNA on the inhibition of DNA replication requires association of SLBP with CTIF. 23286197_Data suggest that oligomerization and SLBP phosphorylation regulate SLBP-SLIP1-histone-mRNA complex formation/disassociation; sequential and ordered assembly is required. 23286197_This paper describes the biophysical characterization of human SLBP and the SLBP-SLIP1 complex. Human SLBP is an intrinsically disordered protein that is phosphorylated at 23 Ser/Thr sites when expressed in a eukaryotic expression system such as baculovirus. Unphosphorylated human SLBP forms a high affinity heterotetramer with SLIP1 and the SLBP-SLIP1 complex is regulated by SLBP phosphorylation. 23329046_the crystal structure of a ternary complex of human SLBP RNA binding domain, human 3'hExo, and a 26-nucleotide stem-loop RNA is reported. 23941746_Alternative splicing allows the synthesis of HBP/SLBP isoforms with different properties that may be important for regulating HBP/SLBP functions during replication stress. 24122909_the S/G2 stable mutant form of SLBP is degraded by proteasome in G1, indicating that indicating that the SLBP degradation in G1 is independent of the previously identified SLBP degradation at S/G2 24255165_C-terminal extension of Lsm4 interacts directly with the histone mRNP, contacting both SLBP and 3'hExo. 25002523_Although the C-terminal tail of dSLBP does not contact the RNA, phosphorylation of the tail promotes SLBP conformations competent for RNA binding and thereby appears to reduce the entropic penalty for the association. 25266719_arsenic, a carcinogenic metal, decreases cellular levels of SLBP by inducing its proteasomal degradation and inhibiting SLBP transcription via epigenetic mechanisms 27203182_CRL4(WDR23) is required for efficient histone mRNA 3' end processing to produce mature histone mRNAs for translation. CRL4(WDR23) binds and ubiquitylates SLBP in vitro and in vivo, and this modification activates SLBP function in histone mRNA 3' end processing without affecting its protein levels. 27254819_CRL4-DCAF11 mediates the degradation of SLBP at the end of S phase and this degradation is essential for the viability of cells. 27454292_SLBP is a potentially important cellular regulator of HIV-1, thereby establishing a link between histone metabolism, inflammation, and HIV-1 infection 27773672_Cyclin F-mediated degradation of SLBP limits H2A.X accumulation and apoptosis upon genotoxic stress in G2 cell cycle checkpoint. 28118078_FEM1 proteins are ancient regulators of Stem-Loop Binding Protein. 28306745_showed that inhibiting the SLBP mRNA and protein levels were rescued by epigenetic modifiers suggesting that nickel's effects on SLBP may be mediated via epigenetic mechanisms 31007083_Knocking down miR-384 promotes growth and metastasis of osteosarcoma MG63 cells by targeting SLBP 32960265_Influenza A virus co-opts ERI1 exonuclease bound to histone mRNA to promote viral transcription. | ENSMUSG00000004642 | Slbp | 1.528101e+03 | 0.9535382 | -0.068637308 | 0.2913199 | 5.525619e-02 | 0.8141572391 | 0.96339664 | No | Yes | 1.453747e+03 | 211.476838 | 1.450640e+03 | 216.339331 | |
| ENSG00000164038 | 133308 | SLC9B2 | protein_coding | Q86UD5 | FUNCTION: Na(+)/H(+) antiporter that extrudes Na(+) or Li(+) in exchange for external protons across the membrane (PubMed:18000046, PubMed:28154142, PubMed:22948142, PubMed:18508966). Contributes to the regulation of intracellular pH, sodium homeostasis, and cell volume. Plays an important role for insulin secretion and clathrin-mediated endocytosis in beta-cells (By similarity). Involved in sperm motility and fertility (By similarity). It is controversial whether SLC9B2 plays a role in osteoclast differentiation or not (By similarity). {ECO:0000250|UniProtKB:Q5BKR2, ECO:0000269|PubMed:18000046, ECO:0000269|PubMed:18508966, ECO:0000269|PubMed:22948142, ECO:0000269|PubMed:28154142}. | Alternative splicing;Antiport;Cell junction;Cell membrane;Cell projection;Cilium;Cytoplasmic vesicle;Endosome;Flagellum;Hydrogen ion transport;Ion transport;Membrane;Mitochondrion;Phosphoprotein;Reference proteome;Sodium;Sodium transport;Synapse;Transmembrane;Transmembrane helix;Transport | Sodium hydrogen antiporters, such as NHEDC2, convert the proton motive force established by the respiratory chain or the F1F0 mitochondrial ATPase into sodium gradients that drive other energy-requiring processes, transduce environmental signals into cell responses, or function in drug efflux (Xiang et al., 2007 [PubMed 18000046]).[supplied by OMIM, Mar 2008]. | hsa:133308; | apical plasma membrane [GO:0016324]; basolateral plasma membrane [GO:0016323]; endosome membrane [GO:0010008]; integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrial membrane [GO:0031966]; plasma membrane [GO:0005886]; sperm principal piece [GO:0097228]; synaptic vesicle membrane [GO:0030672]; identical protein binding [GO:0042802]; lithium:proton antiporter activity [GO:0010348]; monovalent cation:proton antiporter activity [GO:0005451]; sodium:proton antiporter activity [GO:0015385]; clathrin-dependent endocytosis [GO:0072583]; flagellated sperm motility [GO:0030317]; ion transmembrane transport [GO:0034220]; positive regulation of osteoclast development [GO:2001206]; regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0061178]; sodium ion transport [GO:0006814] | 17988971_NHA-oc/NHA2 displays the expected activities of a bona fide cation-proton antiporter and plays a key role(s) in normal osteoclast differentiation and function 18000046_Na+/H+ antiporter genes may contribute to sodium-lithium countertransport activity and salt homeostasis in humans[NHA1 and NHA2] 18508966_restricted to the distal convoluted tubule in the kidney 20053353_cluster of essential, highly conserved titratable residues located in an assembly region made of two discontinuous helices of inverted topology, each interrupted by an extended chain 20332099_Observational study of gene-disease association. (HuGE Navigator) 20713131_mutation in AA residues V161 and F357 reduced ability of transfected BW31a cells to remove intracellular Na and grow in NaCl-medium; yeast expressing F357 F437 cannot grow in 0.4M NaCl, suggesting these residues also essential for antiporter activity 22948142_the ubiquitous tissue distribution of NHA2 suggests that H(+)-coupled transport is more widespread. The coexistence of Na(+) and H(+)-driven chemiosmotic circuits has implications for salt and pH regulation in the kidney. 28154142_The exchange activity of NHA2 (SLC9B2) is electroneutral, despite harboring the 2 conserved aspartic acid residues found in NapA and other bacterial homologues. The equivalent residue to K305 in human NHA2 has been replaced with arginine, which is a mutation that makes NapA electroneutral. A transmembrane embedded lysine residue is essential for electrogenic transport in Na(+)/H(+) antiporters. | ENSMUSG00000037994 | Slc9b2 | 4.446141e+02 | 0.8415532 | -0.248873619 | 0.2794786 | 7.905593e-01 | 0.3739310728 | 0.83844010 | No | Yes | 3.217227e+02 | 36.308351 | 3.741340e+02 | 43.138946 | |
| ENSG00000164073 | 256471 | MFSD8 | protein_coding | Q8NHS3 | FUNCTION: May be a carrier that transport small solutes by using chemiosmotic ion gradients. {ECO:0000305}. | Alternative splicing;Disease variant;Glycoprotein;Lysosome;Membrane;Neurodegeneration;Neuronal ceroid lipofuscinosis;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a ubiquitous integral membrane protein that contains a transporter domain and a major facilitator superfamily (MFS) domain. Other members of the major facilitator superfamily transport small solutes through chemiosmotic ion gradients. The substrate transported by this protein is unknown. The protein likely localizes to lysosomal membranes. Mutations in this gene are correlated with a variant form of late infantile-onset neuronal ceroid lipofuscinoses (vLINCL). [provided by RefSeq, Oct 2008]. | hsa:256471; | integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; transmembrane transporter activity [GO:0022857]; autophagosome maturation [GO:0097352]; lysosome organization [GO:0007040]; neuron development [GO:0048666]; regulation of autophagy [GO:0010506]; regulation of lysosomal protein catabolic process [GO:1905165]; TORC1 signaling [GO:0038202] | 17564970_MFSD8 gene is involved in late-infantile-onset neuronal ceroid lipofuscinose;it was mapped to chromosome 4q28.1-q28.2. 18850119_Results describe a novel mutation in the MFSD8 gene, responsible for neuronal ceroid lipofuscinoses, in a consanguineous Egyptian family 19177532_Study contributes to a better molecular characterization of Italian NCL cases, and will facilitate medical genetic counseling in such families. 19201763_CLN7/MFSD8 defects are not restricted to the Turkish population, as initially anticipated, but are a relatively common cause of NCL in different populations. 19277732_Data show that neuronal ceroid lipofuscinosis in a Saudi family is due to a homozygous novel mutation in the most recently described NCL gene (MFSD8). 20826447_Expression and lysosomal targeting of CLN7 are reported. 24423645_This study showed that Gene disruption of Mfsd8 provides animal model for CLN7 disease. 25227500_In this study, we identified variants in MFSD8 as a novel cause of nonsyndromic autosomal recessive macular dystrophy with central cone involvement. 25270050_A mutation in MFSD8, c.472G>A (p.Gly158Ser), segregates with the disease phenotype in variant late infantile neuronal ceroid lipofuscinosis. 25439737_MFSD8 genetic testing should also be considered in patients with Rett like phenotype at onset and negative MECP2 mutation 28586915_This study highlights a hierarchy of MFSD8 variant severity, predicting three consequences of mutation: (1) nonsyndromic localized maculopathy, (2) nonsyndromic widespread retinopathy, or (3) syndromic neurological disease. 29514215_Quantification revealed that the amounts of 12 different soluble lysosomal proteins were significantly reduced in Cln7 ko MEFs compared with wild-type controls. One of the most significantly depleted lysosomal proteins was Cln5 protein that underlies another distinct neuronal ceroid lipofuscinosis disorder 30144815_We identified a novel homozygous mutation in MFSD8 gene. 30382371_that MFSD8-associated lysosomal dysfunction may contribute to frontotemporal lobar degeneration pathology 31006324_Here and for the first time, we reported on two previously variant late-infantile neuronal ceroid lipofuscinoses-associated variants in MFSD8 but in association with a form of cone-rod dystrophy known as non-syndromic macular dystrophy with central cone involvement. 33226711_Mutation analysis of MFSD8 in an amyotrophic lateral sclerosis cohort from mainland China. 35087090_Aberrant upregulation of the glycolytic enzyme PFKFB3 in CLN7 neuronal ceroid lipofuscinosis. 35216386_A Novel, Apparently Silent Variant in MFSD8 Causes Neuronal Ceroid Lipofuscinosis with Marked Intrafamilial Variability. 35457110_Contribution of Whole-Genome Sequencing and Transcript Analysis to Decipher Retinal Diseases Associated with MFSD8 Variants. | ENSMUSG00000025759 | Mfsd8 | 2.024177e+02 | 0.6429441 | -0.637234750 | 0.4085970 | 2.408238e+00 | 0.1206981536 | 0.75783482 | No | Yes | 1.272331e+02 | 25.749842 | 2.094460e+02 | 42.965272 | |
| ENSG00000164074 | 80167 | ABHD18 | protein_coding | Q0P651 | Alternative splicing;Glycoprotein;Reference proteome;Secreted;Signal | Mouse_homologues mmu:269423; | extracellular region [GO:0005576] | ENSMUSG00000037818 | Abhd18 | 1.246882e+02 | 0.7138061 | -0.486395840 | 0.4388577 | 1.182599e+00 | 0.2768275567 | 0.80469852 | No | Yes | 9.636176e+01 | 22.521402 | 1.390433e+02 | 32.921056 | ||||
| ENSG00000164076 | 79012 | CAMKV | protein_coding | Q8NCB2 | FUNCTION: Does not appear to have detectable kinase activity. | Alternative splicing;Calmodulin-binding;Cell membrane;Cytoplasmic vesicle;Membrane;Phosphoprotein;Reference proteome | hsa:79012; | cytoplasmic vesicle membrane [GO:0030659]; glutamatergic synapse [GO:0098978]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; peptidyl-serine phosphorylation [GO:0018105]; regulation of modification of postsynaptic structure [GO:0099159] | 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) | ENSMUSG00000032936 | Camkv | 8.491207e+02 | 1.0498442 | 0.070175205 | 0.2771945 | 6.537246e-02 | 0.7981976460 | 0.96139092 | No | Yes | 6.701350e+02 | 83.834585 | 6.903887e+02 | 88.794942 | ||
| ENSG00000164323 | 57587 | CFAP97 | protein_coding | Q9P2B7 | Alternative splicing;Coiled coil;Phosphoprotein;Reference proteome | hsa:57587; | ENSMUSG00000031631 | Cfap97 | 1.423072e+02 | 1.0851852 | 0.117941328 | 0.3899097 | 9.008155e-02 | 0.7640735081 | 0.95240548 | No | Yes | 1.566200e+02 | 30.982176 | 1.124265e+02 | 22.942052 | |||||
| ENSG00000164414 | 10559 | SLC35A1 | protein_coding | P78382 | FUNCTION: Transports CMP-sialic acid from the cytosol into Golgi vesicles where glycosyltransferases function (PubMed:15576474). Efficient CMP-sialic acid uptake depends on the presence of free CMP inside the vesicles, suggesting the proteins functions as an antiporter. Binds both CMP-sialic acid and free CMP, but has higher affinity for free CMP (By similarity). {ECO:0000250|UniProtKB:Q61420, ECO:0000269|PubMed:15576474}. | Alternative splicing;Congenital disorder of glycosylation;Golgi apparatus;Membrane;Reference proteome;Sugar transport;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is found in the membrane of the Golgi apparatus, where it transports nucleotide sugars into the Golgi. One such nucleotide sugar is CMP-sialic acid, which is imported into the Golgi by the encoded protein and subsequently glycosylated. Defects in this gene are a cause of congenital disorder of glycosylation type 2F (CDG2F). Two transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Dec 2009]. | hsa:10559; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of Golgi membrane [GO:0030173]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; antiporter activity [GO:0015297]; CMP-N-acetylneuraminate transmembrane transporter activity [GO:0005456]; UDP-galactose transmembrane transporter activity [GO:0005459]; carbohydrate metabolic process [GO:0005975]; carbohydrate transport [GO:0008643]; cellular protein modification process [GO:0006464]; CMP-N-acetylneuraminate transmembrane transport [GO:0015782] | 12682060_substrate binding specificity 15576474_this defect is a new type of congenital disorder of glycosylation (CDG) of type IIf affecting the transport of CMP-sialic acid into the Golgi apparatus. 16343442_this study, we introduced two critical genes encoding human CMP-N-acetylneuraminic acid synthetase and CMP-sialic acid transporter into tobacco suspension-cultured cell to pave a route for sialic biosynthetic pathway. 16923816_CMP-sialic acid transporter is localized in the medial-trans Golgi 23873973_We confirm an autosomal recessive, generalized sialylation defect due to mutations in SLC35A1 25552652_SLC35A1-deficient cells lack of alpha-dystroglycan O-mannosylation, ligand binding and incorporation of sialic acids. 27387429_the SLC35A1 generates additional isoforms through alternative splicing. 28856833_We performed exome sequencing on an individual with a profound neurological presentation and identified rare compound heterozygous mutations, p.Thr156Arg and p.Glu196Lys, in the CMP-sialic acid transporter, SLC35A1. Patient primary fibroblasts and serum showed a considerable decrease in the amount of N- and O-glycans terminating in sialic acid 30115659_Data indicate a congenital deficiency in solute carrier family 35 (CMP-sialic acid transporter), member A1 (SLC35A1) mutation in two siblings born to consanguineous parents and who displayed moderate macrothrombocytopenia. 32303557_Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. 33396746_Novel Insights into Selected Disease-Causing Mutations within the SLC35A1 Gene Encoding the CMP-Sialic Acid Transporter. 34015330_The promiscuous binding pocket of SLC35A1 ensures redundant transport of CDP-ribitol to the Golgi. 34069698_Knockout of the CMP-Sialic Acid Transporter SLC35A1 in Human Cell Lines Increases Transduction Efficiency of Adeno-Associated Virus 9: Implications for Gene Therapy Potency Assays. 34384782_A three-pocket model for substrate coordination and selectivity by the nucleotide sugar transporters SLC35A1 and SLC35A2. | ENSMUSG00000028293 | Slc35a1 | 2.176562e+02 | 0.7479331 | -0.419018806 | 0.3989673 | 1.093966e+00 | 0.2955940456 | 0.81343599 | No | Yes | 1.523917e+02 | 32.933860 | 2.401274e+02 | 52.705733 | |
| ENSG00000164542 | 23366 | KIAA0895 | protein_coding | Q8NCT3 | Alternative splicing;Reference proteome | hsa:23366; | Mouse_homologues 17971504_9530077C05Rik is most abundantly expressed in tissues rich in highly ciliated cells, such as olfactory sensory neurons, and is predicted to be important to cilia. | ENSMUSG00000036411 | 9530077C05Rik | 1.861367e+02 | 0.8462770 | -0.240798118 | 0.4109220 | 3.404357e-01 | 0.5595778566 | 0.89491887 | No | Yes | 1.640571e+02 | 38.218844 | 1.529344e+02 | 36.456403 | ||||
| ENSG00000164654 | 54468 | MIOS | protein_coding | Q9NXC5 | FUNCTION: As a component of the GATOR subcomplex GATOR2, functions within the amino acid-sensing branch of the TORC1 signaling pathway. Indirectly activates mTORC1 and the TORC1 signaling pathway through the inhibition of the GATOR1 subcomplex (PubMed:23723238). It is negatively regulated by the upstream amino acid sensors SESN2 and CASTOR1 (PubMed:25457612, PubMed:27487210). {ECO:0000269|PubMed:23723238, ECO:0000269|PubMed:25457612, ECO:0000269|PubMed:27487210}. | Alternative splicing;Lysosome;Membrane;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:54468; | cell junction [GO:0030054]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; GATOR2 complex [GO:0061700]; lysosomal membrane [GO:0005765]; nucleoplasm [GO:0005654]; cellular response to amino acid starvation [GO:0034198]; positive regulation of TOR signaling [GO:0032008]; protein-containing complex localization [GO:0031503] | ENSMUSG00000042447 | Mios | 3.263836e+02 | 0.7999930 | -0.321940752 | 0.3739930 | 7.285344e-01 | 0.3933586666 | 0.84317192 | No | Yes | 2.976418e+02 | 60.665392 | 3.657058e+02 | 76.163939 | |||
| ENSG00000164707 | 26266 | SLC13A4 | protein_coding | Q9UKG4 | FUNCTION: Sodium/sulfate cotransporter that mediates sulfate reabsorption in the high endothelial venules (HEV). {ECO:0000269|PubMed:10535998, ECO:0000269|PubMed:15607730}. | Ion transport;Membrane;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | hsa:26266; | integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; sodium:sulfate symporter activity [GO:0015382]; anion transmembrane transport [GO:0098656]; sulfate transport [GO:0008272] | 15607730_Here, we characterized the functional properties of the human Na(+)-sulfate cotransporter (hNaS2), determined its tissue distribution, and identified its gene (SLC13A4) structure. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23453247_SLC13A4 and SLC26A2 were the most abundant sulfate transporter mRNAs, which localized to syncytiotrophoblast and cytotrophoblast cells, respectively. 23485456_To investigate the regulation of SLC13A4 gene expression, we analysed the transcriptional activity of the SLC13A4 5'-flanking region in the JEG-3 placental cell line using luciferase reporter assays. 28385533_Study found that despite differential expression of the two SLC13A4 transcripts, no detectable functional difference in the cellular sorting or sulfate transporting was found. However, some variants can influence both mechanism in specific cell membranes. This is like to have clinical implications based on the consequences of impaired sulfate transport during pregnancy in rodent models. 34840533_SLC13A4 Might Serve as a Prognostic Biomarker and be Correlated with Immune Infiltration into Head and Neck Squamous Cell Carcinoma. | ENSMUSG00000029843 | Slc13a4 | 1.538862e+02 | 1.0064832 | 0.009323027 | 0.3611490 | 6.575702e-04 | 0.9795419825 | 0.99658776 | No | Yes | 1.469002e+02 | 22.017843 | 1.319766e+02 | 20.572053 | ||
| ENSG00000164830 | 55074 | OXR1 | protein_coding | Q8N573 | FUNCTION: May be involved in protection from oxidative damage. {ECO:0000269|PubMed:11114193, ECO:0000269|PubMed:15060142}. | Alternative splicing;Disease variant;Epilepsy;Mental retardation;Mitochondrion;Phosphoprotein;Reference proteome;Stress response | hsa:55074; | mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleus [GO:0005634]; oxidoreductase activity [GO:0016491]; adult walking behavior [GO:0007628]; cellular response to hydroperoxide [GO:0071447]; negative regulation of cellular response to oxidative stress [GO:1900408]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of oxidative stress-induced neuron death [GO:1903204]; negative regulation of peptidyl-cysteine S-nitrosylation [GO:1902083]; neuron apoptotic process [GO:0051402]; response to oxidative stress [GO:0006979] | 15060142_Data show that human and yeast oxidation resistance 1 (OXR1) genes are induced by heat and oxidative stress and that their proteins localize to the mitochondria and function to protect against oxidative damage. 17391516_human OXR1 is capable of reducing the DNA damaging effects of reactive oxygen species when expressed in bacteria, indicating the protein has an activity that can contribute to oxidation resistance. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22873401_The protein segment encoded by exon 8 plays an important role in the anti-oxidative function of the human OXR1 protein. 25236744_OXR1 upregulates the expression of antioxidant genes via the p21 signaling pathway to suppress hydrogen peroxide-induced oxidative stress and maintain mtDNA integrity. 25792726_Oxr1 serves as a potential therapeutic target for ALS and other neurodegenerative disorders characterized by TDP-43 or FUS pathology. 26616534_OXR1 may act as a sensor of cellular oxidative stress to regulate the transcriptional networks required to detoxify reactive oxygen species and modulate cell cycle and apoptosis. 29766639_findings provide new insights into the mechanism by which senescent cells are highly resistant to oxidative stress and suggest that OXR1 is a novel senolytic target that can be further exploited for the development of new senolytic agents 30221705_The results of the present study indicated that sevoflurane exerts its neurotoxic effect by regulating the hsamiR302e/OXR1 axis. Therefore, the manipulation of the hsamiR302e/OXR1 pathway will be useful for preventing sevofluraneinduced neurotoxicity. 30852977_Low OXR1 expression is associated with the development of esophageal squamous cell carcinoma. 31502810_MiR-616 promotes the progression of pancreatic carcinoma by targeting OXR1. 31642482_Over-expression of Oxr1 was able to delay neuromuscular abnormalities in the hSOD1G93A amyotrophic lateral sclerosis mouse model. 31785787_Loss of OXR1 is Associated with an Autosomal-Recessive Neurological Disease with Cerebellar Atrophy and Lysosomal Dysfunction. 31845986_Oxidation resistance 1 prevents genome instability through maintenance of G2/M arrest in gamma-ray-irradiated cells. | ENSMUSG00000022307 | Oxr1 | 3.583740e+02 | 0.8119502 | -0.300536777 | 0.2982086 | 1.029044e+00 | 0.3103832592 | 0.81737113 | No | Yes | 3.737472e+02 | 44.821222 | 4.922543e+02 | 59.879043 | ||
| ENSG00000164970 | 203259 | FAM219A | protein_coding | Q8IW50 | Acetylation;Alternative splicing;Phosphoprotein;Reference proteome | The protein encoded by this gene has homologs that have been identified in mouse, macaque, etc organisms. Multiple alternatively spliced transcript variants that encode different protein isoforms have been described for this gene. [provided by RefSeq, Dec 2010]. | hsa:203259; | ENSMUSG00000028439 | Fam219a | 1.826241e+03 | 1.0192692 | 0.027535119 | 0.2958409 | 8.543287e-03 | 0.9263564816 | 0.98598977 | No | Yes | 1.725165e+03 | 162.854244 | 1.642704e+03 | 159.419655 | ||||
| ENSG00000165097 | 221656 | KDM1B | protein_coding | Q8NB78 | FUNCTION: Histone demethylase that demethylates 'Lys-4' of histone H3, a specific tag for epigenetic transcriptional activation, thereby acting as a corepressor. Required for de novo DNA methylation of a subset of imprinted genes during oogenesis. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Demethylates both mono- and di-methylated 'Lys-4' of histone H3. Has no effect on tri-methylated 'Lys-4', mono-, di- or tri-methylated 'Lys-9', mono-, di- or tri-methylated 'Lys-27', mono-, di- or tri-methylated 'Lys-36' of histone H3, or on mono-, di- or tri-methylated 'Lys-20' of histone H4. {ECO:0000269|PubMed:23260659, ECO:0000269|PubMed:23357850}. | 3D-structure;Alternative splicing;Chromatin regulator;Developmental protein;FAD;Flavoprotein;Metal-binding;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | Flavin-dependent histone demethylases, such as KDM1B, regulate histone lysine methylation, an epigenetic mark that regulates gene expression and chromatin function (Karytinos et al., 2009 [PubMed 19407342]).[supplied by OMIM, Oct 2009]. | hsa:221656; | nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; FAD binding [GO:0071949]; flavin adenine dinucleotide binding [GO:0050660]; histone binding [GO:0042393]; histone demethylase activity [GO:0032452]; histone H3-di/monomethyl-lysine-4 FAD-dependent demethylase activity [GO:0140682]; oxidoreductase activity [GO:0016491]; zinc ion binding [GO:0008270]; DNA methylation involved in gamete generation [GO:0043046]; histone H3-K4 demethylation [GO:0034720]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of DNA methylation [GO:0044030]; regulation of gene expression by genetic imprinting [GO:0006349] | 20670891_Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. 23266887_The zinc finger-SWIRM-oxidase domains is required for KDM1B demethylase activity and the binding of FAD. 24924415_These results demonstrate an important role for LSD2 in regulation of DNA methylation and gene silencing in breast cancer. 25036127_regulation of tissue factor pathway inhibitor-2 (TFPI-2) expression by lysine-specific demethylase 1 and 2 25624347_Using transcriptome and chromatin immunoprecipitation-sequencing analyses, the study revealed that LSD2 represses the genes involved in lipid influx and metabolism through demethylation of histone H3K4. 25773598_Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. 28277979_This review focuses on published small-molecule inhibitors targeted at the two flavin adenine dinucleotide-dependent lysine demethylases, lysine-specific demethylases 1 and 2, and how the inhibitors interact with the tertiary structures of the enzymes. 29845195_These results suggest that LSD2 achieves a promoting effect on small cell lung cancer by indirectly regulating TFPI2 expression through the mediation of DNMT3B expression or through the regulation of the demethylation of H3K4me1 in the promoter region of the TFPI2 gene. 30846414_our findings provided new insights into the critical and multifaceted roles of KDM1B in the regulation of cell proliferation and apoptosis, and offered a potentially novel target in preventing the progression of pancreatic cancer. 30970244_While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. 31160694_Reduction in H3K4me patterns due to aberrant expression of methyltransferases and demethylases in renal cell carcinoma: prognostic and therapeutic implications. 32097694_The expression of LSD2 was associated with higher TNM stage and metastasis of the tumor and thus, might serve as a useful marker for Clear cell renal cell carcinoma (ccRCC) progression. 32754863_In vitro evidence of NLRP3 inflammasome regulation by histone demethylase LSD2 in renal cancer: a pilot study. 33067956_[Expression and Clinical Significance of MiR-215 and KDM1B in Patients with Diffuse Large B Cell Lymphoma]. 34329293_Histone demethylase AMX-1 is necessary for proper sensitivity to interstrand crosslink DNA damage. 35327654_The Role of LSD1 and LSD2 in Cancers of the Gastrointestinal System: An Update. | ENSMUSG00000038080 | Kdm1b | 5.239804e+02 | 1.0576414 | 0.080850519 | 0.3064009 | 6.915231e-02 | 0.7925750406 | 0.95966019 | No | Yes | 5.832046e+02 | 101.941696 | 4.418933e+02 | 79.367455 | |
| ENSG00000165119 | 3190 | HNRNPK | protein_coding | P61978 | FUNCTION: One of the major pre-mRNA-binding proteins. Binds tenaciously to poly(C) sequences. Likely to play a role in the nuclear metabolism of hnRNAs, particularly for pre-mRNAs that contain cytidine-rich sequences. Can also bind poly(C) single-stranded DNA. Plays an important role in p53/TP53 response to DNA damage, acting at the level of both transcription activation and repression. When sumoylated, acts as a transcriptional coactivator of p53/TP53, playing a role in p21/CDKN1A and 14-3-3 sigma/SFN induction (By similarity). As far as transcription repression is concerned, acts by interacting with long intergenic RNA p21 (lincRNA-p21), a non-coding RNA induced by p53/TP53. This interaction is necessary for the induction of apoptosis, but not cell cycle arrest. As part of a ribonucleoprotein complex composed at least of ZNF827, HNRNPL and the circular RNA circZNF827 that nucleates the complex on chromatin, may negatively regulate the transcription of genes involved in neuronal differentiation (PubMed:33174841). {ECO:0000250, ECO:0000269|PubMed:16360036, ECO:0000269|PubMed:20673990, ECO:0000269|PubMed:22825850, ECO:0000269|PubMed:33174841}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cell junction;Cell projection;Cytoplasm;DNA-binding;Direct protein sequencing;Glycoprotein;Host-virus interaction;Isopeptide bond;Mental retardation;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Ribonucleoprotein;Spliceosome;Transcription;Transcription regulation;Ubl conjugation;mRNA processing;mRNA splicing | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene is located in the nucleoplasm and has three repeats of KH domains that binds to RNAs. It is distinct among other hnRNP proteins in its binding preference; it binds tenaciously to poly(C). This protein is also thought to have a role during cell cycle progession. Several alternatively spliced transcript variants have been described for this gene, however, not all of them are fully characterized. [provided by RefSeq, Jul 2008]. | hsa:3190; | catalytic step 2 spliceosome [GO:0071013]; cell projection [GO:0042995]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; podosome [GO:0002102]; ribonucleoprotein complex [GO:1990904]; cadherin binding [GO:0045296]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; mRNA binding [GO:0003729]; protein domain specific binding [GO:0019904]; RNA binding [GO:0003723]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of apoptotic process [GO:0043066]; negative regulation of mRNA splicing, via spliceosome [GO:0048025]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of low-density lipoprotein receptor activity [GO:1905599]; positive regulation of receptor-mediated endocytosis [GO:0048260]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of gene expression [GO:0010468]; regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:1902165]; regulation of low-density lipoprotein particle clearance [GO:0010988]; regulation of mRNA splicing, via spliceosome [GO:0048024]; regulation of transcription by RNA polymerase II [GO:0006357]; RNA processing [GO:0006396]; signal transduction [GO:0007165] | 11867641_Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken beta-tropomyosin pre-mRNA. 12052863_c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs(hnRNPK) 12093748_hnRNP K KH3 specifically recognizes a tetrad of sequence 5'd-TCCC. The complex is stabilized by a dense network of methyl-oxygen hydrogen bonds involving the methyl groups of 3 isoleucine residues and the O2 and N3 atoms of the 2 central cytosine bases. 12183465_examination into mechanisms of hnRNP K activities by identifying protein factors that interact with it 12370808_we describe the identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a protein that specifically interacts with Sam68 in vitro and in vivo. 12411317_acts together with Pur(alpha) to repress the transcriptional activity of the CD43 gene promoter during lymphocyte activation 14562022_Nuclear shift of hnRNP K protein in dividing cells may reflect its involvement in signalling multiple events that regulate expression of genes in proliferating cells 15039586_Co-crystallization of the third KH domain of human hnRNP K with a 15-mer ssDNA showed that the crystals contained the complex containing three KH3 domains per 15-mer ssDNA 15486205_ORF57 and hnRNP K interaction may modulate ORF57-mediated regulation of viral gene expression 15514164_hnRNP K is a positive effector of collagen synthesis acting at the post-transcriptional level by interaction with the 3'-untranslated regions (3'-UTRs) of COL1A1, 1A2, and 3A1 mRNAs. 15671036_the mutually antagonistic action of two RNA-binding proteins, Hu and hnRNP K, control the timing of the switch from proliferation to neuronal differentiation through the post-transcriptional regulation of p21 mRNA 15860232_hnRNP K is involved in B cell receptor signalling pathway 16004877_NMR and X-ray crystallographic studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids 16293596_HNRPK down-regulation and interference with HNRPK translation-but not transcription-regulatory activity impairs proliferation, clonogenic potential, and leukemogenic activity of BCR/ABL-expressing myeloid 32Dcl3 and/or primary CD34+ CML-BC cells 16360036_hnRNP K is a HDM2 target and serves as a cofactor for p53. HnRNP K plays key roles in coordinating transcriptional responses to DNA damage. 16404425_Alternative isoform of hnRNPK found in colonic tumor and surrounding mucosa; is first example of RNA editing even in cancer and its surrounding tissue 16492668_arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src 16496041_satellite I RNA binds to hnRNP K protein 16564677_These findings provide a putative mechanism by which transcriptional activity of hnRNP K can be discretely controlled through the regulation of PP1 activity. 17191129_Tandem mass-spectrometric analysis of the peptide at residues 288-303 of heterogeneous nuclear ribonucleoprotein K (hnRNP K) shows that both Arg296 and Arg299 are dimethylated. 17483488_Isolation of antibodies from cells with loss of migration phenotype and identification of their target proteins revealed the involvement of the heterogeneous nuclear ribonucleoprotein K (hnRNP-K), a multifunctional signaling protein, in metastasis 17561226_interacts with Sindbis virus NSP2 and viral subgenomic mRNA 17667925_Heterogeneous nuclear ribonucleoprotein K overexpression in oral squamous cell carcinoma. 17672864_APOBEC3 suppresses HBV replication in hepatocytes by inhibiting hnRNP K-mediated transcription and expression of HBV genes as well as HBV core DNA synthesis. 18441016_hnRNP K binds to the 3'-untranslated region of the c-Src mRNA and inhibits its translation by blocking 80 S ribosome formation 18472002_Inhibition of methylation in hnRNP K attenuated the recruitment of p53 to p21 promoter, and reduced p53 transcriptional activity. 18559600_Cytoplasmic hnRNP K and high thymidine phosphorylase may be potential prognostic and therapeutic markers for nasopharyngeal carcinoma. 18854243_The interaction of hnRNP-K with Nef strongly increased HIV transcription, which depended on Tat and the NF-kB motif in the viral promoter, but not on NF-kB activation. 18854308_HNRNP K and microRNA-16 have roles in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products 19015635_Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. 19101556_The data here provide possible mechanisms of how the non-enzymatic activity of PRMT family protein associates with hnRNP K protein and regulates hnRNP K protein-involved transactivation functions. 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19170760_hnRNP K and RBM42 have a role in the maintenance of cellular ATP level in the stress conditions possibly through protecting their target mRNAs. 19249676_MDM2 released from p53 by RITA promotes degradation of p21 and the p53 cofactor hnRNP K, required for p21 transcription 19258514_Translational inhibition of AR by hnRNP-K may occur in organ-confined tumors but possibly at a reduced level in metastases. HnRNP-K is the first protein identified that directly interacts with and regulates the AR translational apparatus. 19330019_Results collectively establish the regulation and role of ERK-mediated cytoplasmic accumulation of hnRNP K as an upstream modulator of TP, suggesting that hnRNP K may be an attractive candidate as a future therapeutic target for cancer. 19401687_hnRNP K has potential implications at the diagnostic, prognostic and therapeutic levels in prostate cancer. 19520842_hnRNP K has a role in preventing the production of the pro-apoptotic Bcl-x(S) splice isoform 19548310_Cytoplasmic hnRNP K increased significantly from leukoplakia to head-and-neck/oral squamous cell carcinomas. 19609950_Data unveiled an important new signaling pathway that linked by hnRNP K and mutant p53 in pancreatic cancer tumorigenesis. 19653139_The overexpression of hnRNPK, which is regulated by BCR-ABL and Ras-MAPK signaling pathways, may promote the progression of CML. 19747914_proteolytically activated PKC-delta down-regulates hnRNP K protein in a proteasome-dependent manner, which plays an important role in apoptosis induction 19808671_Data demonstrate that expression levels of hnRNP A1, Q, K, R, and U influence HIV-1 production by persistently infected astrocytes, linking these hnRNPs to HIV replication. 19880579_hnRNP-K was identiied as one of the ERK targets essential for IL-2 production and shown to be required for T cell late activation. 20224598_FLIP expression is transcriptionally regulated by hnRNP K and nucleolin, and may be a potential prognostic and therapeutic marker for nasopharyngeal carcinoma. 20371611_heterogeneous nuclear ribonucleoprotein K is a transactivator for human low density lipoprotein receptor gene transcription 20408130_Two human colorectal cancer cell lines of different metastatic potential.Expression of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in LoVo cells was higher than in SW480 cells. 20499280_Higher expression of the heterogeneous nuclear ribonucleoprotein k is associated with melanoma. 20548952_suggested that expanded AUUCU repeats within the spliced intronic sequence strongly bind to hnRNP K. 20623123_Data show that The level of expression of hnRNP K was greater in several myeloid leukemia cell lines such as HL-60, OUN-1, UT-7, and K562 than in PBMCs from healthy individuals. 20888333_Results indicate that hnRNP K has an impact on a late step of herpesviral propagation making it a potential antiviral target. 21184736_these results identify preferred PRMT1 methylation sequences of hnRNP K by direct methylation assay and implicate a role of arginine methylation in regulating intracellular distribution of hnRNP K. 21233203_a model where Pol II transcription-driven recruitment of hnRNP K along the EGR-1 locus compartmentalizes activation of the ERK cascade at these genes, events that regulate synthesis of mature mRNA. 21321982_role of hnRNP K in prostate carcinoma might be played by the interaction with the androgen receptor. 21466159_Heterogeneous nuclear ribonucleoprotein K and nucleolin transcriptionally activate the vascular endothelial growth factor promoter through interaction with secondary DNA structures 21821029_Via phosphorylating hnRNPK Aurora-A participates in regulating p53 activity during DNA damage. 22321252_Elevated cytoplasmic hnRNP K and TP overexpression are associated with poorer survival in oral squamous cell carcinoma patients 22335908_Helicobacter pylori L-form may be associated with up-regulated hnRNPK expression in gastric carcinoma. 22582387_hnRNP K is indispensable for tumor cell viability and targeting of hnRNP K by granzymes contributes to or reinforces the cell death mechanisms by which cytotoxic lymphocytes eliminate tumor cells. 22760167_Overexpression of hnRNP K is associated with early hepatocellular carcinoma in patients with cirrhosis. 22825850_DNA damage-induced Pc2 activation to the p53 transcriptional co-activation through hnRNP K sumoylation. 22879910_hnRNP K, when co-expressed with EBNA2, strongly enhances viral latent membrane protein 2A (LMP2A) expression. 22960638_HNRNPK affects production of the essential NEAT1_2 isoform by negatively regulating its 3'-end polyadenylation by arresting CFIm complex binding near NEAT1_1's alternative polyadenylation site. 23092970_SUMO modification plays a crucial role in the control of hnRNP-K's function as a p53 co-activator in response to DNA damage by UV 23099853_HnRNP K controlled expression of IE2 protein of Human cytomegalovirus during viral replication. 23343766_ATM-dependent phosphorylation of heterogeneous nuclear ribonucleoprotein K promotes p53 transcriptional activation in response to DNA damage. 23455382_Prolonged downregulation of hnRNP K using small interfering RNA significantly decreased cell viability and increased apoptosis in HCC cell lines in a p53-independent manner. 23519117_Caspase-3 cleaves hnRNP K in erythroid differentiation. 23564449_hnRNP-K regulates extracellular matrix, cell motility, and angiogenesis pathways. Involvement of the selected genes (Cck, Mmp-3, Ptgs2, and Ctgf) and pathways was validated by gene-specific expression analysis 23798440_heterogeneous nuclear ribonucleoprotein K (hnRNPK), a protein known to integrate multiple signal transduction pathways with gene expression, as a SERT distal polyadenylation element binding protein 23825951_NS1-BP-hnRNPK complex is a key mediator of influenza A virus gene expression. 23843646_These studies demonstrate hnRNP K to be a multifunctional protein that supports vesicular stomatitis virus infection via its role(s) in suppressing apoptosis of infected cells. 23857582_hnRNPK may play a role in recruitment of XRN2 to gene loci thus regulating coupling 3'-end pre-mRNA processing to transcription termination. 24508256_Data show that SET accumulation up-regulated hnRNPK mRNA and total/phosphorylated protein, promoted hnRNPK nuclear location, and reduced Bcl-x mRNA levels. 24594223_CNBP overexpression caused increase of cell death and suppression of cell metastasis through its induction of G-quadruplex formation in the promoter of hnRNP K resulting in hnRNP K down-regulation 24626777_Results indicate that the interaction between the AR and hnRNP K has an important role in the progression of prostate cancer. 24885469_HnRNP K can induce MMP12 expression and enzyme activity through activating MMP12 promoter, which promotes cell migration and invasion in nasopharyngeal carcinoma cells. 25005557_hnRNP K and PU.1 act synergistically during granulocytic differentiation, hnRNP K seems to have a negative effect on PU.1 activity during monocytic maturation 25281771_hnRNPK is potentially implicated in the radiogenic response of HNSCC. 25410660_Inhibition of CDK2 phosphorylation blocked phosphorylation of hnRNP K, preventing its incorporation into stress granules (SGs). Due to interaction between hnRNP K with TDP-43, the loss of hnRNP K from SGs prevented accumulation of TDP-43. 25497182_Data indicate that twenty proteins were identified as binding partners of the primary activating element in the heterogeneous nuclear ribonucleoprotein K (hnRNP K) promoter. 25569684_HNRNPK might determine efficiency of Hepatitis C virus particle production by limiting the availability of viral RNA for incorporation into virions. 25701787_hnRNP K plays an important role in the mitotic process in colon cancer cells.hnRNP K upregulates NUF2 and promotes the tumorigenicity of colon cancer cells. 25713416_These findings functionally integrate K17, hnRNP K, and gene expression along with RSK and CXCR3 signaling in a keratinocyte-autonomous axis and provide a potential basis for their implication in tumorigenesis 25865411_These results indicate that dengue virus type 2 and Junin virus induce hnRNP K cytoplasmic translocation to favor viral multiplication. 26136337_we investigate the role of hnRNP K in the radioresistance of malignant melanoma cells 26164948_hnRNP K interacts with EV71 5' UTR, which is required for efficient synthesis of viral RNA.[review] 26305187_RTVP-1 regulates glioma cell spreading, migration and invasion and that these effects are mediated via interaction with N-WASP and by interfering with the inhibitory effect of hnRNPK on the function of this protein. 26317903_Data show that proto-oncogene protein c-myc is upregulated by sumoylated heterogeneous nuclear ribonucleoprotein K (hnRNP K) at the translational level in Burkitt's lymphoma cells. 26330540_hnRNP K binds miR-122, a mature liver-specific microRNA required for Hepatitis C virus replication. 26412324_Data implicate hnRNPK in the development of hematological disorders and suggest hnRNPK acts as a tumor suppressor. 26530384_The authors identified two new human proteins that interact with Ehrlichia chaffeensis EtpE-C: CD147 and heterogeneous nuclear ribonucleoprotein K (hnRNP-K). 26586566_It describes the identification of heterogeneous nuclear ribonucleoprotein K (hnRNPK) as one of the composite element binding factors(CEBF) that acts as transactivator of PXR promoter. 26713736_hnRNP K may be a key molecule involved in cell motility in RCC cells 26823606_HhnRNP-K-mediated regulation of NMHC IIA mRNA translation contributes to the control of enucleation in erythropoiesis. 26954065_Our study also suggests that loss of function variants in HNRNPK should be considered as a molecular basis for patients with Kabuki-like syndrome 26972480_Data show that heterogeneous nuclear ribonucleoprotein K (hnRNPK) stabilized of cellular FLICE-inhibitory protein (c-FLIP) protein through inhibition of glycogen synthase kinase 3 beta (GSK3beta) Ser9 phosphorylation during the TNF-related aoptosis-inducing ligand (TRAIL)-induced apoptosis. 27012187_CASC11 can target heterogeneous ribonucleoprotein K (hnRNP-K) to activate WNT/beta-catenin signaling in colorectal cancer cells to promote tumor growth and metastasis. 27049467_hnRNP K is a multifunctional protein that can regulate both oncogenic and tumor suppressive pathways through a plethora of chromatin-, DNA-, RNA-, and protein-mediated activates, suggesting its aberrant expression may have broad-reaching cellular impacts. (Review) 27155326_these findings support a critical role for hnRNP K in the regulation of autophagy in drug-resistant leukemia cells, and this RNP is therefore a potential target of clinical drug-resistance treatment. 27278897_HnRNP K is a promising tissue biomarker for diagnosing gastric cancer. 27292014_These results establish the role of hnRNP K and PCPB1 in the translational control of morphine-induced MOR expression in human neuroblastoma (NMB) cells as well as cells stably expressing MOR (NMB1). 27424288_Nujiangexathone A, a novel compound from Garcinia nujiangensis, down-regulates hnRNPK levels in cervical tumor cells, inducing cell cycle arrest. 27793696_KRAS-mutant colorectal carcinoma shows intrinsic radioresistance along with rapid upregulation of hnRNP K in response to ionizing radiation that can effectively be targeted by MEK inhibition. 27862976_we discovered that hnRNPK plays an important role in bladder cancer, suggesting that it is a potential prognostic marker and a promising target for treating bladder cancer. 28228215_The hnRNPK positively regulates the level of prostate tumor overexpressed 1-antisense 1 which harbors five binding sites for miR-1207-5p. The knockdown of hnRNPK or PTOV1-AS1 increased the enrichment of heme oxygenase-1 mRNA in miR-1207-5p-mediated miRNA-induced silencing complex and thus suppressed the expression of heme oxygenase-1. 28335083_Down-regulation of DAB2IP correlated negatively with hnRNPK and MMP2 expressions in CRC tissues. In conclusion, our study elucidates a novel mechanism of the DAB2IP/hnRNPK/MMP2 axis in the regulation of CRC invasion and metastasis, which may be a potential therapeutic target. 28423622_High HNRNPK expression is associated with pancreatic cancer. 28426877_Tumor cells bearing a p53 mutation showed increased damage levels and delayed repair. Knockdown of hnRNPK applied simultaneously with irradiation reduced colony-forming ability and survival of tumor cells. Taken together, our data shows that hnRNPK is a relevant modifier of DNA damage repair and tumor cell survival. We therefore recommend further studies to evaluate the potential of hnRNPK as a drug target for improvement 28592492_Data suggest that hnRNPK plays role in heat shock response of cells by regulating HSF1; hnRNPK inhibits HSF1 activity, resulting in reduced expression of HSP27 and HSP70 mRNAs; hnRNPK also down-regulates binding of HSF1 to heat shock response element. (hnRNPK = heterogeneous-nuclear ribonucleoprotein K; HSF1 = heat shock transcription factor 1; HSP = heat-shock protein) 28708135_hnRNPK regulates PLK1 expression by competing with the PLK1-targeting miRNAs, miR-149-3p and miR-193b-5p. 28869607_Study findings underscore the biological significance of MRPL33-L and hnRNPK in the tumor formation and identifies hnRNPK as a critical splicing regulator of MRPL33 pre-mRNA in cancer cells. 29409808_Study found that linc00460 physically and specifically interacts with hnRNPK which knockdown diminishes the migratory and invasive capacity of lung cancer cells. 29921878_Data show that heterogeneous nuclear ribonucleoprotein K (hnRNP K) and influenza virus NS1A binding protein (NS1-BP) regulate host splicing events and that viral infection causes mis-splicing of some of these transcripts. 30106132_hnRNPK modulated selective quality-control autophagy modulated selective quality-control autophagy.HnRNPK regulates the expression of HDAC6. 30144205_results position inactivation of HNRNPK and SOCS1 as potential driver events in mycosis fungoides development 30372559_Study revealed a novel lncRNA with a positive function on BM-MSC osteogenic differentiation and proposed a new interaction between hnRNPK and lncRNA. 30397178_As a novel regulator, lnc-LBCS plays an important tumor-suppressor role in BCSCs' self-renewal and chemoresistance, contributing to weak tumorigenesis and enhanced chemosensitivity. The lnc-LBCS-hnRNPK-EZH2-SOX2 regulatory axis may represent a therapeutic target for clinical intervention in chemoresistant bladder cancer. 30444036_This study demonstrates the significance of O-GlcNAcylation on the nuclear translocation of hnRNP-K and its impact on the progression of cholangiocarcinoma. 30468106_this study is the first to identify hnRNP F/H and K as regulators of Mcl-1 alternative splicing. 30771276_data unveiled a new mechanism of regulation of the gelsolin expression by hnRNPK and provides new clues for the discovery of new anti-metastatic therapy 30793470_We studied the HNRNPK gene by Sanger sequencing, and identified a novel splicing variant. We suggest that Okamoto syndrome is identical to Au-Kline syndrome. 30836866_Studies indicate that several post-translational modifications (PTMs) possibly play an important role in modulating heterogeneous nuclear ribonucleoprotein K (hnRNPK) function [Review]. 31061501_Study found that CBFB binds to and enhances the translation of RUNX1 mRNA, which encodes the binding partner of CBFB. CBFB binds and regulates the translation of hundreds of mRNAs through HNRNPK and facilitate translation initiation by eIF4B. Data propose that breast cancer cells evade translation and transcription surveillance simultaneously through downregulating CBFB. 31110205_hnRNPK S379 phosphorylation participates in migration regulation of triple negative MDA-MB-231 cells. 31121493_the upregulated transcriptional activity of HNRNP-K mediated by SERPINA3 promotes HCC cell survival and proliferation and could be an indicator of poor prognosis for HCC patients. 31127648_Role of heterogeneous nuclear ribonucleoprotein K in tumor development. 31178390_The results identify a novel ERK/hnRNPK/DDX3X pathway that influences beta cell survival and is activated under conditions associated with T2D. 31187136_Results suggest that Yes-associated protein 1 (YAP) could be one of the effectors of heterogeneous nuclear ribonucleoprotein K (hnRNPK). 31221168_Study elucidated that LBCS guided hnRNPK to exert its function by directly interacting with the 5'-UTR region of AR mRNA in castration resistant prostate cancer cells. Moreover, the inhibitive effect of LBCS on AR translation was in hnRNPK dependent manner. 31279651_The hnRNPK-lncRNAs interaction is potentially implicated in various pathogenic disorders including tumorigenesis, and Kabuki-like, Au-Kline, and Okamoto syndromes. 31395945_Results provide evidence that HnRNPK mediates the ZRS variant regulating the transcriptional activity of SHH. 31492158_Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is highly expressed in non-small-cell lung cancer (NSCLC), and NSCLC with higher expression of hnRNP K are more frequently rated as high-grade tumors with poor outcome. hnRNP K increases microtubule stability via interacting with microtubule-associated protein 1B light chain and is associated with acetylated a-tubulin during epithelial to mesenchymal transion. 31907279_Taken together, the results show rhythmic protein expression of hnRNP K and provide new insights into its function as a transcriptional amplifier of D site-binding protein. 31998294_LncSSBP1 Functions as a Negative Regulator of IL-6 Through Interaction With hnRNPK in Bronchial Epithelial Cells Infected With Talaromyces marneffei. 32065448_Circular RNA (circ-0075804) promotes the proliferation of retinoblastoma via combining heterogeneous nuclear ribonucleoprotein K (HNRNPK) to improve the stability of E2F transcription factor 3 E2F3. 32385154_RNA-binding motifs of hnRNP K are critical for induction of antibody diversification by activation-induced cytidine deaminase. 32449427_Regulation of the p53 expression profile by hnRNP K under stress conditions. 32581104_hnRNP K Is a Novel Internal Ribosomal Entry Site-Transacting Factor That Negatively Regulates Foot-and-Mouth Disease Virus Translation and Replication and Is Antagonized by Viral 3C Protease. 32866608_Lnc-FAM84B-4 acts as an oncogenic lncRNA by interacting with protein hnRNPK to restrain MAPK phosphatases-DUSP1 expression. 32901844_Heterogeneous nuclear ribonucleoprotein K is overexpressed and contributes to radioresistance irrespective of HPV status in head and neck squamous cell carcinoma. 33619115_A TNFR2-hnRNPK Axis Promotes Primary Liver Cancer Development via Activation of YAP Signaling in Hepatic Progenitor Cells. 33731207_CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated beta-catenin stabilization. 33806648_Heterogeneous Nuclear Ribonucleoprotein K Is Involved in the Estrogen-Signaling Pathway in Breast Cancer. 33931969_Caveolin-1-driven membrane remodelling regulates hnRNPK-mediated exosomal microRNA sorting in cancer. 34314754_SGLT2 promotes pancreatic cancer progression by activating the Hippo signaling pathway via the hnRNPK-YAP1 axis. 34452628_Circ-GALNT16 restrains colorectal cancer progression by enhancing the SUMOylation of hnRNPK. 34575922_Arginine Methylation of hnRNPK Inhibits the DDX3-hnRNPK Interaction to Play an Anti-Apoptosis Role in Osteosarcoma Cells. 34621897_Mass Spectrometry and Computer Simulation Predict the Interactions of AGPS and HNRNPK in Glioma. 34742742_Long noncoding RNA AK023096 interacts with hnRNP-K and contributes to the maintenance of self-renewal in bladder cancer stem-like cells. 34914972_HnRNPK and lysine specific histone demethylase-1 regulates IP-10 mRNA stability in monocytes. 35091468_LCDR regulates the integrity of lysosomal membrane by hnRNP K-stabilized LAPTM5 transcript and promotes cell survival. 35342346_PROX1 promotes breast cancer invasion and metastasis through WNT/beta-catenin pathway via interacting with hnRNPK. 35352475_Heterogeneous nuclear ribonucleoprotein K promotes the progression of lung cancer by inhibiting the p53-dependent signaling pathway. | ENSMUSG00000021546 | Hnrnpk | 2.073227e+04 | 0.8332075 | -0.263252275 | 0.2729019 | 9.462326e-01 | 0.3306801149 | 0.82505234 | No | Yes | 2.184102e+04 | 3917.347818 | 2.010910e+04 | 3698.983349 | |
| ENSG00000165152 | 84302 | PGAP4 | protein_coding | Q9BRR3 | FUNCTION: Golgi-resident glycosylphosphatidylinositol (GPI)-N-acetylgalactosamine transferase involved in the lipid remodeling steps of GPI-anchor maturation. Lipid remodeling steps consist in the generation of 2 saturated fatty chains at the sn-2 position of GPI-anchors proteins (PubMed:29374258). Required for the initial step of GPI-GalNAc biosynthesis, transfers GalNAc to GPI in the Golgi after fatty acid remodeling by PGAP2 (PubMed:29374258). {ECO:0000269|PubMed:29374258}. | Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Transferase;Transmembrane;Transmembrane helix | hsa:84302; | Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; glycosyltransferase activity [GO:0016757]; GPI anchor biosynthetic process [GO:0006506] | ENSMUSG00000039611 | Pgap4 | 1.688456e+03 | 1.2115750 | 0.276883673 | 0.3001275 | 8.581906e-01 | 0.3542455300 | 0.82969965 | No | Yes | 1.661979e+03 | 140.070939 | 1.360026e+03 | 117.648740 | |||
| ENSG00000165209 | 55342 | STRBP | protein_coding | Q96SI9 | FUNCTION: Involved in spermatogenesis and sperm function. Plays a role in regulation of cell growth. Binds to double-stranded DNA and RNA. Binds most efficiently to poly(I:C) RNA than to poly(dI:dC) DNA. Binds also to single-stranded poly(G) RNA. Binds non-specifically to the mRNA PRM1 3'-UTR and adenovirus VA RNA (By similarity). {ECO:0000250}. | 3D-structure;Alternative splicing;Cytoplasm;DNA-binding;Developmental protein;Differentiation;Methylation;RNA-binding;Reference proteome;Repeat;Spermatogenesis | hsa:55342; | cytoplasm [GO:0005737]; manchette [GO:0002177]; nucleus [GO:0005634]; DNA binding [GO:0003677]; double-stranded RNA binding [GO:0003725]; microtubule binding [GO:0008017]; RNA binding [GO:0003723]; single-stranded RNA binding [GO:0003727]; mechanosensory behavior [GO:0007638]; spermatid development [GO:0007286] | 22391137_Low STRBP mRNA is associated with cryptorchidism and Down's syndrome. | ENSMUSG00000026915 | Strbp | 9.353687e+02 | 1.0547618 | 0.076917219 | 0.3119251 | 6.035477e-02 | 0.8059360877 | 0.96141221 | No | Yes | 9.599262e+02 | 158.519646 | 8.109619e+02 | 137.398444 | ||
| ENSG00000165219 | 26130 | GAPVD1 | protein_coding | Q14C86 | FUNCTION: Acts both as a GTPase-activating protein (GAP) and a guanine nucleotide exchange factor (GEF), and participates in various processes such as endocytosis, insulin receptor internalization or LC2A4/GLUT4 trafficking. Acts as a GEF for the Ras-related protein RAB31 by exchanging bound GDP for free GTP, leading to regulate LC2A4/GLUT4 trafficking. In the absence of insulin, it maintains RAB31 in an active state and promotes a futile cycle between LC2A4/GLUT4 storage vesicles and early endosomes, retaining LC2A4/GLUT4 inside the cells. Upon insulin stimulation, it is translocated to the plasma membrane, releasing LC2A4/GLUT4 from intracellular storage vesicles. Also involved in EGFR trafficking and degradation, possibly by promoting EGFR ubiquitination and subsequent degradation by the proteasome. Has GEF activity for Rab5 and GAP activity for Ras. {ECO:0000269|PubMed:16410077}. | Alternative splicing;Endocytosis;Endosome;GTPase activation;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome | hsa:26130; | cytosol [GO:0005829]; endosome [GO:0005768]; plasma membrane [GO:0005886]; cadherin binding [GO:0045296]; GTPase activating protein binding [GO:0032794]; guanyl-nucleotide exchange factor activity [GO:0005085]; endocytosis [GO:0006897]; regulation of GTPase activity [GO:0043087]; regulation of protein transport [GO:0051223]; signal transduction [GO:0007165] | 17545148_EGF-stimulated receptor ubiquitination and trafficking are mediated via GAPex-5: GAPex-5-mediated EGFR ubiquitination is independent of Rab5 activation 31358736_The findings suggest that SPIN90, as an adaptor protein, simultaneously binds inactive Rab5 and Gapex5, thereby altering their spatial proximity and facilitating Rab5 activation. 32321936_CRISPR-mediated gene targeting of CK1delta/epsilon leads to enhanced understanding of their role in endocytosis via phosphoregulation of GAPVD1. 33548618_An interdependence between GAPVD1 gene polymorphism, expression level and response to interferon beta in patients with multiple sclerosis. 33917494_Phosphorylation of GAPVD1 Is Regulated by the PER Complex and Linked to GAPVD1 Degradation. | ENSMUSG00000026867 | Gapvd1 | 1.502363e+03 | 0.9194503 | -0.121156509 | 0.3121778 | 1.492286e-01 | 0.6992736151 | 0.93592030 | No | Yes | 1.206884e+03 | 204.514347 | 1.173319e+03 | 203.904259 | ||
| ENSG00000165283 | 30968 | STOML2 | protein_coding | Q9UJZ1 | FUNCTION: Mitochondrial protein that probably regulates the biogenesis and the activity of mitochondria. Stimulates cardiolipin biosynthesis, binds cardiolipin-enriched membranes where it recruits and stabilizes some proteins including prohibitin and may therefore act in the organization of functional microdomains in mitochondrial membranes. Through regulation of the mitochondrial function may play a role into several biological processes including cell migration, cell proliferation, T-cell activation, calcium homeostasis and cellular response to stress. May play a role in calcium homeostasis through negative regulation of calcium efflux from mitochondria. Required for mitochondrial hyperfusion a pro-survival cellular response to stress which results in increased ATP production by mitochondria. May also regulate the organization of functional domains at the plasma membrane and play a role in T-cell activation through association with the T-cell receptor signaling complex and its regulation. {ECO:0000269|PubMed:17121834, ECO:0000269|PubMed:18641330, ECO:0000269|PubMed:19597348, ECO:0000269|PubMed:19944461, ECO:0000269|PubMed:21746876, ECO:0000269|PubMed:22623988}. | Acetylation;Alternative splicing;Cell membrane;Coiled coil;Cytoplasm;Cytoskeleton;Direct protein sequencing;Lipid-binding;Lipoprotein;Membrane;Mitochondrion;Mitochondrion inner membrane;Phosphoprotein;Reference proteome;Transit peptide | hsa:30968; | cytoskeleton [GO:0005856]; extrinsic component of plasma membrane [GO:0019897]; membrane raft [GO:0045121]; mitochondrial inner membrane [GO:0005743]; mitochondrial intermembrane space [GO:0005758]; cardiolipin binding [GO:1901612]; GTPase binding [GO:0051020]; signaling receptor binding [GO:0005102]; CD4-positive, alpha-beta T cell activation [GO:0035710]; cellular calcium ion homeostasis [GO:0006874]; lipid localization [GO:0010876]; mitochondrial ATP synthesis coupled proton transport [GO:0042776]; mitochondrial calcium ion transmembrane transport [GO:0006851]; mitochondrial protein processing [GO:0034982]; mitochondrion organization [GO:0007005]; positive regulation of cardiolipin metabolic process [GO:1900210]; positive regulation of interleukin-2 production [GO:0032743]; positive regulation of mitochondrial DNA replication [GO:0090297]; positive regulation of mitochondrial membrane potential [GO:0010918]; protein complex oligomerization [GO:0051259]; stress-induced mitochondrial fusion [GO:1990046]; T cell receptor signaling pathway [GO:0050852] | 16533792_overexpression of stomatin-like protein 2 is associated with neoplasms 16671055_SLP-2 is a high abundance protein in several tissues and cells and may play an important biological role; this study uses mass spectrometry to analyze its primary structure 17342323_SLP-2 was overexpressed in endometrial adenocarcinoma compared with their normal counterparts. 17709317_High-level SLP-2 expression was associated with decreased overall survival (P = .011) and was more often found in patients with tumors larger than 20 mm, lymph node metastasis, advanced clinical stage, distant metastasis 18267007_Endosymbiotic origin of paraslipin from an alphaprotoebacterial ancestor (SLP-2) 18641330_SLP-2 is an important player in T cell activation by ensuring sustained TCR signaling 19767238_Familial MGUS and multiple myeloma were associated with a dominant inheritance of hyperphosphorylated paratarg-7 19839737_SLP-2 overexpression is associated with tumour distant metastasis and poor prognosis in pulmonary squamous cell carcinoma. 19944461_negatively modulates mitochondrial sodium-calcium exchange 20877624_Observational study of gene-disease association. (HuGE Navigator) 21209152_plasma concentrations of stomatin (EPB72)-like 2 in early-stage colorectal cancer patients were elevated as compared with those of healthy individuals 21220746_The dominant inheritance of hyperphosphorylated paratarg-7 explains cases of familial IgM monoclonal gammopathy of undetermined significance and Waldenstrom macroglobulinemia 21501885_[review] Stomatin family member STOML2 is oligomeric; it localizes mostly to membrane domains and has been shown to modulate ion channel activity. 21746876_We propose that the function of SLP-2 is to recruit prohibitins to cardiolipin to form cardiolipin-enriched microdomains in which electron transport complexes are optimally assembled. 22081131_SLP-2 and HER2/neu can play a role in lymph node/distant metastases of breast cancers 22158085_investigation of biomarkers for early diagnosis of endometriosis: Data suggest that SLP2, tropomyosin 3, and tropomodulin 3 are autoantigens present in blood of women with endometriosis; immunodominant epitopes were identified. 22623988_SLP-2 facilitates the compartmentalization not only of mitochondrial membranes but also of the plasma membrane into functional microdomains. 23028053_SLP-2 deficiency in T-lymphocytes is associated with abnormal cardiolipin compartmentalization in mitochondrial membranes, defects associated with altered mitochondrial respiration that is increasingly uncoupled from ATP production. 23371255_Increased levels of SLP-2 correlate with lymph node metastasis in gastric cancer. 23667687_Expression of SLP-2 is associated with invasion of esophageal squamous cell carcinoma. 23918306_SLP-2 may play an important role in human GBC tumorigenesis, and SLP-2 might serve as a novel prognostic marker in human GBC. 24190591_SLP-2 was upregulated by TGF-beta1, indicating a possible role of SLP-2 in Papillary thyroid cancer tumorigenesis. 24258357_STOML2 may have a role in progression of gastric adenocarcinoma 25695396_Our study showed that STOML-2 was negatively regulated by miR1207-5p in esophageal carcinoma 25973071_STOML2 was correlated to progression in cervical cancer, and implicated it as a potential predictive factor for the prognosis of cervical cancer. 26487491_Stomatin-like protein 2 is overexpressed in epithelial ovarian cancer and predicts poor patient survival 26750533_The significant association of SLP-2 overexpression with unfavorable clinicopathological characteristics and BRAFV600E mutation indicates that SLP-2 may have a role in aggressiveness of BRAF-mutated papillary thyroid carcinoma. 26932604_The expression of STOML2, a gene that plays a key role in mitochondrial function and T-cell activation, is associated with both IL-6 signaling and asthma risk. 27737933_results reveal an important role of SLP2 membrane scaffolds for the spatial organization of inner membrane proteases regulating mitochondrial dynamics, quality control, and cell survival. 27986413_Slp2 is involved in endometrial stromal cell proliferation and differentiation during decidualization in mice and humans 29364474_The downregulation of SLP-2 by siRNA inhibited cell proliferation, elevated caspase3 activity, and decreased CCBE1 expression 29516570_Data show that SLP-2 is upregulated by high cisplatin concentrations, leading to increased protein turnover. Overexpression of SLP-2 activates the MEK/ERK signaling pathway, and suppresses the mitochondrial apoptosis pathway, indicating that SLP-2 inhibits apoptosis by activating the MEK/ERK pathway and inhibiting the mitochondrial apoptosis pathway in cervical cancer cells. 29556045_SLP-2 promotes non-small cell lung cancer cell proliferation by enhancing survivin expression mediated through the TCF4/beta-catenin pathway. 29951933_TROP-2, SLP-2 and CD56 were effective diagnostic markers for PTC, especially when they were combined to use. 30359340_The knockdown of STOML2 significantly repressed the viability, migration, and invasion of LM3 cells. Authors observed that silencing STOML2 markedly downregulated the expression levels of MMP-2, MMP-9,MTA1, and nuclear factor kappa B (NF-kappaB), and upregulated levels of E-cadherin, tissue inhibitor of metalloproteinases 2 (TIMP2), and the inhibitor of kappa B (IkappaB). 30389319_that SLP-2 may predict a poor prognosis in colorectal cancer patients as a novel marker 30555578_These findings demonstrated a positive feedback loop of SLP2 which leads to acceleration of tumor progression and poor survival of gastric cancer patients. This finding also provided evidence for the reason of SLP2 elevation. 30888245_Hyperphosphorylated paratarg-7 carrier state is a strong molecularly defined risk factor for the development of Waldenstrom's macroglobulinaemia. 32141532_Clinical significance of SLP-2 in epithelial ovarian cancer and its regulatory effect on the Notch signaling pathway. 32814233_Stomatin-like protein 2 (SLP2) regulates the proliferation and invasion of trophoblast cells by modulating mitochondrial functions. 33412331_Characterization of the interactome of c-Src within the mitochondrial matrix by proximity-dependent biotin identification. 33446239_STOML2 potentiates metastasis of hepatocellular carcinoma by promoting PINK1-mediated mitophagy and regulates sensitivity to lenvatinib. 33846782_Stomatinlike protein 2 induces metastasis by regulating the expression of a ratelimiting enzyme of the hexosamine biosynthetic pathway in pancreatic cancer. 34088631_Stomatin-Like Protein-2: A Potential Target to Treat Mitochondrial Cardiomyopathy. | ENSMUSG00000028455 | Stoml2 | 1.401949e+04 | 0.9515802 | -0.071602803 | 0.3065177 | 5.446890e-02 | 0.8154618900 | 0.96373941 | No | Yes | 1.272396e+04 | 1080.281266 | 1.234004e+04 | 1074.568750 | ||
| ENSG00000165494 | 51585 | PCF11 | protein_coding | O94913 | FUNCTION: Component of pre-mRNA cleavage complex II. | 3D-structure;Acetylation;Coiled coil;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;mRNA processing | The protein encoded by this gene binds to CLP1 to form pre-mRNA cleavage factor IIm. The encoded protein is necessary for efficient Pol II transcription termination and may be involved in degradation of the 3' product of polyA site cleavage. [provided by RefSeq, Oct 2016]. | hsa:51585; | cytoplasm [GO:0005737]; mitochondrion [GO:0005739]; mRNA cleavage factor complex [GO:0005849]; nucleoplasm [GO:0005654]; mRNA binding [GO:0003729]; RNA polymerase II complex binding [GO:0000993]; mRNA cleavage [GO:0006379]; mRNA polyadenylation [GO:0006378]; termination of RNA polymerase II transcription [GO:0006369] | 11060040_Human PCf11 is a pre-mRNA processing factor. 17606639_Pcf11 can act as a negative elongation factor to repress RNA Pol II gene expression in eukaryotic cells. 18086705_Pcf11 is required for the efficient degradation of the 3' product of poly(A) site cleavage. 29196535_WNK1 and the associated phosphorylation of the PCF11 CID act to promote transcript release from chromatin-associated Pol II, which in turn facilitates mRNA export to the cytoplasm 30139799_We have reconstituted CF II as a heterodimer of hPcf11 and hClp1. The heterodimer is active in partially reconstituted cleavage reactions, whereas hClp1 by itself is not. Pcf11 moderately stimulates the RNA 5' kinase activity of hClp1; the kinase activity is dispensable for RNA cleavage 30552333_Utilising extensive RNAi-screening authors reveal the landscape and drivers of transcriptome 3'end-diversification, discovering PCF11 as critical regulator, directing alternative polyadenylation (APA) of hundreds of transcripts including a differentiation RNA-operon. 30819644_PCF11 selectively attenuates the expression of other transcriptional regulators by premature cleavage/polyadenylation and termination. 30840896_PCF11 is autoregulated through a conserved IPA site, the removal of which leads to global activation of PASs close to gene promotors. Therefore, PCF11 uses distinct mechanisms to regulate genes of different sizes, and its autoregulation maintains homeostasis of PAS usage in the cell. | ENSMUSG00000041328 | Pcf11 | 1.270348e+03 | 0.9364933 | -0.094659390 | 0.3040691 | 9.819275e-02 | 0.7540092738 | 0.94833080 | No | Yes | 1.220717e+03 | 192.232755 | 1.420323e+03 | 229.046605 | |
| ENSG00000165730 | 219736 | STOX1 | protein_coding | Q6ZVD7 | FUNCTION: Involved in regulating the levels of reactive oxidative species and reactive nitrogen species and in mitochondrial homeostasis in the placenta (PubMed:24738702). Required for regulation of inner ear epithelial cell proliferation via the AKT signaling pathway (By similarity). {ECO:0000250|UniProtKB:B2RQL2, ECO:0000269|PubMed:24738702}.; FUNCTION: [Isoform A]: Involved in cell cycle regulation by binding to the CCNB1 promoter, up-regulating its expression and promoting mitotic entry (PubMed:22253775). Induces phosphorylation of MAPT/tau (PubMed:22995177). {ECO:0000269|PubMed:22253775, ECO:0000269|PubMed:22995177}. | Activator;Alternative splicing;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;DNA-binding;Disease variant;Mitosis;Nucleus;Reference proteome;Transcription;Transcription regulation | The protein encoded by this gene may function as a DNA binding protein. Mutations in this gene are associated with pre-eclampsia/eclampsia 4 (PEE4). Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2009]. | hsa:219736; | cell cortex [GO:0005938]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; fibrillar center [GO:0001650]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; cell cycle [GO:0007049]; cell division [GO:0051301]; cellular response to nitrosative stress [GO:0071500]; inner ear development [GO:0048839]; negative regulation of gene expression [GO:0010629]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cyclin-dependent protein kinase activity [GO:1904031]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of G2/M transition of mitotic cell cycle [GO:0010971]; positive regulation of gene expression [GO:0010628]; positive regulation of otic vesicle morphogenesis [GO:1904120]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of peptidyl-threonine phosphorylation [GO:0010800]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of gene expression [GO:0010468]; regulation of mitochondrial DNA metabolic process [GO:1901858]; regulation of mitochondrial membrane potential [GO:0051881]; regulation of mitochondrion organization [GO:0010821]; regulation of response to oxidative stress [GO:1902882]; regulation of transcription from RNA polymerase II promoter in response to hypoxia [GO:0061418] | 17290274_In conclusion, we are unable to validate STOX1 as a common preeclampsia susceptibility gene. 17290274_Observational study of gene-disease association. (HuGE Navigator) 17617193_Observational study of gene-disease association. (HuGE Navigator) 19079545_Anomalies in STOX1 expression are associated with the onset of preeclampsia. 19577309_role in placental development and preeclampsia (Review) 20110611_The data of this study indicated that STOX1 controls a conserved pathway shared between placenta and brain with overexpression in late-onset Alzheimer's disease. 20400461_The risk allele (Y153H) of the preeclampsia susceptibility gene STOX1 negatively regulates trophoblast invasion by upregulation of the cell-cell adhesion protein a-T-catenin (CTNNA3). 20643876_STOX1 does not show differential expression in deciduas from pregnancies complicated by both pre-eclampsia and FGR as compared with controls. 20716964_methylation is independent of parental origin, but regulates STOX1 expression with the Y153H genotype directing the level of methylation. 21490791_STOX1 is one of the genes that give susceptibility for preeclampsia. 21755018_Upregulation of total tau expression (SFRS7-independent) and tau exon 10 splicing (SFRS7-dependent), as shown in this study to be both affected by STOX1A, is known to have implications in neurodegeneration. 22253775_Mitotic entry is enhanced through the direct upregulation of cyclin B1 expression effectuated by STOX1A. 22728895_Transcription factor STOX1A and its target gene, CNTNAP2, are potentially involved in the etiology of Alzheimer's disease. 22995177_A STOX1A-dependent effect on tau phosphorylation found in neurodegenerative diseases such as Alzheimer's disease. 26758611_The STOX1 mice could help to better understand the endothelial dysfunction in the context of preeclampsia, and guide the search for efficient therapies able to protect the maternal endothelium during the disease and its aftermath. 30955313_expressions of STOX1 is gradually increasing along with the normal pregnancy progression 31189268_STOX1 protein in early onset group, late onset group and control group were 0.78+/-0.04,0.59+/-0.020 and 0.54+/-0.018 respectively, which is higher in early onset group than that in late onset group(P<0.05) 31724315_Investigation of the STOX1 polymorphism on lumbar disc herniation. 32534058_STOX1 gene single nucleotide polymorphism is associated with early-onset preeclampsia. 32807495_circ-ZUFSP regulates trophoblasts migration and invasion through sponging miR-203 to regulate STOX1 expression. 33356399_Functional Evaluation of STOX1 (STORKHEAD-BOX PROTEIN 1) in Placentation, Preeclampsia, and Preterm Birth. | ENSMUSG00000036923 | Stox1 | 9.551774e+01 | 0.7499847 | -0.415066935 | 0.4151259 | 9.907101e-01 | 0.3195688908 | 0.81995823 | No | Yes | 9.624936e+01 | 19.248549 | 1.131632e+02 | 22.944718 | |
| ENSG00000165732 | 9188 | DDX21 | protein_coding | Q9NR30 | FUNCTION: RNA helicase that acts as a sensor of the transcriptional status of both RNA polymerase (Pol) I and II: promotes ribosomal RNA (rRNA) processing and transcription from polymerase II (Pol II) (PubMed:25470060, PubMed:28790157). Binds various RNAs, such as rRNAs, snoRNAs, 7SK and, at lower extent, mRNAs (PubMed:25470060). In the nucleolus, localizes to rDNA locus, where it directly binds rRNAs and snoRNAs, and promotes rRNA transcription, processing and modification. Required for rRNA 2'-O-methylation, possibly by promoting the recruitment of late-acting snoRNAs SNORD56 and SNORD58 with pre-ribosomal complexes (PubMed:25470060, PubMed:25477391). In the nucleoplasm, binds 7SK RNA and is recruited to the promoters of Pol II-transcribed genes: acts by facilitating the release of P-TEFb from inhibitory 7SK snRNP in a manner that is dependent on its helicase activity, thereby promoting transcription of its target genes (PubMed:25470060). Functions as cofactor for JUN-activated transcription: required for phosphorylation of JUN at 'Ser-77' (PubMed:11823437, PubMed:25260534). Can unwind double-stranded RNA (helicase) and can fold or introduce a secondary structure to a single-stranded RNA (foldase) (PubMed:9461305). Together with SIRT7, required to prevent R-loop-associated DNA damage and transcription-associated genomic instability: deacetylation by SIRT7 activates the helicase activity, thereby overcoming R-loop-mediated stalling of RNA polymerases (PubMed:28790157). Involved in rRNA processing (PubMed:14559904, PubMed:18180292). May bind to specific miRNA hairpins (PubMed:28431233). Component of a multi-helicase-TICAM1 complex that acts as a cytoplasmic sensor of viral double-stranded RNA (dsRNA) and plays a role in the activation of a cascade of antiviral responses including the induction of proinflammatory cytokines via the adapter molecule TICAM1 (By similarity). {ECO:0000250|UniProtKB:Q9JIK5, ECO:0000269|PubMed:11823437, ECO:0000269|PubMed:14559904, ECO:0000269|PubMed:18180292, ECO:0000269|PubMed:25260534, ECO:0000269|PubMed:25470060, ECO:0000269|PubMed:25477391, ECO:0000269|PubMed:28431233, ECO:0000269|PubMed:28790157, ECO:0000269|PubMed:9461305}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Antiviral defense;Cytoplasm;Direct protein sequencing;Helicase;Hydrolase;Immunity;Innate immunity;Isopeptide bond;Mitochondrion;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Transcription;Ubl conjugation;rRNA processing | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which is an antigen recognized by autoimmune antibodies from a patient with watermelon stomach disease. This protein unwinds double-stranded RNA, folds single-stranded RNA, and may play important roles in ribosomal RNA biogenesis, RNA editing, RNA transport, and general transcription. [provided by RefSeq, Jul 2008]. | hsa:9188; | B-WICH complex [GO:0110016]; chromosome [GO:0005694]; cytosol [GO:0005829]; membrane [GO:0016020]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; 7SK snRNA binding [GO:0097322]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; double-stranded RNA binding [GO:0003725]; identical protein binding [GO:0042802]; miRNA binding [GO:0035198]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; rRNA binding [GO:0019843]; snoRNA binding [GO:0030515]; defense response to virus [GO:0051607]; innate immune response [GO:0045087]; osteoblast differentiation [GO:0001649]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of myeloid dendritic cell cytokine production [GO:0002735]; positive regulation of transcription by RNA polymerase III [GO:0045945]; R-loop disassembly [GO:0062176]; response to exogenous dsRNA [GO:0043330]; rRNA processing [GO:0006364]; transcription by RNA polymerase II [GO:0006366] | 14559904_RNA helicase II/Gualpha silencing inhibits mammalian ribosomal RNA production 16045751_the function of Gu(alpha)in rRNA processing is at least partially dependent on its ability to interact with ribosomal protein L4. 16385451_Observational study of gene-disease association. (HuGE Navigator) 18180292_in addition to its transcriptional effects, c-Jun regulates rRNA processing and nucleolar compartmentalization of the rRNA processing protein DDX21 20873769_Studies indicated that DDX21, HNRNPC, and RCC2 were isolated from Ku86 multicomponent complex in response to DNA damage. 23419719_Data indicate that DDX21, a nucleolar protein, was confirmed to associate with SET8. 24721576_As sequential interaction of PB1 and NS1 with DDX21 leads to temporal regulation of viral gene expression, influenza A virus likely uses the DDX21-NS1 interaction not only to overcome restriction, but also to regulate the viral life cycle. 25260534_DDX21 expression in breast cancer cells can promote AP-1 activity and rRNA processing, and thus, promote tumorigenesis by two independent mechanisms. 25470060_results uncover the multifaceted role of DDX21 in multiple steps of ribosome biogenesis, and provide evidence implicating a mammalian RNA helicase in RNA modification and Pol II elongation control 25477391_Identification of several late-acting snoRNAs that bind pre-40S particles in human cells and show that their association and function in pre-40S complexes is regulated by the RNA helicase DDX21. 27033607_In dengue virus infected cells, DDX21 translocates from nucleus to cytoplasm to active the innate immune response and thus inhibits DENV replication in the early stages of infection. 28472472_DDX21 can suppress the expression of proteins with G4 qudruplexes in the 3 UTR of its mRNA. 28475895_Results report the biogenesis and function of a box H/ACA snoRNA-ended sno-lncRNA, referred to SLERT (snoRNA-ended lncRNA enhances pre-ribosomal RNA transcription). SLERT is different from Prader-Willi Syndrome (PWS) sno-lncRNAs and plays a crucial role in rRNA biogenesis by dislodging a previously unknown clamp of DDX21 ring-shaped arrangements on Pol I complexes, thereby liberating Pol I for active rRNA transcription. 28705764_DDX21 is both an ATP-dependent and ATP-independent helicase, and both ATPase and ATP-dependent helicase activities are inhibited by Rev in a dose-dependent manner, although ATP-independent helicase activity is not. A conserved binding interaction between DDX protein's DEAD domain and Rev was identified, with Rev's nuclear diffusion inhibitory signal motif playing a significant role in binding. 28790157_Knockdown of SIRT7 leads to the same phenotype as depletion of DDX21 (i.e., increased formation of R loops and DNA double-strand breaks), indicating that SIRT7 and DDX21 cooperate to prevent R-loop accumulation, thus safeguarding genome integrity. 29906500_can drastically reduce DDX21's affinity for quadruplex, indicating that the recognition of quadruplex and specificity for telomeric repeat containing RNA quadruplex is mediated by interactions with the 2'OH of loop nucleotides 30165191_Data indicate that DEAD-box helicase 21 (DDX21) regulated Snail transcription factors (Snail) expression independent of its helicase activity. 30322617_DDX21 induced gastric cancer cell growth by up-regulating levels of Cyclin D1 and CDK2. 31251802_The findings revealed that enhancer-mediated enrichment of novel JMJD3-DDX21 interaction at ENPP2 locus is necessary for nascent transcript synthesis via the resolution of aberrant R-loops formation in response to inflammatory stimulus. 31351877_Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. 31554690_DDX21 knockdown prevented viral late gene transcription and consequently impaired HCMV replication. 31653714_Our work identifies the role of DDX21 in regulation at the translational level through biologically relevant RNA G-quadruplex (rG4) and shows that MAGED2 protein levels are regulated, at least in part, by the potential to form rG4 in their 5'-UTRs. 32231306_RNA helicase DDX21 mediates nucleotide stress responses in neural crest and melanoma cells. 32652076_PRL3-DDX21 Transcriptional Control of Endolysosomal Genes Restricts Melanocyte Stem Cell Differentiation. 33328538_DEAD-box RNA helicase protein DDX21 as a prognosis marker for early stage colorectal cancer with microsatellite instability. 33497018_The RNA-helicase DDX21 upregulates CEP55 expression and promotes neuroblastoma. 33604619_DDX21 interacts with nuclear AGO2 and regulates the alternative splicing of SMN2. 33754909_Identification of Prognostic RBPs in Osteosarcoma. 34125604_Caspase-Dependent Cleavage of DDX21 Suppresses Host Innate Immunity. 34326237_lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. 34578346_DDX21, a Host Restriction Factor of FMDV IRES-Dependent Translation and Replication. 34688145_Identification of MDM2, YTHDF2 and DDX21 as potential biomarkers and targets for treatment of type 2 diabetes. 34903139_Downregulation of DEAD-box helicase 21 (DDX21) inhibits proliferation, cell cycle, and tumor growth in colorectal cancer via targeting cell division cycle 5-like (CDC5L). 34987058_The Roles of RNA Helicases in DNA Damage Repair and Tumorigenesis Reveal Precision Therapeutic Strategies. | ENSMUSG00000020075 | Ddx21 | 3.570800e+03 | 0.9231523 | -0.115359413 | 0.3383487 | 1.150057e-01 | 0.7345156108 | 0.94481142 | No | Yes | 3.800967e+03 | 783.501254 | 3.345707e+03 | 707.385927 | |
| ENSG00000165886 | 80019 | UBTD1 | protein_coding | Q9HAC8 | FUNCTION: May be involved in the regulation of cellular senescence through a positive feedback loop with TP53. Is a TP53 downstream target gene that increases the stability of TP53 protein by promoting the ubiquitination and degradation of MDM2. {ECO:0000269|PubMed:25382750}. | Reference proteome | The degradation of many proteins is carried out by the ubiquitin pathway in which proteins are targeted for degradation by covalent conjugation of the polypeptide ubiquitin. This gene encodes a protein that belongs to the ubiquitin family of proteins. The encoded protein is thought to regulate E2 ubiquitin conjugating enzymes belonging to the UBE2D family. [provided by RefSeq, Mar 2014]. | hsa:80019; | 24211586_Biochemical characterization of recombinant UBTD1 and UBE2D demonstrated that the two proteins form a stable, stoichiometric complex that can be purified to near homogeneity. 25382750_Establish UBTD1 as a regulator of cellular senescence that mediates p53 function, and provide insights into the mechanism of Mdm2 inhibition that impacts p53 dynamics during cellular senescence and tumourigenesis. 30804013_low levels of UBTD1 are associated with poor patient survival, suggesting that biological functions of UBTD1 could be beneficial in limiting cancer progression. 32483419_CXCR4 mediates matrix stiffness-induced downregulation of UBTD1 driving hepatocellular carcinoma progression via YAP signaling pathway. 33884955_UBTD1 regulates ceramide balance and endolysosomal positioning to coordinate EGFR signaling. | ENSMUSG00000025171 | Ubtd1 | 9.053387e+02 | 1.5733539 | 0.653843262 | 0.3399715 | 3.693013e+00 | 0.0546408303 | 0.68299703 | No | Yes | 1.135837e+03 | 191.891549 | 5.799669e+02 | 101.193590 | ||
| ENSG00000166012 | 79101 | TAF1D | protein_coding | Q9H5J8 | FUNCTION: Component of the transcription factor SL1/TIF-IB complex, which is involved in the assembly of the PIC (preinitiation complex) during RNA polymerase I-dependent transcription. The rate of PIC formation probably is primarily dependent on the rate of association of SL1/TIF-IB with the rDNA promoter. SL1/TIF-IB is involved in stabilization of nucleolar transcription factor 1/UBTF on rDNA. Formation of SL1/TIF-IB excludes the association of TBP with TFIID subunits. {ECO:0000269|PubMed:15970593, ECO:0000269|PubMed:17318177}. | DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | TAF1D is a member of the SL1 complex, which includes TBP (MIM 600075) and TAF1A (MIM 604903), TAF1B (MIM 604904), and TAF1C (MIM 604905), and plays a role in RNA polymerase I transcription (Wang et al., 2004 [PubMed 15520167]; Gorski et al., 2007 [PubMed 17318177]).[supplied by OMIM, Jun 2009]. | hsa:79101; | centriolar satellite [GO:0034451]; cytosol [GO:0005829]; mitotic spindle [GO:0072686]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; RNA polymerase transcription factor SL1 complex [GO:0005668]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; regulation of transcription, DNA-templated [GO:0006355] | 15520167_An antigrowth activity and modulations of cell cycle events are identified in cells expressing siRNAMGC5306. 17318177_TAF(I)41 (MGC5306) is integral to transcriptionally active SL1 and plays a role in Pol I recruitment and, therefore, preinitiation complex formation in vivo. | ENSMUSG00000031939 | Taf1d | 1.375946e+03 | 0.8057021 | -0.311681575 | 0.3567448 | 7.375428e-01 | 0.3904491444 | 0.84317192 | No | Yes | 1.014354e+03 | 193.761509 | 1.337561e+03 | 261.726900 | |
| ENSG00000166105 | 112937 | GLB1L3 | protein_coding | Q8NCI6 | Alternative splicing;Glycosidase;Hydrolase;Reference proteome | hsa:112937; | vacuole [GO:0005773]; beta-galactosidase activity [GO:0004565]; carbohydrate metabolic process [GO:0005975] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000031966 | Glb1l3 | 1.334471e+02 | 0.7898427 | -0.340362737 | 0.3525571 | 9.295404e-01 | 0.3349823814 | 0.82561558 | No | Yes | 6.976499e+01 | 16.376691 | 1.033081e+02 | 24.607995 | |||
| ENSG00000166181 | 8539 | API5 | protein_coding | Q9BZZ5 | FUNCTION: Antiapoptotic factor that may have a role in protein assembly. Negatively regulates ACIN1. By binding to ACIN1, it suppresses ACIN1 cleavage from CASP3 and ACIN1-mediated DNA fragmentation. Also known to efficiently suppress E2F1-induced apoptosis. Its depletion enhances the cytotoxic action of the chemotherapeutic drugs. {ECO:0000269|PubMed:10780674, ECO:0000269|PubMed:17112319, ECO:0000269|PubMed:19387494}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Cytoplasm;Direct protein sequencing;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat | This gene encodes an apoptosis inhibitory protein whose expression prevents apoptosis after growth factor deprivation. This protein suppresses the transcription factor E2F1-induced apoptosis and also interacts with, and negatively regulates Acinus, a nuclear factor involved in apoptotic DNA fragmentation. Its depletion enhances the cytotoxic action of the chemotherapeutic drugs. Multiple alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Aug 2011]. | hsa:8539; | cytoplasm [GO:0005737]; membrane [GO:0016020]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; spliceosomal complex [GO:0005681]; fibroblast growth factor binding [GO:0017134]; RNA binding [GO:0003723]; apoptotic process [GO:0006915]; negative regulation of apoptotic process [GO:0043066]; negative regulation of fibroblast apoptotic process [GO:2000270] | 17112319_Apoptosis inhibitor-5/antiapoptosis clone-11 (Api5/Aac11) is a critical determinant of dE2F1-induced apoptosis in vivo and in vitro. 18319255_define a true in vivo target of miR-224 and reaffirm the important role of miRNAs in the dysregulation of cellular processes that may ultimately lead to tumorigenesis 19387494_The authors report here that AAC-11, a survival protein whose expression prevents apoptosis that occurs on deprivation of growth factors, physiologically binds to Acinus and prevents Acinus-mediated DNA fragmentation. 19821157_we infer that Pim-2 could activate API-5 to inhibit the apoptosis of liver cells, and NF-kappaB is the key regulator 20671611_The prognostic significance of the genes casein kinase 2 alpha subunit (CSNK2A1), anti-apoptosis clone-11 (AAC-11), and tumor metastasis suppressor NME1 in completely resected non-small cell lung cancer (NSCLC) patients, was analysed. 22334682_structure and function of API5 23940755_Api5, even if not physically interacting with E2F1, contributes positively to E2F1 transcriptional activity by increasing E2F1 binding to its target promoters. 24769442_API5 acts as an immune escape gene in tumors by rendering them resistant to apoptosis triggered by tumor antigen-specific T cells. 24785373_Low miR-224 and high API5 expression correlated with worse survival of glioblastoma patients. 25070070_API5 overexpression promoted cell proliferation and colony formation in CaSki cancer cells, whereas API5 knockdown inhibited the both properties in HeLa cells; API5 expression is associated with pERK1/2 in a subset of cervical cancer patients and its expression predicts poor overall survival. 25248562_Authors show that API5 increases the metastatic capacity of cervical cancer cells in vitro and in vivo via up-regulation of MMP-9. 25433291_Negative regulation of API-5 expression by miR-1 was demonstrated to promote apoptosis of HepG2 cells. 25477374_miR-143 and miR-145 and the predicted target proteins API5, ERK5, K-RAS, and IRS-1 display regional differences in expression in the colon 25819896_rs17684886 (ZNRF1) and rs599019 (COLEC12) are associated with diabetic retinopathy and rs6427247 (SCYL1BP1) and rs899036 (API5) are associated with severe diabetic retinopathy in Chinese patients with type 2 diabetes 26673663_Authors propose a proapoptotic role for NP in IAV pathogenesis, whereby it suppresses expression of antiapoptotic factor API5, thus potentiating the E2F1-dependent apoptotic pathway and ensuring viral replication. 28336776_study demonstrates that apoptosis inhibitor 5 is an endogenous and direct inhibitor of caspase-2. API5 protein directly binds to the caspase recruitment domain (CARD) of caspase-2 and impedes dimerization and activation of caspase-2. 32383752_Regulation of mRNA export through API5 and nuclear FGF2 interaction. 33319655_Evolution and Structure of API5 and Its Roles in Anti-Apoptosis. 34076992_MicroRNA-224 modulates chemosensitivity of breast cancer cells to docetaxel by apoptosis inhibitor 5. 34372697_DeSUMOylation of Apoptosis Inhibitor 5 by Avibirnavirus VP3 Supports Virus Replication. 34385547_Interplay between p300 and HDAC1 regulate acetylation and stability of Api5 to regulate cell proliferation. | ENSMUSG00000027193 | Api5 | 1.453473e+03 | 0.8458177 | -0.241581352 | 0.3476187 | 4.696310e-01 | 0.4931569937 | 0.87518872 | No | Yes | 1.556787e+03 | 355.171611 | 1.319046e+03 | 308.794329 | |
| ENSG00000166226 | 10576 | CCT2 | protein_coding | P78371 | FUNCTION: Component of the chaperonin-containing T-complex (TRiC), a molecular chaperone complex that assists the folding of proteins upon ATP hydrolysis (PubMed:25467444). The TRiC complex mediates the folding of WRAP53/TCAB1, thereby regulating telomere maintenance (PubMed:25467444). As part of the TRiC complex may play a role in the assembly of BBSome, a complex involved in ciliogenesis regulating transports vesicles to the cilia (PubMed:20080638). The TRiC complex plays a role in the folding of actin and tubulin (Probable). {ECO:0000269|PubMed:20080638, ECO:0000269|PubMed:25467444, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Chaperone;Cytoplasm;Direct protein sequencing;Isopeptide bond;Nucleotide-binding;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene is a molecular chaperone that is a member of the chaperonin containing TCP1 complex (CCT), also known as the TCP1 ring complex (TRiC). This complex consists of two identical stacked rings, each containing eight different proteins. Unfolded polypeptides enter the central cavity of the complex and are folded in an ATP-dependent manner. The complex folds various proteins, including actin and tubulin. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2010]. | hsa:10576; | azurophil granule lumen [GO:0035578]; cell body [GO:0044297]; chaperonin-containing T-complex [GO:0005832]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; microtubule [GO:0005874]; zona pellucida receptor complex [GO:0002199]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; protein folding chaperone [GO:0044183]; ubiquitin protein ligase binding [GO:0031625]; unfolded protein binding [GO:0051082]; binding of sperm to zona pellucida [GO:0007339]; chaperone mediated protein folding independent of cofactor [GO:0051086]; chaperone-mediated protein complex assembly [GO:0051131]; chaperone-mediated protein folding [GO:0061077]; positive regulation of establishment of protein localization to telomere [GO:1904851]; positive regulation of protein localization to Cajal body [GO:1904871]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomerase RNA localization to Cajal body [GO:1904874]; positive regulation of telomere maintenance via telomerase [GO:0032212]; protein folding [GO:0006457]; protein stabilization [GO:0050821]; scaRNA localization to Cajal body [GO:0090666]; toxin transport [GO:1901998] | 19332537_The chaperonin CCT is identified as a novel physiological substrate for p90 ribosomal S6 kinase (RSK) and p70 ribosomal S6 kinase (S6K). 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20573828_PB2 associates with CCT2 as a monomer and the CCT binding site is located in a central region of the PB2 protein. 23190606_Destruction of the beta-tubulin:CCT-beta complex provokes Hsp90-dependent protein ubiquitination and degradation. 23782473_Increased expression of CCT2 is associated with tumor progression and the clinical behavior of gallbladder carcinoma. 24375412_PDCD5 bound the apical domain of the CCTbeta subunit, projecting above the folding cavity without entering it. Like PDCD5, beta-tubulin also interacts with the CCTbeta apical domain, but a second site is found at the sensor loop deep within the folding cavity. 25704758_A role for the TRiC subunits TCP1 and CCT2, and potentially the entire TRiC complex, in breast cancer. 27645772_the novel LCA mutations in CCTbeta and the impact of chaperon disability by these mutations in cellular biology. 29483666_We applied whole-genome sequencing (WGS) on 9 trios where the probands are sporadically affected with the most severe form of the disorder and harbor no coding sequence variants affecting the function of known Hirschsprung disease (HSCR) genes. We found de novo protein-altering variants in three intolerant to change genes-CCT2, VASH1, and CYP26A1-for which a plausible link with the enteric nervous system (ENS) exists 31462707_we suggest that CCT2 correlates with Gli-1 expression and is an important determinant of survival in the colorectal cancer (CRC) patients. The results reveal that CCT2 can regulate the folding of Gli-1 in relation to hypoxia in CRC 31964905_Investigating Chaperonin-Containing TCP-1 subunit 2 as an essential component of the chaperonin complex for tumorigenesis. | ENSMUSG00000034024 | Cct2 | 4.609474e+03 | 0.6477194 | -0.626559048 | 0.2939417 | 4.430899e+00 | 0.0352939102 | 0.61027242 | No | Yes | 3.222813e+03 | 550.126734 | 5.454885e+03 | 954.292069 | |
| ENSG00000166233 | 25820 | ARIH1 | protein_coding | Q9Y4X5 | FUNCTION: E3 ubiquitin-protein ligase, which catalyzes ubiquitination of target proteins together with ubiquitin-conjugating enzyme E2 UBE2L3 (PubMed:15236971, PubMed:21532592, PubMed:24076655, PubMed:27565346, PubMed:23707686). Acts as an atypical E3 ubiquitin-protein ligase by working together with cullin-RING ubiquitin ligase (CRL) complexes and initiating ubiquitination of CRL substrates: associates with CRL complexes and specifically mediates addition of the first ubiquitin on CRLs targets (PubMed:27565346). The initial ubiquitin is then elongated by CDC34/UBE2R1 and UBE2R2 (PubMed:27565346). E3 ubiquitin-protein ligase activity is activated upon binding to neddylated cullin-RING ubiquitin ligase complexes (PubMed:24076655, PubMed:27565346). Plays a role in protein translation in response to DNA damage by mediating ubiquitination of EIF4E2, the consequences of EIF4E2 ubiquitination are however unclear (PubMed:25624349). According to a report, EIF4E2 ubiquitination leads to promote EIF4E2 cap-binding and protein translation arrest (PubMed:25624349). According to another report EIF4E2 ubiquitination leads to its subsequent degradation (PubMed:14623119). Acts as the ligase involved in ISGylation of EIF4E2 (PubMed:17289916). In vitro, controls the degradation of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex member SUN2 and may therefore have a role in the formation and localization of the LINC complex, and as a consequence, nuclear subcellular localization and nuclear morphology (PubMed:29689197). {ECO:0000269|PubMed:14623119, ECO:0000269|PubMed:15236971, ECO:0000269|PubMed:17289916, ECO:0000269|PubMed:21532592, ECO:0000269|PubMed:23707686, ECO:0000269|PubMed:24076655, ECO:0000269|PubMed:25624349, ECO:0000269|PubMed:27565346, ECO:0000269|PubMed:29689197}. | 3D-structure;Acetylation;Cytoplasm;Disease variant;Metal-binding;Nucleus;Reference proteome;Repeat;Transferase;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | hsa:25820; | Cajal body [GO:0015030]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; Lewy body [GO:0097413]; nuclear body [GO:0016604]; nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-like protein transferase activity [GO:0019787]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; ubiquitin-dependent protein catabolic process [GO:0006511] | 15236971_the structure of the RING2 from the RING-IBR-RING motif of HHARI and showed that RING2 has a completely distinct topology from classical RINGs 21532592_UBCH7 exhibits activity with the RING-in-between-RING (RBR) family of E3s that includes parkin (also known as PARK2) and human homologue of ariadne (HHARI; also known as ARIH1) 21590270_Both parkin and HHARI bind the same substrate proteins in cells and induce the formation of aggresomes. 23059369_HHARI is a marker of cellular proliferation associated with nuclear bodies. 24058416_Determined is the three-dimensional solution structure of the catalytic RING2 domain from HHARI. It shows glimpses of a HECT E3 ligase. 24076655_TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. 25624349_DNA damage induces an increase in ARIH1 protein levels and association of ARIH1 with 4EHP. In turn, this causes 4EHP recruitment to the mRNA cap, where it is known to compete with eIF4E. 28552575_Crystal structure of HHARI and UbcH7 reveals UbcH7 approximately Ub binding RING1 domain of auto-inhibited HHARI.HHARI UBA-L domain has Ub binding properties. 28790309_HHARI prevents spurious discharge of Ub from E2 to lysine residues by harboring structural elements that block E2 ~ Ub from adopting a 'closed' conformation. 28930681_ARIH1-mediated mitophagy promotes therapeutic resistance. 29689197_Variants in ARIH1 are associated with developing aortic aneurysms. 30587576_ubiquitin-associated (UBA) domain-containing DCNL1 is monoubiquitylated when bound to CRLs and that this monoubiquitylation depends on the CRL-associated Ariadne RBR ligases TRIAD1 (ARIH2) and HHARI (ARIH1) and strictly requires the DCNL1's UBA domain. 33879767_ARIH1 signaling promotes anti-tumor immunity by targeting PD-L1 for proteasomal degradation. | ENSMUSG00000025234 | Arih1 | 1.574792e+03 | 1.4563367 | 0.542343949 | 0.3017545 | 3.219197e+00 | 0.0727793231 | 0.71859192 | No | Yes | 1.717139e+03 | 284.481233 | 9.993275e+02 | 169.891994 | |
| ENSG00000166479 | 54495 | TMX3 | protein_coding | Q96JJ7 | FUNCTION: Probable disulfide isomerase, which participates in the folding of proteins containing disulfide bonds. May act as a dithiol oxidase. {ECO:0000269|PubMed:15623505}. | Alternative splicing;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Isomerase;Membrane;Redox-active center;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of the disulfide isomerase (PDI) family of endoplasmic reticulum (ER) proteins that catalyze protein folding and thiol-disulfide interchange reactions. The canonical protein encoded by this gene has an N-terminal ER-signal sequence, a catalytically active thioredoxin domain, one transmembrane domain and a C-terminal ER-retention sequence. This gene is expressed in many tissues but has its highest expression in heart and skeletal muscle. It is expressed in the retinal neuroepithelium and lens epithelium in the developing murine eye and haploinsufficiency of this gene in humans and zebrafish is associated with microphthalmia. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Apr 2017]. | hsa:54495; | cell surface [GO:0009986]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; platelet alpha granule membrane [GO:0031092]; protein disulfide isomerase activity [GO:0003756]; protein-disulfide reductase activity [GO:0015035]; peptidyl-cysteine oxidation [GO:0018171] | 15623505_TMX3 is a thioredoxin-related transmembrane protein of the endoplasmic reticulum 17881353_analysis of TMX3 interdomain stabilization of the N-terminal redox-active domain 19240061_Observational study of gene-disease association. (HuGE Navigator) 20485507_Haploinsufficiency for TMX3 results in a small eye phenotype and represents a novel genetic cause of microphthalmia and coloboma. 31304984_Data show that TMX3 expression is upregulated in melanoma cell lines and patient samples. TMX knockdown altered mitochondrial organization, enhanced bioenergetics, and elevated mitochondrial- and NOX4-derived ROS. The TMX-knockdown-induced oxidative stress suppressed melanoma proliferation, migration, and xenograft tumor growth by inhibiting NFAT1. TMX1 is associated with poor disease outcome. | ENSMUSG00000024614 | Tmx3 | 2.810968e+02 | 0.5532003 | -0.854126285 | 0.3794487 | 4.883190e+00 | 0.0271194522 | 0.55891793 | No | Yes | 1.773587e+02 | 35.433139 | 3.276574e+02 | 66.550676 | |
| ENSG00000166483 | 7465 | WEE1 | protein_coding | P30291 | FUNCTION: Acts as a negative regulator of entry into mitosis (G2 to M transition) by protecting the nucleus from cytoplasmically activated cyclin B1-complexed CDK1 before the onset of mitosis by mediating phosphorylation of CDK1 on 'Tyr-15'. Specifically phosphorylates and inactivates cyclin B1-complexed CDK1 reaching a maximum during G2 phase and a minimum as cells enter M phase. Phosphorylation of cyclin B1-CDK1 occurs exclusively on 'Tyr-15' and phosphorylation of monomeric CDK1 does not occur. Its activity increases during S and G2 phases and decreases at M phase when it is hyperphosphorylated. A correlated decrease in protein level occurs at M/G1 phase, probably due to its degradation. | 3D-structure;ATP-binding;Alternative splicing;Cell cycle;Cell division;Kinase;Magnesium;Metal-binding;Mitosis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Tyrosine-protein kinase;Ubl conjugation | This gene encodes a nuclear protein, which is a tyrosine kinase belonging to the Ser/Thr family of protein kinases. This protein catalyzes the inhibitory tyrosine phosphorylation of CDC2/cyclin B kinase, and appears to coordinate the transition between DNA replication and mitosis by protecting the nucleus from cytoplasmically activated CDC2 kinase. [provided by RefSeq, Jul 2008]. | hsa:7465; | cytoplasm [GO:0005737]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase activity [GO:0004672]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; cell division [GO:0051301]; establishment of cell polarity [GO:0030010]; G2/M transition of mitotic cell cycle [GO:0000086]; microtubule cytoskeleton organization [GO:0000226]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; neuron projection morphogenesis [GO:0048812] | 12525641_Wee-1 may serve as a key regulator of both HIV type 1 Vpr- and gamma irradiation-mediated apoptosis 12601350_increases of cyclin D1, cyclin-dependent kinase 4, cyclin E, cyclin A, and Wee1 play an important role in the development of hepatocellular carcinoma from cirrhosis 12669309_Proteasome-dependent degradation of this protein is inhibited in G2-arrested Hep3B cells by TGF beta 1. 14534529_WEE1 is directly transactivated by and increased in association with c-Fos/AP-1. 14760118_Loss of Wee1 expression may have a potential role in promoting tumor progression and may be a significant prognostic indicator in NSCLC. 15070733_beta-TrCP-dependent degradation of Wee1A is important for the normal onset of M-phase in vivo. 15175024_the induction of Cdc2 phosphorylation due to the increase of Wee1 and Myt1 as well as the reduction of Cdc2 and cyclin B1 are involved in 1,25[OH]2VD3-induced G2/M arrest of keratinocytes. 15583029_Inhibiting Hsl7 delayed mitosis but overexpression overrode the replication checkpoint, accelerating Wee1 destruction. Checkpoint activation disrupted Hsl7-Wee1 binding which was restored by polo-like kinase. Hsl7 is a replication checkpoint component 15735666_Level of WEE1 is regulated by KLF2 and enhanced KLF2 expression sensitizes cells to DNA damage-induced apoptosis. 15964826_Akt protein phosphorylation and inactivation of WEE1 promote cell cycle progression at G2/M transition. 16533053_our studies indicate that the 2-ME(2)-induced upregulation of wee1 and subsequent cdc2 phosphorylation are mediated through mitogen-activated protein kinase (MAPK)-ERK-JNK signaling pathways. 17431037_Gamma-irradiated human primary prostate epithelial cells were unable to enforce cell cycle checkpoint arrest and had sustained Cdk2-associated kinase activity because of a lack of inhibitory Cdk phosphorylation by Wee1A tyrosine kinase. 17687495_WEE1 can be introduced to rescue cells from the damage caused by certain antineoplastic agents. 17764157_Significant percentage of the total endogenous Crk II partitions in the nucleus in mammalian cells, where it forms distinct complexes with DOCK180, Wee1, and Abl. 18385244_HIV-1 Vpr binds to the N lobe of the Wee1 kinase domain and enhances kinase activity for CDC2. 18560763_These findings support the view that CK2beta regulates various intracellular processes by modulating the activity of protein kinases that are distinct from CK2. 18820127_HSP90 inhibition abrogates the topoisomerase I poison-induced G2/M checkpoint in p53-null tumor cells by depleting CHK1 and WEE1. 18950845_Observational study of gene-disease association. (HuGE Navigator) 19126550_Cdc34 is a functional target of let-7 and that let-7 induces down-regulation of Cdc34, stabilization of the Wee1 kinase, and an increased fraction of cells in G(2)/M in primary fibroblasts. 19821025_Results identify WEE1 as a potential molecular target in breast cancer. 19858290_These studies identify a novel bifunctional regulatory element in Wee1 that mediates cyclin A/Cdk2 association and nuclear export. 19903823_Abrogation of G(2) arrest by inhibition of ATM and ATR, Chk1, and Wee1 suppressed JCV genome replication. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20038582_multiple regions of Wee1 control its destruction 20372791_analysis of interaction sites between Wee1 kinase and the regulatory beta-subunit of protein kinase CK2 20508983_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20668041_Regulation of Wee1 kinase by miRs may be linked to pituitary tumorigenesis. 20832752_WEE1 is a major regulator of the G(2) checkpoint in glioblastoma cells. 21358821_The over-expression of the naturally under-expressed miR-128 in glioma cells resulted in the inhibition of WEE1 in glioblastoma cells. 21529352_WEE1 inhibition may be a promising strategy to enhance the radiotherapy effect in patients with OS. 21859861_Results demonstrate a novel role of Wee1 in controlling Mus81-Eme1 and DNA replication in human cells. 22112940_Sensitization effects of WEE1 inhibition on TRAIL-mediated apoptosis in breast cancer cell lines. 22289989_Elevated WEE1 expression is associated with acute myeloid leukemia. 22641220_WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. 22692537_Data identify Cdc20, USP44, and Wee1 as relevant Fcp1 targets. 22719872_results reveal the importance of Wee1 as a prognostic biomarker in melanomas, and indicate a potential role for targeted therapy, alone or in combination with other agents 22847610_study shows WEE1 expression in malignant melanoma is directly regulated by miR-195; miR-195-mediated downregulation of WEE1 in metastatic lesions may help to overcome cell cycle arrest under stress conditions in the local tissue microenvironment to allow unrestricted growth of tumour cells 22907750_The results suggested that deregulated CDK1 activity, such as that occurring following inhibition of WEE1 kinase, induces replication stress and loss of genomic integrity through increased firing of replication origins and subsequent nucleotide shortage. 22935698_Wee1 inhibition sensitizes cancer cells to Hsp90 inhibitors. 22942255_Loss-of-function and gain-of-function studies showed that miR-15 family members controlled the expression of WEE1 and CHK1. 23051732_These data support the hypothesis that Cdc14A counteracts Cdk1-cyclin B1 activity through Wee1 dephosphorylation. 23416158_AURKB and WEE1 are targets and biomarkers of therapeutic efficacy, lying downstream of (V600E)B-RAF in melanomas. 23531080_miR-497 is a candidate tumor suppressor in neuroblastoma, through the direct targeting of WEE1. 23538902_Together, these studies have identified Wee1 as a key target of XL888, suggesting novel therapeutic strategies for NRAS-mutant melanoma. 23767999_Our results suggest that Wee1 may be involved in the progression of vulvar carcinomas 24336073_miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. 24377575_High WEE1 expression is associated with breast cancer. 24424027_Regulation of the mitotic inhibitor Wee1 by TOR signalling is a conserved mechanism that helps to couple cell cycle and growth controls. 24536047_A novel mechanism that pancreatic ductal adenocarcinoma cells use to protect against DNA damage in which HuR posttranscriptionally regulates the expression and downstream function of WEE1 upon exposure to DNA-damaging agents. 24626094_Inhibition of WEE1 counters the defective apoptosis of tumor cells expressing high levels of brachyury. 24661910_Taken together, these findings highlight mitotic kinases and, in particular, WEE1 as a rational therapeutic target for medulloblastoma. 24817118_role for CK1delta in controlling the cell cycle 24927813_These results suggest that specific inhibition of Wee1 has deleterious effects on the proliferation and survival of p53 inactive tumors. 25093290_Wee1 is frequently expressed in ovarian serous carcinoma effusions, and its expression is significantly higher following exposure to chemotherapy and it is an independent prognostic marker in serous ovarian carcinoma. 25247294_Study shows that CHD5 is a Nucleosome remodeling and deacetylase complex-associated transcriptional repressor and identifies WEE1 as one of the CHD5-regulated genes that may link CHD5 to tumor suppression. 25428911_Report strong synergism observed by combining Chk1 and Wee1 inhibitors in preclinical models of mantle cell lymphoma. 25649652_This study showed that WEE1 (rs10770042; coding) associated with Alzheimer disease. 25659151_identified the PI3K/Akt pathway, the cell-cycle regulator Wee1 kinase, and protein kinase C (PKC) as prospective regulatory nodes of neuronal excitability through modulation of the FGF14:Nav1.6 complex. 25732734_WEE1 is a valid target of the miR-17-92 cluster in leukemia. 26025928_nasopharyngeal carcinoma cells depend on CHK1 and WEE1 activity for growth 26054341_These data provide a rationale for further evaluation of the combination of Wee1 and Chk1/2 inhibitors in malignant melanoma. 26431163_Data show that proto-oncogene protein Mdm2 inactivation successfully protects tumor suppressor protein (p53)-proficient cells against the cytotoxic effects of Wee1 protein inhibition. 26598692_data indicate that the activity of the DNA replication machinery, beyond TP53 mutation status, determines Wee1 inhibitor sensitivity, and could serve as a selection criterion for Wee1-inhibitor eligible patients 26602815_Data show that H3K36me3-deficient cancers can be targeted by inhibition of WEE1 protein. 26645158_These results suggest that the G2 checkpoint inhibitor MK-1775 can enhance the sensitivity of human NSCLC cells to C ions as well as X rays. 26738845_High nuclear expression of WEE1 protein is associated with all glioma grades and types. 26890070_Consistent with these findings, a genome-scale pooled RNA interference screen revealed that toxic doses of MK-1775 are suppressed by CDK2 or Cyclin A2 knockdown. These findings support G2 exit as the more significant effect of Wee1 inhibition in pancreatic cancers. 27027998_WEE1 is regulated at the translational level by CPEB1 and miR-15b in a coordinated and cell-cycle-dependent manner. 27072241_These results highlight the key role of WEE1 suppression to combat glioblastomas. Moreover, it showed beneficial possibilities of WEE1 suppression with different anticancer approaches for neurological malignancies. 27077811_Wee1 inhibition potentiates Wip1-dependent tumor sensitization effect by reducing levels of Hipk2 kinase, a negative regulator of Wip1 pathway. 27220319_Wee1 staining intensity was a predictor of favorable metastasis-free and overall survival compared to strong intensity and no or weak staining 27363019_WEE1 is over-expressed and could enhance gastric cancer cell proliferation and metastasis 27558135_we overexpressed CKS1B in multiple cell lines and found increased sensitivity to PLK1 knockdown and PLK1 drug inhibition. Finally, combined inhibition of WEE1 and PLK1 results in less apoptosis than predicted based on an additive model of the individual inhibitors, showing an epistatic interaction and confirming a prediction of the yeast data. 28030798_Date show that when Wee1 alone is inhibited, Chk1 suppresses CDC45 loading and thereby limits the extent of unscheduled replication initiation and subsequent S-phase DNA damage, despite very high CDK-activity. 28122647_Wee1 is identified as a novel direct target of miR-194. Ectopic expression of Wee1 at least in part overcomes the suppressive impacts of miR-194 on the malignant phenotypes of human laryngeal squamous cell carcinoma 28178688_the data suggest the importance of WEE1 as an enabler of branching vascularisation in colorectal cancer liver metastases. 28294307_Data suggest that SMURF1 is required for S phase progression; SMURF1 promotes ubiquitination-dependent degradation of WEE1; these functions of SMURF1 appear to be linked and may be important in cell proliferation and tumorigenesis. (SMURF1 = SMAD specific E3 ubiquitin protein ligase 1; WEE1 = wee 1 homolog [S pombe] protein) 28652249_High WEE1 expression is associated with non-small cell lung cancer. 28853983_Study revealed that RNAi mediated co-targeting of WEE1 with AKT3 can synergistically inhibit melanoma in culture as well as in xenograft tumor model and identified that targeting AKT3 and WEE1 leads to deregulation of cell cycle and DNA damage response pathways. 28898169_miR-26b governed temozolomide (TMZ)-resistance-mediate epithelial-mesenchymal transition partly due to governing its target Wee1. 29019284_miR-503 could function as an enhancer of radiation responses in laryngeal carcinoma cells by inhibiting WEE1 29343688_Inhibition of Wee1 by its specific inhibitor MK1775 in combination with sorafenib restored the KRAS mutated cells' response to the multi-target tyrosine kinase inhibitor. 29605721_ATR inhibition synergizes with WEE1 inhibition in TNBC. 29735549_A pulse of gemcitabine and CHK1i followed by WEE1i durably suppressed tumor cell growth. 30089699_Taken together, these data demonstrate a novel mechanism of hepatocyte growth promotion during hepatitis C virus infection involving a miR-373-NORAD-Wee1 axis. 30488993_downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis 30645202_Inhibiting Wee1 and ATR kinases produces tumor-selective synthetic lethality and suppresses metastasis. 30754676_Wee1 is a novel transcription target of C/EBPbeta that is required for the G/M phase of cell cycle progression. 30805988_Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. 30915746_MiR-503-5p regulates cell epithelial-to-mesenchymal transition, metastasis and prognosis of hepatocellular carcinoma through inhibiting WEE1. 31015476_Characterization of Three Novel H3F3A-mutated Giant Cell Tumor Cell Lines and Targeting of Their Wee1 Pathway. 31197522_WEE1 plays a pivotal role in the gastrointestinal stromal tumors proliferation.Upregulated expression of WEE1 is correlated with tumor size, mitotic count. 31238052_Knockdown of SNHG3 upregulated miR-384 expression and that overexpression of miR-384 decreased SNHG3. Inhibition of miR-384 markedly increased WEE1 expression. 31347653_WEE1 inhibition enhances hypoxia/reoxygenation-induced cell death accompanied by mitotic catastrophe and that the process may be mediated by homologous recombination. 31387179_WEE1 expression was increased after EGFR-TKIs resistance. 31390121_Findings suggest that DDX56 can induce intron retention in WEEI, which produces a consequent truncated WEE1 protein that does not function as a tumor suppressor. Expression of WEE1 intron mRNA in colorectal cancer (CRC) tissues was higher than in normal tissues. These observations further indicate that the tumor suppressor WEE1 is suppressed by mRNA splicing abnormalities in CRC. 31738907_Differential properties of mitosis-associated events following CHK1 and WEE1 inhibitor treatments in human tongue carcinoma cells have been reported. 31806643_Identification of Therapeutic Vulnerabilities in Small-cell Neuroendocrine Prostate Cancer. 31866466_WEE1 inhibitor, AZD1775, overcomes trastuzumab resistance by targeting cancer stem-like properties in HER2-positive breast cancer. 31878998_miR-129-1-3p could reverse cisplatin resistance of HNE1/CDDP nasopharyngeal carcinoma cells via inhibiting WEE1 expression. 31919076_WEE1 inhibition induces glutamine addiction in T-cell acute lymphoblastic leukemia. 32027086_Quercetin radiosensitizes non-small cell lung cancer cells through the regulation of miR-16-5p/WEE1 axis. 32120135_WEE1 kinase limits CDK1/CDK2 activities to safeguard DNA replication and mitotic entry in cancer. (Review) 32180106_Targeting MYC-driven replication stress in medulloblastoma with AZD1775 and gemcitabine. 32314355_The Wee1 kinase inhibitor MK1775 suppresses cell growth, attenuates stemness and synergises with bortezomib in multiple myeloma. 32628111_PTEN and DNA-PK determine sensitivity and recovery in response to WEE1 inhibition in human breast cancer. 32633376_Knockdown of long non-coding RNA VIM-AS1 inhibits glioma cell proliferation and migration, and increases the cell apoptosis via modulation of WEE1 targeted by miR-105-5p. 32871104_DNA Damage Promotes TMPRSS2-ERG Oncoprotein Destruction and Prostate Cancer Suppression via Signaling Converged by GSK3beta and WEE1. 32958072_A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. 33020398_Pharmacological Inhibition of WEE1 Potentiates the Antitumoral Effect of the dl922-947 Oncolytic Virus in Malignant Mesothelioma Cell Lines. 33320980_Acquired small cell lung cancer resistance to Chk1 inhibitors involves Wee1 up-regulation. 33542435_The mitotic checkpoint is a targetable vulnerability of carboplatin-resistant triple negative breast cancers. 33564073_MNK1 and MNK2 enforce expression of E2F1, FOXM1, and WEE1 to drive soft tissue sarcoma. 33666372_The lncRNA XIST/miR-125b-2-3p axis modulates cell proliferation and chemotherapeutic sensitivity via targeting Wee1 in colorectal cancer. 33974709_Gene-Gene Interactions of Gemcitabine Metabolizing-Enzyme Genes hCNT3 and WEE1 for Preventing Severe Gemcitabine-Induced Hematological Toxicity. 34171479_Kinome-wide RNAi screening for mediators of ABT-199 resistance in breast cancer cells identifies Wee1 as a novel therapeutic target. 34269904_WEE1 Inhibitor: Clinical Development. 34664776_Circ-SNAP47 (hsa_circ_0016760) and miR-625-5p are regulators of WEE1 in regulation of chemoresistance, growth and invasion of DDP-tolerant NSCLC cells via ceRNA pathway. 34688654_The KRAS-regulated kinome identifies WEE1 and ERK coinhibition as a potential therapeutic strategy in KRAS-mutant pancreatic cancer. 34753061_Long noncoding RNA NORAD acts as a ceRNA mediates gemcitabine resistance in bladder cancer by sponging miR-155-5p to regulate WEE1 expression. 34831326_Pharmacological Inhibition of Wee1 Kinase Selectively Modulates the Voltage-Gated Na(+) Channel 1.2 Macromolecular Complex. 35045293_WEE1 kinase protects the stability of stalled DNA replication forks by limiting CDK2 activity. 35127343_miR-138-5p Inhibits the Growth and Invasion of Glioma Cells by Regulating WEE1. 35315355_WEE1 kinase is a therapeutic vulnerability in CIC-DUX4 undifferentiated sarcoma. | ENSMUSG00000031016 | Wee1 | 1.424414e+03 | 0.7622534 | -0.391657486 | 0.2736476 | 2.030368e+00 | 0.1541832436 | 0.77404149 | No | Yes | 1.195555e+03 | 173.507589 | 1.545371e+03 | 229.740408 | |
| ENSG00000166562 | 90701 | SEC11C | protein_coding | Q9BY50 | FUNCTION: Catalytic component of the signal peptidase complex (SPC) which catalyzes the cleavage of N-terminal signal sequences from nascent proteins as they are translocated into the lumen of the endoplasmic reticulum (PubMed:34388369). Specifically cleaves N-terminal signal peptides that contain a hydrophobic alpha-helix (h-region) shorter than 18-20 amino acids (PubMed:34388369). {ECO:0000269|PubMed:34388369}. | 3D-structure;Endoplasmic reticulum;Hydrolase;Membrane;Protease;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | hsa:90701; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; signal peptidase complex [GO:0005787]; peptidase activity [GO:0008233]; serine-type endopeptidase activity [GO:0004252]; signal peptide processing [GO:0006465] | 34388369_Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. | ENSMUSG00000024516 | Sec11c | 8.625227e+02 | 0.8225812 | -0.281769932 | 0.2775059 | 1.022247e+00 | 0.3119866826 | 0.81775358 | No | Yes | 6.967344e+02 | 82.771913 | 8.775583e+02 | 106.438162 | ||
| ENSG00000166664 | 89832 | CHRFAM7A | protein_coding | Q494W8 | Mouse_homologues FUNCTION: After binding acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane. The channel is blocked by alpha-bungarotoxin. | Membrane;Reference proteome;Transmembrane;Transmembrane helix | The nicotinic acetylcholine receptors (nAChRs) are members of a superfamily of ligand-gated ion channels that mediate fast signal transmission at synapses. The family member CHRNA7, which is located on chromosome 15 in a region associated with several neuropsychiatric disorders, is partially duplicated and forms a hybrid with a novel gene from the family with sequence similarity 7 (FAM7A). Alternative splicing has been observed, and two variants exist, for this hybrid gene. The N-terminally truncated products predicted by the largest open reading frames for each variant would lack the majority of the neurotransmitter-gated ion-channel ligand binding domain but retain the transmembrane region that forms the ion channel. Although current evidence supports transcription of this hybrid gene, translation of the nicotinic acetylcholine receptor-like protein-encoding open reading frames has not been confirmed. [provided by RefSeq, Jul 2008]. | hsa:89832; | acetylcholine-gated channel complex [GO:0005892]; integral component of plasma membrane [GO:0005887]; neuron projection [GO:0043005]; synapse [GO:0045202]; acetylcholine binding [GO:0042166]; extracellular ligand-gated ion channel activity [GO:0005230]; neurotransmitter receptor activity [GO:0030594]; transmembrane signaling receptor activity [GO:0004888]; chemical synaptic transmission [GO:0007268]; ion transmembrane transport [GO:0034220]; nervous system process [GO:0050877]; regulation of membrane potential [GO:0042391]; signal transduction [GO:0007165]; synaptic transmission, cholinergic [GO:0007271] | 11829490_3-Mb map of 15q13-q14 showing that CHRFAM7A is part of a large segmental duplication in the opposite orientation to CHRNA7 and revealing several other duplications 14729617_Human mesothelioma cells and human biopsies of mesothelioma as well as of normal pleural mesothelial cells functionally express CHRNA7. 15885267_Results demonstrate that human and rat nicotinic acetylcholine receptors are senstive targets for volatile organic compounds in industrial products and are used in the risk assessment of these compounds. 16417613_CHRFAM7A was identified as a candidate gene in the D15S165 region in a study of allelic variants at chromosome 15q14 in schizophrenia. 16823804_Observational study of gene-disease association. (HuGE Navigator) 17012698_Observational study of gene-disease association. (HuGE Navigator) 17012698_These observations indicate that episodic memory function is a schizophrenia endophenotype and implicate the CHRFAM7A/CHRNA7 locus in modulating its function. 17192894_In persons with bipolar type schizoaffective disorder, CHRNA7 promoter region allelic variants are linked to the capacity to inhibit the P50 auditory evoked potential and thus are associated with a type of illness similar to schizophrenia. 19082523_In 20 smoking-matched people (n = 10 schizophrenia, n = 10 controls), we found significantly lower CHRFAM7A in cotinine and self-reported smokers versus nonsmokers (p 19149910_No significant associations of 2-bp deletion or CHRFAM7A copy number with antisaccade performance parameters were observed. 19149910_Observational study of gene-disease association. (HuGE Navigator) 19462340_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19631623_Observational study of gene-disease association. (HuGE Navigator) 19641318_Observational study of gene-disease association. (HuGE Navigator) 19641318_polymorphism of CHRFAM7A can be implicated in Alzheimer's disease, dementia with Lewy bodies and Pick's disease 20351714_Observational study of gene-disease association. (HuGE Navigator) 20926142_the partially duplicated alpha7 nAChR subunit gene may specifically participate in the inflammatory response of the innate immune system. 22300029_evidence on the association between variations in CHRNA7 or CHRFAM7A and the risk of dementia is still sparse and inconclusive. Further studies are needed to establish whether some polymorphisms may affect the probability of developing dementia [review] 23553139_lack of CHRFAM7A expression in ADNFLE patients might be an important factor in the pathogenesis of autosomal dominant nocturnal frontal lobe epilepsy 24024466_the involvement of CHRFAM7A in the pathophysiology of the idiopathic generalized epilepsy and indication that c.497-498TG deletion or a nearby polymorphism in the CHRFAM7A gene may play a role in the pathogenesis of this disease 24787912_This association study was replicated in the NIA-LOAD Familial Study dataset. CHRFAM7A is a dominant negative regulator of CHRNA7 function, the receptor that facilitates amyloid-beta1-42 internalization through endocytosis and has been implicated in AD. 25473097_CHRFAM7A, a human-specific and partially duplicated alpha7-nicotinic acetylcholine receptor gene with the potential to specify a human-specific inflammatory response to injury. 25681457_Data show that a 1 kb sequence in the untranslated regions of the alpha7-nicotinic acetylcholine receptor (alpha7nAChR) gene CHRFAM7A that is modulated by lipopolysaccharides (LPS). 26206074_Data show preferential fetal CHRFAM7A expression in the human prefrontal cortex and suggest abnormalities in the CHRFAM7A/CHRNA7 ratios in schizophrenia and bipolar disorder, due mainly to overexpression of CHRFAM7A. 26567012_gp120IIIB promotes the downregulation of CHRFAM7A in neuronal cells. 29305655_Our study observations support the role for CHRFAM7A gene in schizophrenia pathogenesis and suggest a potential novel link between deficient CHRFAM7A expression and negative psychopathology 30089821_Genetic variation CHRFAM7A is associated with nicotine dependence and response to varenicline treatment. 30710073_Study suggests a negative modulatory effect of CHRFAM7A on synaptic transmission (relevance in schizophrenia) and a modulatory effect on Abeta1-42 uptake (relevance to Alzheimer disease). 30924722_Association of a Functional Polymorphism in the CHRFAM7A Gene with Inflammatory Response Mediators and Neuropathic Pain after Spinal Cord Injury. 31048268_The analysis of the CHRFAM7A gene's regulatory region reveals some of the mechanisms driving its expression and responsiveness to LPS in human immune cell models. donepezil differently modulates CHRFAM7A responsiveness to LPS. 31348980_The CHRFAM7A gene contributes to the pathogenesis of neuropsychiatric disorders. 31583530_Acetylcholinesterase inhibitors targeting the cholinergic anti-inflammatory pathway: a new therapeutic perspective in aging-related disorders. 32303780_CHRFAM7A reduces monocyte/macrophage migration and colony formation in vitro. 32890966_Human-specific gene CHRFAM7A mediates M2 macrophage polarization via the Notch pathway to ameliorate hypertrophic scar formation. 33405023_CHRFAM7A Overexpression Attenuates Cerebral Ischemia-Reperfusion Injury via Inhibiting Microglia Pyroptosis Mediated by the NLRP3/Caspase-1 pathway. 33515545_The human-specific duplicated alpha7 gene inhibits the ancestral alpha7, negatively regulating nicotinic acetylcholine receptor-mediated transmitter release. 33956875_Effect of CHRFAM7A Delta2bp gene variant on secondary inflammation after spinal cord injury. 34067314_Structure, Dynamics, and Ligand Recognition of Human-Specific CHRFAM7A (Dupalpha7) Nicotinic Receptor Linked to Neuropsychiatric Disorders. 34088975_Regulation of the acetylcholine/alpha7nAChR anti-inflammatory pathway in COVID-19 patients. 35408823_The Human-Restricted Isoform of the alpha7 nAChR, CHRFAM7A: A Double-Edged Sword in Neurological and Inflammatory Disorders. | ENSMUSG00000030525 | Chrna7 | 3.845321e+02 | 1.0468630 | 0.066072677 | 0.2893751 | 5.251686e-02 | 0.8187402327 | 0.96475641 | No | Yes | 3.054650e+02 | 34.130683 | 3.023818e+02 | 34.804378 | |
| ENSG00000166682 | 80975 | TMPRSS5 | protein_coding | Q9H3S3 | FUNCTION: May play a role in hearing. {ECO:0000269|PubMed:17918732}. | Cell membrane;Disulfide bond;Glycoprotein;Hydrolase;Membrane;Protease;Reference proteome;Serine protease;Signal-anchor;Transmembrane;Transmembrane helix | This gene encodes a protein that belongs to the serine protease family. Serine proteases are known to be involved in many physiological and pathological processes. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]. | hsa:80975; | integral component of membrane [GO:0016021]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; peptidase activity [GO:0008233]; scavenger receptor activity [GO:0005044]; serine-type endopeptidase activity [GO:0004252]; proteolysis [GO:0006508] | 12484779_The purified IGF-binding protein-5 (IGFBP-5) protease fraction secreted by cultured human osteoblasts has been characterized as ADAM9. | ENSMUSG00000032268 | Tmprss5 | 3.220742e+01 | 0.8619222 | -0.214370468 | 0.5858683 | 1.377396e-01 | 0.7105385612 | 0.93853087 | No | Yes | 1.932019e+01 | 10.891545 | 2.538487e+01 | 14.645215 | |
| ENSG00000166797 | 84191 | CIAO2A | protein_coding | Q9H5X1 | FUNCTION: Component of the cytosolic iron-sulfur protein assembly (CIA) complex, a multiprotein complex that mediates the incorporation of iron-sulfur cluster into extramitochondrial Fe/S proteins (PubMed:23891004). As a CIA complex component and in collaboration with CIAO1 specifically matures ACO1 and stabilizes IREB2, connecting cytosolic iron-sulfur protein maturation with cellular iron regulation (PubMed:23891004). May play a role in chromosome segregation through establishment of sister chromatid cohesion. May induce apoptosis in collaboration with APAF1 (PubMed:25716227). {ECO:0000250, ECO:0000269|PubMed:23891004, ECO:0000269|PubMed:25716227}. | 3D-structure;Alternative splicing;Chromosome partition;Cytoplasm;Metal-binding;Reference proteome;Zinc | hsa:84191; | CIA complex [GO:0097361]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; metal ion binding [GO:0046872]; chromosome segregation [GO:0007059]; iron-sulfur cluster assembly [GO:0016226]; protein maturation by [4Fe-4S] cluster transfer [GO:0106035]; protein maturation by iron-sulfur cluster transfer [GO:0097428] | 22683786_putative metal-binding site of bacterial DUF59 proteins is not conserved in Fam96a, but Fam96a interacts tightly in vitro with Ciao1, the cytosolic iron-assembly protein 23891004_CIA2B-CIA1-MMS19 and CIA2A-CIA1 assist different branches of Fe/S protein assembly and intimately link this process to cellular iron regulation via IRP1 Fe/S cluster maturation and IRP2 stabilization. 25716227_FAM96A expression was much reduced in tumorigenic ICC-SCs. 28443470_our data suggest that hTERTR-FAM96A may serve as an efficient anti-tumor agent for the treatment of hepatocellular carcinoma. | ENSMUSG00000032381 | Ciao2a | 2.674284e+02 | 0.5866640 | -0.769393629 | 0.3591462 | 4.619076e+00 | 0.0316182741 | 0.58378765 | No | Yes | 1.870713e+02 | 37.758363 | 3.161448e+02 | 64.838985 | ||
| ENSG00000166831 | 348093 | RBPMS2 | protein_coding | Q6ZRY4 | FUNCTION: RNA-binding protein involved in the regulation of smooth muscle cell differentiation and proliferation in the gastrointestinal system (PubMed:25064856). Binds NOG mRNA, the major inhibitor of the bone morphogenetic protein (BMP) pathway. Mediates an increase of NOG mRNA levels, thereby contributing to the negative regulation of BMP signaling pathway and promoting reversible dedifferentiation and proliferation of smooth muscle cells (By similarity). {ECO:0000250|UniProtKB:Q9W6I1, ECO:0000269|PubMed:25064856}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Developmental protein;RNA-binding;Reference proteome | hsa:348093; | cytoplasm [GO:0005737]; identical protein binding [GO:0042802]; mRNA binding [GO:0003729]; protein homodimerization activity [GO:0042803]; embryonic digestive tract morphogenesis [GO:0048557]; negative regulation of BMP signaling pathway [GO:0030514]; negative regulation of smooth muscle cell differentiation [GO:0051151]; positive regulation of smooth muscle cell proliferation [GO:0048661] | ENSMUSG00000032387 | Rbpms2 | 2.038319e+02 | 1.5281004 | 0.611739321 | 0.3732809 | 2.704672e+00 | 0.1000546441 | 0.75783482 | No | Yes | 2.272530e+02 | 34.973423 | 1.200835e+02 | 19.500525 | |||
| ENSG00000166839 | 348094 | ANKDD1A | protein_coding | Q495B1 | ANK repeat;Alternative splicing;Reference proteome;Repeat | hsa:348094; | signal transduction [GO:0007165] | 30082910_ANKDD1A is a functional tumor suppressor gene, especially in the hypoxia microenvironment. ANKDD1A directly interacts with FIH1 and inhibits the transcriptional activity of HIF1alpha by upregulating FIH1. ANKDD1A decreases the half-life of HIF1alpha by upregulating FIH1, decreases glucose uptake and lactate production, inhibits glioblastoma multiforme (GBM) autophagy, and induces apoptosis in GBM cells under hypoxia 30651372_Results provide evidence that rs34988193 is a deleterious germline variant present in ANKDD1a associated with poor outcomes of patients with low grade glioma. | ENSMUSG00000066510 | Ankdd1a | 4.117512e+01 | 1.8842251 | 0.913971348 | 0.5439930 | 2.955212e+00 | 0.0856010661 | 0.74509483 | No | Yes | 3.636462e+01 | 14.239836 | 1.669969e+01 | 6.810478 | |||
| ENSG00000166971 | 64400 | AKTIP | protein_coding | Q9H8T0 | FUNCTION: Component of the FTS/Hook/FHIP complex (FHF complex) (PubMed:32073997). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). Regulates apoptosis by enhancing phosphorylation and activation of AKT1. Increases release of TNFSF6 via the AKT1/GSK3B/NFATC1 signaling cascade. FHF complex promotes the distribution of AP-4 complex to the perinuclear area of the cell (PubMed:32073997). {ECO:0000269|PubMed:14749367, ECO:0000269|PubMed:18799622, ECO:0000269|PubMed:32073997}. | Alternative splicing;Apoptosis;Cell membrane;Cytoplasm;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | The mouse homolog of this gene produces fused toes and thymic hyperplasia in heterozygous mutant animals while homozygous mutants die in early development. This gene may play a role in apoptosis as these morphological abnormalities are caused by altered patterns of programmed cell death. The protein encoded by this gene is similar to the ubiquitin ligase domain of other ubiquitin-conjugating enzymes but lacks the conserved cysteine residue that enables those enzymes to conjugate ubiquitin to the target protein. This protein interacts directly with serine/threonine kinase protein kinase B (PKB)/Akt and modulates PKB activity by enhancing the phosphorylation of PKB's regulatory sites. Alternative splicing results in two transcript variants encoding the same protein. [provided by RefSeq, Jul 2008]. | hsa:64400; | cytosol [GO:0005829]; FHF complex [GO:0070695]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ubiquitin conjugating enzyme activity [GO:0061631]; apoptotic process [GO:0006915]; early endosome to late endosome transport [GO:0045022]; endosome organization [GO:0007032]; endosome to lysosome transport [GO:0008333]; lysosome organization [GO:0007040]; positive regulation of protein binding [GO:0032092]; positive regulation of protein phosphorylation [GO:0001934]; protein localization to perinuclear region of cytoplasm [GO:1905719]; protein polyubiquitination [GO:0000209]; protein transport [GO:0015031] | 14749367_Ft1 protein interacts directly with PKB, enhancing the phosphorylation of both of its regulatory sites by promoting its interaction with the upstream kinase PDK1 18799622_These data suggest that the FTS/Hook/FHIP complex functions to promote vesicle trafficking and/or fusion via the homotypic vacuolar protein sorting complex. 19270026_Observational study of gene-disease association. (HuGE Navigator) 20132317_Demographic and clinical characteristics and AKT1 single markers and haplotypes, but not AKTIP polymorphisms or interactions between AKT1 and AKTIP, are associated with increased risk for suicidal behavior in bipolar patients. 20635389_Observational study of gene-disease association. (HuGE Navigator) 20945372_These data unraveled the involvement of new oncoprotein FTS in cervical cancer which plays a central role in carcinogenesis. 21424602_Targeted inhibition of FTS led to the shutdown of key elemental characteristics of cervical cancer and could lead to an effective therapeutic strategy. 24022875_*AKTIP single nucleotide polymorphisms are not associated with late-onset depression patients compared to healthy elderly controls. 24971934_FTS silencing reduced EMT and cell migration by EGF treatment. These results demonstrate a novel function for FTS in EGF-mediated EMT process. 25151576_High fused toes homolog expression is associated with radioresistance in cervical cancer. 26110528_AKTIP interacts with telomeric DNA and is required for telomere maintenance. 27342216_results suggest that AKT1, FTO, and AKTIP polymorphisms were not associated with obesity/overweight in Brazilians children. 27512140_Results show that AKTIP co-purifies with A- and B-type lamins and partially co-localizes with lamins in interphase nuclei. In addition, AKTIP depletion lowers lamin A expression and induces senescence hallmarks including telomere homeostasis in human primary fibroblasts. 27535835_FTS is involved in EGFR-mediated repair of DNA damage induced by cisplatin in ME180 cells 34449766_AKTIP interacts with ESCRT I and is needed for the recruitment of ESCRT III subunits to the midbody. | ENSMUSG00000031667 | Aktip | 3.175953e+02 | 0.7763905 | -0.365145588 | 0.3209985 | 1.272461e+00 | 0.2593055443 | 0.79684153 | No | Yes | 2.615654e+02 | 41.380775 | 3.285503e+02 | 53.050491 | |
| ENSG00000167653 | 8000 | PSCA | protein_coding | O43653 | FUNCTION: May be involved in the regulation of cell proliferation. Has a cell-proliferation inhibition activity in vitro. {ECO:0000269|PubMed:18488030}.; FUNCTION: May act as a modulator of nicotinic acetylcholine receptors (nAChRs) activity. In vitro inhibits nicotine-induced signaling probably implicating alpha-3:beta-2- or alpha-7-containing nAChRs. {ECO:0000305|PubMed:25680266}. | Cell membrane;Direct protein sequencing;Disulfide bond;GPI-anchor;Glycoprotein;Lipoprotein;Membrane;Reference proteome;Signal | This gene encodes a glycosylphosphatidylinositol-anchored cell membrane glycoprotein. In addition to being highly expressed in the prostate it is also expressed in the bladder, placenta, colon, kidney, and stomach. This gene is up-regulated in a large proportion of prostate cancers and is also detected in cancers of the bladder and pancreas. This gene includes a polymorphism that results in an upstream start codon in some individuals; this polymorphism is thought to be associated with a risk for certain gastric and bladder cancers. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2010]. | hsa:8000; | anchored component of membrane [GO:0031225]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; plasma membrane [GO:0005886]; acetylcholine receptor binding [GO:0033130]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; regulation of neurotransmitter receptor activity [GO:0099601] | 11752398_This transgenic system helps define the range of cellular changes associated with altered expression of PSCA, shows that transcriptional control is a major component regulating PSCA levels. 12351697_Here we describe the identification of a compact cell-specific and androgen-responsive enhancer between 2.7 and 3 kb upstream of the transcription start site. The enhancer functions autonomously when positioned immediately adjacent to a minimal promoter. 12496358_PSCA is expressed in an intermediate subpopulation of PrEC in transition from a basal to a terminally differentiated secretory phenotype 15814638_Data suggest that PSCA is a promising tumor marker and potential therapeutic target for patients with metastatic prostate cancer. 16957968_Significantly higher prostate stem cell antigen copy number was associated with pancreatic cancer 17492652_The chimeric alpha-PSCA-beta2/CD3zeta-TCR might now be used for arming human cytotoxic T-cells for further studies towards a clinical treatment of prostate cancer. 17503471_data identify prostate stem cell antigen(PSCA) mRNA in initial prostatic intraepithelial neoplasia (PIN) as a significant predictor of subsequent prostate cancer 17549363_Increased levels of IgG antibodies against the prostate stem cell antigen in the plasma is associated with pancreatic cancer 17853904_Result shows PSCA contains the cleavage site of the ER signal peptidase and the resulting cleavage products fail to bind to HLA-A(*)0201 and are not recognized by T lymphocytes. 18076024_This prospective study identifies PSCA mRNA in BPH as a significant predictor of cancer development after TURP 18184265_Overexpression of prostate stem cell antigen (PSCA) is associated with development of gestational trophoblastic neoplasia in hydatidiform mole. 18440837_combination of prostate stem cell antigen enhancer and uroplakin II promoter is feasible in constructing bladder cancer-specific vectors 18488030_Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer 18488030_Observational study of gene-disease association. (HuGE Navigator) 18838214_the expression of PSCA correlates with relevant clinical benchmarks, such as Gleason score and metastasis. 19180924_The expressions of PSCA and Claudin-4 are upregulated in the pancreatic carcinomas. 19343734_Prostate stem cell antigen and prostate-specific membrane antigen are both highly expressed in lymph node and bone metastases of prostate cancer. 19462463_This prospective study identifies PSCA mRNA in preoperatively negative prostatic biopsies as a significant predictor of subsequent cancer after TURP. 19554573_Genetic variations in prostate stem cell antigen is associated with gastric cancer. 19554573_Observational study of gene-disease association. (HuGE Navigator) 19582881_Prostate stem cell antigen polymorphisms are associated with stomach cancer. 19648920_Genetic variation in PSCA confers susceptibility to urinary bladder cancer. 19648920_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19822092_Suggest that PSCA is a marker associated with neoplastic transformation of prostate cells, both in cystoprostatectomies with incidental prostate carcinoma and radical prostatectomies. 19861285_The expression of PSCA can be related to the development of pancreatic cancer, but not to the clinicopathological factors of the tumor. 20083643_Observational study of gene-disease association. (HuGE Navigator) 20083643_rs2294008 polymorphism of PSCA gene may play a role in bladder cancer carcinogenesis. 20085909_Prostate stem cell antigen is highly expressed in non-small cell lung cancer and may be functionally important for this fatal disease. 20230293_Observational study of gene-disease association. (HuGE Navigator) 20230293_PSCA gene polymorphisms may be associated with gastric cancer in Tibetans. 20374648_Correlation between the level of PSCA expression and tumor growth and suggest a role of PSCA in counteracting the natural immune response.in bladder cancer. 20501618_REVIEW: PSCA seems to be a Jekyll and Hyde molecule that plays differential roles, tumor promoting or suppressing, depending on the cellular context. 20502058_We report the first analysis of PSCA, PIWIL1, and TBX2 expression in EAC. Our findings suggest that PSCA and TBX2 might be candidate targets for cancer therapy. 20507324_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20955382_prostate stem cell antigen is a marker associated with neoplastic transformation of prostate cells 21064099_Genetic variant in PSCA is associated with diffuse-type gastric cancer. 21064099_Observational study of gene-disease association. (HuGE Navigator) 21070776_Observational study of gene-disease association. (HuGE Navigator) 21070776_The rs2294008 polymorphism in prostate stem cell antigen increases the risk of noncardia gastric cancer and its precursors in white individuals but protects against proximal cancers. 21070779_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21268123_Data show that the T allele of PSCA rs2294008 is associated with increased risk of gastric cancer. 21429770_Data indicate that RNA silencing of PSCA can inhibit the proliferation and invasiveness properties of prostate cancer PC-3M cells, which may provide a promising therapeutic strategy for prostate cancer. 21497359_Men with the rs1045531 AA genotype of PSCA were at higher risk for prostate cancer. On haplotype analysis CCCAGGTACGG and CGA haplotype carriers showed a significant association with prostate cancer risk 21503583_prostate stem cell antigen is a novel glioma-associated antigen 21538581_This study showed that rs2294008 in the PSCA gene was associated with increased risks of gastric cancer in a Korean population, suggests that rs2294008 might play an important role in gastric carcinogenesis. 21600799_Data indicate that PSA, PSMA, hK2, PSCA, DD3, and their combinations, combined analysis of PSA and/or hK2 expression in pelvic lymph nodes could predict biochemical recurrence free survival (BRFS) following radical prostatectomy (RP). 21681742_results confirm the association between variation in the promoter region of PSCA and GC risk in Caucasians and also indicate that the rs2294008 variant is a similar risk factor for both the diffuse and intestinal-types of GC. 21748756_Integrin alpha(nu) beta(3), NTRS1 and PSCA mRNA expression increased with tumorigenic potential, but mRNA expression levels for these proteins do not translate directly to equivalent expression levels of membrane bound protein. 21914346_PSCA might play an important role in lymph node metastasis in PSCA might play an important role in lymph node metastasis in the breast. Expression of PSCA was correlated with lymph nodes metastasis. 21936014_PSCA is acting as a tumor suppressor in GBC development. 22155405_meta-analysis suggests that PSCA -rs2294008C>T and -rs2976392G>A are potential factors of gastric cancer development for Eastern Asians 22387998_The C allele of rs2294008 at PSCA was associated with increased risk of duodenal ulcer in a recessive model but was associated with decreased risk of gastric cancer. 22416122_A joint effect of two PSCA SNPs, rs2294008 and rs2978974, suggests that both variants may be important for bladder cancer susceptibility. 22426141_The two loci of PSCA (rs2294008 and rs2976392) were both significantly associated with gastric cancer susceptibility and in linkage disequilibrium. 22481254_Findings suggest that prostate stem cell antigen (PSCA) rs2294008 C > T and rs2976392 G > A polymorphisms may contribute to the susceptibility to gastric cancer, particular in non-cardia or diffused gastric cancer. 22502712_Associations exist between PSCA SNPs and breast cancer susceptibility in Korean women 22536409_Two direct YY1 binding sites within the PSCA promoter, an inhibitory upstream site and a downstream site, proximal to the androgen-responsive element, which stimulated PSCA promoter activity. 22778595_intensity of QDs fluorescence remained stable for two weeks (p = 0.083) after conjugation to the PSCA protein, and nearly 93% of positive expression with their fluorescence still could be seen after four weeks 22796266_Results indicate the importance of four gastric cancer susceptibility polymorphisms of IL-10, NOC3L, PSCA and MTRR in the Chinese Han population. 22824379_a genotyping study to assess for associations between the PSCA rs2294008 polymorphism and risk of adenomatous polyps and colorectal cancer 22938426_study suggested rs2294008 in the PSCA gene to be associated with increased risk of gastric cancer and rs2070803 in MUC1 to play a protective role in a Chinese population 22938475_findings suggest that the PSCA rs2294008 C>T polymorphism is a risk factor for gastric cancer, especially in diffuse and noncardia gastric cancer and in Chinese 23391636_Of the 10 target genes, PCA3 and PSCA mRNAs were significantly differently expressed in cancerous tissue than in histologically benign tissue of cancerous prostates 23704932_A C-allele of rs2294008 at PSCA increased the risk of gastric ulcer. 23984394_Overexpression of PSCA and Oct-4 might be closely related to the carcinogenesis, progression, metastasis, or invasive potential and prognosis of gallbladder carcinoma. 23988503_A significant association has been found between the PSCA Trs2294008-Grs2978974 haplotype and higher risk of gallbladder carcinoma in females, whereas this haplotype conferred significantly lower risk to males. 24023815_no clearcut interaction with PSCA SNPs in defining risk of gastric precancerous lesions or cancer 24146278_Our findings demonstrated that rs2294008 and rs2976392 polymorphism of PSCA is a risk-conferring factor associated with increased gastric cancer susceptibility 24183365_expression of PSCA as a cell surface marker increased from benign prostate tissues and High Grade Prostatic Intraepithelial Neoplasias to prostate cancer 24308679_Accumulating evidence shows an association of PSCA with cancers, such as prostate, gastric, bladder, and pancreatic cancer; however, its expression varies with different cancer types. 24320701_PSCA is not a marker for a stem cell population nor is it exclusively expressed in the prostate; the function of PSCA in normal cellular processes or carcinogenesis is currently unknown 24438073_PSCA-chimeric antigen receptor T cells may be developed for treatment of prostate cancer. 24557062_In the Han Chinese population, genetic variation in PSCA was significantly correlated with susceptibility to colorectal cancer. 24654646_the T allele of rs2294008, an intronic variant of the PSCA gene at 8q24 that was previously associated with an increased risk of gastric cancer, was inversely associated with a decreased risk of esophageal squamous cell carcinoma. 25117309_the results suggest that the PSCA rs2294008 (C>T) polymorphism is a risk factor for bladder cancer development. 25374226_rs2294008 polymorphism in the PSCA gene is associated with the risk of bladder cancer. 25503145_PSCA rs2294008 allele A & T was linked with risk of gastric cancer and high-risk gastritis 25582162_The PSCA rs2294008 C>T polymorphism may be acting through induction of gastric mucosal atrophy, finally leading to development of gastric ulcer and gastric cancer in PSCA rs2294008 T allele carriers with Helicobacter infections. 25658482_a significantly increased stomach cancer risk was associated with PSCA SNPs rs2294008 and s2976392 in a Han Chinese population. 25698533_PSCA mRNA is overexpressed in the peripheral blood of prostate cancer patients. 25721731_The PSCA rs2294008 polymorphism is involved in the susceptibility to GC and DU. 25727947_These findings imply that the T allele significantly suppresses PSCA expression in vivo by recruiting YY1 to its promoter, which eventually predisposes gastric epithelial cells to GC development 25964537_PSCA signaling may suppress tumor growth in vivo by modulating immunological characteristics of gallbladder cancer cells. 26006239_These results suggest that PSCA gene variation has a potential effect on its expression and gastric adenocarcinoma risk in the Northwest Chinese population. 26147638_we show that PSCA is a key player in nasopharyngeal carcinoma metastasis 26308216_PSCA rs2294008 Polymorphism is associated with Increased Risk of Cancer 26320491_Genetic Variants of PSCA Gene are not Associated with Colorectal Cancer. 26477693_PSCA has a promoting role in the growth and metastasis of prostate cancer. 26527100_analysis of PSCA level in the peripheral blood of PC patients who underwent radical prostatectomy shows it is related to a GADPH reference level (PSCA/GAPDH ratio) 26554163_Studied six SNP loci: (rs2279115 of BCL2 gene, rs804270 of NEIL2 gene, rs909253 of LTA gene, rs2294008 of PSCA gene, rs3765524 and rs10509670 of PLCE1 gene) to evaluate gastric cancer risk using magnetic nanoparticles and universal tagged arrays. 26785734_our data provide a novel molecular mechanism for the tumor suppressor role of PSCA 26848528_the results indicated that the PSCA rs2294008 T and rs2976392 A alleles were low-penetrate risk factors for gastric cancer in this study population. 26982980_suggest that PSCA is a useful tissue marker for predicting biochemical recurrence in patients with high risk PC receiving NHT and radical prostatectomy 27001215_the PSCA rs2294008 polymorphism may serve as a biomarker of cervical cancer, particularly of early-stage cervical cancer 27050280_Among Chinese Han women, the PSCA rs2294008, rs2978974, and rs2976392 minor alleles are associated with increased breast cancer risk especially in progesterone receptor positive breast cancer patients, with breast cancer risk in postmenopausal women, and with high lymph node metastasis risk, respectively. 27232854_The expression of Ki-67, PSCA, and Cox-2 biomarkers along with other clinicopathologic factors were prognostic factors for biochemical recurrence in patients with clinically localized prostate cancer following radical prostatectomy 28755148_Study shows that PSCA gene is a target of 8q24 locus in Invasive micropapillary carcinoma (IMPC) of the breast, and the PSCA gene amplification results in PSCA protein overexpression. These results demonstrate an independently worse prognosis for PSCA overexpression in IMPC, and suggest that the differential expression of PSCA is associated with cell adhesion molecules in breast cancer. 28845520_PSCA regulates IL-6 expression through p38/NF-kappaB signaling in Prostate cancer. 28971496_PSCA is a novel cell cycle regulator with a key role in prostate cancer cell proliferation. 29332451_Men with the rs1045531 AC genotype of prostate stem cell antigen(PSCA) were at higher risk of prostate cancer in Chinese patients undergoing prostate biopsy. 29855276_PSCA expression is a statistically independent predictor of favorable prognosis in prostate cancer. Although its prognostic impact per se is not very strong, PSCA expression analysis could be considered for inclusion in multi-parametric prognostic tests to distinguish prostate cancers with need for radical therapy. 29892961_The decreased PSCA m-RNA levels were involved in the progress of bladder cancer. T allele takes more responsibility for PSCA m-RNA down-regulation to promote cell proliferation and migration and hinder cell apoptosis, thus leading to a higher risk. 30189721_we confirmed that variants in the PSCA gene showed significant GWAS associations in a Korean population for gastric cancer, especially for females and diffuse type gastric cancer. 30191681_Data imply an upregulated expression of PTGER4 and PSCA as well as a downregulated expression of MBOAT7 in gastric tissue as risk-conferring gastric cancer patho-mechanisms. 30753327_PSCA single nucleotide polymorphisms are associated with Heliobacter pylori-induced gastric atrophy. 31213476_Prostate Stem Cell Antigen Gene Polymorphism Is Associated with H. pylori-related Promoter DNA Methylation in Nonneoplastic Gastric Epithelium. 31416884_Impact of PSCA gene polymorphisms in modulating gastric cancer risk in the Chinese population. 31839644_Impact of PSCA Polymorphisms on the Risk of Duodenal Ulcer. 31983162_Association between PSCA, TNF-alpha, PARP1 and TP53 Gene Polymorphisms and Gastric Cancer Susceptibility in the Brazilian Population. 32640304_Risk of gastric ulcer contributed by genetic polymorphisms of PSCA: A case-control study based on Chinese Han population. 32660489_Association of MUC1 5640G>A and PSCA 5057C>T polymorphisms with the risk of gastric cancer in Northern Iran. 32664326_Evolutionary History of the Risk of SNPs for Diffuse-Type Gastric Cancer in the Japanese Population. 33029516_The Correlation between PSCA Expression and Neuroendocrine Differentiation in Prostate Cancer. 33128686_Polymorphisms PSCA rs2294008, IL-4 rs2243250 and MUC1 rs4072037 are associated with gastric cancer in a high risk population. 33774280_Silencing of the proteasome and oxidative stress impair endoplasmic reticulum targeting and signal cleavage of a prostate carcinoma antigen. 34472665_LncPSCA in the 8q24.3 risk locus drives gastric cancer through destabilizing DDX5. | ENSMUSG00000022598 | Psca | 1.718864e+01 | 1.2399831 | 0.310320476 | 0.7264734 | 1.727235e-01 | 0.6777026082 | No | Yes | 1.604567e+01 | 6.730451 | 1.349998e+01 | 5.922384 | ||
| ENSG00000167733 | 374875 | HSD11B1L | protein_coding | Q7Z5J1 | Alternative splicing;NADP;Oxidoreductase;Reference proteome;Secreted;Signal | This gene is a member of the hydroxysteroid dehydrogenase family. The encoded protein is similar to an enzyme that catalyzes the interconversion of inactive to active glucocorticoids (e.g. cortisone). Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jun 2012]. | hsa:374875; | extracellular region [GO:0005576]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; oxidoreductase activity [GO:0016491] | 19436836_cloning, purification, and characterization of SCDR10B; highly expressed in brain (especially in hippocampal neurons); up-regulated in lung cancer cell lines and lung cancer tissue | 3.226247e+02 | 1.0727144 | 0.101266087 | 0.3200547 | 9.870676e-02 | 0.7533871212 | 0.94816615 | No | Yes | 2.889688e+02 | 40.266563 | 2.394667e+02 | 34.611995 | ||||
| ENSG00000167904 | 137695 | TMEM68 | protein_coding | Q96MH6 | Alternative splicing;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:137695; | integral component of membrane [GO:0016021]; acyltransferase activity [GO:0016746] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000028232 | Tmem68 | 1.415239e+02 | 0.7356510 | -0.442906634 | 0.3743207 | 1.348344e+00 | 0.2455678885 | 0.78892886 | No | Yes | 1.303395e+02 | 25.114159 | 1.610201e+02 | 31.518247 | |||
| ENSG00000168071 | 283234 | CCDC88B | protein_coding | A6NC98 | FUNCTION: Acts as a positive regulator of T-cell maturation and inflammatory function. Required for several functions of T-cells, in both the CD4(+) and the CD8(+) compartments and this includes expression of cell surface markers of activation, proliferation, and cytokine production in response to specific or non-specific stimulation (By similarity). Enhances NK cell cytotoxicity by positively regulating polarization of microtubule-organizing center (MTOC) to cytotoxic synapse, lytic granule transport along microtubules, and dynein-mediated clustering to MTOC (PubMed:25762780). Interacts with HSPA5 and stabilizes the interaction between HSPA5 and ERN1, leading to suppression of ERN1-induced JNK activation and endoplasmic reticulum stress-induced apoptosis (PubMed:21289099). {ECO:0000250|UniProtKB:Q4QRL3, ECO:0000269|PubMed:21289099, ECO:0000269|PubMed:25762780}. | Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Endoplasmic reticulum;Golgi apparatus;Membrane;Phosphoprotein;Reference proteome;Stress response | This gene encodes a member of the hook-related protein family. Members of this family are characterized by an N-terminal potential microtubule binding domain, a central coiled-coiled and a C-terminal Hook-related domain. The encoded protein may be involved in linking organelles to microtubules. [provided by RefSeq, Oct 2009]. | hsa:283234; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; Golgi apparatus [GO:0005794]; membrane [GO:0016020]; dynein light intermediate chain binding [GO:0051959]; microtubule binding [GO:0008017]; cytoplasmic microtubule organization [GO:0031122]; cytoskeleton-dependent intracellular transport [GO:0030705]; defense response to protozoan [GO:0042832]; positive regulation of cytokine production [GO:0001819]; positive regulation of T cell activation [GO:0050870]; positive regulation of T cell proliferation [GO:0042102] | 21289099_Data show that a novel Girdin family protein, Gipie, is expressed in endothelial cells, where it interacts with GRP78, a master regulator of the UPR, and that Gipie/GRP78 interaction controls the IRE1-JNK signaling pathway. 22837380_A genotype dataset found a sarcoidosis risk locus on chromosome 11q13.1, a risk region for Crohn disease. An allele-dependent expression of CCDC88B was found in bronchoalveolar lavage samples of German, Swedish and Czech patients with sarcoidosis. 25762780_results demonstrate an important role for HkRP3 in regulating the clustering of lytic granules and MTOC repositioning during the development of NK cell-mediated killing. 29030607_A critical function of CCDC88B in colonic inflammation and inflammatory bowel disease | ENSMUSG00000047810 | Ccdc88b | 1.422881e+02 | 0.7990903 | -0.323569566 | 0.3366856 | 9.071612e-01 | 0.3408687548 | 0.82750349 | No | Yes | 6.839624e+01 | 17.473898 | 8.693271e+01 | 22.786476 | |
| ENSG00000168283 | 648 | BMI1 | protein_coding | P35226 | FUNCTION: Component of a Polycomb group (PcG) multiprotein PRC1-like complex, a complex class required to maintain the transcriptionally repressive state of many genes, including Hox genes, throughout development. PcG PRC1 complex acts via chromatin remodeling and modification of histones; it mediates monoubiquitination of histone H2A 'Lys-119', rendering chromatin heritably changed in its expressibility (PubMed:15386022, PubMed:16359901, PubMed:26151332, PubMed:16714294, PubMed:21772249, PubMed:25355358, PubMed:27827373). The complex composed of RNF2, UB2D3 and BMI1 binds nucleosomes, and has activity only with nucleosomal histone H2A (PubMed:21772249, PubMed:25355358). In the PRC1-like complex, regulates the E3 ubiquitin-protein ligase activity of RNF2/RING2 (PubMed:15386022, PubMed:26151332, PubMed:21772249). {ECO:0000269|PubMed:15386022, ECO:0000269|PubMed:16359901, ECO:0000269|PubMed:16714294, ECO:0000269|PubMed:16882984, ECO:0000269|PubMed:21772249, ECO:0000269|PubMed:25355358, ECO:0000269|PubMed:26151332, ECO:0000269|PubMed:27827373}. | 3D-structure;Chromatin regulator;Cytoplasm;Metal-binding;Nucleus;Proto-oncogene;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a ring finger protein that is major component of the polycomb group complex 1 (PRC1). This complex functions through chromatin remodeling as an essential epigenetic repressor of multiple regulatory genes involved in embryonic development and self-renewal in somatic stem cells. This protein also plays a central role in DNA damage repair. This gene is an oncogene and aberrant expression is associated with numerous cancers and is associated with resistance to certain chemotherapies. A pseudogene of this gene is found on chromosome X. Read-through transcription also exists between this gene and the upstream COMM domain containing 3 (COMMD3) gene. [provided by RefSeq, Sep 2015]. | hsa:100532731;hsa:648; | cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PcG protein complex [GO:0031519]; PRC1 complex [GO:0035102]; ubiquitin ligase complex [GO:0000151]; promoter-specific chromatin binding [GO:1990841]; RING-like zinc finger domain binding [GO:0071535]; zinc ion binding [GO:0008270]; hemopoiesis [GO:0030097]; heterochromatin assembly [GO:0031507]; histone H2A-K119 monoubiquitination [GO:0036353]; negative regulation of gene expression, epigenetic [GO:0045814]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of ubiquitin-protein transferase activity [GO:0051443]; regulation of gene expression [GO:0010468]; segment specification [GO:0007379] | 12183433_The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. 12482990_Bmi-1 extends the replicative life span of human fibroblasts by suppressing the p16-dependent senescence pathway 12829790_BMI-1 binds the MLL repression domain. Recruitment of BMI-1 to the MLL protein may be able to modulate its function. 14536079_E2a-Pbx1 and Bmi-1 are likely to play a role in the pathogenesis of human lymphoid leukemias through downregulation of the INK4A-ARF gene 14732230_modulation of Bmi-1 protein might be involved in human colorectal carcinogenesis by repressing the INK4a/ARF proteins 15009096_BMI-1 is transiently localized with the centromeric region in interphase. 15029199_BMI1 overexpression is an alternative or additive mechanism in the pathogenesis of medulloblastomas 15563468_Activation or overexpression of MAPKAP kinase 3pK resulted in phosphorylation of Bmi1 and other PcG members and their dissociation from chromatin 15931389_Altered expression is associated with therapy failure and death in patients with multiple types of cancer. 16105758_a subgroup of M0 patients has a high expression level of polycomb group gene BMI-1, which may contribute to leukemogenesis 16107895_CALM-AF10+ T-ALL associatd with elevated expression of subset of HOXA genes and some of there transcriptional and functional regulators, as well as specific overexpression of oncogene BMI1 16314526_EZH2 is essential for BMI1 recruitment to the polycomb bodies. 16501599_Critical for the short-term survival of cancer cells, and inhibition has minimal effect on the survival of normal cells. 16537449_p16(INK4a) expression was regulated by the Polycomb group repressor Bmi-1, which is shown as a direct transcriptional target of c-Myc. 16714294_Bmi-1-Ring1B complex stabilizes the interaction of the ubiquitin ligase complex. 16751658_Bmi1 is required for H2A ubiquitylation; H2A ubiquitylation regulates Bmi1-mediated gene expression 16874656_BMI-1 is expressed in gastrointestinal stem cells and may have a role protein in cellular differentiation and stem cell maintenance 16936260_bmi1 expression caused by oxidative stress may be involved in the pathogenesis of cellular senescence of biliary epithelial cells in primary biliary cirrhosis. 16963837_Advanced cell- & animal imaging, expression profiling, stable siRNA-gene targeting, and TMAs of experimental and clinical samples indicate that activation of the BMI1 oncogene-associated PcG pathway plays an essential role in metastatic prostate cancer. 16982619_Bmi-1 regulates the differentiation and clonogenic self-renewal of I-type neuroblastoma cells 17134822_PcG protein BMI1 is associated with adverse pathological features and clinical PSA recurrence of prostate cancer. 17145810_The oncogenic role of Bmi-1 in human primary breast carcinomas is not determined by its capacity to inhibit INK4a/ARF proteins(p16INK4a, p14ARF, or h-TERT) or to induce telomerase activity. 17145814_Bmi1 overexpression was correlated with the malignant grades of human digestive precancerous tissues, which suggests that advanced Bmi1 dysregulation might predict malignant progression. 17148583_ERK5 probably mediates HOXB9 expression by repressing BMI1 in Hodgkin lymphoma cell lines 17148591_Bmi-1 and LMP1 down-regulate the ataxia telangiectasia-mutated (ATM) tumor suppressor and conclude that Bmi-1 contributes to LMP1-induced oncogenesis in Hodgkin lymphoma 17151361_Our data suggest that Mel-18 regulates Bmi-1 expression during senescence via down-regulation of c-Myc. 17161394_Our results provide new information on the roles of Bmi-1 and HPV-16 E6 in the multi-step process of oral epithelial carcinogenesis. 17179983_The Bmi-1 may act through p16(INK4A)-independent pathways to regulate cellular proliferation during oral cancer progression. 17233832_many human cell types undergo senescence by activation of the p16(INK4a)/Rb pathway, and that introduction of Bmi-1 can inhibit p16(INK4a) expression and extend the life span of human epithelial cells 17298744_The intensive expression of Bmi-1 in breast cancer is related to tumor progression. 17344414_Study shows that the ability of the oncogene BMI1 to repress the INK4A-ARF locus requires its direct association and is dependent on the continued presence of the EZH2-containing Polycomb-Repressive Complex 2 complex. 17360938_In a cohort of 64 CML patients, the level of BMI1 at diagnosis correlated with time to transformation to blast crisis, and the combination of low BMI1 and high proteinase-3 expression was associated with an improved overall survival 17452456_These results suggest that Bmi-1 and Mel-18 may have overlapping functions in cancer cell growth. 17454639_Bmi-1 seems to play a secondary role in chronic myeloid leukemia transformation 17545584_Mel-18 and Bmi-1 may regulate the Akt pathway in breast cancer cells, and that Mel-18 functions as a tumor suppressor by repressing the expression of Bmi-1 and consequently down-regulating Akt activity. 17557835_BMI1 is a target gene for SALL4 in leukemic cells. 17597110_BMI-1 mediated repression of p16(ink4a) may contribute to an increased aggressive behavior of stem cell-like melanoma cells. 17625597_Bmi-1 may promote maintenance of suprabasal keratinocyte survival to prevent premature death during differentiation. 17666329_Bmi-1 mRNA expression is correlated to differentiation and metastasis of gastric carcinoma and may facilitate its prognostic evaluation. 17711569_Elevated Bmi-1 is associated with breast cancer 17917742_Bmi-1 could be a candidate biomarker for long-term survival in HCC 17974970_Examination of various growth-regulatory pathways suggested that Bmi-1 overexpression together with H-Ras promotes human mammary epithelial cell transformation and breast oncogenesis by deregulation of multiple growth-regulatory pathways. 17989714_BMI1 is an oncogenic candidate in a novel TCRB-associated chromosomal aberration in a patient with TCRgammadelta+ T-cell acute lymphoblastic leukemia 18094618_BMI-1 antibody as a potential marker of nasopharyngeal carcinoma may be rational, and could have diagnostic and prognostic value. 18156489_BMI1 is an intrinsic regulator of human stem/progenitor cell self-renewal. 18228133_Bmi-1 expression associated with favorable overall survival only in estrogen receptor-positive breast cancer patients. 18246051_loss of BMI-1 expression was associated with features of aggressive tumors. Low levels of BMI-1 expression were also significantly associated with decreased patient survival. 18264721_Bmi-1 expression in non-small cell lung cancer (NSCLC) in respect to clinicopathological features and therapeutic outcomes 18269588_Neither EGFR nor BMI-1 had significant prognostic impact for basal-like phenotype of breast cancer. 18346113_BMI-1 is a novel independent prognostic marker in oligodendroglial tumours. 18347933_study showed BMI-1 expression was inversely correlated with the differentiation grade of renal clear cell carcinoma; BMI-1 was strongly expressed in both papillary carcinomas and oncocytomas 18427816_BMI1 gene is aberrant at the chromosomal level in a subset of gliomas, and possibly contributes to brain tumour pathogenesis 18438345_Bmi-1 protein is highly expressed in ovarian epithelial cancer tissues, and its expression level correlates with histological grade and clinical phase of the patients. 18439910_Bmi1 can induce multipotency in astrocytes. 18450386_the results suggest that BMI1 does not play a significant prognosticator role ini astrocytic tumors 18475299_Loss of BMI-1 seems to be associated with an aggressive phenotype in endometrial carcinomas. 18565849_BMI-1 measured prior to allo-SCT can serve as a biomarker for predicting outcome in patients with CP-CML receiving allo-SCT 18591938_overexpression associated with the malignant progression of hepatocellular carcinoma 18592462_The relative expression of BMI1 in bladder tumor tissues is greater than 5 times that of nontumor samples and there is a statistically significant correlation between the level of BMI1 expression and the stage of the bladder tumors. 18668134_BMI1 overexpression accompanied with 11q23 rearrangements is associated with acute myeloid leukemias. 18701473_Overexpresion of BMI-1 is associated with ewing sarcoma tumorigenesis 18781299_inhibitors, BMI-1 is presumed to be positively regulated by BCR-ABL and further by posttranscriptional modification in the course of the disease progressio 18817867_Data demonstrate that Bmi1 promotes prostate tumorigenesis, and that Bmi1 expression is associated with a reduction in tumor suppressors p16(INK4A) and p14(ARF). 18829528_An important role for BMI1 in the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. 19012049_candidate for stem cell marker and renewal factor in nasal mucosa, may contribute to tissue homeostasis and differentiation in the epithelium and submucosal glands of normal nasal mucosa, and may play a role in proliferation of nasal polyps 19021148_dysregulated BMI-1 could indeed lead to keratinocytes transformation and tumorigenesis, potentially through promoting cell cycle progression and increasing cell mobility. 19080002_Overexpression of Bmi-1 and Ki67 protein are significantly correlated with tumorigenesis, metastasis and prognosis of colorectal carcinoma. 19080003_Bmi-1 protein is highly expressed in bladder cancer. 19099625_SALL4 and BMI-1 expressions are up-regulated significantly in acute leukemia. 19228380_Expression of Bmi-1 was greater in bladder cancers than in the adjacent normal tissues. Bmi-1 expression was positively correlated with tumor classification and TNM stage, but not with tumor number. 19233177_A polymorphism in Bmi-1 that reduces levels of this important protein. 19269851_transcripotional control of Th2 memory cell generation 19337376_provides evidence that, in the appropriate context, expression of the SYT-SSX2 oncogene leads to loss of Bmi1 polycomb function 19379548_Suppression of Bmi-1 expression is not able to reduce proliferation of K562 cells. 19386347_Distinct expression of BMI1 in hepatocellular carcinoma is reported. 19389366_The more pronounced effects of Bmi-1 over-expression in culture were largely attributable to the attenuated induction of p16(Ink4a) and p19(Arf) in culture, proteins that are generally not expressed by neural stem/progenitor cells in young mice in vivo. 19452584_Increased Bmi1 mRNA expression was significantly associated with esophageal squamous cell carcinoma progression, and the oncoprotein was largely distributed in the cytoplasm of tumor cells. 19543317_BMI1 collaborates with H-RAS to induce an aggressive and metastatic phenotype with the unusual occurrence of brain metastasis. 19556423_BMI1 expression is required for maintenance and self-renewal of normal and leukemic stem and progenitor cells, and expression of BMI1 protects cells against oxidative stress. 19605626_BMI1 is expressed in human GBM tumors and highly enriched in CD133-positive cells. Stable BMI1 knockdown using short hairpin RNA-expressing lentiviruses resulted in inhibition of clonogenic potential in vitro and of brain tumor formation in vivo 19639171_Bmi-1 mRNA might be a new tool to support the diagnosis of breast cancers 19695678_The overexpression of polycomb-group protein Bmi1 is essential for colony formation and cell proliferation, probably by the repression of cellular senescence in intrahepatic cholangiocarcinoma. 19778799_The overexpression of Bmi-1 protein was significantly correlated to the tumorigenesis, metastasis and prognosis of colorectal cancer. 19806792_Inhibition of Bmi-1 expression in human gastric carcinoma cells affects cell proliferation and invasiveness. 19884659_upregulation of Bmi-1 induced epithelial-mesenchymal transition (EMT) and enhanced the motility and invasiveness of nasopharyngeal epithelial cells, whereas silencing endogenous Bmi-1 expression reversed EMT and reduced motility 19903340_Nuclear PTEN reduces BMI1 function independently of its phosphatase activity. 19903841_Bmi-1 protein is downregulated by miR-15a or miR-16 expression and leads to significant reduction in ovarian cancer cell proliferation and clonal growth. 19907431_key role of Bmi-1 downregulation in enforcing aging in primary human keratinocytes 19934271_Bmi1 functions independent of Ink4A/Arf repression in liver cancer development. 19940551_Data show that tumor spheroid cells express ABCG2, Bmi1, WNT5A, CD133, prox1 and VEGFR3. 19956605_Silencing of Bmi1 inhibits drug-induced Top2alpha degradation, increases the persistence of Top2alpha-DNA cleavage complex, and increases Top2 drug efficacy. 19957112_High Bmi1 is associated with colon cancer. 20015867_Although Bmi-1 does not influence the basal level of apoptotic activity, its overexpression protects the tumor cells against the proapoptotic action of (-)-epigallocatechin-3-gallate. 20024662_The expression of Bmi-1 was elevated in colon cancer and might serve as an independent prognostic marker. 20035051_Bmi-1 bestows apoptotic resistance to glioma cells through the IKK-NF-kappaB pathway; Bmi-1 as a useful indicator for glioma prognosis. 20085590_Bmi-1 gene is upregulated in early-stage hepatocellular carcinoma and correlates with ATP-binding cassette transporter B1 expression. 20127006_EZH2 and BMI-1 are upregulated in osteosarcoma. EZH2 and BMI-1 may be useful targets for cancer immunotherapy of osteosarcoma, although knock-down of EZH2 and BMI-1 could not prevent osteosarcoma growth. 20145620_Negative Bmi-1 expression is associated with squamous cell carcinoma of the tongue. 20170541_BMI1 acts as an oncogene and Mel-18 functions as a tumor suppressor via downregulation of BMI1. 20190806_Bmi1 is a MYCN target gene that regulates tumorigenesis through repression of KIF1Bbeta and TSLC1 in neuroblastoma. 20308890_Studies indicate that in the absence of BMI1, reactive oxygen species accumulate, associating with activation of DNA damage response pathways and increased apoptosis. 20377880_Expression of Bmi-1 might be important in the acquisition of an invasive and/or aggressive phenotype of ovarian carcinoma, and serve as a independent biomarker for shortened survival time of patients. 20385206_The study confirmed a direct involvement of Bmi-1 in the regulation of human fetal NSCs self-renewal and proliferation in vitro, and suggested that the Ink4a/Arf locus was a potential downstream target of Bmi-1 pathway. 20426791_BMI1 plays an important role in the late progression of pancreatic cancer and may represent a therapeutic target for treatment of this cancer. 20428793_BMI-1 depletion leads to reduced cell growth and proliferation, and increased cell apoptosis. 20530672_Studies not only highlight Bmi-1 as a cancer-dependent factor in multiple myeloma, but also elucidate a novel antiapoptotic mechanism for Bmi-1 function involving the suppression of Bim. 20551323_the molecular mechanism of Bmi-1-mediated regulation of the p16 gene 20564407_Bmi1 and EZH2 had characteristic and distinctive expression in NSCLCs. 20568112_Loss of BMI1 enhances docetaxel activity and impairs antioxidant response and promotes prostate cancer. 20569464_data suggest that the PS domain of BMI1 is involved in its stability and that it negatively regulates function of BMI1 oncoprotein 20574517_miR-128a has growth suppressive activity in medulloblastoma and that this activity is partially mediated by targeting Bmi-1. 20591222_Bmi-1 downregulation in esophageal carcinoma cells inhibits cell proliferation, cell cycle, and cell migration, while increasing cell apoptosis. 20592341_the BMI-I protein is significantly overexpressed in ovarian, endometrial and cervical cancer 20661663_knockdown of BMI-1 expression can induce cell-cycle arrest and up-regulate p16INK4a, HOXA9 and HOXC13 in HeLa cells 20717685_Our analysis showed correlation between BMI1 and PCGF2 gene's expression and survival in children with medulloblastoma. 20724541_Introduction of BCR-ABL and BMI1 in human cord blood CD34+ cells induces lymphoid leukemia upon transplantation in NOD/SCID mice. 20809956_Overexpression of Bmi-1 is associated with esophageal squamous cell carcinoma. 20818389_Bmi1 and Twist1 act cooperatively to repress expression of both E-cadherin and p16INK4a. 20818602_High BMI1 is associated with lung adenocarcinoma. 20921134_BMI1 contributes to histone ubiquitylation and has a role in maintaining genomic stability. 21039844_A direct correlation analysis showed that ubiquitin-specific protease 22 was strongly correlated with BMI-1 and c-Myc at the mRNA level. 21047978_ESFT that do not overexpress BMI-1 represent a novel subclass with a distinct molecular profile and altered activation of and dependence on cancer-associated biological pathways. 21059209_Decreased Mel-18 and increased Bmi-1 mRNA expression was associated with the carcinogenesis and progression of gastric cancer 21113618_C-MYC and BMI-1 expression was detected in stromal cells only 21125401_Overexpression of the EZH2, RING1, and BMI1 genes is common in MDS and indicate poor prognosis. The products of these genes might participate in epigenetic regulation of Myelodysplastic syndrome. 21152871_Bmi-1 may be a potential molecular target for the therapy of breast carcinoma. 21162745_Bmi-1/Mel-18 ratio can be potentially used as a tool for stratifying women at risk of developing breast malignancy. 21164364_Increase of BMI1 is associated with lung adenocarcinoma. 21170084_a previously unrecognized link between BMI-1, contact inhibition and the Hippo-YAP pathway 21252986_higher expression of BMI-1 and immune responses to BMI-1 protein is associated with improved outcome following allogeneic stem cell transplantation. 21259057_the aim of this study was to investigate the prognosis and clinicopathologic roles of beta-catenin, Wnt1, Smad4, Hoxa9, and Bmi-1 in esophageal squamous cell carcinoma 21276135_WWOX was identified as a Bmi1 target in small cell lung cancer cells 21276221_Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer 21307872_Bmi-1 elicits radioprotective effects on normal human keratino by mitigating the genotoxicity of ionizing radiation through epigenetic mechanisms. 21309753_BMI1 protein human might be an independent prognostic marker in patient diagnosed with uterine cervical cancer. 21370603_Bmi-1 might participate in the development and progression of uterine cervical cancer and have clinical potential not only as a useful predictor of aggressive phenotype but also a promising prognostic predictor. 21373837_Studies indicate that new targets have recently been investigated as potential modulators in myeloid leukemia pathogenesis, including miR-328, BMI1, FOXOs and IL1RAP. 21383063_BMI1 is recruited to sites of DNA DSBs, where it persists for more than 8 h. BMI1 is required for postdamage ubiquitination of histone H2A at lysine 119 and contributes to efficient homology-mediated repair of DNA breaks. 21428211_Bmi-1 siliencing combined treatment of doxorubicin might be a new strategy for biological treatment on breast cancer. 21430439_F box protein betaTrCP regulates BMI1 protein turnover via ubiquitination and degradation. 21496667_results indicate that dMSCs lose their odontogenic differentiation potential during senescence, in part by reduced Bmi-1 expression. 21572997_DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs.( 21638011_A significant association between the expression of Bmi-1 and estrogen receptor was found in primary breast cancers and their metastases. 21656027_Bmi-1 may be associated with the epithelial-mesenchymal transition in lung squamous cell carcinoma 21678481_It has been clearly established that BMI1 is a bonafide oncogene and plays critical roles in promoting cancer stem cell self-renewal and tumorogenesis. 21685941_Knockdown experiments of Bmi1 in vitro and in vivo demonstrate that Hh signaling not only drives Bmi1 expression, but a feedback mechanism exists wherein downstream effectors of Bmi1 may, in turn, activate Hh pathway genes 21725369_correlated the expression of miR-200b, miR-15b and BMI1 with the clinicopathological status and prognosis of tongue squamous cell carcinoma patients 21732356_Data indicate that the dynamics of recognition of UV-damaged chromatin, and the nuclear arrangement of BMI1 protein can be influenced by acetylation and occur as an early event prior to the recruitment of HPbeta to UV-irradiated chromatin. 21735131_simultaneous activation of USP22 and BMI-1 may associate with GC progression and therapy failure 21755556_BMI1 was required to maintain the proliferative capacity of laryngeal Cancer stem cells. 21772249_Crystallography of a complex of the Bmi1/Ring1b RING-RING heterodimer & UbcH5c shows that UbcH5c interacts with Ring1b only. Bmi1/Ring1b interacts with nucleosomal DNA & an acidic patch on histone H4 to achieve specific monoubiquitination of H2A. 21851624_MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. 21856782_dysregulation of the BMI1 gene mediated by MYCN or MYC overexpression, confers increased cell proliferation during neuroblastoma genesis and tumor progression 21865393_Correlating with the loss of H3K27me3, human papillomavirus 16 E6/E7-expressing cells exhibited derepression of specific EZH2-, KMD6A-, and BMI1-targeted HOX genes. 21866458_Bmi1 expression is associated with the clinicopathological characteristics of colorectal cancer. 21883379_Twist1 directly activates Bmi1 expression and that these two molecules function together to mediate cancer stemness and epithelial-mesenchymal transition--{REVIEW} 21919891_Epithelial-mesenchymal transition and cancer stemness: the Twist1-Bmi1 connection 21960247_a c-Myb-Bmi1 transcription-regulatory pathway is required for p190(BCR/ABL) leukemogenesis. 21999765_Overexpression of BMI-1 correlates with drug resistance in B-cell lymphoma cells through the stabilization of survivin expression. 22009787_ATP-binding cassette, G2 subfamily (ABCG2) and BMI-1, predict the transformation of oral leukoplakia to cancer. 22040961_Expressions of ABCB1, BMI-1, HOXB4 were not detected in the patients with non-malignant hematologic diseases, but were higher in de novo acute leukemia patients and lower in patients in complete remission. 22096526_Results suggest that Bmi1 plays a crucial role in the maintenance of the stem cell pool in postnatal skeletal muscle and is essential for efficient muscle regeneration after injury especially after repeated muscle injury. 22120066_Overexpression of BMI1 confers clonal cells resistance to apoptosis and contributes to adverse prognosis in myelodysplastic syndrome. 22120721_report that BMI1 has a critical role in stabilizing cyclin E1 by repressing the expression of FBXW7, a substrate-recognition subunit of the SCF E3 ubiquitin ligase that targets cyclin E1 for degradation 22132147_Measuring BMI-1 autoantibody levels of patients with cervical cancer could have clinical prognostic value as well as a non-tissue specific biomarker for neoplasms expressing BMI-1. 22170051_Regulation of the potential marker for intestinal cells, Bmi1, by beta-catenin and the zinc finger protein KLF4: implications for colon cancer. 22209830_results indicate that Bmi-1 may play an important role in breast cancer radiosensitivity. 22252887_BMI1, which is upregulated in a variety of cancers, has a positive correlation with clinical grade/stage and poor prognosis. [Review] 22265735_Bmi1 expression profiles show a marked elevation in the proneural glioblastoma subtype. 22293918_Bmi-1 plays a part in autoreactive CD4+ T cell proliferative capability and apoptotic resistance in immune thrombocytopenia patients 22366984_Bmi-1 status in breast cancer is an independent prognostic factor, which also is associated with tumor histological grade and basal-like phenotype. 22384090_Bmi1 is down-regulated in the aging brain and displays antioxidant and protective activities in neurons 22420951_Bmi-1 regulates key pathways involved in the metastasis of lung adenocarcinoma cells. 22509111_high expression of p16(INK4a) and low expression of Bmi1 are associated with endothelial cellular senescence in human cornea. 22664532_After cisplatin treatment, Bmi1 is required for the self-renewal of stem-like cells that are important for the expansion of the stem-like cell pool in human A549 cells and that targeting Bmi1 slows down the formation of tumors in vivo. 22725270_expression patterns of ALDH1 and Bmi1 in OE were associated with malignant transformation, suggesting they may be valuable predictors for evaluating risk of oral cancer. 22766604_Both Bmi1 and EZH2, not each alone, could be potent candidates of new target therapy in esophageal squamous cell carcinoma. 22777769_Bmi-1 represses transcription of insulin-like growth factor-1 receptor and abrogates IGF-IR signaling in prostate cancer 22825250_Structural alterations of BMI1 and downstream effectors are rare but they do occur in human myeloproliferative neoplasms. 22842564_These results suggest that Bmi-1 plays an important role in tumor radioadaptive resistance under FIR and may be a potent molecular target for enhancing the efficacy of fractionated RT. 22863087_Coexpression of B-lymphoma Moloney murine leukemia virus insertion region-1 (BMI1) and sex-determining region of Y chromosome-related high mobility group box-2 (SOX2) may promote cervical carcinogenesis. 22872929_Bmi-1 siRNA has a role in inhibiting ovarian cancer cell line growth and decreasing telomerase activity 22898137_Cervical cancer with excessive BMI-1 expression possesses high metastases potential and that BMI-1 may be a promising biomarker for predicting metastasis in cervical cancer. 22912356_We found that overexpression of c-Myc was significantly associated with that of BMI1, and that patients who harbored glioblastomas overexpressing c-Myc and BMI1 showed significantly longer overall survival. 22967049_Knockdown of endogenous Bmi-1 reduced the invasiveness and migration of glioma cells. NF-kappaB transcriptional activity and MMP-9 expression and activity were significantly increased in Bmi-1-overexpressing but reduced in Bmi-1-silenced cells. 22982443_miR-200c overexpression resulted in significant down-regulation of BMI-1. 22994774_There was higher Bmi-1 protein expression in intestinal-type carcinomas. 23023333_Our results suggest that CHMP1A serves as a critical link between cytoplasmic signals and BMI1-mediated chromatin modifications that regulate proliferation of central nervous system progenitor cells. 23046527_There was no significant correlation between p16INK4A expression and BMI1 expression. 23057360_The expressions of Bmi1 and hTERT genes may have a close relation to the proliferative capacity of CD34+ cells. 23078618_This study demonstrates that high expression Bmi-1 is associated with esophageal adenocarcinoma and precancerous lesions, which implies that Bmi-1 plays an important role in early carcinogenesis in esophageal adenocarcinoma. 23092893_Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus. 23138990_High expression of Bmi1 was associated with hepatocarcinogenesis. 23204235_our results link CSCs, EMT, and CIN through the BMI1-AURKA axis 23239878_A positive feedback loop connected by the WNT signaling pathway regulates BMI1 expression. 23242307_Downregulation of the Bmi1 gene by RNAi can inhibit the proliferation and invasivesness of hepatocellular carcinoma cells. 23255074_miR-218 suppressed protein expression of BMI-1 downstream targets of cyclin-dependent kinase 4, a cell cycle regulator, while upregulating protein expression of p53. 23308129_BMI1 could be developed as a dual bio-marker (serum and biopsy) for the diagnosis and prognosis of prostate cancer in Caucasian and African-American men. 23383216_Bmi-1 promotes glioma angiogenesis by activating NF-kappaB signaling. 23437065_These results implicate Bmi1 in the invasiveness and growth of pancreatic cancer and demonstrate its key role in the regulation of pancreatic cancer stem cells. 23468063_The Bmi-1 pathway was downregulated in GBM cells. 23525303_siRNA against Bmi-1 significantly suppresses tumor growth and induces apoptosis in vitro and in vivo. 23559850_BMI-1 might render important oncogenic property of retinoblastomas. 23592496_Authors assessed the functional relevance of two genes, FoxG1 and Bmi1, which were significantly enriched in non-Shh/Wnt MBs and showed these genes to mediate MB stem cell self-renewal and tumor initiation in mice. 23601184_Sp1 and c-Myc are important transcription factors that directly bind to the BMI1 promoter region and regulate gene expression in nasopharyngeal carcinoma. 23671559_Regulation of TCF4-mediated BCL2 gene expression by BMI1 is universal. 23686310_Upregulation of BMI1 and downregulation of miR-16 in mantle cell lymphoma (MCL) side population has a key role in the disease's progression by reducing MCL cell apoptosis. 23700898_Bmi-1 may contribute to malignant cell transformation, and its expression in tissues taken from patients with cervical, breast and ovarian cancer could be a marker for diagnosis and prognosis. 23708658_Data indicate that post-translational phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis. 23712029_BMI-1 expression plays a role in hTERT-induced immortalisation and epithelial-mesenchymal transition. 23727134_BMI-1 and RING1B expression was enhanced with the development of embryos in early pregnancy. 23744494_Single nucleotide polymorphism in BMI1-PIP4K2A is associated with Precursor B-Cell Lymphoblastic Leukemia-Lymphoma. 23807724_These data demonstrate that Bmi-1 plays a vital role in HCC invasion and that Bmi-1 is a potential therapeutic target for HCC 23820733_BMI1 may represent a novel diagnostic marker and a therapeutic target for bladder cancer, and deserves further investigation. 23864317_Bmi1 and FoxF1 may cooperate with hedgehog signaling in non-small-cell lung carcinogenesis. 23873108_Data indicate that B cell-specific Moloney murine leukaemia virus integration site 1 (BMI1) gene was co-regulated with progesterone receptor (PR) in the invasive ductal breast carcinoma. 23873701_Identified a novel t(10;14)(p12;q32)/IGH-BMI1 rearrangement and its IGL variant in six cases of chronic lymphocytic leukemia; this study shows that BMI1 is recurrently targeted by chromosomal aberrations in B-cell leukemia/lymphoma. 23950210_High BMI1 expression is associated with glioma. 23984324_Increased expression of the Bmi-1 in pediatric brain tumors may be important in the acquisition of an aggressive phenotype. 23990442_Bmi-1 was a significant prognostic factor of poor overall survival in lung adenocarcinoma patients. 24039897_MiR-200c and miR-203 overexpression in breast cancer cells downregulated Bmi1 expression accompanied with reversion of resistance to 5-Fu mediated by Bmi1. 24122997_Aberrant EZH2 and BMI1 expression was significantly associated with mode of invasion, but not with lymph node metastasis or survival rate in oral squamous cell carcinoma. 24139839_The data of this study suggest for the first time that the combination of Bmi1 and EZH2 overexpression may be a highly sensitive marker for the prognosis in glioma patients. 24145240_Early detection marker distinguishes cancerous from non-cancerous regions in mouth squamous cell carcinoma 24219349_stem renewal factor Bmi-1 was a direct target of miR-203 24252251_miR-15a inhibits cell proliferation and epithelial-mesenchymal transition in pancreatic ductal adenocarcinoma via the down-regulation of Bmi-1 expression 24292392_Downregulation of BMI-1 inhibits the ability of colorectal cancer-initiating cells to self-renew, resulting in the abrogation of their tumorigenic potential. 24300467_Bmi1 displays higher expression in recurrent than in primary ovarian neoplasms 24312366_findings suggest that TAMs may cause increased Bmi1 expression through miR-30e* suppression, leading to tumor progression 24317363_knockdown of Bmi-1 in PC3 and DU145 cells significantly reduced cell proliferation and upregulated the p16 tumor-suppressor 24337040_Bmi1 knockdown inhibited cell cycle progression through derepression of the p16INK4a/p14ARF locus and caused inhibition of cell proliferation, migration, tumor sphere formation, enhanced sensitivity to cisplatin, and cell cycle arrest in the G0/G1 phase 24342665_The study suggested correlation between increased Bmi-1 expression and clinical progression in ovarian epi | ENSMUSG00000026739 | Bmi1 | 5.480929e+02 | 1.0975728 | 0.134316654 | 0.3680484 | 1.325433e-01 | 0.7158095092 | 0.94026496 | No | Yes | 4.135764e+02 | 85.566530 | 4.497193e+02 | 95.367573 | |
| ENSG00000168300 | 115294 | PCMTD1 | protein_coding | Q96MG8 | Alternative splicing;Cytoplasm;Lipoprotein;Membrane;Myristate;Reference proteome | hsa:115294; | cytoplasm [GO:0005737]; membrane [GO:0016020]; protein-L-isoaspartate (D-aspartate) O-methyltransferase activity [GO:0004719] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23505305_Presence of at least one A allele (AG or AA genotype) for rs1015213 was associated with a shallower anterior chamber depth (-0.07 mm, 95% CI -0.01 to -0.14 mm, p=0.028) after adjusting for age and sex (both p=0.001). 23847314_In our study, rs1015213 (located in the intergenic region between PCMTD1 and ST18) was associated significantly with primary angle closure. 24474268_The three genetic susceptibility loci for primary angle-closure glaucoma did not underlie any major phenotypic diversity in terms of disease severity or progression. 27455018_No significant association of PLEKHA7 rs11024102, COL11A1 rs3753841 and PCMTD1-ST18 rs1015213 with primary angle closure glaucoma was found among ethnic Han Chinese from Sichuan 29310965_In this study, 2 of 8 (primary angle-closure glaucoma) PACG-associated loci were associated significantly with PACS status, the earliest stage in the angle-closure glaucoma disease course. The association of these PACG loci with PACS status suggests that these loci may confer susceptibility to a narrow angle configuration. 30377206_This study has identified an increased percentage of IDH1 and PCMTD1 mutations in Squamous Cell Lung Cancers arising in the Appalachian Kentucky residents versus The Cancer Genome Atlas, with population-specific implications for the personalized treatment of this disease. | ENSMUSG00000051285 | Pcmtd1 | 1.598364e+02 | 0.6777914 | -0.561086780 | 0.4662244 | 1.410191e+00 | 0.2350246315 | 0.78818582 | No | Yes | 9.600195e+01 | 25.267333 | 1.690580e+02 | 45.084811 | |||
| ENSG00000168411 | 55159 | RFWD3 | protein_coding | Q6PCD5 | FUNCTION: E3 ubiquitin-protein ligase required for the repair of DNA interstrand cross-links (ICL) in response to DNA damage (PubMed:21504906, PubMed:21558276, PubMed:26474068, PubMed:28575657, PubMed:28575658). Plays a key role in RPA-mediated DNA damage signaling and repair (PubMed:21504906, PubMed:21558276, PubMed:26474068, PubMed:28575657, PubMed:28575658, PubMed:28691929). Acts by mediating ubiquitination of the RPA complex (RPA1, RPA2 and RPA3 subunits) and RAD51 at stalled replication forks, leading to remove them from DNA damage sites and promote homologous recombination (PubMed:26474068, PubMed:28575657, PubMed:28575658). Also mediates the ubiquitination of p53/TP53 in the late response to DNA damage, and acts as a positive regulator of p53/TP53 stability, thereby regulating the G1/S DNA damage checkpoint (PubMed:20173098). May act by catalyzing the formation of short polyubiquitin chains on p53/TP53 that are not targeted to the proteasome (PubMed:20173098). In response to ionizing radiation, interacts with MDM2 and enhances p53/TP53 ubiquitination, possibly by restricting MDM2 from extending polyubiquitin chains on ubiquitinated p53/TP53 (PubMed:20173098). {ECO:0000269|PubMed:20173098, ECO:0000269|PubMed:21504906, ECO:0000269|PubMed:21558276, ECO:0000269|PubMed:26474068, ECO:0000269|PubMed:28575657, ECO:0000269|PubMed:28575658, ECO:0000269|PubMed:28691929}. | 3D-structure;Coiled coil;Cytoplasm;DNA damage;DNA repair;Disease variant;Fanconi anemia;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation pathway;WD repeat;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:26474068, ECO:0000269|PubMed:28575657, ECO:0000269|PubMed:28575658}. | hsa:55159; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; site of DNA damage [GO:0090734]; MDM2/MDM4 family protein binding [GO:0097371]; metal ion binding [GO:0046872]; p53 binding [GO:0002039]; ubiquitin protein ligase activity [GO:0061630]; cellular response to DNA damage stimulus [GO:0006974]; chromosome breakage [GO:0031052]; double-strand break repair via homologous recombination [GO:0000724]; interstrand cross-link repair [GO:0036297]; mitotic G1 DNA damage checkpoint signaling [GO:0031571]; protein ubiquitination [GO:0016567]; regulation of DNA damage checkpoint [GO:2000001]; replication fork processing [GO:0031297]; response to ionizing radiation [GO:0010212] | 19844255_Observational study of gene-disease association. (HuGE Navigator) 20173098_Study identifies RFWD3 as a positive regulator of p53 stability when the G(1) cell cycle checkpoint is activated and provides an explanation for how p53 is protected from degradation in the presence of high levels of Mdm2. 21504906_RFWD3 and RPA2 functionally interact and participate in replication checkpoint control 21558276_RFWD3 is recruited to sites of DNA damage and facilitates RPA-mediated DNA damage signaling and repair 26474068_RFWD3-dependent ubiquitination of RPA regulates repair at stalled replication forks. 28575657_Single point mutations in the RPA32 subunit of RPA that abolish interaction with RFWD3 also inhibit interstrand crossling repair, demonstrating that RPA-mediated RFWD3 recruitment to stalled replication forks is important for ICL repair. 28575658_E3 ligase RFWD3 functions in timely removal and degradation of RPA and RAD51 to allow homologous recombination progression to subsequent steps following mitomycin C damage. 28666352_The results suggest that RPA phosphorylation enhances the recruitment of PRP19 to RPA-ssDNA and stimulates RPA ubiquitylation through a process requiring both PRP19 and RFWD3, thereby triggering a phosphorylation-ubiquitylation circuitry that promotes ATR activation and homologous recombination. 31571161_Identification of an E3 ligase-encoding gene RFWD3 in non-small cell lung cancer. 32391871_E3 ligase RFWD3 is a novel modulator of stalled fork stability in BRCA2-deficient cells. 32902405_Down-regulation of RFWD3 inhibits cancer cells proliferation and migration in gastric carcinoma. 33044890_The E3 ligase RFWD3 stabilizes ORC in a p53-dependent manner. 33321094_The ubiquitin ligase RFWD3 is required for translesion DNA synthesis. | ENSMUSG00000033596 | Rfwd3 | 4.386479e+03 | 0.6182753 | -0.693678691 | 0.3104508 | 4.948684e+00 | 0.0261105326 | 0.55078242 | No | Yes | 2.800232e+03 | 502.629087 | 4.906781e+03 | 902.606448 | |
| ENSG00000168488 | 11273 | ATXN2L | protein_coding | Q8WWM7 | FUNCTION: Involved in the regulation of stress granule and P-body formation. {ECO:0000269|PubMed:23209657}. | Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Isopeptide bond;Membrane;Methylation;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes an ataxin type 2 related protein of unknown function. This protein is a member of the spinocerebellar ataxia (SCAs) family, which is associated with a complex group of neurodegenerative disorders. Several alternatively spliced transcripts encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:11273; | cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; membrane [GO:0016020]; nuclear speck [GO:0016607]; cadherin binding [GO:0045296]; RNA binding [GO:0003723]; mRNA metabolic process [GO:0016071]; stress granule assembly [GO:0034063] | 17392519_cellular ataxin-2 concentration is important for the assembly of stress granules and P-bodies, ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 20473688_Observational study of gene-disease association. (HuGE Navigator) 25748791_ATXN2L associates with the protein arginine-N-methyltransferase 1 (PRMT1). 30787271_ATXN2L promotes cell invasiveness and oxaliplatin resistance and can be upregulated by EGF via PI3K/Akt signaling. | ENSMUSG00000032637 | Atxn2l | 8.670429e+03 | 1.1482301 | 0.199411719 | 0.2619363 | 5.814238e-01 | 0.4457547672 | 0.85808635 | No | Yes | 8.073932e+03 | 720.717874 | 6.797120e+03 | 622.500158 | |
| ENSG00000168672 | 157638 | LRATD2 | protein_coding | Q96KN1 | Reference proteome | hsa:157638; | cytoplasm [GO:0005737]; plasma membrane [GO:0005886] | 19549893_Observational study of gene-disease association. (HuGE Navigator) 20158306_Observational study of gene-disease association. (HuGE Navigator) 25980316_We demonstrated the decreased of circulating FAM84B mRNA and protein after neoadjuvant CRT may predict pCR, and FAM84B protein is overexpressed in ESCC. 26759717_Data suggest that FAM84B protein and the NOTCH pathway are involved in the progression of esophageal squamous cell carcinoma (ESCC) and may be potential diagnostic targets for ESCC susceptibility. 28186973_High FAM84B expression is associated with prostate cancer progression. 30638658_non-coding RNA (lncRNA) FAM84B-AS promotes resistance of Gastric Cancer to platinum drugs through inhibition of FAM84B expression. 32183428_The Oncogenic Potential of the Centromeric Border Protein FAM84B of the 8q24.21 Gene Desert. 32291380_FAM84B, amplified in pancreatic ductal adenocarcinoma, promotes tumorigenesis through the Wnt/beta-catenin pathway. 32558489_Identification of Hub Genes in Gastric Cancer with High Heterogeneity Based on Weighted Gene Co-Expression Network. 33759248_FAM84B acts as a tumor promoter in human glioma via affecting the Akt/GSK-3beta/beta-catenin pathway. | ENSMUSG00000072568 | Lratd2 | 9.331529e+02 | 1.1381802 | 0.186728973 | 0.2855813 | 4.266200e-01 | 0.5136521592 | 0.88008149 | No | Yes | 9.631157e+02 | 144.647928 | 7.841685e+02 | 120.925093 | |||
| ENSG00000168944 | 153241 | CEP120 | protein_coding | Q8N960 | FUNCTION: Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors and for proper positioning of neurons during brain development. Also implicated in the migration and selfrenewal of neural progenitors. Required for centriole duplication and maturation during mitosis and subsequent ciliogenesis (By similarity). Required for the recruitment of CEP295 to the proximal end of new-born centrioles at the centriolar microtubule wall during early S phase in a PLK4-dependent manner (PubMed:27185865). {ECO:0000250|UniProtKB:Q7TSG1, ECO:0000269|PubMed:27185865}. | 3D-structure;Alternative splicing;Ciliopathy;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Joubert syndrome;Phosphoprotein;Reference proteome;Repeat | This gene encodes a protein that functions in the microtubule-dependent coupling of the nucleus and the centrosome. A similar protein in mouse plays a role in both interkinetic nuclear migration, which is a characteristic pattern of nuclear movement in neural progenitors, and in neural progenitor self-renewal. Mutations in this gene are predicted to result in neurogenic defects. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2009]. | hsa:153241; | centriole [GO:0005814]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; protein C-terminus binding [GO:0008022]; astral microtubule organization [GO:0030953]; centrosome cycle [GO:0007098]; cerebral cortex development [GO:0021987]; interkinetic nuclear migration [GO:0022027]; neurogenesis [GO:0022008]; positive regulation of centriole elongation [GO:1903724]; positive regulation of centrosome duplication [GO:0010825]; positive regulation of cilium assembly [GO:0045724]; positive regulation of establishment of protein localization [GO:1904951] | 17920017_Functional characterization of the homologous mouse gene, and comparison to the human protein. 19584346_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 23810536_CEP120 associates with SPICE1 and CPAP, and depletion of any of these proteins results in short procentrioles. Furthermore, CEP120 or CPAP overexpression results in excessive centriole elongation, a process dependent on CEP120, SPICE1, and CPAP. 23857771_CEP120 is a CPAP-interacting protein that positively regulates centriole elongation. 25361962_We establish a novel locus for Jeune asphyxiating thoracic dystrophy on 5q23.2 by linkage analysis and demonstrate that a mutation in CEP120 within this locus is the most likely cause of the disease. 27208211_The CEP120-associated phenotype ranges from mild classical JS in four patients to more severe conditions in two fetuses. 29741480_These results indicate that Cep120 helps to maintain centrosome homeostasis by inhibiting untimely maturation of the daughter centriole, and defines a potentially new molecular defect underlying the pathogenesis of ciliopathies such as Jeune Asphyxiating Thoracic Dystrophy and Joubert syndrome. 29847808_the ciliopathy-associated centriolar protein CEP120 contains three C2 domains. 30988386_CEP120 interacts with C2CD3 and Talpid3 and is required for centriole appendage assembly and ciliogenesis. 32772081_Exome sequencing links CEP120 mutation to maternally derived aneuploid conception risk. 33297941_Expression patterns of ciliopathy genes ARL3 and CEP120 reveal roles in multisystem development. 33486889_Update of genetic variants in CEP120 and CC2D2A-With an emphasis on genotype-phenotype correlations, tissue specific transcripts and exploring mutation specific exon skipping therapies. 34711653_CEP120-mediated KIAA0753 recruitment onto centrioles is required for timely neuronal differentiation and germinal zone exit in the developing cerebellum. | ENSMUSG00000048799 | Cep120 | 2.293823e+02 | 0.6198877 | -0.689921313 | 0.3568878 | 3.632400e+00 | 0.0566650412 | 0.68299703 | No | Yes | 1.436960e+02 | 25.003837 | 2.304897e+02 | 40.592657 | |
| ENSG00000169221 | 26000 | TBC1D10B | protein_coding | Q4KMP7 | FUNCTION: Acts as GTPase-activating protein for RAB3A, RAB22A, RAB27A, AND RAB35. Does not act on RAB2A and RAB6A. {ECO:0000269|PubMed:16923811, ECO:0000269|PubMed:19077034}. | Alternative splicing;Coiled coil;Cytoplasm;GTPase activation;Methylation;Phosphoprotein;Reference proteome | Small G proteins of the RAB family (see MIM 179508) function in intracellular vesicle trafficking by switching from the GTP-bound state to the GDP-bound state with the assistance of guanine nucleotide exchange factors (GEFs; see MIM 609700) and GTPase-activating proteins (GAPs). TBC1D10B functions as a GAP for several proteins of the Rab family (Ishibashi et al., 2009 [PubMed 19077034]).[supplied by OMIM, Nov 2010]. | hsa:26000; | cytosol [GO:0005829]; plasma membrane [GO:0005886]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; intracellular protein transport [GO:0006886]; regulation of GTPase activity [GO:0043087]; retrograde transport, endosome to Golgi [GO:0042147] | 23248241_Data suggest that EPI64A and B, which are ubiquitously expressed members of the EPI64 subfamily, inactivate Ras and certain Rabs at the periphery of cells. 31527750_Regulation of VEGFR2 trafficking and signaling by Rab GTPase-activating proteins. 32397798_STAT4-mediated down-regulation of miR-3619-5p facilitates stomach adenocarcinoma by modulating TBC1D10B. 34757852_The RabGAPs EPI64A and EPI64B regulate the apical structure of epithelial cells (dagger). | ENSMUSG00000042492 | Tbc1d10b | 7.736876e+03 | 1.4439989 | 0.530069626 | 0.3307741 | 2.568098e+00 | 0.1090387573 | 0.75783482 | No | Yes | 8.889469e+03 | 1274.955969 | 5.076081e+03 | 747.174632 | |
| ENSG00000169627 | 552900 | BOLA2B | protein_coding | Q9H3K6 | FUNCTION: Acts as a cytosolic iron-sulfur (Fe-S) cluster assembly factor that facilitates [2Fe-2S] cluster insertion into a subset of cytosolic proteins (PubMed:26613676, PubMed:27519415). Acts together with the monothiol glutaredoxin GLRX3 (PubMed:26613676, PubMed:27519415). {ECO:0000269|PubMed:26613676, ECO:0000269|PubMed:27519415}. | Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Nucleus;Reference proteome | This gene is located within a region of a segmental duplication on chromosome 16 and is identical to BOLA2 (bolA family member 2). The product of this gene belongs to a family of proteins that are widely conserved and may be involved in iron maturation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:552900;hsa:654483; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; 2 iron, 2 sulfur cluster binding [GO:0051537]; iron-sulfur cluster binding [GO:0051536]; [2Fe-2S] cluster assembly [GO:0044571]; cellular iron ion homeostasis [GO:0006879]; protein maturation by iron-sulfur cluster transfer [GO:0097428] | Mouse_homologues 14718656_NMR structure of BolA2 [BolA2] | ENSMUSG00000047721 | Bola2 | 4.535283e+02 | 0.6319195 | -0.662187300 | 0.3749755 | 3.032719e+00 | 0.0816011539 | 0.73555375 | No | Yes | 3.885420e+02 | 72.344186 | 5.166560e+02 | 98.732337 | |
| ENSG00000169660 | 284004 | HEXD | protein_coding | Q8WVB3 | FUNCTION: Has hexosaminidase activity. Responsible for the cleavage of the monosaccharides N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc) from cellular substrates. Has a preference for galactosaminide over glucosaminide substrates (PubMed:27149221). {ECO:0000269|PubMed:19040401, ECO:0000269|PubMed:23099419, ECO:0000269|PubMed:27149221}. | Alternative splicing;Cytoplasm;Disulfide bond;Glycosidase;Hydrolase;Nucleus;Reference proteome | hsa:284004; | cytoplasm [GO:0005737]; extracellular vesicle [GO:1903561]; nucleus [GO:0005634]; beta-N-acetylhexosaminidase activity [GO:0004563]; hexosaminidase activity [GO:0015929]; N-acetyl-beta-D-galactosaminidase activity [GO:0102148]; carbohydrate metabolic process [GO:0005975] | 21265931_In normoalbuminuric/microalbuminuric patients with essential hypertension renal impairment measured by e-GFR is related to the increased urinary NAG activity and albumin/creatinine ratio rather than elevated concentrations of individual proteins. 21672370_The results suggest that, following coronary angiography and/or percutaneous coronary intervention, contrast-induced nephropathy (CIN) occurs to a certain degree and that NAG may be a useful early CIN marker. 22515481_Studies indicate the most studied biomarkers for acute kidney injury are neutrophil gelatinase-associated lipocalin-2, kidney injury molecule-1, IL-18, cystatin C, N-acetyl-beta-D-glucosaminidase, liver fatty-acid binding protein, and heat shock protein 72. 23099419_the expression and disease relevance of the HEXDC gene in humans demonstrate the expression of this novel enzyme within the joints, and suggests that its activity may significantly contribute to the overall local exoglycosidase activity. 27149221_The catalytically important residues are Asp148 and Glu149, with Glu149 serving as the general acid/base residue and Asp148 as the polarizing residue. HexD is inhibited by Gal-NAG-thiazoline (Ki = 420 nM). | ENSMUSG00000039307 | Hexdc | 1.430489e+03 | 0.9322568 | -0.101200725 | 0.2643710 | 1.470708e-01 | 0.7013505140 | 0.93624959 | No | Yes | 1.012750e+03 | 72.388214 | 1.106893e+03 | 81.342467 | ||
| ENSG00000169679 | 699 | BUB1 | protein_coding | O43683 | FUNCTION: Serine/threonine-protein kinase that performs 2 crucial functions during mitosis: it is essential for spindle-assembly checkpoint signaling and for correct chromosome alignment. Has a key role in the assembly of checkpoint proteins at the kinetochore, being required for the subsequent localization of CENPF, BUB1B, CENPE and MAD2L1. Required for the kinetochore localization of PLK1. Required for centromeric enrichment of AUKRB in prometaphase. Plays an important role in defining SGO1 localization and thereby affects sister chromatid cohesion. Acts as a substrate for anaphase-promoting complex or cyclosome (APC/C) in complex with its activator CDH1 (APC/C-Cdh1). Necessary for ensuring proper chromosome segregation and binding to BUB3 is essential for this function. Can regulate chromosome segregation in a kinetochore-independent manner. Can phosphorylate BUB3. The BUB1-BUB3 complex plays a role in the inhibition of APC/C when spindle-assembly checkpoint is activated and inhibits the ubiquitin ligase activity of APC/C by phosphorylating its activator CDC20. This complex can also phosphorylate MAD1L1. Kinase activity is essential for inhibition of APC/CCDC20 and for chromosome alignment but does not play a major role in the spindle-assembly checkpoint activity. Mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. {ECO:0000269|PubMed:10198256, ECO:0000269|PubMed:15020684, ECO:0000269|PubMed:15525512, ECO:0000269|PubMed:15723797, ECO:0000269|PubMed:16760428, ECO:0000269|PubMed:17158872, ECO:0000269|PubMed:19487456, ECO:0000269|PubMed:20739936}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Cell cycle;Cell division;Centromere;Chromosome;Chromosome partition;Host-virus interaction;Kinase;Kinetochore;Mitosis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Ubl conjugation | This gene encodes a serine/threonine-protein kinase that play a central role in mitosis. The encoded protein functions in part by phosphorylating members of the mitotic checkpoint complex and activating the spindle checkpoint. This protein also plays a role in inhibiting the activation of the anaphase promoting complex/cyclosome. This protein may also function in the DNA damage response. Mutations in this gene have been associated with aneuploidy and several forms of cancer. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:699; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; kinetochore [GO:0000776]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; apoptotic process [GO:0006915]; cell division [GO:0051301]; cell population proliferation [GO:0008283]; meiotic sister chromatid cohesion, centromeric [GO:0051754]; mitotic spindle assembly checkpoint signaling [GO:0007094]; regulation of chromosome segregation [GO:0051983]; regulation of sister chromatid cohesion [GO:0007063] | 11782350_Sequence alterations of mitotic checkpoint genes, hBUB1 and hBUBR1, were examined, and their gene transcripts were quantified using on-line, real-time quantitative reverse transcription-PCR in surgically resected human colorectal cancers 11792804_As part of a common complex in checkpoint-activated cells, Bub1 monitors different spindle events compared to BubR1 and integrates different signals into a single signal that is then relayed to the downstream cell cycle machinery. 11999574_Reduced expression and aberrant transcription of the hBUB1 gene were detected in acute myeloid leukemia specimens. 12096343_mutations in leukemia and lymphoma cells 12375025_Results suggest that mutation of the Bub1 gene might not play a role in human stomach and oral carcinogenesis. 12419313_mitotic checkpoint kinases BubR1 and BuB1, by binding to beta-adaptins, may play novel roles in the regulation of vesicular intracellular traffic 12655561_The checkpoint pathway can be abrogated by mutations in BUB1 in the pancreatic cancer cell line Hs766T. 15013707_No apparent selection trend was established with neoplastic progression or aneuploidy for BUB1 in Barrett esophagus. 15604152_Bub1 protects centromeric cohesion through Shugoshin in mitosis 15723797_Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. 15818396_Bub1 is cleaved during apoptosis 15931389_Altered expression is associated with therapy failure and death in patients with multiple types of cancer. 15933723_Data suggest that Bub1 and Aurora B function in parallel pathways that promote formation of stable bipolar kinetochore-microtubule attachments. 16046481_The two spindle checkpoint arms respond to different spindle cues: whereas the Bub1 arm monitors kinetochore-microtubule attachment, the aurora B arm monitors biorientation. 16449967_expression of wild-type Bub1 suppressed multinuclei formation induced by HTLV-I Tax protein both in A549 and HeLa cells. 16525682_Results indicate that increased expression of the human Bub1 gene is closely linked to abnormal cell proliferation in malignant conditions. 16760428_Phosphorylation of Bub1 at T609 by Cdk1 creates a docking site for the PBD of Plk1 and facilitates the kinetochore recruitment of Plk1. 17158872_Bub1, an anaphase-promoting complex/ cyclosome (APC/C) inhibitor, is also an APC/C substrate. 17210994_Observational study of gene-disease association. (HuGE Navigator) 17488820_Bub1 compromise triggers p53-dependent senescence, which limits the production of aneuploid and potentially cancerous cells. 17617734_Kinetochoe localization of Sgo1 depends on AurB and Bub1. 17620410_BUB1 mediation of caspase-independent mitotic death determines cell fate. 17925231_Bub1 is essential for postimplantation embryogenesis and proliferation of primary embryonic fibroblasts. Bub1 inactivation in adult males inhibits proliferation in seminiferous tubules, reducing sperm production and causing infertility. 18036341_These observations suggest that Bub1 not only regulates the cell cycle, but also may be involved in the cytoskeletal control in interphase cells. 18383837_hBUB1, hBUBR1 and hMAD2 expressions in anaplastic thyroid cancer were significantly higher than those in diffuse cancer. hBUBR1 and hMAD2 expressions in advanced DTCs were significantly higher than those in non-advanced DTCs. 18547097_domain amphiphilicity is of higher importance than amino acid sequence specificity in the determination of protein adsorption and interfacial activity 18791270_BUB1 and BUBR1 overexpression plays a role in cytogenetic and morphologic progression of clear cell renal cell cancer 18922873_data suggest that large T antigen, via Bub1 binding, breaches genome integrity mechanisms, leading to DNA damage responses, p53 stabilization, and tetraploidy. 18995837_The KEN boxes of Bub1 are specifically required for the spindle checkpoint in human cells. 19015261_the spindle checkpoint kinase Bub1 contributes to the maintenance of Sgo1 steady-state protein levels in an APC/C-independent mechanism. 19141287_structure provides insight into the molecular basis of Blinkin-specific recognition by BUB1 and, on a broader perspective, of the mechanism that mediates kinetochore localization of BUB1 in checkpoint-activated cells 19182530_Results suggest a novel molecular mechanism leading to aneuploidy involving interference of TAp73alpha with Bub1 and Bub3 resulting in an altered mitotic checkpoint. 19242126_Depletion of the outer kinetochore protein hBub1 upon activation of SAC primarily triggers early cell death mediated by p53. 19252416_P53 physically interacts with hBub1 at kinetochores in response to mitotic spindle damage suggesting a direct role for hBub1 in the suppression of p53 mediated cell death. 19487456_Bub1 can regulate chromosome segregation in a kinetochore-independent manner. 20468071_Observational study of gene-disease association. (HuGE Navigator) 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20643875_Findings strongly suggest that the loss of spindle assembly checkpoint proteins, such as Bub1 and Mad2, may cause spontaneous miscarriages. 20660191_the interaction between LANA and the kinetochore proteins CENP-F and Bub1 is important for KSHV genome tethering and its segregation to new daughter cells 20929775_findings show phosphorylation of histone H3-threonine 3 mediated by Haspin cooperates with Bub1-mediated histone 2A-serine 121 phosphorylation in targeting the chromosomal passenger complex to the inner centromere in fission yeast and human cells 21092550_Bub1 expression could be regulated by estradiol and paclitaxel in endometrial carcinoma Ishikawa cells. 21199919_h-KNL1 functions to connect Bub1 and BubR1 with the hMis12, Ndc80, and Zwint-1 complexes. 21646403_Results establish that Bub1 has oncogenic properties and suggest that Aurora B is a critical target through which overexpressed Bub1 drives aneuploidization and tumorigenesis. 22071147_the molecular mechanism of DNA damage-induced Bub1 activation and highlight a critical role of Bub1 in DNA damage response 22099307_ATM-mediated Bub1 Ser314 phosphorylation is required for Bub1 activity and is essential for the activation of the spindle checkpoint. 23288590_These results all indicated that BUB1 played important roles in the proliferation and/or progression of the breast carcinoma, and nuclear BUB1 immunohistochemical status is also considered a potent prognostic factor in human breast cancer patients. 23383610_IRAK1, CHEK1 and BUB1 may play an important role in ovarian cancer. 23728478_BUB1 and BUBR1 inhibition decreases proliferation and shows radiosensitizing effects on pediatric glioblastoma cells 23747338_Mutations in BUB1 and BUB3 cause mosaic variegated aneuploidy and increase the risk of colorectal cancer at a young age. 24055156_Bub1-mediated H2A phosphorylation penetrates kinetochores and that this histone mark contributes to a tension-sensitive Sgo1-based molecular switch for chromosome segregation. 24654460_Studied the significance of budding uninhibited by benzimidazoles-1 (Bub1) and mitotic arrest deficient-2 (Mad2) expression in endometrial carcinoma. 24711138_The bioinformatics analysis suggested that this variant may regulate the transcriptional ability of BUB1 24756216_BUB1 is co-expressed with AURKA and AURKB suggesting biological relationship between these spindle cell components in primary and metastatic advanced-stage ovarian serous carcinoma 25308863_Study provides structural and functional insights into the regulation of the key mitotic kinase Bub1 and highlights a mode of substrate-specific kinase activation through intramolecular autophosphorylation of the substrate binding loop. 25564677_The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-beta signaling. 25611342_Bub1, but not BubR1, enhances binding of Bub3 to phosphorylated kinetochores. 25661489_Data show sequential multisite regulation of the microtubule-associated protein KNL1-kinase-adaptor complex BUB1/BUB3 interaction. 25669885_ABBA motifs in BUBR1 and BUB1 are necessary for the spindle assembly checkpoint to work at full strength and to recruit CDC20 to kinetochore 25698537_BUB1B expression was highly correlated to CDC20 and CCNB1 expression in multiple myeloma cells, leading to increased cell proliferation.Bub1 expression promotes cell proliferation and leads to poor survival in multiple myeloma patients 25731686_BubR1 is a component of the Mitotic checkpoint complex and was reported to be overexpressed in Breast cancer. BubR1 overexpression was correlated with poor survival in early stage Breast cancer patients. 26031201_Bub1 coordinates checkpoint signalling by distinct domains for RZZ and BubR1 recruitment and localizes antagonistic checkpoint activities. 26148513_human BUB1 does not associate stably with the MAD2 activator MAD1 (also known as MAD1L1) and, although required for accelerating the loading of MAD1 onto kinetochores, BUB1 is dispensable for the maintenance of steady-state levels of MAD1 there 26223641_Bub1 in complex with LANA recruits PCNA to regulate Kaposi's sarcoma-associated herpesvirus latent replication and DNA translesion synthesis. 26260062_Both Bub1 and BubR1 bind stably to Bub3 that is required for kinetochore localization of the proteins 26287798_MAD2L1 and BUB1 may play important roles in breast cancer progression, and measuring the expression of these genes may assist the prediction of breast cancer prognosis. 26399325_Data provide novel insight into the regulation and kinetochore residency of Bub1 and indicate that its localization is dynamic and tightly controlled through feedback autophosphorylation. 26522589_Bub1 may be associated with cancer stem cell potential. 26658523_Data show that mitotic checkpoint kinase Bub1 is an active kinase regulated by intra-molecular phosphorylation at the P+1 loop. 26885717_Inhibition of Bub1 kinase impaired chromosome arm resolution but exerted only minor effects on mitotic progression or spindle assembly checkpoint function. 26897635_Study shows that BUB1beta and PDZ binding kinase are up-regulated in breast cancer tumors and their expression is associated with improved overall survival (OS) in basal-like tumors but worse OS in luminal and untreated patients. 26912231_Bub1-Plk1-mediated phosphorylation of Cdc20 constitutes an anaphase-promoting complex or cyclosome-inhibitory mechanism that is parallel, but not redundant, to mitotic checkpoint complex formation. 27552964_Chromosomal rearrangement resulting in BUB1 haploinsufficiency identified in a patient with polycystic liver disease. BUB1 may imply germline causes of genetic instability in PLD. 27998540_Results report that BubR1 and RepoMan bind directly to PP2A-B56 using an LSPIxE short linear motif (SLiM), where phosphorylation of the Ser residue enhances binding. RepoMan and BubR1 bind B56 using both hydrophobic and electrostatic interactions. 28072388_Therefore, Mps1 promotes checkpoint activation through sequentially phosphorylating Knl1, Bub1, and Mad1. This sequential multi-target phosphorylation cascade makes the checkpoint highly responsive to Mps1 and to kinetochore-microtubule attachment. 28287092_Finally, the authors show that the kinetochore localization of BUB1 and BUBR1 decreases with the age of the oocyte donors. This could contribute to oocyte aneuploidy. 28604727_Conserved domain 1 (CD1) in human Bub1 binds directly to Mad1 and a phosphorylation site exists in CD1 that stimulates Mad1 binding and SAC signalling. 28810242_MiR-490-5p could regulate TGFbeta/Smad signaling pathways by inhibiting BUB1. 28943088_Small sequence differences in Bub1 and BubR1 direct Bub3 to different phosphorylated targets in the spindle assembly checkpoint signaling cascade. 29727616_The roles for the BUB3-BUB1 complex in S phase. 30205061_Wwhile Bub1 is required for recruitment of BubR1, it is not strictly required for the checkpoint response to unattached kinetochores in diploid human cells. 30429199_Here, we present for the first time the identification and the preclinical profile of the novel BUB1 kinase inhibitor BAY 1816032. In particular, we demonstrate additive and more than additive efficacy upon combination of BUB1 kinase inhibition with taxanes, as well as with PARP inhibitors in cellular and in tumor xenograft models. 30898709_study demonstrated that CDK1 and BUB1 may play a role in Pancreatic ductal adenocarcinoma (PDAC) progression and could be prognostic biomarkers for PDAC patients. 31315522_KNL1 protein expression was also significantly associated with poorer survival. Moreover, there was a significant correlation between KNL1 and BUB1 in colorectal cancer tissues. KNL1 plays an effective role in decreasing apoptosis and promoting the proliferation of colorectal cancer cells, suggesting that its inhibition may represent a promising therapeutic approach in patients with colorectal cancer. 31769059_Histone H2A phosphorylation recruits topoisomerase IIalpha to centromeres to safeguard genomic stability. 32027339_Untangling the contribution of Haspin and Bub1 to Aurora B function during mitosis. 32973131_SET/TAF1 forms a distance-dependent feedback loop with Aurora B and Bub1 as a tension sensor at centromeres. 33248027_BUB1 and CENP-U, Primed by CDK1, Are the Main PLK1 Kinetochore Receptors in Mitosis. 33596090_Differential requirements for the CENP-O complex reveal parallel PLK1 kinetochore recruitment pathways. 33872216_Expression and prognosis analyses of BUB1, BUB1B and BUB3 in human sarcoma. 34013367_KIF4A knockdown suppresses ovarian cancer cell proliferation and induces apoptosis by downregulating BUB1 expression. 34337852_Inhibition of BUB1 suppresses tumorigenesis of osteosarcoma via blocking of PI3K/Akt and ERK pathways. 34419445_Weighted genes associated with the progression of retinoblastoma: Evidence from bioinformatic analysis. 34551298_Bub1 and CENP-U redundantly recruit Plk1 to stabilize kinetochore-microtubule attachments and ensure accurate chromosome segregation. 34884561_Prognostic Value of BUB1 for Predicting Non-Muscle-Invasive Bladder Cancer Progression. 35044816_Biallelic BUB1 mutations cause microcephaly, developmental delay, and variable effects on cohesion and chromosome segregation. | ENSMUSG00000027379 | Bub1 | 1.300067e+03 | 0.7646556 | -0.387118007 | 0.3353252 | 1.291920e+00 | 0.2556943757 | 0.79582240 | No | Yes | 1.267050e+03 | 222.416861 | 1.569657e+03 | 282.439792 | |
| ENSG00000169955 | 65988 | ZNF747 | protein_coding | Q9BV97 | Mouse_homologues NA; + ;NA | Alternative splicing;Metal-binding;Reference proteome;Repeat;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA | hsa:65988; | regulation of transcription, DNA-templated [GO:0006355] | Mouse_homologues NA; + ;NA | ENSMUSG00000045757+ENSMUSG00000078580 | Zfp764+E430018J23Rik | 1.978128e+03 | 1.4818390 | 0.567388713 | 0.3147377 | 3.282753e+00 | 0.0700114040 | 0.71403308 | No | Yes | 2.319774e+03 | 280.943265 | 1.365371e+03 | 170.318403 | |
| ENSG00000170145 | 23235 | SIK2 | protein_coding | Q9H0K1 | FUNCTION: Serine/threonine-protein kinase that plays a role in many biological processes such as fatty acid oxidation, autophagy, immune response or glucose metabolism (PubMed:23322770, PubMed:26983400). Phosphorylates 'Ser-794' of IRS1 in insulin-stimulated adipocytes, potentially modulating the efficiency of insulin signal transduction. Inhibits CREB activity by phosphorylating and repressing TORCs, the CREB-specific coactivators (PubMed:15454081). Phosphorylates EP300 and thus inhibits its histone acetyltransferase activity (PubMed:21084751, PubMed:26983400). In turn, regulates the DNA-binding ability of several transcription factors such as PPARA or MLXIPL (PubMed:21084751, PubMed:26983400). Plays also a role in thymic T-cell development (By similarity). {ECO:0000250|UniProtKB:Q8CFH6, ECO:0000269|PubMed:15454081, ECO:0000269|PubMed:21084751, ECO:0000269|PubMed:23322770, ECO:0000269|PubMed:26983400}. | ATP-binding;Acetylation;Cytoplasm;Endoplasmic reticulum;Kinase;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | hsa:23235; | cytoplasm [GO:0005737]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cellular response to glucose starvation [GO:0042149]; intracellular signal transduction [GO:0035556]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of insulin receptor signaling pathway [GO:0046626] | 20367563_Increased SIK2 expression is associated with diffuse large B-cell lymphoma. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20708153_Depletion of SIK2 also delayed G1/S transition and reduced AKT phosphorylation. Higher expression of SIK2 significantly correlated with poor survival in patients with high-grade serous ovarian cancers 22462548_In adipocytes, Ser358 (not Ser587) is the main site phosphorylated and responsible for the binding of SIK2 to 14-3-3 proteins. 23322770_findings uncover SIK2 as a critical determinant in autophagy progression and further suggest a mechanism in which the interplay among kinase and deacetylase activities contributes to cellular protein pool homeostasis. 23393134_tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases 23577667_These findings revealed a new function of LKB1 and salt-inducible kinases as negative regulators of HTLV-1 transcription. 24129571_a mechanism by which the interplay between SIK2 and p97/VCP contributes to the regulation of ERAD in mammalian cells. 24841198_this study suggests that the tightly linked regulatory loop comprised of the SIK2-PP2A and CaMKI and PME-1 networks may function in fine-tuning cell proliferation and stress response. 25548099_Examination of SIK2 in prostate cancer cells found that it functions both as a positive regulator of cell-cycle progression and a negative regulator of CREB1 activity. Also, the study shows high levels of auto-antibodies against SIK2 in plasma. 26590148_salt-inducible kinase inhibition decreases proinflammatory cytokines in human myeloid cells upon IL-1R stimulation. 27236538_Data show that microRNA miR-203 and the salt-inducible kinase 2 (SIK2) are reverse expressed in colorectal tumors. 27441650_The results suggest that activation of SIK2 is required for the cell viability when proteasome activity is inhibited by peritoneal dialysis solutions. 27478041_Activated SIK2 plays a dual role in augmenting AMPK-induced phosphorylation of acetyl-CoA carboxylase and in activating the PI3K/AKT pathway through p85alpha-S154 phosphorylation. These findings identify SIK2 at the apex of the adipocyte-induced signaling cascades in cancer cells. 27697861_Findings suggest that SIK2 restricts autophagic flux which in the claudin-low subtype is essential for viability of triple-negative breast cancer cells. 27759007_SIK2 controls osteocyte responses to parathyroid hormone. 27807598_Data demonstrate that SIK2 and SIK3 mRNA are downregulated in adipose tissue from obese individuals and that the expression is regulated by weight change. SIK2 is also negatively associated with in vivo insulin resistance (HOMA-IR), independently of BMI and age. 29211348_Data suggest that cAMP/protein kinase A-dependent phosphorylation of SIK1, SIK2, and SIK3 inhibits their catalytic activity by inducing 14-3-3 protein binding. 30004169_results of this study indicate that SIK2 expression can serve as a prognostic biomarker for EOC and that miR-874-3p and miR-874-5p have the potential to enhance clinical treatment of EOC 31235257_Parathyroid hormone can affect the osteogenesis process of adipose tissue stem cells by regulating SIK2 and Wnt4. 31639424_In ovarian cancer (OC) cells SIK2 upregulated the expression of HIF-1alpha by activating the PI3K/AKT signaling pathway, which directly upregulated the transcription of major glycolytic genes to promote glycolysis and promoted mitochondrial fission through phosphorylation of Drp1 at Ser616 site, which inhibited the mitochondrial oxidative phosphorylation. SIK2 promoted growth and metastasis of OC cells. 31849250_High-Throughput Implementation of the NanoBRET Target Engagement Intracellular Kinase Assay to Reveal Differential Compound Engagement by SIK2/3 Isoforms. 31932581_SIK2 enhances synthesis of fatty acid and cholesterol in ovarian cancer cells and tumor growth through PI3K/Akt signaling pathway. 31969556_Salt-inducible kinases (SIKs) regulate TGFbeta-mediated transcriptional and apoptotic responses. 32343771_Salt-inducible Kinases Are Critical Determinants of Female Fertility. 32437091_Tumor-suppressor Fbxw7 targets SIK2 for degradation to interfere with TORC2-AKT signaling in pancreatic cancer. 32966883_JUP/plakoglobin is regulated by salt-inducible kinase 2, and is required for insulin-induced signalling and glucose uptake in adipocytes. 33128264_SIK2 represses AKT/GSK3beta/beta-catenin signaling and suppresses gastric cancer by inhibiting autophagic degradation of protein phosphatases. 33947818_SIK2 orchestrates actin-dependent host response upon Salmonella infection. 34058059_SIK2 kinase synthetic lethality is driven by spindle assembly defects in FANCA-deficient cells. 34302056_Silencing the G-protein coupled receptor 3-salt inducible kinase 2 pathway promotes human beta cell proliferation. 34491613_Salt-inducible kinase 2 functions as a tumor suppressor in hepatocellular carcinoma. 34558647_Role of saltinducible kinase 2 in the malignant behavior and glycolysis of colorectal cancer cells. 34559955_LINC00662 facilitates osteosarcoma progression via sponging miR-103a-3p and regulating SIK2 expression. 35277657_SIK2 maintains breast cancer stemness by phosphorylating LRP6 and activating Wnt/beta-catenin signaling. | ENSMUSG00000037112 | Sik2 | 1.802585e+03 | 1.3917471 | 0.476897125 | 0.2528808 | 3.586779e+00 | 0.0582410415 | 0.68648849 | No | Yes | 1.727570e+03 | 161.817353 | 1.131676e+03 | 109.002943 | ||
| ENSG00000170265 | 8427 | ZNF282 | protein_coding | Q9UDV7 | FUNCTION: Binds to the U5 repressive element (U5RE) of the human T cell leukemia virus type I long terminal repeat. It recognizes the 5'-TCCACCCC-3' sequence as a core motif and exerts a strong repressive effect on HTLV-I LTR-mediated expression. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:8427; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22986521_SUMOylation of ZFP282 potentiates its positive effect on estrogen signaling in breast tumorigenesis. 25373738_ZNF282 is E2F1 co-activator involved in esophageal squamous cell carcinoma | ENSMUSG00000025821 | Zfp282 | 5.497376e+03 | 1.3600163 | 0.443623977 | 0.3012082 | 2.165970e+00 | 0.1410955497 | 0.76841160 | No | Yes | 5.726208e+03 | 656.426324 | 3.702539e+03 | 435.886283 | ||
| ENSG00000170325 | 56980 | PRDM10 | protein_coding | Q9NQV6 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000250}. | 3D-structure;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Methyltransferase;Nucleus;Phosphoprotein;Reference proteome;Repeat;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a transcription factor that contains C2H2-type zinc-fingers. It also contains a positive regulatory domain, which has been found in several other zinc-finger transcription factors including those involved in B cell differentiation and tumor suppression. Studies of the mouse counterpart suggest that this protein may be involved in the development of the central nerve system (CNS), as well as in the pathogenesis of neuronal storage disease. Multiple alternatively spliced transcript variants encoding distinct isoforms have been observed. [provided by RefSeq, Jul 2008]. | hsa:56980; | chromatin [GO:0000785]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; methyltransferase activity [GO:0008168]; methylation [GO:0032259]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of gene expression [GO:0010468] | 19773279_Observational study of gene-disease association. (HuGE Navigator) 20095854_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 25516889_Our results suggest that PRDM10 fusions are present in around 5% of Undifferentiated pleomorphic sarcoma 31313299_PRDM10-fusions represent the critical driver mutations in undifferentiated pleomorphic sarcomas. | ENSMUSG00000042496 | Prdm10 | 2.934070e+02 | 1.1674383 | 0.223346305 | 0.3094611 | 5.177328e-01 | 0.4718104040 | 0.86976192 | No | Yes | 3.383918e+02 | 49.941970 | 2.537717e+02 | 38.626240 | |
| ENSG00000170473 | 84305 | PYM1 | protein_coding | Q9BRP8 | FUNCTION: Key regulator of the exon junction complex (EJC), a multiprotein complex that associates immediately upstream of the exon-exon junction on mRNAs and serves as a positional landmark for the intron exon structure of genes and directs post-transcriptional processes in the cytoplasm such as mRNA export, nonsense-mediated mRNA decay (NMD) or translation. Acts as an EJC disassembly factor, allowing translation-dependent EJC removal and recycling by disrupting mature EJC from spliced mRNAs. Its association with the 40S ribosomal subunit probably prevents a translation-independent disassembly of the EJC from spliced mRNAs, by restricting its activity to mRNAs that have been translated. Interferes with NMD and enhances translation of spliced mRNAs, probably by antagonizing EJC functions. May bind RNA; the relevance of RNA-binding remains unclear in vivo, RNA-binding was detected by PubMed:14968132, while PubMed:19410547 did not detect RNA-binding activity independently of the EJC. {ECO:0000269|PubMed:18026120, ECO:0000269|PubMed:19410547}. | Acetylation;Alternative splicing;Coiled coil;Cytoplasm;Nonsense-mediated mRNA decay;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Translation regulation | hsa:84305; | cell junction [GO:0030054]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; exon-exon junction complex [GO:0035145]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; ribosome binding [GO:0043022]; RNA binding [GO:0003723]; exon-exon junction complex disassembly [GO:1903259]; nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184]; positive regulation of translation [GO:0045727] | 18026120_suggest that PYM functions as a bridge between EJC-bearing spliced mRNAs and the translation machinery to enhance translation of the mRNAs 19410547_PYM is an Exon junction complexes(EJC) disassembly factor that acts both in vitro and in living cells, and that antagonizes important EJC functions. | ENSMUSG00000064030 | Pym1 | 1.141413e+03 | 1.1271245 | 0.172646864 | 0.2914444 | 3.554462e-01 | 0.5510462164 | 0.89250744 | No | Yes | 1.164525e+03 | 114.784312 | 9.181524e+02 | 93.235368 | ||
| ENSG00000170571 | 133418 | EMB | protein_coding | Q6PCB8 | FUNCTION: Plays a role in the outgrowth of motoneurons and in the formation of neuromuscular junctions. Following muscle denervation, promotes nerve terminal sprouting and the formation of additional acetylcholine receptor clusters at synaptic sites without affecting terminal Schwann cell number or morphology. Delays the retraction of terminal sprouts following re-innervation of denervated endplates. May play a role in targeting the monocarboxylate transporters SLC16A1 and SLC16A7 to the cell membrane (By similarity). {ECO:0000250}. | Alternative splicing;Cell junction;Cell membrane;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phosphoprotein;Reference proteome;Repeat;Signal;Synapse;Transmembrane;Transmembrane helix | This gene encodes a transmembrane glycoprotein that is a member of the immunoglobulin superfamily. The encoded protein may be involved in cell growth and development by mediating interactions between the cell and extracellular matrix. A pseudogene of this gene is found on chromosome 1. [provided by RefSeq, Jan 2009]. | hsa:133418; | axon [GO:0030424]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; synapse [GO:0045202]; cell-cell adhesion mediator activity [GO:0098632]; axon guidance [GO:0007411]; dendrite self-avoidance [GO:0070593]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; plasma membrane lactate transport [GO:0035879] | 2963822_Reports the original cloning of the mouse embigin gene, referred to as gp70, from embryonal carcinoma cells. 21787388_Data show that Gp70 (embigin) was negative in all normal, adjacent non-neoplastic and PIN lesions and only a very small percentage of cases. 25773908_Study reports elevated expression of Embigin in pancreatic ductal adenocarcinoma and shows its novel functions in regulating cell proliferation, migration, and invasion. 26090721_embigin is regulated by HOXC8 and its loss plays an important role in the progression of breast tumors 30446621_This study analyzed the direct interaction between CAIV and the two Monocarboxylate transporters (MCTs) chaperones basigin (CD147) and embigin (GP70). 32213169_Rare and common variants analysis of the EMB gene in patients with schizophrenia. | ENSMUSG00000021728 | Emb | 3.236161e+02 | 0.7967154 | -0.327863614 | 0.3698193 | 7.710458e-01 | 0.3798937260 | 0.84161313 | No | Yes | 2.249642e+02 | 42.101606 | 2.985068e+02 | 57.004694 | |
| ENSG00000170832 | 84669 | USP32 | protein_coding | Q8NFA0 | Alternative splicing;Calcium;Golgi apparatus;Hydrolase;Lipoprotein;Membrane;Metal-binding;Methylation;Phosphoprotein;Prenylation;Protease;Reference proteome;Repeat;Thiol protease;Ubl conjugation pathway | hsa:84669; | cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; membrane [GO:0016020]; calcium ion binding [GO:0005509]; thiol-dependent deubiquitinase [GO:0004843]; protein deubiquitination [GO:0016579]; ubiquitin-dependent protein catabolic process [GO:0006511] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20549504_USP32 is overexpressed in breast cancer. 28597490_USP32 silencing arrested cell cycle progression at G0/G1 phase via decreasing CDK4/Cyclin D1 complex and elevating p21. 30926795_the small GTPase Rab7-the logistical centerpiece of late endosome biology-is a substrate of USP32. 32226309_USP32 promotes tumorigenesis and chemoresistance in gastric carcinoma via upregulation of SMAD2. 33744759_Ubiquitin specific peptidase 32 acts as an oncogene in epithelial ovarian cancer by deubiquitylating farnesyl-diphosphate farnesyltransferase 1. 34815782_USP32 confers cancer cell resistance to YM155 via promoting ER-associated degradation of solute carrier protein SLC35F2. | ENSMUSG00000000804 | Usp32 | 6.084833e+02 | 0.7866257 | -0.346250728 | 0.3548649 | 9.477272e-01 | 0.3302984849 | 0.82505234 | No | Yes | 5.870728e+02 | 123.557772 | 6.438894e+02 | 138.940456 | |||
| ENSG00000170835 | 1056 | CEL | protein_coding | P19835 | FUNCTION: Catalyzes the hydrolysis of a wide range of substrates including cholesteryl esters, phospholipids, lysophospholipids, di- and tri-acylglycerols, and fatty acid esters of hydroxy fatty acids (FAHFAs) (PubMed:8471055, PubMed:27509211, PubMed:10220579, PubMed:27650499). Preferentially hydrolyzes FAHFAs with the ester bond further away from the carboxylate. Unsaturated FAHFAs are hydrolyzed more quickly than saturated FAHFAs (By similarity). Has an essential role in the complete digestion of dietary lipids and their intestinal absorption, along with the absorption of fat-soluble vitamins (PubMed:8471055, PubMed:27509211, PubMed:10220579, PubMed:27650499). {ECO:0000250|UniProtKB:Q64285, ECO:0000269|PubMed:10220579, ECO:0000269|PubMed:27509211, ECO:0000269|PubMed:27650499, ECO:0000269|PubMed:8471055}. | 3D-structure;Alternative splicing;Diabetes mellitus;Direct protein sequencing;Disulfide bond;Glycoprotein;Hydrolase;Lipid degradation;Lipid metabolism;Reference proteome;Repeat;Secreted;Serine esterase;Signal | The protein encoded by this gene is a glycoprotein secreted from the pancreas into the digestive tract and from the lactating mammary gland into human milk. The physiological role of this protein is in cholesterol and lipid-soluble vitamin ester hydrolysis and absorption. This encoded protein promotes large chylomicron production in the intestine. Also its presence in plasma suggests its interactions with cholesterol and oxidized lipoproteins to modulate the progression of atherosclerosis. In pancreatic tumoral cells, this encoded protein is thought to be sequestrated within the Golgi compartment and is probably not secreted. This gene contains a variable number of tandem repeat (VNTR) polymorphism in the coding region that may influence the function of the encoded protein. [provided by RefSeq, Jul 2008]. | Mouse_homologues mmu:12613; | cell surface [GO:0009986]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; integral component of postsynaptic specialization membrane [GO:0099060]; presynapse [GO:0098793]; synapse [GO:0045202]; acetylesterase activity [GO:0008126]; catalytic activity [GO:0003824]; heparin binding [GO:0008201]; hydrolase activity [GO:0016787]; neurexin family protein binding [GO:0042043]; signaling receptor activity [GO:0038023]; sterol esterase activity [GO:0004771]; triglyceride lipase activity [GO:0004806]; ceramide catabolic process [GO:0046514]; chemical synaptic transmission [GO:0007268]; cholesterol catabolic process [GO:0006707]; fatty acid catabolic process [GO:0009062]; intestinal cholesterol absorption [GO:0030299]; intestinal lipid catabolic process [GO:0044258]; lipid metabolic process [GO:0006629]; modulation of chemical synaptic transmission [GO:0050804]; neuron cell-cell adhesion [GO:0007158]; pancreatic juice secretion [GO:0030157]; postsynaptic membrane assembly [GO:0097104]; presynaptic membrane assembly [GO:0097105]; protein esterification [GO:0018350]; synaptic vesicle endocytosis [GO:0048488] | 11563913_protein conformation of active site 11945176_transcriptional regulation in monocytes 12110666_determination of functional signifcance of N-terminal cluster 12821548_BSDL was found to be associated with vascular smooth muscle cells [SMCs] but not with macrophages. BSDL was significantly mitogenic for cultured rabbit SMCs 12853459_Liver receptor homolog 1 controls the expression of this enzyme in pancreas. 15114370_Observational study of gene-disease association. (HuGE Navigator) 15841033_Observational study of gene-disease association. (HuGE Navigator) 15841033_The CEL gene polymorphism, especially an increase in the frequency of the L allele, was found to be associated with alcohol-induced pancreatitis. 16266293_Analysis of the ability of human milk to inhibit the attachment of rNV VLPs (recombinant NV-like particles) to their carbohydrate ligands and to characterize potential inhibitors found in milk is presented. 17005819_binds DC-SIGN and inhibits transfer of HIV-1 to CD4+ T lymphocytes. 18037996_BSDL plays a role in optimal platelet activation and thrombus formation by interacting with CXCR4 on platelets. 18544793_Pancreatic enzyme substitution alleviated symptoms and malabsorption and normalized vitamin E levels. The CEL syndrome seems associated with a demyelinating neuropathology. 18803939_Carboxyl ester lipase modulates apo B-lipoprotein in vivo, which results in reduced very-low-density lipoprotein clearance and elevated plasma cholesterol levels. 19760265_Data show that the CEL gene is highly polymorphic, but mutations in CEL are likely to be a rare cause of monogenic diabetes. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21346236_Atypical interactions of pBSDL-J28, expressed on tumoral pancreatic tissue, with dendritic cells may lead to adequate antigen trafficking and processing and result in T cell activation. 21386960_Binding of human milk to pathogen receptor DC-SIGN varies with bile salt-stimulated lipase (BSSL) gene polymorphism. 21784842_CEL-MUT protein has a high propensity to form aggregates found intracellularly and extracellularly; CEL-MODY is a protein misfolding disease 21865348_BSSL and PLRP2 hydrolyzed triglycerides (TG) to free FA and glycerol. 22412885_We identify BSSL as a marker for HIV-1 disease progression and emergence of X4 variants. Additionally, we identified a relation between BSSL genotype and CD4 cell counts prior to infection. 22956586_Monoclonal antibody 16D10 inhibits the proliferation of human pancreatic tumor cells expressing 16D10 plasma membrane antigen. 23395566_This study did not find evidence that supported an association between the common length variations of the CEL VNTR and chronic pancreatitis. 23770712_CEL-MODY patients with a mutation in the variable number of tandem repeat region in exon 11 of the CEL gene displayed severely reduced acinar function and moderately reduced ductal function of the pancreas compared with control subjects. 24062244_subjects with CEL-maturity-onset diabetes of the young develop multiple pancreatic cysts by the time they develop diabetes. upregulated MAPK signaling in the pancreas may reflect the pathophysiological development of pancreatic cysts and diabetes. 25160620_secreted carboxyl ester lipase has a role in a syndrome of diabetes and pancreatic exocrine dysfunction 25774637_Expression of CEL-HYB in originating from a crossover between CEL and its neighboring pseudogene, CELP is associated with chronic pancreatitis. 26498142_Data show that the c.1719C > T transition (single nucleotide polymorphism rs488087) present in bile salt-dependent lipase (BSDL) VNTR may be a useful marker for defining a population at risk of developing pancreatic cancer (PC) 27602750_Data suggest that anti-PRAAHG antibodies were uniquely reactive with a short isoform of BSDL specifically expressed in pre-neoplastic lesions of the pancreas. The detection of truncated BSDL reactive to antibodies against the PRAAHG C-terminal sequence in pancreatic juice or in pancreatic biopsies may be a new tool in the early diagnosis of PDAC. 27650499_CEL deletion mutation increased endoplasmic reticulum stress, activated the unfolded protein response, and caused cell death by apoptosis in chronic pancreatitis. 27773618_We could not demonstrate an association between CEL copy number variants and pancreatic cancer. An association is also unlikely for CEL variable number of tandem repeat lengths (VNTR), although analyses in larger materials are necessary to completely exclude an effect of rare VNTR alleles. 27802312_The CEL gene harbours a variable number of tandem repeats (VNTR) region in exon 11. These results suggest that CEL VNTR lengths might associate with Alcoholic Liver Cirrhosis. 30315106_mucinous domain of pancreatic carboxyl-ester lipase (CEL) contains core 1/core 2 O-glycans that can be modified by ABO blood group determinants 30359675_BAL has inverted stereoselectivity for the surrogates of nerve agent compared to human cholinesterases 31036489_This study found no statistically significant association between CEL-HYB1 and chronic pancreatitis in a cohort of Polish pediatric chronic pancreatitis patients 31561066_Studied association between carboxyl ester lipase (CEL) expression in breast cancer (BC) tissues and the survival of BC patients by analyzing The Cancer Genome Atlas Breast Carcinoma level 3 database. Found high CEL expression was associated with the poor overall survival of breast cancer. 31622635_No statistical significant difference in the copy number of exon 11 repeats, or combinations of, in the Bile-salt stimulate lipase (BSSL) gene is observed when comparing homosexual male cohort composed of Hepatitis C (HCV) multiple exposed uninfected and multiple exposed HCV infected individuals. The exon 11 repeat copy number in the BSSL gene does not affect HCV susceptibility. 31963687_Pathogenic Carboxyl Ester Lipase (CEL) Variants Interact with the Normal CEL Protein in Pancreatic Cells. 32007358_Characterization of CEL-DUP2: Complete duplication of the carboxyl ester lipase gene is unlikely to influence risk of chronic pancreatitis. 32906201_Single nucleotide polymorphisms in CEL-HYB1 increase risk for chronic pancreatitis through proteotoxic misfolding. 33079780_Protein misfolding in combination with other risk factors in CEL-HYB1-mediated chronic pancreatitis. 33417713_Generation of beta Cells from iPSC of a MODY8 Patient with a Novel Mutation in the Carboxyl Ester Lipase (CEL) Gene. 34850019_Two New Mutations in the CEL Gene Causing Diabetes and Hereditary Pancreatitis: How to Correctly Identify MODY8 Cases. | ENSMUSG00000026818 | Cel | 1.205879e+02 | 0.8654836 | -0.208421656 | 0.3556952 | 3.386674e-01 | 0.5605994138 | 0.89534708 | No | Yes | 8.961061e+01 | 16.394477 | 1.104497e+02 | 20.508888 | |
| ENSG00000171103 | 55006 | TRMT61B | protein_coding | Q9BVS5 | FUNCTION: Methyltransferase that catalyzes the formation of N(1)-methyladenine at position 58 (m1A58) in various tRNAs in mitochondrion, including tRNA(Leu) (deciphering codons UUA or UUG), tRNA(Lys) and tRNA(Ser) (deciphering codons UCA, UCU, UCG or UCC) (PubMed:23097428). Catalyzes the formation of 1-methyladenosine at position 947 of mitochondrial 16S ribosomal RNA and this modification is most likely important for mitoribosomal structure and function (PubMed:27631568). In addition to tRNA N(1)-methyltransferase activity, also acts as a mRNA N(1)-methyltransferase by mediating methylation of adenosine residues at the N(1) position of MT-ND5 mRNA, leading to interfere with mitochondrial translation (PubMed:29107537). {ECO:0000269|PubMed:23097428, ECO:0000269|PubMed:27631568, ECO:0000269|PubMed:29107537}. | 3D-structure;Methyltransferase;Mitochondrion;Reference proteome;S-adenosyl-L-methionine;Transferase;Transit peptide;tRNA processing | hsa:55006; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; tRNA (m1A) methyltransferase complex [GO:0031515]; mRNA (adenine-N1-)-methyltransferase activity [GO:0061953]; rRNA (adenine) methyltransferase activity [GO:0016433]; tRNA (adenine-N1-)-methyltransferase activity [GO:0016429]; mitochondrial tRNA methylation [GO:0070901]; mRNA methylation [GO:0080009]; protein homooligomerization [GO:0051260]; tRNA methylation [GO:0030488] | 23097428_Unlike the cytoplasmic tRNA m(1)A58 methyltransferase that consists of an alpha2beta2 heterotetramer formed by Trmt61A and Trmt6, Trmt61B formed a homo-oligomer (presumably a homotetramer) that resembled the bacterial homotetrameric m(1)A58 methyltransferase. 27631568_16S rRNA RNA-DNA differences result from a 1-methyladenosine (m1A) modification introduced by TRMT61B. | 2.113814e+02 | 0.9095782 | -0.136730435 | 0.3796056 | 1.270122e-01 | 0.7215502388 | 0.94084174 | No | Yes | 1.876874e+02 | 33.743844 | 2.086054e+02 | 38.334949 | ||||
| ENSG00000171219 | 55561 | CDC42BPG | protein_coding | Q6DT37 | FUNCTION: May act as a downstream effector of CDC42 in cytoskeletal reorganization. Contributes to the actomyosin contractility required for cell invasion, through the regulation of MYPT1 and thus MLC2 phosphorylation (By similarity). {ECO:0000250|UniProtKB:Q5VT25, ECO:0000269|PubMed:15194684}. | ATP-binding;Coiled coil;Cytoplasm;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Zinc;Zinc-finger | hsa:55561; | cell leading edge [GO:0031252]; centriolar satellite [GO:0034451]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; actin cytoskeleton reorganization [GO:0031532]; actomyosin structure organization [GO:0031032]; intracellular signal transduction [GO:0035556]; peptidyl-threonine phosphorylation [GO:0018107]; protein phosphorylation [GO:0006468] | 15194684_CDC42BPG expression is regulated by promoter methylation and Sp1 binding. 29124443_Findings indicate that CDC42BPG may be a susceptibility locus for hyperuricemia. | ENSMUSG00000024769 | Cdc42bpg | 1.255531e+02 | 1.3172947 | 0.397578100 | 0.3776932 | 1.105089e+00 | 0.2931520140 | 0.81275304 | No | Yes | 1.408767e+02 | 25.133759 | 1.056572e+02 | 19.728266 | ||
| ENSG00000171729 | 55092 | TMEM51 | protein_coding | Q9NW97 | Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:55092; | integral component of membrane [GO:0016021] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000040616 | Tmem51 | 4.501712e+02 | 0.9574361 | -0.062751861 | 0.3174138 | 3.906847e-02 | 0.8433130763 | 0.96897477 | No | Yes | 3.845983e+02 | 33.922422 | 4.154599e+02 | 37.512538 | |||
| ENSG00000171798 | 85442 | KNDC1 | protein_coding | Q76NI1 | FUNCTION: RAS-Guanine nucleotide exchange factor (GEF) that controls the negative regulation of neuronal dendrite growth by mediating a signaling pathway linking RAS and MAP2 (By similarity). May be involved in cellular senescence (PubMed:24788352). {ECO:0000250|UniProtKB:Q0KK55, ECO:0000269|PubMed:24788352}. | Alternative splicing;Cell projection;Coiled coil;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome;Repeat | The protein encoded by this gene is a Ras guanine nucleotide exchange factor that appears to negatively regulate dendritic growth in the brain. Knockdown of this gene in senescent umbilical vein endothelial cells partially reversed the senescence, showing that this gene could potentially be targeted by anti-aging therapies. [provided by RefSeq, Dec 2016]. | hsa:85442; | dendrite [GO:0030425]; guanyl-nucleotide exchange factor complex [GO:0032045]; neuronal cell body [GO:0043025]; perikaryon [GO:0043204]; guanyl-nucleotide exchange factor activity [GO:0005085]; cerebellar granule cell differentiation [GO:0021707]; positive regulation of protein phosphorylation [GO:0001934]; regulation of dendrite development [GO:0050773]; regulation of dendrite morphogenesis [GO:0048814]; small GTPase mediated signal transduction [GO:0007264] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 24788352_KNDC1 knockdown promoted HUVEC proliferation via the extracellular signal-regulated kinase signaling pathway and delayed HUVEC senescence by inhibiting the p53-p21-p16 transduction cascade. 29568929_The results demonstrated that KNDC1 overexpression possibly inhibited HUVEC activity and function and promoted HUVEC senescence. Mechanistic studies demonstrated that KNDC1 triggered a p53ROS positive feedback loop, which serves a crucial role in regulating senescence. | ENSMUSG00000066129 | Kndc1 | 3.294823e+02 | 0.9220563 | -0.117073279 | 0.2925173 | 1.589555e-01 | 0.6901199921 | 0.93352105 | No | Yes | 2.400629e+02 | 41.303561 | 2.715849e+02 | 47.804723 | |
| ENSG00000171953 | 91647 | ATPAF2 | protein_coding | Q8N5M1 | FUNCTION: May play a role in the assembly of the F1 component of the mitochondrial ATP synthase (ATPase). {ECO:0000269|PubMed:11410595}. | Chaperone;Disease variant;Mitochondrion;Primary mitochondrial disease;Reference proteome;Transit peptide | This gene encodes an assembly factor for the F(1) component of the mitochondrial ATP synthase. This protein binds specifically to the F1 alpha subunit and is thought to prevent this subunit from forming nonproductive homooligomers during enzyme assembly. This gene is located within the Smith-Magenis syndrome region on chromosome 17. An alternatively spliced transcript variant has been described, but its biological validity has not been determined. [provided by RefSeq, Jul 2008]. | hsa:91647; | cytosol [GO:0005829]; mitochondrion [GO:0005739]; nuclear speck [GO:0016607]; mitochondrial proton-transporting ATP synthase complex assembly [GO:0033615]; proton-transporting ATP synthase complex assembly [GO:0043461] | 19933271_Data show that wild type human Atp12p rescues the respiratory defect of a yeast ATP12 deletion mutant (Deltaatp12). 20167577_Observational study of gene-disease association. (HuGE Navigator) 20167577_Two possible dementia susceptibility genes including ATPAF2 and TOM1L2 near SREBF1 locus were identified. 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000042709 | Atpaf2 | 1.320870e+03 | 1.0427933 | 0.060453174 | 0.3143060 | 3.633632e-02 | 0.8488226384 | 0.96993871 | No | Yes | 1.181348e+03 | 126.090100 | 1.054101e+03 | 115.740219 | |
| ENSG00000172037 | 3913 | LAMB2 | protein_coding | P55268 | FUNCTION: Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components. | Basement membrane;Cell adhesion;Coiled coil;Disease variant;Disulfide bond;Extracellular matrix;Glycoprotein;Laminin EGF-like domain;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal | Laminins, a family of extracellular matrix glycoproteins, are the major noncollagenous constituent of basement membranes. They have been implicated in a wide variety of biological processes including cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis. Laminins, composed of 3 non identical chains: laminin alpha, beta and gamma (formerly A, B1, and B2, respectively), form a cruciform structure consisting of 3 short arms, each formed by a different chain, and a long arm composed of all 3 chains. Each laminin chain is a multidomain protein encoded by a distinct gene. Several isoforms of each chain have been described. Different alpha, beta and gamma chain isomers combine to give rise to different heterotrimeric laminin isoforms which are designated by Arabic numerals in the order of their discovery, i.e. alpha1beta1gamma1 heterotrimer is laminin 1. The biological functions of the different chains and trimer molecules are largely unknown, but some of the chains have been shown to differ with respect to their tissue distribution, presumably reflecting diverse functions in vivo. This gene encodes the beta chain isoform laminin, beta 2. The beta 2 chain contains the 7 structural domains typical of beta chains of laminin, including the short alpha region. However, unlike beta 1 chain, beta 2 has a more restricted tissue distribution. It is enriched in the basement membrane of muscles at the neuromuscular junctions, kidney glomerulus and vascular smooth muscle. Transgenic mice in which the beta 2 chain gene was inactivated by homologous recombination, showed defects in the maturation of neuromuscular junctions and impairment of glomerular filtration. Alternative splicing involving a non consensus 5' splice site (gc) in the 5' UTR of this gene has been reported. It was suggested that inefficient splicing of this first intron, which does not change the protein sequence, results in a greater abundance of the unspliced form of the transcript than the spliced form. The full-length nature of the spliced transcript is not known. [provided by RefSeq, Aug 2011]. | hsa:3913; | basement membrane [GO:0005604]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; laminin complex [GO:0043256]; laminin-11 complex [GO:0043260]; laminin-3 complex [GO:0005608]; neuromuscular junction [GO:0031594]; synaptic cleft [GO:0043083]; extracellular matrix structural constituent [GO:0005201]; integrin binding [GO:0005178]; structural molecule activity [GO:0005198]; animal organ morphogenesis [GO:0009887]; astrocyte development [GO:0014002]; axon extension involved in regeneration [GO:0048677]; axon guidance [GO:0007411]; basement membrane assembly [GO:0070831]; cell migration [GO:0016477]; metanephric glomerular basement membrane development [GO:0072274]; metanephric glomerular visceral epithelial cell development [GO:0072249]; neuromuscular junction development [GO:0007528]; retina development in camera-type eye [GO:0060041]; Schwann cell development [GO:0014044]; substrate adhesion-dependent cell spreading [GO:0034446]; tissue development [GO:0009888]; visual perception [GO:0007601] | 11891225_Laminin-10/11 and fibronectin differentially prevent apoptosis induced by serum removal via phosphatidylinositol 3-kinase/Akt- and MEK1/ERK-dependent pathways (Laminin 10; separate entry for Laminin 11). 15367484_Deficiency in LAMB2 causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. 15603881_Hepalaminin, an autoantigen from chronic hepatitis C, consists of two domains of laminin beta-2 and a specific domain. 16146715_laminin isoform changes are associated with brain tumor invasion and angiogenesis [review] 16898484_Mutations in the LAMB2 gene encoding laminin beta2, a component of the glomerular basement membrane and the neuro-muscular junction are responsible for the characteristic renal and eye abnormalities of Pierson syndrome. 16912710_Recessive missense mutations in LAMB2 expand the clinical spectrum of LAMB2-associated disorders. 16921188_LAMB2 has to be considered as culprit of milder disorders including nephrosis and variable ocular anomalies. 17371932_Observational study of genotype prevalence. (HuGE Navigator) 17426950_Study summarizes recent progress concerning the molecular mechanisms of laminins in development and disease. 17804866_We demonstrated that overabundance of the beta2 chain of laminin is associated with increased basement membrane thickness and is possibly related to spermatogenic dysfunction 17943323_Milder phenotypes of Pearson Syndrome may be related to hypomorphic LAMB2 alleles. 18058136_Observational study of genotype prevalence. (HuGE Navigator) 18065803_Pierson syndrome is defined by the association of mental retardation, microcoria and DMS caused by mutation in LAMB2 gene 18594871_Study excluded LAMB2 as a candidate gene for Galloway-Mowat syndrome. 18672223_Loss-of-function mutations in laminin beta2 (LAMB2) cause a broad range of ocular pathology, emphasizing the importance of laminin beta2 in eye development. 19048114_LM alpha4 and beta2 have roles in in vitro migration and in vivo tumorigenicity of prostate cancer cells 19147489_beta2 chain-containing laminins (beta2-laminins) bound more avidly to alpha3beta1 and alpha7X2beta1 integrins than beta1 chain-containing laminins (beta1-laminins). 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20507940_Observational study of gene-disease association. (HuGE Navigator) 20556798_All previously reported and several novel LAMB2 mutations in relation to the associated phenotype in patients from 39 unrelated families, are reviewed. 21125408_Novel mutations in steroid-resistant nephrotic syndrome diagnosed in Tunisian children were detected in LAMB2. 21236492_Comprehensive gene sequencing revealed a novel LAMB2 variant (c.440A --> G; His147R) that was homozygous in the 9 living, affected family members, observed at a frequency of 2.1% in the Old Order Mennonite population, and absent in 91 non-Mennonite controls 24856380_No pathogenic LAMB2 mutations were found in the cohort of children with steroid-resistant focal segmental glomerulosclerosis. 24951930_Laminins 411 and 421 differentially promote tumor cell migration via alpha6beta1 integrin and MCAM (CD146). 27614294_Disruption of LAMA4, LAMA5, and LAMB2 or the laminin-receptor interaction occurs in neuromuscular diseases including Pierson syndrome and Lambert-Eaton myasthenic syndrome (LEMS). (Review) 27925579_In conclusion, we reported three Chinese cases with different LAMB2 mutations and different phenotypes, further broadening the range of eye and kidney pathology associated with mutations in LAMB2. 29263159_Collectively, these data show the pathogenicity of LAMB2-S80R and provide the first evidence of genetic modification of Alport phenotypes by variation in another GBM component. This 29599141_Age-related modulation of laminin beta1 versus beta2 chain expression changes the functional properties and phenotype of endothelial cells. The dysregulation of the extracellular matrix during vascular aging may contribute to age-associated impairment of organ function and fibrosis. 33554690_Development of neovascular glaucoma after intraocular surgery in Pierson syndrome. 33749661_Laminin beta2 variants associated with isolated nephropathy that impact matrix regulation. | ENSMUSG00000052911 | Lamb2 | 4.478203e+03 | 0.8469509 | -0.239649808 | 0.2683850 | 7.928167e-01 | 0.3732497846 | 0.83830544 | No | Yes | 3.251931e+03 | 584.045058 | 4.460548e+03 | 821.628692 | |
| ENSG00000172086 | 51315 | KRCC1 | protein_coding | Q9NPI7 | Coiled coil;Reference proteome | hsa:51315; | 29351065_By observing the data obtained from the isothermal titration calorimetry assay, both of the human proteins (KRCC1 and ZFAND6) were demonstrated to bind to their respective Toxoplasma gondii SAG1 and SAG2 proteins. 31908025_KRCC1: A potential therapeutic target in ovarian cancer. | ENSMUSG00000053012 | Krcc1 | 9.485071e+01 | 0.6443098 | -0.634173620 | 0.4029478 | 2.440261e+00 | 0.1182565294 | 0.75783482 | No | Yes | 6.765659e+01 | 11.134434 | 1.136086e+02 | 18.483152 | ||||
| ENSG00000172244 | 375444 | C5orf34 | protein_coding | Q96MH7 | Reference proteome | hsa:375444; | 30771479_Study results firstly revealed that C5orf34 might play a facilitating role in lung adenocarcinoma (LAD) development and progression by regulating MAPK signaling pathway. Furthermore, data implied that C5orf34 may be a potential predictor and treatment target for LAD. | ENSMUSG00000062822 | 4833420G17Rik | 1.182994e+02 | 0.7475041 | -0.419846690 | 0.4512604 | 8.399339e-01 | 0.3594156904 | 0.83257512 | No | Yes | 9.762292e+01 | 22.655838 | 1.174050e+02 | 27.828180 | ||||
| ENSG00000172460 | 124221 | PRSS30P | lncRNA | 3.837187e+02 | 1.0998727 | 0.137336550 | 0.2976293 | 2.192684e-01 | 0.6395979240 | 0.92022661 | No | Yes | 2.921117e+02 | 54.197540 | 2.832621e+02 | 53.677878 | ||||||||||
| ENSG00000172663 | 80194 | TMEM134 | protein_coding | Q9H6X4 | Alternative splicing;Cytoplasm;Host-virus interaction;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:80194; | cytosol [GO:0005829]; integral component of membrane [GO:0016021]; perinuclear region of cytoplasm [GO:0048471] | 22855487_screen identified a novel LMP1-binding protein, transmembrane protein 134; Tmem134 affected LMP1-induced NF-kappaB induction | ENSMUSG00000024845 | Tmem134 | 1.289430e+03 | 1.3895784 | 0.474647211 | 0.3311631 | 2.029741e+00 | 0.1542468538 | 0.77404149 | No | Yes | 1.500915e+03 | 231.717512 | 9.146671e+02 | 145.411854 | |||
| ENSG00000173065 | 55731 | FAM222B | protein_coding | Q8WU58 | Reference proteome | hsa:55731; | nucleoplasm [GO:0005654] | ENSMUSG00000037750 | Fam222b | 1.005259e+03 | 1.4067491 | 0.492365038 | 0.3081649 | 2.593637e+00 | 0.1072937190 | 0.75783482 | No | Yes | 1.140871e+03 | 118.283856 | 7.275261e+02 | 77.994820 | ||||
| ENSG00000173137 | 203054 | ADCK5 | protein_coding | Q3MIX3 | FUNCTION: The function of this protein is not yet clear. It is not known if it has protein kinase activity and what type of substrate it would phosphorylate (Ser, Thr or Tyr). | Kinase;Membrane;Reference proteome;Serine/threonine-protein kinase;Transferase;Transmembrane;Transmembrane helix | hsa:203054; | integral component of membrane [GO:0016021]; protein serine/threonine kinase activity [GO:0004674] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 32277958_aarF domain containing kinase 5 gene promotes invasion and migration of lung cancer cells through ADCK5-SOX9-PTTG1 pathway. | ENSMUSG00000022550 | Adck5 | 3.029239e+02 | 0.9867491 | -0.019244828 | 0.3186866 | 3.639870e-03 | 0.9518917448 | 0.99040425 | No | Yes | 2.274893e+02 | 35.047892 | 2.527798e+02 | 40.027063 | ||
| ENSG00000173226 | 9657 | IQCB1 | protein_coding | Q15051 | FUNCTION: Involved in ciliogenesis. The function in an early step in cilia formation depends on its association with CEP290/NPHP6 (PubMed:21565611, PubMed:23446637). Involved in regulation of the BBSome complex integrity, specifically for presence of BBS2 and BBS5 in the complex, and in ciliary targeting of selected BBSome cargos. May play a role in controlling entry of the BBSome complex to cilia possibly implicating CEP290/NPHP6 (PubMed:25552655). {ECO:0000269|PubMed:23446637, ECO:0000269|PubMed:25552655}. | Alternative splicing;Calmodulin-binding;Ciliopathy;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Leber congenital amaurosis;Nephronophthisis;Phosphoprotein;Reference proteome;Repeat;Senior-Loken syndrome | This gene encodes a nephrocystin protein that interacts with calmodulin and the retinitis pigmentosa GTPase regulator protein. The encoded protein has a central coiled-coil region and two calmodulin-binding IQ domains. It is localized to the primary cilia of renal epithelial cells and connecting cilia of photoreceptor cells. The protein is thought to play a role in ciliary function. Defects in this gene result in Senior-Loken syndrome type 5. Alternative splicing results in multiple transcript variants. A pseudogene of this gene is found on chromosome 6. [provided by RefSeq, Jan 2016]. | hsa:9657; | centriole [GO:0005814]; centrosome [GO:0005813]; cilium [GO:0005929]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; intercellular bridge [GO:0045171]; microtubule cytoskeleton [GO:0015630]; mitotic spindle [GO:0072686]; nucleoplasm [GO:0005654]; photoreceptor connecting cilium [GO:0032391]; photoreceptor outer segment [GO:0001750]; calmodulin binding [GO:0005516]; enzyme binding [GO:0019899]; cilium assembly [GO:0060271]; maintenance of animal organ identity [GO:0048496]; photoreceptor cell maintenance [GO:0045494] | 15723066_nephrocystin-5, RPGR and calmodulin can be coimmunoprecipitated from retinal extracts, and that these proteins localize to connecting cilia of photoreceptors and to primary cilia of renal epithelial cells 20881296_Results show that the onset of renal failure in patients with IQCB1 mutations is highly variable, and that mutations are also found in Leber congenital amaurosis (LCA) patients without nephronophthisis, rendering IQCB1 a new gene for LCA. 21068128_Observational study of gene-disease association. (HuGE Navigator) 21220633_Mutations in NPHP5 can cause Leber congenital amaurosis (LCA)without early-onset renal disease. 21245082_Cone photoreceptors are the main targets for gene therapy of NPHP5 (IQCB1) or NPHP6 (CEP290) blindness: generation of an all-cone Nphp6 hypomorph mouse that mimics the human retinal ciliopathy. 21857984_Data show that the minor allele (N) of I393N in IQCB1 and the common allele (R) of R744Q in RPGRIP1L were associated with severe disease in XlRP with RPGR mutations. 21901789_in a set of consanguineous patient families with Leber congenital amaurosis study identified five putative disease-causing mutations, including four novel alleles, in six families; These five mutations are located in four genes, ALMS1, IQCB1, CNGA3, and MYO7A 22183348_Genetic variation may affect severity of disease for X-linked retinitis pigmentosa. 23446637_NPHP5 mutations impair protein interaction with Cep290 and localize to centrosomes, thereby compromising cilia formation. 24674142_mutation is predicted to introduce a new open reading frame that results in the truncation of the C-terminal 235 amino acids of nephrocystin-5 and its consequent loss of function 25552655_NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. 25851290_High-throughput mutation analysis identified a homozygous truncating mutation (c.1504C>T, p.R502*) in the NPHP5 in 5 families in Iranian children with nephronophthisis. 27328943_that nephrocystin-5 is essential for photoreceptor outer segment formation 27506978_NPHP5-mutant dogs recapitulate the human phenotype of very early loss of rods, and relative retention of the central retinal cone photoreceptors that lack function. 28498859_Dynamic ubiquitination and deubiquitination of NPHP5 plays a crucial role in the regulation of ciliogenesis. NPHP5 directly binds to a deubiquitinating enzyme USP9X/FAM and two E3 ubiquitin ligases BBS11/TRIM32 and MARCH7/axotrophin. 29322253_Study demonstrates the interaction between CNNM4 and IQCB1, which provides the first link between CNNM4 and IQCB1 that causes Leber congenital amaurosis and retinal dystrophy when mutated, providing important insights into the molecular pathogenic mechanisms of retinal dystrophy in Jalili syndrome. 31177295_During ciliogenesis, the mother centriole transforms into a basal body competent to nucleate a cilium. The mother centriole and basal body possess sub-distal appendages (SDAs) and basal feet (BF), respectively. SDAs are distinguishable from BF and the protein NPHP5 is a novel SDA and BF component. NPHP5 regulates BF assembly. 31212307_Cone vision improvement potential in LCA due to CEP290 or NPHP5 mutations is predictable from retinal structure using a machine learning approach. This should allow individual prediction of the maximal efficacy in clinical trials and guide decisions about dosing. 33512896_SENIOR-LOKEN SYNDROME: A Case Series and Review of the Renoretinal Phenotype and Advances of Molecular Diagnosis. 33847778_Genetic variations in the CTLA-4 immune checkpoint pathway are associated with colon cancer risk, prognosis, and immune infiltration via regulation of IQCB1 expression. | ENSMUSG00000022837 | Iqcb1 | 6.594282e+02 | 0.9774358 | -0.032926171 | 0.3584026 | 8.328176e-03 | 0.9272869204 | 0.98610450 | No | Yes | 6.228695e+02 | 119.390522 | 6.327898e+02 | 124.283519 | |
| ENSG00000173692 | 5707 | PSMD1 | protein_coding | Q99460 | FUNCTION: Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair. {ECO:0000269|PubMed:1317798}. | 3D-structure;Acetylation;Alternative splicing;Phosphoprotein;Proteasome;Reference proteome;Repeat | The 26S proteasome is a multicatalytic proteinase complex with a highly ordered structure composed of 2 complexes, a 20S core and a 19S regulator. The 20S core is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. The 19S regulator is composed of a base, which contains 6 ATPase subunits and 2 non-ATPase subunits, and a lid, which contains up to 10 non-ATPase subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides. This gene encodes the largest non-ATPase subunit of the 19S regulator lid, which is responsible for substrate recognition and binding. There is evidence that this proteasome and its subunits interact with viral proteins, including those of coronaviruses. Alternatively spliced transcript variants have been found for this gene.[provided by RefSeq, Aug 2020]. | hsa:5707; | azurophil granule lumen [GO:0035578]; cytosol [GO:0005829]; extracellular region [GO:0005576]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; proteasome accessory complex [GO:0022624]; proteasome complex [GO:0000502]; proteasome regulatory particle [GO:0005838]; proteasome regulatory particle, base subcomplex [GO:0008540]; proteasome storage granule [GO:0034515]; enzyme regulator activity [GO:0030234]; ubiquitin protein ligase binding [GO:0031625]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; regulation of protein catabolic process [GO:0042176] | 16737963_neurotoxic products of inflammation, such as PGJ2, may play a role in neurodegenerative disorders by impairing 26 S proteasome activity and inducing a chain of events that culminates in neuronal cell death 18534977_analysis of how subunit radial displacements open the gate into the proteolytic core in the human 26S proteasome 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21044959_p38 MAPK negatively regulates the proteasome activity by phosphorylating Thr-273 of Rpn2 22031102_The 26S proteasome plays a key role in promoting apoptosis induced by high doses of UV irradiation. 24286321_Increased 26S proteasome non-ATPase regulatory subunit 1 in the aqueous humor of patients with age-related macular degeneration compared to control subjects. 24910440_Together, our findings suggest that the interaction of Psmd1 with Adrm1 is controlled by SUMOylation in a manner that may alter proteasome composition and function. 28442575_RPN13 binds ubiquitin with an affinity similar to that of other proteasome-associated ubiquitin receptors and that RPN2, ubiquitin, and the deubiquitylase UCH37 bind to RPN13 with independent energetics. 30285878_we firstly demonstrate that BCRC-3 is down-regulated in BC tissues and cell lines for the first time. BCRC-3 is capable of functioning as ceRNA for miR-182-5p to regulate the expression of p27. 31703613_The authors demonstrate that PSMD1 and PSMD2 promote the proliferation of HepG2 cells via facilitating cellular lipid droplet accumulation. 32558489_Identification of Hub Genes in Gastric Cancer with High Heterogeneity Based on Weighted Gene Co-Expression Network. 33712704_Proteasome 26S subunit, non-ATPases 1 (PSMD1) and 3 (PSMD3), play an oncogenic role in chronic myeloid leukemia by stabilizing nuclear factor-kappa B. 34572038_26S Proteasome Non-ATPase Regulatory Subunits 1 (PSMD1) and 3 (PSMD3) as Putative Targets for Cancer Prognosis and Therapy. | ENSMUSG00000026229 | Psmd1 | 2.472757e+03 | 0.7548895 | -0.405662536 | 0.3029469 | 1.793088e+00 | 0.1805504198 | 0.77663851 | No | Yes | 2.248319e+03 | 394.372538 | 2.656772e+03 | 477.716114 | |
| ENSG00000173821 | 57674 | RNF213 | protein_coding | Q63HN8 | FUNCTION: Atypical E3 ubiquitin ligase that can catalyze ubiquitination of both proteins and lipids, and which is involved in various processes, such as lipid metabolism, angiogenesis and cell-autonomous immunity (PubMed:21799892, PubMed:26126547, PubMed:26278786, PubMed:26766444, PubMed:30705059, PubMed:32139119, PubMed:34012115). Acts as a key immune sensor by catalyzing ubiquitination of the lipid A moiety of bacterial lipopolysaccharide (LPS) via its RZ-type zinc-finger: restricts the proliferation of cytosolic bacteria, such as Salmonella, by generating the bacterial ubiquitin coat through the ubiquitination of LPS (PubMed:34012115). Also acts indirectly by mediating the recruitment of the LUBAC complex, which conjugates linear polyubiquitin chains (PubMed:34012115). Ubiquitination of LPS triggers cell-autonomous immunity, such as antibacterial autophagy, leading to degradation of the microbial invader (PubMed:34012115). Involved in lipid metabolism by regulating fat storage and lipid droplet formation; act by inhibiting the lipolytic process (PubMed:30705059). Also regulates lipotoxicity by inhibiting desaturation of fatty acids (PubMed:30846318). Also acts as an E3 ubiquitin-protein ligase via its RING-type zinc finger: mediates 'Lys-63'-linked ubiquitination of target proteins (PubMed:32139119, PubMed:33842849). Involved in the non-canonical Wnt signaling pathway in vascular development: acts by mediating ubiquitination and degradation of FLNA and NFATC2 downstream of RSPO3, leading to inhibit the non-canonical Wnt signaling pathway and promoting vessel regression (PubMed:26766444). Also has ATPase activity; ATPase activity is required for ubiquitination of LPS (PubMed:34012115). {ECO:0000269|PubMed:21799892, ECO:0000269|PubMed:26126547, ECO:0000269|PubMed:26278786, ECO:0000269|PubMed:26766444, ECO:0000269|PubMed:30705059, ECO:0000269|PubMed:30846318, ECO:0000269|PubMed:32139119, ECO:0000269|PubMed:33842849, ECO:0000269|PubMed:34012115}. | ATP-binding;Alternative splicing;Angiogenesis;Chromosomal rearrangement;Coiled coil;Cytoplasm;Disease variant;Hydrolase;Immunity;Isopeptide bond;Lipid droplet;Lipid metabolism;Metal-binding;Multifunctional enzyme;Nucleotide-binding;Phosphoprotein;Proto-oncogene;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:32139119, ECO:0000269|PubMed:33842849, ECO:0000305|PubMed:21799892}. | This gene encodes a protein containing a C3HC4-type RING finger domain, which is a specialized type of Zn-finger that binds two atoms of zinc and is thought to be involved in mediating protein-protein interactions. The protein also contains an AAA domain, which is associated with ATPase activity. This gene is a susceptibility gene for Moyamoya disease, a vascular disorder of intracranial arteries. This gene is also a translocation partner in anaplastic large cell lymphoma and inflammatory myofibroblastic tumor cases, where a t(2;17)(p23;q25) translocation has been identified with the anaplastic lymphoma kinase (ALK) gene on chromosome 2, and a t(8;17)(q24;q25) translocation has been identified with the MYC gene on chromosome 8. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2011]. | hsa:57674; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; lipid droplet [GO:0005811]; membrane [GO:0016020]; nucleolus [GO:0005730]; ATP hydrolysis activity [GO:0016887]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; angiogenesis [GO:0001525]; defense response to bacterium [GO:0042742]; lipid droplet formation [GO:0140042]; lipid ubiquitination [GO:0120323]; negative regulation of non-canonical Wnt signaling pathway [GO:2000051]; protein autoubiquitination [GO:0051865]; protein K63-linked ubiquitination [GO:0070534]; protein ubiquitination [GO:0016567]; regulation of lipid metabolic process [GO:0019216]; sprouting angiogenesis [GO:0002040]; ubiquitin-dependent protein catabolic process [GO:0006511]; xenophagy [GO:0098792] | 12112524_KIAA1618 (ALO17) ia a novel fusion partner of anaplastic lymphoma kinase in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor cases. 21048783_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21799892_involvement of RNF213 in genetic susceptibility to moyamoya disease 22377813_The homozygous c.14576G>A variant in RNF213 could be a good DNA biomarker for predicting the severe type of moyamoya disease 22878964_Moyamoya disease is often accompanied by hypertension. RNF213 has been identified as a susceptibility gene for this disease. Associations of p.R4810K (rs112735431, ss179362673) of RNF213 with blood pressure were investigated in moyamoya disease patients. 22931863_A homozygous c.14576G>A variant of RNF213 gene is associated with neurological deficits with vasculopathy in moyamoya disease. 23010677_We propose the existence of a new entity of intracranial major artery stenosis/occlusion caused by the c.14576G>A variant in RNF213. 23110205_RNF213 mutations are associated with MMD susceptibility in Han Chinese. The ischemic type MMD is particularly related to the R4810K mutation. 23466837_There are strong associations between p.R4859K and p.R4810K polymorphisms of the RNF213 gene and Moyamoya disease (Meta-analysis). 23769926_the influences of PDGFRB, MMP-3, and TIMP-2 on MMD may be unremarkable in Chinese Hans. There may be no prominent interaction among these five gene polymorphisms on the occurrence of MMD. 23850618_RNF213 R4810K reduced angiogenic activities of iPSECs from patients with MMD, suggesting that it is a promising in vitro model for MMD. 23970789_A particular subset of patients with various phenotypes of ICASO has a common genetic variant, RNF213 c.14576G>A, indicating that RNF213 c.14576G>A variant is a high-risk allele for ICASO. 23994138_RNF213 R4810K induced mitotic abnormalities and increased risk of genomic instability. 24658080_the moyamoya disease-associated gene product is a unique protein that functions as ubiquitin ligase and AAA+ ATPase, which possibly contributes to vascular development through mechanical processes in the cell. 25043520_Study identified of a novel RNF213 variant in a three-generation family of European ancestry with intracerebral vasculopathy displaying variability in age of onset and clinical severity 25053281_RNF213 was not associated with bipolar disorder or schizophrenia. 25278557_Alterations in RNF213 predispose patients of diverse ethnicities to Moyamoya disease. 25383461_vascular wall was significantly thinner in RNF213-/- mice at 14 days 25817623_Nonatherosclerotic quasi-MMD did not have RNF213 c.14576G>A variant. 25956231_This study demonstrated that the RNF213 mutation should form part of the diagnostic workup for moyamoya in clinical practice. 25964206_Not only p.4810K but also other functional missense variants of RNF213 conferred susceptibility to moyamoya disease(MMD). 26147384_Gene-based association analyses shows nominal significant association with multifocal fibromuscular dysplasia for RNF213. 26277359_We herein report pediatric sibling patients of moyamoya disease who have homozygous wild-type c.14576G>A variant in RNF213, showing different clinical course and disease severity. 26278786_RNF213 plays unique roles in endothelial cells for proper gene expressions in response to inflammatory signals from environments. 26315205_This is the first report, to our knowledge, of different moyamoya disease phenotypes in a familial case involving the same heterozygous c.14429G > A variant in RNF213. 26430847_The findings indicate that the c.14429G>A (p.R4810K) allele of RNF213 is strongly associated with Korean patients with MMD. The homozygous c.14429G>A (p.R4810K) variant is particularly related to early-onset MMD. 26590131_results confirm that the RNF213 p.Arg4810Lys variant is not uncommon in the general Korean population and provide reference data for the association of this variant and MMD 26847828_Results suggested that rs112735431 in RNF213 was associated with increased risk of moyamoya disease, especially among Japanese and Korean compared with Chinese. [meta-analysis]. 26849809_The RNF213 c.14576G>A variant is more common in NF-1 patients who develop moyamoya syndrome than in NF-1 patients without moyamoya syndrome. 27253870_is a susceptibility gene not only for moyamoya disease but also for intracranial atherosclerotic stenosis in East Asians. 27323329_PTP1B/RNF213/alpha-KGDD pathway is critical for survival of HER2(+) breast cancer, and possibly other malignancies, in the hypoxic tumour microenvironment 27462098_Caveolin-1 level was decreased in patients with Moyamoya disease and markedly decreased in RNF213 variant carriers. Path analysis showed that the presence of the RNF213 variant was associated with caveolin-1 levels that could lead to Moyamoya disease. 27476341_RNF213 p.R4810K polymorphism was significantly associated with quasi-moyamoya disease. 27515544_Case-control study and meta-analysis both provide evidence of an association between the rs112735431 polymorphism in the RNF213 gene and moyamoya risk. 27736983_Both RNF213 D4013N and V4146A significantly decreased re-endothelialization in the migration assay compared with RNF213 WT and the control vector. 27745834_We found that RNF213 single nucleotide polymorphism rs6565666 was associated with intracranial aneurysms in French-Canadian individuals. 27748344_RNF213 is not only associated with MMD but also associated with intracranial major artery stenosis. The genotypes of RNF213 correlate with the phenotypes of MMD. 27787485_Moyamoya vasculopathy shows a genetic mutational gradient decreasing from East to West. 28063898_The p.R4810K variant was associated with atherosclerotic and autoimmune quasi-Moyamoya disease in a Chinese population, and a lower prevalence of this variant in patients with quasi-Moyamoya disease compared with patients with Moyamoya disease was observed. 28276505_Data indicate that mysterin/RNF213 is a substrate of ubiquitin specific protease 15 (USP15), and that the conserved skipping of exon 7 significantly decreases its specific affinity for mysterin. 28320162_This study suggests that the rs112735431 polymorphism of the RNF213 may be linked to the hypertension in moyamoya disease. 28414759_The RNF213 p.R4810K variant appears to be significantly associated with coronary artery disease in the Japanese population. 28506590_Genotyping of the p.R4810K missense variant is useful for identifying individuals with an elevated risk for steno-occlusive intracranial arterial diseases in the family members of patients with moyamoya disease. 28617845_These results suggest that, in our cohort of Korean patients, the p.Arg4810Lys is the only variant that is strongly associated with Moyamoya disease among the 30 RNF213 variants listed in the Human Gene Mutation Database. 28635953_Significant association between rare missense RNF213 variants and moyamoya angiopathy was found in European patients. 28686325_Variants in RNF213 are associated with increased susceptibility to moyamoya vasculopathy (MMV). Our findings suggest that RNF213 variants may play a role in the development of MMV in patients with hemangioma syndromes associated with congenital cerebral arterial anomalies 28797616_The major finding of the present study is that genetic variant RNF213 c.14576G>A was significantly associated with anterior circulation of Intracranial Atherosclerosis but not with posterior Circulations of Intracranial Atherosclerosis. 28962888_Peripheral pulmonary artery stenosis in segmental or subsegmental arteries in adulthood with multiple extracranial vasculopathies was found to be associated with homozygosity for RNF213 p.Arg4810Lys. 29160859_RNF213 4810G>A and RNF213 4950G>A were more frequent in MMD patients. We have confirmed that RNF213 4810G>A and 4950G>A are strongly associated with Korean MMD in children and adults as well as for the ischemic and hemorrhagic types. 29174692_in the present study demonstrated, for the first time, that serum sCD163 and CXCL5 levels were significantly elevated in moyamoya diseases(RNF213 mutation) patients compared to those in healthy controls. 29482934_RNF213 p.R4810K polymorphism was associated with an increased risk of intracranial major artery stenosis/occlusion in the East Asian populations. 29500468_We conclude that RNF213 is a gene associated with susceptibility to ICAS in CADASIL patients. MRA follow-up and close observation are necessary for CADASIL patients with the RNF213 variant, as they may be predisposed to ICAS. 29752070_Meta-analysis showed a statistically significant association between RNF213 p.R4810K and moyamoya disease, intracranial major artery stenosis/occlusion, and quasi-moyamoya disease. Apart from the first 2 diseases, no significant association was identified under the recessive, the homozygote, and the heterozygote models in intracranial major artery stenosis/occlusion. 30355208_The RNF213 variant was observed in 49 (63.6%, all heterozygote) patients with Moyamoya disease. The internal carotid artery diameter did not differ between patients with and without this variant 30562119_RNF213 p.Arg4810Lys variant ia a strong risk allele for pulmonary arterial hypertension in Japanese individuals. 30615506_Case Report: Moyamoya disease susceptibility variant RNF213 p.R4810K increases the risk of ischemic stroke attributable to large-artery atherosclerosis. 30705059_findings identify a unique new regulator of cytoplasmic lipid droplets and suggest a potential link between the pathogenesis of moyamoya disease and fat metabolism. 30922903_that susceptibility variants in RNF213 may require additional clinical factors with an effect equivalent to hyperthyroidism in order to develop moyamoya vasculopathy 30925911_Findings show that rare variants of RNF213 are associated with intracranial artery stenosis/occlusion disease (ICASO) in Chinese, but there were no specific clinical characteristics of the RNF213 rare variants carriers. 30992731_Our study demonstrated that the c.14576G>A variant on RNF213 may be a biomarker to good outcome of intracranial major artery stenosis/occlusion in Chinese. 31060437_The RNF213 p.R4810K variant is common in early-onset ischemic stroke with anterior circulation stenosis in Japan. 31064275_RNF213 gene polymorphism may preferentially affect the cerebrovascular lesion in the anterior circulation 31197213_Vascular tortuosity of the internal carotid artery is related to the RNF213 c.14429G > A variant in moyamoya disease. 31290353_Clinical Features and Surgical Outcomes of Patients With Moyamoya Disease and the Homozygous RNF213 p.R4810K Variant. 31542298_Poor outcomes in carriers of the RNF213 variant (p.Arg4810Lys) with pulmonary arterial hypertension. 31578010_The heterozygous p.R4810K variant of RNF213 was associated with better postoperative collateral formation in patients with moyamoya disease. 31590595_Ring Finger Protein 213 Variant and Plaque Characteristics, Vascular Remodeling, and Hemodynamics in Patients With Intracranial Atherosclerotic Stroke: A High-Resolution Magnetic Resonance Imaging and Hemodynamic Study. 31650369_Moyamoya Disease and Spectrums of RNF213 Vasculopathy. 31733606_Role of RNF213 p.4810K variant in the development of intracranial arterial disease in patients treated with nilotinib. 31815282_Association of single nucleotide polymorphisms of MTHFR, TCN2, RNF213 with susceptibility to hypertension and blood pressure. 31949090_Carrying rate of RNF213 p.R4810K gradually decreased when moving from coastal cities in northeast, north, and east China to southern cities or inland areas. Higher frequencies of p.R4810K were observed in patients with moyamoya disease compared with control participants. Onset age of all patients with the GA and AA genotypes were lower than with the GG genotype. 31953610_RNF213 suppresses carcinogenesis in glioblastoma by affecting MAPK/JNK signaling pathway. 32020275_RNF213 polymorphism is associated with the high risk of intracranial arterial stenosis/occlusion. 32139119_Moyamoya disease patient mutations in the RING domain of RNF213 reduce its ubiquitin ligase activity and enhance NFkappaB activation and apoptosis in an AAA+ domain-dependent manner. 32182997_Distribution of Intracranial Major Artery Stenosis/Occlusion According to RNF213 Polymorphisms. 32248732_Mutations of RNF213 are responsible for sporadic cerebral cavernous malformation and lead to a mulberry-like cluster in zebrafish. 32342250_Loss of mitochondrial ClpP, Lonp1, and Tfam triggers transcriptional induction of Rnf213, a susceptibility factor for moyamoya disease. 32954918_Prospective Screening of Extracranial Systemic Arteriopathy in Young Adults with Moyamoya Disease. 33055470_Observation of p.R4810K, a Polymorphism of the Mysterin Gene, the Susceptibility Gene for Moyamoya Disease, in Two Female Japanese Diabetic Patients with Familial Partial Lipodystrophy 1. 33175469_Meta-analysis of genotype and phenotype studies to confirm the predictive role of the RNF213 p.R4810K variant for moyamoya disease. 33200540_RNF213 gene mutation in circulating tumor DNA detected by targeted next-generation sequencing in the assisted discrimination of early-stage lung cancer from pulmonary nodules. 33333224_New insights into TNFalpha/PTP1B and PPARgamma pathway through RNF213- a link between inflammation, obesity, insulin resistance, and Moyamoya disease. 33356381_Role of the RNF213 Variant in Vascular Outcomes in Patients With Intracranial Atherosclerosis. 33370357_Ethnic variation and the relevance of homozygous RNF 213 p.R4810.K variant in the phenotype of Indian Moya moya disease. 33609224_Association of genetic variants of RNF213 with ischemic stroke risk in Koreans. 33706059_RNF213 rare variants and cerebral arteriovenous malformation in a Chinese population. 33769276_Renovascular hypertension and RNF213 p.R4810K variant in Korean children with Moyamoya disease. 33930741_In silico explanation for the causalities of deleterious RNF213 SNPs in Moyamoya disease and insulin resistance. 33960657_A new syndrome of moyamoya disease, kidney dysplasia, aminotransferase elevation, and skin disease associated with de novo variants in RNF213. 34012115_Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. 34013582_An analysis of the demographic history of the risk allele R4810K in RNF213 of moyamoya disease. 34469872_Role of RNF213 polymorphism in defining quasi-moyamoya disease and definitive moyamoya disease. 34482123_Validation and Extension Study Exploring the Role of RNF213 p.R4810K in 2,877 Chinese Moyamoya Disease Patients. 34584155_Expression and clinical significance of IL7R, NFATc2, and RNF213 in familial and sporadic multiple sclerosis. 34599178_Ring finger protein 213 assembles into a sensor for ISGylated proteins with antimicrobial activity. 34624841_Absence of the RNF213 p.R4810K variant may indicate a severe form of pediatric moyamoya disease in Japanese patients. 34666234_First Report: Rare RNF213 Variant Associated with Familial Moyamoya Disease in an African American Family. 34680863_RNF213 c.14576G>A Is Associated with Intracranial Internal Carotid Artery Saccular Aneurysms. 34716882_RNF213 p.Arg4810Lys Heterozygosity in Moyamoya Disease Indicates Early Onset and Bilateral Cerebrovascular Events. 34738993_The association between the Moyamoya disease susceptible gene RNF213 variant and incident cardiovascular disease in a general population: the Nagahama study. 34773068_Ring finger protein 213 c.14576G>A mutation is not involved in internal carotid artery and middle cerebral artery dysplasia. 34991336_Moyamoya Disease Susceptibility Gene RNF213 Regulates Endothelial Barrier Function. 35135845_MMD-associated RNF213 SNPs encode dominant-negative alleles that globally impair ubiquitylation. | ENSMUSG00000070327 | Rnf213 | 1.622270e+03 | 1.3284543 | 0.409748649 | 0.2978418 | 1.874871e+00 | 0.1709181901 | 0.77611497 | No | Yes | 1.299960e+03 | 198.337711 | 1.076388e+03 | 168.638188 |
| ENSG00000173960 | 165324 | UBXN2A | protein_coding | P68543 | Alternative splicing;Reference proteome | hsa:165324; | cis-Golgi network [GO:0005801]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; nucleus [GO:0005634]; acetylcholine receptor binding [GO:0033130]; ubiquitin binding [GO:0043130]; autophagosome assembly [GO:0000045]; cellular response to leukemia inhibitory factor [GO:1990830]; Golgi organization [GO:0007030]; membrane fusion [GO:0061025]; nuclear membrane reassembly [GO:0031468]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; regulation of protein catabolic process [GO:0042176]; regulation of protein ubiquitination [GO:0031396] | 22454508_Ubx2 and Ubxd8 regulates lipid droplet homeostasis. 24625977_Suggest UBXN2A can reconstitute inactive p53-dependent apoptotic pathways in colonic neoplasms. 26188124_veratridine enhances transactivation of UBXN2A, resulting in upregulation of UBXN2A in the cytoplasm, where UBXN2A binds and inhibits the oncoprotein mortalin-2 26634371_UBXN2A binds to mortalin's binding pocket within the substrate-binding domain of mortalin. UBXN2A increases stability of p53 protein targeted by the mortalin-CHIP E3 ubiquitin ligase. 30107089_The existence of a multiprotein complex containing UBXN2A, CHIP, and mot-2 suggests a synergistic tumor suppressor activity of UBXN2A and CHIP in mot-2-enriched tumors. | ENSMUSG00000020634 | Ubxn2a | 2.752220e+02 | 1.0297826 | 0.042339849 | 0.3479370 | 1.459021e-02 | 0.9038574275 | 0.98243436 | No | Yes | 2.718456e+02 | 44.555565 | 2.492503e+02 | 41.826070 | |||
| ENSG00000174442 | 55055 | ZWILCH | protein_coding | Q9H900 | FUNCTION: Essential component of the mitotic checkpoint, which prevents cells from prematurely exiting mitosis. Required for the assembly of the dynein-dynactin and MAD1-MAD2 complexes onto kinetochores. Its function related to the spindle assembly machinery is proposed to depend on its association in the mitotic RZZ complex (PubMed:15824131). {ECO:0000269|PubMed:15824131}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Centromere;Chromosome;Kinetochore;Mitosis;Phosphoprotein;Reference proteome | hsa:55055; | cytosol [GO:0005829]; kinetochore [GO:0000776]; RZZ complex [GO:1990423]; cell division [GO:0051301]; mitotic spindle assembly checkpoint signaling [GO:0007094]; protein localization to kinetochore [GO:0034501] | 12686595_Other name: HZwilch. Homologue of Drosophila Zwilch (CG18729). Localizes to the kinetochore during prometaphase and metaphase of HeLa cells. Complexed with human ROD and human ZW10, similar to situation in Drosophila. 19008095_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000032400 | Zwilch | 5.492200e+02 | 0.6406541 | -0.642382522 | 0.3206698 | 3.866107e+00 | 0.0492706709 | 0.66791998 | No | Yes | 3.817817e+02 | 58.889342 | 6.128330e+02 | 96.560738 | ||
| ENSG00000174516 | 246330 | PELI3 | protein_coding | Q8N2H9 | FUNCTION: E3 ubiquitin ligase catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins. Involved in the TLR and IL-1 signaling pathways via interaction with the complex containing IRAK kinases and TRAF6. Mediates 'Lys-63'-linked polyubiquitination of IRAK1. Can activate AP1/JUN and ELK1. Not required for NF-kappa-B activation. {ECO:0000269|PubMed:12874243, ECO:0000269|PubMed:17675297}. | Alternative splicing;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene is a scaffold protein and an intermediate signaling protein in the innate immune response pathway. The encoded protein helps transmit the immune response signal from Toll-like receptors to IRAK1/TRAF6 complexes. Several transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Jul 2011]. | hsa:246330; | cytosol [GO:0005829]; ubiquitin protein ligase activity [GO:0061630]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of tumor necrosis factor-mediated signaling pathway [GO:0010804]; protein K63-linked ubiquitination [GO:0070534]; protein polyubiquitination [GO:0000209]; regulation of Toll signaling pathway [GO:0008592] | 15917247_Pellino3 is a novel upstream regulator of p38 MAPK that activates CREB in a p38-dependent manner 18326498_suggest that Pellino 3b acts as a negative regulator for IL-1 signaling by regulating IRAK degradation through its ubiquitin protein ligase activity [Pellino 3b] 23892723_Our findings identify RIP2 as a substrate for Pellino3 and Pellino3 as an important mediator in the Nod2 pathway and regulator of intestinal inflammation. 25027698_Peptide PEL3 derived from the interleukin-1 receptor-associated kinase (IRAK)1-binding motif reveals a distinct phosphothreonine peptide binding preference. 26310831_Pellino-3 is involved in endotoxin tolerance and functions as a negative regulator of Toll-like receptor 2/4 signaling. 27302665_The combination of low Pellino3 levels together with high and inducible Pellino1 expression may be an important determinant of the degree of inflammation triggered upon Toll-like receptor 2 engagement by Helicobacter pylori and/or its components, contributing to Helicobacter pylori-associated pathogenesis by directing the incoming signal toward an NF-kB-mediated proinflammatory response. 28011711_Two protective, low-frequency, non-synonymous variants were significantly associated with a decrease in age-related macular degeneration (AMD)risk: A307V in PELI3 and N1050Y in CFH .We also identified a strong protective signal for a common variant (rs8056814) near CTRB1 associated with a decrease in AMD risk (logistic regression: OR = 0.71, P = 1.8 x 10-07). 31520466_PELI3 gene expression in blood and ovarian cancer. 32556677_Long Non-coding RNA MIAT Mediates Non-small Cell Lung Cancer Development Through Regulating the miR-128-3p/PELI3 Axis. 32635799_miR-365a-5p suppresses gefitinib resistance in non-small-cell lung cancer through targeting PELI3. | ENSMUSG00000024901 | Peli3 | 6.244240e+02 | 1.1067244 | 0.146295990 | 0.3199036 | 2.057363e-01 | 0.6501300032 | 0.92370776 | No | Yes | 6.192123e+02 | 82.567549 | 4.957624e+02 | 68.301792 |
| ENSG00000174628 | 124152 | IQCK | protein_coding | Q8N0W5 | Alternative splicing;Reference proteome | This gene belongs to the IQ motif-containing family of proteins. The IQ motif serves as a binding site for different EF-hand proteins such as calmodulin. This gene was identified as a potential candidate gene for obsessive-compulsive disorder in a genome-wide association study. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Feb 2015]. | hsa:124152; | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000073856 | Iqck | 1.566988e+02 | 1.4160817 | 0.501904466 | 0.3342980 | 2.262820e+00 | 0.1325125811 | 0.76149117 | No | Yes | 1.705589e+02 | 23.106713 | 1.148194e+02 | 16.224843 | |||
| ENSG00000174891 | 51319 | RSRC1 | protein_coding | Q96IZ7 | FUNCTION: Has a role in alternative splicing and transcription regulation (PubMed:29522154). Involved in both constitutive and alternative pre-mRNA splicing. May have a role in the recognition of the 3' splice site during the second step of splicing. {ECO:0000269|PubMed:15798186, ECO:0000269|PubMed:29522154}. | Alternative splicing;Coiled coil;Cytoplasm;Disease variant;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;mRNA processing;mRNA splicing | This gene encodes a member of the serine and arginine rich-related protein family. The encoded protein is involved in both constitutive and alternative mRNA splicing. This gene may be associated with schizophrenia. A pseudogene of this gene is located on chromosome 9. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Nov 2012]. | hsa:51319; | cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; alternative mRNA splicing, via spliceosome [GO:0000380]; mRNA splicing, via spliceosome [GO:0000398]; nucleocytoplasmic transport [GO:0006913]; protein phosphorylation [GO:0006468]; response to antibiotic [GO:0046677]; RNA splicing [GO:0008380] | 19065146_Observational study of gene-disease association. (HuGE Navigator) 19065146_This study use Functional MRI and Genome Wide Association Analysis indentic novel gene(RSRC1) associated with schizophrenia. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25937118_Data indicate that arginine/serine-rich coiled-coil 1 (RSRC1) represses estrogen receptor beta (ERbeta) transcriptional activity via ERbeta SUMOylation. 29522154_This study demonstrated that RSRC1 mutation affects intellect and behaviour through aberrant splicing and transcription, downregulating IGFBP3. 29653227_RSRC1 rs6441201A allele was associated with increased neuroblastoma risk in Chinese children. 31257492_RSRC1 knockdown promoted the proliferation and migration of gastric cancer cells. In addition, the knockdown of RSRC1 decreased the expression of phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a potent tumor suppressor gene controlling cellular growth and viability. 32227164_Our findings support the pathogenic role of biallelic loss-of-function RSRC1 variants in autosomal recessive intellectual disability, in addition to contributing to the phenotypic delineation of this emerging condition | ENSMUSG00000034544 | Rsrc1 | 2.554541e+02 | 0.9375767 | -0.092991328 | 0.4160243 | 4.742810e-02 | 0.8276005835 | 0.96748040 | No | Yes | 2.712194e+02 | 66.088649 | 2.213747e+02 | 55.440163 | |
| ENSG00000175104 | 7189 | TRAF6 | protein_coding | Q9Y4K3 | FUNCTION: E3 ubiquitin ligase that, together with UBE2N and UBE2V1, mediates the synthesis of 'Lys-63'-linked-polyubiquitin chains conjugated to proteins, such as IKBKG, IRAK1, AKT1 and AKT2 (PubMed:11057907, PubMed:18347055, PubMed:19713527, PubMed:19465916). Also mediates ubiquitination of free/unanchored polyubiquitin chain that leads to MAP3K7 activation (PubMed:19675569). Leads to the activation of NF-kappa-B and JUN (PubMed:16378096, PubMed:17135271). Seems to also play a role in dendritic cells (DCs) maturation and/or activation (By similarity). Represses c-Myb-mediated transactivation, in B-lymphocytes (PubMed:18093978, PubMed:18758450). Adapter protein that seems to play a role in signal transduction initiated via TNF receptor, IL-1 receptor and IL-17 receptor (PubMed:8837778, PubMed:19825828, PubMed:12140561). Regulates osteoclast differentiation by mediating the activation of adapter protein complex 1 (AP-1) and NF-kappa-B, in response to RANK-L stimulation (By similarity). Together with MAP3K8, mediates CD40 signals that activate ERK in B-cells and macrophages, and thus may play a role in the regulation of immunoglobulin production (By similarity). {ECO:0000250|UniProtKB:P70196, ECO:0000269|PubMed:11057907, ECO:0000269|PubMed:12140561, ECO:0000269|PubMed:16378096, ECO:0000269|PubMed:17135271, ECO:0000269|PubMed:18093978, ECO:0000269|PubMed:18347055, ECO:0000269|PubMed:18758450, ECO:0000269|PubMed:19465916, ECO:0000269|PubMed:19675569, ECO:0000269|PubMed:19713527, ECO:0000269|PubMed:19825828, ECO:0000269|PubMed:8837778}. | 3D-structure;Coiled coil;Cytoplasm;DNA damage;Immunity;Isopeptide bond;Lipid droplet;Metal-binding;Nucleus;Osteogenesis;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene is a member of the TNF receptor associated factor (TRAF) protein family. TRAF proteins are associated with, and mediate signal transduction from, members of the TNF receptor superfamily. This protein has an amino terminal RING domain which is followed by four zinc-finger motifs, a central coiled-coil region and a highly conserved carboxyl terminal domain, known as the TRAF-C domain and mediates signaling from members of the TNF receptor superfamily as well as the Toll/IL-1 family. Signals from receptors such as CD40, TNFSF11/RANCE and IL-1 have been shown to be mediated by this protein. This protein also interacts with various protein kinases including IRAK1/IRAK, SRC and PKCzeta, which provides a link between distinct signaling pathways. This protein functions as a signal transducer in the NF-kappaB pathway that activates IkappaB kinase (IKK) in response to proinflammatory cytokines. The interaction of this protein with UBE2N/UBC13, and UBE2V1/UEV1A, which are ubiquitin conjugating enzymes catalyzing the formation of polyubiquitin chains, has been found to be required for IKK activation by this protein. This protein also interacts with the transforming growth factor (TGF) beta receptor complex and is required for Smad-independent activation of the JNK and p38 kinases. The protein encoded by this gene is a key molecule in antiviral innate and antigen-specific immune responses. [provided by RefSeq, Nov 2021]. | hsa:7189; | CD40 receptor complex [GO:0035631]; cell cortex [GO:0005938]; cytoplasm [GO:0005737]; cytoplasmic side of plasma membrane [GO:0009898]; cytosol [GO:0005829]; endosome membrane [GO:0010008]; lipid droplet [GO:0005811]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; histone deacetylase binding [GO:0042826]; identical protein binding [GO:0042802]; mitogen-activated protein kinase kinase kinase binding [GO:0031435]; protein kinase B binding [GO:0043422]; protein kinase binding [GO:0019901]; protein N-terminus binding [GO:0047485]; thioesterase binding [GO:0031996]; tumor necrosis factor receptor binding [GO:0005164]; tumor necrosis factor receptor superfamily binding [GO:0032813]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; activation of NF-kappaB-inducing kinase activity [GO:0007250]; activation of protein kinase activity [GO:0032147]; antigen processing and presentation of exogenous peptide antigen via MHC class II [GO:0019886]; bone resorption [GO:0045453]; cell development [GO:0048468]; cellular response to cytokine stimulus [GO:0071345]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to lipopolysaccharide [GO:0071222]; cytoplasmic pattern recognition receptor signaling pathway [GO:0002753]; Fc-epsilon receptor signaling pathway [GO:0038095]; I-kappaB kinase/NF-kappaB signaling [GO:0007249]; in utero embryonic development [GO:0001701]; interleukin-1-mediated signaling pathway [GO:0070498]; interleukin-17-mediated signaling pathway [GO:0097400]; MyD88-dependent toll-like receptor signaling pathway [GO:0002755]; myeloid dendritic cell differentiation [GO:0043011]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; neural tube closure [GO:0001843]; odontogenesis of dentin-containing tooth [GO:0042475]; ossification [GO:0001503]; osteoclast differentiation [GO:0030316]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of interleukin-2 production [GO:0032743]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of JNK cascade [GO:0046330]; positive regulation of JUN kinase activity [GO:0043507]; positive regulation of leukocyte adhesion to vascular endothelial cell [GO:1904996]; positive regulation of lipopolysaccharide-mediated signaling pathway [GO:0031666]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of osteoclast differentiation [GO:0045672]; positive regulation of protein ubiquitination [GO:0031398]; positive regulation of T cell cytokine production [GO:0002726]; positive regulation of T cell proliferation [GO:0042102]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription regulatory region DNA binding [GO:2000679]; protein autoubiquitination [GO:0051865]; protein K63-linked ubiquitination [GO:0070534]; protein polyubiquitination [GO:0000209]; regulation of apoptotic process [GO:0042981]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; regulation of immunoglobulin production [GO:0002637]; response to interleukin-1 [GO:0070555]; stimulatory C-type lectin receptor signaling pathway [GO:0002223]; T cell receptor signaling pathway [GO:0050852]; T-helper 1 type immune response [GO:0042088]; tumor necrosis factor-mediated signaling pathway [GO:0033209] | 11751921_the novel zinc finger protein TIZ may play a role during osteoclast differentiation by modulating TRAF6 signaling activity. 12193732_The C-terminal fragment of TRAF6 inhibits lipopolysaccharide-induced NF-kappa B nuclear translocation and c-Jun NH2-terminal kinase activation in endothelial cells. 12270937_Results establish a major role of TRAF3 and -6 in X-linked ectodermal dysplasia receptor (XEDAR) signaling and in the process of ectodermal differentiation. 12496252_Pellino 1 is required for interleukin-1-mediated signaling through its interaction with the interleukin-1 receptor-associated kinase 4-IRAK-tumor necrosis factor receptor-associated factor 6 complex 12714497_direct endothelial-stimulatory role of lipopolysaccharides in initiating angiogenesis through activation of TRAF6-dependent signaling pathways. 12804775_Pellino2 interacts with TRAF6 and activates the MAP kinase pathway. 14530355_TRAF6 interacts with TIR domain-containing adaptor inducing IFN-beta (TRIF) through the TRAF domain of TRAF6 and TRAF6-binding motifs found in the N-terminal portion of TRIF. 14550571_In liver cells, IL-1 stimulates TRAF6 poly-ubiquitination 14593105_STAT3/NF-kappaB p65 cross-talk activated by IL-1 via TRAF6. 14996708_TRAF6 acts as a bifurcation point of the lipopolysaccharide-initiated death and survival signals in endothelial cells. 15125833_TRAF6 ubiquitin ligase kinase mediates IKK activation by BCL10 and MALT1. RNAi-mediated silencing of TRAF6 suppressed TCR-dependent IKK activation and interleukin-2 production in T cells. 15247281_identified a putative TRAF6 interaction site in Mal but not in MyD88 and we demonstrate that Mal can be co-immunoprecipitated with TRAF6 15361868_TLR-mediated IFN-alpha induction requires the formation of a complex consisting of MyD88, TRAF6 and IRF7 as well as TRAF6-dependent ubiquitination. 15456887_Data show that endogenous germinal center kinase is activated by agonists that require TRAF6 for c-Jun N-terminal kinase activation. 15492226_oligomerization and polyubiquitination of TRAF6 induced by TIFA leads to the activation of TAK1 and IKK through a proteasome-independent mechanism 15591054_IL-8-induced NF-kappaB activation proceeds through a TRAF2-independent but TRAF6-dependent pathway, followed by recruitment of IRAK and activation of IKK 15634933_TRAF6 acts as a critical adapter of both the Src/ERK1/2 kinases and IkappaB kinase/NF-kappaB proinflammatory signaling pathways in monocytes and macrophages. 16079148_p62 regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination 16251197_Involvement of the TAB2/TRAF6/TAK1 signalling complex in the Edar signal transduction pathway has important implications for understanding of NF-kappaB activation and anhidrotic ectodermal dysplasia in human. 16260598_TRAF2-dependent CD40 signal transduction requires TRAF6 in nonhemopoietic cells 16280329_LMP1 utilizes two distinct pathways to activate NF-kappaB: a major one through CTAR2/TRAF6/TAK1/IKKbeta (canonical pathway) and a minor one through CTAR1/TRAF3/NIK/IKKalpha (noncanonical pathway) 16354686_Rac1 facilitated the recruitment of Nox2 into the endosomal compartment and subsequent redox-dependent recruitment of TRAF6 to the MyD88/IL-1R1 complex. 16436380_TRAF6 regulates cell fate decisions by inducing caspase 8-dependent apoptosis and the activation of NF-kappaB 16446357_an LMP1-associated complex containing TRAF6, TAB2, and TAK1 plays an essential role in the activation of JNK 16452479_NF-kappa B signaling is induced by the oncoprotein Tio through direct interaction with TRAF6. 16484229_the interaction of IRF-8 with TRAF6 modulates TLR signaling and may contribute to the cross-talk between IFN-gamma and TLR signal pathways 16517750_TGF-beta-activated kinase 1, TNF receptor-associated factor 6, and myeloid differentiation primary response gene (88) are important signal transducers in H. pylori-infected human epithelial cells 16527194_A candidate gene in ectodermal dysplasia. 16635910_Observational study of gene-disease association. (HuGE Navigator) 16920630_TRAF6 is a critical adaptor linking two convergent signaling events; PKCtheta control of CARMA1 phosphorylation, and BCL10-dependent caspase-8 activation. 16932746_knockdown of UL144, TRAF6 or NFkappaB by specific siRNA in infections with UL144-encoding HCMV prevents the activation of CCL22 expression 17135271_An intact RING domain of TRAF6 is required for NF-kappaB activation and biological signaling. 17346928_This study, for the first time, reveals a possible molecular mechanism that the initiation of the IL-17F/IL-17R signaling pathway requires the receptor ubiquitination by TRAF6. 17363736_Inhibition of the MyD88 and TRAF6 adaptor proteins of the TLR pathway blocked not only B7-H1 expression induced by TLR ligands but also that mediated by IFN-gamma 17377523_Sequencing of the promoter region and exons of the TRAF6 gene revealed three sequence variants, one of which was found in three affected members within one family with osteoporosis. 17572386_These data establish a signaling cascade in which regulated Lys63-linked TRAF6 auto-ubiquitination is the critical upstream mediator of osteoclast differentiation. 17608743_Results show that decrease in the ubiquitination of TRAF6 is YopJ-dependent and to prevent or is to remove the K63-polymerized ubiquitin conjugates required for signal transduction. 17724081_These findings demonstrate that Trx, TRAF2, and TRAF6 regulate ASK1 activity by modulating N-terminal homophilic interaction of ASK1. 17878161_IRAK-2 plays a more central role than IRAK-1 in TLR signaling to NFkappaB through TRAF6 ubiquitination 17905570_MyD88, IRAK1 and TRAF6 proteins are crucial early mediators for the IL-1-induced MMP-13 regulation through MAPK pathways and AP-1 activity. 17982039_two independent and indispensable signaling pathways-1) JAK1-associated PI3K signaling and 2) Act1/TRAF6/TAK1-mediated NF-kappaB activation-are stimulated by IL-17A to regulate gene induction in human airway epithelial cells. 18056395_TRAF6-caspase-4 interaction, triggered by LPS, leads to NF-kappaB-dependent transcriptional up-regulation and secretion of important cytokines and chemokines in innate immune signaling in human monocytic cells. 18070982_TRAF6 is involved but with different mechanisms in MyD88-induced and IRAK-induced activation of NF-kappaB and suggest that TRAF6 uses a distinctive mechanism to activate NF-kappaB depending on signals. 18093978_TRAF6 is modified by small ubiquitin-related modifier-1, interacts with histone deacetylase 1, and represses c-Myb-mediated transactivation. 18234474_Suggest that syntenin is a physiological suppressor of TRAF6 and plays an inhibitory role in IL-1R- and TLR4- mediated NF-kappaB activation pathways. 18411265_signals from the IL-1 receptor segregate into at least two separate pathways at the level of IRAK1; one couples through TRAF6 to NFkappaB activation while a second utilizes a TRAF6-independent pathway that is responsible for mRNA stabilization 18457658_In this study, the lysine selection process for TRAF6/p62 ubiquitination was examined. 18474871_Observational study of gene-disease association. (HuGE Navigator) 18617513_The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL 18682563_the HSV U(L)37 virion structural protein can activate NF-kappaB through TRAF6. 18710948_TRAF6 and its E3 ligase activity are required for LMP1-stimulated IRF7 ubiquitination. 18758450_TGF-beta specifically activates TAK1 through interaction of TbetaRI with TRAF6, whereas activation of Smad2 is not dependent on TRAF6. 18759459_Through nuclear magnetic resonance spectroscopy it has been determined that the RING domain of TRAF6 is rigid at the protein core, consistent with the functional requirement that RING domains form a binding scaffold for E2 ubiquitin conjugation enzymes. 18782768_pneumolysin selectively induced expression of MKP1 via a TLR4-dependent MyD88-TRAF6-ERK pathway, which inhibited the PAK4-JNK signaling pathway,leading to up-regulation of MUC5AC mucin production 18922473_These results indicate that TGF-beta activates JNK and p38 through a mechanism similar to that operating in the interleukin-1beta/Toll-like receptor pathway. 18984593_IPS-1 requires TRAF6 and MEKK1 to activate NF-kappaB and mitogen-activated protein kinases that are critical for the optimal induction of type I interferons 18987746_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18996842_TRAF6 and interleukin receptor-associated kinase 2 are novel binding partners for PS1, and IL-1R1 is a new substrate for presenilin-dependent gamma-secretase cleavage 19112497_analysis of TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL 19232518_These results indicate that TRAF6-mediated ubiquitination of IL-1R1 has a decisive role in IL-1R1 signalling and propose a molecular mechanism whereby TRAF6 promotes ubiquitination and RIP of IL-1R1 through its ubiquitin ligase activity. 19254290_Observational study of gene-disease association. (HuGE Navigator) 19331827_BCAR1 is essential for the rapid estrogen effect on osteoclast differentiation, through estrogen receptor alpha and possibly Traf6. 19365808_Data show that NESCA and NEMO interact by their N-terminal region, that NESCA directly associates with TRAF6, which in turn catalyzes NESCA polyubiquitination, and that NESCA overexpression strongly inhibits TRAF6-mediated polyubiquitination of NEMO. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19465916_crystal structures of TRAF6 and its complex with the ubiquitin-conjugating enzyme (E2) Ubc13 19573080_Observational study of gene-disease association. (HuGE Navigator) 19592497_the novel role for Mal in facilitating the direct recruitment of TRAF6 to the plasma membrane, which is necessary for TLR2- and TLR4-induced transactivation of NF-kappaB and regulation of the subsequent pro-inflammatory response. 19713527_TRAF6 was found to be a direct E3 ligase for Akt and was essential for Akt ubiquitination, membrane recruitment, and phosphorylation upon growth-factor stimulation 19716405_DSCR1-1S isoform positively modulates IL-1R-mediated signaling pathways by regulating Tollip/IRAK-1/TRAF6 complex formation. 19773279_Observational study of gene-disease association. (HuGE Navigator) 19786094_show that IRF-5 is phosphorylated by IKKalpha in a MyD88-TRAF6 pathway. 19825828_Act1 mediates IL-17-induced signaling pathways through its E3 ubiquitin ligase activity and TRAF6 is a critical substrate of Act1, which indicates the importance of protein ubiquitination in the IL-17-dependent inflammatory response 19898481_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19996094_Findings suggest that OTUB1 and OTUB2 negatively regulate virus-triggered type I IFN induction and cellular antiviral response by deubiquitinating TRAF3 and TRAF6. 20047764_The binding of TRAF2 and TRAF6 to TICAM-1 cooperatively activates the IFN-inducing pathway through ubiquitination of TICAM-1. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20079715_NUMBL interacts with TRAF6 and promotes the degradation of TRAF6 in vivo, leading to the inhibition of NF-kappaB signaling pathway. 20097753_virus-triggered ubiquitination of TRAF3 and TRAF6 by cIAP1 and cIAP2 is essential for type I IFN induction and cellular antiviral response 20237496_Observational study of gene-disease association. (HuGE Navigator) 20331378_Observational study of gene-disease association. (HuGE Navigator) 20410276_Data identify a new RIG-I/MAVS/TRAF6/IKKbeta/p65Ser536 pathway placed under the control of NOX2, thus characterizing a novel regulatory pathway involved in NF-kappaB-driven proinflammatory response in the context of RSV infection. 20449947_Results indicate that TRAF2 can signal in human B cells, but is not essential for CD40-mediated NF-kappaB activation, and that TRAF2 can compete with TRAF6 for CD40 binding, limiting the capacity of CD40 engagement to induce NF-kappaB activation. 20512936_TRAF auto-ubiquitination is a means of sustaining an open conformation active in downstream signaling. 20529958_Data demonstrate that NEMO/TRAF6 interaction has physiological relevance and might represent a new target for therapeutic purposes. 20542134_In conclusion, Helicobacter pylori-induced miR-146a plays a potential role in a negative feedback loop to modulate the inflammation by targeting IRAK1 and TRAF6. 20568250_Observational study of gene-disease association. (HuGE Navigator) 20588308_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634198_TRAF6 stimulates the accumulation of insoluble and polyubiquitinated mutant DJ-1 into cytoplasmic aggregates. In human post-mortem brains of Parkinson's disease patients, TRAF6 protein colocalizes with aSYN in Lewy bodies. 20644648_Study demonstrates that TRAF6 is not required for atherogenesis in mice and does not associate with clinical disease in humans. 20677014_Observational study of gene-disease association. (HuGE Navigator) 20813000_Meta-analysis of gene-disease association. (HuGE Navigator) 20932475_A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. 21041452_a novel lymphoma-associated mutation in human BAFF-R that results in NF-kappaB activation and reveals TRAF6 as a necessary component of normal BAFF-R signaling. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21078302_findings reveal another signalling molecule affected by S-glutathionylation and uncover a crucial role for GRX-1 in the TRAF6-dependent activation of NF-kappaB by IL-1R/TLRs. 21127049_A20 appeared to be a late-expressed gene in LPS-treated cultures and was associated with TRAF6 degradation and NF-kappaB 21185369_A conserved functional role of the TRAF RING domain dimerization interface and a potentially necessary but insufficient role for RING-dependent TRAF6 K63-linked polyubiquitination towards NF-kappaB activation in cells. 21220427_LPS-induced activation of IRAK4 and TAK1, K63-linked polyubiquitination of IRAK1 and TRAF6, and disrupted IRAK1-TRAF6 and IRAK1-IKKgamma assembly associated with increased A20 expression 21312055_studies suggested that TRAF6 was required for NF-kappaB activation in EC109 cells and it may be a good molecular target for suppressing the survival and proliferation of esophageal cancer cells 21412053_Blocking TRAF6 activity can be used as a therapeutic approach to preserve skeletal muscle mass and function in different disease states and conditions. 21454471_Tumor necrosis factor receptor-associated factor 6 (TRAF6) associates with huntingtin protein and promotes its atypical ubiquitination to enhance aggregate formation. 21460221_Pattern recognition scavenger receptor CD204 attenuates Toll-like receptor 4-induced NF-kappaB activation by directly inhibiting ubiquitination of tumor necrosis factor (TNF) receptor-associated factor 6. 21557901_The TRAF6 mRNA and protein levels in THP-1 cells were also significantly increased with the treatment of anti-beta2GPI/beta2GPI complex. 21629263_TGFbeta, via TRAF6, causes Lys63-linked polyubiquitination of TbetaRI, promoting cleavage of TbetaRI by TNF-alpha converting enzyme (TACE), in a PKCzeta-dependent manner. 21805090_RanBPM influences TRAF6 ubiquitination and the TRAF6-triggered NF-kappaB signaling pathway through RanBPM's interaction with TRAF6. These data suggest that RanBPM participates in gene transcription by binding to TRAF6. 21813202_CD40-mediated apoptosis occurred in the absence of TRAF2 and TRAF3 association, but was significantly reduced when CD40 was deficient in its TRAF6 binding. 21909504_results suggested that TRAF6 may be key molecule to control proinflammatory cytokine production induced by P gingivalis and its LPS. TRAF6 suppression may inhibit inflammatory responses in HPDLCs infected by P gingivalis and its LPS. 21911935_Identified a frequently recurring amplification at chromosome 11 band p13 in NSCLC and SCLC cell lines and tumors. Within this region, only TRAF6 exhibited concomitant mRNA overexpression and gene amplification in lung cancers. 21940676_It is of great importance to elucidate how TRAF6 (as signal mediator) and CLM-3 (as the regulator) cooperate with each other and how they are controlled. 21980489_the protein associates with TGF-beta receptors and components of the TRAF6-TAK1 signaling module 22029577_USP4 deubiquitinates both TRAF2 and TRAF6 in vivo and in vitro in a deubiquitinase activity-dependent manner. 22033216_overexpressed in the peripheral mononuclear cells of patients with Sjogren's syndrome 22033459_Results indicate that stimulation of TLRs induces proteasome-dependent downregulation of TRAF6, and that TRAF6 associated with ubiquitinated IRAK-1 is degraded together by the proteasome. 22140520_TRAF6 can regulate HIV-1 production and expression of IRF7 promotes HIV-1 replication. 22231568_Our data indicate the presence of association of TRAF6 with systemic lupus erythematosus. 22303480_TLR-dependent TRAF6-MKK3-p38 MAPK signaling pathway synergizes with PKCtheta;-MEK-ERK signaling pathway. CARMA1 plays a crucial role in mediating this synergistic effect via TRAF6. 22326918_these findings define a novel pro-angiogenic signaling response in endothelial cells that is regulated by TRAF6. 22447928_the proline-rich Src homology 3 domain-binding motif in TRAF6 interacts directly with activated SFKs to couple LPS engagement of TLR4 to SFK activation and loss of barrier integrity in HMVEC-Ls. 22450746_Traf6 has two important roles in SCC invasion: it promotes cell intrinsic Cdc42-dependent regulation of the actin cytoskeleton and enables production of the paracrine signal, TNFalpha, that enhances the activity of CAFs. 22513239_In psoriatic arthritis patients TRAF6 binding is controlled through TRAF3IP2-variant p.Asp10Asn. 22589256_The data do not support a role of the rs540386 TRAF6 variant as a key component of the genetic network underlying ystemic sclerosis and giant cell arteritis. 22590573_Kaposi's sarcoma associated herpesvirus encoded viral FLICE inhibitory protein K13 activates NF-kappaB pathway independent of TRAF6, TAK1 and LUBAC 22643835_PINK1 positively regulates two key molecules, TRAF6 and TAK1, in the IL-1beta-mediated signaling pathway, consequently up-regulating their downstream inflammatory events 22644879_Interaction of IFNgammaR1 with TRAF6 regulates NF-kappaB activation and IFNlambdaR1 stability. 22656185_Elevated synovial TRAF6 expression correlated with synovitis severity in rheumatoid arthritis 22719861_TRAF6-dependent signaling may be a central pathway in osteoclast differentiation, and that TNF superfamily molecules other than RANKL may modify RANK signaling by interaction with TRAF6-associated signaling 22736025_Data suggest that TRAF6 is a promise target for therapeutic strategies against cancer. 22851693_Beta-TrCP deficiency abolished the translocation TAK1-TRAF6 complex from the membrane to the cytosol, resulting in a diminishment of the IL-1-induced TAK1-dependent pathway. 22851696_The findings define a new role for the IKK-related kinases in suppressing IL-17-mediated NF-kappaB activation through TRAF6-dependent Act1 phosphorylation. 22863753_NLRC3 inhibited Toll-like receptor (TLR)-dependent activation of the transcription factor NF-kappaB by interacting with the TLR signaling adaptor TRAF6 22886393_the overexpression of TRAF6 in osteosarcoma might be related to the tumorigenesis, invasion of osteosarcoma 22901274_findings indicated that common genetic variants in TRAF6 were significantly associated with susceptibility to sepsis-induced ALI in Chinese Han population 22924441_Results show that mutation in TRAF6 protein shows a dominant negative effect against the wild-type TRAF6 protein leading to HED phenotype 23013936_We found a positive correlation between TRAF6 and ubiquitin expression, suggesting that TRAF6 may up regulates ubiquitin activity in cancer cachexia. 23055197_TRAF6 might be involved in the potentiation of growth, proliferation, and invasion of A549 cell line, as well as the inhibition of A549 cell apoptosis by the activation of NF-kappaB 23185365_ubiquitin ligase TRAF6 negatively regulates the JAK-STAT signaling pathway by binding to STAT3 and mediating its ubiquitination 23264041_USP2a plays an important role in TCR signaling by deSUMOylating TRAF6 and mediating TRAF6-MALT1 interaction 23381138_UBE2O negatively regulates TRAF6-mediated NF-kappaB activation by inhibiting TRAF6 polyubiquitination. 23431243_The results indicate that TRAF4 and TRAF6 are overexpressed in IBD. TRAF4 and TRAF6 play different roles in the pathogenesis of IBD 23478027_Human herpesvirus 1 US3 is necessary and sufficient for inhibiting TLR2 signaling at or before the stage of TRAF6 ubiquitination. 23514740_TRAF6 promotes linker for activation of T cell (LAT) ubiquitination and phosphorylation upon T cell receptor (TCR) engagement. 23524951_findings show under non-autophagic conditions, mTOR inhibits AMBRA1 by phosphorylation, whereas on autophagy induction, AMBRA1 is dephosphorylated; in this condition, AMBRA1, interacting with TRAF6, supports ULK1 ubiquitylation by LYS-63-linked chains, and its stabilization, self-association and function 23564640_Inflammatory responses to ischemia are controlled by a balance between TRAF6 ubiquitination and deubiquitination, and Cezanne is a key regulator of this process. 23608757_Parkin suppresses inflammation and cytokine-induced cell death by promoting the proteasomal degradation of TRAF2/6. 23685241_Dengue virus infection significantly induced the expression of miR-146a, which facilitated viral replication by targeting TRAF6 and dampening IFN-beta production. 23707529_TRAF6-deficiency leads to a reduction in presenilin protein levels and reduced PS1 ubiquitination. 23722539_Our results reveal how TRAF6 functions to upregulate HIF-1alpha expression and promote tumor angiogenesis. 23774506_Data indicate that in monocyte- derived macrophages (MDMs) acutely infected with HIV-1 and treated with HCV rCore and HIV-1 rNef, the HIV-1 replication depends on an upstream signal mediated through TRAF2, TRAF5 and TRAF6. 23794009_Significantly lower levels of TRAF-6 was found in CAD patients with the CC genotype. 23794111_TRAF6 inhibited cell apoptosis by downregulation of activated caspase 3 and cleaved poly ADP ribose polymerase and upregulation of c-Jun, Bcl2, and c-Myc. 23818111_These data indicate that overexpression of TRIM22 may negatively regulate the TRAF6-stimulated NF-kappaB pathway by interacting with and degrading TAB2. 23885119_These results indicate that association of PINK1 with SARM1 and TRAF6 is an important step for mitophagy. 23909487_TRAF6-mediated ubiquitination of APPL1 is a vital step for the hepatic actions of insulin through modulation of membrane trafficking and activity of Akt. 23911927_TRAF6 is recruited to and activates mTORC1 through p62 in amino acid-stimulated cells. 24026882_TRAF6 promoted the metastasis of esophageal squamous cell carcinoma. 24128730_TRAF6 mediates ubiquitination of the midbody ring localized protein KIF23/MKLP1. 24301457_Results define CYB5A as a novel prognostic factor for PDAC that exerts its tumor-suppressor function through autophagy induction and TRAF6 modulation. 24301797_CCK8 inhibited TLR9-induced activation of tumor-necrosis factor receptor-associated factor 6. 24302991_miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. 24321786_inhibition of autophagy by chemical or genetic approaches blocked TLR4- or TLR3-induced Lys63 (K63)-linked ubiquitination of TNF receptor-associated factor 6 24337384_Small interfering RNA-mediated silencing of TRAF6 and TAK1. 24360988_miR-125a played a biological function in osteoclastogenesis through a novel TRAF6/ NFATc1/miR-125a regulatory feedback loop. 24399296_Findings indicate that tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) was required for the interaction between TGFbeta type I receptor (TbetaRI) and presenilin 1 (PS1). 24578159_CHIP inhibits NF-kappaB signaling by promoting TRAF6 degradation and plays an important role in osteoclastogenesis and bone remodeling. 24610907_Identified an alternative oncogenic pathway for TRAF6 that uses AEP as its substrate. AEP and TRAF6 protein levels may have prognostic implications in breast cancer patients. Thus, AEP may serve as a biomarker as well as new therapeutic target 24662294_Results suggest that D431E-IRAK2 (interleukin-1 receptor-associated kinase-2) increases NF-kappaB activation by promoting TRAF6 ubiquitination by enhancing TNF Receptor-Associated Factor 6 (TRAF6)-IRAK2 interaction. 24670424_NOD2 downregulates colonic inflammation by IRF4-mediated inhibition of K63-linked polyubiquitination of RICK and TRAF6. 24755241_Together, our data suggest that TRAF6 promotes proliferation of colon cancer cells and it may serve as a potential target for therapy of colon cancer 24812060_TRAF6 and TRAM colocalize, and a TRAF6 binding motif was found in TRAM. Their interaction mediates a novel signaling function for TRAM in TLR4 signaling. 25051892_preventing CD40-TRAF2,3 or CD40-TRAF6 interaction inhibits pro-inflammatory responses in human non-haematopoietic cells. 25061874_phosphorylation of DCBLD2 Y750 recruited TRAF6, leading to increased TRAF6 E3 ubiquitin ligase activity and subsequent activation of AKT, thereby enhancing EGFR-driven tumorigenesis 25134449_PP4R1 could inhibit TRAF6 polyubiquitination. 25202827_identify Syk as an upstream signaling molecule in IL-17A-induced Act1-TRAF6 interaction in keratinocytes, and inhibition of Syk can attenuate CCL20 production 25252821_Over-expression of TRAF6 in pancreatic cancer cells promoted cell proliferation and migration. 25280943_TRAF6 may function as a negative regulator of Notch signaling. 25340740_Tax interacted with and activated TRAF6, and triggered its mitochondrial localization, where it conjugated four carboxyl-terminal lysine residues of MCL-1 with lysine 63-linked polyubiquitin chains 25371197_Data show that the ECSIT (evolutionarily conserved signaling intermediate in Toll pathways) complex, including MEKK7 (TAK1) and TNF receptor-associated factor 6 (TRAF6), plays a role in Toll-like receptor 4 -mediated signals to activate NF-kappa B. 25425640_SUMOylation is a novel mechanism in the regulation of beta-arrestin 2-mediated IL-1R/TRAF6 signaling 25432781_Results suggest that the activated Syk-mediated TRAF6 pathway leads to aberrant activation of B cells in SLE. 25505246_MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. 25605144_High expression of TRAF6 is associated with lung cancer. 25622187_TRAF6 promotes TGFbeta-induced invasion and cell-cycle regulation via Lys63-linked polyubiquitination of Lys178 in TGFbeta type I receptor. 25642822_Findings reveal a mechanism by which TRAF6 is regulated and highlight a role for MST4 in limiting inflammatory responses. 25754842_Data show that toll-like receptor 3/TRIF protein signalling regulates cytokines IL-32 and IFN-beta secretion by activation of receptor-interacting protein-1 (RIP-1) and tumour necrosis factor receptor-associated factor 6 (TRAF6) in cornea epithelial cells. 25907557_The Us3 Protein of Herpes Simplex Virus 1 Inhibits T Cell Signaling by Confining Linker for Activation of T Cells (LAT) Activation via TRAF6 Protein. 25984739_Data suggest MIRN194 (microRNA-194) down-regulates TLR4 (toll-like receptor 4) signal transduction via down-regulation of TRAF6 (TNF receptor-associated factor 6) expression; TLR4 signaling is activated by free fatty acid in inflammatory response. 25999280_In 816 matched pairs of Han Chinese with and without ischemic stroke, TRAF6 rs5030416 was associated with increased susceptibility in various genetic models. No association was found for rs5030411, although it was associated with total cholesterol. 26052093_This study reveals a novel role for TRAF6 in the suppression of autophagy in human pancreatic cancer. 26056944_TRAF-6 mediates S. p-induced PAI-1 expression, and CYLD inhibits PAI-1 expression probably through deubiquitinating TRAF-6 26151128_miR146a has an important promoting effect on the apoptosis of granulosa cells by targeting IRAK1 and TRAF6 via the caspase cascade pathway 26221041_Data indicate that TNF receptor associated factor 6 (TRAF6) activity is regulated by reversible arginine methylation. 26221961_MAVS50, exposing a degenerate TRAF-binding motif within its N-terminus, effectively competed with full-length MAVS for recruiting TRAF2 and TRAF6 26223916_Findings show that TRAF6 is involved in regulating HUVECs proliferation after intermittent hypoxia by modulating cell signaling downstream of AT1R which can be helpful in treating cardiovascular disorders for instance. 26320176_Identify miR-146b-5p as a tumor suppressor and novel prognostic biomarker of gliomas, and suggest miR-146b-5p and TRAF6 as potential therapeutic candidates for malignant gliomas. 26334396_These findings suggest that TRAF6 is required for BLyS-mediated NF-kappaB signaling in myeloma cells 26385923_Transmembrane motif T6BM2-mediated TRAF6 binding is required for MAVS-related antiviral response. 26432169_DK1 inhibits the formation of the TAK1-TAB2-TRAF6 complex and leads to the inhibition of TRAF6 ubiquitination. 26456228_These findings suggest that RNF166 positively regulates RNA virus-triggered IFN-beta production by enhancing the ubiquitination of TRAF3 and TRAF6. 26458771_TIFAB loss increases TRAF6 protein and the dynamic range of TLR4 signaling 26582220_TRAF6 may be involved in cell migration, invasion, and apoptosis of SPC-A-1 cells, possibly through regulating the NF-B-CD24/CXCR4 pathway.. 26627263_The expression of TRAF6 was positively correlated with an advanced N stage and acted as a predictor of a poor prognosis in patients with gastric cancer 26647777_DAT stabilized IkBa by inhibiting the phosphorylation of Ika by the IkB kinase (IKK) complex. DAT induced proteasomal degradation of TRAF6, and DAT suppressed IKKb-phosphorylation through downregulation of TRAF6 26769849_High TRAF6 expression is associated with melanoma invasion and metastasis. 26847475_Study demonstrates that TRAF6 is upregulated in human lung cancer cells, and siRNA-induced TRAF6 knockdown inhibits the invasion of lung cancer cells and promotes apoptosis. 27062898_the polymorphisms in TLR-MyD88-NF-kappaB signaling pathway confer genetic susceptibility to Type 2 diabetes mellitus and diabetic nephropathy. 27315556_The E3 ligase TRAF6 binds to DCP1a and indirectly regulates DCP1a phosphorylation, expression of decapping factors, and gene-specific mRNA decay. 27411454_Peripheral blood levels of S100A8 and TRAF6 in SAE patients were elevated and might be related to the severity of SAE and predict the outcome of SAE. 27468689_CRBN negatively regulates TLR4 signaling via attenuation of TRAF6 and TAB2 ubiquitination. 27473656_These findings show that SopB suppresses host cell apoptosis by binding to TRAF6 and preventing mitochondrial reactive oxygen species generation. 27485827_Low TRAF6 expression is associated with graft-versus-host disease. 27497395_TRAF6 expression was decreased in tympanic membrane of otitis media patients. 27507811_Vav3 is a novel TRAF6 interaction partner that functions in the activation of cooperative signaling between T6BSs and the IVVY motif in the RANK signaling complex. 27681126_HCV infection suppressed the host innate immune response through the induction of autophagic degradation of TRAF6. This finding provided important information for further understanding how HCV evades host immunity to establish persistence 27697099_high expression of TRAF6 is significant for esophageal cancer progression, and TRAF6 indicates poor prognosis in esophageal cancer patients. 27791197_Epigallocatechin-3-gallate is a novel E3 ubiquitin ligase inhibitor that could be used to target TRAF6 activity in melanoma cells, inhibiting cell proliferation/migration. 27808398_We report two siblings with SCID and | ENSMUSG00000027164 | Traf6 | 3.819210e+02 | 1.0847048 | 0.117302492 | 0.3658818 | 9.591967e-02 | 0.7567822505 | 0.94965256 | No | Yes | 3.500632e+02 | 76.487570 | 2.722198e+02 | 60.880872 |
| ENSG00000175322 | 162655 | ZNF519 | protein_coding | Q8TB69 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:162655; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 18987618_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) | 1.670857e+02 | 0.5345831 | -0.903513923 | 0.3601927 | 5.901387e+00 | 0.0151289635 | 0.44119758 | No | Yes | 8.001464e+01 | 16.344773 | 1.680307e+02 | 34.525109 | ||||
| ENSG00000175354 | 5771 | PTPN2 | protein_coding | P17706 | FUNCTION: Non-receptor type tyrosine-specific phosphatase that dephosphorylates receptor protein tyrosine kinases including INSR, EGFR, CSF1R, PDGFR. Also dephosphorylates non-receptor protein tyrosine kinases like JAK1, JAK2, JAK3, Src family kinases, STAT1, STAT3 and STAT6 either in the nucleus or the cytoplasm. Negatively regulates numerous signaling pathways and biological processes like hematopoiesis, inflammatory response, cell proliferation and differentiation, and glucose homeostasis. Plays a multifaceted and important role in the development of the immune system. Functions in T-cell receptor signaling through dephosphorylation of FYN and LCK to control T-cells differentiation and activation. Dephosphorylates CSF1R, negatively regulating its downstream signaling and macrophage differentiation. Negatively regulates cytokine (IL2/interleukin-2 and interferon)-mediated signaling through dephosphorylation of the cytoplasmic kinases JAK1, JAK3 and their substrate STAT1, that propagate signaling downstream of the cytokine receptors. Also regulates the IL6/interleukin-6 and IL4/interleukin-4 cytokine signaling through dephosphorylation of STAT3 and STAT6 respectively. In addition to the immune system, it is involved in anchorage-dependent, negative regulation of EGF-stimulated cell growth. Activated by the integrin ITGA1/ITGB1, it dephosphorylates EGFR and negatively regulates EGF signaling. Dephosphorylates PDGFRB and negatively regulates platelet-derived growth factor receptor-beta signaling pathway and therefore cell proliferation. Negatively regulates tumor necrosis factor-mediated signaling downstream via MAPK through SRC dephosphorylation. May also regulate the hepatocyte growth factor receptor signaling pathway through dephosphorylation of the hepatocyte growth factor receptor MET. Plays also an important role in glucose homeostasis. For instance, negatively regulates the insulin receptor signaling pathway through the dephosphorylation of INSR and control gluconeogenesis and liver glucose production through negative regulation of the IL6 signaling pathways. May also bind DNA. {ECO:0000269|PubMed:10734133, ECO:0000269|PubMed:11909529, ECO:0000269|PubMed:12138178, ECO:0000269|PubMed:12612081, ECO:0000269|PubMed:14966296, ECO:0000269|PubMed:15592458, ECO:0000269|PubMed:18819921, ECO:0000269|PubMed:22080863, ECO:0000269|PubMed:9488479}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Endoplasmic reticulum;Hydrolase;Membrane;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome;S-nitrosylation | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. Members of the PTP family share a highly conserved catalytic motif, which is essential for the catalytic activity. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. Epidermal growth factor receptor and the adaptor protein Shc were reported to be substrates of this PTP, which suggested the roles in growth factor mediated cell signaling. Multiple alternatively spliced transcript variants encoding different isoforms have been found. Two highly related but distinctly processed pseudogenes that localize to chromosomes 1 and 13, respectively, have been reported. [provided by RefSeq, May 2011]. | hsa:5771; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; integrin binding [GO:0005178]; protein kinase binding [GO:0019901]; protein tyrosine phosphatase activity [GO:0004725]; receptor tyrosine kinase binding [GO:0030971]; STAT family protein binding [GO:0097677]; syntaxin binding [GO:0019905]; B cell differentiation [GO:0030183]; erythrocyte differentiation [GO:0030218]; glucose homeostasis [GO:0042593]; insulin receptor signaling pathway [GO:0008286]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of chemotaxis [GO:0050922]; negative regulation of epidermal growth factor receptor signaling pathway [GO:0042059]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of inflammatory response [GO:0050728]; negative regulation of insulin receptor signaling pathway [GO:0046627]; negative regulation of interferon-gamma-mediated signaling pathway [GO:0060336]; negative regulation of interleukin-2-mediated signaling pathway [GO:1902206]; negative regulation of interleukin-4-mediated signaling pathway [GO:1902215]; negative regulation of interleukin-6-mediated signaling pathway [GO:0070104]; negative regulation of lipid storage [GO:0010888]; negative regulation of macrophage colony-stimulating factor signaling pathway [GO:1902227]; negative regulation of macrophage differentiation [GO:0045650]; negative regulation of platelet-derived growth factor receptor-beta signaling pathway [GO:2000587]; negative regulation of positive thymic T cell selection [GO:1902233]; negative regulation of protein tyrosine kinase activity [GO:0061099]; negative regulation of T cell receptor signaling pathway [GO:0050860]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of tumor necrosis factor-mediated signaling pathway [GO:0010804]; negative regulation of type I interferon-mediated signaling pathway [GO:0060339]; negative regulation of tyrosine phosphorylation of STAT protein [GO:0042532]; peptidyl-tyrosine dephosphorylation [GO:0035335]; positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902237]; positive regulation of gluconeogenesis [GO:0045722]; positive regulation of PERK-mediated unfolded protein response [GO:1903899]; regulation of hepatocyte growth factor receptor signaling pathway [GO:1902202]; regulation of interferon-gamma-mediated signaling pathway [GO:0060334]; T cell differentiation [GO:0030217] | 12138178_Results identify TC45 as a protein tyrosine phosphatase responsible for the dephosphorylation of Stat1 in the nucleus. 12171910_TcPTP phosphorylation is regulated by STAT1 12359225_regulates interleukin-6-mediated signaling pathway through STAT3 dephosphorylation 12459463_TCPTP activates p53 and induces caspase-1-dependent apoptosis in a human tumor cell line. 12612081_Insulin makes the TC48-D182A & TC45-D182A 'substrate-trapping' mutants form stable complexes with the tyrosine-phosphorylated IR beta-subunit. Differentially localized TCPTP variants dephosphorylate the IR & downregulate insulin signaling in vivo. 12847239_An important function of TC-PTP is the induction of the nitric oxide pathway that mediates inhibition of T cell proliferation. 14600148_Tc-PTP has a novel role in the regulation of type 1 interferon-stimulated gene expression 15539083_in a STI571-resistant CML sublines, TC-PTP was markedly downregulated. TC-PTP-transduced cells showed dramatic decrease of STAT5 phosphorylation and restored sensitivity to imatinib mesylate as monitored by reduced proliferation and increased apoptosis 16595549_Data suggest that TC48 translocates to the Golgi complex along the secretory pathway, whereas its endoplasmic reticulum localization is maintained by selective retrieval enabled by interactions with p25 and p23. 17554261_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18438405_Observational study of gene-disease association. (HuGE Navigator) 18438405_Single nucleotide polymorphism in PTPN2 gene is associated with ulcerative colitis 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18593762_Observational study of gene-disease association. (HuGE Navigator) 18819921_PTP1B and TCPTP play distinct and non-redundant roles in the regulation of the Met receptor-tyrosine kinase 18840653_matrix-controlled TCPTP phosphatase activity can inhibit VEGFR2 signalling, and the growth, migration and differentiation of human endothelial cells 18936107_Observational study of gene-disease association. (HuGE Navigator) 18948751_TCPTP is a negative regulator of SFK, JAK1 and STAT3 signalling during the cell cycle. 19073967_Observational study of gene-disease association. (HuGE Navigator) 19174780_Observational study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 19264985_Observational study of gene-disease association. (HuGE Navigator) 19336676_PTPN2 regulates cytokine-induced apoptosis and may thereby contribute to the pathogenesis of type 1 diabetes. 19422935_Observational study of gene-disease association. (HuGE Navigator) 19565500_Observational study of gene-disease association. (HuGE Navigator) 19755676_TC-PTP as an important effector of the MYC-driven proliferation program in B-cell lymphomas 19760754_Observational study of gene-disease association. (HuGE Navigator) 19818778_Suggest a functional role for PTPN2 in maintaining the intestinal epithelial barrier and in the pathophysiology of Crohn's disease. 19861958_Observational study of gene-disease association. (HuGE Navigator) 19898481_Observational study of gene-disease association. (HuGE Navigator) 19922411_Data reveal PTPN2, PTPRJ and PTEN as potent regulators of Akt signalling which contribute to ameliorating the consequences of oncogenic K-Ras activity. 19951419_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20059965_PTP1B and TC-PTP are positively implicated in IGF-2-induced migration of MCF-7 cells, suggesting that they could play a role in metastasis development. 20088380_the polymorphic marker C1858T of the PTPN22 gene is associated with T1DM in Russian patients. 20203524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20222910_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20403149_Observational study of gene-disease association. (HuGE Navigator) 20403149_confirmed an association between Crohn disease (CD) and PTPN2, and phenotypic analysis showed an association of this SNP with late age at first diagnosis, inflammatory and penetrating CD behaviour, need for bowel resection and being a smoker at diagnosis 20473312_Our study provides genetic and functional evidence for a tumor suppressor role of PTPN2 and suggests that expression of PTPN2 may modulate response to treatment. 20689057_Silencing of PTPN2 directs epidermal growth factor receptor signaling toward increased phosphatidylinositol 3-kinase activation and increased suppression of epithelial chloride secretory responses. 20886065_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20940300_pCav-1 is a new substrate of TCPTP and that integrin alpha1beta1 acts as a negative regulator of Cav-1 phosphorylation by activating TCPTP. 20962850_Observational study of gene-disease association. (HuGE Navigator) 21072187_Observational study of gene-disease association. (HuGE Navigator) 21115548_data indicate that PTPN2 activity could play a crucial role in the establishment of chronic inflammatory conditions in the intestine, such as Crohn disease. 21179116_Data suggest that decreased expression of PTPN2 may indirectly modulate IL-2 responsiveness. 21220691_IRF-8-induced repression of Toll-like receptor (TLR)3 is specifically mediated by ligand-activated association with Src homology 2 domain-containing protein tyrosine phosphatase. 21246196_We replicated the association of PTPN2 polymorphism with earlier onset of type 1 diabetes and propose that the rs2542151*G allele confers risk to an earlier onset of type 1 diabetes. 21551237_Data suggest association of PTPN2 deletion in T-ALL with activating JAK1 mutations. Data confirm strong association of PTPN2 deletion with TLX1 and NUP214-ABL1 expression. PTPN2 down-regulation reduces lymphoid cell sensitivity to JAK inhibition. 21791476_PTPN2 is a tumor suppressor gene in T-cell malignancies. 21876762_The possibility of cellular phospho-C3G (pC3G) being a substrate of the intracellular T-cell protein tyrosine phosphatase TC-PTP (PTPN2) using the human neuroblastoma cell line, was studied. 21984578_Findings suggest that local IFN production may interact with a genetic factor (PTPN2) to induce aberrant proapoptotic activity of the BH3-only protein Bim, resulting in increased beta-cell apoptosis via JNK activation and the intrinsic apoptotic pathway. 21987459_Loss of PTPN2 is associated with the onset and perpetuation of chronic intestinal inflammation. 22021207_single nucleotide polymorphism in PTPN2 gene is associated with Crohn's disease. 22105363_c-Src is capable of phosphorylating tyr residues of c-Fos whereas the phosphatase TC45 T-cell protein-tyr phosphatase (TC-PTP) dephosphorylates them 22233524_phospho-Ser727 determines the duration of STAT3 activity largely through TC45. 22294642_An association of the PTPN2 and IL2RA genes with juvenile idiopathic arthritis was found. 22377701_A positive correlation was observed between mRNA expression of PTPN2 and NKX2-3 in B cells and in intestinal tissues from both Crohn's disease and ulcerative colitis patients. 22404968_Studies indicate that PTP1B and TCPTP act together in vitro and in vivo to regulate insulin receptor (IR) signaling and glucose homeostasis. 22426692_The study indicates that IL23R-rs11805303 and PTPN2-rs2542151 might contribute to the development of ulcerative colitis and NOD2-P268S might be involved in the etiology of Crohn's disease in the Chinese Han population. 22457781_data confirm the association of PTPN2 variants with susceptibility to both Crohn's disease and ulcerative colitis, suggesting a common disease pathomechanism for these diseases 22671594_Genome-wide association studies have identified single nucleotide polymorphisms within the gene locus encoding protein tyrosine phosphatase nonreceptor type 2 (PTPN2) as a risk factor for the development of chronic inflammatory diseases 22671596_These data identify an important functional role for PTPN2 as a protector of the intestinal epithelial barrier and provide clues as to how PTPN2 mutations may contribute to the pathophysiology of CD. 22960018_Progression to type I diabetes in islet autoimmunity-positive children was associated with the VDR rs2228570 GG genotype and there was an interaction between VDR rs1544410 and PTPN2 rs1893217. 23124138_TC48 dephosphorylates BCR-Abl but not c-Abl and inhibits its activity towards its substrate. 23166300_TCPTP deficiency in human breast cancer cell lines enhances Src family PTKs and STAT3 signaling. 23328081_A yeast two-hybrid screen identified several unique interacting partners of TCPTP(TC48) belonging to two groups - proteins involved in vesicle trafficking and proteins involved in cell adhesion 23454379_the association between TCPTP cleavage and viral replication may have important consequences for the HCV life cycle and HCV-induced liver diseases 23518806_Single nucleotide polymorphisms in the PTPN2 gene is associated with Crohn's disease. 23579275_Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. 23840476_Association of the PTPN2 locus (encoding the T cell protein tyrosine phosphastase) with Caucasian rheumatoid arthritis susceptibility. [Meta-analysis] 24022492_spermidine increased both TCPTP protein levels and enzymatic activity, correlating with a decrease in the phosphorylation of the signal transducers and activators of transcription 1 and 3, downstream mediators of IFN-gamma signaling 24040033_Dtat indicate that activation of protein tyrosine phosphatase non-receptor type 2 (PTPN2) by spermidine ameliorates IFN-gamma-induced inflammatory responses in THP-1 cells. 24127071_Associations were observed between PTPN2 polymorphisms and susceptibility to ulcerative colitis and Crohn's disease. 24480412_PTPN2 variant rs1893217 was associated with risk of Behcet disease development in a Han Chinese population. 24608439_The type 1 diabetes candidate gene BACH2 regulates proinflammatory cytokine-induced apoptotic pathways in pancreatic beta-cells by crosstalk with another candidate gene, PTPN2, and activation of JNK1 and BIM. 24901824_A family wad heterozygous for risk variants of the genes encoding NOD2 and TLR5 and homozygous carriers of PTPN2 risk alleles in Crohn disease. 24936733_The PTPN2 rs1893217 polymorphism is not significantly associated with type 1 diabetes mellitus in Caucasian subjects from Southern Brazil. 25311528_FGFR3 regulation by PTPN1 and PTPN2 depend on FGFR3 localization and A-loop sequence. 25460303_PTPN2 and genes of the vitamin D pathway may have roles in risk of juvenile idiopathic arthritis 25489960_Single-nucleotide polymorphism in PTPN2 gene is associated with Crohn's disease. 25492475_dysfunction of PTPN2 results in aberrant T-cell differentiation and intestinal dysbiosis similar to those observed in human CD. Our findings indicate a novel and crucial role for PTPN2 in chronic intestinal inflammation. 26026268_demonstrate that SIPAR directly interacts with T cell protein tyrosine phosphatase TC45 and enhances its association with STAT3 26090670_Angiopoietin-1 Regulates Brain Endothelial Permeability through PTPN-2 Mediated Tyrosine Dephosphorylation of Occludin 26139109_PTPN2 is linked to the pathogenesis of rheumatoid arthritis via synovial fibroblasts. It is a regulator of lnterleukin-6 production in rheumatoid arthritis synovial fibroblasts. 26208487_Loss of protein tyrosine phosphatase, non-receptor type 2 is associated with activation of AKT and tamoxifen resistance in breast cancer. 26344020_rs2542151 SNP in The PTPN2 gene is associated with T1DM in Chinese Han children 26734582_role of PTPN2 in type 1 diabetes and Crohn's disease (review) 26817397_the molecular mechanisms by which PTP1B and TC-PTP loss co-operates with other genetic aberrations will need to be elucidated to design more effective therapeutic strategies. 26892021_PASD1 serves as a critical nuclear positive regulator of STAT3-mediated gene expression and tumorigenesis. 26928573_Inflammatory bowel disease patients carrying the C-allele of PTPN2 SNP rs1893217 are at greater risk for developing a severe disease course but are more likely to respond to treatment with anti-TNF antibodies. 27342690_ACPA were associated significantly with rs7574865 in STAT-4. The SNP rs2233945 in the PSORS1C1 gene was protective regarding the presence of bone erosions, while rs2542151 in PTPN2 gene was associated with joint damage. 27746364_The increase in miR-210 resulted in down regulation of its target PTPN2 mRNA in pre-eclampsia. 28107378_reported that SNPs in STAT4, PTPN2, PSORS1C1, and TRAF3IP2 are associated with response to TNF-i treatment in RA patients; however, these findings should be validated in a larger population 28145159_PTPN2_rs1893217 CC was associated with a lower risk of having joints with Limitation of motion in patients with Juvenile Idiopathic Arthritis. 28342869_The results suggest that miR-448 might promote Th17 differentiation in multiple sclerosis and thus aggravate the disease through inhibiting PTPN2. 28400551_PTPN2 rs2847297 and rs2847282 may be potential susceptible loci for lung cancer risk. 28804910_In summary, we demonstrate that TCPTP protects the intestinal epithelial barrier by restricting STAT-induced claudin-2 expression. 29247561_The results provide the first indication that PTPN2 variants contribute to the risk of Type 1A diabetes, and possibly induce non-specific Type 1A diabetes. 29423382_RA samples with PTPN2:rs478582 and/or PTPN22:rs2476601 were more positive for MAP than samples without polymorphisms. Combined occurrence of PTPN2:rs478582 and PTPN22:rs2476601 in association with the presence of MAP has significantly increased T-cell response and elevated IFN-gamma expression in RA samples. 29444435_A dual role for myeloid PTPN2 in directly regulating inflammasome activation and IL-1beta production to suppress pro-inflammatory responses during colitis but promote intestinal tumor development, is reported. 29456405_Data suggests that SNPs in PTPN2/22 affect the negative regulation of the immune response in Crohn's disease patients, thus leading to an increase in inflammation/apoptosis and susceptibility of mycobacteria. 29502070_This study shows that a PTPN2-rs7234029 polymorphism is associated with ocular Behcet's disease and is strongly influenced by gender. In addition, our results suggest that the genetic association with PTPN2 may involve the regulation of PTPN2 mRNA expression and cytokine secretion. 29764444_The higher levels of PTPN2 were associated with a worse overall survival both in patients with gliomas and glioblastomas. 29965986_our data indicate a novel role for PTPN2 and PTPN22 in controlling intestinal microbiota composition and further elucidate the complex interplay between genetic risk factors, intestinal microbiota and disease course in IBD patients. 30246029_PTPN2, an anti-inflammatory factor regulated by VDR, was reduced in type 2 diabetics with chronic kidney disease stages 1-2. 30266502_PTPN2 genetic polymorphisms are associated with psoriasis in the Northeastern Chinese population. 30515568_PTPN2 negatively regulates Akt signalling and loss of PTPN2 protein along with increased pAkt-n is a new potential clinical marker of endocrine treatment efficacy in breast cancer 30729132_Further comparison of alanine scanning demonstrates that the reduction in the energy contribution of Arg254 caused by A27S mutation leads to a different inhibitory activity. These observations provide novel insights into the selective binding mechanism of PTP1B inhibitors to TCPTP 31025094_Low PTPN2 protein expression was found in 50.2% of the breast tumours (110/219), gene copy loss in 15.4% (33/214). Low protein expression was associated with a higher relapse rate in patients with Luminal A and HER2-positive tumours, but not triple-negative tumours. 31030572_The association between rs1893217, rs478582 in PTPN2 and T1D risk with different diagnosed age, and related clinical characteristics in Chinese Han population. 31241223_PTPN2 induced by inflammatory response and oxidative stress contributed to glioma progression. 31248982_cell experiments revealed that 1,4-benzoquinone exposure irreversibly inhibits cellular PTPN2 and concomitantly increases tyrosine phosphorylation of STAT1 and expression of STAT1-regulated genes. 31270080_Our study reveals that PTPN2 deletions tend to associate with an improved CIR and OS in the pediatric cohort. Because survival data were only available for 13 PTPN2-deleted pediatric T-ALL, this study may not be powered to identify a significant difference. On the contrary, we did not observe any survival difference according to PTPN2 deletions within the adult population. 31527250_Our findings reveal that PTPN1/2-mediated dephosphorylation of MITA/STING and its degradation by the 20S proteasomal pathway is an important regulatory mechanism of innate immune response to DNA virus. 31735335_Upregulated PTPN2 induced by inflammatory response or oxidative stress stimulates the progression of thyroid cancer. 32286519_Overexpression of TC-PTP in murine epidermis attenuates skin tumor formation. 32373968_Inflammatory response or oxidative stress induces upregulation of PTPN2 and thus promotes the progression of laryngocarcinoma. 32652144_PTPN2 Regulates Interactions Between Macrophages and Intestinal Epithelial Cells to Promote Intestinal Barrier Function. 33001862_Protein tyrosine phosphatase nonreceptor type 2 controls colorectal cancer development. 33122197_PTPN2 regulates the activation of KRAS and plays a critical role in proliferation and survival of KRAS-driven cancer cells. 33193091_Increased Plasma Soluble Interleukin-2 Receptor Alpha Levels in Patients With Long-Term Type 1 Diabetes With Vascular Complications Associated With IL2RA and PTPN2 Gene Polymorphisms. 33411686_PTPN2 negatively regulates macrophage inflammation in atherosclerosis. 33450156_Optogenetic Analysis of Allosteric Control in Protein Tyrosine Phosphatases. 33603165_Proteasome regulation by reversible tyrosine phosphorylation at the membrane. 34057016_Long non-coding RNA TINCR promotes hepatocellular carcinoma proliferation and invasion via STAT3 signaling by direct interacting with T-cell protein tyrosine phosphatase (TCPTP). 34082682_Association of Variants in FCGR2A, PTPN2, and GM-CSF with Cerebral Cavernous Malformation: Potential Biomarkers for a Symptomatic Disease. 34169549_Protein tyrosine phosphatome metabolic screen identifies TC-PTP as a positive regulator of cancer cell bioenergetics and mitochondrial dynamics. 34198814_Gene Polymorphisms of NOD2, IL23R, PTPN2 and ATG16L1 in Patients with Crohn's Disease: On the Way to Personalized Medicine? 34420044_Loss of protein tyrosine phosphatase non-receptor type 2 reduces IL-4-driven alternative macrophage activation. 34623320_T cell protein tyrosine phosphatase protects intestinal barrier function by restricting epithelial tight junction remodeling. 34910875_Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha7 in Phosphatase Activity. 35013194_The catalytic activity of TCPTP is auto-regulated by its intrinsically disordered tail and activated by Integrin alpha-1. 35154122_PTPN2, A Key Predictor of Prognosis for Pancreatic Adenocarcinoma, Significantly Regulates Cell Cycles, Apoptosis, and Metastasis. 35196085_PTPN2 elicits cell autonomous and non-cell autonomous effects on antitumor immunity in triple-negative breast cancer. | ENSMUSG00000024539 | Ptpn2 | 5.253048e+02 | 1.0870573 | 0.120428038 | 0.3291657 | 1.302232e-01 | 0.7182006885 | 0.94076492 | No | Yes | 5.328805e+02 | 86.255316 | 4.571466e+02 | 75.939262 | |
| ENSG00000175414 | 285598 | ARL10 | protein_coding | Q8N8L6 | GTP-binding;Nucleotide-binding;Reference proteome | hsa:285598; | GTP binding [GO:0005525]; GTPase activity [GO:0003924] | ENSMUSG00000025870 | Arl10 | 2.501063e+03 | 1.0269239 | 0.038329268 | 0.2620763 | 2.145005e-02 | 0.8835595736 | 0.97779570 | No | Yes | 2.354851e+03 | 306.695744 | 2.556724e+03 | 340.895622 | ||||
| ENSG00000175482 | 57804 | POLD4 | protein_coding | Q9HCU8 | FUNCTION: As a component of the tetrameric DNA polymerase delta complex (Pol-delta4), plays a role in high fidelity genome replication and repair. Within this complex, increases the rate of DNA synthesis and decreases fidelity by regulating POLD1 polymerase and proofreading 3' to 5' exonuclease activity (PubMed:16510448, PubMed:19074196, PubMed:20334433). Pol-delta4 participates in Okazaki fragment processing, through both the short flap pathway, as well as a nick translation system (PubMed:24035200). Under conditions of DNA replication stress, required for the repair of broken replication forks through break-induced replication (BIR), a mechanism that may induce segmental genomic duplications of up to 200 kb (PubMed:24310611). Involved in Pol-delta4 translesion synthesis (TLS) of templates carrying O6-methylguanine or abasic sites (PubMed:19074196). Its degradation in response to DNA damage is required for the inhibition of fork progression and cell survival (PubMed:24022480). {ECO:0000269|PubMed:16510448, ECO:0000269|PubMed:19074196, ECO:0000269|PubMed:20334433, ECO:0000269|PubMed:24022480, ECO:0000269|PubMed:24035200, ECO:0000269|PubMed:24310611}. | 3D-structure;Alternative splicing;DNA damage;DNA excision;DNA repair;DNA replication;Nucleus;Reference proteome;Ubl conjugation | Mouse_homologues NA; + ;NA | This gene encodes the smallest subunit of DNA polymerase delta. DNA polymerase delta possesses both polymerase and 3' to 5' exonuclease activity and plays a critical role in DNA replication and repair. The encoded protein enhances the activity of DNA polymerase delta and plays a role in fork repair and stabilization through interactions with the DNA helicase Bloom syndrome protein. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Mar 2012]. | hsa:57804; | delta DNA polymerase complex [GO:0043625]; nucleoplasm [GO:0005654]; DNA synthesis involved in DNA repair [GO:0000731]; DNA-dependent DNA replication [GO:0006261] | 16510448_pol delta interacts with PCNA via at least two of its subunits, and p12 could play a role in stabilizing the overall pol delta-PCNA complex as well as pol delta itself 18682526_The DNA polymerase delta enzyme, as well as the isolated p12 subunit, stimulates the DNA helicase activity of Bloom's syndrome helicase. 19931513_these results indicate that POLD4 is required for the in vitro pol delta activity, and that it functions in cell proliferation and maintenance of genomic stability of human cells. 20861182_Low POLD4 is associated with lung cancer. 23233665_The identification of RNF8 allows new insights into the integration of the control of p12 degradation by different DNA damage signaling pathways. 23913683_Data indicate that CRL4(Cdt2) regulates the degradation of the p12 subunit of Pol delta4. 24022480_ubiquitination of p12 through CRL4(Cdt2) and subsequent degradation form one mechanism by which a cell responds to DNA damage to inhibit fork progression. 24035200_Findings provide evidence for the novel concept that Pol delta3 has a role in lagging strand synthesis, and that both forms of Pol delta3 and 4 may participate in DNA replication in higher eukaryotic cells. 24300032_A parallel study of Pol delta4 and Pol delta3 in Okazaki fragment processing provides evidence for a role of Pol delta3 in DNA replication 24691096_the p12/Pol delta is a target as a nuclear substrate of mu-calpain in a calcium-triggered apoptosis and appears to be a potential marker in the study of the chemotherapy of cancer therapies. 30885984_Human Poldelta is a pentameric complex with a dimeric p12 subunit. RKR-mediated dimerization plays a vital role in p12 binding to PCNA and Poldelta5 architecture, and the phenomenon appears to be conserved throughout evolution. 31326365_The role in cellular processes (DNA replication, DNA repair, homologous recombination) and cell cycle regulation of 2 forms of human DNA polymerase delta: delta3 and delta4 was reviewed. (REVIEW) 32855967_Circular RNA circ_0026359 Enhances Cisplatin Resistance in Gastric Cancer via Targeting miR-1200/POLD4 Pathway. | ENSMUSG00000117873+ENSMUSG00000024854 | Gm45928+Pold4 | 4.088871e+02 | 1.3761222 | 0.460608543 | 0.3192799 | 2.079797e+00 | 0.1492600198 | 0.77404149 | No | Yes | 4.553192e+02 | 72.793738 | 2.795203e+02 | 46.364865 |
| ENSG00000175567 | 7351 | UCP2 | protein_coding | P55851 | FUNCTION: UCP are mitochondrial transporter proteins that create proton leaks across the inner mitochondrial membrane, thus uncoupling oxidative phosphorylation from ATP synthesis. As a result, energy is dissipated in the form of heat. | Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | Mitochondrial uncoupling proteins (UCP) are members of the larger family of mitochondrial anion carrier proteins (MACP). UCPs separate oxidative phosphorylation from ATP synthesis with energy dissipated as heat, also referred to as the mitochondrial proton leak. UCPs facilitate the transfer of anions from the inner to the outer mitochondrial membrane and the return transfer of protons from the outer to the inner mitochondrial membrane. They also reduce the mitochondrial membrane potential in mammalian cells. Tissue specificity occurs for the different UCPs and the exact methods of how UCPs transfer H+/OH- are not known. UCPs contain the three homologous protein domains of MACPs. This gene is expressed in many tissues, with the greatest expression in skeletal muscle. It is thought to play a role in nonshivering thermogenesis, obesity and diabetes. Chromosomal order is 5'-UCP3-UCP2-3'. [provided by RefSeq, Jul 2008]. | hsa:7351; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; oxidative phosphorylation uncoupler activity [GO:0017077]; adaptive thermogenesis [GO:1990845]; aging [GO:0007568]; cellular response to amino acid starvation [GO:0034198]; cellular response to glucose stimulus [GO:0071333]; cellular response to insulin stimulus [GO:0032869]; female pregnancy [GO:0007565]; liver regeneration [GO:0097421]; mitochondrial transmembrane transport [GO:1990542]; negative regulation of apoptotic process [GO:0043066]; negative regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0061179]; positive regulation of cell death [GO:0010942]; positive regulation of cold-induced thermogenesis [GO:0120162]; proton transmembrane transport [GO:1902600]; regulation of mitochondrial membrane potential [GO:0051881]; response to cold [GO:0009409]; response to fatty acid [GO:0070542]; response to hypoxia [GO:0001666]; response to superoxide [GO:0000303] | 11381268_Observational study of gene-disease association. (HuGE Navigator) 11466580_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11904148_Glucose induces and leptin decreases expression of uncoupling protein-2 mRNA in human islets. 11920154_microsatellite markers at the UCP2/UCP3 locus on chromosome 11q13 in anorexia nervosa 11935148_evidence for its function as a metabolic regulator (REVIEW) 12030845_hypothesis that recombinant human uncoupling protein-2 (UCP2) ectopically expressed in bacterial inclusion bodies binds nucleotides in a manner identical with the nucleotide-inhibited uncoupling that is observed in kidney mitochondria 12075579_Observational study of gene-disease association. (HuGE Navigator) 12145158_Decreased mitochondrial proton leak and reduced expression in skeletal muscle of obese diet-resistant women 12385772_Observational study of gene-disease association. (HuGE Navigator) 12395215_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12401727_A functional polymorphism in the promoter enhances obesity risk but reduces type 2 diabetes risk in obese middle-aged humans 12401727_Observational study of gene-disease association. (HuGE Navigator) 12670931_purine nucleotides must be the physiological inhibitors of UCP2-mediated uncoupling in vivo. 12690079_Observational study of gene-disease association. (HuGE Navigator) 12690079_there is a unique, near-uniform distribution of the uncoupling protein 2 insertion/deletion polymorphism in Tonga which may be relevant to the prevalence of obesity and type 2 diabetes 12716765_Observational study of gene-disease association. (HuGE Navigator) 12716765_the common -866G/A polymorphism in the UCP2 gene may contribute to the biological variation of insulin secretion 12720538_UCPs in adipose tissue may play a role in the reduction in 24-h energy expenditure observed in post-obese individuals. 12734183_study of isolation, refolding, transport properties, and regulation of recombinant UCP2 12746756_No evidence for involvement of the promoter polymorphism -866 G/A of the UCP2 gene in childhood-onset obesity in humans 12746756_Observational study of gene-disease association. (HuGE Navigator) 12787871_the recent advancements on the role of UCP2 in the brain and portray this uncoupler as an important player in normal neuronal function as well as a key cell death-suppressing device. 12797456_Observational study of gene-disease association. (HuGE Navigator) 12797456_These data are the first to suggest that polymorphisms in the UCP2 gene may be genetic risk factors of spina bifida. 12855674_Overexpression of human UCP2 attenuates reactive oxygen species generation and prevents mitochondrial Ca2+ overload in cultured neonatal rat cardiomyocytes revealing a novel mechanism of cardioprotection 12858170_Data suggest that uncoupling protein-2 is an inducible protein that is neuroprotective by activating cellular redox signaling or by inducing mild mitochondrial uncoupling that prevents the release of apoptogenic proteins. 12915397_Observational study of gene-disease association. (HuGE Navigator) 12915397_Uncoupling protein-2 variants show significant effects on insulin secretion in interaction with family-specific factors. However, the associated allele and the effects on gene expression are opposite to those reported previously. 12960023_Mitochondrial UCPs precondition neurons by dissociating cellular energy production from that of free radicals to withstand the harmful effects of cellular stress occurring in a variety of neurodegenerative disorders, including epilepsy. 14627764_Observational study of gene-disease association. (HuGE Navigator) 14627764_variation of the uncoupling protein 2 promoter is not associated with obesity or obesity-related intermediary phenotypes in Danish subjects 14659466_Uncoupling protein 2 has a role in neuroprotection [review] 14693721_Juvenile obese subjects who are homozygous for the A variant have an increased ratio (3.6 +/- 1.2) of calories derived from carbohydrates to those from lipids. 14693721_Observational study of gene-disease association. (HuGE Navigator) 14730379_In type 2 diabetes there is a significant gene to gene interaction between the Ala55Val polymorphism in the uncoupling protein 2 gene ( UCP2) and the 161C>T polymorphism in the exon 6 of ppargamma. 14747301_UCP2 promoter polymorphism -866G/A does not affect obesity in Japanese type 2 diabetic patients but affects its transcription in beta-cells. 14759071_Observational study of gene-disease association. (HuGE Navigator) 14759071_UCP2 allelic variants may not have a direct role in the pathogenesis and development of obesity. 14974928_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15016641_Mitochondrial UCP2 in circulating monocytes may prevent excessive accumulation of monocytes/macrophages in the arterial wall, thereby reducing atherosclerotic plaque formation. 15039126_Observational study of gene-disease association. (HuGE Navigator) 15045692_Observational study of gene-disease association. (HuGE Navigator) 15061987_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15106800_Observational study of gene-disease association. (HuGE Navigator) 15106800_Polymorphism of uncoupling protein 2 gene is associated with hypertension, and suggests the possibility of uncoupling protein 2 gene as a target molecule for studies on the etiology and treatment of hypertension. 15120704_No association between the different polymorphisms and diabetic nephropathy. 15149891_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 15165610_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15192823_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15220218_Observational study of gene-disease association. (HuGE Navigator) 15220218_The -866A/A genotype was associated with diabetes in women, but not in men. The -866A/A genotype of the UCP2 gene may contribute to diabetes susceptibility by affecting insulin sensitivity. 15308133_Observational study of gene-disease association. (HuGE Navigator) 15448008_UCP2 may be part of a novel adaptive response by which oxidative stress is modulated in colon cancer 15475368_UCP2 has a role in hydroperoxy fatty acid cycling 15520825_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15527803_anion channel structure of UCP2 protein is oligomeric and the second transmembrane domain is essential for the voltage-dependence of this anion channel 15562023_Observational study of gene-disease association. (HuGE Navigator) 15562023_Single nucleotid polymorphism is associated with type 2 diabetes risk in Italy. 15564896_Observational study of gene-disease association. (HuGE Navigator) 15564896_The hypothesis that differences in the UCP-2 genes influence the susceptibility to anorexia nervossa was not supported. 15604415_Observational study of gene-disease association. (HuGE Navigator) 15604415_Our results suggest a role of UCP2 in atherogenesis 15634780_The present results expose the critical importance of UCP2 in normal nigral dopamine cell metabolism. 15827742_UCP-2 can modify atherosclerotic processes inhuman vascular muscle smooth cells in response to high glucose and Angiotensin II. 15833942_reduction of free fatty acids(FFAs) induced by bilio-pancreatic diversion acts in inhibiting FFA transportation to the mitochondria uncoupling protein 2 (UCP-2), contributing to the decreased lipid oxidation inside the adipose tissue 15905464_UCP2 functions as a physiologic regulator of ROS generation in endothelial cells. 15910756_Our results suggest that the LEP and UCP2/UCP3 genes are unlikely to have a substantial effect on variation in obesity phenotypes in this particular US Caucasian population. 15951317_Observational study of gene-disease association. (HuGE Navigator) 15959859_Observational study of gene-disease association. (HuGE Navigator) 15985484_Observational study of gene-disease association. (HuGE Navigator) 16021520_Observational study of gene-disease association. (HuGE Navigator) 16021520_UCP2 promoter polymorphism may contribute to multiple sclerosis susceptibility by regulating the level of UCP2 protein in the central nervous and/or the immune systems. 16054055_neuronal-specific expression of hUCP2 in adult flies decreases cellular oxidative damage and is sufficient to extend life span 16167150_Observational study of gene-disease association. (HuGE Navigator) 16167150_variation in UCP2 may play a role in energy metabolism, but this gene does not contribute significantly to the aetiology of type 2 diabetes and/or obesity in Pima Indians. 16282353_The principal mechanisms regulating UCP2 gene expression are similar in rats and humans, being consistent with a role for UCP2 as a modulator of insulin secretion in humans. 16373902_G-866A polymorphism in the UCP2 gene is associated with a reduced risk of diabetic neuropathy in type 1 diabetes. 16373902_Observational study of gene-disease association. (HuGE Navigator) 16562813_Observational study of gene-disease association. (HuGE Navigator) 16567833_Observational study of gene-disease association. (HuGE Navigator) 16567833_UCP2 promoter gene polymorphism -866 G/A was significantly associated with nerve conduction slowing and vasomotor sympathetic functions suggesting higher UCP2 activity related to A allele has energy-depleting effect on peripheral nerve function in NIDDM 16622580_Observational study of gene-disease association. (HuGE Navigator) 16644712_Functional promoter variants UCP2 and UCP3 increase the prospective risk of type 2 diabetes, and is exacerbated by obesity. 16644712_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16741267_Observational study of gene-disease association. (HuGE Navigator) 16982633_Observational study of gene-disease association. (HuGE Navigator) 16982633_UCP2 may have a role in dissipating the excess energy of a high-glucose environment 17052689_In conclusion, our results provide correlative evidence for the 'PPARgamma-->UCP2-->neuroprotection' cascade in ischemic brain injury. 17066476_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17066476_These findings suggest that UCP2 and UCP4 have a modest but important involvement in the genetic etiology of schizophrenia. 17130180_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17170091_UCP2 overexpression can negatively impact proinsulin processing 17240372_UCP2 is very unstable, with a half-life close to 30min, compared to 30h for its homologue UCP1, a difference that may highlight different physiological functions. 17242157_human uncoupling proteins 1 and 2 are activated by polyunsaturated fatty acids in planar lipid bilayers 17316620_High UCP2 transcript levels were detected in reticulocytes and other maturating erythroid cells in mice exposed to hypoxia, and in umbilical cord blood of human neonates and peripheral blood of adults, suggesting involvement of UCP2 in erythropoiesis 17342078_Observational study of gene-disease association. (HuGE Navigator) 17342078_These data suggest that the A55V polymorphism of the UCP2 gene directly affected the levels of leptin but not via an effect on insulin. 17351641_Results show that UCP2 is essential for mitochondrial Ca(2+) sequestration in response to cell stimulation under physiological conditions. 17397545_Observational study of gene-disease association. (HuGE Navigator) 17462780_UCP-2 can possibly modify atherosclerotic processes initiated in vascular cells and agents that increase UCP-2 expression in vascular may prevent atherosclerosis. 17463068_Link between UCP2 single-nucleotide polymorphism and multiple sclerosis and a slight relation with muscle mitochondrial haplotypes. 17463068_Observational study of genotype prevalence. (HuGE Navigator) 17502873_Observational study of gene-disease association. (HuGE Navigator) 17502873_UCP2 A55V variant might predispose to obesity and Val55 allele to confer population-attributable risk for 9.5% of obese disorders and increase insulin. V-A-T haplotype within UCP2-UCP3 gene cluster is significantly associated with obesity in Paiwanese. 17512314_2 SNPs in UCP2, -866G>A and +4787C>T (A55V) that were tightly linked (r(2) = 0.97), were significantly associated with decreased HDL cholesterol levels in Korean women 17512314_Observational study of gene-disease association. (HuGE Navigator) 17544366_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17544366_UCP2-3 polymorphisms were important genetic factor for the very low calorie diet-induced reduction of body fat mass 17555951_Demonstrate an interaction between the UCP2 -866G>A variant and smoking to increase oxidative stress in diabetics. 17555951_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17570749_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17601350_Observational study of gene-disease association. (HuGE Navigator) 17666355_the expression of uncoupling protein-2 (UCP2) in the granular cell of women undergoing in vitro fertilization (IVF) and its role in embryo development 17701054_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17701054_comparison of risk genotype combinations of: UCP2-866GG, mtDNA 10398A and PGC1alpha p.Thr394Thr or p.Gly482Ser for NIDDM 17718790_Observational study of gene-disease association. (HuGE Navigator) 17718790_the association of plasma fatty acids with the -866 G/A polymorphism of uncoupling protein 2 (UCP2) in obese children 17870627_Observational study of gene-disease association. (HuGE Navigator) 17894153_Ala55Val polymorphism on UCP2 gene predicts greater weight loss in morbidly obese patients undergoing gastric banding. 17894153_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17985659_Observational study of genotype prevalence. (HuGE Navigator) 17985659_a single nucleotide polymorphism in the promoter region of the UCP2 gene has a significant association with HDL cholesterol level in Iranian nonobese nondiabetic subjects 18006489_temperature heterogeneity in atherosclerotic plaques is at least in part attributed to UCP2 expression in macrophages. The heat generated might be used to detect unstable, macrophage-rich, atherosclerotic plaques via thermography. 18018477_The brain may respond to neuroprotection through the increased expression of UCP2 under ischemic conditions. 18031357_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18031357_body fat was reduced significantly less in the ValVal type compared with the other types (p=0.016), whereas the changes in lean body mass, protein, mineral, and water contents were not significantly different according to the Ala55Val polymorphism 18059082_Observational study of gene-disease association. (HuGE Navigator) 18167556_Increased expression of UCP-2 and decreased expression of UCP-3 in humans with chronic hyperglycemia may contribute to impaired glucose-stimulated insulin secretion 18192542_Observational study of gene-disease association. (HuGE Navigator) 18192542_The A allele of the -866G>A variant of UCP2 was associated with reduced risk of coronary artery disease in men with type 2 diabetes in a 6-year prospective study. 18223008_Findings suggest a role of UCP2-UCP3 gene cluster haplotypes in diabetes; in particular, effects of the high-risk haplotypes were more apparent in overweight Caucasian women. 18223008_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18242170_Observational study of gene-disease association. (HuGE Navigator) 18301352_Observational study of gene-disease association. (HuGE Navigator) 18301352_The -866 A/A genotype may contribute to reduced LDL particle size levels, a significant risk factor for the development of coronary artery disease. 18301432_neuroprotection in the thalamus correlated with a high expression of UCP2, which is neuroprotective in a number of models of neurodegenerative diseases 18305572_Significant expression of UCP2 and UCP3 was observed also in cultured keratinocytes and fibroblasts. 18413749_Overexpression of UCP2 in HCT116 human colon cancer cells inhibits reactive oxygen species accumulation and apoptosis after exposure to chemotherapeutic agents. 18426866_Observational study of gene-disease association. (HuGE Navigator) 18460338_Observational study of gene-disease association. (HuGE Navigator) 18460338_Subjects with a 45bp insertion allele of UCP2+3474 ins/del might have a higher risk of developing obesity, although the biological effects of this variant are not completely known. 18496642_Observational study of gene-disease association. (HuGE Navigator) 18496642_The Ala55Val polymorphism of UCP2 was not associated with incident T2DM in the Atherosclerosis Risk cohort in Communities (ARIC). 18593920_Microenvironment activation of highly conserved mammalian UCPs may facilitate the Warburg effect in the absence of permanent respiratory impairment. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18720901_Observational study of genotype prevalence. (HuGE Navigator) 18755175_Observational study of gene-disease association. (HuGE Navigator) 18839467_Observational study of gene-disease association. (HuGE Navigator) 18846338_Down-regulation of the UCP-2 gene expression in steatotic hepatocytes could alleviate the ischemia-reperfusion injury of liver cells. 18924534_Observational study of gene-disease association. (HuGE Navigator) 18940651_Observational study of gene-disease association. (HuGE Navigator) 18940651_The results suggest that the del/del polymorphism of the UCP2 gene is not associated with lower resting energy expenditure in nondialyzed CKD patients 18956255_Nonsynonymous rs660339 in the human UCP2 gene in men, and the haplotype (UCP3 rs2075577-UCP2 rs660339) in women might be good obesity markers. 18956255_Observational study of gene-disease association. (HuGE Navigator) 18977241_Observational study of gene-disease association. (HuGE Navigator) 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19039313_The UCP2-866G>A and UCP3-55C>T gene variants showed significant associations with BMI level, waist circumference, and body weight in the children but not in the adults. 19056482_Observational study of gene-disease association. (HuGE Navigator) 19056576_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19065272_These results show for the first time a direct association between UCP2 amino acid alteration and human disease and highlight a role for mitochondria in hormone secretion. 19137581_In our Irish study population, UCP2 polymorphisms did not influence neural tube defects risk. 19137581_Observational study of gene-disease association. (HuGE Navigator) 19140314_Observational study of gene-disease association. (HuGE Navigator) 19155787_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19155787_UCP2 polymorphisms are associated with heart rate variability in young and healthy states. 19336475_Observational study of gene-disease association. (HuGE Navigator) 19368944_Observational study of gene-disease association. (HuGE Navigator) 19368944_acute insulin response to glucose is associated with the -55C/T UCP2 homozygous mutation and the presence of this mutation could alter postchallenge insulin concentration 19387457_Observational study of gene-disease association. (HuGE Navigator) 19387457_UCP2 -866 G/A polymorphism was associated with chronic inflammatory diseases. 19413708_polymorphisms associated with fat metabolism, obesity and diabetes 19420105_Observational study of gene-disease association. (HuGE Navigator) 19491387_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19527523_Diabetic patients in a post-myocardial infarction cohort with UCP2-866 AA/GA genotype have poorer survival and higher myeloperoxidase levels than their GG counterparts. 19527523_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19531977_Observational study of gene-disease association. (HuGE Navigator) 19531977_Results contribute to explain the reduced cardiovascular risk associated with the A allele of the UCP2 -866G>A polymorphism. 19556617_In the prospective group 120 non-diabetic patients who were carriers of the G allele had significantly higher maximum blood glucose recordings than non-carriers. In the retrospective study 103 non-diabetic patients showed a similar relationship. 19556617_Observational study of gene-disease association. (HuGE Navigator) 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19653005_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19659999_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19680556_Observational study of gene-disease association. (HuGE Navigator) 19681913_Data show that UCP2 haplotype [A Val Ins] seems to be an important risk factor associated with PDR in both type 2 and 1 diabetic groups. 19681913_Observational study of gene-disease association. (HuGE Navigator) 19727213_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19727213_The 866A/G polymorphism of UCP2 gene was not a risk factor for abdominal obesity. 19753844_The -2723T/A, -1957G/A, -866G/A alleles, and ID 45 haplotype are associated with an increased risk of type 2 diabetes in a northwestern Colombian population. 19760975_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19769793_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 19769793_Genetic variation in the UCP2-UCP3 gene cluster may act as a modifier increasing serum lipid levels and indices of abdominal obesity 19833146_CNTF and IL6 polymorphisms in interaction with UCP2 polymorphisms had similar strong effects on weight gain in women and men. 19833146_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19846869_these results suggest that UCP2 can regulate mast cell activation. 19876004_Observational study of gene-disease association. (HuGE Navigator) 19889414_Observational study of gene-disease association. (HuGE Navigator) 19889414_The association of the UCP2 functional promoter variant with the leukocyte telomere length implies a link between mitochondrial production of reactive oxygen species and shorter telomere length in T2D. 19895332_Observational study of gene-disease association. (HuGE Navigator) 19908126_Observational study of gene-disease association. (HuGE Navigator) 19908126_The -866 AA genotype and A allele of the UCP2 gene is associated with obesity and A allele associated with hyperinsulinemia in obese subjects. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19946892_analysis of the nutritional and hormonal regulation of UCP2 and on its physiological roles [review] 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 19950601_Observational study of gene-disease association. (HuGE Navigator) 19950601_The UCP2 gene mutation might be a risk factor of obesity in Chengdu area. However, this gene mutation may not be an impact factor on the alternation of gut bacteria. 20056920_Expression of UCP2 during disease processes associated with the generation of ROS, such as heart failure, could exacerbate defects in excitation contraction coupling and cause the loss of contractile function, enhancing arrhythmogenic potential. 20142250_prostate cancer was inversely associated with the UCP2 rs660339 Val55 allele 20145583_An interaction between -866G>A and beta-blocker treatment was found in individuals with diabetes (P=0.002) but not those without (P=0.79). 20145583_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20332018_No indication for protonophoric reduction of mitochondrial membrane potential was found in UCP2 overexpressing HeLa cells and only little in HUVEC. ROS levels increased instead of being reduced in these cells. 20359253_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20359253_The results suggest that the UCP2 gene may contribute to weight loss and fat change in response to sibutramine therapy in obese Taiwanese patients. 20384434_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20416077_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20427509_hypothyroidism is associated with a profound decrease in UCP2 mRNA expression. UCP2 is a determinant of fat oxidation pathways and may be involved in the changes seen in the metabolic pathways in thyroid disease. 20468064_Observational study of gene-disease association. (HuGE Navigator) 20538960_Observational study of gene-disease association. (HuGE Navigator) 20595066_The UCP2 knockdown participates in the mechanism of action of cisplatin, thus providing evidence that targeting UCP2 may offer clinical benefit in the treatment of cancer. 20602615_Observational study of gene-disease association. (HuGE Navigator) 20608355_UCP2 plays an important protective role against oxidative stress damage to human sperm by diminishing reactive oxygen species production. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20734064_Observational study of gene-disease association. (HuGE Navigator) 20802238_Observational study of gene-disease association. (HuGE Navigator) 20804304_In the Iranian population, there are no associations between -866G/A polymorphism in the uncoupling protein 2 (UCP2)gene promoter with type 2 diabetes. In obese patients, the A allele showed some protective effect for obesity. 20804304_Observational study of gene-disease association. (HuGE Navigator) 20813684_UCP2 may play an important role in the regulation of macrophage activity and cytokine secretion to contribute to spontaneous abortion. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20937264_Observational study of gene-disease association. (HuGE Navigator) 21029457_Observational study of gene-disease association. (HuGE Navigator) 21029457_Study in type 2 diabetic patients shows a significant association of C-reactive protein levels with the G(-866)A polymorphism of UCP2 beyond the effect of major confounders. 21074158_Observational study of gene-disease association. (HuGE Navigator) 21074158_the UCP gene cluster variation (UCP2-UCP3) may not have a role in development of type 2 diabetes mellitus in women 21128330_UCP2 is highly expressed in human colon cancer tissue and may be involved in colon cancer metastasis. 21175267_The genotype and the allele frequencies of Ala55Val of the UCP2 gene showed a significant protective effect against the development of type 2 diabetes. 21254885_We found a suggestive association between NOS3 (-786 T/C; G298A) and UCP2 (A55V) polymorphisms and power athletes in Italy. 21266666_The result suggested that the polymorphism in uncoupling protein 2 may be a potential genetic risk factor for neural tube defects in a high-risk area of China. 21340542_A-IGF1R/Asp-IRS2/Val-UCP2 allele combination is associated with a decreased all-cause mortality risk and with an increased chance of longevity. 21374988_The use of genipin in combination with VitE can increase mitochondrial membrane potential, increase UCO2 expression, and markedly relieve the adipose degeneration of liver cells. 21557918_In PCOS patients, there was a correlation between UCP-2 and CYP11A1 expression, which was significantly higher than in the control group. 21584501_No association exists between the -866G/A polymorphism in the promoter of the UCP2 gene and Polycystic ovary syndrome. 21613221_Letm1 and UCP2/3 independently contribute to two distinct, mitochondrial Ca(2+) uptake pathways in intact endothelial cells. 21720911_The genetic variants in the ESR, FTO, and UCP2 genes may be considered as a screening tool prior to bariatric surgery to help clinicians predict weight loss or glycemic control outcomes for severely obese patients. 21751002_UCP2 -866G/A polymorphism is unlikely to be associated with increased type 2 diabetes risk; UCP2 Ala55Val polymorphism may be risk factor for susceptibility to type 2 diabetes in individuals of Asian descent (Meta-Analysis) 21873561_Serological N-acetyl-glucosaminidase, telomere length, and the UCP2-886G>A variant are independent risk factors for type 2 diabetes. 21883184_Polymorphisms of the UCP2 gene (rs660339 and rs659366) were associated with central obesity. 21935467_UCP2 is over-expressed in breast and many other cancers. UCP2 promotes tumorigenic properties in vitro and in vivo and genipin suppresses the tumor promoting function of UCP2. 22001364_The results of this study found that the UCP2 mRNA levels were significantly lower in patients with bipolar disorder and schizophrenia. 22070905_This study demonistrated that DD genotype of UCP-2 might result in mitochondrial dysfunction, which is also a source of oxidative stress and contribute obsessive compulsive disorders. 22085932_human pluripotent stem cells contain active mitochondria and require UCP2 repression for full differentiation potential. 22134120_This is the first report suggesting that the presence of the -866A/55Val/Ins haplotype is associated with decreased UCP2 gene expression in human retina. 22241057_common genetic variation at the UCP3/2 gene locus is associated with training-related improvements in delta efficiency, an index of skeletal muscle performance 22266335_Human UPC2 has a neuroprotective role in dopaminergic neurons in a Drosophila sporadic Parkinson disease model. 22292025_Over-expression of UCP2 has anti-apoptotic properties by inhibiting ROS-mediated apoptosis in A549 cells under hypoxic conditions. 22349573_The UCP2 -866G-allele is associated with decreased insulin sensitivity in Danish subjects and is associated with obesity in a combined meta-analysis. 22490607_UCP2 could play a role as a determinant of the neurotoxicity mediated by NMDAR through a mechanism related to the unidentified interaction with the essential GluN1 subunit toward modulation of mitochondrial Ca(2+) levels in neurons 22524567_Neuronal UCP2 exhibits transmembrane chloride transport activity. 22533685_Report the importance of environmental settings in studying the involvement of UCP2 gene polymorphisms in the development of obesity in a Balinese population. 22568573_Uncoupling protein-2 45-base pair insertion/deletion polymorphism is not associated with severe obesity and weight loss in morbidly obese subjects 22613561_The A allele variant of UCP2 was associated with increased risk of ischemic stroke in Chinese diabetic women with type 2 diabetes 22634308_These findings question an impact of moderately elevated UCP-2 levels in beta cells as seen in diabetes. 22733179_No association is found between UCP2-45-bp insertion/deletion and body mass index variation in the Chinese population. 22807616_Uncoupling protein 2 regulates glucagon-like peptide-1 secretion in L-cells 22847181_Low UCP-2 expression is associated with low treatment response in lung cancer. 22995745_Genetic polymorphisms within the UCP2 gene are associated with predisposition to recurrent miscarriage in non-obese women. 23008094_Increased ovarian UCP2 expression in response to T3 treatment in PCOS may alter pregnenolone synthesis by influencing P450scc expression, thus alte | ENSMUSG00000033685 | Ucp2 | 2.290278e+03 | 1.8718008 | 0.904426878 | 0.3394480 | 6.902462e+00 | 0.0086077133 | 0.33744247 | No | Yes | 3.168006e+03 | 513.706146 | 1.293262e+03 | 215.785678 | |
| ENSG00000176248 | 29882 | ANAPC2 | protein_coding | Q9UJX6 | FUNCTION: Together with the RING-H2 protein ANAPC11, constitutes the catalytic component of the anaphase promoting complex/cyclosome (APC/C), a cell cycle-regulated E3 ubiquitin ligase that controls progression through mitosis and the G1 phase of the cell cycle. The APC/C complex acts by mediating ubiquitination and subsequent degradation of target proteins: it mainly mediates the formation of 'Lys-11'-linked polyubiquitin chains and, to a lower extent, the formation of 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains. The CDC20-APC/C complex positively regulates the formation of synaptic vesicle clustering at active zone to the presynaptic membrane in postmitotic neurons. CDC20-APC/C-induced degradation of NEUROD2 drives presynaptic differentiation. {ECO:0000269|PubMed:11739784, ECO:0000269|PubMed:18485873}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Differentiation;Mitosis;Neurogenesis;Phosphoprotein;Reference proteome;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | A large protein complex, termed the anaphase-promoting complex (APC), or the cyclosome, promotes metaphase-anaphase transition by ubiquitinating its specific substrates such as mitotic cyclins and anaphase inhibitor, which are subsequently degraded by the 26S proteasome. Biochemical studies have shown that the vertebrate APC contains eight subunits. The composition of the APC is highly conserved in organisms from yeast to humans. The product of this gene is a component of the complex and shares sequence similarity with a recently identified family of proteins called cullins, which may also be involved in ubiquitin-mediated degradation. [provided by RefSeq, Jul 2008]. | hsa:29882; | anaphase-promoting complex [GO:0005680]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; ubiquitin protein ligase binding [GO:0031625]; cell cycle [GO:0007049]; cell division [GO:0051301]; negative regulation of gene expression [GO:0010629]; positive regulation of axon extension [GO:0045773]; positive regulation of dendrite morphogenesis [GO:0050775]; positive regulation of synapse maturation [GO:0090129]; positive regulation of synaptic plasticity [GO:0031915]; protein K11-linked ubiquitination [GO:0070979]; ubiquitin-dependent protein catabolic process [GO:0006511] | 11739784_APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. 16364912_Results describe the locations of Cdh1 and Apc2 in the anaphase-promoting complex/cyclosome of human and Xenopus laevis. 20826619_The authors showed, in vitro, a direct interaction between Orf virus anaphase promoting complex regulator and APC2 and its interference with interactions between APC11 and APC2. 21768287_It proposes a role for APC/C(Cdh1) in modulating the status of PCNA monoubiquitination and UV DNA repair before S phase entry. 21903591_anaphase-promoting complex (APC)-2-cell cycle and apoptosis regulatory protein (CARP)-1 interaction antagonists are novel regulators of cell growth and apoptosis 24804778_The regulation of Mdm2 by the E3 ubiquitin ligase APC/C is shown. It has important therapeutic implications for tumors with Mdm2 overexpression. 25257309_A transcriptome-wide analysis revealed that SNW1 or PRPF8 depletion affects the splicing of specific introns in a subset of pre-mRNAs, including pre-mRNAs encoding the cohesion protein sororin and the APC/C subunit APC2. 26046517_The results showed that APC2 and APC7 subunits were both over expressed in cancer cell lines | ENSMUSG00000026965 | Anapc2 | 4.308488e+03 | 1.0578939 | 0.081194993 | 0.2757663 | 8.660715e-02 | 0.7685356046 | 0.95414161 | No | Yes | 3.773305e+03 | 395.660916 | 3.776980e+03 | 406.430341 |
| ENSG00000176273 | 159371 | SLC35G1 | protein_coding | Q2M3R5 | FUNCTION: May play a role in intracellular calcium sensing and homeostasis. May act as a negative regulator of plasma membrane calcium-transporting ATPases preventing calcium efflux from the cell. {ECO:0000269|PubMed:22084111}. | Alternative splicing;Cell membrane;Endoplasmic reticulum;Membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein which is a member of the drug/metabolite transporter protein superfamily. The encoded protein may play a role in the regulation of calcium levels inside the cell. [provided by RefSeq, Sep 2016]. | hsa:159371; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; calcium ion export across plasma membrane [GO:1990034]; regulation of cytosolic calcium ion concentration [GO:0051480] | 28370613_This study did not detect the identified familial PD-linked *184ProGlyext*5 mutation. 29666234_results suggest that ubiquitinated proteins are exported from the nucleus to the cytosol in the UBIN-POST complex-dependent manner for the maintenance of nuclear protein homeostasis. | ENSMUSG00000044026 | Slc35g1 | 2.220958e+02 | 0.9488925 | -0.075683398 | 0.3203382 | 5.661079e-02 | 0.8119352150 | 0.96274656 | No | Yes | 2.495682e+02 | 33.692497 | 2.446524e+02 | 33.745599 | |
| ENSG00000176890 | 7298 | TYMS | protein_coding | P04818 | FUNCTION: Catalyzes the reductive methylation of 2'-deoxyuridine 5'-monophosphate (dUMP) to thymidine 5'-monophosphate (dTMP), using the cosubstrate, 5,10- methylenetetrahydrofolate (CH2H4folate) as a 1-carbon donor and reductant and contributes to the de novo mitochondrial thymidylate biosynthesis pathway. {ECO:0000269|PubMed:11278511, ECO:0000269|PubMed:21876188}. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Isopeptide bond;Membrane;Methyltransferase;Mitochondrion;Mitochondrion inner membrane;Nucleotide biosynthesis;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation | PATHWAY: Pyrimidine metabolism; dTTP biosynthesis. {ECO:0000305|PubMed:11278511}. | Thymidylate synthase catalyzes the methylation of deoxyuridylate to deoxythymidylate using, 10-methylenetetrahydrofolate (methylene-THF) as a cofactor. This function maintains the dTMP (thymidine-5-prime monophosphate) pool critical for DNA replication and repair. The enzyme has been of interest as a target for cancer chemotherapeutic agents. It is considered to be the primary site of action for 5-fluorouracil, 5-fluoro-2-prime-deoxyuridine, and some folate analogs. Expression of this gene and that of a naturally occurring antisense transcript, mitochondrial enolase superfamily member 1 (GeneID:55556), vary inversely when cell-growth progresses from late-log to plateau phase. Polymorphisms in this gene may be associated with etiology of neoplasia, including breast cancer, and response to chemotherapy. [provided by RefSeq, Aug 2017]. | hsa:7298; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrial inner membrane [GO:0005743]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleus [GO:0005634]; folic acid binding [GO:0005542]; protein homodimerization activity [GO:0042803]; sequence-specific mRNA binding [GO:1990825]; thymidylate synthase activity [GO:0004799]; translation repressor activity, mRNA regulatory element binding [GO:0000900]; aging [GO:0007568]; cartilage development [GO:0051216]; circadian rhythm [GO:0007623]; developmental growth [GO:0048589]; DNA biosynthetic process [GO:0071897]; dTMP biosynthetic process [GO:0006231]; dTTP biosynthetic process [GO:0006235]; intestinal epithelial cell maturation [GO:0060574]; liver regeneration [GO:0097421]; methylation [GO:0032259]; negative regulation of translation [GO:0017148]; response to cytokine [GO:0034097]; response to ethanol [GO:0045471]; response to folic acid [GO:0051593]; response to glucocorticoid [GO:0051384]; response to organophosphorus [GO:0046683]; response to progesterone [GO:0032570]; response to toxic substance [GO:0009636]; response to vitamin A [GO:0033189]; response to xenobiotic stimulus [GO:0009410]; tetrahydrofolate interconversion [GO:0035999]; uracil metabolic process [GO:0019860] | 11102983_Observational study of genotype prevalence. (HuGE Navigator) 11251009_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11445856_Observational study of gene-environment interaction. (HuGE Navigator) 11697906_liganded crystal structures with pyrrolo(2,3-d)pyrimidine-based antifolate compounds 11804689_high level of expression associated with poor prognosis in p-stage I adenocarcinoma of the lung 11937185_Observational study of gene-disease association. (HuGE Navigator) 11989786_Length polymorphism of thymidylate synthase regulatory region in Chinese populations and evolution of the novel alleles 11989786_Observational study of genotype prevalence. (HuGE Navigator) 11992400_TK1 gene expression together with TS, TP and DPD gene expression may play important roles in influencing the malignant behavior of epithelial ovarian cancer. 12018454_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12067974_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12084458_Mutants of TS that confer resistance to MTX as well as 5-FU by random as well as site-directed mutagenesis. Review. 12084459_Thymidylate synthase as a translational regulator of cellular gene expression. Review. 12084460_rTS gene expression likely plays a role in down-regulating TS through a natural RNA-based antisense mechanism. Review. 12084461_Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Review. 12084462_Structure-based studies on species-specific inhibition of thymidylate synthase. Review. 12147691_mutations in TS result in conformational changes that confer 5-fluordeoxyuridine resistance 12215845_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12215845_TYMS and MTHFR compete for limiting supplies of folate required for the remethylation of homocysteine. 12220503_the identified highly conserved protein domains, which occur at the homodimeric interface of thymidylate synthase(TS), represent potential participating sites for binding of TS protein to its mRNA. 12239455_Enhanced expression mediates resistance of uterine cervical cancer cells to radiation 12466474_cytomegalovirus infection induces cellular thymidylate synthase gene expression in quiescent human embryonic lung fibroblasts 12530000_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12576451_Gene expression is a predictor for the efficacy of fluoropyrimidine-based chemotherapy for metastatic colorectal cancer. 12576452_Expression predicts the reponse of metastatic colorectal cancer to raltitrexel. 12604405_Associations between polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and susceptibility to malignant lymphoma. 12640689_polymorphism and overexpression of Thymidylate synthase mrna is associated with non-small-cell lung cancer 12684695_Results suggest that the genotyping for thymidylate synthase and methylenetetrahydrofolate reductase polymorphisms may be a useful indicator in determining the appropriate dose of methotrexate in patients with rheumatoid arthritis. 12782596_Observational study of genotype prevalence. (HuGE Navigator) 12845668_Overexpression of thymidylate synthase mediates desensitization for 5-fluorouracil of cervical cancer cells 12859954_Results describe the kinetic parameters of human recombinant thymidylate synthase (hrTS) with its natural substrate, dUMP, and E-5-(2-bromovinyl)-2(')-deoxyuridine monophosphate (BVdUMP). 12907731_Findings identify Cys-180 as a critical residue for the in vitro and in vivo translational regulatory effects of human thymidylate synthase. 12972956_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14519634_TS and DPD quantitation may be helpful to evaluate prognosis of patients receiving adjuvant 5-FU and that patients with high TS and low DPD may benefit from adjuvant 5-FU chemotherapy in colorectal cancer. 14519641_High TS expression is a marker of poor prognosis in resected pancreatic cancer. Patients with high intratumoral TS expression benefit from adjuvant therapy. 14576935_These results indicate that postoperative chemotherapy with 5-FU could be effective for patients with the G-phenotype tumor, since the incidence of intratumoral expression of TS, the target enzyme of 5-FU, is significantly low in G-phenotype tumors 14578129_Observational study of gene-disease association. (HuGE Navigator) 14586640_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14656021_Observational study of gene-disease association. (HuGE Navigator) 14702180_There is prognostic significance to TS mRNA expression in breast neoplasms. 14745930_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 14760062_thymidylate synthase loss of heterozygosity has a role in progression of colorectal cancer 14967037_Molecular features of thymidylate synthase that control its degradation are examined; the carboxyl-terminal conformational shift is not required for ligand-mediated stabilization; the amino-terminus governs TS stability and its response to ligands. 14970324_TYMS gene amplification in 23% of 31 5-FU-treated cancers, whereas no amplification was observed in metastases of patients that had not been treated with 5-FU. 14977639_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 15093541_Over expression of Thymidylate synthase exhibits oncogene-like activity and suggest a link between DNA synthesis and the induction of a neoplastic phenotype 15115918_6 bp/1494 polymorphism varies greatly within different ethnic populations and is in linkage disequilibrium with the thymidylate synthase 5' tandem repeat enhancer polymorphism 15161716_thymidylate synthase and p53 have roles in regulating Fas-mediated apoptosis in response to antimetabolites 15198953_Observational study of gene-disease association. (HuGE Navigator) 15213713_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15244514_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15244514_Polymorphism of TYMS partially modifies the risk of esophageal and stomach cancer in patients with smoking habit. 15251465_Thymidylate synthase inhibition triggers glucose-dependent apoptosis in p53-negative leukemic cells 15259039_Observational study of gene-disease association. (HuGE Navigator) 15260847_The combination of low expression of TS and induction of p16(INK4a) after chemotherapy can be important indicators of the sensitivity to 5-FU-based chemotherapy in colorectal cancers. 15284183_Observational study of gene-disease association. (HuGE Navigator) 15284183_Thymidylate synthase polymorphisms are associated with esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma 15316940_TS in malignant tissue was statistically higher than in normal stromal tissue in both differentiated and undifferentiated types (p < 0.0001). 15355920_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15386366_Observational study of gene-disease association. (HuGE Navigator) 15386366_Polymorphisms in the TS gene may contribute to gastric cancer susceptibility. 15386371_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15457444_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15457444_Polymorphisms of reduced folate carrier,aminoimidazole carboxamide ribonucleotide transformylase,and thymidylate synthase genes contribute to the therapeutic response in rheumatoid arthritis patients to methotrexate. 15459215_Observational study of gene-disease association. (HuGE Navigator) 15510613_Observational study of gene-disease association. (HuGE Navigator) 15510613_The common MTHFR C677T and TS enhancer region polymorphisms were not risk factors for breast cancer in this patient cohort nor were they associated with phenotypic features or with prognosis. 15571262_Thymidylate synthase polymorphism may have a role in colorectal cancer 15571263_Observational study of genotype prevalence. (HuGE Navigator) 15571263_there is a high level of homology in polymorphic status of thymidylate synthase between normal and malignant colonic tissues 15571283_thymidylate synthase is inhibited by trifluorothymidine in human tumor cells 15579479_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15585623_Observational study of gene-disease association. (HuGE Navigator) 15585623_Thymidylate synthase 5'- and 3'-untranslated region polymorphisms are associated with risk and progression of squamous cell carcinoma of the head and neck 15598787_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15677700_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15682292_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15682292_Our findings suggest that the TYMS 3'UTR del/del genotype is a significant determinant of elevated RBC folate concentration in a non-smoking population 15713801_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15735113_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15736425_Investigators found a relationship between the TSmRNA expression, evaluated by real-time PCR, and with the TS level determined by immunohistochemical assay in colorectal cancer. 15788669_High TS1 gene expression is associated with chemotherapy resistance in esophageal cancer 15797993_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15817609_Important role for folate deficiency and impaired TS activity in the etiology of esophagel and gastric cancer. 15817609_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15897576_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15930032_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15930032_Polymorphisms in the 5'- and 3'-untranslated regions are a risk factor for gastric cancer in China. 15952572_The nasopharyngeal carcinoma patients with positive LRP and TS expression may be less sensitive to chemotherapy with DDP + 5-FU. 15953655_Observational study of gene-disease association. (HuGE Navigator) 15985285_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15985285_TS polymorphism may be a genetic determinant of plasma homocysteine level in Korean patients with recurrent spontaneous abortion. 15993511_thymidylate synthase and dihydropyrimidine dehydrogenase are involved in resistance to 5-fluorouracil in human lung cancer cells 15999119_thymidylate synthase and orotate phosphoribosyl transferase, but not dihydropyrimidine dehydrogenase, are more highly expressed in prostate cancer than in benign prostatic hyperplasia 16030402_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16045580_Observational study of gene-disease association. (HuGE Navigator) 16051637_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16077970_Thymidylate synthase expression levels might be used both as a prognostic marker and to help identify patients who would benefit from 5-FU based regime. 16130010_Observational study of gene-disease association. (HuGE Navigator) 16130010_analysis of TS polymorphisms in childhood acute lymphoblastic leukemia patients 16132996_Association of mRNA expresion patterns with tumor stage and suggested new prognostic and predictive markers for patients with colorectal cancer. 16141798_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16162288_the rTS signaling pathway may be active within human mitochondria 16182121_Observational study of gene-disease association. (HuGE Navigator) 16188144_Observational study of gene-disease association. (HuGE Navigator) 16219137_Observational study of gene-disease association. (HuGE Navigator) 16234002_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16242255_Observational study of gene-disease association. (HuGE Navigator) 16259621_This study demonstrates the ubiquitin-independent nature of TS proteolysis by showing that a 'lysine-less' polypeptide, in which all lysine residues were replaced by arginine, is still subject to proteasome-mediated degradation. 16284371_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16317430_Observational study of gene-disease association. (HuGE Navigator) 16328059_Observational study of gene-disease association. (HuGE Navigator) 16328315_Observational study of genotype prevalence. (HuGE Navigator) 16333305_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16334126_Observational study of gene-disease association. (HuGE Navigator) 16365025_Observational study of gene-disease association. (HuGE Navigator) 16424979_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16447238_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16456808_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16467096_The Thymidylate Synthase expression increased significantly between baseline and biopsy 2 and dropped to near baseline levels at biopsy 3. 16575011_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16580699_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16596248_Expression level and polymorphisms predict disease-free survival in 5-fluorouracil treated colorectal cancer. 16609021_The Thymidylate Synthase(TS) mRNA in plasma originated from tumors, it may indicate poor prognosis and might help to classify tumors in Dukes' stages B and C. The TS promoter enhancer region genotype may influence TS mRNA expression in plasma. 16617381_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16622263_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16675565_Expression is not prognostic of survival in colon cancer. 16722845_Observational study of genotype prevalence. (HuGE Navigator) 16723031_Observational study of gene-disease association. (HuGE Navigator) 16773218_RTQ-LDA and RTQ identified thymidylate synthetase as significantly over-expressed in dead-of-disease compared to long-term survival glioblastoma multiforme patients. 16920564_Observational study of gene-disease association. (HuGE Navigator) 16929515_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16929515_TS tandem repeat polymorphism was significantly related to survival. 17000685_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17018589_5FU sensitivity of a tumor is modified by aneuploidy-producing copy-number changes of TS alleles by one or more of the following: LOH, amplification, and copy-number changes due to polysomy. 17065089_analysis of Ki67 and thymidylate synthase expression in primary tumour compared with metastatic nodes in breast cancer patients 17181924_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17187508_Observational study of genotype prevalence. (HuGE Navigator) 17201138_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17203168_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17203168_thymidylate synthase and dihydropyrimidine dehydrogenase gene polymorphisms have a role in response and toxicity to capecitabine-raltitrexed treatment in colorectal cancer 17220568_Findings provide fundamental and useful information for genotyping TYMS in the Japanese and probably other Asian populations. 17220568_Observational study of genotype prevalence. (HuGE Navigator) 17273745_Observational study of gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17273745_genetic polymorphisms of TS as a prognostic factor in advanced colorectal cancer patients treated with 5-fluorouracil plus oxaliplatin or irinotecan. 17290389_Observational study of gene-disease association. (HuGE Navigator) 17311259_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17317154_a significant association between the presence of the 3G allele and TYMS mRNA expression and catalytic activity, particularly in p53-mutated cell lines 17367411_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17369151_Observational study of genetic testing. (HuGE Navigator) 17372271_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17372271_increased risk of breast cancer associated with the double repeat (2R) allele in the thymidylate synthase (TYMS) 5'-untranslated region (UTR) (P, trend = 0.07) 17377791_Gene expression may be a response to fluorouracil sensitivity in stomach cancers of different histologic origin. 17385677_thymidylate synthetase 1494del6 polymorphism is associated with a significantly lower risk of breast cancer and may be a potential genetic marker 17395259_No differences of these polymorphisms were found between ALL cells and normal blood cells, indicating an ethnic rather than leukemic origin. 17395259_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17410198_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17417073_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17417073_findings suggest that there may be an important relationship between the TYMS haplotypes examined and 5-fluorouracil toxicity 17439323_Observational study of gene-disease association. (HuGE Navigator) 17449906_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17454638_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17512053_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17546637_Observational study of gene-disease association. (HuGE Navigator) 17549369_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17551301_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17581305_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17596206_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17655928_Observational study of gene-disease association. (HuGE Navigator) 17655928_similar frequencies of the MTHFR, the MTRR and the TYMS genotypes were seen in patients and controls. 17659576_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17659576_This study supports the hypothesis that reduced MTHFR activity and enhanced TYMS activity, both of which are essential elements in minimizing uracil misincorporation into DNA, may protect against the development of hepatocellular carcinoma. 17661145_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17661145_Patients with thymidylate synthase genotype 2/2 had a significantly higher rate of complete pathologic response with 53 percent (8/15) compared with 22 percent in the 2/3 or 3/3 group. 17684410_study found that functional polymorphisms in methylenetetrahydrofolate reductase and thymidylate synthase were significantly associated with increased risk of esophageal squamous cell carcinoma, gastric cardia carcinoma, and pancreatic carcinoma 17684476_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17688376_Ts mRNA is induced by preoperative chemoradiotherapy in both responders and nonresponders to combined modality treatment in rectal carcinoma. 17716232_single nucleotide polymorphism of thymidylate synthase in colorectal cancer (Review) 17854149_Expression of thymidylate synthase and thymidine phosphorylase mRNA is a useful predictive parameter for the survival of postoperative gastric cancer patients after 5-fluorouracil-based adjuvant chemotherapy. 17891500_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17971770_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18095031_Observational study of gene-disease association. (HuGE Navigator) 18098291_Common variation in MTHFR and TYMS genes may be associated with RCC risk, particularly when vegetable intake is low. 18174236_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18177869_Examined association between MTHFR/thymidylate synthase gene polymorphisms and methylation of p16(INK4A) and hMLH1 genes in spontaneously aborted embryos with normal chromosomal integrity. 18177869_Observational study of gene-disease association. (HuGE Navigator) 18192902_Observational study of gene-disease association. (HuGE Navigator) 18203297_Observational study of gene-disease association. (HuGE Navigator) 18203297_There is a relationship between TS genotype and TS protein expression in clinical specimens of colon adenocarcinoma 18221821_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18245544_Genetic variation is not useful for predicting toxicity from capecitabine in patients with advanced colorectal cancer. 18245544_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18267032_Observational study of gene-disease association. (HuGE Navigator) 18274813_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18274813_findings do not suggest a significant association of MTHFR/TS allele/genotype with methotrexate response in our ethnically distinct Indian (Asian) rheumatoid arthritis patient 18281538_This study reports the differential TS expression in the spectrum of neuroendocrine tumors and indicates TS as a possible marker of treatment efficacy in well-differentiated gastroenteropancreatic neuroendocrine carcinomas patients treated with 5-FU. 18285546_Observational study of gene-disease association. (HuGE Navigator) 18299612_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18322994_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18336674_Observational study of gene-disease association. (HuGE Navigator) 18365142_increased expression of the TYMS oncogene may be of importance for the development of human intracranial meningiomas 18370400_at least one-third of hTS populates the inactive conformer 18381794_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18406541_Observational study of gene-disease association. (HuGE Navigator) 18427977_Observational study of genetic testing. (HuGE Navigator) 18430179_demonstrates increased survival for neoadjuvant treated esophageal adenocarcinoma with TS, ERCC1, or GSTP-1 mRNA level at or below the median 18448328_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18458991_Observational study of gene-disease association. (HuGE Navigator) 18458991_Thymidylate synthase genotypes were not associated with thrombosis and serum homocysteine concentration in Behcets patients; mean serum homocysteine level in patients with thrombosis was significantly higher than in patients without thrombosis 18498133_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18505590_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18510611_Observational study of gene-disease association. (HuGE Navigator) 18540691_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18584349_Microsatellite instability and the association with plasma homocysteine and thymidylate synthase was determined in colorectal cancer. 18584349_Observational study of gene-disease association. (HuGE Navigator) 18589584_Observational study of gene-disease association. (HuGE Navigator) 18589584_The genotype 6bp-/6bp- in the region untranslated 3' of TS gene, in the analyzed sample participates in important way in the development of CRC of the Mexican population. 18593893_The transcriptional status of four key genes, thymidylate synthase (TYMS), MORF-related gene X (MRGX), Bcl2-antagonist/killer (BAK), and ATPase, Cu(2+) transporting beta polypeptide (ATP7B), can accurately predict response to 5-FU. 18597678_prognostic factor for survival in colorectal carcinoma with radical resection and 5-FU chemotherapy; postoperative survival was significantly better in patients with low TS activity 18600534_functional inactivity and mutations of p53 differentially affect TS, potentially influencing response to TS inhibitor-based treatment 18607581_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18629514_findings indicated that the TS gene polymorphism holds important prognostic information for stage II colon cancer; 5 year survival rate was significantly higher in patients with TS homozygocity than in heterozygous patients 18635682_Observational study of gene-disease association. (HuGE Navigator) 18636124_Observational study of gene-disease association. (HuGE Navigator) 18642643_No association was detected between the Single Nucleotide Polymorphism in the Thymidylate synthase and susceptibility to hemifacial spasm. 18642643_Observational study of gene-disease association. (HuGE Navigator) 18669903_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18669903_no association between either green tea intake or gene polymorphisms of MTHFR (C677T and A1298C) and TYMS (1494 ins/del) and breast cancer risk. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18676755_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18677108_Analysis of in vivo C-MYC interactions with TS, IMPDH2 and PRPS2 genes confirmed that they are direct C-MYC targets. 18704422_The polymorphisms of TS 3'-UTR ins6/del6 are associated with gastric cancer patients treated with fluorouracil-based adjuvant chemotherapy. 18722050_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18728661_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18785313_Observational study of gene-disease association. (HuGE Navigator) 18830263_Observational study of gene-disease association. (HuGE Navigator) 18855534_Copy-number analysis of TYMS genes in frozen and FFPE DNAs of colorectal cancers is reported. 18976010_The relationship of nuclear to cytoplasmic thymidylate synthase expression, given as a ratio of continuous AQUA scores, is a strong predictor of colon cancer survival. 18983896_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18985814_Bcl-2 expression significantly correlates with thymidylate synthase expression in colorectal cancer patients. 18988749_Observational study of genetic testing. (HuGE Navigator) 18989779_Meta-analysis of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18989779_polymorphisms in the 5'UTR and 3'UTR of thymidylate synthase may be associated with gastric cancer susceptibility 18990369_Mutations of MTHFR and TSER are not likely significant risk factors of idiopathic recurrent spontaneous abortion in Korean women. 18990369_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18992148_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19020309_MTHFR and TS gene variants are associated with decreased acute lymphoblastic leukemia risk 19020309_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19020759_Measurement of TS and ERCC1 mRNA levels in primary colorectal cancer can predict those in synchronous liver metastases, but not in metachronous ones. 19020767_head & neck, gastric, colorectal, breast, lung & pancreatic cancer were examined; findings show mRNA expression & protein level of thymidylate synthase, dihydropyrimidine dehydrogenase & orotate phosphoribosyltransferase differed according to cancer type 19048631_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19064578_Observational study of gene-disease association. (HuGE Navigator) 19074750_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19082493_Combination of polymorphisms within 5' and 3' untranslated regions of thymidylate synthase gene modulates response to fluorouracil in colorectal cancer. 19082493_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19147506_No correlation was found between thymidylate synthase protein expression and gene polymorphism in patients with colorectal carcinoma 19147506_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19154585_Microsatellite instability was correlated with TS gene expression but not significantly with DPD. 19159907_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19162321_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19190136_Observational study of gene-disease association. (HuGE Navigator) 19190136_rs3819102, an htSNP located in the 3'-flanking region of the TYMS gene and an intron of the ENOSF1 gene, was found to be associated with an increased risk of endometrial cancer. 19295264_recurrence-free survival time curve showed significant differences when stratified by tumor diameter, ER expression, and TS levels, but not by menopausal status, nodal status, surgical method, p53 expression, DPD activity, or HER2 expression 19306093_Allelic im | ENSMUSG00000025747 | Tyms | 4.535124e+03 | 0.9088274 | -0.137921835 | 0.2682859 | 2.713606e-01 | 0.6024205544 | 0.90922991 | No | Yes | 4.155446e+03 | 403.116573 | 4.301796e+03 | 427.902515 |
| ENSG00000176945 | 200958 | MUC20 | protein_coding | Q8N307 | FUNCTION: May regulate MET signaling cascade. Seems to decrease hepatocyte growth factor (HGF)-induced transient MAPK activation. Blocks GRB2 recruitment to MET thus suppressing the GRB2-RAS pathway. Inhibits HGF-induced proliferation of MMP1 and MMP9 expression. {ECO:0000269|PubMed:15314156}. | Alternative splicing;Cell membrane;Cell projection;Glycoprotein;Membrane;Reference proteome;Repeat;Secreted;Signal | This gene encodes a member of the mucin protein family. Mucins are high molecular weight glycoproteins secreted by many epithelial tissues to form an insoluble mucous barrier. The C-terminus of this family member associates with the multifunctional docking site of the MET proto-oncogene and suppresses activation of some downstream MET signaling cascades. The protein features a mucin tandem repeat domain that varies between two and six copies in most individuals. Multiple variants encoding different isoforms have been found for this gene. A related pseudogene, which is also located on chromosome 3, has been identified. [provided by RefSeq, Apr 2014]. | hsa:200958; | apical plasma membrane [GO:0016324]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; extracellular region [GO:0005576]; Golgi lumen [GO:0005796]; microvillus membrane [GO:0031528]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; hepatocyte growth factor receptor signaling pathway [GO:0048012] | 15314156_Results suggest that MUC20 is a novel regulator of the Met signaling cascade which has a role in suppression of the Grb2-Ras pathway. 16029633_Observational study of gene-disease association. (HuGE Navigator) 16508246_Observational study of gene-disease association. (HuGE Navigator) 18834073_Reflux laryngitis is associated with down-regulation of mucin gene expression. 23787019_MUC20 is expressed in recurrent colorectal neoplasms and may have a role in poor outcome 25168902_MUC20 is a novel transmembrane mucin expressed by the human corneal and conjunctival epithelia, and suggest that differential expression of MUC20 during differentiation has a role in maintaining ocular surface homeostasis. 26323930_A combination of MUC13/MUC20 expression was a potential prognostic marker for patients with ESCC, who received neoadjuvant chemotherapy followed by surgery. 26616226_Results suggest that MUC20 enhances aggressive behaviors of EOC cells by activating integrin beta1 signaling and provide novel insights into the role of MUC20 in ovarian cancer metastasis. 26673820_High MUC20 expression is associated wit response to chemotherapy in esophageal squamous cell carcinoma. 26770020_increased gene expression of MUC16 and MUC20 was found in patients with remission ulcerative colitis 29993037_MUC20 knockdown suppresses the malignant phenotypes of PDAC cells at least partially through the inhibition of the HGF/MET pathway. 30236127_the combination of MUC4, MUC16 and MUC20 signature is associated with statistically significant reduced overall survival and increased hazard ratio in pancreatic, colon and stomach cancer. 31519421_MUC20 expression marks the receptive phase of the human endometrium. 31898364_Lnc-MUC20-9 binds to ROCK1 and functions as a tumor suppressor in bladder cancer. 33620575_Identification of oral squamous cell carcinoma markers MUC2 and SPRR1B downstream of TANGO. 34651428_Mucin 20 modulates proteasome capacity through c-Met signalling to increase carfilzomib sensitivity in mantle cell lymphoma. 35065708_A flexible summary statistics-based colocalization method with application to the mucin cystic fibrosis lung disease modifier locus. 35166958_Intracellular MUC20 variant 2 maintains mitochondrial calcium homeostasis and enhances drug resistance in gastric cancer. | ENSMUSG00000035638 | Muc20 | 1.783117e+01 | 1.0559778 | 0.078579483 | 0.7061497 | 1.215012e-02 | 0.9122288810 | 0.98338795 | No | Yes | 1.047431e+01 | 5.307104 | 1.070744e+01 | 5.399521 | |
| ENSG00000177427 | 125170 | MIEF2 | protein_coding | Q96C03 | FUNCTION: Mitochondrial outer membrane protein involved in the regulation of mitochondrial organization (PubMed:29361167). It is required for mitochondrial fission and promotes the recruitment and association of the fission mediator dynamin-related protein 1 (DNM1L) to the mitochondrial surface independently of the mitochondrial fission FIS1 and MFF proteins. Regulates DNM1L GTPase activity. {ECO:0000269|PubMed:21508961, ECO:0000269|PubMed:23283981, ECO:0000269|PubMed:23530241, ECO:0000269|PubMed:23921378, ECO:0000269|PubMed:29361167, ECO:0000269|PubMed:29899447}. | 3D-structure;Alternative splicing;Disease variant;Membrane;Mitochondrion;Mitochondrion outer membrane;Phosphoprotein;Primary mitochondrial disease;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes an outer mitochondrial membrane protein that functions in the regulation of mitochondrial morphology. It can directly recruit the fission mediator dynamin-related protein 1 (Drp1) to the mitochondrial surface. The gene is located within the Smith-Magenis syndrome region on chromosome 17. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jun 2011]. | hsa:125170; | integral component of membrane [GO:0016021]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; dynamin family protein polymerization involved in mitochondrial fission [GO:0003374]; mitochondrion organization [GO:0007005]; positive regulation of mitochondrial fission [GO:0090141]; positive regulation of protein targeting to membrane [GO:0090314]; regulation of mitochondrion organization [GO:0010821] | 15517828_Smith-Magenis syndrome is caused by a de novo deletion on chromosome 17. 21508961_MiD49/51 are new mediators of mitochondrial division affecting Drp1 action at mitochondria. 21508961_Mitochondrial outer membrane protein that recruits fission mediator Drp1 to the mitochondrial surface. 23283981_we find that either MiD49 or MiD51 can mediate Drp1 recruitment and mitochondrial fission in the absence of Fis1 and Mff. 23880462_MIEF1 and MIEF2 are differentially expressed in human tissues during development 23921378_MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and suggest that they provide specificity to the division of mitochondria. 26564796_These findings and data showing MARCH5-dependent degradation of MiD49 upon stress support the possibility that MARCH5 regulation of MiD49 is a novel mechanism controlling mitochondrial fission and, consequently, the cellular response to stress. 26903540_The results indicate that Drp1-dependent mitochondrial fission through MiD49/MiD51 regulates cristae remodeling during intrinsic apoptosis. 27660309_MiD49 and MiD51 recruit inactive forms of Drp1 in mitochondrial fission. [review] 28137654_Knockdown of MIEF2 reduces DOX-induced mitochondrial fission and apoptosis in cardiomyocytes and in vivo. Also, knockdown of MIEF2 protects heart from DOX-induced cardiotoxicity. Our study identifies a novel pathway composed of Foxo3a and MIEF2 that mediates DOX cardiotoxicity. 29431643_In health, MiD49 regulate Drp1-mediated fission, whereas in disease, epigenetic upregulation of MiD49 increases mitotic fission, which drives pathological proliferation and apoptosis resistance 29899447_cryo-electron microscopy structure of full-length human DRP1 co-assembled with MID49 and an analysis of structure- and disease-based mutations 32068312_An epigenetic increase in mitochondrial fission by MiD49 and MiD51 regulates the cell cycle in cancer: Diagnostic and therapeutic implications. 32236517_Mitochondrial fragmentation enables localized signaling required for cell repair. 33317572_MIEF2 over-expression promotes tumor growth and metastasis through reprogramming of glucose metabolism in ovarian cancer. 33414447_MIEF2 reprograms lipid metabolism to drive progression of ovarian cancer through ROS/AKT/mTOR signaling pathway. | ENSMUSG00000018599 | Mief2 | 1.147882e+03 | 1.0220907 | 0.031523185 | 0.3063595 | 1.038000e-02 | 0.9188501154 | 0.98472851 | No | Yes | 1.010588e+03 | 150.062411 | 9.663807e+02 | 147.317686 | |
| ENSG00000177728 | 9772 | TMEM94 | protein_coding | Q12767 | Alternative splicing;Glycoprotein;Membrane;Mental retardation;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:9772; | integral component of membrane [GO:0016021] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 32825426_Fetal Anomalies Associated with Novel Pathogenic Variants in TMEM94. | ENSMUSG00000020747 | Tmem94 | 4.054897e+03 | 1.1676848 | 0.223650873 | 0.2771097 | 6.478352e-01 | 0.4208877146 | 0.84954815 | No | Yes | 3.526586e+03 | 503.497163 | 3.167170e+03 | 463.980968 | |||
| ENSG00000178188 | 25970 | SH2B1 | protein_coding | Q9NRF2 | FUNCTION: Adapter protein for several members of the tyrosine kinase receptor family. Involved in multiple signaling pathways mediated by Janus kinase (JAK) and receptor tyrosine kinases, including the receptors of insulin (INS), insulin-like growth factor I (IGF1), nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), glial cell line-derived neurotrophic factor (GDNF), platelet-derived growth factor (PDGF) and fibroblast growth factors (FGFs). In growth hormone (GH) signaling, autophosphorylated ('Tyr-813') JAK2 recruits SH2B1, which in turn is phosphorylated by JAK2 on tyrosine residues. These phosphotyrosines form potential binding sites for other signaling proteins. GH also promotes serine/threonine phosphorylation of SH2B1 and these phosphorylated residues may serve to recruit other proteins to the GHR-JAK2-SH2B1 complexes, such as RAC1. In leptin (LEP) signaling, binds to and potentiates the activation of JAK2 by globally enhancing downstream pathways. In response to leptin, binds simultaneously to both, JAK2 and IRS1 or IRS2, thus mediating formation of a complex of JAK2, SH2B1 and IRS1 or IRS2. Mediates tyrosine phosphorylation of IRS1 and IRS2, resulting in activation of the PI 3-kinase pathway. Acts as positive regulator of NGF-mediated activation of the Akt/Forkhead pathway; prolongs NGF-induced phosphorylation of AKT1 on 'Ser-473' and AKT1 enzymatic activity. Enhances the kinase activity of the cytokine receptor-associated tyrosine kinase JAK2 and of other receptor tyrosine kinases, such as FGFR3 and NTRK1. For JAK2, the mechanism seems to involve dimerization of both, SH2B1 and JAK2. Enhances RET phosphorylation and kinase activity. Isoforms seem to be differentially involved in IGF-I and PDGF-induced mitogenesis (By similarity). {ECO:0000250, ECO:0000269|PubMed:11827956, ECO:0000269|PubMed:14565960, ECO:0000269|PubMed:15767667, ECO:0000269|PubMed:16569669, ECO:0000269|PubMed:17471236, ECO:0000269|PubMed:9694882, ECO:0000269|PubMed:9742218}. | 3D-structure;Alternative splicing;Cytoplasm;Membrane;Methylation;Nucleus;Phosphoprotein;Reference proteome;SH2 domain | This gene encodes a member of the SH2-domain containing mediators family. The encoded protein mediates activation of various kinases and may function in cytokine and growth factor receptor signaling and cellular transformation. Alternatively spliced transcript variants have been described. [provided by RefSeq, Mar 2009]. | hsa:25970; | cytosol [GO:0005829]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; transmembrane receptor protein tyrosine kinase adaptor activity [GO:0005068]; intracellular signal transduction [GO:0035556]; positive regulation of SMAD protein signal transduction [GO:0060391] | 15767667_SH2-B or APS homodimerization and SH2-B/APS heterodimerization thus provide direct mechanisms for activating and inhibiting Janus kinase 2 17228025_Observational study of gene-disease association. (HuGE Navigator) 17471236_overexpression of SH2B1beta, by enhancing phosphorylation/activation of RET transducers, potentiates the cellular differentiation and the neoplastic transformation thereby induced, and counteracts the action of RET inhibitors. 19079260_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19079261_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 19164386_Observational study of gene-disease association. (HuGE Navigator) 19342444_SH2B1beta functions as an adapter protein that cross-links actin filaments, leading to modulation of cellular responses in response to JAK2 activation. 19692490_Observational study of gene-disease association. (HuGE Navigator) 19746409_Observational study of gene-disease association. (HuGE Navigator) 19851340_Observational study of gene-disease association. (HuGE Navigator) 19910938_Observational study of gene-disease association. (HuGE Navigator) 20215397_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20386550_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20571754_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20616199_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20712903_Observational study of gene-disease association. (HuGE Navigator) 20724581_Observational study of gene-disease association. (HuGE Navigator) 20725061_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20808231_Deletions of the 16p11.2 SH2B1-containing region were identified in 31 patients with developmental delay and obesity. 20816152_Observational study of gene-disease association. (HuGE Navigator) 20816195_Observational study of gene-disease association. (HuGE Navigator) 21045733_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 21566085_Adapter protein SH2B1beta binds filamin A to regulate prolactin-dependent cytoskeletal reorganization and cell motility 21750520_Meta-analysis of 4992 subjects revealed seven SNPs near four loci, including NEGR1, TMEM18, SH2B1 /ATP2A1 and MC4R, showing significant association at 0.005 21796141_Our results suggest that there is a visceral fat area (VFA)-specific genetic factor and that a polymorphism in the SH2B1 gene influences the risk of visceral fat accumulation. 21907990_Variability at the SH2B1 obesity locus is associated with myocardial infarction in type 2 diabetic patients and with reduced insulin-stimulated nitric oxide synthase activity in human endothelial cells. 22248999_With the current study we were able to replicate and confirm that the SH2B1 gene locus is significantly associated with complex obesity in a Caucasian population. 22443470_Our study supports earlier reports of SH2B1 to be of importance in insulin sensitivity and, in addition, suggests potential roles of NEGR1 and MTCH2. 22901222_High SH2B1 expression was associated with lymph node metastasis, and recurrence in non-small cell lung cancer. 22942098_The known, obesity related, sh2b1 gene single nucleotide polymorphisms rs4788102 and rs7498665 are associated with plasma triglyceride levels. 23054017_Demonstration of the additive effect of four polymorphisms on the LRP5, LEPR, near MC4R and SH2B1 genes on metabolic syndrome risk. 23121087_Data show the synthetic effect of SNPs on the indices of adiposity and risk of obesity in Chinese girls, but failed to replicate the effect of five separate variants of SEC16B rs10913469, SH2B1 rs4788102, PCSK1 rs6235, KCTD15 rs29941 and BAT2 rs2844479. 23160192_SH2B1 plays a critical role in the control of human food intake and body weight and is implicated in maladaptive human behavior. 23190452_Studies indicate that insulin receptor (IR) and IGF Type 1 Receptor (IGFR) have been identified as important partners of Grb10/14 and SH2B1/B2 adaptors. 23270367_The rare coding mutation betaThr656Ile/gammaPro674Ser (g.9483C/T) in SH2B1 was exclusively detected in overweight or obese individuals. 23519644_The obesity risk alleles of non-synonymous SNPs at SH2B1 and APOB48R have no strong effect on weight loss-related phenotypes in overweight children after a 1-year lifestyle intervention. 23640704_Common variants near BDNF and SH2B1 show nominal evidence of association with snacking behavior in European populations. 23825611_rare missense mutations of FTO and SH2B1 did not confer risks of obesity in Chinese Han children in our cohort 24103803_Data (from in excess of six genetic association studies) suggest that an SNP in SH2B1 (rs4788102) is not associated with abnormal glucose homeostasis in obese subjects of European ancestry. [META-ANALYSIS] 24621099_This study highlighted the importance of two candidate genes, SH2B1 and FAIM2, in the risk of overweight/obesity. 24736401_SH2B1 can enhance neurite outgrowth and accelerate the maturation of human induced neurons under defined conditions. 24971614_4 novel variants in SH2B1 were identified in individuals with obesity and insulin resistance. 25234362_We report evidence that the 16p11.2 deletion may influence specific obesity-associated disinhibited eating behaviors 25471250_The rs7359397 (SH2B1) was associated with the body weight, body mass index, and truncal fat mass reduction. 26031769_Mutation analysis has demonstrated that variation in the SH2B1 gene is frequent in both lean and obese groups, with distinctive variations being present on either side of the weight spectrum. 26077624_that SH2B1 is a key positive mediator of pathological cardiac hypertrophy 27164951_Results identified SH2B as a functional target of miR-361 which down-regulation suppresses lung cancer progression and metastasis through regulation of SH2B1. 27530450_SH2B1 polymorphisms are associated with HbA1c, largely independent of BMI, in European American young adults. 27802221_This study is the first to show that neuronal SH2B1, a key protein in insulin signaling, may have a role in the accumulation of Abeta42 in an animal model of Alzheimer's Disease. 28039048_SH2B1 fine-tunes global-local chromatin states. 28544142_Chromosome microarray analysis was performed on both twins and their parents. An identical 244 kb microdeletion on 16p11.2 including 9 Refseq genes, including SH2B1, was identified in the twins. 28694205_SH2B1 and RABEP1 genetic variants are associated with worsening of LDL and glucose parameters in patients treated with psychotropic drugs 29127727_The high-resolution structure of the SH2 domain of SH2B1 further reveals conformationally plastic protein loops that may contribute to the ability of the protein to recognize dissimilar ligands. Together, numerous hydrophobic and electrostatic interactions, in addition to backbone conformational flexibility, permit the recognition of diverse peptides by SH2B1. 29380446_Data reported that SH2B1 expression levels were significantly upregulated and positively associated with EMT markers and poor patient survival in lung adenocarcinoma (LADC) specimens. Further results reveal that SH2B1 has a major role in LADC progression and that SH2B1 is a critical activator of Wnt/beta-catenin signaling. 29631267_The increased expression of genes in leptin (JAK/STAT or AKT) signaling implies that the main mode of action for human SH2B1 mutations might affect leptin signaling rather than insulin signaling. 30370521_Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. 30518945_Copy number variations in SH2B1 gene is associated with distal syndromes with intellectual disability. 31439647_These studies demonstrate that the PH domain plays a crucial role in how SH2B1 controls energy balance and glucose homeostasis. 31739166_provide a mini overview of the roles of SH2B1 in cancer. 32251290_Mice with LepR neuron-specific or adult-onset, hypothalamus-specific ablation of Sh2b1 develop obesity, insulin resistance, and liver steatosis. Hypothalamic overexpression of human SH2B1 protects against high fat diet-induced obesity and metabolic syndromes. Results unravel an unrecognized LepR neuron Sh2b1/ sympathetic nervous system/brown adipose tissue /thermogenesis axis that combats obesity and metabolic disease. 32365683_Association of the SH2B1 rs7359397 Gene Polymorphism with Steatosis Severity in Subjects with Obesity and Non-Alcoholic Fatty Liver Disease. 33356989_Hsa_circ_0075960 Serves as a Sponge for miR-361-3p/SH2B1 in Endometrial Carcinoma. | ENSMUSG00000030733 | Sh2b1 | 3.693255e+03 | 1.0028701 | 0.004134705 | 0.2656663 | 2.439735e-04 | 0.9875378282 | 0.99716006 | No | Yes | 2.930564e+03 | 292.039587 | 3.070414e+03 | 313.946295 | |
| ENSG00000178234 | 63917 | GALNT11 | protein_coding | Q8NCW6 | FUNCTION: Polypeptide N-acetylgalactosaminyltransferase that catalyzes the initiation of protein O-linked glycosylation and is involved in left/right asymmetry by mediating O-glycosylation of NOTCH1. O-glycosylation of NOTCH1 promotes activation of NOTCH1, modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO). Polypeptide N-acetylgalactosaminyltransferases catalyze the transfer of an N-acetyl-D-galactosamine residue to a serine or threonine residue on the protein receptor. Displays the same enzyme activity toward MUC1, MUC4, and EA2 than GALNT1. Not involved in glycosylation of erythropoietin (EPO). {ECO:0000269|PubMed:11925450, ECO:0000269|PubMed:24226769}. | Alternative splicing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Heterotaxy;Lectin;Manganese;Membrane;Metal-binding;Notch signaling pathway;Phosphoprotein;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | hsa:63917; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; carbohydrate binding [GO:0030246]; metal ion binding [GO:0046872]; Notch binding [GO:0005112]; polypeptide N-acetylgalactosaminyltransferase activity [GO:0004653]; cilium assembly [GO:0060271]; determination of left/right symmetry [GO:0007368]; Notch receptor processing [GO:0007220]; Notch signaling involved in heart development [GO:0061314]; O-glycan processing [GO:0016266]; protein O-linked glycosylation via threonine [GO:0018243]; regulation of Notch signaling pathway [GO:0008593] | 20422447_GALNT11, the human ortholog to Drosophila melanogaster pgant35A, was shown not to support rescue of the l(2)35Aa lethality during Drosophila embryogenesis. 20547088_Observational study of genotype prevalence. (HuGE Navigator) 20547088_Single nucleiotide polymorphism in the GALNT11 genes were investigated to assess allele frequencies in different populations. This SNP would be useful marker for forensic individualization, in particular, as ancestry-informative markers. 24076351_evidence suggests that CLL patient samples harbor aberrant O-glycosylation highlighted by Tn antigen expression and that the over-expression of GALNT11 constitutes a new molecular marker for CLL 25493955_possible role in the deterioration of kidney function [meta-analysis] | ENSMUSG00000038072 | Galnt11 | 1.148259e+03 | 0.7464930 | -0.421799385 | 0.2833631 | 2.219002e+00 | 0.1363211242 | 0.76593507 | No | Yes | 9.136449e+02 | 121.251562 | 1.279385e+03 | 173.615755 | |
| ENSG00000178538 | 767 | CA8 | protein_coding | P35219 | FUNCTION: Does not have a carbonic anhydrase catalytic activity. | 3D-structure;Disease variant;Mental retardation;Metal-binding;Phosphoprotein;Reference proteome;Zinc | The protein encoded by this gene was initially named CA-related protein because of sequence similarity to other known carbonic anhydrase genes. However, the gene product lacks carbonic anhydrase activity (i.e., the reversible hydration of carbon dioxide). The gene product continues to carry a carbonic anhydrase designation based on clear sequence identity to other members of the carbonic anhydrase gene family. The absence of CA8 gene transcription in the cerebellum of the lurcher mutant in mice with a neurologic defect suggests an important role for this acatalytic form. Mutations in this gene are associated with cerebellar ataxia, mental retardation, and dysequilibrium syndrome 3 (CMARQ3). Polymorphisms in this gene are associated with osteoporosis, and overexpression of this gene in osteosarcoma cells suggests an oncogenic role. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2016]. | hsa:767; | cytoplasm [GO:0005737]; carbonate dehydratase activity [GO:0004089]; hydro-lyase activity [GO:0016836]; zinc ion binding [GO:0008270]; one-carbon metabolic process [GO:0006730]; phosphatidylinositol-mediated signaling [GO:0048015] | 17219437_Overexpression of Carbonic anhydrase-related protein VIII promotes colon cancer cell growth 19172221_Observational study of gene-disease association. (HuGE Navigator) 19172221_The results suggest that the variations of CA8 and CA10 loci may be important determinants of osteoporosis in Japanese women. 19360879_Crystal structure of human carbonic anhydrase-related protein VIII reveals the basis for catalytic silencing 19461874_Consanguineous Iraqi family in which affected siblings had mild mental retardation and congenital ataxia characterized by quadrupedal gait. The mutation S100P is associated with proteasome-mediated degradation, and presumably represents a null mutation. 20819067_review article describes the previous data on CARP VIII, including its structure, role in neurodegeneration and cancer; and bioinformatic and expression analyses. 21812104_This report expands the neurological and radiological phenotype associated with CA8 mutations. 24476000_Overexpression of CA8 in MERRF cybrids significantly decreases cell death. 24794067_CA8 overexpression desensitizes neuronal cells to STS induced apoptotic stress and increases cell migration and invasion ability in neuronal cells. 26711783_we observed increased expression of CA8 in more aggressive types of human osteosarcoma (OS) cells and found that CA8 expression is correlated with disease stages, such that more intense expression occurs in the disease late stage 29407793_results suggest that Sp1 transactivates hCA8 gene through the proximal GC box element in the promoter region 29792187_The CARP VIII and XI proteins were associated to non-hypoxic conditions and CARP XI also to the expression of cytoplasmic CA II staining suggesting that the CARPs play a role in tumorigenesis of diffusively infiltrating gliomas. 31199789_These genomic studies significantly advance the literature regarding structure-function studies on CA8-ITPR1 critical to calcium signaling pathways, synaptic functioning, neuronal excitability and analgesic responses. 31693170_Cerebellar ataxia with normal intellect associated with a homozygous truncating variant in CA8. 31715371_CA8 promotes renal cell carcinoma proliferation and migration though its expression level is lower in tumor compared to adjacent normal tissue 32583043_Expression and Functional Study of Single Mutations of Carbonic Anhydrase 8 in Neuronal Cells. 33247772_Reversion mutation of cDNA CA8-204 minigene construct produces a truncated functional peptide that regulates calcium release in vitro and produces profound analgesia in vivo. | ENSMUSG00000041261 | Car8 | 5.670632e+02 | 0.7787226 | -0.360818543 | 0.2876352 | 1.597047e+00 | 0.2063222458 | 0.78763590 | No | Yes | 5.684995e+02 | 80.260804 | 6.332466e+02 | 91.559019 | |
| ENSG00000178921 | 5198 | PFAS | protein_coding | O15067 | FUNCTION: Phosphoribosylformylglycinamidine synthase involved in the purines biosynthetic pathway. Catalyzes the ATP-dependent conversion of formylglycinamide ribonucleotide (FGAR) and glutamine to yield formylglycinamidine ribonucleotide (FGAM) and glutamate. {ECO:0000305|PubMed:10548741}. | ATP-binding;Cytoplasm;Glutamine amidotransferase;Ligase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Purine biosynthesis;Reference proteome | PATHWAY: Purine metabolism; IMP biosynthesis via de novo pathway; 5-amino-1-(5-phospho-D-ribosyl)imidazole from N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide: step 1/2. {ECO:0000305|PubMed:10548741}. | Purines are necessary for many cellular processes, including DNA replication, transcription, and energy metabolism. Ten enzymatic steps are required to synthesize inosine monophosphate (IMP) in the de novo pathway of purine biosynthesis. The enzyme encoded by this gene catalyzes the fourth step of IMP biosynthesis. [provided by RefSeq, Jul 2008]. | hsa:5198; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; phosphoribosylformylglycinamidine synthase activity [GO:0004642]; 'de novo' AMP biosynthetic process [GO:0044208]; 'de novo' IMP biosynthetic process [GO:0006189]; 'de novo' XMP biosynthetic process [GO:0097294]; anterior head development [GO:0097065]; glutamine metabolic process [GO:0006541]; GMP biosynthetic process [GO:0006177]; purine ribonucleoside monophosphate biosynthetic process [GO:0009168]; response to xenobiotic stimulus [GO:0009410] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 29337348_Study observed FGAMS as distinct puncta throughout neuronal processes, where it co-localized with neurofilament heavy chain, tubulin, and mitochondria. Data provide insight into potential purine biosynthetic mechanisms utilized within neurons during homeostasis as well as viral infection. 30987822_A total of 429 proteins are newly identified to interact with PFAS. PFAS interacts with CAD, CCT2, PRDX1, and PHGDH. 32485148_ERK2 Phosphorylates PFAS to Mediate Posttranslational Control of De Novo Purine Synthesis. | ENSMUSG00000020899 | Pfas | 9.204673e+03 | 1.1985742 | 0.261319196 | 0.2909548 | 8.051908e-01 | 0.3695459114 | 0.83664500 | No | Yes | 9.073371e+03 | 826.781087 | 7.280891e+03 | 680.777837 |
| ENSG00000179094 | 5187 | PER1 | protein_coding | O15534 | FUNCTION: Transcriptional repressor which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. Regulates circadian target genes expression at post-transcriptional levels, but may not be required for the repression at transcriptional level. Controls PER2 protein decay. Represses CRY2 preventing its repression on CLOCK/ARNTL target genes such as FXYD5 and SCNN1A in kidney and PPARA in liver. Besides its involvement in the maintenance of the circadian clock, has an important function in the regulation of several processes. Participates in the repression of glucocorticoid receptor NR3C1/GR-induced transcriptional activity by reducing the association of NR3C1/GR to glucocorticoid response elements (GREs) by ARNTL:CLOCK. Plays a role in the modulation of the neuroinflammatory state via the regulation of inflammatory mediators release, such as CCL2 and IL6. In spinal astrocytes, negatively regulates the MAPK14/p38 and MAPK8/JNK MAPK cascades as well as the subsequent activation of NFkappaB. Coordinately regulates the expression of multiple genes that are involved in the regulation of renal sodium reabsorption. Can act as gene expression activator in a gene and tissue specific manner, in kidney enhances WNK1 and SLC12A3 expression in collaboration with CLOCK. Modulates hair follicle cycling. Represses the CLOCK-ARNTL/BMAL1 induced transcription of BHLHE40/DEC1. {ECO:0000269|PubMed:24005054}. | Alternative splicing;Biological rhythms;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | This gene is a member of the Period family of genes and is expressed in a circadian pattern in the suprachiasmatic nucleus, the primary circadian pacemaker in the mammalian brain. Genes in this family encode components of the circadian rhythms of locomotor activity, metabolism, and behavior. This gene is upregulated by CLOCK/ARNTL heterodimers but then represses this upregulation in a feedback loop using PER/CRY heterodimers to interact with CLOCK/ARNTL. Polymorphisms in this gene may increase the risk of getting certain cancers. Alternative splicing has been observed in this gene; however, these variants have not been fully described. [provided by RefSeq, Jan 2014]. | hsa:5187; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin DNA binding [GO:0031490]; DNA-binding transcription factor binding [GO:0140297]; E-box binding [GO:0070888]; kinase binding [GO:0019900]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; transcription cis-regulatory region binding [GO:0000976]; transcription corepressor binding [GO:0001222]; ubiquitin protein ligase binding [GO:0031625]; circadian regulation of gene expression [GO:0032922]; circadian regulation of translation [GO:0097167]; circadian rhythm [GO:0007623]; entrainment of circadian clock [GO:0009649]; entrainment of circadian clock by photoperiod [GO:0043153]; histone H3 acetylation [GO:0043966]; histone H3 deacetylation [GO:0070932]; histone H4 acetylation [GO:0043967]; negative regulation of glucocorticoid receptor signaling pathway [GO:2000323]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of JNK cascade [GO:0046329]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of transcription by RNA polymerase II [GO:0045944]; posttranscriptional regulation of gene expression [GO:0010608]; regulation of circadian rhythm [GO:0042752]; regulation of cytokine production involved in inflammatory response [GO:1900015]; regulation of hair cycle [GO:0042634]; regulation of p38MAPK cascade [GO:1900744]; regulation of sodium ion transport [GO:0002028]; response to cAMP [GO:0051591] | 11306557_Observational study of gene-disease association. (HuGE Navigator) 12916719_human Per1 (hPER1) reporter gene activity shows circadian rhythmicity in a human neuroblastoma, but not in a astrocytoma or a hepatoma cell line. 14712925_Circadian oscillations of Per1, Per2, and Per3 mRNA expression were observed in serum-stimulated normal human fibroblasts. 14750904_Circadian hPER1 degradation through a proteasomal pathway can be regulated through phosphorylation by CKI, but not by subcellular localization. The mRNA levels reached a maximum at 1 h & minimal levels at 12 h. 15781181_physical chemistry and three-dimensional structure of the protein transduction domain 15790588_disturbances in PER gene expression may result in disruption of the control of the normal circadian clock, thus benefiting the survival of cancer cells and promoting carcinogenesis 15917222_RNA interference against beta-TRCP greatly decreases Clock-dependent gene expression in tissue culture cells, indicating that beta-TRCP controls endogenous Per1 activity and the circadian clock by directly targeting Per1 for degradation 16528748_Observational study of gene-disease association. (HuGE Navigator) 16678109_Per1 provides an important link between the circadian system and the cell cycle system. 16757810_These results together suggested that RACK1 might act as a novel signal molecule to mediate or regulate the functions of PER1 through protein interaction. 17051316_Observational study of gene-disease association. (HuGE Navigator) 17051316_This is the first reported association between a PER1 polymorphism and extreme diurnal preference 17106427_Observational study of gene-disease association. (HuGE Navigator) 17264841_Haplotype analysis of clock gene variants with autistic disorder resulted in per1 having a single significant result for the markers rs2253820-rs885747. 17264841_Observational study of gene-disease association. (HuGE Navigator) 17274950_We have examined the circadian expression of clock genes in human leukocytes and found that Per1 mRNA exhibits a robust circadian expression 17440215_clear circadian rhythm was shown for hPer1, hPer2, and hCry2 expression in CD34(+) cells and for hPer1 in the whole bone marrow, with maxima from early morning to midday. 17592726_Our data indicate that PER1 transcription is mainly uncoupled from promoter methylation but probably involves availability. 17621597_Photoreceptor layers obtained from Period1-luciferase transgenic rats express a robust circadian rhythm in bioluminescence, and demonstrate that mammalian photoreceptors contain a circadian pacemaker that can drive rhythmic melatonin synthesis. 17699798_Results revealed a disturbed transcription of Per1 during tumor progression, which might be the cause for disrupted daily oscillation of DPD in undifferentiated colon carcinoma cells. 17718421_direct interaction between RACK1 and PER1 17971899_a role for both Per1 and Per2 in normal breast function and show for the first time that deregulation of the circadian clock may be an important factor in the development of familial breast cancer 17984998_Observational study of gene-disease association. (HuGE Navigator) 17994337_CLOCK/BMAL1-mediated activation of PER1 by AP1 and E-Box elements is distinct from peripheral transcriptional modulation via cAMP-induced CREB and C/EBP. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18228528_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18321934_obstructive sleep apnoea syndrome caused disruptions of periodic expression of PER1(period homolog 1) mRNA in peripheral blood leukocytes. 18444243_The expression level of PER1 was decreased in hepatocellular carcinoma. 18480551_Involved in the inhibition of proliferation of MIA-PaCa2 pancreatic cancer cells by TNF-alpha. 18821565_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18990770_Observational study of gene-disease association. (HuGE Navigator) 18990770_Variations in circadian genes are associated with serum levels of androgens and IGF markers, particularly PER1 rs2585405:G>C(Ala962Pro). 19013183_There was no significant age-related phase difference in PER1 or PER2 rhythm with respect to sleep timing; however, PER3 expression pattern was altered in the older subjects. 19058789_Observational study of gene-disease association. (HuGE Navigator) 19148895_data demonstrate a correlated decrease of Per1 and ER-beta in colorectal tumors, mediated probably by epigenetic mechanisms 19296127_expression of PER2 is significantly associated with lymph node metastasis and poor prognosis in breast cancer 19675098_PER1 acts as an anti-apoptotic factor in pancreatic cancer cells 19693801_Observational study of gene-disease association. (HuGE Navigator) 19752089_Overexpression of Per1 in prostate cancer cells resulted in significant growth inhibition and apoptosis. Results support the emerging role of circadian genes as key players in malignant transformation. 19839995_Observational study of gene-disease association. (HuGE Navigator) 19861640_Sleep-wake cycles have been shown to influence the rhythmic mRNA expression of clock genes in peripheral blood cells of healthy participants 19912323_We found that overnight expression of BMAL1, but not PER1, was reduced in Parkinson's disease. Because our sampling span was limited to 12 h, we cannot rule out the possibility that PER1 is expressed differently during the unsampled part of the day. 19923301_A small population of rhythmic neurons in Per1-deficient suprachiasmatic nuclei explants is sufficient to control wheel-running activity. 19934327_Observational study of gene-disease association. (HuGE Navigator) 20072116_Observational study of gene-disease association. (HuGE Navigator) 20174623_Observational study of gene-disease association. (HuGE Navigator) 20481271_The expression levels of Per1 in glioma cells were significantly different from the surrounding non-glioma cells. The difference in the expression rate of Per1 in high-grade (grade III and IV) and low-grade (grade 1 and II) gliomas was insignificant. 20541418_There were no significant differences in PER1 expression between patients with Alzheimer's dementia, those with mild cognitive impairment, and controls. An increase in PER1 expression during the wake stage is observed for all three groups. 20861012_ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression 20978934_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21380491_Overexpression of the Bmal1 gene and reduced expression of the Per1 gene may thus be useful predictors of liver metastasis. 21411511_Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBalpha, RIP140, and PGC1alpha) genes. 21459569_PER1 and PER3 may modulate apoptotic reactions to cisplatin in gingival cancer cells 22217103_treatment with isoprenaline or dexamethasone induces circadian expression of hPer1, hPer2, hPer3, and hBMAL1 22801371_dose-specific glucocorticoid responses are specific, able to distinctly express a single gene,PER1; mapped PER1 response to a single GR binding site; overexpression of PER1 led to regulation of additional circadian rhythm genes suggesting hypersensitive expression of PER1 impacts circadian gene expression 23034908_A common polymorphism near PER1 is associated with the timing of human behavioral rhythms, and shows evidence of association with time of death. 23129285_Expression of the CLOCK, BMAL1, and PER1 circadian genes in human oral mucosa cells as dependent on CLOCK gene polymorphic variants. 23133559_type II protein arginine methyltransferase 5 has a role in the regulation of Circadian Per1 and CRY1 genes 23242607_Loss of Per1 expression is associated with skin tumors. 23405233_Loss of PER1 expression is associated with proliferation and metastasis of buccal squamous cell carcinoma. 23546644_Studies indicate that in the cytoplasm, PER3 protein heterodimerizes with PER1, PER2, CRY1, and CRY2 proteins and enters into the nucleus, resulting in repression of CLOCK-BMAL1-mediated transcription. 23584858_Data suggest that clock genes Per1, Cry1, Clock, and Bmal1 and their protein products may be directly involved in the daytime-dependent regulation and adaptation of hormone synthesis. 23752693_we identified the circadian clock PER1 mRNA as a novel substrate of the endoribonuclease activity of the unfolded protein response sensor IRE1alpha 24005054_Knockdown of either BMAL1 or Period1 in human anagen hair follicles significantly prolonged anagen. 24154698_Per1 regulates aldosterone levels, and Per1 plays an integral role in the regulation of Na(+) retention. 24449901_Casein kinase 1 primarily regulates the accumulating phase of the PER-CRY repressive complex by controlling the nuclear import rate. 24551282_Per1 and Per2 may play important roles in tumor development, invasion and prognosis, and Per2 may serve as a novel prognostic biomarker of human gastric cancer. 24578160_findings suggested that the expression of hPER1 is regulated by miR-29a/b/c, which may also provide a new clue for the function of hPER1 24678593_The PER1 c.2247C> T and c.2884C> G polymorphisms singly and in combination were found to be related to the lumbar spine bone mineral density in postmenopausal Korean women. 24857656_S714G mutation in hPER1 is associated with feeding rhythms and obesity in mice. 25070164_There is not a significant difference in the expression of CLOCK, BMAL1, and PER1 in buccal epithelial cells of patients with essential arterial hypertension regardless of patient genotype. 25310406_PER1 and BMAL1 operate as cell-autonomous modulators of human pigmentation and may be targeted for future therapeutic strategies 25344870_Functional SNPs found in PER1 were significantly associated with recurrence-free survival in a Chinese population with hepatocellular carcinoma. 25550826_In patients with non-small cell lung cancer, loss of Per1 was correlated with poor differentiation, tumor status, high p-TNM stage and lymph node metastasis. Patients with lower expression of Per1, Per2 and Per3 had a shorter survival time. 25863084_Fourteen proteins were identified to interact with per1 in tumor. 26168277_In adipose tissue, acute sleep deprivation vs sleep increased methylation in two promoter-interacting enhancer regions of PER1 26319354_Collectively, these data show that KPNB1 is required for timely nuclear import of PER/CRY in the negative feedback regulation of the circadian clock. 26339386_Period 1 and estrogen receptor-beta are downregulated in Chinese colon cancers. 26377793_Data demonstrate a role for Per1 in the transcriptional regulation of NHE3 and SGLT1 in the kidney. 26507264_ARNTL and PER1 were associated with PD. 26624862_The PER1 c.2884C > G polymorphism and PER3 54bp VNTR were associated with annual percent changes in bone mineral density of femoral neck after 1 year of hormone therapy. PER1 c.2884C > G polymorphism may be associated with risk of non-response to HT in postmenopausal Korean women. 26850841_when overexpressed, c-MYC is able to repress Per1 transactivation by BMAL1/CLOCK via targeting selective E-box sequences. Importantly, upon serum stimulation, MYC was detected in BMAL1 protein complexes 26923637_Per1 is regulated by microRNA-34a, a small non-coding RNA that directly binds to genes and inhibit gene expression. 26943040_Low PER1 expression is associated with oral squamous cell carcinoma. 27492458_Low Per1 gene expression is associated with colorectal liver metastases. 27602964_the role of PER1 in carcinogenesis is exerted not only by regulating downstream genes, but also through the synergistic dysregulation of many other clock genes in the clock gene network. 29377895_TNPO1-mediated nuclear import may constitute a novel input pathway of how cellular redox state signals to the clock, since redox stress increases binding of TNPO1 to PER1 and decreases its nuclear localization. TNPO1 is one of the novel players essential for normal circadian periods and potentially for redox regulation of the clock. 30110692_the strongest correlation was found between a decrease in placental PER1 expression and increased anxiety scores. Labour status was found to have a profound effect on mRNA expression. 30121446_Our findings suggest a putative over-representation of PER3 polymorphisms in bipolar patients with alcohol abuse. 30195196_A two-way ANOVA revealed significant differences between the clock genes profiles of controls and RBD patients for hPer1 (P = 0.0324), hPer2 (P = 0.0039), hBmal1 (P = 0.0160), and hNr1d1 (P = 0.0046). 30665264_Study reported an association between the risk of prostate cancer and the circadian pathway where significant associations were found for the genes NPAS2 and PER1. 30774045_These data together suggest that Per-1 is a novel negative regulator of HIV-1 transcription. This restrictive activity of Per-1 to HIV-1 replication may contribute to HIV-1 latency in resting CD4+ T-cells. 32086839_The clock gene Period 1 (Per1) is downregulated in oral squamous cell carcinoma (OSCC) and low Per1 expression is associated with TNM clinical stage and poor prognosis of OSCC patients. Per1 promotes OSCC progression by inhibiting autophagy-mediated cell apoptosis and enhancing cell proliferation in an AKT/mTOR pathway-dependent manner. 32541172_PER1 rs3027188 Polymorphism and its Association with the Risk of Colorectal Cancer in the Japanese Population. 32805476_Comparing expression levels of PERIOD genes PER1, PER2 and PER3 in chronic insomnia patients and medical staff working in the night shift. 32849922_Period Family of Clock Genes as Novel Predictors of Survival in Human Cancer: A Systematic Review and Meta-Analysis. 33723221_PER1 suppresses glycolysis and cell proliferation in oral squamous cell carcinoma via the PER1/RACK1/PI3K signaling complex. 33804124_The Circadian Protein PER1 Modulates the Cellular Response to Anticancer Treatments. 34730281_The promoter hypermethylation of SULT2B1 accelerates esophagus tumorigenesis via downregulated PER1. 35255118_Disrupting Circadian Rhythm via the PER1-HK2 Axis Reverses Trastuzumab Resistance in Gastric Cancer. | ENSMUSG00000020893 | Per1 | 2.791748e+03 | 1.0448259 | 0.063262510 | 0.3082751 | 4.130129e-02 | 0.8389574687 | 0.96860786 | No | Yes | 2.591906e+03 | 365.849286 | 2.415796e+03 | 350.199706 | |
| ENSG00000179195 | 144348 | ZNF664 | protein_coding | Q8N3J9 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:144348; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 19936222_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) | ENSMUSG00000079215 | Zfp664 | 4.082379e+03 | 0.9132203 | -0.130965209 | 0.3123184 | 1.771964e-01 | 0.6737937767 | 0.93064572 | No | Yes | 3.984180e+03 | 673.889886 | 3.924350e+03 | 680.649115 | ||
| ENSG00000179523 | 645212 | EIF3J-DT | lncRNA | 31421822_EIF3J-AS1 promotes CTNND2 expression via sponging miR-122-5p. Hypoxia-induced EIF3J-AS1 facilitates hepatocellular carcinoma progression via regulating CTNND2. 31709617_H3K27 acetylation-induced lncRNA EIF3J-AS1 improved proliferation and impeded apoptosis of colorectal cancer through miR-3163/YAP1 axis. 32811869_LncRNA EIF3J-AS1 enhanced esophageal cancer invasion via regulating AKT1 expression through sponging miR-373-3p. 33764843_Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. | 8.535866e+01 | 0.7425887 | -0.429364650 | 0.4231275 | 1.032320e+00 | 0.3096144386 | 0.81718332 | No | Yes | 7.305441e+01 | 14.785149 | 9.098625e+01 | 18.691461 | |||||||||
| ENSG00000179818 | 400960 | PCBP1-AS1 | lncRNA | 34107009_LncRNA PCBP1-AS1 correlated with the functional states of cancer cells and inhibited lung adenocarcinoma metastasis by suppressing the EMT progression. | 9.650334e+02 | 0.8135756 | -0.297651728 | 0.2695044 | 1.223156e+00 | 0.2687422060 | 0.79897038 | No | Yes | 6.260067e+02 | 86.863751 | 8.737139e+02 | 123.960379 | |||||||||
| ENSG00000179832 | 727957 | MROH1 | protein_coding | Q8NDA8 | Alternative splicing;Reference proteome;Repeat | hsa:727957; | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000022558 | Mroh1 | 7.141733e+02 | 1.1876391 | 0.248096480 | 0.2831810 | 7.606711e-01 | 0.3831185859 | 0.84285514 | No | Yes | 3.458150e+02 | 43.442495 | 2.437238e+02 | 31.421060 | ||||
| ENSG00000179846 | 284353 | NKPD1 | protein_coding | Q17RQ9 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | Mouse_homologues mmu:69547; | integral component of membrane [GO:0016021] | 15935074_altered membrane NTPase activity is a biochemical hallmark of HPRT deficiency, but species and cell-type differences have to be considered 27745872_Study suggests that nonsynonymous variation in the gene NKPD1 affects depressive symptoms in the general population. | ENSMUSG00000060621 | Nkpd1 | 3.256109e+01 | 1.2124743 | 0.277954207 | 0.5292566 | 2.771215e-01 | 0.5985941288 | 0.90859245 | No | Yes | 2.472706e+01 | 9.002843 | 2.170464e+01 | 8.271717 | |||
| ENSG00000180035 | 197407 | ZNF48 | protein_coding | Q96MX3 | FUNCTION: May be involved in transcriptional regulation. | Acetylation;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:197407; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000045598 | Zfp553 | 2.479222e+03 | 1.3178206 | 0.398153934 | 0.3314116 | 1.450524e+00 | 0.2284439304 | 0.78792184 | No | Yes | 2.775800e+03 | 364.945196 | 1.805056e+03 | 243.980024 | |||
| ENSG00000180098 | 54952 | TRNAU1AP | protein_coding | Q9NX07 | FUNCTION: Involved in the early steps of selenocysteine biosynthesis and tRNA(Sec) charging to the later steps resulting in the cotranslational incorporation of selenocysteine into selenoproteins. Stabilizes the SECISBP2, EEFSEC and tRNA(Sec) complex. May be involved in the methylation of tRNA(Sec). Enhances efficiency of selenoproteins synthesis (By similarity). {ECO:0000250, ECO:0000269|PubMed:16508009}. | 3D-structure;Alternative splicing;Cytoplasm;Nucleus;Protein biosynthesis;RNA-binding;Reference proteome;Repeat | hsa:54952; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; RNA binding [GO:0003723]; tRNA binding [GO:0000049]; selenocysteine incorporation [GO:0001514] | 16508009_our studies delineating the multiple, coordinated protein-nucleic acid interactions between SECp43 and the selenoprotein cotranslational factors. 28414460_Data confirm interactions among components of the early steps of the selenocysteine biosynthesis pathway (SEPSECS, SECP43, SEPHS1, and SEPHS2); SECP43, which interacts with SEPSECS and SEPHS1, is a globular protein that forms oligomers in vivo. 29758194_Knockdown of Trnau1ap reduced cell migration and proliferation. Knockdown of Trnau1ap blocked the PI3K/Akt signaling pathway. | ENSMUSG00000028898 | Trnau1ap | 1.135477e+03 | 0.7197316 | -0.474469137 | 0.3089150 | 2.383790e+00 | 0.1225997712 | 0.75814553 | No | Yes | 8.581900e+02 | 92.910026 | 1.205128e+03 | 133.096813 | ||
| ENSG00000180104 | 11336 | EXOC3 | protein_coding | O60645 | FUNCTION: Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane. | Acetylation;Cell projection;Cytoplasm;Exocytosis;Golgi apparatus;Protein transport;Reference proteome;Transport | The protein encoded by this gene is a component of the exocyst complex, a multiple protein complex essential for targeting exocytic vesicles to specific docking sites on the plasma membrane. Though best characterized in yeast, the component proteins and functions of exocyst complex have been demonstrated to be highly conserved in higher eukaryotes. At least eight components of the exocyst complex, including this protein, are found to interact with the actin cytoskeletal remodeling and vesicle transport machinery. The complex is also essential for the biogenesis of epithelial cell surface polarity. [provided by RefSeq, Jul 2008]. | hsa:11336; | cytosol [GO:0005829]; exocyst [GO:0000145]; Golgi apparatus [GO:0005794]; growth cone [GO:0030426]; midbody [GO:0030496]; perinuclear region of cytoplasm [GO:0048471]; presynaptic membrane [GO:0042734]; secretory granule membrane [GO:0030667]; cadherin binding [GO:0045296]; SNARE binding [GO:0000149]; exocyst localization [GO:0051601]; exocytosis [GO:0006887]; protein transport [GO:0015031] | 22381337_Data demonstrate that the expression of alpha-E-catenin is increased by Sec6 siRNAs, and E-cadherin and beta-catenin localize mainly at the cell-cell contact region in HSC3 cells, which were transfected with Sec6 siRNA. 24949832_Sec6 regulates cytoplasmic translocation of p27 through p27 phosphorylation at Thr157, thereby promoting p27 degradation in the cytoplasm via interaction with Jab1 and Siah1 and suppressing cell cycle progression. 26247921_Sec6 regulates NF-kappaB transcriptional activity via the control of the phosphorylation of IkappaBalpha, p90RSK1, and ERK 26283729_The study explores the role of the exocyst complex component Sec6/8 in genomic stability. 26892009_Sec6 regulate Bcl-2 and Mcl-1 expressions but not Bcl-xl in malignant peripheral nerve sheath tumor cells. 29729335_Sec6 increased the phosphorylation of p38 MAPK through the activation of MAPK kinase 3/6 (MKK3/6). Moreover, Sec6 knockdown suppressed the phosphorylation of HSP27 at Ser(78) and Ser(82) sites via suppression of activated MK2. | ENSMUSG00000034152 | Exoc3 | 2.356329e+03 | 1.0099962 | 0.014349926 | 0.2770212 | 2.666679e-03 | 0.9588156282 | 0.99162977 | No | Yes | 2.056689e+03 | 133.979535 | 1.902178e+03 | 127.422530 | |
| ENSG00000180182 | 9282 | MED14 | protein_coding | O60244 | FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. {ECO:0000269|PubMed:15340088, ECO:0000269|PubMed:15625066, ECO:0000269|PubMed:16595664}. | 3D-structure;Activator;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | The activation of gene transcription is a multistep process that is triggered by factors that recognize transcriptional enhancer sites in DNA. These factors work with co-activators to direct transcriptional initiation by the RNA polymerase II apparatus. The protein encoded by this gene is a subunit of the CRSP (cofactor required for SP1 activation) complex, which, along with TFIID, is required for efficient activation by SP1. This protein is also a component of other multisubunit complexes e.g. thyroid hormone receptor-(TR-) associated proteins which interact with TR and facilitate TR function on DNA templates in conjunction with initiation factors and cofactors. This protein contains a bipartite nuclear localization signal. This gene is known to escape chromosome X-inactivation. [provided by RefSeq, Jul 2008]. | hsa:9282; | core mediator complex [GO:0070847]; mediator complex [GO:0016592]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; nuclear receptor coactivator activity [GO:0030374]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; vitamin D receptor binding [GO:0042809]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription initiation from RNA polymerase II promoter [GO:0060261]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357]; stem cell population maintenance [GO:0019827] | 12509459_DRIP150 binds to ISGF3 and regulates transcription 12825353_CRSP2 gene is expressed in the retina and its exact genomic location is on Xp11.4 between DXS1368 and DXS993 15625066_Coactivation of ERalpha by DRIP150 in ZR-75 cells is NR box-independent and requires a novel sequence with putative alpha-helical structure. 16239257_MED14 and MED1 are used by glucocorticoid receptor in a gene-specific manner, providing a mechanism for promoter selectivity by glucocorticoid receptor 17306756_Coactivator that enhances estrogen receptor alpha- and specificity protein (SP)-1-mediated transactivation in breast cancer cells. 23572530_VitD-mediated stimulation of GC anti-inflammatory affects human monocytes in a process involving GM-CSF and MED14 25383669_This results from a dramatically enhanced ability of MED14-containing complexes to associate with Pol II. 28813667_Functional analysis implicates TNRC6A, NAT10, MED14, and WDR5 in RNA-mediated transcriptional activation. | ENSMUSG00000064127 | Med14 | 2.369939e+03 | 1.0698892 | 0.097461353 | 0.2691444 | 1.295402e-01 | 0.7189092768 | 0.94084174 | No | Yes | 2.573100e+03 | 324.904903 | 2.133919e+03 | 276.492311 | |
| ENSG00000180806 | 3225 | HOXC9 | protein_coding | P31274 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | 3D-structure;DNA-binding;Developmental protein;Homeobox;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene belongs to the homeobox family of genes. The homeobox genes encode a highly conserved family of transcription factors that play an important role in morphogenesis in all multicellular organisms. Mammals possess four similar homeobox gene clusters, HOXA, HOXB, HOXC and HOXD, which are located on different chromosomes and consist of 9 to 11 genes arranged in tandem. This gene is one of several homeobox HOXC genes located in a cluster on chromosome 12. [provided by RefSeq, Jul 2008]. | hsa:3225; | aggresome [GO:0016235]; chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; embryonic skeletal system morphogenesis [GO:0048704]; proximal/distal pattern formation [GO:0009954]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription, DNA-templated [GO:0006351] | 17588684_Up-regulation of HOXC9 was detected in a set of 54 astrocytomas of different grades and significantly associated with malignancy. 19506903_Methylated state of this set of genes may be more specific to the late rather than the early stage of NSCLC. 21493894_The data identify HOXC9 as an endothelial cell active transcriptional repressor promoting the resting, antiangiogenic endothelial cell phenotype in an interleukin 8-dependent manner 21507931_HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. 22106857_We did not find any potential pathological mutations in the Hoxc9 gene among Chinese patients with congenital heart disease. 23579273_HoxC9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. 24274069_HOXC9 coordinates diverse cellular processes associated with differentiation by directly activating and repressing the transcription of distinct sets of genes. 25444900_Results provide a set of the essential genes in the miR-193a-3p/HOXC9/DNA damage response/oxidative stress pathway axis as the diagnostic targets for the guided anti-bladder cancer chemotherapy. 26582930_downregulation of HOXC9 releases its transcriptional inhibition of DAPK1, resulting in the activation of the DAPK1-Beclin1 pathway, which induces autophagy in glioblastoma cells 26647900_HOXC9 and HOXC10 may play an important role in the development of obesity, adverse fat distribution, and subsequent alterations in whole-body metabolism and adipose tissue function. 30205370_HOXC9 knockdown inhibited the metastasis and stem cell-like phenotype of GC cells without significant effects on cell proliferation as a direct target of miR-26a. 31414766_Transcription factor homeobox C9 (HOXC9) may play a critical role in colorectal cancer (CRC) progression and serve as a potential marker of poor prognosis in CRC. 32816159_HOXC9 overexpression is associated with gastric cancer progression and a prognostic marker for poor survival in gastric cancer patients. 34159686_Upregulation of HOXC9 generates interferon-gamma resistance in gastric cancer by inhibiting the DAPK1/RIG1/STAT1 axis. | ENSMUSG00000036139 | Hoxc9 | 4.973697e+02 | 1.2946020 | 0.372508642 | 0.2888250 | 1.661655e+00 | 0.1973799653 | 0.78383913 | No | Yes | 5.314947e+02 | 56.906302 | 3.712913e+02 | 41.239163 | |
| ENSG00000181019 | 1728 | NQO1 | protein_coding | P15559 | FUNCTION: The enzyme apparently serves as a quinone reductase in connection with conjugation reactions of hydroquinons involved in detoxification pathways as well as in biosynthetic processes such as the vitamin K-dependent gamma-carboxylation of glutamate residues in prothrombin synthesis. | 3D-structure;Alternative splicing;Cytoplasm;FAD;Flavoprotein;Isopeptide bond;NAD;NADP;Oxidoreductase;Phosphoprotein;Reference proteome;Ubl conjugation | This gene is a member of the NAD(P)H dehydrogenase (quinone) family and encodes a cytoplasmic 2-electron reductase. This FAD-binding protein forms homodimers and reduces quinones to hydroquinones. This protein's enzymatic activity prevents the one electron reduction of quinones that results in the production of radical species. Mutations in this gene have been associated with tardive dyskinesia (TD), an increased risk of hematotoxicity after exposure to benzene, and susceptibility to various forms of cancer. Altered expression of this protein has been seen in many tumors and is also associated with Alzheimer's disease (AD). Alternate transcriptional splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jul 2008]. | hsa:1728; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendrite [GO:0030425]; neuronal cell body [GO:0043025]; synapse [GO:0045202]; cytochrome-b5 reductase activity, acting on NAD(P)H [GO:0004128]; identical protein binding [GO:0042802]; NAD(P)H dehydrogenase (quinone) activity [GO:0003955]; NADH dehydrogenase (quinone) activity [GO:0050136]; NADPH dehydrogenase (quinone) activity [GO:0008753]; RNA binding [GO:0003723]; superoxide dismutase activity [GO:0004784]; aging [GO:0007568]; cell redox homeostasis [GO:0045454]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to metal ion [GO:0071248]; NADH oxidation [GO:0006116]; NADPH oxidation [GO:0070995]; negative regulation of apoptotic process [GO:0043066]; negative regulation of catalytic activity [GO:0043086]; nitric oxide biosynthetic process [GO:0006809]; positive regulation of neuron apoptotic process [GO:0043525]; response to alkaloid [GO:0043279]; response to amine [GO:0014075]; response to carbohydrate [GO:0009743]; response to electrical stimulus [GO:0051602]; response to estradiol [GO:0032355]; response to ethanol [GO:0045471]; response to flavonoid [GO:1905395]; response to hormone [GO:0009725]; response to hydrogen sulfide [GO:1904880]; response to ischemia [GO:0002931]; response to L-glutamine [GO:1904844]; response to nutrient [GO:0007584]; response to oxidative stress [GO:0006979]; response to testosterone [GO:0033574]; response to tetrachloromethane [GO:1904772]; response to toxic substance [GO:0009636]; synaptic transmission, cholinergic [GO:0007271]; xenobiotic metabolic process [GO:0006805] | 11051261_Observational study of gene-disease association. (HuGE Navigator) 11219774_Observational study of gene-disease association. (HuGE Navigator) 11319169_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11520401_Observational study of gene-disease association. (HuGE Navigator) 11551408_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11679176_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11688992_Observational study of gene-disease association. (HuGE Navigator) 11688992_polymorphisms associated with idiopathic Parkinson's disease 11722480_Observational study of gene-disease association. (HuGE Navigator) 11766168_Observational study of gene-disease association. (HuGE Navigator) 11773862_Observational study of gene-disease association. (HuGE Navigator) 11774269_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11810042_Observational study of gene-disease association. (HuGE Navigator) 11821413_Interaction of the molecular chaperone Hsp70 with human NAD(P)H:quinone oxidoreductase 1. 11840286_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11894133_Lack of NQO1 induction in human tumors is not due to changes in promoter region. 11895912_Observational study of gene-disease association. (HuGE Navigator) 11943609_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11948402_Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene 11956078_Glutathione S-transferase P1 and NADPH quinone oxidoreductase polymorphisms are associated with aberrant promoter methylation of P16(INK4a) and O(6)-methylguanine-DNA methyltransferase in sputum. 11956078_Observational study of gene-disease association. (HuGE Navigator) 11967624_Observational study of gene-disease association. (HuGE Navigator) 12018106_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12036913_Observational study of gene-disease association. (HuGE Navigator) 12037698_Observational study of gene-disease association. (HuGE Navigator) 12163326_NQO1-Pro/Pro genotype is a risk factor for lung adenocarcinoma development 12163326_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12164325_Observational study of genotype prevalence. (HuGE Navigator) 12171070_A novel plasma membrane quinone reductase and NAD(P)H:quinone oxidoreductase 1 are upregulated by serum withdrawal in human promyelocytic HL-60 cells as a model to analyze cell responses to oxidative stress. 12171760_Observational study of gene-disease association. (HuGE Navigator) 12172217_The 465 SNP was the major cause of increased alternative splicing and decreased expression of NQO1 protein in HCT-116R30A cells. 12232053_NQO1 regulates p53 proteasomal degradation independently of MDM2 and ubiquitin. 12370194_laminar flow-induced promoter activation in endothelial cells is inhibited by expression of antisense Nrf2 12383707_Benzo(a)pyrene activates extracellular signal-related and p38 mitogen-activated protein kinases in HT29 colon adenocarcinoma cells: involvement in NAD(P)H:quinone reductase activity and cell proliferation 12393447_The genotype distribution of the NQO1 gene does not indicate a role for base excision repair in the development of therapy-related acute myeloblastic leukemia 12393620_inactivating NQO1 polymorphism is associated with an increased risk of de novo leukemia with MLL translocations in infants and children 12397416_Observational study of gene-disease association. (HuGE Navigator) 12419832_Observational study of gene-disease association. (HuGE Navigator) 12439220_Observational study of gene-disease association. (HuGE Navigator) 12460800_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12468438_Observational study of gene-disease association. (HuGE Navigator) 12480594_Observational study of gene-disease association. (HuGE Navigator) 12529318_association between p53 and NQO1 that may represent an alternate mechanism of p53 stabilization by NQO1 in a wide variety of human cell types. 12694753_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12694753_the NQO1 genetic polymorphism elevates bladder cancer risk, especially in male Caucasian smokers 12711112_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12713578_Observational study of gene-disease association. (HuGE Navigator) 12718576_Observational study of genotype prevalence. (HuGE Navigator) 12718704_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 12771035_NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T polymorphism is associated with esophageal squamous cell carcinoma in a German Caucasian and a northern Chinese population 12771035_Observational study of gene-disease association. (HuGE Navigator) 12777965_CYP2E1 and NQO1 genotypes may play an important role in development of smoking related bladder cancer among Korean men 12777965_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12834817_Observational study of gene-disease association. (HuGE Navigator) 12854127_Observational study of gene-disease association. (HuGE Navigator) 12867492_...NQO1 expression is up-regulated by B[a]P treatment. P. 1040 12960511_Observational study of gene-disease association. (HuGE Navigator) 13130177_Observational study of gene-disease association. (HuGE Navigator) 14506737_Observational study of gene-disease association. (HuGE Navigator) 14568289_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 14592434_These results provide the first evidence of a proximal repressor region exerting a negative role on the regulation of the hNQO1 promoter in small cell lung carcinoma. 14634213_NQO1 has an important role in stabilizing hot-spot p53 mutant proteins in cancer 14634838_Observational study of genotype prevalence. (HuGE Navigator) 14669229_Observational study of gene-disease association. (HuGE Navigator) 14675732_This experiment conclude that NQO1 activity co-localizes closely with AD pathology supporting a presumed role as an antioxidant system upregulated in response to the oxidative stress of the AD process. 14688016_Observational study of gene-disease association. (HuGE Navigator) 14694720_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14694720_The subjects carrying NQO1c. 609 T/T genotype and together with the habit of smoking or drinking may be more susceptible to benzene poisoning. 14716779_Observational study of genotype prevalence. (HuGE Navigator) 14720419_Observational study of gene-disease association. (HuGE Navigator) 14729580_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 14985350_NO signals the transcriptional up-regulation of NQO1 and other detoxifying enzyme and protective genes through Nrf2 via the ARE to counteract NO-induced apoptosis of neuroblastoma cells 15061915_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15079792_Observational study of gene-disease association. (HuGE Navigator) 15111988_Observational study of genotype prevalence. (HuGE Navigator) 15116053_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15138483_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15151706_NQO1 gene polymorphism may confer susceptibility to the development of tardive dyskinesia in schizophrenia 15151706_Observational study of gene-disease association. (HuGE Navigator) 15184245_In smokers the variant NQO1 genotype may confer an increased risk for squamous cell carcinoma. 15184245_Observational study of gene-disease association. (HuGE Navigator) 15196853_Observational study of gene-disease association. (HuGE Navigator) 15219943_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15226677_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15279067_Observational study of healthcare-related. (HuGE Navigator) 15280903_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15298951_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15298951_The association between oral contraceptives use and breast cancer risk is dependent on NQO1 genotype, age and menopausal status. 15312971_While overt NQO1 immunoreactivity was absent in the surrounding nervous tissue, in the Parkinsonian SNpc a marked increase in the astroglial and neuronal expression of NQO1 was consistently observed. 15334552_NQO1 may have a role in familial acute myeloid leukemia 15352038_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15355699_Observational study of gene-disease association. (HuGE Navigator) 15365958_Observational study of gene-disease association. (HuGE Navigator) 15370874_Observational study of gene-disease association. (HuGE Navigator) 15382274_Observational study of gene-disease association. (HuGE Navigator) 15385560_Nrf3 is a negative regulator of ARE-mediated gene expression of NQO1 15466980_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15498787_Observational study of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 15498787_The observed risk reductions of lung cancer may be attributable to the greatly reduced procarcinogen activating of NAD(P)H:quinone oxidoreductase 1 in individuals with at least one copy of the T allele. 15590400_Observational study of gene-disease association. (HuGE Navigator) 15590400_There may be a modulating role for NQO1 in the pathogenesis of pediatric sporadic Burkitt's lymphoma. 15618957_Observational study of gene-disease association. (HuGE Navigator) 15618957_Possible role for NQO1 gene as an MLL-independent risk factor, in the leukemogenic process of this subtype of infant acute lymphoblastic leukemia. 15640066_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15661231_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15694256_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15714076_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, gene-gene interaction, gene-environment interaction, pharmacogenomic / toxicogenomic, and healthcare-related. (HuGE Navigator) 15727169_Observational study of gene-disease association. (HuGE Navigator) 15731166_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15731166_The joint carriage of CYP1A1 Val(462) and NQO1 Ser(187) alleles, particularly in smokers, was related to colorectal adenoma risk, with a propensity for formation of multiple lesions 15734732_Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants 15746160_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15747169_Observational study of gene-disease association. (HuGE Navigator) 15748501_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15748501_The genetic polymorphisms of NQO1, GSTT1 and GSTM1 led to declining of detoxifying ability in benzene metabolism, so the individual with NQO1 C609T T/T genotype, GSTT1 null genotype and GSTM1 null genotype is most susceptive to benzene poisoning. 15749015_NQO1 has a role in regulating ubiquitin-independent degradation of ornithine decarboxylase by the 20S proteasome 15763338_NQO1 transcription might be inappropriately suppressed by promoter hypermethylation in a subset of hepatocellular carcinoma, as well as GSTP1 gene 15767364_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15781212_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15808404_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15829318_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15838728_Observational study of gene-disease association. (HuGE Navigator) 15938845_Observational study of gene-disease association. (HuGE Navigator) 16003741_Immunohistochemistry of resected pancreatic specimens demonstrated an increased immunoreactivity for NQO1 in pancreatic cancer 16006997_Observational study of gene-disease association. (HuGE Navigator) 16038261_Observational study of gene-disease association. (HuGE Navigator) 16039674_Observational study of gene-disease association. (HuGE Navigator) 16054862_Observational study of gene-disease association. (HuGE Navigator) 16079101_Low/null activity polymorphisms of this enzyme is not with the risk of developing aplastic anemia in Caucasian patients. 16079101_Observational study of gene-disease association. (HuGE Navigator) 16157195_Observational study of gene-disease association. (HuGE Navigator) 16170238_Observational study, meta-analysis, and HuGE review of genotype prevalence, gene-disease association, gene-gene interaction, gene-environment interaction, and healthcare-related. (HuGE Navigator) 16172237_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16235982_Observational study of gene-disease association. (HuGE Navigator) 16235998_Observational study of genetic testing. (HuGE Navigator) 16266898_Observational study of gene-disease association. (HuGE Navigator) 16284498_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16297214_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16321221_Observational study of gene-disease association. (HuGE Navigator) 16343742_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16369108_Observational study of genetic testing. (HuGE Navigator) 16385446_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16413497_Observational study of gene-disease association. (HuGE Navigator) 16520888_Observational study of gene-disease association. (HuGE Navigator) 16532285_The increased expression in noncancer pancreatic tissue from smokers suggests that NQO1 expression may be a good candidate as a biomarker for pancreatic cancer, especially in risk groups such as smokers. 16595077_NAD(P)H:quinone acceptor oxidoreductase (NQO) gene family belongs to the flavoprotein clan and, in the human genome, consists of two genes (NQO1 and NQO2).(review) 16598069_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16610002_Up-regulation of NQO1 may represent an adaptive stress response to limit further disease progression by detoxifying reactive species. 16620556_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16702380_Meta-analysis of gene-disease association. (HuGE Navigator) 16775388_Observational study of gene-disease association. (HuGE Navigator) 16818284_In Israeli patients, NQO1*2 does not predispose to de novo AML; analysis showed significant differences in the frequencies of NQO1*2 by ethnic group 16818284_Observational study of gene-disease association. (HuGE Navigator) 16978807_transfection of SK-N-MC cells with NQO1 protects against dopamine-induced toxicity 16985026_Observational study of gene-disease association. (HuGE Navigator) 17011189_From in silico docking and comparative analysis, novel inhibitors of human NAD(P)H quinone oxidoreductase (NQO1) have been identified. 17027152_Observational study of gene-disease association. (HuGE Navigator) 17027152_Our findings suggested that this polymorphism might not represent additional genetic risk factor for LOAD. 17082176_NQO1 gene polymorphisms is associated with sporadic distal colorectal adenomas 17082176_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17113562_Observational study of genotype prevalence. (HuGE Navigator) 17118447_NQO1 polymorphism confers interindividual variability of response to treatment in patients with acute myeloid leukemia 17118447_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17119198_Observational study of gene-disease association. (HuGE Navigator) 17164365_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17166422_Observational study of gene-disease association. (HuGE Navigator) 17166422_The NQO1 C609T polymorphisms should be associated with the genetic susceptibility of esophageal cancer 17178637_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17179690_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17188257_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17332305_the NQO1 C609T single-nucleotide polymorphism is associated with approximately 7-fold lower NQO1 activity. 17332311_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17332311_This is the first study showing robust (to bias due to population structure) evidence that the NQO1 C609T variant is a risk factor for childhood leukemia. 17337051_Our preliminary report confirmed in multiple myeloma patients the trend, for a worse response to therapy in patients positive for NQO1*2. 17363580_Observational study of gene-disease association. (HuGE Navigator) 17367411_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17372252_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17378176_Observational study of gene-disease association. (HuGE Navigator) 17395013_NQO1 has an important role as a mediator of neutrophil elastase-regulated oxidant stress and MUC5AC mucin gene expression. 17400324_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17405841_role for NQO1 in the metabolic complications of human obesity 17424838_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17424838_Subjects carrying NQO1 rs1800566 TT genotype and together with the habit of smoking or drinking could be more susceptible to chronic benzene poisoning. 17428572_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17449559_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17469025_Data are consistent with the notion that NQO1 polymorphisms influence the course and clinical outcomes of urinary bladder neoplasms. 17469025_Observational study of gene-disease association. (HuGE Navigator) 17476281_Observational study of gene-disease association. (HuGE Navigator) 17496311_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17507624_Observational study of gene-disease association. (HuGE Navigator) 17541156_The chemopreventive activity of taxifolin was determined by measuring the activity of NQO1 in HCT116 cells. 17575500_NQO1 TT genotype may offer protection from reflux complications such as Barrett esophagus and esophageal adenocarcinoma. 17575500_Observational study of gene-disease association. (HuGE Navigator) 17581325_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17603928_Overexpression of NQO1 in various tumors suggest the feasibility of developing diaminophenothiazinium-based redox cyclers into anticancer agents. 17619904_NQO1 polymorphism is associated with urothelial cancer 17619904_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17644186_Case-control study of the possible relationship between the NQO1 gene polymorphism and mood disorders (patients with major depressive disorder, patients with bipolar I disorder, controls was carried out using PCR-based techniques. 17644186_Observational study of gene-disease association. (HuGE Navigator) 17652311_Observational study of gene-disease association. (HuGE Navigator) 17669387_down-regulation of NQO1 effectively suppresses TNF-alpha-induced human aortic smooth muscle cell migration through inhibition of MMP-9 expression 17726138_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17726138_potentially higher levels of iron-generated oxidative stress related gene alleles may be at increased risk of breast cancer 17869325_Observational study of genotype prevalence. (HuGE Navigator) 17885617_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17901563_The highly expressed and inducible endogenous NQO1 in cardiovascular cells may act as a potential O2(.-) scavenger. 18024413_Observational study of gene-disease association. (HuGE Navigator) 18024413_Patients carrying a variant low-activity NQO1 allele had a significantly increased risk of developing a SMN. The observed effect was restricted to solid tumors 18034618_NQO1 protein genotype was not associated with warfarin dose requirements in the African-Americans population. 18061666_Observational study of gene-disease association. (HuGE Navigator) 18061666_Polymorphisms in NQO1 is not associated with multiple myeloma 18061941_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18074351_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18074351_The NQO1 609CT genotype is associated with increased adenoma risk among smokers, not diminished by high fruit and vegetable consumption. The observed gene-gene interactions may point to a role for NFE2L2 polymorphisms in NQO1-related adenoma formation 18091324_study shows that NQO1 is consistently overexpressed in pancreatic ductal adenocarcinoma and may be a clinically useful diagnostic adjunct for detection of ancreatic ductal adenocarcinoma 18098117_Non-laminar flow during cardiopulmonary bypass may diminish the transcriptional activation of the NQO1 in T carriers 18156703_Observational study of gene-disease association. (HuGE Navigator) 18203021_Observational study of gene-disease association. (HuGE Navigator) 18203021_gene polymorphisms in CYP1A1, GSTT1, GSTP1, GSTM1, and NQO1 were characterized in Saudi individuals with a diagnosis of DLBCL; CYP1A1, GSTT1, GSTP1 demonstrated significant association of DLBCL risk; GSTM1 and NQO1 did not. 18214807_Benzene exposed workers with the T/T genotype for NQO1 (NAD(P)H dehydrogenase quinone 1) showed significant increases in Micronuclei and Chromosomal aberrations 18214807_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18218609_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18218609_The researchers found an association between the NQO1 homozygous wild-type allele and reduced birth weight, birth length, and birth head circumference in babies born to smoking mothers. 18225579_Observational study of gene-disease association. (HuGE Navigator) 18225579_Study of Turkish prostate cancer patients suggests that mutation of the NQO1 gene may effect the serum PSA and alkaline phosphatase levels. 18253865_Observational study of gene-disease association. (HuGE Navigator) 18253865_These results indicated that the C/C genotype had a possible protective effect against AD development, and the T allele might be a weak risk factor for late onset AD. 18388957_Data show that p33(ING1b) is degraded in the 20S proteasome and that NAD(P)H quinone oxidoreductase 1 (NQO1), previously shown to modulate the degradation of p53 in the 20S proteasome, inhibits the degradation of p33(ING1b). 18393254_Observational study of genetic testing. (HuGE Navigator) 18404535_chlorophyllin exerts antioxidant effect by inducing HO-1 and NQO1 expression mediated by PI3K/Akt and Nrf2 18407955_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18413200_Observational study of gene-disease association. (HuGE Navigator) 18416817_ATBF1 and NQO1 as candidate targets for allelic loss at chromosome arm 16q in breast cancer: absence of somatic ATBF1 mutations and no role for the C609T NQO1 polymorphism. 18444152_Observational study of gene-disease association. (HuGE Navigator) 18444152_study concluded that genetic variation, especially the NQO1 609C > T polymorphism, is a more important predictor of rectal NQO1 phenotype than fruit and vegetable consumption 18444911_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18444911_The NQO1 C/C genotype was significantly higher among Filipino children with acute lymphoblastic leukemia. This suggests a possible role for the NQO1 C/C genotype in the susceptibility of paediatric ALL cases in the Philippines. 18457324_Analyses indicated no association between the NQO1*2 polymorphism and the risk of anthracycline-related CHF 18457324_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18510611_Observational study of gene-disease association. (HuGE Navigator) 18511948_NQO1 genotype is a prognostic and predictive marker for breast cancer. 18511948_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18569591_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18587252_Significantly different distribution of the HO-1 genotype was found in subjects with different-stage UCs; however, it was not related to the NAD(P)H:quinone oxidoreductase 1 genotype. 18601742_Observational study of gene-disease association. (HuGE Navigator) 18632753_Observational study of gene-disease association. (HuGE Navigator) 18636124_Observational study of gene-disease association. (HuGE Navigator) 18676018_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18720901_Observational study of genotype prevalence. (HuGE Navigator) 18784359_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18798003_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18798003_genetic polymorphisms for NQO1, CYP2E1, and ALDH2 synergistically with cumulative smoking amounts and alcohol drinking levels interact in the carcinogenesis of lung cancer in Koreans. 18813798_alpha-lipoic acid controls cellular redox status, and upregulates quinone reductase NQO1 via Nrf2 activation in human leukemia HL-60 cells 18820947_Suppression of NQO1 may contribute to differential susceptibility of biliary epithelial cells to chemical-induced cytotoxicity and carcinogenesis. 18836923_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18936436_Observational study of genotype prevalence. (HuGE Navigator) 18945694_Meta-analysis of gene-disease association. (HuGE Navigator) 18950733_A lack of activity in the detoxification enzymes NQO1 and GSTM, and biomarker levels are strongly associated with coronary heart disease with sex as a mitigating factor. 18950733_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18977034_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18977034_The combined genotypes of T/T in NQO1 Pro187Ser and Val/Val in MnSOD Ala-9Val polymorphisms were found to be independently associated with a significantly higher risk of TD. 18981090_LPS induces NQO1 and HO-1 expression in human monocytes via Nrf2 to modulate their inflammatory responsiveness 18986377_Observational study of gene-disease association. (HuGE Navigator) 18992148_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19012493_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19012698_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19017358_Observational study of gene-disease association. (HuGE Navigator) 19017358_study demonstrated that the AC genotype at position -1221 in the NQO1 gene caused decreased transcription and was associated with a lower incidence of ALI following major trauma 19027876_Asthmatic children with a functional polymorphism of NQO1 may require more intensive pharmaceutical treatment to effectively control their asthma. 19027876_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19027952_Frequencies of polymorphic variants of RAD51, XRCC3, NQO1, GSTA1, GSTM1, GSTT1, CYP3A4 and XPD enzymes were similar in patients and controls. 19027952_Observational study of gene-disease association. (HuGE Navigator) 19059883_NQO1 modulates cellular redox status and influences the biologic and physiologic effects of ozone. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19096102_in Burkitt's lymphoma cell lines the NQO1 gene is not efficiently translated and this effect is not related to (C609T) polymorphism. 19138946_Resveratrol can prevent breast cancer initiation by blocking multiple sites in the estrogen genotoxicity pathway. 19151739_Observational study of gene-disease association. (HuGE Navigator) 19162321_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19162321_Polymorphisms in genes NQO1 and TS were associated with a significantly slow response to induction chemotherapy. 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19174490_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19179423_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19214744_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19229058_Activation of NAD(P)H:quinone oxidoreductase 1 prevents arterial restenosis by suppressing vascular smooth muscle cell proliferation. 19251446_Observational study of gene-disease association. (HuGE Navigator) 19251446_These results suggest that the C609T polymorphism of NQO1 is associated with carotid artery plaques in type 2 diabetic patients. 19330589_Observational study of genotype prevalence. (HuGE Navigator) 19336370_Observational study of gene-disease association. (HuGE Navigator) 19339270_Observational study of gene-disease association. (HuGE Navigator) 19360290_Findings suggest that mtDNA 4,977-bp deletion associated with NQO1 deficiency is involved in carcinogenesis and progression of breast cancer. 19360290_Observational study of gene-disease association and gene-gene int | ENSMUSG00000003849 | Nqo1 | 1.238660e+02 | 0.8537036 | -0.228192854 | 0.4186789 | 2.940229e-01 | 0.5876540056 | 0.90567577 | No | Yes | 1.406988e+02 | 22.712899 | 1.412042e+02 | 23.560090 | |
| ENSG00000181085 | 225689 | MAPK15 | protein_coding | Q8TD08 | FUNCTION: Atypical MAPK protein that regulates several process such as autophagy, ciliogenesis, protein trafficking/secretion and genome integrity, in a kinase activity-dependent manner (PubMed:22948227, PubMed:24618899, PubMed:29021280, PubMed:21847093, PubMed:20733054). Controls both, basal and starvation-induced autophagy throught its interaction with GABARAP, MAP1LC3B and GABARAPL1 leading to autophagosome formation, SQSTM1 degradation and reduced MAP1LC3B inhibitory phosphorylation (PubMed:22948227). Regulates primary cilium formation and the localization of ciliary proteins involved in cilium structure, transport, and signaling (PubMed:29021280). Prevents the relocation of the sugar-adding enzymes from the Golgi to the endoplasmic reticulum, thereby restricting the production of sugar-coated proteins (PubMed:24618899). Upon amino-acid starvation, mediates transitional endoplasmic reticulum site disassembly and inhibition of secretion (PubMed:21847093). Binds to chromatin leading to MAPK15 activation and interaction with PCNA, that which protects genomic integrity by inhibiting MDM2-mediated degradation of PCNA (PubMed:20733054). Regulates DA transporter (DAT) activity and protein expression via activation of RhoA (PubMed:28842414). In response to H(2)O(2) treatment phosphorylates ELAVL1, thus preventing it from binding to the PDCD4 3'UTR and rendering the PDCD4 mRNA accessible to miR-21 and leading to its degradation and loss of protein expression (PubMed:26595526). Also functions in a kinase activity-independent manner as a negative regulator of growth (By similarity). Phosphorylates in vitro FOS and MBP (PubMed:11875070, PubMed:16484222, PubMed:20638370, PubMed:19166846). During oocyte maturation, plays a key role in the microtubule organization and meiotic cell cycle progression in oocytes, fertilized eggs, and early embryos (By similarity). Interacts with ESRRA promoting its re-localization from the nucleus to the cytoplasm and then prevents its transcriptional activity (PubMed:21190936). {ECO:0000250|UniProtKB:Q80Y86, ECO:0000250|UniProtKB:Q9Z2A6, ECO:0000269|PubMed:11875070, ECO:0000269|PubMed:16484222, ECO:0000269|PubMed:19166846, ECO:0000269|PubMed:20638370, ECO:0000269|PubMed:20733054, ECO:0000269|PubMed:21190936, ECO:0000269|PubMed:21847093, ECO:0000269|PubMed:22948227, ECO:0000269|PubMed:24618899, ECO:0000269|PubMed:26595526, ECO:0000269|PubMed:28842414, ECO:0000269|PubMed:29021280}. | ATP-binding;Alternative splicing;Cell junction;Cell projection;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Golgi apparatus;Kinase;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Tight junction;Transferase;Ubl conjugation | hsa:225689; | autophagosome [GO:0005776]; bicellular tight junction [GO:0005923]; cell-cell junction [GO:0005911]; centriole [GO:0005814]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; extracellular region [GO:0005576]; Golgi apparatus [GO:0005794]; meiotic spindle [GO:0072687]; nucleus [GO:0005634]; ATP binding [GO:0005524]; chromatin binding [GO:0003682]; kinase activity [GO:0016301]; MAP kinase activity [GO:0004707]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; SH3 domain binding [GO:0017124]; cellular response to DNA damage stimulus [GO:0006974]; dopamine uptake [GO:0090494]; endoplasmic reticulum organization [GO:0007029]; intracellular signal transduction [GO:0035556]; negative regulation of cell migration [GO:0030336]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of metaphase/anaphase transition of meiosis I [GO:1905188]; positive regulation of spindle assembly [GO:1905832]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomere capping [GO:1904355]; positive regulation of telomere maintenance via telomerase [GO:0032212]; protein autophosphorylation [GO:0046777]; protein localization to ciliary transition zone [GO:1904491]; regulation of autophagy [GO:0010506]; regulation of cilium assembly [GO:1902017]; regulation of COPII vesicle coating [GO:0003400] | 11875070_ERK8, a new member of the mitogen-activated protein kinase family. 16336213_The activity of ERK8 in transfected HEK-293 cells depends on the relative rates of ERK8 autophosphorylation and dephosphorylation by one or more members of the PPP family of protein serine/threonine phosphatases. 16484222_Erk8 has a role as a novel effector of RET/PTC3 and, therefore, RET biological functions 16624805_human ERK8 has a role as a negative regulator of human GRalpha, acting through Hic-5 19166846_Regulation of the activity and expression of DERK8 by DNA damage are reported. 20395206_Extracellular signal-regulated kinase 8-mediated c-Jun phosphorylation increases tumorigenesis of human colon cancer 20733054_Data show that ERK8 prevents HDM2-mediated PCNA destruction by inhibiting the association of PCNA with HDM2, and implicate ERK8 in the regulation of genomic stability. 21190936_a novel function for ERK8 as a bona fide ERRalpha corepressor, involved in control of its cellular localization by nuclear exclusion, and suggest a key role for this MAP kinase in the regulation of the biological activities of this nuclear receptor. 22948227_ATG8-like proteins (MAP1LC3B, GABARAP and GABARAPL1) are novel interactors of MAPK15/ERK8, a MAP kinase involved in cell proliferation and transformation. 23326322_Data suggest that the model coulb be a tool for the development of specific ERK8 kinase inhibitors. 24618899_ERK8 appears as a constitutive brake on N-Acetylgalactosaminyltransferase relocalisation, and the loss of its expression could drive cancer aggressivity through increased cell motility. 26035356_The present study suggests that MAPK15 overexpression may contribute to the malignant transformation of gastric mucosa by prolonging the stability of c-Jun. 26291129_depletion of endogenous MAPK15 expression inhibited BCR-ABL1-dependent cell proliferation, in vitro 26595526_In HeLa cells, phosphorylation of HuR by ERK8 prevents it from binding to PDCD4 mRNA and allows miR-21-mediated degradation of PDCD4. 26988910_High MAPK15 expression is associated with male germ cell tumors. 29021280_Our results describe a primary cilia-related role for this poorly studied member of the MAPK family in vivo, and indicate a broad requirement for MAPK15 in the formation of multiple ciliary classes across species. 30070699_As multiple roles of MAPK15 were observed among these studies, therefore, it remains unclear whether MAPK15 acts as a proto-oncogene or tumor suppressor. Here, the recent literature on human MAPK15 and the resulting functions will be discussed. 30131341_MAPK15 stimulates ULK1, is part of the ULK complex, colocalizes in autophagosomes, and is involved in autophagosome biogenesis. 34224814_MAPK15-ULK1 signaling regulates mitophagy of airway epithelial cell in chronic obstructive pulmonary disease. | ENSMUSG00000063704 | Mapk15 | 3.153518e+02 | 0.6045084 | -0.726165781 | 0.3058561 | 5.579514e+00 | 0.0181717651 | 0.48162965 | No | Yes | 1.434694e+02 | 44.037915 | 2.870537e+02 | 90.042507 | ||
| ENSG00000181192 | 55526 | DHTKD1 | protein_coding | Q96HY7 | FUNCTION: The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components: 2-oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3) (By similarity). {ECO:0000250}. | 3D-structure;Charcot-Marie-Tooth disease;Disease variant;Glycolysis;Mitochondrion;Neurodegeneration;Neuropathy;Oxidoreductase;Reference proteome;Thiamine pyrophosphate;Transit peptide | This gene encodes a component of a mitochondrial 2-oxoglutarate-dehydrogenase-complex-like protein involved in the degradation pathways of several amino acids, including lysine. Mutations in this gene are associated with 2-aminoadipic 2-oxoadipic aciduria and Charcot-Marie-Tooth Disease Type 2Q. [provided by RefSeq, May 2013]. | hsa:55526; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; oxoglutarate dehydrogenase (succinyl-transferring) activity [GO:0004591]; thiamine pyrophosphate binding [GO:0030976]; generation of precursor metabolites and energy [GO:0006091]; glycolytic process [GO:0006096]; hematopoietic progenitor cell differentiation [GO:0002244]; tricarboxylic acid cycle [GO:0006099] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 23141293_DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. 23141294_A nonsense mutation in DHTKD1 causes Charcot-Marie-Tooth disease type 2 in a large Chinese pedigree. 24076469_DHTKD1 contributes to mitochondrial biogenesis and function maintenance. 25860818_DHTKD1 encodes the E1 subunit of the alpha-ketoadipic acid dehydrogenase complex 32024885_Synthetic analogues of 2-oxo acids discriminate metabolic contribution of the 2-oxoglutarate and 2-oxoadipate dehydrogenases in mammalian cells and tissues. 32160276_DHTKD1 and OGDH display substrate overlap in cultured cells and form a hybrid 2-oxo acid dehydrogenase complex in vivo. 32303640_Structure-function analyses of the G729R 2-oxoadipate dehydrogenase genetic variant associated with a disorder of l-lysine metabolism. 32633484_Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex. 34484123_Knock-Out of DHTKD1 Alters Mitochondrial Respiration and Function, and May Represent a Novel Pathway in Cardiometabolic Disease Risk. 35052424_Heterozygous DHTKD1 Variants in Two European Cohorts of Amyotrophic Lateral Sclerosis Patients. | ENSMUSG00000025815 | Dhtkd1 | 2.063425e+03 | 0.9614873 | -0.056660363 | 0.2557214 | 4.947076e-02 | 0.8239867643 | 0.96654947 | No | Yes | 1.916731e+03 | 175.684824 | 1.981715e+03 | 186.281295 | |
| ENSG00000181450 | 339500 | ZNF678 | protein_coding | Q5SXM1 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA; + ;NA; + ;NA; + ;NA | Mouse_homologues mmu:629016;;NA;NA;NA;NA | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20546612_Observational study of gene-disease association. (HuGE Navigator) 20846217_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000098905+ENSMUSG00000114923+ENSMUSG00000055341+ENSMUSG00000057842+ENSMUSG00000048280 | Zfp953+Gm49345+Zfp457+Zfp595+Zfp738 | 1.412876e+02 | 1.4450801 | 0.531149485 | 0.3471013 | 2.404492e+00 | 0.1209873653 | 0.75783482 | No | Yes | 1.990620e+02 | 29.603290 | 1.317969e+02 | 19.934001 | |
| ENSG00000182149 | 9798 | IST1 | protein_coding | P53990 | FUNCTION: ESCRT-III-like protein involved in cytokinesis, nuclear envelope reassembly and endosomal tubulation (PubMed:19129479, PubMed:26040712, PubMed:28242692). Is required for efficient abscission during cytokinesis (PubMed:19129479). Involved in recruiting VPS4A and/or VPS4B to the midbody of dividing cells (PubMed:19129480, PubMed:19129479). During late anaphase, involved in nuclear envelope reassembly and mitotic spindle disassembly together with the ESCRT-III complex: IST1 acts by mediating the recruitment of SPAST to the nuclear membrane, leading to microtubule severing (PubMed:26040712). Recruited to the reforming nuclear envelope (NE) during anaphase by LEMD2 (PubMed:28242692). Regulates early endosomal tubulation together with the ESCRT-III complex by mediating the recruitment of SPAST (PubMed:23897888). {ECO:0000269|PubMed:19129479, ECO:0000269|PubMed:19129480, ECO:0000269|PubMed:23897888, ECO:0000269|PubMed:26040712, ECO:0000269|PubMed:28242692}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a protein with MIT-interacting motifs that interacts with components of endosomal sorting complexes required for transport (ESCRT). ESCRT functions in vesicle budding, such as that which occurs during membrane abscission in cytokinesis. There is a pseudogene for this gene on chromosome 19. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2012]. | hsa:9798; | azurophil granule lumen [GO:0035578]; centrosome [GO:0005813]; cytosol [GO:0005829]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; Flemming body [GO:0090543]; intracellular membrane-bounded organelle [GO:0043231]; midbody [GO:0030496]; nuclear envelope [GO:0005635]; cadherin binding [GO:0045296]; identical protein binding [GO:0042802]; MIT domain binding [GO:0090541]; protein domain specific binding [GO:0019904]; protein-containing complex binding [GO:0044877]; abscission [GO:0009838]; cell division [GO:0051301]; cytoskeleton-dependent cytokinesis [GO:0061640]; ESCRT III complex disassembly [GO:1904903]; establishment of protein localization [GO:0045184]; multivesicular body assembly [GO:0036258]; positive regulation of collateral sprouting [GO:0048672]; positive regulation of proteolysis [GO:0045862]; protein localization [GO:0008104]; protein transport [GO:0015031]; viral capsid secondary envelopment [GO:0046745]; viral release from host cell [GO:0019076] | 19001599_OLC1 (KIAA0174) is a candidate oncogene in lung cancer whose expression may be regulated by exposure to cigarette smoke. 19129479_IST1 and CHMP1 act together to recruit and modulate specific VPS4 activities required during the final stages of cell division. 19129480_hIST1 is essential for cytokinesis in mammalian cells. 19525971_Data show that the N-terminal core domains of increased sodium tolerance-1 (IST1) and charged multivesicular body protein-3 (CHMP3) form equivalent four-helix bundles, revealing that IST1 is a previously unrecognized ESCRT-III family member. 20719964_These data suggest that Ist1 interaction is important for spartin recruitment to the midbody and that spartin participates in cytokinesis. 20849418_Results demonstrate that human calpain 7 is proteolytically active, and imply that calpain 7 is activated by ESCRT-III-related protein IST1. 21439932_These results demonstrate that cigarette smoke regulates OLC1 expression in lung cancer cells by compromising its ubiquitination and subsequent degradation through the ubiquitin E3 ligase APC. 21616915_The interaction between CL7MIT and CHMP1B and between CL7MIT and IST1 became stronger when IST1 or CHMP1B was additionally coexpressed, suggesting formation of ternary complex of calpain-7, IST1 and CHMP1B. 23609236_OLC1 over-expression is an important factor in epithelial ovarian carcinoma prognosis and can be a potential biomarker for ovarian carcinoma. 23649269_the binding of ALG-2 to IST1 is Ca(2+)-dependent 23897888_The results suggest that inclusion of IST1 into the ESCRT complex allows recruitment of spastin to promote fission of recycling tubules from the endosome. 24403489_Over-expression of the OCL1 is associated with colorectal cancer. 24608342_Overexpression of OLC1 promotes tumorigenesis of human esophageal squamous cell carcinoma. 24880589_Findings indicate that a high expression level of overexpressed in lung cancer 1 (OLC1) serves as a biomarker for poor prognosis for breast cancer, suggesting that OLC1 may be a potential target of antiangiogenic therapy for breast cancer. 25657007_Crystal structures of three molecular complexes reveal that IST1 binds to the MIT domains of VPS4 and LIP5. 26011858_Structural and biochemical studies reveal an unusually tight interaction between ULK3 and IST1, an ESCRT-III subunit required for cytokinetic abscission. 28038462_Nuclear overexpression of OLC1 was associated with clinicopathological parameters and could predict a poor overall survival in gastric adenocarcinoma. 28532543_The expression of OLC1 in the tumor tissues of lung adenocarcinoma is higher than that in squamous cell carcinoma. 30110633_Visualization of IST1 structures in cells lacking the microtubule-severing enzyme spastin and in cells depleted of specific ESCRT-III components or the ATPase VPS4 demonstrated the contribution of these components to the organization and function of ESCRTs in cells. 32251413_To learn how certain ESCRT-IIIs shape positively curved membranes, we determined structures of human membrane-bound CHMP1B-only, membrane-bound CHMP1B + IST1, and IST1-only filaments by cryo-EM | ENSMUSG00000031729 | Ist1 | 4.797712e+03 | 0.8010544 | -0.320027848 | 0.2824723 | 1.296619e+00 | 0.2548317138 | 0.79582240 | No | Yes | 3.582156e+03 | 363.820903 | 4.581453e+03 | 476.709613 | |
| ENSG00000182173 | 283989 | TSEN54 | protein_coding | Q7Z6J9 | FUNCTION: Non-catalytic subunit of the tRNA-splicing endonuclease complex, a complex responsible for identification and cleavage of the splice sites in pre-tRNA. It cleaves pre-tRNA at the 5' and 3' splice sites to release the intron. The products are an intron and two tRNA half-molecules bearing 2',3' cyclic phosphate and 5'-OH termini. There are no conserved sequences at the splice sites, but the intron is invariably located at the same site in the gene, placing the splice sites an invariant distance from the constant structural features of the tRNA body. The tRNA splicing endonuclease is also involved in mRNA processing via its association with pre-mRNA 3'-end processing factors, establishing a link between pre-tRNA splicing and pre-mRNA 3'-end formation, suggesting that the endonuclease subunits function in multiple RNA-processing events. {ECO:0000269|PubMed:15109492}. | Acetylation;Alternative splicing;Disease variant;Methylation;Neurodegeneration;Nucleus;Phosphoprotein;Pontocerebellar hypoplasia;Reference proteome;mRNA processing;tRNA processing | This gene encodes a subunit of the tRNA splicing endonuclease complex, which catalyzes the removal of introns from precursor tRNAs. The complex is also implicated in pre-mRNA 3-prime end processing. Mutations in this gene result in pontocerebellar hypoplasia type 2.[provided by RefSeq, Oct 2009]. | hsa:283989; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; tRNA-intron endonuclease complex [GO:0000214]; mRNA processing [GO:0006397]; tRNA splicing, via endonucleolytic cleavage and ligation [GO:0006388]; tRNA-type intron splice site recognition and cleavage [GO:0000379] | 18711368_In two subtypes, PCH2 and PCH4, we identified mutations in three of the four different subunits of the tRNA-splicing endonuclease complex. 20956791_We confirm that the common p.A307S mutation in TSEN54 is responsible for most of the patients with a PCH2 phenotype. 21383226_The results demonistrated that not all cases of clinically defined pontocerebellar hypoplasia-4 result from mutations in TSEN54. 21468723_TSEN54 mutation causes a severe form of pontocerebellar hypoplasia type 1 in a family. 24938831_A novel heterozygous mutation was found in the TSEN54 gene by c.254A > T(+) (p.E85V), which may be a new subtype of hereditary ataxia 26701950_TSEN54 gene-related pontocerebellar hypoplasia type 2 presented with exaggerated startle response in cousins. 32697043_A homozygote variant in the tRNA splicing endonuclease subunit 54 causes pontocerebellar hypoplasia in a consanguineous Iranian family. | ENSMUSG00000020781 | Tsen54 | 3.817400e+03 | 1.2727971 | 0.348002472 | 0.3031782 | 1.329718e+00 | 0.2488553218 | 0.79042389 | No | Yes | 3.954354e+03 | 521.543250 | 2.595825e+03 | 351.574661 | |
| ENSG00000182185 | 5890 | RAD51B | protein_coding | O15315 | FUNCTION: Involved in the homologous recombination repair (HRR) pathway of double-stranded DNA breaks arising during DNA replication or induced by DNA-damaging agents. May promote the assembly of presynaptic RAD51 nucleoprotein filaments. Binds single-stranded DNA and double-stranded DNA and has DNA-dependent ATPase activity. Part of the RAD21 paralog protein complex BCDX2 which acts in the BRCA1-BRCA2-dependent HR pathway. Upon DNA damage, BCDX2 acts downstream of BRCA2 recruitment and upstream of RAD51 recruitment. BCDX2 binds predominantly to the intersection of the four duplex arms of the Holliday junction and to junction of replication forks. The BCDX2 complex was originally reported to bind single-stranded DNA, single-stranded gaps in duplex DNA and specifically to nicks in duplex DNA. The BCDX2 subcomplex RAD51B:RAD51C exhibits single-stranded DNA-dependent ATPase activity suggesting an involvement in early stages of the HR pathway. {ECO:0000269|PubMed:11751635, ECO:0000269|PubMed:11751636, ECO:0000269|PubMed:11842113, ECO:0000269|PubMed:12441335, ECO:0000269|PubMed:23108668, ECO:0000269|PubMed:23149936}. | ATP-binding;Alternative splicing;Chromosomal rearrangement;DNA damage;DNA recombination;DNA repair;DNA-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene is a member of the RAD51 protein family. RAD51 family members are evolutionarily conserved proteins essential for DNA repair by homologous recombination. This protein has been shown to form a stable heterodimer with the family member RAD51C, which further interacts with the other family members, such as RAD51, XRCC2, and XRCC3. Overexpression of this gene was found to cause cell cycle G1 delay and cell apoptosis, which suggested a role of this protein in sensing DNA damage. Rearrangements between this locus and high mobility group AT-hook 2 (HMGA2, GeneID 8091) have been observed in uterine leiomyomata. [provided by RefSeq, Mar 2016]. | hsa:5890; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; Rad51B-Rad51C-Rad51D-XRCC2 complex [GO:0033063]; replication fork [GO:0005657]; ATP binding [GO:0005524]; ATP-dependent activity, acting on DNA [GO:0008094]; DNA binding [GO:0003677]; double-stranded DNA binding [GO:0003690]; single-stranded DNA binding [GO:0003697]; blastocyst growth [GO:0001832]; DNA recombination [GO:0006310]; DNA repair [GO:0006281]; double-strand break repair via homologous recombination [GO:0000724]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of G2/M transition of mitotic cell cycle [GO:0010971]; reciprocal meiotic recombination [GO:0007131]; somite development [GO:0061053] | 11744692_This work describes the in vitro and in vivo identification of the RAD51B/RAD51C heterocomplex 11978964_involved in the frequently occurring t(6;14) (p21;q23-->q24) in pulmonary chondroid hamartomas 12427746_Rad51B and Rad51C function through interactions with the human Rad51 recombinase and play a crucial role in the homologous recombinational repair pathway 12441335_Rad51B protein may have a specific function in Holliday junction processing in the homologous recombinational repair pathway in humans 14704354_a fragment of Rad51B containing amino acid residues 1-75 interacts with the C-terminus and linker of Rad51C, residues 79-376, and this region of Rad51C also interacts with mRad51D and Xrcc3 15701685_motif in the N-terminus of Rad51B serves as an NLS that allows Rad51B to localize to the nucleus independent of Rad51C or BRCA2 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19329439_EVL protein is a novel recombination factor that may be required for repairing specific DNA lesions, and that may cause tumor malignancy by its inappropriate expression. 19330030_A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). 19330030_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19657362_BCR/ABL fragments were used for identifying the sites of BCR/ABL interaction with RAD51B 19714462_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20095854_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20195514_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20237344_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20610542_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20610542_polymorphisms in and haplotypes of the RAD51L1 gene, which is involved in the double-strand break repair pathway, modulate gamma-radiation-induced mutagen sensitivity. 21368091_findings support the notion that DNA repair genes, in particular RAD51L1, play a role in nasopharyngeal carcinoma etiology and development 21533530_our results suggest that RAD51L1 is unlikely to represent a high-penetrance breast cancer susceptibility gene. 21852249_rs11249433 at 1p.11.2, and two highly correlated single-nucleotide polymorphisms rs999737 and rs10483813 (r(2)= 0.98) at 14q24.1 (RAD51L1), for up to 46 036 invasive breast cancer cases and 46 930 controls from 39 studies, were genotyped. 22017238_Single Nucleotide Polymorphisms in RAD51L1 gene is associated with glioblastoma. 22454379_Single nucleotide polymorphism in RAD51L1 is associated with breast cancer. 23001122_SNP in RAD51B at 14q24.1 was significantly associated with male breast cancer risk 23108668_Study observed centrosome defects in the absence of XRCC3. While RAD51B and RAD51C act early in homologous recombination, XRCC3 functions jointly with GEN1 later in the pathway at the stage of Holliday junction resolution. 23810717_The HsRAD51B-HsRAD51C complex plays a role in stabilizing the HsRAD51 nucleoprotein filament during the presynaptic phase of homologous recombination. 24022229_This study provides robust evidence for an association of rheumatoid arthritis susceptibility with genes involved in B cell differentiation (BACH2) and DNA repair (RAD51B). 24139550_It confirms that RAD51 paralog mutations confer breast and ovarian cancer predisposition and are rare events. 24498017_Data indicate that complement factor H (CFH) R1210C and common variants in COL8A1 and RAD51B plus six genes contribute predictive information for advanced macular degeneration (AMD) beyond macular and behavioral phenotypes. 24526414_the risk of developing AMD exhibits dose dependency as well as an epistatic combined effect in rs17105278 T>C and rs4902566 C>T carriers and that the elevated risk for rs17105278 T>C carriers may be due to decreased transcription of RAD51B. 25255808_Relative excess risk of breast cancer due to interaction between RAD51L1 single-nucleotide polymorphism and BMI. 25600502_a novel germ line RAD51B nonsense mutation, and reduced expression of RAD51B in melanoma cells indicating inactivation of RAD51B 26261251_Mutations in epithelial ovarian cancer cases were more frequent in RAD51C (14 occurrences, 0.41%) and RAD51D (12 occurrences, 0.35%) than in RAD51B (two occurrences, 0.06%). 26339569_The aim of the present study was to evaluate the relationship between prostate cancer risk and the presence of single nucleotide polymorphisms in the genes involved in Homologous recombination repair, RAD51, RAD51B, XRCC2 and XRCC3. 27149063_common variation is significantly associated with familial breast cancer risk 27334422_our study suggested that miRNA-binding site genetic variants of RAD51B may modify the susceptibility to cervical cancer, which is important to identify individuals with differential risk for this malignancy and to improve the effectiveness of preventive intervention. 27651161_over-expression of RAD51B promoted cell proliferation, aneuploidy, and drug resistance, while RAD51B knockdown led to G1 arrest and sensitized cells to 5-fluorouracil 27683114_hypermethylation of homologous recombination DNA repair genes including RAD51B and XRCC3 is associated with an inflamed phenotype in squamous cell cancers of the head and neck, lung and cervix. 28361912_We successfully identified a common variant, rs911263, as being significantly associated with the disease status . In addition, this SNP was shown to be related to erosion, a clinical assessment of disease severity in RA (P = 2.89 x 10(-5), OR = 0.52). These findings shed light on the role of RAD51B in the onset and severity of Rheumatoid arthritis (RA). 31584931_Here we provide three coherent sets of isogenic mutants, both in transformed and non-transformed human cells. Importantly, using these mutant lines, we report the unanticipated result that RAD51B has a less crucial role in homologous recombination than the other four paralogs, and find that all RAD51 paralogs are critically important for early functions during homologous recombination 32669601_Sequential role of RAD51 paralog complexes in replication fork remodeling and restart. 33622874_TBX15 rs98422, DNM3 rs1011731, RAD51B rs8017304, and rs2588809 Gene Polymorphisms and Associations With Pituitary Adenoma. 34060991_Serum Levels of ARMS2, COL8A1, RAD51B, and VEGF and their Correlations in Age-related Macular Degeneration. | ENSMUSG00000059060 | Rad51b | 9.340811e+01 | 0.8070239 | -0.309316632 | 0.4360727 | 5.334416e-01 | 0.4651635321 | 0.86741238 | No | Yes | 7.429485e+01 | 13.535379 | 9.264065e+01 | 17.123129 | |
| ENSG00000182224 | 124637 | CYB5D1 | protein_coding | Q6P9G0 | Alternative splicing;Heme;Iron;Metal-binding;Reference proteome | hsa:124637; | metal ion binding [GO:0046872] | Mouse_homologues 17971504_Cyb5d1 is most abundantly expressed in tissues rich in highly ciliated cells, such as olfactory sensory neurons, and is predicted to be important to cilia. | ENSMUSG00000044795 | Cyb5d1 | 1.169386e+03 | 0.8529239 | -0.229511010 | 0.3235584 | 5.148943e-01 | 0.4730278087 | 0.87032151 | No | Yes | 1.451385e+03 | 232.267397 | 1.906183e+03 | 312.716130 | |||
| ENSG00000182378 | 55344 | PLCXD1 | protein_coding | Q9NUJ7 | Cytoplasm;Reference proteome | This gene is the most terminal protein-coding gene in the pseudoautosomal (PAR) region on chromosomes X and Y. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Mar 2010]. | hsa:55344; | cytoplasm [GO:0005737]; phosphoric diester hydrolase activity [GO:0008081]; lipid metabolic process [GO:0006629] | ENSMUSG00000064247 | Plcxd1 | 3.435814e+03 | 1.0722815 | 0.100683756 | 0.2632312 | 1.457744e-01 | 0.7026066042 | 0.93624959 | No | Yes | 2.731683e+03 | 372.118445 | 2.690654e+03 | 375.817801 | |||
| ENSG00000182400 | 122553 | TRAPPC6B | protein_coding | Q86SZ2 | FUNCTION: Component of a transport protein particle (TRAPP) complex that may function in specific stages of inter-organelle traffic (PubMed:16025134, PubMed:16828797). Specifically involved in the early development of neural circuitry, likely by controlling the frequency and amplitude of intracellular calcium transients implicated in the regulation of neuron differentiation and survival (Probable). {ECO:0000269|PubMed:16025134, ECO:0000269|PubMed:16828797, ECO:0000305|PubMed:28626029}. | 3D-structure;Alternative splicing;Disease variant;Endoplasmic reticulum;Epilepsy;Golgi apparatus;Mental retardation;Neurogenesis;Reference proteome | TRAPPC6B is a component of TRAPP complexes, which are tethering complexes involved in vesicle transport (Kummel et al., 2005 [PubMed 16025134]).[supplied by OMIM, Mar 2008]. | hsa:122553; | cis-Golgi network [GO:0005801]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; trans-Golgi network [GO:0005802]; TRAPP complex [GO:0030008]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; nervous system development [GO:0007399]; regulation of GTPase activity [GO:0043087] | 27569842_This study identified a TRAPPC6B gene mutation associated to the RLS locus on chromosome 14. 28626029_This study provides clinical and functional evidence of the role of TRAPPC6B in brain development and function. | ENSMUSG00000020993 | Trappc6b | 2.577151e+02 | 0.7275301 | -0.458921134 | 0.3643977 | 1.539326e+00 | 0.2147181633 | 0.78763590 | No | Yes | 2.110927e+02 | 39.236392 | 2.686427e+02 | 50.997218 | |
| ENSG00000182511 | 2242 | FES | protein_coding | P07332 | FUNCTION: Tyrosine-protein kinase that acts downstream of cell surface receptors and plays a role in the regulation of the actin cytoskeleton, microtubule assembly, cell attachment and cell spreading. Plays a role in FCER1 (high affinity immunoglobulin epsilon receptor)-mediated signaling in mast cells. Acts down-stream of the activated FCER1 receptor and the mast/stem cell growth factor receptor KIT. Plays a role in the regulation of mast cell degranulation. Plays a role in the regulation of cell differentiation and promotes neurite outgrowth in response to NGF signaling. Plays a role in cell scattering and cell migration in response to HGF-induced activation of EZR. Phosphorylates BCR and down-regulates BCR kinase activity. Phosphorylates HCLS1/HS1, PECAM1, STAT3 and TRIM28. {ECO:0000269|PubMed:11509660, ECO:0000269|PubMed:15302586, ECO:0000269|PubMed:15485904, ECO:0000269|PubMed:16455651, ECO:0000269|PubMed:17595334, ECO:0000269|PubMed:18046454, ECO:0000269|PubMed:19001085, ECO:0000269|PubMed:19051325, ECO:0000269|PubMed:20111072, ECO:0000269|PubMed:2656706, ECO:0000269|PubMed:8955135}. | 3D-structure;ATP-binding;Alternative splicing;Cell junction;Cell membrane;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Golgi apparatus;Kinase;Lipid-binding;Membrane;Nucleotide-binding;Phosphoprotein;Proto-oncogene;Reference proteome;SH2 domain;Transferase;Tumor suppressor;Tyrosine-protein kinase | This gene encodes the human cellular counterpart of a feline sarcoma retrovirus protein with transforming capabilities. The gene product has tyrosine-specific protein kinase activity and that activity is required for maintenance of cellular transformation. Its chromosomal location has linked it to a specific translocation event identified in patients with acute promyelocytic leukemia but it is also involved in normal hematopoiesis as well as growth factor and cytokine receptor signaling. Alternative splicing results in multiple variants encoding different isoforms.[provided by RefSeq, Jan 2009]. | hsa:2242; | cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; focal adhesion [GO:0005925]; Golgi apparatus [GO:0005794]; microtubule cytoskeleton [GO:0015630]; ATP binding [GO:0005524]; immunoglobulin receptor binding [GO:0034987]; microtubule binding [GO:0008017]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; phosphatidylinositol binding [GO:0035091]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; signaling receptor binding [GO:0005102]; cell adhesion [GO:0007155]; cell differentiation [GO:0030154]; cellular response to vitamin D [GO:0071305]; centrosome cycle [GO:0007098]; chemotaxis [GO:0006935]; innate immune response [GO:0045087]; microtubule bundle formation [GO:0001578]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of actin cytoskeleton reorganization [GO:2000251]; positive regulation of microtubule polymerization [GO:0031116]; positive regulation of monocyte differentiation [GO:0045657]; positive regulation of myeloid cell differentiation [GO:0045639]; positive regulation of neuron projection development [GO:0010976]; protein autophosphorylation [GO:0046777]; regulation of cell adhesion [GO:0030155]; regulation of cell differentiation [GO:0045595]; regulation of cell motility [GO:2000145]; regulation of cell population proliferation [GO:0042127]; regulation of cell shape [GO:0008360]; regulation of mast cell degranulation [GO:0043304]; regulation of vesicle-mediated transport [GO:0060627]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11994747_Closing in on the biological functions of Fps/Fes and Fer. A review. 12653561_Fes naturally adopts an inactive conformation in vivo, and maintenance of the inactive structure requires the coiled-coil and SH2 domains. 12871378_Fps/Fes and Fer are expressed in human and mouse platelets, and are activated following stimulation with collagen and collagen-related peptide (CRP), suggesting a role in GPVI receptor signaling 15003822_Fes transduces inductive signals for terminal macrophage and granulocyte differentiation, and this biological activity is mediated through the activation of lineage-specific transcription factors. 15099290_FPS mediates enhanced sensitization to VEGF and PDGF signaling in ECs; this hypersensitization contributes to excessive angiogenic signaling and underlies the observed hypervascular phenotype of human myristoylated FPS expressed in transgenic mice. 15485904_c-Fes is a regulator of the tubulin cytoskeleton and may contribute to Fes-induced morphological changes in myeloid hematopoietic and neuronal cells 15630569_NMR assignment of the SH2 domain 15713745_PEDF downregulates Fyn through Fes, resulting in inhibition of FGF-2-induced capillary morphogenesis of endothelial cells 15929003_NMR analysis of Src homology 2 domain from the human feline sarcoma oncogene Fes 16455651_FES has a growth suppressive function in colorectal neoplasms. 16792528_A novel Fes-KAP-1 interaction is reported, suggesting a dual role for KAP-1 as both a Fes activator and downstream effector. 17521372_These novel results indicate the involvement of Fes in VEGF-A-induced cellular responses by cultured endothelial cells. 17595334_shows a major function of FES downstream of activated KIT receptor and thereby points to FES as a novel target in KIT-related pathologi 18046454_Ezrin/Fes interaction at cell-cell contacts plays an essential role in hepatocyte growth factor-induced cell scattering and implicates Fes in the cross-talk between cell-cell and cell-matrix adhesion. 18775312_The SH2 and catalytic domains of active Fes and Abl pro-oncogenic kinases form integrated structures essential for effective tyrosine kinase signaling. 19001085_study shows Fes phosphorylates C-terminal tyrosine residues in HS1 implicated in actin stabilization; coordinated action of F-BAR & SH2 domains of Fes allow for coupling to FcepsilonRI signaling & potential regulation of actin reorganization in mast cells 19051325_Study of promoter methylation as important mechanism responsible for downregulation of FES gene expression in colorectal cancer cells. Treatment with DNA methyltransferase inhibitor resulted in expression of functional FES transcripts in CRC cell lines. 19082481_Downregulation of the c-Fes protein-tyrosine kinase inhibits the proliferation of renal carcinoma. 19382747_c-Fes oligomerization is independent of activation; data suggest that conformational changes, rather than oligomerization, govern c-Fes kinase activation and downstream signaling in vivo. 19464057_Observational study of gene-disease association. (HuGE Navigator) 19865112_Observational study of gene-disease association. (HuGE Navigator) 19885553_FGF-2 activates Fes via the second coiled-coil domain, leading to lamellipodium formation and chemotaxis by endothelial cells 19913121_Observational study of gene-disease association. (HuGE Navigator) 20030920_c-fes gene expression was found in myeloid leukemias, whereas low or no expression in lymphocytic leukemias. 20117079_FES kinase is a mediator of wild-type KIT signalling implicated in cell migration. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23404507_analysis of the JAK-Fes-phospholipase D signaling pathway that is enhanced in highly proliferative breast cancer cells 31573955_Identification of FES as a Novel Radiosensitizing Target in Human Cancers. 31746983_This study supports the critical role of FER and FES tyrosine kinase fusions in the pathogenesis of follicular T-cell lymphoma and provides additional evidence that these can drive follicular T-cell lymphoma in the absence of RHOA mutations. | ENSMUSG00000053158 | Fes | 3.118662e+01 | 0.9393523 | -0.090261759 | 0.5618458 | 2.423606e-02 | 0.8762857681 | 0.97687083 | No | Yes | 2.802579e+01 | 6.549749 | 2.482325e+01 | 6.192950 | |
| ENSG00000182544 | 84975 | MFSD5 | protein_coding | Q6N075 | FUNCTION: Mediates high-affinity intracellular uptake of the rare oligo-element molybdenum. {ECO:0000269|PubMed:21464289}. | Alternative splicing;Cell membrane;Ion transport;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:84975; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; molybdate ion transmembrane transporter activity [GO:0015098] | Mouse_homologues 27272503_Results show that Mfsd5 and Mfsd11 are both affected by altered energy homeostasis, suggesting plausible involvement in the energy regulation. Moreover, the first histological mapping of MFSD5 and MFSD11 shows ubiquitous expression in the periphery and the central nervous system of mice, where the proteins are expressed in excitatory and inhibitory mouse brain neurons. | ENSMUSG00000045665 | Mfsd5 | 2.245940e+03 | 1.1162511 | 0.158661563 | 0.3260775 | 2.372687e-01 | 0.6261858045 | 0.91646120 | No | Yes | 2.064302e+03 | 226.676278 | 1.813570e+03 | 204.512204 | ||
| ENSG00000182872 | 8241 | RBM10 | protein_coding | P98175 | FUNCTION: May be involved in post-transcriptional processing, most probably in mRNA splicing. Binds to RNA homopolymers, with a preference for poly(G) and poly(U) and little for poly(A) (By similarity). May bind to specific miRNA hairpins (PubMed:28431233). {ECO:0000250|UniProtKB:P70501, ECO:0000269|PubMed:18315527, ECO:0000269|PubMed:28431233}. | 3D-structure;Acetylation;Alternative splicing;Metal-binding;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Zinc;Zinc-finger;mRNA processing;mRNA splicing | This gene encodes a nuclear protein that belongs to a family proteins that contain an RNA-binding motif. The encoded protein associates with hnRNP proteins and may be involved in regulating alternative splicing. Defects in this gene are the cause of the X-linked recessive disorder, TARP syndrome. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Mar 2011]. | hsa:8241; | nuclear speck [GO:0016607]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; miRNA binding [GO:0035198]; RNA binding [GO:0003723]; 3'-UTR-mediated mRNA stabilization [GO:0070935]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of mRNA splicing, via spliceosome [GO:0048025]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of smooth muscle cell apoptotic process [GO:0034393]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381] | 16552754_From these results, it seems that the X-chromosome, through its RBM genes, plays a formerly unknown role in the regulation of programmed cell death (apoptosis) in breast cancer. 18315527_S1-1 constitutes hundreds of nuclear domains, which dynamically change their structures in a reversible manner; upon globally reducing RNA polymerase II transcription, S1-1 nuclear bodies enlarge and decrease in number. 18820371_results indicate that very few genes are involved in the last steps of the apoptotic cascade in breast cancer, among them one of the X-chromosome RBM family 20451169_Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. 23294349_S1-1 contains multiple nuclear localisation sequence that act cooperatively. 24000153_This study established RBM10 as an important regulator of alternative splicing, presented a mechanistic model for RBM10-mediated splicing regulation and provided a molecular link to understanding a human congenital disorder. 24332178_Antagonizes the effects of RBM5, RBM6, and RBM10 in cell colony formation. 24530524_RBM10 regulates alternative splicing of Fas and Bcl-x genes. 25889998_The ability of RBM10v1 to regulate alternative splicing depends, at least in part, on a structural alteration within the second RNA recognition motif domain, and correlates with preferential expression of the NUMB exon 11 inclusion variant. 26751795_Src family tyrosine kinase signaling may regulate FilGAP through association with RBM10 26853560_RBM10 is a tumor suppressor that represses Notch signaling and cell proliferation through the regulation of NUMB alternative splicing 26998913_RBM10-TFE3 fusion variant (from chromosome X paracentric inversion), therefore, appears to be a recurrent molecular event in Xp11.2 RCCs. RBM10-TFE3 fusion should be added in the list of screened fusion transcripts in targeted molecular diagnostic multiplex RT-PCR 28091594_RBM10 mutations contribute to lung adenocarcinoma pathogenesis. 28288037_Xp11 translocation renal cell carcinomas with RBM10-TFE3 gene fusion demonstrating melanotic features and overlapping morphology with t(6;11) RCC. 28296677_RBM10-TFE3 is a recurrent gene fusion in Xp11 translocation renal cell carcinoma. 28347232_RNA-binding motif 10 messenger RNA and protein were reduced in lung adenocarcinoma tissues, and RNA-binding motif 10 overexpression inhibited lung adenocarcinoma cancer cell malignant behavior in vitro. Molecularly, RNA-binding motif 10 regulates many gene pathways involving in the tumor development or progression. 28379442_RBM10 functions as a splicing regulator using two RNA-binding units with different specificities to promote exon skipping. 28586478_RNA binding motif protein 10 (RBM10) negatively regulates its own mRNA and protein expression and that of RNA binding motif protein 5 (RBM5) by promoting alternative splicing-coupled nonsense-mediated mRNA decay (AS-NMD). 28634282_The high rate of TERT promoter mutations, MED12 mutations, RBM10 mutations, and chromosome 1q gain highlight their likely association with tumor virulence 28662214_results provide evidence that RBM10 expression, in RBM5-null tumors, may contribute to tumor growth and metastasis. Measurement of both RBM10 and RBM5 expression in clinical samples may therefore hold prognostic and/or potentially predictive value 28728573_Our work has not only expanded the number of pre-mRNA targets for RBM10, but identified RBM10 as a novel regulator of SMN2 alternative inclusion. 29274279_RBM10: Harmful or helpful-many factors to consider. 29450990_The well-known high-fidelity RNA splice site recognition by RBM10, and probably by RBM5 and RBM6, can thus be largely rationalized by a cooperative binding action of RRM and ZnF domains 30257214_RBM10 binds the pre-mRNA UTR, assembles the Star-PAP complex, and guides this complex specifically to mRNAs encoding regulators of cardiac hypertrophy. 30403180_RBM10 acts as a tumor suppressor in osteosarcoma. This could enable to define a new strategy for diagnosis and treatment of patients with osteosarcoma. 30483773_we found that RBM10 activated key proliferative signaling pathways [such as the epidermal growth factor receptor (EGFR), mitogenactivated protein kinase (MAPK) and phosphoinositide 3kinase (PI3K)AKT pathways] and inhibited apoptotic pathways. In addition, we demonstrated that a high expression of RBM10 protein in patient tissue samples was associated with a shorter overall survival time and a poor prognosis 30908700_The present study suggests that the carcinogenesis of RBM10-TFE3 RCC in some, but not all, patients may be associated with chronic kidney disease. 30955253_RBM10 reduces the phosphorylation of CREB via the AKT signalling pathway, suggesting that RBM10 exhibits its effect on lung adenocarcinoma cell proliferation via the RAP1/AKT/CREB signalling pathway. 31591476_RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53. 31820547_RBM10 regulates centriole duplication in HepG2 cells by ectopically assembling PLK4-STIL complexes in the nucleus. 32432721_Dengue virus targets RBM10 deregulating host cell splicing and innate immune response. 32572914_Protective effect of the RNA-binding protein RBM10 in hepatocellular carcinoma. 32812661_A novel missense variant in RBM10 can cause a mild form of TARP syndrome with developmental delay and dysmorphic features. 32947864_RBM10, a New Regulator of p53. 33064970_The role of RBM10 mutations in the development, treatment, and prognosis of lung adenocarcinoma. 33130397_RNA binding motif protein 10 suppresses lung cancer progression by controlling alternative splicing of eukaryotic translation initiation factor 4H. 33340101_Phenotypic spectrum of the RBM10-mediated intellectual disability and congenital malformation syndrome beyond classic TARP syndrome features. 33515724_RBM10: Structure, functions, and associated diseases. 34576144_Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association. 34638866_Sequestration of RBM10 in Nuclear Bodies: Targeting Sequences and Biological Significance. | ENSMUSG00000031060 | Rbm10 | 8.071920e+03 | 1.1708380 | 0.227541470 | 0.3066738 | 5.495681e-01 | 0.4584942322 | 0.86366237 | No | Yes | 8.355051e+03 | 876.294253 | 6.360851e+03 | 684.538793 | |
| ENSG00000182957 | 221178 | SPATA13 | protein_coding | Q96N96 | FUNCTION: Acts as guanine nucleotide exchange factor (GEF) for RHOA, RAC1 and CDC42 GTPases. Regulates cell migration and adhesion assembly and disassembly through a RAC1, PI3K, RHOA and AKT1-dependent mechanism. Increases both RAC1 and CDC42 activity, but decreases the amount of active RHOA. Required for MMP9 up-regulation via the JNK signaling pathway in colorectal tumor cells. Involved in tumor angiogenesis and may play a role in intestinal adenoma formation and tumor progression. {ECO:0000269|PubMed:17145773, ECO:0000269|PubMed:17599059, ECO:0000269|PubMed:19151759, ECO:0000269|PubMed:19893577, ECO:0000269|PubMed:19934221}. | Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome;SH3 domain | hsa:221178; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; filopodium [GO:0030175]; lamellipodium [GO:0030027]; nucleoplasm [GO:0005654]; ruffle membrane [GO:0032587]; guanyl-nucleotide exchange factor activity [GO:0005085]; identical protein binding [GO:0042802]; cell migration [GO:0016477]; filopodium assembly [GO:0046847]; lamellipodium assembly [GO:0030032]; regulation of cell migration [GO:0030334]; regulation of small GTPase mediated signal transduction [GO:0051056] | 17599059_These results suggest that similar to Asef, Asef2 plays an important role in cell migration, and that Asef2 activated by truncated mutant APC is required for aberrant migration of colorectal tumor cells. 19151759_Asef2, Neurabin2 and APC cooperatively regulate actin cytoskeletal organization and are required for HGF-induced cell migration. 19910543_Observational study of gene-disease association. (HuGE Navigator) 19934221_Asef2 activates Rac1 to modulate adhesion and actin dynamics and thereby regulate cell migration. 24144700_A new role for Rac activation, promoted by Asef2, in modulating actomyosin contractility. 24874604_Phosphorylation of S106 modulates Asef2 guanine nucleotide exchange factor activity and Asef2-mediated cell migration and adhesion turnover. 32339198_Mutations in SPATA13 cause primary angle closure glaucoma. | ENSMUSG00000021990 | Spata13 | 1.964893e+02 | 0.9988632 | -0.001640990 | 0.3371031 | 2.385263e-05 | 0.9961032146 | 0.99926282 | No | Yes | 2.048793e+02 | 31.153421 | 2.121267e+02 | 33.122526 | ||
| ENSG00000182979 | 9112 | MTA1 | protein_coding | Q13330 | FUNCTION: Transcriptional coregulator which can act as both a transcriptional corepressor and coactivator. As a part of the histone-deacetylase multiprotein complex (NuRD), regulates transcription of its targets by modifying the acetylation status of the target chromatin and cofactor accessibility to the target DNA. In conjunction with other components of NuRD, acts as a transcriptional corepressor of BRCA1, ESR1, TFF1 and CDKN1A. Acts as a transcriptional coactivator of BCAS3, PAX5 and SUMO2, independent of the NuRD complex. Stimulates the expression of WNT1 by inhibiting the expression of its transcriptional corepressor SIX3. Regulates p53-dependent and -independent DNA repair processes following genotoxic stress. Regulates the stability and function of p53/TP53 by inhibiting its ubiquitination by COP1 and MDM2 thereby regulating the p53-dependent DNA repair. Plays an important role in tumorigenesis, tumor invasion, and metastasis. Involved in the epigenetic regulation of ESR1 expression in breast cancer in a TFAP2C, IFI16 and HDAC4/5/6-dependent manner. Plays a role in the regulation of the circadian clock and is essential for the generation and maintenance of circadian rhythms under constant light and for normal entrainment of behavior to light-dark (LD) cycles. Positively regulates the CLOCK-ARNTL/BMAL1 heterodimer mediated transcriptional activation of its own transcription and the transcription of CRY1. Regulates deacetylation of ARNTL/BMAL1 by regulating SIRT1 expression, resulting in derepressing CRY1-mediated transcription repression. Isoform Short binds to ESR1 and sequesters it in the cytoplasm and enhances its non-genomic responses. With TFCP2L1, promotes establishment and maintenance of pluripotency in embryonic stem cells (ESCs) and inhibits endoderm differentiation (By similarity). {ECO:0000250|UniProtKB:Q8K4B0, ECO:0000269|PubMed:16617102, ECO:0000269|PubMed:17671180, ECO:0000269|PubMed:17922032, ECO:0000269|PubMed:19837670, ECO:0000269|PubMed:21965678, ECO:0000269|PubMed:24413532}. | 3D-structure;Acetylation;Activator;Alternative splicing;Biological rhythms;Cytoplasm;Cytoskeleton;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein that was identified in a screen for genes expressed in metastatic cells, specifically, mammary adenocarcinoma cell lines. Expression of this gene has been correlated with the metastatic potential of at least two types of carcinomas although it is also expressed in many normal tissues. The role it plays in metastasis is unclear. It was initially thought to be the 70kD component of a nucleosome remodeling deacetylase complex, NuRD, but it is more likely that this component is a different but very similar protein. These two proteins are so closely related, though, that they share the same types of domains. These domains include two DNA binding domains, a dimerization domain, and a domain commonly found in proteins that methylate DNA. The profile and activity of this gene product suggest that it is involved in regulating transcription and that this may be accomplished by chromatin remodeling. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2011]. | hsa:9112; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; microtubule [GO:0005874]; nuclear envelope [GO:0005635]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; NuRD complex [GO:0016581]; chromatin binding [GO:0003682]; histone deacetylase binding [GO:0042826]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription coactivator activity [GO:0003713]; transcription corepressor activity [GO:0003714]; zinc ion binding [GO:0008270]; chromatin remodeling [GO:0006338]; circadian regulation of gene expression [GO:0032922]; double-strand break repair [GO:0006302]; entrainment of circadian clock by photoperiod [GO:0043153]; histone deacetylation [GO:0016575]; locomotor rhythm [GO:0045475]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of protein autoubiquitination [GO:1902499]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; regulation of gene expression, epigenetic [GO:0040029]; response to ionizing radiation [GO:0010212]; signal transduction [GO:0007165] | 11804687_expression is closely related to invasiveness and metastasis in non-small-cell lung carcinoma 11948399_metastasis-associated protein (MTA)1 enhances migration, invasion, and anchorage-independent survival of immortalized human keratinocytes 12167865_naturally occurring short form that contains a previously unknown sequence of 33 amino acids with an ER-binding motif, Leu-Arg-Ile-Leu-Leu (LRILL); MTA1s localizes in the cytoplasm, sequesters ER in the cytoplasm, and enhances non-genomic responses of ER 12431981_MTA1 binds to the MMP-9 promoter, thereby repressing expression of this type IV collagenase via histone-dependent and independent mechanisms 12527756_interaction with MAT1 and regulation of estrogen receptor transactivation functions 12650603_The results suggest that the MTA1 protein may serve multiple functions in cellular signaling, chromosome remodeling and transcription processes that are important in the progression, invasion and growth of metastatic epithelial cells. 12684630_High expression of the MTA1 gene is associated with hepatocellular carcinoma 12920132_MTA1 and MTA2 repress transcription specifically, are located in the nucleus, and contain associated histone deacetylase activity; MTA1 associates with a different set of transcription factors from MTA2 14735193_enhanced expression of MTA1 promotes the acquisition of an invasive, metastatic phenotype, and thus enhances the malignancy of pancreatic adenocarcinoma cells by modulation of the cytoskeleton 14871807_This study identified an association of MTA1 expression and prostate cancer progression. 15077195_MTA1s interacts with CKI-gamma2 in vitro and in vivo and colocalizes in the cytoplasm 15095300_overexpression of MTA1 protein and acetylation level of histone H4 protein are closely related 15254226_Metastasis-associated protein 1 interacts with NRIF3, an estrogen-inducible nuclear receptor coregulator 16172399_MTA1 is one of the first downstream targets of c-MYC function that are essential for the transformation potential of c-MYC. 16244788_MTA1 overexpression is associated with high recurrence of breast cancer 16502042_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16617102_role for MTA1-BCAS3 pathway in promoting cancerous phenotypes in breast tumor cells 16646402_Overexpression of the MTA1 gene correlates with lymph node metastasis of carcinomas of the larynx. 16807247_structure and antiestrogenic activity of the unique C-terminal, NR-box motif-containing region of MTA1 16831056_These studies demonstrate that MTA1 is expressed in both benign and malignant neoplasms. While its expression is associated with tissue invasion it may not be sufficient for the progression of neoplasms to metastatic stages. 16855396_BCAS3 overexpression in hormone receptor-positive premenopausal breast cancer seemed to be associated with impaired responses to tamoxifen 16969516_MTA1 enhances angiogenesis by stabilization of the HIF-1alpha protein, which is closely related to the increased metastatic potential of cancer cells with high MTA1 expression 17666527_role for the MTA1 as an upstream modifier of Six3 and indicate that Six3 is a direct stimulator of rhodopsin expression. 17868030_Results suggest that MTA1 promotes the metastatic ability of B16F10 cancer cells. 17922032_These findings strongly implicate MTA1 in the transcriptional repression of BRCA1 leading to abnormal centrosome number and chromosomal instability. 17922035_study shows HSF1 binds to MTA1 in vitro & in breast carcinoma; repression of estrogen-dependent transcription may contribute to role of HSF1 in cancer 18196870_MTA1 expression was significantly higher in noninvasive breast cancer cell lines than in invasive ones. 18306220_MTA1 is closely associated with microvascular invasion, frequent postoperative recurrence, and poor survival of HCC patients, especially in those with HBV-associated HCC. 18362831_HIF-1alpha expression may be regulated through HDAC1/MTA1, which is associated with a poor prognosis for pancreatic carcinoma 18640824_Histologically, MTA1 protein production was strongly associated with cancer cell invasion, and clinically there was a correlation between lymph node metastasis and MTA1 protein production. 18719363_MTA1 is up-regulated in advanced ovarian cancer, represses ERbeta, and enhances expression of GRO. 18769059_This study was undertaken to explore the potential role of MTA1 in mouse liver. 19363681_the high expression level of MTA proteins in human chorionic cells might facilitate trophoblast cell migration and neoangiogenesis 19366061_ezrin and metastatic tumor antigen positivity can be additional prognostic markers in osteosarcoma of the jaw. 19377441_Metastasis-associated gene 1 expression is significantly associated with malignant behavior in pancreatic endocrine tumors. 19403384_MTA1 may promote lung carcinogenesis by enhancing HIF-1alpha protein activity. 19584269_Results suggest that miR-661 be further investigated for therapeutic use in down-regulating the expression of MTA1 in cancer cells. 19805145_MTA1 is required for optimum DNA double-strand break repair after ionizing radiation. 20071335_there is a p53-independent function of MTA1 in DNA damage response via modulation of the p21 WAF1-proliferating cell nuclear antigen pathway 20427275_Findings suggest that, in addition to its role in the repair of double strand breaks caused by ionizing radiation, MTA1 also participates in the UV-induced ATR-mediated DNA damage checkpoint pathway. 20547755_identify MTA1, a subunit of the NuRD complex, as a new HIC1 corepressor. 20619094_Expression of Mta-1 expression and VEGF is positively associated with the development of endometrial cancer. 20651739_Data show that induced levels of metastasis-associated protein 1 (MTA1) are sufficient to transform Rat1 fibroblasts and that the transforming potential of MTA1 is dependent on its acetylation at Lys626. 20661085_High metastasis-associated protein 1 nuclear expression is associated with non-small cell lung cancer. 20697987_Increased expression of MTA1 is associated with invasive ductal breast carcinoma. 20717904_MTA1 pro-angiogenic and pro-invasive functions create permissive environment for prostate tumor growth and likely support metastasis. 20935042_Mta1 protein overexpression is an independent prognostic factor for patients with non-small-cell lung cancer. 21047798_RbAp48 interacts with the NuRD subunit MTA-1 via a surface that is distinct from its FOG-binding pocket, providing a first glimpse into the way in which NuRD assembly facilitates interactions with cofactors 21240515_There was a significant correlation between the expression of MTA1 and lymph node metastasis in tonsil cancer. 21258411_findings suggest that TGF-beta1 regulates EMT via stimulating expression of MTA1, which in turn, induces FosB to repress E-cadherin expression and thus, revealed an inherent function of MTA1 as a target and effector of TGF-beta1 signaling in epithelial cells 21290196_Overexpression of metastasis-associated protein 1 is significantly correlated with tumor angiogenesis in patients with early-stage non-small cell lung cancer. 21336715_MTA1 plays an important role in the migration and invasion of cervical cancer cells. 21445634_in ileal neuroendocrine cancer, HER-2/neu overexpression plays a role in the carcinogenetic process and by triggering the altered expression of c-Met and MTA-1, may activate the molecular pathway(s) promoting tumor progression and metastasis development. 21448429_Data suggest that MTA1, as a regulator of tumor-associated lymphangiogenesis, promotes lymphangiogenesis in colorectal cancer by mediating the VEGF-C expression. 21555589_MTA1-mediated activation of ARF and ARF-mediated functional inhibition of MTA1 represent a p53-independent bidirectional autoregulatory mechanism in which these two opposites act in concert to regulate cell homeostasis and oncogenesis 21595884_Silencing effect of MTA1 could efficiently inhibit the invasion and proliferation in MDA-MB-231 cells. 21617866_HDAC1 and MTA1 expression levels are potential prognostic indicators for colon cancer. 21661415_Barrett's esophagus (BE) with HDAC-1 and MTA-1 expression is considered to be a precancerous lesion re quiring curative treatment. 21725997_immunohistochemical analysis of approximately 300 liver tissue cores from confirmed cases of Opisthorchis viverrini-induced cholangiocarcinoma showed that MTA1 expression was elevated in >80% of the specimens 21813470_The data suggest that EIF5A2 plays an important oncogenic role in CRC aggressiveness by the upregulation of MTA1 to induce EMT. 21835429_Metastasis tumor antigen 1 overexpression can be used as a predictor of clinical outcome in patients with ovarian cancer and therefore may represent a new prognostic marker. 21837953_MTA1 siRNA can inhibit the anchorage-independent growth of prostate cancer cells by inducing anoikis. 21892752_MTA1may play an important role in the smoked-related progress of non-small cell lung cancer 21949998_Aberrant expression of MTA1 and RECK gene may be involved in the invasion and metastasis of nasopharyngeal carcinoma. 21965678_SUMOylation and SUMO-interacting motif (SIM) of metastasis tumor antigen 1 (MTA1) synergistically regulate its transcriptional repressor function 22022494_The fine coordination between MTA1 and p53 in pachytene spermatocytes upon hyperthermal stimulation, was investigated. 22098246_The genetic polymorphism IVS4-81G/A in MTA1 were correlated with MTA1 overexpression and may be an important risk factor for the recurrence of hepatocellular carcinoma. 22139444_MiR-30c was directly bound to the 3'-untranslated regions of MTA1. 22176993_MTA1 may play a role in promoting cervical cancer cell invasion, migration, adhesion, as well as cell growth and colony formation. 22253283_Study reveals several new physiologic functions of MTA1 related to DNA damage, inflammatory responses, and infection, in which MTA1 functions as a permissive 'gate keeper' for cancer-causing parasites. 22270359_Leptin-induced epithelial-mesenchymal transition in breast cancer cells requires beta-catenin activation via Akt/GSK3- and MTA1/Wnt1 protein-dependent pathways. 22270988_High expression of MTA1 protein is closely associated with esophageal tumor progression, increased tumor angiogenesis, and poor survival. 22286760_the MTA1 gene is a target of p53-mediated transrepression 22537306_results of this study suggest that nuclear overexpression of MTA1 correlates significantly with poorer disease free survival and poorer overall survival in nasopharyngeal carcinoma 22547301_MTA1 gene overexpression can be used as a molecular biological marker to predict the prognosis of middle esophageal squamous cell carcinoma. 22575258_Metastasis-associated protein 1 staining is not ideal for investigating the possible malignant nature of smaller metastases because of the relatively low concordance between the primary tumor and metastases. 22576802_MTA1 functions in regulating the invasive phenotype of lung cancer cells and this regulation may be through altered miRNA expression 22700976_Metastasis-associated protein 1/histone deacetylase 4-nucleosome remodeling and deacetylase complex regulates phosphatase and tensin homolog gene expression and function. 22844865_The conversely abnormal expression levels of MTA1 and RECK may be collectively involved in progression of malignancies and may serve as molecular predictors for metastasis, recurrence, and prognosis of nasopharyngeal carcinoma. 22864797_Overexpression of MTA1 is associated with Breast Cancer. 23227138_results demonstrate that MTA1 plays an important role in controlling the malignant transformation of prostate cancer cells through the p-AKT/E-cadherin pathway 23283219_MTA1 overexpression is frequently observed in node-negative gastric cancer patients and is significantly associated with increased angiogenesis and poor prognosis. 23343746_These findings demonstrate that MTA1 protein plays important roles in regulating the migration, invasion, and angiogenesis potentials of 95D cells. 23352453_MTA1 possesses an inherent histone amplifier activity with an instructive role in impacting the epigenetic landscape. 23371285_High MTA-1 expression is associated with metastasis and epithelial to mesenchymal transition in colorectal cancer. 23592142_These results demonstrated that MTA1 played an important role in the cell metastasis in ovarian cancer. 23618874_we have demonstrated a critical role of MTA1 in regulating NPC metastasis. This kind of role is associated with its influence on cytoskeleton organization. Rho GTPases and Hedgehog signaling are involved in MTA1-mediated metastasis promotion. 23718732_MTA1 and miR-125b have antagonistic effects on the migration and invasion of NSCLC cells. 23791785_The structure of HDAC1 in complex with MTA1 from the NuRD complex is reported. 23831020_MTA1 acts as a potent corepressor of the nuclear receptor NR4A1 transcription by interacting with HDAC2 and may serve as a novel DTX-resistance promoter in PC-3 cells 23866297_Metastasis-associated protein 1 is a novel marker predicting survival and lymph nodes metastasis in cervical cancer. 23900262_We concluded that MTA1 nuclear overexpression may be a prognostic indicator and a future therapeutic target for aggressive PCa in African American men 23941622_MTA1 promotes nasopharyngeal carcinoma cell proliferation via enhancing G1 to S phase transition, leading to increased tumor growth. 24089055_MTA1 has an important role in the maintenance of circadian rhythmicity. 24214543_MTA1 gene plays an important role in progressiona and metastasis of ovarian cancers. 24265228_Crosstalk between VEGF and MTA1 signaling pathways contribute to aggressiveness of breast carcinoma. 24413532_MTA1-TFAP2C or the MTA1-IFI16 complex may contribute to the epigenetic regulation of ESR1 expression. 24424621_These results demonstrate a novel role for MTA1 in the regulation of esophageal squamous cell carcinoma invasion and provide insight into the mechanisms involved in this process. 24920672_RbAp48 recognizes MTA1 using the same site that it uses to bind histone H4, showing that assembly into NuRD modulates RbAp46/48 interactions with histones. 25205035_MTA1 drove a global decondensation of chromatin structure, it changed the expression of only a small proportion of genes. 25245523_MTA1 plays a critical role in promoting cisplatin resistance in nasopharyngeal carcinoma cells by regulating cancer stem cell properties via the PI3K/Akt signaling pathway. 25301048_High expression of the MTA1 protein was seen in oral squamous cell carcinoma, and was closely associated with tumor progression and increased tumor angiogenesis. 25315816_In this review, we will briefly discuss the researches for the identification and characterization of the mta1 gene, its human counterpart MTA1, and their protein products 25315817_In this review, we described various molecular studies of osteosarcoma, especially associated with MTA1. 25319202_This review focuses on the current knowledge about the function and regulation of MTA1 and MTA3 proteins in gynecological cancer, including ovarian, endometrial, and cervical tumors. 25332143_MTA1 is involved in prostate tumor angiogenesis by regulating several pro-angiogenic factors. Evidence for MTA1 as a prognostic marker for aggressive prostate cancer and disease recurrence has been described. 25332144_Therefore, targeting MTA1 would not only suppress tumor progression but also greatly sensitize cancer cells to DNA damage-based radio- and chemotherapies 25332145_propose that MTA1 is a stress response protein that is upregulated in various stress-related situations such as heat shock, hypoxia, and ironic radiation. 25332146_Recent studies demonstrating the bi-regulatory nature of MTA1 challenged the conventional paradigm of MTA function and raised many questions in fully understanding their roles in cancer progression 25344802_the concept of the dynamic nature of corepressor versus coactivator complexes and the MTA1 proteome as a function of time to signal is likely to be generally applicable to other multiprotein regulatory complexes in living systems. 25352341_In this review, we summarise the structure and function of the four domains of MTA1 and discuss the possible functions of less well-characterised regions of the protein. 25359583_This review gives a brief account on the existing biological and molecular data in the context of head and neck cancer invasion and metastasis in relation to MTA1. 25374266_Taken together, current literature suggests that MTA proteins, especially MTA1, act as a master co-regulatory molecule involved in the carcinogenesis and progression of various malignant tumors 25416046_MTA1 expression was significantly associated with poor metastasis-free survival in nasopharyngeal carcinoma patients. 25501238_Results show that MTA1 promotes the proliferation of epithelial ovarian cancer cells by enhancing DNA repair. 25502548_MTA1 plays a critical role in regulating the malignant behaviors of small cell lung cancer 25688501_MTA1 might be important biological marker involved in the carcinogenesis, metastasis, and invasion of gallbladder adenocarcinoma, and MTA1 is an independent factor of prognosis. 25797255_protein abundance of YB-1 and MTA1, irrespective of DNA or mRNA status, can predict for prostate cancer relapse and uncover a vast underappreciated repository of biomarkers regulated at the level of protein expression. 25977170_MTA1 is associated with the aggressive nature of pituitary tumors and may be a potential therapeutic target in this tumor type. 25998575_report demonstrates that miR-125a-3p inhibits the proliferation, migration, and invasion of NSCLC cells through down-regulation of MTA1 26168722_MTA1 dysregulation in a subset of salivary gland cancer might promote aggressive phenotypes by compromising the tumor suppressor activity of ERbeta 26503703_Inhibition of MTA1 by ERalpha contributes to protection hepatocellular carcinoma from tumor proliferation and metastasis 26543080_MTA1 is up-regulated in CRC; its expression is inversely associated with lymphatic metastases and the expression of VEGFC, VEGFD and VEGFR3 26689197_MTA1 plays an important role in Epithelial-to-mesenchymal transition (EMT) to promote metastasis via suppressing E-cadherin expression, resulting in a poor prognosis in MPM. MTA1 is a novel biomarker and indicative of a poor prognosis in MPM patients 26698569_We revealed a new molecular mechanism of MTA1-mediated invasion and metastasis in lung cancer through downstream target EpCAM, and interfering with EpCAM function may be a novel therapeutic strategy for treatment of MTA1-overexpressing lung carcinoma 26728277_the augmentation of endogenous MTA1 expression during neuronal ischemic injury acts additionally to an endocrinous cascade orchestrating intimate interactions between ERalpha and BCL2 pathways. 26797758_High MTA1 expression is associated with osteosarcoma progression. 26943043_Findings highlight MTA1 as a key upstream regulator of prostate tumorigenesis and cancer progression. 26968810_Data show that KRAS, MTA1 and HMGA2 are direct targets of miR-543. 27026268_show that AR might be an additional marker for endocrine responsiveness in ER(+) cancers and suggests that blocking MTA1 might be an effective way to inhibit AR/HER2 signaling in ER(-) breast cancer 27035126_There is no interaction between IGFBP3 and MTA1 in ESCC, and they are not independent risk factors for esophageal squamous cell carcinoma prognosis. 27044752_Results show that metastasis-associated protein 1 (MTA1) and tyrosine hydroxylase (TH) levels were significantly down-regulated in Parkinson disease (PD) samples as compared with normal brain tissue. 27048111_RNAi aiming at MTA1 can effectively inhibit the expression of MTA1 in endometrial carcinoma Ishikawa cells 27052252_Results showed that in the development of colon cancer, MTA1 is linked to certain signal pathways, such as Wnt/Notch/nucleotide excision repair pathways. The findings also suggested that MTA1 demonstrates the closest relationship in a coregulation process with the key molecules AKT1, EP300, CREBBP, SMARCA4, RHOA, and CAD. 27098840_The crystal structure reveals an extensive interface between MTA1 and RBBP4. 27144666_The MTA1 subunit of the nucleosome remodeling and deacetylase complex can recruit two copies of RBBP4/RBBP7. 27320813_MTA1 expression is upregulated in tumours compared to normal colon cancer samples. 27323185_the significance of MTA1 expression in the invasion and metastasis of medulloblastoma 27506865_Metastasis-associated protein-1 expression levels in HPV-infected non-small cell lung cancer tissues correlated positively with tumor stage and nodal metastasis. 27574100_elevated expression of MTA1 was significantly correlated with recurrence, and was an independent risk factor for lymph node metastasis in gastric cancer 27583980_Suppression of breast cancer metastasis occurs following miR-421 inhibition of MTA1 expression. 27603575_Data show that metastasis associated 1 (MTA1) represses neuronal nitric oxide synthase (nNOS) expression upon oxygen glucose deprivation (OGD)-induced oxidative stress. 28231399_Data indicate a positive correlation between metastasis-associated protein 1 (MTA1) and microRNA miR-22, supporting their inhibitory effect on E-cadherin expression. 28288133_Downregulation of miR-30e can increase levels of MTA1 in human hepatocellular carcinoma, and furthermore promote cell invasion and metastasis by promoting ErbB2. 28393842_Together findings presented here recognize an inherent role of MTA1 as a modifier of DNMT3a and IGFBP3 expression, and consequently, the role of MTA1-DNMT3a-IGFBP3 axis in breast cancer progression. 28418915_High Expressions of MTA1 is associated with epithelial to mesenchymal transition and metastasis in non-small-cell lung cancer. 28443468_our studies indicate that Curcumin increases the sensitivity of Paclitaxel-resistant non-small-cell lung cancer cells to Paclitaxel through microRNA-30c-mediated MTA1 reduction. Curcumin might be a potential adjuvant for non-small-cell lung cancer patients during Paclitaxel treatment. 28504714_Findings illustrate how cancer cells utilize a chromatin remodeling factor to engage a core survival pathway to support its cancerous phenotypes, and reveal new facets of MTA1-SGK1 axis by a physiologic signal in cancer progression. 28506766_Our findings revealed that miR-183 upregulation and MTA1 gene silence significantly repressed the epithelial-mesenchymal transition, migration and invasion of human pancreatic cancer cells. 28570554_In subgroup analyses based on study quality and tumor type, MTA1 overexpression was obviously related to clinical parameters, such as lymph node metastasis and TNM stage, and was also associated with prognosis of patients with gastrointestinal or esophageal cancer 28589145_MTA1 contributed to neovascularization of residual tumors. Cellular experiments further revealed that MTA1 increased the stability and the expression of HIF-1alpha, and overexpression of MTA1 enhanced tube formation and neovessels of chick embryos. 28631568_In this study, we aimed to analyze the expression of miR-183 and MTA1 in nasopharyngeal carcinoma tissues and cells; explore the role of miR-183 in NPC cell proliferation, invasion, as well as DDP-induced apoptosis; and investigate the relationship between miR-183 and MTA1 in nasopharyngeal carcinoma cells. 28635511_MTA 1 protein may constitute an important prognostic marker in pancreatic cancer and could improve prognosis and treatment. 28686969_Inhibit the metastasis and EMT via MTA1. 28739699_MTA1 expression was positively correlated with LAT1 (p=0.013) and CD34 (p=0.034) expression, but not with Ki-67 (p=0.078). MTA1 shows promise as a diagnostic and prognostic marker in esophageal cancer, and we anticipate that the gene will also prove to be a good therapeutic target. 28861931_TRIM25 inhibits hepatocellular carcinoma progression by targeting MTA1. 28901313_MiR-30c and MTA1 abnormally expressed in ovarian cancer (OC), which may be related to metastasis of OC. In MiR-30c as a tumor suppressor gene, its expression in OC could lead to reduced expression of MTA1, which may be one of the mechanisms of metastasis of OC cells. 29061215_Overexpression of MTA1 could be a marker of poor prognosis in Chinese NSCLC patients, but not in lung cancer or small cell lung cancer. 29125886_MTA1 participates in Nasopharyngeal carcinoma malignant transformation via regulating IQGAP1 expression and PI3K/Akt signaling pathway. 29130361_MTA1 induces AMPK activation and subsequent autophagy that could contribute to tamoxifen resistance in breast cancer. 29465084_Data found that MTA1 is a PKD1-interacting substrate, and that PKD1 phosphorylates MTA1, supports its nucleus-to-cytoplasmic redistribution and utilises its N-terminal and kinase domains to effectively inhibit the levels of MTA1 via polyubiquitin-dependent proteosomal degradation. 29573865_MTA1 immunopositivity was significantly associated with progression of gastric cancer, and may be helpful in gastric cancer prognosis. 29654742_data indicate that suppression of TRIM25 expression by high levels of miR-873 dictates MTA1 protein upregulation in hepatocellular carcinoma 30013189_High expression of FMNL1, resulted from decreased miR-16 and/or MTA1 amplification. 30027683_MTA1 promotes prostate tumor growth and formation of bone metastasis by positively regulating CTSB expression in the primary tumor cells. MTA1 is an attractive target for the treatment of prostate cancer and prevention of bone metastasis. 30118695_It has been shown that TRIM25 ubiquitinates MTA-1 at lysine 98 and degrades it normal liver cells. 30170442_MTA1 is closely associated with the occurrence and development of endometriosis. Thus, MTA1 level may be used as a new indicator to predict the progression of endometriosis 30268463_this study shows relationship between MTA1 and spread through air space and their joint influence on prognosis of patients with stage I-III lung adenocarcinoma 30313027_Meta-analysis demonstrated that increased MTA1 expression was an effective predictor of a worse prognosis in solid tumor patients. 30314706_The circ-UBAP2 was mainly observed in the cytoplasm and was capable of sponging miRNA-661 to increase the expression of the oncogene MTA1. 30327463_MTA1 binding to PWWP2A during recruitment of the entire MHR complex . 30396299_These results indicated that MTA1 potentially promoted pancreatic cancer metastasis via HIF-alpha/VEGF pathway. This research supplies a new molecular mechanism for MTA1 in the pancreatic cancer progression and metastasis. MTA1 may be an effective therapy target in pancreatic cancer. 30610787_High MTA1 expression is associated with proliferation and metastasis of colorectal cancer. 30631898_Upregulation of MTA1 expression by human papillomavirus infection promotes cisplatin resistance in cervical cancer cells via modulation of NF-kappaB/APOBEC3B cascade 30642362_MTA1 and MTA2 play opposing roles in the metastasis of ZR-75-30 luminal B breast cancer cells in vitro. 30651530_MTA1 silencing with si-RNA significantly reduced the tumor growth rate in nude mice (p<0.01), reduced tumor MVD, and 70% of mice survived for more than 30 days. MTA1 overexpression resulted in the death of all mice at 30 days after tumor inoculation and upregulated the expression of COX-2, Ang1/2, HIF-1a and VEGF, which were down-regulated by MTA1 silencing. 30745574_MTA1 and S100A4 are involved in angiogenesis for promoting tumor growth. 30782165_This is the first report showing that breast cancer exosomes contain MTA1 and can transfer it to other cells resulting in changes to hypoxia and estrogen receptor signaling in the tumor microenvironment. 30802976_overexpression of exon 4-excluded form in tumor associated with early recurrence of hepatitis B virus-associated hepatocellular carcinoma 30979588_Overexpression of MTA1 contributed to EC tumor growth, while knockdown of MTA1 resulted in tumor growth inhibition. 30979652_MTA1 constitutes potential tumor biomarkers in LSCC and could be integrated into a multiparametric prognostic model. 31152708_MTA1 expression was upregulated and positively related to the metastasis in oral squamous carcinoma (OSCC) tissues and cell lines. MTA1 overexpression promoted OSCC invasion, migration, and induced EMT, while its silencing had the opposite effect both in vitro and in xenografts in vivo. MTA1 promotes EMT by activating its downstream hedgehog signaling in OSCC. 31570164_MTA1 coregulator has a role in regulating LDHA expression and function in breast cancer 31783401_MTA1 promotes tumorigenesis and development of esophageal squamous cell carcinoma via activating the MEK/ERK/p90RSK signaling pathway. 31811272_A TGF-beta-MTA1-SOX4-EZH2 signaling axis drives epithelial-mesenchymal transition in tumor metastasis. 32252197_Inhibition of MTA1 expression can inhibit the proliferation of Eca109 cells and induce apoptosis. This process may be related to the down-regulation of PCNA [proliferating cell nuclear antigen ] protein expression and activation of caspase-3 protein expression. 32410569_MiR-543 promotes tumorigenesis and angiogenesis in non-small cell lung cancer via modulating metastasis associated protein 1. 32632610_Leptin promotes endothelial dysfunction in chronic kidney disease by modulating the MTA1-mediated WNT/beta-catenin pathway. 32678829_Downregulation of Jumonji-C domain-containing protein 5 inhibits proliferation by silibinin in the oral cancer PDTX model. 32901005_Chromatin modifier MTA1 regulates mitotic transition and tumorigenesis by orchestrating mitotic mRNA processing. 33264408_The topology of chromatin-binding domains in the NuRD deacetylase complex. 34019948_O-GlcNAc modification regulates MTA1 transcriptional activity during breast cancer cell genotoxic adaptation. 34182519_High Expression of MTA1 Predicts Unfavorable Survival in Patients With Oral Squamous Cell Carcinoma. 34337713_A long non-coding RNA, ELFN1-AS1, sponges miR-1250 to upregulate MTA1 to promote cell proliferation, migration and invasion, and induce apoptosis in colorectal cancer. 34502289_Molecular Mechanisms and Animal Models of HBV-Related Hepatocellular Carcinoma: With Emphasis on Metastatic Tumor Antigen 1. 35369814_Role of lncRNA AGAP2-AS1 in Breast Cancer Cell Resistance to Apoptosis by the Regulation of MTA1 Promoter Activity. | ENSMUSG00000021144 | Mta1 | 9.080604e+03 | 1.1927949 | 0.254346025 | 0.2592577 | 9.690242e-01 | 0.3249236617 | 0.82378328 | No | Yes | 8.392177e+03 | 727.881399 | 7.297544e+03 | 649.497214 | |
| ENSG00000183048 | 1468 | SLC25A10 | protein_coding | Q9UBX3 | FUNCTION: Involved in translocation of malonate, malate and succinate in exchange for phosphate, sulfate, sulfite or thiosulfate across mitochondrial inner membrane. | Alternative splicing;Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Primary mitochondrial disease;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of a family of proteins that translocate small metabolites across the mitochondrial membrane. The encoded protein exchanges dicarboxylates, such as malate and succinate, for phosphate, sulfate, and other small molecules, thereby providing substrates for metabolic processes including the Krebs cycle and fatty acid synthesis. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2012]. | hsa:1468; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; antiporter activity [GO:0015297]; dicarboxylic acid transmembrane transporter activity [GO:0005310]; malate transmembrane transporter activity [GO:0015140]; oxaloacetate transmembrane transporter activity [GO:0015131]; succinate transmembrane transporter activity [GO:0015141]; sulfate transmembrane transporter activity [GO:0015116]; thiosulfate transmembrane transporter activity [GO:0015117]; dicarboxylic acid transport [GO:0006835]; gluconeogenesis [GO:0006094]; ion transport [GO:0006811]; malate transmembrane transport [GO:0071423]; mitochondrial transport [GO:0006839]; oxaloacetate transport [GO:0015729]; phosphate ion transmembrane transport [GO:0035435]; succinate transmembrane transport [GO:0071422]; sulfate transport [GO:0008272]; sulfide oxidation, using sulfide:quinone oxidoreductase [GO:0070221]; thiosulfate transport [GO:0015709] | 16027120_Slc25a10 plays an important role in supplying malate for citrate transport required for fatty acid synthesis and its inhibition might effectively reduce lipid accumulation in adipose tissues 20877624_Observational study of gene-disease association. (HuGE Navigator) 21555518_SLC25A10 is a target gene of the BACH1 transcription factor according to ChIP-seq analysis in HEK 293 cells. 23266187_Compares and contrasts all the known human SLC25A* genes and includes functional information. 25797253_the SLC25A10 carrier plays an important role in regulating redox homeostasis to protect confluent cells against oxidative stress. 28570579_study identifies novel genes associated with insulin sensitivity in adipocytes in women independently of obesity. KFL15 and SLC25A10 are inhibitors of insulin-stimulated lipogenesis under conditions when glucose transport is the rate limiting step 29211846_Study report the first recessive mutations of SLC25A10 associated to an inherited severe mitochondrial neurodegenerative disorder. We propose that SLC25A10 loss-of-function causes pathological disarrangements in respiratory-demanding conditions and oxidative stress vulnerability. 30205167_Data show that targeting of mitochondrial dicarboxylate carrier (SLC25A10) was effective in overcoming chronic-cycling hypoxia-induced enhanced death resistance in vitro and in vivo by disturbing increased antioxidant capacity. 30943427_identified a new protein-protein interaction mechanism in which CLOCK can directly regulate cell metabolism via the mitochondrial membrane transporter SLC25A10. | ENSMUSG00000025792 | Slc25a10 | 5.687510e+03 | 1.6393666 | 0.713138477 | 0.3398633 | 4.366360e+00 | 0.0366552075 | 0.61802832 | No | Yes | 7.313470e+03 | 1221.536781 | 3.434705e+03 | 589.012508 | |
| ENSG00000183474 | 728340 | GTF2H2C | protein_coding | Q6P1K8 | FUNCTION: Component of the core-TFIIH basal transcription factor involved in nucleotide excision repair (NER) of DNA and, when complexed to CAK, in RNA transcription by RNA polymerase II. {ECO:0000250}. | DNA damage;DNA repair;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:728340;hsa:730394; | nuclear speck [GO:0016607]; transcription factor TFIIH core complex [GO:0000439]; transcription factor TFIIH holo complex [GO:0005675]; zinc ion binding [GO:0008270]; nucleotide-excision repair [GO:0006289]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription, DNA-templated [GO:0006351] | Mouse_homologues 14980221_p44 of TFIIH has a role in binding to the nonstructural protein of Rift Valley fever virus to form nuclear filamentous structures | ENSMUSG00000021639 | Gtf2h2 | 5.347025e+02 | 1.0681103 | 0.095060572 | 0.3898836 | 5.969206e-02 | 0.8069833162 | 0.96141221 | No | Yes | 4.275566e+02 | 91.731438 | 4.460020e+02 | 97.940341 | ||
| ENSG00000183579 | 84133 | ZNRF3 | protein_coding | Q9ULT6 | FUNCTION: E3 ubiquitin-protein ligase that acts as a negative regulator of the Wnt signaling pathway by mediating the ubiquitination and subsequent degradation of Wnt receptor complex components Frizzled and LRP6. Acts on both canonical and non-canonical Wnt signaling pathway. Acts as a tumor suppressor in the intestinal stem cell zone by inhibiting the Wnt signaling pathway, thereby resticting the size of the intestinal stem cell zone (PubMed:22575959). Along with RSPO2 and RNF43, constitutes a master switch that governs limb specification (By similarity). {ECO:0000250|UniProtKB:Q08D68, ECO:0000269|PubMed:22575959}. | Alternative splicing;Cell membrane;Developmental protein;Membrane;Metal-binding;Reference proteome;Signal;Transferase;Transmembrane;Transmembrane helix;Ubl conjugation pathway;Wnt signaling pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | hsa:84133; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; frizzled binding [GO:0005109]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; limb development [GO:0060173]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of non-canonical Wnt signaling pathway [GO:2000051]; protein ubiquitination [GO:0016567]; regulation of Wnt signaling pathway, planar cell polarity pathway [GO:2000095]; stem cell proliferation [GO:0072089]; ubiquitin-dependent protein catabolic process [GO:0006511]; Wnt receptor catabolic process [GO:0038018]; Wnt signaling pathway [GO:0016055] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20935629_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22575959_study provides new mechanistic insights into the regulation of Wnt receptor turnover, and reveals ZNRF3 as a tractable target for therapeutic exploration 23504200_ZNRF3 inhibits gastric cancer cell growth and promotes cell apoptosis by affecting the Wnt/beta-catenin/TCF4 signalling pathway. 23670221_Genes within recently identified loci associated with waist-hip ratio (WHR) exhibit fat depot-specific mRNA expression, which correlates with obesity-related traits. Adipose tissue (AT) mRNA expression of 6 genes (TBX15/WARS2, STAB1, PIGC, ZNRF3, GRB14 24225776_Signalling potency of R-spondin proteins depends on their ability to recruit ZNRF3 or RNF43 via Fu1 into a complex with LGR receptors, which interact with Rspo via Fu2 24349440_ZNRF3 binds RSPO1 and LGR5-RSPO1 with micromolar affinity via RSPO1 furin-like 1 (Fu1) domain. 25891077_Data indicate Dishevelled (DVL) as a dual function adaptor to recruit negative regulators ZNRF3/RNF43 to Wnt receptors to ensure proper control of pathway activity. 25923840_ZnRF3 negatively influences both the Wnt and Hedgehog proliferative pathways and probably this way it negatively regulates cancer progression 26423400_miR-93/ZNRF3/Wnt/beta-catenin regulatory network contributes to the growth of lung carcinoma. 26549292_Report shows that miR-146b participated in migration, invasion and chemoresistance in osteosarcoma via downregulation of ZNRF3. 27352324_ZnRF3 negatively influences both the Wnt and Hedgehog proliferative pathways, and probably this way it negatively regulates cancer progression. 27448298_Data indicate that clinical specimens showed a significant inverse correlation between zinc and ring finger 3 (ZNRF3) and beta-catenin mRNA levels. 27661107_Data suggest that inactivation of RNF43 and ZNRF3 is important in serrated tumorigenesis and has identified a potential therapeutic strategy for this cancer subtype. 27733215_ZNRF3 inhibited the metastasis and tumorigenesis via suppressing the Wnt/beta-catenin signaling pathway in NPC cells. 29497989_The ZNRF3 is ubiquitinated by beta-TRCP in both CKI-phosphorylation- and degron-dependent manners. 29735715_Missense variants of ZNRF3 are associated with Disorders of Sex Development. 29769720_results establish that RSPO2, without the LGR4/5/6 receptors, serves as a direct antagonistic ligand to RNF43 and ZNRF3, which together constitute a master switch that governs limb specification; these findings have direct implications for regenerative medicine and WNT-associated cancers 30348125_The present study suggests rs7290117 in ZNRF3 may be involved in the regulation of axial length, though our results do not support a contribution of the SNPs we tested in ZNRF3, HGF and MFRP to primary angle-closure glaucoma in northern Chinese people 30816445_miR106b3p promotes cell proliferation and invasion, partially by downregulating ZNRF3 and inducing EMT via Wnt/betacatenin signaling in esophageal squamous cell carcinoma cells. 31934854_The tumor suppressor PTPRK promotes ZNRF3 internalization and is required for Wnt inhibition in the Spemann organizer. 33367931_MicroRNA301a/ZNRF3/wnt/betacatenin signal regulatory crosstalk mediates glioma progression. 33513905_Low Protein Expression of both ATRX and ZNRF3 as Novel Negative Prognostic Markers of Adult Adrenocortical Carcinoma. 34273374_RSPO2 silence inhibits tumorigenesis of nasopharyngeal carcinoma by ZNRF3/Hedgehog-Gli1 signal pathway. 34590584_A MET-PTPRK kinase-phosphatase rheostat controls ZNRF3 and Wnt signaling. 34657141_A positive feedback loop of lncRNA-RMRP/ZNRF3 axis and Wnt/beta-catenin signaling regulates the progression and temozolomide resistance in glioma. 34716314_Somatic driver mutation prevalence in 1844 prostate cancers identifies ZNRF3 loss as a predictor of metastatic relapse. 35039505_RNF43/ZNRF3 loss predisposes to hepatocellular-carcinoma by impairing liver regeneration and altering the liver lipid metabolic ground-state. | ENSMUSG00000041961 | Znrf3 | 1.694050e+03 | 1.2131794 | 0.278792908 | 0.2659196 | 1.096612e+00 | 0.2950109113 | 0.81312088 | No | Yes | 1.629231e+03 | 113.163815 | 1.333968e+03 | 95.277452 | |
| ENSG00000183597 | 128989 | TANGO2 | protein_coding | Q6ICL3 | Alternative splicing;Cytoplasm;Disease variant;Golgi apparatus;Mitochondrion;Neurodegeneration;Reference proteome | This gene belongs to the transport and Golgi organization family, whose members are predicted to play roles in secretory protein loading in the endoplasmic reticulum. Depletion of this gene in Drosophila S2 cells causes fusion of the Golgi with the ER. In mouse tissue culture cells, this protein co-localizes with a mitochondrially targeted mCherry protein and displays very low levels of co-localization with Golgi and peroxisomes. Allelic variants of this gene are associated with rhabdomyolysis, metabolic crises with encephalopathy, and cardiac arrhythmia. [provided by RefSeq, Apr 2016]. | hsa:128989; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; mitochondrion [GO:0005739]; Golgi organization [GO:0007030]; protein secretion [GO:0009306] | 26805781_Exons 3-9 heterozygous deletion in TANGO2 are recurrent pathogenic alleles present in the Latino/Hispanic and European populations, respectively, causing considerable morbidity in the homozygotes in these populations. 30245509_Variants from the 14 patients described are shown above the gene depiction with red bars for deletions and lines for single nucleotide variants (SNV), and pathogenic variants from prior studies are shown below (light blue bars for deletions and lines for SNV) based on TANGO2 transcript NM152906.6 (2623 bp). 31276219_TANGO2 deficiency as a cause of neurodevelopmental delay with indirect effects on mitochondrial energy metabolism. 31339582_Clinical presentation and proteomic signature of patients with TANGO2 mutations. 32909282_The phenotype associated with variants in TANGO2 may be explained by a dual role of the protein in ER-to-Golgi transport and at the mitochondria. 32929747_Clinical and biological characterization of 20 patients with TANGO2 deficiency indicates novel triggers of metabolic crises and no primary energetic defect. 33342685_Clinical phenotype associated with TANGO2 gene mutation. 35197517_Mitochondrial dysfunction associated with TANGO2 deficiency. | ENSMUSG00000013539 | Tango2 | 1.415615e+03 | 1.2343432 | 0.303743534 | 0.3090509 | 9.610504e-01 | 0.3269223621 | 0.82378328 | No | Yes | 1.410674e+03 | 205.988685 | 1.088966e+03 | 163.499307 | ||
| ENSG00000183605 | 119559 | SFXN4 | protein_coding | Q6P4A7 | FUNCTION: Mitochondrial amino-acid transporter (By similarity). Does not act as a serine transporter: not able to mediate transport of serine into mitochondria (PubMed:30442778). {ECO:0000250|UniProtKB:Q9H9B4, ECO:0000269|PubMed:30442778}. | Acetylation;Alternative splicing;Amino-acid transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Primary mitochondrial disease;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the sideroflexin family. The encoded protein is a transmembrane protein of the inner mitochondrial membrane, and is required for mitochondrial respiratory homeostasis and erythropoiesis. Mutations in this gene are associated with mitochondriopathy and macrocytic anemia. Alternatively spliced transcript variants have been found in this gene. [provided by RefSeq, Jan 2014]. | hsa:119559; | integral component of mitochondrial inner membrane [GO:0031305]; mitochondrion [GO:0005739]; ion transmembrane transporter activity [GO:0015075]; transmembrane transporter activity [GO:0022857]; amino acid transport [GO:0006865]; mitochondrial transmembrane transport [GO:1990542] | 14756423_we isolated a cDNA encoding a novel sideroflexin protein (SFXN4), which showed 59% identity and 71% similarity to mouse sideroflexin4. 16385451_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 24119684_Our findings establish mutations in SFXN4 as a cause of mitochondriopathy and macrocytic anemia. 31059822_Prenatal onset of mitochondrial disease is associated with sideroflexin 4 deficiency. 31873120_Sideroflexin 4 affects Fe-S cluster biogenesis, iron metabolism, mitochondrial respiration and heme biosynthetic enzymes. 35333655_Sideroflexin 4 is a complex I assembly factor that interacts with the MCIA complex and is required for the assembly of the ND2 module. | ENSMUSG00000063698 | Sfxn4 | 1.954367e+03 | 0.5722366 | -0.805316216 | 0.3349429 | 5.780445e+00 | 0.0162054399 | 0.45005072 | No | Yes | 1.281100e+03 | 203.612533 | 2.273796e+03 | 369.866237 | |
| ENSG00000183666 | 728411 | GUSBP1 | transcribed_unprocessed_pseudogene | 2.805410e+02 | 0.7851047 | -0.349043061 | 0.3366182 | 1.088753e+00 | 0.2967477263 | 0.81383297 | No | Yes | 2.172211e+02 | 37.528407 | 3.039178e+02 | 53.421327 | ||||||||||
| ENSG00000183718 | 84851 | TRIM52 | protein_coding | Q96A61 | FUNCTION: E3 ubiquitin-protein ligase (PubMed:27667714). Positively regulates the NF-kappa-B signaling pathway (PubMed:27667714, PubMed:28073078). {ECO:0000269|PubMed:27667714, ECO:0000269|PubMed:28073078}.; FUNCTION: (Microbial infection) Exhibits antiviral activity against Japanese encephalitis virus (JEV). Ubiquitinates the viral non-structural protein 2 (NS2A) and targets it for proteasome-mediated degradation. {ECO:0000269|PubMed:27667714}. | Alternative splicing;Antiviral defense;Cytoplasm;Metal-binding;Nucleus;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | hsa:84851; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleus [GO:0005634]; transcription coactivator activity [GO:0003713]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; defense response to virus [GO:0051607]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; protein autoubiquitination [GO:0051865]; protein ubiquitination [GO:0016567] | 28073078_these data suggested that TRIM52 was a positive regulator of the NF-kappaB pathway. 29089476_TRIM52 can promote cell proliferation and HBx may regulate TRIM52 expression via the NF-kappaB signaling pathway in Hepatitis B Virus-Associated Hepatocellular Carcinoma. 29898761_Our findings suggest that TRIM52 up-regulation promotes proliferation, migration and invasion of HCC cells through the ubiquitination of PPM1A. 30185771_TRIM52 plays an oncogenic role in ovarian cancer development. 31002351_Based on these data, it was speculated that TRIM52 is critical for lung cancer progression and that downregulation of TRIM52 could inhibit cell proliferation by blocking cell cycle progression. 31133683_A repetitive acidic region contributes to the extremely rapid degradation of the cell-context essential protein TRIM52. 32898524_TRIM52 positively mediates NF-kappaB to promote the growth of human benign prostatic hyperplasia cells through affecting TRAF2 ubiquitination. | ENSMUSG00000022113 | Trim52 | 7.105530e+02 | 0.7724310 | -0.372522102 | 0.3126452 | 1.393307e+00 | 0.2378474335 | 0.78818582 | No | Yes | 4.937424e+02 | 91.781388 | 7.452570e+02 | 141.446773 | |
| ENSG00000183814 | 286826 | LIN9 | protein_coding | Q5TKA1 | FUNCTION: Acts as a tumor suppressor. Inhibits DNA synthesis. Its ability to inhibit oncogenic transformation is mediated through its association with RB1. Plays a role in the expression of genes required for the G1/S transition. {ECO:0000269|PubMed:15538385, ECO:0000269|PubMed:16730350}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Coiled coil;DNA synthesis;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Tumor suppressor;Ubl conjugation | This gene encodes a tumor suppressor protein that inhibits DNA synthesis and oncogenic transformation through association with the retinoblastoma 1 protein. The encoded protein also interacts with a complex of other cell cycle regulators to repress cell cycle-dependent gene expression in non-dividing cells. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jul 2012]. | hsa:286826; | nucleoplasm [GO:0005654]; transcription repressor complex [GO:0017053]; DNA binding [GO:0003677]; cell cycle [GO:0007049]; DNA biosynthetic process [GO:0071897]; regulation of cell cycle [GO:0051726]; regulation of transcription by RNA polymerase II [GO:0006357]; reproduction [GO:0000003]; transcription, DNA-templated [GO:0006351] | 15538385_Human Lin-9 has tumor-suppressing activities and the ability of hLin-9 to inhibit transformation is mediated through its association with pRB. 16730350_Mutation of BARA/LIN-9 restores the expression of E2F target genes. 17098733_Mip/LIN-9 is required for the expression of B-Myb, and both proteins collaborate in the control of the cell cycle progression via the regulation of S phase and cyclin A, cyclin B, and CDK1 17159899_human LIN-9, together with B-MYB, has a critical role in the activation of genes that are essential for progression into mitosis 17563750_The repressor complex that Mip/LIN-9 forms with p107 takes functional precedence over the transcriptional activation linked to the Mip/LIN-9 and B-Myb interaction. 20360844_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21860417_inactivation of LIN9, a subunit of DREAM, results in premature senescence, which can be overcome by the SV40 large T (LT) antigen 23381944_Study links hTRM9L and tRNA modifications to inhibition of tumour growth via LIN9 and HIF1-alpha-dependent mechanisms. 24141769_Results show that E7 interacts with the B-Myb, FoxM1 and LIN9 components of this activator complex, leading to cooperative transcriptional activation of mitotic genes in primary cells and E7 recruitment to the corresponding promoters. 24475316_LIN-9 phosphorylation on threonine-96 is required for transcriptional activation of LIN-9 target genes and promotes cell cycle progression. 28061449_The MuvB multiprotein complex, together with B-MYB and FOXM1 (MMB-FOXM1) regulate the expression of mitotic kinesins in breast cancer cells. 28807940_High LIN9 expression is associated with Triple-negative breast cancers. 32054769_LIN9 and NEK2 Are Core Regulators of Mitotic Fidelity That Can Be Therapeutically Targeted to Overcome Taxane Resistance. | ENSMUSG00000058729 | Lin9 | 1.952796e+02 | 0.9455329 | -0.080800489 | 0.3866242 | 4.385960e-02 | 0.8341150424 | 0.96760164 | No | Yes | 1.641047e+02 | 29.941347 | 1.804412e+02 | 33.562112 | |
| ENSG00000184007 | 8073 | PTP4A2 | protein_coding | Q12974 | FUNCTION: Protein tyrosine phosphatase which stimulates progression from G1 into S phase during mitosis. Promotes tumors. Inhibits geranylgeranyl transferase type II activity by blocking the association between RABGGTA and RABGGTB. {ECO:0000269|PubMed:14643450}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Disulfide bond;Endosome;Hydrolase;Lipoprotein;Membrane;Methylation;Prenylation;Protein phosphatase;Reference proteome | The protein encoded by this gene belongs to a small class of the protein tyrosine phosphatase (PTP) family. PTPs are cell signaling molecules that play regulatory roles in a variety of cellular processes. PTPs in this class contain a protein tyrosine phosphatase catalytic domain and a characteristic C-terminal prenylation motif. This PTP has been shown to primarily associate with plasmic and endosomal membrane through its C-terminal prenylation. This PTP was found to interact with the beta-subunit of Rab geranylgeranyltransferase II (beta GGT II), and thus may function as a regulator of GGT II activity. Overexpression of this gene in mammalian cells conferred a transformed phenotype, which suggested its role in tumorigenesis. Alternatively spliced transcript variants have been described. Related pseudogenes exist on chromosomes 11, 12 and 17. [provided by RefSeq, Aug 2010]. | hsa:8073; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; early endosome [GO:0005769]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; prenylated protein tyrosine phosphatase activity [GO:0004727]; protein tyrosine phosphatase activity [GO:0004725]; protein tyrosine/serine/threonine phosphatase activity [GO:0008138] | 14643450_PRL phosphatases increase cell proliferation by stimulating progression from G1 into S phase 16832410_PRL-2 mRNA expression was not significantly higher in malignant compared to benign breast tissue. 16957164_PRL-2 was expressed heavily in almost every tissue and cell type examined. Widespread expression of PRL-2 in multiple organ systems suggests an important functional role for these enzymes in normal tissue homeostasis. 17440740_Colonic adenocarcinoma cells have the ability to produce PTP4A1, PTP4a2, and PTP4A3, which may relate to the lymph node metastasis of colonic adenocarcinoma. 17666324_PRL-2 can promote cell adhesion and invasion activity of human hepatocellular carcinoma cells. 20226699_Mouse pre-B-cells transfected with human PRL-2 had higher growth responses to Epo or IL-3, shorter cell cycle, less Epo requirement for survival, more cell migration, less cell adhesion, change to an immature cell morphology, & more Bmi-1 expression. 20841483_Overexpression of the protein tyrosine phosphatase PRL-2 correlates with breast tumor formation and progression. 21765462_Results suggest a model in which PRL-2 promotes cell migration and invasion through an ERK1/2-dependent signaling pathway. 21993571_PRL-2 plays an important role in lung cancer and can be a biomarker of lung cancer, substituting for the function of other PRLs. 22413991_Studies indicate that PRL-1 and PRL-2 and PRL-3 are oncogenes and belong to the few phosphatases that lead to the development of cancer. 23362275_Results suggest that TRP32 maintains the reduced state of PRL and thus regulates the biological function of PRL. 23568563_PTP4A2 expression is correlated with overall survival in progestin receptor-positive breast carcinomas. 25193986_Our findings highlight the important role of miR-29c in regulating CRC EMT via GSK-3b/b-catenin signaling by targeting GNA13 and PTP4A and provide new insights into the metastatic basis of colorectal cancer 27872499_study identified a critical role for PRL2 phosphatase in the proliferation and survival of human T-ALL cells; demonstrated that PRL2 is important for the leukemogenic potential of oncogenic NOTCH1 in vivo 28220038_identified a critical role for PRL2 phosphatase in the proliferation and survival of human AML cells. Further, we demonstrated that PRL2 is essential for the leukemogenic potential of AML1-ETO9a in vivo 30718434_altered PRL-2 expression leads to metabolic reprogramming of the cells. These findings uncover a magnesium-sensitive mechanism controlling PRL expression, which plays a role in cellular bioenergetics 32733084_A FRET-based screening method to detect potential inhibitors of the binding of CNNM3 to PRL2. 32788364_Mechanism of PRL2 phosphatase-mediated PTEN degradation and tumorigenesis. 34992672_Downregulated Expression of miRNA-130a-5p Aggravates Hepatoma Progression via Targeting PTP4A2. | ENSMUSG00000028788 | Ptp4a2 | 4.483481e+03 | 0.7633766 | -0.389533174 | 0.3016572 | 1.646815e+00 | 0.1993929711 | 0.78383913 | No | Yes | 3.986395e+03 | 696.351239 | 4.416108e+03 | 790.938961 | |
| ENSG00000184014 | 23258 | DENND5A | protein_coding | Q6IQ26 | FUNCTION: Guanine nucleotide exchange factor (GEF) which may activate RAB6A and RAB39A and/or RAB39B. Promotes the exchange of GDP to GTP, converting inactive GDP-bound Rab proteins into their active GTP-bound form. Involved in the negative regulation of neurite outgrowth (By similarity). {ECO:0000250|UniProtKB:G3V7Q0, ECO:0000269|PubMed:20937701}. | Alternative splicing;Disease variant;Epilepsy;Golgi apparatus;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome;Repeat | This gene encodes a DENN-domain-containing protein that functions as a RAB-activating guanine nucleotide exchange factor (GEF). This protein catalyzes the conversion of GDP to GTP and thereby converts inactive GDP-bound Rab proteins into their active GTP-bound form. The encoded protein is recruited by RAB6 onto Golgi membranes and is therefore referred to as RAB6-interacting protein 1. This protein binds with RAB39 as well. Alternative splicing results in multiple transcript variants encoding distinct isoforms. Mutations in this gene are associated with early infantile epileptic encephalopathy-49. [provided by RefSeq, Feb 2017]. | hsa:23258; | cytosol [GO:0005829]; Golgi membrane [GO:0000139]; trans-Golgi network [GO:0005802]; guanyl-nucleotide exchange factor activity [GO:0005085]; small GTPase binding [GO:0031267]; negative regulation of neuron projection development [GO:0010977]; retrograde transport, endosome to Golgi [GO:0042147] | 19141279_crystal structure of Rab6a(GTP) in complex with a 378-residue internal fragment of the effector Rab6IP1 was solved at 3.2 angstroms resolution. This Rab6IP1 region encompasses an all alpha-helical RUN domain followed in tandem by a PLAT domain 22558185_A RUN Domain from DENND5/Rab6IP1 binds sorting Nexin 1 and is involved in the trafficking of vesicles at the level of Golgi. 27866705_identification of an epileptic encephalopathy additionally featuring cerebral calcifications and coarse facial features caused by recessive loss-of-function mutations in DENND5A 34906508_Novel loss-of-function variant in DENND5A impedes melanosomal cargo transport and predisposes to familial cutaneous melanoma. | ENSMUSG00000035901 | Dennd5a | 3.721603e+03 | 0.8068054 | -0.309707271 | 0.2791164 | 1.243614e+00 | 0.2647757179 | 0.79788700 | No | Yes | 3.491699e+03 | 356.647751 | 4.447506e+03 | 465.380659 | |
| ENSG00000184058 | 6899 | TBX1 | protein_coding | O43435 | FUNCTION: Probable transcriptional regulator involved in developmental processes (By similarity). Binds to the palindromic T site 5'-TTCACACCTAGGTGTGAA-3' DNA sequence (PubMed:11111039). Is required for normal development of the pharyngeal arch arteries (By similarity). {ECO:0000250|UniProtKB:P70323, ECO:0000269|PubMed:11111039}. | 3D-structure;Alternative splicing;DNA-binding;Developmental protein;Disease variant;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene is a member of a phylogenetically conserved family of genes that share a common DNA-binding domain, the T-box. T-box genes encode transcription factors involved in the regulation of developmental processes. This gene product shares 98% amino acid sequence identity with the mouse ortholog. DiGeorge syndrome (DGS)/velocardiofacial syndrome (VCFS), a common congenital disorder characterized by neural-crest-related developmental defects, has been associated with deletions of chromosome 22q11.2, where this gene has been mapped. Studies using mouse models of DiGeorge syndrome suggest a major role for this gene in the molecular etiology of DGS/VCFS. Several alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:6899; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; protein dimerization activity [GO:0046983]; protein homodimerization activity [GO:0042803]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; angiogenesis [GO:0001525]; anterior/posterior pattern specification [GO:0009952]; aorta morphogenesis [GO:0035909]; artery morphogenesis [GO:0048844]; blood vessel development [GO:0001568]; blood vessel morphogenesis [GO:0048514]; cell fate specification [GO:0001708]; cell population proliferation [GO:0008283]; cellular response to fibroblast growth factor stimulus [GO:0044344]; cellular response to retinoic acid [GO:0071300]; cochlea morphogenesis [GO:0090103]; coronary artery morphogenesis [GO:0060982]; determination of left/right symmetry [GO:0007368]; ear morphogenesis [GO:0042471]; embryonic cranial skeleton morphogenesis [GO:0048701]; embryonic viscerocranium morphogenesis [GO:0048703]; enamel mineralization [GO:0070166]; epithelial cell differentiation [GO:0030855]; face morphogenesis [GO:0060325]; heart development [GO:0007507]; heart morphogenesis [GO:0003007]; inner ear morphogenesis [GO:0042472]; lymph vessel development [GO:0001945]; mesenchymal cell apoptotic process [GO:0097152]; mesoderm development [GO:0007498]; middle ear morphogenesis [GO:0042474]; muscle cell fate commitment [GO:0042693]; muscle organ development [GO:0007517]; muscle organ morphogenesis [GO:0048644]; muscle tissue morphogenesis [GO:0060415]; negative regulation of cell differentiation [GO:0045596]; negative regulation of mesenchymal cell apoptotic process [GO:2001054]; neural crest cell migration [GO:0001755]; odontogenesis of dentin-containing tooth [GO:0042475]; outer ear morphogenesis [GO:0042473]; outflow tract morphogenesis [GO:0003151]; outflow tract septum morphogenesis [GO:0003148]; parathyroid gland development [GO:0060017]; pattern specification process [GO:0007389]; pharyngeal system development [GO:0060037]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of mesenchymal cell proliferation [GO:0002053]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of tongue muscle cell differentiation [GO:2001037]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of animal organ morphogenesis [GO:2000027]; regulation of transcription by RNA polymerase II [GO:0006357]; retinoic acid receptor signaling pathway [GO:0048384]; semicircular canal morphogenesis [GO:0048752]; sensory perception of sound [GO:0007605]; social behavior [GO:0035176]; soft palate development [GO:0060023]; thymus development [GO:0048538]; thyroid gland development [GO:0030878]; tongue morphogenesis [GO:0043587]; vagus nerve morphogenesis [GO:0021644] | 11748311_mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects 12533514_identified a single cis-element upstream of Tbx1 that recognized winged helix/forkhead box (Fox)-containing transcription factors 12858556_Genetic dissection of the DiGeorge syndrome phenotype. 15337468_Mutations in TBX1 are not likely to be involved in the cardiac phenotype observed in del22q11 patients. 15337468_Observational study of gene-disease association. (HuGE Navigator) 15703190_a novel nuclear localization signal in Tbx1 is deleted in DiGeorge syndrome patients harboring the 1223delC mutation 16586352_role for Tbx1 in mediating epithelial-mesenchymal signalling in regions of the developing face 16684884_Data show that deficits in prepulse inhibition, a behavioral abnormality and schizophrenia endophenotype, in Df1/+ mice are caused by haploinsufficiency of two genes, Tbx1 and Gnb1l. 16854283_Observational study of gene-disease association. (HuGE Navigator) 17273972_TBX1 missense mutations cause gain of function resulting in Shprintzen syndrome 17377518_screen for TBX1 gene mutations identified 2 mutations in patients with some features compatible with 22q11.2-deletion syndrome but with no deletions. 17438107_T-box transcription factor and a molecule implicated in mesodermal developmecan may be a potential target for human T-cell-mediated cancer immunotherapy. 17479646_Observational study of gene-disease association. (HuGE Navigator) 17622328_Observational study of gene-disease association. (HuGE Navigator) 17622328_TBX1 variation does not make a strong contribution to the genetic etiology of nonsyndromic forms of psychiatric disorders commonly seen in patients with 22q11DS. 17850965_Observational study of gene-disease association. (HuGE Navigator) 17850965_Our data suggest that the genetic polymorphisms within TBX1 do not confer an increased susceptibility to schizophrenia in the Chinese population. 19050236_T-bet expression does not inhibit interferon-alpha-dependent interleukin-2 secretion in human T(central memory) cells. 19336370_Observational study of gene-disease association. (HuGE Navigator) 19407855_Fluorescent in situ hybridisation analysis on FGFR4, ETS2 and brachyury failed to show either amplification or translocation for ERG and ETS2 loci 19467348_Atypical deletion of 22q11.2 detection using the FISH TBX1 probe and molecular characterization with high-density SNP arrays is reported. 19645056_Observational study of gene-disease association. (HuGE Navigator) 19948535_Observational study of gene-disease association. (HuGE Navigator) 20071775_Brachyury is overexpressed in various human tumor tissues and tumor cell lines compared with normal tissues. 20497193_Studies indicate that mutations in the TBX1 gene have been found in patients with phenotypes reminiscent of 22q11.2 syndromes. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20937753_Observational study of gene-disease association. (HuGE Navigator) 20937753_This is the first comprehensive investigation of common and rare TBX1 genetic variants in non-syndromic tetralogy of Fallot cases and it has identified a rare novel functional genetic variant that is a likely susceptibility factor to tetralogy of Fallot. 21041608_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21796729_common DNA variations in TBX1 do not explain variable cardiovascular expression in 22q11DS patients, implicating existence of modifiers in other genes on 22q11.2 or elsewhere in the genome (TBX1 ) 22095455_TBX1 T-box domain binds DNA as two distinct monomers. 22164283_Brachyury and related Tbx proteins interact with the Mixl1 homeodomain protein and negatively regulate Mixl1 transcriptional activity 22185286_TBX1 genetic variants may be associated with conotruncal heart defects. 22801995_The sequence variants within TBX1 gene promoter may contribute to the ventricular septal defect etiology by altering the expression levels of TBX1 gene. 22842189_TBX1 can alter TGF-beta/BMP, an important signaling pathway, through interacting with HOXD10. Above findings may shed light on the mechanism of TBX1 mutations leading to renal malformations found in patients carrying a 22q11 deletion. 22893440_We describe eight patients with variable phenotype features harboring atypical distal deletions of chromosome 22q11.2 not encompassing the TBX1 gene. 22931165_shRNA silencing of the T-box transcription factor Brachyury resulted in downregulation of the EMT and stem cell markers in adenoid cystic carcinoma cell lines. Brachyury expression in clinical samples of AdCC was extremely high and closely related to EMT. 23034814_common DNA variants in TBX1 may be nominally causative for CP in patients with 22q11DS. This raises the possibility that genes elsewhere on the remaining allele of 22q11.2 or in the genome could be relevant. 23828768_Findings indicate an association between TBX1 variations and fetal CTD. The results also demonstrate the power of array CGH to further scrutinize the critical gene(s) of del22q11.2 syndrome responsible for heart defects. 23945394_Results show that TBX1 regulates brain angiogenesis through the DLL4/Notch1-VEGFR3 regulatory axis. 24295890_DNA sequence variants within the TBX1 gene promoter may change TBX1 level, contributing to indirect inguinal hernia development as a rare risk factor 24637876_TBX1 isoform C is the biologically essential variant and that TBX1 mutations are associated with a wide phenotypic spectrum, including most of 22q11.2DS phenotypes. 24998776_Observations suggest that TBX1 loss-of-function mutation may be involved in the pathogenesis of isolated conotrucal heart defects (CTDs)in patients without 22q11.2 deletion. 25168891_SNPs in three genes CYP26B1 rs2241057, CISD1 rs2251039, rs2590370, and TBX1 rs4819522 were involved in six potential pathways to influence serum prostate-specific antigen levels. 25860641_TBX1 loss-of-function mutation with enhanced susceptibility to double outlet right ventricle (DORV) and ventricular septal defect (VSD)in humans, which provides novel insight into the molecular mechanism underlying Congenital heart disease (CHD). 26036351_The results clearly suggest a possible etiologic association between the TBX1 deletion and Tetralogy of Fallot. 27576016_A genome wide are study to identify acute kidney injury risk in critically ill patients identified a locus on chromosome 22 found 140kb upstream of TBX1, and may affect pathways that contribute to AKI pathophysiology. 28272434_a mutation, c.303-305delGAA, located in the third exon of TBX1 that does not disrupt TBX1 mRNA expression or DNA binding activity, but results in decreased TBX1 protein levels and transcriptional activity. 28920943_Studied expression, function, and regulation of T-box transcription factor (TBX1), in human parathyroid adult normal and tumor tissues. 29500247_We identified rare damaging variants in four genes known to be mutated in syndromic lip and/or cleft palate (syCL/P) : TP63 (one family), TBX1 (one family), LRP6 (one family) and GRHL3 (two families), and clinical reassessment confirmed the isolated nature of their lip and/or cleft palate (CL/P). 29568912_PCR and western blotting demonstrated that TBX1 expression may be associated with congenital heart disease. 29596833_The screening of TBX1 coding sequence identified a novel missense mutation c.569C>A (p.P190Q) in six unrelated patients with syndromic congenital heart defects. 30137364_Study of dominantly transmitted isolated hypoparathyroidism in two multigenerational families with 14 affected family members revealed two splice-altering mutations in TBX1(c.1009+1G>C and c.1009+2T>C) leading to skipping of exon 8. TBX1 splice-altering mutations can have incomplete penetrance and variable expressivity, and are the bases for hypoparathyroidism in patients with the common 22q11 deletion syndrome. 30543152_TBX1 is frequently inactivated by promoter methylation and functions as a potential tumor suppressor in thyroid cancer through inhibiting the activities of the PI3K/AKT and MAPK/ERK signaling pathways 31713604_Transactivation of SOX5 by Brachyury promotes breast cancer bone metastasis. 31869684_Generation of human iPSC line from a patient with Tetralogy of Fallot, YAHKMUi001-A, carrying a mutation in TBX1 gene. 31922246_MicroRNA3651 promotes colorectal cancer cell proliferation through directly repressing Tbox transcription factor 1. 31963474_TBX1 and Basal Cell Carcinoma: Expression and Interactions with Gli2 and Dvl2 Signaling. 32947497_Genetic Variant of TBX1 Gene Is Functionally Associated With Adolescent Idiopathic Scoliosis in the Chinese Population. 32949294_Relationship Between Severity of T Cell Lymphopenia and Immune Dysregulation in Patients with DiGeorge Syndrome (22q11.2 Deletions and/or Related TBX1 Mutations): a USIDNET Study. 33557670_T-box transcription factor TBX1, targeted by microRNA-6727-5p, inhibits cell growth and enhances cisplatin chemosensitivity of cervical cancer cells through AKT and MAPK pathways. 34332615_Variants in a cis-regulatory element of TBX1 in conotruncal heart defect patients impair GATA6-mediated transactivation. 34612069_SNP-mediated binding of TBX1 to the enhancer element of IL-10 reduces the risk of Behcet's disease. 34919666_TBX1 functions as a putative oncogene of breast cancer through promoting cell cycle progression. 35095830_Case Report: Unmanipulated Matched Sibling Donor Hematopoietic Cell Transplantation In TBX1 Congenital Athymia: A Lifesaving Therapeutic Approach When Facing a Systemic Viral Infection. | ENSMUSG00000009097 | Tbx1 | 1.622576e+03 | 1.2964781 | 0.374597870 | 0.3040571 | 1.508669e+00 | 0.2193423436 | 0.78763590 | No | Yes | 1.817581e+03 | 197.562298 | 1.307621e+03 | 146.516472 | |
| ENSG00000184178 | 152579 | SCFD2 | protein_coding | Q8WU76 | FUNCTION: May be involved in protein transport. | Alternative splicing;Protein transport;Reference proteome;Transport | hsa:152579; | plasma membrane [GO:0005886]; secretory granule [GO:0030141]; syntaxin binding [GO:0019905]; intracellular protein transport [GO:0006886]; vesicle docking involved in exocytosis [GO:0006904]; vesicle-mediated transport [GO:0016192] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 32213542_PSF Promotes ER-Positive Breast Cancer Progression via Posttranscriptional Regulation of ESR1 and SCFD2. | ENSMUSG00000062110 | Scfd2 | 1.082094e+03 | 0.8134792 | -0.297822643 | 0.2919088 | 1.057337e+00 | 0.3038233399 | 0.81397501 | No | Yes | 1.015629e+03 | 100.906306 | 1.154384e+03 | 117.397068 | ||
| ENSG00000184221 | 116448 | OLIG1 | protein_coding | Q8TAK6 | FUNCTION: Promotes formation and maturation of oligodendrocytes, especially within the brain. Cooperates with OLIG2 to establish the pMN domain of the embryonic neural tube (By similarity). {ECO:0000250, ECO:0000269|PubMed:10719889}. | DNA-binding;Developmental protein;Nucleus;Reference proteome;Transcription;Transcription regulation | hsa:116448; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; neuron differentiation [GO:0030182]; neuron fate commitment [GO:0048663]; oligodendrocyte development [GO:0014003]; regulation of transcription by RNA polymerase II [GO:0006357] | 16267213_Olig1 and Olig2 transcription factors in the human central nervous system are important not only for differentiation of the oligodendrocyte lineage, but they may also have a role in neural cell specification. 16820418_identified novel coding variants in the Olig1 gene, including a trinucleotide repeat, but found no evidence to support the hypothesis that genetic variation in Olig1 influences susceptibility to multiple sclerosis 17171653_Mutations in OLIG1 and OLIG2 are not likely to be associated with this subgroup of hypomyelinating disorders. 17283288_Observational study of gene-disease association. (HuGE Navigator) 17388669_OLIG1 protein expression significantly correlates with overall survival in non-small cell lung cancer patients 17690840_No significan correlation was found between proliferation index in pilocytic astrocytomas and Olig-1 expression. 21206754_analysis of conserved and non-conserved functional elements at the Olig1 and Olig2 locus 21446039_Olig1 was not expressed in freshly isolated oligodendrocytes, but is expressed from the beginning of process extension until membrane maintenance. 23165259_This study review OLIG1 have developmental functions in patterning, neuron subtype specification and the formation of oligodendrocytes and play the role in the postnatal brain during repair processes and in neurological disease states. 23720758_Olig1 is a Smad cofactor involved in TGF-beta-induced cell motility 25930075_This study demonstrated that increased expression of mRNA of OLIG1 in ventral prefrontal white matter in major depressive disorder. 28253550_Olig1 has a critical function in regulation of postnatal neural progenitor cell production in response to Noggin. | ENSMUSG00000046160 | Olig1 | 7.778754e+01 | 0.7907429 | -0.338719439 | 0.4667313 | 4.876823e-01 | 0.4849629871 | 0.87421307 | No | Yes | 5.132950e+01 | 14.020374 | 6.078510e+01 | 17.203532 | ||
| ENSG00000184319 | 118433 | RPL23AP82 | transcribed_unprocessed_pseudogene | 5.939691e+02 | 0.7212645 | -0.471399707 | 0.3064202 | 2.419008e+00 | 0.1198707958 | 0.75783482 | No | Yes | 5.206144e+02 | 75.003873 | 7.173456e+02 | 105.614833 | ||||||||||
| ENSG00000184508 | 374659 | HDDC3 | protein_coding | Q8N4P3 | FUNCTION: ppGpp hydrolyzing enzyme involved in starvation response. {ECO:0000269|PubMed:20818390}. | 3D-structure;Acetylation;Alternative splicing;Hydrolase;Manganese;Metal-binding;Reference proteome | hsa:374659; | guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity [GO:0008893]; metal ion binding [GO:0046872] | 20818390_Mesh1, encoded by HDDC3 in humans, contains an active site for ppGpp hydrolysis and a conserved His-Asp-box motif for Mn(2+) binding. It catalyzes hydrolysis of ppGpp both in vitro and in vivo. 32462112_MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis. | ENSMUSG00000030532 | Hddc3 | 1.669955e+03 | 1.1830213 | 0.242475995 | 0.2985174 | 6.602206e-01 | 0.4164820718 | 0.84868329 | No | Yes | 1.763831e+03 | 197.998478 | 1.251844e+03 | 144.651801 | ||
| ENSG00000184640 | 10801 | SEPTIN9 | protein_coding | Q9UHD8 | FUNCTION: Filament-forming cytoskeletal GTPase (By similarity). May play a role in cytokinesis (Potential). May play a role in the internalization of 2 intracellular microbial pathogens, Listeria monocytogenes and Shigella flexneri. {ECO:0000250, ECO:0000305}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Chromosomal rearrangement;Cytoplasm;Cytoskeleton;Disease variant;GTP-binding;Nucleotide-binding;Phosphoprotein;Reference proteome | This gene is a member of the septin family involved in cytokinesis and cell cycle control. This gene is a candidate for the ovarian tumor suppressor gene. Mutations in this gene cause hereditary neuralgic amyotrophy, also known as neuritis with brachial predilection. A chromosomal translocation involving this gene on chromosome 17 and the MLL gene on chromosome 11 results in acute myelomonocytic leukemia. Multiple alternatively spliced transcript variants encoding different isoforms have been described.[provided by RefSeq, Mar 2009]. | hsa:10801; | actin cytoskeleton [GO:0015629]; axoneme [GO:0005930]; cell division site [GO:0032153]; cytoplasm [GO:0005737]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; non-motile cilium [GO:0097730]; perinuclear region of cytoplasm [GO:0048471]; septin complex [GO:0031105]; septin ring [GO:0005940]; stress fiber [GO:0001725]; cadherin binding [GO:0045296]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; molecular adaptor activity [GO:0060090]; cellular protein localization [GO:0034613]; cytoskeleton-dependent cytokinesis [GO:0061640]; positive regulation of non-motile cilium assembly [GO:1902857] | 11593400_Describes alternative splicing of this gene. 12095151_Fusion of MLL and MSF in adult de novo acute myelomonocytic leukemia (M4) with t(11;17)(q23;q25). 12388755_These results reveal that MSF is required for the completion of cytokinesis and suggest a role that is distinct from that of Nedd5. 12626509_Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules 15485874_Sept7/9b/11 form a complex that has effects on filament elongation, bundling, or disruption 15782116_overexpression of SEPT9 in neoplasia is not simply a proliferation-associated phenomenon, despite its role in cytokines 16161048_SEPT9_v1 is also upregulated in both serous and mucinous carcinomas 16186812_Three mutations in the gene septin 9 (SEPT9) in six families with hereditary neuralgic amyotrophy linked to chromosome 17q25 were reported. 16424018_MSF-A stabilizes HIF-1alpha protein by preventing its ubiquitination and, consequently, activates HIF downstreatm survival genes to promotor tumor progression and angiogenesis 17546647_SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling 18075300_SEPT9_V1 confers resistance to microtubule-mediated HIF-1 inhibitors. 18492087_Both children with dysmorphic syndrome of hereditary neuralgic amyotrophy were shown to have inherited the paternal SEPT9 mutation. 18525421_In this report confirming SEPT9 mutation in a family with suspected hereditary neuralgic amyotrophy, electrophysiological, clinical phenotype, and molecular genetic data of three members are presented. 18642054_monocytic differentiation and a poor prognosis may also be associated with acute myeloid leukemia with the variant MLL/SEPT9 fusion transcript 19018278_SEPT9 DNA methylation may have a role in colorectal cancer 19071215_In mammary epithelial cells, up-regulation of SEPT9_v1 stabilizes JNK by delaying its degradation, thereby activating the JNK transcriptome and suggesting a a novel functional role of SEPT9_v1 in driving cellular proliferation of mammary epithelial cells. 19139049_An intragenic 38 Kb SEPT9 duplication that is linked to hereditary neuralgic amyotrophy in 12 North American families that share the common founder haplotype, is reported. 19204161_Observational study of gene-disease association. (HuGE Navigator) 19204161_Rarely, individuals with sporadic brachial plexopathy may have the same conserved 17q25 sequence found in many North American kindreds with the hereditary version. 19251694_a new mechanism of oxygen-independent activation of HIF-1 has been identified that is mediated by SEPT9_v1 blockade of RACK1 activity on HIF-1alpha degradation 19406918_SEPT9 assay successfully identified 72% of cancers at a specificity of 93% in the training study and 68% of cancers at a specificity of 89% in the testing study 19451530_Results suggest that mutation of the SEPT9 gene is the molecular basis of some cases of hereditary neuralgic amyotrophy (HNA). 19851296_Observational study of gene-disease association. (HuGE Navigator) 19939853_A total of seven heterogeneous SEPT9 duplications have been identified in this study as a causative factor of hereditary neuralgic amyotrophy. 20019224_missense mutation c.262C>T results in a phenotypic spectrum of hereditary neuralgic amyotrophy in a large Japanese family 20029986_Results verify IRX1, EBF3, SLC5A8, SEPT9, and FUSSEL18 as valid methylation markers in two separate sets of HNSCC specimens; also preliminarily show a trend between HPV16 positivity and target gene hypermethylation of IRX1, EBF3, SLC5A8, and SEPT9. 20113838_Coexistence of alternative MLL-SEPT9 fusion transcripts in an acute myeloid leukemia with t(11;17)(q23;q25). 20140221_Data show that methylated DNA from advanced precancerous colorectal lesions can be detected using a panel of two DNA methylation markers, ALX4 and SEPT9. 20198315_Observational study of gene-disease association. (HuGE Navigator) 20407014_New insights and validation are provided for applying SEPT9 transcript variant 1 as a potential target for antitumor therapy via interruption of the HIF-1 pathway. 20682395_Case represents an additional MLL-SEPT9-positive AML that was considered to be related to therapy. RT-PCR and sequencing analyses demonstrated MLL-SEPT9 fusion transcripts with the breakpoint of MLL exon 8/SEPT9 exon 2 and MLL exon 9/SEPT9 exon 2. 21059847_Data demonstrate that SEPT9 mediates the localization of the vesicle-tethering exocyst complex to the midbody, providing mechanistic insight into the role of SEPT9 during cell division. 21267688_Increased methylation of septin 9 resulting in decreased mRNA and protein expression is associated with colorectal cancer. 21737677_uneven distribution of SEPT9 among core septin heteromers causes heterogeneity with respect to both subunit number and polymerization interfaces 21767235_Data illustrated roles of SEPT9 that might contribute to hetero-trimeric septin complex formation. SEPT9 can substitute for septins of the SEPT2 group and partially for SEPT7. 21831286_SEPT9 gene amplification and overexpression during human breast tumorigenesis 22123865_SEPT9 holds a terminal position in the septin octamers, mediating abscission-specific polymerization during cytokinesis. 22278362_SEPT9_v4 expression may be clinically relevant and contribute to some forms of drug resistance. 22956766_Myeloid K562 cells express three SEPT9 isoforms, all of which have an equal propensity to hetero-oligomerize with SEPT7-containing hexamers to generate octameric heteromers. 22981636_The identification of a SEPT9 mutation in a neonate with respiratory distress due to vocal cord paralysis expands the differential diagnosis in these patients. 23049919_SEPT9 in plasma has a role in both left- and right-sided colon cancers 23118862_Matrix stiffness regulates endothelial cell proliferation through septin 9 23572511_SEPT2 forms a 1:1:1 complex with SEPT7 and SEPT9. 23862763_Serum methylation levels of TAC1, SEPT9, and EYA4 were significant discriminants between stage I colorectal cancer and healthy controls. 23988185_epigenetic deregulation of SEPT9 plays a role in the development of colorectal cancer. Aberrant hypermethylation of this gene occurs only in one of its CpG islands and this hypermethylation likely is an early event in the adenoma-carcinoma sequence. 23990466_SEPT9 plays multiple roles in abscission, one of which is regulated by the action of Cdk1 and Pin1. 24067372_SEPT9 isoform 1 is required for the association between HIF-1alpha and importin-alpha to promote efficient nuclear translocation. 24127542_The study shows increased SEPT9 expression as a consequence of genomic amplification and is the first study to profile SEPT9_v1 through SEPT9_v7 isoform-specific mRNA expression in tumor and nontumor tissues from patients with breast cancer. 24344182_SEPT9 repeat motifs bind and bundle MTs, and thereby promote asymmetric neurite growth. 24386354_prognostic value of SHOX2 and SEPT9 DNA methylation in benign, paramalignant and malignant pleural effusions 24535900_Our results suggest that SEPT9 gene methylation is a valuable biomarker for screening CRC in the Chinese population 24633736_SEPT9 promoter methylation is detected in free circulating DNA of lung cancer patients 25293760_Authors report here that the septins SEPT2, -9, -11, and probably -7 form fibrillar structures around the chlamydial inclusion. 25472714_we demonstrate that SEPT9 negatively regulates EGFR degradation by preventing the association of the ubiquitin ligase Cbl with CIN85, resulting in reduced EGFR ubiquitylation 25526039_Results show that in plasma samples, elevated methylated SEPT9 (mSEPT9) values were detected in colorectal cancer, but not in adenomas. Tissue levels of mSEPT9 alone are not sufficient to predict mSEPT9 levels in plasma. 25898316_SEPT9_i1 is expressed in high-grade prostate tumors suggesting it has a significant role in prostate tumorigenesis. 25946211_KRAS mutations and SEPT9 promoter methylation were present in 34% (29/85) and in 82% (70/85) of primary tumor tissue samples. 26471083_provide a full overview of the theoretical basis, development, validation, and clinical applications of the SEPT9 assay for both basic science researchers and clinical practitioners 26633373_SEPT9 promoter methylation and MN frequency, both measured in peripheral blood, occur at an early stage compared to carcinoma development, indicating that the approach might be suitable to monitor CRC development. 26823018_The first evidence of an interaction between septins and a nonmitotic kinesin is provided and it is suggested that SEPT9 modulates the interactions of KIF17 with membrane cargo. 27133379_our study has validated a new SEPT9 assay and combined testing as an aid in cancer detection, providing a new approach for opportunistic CRC screening. 27417143_Septin 9 regulates lipid droplet growth through binding to phosphatidylinositol-5-phosphate in Hepatitis C virus infected cells.Septin 9 regulates lipid droplets growth and perinuclear accumulation in a manner dependent on dynamic microtubules.Septin 9 regulates Hepatitis C virus replication. 27499429_These results indicate that SEPT9_v2 promoter hypermethylation, which silences the expression of SEPT9_v2 mRNA, is observed in a significant proportion of breast tumors, and that methylated SEPT9_v2 may serve as a novel tumor marker for breast cancer. 27660666_Study shows a stepwise increase of SEPT9 methylation from non-cancerous to cancerous tissue in colorectal adenocarcinoma. 27753040_Studies indicate that methylated Septin 9 ((m)SEPT9) can be consistently detected in plasma samples derived from whole blood samples collected with S-Monovette(R) K3E and BD Vacutainer (R) K2EDTA tubes stored at 2-8 degrees C for a maximum of 24 h and for samples collected in S-Monovette CPDA tubes stored at 18-25 degrees C for up to 48 h. 27999621_Study found SHOX2 and SEPT9 frequently methylated in biliary tract cancers. 28128742_The SEPT9 assay exhibited satisfactory performance in colorectal cancer diagnosis and screening, while more evidence is needed for therapeutic effect monitoring and prognosis prediction. 28338090_Thus our data indicate that Sept9_i2 is a negative regulator of breast tumorigenesis. We propose that Sept9 tumorigenic properties depend on the balance between Sept9_i1 and Sept9_i2 expression levels. 29610456_Post-therapeutic SHOX2 and SEPT9 circulating cell-free DNA(ccfDNA) methylation levels correlated with UICC stage (all P <0.01). SEPT9 ccfDNA methylation further allowed for an accurate pre- and post-therapeutic detection of distant metastases. 29627389_The primary aim of this study was to evaluate the diagnostic accuracy of a PCR-based assay for the analysis of SEPT9 promoter methylation in circulating cell-free DNA (mSEPT9) for diagnosing hepatocellular carcinoma. 29724999_Repression of SEPTIN2 and SEPTIN9 suppresses tumor growth of human glioblastoma cells and tumor progression in a mouse model. 29970704_The plasma levels of septin-9 and clusterin in ovarian cancer patients were abnormally elevated, which might be used as potential candidates of peripheral blood tumor biomarkers for early diagnosis of EOC and septin-9 might be related to distal metastases of EOC. 30043648_mSEPT9 is effective for colorectal cancer postsurgical assessment and prognosis prediction. 30158637_The authors propose that septins in general and SEPT9 in particular play a previously unappreciated role in osteoclastic bone resorption. 30273999_SEPT9 DNA methylation was detected in untreated colorectal carcinomas but not in adenomas. 30426784_The quantitative profiling of blood mSEPT9 determines the detection performance on colorectal tumors. 30670682_Authors show that coupling septins (including SEPT9_i1) overexpression together with long-chain tubulin polyglutamylation induce significant paclitaxel resistance in several naive (taxane-sensitive) cell lines and accordingly stimulate the binding of CLIP-170 and MCAK to microtubules. 31088406_Study indicated that peripheral SEPT9 methylated DNA may be useful for the screening, early diagnosis, and recurrence monitoring of colorectal cancer. 31114136_methylated septin 9 is associated with advanced stages in Chinese colorectal cancer patients 31285548_results indicate that SEPT9 promotes upregulation and both trafficking and secretion of MMPs near FAs, thus enhancing migration and invasion of breast cancer cells 31316143_Methylated Septin 9 and Carcinoembryonic Antigen for Serological Diagnosis and Monitoring of Patients with Colorectal Cancer After Surgery. 31426855_Study found that hypermethylation of SEPT9 exhibits a high sensitivity and specificity in the diagnosis of cervical cancer. Up- and downregulation of SEPT9 affected the biological behavior of cervical cancer cells through the regulation of HMGB1. Moreover, SEPT9 mediated miR-375 could enhance the cell resistance to ionizing radiation via affecting the tumor-associated macrophages (TAMs) polarization. 31558699_SEPT9_i1 regulates human breast cancer cell motility through cytoskeletal and RhoA/FAK signaling pathway regulation. 32106387_Down-regulation of SEPT9 inhibits glioma progression through suppressing TGF-beta-induced epithelial-mesenchymal transition (EMT). 32122354_First description of a SEPT9 variant associated to a Charcot-Marie-Tooth Disease (CMT) phenotype; this suggests that SEPT9 is a new sufficient candidate gene in CMT. 32138771_SEPT9_v2, frequently silenced by promoter hypermethylation, exerts anti-tumor functions through inactivation of Wnt/beta-catenin signaling pathway via miR92b-3p/FZD10 in nasopharyngeal carcinoma cells. 32378260_Comprehensive analysis on the whole Rho-GAP family reveals that ARHGAP4 suppresses EMT in epithelial cells under negative regulation by Septin9. 32454907_Septin 9 Methylation in Nasopharyngeal Swabs: A Potential Minimally Invasive Biomarker for the Early Detection of Nasopharyngeal Carcinoma. 32566042_A First Step to a Biomarker of Curative Surgery in Colorectal Cancer by Liquid Biopsy of Methylated Septin 9 Gene. 32945374_Plasma levels of methylated septin 9 are capable of detecting hepatocellular carcinoma and hepatic cirrhosis. 33178361_SEPT9 Gene Methylation as a Noninvasive Marker for Hepatocellular Carcinoma. 33915163_CircRNA SEPT9 contributes to malignant behaviors of glioma cells via miR-432-5p-mediated regulation of LASP1. 33931320_Role of methylated septin 9 as an adjunct diagnostic and prognostic biomarker in hepatocellular carcinoma. 34350965_Insights into animal septins using recombinant human septin octamers with distinct SEPT9 isoforms. 34409731_Proteomic profiling of the oncogenic septin 9 reveals isoform-specific interactions in breast cancer cells. 34524873_ARHGAP4-SEPT2-SEPT9 complex enables both up- and down-modulation of integrin-mediated focal adhesions, cell migration, and invasion. 34854883_Septin-microtubule association via a motif unique to isoform 1 of septin 9 tunes stress fibers. 34873218_Application of droplet digital polymerase chain reaction of plasma methylated septin 9 on detection and early monitoring of colorectal cancer. 35089073_Noninvasive Detection of Esophageal Cancer by the Combination of mSEPT9 and SNCG. 35481971_Distinct Performance of Methylated SEPT9 in Upper and Lower Gastrointestinal Cancers and Combined Detection with Protein Markers. | ENSMUSG00000059248 | Septin9 | 4.011228e+03 | 1.4076176 | 0.493255493 | 0.3302872 | 2.222824e+00 | 0.1359841074 | 0.76593507 | No | Yes | 4.673986e+03 | 608.006953 | 2.793092e+03 | 373.266269 | |
| ENSG00000184708 | 56478 | EIF4ENIF1 | protein_coding | Q9NRA8 | FUNCTION: EIF4E-binding protein that regulates translation and stability of mRNAs in processing bodies (P-bodies) (PubMed:16157702, PubMed:24335285, PubMed:27342281, PubMed:32354837). Plays a key role in P-bodies to coordinate the storage of translationally inactive mRNAs in the cytoplasm and prevent their degradation (PubMed:24335285, PubMed:32354837). Acts as a binding platform for multiple RNA-binding proteins: promotes deadenylation of mRNAs via its interaction with the CCR4-NOT complex, and blocks decapping via interaction with eIF4E (EIF4E and EIF4E2), thereby protecting deadenylated and repressed mRNAs from degradation (PubMed:27342281, PubMed:32354837). Component of a multiprotein complex that sequesters and represses translation of proneurogenic factors during neurogenesis (By similarity). Promotes miRNA-mediated translational repression (PubMed:24335285, PubMed:27342281, PubMed:28487484). Required for the formation of P-bodies (PubMed:16157702, PubMed:22966201, PubMed:27342281, PubMed:32354837). Involved in mRNA translational repression mediated by the miRNA effector TNRC6B by protecting TNRC6B-targeted mRNAs from decapping and subsequent decay (PubMed:32354837). Also acts as a nucleoplasmic shuttling protein, which mediates the nuclear import of EIF4E and DDX6 by a piggy-back mechanism (PubMed:10856257, PubMed:28216671). {ECO:0000250|UniProtKB:Q9EST3, ECO:0000269|PubMed:10856257, ECO:0000269|PubMed:16157702, ECO:0000269|PubMed:22966201, ECO:0000269|PubMed:24335285, ECO:0000269|PubMed:27342281, ECO:0000269|PubMed:28216671, ECO:0000269|PubMed:28487484, ECO:0000269|PubMed:32354837}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Isopeptide bond;Nucleus;Phosphoprotein;Protein transport;RNA-mediated gene silencing;Reference proteome;Translation regulation;Transport;Ubl conjugation | The protein encoded by this gene is a nucleocytoplasmic shuttle protein for the translation initiation factor eIF4E. This shuttle protein interacts with the importin alpha-beta complex to mediate nuclear import of eIF4E. It is predominantly cytoplasmic; its own nuclear import is regulated by a nuclear localization signal and nuclear export signals. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2009]. | hsa:56478; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; P-body [GO:0000932]; PML body [GO:0016605]; kinase binding [GO:0019900]; mRNA binding [GO:0003729]; nuclear export signal receptor activity [GO:0005049]; RNA binding [GO:0003723]; gene silencing by RNA [GO:0031047]; mRNA stabilization [GO:0048255]; negative regulation of deadenylation-dependent decapping of nuclear-transcribed mRNA [GO:0106289]; negative regulation of neuron differentiation [GO:0045665]; negative regulation of translation [GO:0017148]; P-body assembly [GO:0033962]; positive regulation of miRNA mediated inhibition of translation [GO:1905618]; positive regulation of nuclear-transcribed mRNA poly(A) tail shortening [GO:0060213]; protein import into nucleus [GO:0006606]; stem cell population maintenance [GO:0019827] | 15840819_human P bodies contain the cap-binding protein eIF4E and the related factor eIF4E-transporter (eIF4E-T), suggesting novel roles for these proteins in targeting mRNAs for 5' --> 3' degradation 16157702_A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay is described. 18343217_Overexpression of eIF4E-T triggers the movement of eIF4E into the processing bodies. 19850929_Hsp90 probably contributes to the correct localization of eIF4E and 4E-T to stress granules and also to the interaction between eIF4E and eIF4G, both of which may be needed for eIF4E to acquire the physiological functionality 22966201_The c-Jun N-terminal kinase (JNK) is targeted to processing bodies in response to oxidative stress and promotes the phosphorylation of 4E-T. 23902945_Data demonstrate that EIF4ENIF1 is associated with dominantly inherited primary ovarian insufficiency. 23991149_that while both eIF4E1 and eIF4E2 bind 4E-T via the canonical YX 4Lvarphi sequence, nearby downstream sequences also influence eIF4E:4E-T interactions 24335285_This work has demonstrated the conserved yet unpredicted and surprising translational control of bound mRNAs by 4E-T, which does not involve eIF4E, nor P-body components. 25456498_neural precursors are transcriptionally primed to generate neurons, but an eIF4E/4E-T complex sequesters and represses translation of proneurogenic proteins to determine appropriate neurogenesis. 26027925_our data support a model where 4E-T promotes mRNA turnover by physically linking the 3'-terminal mRNA decay machinery to the 5' cap 27342281_we demonstrate that joint deletion of two short conserved motifs that bind UNR and DDX6 relieves repression of 4E-T-bound mRNA, in part reliant on the 4E-T-DDX6-CNOT1 axis. | ENSMUSG00000020454 | Eif4enif1 | 2.072803e+03 | 0.9887488 | -0.016324093 | 0.2758275 | 3.593740e-03 | 0.9521971996 | 0.99040425 | No | Yes | 1.893629e+03 | 151.588193 | 1.788111e+03 | 146.876778 | |
| ENSG00000184922 | 752 | FMNL1 | protein_coding | O95466 | FUNCTION: May play a role in the control of cell motility and survival of macrophages (By similarity). Plays a role in the regulation of cell morphology and cytoskeletal organization. Required in the cortical actin filament dynamics and cell shape. {ECO:0000250, ECO:0000269|PubMed:21834987}. | 3D-structure;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Lipoprotein;Membrane;Myristate;Phosphoprotein;Reference proteome | This gene encodes a formin-related protein. Formin-related proteins have been implicated in morphogenesis, cytokinesis, and cell polarity. An alternative splice variant has been described but its full length sequence has not been determined. [provided by RefSeq, Jul 2008]. | hsa:752; | bleb [GO:0032059]; cell cortex [GO:0005938]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; membrane [GO:0016020]; phagocytic vesicle [GO:0045335]; plasma membrane [GO:0005886]; actin filament binding [GO:0051015]; GTPase activating protein binding [GO:0032794]; small GTPase binding [GO:0031267]; actin filament severing [GO:0051014]; cell migration [GO:0016477]; cortical actin cytoskeleton organization [GO:0030866]; regulation of cell shape [GO:0008360] | 14592423_Western blot analysis demonstrated that the protein encoded by this gene is overexpressed in lymphoid malignancies, cancer cell lines and peripheral blood leukocyte from chronic lymphocytic leukemia (CLL) patients. 19815554_Results describe N-myristoylation as a regulative mechanism of FMNL1 responsible for membrane trafficking potentially involved in a diversity of polarized processes of hematopoietic lineage-derived cells. 20617518_Our data suggest that FRL1 is responsible for modifying actin at the macrophage podosome and may be involved in actin cytoskeleton dynamics during adhesion and migration within tissues. 21148482_after Rac-dependent activation of FMNL1, srGAP2 mediates a potent mechanism to limit the duration of Rac action and inhibit formin activity during rapid actin dynamics. 23182705_constitutively activated form of FMNL1 (FMNL1gamma) induces localization of AHNAK1 to the cell membrane. 23460512_Results suggest that macrophage coiling phagocytosis is a complex process involving several actin nucleation/regulatory factors. They also point specifically to the formins mDia1 and FMNL1 as novel regulators of spirochete uptake by human immune cells. 23801653_FMNL1 contributes to leukemogenesis and could act in part through Rac1 regulation. 29283461_Results indicated that FMNL1 is a susceptibility gene for leprosy. 30013189_High expression of FMNL1, resulted from decreased miR-16 and/or MTA1 amplification. 30977161_The carboxy-terminus of the formin FMNL1 bundles actin to potentiate adenocarcinoma migration. 31001854_miR-143 inhibits proliferation and metastasis of nasopharyngeal carcinoma cells via targeting FMNL1 based on clinical and radiologic findings. 31387165_Suppressing FMNL1 expression could inhibit bone metastasis in NSCLC through blocking TGF-beta1 signaling. 31861134_Formin-like 1 protein (FMNL1) is a promising therapeutic target for glioblastoma multiforme (GBM) progression. | ENSMUSG00000055805 | Fmnl1 | 4.991403e+01 | 1.3479888 | 0.430808559 | 0.4908223 | 7.545163e-01 | 0.3850500991 | 0.84317192 | No | Yes | 5.374241e+01 | 12.941079 | 3.035496e+01 | 7.928724 | |
| ENSG00000185163 | 317781 | DDX51 | protein_coding | Q8N8A6 | FUNCTION: ATP-binding RNA helicase involved in the biogenesis of 60S ribosomal subunits. {ECO:0000250}. | ATP-binding;Acetylation;Helicase;Hydrolase;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribosome biogenesis;rRNA processing | hsa:317781; | membrane [GO:0016020]; nucleolus [GO:0005730]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; rRNA processing [GO:0006364] | 27198888_these data suggest that DDX51 aids cell cancer proliferation by regulating multiple signaling pathways | ENSMUSG00000029504 | Ddx51 | 3.292201e+03 | 1.1644943 | 0.219703616 | 0.2708865 | 6.672151e-01 | 0.4140242260 | 0.84733931 | No | Yes | 3.047634e+03 | 276.166825 | 2.641530e+03 | 245.752667 | ||
| ENSG00000185359 | 9146 | HGS | protein_coding | O14964 | FUNCTION: Involved in intracellular signal transduction mediated by cytokines and growth factors. When associated with STAM, it suppresses DNA signaling upon stimulation by IL-2 and GM-CSF. Could be a direct effector of PI3-kinase in vesicular pathway via early endosomes and may regulate trafficking to early and late endosomes by recruiting clathrin. May concentrate ubiquitinated receptors within clathrin-coated regions. Involved in down-regulation of receptor tyrosine kinase via multivesicular body (MVBs) when complexed with STAM (ESCRT-0 complex). The ESCRT-0 complex binds ubiquitin and acts as sorting machinery that recognizes ubiquitinated receptors and transfers them to further sequential lysosomal sorting/trafficking processes. May contribute to the efficient recruitment of SMADs to the activin receptor complex. Involved in receptor recycling via its association with the CART complex, a multiprotein complex required for efficient transferrin receptor recycling but not for EGFR degradation. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Endosome;Membrane;Metal-binding;Phosphoprotein;Protein transport;Reference proteome;Transport;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene regulates endosomal sorting and plays a critical role in the recycling and degradation of membrane receptors. The encoded protein sorts monoubiquitinated membrane proteins into the multivesicular body, targeting these proteins for lysosome-dependent degradation. [provided by RefSeq, Dec 2010]. | hsa:9146; | cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; endosome [GO:0005768]; ESCRT-0 complex [GO:0033565]; extracellular exosome [GO:0070062]; intracellular membrane-bounded organelle [GO:0043231]; lysosome [GO:0005764]; multivesicular body membrane [GO:0032585]; phagocytic vesicle lumen [GO:0097013]; metal ion binding [GO:0046872]; phosphatidylinositol binding [GO:0035091]; protein domain specific binding [GO:0019904]; ubiquitin binding [GO:0043130]; ubiquitin-like protein ligase binding [GO:0044389]; endosomal transport [GO:0016197]; macroautophagy [GO:0016236]; membrane invagination [GO:0010324]; multivesicular body assembly [GO:0036258]; negative regulation of angiogenesis [GO:0016525]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of platelet-derived growth factor receptor signaling pathway [GO:0010642]; negative regulation of receptor signaling pathway via JAK-STAT [GO:0046426]; negative regulation of vascular endothelial growth factor receptor signaling pathway [GO:0030948]; positive regulation of exosomal secretion [GO:1903543]; positive regulation of gene expression [GO:0010628]; protein localization to membrane [GO:0072657]; protein targeting to lysosome [GO:0006622]; regulation of MAP kinase activity [GO:0043405]; regulation of protein catabolic process [GO:0042176]; signal transduction [GO:0007165] | 12444101_The HRS domain required for merlin binding is narrowed to a region (residues 470-497) containing the predicted coiled-coil domain whereas the major domain responsible for HRS growth suppression is distinct (residues 498-550). 12444102_HRS inhibits Stat3 activation in schwannoma cells. 12554698_Overexpression Hgs in T cells yielded a dose-dependent decrease in cotransfected reporter gene expression, indicating an inhibitory function of this molecule. 15113837_Data suggested that association with Hrs is a prerequisite for signal transducing adaptor molecule function. 15212941_Hrs regulates the sorting of ligand-stimulated and unstimulated growth factor receptors on early endosomes, and the FYVE domain, which is required for Hrs to reside in a microdomain of early endosomes. 15772161_the endosome-associated protein hrs is a subunit of a protein complex containing actinin-4, BERP, and myosin V that is necessary for efficient TfR recycling but not for EGFR degradation 15828871_Analysis with phospho-specific antibodies indicates that 3 kinases generate a signal-specific, combinatorial phosphorylation profile of the Hrs-STAM complex, with the potential of diversifying tyrosine kinase receptor signalling through a common element. 15944737_Hrs plays role in a cargo-specific recycling mechanism, which is critical to controlling functional activity of the largest known family of signaling receptors. 16352611_role of HRS in up-regulating MAPK, presumably involving interaction with PELP1. 16448788_PIKfyve is distributed in microdomains that are distinct from those occupied by EEA1 and Hrs 16516194_Hgs is a novel Smad5 interactor and an inhibitor of bone morphogenetic protein (BMP) signaling. 16707569_These results indicate that Tsg101 is required for the formation of stable vacuolar domains within the early endosome that develop into multivesicular body (MVBs) and Hrs is required for the accumulation of internal vesicles within MVBs. 16832584_HGS and GUK1 were significantly over expressed in GH-secreting adenomas, compared with ACTH-secreting adenomas and nonfunctioning tumors, and with PRL-secreting adenomas, respectively. 17014699_Four proteins (TSG101,Hrs,Aip1/Alix, and Vps4B) of the ESCRT (endosomal sorting complex required for transport) machinery were localized in T cells and macrophages by quantitative electron microscopy. 17138565_Specific sequence in the beta2-adrenergic receptor cytoplasmic tail confers Hrs dependence on receptor recycling. 17320394_hSpry2 binds to the endocytic regulatory protein, hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and blocks intracellular signal propagation. 17445799_We conclude that Hrs is a positive regulator of VEGF-R2 and IR signaling and that ectopic expression of Hrs protects both VEGF-R2 and IR from degradation. 17545595_Targeted disruption of Hrs by small interfering RNA effectively attenuated the proliferation, anchorage-independent growth, tumorigenesis, and metastatic potential of HeLa cells in vitro and in vivo. 17624298_propose a novel function of Hrs, as a crucial player in the maturation of autophagosomes 17675298_HRS mediates post-endocytic trafficking of protease-activated receptor 2 and calcitonin receptor-like receptor 17804729_PELP1 and HRS reallocate to autophagosomes in response to resveratrol treatment, which might be important in the process of autophagy in the cancer cells. 18362181_These results indicate that Eps15b is an endosomally localized isoform of Eps15 that is present in the Hrs complex via direct Hrs interaction and important for the sorting function of this complex. 18445679_these results indicate that precise sorting of IL-2Rbeta from early to late endosomes is mediated by Hrs, a known sorting component of the ubiquitin-dependent machinery, in a manner that is independent of UIM-ubiquitin binding 18675823_Data show that Trak1 interacts with hepatocyte-growth-factor-regulated tyrosine kinase substrate (Hrs), an essential component of the endosomal sorting and trafficking machinery. 18767904_Data show that Hrs is essential for lysosomal targeting but not multivesicular body biogenesis and transport to late endosomes, while SNX3 is required for multivesicular body formation, but not for EGF receptor degradation. 20812348_Plasma hepatocyte growth factor is associated with periampullary cancer. 21304933_ESCRT-0 component HRS is required for HIV-1 Vpu-mediated BST-2/tetherin down-regulation. 21748597_Hrs inhibits HIV-1 production by inhibiting citron kinase-mediated exocytosis. 22138215_These results suggested that HCV secretion from host cells requires Hrs-dependent exosomal pathway in which the viral assembly is also involved. 22800866_Hrs tyrosine phosphorylation detected upon EGF-stimulation. 22832105_Hrs is a regulator of endosomal cholesterol trafficking. 23867513_the role for tumoral c-Met expression or sMet/HGF levels as upfront selection criterion or predictive biomarkers deserve further study in this emerging area of therapeutic focus in RCC 24204276_we identified an interaction between EsxH, which is secreted by the Esx-3 TSSS, and human hepatocyte growth factor-regulated tyrosine kinase substrate (Hgs/Hrs), a component of the endosomal sorting complex required for transport (ESCRT). 25296754_ESCRT-0 protein hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) is targeted to endosomes independently of signal-transducing adaptor molecule (STAM) and the complex formation with STAM promotes its endosomal dissociation. 26715116_Results demonstrate the existence of crosstalk between beta-catenin signaling and HGS in two different types of cancer (hepatoblastoma and colorectal). 26819309_detailed analysis of the subcellular localization and functional significance of Hrs in macropinocytosis-mediated entry of Kaposi's Sarcoma-Associated Herpesvirus 28959041_Degradation of CLR-RAMP2 was sensitive to overexpression of hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), but not to HRS knockdown, whereas CLRDelta9KR-RAMP2 degradation was unaffected. Overexpression, but not knockdown of HRS, promoted hyperubiquitination of CLR under basal conditions. These results propose a role for ubiquitin and HRS in the regulation of AM-induced degradation of CLR-RAMP2. 30456986_Post-traumatic stress disorder (PTSD) is associated with differential methylation, measured in blood, within HGS and NRG1 across three civilian cohorts. 31597781_The ESCRT-0 protein HRS interacts with the HTLV-II antisense protein APH-2 and suppresses viral replication. [APH-2] 31744816_Study provides in vivo evidence that Beclin 1 suppresses tumor proliferation by regulating the endocytic trafficking and degradation of the EGFR and transferrin (TFR1) receptors. Beclin 1 promoted endosomal recruitment of hepatocyte growth factor tyrosine kinase substrate (HRS), which was necessary for sorting surface receptors to intraluminal vesicles for signal silencing and lysosomal degradation. 32044971_Hepatocyte growth factor-regulated tyrosine kinase substrate is essential for endothelial cell polarity and cerebrovascular stability. 32546618_Epstein-Barr Virus LMP1 Promotes Syntenin-1- and Hrs-Induced Extracellular Vesicle Formation for Its Own Secretion To Increase Cell Proliferation and Migration. | ENSMUSG00000025793 | Hgs | 5.468365e+03 | 1.0069772 | 0.010030948 | 0.2630699 | 1.463551e-03 | 0.9694832619 | 0.99408977 | No | Yes | 4.670988e+03 | 519.865695 | 4.802640e+03 | 548.345905 | |
| ENSG00000185480 | 55010 | PARPBP | protein_coding | Q9NWS1 | FUNCTION: Required to suppress inappropriate homologous recombination, thereby playing a central role DNA repair and in the maintenance of genomic stability. Antagonizes homologous recombination by interfering with the formation of the RAD51-DNA homologous recombination structure. Binds single-strand DNA and poly(A) homopolymers. Positively regulate the poly(ADP-ribosyl)ation activity of PARP1; however such function may be indirect. {ECO:0000269|PubMed:20931645, ECO:0000269|PubMed:22153967}. | Alternative splicing;Cytoplasm;DNA damage;DNA repair;DNA-binding;Nucleus;Reference proteome | hsa:55010; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; DNA binding [GO:0003677]; DNA repair [GO:0006281]; negative regulation of double-strand break repair via homologous recombination [GO:2000042] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20931645_Demonstrated that C12orf48 protein could directly interact with Poly(ADP-ribose) Polymerase-1 (PARP-1). Knockdown of C12orf48 by siRNA in PDAC cells significantly suppressed their growth. 22153967_PARI suppresses inappropriate recombination events at mammalian replication forks. 23039964_Knockdown of C12orf48 by siRNA in HeLa cells significantly suppressed their growth 23436799_PARI overexpression promotes genomic instability and pancreatic tumorigenesis. 26792895_PARI inhibits homologous recombination in vivo, and its knockdown suppresses the UV sensitivity of RAD18-depleted cells 28894029_PARI is a latent modulator of stalled fork processing, which is required for stable genome inheritance under both endogenous and exogenous replication stress 29805304_PARI plays potential oncogenic roles and functions as a transcriptional target and effector of FOXM1 in gastric cancer development 30949905_PARP1-binding protein was significantly upregulated in Hepatocellular Carcinoma tissues compared with normal liver. High PARPBP expression was associated with elevated serum AFP level, vascular invasion, poor tumor differentiation, and advanced TNM stage. 30967629_show that PARI expression negatively correlates with expression of differentiation markers in clinical myeloid leukemia samples, suggesting that targeting PARI may restore differentiation ability of leukemia cells and antagonize their proliferation 31611199_PARPBP expression is enhanced in lung adenocarcinoma tissues and is a potential factor in the progression of lung adenocarcinoma. | ENSMUSG00000035365 | Parpbp | 1.446344e+02 | 0.6323529 | -0.661198233 | 0.4149114 | 2.503104e+00 | 0.1136221633 | 0.75783482 | No | Yes | 1.033454e+02 | 20.403928 | 1.702276e+02 | 33.995805 | ||
| ENSG00000185485 | 255812 | SDHAP1 | transcribed_unprocessed_pseudogene | 32211849_LncRNA SDHAP1 confers paclitaxel resistance of ovarian cancer by regulating EIF4G2 expression via miR-4465. | 3.277772e+02 | 0.7868074 | -0.345917564 | 0.2932547 | 1.390496e+00 | 0.2383213101 | 0.78818582 | No | Yes | 2.125394e+02 | 46.270906 | 3.185071e+02 | 70.965755 | |||||||||
| ENSG00000185504 | 80233 | FAAP100 | protein_coding | Q0VG06 | FUNCTION: Plays a role in Fanconi anemia-associated DNA damage response network. Regulates FANCD2 monoubiquitination and the stability of the FA core complex. Induces chromosomal instability as well as hypersensitivity to DNA cross-linking agents, when repressed. {ECO:0000269|PubMed:17396147}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;DNA-binding;Nucleus;Phosphoprotein;Reference proteome | FAAP100 is a component of the Fanconi anemia (FA; MIM 277650) core complex and is required for core complex stability and FANCD2 (see MIM 227646) monoubiquitination (Ling et al., 2007 [PubMed 17396147]).[supplied by OMIM, Mar 2008]. | hsa:80233; | cytosol [GO:0005829]; Fanconi anaemia nuclear complex [GO:0043240]; nucleoplasm [GO:0005654]; DNA binding [GO:0003677]; interstrand cross-link repair [GO:0036297] | 17396147_FAAP100 as a new critical component of the FA-BRCA DNA damage response network. | ENSMUSG00000025384 | Faap100 | 3.186320e+03 | 1.4949739 | 0.580120256 | 0.3347182 | 3.013654e+00 | 0.0825659589 | 0.73907303 | No | Yes | 3.668353e+03 | 526.630391 | 2.165734e+03 | 319.470231 | |
| ENSG00000185596 | WASH3P | transcribed_unprocessed_pseudogene | 1.375780e+03 | 1.0244658 | 0.034871862 | 0.2738151 | 1.649622e-02 | 0.8978026673 | 0.98099463 | No | Yes | 1.195147e+03 | 152.733007 | 1.157716e+03 | 151.715391 | |||||||||||
| ENSG00000185986 | 728609 | SDHAP3 | transcribed_unprocessed_pseudogene | 7.516226e+02 | 0.9200875 | -0.120157008 | 0.3057332 | 1.562277e-01 | 0.6926535923 | 0.93378797 | No | Yes | 5.341306e+02 | 113.822529 | 6.826297e+02 | 149.012933 | ||||||||||
| ENSG00000186073 | 84529 | CDIN1 | protein_coding | Q9Y2V0 | FUNCTION: Plays a role in erythroid cell differentiation. {ECO:0000269|PubMed:31191338}. | Alternative splicing;Congenital dyserythropoietic anemia;Cytoplasm;Disease variant;Hereditary hemolytic anemia;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a protein with two predicted helix-turn-helix domains. Mutations in this gene were found in families with congenital dyserythropoietic anemia type Ib. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Mar 2014]. | hsa:84529; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; erythrocyte differentiation [GO:0030218] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 29031773_Mutation in C!%ORF41 gene is associated with congenital dyserythropoietic anemia. 32239177_A complex comprising C15ORF41 and Codanin-1: the products of two genes mutated in congenital dyserythropoietic anaemia type I (CDA-I). 32293259_Characterization of the interactions between Codanin-1 and C15Orf41, two proteins implicated in congenital dyserythropoietic anemia type I disease. 32518175_Genetic and functional insights into CDA-I prevalence and pathogenesis. 33159567_Congenital dyserythropoietic anemia types Ib, II, and III: novel variants in the CDIN1 gene and functional study of a novel variant in the KIF23 gene. | ENSMUSG00000040282 | Cdin1 | 1.336534e+02 | 0.8941015 | -0.161489505 | 0.3728211 | 1.903299e-01 | 0.6626422206 | 0.92880790 | No | Yes | 1.140069e+02 | 21.808296 | 1.244231e+02 | 24.358780 | |
| ENSG00000186184 | 51082 | POLR1D | protein_coding | P0DPB5 | Mouse_homologues FUNCTION: DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Common core component of RNA polymerases I and III which synthesize ribosomal RNA precursors and small RNAs, such as 5S rRNA and tRNAs, respectively. | Acetylation;Alternative splicing;Direct protein sequencing;Phosphoprotein;Reference proteome | The protein encoded by this gene is a component of the RNA polymerase I and RNA polymerase III complexes, which function in the synthesis of ribosomal RNA precursors and small RNAs, respectively. Mutations in this gene are a cause of Treacher Collins syndrome (TCS), a craniofacial development disorder. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2011]. | Mouse_homologues mmu:20018; | cytosol [GO:0005829]; nucleoplasm [GO:0005654] | 8955128_Characterization of the homologous mouse protein. 12446911_analyzed the kinetics of assembly and elongation of the RNA polymerase I complex on endogenous ribosomal genes in the nuclei of living cells with the use of in vivo microscopy 19724895_Observational study of gene-disease association. (HuGE Navigator) 19834535_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 21131976_heterozygous mutations of POLR1D in 252 individuals with Treacher Collins syndrome 24603435_Autosomal recessive POLR1D mutation with decrease of TCOF1 mRNA is responsible for Treacher Collins syndrome. 24690222_Mutations in TCOF1, POLR1C and POLR1D have all been implicated in causing TCS 25348728_Whole exome sequencing identified a nonsynonymous mutation in POLR1D (subunit of RNA polymerase I and II): exon2:c.T332C:p.L111P. 25790162_We report a clinical and extensive molecular study, including TCOF1, POLR1D, POLR1C, and EFTUD2 genes, in a series of 146 patients with TCS. 30582221_High POLR1D expression is associated with colorectal cancer progression. 31649276_POLR1B and neural crest cell anomalies in Treacher Collins syndrome type 4. 31722331_High POLR1D expression is an independent prognostic factor for poor overall survival in colorectal cancer. 32087735_Cell-free DNA analysis reveals POLR1D-mediated resistance to bevacizumab in colorectal cancer. | ENSMUSG00000029642 | Polr1d | 3.178356e+03 | 0.7480252 | -0.418841294 | 0.2661084 | 2.473709e+00 | 0.1157644452 | 0.75783482 | No | Yes | 2.580503e+03 | 251.974155 | 3.052133e+03 | 305.623914 | |
| ENSG00000186815 | 53373 | TPCN1 | protein_coding | Q9ULQ1 | FUNCTION: Intracellular channel initially characterized as a non-selective Ca(2+)-permeable channel activated by NAADP (nicotinic acid adenine dinucleotide phosphate), it is also a voltage-gated highly-selective Na(+) channel activated directly by PI(3,5)P2 (phosphatidylinositol 3,5-bisphosphate) that senses pH changes and confers electrical excitability to organelles (PubMed:19620632, PubMed:23063126, PubMed:24776928, PubMed:23394946). Localizes to the early and recycling endosomes membranes where it plays a role in the uptake and processing of proteins and regulates organellar membrane excitability, membrane trafficking and pH homeostasis (PubMed:23394946) (Probable). Ion selectivity is not fixed but rather agonist-dependent and under defined ionic conditions, can be readily activated by both NAADP and PI(3,5)P2 (Probable). Required for mTOR-dependent nutrient sensing (PubMed:23394946) (Probable). {ECO:0000269|PubMed:19620632, ECO:0000269|PubMed:23063126, ECO:0000269|PubMed:23394946, ECO:0000269|PubMed:24776928, ECO:0000305|PubMed:32679067}.; FUNCTION: (Microbial infection) During Ebola virus (EBOV) infection, controls the movement of endosomes containing virus particles and is required by EBOV to escape from the endosomal network into the cell cytoplasm. {ECO:0000269|PubMed:25722412}. | Alternative splicing;Calcium;Calcium channel;Calcium transport;Coiled coil;Endosome;Glycoprotein;Ion channel;Ion transport;Lysosome;Membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport;Voltage-gated channel | Voltage-gated Ca(2+) and Na+ channels have 4 homologous domains, each containing 6 transmembrane segments, S1 to S6. TPCN1 is similar to these channels, but it has only 2 domains containing S1 to S6 (Ishibashi et al., 2000 [PubMed 10753632]).[supplied by OMIM, Mar 2008]. | hsa:53373; | early endosome membrane [GO:0031901]; endolysosome [GO:0036019]; endosome [GO:0005768]; endosome membrane [GO:0010008]; integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; recycling endosome membrane [GO:0055038]; identical protein binding [GO:0042802]; intracellular phosphatidylinositol-3,5-bisphosphate-sensitive cation channel activity [GO:0097682]; ligand-gated sodium channel activity [GO:0015280]; NAADP-sensitive calcium-release channel activity [GO:0072345]; phosphatidylinositol-3,5-bisphosphate binding [GO:0080025]; protein homodimerization activity [GO:0042803]; syntaxin binding [GO:0019905]; voltage-gated calcium channel activity [GO:0005245]; voltage-gated sodium channel activity [GO:0005248]; endocytosis involved in viral entry into host cell [GO:0075509]; ion transmembrane transport [GO:0034220]; positive regulation of autophagy [GO:0010508]; regulation of ion transmembrane transport [GO:0034765] | 19620632_Data show that human two-pore channel TPC1 is critical for nicotinic acid adenine dinucleotide phosphate action and is likely the long sought after target channel for NAADP. 22270358_OsTPC1 plays a crucial role in TvX-induced Ca(2+) influx as a plasma membrane Ca(2+)-permeable channel consequently required for the regulation of phytoalexin biosynthesis 22500018_The concerted regulation of TPC1 activity by luminal Ca(2+) and by membrane potential thus provides a potential mechanism to explain NAADP-induced Ca(2+) oscillations. 24776928_TPC1 is a member of a new family of voltage-gated Na(+) channels that senses pH changes and confers electrical excitability to organelles. 24847115_NAADP triggers H+ release from lysosomes and endolysomes through activation of TPC1, but that the Ca2+ -releasing ability of TPC1 will depend on the ionic composition of the acidic stores 25157141_TPC2, but not TPC1, caused a proliferation of endolysosomal structures, dysregulating intracellular trafficking, and cellular pigmentation. 25451935_NAADP induced marked Ca(2+) transients in HEK293 cells that stably coexpressed hTPC2 with hTPC1 or cTPC3, but failed to evoke any such response in cells that coexpressed interacting hTPC2 and rTPC3 subunits 25665131_The TPC1 was shown to interact with citron kinase, with TPC1 overexpression affecting RhoA activity and myosin light chain phosphorylation levels in cytokinesis. 25722412_TPC1 and TPC2 proteins play a key role in Ebola virus infection and may be effective targets for antiviral therapy. 26152696_Studies suggest that both two-pore channels TPC1 and TPC2 as nicotinic acid adenine dinucleotide phosphate (NAADP) targets. 27353380_These findings indicate potential differential regulation of signaling processes by TPC1 and TPC2 in breast cancer cells. 28252105_These data point to a process in which Ca(2+) permeation in human TPC1 has a positive feedback on channel activity while Na(+) acts as a negative regulator. 29129203_a cluster of arginine residues in the first domain required for selective voltage-gating of TPC1 map not to the voltage-sensing fourth transmembrane region (S4) but to a cytosolic downstream region (S4-S5 linker). 30860481_cryo-EM structure of human TPC2 provides insights into the mechanism of PI(3,5)P2-regulated gating of TPC2, which is distinct from that of TPC1 31557916_TPC1 is involved in the norepinephrine (NE)-stimulated [Ca(2+)]i rise in Smooth muscle cells. Inhibition of TPC1 activity by NED 19 could be the reason for partial inhibition of aortic rings contraction in response to NE. | ENSMUSG00000032741 | Tpcn1 | 2.149063e+03 | 1.1285484 | 0.174468293 | 0.2958037 | 3.527753e-01 | 0.5525462902 | 0.89296216 | No | Yes | 1.747686e+03 | 183.622902 | 1.638133e+03 | 176.650740 | |
| ENSG00000187079 | 7003 | TEAD1 | protein_coding | P28347 | FUNCTION: Transcription factor which plays a key role in the Hippo signaling pathway, a pathway involved in organ size control and tumor suppression by restricting proliferation and promoting apoptosis. The core of this pathway is composed of a kinase cascade wherein MST1/MST2, in complex with its regulatory protein SAV1, phosphorylates and activates LATS1/2 in complex with its regulatory protein MOB1, which in turn phosphorylates and inactivates YAP1 oncoprotein and WWTR1/TAZ. Acts by mediating gene expression of YAP1 and WWTR1/TAZ, thereby regulating cell proliferation, migration and epithelial mesenchymal transition (EMT) induction. Binds specifically and cooperatively to the SPH and GT-IIC 'enhansons' (5'-GTGGAATGT-3') and activates transcription in vivo in a cell-specific manner. The activation function appears to be mediated by a limiting cell-specific transcriptional intermediary factor (TIF). Involved in cardiac development. Binds to the M-CAT motif. {ECO:0000269|PubMed:18579750, ECO:0000269|PubMed:19324877}. | 3D-structure;Acetylation;Activator;Alternative splicing;DNA-binding;Direct protein sequencing;Disease variant;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene encodes a ubiquitous transcriptional enhancer factor that is a member of the TEA/ATTS domain family. This protein directs the transactivation of a wide variety of genes and, in placental cells, also acts as a transcriptional repressor. Mutations in this gene cause Sveinsson's chorioretinal atrophy. Additional transcript variants have been described but their full-length natures have not been experimentally verified. [provided by RefSeq, May 2010]. | hsa:7003; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; TEAD-YAP complex [GO:0140552]; transcription regulator complex [GO:0005667]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; embryonic organ development [GO:0048568]; hippo signaling [GO:0035329]; positive regulation of cell growth [GO:0030307]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein-containing complex assembly [GO:0065003]; regulation of transcription by RNA polymerase II [GO:0006357] | 1851669_TEAD1 has non-AUG translation initiation 11751932_These results are consistent with two plausible models of cryptic MCAT enhancer regulation by Pur alpha, Pur beta, and MSY1 involving either competitive single-stranded DNA binding or masking of MCAT-bound transcription enhancer factor-1. 12376544_TEAD1 (TEF-1) interacts with a muscle-specific cofactor to promote skeletal muscle gene expression. 12861002_Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions 15016762_A missense mutation (Y421H) in TEAD1 is tightly linked to Sveinsson's chorioretinal atrophy (SCRA), an autosomal dominant eye disease characterized by symmetrical lesions radiating from the optic disc involving the retina and the choroid. 15016762_the mutation in the TEAD1 gene is the cause of Sveinsson's chorioretinal atrophy. 18177740_regulation of IFITM3 by TEF-1 demonstrates that TEF-1 dependent regulation is more widespread than its previously established role in the expression of muscle specific genes 18775765_TEAD1 inhibits prolactin gene expression in cultured human uterine decidual cells. 19002168_TEAD1 and the ubiquitin ligase c-Cbl were identified as novel basal cell markers in prostate cancer 19015275_Here we show that during vertebrate neural tube development, the TEA domain transcription factor (TEAD) is the cognate DNA-binding partner of YAP. 19410955_The primary cellular origin of circumpapillary dysgenesis of the pigment epithelium is within the choroid instead of the pigment epithelium. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23029054_our data reveal a new, Livin-dependent, apoptotic role for TEAD1 in mammals and provide mechanistic insight downstream of TEAD1 deregulation in cancers. 23576552_Data indicate that knockdown of TEAD1/3/4 induces an almost identical cellular senescent phenotype as YAP silencing. 23994529_identified an intronic region of the NAIP gene responding to TEAD1/YAP activity, suggesting that regulation of NAIP by TEAD1/YAP is at the transcriptional level 24344135_the first evidence demonstrating that TEAD1 is a novel general repressor of smooth muscle-specific gene expression through interfering with myocardin binding to SRF. 24953189_TEAD1 causes the nuclear sequestration of p65 leading to a novel TEAD1/p65 complex that associates with the intronic enhancer and is necessary for cytokine induction of MnSOD. 25384421_the YAP-TEAD interaction can be disrupted using cyclic YAP-like peptides, which targets the HIPPO pathway 25628125_Our findings suggest that genetic variants of Hippo pathway genes, particularly YAP1 rs11225163, TEAD1 rs7944031 and TEAD4 rs1990330, may independently or jointly modulate survival of CM patients. 25865119_Melanoma reprogramming involves thousands of genomic regulatory regions underlying the proliferative and invasive states, identifying SOX10/MITF and AP-1/TEAD as regulators, respectively. 25915126_Suggest central role for TEAD and YAP as signal-responsive regulators of multipotent pancreatic progenitors. 25995450_TAZ negatively regulate transcription of DeltaNp63 through TEAD1,2,3 and 4 transcription factors. 26091538_Our data suggest that AIC is a genetically heterogeneous disease and is not restricted to the X chromosome, and that TEAD1 mutations may be present in male patients. 26173433_show that the proangiogenic microfibrillar-associated protein 5 (MFAP5) is a direct transcriptional target of YAP/TEAD in cholangiocarcinoma cells transcription factors. 26295846_TEAD1 mediates YAP1 chromatin-binding genome-wide. 27359056_Yap1 post-translational modifications favoring its ubiquitination and apoptosis characterize hepatocellular carcinoma (HCC) with better prognosis, whereas conditions favoring the formation of YAP1-TEAD complexes are associated with aggressiveness and acquisition of stemness features by HCC cells 27433809_MYC and TEAD activity is able to stratify different breast cancer subtypes in large panels of breast cancer patients. 27434865_TEAD1 could enhance the expression levels of SP1, by directly binding to its promoter. 27725085_Here, the authors show that TEAD1-expressing skeletal muscle of transgenic mice features a dramatic hyperplasia of muscle stem cells (i.e. satellite cells, SCs) but surprisingly without affecting muscle tissue size. 28028053_The authors show MRTF family proteins bind YAP via a conserved PPXY motif that interacts with the YAP WW domain. This interaction allows MRTF to recruit NcoA3 to the TEAD-YAP transcriptional complex and potentiate its transcriptional activity. 28077648_Collectively, these results indicate that human papillomavirus 16 E6 induces upregulation of APOBEC3B through increased levels of TEADs, highlighting the importance of the TEAD-APOBEC3B axis in carcinogenesis. 28349829_Upregulation of transcriptional enhancer activator domain 1 was found in hepatocellular carcinoma tissues and inversely correlated with miR-590-3p. Our results indicate a tumor suppressor role of miR-590-3p in hepatocellular carcinoma through targeting transcriptional enhancer activator domain 1 and suggest its use in the diagnosis and prognosis of liver cancer. 28483529_This identifies the YAP1/TEAD1 complex as the representative dysregulated profile of Hippo signaling in OS and provides proof-of-principle that targeting TEAD1 may be a therapeutic strategy of osteosarcoma. 28759040_TEAD1 and TEAD4 are oncogenic factors, whose aberrant activation are, in part, mediated by the silence of miR-377-3p, miR-1343-3p and miR-4269. 28892790_YAP1 interacted with TEAD1, exerted their transcriptional control of the functional target, glucose transporter 1 (Glut1). 29154888_adult human and mouse hearts had more Taz than Yap1 by mRNA and protein expression and their increases in diseased hearts were proportional and did not change Yap1/Taz ratio. Yap1, Taz, and Tead1 were accumulated in the nuclear fraction and cardiomyocyte nuclei of diseased hearts 29680477_YAP1-TEAD1 signaling induces mitochondrial biogenesis in endothelial cells and stimulates angiogenesis through PGC1alpha. 29739039_TEA domain family member 1 (TEAD1) expression is highly expressed in hepatocellular carcinoma (HCC). 30206136_Data show that hyperactivated WWTR1 (TAZ) protein induced substantial myeloid cell infiltration into the liver and the secretion of proinflammatory cytokines through a TEA domain transcription factor 1 (TEAD)-dependent mechanism. 30275445_TEA domain transcription factor1 (TEAD1) trans occupies at accessibility sites within AQP4, EGFR, and CDH4. TEAD1 knockout in patient-derived glioblastoma (GBM) lines diminishes migration, in vitro and in vivo, and alters migratory and epithelial-to-mesenchymal transition (EMT) transcriptome signatures with downregulation of its target AQP4. 30417565_High TEAD1 expression is associated with bladder cancer progression. 30771694_TEAD1 is a novel mechano-responsive gene and plays an important role in force-induced osteoclastogenesis, which is dependent, as least partially, on transcriptional regulation of Osteoprotegerin. 30903741_combining biophysical, structural and molecular mod-elling data, we analyse the effect of this mutation onthe YAP:TEAD interaction at the atomic level 31043565_activity tightly controlled through the regulation of palmitoylation and stability via the orchestration of FASN, depalmitoylases, and E3 ubiquitin ligase in response to cell contact 31138678_Interaction proteomics revealed that VGLL3 bound TEAD1, TEAD3 and TEAD4 in myoblasts and/or myotubes. 31852972_AR facilitates YAP-TEAD interaction with the AM promoter to enhance mast cell infiltration into cutaneous neurofibroma. 32119877_Nuclear actin regulates cell proliferation and migration via inhibition of SRF and TEAD. 32142794_TEAD1 and TEAD3 Play Redundant Roles in the Regulation of Human Epidermal Proliferation. 32280078_Arsenic trioxide-induced upregulation of miR-1294 suppresses tumor growth in hepatocellular carcinoma by targeting TEAD1 and PIM1. 33060790_A new perspective on the interaction between the Vg/VGLL1-3 proteins and the TEAD transcription factors. 33116297_YAP-TEAD1 control of cytoskeleton dynamics and intracellular tension guides human pluripotent stem cell mesoderm specification. 33333006_Motif orientation matters: Structural characterization of TEAD1 recognition of genomic DNA. 33760147_Circular RNA circCCT3 promotes hepatocellular carcinoma progression by regulating the miR12875p/TEAD1/PTCH1/LOX axis. 33862464_YTHDF1-regulated expression of TEAD1 contributes to the maintenance of intestinal stem cells. 33864784_Novel TEAD1 gene variant in a Serbian family with Sveinsson's chorioretinal atrophy. 34048786_RNA methyltransferase NSUN2 promotes hypopharyngeal squamous cell carcinoma proliferation and migration by enhancing TEAD1 expression in an m(5)C-dependent manner. 35121738_ROCK1 mechano-signaling dependency of human malignancies driven by TEAD/YAP activation. 35169903_MiR-597-5p suppresses the progression of hepatocellular carcinoma via targeting transcriptional enhancer associate domain transcription factor 1 (TEAD1). | ENSMUSG00000055320 | Tead1 | 1.211380e+03 | 0.9779348 | -0.032189880 | 0.2797651 | 1.312141e-02 | 0.9088028403 | 0.98338795 | No | Yes | 1.337117e+03 | 210.141019 | 1.200617e+03 | 193.558586 | |
| ENSG00000187109 | 4673 | NAP1L1 | protein_coding | P55209 | FUNCTION: Histone chaperone that plays a role in the nuclear import of H2A-H2B and nucleosome assembly (PubMed:20002496, PubMed:26841755). Participates also in several important DNA repair mechanisms: greatly enhances ERCC6-mediated chromatin remodeling which is essential for transcription-coupled nucleotide excision DNA repair (PubMed:28369616). Stimulates also homologous recombination (HR) by RAD51 and RAD54 which is essential in mitotic DNA double strand break (DSB) repair (PubMed:24798879). Plays a key role in the regulation of embryonic neurogenesis (By similarity). Promotes the proliferation of neural progenitors and inhibits neuronal differentiation during cortical development (By similarity). Regulates neurogenesis via the modulation of RASSF10; regulates RASSF10 expression by promoting SETD1A-mediated H3K4 methylation at the RASSF10 promoter (By similarity). {ECO:0000250|UniProtKB:P28656, ECO:0000269|PubMed:20002496, ECO:0000269|PubMed:24798879, ECO:0000269|PubMed:26841755, ECO:0000269|PubMed:28369616}.; FUNCTION: (Microbial infection) Positively regulates Epstein-Barr virus reactivation in epithelial cells through the induction of viral BZLF1 expression. {ECO:0000269|PubMed:23691099}.; FUNCTION: (Microbial infection) Together with human herpesvirus 8 protein LANA1, assists the proper assembly of the nucleosome on the replicated viral DNA. {ECO:0000269|PubMed:27599637}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Lipoprotein;Methylation;Neurogenesis;Nucleus;Phosphoprotein;Prenylation;Reference proteome | This gene encodes a member of the nucleosome assembly protein (NAP) family. This protein participates in DNA replication and may play a role in modulating chromatin formation and contribute to the regulation of cell proliferation. Alternative splicing results in multiple transcript variants encoding different isoforms; however, not all have been fully described. [provided by RefSeq, Apr 2015]. | hsa:4673; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; melanosome [GO:0042470]; membrane [GO:0016020]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; RNA binding [GO:0003723]; DNA replication [GO:0006260]; nucleosome assembly [GO:0006334]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of neural precursor cell proliferation [GO:2000179]; positive regulation of neurogenesis [GO:0050769] | 14966293_These results suggest that the binding of sequence-specific DNA binding proteins to human nucleosome assembly protein 1 may be an important step contributing to the activation of transcription. 16287874_both the acidic nature of nucleosome assembly protein I and its other functional structure(s) may be essential to mediate the assembly and disassembly of the dimers in nucleosome core particles 17339334_Data show that Alien binds in vivo and in vitro to NAP1 and modulates its activity by enhancing NAP1-mediated nucleosome assembly on DNA. 20002496_The biochemical properties of two human NAP1-like proteins, hNAP1L1 and hNAP1L4, were characterized. 21333655_Results identified several proteins interacting with NAP1L2, including the ubiquitously expressed members of the nucleosome assembly protein family, NAP1L1 and NAP1L4. 22134777_NAP1L1 was down-expressed following antiproliferative agent treatment in AuL12-treated cells in comparison with control and with Au(2)Phen-treated ovarian cancer cells. 23691099_TAF-Ibeta promotes BZLF1 expression and subsequent lytic infection by affecting chromatin at the BZLF1 promoter 24798879_Nap1 stimulates homologous recombination by RAD51 and RAD54 in higher-ordered chromatin containing histone H1. 28369616_NAP1L1 increases CSB processivity by decreasing the pausing probability during translocation. Our study, therefore, uncovers the different steps of CSB-mediated chromatin remodeling that can be regulated by NAP1L1. 28659470_Here the authors confirmed the hepatitis C virus NS5A-NAP1L1 interaction and mapped it to the C terminus of NS5A of both hepatitis C virus genotype 1 and 2. 28687276_knockdown of NAP1L1 suppresses IkappaBalpha degradation and nuclear transport of p65 subunit after treatment with TNF-a stimulation, leading to attenuation of the NF-kappaB transcriptional activity, whereas NAP1L4 knockdown remains silent.results of this study suggest that NAP1L1 downregulation renders the cell vulnerable to apoptotic cell death through attenuation of NF-kappaB transcriptional activity on the anti-apop... 29541944_In summary, the authors identified NAP1L1 as a novel binding partner of hepatitis c virus NS3 and found that the protease domain of NS3 is responsible for interactions with NAP1L1. They demonstrated the functional role of NAP1L1 in hepatitis c virus entry through regulating the protein expression of SR-B1. 29572888_Through regulating NAP1L1, PRDM8 inhibits PI3K/AKT/mTOR signaling in hepatocellular carcinoma. 30165194_Long non-coding RNAs (lncRNAs) CDKN2B-AS1 promoted nucleosome assembly protein 1 like 1 (NAP1L1) expression by sponging let-7c-5p in hepatocellular carcinoma (HCC) cells. 30459273_evaluated the currently known TBK1-mediated phosphorylation sites in the SKICH domains of NDP52 and TAX1BP1 on the basis of their interactions with NAP1 31634504_The NAP1L1 regulates cell fate by controlling the expression of p53-responsive proarrest and proapoptotic genes through selective modulation of p53 acetylation at specific sites during normal homeostasis and in stress-induced responses. 32522932_Prognostic significance of NAP1L1 expression in patients with early lung adenocarcinoma. 34563951_NAP1L1 targeting suppresses the proliferation of nasopharyngeal carcinoma. 35351053_NAP1L1 promotes tumor proliferation through HDGF/C-JUN signaling in ovarian cancer. | ENSMUSG00000058799 | Nap1l1 | 9.743215e+03 | 1.1467159 | 0.197508027 | 0.3219914 | 3.685187e-01 | 0.5438128120 | 0.89063840 | No | Yes | 1.189809e+04 | 2221.264427 | 8.397799e+03 | 1608.096279 | |
| ENSG00000187595 | 201181 | ZNF385C | protein_coding | Q66K41 | Alternative splicing;Metal-binding;Nucleus;Reference proteome;Repeat;Zinc;Zinc-finger | Mouse_homologues mmu:278304; | nucleus [GO:0005634]; nucleic acid binding [GO:0003676]; zinc ion binding [GO:0008270] | ENSMUSG00000014198 | Zfp385c | 3.371128e+01 | 1.3180786 | 0.398436387 | 0.5219374 | 5.861432e-01 | 0.4439144147 | 0.85798841 | No | Yes | 3.106412e+01 | 7.220064 | 2.198292e+01 | 5.398964 | ||||
| ENSG00000187624 | 400566 | C17orf97 | protein_coding | Q6ZQX7 | FUNCTION: May be involved in ATE1-mediated N-terminal arginylation. {ECO:0000250|UniProtKB:Q810M6}. | Alternative splicing;Reference proteome;Repeat | hsa:400566; | protein arginylation [GO:0016598] | 33443146_The Ligand of Ate1 is intrinsically disordered and participates in nucleolar phase separation regulated by Jumonji Domain Containing 6. | ENSMUSG00000053783 | 1700016K19Rik | 1.326784e+02 | 1.3821799 | 0.466945451 | 0.4285634 | 1.205851e+00 | 0.2721554431 | 0.80044804 | No | Yes | 1.437422e+02 | 26.242976 | 8.384658e+01 | 16.199539 | ||
| ENSG00000187688 | 51393 | TRPV2 | protein_coding | Q9Y5S1 | FUNCTION: Calcium-permeable, non-selective cation channel with an outward rectification. Seems to be regulated, at least in part, by IGF-I, PDGF and neuropeptide head activator. May transduce physical stimuli in mast cells. Activated by temperatures higher than 52 degrees Celsius; is not activated by vanilloids and acidic pH. {ECO:0000269|PubMed:10201375}. | 3D-structure;ANK repeat;Calcium;Calcium channel;Calcium transport;Cell membrane;Cytoplasm;Glycoprotein;Ion channel;Ion transport;Membrane;Phosphoprotein;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene encodes an ion channel that is activated by high temperatures above 52 degrees Celsius. The protein may be involved in transduction of high-temperature heat responses in sensory ganglia. It is thought that in other tissues the channel may be activated by stimuli other than heat. [provided by RefSeq, Jul 2008]. | hsa:51393; | axonal growth cone [GO:0044295]; cell body [GO:0044297]; cell surface [GO:0009986]; growth cone membrane [GO:0032584]; integral component of plasma membrane [GO:0005887]; melanosome [GO:0042470]; plasma membrane [GO:0005886]; calcium channel activity [GO:0005262]; cation channel activity [GO:0005261]; ion channel activity [GO:0005216]; ion transmembrane transporter activity [GO:0015075]; calcium ion import across plasma membrane [GO:0098703]; calcium ion transmembrane transport [GO:0070588]; positive regulation of axon extension [GO:0045773]; positive regulation of calcium ion import [GO:0090280]; positive regulation of cold-induced thermogenesis [GO:0120162]; response to heat [GO:0009408]; response to temperature stimulus [GO:0009266]; sensory perception [GO:0007600] | 15249591_TRPV2 and PKA have roles as a signaling module for transduction of physical stimuli in mast cells 16533525_It is concluded that PI3-kinase induces or modulates the activity of recombinant TRPV2 channels; in contrast to the previously proposed mechanism, activation of TRPV2 channels by PI3-kinase is not due to channel translocation to the plasma membrane. 16777226_Both TRPV1 and TRPV2 are found in human peripheral blood lymphocytes. 16882997_The TRPV2 ARD provides the first structural insight into a domain that coordinates nociceptive sensory transduction and is likely to be a prototype for other TRPV channel ARDs 17977643_A progressive loss of s-TRPV2 accompanied by an increase of hTRPV2 expression was found in high-grade and -stage UC. 18787888_Messenger RNA transcripts of TRPVs 1 through 4 are detected for the first time in human pulmonary artery smooth muscle cells. 18983665_relative gene expression ofTRPV1-4 in leukocytes is: TRPV3 < TRPV4, TRPV1 and TRPV2; leukocytes in hyposensitive subjects showed up-regulation of TRPV1, which was almost doubly expressed 19482060_The reduction of TRPV2 and TRPM8 immunoreactive nerve fibers in skin from individuals with Norrbottnian congenital insensitivity to pain further suggests that these ion channels are expressed primarily on nociceptive primary sensory neurons. 19619644_TRPV1-immunoreactivity was significantly higher and TRPV2-immunoreactivity was significantly lower in peripheral blood mononuclear cells from end-stage kidney disease patients compared to controls. 20093382_TRPV2 negatively controls glioma cell survival and proliferation, as well as resistance to Fas-induced apoptotic cell death in an ERK-dependent manner. 20103638_Findings establish a role for TRPV2 in PCa progression to the aggressive castration-resistant stage, prompting evaluation of TRPV2 as a potential prognostic marker and therapeutic target in the setting of advanced PCa. 20113837_TRPV2 plays a role in human hepatocarcinogenesis and might be a prognostic marker of patients with hepatocellular carcinoma (HCC). 20686800_A calmodulin binding site was located in the C-termini of TRPV2 (654-683) and TRPV5 (587-616). 20926660_Decrease in membrane phosphatidylinositol 4,5-bisphosphate (PIP2) levels on channel activation underlies a major component of divalent calcium ion-dependent desensitization of TRPV2 and may play a similar role in other TRP channels. 21506114_Human corneal epithelial cells possess thermosensitive TRPV2 activity. 21574765_TRPV2 plays a key role in mast-cell degranulation in response to mechanical, heat and red laser-light stimulation. 22327830_Cultivated HCjE cells and human conjunctiva express TRPV2, mRNA. 23294145_The increased expression of TRPV2 gene in peripheral lymphocytes is closely correlated with childhood asthma in the north of China. 23426965_Duchenne muscular dystrophy primary human myotubes exhibit abnormal elevation of TRPV2-dependant cation entry. 23453732_TRPV1 and TRPV2 are potential targets for potential therapeutic interventions in cardiovascular disease. 23542034_TRPV2-coupled signaling as a key player in mediating the cellular actions of heat shock on DCs 23741410_TRPV2 mediates adrenomedullin stimulation of prostate and urothelial cancer cell adhesion, migration and invasion. 23786999_Sarcolemmal TRPV2 accumulation appears to have considerable pathological impact on Dilated cardiomyopathy (DCM) progression. 23964684_The addition of TRPV2 inhibitor Tranilast in the incubation medium prevent the calcium influx triggered by lysoPC and reduced lysoPC-induced cytotoxicity,whereas TRPV4 inhibitor RN 1734 was without effect 24475260_An upregulation in the expression of vanilloid transient potential channels 2 enhances hypotonicity-induced cytosolic Ca(2) rise in human induced pluripotent stem cell model of Hutchinson-Gillford Progeria. 24764033_TRPV1-, TRPV2-, P2X3-, and parvalbumin-immunoreactivity neurons in the human nodose ganglion innervate the pharynx and epiglottis through the pharyngeal branch and superior laryngeal nerve 24878697_High expression of TRPV2 mRNA was closely associated with advanced pathological stage and worse survival of patients with esophageal squamous cell carcinoma. 25001513_Aim of this review is to study the role of the TRPV2 channel, a member of the TRPV subfamily of TRP channels, in tumor progression. [review] 25268680_Data indicate that transient receptor potential vanilloid 2 (TRPV2) siRNA virtually abolished stretch-activated current. 25333484_Using multiple sequence alignments as source for evolutionary, bioinformatics and statistical analysis, we have analyzed the evolutionary profiles for TRPV1, TRPV2, TRPV3 and TRPV4 25641129_There appears to be TRPV1- or TRPV2-IR sympathetic pathway in the human stellate ganglion and spinal cord. 25869297_This study evaluated the role of Trpv2 in arthritis and the pharmacological effects of Trpv2 agonists. 26416880_transient receptor potential vanilloid 2 is an intracellular Ca(2+)-permeable transient receptor potential vanilloid channel upregulated by nerve growth factor via the mitogen-activated protein kinase signaling pathway to augment neurite outgrowth. 26865051_both TRPV2 and TRPV4 are involved in migration of human cardiac c-kit(+) progenitor cells. 26993604_High TRPV2 expression is associated with breast cancer. 27079220_The results of the study show that polymorphism of TRPV2 influenced the susceptibility to FM. 27298359_Binding of capsaicin to TRPV2_Quad antagonizes resiniferatoxin-induced activation likely through competition for the same binding sites. 27553072_CGRP- and TRPV2-containing trigeminal neurons probably innervate the paranasal sinus mucosae, and project into spinal and principal trigeminal sensory nuclei. 28782273_Study shows that despite lacking gastric mucosal inflammation, up-regulation of TRPV1 and TRPV2, down-regulation of BDNF were observed in functional dyspepsia (FD) patients. These suggest that immune alteration may contribute to the pathogenesis of FD without any previous infection. 28905239_the expression of transient receptor potential vanilloid-1 (TRPV1), transient receptor potential vanilloid-2 (TRPV2) and transient receptor potential vanilloid-3 (TRPV3) channels in native human odontoblasts, was examined. 29393104_TRPV2 expression was inversely correlated with CRP and TpI and directly correlated with serum IGF-1. We assume that peripheral TRPV2 downregulation occurs concomitantly with the accumulation of TRPV2-white blood cells in the peri-infarct zone. TRPV2 may thus represent a novel target for treatment in the acute phase after MI. 29523858_TRPV2 recruitment to lipid rafts and TRPV2-mediated calcium influx are required for phagocytosis in macrophages from cystic fibrosis patients. 30235802_TRPV2 mediates IL-8 secretion and calcium signaling in testicular peritubular cells. 30326911_TRPV2-induced Ca(2+)-calcineurin-NFAT signaling regulates differentiation of osteoclast in multiple myeloma 30851707_TRPV2 regulates key intracellular processes implicated in cell invasion in arthritis 31653969_The TRPV2 cation channels: from urothelial cancer invasiveness to glioblastoma multiforme interactome signature. 31690728_The expression and role of TRPV2 in esophageal squamous cell carcinoma. 31719194_A methionine-dependent redox sensitivity of TRPV2 which may be an important endogenous mechanism for regulation of TRPV2 activity and account for its pivotal role for phagocytosis in macrophages. 31783610_Trafficking of Stretch-Regulated TRPV2 and TRPV4 Channels Inferred Through Interactomics. 31857697_TRPV2 channel as a possible drug target for the treatment of heart failure. 32751388_The Effects of Cannabidiol and Prognostic Role of TRPV2 in Human Endometrial Cancer. 32985655_TRPV2 interacts with actin and reorganizes submembranous actin cytoskeleton. 34158856_TRPV2-spike protein interaction mediates the entry of SARS-CoV-2 into macrophages in febrile conditions. 34855064_TRPV2 Promotes Cell Migration and Invasion in Gastric Cancer via the Transforming Growth Factor-beta Signaling Pathway. 35106663_ASO Visual Abstract: TRPV2 Promotes Cell Migration and Invasion in Gastric Cancer via the TGF-beta-Signaling Pathway. 35406761_Oxidative Stress-Induced TRPV2 Expression Increase Is Involved in Diabetic Cataracts and Apoptosis of Lens Epithelial Cells in a High-Glucose Environment. | ENSMUSG00000018507 | Trpv2 | 1.200838e+01 | 1.8732160 | 0.905517228 | 0.9093309 | 8.800090e-01 | 0.3481992109 | No | Yes | 8.385962e+00 | 3.936609 | 4.670097e+00 | 2.405756 | ||
| ENSG00000187741 | 2175 | FANCA | protein_coding | O15360 | FUNCTION: DNA repair protein that may operate in a postreplication repair or a cell cycle checkpoint function. May be involved in interstrand DNA cross-link repair and in the maintenance of normal chromosome stability. | 3D-structure;Alternative splicing;Cytoplasm;DNA damage;DNA repair;Disease variant;Fanconi anemia;Nucleus;Phosphoprotein;Reference proteome | The Fanconi anemia complementation group (FANC) currently includes FANCA, FANCB, FANCC, FANCD1 (also called BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ (also called BRIP1), FANCL, FANCM and FANCN (also called PALB2). The previously defined group FANCH is the same as FANCA. Fanconi anemia is a genetically heterogeneous recessive disorder characterized by cytogenetic instability, hypersensitivity to DNA crosslinking agents, increased chromosomal breakage, and defective DNA repair. The members of the Fanconi anemia complementation group do not share sequence similarity; they are related by their assembly into a common nuclear protein complex. This gene encodes the protein for complementation group A. Alternative splicing results in multiple transcript variants encoding different isoforms. Mutations in this gene are the most common cause of Fanconi anemia. [provided by RefSeq, Jul 2008]. | hsa:2175; | cytoplasm [GO:0005737]; Fanconi anaemia nuclear complex [GO:0043240]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA repair [GO:0006281]; female gonad development [GO:0008585]; interstrand cross-link repair [GO:0036297]; male gonad development [GO:0008584]; male meiotic nuclear division [GO:0007140]; protein-containing complex assembly [GO:0065003]; regulation of CD40 signaling pathway [GO:2000348]; regulation of cell population proliferation [GO:0042127]; regulation of DNA-binding transcription factor activity [GO:0051090]; regulation of inflammatory response [GO:0050727]; regulation of regulatory T cell differentiation [GO:0045589] | 12031647_used to subtype Fanconi anemia T cells 12210728_FANCA may function to recruit IKK2, thus providing the cell a means of rapidly responding to stress. 12444097_Mutant FANCA proteins complement the sensitivity of DNA crosslinker mitomycin C at different grades: five proteins (group I) behave like wild-type FANCA, whereas the other proteins are either mildly (group II, n=4) or severely (group III, n=12) impaired. 12637330_Leukemic cells bearing both characteristic complex cytogenetic defects and marked decrease in nuclear FANCA, were analyzed for possible role of RANCA in cytogenetic instability and clonal progression to AML. 14749703_Deletion and reduced expression of the Fanconi anemia FANCA gene is associated with sporadic acute myeloid leukemia 15059067_2 unique Fanconi-anemia-causing mutations were found: FANCA gross deletion of exons 6-31 & FANCA splice-site mutation IVS 42-2A>C. The gross deletion had recombination between 2 highly homologous Alu elements. The splice mutation had intron 42 retention. 15138265_FANCA and FANCG uniquely respond to oxidative damage by forming complexes via intermolecular disulfide linkages 15256425_FA proteins function at the level of chromatin during S phase to regulate and maintain genomic stability. 15591268_determination of whether FANCA gene mutations predispose to the development of familial pancreatic cancer 15790592_can be actively exported out of the nucleus by CRM1 15860134_Observational study of gene-disease association. (HuGE Navigator) 15860134_the FANCA allele with the tandem duplication does not appear to modify breast cancer risk but may act as a low penetrance protective allele for ovarian cancer 15917947_Observational study of genotype prevalence. (HuGE Navigator) 16946016_FANCA functions as a GnRH-induced signal transducer. 18056155_Observational study of gene-disease association. (HuGE Navigator) 18224251_Results describe upregulated ATM gene expression and activated DNA crosslink-induced damage response checkpoints in Fanconi anemia with FANCA mutations, and the implications for carcinogenesis. 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18457264_FANCA proteins are defective in Fanconi anemia patients. The disease-associated FANCA mutant was defective in binding to FANCG. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18950845_Observational study of gene-disease association. (HuGE Navigator) 19012493_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19015634_FANCA deficiencies may contribute to the high risk of FA patients for developing HPV-associated squamous cell carcinoma. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19109555_Phosphorylation of FANCA on serine 1449 is a DNA damage-specific event that is downstream of ATR and is functionally important in Fanconi anemia 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19491337_Fanca-/- hematopoietic stem cells have a mobilization defect which can be overcome by administration of the Rac inhibitor NSC23766 19625176_Observational study of gene-disease association. (HuGE Navigator) 19690177_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19714462_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20425471_Thirteen genetic subtypes have been described (A, B, C, D1, D2, E, F, G, I, J, L, M, and N), of which FANCA, FANCC, and FANCG are the three most common disease-causing genes 20453000_Observational study of gene-disease association. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 20864535_Cytoplasmic FANCA-FANCC complex was essential for NPMc stability. 21236561_FANCA deletions might contribute to breast cancer susceptibility, potentially in combination with other germline mutations 21273304_All missense mutations studied lead to an altered FANCA protein that is unable to relocate to the nucleus and activate the FA/BRCA pathway. 22194614_the nucleic acid-binding domain of FANCA is located primarily at its C terminus, where most disease-causing mutations are found. 23021409_Sequence variants in FANCA could therefore be potential spoilers of the Fanconi-BRCA pathway and as a result, they could in turn have an impact in non-BRCA1/2 breast cancer families. 23067021_FANCA and FANCG are the major Fanconi anemia genes in the Korean population. 23898106_A total of 13/166 patients were diagnosed with FA and 8/13 belonged to the FA-A subtype. A novel point mutation was identified in exon 26 of FANCA gene. 24045675_Results suggest that the nonsynonymous single nucleotide polymorphism (rs2239359) in the FANCA gene or other causal variations coexisting with the GGGAGG haplotype may increase risk of premature ovarian failure in Korean women. 24046015_Data indicate that TLR-induced IL-1beta overproduction in FANCA- and FANCC-deficient mononuclear phagocyte cell lines and primary cells requires activation of the inflammasome. 24349332_Human Fanconi anemia complementation group a protein stimulates the 5' flap endonuclease activity of FEN1. 24356203_c.190-256_283 + 1680del2040 dupC mutation in the FANCA gene is a founder mutation in Macedonian Fanconi anemia patients of Gypsy-like ethnic origin. 24486548_miR-503 gene is methylated in non-small cell lung cancer cells. miR-503 targets a homologous DNA region in the 3'-UTR region of the Fanconi anemia complementation group A protein (FANCA) gene and represses its expression at the transcriptional level. 24704046_Identified two unpredictable splicing mutations that act on either sides of FANCA exon 8. 25015289_FANCA-modulated neddylation pathway involved in CXCR5 membrane targeting and cell mobility. 25108529_Proliferation is compromised in FANCA-deficient pluripotent embryonic stem cells. 25243787_BRCA2 rs10492396 (AG vs. GG: adjusted hazard ratio (adjHR)=1.85, 95% confidence interval (CI)=1.16-2.95, P=0.010), rs206118 (CC vs. TT+TC: adjHR=2.44, 95% CI=1.27-4.67, P=0.007), rs3752447 and FANCA rs62068372 25751062_The I939S point mutation prevented binding to the FAAP20 subunit of the FA core complex, caused SUMOylation at K921, RNF4-mediated polyubiquitination and degradation. 25863087_A frameshifting mutation and a truncating mutation of FANCA are associated with Fanconi anemia. 26366677_FANCA safeguards interphase and mitosis during hematopoiesis 26584049_Using human and murine cells defective in FANCD2 or FANCA and primary bone marrow cells derived from FANCD2 deficient mice, we show that the FA pathway removes R loops and that many DNA breaks accumulated in FA cells are R loop-dependent. 26841305_Results identified homozygous mutations in FANCA and FANCP/SLX4 genes, both located on chromosome 16, were the affected recessive FA genes in three and one family respectively. Genotyping with short tandem repeat markers and SNP arrays revealed uniparental disomy of the entire mutation-carrying chromosome 16 in all four patients. 27121516_High resolution melting curve analysis was used to screen FANCA, and LinReg software version 1.7 was utilized for analysis of expression. RESULTS: In total, six sequence alterations were identified, which included two stop codons, two frames-shift mutations, one large deletion and one amino acid exchange. FANCA expression was downregulated in patients who had sequence alterations. 27867017_FancA amplification and overexpression appear to be crucial for radiotherapeutic failure in head and neck squamous cell carcinomas. 28440412_the current study was the first to report a promoter region variation in the FANCA gene among women with breast cancer in Iran. The results of the present study confirmed the allelic variants in the FANCA promoter region as a tumor suppressor gene. 28864460_we show that although FANCA S1088F protein properly localizes to the nucleus, it alters FANC complex function, enhances sensitivity to DNA damaging agents, and sensitizes cells to PARP inhibitors in vitro and in vivo. 29269525_variants at residues His913 and Arg951 are hypomorphic mutations. Consistent with these findings, the clinical phenotype of most of the patients carrying these mutations is mild. These data further support the recent finding that the Fanconi anemia proteins play a role in mitochondria, and open up possibilities for genotype/phenotype studies based on novel mitochondrial criteria. 29621589_Mutations in the FANCA gene are associated with esophageal atresia and tracheoesophageal fistula in humans. 29702541_Novel Variations of FANCA Gene Provokes Fanconi Anemia: Molecular Diagnosis in a Special Chinese Family. 29904161_We report an extraordinarily high frequency of Fanconi anemia (FA) in a specific subgroup of azoospermic patients (7.1%). The screening for FANCA pathogenic variants in such patients has the potential to identify undiagnosed FA before the appearance of other severe clinical manifestations of the disease. 30057198_FANCG stimulates FANCA-mediated strand annealing and strand exchange. 30339652_unique consanguineous family with 2 genomic instability disorders, Fanconi anemia and ataxia telangiectasia, revealed exceptional combinations of null mutations in the FANCA and ATM genes 30809872_Sanger's sequencing of the nine selected exons of FANCA gene in Fanconi anemia (FA) cases revealed 19 genetic alterations of which 15 were single nucleotide variants, three were insertions and one was microdeletion. 31251079_Elevated FANCA expression interferes with p53 function and is associated with chronic lymphocytic leukemia. 31535215_Variants in FANCA are associated with premature ovarian insufficiency. 31586946_Optimised molecular genetic diagnostics of Fanconi anaemia by whole exome sequencing and functional studies. 32466131_The FANC/BRCA Pathway Releases Replication Blockades by Eliminating DNA Interstrand Cross-Links. 32947577_Clinical and Genetic Features of Patients With Fanconi Anemia in Lebanon and Report on Novel Mutations in the FANCA and FANCG Genes. 33172906_Esophageal cancer as initial presentation of Fanconi anemia in patients with a hypomorphic FANCA variant. 33394227_Severe telomere shortening in Fanconi anemia complementation group L. 33523834_Fanconi anemia A protein participates in nucleolar homeostasis maintenance and ribosome biogenesis. 34058059_SIK2 kinase synthetic lethality is driven by spindle assembly defects in FANCA-deficient cells. 34864095_In silico study of missense variants of FANCA, FANCC and FANCG genes reveals high risk deleterious alleles predisposing to Fanconi anemia pathogenesis. | ENSMUSG00000032815 | Fanca | 6.051561e+03 | 0.9448936 | -0.081776272 | 0.2712387 | 9.056725e-02 | 0.7634572497 | 0.95231693 | No | Yes | 4.690608e+03 | 296.087527 | 5.161684e+03 | 334.079091 | |
| ENSG00000187792 | 7621 | ZNF70 | protein_coding | Q9UC06 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7621; | cytoplasm [GO:0005737]; megasporocyte nucleus [GO:0043076]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; regulation of transcription by RNA polymerase II [GO:0006357] | 27353377_We demonstrated that ZNF70 interacts with ZFP64 and activates HES1 transcription by binding to the HES1 promoter. In addition, HES1 gene expression is increased in ILDR2-knockdown HepG2 cells, in which ZNF70 is translocated from the cytoplasm to the nucleus, suggesting that ZNF70 migration to the nucleus after dissociating from the ILDR2-ZNF70 complex activates HES1 transcription. These results support a novel link betwee 32053575_miRNA-574-5p downregulates ZNF70 and influences the progression of human esophageal squamous cell carcinoma through reactive oxygen species generation and MAPK pathway activation. | 9.944848e+02 | 1.1797274 | 0.238453567 | 0.2774512 | 7.426592e-01 | 0.3888103691 | 0.84317192 | No | Yes | 8.648072e+02 | 116.987911 | 7.876411e+02 | 109.420636 | ||||
| ENSG00000188191 | 5575 | PRKAR1B | protein_coding | P31321 | FUNCTION: Regulatory subunit of the cAMP-dependent protein kinases involved in cAMP signaling in cells. {ECO:0000269|PubMed:20819953}. | 3D-structure;Cell membrane;Disulfide bond;Membrane;Methylation;Nitration;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;cAMP;cAMP-binding | The protein encoded by this gene is a regulatory subunit of cyclic AMP-dependent protein kinase A (PKA), which is involved in the signaling pathway of the second messenger cAMP. Two regulatory and two catalytic subunits form the PKA holoenzyme, disbands after cAMP binding. The holoenzyme is involved in many cellular events, including ion transport, metabolism, and transcription. Several transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Aug 2015]. | hsa:5575; | cAMP-dependent protein kinase complex [GO:0005952]; ciliary base [GO:0097546]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; glutamatergic synapse [GO:0098978]; hippocampal mossy fiber to CA3 synapse [GO:0098686]; multivesicular body [GO:0005771]; plasma membrane [GO:0005886]; Schaffer collateral - CA1 synapse [GO:0098685]; cAMP binding [GO:0030552]; cAMP-dependent protein kinase inhibitor activity [GO:0004862]; cAMP-dependent protein kinase regulator activity [GO:0008603]; protein kinase A catalytic subunit binding [GO:0034236]; learning or memory [GO:0007611]; modulation of chemical synaptic transmission [GO:0050804]; negative regulation of cAMP-dependent protein kinase activity [GO:2000480]; protein phosphorylation [GO:0006468]; regulation of synaptic vesicle cycle [GO:0098693] | 11943777_The paired-like homeodomain protein, Arix, mediates protein kinase A-stimulated dopamine beta-hydroxylase gene transcription through its phosphorylation status 12634056_RIalpha and RIbeta homodimers as well as an RIalpha:RIbeta heterodimer and several of the mutants were able to bind to the R-binding domain of AKAP149/D-AKAP1 20379146_Observational study and genome-wide association study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20588308_Observational study of gene-disease association. (HuGE Navigator) 24722252_This study demonistrated that PRKAR1B mutation associated with a new neurodegenerative disorder with unique pathology. 25108559_No pathogenic PRKAR1B mutations were found in early onset familial dementia patients in a Dutch cohort. 32895490_Genomic and sequence variants of protein kinase A regulatory subunit type 1beta (PRKAR1B) in patients with adrenocortical disease and Cushing syndrome. 33833410_Variants in PRKAR1B cause a neurodevelopmental disorder with autism spectrum disorder, apraxia, and insensitivity to pain. 34716310_EIF4A3-induced circular RNA PRKAR1B promotes osteosarcoma progression by miR-361-3p-mediated induction of FZD4 expression. | ENSMUSG00000025855 | Prkar1b | 1.628927e+03 | 1.5808132 | 0.660666927 | 0.3500309 | 3.637134e+00 | 0.0565041222 | 0.68299703 | No | Yes | 1.838149e+03 | 284.215904 | 9.228456e+02 | 146.980752 | |
| ENSG00000188706 | 51114 | ZDHHC9 | protein_coding | Q9Y397 | FUNCTION: Palmitoyltransferase that could catalyze the addition of palmitate onto various protein substrates (Probable). The ZDHHC9-GOLGA7 complex is a palmitoyltransferase specific for HRAS and NRAS (PubMed:16000296). May have a palmitoyltransferase activity toward the beta-2 adrenergic receptor/ADRB2 and therefore regulate G protein-coupled receptor signaling (PubMed:27481942). {ECO:0000269|PubMed:16000296, ECO:0000269|PubMed:27481942}.; FUNCTION: (Microbial infection) Through a sequential action with ZDHHC20, rapidly and efficiently palmitoylates SARS coronavirus-2/SARS-CoV-2 spike protein following its synthesis in the endoplasmic reticulum (ER). In the infected cell, promotes spike biogenesis by protecting it from premature ER degradation, increases half-life and controls the lipid organization of its immediate membrane environment. Once the virus has formed, spike palmitoylation controls fusion with the target cell. {ECO:0000269|PubMed:34599882}. | Acyltransferase;Disease variant;Endoplasmic reticulum;Golgi apparatus;Lipoprotein;Membrane;Mental retardation;Palmitate;Reference proteome;Transferase;Transmembrane;Transmembrane helix | This gene encodes an integral membrane protein that is a member of the zinc finger DHHC domain-containing protein family. The encoded protein forms a complex with golgin subfamily A member 7 and functions as a palmitoyltransferase. This protein specifically palmitoylates HRAS and NRAS. Mutations in this gene are associated with X-linked cognitive disability. Alternate splicing results in multiple transcript variants that encode the same protein.[provided by RefSeq, May 2010]. | hsa:51114; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; intrinsic component of Golgi membrane [GO:0031228]; palmitoyltransferase complex [GO:0002178]; palmitoyltransferase activity [GO:0016409]; protein-cysteine S-palmitoyltransferase activity [GO:0019706]; Ras palmitoyltransferase activity [GO:0043849]; MAPK cascade [GO:0000165]; peptidyl-L-cysteine S-palmitoylation [GO:0018230]; protein palmitoylation [GO:0018345]; protein targeting to membrane [GO:0006612] | 16000296_Data show that H- and N-Ras are palmitoylated by a human protein palmitoyltransferase encoded by the ZDHHC9 and GCP16 genes. 17519897_DHHC9 is a gastrointestinal-related protein highly expressed in microsatellite stable colorectal cancers. 21388813_Studies indicate that mutations in DHHC9 were associated with X-linked mental retardation. 22230506_MMSA-1 may play a pivotal role in multiple myeloma proliferation and osteolysis destruction. 24248599_Data indicate that sp-Erf2/zDHHC9 palmitoylates Ras proteins in a highly selective manner in the trans-Golgi compartment to facilitate PM targeting via the trans-Golgi network, a role that is most certainly critical for Ras-driven tumorigenesis. 24811172_Two missense mutation, R148W and P150S,of zDHHC9, affecting the autopalmitoylation is associated with the X-linked intellectual disability. 26358559_ZDHHC9 gene mutation is associated with Lujan-Fryns syndrome. 26493349_Report demonstrated that MMSA-1 is specifically expressed in multiple myeloma patients and its upregulation is associated with unfavorable clinical features and poor prognosis. 26493479_studies suggest that ZDHHC9 may serve as a safe and effective target for developing therapies against NRAS-driven cancers 27747153_Data demonstrate that ZDHHC9 mutations are associated with reductions in cortical thickness and white matter microstructural integrity, particularly in regions and networks known to contribute to language function. 28168288_The results demonstrate that a mutation in a ZDHHC9 mutation impacts upon white matter organization across the whole-brain, but also shows regionally specific effects, according to variation in gene expression. 28687527_De novo ZDHHC9 mutation was identified in a patient with X-linked intellectual disability. 34620861_DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. | ENSMUSG00000036985 | Zdhhc9 | 5.332056e+03 | 1.3388507 | 0.420995077 | 0.3003698 | 1.986853e+00 | 0.1586702938 | 0.77593452 | No | Yes | 5.357089e+03 | 474.122047 | 3.816213e+03 | 346.850423 | |
| ENSG00000189159 | 51155 | JPT1 | protein_coding | Q9UK76 | FUNCTION: Modulates negatively AKT-mediated GSK3B signaling (PubMed:21323578, PubMed:22155408). Induces CTNNB1 'Ser-33' phosphorylation and degradation through the suppression of the inhibitory 'Ser-9' phosphorylation of GSK3B, which represses the function of the APC:CTNNB1:GSK3B complex and the interaction with CDH1/E-cadherin in adherent junctions (PubMed:25169422). Plays a role in the regulation of cell cycle and cell adhesion (PubMed:25169422, PubMed:25450365). Has an inhibitory role on AR-signaling pathway through the induction of receptor proteosomal degradation (PubMed:22155408). {ECO:0000269|PubMed:21323578, ECO:0000269|PubMed:22155408, ECO:0000269|PubMed:25169422, ECO:0000269|PubMed:25450365}. | Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome | hsa:51155; | cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634] | 19145478_These data suggest a role for Hn1 in the biology of malignant brain tumors. 21323578_EGF and kinase inhibitors increase HN1 expression, and Silencing of HN1 results in cell cycle alterations in prostate cells. 22155408_HN1 maintains a balance between the androgen-regulated nuclear translocation of androgen receptor and steady-state Akt phosphorylation, predominantly in the absence of androgens. 25169422_Data report that HN1 is an essential factor for beta-catenin turnover and signaling, augments cell growth and migration in prostate cancer cells. 25450365_miR-132 is significantly down-regulated in breast cancer tissues and cancer cell lines. Additional study identifies HN1 as a novel direct target of miR-132. MiR-132 down-regulates HN1 expression by binding to the 3' UTR of HN1. 28490334_Downregulation of MYC abrogated the effect of HN1 overexpression in breast cancer cell lines. Taken together, these data reveal that HN1 promotes the progression of breast cancer by upregulating MYC expression, and might be a therapeutic target for breast cancer. 31257225_HNRNPA1-mediated alternative polyadenylation of HN1 contributes to cancer- and senescence-related phenotypes. 31808620_Jupiter microtubule-associated homolog 1 (JPT1): A predictive and pharmacodynamic biomarker of metformin response in endometrial cancers. 32700728_HN1 as a diagnostic and prognostic biomarker for liver cancer. 32828320_HN1 promotes tumor associated lymphangiogenesis and lymph node metastasis via NF-kappaB signaling activation in cervical carcinoma. 34337714_LncRNA HOXA11-AS promotes cell growth by sponging miR-24-3p to regulate JPT1 in prostate cancer. | ENSMUSG00000020737 | Jpt1 | 8.758937e+03 | 1.4537113 | 0.539740834 | 0.3274627 | 2.709229e+00 | 0.0997691803 | 0.75783482 | No | Yes | 1.292581e+04 | 1787.529457 | 7.479651e+03 | 1061.312394 | ||
| ENSG00000196155 | 25894 | PLEKHG4 | protein_coding | Q58EX7 | FUNCTION: Possible role in intracellular signaling and cytoskeleton dynamics at the Golgi. | Alternative splicing;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome | The protein encoded by this gene can function as a guanine nucleotide exchange factor (GEF) and may play a role in intracellular signaling and cytoskeleton dynamics at the Golgi apparatus. Polymorphisms in the region of this gene have been found to be associated with spinocerebellar ataxia in some study populations. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2015]. | hsa:25894; | cytosol [GO:0005829]; guanyl-nucleotide exchange factor activity [GO:0005085]; regulation of small GTPase mediated signal transduction [GO:0051056] | 12796826_Spinocerebellar ataxia type 4 (SCA4) is mapped to chromosome 16q22.1 in northern germany.Haplotype analyses refined the gene locus to a 3.69 cM interval between D16S3019 and D16S512. 15148151_Observational study of genotype prevalence. (HuGE Navigator) 15455264_the autosomal dominant cerebellar ataxia that we have characterized is allelic with SCA4 and Japanese 16q-linked ADCA type III. 16001362_puratrophin-1 has a role in intracellular signaling and actin dynamics at the Golgi apparatus 16491300_Mutations of the puratrophin1 gene on chromosome 16q22.1 are not a common genetic cause of cerebellar ataxia in a European population. 16491300_Observational study of genotype prevalence. (HuGE Navigator) 16780885_We found the C-to-T substitution in the puratrophin-1 gene in 20 patients with ataxia (16 heterozygotes and four homozygotes) and four asymptomatic carriers in 9 of 24 families with an unknown type of ADCA. 17357132_among 686 autosomal dominant spinocerebellar ataxia families in our cohort, 57 families were identified to have 65 affected individuals, who carried the C-to-T substitution of the puratrophin-1 gene 17611710_Disease locus of 16q-autosomal dominant cerebellar ataxia was definitely confined to a 900-kb genomic region between the SNP04 and the -16C>T substitution in the puratrophin-1 gene in 16q22.1. 18482007_Rac1 activation specifically in membrane ruffles by the guanine-nucleotide-exchange factor FLJ00068 is sufficient for insulin induction of glucose uptake into skeletal-muscle cells. 19065522_Observational study of gene-disease association. (HuGE Navigator) 19065522_The mutation of c.-16C to T of the PURATROPHIN-1 gene might be rare in SCA patients in China. 20424877_(TGGAA)(n) repeats in the insertion mutation of PLEKHG4 are related to the pathogenesis of SCA31 21357611_This letter suggested cerebellar ataxia due to a pentanucleotide repeat (TAGAA) expansion on the puratrophin-1 (PLEKHG4) gene on chromosome 16q-22.1. 25025572_Role of the guanine nucleotide exchange factor in Akt2-mediated plasma membrane translocation of GLUT4 in insulin-stimulated skeletal muscle. | ENSMUSG00000014782 | Plekhg4 | 1.834558e+03 | 1.2238568 | 0.291434819 | 0.2566179 | 1.308158e+00 | 0.2527284901 | 0.79332654 | No | Yes | 1.625781e+03 | 189.432122 | 1.372275e+03 | 164.447914 | |
| ENSG00000196233 | 84458 | LCOR | protein_coding | Q96JN0 | FUNCTION: May act as transcription activator that binds DNA elements with the sequence 5'-CCCTATCGATCGATCTCTACCT-3' (By similarity). Repressor of ligand-dependent transcription activation by target nuclear receptors. Repressor of ligand-dependent transcription activation by ESR1, ESR2, NR3C1, PGR, RARA, RARB, RARG, RXRA and VDR. {ECO:0000250, ECO:0000269|PubMed:12535528}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | LCOR is a transcriptional corepressor widely expressed in fetal and adult tissues that is recruited to agonist-bound nuclear receptors through a single LxxLL motif, also referred to as a nuclear receptor (NR) box (Fernandes et al., 2003 [PubMed 12535528]).[supplied by OMIM, Mar 2008]. | hsa:84458; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; estrogen receptor binding [GO:0030331]; histone deacetylase binding [GO:0042826]; histone methyltransferase binding [GO:1990226]; transcription corepressor activity [GO:0003714]; transcription corepressor binding [GO:0001222]; ubiquitin-specific protease binding [GO:1990381]; cellular response to estradiol stimulus [GO:0071392]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 12535528_Data show that LCoR (ligand-dependent nuclear receptor corepressor) represents a class of corepressor that attenuates agonist-activated nuclear receptor signaling by multiple mechanisms [Ligand-dependent nuclear receptor corepressor]. 19744931_HDAC6 can function as a cofactor of LCoR but they can act to enhance expression of specific estrogen-regulated genes 19744932_role for LCoR and CtBP1 as attenuators of progesterone-regulated transcription, but LCoR and CtBP1 can act to enhance transcription of some genes 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21565892_These results showed that the presence of miR-615-3p repressed the expression of LCoR, a derepressor of peroxisome proliferator-activated receptor gamma (PPARgamma). 21856747_Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. 22277651_LCoR recruitment by KLF6 regulates expression of the cyclin-dependent kinase inhibitor CDKN1A gene. 28414308_Taken together, these results provide new insights into the mechanism of action of LCoR and RIP140 and highlight their strong interplay for the control of gene expression and cell proliferation in breast cancer cells. 28530657_miR-199a promotes stem cell properties in mammary stem cells and breast cancer stem cells by directly repressing nuclear receptor corepressor LCOR, which primes interferon responses. 31186533_C10ORF12 protein functions as a positive regulator of PRC2 and facilitates PRC2-mediated H3K27me3 modification of chromatin. 32157438_A decrease of nuclear LCoR expression in line with an increase of dedifferentiation of CIN can be observed. Nuclear LCoR overexpression correlates with CIN II progression indicating a prognostic value of LCoR in cervical intraepithelial neoplasia 34410950_The Expression of NRIP1 and LCOR in Endometrioid Endometrial Cancer. | ENSMUSG00000025019 | Lcor | 1.842184e+03 | 0.6950644 | -0.524781495 | 0.2777492 | 3.478255e+00 | 0.0621802773 | 0.69776574 | No | Yes | 1.554582e+03 | 291.381337 | 1.675563e+03 | 322.042317 | |
| ENSG00000196497 | 79711 | IPO4 | protein_coding | Q8TEX9 | FUNCTION: Functions in nuclear protein import as nuclear transport receptor. Serves as receptor for nuclear localization signals (NLS) in cargo substrates. Is thought to mediate docking of the importin/substrate complex to the nuclear pore complex (NPC) through binding to nucleoporin and the complex is subsequently translocated through the pore by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to the importin, the importin/substrate complex dissociates and importin is re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus (By similarity). Mediates the nuclear import of RPS3A. In vitro, mediates the nuclear import of human cytomegalovirus UL84 by recognizing a non-classical NLS. {ECO:0000250, ECO:0000269|PubMed:11823430}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Nucleus;Protein transport;Reference proteome;Repeat;Transport | hsa:79711; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; membrane [GO:0016020]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; nuclear import signal receptor activity [GO:0061608]; nuclear localization sequence binding [GO:0008139]; small GTPase binding [GO:0031267]; DNA replication-dependent chromatin assembly [GO:0006335]; DNA replication-independent chromatin assembly [GO:0006336]; protein import into nucleus [GO:0006606] | Mouse_homologues 16166738_Metallothionein deficient mice have a marked decrease in Importin 4 transcripts during acute lung injury 20805509_report a function of IPO4 and nuclear import in the Fanconi anemia pathway of DNA repair | ENSMUSG00000002319 | Ipo4 | 1.199887e+04 | 1.4033604 | 0.488885604 | 0.3178778 | 2.371582e+00 | 0.1235617817 | 0.75871535 | No | Yes | 1.379340e+04 | 1790.870144 | 8.416388e+03 | 1121.195275 | ||
| ENSG00000196550 | 729533 | FAM72A | protein_coding | Q5TYM5 | FUNCTION: May play a role in the regulation of cellular reactive oxygen species metabolism. May participate in cell growth regulation. {ECO:0000269|PubMed:21317926}. | Alternative splicing;Cytoplasm;Mitochondrion;Reference proteome | hsa:729533; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; mitochondrion [GO:0005739] | 18676834_Using deletion constructs, the authors find that Ugene binds to the first 25 amino acids of the UNG2 NH(2) terminus. They suggest that Ugene induction in cancer may contribute to the cancer phenotype by interacting with the BER pathway. 19755123_this study provides an additional possible mechanism of neurotoxicity in Alzheimer's disease, the induction of p17(FAM72B), through which Abeta acts to induce apoptosis and exhibit other Alzheimer's disease characteristics. 21317926_It was found that Ugene, designated herein as LMP1-induced protein (LMPIP), was induced, in a time-dependent manner, in EBV-infected peripheral blood mononuclear cells and LMP1-transfected 293 cells. 23900679_Data indicate that p17 (p17 amyloid-beta peptide-induced protein; known as Ugene, LMPIP, or FAM72A/B) drives the cell cycle into the G0/G1 phase and enhances survival of proliferating cells. 26206078_An epistemological characterization of the human tumorigenic neuronal paralogous FAM72 gene loci (FAM72A, FAM72B, FAM72C, FAM72D). 34819670_FAM72A antagonizes UNG2 to promote mutagenic repair during antibody maturation. | ENSMUSG00000055184 | Fam72a | 4.318630e+02 | 1.0040677 | 0.005856560 | 0.3180752 | 3.396701e-04 | 0.9852957074 | 0.99716006 | No | Yes | 3.431844e+02 | 56.117601 | 3.688659e+02 | 61.762037 | ||
| ENSG00000196605 | 162993 | ZNF846 | protein_coding | Q147U1 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000250}. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:162993; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 4.171289e+01 | 1.1258016 | 0.170952551 | 0.5807966 | 8.673921e-02 | 0.7683642434 | 0.95414161 | No | Yes | 3.768088e+01 | 12.342942 | 3.690449e+01 | 12.082638 | |||||
| ENSG00000196689 | 7442 | TRPV1 | protein_coding | Q8NER1 | FUNCTION: Ligand-activated non-selective calcium permeant cation channel involved in detection of noxious chemical and thermal stimuli. Seems to mediate proton influx and may be involved in intracellular acidosis in nociceptive neurons. Involved in mediation of inflammatory pain and hyperalgesia. Sensitized by a phosphatidylinositol second messenger system activated by receptor tyrosine kinases, which involves PKC isozymes and PCL. Activation by vanilloids, like capsaicin, and temperatures higher than 42 degrees Celsius, exhibits a time- and Ca(2+)-dependent outward rectification, followed by a long-lasting refractory state. Mild extracellular acidic pH (6.5) potentiates channel activation by noxious heat and vanilloids, whereas acidic conditions (pH <6) directly activate the channel. Can be activated by endogenous compounds, including 12-hydroperoxytetraenoic acid and bradykinin. Acts as ionotropic endocannabinoid receptor with central neuromodulatory effects. Triggers a form of long-term depression (TRPV1-LTD) mediated by the endocannabinoid anandamine in the hippocampus and nucleus accumbens by affecting AMPA receptors endocytosis. {ECO:0000250|UniProtKB:O35433, ECO:0000269|PubMed:11050376, ECO:0000269|PubMed:11226139, ECO:0000269|PubMed:11243859, ECO:0000269|PubMed:12077606}. | 3D-structure;ANK repeat;ATP-binding;Alternative splicing;Calcium;Calcium channel;Calcium transport;Calmodulin-binding;Cell junction;Cell membrane;Cell projection;Glycoprotein;Ion channel;Ion transport;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Postsynaptic cell membrane;Reference proteome;Repeat;Synapse;Transmembrane;Transmembrane helix;Transport | Capsaicin, the main pungent ingredient in hot chili peppers, elicits a sensation of burning pain by selectively activating sensory neurons that convey information about noxious stimuli to the central nervous system. The protein encoded by this gene is a receptor for capsaicin and is a non-selective cation channel that is structurally related to members of the TRP family of ion channels. This receptor is also activated by increases in temperature in the noxious range, suggesting that it functions as a transducer of painful thermal stimuli in vivo. Four transcript variants encoding the same protein, but with different 5' UTR sequence, have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:7442; | cytosol [GO:0005829]; dendritic spine membrane [GO:0032591]; external side of plasma membrane [GO:0009897]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; intrinsic component of plasma membrane [GO:0031226]; mitochondrion [GO:0005739]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; postsynaptic membrane [GO:0045211]; ATP binding [GO:0005524]; calcium channel activity [GO:0005262]; calcium-release channel activity [GO:0015278]; calmodulin binding [GO:0005516]; chloride channel regulator activity [GO:0017081]; excitatory extracellular ligand-gated ion channel activity [GO:0005231]; extracellular ligand-gated ion channel activity [GO:0005230]; identical protein binding [GO:0042802]; ion channel activity [GO:0005216]; metal ion binding [GO:0046872]; phosphatidylinositol binding [GO:0035091]; phosphoprotein binding [GO:0051219]; temperature-gated ion channel activity [GO:0097603]; transmembrane signaling receptor activity [GO:0004888]; voltage-gated calcium channel activity [GO:0005245]; behavioral response to pain [GO:0048266]; calcium ion import across plasma membrane [GO:0098703]; calcium ion transmembrane transport [GO:0070588]; cell surface receptor signaling pathway [GO:0007166]; cellular response to acidic pH [GO:0071468]; cellular response to alkaloid [GO:0071312]; cellular response to ATP [GO:0071318]; cellular response to heat [GO:0034605]; cellular response to nerve growth factor stimulus [GO:1990090]; cellular response to temperature stimulus [GO:0071502]; cellular response to tumor necrosis factor [GO:0071356]; chemosensory behavior [GO:0007635]; detection of chemical stimulus involved in sensory perception of pain [GO:0050968]; detection of temperature stimulus involved in sensory perception of pain [GO:0050965]; detection of temperature stimulus involved in thermoception [GO:0050960]; diet induced thermogenesis [GO:0002024]; fever generation [GO:0001660]; glutamate secretion [GO:0014047]; lipid metabolic process [GO:0006629]; microglial cell activation [GO:0001774]; negative regulation of establishment of blood-brain barrier [GO:0090212]; negative regulation of heart rate [GO:0010459]; negative regulation of mitochondrial membrane potential [GO:0010917]; negative regulation of systemic arterial blood pressure [GO:0003085]; negative regulation of transcription by RNA polymerase II [GO:0000122]; peptide secretion [GO:0002790]; positive regulation of apoptotic process [GO:0043065]; positive regulation of gastric acid secretion [GO:0060454]; positive regulation of nitric oxide biosynthetic process [GO:0045429]; protein homotetramerization [GO:0051289]; response to capsazepine [GO:1901594]; response to peptide hormone [GO:0043434]; sensory perception of mechanical stimulus [GO:0050954]; smooth muscle contraction involved in micturition [GO:0060083]; thermoception [GO:0050955] | 11884385_Direct phosphorylation of capsaicin receptor VR1 by protein kinase Cepsilon and identification of two target serine residues 12393854_ASICs are leading acid sensors in human nociceptors and VR1 participates in the nociception mainly under extremely acidic conditions. 12573376_In rectal hypersensitivity, nerve fibres immunoreactive to TRPV1 were increased in muscle, submucosal, and mucosal layers. 12808128_calmodulin binds to a 35-aa segment in the C terminus of TRPV1, and disruption of the calmodulin-binding segment prevents TRPV1 desensitization 12812762_Proposal that residue Y671 is critical for the high relative Ca(2+) permeability of TRPV1 and participates in the structural rearrangements of the channel protein leading to Ca(2+)-dependent desensitization. 14506258_Ser-116 and possibly Thr-370 are the most important residues involved in the cyclic AMP-dependent protein kinase pathway reduction of desensitization of capsaicin-activated currents. 14506258_Ser-116 and possibly Thr-370 of vanilloid receptor TRPV1 are the most important residues involved in the mechanism of PKA-dependent reduction of desensitization of capsaicin-activated currents 14520770_in fibers of human tooth pulp ... may play a role in perception of dental pain 14630912_vanilloid receptor 1 has a role in regulating vanilloid binding with Ca2+/calmodulin-dependent kinase II 14987252_VR1 is widely distributed in the skin, suggesting a major role for this receptor, e.g. in nociception and neurogenic inflammation. 15051629_Review notes that high expression of TRPV1 has been detected in inflammatory diseases of the colon and ileum, whereas neuropeptides released upon sensory nerve stimulation triggered by TRPV1 activation may play a role in intestinal motility disorders. 15190102_Our observations suggest that the homologous TRP domain in the TRP protein family may function as a general, evolutionary conserved AD involved in subunit multimerization 15306801_temperature sensing is tightly linked to voltage-dependent gating in the cold-sensitive channel TRPM8 and the heat-sensitive channel TRPV1 15574747_There is a minor role for transient receptor potential vanilloid receptor-1 as a mediator of cutaneous acid-induced pain. 15644492_TRPV1b receptors are expressed in trigeminal ganglion neurons and contribute to thermal nociception. 15685214_Piperine was agonistic to TRPV1 as measured by patch-clamp techniques. TRPV1 was antagonized by the competitive antagonist capsazepine and the non-competitive TRPV1 blocker ruthenium red. 15691846_Ca2+-dependent desensitization of TRPV1 might be in part regulated through channel dephosphorylation by calcineurin 15708075_Increased urothelial TRPV1 in patients with neurogenic detrusor overactivity may play a role in the pathophysiology, in concert with increased TRPV1 nerve fibers. 15738111_expression of vanilloid receptor subtype-1 and acid-sensing ion channel genes in the human trigeminal ganglion neurons 15793280_TRPV1 as a significant novel player in human hair growth control. 15992990_A progressive loss of TRPV1 expression in the urothelium as TCC stage increased and cell differentiation was lower. 16081411_several endogenous non-cannabinoid N-acylethanolamines activate TRPV(1) 16099171_Gadolinium activates and potentiates the TRPV1 by neutralizing two specific proton-sensitive sites on the extracellular side of the pore-forming loop. 16301147_may play a role in maintenance of the physiologic condition of the TMJ 16319926_NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels 16406364_Results describe the modification of 12-phenylacetyl-ricinoleoyl-vanillamide and its activity at TRPV1 and CB2 receptors. 16777226_Both TRPV1 and TRPV2 are found in human peripheral blood lymphocytes. 16793902_PAR2 activates PKCepsilon and PKA in sensory neurons, and thereby sensitizes TRPV1 to cause thermal hyperalgesia 16842630_Opioid receptor agonist morphine acts via inhibition of adenylate cyclase to inhibit protein kinase A-potentiated TRPV1 responses. 16996476_TRPV1 antagonists, including TRPV1 siRNAs, have potential in the treatment of both, neuropathic and visceral pain 16996800_Here, we review the potential therapeutic indications and adverse effects of TRPV1 antagonists. 17016800_TRPV1 may play a role in urinary sensory urgency and premature first bladder sensation on filling. 17018028_data support the hypothesis that the TRPV1b splice variant is a naturally existing inhibitory modulator of TRPV1. 17074976_We propose a new model for nerve growth factor (NGF)-mediated hyperalgesia in which physical coupling of TRPV1 and PI3K in a signal transduction complex facilitates trafficking of TRPV1 to the plasma membrane. 17109831_These data do not rule out involvement of TRPV1 in the aetiology of burning dysaesthesia following lingual nerve injury but suggest that TRPV1 at the injury site does not play a primary role. 17176640_Gingerols and shogaols activated TRPV1 and increased adrenaline secretion. 17184838_we assigned a novel role to capsaicin-sensitive TRPV1 channels. They are important Ca2+ influx channels required for cell migration. 17217056_TRPV1 is a nonselective cation channel with significant permeability to calcium, protons, and large polyvalent cations--{REVIEW} 17347480_Capsaicin-induced calcium influx through TRPV1 channels prevents adipogenesis, prevents downregulation of TRPV1 expression and prevents obesity. 17385724_IGF-I and insulin enhance TRPV1 protein expression and activity, and impaired pain sensation might result from distorted TRPV1 regulation in the peripheral nervous system 17392452_Together, these results indicate that tonic TRPV1 activation regulates body temperature. TRPV1 function in thermoregulation is conserved from rodents to primates. 17428642_Recent studies indicate that primary sensory neurons of the pancreas express transient receptor potential V1 (TRPV1) channels whose activation induces pancreatic inflammation 17442041_TRPV1 gene and protein expression inversely correlated with glioma grading, with marked loss of TRPV1 expression in the majority of grade IV glioblastoma multiforme. 17442052_The advantage of SH-SY5Y(hTRPV1) is the ability of hTRPV1 to couple to neuronal biochemical machinery and produce large quantities of cells. 17508023_Heat-shock-induced MMP-1 expression is mediated by activation of TRPV1 and is dependent on a calcium-dependent signaling process in human epidermal keratinocytes. 17508360_TRPV1 activation induces inflammatory cytokine release in corneal epithelium through MAPK signaling. 17521436_accumulation of TRPV1 and TRPV3 in peripheral nerves after injury, in spared axons, matches our previously reported changes in avulsed DRG. 17560936_Here, we demonstrate the expression of TRPV1 in synovial fibroblasts (SF) from patients with symptomatic osteoarthritis (OA) and rheumatoid arthritis (RA). 17567713_a novel group of compounds that activate TRPV1 and, consequently, provide a molecular mechanism that may account for off tastes of sweeteners and metallic tasting salts 17582772_TRPV1-positive fibres were overall significantly increased in tongue of Burning mouth syndrome. 17596456_We conclude that phosphoinositides have both inhibitory and activating effects on TRPV1, resulting in complex and distinct regulation at various stimulation levels. 17626206_Tonic activation of TRPV1 channels in the abdominal viscera by yet unidentified nonthermal factors inhibits skin vasoconstriction and thermogenesis, thus having a suppressive effect on body temperature. 17659837_These data provide the first clinical evidence that a TRPV1 antagonist may alleviate pain and hyperalgesia associated with inflammation and tissue injury. 17913835_Incorporation of multiple nonsynonymous polymorphisms, informed by the population-specific haplotype block structure of the TRPV1 gene 17927589_TRPV1 expression was significantly increased in distal small nerve fibre neuropathy compared with controls. 18076848_Increased expression within the pulps of hypomineralised teeth may be indicative of an underlying pulpal inflammation 18222434_Testicular hyperthermia in mice lacking transient receptor potential vanilloid receptor-1 results in a much more rapid and massive germ cell depletion from the seminiferous tubules than in wild-type animals. 18230619_several proton-sensitive sites on the extracellular side of the pore-forming loop of the TRPV1 channel may play an important role as a brake to suppress the excessive activity of this channel under physiological conditions. 18267041_TRPV1 may be involved in pain mechanisms associated with cervical ripening and labor. 18414789_Ara C treatment blocked insertion of TRPV1 in the cell membrane, resulting in accumulation of the receptors in the cytoplasm, loss of capsaicin sensitivity, and membrane-bound immunostaining, which was restored with a rebound on withdrawal of Ara C. 18482991_Increased translocation and multimerization of TRPV1 channels provide a cellular mechanism for fine-tuning of nociceptive responses that allow for rapid modulation of TRPV1 responses independent of transcriptional changes. 18499726_The results of this study support the hypothesis that 4-ONE is a relevant endogenous activator of vagal C-fibres via an interaction with TRPA1, and at less relevant concentrations, it may activate nerves via TRPV1. 18503554_TRPV1 mediates heat shock-induced MMP-1 expression via calcium-dependent PKCalpha signaling in HaCaT cells 18552442_expressed in airway C fibers and activated by neurogenic inflammation which leads to provocaion of cough reflex. 18559878_findings suggest that modification of I696, W697, & R701 to alanine altered channel function by affecting events downstream of initial stimuli-sensing step & imply that intersubunit interactions within the TRP box play an important role in TRPV1 gating. 18574245_PI(4,5)P(2), and not other phosphoinositides or other lipids, is the endogenous phosphoinositide regulating TRPV1 channels 18584893_TRPV1 regulates the expression and secretion of endothelial cell-derived CGRP. 18769453_TRPV1 signaling is a significant, previously unreported player in human sebocyte biology 18786439_Disease-free survival was significantly better in HCC patients with high versus those with low VR1 expression levels. 18787888_May be a mediator or part of a critical pathway in chronic hypoxia-induced proliferation of human pulmonary artery smooth muscle cells. 18791833_Full-length TRPV1 has been modeled by assembly of its major modules: the cytosolic N-terminal, C-terminal, and membrane-spanning region, by modeling of the vanilloid receptor in the closed and desensitized states. 18830626_Increased genetic expression may serve as a prognostic factor in prostate cancer. 18852901_Although keratinocytes express TRPV1 mRNA, neither responded to vanilloids with Ca(2+)-cytotoxicity 18983665_relative gene expression ofTRPV1-4 in leukocytes is: TRPV3 < TRPV4, TRPV1 and TRPV2; leukocytes in hyposensitive subjects showed up-regulation of TRPV1, which was almost doubly expressed 19036069_presence of TRPV1 may provide a link between inflammatory mediators and platelet activation 19056576_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19074743_Together with TRPV1, the capsaicin receptor, transient receptor potential cation channel subfamily A member 1 ankyrin may contribute to chemical hypersensitivity, chronic cough, and airway inflammation --{REVIEW} 19161409_expression of TRPV1 is affected by both the intrinsic ageing and photoaging processes 19328632_Moreover, TRPV1 activation triggers apoptotic neuronal death of rat cortical cultures, which may be responsible, at least in part, for the volume loss of neocortex in chronic epilepsy. 19333172_The TRPV1 was increase in the peritoneum of women with chronic pelvic pain. 19397909_These data introduce TRPV1-coupled signaling as a novel player in human monocyte-derived DC biology with anti-inflammatory actions. 19482060_Nerve fibers in human skin express TRPV1, TRPV2 and TRPM8 that co-localize with the sensory neuropeptides CGRP and SP, but not with NF200, VIP or TH. 19598235_Observational study of gene-disease association. (HuGE Navigator) 19608651_Human spermatozoa exhibit a completely functional endocannabinoid system related to anandamide, and the anandamide-binding TRPV1 receptor could be involved in the sperm fertilizing ability. 19619644_TRPV1-immunoreactivity was significantly higher and TRPV2-immunoreactivity was significantly lower in peripheral blood mononuclear cells from end-stage kidney disease patients compared to controls. 19651168_The results show that TRPV1 protein expression increases during RA-induced differentiation in vitro, which generates an altered intracellular Ca(2+) homeostasis. 19656659_Endogenous expression of TRPV1 channel in cultured human melanocytes. (Letter) 19749171_REVIEW: data on the distribution of TRPV1 channels in the body and on thermoregulatory responses to TRPV1 agonists and antagonists 19770677_Homozygous TRPV1 315C influences the susceptibility to functional dyspepsia in a Japanese population through altering the upper gastrointestinal sensation 19770677_Observational study of gene-disease association. (HuGE Navigator) 19778904_endoplasmic reticulum of dorsal root ganglion neurons contains functional TRPV1 channels 19877503_Observational study of gene-disease association. (HuGE Navigator) 19877503_UTR-3 polymorphisms in TRPV1 could be associated with the susceptibility to childhood asthma of Han Nation ethnic group in Beijing. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19923855_Since the number of TRPV1 receptor-positive nonneural cells is increased in inflammatory conditions, we hypothesize that TRPV1-receptor-mediated processes might play role in the pathogenesis of OLP. 19959817_esophageal epithelial cells produce PAF in response to TRPV1-mediated calcium elevation 20034385_Observational study of gene-disease association. (HuGE Navigator) 20034385_Polymorphisms in gene encoding TRPV1-receptor involved in pain perception are unrelated to chronic pancreatitis. 20060270_One of the mechanisms of glycolic acid(GA)-induced epidermal proliferation is a growth response of basal keratinocytes to the local elevation of H(+)-ion concentration by infiltrated GA. This response is mediated by TRPV1 activation and ATP release. 20096818_OTR and TRPV1 may be involved in dysmenorrhea and its severity in adenomyosis 20145248_analysis of the structural basis for proton and magnesium modulation of fully capsaicin-bound human TRPV1 receptors and comparison to rat receptors 20190512_PKC-dependent potentiation of TRPV1 activities under hypoxia and hyperglycemia might be involved in early diabetic neuropathy. 20351268_the turret is part of the temperature-sensing apparatus in thermoTRP channels, and its conformational change may give rise to the large entropy that defines high temperature sensitivity 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20456759_Increased TRPV1 in the esophageal mucosa may contribute to symptoms both in non-erosive reflux disease and erosive esophagitis patients 20515731_Studies indicate that there is evidence that TRPV1 forms heteromeric channel complexes. 20518854_Transient receptor potential vanilloid-1 mRNA level and its protein expression were significantly greater in patients with erosive esophagitis than asymptomatic patients and healthy controls. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20639579_Genetically determined level of TRPV1 activity is relevant for asthma pathophysiology. 20639579_Observational study of gene-disease association. (HuGE Navigator) 20691059_These findings and those of others reported in the literature indicate reciprocal interactions between TRPV1 and ROS play critical roles in the pathological and nociceptive responses active during arthritic inflammation. 20696376_The proper function of transgenic NompC, a putative Drosophila mechanosensitive TRP channel, is not only required for fly locomotion, but also crucial for larval crawling. 20807792_A portion of the NO component of thermal hyperaemia may be due to activation of TRPV-1 channels. 21037510_Observational study of gene-disease association. (HuGE Navigator) 21044960_the charged residues in S4 and the S4-S5 linker contribute to voltage sensing in TRPV1 and, despite their highly conserved nature, regulate the temperature and chemical gating in the various TRPV channels in different ways 21076423_Activation of TRPV1 by endogenous anandamide mediates regulation of synaptic strength at central synapses. 21076424_TRPV1 channels modulate synaptic strength in a cell type-specific manner in the nucleus accumbens. 21083604_Downregulation of TRPV1 mRNA expression may represent an independent negative prognostic factor for bladder cancer patients. 21160085_The present study shows that TRPV1 expression in peritoneal endometriosis foci is related to chronic pelvic pain in women. 21168271_Western blotting confirms gene and protein expression of functional TRPV1 channel, which plays a crucial role in mediating thermal sensation in teeth. 21285946_The authors conclude that proton activation and potentiation of TRPV1 are both voltage dependent and that amino acid 660 is essential for proton-mediated gating of TRPV1. 21310942_Loss of TRPV1 is associated with urothelial cancer. 21345654_blockade of TRPV1 activation can suppress the atopic dermatitis-like symptoms by accelerating skin barrier recovery 21351130_in female patients with overactive bladder symptoms, urothelial cells demonstrated augmented trpv1 signaling. 21451044_TRPV1 expression reveals a high density of receptors in discrete brain regions of Trpv1 reporter mice, most notably in a contiguous band of cells within and adjacent to the caudal hypothalamus, and in arteriolar smooth muscle cells. 21493704_TRPV1 activation in endothelial cells may trigger Ca(2+)-dependent PI3K/Akt/CaMKII signalling, which leads to enhanced phosphorylation of TRPV1, increased TRPV1-eNOS complex formation, eNOS activation and, ultimately, NO production. 21506114_Human corneal epithelial cells possess thermosensitive TRPV1 activity. 21546516_This is the first study to report that the 57 kb deletion extends into the TRPV1 gene causing dysregulation of transcription in peripheral blood mononuclear cells isolated from cystinosis patients. 21555515_there is a cholesterol-binding site in TRPV1 and that the functions of TRPV1 depend on the genetic variant and membrane cholesterol content. 21616913_This indicates a role for the pro-nociceptive gene TRPV1 in genetic susceptibility to symptomatic knee OA, which may also be influenced by a role for this molecule in cartilage function. 21654202_This review summarizes the current knowledge of the molecular determinants of TRPV1 channel activation by heat, protons and capsaicin. 21911503_The chimeric approach reveals that differences in the TRPV1 pore domain determine species-specific sensitivity to block of heat activation. 21926175_biochemical evidence of dimerization of TRPV1 subunits upon exposure to phenylarsine oxide and hydrogen peroxide 21945155_Activation of TRPV1 translates into changes of intracellular Ca(2+), a known mechanism triggering the secretion of chromogranin A 21951314_These findings may indicate that TRPV1 is not a key mediator of the symptoms in seasonal allergic rhinitis 21956871_These results suggest that TRPV1 is expressed in myoepithelial cells of the human submandibular gland and mediates myoepithelial contraction via a mechanism involving MLC(20) phosphorylation. 21960521_HCl-induced activation of TRPV1 causes ATP release from esophageal epithelial cells that causes release of CGRP and SP from esophageal submucosal neurons. 21962912_Agonist-induced activation of nociceptors by TRPA1 and TRPV1 elicits painful sensations, whereas nonneuronal tissue cells respond with differential release of inflammatory mediators, thus influencing local vasodilatation and neuronal sensitization. 21981454_Agonists of TRPV1 and TRPA1 induced MUC5B release in the human nasal airways in vivo. 22089942_In addition, the present study demonstrates that capsaicin's inhibitory effect on collagen-induced canine platelet aggregation is not mediated by TRPV1. 22162417_Nominal association was confirmed for TRPV3 rs7217270 in migraine with aura and TRPV1 rs222741 in the overall migraine group. 22171160_TRPV1 induced increases in IL-6/IL-8 release occur through TAK1 activation of JNK1-dependent and JNK1-independent signaling pathways. 22242698_TRPV1 can be expressed on bronchial fibroblasts in situations where an underlying inflammatory stimulus exists, as is the case in airway diseases such as asthma and COPD. 22260796_data illustrate that presynaptic calcineurin activity is required for both TRPV1-induced long-term depression(LTD)and TRPV1 agonist-induced depression; finding is the first to demonstrate the TRPV1-induced signaling mechanism in CA1 hippocampus 22262838_role of TRPV1 in modulating microtubule assembly/disassembly and suggests the participation of these two players in a feedback cycle linking nociception and cytoskeletal remodeling. 22262885_The results of this study provided the first evidence that TRPV1 channels regulate cortical excitability to paired-pulse stimulation in humans. 22327830_Cultivated HCjE cells and human conjunctiva express TRPV1, mRNA. 22389490_20-HETE both activates and sensitizes mouse and human TRPV1, in a kinase-dependent manner, involving the residue Ser(502) in heterologously expressed hTRPV1 22401740_Increased frequency of the Met315Ile TRPV1 genotype in females diagnosed with neuropathic pain suggests that this polymorphism, together with other physiological factors such as sex, may influence individual susceptibility to neuropathic pain. 22443337_TRPV1 single nucleotide polymorphisms may enhance susceptibility to cough in current smokers and in subjects with a history of workplace exposures. 22493457_TRPV1 agonists induce long-term receptor down-regulation by modulating the expression level of the channel through a mechanism that promotes receptor endocytosis and degradation 22518827_TRPV-1 channels participate in reflex cutaneous vasodilation. 22556011_The results suggested that the increased TRPV1-positive nerve fibers may integrate various stimuli on peripheral terminals or primary sensory neurons and generate hyperalgesia in endometriosis. 22566504_Data show the expression of TRPC6 and TRPV1 was significantly increased after 24 hours of exposure to an atheroprone flow, whereas the expression of TRPC3 and TRPM7 was significantly higher in endothelial cells exposed to shear stress. 22568077_TRPV1 mRNA was detected in hPDL cells and the TRPV1 expression was observed by confocal laser scanning microscope 22570472_N-glycosylation determines ionic permeability and desensitization of the TRPV1 capsaicin receptor 22647236_The over-expression of TRPV1 in cortical lesions of tuberous sclerosis complex and focal cortical dysplasia type suggested the possible involvement of TRPV1 in the intrinsic and increased epileptogenicity of malformations of cortical development 22691178_The increase in capsaicin sensitivity correlated with increased urothelial TRPV1 expression in overactive bladder cells. 22792722_Interaction of sea amemone Heteractis crispa Kunitz type polypeptides with pain vanilloid receptor TRPV1 22809607_TRPV1 gene polymorphisms at positions rs222748 and rs8065080 may be unrelated to nonspecific chronic cough in children. 22814029_This is the first evidence that CB(2) and TRPV(1) receptors cooperate to modulate MDR1 expression. 22820645_TRPV1 is highly expressed in tumor & weakly expressed in tumor-free brain. Endovanilloidsfrom neural precursor cells activates TRPV1 in high-grade astrocytomas. TRPV1 stimulation triggers tumor cell death via the endoplasmic reticulum stress pathway. 22829138_particulate matter induces TRPV1 channel opening, resulting in increase of Ca2+ concentration and cAMP level, and all these responses lead to mucin protein secreted from human bronchial epithelial cells. 22870787_Results suggest that the single nucleotide polymorphisms of the TRPV1 gene may not be associated with irritable bowel syndrome in Korean populations. 22883519_The expression of TRPV1 was correlated with the degree of pain in patients with endometriosis. 22935106_findings strongly suggest that TRPV1 sensitization influences the hypersecretion of mucus and inflammatory cytokines 22936245_mRNA and protein levels of TRPV1 are upupregulated in the brain from patients with mesial temporal lobe epilepsy 22942745_This study is the first to shed light on the structural determinants required for the interaction between TRPV1 and evodiamine, and gives new suggestions for the rational design of novel TRPV1 ligands. 23047628_Single nucleotide polymorphism of TRPV1 315G>C, rs5981521 of pri-miR-325 and SCN10A is related to the development of functional dyspepsia. This involvement differed between Helicobater pylori-positive and -negative patients. 23074220_analysis of the functional interactions between MRGPR-X1 and TRPV1 23123284_These results indicate that CGRP may participate in trigeminal nociceptive system sensitization by induced increase in TRPV1 23139219_Capsaicin inhibits chloride secretion in part by causing NKCC1 internalization, but by a mechanism that appears to be independent of TRPV1. 23183769_an inward calcium current via the TRPV1 channels of endothelial cells correlates with a stronger adhesion between monocytes and endothelial cells 23264624_the direct anchoring of both PKA and AC to TRPV1 by AKAP79/150 facilitates the response to inflammatory mediators and may be critical in the pathogenesis of thermal hyperalgesia. 23279936_these results propose a novel mechanism by which cellular cholesterol depletion modulates the function of TRPV1. 23398938_This study demonstrated an increased activity of TRPV1 in DRG neurons as a new mechanism contributing to opioid withdrawal-induced hyperalgesia. 23453732_TRPV1 and TRPV2 are potential targets for potential therapeutic interventions in cardiovascular disease. 23458684_the activation of TRPM2 channel, which mediates ATP release, and TRPV1 channel plays significant roles in the cellular responses to DNA damage induced by gamma-irradiation and UVB irradiation. 23508958_Excitation and modulation of TRPA1, TRPV1, and TRPM8 channel-expressing sensory neurons by the pruritogen chloroquine. 23510405_This study evaluated the presence of TRPV1 in human and canine mammary cancer cells and the role of TRPV1 in regulating cell proliferation. 23616546_Disruption of the interaction of TRPV1 and an anchoring protein AKAP79 inhibits inflammatory hyperalgesia. 23689362_TRPV1 is a critical disease modifier in experimental autoimmune encephalomyelitis, and we identify a predictor of severe disease course and a novel target for MS therapy. 23699529_The results of this study suggested that antagonizing the TRPV1-AKAP79 interaction will be a useful strategy for inhibiting inflammatory hyperalgesia. 23709255_TRPV1 is expressed in cells that are particularly active in Ca(2) exchange as well as in cells with significant water transport activity in eyes of New Zealand White rabbits and humans 23820422_Data suggest that expression of neuronal markers VR1 (vanilloid receptor 1), PGP9.5 (ubiquitin C-terminal hydroxylase-L1, human), and NGFp75 (p75 neurotrophin receptor) in endometrium of women is independent of presence of endometriosis. 23902373_Evodiamine qualifies as structurally unique lead structure to develop new potent TRPV1 agonists/desensitizers. 24002057_Rhinovirus infection causes upregulation of neuronal TRPV1 expression mediated indirectly by virus-induced soluble factors. 24073800_The degradation of claudin-3 and claudin-4 induced by acidic stress could be attenuated by specific TRPV1 blockers. 24084605_In uveal melanoma cells, TRPV1, TRPA1 and TRPM8 gene expression were identified. 24098582_TRPV1 potentiates TGFbeta-induction of corneal myofibroblast development through an oxidative stress-mediated p38-SMAD2 signaling loop. 24139494_IR features an overexpression of TRPV1 in the nasal mucosa and increased SP levels in nasal secretions. Capsaicin exerts its therapeutic action by ablating the TRPV1-SP nociceptive signaling pathway in the nasal mucosa. 24152419_There is increased TRPV1 immunoreactivity in human osteoarthritis synovium, confirming the diseased joint as a potential therapeutic target for TRPV1-mediated analgesia. 24189713_TRPV1 mRNA or protein expression is not increased in the rectal mucosa of Irritable bowel syndrome patients,including visceral hypersensitivity. 24190005_In Burning mouth syndrome patients was increased TRPV1, decreased CB1 and increased CB2 expression in tongue epithelial cells also associated with a change in their distribution. 24275229_sensitization of TRPV1 via activation of TRPA1, which involves adenylyl cyclase, increased cAMP, subsequent translocation and activation of PKA, and phosphorylation of TRPV1 at PKA phosphorylation residues. 24296694_Laryngeal TRPV1 plays an important role in cough sensitivity. 24312564_intact 2-arachidonoylglycerol and 1-arachidonoylglycerol are endogenous TRPV1 activators 24340836_Our study demonstrates that TRPV1 may be a susceptible gene for type 1 diabetes in an Ashkenazi Jewish population 24458144_TRPV1-immunoreactive nerve fibers and NGF have a role in inflammation in interstitial cystitis/bladder pain syndrome 24564660_Comparison of the effects induced by mutations in homologous positions in different TRP receptors (or more generally in other distantly related ion channels) may elucidate the gating mechanisms conserved during evolution. [review] 24564662_There is now mounting evidence that TRPV1 receptors may be an important element in modulation of nociceptive information at the spinal cord level and represent an interesting target for analgesic therapy. [review] 24569998_furanocoumarins represent a novel group of TRPV1 modulators that may become important lead compounds in the drug discovery process aimed at developing new treatments for pain management. 24599956_the signaling lipid phosphoinositide 4,5-bisphosphate (PI(4,5)P2) has opposite effects on the function of TRPV1 ion channels depending on which leaflet of the cell membrane it resides in. 24658385_Degradation of TRPV1 is mediated by autophagy, and that this pathway can be enhanced by cortisol. 24720453_genetic polymorphism of TRPV1 945G>C could be one of the pathophysiological factors of FD, especially in the case of H. pyloripositive patients in Korea. 24764033_TRPV1-, TRPV2-, P2X3-, and parvalbumin-immunoreactivity neurons in the human nodose ganglion innervate the pharynx and epiglottis through the pharyngeal branch and superior laryngeal nerve 24798548_TRPV1 channels are activated by intracellular long-chain acyl CoAs 24868547_sensory proteins P2X3 and TRPV1 are in correlation with urothelial differentiation, while P2X5 and TRPV4 have unique expression patterns 24871046_Demonstrated how and why these TRP channels TRPV1, TRPA1, and TRPM8 are important to start up a systemic response of energy expenditure, energy allocation, and water retention and how this is linked to a continuously activated immune system in chronic inflammatory diseases. [Review] 24889371_TRPV1 over-expression may be an effective way to induce necrosis and that this approach may not always be associated with enhancement of cancer cell proliferation. 24931369_airway epithelial cells TRPV1 activation potently induces allergic cytokine thymic stromal lymphopoietin (TSLP) release 25170901_The described L-carnitine osmoprotective effect is elicited through suppression of hypertonic-induced TRPV1 activation leading to increases in L-carnitine uptake through a described Na(+)-dependent L-carnitine transporter 25210497_TRPV1 may play a crucial role in 14,15-epoxyeicosatrienoic acid -induced calcium influx, nitric oxide production and angiogenesis. 25216685_High TRPV1 expression is associated with osteoporosis in thalassemia major. 25257701_Genetic variation in TRPV1 and TAS2Rs influence sensations from sampled EtOH and may potentially influence how individuals initially respond to alcoholic beverages. 25266715_fibroblast growth factor-17 and neuregulin 2 were significantly up-regulated in capsaicin-treat | ENSMUSG00000005952 | Trpv1 | 9.938270e+02 | 1.0328053 | 0.046568264 | 0.2727798 | 2.877302e-02 | 0.8653041854 | 0.97426955 | No | Yes | 7.661708e+02 | 85.726594 | 7.739037e+02 | 88.783404 | |
| ENSG00000196776 | 961 | CD47 | protein_coding | Q08722 | FUNCTION: Has a role in both cell adhesion by acting as an adhesion receptor for THBS1 on platelets, and in the modulation of integrins. Plays an important role in memory formation and synaptic plasticity in the hippocampus (By similarity). Receptor for SIRPA, binding to which prevents maturation of immature dendritic cells and inhibits cytokine production by mature dendritic cells. Interaction with SIRPG mediates cell-cell adhesion, enhances superantigen-dependent T-cell-mediated proliferation and costimulates T-cell activation. May play a role in membrane transport and/or integrin dependent signal transduction. May prevent premature elimination of red blood cells. May be involved in membrane permeability changes induced following virus infection. {ECO:0000250, ECO:0000269|PubMed:11509594, ECO:0000269|PubMed:15383453, ECO:0000269|PubMed:7691831}. | 3D-structure;Alternative splicing;Cell adhesion;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phosphoprotein;Pyrrolidone carboxylic acid;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene encodes a membrane protein, which is involved in the increase in intracellular calcium concentration that occurs upon cell adhesion to extracellular matrix. The encoded protein is also a receptor for the C-terminal cell binding domain of thrombospondin, and it may play a role in membrane transport and signal transduction. This gene has broad tissue distribution, and is reduced in expression on Rh erythrocytes. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jul 2010]. | hsa:961; | cell surface [GO:0009986]; extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; specific granule membrane [GO:0035579]; tertiary granule membrane [GO:0070821]; cell-cell adhesion mediator activity [GO:0098632]; protein binding involved in heterotypic cell-cell adhesion [GO:0086080]; thrombospondin receptor activity [GO:0070053]; ATP export [GO:1904669]; cell migration [GO:0016477]; cellular response to interferon-gamma [GO:0071346]; cellular response to interleukin-1 [GO:0071347]; cellular response to interleukin-12 [GO:0071349]; integrin-mediated signaling pathway [GO:0007229]; negative regulation of Fc-gamma receptor signaling pathway involved in phagocytosis [GO:1905450]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cell-cell adhesion [GO:0022409]; positive regulation of inflammatory response [GO:0050729]; positive regulation of monocyte extravasation [GO:2000439]; positive regulation of phagocytosis [GO:0050766]; positive regulation of stress fiber assembly [GO:0051496]; positive regulation of T cell activation [GO:0050870]; regulation of interferon-gamma production [GO:0032649]; regulation of interleukin-10 production [GO:0032653]; regulation of interleukin-12 production [GO:0032655]; regulation of interleukin-6 production [GO:0032675]; regulation of nitric oxide biosynthetic process [GO:0045428]; regulation of tumor necrosis factor production [GO:0032680] | 11907074_The interaction of CD47 with SHPS-1 may contribute to the recruitment of B lymphocytes via endothelial cells under steady state conditions. 11980922_Interactions of thrombospondins with alpha4beta1 integrin and CD47 differentially modulate T cell behavior 12218055_regulates integrin function. Differential effects on alpha vbeta 3 and alpha 4beta1 integrin-mediated adhesion. 12351399_mechanism of antigen-induced caspase-independent cell death in normal and leukemic cells 12370491_An autocrine loop of the integrin alpha(V)beta(3)/CD47 receptor complex and thrombospondin-1 is identified as the molecular coupling device between mechanical loading and apoptosis. 12393442_distribution of CD47, most restrained by cytoskeleton linkages plus a smaller fraction able to diffuse in the membrane, and colocalization with Rh protein complexes 12393467_Evidence protein 4.2 interacts with the Rh membrane complex member CD47 obtained from red cells of patients with hereditary spherocytosis associated with complete protein 4.2 deficiency 12609828_CD47 expressed on platelets significantly contributes to platelet adhesion on tumor necrosis factor-alpha (TNF-alpha)-stimulated vascular endothelial cells 12646616_Ligation of CD47 by any of its known ligands rapidly induces heterotrimeric Gi alpha-dependent but caspase-independent apoptosis in Jurkat T cells and, importantly, in activated primary T cells. 12736272_the extracellular Ig domain of IAP(CD47), when bound to thrombospondin, interacts with integrin alphaIIbbeta3 and can change alphaIIbbeta3 in a high affinity state without the requirement of intracellular signaling 12902472_Interaction of CD47 with thrombospondin-1 contributes to the perpetuation of inflammation in rheumatoid synovitis. 14551146_protein 4.2 strongly influences CD47 levels as well as the extent of membrane skeleton attachment in erythrocytes 14747477_IAP gene expression is regulated by alpha-Pal 14966135_CD47 recruits alpha(v)beta(3) and its associated signaling molecules to these domains 15292185_CD47 has a role in inducing alpha4beta1 integrin activation and adhesion in sickle reticulocytes 15359629_Review. The interaction between CD47 and SIRPalpha seems to be important to limit destruction of host cells in autoimmune diseases like autoimmune hemolytic anemia (AIHA), where macrophages destroy antibody or complement opsonized cells. 15498560_CD47 short interferint RNA inhibited cell proliferation in cultures of hepatoma cells. 15725479_Apoptosis inducing single-chain antibody fragments against cd47 antigens will be effective as a novel therapy for multiple myeloma. 15842360_SHPS-1 negatively regulates platelet function via CD47, especially alpha(IIb)beta(3)-mediated outside-in signaling 15880429_Novel CD47-dependent intercellular adhesion modulates cell movement. 15917238_CD47 associates with Fas upon its activation and augments Fas-mediated apoptosis. 16291597_SIRPalpha-CD47 interactions, which reportedly define self, exhibit cell type specificity and limited cross-species reactivity 16581280_RhD co-localizeS with CD47 but not calreticulin or glycophorin-A on human RBCs. 16697668_highlight major species differences in CD47-SIRPalpha interactions and CD47 integration, suggesting that signaling by CD47 in man may be qualitatively different from mouse. 16951312_CD47 stimulation by a thrombospondin-1 peptide induces naive or memory CD4+CD25- T cells to become suppressive. 17098740_analysis of species-specific CD47 interactions with signal regulatory protein alpha 17360380_Genetic induction of human CD47 on porcine cells could provide inhibitory signaling to SIRPalpha on human macrophages, providing a novel approach to preventing macrophage-mediated xenograft rejection. 17363705_In this review the authors provide mechanistic insight into the signaling pathways used by thrombospondin-1 and CD47 at the cellular and molecular level to prevent nitric oxide signaling through cyclic GMP, thereby exacerbating tissue ischemia. 17572512_Upregulation of CD47 in sporadic inclusion body myositis. 18186923_CD47/IAP is necessary for chondrocyte mechanotransduction. Through interactions with alpha5beta1 integrin and thrombospondin, CD47/IAP may modulate chondrocyte responses to mechanical signals. 18332220_On both RBCs and microbeads, human CD47 potently inhibits phagocytosis as does direct inhibition of myosin. CD47-SIRPalpha interaction initiates a dephosphorylation cascade directed in part at phosphotyrosine in myosin. 18497546_CD47 may play an inhibitory role in NK cell-mediated cytotoxicity against cancer cells, implying a possible mechanism of immune escape in human cancer. 18523271_A T cell-negative sensor that may serve to dampen unchecked proinflammatory T helper cell type 1 (Th1) responses, such as occur during contact hypersensitivity in transgenic mice. 18524990_CD47 is enriched at endothelial junctions, and its interaction with SIRPgamma is required for human T-cell transendothelial migration 18657508_These results explain the specificity of CD47 for the SIRP family of paired receptors in atomic detail. 18675774_TSP-1's C-terminal domain opens to interact with the CD-47 receptor 18855618_In this review, the CD47/thrombospondin-1/signal-regulatory protein (SIRP)-alpha inhibitory axis is proposed as an important sensor to maintain homeostasis and regulate innate and adaptive immune responses. 18954403_Expression levels of CD47, CD35, CD55, and CD59 on red blood cells and signal-regulatory protein-alpha,beta on monocytes from patients with warm autoimmune hemolytic anemia. 18979398_Down-regulation of the IAP antagonistis XAF1, Smac/DIABLO and HtrA2, has been related to the onset and progression of various malignancies. 19144568_expression downregulated in multiple slerosis lesions, suggesting role in CNS inflammation 19360900_CD47 signaling can negatively affect the viability and function of cerebral endothelial cells, further supporting the notion that CD47 may be a potential neurovascular target for stroke and brain injury 19628875_Data show show that the CD47-SIRPalpha interaction will span a distance of around 14 nm between interacting cells, comparable with that of an immunological synapse. 19632178_Study concludes that CD47 upregulation is an important mechanism that provides protection to normal HSCs during inflammation-mediated mobilization, and that leukemic progenitors co-opt this ability in order to evade macrophage killing. 19661269_Disruption of CD47-TSP-1 interactions or preventing the recruitment of DCs to tumors may provide novel approaches to therapy of myeloma bone disease and osteoporosis. 19748659_CD47/SIRP-alpha interactions are implicated in the pathogenesis of DC-driven allergic airway inflammation. 19794081_Reduced levels of CD47 are expected to result in a reduced level of phosphatidylserine exposure following ligation. 19952055_Our findings suggest that microRNA dysregulated in multiple sclerosis lesions reduce CD47 in brain resident cells, releasing macrophages from inhibitory control, thereby promoting phagocytosis of myelin. 20033473_Data show that cartilage oligomeric matrix protein (COMP) promotes cell attachment via independent mechanisms involving cell surface CD47 and alphaVbeta3 integrin and that cell attachment to COMP induces formation of fascin-stabilized actin microspikes. 20299253_Inhibition of engulfment correlates with affinity of CD47 for SIRPA - but only at low levels of CD47. 20364320_CD47 signaling is active within cells of the neurovascular interface. 20675593_Escherichia coli upregulation of CD47 in dendritic cells (DC) triggers production of DC natural ligand thrombospondin-1 and prevents the cell maturation process. 20705613_Poor prognosis of breast cancer patients with high expression of CD47 is due to an active CD47/SIRPA signaling pathway in circulating cells. 20826801_The role of cis dimerization of signal regulatory protein alpha (SIRPalpha) in binding to CD47. 20923780_Thrombospondin-1 inhibits VEGF receptor-2 signaling by disrupting its association with CD47 21178137_Increased CD47 expression correlated with high amounts of calreticulin on cancer cells and was necessary for protection from calreticulin-mediated phagocytosis. 21203512_Amyloid-beta inhibits No-cGMP signaling in a CD36- and CD47-dependent manner 21211546_Results demonstrate that the CD47 pathway in ITP patients abnormally modulates platelet homeostasis. 21343308_inhibition of T cell receptor signaling by thrombospondin-1 is mediated by CD47 and requires its modification at Ser(64). 22014686_vitamin D binding protein does not bind to TSP-1, CD36 or CD47 22079249_a specific and age-dependent interaction of soluble fibrinogen with human RBC membrane; additionally we present CD47 as a putative mediator in this process. This interaction may contribute to RBC hyperaggregation in inflammation. 22215724_Activated CD47 promotes pulmonary arterial hypertension through targeting caveolin-1. 22427202_CD47 on experimentally aged erythrocytes is recognized by SIRPalpha. 22451913_data,show that CD47 is a commonly expressed molecule on all cancers, its function to block phagocytosis is known, and blockade of its function leads to tumor cell phagocytosis and elimination 22511785_Surfactant protein D (Sp-D) binds to membrane-proximal domain (D3) of signal regulatory protein alpha (SIRPalpha), a site distant from binding domain of CD47, while also binding to analogous region on signal regulatory protein beta (SIRPbeta). 22535707_ectopic expression of murine Cd47 in human hepatocytes selectively favors engraftment upon transplantation into mice, a finding that should have a profound impact on the generation of robust humanized small animal models. 22641236_CD47 expression is a novel prognostic marker in esophageal squamous cell carcinoma that is directly inhibited by the miR-133a tumor suppressor. 22734047_Data show that CD47 is expressed in normal myelin and in foamy macrophages and reactive astrocytes within active multiple sclerosis (MS)lesions. 22815286_Both human endothelial and leukocyte CD47 are required for T cell transendothelial migration at sites of inflammation. 22870271_CD47(low) status on CD4 effectors is necessary for the contraction/resolution of the immune response in humans and mice. 22874555_CD47 deficiency confers cell survival through the activation of autophagic flux and identifies CD47 blockade as a pharmacological route to modulate autophagy for protecting tissue from radiation injury. 22990013_Engulfment of hematopoietic stem cells caused by down-regulation of CD47 is critical in the pathogenesis of hemophagocytic lymphohistiocytosis. 23011546_The eSNP (single nucleotide polymorphism associated with expression) rs12695175 in CD47 associated with CRC specific survival (HR = 2.18, 95 % CI 1.10-4.33, CC versus AA) and with overall survival. 23206327_This study analysis revealed up-regulation of CD14, CD18, CD45, TLR2, TLR4, CD74, C1q, C3, CCL2, and down-regulation of CD36, CX3CR1, CX3CL1 and CD200 in postmenopausal women. 23275336_Inhibitor of apoptosis proteins (IAPs) and their antagonists regulate spontaneous and tumor necrosis factor (TNF)-induced proinflammatory cytokine and chemokine production 23642736_Data indicate that the level of CD47 on erythroblast of myelodysplastic syndrome (MDS) patients had a significant positive correlation with their peripheral RBC count. 23669395_An avidity-improved CD47 fusion protein (CD47-Var1) suppresses the release of a wide array of inflammatory cytokines by CD172a(+) cells. 23690610_Anti-CD47 antibody-mediated phagocytosis of cancer cells by macrophages results in increased priming of transgenic CD8+ OT-I T cells but decreased priming of CD4+ OT-II T cells. 23774794_Findings indicate CD47 as a potential prognostic marker for melanoma development as well as a target for therapeutic intervention with RNAi-based nanomedicines. 24006483_Data indicate that CD47 in cis interactions regulate LFA-1 (integrin alphaLbeta2) and VLA-4 (integrin alpha4beta1) integrin affinity, and this process plays a substantial role in the adhesion of T-cells. 24143245_'clustering' SIRPalpha into plasma membrane microdomains is essential for activated monocytes and macrophages to effectively interact with CD47 and initiate intracellular signaling 24269920_Studies suggest that the mouse strain effect on xenogeneic engraftment might be ascribed mainly to the binding affinity of strain-specific polymorphic SIRPA with human CD47. 24523067_Suppression of CD47 by morpholino approach suppressed growth of hepatocellular carcinoma in vivo and exerted a chemosensitization effect through blockade of CTSS/PAR2 signaling. 24550402_Polymorphisms in the human inhibitory signal-regulatory protein alpha do not affect binding to its ligand CD47. 24631171_CD24/CD44/CD47 are differentially expressed in cells of urothelial BCa in patients undergoing RC and influence cancer-specific survival of patients 24726056_CD47 plays the pivotal role in the immune evasion of primary effusion lymphoma cells in body cavities. Therapeutic antibody targeting of CD47 could be an effective therapy for PEL. 24840181_Staphylococcal SElX and SSL6 proteins bind cell surface receptors PSGL-1 and CD47, respectively. 24848268_the TSP-derived 4N1K peptide effects on cell adhesion and integrin activation are independent of CD47 24887393_The thrombospondin-1 receptor CD47 directly or indirectly regulates intercellular communication mediated by the transfer of extracellular vesicles between vascular cells. 24911792_expression is induced following EBV infection of B cells; ligation cases G1 arres 25001859_Elevated postinjury thrombospondin 1-CD47 triggering aids differentiation of patients' defective inflammatory CD1a+dendritic cells. 25200950_CD47 associates with and regulates VEGFR2 in T cells. CD47 signaling modulates the ability of VEGF to regulate proliferation and TCR signaling, and autocrine production of vascular endothelial growth factor by T cells contributes to this regulation. 25230070_Report poor prognosis/survival in luminal breast cancer patients with circulating tumor cells co-expressing MET and CD47. 25550546_CD47 regulates the epigenetic code by targeting UHRF1. 25593301_study demonstrates that antigen-presenting cells(CD47+) in coculture with human macrophages show a CD47 concentration-dependent inhibition of phagocytosis 25658794_Expression of CD44, CD47 and c-met was upregulated in OCCC and pairwise correlation. CD44, CD47 and c-met may have synergistic effects on the development of OCCC and are prognostic factors for ovarian cancer. 25717063_CD47 is highly expressed on cancer stem cells, but not on other nonmalignant cells in the pancreas. 25747479_CD47 has a central role in hydrogen sulfate biosynthesis, regulation and signaling in T cells. 25837251_Velcro' engineering of high affinity CD47 ectodomain as signal regulatory protein alpha (SIRPalpha) antagonists that enhance antibody-dependent cellular phagocytosis 25896326_during translation of CD47, the scaffold function of the 3' UTR facilitates binding of proteins to nascent proteins, to direct their transport or function--and this role of 3' UTRs can be regulated by polyadenylation 25914268_CD47 expression is decreased on the surface of erythrocytes in obese subjects. These changes in CD47 expression on the erythrocytes surface may be an adaptive response to hyperfibrinogenemia associated with obesity. 25943033_While CD47 expression on circulating AML blasts has been shown to be a negative prognostic marker for a very defined population of AML patients with NK AML, CD47 expression on AML BM blasts is not 26077800_CD47 is an adverse prognostic factor and promising therapeutic target in gastric cancer. 26085683_Loss of cell surface CD47 clustering formation and binding avidity to SIRPalpha facilitate apoptotic cell clearance by macrophages. 26093091_CD47 is a critical regulator in the metastasis of osteosarcoma and results suggest that targeted inhibition of this antigen by anti-CD47 may be a novel immunotherapeutic approach in the management of this tumor. 26116271_agents that block the CD47:SIRP-alpha engagement are attractive therapeutic targets as a monotherapy or in combination with additional immune-modulating agents for activating antitumor T cells in vivo 26311851_CD47 mediates signaling from the extracellular matrix that coordinately regulates basal metabolism and cytoprotective responses to radiation injury 26446381_CD47 is a promising cancer biomarker, and targeting CD47 presents an effective and potential therapeutic strategy through synthesized mechanisms 26512116_CD47 expression contributes to the lethal breast cancer phenotype that is mediated by HIF-1. 26573233_Data suggest a reduction in the CD47 antigen/signal-regulatory protein alpha (SIRPalpha) pathway by programmed cell death protein 1 (PD-1) blockade, which regulates Myeloid-derived suppressor cells (MDSCs) and tumor associated macrophages (TAMs). 26840086_Data show that anti-human CD47 antibody B6H12 decreased expression of epidermal growth factor receptor (EGFR) and the stem cell transcription factors. 27091975_Our results highlight an underappreciated contribution of the adaptive immune system to anti-CD47 adjuvant therapy and suggest that targeting both innate and adaptive immune checkpoints can potentiate the vaccinal effect of antitumor antibody therapy. 27095555_suggest that microglial activation may be partially caused by CD47/signal regulatory protein-alpha- and CD200/CD200R-mediated reductions in the immune inhibitory pathways 27232934_Prolongation of transient porcine chimerism via transgenic expression of human CD47 in a primate model is associated with an immune modulating effect, leading to markedly prolonged survival of donor swine skin xenografts. 27259369_thrombospondin-1 via CD47 inhibits renal tubular epithelial cell recovery after ischemia reperfusion injury through inhibition of proliferation/self-renewal. 27322955_Results show that the stronger expression of CD47 by cancer cells and larger number of total macrophages/M2 were independently related to shorter survivals. 27349907_the results obtained by combining bioinformatics and preclinical studies strongly suggest that targeting TSP-1/CD47 axis may represent a valuable therapeutic alternative for hampering melanoma spreading. 27411490_CD47 is overexpressed in primary non-small cell lung cancer (NSCLC) tissues and cell lines, suggesting that CD47 is a promising therapeutic target for NSCLC. 27437576_atherogenesis is associated with upregulation of CD47, a key anti-phagocytic molecule that is known to render malignant cells resistant to programmed cell removal, or 'efferocytosis' 27714815_CD47 seems to mediate fusion mostly through broad contact surfaces between the partners' cell membrane while syncytin-1 mediate fusion through phagocytic-cup like structure. 27742621_In pulmonary hypertension TSP1-CD47 is upregulated, and contributes to pulmonary arterial vasculopathy and dysfunction. 27855370_The results suggested a critical role of CD47 in laryngeal squamous cell carcinoma development. 27856600_TTI-621 (SIRPalphaFc) is a fully human recombinant fusion protein that blocks the CD47-SIRPalpha axis by binding to human CD47 and enhancing phagocytosis of malignant cells..These data indicate that TTI-621 is active across a broad range of human tumors. 27863402_Blocking CD47 using antibodies could efficiently induce macrophage-mediated phagocytosis of tumor cells and treat cancers. 28076333_Results indicated that surgical resection combined with anti-CD47 blocking immunotherapy promoted an inflammatory response and prolonged survival in xenograft animal model, and is therefore an attractive strategy for clinical translation. 28077173_this review highlights the various functions of CD47, discusses anti-tumor responses generated by both the innate and adaptive immune systems as a consequence of administering anti-CD47 blocking antibody, and finally elaborates on the clinical potential of CD47 blockade. 28100392_we observed neutrophilic infiltration was slightly increased in anti-CD47 treated tumors compared to IgG control. 28378740_results suggest that cancers can evolve SE to drive CD47 overexpression to escape immune surveillance 28393401_Among the various candidate genes involved in acute rejection, CD47 inhibits monocyte/macrophage-mediated phagocytosis by identifying the CD47 signal regulatory protein alpha (SIRP-alpha) as self/non-self. Tissue factor pathway inhibitor (TFPI) is involved in the regulation of the coagulation pathway and is able to bind to another ligand of CD47, thrombospondin-1 (TSP-1). 28522599_Engineering macrophages to eat cancer: from 'marker of self' CD47 and phagocytosis to differentiation 28801364_High CD47 expression is not associated with Fibrolamellar Hepatocellular Carcinoma. 28968355_overexpression of human CD200 in donor pigs could constitute a promising strategy for overcoming xenograft rejection. 29042481_Results show that the thrombospondin 1 (TSP1) and its receptor CD47 (CD47) axis selectively regulates NADPH oxidase 1 (Nox1) in the regulation of endothelial senescence and suggest potential targets for controlling the aging process at the molecular level. 29321087_These results suggest that CD47 might be a useful predictor of poor prognosis and metastasis and a potential target for treating glioblastomas. 29367423_High CD47 expression is associated with the increase in the ability of surviving breast cancer cells to evade innate and adaptive immunity. 29748606_CD47-ligation induced cell death in T-acute lymphoblastic leukemia. 30056566_Therefore, the differential responses observed with CD47 blockade are due to autonomous activation of protective autophagy in normal tissue and enhancement immune cytotoxicity against cancer cells. 30466764_PD1/PDL1 and CD47 may be involved in the disease progression and prognosis of T-lymphoblastic lymphoma/leukemia 30525058_this study shows that CD47 blockade inhibits tumor progression through promoting phagocytosis of tumor Cells by M2 polarized macrophages in endometrial cancer 30710089_cancer cell expression of SLAMF7 is not required for phagocytosis and, in contrast to CD47 expression, should not be used as selection criterion for CD47-targeted therapy. 30713045_Decreased CD47 expression along the storage period was statistically significant in both groups, group 1 (unfiltered packed red cell units), and group 2 (filtered 'leuco-reduced' red cell units). The expression of CD47 was significantly higher in group 2 than group 1 on day zero, 1st and 21st days, and this correlated with 2,3-DPG decreases on day 21 of group 1 but not group 2. 30741466_The stemness index determined by the expression of CD47 and CD133 is a promising prognostic predictor, and CD47 is a potential therapeutic target for CSCs in ESCC patients. 30833751_QPCTL is critical for pyroglutamate formation on CD47 at the SIRPalpha binding site shortly after biosynthesis. 30878596_CD47 and PD-L1 serve as critical innate and adaptive checkpoints, respectively, both of them are critical to the immune system. Therefore, we believe this dual-targeting strategy will provide insight into tumor immunotherapy for better tumor control. 30938231_Targeting CD47 in Anaplastic Thyroid Carcinoma Enhances Tumor Phagocytosis by Macrophages and Is a Promising Therapeutic Strategy. 31025548_Syncytin 1, CD9, and CD47 regulating cell fusion to form PGCCs associated with cAMP/PKA and JNK signaling pathway. 31089204_Transmembrane protein CD47 is highly expressed on many types of cancer cells and can directly bind to the receptor signal regulatory protein alpha (SIRPalpha), which is highly expressed on phagocytic cells. 31132784_CD47 promotes the malignancy of colorectal cancer in association with EMT and enhances the stemness of cancer cells. All recurrent tumors highly expressed CD47 and CD44 and showed the epithelial-mesenchymal transition phenotype. 31359339_Expression of CD47 antigen in Reed-Sternberg cells as a new potential biomarker for classical Hodgkin lymphoma. 31409608_CD47 Expression Defines Efficacy of Rituximab with CHOP in Non-Germinal Center B-cell (Non-GCB) Diffuse Large B-cell Lymphoma Patients (DLBCL), but Not in GCB DLBCL. 31493748_CD47 in endometriosis-associated ovarian cancer may be a useful surface marker. 31495971_Transgenic expression of human CD47 reduces phagocytosis of porcine endothelial cells and podocytes by baboon and human macrophages. 31522278_PD-L1 expression correlated with CD47 expression. 31534047_Dysregulated integrin alphaVbeta3 and CD47 signaling promotes joint inflammation, cartilage breakdown, and progression of osteoarthritis. 31712778_Thrombospondin-1 promotes tumor progression in cutaneous T-cell lymphoma via CD47. 31861233_CD47-SIRPalpha Signaling Induces Epithelial-Mesenchymal Transition and Cancer Stemness and Links to a Poor Prognosis in Patients with Oral Squamous Cell Carcinoma. 32043750_Association between CD47 expression, clinical characteristics and prognosis in patients with advanced non-small cell lung cancer. 32064335_Intratumoral delivery of CCL25 enhances immunotherapy against triple-negative breast cancer by recruiting CCR9(+) T cells. 32082304_Enhanced Expression of CD47 Is Associated With Off-Target Resistance to Tyrosine Kinase Inhibitor Gefitinib in NSCLC. 32149101_MicroRNA-200a Promotes Phagocytosis of Macrophages and Suppresses Cell Proliferation, Migration, and Invasion in Nasopharyngeal Carcinoma by Targeting CD47. 32196807_TSP1-CD47-SIRPalpha signaling facilitates the development of endometriosis by mediating the survival of ectopic endometrium. 32226539_Tumor-intrinsic CD47 signal regulates glycolysis and promotes colorectal cancer cell growth and metastasis. 32240171_We also demonstrate the utility of the assay to characterize the activity of the first reported small molecule antagonists of the SIRPa-CD47 interaction. 32296041_The identification of a CD47-blocking ''hotspot'' and design of a CD47/PD-L1 dual-specific antibody with limited hemagglutination. 32433947_The CD47-SIRPalpha Immune Checkpoint. 32576628_Clinicopathological significance of CD47 expression in hepatocellular carcinoma. 32576895_CD47 is a negative regulator of intestinal epithelial cell self-renewal following DSS-induced experimental colitis. 32677994_Role of CD47 in Hematological Malignancies. 32748759_Targeting CD47 Inhibits Tumor Development and Increases Phagocytosis in Oral Squamous Cell Carcinoma. 32814713_CD47 prevents the elimination of diseased fibroblasts in scleroderma. 32830120_Positive tumour CD47 expression is an independent prognostic factor for recurrence in resected non-small cell lung cancer. 32923164_Immune vulnerability of ovarian cancer stem-like cells due to low CD47 expression is protected by surrounding bulk tumor cells. 32929084_Dual blockade of CD47 and HER2 eliminates radioresistant breast cancer cells. 33084148_CD24, not CD47, negatively impacts upon response to PD-1/L1 inhibitors in non-small-cell lung cancer with PD-L1 tumor proportion score < 50. 33389016_CD47 expression in gastric cancer clinical correlates and association with macrophage infiltration. 33406263_CD47 promotes T-cell lymphoma metastasis by up-regulating AKAP13-mediated RhoA activation. 33429083_A bispecific antibody targeting GPC3 and CD47 induced enhanced antitumor efficacy against dual antigen-expressing HCC. 33582561_METTL3/IGF2BP1/CD47 contributes to the sublethal heat treatment induced mesenchymal transition in HCC. 33765961_CD47 expression and CD163(+) macrophages correlated with prognosis of pancreatic neuroendocrine tumor. 33799989_Expression and Prognostic Significance of CD47-SIRPA Macrophage Checkpoint Molecules in Colorectal Cancer. 33815356_Immune Checkpoint-Associated Locations of Diffuse Gliomas Comparing Pediatric With Adult Patients Based on Voxel-Wise Analysis. 33833053_Overexpression of CD47 is associated with brain overgrowth and 16p11.2 deletion syndrome. 33836556_CD47 blockade reduces the pathologic features of experimental cerebral malaria and promotes survival of hosts with Plasmodium infection. 33917174_Transcriptomic Profiles of CD47 in Breast Tumors Predict Outcome and Are Associated with Immune Activation. 33920030_Thrombospondin-1 CD47 Signalling: From Mechanisms to Medicine. 33979899_PD-L1 and CD47 co-expression predicts survival and enlightens future dual-targeting immunotherapy in non-small cell lung cancer. 33999416_Anti-CD47 antibody synergizes with cisplatin against laryngeal cancer by enhancing phagocytic ability of macrophages. 34016744_Exosomal CD47 Plays an Essential Role in Immune Evasion in Ovarian Cancer. 34068752_CD47 Potentiates Inflammatory Response in Systemic Lupus Erythematosus. 34195295_Clinicopathological and Prognostic Significance of CD47 Expression in Lung Neuroendocrine Tumors. 34203368_CD47 in the Brain and Neurodegeneration: An Update on the Role in Neuroinflammatory Pathways. 34425695_HIV-1 Vpu Promotes Phagocytosis of Infected CD4(+) T Cells by Macrophages through Downregulation of CD47. 34454175_Absence of CD47 maintains brown fat thermogenic capacity and protects mice from aging-related obesity and metabolic disorder. 34471125_Structure of the human marker of self 5-transmembrane receptor CD47. 34512146_CD47/SIRPalpha pathway mediates cancer immune escape and immunotherapy. 34559187_CD47 blockade enhances the efficacy of intratumoral STING-targeting therapy by activating phagocytes. 34654058_Expression of CD47 in the pancreatic beta-cells of people with recent-onset type 1 diabetes varies according to disease endotype. 34670667_[Overexpression of CD47 inhibits apoptosis of SW480 human colon cancer cells by blocking Fas/FasL pathway]. 34846059_Less phagocytosis of viral vectors by tethering with CD47 ectodomain. 34858730_Cancer cells under immune attack acquire CD47-mediated adaptive immune resistance independent of the myeloid CD47-SIRPalpha axis. 34966389_Significance of CD47 and Its Association With Tumor Immune Microenvironment Heterogeneity in Ovarian Cancer. 34998671_Immune inactivation by CD47 expression predicts clinical outcomes and therapeutic responses in clear cell renal cell carcinoma patients. 35048182_ZBTB28 inhibits breast cancer by activating IFNAR and dual blocking CD24 and CD47 to enhance macrophages phagocytosis. 35084991_Clonal Expansion of Stem/Progenitor Cells in Cancer, Fibrotic Diseases, and Atherosclerosis, and CD47 Protection of Pathogenic Cells. 35317832_SIRPalpha and PD1 expression on tumor-associated macrophage predict prognosis of intrahepatic cholangiocarcinoma. | ENSMUSG00000055447 | Cd47 | 8.299679e+02 | 0.6448600 | -0.632942143 | 0.2846460 | 4.922940e+00 | 0.0265023808 | 0.55176060 | No | Yes | 5.717559e+02 | 99.165196 | 9.777400e+02 | 173.150515 | |
| ENSG00000196792 | 29966 | STRN3 | protein_coding | Q13033 | FUNCTION: Binds calmodulin in a calcium dependent manner. May function as scaffolding or signaling protein. | 3D-structure;Acetylation;Alternative splicing;Calmodulin-binding;Coiled coil;Cytoplasm;Membrane;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:29966; | cytoplasm [GO:0005737]; dendrite [GO:0030425]; FAR/SIN/STRIPAK complex [GO:0090443]; Golgi apparatus [GO:0005794]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; armadillo repeat domain binding [GO:0070016]; calmodulin binding [GO:0005516]; protein phosphatase 2A binding [GO:0051721]; protein-containing complex binding [GO:0044877]; small GTPase binding [GO:0031267]; negative regulation of intracellular estrogen receptor signaling pathway [GO:0033147]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of transcription by RNA polymerase II [GO:0045944]; response to estradiol [GO:0032355] | 23678272_STRN3 (rs2273171)is associated with single nucleotide polymorphisms in Korean patients, either non-segmental or segmental type. 24550388_This result combined with a number of biophysical analyses provide evidence that the coiled coil domain of striatin 3 and the PP2A A subunit form a stable core complex with a 2:2 stoichiometry 25015106_The structure and protein-binding domains of the SG2NA protein variants have been described. 25531779_this study propose that the STRIPAK complex, FAM40A, FAM40B and STRN3, regulates the mode of cancer cell migration by controlling the activity of MST3 and 4, which locally coordinate the phosphorylation of ERM proteins and inhibit the dephosphorylation of MLC leading to increased actin-membrane linkage. 26022125_results demonstrate that XBP1 mRNA splicing plays an important role in maintaining the function of bone marrow-derived macrophages and provide new insight into the study and treatment of atherosclerosis 30520818_we detected novel and related STRN-NTRK3 and STRN3-NTRK3 fusions in 2 fibrosarcomas that occurred in the bone and soft tissue of young adult patients 32589942_Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer. | ENSMUSG00000020954 | Strn3 | 2.819685e+02 | 1.0103144 | 0.014804296 | 0.3628089 | 1.620646e-03 | 0.9678880394 | 0.99408977 | No | Yes | 2.979408e+02 | 58.196724 | 2.501594e+02 | 50.230099 | ||
| ENSG00000196793 | 8187 | ZNF239 | protein_coding | Q16600 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | MOK2 proteins are DNA- and RNA-binding proteins that are mainly associated with nuclear RNP components, including the nucleoli and extranucleolar structures (Arranz et al., 1997 [PubMed 9121460]).[supplied by OMIM, Mar 2008]. | hsa:8187; | nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 12409453_In this study, we identify a novel interaction between lamin A/C and hsMOK2 by using the yeast two-hybrid system 16385451_Observational study of gene-disease association. (HuGE Navigator) 17760566_Results indicate that pathogenic mutations in lamin A/C lead to sequestration of hsMOK2 into nuclear aggregates, which may deregulate MOK2 target genes. 19490114_Data identified Ser38 and Ser129 of hsMOK2 as phosphorylation sites of JNK3 kinase, and Ser46 as a phosphorylation site of Aurora A and protein kinase A. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20641033_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) | ENSMUSG00000042097 | Zfp239 | 1.177048e+02 | 0.8419862 | -0.248131582 | 0.3960321 | 3.991298e-01 | 0.5275390222 | 0.88668779 | No | Yes | 1.002725e+02 | 20.148718 | 1.250095e+02 | 25.482262 | |
| ENSG00000197008 | 7697 | ZNF138 | protein_coding | P52744 | FUNCTION: May be involved in transcriptional regulation as a repressor. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7697; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 1.775867e+02 | 1.0811230 | 0.112530670 | 0.3417267 | 1.041957e-01 | 0.7468515204 | 0.94694596 | No | Yes | 1.929085e+02 | 25.600469 | 1.724988e+02 | 23.044741 | |||||
| ENSG00000197050 | 147923 | ZNF420 | protein_coding | Q8TAQ5 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a KRAB-type zinc finger protein that negatively-regulates p53-mediated apoptosis. Under stress conditions, the encoded protein is phosphorylated by ATM and dissociates from p53, which activates p53 and initiates apoptosis. [provided by RefSeq, Jul 2016]. | hsa:147923; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 19377469_Apak (ZNF420 or FLJ32191) is a negative regulator of p53-mediated apoptosis. 19629643_four of seven types of DNA damage signals examined resulted in significant Apak phosphorylation and dissociation of Apak from p53, releasing the inhibition of p53 transcriptional activity 20713054_Apak is differentially regulated in the ARF and ATM-dependent manner in response to oncogenic stress and DNA damage, respectively. 22334068_Apak competes with p53 for binding to inhibit p53AIP1 expression. 25691462_ARF coordinates Apak to regulate ribosome biogenesis upon cellular stress. 25961932_Functional importance of H3K9me3 in hypoxia, apoptosis and repression of APAK. | ENSMUSG00000063047 | Zfp780b | 8.553024e+01 | 0.6836577 | -0.548654019 | 0.4543661 | 1.363095e+00 | 0.2430017551 | 0.78892886 | No | Yes | 6.245959e+01 | 14.079031 | 8.201712e+01 | 18.903755 | |
| ENSG00000197122 | 6714 | SRC | protein_coding | P12931 | FUNCTION: Non-receptor protein tyrosine kinase which is activated following engagement of many different classes of cellular receptors including immune response receptors, integrins and other adhesion receptors, receptor protein tyrosine kinases, G protein-coupled receptors as well as cytokine receptors. Participates in signaling pathways that control a diverse spectrum of biological activities including gene transcription, immune response, cell adhesion, cell cycle progression, apoptosis, migration, and transformation. Due to functional redundancy between members of the SRC kinase family, identification of the specific role of each SRC kinase is very difficult. SRC appears to be one of the primary kinases activated following engagement of receptors and plays a role in the activation of other protein tyrosine kinase (PTK) families. Receptor clustering or dimerization leads to recruitment of SRC to the receptor complexes where it phosphorylates the tyrosine residues within the receptor cytoplasmic domains. Plays an important role in the regulation of cytoskeletal organization through phosphorylation of specific substrates such as AFAP1. Phosphorylation of AFAP1 allows the SRC SH2 domain to bind AFAP1 and to localize to actin filaments. Cytoskeletal reorganization is also controlled through the phosphorylation of cortactin (CTTN) (Probable). When cells adhere via focal adhesions to the extracellular matrix, signals are transmitted by integrins into the cell resulting in tyrosine phosphorylation of a number of focal adhesion proteins, including PTK2/FAK1 and paxillin (PXN) (PubMed:21411625). In addition to phosphorylating focal adhesion proteins, SRC is also active at the sites of cell-cell contact adherens junctions and phosphorylates substrates such as beta-catenin (CTNNB1), delta-catenin (CTNND1), and plakoglobin (JUP). Another type of cell-cell junction, the gap junction, is also a target for SRC, which phosphorylates connexin-43 (GJA1). SRC is implicated in regulation of pre-mRNA-processing and phosphorylates RNA-binding proteins such as KHDRBS1 (Probable). Also plays a role in PDGF-mediated tyrosine phosphorylation of both STAT1 and STAT3, leading to increased DNA binding activity of these transcription factors (By similarity). Involved in the RAS pathway through phosphorylation of RASA1 and RASGRF1 (PubMed:11389730). Plays a role in EGF-mediated calcium-activated chloride channel activation (PubMed:18586953). Required for epidermal growth factor receptor (EGFR) internalization through phosphorylation of clathrin heavy chain (CLTC and CLTCL1) at 'Tyr-1477'. Involved in beta-arrestin (ARRB1 and ARRB2) desensitization through phosphorylation and activation of GRK2, leading to beta-arrestin phosphorylation and internalization. Has a critical role in the stimulation of the CDK20/MAPK3 mitogen-activated protein kinase cascade by epidermal growth factor (Probable). Might be involved not only in mediating the transduction of mitogenic signals at the level of the plasma membrane but also in controlling progression through the cell cycle via interaction with regulatory proteins in the nucleus (PubMed:7853507). Plays an important role in osteoclastic bone resorption in conjunction with PTK2B/PYK2. Both the formation of a SRC-PTK2B/PYK2 complex and SRC kinase activity are necessary for this function. Recruited to activated integrins by PTK2B/PYK2, thereby phosphorylating CBL, which in turn induces the activation and recruitment of phosphatidylinositol 3-kinase to the cell membrane in a signaling pathway that is critical for osteoclast function (PubMed:8755529, PubMed:14585963). Promotes energy production in osteoclasts by activating mitochondrial cytochrome C oxidase (PubMed:12615910). Phosphorylates DDR2 on tyrosine residues, thereby promoting its subsequent autophosphorylation (PubMed:16186108). Phosphorylates RUNX3 and COX2 on tyrosine residues, TNK2 on 'Tyr-284' and CBL on 'Tyr-731' (PubMed:20100835, PubMed:21309750). Enhances DDX58/RIG-I-elicited antiviral signaling (PubMed:19419966). Phosphorylates PDPK1 at 'Tyr-9', 'Tyr-373' and 'Tyr-376' (PubMed:14585963). Phosphorylates BCAR1 at 'Tyr-128' (PubMed:22710723). Phosphorylates CBLC at multiple tyrosine residues, phosphorylation at 'Tyr-341' activates CBLC E3 activity (PubMed:20525694). Involved in anchorage-independent cell growth (PubMed:19307596). Required for podosome formation (By similarity). Mediates IL6 signaling by activating YAP1-NOTCH pathway to induce inflammation-induced epithelial regeneration (PubMed:25731159). {ECO:0000250|UniProtKB:P05480, ECO:0000269|PubMed:11389730, ECO:0000269|PubMed:12615910, ECO:0000269|PubMed:14585963, ECO:0000269|PubMed:16186108, ECO:0000269|PubMed:18586953, ECO:0000269|PubMed:19307596, ECO:0000269|PubMed:19419966, ECO:0000269|PubMed:20100835, ECO:0000269|PubMed:20525694, ECO:0000269|PubMed:21309750, ECO:0000269|PubMed:21411625, ECO:0000269|PubMed:22710723, ECO:0000269|PubMed:25731159, ECO:0000269|PubMed:7853507, ECO:0000269|PubMed:8755529, ECO:0000269|PubMed:8759729, ECO:0000305|PubMed:11964124, ECO:0000305|PubMed:8672527, ECO:0000305|PubMed:9442882}. | 3D-structure;ATP-binding;Alternative splicing;Cell adhesion;Cell cycle;Cell junction;Cell membrane;Cytoplasm;Cytoskeleton;Disease variant;Host-virus interaction;Immunity;Kinase;Lipoprotein;Membrane;Mitochondrion;Mitochondrion inner membrane;Myristate;Nucleotide-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;SH2 domain;SH3 domain;Transferase;Tyrosine-protein kinase;Ubl conjugation | This gene is highly similar to the v-src gene of Rous sarcoma virus. This proto-oncogene may play a role in the regulation of embryonic development and cell growth. The protein encoded by this gene is a tyrosine-protein kinase whose activity can be inhibited by phosphorylation by c-SRC kinase. Mutations in this gene could be involved in the malignant progression of colon cancer. Two transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:6714; | actin filament [GO:0005884]; caveola [GO:0005901]; cell junction [GO:0030054]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; focal adhesion [GO:0005925]; glutamatergic synapse [GO:0098978]; late endosome [GO:0005770]; lysosome [GO:0005764]; membrane raft [GO:0045121]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; neuron projection [GO:0043005]; nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; podosome [GO:0002102]; postsynaptic density [GO:0014069]; postsynaptic specialization, intracellular component [GO:0099091]; ruffle membrane [GO:0032587]; ATP binding [GO:0005524]; ATPase binding [GO:0051117]; BMP receptor binding [GO:0070700]; cadherin binding [GO:0045296]; connexin binding [GO:0071253]; enzyme binding [GO:0019899]; ephrin receptor binding [GO:0046875]; estrogen receptor binding [GO:0030331]; growth factor receptor binding [GO:0070851]; heme binding [GO:0020037]; insulin receptor binding [GO:0005158]; integrin binding [GO:0005178]; kinase binding [GO:0019900]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; phospholipase activator activity [GO:0016004]; phospholipase binding [GO:0043274]; phosphoprotein binding [GO:0051219]; protein C-terminus binding [GO:0008022]; protein kinase activity [GO:0004672]; protein kinase C binding [GO:0005080]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; scaffold protein binding [GO:0097110]; SH2 domain binding [GO:0042169]; signaling receptor binding [GO:0005102]; transmembrane transporter binding [GO:0044325]; activation of protein kinase B activity [GO:0032148]; adherens junction organization [GO:0034332]; angiotensin-activated signaling pathway involved in heart process [GO:0086098]; bone resorption [GO:0045453]; branching involved in mammary gland duct morphogenesis [GO:0060444]; cell adhesion [GO:0007155]; cell cycle [GO:0007049]; cell differentiation [GO:0030154]; cell population proliferation [GO:0008283]; cell-cell adhesion [GO:0098609]; cellular response to fatty acid [GO:0071398]; cellular response to fluid shear stress [GO:0071498]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to hypoxia [GO:0071456]; cellular response to insulin stimulus [GO:0032869]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to peptide hormone stimulus [GO:0071375]; cellular response to platelet-derived growth factor stimulus [GO:0036120]; cellular response to progesterone stimulus [GO:0071393]; entry of bacterium into host cell [GO:0035635]; ephrin receptor signaling pathway [GO:0048013]; epidermal growth factor receptor signaling pathway [GO:0007173]; ERBB2 signaling pathway [GO:0038128]; Fc-gamma receptor signaling pathway involved in phagocytosis [GO:0038096]; focal adhesion assembly [GO:0048041]; forebrain development [GO:0030900]; innate immune response [GO:0045087]; integrin-mediated signaling pathway [GO:0007229]; interleukin-6-mediated signaling pathway [GO:0070102]; intestinal epithelial cell development [GO:0060576]; intracellular signal transduction [GO:0035556]; leukocyte migration [GO:0050900]; macroautophagy [GO:0016236]; negative regulation of anoikis [GO:2000811]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of focal adhesion assembly [GO:0051895]; negative regulation of inflammatory response to antigenic stimulus [GO:0002862]; negative regulation of intrinsic apoptotic signaling pathway [GO:2001243]; negative regulation of mitochondrial depolarization [GO:0051902]; negative regulation of protein-containing complex assembly [GO:0031333]; negative regulation of telomerase activity [GO:0051974]; negative regulation of telomere maintenance via telomerase [GO:0032211]; negative regulation of transcription, DNA-templated [GO:0045892]; neurotrophin TRK receptor signaling pathway [GO:0048011]; odontogenesis [GO:0042476]; oogenesis [GO:0048477]; osteoclast development [GO:0036035]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-tyrosine autophosphorylation [GO:0038083]; peptidyl-tyrosine phosphorylation [GO:0018108]; platelet activation [GO:0030168]; positive regulation of apoptotic process [GO:0043065]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0045737]; positive regulation of cytokine production [GO:0001819]; positive regulation of dephosphorylation [GO:0035306]; positive regulation of DNA biosynthetic process [GO:2000573]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of glucose metabolic process [GO:0010907]; positive regulation of insulin receptor signaling pathway [GO:0046628]; positive regulation of integrin activation [GO:0033625]; positive regulation of lamellipodium morphogenesis [GO:2000394]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of non-membrane spanning protein tyrosine kinase activity [GO:1903997]; positive regulation of Notch signaling pathway [GO:0045747]; positive regulation of ovarian follicle development [GO:2000386]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of phosphatidylinositol 3-kinase activity [GO:0043552]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of platelet-derived growth factor receptor-beta signaling pathway [GO:2000588]; positive regulation of podosome assembly [GO:0071803]; positive regulation of protein autophosphorylation [GO:0031954]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of protein localization to nucleus [GO:1900182]; positive regulation of protein processing [GO:0010954]; positive regulation of protein serine/threonine kinase activity [GO:0071902]; positive regulation of protein transport [GO:0051222]; positive regulation of small GTPase mediated signal transduction [GO:0051057]; positive regulation of smooth muscle cell migration [GO:0014911]; positive regulation of transcription, DNA-templated [GO:0045893]; primary ovarian follicle growth [GO:0001545]; progesterone receptor signaling pathway [GO:0050847]; protein autophosphorylation [GO:0046777]; protein destabilization [GO:0031648]; regulation of bone resorption [GO:0045124]; regulation of caveolin-mediated endocytosis [GO:2001286]; regulation of cell projection assembly [GO:0060491]; regulation of cell-cell adhesion [GO:0022407]; regulation of early endosome to late endosome transport [GO:2000641]; regulation of epithelial cell migration [GO:0010632]; regulation of intracellular estrogen receptor signaling pathway [GO:0033146]; regulation of postsynaptic neurotransmitter receptor activity [GO:0098962]; regulation of protein binding [GO:0043393]; regulation of vascular permeability [GO:0043114]; response to acidic pH [GO:0010447]; response to electrical stimulus [GO:0051602]; response to interleukin-1 [GO:0070555]; response to mechanical stimulus [GO:0009612]; response to mineralocorticoid [GO:0051385]; response to nutrient levels [GO:0031667]; response to virus [GO:0009615]; response to xenobiotic stimulus [GO:0009410]; signal complex assembly [GO:0007172]; signal transduction [GO:0007165]; stimulatory C-type lectin receptor signaling pathway [GO:0002223]; stress fiber assembly [GO:0043149]; substrate adhesion-dependent cell spreading [GO:0034446]; T cell costimulation [GO:0031295]; transcytosis [GO:0045056]; transforming growth factor beta receptor signaling pathway [GO:0007179]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; uterus development [GO:0060065]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11940607_Coordinate interactions of Csk, Src, and Syk kinases with [alpha]IIb[beta]3 initiate integrin signaling to the cytoskeleton. 11994743_v-Src's hold over actin and cell adhesions. 12014986_PLD1 is threonine-phosphorylated in human-airway epithelial cells by a PKCdelta and src dependent mechanism 12060669_The Src-cortactin pathway is required for clustering of E-selectin and ICAM-1 in endothelial cells 12063167_Activation of a Src-dependent Raf-MEK1/2-ERK signaling pathway is required for IL-1alpha-induced upregulation of beta-defensin 2 in human middle ear epithelial cells. 12162444_REVIEW: Arsenic carcinogenicity: relevance of c-Src activation 12216109_Functional involvement of src and focal adhesion kinase in a CD99 splice variant-induced motility of human breast cancer cells 12393736_The p85 subunit of PI 3-kinase activates Src independently of the enzymatic activity of PI 3- kinase. Ligand binding of GPIb, without receptor clustering, is sufficient to activate Src. 12416028_pp60c-src may be involved in stabilization of dynamic HT-29 cell adhesion to ECM components, and this kinase appears to be part of a mechanosensory protein complex during integrin-mediated cell adhesion 12420216_Src activation may contribute to colon tumor progression and metastasis in part by activating Akt-mediated survival pathways that decrease sensitivity of detached cells to anoikis. 12429742_mediation of synergism with epidermal growth factor receptor by STAT5b 12429837_Data report a novel positioning of Src, mediating signals from insulin to phosphatidylinositol 3-kinase and to beta(2)-adrenergic receptor trafficking. 12456372_Mutual interaction of c-Src with EGFR is required for many EGFR-mediated cellular functions including proliferation, migration, survival and EGFR endocytosis, as discussed in this review. 12462387_c-Src activation is associated with early stages of breast carcinomas with low aggressiveness. 12604776_src has a role in promoting destruction of c-Cbl and enabling EGFR to evade desensitization as part of an oncogenesis pathway 12618767_c-Src, lacking cysteine at position 520, is resistant to 1 microM N-(9-acridinyl) maleimide (NAM) but sensitive to 5 microM. Changing Phe520 to cysteine made it sensitive to 1 microM. Double mutation in clustered cysteines restored resistance to 5 microM. 12631284_Results demonstrate that c-Src and TRAF6 are key mediators of interleukin-1-induced AP-1 activation and provide evidence of cross talk between c-Src and TRAF6 molecules through PI3 kinase-Akt-JNK pathways. 12663375_c-Src regulates NAD(P)H oxidase-derived *O2- generation acutely by stimulating p47phox phosphorylation and translocation and chronically by increasing protein content of gp91phox, p22phox, and p47phox in Ang II-stimulated cells. 12829810_stimulated by HBX protein acting on the mitochondrial transition pore and effecting hepatitis B virus replication. 12842760_involvement of G(i), Src tyrosine kinase and PI3 Kinase, respectively, in TNFalpha production 12893822_v-SRC regulates the nucleo-cytoplasmic delocalization of the major isoform of TEL-ETV6 12907754_role of c-Src on PRL-stimulated proliferation of T47D and MCF7 breast cancer cells 12939401_FAK and Src are both important survival factors, playing a role in protecting colon cancer cell lines from Adenovirus-containing FAK-CD (Ad-FAK-CD)-induced apoptosis 12947321_Inhibition of Src signaling results in a marked reduction of pancreatic cancer cellular invasiveness. Src may represent a novel therapeutic target for this deadly cancer. 12975382_Src has an appreciable role in the organization of the Golgi apparatus, which may be linked to its involvement in protein transport from the Golgi apparatus to the endoplasmic reticulum 14504278_SRC tyrosine kinases are activated and have a role in tyrosine phosphorylation of ezrin and activation of p38 14523024_c-src activation of PLCgamma is mediated by GIT1 14562045_The data indicate an increase in the expression and total activity of endogenous p60(c-Src) in several GM-CSF-independent 1 cultured leukemia cell mutants. 14617636_focal adhesion kinase and src are stimulated by G alpha q and platelet activating factor receptor in vascular endothelium 14627343_role of IGF-1R and c-Src in human pancreatic carcinogenesis. Coexpression of both these molecules may play important role in transformation of pancreatic ductal cells. 14670955_data suggest that agonist-induced binding of Src kinase to the Src homology 3 binding sites in the P2Y(2) purinergic receptor facilitates Src activation and allows Src to efficiently phosphorylate the epidermal growth factor receptor 14676843_A novel regulatory network RAFTK/Pyk2, Src and p38 appears to be critical for VEGF-induced endothelial cell migration. 14706618_v-Src and receptor tyrosine kinase ErbB2 activate beta-catenin-TCF-mediated transcription. 14766744_Ret tyrosine 981 constitutes the major binding site of the Src homology 2 domain of Src and therefore the primary residue responsible for Src activation upon Ret engagement 14963038_EGFR and c-Src-mediated Stat-3 activation is facilitated by Pyk2 15033452_src and protein kinase C have roles in inducing phosphorylation of p38 mitogen-activated protein kinase in epithelial cells after mechanical pressure 15060621_mechanisms of c-Src activation in human cancer (review) 15062576_both the androgen receptor interacting domains of the coactivator SRC1 and of the corepressor SMRT compete for interaction with the androgen receptor N-terminus 15075377_SH2 and SH3 domains of Src mediate peripheral accumulation of phospho-myosin, leading to integrin adhesion complex assembly, whereas loss of SH2 or SH3 function restores normal regulation of E-cadherin and inhibits vimentin expression 15094065_hTAF(II)68-mediated transactivation is linked to the cytoplasmic Src signal transduction pathway. The hTAF(II)68 protein can associate with the SH3 domains of several cell signaling proteins, including v-Src. 15117969_data suggest that colony-stimulating factor receptor-1R (CSF-1R) disrupts cell adhesion by uncoupling adherens junction complexes from the cytoskeleton and promoting cadherin internalization through a Src-dependent mechanism 15254245_Src kinase activity is suppressed by RAC1, which regulates G1/S progression 15271991_Src plays an obligatory role in the mechanism for receptor and diacylglycerol activation of TRPC3 15276210_Results provide important insight to the mechanism of SRC transcriptional activation in liver cancer cells. 15286700_A prolactin signalling cascade in W53 cells involves Src kinases that mediate stimulation of c-Myc expression. 15297464_colony stimulating factor 1 receptor phosphorylation was not markedly affected by Src-family kinases (SFK) inhibition, indicating that lack of SFK binding is not responsible for diminished Y559F phosphorylation. 15456896_Src inhibitors inhibited COX-2 induction in Helicobacter pylori infected gastric mucosa cells. 15467744_Fibronectin and type IV collagen activate ERalpha AF-1 by the c-Src pathway in breast cancer 15485865_G16-mediated activation of nuclear factor kappaB by the adenosine A1 receptor requires c-Src, protein kinase C, and ERK signaling 15494217_c-Src participates in regulating the endocytosis of PDGFbeta receptor-GPCR complexes in response to PDGF. 15498833_c-Src has a role in regulating the dissociation of AP-2 from agonist-occupied AT1R and beta-arrestin during the clathrin-mediated internalization of receptors 15522237_Data demonstrate that type I collagen synergistically enhances platelet-derived growth factor (PDGF)-induced smooth muscle cell proliferation through Src-dependent crosstalk between the alpha2beta1 integrin and the PDGF receptor beta. 15528270_src kinase potentiates tamoxifen agonist action through serine 167-dependent stabilization of estrogen receptor promoter interaction and through elevation of SRC-1 and cAMP response element binding protein-binding protein coactivation of ER 15532711_ERK activation mediated pure pressure-induced proliferative response of aortic smooth muscle cells. This activation was partly mediated by c-Src. 15564375_FRS2-dependent SRC activation is required for FGFR-induced phosphorylation of Sprouty and suppression of ERK activity 15583006_there is a complex interplay between PKCalpha, Syk, and Src involving physical interaction, phosphorylation, translocation within the cell, and functional activity regulation 15618223_PMA transactivates the EGFR and increases cell proliferation by activating the PKCdelta/c-Src pathway in glioblastoma cells 15623523_Csk achieves a low K(m) for the substrate Src, not by stabilizing protein-protein interactions but rather by facilitating a fast phosphoryl transfer step 15623525_Src phosphorylates ezrin at tyrosine 477 15637072_Involvement of c-Src and protein kinase C delta in the inhibition of Cl(-)/OH- exchange activity in Caco-2 cells by serotonin. 15647376_Src-dependent ezrin phosphorylation has a role in adhesion-mediated events in epithelial cells 15688032_SRC-1 is a novel Smad3/4 transcriptional partner, facilitating the functional link between Smad3 and p300/CBP. 15689620_Data show that ionizing radiation causes stress-induced activation of insulin-like growth factor-1 receptor-Src-Mek-Erk-Egr-1 signaling that regulates the clusterin pro-survival cascade. 15735682_HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE regulate Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas 15746183_data demonstrate that Src tyrosine kinase modulates human airway smooth muscle (ASM) cell proliferation and migration 15774483_results indicated that Src kinase-dependent tyrosine phosphorylation of p47(phox) regulates hyperoxia-induced NADPH oxidase activation and reactive oxygen species production in human pulmonary artery endothelial cells 15777850_a specific cross-talk between TGF-beta1 and fibronectin-binding integrin signal pathways leads to the activation of c-Src/Rac1/actin-organization, leading to changes in cell cycle regulator levels in hepatoma cells 15818752_c-src codon 531 mutation in colorectal cancer is not the cause of c-src activation. 15853660_This review summarizes some of the latest evidence for the role of c-Src in tumorigenesis and particularly in human tumor progression. 15872086_calcium activates PLC-gamma1 via increased PIP3 formation mediated by c-src- and fyn-activated PI3K 15872089_dynamin, Cbl, and Src coordinately participate in signaling complexes that are important in the assembly and remodeling of the actin cytoskeleton, leading to changes in osteoclast adhesion, migration, and resorption 16002577_c-Src tyrosine kinase was identified as a binding partner of the BMPR-II C-terminus. 16055703_Semaphorin 4D/plexin-B1 induces endothelial cell migration through the activation of PYK2, Src, and the phosphatidylinositol 3-kinase-Akt pathway 16158055_MUC1 is delivered to mitochondria by a mechanism involving activation of the ErbB receptor-->c-Src pathway and transport by the molecular chaperone HSP70/HSP90 complex 16158247_The Src, a proto-oncogene, has been strongly implicated in the growth, progression and metastasis of a number of human cancers. 16163059_Src activity contributes to constitutive and EGF-induced VEGF expression and angiogenic potential in pancreatic cancer cells 16168436_Data report three crystal structures of a dimeric active c-Src kinase domain, in an apo and two ligand complexed forms, with resolutions ranging from 2.9A to 1.95A. 16260653_FAK regulates integrin-dependent MMP-2 and MMP-9 expression and release by T lymphoid cells through src kinase 16317091_rhMIF induced VCAM-1 and ICAM-1 up-regulation in 12 hours via Src, PI3K, and NFkappaB 16322069_LPC-induced Flk-1/KDR transactivation via c-Src may have important implications for the progression of atherosclerosis 16377083_Stat3 downregulation in vSrc transformed NIH3T3 cells or in breast cancer lines with activated Src induces apoptosis, but in normal cells Stat3 inhibition at post-confluence causes apoptosis while in sparsely growing cells it induces growth retardation 16400523_Cells with single Src family kinase knockdown show that Src, Fyn and Yes kinases are all required for vascular endothelial growth factor (VEGF) mitogenic signaling. 16407214_Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking 16412380_These studies also raise the possibility that fully active forms of c-Src and Fak in breast tumors. 16414009_IR-induced ERK1/2 activation involves EGFR through a Src-dependent pathway that is distinct from EGFR ligand activation 16440311_The results demonstrate that collagen-evoked ectodomain cleavage of DDR1 is mediated in part by Src-dependent activation 16441665_Data show that the interaction between Src and major vault protein (MVP) is critically dependent on Src activity and protein (MVP) tyrosyl phosphorylation, which are induced by epidermal growth factor stimulation. 16465417_Results suggest that c-Src may be involved in the formation of giant cells in cherubism, central giant cell granuloma and giant cell tumors. 16492668_arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src 16523241_These data suggest that FAK might differently regulate apoptosis and focal adhesion formation through site-specific, Src-dependent tyrosine phosphorylation in senescent cells. 16543952_nephrin phosphorylation results in Src kinase activation, recruitment of Nck, and assembly of actin filaments in an Nck-dependent fashion 16569657_Although a key factor in TRAF6-dependent activation of PI 3-kinase, ectopic expression of Src was insufficient for NF-kappaB activation and, in contrast to NF-kappaB, was not inhibited by IRAK2. 16619028_Src activation triggers Caspase-8 phosphorylation on Tyr380 and impairs Fas-induced apoptosis. 16636672_Study shows EGF-stimulation-induced Csk-binding protein (Cbp) tyrosine phosphorylation followed by Cbp-Csk association, in a SFK-dependent manner. 16690617_nNOS is a new p60(src) kinase substrate essential for SST5-mediated anti-proliferative action 16728403_the Src/Fak complex has a role in breast cancer tumorigenesis 16825188_Upon IGF-I stimulation, a complex assembles on SHPS-1 that contains SHP-2, c-Src, and Shc wherein Src phosphorylates Shc, a signaling step that is necessary for an optimal mitogenic response 16837460_v-Src-dependent down-regulation of the Ste20-like kinase SLK through casein kinase II. 16844778_Hsp90 inhibition transiently activates Src kinase and promotes Src-dependent p85 PI3K and Akt and Erk activation 16862215_Nicotine induces cell proliferation by beta-arrestin-mediated activation of the Src and Rb-Raf-1 pathways om ;img camcer. 16882656_extracellular matrix fragments produced by apoptotic EC initiate a state of resistance to apoptosis in fibroblasts via an alpha2beta1 integrin/SFK (Src and Fyn)/phosphatidylinositol 3-kinase (PI3K)-dependent pathway 16891307_A key coactivator for the proper function of HNF4 alpha in human liver and for an integrative control of multiple hepatic genes involved in metabolism and homeostasis. 16894533_Genetic alterations of SRC1 are rare in prostate cancer. The nuclear protein accumulation of SRC1 seems to be mildly increased in androgen ablation resistant prostate cancers. . 16951177_inhibition may be an attractive therapeutic approach for patients with ovarian carcinoma 16959222_Curcumin treatment inhibited v-src gene expression in K562 chronic leukaemia cells. 17018524_steps involving Src association, phosphorylation, and product release are fast, and a structural change in Csk participates in limiting the catalytic cycle 17030621_HSP90 binding to ErbB2 participates in regulation of src kinase activity as well as kinase stability 17038311_data delineate multiple PTH1R structural determinants for ERK1/2 activation and identify mechanism involving proline-rich motifs in receptor C terminus for reciprocal scaffolding of c-Src and beta-arrestin2 with a class II G-protein-coupled receptor 17046078_SHP1 activation, association with Src and dephosphorylation of specific proteins were dependent on extracellular calcium and maintenance of a higher cytosolic calcium plateau. 17056000_These results suggest that dimerization of Src plays an important role in the regulation of Src tyrosine kinase activity. 17062641_Serum-independent growth of 5637 cells involves the transmembrane signaling cascade via EGFR ligand(s) (but not HGF), EGFR, Src and p145(met) 17088251_Src and Rac1 have roles in focal adhesion kinase and ERK mitogenic signaling in epithelial cells 17135298_Stimulation of collagen type I secretion by TGF-beta requires a PKCdelta-Src-ERK1/2 signaling motif. 17215324_Src and focal adhesion kinase mediate cyclic mechanical strain-induced extracellular regulated kinase phosphorylation and proliferation of pulmonary epithelial cells. 17224200_P60-c-src suppresses apoptosis through inhibition of caspase 8 activation in hepatoma cells. 17307333_These lines of evidence suggested that a Gq-coupled receptor activates specifically p38 MAPK through lipid rafts and Src kinase activation, in which flotillins positively modulate the Gq signaling. 17317726_results delineate a novel force-activated inside-out Src/PI3K/FAK/Akt pathway by which cancer cells regulate their own adhesion. 17325034_study elucidated a nongenomic extranuclear signal mediated by the RAR-SRC interaction that is negatively regulated by CSK and is required for RA-induced neuronal differentiation 17369846_Src plays a crucial role in CUTL1-induced tumor cell migration 17443665_Fak/Src signaling to the PI3-K/Akt-1 and MEK/Erk pathways undergoes a differentiation state-specific uncoupling in enterocytes. 17471235_CD99 isoforms dictate opposite functions in tumour malignancy and metastases by activating or repressing c-Src kinase activity. 17510314_These data suggest that Src-induced u-PAR gene expression and invasion/intravasation in vivo is also mediated via AP-1 region -190/-171, especially bound with c-Jun phosphorylated at Ser(73/63). 17537434_The interplay between palladin, SPIN90 and Src and characterized the role of palladin and SPIN90 in platelet derived growth factor and Src-induced cytoskeletal remodeling, was analyzed. 17537435_These results suggest that palmitoylation of Src-family kinases (SFKs) is critical for SFK localization and trafficking and implicate that two distinct trafficking pathways for SFKs may be involved in SFKs' specific functions. 17553930_E-cadherin signaling is an important activator of c-Src at cell-cell contacts, providing a key input into a signaling pathway where quantitative changes in signal strength may result in qualitative differences in functional outcome. 17574549_These findings support a novel role for RACK1 as a key regulator of cell migration and adhesion dynamics through the regulation of Src activity, and the modulation of paxillin phosphorylation at early adhesions. 17593353_Indirect evidence for an impaired src kinase regulation of I (Ca,L) together with an increased phosphatase activity suggests that a complex alteration in the kinase/phosphatase balance leads to I (Ca,L) dysregulation in chronic AF. 17620427_Activated c-Src may play a role in survival, metastasis, and invasion of malignant pleural mesothelioma 17641225_c-Src controls functional association between integrin alphav-beta3 and VEGFR-2 via integrin beta3 phosphorylation. 17691821_These results demonstrate that the chimeric strategy is useful for detailed dissection of the mechanistic basis of substrate specificity and regulation of protein tyrosine kinases. 17716980_norepinephrine stimulation activates Src tyrosine kinase and this activation is required for increased IL-6 expression 17785844_Endothelial Src activity is required for ICAM-mediated tyrosine phosphorylation of vascular endothelial cadherin and for efficient neutrophil migration during passage of leukocytes across the vascular endothelial monolayer. 17803936_Src was identified as Death-associated protein kinase (DAPK) regulator through their reciprocal modification of DAPK Y491/492 residues and establishes a functional link of this DAPK-regulatory circuit to tumor progression. 17804738_Colorectal cancer cells may develop acquired resistance to cetuximab via altering EGFR levels through ubiquitination and degradation and using Src kinase-mediated cell signaling for cell growth and survival. 17849451_TAE226-induced apoptosis in breast cancer cells with overexpressed SRC. 17855352_analysis of Src and C-terminal Src kinase interactions in integrin alphaIIbbeta3-mediated signaling to the cytoskeleton 17933561_These observations suggest that within in vivo-like conditions Src may have a leading role in the induction of sustained ERK1/2 activation through the Src/Ras/Raf pathway. 17934522_These results reveal a potentially important role for CHK in Src activation and tumorigenicity in colon cancer cells. 17967179_human breast cells are a uniquely permissive environment for HGF transactivation by Src/Stat3 which may allow for the inappropriate activation of HGF transcription during the early stages of breast transformation 17974954_This study suggests that (a) PTP1B can act as an important activator of Src in colon cancer cells via dephosphorylation at Y530 of Src and (b) elevated levels of PTP1B can increase tumorigenicity of colon cancer cells by activating Src. 17997837_Brk-mediated STAT5b phosphorylation leads to STAT5b transcriptional activity, and this activity is further increased by kinase active c-Src. 18024423_These data suggest that phosphorylation of PDK1 on Tyr(9), distinct from Tyr(373/376), is important for PDK1/Src complex formation, leading to PDK1 activation. 18057010_Silencing Cdc42 blocks activation of EGFR and Src induced by Ca2+ depletion, resulting in a reduction in E-cadherin degradation 18061419_Col-IV regulates the secretion of MMP-9 via a Src and FAK dependent pathway in MCF-7 cells 18063684_The SRC-STAT5 pathway is essential for decidualization of estrogen-primed human endometrial stromal cells. 18069897_novel mechanism of c-SRC regulation whereby in response to stress c-SRC activity is regulated, at least in part, through loss of the interaction with its inhibitor, PR55gamma 18070925_there is functional conservation between Qr domains of yeast Gal11p and mammalian SRC proteins as direct targets of activator proteins in yeast. 18080320_TNF activated ERK in an EGFR and Src dependent and an EGFR and Src independent modes 18086662_attenuation of the Erk1/2 signaling pathway is essential for andrographolide-mediated inhibition of v-Src transformation 18089800_Association of caspase-8 and Src may help reconcile why a potential tumor suppressor such as caspase-8 is rarely absent in cancers. 18095097_c-Src is a mediator of substance P-stimulated ERK1/2 phosphorylation in human U373 MG glioblastoma cells. 18156174_Src-dependent phosphorylation at Tyr-529 facilitates inactive ERK binding to RSK2, which might be a general requirement for RSK2 activation by EGF through the MEK/ERK pathway. 18180305_VE-cadherin has a role in modulating c-Src activation in VEGF signaling 18209475_This study analysed whether the serotonin transport mediated by the plasma-membrane transporter SERT is regulated by its Tyr-phosphorylation. 18211905_CD63 plays a role in the regulation of ROMK channels through its association with RPTPalpha, which in turn interacts with and activates Src family PTK, thus reducing ROMK activity. 18223692_Src, a nonreceptor tyrosine kinase, is overexpressed in androgen-independent prostate carcinoma 18251740_These data provide direct evidence for increased activation of Src-family tyrosine kinases in the pathogenesis of hyperproliferative epidermal disorders. 18252715_a new signaling connection from ILK to cofilin for dynamic actin polymerization during cell adhesion, depending on the activity of ILK-associated c-Src. 18322799_Anoikis causes a down-activation of Fak, Src, Akt-1 and Erk1/2, a loss of Fak-Src association, and a sustained/enhanced activation of p38b, which is required as apoptosis/anoikis driver 18327819_results indicate that Src and E-cadherin may play an important role in epithelial-mesenchymal transformations, invasion, and aggressive clinicopathologic features of head and neck squamous carcinoma 18353785_results demonstrate that TGFbeta1 induces alphavbeta3 integrin expression via a beta3 integrin-, c-Src-, and p38 MAPK-dependent pathway 18388244_Role of Src kinase in the early action of TCDD through a nongenomic pathway in MCF10A cells is reported. 18397177_Rac1-dependent NADPH oxidases play a critical role in activating c-Src following hypoxia/reoxygenation injury 18403635_These findings suggest a novel MUC1-Src-CrkL-Rac1/Cdc42 signaling cascade following ICAM-1 ligation, through which MUC1 regulates cytoskeletal reorganization and directed cell motility during cell migration. 18434302_Within the endothelial cell, p11 is required for Src kinase-mediated tyrosine phosphorylation of A2, which signals translocation of both proteins to the cell surface. 18441016_hnRNP K binds to the 3'-untranslated region of the c-Src mRNA and inhibits its translation by blocking 80 S ribosome formation 18445593_Csk serves to maintain the constancy of baseline excitatory synaptic transmission by inhibiting Src kinase-dependent synaptic plasticity in the hippocampus 18451158_A Met/c-Src-mediated signaling pathway is a mediator of EGFR tyrosine phosphorylation and cell growth in the presence of EGFR tyrosine kinase inhibitors. 18457834_These results suggest that Src-family tyrosine kinases, including c-Src and Lyn, are involved in formation of the cortical actin cap, which may serve as a platform of tyrosine phosphorylation signaling. 18463161_c-Src is an important regulator of Nox1 activity 18467332_tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases are regulated by phosphorylation 18474606_Src phosphorylation of Bif-1 suppresses the interaction between Bif-1 and Bax, resulting in the inhibition of Bax activation during anoikis. 18493848_Src kinase, in conjunction with antihormone therapies, effectively prevents antihormone resistance in breast cancer cells in vitro 18503159_a critical role for a EGF-R-connected p60c-src-kinase-mediated pathway involving STAT3 and contributing to cell survival and proliferation within colorectal carcinomatumours. 18509267_The effect of OGP(10-14) on differentiation of a cancer megakaryoblast cell line and its involvement on RhoA and Src family kinases signaling pathway, was evaluated. 18524994_pp60c-Src TK-activity was selectively induced by VEGF in MM tumor and ECs 18550772_These studies demonstrate positive cross talk between ER, c-Src, EGFR, and STAT5b in ER+ breast cancer cells. 18562041_the Src-specific sites are all required for overexpressed Focal adhesion kinase to inhibit invadopodia formation. 18566211_This study reveals for the first time that alpha-catenin is a key regulator of beta-catenin transcriptional activity and that the status of alpha-catenin expression in tumor tissues might have prognostic value for Src targeted therapy 18594011_expression and activation of c-Src as well as EGF receptor promotes head and neck squamous cell carcinoma progression 18594017_PLCgamma-1 and c-Src activation contribute to HNSCC invasion downstream of EGF receptor 18606683_study shows that TNFR1 associates with Jak2, c-Src, and PI3K in various cell types to engage signaling pathways, activate transcription factors, and modulate gene expression 18617000_Regulation of hERG channels by protein tyrosine kinases modifies the channel activity and thus likely alters electrophysiological properties including action potential duration and cell excitability in human heart and neurons. 18632638_analysis of PAFR activation and pleiotropic effects on tyrosine phospho-EGFR/Src/FAK/paxillin in ovarian cancer 18644869_This study demonstrates an important role of the dynamic raft reorganization induced by CD44 clustering in eliciting the matrix-derived survival signal through the CD44-SRC-integrin beta1 axis . 18662984_PI3K and Src-ELMO-Dock2 pathways work in parallel to activate Rac2 and modulate chemotaxis in response to a CXCL8 gradient in neutrophils. 18667434_In vascular smooth muscle cells shedding of membrane-bound epidermal growth factor (EGF) receptor ligands and activation of the nonreceptor tyrosine kinases Pyk2 and Src contributed to the thrombin-induced ERK1/2 phosphorylation. 18694941_The Diaphanous-related Formin FHOD1 associates with ROCK1 and promotes Src-dependent plasma membrane blebbing. 18802114_c-Src is involved in lipoteichoic acid-induced heme oxygenase-1 expression in human cultured tracheal smooth muscle cells. 18832467_data are compatible with a model wherein interaction of MDA-9/syntenin with c-Src promotes the formation of an active FAK/c-Src signaling complex, leading to enhanced tumor cell invasion and metastatic spread 18839319_Src overexpression downregulates alpha3 integrin total protein expression and localization to the cell surface of HCT116 colon cancer cells. This indicates that Src activity may enhance metastasis by altering alpha3 integrin expression. 18922903_Src and ADAM-17-mediated shedding of transforming growth factor-alpha is a mechanism of acute resistance to TRAIL. 18924149_Src and Src-associated signaling pathways are activated in relation to hypoxia in neoplasms 18948751_TCPTP is a negative regulator of SFK, JAK1 and STAT3 signalling during the cell cycle. 19035465_KV11 peptide (from human Apolipoprotein A) suppresses angiogenesis and tumor progression by targeting the c-Src/ERK signaling pathways in VEGF induced cells 19047046_as a mediator of SF- and Src-stimulated NF-kappaB activity. Finally, the Src/Rac1/MKK3/6/p38 and Src/TAK1/NF-kappaB-inducing kinase pathways exhibited cross-talk at the level of MKK3. 19048118_Src family kinases mediate betel quid-induced oral cancer cell motility 19066724_Data suggest that upon phosphorylation, Src is directly translocated from focal adhesions to membrane ruffles, thereby promoting formation of new adhesion complexes. 19074832_the c-Src-Rac1-p38 MAPK pathway is required for activation of Akt in response to radiation and plays a cytoprotective role against radiation in human cancer cells 19111446_Mechanical wounding induces significant ROS generation at the wound edge which, in turn, induced ligand-independent KGFR and FRS2 activation via c-Src kinase signaling. 19112177_HINT1 up-regulates cellular levels of p27(KIP1) by two mechanisms: 1) it inhibits its ubiquitylation by targeting the SCF(SKP2) ubiquitin ligase complex, and 2) it inhibits the phosphorylation of p27(KIP1) by Src via inhibiting Src expression. 19130504_Nef regulates the NADPH oxidase p47(phox)activity through the activation of the Src kinases and PI3K 19147496_S100B interacts with Src kinase, thereby stimulating the PI3K/Akt and PI3K/RhoA pathways. 19160018_Rho mediates various phenotypes of malignant transformation by Ras and Src through its effectors, ROCK and mDia [review] 19176482_Presenilin 1 affects focal adhesion site formation and cell force generation via c-Src transcriptional and posttranslational regulation 19199380_Increased PRL1 expression results in activation of Src and ERK1/2, which stimulates MMP2 and MMP9 production, leading to increased cell migration and invasion. 19223541_Sustained Src inhibition results in signal transducer and activator of transcription 3 (STAT3) activation and cancer cell survival via altered Janus-activated kinase-STAT3 binding. 19258394_These results suggest that SFK trafficking is specified by the palmitoylation state in the SH4 domain. 19262138_P-selectin cross-links PSGL-1 and enhances neutrophil adhesion to fibrinogen and ICAM-1 in a Src kinase-dependent, but GPCR-independent mechanism. 19262695_beta-arrestins and c-Src are implicated in the activation of Akt in response to ghrelin through the GHS-R1a 19265199_the signal transduction pathways mediated by c-Kit/D816V are markedly different from those activated by wild-type c-Kit and altered substrate specificity of c-Kit circumvents a need for Src family kinases in signaling of growth and survival 19302982_activated c-Src played a critical role in the HBx-induced epithelial-to-mesenchymal transition in hepatoma cells. 19305428_mutant EGFR-Src interaction and cooperativity play critical roles in constitutive eng | ENSMUSG00000027646 | Src | 1.314802e+03 | 1.2908794 | 0.368354210 | 0.3036209 | 1.450175e+00 | 0.2284999304 | 0.78792184 | No | Yes | 1.438951e+03 | 158.526751 | 1.048124e+03 | 119.225641 | |
| ENSG00000197170 | 5718 | PSMD12 | protein_coding | O00232 | FUNCTION: Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair. {ECO:0000269|PubMed:1317798}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Isopeptide bond;Mental retardation;Proteasome;Reference proteome;Ubl conjugation | The 26S proteasome is a multicatalytic proteinase complex with a highly ordered structure composed of 2 complexes, a 20S core and a 19S regulator. The 20S core is composed of 4 rings of 28 non-identical subunits; 2 rings are composed of 7 alpha subunits and 2 rings are composed of 7 beta subunits. The 19S regulator is composed of a base, which contains 6 ATPase subunits and 2 non-ATPase subunits, and a lid, which contains up to 10 non-ATPase subunits. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides. This gene encodes a non-ATPase subunit of the 19S regulator. A pseudogene has been identified on chromosome 3. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2015]. | hsa:5718; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; membrane [GO:0016020]; nuclear proteasome complex [GO:0031595]; nucleoplasm [GO:0005654]; proteasome accessory complex [GO:0022624]; proteasome complex [GO:0000502]; proteasome regulatory particle [GO:0005838]; proteasome regulatory particle, lid subcomplex [GO:0008541]; secretory granule lumen [GO:0034774] | 28132691_we identified six de novo genomic deletions and four de novo point mutations in unrelated individuals with intellectual disability, congenital malformations, ophthalmologic anomalies, feeding difficulties, deafness, and subtle dysmorphic facial features 30421579_We performed WES on six affected siblings from a multiplex family with ID and autistic features. We identified an inherited heterozygous nonsense mutation in PSMD12 (NM_002816: c.367C>T: p.R123X) in the multiplex family and a de novo nonsense mutation in the same gene (NM_002816: c.601C>T: p.R201X) in the simplex family. 32222279_PSMD12 promotes breast cancer growth via inhibiting the expression of pro-apoptotic genes. | ENSMUSG00000020720 | Psmd12 | 1.174836e+03 | 1.3425163 | 0.424939562 | 0.3621849 | 1.356057e+00 | 0.2442220486 | 0.78892886 | No | Yes | 1.497168e+03 | 294.633701 | 1.013406e+03 | 204.624177 | |
| ENSG00000197183 | 140688 | NOL4L | protein_coding | Q96MY1 | Alternative splicing;Phosphoprotein;Reference proteome | hsa:140688; | cytosol [GO:0005829]; nucleoplasm [GO:0005654] | 20520637_report characterization of two chimeric transcripts identified in AML translocation cases involving CBFA2T2 and C20orf112 21765475_the leukemogenic PAX5-C20S fusion protein is a tetramer, which interacts extraordinarily stably with chromatin as determined by Fluorescence Recovery After Photobleaching in living cells 32522628_LncRNA double homeobox A pseudogene 8 (DUXAP8) facilitates the progression of neuroblastoma and activates Wnt/beta-catenin pathway via microRNA-29/nucleolar protein 4 like (NOL4L) axis. 33940382_NOL4L, a novel nuclear protein, promotes cell proliferation and metastasis by enhancing the PI3K/AKT pathway in ovarian cancer. | ENSMUSG00000061411 | Nol4l | 2.284358e+02 | 1.2366200 | 0.306402264 | 0.3675342 | 7.062524e-01 | 0.4006907285 | 0.84538413 | No | Yes | 2.479722e+02 | 56.444851 | 1.968595e+02 | 46.262722 | |||
| ENSG00000197223 | 10438 | C1D | protein_coding | Q13901 | FUNCTION: Plays a role in the recruitment of the RNA exosome complex to pre-rRNA to mediate the 3'-5' end processing of the 5.8S rRNA; this function may include MPHOSPH6. Can activate PRKDC not only in the presence of linear DNA but also in the presence of supercoiled DNA. Can induce apoptosis in a p53/TP53 dependent manner. May regulate the TRAX/TSN complex formation. Potentiates transcriptional repression by NR1D1 and THRB (By similarity). {ECO:0000250, ECO:0000269|PubMed:10362552, ECO:0000269|PubMed:11801738, ECO:0000269|PubMed:17412707, ECO:0000269|PubMed:9679063}. | Apoptosis;Cytoplasm;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;rRNA processing | The protein encoded by this gene is a DNA binding and apoptosis-inducing protein and is localized in the nucleus. It is also a Rac3-interacting protein which acts as a corepressor for the thyroid hormone receptor. This protein is thought to regulate TRAX/Translin complex formation. Alternate splicing results in multiple transcript variants that encode the same protein. Multiple pseudogenes of this gene are found on chromosome 10.[provided by RefSeq, Jun 2010]. | hsa:10438; | cytoplasm [GO:0005737]; exosome (RNase complex) [GO:0000178]; nuclear exosome (RNase complex) [GO:0000176]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; DNA binding [GO:0003677]; nuclear receptor binding [GO:0016922]; RNA binding [GO:0003723]; transcription corepressor activity [GO:0003714]; apoptotic process [GO:0006915]; maturation of 5.8S rRNA [GO:0000460]; regulation of gene expression [GO:0010468] | 9679063_The C1D protein interacts with the catalytic subunit of DNA-PK and is a very effective substrate for DNA-PK in vitro. Moreover, C1D directs the activation of DNA-PK in a manner that does not require DNA termini, suggesting a role for C1D in DNA repair. 11801738_TRAX enhances the DNA binding capacity of Translin, that binds to recombination regions in some malignancies. C1D interacts with TRAX following g-irradiation and prevents formation of TRAX/Translin complex, thereby inhibits any unwanted recombination. 17412707_Results are consistent with a role for the exosome-associated protein C1D in the recruitment of the exosome to pre-rRNA to mediate the 3' end processing of the 5.8S rRNA. 17599775_Anti-C1D autoantibodies were observed in patients with polymyositis-scleroderma overlap syndrome. 19423540_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20530579_C1D is associated with the DNA repair complex and may promote repair of ultraviolet irradiation-induced DNA damage. 30165670_Data confirm that C1D is not directly involved in repair of UV-damaged DNA; C1D appears to protects cells from oxidative stress (induced by UV irradiation or hydrogen peroxide oxidation) by regulating expression of DNA repair/transcription enzymes such as APEX1 or DDIT3. (C1D = nuclear receptor corepressor C1D; APEX1 = apurinic/apyrimidinic endodeoxyribonuclease-1; DDIT3 = DNA damage inducible transcript-3) | ENSMUSG00000000581 | C1d | 1.316947e+02 | 0.6465132 | -0.629248210 | 0.3892185 | 2.569858e+00 | 0.1089175236 | 0.75783482 | No | Yes | 9.326810e+01 | 18.227948 | 1.419980e+02 | 28.095234 | |
| ENSG00000197343 | 79027 | ZNF655 | protein_coding | Q8N720 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a zinc finger protein. The zinc finger proteins are involved in DNA binding and protein-protein interactions. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:79027; | cytoplasm [GO:0005737]; nucleolus [GO:0005730]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; regulation of transcription by RNA polymerase II [GO:0006357] | 34144060_ZNF655 is involved in development and progression of non-small-cell lung cancer. | ENSMUSG00000007812 | Zfp655 | 1.163257e+03 | 0.8998015 | -0.152321255 | 0.2611921 | 3.352393e-01 | 0.5625901755 | 0.89583618 | No | Yes | 9.460847e+02 | 98.593234 | 1.126998e+03 | 119.512481 | |
| ENSG00000197355 | 91373 | UAP1L1 | protein_coding | Q3KQV9 | Alternative splicing;Nucleotidyltransferase;Reference proteome;Transferase | hsa:91373; | UDP-N-acetylglucosamine diphosphorylase activity [GO:0003977]; UDP-N-acetylglucosamine biosynthetic process [GO:0006048] | 30097606_UAP1L1 is upregulated in a distinct subset of HCC tissues. Patients with upregulated expression of UAP1L1 have a poor prognosis. UAP1L1 overexpression promoted, and UAP1L1 knockdown reduced the proliferation of human hepatoma cells both in vitro and in vivo. UAP1L1 directly interacts with OGT but is not a substrate and cannot activate it alone.UAP1L1 knockdown attenuated c-MYC O-GlcNAcylation and protein stability. 32310823_Identification of UAP1L1 as tumor promotor in gastric cancer through regulation of CDK6. 33434300_Silencing of UAP1L1 inhibits proliferation and induces apoptosis in esophageal squamous cell carcinoma. | ENSMUSG00000026956 | Uap1l1 | 9.225965e+02 | 1.1398107 | 0.188794228 | 0.2832023 | 4.426010e-01 | 0.5058697211 | 0.87772259 | No | Yes | 8.256540e+02 | 89.881747 | 7.344969e+02 | 82.118559 | |||
| ENSG00000197380 | 147906 | DACT3 | protein_coding | Q96B18 | FUNCTION: May be involved in regulation of intracellular signaling pathways during development. Specifically thought to play a role in canonical and/or non-canonical Wnt signaling pathways through interaction with DSH (Dishevelled) family proteins. {ECO:0000269|PubMed:18538736}. | Coiled coil;Methylation;Phosphoprotein;Reference proteome;Wnt signaling pathway | hsa:147906; | cytoplasm [GO:0005737]; delta-catenin binding [GO:0070097]; identical protein binding [GO:0042802]; protein kinase A binding [GO:0051018]; protein kinase C binding [GO:0005080]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of cell growth [GO:0030308]; negative regulation of epithelial to mesenchymal transition [GO:0010719]; negative regulation of Wnt signaling pathway [GO:0030178]; Wnt signaling pathway [GO:0016055] | 18538736_Epigenetic repression of DACT3 leads to aberrant Wnt-beta-catenin signaling in colorectal cancer cells. 33054518_Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. | ENSMUSG00000078794 | Dact3 | 3.021846e+02 | 1.0525500 | 0.073888742 | 0.3311618 | 4.956587e-02 | 0.8238204226 | 0.96654947 | No | Yes | 3.481144e+02 | 58.203643 | 3.546461e+02 | 61.861623 | ||
| ENSG00000197381 | 104 | ADARB1 | protein_coding | P78563 | FUNCTION: Catalyzes the hydrolytic deamination of adenosine to inosine in double-stranded RNA (dsRNA) referred to as A-to-I RNA editing. This may affect gene expression and function in a number of ways that include mRNA translation by changing codons and hence the amino acid sequence of proteins; pre-mRNA splicing by altering splice site recognition sequences; RNA stability by changing sequences involved in nuclease recognition; genetic stability in the case of RNA virus genomes by changing sequences during viral RNA replication; and RNA structure-dependent activities such as microRNA production or targeting or protein-RNA interactions. Can edit both viral and cellular RNAs and can edit RNAs at multiple sites (hyper-editing) or at specific sites (site-specific editing). Its cellular RNA substrates include: bladder cancer-associated protein (BLCAP), neurotransmitter receptors for glutamate (GRIA2 and GRIK2) and serotonin (HTR2C), GABA receptor (GABRA3) and potassium voltage-gated channel (KCNA1). Site-specific RNA editing of transcripts encoding these proteins results in amino acid substitutions which consequently alter their functional activities. Edits GRIA2 at both the Q/R and R/G sites efficiently but converts the adenosine in hotspot1 much less efficiently. Can exert a proviral effect towards human immunodeficiency virus type 1 (HIV-1) and enhances its replication via both an editing-dependent and editing-independent mechanism. The former involves editing of adenosines in the 5'UTR while the latter occurs via suppression of EIF2AK2/PKR activation and function. Can inhibit cell proliferation and migration and can stimulate exocytosis. {ECO:0000269|PubMed:18178553, ECO:0000269|PubMed:19908260, ECO:0000269|PubMed:21289159}.; FUNCTION: [Isoform 1]: Has a lower catalytic activity than isoform 2. {ECO:0000269|PubMed:9149227}.; FUNCTION: [Isoform 2]: Has a higher catalytic activity than isoform 1. {ECO:0000269|PubMed:9149227}. | 3D-structure;Alternative promoter usage;Alternative splicing;Antiviral defense;Disease variant;Hydrolase;Immunity;Innate immunity;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Zinc;mRNA processing | This gene encodes the enzyme responsible for pre-mRNA editing of the glutamate receptor subunit B by site-specific deamination of adenosines. Studies in rat found that this enzyme acted on its own pre-mRNA molecules to convert an AA dinucleotide to an AI dinucleotide which resulted in a new splice site. Alternative splicing of this gene results in several transcript variants, some of which have been characterized by the presence or absence of an ALU cassette insert and a short or long C-terminal region. [provided by RefSeq, Jul 2008]. | hsa:104; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; synapse [GO:0045202]; double-stranded RNA adenosine deaminase activity [GO:0003726]; double-stranded RNA binding [GO:0003725]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; tRNA-specific adenosine deaminase activity [GO:0008251]; adenosine to inosine editing [GO:0006382]; base conversion or substitution editing [GO:0016553]; defense response to virus [GO:0051607]; facial nerve morphogenesis [GO:0021610]; hypoglossal nerve morphogenesis [GO:0021618]; innate immune response [GO:0045087]; innervation [GO:0060384]; motor behavior [GO:0061744]; motor neuron apoptotic process [GO:0097049]; mRNA processing [GO:0006397]; multicellular organism growth [GO:0035264]; muscle tissue morphogenesis [GO:0060415]; negative regulation of cell migration [GO:0030336]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of protein kinase activity by regulation of protein phosphorylation [GO:0044387]; neuromuscular process controlling posture [GO:0050884]; neuromuscular synaptic transmission [GO:0007274]; positive regulation of viral genome replication [GO:0045070]; regulation of cell cycle [GO:0051726]; RNA processing [GO:0006396]; spinal cord ventral commissure morphogenesis [GO:0021965] | 11907222_overexpression inhibits HDV RNA replication and compromises virus viability 12163487_Adenosine to inosine RNA editing requires formation of a ternary complex on the GluR-B R/G site 12414985_short inhibitory RNA-mediated knockdown of host ADAR1 expression but not that of ADAR2 led to decreased HDV amber/W editing and virus production 12859334_the Q/R site of GluRs editing is regulated in a regional manner and the GluR2 Q/R site editing is critically regulated by ADAR2 in human brain 15383678_assayed enzymatic activity of N-terminal deletion constructs of hADAR2 to determine the role of the double-stranded RNA binding motifs and the intervening linker peptide 16141067_Inositol hexakisphosphate is buried within the catalytic domain of ADAR2 and is required for editing of transfer RNA. 16733555_results show that bipolar affective disorder may not be caused by mutations in ADARB1. 16961545_Serum adenosine deaminase (ADA) activity of active Behcet's disease (BD) was higher than that of inactive BD (P < 0.01), but erythrocyte ADA activity was found to be lower in active BD than inactive BD (P < 0.01). 17071493_CD26 and cell surface adenosine deaminase are selectively expressed on ALK-positive, but not on ALK-negative, anaplastic large cell lymphoma and Hodgkin's lymphoma 17525170_The CTD of POLR2A and ADAR2 function together to enforce the order of events that allows editing to precede splicing, and they furthermore suggest a new role for the CTD as a coordinator of two interdependent pre-mRNA processing events. 17728516_Reference values of serum adenosine deaminase (ADA) in normal pregnancy may provide important database for making clinical decisions in pregnancies complicated by conditions where cellular immunity has been altered. 18178553_elevated levels of ADAR1, as found in astrocytomas, do indeed interfere with ADAR2 specific editing activity, and the endogenous ADAR1 can form heterodimers with ADAR2 in astrocytes 19156214_analysis of a splice variant that extends the open reading frame of ADAR2 by 49 amino acids through the utilization of an exon located 18 kilobases upstream of the previously annotated first coding exon and driven by a candidate alternative promoter 19642681_our understanding of the importance of functional groups found in the edited nucleotide and the role of specific active site residues of ADAR2 19713932_Results imply that ADAR1 and ADAR2 have biological functions as RNA-binding proteins that extend beyond editing per se and that even genomically encoded ADARs that are catalytically inactive may have such functions. 20011587_Observational study of gene-disease association. (HuGE Navigator) 20215858_The high conservation of the novel ADAR2 alternative exon in mammals indicates a physiological importance for this exon. 20372915_Elucidation of the molecular mechanism underlying the co-occurrence of reduced ADAR2 activity and abnormal TDP-43 pathology in the same motor neurons may provide a clue to the neurodegenerative process of sporadic amyotrophic lateral sclerosis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21289159_The authors found that, analogously to ADAR1, ADAR2 enhances the release of progeny virions by an editing-dependent mechanism. 21622951_The strong functional similarity of human ADAR2 and Drosophila Adar suggests rather that these are true orthologs. 22226999_These results indicated that ADAR2 downregulation is a profound pathological change relevant to death of motor neurons in ALS. 22366356_ADAR2 activity at the GluA2 pre-mRNA Q/R site correlates with the ADAR2 mRNA level relative to the GluA2 pre-mRNA in different cultured cell lines. 22414730_ADAR2 activity does not consistently change due to the overexpression or knockdown of TDP-43 or the expression of abnormal TDP-43 in amyotrophic lateral sclerosis (ALS) motor neurons. 22525274_Findings demonstrate that post-transcriptional A-to-I RNA editing might be crucial for glioblastoma pathogenesis, identify ADAR2-editing enzyme as a candidate tumor suppressor gene and provide proof that ADAR2 may represent a suitable target. 23621630_The results represent the first evidence that the ADAR1 p150 isoform is the determinant of DSH and may give insight into the currently unknown mechanisms involved in the development of DSH. 24386085_ADAR2-mediated editing of the complementary antisense transcripts is a novel mechanism for regulating the biogenesis of specific miRNAs during hepatocarcinogenesis. 24443933_altered RNA editing efficiency of AMPA receptors due to down-regulation of ADAR2 has a possible role in the pathophysiology of mental disorders. 24509948_The ADAR2 alternative splicing variants may be correlated with the invasiveness of gliomas. 25564529_Characterization of the ADAR2 catalytic domain-RNA interaction. 25582055_ADAR2 is a key factor for maintaining edited-miRNA population and balancing the expression of several essential miRNAs involved in cancer. 25673044_Therefore, the expression of ADAR1 and ADAR2 was analyzed in chordoma tissues. It was found that ADAR1 was significantly overexpressed, which was accompanied by enhanced pre-miR-10a and pri-miR-125a A-to-I editing 25692240_Data show that a large fraction of the edited genes are positively correlated ADAR (ADAR1) and ADARB1 (ADAR2). 25732952_ADARB1 rs9983925 and rs4819035 and HTR2C rs6318 were associated with suicide attempt risk. 25873329_we conclude that this aberrant alternative splicing pattern of ADAR2 downregulates A-to-I editing in glioma 26252972_Detailed structural analysis indicates that the minor groove width of dsRNA and global shape of RNA may play an important role in the specific reading mechanism of ADAR2. 26485095_These findings suggest that adenosine deaminase acting on RNA 2 is subject to different regulations by DNA methyltransferase and histone deacetylase enzymes in neuronal SH-SY5Y cells. 26655226_A-to-I RNA editing levels catalyzed by ADAR1 and ADARB1 decreased in Alzheimer's disease patients' brain tissues, mainly in the hippocampus and to a lesser degree in the temporal and frontal lobes. 26712564_These data suggest that, like ADAR2, underlying sequences in dsRNA may influence how NF90 recognizes its target RNAs 27065196_Four crystal structures of the human ADAR2 deaminase domain bound to RNA duplexes bearing a mimic of the deamination reaction intermediate. 27614075_we determined the importance of specific amino acids at 19 different positions in the ADAR2 5' binding loop and revealed six residues that provide essential structural elements supporting the fold of the loop and key RNA-binding functional groups. This work provided new insight into RNA recognition by ADAR2 and established a new tool for defining structure-function relationships in ADAR reactions. 28035363_Data indicate that ADAR2 suppresses tumor growth and induces apoptosis by editing and stabilizing IGFBP7 in esophageal squamous cell carcinoma. 28084537_2-way interaction between TPH2 rs4290270 and general traumas revealed that TT homozygotes with history of general traumas had an increased risk for suicide attempt. 3-way interaction of general traumas, TPH2 rs4290270 and ADARB1 rs4819035 indicated that highest predisposition to suicide attempt was observed in individuals who experienced general traumas and were TT homozygote for rs4290270 and TT homozygote for rs4819035. 28217931_The deaminase domain-RNA contact surfaces are reviewed and models of how full length ADARs, bearing double stranded RNA-binding domains (dsRBDs) and deaminase domains, could process naturally occurring substrate RNAs are presented. 28550310_Adenosine-to-inosine editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme, which correlated with downregulation of ADAR2. 28913566_The range of human disease associated with ADAR1 mutations may extend further to include other inflammatory conditions while ADAR2 mutations may affect psychiatric conditions. 29637273_Changes in expression levels of ADAR1 and ADAR2 may represent an important regulatory mechanism in idiopathic pulmonary fibrosis, by regulating the processing of key miRNAs such as miRNA-21. 29950133_This study investigates the genome-wide binding preferences of the nuclear constitutive isoforms ADAR1-p110 and ADAR2 on human miRNA species by RNA immunoprecipitation of ADAR-bound small RNAs . 29956457_The findings illustrate that lipopolysaccharide participates in Hirschsprung's disease through the ADAR2-miR-142-STAU1 axis. 30590609_Stably introduced into cancer cell lines, the system reports on elevated endogenous ADAR1 editing activity induced by interferon as well as knockdown of ADAR1 and ADAR2. In a single-well setup we used the reporter in HeLa cells to screen a small molecule library of 33 000 compounds 30945056_that the mislocalization of ADAR2 in C9orf72 mediated amyotrophic lateral sclerosis/frontotemporal dementia is responsible for the alteration of RNA processing events that may impact vast cellular functions 31039163_Strong downregulation of ADAR2 and increase in ADAR1 expression was observed in blood samples from congenital heart disease (CHD) patients. 31095429_AKT-1, -2, and -3 interact with both ADAR1p110 and ADAR2 and phosphorylate these RNA editases. Using site-directed mutagenesis of suspected AKT phosphorylation sites, AKT was found to primarily phosphorylate ADAR1p110 and ADAR2 on T738 and T553, respectively, and overexpression of the phosphomimic mutants ADAR1p110 (T738D) and ADAR2 (T553D) resulted in a 50-100% reduction in editase activity. 31270214_adenosine deaminase acting on RNA 1 (ADAR1) and ADAR2, which catalyze adenosine-to-inosine RNA editing, downregulate the expression of constitutive androstane receptor (CAR) in human liver-derived cells by attenuating splicing 31491024_In lung adenocarcinoma, downregulation of ADARB1 was related to shorter first progression (FP), overall survival time (OS) and post-progression survival time (PPS). 31852792_ADAR2 Is Involved in Self and Nonself Recognition of Borna Disease Virus Genomic RNA in the Nucleus. 32034135_ADAR2 binds to double-stranded RNA formed between GA-rich sequences and polypyrimidine-tract and precludes access of U2AF65 to 3' splice site. 32132027_Overexpression of ADAR2, that was incapable of binding to RNA, suppressed growth, motility, and invasion of MPM cells. However, overexpression of ADAR2 that had no enzyme activity did not alter the malignant properties of MPM cells. 32203492_We also observe a dominance of long-range interactions within ADAR substrates and analyze differences between ADAR1 and ADAR2 editing substrates. Moreover, we unexpectedly discovered that ADAR proteins bind dsRNA substrates tandemly in vivo, each with a 50-bp footprint. 32220291_ADARB1 as a genetic etiology in individuals with intellectual disability, microcephaly, and epilepsy. 32433965_Unbiased Identification of trans Regulators of ADAR and A-to-I RNA Editing. 32597966_Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition. 32603639_What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs. 32719099_Biallelic variants in ADARB1, encoding a dsRNA-specific adenosine deaminase, cause a severe developmental and epileptic encephalopathy. 33170095_Short-Chain Guide RNA for Site-Directed A-to-I RNA Editing. 34380502_Developing PspCas13b-based enhanced RESCUE system, eRESCUE, with efficient RNA base editing. 34535666_ADAR-mediated RNA editing of DNA:RNA hybrids is required for DNA double strand break repair. 35044296_Comprehensive interrogation of the ADAR2 deaminase domain for engineering enhanced RNA editing activity and specificity. | ENSMUSG00000020262 | Adarb1 | 1.892582e+03 | 0.9317587 | -0.101971702 | 0.2671534 | 1.471778e-01 | 0.7012470563 | 0.93624959 | No | Yes | 1.409328e+03 | 182.435594 | 1.690810e+03 | 224.211092 | |
| ENSG00000197467 | 1305 | COL13A1 | protein_coding | Q5TAT6 | FUNCTION: Involved in cell-matrix and cell-cell adhesion interactions that are required for normal development. May participate in the linkage between muscle fiber and basement membrane. May play a role in endochondral ossification of bone and branching morphogenesis of lung. Binds heparin. At neuromuscular junctions, may play a role in acetylcholine receptor clustering (PubMed:26626625). {ECO:0000250|UniProtKB:Q9R1N9, ECO:0000269|PubMed:10865988, ECO:0000269|PubMed:11956183, ECO:0000269|PubMed:26626625}. | Alternative splicing;Cell adhesion;Cell junction;Cell membrane;Collagen;Congenital myasthenic syndrome;Developmental protein;Differentiation;Disulfide bond;Heparin-binding;Membrane;Osteogenesis;Postsynaptic cell membrane;Reference proteome;Signal-anchor;Synapse;Transmembrane;Transmembrane helix | This gene encodes the alpha chain of one of the nonfibrillar collagens. The function of this gene product is not known, however, it has been detected at low levels in all connective tissue-producing cells so it may serve a general function in connective tissues. Unlike most of the collagens, which are secreted into the extracellular matrix, collagen XIII contains a transmembrane domain and the protein has been localized to the plasma membrane. The transcripts for this gene undergo complex and extensive splicing involving at least eight exons. Like other collagens, collagen XIII is a trimer; it is not known whether this trimer is composed of one or more than one alpha chain isomer. A number of alternatively spliced transcript variants have been described, but the full length nature of some of them has not been determined. [provided by RefSeq, Jul 2008]. | hsa:1305; | cell-cell junction [GO:0005911]; collagen trimer [GO:0005581]; collagen type XIII trimer [GO:0005600]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; plasma membrane [GO:0005886]; postsynaptic membrane [GO:0045211]; extracellular matrix structural constituent [GO:0005201]; heparin binding [GO:0008201]; cell differentiation [GO:0030154]; cell-cell adhesion [GO:0098609]; cell-matrix adhesion [GO:0007160]; collagen fibril organization [GO:0030199]; endochondral ossification [GO:0001958]; extracellular matrix organization [GO:0030198]; morphogenesis of a branching structure [GO:0001763]; notochord development [GO:0030903]; skeletal system development [GO:0001501] | 11956183_The type XIII collagen ectodomain is a 150-nm rod and capable of binding to fibronectin, nidogen-2, perlecan, and heparin. 12832406_two widely separated coiled-coil domains of type XIII and related collagens function as independent oligomerization domains participating in the folding of distinct areas of the molecule. 16385451_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20708005_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 26626625_Congenital myasthenic syndrome type 19 is caused by mutations in COL13A1. 27245701_We identified overexpression of collagen type XIII alpha 1 in active Thyroid-associated ophthalmopathy affected fat. 27580012_The combination of constitutively low expression of COL13A1, high physiological and metabolic demands, and consequentially relatively high exposure to stressors may explain the particular vulnerability of inferior rectus to thyroid-associated ophthalmopathy. 28415608_this study shows that COL13A1 production by urothelial carcinoma of the bladder plays a pivotal role in tumor invasion through the induction of tumor budding 28837258_Urine levels of COL4A1, COL13A1, the combined values of COL4A1 and COL13A1 (COL4A1 + COL13A1), and CYFRA21-1 were significantly elevated in urine from patients with BCa compared to the controls. A high urinary COL4A1 + COL13A1 was found to be an independent risk factor for intravesical recurrence. 29307798_Findings suggest a significant association between variants in COL13A1, ADIPOQ, SAMM50, and PNPLA3, and risk of NAFLD/elevated transaminase levels in Mexican adults with an admixed ancestry. 29363764_The authors report a congenital myasthenic syndrome due to mutations in COL13A1, which encodes an extracellular matrix protein that is concentrated at the neuromuscular junction and highlights a role for these extracellular matrix proteins in maintaining synaptic stability that is independent of the AGRN/MuSK clustering pathway. 29369589_It was established that the frequency of individuals with the COL13A1*D/*D genotype was higher in the senile age period. The LAMA2*I/*D genotype was predisposing to longevity among women. 30285809_Results indicate a function of collagen XIII in promoting cancer metastasis, cell invasion, and anoikis resistance. 30767057_The data of this study support the causality of COL13A1 variants for Congenital myasthenic syndrome. 31081514_patients with COL13A1 mutations present mostly with severe early-onset myasthenic syndrome with feeding and breathing difficulties 31220558_Data suggest that ColXIII has a role in age-dependent cortical bone deterioration with possible implications for osteoporosis and fracture risk. 34404755_TGF-beta2 and collagen play pivotal roles in the spheroid formation and anti-aging of human dermal papilla cells. | ENSMUSG00000058806 | Col13a1 | 2.012200e+02 | 1.0546490 | 0.076762914 | 0.3181427 | 5.771442e-02 | 0.8101455498 | 0.96238102 | No | Yes | 1.749039e+02 | 19.548098 | 1.749787e+02 | 20.162921 | |
| ENSG00000197837 | 121504 | H4-16 | protein_coding | P62805 | FUNCTION: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. | 3D-structure;Acetylation;Chromosomal rearrangement;Chromosome;Citrullination;DNA-binding;Direct protein sequencing;Hydroxylation;Isopeptide bond;Methylation;Nucleosome core;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H4 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. [provided by RefSeq, Aug 2015]. | hsa:121504;hsa:554313;hsa:8294;hsa:8359;hsa:8360;hsa:8361;hsa:8362;hsa:8363;hsa:8364;hsa:8365;hsa:8366;hsa:8367;hsa:8368;hsa:8370; | CENP-A containing nucleosome [GO:0043505]; chromosome, telomeric region [GO:0000781]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; membrane [GO:0016020]; nuclear chromosome [GO:0000228]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; DNA binding [GO:0003677]; protein domain specific binding [GO:0019904]; protein heterodimerization activity [GO:0046982]; RNA binding [GO:0003723]; DNA replication-dependent chromatin assembly [GO:0006335]; DNA replication-independent chromatin assembly [GO:0006336]; DNA-templated transcription, initiation [GO:0006352]; negative regulation of megakaryocyte differentiation [GO:0045653]; nucleosome assembly [GO:0006334]; telomere organization [GO:0032200] | 12385581_low levels of histone acetylation is associated with the development and progression of gastric carcinomas, possibly through alteration of gene expression 15095300_overexpression of MTA1 protein and acetylation level of histone H4 protein are closely related 15345777_peptidylarginine deiminase 4 regulates histone Arg methylation by converting methyl-Arg to citrulline and releasing methylamine; data suggest that PAD4 mediates gene expression by regulating Arg methylation and citrullination in histones 16177192_lack of biotinylation of K12 in histone H4 is an early signaling event in response to double-strand breaks 16322686_The decrease in trimethylation of lysine 20 of histone H4 in breast cancer cells was accompanied by diminished expression of Suv4-20h2 histone methyltransferase. 16469925_incorporation of acetylated histone H4-K16 into nucleosomal arrays inhibits the formation of compact 30-nanometer-like fibers and impedes the ability of chromatin to form cross-fiber interactions 16531610_Apoptosis is associated with global DNA hypomethylation and histone deacetylation events in leukemia cells. 16782888_BTG2 contributes to retinoic acid activity by favoring differentiation through a gene-specific modification of histone H4 arginine methylation and acetylation levels. 17522015_Relationship between histone H4 modification, epigenetic regulation of BDNF gene expression, and long-term memory for extinction of conditioned fear. 17548343_H4 tail and its acetylation have novel roles in mediating recruitment of multiple regulatory factors that can change chromatin states for transcription regulation 17848202_Brd2 bromodomain 2 is monomeric in solution and dynamically interacts with H4-AcK12; additional secondary elements in the long ZA loop may be a common characteristic of BET bromodomains. 18001726_Spermatids Hypac-H4 impairment in mixed atrophy did not deteriorate further by AZFc region deletion. 18319261_the SET8 and PCNA interaction couples H4-K20 methylation with DNA replication 18408754_H4K20 monomethylation and PR-SET7 are important for L3MBTL1 function 18671804_High expression of acetylated H4 is more common in aggressive than indolent cutaneous T-cell lymphoma. 18684714_Jade-1/1L are crucial co-factors for HBO1-mediated histone H4 acetylation 18974389_Our findings indicate an important role of histone H4 modifications in bronchial carcinogenesis 19348949_results indicate, by acetylation of histone H4 K16 during S-phase, early replicating chromatin domains acquire the H4K16ac-K20me2 epigenetic label that persists on the chromatin throughout mitosis & is deacetylated in early G1-phase of the next cell cycle 19438744_acetylated H4 is overexpressed in diffuse large B-cell lymphoma and peripheral T-cell lymphoma relative to normal lymphoid tissue. 19536143_The release of histone H4 by holocrine secretion from the sebaceous gland may play an important role in innate immunity. 19578722_histone modification including PRC2-mediated repressive histone marker H3K27me3 and active histone marker acH4 may involve in CD11b transcription during HL-60 leukemia cells reprogramming to terminal differentiation 19667075_A role of Cdk7 in regulating elongation is further suggested by enhanced histone H4 acetylation and diminished histone H4 trimethylation on lysine 36-two marks of elongation-within genes when the kinase was inhibited. 19747413_Downregulated by zinc and upregulated by docosahexaenoate in a neuroblastoma cell line. 19805290_Data showed the dynamic fluctuation of histone H4 acetylation levels during mitosis, as well as acetylation changes in response to structurally distinct histone deacetylase inhibitors. 19818714_Data directly implicate BBAP in the monoubiquitylation and additional posttranslational modification of histone H4 and an associated DNA damage response. 20512922_Our findings reveal the molecular mechanisms whereby the DNA sequences within specific gene bodies are sufficient to nucleate the monomethylation of histone H4 lysine 200 which, in turn, reduces gene expression by half. 20949922_the imatinib-induced hemoglobinization and erythroid differentiation in K562 cells are associated with global histone H4 21478274_The crystal structure of an HJURP-CENP-A-histone H4 complex shows that HJURP binds a CENP-A-H4 heterodimer 21724829_phosphorylation of histone H4 Ser 47 catalyzed by the PAK2 kinase, promotes nucleosome assembly of H3.3-H4 and inhibits nucleosome assembly of H3.1-H4 by increasing the binding affinity of HIRA to H3.3-H4 and reducing association of CAF-1 with H3.1-H4 21973049_TNF-alpha inhibition of AQP5 expression in human salivary gland acinar cells is due to the epigenetic mechanism by suppression of acetylation of histone H4. 22228774_HAT1 differentially impacts nucleosome assembly of H3.1-H4 and H3.3-H4. 22360506_our data suggest that global histone H3 and H4 modification patterns are potential markers of tumor recurrence and disease-free survival in non-small cell lung cancer 22368283_Human PIH1 domain-containing protein 1 (PIH1) interacts directly with histone H4 and recruits the Brg1-SWI/SNF complex via SNF5 to human rRNA genes. 22894908_This study focused on the distribution of a specific histone modification, namely H4K12ac, in human sperm and characterized its specific enrichment sites in promoters throughout the whole human genome. 23048028_SRP68/72 heterodimers as major nuclear proteins whose binding of histone H4 tail is inhibited by H4R3 methylation. 23363600_Data indicate that G1-phase histone assembly is restricted to CENP-A and H4. 24481548_An increase in histone H4 acetylation caused by hypoxia in human neuroblastoma cell lines corresponds to increased levels of N-myc transcription factor in these cells. 25283865_Acetylation at lysine 5 of histone H4 associated with lytic gene promoters during reactivation of Kaposi's sarcoma-associated herpesvirus. 25294883_Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. 25611806_Systemic lupus erythematosus appears to be associated with an imbalance in histone acetyltransferases and histone deacetylase enzymes favoring pathologic H4 acetylation. 25713080_Data indicate that MEP50 WD repeat protein is essential for methylation of histones H4 and H2A by PRMT5 arginine methyltransferase. 25788266_H4K12ac is regulated by estrogen receptor-alpha and is associated with BRD4 function and inducible transcription 26563484_Data show that Omomyc protein co-localized with proto-oncogene protein c-myc (c-Myc), protein arginine methyltransferase 5 (PRMT5) and histone H4 H4R3me2s-enriched chromatin domains. 28450392_Data suggest that O-GlcNAc transferase 1 (OGT1) specifically binds to, O-GlcNAcylates, and stabilizes nonspecific lethal protein3 (NSL3); stabilization of NSL3 by OGT1 up-regulates global acetylation levels of histone 4 at Lys-5, Lys-8, and Lys-16. 28546430_Data suggest post-translational modifications of histones, trimethylation of lysine 36 in H3 (H3K36me3) and acetylation of lysine 16 in H4 (H4K16ac), have roles in DNA damage repair; H3K36me3 stimulates H4K16ac upon DNA double-strand break; SETD2, LEDGF, and KAT5 are required for these epigenetic changes. (SETD2 = SET domain containing 2; LEDGF = lens epithelium-derived growth factor; KAT5 = lysine acetyltransferase 5) 28977641_ata show that PP32 and SET/TAF-Ibeta proteins block HAT1-mediated H4 acetylation. | ENSMUSG00000069274 | H4c6 | 1.222117e+01 | 0.9774756 | -0.032867358 | 0.8310886 | 1.587265e-03 | 0.9682202887 | No | Yes | 5.344109e+00 | 2.295897 | 5.625932e+00 | 2.400515 | ||
| ENSG00000197863 | 388536 | ZNF790 | protein_coding | Q6PG37 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:388536; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 4.492974e+01 | 0.8188387 | -0.288348731 | 0.4733990 | 3.767964e-01 | 0.5393227811 | 0.89033215 | No | Yes | 4.719004e+01 | 10.171753 | 5.694009e+01 | 12.580846 | |||||
| ENSG00000197879 | 4641 | MYO1C | protein_coding | O00159 | FUNCTION: Myosins are actin-based motor molecules with ATPase activity. Unconventional myosins serve in intracellular movements. Their highly divergent tails are presumed to bind to membranous compartments, which would be moved relative to actin filaments. Involved in glucose transporter recycling in response to insulin by regulating movement of intracellular GLUT4-containing vesicles to the plasma membrane. Component of the hair cell's (the sensory cells of the inner ear) adaptation-motor complex. Acts as a mediator of adaptation of mechanoelectrical transduction in stereocilia of vestibular hair cells. Binds phosphoinositides and links the actin cytoskeleton to cellular membranes. {ECO:0000269|PubMed:24636949}.; FUNCTION: Isoform 3 is involved in regulation of transcription. Associated with transcriptional active ribosomal genes. Appears to cooperate with the WICH chromatin-remodeling complex to facilitate transcription. Necessary for the formation of the first phosphodiester bond during transcription initiation (By similarity). {ECO:0000250}. | 3D-structure;ATP-binding;Acetylation;Actin-binding;Alternative splicing;Calmodulin-binding;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Membrane;Methylation;Motor protein;Myosin;Nuclear pore complex;Nucleotide-binding;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Translocation;Transport;mRNA transport | This gene encodes a member of the unconventional myosin protein family, which are actin-based molecular motors. The protein is found in the cytoplasm, and one isoform with a unique N-terminus is also found in the nucleus. The nuclear isoform associates with RNA polymerase I and II and functions in transcription initiation. The mouse ortholog of this protein also functions in intracellular vesicle transport to the plasma membrane. Multiple transcript variants encoding different isoforms have been found for this gene. The related gene myosin IE has been referred to as myosin IC in the literature, but it is a distinct locus on chromosome 19. [provided by RefSeq, Jul 2008]. | hsa:4641; | actin cytoskeleton [GO:0015629]; B-WICH complex [GO:0110016]; basal plasma membrane [GO:0009925]; brush border [GO:0005903]; cytoplasm [GO:0005737]; cytoplasmic vesicle membrane [GO:0030659]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; filamentous actin [GO:0031941]; lateral plasma membrane [GO:0016328]; membrane [GO:0016020]; membrane raft [GO:0045121]; microvillus [GO:0005902]; nuclear body [GO:0016604]; nuclear pore [GO:0005643]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; phagocytic vesicle [GO:0045335]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; stereocilium membrane [GO:0060171]; unconventional myosin complex [GO:0016461]; vesicle [GO:0031982]; actin filament binding [GO:0051015]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; microfilament motor activity [GO:0000146]; protein C-terminus binding [GO:0008022]; signaling receptor binding [GO:0005102]; small GTPase binding [GO:0031267]; actin filament organization [GO:0007015]; cellular response to interferon-gamma [GO:0071346]; mRNA transport [GO:0051028]; positive regulation of cell migration [GO:0030335]; positive regulation of cell migration by vascular endothelial growth factor signaling pathway [GO:0038089]; positive regulation of cellular response to insulin stimulus [GO:1900078]; positive regulation of protein targeting to membrane [GO:0090314]; positive regulation of transcription by RNA polymerase III [GO:0045945]; positive regulation of vascular endothelial growth factor signaling pathway [GO:1900748]; protein targeting [GO:0006605]; protein targeting to membrane [GO:0006612]; regulation of bicellular tight junction assembly [GO:2000810]; vesicle transport along actin filament [GO:0030050] | 16514417_NM1 cooperates with a chromatin remodelling complex containing WSTF (Williams syndrome transcription factor) and SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A member 5 protein (SNF2h). 16603771_WSTF-SNF2h-NM1 forms a platform in transcription while providing chromatin remodeling 18599791_Sensing molecular tension is crucial for a wide array of cellular processes. Myosin I dramatically alters its motile properties in response to tension. 19027848_Observational study of gene-disease association. (HuGE Navigator) 19027848_identification of 6 heterozygous missense mutations in MYO1C and additional 5 heterozygous missense mutations in MYO1F in bilateral sensorineural hearing loss 19729515_NM1 facilitates maturation and accompanies export-competent preribosomal subunits to the nuclear pore complex, thus modulating export. 21402783_Myo1c plays a critical role in the translocation of Neph1 complexes in podocytes. Myo1c's ability to interact with membranes, F-actin, Neph1, and nephrin indicates that it actively contributes to the dynamic organization of the filtration slit. 22328521_Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. 23262137_This is the first report demonstrating that Myo1c is an important mediator of VEGF-induced VEGFR2 delivery to the cell surface and plays a role in angiogenic signaling. 23438938_Both nucleolar localization signals are functional and necessary for nucleolar localization of specifically myosin IC isoform B. 23555303_These results suggest a unique structural role for NM1 in which the interaction with SNF2h stabilizes B-WICH at the gene promoter and facilitates recruitment of the HAT PCAF 24523293_myosin 1c manipulations lead to loss of the actin filaments and to similar endoplasmic reticulum phenotype as observed after actin depolymerization. 24636949_The structural context and the chemical environment of Myo1c mutations that are involved in sensorineural hearing loss in humans are described and their impact on motor function is discussed. 24698832_The relationship between MYO1C and KAT6B suggests that the two are interacting in chromatin remodelling for gene expression in human masseter muscle. This is the nuclear myosin1 (NM1) function of MYO1C. 24901984_NM1 phosphorylation by GSK3beta blocks NM1 ubiquitination by UBR5 and degradation by the proteasome, leads to NM1 association with the chromatin and promotes rDNA transcription activation at G1. 25551774_Ablating MYO1C function causes abnormal cholesterol distribution, which has a major selective impact on the autophagy pathway 25660542_Myo1c significantly increases the frequency of kinesin-1-driven microtubule-based runs that begin at actin/microtubule intersections. The actin-binding protein tropomyosin 2 abolishes Myo1c-specific effects on both run initiation and run termination. 27044863_Study presents structural demonstration of a cargo protein, Neph1, attached to Myo1c, providing novel insights into the role of Myo1c in intracellular movements of this critical slit diaphragm protein. 27192697_The results establish a mechanistic connection between the calcium regulation of the motor function of myosin IC in the cytoplasm and the induction of its import into the nucleus. 27246739_In glioblastoma 1321 N1 cells, we recently identified Myo1c as a new interactor of SHIP2. SHIP2 localization at lamellipodia and ruffles is impaired in Myo1c depleted cells. In the absence of Myo1c, N1 cells tend to associate to form clusters. 27365048_Upon DNA damage, an increase in the levels of chromatin bound motor protein nuclear myosin 1 (NM1) ensues, which appears to be functionally linked to Upsilon-H2AX signaling. 27468717_Overexpression of MYO1C is associated with gastric cancer. 27716847_Cells expressing excess of MYO1C had low basal level of phosphorylated protein kinase B. 28893906_NTR(35), which harbors the R21G mutation, was unable to confer MYO1C(35)-like kinetic behavior. Thus, the NTRs affect the specific nucleotide-binding properties of MYO1C isoforms, adding to their kinetic diversity. We propose that this level of fine-tuning within MYO1C broadens its adaptability within cells. 29179037_E2 and NM1 associate via their N-terminal domains and this interaction is ATP dependent. 30872458_MYO1C stabilizes actin at the Golgi complex, facilitating the arrival of incoming transport carriers at the Golgi. 33201534_Isolation and identification in human blood serum of the proteins possessing the ability to bind with 48 kDa form of unconventional myosin 1c and their possible diagnostic and prognostic value. 34019593_Myosin 1C isoform A is a novel candidate diagnostic marker for prostate cancer. 34380438_SH3BGRL3 binds to myosin 1c in a calcium dependent manner and modulates migration in the MDA-MB-231 cell line. | ENSMUSG00000017774 | Myo1c | 3.687320e+03 | 1.3118142 | 0.391563420 | 0.2931500 | 1.797550e+00 | 0.1800089739 | 0.77663851 | No | Yes | 4.059698e+03 | 451.191548 | 2.714178e+03 | 310.054067 | |
| ENSG00000197905 | 7004 | TEAD4 | protein_coding | Q15561 | FUNCTION: Transcription factor which plays a key role in the Hippo signaling pathway, a pathway involved in organ size control and tumor suppression by restricting proliferation and promoting apoptosis. The core of this pathway is composed of a kinase cascade wherein MST1/MST2, in complex with its regulatory protein SAV1, phosphorylates and activates LATS1/2 in complex with its regulatory protein MOB1, which in turn phosphorylates and inactivates YAP1 oncoprotein and WWTR1/TAZ. Acts by mediating gene expression of YAP1 and WWTR1/TAZ, thereby regulating cell proliferation, migration and epithelial mesenchymal transition (EMT) induction. Binds specifically and non-cooperatively to the Sph and GT-IIC 'enhansons' (5'-GTGGAATGT-3') and activates transcription. Binds to the M-CAT motif. {ECO:0000269|PubMed:18579750, ECO:0000269|PubMed:19324877}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene product is a member of the transcriptional enhancer factor (TEF) family of transcription factors, which contain the TEA/ATTS DNA-binding domain. It is preferentially expressed in the skeletal muscle, and binds to the M-CAT regulatory element found in promoters of muscle-specific genes to direct their gene expression. Alternatively spliced transcripts encoding distinct isoforms, some of which are translated through the use of a non-AUG (UUG) initiation codon, have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:7004; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-DNA complex [GO:0032993]; transcription regulator complex [GO:0005667]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; cell fate specification [GO:0001708]; embryo implantation [GO:0007566]; embryonic organ development [GO:0048568]; hippo signaling [GO:0035329]; muscle organ development [GO:0007517]; positive regulation of stem cell population maintenance [GO:1902459]; regulation of transcription by RNA polymerase II [GO:0006357]; skeletal system development [GO:0001501]; transcription, DNA-templated [GO:0006351]; trophectodermal cell fate commitment [GO:0001830] | 8702974_The paper described that the ORF of TEF-3 initiates with an ATT codon encoding isoleucine. 8921372_The paper described several reasons to designate the putative translation initiation codon as the leucine (TTG) codon, which is 7 codons upstream from the isoleucine (ATT) codon. 15520314_Constitutive activation of alpha1-adrenergic signaling through the RTEF-1 transcription factor results in chronic elevation of PP1beta expression and connexin dephosphorylation. This mechanism may underlie some defects in cardiac conduction. 17652751_Novel RTEF-1 transcripts are present within human ocular vascular endothelial cells and mouse neural retina during normal and retinopathy of prematurity development, and alternatively spliced products are produced under hyperoxic and hypoxic conditions 18579725_The gain of function studies indicated that TEA domain family member 4 activate NR5A1 gene expression. 18840614_TEF3 mediates the expression of Down syndrome candidate region 1 isoform 1 (DSCR1-1L) in endothelial cells 21169295_the RTEF-1-driven increase of VEGF-B plays an important role in communication between the endothelium and myocardium 21169383_TEF3, mainly its nuclear localization, is required for VEGF-A(165)-induced endothelial proliferation, migration, tube formation, and in vivo Matrigel angiogenesis. 21540178_RTEF-1 as a regulator of HIF-1alpha transcription 22433836_RTEF-1 plays an important role in FGFR1- stimulated vasodilatation. 22652601_Blocking connexin 43 function inhibited RTEF-1-induced endothelial cell connections and aggregation 22761647_High TEAD4 expression is associated with Age-Related Macular Degeneration. 22843786_These results show that RTEF-1-stimulated IGFBP-1 expression may be central to the mechanism by which RTEF-1 attenuates blood glucose levels. 22996285_These data suggest that TFF3 and survivin expressions play a vital role in gastric cancer development, and these two proteins are important markers for prognosis in gastric cancer. 23576552_Data indicate that knockdown of TEAD1/3/4 induces an almost identical cellular senescent phenotype as YAP silencing. 23780915_convergent optimization of the YAP/TAZ TEAD binding site suggests that the similarity in the affinities of binding of YAP to TEAD and of TAZ to TEAD is important for Hippo pathway functionality. 24325916_the multilevel perturbations of TEAD4 at epigenetic, transcriptional and posttranslational levels may contribute to GC development. 24520353_Edg-1 is a potential target gene of RTEF-1 and is involved in RTEF-1-induced angiogenesis in endothelial cells. 25628125_Our findings suggest that genetic variants of Hippo pathway genes, particularly YAP1 rs11225163, TEAD1 rs7944031 and TEAD4 rs1990330, may independently or jointly modulate survival of CM patients. 25687649_the peptides TEF3-11-66 and TEF3-1197-434 functioned as two independent activation domains, suggesting that N-terminal domain of TEF3-1 also has transcriptional activation capacity 25970772_TEAD4 and KLF5, in collaboration, promoted triple negative breast cancer cell proliferation and tumor growth in part by inhibiting p27 gene transcription 25995450_TAZ negatively regulate transcription of DeltaNp63 through TEAD1,2,3 and 4 transcription factors. 26041389_potential anti-oxidation gene and can prevent H2O2-induced endothelial cell oxidative damage by activating Klotho 26387538_The transcription factor TEAD4 regulates a pro-metastasis transcription program in a YAP-independent manner in CRC, thus providing a novel mechanism of TEAD4 transcriptional regulation and its oncogenic role in CRC, independently of the Hippo pathway. 26487755_TEAD4 overexpression induced p16 in HAoSMCs homozygous for the nonrisk coronary disease allele, but not for the risk allele. 26832411_Tead4 cooperates with AP1 transcription factors to coordinate target gene transcription. 26885617_High TEF3 expression is associated with cell cycle progression and angiogenesis in colon cancer. 27291620_TEAD4, the transcription factor that mediates Hippo-YAP signalling, undergoes alternative splicing facilitated by the tumour suppressor RBM4. 27966820_Our results suggest that TEAD4 plays a role in the pathophysiology of atypical teratoid/rhabdoid tumor, which represents a new insight into the biology of this aggressive tumor 28051067_Hippo pathway transcription factor TEAD4 directly associates with the Wnt pathway transcription factor TCF4 via their DNA-binding domains, forming a complex on target genes. VGLL4 binds to this TEAD4-TCF4 complex to inhibit transactivation of both TCF4 and TEAD4. 28077648_Collectively, these results indicate that human papillomavirus 16 E6 induces upregulation of APOBEC3B through increased levels of TEADs, highlighting the importance of the TEAD-APOBEC3B axis in carcinogenesis. 28315328_It was found that the TEAD4-YAP complex in the nuclei may be related closely to transcriptions of G1 arrest-related genes. 28368398_our work provides a structural basis for understanding the regulatory mechanism of TEAD4-mediated gene transcription 28430104_Combining single site-directed mutagenesis and double mutant analyses, the authors conduct a detailed analysis on the role of several residues located at the YAP:TEAD interface. The results provide quantitative understanding of the interactions taking place at the YAP:TEAD interface and give insights into the formation of the YAP:TEAD complex and more particularly on the interaction between TEAD and the ohm-loop found ... 28752853_Osmotic stress promotes TEAD4 cytoplasmic translocation via p38 MAPK in a Hippo-independent manner. Stress-induced TEAD inhibition predominates YAP-activating signals and selectively suppresses YAP-driven cancer cell growth. 28759040_TEAD1 and TEAD4 are oncogenic factors, whose aberrant activation are, in part, mediated by the silence of miR-377-3p, miR-1343-3p and miR-4269. 28960584_Studied the effect of TEAD4 acylation on its interaction with YAP and TAZ; found YAP and TAZ bind in a similar manner to both acylated and non-acylated TEAD4. Also found TEAD4 acylation significantly enhances its stability. 29157094_TEAD4 plays an important tumor-promoting role in colorectal cancer by directly targeting the YAP1. 29667772_TEAD4 is up-regulated in lung adenocarcinoma and its expression is an unfavourable prognostic factor.TEAD4 expression is regulated by miR-6839-3p. 29766649_LOX nuclear localization was significantly associated with poor survival in patients with Colorectal cancer (CRC). Nuclear LOX expression was correlated with synchronous or postoperative lung/hepatic metastasis. LOX may prove to be a potential target gene of YAP and TEAD4. 30392265_To analyze the clinical characteristics, treatment methods and prognosis of renal cell carcinoma associated with Xp11.2 translocation/TFE3 gene fusions 30794805_High TEAD4 expression is associated with Hepatoblastoma. 31138678_Interaction proteomics revealed that VGLL3 bound TEAD1, TEAD3 and TEAD4 in myoblasts and/or myotubes. 31289134_The activation of TEAD4 by glucocorticoids promoted breast cancer stem cells maintenance, cell survival, metastasis, and chemoresistance.TEAD4 expression positively correlates with glucocorticoid receptor expression in breast cancer. 31303470_data reveal a non-canonical function of YAP1 and TEAD4 as ERalpha cofactors in regulating cancer growth, highlighting the potential of YAP/TEAD as possible actionable drug targets for ERalpha(+) breast cancer. 31733305_YAP1, TEF3 and DSCR1-1L is a common signaling pathway downstream of several angiogenic factors that regulates angiogenesis. 31777916_The results demonstrated that the genomic interval on chromosome 19 is required for the appropriate level of Tead4 expression in blastocysts and suggested that an inter-chromosomal enhancer-promoter interaction may be the underlying mechanism. 31924214_TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. 32286937_Rational Design and Intramolecular Cyclization of Hotspot Peptide Segments at YAP-TEAD4 Complex Interface. 32378727_Expression and prognostic significance of YAP, TAZ, TEAD4 and p73 in human laryngeal cancer. 32800942_TEAD4 promotes tumor development in patients with lung adenocarcinoma via ERK signaling pathway. 32962824_TEAD4 transcriptional regulates SERPINB3/4 and affect crosstalk between keratinocytes and T cells in psoriasis. 33069758_VGLL1 phosphorylation and activation promotes gastric cancer malignancy via TGF-beta/ERK/RSK2 signaling. 33517828_The interaction of TEA domain transcription factor 4 (TEAD4) and Yes-associated protein 1 (YAP1) promoted the malignant process mediated by serum/glucocorticoid regulated kinase 1 (SGK1). 33713163_RTEF-1 Inhibits Vascular Smooth Muscle Cell Calcification through Regulating Wnt/beta-Catenin Signaling Pathway. 34003522_YAP/TEAD4-induced KIF4A contributes to the progression and worse prognosis of esophageal squamous cell carcinoma. 34030484_TEAD4 is an Immune Regulating-Related Prognostic Biomarker for Bladder Cancer and Possesses Generalization Value in Pan-Cancer. 34075638_Biochemical properties of VGLL4 from Homo sapiens and Tgi from Drosophila melanogaster and possible biological implications. 34196069_The transcription factor TEAD4 enhances lung adenocarcinoma progression through enhancing PKM2 mediated glycolysis. 34396425_HOXB13 suppresses proliferation, migration and invasion, and promotes apoptosis of gastric cancer cells through transcriptional activation of VGLL4 to inhibit the involvement of TEAD4 in the Hippo signaling pathway. 34569056_TEA domain transcription factor 4 modulates repression of fetal haemoglobin by direct binding to the gamma-globin gene promoters. 34951409_TEAD4 overexpression suppresses thyroid cancer progression and metastasis in vitro by modulating Wnt signaling. 35062042_TAZ promotes vasculogenic mimicry in gastric cancer through the upregulation of TEAD4. 35065165_Pan-cancer analysis, cell and animal experiments revealing TEAD4 as a tumor promoter in ccRCC. | ENSMUSG00000030353 | Tead4 | 1.690102e+03 | 1.3521133 | 0.435216044 | 0.3131728 | 1.921616e+00 | 0.1656786641 | 0.77593452 | No | Yes | 1.967021e+03 | 263.162655 | 1.204627e+03 | 165.833085 | |
| ENSG00000197943 | 5336 | PLCG2 | protein_coding | P16885 | FUNCTION: The production of the second messenger molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) is mediated by activated phosphatidylinositol-specific phospholipase C enzymes. It is a crucial enzyme in transmembrane signaling. {ECO:0000269|PubMed:23000145}. | 3D-structure;Calcium;Disease variant;Hydrolase;Lipid degradation;Lipid metabolism;Phosphoprotein;Reference proteome;Repeat;SH2 domain;SH3 domain;Transducer | The protein encoded by this gene is a transmembrane signaling enzyme that catalyzes the conversion of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate to 1D-myo-inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) using calcium as a cofactor. IP3 and DAG are second messenger molecules important for transmitting signals from growth factor receptors and immune system receptors across the cell membrane. Mutations in this gene have been found in autoinflammation, antibody deficiency, and immune dysregulation syndrome and familial cold autoinflammatory syndrome 3. [provided by RefSeq, Mar 2014]. | hsa:5336; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; intracellular vesicle [GO:0097708]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; phosphatidylinositol phospholipase C activity [GO:0004435]; phospholipase C activity [GO:0004629]; phosphorylation-dependent protein binding [GO:0140031]; phosphotyrosine residue binding [GO:0001784]; protein kinase binding [GO:0019901]; protein tyrosine kinase binding [GO:1990782]; scaffold protein binding [GO:0097110]; activation of store-operated calcium channel activity [GO:0032237]; antifungal innate immune response [GO:0061760]; B cell differentiation [GO:0030183]; B cell receptor signaling pathway [GO:0050853]; calcium-mediated signaling [GO:0019722]; cell activation [GO:0001775]; cellular response to calcium ion [GO:0071277]; cellular response to lectin [GO:1990858]; cellular response to lipid [GO:0071396]; Fc-epsilon receptor signaling pathway [GO:0038095]; follicular B cell differentiation [GO:0002316]; inositol trisphosphate biosynthetic process [GO:0032959]; intracellular signal transduction [GO:0035556]; lipopolysaccharide-mediated signaling pathway [GO:0031663]; macrophage activation involved in immune response [GO:0002281]; negative regulation of programmed cell death [GO:0043069]; phosphatidylinositol biosynthetic process [GO:0006661]; phospholipid catabolic process [GO:0009395]; platelet activation [GO:0030168]; positive regulation of calcium-mediated signaling [GO:0050850]; positive regulation of cell cycle G1/S phase transition [GO:1902808]; positive regulation of dendritic cell cytokine production [GO:0002732]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of gene expression [GO:0010628]; positive regulation of I-kappaB phosphorylation [GO:1903721]; positive regulation of interleukin-10 production [GO:0032733]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of interleukin-2 production [GO:0032743]; positive regulation of interleukin-23 production [GO:0032747]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of macrophage cytokine production [GO:0060907]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of neuroinflammatory response [GO:0150078]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NLRP3 inflammasome complex assembly [GO:1900227]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of phagocytosis, engulfment [GO:0060100]; positive regulation of reactive oxygen species biosynthetic process [GO:1903428]; positive regulation of receptor internalization [GO:0002092]; positive regulation of tumor necrosis factor production [GO:0032760]; positive regulation of type I interferon production [GO:0032481]; regulation of calcineurin-NFAT signaling cascade [GO:0070884]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; regulation of lipid metabolic process [GO:0019216]; release of sequestered calcium ion into cytosol [GO:0051209]; response to axon injury [GO:0048678]; response to yeast [GO:0001878]; stimulatory C-type lectin receptor signaling pathway [GO:0002223]; T cell receptor signaling pathway [GO:0050852]; toll-like receptor signaling pathway [GO:0002224]; Wnt signaling pathway [GO:0016055] | 12049640_collagen receptor glycoprotein VI and alphaIIbbeta3 trigger distinct patterns of receptor signalling in platelets, leading to tyrosine phosphorylation of PLCgamma2 (integrin alphaiibbeta3) 12181444_Two tyrosine residues in regulating the activity of PLCgamma2 12359094_full-length cDNA for human PLCgamma2 and expressed it in E. coli using the expression vector pT5T 12813055_PLCG2 has a signaling role in platelet glycoprotein Ib alpha calcium flux and cytoskeletal reorganization 14606067_in gastric cancer, protein translocation of PLCgamma2 and PKCalpha is critical event in the process of apoptosis induction. 15509800_PLC-gamma2 is phosphorylated on Y753, Y759, and Y1217 in response to engagement of the B-cell receptor 15744341_The PLCgamma2 is present in the majority of mediastinal B cell lymphomas. 15972651_PLC-gamma1 and PLC-gamma2 both regulate the functions of ITAM-containing receptors, whereas only PLC-gamma2 regulates the function of DAP10-coupled receptors. 16172125_novel mechanism of PLCgamma(2) activation by Rac GTPases involving neither protein tyrosine phosphorylation nor PI3K-mediated generation of PtdInsP(3) 17023658_observations suggest a model in which TFII-I suppresses agonist-induced calcium entry by competing with TRPC3 for binding to phospholipase C-gamma 18000612_intracellular mediators and pathways activated by leptin downstream of JAK2 were found to include phosphatidylinositol-3 kinase, phospholipase Cgamma2 and protein kinase C, as well as the p38 MAP kinase-phospholipase A(2) axis. 18022864_Plasmacytoid dendritic cells express a signalosome consisting of Lyn, Syk, Btk, Slp65 (Blnk) and PLCgamma2. Triggering CD303 leads to tyrosine phosphorylation of Syk, Slp65, PLCgamma2 & cytoskeletal proteins. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18728011_rac regulates its effector phospholipase Cgamma2 through interaction with a split pleckstrin homology domain 19086053_Observational study of gene-disease association. (HuGE Navigator) 19308021_Observational study of gene-disease association. (HuGE Navigator) 19965664_Data show that RTX treatment results in a time-dependent inhibition of the BCR-signaling cascade involving Lyn, Syk, PLC gamma 2, Akt, and ERK, and calcium mobilization. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20068106_SYK, together with phospholipase Cgamma2, may serve as potential biomarkers to predict dasatinib therapeutic response in patients. 20086178_Data show that bile acid reflux present in patients with BE may increase reactive oxygen species production and cell proliferation via activation of PI-PLCgamma2, ERK2 MAP kinase, and NADPH oxidase NOX5-S, thereby contributing to the development of EA. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21245382_Rac2 binding in the absence of lipid surfaces was not able to activate phospholipase C gamma 2. 21506110_Data indicate a role for PLCgamma2 and Ca(2+) signaling through the modulation of MEK/ERK in IL3/GM-csf stimulated human hematopoietic stem/progenitor cells. 22236196_Genomic deletions in PLCG2 cause gain of PLCgamma(2) function, leading to signaling abnormalities in multiple leukocyte subsets and a phenotype encompassing both excessive and deficient immune function. 22837484_PLCgamma2 participates in T cell receptor (TCR) signal transduction and plays a role in T cell selection in a transgenic mouse model. 23000145_Overexpression of the altered p.Ser707Tyr protein and ex vivo experiments using affected individuals' leukocytes showed clearly enhanced PLCgamma2 activity. 23039362_Associations between treatment response and Lyn, Syk, PLCgamma2 and ERK were not found. 23555801_BANK1 and BLK have roles in B-cell signaling through phospholipase C gamma 2 24080446_Single-nucleotide polymorphisms in PLCG2 gene is associated with breast cancer risk after menopausal hormone replacement therapy. 24127488_down-regulation of PLCgamma2-beta-catenin pathway occurs in mice and humans and leads to myeloid-derived suppressor cells-mediated tumor expansion. 24166973_early Ca(2+) fluxing provides feed-forward signal amplification by promoting anchoring of the PLCgamma2 C2 domain to phospho-SLP65. 24489640_The relationship between upstream tyrosine kinase SYK and its target, PLCgamma2, is maximally predictive and sufficient to distinguish chronic lymphocytic leukemia from healthy controls. 24868545_amarogentin prevents platelet activation through the inhibition of PLC gamma2-PKC cascade and MAPK pathway 24869598_identified three distinct mutations in PLCgamma2 in two patients resistant to ibrutinib 25012946_Data show that phospholipase Cgamma2 (PLCgamma2) is strongly expressed in B cell non-Hodgkin lymphoma and especially in a large subset of Diffuse large B-cell lymphoma (DLBCL). 25227611_The autoinhibitory C-terminal SH2 domain of phospholipase C-gamma2 stabilizes B cell receptor signalosome assembly. 25349203_PLCG2 missense mutation is a risk factor in the development of steroid sensitive nephrotic syndrome in childhood. 25972157_Characterization of the effect of missense point-mutation at R665W in PLCG2 on signaling mechanisms of ibrutinib resistance in chronic lymphocytic leukemia cells. 27196803_The results suggest a new mechanism of PLCgamma activation with unique thermodynamic features and assign a novel regulatory role to its spPH domain. 27442322_Ocular manifestations of phospholipase-Cgamma2-associated antibody deficiency and immune dysregulation show mutations in the PLC[gamma]2 gene leading to aberrant function of immune cells and overproduction of interleukin-1 [beta] (IL-1[beta]). 27542411_R665W and L845F be referred to as allomorphic rather than hypermorphic mutations of PLCG2 Rerouting of the transmembrane signals emanating from BCR and converging on PLCgamma2 through Rac in ibrutinib-resistant CLL cells may provide novel drug treatment strategies to overcome ibrutinib resistance mediated by PLCG2 mutations or to prevent its development in ibrutinib-treated CLL patients. 28366935_finding that mutations or polymorphisms in two putative calcium-regulated domains of PLCG2 are associated with ibrutinib-resistant CLL adds to the evidence supporting complex regulatory shifts in the PLCG2 protein likely occurring during the development of resistance 28714976_Data show that protein-altering changes are in PLCG2, ABI3, and TREM2 genes highly expressed in microglia and highlight an immune-related protein-protein interaction network in Alzheimer's disease. 28786489_Syk-induced signals in bone marrow stromal cell lines are mediated by phospholipase C gamma1 (PLCgamma1) in osteogenesis and PLCgamma2 in adipogenesis. 29381098_While BTK/PLCG2 mutations have characteristics suggesting that they can drive ibrutinib resistance, this conclusion remains formally unproven until specific inhibition of such mutations is shown to cause regression of ibrutinib-resistant chronic lymphocytic leukemia . Data suggest that alternative mechanisms of resistance do exist in some patients. 30326945_suggest distinct effects of the microglial genes, ABI3 and PLCG2 in neurodegenerative diseases that harbor significant vs. low/no amyloid ss pathology 30705288_Study shows that rare coding variants in TREM2, PLCG2, and ABI3 modulate susceptibility to Alzheimer disease in populations from Argentina, and they may have a European heritage. 31131421_The effect of the PLCG2 rs72824905-G on 7 neurodegenerative diseases and longevity, was studied in 53,627 patients, 3,516 long-lived individuals and 149,290 study-matched controls. It was associated with reduced Alzheimer disease, Lewy-body dementia, and frontotemporal dementia. It had no effect on Parkinson disease, amyotrophic lateral sclerosis or multiple sclerosis. It was also associated with longevity. 31560769_We performed pathway analyses on the largest available collection of advanced age-related macular cases and controls in the world. Eight genes strongly contributed to significant pathways from the three larger databases, and one gene (PLCG2) was central to significant pathways from all four databases. This is, to our knowledge, the first study to identify PLCG2 as a candidate gene for AMD based solely on genetic burden. 32014489_A new report of autoinflammation and PLCG2-associated antibody deficiency and immune dysregulation (APLAID) with a homozygous pattern from Iran. 32166339_PLCG2 protective variant p.P522R modulates tau pathology and disease progression in patients with mild cognitive impairment. 32184360_Noncatalytic Bruton's tyrosine kinase activates PLCgamma2 variants mediating ibrutinib resistance in human chronic lymphocytic leukemia cells. 32232486_Novel BCL2 mutations in venetoclax-resistant, ibrutinib-resistant CLL patients with BTK/PLCG2 mutations. 32514138_Alzheimer's-associated PLCgamma2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. 32671674_Severe Autoinflammatory Manifestations and Antibody Deficiency Due to Novel Hypermorphic PLCG2 Mutations. 32894242_Examination of the Effect of Rare Variants in TREM2, ABI3, and PLCG2 in LOAD Through Multiple Phenotypes. 32917267_The Alzheimer's disease-associated protective Plcgamma2-P522R variant promotes immune functions. 33089525_A novel somatic PLCG2 variant associated with resistance to BTK and SYK inhibition in chronic lymphocytic leukemia. 33092647_Association of ABI3 and PLCG2 missense variants with disease risk and neuropathology in Lewy body disease and progressive supranuclear palsy. 33523007_PLCG2 rs72824905 Variant Reduces the Risk of Alzheimer's Disease and Multiple Sclerosis. 33645887_Neutrophil Phospholipase Cgamma2 Drives Autoantibody-Induced Arthritis Through the Generation of the Inflammatory Microenvironment. 33823896_TREM2/PLCgamma2 signalling in immune cells: function, structural insight, and potential therapeutic modulation. 34093563_Case Report: A Rare Case of Autoinflammatory Phospholipase Cgamma2 (PLCgamma2)-Associated Antibody Deficiency and Immune Dysregulation Complicated With Gangrenous Pyoderma and Literature Review. 34157287_The role of PLCgamma2 in immunological disorders, cancer, and neurodegeneration. 34607960_Phospholipase Cgamma2 regulates endocannabinoid and eicosanoid networks in innate immune cells. 34615897_PLCgamma2 regulates TREM2 signalling and integrin-mediated adhesion and migration of human iPSC-derived macrophages. 34653364_Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. 35180881_PLCG2 is associated with the inflammatory response and is induced by amyloid plaques in Alzheimer's disease. 35196427_Mechanisms of Resistance to Noncovalent Bruton's Tyrosine Kinase Inhibitors. | ENSMUSG00000034330 | Plcg2 | 6.018773e+02 | 0.8288997 | -0.270730528 | 0.2866632 | 8.945255e-01 | 0.3442538886 | 0.82878860 | No | Yes | 4.177363e+02 | 45.626708 | 5.207796e+02 | 58.350820 | |
| ENSG00000197965 | 9019 | MPZL1 | protein_coding | O95297 | FUNCTION: Cell surface receptor, which is involved in signal transduction processes. Recruits PTPN11/SHP-2 to the cell membrane and is a putative substrate of PTPN11/SHP-2. Is a major receptor for concanavalin-A (ConA) and is involved in cellular signaling induced by ConA, which probably includes Src family tyrosine-protein kinases. Isoform 3 seems to have a dominant negative role; it blocks tyrosine phosphorylation of MPZL1 induced by ConA. Isoform 1, but not isoform 2 and isoform 3, may be involved in regulation of integrin-mediated cell motility. {ECO:0000269|PubMed:11751924, ECO:0000269|PubMed:12410637}. | 3D-structure;Alternative splicing;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phosphoprotein;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:9019; | cell surface [GO:0009986]; focal adhesion [GO:0005925]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; structural molecule activity [GO:0005198]; cell-cell signaling [GO:0007267]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11751924_PZR is a major receptor of ConA and has an important role in cell signaling via c-Src. Considering the various biological activities of ConA, the study of PZR may have major therapeutic implications 12684038_Characterization of PZR1b, an alternative spliced isoform of PZR. 16702974_Observational study of gene-disease association. (HuGE Navigator) 16702974_the MPZL1/PZR gene may be important in the predisposition to schizophrenia among Han Chinese 18568953_Phosphorylation and localization of PZR in cultured endothelial cells is reported. 19064610_Observational study of gene-disease association. (HuGE Navigator) 19536175_Observational study of gene-disease association. (HuGE Navigator) 19629567_Clinico-electrophysiological features and MRI findings are described in leg musculature from three patients belonging to a CMT2J pedigree due to MPZ Thr124Met mutation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24296779_MPZL1 is a target gene within the 1q24.1-24.2 amplicon that plays a pivotal role in HCC cell migration and tumor metastasis, and a novel MPZL1/Src/cortactin signaling cascade. 28912526_These data demonstrate that the formation of this MPZL1-PTPN11-GRB2 complex is triggered by the attachment of HER2+ breast cancer cells to fibronectin. 30392906_dimerization of MPZL1 participates in control of its signal transmission in cell adhesion. 30877754_Data indicate that myelin protein zero-like 1 (PZR) may promote the invasion and migration of colorectal cancer (CRC) cells through increasing the phosphorylation of focal adhesion kinase (FAK) and protooncogene SRC (Src). 31233194_High MPZL1 expression is associated with proliferation and metastasis of ovarian cancer. 31322261_The present study showed that the expression and protein levels of MPZL1 were significantly higher in gallbladder carcinoma tissues, especially in patients diagnosed with advanced tumor stages. | ENSMUSG00000026566 | Mpzl1 | 4.117349e+03 | 0.8761829 | -0.190696103 | 0.2603498 | 5.417120e-01 | 0.4617240984 | 0.86557407 | No | Yes | 3.308946e+03 | 279.661688 | 3.816578e+03 | 330.389510 | ||
| ENSG00000198176 | 7027 | TFDP1 | protein_coding | Q14186 | FUNCTION: Can stimulate E2F-dependent transcription. Binds DNA cooperatively with E2F family members through the E2 recognition site, 5'-TTTC[CG]CGC-3', found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication (PubMed:8405995, PubMed:7739537). The E2F1:DP complex appears to mediate both cell proliferation and apoptosis. Blocks adipocyte differentiation by repressing CEBPA binding to its target gene promoters (PubMed:20176812). {ECO:0000269|PubMed:20176812, ECO:0000269|PubMed:7739537, ECO:0000269|PubMed:8405995}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cell cycle;Cytoplasm;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a member of a family of transcription factors that heterodimerize with E2F proteins to enhance their DNA-binding activity and promote transcription from E2F target genes. The encoded protein functions as part of this complex to control the transcriptional activity of numerous genes involved in cell cycle progression from G1 to S phase. Alternative splicing results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 1, 15, and X.[provided by RefSeq, Jan 2009]. | hsa:7027; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; Rb-E2F complex [GO:0035189]; RNA polymerase II transcription regulator complex [GO:0090575]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; protein domain specific binding [GO:0019904]; anoikis [GO:0043276]; epidermis development [GO:0008544]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of fat cell proliferation [GO:0070345]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of DNA biosynthetic process [GO:2000278]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription involved in G1/S transition of mitotic cell cycle [GO:0000083]; transcription by RNA polymerase II [GO:0006366] | 12029633_TFDP1, CUL4A, and CDC16 are probable targets of an amplification mechanism and therefore may be involved, together or separately, in development and/or progression of some hepatocellular carcinomas 12607600_expression of the transcription factor DP-1 and its heterodimeric partner E2F-1 in non-Hodgkin lymphoma 14618416_TFDP1 may have a role in progression of some hepatocellular carcinomas by promoting growth of the tumor cells 15863509_DP-1alpha is a novel isoform of DP-1 that acts as a dominant-negative regulator of cell cycle progression 16135794_DP1 is a critical direct target of ARF. 18687693_SOCS-3 acts as a negative regulator of the cell cycle progression under E2F/DP-1 control by interfering with heterodimer formation between DP-1 and E2F 19738611_Observational study of gene-disease association. (HuGE Navigator) 19995430_13q34 amplification may be of relevance in tumor progression of breast cancers by inducing overexpression of CUL4A and TFDP1, important in cell cycle regulation. These genes were also overexpressed in non-basal-like tumor samples. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20513349_the DP-1 'Stabilon' domain was a C-terminal acidic motif and was quite important for DP-1 stability. 21715488_The authors demonstrate that adenovirus E1A binds to E2F/DP-1 complexes through a direct interaction with DP-1 and may selectively activate a subset of E2F-regulated cellular genes during infection. 23934193_somatic mutations in DP-1 uncouple normal control of the E2F pathway, and thus define a new mechanism that could contribute to aberrant proliferation in tumor cells 25133581_The TFDP1 indel84 mutation generates a gain-of-function phenotype by increasing cell proliferation, migration, and invasion of colorectal cancer cells. 26684807_Amplification of CUL4A, IRS2, and TFDP1 genes showed a significant difference in disease-free survival by both univariate and multivariate survival analyses in intrahepatic cholangiocarcinoma. 27323154_According to our study results, the gene TFDP1 and the cell cycle pathway are strongly associated with high-grade glioblastoma multiforme (GBM); this result may provide new insights into the pathogenesis of GBM. 27802335_role for E2F1 and TFDP1 in the transcriptional regulation of PITX1 in articular chondrocytes 27825926_Here, the authors show that an acidic region of DP1, whose function has remained elusive, binds to the plekstrin homology (PH) domain of the p62 subunit of TFIIH that contributes to transcriptional activation. 27871936_COMMD9 participates in TFDP1/E2F1 activation and plays a critical role in non-small cell lung cancer. 30365067_DILC may mediate the crosstalk between the cascades of IL6/STAT3 and TNFalpha signaling, indicating that DILC may act as a prognostic biomarker of sepsis, and may serve as a potential therapeutic target for the treatment of sepsis. 30638096_We observed a downregulation of TFDP1 in the endometrium cells of women with deep infiltrating endometriosis when compared to the controls 31783876_It uncovered E2F1 and TFDP1 as transport substrates of KPNA2 being retained in the cytoplasm upon KPNA2 ablation, thereby resulting in reduced STMN1 expression. 32066912_miR-4711-5p regulates cancer stemness and cell cycle progression via KLF5, MDM2 and TFDP1 in colon cancer cells. | ENSMUSG00000038482 | Tfdp1 | 2.910331e+03 | 1.2136008 | 0.279293912 | 0.2662184 | 1.109302e+00 | 0.2922336358 | 0.81275304 | No | Yes | 3.349682e+03 | 270.996287 | 2.567301e+03 | 213.003360 | |
| ENSG00000198252 | 6815 | STYX | protein_coding | Q8WUJ0 | FUNCTION: Catalytically inactive phosphatase (PubMed:23847209). Acts as a nuclear anchor for MAPK1/MAPK3 (ERK1/ERK2) (PubMed:23847209). Modulates cell-fate decisions and cell migration by spatiotemporal regulation of MAPK1/MAPK3 (ERK1/ERK2) (PubMed:23847209). By binding to the F-box of FBXW7, prevents the assembly of FBXW7 into the SCF E3 ubiquitin-protein ligase complex, and thereby inhibits degradation of its substrates (PubMed:28007894). Plays a role in spermatogenesis (By similarity). {ECO:0000250|UniProtKB:Q60969, ECO:0000269|PubMed:23847209, ECO:0000269|PubMed:28007894}. | 3D-structure;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome | Mouse_homologues NA; + ;NA | The protein encoded by this gene is a pseudophosphatase, able to bind potential substrates but lacking an active catalytic loop. The encoded protein may be involved in spermiogenesis. Two transcript variants encoding the same protein have been found for these genes. [provided by RefSeq, Oct 2011]. | hsa:6815; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; F-box domain binding [GO:1990444]; protein tyrosine/serine/threonine phosphatase activity [GO:0008138]; pseudophosphatase activity [GO:0001691]; MAPK export from nucleus [GO:0045204]; negative regulation of protein binding [GO:0032091]; negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process [GO:0062026]; regulation of ERK1 and ERK2 cascade [GO:0070372] | 19369647_Observational study of gene-disease association. (HuGE Navigator) 23847209_results identify STYX as an important regulator of ERK1/2 signaling critical for cell migration 28007894_The authors show that STYX binds to the F-box domain of FBXW7 and disables its recruitment into the SCF ubiquitin ligase complex. 28408485_Results suggest that STYX is a pseudophosphatase that uses the 'competitor' and 'anchor' mode of action to exert its biologic roles. [review] 30981757_STYX bound to the F-box and WD repeat domain-containing7 (FBXW7) protein and inhibited its function. Co-regulation of STYX and FBXW7 expression reversed the biological changes mediated by regulation of STYX expression alone in CRC cells. 32239181_STYX/FBXW7 axis participates in the development of endometrial cancer cell via Notch-mTOR signaling pathway. | ENSMUSG00000053205+ENSMUSG00000071748 | Styx+Styx-ps | 1.902700e+02 | 1.0550375 | 0.077294328 | 0.3642766 | 4.553242e-02 | 0.8310280197 | 0.96751127 | No | Yes | 2.117357e+02 | 43.768487 | 1.783160e+02 | 37.834969 |
| ENSG00000198586 | 9874 | TLK1 | protein_coding | Q9UKI8 | FUNCTION: Rapidly and transiently inhibited by phosphorylation following the generation of DNA double-stranded breaks during S-phase. This is cell cycle checkpoint and ATM-pathway dependent and appears to regulate processes involved in chromatin assembly. Isoform 3 phosphorylates and enhances the stability of the t-SNARE SNAP23, augmenting its assembly with syntaxin. Isoform 3 protects the cells from the ionizing radiation by facilitating the repair of DSBs. In vitro, phosphorylates histone H3 at 'Ser-10'. {ECO:0000269|PubMed:10523312, ECO:0000269|PubMed:10588641, ECO:0000269|PubMed:11314006, ECO:0000269|PubMed:11470414, ECO:0000269|PubMed:12660173, ECO:0000269|PubMed:9427565}. | ATP-binding;Acetylation;Alternative splicing;Cell cycle;Chromatin regulator;Coiled coil;DNA damage;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene is a serine/threonine kinase that may be involved in the regulation of chromatin assembly. The encoded protein is only active when it is phosphorylated, and this phosphorylation is cell cycle-dependent, with the maximal activity of this protein coming during S phase. The catalytic activity of this protein is diminished by DNA damage and by blockage of DNA replication. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2011]. | hsa:9874; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cell cycle [GO:0007049]; cellular response to DNA damage stimulus [GO:0006974]; chromatin organization [GO:0006325]; chromosome segregation [GO:0007059]; intracellular protein transport [GO:0006886]; intracellular signal transduction [GO:0035556]; peptidyl-serine phosphorylation [GO:0018105]; protein phosphorylation [GO:0006468]; regulation of chromatin assembly or disassembly [GO:0001672] | 12955071_we show that Chk1 is essential for the suppression of TLK activity after replication block, but that ATR, Chk2 and BRCA1 are dispensable for TLK suppression. 14732354_Both eIF4E and TLK1B are elevated in breast cancer specimens but not in benign breast specimens from noncancer patients. Degree of TLK1B elevation is correlated with degree of IF4E overexpression. 15950181_In addition, it could be demonstrated that increasing the Tlk1 activity in HT1080 cells by forced Tlk1 overexpression leads to an increased phosphorylation of endogenous p68. 16156902_TLK1B promotes the repair of double strand breaks, likely as a consequence of a change in chromatin remodeling capacity that must precede the assembly of repair complexes at the sites of damage 17054786_Studies provide evidence for TLK1B-mediated phosphorylation of Asf1 triggering DNA repair. 18838128_Our data suggest that human PKU-beta/TLK1 plays an important role in chromosome integrity via the regulation of myosin II dynamics by phosphorylating MRLC during mitosis. 20016786_ASF1 cellular levels are tightly controlled by distinct pathways and provide a molecular mechanism for post-translational regulation of dASF1 and hASF1a by TLK kinases. 20381954_Silencing of TLK1 enhanced DNA damage induced by cisplatin treatment, suggesting that TLK1 plays a pivotal role for the repair of cisplatin-induced DNA damage. 21048794_Adenoviral delivery of Tousled kinase protects salivary glands against ionizing radiation damage. 21647934_TLKs appear to be intimately linked to the pattern of resistance to DNA damage, and specifically double-strand breaks. 24376897_Tousled-like kinase-dependent phosphorylation of Rad9 plays a role in cell cycle progression and G2/M checkpoint exit. 24598821_Data indicate Tousled-like kinases (TLK1) phosphorylation has an impact on cell cycle proteins Asf1a and Asf1b function. 26855419_Our results suggest that GA-mediated transient modulation of TLK1 activity promotes DNA repair and suppresses radiation cytoxicity in salivary gland cells. 26860083_TLK1B mediated phosphorylation of Rad9 regulates its nuclear/cytoplasmic localization and cell cycle checkpoint 28426283_Following DNA damage, addition of the TLK1 inhibitor, THD, or overexpression of NEK1-T141A mutant impaired ATR and Chk1 activation, indicating the existence of a TLK1>NEK1>ATR>Chk1 pathway. Indeed, overexpression of the NEK1-T141A mutant resulted in an altered cell cycle response after exposure of cells to oxidative stress, including bypass of G1 arrest and implementation of an intra S-phase checkpoint. 30252587_Overexpression of TLK1 substantially reduces DNA damage and G2/M arrest by activation of TLK1-dependent cell cycle checkpoint response. 30737777_Targeting the TLK1/NEK1 axis might be a novel therapy for PCa. 30928383_Authors believe that this TLK1-Nek1 mediated DDR axis is likely to be a common adaptive response during the transition of PCa cells toward androgen-insensitive growth, and hence CRPC progression. 31311824_The levels of circular RNA TLK1 were significantly increased in the brain tissue and plasma of ischemic stroke patients. 31362519_Variation analysis of tousled like kinase 1 gene in patients with sporadic premature ovarian insufficiency. 31914854_The TLK1/Nek1 axis contributes to mitochondrial integrity and apoptosis prevention via phosphorylation of VDAC1. 32468002_Knockdown of Tousledlike kinase 1 inhibits survival of glioblastoma multiforme cells. 32755577_Tousled-Like Kinases Suppress Innate Immune Signaling Triggered by Alternative Lengthening of Telomeres. 34410682_CircTLK1 modulates sepsis-induced cardiomyocyte apoptosis via enhancing PARP1/HMGB1 axis-mediated mitochondrial DNA damage by sponging miR-17-5p. | ENSMUSG00000041997 | Tlk1 | 4.683178e+02 | 1.3163028 | 0.396491389 | 0.3438457 | 1.355894e+00 | 0.2442504048 | 0.78892886 | No | Yes | 5.193169e+02 | 110.194360 | 3.240868e+02 | 70.692747 | |
| ENSG00000198626 | 6262 | RYR2 | protein_coding | Q92736 | FUNCTION: Calcium channel that mediates the release of Ca(2+) from the sarcoplasmic reticulum into the cytoplasm and thereby plays a key role in triggering cardiac muscle contraction. Aberrant channel activation can lead to cardiac arrhythmia. In cardiac myocytes, calcium release is triggered by increased Ca(2+) levels due to activation of the L-type calcium channel CACNA1C. The calcium channel activity is modulated by formation of heterotetramers with RYR3. Required for cellular calcium ion homeostasis. Required for embryonic heart development. {ECO:0000269|PubMed:10830164, ECO:0000269|PubMed:20056922, ECO:0000269|PubMed:27733687, ECO:0000269|PubMed:33536282}. | 3D-structure;Alternative splicing;Calcium;Calcium channel;Calcium transport;Calmodulin-binding;Cardiomyopathy;Coiled coil;Developmental protein;Disease variant;Ion channel;Ion transport;Ligand-gated ion channel;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Sarcoplasmic reticulum;Transmembrane;Transmembrane helix;Transport | This gene encodes a ryanodine receptor found in cardiac muscle sarcoplasmic reticulum. The encoded protein is one of the components of a calcium channel, composed of a tetramer of the ryanodine receptor proteins and a tetramer of FK506 binding protein 1B proteins, that supplies calcium to cardiac muscle. Mutations in this gene are associated with stress-induced polymorphic ventricular tachycardia and arrhythmogenic right ventricular dysplasia. [provided by RefSeq, Jul 2008]. | hsa:6262; | calcium channel complex [GO:0034704]; cytoplasmic vesicle membrane [GO:0030659]; junctional sarcoplasmic reticulum membrane [GO:0014701]; membrane [GO:0016020]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; sarcolemma [GO:0042383]; sarcoplasmic reticulum [GO:0016529]; sarcoplasmic reticulum membrane [GO:0033017]; smooth endoplasmic reticulum [GO:0005790]; Z disc [GO:0030018]; calcium channel activity [GO:0005262]; calcium ion binding [GO:0005509]; calcium-induced calcium release activity [GO:0048763]; calcium-release channel activity [GO:0015278]; calmodulin binding [GO:0005516]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; protein kinase A catalytic subunit binding [GO:0034236]; protein kinase A regulatory subunit binding [GO:0034237]; protein self-association [GO:0043621]; ryanodine-sensitive calcium-release channel activity [GO:0005219]; suramin binding [GO:0043924]; transmembrane transporter binding [GO:0044325]; calcium ion transport [GO:0006816]; calcium ion transport into cytosol [GO:0060402]; calcium-mediated signaling [GO:0019722]; calcium-mediated signaling using intracellular calcium source [GO:0035584]; cardiac muscle contraction [GO:0060048]; cardiac muscle hypertrophy [GO:0003300]; cell communication by electrical coupling involved in cardiac conduction [GO:0086064]; cellular calcium ion homeostasis [GO:0006874]; cellular response to caffeine [GO:0071313]; cellular response to epinephrine stimulus [GO:0071872]; detection of calcium ion [GO:0005513]; embryonic heart tube morphogenesis [GO:0003143]; establishment of protein localization to endoplasmic reticulum [GO:0072599]; left ventricular cardiac muscle tissue morphogenesis [GO:0003220]; positive regulation of ATPase-coupled calcium transmembrane transporter activity [GO:1901896]; positive regulation of heart rate [GO:0010460]; positive regulation of sequestering of calcium ion [GO:0051284]; positive regulation of the force of heart contraction [GO:0098735]; Purkinje myocyte to ventricular cardiac muscle cell signaling [GO:0086029]; regulation of atrial cardiac muscle cell action potential [GO:0098910]; regulation of AV node cell action potential [GO:0098904]; regulation of cardiac muscle contraction [GO:0055117]; regulation of cardiac muscle contraction by calcium ion signaling [GO:0010882]; regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion [GO:0010881]; regulation of cytosolic calcium ion concentration [GO:0051480]; regulation of heart rate [GO:0002027]; regulation of SA node cell action potential [GO:0098907]; regulation of ventricular cardiac muscle cell action potential [GO:0098911]; release of sequestered calcium ion into cytosol [GO:0051209]; release of sequestered calcium ion into cytosol by sarcoplasmic reticulum [GO:0014808]; response to caffeine [GO:0031000]; response to hypoxia [GO:0001666]; response to muscle activity [GO:0014850]; response to muscle stretch [GO:0035994]; response to redox state [GO:0051775]; sarcoplasmic reticulum calcium ion transport [GO:0070296]; type B pancreatic cell apoptotic process [GO:0097050]; ventricular cardiac muscle cell action potential [GO:0086005] | 12015469_Mutations in the gene coding for cardiac ryanodine receptor (hRYR2) may be a key step in molecular pathogenesis of ARVDs, and catecholaminergic ventricular arrhythmias or with familial ventricular tachyarrhythmia. 12106942_RyR2 mutations are associated with polymorphic ventricular arrhythmia, syncope and sudden death in response to physical or emotional stress. 12446682_identification of binding site for FKBP12.6 protein 12459180_Data show that VTSIP- and ARVD2-associated point mutations influence positively and negatively, respectively, the binding of RyR2 to its gating protein FKBP12.6. 12576471_the DR3 region of Ryanodine Receptor 2 has a role in excitation-contraction coupling and in channel regulation 12754204_data demonstrate that defective regulation of ryanodine receptor 2 causes altered cellular phenotype via profound perturbations in intracellular calcium signaling and highlight a key modulatory role of FK506 binding protein 12.6 12919952_RYR2 mutations associated with ventricular tachycardia mediate increased calcium release in stimulated cardiomyocytes. 12959641_The RyR2 C-terminal tail may be important for the oligomeric assembly of the native channel. 14593104_the TM10 sequence constitutes an essential determinant for channel activation and gating; TM10 is an inner helix of RyR 14722100_there is a specific binding subdomain in RyR1 and RyR2 participating in RyR-RyR interaction 15016728_CaMKII-mediated phosphorylation of RyR2 at Ser-2815 may contribute to the enhanced contractility observed at higher heart rates. 15033925_This is the 1st evidence that RyRs directly control primary beta cell insulin secretion independently of glucose & by 2 mechanisms, including a novel cytosolic Ca2+-independent mechanism likely involving changes in Ca2+ in the lumens of non-ER organelles. 15044459_RyR2 activity seems to play an essential role in beta-cell survival in vitro by suppressing a death pathway mediated by calpain-10 15047862_compelling evidence that specific interaction between cytoplasmic and transmembrane domains is an important mechanism in the intrinsic modulation of RyR Ca(2+) release channels 15147738_CLIC2 inhibited cardiac ryanodine receptor Ca2+ release channels in lipid bilayers when added to the cytoplasmic side of the channels and inhibited Ca2+ release from cardiac sarcoplasmic reticulum vesicles 15197150_3 mutations (P2328S, Q4201R, & V4653F)had decreased FKBP12.6 binding, that stabilizes the channel's closed state. After PKA phosphorylation, mutant channels had a gain-of-function defect (leaky Ca2+ release) & a rightward shift in the Mg2+ IC50. 15215235_five non-conserved amino acids in the C-terminal region flanking the CaM-binding domain have a key role in CaM inhibition of RyR2 15466642_9 new RYR2 mutations were found among people with long-QT syndrome revealed by swimming-triggered arrhythmias. 15591045_novel interaction site for FKBP12.6 may be present at the RyR2 C terminus, proximal to the channel pore, a sterically appropriate location that would enable this protein to play a central role in the modulation of this critical ion channel 15749201_The R2401H mutation can be expected to alter Ca-induced Ca release and E-C coupling resulting in CPVT. 15851612_Atrial tissue from both dogs and humans with chronic atrial fibrillation showed a significant increase in PKA phosphorylation of RyR2, with a corresponding decrease in calstabin2 binding to the channel 15887426_RyR2-encoded cardiac ryanodine receptor/calcium release channel mutations are responsible for catecholaminergic polymorphic ventricular tachycardia (CPVT)- a novel pathogenic basis for unexplained drownings. 16188589_Observational study of gene-disease association. (HuGE Navigator) 16188589_Putative pathogenic type 1 catecholaminergic polymorphic ventricular tachycardia-causing mutations in RyR2 were detected in 6% of unrelated, genotype-negative long QT syndrome referrals. 16272262_RYR2 mutations may have a role in catecholaminergic polymorphic ventricular tachycardia 16483256_Ser-2030, but not Ser-2808, is the major PKA phosphorylation site in RyR2 responding to PKA activation upon beta-adrenergic stimulation in both normal and failing hearts, and that RyR2 is not hyperphosphorylated by PKA in heart failure. 16601229_Intracellular Ca2+ cycling in normal heart relies on intricate interplay of CASQ2 with proteins of RyR2 channel complex, and disruption of these interactions can lead to cardiac arrhythmia. 16769042_Observational study of gene-disease association. (HuGE Navigator) 16818210_Putative CPVT1-causing mutations in RyR2 were seen in <40% of unrelated patients referred with a diagnosis of CPVT and preferentially in males. 16825580_The knock-in mouse harboring the RyR2 R4497C mutation(also found in patients with familial sudden death) cause cardiac arrhythmias through delayed afterdepolarization and triggered activity due to altered intracellular calcium handling. 17052226_Cardiac RyR2 plasmids with various CPVT mutations enable expression and analysis of Ca2+ release mediated by the wild-type and mutated RyR2. 17062961_Postmortem molecular screening of the RyR2 gene could be useful for investigation for cause of death in sudden unexplained death (SUD). The possible association of the RyR2 mutation with status thymico-lymphaticus is discussed. 17200109_the redox state of the RyR is intimately connected with FKBP binding affinity. 17259277_together, our results demonstrate that RyR1 differs markedly from RyR2 with respect to their responses to Ca(2+) overload and luminal Ca(2+). 17313373_Data show that K201 abolished spontaneous calcium release in cardiac myocytes, and that treating ventricular myocytes with FK506 to dissociate FKBP12.6 from ryanodine receptor RyR2 did not affect the suppression of spontaneous Ca2+ release by K201. 17322175_We provide the first evidence that RyR2 splice variants exquisitely modulate intracellular Ca(2+) signaling and are key determinants of cardiomyocyte apoptotic susceptibility. 17330843_P2X(7) receptor has an antiapoptotic function in melanoma cells 17556193_Finds a link between RyR2 mutations and the alterations of ion channels that may trigger cardiac arrhythmias associated with SIDS. 17558603_No disease-causing mutations in the RyR2 were found in Finish patients with sporadic arrhythmogenic right ventricular cardiomyopathy; an allelic variant A1136V was found in patients and health controls. 17693412_PKA-dependent phosphorylation enhances the response of RyR2 to luminal Ca(2+) and reduces the threshold for SOICR and this effect of PKA is largely mediated by phosphorylation at Ser-2,030 17875969_large genomic deletion in RYR2 leads to extended clinical phenotypes eg, sinoatrial node and atrioventricular node dysfunction, atrial fibrillation, atrial standstill, and dilated cardiomyopathy 17921453_the loss of FKBP12.6 has no significant effect on the conduction and activation of RyR2 or the propensity for spontaneous Ca(2+) release and stress-induced ventricular arrhythmias 17967164_On the basis of the three-dimensional localizations of a number of residues or regions, a model for the subunit organization in the structure of RyR2 is proposed. 18262818_We found a novel mutation (V2321M) in exon 46 of the RyR2 gene in a sudden unexplained death case. 18586264_May play a role in calcium-dependent nuclear processes. 18618700_The results suggest major differences in the energetics of calmodulin binding to and dissociation from RyR1 (in skeletal muscle) and RyR2 (in cardiac muscle). 18929323_Patients with a RyR2 mutation show increased TPE at high heart rates 19216760_We report two novel CPVT-causing RyR2 mutations and a novel RyR2 variant of uncertain clinical significance in a patient with abundant resting ventricular premature complexes 19226252_The findings suggest that FKBP12.6 regulation of RyR2 is unlikely to be the primary defect in inherited arrhythmogenic cardiac disease. 19398665_Cardiac and fatal or near-fatal events were not rare in both catecholaminergic polymorphic ventricular tachycardia RYR2 and CASQ2 mutation probands and affected family members during the long-term follow-up 19626040_Observational study of gene-disease association. (HuGE Navigator) 19781797_We identified two novel RyR2 missense mutations (G2145R and R3570W) in three victims of sudden cardiac death 19913121_Observational study of gene-disease association. (HuGE Navigator) 19926015_Observational study of gene-disease association. (HuGE Navigator) 20045464_The domain analysis of the N-terminal region (residues 1-759) of the human cardiac ryanodine receptor, is reported. 20080988_The elevation of SR Ca(2+) load--in the absence of beta-adrenergic stimulation--is sufficient to increase the propensity for triggered arrhythmias in RyR2(R4496C+/-) cardiomyocytes 20106799_Exercise test revealed a high prevalence of arrhythmias in catecholaminergic polymorphic ventricular tachycardia mutation carriers diagnosed by cascade genetic screening 20132818_mutations in RyR2 are causative of inherited disorder which often results in sudden cardiac death; review of advances in the field, description of controversy surrounding the exact consequences of RyR2 mutation & how disparate data may be reconciled 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20408814_Observational study of gene-disease association. (HuGE Navigator) 20424473_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20676041_Identify a novel CPVT RyR2 mutation, E189D that results in catecholaminergic polymorphic ventricular tachycardia. 20716843_ryanodine receptor-2 mrna levels were reduced in cumulative rejection episodes 21151189_This study demonistrated that use joint test to identified that RYR22 association to Autism Spectrum Disorder. 21270101_RyR2 cardiac Ca(2+) release channel inhibition appears to determine efficacy of class I drugs for the prevention of catecholaminergic polymorphic ventricular tachycardia. 21315846_Mutations in the gene encoding the cardiac ryanodine receptor type 2 (RyR2) can be found in 50% to 55% of patients with clinical CPVT. 21531043_Alterations in RyR2 expression at early disease stages may reflect the onset of pathologic mechanisms leading to later neurodegeneration. 21652165_Genetic analysis revealed a novel missense mutation in exon 90 of the ryanodine receptor (RyR2) gene resulting in substitution of arginine for serine at residue 4153 (S4153R). 21659649_The RyR2-G230C mutant exhibits similar biophysical defects compared with previously characterized catecholaminergic polymorphic ventricular tachycardia mutations 21768539_S2246L causes an abnormally tight local subdomain-subdomain interaction in the central domain & unzipping of the N-terminal domain, activating RyR2 in a diastolic state reflecting on the increased Ca+2 spark frequency, causing lethal arrhythmia. 21872879_Two causative genes of CPVT have been identified: RYR2, encoding the cardiac ryanodine receptor (RyR2) Ca(2+) release channel, and CASQ2, encoding cardiac calsequestrin Their mutations have been found in 60% of patients with CPVT. 21881589_RyR expressed in epidermal keratinocytes is associated with both differentiation of keratinocytes and epidermal barrier homeostasis. 22178870_the suitability of iPS cells in modeling RYR2-related cardiac disorders 22222782_The results show that one variant (p.H4579Y) of the RYR2 gene co-segregates with catecholaminergic polymorphic ventricular tachycardia and is presumed to be pathogenic in a cohort of sudden unexplained death cases. 22374134_NH2-terminal region of RyR2 is an important determinant of Ca2+ release termination. Abnormal fractional Ca2+ release attributable to aberrant termination of Ca2+ release is a common defect in RyR2-associated cardiomyopathies. 22456474_Enhanced SR Ca(2+) leak thru CaMKII-hyperphosphorylated RyR2, plus larger I(NCX) for a given SR Ca(2+) release & increased diastolic [Ca(2+)](i)-voltage coupling gain, causes AF-promoting atrial delayed afterdepolarizations/triggered activity in cAF. 22511749_Suggest that increased CaMKII phosphorylation of RyR2 plays a role in the development of pathological sarcoplasmic reticulum Ca(2+) leak and heart failure development. 22519458_In addition, RyR2 gene coding variants were found in 50% of affected individuals. 22608700_catecholaminergic polymorphic ventricular tachycardia associated with a novel mutation of the ryanodine receptor 2 22787013_Familial evaluation in catecholaminergic polymorphic ventricular tachycardia showed relatives carrying an Ryr2 mutation have a marked phenotypic diversity. 22802361_Results demonstrate several novel aspects of RyR2 gating behavior. 22948152_RyR1, RyR2, and RyR3 transcripts were detected in human T cells, RyR1/2 transcript levels increased, whereas those of RyR3 decreased after T cell activation. 23094885_The cardiac ryanodine receptor gene contains most of the catecholaminergic polymorphic ventricular tachycardia mutations. 23152493_A novel RyR2-Val2475Phe mutation associated with catecholaminergic polymorphic ventricular tachycardia in humans triggers calcium ion (Ca2+)-dependent arrhythmias in whole hearts and intact mice. 23258540_Ligand-dependent conformational changes in the clamp region of the cardiac ryanodine receptor. 23498838_S4153R mutation is a gain-of-function RYR2 mutation associated with a clinical phenotype characterized by both catecholaminergic polymorphic ventricular tachycardia and atrial fibrillation. 23516528_the chronic gain-of-function defect in RyR2 due to CaMKII hyperphosphorylation is a novel mechanism that contributes to pathogenesis of type 2 diabetes 23595086_Genetic background of catecholaminergic polymorphic ventricular tachycardia in Japan. 23632022_introducing alanine at position 4864 produces no significant change in RyR2 function. In contrast, function is altered when glycine 4864 is replaced by either valine or proline, the former preventing channel opening. 23651034_Of a total of 715 Sudden cardiac death cases, seven (1.0%) carried one of the ten mutations assayed: three carried KCNH2 R176W, one KCNH2 L552S, two PKP2 Q59L, and one RYR2 R3570W. 23829686_Rank-based genome-wide analysis revealed for the first time an association of RYR2 variants with asthma. 23848845_Soluble hRyR2(1-606) was expressed in Escherichia coli. Purification conditions were optimized using thermal shift assay. The quality and stability of the sample was probed by dynamic light scattering. 23943880_RyR2 N-terminal fragment possesses the intrinsic ability to oligomerize, enabling apparent tetramer formation. 24147812_Mutation in RyR2 gene causes diastolic leakage of calcium cytosol and diastolic depolarization of cell's membrane, triggering polymorphic ventricular tachycardia. 24285081_It mediates calcium release from intracellular calcium stores such as the ER into the cytoplasm.(review) 24394973_RYR2 exon 3 deletion is frequently associated with left ventricular non-compaction. 24445321_Calcium-dependent cardiomyopathy is exacerbated by genetic ablation of ryanodine receptor 2. 24447446_Previously reported plausible pathogenic missense polymorphism G1886S may not be an independent predisposition factor of sudden unexplained nocturnal death syndrome in the southern Chinese Han population. 24786399_Dual-tilt electron tomography showed ventricular RYR2 tetramer packing within a dyad was nonuniform containing a mix of checkerboard and side-by-side arrangements, as well as isolated tetramers. 24793461_Mutations in genes encoding cardiac ryanodine receptor 2 (RyR2) have been identified in several patients and are recognized as causing the autosomal dominant and recessive forms of CPVT 24805197_The S96A HRC mutation disrupts the Ca2+ -microdomain around the RyR2, as it alters the Ca2+ -dependent association of RyR2 and HRC. 24837260_CPVT is an inherited cardiac disorder in which patients have structurally normal hearts but increased risk of stress-induced ventricular arrhythmias because of mutations in the cardiac 'ryanodine receptor 2' (RyR2; CPVT1) 24929545_Rotavirus mimics human RYR2 by producing VP6 protein having high degree of homology towards RYR2 and significant antigenicity with respect to myasthenia gravis associated HLA haplotypes. 24950728_Data showed RyR2 on two novel familial compound mutations, c.6224T>C and c.13781A>G, with the clinical presentation of idiopathic ventricular fibrillation. 24978818_Three novel RYR2 missense mutations have been described in a Kazakh idiopathic ventricular tachycardia study cohort. 25036739_dysregulation of RyR2-mediated Ca(2+) release via aberrant CaM(F90L)-RyR2 interaction is a potential mechanism that underlies familial IVF 25041964_RYR2 mutations are frequent (9% of ARVC/D probands) and are associated with a conventional phenotype of ARVC/D. 25370123_SPRY2 domain forms two antiparallel beta sheets establishing a core, and four additional modules of which several are required for proper folding 25372681_Here the solution and crystal structures determined under near-physiological conditions, as well as a homology model of the human RyR2 N-terminal region, are presented. 25389315_Suggest that miR-106b-25 cluster-mediated post-transcriptional regulation of RyR2 is a potential molecular mechanism involved in paroxysmal atrial fibrillation pathogenesis. 25435091_RYR2 C2277R mutation is a cause of catecholaminergic polymorphic ventricular tachycardia in a family with high lethality in younger individuals. 25440180_functional heterologous expression studies suggest that the RyR2(R420Q) behaves as an aberrant channel, as a loss- or gain-of-function mutation depending on cytosolic intracellular Ca(2+) concentration. 25456695_Catecholaminergic polymorphic ventricular tachycardia (CPVT) initially diagnosed as idiopathic ventricular fibrillation: the importance of thorough diagnostic work-up and follow-up. 25627681_data shed new insights into the structure-function relationship of the NH2-terminal domains of RyR2 and the action of NH2-terminal disease mutations 25648700_The principal action of flecainide in CPVT is not via a direct interaction with RyR2. Suggest model of flecainide action in which Na(+)-dependent modulation of intracellular Ca(2+) handling attenuates RyR2 dysfunction in CPVT. 25753936_The results of our pilot study suggest that the rs2819742 variant within the gene for the RYR2 receptor could be associated with statin-induced myalgia/myopathy in patients on low doses of common statins. 25773045_Our study identifies a significant role of RyR2 rs3766871 minor allele for increased susceptibility to VT/VF in a population of implanted with a cardioverter defibrillator patients with heart failure. 25775566_analysis of arrhythmia mechanisms in a RyR2-linked CPVT mutation (RyR2-A4860G) that depresses channel activity 25814417_G357S_RyR2 mutation in the cardiac ryanodine receptor was identified in 179 family members with Catecholaminergic polymorphic ventricular tachycardia and in 6 Sudden cardiac death cases. 25835811_A deletion of exon 3 of the RYR2 gene was found in a family with catecholaminergic polymorphic ventricular tachycardia. 25844899_In transgenic mice, CPVT-associated RyR2 impaired glucose homeostasis. Pancreatic islets had intracellular Ca2+ leak via oxidized and nitrosylated RyR2 channels, activated ER stress response, mitochondrial dysfunction, and decreased insulin release. 25925909_We propose a mechanism in which RYR2 sequence variants result in aberrant stress-induced calcium release into the mitochondria of autonomic neurons, resulting in an increased risk to develop autonomic/functional disease such as cyclic vomiting syndrome. 25966694_Studies indicate that the ryanodine receptors (RyRs: RyR1, RyR2, RyR3) and inositol 1,4,5-trisphosphate receptors (IP3Rs: IP3R1, IP3R2, IP3R3) are the major Ca(2+) release channels (CRCs) on the endo/sarcoplasmic reticulum (ER/SR). 26009179_unlike RyR1 normal WT RyR2 does not bind dantrolene in spite of the fact that the RyR1 dantrolene-binding site (residues 590-609) is conserved in RyR2 26009186_How phosphorylation of RyR affects channel activity and whether proteins such as the FK-506 binding proteins (FKBP12 and FKBP12.6) are involved in heart failure 26114861_Half of the RYR2 mutations in catecholaminergic polymorphic ventricular tachycardia cohort were de novo, and most of the remaining mutations were inherited from mothers. 26153920_induced Pluripotent Stem Cell-derived cardiomyocytes are useful for investigating the similarities/differences in the pathophysiological consequences of RyR2 versus CASQ2 mutations underlying Catecholaminergic polymorphic ventricular tachycardia. 26309258_results support aberrant RyR2 regulation as the disease mechanism for CPVT associated with CaM mutations and shows that CaM mutations not associated with CPVT can also affect RyR2 26663082_although the EF-hand domain is not required for RyR2 activation by cytosolic Ca(2+), it plays an important role in luminal Ca(2+) activation and SOICR 26742492_RYR2, PTDSS1 and AREG are autism susceptibility genes that are implicated in a Lebanese population-based study of copy number variations in this disease. 27005929_RYR2 variants show possible pathogenic Fibrosis of the Cardiac Conduction system. 27157848_The most common form of CPVT is due to autosomal dominant variants in the cardiac ryanodine receptor gene (RYR2). 27226555_CaM and S100A1 can concurrently bind to and functionally modulate RyR1 and RyR2, but this does not involve direct competition at the RyR CaM binding site. 27452199_Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT) Associated With Ryanodine Receptor (RyR2) Gene Mutations- Long-Term Prognosis After Initiation of Medical Treatment. 27494721_The left atrium / right atrium expression ratio was significantly increased in Atrial fibrillation for ryanodine receptor 2 - gene related to calcium uptake and release, and located on the sarcoplasmic reticulum membrane. 27523319_Long-Term Prognosis of Catecholaminergic Polymorphic Ventricular Tachycardia Patients With Ryanodine Receptor (RYR2) Mutations. 27609834_a direct interaction exists between RyR2 and CSQ2, is reported. 27663074_Common variants rs790899 and rs1891246 of RYR2 were significantly associated with HG and weight loss. 27733687_These results also suggest that altered cytosolic Ca(2+) activation of RyR2 represents a common defect of RyR2 mutations associated with catecholaminergic polymorphic ventricular tachycardia and atrial fibrillation, which could potentially be suppressed by carvedilol or (R)-carvedilol. 27927985_The unique properties of the CaM-F142L mutation may provide novel clues on how to suppress excessive RyR2 Ca(2+) release by manipulating the CaM-RyR2 interaction. 27987400_Our findings provided evidence that Indel polymorphism rs10692285 might contribute to sudden unexplained death (SUD) susceptibility through affecting the expression of RYR2, which suggest that abnormal ion channel activity is very likely to be the underlying mechanism of SUD. 27988446_Genetic analysis of the index case identified only one rare novel variant p.Ile11Ser (c.32T>G) in the RyR2 gene. Subsequent familial analysis identified segregation of the genetic variant with the disease. To our knowledge, there has been no previous case report of catecholaminergic polymorphic ventricular tachycardia associated to this missense variant. 28065668_Cardiac adrenergic response and progression towards HF proceed unaltered in mice harboring the RyR2-S2808A mutation. Preventing RyR2-S2808 phosphorylation does not preclude a normal sympathetic response nor mitigates the pathophysiological consequences of MI. 28084961_Genotype may predict phenotype in catecholaminergic polymorphic ventricular tachycardia, including a higher risk of life-threatening cardiac events in subjects with variants in the C-terminus of ryanodine receptor-2 (RyR2). [review] 28086167_We sought to identify genetic alterations in cardiac ion channels in patients with micro-ischemic disease. Genetic analysis with Sanger technology and posterior bioinformatic assessment identified two rare variations with potential pathogenic effects (RyR2_p.M4019T and SCN5A_p.H445D) in two individuals. 28202948_Five of the 19 patients (26.3%) had either a pathogenic variant or a likely pathogenic variant in MYBPC3 (n=1), MYH7 (n=1), RYR2 (n=2), or TNNT2 (n=1). All five variants were missense variants that have been reported previously in patients with channelopathies or cardiomyopathies 28237968_In a national cohort of RyR2 mutation-positive CPVT patients, SCD, ASCD and syncope were presenting events in the majority of probands and also occurred in 36% of relatives identified through family screening. 28405885_We identified a variant in the RYR2 gene (NM_001035) which involved a change of a glycine to an arginine in position155 of the gene product (c.463G > A, p.Gly155Arg, p.G155R). This RYR2 gene mutation is a novel rare genetic variant as it was not present in any of the international databases consulted. 28476886_Data suggest that post-translational modifications (phosphorylation, oxidation, and nitrosylation) of RyR2 (ryanodine receptor 2) occur downstream of production of amyloid beta-peptides through ADRB2 (beta2-adrenergic receptor) Ca2+ signaling cascade that activates PKA (protein kinase A). 29477366_Structural analysis revealed that the A165D mutation is located in a loop that is involved in inter-subunit interactions in the RyR2 tetrameric structure. 30496756_Ryr2 expression is upregulated by mir106-25 during atrial fibrillation. 30530841_results support a binding model where the CaM C-domain is anchored to RyR2 CaMBD2 and saturated with Ca(2+) during Ca(2+) oscillations, while the CaM N-domain functions as a dynamic Ca(2+) sensor that can bridge noncontiguous regions of RyR2 or clamp down onto CaMBD2. 30542613_review of the molecular environment of individual amino acid residues that form binding sites for essential modulators of ion channel function and determine its role in Ca 2+ signalling. 30835254_Cardiac hypertrophy and arrhythmia in mice induced by a mutation in ryanodine receptor 2. 30928430_esults challenge the current concept that CaM universally functions as a canonical inhibitor of RyR2 across species. Rather, CaM's biological action on human RyR2 appears to be more nuanced, with inhibitory activity only on phosphorylated RyR2 channels, which occurs during exercise or in patients with heart failure. 31112425_72 distinct RYR2 variants were identified in catecholaminergic polymorphic ventricular tachycardia patients. 31135957_Detection of RYR2 expression and/or promoter methylation might enable risk assessment for malignant conversion of dysplastic lesions. 31289932_A novel and de novo occurring mutation (both parents tested negative) in the cardiac ryanodine receptor gene RyR2 was detected.Mutations in this channel may result in the catecholaminergic polymorphic ventricular tachycardia (CPVT), a cardiac arrhythmia that can lead to syncope and sudden cardiac death. 31440994_RYR2 rs16835904 TC-TT genotype facilitated the risk of astrocytoma in male. RYR2 rs12594 AA genotype and AG genotype were associated with overall survival of astrocytoma. 31663850_The authors discovered that a significant role of Kv2.1 at neuronal endoplasmic reticulum-plasma membrane junctions is to promote the clustering and functional coupling of plasma membrane L-type Ca(2+) channels (LTCCs) to ryanodine receptor (RyR) endoplasmic reticulum Ca(2+) release channels. 31763755_Role of cardiac ryanodine receptor calmodulin-binding domains in mediating the action of arrhythmogenic calmodulin N-domain mutation N54I. 31875585_Co-Phenotype of Left Ventricular Non-Compaction Cardiomyopathy and Atypical Catecholaminergic Polymorphic Ventricular Tachycardia in Association With R169Q, a Ryanodine Receptor Type 2 Missense Mutation. 31913406_Identification of a Novel Homozygous Multi-Exon Duplication in RYR2 Among Children With Exertion-Related Unexplained Sudden Deaths in the Amish Community. 31934898_Catecholaminergic Polymorphic Ventricular Tachycardia. 31994352_Cardiac arrest in a mother and daughter and the identification of a novel RYR2 variant, predisposing to low penetrant catecholaminergic polymorphic ventricular tachycardia in a four-generation Canadian family. 32077934_Identification of an amino-terminus determinant critical for ryanodine receptor/Ca2+ release channel function. 32152366_Classification and correlation of RYR2 missense variants in individuals with catecholaminergic polymorphic ventricular tachycardia reveals phenotypic relationships. 32220801_SIDS associated RYR2 p.Arg2267His variant may lack pathogenicity. 32572717_Impact of NR5A2 and RYR2 3'UTR polymorphisms on the risk of breast cancer in a Chinese Han population. 32612034_[RyR2 mutation-linked arrhythmogenic diseases and its therapeutic strategies]. 32663189_Molecular characterization of the calcium release channel deficiency syndrome. 32669538_Rieske iron-sulfur protein induces FKBP12.6/RyR2 complex remodeling and subsequent pulmonary hypertension through NF-kappaB/cyclin D1 pathway. 32681117_An NGS-based genotyping in LQTS; minor genes are no longer minor. 32683896_Loss of SPEG Inhibitory Phosphorylation of Ryanodine Receptor Type-2 Promotes Atrial Fibrillation. 32866913_Rare RYR2 p.Thr85Ile variant is associated with catecholaminergic polymorphic ventricular tachycardia. 32878990_The central domain of cardiac ryanodine receptor governs channel activation, regulation, and stability. 32897880_Role of defective calcium regulation in cardiorespiratory dysfunction in Huntington's disease. 32899693_Structure and Function of the Human Ryanodine Receptors and Their Association with Myopathies-Present State, Challenges, and Perspectives. 32931925_Calcium signaling consequences of RyR2 mutations associated with CPVT1 introduced via CRISPR/Cas9 gene editing in human-induced pluripotent stem cell-derived cardiomyocytes: Comparison of RyR2-R420Q, F2483I, and Q4201R. 32980662_The functional significance of redox-mediated intersubunit cross-linking in regulation of human type 2 ryanodine receptor. 33315912_Genealogy and clinical course of catecholaminergic polymorphic ventricular tachycardia caused by the ryanodine receptor type 2 P2328S mutation. 33664309_A SPRY1 domain cardiac ryanodine receptor variant associated with short-coupled torsade de pointes. 33686871_Novel RyR2 Mutation (G3118R) Is Associated With Autosomal Recessive Ventricular Fibrillation and Sudden Death: Clinical, Functional, and Computational Analysis. 33825858_Identification of loss-of-function RyR2 mutations associated with idiopathic ventricular fibrillation and sudden death. 33902292_cAMP Imaging at Ryanodine Receptors Reveals beta2-Adrenoceptor Driven Arrhythmias. 33984427_Early Lethal Noncompaction Cardiomyopathy in Siblings With Compound Heterozygous RYR2 Variant. 34088380_Phenotype/Genotype Relationship in Left Ventricular Noncompaction: Ion Channel Gene Mutations Are Associated With Preserved Left Ventricular Systolic Function and Biventricular Noncompaction: Phenotype/Genotype of Noncompaction. 34111951_Impaired Binding to Junctophilin-2 and Nanostructural Alteration in CPVT Mutation. 34533024_Computational Analysis of Binding Interactions between the Ryanodine Receptor Type 2 and Calmodulin. 34546788_Human RyR2 (Ryanodine Receptor 2) Loss-of-Function Mutations: Clinical Phenotypes and In Vitro Characterization. 34661651_Loss-of-function mutations in cardiac ryanodine receptor channel cause various types of arrhythmias including long QT syndrome. 34730774_Clinical and Functional Characterization of Ryanodine Receptor 2 Variants Implicated in Calcium-Release Deficiency Syndrome. 34813985_Mutation in RyR2-FKBP Binding site alters Ca(2+) signaling modestly but increases ''arrhythmogenesis'' in human stem cells derived cardiomyocytes. 34877784_RYR2 mutation in non-small cell lung cancer prolongs survival via down-regulation of DKK1 and up-regulation of GS1-115G20.1: A weighted gene Co-expression network analysis and risk prognostic models. 34888385_Bioinformatic Analysis of Immune Significance of RYR2 Mutation in Breast Cancer. 35439358_Characterization of the mechanism by which a nonsense variant in RYR2 leads to disordered calcium handling. | ENSMUSG00000021313 | Ryr2 | 8.651003e+01 | 0.3373688 | -1.567601401 | 0.4435671 | 1.218209e+01 | 0.0004825068 | 0.07700508 | No | Yes | 4.207199e+01 | 15.795274 | 1.012468e+02 | 38.941193 | |
| ENSG00000198715 | 112770 | GLMP | protein_coding | Q8WWB7 | FUNCTION: Required to protect lysosomal transporter MFSD1 from lysosomal proteolysis and for MFSD1 lysosomal localization. {ECO:0000250|UniProtKB:Q9JHJ3}. | Alternative splicing;Glycoprotein;Lysosome;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:112770; | cytosol [GO:0005829]; integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; nucleus [GO:0005634]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein localization to lysosome [GO:0061462]; protein stabilization [GO:0050821] | 18021396_NCU-G1 is a dual-function protein capable of functioning as a transcription factor as well as a nuclear receptor co-activator. | ENSMUSG00000001418 | Glmp | 3.681655e+03 | 1.2277264 | 0.295989076 | 0.3149173 | 8.823029e-01 | 0.3475717146 | 0.82969965 | No | Yes | 3.818334e+03 | 393.929493 | 2.832106e+03 | 300.197770 | ||
| ENSG00000198718 | 23116 | TOGARAM1 | protein_coding | Q9Y4F4 | FUNCTION: Involved in ciliogenesis (PubMed:32453716). It is required for appropriate acetylation and polyglutamylation of ciliary microtubules, and regulation of cilium length (PubMed:32453716). Interacts with microtubules and promotes microtubule polymerization via its HEAT repeat domains, especially those in TOG region 2 and 4 (By similarity). {ECO:0000250|UniProtKB:Q17423, ECO:0000250|UniProtKB:Q6A070, ECO:0000269|PubMed:32453716}. | Alternative splicing;Cell projection;Ciliopathy;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Disease variant;Joubert syndrome;Reference proteome;Repeat | hsa:23116; | axoneme [GO:0005930]; ciliary basal body [GO:0036064]; cilium [GO:0005929]; cytoplasmic microtubule [GO:0005881]; microtubule organizing center [GO:0005815]; mitotic spindle [GO:0072686]; spindle microtubule [GO:0005876]; microtubule binding [GO:0008017]; axoneme assembly [GO:0035082]; cilium assembly [GO:0060271]; microtubule cytoskeleton organization [GO:0000226]; mitotic spindle assembly [GO:0090307]; non-motile cilium assembly [GO:1905515]; positive regulation of microtubule polymerization [GO:0031116] | Mouse_homologues 28063167_This study reveals that the cell-specific loss of TOG in oligodendrocytes severely reduced myelination in the CNS. | ENSMUSG00000035614 | Togaram1 | 9.611900e+01 | 0.6774075 | -0.561904080 | 0.5088381 | 1.135860e+00 | 0.2865288968 | 0.80925093 | No | Yes | 6.300519e+01 | 18.583201 | 1.087434e+02 | 32.752275 | ||
| ENSG00000198920 | 9851 | KIAA0753 | protein_coding | Q2KHM9 | FUNCTION: Involved in centriole duplication. Positively regulates CEP63 centrosomal localization. Required for WDR62 centrosomal localization and promotes the centrosomal localization of CDK2 (PubMed:24613305, PubMed:26297806). {ECO:0000269|PubMed:26297806}. | Alternative splicing;Ciliopathy;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | This gene encodes a subunit of a protein complex that regulates ciliogenesis and cilia maintenance. The encoded protein has also been shown to regulate centriolar duplication. Mutations in this gene cause an orofaciodigital syndrome and a form of Joubert syndrome in human patients. [provided by RefSeq, May 2017]. | hsa:9851; | centriolar satellite [GO:0034451]; centriole [GO:0005814]; centrosome [GO:0005813]; cytosol [GO:0005829]; centriole replication [GO:0007099]; protein localization to centrosome [GO:0071539] | 28220259_Whole exome sequencing of the family identified compound heterozygous mutations in KIAA0753, i.e., a missense mutation (p.Arg257Gly) and an intronic mutation (c.2359-1G>C). The intronic mutation alters normal splicing by activating a cryptic acceptor splice site in exon 16 29138412_We demonstrate that KIAA0753 is expressed in normal fetal human growth plate and show that the affected fetus, with a compound heterozygous frameshift and a nonsense mutation in KIAA0753, has an abnormal proliferative zone and a broad hypertrophic zone. 34241634_A ciliopathy complex builds distal appendages to initiate ciliogenesis. 34523780_Genetic and phenotypic heterogeneity in KIAA0753-related ciliopathies. 34711653_CEP120-mediated KIAA0753 recruitment onto centrioles is required for timely neuronal differentiation and germinal zone exit in the developing cerebellum. | ENSMUSG00000020807 | 4933427D14Rik | 6.439012e+02 | 0.9450719 | -0.081503966 | 0.2813554 | 8.335444e-02 | 0.7728020158 | 0.95442153 | No | Yes | 5.709991e+02 | 90.178753 | 6.195996e+02 | 100.235248 | |
| ENSG00000203485 | 64423 | INF2 | protein_coding | Q27J81 | FUNCTION: Severs actin filaments and accelerates their polymerization and depolymerization. {ECO:0000250}. | Acetylation;Actin-binding;Alternative splicing;Charcot-Marie-Tooth disease;Coiled coil;Cytoplasm;Direct protein sequencing;Disease variant;Neurodegeneration;Neuropathy;Phosphoprotein;Reference proteome | This gene represents a member of the formin family of proteins. It is considered a diaphanous formin due to the presence of a diaphanous inhibitory domain located at the N-terminus of the encoded protein. Studies of a similar mouse protein indicate that the protein encoded by this locus may function in polymerization and depolymerization of actin filaments. Mutations at this locus have been associated with focal segmental glomerulosclerosis 5.[provided by RefSeq, Aug 2010]. | hsa:64423; | perinuclear region of cytoplasm [GO:0048471]; actin binding [GO:0003779]; small GTPase binding [GO:0031267]; actin cytoskeleton organization [GO:0030036]; regulation of mitochondrial fission [GO:0090140] | 20023659_Study identified nine independent nonconservative missense mutations in INF2, which encodes a member of the formin family of actin-regulating proteins. 20803156_In conclusion, we described an additional familial case of the autosomal dominant form of focalsegmental glomerulosclerosis associated with INF2 mutations. 21258034_Six of the seven distinct altered residues localized to an INF2 region that corresponded to a subdomain of the mDia1 diaphanous inhibitory domain reported to co-immunoprecipitate with IQ motif-containing GTPase-activating protein 1 21278336_The effects of disease-causing INF2 mutations suggest an important role for this protein and its interaction with other formins in modulating glomerular podocyte phenotype and function. 21866090_INF2 mutations were responsible for 16% of all cases of autosomal dominant focal and segmental glomerulosclerosis, with these mutations clustered in exon 4. 21998196_Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. 21998204_Actin monomers inhibit microtubule binding/bundling by INF2 22187985_INF2 mutations appear to cause many cases of FSGS-associated Charcot-Marie-Tooth neuropathy, showing that INF2 is involved in a disease affecting both the kidney glomerulus and the peripheral nervous system. 22879592_Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. 22961558_Our study confirms the link between INF2 mutations and Charcot-Marie-Tooth-associated glomerulopathy and widens the spectrum of pathogenic mutations. 22971997_A novel mutation, outside of the candidate region for diagnosis, in the inverted formin 2 gene can cause focal segmental glomerulosclerosis. 22986496_Formation of stabilized, detyrosinated microtubules required the formin INF2. 23014460_INF2 mutations were found in 2 of 281 individuals with sporadicfocal and segmental glomerulosclerosis 23349293_study found actin polymerization through ER-localized INF2 was required for efficient mitochondrial fission; INF2-induced actin filaments may drive initial mitochondrial constriction, which allows Drp1-driven secondary constriction 23521651_This study showed that INF2 mutations in Charcot-Marie-Tooth disease complicated with focal segmental glomerulosclerosis. 23620398_In podocytes, INF2 appears to be an important modulator of actin-dependent behaviors that are under the control of Rho/mDia signaling. 23847988_INF2 mutation was detected both father and his son 23921379_actin monomer binding to the DAD of INF2 competes with the DID/DAD interaction, thereby activating actin polymerization 24174593_this study identifed three novel mutations of INF likely efect hereditary neuropathy with glomerulopathy. 25165188_INF2 mutations are associated with focal segmental glomerulosclerosis. 26039629_Report novel mutations in the inverted formin 2 gene of Chinese families contributing to focal segmental glomerulosclerosis. 26124273_Assembly and turnover of short actin filaments by the formin INF2 and profilin. 26305500_The authors propose Spire1C isoform cooperates with INF2 to regulate actin assembly at endoplasmic reticulum-mitochondrial contacts. 26383224_Propose that examination of INF2 expression may help to differentiate minimal change disease from focal segmental glomerulosclerosis and evaluate the clinical severity of steroid resistance nephrotic syndrome in children. 26621033_FHOD1 and INF2 are novel regulators of inter- and intra-structural contractility of podosomes. 26764407_Disease causing mutations in inverted formin 2 regulate its binding to G-actin, F-actin capping protein (CapZ alpha-1) and profilin 2. 27974406_All individuals with INF2 mutations presenting with a thrombotic microangiopathy also had atypical hemolytic uremic syndrome risk haplotypes, potentially accounting for the genetic pleiotropy 28448495_hese findings reveal novel molecular events underlying the regulation of INF2 function and localization, and provided insights in understanding the relationship between SPOP mutations and dysregulation of mitochondrial dynamics in prostate cancer. 29142021_INF2-mediated actin polymerization on the endoplasmic reticulum stimulates mitochondrial division by two independent mechanisms: (1) mitochondrial calcium uptake, leading to inner mitochondrial membrane constriction; and (2) Drp1 oligomerization, leading to outer mitochondrial membrane constriction. 29947928_Studies indicate that INF2, a formin, that is mutated in hereditary renal and neurodegenerative disorders. 30126379_INF2 seems to be not only the cause of FSGS, but also of ESRD of unknown etiology. 30579254_INF2 is a novel pro-apoptotic inducer of oxidative injury in epidermal cells. 31354004_our results indicated that oxidative stress-mediated HaCaT cells apoptosis could be reversed by Tan IIA treatment via reducing INF2-related mitochondrial stress in a manner dependent on the ERK signaling pathway. 31515790_The phenotypic feature of the pedigree is autosomal dominant intermediate Charcot-Marie-Tooth disease and focal segmental glomerulosclerosis, which may be attributed to the c.341G>A mutation of the INF2 gene. 31871199_Study used lysine-to-glutamine mutations to map the relevant lysines on actin for INF2 regulation. K50Q- and K61Q-actin, when bound to CAP2, inhibit full-length INF2 but not INF2 lacking inhibitory domain (DID). CAP WH2 domain binds INF2-DID with submicromolar affinity but has weak affinity for actin monomers, while INF2-C-terminal diaphanous autoregulatory domain binds CAP/K50Q-actin 5-fold better than CAP/WT-actin. 31924668_FSGS-Causing INF2 Mutation Impairs Cleaved INF2 N-Fragment Functions in Podocytes. 32727404_Multiple formin proteins participate in glioblastoma migration. 32844773_Steroid Resistant Nephrotic Syndrome with Clumsy Gait Associated With INF2 Mutation. 34698992_Role of formin INF2 in human diseases. | ENSMUSG00000037679 | Inf2 | 2.885089e+03 | 1.2806268 | 0.356850110 | 0.2981012 | 1.447410e+00 | 0.2289439233 | 0.78818582 | No | Yes | 2.990319e+03 | 364.079097 | 2.223993e+03 | 278.214651 | |
| ENSG00000204120 | 26058 | GIGYF2 | protein_coding | Q6Y7W6 | FUNCTION: Key component of the 4EHP-GYF2 complex, a multiprotein complex that acts as a repressor of translation initiation (PubMed:22751931, PubMed:31439631). In the 4EHP-GYF2 complex, acts as a factor that bridges EIF4E2 to ZFP36/TTP, linking translation repression with mRNA decay (PubMed:31439631). Also recruits and bridges the association of the 4EHP complex with the decapping effector protein DDX6, which is required for the ZFP36/TTP-mediated down-regulation of AU-rich mRNA (PubMed:31439631). May act cooperatively with GRB10 to regulate tyrosine kinase receptor signaling, including IGF1 and insulin receptors (PubMed:12771153). {ECO:0000269|PubMed:12771153, ECO:0000269|PubMed:22751931, ECO:0000269|PubMed:31439631}. | 3D-structure;Acetylation;Alternative splicing;Disease variant;Isopeptide bond;Methylation;Neurodegeneration;Parkinson disease;Parkinsonism;Phosphoprotein;Reference proteome;Ubl conjugation | This gene contains CAG trinucleotide repeats and encodes a protein containing several stretches of polyglutamine residues. The encoded protein may be involved in the regulation of tyrosine kinase receptor signaling. This gene is located in a chromosomal region that was genetically linked to Parkinson disease type 11, and mutations in this gene were thought to be causative for this disease. However, more recent studies in different populations have been unable to replicate this association. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2013]. | hsa:26058; | cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endosome [GO:0005768]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; perikaryon [GO:0043204]; protein-containing complex [GO:0032991]; proximal dendrite [GO:1990635]; vesicle [GO:0031982]; cadherin binding [GO:0045296]; proline-rich region binding [GO:0070064]; RNA binding [GO:0003723]; adult locomotory behavior [GO:0008344]; cellular protein metabolic process [GO:0044267]; feeding behavior [GO:0007631]; homeostasis of number of cells within a tissue [GO:0048873]; insulin-like growth factor receptor signaling pathway [GO:0048009]; mitotic G1 DNA damage checkpoint signaling [GO:0031571]; mRNA destabilization [GO:0061157]; multicellular organism growth [GO:0035264]; musculoskeletal movement [GO:0050881]; negative regulation of translation [GO:0017148]; neuromuscular process controlling balance [GO:0050885]; post-embryonic development [GO:0009791]; posttranscriptional gene silencing [GO:0016441]; spinal cord motor neuron differentiation [GO:0021522] | 18358451_Observational study of gene-disease association. (HuGE Navigator) 18358451_These data strongly support GIGYF2 as a PARK11 gene with a causal role in familial Parkinson disease. 18923002_Mutations in GIGYF2 are not strongly related to the development of the Parkinson's disease in Portuguese and North American populations. 18923002_Observational study of gene-disease association. (HuGE Navigator) 19117363_Observational study of gene-disease association. (HuGE Navigator) 19133664_Observational study of gene-disease association. (HuGE Navigator) 19133664_This study suggested that reported mutations in GIGYF2 are not a common cause of Parkinson's disease in Norway and United States. 19250854_Observational study of gene-disease association. (HuGE Navigator) 19250854_The results of this study concluded that neither of Asn56Ser and Asn457Thr variants plays a major role in the pathogenesis of parkinson disease. 19279319_Observational study of gene-disease association. (HuGE Navigator) 19279319_We believe that variation in a gene other than GIGYF2 accounts for the previously reported linkage finding on chromosome 2q36-37. 19321232_Observational study of gene-disease association. (HuGE Navigator) 19321232_The results of this study did not support a role for GIGYF2 in the genetic etiology of Belgian Parkinson disease. 19348706_The current genetic evidence suggestes that GIGYF2 is not a common cause od parkinson disease in several Caucasian. 19429085_GIGYF2 Asn56Ser and Asn457Thr mutations are a rare cause of PD in North American Caucasian population 19429085_Observational study of gene-disease association. (HuGE Navigator) 19449032_analysis of GIGYF2 variants in Parkinson's disease from two Asian populations 19482505_GIGYF2 mutations are not a frequent cause of Parkinson's disease 19482505_Observational study of gene-disease association. (HuGE Navigator) 19562763_This letter suggested that GIGYF2 is neither responsible for PARK11 nor a gene implicated in Parkinson disease. 19638301_Observational study of gene-disease association. (HuGE Navigator) 19638301_The GIGYF2 Asn56Ser mutation is rare in Chinese PD patients. 19845746_Observational study of gene-disease association. (HuGE Navigator) 19845746_our findings suggest that GIGYF2 variants are not a frequent cause of Parkinson's disease in the Spanish population, since we found no clearly segregating GIGYF2 variants 20004041_Observational study of gene-disease association. (HuGE Navigator) 20004041_The results of this study do not support a major role of GIGYF2 in parkinson disease. 20044296_GIGYF2 variants are not associated with Parkinson's disease in the mainland Chinese Population. 20044296_Observational study of gene-disease association. (HuGE Navigator) 20060621_Observational study of gene-disease association. (HuGE Navigator) 20060621_These data, together with those recently reported by other groups, suggest that GIGYF2 is unlikely to be the PARK11 gene. 20178831_Observational study of gene-disease association. (HuGE Navigator) 20178831_We identified nine novel variants in GIGYF2 gene, which might be associated with PD in the Chinese population. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20641165_GIGYF2 is unlikely to play a major role in PD in Japanese patients, similar to other populations. 20816920_No clearly pathogenic mutations are identified in GIGYF2 and ATP13A2 in Brazilian patients with early-onset Parkinson's disease. 20816920_Observational study of gene-disease association. (HuGE Navigator) 22115759_within the Chinese population, the c.297T>C p.Ala99Ala polymorphism of the GIGYF2 gene may be associated with an increased risk of developing Parkinson disease. 22503729_Our result indicated that SCNA, LRRK2, UCHL1, HtrA2 and GIGYF2 genes' mutations might not be a main reason for Chinese Autosomal dorminant Parkinson's disease 22751931_GIGYF2 and the zinc finger protein 598 (ZNF598) are identified as components of the 4EHP complex. 26134514_required, this finding may shed light on the GIGYF2-associated mechanisms that lead to PD and suggests insulin dysregulation as a disease-specific mechanism for both PD and cognitive dysfunction. 26152800_Results suggest that the N56S and N457T of GIGYF2 are risk factors for Parkinson's disease in Caucasians, but not in Asians 27157137_Full-length GIGYF2 coimmunoprecipitates with AGO2 in human cells, and upon tethering to a reporter mRNA, GIGYF2 exhibits strong, dose-dependent silencing activity, involving both mRNA destabilization and translational repression. 28873462_genetic analyses of indel loci in ACE, DJ-1, and GIGYF2 genes, was performed to explore the potential contribution of insertion (I)/deletion (D) polymorphisms (indels) to the risk of PD in a Chinese population. 29554310_GIGYF2 has two distinct mechanisms of repression: one depends on 4EHP binding and mainly affects translation; the other is 4EHP-independent and involves the CCR4/NOT complex and its deadenylation activity. 32726578_GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control. 33053355_4EHP and GIGYF1/2 Mediate Translation-Coupled Messenger RNA Decay. 33239198_Association study of DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1 with Parkinson's disease. 34665823_Translational repression of NMD targets by GIGYF2 and EIF4E2. | ENSMUSG00000048000 | Gigyf2 | 2.537111e+03 | 0.8133680 | -0.298019802 | 0.2779646 | 1.149978e+00 | 0.2835536513 | 0.80801941 | No | Yes | 1.878067e+03 | 245.220943 | 2.210107e+03 | 295.673750 | |
| ENSG00000204305 | 177 | AGER | protein_coding | Q15109 | FUNCTION: Mediates interactions of advanced glycosylation end products (AGE). These are nonenzymatically glycosylated proteins which accumulate in vascular tissue in aging and at an accelerated rate in diabetes. Acts as a mediator of both acute and chronic vascular inflammation in conditions such as atherosclerosis and in particular as a complication of diabetes. AGE/RAGE signaling plays an important role in regulating the production/expression of TNF-alpha, oxidative stress, and endothelial dysfunction in type 2 diabetes. Interaction with S100A12 on endothelium, mononuclear phagocytes, and lymphocytes triggers cellular activation, with generation of key proinflammatory mediators. Interaction with S100B after myocardial infarction may play a role in myocyte apoptosis by activating ERK1/2 and p53/TP53 signaling (By similarity). Receptor for amyloid beta peptide. Contributes to the translocation of amyloid-beta peptide (ABPP) across the cell membrane from the extracellular to the intracellular space in cortical neurons. ABPP-initiated RAGE signaling, especially stimulation of p38 mitogen-activated protein kinase (MAPK), has the capacity to drive a transport system delivering ABPP as a complex with RAGE to the intraneuronal space. Can also bind oligonucleotides. {ECO:0000250, ECO:0000269|PubMed:19906677, ECO:0000269|PubMed:20943659, ECO:0000269|PubMed:21559403, ECO:0000269|PubMed:21565706}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Glycoprotein;Immunoglobulin domain;Inflammatory response;Membrane;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | The advanced glycosylation end product (AGE) receptor encoded by this gene is a member of the immunoglobulin superfamily of cell surface receptors. It is a multiligand receptor, and besides AGE, interacts with other molecules implicated in homeostasis, development, and inflammation, and certain diseases, such as diabetes and Alzheimer's disease. Many alternatively spliced transcript variants encoding different isoforms, as well as non-protein-coding variants, have been described for this gene (PMID:18089847). [provided by RefSeq, May 2011]. | hsa:177; | apical plasma membrane [GO:0016324]; cell junction [GO:0030054]; cell surface [GO:0009986]; extracellular region [GO:0005576]; fibrillar center [GO:0001650]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; postsynapse [GO:0098794]; advanced glycation end-product receptor activity [GO:0050785]; amyloid-beta binding [GO:0001540]; identical protein binding [GO:0042802]; protein-containing complex binding [GO:0044877]; S100 protein binding [GO:0044548]; scavenger receptor activity [GO:0005044]; signaling receptor activity [GO:0038023]; transmembrane signaling receptor activity [GO:0004888]; astrocyte activation [GO:0048143]; cell surface receptor signaling pathway [GO:0007166]; cellular response to amyloid-beta [GO:1904646]; glucose mediated signaling pathway [GO:0010255]; induction of positive chemotaxis [GO:0050930]; inflammatory response [GO:0006954]; learning or memory [GO:0007611]; microglial cell activation [GO:0001774]; modulation of age-related behavioral decline [GO:0090647]; negative regulation of blood circulation [GO:1903523]; negative regulation of interleukin-10 production [GO:0032693]; negative regulation of long-term synaptic depression [GO:1900453]; negative regulation of long-term synaptic potentiation [GO:1900272]; neuron projection development [GO:0031175]; positive regulation of activated T cell proliferation [GO:0042104]; positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process [GO:1902961]; positive regulation of chemokine production [GO:0032722]; positive regulation of dendritic cell differentiation [GO:2001200]; positive regulation of endothelin production [GO:1904472]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of heterotypic cell-cell adhesion [GO:0034116]; positive regulation of interleukin-1 beta production [GO:0032731]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of JNK cascade [GO:0046330]; positive regulation of JUN kinase activity [GO:0043507]; positive regulation of monocyte chemotactic protein-1 production [GO:0071639]; positive regulation of monocyte extravasation [GO:2000439]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of p38MAPK cascade [GO:1900745]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of tumor necrosis factor production [GO:0032760]; protein localization to membrane [GO:0072657]; regulation of CD4-positive, alpha-beta T cell activation [GO:2000514]; regulation of DNA binding [GO:0051101]; regulation of inflammatory response [GO:0050727]; regulation of long-term synaptic potentiation [GO:1900271]; regulation of NIK/NF-kappaB signaling [GO:1901222]; regulation of p38MAPK cascade [GO:1900744]; regulation of spontaneous synaptic transmission [GO:0150003]; regulation of synaptic plasticity [GO:0048167]; regulation of T cell mediated cytotoxicity [GO:0001914]; response to amyloid-beta [GO:1904645]; response to hypoxia [GO:0001666]; response to wounding [GO:0009611]; transcytosis [GO:0045056]; transport across blood-brain barrier [GO:0150104] | 11700025_mRNA and protein expression in Caco-2 cells 11739380_this study we have investigated a possible role of RAGE and amphoterin in the retinoic acid-induced differentiation of neuroblastoma cells. 11811511_susceptibility to the development of chronic periodontitis could be influenced by the 1704G/T polymorphism of the RAGE gene, independently of diabetes 11884895_Polymorphisms 1704G/T, 2184A/G, and 2245G/A in the rage gene are not associated with diabetic retinopathy in NIDDM 12029499_allele frequencies and genotype distribution combinations of four polymorphisms in the RAGE gene (6p21.3, G82S, 1704G/T, 2184A/G and 2245A/G) of 272 subjects (130 patients with plaque psoriasis and 142 healthy control). 12070776_increased prevalence of the 82S allele in patients with rheumatoid arthritis compared with control subjects 12477623_There is an association of Gly82Ser polymorphism in this gene with diabetic retinopathy in type II diabetic Asian Indian patients. 12495433_Data show that vascular endothelial cells and pericytes express three novel splice variants of receptor for advanced glycation end-products (RAGE) mRNA. 12579287_co-expression of RAGE and amphoterin is closely associated with invasion and metastasis of colorectal cancer 12598893_These data reinforce the importance of receptor for advanced glycation end products (RAGE)-ligand interactions in modulating properties of CD4+ T cells that infiltrate the central nervous system 12606536_association between -374 T/A RAGE polymorphism and cardiovascular disease and albumin excretion in type 1 diabetics with poor metabolic control suggests gene-environment interaction in development of diabetic nephropathy and cardiovascular complications 12618340_The RAGE is expressed in dying neurons and suggest that RAGE may have a role in neuronal cell death mediated by ischemic stress. 12651613_Advanced glycation end products and receptor for advanced glycation end products are found in AA amyloidosis. 12837757_AGEs can augment inflammatory responses by up-regulating COX-2 via RAGE and multiple signaling pathways, thereby leading to monocyte activation and vascular cell dysfunction 12859967_Results suggest a possible role of S100 protein- and RAGE-mediated signal transduction in the development of specific cancers. 12935895_In vitro binding studies using a series of C-terminal deletion mutants of human RAGE revealed the importance of the membrane-proximal cytoplasmic region of RAGE for the direct ERK-RAGE interaction. 12941744_No association betwween -429T/C and -37T/A polymorphism in RAGE gene promoter with diabetic retinopathy in NIDDM in Chinese patients 14580673_identified three novel RAGE transcripts all encoding truncated soluble forms of RAGE. The relative expression ratios for the full-length RAGE transcript to the sum of its splice-variants encoding the soluble variants varied strongly among tissues. 14595542_Oleate, not ligands of the receptor for advanced glycation end-products, acts as an enhancer of human smooth muscle cell proliferation. 14704946_Our study failed to demonstrate an association between either - 429 T/C or - 374 T/A gene polymorphism of the RAGE gene and diabetic retinopathy in Caucasians with type 2 diabetes 14747204_RAGE G1704T polymorphisms may be useful in identifying the risk for developing diabetic nephropathy in type 2 diabetic patients. 15009731_AGE might be involved in the growth and invasion of melanoma through interactions with RAGE and represent promising candidates for assessing the future therapeutic potential of this therapy in treating patients with melanoma. 15019601_results suggest that the decrease in monocyte RAGE expression can be at least partly accounted for by the ligand engagement and may be a factor contributing to the development of diabetic vascular complications 15033494_AGER mediates S100A13 protein translocation in response to extracellular S100 15052533_Patients with type 2 diabetes and the 63bp deletion in the promoter of RAGE seem to be protected from diabetic nephropathy. 15053925_Only cells expressing RAGE at the cell surface showed hypersensitivity to Abeta. 15127201_Findings using advanced glycosylation end products (AGEs) with strong RAGE-binding properties indicate that AGEs may not uniformly play a role in cellular activation. 15155381_Review. RAGE is an amplification step in vascular inflammation and acceleration of atherosclerosis. 15170618_the RAGE pathway plays a critical proinflammatory role in vasculitic neuropathy. 15180953_Advanced glycation end products (AGE) by binding to AGE receptor (RAGE) per se could control mesangial cell growth. 15203887_Corellation of expression of tissue factor (TF) and the receptor for advanced glycation end products (RAGEs) and vascular complications in diabetic patients 15213278_RAGE expression is upregulated on monocytes from patients with chronic kidney disease 15239215_RAGE and MMP-9 are expressed concordant with the metastatic ability of the human pancreatic cancer cells. Could be key to regulating metastatic ability of pancreatic cancer. 15247020_AGER has a role as a pleiotropic antagonistic gene (review) 15289604_AGER1 suppressed AGE mediated NF-kappaB and MAPK/p44/p42 activities and suppressed AGE-mediated mesangial cell inflammatory injury through negative regulation of RAGE 15448098_Thiazolidinedione antidiabetics modulate vascular endothelial RAGE expression. 15539404_Down-regulation of RAGE may be considered as a critical step in tissue reorganization and the formation of lung tumours. 15547674_374T/A polymorphism of the RAGE gene may reduce susceptibility to coronary artery disease 15555779_RAGE may have multiple functions in the human brain, mediated by the individual or coordinated efforts of the different RAGE isoforms 15599399_data demonstrate that the RAGE-NF-kappaB axis operates in diabetic neuropathy, by mediating functional sensory deficits, and that its inhibition may provide new therapeutic approaches 15663911_RAGE, via its interaction with ligands, serves as a cofactor exacerbating diabetic vascular disease. (review) 15666359_DU145 cells, a hormone-independent prostate cancer cell line, showed the highest RAGE mRNA expression 15790669_The RAGE2 haplotype is associated with diabetic nephropathy (DN) in type 2 diabetics and with earlier DN onset and can be regarded as a marker for DN. 15798956_data suggest that G1704T and G82S polymorphisms of the advanced glycation end product receptor (RAGE) gene are not related to microalbuminuria in Japanese type 2 diabetic patients 15803111_82 Serine allele of the RAGE gene is a risk allele for developing advanced nephropathy in diabettes mellitus. 15893388_Reactive oxygen species generated by the TNF-alpha-stimulated umbilical vein endothelial cells induce RAGE expression. 15896660_results show that the -374A allele (-374T>A polymorphism) in the RAGE gene is related to the susceptibility of developing ischemic heart disease in African-Brazilians with type 2 diabetes 15911356_Antibodies against RAGE receptor blocked Abeta-induced activation of the p38, JNK pathways, and NF-kappaB in CTL cybrids and offered protection against the neurotoxic effects of Abeta. 15915542_results reveal HMGB1 and RAGE as the first known autocrine loop modulating the maturation of plasmacytoid dendritic cells 15926115_data suggest that activation of the receptor for advanced glycation end products (RAGE) pathway may be one of the first steps in the pathogenesis of diabetic polyneuropathies 15930093_ACE inhibition reduces the accumulation of AGE in diabetes partly by increasing the production and secretion of sRAGE into plasma 15942078_polymorphisms G1704T and G82S of the RAGE gene are not related to DR in Japanese type 2 diabetic patients. 15942086_polymorphisms G1704T and G82S of the RAGE gene are not related to DR in Japanese type 2 diabetic patients. 15944249_Advanced glycosylation end-product receptor (RAGE) is required for the effect of high mobility group box 1 (HMGB1) on dendritic cells; HMGB1/RAGE interaction results in downstream activation of MAP kinases and NF-kappa B. 15986224_data suggests that the CML-RAGE-NF-kappaB pathway is an evident proinflammatory pathomechanism in mononuclear effector cells in polymyositis and dermatomyositis 16037279_RAGE, an endogenous decoy receptor, may be related to individual variations in resistance to the development of diabetic vascular complications, observed in transgenic mice. 16052547_The increase in RAGE noted in osteoarthritis(OA) cartilage and the ability of RAGE ligands to stimulate chondrocyte MAP kinase and NF-kappaB activity and to stimulate MMP-13 production suggests that chondrocyte RAGE signaling could play a role in OA. 16061848_S100P plays a major role in the aggressiveness of pancreatic cancer that is likely mediated by its ability to activate RAGE 16127291_These results suggest that RAGE expression appears to be closely associated with the invasiveness of oral squamous cell carcinoma and represents a promising candidate for assessing the future therapeutic potential in treating patients with oral carcinoma. 16159602_Analysis of specific manifestations of cardiovascular disease, including coronary heart disease (CHD), cardiovascular disease (CVD), myocardial infarction (MI) and ischemic disease (ISD) revealed no association with RAGE genotype. 16166741_Data suggest that re-expression of the receptor for advanced glycation end-products in lung cancer cells impairs the proliferative stimulus mediated by fibroblasts. 16271939_a cross-talk exists between the AGE-RAGE system and the angiotensin II receptor (renin-angiotensin system), and serum levels of sRAGE may reflect endothelial RAGE expression 16286548_Patients with Alzheimer disease have reduced levels of sRAGE in plasma compared with patients with vascular dementia and controls. 16374460_RAGE is mainly expressed in fibroblasts, dendrocytes, and keratinocytes and to a minor extent in endothelial and mononuclear cells. 16440015_study found a modest association with the -RAGE 374T/A polymorphism in the nonproliferative diabetic retinopathy subgroup 16493495_The existence of splice variants of RAGE in endothelial cells might explain the lack of interaction of extracellular AGEs with endothelial cells. 16644647_Association of level in serum with albuminuria may be a marker of microvascular damage in type 2 diabetes. 16728681_3 AGER genetic variants: -429T>C, -374T>A, and Gly82Ser were determined in cerebral ischemia and stroke patients. Specific haplotypes were associated with reduced risk of incident myocardial infarction & ischemic stroke, independent of diabetes status. 16728681_Observational study of gene-disease association. (HuGE Navigator) 16847147_CRP at concentrations known to predict future vascular events upregulates RAGE expression in human endothelial cells at both the protein and mRNA level. Silencing of the RAGE gene prevents CRP-induced macrophage chemoattractant protein 1 activation. 16899960_adhesion of red blood cells from type 2 diabetes patients was mediated by RAGE 16954185_AGER1 negatively regulates AGE-mediated oxidant stress-dependent signaling via the EGFR and Shc/Grb2/Ras pathway. 16954682_observation suggests that the RAGE gene polymorphism is not associated with in-stent restenosis after coronary artery stenting in non-diabetic patients in the Korean population 16969646_results show an association between the AGER -374 T/A polymorphism & type 1 diabetes; the polymorphism was associated with diabetic nephropathy in both type 1 & type 2 diabetes & with sight-threatening retinopathy in type 1 diabetic patients 16969649_Serum levels are associated with the severity of nephropathy in type 2 diabetic patients. 17004092_Telmisartan: it may work as an anti-inflammatory agent against AGE by suppressing RAGE expression via PPAR-gamma activation in the liver. 17024691_Serum levels are significantly higher in type 2 diabetic patients than in non-diabetics and are positively associated with the presence of coronary artery disease. 17035340_data suggest that the autocrine/paracrine release of HMGB1 and the integrity of the HMGB1/RAGE pathway are required for the migratory function of dendritic cells 17158877_Soluble RAGE forms tetramers that bind to hexamers of Ca2+-calgranulin C, creating a large platform for effectively transmitting RAGE-dependent signals from extracellular S100 proteins to the cytoplasmic signaling complexes. 17170388_association of RAGE and its ligands with sarcoidosis and suggest that an intrinsic genetic factor could be in part involved in its expression. In Italian patients, the -374 T/A polymorphism seems to be significantly associated with this disease. 17172923_The results of this large population study demonstrate that the AA/GA genotypes of the RAGE +557G>A polymorphism are associated with a significantly decreased risk of significant coronary artery disease. 17218539_study demonstrates that HOCl-modified albumin acts as a ligand for RAGE and promotes RAGE-mediated inflammatory complications 17224333_subjects with homozygosity for the minor S allele of the G82S polymorphism of RAGE had higher risk factors for cardiovascular disease 17237617_We demonstrated that after successful TX, PAPP-A and sRAGE decrease and early chronic vascular changes in the kidney TX are associated with elevation of their serum levels. 17343760_Soluble but not sescretory protein is associated with albuminuria in type 2 diabetic paaatients, whil neither is associated with markers of glucose control or macrovascular disease. 17345061_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17355942_Observational study of gene-disease association. (HuGE Navigator) 17405935_evidence provided for activation of the NF-kappaB system in intervertebral discs in vivo which correlates with accumulated oxidative stress & increases in age & disc degeneration; oxidative stress may lead to RAGE activation & NF-kappaB translocation 17425804_the RAGE Ser82 allele does not predispose to cardiovascular events in rheumatoid arthritis. 17434489_Accumulation of AGE may have a role in the development of osteoarthritis. 17497647_Expression of Endogenous secretory receptor for advanced glycation endproducts was significantly higher in chondrosarcomas 17508727_High level bacterial expression systems and purification protocols were generated for the extracellular region of RAGE (sRAGE) and the five permutations of single and tandem domain constructs. 17512509_The -429 T>C polymorphism in the RAGE promoter is associated with type 1 diabetes in a Brazilian population, with a 2-fold increase of C-allele frequency compared to non-diabetic subjects 17536039_Data show that soluble RAGE blocks scavenger receptor CD36-mediated uptake of hypochlorite-modified low-density lipoprotein. 17549666_it is concluded that CDK5RAP3, CCNB2, and RAGE genes may be used as a very reliable biomarkers of lung adenocarcinoma 17560613_Olmesartan may play a protective role against proliferative diabetic retinopathy by attenuating the deleterious effects of AGEs via down-regulation of RAGE. 17588956_RAGE was found throughout the testis, caput epididymis, particularly the principle cells apical region, and on sperm acrosomes in men with diabetes mellitus. 17592553_Serum levels of sRAGE are associated with inflammatory markers in patients with type 2 diabetes. 17593216_Multivariate analysis showed high-grade expression of RAGE to be an independent prognostic factor for disease-free survival in oral squamous cell carcinoma. 17601350_Observational study of gene-disease association. (HuGE Navigator) 17613396_Higher levels of plasma RAGE measured shortly after reperfusion predicted poor short-term outcomes from lung transplantation. Elevated plasma RAGE levels may have both pathogenetic and prognostic value in patients after lung transplantation. 17639302_Treating type 2 diabetic patients with thiazolidinedione can increase circulating levels of RAGE. 17640970_Data suggest that deregulation of RAGE expression in myoblasts might concur in rhabdomyosarcomagenesis and that increasing RAGE expression in rhabdomyosarcoma cells might reduce their tumor potential. 17660747_Structural and the binding data suggest that tetrameric S100B triggers RAGE activation by receptor dimerisation. 17661837_The receptor of advanced glycation end product (RAGE) -374 T/A single nucleotide polymorphism affected the dialysate-to-plasma ratio of creatinine at baseline, suggesting that the RAGE polymorphism may effect the basal peritoneal solute transport rate. 17700210_Observational study of gene-disease association. (HuGE Navigator) 17714874_De-N-glycosylation or G82S mutation of RAGE increases affinity for AGE ligands, and may sensitize cells or conditions with it to AGE. 17924846_result confirms that the -374AA genotype of the RAGE gene promoter is a protective factor against the severity of coronary artery disease lesions in type 2 diabetic patients 18032526_Suggest key mechanisma by which AGER1 maintains cellular resistance against AGE-induced oxidative stress. 18046662_Plasma soluble RAGE and endogenous secretory RAGE are not associated with measures of diabetic neuropathy in type 2 diabetes patients. 18050248_Calculating the S100A12:sRAGE ratio might help to detect patients with KD who are at risk of being unresponsive to IVIG therapy. 18058469_serum sRAGE levels were influenced by genetic polymorphisms (-429 T/C, Gly82Ser and 2184 A/G) of the RAGE gene in breast cancer. 18078623_HMGB-1 and BSA-AGE stimulated the invasiveness of fibroblast-like synoviocytes in rheumatoid arthritis by activation of RAGE. 18079485_Single nucleotide polymorphisms may contribute to development of glucose intolerance and type 2 diabetes. 18079965_endothelial RAGE and its ligands mediate vascular and inflammatory stresses that culminate in atherosclerosis in the vulnerable vessel wall 18080716_HCC during the early stage of tumorigenesis with less blood supply may acquire resistance to stringent hypoxic milieu by hypoxia-induced RAGE expression. 18089847_RAGE_v1 variant is the primary secreted soluble isoform of RAGE. identification of functional splice variants of RAGE underscores biological diversity of the RAGE gene & will aid in understanding of the gene in the normal & pathological state. 18179750_the -374AA genotype of the RAGE gene promoter could be associated with a reduced risk of in-stent restenosis after coronary stent implantation 18231794_This preliminary study supports the hypothesis that the RAGE system might participate in the disease pathway of primary Sjogren's syndrome (SS), and that sRAGE may be a potential biomarker to aid in the diagnosis of primary SS. 18245812_Loss of RAGE contributes to idiopathic pulmonary fibrosis pathogenesis. 18256374_Endothelial dysfunction in chronic kidney disease may be partly mediated by advanced glycation end product-induced inhibition of endothelial nitric oxide synthase through RAGE activation. 18279703_RAGE could also behave as a receptor for Mycobacterium tuberculosis 18279705_-374T/A RAGE polymorphism is an independent protective factor for cardiac events in nondiabetic patients with coronary artery disease 18302220_study identified advanced glycation end products as the ligand for RAGE on both invasive and non-invasive prostate cancer cells 18322992_The SAA-RAGE-stimulated NF-kappaB signaling pathway has an important role in the pathogenesis of rheumatoid arthritis. 18339893_S100A8/A9-promoted cell growth occurs through RAGE signaling and activation of NF-kappaB. 18355449_These findings reveal new aspects of RAGE regulation and signaling and also provide a new interaction between RAGE and human pathologies. 18421017_results indicate that RAGE serves a protective role in the lung, and that loss of the receptor is related to functional changes of pulmonary cell types, with the consequences of fibrotic disease 18427074_results show significant downregulation of the total RAGE mRNA transcripts in peripheral blood mononuclear cells of patients with probable Alzheimer disease 18431028_low expression of endogenous secretory RAGE in the hippocampus would be associated with the development of Alzheimer's disease 18438937_shedding of RAGE might occur as reactive oxygen species accumulate in brain cells and be part of the process of neurodegeneration 18458846_AGEs induce ROS generation and intensify the proliferation and activation of HSCs, supporting the possibility that antioxidants may represent a promising treatment for prevention of the development of hepatic fibrosis in NASH. 18477569_the peritoneal membrane of the uraemic patients showed an increased submesothelial thickness and a marked induction of RAGE expression and NFkappaB-binding activity 18480271_Distinct regions of RAGE are involved in Abeta-induced cellular and neuronal toxicity with respect to the Abeta aggregation state, suggesing the blockage of particular sites of the receptor as a therapeutic strategy to attenuate neuronal death. 18519797_RAGE Gly82Ser polymorphism may confer not only an increased risk of gastric cancer but also with invasion of gastric cancer in the Chinese population. 18550199_Serum AGER levels are associated with the severity of renal dysfunction and duration of diabetes in type 2 diabetic patients/ 18575614_Observational study of gene-disease association. (HuGE Navigator) 18575614_association between diabetic complications and LTA, TNF and AGER polymorphisms is complex, with partly different alleles conferring susceptibility in type 1 and type 2 diabetic patients 18576917_Advanced oxidation protein products might be new ligands of endothelial RAGE. AOPPs-HSA activates vascular ECs via RAGE-mediated signals. 18595673_Circulating soluble receptor for advanced glycation end products is inversely associated with body mass index and waist/hip ratio in the general population. 18603587_The sheddase ADAM10 was identified as a membrane protease responsible for RAGE cleavage. 18606705_study defines RAGE (receptor for advanced glycation end products) as the hS100A7 receptor, whereas hS100A15 functions through a Gi protein-coupled receptor; hS100A7-RAGE binding, signaling, and chemotaxis are zinc-dependent in vitro 18615900_GFR is a principal determinant of sRAGE concentration and gradual sRAGE increase in subjects with advancing impairment of renal function is paralleled by AGE and its receptor 18615900_Observational study of gene-disease association. (HuGE Navigator) 18645306_Mediation of RAGE affects the balance of cellular proliferation and apoptosis 18657529_RAGE levels increase in conjunction with the onset of Alzheimer's disease, and continue to increase linearly as a function of pathologic severity 18660489_Observational study of gene-disease association. (HuGE Navigator) 18667420_Structural basis for pattern recognition by the receptor for advanced glycation end products (RAGE). 18689872_RAGE, carboxylated glycans and S100A8/A9 play essential roles in tumor-stromal interactions, leading to inflammation-associated colon carcinogenesis. 18698218_maternal esRAGE concentrations are significantly increased in patients with preeclampsia during pregnancy 18701333_association of low soluble isoform of RAGE (sRAGE) with high CML-protein levels in diabetic patients who developed severe diabetic complications supports the hypothesis that sRAGE protects vessels AGE-mediated diabetic microvascular damage 18775683_RAGE was markedly and diffusely expressed in blood vessels, neurons, and the choroid plexus in the brains of two patients who died as the result of brain edema that developed during the treatment of severe diabetic ketoacidosis 18777492_likely that circulating soluble RAGE and endogenous secretory RAGE are distinct markers and that circulating esRAGE levels are associated with the status of early-stage atherosclerosis. 18789567_Advanced glycation end products (AGE) and circulating receptor for AGE (RAGE) levels are independently associated with decreased glomerular filtration rate (GFR) and seem to predict decreased GFR. 18796298_Observational study of gene-disease association. (HuGE Navigator) 18796298_We found no consistent association between prevalent type 2 diabetes mellitus and insulin indices and AGER polymorphisms 18825489_elevated serum HMGB-1 and sRAGE levels are associated with the disease severity and immunological abnormalities in systemic sclerosis. 18854308_HNRNP K and microRNA-16 have roles in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products 18922799_the interaction of the RAGE cytoplasmic domain with Dia-1 is required to transduce extracellular environmental cues evoked by binding of RAGE ligands to their cell surface receptor, resulting in Rac-1 and Cdc42 activation and cellular migration 18926539_Serum endogenous secretory RAGE level is an independent risk factor for the progression of carotid atherosclerosis in type 1 diabetes. 18948101_RAGE signaling contributes to neuroinflammation in infantile neuronal ceroid lipofuscinosis. 18952609_ADAM10 and MMP9 to be involved in RAGE shedding. 18977241_Observational study of gene-disease association. (HuGE Navigator) 18987644_Observational study of gene-disease association. (HuGE Navigator) 19005067_Thus, HMGB1-RAGE signaling links necrosis with macrophage activation and may provide a target for anti-inflammatory therapy in stroke. 19023628_Negative RAGE expression correlates with deeper tumor invasion and venous invasion in patients with esophageal squamous cell carcinoma. 19032093_new insight into AGE-RAGE interaction 19061941_A set of tumours, healthy tissues and various cancer cell lines were screened for RAGE splicing variants and analysed their structure. 19071026_These results indicate that RAGE signaling directly contributes to pathology in cerebral ischemia. 19087540_The increased expression of HMGB1 and RAGE in the placenta may play an important role in the pathogenesis of pre-eclampsia. 19088375_Increased soluble RAGE is localized in ectopic endometrial cells of both adenomyosis and ovarian endometriomas; RAGE may contribute to the pathogenesis of endometriosis. 19103440_Our results provide support for a reduction of S100B levels during reconvalescence from acute paranoid schizophrenia that is regulated by its scavenger RAGE 19129693_Angiotensin II induced RAGE mRNA and protein expression through AT2 receptors. 19133252_No association of RAGE selected gene polymorphisms with 12-months outcome of renal transplants was shown in study. 19143681_several differences in the levels of advanced glycation end products, sRAGE, and proinflammatory cytokines between euglycemic and diabetic pregnancies 19143821_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19167759_Observational study of gene-disease association. (HuGE Navigator) 19183936_sRAGE is associated with greater prevalence of cardiovascular disease in type 1 diabetes, which may be explained by endothelial and renal dysfunction. 19201044_treatment of THP-1 cells with 100 microg of CRP/ml/10(6) cells for 24 h, augmented the expression of RAGE and EN-RAGE genes 19204726_Observational study of gene-disease association. (HuGE Navigator) 19217461_No significant correlation between serum levels off RAGE in hypertensive agents. 19217891_Plasma levels of sRAGE are significantly reduced in definite and borderline nonalcoholic fatty liver disease. 19248116_IL-7 stimulates chondrocyte secretion of S100A4 via activation of JAK/STAT signaling, and then S100A4 acts in an autocrine manner to stimulate MMP-13 production via RAGE. 19252206_Elevated serum advanced glycation end products and their circulating receptors are associated with anaemia in older community-dwelling women. 19275767_a novel biological and genetic marker for vascular disease (Review) 19277685_The up-regulation of RAGE in the Alzheimer Disease optic nerves indicates that RAGE may play a role in the pathophysiology of Alzheimer Disease optic neuropathy. 19284577_RAGE activity influences co-development of joint and vascular disease in rheumatoid arthritis patients. 19285601_G allele at RAGEG82S may be more associated with inflammatory markers under obese status than nonobese conditions. 19332646_the role of recombinant human RAGE in Abeta production was examined in the brain tissue of an AD animal model as well as in cultured cells 19376511_results suggest a possibility that RAGE-VEGF regulation may be related to reproductive dysfunction in aging women 19380603_AGE-2 and AGE-3 activate monocytes via the receptor for AGE (RAGE), leading to the up-regulation of adhesion molecule expression and cytokine production. 19380826_Amyloid beta protein-induced CC-type chemokine receptor 5 (CCR5) up-regulation in human brain microvascular endothelial cells depends on RAGE. 19405075_Stimulation with advanced glycation end products up-regulated endothelial RAGE expression in saphenous vein endothelial cells cultured from diabetic patients. 19439163_This study demonstrated an association of decreased serum endogenous secretory receptor for advanced glycation endproducts (esRAGE) level with coronary plaque progression in patients with diabetes. 19440065_Plasma levels of soluble receptor for advanced glycation end products are associated with endothelial function and predict cardiovascular events in nondiabetic patients. 19448391_High circulating AGEs and RAGE predict cardiovascular disease mortality among older community-dwelling women 19451748_The plasma concentration of sRAGE was highest in patients with sepsis (2,210 +/- 252 pg/ml), while the levels of sRAGE correlated inversely with that of HMGB1 in patients with acute pancreatitis. 19501570_Proteolysis with thrombin or factor Xa revealed several protease sensitive sites in RAGE. 19530996_expression detected in synovial tissue and on the immune cells of rheumatoid arthritis patients; genetic polymorphism is associated with disease 19542745_G82S polymorphism in the RAGE gene is associated with diabetic retinopathy and G-A haplotype containing 1704G and 82S allele might be a genetic marker of diabetic retinopathy in Chinese T2DM patients. 19571577_Advanced glycation end products induce calcification of vascular smooth muscle cells through RAGE/p38 MAPK. 19576587_Report increased expression of RAGE and EN-RAGE in non-diabetic pre-mature coronary artery disease. 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19587357_Meta-analysis of gene-disease association. (HuGE Navigator) 19591173_Extracellular domain of RAGE down-regulates RAGE expression and inhibits p65 and p52 activation in human-salivary gland-cell-lines. 19616266_advanced glycosylation end-product receptor may capture and eliminate circulating serum high mobility group box 1 in humans 19635564_Blockade of the renin angiotensin system by irbesartan may play a protective role against tubular injury in diabetes by attenuating the deleterious effects of AGEs via down-regulation of RAGE. 19659809_Observational study of gene-disease association. (HuGE Navigator) 19672558_Studies indicate that RAGE is subject to ectodomain shedding by ADAM10 and MMP9 and the derived sRAGE can sequester Abeta for systemic clearance and antagonize Abeta binding to cell surface RAGE. 19679874_may be important mediator of cellular injury in fetuses delivered in the setting of inflammation-induced preterm birth 19766904_G82S and -429 T/C polymorphisms of RAGE were associated with the circulating levels of esRAGE but not with coronary artery disease in Chinese patients with Diabetes Melitus, Type 2. 19786934_Data demonstrate that sRAGE level, but not HMGB1 level, is significantly higher in AP patients who develop organ failure than in AP patients without organ failure who recover. 19815443_findings show receptor engagement by distinct advanced glycation end-products differentially enhances expression of RAGE isoforms and adhesion of red blood cells from diabetic patients 19820033_Reduction of advenced glycation end products in normal diets may lower oxidant stress/inflammation and restore levels of AGER an antioxidant, in healthy and aging subjects. 19822091_Molecular study of receptor for advanced glycation endproduct gene promoter and identification of specific HLA haplotypes possibly involved in chronic fatigue syndrome. 19834494_Our data suggest that targeted inhibition of RAGE or its ligands may serve as novel targets to enhance current cancer therapies. 19851445_Observational study of gene-di | ENSMUSG00000015452 | Ager | 8.390195e+01 | 0.4960090 | -1.011561685 | 0.3993058 | 6.211551e+00 | 0.0126919376 | 0.40889816 | No | Yes | 3.325791e+01 | 9.551810 | 7.652771e+01 | 21.910258 | |
| ENSG00000204348 | 1797 | DXO | protein_coding | O77932 | FUNCTION: Decapping enzyme for NAD-capped RNAs: specifically hydrolyzes the nicotinamide adenine dinucleotide (NAD) cap from a subset of RNAs by removing the entire NAD moiety from the 5'-end of an NAD-capped RNA (PubMed:28283058). The NAD-cap is present at the 5'-end of some RNAs and snoRNAs (PubMed:28283058). In contrast to the canonical 5'-end N7 methylguanosine (m7G) cap, the NAD cap promotes mRNA decay (PubMed:28283058). Preferentially acts on NAD-capped transcripts in response to environmental stress (PubMed:31101919). Also acts as a non-canonical decapping enzyme that removes the entire cap structure of m7G capped or incompletely capped RNAs and mediates their subsequent degradation (By similarity). Specifically degrades pre-mRNAs with a defective 5'-end m7G cap and is part of a pre-mRNA capping quality control (By similarity). Has decapping activity toward incomplete 5'-end m7G cap mRNAs such as unmethylated 5'-end-capped RNA (cap0), while it has no activity toward 2'-O-ribose methylated m7G cap (cap1) (PubMed:29601584). In contrast to canonical decapping enzymes DCP2 and NUDT16, which cleave the cap within the triphosphate linkage, the decapping activity releases the entire cap structure GpppN and a 5'-end monophosphate RNA (By similarity). Also has 5'-3' exoribonuclease activities: The 5'-end monophosphate RNA is then degraded by the 5'-3' exoribonuclease activity, enabling this enzyme to decap and degrade incompletely capped mRNAs (PubMed:29601584). Also possesses RNA 5'-pyrophosphohydrolase activity by hydrolyzing the 5'-end triphosphate to release pyrophosphates (By similarity). Exhibits decapping activity towards FAD-capped RNAs (PubMed:32374864). Exhibits decapping activity towards dpCoA-capped RNAs in vitro (By similarity). {ECO:0000250|UniProtKB:O70348, ECO:0000269|PubMed:28283058, ECO:0000269|PubMed:29601584, ECO:0000269|PubMed:31101919, ECO:0000269|PubMed:32374864}. | Exonuclease;Hydrolase;Magnesium;Metal-binding;Nuclease;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome | This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. The function of its protein product is unknown, but its ubiquitous expression and conservation in both simple and complex eukaryotes suggests that this may be a housekeeping gene. [provided by RefSeq, Jul 2008]. | hsa:1797; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; 5'-3' exonuclease activity [GO:0008409]; magnesium ion binding [GO:0000287]; mRNA binding [GO:0003729]; nucleotide binding [GO:0000166]; RNA NAD-cap (NAD-forming) hydrolase activity [GO:0110152]; RNA pyrophosphohydrolase activity [GO:0034353]; mRNA catabolic process [GO:0006402]; NAD-cap decapping [GO:0110155]; nuclear mRNA surveillance [GO:0071028]; nuclear-transcribed mRNA catabolic process [GO:0000956]; nucleic acid phosphodiester bond hydrolysis [GO:0090305]; RNA destabilization [GO:0050779] | 19204726_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 29601584_Study showed that DXO is a multifunctional enzyme that possesses both decapping and 5'-3' exoribonuclease activities toward non-2'-O-methylated RNA transcripts. DXO is directly involved in the mRNA quality control mechanism by degrading mRNAs with abnormal cap structures. 29672716_DOM3Z limits both miR-122-dependent and miR-122-independent Hepatitis C RNA accumulation. | ENSMUSG00000040482 | Dxo | 2.085428e+03 | 1.1811967 | 0.240249267 | 0.2812798 | 7.323499e-01 | 0.3921225422 | 0.84317192 | No | Yes | 2.104627e+03 | 217.594884 | 1.722306e+03 | 183.010075 | |
| ENSG00000204371 | 10919 | EHMT2 | protein_coding | Q96KQ7 | FUNCTION: Histone methyltransferase that specifically mono- and dimethylates 'Lys-9' of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones. Also mediates monomethylation of 'Lys-56' of histone H3 (H3K56me1) in G1 phase, leading to promote interaction between histone H3 and PCNA and regulating DNA replication. Also weakly methylates 'Lys-27' of histone H3 (H3K27me). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these 2 activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX and/or DP1. May also methylate histone H1. In addition to the histone methyltransferase activity, also methylates non-histone proteins: mediates dimethylation of 'Lys-373' of p53/TP53. Also methylates CDYL, WIZ, ACIN1, DNMT1, HDAC1, ERCC6, KLF12 and itself. {ECO:0000269|PubMed:11316813, ECO:0000269|PubMed:18438403, ECO:0000269|PubMed:20084102, ECO:0000269|PubMed:20118233, ECO:0000269|PubMed:22387026, ECO:0000269|PubMed:8457211}. | 3D-structure;ANK repeat;Acetylation;Alternative splicing;Chromatin regulator;Chromosome;Isopeptide bond;Metal-binding;Methylation;Methyltransferase;Nucleus;Phosphoprotein;Reference proteome;Repeat;S-adenosyl-L-methionine;Transferase;Ubl conjugation;Zinc | This gene encodes a methyltransferase that methylates lysine residues of histone H3. Methylation of H3 at lysine 9 by this protein results in recruitment of additional epigenetic regulators and repression of transcription. This gene was initially thought to be two different genes, NG36 and G9a, adjacent to each other in the HLA locus. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:10919; | chromatin [GO:0000785]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; C2H2 zinc finger domain binding [GO:0070742]; histone methyltransferase activity (H3-K27 specific) [GO:0046976]; histone methyltransferase activity (H3-K9 specific) [GO:0046974]; histone-lysine N-methyltransferase activity [GO:0018024]; p53 binding [GO:0002039]; promoter-specific chromatin binding [GO:1990841]; protein-lysine N-methyltransferase activity [GO:0016279]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; transcription corepressor binding [GO:0001222]; zinc ion binding [GO:0008270]; behavioral response to cocaine [GO:0048148]; cellular response to cocaine [GO:0071314]; cellular response to starvation [GO:0009267]; cellular response to xenobiotic stimulus [GO:0071466]; chromatin organization [GO:0006325]; DNA methylation [GO:0006306]; DNA methylation on cytosine within a CG sequence [GO:0010424]; fertilization [GO:0009566]; histone lysine methylation [GO:0034968]; histone methylation [GO:0016571]; long-term memory [GO:0007616]; negative regulation of autophagosome assembly [GO:1902902]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neuron fate specification [GO:0048665]; organ growth [GO:0035265]; peptidyl-lysine dimethylation [GO:0018027]; phenotypic switching [GO:0036166]; regulation of DNA methylation [GO:0044030]; regulation of DNA replication [GO:0006275]; regulation of histone H3-K4 methylation [GO:0051569]; regulation of histone H3-K9 methylation [GO:0051570]; response to ethanol [GO:0045471]; response to fungicide [GO:0060992]; spermatid development [GO:0007286]; synaptonemal complex assembly [GO:0007130] | 15269344_interaction of G9a with a sequence-specific transcription factor that regulates gene repression through CDP/cut. 15590646_Phe at the position equivalent to Phe(281) of Neurospora crassa DIM-5 or Phe(1205) of human G9a allows the enzyme to perform di and tri-methylation, whereas a Tyr at this position is restrictive, inhibiting tri-methylation and yielding a mono- or di-MTase 16287849_Gfi1 associates with G9a and HDAC1 on the promoter of the cell cycle regulator p21Cip/WAF1, resulting in an increase in K9 dimethylation at histone H3 16501248_Observational study of gene-disease association. (HuGE Navigator) 17005678_results show human cytomegalovirus IE86 interacts with HDAC1 and histone methyltransferases G9a and Suvar(3-9)H1 and that coexpression of these chromatin remodeling enzymes with IE86 increases autorepression of the major immediate-early promoter 17085482_Direct cooperation between DNMT1 and G9a provides a mechanism of coordinated DNA and H3K9 methylation during cell division. 17145766_These studies establish a critical role for G9a methyltransferase, histone deacetylases, and the Swi/Snf-Brm complex in the SHP-mediated inhibition of hepatic bile acid synthesis via coordinated chromatin modification at target genes. 17584299_ZNF200 is a novel binding partner of G9a. 17707230_The results reveal the mechanism underlying CSB-mediated activation of ribosomal DNA transcription and link G9a-dependent histone methylation to Pol I transcription elongation through chromatin. 18446223_These data suggest that the 2 HMTs, SUV39H1 and G9a are required to perpetuate the malignant phenotype. 18647749_/EBPbeta as a direct substrate of G9a-mediated post-translational modification that alters the functional properties of C/EBPbeta during gene regulation. 18809684_G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance 18850007_Hypoxia upregulates G9a histone methyltransferase & HDAC1. Overexpression of G9a & HDAC1 attenuated RUNX3 expression. The overexpression of G9a & HDAC1, but not their mutants, inhibited the nuclear localization & expression of RUNX3. 19061646_Chromodomain on Y-like (CDYL) is identified as a REST corepressor that physically bridges REST and the histone methylase G9a to repress transcription. 19124506_Observational study of gene-disease association. (HuGE Navigator) 19144645_Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins 19531572_We found robust DNA hypomethylation in G9a/GLP knockdown of murine ES cells but a lack of DNA methylation changes in G9a/GLP knockdown human cancer cells; intriguingly, this distinction also extended to markers of global DNA methylation. 19690169_Data show that RelB induces facultative heterochromatin formation by directly interacting with the histone H3 lysine 9 methyltransferase G9a. 19776757_EVI-1 interacts with histone methyltransferases SUV39H1 and G9a for transcriptional repression and bone marrow immortalization. 19843671_EHMT2 polymorphisms are associated with colorectal cancer. 19843671_Observational study of gene-disease association. (HuGE Navigator) 19851445_Observational study of gene-disease association. (HuGE Navigator) 20118233_G9a and Glp methylate lysine 373 in the tumor suppressor p53 20335163_G9a is responsible for the transcriptional quiescence of latent HIV-1 provirus 20421388_Genome-wide analysis and microarray can identify genes that are specifical regulated by G9a. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20940408_Findings establish a functional contributio of G9a overexpression with concomitant dysregulation of epigenetic pathways in lung cancer progression. 20960050_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21041608_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21847359_Our results suggest that dysregulation of EHMT2 plays an important role in the growth regulation of cancer cells. 21984853_the coactivator and corepressor functions of G9a involve different G9a domains and different molecular mechanisms 22389001_study concludes that a subset of cancer-related Runx2 target genes require recruitment of G9a for their expression, but do not depend on its histone methyltransferase activity 22406531_Snail interacted with G9a, a major euchromatin methyltransferase responsible for H3K9me2, and recruited G9a and DNA methyltransferases to the E-cadherin promoter for DNA methylation. 22801367_data show that HMTase G9a negatively regulates JAK2 transcription and H3Y41 phosphorylation on the lmo2 promoter; and study provides new insights into G9a function in the regulation of hematopoiesis and leukemogenesis 23151507_findings indicate distinct mechanisms of G9a coactivator vs. corepressor functions in transcriptional regulation and provide insight into the molecular mechanisms of G9a coactivator function 23244065_With G9a and GLP1 as representative Protein methyltransferases (PMTs), it demonstrated the use of methionine analogues, a MAT variant and engineered PMTs for substrate labeling in vitro and genome-wide chromatin modifications within living cells. 23283977_G9a mediates E4BP4-dependent suppression of hepatic Fgf21 by enhancing histone methylation (H3K9me2) of the Fgf21 promoter 23466651_Histone-lysine methyltransferase EHMT2 is involved in proliferation, apoptosis, cell invasion, and DNA methylation of human neuroblastoma cells. 23481705_H3K9me3-positive grade II oligodendrogliomas, but not other tumor subtypes, show improved overall survival compared with H3K9me3-negative cases. 23629948_These findings suggest that h-CDYLb and G9a are cooperatively involved in hepatocellular carcinomas 23637228_Sumoylation of Sharp-1 exerts an impact on chromatin structure and transcriptional repression of muscle gene expression through recruitment of G9a. 23760081_Genome association studies identified EHMT2 as a new risk-associated loci for chronic hepatitis B on the HLA region of chromosome 6. 23807219_G9a inhibition may help overcome drug resistance in certain cancer cells. 23918802_G9a directly represses genes known to participate in the autophagic process. 24019522_all of the H3K36-specific methyltransferases, including ASH1L, HYPB, NSD1, and NSD2 were inhibited by ubH2A, whereas the other histone methyltransferases, including PRC2, G9a, and Pr-Set7 were not affected by ubH2A. 24315373_G9A inactivation depletes serine and its downstream metabolites, triggering cell death with autophagy in cancer cell lines of different tissue origins. 24389103_PRC2 and G9a/GLP interact physically and functionally. 24492005_G9a functions as a coactivator for p21 transcription, and directs cells to undergo apoptosis. 24652950_G9a- and EZH2-mediated histone hypermethylation. 24658378_overexpression of EHMT2 might be a prognostic marker in patients with melanoma. 24805087_MCM7 and G9a may serve as effective prognostic factors and could also be used as biomarkers for predicting various clinical outcomes of oesophageal squamous cell carcinoma 24833465_G9a-dependent H3K9me2 repression on CD133 and Sox2 was one of the main switches of the self-renewal in glioma cancer stem cells. 25027955_Genetic or pharmacological inhibition of G9a reduced cell proliferation without inducing necrosis or apoptosis. 25071150_Results show that G9a acts positively to promote tumor formation. 25079219_The expression level of EHMT1 and EHMT2 inversely correlates with the type I interferon responsiveness in chronic myeloid leukemia cell lines. 25115793_G9a contributes to multiple steps of ovarian cancer metastasis. 25365549_The current knowledge on the mechanisms of action and function of EHMT2, with particular emphasis on their interplay in the regulation of chromatin states and biological processes. 25595900_Down-regulation of G9a triggers DNA damage response and inhibits colorectal cancer cells proliferation. 25613390_High G9a contributes to endometrial cancer progression. 25634693_H3K9 methyltransferase G9a plays role in the apoptosis and autophagy in oral squamous cell carcinoma. 25749385_G9a protein is essential for the induction of epithelial-to-mesenchymal transition and cancer stem cell-like properties in HNSCC 25789554_Data indicate zinc finger proteins ZNF644 and WIZ as two core subunits in the histone-lysine N-methyltransferase G9a/GLP complex, and interact with the transcription activation domain of G9a and GLP. 25840984_we defined a role for activated STAT3 and G9a histone methyltransferase in epigenetic silencing of miR-200c, which promotes the formation of breast CSCs defined by elevated cell surface levels of the leptin receptor 25843229_KMT1c (G9a) is markedly up regulated after differentiation of monocytes into immature dendritic cells. 25935252_This study demonstrate that the increases in a restrictive epigenome seen in schizophrenia are sex dependent. Specifically,G9A were significantly increased in lymphocytes from men with schizophrenia. 25982273_LASP-1 associated with UHRF1, G9a, Snail1 and di- and tri-methylated histoneH3 in a CXCL12-dependent manner based on immunoprecipitation and proximity ligation assays 26192994_Homocysteine Induces Collagen I Expression by Downregulating Histone Methyltransferase G9a 26320100_data provide genetic and pharmacologic evidence that EHMT1 and EHMT2 are epigenetic regulators involved in gamma-globin repression and represent a novel therapeutic target for SCD. 26397365_The present study aimed at examining the potential role of autophagy in the anti-proliferation effect of G9a inhibition on transitional cell carcinoma T24 and UMUC-3 cell lines in vitro. 26551542_G9a has a dominant function in transcriptional repression of potassium channels and in acute-to-chronic pain transition. 26688070_G9a promotes H3K27 methylation of the E-cadherin promoter by upregulating PCL3 to increase PRC2 promoter recruitment and by downregulating the H3K27 demethylase KDM7A to silence E-cadherin gene 26725954_IFNgamma induced PPAR gamma coactivator-1 alpha (PGC-1alpha) positively regulated RIG-I; with PRMT-1 and G9a affecting PGC-1alpha in a counter-regulatory manner. 26765460_high expression in hepatocellular carcinoma associated with worse outcome 26960573_Along with the PHF20/MOF complex, G9a links the crosstalk between ERalpha methylation and histone acetylation that governs the epigenetic regulation of hormonal gene expression. 26997278_The G9a exon 10(+) isoform is necessary for neuron differentiation and regulates the alternative splicing pattern of its own pre-mRNA, enhancing exon 10 inclusion. 27052169_G9a and GLP are required for stable maintenance of imprinted DNA methylation in embryonic stem cells. 27081761_Depletion of G9a inhibits cell growth and induces cells apoptosis in gastric cancer. 27174920_EHMT2 can directly repress Beclin-1 and the inhibition of EHMT2 may be a useful therapeutic approach for cancer prevention by activating autophagy 27431310_G9a inhibition potentiates the anti-tumour activity of DNA double-strand break inducing agents by impairing DNA repair independent of p53 status. 27452519_Low G9a expression is associated with lung and colonic cancer. 27531902_interleukin-8 is a downstream effector of G9a to increase gemcitabine resistance in pancreatic cancer. Overexpression of G9a correlated with poor survival and early recurrence in pancreatic cancer patients. 27588951_we find an estrogen receptor-independent synthetic lethal interaction between a GATA3 frameshift mutant with an extended C-terminus and the histone methyltransferases G9A and GLP, indicating perturbed epigenetic regulation 27748874_High expression of G9a and H3K9me2 in glioma tissue samples was associated with the WHO grade, which indicated that G9a and H3K9me2 may promote generation and development of glioma. BIX01294 may be a potential novel therapeutic agent in the treatment of glioma. 28246360_Inhibition of EZH2 (by GSK-343 or EPZ-6438) or inhibition of EHMT2 (by UNC-0638) in the Th17 primary cell model of HIV latency or resting memory T cells isolated from HIV-1-infected patients receiving highly active antiretroviral therapy, was sufficient to induce the reactivation of latent proviruses. 28265008_Taken together, our findings provide evidence that G9a protects head and neck squamous cell carcinomas (HNSCC)cells against chemotherapy by increasing the synthesis of GSH, and imply G9a as a promising target for overcoming cisplatin resistance in HNSCC 28278257_G9A overexpression is associated with peritoneal fibrosis. 28351524_Knockdown of G9a increased the sensitivity of cells to radiation treatment and sensitized cells to DNA damage agents through PP2A-RPA axis. 28383547_study uncovers a novel mechanism of G9A promoting tumor cell growth and invasion by silencing CASP1, and implies that G9A may serve as a therapeutic target in treating Non-small-cell lung cancer. 28423509_G9a inhibition impairs anchorage-dependent and -independent cell growth in hepatocellular carcinoma cells 28455378_G9a is an important regulator in placental diseases caused by defective vascular maturation. 28483947_these findings reveal a role for REST-associated G9a and histone H3K9 methylation in the repression of USP37 expression in medulloblastoma. Reactivation of USP37 by G9a inhibition has the potential for therapeutic applications in REST-expressing medulloblastomas 28532996_Identified the tumor suppressor RARRES3 as a critical target of G9a. Epigenetic silencing of RARRES3 contributed to the tumor-promoting function of G9a 28532996_Loss of miR-1 and gene copy number gain of G9a contributed to its frequent upregulation in liver cancer, which epigenetically silenced the expression of tumor suppressor RARRES3, and ultimately contributed to hepatocellular carcinoma (HCC) cell proliferation and migration. 28615290_Regulated methylation and phosphorylation serve as a switch controlling G9a and GLP coactivator function, suggesting that this mechanism may be a general paradigm for directing specific transcription factor and coregulator actions on different genes. 28630300_G9a plays a role as an epigenetic mediator of hypoxic response in breast cancer 28667416_The study identified that decreased expression of SATB2 was correlated with tumor progression and poor prognosis in non-small cell lung cancer (NSCLC) patients. Furthermore, SATB2 suppressed lung cancer cell invasion and metastasis and regulated the expression of EMT-related proteins and histone methylation by G9a. 28698370_regulatory pathway based on casein kinase 2-G9a-RPA permits homologous recombination in cancer cells 28803780_A histone H3K9-like mimic within LIG1 is methylated by G9a and GLP and avidly binds UHRF1. Interaction with methylated LIG1 promotes the recruitment of UHRF1 to DNA replication sites and is required for DNA methylation maintenance. 28819251_G9a promotes breast cancer by regulating iron metabolism through the repression of ferroxidase hephaestin. 29053336_Suggest that a novel and functionally interdependent interplay between EZH2 and G9a regulates histone methylation-mediated epigenetic repression of the antifibrotic CXCL10 gene in idiopathic pulmonary fibrosis. 29113759_Our results demonstrate of the central role of G9a HMT in the promotion of endothelial cells proliferation, and suggest that endothelial G9a HMT may be a target in the treatment of vascular proliferative disorders and tumor neovascularization. 29133140_that G9a contributes to biliary tract cancer carcinogenesis and may represent a potential prognostic factor as well as a therapeutic target 29238036_EHMT2 rs35875104 showed a strong association with the risk of chronic hepatitis B. 29259012_High G9A expression is associated with Ovarian Cancer metastasis. 29337058_In this study, EHMT2 (a histone methyltransferase) was determined to be significantly overexpressed in breast cancer tissues and in Oncomine data. In addition, knockdown of EHMT2 reduced cell migration and invasion. 29374157_EHMT2 expression and enzymatic activity levels were elevated in EGFR-tyrosine kinase inhibitor-resistant non-small cell lung cancer cells. 29449539_G9A promotes gastric cancer peritoneal metastasis by upregulating ITGB3 in a SET domain-independent manner. 29467226_unveil E4BP4 as a critical transcriptional modulator repressing RASSF8 expression through histone methyltransferases, G9a and SUV39H1. 29807539_Results establish a direct functional link between G9a and MMP-9 in the context of pro-osteoclastogenic transcriptional programs. The model suggests that G9a mediated H3K27me1 serves as an essential mark for MMP-9 recruitment and proteolytic activity at genes encoding positive regulators of osteoclast differentiation, thus keeping them active. 29855824_In summary, it was demonstrated here that the overexpression of GLP is associated with a worse prognosis in Chronic Lymphocytic Leukemia (CLL) and that the inhibition of GLP/G9a complex leads to CLL cell death. These results suggest a potential use for GLP/G9a as markers for disease progression, as well as a promising epigenetic target for CLL treatment and prevention of disease evolution. 29860315_G9a and GLP directly bound to the alpha subunit of HIF-1 (HIF-1alpha) and catalyzed mono- and di-methylation of HIF-1alpha. The methylation suppressed HIF-1 transcriptional activity. 30158521_Kaempferol activates the IRE1-JNK-CHOP signaling from cytosol to nucleus, and G9a inhibition activates autophagic cell death in GC cells. 30241941_Results show that casein kinase 2 alpha (CK2 )downregulation leads to H3K9me3 and senescence-associated heterochromatic foci (SAHF) formation by increasing H3K9 trimethylase SUV39h1 (SUV39h1) and decreasing H3K9 dimethylase G9a (G9a). 30305606_Inhibiting G9a/GLP demethylation potentially represents a novel method to restore sensitivity of treatment-resistant B-ALL tumors to GC-induced cell death. 30365075_Data found that EHMT2 expression was upregulated in breast cancer (BRC) tissues and cell lines, which indirectly controlled HSPD1 expression and its localization, thus inhibiting cell apoptosis. These results identified EHMT2 as a novel BRC prognostic marker. 30535125_we reported that FOXO1 is methylated by G9a at K273 residue in vitro and in vivo. Methylation of FOXO1 by G9a increased interaction between FOXO1 and a specific E3 ligase, SKP2, and decreased FOXO1 protein stability. 30718920_Cyclin D1 is required for recruitment of G9a to target genes in chromatin. 30747104_The most compelling new results revealed a consistent spontaneous network hyperactivity in neurons deficient for CNTN5 or EHMT2. 30833420_High G9a expression is associated with Alveolar Rhabdomyosarcoma. 30867409_Endogenous G9a is SUMOylated in proliferating skeletal myoblasts. There are four potential SUMOylation consensus motifs in G9a. Mutation of all four acceptor lysine residues inhibits SUMOylation. In contrast, SUMO-defective G9a is unable to enhance proliferation of myoblasts. The proliferation defect of primary myoblasts from conditional G9a-deficient mice is rescued by wild-type, but not SUMOylation-defective, G9a. 31095524_CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. 31271097_Overexpression of Snail2 enhanced the migration and invasion ability in lung carcinoma, whereas G9a and Histone Deacetylase inhibition significantly reversed this effect. 31599405_EHMT2 promotes the development of prostate cancer by inhibiting PI3K/AKT/mTOR pathway. 31770281_G9a expression was upregulated in Gastric Cancer tissues, was significantly correlated with patients' lymph node metastasis, had shorter survival time and could be an effective prognostic biomarker for Gastric Cancer . 31775874_Results show that EHMT2 is overexpressed in PARP inhibitors (PARPi)-resistant high-grade serous ovarian carcinoma (HGSOC) cell lines and patient-derived ascites. EHMT2 correlates with HGSOC progression and chemoresistance, and combined EHMT1/2 high expression level correlates with poor patient outcomes. Knockdown or inhibition of EHMT1/2 restores PARPi sensitivity suggesting their implication in PARPi resistance. 31990985_The Histone Methyltransferase G9a Promotes Cholangiocarcinogenesis Through Regulation of the Hippo Pathway Kinase LATS2 and YAP Signaling Pathway. 32015216_Abnormal overexpression of G9a in melanoma cells promotes cancer progression via upregulation of the Notch1 signaling pathway. 32033749_RNA-seq based transcriptome analysis of EHMT2 functions in breast cancer. 32060676_Towards the understanding of the activity of G9a inhibitors: an activity landscape and molecular modeling approach. 32292512_G9a-mediated repression of CDH10 in hypoxia enhances breast tumour cell motility and associates with poor survival outcome. 32327527_Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. 32512705_G9a Is SETting the Stage for Colorectal Oncogenesis. 32684241_Euchromatin histone methyltransferase II (EHMT2) regulates the expression of ras-related GTP binding C (RRAGC) protein. 33077425_Histone Methyltransferase G9a Regulates Expression of Nuclear Receptors and Cytochrome P450 Enzymes in HepaRG Cells at Basal Level and in Fatty Acid Induced Steatosis. 33113380_G9a Plays Distinct Roles in Maintaining DNA Methylation, Retrotransposon Silencing, and Chromatin Looping. 33121982_Whole-proteome analysis of mesonephric-derived cancers describes new potential biomarkers. 33283949_Targeting H3K9 methyltransferase G9a and its related molecule GLP as a potential therapeutic strategy for cancer. 33298547_G9a Promotes Invasion and Metastasis of Non-Small Cell Lung Cancer through Enhancing Focal Adhesion Kinase Activation via NF-kappaB Signaling Pathway. 33323965_G9a controls pluripotent-like identity and tumor-initiating function in human colorectal cancer. 33530253_Prognostic value of lymphocyte-to-monocyte ratio and histone methyltransferase G9a histone methyltransferase in patients with double expression lymphoma: A retrospective observational study. 33931742_Targeting protein lysine methyltransferase G9A impairs self-renewal of chronic myelogenous leukemia stem cells via upregulation of SOX6. 34168040_A disproportionate impact of G9a methyltransferase deficiency on the X chromosome. 34519223_H3K9 methyltransferase EHMT2/G9a controls ERVK-driven noncanonical imprinted genes. 34588438_SPOP mutation induces DNA methylation via stabilizing GLP/G9a. 34619147_Heterodimerization of H3K9 histone methyltransferases G9a and GLP activates methyl reading and writing capabilities. 34681949_EHMT2/G9a as an Epigenetic Target in Pediatric and Adult Brain Tumors. 34954891_EHMT1/EHMT2 in EMT, cancer stemness and drug resistance: emerging evidence and mechanisms. 34972816_Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. 35054776_G9a Knockdown Suppresses Cancer Aggressiveness by Facilitating Smad Protein Phosphorylation through Increasing BMP5 Expression in Luminal A Type Breast Cancer. | ENSMUSG00000013787 | Ehmt2 | 5.395625e+03 | 1.2875910 | 0.364674422 | 0.2969999 | 1.515990e+00 | 0.2182273517 | 0.78763590 | No | Yes | 5.638700e+03 | 620.917777 | 3.994511e+03 | 451.593087 | |
| ENSG00000204580 | 780 | DDR1 | protein_coding | Q08345 | FUNCTION: Tyrosine kinase that functions as cell surface receptor for fibrillar collagen and regulates cell attachment to the extracellular matrix, remodeling of the extracellular matrix, cell migration, differentiation, survival and cell proliferation. Collagen binding triggers a signaling pathway that involves SRC and leads to the activation of MAP kinases. Regulates remodeling of the extracellular matrix by up-regulation of the matrix metalloproteinases MMP2, MMP7 and MMP9, and thereby facilitates cell migration and wound healing. Required for normal blastocyst implantation during pregnancy, for normal mammary gland differentiation and normal lactation. Required for normal ear morphology and normal hearing (By similarity). Promotes smooth muscle cell migration, and thereby contributes to arterial wound healing. Also plays a role in tumor cell invasion. Phosphorylates PTPN11. {ECO:0000250, ECO:0000269|PubMed:12065315, ECO:0000269|PubMed:16234985, ECO:0000269|PubMed:16337946, ECO:0000269|PubMed:19401332, ECO:0000269|PubMed:20093046, ECO:0000269|PubMed:20432435, ECO:0000269|PubMed:20884741, ECO:0000269|PubMed:21044884, ECO:0000269|PubMed:9659899}. | 3D-structure;ATP-binding;Alternative splicing;Calcium;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Kinase;Lactation;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Pregnancy;Receptor;Reference proteome;Secreted;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | Receptor tyrosine kinases play a key role in the communication of cells with their microenvironment. These kinases are involved in the regulation of cell growth, differentiation and metabolism. The protein encoded by this gene belongs to a subfamily of tyrosine kinase receptors with homology to Dictyostelium discoideum protein discoidin I in their extracellular domain, and that are activated by various types of collagen. Expression of this protein is restricted to epithelial cells, particularly in the kidney, lung, gastrointestinal tract, and brain. In addition, it has been shown to be significantly overexpressed in several human tumors. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Feb 2011]. | hsa:780; | extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; collagen binding [GO:0005518]; metal ion binding [GO:0046872]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase collagen receptor activity [GO:0038062]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; axon development [GO:0061564]; branching involved in mammary gland duct morphogenesis [GO:0060444]; cell adhesion [GO:0007155]; collagen-activated tyrosine kinase receptor signaling pathway [GO:0038063]; ear development [GO:0043583]; embryo implantation [GO:0007566]; lactation [GO:0007595]; mammary gland alveolus development [GO:0060749]; negative regulation of cell population proliferation [GO:0008285]; neuron projection extension [GO:1990138]; peptidyl-tyrosine autophosphorylation [GO:0038083]; positive regulation of kinase activity [GO:0033674]; protein autophosphorylation [GO:0046777]; regulation of cell growth [GO:0001558]; regulation of cell-matrix adhesion [GO:0001952]; regulation of extracellular matrix disassembly [GO:0010715]; smooth muscle cell migration [GO:0014909]; smooth muscle cell-matrix adhesion [GO:0061302]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; wound healing, spreading of cells [GO:0044319] | 12935821_gene expression analysis with DDR1 overexpression, using microarrays specific for human and mouse genes coding for extracellular matrix proteins or ECM-interacting factors 14764702_The interaction of DDR1b isoform with collagen up-regulates the production of IL-8, macrophage inflammatory protein-1 alpha, and monocyte chemoattractant protein-1 in macrophages in a p38 mitogen-activated protein kinase- and NF-kappa B-dependent manner. 15111304_DDR1 and DDR2 have roles in the regulation of collagen turnover mediated by SMCs in obstructive diseases of blood vessels and the lung 15136580_several residues within loop 1 (Ser-52-Thr-57) and loop 3 (Arg-105-Lys-112) as well as Ser-175 in loop 4 of DDR1 are critically involved in collagen binding 15213330_Observational study of gene-disease association. (HuGE Navigator) 15240533_up-regulation of DDR1, CLDN3, and epithelial cell adhesion molecule are early events in the development of epithelial ovarian cancer 16440311_The results demonstrate that collagen-evoked ectodomain cleavage of DDR1 is mediated in part by Src-dependent activation 16774916_dimeric receptor tyrosine kinase DDR1 requires a transmembrane leucine zipper for activation 16912190_DDR1 signaling provides a novel target for therapeutic intervention with the prosurvival/antiapoptotic machinery of tumor cells. 17001518_Results strongly suggest that DDR1 mediates cell invasion-related signaling between collagen type I and MMP-2 and -9 in pituitary adenoma cells. 17027969_Thus, DARPP-32 signaling downstream of DDR1 is a potential new target for effective anti-metastatic breast cancer therapy 17101694_DDR1 knockdown decreased melanocyte adhesion to collagen IV and shifted melanocyte localization in a manner similar to CCN3 knockdown. 17299390_altered expression of DDR1 may contribute to malignant progression of non-small cell lung carcinoma 17440435_In the present study we screened for single nucleotide polymorphisms (SNPs) in the DNA from 100 schizophrenia patients. We identified mutation within exon 10 that produces the amino-acid substitution N502S in the a-d isoforms, and M475V in the e isoform. 17440435_Observational study of gene-disease association. (HuGE Navigator) 17704737_DDR1 gene may be regarded as a potential marker for some types of endometrial cancer. 17970783_PCA-1-DDR-1 signaling is a new important axis involved in malignant potential prostate cancer associated with hormone-refractory status. 17982627_There are significantly higher number of invading cells in DDR1a expressing hepatocellular carcinoma cells. 18023033_DDR2 was differentially upregulated in nasopharyngeal carcinoma and modulated by EBV Zta protein 18065762_The identification of DDR1 dimers provides new insights into the molecular structure of receptor tyrosine kinases and suggests distinct signaling mechanisms of each receptor subfamily. 18190796_KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. 18270328_Observational study of gene-disease association. (HuGE Navigator) 18362184_Initiation of the signal requires two collagen receptors, alpha2beta1 integrin and discoidin domain receptor (DDR). Each receptor propagates signals through separate pathways that converge to up-regulate N-cadherin. 18836851_Differentiation of a human oligodendroglial cell line, HOG16, was associated with an increase in mRNA expression of DDR1 and several myelin proteins but not other proteins. DDR1 is upregulated when oligodendrocyte myelinating machinery is activated. 19262089_transfection of the vector encoding full-length DDR1 or siRNA targeting DDR1 up- or downregulated respectively secretion of MMP-2 and -9, and cell invasion 19401332_The collagen receptor DDR1 regulates cell spreading and motility by associating with myosin IIA. 19752756_acts as a costimulatory receptor in T-cell activation 19837266_Observational study of gene-disease association. (HuGE Navigator) 19851445_Observational study of gene-disease association. (HuGE Navigator) 20007060_We failed to show a strong association between the DDR1 gene SNPs and NSV. These facts indicate that there is no genetic association between DDR1 and NSV. 20093046_Type I collagen induces smooth muscle cell migration through DDR1 20182441_Observational study of gene-disease association. (HuGE Navigator) 20182441_we propose DDR1 as a susceptibility gene for vitiligo, possibly implicating a defective cell adhesion in vitiligo pathogenesis 20219323_Astaxanthin attenuates the UVA-induced up-regulation of matrix-metalloproteinase-1 and skin fibroblast elastase in human dermal fibroblasts 20353877_Results suggest that DDR1 can represent an additional receptor regulating T cell movement in the tissues. 20372823_Observational study of gene-disease association. (HuGE Navigator) 20372823_results suggest that the discoidin domain receptor 1 gene is associated with increased susceptibility, pathological advancement, and the development of proteinuria in childhood IgA nephropathy. 20380825_These observations suggest that DDR1 could be an important constituent of myelin. 20487506_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20587610_Observational study of gene-disease association. (HuGE Navigator) 20596615_up-regulation of DDR1 may contribute to the progression and poor prognosis of NSCLC and this effect may be associated with increased invasiveness. 20799954_Decreased expression of miR-199a-5p contributes to increased cell invasion by functional deregulation of DDR1 activity in hepatocellular carcinoma. 20884741_These data suggest that extracellular matrix signalling via DDR1 regulates aspects of bronchial epithelial repair, integrity and MMP expression in the airways. 20920467_MMTR is an intrinsic negative cell cycle regulator that modulates the CAK kinase activity via interaction with MAT1. 21044884_Discoidin domain receptor 1 transmembrane signalling, collagen binding and receptor activation occurred with triple-helical peptides containing the GVMGFO motif 21170030_Data show that DDR1 coordinates the Par3/Par6 cell-polarity complex through its carboxy terminus, binding PDZ domains in Par3 and Par6. 21335558_Collagen I induces discoidin domain receptor (DDR) 1 expression through DDR2 and a JAK2-ERK1/2-mediated mechanism in primary human lung fibroblasts. 21356365_These data elucidate DDR1 function pertinent to cirrhosis and indicate the importance of epithelial cell-collagen interactions in chronic liver injury. 21461859_The DDR1 mediates MMP-2 and -9 secretions and invasion induced by native type IV collagen in MDA-MB-231 breast cancer cells. 21541037_The expression of the DDR1 protein significantly correlated with poor disease-free survival in patients with serous ovarian cancer. 21551363_malfunction of neprilysin in infected macrophages may contribute to acceleration of beta amyloidosis in HIV-inflicted brains 22057045_The map of phosphotyrosine mediated interactors of DDR1 created in this study will serve as a starting point for functional investigations which will enhance our knowledge on the role of the DDR1 receptor in health and disease. 22077590_The results of this study suggested that the DDR1 A2RE sequence is functionally involved in the hnRNP A2/B1-mediated splicing and transport of the DDR1c mRNA. 22223527_Disruption of DDR1 hampers tumor cell survival, leading to impaired early tumor-bone engagement during skeletal homing. 22374871_Most AML patients had a significant decrease in miR-199b-5p levels with elevated PODXL & DDR1 expressions. Both PODXL & DDR1 are targets of miR-199b-5p. 22380734_DDR1 polymorphisms are not likely to contribute to predispositions of aspirin-exacerbated respiratory disease, but may be potentially associated with FEV(1) decline by aspirin provocation in asthmatics. 22421314_This study found a statistically significant increased expression of DDR1c in the prefrontal cortex from the schizophrenia group compared to the control group 22507556_Expression of DDR1 was decreased in lesional versus perilesional vitiligo skin in the majority of patients. 22752569_DD1 mRNA and protein levels were higher in patients with recurrent Hepatocellular carcinoma , suggesting this gene maybe involved in tumor invasion and metastasis. 22895534_these data suggest that NEP can augment taxane-induced apoptosis through inhibition of Akt/Bad signaling 23284937_Discoidin domain receptors promote alpha1beta1- and alpha2beta1-integrin mediated cell adhesion to collagen by enhancing integrin activation. 23307244_DDR1 expression was not predictive for patient survival in human breast cancer 23335507_Discoidin domain receptors are unique receptor tyrosine kinases in collagen-mediated signaling [review] 23382212_results identify the ARCH domain of XPD as a platform for the recruitment of CAK and as a molecular switch that might control TFIIH composition and play a role in conversion of TFIIH from a factor active in transcription to a factor involved in DNA repair 23519472_a role for the collagenase of membrane-type MMPs in regulation of DDR1 cleavage and activation at the cell-matrix interface. 23530036_the expression of the novel collagen receptor discoidin domain receptor 1 (DDR1) by human MKs at both mRNA and protein levels and provide evidence of DDR1 involvement in the regulation of MK motility on type I collagen 23761027_Upregulation of DDR1 induced by collagen I may contribute to the development and progression of NSCLC. 23810922_The DDR1 extracellular domain plays a crucial role in receptor oligomerization which mediates high-affinity interactions with its ligand. 24018687_regulation of DDRs by glucose 24136166_We show that expression of the latent LMP1 in primary human germinal center B cells, the presumed progenitors of HRS cells, upregulates discoidin domain receptor 1 (DDR1), a receptor tyrosine kinase activated by collagen. 24361528_In this review we highlight the mechanisms whereby DDRs(DDR1 and DDR2) regulate two important processes, namely inflammation and tissue fibrosis. 24509848_N-glycosylation at the highly conserved (211)NDS motif evolved to act as a negative repressor of DDR1 phosphorylation in the absence of ligand 24671415_These findings indicate that the extracellular juxtamembrane region of DDR1 is exceptionally flexible and does not constrain the basal or ligand-activated state of the receptor. 24723326_High DDR1 protein expression is associated with recurrence in ameloblastomas. 24768818_Crystal structures of DDR1 reveal a DFG-out conformation (DFG, Asp-Phe-Gly) of the kinase domain that is stabilized by a salt bridge between the activation loop and alphaD helix. Differences to Abelson kinase are observed in the DDR1 P-loop's beta-hairpin. 25064398_Report aberrant methylation of DDR1 in men with nonobstructive azoospermia. 25155634_found an inverse correlation between ZEB1 and DDR1 expression in various cancer cell lines and in human breast carcinoma tissues 25369402_Silencing of DDR1 inhibited tumor cell growth and motility, and induced TGFBI expression, both in vitro and in vivo. 25422375_DDR1 depletion blocked cell invasion in a collagen gel 25427824_We uncovered ten new mutations in TNK2 and DDR1 within serous and endometrioid ECs, thus providing novel insights into the mutation spectrum of each gene in EC. 25470979_Defective Ca(2+) binding in a conserved binding site causes incomplete N-glycan processing and endoplasmic reticulum trapping of DDR1 and DDR2 receptors. 25667101_A DDR1(Low)/DDR2(High) protein profile is associated with TNBC and may identify invasive carcinomas with worse prognosis. 25673773_suggest that DDR1 suppression may enhance adipose-derived stem cell chondrogenesis by enhancing the expression of chondrogenic genes and cartilaginous matrix deposition. 25774665_MT1-MMP-discoidin domain receptor 1 axis regulates collagen-induced apoptosis in breast cancer cells 25992553_alpha5(IV), but not alpha1(IV), promotes lung cancer cell proliferation and tumor angiogenesis through non-integrin collagen receptor DDR1-mediated ERK activation. 26091274_Reduced DDR1 expression may be implicated in impaired melanocyte adhesion process involved in vitiligo pathogenesis 26286316_Hypoxia can increase DDR1 expression in pituitary adenoma cells, leading to improved MMP-2 and MMP-9 secretion, and promoting pituitary adenoma cell proliferation and invasion. 26297342_DDR1 expression is a prognostic indicator in pancreatic ductal carcinoma. 26589794_Knockdown DDR1 reversed the effects of Galpha13 knockdown on cell-cell adhesion and proteolytic invasion in three-dimensional collagen. 26655502_Upregulation of DDR1 collagen receptor is associated with breast cancer. 26702058_Data suggest SCl2 (Streptococcus pyogenes) binds to DDR (DDR1, DDR2) ectodomain w/o stimulating receptor signaling; here, protein engineering was used to construct SCl-like proteins that inhibit collagen-DDR interactions and macrophage migration. 26719540_DDR1 is a key modulator of RIT activity. 27020590_Our results suggest that DDR1 is both a prognostic marker for renal clear cell carcinoma and a potential functional target for therapy 27179963_DDR1 overexpression promoted GC cell proliferation (p < 0.05), migration (p < 0.01), and invasion (p < 0.01), and accelerated the growth (p < 0.05) as well as the microvessel formation (p < 0.01) of transplantation tumor in nude mice. 27368100_Study concludes that non-canonical DDR1 signaling enables breast cancer cells to exploit the ubiquitous interstitial matrix component collagen I to undergo metastatic reactivation in multiple target organs. 27384677_Data suggest that IGF-I/IGF-IR system triggers stimulatory actions through both GPER and DDR1 in aggressive tumors as mesothelioma and lung tumors. 27605668_Isoform b of DDR1 is responsible for collagen I-induced up-regulation of N-cadherin and tyrosine 513 of DDR1b is necessary. 27720259_data demonstrate that TGF-beta1 favors linear invadosome formation through the expressions of both the inducers, such as collagen and LOXL2, and the components such as DDR1 and MT1-MMP of linear invadosomes in cancer cells. Meanwhile, our data uncover a new TGF-beta1-dependent regulation of DDR1 expression. 28060374_the tyrosine kinase DDR1 is expressed at the cell surface of circulating B-CLL cells, although with a remarkable heterogeneity among individual cases. DDR1 gene expression correlates with that of the ZAP70 tyrosine kinase and with the IGVH mutational status, which are regarded as prognostic markers in CLL 28368050_our study provides a novel regulatory pathway involving TM4SF1, DDR1, MMP2 and MMP9, which promotes the formation and function of invadopodia to support cell migration and invasion in pancreatic cancer. 28590245_These results support an activation mechanism of DDR1 whereby collagen induces lateral association of DDR1 dimers and phosphorylation between dimers. 28591735_findings demonstrate that, in human breast cancer cells, DDR1 regulates IR expression and ligand dependent biological actions. This novel functional crosstalk is likely clinically relevant 28743276_These findings demonstrate a critical role of miR-199a-3p/DDR1 pathway in ovarian cancer development. 28863860_This study suggested that DDR1 and DDR2 knockdown alters brain immunity and significantly reduces the level of triggering receptor expressed on myeloid cells (TREM)-2 and microglia. 28864681_Furthermore, the inhibition of DDR1 with 7rh showed striking efficacy in combination with chemotherapy in orthotopic xenografts and autochthonous pancreatic tumors where it significantly reduced DDR1 activation and downstream signaling, reduced primary tumor burden, and improved chemoresponse. 28887161_we further analyzed the CpG methylation levels at the DDR1 promoter in EOC cells and found that the CpG methylation levels of DDR1 promoter correlated negatively with the expression of DDR1 along the EMT spectrum. Therefore, EMT stratification could be a potential biomarker to predict patient response to DDR1-targeting drugs. 29039472_E2F1 knockdown decreased the expression of discoidin domain receptor 1 (DDR1) which plays a crucial role in many fundamental processes such as cell differentiation, adhesion, migration and invasion. 29298894_Data (including data from studies using knockout mice) suggest that DDR1 plays role in promoting mammary tumor growth; tumor-extrinsic DDR1 appears to be required for promotion of mammary tumors by interleukin-6. 29429150_The three well-conserved seed matched sites for miR-199a/b-5p in the discoidin domain receptor 1 (DDR1) 3'-UTR were targeted, and miRNA binding to at least two sites was required for DDR1 inhibition. 29559654_this study shows that complement C1q stimulates the progression of hepatocellular tumor through the activation of DDR1 29616590_These results show a deleterious effect of high co-expression of DDR1 and DDR2 and a physical interaction between the two receptors. 29733741_differential collagen degradation by MT1-MMP induced a structural disorganization of adult collagen and inhibits DDR1 activation. 29866925_This study highlights that CAF-induced elevation of DDR1 expression in gastric carcinoma cells enhances peritoneal tumorigenesis, and that inhibition of DDR1 is an attractive strategy for the treatment of gastric carcinoma peritoneal metastasis 30121624_In poorly differentiated thyroid cancer, DDR1 silencing or downregulation blocks the IGF-2/IR-A autocrine loop and induces cellular differentiation. These results may open novel therapeutic approaches for thyroid cancer. 30597424_DDR1 variants may confer a risk of SZ through WM microstructural alterations leading to cognitive dysfunction. 30691832_high level of phosphorylated DDR1 predicts shorter survival of esophageal squamous cell carcinoma after treatment 30760555_DDR1 regulates actomyosin contractility at cell-cell junctions and thereby contribute to epithelial polarization and morphogenesis. 30890477_Results of stratified analysis revealed a correlation between DDR1 CC genotype and early age of onset, clinical type of vitiligo and absence of family history of autoimmune disorders. 30917320_critical function of discoidin domain receptor 1 (DDR1) in modulating glioblastoma therapy resistance 30963567_Thus, this study identifies DDR1 as an important target for modulating epithelial-mesenchymal transition and induction of apoptosis in prostate cancer cells. 31018949_Decreased DDR1 Expression Is Associated With Clear Cell Renal Cell Carcinoma. 31086186_Study showed that bladder tumor cells expressing the collagen receptor, CD167a or Discoidin domain receptor 1, responded to collagen I stimulation at the primary tumor to promote local invasion and utilized the same receptor to preferentially colonize at airway smooth muscle cells - a rich source of collagen III in lung. 31253192_Betel nut alkaloid may recruit DNMT3B to regulate miR-486-3p/DDR1 axis in oral cancer andmiR-486-3p and DDR1 may serve as potential therapeutic targets of oral cancer. 31271515_High DDR1 expression is associated with melanoma. 31351093_Live cell measurements of interaction forces and binding kinetics between Discoidin Domain Receptor 1 (DDR1) and collagen I with atomic force microscopy. 31383731_we provide evidence that activated DDR1 translocates to the nucleus, localizes to chromatin, and regulates the transcription of profibrotic genes, including collagen IV. 31578591_To the best of our knowledge, the results of the present study demonstrated for the first time that DDR1 expressed on the tumor cells promoted breast tumor growth by suppressing antitumor immunity. The present findings indicated that DDR1 may not only have a critical role in the progression of breast cancer, but may also serve as a potential therapeutic target for breast cancer, particularly Triplenegative breast cancer 31659178_Identification of a novel synthetic lethal vulnerability in non-small cell lung cancer by co-targeting TMPRSS4 and DDR1. 31745115_DDR1 autophosphorylation is a result of aggregation into dense clusters. 32047176_Discoidin Domain Receptors, DDR1b and DDR2, Promote Tumour Growth within Collagen but DDR1b Suppresses Experimental Lung Metastasis in HT1080 Xenografts. 32330138_Postn is signaling though DDR1 is mechanistically involved in OA pathophysiology. 32492656_Influences of TP53 and the anti-aging DDR1 receptor in controlling Raf/MEK/ERK and PI3K/Akt expression and chemotherapeutic drug sensitivity in prostate cancer cell lines. 32668815_Targeting Discoidin Domain Receptor 1 (DDR1) Signaling and Its Crosstalk with beta1-integrin Emerges as a Key Factor for Breast Cancer Chemosensitization upon Collagen Type 1 Binding. 32716315_The cross-talk between DDR1 and STAT3 promotes the development of hepatocellular carcinoma. 32776088_Discoidin Domain Receptor 1 is a therapeutic target for neurodegenerative diseases. 32839343_Two-step release of kinase autoinhibition in discoidin domain receptor 1. 32977456_Discoidin Domain Receptors 1 Inhibition Alleviates Osteoarthritis via Enhancing Autophagy. 33234027_Overexpression of DDR1 Promotes Migration, Invasion, Though EMT-Related Molecule Expression and COL4A1/DDR1/MMP-2 Signaling Axis. 33239676_miR-199b-5p-DDR1-ERK signalling axis suppresses prostate cancer metastasis via inhibiting epithelial-mesenchymal transition. 33309038_Biological information and functional analysis reveal the role of discoidin domain receptor 1 in oral squamous cell carcinoma. 33340991_Discoidin domain receptor 1 activation links extracellular matrix to podocyte lipotoxicity in Alport syndrome. 33634432_Complex roles of discoidin domain receptor tyrosine kinases in cancer. 33976105_Activation of transmembrane receptor tyrosine kinase DDR1-STAT3 cascade by extracellular matrix remodeling promotes liver metastatic colonization in uveal melanoma. 34071309_Association between Interferon-Lambda-3 rs12979860, TLL1 rs17047200 and DDR1 rs4618569 Variant Polymorphisms with the Course and Outcome of SARS-CoV-2 Patients. 34108622_Combined inhibition of DDR1 and CDK4/6 induces synergistic effects in ER-positive, HER2-negative breast cancer with PIK3CA/AKT1 mutations. 34206590_DDR1 Affects Metabolic Reprogramming in Breast Cancer Cells by Cross-Talking to the Insulin/IGF System. 34323026_Coexpression of the discoidin domain receptor 1 gene with oligodendrocyte-related and schizophrenia risk genes in the developing and adult human brain. 34494537_[Research Progress of Discoidin Domain Receptor 1 in Breast Cancer and Other Malignant Tumors]. 34732895_Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. 34815375_Discoidin Domain Receptor Tyrosine Kinase 1 (DDR1): A Novel Predictor for Recurrence of Hepatocellular Carcinoma After Curative Resection. 34893587_Breast tissue regeneration is driven by cell-matrix interactions coordinating multi-lineage stem cell differentiation through DDR1. 35089546_Discoidin domain receptor 1 promotes hepatocellular carcinoma progression through modulation of SLC1A5 and the mTORC1 signaling pathway. 35140331_DDR1 promotes hepatocellular carcinoma metastasis through recruiting PSD4 to ARF6. | ENSMUSG00000003534 | Ddr1 | 8.123403e+02 | 1.3178682 | 0.398206075 | 0.3259607 | 1.486077e+00 | 0.2228261052 | 0.78763590 | No | Yes | 7.698366e+02 | 95.092675 | 5.677548e+02 | 72.315537 | |
| ENSG00000204588 | 440894 | LINC01123 | lncRNA | 31488218_Functional assays showed that LINC01123 promoted NSCLC cell proliferation and aerobic glycolysis. Mechanistic investigations revealed that LINC01123 was a direct transcriptional target of c-Myc. LINC01123 increased c-Myc mRNA expression by sponging miR-199a-5p. 32700743_LINC01123 facilitates proliferation, invasion and chemoresistance of colon cancer cells. 33191397_ZEB1-activated LINC01123 accelerates the malignancy in lung adenocarcinoma through NOTCH signaling pathway. 33837611_LINC01123 enhances osteosarcoma cell growth by activating the Hedgehog pathway via the miR-516b-5p/Gli1 axis. 34403009_LINC01123 promotes cell proliferation and migration via regulating miR-1277-5p/KLF5 axis in ox-LDL-induced vascular smooth muscle cells. 34511356_LINC01123 is associated with prognosis of oral squamous cell carcinoma and involved in tumor progression by sponging miR-34a-5p. 34519373_LINC01123 potentially correlates with radioresistance in glioma through the miR-151a/CENPB axis. 35115487_LINC01123 promotes immune escape by sponging miR-214-3p to regulate B7-H3 in head and neck squamous-cell carcinoma. 35325644_Long non-coding RNA LINC01123 promotes cell proliferation, migration and invasion via interacting with SRSF7 in colorectal cancer. | 5.665549e+02 | 1.0729568 | 0.101591957 | 0.2982201 | 1.182265e-01 | 0.7309660407 | 0.94341231 | No | Yes | 5.590239e+02 | 69.624027 | 5.606232e+02 | 71.558831 | |||||||||
| ENSG00000204642 | 3134 | HLA-F | protein_coding | P30511 | FUNCTION: Non-classical major histocompatibility class Ib molecule postulated to play a role in immune surveillance, immune tolerance and inflammation. Functions in two forms, as a heterotrimeric complex with B2M/beta-2 microglobulin and a peptide (peptide-bound HLA-F-B2M) and as an open conformer (OC) devoid of peptide and B2M (peptide-free OC). In complex with B2M, presents non-canonical self-peptides carrying post-translational modifications, particularly phosphorylated self-peptides. Peptide-bound HLA-F-B2M acts as a ligand for LILRB1 inhibitory receptor, a major player in maternal-fetal tolerance. Peptide-free OC acts as a ligand for KIR3DS1 and KIR3DL2 receptors (PubMed:28636952). Upon interaction with activating KIR3DS1 receptor on NK cells, triggers NK cell degranulation and anti-viral cytokine production (PubMed:27455421). Through interaction with KIR3DL2 receptor, inhibits NK and T cell effector functions (PubMed:24018270). May interact with other MHC class I OCs to cross-present exogenous viral, tumor or minor histompatibility antigens to cytotoxic CD8+ T cells, triggering effector and memory responses (PubMed:23851683). May play a role in inflammatory responses in the peripheral nervous system. Through interaction with KIR3DL2, may protect motor neurons from astrocyte-induced toxicity (PubMed:26928464). {ECO:0000269|PubMed:23851683, ECO:0000269|PubMed:24018270, ECO:0000269|PubMed:26928464, ECO:0000269|PubMed:27455421, ECO:0000269|PubMed:28636952}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Endosome;Glycoprotein;Immunity;Lysosome;MHC I;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene belongs to the HLA class I heavy chain paralogues. It encodes a non-classical heavy chain that forms a heterodimer with a beta-2 microglobulin light chain, with the heavy chain anchored in the membrane. Unlike most other HLA heavy chains, this molecule is localized in the endoplasmic reticulum and Golgi apparatus, with a small amount present at the cell surface in some cell types. It contains a divergent peptide-binding groove, and is thought to bind a restricted subset of peptides for immune presentation. This gene exhibits few polymorphisms. Multiple transcript variants encoding different isoforms have been found for this gene. These variants lack a coding exon found in transcripts from other HLA paralogues due to an altered splice acceptor site, resulting in a shorter cytoplasmic domain. [provided by RefSeq, Jul 2008]. | hsa:3134; | cell surface [GO:0009986]; early endosome membrane [GO:0031901]; endoplasmic reticulum [GO:0005783]; ER to Golgi transport vesicle membrane [GO:0012507]; external side of plasma membrane [GO:0009897]; extracellular space [GO:0005615]; Golgi membrane [GO:0000139]; integral component of lumenal side of endoplasmic reticulum membrane [GO:0071556]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; MHC class I protein complex [GO:0042612]; MHC class Ib protein complex [GO:0032398]; phagocytic vesicle membrane [GO:0030670]; plasma membrane [GO:0005886]; recycling endosome membrane [GO:0055038]; 14-3-3 protein binding [GO:0071889]; beta-2-microglobulin binding [GO:0030881]; peptide antigen binding [GO:0042605]; TAP1 binding [GO:0046978]; TAP2 binding [GO:0046979]; antigen processing and presentation of endogenous peptide antigen via MHC class Ib [GO:0002476]; antigen processing and presentation of exogenous peptide antigen via MHC class Ib [GO:0002477]; antigen processing and presentation of peptide antigen via MHC class I [GO:0002474]; negative regulation of natural killer cell cytokine production [GO:0002728]; negative regulation of natural killer cell degranulation [GO:0043322]; negative regulation of natural killer cell mediated cytotoxicity [GO:0045953]; negative regulation of neuron death [GO:1901215]; negative regulation of T cell cytokine production [GO:0002725]; positive regulation of natural killer cell cytokine production [GO:0002729]; positive regulation of natural killer cell degranulation [GO:0043323]; positive regulation of T cell mediated cytotoxicity [GO:0001916] | 12874228_This is the first example of placental HLA-F expression--primarily in trophoblasts that have invaded the maternal decidua--in the same cells that simultaneously express the other two nonclassical class I antigens HLA-E and HLA-G. 14607927_HLA-F surface expression on B cell and monocyte cell lines correlates with the presence of a limited amount of endoglycosidase H (Endo H)-resistant HLA-F; however, clearly not all surface-expressed HLA-F is fully glycosylated. 16570139_The results of this analysis confirmed several previously reported coding sequence variants, identified several new allelic variants, and also defined extensive variation in intron and flanking sequences. 16709803_HLA-F is entirely dependent on its cytoplasmic tail for export from the endoplasmic reticulum. 17157219_strong positive directional selection is acting for maintaining the observed low polymorphism on HLA-E, -F and -G loci 17971048_Observational study of gene-disease association. (HuGE Navigator) 18658158_the role of NKG2D and 2B4 is not focussed on trophoblast recognition in normal pregnancy, but is more likely involved in cross-talk among maternal cells of the placental bed 18941505_Across HIV Gag protein, the rise of polymorphisms from independent origin during the last twenty years of epidemic was related to an association with an HLA allele accumulated in one of either B or F subtypes 19622345_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19664746_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19851445_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20483783_These data suggest that HLA-F is expressed independently of peptide and that a physical interaction specific to MHC-I HC plays a role in the function of MHC-I HC expression in activated lymphocytes. 20593013_Observational study of gene-disease association. (HuGE Navigator) 20865824_surface marker on activated lymphocytes 22490650_Mifepristone can inhibit the effects of progesterone by down-regulating the expressions of HLA-G, HLA-E and HLA-F mRNA in trophoblasts during the first trimester. 22544725_Upregulated HLA-F expression (p = 0.026) and downregulated HLA I expression (p = 0.013) could be an independent unfavorable prognostic factor. 23542057_expression in gastric cancer lesion was unrelated to patient prognosis 23851683_A previously unrecognized model of Ag cross-presentation mediated by HLA-F & MHC-I open conformers on activated lymphocytes & monocytes may significantly contribute to the regulation of immune system functions & the immune defense. 24018270_HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. 25413105_This is the first study regarding HLA-F polymorphisms in a Euro-Brazilian population contributing to the Southern Brazilian genetic characterization. 25420801_Review of the impact of human leukocyte antigen molecules E, F, and G on the outcome of transplantation. 25461528_The effect of Japanese encephalitis virus and TNF-alpha exposure on NFkappaB-mediated induction of HLA-F is reported. 25862890_We identified that the HLA-E and HLA-F in gastric cancer independently affected clinical factors, including postoperative outcome 26332651_provides substantial evidence that the rs7903146 variant is significantly associated with the risk of diabetic retinopathy in Caucasian populations 26928464_Overexpression of a single MHCI molecule, HLA-F, protects human MNs from ALS astrocyte-mediated toxicity, whereas knockdown of its receptor, the killer cell immunoglobulin-like receptor KIR3DL2, on human astrocytes results in enhanced motor neurons death 27447835_Two eSNPs were associated with fecundability at a FDR of 5%; both were in the HLA region and were eQTLs for the TAP2 gene (P = 1.3x10-4) and the HLA-F gene (P = 4.0x10-4), respectively. 27455421_this study established HLA-F as a ligand of KIR3DS1 and have demonstrated cell-context-dependent expression of HLA-F that might explain the widespread influence of KIR3DS1 in human disease, including delayed progression of disease caused by human immunodeficiency virus type 1 27522114_Data suggest that the immunoproteasome is involved in pathologic MHC class I (HLA-A, B, C, F and G) expression and maintenance of myokine production in Idiopathic inflammatory myopathies (IIMs). 28185362_In this study, we describe and confirm the distinct expression of HLA-F, HLA-G, HLA-E, and HLA-C in placental tissue 28636952_the results of this study provide a blueprint of the molecular details of HLA-F, which will inform future exploration of its roles in human health and allow for the development of additional, targeted therapeutics 30031767_interactions between KIR3DS1 and HLA-F contribute to NK cell-mediated control of HCV. 30245028_The rs2523393 A allele creates a GATA2 binding site in a progesterone-responsive distal enhancer that loops to the HLA-F promoter. 30510890_high HLA-F expression is associated with Nasopharyngeal Carcinoma (NPC) local recurrence and distant metastasis and may be regarded as a poor prognostic factor for NPC patients. 30755240_This study showed that the HLA-F expression was positively correlated with malignant phenotype and negatively correlated with overall survival. 31717259_HLA-F variants bound to selected peptides, were structurally compared. 31863778_HLA-F-AS1 also enhanced the expressions of PFN1, which was validated as a target gene of miR-330-3p. CONCLUSION: HLA-F-AS1 promoted colorectal cancer progression via regulating miR-330-3p/PFN1 axis 32020202_Variation in the HLA-F gene locus with functional impact is associated with pregnancy success and time-to-pregnancy after fertility treatment. 34257557_Development of a Novel Prognostic Signature Based on Antigen Processing and Presentation in Patients with Breast Cancer. 35181585_HLA-E and HLA-F Are Overexpressed in Glioblastoma and HLA-E Increased After Exposure to Ionizing Radiation. | 1.667549e+02 | 1.0758982 | 0.105541522 | 0.3220505 | 1.073134e-01 | 0.7432237839 | 0.94662178 | No | Yes | 1.298659e+02 | 17.097454 | 1.278343e+02 | 17.292077 | |||
| ENSG00000204852 | 79600 | TCTN1 | protein_coding | Q2MV58 | FUNCTION: Component of the tectonic-like complex, a complex localized at the transition zone of primary cilia and acting as a barrier that prevents diffusion of transmembrane proteins between the cilia and plasma membranes. Regulator of Hedgehog (Hh), required for both activation and inhibition of the Hh pathway in the patterning of the neural tube. During neural tube development, it is required for formation of the most ventral cell types and for full Hh pathway activation. Functions in Hh signal transduction to fully activate the pathway in the presence of high Hh levels and to repress the pathway in the absence of Hh signals. Modulates Hh signal transduction downstream of SMO and RAB23 (By similarity). {ECO:0000250}. | Alternative splicing;Cell projection;Ciliopathy;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Developmental protein;Glycoprotein;Joubert syndrome;Reference proteome;Secreted;Signal | This gene encodes a member of a family of secreted and transmembrane proteins. The orthologous gene in mouse functions downstream of smoothened and rab23 to modulate hedgehog signal transduction. This protein is a component of the tectonic-like complex, which forms a barrier between the ciliary axoneme and the basal body. A mutation in this gene was found in a family with Joubert syndrome-13. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:79600; | cytoskeleton [GO:0005856]; cytosol [GO:0005829]; extracellular space [GO:0005615]; membrane [GO:0016020]; MKS complex [GO:0036038]; central nervous system interneuron axonogenesis [GO:0021956]; cilium assembly [GO:0060271]; dorsal/ventral neural tube patterning [GO:0021904]; in utero embryonic development [GO:0001701]; neural tube formation [GO:0001841]; protein localization to ciliary transition zone [GO:1904491]; regulation of smoothened signaling pathway [GO:0008589]; somatic motor neuron differentiation [GO:0021523]; telencephalon development [GO:0021537] | 21725307_Mutations in Tctn1 is associated with ciliopathies. 25304031_TCTN1 may serve as a novel prognostic factor and a potential therapeutic target for glioblastoma. 25737023_These data suggest TCTN1 is essential for glioma cell viability, and dysregulation of TCTN1 may play a key role in glioma tumorigenesis. 26310786_These findings confirmed the direct association between the TCTN1 gene and prostate cancer growth in vitro. 28123172_Flow cytometry analysis showed that depletion of TCTN1 could cause cell cycle arrest at the G2/M phase. This indicates that TCTN1 may be crucial for CRC cell growth. 28631893_we describe the phenotype of a patient with Varadi syndrome who is homozygous for a previously reported mutation in TCTN1 (NM_001082538.2:c.342-2A>G, p.Gly115Lysfs*8) and suggest that allelic disorders linked to TCTN1 include Varadi syndrome, in addition to Joubert syndrome and Meckel-Gruber syndrome. 31302911_Two novel TCTN1 mutations were identified in a family with Joubert syndrome 34859261_Function and transcriptional regulation of TCTN1 in oral squamous cell carcinoma. | ENSMUSG00000038593 | Tctn1 | 1.045667e+03 | 0.7470772 | -0.420670848 | 0.2722067 | 2.387631e+00 | 0.1222988156 | 0.75814553 | No | Yes | 7.301812e+02 | 72.004777 | 1.040476e+03 | 104.456711 | |
| ENSG00000205060 | 84912 | SLC35B4 | protein_coding | Q969S0 | FUNCTION: Sugar transporter that specifically mediates the transport of UDP-xylose (UDP-Xyl) and UDP-N-acetylglucosamine (UDP-GlcNAc) from cytosol into Golgi. {ECO:0000269|PubMed:15911612}. | Alternative splicing;Golgi apparatus;Membrane;Reference proteome;Sugar transport;Transmembrane;Transmembrane helix;Transport | Glycosyltransferases, such as SLC35B4, transport nucleotide sugars from the cytoplasm where they are synthesized, to the Golgi apparatus where they are utilized in the synthesis of glycoproteins, glycolipids, and proteoglycans (Ashikov et al., 2005 [PubMed 15911612]).[supplied by OMIM, Mar 2008]. | hsa:84912; | endoplasmic reticulum [GO:0005783]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of Golgi membrane [GO:0030173]; transmembrane transporter activity [GO:0022857]; UDP-N-acetylglucosamine transmembrane transporter activity [GO:0005462]; UDP-xylose transmembrane transporter activity [GO:0005464]; carbohydrate transport [GO:0008643]; regulation of gluconeogenesis [GO:0006111]; UDP-N-acetylglucosamine transmembrane transport [GO:1990569]; UDP-xylose transmembrane transport [GO:0015790] | 15911612_Transport capabilities for UDP-Glc and UDP-Xyl were determined with Golgi vesicles isolated from transformed cells 21507882_Decreased SLC35B4 expression led to increased gluconeogenesis in HepG2 cells. 21918738_subcellular localization of SLC35B4 29682886_SLC35B4 single nucleotide polymorphism rs1646724 was associated with recurrence in patients with localized disease and survival in patients with advanced prostate cancer. 29867058_Glucose levels activate SLC35B4 expression. This triggers a downstream effect via Hsp60 and other proteins. We hypothesize that the downstream effect on the proteins is mediated via altering the glycosylation pattern inside liver cells 30458018_Our data showed that knock-out of the SLC35B4 gene does not affect major UDP-Xyl- and UDP-GlcNAc-dependent glycosylation pathways. 31175271_A novel YAP1/SLC35B4 axis promotes gastric cancer (GC) development and progression, and this axis could be a potential candidate for prognosis and therapeutics in GC. | ENSMUSG00000018999 | Slc35b4 | 3.335400e+03 | 0.6597123 | -0.600091059 | 0.2852615 | 4.307306e+00 | 0.0379490211 | 0.62366758 | No | Yes | 2.197531e+03 | 264.301772 | 3.469550e+03 | 427.100332 | |
| ENSG00000205084 | 79583 | TMEM231 | protein_coding | Q9H6L2 | FUNCTION: Transmembrane component of the tectonic-like complex, a complex localized at the transition zone of primary cilia and acting as a barrier that prevents diffusion of transmembrane proteins between the cilia and plasma membranes. Required for ciliogenesis and sonic hedgehog/SHH signaling (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Disease variant;Glycoprotein;Joubert syndrome;Meckel syndrome;Membrane;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein, which is a component of the B9 complex involved in the formation of the diffusion barrier between the cilia and plasma membrane. Mutations in this gene cause Joubert syndrome (JBTS). Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jan 2013]. | hsa:79583; | ciliary membrane [GO:0060170]; ciliary transition zone [GO:0035869]; integral component of membrane [GO:0016021]; MKS complex [GO:0036038]; camera-type eye development [GO:0043010]; cilium assembly [GO:0060271]; embryonic digit morphogenesis [GO:0042733]; in utero embryonic development [GO:0001701]; neuroepithelial cell differentiation [GO:0060563]; regulation of protein localization [GO:0032880]; smoothened signaling pathway [GO:0007224]; vasculature development [GO:0001944] | 23012439_mutations in TMEM231 cause JBTS, reinforcing the relationship between this condition and the disruption of the barrier at the ciliary transition zone. 23349226_TMEM231 represents a novel MKS locus. The very recent identification of TMEM231 mutations in Joubert syndrome supports the growing appreciation of the overlap in the molecular pathogenesis between these two ciliopathies. 25869670_Tmem231 is critical for organizing the Meckel syndrome complex and controlling ciliary composition, defects in which cause OFD3 and MKS. 27449316_Results identified a rare gene conversion event in TMEM231, leading to loss of exon 4, which in combination with c.712G>A missense mutation caused Joubert syndrome and in combination with c.334T>G missense mutation caused Meckel-Gruber syndrome. 31663672_Long-read nanopore sequencing resolves a TMEM231 gene conversion event causing Meckel-Gruber syndrome. | ENSMUSG00000031951 | Tmem231 | 7.982224e+02 | 1.0090233 | 0.012959534 | 0.3295070 | 1.560029e-03 | 0.9684939854 | 0.99408977 | No | Yes | 6.290497e+02 | 79.893715 | 6.707312e+02 | 87.165184 | |
| ENSG00000205352 | 54458 | PRR13 | protein_coding | Q9NZ81 | FUNCTION: Negatively regulates TSP1 expression at the level of transcription. This down-regulation was shown to reduce taxane-induced apoptosis. {ECO:0000269|PubMed:16847352}. | Alternative splicing;Nucleus;Reference proteome;Transcription;Transcription regulation | hsa:54458; | cytosol [GO:0005829]; nucleoplasm [GO:0005654] | 3.421338e+03 | 1.3469961 | 0.429745681 | 0.2945831 | 2.181908e+00 | 0.1396412137 | 0.76815104 | No | Yes | 3.938427e+03 | 380.141076 | 2.505114e+03 | 248.821515 | |||||
| ENSG00000205423 | 255919 | CNEP1R1 | protein_coding | Q8N9A8 | FUNCTION: Forms with the serine/threonine protein phosphatase CTDNEP1 an active complex which dephosphorylates and may activate LPIN1 and LPIN2. LPIN1 and LPIN2 are phosphatidate phosphatases that catalyze the conversion of phosphatidic acid to diacylglycerol and control the metabolism of fatty acids at different levels. May indirectly modulate the lipid composition of nuclear and/or endoplasmic reticulum membranes and be required for proper nuclear membrane morphology and/or dynamics. May also indirectly regulate the production of lipid droplets and triacylglycerol. {ECO:0000269|PubMed:22134922}. | Acetylation;Alternative splicing;Cytoplasm;Lipid metabolism;Membrane;Nucleus;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein that belongs to the Tmemb_18A family. A similar protein in yeast is a component of an endoplasmic reticulum-associated protein phosphatase complex and is thought to play a role in the synthesis of triacylglycerol. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Aug 2013]. | hsa:255919; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; integral component of membrane [GO:0016021]; Nem1-Spo7 phosphatase complex [GO:0071595]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; lipid metabolic process [GO:0006629]; positive regulation of protein dephosphorylation [GO:0035307]; positive regulation of triglyceride biosynthetic process [GO:0010867]; protein localization to nucleus [GO:0034504] | 22134922_Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. 25548215_The endoplasmic reticulum adaptor protein ERAdP initiates NK cell activation via the Ubc13-mediated NF-kappaB pathway. | ENSMUSG00000036810 | Cnep1r1 | 1.152674e+02 | 0.6928917 | -0.529298172 | 0.4031411 | 1.657249e+00 | 0.1979751761 | 0.78383913 | No | Yes | 8.241093e+01 | 16.224652 | 1.162767e+02 | 23.176925 | |
| ENSG00000205572 | 8293 | SERF1B | protein_coding | O75920 | FUNCTION: Positive regulator of amyloid protein aggregation and proteotoxicity (PubMed:20723760, PubMed:22854022, PubMed:31034892). Induces conformational changes in amyloid proteins, such as APP, HTT, and SNCA, driving them into compact formations preceding the formation of aggregates (PubMed:20723760, PubMed:22854022, PubMed:31034892). {ECO:0000269|PubMed:20723760, ECO:0000269|PubMed:22854022, ECO:0000269|PubMed:31034892}. | Alternative splicing;Cytoplasm;Nucleus;Reference proteome | This gene is part of a 500 kb inverted duplication on chromosome 5q13. This duplicated region contains at least four genes and repetitive elements which make it prone to rearrangements and deletions. The repetitiveness and complexity of the sequence have also caused difficulty in determining the organization of this genomic region. This gene is the centromeric copy which is identical to the telomeric copy. Alternatively spliced transcripts have been documented but it is unclear whether alternative splicing occurs for both the centromeric and telomeric copies of the gene. The gene encodes a protein of unknown function which bears low-level homology with the RNA-binding domain of matrin-cyclophilin, a protein which colocalizes with small nuclear ribonucleoproteins (snRNPs) and the SMN1 gene product. [provided by RefSeq, Jul 2008]. | hsa:728492;hsa:8293; | cytosol [GO:0005829]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; amyloid fibril formation [GO:1990000]; nervous system development [GO:0007399]; protein destabilization [GO:0031648] | ENSMUSG00000021643 | Serf1 | 4.286477e+02 | 0.9281626 | -0.107550460 | 0.2914826 | 1.379345e-01 | 0.7103430616 | 0.93850268 | No | Yes | 3.262080e+02 | 95.314181 | 3.535267e+02 | 105.829194 | ||
| ENSG00000205726 | 6453 | ITSN1 | protein_coding | Q15811 | FUNCTION: Adapter protein that provides a link between the endocytic membrane traffic and the actin assembly machinery (PubMed:11584276, PubMed:29887380). Acts as guanine nucleotide exchange factor (GEF) for CDC42, and thereby stimulates actin nucleation mediated by WASL and the ARP2/3 complex (PubMed:11584276). Plays a role in the assembly and maturation of clathrin-coated vesicles (By similarity). Recruits FCHSD2 to clathrin-coated pits (PubMed:29887380). Involved in endocytosis of activated EGFR, and probably also other growth factor receptors (By similarity). Involved in endocytosis of integrin beta-1 (ITGB1) and transferrin receptor (TFR); internalization of ITGB1 as DAB2-dependent cargo but not TFR may involve association with DAB2 (PubMed:22648170). Promotes ubiquitination and subsequent degradation of EGFR, and thereby contributes to the down-regulation of EGFR-dependent signaling pathways. In chromaffin cells, required for normal exocytosis of catecholamines. Required for rapid replenishment of release-ready synaptic vesicles at presynaptic active zones (By similarity). Inhibits ARHGAP31 activity toward RAC1 (PubMed:11744688). {ECO:0000250|UniProtKB:Q9WVE9, ECO:0000250|UniProtKB:Q9Z0R4, ECO:0000269|PubMed:11584276, ECO:0000269|PubMed:11744688, ECO:0000269|PubMed:22648170, ECO:0000269|PubMed:29887380}.; FUNCTION: [Isoform 1]: Plays a role in synaptic vesicle endocytosis in brain neurons. {ECO:0000250|UniProtKB:Q9Z0R4}. | 3D-structure;Alternative splicing;Calcium;Cell junction;Cell membrane;Cell projection;Coated pit;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Endocytosis;Endosome;Exocytosis;Host-virus interaction;Membrane;Metal-binding;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;SH3 domain;Synapse;Synaptosome;Transport | The protein encoded by this gene is a cytoplasmic membrane-associated protein that indirectly coordinates endocytic membrane traffic with the actin assembly machinery. In addition, the encoded protein may regulate the formation of clathrin-coated vesicles and could be involved in synaptic vesicle recycling. This protein has been shown to interact with dynamin, CDC42, SNAP23, SNAP25, SPIN90, EPS15, EPN1, EPN2, and STN2. Multiple transcript variants encoding different isoforms have been found for this gene, but the full-length nature of only two of them have been characterized so far. [provided by RefSeq, Jul 2008]. | hsa:6453; | clathrin-coated pit [GO:0005905]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; lamellipodium [GO:0030027]; neuron projection [GO:0043005]; nuclear envelope [GO:0005635]; plasma membrane [GO:0005886]; presynapse [GO:0098793]; recycling endosome [GO:0055037]; calcium ion binding [GO:0005509]; guanyl-nucleotide exchange factor activity [GO:0005085]; molecular adaptor activity [GO:0060090]; proline-rich region binding [GO:0070064]; cellular protein localization [GO:0034613]; clathrin-dependent synaptic vesicle endocytosis [GO:0150007]; endocytosis [GO:0006897]; endosomal transport [GO:0016197]; exocytosis [GO:0006887]; intracellular signal transduction [GO:0035556]; protein transport [GO:0015031]; regulation of small GTPase mediated signal transduction [GO:0051056] | 12960435_intersectin interaction with dynamin was important in regulating the fission and internalization of caveolae 16442855_review of roles of ITSN1 and DSCR1 in Down syndrome, Alzheimer's disease, endocytosis and vesicle trafficking 16874303_Results extend the current role of intersectin-1L in endocytosis to a function in exocytosis and support the idea that intersectin-1L is an adaptor that coordinates exo-endocytotic membrane trafficking in secretory cells. 16914641_ITSN forms a complex with Cbl in vivo mediated by the Src homology (SH) 3 domains binding to the Pro-rich COOH terminus of Cbl. This interaction stimulates the ubiquitylation and degradation of the activated EGFR. 17405881_findings demonstrate a novel role of ITSN-1s as a negative regulator of the mitochondrial pathway-dependent apoptosis secondary to activation of the Erk1/2 survival signaling pathway 18539136_These results demonstrate that alternative splicing leads to the formation of two pools of ITSN1 with potentially different properties in neurons, affecting ITSN1 function as adaptor protein. 18692052_These data provide a molecular link between SHIP2 and ITSN1 which are involved in receptor endocytosis regulation. 19166927_Intersectin 1 forms a complex with adaptor protein (SH3KBP1 protein), implicated in downregulation of receptor tyrosine kinases. 19356586_The higher exchange activity of ITSN1L towards the GDP-bound conformation of Cdc42 could represent an evolutionary adaptation of this GEF that ensures nucleotide exchange towards the formation of the signalling-active GTP-bound form of Cdc42. 19777371_The identification of fifteen novel transcriptional isoforms of the human ITSN1 gene with full-length coding sequences , is reported. 20493827_Downregulation of intersectin-1s induces apoptosis of brain glioblastoma cells. 20659428_microexons provide a mechanism for the control of tissue-specific interactions of ITSN1 and Src with their partners. 20842712_Studied the inhibitory mechanism of ITSN1L, & id'd a novel short amino acid motif which mediates autoinhibition. Found this motif is located in the SH3 DH linker region, and show that W1221 acts as key residue in establishing inhibitory interaction. 20946875_These findings expand the role of ITSN1 as a scaffolding molecule bringing together components of endocytic complexes. 21129155_ITSN-1s, via its SH3A domain has the unique ability to regulate dyn2 assembly-disassembly and function during endocytosis 21503949_ITSN1 has two isoforms: ITSN1-long and ITSN1-short. siRNA-mediated down regulation of ITSN1-short inhibits migration and invasion of glioma cells. 21712076_This novel mammalian ITSN1 isoform possesses a significantly altered domain structure and performs specific protein-protein interactions. 21876463_study reveals a link between overexpression of specific ITSN1 isoforms and behavioral phenotypes 22266851_Silencing ITSN1 significantly inhibits the anchorage independent growth of tumor cells in vitro. 22750298_the neuron-specific isoform of the stable tubule-only polypeptide (STOP) interacts with SH3A domain of ITSN1. 23936226_ITSN1 and ITSN2 bind similar proline-rich ligands but are differentially recognized by SH2 domain-containing proteins. 24284073_Our observations suggest that ITSN1 is an important general regulator of Cdc42-, Nck- and N-WASP-dependent actin polymerisation 25783631_Abnormal expression of the intersectin1-L protein in epileptic brain tissue may play an important role in epilepsy, especially refractory epilepsy. 25832561_Study suggests critical roles of ITSN1-S in malignant glioma proliferation, indicating a potential usage of ITSN1-S in the therapeutic intervention as a novel molecular target 27670116_study reports that vaccinia virus A36 contains three NPF motifs that interact with the Eps15 homology (EH) domains in intersectin-1 and Eps15 to promote the release of virus from infected cells and their subsequent spread 28161632_These findings provide the basis for further functional investigations of the ITSN/CR16 complex that may play an important role in actin remodeling and cellular invasion. 28787396_Silencing ITSN1 in neuroblastoma cells led to decreased tumor growth in an orthotopic mouse model. Orthotopic animal models can provide insight into the role of ITSN1 pathways in neuroblastoma tumorigenesis. 29030480_Here, we discover that intersectin-s binds DENND2B, a guanine nucleotide exchange factor for the exocytic GTPase Rab13, and this interaction promotes recycling of ligand-free EGFR to the cell surface. Our study thus reveals a novel mechanism controlling the fate of internalized EGFR with important implications for cancer. 29851086_Different modes of ubiquitination regulated by AIP4 have opposite effects on ITSN1 isoform stability. 29958948_WIP enhances the interaction of N-WASP with ITSN1 and promotes ITSN1/beta-actin association. 30540523_the long isoform of intersectin-1 (ITSN-1), a guanine nucleotide exchange factor for Cdc42, is identified as a novel Golgi component and an interaction partner of GCC88 responsible for mediating the actin-dependent dispersal of the Golgi ribbon. 31160551_two isoforms of intersectin1 ITSN1-S and ITSN1-L produced by alternative splicing exerted opposite functions in glioma development. 32780150_ITSN1 regulates SAM68 solubility through SH3 domain interactions with SAM68 proline-rich motifs. 34625530_Endocytic protein intersectin1-S shuttles into nucleus to suppress the DNA replication in breast cancer. 34707297_ITSN1: a novel candidate gene involved in autosomal dominant neurodevelopmental disorder spectrum. | ENSMUSG00000022957 | Itsn1 | 1.394095e+03 | 0.9118029 | -0.133206020 | 0.2687955 | 2.424610e-01 | 0.6224342431 | 0.91561964 | No | Yes | 1.391120e+03 | 162.735483 | 1.428273e+03 | 171.501077 | |
| ENSG00000205771 | 440278 | CATSPER2P1 | transcribed_unprocessed_pseudogene | Catsper genes belong to a family of putative cation channels that are specific to spermatozoa and localize to the flagellum. This gene is part of a tandem repeat on chromosome 15q15; this copy of the gene is thought to be a pseudogene. [provided by RefSeq, Oct 2008]. | 2.428097e+01 | 1.0347129 | 0.049230556 | 0.5976225 | 6.788025e-03 | 0.9343370073 | 0.98790885 | No | Yes | 3.667121e+01 | 9.297352 | 3.598716e+01 | 9.747191 | |||||||||
| ENSG00000206560 | 23243 | ANKRD28 | protein_coding | O15084 | FUNCTION: Putative regulatory subunit of protein phosphatase 6 (PP6) that may be involved in the recognition of phosphoprotein substrates. Involved in the PP6-mediated dephosphorylation of NFKBIE opposing its degradation in response to TNF-alpha. Selectively inhibits the phosphatase activity of PPP1C. Targets PPP1C to modulate HNRPK phosphorylation. {ECO:0000269|PubMed:16564677, ECO:0000269|PubMed:18186651}. | ANK repeat;Alternative promoter usage;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Repeat | hsa:23243; | cytosol [GO:0005829]; nucleoplasm [GO:0005654] | 16564677_These findings provide a putative mechanism by which transcriptional activity of hnRNP K can be discretely controlled through the regulation of PP1 activity. 17023142_CaMKII and GSK3 mediate regulation of PITK. 19118547_ANKRD28, a novel binding partner of DOCK180, promotes cell migration by regulating focal adhesion formation. 25902538_hnRNP K and hnRNP L may serve as A1CF-like cofactors in AID-mediated class switch recombination and somatic hypermutation 27026398_findings suggest that BRCA1 is a novel modulator of PP6 signalling via its interaction with ANKRD28. 30572391_HPV16 infection and over-expression of hnRNP K protein were associated with the increased risk of cervical intraepithelial neoplasia. There might be interaction between hnRNP K protein overexpression and HPV16 infection existed on the progress of CIN/ [ cervical intraepithelial neoplasia II/III]. | ENSMUSG00000014496 | Ankrd28 | 6.944278e+02 | 0.8278214 | -0.272608635 | 0.3066300 | 7.918998e-01 | 0.3735262965 | 0.83844010 | No | Yes | 6.342756e+02 | 99.310407 | 7.094879e+02 | 113.807354 | ||
| ENSG00000211584 | 55652 | SLC48A1 | protein_coding | Q6P1K1 | FUNCTION: Heme transporter that regulates intracellular heme availability through the endosomal or lysosomal compartment. {ECO:0000269|PubMed:18418376}. | Alternative splicing;Endosome;Lysosome;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:55652; | endosome membrane [GO:0010008]; integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; plasma membrane [GO:0005886]; heme binding [GO:0020037]; heme transmembrane transporter activity [GO:0015232]; heme transport [GO:0015886] | 18418376_Human and worm proteins localize together, and bind and transport haem, thus establishing an evolutionarily conserved function for HRG-1. 19875448_HRG-1 regulates V-ATPase activity, which is essential for endosomal acidification, heme binding, and receptor trafficking in mammalian cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21555518_SLC48A1 is a target gene of the BACH1 transcription factor according to ChIP-seq analysis in HEK 293 cells. 22174408_a model in which heme is translocated across membranes facilitated by conserved amino acids positioned on the exoplasmic, cytoplasmic, and transmembrane regions of HRG-1-related proteins. 23395172_These results reveal HRG1 as the long-sought heme transporter for heme-iron recycling in macrophages and suggest that genetic variations in HRG1 could be modifiers of human iron metabolism. 24141772_HRG-1 may represent a novel target for selectively disrupting V-ATPase activity and the metastatic potential of cancer cells. | ENSMUSG00000081534 | Slc48a1 | 7.980817e+02 | 1.1048482 | 0.143848139 | 0.3166519 | 2.035878e-01 | 0.6518403175 | 0.92370776 | No | Yes | 7.620082e+02 | 81.908752 | 6.591121e+02 | 72.956840 | ||
| ENSG00000213380 | 84342 | COG8 | protein_coding | Q96MW5 | FUNCTION: Required for normal Golgi function. {ECO:0000250}. | Congenital disorder of glycosylation;Golgi apparatus;Membrane;Protein transport;Reference proteome;Transport | This gene encodes a protein that is a component of the conserved oligomeric Golgi (COG) complex, a multiprotein complex that plays a structural role in the Golgi apparatus, and is involved in intracellular membrane trafficking and glycoprotein modification. Mutations in this gene cause congenital disorder of glycosylation, type IIh, a disease that is characterized by under-glycosylated serum proteins, and whose symptoms include severe psychomotor retardation, failure to thrive, seizures, and dairy and wheat product intolerance. [provided by RefSeq, Jul 2008]. | hsa:84342; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; Golgi transport complex [GO:0017119]; membrane [GO:0016020]; trans-Golgi network membrane [GO:0032588]; glycosylation [GO:0070085]; Golgi organization [GO:0007030]; intra-Golgi vesicle-mediated transport [GO:0006891]; protein transport [GO:0015031]; retrograde transport, vesicle recycling within Golgi [GO:0000301] | 21805148_wthe origin, evolution and preservation of the COG8/PDF same-strand overlap follow similar mechanistic steps as those documented for antisense overlaps where gain and/or loss of splice sites and polyadenylation signals seems to drive the process. 25179963_Targeted silencing of components of lobe B of the COG complex, namely COG5, COG6, COG7 and COG8, inhibited HIV-1 replication 26045774_TMED6-COG8 chimera might act as a novel diagnostic marker in TFE3 translocation renal cell carcinoma. 28619360_The exome sequencing was performed to screen all CDG type II-related genes, and two novel frameshift mutations were found: c.171dupG (p.Leu58Alafs*29) and c.1656dupC (p.Ala553Argfs*15) in COG8. 29467253_TMEM-COG4, COG7 and COG8 subunits restore endogenous COG localization to the Golgi membranes | ENSMUSG00000031916 | Cog8 | 3.348595e+03 | 1.0810378 | 0.112417023 | 0.2878461 | 1.531819e-01 | 0.6955129534 | 0.93507407 | No | Yes | 3.290078e+03 | 244.425644 | 2.855299e+03 | 218.009113 | |
| ENSG00000213397 | 55559 | HAUS7 | protein_coding | Q99871 | FUNCTION: Contributes to mitotic spindle assembly, maintenance of centrosome integrity and completion of cytokinesis as part of the HAUS augmin-like complex. {ECO:0000269|PubMed:19369198, ECO:0000269|PubMed:19427217}. | Alternative splicing;Cell cycle;Cell division;Coiled coil;Cytoplasm;Cytoskeleton;Microtubule;Mitosis;Phosphoprotein;Reference proteome | This gene encodes a subunit of the augmin complex, which regulates centrosome and mitotic spindle integrity, and is necessary for the completion of cytokinesis. The encoded protein was identified by interaction with ubiquitin C-terminal hydrolase 37. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2012]. | Mouse_homologues mmu:73738; | centrosome [GO:0005813]; cytosol [GO:0005829]; HAUS complex [GO:0070652]; mitotic spindle microtubule [GO:1990498]; microtubule minus-end binding [GO:0051011]; thioesterase binding [GO:0031996]; cell division [GO:0051301]; centrosome cycle [GO:0007098]; spindle assembly [GO:0051225] | 11278605_Discusses transcript evidence for TREX1 and TREX2, as well as readthrough transcripts with upstream loci ATRIP and HAUS7. 29017965_HAUS7 mutation may be associated with chromosome misalignment, resulting in severe oligozoospermia. | ENSMUSG00000031371 | Haus7 | 4.418627e+03 | 0.9790010 | -0.030617763 | 0.3020594 | 1.025310e-02 | 0.9193459906 | 0.98484331 | No | Yes | 4.425638e+03 | 494.852913 | 3.924986e+03 | 450.581313 | |
| ENSG00000213995 | 55739 | NAXD | protein_coding | Q8IW45 | FUNCTION: Catalyzes the dehydration of the S-form of NAD(P)HX at the expense of ATP, which is converted to ADP. Together with NAD(P)HX epimerase, which catalyzes the epimerization of the S- and R-forms, the enzyme allows the repair of both epimers of NAD(P)HX, a damaged form of NAD(P)H that is a result of enzymatic or heat-dependent hydration. {ECO:0000255|HAMAP-Rule:MF_03157, ECO:0000269|PubMed:30576410}. | ATP-binding;Alternative splicing;Disease variant;Glycoprotein;Lyase;Mitochondrion;NAD;NADP;Neurodegeneration;Nucleotide-binding;Phosphoprotein;Reference proteome | hsa:55739; | mitochondrial matrix [GO:0005759]; ADP-dependent NAD(P)H-hydrate dehydratase activity [GO:0052855]; ATP binding [GO:0005524]; ATP-dependent NAD(P)H-hydrate dehydratase activity [GO:0047453]; metabolite repair [GO:0110051]; NAD biosynthesis via nicotinamide riboside salvage pathway [GO:0034356] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 24611804_Wide tissue distribution of dehydratase and epimerase is consistent with mRNA data for Carkd and Aibp, and their presence in both mitochondria and cytosol is explained by the existence of differently targeted forms of CARKD and AIBP. 30576410_results show that NAXD deficiency can be classified as a metabolite repair disorder in which accumulation of damaged metabolites likely triggers devastating effects in tissues such as the brain and the heart, eventually leading to early childhood death. | ENSMUSG00000031505 | Naxd | 1.254540e+03 | 0.9296037 | -0.105312326 | 0.2750177 | 1.495256e-01 | 0.6989891258 | 0.93592030 | No | Yes | 9.331947e+02 | 87.400278 | 1.072924e+03 | 102.721456 | ||
| ENSG00000214078 | 8904 | CPNE1 | protein_coding | Q99829 | FUNCTION: Calcium-dependent phospholipid-binding protein that plays a role in calcium-mediated intracellular processes (PubMed:14674885). Involved in the TNF-alpha receptor signaling pathway in a calcium-dependent manner (PubMed:14674885). Exhibits calcium-dependent phospholipid binding properties (PubMed:9430674, PubMed:19539605). Plays a role in neuronal progenitor cell differentiation; induces neurite outgrowth via a AKT-dependent signaling cascade and calcium-independent manner (PubMed:23263657, PubMed:25450385). May recruit target proteins to the cell membrane in a calcium-dependent manner (PubMed:12522145). May function in membrane trafficking (PubMed:9430674). Involved in TNF-alpha-induced NF-kappa-B transcriptional repression by inducing endoprotease processing of the transcription factor NF-kappa-B p65/RELA subunit (PubMed:18212740). Also induces endoprotease processing of NF-kappa-B p50/NFKB1, p52/NFKB2, RELB and REL (PubMed:18212740). {ECO:0000269|PubMed:12522145, ECO:0000269|PubMed:14674885, ECO:0000269|PubMed:18212740, ECO:0000269|PubMed:19539605, ECO:0000269|PubMed:23263657, ECO:0000269|PubMed:25450385, ECO:0000269|PubMed:9430674}. | Acetylation;Calcium;Cell membrane;Cytoplasm;Differentiation;Membrane;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation | Calcium-dependent membrane-binding proteins may regulate molecular events at the interface of the cell membrane and cytoplasm. This gene encodes a calcium-dependent protein that also contains two N-terminal type II C2 domains and an integrin A domain-like sequence in the C-terminus. However, the encoded protein does not contain a predicted signal sequence or transmembrane domains. This protein has a broad tissue distribution and it may function in membrane trafficking. This gene and the gene for RNA binding motif protein 12 overlap at map location 20q11.21. Alternate splicing results in multiple transcript variants encoding different proteins. [provided by RefSeq, Aug 2008]. | hsa:8904; | azurophil granule membrane [GO:0035577]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; membrane [GO:0016020]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; calcium ion binding [GO:0005509]; calcium-dependent phospholipid binding [GO:0005544]; endopeptidase activity [GO:0004175]; identical protein binding [GO:0042802]; NF-kappaB binding [GO:0051059]; phosphatidylserine binding [GO:0001786]; transporter activity [GO:0005215]; cellular response to calcium ion [GO:0071277]; lipid metabolic process [GO:0006629]; negative regulation of DNA binding [GO:0043392]; negative regulation of gene expression [GO:0010629]; negative regulation of NIK/NF-kappaB signaling [GO:1901223]; neuron projection extension [GO:1990138]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of tumor necrosis factor-mediated signaling pathway [GO:1903265]; proteolysis [GO:0006508]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; vesicle-mediated transport [GO:0016192] | 14674885_Copine I regulates tumour necrosis factor-alpha receptor signaling pathways. 18523666_Copine1 is upregulated in plasma membranes of TM cells in individuals with primary open-angle glaucoma. 18831769_CPNE1 shares a promoter and 5'UTR exons with RBM12. This genomic structure is conserved among multiple species. 29151113_CPNE1 overexpression can upregulate TRAF2 expression in prostate cancer DU-145 cells as determined by Western blotting and immunofluorescence assays. 29207139_Results demonstrated that CPNE1 was highly expressed in the osteosarcoma (OS) tissues and cell lines. Its knockdown significantly inhibited cell proliferation, colony formation, invasion and metastasis in OS cell lines. 29448099_Collectively, our findings suggest that JAB1 activates the neuronal differentiation ability of CPNE1 through the binding of C2A domain in CPNE1 with MPN domain in JAB1. 29970127_We observed that knockdown of CPNE1 and increased expression of miR-335-5p inhibits cell proliferation and motility in NSCLC cells, and found that CPNE1 was a target of miR-335-5p. 30221693_Study find that CPNE1 expression is upregulated in non-small cell lung cancer (NSCLC) and associated with advanced TNM stage, lymph node and distant metastasis. Furthermore, its knockdown inhibits the cell cycle in NSCLC cells. These data strongly suggest that CPNE1 is an oncogene in NSCLC and serves an important role in tumorigenesis of NSCLC progression. 32181526_High CPNE1 expression is associated with tumorigenesis and radioresistance via the AKT singling pathway in triple-negative breast cancer. 34969745_Prognostic Value of Copine 1 in Patients With Renal Cell Carcinoma. 35101055_CPNE1 promotes non-small cell lung cancer progression by interacting with RACK1 via the MET signaling pathway. | ENSMUSG00000074643 | Cpne1 | 6.230538e+03 | 1.1707291 | 0.227407279 | 0.2818108 | 6.595431e-01 | 0.4167212973 | 0.84868329 | No | Yes | 6.485542e+03 | 477.876129 | 5.410678e+03 | 409.498083 | |
| ENSG00000214174 | 201283 | AMZ2P1 | transcribed_unprocessed_pseudogene | 1.722160e+02 | 0.7091471 | -0.495843143 | 0.3431165 | 2.048147e+00 | 0.1523915562 | 0.77404149 | No | Yes | 1.295715e+02 | 22.006347 | 1.862129e+02 | 32.083185 | ||||||||||
| ENSG00000214212 | 255809 | C19orf38 | protein_coding | A8MVS5 | Glycoprotein;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:255809; | integral component of membrane [GO:0016021] | ENSMUSG00000057191 | AB124611 | 6.386421e+01 | 1.1474556 | 0.198438317 | 0.4337193 | 2.101588e-01 | 0.6466429696 | 0.92280718 | No | Yes | 6.118326e+01 | 13.208154 | 5.495353e+01 | 12.619665 | ||||
| ENSG00000214725 | 440356 | CDIPTOSP | transcribed_unitary_pseudogene | 3.980891e+01 | 1.1939862 | 0.255786125 | 0.5307061 | 2.352207e-01 | 0.6276794609 | 0.91646120 | No | Yes | 4.570610e+01 | 11.436523 | 3.058064e+01 | 8.154983 | ||||||||||
| ENSG00000214960 | 729920 | CRPPA | protein_coding | A4D126 | FUNCTION: Cytidylyltransferase required for protein O-linked mannosylation (PubMed:22522420, PubMed:27130732, PubMed:27601598, PubMed:26687144, PubMed:22522421, PubMed:26923585). Catalyzes the formation of CDP-ribitol nucleotide sugar from D-ribitol 5-phosphate (PubMed:27130732, PubMed:26687144, PubMed:26923585). CDP-ribitol is a substrate of FKTN during the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1), which is required for binding laminin G-like domain-containing extracellular proteins with high affinity (PubMed:27130732, PubMed:26687144, PubMed:26923585). Shows activity toward other pentose phosphate sugars and mediates formation of CDP-ribulose or CDP-ribose using CTP and ribulose-5-phosphate or ribose-5-phosphate, respectively (PubMed:26687144). Not Involved in dolichol production (PubMed:26687144). {ECO:0000269|PubMed:22522420, ECO:0000269|PubMed:22522421, ECO:0000269|PubMed:26687144, ECO:0000269|PubMed:26923585, ECO:0000269|PubMed:27130732, ECO:0000269|PubMed:27601598}. | 3D-structure;Alternative splicing;Congenital muscular dystrophy;Cytoplasm;Disease variant;Dystroglycanopathy;Limb-girdle muscular dystrophy;Lissencephaly;Nucleotidyltransferase;Reference proteome;Transferase | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:26687144, ECO:0000269|PubMed:26923585, ECO:0000269|PubMed:27130732}. | This gene encodes a 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase-like protein. Mutations in this gene are the cause of Walker-Warburg syndrome. Alternate splicing results in multiple transcript variants. [provided by RefSeq, May 2012]. | hsa:729920; | cytosol [GO:0005829]; cytidylyltransferase activity [GO:0070567]; D-ribitol-5-phosphate cytidylyltransferase activity [GO:0047349]; protein homodimerization activity [GO:0042803]; axon guidance [GO:0007411]; isoprenoid biosynthetic process [GO:0008299]; protein O-linked mannosylation [GO:0035269] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22522420_ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. 22522421_Mutations in ISPD cause Walker-Warburg syndrome and defective glycosylation of alpha-dystroglycan. 23217329_TMEM5 mutations were frequently associated with gonadal dysgenesis and neural tube defects, and ISPD mutations were frequently associated with brain vascular anomalies. 23288328_study report the involvement of the ISPD gene in milder dystroglycanopathy phenotypes ranging from congenital muscular dystrophy to limb-girdle muscular dystrophy and identified allelic ISPD variants in nine cases belonging to seven families 23390185_we identified a novel homozygous c.161G>C/p.G54A variant in ISPD in patients with limb-girdle muscular dystrophy 25444434_study describes a new homozygous missense mutation c.367G>A (p.Gly123Arg) in the ISPD gene in a family of Pakistani origin with 2 cousins from consanguineous parents affected with a congenital muscular dystrophy (CMD) /early limb-girdle muscular dystrophy intermediate phenotype and CMD respectively 26087224_ISPD gene homozygous deletion as a prenatal manifestation of Walker-Warburg syndrome has been found in 3 female fetuses of one family. 26220087_Reduced levels of GYLTL1B and ISPD mRNA associated with increased patient mortality and are the likely cause of alphaDG hypoglycosylation in ccRCC. 26404900_data suggest that the genetic heterogeneity of Limb Girdle Muscular Dystrophy with and without alpha-DG defects is greater than previously realized. 27194101_ISPD and FKTN are essential for the incorporation of ribitol into alpha-dystroglycan. 31909476_A splice site mutation c.1251G>A of ISPD gene is a common cause of congenital muscular dystrophy in Chinese patients. 33290265_Hsa_circ_0079480 promotes tumor progression in acute myeloid leukemia via miR-654-3p/HDGF axis. | ENSMUSG00000043153 | Crppa | 1.372165e+01 | 1.6824945 | 0.750601765 | 0.7903275 | 8.811920e-01 | 0.3478754155 | No | Yes | 1.612945e+01 | 5.105981 | 9.533320e+00 | 3.308979 | |
| ENSG00000215041 | 84461 | NEURL4 | protein_coding | Q96JN8 | FUNCTION: Promotes CCP110 ubiquitination and proteasome-dependent degradation. By counteracting accumulation of CP110, maintains normal centriolar homeostasis and preventing formation of ectopic microtubular organizing centers. {ECO:0000269|PubMed:22261722, ECO:0000269|PubMed:22441691}. | 3D-structure;Alternative splicing;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;Ubl conjugation pathway | The protein encoded by this gene is predicted and it includes two isoforms resulting from two alternatively spliced transcript variants. [provided by RefSeq, Jul 2008]. | hsa:84461; | centriole [GO:0005814]; cytoplasm [GO:0005737]; ubiquitin protein ligase activity [GO:0061630] | 22261722_the NEURL4-HERC2 complex participates in the ubiquitin-dependent regulation of centrosome architecture 22441691_Neurl4 counteracts accumulation of CP110, thereby maintaining normal centriolar homeostasis and preventing formation of ectopic microtubular organizing centres 28385950_The data support a model in which the daughter centriole promotes ciliogenesis through Neurl-4-dependent regulation of CP110 levels at the mother centriole. 35157000_Neuralized-like protein 4 (NEURL4) mediates ADP-ribosylation of mitochondrial proteins. | ENSMUSG00000047284 | Neurl4 | 4.811753e+03 | 0.8134036 | -0.297956635 | 0.2524069 | 1.372510e+00 | 0.2413811549 | 0.78892886 | No | Yes | 3.324346e+03 | 467.928522 | 4.589399e+03 | 662.448265 | |
| ENSG00000215067 | 100506713 | ALOX12-AS1 | lncRNA | 34174394_Antisense overlapping long non-coding RNA regulates coding arachidonate 12-lipoxygenase gene by translational interference. | 2.196753e+02 | 1.0751391 | 0.104523298 | 0.3170801 | 1.097141e-01 | 0.7404698777 | 0.94589933 | No | Yes | 1.935059e+02 | 20.526220 | 1.878238e+02 | 20.262458 | |||||||||
| ENSG00000215193 | 55670 | PEX26 | protein_coding | Q7Z412 | FUNCTION: Probably required for protein import into peroxisomes. Anchors PEX1 and PEX6 to peroxisome membranes, possibly to form heteromeric AAA ATPase complexes required for the import of proteins into peroxisomes. Involved in the import of catalase and proteins containing a PTS2 target sequence, but not in import of proteins with a PTS1 target sequence. {ECO:0000269|PubMed:12717447, ECO:0000269|PubMed:12851857}. | Alternative splicing;Disease variant;Membrane;Peroxisome;Peroxisome biogenesis disorder;Protein transport;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix;Transport;Zellweger syndrome | This gene belongs to the peroxin-26 gene family. It is probably required for protein import into peroxisomes. It anchors PEX1 and PEX6 to peroxisome membranes, possibly to form heteromeric AAA ATPase complexes required for the import of proteins into peroxisomes. Defects in this gene are the cause of peroxisome biogenesis disorder complementation group 8 (PBD-CG8). PBD refers to a group of peroxisomal disorders arising from a failure of protein import into the peroxisomal membrane or matrix. The PBD group is comprised of four disorders: Zellweger syndrome (ZWS), neonatal adrenoleukodystrophy (NALD), infantile Refsum disease (IRD), and classical rhizomelic chondrodysplasia punctata (RCDP). Alternatively spliced transcript variants have been identified for this gene. [provided by RefSeq, Dec 2010]. | hsa:55670; | cytosol [GO:0005829]; integral component of peroxisomal membrane [GO:0005779]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; ATPase binding [GO:0051117]; protein C-terminus binding [GO:0008022]; protein-containing complex binding [GO:0044877]; protein import into peroxisome matrix [GO:0016558]; protein import into peroxisome membrane [GO:0045046] | 12851857_degree of temperature sensitivity in pex26 cell lines is predictive of the clinical phenotype in patients with PEX26 deficiency 15858711_PEX26 deficiency impairs peroxisomal import of both PTS1- and PTS2-targeted matrix proteins. It undergoes alternative splicing to produce several splice forms, including PEX26-delta, which rescues peroxisome biogenesis in PEX26-deficient cells. 16257970_Pex26p functions in recruiting to peroxisomes the complexes of the AAA ATPase peroxins. 16763195_We analyzed targeting of human PEX26. Its C-terminal-targeting signal contains two binding sites for PEX19 and we conclude C-terminal PEX19-binding sites mark tail-anchored proteins for delivery to peroxisomes. 19105186_the relative fraction of disease-causing alleles that occur in the coding and splice junction sequences of PEX26 gene. 23460677_PEX19 formed a complex with the peroxisomal tail anchored protein PEX26 in the cytosol and translocated it directly to peroxisomes by a TRC40-independent class I pathway. 25016021_results suggested that peroxisome biogenesis requires Pex1p- and Pex6p-regulated dissociation of Pex14p from Pex26p 26627908_In yeast, PEX26 follows the pathway that also ensures correct targeting of Pex15: PEX26 enters the endoplasmic reticulum (ER) in a GET-dependent and Pex19-independent manner. 30366024_Data support an alternative PEX14-dependent mechanism of peroxisomal membrane association for the splice variant, which lacks a transmembrane domain. Structure-function relationships of PEX26 isoforms explain an extended function in peroxisomal homeostasis and these findings may improve our understanding of the broad phenotype of PEX26-associated human disorders. 30446579_An autosomal recessive missense variant, c.153C>A (p.F51L) in PEX26 was identified in Ashkenazi Jewish individual with a milder form of Zellweger spectrum and hearing loss. Binding of Pex26-F51L to Pex1 and Pex6 is severely impaired and affects peroxisome assembly. Pex26 in the patient's fibroblasts is reduced to approximately 30% of control, suggesting that Pex26-F51L is unstable in cells. There are also other changes... 31831025_There are no significant differences between PEX1-, PEX6-, and PEX26-associated phenotypes inclinical and genetic spectrum of Heimler syndrome. 34074205_Silencing PEX26 as an unconventional mode to kill drug-resistant cancer cells and forestall drug resistance. | ENSMUSG00000067825 | Pex26 | 3.806613e+03 | 1.1579409 | 0.211561592 | 0.2911364 | 5.245515e-01 | 0.4689065916 | 0.86836012 | No | Yes | 3.698879e+03 | 293.551253 | 3.017245e+03 | 245.886131 | |
| ENSG00000215302 | WHAMMP4 | transcribed_unprocessed_pseudogene | 5.272034e+01 | 0.6782122 | -0.560191325 | 0.5632522 | 8.749191e-01 | 0.3495970717 | 0.82969965 | No | Yes | 3.265293e+01 | 15.426152 | 5.672962e+01 | 27.352103 | |||||||||||
| ENSG00000223875 | NBEAP3 | unprocessed_pseudogene | 1.176706e+01 | 0.2654801 | -1.913324177 | 0.9190519 | 4.315628e+00 | 0.0377638377 | No | Yes | 3.578756e+00 | 2.219883 | 1.615415e+01 | 8.677610 | ||||||||||||
| ENSG00000224389 | 721 | C4B | protein_coding | P0C0L5 | FUNCTION: Non-enzymatic component of the C3 and C5 convertases and thus essential for the propagation of the classical complement pathway. Covalently binds to immunoglobulins and immune complexes and enhances the solubilization of immune aggregates and the clearance of IC through CR1 on erythrocytes. C4A isotype is responsible for effective binding to form amide bonds with immune aggregates or protein antigens, while C4B isotype catalyzes the transacylation of the thioester carbonyl group to form ester bonds with carbohydrate antigens.; FUNCTION: Derived from proteolytic degradation of complement C4, C4a anaphylatoxin is a mediator of local inflammatory process. It induces the contraction of smooth muscle, increases vascular permeability and causes histamine release from mast cells and basophilic leukocytes. | 3D-structure;Blood group antigen;Cell junction;Cell projection;Cleavage on pair of basic residues;Complement pathway;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunity;Inflammatory response;Innate immunity;Phosphoprotein;Reference proteome;Secreted;Signal;Sulfation;Synapse;Systemic lupus erythematosus;Thioester bond | This gene encodes the basic form of complement factor 4, and together with the C4A gene, is part of the classical activation pathway. The protein is expressed as a single chain precursor which is proteolytically cleaved into a trimer of alpha, beta, and gamma chains prior to secretion. The trimer provides a surface for interaction between the antigen-antibody complex and other complement components. The alpha chain may be cleaved to release C4 anaphylatoxin, a mediator of local inflammation. Deficiency of this protein is associated with systemic lupus erythematosus. This gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. Varying haplotypes of this gene cluster exist, such that individuals may have 1, 2, or 3 copies of this gene. In addition, this gene exists as a long form and a short form due to the presence or absence of a 6.4 kb endogenous HERV-K retrovirus in intron 9. [provided by RefSeq, May 2020]. | hsa:100293534;hsa:721; | axon [GO:0030424]; blood microparticle [GO:0072562]; classical-complement-pathway C3/C5 convertase complex [GO:0005601]; dendrite [GO:0030425]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; plasma membrane [GO:0005886]; symbiont cell surface [GO:0106139]; synapse [GO:0045202]; carbohydrate binding [GO:0030246]; complement binding [GO:0001848]; endopeptidase inhibitor activity [GO:0004866]; complement activation [GO:0006956]; complement activation, classical pathway [GO:0006958]; detection of molecule of bacterial origin [GO:0032490]; inflammatory response [GO:0006954]; innate immune response [GO:0045087]; opsonization [GO:0008228]; positive regulation of apoptotic cell clearance [GO:2000427] | 11062289_Observational study of gene-disease association. (HuGE Navigator) 11168010_Observational study of gene-disease association. (HuGE Navigator) 11803045_Allelic distribution of complement components BF, C4A, C4B, and C3 in Psoriasis vulgaris. 12224044_C4A and C4B gene-dosage variations play in infectious and autoimmune diseases. 12226794_Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. 12480675_Observational study of gene-disease association. (HuGE Navigator) 12893820_C4b and C3b do not undergo the same conformational changes upon binding to the C4BP mutants as during the interaction with the wild type C4BP, which then results in an observed loss of the cofactor activity 12907438_complex of C4b and protein S could act as a bridge between coagulation and inflammation due to the involvement of C4BP in regulating complement activation. 15033778_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15033778_negative effect of C4B(*)Q0 on health or survival 15096498_C4b-binding protein-protein S complex inhibits the phagocytosis of apoptotic cells 15456488_Plasma-derived PROS-C4BP complex has direct anticoagulant activity; enhanced direct activity of PROS-Heerlen-C4BP may compensate for low free protein S levels and low cofactor activity in individuals with protein S-Heerlen. 15665772_Acute rejection of kidney transpl with C4d expression was diffuse and showed a higher proportionate elevation of serum creatinine at biopsy and 4 weeks after diagnosis. 15787745_C4 null alleles were significantly more common in Henoch-Schonlein purpura patients than in controls 15816885_multicenter analysis of C4d staining in protocol biopsies from renal allografts 15998580_Observational study of gene-disease association. (HuGE Navigator) 16098595_The tertiary structures of C4A and C4B were compared using near and far-UV circular dichroism, ANS fluorescence, site-specific monoclonal antibodies and isoelectric focusing. 16386506_C4d is a possibe marker for the identification of humoral rejection in any clinical setting after kidney transplantation. 16504674_C4d peritubular capillary expression did not differentiate patients after kidney transplantation immunosuppression , but it predisposes to progression of chronic morphological findings during 1-year observation. 16889542_C3d was somewhat more predictive of margination than C4d in ABO-incompatible renal allografts 16893076_Results describe three distinct profiles of serum complement C4 proteins in pediatric systemic lupus erythematosus (SLE) patients, and show tight associations of complement C4 and C3 protein levels in SLE but not in healthy subjects. 16908004_47% of the C4 genes adjacent to the RP2 gene were the short gene and 53% were the long gene 16980082_C4d could be used as a marker for rejection following hepatic transplantation. 17015733_Both galactose-specific and mannose-specific mannose-binding lectins isolated from common carp were found to associate with a serine protease that cleaves native human C4 into C4b but not C4i 17202363_suggest a direct role of lgtC expression in the inhibition of C4b deposition and consequent serum resistance of R2866 17212707_Observational study of gene-disease association. (HuGE Navigator) 17257223_C4 gene deficiencies are associated with predisposition to chronic periodontitis. 17318071_demonstrated a significant association between diffuse C4d staining, production of donor-specific antibodies, and graft failure 17425651_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17425651_findings indicate that the C4B*Q0 genotype can be considered as a major covariate of smoking in precipitating the risk for acute myocardial infarction and associated deaths 17503323_Gene dosage variation and associated polymorphisms of C4B in systemic lupus erythematosus were studied. 17728371_C4B*Q0 was present in two out of the 130 SLE patients; overall, our results do not demonstrate a significant association to these known C4 mutations in the Malaysian scenario 17728371_Observational study of gene-disease association. (HuGE Navigator) 17915330_the binding of Factor H and C4bp to Aspergillus spp. appears to be even stronger than to Candida spp. and different, albeit possibly nearby, binding moieties mediate this surface attachment. 17921792_partial or complete C4b deficiencies were found in oligoarthritis and polyarthritis patients 17971360_electron dense C4d (complement 4d) deposition in peritubular capillaries was observed in most Lupus Nephritis patients 17984207_The binding of complement components C4b and C3b to the proteins of Neisseria gonorrhoeae and Neisseria meningitidis is reported. 18032375_This observation indicates that low C4B copy number is a strong risk factor for short-term mortality after acute myocardial infarction (AMI) in smoking Icelandic patients. 18065805_C4d immunostaining for the diagnosis of acute antibody-mediated rejection in renal transplant recipients 18085389_Establish reproducible procedure for C4d detection with a polyclonal antibody. 18091514_C4d positive chronic rejection is very common, associated with proteinuria, and has a poor outcome. 18179706_In 2,250 genetic typings of autistic subjects, only one individual carried a chromosome containing both C4B null allele and CYP21A2 mutations. 18315707_Confirmed the independent prognostic value of peritubular capillary C4d staining on renal allograft survival in Chinese. 18347532_omplement C4d region staining in peritubular capillaries in kidney allograft biopsies is a hallmark of antibody-mediated rejection. 18360261_The finding of diffuse C4d on follow-up biopsy is significantly associated with kidney graft loss at 1 year, regardless of index biopsy C4d results. [C4d, complement 4d] 18365397_Complement 4d in renal transplant biopsy is indicative of chronic graft rejection. 18645500_HLA-specific antibodies are associated with vascular C4d deposition and soluble C4d in broncho-alveolar lavage of lung allografts 18682851_C4BP binds to jeopardized cardiomyocytes early after acute myocardial infarct and co-localizes to other well known markers such as complemenb 3b. 18927458_platelet C4d is associated with severe acute ischemic stroke; platelet C4d may be a biomarker as well as pathogenic clue that links cerebrovascular inflammation and thrombosis 18929826_Measurement of the E-C4d/erythrocyte complement receptor 1 ratio may be a noninvasive method for detecting acute rejection after cardiac transplantation. 19062096_Observational study of gene-disease association. (HuGE Navigator) 19135723_demonstration of diversity associated with gene copy-number variation of complement C4, CYP21 & tenascin; also offers an explanation for low prevalence of systemic lupus erythematosus but high incidence of congenital adrenal hyperplasia in Asian-Indians 19137635_Observational study of gene-disease association. (HuGE Navigator) 19150565_improved survival is seen in patients with C4A or C4B deficiency and renal cell carcinoma treated with cytokine therapy with or without surgery 19167759_Observational study of gene-disease association. (HuGE Navigator) 19505723_Observational study of genotype prevalence. (HuGE Navigator) 19900552_deposits are associated with plasma cells and donor-specific antibodies in renal transplants developing chronic rejection 19913121_Observational study of gene-disease association. (HuGE Navigator) 20022265_Increased erythrocyte C4D is associated with known alloantibody and autoantibody markers of antibody-mediated rejection in human lung transplant recipients. 20139276_Data show that C1q, C4, C3, and C9 bind to thrombin receptor-activating peptide-activated platelets in lepirudin-anticoagulated platelet-rich plasma (PRP) and whole blood. 20452682_C4B null allele has a significant risk for association with autism and may indicate its possible contributing role to autoimmunity in autism. 20452682_Observational study of gene-disease association. (HuGE Navigator) 20506482_Observational study of gene-disease association and genetic testing. (HuGE Navigator) 20580617_Homozygous complement C4B deficiency is described in a female patient with membranoproliferative glomerulonephritis type III characterized by renal biopsies with strong complement C4 and IgG deposits. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20795316_The physiology and molecular basis of the Chido blood-group system is discussed. Review. 21085669_Data indicate that the deposition of both C4 and C3 showed a significant positive correlation with the serum concentration of Ficolin-3. 21345967_C4 mRNA levels of the two isoforms (C4A and C4B) were significantly reduced in hepatocytes transfected with RNA from HCV genotype 1a or 2a. 21620080_Findings suggest that significant allograft dysfunction is not related to C4d positivity in heart transplant recipients. 21642539_Flavivirus soluble nonstructural protein NS1 recruits human complement 4b-binding protein (C4BP) to the surface of cells in order to inactivate C4b on the plasma membrane. 21857912_Data show that in the UK cohort, total C4 GCN ranged from 2 to 6, with copy numbers from 0 to 4 observed for both C4A and C4B, while in the Spanish cohort, C4A GCN from 0 to 6 and C4B GCN from 0 to 5. 21967755_The C4B*Q0 genotype may be associated with hyperreactivity of the HPA axis (manifested as an increased responsiveness to ACTH-stimulation), probably through enhanced function of steroid 21-hydroxylase. 22076784_Our findings indicate a relationship between C4B copy number variation and rheumatoid arthritis. 22132894_Purified C4b inhibited IFN-beta-induced CXCL10 production & STAT1 phosphorylation. 22151770_C4B is correlated with the end of disease remission at 9-month post diagnosis in new onset type 1 diabetes. 22169613_There is a connection between elevated concentrations of both complement C4b and TTR and the pathogenesis of proliferative vitreoretinopathy. 22387014_Although complete homozygous deficiency of complement C4 is one of the strongest genetic risk factors for SLE, partial C4 deficiency states do not independently predispose to the disease. 22393059_all analyzed cofactors form similar trimolecular complexes with FI and C3b/C4b, and the accessibility of FIMAC and SP domains is crucial for the function of FI 22737222_analysis of gene copy number of complement C4A, C4B and C4A silencing mutation by real-time quantitative polymerase chain reaction 22785613_The precise order and size of all C4 genes were determined in RCCX, a multiallelic copy number variation locus. 22841245_Graft loss was significantly associated with late humoral rejection and C4d positivity in kidney transplant recipients. 22878256_Systemic lupus erythematosus patients have elevated complement C4d deposition on platelets. 23006730_Data show that in preeclamptic women, diffuse placental C4d was associated with a significantly lower gestational age at delivery, and the mRNA expression of the complement regulatory proteins CD55 and CD59 was significantly upregulated. 23026593_Results suggest that detection of C4d staining in acute 'cell-mediated' rejection does not imply a worse renal prognosis. 23199209_Data indicate that the increasing intensity of C4d staining was associated with worsened outcome. 23274969_Minimal peritubular capillary C4d+ staining is associated with antibody-mediated rejection, circulating donor specific antibodies and immune-response-related gene activation in kidney transplantation. 23447068_C4d staining in post-reperfusion biopsies and an early rise in donor specific antibodies after transplantation are risk factors for rejection in moderately sensitized patients. 23698598_Arteriolar C4d deposition may be a pathologic marker of transplantation-associated thrombotic microangiopathy implicating localized complement fixation in hematopoietic stem cell transplant recipients with kidney disease secondary to small vessel injury. 23715124_Low C4 gene copy numbers are associated with superior graft survival in patients transplanted with a deceased donor kidney. 23911397_Studies indicate that initiation of lectin compleme pathway leads to activation of the serine proteases MASP-1 and MASP-2 resulting in deposition of C4 on the activator and assembly of the C3 convertase. 23918728_Copy number variations of C4 isotypes (C4A and C4B) were detected by real-time PCR in 905 patients with Behcet disease. 23979690_The active site in monomeric C4b for suppressing Th1 cytokine production was determined. 23981509_Complement activation products C3d and C4d binding to lymphocytes can reflect the disease activity of systemic lupus erythematosus and can be used as biomarkers for SLE. 24030736_Report clinical/genomic significance of donor-specific antibody-positive/C4d-negative and donor-specific antibody-negative/C4d-negative transplant glomerulopathy. 24166212_C4d can be an appropriate marker of antibody response and complement activation in patients with primary Sjogren's syndrome. 24484408_Serum C4b, FN, and PEPD are associated with the pathological changes of pulmonary tuberculosis. 24551304_C4d deposition in liver allografts is independent of the crossmatch results and occurs with a variety of pathologic abnormalities and underlying liver diseases. 24638111_our study indicates that Finnish NTM patients had significantly more often C4 deficiencies than the healthy control subjects. 25073036_Transplant glomerulopathy is associated with poor prognosis, independently of C4d detection. 25082343_no statistically significant difference in C4d expression in liver biopsies from acute graft rejection patients as compared with HCV infection recurrence 25420802_Kidney transplantation graft rejection time in patients with positive C4d was 15 months in average versus 8 months with negative C4d. 25706274_There was a significant association between C4d deposition on allograft endothelial cells and presence of class II DSA I chronic liver allograft failure. 25778989_C4d immunostaining was a significant predictor of coronary allograft vasculopathy and death but not subsequent episodes of cell-mediated rejection. There was also a trend toward increased graft failure. 25799154_Elevated levels of the complement activation product C4d in bronchial fluids is associated with lung cancer. 25894167_Late antibody/C4d-mediated rejection can emerge soon after the modification of immunosuppressive drug dosages and may be responsible for graft dysfunction or loss. 26031580_C4d-negative acute antibody-mediated graft rejection resembles C4d-positive aAMR in terms of clinical and pathological features. C4d positivity has no influence on short-term outcome. 26071493_Suggest a prominent prognostic value of C4d staining as a rejection marker in ABO compatible kidney transplantation. 26151608_Isolated C4d deposition and isolated interstitial inflammation appear to be benign lesions, but C4d deposition in association with interstitial inflammation in the biopsy is strongly associated with the development of chronic graft dysfunction. 26305533_Carrying no copies of C4B significantly increases the risk of cCSC, whereas carrying three C4B copies is protective. 26517116_The interaction between C4BP and Leptospira interrogans LcpA, LigA and LigB is sensitive to ionic strength and inhibited by heparin. 26678451_Data indicate Ig-like transcript 4 (ILT4) as a cellular receptor for complement split products (CSPs) complement component 4d (C4d). 26800705_C4A and C4B gene copy numbers are stronger risk factors for juvenile-onset than for adult-onset systemic lupus erythematosus 26814708_The aims of this study were to elucidate the copy number variations of C4A and C4B in relation to disease risk in systemic lupus erythematosus, and to compare the basis of race-specific C4A deficiency between East Asians and individuals of European descent 26814963_Schizophrenia risk from complex variation of complement component 4 27630295_C4d is expressed in esophageal squamous cell carcinoma 27758680_An elevated number of C4 genes was observed in Alzheimer's disease (AD) patients as compared with healthy controls. The presence of high C4A and C4B copy numbers in AD patients could explain the increased C4 protein expression observed in AD patients, thus highlighting a possible role for C4A and C4B copy number variations in the risk of developing AD. 27846156_Quantification of plasma C4d+ microvesicles provides information about presence of antibody-mediated rejection, its severity and response to treatment in transplant patients. 28132614_Comparative analysis of C4 alone, C4-Tryp, and C4-MASP2 revealed the impact of Tryp on C4 was similar as MASP2. 28512867_The structural basis for inhibition of the classical and lectin complement pathways by S. aureus extracellular adherence protein binding to C4b has been presented. 28832994_results suggest that the C4B gene number associates positively with inflammation in patients with PIBD. Multiple copies of the C4B gene may thus aggravate the IBD-associated dysbiosis through escalated complement reactivity towards the microbiota. 28833936_Persistent C4d staining on kidney transplant follow-up biopsies was associated with worse clinical outcomes in patients with antibody-mediated rejection. 29080553_for the first time, a complete overview of C4 in SLE from genetic variation to binding capacity using a novel test. As this test detects crossing over of Rodgers and Chido antigens, it will allow for more accurate measurement of C4 in future studies. 29184132_Our findings suggest that circulating plasma C4d is a promising new prognostic biomarker in patients with MPM and, moreover, helps to select patients for surgery following induction chemotherapy. 29340789_Our study suggests that A2160 [complement 4-binding protein ]may be a useful prognostic biomarker for epithelial ovarian cancer, and higher pretreatment levels of A2160 predicts poor survival outcome. 29928053_This reported cohort of homozygous deficiencies of C4A or C4B suggests that C4 deficiencies may have various unrecorded disease associations. 30026462_Exploratory analysis of C4B GCN (gene copy numbers)showed positive correlation with neuropil contraction in the cerebellum and superior temporal gyrus among YASZ while AOSZ showed neuropil contraction in the prefrontal and subcortical structures. Thus, C4A and C4B GCN are associated with neuropil contraction in regions often associated with schizophrenia, and may be neuromaturationally dependent. 30041577_Low C4 in systemic lupus erythematosus patients is due to consumption rather than deficient synthesis related to lower C4A & B gene copy numbers. 30323030_The authors conclude that Streptococcus pneumoniae PspA and PspC help the pneumococcus to evade complement attack by binding C4BP and so inactivating C4b. 30465166_The purpose of this study was to evaluate C4A and C4B in patients with congenital adrenal hyperplasia in relation to CYP21A2 genotype and psychiatric and autoimmune comorbidity. We determined the copy numbers of C4A and C4B in 145 patients with CAH .No association was found between C4 copy number and autoimmune disease. 30926239_Capsid-deposited C4b neutralizes infection by human adenovirus independent of C2 and C3 but requires C1q antibody engagement. C4b inhibits capsid disassembly, preventing endosomal escape and cytosolic access. 31315998_Employing a range of biochemical approaches, the authors showed (i) a direct association of C1q to Chandipura virus, (ii) deposition of complement proteins C3b, C4b, and C1q on Chandipura virus, and (iii) virus aggregation. 32499649_Complement genes contribute sex-biased vulnerability in diverse disorders. 32691186_Association between complement 4 copy number variation and systemic lupus erythematosus: a meta-analysis. 33433412_Glomerular C4d deposition in proliferative glomerular diseases. 34299197_Complement C4 Is Reduced in iPSC-Derived Astrocytes of Autism Spectrum Disorder Subjects. 34358307_The BS variant of C4 protects against age-related loss of white matter microstructural integrity. 34480088_Integrative brain transcriptome analysis links complement component 4 and HSPA2 to the APOE epsilon2 protective effect in Alzheimer disease. | ENSMUSG00000073418 | C4b | 3.182185e+01 | 0.7016888 | -0.511096671 | 0.6327734 | 6.413638e-01 | 0.4232173850 | 0.85072957 | No | Yes | 1.668728e+01 | 7.755167 | 2.620589e+01 | 12.181894 | |
| ENSG00000224877 | 284184 | NDUFAF8 | protein_coding | A1L188 | FUNCTION: Involved in the assembly of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I, MT-ND1) (PubMed:27499296). Required to stabilize NDUFAF5 (PubMed:27499296). {ECO:0000269|PubMed:27499296}. | Disease variant;Disulfide bond;Mitochondrion;Primary mitochondrial disease;Reference proteome | hsa:284184; | mitochondrion [GO:0005739]; mitochondrial respiratory chain complex I assembly [GO:0032981] | 31866046_Pathogenic Bi-allelic Mutations in NDUFAF8 Cause Leigh Syndrome with an Isolated Complex I Deficiency. | ENSMUSG00000078572 | Ndufaf8 | 3.456745e+03 | 1.0155469 | 0.022256864 | 0.3198912 | 4.791482e-03 | 0.9448140832 | 0.98943389 | No | Yes | 3.713828e+03 | 475.244491 | 2.994212e+03 | 393.633733 | ||
| ENSG00000224892 | 220433 | RPS4XP16 | transcribed_processed_pseudogene | 1.023450e+01 | 0.4955589 | -1.012871656 | 0.8726906 | 1.340087e+00 | 0.2470186313 | No | Yes | 4.946316e+00 | 2.630933 | 1.194652e+01 | 6.162813 | |||||||||||
| ENSG00000225177 | lncRNA | 2.384455e+02 | 0.9799852 | -0.029168138 | 0.3316961 | 7.718042e-03 | 0.9299940015 | 0.98676673 | No | Yes | 2.259598e+02 | 34.389535 | 2.001159e+02 | 31.283102 | ||||||||||||
| ENSG00000225791 | 401264 | TRAM2-AS1 | lncRNA | 5.897846e+01 | 0.9397365 | -0.089671873 | 0.4440609 | 4.038582e-02 | 0.8407281048 | 0.96860786 | No | Yes | 4.396981e+01 | 9.428359 | 5.309867e+01 | 11.523487 | ||||||||||
| ENSG00000225921 | 51406 | NOL7 | protein_coding | Q9UMY1 | 3D-structure;Alternative splicing;Coiled coil;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene localizes to the nucleolus, where it maintains nucleolar structure and cell growth rates. The encoded protein also functions as a tumor suppressor and regulator of angiogenesis. The RB tumor suppressor gene recruits transcription factors to this gene and positively regulates its expression. Two transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Dec 2015]. | hsa:51406; | chromosome [GO:0005694]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; RNA binding [GO:0003723] | 20206243_determined that the essential elements required for the optimal promoter activity of NOL7 were 560 bp upstream of its translation start site. 21818416_Three alternatively spliced forms of the human NOL7 gene coding for relatively small proteins were identified 22123719_NOL7 is the legitimate tumor suppressor gene located on 6p23. 23085760_NOL7 significantly alters TSP-1 expression and may be a master regulator that coordinates the post-transcriptional expression of key signaling factors critical for the regulation of the angiogenic phenotype. 34642294_NOL7 facilitates melanoma progression and metastasis. | ENSMUSG00000063200 | Nol7 | 1.686657e+03 | 1.2015099 | 0.264848488 | 0.3099649 | 7.169518e-01 | 0.3971456080 | 0.84483554 | No | Yes | 1.865709e+03 | 291.263228 | 1.426319e+03 | 228.641351 | ||
| ENSG00000226696 | 104355426 | LENG8-AS1 | lncRNA | 7.324298e+01 | 1.0215800 | 0.030802229 | 0.4064994 | 5.755437e-03 | 0.9395268243 | 0.98870361 | No | Yes | 4.721046e+01 | 11.274593 | 5.120897e+01 | 12.446302 | ||||||||||
| ENSG00000226823 | SUGT1P1 | unprocessed_pseudogene | 9.555739e+00 | 1.2841509 | 0.360814783 | 1.1110023 | 1.169285e-01 | 0.7323899854 | No | Yes | 7.186726e+00 | 4.645582 | 6.242450e+00 | 4.182592 | ||||||||||||
| ENSG00000226950 | 57291 | DANCR | lncRNA | This gene produces a long non-coding RNA that functions as a negative regulator of cell differentiation. This transcript associates with enhancer of zeste homolog 2 to repress expression of the runt related transcription factor 2 gene. Increased expression of this transcript may be associated with cancer. Alternatively spliced transcript variants have been observed. [provided by RefSeq, Dec 2016]. | 22302877_ANCR lncRNA is thus required to enforce the undifferentiated cell state within epidermis 23438432_These data suggest that ANCR is an essential mediator of osteoblast differentiation. 25660720_Significant upregulation of long non-coding RNA-DANCR in circulating monocytes in the postmenopausal osteoporosis patients. 26514989_Sox4 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cell via activation of long noncoding RNA DANCR 26617879_Our data indicated that lncRNA DANCR expression might be a novel potential biomarker for colorectal cancer prognosis. 26986815_High ANCR long noncoding RNA expression is associated with Osteosarcoma. 27716745_Knockdown of ANCR induced an epithelial-mesenchymal transition program and promoted cell migration and invasion in MCF10A, whereas ectopic expression of ANCR repressed breast cancer cells migration and invasion. 27919960_This study revealed, for the first time, the importance of DANCR in clinical diagnosis and disease progression of HCC 28618943_according to the results of this study,DANCR is dramatically upregulated in Gastric cancer tissues and the high expression of DANCR is significantly associated with worse overall survival in Gastric cancer patients 28642170_results suggest that DANCR is a crucial upregulator of osteosarcoma and an independent predictor of prognosis. DANCR increases cancer stem cells function by upregulating AXL via competitively binding to miR-33a-5p, and this function is sequentially performed through the PI3K-Akt signaling pathway. 28674107_results characterized miR-1305-Smad4 axis as a major downstream functional mechanism of lncRNA DANCR in promoting the chondrogenesis in synovium-derived mesenchymal stem cells. 28679390_inhibited the cell proliferation, migration, and invasion of osteosarcoma cells, possibly through interacting with EZH2 and regulating the expression of p21 and p27 29115577_DANCR was found to mediate the proliferation and osteogenic differentiation of HBMSCs via p38 MAPK inactivation, but not via extracellular signal-regulated protein kinase (ERK)1/2 or c-Jun N-terminal kinase (JNK) MAPKs, but. 29180471_DANCR is a critical lncRNA widely overexpressed in human cancers.MYC directly regulates DANCR. 29266795_Long non-coding RNA DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. 29301870_he present study demonstrated that DANCR/miR-634/RAB1A axis plays crucial roles in the progression of glioma, and DANCR might potentially serve as a therapeutic target for the treatment of glioma patients. 29338713_LncRNA DANCR could inhibit osteoblast differentiation by regulating FOXO1 expression. 29572052_DANCR promotes cisplatin resistance via activating AXL/PI3K/Akt/NF-kappaB signaling pathway in glioma. 29602127_DANCR positively affected glioma progression via activating Wnt/beta-catenin signaling pathway. 29635134_DANCR promotes non-small cell lung cancer cell proliferation, migration and invasion by regulating the tumor suppressor miR-758-3p. 29651883_DANCR upregulation was seen in lung cancer, particularly in high-grade lung cancer tissues and aggressive cancer cells. Ectopic DANCR expression induced lung cancer cell proliferation and colony formation, whereas DANCR silencing induced opposing effects. 29678573_the study revealed that lncRNA DANCR might promote the proliferation of osteoarthritis chondrocytes and reduce apoptosis through the miR-577/SphK2 axis. 29717105_DANCR promoted HSP27 expression and its mediation of proliferation/metastasis via miR-577 sponging. 29728310_Long non-coding RNAs-DANCR likely serves as a useful disease biomarker or therapeutic cancer target. 29913147_Data indicate that miR-758 is the direct target of long noncoding RNAs (lncRNAs) ANCR. 29940760_knockdown of DANCR increased the expression of miR-33a-5p, reduced Epithelial-Mesenchymal Transition, and increased apoptosis in glioma 30361290_DANCR promoted the proliferation, inflammation, and reduced cell apoptosis in osteoarthritis chondrocytes through regulating miR-216a-5p/JAK2/STAT3 signaling pathway. 30362591_DANCR promotes cervical cancer progression by functioning as a competing endogenous RNA (ceRNA) to regulate ROCK1 expression via sponging miR-335-5. 30419948_Authors found that DANCR was distributed mostly in the cytoplasm and DANCR functioned as a miRNA sponge to positively regulate the expression of musashi RNA binding protein 2 (MSI2) through sponging miR-149 and subsequently promoted malignant phenotypes of bladder cancer cells, thus playing an oncogenic role in bladder cancer pathogenesis. 30430564_DANCR also strongly suppressed hepatocellular carcinoma tumor growth in vivo via targeting miR-216a-5p and KLF12. 30518934_Long non-coding RNA DANCR upregulates PIK3CA/AKT signaling through activating serine phosphorylation of RXRA. 30535487_lncRNA DANCR was upregulated in lung adenocarcinoma (LAD) tissue and cell lines, compared with paratumor tissue and a normal lung cell line. Additionally, elevated DANCR expression was associated with poor prognosis in the patients with LAD. In LAD cell lines DANCR promoted the invasion of lung adenocarcinoma cells by positively regulating HMGA2. 30555573_Taken together, these results suggest that DANCR acts as a prognostic biomarker and increases HIF-1alpha mRNA stability by interacting with NF90/NF45, leading to metastasis and disease progression of nasopharyngeal carcinoma. 30582654_lnc-DANCR-mediated miR-665 downregulation regulates the malignant phenotype of CC cells. 30720067_lncRNA DANCR was upregulated in NPC cells. DANCR promoted cell proliferation and migration, and inhibited apoptosis in NPC cells. Mechanistically, DANCR regulated the levels of p-AKT, PTEN, BAX and BCL2 in NPC cells. 30849642_The present study demonstrates that DANCR, acting as an oncogene promotes nasopharyngeal carcinoma progression. 30910839_The present study demonstrated that DANCR was associated with papillary thyroid cancer (PTC) aggressive clinical features and may serve as a diagnostic biomarker for detecting PTC patients. 30910842_DANCR might act as a tumor promoter by targetting miRNA-216a-5p, which might provide a potential therapy target for breast cancer treatment. 31002130_LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. 31038266_DANCR acted as a ceRNA to decoy miR-27a-3p. 31074173_LncRNA ANCR is positively correlated with transforming growth factor-beta1 in patients with osteoarthritis. 31114986_Long non-coding RNA DANCR upregulates IGF2 expression and promotes ovarian cancer progression. 31161373_DANCR played an important role in promoting tumorigenesis of endometrial cancer via sponging miR-214. 31213122_Knockdown of DANCR reduces osteoclastogenesis and root resorption induced by compression force via Jagged1. 31213582_DANCR knockdown inhibited growth and metastasis of PC cells. Furthermore, DANCR acted as sponge to regulate miR-214-5p, and miR-214-5p inhibitor reversed the effects of DANCR knockdown on PC cells. DANCR positively modulated E2F2 expression through miR-214-5p in PC cells. CONCLUSIONS Collectively, Findings demonstrated that lncRNA DANCR/miR-214-5p/E2F2 axis acts as an oncogene in PC development. 31235689_Long noncoding RNA ANCR inhibits the differentiation of mesenchymal stem cells toward definitive endoderm by facilitating the association of PTBP1 with ID2. 31383847_This study showed that upregulated DANCR could promote cholangiocarcinoma progress through transcriptional inactivation of the target tumor suppressor gene FBP1 epigenetically, which revealed that DANCR could provide a theoretical basis for clinical diagnosis and treatment of cholangiocarcinoma. 31401160_MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1 31515968_LncRNA DANCR promotes proliferation and metastasis in pancreatic cancer by regulating miRNA-33b. 31545000_Results indicate that non-protein coding RNA DANCR (DANCR) plays a promotional role in tumor angiogenesis in ovarian cancer through regulation of miR-145/vascular endothelial growth factor (VEGF) axis. 31727012_Silencing of ANCR inhibited the migration and invasion of Osteosarcoma cells through activation of the p38MAPK signalling pathway 31746351_LncRNA ANCR downregulates hypoxiainducible factor 1alpha and inhibits the growth of HPVnegative cervical squamous cell carcinoma under hypoxic conditions. 31804607_IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. 31805275_LncRNA DANCR sponges miR-216a to inhibit odontoblast differentiation through upregulating c-Cbl. 31841200_Long non-coding RNA DANCR induces chondrogenesis by regulating the miR-1275/MMP-13 axis in synovial fluid-derived mesenchymal stem cells. 31858532_LncRNA DANCR aggravates the progression of ovarian cancer by downregulating UPF1. 31860165_The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. 31863900_Long non-coding RNA DANCR promotes colorectal tumor growth by binding to lysine acetyltransferase 6A. 31918278_LncRNA DANCR silence inhibits SOX5-medicated progression and autophagy in osteosarcoma via regulating miR-216a-5p. 32196584_LncRNA DANCR affected cell growth, EMT and angiogenesis by sponging miR-345-5p through modulating Twist1 in cholangiocarcinoma. 32196604_Upregulation of lncRNA DANCR functions as an oncogenic role in non-small lung cancer by regulating miR-214-5p/CIZ1 axis. 32222722_Long Noncoding RNA DANCR Suppressed Lipopolysaccharide-Induced Septic Acute Kidney Injury by Regulating miR-214 in HK-2 Cells. 32417846_Knockdown of DANCR Suppressed the Biological Behaviors of Ovarian Cancer Cells Treated with Transforming Growth Factor-beta (TGF-beta) by Sponging MiR-214. 32423468_Roles of DANCR/microRNA-518a-3p/MDMA ceRNA network in the growth and malignant behaviors of colon cancer cells. 32505219_MicroRNA-33a-5p sponges to inhibit pancreatic beta-cell function in gestational diabetes mellitus LncRNA DANCR. 32633342_LncRNA DANCR regulates osteosarcoma migration and invasion by targeting miR-149/MSI2 axis. 32778797_LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/beta-catenin signaling pathway. 32860589_LncRNA DANCR regulates the growth and metastasis of oral squamous cell carcinoma cells via altering miR-216a-5p expression. 32964966_LncRNA DANCR promotes ATG7 expression to accelerate hepatocellular carcinoma cell proliferation and autophagy by sponging miR-222-3p. 32971057_Long Noncoding RNA DANCR Regulates Cell Proliferation by Stabilizing SOX2 mRNA in Nasopharyngeal Carcinoma. 33058834_LncRNA DANCR upregulation induced by TUFT1 promotes malignant progression in triple negative breast cancer via miR-874-3p-SOX2 axis. 33089960_SP1-induced lncRNA DANCR contributes to proliferation and invasion of ovarian cancer. 33272103_Crosstalk between lncRNA DANCR and miR-125b-5p in HCC cell progression. 33302540_Long Noncoding RNA DANCR Activates Wnt/beta-Catenin Signaling through MiR-216a Inhibition in Non-Small Cell Lung Cancer. 33317921_Silencing LncRNA-DANCR attenuates inflammation and DSS-induced endothelial injury through miR-125b-5p. 33414433_LncRNA DANCR represses Doxorubicin-induced apoptosis through stabilizing MALAT1 expression in colorectal cancer cells. 33481014_Long non-coding RNA DANCR accelerates colorectal cancer progression via regulating the miR-185-5p/HMGA2 axis. 33577030_Long non-coding RNA DANCR promoted non-small cell lung cancer cells metastasis via modulating of miR-1225-3p/ErbB2 signal. 33629241_Long non-coding RNA DANCR modulates osteogenic differentiation by regulating the miR-1301-3p/PROX1 axis. 33629310_Emerging role of lncRNA DANCR in progenitor cells: beyond cancer. 33638615_Long noncoding RNA DANCR confers cytarabine resistance in acute myeloid leukemia by activating autophagy via the miR-874-3P/ATG16L1 axis. 33885171_Long non-coding DANCR targets miR-185-5p to upregulate LIM and SH3 protein 1 promoting prostate cancer via the FAK/PI3K/AKT/GSK3beta/snail pathway. 33956301_Assessment of lncRNA DANCR, miR-145-5p and NRAS axis as biomarkers for the diagnosis of colorectal cancer. 34003569_LncRNA DANCR improves the dysfunction of the intestinal barrier and alleviates epithelial injury by targeting the miR-1306-5p/PLK1 axis in sepsis. 34050113_Long noncoding RNA differentiation antagonizing nonprotein coding RNA promotes the proliferation, invasion and migration of neuroblastoma cells via targeting beta-1, 4-galactosyltransferase III by sponging miR-338-3p. 34233586_Kruppel-like factor 5-induced overexpression of long non-coding RNA DANCR promotes the progression of cervical cancer via repressing microRNA-145-3p to target ZEB1. 34278459_Long noncoding RNA DANCR represses the viability, migration and invasion of multiple myeloma cells by sponging miR135b5p to target KLF9. 34414934_Expression of ANCR in nasopharyngeal carcinoma patients and its clinical significance. 34515297_Inhibition of the lncRNA DANCR attenuates cardiomyocyte injury induced by oxygen-glucose deprivation via the miR-19a-3p/MAPK1 axis. 34652251_lncRNA DANCR promotes the migration an invasion and of trophoblast cells through microRNA-214-5p in preeclampsia. 34783641_Long non-coding RNA anti-differentiation non-coding RNA affects proliferation, invasion, and migration of breast cancer cells by targeting miR-331. 35147200_Expression of SOX2OT, DANCR and TINCR long noncoding RNAs in papillary thyroid cancer and its effects on clinicopathological features. 35235755_Long noncoding RNA DANCR expression and its predictive value in patients with atherosclerosis. | 4.694103e+03 | 0.9221189 | -0.116975291 | 0.2921339 | 1.612102e-01 | 0.6880447763 | 0.93352105 | No | Yes | 4.422826e+03 | 349.795030 | 4.350359e+03 | 353.043500 | ||||||||
| ENSG00000227372 | 57212 | TP73-AS1 | transcribed_unitary_pseudogene | 20477830_PDAM knockdown induces cisplatin resistance in glioma cells harboring wild-type p53. The finding suggests that PDAM may possess the capacity to modulate apoptosis via regulation of p53-dependent anti-apoptotic genes (BCL2 and BCL2L1). 20477830_The study suggested that PDAM might function as a non-protein-coding RNA.And PDAM deregulation play a role in oligodendroglial tumors development, PDAM may possess the capacity to modulate apoptosis via regulation of p53-dependent anti-apoptotic genes. 22388545_Report expression of TP73 isoforms in human cell lines. 26410378_KIAA0495 methylation was detected in none of both primary myeloma samples at diagnosis (N = 61) and at relapse/progression (N = 16). KIAA0495 methylation appeared unimportant in the pathogenesis or progression of myeloma. 26799587_High expression level of long non-coding RNA TP73-AS1 is associated with esophageal squamous cell carcinoma. 28379612_Study showed that TP73-AS1 was specifically upregulated in brain glioma tissues and cell lines, and was associated with poorer prognosis in patients with glioma. TP73-AS1 knock down suppressed human brain glioma cell proliferation and invasion in vitro. TP73-AS1 seems to act as an oncogenic lncRNA promoting brain glioma proliferation and invasion. 28639399_Study showed that TP73-AS1 was specifically upregulated in BC tissues and breast cancer (BC) cell lines and was correlated to a poor prognosis. Its knocking down suppressed BC cell proliferation in vitro through regulation of TFAM. Also, data revealed that TP73-AS1 could regulate miR-200a through direct targeting. 28857253_Data show a direct binding between TP73-AS1, miR-200a and ZEB1. TP73-AS1 promoted ZEB1 expression via competing with its 3'UTR for miR-200a binding. ZEB1 binds the promoter region of TP73-AS1 activating its expression. TP73-AS1 and ZEB1 expression was increased, whereas miR-200a expression was low in breast cancer (BC). These regulating loop TP73-AS1/miR-200a/ZEB1 in BC promotes cancer cell invasion and migration. 29412778_TP73-AS1 inhibited the brain glioma growth and metastasis as a competing endogenous RNA (ceRNA) through miR-124-dependent iASPP regulation. 29474580_miR-941 sponge region of nine high-affinity miR-941 binding sites clustering within 1 kb 29625110_lncRNA TP73-AS1 may serve as a tumor suppressor participated in bladder cancer progression, which provided a promising therapy strategy for patients with bladder cancer. 29750302_TP73-AS1 may be an oncogenic lncRNA that promotes the proliferation of ovarian cancer cells 29803931_An explicit oncogenic role of TP73-AS1 in the NSCLC tumorigenesis, suggesting a TP73-AS1-miR-449a-EZH2 axis and providing new insight for NSCLC tumorigenesis. 29864904_TP73-AS1 could function as a sponge of miR-142 to positively regulate Rac1 in osteosarcoma cells. 29904939_Study found that TP73-AS1 was upregulated in the ovarian cancer tissues and ovarian cancer cells, and correlated with poor prognosis. it promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. 29966969_TP73-AS1 was upregulated in cholangiocarcinoma, facilitating migration and invasive potential of tumor cells. 30010111_this study demonstrated that TP73-AS1 regulated Colorectal cancer (CRC)progression by acting as a competitive endogenous RNA to sponge miR-194 to modulate the expression of TGFa 30016766_TP73-AS1 knockdown activated PI3K/Akt/mTOR signaling pathway, while overexpression of TP73-AS1 induced inhibition of PI3K/Akt/mTOR pathway and these effects could be partly abolished by overexpression of KISS1. 30118890_Data showed that TP73-AS1 was enhanced in gastric cancer (GC) tissues and cells, and tightly associated with tumor size, TNM stage, and overall survival. Decreased TP73-AS1 could restrain cell growth and promoted apoptosis partly by regulating Bcl-2/caspase-3 pathway. Importantly, TP73-AS1 could promote xenograft growth in vivo. 30193732_Twist-related protein 1 (TWIST1) was a target of miR-490-3p and participated in long non-coding RNA TP73 antisense RNA 1 (TP73-AS1)/miR-490-3p-modulated MDA-MB-231cell vasculogenic mimicry (VM) formation. 30279010_TP73-AS1 was upregulated in gastric cancer tissues and cell lines. Furthermore, TP73-AS1 exerted oncogenic role in gastric cancer through promoting cell growth and metastasis. In addition, TP73-AS1 was certified as a ceRNA by regulating miR-194-5p/SDAD1 axis. 30472379_we showed that TP73AS1 inhibits CRC cell growth by functioning as a ceRNA (competing endogenous RNAs) to regulate PTEN levels. Our findings provide new insights into the underlying molecular mechanisms of TP73AS1-mediated CRC. 30541897_LncRNA TP73-AS1 promoted the progression of lung adenocarcinoma via PI3K/AKT pathway. 30643007_LncRNA TP73 antisense RNA 1T (TP73-AS1) positively regulates 3-hydroxybutyrate dehydrogenase type 2 (BDH2) by regulating miR-141. 30867410_results reveal that high TP73-AS1 predicts poor prognosis in primary glioblastoma multiform cohorts and that this lncRNA promotes tumor aggressiveness and temozolomide resistance in glioblastoma cancer stem cells. 31015368_LncRNA TP73-AS1 down-regulates miR-139-3p to promote retinoblastoma cell proliferation. 31081944_The lncRNA TP73-AS1 is a prognostic marker. 31129247_Results found that P73-AS1 was upregulated in non-small cell lung cancer (NSCLC) tissues and predicted poor survival. TP73-AS1 is a potential upstream positive regulator of miRNA to promote NSCLC cell migration and invasion. 31210297_LncRNA TP73-AS1 promotes malignant progression of hepatoma by regulating microRNA-103. 31232491_LncRNA TP73-AS1 is a novel regulator in cervical cancer via miR-329-3p/ARF1 axis. 31432138_the findings suggested that upregulation of TP73AS1 promoted cervical cancer progression by promoting CCND2 via the suppression of miR607 expression. 31549851_Association of TP73-AS1 gene polymorphisms with the risk and survival of gastric cancer in a Chinese Han Population 31652459_Role of long non-coding RNA TP73-AS1 in cancer. 31663517_LncRNA TP73-AS1 promoted proliferation of cervical cancer cell lines by targeting miR-329-3p to regulate the expression of the SMAD2 gene. A regulatory network was formed between lncRNA TP73-AS1, miR-329-3p, and SMAD2. 31678571_role of TP73-AS1 lncRNA in human cancers 31901156_Knockdown of long noncoding RNA TP73-AS1 suppresses the malignant progression of breast cancer cells in vitro through targeting miRNA-125a-3p/metadherin axis. 31978409_Long non-coding RNA TP73-AS1 in cancers. 32412786_lncRNA TP73-AS1 Regulates miR-21/PTEN Axis to Affect Cell Proliferation in Acute Myeloid Leukemia. 32818670_LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-beta1 signaling. 32901838_Long noncoding RNA TP73AS1 accelerates the progression and cisplatin resistance of nonsmall cell lung cancer by upregulating the expression of TRIM29 via competitively targeting microRNA34a5p. 33400379_LncRNA TP73-AS1 regulates miR-495 expression to promote migration and invasion of nasopharyngeal carcinoma cells through junctional adhesion molecule A. 33589576_Long non-coding RNA TP73-AS1 is a potential immune related prognostic biomarker for glioma. 33683827_LncRNA TP73-AS1 enhances the malignant properties of pancreatic ductal adenocarcinoma by increasing MMP14 expression through miRNA -200a sponging. 34007135_Long non-coding RNA TP73-AS1 promotes pancreatic cancer growth and metastasis through miRNA-128-3p/GOLM1 axis. 34103010_LncRNA TP73-AS1 promotes oxidized low-density lipoprotein-induced apoptosis of endothelial cells in atherosclerosis by targeting the miR-654-3p/AKT3 axis. 34115613_TP73-AS1 is induced by YY1 during TMZ treatment and highly expressed in the aging brain. | 3.351801e+03 | 1.1547753 | 0.207612110 | 0.2707811 | 5.926594e-01 | 0.4413925667 | 0.85687843 | No | Yes | 2.980289e+03 | 246.908828 | 2.628675e+03 | 223.373713 | |||||||||
| ENSG00000228049 | 246721 | POLR2J2 | protein_coding | Q9GZM3 | FUNCTION: DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB11 is part of the core element with the central large cleft (By similarity). {ECO:0000250}. | Alternative splicing;DNA-directed RNA polymerase;Nucleus;Reference proteome;Transcription | This gene is a member of the RNA polymerase II subunit 11 gene family, which includes three genes in a cluster on chromosome 7q22.1 and a pseudogene on chromosome 7p13. The founding member of this family, DNA directed RNA polymerase II polypeptide J, has been shown to encode a subunit of RNA polymerase II, the polymerase responsible for synthesizing messenger RNA in eukaryotes. This locus produces multiple, alternatively spliced transcripts that potentially express isoforms with distinct C-termini compared to DNA directed RNA polymerase II polypeptide J. Most or all variants are spliced to include additional non-coding exons at the 3' end which makes them candidates for nonsense-mediated decay (NMD). Consequently, it is not known if this locus expresses a protein or proteins in vivo. [provided by RefSeq, Jul 2008]. | hsa:246721;hsa:548644; | RNA polymerase II, core complex [GO:0005665]; DNA binding [GO:0003677]; protein dimerization activity [GO:0046983]; RNA polymerase II activity [GO:0001055] | 1.152958e+02 | 0.9729965 | -0.039493528 | 0.3509858 | 1.252829e-02 | 0.9108790429 | 0.98338795 | No | Yes | 8.721044e+01 | 26.249270 | 9.994517e+01 | 30.711452 | ||||
| ENSG00000228643 | lncRNA | 9.541653e+00 | 0.6636389 | -0.591529609 | 0.9119122 | 4.046718e-01 | 0.5246863654 | No | Yes | 5.406946e+00 | 2.751593 | 1.032635e+01 | 5.121771 | |||||||||||||
| ENSG00000229043 | 101927021 | ZFAND2A-DT | lncRNA | 2.926031e+02 | 1.2938098 | 0.371625555 | 0.3117096 | 1.439306e+00 | 0.2302516687 | 0.78818582 | No | Yes | 2.811795e+02 | 25.906239 | 2.092634e+02 | 20.222862 | ||||||||||
| ENSG00000229124 | 100507347 | VIM-AS1 | lncRNA | 29656793_VIM-AS1 was significantly upregulated in high-grade colorectal cancer. 30506719_lncRNA VIM-AS1 and lncRNA CTBP1-AS2 expression levels are associated with type 2 diabetes susceptibility. 31786880_lncRNA VIM-AS1 was overexpressed in prostate cancer (PCa) tissues and cell lines and promoted PCa proliferation and metastasis via epithelial-mesenchymal transition through regulating vimentin, which might provide a novel target for the diagnosis and therapy for PCa. 32447557_Long non-coding RNA VIM Antisense RNA 1 (VIM-AS1) sponges microRNA-29 to participate in diabetic retinopathy. 32633376_Knockdown of long non-coding RNA VIM-AS1 inhibits glioma cell proliferation and migration, and increases the cell apoptosis via modulation of WEE1 targeted by miR-105-5p. 33173977_lncRNA VIMAS1 promotes cell proliferation, metastasis and epithelialmesenchymal transition by activating the Wnt/betacatenin pathway in gastric cancer. 35202907_Clinical significance of Vimentin Antisense RNA 1 and its correlation with other epithelial to mesenchymal transition markers in oral cancers. | 3.588460e+01 | 0.5900057 | -0.761199175 | 0.5238491 | 2.033201e+00 | 0.1538961339 | 0.77404149 | No | Yes | 3.547729e+01 | 7.938792 | 5.654210e+01 | 12.723176 | |||||||||
| ENSG00000229320 | 90133 | KRT8P12 | transcribed_processed_pseudogene | 8.335429e+01 | 1.2925682 | 0.370240365 | 0.4014322 | 8.689384e-01 | 0.3512493691 | 0.82969965 | No | Yes | 1.167332e+02 | 24.362343 | 9.558911e+01 | 20.884286 | ||||||||||
| ENSG00000229833 | 100131801 | PET100 | protein_coding | P0DJ07 | FUNCTION: Plays an essential role in mitochondrial complex IV maturation and assembly. {ECO:0000269|PubMed:24462369, ECO:0000269|PubMed:25293719}. | Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Primary mitochondrial disease;Reference proteome;Transit peptide;Transmembrane;Transmembrane helix | Mitochondrial complex IV, or cytochrome c oxidase, is a large transmembrane protein complex that is part of the respiratory electron transport chain of mitochondria. The small protein encoded by this gene plays a role in the biogenesis of mitochondrial complex IV. This protein localizes to the inner mitochondrial membrane and is exposed to the intermembrane space. Mutations in this gene are associated with mitochondrial complex IV deficiency. This gene has a pseudogene on chromosome 3. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2014]. | hsa:100131801; | integral component of mitochondrial inner membrane [GO:0031305]; unfolded protein binding [GO:0051082]; mitochondrial cytochrome c oxidase assembly [GO:0033617] | 22356826_COX assembly candidate, ortholog of fungal PET100 22356826_Protein identified in mammalian mitochondria 24462369_We identify PET100 as a complex IV biogenesis factor in humans and characterize its location and role in mitochondria.We found PET100 (MIM 614770) mutations in ten Lebanese individuals with Leigh syndrome and isolated complex IV deficiency. 28199844_The short isoform of the myofibrillogenesis regulator 1 (MR-1S) as a new COX assembly factor, which works with the highly conserved PET100 and PET117 chaperones to assist COX biogenesis in higher eukaryotes. | ENSMUSG00000087687 | Pet100 | 1.825897e+03 | 0.8352573 | -0.259707334 | 0.3057392 | 7.129804e-01 | 0.3984561619 | 0.84485965 | No | Yes | 1.827686e+03 | 214.718134 | 1.845765e+03 | 222.632373 | |
| ENSG00000230606 | lncRNA | 6.417708e+02 | 0.6969316 | -0.520910975 | 0.3763141 | 1.972664e+00 | 0.1601654123 | 0.77593452 | No | Yes | 3.626153e+02 | 161.278925 | 6.138379e+02 | 280.028450 | ||||||||||||
| ENSG00000231584 | FAHD2CP | transcribed_unprocessed_pseudogene | 7.065936e+02 | 1.1338794 | 0.181267212 | 0.3031577 | 3.557520e-01 | 0.5508750101 | 0.89250744 | No | Yes | 6.466516e+02 | 67.995978 | 5.540235e+02 | 60.049543 | |||||||||||
| ENSG00000231663 | 101927765 | COA6-AS1 | lncRNA | 8.179973e+01 | 1.0173417 | 0.024804263 | 0.3940811 | 3.961090e-03 | 0.9498165222 | 0.99022718 | No | Yes | 7.268166e+01 | 13.384802 | 6.568611e+01 | 12.505308 | ||||||||||
| ENSG00000232098 | lncRNA | 1.071117e+03 | 1.3018659 | 0.380580865 | 0.2941887 | 1.642628e+00 | 0.1999652446 | 0.78383913 | No | Yes | 1.346218e+03 | 194.310007 | 1.078300e+03 | 160.251448 | ||||||||||||
| ENSG00000232956 | 285958 | SNHG15 | lncRNA | This gene represents a snoRNA host gene that produces a short-lived long non-coding RNA. This non-coding RNA is upregulated in tumor cells and may contribute to cell proliferation by acting as a sponge for microRNAs. [provided by RefSeq, Dec 2017]. | 26662309_Our findings present that elevated lncRNA SNHG15 could be identified as a poor prognostic biomarker in GC and regulate cell invasion. 29048682_SNHG15 and miR-153 are new potential therapeutic targets for anti-angiogenesis treatment of glioma 29217194_Long noncoding RNA SNHG15 was highly expressed in breast cancer tissues and cell lines. SNHG15 knockdown significantly inhibited the proliferation and induced apoptosis in breast cancer cells in vitro and in vivo. In mechanism, we found that SNHG15 acted as a competing endogenous RNA to sponge miR-211-3p, which was downregulated in breast cancers and inhibited cell proliferation and migration. 29604394_Results show that enhanced expression of lncRNA SNHG15 promoted colon cancer cell progression and was correlated with poor prognosis in colon cancer patients. Mechanistically, SNHG15 maintains Slug stability in living cells by impeding its ubiquitination and degradation through interaction with the zinc finger domain of Slug. 29630731_the ectopic overexpression of SNHG15 contribute to the non-small cell lung cancer tumorigenesis by regulating CDK14 protein via sponging miR-486. 29750422_the present study suggested that SNHG15 may be involved in the nuclear factorkappaB signaling pathway, induce the epithelialmesenchymal transition process, and promote renal cell carcinoma invasion and migration. 29771418_The expression of SNHG15 is up-regulated on non-small lung cancer cells, promoting promotes cell proliferation and invasion. 30280769_SNHG15 levels were significantly up-regulated in both sera and tumors tissues from pancreatic ductal adenocarcinoma (PDAC) patients. Clinicopathologic analysis revealed that high SNHG15 expression was associated with tumor differentiation, lymph node metastasis, tumor stage and shorter overall survival. Cox multivariate analyses confirmed that SNHG15 expression was an independent prognostic factor in PDAC. 30317592_Lnc-SNHG15 overexpression was significantly associated with colorectal liver metastasis and poor survival. 30402848_SNHG15 is highly expressed in lung cancer (LC) tissues, which promotes the occurrence and progression of LC via regulating proliferation and migration of LC cells by targeting microRNA-211-3p. 30840276_Overexpression of lncSNHG15 remarkably promoted the proliferation and migration of non-small cell lung cancer cells. lncSNHG15 could bind to miR-211-3p. 30945457_SNHG15 served as a ceRNA and reversed the activity of miR-338-3p, thus facilitating CRC tumorigenesis by enhancing the expression of FOS and RAB14. 30981837_Results show that SNHG15 is upregulated in prostate cancer (PCa) cells and promoted cell proliferation. Its downregulation inhibited PCa cell migration and invasion. SNHG15 was located in the cytoplasm of PCa cells and acted as a molecular sponge of miR-338 resulting in post-transcriptional regulated FKBP1A. These study suggested that SNHG15 functions as an oncogene by modulating miR-338/FKBP1A axis in prostate cancer. 31014355_Several of these genes are functionally related to AIF, a protein that we found to specifically interact with SNHG15, suggesting that the SNHG15 acts, at least in part, by regulating the activity of AIF. 31310393_LncRNA SNHG15 promotes hepatocellular carcinoma progression by sponging miR-141-3p. 31636472_Up-regulation of SNHG15 was found in hepatocellular carcinoma (HCC) and was related to aggressive behaviors in HCC patients. Knockdown of SNHG15 restrained HCC cell proliferation, migration and invasion. SNHG15 served as a molecular sponge for miR-490-3p which directly targets HDAC2. HDAC2 was involved in HCC progression by interacting with the SNHG15/miR-490-3p axis. 31696491_Elevated expression of lncRNA SNHG15 in spinal tuberculosis: preliminary results. 31774607_LncRNA SNHG15 may serve as a prospective and novel biomarker for molecular diagnosis and therapeutics in patients with cancer. 32141559_Up-regulation of SNHG15 facilitates cell proliferation, migration, invasion and suppresses cell apoptosis in breast cancer by regulating miR-411-5p/VASP axis. 32247266_Long non-coding RNA SNHG15 is a competing endogenous RNA of miR-141-3p that prevents osteoarthritis progression by upregulating BCL2L13 expression. 32633324_High lncSNHG15 expression may predict poor cancer prognosis: a meta-analysis based on the PRISMA and the bio-informatics analysis. 32633365_LncRNA SNHG15 promotes the proliferation of nasopharyngeal carcinoma via sponging miR-141-3p to upregulate KLF9. 32643707_Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 15 (SNHG15) Alleviates Osteoarthritis Progression by Regulation of Extracellular Matrix Homeostasis. 32655137_LncRNA SNHG15 regulates EGFR-TKI acquired resistance in lung adenocarcinoma through sponging miR-451 to upregulate MDR-1. 33372376_Long noncoding RNA small nucleolar RNA host gene 15 deteriorates liver cancer via microRNA-18b-5p/LIM-only 4 axis. 33572758_The Expression of the Cancer-Associated lncRNA Snhg15 Is Modulated by EphrinA5-Induced Signaling. 33899079_LncRNA SNHG15 modulates gastric cancer tumorigenesis by impairing miR-506-5p expression. 34016097_Down-regulation LncRNA-SNHG15 contributes to proliferation and invasion of bladder cancer cells. 34551140_Increased serum exosomal long non-coding RNA SNHG15 expression predicts poor prognosis in non-small cell lung cancer. 34734088_lncRNA SNHG15 Promotes Ovarian Cancer Progression through Regulated CDK6 via Sponging miR-370-3p. 34868528_LncRNA SNHG15 Promotes Oxidative Stress Damage to Regulate the Occurrence and Development of Cerebral Ischemia/Reperfusion Injury by Targeting the miR-141/SIRT1 Axis. | 4.168580e+03 | 0.9921451 | -0.011377018 | 0.2874050 | 1.585921e-03 | 0.9682337431 | 0.99408977 | No | Yes | 3.589015e+03 | 317.959128 | 3.376226e+03 | 306.261068 | ||||||||
| ENSG00000234684 | 100507495 | SDCBP2-AS1 | lncRNA | 34218800_Long non-coding RNA SDCBP2-AS1 delays the progression of ovarian cancer via microRNA-100-5p-targeted EPDR1. | 1.043823e+02 | 1.0444786 | 0.062782940 | 0.3612916 | 3.051335e-02 | 0.8613305288 | 0.97337236 | No | Yes | 1.156852e+02 | 18.259563 | 1.101602e+02 | 17.640355 | |||||||||
| ENSG00000234912 | 654434 | SNHG20 | lncRNA | 27543107_Results show that LncRNA SNHG20 is up-regulated in human colorectal cancer (CRC) tissues and cell lines, correlates with poor prognosis and provide evidence that it participates in CRC progression. 28282787_These results showed that the SNHG20/EZH2/E-cadhein regulator pathway might contribute to the development of novel therapeutic strategies for hepatocellular carcinoma patients. 28981099_rescue experiments indicated that SNHG20 functioned as an oncogene partly via repressing p21 in non-small cell lung cancer (NSCLC) cells. Taken together, our findings demonstrate that SNHG20 is a new candidate for use in NSCLC diagnosis, prognosis and therapy. 29101241_our results also revealed that lncRNA SNHG20 knockdown inhibited Wnt/b catenin signaling activity by suppressing beta-catenin expression and reversing the downstream target gene expression. Taken together, lncRNA SNHG20 plays an pivotal role in ovarian cancer progression by regulating Wnt/b-catenin signaling 29604594_SNHG20 could function as an oncogenic long non-coding RNA by regulating miR-140-5p-ADAM10 axis and MEK/ERK signaling pathway in cervical cancer. 30072099_The lncRNA SNHG20/miR-139/RUNX2 axis modulates the osteosarcoma tumorigenesis and apoptosis via mitochondrial apoptosis pathway, providing a novel insight for the pathophysiological process. 30120876_High SNHG20 expression is associated with enhanced cell migration and invasion in osteosarcoma. 30394668_The important function of SNHG20/miR-197/LIN28 axis in the oncogenesis. 30520073_LncRNA SNHG20 contributes to cell proliferation and invasion by upregulating ZFX expression sponging miR-495-3p in gastric cancer 30657567_Expression level of SNHG20 mRNA in glioma tissues was significantly higher than that in para-carcinoma tissues. After SNHG20 knockout, the protein expressions in the PTEN/PI3K/AKT signaling pathway were inhibited. SNHG20 siRNA transfection significantly inhibited proliferation of U118 and U251 cells with expression of Bax up-regulation and that of Bcl-2 down-regulation. 30690477_HBx promoted the proliferation of hepatocellular carcinoma (HCC) cell and reduced the apoptosis of HCC cells through the SNHG20/PTEN signalling pathway 30780105_Knockdown of SNHG20 suppresses cell proliferation, migration and invasion in non-small cell lung cancer tissue. 30846486_These results suggest that SNHG20 may serve as an independent prognostic predictor and function as a noncoding oncogene in epithelial ovarian cancer progression, which might be a possible novel diagnostic marker and treatment target. 31081112_LncRNA SNHG20 promotes the development of laryngeal squamous cell carcinoma by regulating miR-140. 31298384_LncRNA SNHG20 promoted the proliferation of glioma cells via sponging miR-4486 to regulate the MDM2-p53 pathway. 31408278_SNHG20 expression was decreased in the nonalcoholic fatty liver disease but increased in nonalcoholic fatty liver disease-hepatocellular carcinoma livers.SNHG20 may facilitate the progression of nonalcoholic fatty liver disease to the hepatocellular carcinoma via inducing liver Kupffer cells M2 polarization via STAT6 activation. 31957836_Effects of lncRNA SNHG20 on proliferation and apoptosis of non-small cell lung cancer cells through Wnt/beta-catenin signaling pathway. 32118721_Up-regulated SNHG20 predicts unfavorable prognosis for multiple kinds of cancers. [meta-analysis] 32456448_Knockdown SNHG20 Suppresses Nonsmall Cell Lung Cancer Development by Repressing Proliferation, Migration and Invasion, and Inducing Apoptosis by Regulating miR-2467-3p/E2F3. 32495922_LncRNA SNHG20 enhances the progression of oral squamous cell carcinoma by regulating the miR-29a/DIXDC1/Wnt regulatory axis. 33089952_LncRNA SNHG20 promoted proliferation, invasion and inhibited cell apoptosis of lung adenocarcinoma via sponging miR-342 and upregulating DDX49. 33179110_Long noncoding RNA SNHG20 promotes colorectal cancer cell proliferation, migration and invasion via miR495/STAT3 axis. 34080033_Long noncoding RNA SNHG20 promotes cell proliferation, migration and invasion in retinoblastoma via the miR3355p/E2F3 axis. 34282711_Long noncoding RNA SNHG20 regulates cell migration, invasion, and proliferation via the microRNA-19b-3p/RAB14 axis in oral squamous cell carcinoma. 34302635_LncRNA SNHG20 promotes cell proliferation and invasion by suppressing miR-217 in ovarian cancer. 34836544_LncRNA SNHG20 promotes migration and invasion of ovarian cancer via modulating the microRNA-148a/ROCK1 axis. | 1.463249e+03 | 0.8961844 | -0.158132537 | 0.2639133 | 3.588547e-01 | 0.5491429905 | 0.89250744 | No | Yes | 1.069567e+03 | 83.799283 | 1.252541e+03 | 100.496327 | |||||||||
| ENSG00000235034 | 342918 | C19orf81 | protein_coding | C9J6K1 | Reference proteome | hsa:342918; | ENSMUSG00000008028 | 1700008O03Rik | 3.370872e+02 | 1.2875638 | 0.364643880 | 0.3554918 | 1.046576e+00 | 0.3062970624 | 0.81492114 | No | Yes | 3.551829e+02 | 57.403350 | 2.481511e+02 | 41.783839 | |||||
| ENSG00000236088 | 100874058 | COX10-DT | lncRNA | 34323174_Regulating COX10-AS1 / miR-142-5p / PAICS axis inhibits the proliferation of non-small cell lung cancer. | 3.157823e+02 | 0.6965021 | -0.521800451 | 0.3151367 | 2.673312e+00 | 0.1020434973 | 0.75783482 | No | Yes | 2.092947e+02 | 35.422219 | 2.980137e+02 | 51.252588 | |||||||||
| ENSG00000237296 | 641298 | SMG1P1 | transcribed_unprocessed_pseudogene | 3.880428e+02 | 1.1415997 | 0.191056859 | 0.3222921 | 3.408647e-01 | 0.5593305382 | 0.89480137 | No | Yes | 3.325551e+02 | 68.780133 | 3.322043e+02 | 70.500962 | ||||||||||
| ENSG00000237441 | 5863 | RGL2 | protein_coding | O15211 | FUNCTION: Probable guanine nucleotide exchange factor. Putative effector of Ras and/or Rap. Associates with the GTP-bound form of Rap 1A and H-Ras in vitro (By similarity). {ECO:0000250}. | Alternative splicing;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome | hsa:5863; | cytosol [GO:0005829]; guanyl-nucleotide exchange factor activity [GO:0005085]; negative regulation of cardiac muscle cell apoptotic process [GO:0010667]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; Ras protein signal transduction [GO:0007265]; regulation of Ral protein signal transduction [GO:0032485] | 18540861_RGL2 is phosphorylated by PKA and phosphorylation reduces the ability of RGL2 to bind H-Ras. results of the present study indicate that Ras may distinguish between the different RalGDS family members by their phosphorylation by PKA. 19851445_Observational study of gene-disease association. (HuGE Navigator) 20801877_Rgl2 and RalB both localized to the leading edge, and this localization of RalB was dependent on endogenous Rgl2 expression. 21160498_SAMD9, an IFN-gamma-responsive protein, interacts with RGL2 to diminish the expression of EGR1, a protein of direct relevance to the pathogenesis of ectopic calcification and inflammation. | ENSMUSG00000041354 | Rgl2 | 9.295393e+02 | 1.2889653 | 0.366213416 | 0.2750532 | 1.810200e+00 | 0.1784842731 | 0.77663851 | No | Yes | 9.837666e+02 | 104.413616 | 7.476297e+02 | 82.196484 | ||
| ENSG00000238105 | 55592 | GOLGA2P5 | transcribed_unprocessed_pseudogene | 2.608440e+02 | 0.5271284 | -0.923773684 | 0.3107212 | 8.409151e+00 | 0.0037333704 | 0.22100826 | No | Yes | 1.161717e+02 | 25.946188 | 2.678403e+02 | 60.550214 | ||||||||||
| ENSG00000239467 | 285141 | lncRNA | 4.477066e+01 | 0.7962260 | -0.328750163 | 0.4813721 | 4.530594e-01 | 0.5008856660 | 0.87604856 | No | Yes | 3.492542e+01 | 8.206456 | 4.595635e+01 | 10.831480 | |||||||||||
| ENSG00000239523 | 100506826 | MYLK-AS1 | lncRNA | 33168027_LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. 34181498_LncRNA MYLK-AS1 acts as an oncogene by epigenetically silencing large tumor suppressor 2 (LATS2) in gastric cancer. | 1.028544e+02 | 0.9225440 | -0.116310441 | 0.4000899 | 8.597911e-02 | 0.7693525077 | 0.95414161 | No | Yes | 9.288591e+01 | 13.949144 | 1.028835e+02 | 15.752117 | |||||||||
| ENSG00000239791 | lncRNA | 7.195354e+01 | 0.8238200 | -0.279598968 | 0.4070753 | 4.614438e-01 | 0.4969499979 | 0.87548575 | No | Yes | 7.269946e+01 | 18.108595 | 9.094794e+01 | 22.949222 | ||||||||||||
| ENSG00000239900 | 158 | ADSL | protein_coding | P30566 | FUNCTION: Catalyzes two non-sequential steps in de novo AMP synthesis: converts (S)-2-(5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinate (SAICAR) to fumarate plus 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide, and thereby also contributes to de novo IMP synthesis, and converts succinyladenosine monophosphate (SAMP) to AMP and fumarate. {ECO:0000269|PubMed:10888601}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Epilepsy;Isopeptide bond;Lyase;Purine biosynthesis;Reference proteome;Ubl conjugation | PATHWAY: Purine metabolism; AMP biosynthesis via de novo pathway; AMP from IMP: step 2/2.; PATHWAY: Purine metabolism; IMP biosynthesis via de novo pathway; 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide from 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate: step 2/2. | The protein encoded by this gene belongs to the lyase 1 family. It is an essential enzyme involved in purine metabolism, and catalyzes two non-sequential reactions in the de novo purine biosynthetic pathway: the conversion of succinylaminoimidazole carboxamide ribotide (SAICAR) to aminoimidazole carboxamide ribotide (AICAR) and the conversion of adenylosuccinate (S-AMP) to adenosine monophosphate (AMP). Mutations in this gene are associated with adenylosuccinase deficiency (ADSLD), a disorder marked with psychomotor retardation, epilepsy or autistic features. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Dec 2015]. | hsa:158; | cytosol [GO:0005829]; protein-containing complex [GO:0032991]; (S)-2-(5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinate AMP-lyase (fumarate-forming) activity [GO:0070626]; identical protein binding [GO:0042802]; N6-(1,2-dicarboxyethyl)AMP AMP-lyase (fumarate-forming) activity [GO:0004018]; 'de novo' AMP biosynthetic process [GO:0044208]; 'de novo' IMP biosynthetic process [GO:0006189]; 'de novo' XMP biosynthetic process [GO:0097294]; aerobic respiration [GO:0009060]; AMP biosynthetic process [GO:0006167]; AMP salvage [GO:0044209]; GMP biosynthetic process [GO:0006177]; purine nucleotide biosynthetic process [GO:0006164]; response to hypoxia [GO:0001666]; response to muscle activity [GO:0014850]; response to nutrient [GO:0007584]; response to starvation [GO:0042594] | 12016589_Mutation of a nuclear respiratory factor 2 binding site in the 5' untranslated region of the ADSL gene in three patients with adenylosuccinate lyase deficiency. 12590570_Mutations at position 276 result in structurally impaired adenylosuccinate lyases which are assembled into the defective tetramers associated with the mild variant of ADSL deficiency in humans. 12833398_Variable expression of ADSL deficiency is reported in three patients belonging to a family which originates from Portugal. 15471876_a mutation in adenylosuccinate lyase may be associated with autism 15571235_case report of adenylosuccinate lyase deficiency shows a mutation in ASDL 16973378_cloning, expression and purification of catalytically active human adenylosuccinate lyase 17188615_ADSL deficiency may present with prenatal growth restriction, fetal and neonatal hypokinesia, and rapidly fatal neonatal encephalopathy. 18524658_Analysis of the ADSL gene showed a R426H mutation in four unrelated patients with metabolic diseases. 18649008_D-ribose administration in Polish patients with adenylosuccinate lyase deficiency was accompanied by neither reduction in seizure frequency nor growth enhancement. 19405474_Biochemical and biophysical analysis of five disease-associated human adenylosuccinate lyase mutants. 20177786_Case Report: Malaysian patient compound heterozygous for two novel ADSL mutations giving rise to adenylosuccinate lyase deficiency. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20933180_the cases of the only three children diagnosed to date in the United Kingdom with adenylosuccinate lyase deficiency 22180458_Results proved in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency that various mutations of ADSL destabilize to various degrees purinosome assembly and found that the ability to form purinosomes correlates with clinical phenotypes of individual ADSL patients. 22812634_structural and biochemical characterization data of WT and mutant R303C ADSL by enzyme kinetics, product binding by isothermal titration calorimetry and X-ray crystallography to reveal the effects of the R303C mutation that results in a nonparallel reduction in enzyme activity 23714113_Missense mutations in the adenylosuccinate lyase is associated with Adenylosuccinate lyase deficiency, an inborn error of purine metabolism characterized by neurological and physiological symptoms. 29467457_involved in endometrial cancer aggressiveness by regulating expression of killer cell lectin-like receptor C3 30573755_ADSL undergoes conformational changes during catalysis which, together with the crystal structure of a hitherto undetermined product bound conformation, explains the molecular origin of disease for several modern human ADSL mutants. 31729379_Adenylosuccinate lyase (ADSL) is an EglN2 hydroxylase substrate in triple negative breast cancer. | ENSMUSG00000022407 | Adsl | 5.366537e+03 | 0.4411079 | -1.180796510 | 0.3470579 | 1.139890e+01 | 0.0007348777 | 0.09944063 | No | Yes | 2.860712e+03 | 694.143234 | 6.665450e+03 | 1658.176338 |
| ENSG00000240204 | 100287482 | SMKR1 | protein_coding | H3BMG3 | Reference proteome | hsa:100287482; | 3.740355e+01 | 0.7409014 | -0.432646481 | 0.5100897 | 7.272556e-01 | 0.3937742132 | 0.84317192 | No | Yes | 3.857199e+01 | 11.111573 | 5.522352e+01 | 16.430935 | |||||||
| ENSG00000240344 | 53938 | PPIL3 | protein_coding | Q9H2H8 | FUNCTION: PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. May be involved in pre-mRNA splicing. | 3D-structure;Acetylation;Alternative splicing;Isomerase;Methylation;Reference proteome;Rotamase;Spliceosome;mRNA processing;mRNA splicing | This gene encodes a member of the cyclophilin family. Cyclophilins catalyze the cis-trans isomerization of peptidylprolyl imide bonds in oligopeptides. They have been proposed to act either as catalysts or as molecular chaperones in protein-folding events. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2008]. | hsa:53938; | catalytic step 2 spliceosome [GO:0071013]; peptidyl-prolyl cis-trans isomerase activity [GO:0003755]; mRNA splicing, via spliceosome [GO:0000398]; protein folding [GO:0006457]; protein peptidyl-prolyl isomerization [GO:0000413] | 15989758_The novel full-length gene of human PPIL3 may be correlated with the formation of human glioma. 16510998_Human cyclophilin J, a new member of the cyclophilin family, has been expressed and crystallized. 18474220_Human peptidyl-prolyl isomerase-like 3 (Ppil3) is one of the Apoptin-associated proteins. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 26020957_Suppression of CYPJ could repress the growth of HCC, which makes CYPJ a potential target for the development of new strategies to treat this malignancy. 26977013_CYPJ structurally resembles CYPA. It is sensitive to inhibition by CsA and plays a role in regulating cell growth, proliferation, and apoptosis. 28739742_Cyclophilin J expression was up-regulated in gastric carcinoma compared to normal gastric tissues. | ENSMUSG00000026035 | Ppil3 | 1.937443e+02 | 0.9955443 | -0.006442625 | 0.3381326 | 3.728063e-04 | 0.9845952482 | 0.99716006 | No | Yes | 2.064741e+02 | 33.953193 | 2.408180e+02 | 40.240503 | |
| ENSG00000240583 | 358 | AQP1 | protein_coding | P29972 | FUNCTION: Forms a water-specific channel that provides the plasma membranes of red cells and kidney proximal tubules with high permeability to water, thereby permitting water to move in the direction of an osmotic gradient. {ECO:0000269|PubMed:1373524}. | 3D-structure;Alternative splicing;Blood group antigen;Cell membrane;Direct protein sequencing;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene encodes a small integral membrane protein with six bilayer spanning domains that functions as a water channel protein. This protein permits passive transport of water along an osmotic gradient. This gene is a possible candidate for disorders involving imbalance in ocular fluid movement. [provided by RefSeq, Aug 2016]. | hsa:358; | apical part of cell [GO:0045177]; apical plasma membrane [GO:0016324]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; brush border [GO:0005903]; brush border membrane [GO:0031526]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; nuclear membrane [GO:0031965]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; sarcolemma [GO:0042383]; ammonium transmembrane transporter activity [GO:0008519]; carbon dioxide transmembrane transporter activity [GO:0035379]; glycerol transmembrane transporter activity [GO:0015168]; identical protein binding [GO:0042802]; intracellular cGMP-activated cation channel activity [GO:0005223]; nitric oxide transmembrane transporter activity [GO:0030184]; potassium channel activity [GO:0005267]; potassium ion transmembrane transporter activity [GO:0015079]; transmembrane transporter activity [GO:0022857]; water channel activity [GO:0015250]; water transmembrane transporter activity [GO:0005372]; ammonium transport [GO:0015696]; carbon dioxide transmembrane transport [GO:0035378]; carbon dioxide transport [GO:0015670]; cell volume homeostasis [GO:0006884]; cellular homeostasis [GO:0019725]; cellular hyperosmotic response [GO:0071474]; cellular response to cAMP [GO:0071320]; cellular response to copper ion [GO:0071280]; cellular response to dexamethasone stimulus [GO:0071549]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to hypoxia [GO:0071456]; cellular response to inorganic substance [GO:0071241]; cellular response to mechanical stimulus [GO:0071260]; cellular response to mercury ion [GO:0071288]; cellular response to nitric oxide [GO:0071732]; cellular response to retinoic acid [GO:0071300]; cellular response to salt stress [GO:0071472]; cellular response to UV [GO:0034644]; cellular water homeostasis [GO:0009992]; cerebrospinal fluid secretion [GO:0033326]; cGMP-mediated signaling [GO:0019934]; defense response to Gram-negative bacterium [GO:0050829]; establishment or maintenance of actin cytoskeleton polarity [GO:0030950]; glycerol transport [GO:0015793]; hyperosmotic response [GO:0006972]; lateral ventricle development [GO:0021670]; multicellular organismal water homeostasis [GO:0050891]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; nitric oxide transport [GO:0030185]; odontogenesis [GO:0042476]; pancreatic juice secretion [GO:0030157]; positive regulation of angiogenesis [GO:0045766]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of saliva secretion [GO:0046878]; potassium ion transport [GO:0006813]; renal water homeostasis [GO:0003091]; renal water transport [GO:0003097]; transepithelial water transport [GO:0035377]; water transport [GO:0006833] | 11884383_transmembrane biogenesis is cotranslational in intact mammalian cells 11909995_A novel role for aquaporin-1 as a gated ion channel reshapes our current views of this ancient family of transmembrane channel proteins. 11922632_These data suggest that the transcription of the AQP1 by hypertonicity in renal cells is upregulated by the interaction with putative DNA binding proteins to a novel HRE located at -54 to -46 in the AQP1 gene. 12002613_This study showed the distinct localization of AQP1 in the mesangial cells of human glomeruli, suggesting its role in water movement through these cells. 12027013_visualization of the water-selective pathway at 3.7A resolution in a three-dimenional density map (REVIEW) 12237771_In astrocytomas, aquaporin 1 expressed in microvessel endothelia and neoplastic astrocytes. In metastatic carcinomas, aquaporin 1 present in microvessel endothelia and reactive astrocytes. Aquaporin 1 may participate in formation of brain tumour oedema. 12399631_Data suggest that variations found in plasma osmolarity during hemodialysis may induce aquaporin 1 expression on the membrane of intact red blood cells. 12498798_Data present a detailed comparison between the cryo-electron microscopy and X-ray crystallography model structures of the human and bovine water channel aquaporin-1 (AQP1). 12781664_Transmembrane helices at the periphery of the hAQP1 tetramer exhibited smaller extraction forces than helices at the interface between hAQP1 monomers. 14514735_substantial and striking upregulation of AQP-1 in the glomeruli of most diseased kidneys. 14592814_presence of AQP-1 in endothelia and water-transporting epithelia and new locations: mammary epithelium, articular chondrocytes, synoviocytes, and synovial microvessels where it may be involved in milk, chondrocyte volume, synovial fluid, and homeostasis. 14701836_lack of significant cyclic guanine nucleotide ion channel activity rules out a secondary role of aquaporin 1 water channels in cellular signal transduction 14753493_Increased expression. Over-expression occurred on astrocytic processes. Induction of AQP1 and AQP4 on reactive astrocytes in subarachnoid hemorrhage. May be involved in brain edema formation or resolution. 14753494_Intense upregulation of AQP1 expression was found in all glioblastomas. Expression of aquaporins in glioblastomas suggests pathologic role. Selective AQP inhibition might be new therapeutic option for tumor-associated cerebral edema. 15024704_AQP1 has a role in the movement of extracellular matrix and metabolic water across the membranes of chondrocytes and synoviocytes 15135660_AQP1 co-localizes with t-tubular and caveolar proteins 15502805_Aquaporin-1 is a reliable marker for clear cell renal cell carcinomas of lower grades but not for higher grades. 15563082_The expression of AQP-1 mRNA was positively correlated with Bcl-2 mRNA expression in nasal polyps. AQP-1 contributes to the survival of eosinophils in nasal polyps. 15667881_AQP-1 may be a possible critical reabsorption factor, acting to reduce abnormal fluid retention in endotubular cells and the extracellular matrix and, to a lesser extent, in Leydig cells. 15783300_Observational study of genotype prevalence. (HuGE Navigator) 15809704_increased AQP1 expression in some human adenocarcinomas may be a consequence of angiogenesis and important for the formation or clearance of tumor edema 15847654_Observational study of genetic testing. (HuGE Navigator) 16300893_AQP1 expression heavely in cystic hemangioblastomas. 16481371_Aquaporin (AQP) 1 is predominantly situated in the apical plasma membrane domain of the human choroid plexus, although distinct basolateral and endothelial immunoreactivity is also observed. 16515633_high AQP1 expression may play an important role in ovarian carcinogenesis, disease progression, and ascites formation. 16534779_AQPs are differentially expressed in the peripheral versus central nervous system and that channel-mediated water transport mechanisms may be involved in peripheral neuronal activity by regulating water homeostasis in nerve plexuses and bundles. 16574458_Review. In Colton (null) RBC ghosts, lack of AQP1 resulted in about 30% reduction of the alkalinization rates. 16698771_aquaporin-1-mediated CO(2) permeation is to be expected only in membranes with a low intrinsic CO(2) permeability 16814974_These results in human fetal brain lend morphological support to the previous findings that aquaporin-1 and aquaporin-4 play different roles in the regulation of the water homeostasis of the brain. 16871401_Significant increased expression levels of AQP1 and AQP4 were seen in Creutzfeldt-Jakob disease, but not in advanced Alzheimer's disease and diffuse Lewy body disease. 17012249_AQP1 is responsible for 60% of the high P(CO2) of red cells and that another, so far unidentified, CO2 pathway is present in this membrane that may account for at least 30% of total P(CO2). 17077939_In hemangioblastomas, expression of AQP1 was predominantly localized on membranes of stromal cells. The expression level of AQP1 in cystic group of hemangioblastomas is much higher than that of solid group. 17219999_Inhibiting AQP-1 with acetazolamide may significantly induce apoptosis of Hep-2 cells. 17273788_The deregulation of aquaporin-1 in menorrhagia may be involved in abnormal endometrial vascular growth and permeability. 17408468_Observational study of gene-disease association. (HuGE Navigator) 17409744_AQP1 is expressed at the endomysial capillary endothelial cell and further AQP1 may be expressed at the human skeletal myofiber plasma membrane. 17511167_AQP1 mRNA and protein expression level in laryngeal tumor tissues is remarkably stronger than that in normal tissues. 17545093_Data show that AQP-1, 3, 8, 9 mRNA expression was detected in both amnion and chorion and can be associated with intramembranous transport and volume regulation of amniotic fluid. 17549682_Only the presence of AQP1, but not AQP4, enhanced cell growth and migration, typical properties of gliomas, while AQP4 enhanced cell adhesion suggesting differential biological roles for AQP1 and AQP4 in glioma cell biology. 17632520_In AQP1, Asn49 and Lys51 interact with Asp185 at the C terminus of TM5 to form a polar, quaternary structural motif that influences multiple stages of folding. 17854859_AQP-1 is up-regulated in biliary dysplasia...and down-regulation of AQP-1 is associated with mucin production and aggressive progression of intrahepatic cholangiocarcinoma. 17890385_These results establish the nature and determinants of AQP1 diffusion in cell plasma membranes and demonstrate long-range nonanomalous diffusion of AQP1. 17894331_a detailed mechanism for ion exclusion in aquaporin-1 (AQP1) at an atomistic level is investigated by calculating the free energy for transport of ions in AQP1 17898873_Increased AQP1 expression may relieve intracellular acidosis and edema in highly glycolytic glioma cells. 18052958_Aquaporin 1 may be involved in the pathophysiology of migraine. 18067501_Observational study of genetic testing. (HuGE Navigator) 18080132_These data illustrate the potential of the peritoneal membrane as an experimental model in the investigation of the role of AQP1--REVIEW 18202181_For small solutes permeating through AQP1, a remarkable anticorrelation between permeability and solute hydrophobicity is observed, rendering AQP1 a selective filter for small polar solutes. 18247144_The aim of this study was to use immunohistochemsitry to investigate the expression of aquaporins 1, 2 and 3 within the human intervertebral disc. 18275976_AQP1 is involved in hypoxia-inducible angiogenesis in retinal vascular endothelial cells through a mechanism that is independent of the VEGF signaling pathway. 18280225_AQP1 is not expressed in human airway epithelial cells from the A549 adeoncarcinoma cell line. 18282122_The genomic, structural and functional aspects of AQP1 are briefly described. Its role in human tumors and, in particular, those of the kidney is discussed. Review. 18313673_AQP1 may be involved in the tumorigenesis and progression of endometrioid adenocarcinoma by promoting angiogenesis, and AQP1 level may be both a tumor indicator and a new therapeutic target. 18392839_Our study shows that AQP4 downregulation can occur in muscular dystrophies with either normal or disrupted expression of dystrophin-associated proteins, and that this might be associated with upregulation of AQP1. 18509662_These observations suggest the possible association of astrocytic AQP1 with Abeta deposition in Alzheimer disease brains. 18510579_Observational study of genetic testing. (HuGE Navigator) 18538351_Modulation of AQP1 expression by maternal hormones may regulate invasion and fetal-placental-amnion water homeostasis during gestation. 18544259_We show high expression of AQP1 water channels in intractable epilepsy and suggest two mechanisms to explain this finding. Increased AQP1 expression of astrocytes may be a cause or a consequence of IE. 18563339_AQP1 and HIF1 interact each other and regulate the oncogenesis of breast cancer 18575775_The objective of this study was to evaluate the AQP1 expression in endometrial blood vessels during normal cycle and after mifepristone treatment. 18841368_AQP1 expression in invasive breast carcinomas is associated with a basal-like phenotype and poor prognosis. 19080511_The expression level of AQP1 of patients with preeclampsia increases in placenta and peritoneum and decreases in embryolemma. 19253825_Hypoxia regulates the expression of AQP1 in vascular endothelial cells. 19306058_AQP1 expression is a new characteristic feature of a particularly aggressive subgroup of basal-like breast carcinomas. 19424603_AQP1-specific siRNA knockdown impaired water permeability of ARPE-19 cells. 19472194_These results provide evidence that NPA motifs are important for water permeation but not essential for the expression, intracellular processing and the basic structure of human aquaporin 1. 19522191_The expression of AQP1 in tumor cells and micro-angiogenesis of primary laryngeal carcinoma are higher than normal. 19545896_Alteration of aquaporin 1 and aquaporin 3 expression in fetal membranes and placenta may be important in the pathophysiology of isolated oligohydramnios. 19619954_AQP-1 was overexpressed in hepatitis B virus associated cirrhotic liver tissues. AQP-1, similar to CK19, might be a more specific and more sensitive marker than CK7 for the identification of HPCs. 19670620_The expression of AQP1 and VEGF in laryngeal carcinoma was significantly higher than that in vocal cord polyps and normal controls. 19726340_The expression of AQP1 mRNA was significantly lower in oligohydramnios placenta than in normal pregnancy placenta at term. 19772916_TXA(2) receptor mediates water influx through aquaporins in astrocytoma cells via TXA(2) receptor-mediated activation of G alpha(12/13), Rho A, Rho kinase and Na(+)/H(+)-exchanger. 19787701_AQP1 activity of cell membrane affects HT20 colon cancer cell migration. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20063900_An increase in the level of water transport in response to changing osmotic conditions in the cellular environment may be due to a protein kinase C-dependent increase in AQP1 membrane localization. 20101282_Observational study of gene-disease association. (HuGE Navigator) 20101282_There was no association between common sequence variants in the AQP1 or SLC4A10 genes and primary open-angle glaucoma in the Caucasian population. 20137115_Inhibiting AQP1 expression with siRNA can inhibit the proliferation and induce apoptosis of K562 cells. 20149606_Potential role for synovial AQP1 and other aquaporins in joint swelling and vasogenic edema. 20360993_Observational study of gene-disease association. (HuGE Navigator) 20409716_Aquaporin 1 is localized in the dura mater following chronic subdural hematoma; the outer membrane might be the source of increased fluid accumulation responsible for chronic hematoma enlargement. 20424473_Observational study of gene-disease association. (HuGE Navigator) 20431033_Observational study of genetic testing. (HuGE Navigator) 20461409_Expression of AQP1, AQP4, or AQP6 mRNA did not differ in vestibular endorgans from patients with Meniere's disease. 20578142_AQP-1 enhances osmotic water permeability and FGF-induced dynamic membrane blebbing in liver endothelial cell and thereby drives invasion and pathological angiogenesis during cirrhosis. 20628061_Data identify miR-320a as a potential modulator of aquaporin 1 and 4 and explore the possibility of using miR-320a to alter the expression of aquaporin 1 and 4 in normal and ischemic conditions. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20739606_Compared with human AQP1, zebrafish Aqp1a has about twice the selectivity for CO(2) over NH(3). 20795314_The physiology and molecular basis of the Colton blood group antigen AQP1 is discussed. Review. 20806077_The purpose of this study was to determine the effects of down-regulation of Aquaporin 1 (AQP1) and Aquaporin 5 (AQP5) on cell proliferation and migration in human corneal endothelial (HCEC) and human corneal epithelial (CEPI17) cell lines, respectively. 20828513_The expression level of aquaporin-1 in nasopharyngeal cancer tissue could be involved in tumour migration. 20965731_Expression of AQP1 in pediatric brain tumors. Increased in ependymomas in posterior fossa and in pilocytic astrocytomas. Not expressed in medulloblastoma, primitive neuroectodermal tumor, germinoma. 20969805_This review outlines newly emerging evidence indicating that AQP-1 plays an important role in pain signal transduction and migraine. 21063116_After sequencing analysis of the coding regions and exon-intron boundaries one single variation, no significant mutation in AQP 1 was found. 21063116_Observational study of gene-disease association. (HuGE Navigator) 21107133_No difference is found between aquaporin 1 expression in Alzheimer's disease brain and its expression in cerebral amyloid angiopathy. 21237499_Expression patterns of aquaporin 1, 3, and 5 in lung cancer cells are mostly associated with cellular differentiation. 21244858_AQP1 and AQP5 might play an important role in the development of lung edema and lung injury 21252246_The late rise of AQP1 suggests a role in corpus luteum formation. 21271497_The researchers found an association between carriers of the AQP1 single nucleotide polymorphism and greater fluid loss in long-distance running. 21360438_An important role of AQP1 in tumor angiogenesis is sustained by the abundant expression of this protein in the endothelia of tumor capillaries. 21373963_Water flux through human aquaporin 1 was studied under osmotic challenges and the inhibition by intracellular furosemide. 21395179_AQP1 and VEGF had a higher expression in nasopharyngeal carcinoma tissues than in non-tumor tissues. 21538271_AQP-1 expression is increased in colorectal carcinoma while the expression of AQP-3 is not. There is no correlation between the expression of AQP-1 and AQP-3 in CRC. 21551254_AQP1 is a promising oncogene candidate for ACC and is transcriptionally regulated by promoter hypomethylation. 21612401_hypoxia-induced expression of AQP1 requires transcriptional activation, and the HIF-1 binding site of the 5'-promoter is necessary for transcriptional activation 21760919_Negative stain transmission electron microscopy and single particle analysis of KCC4 and the aquaporin-1 AQP1 water channel, revealed the expected quaternary structures within homogeneous preparations, and thus correct protein folding and assembly. 21784068_REVIEW: summarizes literature data concerning the involvement of AQP-1 and -4 in human brain tumor growth and edema formation 21793635_our results suggest, for the first time, that the rs1049305 (C/G, UTR3) AQP1 polymorphism could be involved in the genetic susceptibility to develop water retention in patients with liver cirrhosis. 21839760_This study demonistrated that AQP1 in inner ear plays an important role in the development of motion sickmness, and might be a potential target for the prevention or management of motion sickness. 21896312_The mean concentration of AQP1 in CSF was significantly elevated in patients with BM (BM: 3.8+/-3.4ng/ml, controls 22006723_Phosphorylation of tyrosine Tyr253 in the carboxyl terminal domain, acts as a master switch regulating responsiveness of AQP1 ion channels to cGMP, and the tetrameric central pore is the ion permeation pathway. 22093331_Aberrant expressions of AQP1 in periportal sinusoidal regions in human cirrhotic liver indicate the proliferation of arterial capillaries directly connected to the sinusoids, contributing to microvascular resistance in cirrhosis. 22269467_AQP1 is normally expressed in the temporomandibular joint (TMJ) disc with a role in the maintenance of TMJ homeostasis 22310126_regulates microvessel permeability and barrier function and contributes to blood vessel formation 22334691_novel pathway in mammalian cells whereby a hypotonic stimulus directly induces intracellular calcium elevations through transient receptor potential channels, which trigger AQP1 translocation 22348807_A new AQP1 null allele identified in a Gypsy woman who developed an anti-CO3 during her first pregnancy. 22372348_We found AQP1 and AQP4 in lung cancer cell extravasation and spread, which may provide a functional explanation for the expression of AQP1 and AQP4 in lung cancer tissues and lung cancer cell lines. 22472942_expression of aquaporin 1 and 5 was higher in uterine leiomyomata than in unaffected uteri 22901156_AQP1 expression significantly increased in advanced stage of cervical cancer, deeper infiltration, metastatic lymph nodes and larger tumor volume. 22901921_Exosomal AQP1 is downregulated after the release of ureteropelvic junction obstruction. This may be due to locally increased TGF-beta-1 in the postobstructed kidney. 22964306_Data indicate reconstitution of AQP-1 into cholesterol-containing vesicles leads to drastic increases in P(CO2). 23029502_Selective expression of AQP1 in the trigeminal neurons innervating the oral mucosa indicates an involvement of AQP1 in oral sensory transduction. 23219802_Stomatin interacts with GLUT1/SLC2A1, band 3/SLC4A1, and aquaporin-1 in human erythrocyte membrane domains 23220481_This study demonstrated that laminar shear stress stimulates the endothelial expression of AQP1 that plays a role in wound healing. 23268390_K562 cells show a significant increase of AQP1 expression after retinoic acid-induced erythroid differentiation. 23276695_AQP1(+) cells immediately beneath the epithelial basement membrane may be stromal niche-like cells that directly interact with N-cad(+) limbal basal epithelial progenitor cells. 23313295_High AQP1 expression is associated with malignant pleural mesothelioma. 23317544_These findings suggest that NKCC1 and AQP1 participate in meningioma biology and invasion 23332061_hAQP1 is a constitutively open channel that closes mediated by membrane-tension increments. 23361277_AQP1-immunoexpression had a good correlation with high-grade tumors. 23393355_AQP1 expression correlates with the grade of malignancy in astrocytoma and is associated with angiogenesis, as well as with invasion of grade IV tumour in areas of tumour infiltration 23450058_Estrogen induces AQP1 expression by activating ERE in the promoter of the Aqp1 gene, resulting in tubulogenesis of vascular endothelial cells. 23549977_AQP-1 appears to be involved in the arterial capillary proliferation in the cirrhotic liver. [Review] 23928039_AQP1 and 4 gene expression levels did not differ between mesial temporal sclerosis patients and control groups 23949237_Aquaporin-1 is associated with arterial capillary proliferation and hepatic sinusoidal transformation contributing to portal hypertension in primary biliary cirrhosis. 23974882_The data suggest that the increased expression of AQP1 and AQP3 in pterygial tissues may be involved in the pathogenesis of pterygia 24014128_Cox-regression analyses revealed the AQP1 -783G/C genotype status as an independent prognostic-factor when jointly considering other predictors of survivalin glioblastoma multiforme. 24028651_Aquaporin-1 is induced in leukocytes of patients with sepsis and exhibits higher expression in septic shock. 24086369_Anti-AQP1 autoantibodies are present in a subgroup of patients with chronic demyelination in the central nervous sytem and similarities with anti-AQP4-seronegative neuromyelitis optica spectrum disorders. 24169407_Over expression of aquaporin 1 causes increased intracranial pressure, and downregulation reduces pressure and alleviates the symptomatology and complications of idiopathic intracranial hypertension. 24252214_Report is the first to establish astrocytic water channel loss in a subset of human central pontine myelinolysis (CPM) cases and suggests AQP1 and AQP4 may be involved in the pathogenesis of CPM 24333057_the prevalence of this AQP1 c.601delG allele in the ethnic minority of Romani 24493792_AQP1 promoter hypomethylation is common in ACC, and AQP1 tends to be overexpressed in these tumors. Increased AQP1 methylation is associated with improved prognosis. 24688444_the expression of AQP1, at both the mRNA and protein levels, in membranes from PVR and ERM. 24777974_Results show that in AQP1, helix 3 inverts its orientation in the membrane after the initial insertion, whereas this does not occur in the homologous AQP4. 24803718_Freshly obtained urine samples from renal cell carcinoma patients are positive for AQP1 whereas archived samples and fresh healthy control samples are negative. 24918928_Aquaporin 1 and 3 were upregulated in cervical cancer compared to mild cervicitis and cervical intraepithelial neoplasia 2-3 (P<0.05). 25252847_AQP1 over-expression effectively inhibited cell proliferation and induced cell growth arrest in G1 phase of K562 cells. 25299207_the reduction of aquaporins 1 and 3 may be a factor resulting in lumbar intervertebral disc degeneration. 25495276_The AQP1 rs1049305 single nucleotide polymorphism is associated with running performance, but not relative body weight change, during South African Ironman Triathlons. 25582271_Through molecular genotyping we also identified polymorphisms in RhCE, Kell, Duffy, Colton, Lutheran and Scianna loci in donors and patients. 25609088_CAII increases water conductance through AQP1 by a physical interaction between the two proteins. 25721378_Study found a significant correlation between AQP1, AQP3, and AQP5 overexpression and lymph node metastasis in patients with surgically resected colon cancer. 25987071_High AQP1 expression is associated with recurrence in bladder uroepithelial cell carcinoma . 26074259_AQP1 was found to be a marker of a subgroup of aggressive basaloid-like squamous cell carcinomas 26115524_the benign subependymomas aquaporins 1 and 4 are dramatically redistributed and upregulated. 26151179_The results suggest that AQP1 may facilitate lung cancer cell proliferation and migration in an MMP-2 and-9-dependent manner 26176849_AQP1 knockdown can effectively inhibit cell proliferation, adhesion, invasion and tumorigenesis by targeting TGF-beta signaling pathway and focal adhesion genes. 26178576_PBEF regulates the expression of inflammatory factors and AQP1 through the MAPK pathways. 26181025_Results demonstrate the clinical utility, specificity, and sensitivity of urine AQP1 and PLIN2 to diagnose renal cell carcinoma. 26261083_The study demonstrated the abnormal expression pattern of AQP1, AQP2, AQP3, and AQP4 in the kidney tissues of patients with nephrotic syndrome, providing a basis for an improved understanding of the role of aquaporins in the pathogenesis of this disease. 26549133_the downregulation of AQP1 impedes extracellular to intracellular fluid transportation. 26707370_No significant relationship was found between gallbladder mucosa aquaporin expression and the presence or absence of gallbladder stones. 26717516_These data provide support for a continuous role of KLF2 in stabilizing the vessel wall via co-temporal expression of eNOS and AQP1 both preceding and during the pathogenesis of atherosclerosis. 26773069_The role of AQP1 in malignant pleural mesothelioma. 26786101_Based on a comparative mutagenic analysis of AQP1, AQP 3, and AQP 4, the results suggest that loop D interactions may provide a general structural framework for tetrameric assembly within the AQP family. 26812884_High cytoplasmic expression of AQP1 is associated with breast cancer progression. 26823734_Suggest that sputum AQP1 and AQP5 could be used as diagnostic markers in mild-to-moderate adult-onset asthma. 26838488_AQP1 was overexpressed in cystic cholangiocytes of patients with polycystic liver disease. A tendency of increased AQP1 protein expression in correlation with the cyst size was also found. 26848795_The BRAF V600 mutations were significantly associated with AQP1 expression (P=0.014). Long-term follow-up indicated a reduced progression-free survival (P=0.036) and overall survival (P=0.017) for the AQP1-positive cutaneous melanoma patients. 26919570_Findings suggest that a defined population of AQP4- and AQP1-expressing reactive astrocytes may modify alpha-syn deposition in the neocortex of patients with Parkinson's Disease. 26923194_Study provides evidence that Mef2c cooperated with Sp1 to activate human AQP1 transcription by binding to its proximal promoter in human umbilical cord vein endothelial cells indicating that AQP1 is a direct target of Mef2c in regulating angiogenesis and vasculogenesis of endothelial cells. 26972451_We demonstrated that AQP1-like immunoreactivity and expression are significantly increased in plaques of Peyronie's disease patients Vs controls, implying that AQP1 overexpression might be the consequence of a localized maladaptive response of the connective tissue to repeated mechanical trauma 27143047_Strong aquaporin 1 protein expression was an independent adverse prognosticator in hilar cholangiocarcinoma. 27157540_Aquaporin1 overexpression restored anti-cell proliferation and metastatic effect of si-Gli1. 27295912_Urinary AQP1 is significantly lower in patients with clear cell renal carcinoma than in controls. Urinary AQP1 increases after nephrectomy in clear cell renal carcinoma patients. 27465156_AQP1 expression was significantly lower in hereditary spherocytosis patients and silent carriers than in the simultaneously processed normal control, while no AQP1 decrease was detected in the autoimmune hemolytic anemia sample. The decreased AQP1 expression could contribute to explain variable degrees of anemia in hereditary spherocytosis. 27497833_results suggest regulation of AQP1 and AQP5 at the post-translational level and support previous observations on the implication of AQP1 and 5 in maintenance of lens transparency 27817002_AQP-1 overexpression is associated with prostate cancer progression. 27835672_Data show that the expression of aquaporin (AQP) 1, AQP3, AQP5, epithelial Na+ channel (ENaC) and sodium potassium ATPase (Na-K-ATPase) are altered in patients with acute respiratory failure (ARF) due to diffuse alveolar damage (DAD), and the cause of DAD does not seem to influence the level of impairment of these channels. 28072927_The native AQP1 gene, normally latent in human salivary gland cells, can be activated by both promoter-independent artificial transcription and epigenetic editing of the promoter. 28129470_AQP1 is expressed in atherosclerotic lesion neovasculature in human and mouse arteries and AQP1 deficiency augments lesion development in angiotensin II-promoted atherosclerosis in mice. 28409279_data suggest an important functional role of AQP1 in the pathobiology of hypoxia-induced pulmonary hypertension 28596233_function of AQP1 in tonicity response could be coupled or correlated to its function in band 3-mediated CO2/HCO3(-) exchange 28708257_The expression of AQP1 and 3 is a manifestation of physiological adaptation of functionally mature chondrocytes able to respond to the change of their internal environment influenced by extracellular matrix. 28766180_AQP1, 3 and 5 expression through the stages of human salivary gland morphogenesis, was examined. 28827734_Integrative epigenomics, transcriptomics and proteomics of patients chondrocytes from hip or knee osteoarthritis (OA) identified AQP1, COL1A1 and CLEC3B as significantly and differentially regulated suggesting they play an important role in OA pathogenesis. 29087027_Adenovirus-mediated human AQP1 expression in hepatocytes improves lipopolysaccharide-induced cholestasis in rats. 29104239_AQP1 may interact with VEGFA and play a role in vasculogenic mimicry, especially under hypoxic conditions, but the heterogeneity of malignant mesothelioma cells may result in different dominant pathways between patients. 29257313_AQP1 may serve a role in glioblastoma multiforme migration, invasion and vasculogenic mimicry formation 29258764_indicate pivotal roles for aquaporin-1 and -5 in the aggressive growth and metastatic potential of soft tissue sarcomas 29495596_The present study represents further confirmation of the hypothesized prognostic role of AQP1, which seems a reliable prognostic indicator. 29538612_Data show that miR-320 negatively regulates aquaporin 1 (AQP1) expression by targeting its 3' untranslated regions (3'-UTR). 29554889_Study demonstrated that a region upstream of the AQP1 gene (- 2300 to - 2200 bp) contains an enhancer required for pH-mediated regulation of AQP1 gene expression. 29650961_Case-control analyses reveal significant overrepresentation of rare variants in ATP13A3, AQP1 and SOX17, and provide independent validation of a critical role for GDF2 in heritable pulmonary arterial hypertension. 29769447_conclude that AQP1 is a major regulator of the platelet procoagulant response, able to modulate coagulation after injury or pathologic stimuli without affecting other platelet functional responses or normal hemostasis 29916035_used solid-state MAS NMR to investigate the binding of a lead compound [1-(4-methylphenyl)1H-pyrrole-2,5-dione] to AqpZ, through mapping of chemical shift perturbations in the presence of the compound. 30021355_H19 acted as AQP1 ceRNA in regulating miR-874. 30270117_Dermal fibroblasts and endothelial cells highly expressed AQP1 in systemic sclerosis lesional skin, and AQP1 expression in dermal fibroblasts and endothelial cells positively correlated with the degrees of tissue fibrosis and edema, respectively. 30378011_Aquaporin-1 Protein Expression of the Primary Tumor May Predict Cerebral Progression of Cutaneous Melanoma. 30467901_Aquaporin 1 and perilipin 2 possess high sensitivity and specificity for detecting clear cell and papillary renal cell carcinoma. Use of these markers might compliment renal mass biopsy in the characterization of small renal masses. 30531392_positive AQP1 expression is associated with the tumorigenesis and progression of pancreatic ductal adenocarcinoma; AQP1 is a diagnostic marker of pancreatic ductal adenocarcinoma and a predictive marker of poor prognosis in pancreatic ductal adenocarcinoma patients 30739527_AQP1 transcript expression was decreased in colorectal carcinoma (CRC) compared to normal mucosa, and this was associated with AQP1 promoter hypermethylation. AQP1 transcript expression increased with advanced disease but was not an independent prognostic indicator. 30840592_AQP1 may play a role in the risk of malignant mesothelioma. 31039249_We defined histologically the expression of AQP1 in the ChP from 6 nonsurvival preterm-pregnancy infants ranging ages between 20 and 25 gestational weeks in which AQP1 was mainly expressed at the apical pole of the We also defined histologically the expression of AQP1 in the ChP from 6 nonsurvival preterm-pregnancy infants ranging ages between 20 and 25 gestational epithelium in control and lissencephalic patients 31254364_homeostasis and physiological function of AQP1 in endothelial health are maintained by the MEF2C and miR-133a-3p.1 regulatory circuit 31261369_Aquaporin1-3 expression in normal and hydronephrotic kidneys in the human fetus. 31305898_To our knowledge, this work represents the first demonstration of AQP1/AQP3 expression in human melanocytic skin tumors 31311397_The expression AQP-1 protein was significantly reduced after transfected with AQP-1 silencing vector. Inhibition of miR-223 expression could suppress proliferation and promote apoptosis of SW579, and its mechanism i | ENSMUSG00000004655 | Aqp1 | 3.519449e+01 | 0.9384390 | -0.091665103 | 0.5585208 | 2.625005e-02 | 0.8712911442 | 0.97584187 | No | Yes | 2.781410e+01 | 6.903149 | 3.210135e+01 | 8.383171 | |
| ENSG00000240771 | 115557 | ARHGEF25 | protein_coding | Q86VW2 | FUNCTION: May play a role in actin cytoskeleton reorganization in different tissues since its activation induces formation of actin stress fibers. It works as a guanine nucleotide exchange factor for Rho family of small GTPases. Links specifically G alpha q/11-coupled receptors to RHOA activation. May be an important regulator of processes involved in axon and dendrite formation. In neurons seems to be an exchange factor primarily for RAC1. Involved in skeletal myogenesis (By similarity). {ECO:0000250, ECO:0000269|PubMed:11861769, ECO:0000269|PubMed:12547822, ECO:0000269|PubMed:15069594, ECO:0000269|PubMed:15632174, ECO:0000269|PubMed:16314529, ECO:0000269|PubMed:17606614}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Guanine-nucleotide releasing factor;Membrane;Reference proteome | Rho GTPases alternate between an inactive GDP-bound state and an active GTP-bound state, and GEFs facilitate GDP/GTP exchange. This gene encodes a guanine nucleotide exchange factor (GEF) which interacts with Rho GTPases involved in contraction of vascular smooth muscles, regulation of responses to angiotensin II and lens cell differentiation. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2012]. | hsa:115557; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extrinsic component of membrane [GO:0019898]; myofibril [GO:0030016]; plasma membrane [GO:0005886]; sarcomere [GO:0030017]; guanyl-nucleotide exchange factor activity [GO:0005085]; axon guidance [GO:0007411]; regulation of Rho protein signal transduction [GO:0035023]; regulation of small GTPase mediated signal transduction [GO:0051056] | 11861769_Results show that p63RhoGEF is a RhoA-specific GEF that may play a key role in actin cytoskeleton reorganization in different tissues, especially in heart cellular morphology. 15322108_GEFT enhances dendritic spine formation and neurite outgrowth in primary neurons and neuroblastoma cells, respectively, through the activation of Rac/Cdc42-PAK signaling pathways. 15632174_a new signaling pathway by which G alpha(q/11)-coupled receptors specifically induce Rho signaling through a direct interaction of activated G alpha(q/11) subunits with p63RhoGEF. 16314529_Endogenous GEFT is transcriptionally upregulated during myogenic differentiation and downregulated during adipogenic differentiation. 16496360_Together, our data provide new evidence suggesting that GEFT is an important regulator of multiple processes involved in axon and dendrite formation. 18096806_the crystal structure of the Galphaq-p63RhoGEF-RhoA complex, detailing the interactions of Galphaq with the Dbl and pleckstrin homology (DH and PH) domains of p63RhoGEF was determined 18851832_MLK3 functions in a regulated way to limit levels of the activated GTPase Rho by binding to the Rho activator, p63RhoGEF/GEFT, which, in turn, prevents its activation by Galphaq. 20739613_Specifically mediates angiotensin II- Gq/11-dependent RhoA activation and downstream signaling and processes in vascular smooth muscle cells 21832057_p63RhoGEF is regulated chiefly through allosteric control by Galpha(q), as opposed to other known Galpha-regulated RhoGEFs, which are instead sequestered in the cytoplasm, perhaps because of their high basal activity. 21885830_Signaling through p63RhoGEF provides a novel mechanism for selective regulation of blood pressure. 23380069_p63RhoGEF-mediated formation of a single polarized lamellipodium is required for chemotactic migration in breast carcinoma cells. 23884432_Signaling efficiency of Galphaq through its effectors p63RhoGEF and GEFT depends on their subcellular location. 24356540_assessed p63RhoGEF gene and protein expression and RhoA/Rho kinase activity in essential hypertensive and Bartter's and Gitelman's syndrome patients, a model opposite to hypertension 26435194_p63RhoGEF can rapidly activate RhoA through endogenous G-protein coupled receptors. 27833100_p63RhoGEF(619) relocates to the plasma membrane upon activation of Galphaq coupled GPCRs, resembling the well-known activation mechanism of RhoGEFs activated by Galpha12/13. | ENSMUSG00000019467 | Arhgef25 | 1.103023e+03 | 0.9985000 | -0.002165629 | 0.2691919 | 6.569035e-05 | 0.9935332496 | 0.99877177 | No | Yes | 9.234703e+02 | 86.206336 | 9.332077e+02 | 89.509111 | |
| ENSG00000240849 | 387521 | PEDS1 | protein_coding | A5PLL7 | FUNCTION: Plasmanylethanolamine desaturase involved in plasmalogen biogenesis in the endoplasmic reticulum membrane (PubMed:31604315, PubMed:32209662). Plasmalogens are glycerophospholipids with a hydrocarbon chain linked by a vinyl ether bond at the glycerol sn-1 position, and are involved in antioxidative and signaling mechanisms (PubMed:31604315). {ECO:0000269|PubMed:31604315, ECO:0000269|PubMed:32209662, ECO:0000303|PubMed:31604315}. | Alternative splicing;Endoplasmic reticulum;Fatty acid metabolism;Lipid metabolism;Membrane;Oxidoreductase;Reference proteome;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; fatty acid metabolism. {ECO:0000269|PubMed:31604315}. | Co-transcription of this gene and the neighboring downstream gene (ubiquitin-conjugating enzyme E2 variant 1) generates a rare read-through transcript, which encodes a fusion protein comprised of sequence sharing identity with each individual gene product. The protein encoded by this individual gene lacks a UEV1 domain but includes three transmembrane regions. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jun 2009]. | hsa:387521;hsa:387522; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; oxidoreductase activity [GO:0016491]; plasmanylethanolamine desaturase activity [GO:0050207]; ether lipid biosynthetic process [GO:0008611]; fatty acid metabolic process [GO:0006631] | 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21555518_TMEM189 is a target gene of the BACH1 transcription factor according to ChIP-seq analysis in HEK 293 cells. 31604315_TMEM189 could functionally replace CarF in M. xanthus, and knockout of TMEM189 in a human cell line eliminated plasmalogens; study reveals TMEM189 as the long-sought desaturase for animal plasmalogen biosynthesis 32209662_these results assign the TMEM189 gene to plasmanylethanolamine desaturase and suggest that the previously characterized phenotype of Tmem189-deficient mice may be caused by a lack of plasmalogens. | ENSMUSG00000090213+ENSMUSG00000089739 | Peds1+Gm20431 | 3.325812e+03 | 1.1695320 | 0.225931342 | 0.3090203 | 5.330998e-01 | 0.4653065538 | 0.86741238 | No | Yes | 3.099100e+03 | 337.339528 | 2.582727e+03 | 288.560268 |
| ENSG00000242485 | 55052 | MRPL20 | protein_coding | Q9BYC9 | 3D-structure;Alternative splicing;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein;Transit peptide | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 39S subunit protein. A pseudogene corresponding to this gene is found on chromosome 21q. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jan 2016]. | hsa:55052; | mitochondrial inner membrane [GO:0005743]; mitochondrial large ribosomal subunit [GO:0005762]; mitochondrial ribosome [GO:0005761]; mitochondrion [GO:0005739]; RNA binding [GO:0003723]; rRNA binding [GO:0019843]; structural constituent of ribosome [GO:0003735]; ribosomal large subunit assembly [GO:0000027]; translation [GO:0006412] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000029066 | Mrpl20 | 6.170541e+03 | 0.9710550 | -0.042375020 | 0.2869581 | 2.175590e-02 | 0.8827383248 | 0.97743725 | No | Yes | 6.029955e+03 | 514.143458 | 5.468200e+03 | 478.485696 | ||
| ENSG00000243364 | 1945 | EFNA4 | protein_coding | P52798 | FUNCTION: Cell surface GPI-bound ligand for Eph receptors, a family of receptor tyrosine kinases which are crucial for migration, repulsion and adhesion during neuronal, vascular and epithelial development. Binds promiscuously Eph receptors residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. May play a role in the interaction between activated B-lymphocytes and dendritic cells in tonsils. | Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;GPI-anchor;Glycoprotein;Lipoprotein;Membrane;Reference proteome;Secreted;Signal | This gene encodes a member of the ephrin (EPH) family. The ephrins and EPH-related receptors comprise the largest subfamily of receptor protein-tyrosine kinases and have been implicated in mediating developmental events, especially in the nervous system and in erythropoiesis. Based on their structures and sequence relationships, ephrins are divided into the ephrin-A (EFNA) class, which are anchored to the membrane by a glycosylphosphatidylinositol linkage, and the ephrin-B (EFNB) class, which are transmembrane proteins. This gene encodes an EFNA class ephrin. Three transcript variants that encode distinct proteins have been identified. [provided by RefSeq, Jul 2008]. | hsa:1945; | anchored component of membrane [GO:0031225]; extracellular region [GO:0005576]; intrinsic component of plasma membrane [GO:0031226]; plasma membrane [GO:0005886]; ephrin receptor binding [GO:0046875]; transmembrane-ephrin receptor activity [GO:0005005]; axon guidance [GO:0007411]; cell-cell signaling [GO:0007267]; ephrin receptor signaling pathway [GO:0048013] | 12030849_Characterization of the promoter 16540516_This provides genetic evidence that Twist1, Msx2 and Efna4 function together in boundary formation and the pathogenesis of coronal synostosis. 18819711_CLL B-cells showed a more heterogeneous Eph/EFN profile, specially EFNA4, EphB6 and EphA10. EphB6 and EFNA4 were further related with the clinical course of CLL. 19828693_EFNA4-EphA2 interactions are involved in Chronic lymphocytic leukemia cell trafficking between blood and the tissues 20071790_The cytoplasmic pattern of ephrin A4 could identify a subgroup of primary osteosarcoma patients with a high liability for progression, poor prognosis, and inferior response to chemotherapy. 20643727_Observational study of gene-disease association. (HuGE Navigator) 23686814_The interaction between ephrin-As, Eph receptors and integrin alpha3 is plausibly important for the crosstalk between Eph and integrin signalling pathways at the membrane protrusions and in the migration of brain cancer cells. 25070915_The present study provides evidence that microglia upregulates endothelial ephrin-A3 and ephrin-A4 to facilitate in vitro angiogenesis of brain endothelial cells, which is mediated by microglia-released TNF-alpha. 27374180_Study present for the first time in vitro and in vivo evidence suggesting that the major role of two ephrin A4 isoforms in chronic lymphocytic leukemia could be related with a non-previously described mechanism of survival linked to extravasation strongly dependent on integrin signaling. 33436772_Ephrin A4-ephrin receptor A10 signaling promotes cell migration and spheroid formation by upregulating NANOG expression in oral squamous cell carcinoma cells. | ENSMUSG00000028040 | Efna4 | 6.525456e+02 | 1.1597053 | 0.213758262 | 0.3304623 | 4.223220e-01 | 0.5157806771 | 0.88131185 | No | Yes | 6.490316e+02 | 78.673449 | 5.243177e+02 | 65.590537 | |
| ENSG00000243701 | 344595 | DUBR | lncRNA | 33758355_DUBR suppresses migration and invasion of human lung adenocarcinoma cells via ZBTB11-mediated inhibition of oxidative phosphorylation. | 1.384496e+02 | 1.0221247 | 0.031571172 | 0.3583525 | 7.603456e-03 | 0.9305142880 | 0.98676673 | No | Yes | 1.139196e+02 | 24.894907 | 1.209229e+02 | 26.916994 | |||||||||
| ENSG00000243716 | 100132247 | NPIPB5 | protein_coding | Q92617 | Alternative splicing;Membrane;Reference proteome;Transmembrane;Transmembrane helix | integral component of membrane [GO:0016021]; nucleoplasm [GO:0005654] | 2.069657e+03 | 0.7860781 | -0.347255514 | 0.2573844 | 1.785348e+00 | 0.1814939930 | 0.77709619 | No | Yes | 1.473512e+03 | 276.357241 | 2.246593e+03 | 431.992751 | |||||||
| ENSG00000243970 | 728448 | PPIEL | transcribed_unprocessed_pseudogene | This transcribed pseudogene is related to PPIE (Gene ID: 10450). Expression of this pseudogene may be downregulated in non-small cell lung cancer (NSCLC). Differential DNA methylation of this locus may be associated with intellectual disability and bipolar disorder in human patients. [provided by RefSeq, Sep 2016]. | 4.258039e+02 | 0.8406671 | -0.250393563 | 0.2899493 | 7.568073e-01 | 0.3843295277 | 0.84308088 | No | Yes | 2.828742e+02 | 46.277010 | 3.859503e+02 | 64.373698 | |||||||||
| ENSG00000244879 | GABPB1-AS1 | lncRNA | 6.479767e+02 | 0.5704304 | -0.809877208 | 0.3058412 | 6.892731e+00 | 0.0086546947 | 0.33746753 | No | Yes | 3.453656e+02 | 70.610324 | 6.870873e+02 | 142.918253 | |||||||||||
| ENSG00000245958 | SEPTIN7P14 | transcribed_unprocessed_pseudogene | 2.956910e+02 | 0.6335871 | -0.658385218 | 0.3312923 | 3.846447e+00 | 0.0498514867 | 0.66791998 | No | Yes | 1.845026e+02 | 31.860315 | 3.189629e+02 | 55.723223 | |||||||||||
| ENSG00000246067 | RAB30-DT | lncRNA | 1.997446e+02 | 1.0932964 | 0.128684535 | 0.3309520 | 1.479029e-01 | 0.7005475208 | 0.93624959 | No | Yes | 1.530393e+02 | 24.237075 | 1.552117e+02 | 24.944630 | |||||||||||
| ENSG00000249069 | 104355136 | LINC01033 | lncRNA | 3.292018e+01 | 0.5713973 | -0.807433876 | 0.5521254 | 2.163715e+00 | 0.1413026616 | 0.76841160 | No | Yes | 3.775402e+01 | 10.034111 | 5.020987e+01 | 13.737486 | ||||||||||
| ENSG00000249087 | 148898 | ZNF436-AS1 | lncRNA | 1.585582e+02 | 0.8093457 | -0.305172041 | 0.3248721 | 8.788910e-01 | 0.3485055930 | 0.82969965 | No | Yes | 9.407136e+01 | 13.658461 | 1.158500e+02 | 17.020885 | ||||||||||
| ENSG00000249196 | 283352 | TMEM132D-AS1 | lncRNA | 1.506831e+01 | 1.1826390 | 0.242009707 | 0.7767049 | 8.904179e-02 | 0.7653988711 | No | Yes | 2.603838e+01 | 9.090136 | 1.893516e+01 | 7.357829 | |||||||||||
| ENSG00000250366 | 100507043 | TUNAR | protein_coding | A0A1B0GTB2 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | integral component of membrane [GO:0016021] | 3.407264e+01 | 0.6952635 | -0.524368187 | 0.5598425 | 8.731593e-01 | 0.3500821665 | 0.82969965 | No | Yes | 2.700436e+01 | 6.128754 | 3.446245e+01 | 7.831980 | |||||||
| ENSG00000250673 | 345051 | REELD1 | protein_coding | A0A1B0GV85 | Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | integral component of membrane [GO:0016021] | 2.697211e+01 | 0.2174463 | -2.201268708 | 0.6584233 | 1.001712e+01 | 0.0015509179 | 0.15104296 | No | Yes | 1.301706e+01 | 7.520838 | 6.748453e+01 | 39.560147 | |||||||
| ENSG00000250903 | 100508120 | GMDS-DT | lncRNA | 31860169_LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. 34981821_lncRNA GMDSAS1 upregulates IL6, TNFalpha and IL1beta, and induces apoptosis in human monocytic THP1 cells via miR965p/caspase 2 signaling. | 7.250404e+01 | 0.9225279 | -0.116335507 | 0.4388656 | 7.118419e-02 | 0.7896203415 | 0.95778572 | No | Yes | 6.030093e+01 | 16.874763 | 6.976784e+01 | 19.928283 | |||||||||
| ENSG00000251136 | 101929709 | RIPK2-DT | lncRNA | 2.715029e+02 | 0.6314952 | -0.663156385 | 0.2970655 | 4.869553e+00 | 0.0273345736 | 0.55921720 | No | Yes | 1.437912e+02 | 29.752662 | 2.725244e+02 | 57.080389 | ||||||||||
| ENSG00000251141 | MRPS30-DT | lncRNA | 1.221518e+01 | 0.6004482 | -0.735888380 | 0.8555544 | 7.565393e-01 | 0.3844137200 | No | Yes | 9.589420e+00 | 3.914848 | 1.188641e+01 | 4.639543 | ||||||||||||
| ENSG00000251192 | 641339 | ZNF674 | protein_coding | Q2M3X9 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a zinc finger protein with an N-terminal Kruppel-associated box-containing (KRAB) domain and 11 Kruppel-type C2H2 zinc finger domains. Like other zinc finger proteins, this gene may function as a transcription factor. This gene resides on an area of chromosome X that has been implicated in nonsyndromic X-linked cognitive disabilities. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2017]. | hsa:641339; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 16385466_Identification of ZNF674 as the third X-linked mental retardation gene in this cluster may indicate a common role for these zinc-finger genes that is crucial to human cognitive functioning. 22126752_These findings question the responsibility of ZNF674 deletions in intellectual disability (ID) associated with X-linked retinal dystrophy. | ENSMUSG00000055150 | Zfp78 | 6.168656e+01 | 0.9055517 | -0.143131054 | 0.4238419 | 1.117962e-01 | 0.7381083683 | 0.94494829 | No | Yes | 5.914802e+01 | 14.058039 | 5.482044e+01 | 13.071970 | |
| ENSG00000251580 | 105374366 | LINC02482 | lncRNA | 5.616574e+01 | 0.9338437 | -0.098746940 | 0.4302681 | 5.252374e-02 | 0.8187285712 | 0.96475641 | No | Yes | 4.197674e+01 | 8.079085 | 4.485919e+01 | 8.797240 | ||||||||||
| ENSG00000254852 | 642799 | NPIPA2 | protein_coding | E9PIF3 | Reference proteome | nucleoplasm [GO:0005654] | 4.165932e+02 | 0.6296546 | -0.667367345 | 0.2913725 | 5.212762e+00 | 0.0224216858 | 0.52906651 | No | Yes | 2.037629e+02 | 50.675965 | 3.867314e+02 | 98.057649 | |||||||
| ENSG00000256294 | 7768 | ZNF225 | protein_coding | Q9UK10 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7768; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 7.752620e+01 | 0.8615296 | -0.215027810 | 0.4002064 | 2.846590e-01 | 0.5936636482 | 0.90759816 | No | Yes | 6.420281e+01 | 12.313071 | 7.883132e+01 | 15.028603 | |||||
| ENSG00000256673 | unprocessed_pseudogene | 2.558077e+02 | 0.7273454 | -0.459287478 | 0.3892673 | 1.388247e+00 | 0.2387013599 | 0.78818582 | No | Yes | 1.403525e+02 | 41.538828 | 2.396580e+02 | 72.275943 | ||||||||||||
| ENSG00000257218 | 283459 | GATC | protein_coding | O43716 | FUNCTION: Allows the formation of correctly charged Gln-tRNA(Gln) through the transamidation of misacylated Glu-tRNA(Gln) in the mitochondria. The reaction takes place in the presence of glutamine and ATP through an activated gamma-phospho-Glu-tRNA(Gln). {ECO:0000255|HAMAP-Rule:MF_03149, ECO:0000269|PubMed:19805282}. | ATP-binding;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Primary mitochondrial disease;Protein biosynthesis;Reference proteome | hsa:283459; | glutamyl-tRNA(Gln) amidotransferase complex [GO:0030956]; mitochondrion [GO:0005739]; ATP binding [GO:0005524]; glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity [GO:0050567]; glutaminyl-tRNAGln biosynthesis via transamidation [GO:0070681]; mitochondrial translation [GO:0032543]; regulation of translational fidelity [GO:0006450] | 19805282_Studies showed in vitro Gln-tRNA(Gln) formation catalyzed by the recombinant mtGluRS and hGatCAB. 20877624_Observational study of gene-disease association. (HuGE Navigator) 24579914_The mitochondrial defective phenotype provoked by the absence of gatA in human cells is confirmed by means of a deficient ability to grow when galactose is used as a carbon source. | ENSMUSG00000029536 | Gatc | 2.244807e+03 | 0.9732175 | -0.039165834 | 0.2667061 | 2.217263e-02 | 0.8816287694 | 0.97741608 | No | Yes | 2.241064e+03 | 192.848475 | 2.250699e+03 | 198.147000 | ||
| ENSG00000257365 | 100529261 | FNTB | protein_coding | P49356 | FUNCTION: Essential subunit of the farnesyltransferase complex. Catalyzes the transfer of a farnesyl moiety from farnesyl diphosphate to a cysteine at the fourth position from the C-terminus of several proteins having the C-terminal sequence Cys-aliphatic-aliphatic-X. {ECO:0000269|PubMed:12036349, ECO:0000269|PubMed:12825937, ECO:0000269|PubMed:16893176, ECO:0000269|PubMed:19246009, ECO:0000269|PubMed:8494894}. | 3D-structure;Alternative splicing;Lipid metabolism;Metal-binding;Phosphoprotein;Prenyltransferase;Reference proteome;Repeat;Transferase;Zinc | This locus represents naturally occurring read-through transcription between the neighboring CHURC1 (churchill domain containing 1) and FNTB (farnesyltransferase, CAAX box, beta) on chromosome 14. The read-through transcript produces a fusion protein that shares sequence identity with each individual gene product. [provided by RefSeq, Feb 2011]. | hsa:100529261;hsa:2342; | cytosol [GO:0005829]; microtubule associated complex [GO:0005875]; protein farnesyltransferase complex [GO:0005965]; protein farnesyltransferase activity [GO:0004660]; zinc ion binding [GO:0008270]; lipid metabolic process [GO:0006629]; protein farnesylation [GO:0018343]; protein prenylation [GO:0018342] | Mouse_homologues 15837621_FTase is essential for embryonic development, but dispensable for adult homeostasis. Elimination of FTase during tumor progression had a limited but significant inhibitory effect on tumor development. 20106865_Both Fntb and Pggt1b are required for the homeostasis of skin keratinocytes. 22039581_deficiencies in either FTase or GGTase-I lead to severe hepatocellular disease. Our studies lay to rest the earlier notion that FTase is dispensable in adult tissues 24622074_Data suggest that farnesyltransferase might constitute a target in the management of severe acute pancreatitis (AP). 25853815_the catalytic b subunit of farnesyl transferase associated with a cytoskeletal protein...lonafarnib remarkably inhibited he expression of the cytoskeletal protein and interrupted its interaction with farnesyl transferase. 34315531_Protein farnesylation is upregulated in Alzheimer's human brains and neuron-specific suppression of farnesyltransferase mitigates pathogenic processes in Alzheimer's model mice. | ENSMUSG00000033373 | Fntb | 4.711479e+02 | 1.0047630 | 0.006855172 | 0.3336764 | 4.212735e-04 | 0.9836246101 | 0.99700442 | No | Yes | 2.475289e+02 | 38.371318 | 2.664384e+02 | 43.113611 | |
| ENSG00000257727 | 10330 | CNPY2 | protein_coding | Q9Y2B0 | FUNCTION: Positive regulator of neurite outgrowth by stabilizing myosin regulatory light chain (MRLC). It prevents MIR-mediated MRLC ubiquitination and its subsequent proteasomal degradation. | Alternative splicing;Disulfide bond;Endoplasmic reticulum;Phosphoprotein;Reference proteome;Signal | hsa:10330; | endoplasmic reticulum [GO:0005783]; negative regulation of gene expression [GO:0010629]; positive regulation of low-density lipoprotein receptor activity [GO:1905599]; regulation of low-density lipoprotein particle clearance [GO:0010988] | 12826659_MSAP is a novel MIR-interacting protein that enhances neurite outgrowth and increases myosin regulatory light chain in fetal and adult brain. 19240061_Observational study of gene-disease association. (HuGE Navigator) 22378787_Our results are consistent with an important role of FGF21 and Cnpy2/Msap in the regulation of LDLRs in cultured cells. 25589425_CNPY2 is a HIF-1alpha-regulated, secreted angiogenic growth factor that promotes smooth muscle cell migration, proliferation, and tissue revascularization. 26835537_CNPY2 may play a critical role in colorectal cancer development by enhancing cell proliferation, migration, and angiogenesis and by inhibiting apoptosis through negative regulation of the p53 pathway 28235487_These findings suggested that CNPY2 promoted cancer cell growth in renal cell carcinoma cells through regulating TP53 gene expression. 29135454_CNPY2 isoform2 represents as a novel and valuable prognostic indicator for colorectal cancer patients. 29569784_Overexpression of CNPY2 can activate the AKT/GSK3beta pathway, which leads to the inactivation of GSK-3beta. The inactivation of GSK-3beta increases the level of Snail, and then decreases the expression of E-cadherin to promote EMT. 29864955_CNPY2 can serve as a therapeutic target to promote the effect of chemotherapy in non-small cell lung cancer. 30070972_This study showed that serum CNPY2 isoform 2 may be a valuable biomarker for the early detection of colorectal cancer and presents an improvement in the diagnostic efficiency by combination of CEA and CA19-9. 30415246_CNPY2 knockout resulted in the significant suppression of MHCC97H cell proliferation, tumor growth, and hemorrhage. 31047986_Knockout of Cnpy2 resulted in up-regulation of p16(INK4a). 31894275_miR30a3p suppresses the proliferation and migration of lung adenocarcinoma cells by downregulating CNPY2. 33482578_Sleeping Beauty insertional mutagenesis screen identifies the pro-metastatic roles of CNPY2 and ACTN2 in hepatocellular carcinoma tumor progression. 33721832_Hypoxia-induced CNPY2 upregulation promotes glycolysis in cervical cancer through activation of AKT pathway. | ENSMUSG00000025381 | Cnpy2 | 2.292895e+03 | 0.6630751 | -0.592755918 | 0.2736704 | 4.597941e+00 | 0.0320103793 | 0.58475998 | No | Yes | 1.669207e+03 | 165.860772 | 2.550569e+03 | 258.977734 | ||
| ENSG00000258366 | 51750 | RTEL1 | protein_coding | Q9NZ71 | FUNCTION: ATP-dependent DNA helicase implicated in telomere-length regulation, DNA repair and the maintenance of genomic stability. Acts as an anti-recombinase to counteract toxic recombination and limit crossover during meiosis. Regulates meiotic recombination and crossover homeostasis by physically dissociating strand invasion events and thereby promotes noncrossover repair by meiotic synthesis dependent strand annealing (SDSA) as well as disassembly of D loop recombination intermediates. Also disassembles T loops and prevents telomere fragility by counteracting telomeric G4-DNA structures, which together ensure the dynamics and stability of the telomere. {ECO:0000255|HAMAP-Rule:MF_03065, ECO:0000269|PubMed:18957201, ECO:0000269|PubMed:23453664, ECO:0000269|PubMed:24009516}. | 4Fe-4S;ATP-binding;Alternative splicing;DNA damage;DNA repair;DNA-binding;Disease variant;Dyskeratosis congenita;Helicase;Hydrolase;Iron;Iron-sulfur;Metal-binding;Nucleotide-binding;Nucleus;Reference proteome | This gene encodes a DNA helicase which functions in the stability, protection and elongation of telomeres and interacts with proteins in the shelterin complex known to protect telomeres during DNA replication. Mutations in this gene have been associated with dyskeratosis congenita and Hoyerall-Hreidarsson syndrome. Read-through transcription of this gene into the neighboring downstream gene, which encodes tumor necrosis factor receptor superfamily, member 6b, generates a non-coding transcript. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Sep 2013]. | hsa:51750; | chromosome, telomeric region [GO:0000781]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 4 iron, 4 sulfur cluster binding [GO:0051539]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; DNA polymerase binding [GO:0070182]; metal ion binding [GO:0046872]; DNA duplex unwinding [GO:0032508]; DNA repair [GO:0006281]; mitotic telomere maintenance via semi-conservative replication [GO:1902990]; negative regulation of DNA recombination [GO:0045910]; negative regulation of t-circle formation [GO:1904430]; negative regulation of telomere maintenance in response to DNA damage [GO:1904506]; positive regulation of telomere capping [GO:1904355]; positive regulation of telomere maintenance [GO:0032206]; positive regulation of telomere maintenance via telomere lengthening [GO:1904358]; positive regulation of telomeric loop disassembly [GO:1904535]; regulation of double-strand break repair via homologous recombination [GO:0010569]; replication fork processing [GO:0031297]; strand displacement [GO:0000732]; telomere maintenance [GO:0000723]; telomere maintenance in response to DNA damage [GO:0043247]; telomeric loop disassembly [GO:0090657] | 18957201_Study finds that rtel-1 mutant worms and RTEL1-depleted human cells share characteristic phenotypes with yeast srs2 mutants: lethality upon deletion of the sgs1/BLM homolog, hyperrecombination, and DNA damage sensitivity.[RTEL1] 19578366_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19578366_On 9p21, rs1412829 near CDKN2B had discovery P = 3.4 x 10(-8), replication P = 0.0038 and combined P = 1.85 x 10(-10). On 20q13.3, rs6010620 intronic to RTEL1 had discovery P = 1.5 x 10(-7), replication P = 0.00035 and combined P = 3.40 x 10(-9). 19578367_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20368557_Polymorphisms in the LIG4, BTBD2, HMGA2, and RTEL1 genes, which are involved in the double-strand break repair pathway, are associated with glioblastoma multiforme survival. 20462933_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20610542_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20847058_Observational study of gene-disease association. (HuGE Navigator) 21350045_SUSCEPTIBILITY LOCI FOR GLIOMA AND GLIOBLASTOMA RISK IN A CHINESE POPULATION INCLUDED RTEL1 21920947_rs6010620 a genetic variant of RTEL1, was one of 3 genetic variants implicated in a pool of US epidemiologic studies of glioma risk 23115063_rs6010620 (RTEL1) was associated with an increased risk of glioma when restricting to cases with family history of brain tumours. 23329068_The nonsense mutations both cause truncation of the RTEL1 protein, resulting in loss of the PIP box; this may abrogate an important protein-protein interaction. These findings implicate a new telomere biology gene, RTEL1, in the etiology of DC. 23453664_These biallelic RTEL1 mutations are responsible for a major subgroup ( approximately 29%) of HHS. 23591994_results identify RTEL1 as a novel Hoyeraal-Hreidarsson syndrome-causing gene and highlight its role as a genomic caretaker in humans. 23683922_Two significant Ten candidate tagging SNPs in RTEL1 gene were observed to be associated with glioma risk 23812731_Results indicate the potential roles of regulator of telomere elongation helicase 1 (RTEL1) and telomerase reverse transcriptase (TERT) in astrocytoma development. 23959892_Data show that RTEL1 interacts with the shelterin protein TRF1, indicating a potential recruitment mechanism of RTEL1 to telomeres. 24009516_The molecular data and the patterns of inheritance are consistent with a hypomorphic mutation in RTEL1 as the underlying basis of the clinical and cellular phenotypes. 24130156_The C-terminal extension of RTEL1, downstream of its catalytic domain and including several HHS-associated mutations, contains a yet unidentified tandem of harmonin-N-like domains. 24523019_suggested that RTEL1 rs6010620 polymorphism is likely to be associated with increased glioma risk, which lends further biological plausibility to these findings 24561255_Authors propose that RTEL1 serves as a human analog of Srs2 to inhibit (CTGCAG) repeat expansions and fragility, likely by unwinding problematic hairpins. 25227808_rs6010620 polymorphism in the RTEL1 gene is associated with increased risk of glioma in both Caucasians and Asians. [Meta-Analysis] 25556444_Association between the RTEL1 rs6010620 polymorphism and glioma risk was significant. [Meta-Analysis] 25607374_Rare loss-of-function variants in RTEL1 represent a newly defined genetic predisposition for FIP, supporting the importance of telomere-related pathways in pulmonary fibrosis. 25620558_The shelterin protein TRF2 recruits RTEL1 to telomeres in S phase, which is required to prevent catastrophic t-loop processing by structure-specific nucleases. 25628358_This work unravels completely unanticipated roles for RTEL1 in RNP trafficking and strongly suggests that defects in RNP biogenesis pathways contribute to the pathology of Hoyeraal-Hreidarsson syndrome 25848748_PARN and RTEL1 mutation carriers had shortened leukocyte telomere lengths. 26014354_RTEL1 single nucleotide polymorphisms are associated with decreased susceptibility to pediatric brain astrocytoma. 26022962_Heterozygous RTEL1 mutations are responsible for familial pulmonary fibrosis (FPF) and, thereby, extend the clinical spectrum of RTEL1 deficiency. Thus, RTEL1 enlarges the number of telomere-associated genes implicated in FPF. 26025130_A homozygous mutation of RTEL1 in a child presenting with an apparently isolated natural killer cell deficiency. 26156397_results suggest a significant association between the RETL1, TREH, and PHLDB1 genes and GBM development in the Han Chinese population 26581417_Telomere length is associated with Esophageal squamous cell carcinoma risk in a U-shaped pattern and demonstrates that TL-SNPs may not be important in carcinogenesis in Chinese population. 26803811_Deletion in the RTEL1 gene is associated with metastatic glioblastoma. 26839018_Genetic risk variants in the RTEL1 gene is associated with somatic biomarkers in glioma. 26939676_This meta-analysis demonstrates that the RTEL1 rs2297440 polymorphism plays a moderate, but significant role in the risk of glioma. 27485611_promoter methylated in 51.4% of lung cancer patients and in 8.8% of healthy individuals 27540018_Pulmonary fibrosis patients with mutations in telomerase reverse transcriptase, telomerase RNA component, regulator of telomere elongation helicase 1 and poly(A)-specific ribonuclease were identified and clinical data were analysed. Genetic mutations in telomere related genes lead to a variety of interstitial lung disease diagnoses that are universally progressive. 27765928_Findings suggest a potential association between regulator of telomere elongation helicase 1 (RTEL1) polymorphisms and lung cancer (LC) risk in a Chinese Han population. 28360516_SNPs in the RTEL1 are associated with COPD in a Chinese Han population. It is possible that these variants are COPD risk factors. 28495916_Observation are firstly, heterozygous LOF RTEL1 variants are associated with myelodysplasia and liver disease in adulthood. Secondly, biallelic RTEL1 variants can present with just bone marrow failure in adulthood. Thirdly, many heterozygous variants, and even some biallelic RTEL1 variants, are bystanders. 28953687_The allele 'G' of rs6089953 and rs6010621 and the allele 'A' of rs2297441 were associated with decreased risk of High altitude pulmonary edema, haplotype 'GG, GT, AT' of rs6089953-rs6010621 were detected significantly associated with High altitude pulmonary edema risk in the Chinese population. 29151059_a novel association signal in the RTEL1 gene (intronic single nucleotide poly morphism (SNP) rs2297439; P=2.82x10(-7)) that is independent of previously reported Telomere-associated SNPs in this region. 29344583_heterozygous RTEL1 variants were associated with marrow failure, and telomere length measurement alone may not identify patients with telomere dysfunction carrying RTEL1 variants. 29522136_hRTEL1 contributes to the maintenance of long telomeres by preserving long G-overhangs, thereby facilitating POT1 binding and elongation by telomerase. 30345460_RTEL1 SNPs were associated with relative telomere length. Shorter relative telomere length was associated with an increased risk of stroke. 30462709_Study demonstrates that previously identified loci in RTEL1 are confirmed to have an association with increased risk of adult gliomas. Moreover, two coding variants (rs6062302 and rs115303435) were found to confer independent risk for glioma in RTEL1. A novel missense SNP (rs77086616, T434M) was also observed only in Korean glioma samples. 30523160_Regulator of telomere length 1 (RTEL1) mutations are associated with heterogeneous pulmonary and extra-pulmonary phenotypes. 30623606_RTEL1 SNPs may have a protective role against coronary heart disease risk. 31677132_Homozygous RTEL1 variant is associated with telomeropathies. 31721021_The rs6010620 (RTEL1), rs4977756 (CDKN2A/B), and rs498872 (PHLDB1) are associated with glioma risk in the Portuguese population. The GA genotype of the rs6010620 (RTEL1) was associated with a decreased risk of glioblastomas (OR 0.45). 32398827_RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. 32398829_SLX4 interacts with RTEL1 to prevent transcription-mediated DNA replication perturbations. 32460026_Synthetic Lethality between DNA Polymerase Epsilon and RTEL1 in Metazoan DNA Replication. 32542379_Full length RTEL1 is required for the elongation of the single-stranded telomeric overhang by telomerase. 32561545_Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. 33200790_miR-4530 inhibits the malignant biological behaviors of human glioma cells by directly targeting RTEL1. 34021146_RTEL1 influences the abundance and localization of TERRA RNA. 34479523_Pulmonary fibrosis in dyskeratosis congenita: a case report with a PRISMA-compliant systematic review. | ENSMUSG00000038685 | Rtel1 | 2.176983e+03 | 1.0510279 | 0.071800916 | 0.2581117 | 7.786665e-02 | 0.7802095476 | 0.95675823 | No | Yes | 1.772586e+03 | 204.831444 | 1.799619e+03 | 213.539550 | |
| ENSG00000258643 | 100529063 | BCL2L2-PABPN1 | protein_coding | Q92843 | FUNCTION: Promotes cell survival. Blocks dexamethasone-induced apoptosis. Mediates survival of postmitotic Sertoli cells by suppressing death-promoting activity of BAX. {ECO:0000269|PubMed:8761287}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Membrane;Methylation;Mitochondrion;Phosphoprotein;Reference proteome | This locus represents naturally occurring read-through transcription between the neighboring BCL2L2 (BCL2-like 2) and PABPN1 (poly(A) binding protein, nuclear 1) genes on chromosome 14. The read-through transcript encodes a fusion protein that shares sequence identity with each individual gene product. [provided by RefSeq, Dec 2010]. | hsa:599; | Bcl-2 family protein complex [GO:0097136]; cytosol [GO:0005829]; mitochondrial outer membrane [GO:0005741]; BH domain binding [GO:0051400]; disordered domain specific binding [GO:0097718]; identical protein binding [GO:0042802]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; protein-containing complex binding [GO:0044877]; brain development [GO:0007420]; cellular response to amyloid-beta [GO:1904646]; cellular response to estradiol stimulus [GO:0071392]; cellular response to glycine [GO:1905430]; extrinsic apoptotic signaling pathway in absence of ligand [GO:0097192]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; negative regulation of apoptotic process [GO:0043066]; negative regulation of intrinsic apoptotic signaling pathway [GO:2001243]; negative regulation of mitochondrial membrane permeability [GO:0035795]; negative regulation of release of cytochrome c from mitochondria [GO:0090201]; regulation of apoptotic process [GO:0042981]; response to ischemia [GO:0002931]; Sertoli cell proliferation [GO:0060011]; spermatogenesis [GO:0007283] | ENSMUSG00000092232 | Gm20521 | 5.651537e+02 | 0.6846282 | -0.546607411 | 0.2786010 | 3.788238e+00 | 0.0516139475 | 0.67425631 | No | Yes | 4.178001e+02 | 55.414822 | 6.078152e+02 | 82.230640 | ||
| ENSG00000258674 | protein_coding | E7ENQ6 | FUNCTION: Complex I functions in the transfer of electrons from NADH to the respiratory chain. Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. {ECO:0000256|RuleBase:RU368034}. | Coiled coil;Electron transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Respiratory chain;Transmembrane;Transmembrane helix;Transport | mitochondrial respiratory chain complex I [GO:0005747]; NADH dehydrogenase (ubiquinone) activity [GO:0008137] | ENSMUSG00000048967 | Yjefn3 | 3.742710e+02 | 0.8579220 | -0.221081578 | 0.2964692 | 5.482965e-01 | 0.4590145874 | 0.86403075 | No | Yes | 2.752426e+02 | 33.919787 | 3.397690e+02 | 42.705737 | ||||||
| ENSG00000258857 | lncRNA | 2.049594e+01 | 0.9419658 | -0.086253418 | 0.6551434 | 1.651665e-02 | 0.8977397435 | 0.98099463 | No | Yes | 2.021958e+01 | 5.411178 | 1.813376e+01 | 5.110985 | ||||||||||||
| ENSG00000259066 | protein_coding | G3V3P5 | Mouse_homologues FUNCTION: E3 ubiquitin-protein ligase which is a component of the N-end rule pathway. Recognizes and binds to proteins bearing specific N-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation. {ECO:0000250}. | Reference proteome | Mouse_homologues PATHWAY: Protein modification; protein ubiquitination. | Mouse_homologues mmu:66622; | membrane [GO:0016020] | Mouse_homologues 24664117_These data provide the first evidence of ubiquitin ligase activity in mammalian spermatozoa and indicate UBR7 involvement in spermiogenesis. | ENSMUSG00000041712 | Ubr7 | 2.917097e+01 | 0.0594252 | -4.072781326 | 1.6634487 | 4.376038e+00 | 0.0364476238 | 0.61802832 | No | Yes | 7.395144e+00 | 3.335068 | 5.387782e+01 | 21.586232 | |||
| ENSG00000259209 | lncRNA | 1.690563e+01 | 1.1882395 | 0.248825713 | 0.6957615 | 1.273648e-01 | 0.7211801574 | No | Yes | 1.144907e+01 | 6.733762 | 1.041354e+01 | 6.214131 | |||||||||||||
| ENSG00000260596 | 100288687 | DUX4 | protein_coding | Q9UBX2 | FUNCTION: [Isoform 1]: Transcription factor that is selectively and transiently expressed in cleavage-stage embryos (PubMed:28459457). Binds to double-stranded DNA elements with the consensus sequence 5'-TAATCTAATCA-3' (PubMed:28459457, PubMed:28459454, PubMed:29572508, PubMed:30540931, PubMed:30315230). Binds to chromatin containing histone H3 acetylated at 'Lys-27' (H3K27ac) and promotes deacetylation of H3K27ac. In parallel, binds to chromatin that lacks histone H3 acetylation at 'Lys-27' (H3K27ac) and recruits EP300 and CREBBP to promote acetylation of histone H3 at 'Lys-27' at new sites (PubMed:26951377). Involved in transcriptional regulation of numerous genes, primarily as transcriptional activator, but mediates also repression of a set of target genes (PubMed:17984056, PubMed:27378237, PubMed:26951377, PubMed:28459457, PubMed:28459454, PubMed:29618456, PubMed:30540931, PubMed:29572508). Promotes expression of ZSCAN4 and KDM4E, two proteins with essential roles during early embryogenesis (PubMed:27378237, PubMed:26951377, PubMed:28459457, PubMed:29618456). Heterologous expression in cultured embryonic stem cells mediates also transcription of HERVL retrotransposons and transcripts derived from ACRO1 and HSATII satellite repeats (PubMed:28459457). May activate expression of PITX1 (PubMed:17984056). May regulate microRNA (miRNA) expression (PubMed:24145033). Inappropriate expression can inhibit myogenesis and promote apoptosis (PubMed:26951377, PubMed:28935672, PubMed:29618456). {ECO:0000269|PubMed:17984056, ECO:0000269|PubMed:24145033, ECO:0000269|PubMed:26951377, ECO:0000269|PubMed:27378237, ECO:0000269|PubMed:28459454, ECO:0000269|PubMed:28459457, ECO:0000269|PubMed:28935672, ECO:0000269|PubMed:29572508, ECO:0000269|PubMed:29618456, ECO:0000269|PubMed:30315230, ECO:0000269|PubMed:30540931}.; FUNCTION: [Isoform 2]: Probably inactive as a transcriptional activator, due to the absence of the C-terminal region that is important for transcriptional activation. Can inhibit transcriptional activation mediated by isoform 1. Heterologous expression of isoform 2 has no deleterious effect on cell survival. {ECO:0000269|PubMed:29618456}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | This gene is located within a D4Z4 repeat array in the subtelomeric region of chromosome 4q. The D4Z4 repeat is polymorphic in length; a similar D4Z4 repeat array has been identified on chromosome 10. Each D4Z4 repeat unit has an open reading frame (named DUX4) that encodes two homeoboxes; the repeat-array and ORF is conserved in other mammals. The encoded protein has been reported to function as a transcriptional activator of paired-like homeodomain transcription factor 1 (PITX1; GeneID 5307). Contraction of the macrosatellite repeat causes autosomal dominant facioscapulohumeral muscular dystrophy (FSHD). Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2015]. | hsa:100288687; | cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; apoptotic process [GO:0006915]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of G0 to G1 transition [GO:0070317]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 23966205_Results show that both DUX4-FL isoforms are expressed in facioscapulohumeral muscular dystrophy (FSHD) myotubes; DUX4-FL expression level is much lower in trapezius than in quadriceps myotubes, which is confirmed by the level of expression of DUX4 downstream genes 24723486_The distinct gene signature and immunoprofile of CIC-DUX4 sarcomas suggest a distinct pathogenesis from Ewing sarcoma. 24838473_There is a special role of the 4q/10q D4Z4 chromatin and the DUX4 open reading frame in facioscapulohumeral muscular dystrophy. 24861551_These findings demonstrate that the expression of DUX4 accounts for the majority of the gene expression changes in facioscapulohumeral dystrophy skeletal muscle together with an immune cell infiltration. 25326393_Our results demonstrate that FRG1 is a direct DUX4 transcriptional target uncovering a novel regulatory circuit contributing to Facioscapulohumeral muscular dystrophy. 25564732_This feedback loop illustrates an unexpected mode of autoregulatory behavior of a transcription factor, is consistent with 'bursts' of DUX4 expression in facioscapulohumeral muscular dystrophy muscle. 26113644_Loss of D4Z4 repression in facioscapulohumeral muscular dystrophy is observed as hypomethylation of the array accompanied by loss of repressive chromatin marks. [Review] 26192274_DUX4 mRNAs were induced during the differentiation of hMSCs into osteoblasts and that this process involved DUX4 and new longer protein forms. 26246499_Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. 26356006_Recent studies indicate that a combination of genetic and epigenetic factors that act on the D4Z4 repeat array determine the probability of DUX4 expression in skeletal muscle and disease penetrance and progression 26800124_CIC-DUX4 gene fusion is associated with Round cell sarcoma. 26816005_Interactions between DUX4 and DUX4c with cytoplasmic proteins play a major role during muscle differentiation. 26823969_Report DNA-binding sequence preferences of DUX4. 26872601_selective loss of H3K9me3 from the DUX4 locus is associated with expression of DUX4 in late-phase squamous differentiation of human keratinocytes in vitro and in vivo. 26951377_We show that a DUX4 minigene, bearing only the homeodomains and C-terminus, is transcriptionally functional and cytotoxic, and that overexpression of a nuclear targeted C-terminus impairs the ability of WT DUX4 to interact with p300 and to regulate target genes. 27019113_data shows that DUX4 can become an oncogenic driver as a result of somatic chromosomal rearrangements and that acute lymphoblastic leukemia in adolescents and young adults may be a clinical entity distinct from ALL at other ages 27079694_The aim of this study was to describe seven cases of CIC-DUX4 fusion-positive sarcomas, including the first reported example arising primarily in bone. Our series confirms that CIC-DUX4 fusion-positive sarcomas are aggressive tumours with an adverse prognosis 27245141_These novel inhibitors of DUX4 transcriptional activity may thus act on pathways or cofactors needed by DUX4 for transcriptional activation in these cells. 27265895_recurrent IGH-DUX4 or ERG-DUX4 fusions, ETV6-RUNX1-like gene-expression profile in B-cell precursor acute lymphoblastic leukaemia, is reported. 27519269_It has been proposed that the induction of DNA damage is a novel function of the DUX4 protein affecting myogenic differentiation of facioscapulohumeral dystrophy myoblasts. 27556182_We propose that DUX4 controls the cellular migration of mesenchymal stem cells through the CXCR4 receptor. 27664537_Targeted next-generation sequencing of CIC-DUX4 soft tissue sarcomas demonstrates low mutational burden and recurrent chromosome 1p loss. 27744317_Transcriptomic analysis showed that DUX4 operates through both target gene activation and repression to orchestrate a transcriptome characteristic of a less-differentiated cell state. 27776115_DUX4 rearrangement and overexpression is associated with acute lymphoblastic leukemia. 28171552_The study describes a model system for inducible DUX4 expression that enables reproducible and synchronized experiments and validates the fidelity and facioscapulohumeral dystrophy (FSHD) relevance of multiple distinct models of DUX4 expression. 28173143_DUX4 and Dux may regulate some common pathways, and despite diverging from a common progenitor under different selective pressures for millions of years, the two genes maintain partial functional homology. 28263188_Estrogens antagonize DUX4 transcriptional activity and its differentiation inhibitory function and support the protective role of these hormones toward facioscapulohumeral muscular dystrophy myoblast in in vitro differentiation. 28273136_MYC, DUX4, and EIF4A3 might contribute to facioscapulohumeral dystrophy pathophysiology 28346326_Findings show CIC-DUX4 sarcomas occur mostly in young adults within the somatic soft tissues, having a wide spectrum of morphology including round, epithelioid and spindle cells, and associated with an aggressive clinical course, with an inferior survival compared with Ewing sarcoma. Results support classification of CIC-rearranged tumors as an independent molecular and clinical subset of small blue round cell tumors. 28404587_Gene silencing of CIC-DUX4 as well as Ccnd2, Ret, and Bcl2 effectively inhibited CDS tumor growth in vitro 28459454_DUX4 activate genes associated with a cleavage-stage embryos in muscle cells. 28459457_DUX4 role in activating cleavage-stage genes and MERVL/HERVL retrotransposons. 28540412_results underscore the complexity of the region immediately downstream of the D4Z4 and uncover a previously unknown function for the beta-satellite region in Dux4 cleavage and polyadenylation. 28645808_Case Report: t(10;19) CIC-DUX4 undifferentiated small round cell sarcoma of the abdominal wall. 28744936_We discuss the involvement of this rearrangement in Facioscapulohumeral dystrophy (FSHD), since all mutations in SMCHD1 are not associated with D4Z4 hypomethylation and do not always segregate with the disease 28935672_The DUX4 homeodomains mediate inhibition of myogenesis and are functionally exchangeable with the Pax7 homeodomain. 29162933_Biallelic DUX4 expression lowers the threshold for disease presentation and is a modifier for disease severity in FSHD2. 29478599_The recent identification of aberrant activation of DUX4 transcription in Facioscapulohumeral muscular dystrophy. 29533181_These and other approaches identified the Nucleosome Remodeling Deacetylase (NuRD) and Chromatin Assembly Factor 1 (CAF-1) complexes as necessary for DUX4 repression in human skeletal muscle cells and induced pluripotent stem (iPS) cells.Furthermore, DUX4-induced expression of MBD3L proteins partly relieved this repression in FSHD muscle cells. 29741619_Dux4 regulates the expression of small RNAs in the facioscapulohumeral muscular dystrophy muscle cells. 29759937_Expression of DUX4-fl is regulated by multiple epigenetic pathways. 29899111_Cyclic AMP production reduces DUX4 expression through a protein kinase A-dependent mode of action in facioscapulohumeral muscular dystrophy patient myotubes. 30107443_We generated FSHD2-derived isogenic control clones with SMCHD1 mutation corrected by clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated 9 (Cas9) and homologous recombination and found in the myocytes obtained from these clones that DUX4 basal expression and the OS-induced upregulation were markedly suppressed due to an increase in the heterochromatic state at 4q35. 30122154_Sporadic DUX4 expression seen in FSHD myocytes is due to the incomplete repression by the PRC2 complex. 30281091_cis duplications of D4Z4 repeats explain DUX4 expression and disease presentation in FSHD2 families with unusual long D4Z4 repeats on 4qA chromosomes. 30312408_Authors examined the correlation between MRI characteristics, muscle pathology and expression of DUX4 target genes. Results show that the presence of elevated MRI short tau inversion recovery signal has substantial predictive value in identifying muscles with active disease as determined by histopathology and DUX4 target gene expression. 30322619_The study revealed the basis for target DNA recognition by DUX4. 30446688_Data indicate the anti-myogenic properties of double homeobox 4 protein (DUX4) in myogenic progenitor cells. 30540931_the crystal structure of the tandem homeodomains of DUX4 bound to DNA, is reported. 31018108_When misexpressed in facioscapulohumeral muscular dystrophy skeletal muscle, the DUX4 leads to accumulated muscle pathology. (Review) 31099776_Homeotic DUX4 Genes that Control Human Embryonic Development at the Two-Cell Stage Are Surrounded by Regions Contacting with rDNA Gene Clusters 31209064_For tracking reductions in DUX4 mRNA. 31243274_IGH-DUX4 translocation occurs on the silenced IGH allele in B-cell acute lymphoblastic leukemia. 31327741_DUX4-expressing cancers are characterized by low anti-tumor immune activity. DUX4 blocks interferon-gamma-mediated induction of MHC class I and antigen presentation. 31630170_DUX4-induced bidirectional HSATII satellite repeat transcripts form intranuclear double-stranded RNA foci in human cell models of FSHD. 31676591_Furthermore, the efficient suppression of DUX4 after restoring SMCHD1 levels by genome editing of the mutant allele provides further guidance for therapeutic strategies 31693889_NFE2L3 Controls Colon Cancer Cell Growth through Regulation of DUX4, a CDK1 Inhibitor. 31722199_DUX4-Induced Histone Variants H3.X and H3.Y Mark DUX4 Target Genes for Expression. 31844661_4-methylumbelliferone does not disrupt DUX4-C1QBP binding and has only a limited effect on DUX4 transcriptional activity, establishing that HA signaling has a central function in pathology and is a target for Facioscapulohumeral muscular dystrophy (FSHD) therapeutics 31886887_Expanding the differential of superficial tumors with round-cell morphology: Report of three cases of CIC-rearranged sarcoma, a potentially under-recognized entity. 32048619_these are relatively aggressive tumours, especially CIC-DUX4-positive sarcomas. 32083293_Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. 32086799_Consequences of epigenetic derepression in facioscapulohumeral muscular dystrophy. 32182342_Identification of G-quadruplex structures as potential regulators of DUX4 expression. 32242220_DUX4 expressing immortalized FSHD lymphoblastoid cells express genes elevated in FSHD muscle biopsies, correlating with the early stages of inflammation. 32278354_Transgenic mice expressing tunable levels of DUX4 develop characteristic facioscapulohumeral muscular dystrophy-like pathophysiology ranging in severity. 32347924_PAX7 target gene repression associates with FSHD progression and pathology over 1 year. 32576599_p38alpha Regulates Expression of DUX4 in a Model of Facioscapulohumeral Muscular Dystrophy. 32759720_Membrane Repair Deficit in Facioscapulohumeral Muscular Dystrophy. 32840647_Imaging features and clinical course of undifferentiated round cell sarcomas with CIC-DUX4 and BCOR-CCNB3 translocations. 33436523_Chromosome 10q-linked FSHD identifies DUX4 as principal disease gene. 33987655_Systemic antisense therapeutics inhibiting DUX4 expression ameliorates FSHD-like pathology in an FSHD mouse model. 34131248_CRISPR mediated targeting of DUX4 distal regulatory element represses DUX4 target genes dysregulated in Facioscapulohumeral muscular dystrophy. 34267371_p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. 34547766_Distinct clinical characteristics of DUX4- and PAX5-altered childhood B-lymphoblastic leukemia. 34880230_Human miRNA miR-675 inhibits DUX4 expression and may be exploited as a potential treatment for Facioscapulohumeral muscular dystrophy. 34880314_A proteomics study identifying interactors of the FSHD2 gene product SMCHD1 reveals RUVBL1-dependent DUX4 repression. 35082321_Facioscapulohumeral dystrophy transcriptome signatures correlate with different stages of disease and are marked by different MRI biomarkers. 35468884_Antiapoptotic Protein FAIM2 is targeted by miR-3202, and DUX4 via TRIM21, leading to cell death and defective myogenesis. | ENSMUSG00000048502 | Duxbl1 | 1.542409e+01 | 0.1394289 | -2.842397957 | 0.8428828 | 9.946400e+00 | 0.0016116423 | No | Yes | 4.941759e+00 | 4.954798 | 4.653200e+01 | 47.616491 | ||
| ENSG00000261787 | 100129654 | TCF24 | protein_coding | Q7RTU0 | FUNCTION: Putative transcription factor. {ECO:0000250}. | DNA-binding;Nucleus;Reference proteome;Transcription;Transcription regulation | hsa:100129654; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; protein dimerization activity [GO:0046983]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; developmental process [GO:0032502]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000099032 | Tcf24 | 2.806469e+01 | 0.9889289 | -0.016061335 | 0.5803218 | 7.445250e-04 | 0.9782316348 | 0.99623234 | No | Yes | 2.740291e+01 | 7.455088 | 2.514156e+01 | 7.196779 | |||
| ENSG00000262583 | TMEM231P1 | transcribed_unprocessed_pseudogene | 1.494094e+01 | 1.1324572 | 0.179456584 | 0.7421579 | 5.594774e-02 | 0.8130193048 | No | Yes | 1.974613e+01 | 11.969687 | 2.247861e+01 | 13.773477 | ||||||||||||
| ENSG00000262919 | 92002 | CCNQ | protein_coding | Q8N1B3 | FUNCTION: Activating cyclin for the cyclin-associated kinase CDK10. {ECO:0000269|PubMed:18297069, ECO:0000269|PubMed:24218572}. | Acetylation;Alternative splicing;Cyclin;Reference proteome | Mutations in this gene have been shown to cause an X-linked dominant STAR syndrome that typically manifests syndactyly, telecanthus and anogenital and renal malformations. The protein encoded by this gene contains a cyclin-box-fold domain which suggests it may have a role in controlling nuclear cell division cycles. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Oct 2008]. | hsa:92002; | cyclin-dependent protein kinase holoenzyme complex [GO:0000307]; nucleus [GO:0005634]; cyclin-dependent protein serine/threonine kinase regulator activity [GO:0016538]; regulation of cell cycle G2/M phase transition [GO:1902749]; regulation of transcription by RNA polymerase II [GO:0006357] | 18297069_Mutations in the cyclin family member FAM58A cause an X-linked dominant disorder characterized by syndactyly, telecanthus and anogenital and renal malformations. 26882209_These ophthalmic findings are the first reported to our knowledge in association with STAR syndrome. The literature frequently demonstrates that patients with developmental anomalies often have ocular manifestations, warranting a full ophthalmic examination when the diagnosis of STAR syndrome has been made or is being considered. 28322501_this is the first occurrence of a nonsense variant in FAM58A described in individuals with STAR syndrome and the phenotype in this pedigree suggests that tethered cord and hearing loss are features of STAR syndrome. 34369103_Functional characterization of CDK10 and cyclin M truncated variants causing severe developmental disorders. 35291876_Functional characterization of the human Cdk10/Cyclin Q complex. | ENSMUSG00000049489 | Ccnq | 3.561228e+03 | 1.1427274 | 0.192481308 | 0.3242223 | 3.494926e-01 | 0.5544005212 | 0.89296216 | No | Yes | 3.820303e+03 | 475.953917 | 2.859195e+03 | 365.870439 | |
| ENSG00000263001 | 2969 | GTF2I | protein_coding | P78347 | FUNCTION: Interacts with the basal transcription machinery by coordinating the formation of a multiprotein complex at the C-FOS promoter, and linking specific signal responsive activator complexes. Promotes the formation of stable high-order complexes of SRF and PHOX1 and interacts cooperatively with PHOX1 to promote serum-inducible transcription of a reporter gene deriven by the C-FOS serum response element (SRE). Acts as a coregulator for USF1 by binding independently two promoter elements, a pyrimidine-rich initiator (Inr) and an upstream E-box. Required for the formation of functional ARID3A DNA-binding complexes and for activation of immunoglobulin heavy-chain transcription upon B-lymphocyte activation. {ECO:0000269|PubMed:10373551, ECO:0000269|PubMed:11373296, ECO:0000269|PubMed:16738337}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;DNA-binding;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Williams-Beuren syndrome | This gene encodes a phosphoprotein containing six characteristic repeat motifs. The encoded protein binds to the initiator element (Inr) and E-box element in promoters and functions as a regulator of transcription. This locus, along with several other neighboring genes, is deleted in Williams-Beuren syndrome. There are many closely related genes and pseudogenes for this gene on chromosome 7. This gene also has pseudogenes on chromosomes 9, 13, and 21. Alternatively spliced transcript variants encoding multiple isoforms have been observed. [provided by RefSeq, Jul 2013]. | hsa:2969; | cytoplasm [GO:0005737]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; negative regulation of angiogenesis [GO:0016525]; positive regulation of DNA repair by transcription from RNA polymerase II promoter [GO:0100026] | 12082086_G-kinase I beta interacted specifically with TFII-I, an unusual transcriptional regulator that associates with multiple proteins to modulate both basal and signal-induced transcription 12865760_GTF2IRD1 and GTF2I have roles in causing deficits on visual spatial functioning 14556246_Comparison of these five families with reports of other individuals with partial deletions of the WS region most strongly implicates GTF2I in the mental retardation of WS. 15664986_TFII-I is required for optimal induction of Grp78 by ER stress 15767439_These results demonstrate that USF1/USF2 and TFII-I interact cooperatively at the upstream RBEIII element and are necessary for the induction of latent HIV-1 in response to T-cell activation signals. 15941713_human VEGFR-2 promoter is functionally counter-regulated by TFII-I and TFII-IRD1. 16166082_cGMP-dependent protein kinase Ibeta binds to TFII-I and IRAG through a common interaction motif 16314517_TFII-I is recruited to the cyclin D1 promoter and transcriptionally activates this gene. 16738337_TFII-I directly interacts with Bright through amino acids in Bright's protein interaction domain 16943425_The data suggest that TFII-I and USF regulate chromatin structure accessibility and recruitment of transcription complexes in the beta-globin gene locus. 17023658_observations suggest a model in which TFII-I suppresses agonist-induced calcium entry by competing with TRPC3 for binding to phospholipase C-gamma 18445785_TFII-I, PARP1, and SFPQ proteins, each previously implicated in gene regulation, form a complex controlling transcription of DYX1C1. Allelic differences in the promoter or 5'UTR of DYX1C1 may affect factor binding and thus regulation of the gene. 18579769_Bioinformatics and microarray results were combined to identify TFII-I downstream targets in the vertebrate genome. 18976654_These results demonstrate an essential role of TFII-I bound at an upstream LTR element for viral replication. 19097122_alpha(2)-macroglobulin induced increase in GRP78 synthesis is caused by transcriptional upregulation of TFII-I 19111598_analyzed promoter regions of TFII-I genes and described their additional exons, as well as tested tissue specificity of both previously reported and novel alternatively spliced isoforms 19182516_Results suggest that TFII-I appears to have distinct roles in distinct phases of the mammalian cell cycle. 19701889_these results provide an initial step in understanding the biological role of Itk-TFII-I signaling in T-cell function. 19880526_TFII-I plays an inhibitory role in regulating genes that are essential in osteogenesis and intersects with the bone-specific transcription factor Runx2 and the retinoblastoma protein, pRb. 19897463_functional hemizygosity for the GTF2I and GTF2IRD1 genes is the main cause of the neurocognitive profile and some aspects of the gestalt phenotype of Williams-Beuren syndrome 21328569_These data indicate that Gtf2i is involved in several aspects of embryonic development and the development of social neurocircuitry and that GTF2I haploinsufficiency could be a contributor to the hypersociability in WBS patients. 21407215_TFII-I may modulate the cellular functions of BRCA1. 21503958_Data demonstrate that not only does cell surface GRP78 regulate apoptosis, but it also regulates Ca(2+) homeostasis by controlling cell surface localization of TFII-I. 21549311_Data show that Igh 3' enhancer-bound OCA-B and promoter-bound TFII-I mediate promoter-enhancer interactions, in both cis and trans, that are important for Igh transcription. 21813151_These data indicate that an E-box motif (RBE1) within the core promoter in the long terminal repeat of HIV-1 is a bona fide binding site for the RBF-2 transcription factor complex USF1, USF2, and TFII-I. 22048961_Our findings suggest the GTF2i gene is important in the etiology of autism in individuals with this duplication and in non-duplication cases with severe social interaction problems and repetitive behaviors. 22566418_TFII-I gene deletion may explain the Williams-Beuren syndrome phenotype because it acts as a negative regulator of TRPC3 expression in human B lymphocytes. 22578324_GTF2I duplication results in separation anxiety in mice and humans 22608712_CLIP2 haploinsufficiency by itself does not lead to the physical or cognitive characteristics of the Williams-Beuren syndrome; GTF2IRD1 and GTF2I are the main genes causing the cognitive defects 24231951_Results reveal novel mechanisms by which TFII-I and DBC1 can modulate cellular fate by affecting cell-cycle control as well as the homologous recombination pathway. 24922507_TFII-I bridges Proliferating Cell Nuclear Antigen and Polzeta to promote Translesion synthesis 24974848_GTF2I mutation correlated with better survival. 25429715_Findings implicate the GTF2I gene in the neurogenetic basis of social communication and social anxiety, both in Williams syndrome and among individuals in healthy populations 25480810_The GTF2I rs117026326 polymorphism is associated with anti-SSA-positive primary Sjogren's syndrome. 25501393_A proportion of this transcriptional dysregulation is caused by dosage imbalances in GTF2I, which encodes a key transcription factor at 7q11.23 that is associated with the LSD1 repressive chromatin complex and silences its dosage-sensitive targets. 25869096_show that SUMOylation is critical for TFII-I to promote cell proliferation and colony formation. Our findings contribute to understanding the role of SUMOylation in liver cancer development 25926634_Rather than contributing positively to promoter activity, a putative initiator element at the transcription start site acts as a target for negative regulation imposed on the L4P promoter of human adenovirus Type 5 by cellular TFII-I. 26285132_Copy-number variation in the general transcription factor gene, GTF2I is associated with gene-dose-dependent anxiety in mouse models and in both Williams syndrome and Dup7. 26320362_Study demonstrates a significant association between SLE in Chinese Han population and the GTF2I rs117026326 T allele/GTF2IRD1 rs4717901 C allele. 26656605_A novel interaction between TFII-I and Mdm2 with a negative effect on TFII-I transcriptional activity has been documented. 26814176_The authors found that human adenovirus 5 infection or ectopic E4-ORF3 expression leads to SUMOylation of TFII-I that precedes a rapid decline in TFII-I protein levels. 26853120_A common polymorphism in the Williams syndrome gene GTF2I associated with reduced social anxiety predicts decreased threat-related amygdala reactivity, which mediates an association between genotype and increased warmth in women. 27272985_we identified largest-ever effect on Asian rheumatoid arthritis across human non-HLA regions at GTF2I by heterogeneity mapping followed by replication studies, and pinpointed a possible causal variant 27503288_We identified rs117026326 on GTF2I with GWAS significance (P = 1.10 x 10(-15)) and rs13079920 on RBMS3 with suggestive significance (P = 2.90 x 10(-5)) associating with Primary Sjogren's syndrome in women. 28085512_TFII-I factors and the neural crest master regulator AP2alpha associate with the promoters of key genes important in the development of facial and dental structures. GTF2I and GTF2IRD1 are deleted in Williams-Beuren syndrome. 28424317_the results reported here support a model whereby common genetic variation in GTF2I mediates human sociality and anxiety via effects on oxytocin reactivity. 28448514_a new BRAF fusion in pilocytic astrocytoma 28461154_GTF2I/Gtf2i, its physiologic role in human disorders was relatively unknown until recently. Novel studies show that it is involved in an array of human diseases including neurocognitive disorders, systemic lupus erythematosus (SLE), and cancer. 28676218_the frequency of GTF2I mutation is higher in more indolent TETs and correlated with better prognosis. 28982856_GTF2I mutations common in thymic epithelial tumors are not present/uncommon in hematological malignancies. 29438696_demonstrate a marked prevalence of a thymoma-specific mutated oncogene, GTF2I 31374066_A case-control study identified the involvement of GTF2I gene polymorphisms in development of systemic lupus erythematosus, particularly in renal involvement. 31520790_A significant genetic association in the variant rs117026326 of GTF2I and neuromyelitis optica spectrum disorder was observed in a Norther Han Chinese population. 32349160_A small 7q11.23 microduplication involving GTF2I in a family with intellectual disability. 32544250_TFII-I-mediated polymerase pausing antagonizes GLI2 induction by TGFbeta. 32700803_Multiomics data reveals the influences of myasthenia gravis on thymoma and its precision treatment. 32907914_Gene of the month: GTF2I. 33636181_Binding of Gtf2i-beta/delta transcription factors to the ARMS2 gene leads to increased circulating HTRA1 in AMD patients and in vitro. 34036345_Sjogren's syndrome-associated SNPs increase GTF2I expression in salivary gland cells to enhance inflammation development. 34248934_Association of GTF2I, NFKB1, and TYK2 Regional Polymorphisms With Systemic Sclerosis in a Chinese Han Population. 34280590_Role of the multifunctional transcription factor TFII-I in DNA damage repair. 34497477_GTF2I Mutation in Thymomas: Independence From Racial-Ethnic Backgrounds. An Indian/German Comparative Study. | ENSMUSG00000060261 | Gtf2i | 8.254759e+03 | 1.1754166 | 0.233172192 | 0.2528106 | 8.470102e-01 | 0.3573995215 | 0.83189236 | No | Yes | 9.166215e+03 | 1108.373817 | 6.994586e+03 | 867.672405 | |
| ENSG00000264247 | 400657 | ZNF407-AS1 | lncRNA | 31132669_LINC00909 could act as a ceRNA to interact with miR-194 and thereby up-regulate the expression of MUC1-C. | 1.113433e+02 | 0.7765520 | -0.364845526 | 0.4304041 | 6.938051e-01 | 0.4048731215 | 0.84651196 | No | Yes | 9.601789e+01 | 20.257845 | 1.152278e+02 | 24.916723 | |||||||||
| ENSG00000266074 | 57597 | BAHCC1 | protein_coding | Q9P281 | Acetylation;Coiled coil;Phosphoprotein;Reference proteome | hsa:57597; | chromatin binding [GO:0003682] | 27928163_variants. We identified two candidate homozygous missense variants, R942Q in the tubulin-folding cofactor D (TBCD) gene and H250Q in the bromo-adjacent homology domain and coiled-coil containing 1 (BAHCC1) gene, located on chromosome 17q25.3 with an interval of 1.4 Mbp. 33139953_BAHCC1 binds H3K27me3 via a conserved BAH module to mediate gene silencing and oncogenesis. | ENSMUSG00000039741 | Bahcc1 | 1.528588e+03 | 1.0001723 | 0.000248536 | 0.2766262 | 7.393031e-07 | 0.9993139571 | 0.99979695 | No | Yes | 1.367516e+03 | 203.493488 | 1.441392e+03 | 220.246113 | |||
| ENSG00000267374 | 647946 | MIR924HG | lncRNA | 32831137_LINC00669 insulates the JAK/STAT suppressor SOCS1 to promote nasopharyngeal cancer cell proliferation and invasion. | 8.210252e+01 | 0.9247470 | -0.112869386 | 0.3916115 | 8.255968e-02 | 0.7738581175 | 0.95445095 | No | Yes | 8.728035e+01 | 13.198793 | 8.567844e+01 | 13.104282 | |||||||||
| ENSG00000267680 | 7767 | ZNF224 | protein_coding | Q9NZL3 | FUNCTION: May be involved in transcriptional regulation as a transcriptional repressor. The DEPDC1A-ZNF224 complex may play a critical role in bladder carcinogenesis by repressing the transcription of the A20 gene, leading to transport of NF-KB protein into the nucleus, resulting in suppression of apoptosis of bladder cancer cells. {ECO:0000269|PubMed:20587513}. | 3D-structure;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the Krueppel C2H2-type zinc-finger family of proteins. The encoded protein represses transcription of the aldolase A gene, which encodes a key enzyme in glycolysis. The encoded zinc-finger protein may also function as a transcriptional co-activator with Wilms' tumor protein 1 to regulate apoptotic genes in leukemia. [provided by RefSeq, Jul 2016]. | hsa:7767; | nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357] | 12239212_Results suggest that bone marrow zinc finger 2 (BMZF2) interferes with the transactivation potential of Wilms tumor suppressor gene (WT1). 12527367_We demonstrate that ZNF224, a Kruppel-like zinc finger transcription factor, is the repressor protein that specifically binds to the negative cis-element AldA-NRE and affects the AldA-NRE-mediated transcription 17900823_Differential expression and cellular localization of ZNF224 and ZNF255, two isoforms of the Kruppel-like zinc-finger protein family. 19505435_CIC silencer activity extends over 26 bp (-595/-569), which specifically bind a protein ZNF224 present in HepG2 cell nuclear extracts. 19741270_ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20574532_Observational study of gene-disease association. (HuGE Navigator) 20587513_Coimmunoprecipitation and immunocytochemistry revealed that DEPDC1 interacted and colocalized with zinc finger transcription factor ZNF224, a known transcriptional repressor. 20591825_ZNF224 acts as a transcriptional co-regulator of WT1. 21187159_REVIEW: ZNF224 is a multifunctional protein and alternative splicing, sub-cellular compartmentalization and isoform-specific interactions may modulate its activity 23362234_ZNF224 acts in fine tuning of WT1-dependent control of gene expression, acting as a co-activator of WT1 in the regulation of proapoptotic genes and suppressing WT1 mediated transactivation of antiapoptotitc genes. 26320177_Data show that Wilms' tumor 1 protein (WT1)binds to the zinc finger protein 224 (ZNF224) promoter and represses ZNF224 expression. 27105517_The analyses using human breast ductal carcinoma tissues exhibited that the expression of ZNF224 and miR-663a was increased in cancer compared to non-cancer region. 28040726_we showed that ZNF224 positively modulates cyclin D3 gene expression. Consistently, we observed that alteration of ZNF224 expression leads to defects in cell cycle control. All together, our results strongly suggest that in Chronic lymphocytic leukaemia (CLL)cells high expression level of ZNF224 can lead to inappropriate cell growth and apoptosis resistance, thus contributing to CLL progression 28215224_Increasing information on the mechanism through which ZNF224 can operate could lead to the identification of agents capable of modulating ZNF224 function, thus potentially paving the way to new therapeutic strategies for treatment of cancer 30176265_identify the receptor tyrosine kinase Axl as a novel target of ZNF224 transcriptional repression activity. 34181020_ZNF224 is a mediator of TGF-beta pro-oncogenic function in melanoma. 34684876_ZNF224 Protein: Multifaceted Functions Based on Its Molecular Partners. | 2.594120e+02 | 0.5950305 | -0.748964392 | 0.3656573 | 4.022144e+00 | 0.0449065834 | 0.64873770 | No | Yes | 1.332680e+02 | 31.840850 | 2.712139e+02 | 65.798387 | |||
| ENSG00000267750 | RUNDC3A-AS1 | lncRNA | 5.032251e+01 | 0.5455694 | -0.874165450 | 0.4694151 | 3.224642e+00 | 0.0725376386 | 0.71859192 | No | Yes | 1.576245e+01 | 6.646479 | 3.284271e+01 | 14.155235 | |||||||||||
| ENSG00000268750 | protein_coding | M0QXF5 | Reference proteome | 9.914538e+01 | 0.6725815 | -0.572219077 | 0.5183083 | 1.251793e+00 | 0.2632102758 | 0.79788700 | No | Yes | 4.958111e+01 | 29.489150 | 8.882934e+01 | 54.121209 | ||||||||||
| ENSG00000268858 | 112268269 | lncRNA | 4.959015e+02 | 1.1301864 | 0.176560741 | 0.3020525 | 3.441046e-01 | 0.5574695205 | 0.89423300 | No | Yes | 5.221241e+02 | 111.150548 | 4.902332e+02 | 107.630013 | |||||||||||
| ENSG00000269293 | 100129195 | ZSCAN16-AS1 | lncRNA | 34097562_ZSCAN16-AS1 expedites hepatocellular carcinoma progression via modulating the miR-181c-5p/SPAG9 axis to activate the JNK pathway. 34498716_Long noncoding RNA ZSCAN16AS1 promotes the malignant properties of hepatocellular carcinoma by decoying microRNA451a and consequently increasing ATF2 expression. | 7.364346e+02 | 1.3018222 | 0.380532415 | 0.3198976 | 1.406420e+00 | 0.2356515383 | 0.78818582 | No | Yes | 8.685997e+02 | 121.594367 | 5.273080e+02 | 76.185848 | |||||||||
| ENSG00000269386 | 100507567 | RAB11B-AS1 | lncRNA | 31900259_HIF2-Induced Long Noncoding RNA RAB11B-AS1 Promotes Hypoxia-Mediated Angiogenesis and Breast Cancer Metastasis. | 9.338977e+01 | 1.1055045 | 0.144704897 | 0.3798945 | 1.420623e-01 | 0.7062393365 | 0.93718130 | No | Yes | 8.364251e+01 | 13.141666 | 7.746527e+01 | 12.591411 | |||||||||
| ENSG00000269858 | 112398 | EGLN2 | protein_coding | Q96KS0 | FUNCTION: Prolyl hydroxylase that mediates hydroxylation of proline residues in target proteins, such as ATF4, IKBKB, CEP192 and HIF1A (PubMed:11595184, PubMed:12039559, PubMed:15925519, PubMed:16509823, PubMed:17114296, PubMed:23932902). Target proteins are preferentially recognized via a LXXLAP motif (PubMed:11595184, PubMed:12039559, PubMed:15925519). Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins (PubMed:11595184, PubMed:12039559, PubMed:12181324, PubMed:15925519, PubMed:19339211). Hydroxylates a specific proline found in each of the oxygen-dependent degradation (ODD) domains (N-terminal, NODD, and C-terminal, CODD) of HIF1A (PubMed:11595184, PubMed:12039559, PubMed:12181324, PubMed:15925519). Also hydroxylates HIF2A (PubMed:11595184, PubMed:12039559, PubMed:15925519). Has a preference for the CODD site for both HIF1A and HIF2A (PubMed:11595184, PubMed:12039559, PubMed:15925519). Hydroxylated HIFs are then targeted for proteasomal degradation via the von Hippel-Lindau ubiquitination complex (PubMed:11595184, PubMed:12039559, PubMed:15925519). Under hypoxic conditions, the hydroxylation reaction is attenuated allowing HIFs to escape degradation resulting in their translocation to the nucleus, heterodimerization with HIF1B, and increased expression of hypoxy-inducible genes (PubMed:11595184, PubMed:12039559, PubMed:15925519). EGLN2 is involved in regulating hypoxia tolerance and apoptosis in cardiac and skeletal muscle (PubMed:11595184, PubMed:12039559, PubMed:15925519). Also regulates susceptibility to normoxic oxidative neuronal death (PubMed:11595184, PubMed:12039559, PubMed:15925519). Links oxygen sensing to cell cycle and primary cilia formation by hydroxylating the critical centrosome component CEP192 which promotes its ubiquitination and subsequent proteasomal degradation (PubMed:23932902). Hydroxylates IKBKB, mediating NF-kappa-B activation in hypoxic conditions (PubMed:17114296). Also mediates hydroxylation of ATF4, leading to decreased protein stability of ATF4 (By similarity). {ECO:0000250|UniProtKB:Q91YE2, ECO:0000269|PubMed:11595184, ECO:0000269|PubMed:12039559, ECO:0000269|PubMed:12181324, ECO:0000269|PubMed:15925519, ECO:0000269|PubMed:16509823, ECO:0000269|PubMed:17114296, ECO:0000269|PubMed:19339211, ECO:0000269|PubMed:23932902}. | 3D-structure;Alternative initiation;Dioxygenase;Iron;Metal-binding;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Ubl conjugation;Vitamin C | The hypoxia inducible factor (HIF) is a transcriptional complex that is involved in oxygen homeostasis. At normal oxygen levels, the alpha subunit of HIF is targeted for degration by prolyl hydroxylation. This gene encodes an enzyme responsible for this post-translational modification. Alternative splicing results in multiple transcript variants. Read-through transcription also exists between this gene and the upstream RAB4B (RAB4B, member RAS oncogene family) gene. [provided by RefSeq, Feb 2011]. | hsa:112398; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 2-oxoglutarate-dependent dioxygenase activity [GO:0016706]; ferrous iron binding [GO:0008198]; L-ascorbic acid binding [GO:0031418]; oxygen sensor activity [GO:0019826]; peptidyl-proline 4-dioxygenase activity [GO:0031545]; peptidyl-proline dioxygenase activity [GO:0031543]; cell redox homeostasis [GO:0045454]; cellular response to hypoxia [GO:0071456]; intracellular estrogen receptor signaling pathway [GO:0030520]; peptidyl-proline hydroxylation to 4-hydroxy-L-proline [GO:0018401]; positive regulation of protein catabolic process [GO:0045732]; regulation of cell growth [GO:0001558]; regulation of neuron apoptotic process [GO:0043523]; response to hypoxia [GO:0001666] | 12351678_identified a human homolog of Caenorhabditis elegans Egl9 as a HIF prolyl hydroxylase 15156561_effect of WT or mutated VHL on PHD 1, 2, and 3 15247232_PHD1, PHD2, and PHD3 have roles in the regulation of hypoxia-inducible factor 16509823_defines the existence of two species of PHD1 and provide evidence that they are generated by alternative translational initiation 16649251_Understanding the mechanisms by which nickel can inhibit HIF-PHD's and stabilize HIF-1alpha may be important in the treatment of cancer and ischemic diseases. 17101781_Multiple mitochondrial products, including tricarboxylic acid intermediates and reactive oxygen species, can coordinate PHD activity, HIF stabilization, and cellular responses to O(2) depletion. 17114296_hypoxia releases repression of NFkappaB activity through decreased prolyl hydroxylase-dependent hydroxylation of IKKbeta 18710826_Results describe the expression, purification and characterization of the human prolyl hydroxylase PHD1 in Escherichia coli. 18773095_The identification of EGLN2 as a significantly deregulated gene that maps within the paired chromosome region directly implicates defects in the oxygen-sensing network to the biology of renal oncocytoma. 18927305_Overexpression of the oxygen sensor PHD-1 is associated with tumor aggressiveness in pancreatic endocrine tumors. 19134330_The expression imbalance of HPH1 and FIH-1 in placenta may play an important role in the pathogenesis and development of severe pre-eclampsia through inhibiting HIF-1alpha. 19147576_Authors identify an interaction between melanoma antigen-11 (MAGE-11) cancer-testis antigen and the major HIF-alpha hydroxylating enzyme PHD2. 19229863_Prolyl hydroxylase inhibitor reduced mitochondrial cytochrome c release, nuclear translocation of apoptosis inducing factor (AIF), and promoted Akt phosphorylation 19339211_Results describe the localization signal of HIF-prolyl hydroxylases (PHDs) 1-3, and their intracellular localization in response to hypoxia. 19631610_these findings provide new insights into the mechanisms of the regulation of the oxygen sensor cascade of PHD1 and PHD2 in different cellular compartments. 20418890_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20574001_Prolyl hydroxylase-dependent (but hypoxia inducible factor HIF-1alpha and -2alpha-independent) activation of hypoxia-induced monocyte-endothelial adhesion assigns a new function to monocytic ICAM-1 under acute hypoxic conditions. 20600011_role for PHD1 as a positive regulator of intestinal epithelial cell apoptosis in the inflamed colon 20727020_Methylation-induced epigenetic silencing of PHD1, PHD2, PHD3 and FIH is unlikely to underlie up-regulated HIF-1alpha expression in human breast cancer but may play a role in other tumour types. 20840881_human prolyl hydroxylase might play an important role in determining the physiology and structure of the corpora lutea during the menstrual cycle and early pregnancy 20959442_Observational study of gene-disease association. (HuGE Navigator) 20978146_Elevated PHD1 concomitant with decreased PHD2 are causatively related to Rpb1 hydroxylation and oncogenesis in human renal clear cell carcinomas with WT VHL gene. 21748337_The expression of PHD genes and their relationship with the tumor behavior and apoptosis-associated factors in non-small cell lung cancer, was investigated. 21877141_PHD1 expression correlated with high proliferation, and these tumors were mainly estrogen receptor-negative. 21887331_PHD1 and PHD2 are independent negative prognostic factors in NSCLC. 21951999_Coexistence of PHD1 stabilized ATF4, as opposed to the destabilization of ATF4 by PHD3. 22343896_(R)-2HG, but not (S)-2HG, stimulates EGLN activity, leading to diminished HIF levels, which enhances the proliferation and soft agar growth of human astrocytes 22395314_Here, we outline specific functions of PHD enzymes in surgically relevant pathological conditions, and discuss how these functions might be exploited in order to support the treatment of surgically relevant diseases. 22859986_miR-205 serves a protective role against both oxidative and endoplasmic reticulum stresses via the suppression of EGLN2 and subsequent decrease in intracellular reactive oxygen species. 23026137_Findings provided strong evidence for the hypothesis that rs10680577 contributes to hepatocarcinogenesis, possibly by affecting RERT-lncRNA structure and subsequently EGLN2 expression. 23077531_Principal component analysis of the covariance matrix of free AIRE-PHD1 highlights the presence of a 'flapping' movement, which is blocked in an open conformation upon binding to H3K4me0. 23531419_Onconeuronal antigen Cdr2 correlates with HIF prolyl-4-hydroxylase PHD1 and worse prognosis in renal cell carcinoma. 23932902_By modulating Cep192 levels, PHD1 thereby affects the processes of centriole duplication and centrosome maturation and contributes to the regulation of cell-cycle progression. 24045616_Results show that variants in two adjacent genes, EGLN2 and CYP2A6, influence smoking behavior related to disease risk. 24195777_The diminished expression of PHD1 and PHD2 and elevated level of FIH protein in cancerous tissue compared to histopathologically unchanged colonic mucosa was not associated with DNA methylation within the CpG islands of the PHD1, PHD2 and FIH genes. 24517638_study conducted to investigate the association between gastric cancer (GC) susceptibility with a 4-bp insertion/deletion polymorphism (rs10680577) in the proximal promoter of EGLN2; findings showed that the heterozygote and the homozygote 4-bp del/del confer a significantly increased risk of GC 24644426_PHD-1 played an important role in hypoxic response pathway of trophoblast through modulating the level of HIF-2alpha. 24894671_Data indicate that the prolyl hydrolase 1 (PHD1) rs10680577 polymorphism is associated with the risk of non-small cell lung cancer in a Chinese population. 24935227_PHD1 could induce cell cycle arrest in lung cancer cells, resulting in the suppression of cell proliferation. 25263965_Germ-line PHD1 and PHD2 mutations detected in patients with pheochromocytoma/paraganglioma-polycythemia 25609945_rs3733829 in the EGLN2 gene is significantly associated with the risk of COPD in Chinese populations of Hainan province. 26492917_EglN2 associates with the NRF1-PGC1alpha complex and controls mitochondrial function in breast cancer 26644182_PHD1 is phosphorylated by CDK2, CDK4 and CDK6 at Serine 130. 26997627_siRNA-mediated knockdown of PHD1 inhibited glucose-stimulated insulin secretion in pancreatic Beta cells. 27130823_This study provides new information relating to the possible mechanism of therapeutic action of hydroxylase inhibitors that has been reported in pre-clinical models of intestinal and hepatic disease. 27502280_Tuning the Transcriptional Response to Hypoxia by Inhibiting Hypoxia-inducible Factor (HIF) Prolyl and Asparaginyl Hydroxylases. 28036276_EglN2 might act as an FBW7 ubiquitin ligase substrate contributing to the progression of triple negative breast cancer. 28218358_Our study provided initial evidence that the insertion/deletion polymorphism rs10680577 may play a functional role in the development of CRC in the Chinese population 29277791_In advanced-stage Hodgkin's Lymphoma patients strong cytoplasmic PHD1 expression in Reed-Sternberg cells was associated with poor relapse-free survival among patients treated with involved-field radiotherapy and advanced-stage patients treated with doxorubicin, bleomycin, vinblastine and darcabazine and involved-field radiotherapy. 29476053_TCF7L2 positively regulated aerobic glycolysis by suppressing Egl-9 family hypoxia inducible factor 2 (EGLN2), leading to upregulation of hypoxia inducible factor 1 alpha subunit (HIF-1alpha). 29693343_4-bp Insertion/deletion Polymorphism within the Promoter of EGLN2 is associated with breast cancer. 30248340_High PHD1 expression is associated with liver fibrosis. 30665327_Study consistently found that hypermethylation at cg25923056-EGLN2 downregulated the corresponding gene EGLN2 expression in lung adenocarcinomas (LUAD) tumor tissues. Meanwhile, EGLN2 expression was negatively correlated with HIF1A expression in tumor tissues and significantly interacted with HIF1A expression on overall survival. 31414584_Meta-analysis showed that EGLN2 rs10680577 polymorphism was significantly associated with cancer risk. 31500697_The authors did not detect prolyl-hydroxylase activity on any reported non-HIF protein or peptide, using conditions supporting robust HIF-alpha hydroxylation. 31729379_Adenylosuccinate lyase (ADSL) is an EglN2 hydroxylase substrate in triple negative breast cancer. 32458625_Association of EGLN2 rs10680577 Polymorphism with the Risk and Clinicopathological Features of Patients with Prostate Cancer. | ENSMUSG00000058709 | Egln2 | 4.054927e+03 | 1.1934756 | 0.255169114 | 0.3121942 | 6.712195e-01 | 0.4126267668 | 0.84733931 | No | Yes | 4.195570e+03 | 529.566522 | 3.107598e+03 | 402.733749 | |
| ENSG00000270647 | 8148 | TAF15 | protein_coding | Q92804 | FUNCTION: RNA and ssDNA-binding protein that may play specific roles during transcription initiation at distinct promoters. Can enter the preinitiation complex together with the RNA polymerase II (Pol II). {ECO:0000269|PubMed:19124016}. | 3D-structure;ADP-ribosylation;Acetylation;Alternative splicing;Chromosomal rearrangement;Cytoplasm;DNA-binding;Direct protein sequencing;Isopeptide bond;Metal-binding;Methylation;Nucleus;Phosphoprotein;Proto-oncogene;RNA-binding;Reference proteome;Repeat;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the TET family of RNA-binding proteins. The encoded protein plays a role in RNA polymerase II gene transcription as a component of a distinct subset of multi-subunit transcription initiation factor TFIID complexes. Translocations involving this gene play a role in acute leukemia and extraskeletal myxoid chondrosarcoma, and mutations in this gene may play a role in amyotrophic lateral sclerosis. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, May 2012]. | hsa:8148; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; mRNA 3'-UTR binding [GO:0003730]; RNA binding [GO:0003723]; transcription coregulator activity [GO:0003712]; mRNA stabilization [GO:0048255]; positive regulation of transcription, DNA-templated [GO:0045893]; RNA splicing [GO:0008380] | 12359745_The transcription factor gene CIZ/NMP4 is recurrently involved in acute leukemia through fusion with either EWSR1 or TAF15. 15094065_hTAF(II)68-mediated transactivation is linked to the cytoplasmic Src signal transduction pathway. The hTAF(II)68 protein can associate with the SH3 domains of several cell signaling proteins, including v-Src. 18330902_The oncogenic effect of the t(9;17) translocation may be due to the hTAF(II)68-TEC chimeric protein and that fusion of the hTAF(II)68 NTD to the TEC protein produces a gain of function chimeric product. 18620564_FUS, EWS and TAF15 proto-oncoproteins were targeted to stress granules induced by heat shock and oxidative stress 19124016_Data demonstrate that arginine methylation of TAF15 by PRMT1 is a crucial event determining its proper localization and gene regulatory function. 19282884_Data show that a fraction of human U1 snRNA specifically associates with the nuclear RNA-binding protein TBP-associated factor 15 (TAF15). 19426707_These results suggest that caspase-mediated degradation may represent a novel regulatory mechanism that controls TAF15 and TAF15-CIZ/NMP4 activities. 20048082_Our findings define a role for a tumor-specific TAF15 antigen in malignant processes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21344536_Elevated TAF15 mRNA levels did not translate to strongly elevated protein levels, consistent with its infrequent occurrence as translocation partner in tumors. 21438137_These results suggest that additional studies are needed to determine whether mutations in the TAF15 gene represent a cause of familial amyotrophic lateral sclerosis. 21504714_Rare translocation t(12;17)(p13;q12), this translocation has been reported in 25 cases and its putative molecular consequence, the formation of a TAF15-ZNF384 fusion gene, in only six cases. 21856723_REsults suggest the possibility that alterations of TAF15 and EWS might also be involved in the pathogenesis of FUS proteinopathies such as ALS and FTLD. 22019700_The existence of a functionally discrete subset of U1 snRNP in association with TAF15 was suggested and provided further support for the involvement of U1 snRNP components in early steps of coordinated gene expression. 22065782_Missense mutations of TAF-15, an RNA-binding protein, were found in patients with amyotrophic lateral sclerosis, and gene conferred neurodegeneration when expressend in Drosophila. 22771914_TAF15 plays a role in RNA transport and/or local RNA translation 23049996_Data show that FUS and TAF15 locate to cellular stress granules to a larger extend than EWS. FET-protein stress granule association most likely is a downstream response to cellular stress. 23128393_TAF15 depletion inhibits growth & increases apoptosis. Its knockdown affects many genes involved in cell cycle & cell death. Among these, targets of microRNAs generated from the onco-miR-17 locus were overrepresented. 24055347_Data indicate that distinct differences in proteins becoming Poly(ADP-ribose) PARylated upon various genotoxic insults are observed, exemplified by the PARylation of RNA-processing factors THRAP3 and TAF15 under oxidative stress. 24474660_our data suggest that TAF15 and TLS/FUS operate within similar but not identical hnRNP M-TET protein complexes to influence the transcriptional or post-transcriptional output of a particular cell type. 26612539_RNA binding by TAF-15 is dependent upon structural elements in the RNA rather than sequence. 27311318_Aggregation of FET proteins FUS, EWSR1, and TAF15 mediate a pathological change in amyotrophic lateral sclerosis. (Review) 27378374_In human stem cell-derived motor neurons, the RNA profile associated with concomitant loss of both TAF15 and FUS resembles that observed in the presence of the amyotrophic lateral sclerosis (ALS)-associated mutation FUS R521G, but contrasts with late-stage sporadic ALS patients. 27415968_Studies provide evidence that FUS/TLS, EWS and TAF15 proteins play a major role in neurodegenerative disorders. (review). 27903134_O-GlcNAc glycosylation stoichiometry of TAF15 28291257_In a cohort of youth at risk for bipolar disorder, pathway analysis showed an enrichment of the glucocorticoid receptor (GR) pathway with the genes MED1, HSPA1L, GTF2A1 and TAF15, which might underlie the previously reported role of stress response in the risk for bipolar disorder in vulnerable populations. 28945358_weak, multivalent interactions between TAF15 fibrils and heptads throughout RNA pol II CTD collectively mediate complex formation. 29193371_Results find that the Asp residue in TAF15 YGGDR(S/G)G repeats confers poor binding to PRMT1 indicating that YGGDR(S/G)G repeats may contribute to TAF15 specialization by enabling differential interactions with PRMT1 and reduced overall levels of TAF15 methylation compared with other FET proteins. 30398641_FUS, EWSR1, TAF15 and MATR3 within the RNAP II/U1 snRNP machinery play distinct roles in the development of amyotrophic lateral sclerosis and spinal muscular atrophy that are more intimately tied to one another than previously thought. 31020999_NR4A3 fusion proteins trigger an axon guidance switch that marks the difference between EWSR1 and TAF15 translocated extraskeletal myxoid chondrosarcomas cells. 31887411_TRPM2-AS promotes cancer cell proliferation through control of TAF15. 32871048_LncRNA PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA. 32915460_SCF-Slimb is critical for Glycogen synthase kinase-3beta-mediated suppression of TAF15-induced neurotoxicity in Drosophila. 33674598_Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. 33875793_LncRNA GAS5 activates the HIF1A/VEGF pathway by binding to TAF15 to promote wound healing in diabetic foot ulcers. | ENSMUSG00000020680 | Taf15 | 8.141901e+03 | 1.3263191 | 0.407427864 | 0.2635862 | 2.379901e+00 | 0.1229053363 | 0.75814553 | No | Yes | 9.361221e+03 | 1303.756026 | 5.787894e+03 | 826.992491 | |
| ENSG00000272391 | 100101267 | POM121C | protein_coding | A8CG34 | FUNCTION: Essential component of the nuclear pore complex (NPC). The repeat-containing domain may be involved in anchoring components of the pore complex to the pore membrane. When overexpressed in cells induces the formation of cytoplasmic annulate lamellae (AL). {ECO:0000269|PubMed:17900573}. | Alternative splicing;Endoplasmic reticulum;Membrane;Nuclear pore complex;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Translocation;Transmembrane;Transmembrane helix;Transport;mRNA transport | Mouse_homologues mmu:107939; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; nuclear pore [GO:0005643]; nuclear localization sequence binding [GO:0008139]; structural constituent of nuclear pore [GO:0017056]; mRNA transport [GO:0051028]; protein import into nucleus [GO:0006606]; RNA export from nucleus [GO:0006405] | 29487953_Gene expression and adipocyte functional studies support the notion that FAM13A and POM121C control adipocyte lipolysis and adipogenesis, respectively, and might thereby be involved in genetic control of systemic insulin sensitivity | ENSMUSG00000053293 | Pom121 | 8.952587e+03 | 1.2263022 | 0.294314583 | 0.2921131 | 1.010355e+00 | 0.3148178344 | 0.81928722 | No | Yes | 8.661188e+03 | 899.145136 | 6.834528e+03 | 727.975844 | ||
| ENSG00000272438 | lncRNA | 2.887459e+01 | 0.6620331 | -0.595024766 | 0.6552002 | 7.614983e-01 | 0.3828600303 | 0.84285514 | No | Yes | 1.010850e+01 | 4.696248 | 1.529733e+01 | 7.311916 | ||||||||||||
| ENSG00000272645 | GTF2IP20 | transcribed_unprocessed_pseudogene | 1.568300e+03 | 0.8312618 | -0.266625144 | 0.2570557 | 1.072440e+00 | 0.3003949968 | 0.81383297 | No | Yes | 9.394614e+02 | 170.726851 | 1.339870e+03 | 249.450505 | |||||||||||
| ENSG00000272779 | BMS1P20 | transcribed_unprocessed_pseudogene | 4.300030e+02 | 1.0707163 | 0.098576336 | 0.2811507 | 1.231016e-01 | 0.7256946425 | 0.94139721 | No | Yes | 4.545027e+02 | 50.383933 | 3.868046e+02 | 44.143478 | |||||||||||
| ENSG00000273154 | 84619 | protein_coding | A0A0J9YYD9 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | B cell receptor complex [GO:0019815]; integral component of membrane [GO:0016021]; protein kinase binding [GO:0019901]; B cell receptor signaling pathway [GO:0050853]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; regulation of MAP kinase activity [GO:0043405]; regulation of NIK/NF-kappaB signaling [GO:1901222]; regulation of phosphatidylinositol 3-kinase signaling [GO:0014066]; regulation of release of sequestered calcium ion into cytosol [GO:0051279]; regulation of transcription by RNA polymerase II [GO:0006357]; T cell receptor signaling pathway [GO:0050852] | 6.130059e+02 | 0.7365231 | -0.441197294 | 0.2800815 | 2.443433e+00 | 0.1180176866 | 0.75783482 | No | Yes | 3.864591e+02 | 72.541583 | 5.943771e+02 | 113.994389 | ||||||||
| ENSG00000274333 | lncRNA | 4.436129e+02 | 0.6273679 | -0.672616394 | 0.3350959 | 4.018988e+00 | 0.0449907009 | 0.64873770 | No | Yes | 2.601266e+02 | 66.928773 | 4.667525e+02 | 122.660935 | ||||||||||||
| ENSG00000274425 | lncRNA | 7.047292e+02 | 1.2848111 | 0.361556264 | 0.3209441 | 1.271787e+00 | 0.2594317045 | 0.79684153 | No | Yes | 1.354114e+03 | 228.927241 | 1.118746e+03 | 203.533147 | ||||||||||||
| ENSG00000275066 | 11276 | SYNRG | protein_coding | Q9UMZ2 | FUNCTION: Plays a role in endocytosis and/or membrane trafficking at the trans-Golgi network (TGN) (PubMed:15758025). May act by linking the adapter protein complex AP-1 to other proteins (Probable). Component of clathrin-coated vesicles (PubMed:15758025). Component of the aftiphilin/p200/gamma-synergin complex, which plays roles in AP1G1/AP-1-mediated protein trafficking including the trafficking of transferrin from early to recycling endosomes, and the membrane trafficking of furin and the lysosomal enzyme cathepsin D between the trans-Golgi network (TGN) and endosomes (PubMed:15758025). {ECO:0000269|PubMed:15758025, ECO:0000305|PubMed:12538641}. | 3D-structure;Acetylation;Alternative splicing;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Endocytosis;Golgi apparatus;Membrane;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | This gene encodes a protein that interacts with the gamma subunit of AP1 clathrin-adaptor complex. The AP1 complex is located at the trans-Golgi network and associates specific proteins with clathrin-coated vesicles. This encoded protein may act to connect the AP1 complex to other proteins. Alternatively spliced transcript variants that encode different isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:11276; | AP-1 adaptor complex [GO:0030121]; clathrin coat of trans-Golgi network vesicle [GO:0030130]; cytoplasm [GO:0005737]; Golgi apparatus [GO:0005794]; perinuclear region of cytoplasm [GO:0048471]; endocytosis [GO:0006897]; intracellular protein transport [GO:0006886] | 15758025_the aftiphilin/p200/gamma-synergin complex facilitates AP-1 function 16162817_AP-3 and AP-1 function in partially redundant pathways to transfer tyrosinase from distinct endosomal subdomains to melanosomes 18815278_Data show that by recruiting aftiphilin/gamma-synergin in addition to clathrin, AP-1 coordinates formation of Weibel-Palade bodies with their acquisition of a regulated secretory phenotype. 35090779_Interacting with AP1 complex mutated synergin gamma (SYNRG) reveals a novel coatopathy in the form of complicated hereditary spastic paraplegia. | ENSMUSG00000034940 | Synrg | 7.159027e+02 | 0.8431505 | -0.246137952 | 0.3123607 | 6.329754e-01 | 0.4262659958 | 0.85078492 | No | Yes | 7.000422e+02 | 112.473620 | 7.274946e+02 | 119.635586 | |
| ENSG00000275496 | 102724701 | lncRNA | 5.261232e+01 | 0.4740608 | -1.076855906 | 1.5781825 | 4.822612e-01 | 0.4873998704 | 0.87510946 | No | Yes | 3.451871e+01 | 25.224474 | 9.039017e+01 | 67.122620 | |||||||||||
| ENSG00000275778 | 100533464 | protein_coding | A0A087WY73 | Reference proteome | This locus represents naturally occurring read-through transcription between the neighboring PRH1 (proline-rich protein HaeIII subfamily 1) and PRR4 (proline rich 4, lacrimal) genes on chromosome 12. The read-through transcript is a candidate for nonsense-mediated mRNA decay (NMD), and is thus unlikely to produce a protein product. [provided by RefSeq, Feb 2011]. | 3.184207e+01 | 0.5997117 | -0.737658899 | 0.5756763 | 1.544011e+00 | 0.2140217592 | 0.78763590 | No | Yes | 1.273362e+01 | 3.288656 | 1.654118e+01 | 4.177473 | ||||||||
| ENSG00000276234 | 6871 | TADA2A | protein_coding | A0A087WZS1 | Mouse_homologues FUNCTION: Component of the ATAC complex, a complex with histone acetyltransferase activity on histones H3 and H4 (By similarity). Required for the function of some acidic activation domains, which activate transcription from a distant site (By similarity). Binds double-stranded DNA (PubMed:16299514). Binds dinucleosomes, probably at the linker region between neighboring nucleosomes (PubMed:16299514). Plays a role in chromatin remodeling (By similarity). May promote TP53/p53 'Lys-321' acetylation, leading to reduced TP53 stability and transcriptional activity (By similarity). May also promote XRCC6 acetylation thus facilitating cell apoptosis in response to DNA damage (By similarity). {ECO:0000250|UniProtKB:O75478, ECO:0000269|PubMed:16299514}. | Metal-binding;Proteomics identification;Reference proteome;Zinc;Zinc-finger | Many DNA-binding transcriptional activator proteins enhance the initiation rate of RNA polymerase II-mediated gene transcription by interacting functionally with the general transcription machinery bound at the basal promoter. Adaptor proteins are usually required for this activation, possibly to acetylate and destabilize nucleosomes, thereby relieving chromatin constraints at the promoter. The protein encoded by this gene is a transcriptional activator adaptor and has been found to be part of the PCAF histone acetylase complex. Several alternatively spliced transcript variants encoding different isoforms of this gene have been described, but the full-length nature of some of these variants has not been determined. [provided by RefSeq, Oct 2009]. | Mouse_homologues mmu:217031; | Mouse_homologues ATAC complex [GO:0140672]; chromosome [GO:0005694]; mitotic spindle [GO:0072686]; nucleus [GO:0005634]; SAGA complex [GO:0000124]; SAGA-type complex [GO:0070461]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; transcription coactivator activity [GO:0003713]; chromatin remodeling [GO:0006338]; histone H3 acetylation [GO:0043966]; mitotic cell cycle [GO:0000278]; positive regulation of histone acetylation [GO:0035066]; regulation of cell cycle [GO:0051726]; regulation of cell division [GO:0051302]; regulation of embryonic development [GO:0045995]; regulation of histone deacetylation [GO:0031063]; regulation of protein phosphorylation [GO:0001932]; regulation of protein stability [GO:0031647]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of tubulin deacetylation [GO:0090043] | 16299514_The three-dimensional solution structure of the SWIRM domain from the human transcriptional adaptor ADA2alpha was reported. 18059173_hADA2a and hADA3 as crucial cofactors of beta-catenin that are likely involved in the assembly of transactivation-competent beta-catenin complexes at Wnt target genes. 20602615_Observational study of gene-disease association. (HuGE Navigator) 22644376_CCDC134 increased the PCAF-dependent K320 acetylation of p53 and p53 protein stability in the presence of hADA2a overexpression. 25492086_Single nucleotide polymorphism in ADA2 gene is associated with type 1 diabetes. 26468280_results thus demonstrate that the catalytic activity of GCN5 is stimulated by subunits of the ADA2a- or ADA2b-containing HAT modules and is further increased by incorporation of the distinct HAT modules in the ATAC or SAGA holo-complexes 34296306_Circular RNA TADA2A promotes proliferation and migration via modulating of miR638/KIAA0101 signal in nonsmall cell lung cancer. | ENSMUSG00000018651 | Tada2a | 3.823057e+02 | 0.6989215 | -0.516797656 | 0.3182243 | 2.595799e+00 | 0.1071474258 | 0.75783482 | No | Yes | 3.238401e+02 | 59.440080 | 3.708740e+02 | 69.610001 | |
| ENSG00000277053 | 2970 | GTF2IP1 | transcribed_unprocessed_pseudogene | 1.488336e+03 | 0.9116925 | -0.133380824 | 0.2631228 | 2.519418e-01 | 0.6157111222 | 0.91334267 | No | Yes | 1.319276e+03 | 172.945052 | 1.358159e+03 | 182.558588 | ||||||||||
| ENSG00000277117 | 102723996 | protein_coding | O75144 | FUNCTION: Ligand for the T-cell-specific cell surface receptor ICOS. Acts as a costimulatory signal for T-cell proliferation and cytokine secretion; induces also B-cell proliferation and differentiation into plasma cells. Could play an important role in mediating local tissue responses to inflammatory conditions, as well as in modulating the secondary immune response by co-stimulating memory T-cell function (By similarity). {ECO:0000250}. | 3D-structure;Adaptive immunity;Alternative splicing;B-cell activation;Cell membrane;Disulfide bond;Glycoprotein;Immunity;Immunoglobulin domain;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:23308; | cytoplasmic ribonucleoprotein granule [GO:0036464]; external side of plasma membrane [GO:0009897]; extracellular exosome [GO:0070062]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; signaling receptor binding [GO:0005102]; adaptive immune response [GO:0002250]; B cell activation [GO:0042113]; defense response [GO:0006952]; hyperosmotic response [GO:0006972]; positive regulation of activated T cell proliferation [GO:0042104]; regulation of cytokine production [GO:0001817]; signal transduction [GO:0007165]; T cell activation [GO:0042110]; T cell receptor signaling pathway [GO:0050852] | Mouse_homologues 11418641_This molecule acts as a costimulatory counterreceptor by augmenting the recall responses of CD8+ T cells. 12145647_These studies demonstrate that T(R) cells and the ICOS-ICOS-ligand signaling pathway are critically involved in respiratory tolerance and in downregulating pulmonary inflammation in asthma. 12370365_The involvement of the B7h-Icos costimulatory pathway has been demonstrated in the development and progression of collagen-induced arthritis. 12456022_ICOS-L is a ligand for ICOS and plays an important functional role in the activation of memory T cells by endothelial cells [review] 12594252_While interactions of B7RP-1 with ICOS have no effect on the migration of T cells into B cell follicles, they are required for Th1 and Th2 cells to support fulminant B cell responses in vivo. 12714510_B7-H2 on B cells is important for recruiting T-cell help via its interaction with ICOS and plays a critical role in costimulating humoral immune responses. 12853164_data suggest that ICOS/B7RP-1 interactions may not affect the organogenesis, but involve in the functional development of Peyer's patches 12865411_the ICOS-B7h pathway has roles in regulating alloimmune responses in vivo 14615582_Th cells, when activated with B7h-deficient APC, exhibited reduced proliferation and IL-2 production. 15636609_ICOS-B7h has a role in transplantation tolerance to MHC class II mismatched skin grafts 15728513_Experiments with mice genetically engineered for the absence of B7-related protein 1 (B7RP-1KO) demonstrate that efficient T helper 2 (TH2) cell sensitization and effector function occur in the absence of the ICOS-B7RP-1 costimulatory pathway. 15880041_ICOS-B7h pathway is critical in the activation of effector/memory T cells that are necessary for the progression of chronic rejection. 16081804_The B7RP-1/ICOS interaction plays an essential role in the development of CXCR-positive helper T cells in vivo, which preferentially migrate to the germinal center B cell zone where they provide cognate help to B cells. 16540559_ICOSL is upregulated in nephritic glomeruli, where it locally reduces accumulation of T cells and macrophages and attenuates renal injury. 16621988_critical role played by ICOS-L-expressing and interleukin 10-producing dendritic cells from Chlamydia-infected mice in the infection-mediated inhibition of allergic responses 16887997_B7h shedding defines a novel immunoregulatory mechanism by which toll-like receptor (TLR)7/8 and TLR9 signals can enhance T cell help during antibody responses. 17039566_These results suggest that the ICOS/B7RP-1 interaction plays a critical role in the pathogenesis of uveitis. 17975322_The low costimulatory molecule expression (PD-L1, ICOS-L and CD40) may explain the suboptimal T cell activation by renal proximal tubular epithelial cells. 18294651_The generated a 3T3 cellular library retrovirally expressing mutants of the murine costimulatory B7h gene. Screening of this unbiased cellular library identified six residues of murine B7h that are critical for binding to the ICOS receptor. 18390703_a novel mechanism for the regulation of ICOSL expression in vivo and suggest that the ICOS costimulatory pathway is subject to negative feedback regulation by ICOSL down-regulation in response to ICOS expression. 19155489_Levels of follicular T helper cells and germinal center B cells in two different models of autoimmunity are shown to be dependent on the maintenance of the inducible T-cell co-stimulator (ICOS)/B7-related protein-1 (B7RP-1) pathway. 19208362_These findings implicate ICOSL-induced IL-10, but not IDO in the regulation of BM-derived pDC function. 19249753_compared T cell responses of WT, CD28 knockout (KO), B7-H2 KO, and CD28-B7-H2 double KO (DKO) mice in a model of delayed-type hypersensitivity 20190137_The data suggest that ICOSL plays a significant role in immunoregulation and protective immunity against Chlamydia infections 21364749_A role for ICOS costimulation in the maintenance of effector memory but not central memory CD4 T cells. 21402623_expression is essential for allergic airway hyperresponsiveness 21636296_demonstrate that inducible costimulator (ICOS) provides a critical early signal to induce the transcription factor Bcl6, and Bcl6 then induces CXCR5, the canonical feature of follicular helper CD4(+) T cells 21768353_Findings suggest that the noncanonical NF-kappaB pathway regulates the development of Tfh cells by mediating ICOSL gene expression in B cells. 21958057_Results indicate that ICOSLG blockade may enhance cytotoxity in allogeneic mixed lymphocyte-hematologic neoplasm cell reactions. 22154192_Induction of IL-10 producing regulatory T cells mainly relied on the inducible costimulator ligand (ICOSL)/ICOS axis 22172879_ICOS/B7h signal plays an important role in direct allorecognition, eliciting allogeneic responses in vitro. 22686515_lineage-specific transgenic expression on dendritic and plasma cells generates different outcome upon ICOS costimulation 22732700_Data suggest that knockdown of Icosl in mouse leukemic cells may significantly enhance graft versus leukemia effect after allogeneic bone marrow transplantation. 23498793_In cardiac transplantation apoptosis of CD8(+) T cells in recipient draining lymph nodes was enhanced by pretreatment with donor specific transfusion and impaired ICOS/B7h allorecognition. 23578385_ICOS-L maintains tolerance at the fetomaternal interface. 24449576_Data indicate that the signals provided by ICOSL-expressing B cells to Teff cells and Tfh cells are necessary for the development of arthritis. 24610013_Findings indicate the separable roles of delivery of antigens and ICOS-L by cognate B cells for follicular Th (Tfh) cell maturation and function. 24729612_the B7h-ICOS interaction may modulate the spread of cancer metastases 25002484_The need for additional immune suppression in the intestine reflects commensal microbe-driven T-cell activation through the accessory costimulation molecules ICOSL and OX40L in B7 deprived mice. 25108021_the costimulatory ligand ICOS ligand (ICOSL) is selectively downregulated on the surface of B cells in an ADAM17-specific manner, although it is not proteolitically processed by recombinant ADAM17 in vitro. 25317561_ICOSL is a molecular linkage between T-B interactional dynamics and positive selection for high-affinity bone-marrow plasma cell formation; study reveals a pathway by which follicular T-helper cells control the quality of long-lived humoral immunity 25552357_restoring control of the T follicular helper-germinal center B-cell axis by blocking the ICOS-ICOSL pathway reduced the development of atherosclerosis and the formation of tertiary lymphoid organs. 25590646_There was a clearly positive correlation between the presence of IL-17-producing cells and ICOS expression in ICOSL KO mice; it also showed that Th17 was involved in the pathological tissue remodeling in liver fibrosis induced by schistosomiasis. 25769613_ICOS-ICOS-L interaction promoted cytokine production and survival in type 2 innate lymphoid cells through STAT5 signaling in asthma. 25786178_Selective ablation of ICOSL in CD11c+ cells, but not in B cells, dramatically ameliorates kidney and lung inflammation in lupus-prone transgenic mice. 27138044_this study shows that sICOSL in the serum of healthy donors increases in an age-dependent manner and that the matrix metalloproteinase inhibitor (MMPI) could suppress sICOSL production 27183569_study concludse that lymphopenia-driven autoimmunity and the development of Tfh17 cells in Rasgrp1-deficient mice occur independently of CD275(B7-H2) 27798154_results showed that monocyte-derived osteoclast (OC)-like cells (MDOCs) express B7h during their differentiation, and that B7h triggering reversibly inhibits OC differentiation and function both in vitro and in vivo 28002569_The expression of ICOSL in the cornea and the ICOS-mediated induction of Foxp3+ CD4+ regulatory T cells may contribute to successful corneal allograft survival. 28814605_study confirms the importance of ICOSL shedding in ICOS/ICOSL function and expression and it identifies ADAM10 as the most important sheddase for controlling ICOSL levels 30747722_work identified a potentially novel role for ICOSL, which serves as an endogenous alphavbeta3-selective antagonist to maintain glomerular filtration. 33753483_ICOS ligand and IL-10 synergize to promote host-microbiota mutualism. 34710738_Attenuate ICOSL and IL-27 in Aire-overexpressing DC2.4 cells suppress TFH cell differentiation. | ENSMUSG00000000732 | Icosl | 1.083286e+02 | 1.5996834 | 0.677786398 | 0.5637320 | 1.266956e+00 | 0.2603385270 | 0.79684153 | No | Yes | 1.191772e+02 | 36.122676 | 7.107665e+01 | 22.323379 | |||
| ENSG00000278175 | 389741 | GLIDR | lncRNA | 28211799_Increased GLIDR expression is associated with prostate cancer. 34369267_Long non-coding RNA GLIDR accelerates the tumorigenesis of lung adenocarcinoma by miR-1270/TCF12 axis. | 7.010697e+02 | 1.0228919 | 0.032653736 | 0.2971445 | 1.211599e-02 | 0.9123517619 | 0.98338795 | No | Yes | 7.111045e+02 | 64.051642 | 6.333473e+02 | 58.697457 | |||||||||
| ENSG00000278311 | 79893 | GGNBP2 | protein_coding | Q9H3C7 | FUNCTION: May be involved in spermatogenesis. | Alternative splicing;Cytoplasmic vesicle;Developmental protein;Differentiation;Phosphoprotein;Reference proteome;Spermatogenesis | hsa:79893; | cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; nucleus [GO:0005634]; cell differentiation [GO:0030154]; labyrinthine layer blood vessel development [GO:0060716]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of gene expression [GO:0010629]; negative regulation of peptidyl-serine phosphorylation of STAT protein [GO:0033140]; negative regulation of protein tyrosine kinase activity [GO:0061099]; negative regulation of tyrosine phosphorylation of STAT protein [GO:0042532]; spermatogenesis [GO:0007283] | 15145523_LCRG1 is a novel gene localized to the tumor suppressor locus D17S800-D17S930 involved in laryngeal carcinoma. 22350815_There is an essential role for ZNF403 in cell proliferation and G2/M cell-cycle transition. 24842157_Results show that downregulation of RNASET2 and GGNBP2 in drug-resistant ovarian cancer tissues/cells contributes to the regulation of drug resistance in ovarian cancer. 27357812_Results identified GGNBP2 as a novel tumor suppressor which is downregulated in human breast tumors and cell lines and suggest that GGNBP2 may function as a corepressor to inhibit ERa's transcriptional activity and consequently inhibit tumorigenic potential of breast cancer cells. 28864131_The effects of ZFP403 on cell proliferation and metastasis suggest that it may serve as a tumor suppressor in ovarian cancer. 30450530_Our data demonstrate that GGNBP2 suppresses cancer aggressiveness by inhibition of IL-6/STAT3 activation in triple-negative breast cancer 31797427_Effects of gametogenetin-binding protein 2 on proliferation, invasion and migration of prostate cancer PC-3 cells. 33125130_Effects of zinc finger protein 403 on the proliferation, migration and invasion abilities of prostate cancer cells. | ENSMUSG00000020530 | Ggnbp2 | 7.335514e+02 | 0.9049765 | -0.144047694 | 0.3173640 | 2.043099e-01 | 0.6512642546 | 0.92370776 | No | Yes | 6.412597e+02 | 95.943867 | 7.456585e+02 | 114.267216 | ||
| ENSG00000278390 | 101929140 | lncRNA | 2.197642e+01 | 0.6005759 | -0.735581610 | 0.6355707 | 1.322324e+00 | 0.2501753359 | 0.79158962 | No | Yes | 1.761712e+01 | 5.282594 | 3.211899e+01 | 9.617014 | |||||||||||
| ENSG00000282988 | protein_coding | A0A0U1RRH7 | Acetylation;Chromosome;DNA-binding;Nucleosome core;Nucleus;Reference proteome;Ubl conjugation | nucleosome [GO:0000786]; nucleus [GO:0005634]; DNA binding [GO:0003677]; protein heterodimerization activity [GO:0046982] | 1.974037e+01 | 1.7549494 | 0.811429412 | 0.7044374 | 1.398773e+00 | 0.2369291067 | 0.78818582 | No | Yes | 3.243069e+01 | 14.270025 | 1.238161e+01 | 5.837288 | |||||||||
| ENSG00000285725 | lncRNA | 5.115553e+02 | 1.6941190 | 0.760535228 | 0.3276326 | 5.359978e+00 | 0.0206040374 | 0.51149026 | No | Yes | 6.954180e+02 | 106.191392 | 3.291598e+02 | 52.232418 | ||||||||||||
| ENSG00000286522 | 8358 | H3C2 | protein_coding | P68431 | FUNCTION: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. | 3D-structure;ADP-ribosylation;Acetylation;Chromosome;Citrullination;DNA-binding;Direct protein sequencing;Disease variant;Hydroxylation;Methylation;Nucleosome core;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H3 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6p22-p21.3. [provided by RefSeq, Aug 2015]. | hsa:8350;hsa:8351;hsa:8352;hsa:8353;hsa:8354;hsa:8355;hsa:8356;hsa:8357;hsa:8358;hsa:8968; | extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; membrane [GO:0016020]; nuclear chromosome [GO:0000228]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; cadherin binding [GO:0045296]; DNA binding [GO:0003677]; protein heterodimerization activity [GO:0046982]; DNA replication-dependent chromatin assembly [GO:0006335]; nucleosome assembly [GO:0006334]; regulation of gene expression, epigenetic [GO:0040029]; telomere organization [GO:0032200] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20442396_a catalytic and inhibitory loop mechanism may better describe the cross-talk relationship between H2B ubiquitination and H3 Lys-79 methylation 20587610_Observational study of gene-disease association. (HuGE Navigator) 20864036_MSK1- and MSK2-mediated H3K27me3S28 phosphorylation serves as a mechanism to activate a subset of PcG target genes determined by the biological stimuli and thereby modulate the gene expression program determining cell fate. 20920473_These findings suggested that at metaphase Thr 3 phosphorylated histone H3 may also participate in kinetochore assembly to promote faithful chromosome segregation and serve as another recognition code for kinetochore proteins. 24242757_These data suggest that adult brainstem gliomas differ from adult supratentorial gliomas. In particular, histone genes (H3F3A (K27M) mutations are frequent in adult brainstem gliomas. 26399631_Study examined the relationship of K27M mutations in the distinct histone H3 variants (i.e. HIST1H3B and H3F3A) with specific pontine glioma biology 26727948_This study showed that HIST1H3B (n = 3) across all primary, contiguous, and metastatic tumor sites in all Diffuse intrinsic pontine glioma. 29063957_H3K27M mutation in H3F3A occurs exclusively in pediatric high-grade gliomas arising from the midline and presents with varied histomorphological features ranging from low-grade pilomyxoid astrocytoma to highly pleomorphic glioblastoma along 32433577_Histone H3K27M Mutation Overrides Histological Grading in Pediatric Gliomas. | 3.690787e+01 | 1.1202927 | 0.163875704 | 0.5215048 | 1.017294e-01 | 0.7497640319 | 0.94694596 | No | Yes | 1.831582e+01 | 5.722095 | 9.767417e+00 | 3.199285 | |||
| ENSG00000288596 | 56260 | C8orf44 | lncRNA | 6.782262e+01 | 0.4051268 | -1.303554448 | 0.4379029 | 8.635391e+00 | 0.0032969453 | 0.21440446 | No | Yes | 3.379855e+01 | 7.064154 | 8.970051e+01 | 18.412461 | ||||||||||
| ENSG00000288674 | 5664 | protein_coding | Q8NI60 | FUNCTION: Atypical kinase involved in the biosynthesis of coenzyme Q, also named ubiquinone, an essential lipid-soluble electron transporter for aerobic cellular respiration (PubMed:25498144, PubMed:21296186, PubMed:25540914, PubMed:27499294). Its substrate specificity is unclear: does not show any protein kinase activity (PubMed:25498144, PubMed:27499294). Probably acts as a small molecule kinase, possibly a lipid kinase that phosphorylates a prenyl lipid in the ubiquinone biosynthesis pathway, as suggested by its ability to bind coenzyme Q lipid intermediates (PubMed:25498144, PubMed:27499294). Shows an unusual selectivity for binding ADP over ATP (PubMed:25498144). {ECO:0000269|PubMed:25498144, ECO:0000269|PubMed:27499294, ECO:0000305|PubMed:21296186, ECO:0000305|PubMed:25540914}. | 3D-structure;ATP-binding;Alternative splicing;Direct protein sequencing;Disease variant;Kinase;Membrane;Mitochondrion;Neurodegeneration;Nucleotide-binding;Primary mitochondrial disease;Reference proteome;Transferase;Transit peptide;Transmembrane;Transmembrane helix;Ubiquinone biosynthesis | PATHWAY: Cofactor biosynthesis; ubiquinone biosynthesis. {ECO:0000269|PubMed:25498144}. | hsa:56997; | extrinsic component of mitochondrial inner membrane [GO:0031314]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; ADP binding [GO:0043531]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; phosphorylation [GO:0016310]; ubiquinone biosynthetic process [GO:0006744] | Mouse_homologues 11756438_These data reveal that a novel PS1/2-independent mechanism plays a partial role in Notch signal transduction 11817902_inhibits protein synthesis 11904448_Wild-type and mutated presenilins 2 trigger p53-dependent apoptosis and down-regulate presenilin 1 expression in HEK293 human cells and in murine neurons 12198112_PS2/gamma-secretase contains PEN-2 and requires it for presenilin expression 12646573_role in stability and maturation of nicastrin in mammalian brain 12684521_gamma-secretase activity in blastocysts from normal, heterozygote and knockout mice 12885769_presenilins are multifunctional proteins with catalytic activity as well as roles in the generation, stabilization, and transport of the gamma-secretase complex 14566063_gamma-secretase contains a presenilin dimer in its catalytic core, that binding of substrate is at a site separate from the active site, and that substrate is cleaved at the interface of two PS molecules 15122901_Results establish a critical role of presenilin 2 in myelopoiesis; partial loss of PS2 in mice leads to age-dependent myeloproliferative disease, mediated through its gamma-secretase activity. 15148382_presenilins are essential for the ongoing maintenance of cortical structures and function 15264228_Expression of mutant presenilin 2(M239V-PS2) caused neuronal cytotoxicity which could be inhibited by the combination of two clinically usable inhibitors of superoxide-generating enzymes, apocynin and oxypurinol 15287885_Our data demonstrate that presenilin-directed gamma-secretase inhibitors affect caspase 3 activity in a presenilin-independent manner 15322084_presenilin 1 (PS1)-derived fragments, mature nicastrin, APH-1, and PEN-2, associate with cholesterol-rich detergent insoluble membrane (DIM) domains of non-neuronal cells and neurons 15537629_results suggest that the proximal two-thirds of the PEN-2 TMD1 is functionally important for endoproteolysis of PS1 holoproteins and the generation of PS1 fragments, essential components of the gamma-secretase complex 15613480_PS1 and PS2 affect caveolin 1 trafficking in an indirect manner and are required for caveolae formation by controlling transport of intracellular caveolin 1 to the plasma membrane. 15649694_The metabolic profile, however, showed clear indicators of hypometabolism with age in the PS2APP mice: both N-acetyl-aspartate and glutamate were significantly reduced in the older animals. 15858415_presenilin 2 may be implicated in neuronal cell death by altering the antiapoptotic activity of the transcription factors 15866159_animals lacking all presenilin alleles have no somites 15922300_presenilin 1 and 2 in mouse embryonic fibroblasts have roles in responding to invasion and replication of Salmonella typhimurium 16103123_presenilin, nicastrin, APH-1, and PEN-2, are present and enriched on phagosome membranes from both murine macrophages and Drosophila S2 phagocytes 16204356_PS2 plays an important role in cardiac excitation-contraction coupling by interacting with RyR2 16384915_tyrosinase and related proteins as physiological substrates of presenilins and link gamma-secretase activity with intracellular protein transport 16641999_TMP21, a member of the p24 cargo protein family, is a component of presenilin complexes and differentially regulates gamma-secretase cleavage without affecting epsilon-secretase activity 16887746_the promoter of the mouse Presenilin-2 gene is bound and activated by CLOCK and BMAL1, transcription factors of the mammalian circadian clock 17121184_Findings indicate that PS2 is involved in the abnormalities of the lipid profile, which could cause or result in Alzheimer's disease. 17292944_C-terminal fragment of presenillin 2 is released by shedding into the extracellular compartment as a soluble form. This release is 4.07-fold increased during apoptosis. 17556541_PS2 is necessary for the proteolytic activity of gamma-secretase. 17614943_environmental enrichment effectively enhanced memory and partially rescued the forebrain atrophy of the Presenilin 1 and 2 conditional double knockout mice 17698590_These findings suggest a model in which PS-dependent Notch signaling influences positive selection and the development of alphabeta T cells by modifying T-cell receptor signal transduction. 17728018_Thus these results demonstrate up-regulation of PS2 protein expression by sex steroids, which in turn may influence PS2 associated brain functions. 17874292_Age and sex dependent alteration in presenilin expression. 18195359_B cells deficient in both PS1 and PS2 function have an unexpected and substantial deficit in both lipopolysaccharide and B cell antigen receptor-induced proliferation and signal transduction events 18293935_Mature integrin beta1 with increased expression level is delivered to the cell surface, which results in an increased cell surface expression level of mature integrin beta 1 in presenilin (PS)1 and PS2 double-deficient fibroblasts. 18440065_Data demonstrate facilitation of ryanodine receptor gating by presenilin-2 NTF1-87. 18803281_PS2 mutation causes reduction of anxiety 18822370_the oxidative stress status in PS1 and PS2 double-knockout (PS cDKO) mice using F(2)-isoprostanes (iPF(2alpha)-III) as the marker of lipid peroxidation 19297528_Met is processed in epithelial cells by presenilin-dependent regulated intramembrane proteolysis (PS-RIP) independently of ligand stimulation. 19376115_PS2beta may inhibit gamma-secretase activity by affecting the gamma-secretase complex assembly 19382908_Presenilin-2 dampens intracellular Ca2+ stores by increasing Ca2+ leakage and reducing Ca2+ uptake 19573580_Egr-1 over-expression and down-regulation, as well as in-vivo examination of Egr-1 and Psen2 expression during fear conditioning, showed that Egr-1 does not regulate the Psen2 promoter. 19834068_Data show that presenilins 1 and 2 are highly enriched in a subcompartment of the endoplasmic reticulum associated with mitochondria that forms a physical bridge between the two organelles. 20573903_These results indicate that disruption of ER Ca(2+) leak function of presenilin 1 and 2 may play an important role in Alzheimer disease pathogenesis. 20875805_partial, but highly significant, loss of mGlur2 receptors in amyloid-affected discrete brain regions of Alzheimer cases and PS2APP mice 20953898_These findings suggest that inflammation in the presenilin 1 (PS1) and presenilin 2 (PS2) double knockout mice could expand to the periphery, including the oral tissue, which could ultimately induce abnormalities in the periodontal and salivary tissues. 20974250_4-O-Methylhonokiol attenuates memory impairment in presenilin 2 mutant mice . 21084199_This study confirms the regulatory role of CXCR2 in APP processing, and poses it as a potential target for developing novel therapeutics for intervention in Alzheimer's disease. 21146496_this study shows that the contribution of PS2 on gamma-secretase activity is of less importance, explaining the mild phenotype of PS2-deficient mice. 21206757_PS2 is the predominant gamma-secretase in microglia and modulates release of proinflammatory cytokines 21296884_Study supports the hypothesis that Pen-2 is more than a structural component of the gamma-secretase complex and may contribute to the catalytic mechanism of the enzyme. 21414900_Endogenous wild-type presenilin 2 is necessary for proper protein degradation through the autophagosome-lysosome system by functioning at the lysosomal level. 21910732_Presenilin 2 has the ability to regulate nectin-1 processing. 21914807_Polar transmembrane-based amino acids in presenilin 1 are involved in endoplasmic reticulum localization, Pen2 protein binding, and gamma-secretase complex stabilization 22140537_Upregulation of PS1/gamma-secretase activity may be a risk factor for late onset sporadic Alzheimer's disease. 22170863_Inhibition of APP cleavage by gamma-secretase did not ameliorate synaptic/memory deficits of familial Danish dementia mice. 22311977_lack of evidence for presenilins as endoplasmic reticulum Ca2+ leak channels 22715045_Double knockout Psen1Psen2 leads to a decrease in immunoreactivity for drebrin A at both synaptic and nonsynaptic areas of CA1 hippocampus. 22723704_transcriptome studies of presenilin knockout mise mouse brains reveal, for the first time, a role for presenilins in regulating lysosomal biogenesis 22796216_Distribution and expression profile of Psen2 differs from Psen1 in the cerebral cortex during development. 23103503_These data indicate that alterations in lysosomal calcium in the absence of presenilins might be leading to disruptions in autophagy. 23359614_data delineate a promoter responsive element targeted by parkin that drives differential regulation of presenilin-1 and presenilin-2 transcription with functional consequences for gamma-secretase activity and cell death. 23370287_PS2 mutation significantly accelerates the onset of cognitive impairment in associative trace eyeblink memory. 23589300_Interactome analyses of mature gamma-secretase complexes reveal distinct molecular environments of presenilin (PS) paralogs and preferential binding of signal peptide peptidase to PS2. 23918386_mechanism by which PS regulates synaptic function and calcium homeostasis using acute hippocampal slices from PS1 and PS2 conditional knockouts and primary cultured postnatal hippocampal neurons 23952003_One mechanism by which PS2 works to reign in proinflammatory microglial behavior and PS2 dysfunction or deficiency could result in unchecked proinflammatory activation. contributing to neurodegeneration. 24145027_At the transcriptional level, Psen1/2 removal induced cyclic AMP response element-binding protein (CREB)/CREB-binding protein binding. 24858037_The loss of PS2 could have a critical role in lung tumor development through the upregulation of iPLA2 activity by reducing gamma-secretase. 25204494_This study presents novel evidence for the differential expression of PS proteins in a nongenetic model for aging, resulting in an overall increase of the PS2 to PS1 ratio. 25652771_Results showed that epigenetic mechanisms play a pivotal role in transcriptional regulation of PS1 and PS2 during cerebral cortical development 26081153_These observations illustrate a novel PS2-dependent means of modulating LPS-mediated immune responses and identify a functional distinction between PS1 and PS2 in innate immunity. 26593275_Results suggest that mutation of PS2 can lead to NF-kappaB mediate amyloidogensis, and this effect can be amplified by the absence of estrogen. 27177724_Study shows that the binding of HSF-1, Cdx1, Ets-1 and Sp1 to Presenilin 1 promoter and that of Nkx2.2, HFH-2, Cdx1 and NF-kappaB to Presenilin 2 promoter regulate their differential expression during brain development. 27239030_Presenilin 2 (PS2), mutations in which underlie familial Alzheimer's disease (FAD), promotes endoplasmic reticulum-mitochondria coupling only in the presence of mitofusin 2 (Mfn2). 27354375_ARF4 is required for Presenilin basal body localization, Notch signaling, and subsequent epidermal differentiation. 27889678_Although hyperactivity and hypersynchronicity were respectively detected in mice expressing the PS2-N141I or the APP Swedish mutant alone, the increase in cross-frequency coupling specifically characterized the 6-month-old PS2APP mice, just before the surge of the cognitive decline 28879407_The altered expressions of hippocampal microRNAs were associated to the imbalance between neurotoxic and neuroprotective functions and seemed to affect neurodegeneration in PSEN1/PSEN2 double knockout mice more severely than in wild-type mice. 28994238_PS1- and PS2-dependent substrate processing in murine cells lacking presenilins (PSs) or stably re-expressing human PS1 or PS2 in an endogenous PS-null (PSdKO) background, were studied. 30429473_Study uncovers a role of PS in presynaptic mechanisms, through APP cleavage and regulation of Syt7, that highlights aberrant synaptic vesicle processing as a possible new pathway in Alzheimer's disease. 31862541_Loss of presenilin 2 age-dependently alters susceptibility to acute seizures and kindling acquisition. 32961023_Neuron-specific deletion of presenilin enhancer2 causes progressive astrogliosis and age-related neurodegeneration in the cortex independent of the Notch signaling. 33494218_Calcium Signaling and Mitochondrial Function in Presenilin 2 Knock-Out Mice: Looking for Any Loss-of-Function Phenotype Related to Alzheimer's Disease. | ENSMUSG00000010609 | Psen2 | 1.824696e+02 | 0.9450267 | -0.081572949 | 0.3274234 | 6.124159e-02 | 0.8045442065 | 0.96141221 | No | Yes | 1.471140e+02 | 34.926572 | 1.888998e+02 | 45.921727 | ||
| ENSG00000288743 | 105370203 | lncRNA | 5.959885e+01 | 0.9347228 | -0.097389528 | 0.4304834 | 5.209218e-02 | 0.8194619086 | 0.96538577 | No | Yes | 4.345682e+01 | 8.501476 | 3.959581e+01 | 8.014711 | |||||||||||
| ENSG00000289483 | lncRNA | 1.875580e+01 | 1.5779710 | 0.658070686 | 0.6956008 | 8.960158e-01 | 0.3438522774 | 0.82878860 | No | Yes | 1.941392e+01 | 6.340596 | 9.868056e+00 | 3.421983 | ||||||||||||
| ENSG00000289486 | lncRNA | 9.385896e+01 | 1.4341973 | 0.520243551 | 0.4192568 | 1.583071e+00 | 0.2083188552 | 0.78763590 | No | Yes | 8.540093e+01 | 19.918499 | 5.612820e+01 | 13.668074 | ||||||||||||
| ENSG00000289586 | 105377294 | lncRNA | 5.988096e+01 | 1.5174615 | 0.601659927 | 0.4378486 | 1.854939e+00 | 0.1732099992 | 0.77611497 | No | Yes | 6.150560e+01 | 12.661272 | 3.850165e+01 | 8.152409 |
- Note:
- The spreadsheet above contains the non-differential expressed genes but with significantly changes in alternative splicing (q < 0.05 and dif > 0.05). Please Click HERE to check all mapped genes in a Microsoft .excel file.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| lncRNA | 52 |
| protein_coding | 837 |
| transcribed_processed_pseudogene | 2 |
| transcribed_unitary_pseudogene | 3 |
| transcribed_unprocessed_pseudogene | 18 |
| unprocessed_pseudogene | 3 |
MA plot
PCA plot
Co-expression Analysis
The co-expression pattern was determined using weighted gene co-expression network analysis (WGCNA) through WGCNA R software.
Choosing the soft-thresholding power: analysis of network topology
Quantifying module-trait associations
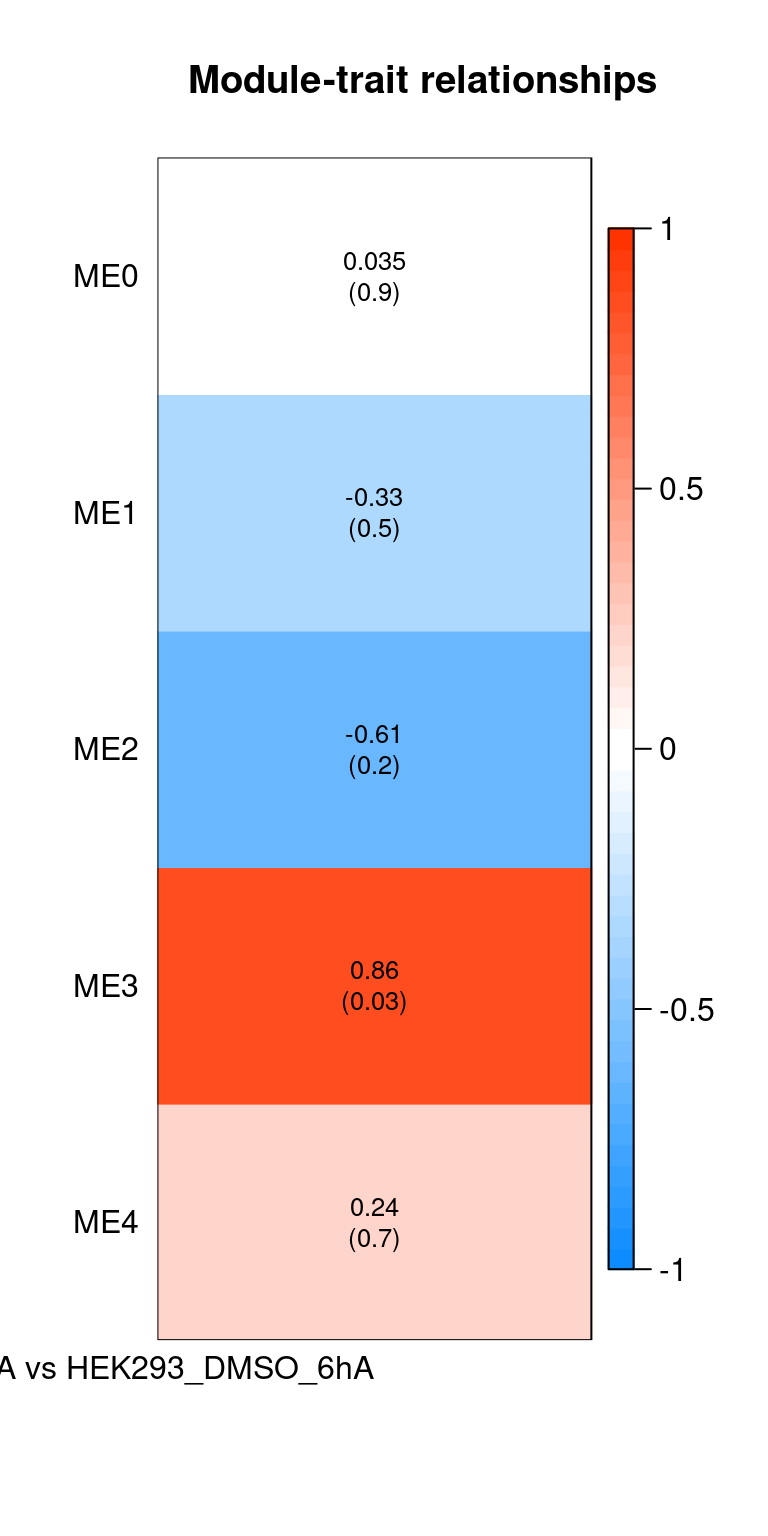
In the above figure, each cell contains the corresponding correlation (upper) and p-value (lower). ME, Module Eigengene.
Spreadsheet
| Cluster ID | Correlation | P-value | DEGs ID | DEGs Symbol |
|---|---|---|---|---|
| ME0 | 0.03540522 | 0.94691436 | ENSG00000277991||ENSG00000280893 | ENSG00000277991||ENSG00000280893 |
| ME1 | -0.33015632 | 0.52275957 | ENSG00000249967||ENSG00000260566 | ENSG00000249967||ENSG00000260566 |
| ME2 | -0.61484207 | 0.19395151 | ENSG00000083457||ENSG00000090263||ENSG00000100372||ENSG00000111843||ENSG00000125037||ENSG00000143106||ENSG00000158164||ENSG00000163541||ENSG00000164032||ENSG00000165775||ENSG00000186567||ENSG00000189043||ENSG00000255513||ENSG00000261326||ENSG00000273373||ENSG00000274290 | ITGAE||MRPS33||SLC25A17||TMEM14C||EMC3||PSMA5||TMSB15A||SUCLG1||H2AZ1||FUNDC2||CEACAM19||NDUFA4||ENSG00000255513||ENSG00000261326||ENSG00000273373||H2BC6 |
| ME3 | 0.85656789 | 0.02938376 | ENSG00000021826||ENSG00000070718||ENSG00000079332||ENSG00000087995||ENSG00000100814||ENSG00000102158||ENSG00000113811||ENSG00000117748||ENSG00000117862||ENSG00000129315||ENSG00000131238||ENSG00000132196||ENSG00000134419||ENSG00000139131||ENSG00000142396||ENSG00000146530||ENSG00000147162||ENSG00000151779||ENSG00000155561||ENSG00000162813||ENSG00000163682||ENSG00000164199||ENSG00000167447||ENSG00000168036||ENSG00000173915||ENSG00000176225||ENSG00000181610||ENSG00000198408||ENSG00000198522||ENSG00000198743||ENSG00000198843||ENSG00000215695||ENSG00000230673||ENSG00000237550||ENSG00000248101||ENSG00000259494||ENSG00000259522||ENSG00000261740||ENSG00000263244||ENSG00000264112 | CPS1||AP3M2||SAR1A||METTL2A||CCNB1IP1||MAGT1||SELENOK||RPA2||TXNDC12||CCNT1||PPT1||HSD17B7||RPS15A||YARS2||ERVK3-1||VWDE||OGT||NBAS||NUP205||BPNT1||RPL9||ADGRV1||SMG8||CTNNB1||ATP5MK||RTTN||MRPS23||OGA||GPN1||SLC5A3||SELENOT||RSC1A1||ENSG00000230673||ENSG00000237550||ENSG00000248101||MRPL46||ENSG00000259522||BOLA2-SMG1P6||ENSG00000263244||ENSG00000264112 |
| ME4 | 0.23522263 | 0.65367345 | ENSG00000229186||ENSG00000270149 | ENSG00000229186||ENSG00000270149 |
Help
still working on
2. Functional Enrichment Analysis
2-1. Spreadsheet
GSEA
Please Click HERE to download a Microsoft .excel that contains all GSEA results.
GO: BP
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0003180 | aortic valve morphogenesis | 28 | 0.6271806 | 2.304789 | 4.458257e-05 | 0.008095331 | 0.007596445 | 3493 | tags=61%, list=21%, signal=48% | SLIT3||NKX2-5||EMILIN1||TGFB1||HEY2||SOX9||GATA4||SNAI2||NFATC1||JAG1||SLIT2||GATA3||ROCK1||NOTCH1||HEYL||TWIST1||HEY1 | 3.046823 | 3.8491773 | 3.0562751 | 3.756465 | 3.098051 | 3.3287022 | 3.8262681 | 3.6637935 | 2.986013 | 3.058380 | 3.093985 | 3.8077449 | 3.8876066 | 3.8510733 | 3.259762 | 2.93317024 | 2.9520621 | 3.6924654 | 3.7534931 | 3.820587 | 3.088966 | 2.976930 | 3.218143 | 3.317619 | 3.379931 | 3.28699658 | 3.812674 | 3.8295385 | 3.8364885 | 3.6174662 | 3.697463 | 3.6752739 |
| GO:0045746 | negative regulation of Notch signaling pathway | 32 | 0.6179029 | 2.344795 | 1.173532e-04 | 0.015013207 | 0.014087997 | 3904 | tags=62%, list=23%, signal=48% | CHAC1||BEND6||AKT1||RITA1||HEY2||BCL6||ZBTB7A||NIBAN2||NEURL1||YTHDF2||ARRB1||GATA2||DLX2||DLK2||EGFL7||ARRDC1||HEY1||WWP2||BMP7||CBFA2T2 | 4.355973 | 4.6517888 | 4.5236063 | 4.468472 | 4.399548 | 4.6462843 | 4.6393175 | 4.4439089 | 4.312496 | 4.336293 | 4.417036 | 4.6755778 | 4.6686034 | 4.6102988 | 4.567327 | 4.54748616 | 4.4534598 | 4.5204280 | 4.4076445 | 4.475118 | 4.393786 | 4.265706 | 4.527295 | 4.520475 | 4.625189 | 4.78119635 | 4.668632 | 4.6212928 | 4.6275677 | 4.4274066 | 4.490482 | 4.4126482 |
| GO:0002689 | negative regulation of leukocyte chemotaxis | 10 | 0.7934100 | 2.143113 | 5.516190e-04 | 0.042535116 | 0.039913832 | 2295 | tags=70%, list=14%, signal=60% | NBL1||RIN3||PADI2||MIF||C5||SLIT2||DDT | 9.438425 | 8.5623805 | 8.9732226 | 8.370668 | 9.519282 | 9.6094660 | 8.3753996 | 8.2078402 | 9.405721 | 9.184429 | 9.681903 | 8.7418088 | 8.4458263 | 8.4810368 | 8.613737 | 9.18586864 | 9.0596609 | 8.6002663 | 8.2496806 | 8.231326 | 9.538188 | 9.247543 | 9.731525 | 9.523222 | 9.884383 | 9.37177844 | 8.384216 | 8.3640423 | 8.3778668 | 8.2299787 | 8.200008 | 8.1932687 |
| GO:1902895 | positive regulation of pri-miRNA transcription by RNA polymerase II | 36 | 0.4957456 | 1.933068 | 7.687225e-04 | 0.051513559 | 0.048338966 | 2904 | tags=47%, list=17%, signal=39% | WT1||JUN||NGFR||MRTFA||TGFB1||SMARCA4||BMP2||RELA||TP53||KLF4||KLF5||SRF||GATA3||SMAD4||GATA2||STAT3||SREBF2 | 4.183584 | 4.8300483 | 4.3951332 | 4.897816 | 4.207891 | 4.5718848 | 4.9209201 | 4.9830600 | 4.129479 | 4.195087 | 4.224551 | 4.8199548 | 4.8269531 | 4.8431389 | 4.559787 | 4.34384828 | 4.2654415 | 4.8596219 | 4.8852325 | 4.947185 | 4.222982 | 4.055211 | 4.332096 | 4.504181 | 4.557585 | 4.65009511 | 4.920693 | 4.9024827 | 4.9393492 | 4.9627922 | 5.016616 | 4.9691701 |
| GO:0023019 | signal transduction involved in regulation of gene expression | 13 | 0.7054338 | 2.071609 | 7.923081e-04 | 0.052469442 | 0.049235942 | 1857 | tags=38%, list=11%, signal=34% | FGF8||MESP1||TRAF3IP2||GSC||TRAF2 | 4.940753 | 5.1910593 | 5.1281275 | 5.234818 | 4.831014 | 5.1506231 | 5.2114150 | 5.2272277 | 4.930468 | 5.134901 | 4.728278 | 5.1581468 | 5.2116500 | 5.2028128 | 5.364982 | 4.91242964 | 5.0699095 | 5.2480034 | 5.1678872 | 5.286053 | 4.864877 | 4.892677 | 4.730318 | 5.327285 | 5.170984 | 4.92549797 | 5.233708 | 5.1620695 | 5.2372276 | 5.2210306 | 5.243721 | 5.2167855 |
| GO:0019933 | cAMP-mediated signaling | 38 | 0.5428726 | 2.139793 | 9.788781e-04 | 0.062005045 | 0.058183901 | 2034 | tags=37%, list=12%, signal=32% | AKAP6||EPHA5||OPRL1||RIMS2||RAPGEF3||MGRN1||ADGRG6||SLC9A3R1||MAPK7||CDC34||ADCY1||SOX9||GAL||RNF157 | 4.159568 | 4.0032661 | 4.0715152 | 3.946633 | 4.139292 | 4.3049979 | 3.9964603 | 3.9139478 | 4.115763 | 4.117372 | 4.241905 | 4.0189065 | 3.9829448 | 4.0077125 | 4.022294 | 4.10957770 | 4.0813036 | 3.9962510 | 3.8819431 | 3.959358 | 4.157219 | 4.039239 | 4.215849 | 4.287293 | 4.367713 | 4.25772991 | 4.042813 | 3.9676664 | 3.9777424 | 3.9002002 | 3.928073 | 3.9134357 |
| GO:0034112 | positive regulation of homotypic cell-cell adhesion | 10 | 0.7524047 | 2.032351 | 1.431614e-03 | 0.079005437 | 0.074136620 | 1635 | tags=50%, list=10%, signal=45% | CCL5||JAK2||IL6ST||LGALS1||DMTN | 2.983317 | 3.4065727 | 3.1102311 | 3.523235 | 3.060195 | 3.1822949 | 3.4242245 | 3.4962381 | 2.880817 | 2.998136 | 3.065008 | 3.3330630 | 3.4868424 | 3.3956550 | 3.269752 | 3.02095951 | 3.0256938 | 3.3506790 | 3.5765276 | 3.627708 | 3.109859 | 2.965183 | 3.101035 | 3.202646 | 3.275997 | 3.05994381 | 3.376802 | 3.4921023 | 3.4011936 | 3.5156672 | 3.525914 | 3.4458242 |
| GO:0003188 | heart valve formation | 12 | 0.6922719 | 1.978777 | 1.688506e-03 | 0.088005535 | 0.082582075 | 3493 | tags=75%, list=21%, signal=59% | HEY2||SOX9||GATA4||SMAD4||NOTCH1||DCHS1||ZFPM1||TWIST1||HEY1 | 2.946298 | 4.0699746 | 3.0744635 | 3.906515 | 3.011390 | 3.1969788 | 4.0090328 | 3.8154026 | 2.925879 | 3.075717 | 2.826318 | 4.0782954 | 4.0956811 | 4.0352789 | 3.289676 | 2.90800502 | 2.9975718 | 3.8884157 | 3.8878356 | 3.942605 | 2.955384 | 3.096345 | 2.978445 | 3.147544 | 3.043518 | 3.37915507 | 3.980700 | 4.0195191 | 4.0264586 | 3.7239068 | 3.883561 | 3.8341408 |
| GO:0001701 | in utero embryonic development | 310 | 0.2486092 | 1.374831 | 1.815270e-03 | 0.092892339 | 0.087167723 | 4190 | tags=36%, list=25%, signal=28% | APBA1||ACVRL1||PTH1R||SOX18||NLE1||ETV2||EPN1||NR2F2||FBLL1||PHLDA2||PLCD3||B9D1||RRP7A||GPI||CEBPB||ZFP36L1||AKT1||CITED1||WDTC1||HEY2||C5||RPL13||CDX2||BMP2||SCO2||TET1||TAB1||GATA6||CNOT3||PCGF2||FURIN||CCDC24||TP53||ZMIZ1||ARHGDIG||MBD3||KLF4||WNT3A||CMTM3||FOXC1||ASF1B||ENDOG||NXN||ADCY9||ANKS6||KEAP1||LATS1||GLI3||SRF||CITED2||DBN1||GATA3||TRIM28||ETNK2||XAB2||SMAD4||BPTF||VASH2||SH3PXD2A||TANC2||JUNB||HES1||GATA2||YBX3||PTCH1||ARNT2||VASH1||MYH9||NOTCH1||PEMT||BCOR||EMX1||CMIP||HAND1||SLC39A3||CELF4||SOX8||KRT8||LY6E||NOG||BYSL||IHH||TBX3||APBA3||FBXW8||CAPN2||MAFG||CCM2||LMX1B||INPP5K||TWIST1||PRMT1||HEY1||ERCC2||WDR74||KIFBP||ELL||SEC24C||CEBPA||SLC39A1||FKBP10||BMP7||SPINT2||OTUD7B||AKAP3||TFEB||MGAT1||ZNF335||MAFF||E2F8||SUPT6H||BTF3||BCL2L1 | 6.082598 | 6.1328644 | 6.2704657 | 6.210872 | 6.060181 | 6.3510879 | 6.1487458 | 6.1578068 | 6.056074 | 6.054621 | 6.135602 | 6.0441227 | 6.1911115 | 6.1592502 | 6.239516 | 6.32907637 | 6.2409686 | 6.2039661 | 6.2777530 | 6.147953 | 6.085691 | 5.957501 | 6.131751 | 6.278166 | 6.365826 | 6.40631279 | 6.155475 | 6.1571653 | 6.1334758 | 6.1882815 | 6.156014 | 6.1285046 |
| GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 56 | 0.4237660 | 1.823864 | 2.012064e-03 | 0.100790799 | 0.094579430 | 3956 | tags=46%, list=24%, signal=36% | EMILIN1||SLC9A3R1||GPER1||CSK||PTEN||DUSP9||TNIP1||DAB2IP||EPHB2||PIN1||KLF4||ABL1||GSTP1||WNK2||SMAD4||ARRB1||FBLN1||CNKSR3||SIRPA||EIF3A||SEMA6A||SPRY1||PSCA||PTPN1||RANBP9||NDRG2 | 5.985765 | 5.4148713 | 5.8697030 | 5.487997 | 6.087991 | 6.1456129 | 5.3778395 | 5.4837327 | 5.926974 | 5.859555 | 6.154017 | 5.4357876 | 5.4220069 | 5.3863696 | 5.694956 | 5.96605765 | 5.9332179 | 5.5468664 | 5.4072217 | 5.506350 | 6.064348 | 5.972592 | 6.216452 | 6.080219 | 6.278049 | 6.06883421 | 5.405245 | 5.3606042 | 5.3672665 | 5.4829995 | 5.500700 | 5.4673048 |
| GO:1902894 | negative regulation of pri-miRNA transcription by RNA polymerase II | 14 | 0.6442249 | 1.931694 | 2.218027e-03 | 0.106382524 | 0.099826557 | 3189 | tags=50%, list=19%, signal=41% | NFIB||TGFB1||SOX9||RELA||SRF||PPARD||HDAC4 | 3.749870 | 4.0818522 | 3.8317970 | 3.951751 | 3.757235 | 4.0469574 | 4.1102347 | 3.9902978 | 3.696126 | 3.718980 | 3.830869 | 4.0425327 | 4.1239130 | 4.0779555 | 3.820621 | 3.90193577 | 3.7697423 | 3.9983170 | 3.8566849 | 3.995748 | 3.762240 | 3.616877 | 3.880543 | 4.000918 | 4.108569 | 4.02921506 | 4.110020 | 4.0919340 | 4.1285181 | 3.9782038 | 4.044400 | 3.9465548 |
| GO:0071676 | negative regulation of mononuclear cell migration | 10 | 0.7276548 | 1.965498 | 2.877367e-03 | 0.125787900 | 0.118036050 | 2134 | tags=50%, list=13%, signal=44% | NBL1||PLCB1||PADI2||AKT1||SLIT2 | 4.127024 | 4.4157875 | 4.2282433 | 4.487945 | 4.208406 | 4.6074370 | 4.3955473 | 4.5085954 | 3.992794 | 4.120148 | 4.256109 | 4.3923586 | 4.4571280 | 4.3969663 | 4.323952 | 4.25875075 | 4.0922427 | 4.4869633 | 4.4660446 | 4.510486 | 4.243671 | 3.920166 | 4.417990 | 4.550722 | 4.664691 | 4.60464328 | 4.423890 | 4.3784197 | 4.3839040 | 4.5257777 | 4.547289 | 4.4509570 |
| GO:0033234 | negative regulation of protein sumoylation | 10 | -0.8020282 | -1.872190 | 3.479361e-03 | 0.142764990 | 0.133966905 | 1178 | tags=40%, list=7%, signal=37% | CAPN3||PARK7||CTNNB1||MAGEA2B | 6.105625 | 6.6249125 | 6.5813026 | 6.773236 | 5.899356 | 5.8640256 | 6.6274852 | 6.7749628 | 6.216591 | 6.324840 | 5.701986 | 6.6488955 | 6.6278377 | 6.5975429 | 6.732738 | 6.33096742 | 6.6498014 | 6.7943781 | 6.7348840 | 6.789692 | 5.781097 | 6.146362 | 5.734406 | 5.772491 | 5.753933 | 6.04665113 | 6.648194 | 6.6003794 | 6.6334670 | 6.7570767 | 6.786055 | 6.7815879 |
| GO:0060343 | trabecula formation | 16 | 0.6142177 | 1.920332 | 3.508024e-03 | 0.142764990 | 0.133966905 | 3493 | tags=56%, list=21%, signal=45% | MMP2||ADGRG6||WNT10B||COL1A1||HEY2||CAV3||SRF||ADAMTS1||HEY1 | 5.156899 | 5.0931175 | 5.3323914 | 5.068463 | 5.214049 | 5.3183533 | 5.1190163 | 5.0686200 | 5.151727 | 5.252006 | 5.060606 | 5.0986802 | 5.1034180 | 5.0771183 | 5.307608 | 5.36260474 | 5.3264194 | 5.0719544 | 5.0200676 | 5.111898 | 5.248885 | 5.163963 | 5.227948 | 5.314113 | 5.101921 | 5.51022392 | 5.117654 | 5.1250879 | 5.1142857 | 5.0538570 | 5.064990 | 5.0868177 |
| GO:0008360 | regulation of cell shape | 119 | 0.3327203 | 1.624478 | 3.702014e-03 | 0.148847409 | 0.139674486 | 5560 | tags=50%, list=33%, signal=33% | PLXNC1||CDC42EP2||PLXNA2||PARVA||CDC42EP1||ARHGDIA||PARVB||ALDOA||SLC9A3R1||SH3KBP1||FGD1||RHOG||RAC3||PRAG1||RHOC||CDC42EP4||DMTN||MARK2||DLC1||FBLIM1||RND2||BAIAP2||DAPK3||FMNL1||CFAP410||CORO1A||MYH9||RHOF||FMNL3||PALM||C15orf62||ARHGEF18||PLXNA4||FGD3||MYH14||WDR1||DNMBP||PLXND1||FYN||SHROOM3||FGD4||CDC42EP3||ITGB2||DAG1||ARAP1||PHIP||PLXNA1||FAM171A1||SYNE3||SEMA4A||RASA1||PLXNB2||EPB41L3||RHOQ||WASF3||RAC1||RHOBTB2||CFDP1||RHOB | 5.568858 | 5.3652403 | 5.3945434 | 5.379409 | 5.593074 | 5.7265355 | 5.3541811 | 5.3730291 | 5.530653 | 5.490541 | 5.678516 | 5.3671361 | 5.3720956 | 5.3564450 | 5.317197 | 5.45727834 | 5.4056910 | 5.4088200 | 5.3026966 | 5.423716 | 5.595652 | 5.498165 | 5.679696 | 5.703571 | 5.838667 | 5.62952438 | 5.371415 | 5.3327720 | 5.3580898 | 5.3653910 | 5.393597 | 5.3598715 |
| GO:0007266 | Rho protein signal transduction | 101 | 0.3449992 | 1.651129 | 4.085750e-03 | 0.159713099 | 0.149870563 | 2792 | tags=29%, list=17%, signal=24% | CDC42EP2||RASGRF1||CDC42EP1||GPR199P||ARHGDIA||NGFR||TAX1BP3||ARHGAP1||RHOG||SCAI||PRAG1||BCL6||RASGRF2||RIPOR1||RHOV||WAS||CDC42EP4||ARHGDIG||DLC1||RTN4R||ABL1||LIMK1||F2R||ROCK1||ARRB1||CFL1||BCR||ARHGEF2||C15orf62 | 5.418653 | 5.3464475 | 5.4128693 | 5.387374 | 5.443326 | 5.5725215 | 5.3701101 | 5.4142448 | 5.376848 | 5.425819 | 5.452281 | 5.3329609 | 5.3487918 | 5.3574828 | 5.403626 | 5.41704126 | 5.4178967 | 5.3852119 | 5.3434852 | 5.432063 | 5.446538 | 5.384317 | 5.496920 | 5.567112 | 5.624864 | 5.52380442 | 5.375301 | 5.3565353 | 5.3783974 | 5.4076983 | 5.425625 | 5.4093429 |
| GO:0034143 | regulation of toll-like receptor 4 signaling pathway | 15 | 0.6180871 | 1.895922 | 4.136781e-03 | 0.160592698 | 0.150695956 | 4665 | tags=60%, list=28%, signal=43% | NR1D1||IFI35||DAB2IP||NINJ1||TNFAIP3||NR1H3||TIRAP||PELI1||APPL1 | 4.494065 | 5.9699951 | 4.4926917 | 5.978728 | 4.341320 | 4.2080642 | 6.1213735 | 6.0081781 | 4.550344 | 4.620438 | 4.290951 | 5.9192344 | 5.9585156 | 6.0300399 | 5.059880 | 4.14090458 | 4.0480253 | 5.8974196 | 5.9762180 | 6.058073 | 4.176316 | 4.519362 | 4.307347 | 4.086680 | 4.176289 | 4.34881514 | 6.111378 | 6.1096492 | 6.1428501 | 6.0738663 | 5.884808 | 6.0583316 |
| GO:0060644 | mammary gland epithelial cell differentiation | 16 | 0.6061348 | 1.895061 | 4.181551e-03 | 0.161218836 | 0.151283507 | 2862 | tags=50%, list=17%, signal=41% | ERBB4||ZNF703||CEBPB||AKT1||FOXB1||LATS1||PTCH1||AKT2 | 4.800369 | 5.1157896 | 4.9356951 | 4.908921 | 4.858614 | 5.2484526 | 5.1258140 | 4.9152376 | 4.756352 | 4.715455 | 4.920999 | 5.1305819 | 5.1323498 | 5.0839161 | 4.948951 | 5.00690719 | 4.8466894 | 4.9706808 | 4.8334406 | 4.919324 | 4.855126 | 4.691859 | 5.011215 | 5.161604 | 5.312703 | 5.26691727 | 5.131544 | 5.1085348 | 5.1372040 | 4.8959834 | 4.952856 | 4.8961250 |
| GO:0060039 | pericardium development | 16 | 0.5965507 | 1.865097 | 5.487805e-03 | 0.199295830 | 0.187013955 | 3882 | tags=44%, list=23%, signal=34% | WT1||SOS1||BMP2||NOTCH1||CCM2||FLRT3||BMP7 | 2.638406 | 3.2346428 | 2.9009791 | 3.393027 | 2.673485 | 3.0115760 | 3.3009752 | 3.3972917 | 2.545715 | 2.611363 | 2.750502 | 3.1601402 | 3.3009450 | 3.2393971 | 3.022449 | 2.86488615 | 2.8068914 | 3.3726647 | 3.3920780 | 3.414041 | 2.687465 | 2.528005 | 2.792754 | 2.910154 | 3.030051 | 3.08881634 | 3.333357 | 3.2836658 | 3.2853495 | 3.4008116 | 3.421880 | 3.3686866 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | 241 | 0.2553247 | 1.383016 | 5.653451e-03 | 0.202696037 | 0.190204620 | 4130 | tags=34%, list=25%, signal=26% | ERBB4||IL12RB2||MVP||CCL5||JAK2||GPRC5A||EPHA5||EFEMP1||ACE||FGF8||IL6ST||MIF||CRLF1||MLST8||SHC1||HIPK2||MATK||SRCIN1||TMEM102||HSF1||EPHB3||CSK||MAP2K2||HES5||TGFB1||ERBB3||CHMP6||MAP2K7||MAP2K3||CSPG4||TTBK1||TP53||DMTN||EPHB2||WNT3A||PRKCE||ABL1||PPP2R1A||ROR1||NEURL1||DDR1||MAPK3||EPHA8||HES1||BCR||EGFR||ARHGEF2||RIPK2||IGF1R||ABI2||CD81||SRC||SH3BP5L||ARRB2||DYRK1B||ZGPAT||CBL||BTK||CTF1||TYRO3||EIF2AK2||IL12A||ERBB2||PRKCZ||PPP2R5B||FLT4||PTPN1||CBLB||DVL2||TNFRSF1A||HCLS1||SOCS1||MAP2K5||LTK||EPHB4||NTRK2||PRKCD||NF2||RACK1||NEK1||FYN | 6.036117 | 5.7201006 | 5.8494756 | 5.636387 | 6.093850 | 6.1970198 | 5.6559852 | 5.6075929 | 5.983658 | 5.909318 | 6.199420 | 5.7860266 | 5.6987462 | 5.6730842 | 5.681946 | 5.95599835 | 5.8963213 | 5.7159780 | 5.5773050 | 5.612248 | 6.105611 | 5.941866 | 6.220562 | 6.167869 | 6.346519 | 6.06225563 | 5.663290 | 5.6551698 | 5.6494622 | 5.6037137 | 5.624123 | 5.5947849 |
| GO:0031065 | positive regulation of histone deacetylation | 16 | 0.5927423 | 1.853190 | 5.792683e-03 | 0.206330737 | 0.193615326 | 5258 | tags=62%, list=31%, signal=43% | JDP2||BCL6||TP53||SREBF1||ZBTB7B||PINK1||CTBP1||ING2||PRKD2||AKAP8L | 4.987732 | 4.9587835 | 5.1499679 | 4.946496 | 4.974599 | 5.2588229 | 4.9509107 | 4.9534824 | 4.906926 | 5.009103 | 5.043687 | 4.9260764 | 4.9857982 | 4.9638445 | 5.142663 | 5.19774986 | 5.1080696 | 4.9562312 | 4.9185426 | 4.964301 | 5.007052 | 4.862014 | 5.048175 | 5.226824 | 5.198950 | 5.34640406 | 4.964227 | 4.9629304 | 4.9252352 | 4.9122853 | 5.011441 | 4.9348382 |
| GO:0030522 | intracellular receptor signaling pathway | 214 | 0.2658404 | 1.425657 | 6.092530e-03 | 0.208019217 | 0.195199751 | 3451 | tags=31%, list=21%, signal=25% | JAK2||ESRRG||OAS3||PMEPA1||NR4A3||TRIM24||RXRG||FOXA1||TRERF1||PADI2||HSPA1A||NOP53||NR1D1||NR2F2||HSPA1B||USP15||RARG||EP300||WBP2||CARM1||GPER1||PKN1||SEC14L1||ZBTB7A||PML||VPS18||SMARCA4||RXRA||BMP2||VDR||PAGR1||DDX54||RELA||GPATCH3||ZMIZ1||TKFC||SREBF1||NCOR2||ESRRA||ARID1A||SNAI2||NR2F6||LATS1||CITED2||RNF6||PLIN5||PUM1||DAB2||RIPK2||NR2F1||TBX1||STAT3||SRC||NLRX1||FOXP1||THRA||PPARD||ALOXE3||TADA3||UBE3A||RNF135||CLPB||TNFAIP3||NR1H3||HEYL||TWIST1||AHR | 5.499700 | 5.9385058 | 5.6030206 | 5.769635 | 5.494139 | 5.4986338 | 5.9115002 | 5.7793126 | 5.504670 | 5.598008 | 5.388833 | 5.9575737 | 5.9449562 | 5.9126155 | 5.707910 | 5.50391755 | 5.5899407 | 5.7582348 | 5.7329044 | 5.816487 | 5.524430 | 5.498810 | 5.458410 | 5.501150 | 5.447919 | 5.54519000 | 5.925176 | 5.8983208 | 5.9108789 | 5.7638104 | 5.799797 | 5.7740916 |
| GO:0034505 | tooth mineralization | 16 | 0.5898212 | 1.844057 | 6.097561e-03 | 0.208019217 | 0.195199751 | 6041 | tags=88%, list=36%, signal=56% | COL1A1||WNT6||TFAP2A||NECTIN1||FAM20C||BCOR||TBX1||TSPEAR||ALPL||STIM1||CNNM4||FOXO1||SLC4A2||TCIRG1 | 3.861479 | 4.1335009 | 4.3632204 | 4.188423 | 3.874391 | 4.4085297 | 4.1350091 | 4.1495047 | 3.800162 | 3.732780 | 4.033909 | 4.0590935 | 4.2258329 | 4.1104891 | 4.319250 | 4.47335293 | 4.2902688 | 4.2325287 | 4.1723023 | 4.159378 | 3.805603 | 3.744330 | 4.054245 | 4.157601 | 4.335250 | 4.68254694 | 4.175927 | 4.1612578 | 4.0653622 | 4.1382624 | 4.232944 | 4.0728009 |
| GO:2000352 | negative regulation of endothelial cell apoptotic process | 14 | 0.6251534 | 1.874509 | 6.163780e-03 | 0.209011567 | 0.196130947 | 5151 | tags=79%, list=31%, signal=54% | PAK4||MAPK7||ABL1||GATA3||NDNF||GATA2||TNFAIP3||NFE2L2||GAS6||RAMP2||TNIP2 | 3.473572 | 3.8592486 | 3.7244759 | 3.788249 | 3.495003 | 3.9101617 | 3.8513141 | 3.7987318 | 3.348262 | 3.515391 | 3.549149 | 3.8540473 | 3.8738264 | 3.8497578 | 3.759325 | 3.76626292 | 3.6446359 | 3.7897293 | 3.7387380 | 3.834685 | 3.566105 | 3.225869 | 3.658068 | 3.770355 | 3.925319 | 4.02359169 | 3.873867 | 3.8814268 | 3.7971504 | 3.8057528 | 3.809277 | 3.7810016 |
| GO:0003209 | cardiac atrium morphogenesis | 24 | 0.5488320 | 1.932126 | 6.643357e-03 | 0.217415433 | 0.204016913 | 3029 | tags=42%, list=18%, signal=34% | MESP1||NKX2-5||SOS1||HEY2||BMP2||GATA4||NOTCH1||SOX4||NOG||ZFPM1 | 3.070474 | 3.4887095 | 3.4385993 | 3.595256 | 3.104493 | 3.6228377 | 3.5029623 | 3.5811595 | 2.968414 | 3.036721 | 3.196685 | 3.4458635 | 3.5666865 | 3.4502958 | 3.422135 | 3.52292243 | 3.3663553 | 3.5376531 | 3.5505169 | 3.692430 | 3.115171 | 2.891920 | 3.280272 | 3.473666 | 3.632320 | 3.74933157 | 3.514856 | 3.5002873 | 3.4936616 | 3.4765865 | 3.690923 | 3.5679250 |
| GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 27 | 0.5268685 | 1.909290 | 7.076350e-03 | 0.225124265 | 0.211250678 | 3253 | tags=48%, list=19%, signal=39% | HIPK2||EP300||CREBBP||C20orf27||CITED1||TGFB1I1||GIPC1||CITED2||SMAD4||DAB2||MEN1||FERMT1||STK11 | 5.960663 | 6.3247321 | 5.9406764 | 6.463523 | 5.903785 | 5.9958109 | 6.3621383 | 6.4853672 | 5.921308 | 6.027798 | 5.930450 | 6.3360813 | 6.3189145 | 6.3191332 | 6.153019 | 5.73691190 | 5.9014113 | 6.4684644 | 6.4031813 | 6.516678 | 5.923527 | 5.911342 | 5.876067 | 6.128097 | 6.087956 | 5.74086263 | 6.368141 | 6.3590659 | 6.3591892 | 6.4925056 | 6.481372 | 6.4821971 |
| GO:0060074 | synapse maturation | 19 | 0.5609792 | 1.846752 | 7.083065e-03 | 0.225124265 | 0.211250678 | 3333 | tags=58%, list=20%, signal=46% | PTEN||DAB2IP||BCAN||PFN1||ADGRL1||NEFL||NEURL1||SHANK1||PALM||CDC20||CAMK2B | 7.051865 | 6.7051873 | 6.8837023 | 6.697526 | 7.068263 | 7.2387229 | 6.6879628 | 6.6950223 | 6.969530 | 7.033142 | 7.147273 | 6.7521888 | 6.6740427 | 6.6881220 | 6.700168 | 7.02105928 | 6.9116608 | 6.7871163 | 6.5976030 | 6.701632 | 7.105573 | 6.929748 | 7.159587 | 7.218830 | 7.315119 | 7.17879723 | 6.689175 | 6.6657004 | 6.7086922 | 6.6967052 | 6.707895 | 6.6803337 |
| GO:0043433 | negative regulation of DNA-binding transcription factor activity | 124 | 0.3133381 | 1.550005 | 7.410678e-03 | 0.230123299 | 0.215941640 | 4091 | tags=43%, list=24%, signal=33% | HMOX1||FOXP3||ID3||NLRC5||NUPR1||IRAK1||PIAS4||NFKBIB||SUFU||GLIS2||DAP||DAB2IP||TAF3||ZNF431||HES6||KLF4||ID1||NFKBIL1||BHLHE40||CDKN2A||PRDX2||KEAP1||SFRP5||ARRB1||HES1||PTCH1||MAD2L2||PEX14||TRAF3||HAND1||MEN1||FLNA||ARRB2||CYP1B1||CCDC22||SIGIRR||PARP10||HDAC4||TNFAIP3||BRMS1||CACTIN||HEYL||TWIST1||NFKBIE||WFS1||WWP2||BMP7||MAP2K5||ZC3H12A||TRIB1||RNF2||CHP1||MAP3K10 | 5.392161 | 5.4264760 | 5.3346160 | 5.362249 | 5.396606 | 5.4941105 | 5.4001138 | 5.3391095 | 5.365065 | 5.357344 | 5.452141 | 5.4616343 | 5.4172537 | 5.3998341 | 5.328626 | 5.35568483 | 5.3192891 | 5.3907400 | 5.3077388 | 5.386757 | 5.395703 | 5.311025 | 5.478247 | 5.449175 | 5.558507 | 5.47233471 | 5.415355 | 5.3801173 | 5.4046434 | 5.3373444 | 5.345098 | 5.3348660 |
| GO:0002115 | store-operated calcium entry | 12 | 0.6703131 | 1.916010 | 7.430694e-03 | 0.230123299 | 0.215941640 | 2533 | tags=42%, list=15%, signal=35% | HOMER3||ORAI2||CRACR2B||ORAI3||ORAI1 | 3.437469 | 4.2779164 | 3.6612765 | 4.272578 | 3.440235 | 3.6351000 | 4.2083576 | 4.1892402 | 3.406800 | 3.488703 | 3.415493 | 4.2188903 | 4.3754852 | 4.2341505 | 3.802025 | 3.60093557 | 3.5696827 | 4.2319507 | 4.3232313 | 4.261040 | 3.385200 | 3.402713 | 3.528515 | 3.513605 | 3.590680 | 3.78707024 | 4.226193 | 4.2268808 | 4.1712966 | 4.1786089 | 4.213523 | 4.1752768 |
| GO:0003184 | pulmonary valve morphogenesis | 15 | 0.5911241 | 1.813216 | 7.440476e-03 | 0.230123299 | 0.215941640 | 5306 | tags=73%, list=32%, signal=50% | HEY2||NFATC1||JAG1||SLIT2||NOTCH1||HEYL||HEY1||SMAD6||NOTCH2||ROBO1||ROBO2 | 3.157793 | 4.1431887 | 3.3824405 | 4.134366 | 3.187832 | 3.5188174 | 4.1158221 | 3.9980430 | 3.095417 | 3.273777 | 3.096794 | 4.0718243 | 4.2255684 | 4.1279595 | 3.723971 | 3.14673398 | 3.2039564 | 4.0616256 | 4.1463102 | 4.192138 | 3.190065 | 3.148198 | 3.224230 | 3.484646 | 3.457389 | 3.60979608 | 4.115808 | 4.1231756 | 4.1084452 | 3.9277611 | 4.022149 | 4.0416654 |
| GO:0021772 | olfactory bulb development | 22 | 0.5437678 | 1.869770 | 7.786053e-03 | 0.238258499 | 0.223575497 | 3438 | tags=45%, list=21%, signal=36% | ERBB4||SKI||SALL1||SLIT2||SRF||DLX2||OGDH||LHX2||UNCX||ATF5 | 3.929605 | 4.6053667 | 3.9592380 | 4.485733 | 3.924908 | 4.1258326 | 4.6492032 | 4.4583737 | 3.893819 | 3.910195 | 3.983222 | 4.5977082 | 4.6283180 | 4.5897862 | 4.111198 | 3.91271587 | 3.8399696 | 4.4472569 | 4.4498400 | 4.557343 | 3.916611 | 3.833286 | 4.018829 | 4.112918 | 4.155404 | 4.10871177 | 4.648614 | 4.6514263 | 4.6475671 | 4.4524065 | 4.459852 | 4.4628430 |
| GO:2000310 | regulation of NMDA receptor activity | 17 | 0.5775444 | 1.836220 | 7.788162e-03 | 0.238258499 | 0.223575497 | 3166 | tags=53%, list=19%, signal=43% | RASGRF1||PPARGC1A||RASGRF2||CNIH2||EPHB2||SLC6A9||CAPN1||MAPK8IP2||PINK1 | 4.040556 | 4.9489480 | 4.3926948 | 4.851130 | 4.034159 | 4.3287839 | 4.8139188 | 4.7593930 | 3.981085 | 4.122092 | 4.014704 | 4.9710280 | 4.9998836 | 4.8728822 | 4.524756 | 4.30367582 | 4.3397700 | 4.8785909 | 4.8597891 | 4.814253 | 4.078897 | 3.985384 | 4.036677 | 4.293637 | 4.234526 | 4.44955220 | 4.877846 | 4.8060383 | 4.7552356 | 4.7477288 | 4.811940 | 4.7168745 |
| GO:0043087 | regulation of GTPase activity | 271 | 0.2387344 | 1.302209 | 7.876093e-03 | 0.239635244 | 0.224867398 | 4107 | tags=32%, list=25%, signal=25% | PLXNC1||CCL5||PROM2||EPHA5||RASGRF1||PLXNA2||RAPGEF3||MLST8||SNX18||MTSS2||ADAP1||GMIP||TRAPPC6A||SBF1||TAX1BP3||RAPGEF1||GNAO1||EPHB3||FGD1||RHOG||S100A10||PREX1||SGSM1||TBC1D10B||ARL2||IQSEC1||SIPA1L3||HRAS||BCL6||ASAP1||DAB2IP||SYDE1||SLC27A4||TBC1D25||RGS7||TBC1D16||PIN1||RGMA||APC2||GPSM1||ADAP2||RTN4R||SH3BP1||RRP1B||MAPRE2||ENSG00000285162||POLDIP2||F2R||EVI5L||TBC1D2||ARHGAP11A||ARRB1||BCR||ARHGAP22||TBC1D13||DVL3||TBC1D17||RASGRP3||AKT2||ARRB2||TBC1D22A||ELMOD1||PLXNA4||ARHGAP12||ABR||RANGAP1||SPRY1||SIPA1||FGD3||TBC1D10A||ERBB2||SCRIB||RAP1GAP||DENND1B||DENND1A||TBC1D9B||WNK1||CBLB||DVL2||USP6NL||RGP1||GPR137B||TBC1D5||NTRK2||ARAP3||RACK1||ARHGEF19||PLXND1 | 5.316111 | 5.3272608 | 5.3040868 | 5.314249 | 5.346186 | 5.4461884 | 5.3406130 | 5.3195001 | 5.274442 | 5.280650 | 5.390273 | 5.3279740 | 5.3361474 | 5.3176010 | 5.280104 | 5.31966407 | 5.3121872 | 5.3242125 | 5.2754917 | 5.342219 | 5.346297 | 5.291652 | 5.398628 | 5.455040 | 5.508363 | 5.37189587 | 5.347425 | 5.3383187 | 5.3360700 | 5.3070490 | 5.337252 | 5.3140257 |
| GO:0031589 | cell-substrate adhesion | 265 | 0.2529416 | 1.373910 | 7.960879e-03 | 0.239635244 | 0.224867398 | 2993 | tags=25%, list=18%, signal=21% | LAMB3||ACVRL1||COL5A3||NTNG1||JAK2||PARVA||PPFIA2||NTN4||TNXB||PARVB||ZYX||PPM1F||SORBS3||EMILIN1||ATRNL1||SRCIN1||NID1||LGALS1||EPHB3||COL1A1||RAC3||S100A10||PTEN||PREX1||ARL2||BCL6||TRIP6||ACTG1||RREB1||JUP||TMEM8B||DMTN||ACTN1||PXN||TRIOBP||ID1||DLC1||DAPK3||PRKCE||CDKN2A||SPOCK1||ABL1||RRAS||MELTF||JAG1||SRF||POLDIP2||L1CAM||DDR1||ROCK1||NDNF||HOXD3||COL26A1||CIB1||FBLN1||BCR||CORO1A||NOTCH1||ADAM9||SRC||MEN1||FLNA||DNM2||FERMT1||UTRN||FBLN2||PPARD | 5.494837 | 5.5271240 | 5.4241855 | 5.389540 | 5.487560 | 5.5090963 | 5.4597378 | 5.3658344 | 5.462460 | 5.521034 | 5.500407 | 5.5439744 | 5.5424018 | 5.4944487 | 5.363365 | 5.45811339 | 5.4491939 | 5.4074908 | 5.3527668 | 5.407671 | 5.536995 | 5.409612 | 5.512925 | 5.532338 | 5.468494 | 5.52560776 | 5.474357 | 5.4528544 | 5.4518899 | 5.3591931 | 5.390148 | 5.3478293 |
| GO:0061469 | regulation of type B pancreatic cell proliferation | 11 | 0.6800946 | 1.894768 | 8.082497e-03 | 0.242002004 | 0.227088304 | 2749 | tags=55%, list=16%, signal=46% | NR4A3||NUPR1||NR1D1||WDR13||WNT3A||MEN1 | 4.357384 | 4.3681427 | 4.6481123 | 4.238546 | 4.504310 | 4.7518082 | 4.2172858 | 4.1119740 | 4.265362 | 4.216469 | 4.565088 | 4.4152941 | 4.3813442 | 4.3056145 | 4.482536 | 4.78419227 | 4.6617711 | 4.3192598 | 4.1804176 | 4.212266 | 4.476914 | 4.352281 | 4.666296 | 4.584497 | 4.780871 | 4.87501036 | 4.258540 | 4.1893681 | 4.2030150 | 4.0847735 | 4.174214 | 4.0748441 |
| GO:0045599 | negative regulation of fat cell differentiation | 44 | 0.4452379 | 1.838024 | 8.166969e-03 | 0.242418848 | 0.227479459 | 2479 | tags=36%, list=15%, signal=31% | JDP2||MKX||CCDC85B||LRP3||ZFP36L2||WNT10B||GPER1||TGFB1||TRIB3||TRPV4||WNT3A||TGFB1I1||JAG1||GATA3||GATA2||TLCD3B | 4.014553 | 4.0039977 | 4.0137960 | 3.997200 | 3.976498 | 4.2033100 | 3.9948104 | 3.9409584 | 3.975842 | 3.908796 | 4.148324 | 3.9711368 | 4.0261805 | 4.0140976 | 4.003362 | 4.06749543 | 3.9687860 | 4.0068111 | 3.9841526 | 4.000542 | 4.017307 | 3.769888 | 4.120204 | 4.119082 | 4.265197 | 4.22177500 | 4.025332 | 3.9758601 | 3.9827393 | 3.9286001 | 3.986610 | 3.9064733 |
| GO:0071364 | cellular response to epidermal growth factor stimulus | 43 | 0.4491047 | 1.827848 | 8.260670e-03 | 0.242418848 | 0.227479459 | 3697 | tags=44%, list=22%, signal=35% | ERBB4||ZFP36L2||ZFP36L1||COL1A1||AKT1||DAB2IP||SOX9||ID1||FOXC1||BAIAP2||SNAI2||GSTP1||EGFR||NCL||CBL||PPP1R9B||INPP5K||ERBB2||ZFP36 | 7.282795 | 8.2592573 | 7.2936654 | 8.241285 | 7.153191 | 7.0133766 | 8.2424957 | 8.2470340 | 7.344702 | 7.375344 | 7.114514 | 8.3708688 | 8.2095209 | 8.1904907 | 7.559832 | 6.92224471 | 7.3284034 | 8.2684023 | 8.1870053 | 8.266953 | 7.096420 | 7.288388 | 7.064452 | 6.985069 | 7.049961 | 7.00432775 | 8.188482 | 8.2677607 | 8.2697641 | 8.2915678 | 8.122596 | 8.3191933 |
| GO:1990090 | cellular response to nerve growth factor stimulus | 43 | 0.4446553 | 1.809739 | 8.260670e-03 | 0.242418848 | 0.227479459 | 4465 | tags=51%, list=27%, signal=38% | MICALL1||MAPT||EEF2||EHD1||RAPGEF1||USP8||AKT1||PTEN||TMEM108||WASF1||ID1||BPTF||CREB1||CIB1||CORO1A||UBE3A||CBL||HSPA5||NTRK2||E2F1||NTRK1||ARF6 | 6.378992 | 6.6783741 | 6.2979068 | 6.574629 | 6.407963 | 6.7391153 | 6.6335772 | 6.5858349 | 6.300030 | 6.382919 | 6.450117 | 6.6681392 | 6.7136402 | 6.6526439 | 6.238461 | 6.36821777 | 6.2840254 | 6.6319947 | 6.5043606 | 6.584657 | 6.463431 | 6.242170 | 6.504717 | 6.797583 | 6.851159 | 6.55121496 | 6.648979 | 6.6424251 | 6.6090101 | 6.5697291 | 6.620575 | 6.5665610 |
| GO:0060973 | cell migration involved in heart development | 13 | 0.6334120 | 1.860106 | 8.333333e-03 | 0.242418848 | 0.227479459 | 3882 | tags=62%, list=23%, signal=47% | SEMA3C||MESP1||SNAI2||NOTCH1||DCHS1||TWIST1||FLRT3||BMP7 | 2.527097 | 2.7301331 | 2.6408160 | 2.652418 | 2.563095 | 2.8762845 | 2.6236144 | 2.5502135 | 2.505761 | 2.459137 | 2.612106 | 2.6909941 | 2.8117169 | 2.6840843 | 2.591797 | 2.71592542 | 2.6116194 | 2.6807988 | 2.6704461 | 2.604839 | 2.607299 | 2.352519 | 2.706742 | 2.786165 | 2.898692 | 2.93965252 | 2.631641 | 2.5697553 | 2.6677495 | 2.4885768 | 2.661152 | 2.4941613 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 133 | 0.3075185 | 1.530252 | 8.494960e-03 | 0.244204030 | 0.229154627 | 3970 | tags=38%, list=24%, signal=29% | NTNG1||DAB1||PCDHGC3||MMP24||GRID2||CDH22||PCDHGA7||PCDH19||SDK2||FXYD5||CDH23||PCDHGB5||LRFN3||MAPK7||IGSF9||BMP2||LRFN4||ADGRL1||PCDH10||CLDN9||KLF4||SCARF2||VSTM2L||NECTIN1||OBSL1||PALLD||L1CAM||PCDHB2||PCDH17||SDK1||DCHS1||LRFN5||CLDN4||PCDH1||CDH4||BSG||PTPRS||PTPN23||NECTIN2||FLRT3||KIFAP3||CDHR1||WNK1||MBP||PVR||PTPRF||PCDH18||MAP2K5||CLSTN2||ARVCF | 3.728390 | 4.3220616 | 4.1835515 | 4.283775 | 3.781500 | 4.2287676 | 4.1861105 | 4.2491240 | 3.647671 | 3.660463 | 3.866427 | 4.3320619 | 4.3696002 | 4.2624827 | 4.126244 | 4.31225226 | 4.1029073 | 4.2873565 | 4.3007336 | 4.262981 | 3.782320 | 3.620694 | 3.925424 | 4.016445 | 4.174832 | 4.45964532 | 4.222235 | 4.1891943 | 4.1458874 | 4.2428853 | 4.323983 | 4.1767279 |
| GO:0050907 | detection of chemical stimulus involved in sensory perception | 13 | -0.7226637 | -1.801681 | 8.892978e-03 | 0.250292855 | 0.234868219 | 1036 | tags=38%, list=6%, signal=36% | TAS2R4||TAS2R20||OR2L2||ASIC3||OR2B6 | -1.292352 | -0.4116952 | 0.1677676 | -0.524191 | -1.359432 | -0.6513143 | -0.3557263 | -0.2868617 | -1.201882 | -1.325799 | -1.353954 | -0.3487589 | -0.4382116 | -0.4502611 | 0.510064 | 0.04795625 | -0.1328246 | -0.6522993 | -0.6629499 | -0.289482 | -1.427704 | -1.573840 | -1.115372 | -1.330115 | -1.136263 | 0.08055799 | -0.237695 | -0.3333873 | -0.5091049 | -0.5495356 | 0.042043 | -0.4222907 |
| GO:0060563 | neuroepithelial cell differentiation | 17 | 0.5654094 | 1.797638 | 9.034268e-03 | 0.253004448 | 0.237412707 | 2744 | tags=59%, list=16%, signal=49% | FGF8||DLX3||B9D1||CEBPB||BMP2||ABL1||FAM20C||HES1||EMX1||SOX4 | 3.789753 | 4.6397903 | 3.7592479 | 4.614715 | 3.801944 | 3.9928646 | 4.6096266 | 4.6024314 | 3.768533 | 3.781482 | 3.818769 | 4.6698833 | 4.6153762 | 4.6335765 | 4.016150 | 3.66421314 | 3.5564111 | 4.5880753 | 4.5614111 | 4.691369 | 3.781753 | 3.703721 | 3.912594 | 3.974353 | 4.061709 | 3.93978004 | 4.610822 | 4.5756532 | 4.6416492 | 4.6174509 | 4.597599 | 4.5921206 |
| GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 45 | 0.4328736 | 1.788443 | 9.225092e-03 | 0.256446872 | 0.240642987 | 4288 | tags=49%, list=26%, signal=36% | ERBB4||CCL5||JAK2||GPER1||MAZ||ERBB3||SOX9||ROR1||F2R||IGF1R||SRC||RGL2||PPARD||PRR5||UBE3A||CBL||PRR5L||UNC5B||HCLS1||NTRK2||FYN||NTRK1 | 5.211792 | 5.0840038 | 5.2114839 | 4.954311 | 5.316445 | 5.4855505 | 5.0573940 | 5.0080892 | 5.104087 | 5.119414 | 5.393163 | 5.0904928 | 5.1196465 | 5.0407724 | 5.102735 | 5.31151762 | 5.2126511 | 4.9959748 | 4.9039055 | 4.961556 | 5.294341 | 5.185598 | 5.456450 | 5.428434 | 5.594502 | 5.42721619 | 5.081327 | 5.0536177 | 5.0368878 | 5.0165962 | 5.023841 | 4.9835110 |
| GO:0048096 | chromatin-mediated maintenance of transcription | 11 | 0.6739591 | 1.877674 | 9.476031e-03 | 0.261473430 | 0.245359776 | 5325 | tags=91%, list=32%, signal=62% | PADI2||ARID1B||ZMIZ1||ELOF1||ARID1A||SMARCD1||ZMIZ2||SERTAD2||SERTAD1||CHEK1 | 5.367097 | 5.2768361 | 5.4086186 | 5.165860 | 5.411677 | 5.6122556 | 5.2944240 | 5.2377883 | 5.281379 | 5.326341 | 5.485507 | 5.3086931 | 5.2562666 | 5.2650000 | 5.315582 | 5.50807206 | 5.3956992 | 5.1993178 | 5.0811922 | 5.213448 | 5.410978 | 5.274474 | 5.537598 | 5.548756 | 5.642183 | 5.64379140 | 5.318173 | 5.2573947 | 5.3069813 | 5.2598645 | 5.226942 | 5.2263027 |
| GO:0045666 | positive regulation of neuron differentiation | 66 | 0.3913741 | 1.744802 | 1.002950e-02 | 0.265706868 | 0.249332323 | 5501 | tags=52%, list=33%, signal=35% | NBL1||DAB1||FOXA1||NKX2-5||ETV5||BEND6||PTEN||BCL6||BMP2||TCF3||EIF4G1||MAP1B||HOXD3||GATA2||DLX2||ARHGEF2||NKX2-2||NKX6-1||ADRA2C||IRX3||HEYL||KCTD11||BMP7||FOXG1||CXCL12||TIMP2||RARA||CPNE1||CSNK1E||TGIF2||CYB5D2||PRPF19||IMPACT||CSNK1D | 5.117058 | 5.1399974 | 5.3345408 | 5.119742 | 5.108724 | 5.3923401 | 5.1663334 | 5.1648982 | 5.062064 | 5.115354 | 5.171673 | 5.1292094 | 5.1544459 | 5.1362193 | 5.310622 | 5.36617418 | 5.3262555 | 5.1314658 | 5.0630065 | 5.162951 | 5.120197 | 4.995242 | 5.203178 | 5.301695 | 5.340595 | 5.52475703 | 5.197435 | 5.1397548 | 5.1612198 | 5.1413486 | 5.199151 | 5.1535493 |
| GO:0032836 | glomerular basement membrane development | 10 | 0.6818157 | 1.841681 | 1.032889e-02 | 0.265706868 | 0.249332323 | 1598 | tags=50%, list=10%, signal=45% | WT1||SULF1||NID1||COL4A4||COL4A3 | 4.373399 | 4.6093139 | 4.8829030 | 4.456527 | 4.417680 | 4.8577464 | 4.5253930 | 4.3447717 | 4.330139 | 4.319946 | 4.465489 | 4.6202302 | 4.6389964 | 4.5677739 | 4.937169 | 4.94660982 | 4.7571442 | 4.4517555 | 4.5167036 | 4.398697 | 4.412440 | 4.286046 | 4.543106 | 4.636767 | 4.752337 | 5.13570293 | 4.568416 | 4.5271097 | 4.4792749 | 4.3508258 | 4.339661 | 4.3438058 |
| GO:0002076 | osteoblast development | 13 | 0.6248123 | 1.834852 | 1.034483e-02 | 0.265706868 | 0.249332323 | 3648 | tags=46%, list=22%, signal=36% | PTH1R||JUND||LRP5||MEN1||HDAC4||SATB2 | 3.775990 | 3.8644834 | 4.0916649 | 3.750017 | 3.781670 | 4.3337075 | 3.7590481 | 3.5940287 | 3.707144 | 3.740520 | 3.874795 | 3.8722843 | 3.9147962 | 3.8042197 | 3.949365 | 4.23462071 | 4.0768179 | 3.8392317 | 3.7115156 | 3.694939 | 3.793887 | 3.557944 | 3.964545 | 4.197087 | 4.320759 | 4.47027640 | 3.820859 | 3.7351933 | 3.7190019 | 3.5786827 | 3.654068 | 3.5472361 |
| GO:0042219 | cellular modified amino acid catabolic process | 23 | 0.5284025 | 1.834592 | 1.038422e-02 | 0.265706868 | 0.249332323 | 2926 | tags=43%, list=17%, signal=36% | DIO2||CHAC1||ALDH4A1||AHCY||GGACT||DPEP1||DIO3||SARDH||NA||HOGA1 | 5.225075 | 5.1224744 | 5.2627731 | 5.017458 | 5.270010 | 5.4755994 | 5.0454079 | 4.9797486 | 5.147122 | 5.210067 | 5.313146 | 5.1925634 | 5.1144739 | 5.0571775 | 5.156533 | 5.34054464 | 5.2851160 | 5.0649253 | 4.9593406 | 5.026137 | 5.333833 | 5.156133 | 5.313589 | 5.493867 | 5.472448 | 5.46028215 | 5.064182 | 5.0490689 | 5.0226669 | 4.9627232 | 5.013548 | 4.9623709 |
| GO:0043030 | regulation of macrophage activation | 27 | 0.4956103 | 1.796015 | 1.042831e-02 | 0.265706868 | 0.249332323 | 5386 | tags=63%, list=32%, signal=43% | MIF||NR1D1||JUND||TTBK1||SHPK||SPHK1||MYO18A||LRFN5||NR1H3||CEBPA||GPR137B||ZC3H12A||GRN||LDLR||IL4R||TNIP2||HSPD1 | 7.814790 | 7.2987439 | 7.3895952 | 7.241303 | 7.877124 | 7.9818594 | 7.1638075 | 7.1450284 | 7.789800 | 7.587207 | 8.032774 | 7.4561849 | 7.2048635 | 7.2212369 | 7.141285 | 7.56633707 | 7.4291402 | 7.3820890 | 7.1495009 | 7.181123 | 7.888421 | 7.625339 | 8.081617 | 7.886761 | 8.243023 | 7.77320876 | 7.154801 | 7.1625700 | 7.1739867 | 7.1661452 | 7.135914 | 7.1327906 |
| GO:0030513 | positive regulation of BMP signaling pathway | 29 | 0.4877706 | 1.810401 | 1.052632e-02 | 0.265706868 | 0.249332323 | 2569 | tags=34%, list=15%, signal=29% | ACVRL1||SULF1||FKBP8||HES5||GATA4||FBXL15||UBE2O||SMAD4||HES1||NOTCH1 | 4.597959 | 4.4842683 | 4.6606882 | 4.467502 | 4.622056 | 4.9006111 | 4.4339713 | 4.4679434 | 4.558295 | 4.499403 | 4.726451 | 4.4927869 | 4.5027957 | 4.4568182 | 4.569588 | 4.74161708 | 4.6657214 | 4.5010559 | 4.4276624 | 4.472838 | 4.609678 | 4.530973 | 4.719297 | 4.832568 | 4.962254 | 4.90409109 | 4.477358 | 4.4249741 | 4.3984611 | 4.4569571 | 4.498521 | 4.4478444 |
| GO:0046321 | positive regulation of fatty acid oxidation | 13 | 0.6224871 | 1.828024 | 1.063218e-02 | 0.265993614 | 0.249601398 | 4133 | tags=62%, list=25%, signal=46% | NR4A3||PPARGC1A||MTLN||PLIN5||AKT2||PPARD||TWIST1||IRS2 | 3.614846 | 3.4879187 | 3.9640389 | 3.354400 | 3.704020 | 4.1381131 | 3.4115377 | 3.2719611 | 3.449182 | 3.531327 | 3.834840 | 3.5587018 | 3.4689127 | 3.4332281 | 3.905227 | 4.09293485 | 3.8846964 | 3.4461803 | 3.2590124 | 3.351938 | 3.717058 | 3.453156 | 3.906318 | 3.930078 | 4.206240 | 4.25704344 | 3.503319 | 3.3078647 | 3.4168001 | 3.2189347 | 3.324735 | 3.2702732 |
| GO:0022038 | corpus callosum development | 14 | 0.5940310 | 1.781189 | 1.085999e-02 | 0.268126480 | 0.251602823 | 4759 | tags=71%, list=28%, signal=51% | EPHB3||EPHB2||C12orf57||RTN4R||NSUN5||NIN||TSKU||PTPRS||CDK5||RTN4RL1 | 5.231635 | 5.2114457 | 5.2560120 | 4.908348 | 5.338507 | 5.5125989 | 5.0912754 | 4.8648145 | 5.187550 | 5.008877 | 5.462047 | 5.3096787 | 5.2234477 | 5.0930079 | 5.136024 | 5.37059393 | 5.2518846 | 4.9139483 | 4.8950893 | 4.915916 | 5.349899 | 5.172928 | 5.476669 | 5.424190 | 5.623400 | 5.48288334 | 5.135680 | 5.1031557 | 5.0330934 | 4.8042020 | 4.910315 | 4.8778863 |
| GO:0048665 | neuron fate specification | 19 | 0.5339043 | 1.757620 | 1.094656e-02 | 0.268126480 | 0.251602823 | 7157 | tags=95%, list=43%, signal=54% | TLX3||FOXA1||SUFU||GLI3||NKX2-2||EHMT2||ISL2||DMRTA2||SIX1||DLL1||ISL1||GLI2||MNX1||ATOH1||HOXD10||POU3F2||POU4F1||EYA1 | 2.787442 | 2.9739603 | 3.3680867 | 3.216943 | 2.857432 | 3.4113231 | 3.0765870 | 3.2648930 | 2.743412 | 2.704287 | 2.906555 | 2.9466247 | 3.0415919 | 2.9311684 | 3.500709 | 3.34983637 | 3.2419570 | 3.2246391 | 3.2197212 | 3.206408 | 2.824476 | 2.722733 | 3.010231 | 3.187186 | 3.346512 | 3.65964194 | 3.101553 | 3.0837621 | 3.0438421 | 3.2334585 | 3.284636 | 3.2760653 |
| GO:0032691 | negative regulation of interleukin-1 beta production | 10 | 0.6737839 | 1.819986 | 1.114433e-02 | 0.269233686 | 0.252641796 | 3937 | tags=70%, list=24%, signal=54% | CPTP||PML||GSTP1||ARRB2||TNFAIP3||GIT1||ZC3H12A | 7.342747 | 6.2572276 | 7.1181222 | 6.280551 | 7.477900 | 7.4909296 | 6.1700864 | 6.2541510 | 7.276360 | 7.146937 | 7.571585 | 6.3108430 | 6.2424217 | 6.2167718 | 6.843690 | 7.28047993 | 7.1941475 | 6.3997661 | 6.1613570 | 6.270639 | 7.431241 | 7.350072 | 7.637033 | 7.424209 | 7.692976 | 7.33067216 | 6.211465 | 6.1363676 | 6.1614100 | 6.2643340 | 6.247388 | 6.2506751 |
| GO:0001964 | startle response | 13 | 0.6139490 | 1.802951 | 1.149425e-02 | 0.273650948 | 0.256786838 | 1603 | tags=46%, list=10%, signal=42% | GRID2||GLRB||GRIN2D||NPAS1||PTEN||DVL1 | 3.915743 | 3.8376853 | 4.1505815 | 3.930827 | 3.908704 | 4.3267696 | 3.8832933 | 3.9031226 | 3.821976 | 3.880225 | 4.036421 | 3.8328236 | 3.8707821 | 3.8087716 | 4.162093 | 4.17800375 | 4.1107963 | 3.9482753 | 3.9221259 | 3.921921 | 3.915351 | 3.776454 | 4.023703 | 4.240537 | 4.363647 | 4.37239392 | 3.942917 | 3.8896771 | 3.8144037 | 3.8703331 | 3.957409 | 3.8800379 |
| GO:0097107 | postsynaptic density assembly | 13 | 0.6142199 | 1.803746 | 1.149425e-02 | 0.273650948 | 0.256786838 | 3332 | tags=54%, list=20%, signal=43% | GRID2||ZDHHC12||PTEN||LRFN4||LRFN1||SHANK3||PTPRS | 3.585886 | 3.7964610 | 3.8458100 | 3.583129 | 3.650486 | 4.0639215 | 3.6652742 | 3.4930417 | 3.540606 | 3.402052 | 3.788195 | 3.7741627 | 3.8540170 | 3.7593962 | 3.788951 | 4.01820986 | 3.7124625 | 3.6329420 | 3.5926397 | 3.521612 | 3.671709 | 3.404092 | 3.842275 | 3.849892 | 4.086742 | 4.22990496 | 3.714262 | 3.6576724 | 3.6223961 | 3.4663094 | 3.599382 | 3.4066339 |
| GO:0060740 | prostate gland epithelium morphogenesis | 20 | 0.5323409 | 1.772386 | 1.157025e-02 | 0.273650948 | 0.256786838 | 2951 | tags=40%, list=18%, signal=33% | SULF1||FOXA1||HOXA13||RARG||SOX9||HOXB13||NOTCH1||NOG | 2.312092 | 3.0651056 | 2.6027561 | 3.141810 | 2.327158 | 2.6693923 | 3.0691610 | 3.1158177 | 2.301009 | 2.351750 | 2.282625 | 3.0449280 | 3.1330939 | 3.0146550 | 2.715845 | 2.52178257 | 2.5633264 | 3.0891363 | 3.1729834 | 3.161881 | 2.349602 | 2.253279 | 2.375730 | 2.601037 | 2.621147 | 2.77930561 | 3.039369 | 3.0509791 | 3.1159482 | 3.1055763 | 3.130041 | 3.1117236 |
| GO:0097062 | dendritic spine maintenance | 16 | 0.5579916 | 1.744543 | 1.189024e-02 | 0.275003892 | 0.258056405 | 4683 | tags=50%, list=28%, signal=36% | ZNF804A||ZMYND8||ABHD17B||IGF1R||ITPKA||PICK1||FYN||PRNP | 3.716334 | 4.7130718 | 4.1961942 | 4.726553 | 3.701250 | 4.1456806 | 4.7499653 | 4.7303468 | 3.680094 | 3.757292 | 3.710567 | 4.6971478 | 4.7459423 | 4.6955544 | 4.426785 | 4.13653230 | 3.9906236 | 4.6804253 | 4.6978688 | 4.798527 | 3.698759 | 3.618420 | 3.781935 | 3.969581 | 4.039023 | 4.39199138 | 4.741239 | 4.7673201 | 4.7411788 | 4.6920188 | 4.786434 | 4.7108467 |
| GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | 12 | 0.6325183 | 1.807978 | 1.200343e-02 | 0.275784918 | 0.258789300 | 2861 | tags=50%, list=17%, signal=41% | PLCB1||GRK2||CHRM4||GNA11||CHRM3||HRH3 | 4.893457 | 5.1156089 | 5.0886062 | 5.059752 | 4.886372 | 5.1611386 | 5.1653610 | 5.0573159 | 4.829588 | 4.948236 | 4.900084 | 5.0405341 | 5.1890510 | 5.1134186 | 5.073886 | 5.12932072 | 5.0617077 | 5.1027101 | 4.9975075 | 5.076965 | 4.928269 | 4.768477 | 4.955390 | 5.070630 | 5.061744 | 5.33411763 | 5.189280 | 5.1591097 | 5.1473689 | 5.0254094 | 5.081349 | 5.0646191 |
| GO:0039702 | viral budding via host ESCRT complex | 21 | 0.5211070 | 1.763785 | 1.269205e-02 | 0.285774215 | 0.268162993 | 4648 | tags=67%, list=28%, signal=48% | MVB12A||VPS4A||VPS37D||CHMP6||CHMP4C||CHMP2B||VPS37B||PDCD6IP||CHMP1A||VPS37C||CHMP4B||VPS28||VPS37A||CHMP2A | 5.643077 | 5.7816533 | 5.5970185 | 5.665576 | 5.667216 | 5.7853546 | 5.7691573 | 5.6934196 | 5.588336 | 5.631965 | 5.706449 | 5.7848528 | 5.7851682 | 5.7749154 | 5.623576 | 5.59349166 | 5.5735474 | 5.6680739 | 5.6106778 | 5.716050 | 5.679713 | 5.595999 | 5.723058 | 5.798162 | 5.818810 | 5.73787033 | 5.775773 | 5.7487372 | 5.7827382 | 5.6808850 | 5.721504 | 5.6774529 |
| GO:0001501 | skeletal system development | 387 | 0.2170638 | 1.230916 | 1.312293e-02 | 0.288996949 | 0.271187121 | 3542 | tags=29%, list=21%, signal=24% | ACVRL1||MKX||NOTUM||SULF1||PTH1R||MMP2||EFEMP1||FGF8||PAPSS2||HOXA13||NLE1||MEX3C||HSD17B1||PHOSPHO1||RARG||EP300||WNT10B||COL9A2||ZNF219||DLL3||IRX5||COL1A1||DEAF1||COL5A2||RUNX1||HES5||SNX10||SUFU||NFIB||SKI||TGFB1||HOXB9||PITX1||TFAP2A||BCAN||RPL13||SOX9||FUZ||LRP5||PAX1||BMP2||MEF2D||VDR||TRPV4||PCGF2||TP53||TCOF1||HAPLN3||ALX4||HOXA10||FLI1||FOXC1||GNA11||KIAA1217||GSC||ZNF385A||ZBTB16||ESRRA||TEAD4||IFT140||SNAI2||TLE5||ANKH||MAF||TULP3||GLI3||GNAS||SRF||CITED2||COL19A1||WASF2||HOXA3||MAPK3||CSRNP1||SNORC||HOXD3||MED12||CREB3L2||DLX2||NDST1||TBX1||PIP4K2A||HOXC9||HAND1||EVC||SOX4||SRC||SLC39A3||DCHS1||NCAN||SERPINH1||FOXP1||GDF11||ATP6AP1||NOG||ANKRD11||THRA||ZFPM1||IHH||TBX3||HOXB8||HOXB2||HDAC4||SIX2||TIMP1||TSKU||UNCX||RAI1||TWIST1||VKORC1||HOXA6||FOXN3||SLC38A10||NAB2 | 5.342422 | 5.2870595 | 5.2245714 | 5.225465 | 5.268150 | 5.3296623 | 5.2436226 | 5.1931377 | 5.370132 | 5.334859 | 5.321840 | 5.3110217 | 5.2991820 | 5.2502580 | 5.181010 | 5.25505938 | 5.2366193 | 5.2593296 | 5.1969942 | 5.219377 | 5.310390 | 5.209377 | 5.282802 | 5.321107 | 5.408456 | 5.25532492 | 5.262244 | 5.2405188 | 5.2278961 | 5.1869014 | 5.210186 | 5.1821690 |
| GO:0051547 | regulation of keratinocyte migration | 10 | 0.6637338 | 1.792839 | 1.331884e-02 | 0.288996949 | 0.271187121 | 2710 | tags=50%, list=16%, signal=42% | PTEN||IQSEC1||EPPK1||MAPRE2||ADAM9 | 3.182555 | 4.2996178 | 3.5918022 | 4.405071 | 3.069704 | 3.6170130 | 4.3589012 | 4.4686556 | 3.138002 | 3.305728 | 3.095253 | 4.2282198 | 4.3477260 | 4.3202108 | 3.941028 | 3.41203063 | 3.3458868 | 4.3304483 | 4.4262887 | 4.455530 | 3.139274 | 2.973880 | 3.090983 | 3.600589 | 3.529943 | 3.71447144 | 4.346142 | 4.3618839 | 4.3685859 | 4.4488469 | 4.498862 | 4.4577633 |
| GO:1904779 | regulation of protein localization to centrosome | 10 | 0.6660748 | 1.799162 | 1.331884e-02 | 0.288996949 | 0.271187121 | 386 | tags=30%, list=2%, signal=29% | MARK4||PARD6A||BICD1 | 4.422282 | 4.6435607 | 4.6226154 | 4.604298 | 4.438107 | 4.7381448 | 4.6671837 | 4.6022579 | 4.343493 | 4.506097 | 4.412627 | 4.6546445 | 4.6578759 | 4.6178194 | 4.641059 | 4.64141120 | 4.5846395 | 4.5974789 | 4.5188547 | 4.691381 | 4.538967 | 4.313638 | 4.452814 | 4.726314 | 4.669441 | 4.81494035 | 4.713991 | 4.6435155 | 4.6428799 | 4.5866395 | 4.607693 | 4.6123115 |
| GO:0150105 | protein localization to cell-cell junction | 19 | 0.5277658 | 1.737413 | 1.352222e-02 | 0.288996949 | 0.271187121 | 3429 | tags=53%, list=20%, signal=42% | TJP3||LSR||ACTG1||ACTB||TJP1||FLNA||DLG5||ARHGEF18||CGNL1||SCRIB | 8.542217 | 7.8198399 | 7.9454093 | 7.731939 | 8.569403 | 8.4169615 | 7.7571394 | 7.7176592 | 8.491791 | 8.530894 | 8.601802 | 7.7992127 | 7.8387733 | 7.8212616 | 7.663708 | 8.08247410 | 8.0533836 | 7.8225348 | 7.6264378 | 7.740151 | 8.676999 | 8.434464 | 8.586413 | 8.590148 | 8.377884 | 8.26363822 | 7.759516 | 7.7766923 | 7.7349043 | 7.7335091 | 7.720877 | 7.6983727 |
| GO:0035930 | corticosteroid hormone secretion | 10 | 0.6611805 | 1.785942 | 1.359065e-02 | 0.288996949 | 0.271187121 | 3207 | tags=40%, list=19%, signal=32% | KCNQ1||SELENOM||GAL||C1QTNF1 | 5.001241 | 4.7197821 | 4.7198913 | 4.635498 | 5.005440 | 5.0592595 | 4.6649355 | 4.6112053 | 5.009393 | 4.841442 | 5.137672 | 4.7575292 | 4.6917529 | 4.7092571 | 4.580552 | 4.86299838 | 4.7021593 | 4.6474844 | 4.6045733 | 4.653939 | 5.058174 | 4.831234 | 5.111809 | 5.019755 | 5.206570 | 4.93819386 | 4.701077 | 4.6404170 | 4.6525962 | 4.5908707 | 4.665285 | 4.5758617 |
| GO:0045820 | negative regulation of glycolytic process | 10 | 0.6613812 | 1.786484 | 1.359065e-02 | 0.288996949 | 0.271187121 | 3879 | tags=70%, list=23%, signal=54% | NUPR1||PPARGC1A||DDIT4||STAT3||CBFA2T3||HDAC4||GIT1 | 3.550037 | 3.8667496 | 4.1211744 | 4.323986 | 3.671900 | 4.4420887 | 3.8737919 | 4.2520533 | 3.512541 | 3.514031 | 3.620845 | 3.8288269 | 3.8773624 | 3.8932809 | 4.058858 | 4.21387770 | 4.0859920 | 4.4112245 | 4.2049906 | 4.348074 | 3.619473 | 3.599499 | 3.789108 | 4.495469 | 4.475507 | 4.35108903 | 3.876774 | 3.8210744 | 3.9217662 | 4.2415965 | 4.282369 | 4.2316933 |
| GO:0090025 | regulation of monocyte chemotaxis | 11 | 0.6418337 | 1.788171 | 1.365663e-02 | 0.288996949 | 0.271187121 | 101 | tags=18%, list=1%, signal=18% | NBL1||CCL5 | 5.058869 | 6.3717081 | 5.1154005 | 6.393448 | 4.912986 | 4.8723209 | 6.4867489 | 6.4011555 | 5.105152 | 5.210995 | 4.834877 | 6.3345911 | 6.3549995 | 6.4240045 | 5.596238 | 4.80230500 | 4.7926311 | 6.3176177 | 6.3937421 | 6.465210 | 4.817062 | 5.042351 | 4.869809 | 4.797562 | 4.768995 | 5.03532299 | 6.481184 | 6.4669132 | 6.5117847 | 6.4633971 | 6.293532 | 6.4407117 |
| GO:0061082 | myeloid leukocyte cytokine production | 22 | 0.5105437 | 1.755527 | 1.387949e-02 | 0.290437262 | 0.272538673 | 1181 | tags=36%, list=7%, signal=34% | HMOX1||NR4A3||HLA-G||SEMA7A||TICAM1||TGFB1||CUEDC2||BCL6 | 3.648231 | 4.6028230 | 3.4987612 | 4.651194 | 3.605763 | 3.7210074 | 4.6923295 | 4.6882042 | 3.620988 | 3.640144 | 3.682862 | 4.5755846 | 4.6048399 | 4.6275740 | 3.775132 | 3.37567620 | 3.2988984 | 4.5701707 | 4.6242569 | 4.752994 | 3.577825 | 3.515722 | 3.716371 | 3.725562 | 3.843159 | 3.58253530 | 4.656896 | 4.6919469 | 4.7272868 | 4.6883759 | 4.682377 | 4.6938367 |
| GO:0003215 | cardiac right ventricle morphogenesis | 20 | 0.5198450 | 1.730781 | 1.421488e-02 | 0.294683990 | 0.276523691 | 2744 | tags=50%, list=16%, signal=42% | SEMA3C||NKX2-5||HEY2||PPP1R13L||GATA4||JAG1||GATA3||NOTCH1||HAND1||SOX4 | 3.004774 | 3.2344474 | 3.1730063 | 3.291343 | 3.023615 | 3.3490884 | 3.2460637 | 3.2431100 | 2.958093 | 2.922400 | 3.125572 | 3.2629955 | 3.2117865 | 3.2280849 | 3.296775 | 3.15564977 | 3.0564417 | 3.2416954 | 3.2856494 | 3.344824 | 3.021062 | 2.817281 | 3.206349 | 3.270737 | 3.377273 | 3.39611456 | 3.240641 | 3.2688568 | 3.2283948 | 3.3365973 | 3.138565 | 3.2473681 |
| GO:0045824 | negative regulation of innate immune response | 45 | 0.3997318 | 1.651516 | 1.476015e-02 | 0.299945382 | 0.281460842 | 4627 | tags=51%, list=28%, signal=37% | OAS3||NLRC5||HLA-G||USP15||SUSD4||YTHDF2||NMI||TTLL12||LGALS9||NLRX1||ARRB2||FAM3A||TNFAIP3||CACTIN||TYRO3||NR1H3||HLA-A||HLA-E||GRN||NR1H2||LYAR||CNOT7||SAMHD1 | 6.922903 | 6.5084994 | 6.7874384 | 6.342917 | 6.917807 | 6.8487571 | 6.4051643 | 6.3076105 | 6.948789 | 6.852617 | 6.964772 | 6.5677579 | 6.5044508 | 6.4509158 | 6.641651 | 6.88242481 | 6.8273608 | 6.4060031 | 6.2807089 | 6.339311 | 6.930322 | 6.822085 | 6.995711 | 6.870350 | 6.921367 | 6.74917456 | 6.447625 | 6.3786958 | 6.3882008 | 6.3035401 | 6.313725 | 6.3055462 |
| GO:0005513 | detection of calcium ion | 10 | -0.7442469 | -1.737310 | 1.486636e-02 | 0.300886017 | 0.282343509 | 878 | tags=40%, list=5%, signal=38% | CALM2||KCNIP2||RYR2||CASR | 6.116351 | 6.5304833 | 6.1409523 | 6.556360 | 6.096110 | 6.1787065 | 6.6094619 | 6.6370964 | 6.114860 | 6.214044 | 6.013161 | 6.4933144 | 6.5182221 | 6.5785763 | 6.227579 | 6.08649463 | 6.1046624 | 6.5726848 | 6.5226827 | 6.573133 | 6.067436 | 6.124954 | 6.095365 | 6.109511 | 6.221840 | 6.20229087 | 6.580465 | 6.6075888 | 6.6397213 | 6.6176182 | 6.657742 | 6.6356488 |
| GO:0001974 | blood vessel remodeling | 29 | 0.4555114 | 1.690668 | 1.503759e-02 | 0.301233511 | 0.282669588 | 2819 | tags=41%, list=17%, signal=34% | ACVRL1||SEMA3C||ACE||FGF8||TGFB1||BAK1||FOXC1||JAG1||BAX||HOXA3||CST3||FLNA | 5.085654 | 4.6193378 | 5.0506620 | 4.585064 | 5.092476 | 5.3133516 | 4.5514578 | 4.4958129 | 5.071747 | 4.948871 | 5.223221 | 4.6140507 | 4.6319382 | 4.6119409 | 4.900314 | 5.19556169 | 5.0409825 | 4.6466652 | 4.5387053 | 4.567647 | 5.075409 | 4.955918 | 5.232698 | 5.220442 | 5.398330 | 5.31579912 | 4.626748 | 4.5216145 | 4.5028972 | 4.4990904 | 4.527642 | 4.4599058 |
| GO:0051156 | glucose 6-phosphate metabolic process | 18 | 0.5298058 | 1.714466 | 1.530124e-02 | 0.305428008 | 0.286605594 | 2727 | tags=50%, list=16%, signal=42% | PGLS||GPI||G6PD||HK2||SHPK||TP53||TKT||G6PC3||HK1 | 7.294661 | 6.9175962 | 7.2788327 | 7.002399 | 7.322979 | 7.4462376 | 6.8837697 | 6.9646767 | 7.275784 | 7.354180 | 7.252027 | 6.9290232 | 6.9327686 | 6.8906207 | 7.091723 | 7.38294385 | 7.3448248 | 7.0698231 | 6.9003145 | 7.031634 | 7.413769 | 7.266342 | 7.284297 | 7.519362 | 7.342939 | 7.47070670 | 6.909143 | 6.8519406 | 6.8896406 | 6.9669899 | 6.987576 | 6.9390537 |
| GO:0033599 | regulation of mammary gland epithelial cell proliferation | 15 | 0.5609951 | 1.720798 | 1.577381e-02 | 0.306786080 | 0.287879973 | 3140 | tags=47%, list=19%, signal=38% | ZNF703||GPX1||DEAF1||RREB1||BAX||GATA3||PYGO2 | 7.279535 | 6.6606667 | 6.9856553 | 6.566897 | 7.376302 | 7.4335782 | 6.5444599 | 6.5147937 | 7.235708 | 7.121787 | 7.460318 | 6.7587438 | 6.6281083 | 6.5896534 | 6.805285 | 7.09329047 | 7.0423259 | 6.6539211 | 6.4693396 | 6.571520 | 7.372343 | 7.225977 | 7.516024 | 7.465195 | 7.607848 | 7.19817829 | 6.604279 | 6.4815028 | 6.5449863 | 6.5039046 | 6.531130 | 6.5092020 |
| GO:0045773 | positive regulation of axon extension | 38 | 0.4201763 | 1.656172 | 1.589347e-02 | 0.306786080 | 0.287879973 | 4327 | tags=39%, list=26%, signal=29% | TRPV2||DBNL||MAPT||SEMA7A||TWF2||SRF||LIMK1||L1CAM||DBN1||MAP1B||CDH4||GDI1||MEGF8||NTN1||CXCL12 | 4.797363 | 5.2896377 | 4.9282898 | 5.298149 | 4.798995 | 5.0435798 | 5.3256403 | 5.4031372 | 4.707227 | 4.828739 | 4.851976 | 5.2854137 | 5.3028857 | 5.2805176 | 5.079065 | 4.88037167 | 4.8119157 | 5.2622699 | 5.2753526 | 5.355066 | 4.813389 | 4.685932 | 4.890322 | 4.961543 | 5.091545 | 5.07422841 | 5.327958 | 5.3360415 | 5.3128247 | 5.3478508 | 5.495501 | 5.3613949 |
| GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 16 | -0.6659216 | -1.748448 | 1.591788e-02 | 0.306786080 | 0.287879973 | 1634 | tags=38%, list=10%, signal=34% | CXCL3||LEAP2||RPL39||RPL30||SPINK5||H2BC6 | 10.371731 | 9.8624093 | 10.1298141 | 9.810150 | 10.335840 | 10.0393515 | 9.8411751 | 9.7143275 | 10.432173 | 10.425709 | 10.250011 | 9.9151395 | 9.8298207 | 9.8407654 | 10.033233 | 10.09066220 | 10.2561607 | 9.8415580 | 9.7842222 | 9.804080 | 10.320164 | 10.483923 | 10.188174 | 10.192761 | 10.071978 | 9.83001075 | 9.826680 | 9.8520789 | 9.8446481 | 9.8165832 | 9.419271 | 9.8668471 |
| GO:0002011 | morphogenesis of an epithelial sheet | 49 | 0.3965725 | 1.671217 | 1.625616e-02 | 0.309378304 | 0.290312447 | 4924 | tags=51%, list=29%, signal=36% | ACVRL1||SOS1||MRTFA||PTEN||RREB1||RHOC||DVL1||JAG1||SRF||DDR1||NOTCH1||CARMIL2||CD151||FLNA||FERMT1||ARHGAP12||HOXB2||FLRT3||MEGF8||DVL2||BMP7||DAG1||TMEFF2||PHLDB2||NOTCH2 | 4.840769 | 4.8174554 | 5.0509551 | 4.860269 | 4.848841 | 5.1080609 | 4.7969694 | 4.8163776 | 4.778725 | 4.846204 | 4.895019 | 4.7960156 | 4.8546104 | 4.8010058 | 4.999782 | 5.10366746 | 5.0475402 | 4.8684053 | 4.8308146 | 4.881114 | 4.866973 | 4.747829 | 4.926047 | 5.001935 | 5.058189 | 5.25197453 | 4.827478 | 4.7764200 | 4.7865018 | 4.8000464 | 4.843881 | 4.8048028 |
| GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 23 | 0.4973007 | 1.726608 | 1.626861e-02 | 0.309378304 | 0.290312447 | 4190 | tags=52%, list=25%, signal=39% | HSPA1A||HSPA1B||AKT1||MAPK7||EYA2||PRDX2||SNAI2||EYA4||UNC5B||MAP2K5||FYN||BCL2L1 | 6.950029 | 6.9456919 | 6.4760358 | 6.228188 | 7.151248 | 6.8894234 | 6.8489331 | 6.1765076 | 6.932394 | 6.945032 | 6.972372 | 6.9849412 | 6.9416766 | 6.9094625 | 6.326481 | 6.55770185 | 6.5329414 | 6.2870957 | 6.1441980 | 6.249495 | 7.254270 | 7.007523 | 7.180949 | 6.918788 | 6.965801 | 6.77705714 | 6.903099 | 6.8284343 | 6.8136642 | 6.1697157 | 6.194472 | 6.1651628 |
| GO:0051591 | response to cAMP | 66 | 0.3739383 | 1.667071 | 1.651917e-02 | 0.312145716 | 0.292909314 | 2032 | tags=30%, list=12%, signal=27% | AKAP6||WT1||KCNQ1||RAPGEF3||VGF||ALDH3A1||JUN||RAPGEF1||CARM1||WNT10B||ZFP36L1||COL1A1||HCN2||CITED1||THBD||RELA||DMTN||SREBF1||CDO1||HCN4 | 5.599465 | 6.0394567 | 5.7829015 | 6.077093 | 5.564898 | 5.7008034 | 5.9431965 | 6.0330608 | 5.619869 | 5.669965 | 5.503546 | 6.0863924 | 6.0434907 | 5.9867586 | 5.819717 | 5.75979649 | 5.7684612 | 6.0913971 | 6.0413966 | 6.097825 | 5.580056 | 5.596465 | 5.516956 | 5.675947 | 5.598249 | 5.81942774 | 5.958679 | 5.9329546 | 5.9378260 | 6.0165367 | 6.064128 | 6.0180081 |
| GO:0061430 | bone trabecula morphogenesis | 10 | 0.6492384 | 1.753685 | 1.658059e-02 | 0.312145716 | 0.292909314 | 1479 | tags=40%, list=9%, signal=36% | MMP2||WNT10B||COL1A1||SBNO2 | 2.156895 | 2.4750438 | 2.9428117 | 2.584817 | 2.135206 | 2.8629805 | 2.5280031 | 2.5041092 | 2.010246 | 2.181153 | 2.267522 | 2.3693255 | 2.6163671 | 2.4277376 | 3.018928 | 3.00955296 | 2.7883620 | 2.5379491 | 2.6154616 | 2.599880 | 2.198587 | 1.888392 | 2.288794 | 2.605613 | 2.760190 | 3.16416720 | 2.601141 | 2.5276348 | 2.4513465 | 2.4695208 | 2.586007 | 2.4531289 |
| GO:0070527 | platelet aggregation | 45 | 0.3930072 | 1.623733 | 1.660517e-02 | 0.312145716 | 0.292909314 | 3966 | tags=40%, list=24%, signal=31% | CSRP1||JAK2||IL6ST||HSPB1||ACTG1||ACTB||DMTN||WNT3A||GNAS||STXBP1||TLN1||MYH9||FLNA||FIBP||C1QTNF1||TYRO3||PRKCA||PRKCD | 7.795761 | 7.2819465 | 7.4445030 | 7.309653 | 7.809562 | 7.6828070 | 7.2336977 | 7.2694999 | 7.757095 | 7.823928 | 7.805438 | 7.3026217 | 7.2679873 | 7.2749977 | 7.279709 | 7.51168153 | 7.5289461 | 7.3680025 | 7.2242580 | 7.332836 | 7.879016 | 7.753444 | 7.793359 | 7.797539 | 7.661885 | 7.58062618 | 7.237720 | 7.2316202 | 7.2317439 | 7.3135888 | 7.162772 | 7.3264662 |
| GO:0042593 | glucose homeostasis | 176 | 0.2608443 | 1.362026 | 1.712230e-02 | 0.318090594 | 0.298487831 | 4568 | tags=39%, list=27%, signal=29% | UCP2||EPHA5||FOXA1||PIK3R2||NR1D1||VGF||NGFR||GPI||LGALS1||GPER1||SESN2||PPARGC1A||AKT1||PLSCR3||HK2||SMARCA4||LRP5||FOXK2||CAV3||PRKACA||BAD||TRPV4||FOXK1||PIM3||GATA4||MBD5||NMB||PRKCE||ENDOG||FOXA3||PCK2||SIDT2||SRF||ASPSCR1||RAB11B||PTCH1||GCLM||IGF1R||STAT3||HK1||SOX4||MEN1||CYBA||PDK2||ALOX5||ABCG1||PPARD||RAB11FIP5||STK11||INPP5K||KLF7||C2CD2L||FIS1||BACE2||WFS1||CEBPA||PPP3CA||LRRC8D||RACK1||KLF15||IRS2||PHPT1||SERPINF1||BRSK2||POLG||IGFBP5||PPP1R3G||ZNF236||HIF1A | 6.684415 | 6.8286258 | 6.5848726 | 6.746188 | 6.661799 | 6.5726488 | 6.7912244 | 6.7703857 | 6.697844 | 6.716210 | 6.638038 | 6.9037754 | 6.7904798 | 6.7885799 | 6.550039 | 6.53576885 | 6.6652833 | 6.7709567 | 6.6983970 | 6.768048 | 6.653589 | 6.712831 | 6.617363 | 6.647903 | 6.578929 | 6.48658696 | 6.785639 | 6.7873386 | 6.8006484 | 6.7439692 | 6.783329 | 6.7835006 |
| GO:0007595 | lactation | 37 | 0.4191193 | 1.641811 | 1.733615e-02 | 0.320353761 | 0.300611527 | 2453 | tags=35%, list=15%, signal=30% | ERBB4||APRT||HK2||VDR||NCOR2||FOXB1||CDO1||USF2||MTCO2P12||NEURL1||DDR1||MTX1||CREB1 | 9.281733 | 8.5412210 | 9.6058172 | 8.908619 | 9.366476 | 9.6023925 | 8.6645583 | 8.7783728 | 9.147691 | 9.217333 | 9.461093 | 7.9913668 | 8.7453998 | 8.7614028 | 9.397407 | 9.79705447 | 9.5953445 | 8.8257574 | 9.1545708 | 8.707610 | 9.410510 | 9.256088 | 9.426757 | 9.464278 | 9.486044 | 9.82738876 | 8.648123 | 8.6945116 | 8.6505663 | 8.8570686 | 8.746523 | 8.7281275 |
| GO:0090083 | regulation of inclusion body assembly | 15 | 0.5586389 | 1.713570 | 1.755952e-02 | 0.321962735 | 0.302121346 | 1177 | tags=33%, list=7%, signal=31% | HSPA1A||HSPA1B||HSF1||SNCAIP||CDC34 | 7.127172 | 7.4494596 | 6.5072683 | 6.571986 | 7.351779 | 6.8961812 | 7.3775696 | 6.5547249 | 7.136223 | 7.190984 | 7.050866 | 7.4815831 | 7.4414713 | 7.4247314 | 6.448704 | 6.51047867 | 6.5604536 | 6.5950720 | 6.5034313 | 6.615016 | 7.487567 | 7.230199 | 7.325759 | 6.961417 | 6.922100 | 6.80017088 | 7.435601 | 7.3479408 | 7.3473676 | 6.5277258 | 6.570585 | 6.5654854 |
| GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 26 | 0.4727032 | 1.698784 | 1.784414e-02 | 0.325063582 | 0.305031099 | 4505 | tags=46%, list=27%, signal=34% | HSPA1A||TLR5||HSPA1B||IFI35||USP15||NINJ1||PUM1||CYBA||NR1H3||TIRAP||FLOT1||PELI1 | 6.393924 | 6.9874664 | 5.9180296 | 6.474349 | 6.581998 | 6.1826501 | 6.9678724 | 6.4398827 | 6.421453 | 6.435213 | 6.322505 | 6.9945693 | 6.9940256 | 6.9737064 | 5.987508 | 5.91866501 | 5.8443661 | 6.4566963 | 6.4390464 | 6.525839 | 6.689318 | 6.472774 | 6.575764 | 6.195349 | 6.189683 | 6.16270812 | 7.009850 | 6.9514609 | 6.9413540 | 6.4563506 | 6.401227 | 6.4613032 |
| GO:0007212 | dopamine receptor signaling pathway | 30 | 0.4468750 | 1.673048 | 1.829268e-02 | 0.328448796 | 0.308207695 | 2863 | tags=33%, list=17%, signal=28% | GNAL||CALY||KLF16||SLC9A3R1||GNAO1||GNA11||GNAS||PALM||FLNA||ARRB2 | 5.878103 | 6.0931219 | 5.9568093 | 5.904357 | 5.877165 | 6.1389491 | 6.0806295 | 5.8870248 | 5.813155 | 5.891755 | 5.927054 | 6.0999684 | 6.1105523 | 6.0685143 | 5.924257 | 6.00267759 | 5.9423191 | 5.9346646 | 5.8548411 | 5.922292 | 5.878808 | 5.772743 | 5.973000 | 6.100194 | 6.143641 | 6.17210475 | 6.099534 | 6.0880540 | 6.0539115 | 5.8738344 | 5.893469 | 5.8936816 |
| GO:0097320 | plasma membrane tubulation | 17 | 0.5312314 | 1.688974 | 1.838006e-02 | 0.328448796 | 0.308207695 | 4075 | tags=59%, list=24%, signal=45% | MICALL1||PACSIN3||ASAP1||EHD2||PLEKHM2||PACSIN1||SNX33||DNM2||WASL||SH3GLB1 | 4.220128 | 4.3293067 | 4.3484193 | 4.301932 | 4.264033 | 4.5498353 | 4.3435500 | 4.2941725 | 4.133010 | 4.189961 | 4.330213 | 4.3074936 | 4.3455556 | 4.3346053 | 4.352999 | 4.39323094 | 4.2974273 | 4.3531057 | 4.2209218 | 4.328375 | 4.266846 | 4.150832 | 4.366375 | 4.516434 | 4.560085 | 4.57238914 | 4.368816 | 4.3225732 | 4.3388787 | 4.2753607 | 4.339937 | 4.2660929 |
| GO:0046597 | negative regulation of viral entry into host cell | 14 | 0.5599683 | 1.679053 | 1.849134e-02 | 0.329391647 | 0.309092441 | 3490 | tags=57%, list=21%, signal=45% | PTX3||IFITM2||TRIM8||GSN||TRIM11||LY6E||TRIM59||TRIM26 | 5.114276 | 4.9603936 | 5.2083105 | 5.006592 | 5.154819 | 5.3758896 | 4.9214211 | 4.9420226 | 5.061155 | 5.005401 | 5.263327 | 4.9847423 | 4.9881159 | 4.9068673 | 5.058814 | 5.35525692 | 5.1955749 | 5.0902659 | 4.9692664 | 4.956428 | 5.149371 | 5.015813 | 5.286573 | 5.241465 | 5.419183 | 5.45791586 | 4.925077 | 4.9177725 | 4.9214047 | 4.9589363 | 4.974587 | 4.8911898 |
| GO:0042246 | tissue regeneration | 44 | 0.3836916 | 1.583950 | 1.905626e-02 | 0.332385445 | 0.311901743 | 2684 | tags=36%, list=16%, signal=31% | ERBB4||GAP43||MYOZ1||SOX2||GPX1||WNT10B||RUNX1||CDKN1A||SELENON||GATA4||FZD9||NINJ1||EPPK1||KLF5||NOTCH1||CD81 | 5.901740 | 5.7691850 | 5.7999685 | 5.624109 | 5.989419 | 6.1138353 | 5.6468165 | 5.5951946 | 5.845881 | 5.755118 | 6.083942 | 5.8499265 | 5.7694082 | 5.6834187 | 5.640904 | 5.91592653 | 5.8295276 | 5.6765740 | 5.5868720 | 5.607343 | 5.963364 | 5.867149 | 6.125816 | 6.073765 | 6.251257 | 6.00518416 | 5.675986 | 5.6248928 | 5.6390870 | 5.5962665 | 5.629464 | 5.5589921 |
| GO:0045472 | response to ether | 10 | 0.6375986 | 1.722244 | 1.929872e-02 | 0.332385445 | 0.311901743 | 4744 | tags=80%, list=28%, signal=57% | FOXP3||PPARGC1A||HNRNPD||LARP1||CDK4||ZC3H12A||GOLPH3||PTAFR | 6.856272 | 6.9919077 | 6.6373252 | 6.840446 | 6.913275 | 6.9730348 | 6.9982065 | 6.7978211 | 6.764319 | 6.903328 | 6.896945 | 6.9944651 | 6.9534006 | 7.0269174 | 6.700901 | 6.63670015 | 6.5714718 | 6.7663177 | 6.8167381 | 6.933170 | 6.932710 | 6.820783 | 6.981650 | 7.033968 | 7.059952 | 6.81260935 | 6.954760 | 7.0069165 | 7.0318728 | 6.8037114 | 6.802719 | 6.7869721 |
| GO:0061620 | glycolytic process through glucose-6-phosphate | 10 | 0.6339196 | 1.712306 | 1.929872e-02 | 0.332385445 | 0.311901743 | 2727 | tags=70%, list=16%, signal=59% | GALK1||HK2||FOXK2||PFKL||FOXK1||PFKP||HK1 | 8.489739 | 7.9389278 | 8.4731815 | 7.970359 | 8.513237 | 8.5648022 | 7.8920739 | 7.9812493 | 8.423672 | 8.428957 | 8.608785 | 8.0045098 | 7.9273911 | 7.8822232 | 8.316673 | 8.57101310 | 8.5195222 | 8.0469081 | 7.8748702 | 7.984074 | 8.583433 | 8.427084 | 8.524890 | 8.542339 | 8.525646 | 8.62447010 | 7.902957 | 7.8618190 | 7.9109654 | 7.9663303 | 7.989997 | 7.9873044 |
| GO:0030851 | granulocyte differentiation | 23 | 0.4833482 | 1.678166 | 1.973001e-02 | 0.333129488 | 0.312599933 | 5118 | tags=65%, list=31%, signal=45% | FASN||RUNX1||L3MBTL3||CITED2||GATA2||CBFA2T3||ZFPM1||TESC||CEBPA||HCLS1||TRIB1||ZBTB46||RARA||HAX1||LEF1 | 5.118284 | 5.3503876 | 5.1303409 | 5.320846 | 5.172784 | 5.3309990 | 5.3280236 | 5.2834685 | 5.056748 | 5.057436 | 5.233391 | 5.3571560 | 5.3537715 | 5.3401793 | 5.082409 | 5.19748956 | 5.1085862 | 5.3024208 | 5.3095945 | 5.350064 | 5.171391 | 5.042447 | 5.293598 | 5.290026 | 5.402885 | 5.29729823 | 5.328789 | 5.3163637 | 5.3388300 | 5.2621922 | 5.319517 | 5.2680037 |
| GO:0042769 | DNA damage response, detection of DNA damage | 13 | 0.5709309 | 1.676621 | 1.982759e-02 | 0.333129488 | 0.312599933 | 4509 | tags=46%, list=27%, signal=34% | MRPS35||CFAP410||MRPS26||SOX4||NEK1||MRPS9 | 9.113267 | 8.7176422 | 8.8603849 | 8.699576 | 9.107757 | 8.9111707 | 8.6650922 | 8.7159165 | 9.133971 | 9.083994 | 9.121369 | 8.7961892 | 8.7092940 | 8.6433620 | 8.734531 | 8.90352549 | 8.9351423 | 8.7729189 | 8.6439045 | 8.678799 | 9.146265 | 9.066261 | 9.109635 | 9.099028 | 8.917207 | 8.68809386 | 8.678904 | 8.6647019 | 8.6515406 | 8.6972919 | 8.716218 | 8.7340061 |
| GO:0042119 | neutrophil activation | 16 | 0.5400520 | 1.688455 | 2.012195e-02 | 0.333129488 | 0.312599933 | 4744 | tags=62%, list=28%, signal=45% | CCL5||PREX1||TRAF3IP2||DNASE1||PRKCD||KMT2E||GRN||STXBP2||ITGB2||PTAFR | 3.863190 | 4.4406076 | 4.4153681 | 4.389326 | 3.961053 | 4.4071671 | 4.2967306 | 4.3442853 | 3.782808 | 3.778481 | 4.015332 | 4.4508632 | 4.4975795 | 4.3705288 | 4.286499 | 4.59554751 | 4.3450490 | 4.4531581 | 4.3180613 | 4.393587 | 3.916421 | 3.830859 | 4.120366 | 4.151175 | 4.264248 | 4.73704206 | 4.341375 | 4.2877425 | 4.2598823 | 4.3540824 | 4.426426 | 4.2467037 |
| GO:0035108 | limb morphogenesis | 120 | 0.3007555 | 1.474038 | 2.016129e-02 | 0.333129488 | 0.312599933 | 3057 | tags=29%, list=18%, signal=24% | AFF3||SEMA3C||ZNF358||FGF8||PLXNA2||HOXA13||RARG||B9D1||CREBBP||ZNF219||FBXW4||CRABP2||TBX2||SKI||PITX1||TFAP2A||SOX9||LRP5||BAK1||ALX4||HOXA10||ZBTB16||IFT140||BAX||TULP3||SALL1||GLI3||SMAD4||PTCH1||NOTCH1||SOX4||IQCE||NOG||IHH||TBX3 | 3.724979 | 4.4859022 | 4.0957183 | 4.544483 | 3.709382 | 4.0203493 | 4.4788015 | 4.5458002 | 3.696412 | 3.773353 | 3.703920 | 4.4931433 | 4.5125004 | 4.4513884 | 4.262894 | 3.99845312 | 4.0100351 | 4.5202513 | 4.5376092 | 4.575045 | 3.700576 | 3.662340 | 3.763419 | 3.910041 | 3.961594 | 4.17547401 | 4.496108 | 4.4680494 | 4.4720869 | 4.5223548 | 4.576459 | 4.5380490 |
| GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 13 | 0.5695293 | 1.672506 | 2.040230e-02 | 0.333129488 | 0.312599933 | 3959 | tags=62%, list=24%, signal=47% | NFKBIL1||PRDX2||SASH1||SIGIRR||TNFAIP3||CACTIN||PRKCA||TRIB1 | 6.597514 | 5.9402867 | 6.4408712 | 5.895262 | 6.675250 | 6.8007402 | 5.8391552 | 5.8621185 | 6.555523 | 6.425354 | 6.788031 | 6.0190397 | 5.9106242 | 5.8877595 | 6.221119 | 6.58437161 | 6.4928771 | 5.9774767 | 5.8130891 | 5.890530 | 6.667023 | 6.546471 | 6.801009 | 6.760965 | 6.942164 | 6.68701718 | 5.856902 | 5.8380605 | 5.8222947 | 5.8504869 | 5.878027 | 5.8577004 |
| GO:0002067 | glandular epithelial cell differentiation | 42 | 0.3967876 | 1.604214 | 2.041742e-02 | 0.333129488 | 0.312599933 | 2749 | tags=33%, list=16%, signal=28% | FGF8||FOXA1||FASN||RARG||AKT1||BMP2||GATA6||BAD||SIDT2||HES1||NOTCH1||NKX2-2||NKX6-1||MEN1 | 4.704469 | 4.8146153 | 4.9569853 | 4.898530 | 4.750276 | 5.0980211 | 4.8210733 | 4.8699851 | 4.635360 | 4.682586 | 4.791019 | 4.7990423 | 4.8511684 | 4.7929223 | 4.946992 | 4.98954108 | 4.9338334 | 4.9082882 | 4.8656424 | 4.921076 | 4.738120 | 4.634149 | 4.868939 | 4.965933 | 5.114827 | 5.20339528 | 4.847556 | 4.8182864 | 4.7969293 | 4.8615571 | 4.902625 | 4.8451649 |
| GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 13 | 0.5689727 | 1.670871 | 2.068966e-02 | 0.336595575 | 0.315852418 | 4334 | tags=77%, list=26%, signal=57% | GDF15||RAPGEF3||EHD1||EHD2||ADAM9||NFATC2||CAPN2||CAMK1||ADGRB1||FLOT1 | 4.393911 | 4.6080489 | 4.3773299 | 4.466575 | 4.408433 | 4.5388517 | 4.5689797 | 4.4369968 | 4.393064 | 4.389035 | 4.399615 | 4.6186992 | 4.6033946 | 4.6019932 | 4.444951 | 4.34353387 | 4.3410502 | 4.4832264 | 4.3957404 | 4.518022 | 4.411257 | 4.333426 | 4.477039 | 4.520418 | 4.565550 | 4.53019500 | 4.615275 | 4.5294191 | 4.5609340 | 4.4311976 | 4.479143 | 4.3995348 |
| GO:0060421 | positive regulation of heart growth | 30 | 0.4397129 | 1.646234 | 2.096037e-02 | 0.339039941 | 0.318146146 | 1761 | tags=27%, list=11%, signal=24% | ERBB4||AKAP6||WT1||TBX2||HEY2||GATA6||PIN1||GLI1 | 5.221676 | 5.6180189 | 5.3087893 | 5.534947 | 5.206439 | 5.2120300 | 5.6552676 | 5.6309509 | 5.189759 | 5.275095 | 5.198639 | 5.6713761 | 5.5972753 | 5.5838566 | 5.466689 | 5.25097951 | 5.1941561 | 5.5012052 | 5.5222340 | 5.580236 | 5.238633 | 5.177020 | 5.202998 | 5.123918 | 5.162260 | 5.34052996 | 5.650780 | 5.6559028 | 5.6591075 | 5.6292103 | 5.645529 | 5.6179806 |
| GO:0070102 | interleukin-6-mediated signaling pathway | 14 | 0.5514516 | 1.653515 | 2.113296e-02 | 0.340852265 | 0.319846783 | 3809 | tags=43%, list=23%, signal=33% | JAK2||IL6ST||SMAD4||STAT3||SRC||CEBPA | 2.675963 | 4.0888717 | 3.0165669 | 4.244190 | 2.609241 | 3.0234834 | 4.1473982 | 4.2964470 | 2.637339 | 2.783073 | 2.600975 | 4.0704774 | 4.0846576 | 4.1111838 | 3.389314 | 2.84068598 | 2.7301868 | 4.1171081 | 4.2764999 | 4.330525 | 2.618191 | 2.516428 | 2.687965 | 3.061100 | 2.969940 | 3.03786251 | 4.113909 | 4.1533119 | 4.1743229 | 4.2595598 | 4.336505 | 4.2922415 |
| GO:0003161 | cardiac conduction system development | 31 | 0.4324859 | 1.628139 | 2.128483e-02 | 0.342320876 | 0.321224889 | 4204 | tags=52%, list=25%, signal=39% | MSC||MESP1||NKX2-5||HEY2||GATA6||GATA4||HCN4||SCN5A||SMAD4||NOTCH1||TBX3||IRX3||HEY1||CACNA1G||MAML1||MSX2 | 3.718805 | 4.9436297 | 3.8413935 | 4.851481 | 3.685349 | 3.7611227 | 4.9452201 | 4.8658629 | 3.737959 | 3.859162 | 3.541670 | 4.9632500 | 4.9420697 | 4.9253187 | 4.151053 | 3.57093701 | 3.7390643 | 4.8000915 | 4.8366624 | 4.915278 | 3.621463 | 3.763668 | 3.667244 | 3.732740 | 3.701553 | 3.84509356 | 4.931671 | 4.9512026 | 4.9526920 | 4.8361006 | 4.900939 | 4.8598017 |
| GO:0036302 | atrioventricular canal development | 11 | 0.6124854 | 1.706406 | 2.173913e-02 | 0.345328596 | 0.324047254 | 3057 | tags=64%, list=18%, signal=52% | TBX2||BMP2||GATA6||GATA4||SMAD4||FOXN4||TBX3 | 2.393474 | 4.0018383 | 2.9006416 | 4.195665 | 2.407640 | 3.0296827 | 4.0913332 | 4.2782441 | 2.276757 | 2.493731 | 2.401749 | 3.9364011 | 4.0169981 | 4.0497687 | 3.276116 | 2.66337594 | 2.6735957 | 4.0611800 | 4.1666608 | 4.344804 | 2.431603 | 2.264323 | 2.515753 | 3.057415 | 3.025537 | 3.00562247 | 4.075148 | 4.1024906 | 4.0962193 | 4.2610767 | 4.288488 | 4.2850139 |
| GO:0061001 | regulation of dendritic spine morphogenesis | 44 | 0.3796798 | 1.567389 | 2.177858e-02 | 0.345328596 | 0.324047254 | 3333 | tags=39%, list=20%, signal=31% | DBNL||PPFIA2||DNM3||CUX2||SRCIN1||ARC||PTEN||BAIAP2||KIF1A||DBN1||TANC2||CFL1||ABI2||SHANK3||ITPKA||UBE3A||CAMK2B | 5.566168 | 5.3214221 | 5.4886381 | 5.445423 | 5.637284 | 5.7636231 | 5.3327824 | 5.4682980 | 5.483560 | 5.529407 | 5.678281 | 5.3004354 | 5.3390185 | 5.3245495 | 5.380528 | 5.58847833 | 5.4894211 | 5.4526530 | 5.3990075 | 5.483348 | 5.647482 | 5.516804 | 5.738966 | 5.739923 | 5.831996 | 5.71634350 | 5.366074 | 5.3027208 | 5.3288491 | 5.4708803 | 5.483432 | 5.4503890 |
| GO:0010875 | positive regulation of cholesterol efflux | 18 | 0.5118305 | 1.656298 | 2.263309e-02 | 0.349540487 | 0.327999582 | 5957 | tags=61%, list=36%, signal=39% | RXRA||ABCA3||PTCH1||ABCG1||NR1H3||PLTP||ZDHHC8||NR1H2||GPS2||LRP1||ABCA7 | 3.677244 | 3.9892967 | 4.1905559 | 3.893257 | 3.713382 | 4.1838900 | 3.9856908 | 3.8470319 | 3.578255 | 3.616571 | 3.824546 | 3.9315249 | 4.0298994 | 4.0046656 | 4.195928 | 4.22437395 | 4.1504028 | 3.9373396 | 3.8939390 | 3.847081 | 3.682666 | 3.572379 | 3.869361 | 3.912287 | 4.083527 | 4.49283683 | 4.051760 | 3.9694402 | 3.9333050 | 3.8295628 | 3.906140 | 3.8034071 |
| GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 44 | 0.3772992 | 1.557561 | 2.268603e-02 | 0.349540487 | 0.327999582 | 3347 | tags=34%, list=20%, signal=27% | CCL5||OAS3||SLIT3||NLRC5||PADI2||PIAS4||PALM3||GSTP1||SLIT2||YTHDF2||TTLL12||SIGIRR||CCDC3||CACTIN||NR1H3 | 5.886372 | 5.3555046 | 5.8330665 | 5.387238 | 5.990469 | 6.0665320 | 5.3248891 | 5.3993865 | 5.817538 | 5.746280 | 6.074305 | 5.3730315 | 5.3614678 | 5.3317000 | 5.707346 | 5.93053828 | 5.8524931 | 5.4328441 | 5.3172592 | 5.409047 | 5.959314 | 5.870762 | 6.129268 | 5.982724 | 6.204039 | 6.00226890 | 5.360994 | 5.2978846 | 5.3150486 | 5.3877031 | 5.426382 | 5.3836884 |
| GO:0045668 | negative regulation of osteoblast differentiation | 36 | 0.4067872 | 1.586192 | 2.287854e-02 | 0.349555991 | 0.328014131 | 3755 | tags=44%, list=22%, signal=35% | ID3||TWIST2||CITED1||SUFU||SKI||SOX9||LRP5||PTCH1||NOTCH1||MEN1||TOB1||NOG||HDAC4||TWIST1||HDAC7||VEGFC | 3.881577 | 4.0516006 | 3.8236242 | 3.747163 | 3.989378 | 4.1607994 | 3.9467058 | 3.6719042 | 3.768779 | 3.811687 | 4.048289 | 4.0609687 | 4.0849398 | 4.0078168 | 3.732355 | 3.91824014 | 3.8142486 | 3.7704903 | 3.7232373 | 3.747374 | 4.007676 | 3.792340 | 4.146286 | 4.105152 | 4.222357 | 4.15247295 | 3.982493 | 3.9271161 | 3.9298301 | 3.6808324 | 3.685565 | 3.6490422 |
| GO:0045600 | positive regulation of fat cell differentiation | 47 | 0.3771702 | 1.574108 | 2.293359e-02 | 0.349555991 | 0.328014131 | 3988 | tags=51%, list=24%, signal=39% | LPL||CARM1||CEBPB||ZFP36L1||AKT1||ZBTB7C||LRP5||BMP2||RREB1||TFE3||ZNF385A||ZBTB16||KLF5||CREB1||PPARD||FNDC5||ZBTB7B||CCDC3||HNRNPU||ZFP36||CEBPA||BMP7||ZC3H12A||METRNL | 4.358981 | 4.9766949 | 4.3478577 | 4.915778 | 4.302130 | 4.4641878 | 5.0082699 | 4.9365109 | 4.331826 | 4.442673 | 4.298464 | 4.9625937 | 4.9902139 | 4.9771447 | 4.571074 | 4.23964793 | 4.2037330 | 4.9026578 | 4.8748992 | 4.968180 | 4.313039 | 4.258843 | 4.333481 | 4.455480 | 4.494655 | 4.44190752 | 4.989780 | 5.0153481 | 5.0195023 | 4.9375813 | 4.958892 | 4.9126892 |
| GO:0048167 | regulation of synaptic plasticity | 137 | 0.2842896 | 1.417594 | 2.305825e-02 | 0.349555991 | 0.328014131 | 4584 | tags=42%, list=27%, signal=30% | MCTP1||GRID2||SYT12||RASGRF1||GRIN2D||RIMS2||MAPT||VGF||SORCS2||MAP1A||NRGN||ARC||PTEN||CRTC1||HRAS||RASGRF2||ADCY1||MPP2||EPHB2||BAIAP2||NCDN||ABL1||JPH3||GIPC1||ITPR3||SYNGR1||TSHZ3||NEURL1||SRF||STXBP1||DBN1||MAP1B||CREB1||SHANK3||ARF1||RAB3A||ITPKA||PICK1||CDC20||SYT7||CAMK2B||PRKCZ||ERC1||SLC24A2||NTRK2||MECP2||GFAP||ADGRB1||CDK5||CLN3||BRSK1||SLC24A1||CPEB3||SQSTM1||SYAP1||RARA||ADORA1 | 4.980935 | 5.0138765 | 5.1008337 | 5.046108 | 4.997182 | 5.1911857 | 4.9893111 | 5.0763184 | 4.936023 | 5.022704 | 4.982773 | 5.0320041 | 5.0245386 | 4.9846387 | 5.088820 | 5.11555789 | 5.0979954 | 5.0847311 | 4.9720184 | 5.078809 | 5.009474 | 4.951402 | 5.029532 | 5.173382 | 5.146545 | 5.25155674 | 5.016864 | 4.9763296 | 4.9743393 | 5.0715714 | 5.097768 | 5.0593483 |
| GO:1901984 | negative regulation of protein acetylation | 22 | -0.5971440 | -1.683362 | 2.312713e-02 | 0.349555991 | 0.328014131 | 552 | tags=14%, list=3%, signal=13% | PARK7||ZNF451||MAGEA2B | 6.107664 | 6.7622770 | 6.3496616 | 6.747961 | 6.006187 | 6.0489983 | 6.8022912 | 6.7610123 | 6.146278 | 6.233194 | 5.926456 | 6.7530978 | 6.7634222 | 6.7702594 | 6.502963 | 6.20000494 | 6.3299404 | 6.7400428 | 6.7021834 | 6.799968 | 5.952070 | 6.122889 | 5.936073 | 5.989530 | 5.955907 | 6.19026141 | 6.793853 | 6.7867393 | 6.8259774 | 6.7334923 | 6.773956 | 6.7751997 |
| GO:0034122 | negative regulation of toll-like receptor signaling pathway | 21 | 0.4823626 | 1.632647 | 2.338009e-02 | 0.349555991 | 0.328014131 | 5520 | tags=62%, list=33%, signal=42% | NR1D1||DAB2IP||NFKBIL1||ARRB2||TNFAIP3||PTPRS||CACTIN||TYRO3||IRF4||SARM1||LRRC14||GRAMD4||GPS2 | 4.098321 | 4.1661472 | 4.4773877 | 4.146204 | 4.142195 | 4.5190664 | 4.1682948 | 4.1130118 | 4.001687 | 4.018822 | 4.259759 | 4.1360057 | 4.2009713 | 4.1607181 | 4.479630 | 4.49569795 | 4.4565667 | 4.1582784 | 4.1211698 | 4.158843 | 4.132752 | 3.994304 | 4.284877 | 4.319999 | 4.472994 | 4.73358529 | 4.226099 | 4.1518330 | 4.1250430 | 4.0621969 | 4.147232 | 4.1282324 |
| GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 21 | 0.4835251 | 1.636582 | 2.338009e-02 | 0.349555991 | 0.328014131 | 4814 | tags=48%, list=29%, signal=34% | CHST9||B3GAT1||CHST12||CHPF||B3GALT6||B3GAT3||CHST13||PXYLP1||CHST3||XYLT1 | 3.851166 | 4.0549971 | 4.2067005 | 3.784315 | 3.916393 | 4.2622332 | 3.8618765 | 3.5368637 | 3.793225 | 3.721163 | 4.021778 | 4.0548901 | 4.1306658 | 3.9752518 | 4.061554 | 4.36178476 | 4.1808269 | 3.8439906 | 3.7700904 | 3.736768 | 3.889455 | 3.756864 | 4.083984 | 4.017242 | 4.250683 | 4.48151436 | 3.909304 | 3.8781017 | 3.7958564 | 3.5473188 | 3.609825 | 3.4489050 |
| GO:0060998 | regulation of dendritic spine development | 59 | 0.3722375 | 1.622063 | 2.338530e-02 | 0.349555991 | 0.328014131 | 3333 | tags=39%, list=20%, signal=31% | DBNL||FSTL4||PPFIA2||DNM3||CUX2||PTEN||ASAP1||ZMYND8||BAIAP2||KIF1A||NEURL1||DBN1||TANC2||SHANK1||SHANK3||SDK1||FOXO6||ARF1||ITPKA||DLG5||UBE3A||PTPRS||CAMK2B | 4.915441 | 5.5873584 | 4.9161841 | 5.534195 | 4.940800 | 5.1073596 | 5.6100746 | 5.6287436 | 4.853228 | 4.980914 | 4.909338 | 5.5806237 | 5.5967702 | 5.5846322 | 5.011706 | 4.91804597 | 4.8118852 | 5.5003839 | 5.4913953 | 5.607870 | 4.965403 | 4.831321 | 5.019244 | 5.134929 | 5.114314 | 5.07212685 | 5.601197 | 5.6031352 | 5.6257625 | 5.5933969 | 5.688552 | 5.6023560 |
| GO:0033262 | regulation of nuclear cell cycle DNA replication | 11 | 0.6071378 | 1.691507 | 2.341137e-02 | 0.349555991 | 0.328014131 | 1181 | tags=36%, list=7%, signal=34% | CDT1||DACH1||WIZ||BCL6 | 4.254663 | 5.2608971 | 4.4288905 | 5.353067 | 4.287066 | 4.7846520 | 5.2846587 | 5.3843028 | 4.174640 | 4.244500 | 4.340055 | 5.2084500 | 5.3060916 | 5.2664817 | 4.531681 | 4.40346958 | 4.3451646 | 5.2757862 | 5.3684723 | 5.411623 | 4.296202 | 4.125559 | 4.423988 | 4.759486 | 4.945820 | 4.63120583 | 5.272781 | 5.3100516 | 5.2708037 | 5.3553187 | 5.447759 | 5.3476636 |
| GO:0007411 | axon guidance | 183 | 0.2558033 | 1.343838 | 2.367902e-02 | 0.350518478 | 0.328917303 | 5005 | tags=44%, list=30%, signal=31% | GAP43||PLXNC1||SEMA3C||SLIT3||EPHA5||FGF8||CRPPA||PLXNA2||LAMA2||SEMA7A||NGFR||SOS1||EPHB3||RHOG||DPYSL5||TUBB3||NRXN3||NFIB||IGSF9||NOTCH3||NIBAN2||SEMA6B||TUBB2B||DVL1||EPHB2||SEMA4G||WNT3A||ENAH||VSTM2L||NECTIN1||PALLD||SLIT2||GLI3||L1CAM||GATA3||SMAD4||VAX1||EPHA8||PTCH1||NOTCH1||ISL2||SEMA6A||NOG||LHX2||PLXNA4||CDH4||BSG||ERBB2||KLF7||UNC5A||FLRT3||UNC5B||MEGF8||FOXD1||BMP7||SEMA4F||EPHB4||FOXG1||EFNB3||PLXND1||SLIT1||FYN||SEMA3F||NTN1||CDK5||NTRK1||CXCL12||VASP||GBX1||DPYSL2||DAG1||CRMP1||LYPLA2||SMO||PLXNA1||PTPRM||EFNA2||NOTCH2||EFNA4||SEMA4A | 4.625507 | 5.1335028 | 4.5639284 | 5.097591 | 4.559966 | 4.6050550 | 5.0656632 | 5.0782546 | 4.669895 | 4.670432 | 4.531819 | 5.2142008 | 5.0921197 | 5.0906707 | 4.616517 | 4.50686081 | 4.5663214 | 5.1225587 | 5.0587128 | 5.110706 | 4.498102 | 4.626004 | 4.552934 | 4.566733 | 4.627933 | 4.61973677 | 5.078867 | 5.0327103 | 5.0848495 | 5.0552432 | 5.088588 | 5.0906595 |
| GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 35 | 0.4108565 | 1.595610 | 2.397399e-02 | 0.350518478 | 0.328917303 | 3962 | tags=46%, list=24%, signal=35% | NR1D1||TNIP1||DAB2IP||ABL1||GSTP1||NLRX1||CARD19||OTUD7A||CCDC22||USP10||TNFAIP3||CACTIN||TRIM59||TLE1||ZC3H12A||OTUD7B | 6.415228 | 5.8244054 | 6.3233268 | 5.895901 | 6.487971 | 6.5266567 | 5.8131848 | 5.9065762 | 6.364535 | 6.324755 | 6.546570 | 5.8504128 | 5.8189652 | 5.8034407 | 6.200710 | 6.38819898 | 6.3736373 | 5.9438196 | 5.8110326 | 5.929210 | 6.467371 | 6.394950 | 6.594479 | 6.476689 | 6.646598 | 6.44862859 | 5.825914 | 5.7978356 | 5.8156647 | 5.8932892 | 5.911580 | 5.9147663 |
| GO:0090494 | dopamine uptake | 10 | -0.7158592 | -1.671044 | 2.451368e-02 | 0.356247570 | 0.334293332 | 1935 | tags=40%, list=12%, signal=35% | PRKN||MAPK15||TOR1A||PARK7 | 5.202591 | 5.5476114 | 5.6411927 | 5.634910 | 4.918278 | 4.5542390 | 5.5530194 | 5.5653871 | 5.381368 | 5.528267 | 4.500838 | 5.5534621 | 5.5345972 | 5.5546871 | 5.768539 | 5.33444968 | 5.7777160 | 5.7006072 | 5.5592264 | 5.641412 | 4.717910 | 5.358830 | 4.545973 | 4.540162 | 4.254995 | 4.81370057 | 5.584825 | 5.5161790 | 5.5572282 | 5.5564916 | 5.546322 | 5.5929303 |
| GO:0035909 | aorta morphogenesis | 27 | 0.4445446 | 1.610961 | 2.458101e-02 | 0.356247570 | 0.334293332 | 2744 | tags=41%, list=16%, signal=34% | ACVRL1||FGF8||NPRL3||TBX2||HEY2||JAG1||SRF||HES1||NOTCH1||TBX1||SOX4 | 3.776900 | 4.4489883 | 3.9940933 | 4.258495 | 3.806695 | 4.1475328 | 4.3709795 | 4.1835884 | 3.727922 | 3.737213 | 3.861662 | 4.4291030 | 4.4972571 | 4.4193493 | 4.115342 | 3.97795558 | 3.8791953 | 4.2710329 | 4.2209982 | 4.282713 | 3.758505 | 3.724397 | 3.928769 | 4.028569 | 4.145932 | 4.25891122 | 4.401011 | 4.3695960 | 4.3417211 | 4.1775212 | 4.223168 | 4.1491068 |
| GO:0038084 | vascular endothelial growth factor signaling pathway | 32 | 0.4236391 | 1.607610 | 2.461899e-02 | 0.356247570 | 0.334293332 | 6184 | tags=59%, list=37%, signal=38% | PTP4A3||HSPB1||VEGFB||DAB2IP||FOXC1||MYO1C||SEMA6A||IL12A||FLT4||VEGFC||GAB1||ADGRA2||PRKD2||FLT1||ROBO1||SPRY2||PGF||DLL1||PRKD1 | 4.546597 | 4.4531573 | 4.5002040 | 4.411254 | 4.603186 | 4.6155537 | 4.4056759 | 4.3860479 | 4.563961 | 4.426670 | 4.641047 | 4.4602200 | 4.4749794 | 4.4237925 | 4.439628 | 4.55350205 | 4.5052222 | 4.3970363 | 4.4325216 | 4.403958 | 4.559059 | 4.533981 | 4.710150 | 4.544099 | 4.639802 | 4.66011923 | 4.416849 | 4.4318771 | 4.3675182 | 4.3765661 | 4.435375 | 4.3447326 |
| GO:0099532 | synaptic vesicle endosomal processing | 10 | 0.6195930 | 1.673608 | 2.473498e-02 | 0.357008245 | 0.335007129 | 979 | tags=20%, list=6%, signal=19% | VAMP4||AP3D1 | 4.110501 | 4.7356853 | 4.4547358 | 4.955598 | 4.094381 | 4.6306784 | 4.8193072 | 4.9608608 | 4.020789 | 4.067623 | 4.234264 | 4.6375985 | 4.7880945 | 4.7765548 | 4.630378 | 4.43558071 | 4.2764015 | 4.9448254 | 4.8964415 | 5.022704 | 4.086443 | 3.951161 | 4.231872 | 4.536659 | 4.636574 | 4.71337557 | 4.846601 | 4.8105899 | 4.8003200 | 4.9680226 | 5.006861 | 4.9059062 |
| GO:0048557 | embryonic digestive tract morphogenesis | 10 | 0.6182142 | 1.669884 | 2.500680e-02 | 0.360008314 | 0.337822315 | 3200 | tags=50%, list=19%, signal=40% | RBPMS2||GLI3||HLX||IHH||SIX2 | 3.296015 | 4.5916933 | 3.3387840 | 4.179042 | 3.276695 | 3.2781378 | 4.6063467 | 4.1814076 | 3.359506 | 3.333412 | 3.189373 | 4.6345994 | 4.5837256 | 4.5556420 | 3.634185 | 3.19594990 | 3.1329479 | 4.1461997 | 4.1616560 | 4.227957 | 3.198486 | 3.341470 | 3.286537 | 3.220438 | 3.262781 | 3.34824275 | 4.595113 | 4.6005867 | 4.6231863 | 4.1801507 | 4.157969 | 4.2057070 |
| GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 20 | 0.4887412 | 1.627224 | 2.545455e-02 | 0.363664052 | 0.341252763 | 3491 | tags=50%, list=21%, signal=40% | DNAI3||CORO1B||WASF1||WAS||WASF2||ABI2||ARF1||PICK1||FCHSD2||WASL | 6.333057 | 6.2431397 | 6.4415992 | 6.162252 | 6.407838 | 6.5430447 | 6.2201552 | 6.2059625 | 6.276658 | 6.377359 | 6.343342 | 6.2429019 | 6.2514928 | 6.2349771 | 6.329931 | 6.51964761 | 6.4686030 | 6.1950906 | 6.0852590 | 6.203418 | 6.438767 | 6.317327 | 6.463229 | 6.554926 | 6.473723 | 6.59774660 | 6.229559 | 6.2043254 | 6.2264500 | 6.1924250 | 6.223303 | 6.2019860 |
| GO:0035066 | positive regulation of histone acetylation | 26 | 0.4458058 | 1.602121 | 2.585579e-02 | 0.363959653 | 0.341530148 | 2454 | tags=38%, list=15%, signal=33% | FOXP3||WBP2||PPARGC1A||RPS6KA4||RUVBL2||GATA3||SMAD4||MAPK3||SPHK2||ARRB1 | 5.471007 | 5.3604121 | 5.5983259 | 5.350899 | 5.511048 | 5.7497441 | 5.3896532 | 5.3762661 | 5.380978 | 5.434801 | 5.589073 | 5.3627379 | 5.3683997 | 5.3500375 | 5.563934 | 5.67040656 | 5.5578366 | 5.3867356 | 5.2845186 | 5.379209 | 5.534115 | 5.358137 | 5.628011 | 5.711706 | 5.756590 | 5.78010179 | 5.430071 | 5.3764961 | 5.3614878 | 5.3685604 | 5.401624 | 5.3582568 |
| GO:0006337 | nucleosome disassembly | 15 | 0.5389071 | 1.653045 | 2.589286e-02 | 0.363959653 | 0.341530148 | 5171 | tags=73%, list=31%, signal=51% | HMGA1||GRWD1||SMARCA4||H2AW||ARID1A||SMARCE1||SMARCD2||SMARCD1||SMARCC2||SMARCD3||SMARCB1 | 6.608045 | 6.9213047 | 6.5415188 | 6.852027 | 6.596010 | 6.7762350 | 6.9856455 | 6.9124850 | 6.549771 | 6.614854 | 6.657476 | 6.8780997 | 6.9446105 | 6.9402491 | 6.596323 | 6.53913841 | 6.4870225 | 6.8549752 | 6.7930998 | 6.905802 | 6.624580 | 6.504340 | 6.654760 | 6.811680 | 6.817722 | 6.69607556 | 6.961624 | 6.9842143 | 7.0106804 | 6.8970409 | 6.928962 | 6.9112752 |
| GO:0090288 | negative regulation of cellular response to growth factor stimulus | 69 | 0.3461522 | 1.558781 | 2.601331e-02 | 0.363959653 | 0.341530148 | 3394 | tags=32%, list=20%, signal=26% | NBL1||SULF1||HIPK2||EMILIN1||RBPMS2||NGFR||SHISA2||SKI||DAB2IP||NIBAN2||FUZ||ABL1||CREB3L1||SLIT2||GATA3||NOTCH1||TOB1||SEMA6A||NOG||SPRY1||FSTL3||IL12A | 3.393082 | 4.2384050 | 3.7839188 | 4.215798 | 3.382413 | 3.7058149 | 4.2388948 | 4.2437934 | 3.335520 | 3.461905 | 3.378950 | 4.2407412 | 4.2570078 | 4.2171885 | 3.954760 | 3.70988785 | 3.6703599 | 4.1789679 | 4.2018459 | 4.265191 | 3.365024 | 3.323053 | 3.455946 | 3.569284 | 3.637945 | 3.89009564 | 4.254202 | 4.2259566 | 4.2363843 | 4.2211482 | 4.263904 | 4.2460092 |
| GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 43 | 0.3806026 | 1.549046 | 2.615879e-02 | 0.363959653 | 0.341530148 | 2951 | tags=33%, list=18%, signal=27% | GDF15||ACVRL1||PMEPA1||RBPMS||EMILIN1||GDF1||TGFB1||BMP2||PIN1||SMAD4||DAB2||CSNK2B||GDF11||NOG | 4.183134 | 4.1852991 | 4.0311397 | 4.140529 | 4.228460 | 4.3208437 | 4.1636915 | 4.1660666 | 4.156196 | 4.082711 | 4.301810 | 4.2154466 | 4.1916491 | 4.1479921 | 3.936535 | 4.12175978 | 4.0291812 | 4.1638794 | 4.1045050 | 4.152516 | 4.230181 | 4.112888 | 4.333857 | 4.280378 | 4.436119 | 4.23843191 | 4.173764 | 4.1659561 | 4.1512639 | 4.1450737 | 4.229621 | 4.1212429 |
| GO:0038065 | collagen-activated signaling pathway | 12 | 0.5829863 | 1.666397 | 2.629323e-02 | 0.363959653 | 0.341530148 | 4671 | tags=50%, list=28%, signal=36% | JAK2||COL1A1||COL4A3||DDR1||COL4A2||COL4A1 | 2.335072 | 2.5868029 | 2.7100800 | 2.428644 | 2.368605 | 2.7507742 | 2.4826175 | 2.3384368 | 2.294678 | 2.279542 | 2.426427 | 2.5085994 | 2.6821191 | 2.5642135 | 2.757984 | 2.79999573 | 2.5611676 | 2.4193409 | 2.4605576 | 2.405461 | 2.368934 | 2.215481 | 2.506726 | 2.601362 | 2.714323 | 2.91855844 | 2.535694 | 2.4405368 | 2.4699699 | 2.3008817 | 2.443755 | 2.2643809 |
| GO:0060841 | venous blood vessel development | 12 | 0.5823066 | 1.664454 | 2.629323e-02 | 0.363959653 | 0.341530148 | 3348 | tags=33%, list=20%, signal=27% | ACVRL1||SEMA3C||NOTCH1||CCM2 | 2.138388 | 2.6999943 | 2.7207196 | 2.739687 | 2.232266 | 2.6820190 | 2.7176595 | 2.7806316 | 2.069811 | 2.072109 | 2.264467 | 2.6977375 | 2.6924049 | 2.7097855 | 2.893924 | 2.66335244 | 2.5869132 | 2.7146346 | 2.7029418 | 2.799545 | 2.235305 | 2.078123 | 2.368755 | 2.458832 | 2.570063 | 2.96652857 | 2.753623 | 2.7239369 | 2.6743080 | 2.7602342 | 2.860775 | 2.7170958 |
| GO:0071276 | cellular response to cadmium ion | 23 | 0.4699454 | 1.631631 | 2.630668e-02 | 0.363959653 | 0.341530148 | 2552 | tags=43%, list=15%, signal=37% | HMOX1||MT1X||JUN||HSF1||MT2A||AKT1||MAPK3||OGG1||EGFR||MT1F | 5.824901 | 6.0543052 | 5.8457708 | 6.079619 | 5.859371 | 5.9229389 | 6.0701323 | 6.1085217 | 5.805773 | 5.830106 | 5.838624 | 6.0631039 | 6.0556748 | 6.0440732 | 5.955335 | 5.77119991 | 5.8040173 | 6.0787733 | 6.0465562 | 6.112766 | 5.826634 | 5.837951 | 5.912029 | 5.872585 | 5.960187 | 5.93464270 | 6.083124 | 6.0646833 | 6.0625009 | 6.0845764 | 6.123264 | 6.1174242 |
| GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 27 | 0.4399751 | 1.594401 | 2.644320e-02 | 0.364825465 | 0.342342602 | 3809 | tags=44%, list=23%, signal=34% | NR2F2||PLK1||PTEN||CDKN1A||HEXIM2||CDKN2A||LATS1||LATS2||MEN1||INCA1||TNFAIP3||CEBPA | 5.381282 | 5.4489487 | 5.3586767 | 5.504292 | 5.332553 | 5.5405503 | 5.4428756 | 5.4887443 | 5.344759 | 5.371962 | 5.425939 | 5.5018912 | 5.4331915 | 5.4101778 | 5.320380 | 5.40926293 | 5.3449203 | 5.5318688 | 5.4618802 | 5.518178 | 5.382696 | 5.148669 | 5.449320 | 5.488962 | 5.607027 | 5.52309065 | 5.469662 | 5.4111407 | 5.4472204 | 5.4821950 | 5.512330 | 5.4713953 |
| GO:0030308 | negative regulation of cell growth | 149 | 0.2747911 | 1.390581 | 2.675159e-02 | 0.367279789 | 0.344645675 | 3388 | tags=29%, list=20%, signal=23% | CDA||MAP2||ACVRL1||WT1||SEMA3C||SLIT3||FSTL4||CCDC85B||HSPA1A||HSPA1B||SEMA7A||G6PD||SESN2||CDKN1A||TGFB1||BCL6||PML||SMARCA4||SEMA6B||CAV3||TP53||SEMA4G||RGMA||WNT3A||OSGIN1||CDKN2A||RTN4R||BCL11A||PPP2R1A||SLIT2||RNF6||NPR1||SMAD4||SPHK2||PRDM11||SEMA6A||PPARD||DNAJB2||STK11||PPP1R9B||PTPRS||SIPA1||CTDP1 | 6.690414 | 6.4223152 | 6.5744850 | 6.285862 | 6.755075 | 6.7369891 | 6.3730327 | 6.2780094 | 6.649341 | 6.639709 | 6.778018 | 6.4682690 | 6.4182915 | 6.3789958 | 6.430045 | 6.64939753 | 6.6338018 | 6.3394926 | 6.2263472 | 6.289522 | 6.795317 | 6.665728 | 6.800189 | 6.750094 | 6.771402 | 6.68818196 | 6.389224 | 6.3664529 | 6.3632818 | 6.2690237 | 6.292282 | 6.2726136 |
| GO:0045662 | negative regulation of myoblast differentiation | 14 | 0.5415732 | 1.623895 | 2.700323e-02 | 0.368041584 | 0.345360523 | 3057 | tags=57%, list=18%, signal=47% | ID3||TGFB1||SOX9||MBNL3||NOTCH1||SOX8||PPARD||TBX3 | 4.451502 | 4.2624355 | 4.2149315 | 3.738922 | 4.605110 | 4.6243842 | 4.1013988 | 3.6509333 | 4.317688 | 4.377863 | 4.638437 | 4.3494891 | 4.2329339 | 4.2005971 | 4.092003 | 4.30355698 | 4.2411249 | 3.8066928 | 3.6441452 | 3.761094 | 4.618938 | 4.391134 | 4.779148 | 4.590266 | 4.764549 | 4.50619020 | 4.147314 | 4.0585116 | 4.0969945 | 3.6545813 | 3.692472 | 3.6043961 |
| GO:0030033 | microvillus assembly | 13 | 0.5520284 | 1.621112 | 2.816092e-02 | 0.379619639 | 0.356225065 | 3440 | tags=46%, list=21%, signal=37% | FXYD5||SLC9A3R1||KLF5||FSCN1||PODXL||RAP1GAP | 4.125803 | 4.2982244 | 3.9807329 | 4.215972 | 4.159613 | 4.2629830 | 4.3067357 | 4.1825123 | 4.134953 | 4.039338 | 4.198678 | 4.2752383 | 4.2944957 | 4.3245114 | 3.957374 | 3.96711685 | 4.0169963 | 4.2009679 | 4.1719042 | 4.273152 | 4.140523 | 4.114683 | 4.221471 | 4.265951 | 4.348476 | 4.16893404 | 4.346105 | 4.2576834 | 4.3150282 | 4.1768243 | 4.224339 | 4.1452739 |
| GO:0010664 | negative regulation of striated muscle cell apoptotic process | 20 | 0.4786747 | 1.593708 | 2.876033e-02 | 0.381038516 | 0.357556502 | 4200 | tags=55%, list=25%, signal=41% | JAK2||NUPR1||NKX2-5||HSF1||HEY2||GATA4||BAG3||RGL2||NFE2L2||BMP7||MDK | 5.150259 | 5.1744317 | 5.1901262 | 4.854238 | 5.212511 | 5.3617336 | 5.0527536 | 4.8177764 | 5.089432 | 5.056781 | 5.293031 | 5.2117787 | 5.1886835 | 5.1213085 | 5.055294 | 5.30172084 | 5.2027699 | 4.8920857 | 4.8652483 | 4.803972 | 5.209721 | 5.113728 | 5.307573 | 5.275099 | 5.379418 | 5.42654649 | 5.099384 | 5.0543819 | 5.0028798 | 4.8349269 | 4.867599 | 4.7481788 |
| GO:0072111 | cell proliferation involved in kidney development | 14 | 0.5395053 | 1.617695 | 2.876431e-02 | 0.381038516 | 0.357556502 | 2488 | tags=36%, list=15%, signal=30% | WT1||MYC||BMP2||GATA3||PTCH1 | 3.487718 | 3.9895074 | 3.8063093 | 4.158393 | 3.481034 | 3.7970621 | 3.9378922 | 4.0625332 | 3.492143 | 3.550841 | 3.417061 | 3.9667320 | 4.0422029 | 3.9580946 | 3.904631 | 3.78364024 | 3.7247914 | 4.2279699 | 4.0867992 | 4.156955 | 3.474374 | 3.451490 | 3.516483 | 3.671338 | 3.656800 | 4.03107479 | 3.951719 | 3.9105069 | 3.9510654 | 4.0030659 | 4.117891 | 4.0643567 |
| GO:0048670 | regulation of collateral sprouting | 16 | 0.5214431 | 1.630275 | 2.926829e-02 | 0.384718415 | 0.361009623 | 1815 | tags=31%, list=11%, signal=28% | FSTL4||CRABP2||RGMA||WNT3A||RND2 | 3.351907 | 4.1161044 | 3.9907082 | 4.170986 | 3.324255 | 3.7788388 | 4.1198203 | 4.2142709 | 3.269622 | 3.466680 | 3.311879 | 4.1086800 | 4.1118520 | 4.1277093 | 4.126631 | 3.96738171 | 3.8660844 | 4.1481856 | 4.1809937 | 4.183511 | 3.402072 | 3.183845 | 3.377112 | 3.597496 | 3.568250 | 4.10529999 | 4.150135 | 4.0615967 | 4.1460102 | 4.2200224 | 4.228791 | 4.1937697 |
| GO:0071294 | cellular response to zinc ion | 12 | 0.5760218 | 1.646490 | 2.943698e-02 | 0.384718415 | 0.361009623 | 3662 | tags=58%, list=22%, signal=46% | MT1X||ATP13A2||MT2A||TSPO||CREB1||MT1F||KCNK3 | 6.346331 | 6.0080889 | 6.2483116 | 6.067417 | 6.374148 | 6.5095977 | 5.9774799 | 6.0378648 | 6.304286 | 6.307245 | 6.424190 | 5.9776088 | 6.0377505 | 6.0082805 | 6.267019 | 6.22495995 | 6.2526395 | 6.1115789 | 5.9769043 | 6.109679 | 6.381949 | 6.301095 | 6.436204 | 6.588597 | 6.639401 | 6.27440233 | 6.012486 | 5.9306759 | 5.9880596 | 6.0390250 | 6.060600 | 6.0135853 |
| GO:0048661 | positive regulation of smooth muscle cell proliferation | 62 | 0.3559869 | 1.564447 | 2.959590e-02 | 0.384718415 | 0.361009623 | 1324 | tags=21%, list=8%, signal=19% | HMOX1||CCL5||JAK2||NR4A3||MMP2||IRAK1||RBPMS2||JUN||PPARGC1A||AKT1||HES5||TGFB1||MEF2D | 5.617010 | 5.5017311 | 5.6374278 | 5.470795 | 5.655563 | 5.7188775 | 5.4806904 | 5.4503786 | 5.564180 | 5.590479 | 5.693128 | 5.5169647 | 5.5197560 | 5.4678847 | 5.579560 | 5.69393780 | 5.6365183 | 5.5036455 | 5.4277696 | 5.479928 | 5.677667 | 5.569699 | 5.715390 | 5.681788 | 5.689129 | 5.78346997 | 5.499888 | 5.4883682 | 5.4534100 | 5.4442886 | 5.468810 | 5.4378520 |
| GO:1905209 | positive regulation of cardiocyte differentiation | 13 | 0.5486635 | 1.611230 | 2.959770e-02 | 0.384718415 | 0.361009623 | 2863 | tags=54%, list=17%, signal=45% | NKX2-5||GPER1||TGFB1||GATA6||WNT3A||GATA4||ARRB2 | 3.996057 | 4.0882071 | 4.0957826 | 4.194570 | 4.042780 | 4.3129458 | 4.1069601 | 4.1760877 | 3.905112 | 3.897500 | 4.168705 | 4.0895311 | 4.1024198 | 4.0725145 | 4.091115 | 4.13998459 | 4.0549853 | 4.2024108 | 4.1652116 | 4.215615 | 4.037939 | 3.821811 | 4.238583 | 4.218085 | 4.377573 | 4.33843187 | 4.127125 | 4.0923503 | 4.1011781 | 4.1672135 | 4.198403 | 4.1623815 |
| GO:0032355 | response to estradiol | 101 | 0.3068185 | 1.468400 | 2.973662e-02 | 0.384718415 | 0.361009623 | 3287 | tags=35%, list=20%, signal=28% | WNT8B||FOXA1||ZNF703||OPRL1||NR2F2||EEF2||GPX4||GPI||HSF1||GPER1||PPARGC1A||COL1A1||PTEN||TGFB1||HNRNPD||ARPC1B||IGFBP2||BAD||MBD3||NCOR2||ENDOG||ESRRA||RUVBL2||GSTP1||STXBP1||MAP1B||PTCH1||OGG1||EGFR||ARNT2||STAT3||NA||IHH||ASS1||PPP1R9B | 6.705869 | 6.7146935 | 6.6010196 | 6.561242 | 6.730425 | 6.8364284 | 6.6499022 | 6.5614292 | 6.668979 | 6.699331 | 6.748188 | 6.7305045 | 6.7234510 | 6.6897977 | 6.491638 | 6.64258910 | 6.6628399 | 6.6041712 | 6.4994260 | 6.578079 | 6.726367 | 6.667449 | 6.794647 | 6.852974 | 6.966212 | 6.67531717 | 6.660034 | 6.6474939 | 6.6421204 | 6.5530807 | 6.583835 | 6.5471026 |
| GO:0001945 | lymph vessel development | 22 | 0.4671139 | 1.606192 | 3.012864e-02 | 0.386294977 | 0.362489027 | 3755 | tags=36%, list=22%, signal=28% | ACVRL1||SOX18||NR2F2||FOXC1||VASH1||TBX1||FLT4||VEGFC | 3.221393 | 3.7680592 | 3.8415912 | 3.775032 | 3.211471 | 3.7923584 | 3.7849504 | 3.7707848 | 3.132426 | 3.227831 | 3.299090 | 3.7234353 | 3.8046238 | 3.7749516 | 4.044249 | 3.82556595 | 3.6243859 | 3.7719938 | 3.7536630 | 3.799076 | 3.194327 | 3.041076 | 3.379103 | 3.543228 | 3.666601 | 4.10510937 | 3.826123 | 3.7963029 | 3.7307828 | 3.7437580 | 3.851770 | 3.7131212 |
| GO:1902473 | regulation of protein localization to synapse | 19 | 0.4903582 | 1.614266 | 3.026401e-02 | 0.386294977 | 0.362489027 | 6830 | tags=63%, list=41%, signal=37% | MAPT||HRAS||DVL1||ABHD17B||IQSEC2||PRKCZ||DAG1||RAP1A||WNT5A||ARHGAP44||NLGN2||GPC6 | 4.365457 | 4.4597491 | 4.5687443 | 4.486697 | 4.385079 | 4.7576371 | 4.3886425 | 4.3795763 | 4.280198 | 4.291181 | 4.512851 | 4.4221842 | 4.5202903 | 4.4347848 | 4.513720 | 4.65505493 | 4.5333572 | 4.5188000 | 4.4724738 | 4.468270 | 4.393096 | 4.245034 | 4.505346 | 4.621048 | 4.773661 | 4.86755124 | 4.423078 | 4.4033042 | 4.3381841 | 4.3716160 | 4.430425 | 4.3350796 |
| GO:0050728 | negative regulation of inflammatory response | 78 | 0.3298964 | 1.512538 | 3.050847e-02 | 0.387657344 | 0.363767436 | 4699 | tags=44%, list=28%, signal=32% | FOXP3||MVK||NR1D1||GPX1||GPER1||FNDC4||MAPK7||FURIN||KLF4||GSTP1||GATA3||SRC||NLRX1||SIRPA||ALOX5||PPARD||LRFN5||TNFAIP3||TYRO3||NR1H3||ZFP36||TNFRSF1A||FEM1A||GIT1||PRKCD||METRNL||FYN||MDK||SERPINF1||SHARPIN||GRN||LDLR||ADORA1||ADA | 6.904130 | 6.2876778 | 6.7146594 | 6.170181 | 6.942084 | 6.8615121 | 6.1861341 | 6.1310013 | 6.910953 | 6.802842 | 6.992351 | 6.3559444 | 6.2715334 | 6.2327939 | 6.536188 | 6.81257888 | 6.7797242 | 6.2477459 | 6.1072635 | 6.151942 | 6.934451 | 6.858710 | 7.028093 | 6.880006 | 6.970732 | 6.72299703 | 6.228981 | 6.1576449 | 6.1707732 | 6.1326871 | 6.129474 | 6.1308407 |
| GO:2000737 | negative regulation of stem cell differentiation | 21 | 0.4672537 | 1.581508 | 3.072812e-02 | 0.389568928 | 0.365561216 | 3687 | tags=57%, list=22%, signal=45% | ZFP36L2||HES5||WNT3A||NELFB||JAG1||YTHDF2||HES1||NOTCH1||STAT3||NFE2L2||ESRRB||HNRNPU | 4.913080 | 5.9088565 | 4.7675772 | 5.893066 | 4.788015 | 4.9675868 | 5.9909487 | 5.9816456 | 4.897812 | 5.020589 | 4.813276 | 5.8838223 | 5.9128583 | 5.9295187 | 5.250035 | 4.46287825 | 4.4334566 | 5.8330250 | 5.8614645 | 5.980432 | 4.771786 | 4.774278 | 4.817521 | 5.070469 | 5.089078 | 4.71299642 | 5.952187 | 6.0017069 | 6.0181380 | 5.9826396 | 5.990609 | 5.9716249 |
| GO:0061050 | regulation of cell growth involved in cardiac muscle cell development | 13 | 0.5467978 | 1.605751 | 3.103448e-02 | 0.389939963 | 0.365909386 | 1690 | tags=31%, list=10%, signal=28% | AKAP6||G6PD||CAV3||PIN1 | 6.335921 | 6.1874524 | 6.4133588 | 5.990772 | 6.344684 | 6.4155656 | 6.2085797 | 6.0375289 | 6.265411 | 6.353183 | 6.386475 | 6.2119330 | 6.1859593 | 6.1640666 | 6.411649 | 6.45503042 | 6.3722066 | 6.0160439 | 5.9412792 | 6.013746 | 6.393759 | 6.239854 | 6.394991 | 6.307490 | 6.394383 | 6.53558562 | 6.229453 | 6.2029889 | 6.1930520 | 6.0252105 | 6.091668 | 5.9939765 |
| GO:0003207 | cardiac chamber formation | 10 | 0.6074573 | 1.640828 | 3.153031e-02 | 0.395287525 | 0.370927397 | 2744 | tags=60%, list=16%, signal=50% | MESP1||TBX2||BMP2||NOTCH1||HAND1||SOX4 | 3.298086 | 3.0694736 | 3.3870164 | 3.057278 | 3.313796 | 3.6515888 | 3.0564383 | 3.0402152 | 3.291630 | 3.205444 | 3.391189 | 3.1135470 | 3.0553918 | 3.0384011 | 3.391267 | 3.49338669 | 3.2675582 | 3.0745768 | 3.0260927 | 3.070664 | 3.341936 | 3.097269 | 3.476868 | 3.585601 | 3.689061 | 3.67788606 | 3.054892 | 3.0384725 | 3.0757090 | 3.0126760 | 3.114183 | 2.9907579 |
| GO:0051154 | negative regulation of striated muscle cell differentiation | 24 | 0.4501964 | 1.584886 | 3.181818e-02 | 0.396198370 | 0.371782110 | 3388 | tags=42%, list=20%, signal=33% | NKX2-5||G6PD||BMP2||CAV3||PLPP7||SMAD4||NOTCH1||HDAC5||HDAC4||CTDP1 | 5.586110 | 6.0489589 | 5.7431552 | 6.068751 | 5.443228 | 5.5859093 | 6.0813261 | 6.1139529 | 5.600890 | 5.771706 | 5.355738 | 6.0744299 | 6.0540785 | 6.0177989 | 5.868732 | 5.64143184 | 5.7097885 | 6.1302994 | 5.9957246 | 6.077057 | 5.464525 | 5.500030 | 5.361569 | 5.473393 | 5.427351 | 5.82360357 | 6.089843 | 6.0657630 | 6.0882469 | 6.0995621 | 6.120771 | 6.1214185 |
| GO:0060285 | cilium-dependent cell motility | 70 | -0.4324858 | -1.504819 | 3.188821e-02 | 0.396198370 | 0.371782110 | 4560 | tags=46%, list=27%, signal=33% | BBS2||TTC12||TEKT4||SPEF2||INPP5B||EFHC1||SORD||DPCD||DNAH1||LDHC||ROPN1L||DNAH11||TTLL3||CCSAP||MKKS||VPS13A||TCTE1||CFAP251||DNAH2||BBS4||TTLL9||DNAH17||CATSPER2||DNAH6||SLIRP||CFAP20||FSIP2||CFAP44||CFAP69||QRICH2||TEKT5||CFAP54 | 3.847601 | 4.3441322 | 4.0759797 | 4.401371 | 3.633671 | 3.5678359 | 4.3808125 | 4.3673135 | 3.970617 | 3.982482 | 3.548909 | 4.3505740 | 4.3316914 | 4.3500512 | 4.345025 | 3.88093848 | 3.9579444 | 4.3621447 | 4.4088108 | 4.432275 | 3.672599 | 3.753511 | 3.459110 | 3.456703 | 3.401255 | 3.81051509 | 4.391824 | 4.3764039 | 4.3741459 | 4.3674285 | 4.385204 | 4.3490819 |
| GO:0001756 | somitogenesis | 51 | 0.3546916 | 1.498991 | 3.215926e-02 | 0.396475811 | 0.372042453 | 2569 | tags=31%, list=15%, signal=27% | SEMA3C||MESP2||MESP1||PLXNA2||NLE1||EP300||DLL3||COBL||CDX2||TP53||WNT3A||FOXC1||FOXB1||SMAD4||MED12||NOTCH1 | 3.531672 | 4.4264062 | 3.8617342 | 4.453282 | 3.536208 | 3.8889778 | 4.4207984 | 4.4471153 | 3.460097 | 3.620835 | 3.509353 | 4.3805413 | 4.4632521 | 4.4342082 | 4.080391 | 3.73757767 | 3.7395724 | 4.3988923 | 4.4697042 | 4.489679 | 3.541749 | 3.427011 | 3.632533 | 3.936522 | 3.872514 | 3.85665176 | 4.435699 | 4.4174966 | 4.4090707 | 4.4228868 | 4.473186 | 4.4448321 |
| GO:0002385 | mucosal immune response | 10 | -0.6940434 | -1.620119 | 3.242132e-02 | 0.396475811 | 0.372042453 | 1058 | tags=30%, list=6%, signal=28% | XCL1||RPL39||H2BC6 | 8.950108 | 8.4060391 | 8.6680879 | 8.447445 | 8.803734 | 7.9754527 | 8.4195906 | 7.9881955 | 9.128236 | 9.131469 | 8.505201 | 8.4555042 | 8.3430590 | 8.4172974 | 8.715069 | 8.40155431 | 8.8515230 | 8.4093183 | 8.5094529 | 8.421479 | 8.727911 | 9.229385 | 8.305597 | 8.422383 | 7.714652 | 7.65755572 | 8.355500 | 8.4697401 | 8.4312003 | 8.4342248 | 5.508159 | 8.5326556 |
| GO:0021532 | neural tube patterning | 34 | 0.4027664 | 1.552323 | 3.264813e-02 | 0.396475811 | 0.372042453 | 2488 | tags=35%, list=15%, signal=30% | FGF8||FOXA1||FKBP8||SUFU||PRKACA||WNT3A||GSC||IFT140||TULP3||GLI3||HES1||PTCH1 | 4.379367 | 4.6544456 | 4.4748763 | 4.685998 | 4.416036 | 4.7065165 | 4.6100737 | 4.6712081 | 4.308272 | 4.320208 | 4.501391 | 4.6668468 | 4.6863244 | 4.6090506 | 4.458955 | 4.52754793 | 4.4365606 | 4.7151028 | 4.6606165 | 4.681752 | 4.423115 | 4.301578 | 4.515467 | 4.630555 | 4.780100 | 4.70501981 | 4.632299 | 4.5984828 | 4.5991802 | 4.6462596 | 4.729963 | 4.6355375 |
| GO:0032570 | response to progesterone | 27 | 0.4293361 | 1.555847 | 3.277467e-02 | 0.396475811 | 0.372042453 | 3347 | tags=52%, list=20%, signal=42% | TRERF1||WBP2||GPI||TGFB1||TSPO||BAD||RELA||SREBF1||NKX2-2||SRC||FOSB||CLDN4||UBE3A||NR1H3 | 5.452584 | 5.1482590 | 5.3193086 | 5.003279 | 5.531188 | 5.7021600 | 5.1185767 | 4.9629023 | 5.379432 | 5.370342 | 5.596437 | 5.1351063 | 5.1529436 | 5.1566353 | 5.161410 | 5.42336810 | 5.3603787 | 5.0512845 | 4.8955299 | 5.057265 | 5.533508 | 5.391998 | 5.655994 | 5.665484 | 5.816965 | 5.61637594 | 5.157040 | 5.0802290 | 5.1174379 | 4.9651146 | 4.996412 | 4.9263270 |
| GO:0060251 | regulation of glial cell proliferation | 22 | 0.4612551 | 1.586046 | 3.283683e-02 | 0.396475811 | 0.372042453 | 4177 | tags=50%, list=25%, signal=38% | ETV5||SKI||TSPO||CREB1||HES1||EGFR||NOTCH1||IDH2||MYB||GFAP||E2F1 | 5.292344 | 5.4918933 | 5.4303958 | 5.582516 | 5.335744 | 5.5649567 | 5.4998969 | 5.5491155 | 5.238884 | 5.249139 | 5.384389 | 5.4812552 | 5.5289611 | 5.4646891 | 5.347995 | 5.52886264 | 5.4084163 | 5.5579263 | 5.5452488 | 5.642427 | 5.351241 | 5.193673 | 5.450755 | 5.536995 | 5.526648 | 5.62901038 | 5.491370 | 5.4877225 | 5.5203754 | 5.5225732 | 5.582870 | 5.5412411 |
| GO:0002021 | response to dietary excess | 15 | 0.5223459 | 1.602245 | 3.303571e-02 | 0.396475811 | 0.372042453 | 1522 | tags=33%, list=9%, signal=30% | GDF15||VGF||PCSK1N||PPARGC1A||TRPV4 | 3.070921 | 3.1151029 | 2.9449196 | 2.804029 | 2.732502 | 3.2166522 | 2.9350516 | 2.6351896 | 3.174294 | 2.989186 | 3.042949 | 3.1827205 | 3.0883400 | 3.0717468 | 2.891715 | 3.12265261 | 2.8010717 | 2.8589597 | 2.7866456 | 2.764788 | 2.844931 | 2.375689 | 2.919085 | 3.048381 | 3.348834 | 3.23691222 | 2.974066 | 2.9152985 | 2.9149844 | 2.6306933 | 2.703658 | 2.5680198 |
| GO:0097186 | amelogenesis | 13 | 0.5444357 | 1.598815 | 3.304598e-02 | 0.396475811 | 0.372042453 | 6041 | tags=77%, list=36%, signal=49% | NECTIN1||FAM20C||TBX1||PERP||STIM1||CNNM4||FOXO1||RELT||SLC4A2||TCIRG1 | 3.963158 | 4.3828420 | 4.4287293 | 4.345776 | 3.976719 | 4.4133201 | 4.3535413 | 4.2903707 | 3.920235 | 3.812124 | 4.137833 | 4.3240332 | 4.4583356 | 4.3628263 | 4.337778 | 4.57043453 | 4.3666584 | 4.3621655 | 4.3574074 | 4.317335 | 3.899354 | 3.885180 | 4.132070 | 4.141290 | 4.340453 | 4.70150134 | 4.381983 | 4.3851939 | 4.2915003 | 4.2811084 | 4.363112 | 4.2234696 |
| GO:0010665 | regulation of cardiac muscle cell apoptotic process | 26 | 0.4351451 | 1.563809 | 3.313911e-02 | 0.396475811 | 0.372042453 | 4200 | tags=50%, list=25%, signal=38% | JAK2||NUPR1||NKX2-5||HSF1||PTEN||HEY2||TP53||GATA4||RGL2||CAPN2||NFE2L2||LTK||MDK | 5.044873 | 5.2122753 | 5.0613899 | 5.019154 | 5.091460 | 5.2451800 | 5.1413735 | 5.0048745 | 4.991226 | 4.991359 | 5.146411 | 5.2235984 | 5.2288873 | 5.1839229 | 5.013977 | 5.13318138 | 5.0341690 | 5.0260192 | 5.0084307 | 5.022952 | 5.087372 | 5.008512 | 5.173759 | 5.200189 | 5.250054 | 5.28406542 | 5.164752 | 5.1388837 | 5.1201370 | 5.0118457 | 5.036182 | 4.9657098 |
| GO:0015701 | bicarbonate transport | 16 | -0.6211321 | -1.630848 | 3.317465e-02 | 0.396475811 | 0.372042453 | 3505 | tags=50%, list=21%, signal=40% | SLC39A8||SLC26A7||BEST1||SLC4A5||SLC4A7||SLC26A2||SLC4A8||SLC26A10 | 3.417937 | 3.7952527 | 4.1430982 | 3.988435 | 3.449358 | 3.8762217 | 3.7934808 | 3.8927417 | 3.386673 | 3.297536 | 3.557414 | 3.7274959 | 3.8673158 | 3.7875292 | 4.131673 | 4.21855138 | 4.0754555 | 3.9929073 | 3.9987791 | 3.973497 | 3.357233 | 3.407123 | 3.574628 | 3.495682 | 3.668908 | 4.32586377 | 3.826092 | 3.8179476 | 3.7346387 | 3.8624730 | 3.992709 | 3.8172411 |
| GO:0048048 | embryonic eye morphogenesis | 29 | 0.4198100 | 1.558160 | 3.345865e-02 | 0.399022716 | 0.374432401 | 3882 | tags=41%, list=23%, signal=32% | EFEMP1||HIPK2||RARG||TBX2||TFAP2A||MFAP2||CITED2||SIX3||IHH||TWIST1||VAX2||BMP7 | 2.744012 | 3.3286562 | 3.1492310 | 3.459007 | 2.727731 | 3.1730806 | 3.4063227 | 3.5132569 | 2.682838 | 2.789460 | 2.757672 | 3.2496048 | 3.4025539 | 3.3297575 | 3.293982 | 3.10405574 | 3.0372053 | 3.4551275 | 3.4370134 | 3.484483 | 2.759627 | 2.604654 | 2.811004 | 3.064770 | 3.095137 | 3.34290182 | 3.425078 | 3.4008880 | 3.3928067 | 3.5027388 | 3.539985 | 3.4966649 |
| GO:2001224 | positive regulation of neuron migration | 14 | 0.5301566 | 1.589663 | 3.375404e-02 | 0.400162383 | 0.375501835 | 4833 | tags=57%, list=29%, signal=41% | DAB2IP||ZNF609||ARHGEF2||FLNA||SEMA6A||FBXO31||MDK||NSMF | 5.405551 | 5.2022180 | 5.5548256 | 4.983463 | 5.454050 | 5.6879405 | 5.1496524 | 4.9620628 | 5.344822 | 5.295424 | 5.562224 | 5.2108244 | 5.2416633 | 5.1527584 | 5.381083 | 5.69862025 | 5.5672552 | 5.0343800 | 4.9788544 | 4.935448 | 5.443055 | 5.339108 | 5.570643 | 5.558467 | 5.685044 | 5.80940793 | 5.232255 | 5.1359772 | 5.0764221 | 4.9990715 | 5.001321 | 4.8826309 |
| GO:0048488 | synaptic vesicle endocytosis | 55 | 0.3455144 | 1.480815 | 3.391627e-02 | 0.400162383 | 0.375501835 | 3615 | tags=35%, list=22%, signal=27% | VAMP4||SNAP91||DNM3||SNCB||SYNJ2||SH3GL1||ACTG1||ACTB||PACSIN1||ROCK1||PIP5K1C||SNCG||SYT1||CALM1||DNM2||DNAJC6||CALM3||SCRIB||DENND1A | 7.173212 | 6.7724491 | 6.7031161 | 6.667913 | 7.199189 | 7.0978624 | 6.7099401 | 6.6643891 | 7.124122 | 7.161808 | 7.231634 | 6.7541324 | 6.7950863 | 6.7678270 | 6.490042 | 6.81105225 | 6.7866593 | 6.7315689 | 6.5887773 | 6.679787 | 7.295081 | 7.076330 | 7.217693 | 7.240308 | 7.067338 | 6.97310945 | 6.707745 | 6.7232221 | 6.6987469 | 6.6773127 | 6.673383 | 6.6422163 |
| GO:0021542 | dentate gyrus development | 10 | 0.6017121 | 1.625309 | 3.397662e-02 | 0.400162383 | 0.375501835 | 5118 | tags=60%, list=31%, signal=42% | PTEN||TMEM108||EMX2||MDK||SMO||LEF1 | 5.378287 | 5.3447795 | 5.4991869 | 4.933187 | 5.439649 | 5.5784281 | 5.2253468 | 4.9019384 | 5.337473 | 5.242190 | 5.539100 | 5.3945752 | 5.3834307 | 5.2520217 | 5.361338 | 5.61175406 | 5.5135086 | 4.9735198 | 4.9462675 | 4.878105 | 5.409606 | 5.388443 | 5.517549 | 5.429832 | 5.556808 | 5.73255210 | 5.286731 | 5.2271164 | 5.1593823 | 4.9430037 | 4.946405 | 4.8124022 |
| GO:0007519 | skeletal muscle tissue development | 117 | 0.2924211 | 1.426285 | 3.403403e-02 | 0.400162383 | 0.375501835 | 5159 | tags=48%, list=31%, signal=33% | MYOZ1||MSC||HDAC9||VGLL2||NUPR1||NR2F2||EEF2||EMD||EP300||GPX1||WNT10B||POPDC3||SKI||TGFB1||PITX1||NIBAN2||MEF2D||SELENON||WNT3A||MYL6B||KLF5||NEURL1||CITED2||COL19A1||VAX1||YBX3||NOTCH1||TBX1||HIVEP3||SOX8||ZNF689||P2RX2||HLX||PLEC||HDAC4||GTF3C5||DNER||HEYL||TWIST1||MYH14||DMRTA2||USP19||PPP3CA||MAFF||CDK5||NR4A1||FLOT1||FLNB||UQCC2||SMO||GPC1||MYORG||BCL9||IGF2||CFL2||RYR1 | 6.189185 | 5.9167919 | 6.1467554 | 5.916937 | 6.245629 | 6.3768997 | 5.8685199 | 5.9379056 | 6.137019 | 6.143593 | 6.282231 | 5.9598337 | 5.9099490 | 5.8794490 | 6.098676 | 6.17985085 | 6.1604992 | 5.9664639 | 5.8400259 | 5.941240 | 6.251089 | 6.150127 | 6.330051 | 6.399615 | 6.474775 | 6.24706222 | 5.883513 | 5.8649409 | 5.8569771 | 5.9191058 | 5.969352 | 5.9247315 |
| GO:0048593 | camera-type eye morphogenesis | 90 | 0.3141421 | 1.480900 | 3.403933e-02 | 0.400162383 | 0.375501835 | 4011 | tags=39%, list=24%, signal=30% | SDK2||PITX3||HIPK2||TBX2||SKI||TFAP2A||SOX9||LRP5||BAK1||EPHB2||C12orf57||NECTIN1||OBSL1||NAGLU||JAG1||BAX||SHROOM2||GLI3||CITED2||SIX3||DIO3||ABI2||RDH13||SDK1||FOXN4||SOX8||IHH||TSKU||TWIST1||HMGN1||BMP7||ZHX2||RING1||NTRK2||MEGF11 | 4.848711 | 5.0142957 | 4.9568162 | 5.217092 | 4.854937 | 5.0189160 | 5.0250918 | 5.2298023 | 4.836577 | 4.887697 | 4.821013 | 5.0401128 | 5.0135887 | 4.9887278 | 5.052829 | 4.88827541 | 4.9241063 | 5.2290100 | 5.1553312 | 5.264786 | 4.815472 | 4.876119 | 4.872422 | 4.985268 | 5.033496 | 5.03740108 | 5.043481 | 5.0052035 | 5.0263362 | 5.2288098 | 5.229313 | 5.2312834 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 109 | 0.2987890 | 1.442607 | 3.426510e-02 | 0.400162383 | 0.375501835 | 3454 | tags=37%, list=21%, signal=29% | NOTUM||MESP1||NKX2-5||EMD||SOX2||TLE6||SLC9A3R1||SHISA3||IGFBP6||DAB2IP||SOX9||FUZ||AXIN1||BMP2||TPBGL||IGFBP2||HECW1||APC2||GLI1||RUVBL2||SNAI2||TLE5||LATS1||GLI3||LATS2||FRMD8||SFRP5||TLE3||DAB2||NOTCH1||MAD2L2||TLE2||FERMT1||NOG||NKD2||TLE4||STK11||PTPRU||LZTS2||TLE1 | 4.963840 | 4.9190308 | 5.0056905 | 4.915552 | 4.957601 | 5.0212813 | 4.8987124 | 4.9255712 | 4.974972 | 4.961597 | 4.954877 | 4.9558969 | 4.9222247 | 4.8779117 | 4.962647 | 5.02175618 | 5.0317066 | 4.9418938 | 4.8720197 | 4.931758 | 4.952144 | 4.973701 | 4.946817 | 4.980151 | 5.018708 | 5.06376973 | 4.924659 | 4.8867669 | 4.8843557 | 4.9209128 | 4.952692 | 4.9026637 |
| GO:0008637 | apoptotic mitochondrial changes | 89 | 0.3133068 | 1.471358 | 3.474320e-02 | 0.404012789 | 0.379114954 | 3524 | tags=34%, list=21%, signal=27% | ERBB4||BIK||HSPA1A||CHCHD10||JUN||BOK||AIFM2||GPX1||TMEM102||GPER1||AKT1||ZNF205||PLSCR3||HK2||BAD||BAK1||TP53||FZD9||EYA2||BAX||MFF||ATG3||GCLM||ARRB2||PRELID1||TIMM50||PINK1||PDCD5||RTL10||FIS1 | 6.481848 | 6.4808101 | 6.3738094 | 6.478250 | 6.467865 | 6.4739396 | 6.4444399 | 6.4691983 | 6.484349 | 6.493223 | 6.467857 | 6.5394204 | 6.4418450 | 6.4592768 | 6.404664 | 6.33536322 | 6.3805449 | 6.4970174 | 6.4261030 | 6.510218 | 6.451955 | 6.450283 | 6.500784 | 6.472861 | 6.568537 | 6.37385757 | 6.453323 | 6.4202896 | 6.4594005 | 6.4659552 | 6.473974 | 6.4676532 |
| GO:0048143 | astrocyte activation | 16 | -0.6160947 | -1.617622 | 3.510860e-02 | 0.405287150 | 0.380310781 | 1059 | tags=25%, list=6%, signal=23% | PSEN1||C5AR1||AGER||CNTF | 4.433361 | 5.4735831 | 5.0338018 | 5.480401 | 4.465865 | 4.9462777 | 5.2807638 | 5.3693667 | 4.332158 | 4.480803 | 4.482034 | 5.5144883 | 5.5397301 | 5.3600536 | 5.033452 | 5.09199895 | 4.9735230 | 5.5181194 | 5.4889013 | 5.432885 | 4.475795 | 4.368303 | 4.547862 | 4.774102 | 4.787277 | 5.22943082 | 5.343694 | 5.2733029 | 5.2227310 | 5.3927893 | 5.428184 | 5.2831967 |
| GO:0060396 | growth hormone receptor signaling pathway | 16 | 0.5086474 | 1.590270 | 3.536585e-02 | 0.405287150 | 0.380310781 | 2681 | tags=31%, list=16%, signal=26% | GDF15||JAK2||PXN||MBD5||STAT3 | 2.816489 | 3.7346166 | 3.2916469 | 3.749456 | 2.760814 | 3.2564332 | 3.7704024 | 3.7666456 | 2.716742 | 2.981491 | 2.735845 | 3.7234870 | 3.7611448 | 3.7188439 | 3.485975 | 3.21463357 | 3.1520788 | 3.7075039 | 3.7123699 | 3.825385 | 2.842936 | 2.661792 | 2.771965 | 3.161414 | 3.094294 | 3.48299270 | 3.810615 | 3.7464153 | 3.7533124 | 3.7620506 | 3.807748 | 3.7290541 |
| GO:0051127 | positive regulation of actin nucleation | 13 | 0.5400725 | 1.586001 | 3.591954e-02 | 0.410123919 | 0.384849478 | 3491 | tags=62%, list=21%, signal=49% | WASF1||WAS||WASF2||GSN||ABI2||FCHSD2||MAGEL2||WASL | 5.030788 | 5.3421116 | 5.3425933 | 5.450190 | 5.014210 | 5.0676515 | 5.3999573 | 5.4711343 | 5.042751 | 5.154310 | 4.882439 | 5.3087691 | 5.3372729 | 5.3794160 | 5.352980 | 5.30496713 | 5.3690646 | 5.4320442 | 5.4063357 | 5.510153 | 5.024061 | 5.080056 | 4.934809 | 5.067854 | 4.873989 | 5.23818718 | 5.393342 | 5.3855222 | 5.4207701 | 5.4609776 | 5.450215 | 5.5016980 |
| GO:0045806 | negative regulation of endocytosis | 40 | 0.3735907 | 1.490768 | 3.626714e-02 | 0.411588150 | 0.386223473 | 6184 | tags=60%, list=37%, signal=38% | MCTP1||PROM2||RIN3||ATXN2||PACSIN3||UBQLN2||WDR54||PACSIN1||LRSAM1||SNX33||NR1H3||ANKRD13B||NECAB2||ARF6||NR1H2||UNC119||APOC1||EPHA3||RAC1||ABCA2||PICALM||ABCA7||RUBCN||PRKD1 | 4.650917 | 4.9379666 | 4.8413111 | 4.847615 | 4.669196 | 4.8478870 | 4.8783499 | 4.8176948 | 4.603864 | 4.660641 | 4.686999 | 4.9430428 | 4.9554746 | 4.9150863 | 4.814686 | 4.90180866 | 4.8054661 | 4.8618398 | 4.8219821 | 4.858684 | 4.668508 | 4.617970 | 4.719331 | 4.764865 | 4.799658 | 4.97062448 | 4.912160 | 4.8429838 | 4.8790766 | 4.8304531 | 4.849110 | 4.7724167 |
| GO:0002890 | negative regulation of immunoglobulin mediated immune response | 11 | 0.5763531 | 1.605740 | 3.651059e-02 | 0.412393967 | 0.386979631 | 2455 | tags=45%, list=15%, signal=39% | FOXP3||CR2||SUSD4||BCL6||CR1L | 2.626419 | 4.3902005 | 3.2008072 | 4.533652 | 2.580931 | 2.7975878 | 4.3217206 | 4.5201460 | 2.623594 | 2.793277 | 2.440920 | 4.4342697 | 4.4045808 | 4.3297503 | 3.648182 | 3.00244783 | 2.8141536 | 4.4578312 | 4.6022872 | 4.537216 | 2.509187 | 2.650330 | 2.579826 | 2.522861 | 2.583485 | 3.18859739 | 4.313571 | 4.3058515 | 4.3454334 | 4.4825189 | 4.582264 | 4.4935685 |
| GO:0035372 | protein localization to microtubule | 17 | 0.4994353 | 1.587883 | 3.676012e-02 | 0.412580817 | 0.387154966 | 3687 | tags=59%, list=22%, signal=46% | GAS2L1||CHAMP1||MAP1A||DVL1||ABHD17A||MAPRE2||ABHD17B||MID1||MAPRE3||HNRNPU | 5.343364 | 5.9339931 | 5.2976749 | 5.971653 | 5.285767 | 5.5082159 | 5.9846394 | 5.9773191 | 5.304004 | 5.414290 | 5.309089 | 5.8927165 | 5.9457281 | 5.9626160 | 5.608181 | 5.11869258 | 5.1075528 | 5.9310115 | 5.9359768 | 6.045066 | 5.294156 | 5.221763 | 5.338966 | 5.563052 | 5.591724 | 5.35888340 | 5.940847 | 5.9999629 | 6.0121058 | 5.9842247 | 5.987605 | 5.9599704 |
| GO:0002719 | negative regulation of cytokine production involved in immune response | 12 | 0.5621087 | 1.606721 | 3.686768e-02 | 0.412580817 | 0.387154966 | 1181 | tags=42%, list=7%, signal=39% | HMOX1||FOXP3||TGFB1||CUEDC2||BCL6 | 3.914675 | 3.2747529 | 3.4698780 | 2.872138 | 3.971183 | 3.9567603 | 3.2180758 | 2.7015961 | 3.887140 | 3.768508 | 4.072050 | 3.2854908 | 3.2747730 | 3.2639142 | 3.386878 | 3.54890930 | 3.4692994 | 2.9287477 | 2.7538675 | 2.926902 | 3.940067 | 3.861331 | 4.101657 | 3.929313 | 4.138964 | 3.77925218 | 3.267885 | 3.1699668 | 3.2147085 | 2.6526778 | 2.761126 | 2.6888627 |
| GO:1902074 | response to salt | 12 | 0.5616393 | 1.605379 | 3.686768e-02 | 0.412580817 | 0.387154966 | 4329 | tags=58%, list=26%, signal=43% | PITX3||HSF1||HNRNPD||NEFL||HSPA5||ZC3H12A||HNRNPA1 | 7.523219 | 8.4451354 | 7.3765119 | 8.298867 | 7.454741 | 7.6165849 | 8.4821350 | 8.2678579 | 7.486230 | 7.682376 | 7.385060 | 8.4123099 | 8.4379623 | 8.4842116 | 7.795488 | 7.07396384 | 7.1457798 | 8.2529064 | 8.3007890 | 8.341542 | 7.417766 | 7.492258 | 7.453237 | 7.692401 | 7.713148 | 7.42693647 | 8.458903 | 8.4751897 | 8.5118027 | 8.2647347 | 8.268473 | 8.2703606 |
| GO:0098885 | modification of postsynaptic actin cytoskeleton | 10 | 0.5964290 | 1.611039 | 3.723838e-02 | 0.414038589 | 0.388522901 | 1924 | tags=40%, list=11%, signal=35% | PFN1||WASF1||BAIAP2||CTTNBP2 | 8.120573 | 7.4395009 | 7.9504619 | 7.473235 | 8.126902 | 8.2785193 | 7.4152147 | 7.4335463 | 8.040580 | 8.110468 | 8.205886 | 7.4795008 | 7.4053727 | 7.4326528 | 7.759712 | 8.06420919 | 8.0095606 | 7.5897665 | 7.3513282 | 7.468760 | 8.185093 | 7.980621 | 8.204506 | 8.263537 | 8.358163 | 8.20993691 | 7.416248 | 7.3909070 | 7.4381029 | 7.4389227 | 7.444021 | 7.4175588 |
| GO:0021510 | spinal cord development | 78 | 0.3201298 | 1.467759 | 3.728814e-02 | 0.414038589 | 0.388522901 | 3823 | tags=37%, list=23%, signal=29% | DAB1||MPST||ACTL6B||LBX1||AKT1||SUFU||NEFL||WNT3A||FOXB1||PHGDH||TULP3||BAG3||SOX12||GLI3||GATA2||MED12||PTCH1||NOTCH1||NKX2-2||SOX4||FOXN4||ISL2||NOG||HOXB8||PTPRS||UNCX||ERCC2||DCTN1||DAAM2 | 4.487962 | 4.5164533 | 4.6339697 | 4.385549 | 4.541751 | 4.8205356 | 4.4996127 | 4.4125880 | 4.388358 | 4.434824 | 4.629230 | 4.5106590 | 4.5523803 | 4.4855290 | 4.605679 | 4.68611157 | 4.6086691 | 4.4264376 | 4.3351170 | 4.393613 | 4.571844 | 4.381464 | 4.658215 | 4.739219 | 4.870104 | 4.84889274 | 4.535744 | 4.4810744 | 4.4813297 | 4.4071723 | 4.449624 | 4.3801151 |
| GO:0150076 | neuroinflammatory response | 12 | 0.5599517 | 1.600556 | 3.743927e-02 | 0.414038589 | 0.388522901 | 1985 | tags=50%, list=12%, signal=44% | NUPR1||NR1D1||ADCY1||TTBK1||SPHK1||DAGLA | 3.995487 | 4.7602333 | 4.5681092 | 4.688778 | 4.085002 | 4.5323893 | 4.5478086 | 4.5805524 | 3.898089 | 3.920979 | 4.153328 | 4.7882064 | 4.8216989 | 4.6661941 | 4.392611 | 4.76170518 | 4.5255450 | 4.7224380 | 4.6693728 | 4.673923 | 4.065630 | 3.944366 | 4.230639 | 4.286078 | 4.372726 | 4.86777744 | 4.592862 | 4.5227622 | 4.5267214 | 4.6396567 | 4.614381 | 4.4827514 |
| GO:0010878 | cholesterol storage | 14 | 0.5245175 | 1.572754 | 3.815674e-02 | 0.419500523 | 0.393648236 | 3347 | tags=50%, list=20%, signal=40% | EHD1||LPL||SCARB1||ABCG1||SREBF2||PPARD||NR1H3 | 4.013131 | 4.2762969 | 4.2047250 | 4.213285 | 4.137477 | 4.4818338 | 4.1527098 | 4.1316292 | 3.874052 | 4.022770 | 4.131089 | 4.2993948 | 4.3243921 | 4.2022385 | 4.077102 | 4.37644229 | 4.1432190 | 4.2491184 | 4.2334753 | 4.155530 | 4.191736 | 3.905323 | 4.288440 | 4.367559 | 4.503324 | 4.56745696 | 4.211877 | 4.1360778 | 4.1081686 | 4.1248426 | 4.164509 | 4.1048959 |
| GO:0043502 | regulation of muscle adaptation | 60 | 0.3386198 | 1.480622 | 3.880765e-02 | 0.422384272 | 0.396354270 | 3998 | tags=38%, list=24%, signal=29% | AKAP6||NR4A3||IL6ST||G6PD||PPARGC1A||FBXO32||CAV3||PIN1||KLF4||SELENON||SCN5A||SMAD4||ROCK1||CDK9||NOTCH1||GTF2IRD1||HDAC4||CAMK2B||CTDP1||TNFRSF1A||PPP3CA||PRKCA||LMNA | 5.094809 | 5.4775753 | 5.2912689 | 5.438833 | 5.072332 | 5.2676083 | 5.4966208 | 5.4704649 | 5.050320 | 5.166936 | 5.064340 | 5.4754613 | 5.4919516 | 5.4651867 | 5.415085 | 5.24701150 | 5.2029184 | 5.4163202 | 5.4080141 | 5.490719 | 5.103451 | 5.027311 | 5.085143 | 5.234546 | 5.233252 | 5.33275194 | 5.499159 | 5.4917022 | 5.4989891 | 5.4666662 | 5.496256 | 5.4480622 |
| GO:1904894 | positive regulation of receptor signaling pathway via STAT | 25 | 0.4311706 | 1.534887 | 3.884533e-02 | 0.422384272 | 0.396354270 | 2929 | tags=36%, list=17%, signal=30% | ERBB4||CCL5||JAK2||HES5||TGFB1||F2R||HES1||NOTCH1||CYP1B1 | 4.872603 | 5.0343943 | 4.9896777 | 4.986229 | 4.890154 | 4.9513786 | 4.9873180 | 4.9680636 | 4.829891 | 4.885779 | 4.901168 | 5.0617828 | 5.0369679 | 5.0038472 | 4.941748 | 5.04082966 | 4.9847415 | 4.9598559 | 4.9829749 | 5.015319 | 4.925191 | 4.833526 | 4.910080 | 5.018271 | 4.833088 | 4.99579144 | 5.014348 | 4.9790541 | 4.9681469 | 4.9569523 | 4.990628 | 4.9563435 |
| GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 12 | 0.5577293 | 1.594203 | 3.886825e-02 | 0.422384272 | 0.396354270 | 5957 | tags=67%, list=36%, signal=43% | ITM2C||BACE2||NECAB3||ITM2B||NECAB2||AGO2||ABCA2||ABCA7 | 4.695180 | 5.3832550 | 5.0678872 | 5.284188 | 4.722511 | 5.0041527 | 5.3160254 | 5.2588205 | 4.648053 | 4.692991 | 4.742934 | 5.3673202 | 5.4353240 | 5.3455949 | 5.188745 | 5.07321820 | 4.9301070 | 5.2237545 | 5.3358831 | 5.290724 | 4.685490 | 4.675851 | 4.802705 | 4.806035 | 4.890267 | 5.27224828 | 5.357275 | 5.3279746 | 5.2611507 | 5.2600013 | 5.295078 | 5.2204160 |
| GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 10 | 0.5923989 | 1.600153 | 3.886926e-02 | 0.422384272 | 0.396354270 | 4665 | tags=60%, list=28%, signal=43% | TICAM1||H19||IL17RA||STAT3||HIF1A||APPL1 | 2.320931 | 3.5698136 | 2.9273172 | 3.681852 | 2.255751 | 2.8997615 | 3.6526128 | 3.7678922 | 2.280488 | 2.498458 | 2.163630 | 3.5017030 | 3.5824764 | 3.6226437 | 3.064497 | 2.93928237 | 2.7623015 | 3.6370825 | 3.6716521 | 3.735103 | 2.324053 | 2.197476 | 2.242862 | 2.839725 | 2.619902 | 3.18319648 | 3.667716 | 3.6280885 | 3.6617182 | 3.7085184 | 3.841503 | 3.7504346 |
| GO:0098815 | modulation of excitatory postsynaptic potential | 33 | 0.3940841 | 1.503298 | 3.912176e-02 | 0.422543913 | 0.396504073 | 2997 | tags=36%, list=18%, signal=30% | RIMS2||CUX2||PTEN||TMEM108||SH3GL1||DVL1||ZMYND8||BAIAP2||SHANK1||SHANK3||CELF4||S1PR2 | 4.148057 | 4.7485709 | 4.3610754 | 4.672347 | 4.099068 | 4.3545289 | 4.6947812 | 4.6372522 | 4.096684 | 4.232156 | 4.111467 | 4.7726179 | 4.7877467 | 4.6831486 | 4.452584 | 4.28233387 | 4.3431218 | 4.7041922 | 4.6508659 | 4.661428 | 4.105413 | 4.048470 | 4.141791 | 4.313009 | 4.329055 | 4.41923476 | 4.759191 | 4.6748304 | 4.6479745 | 4.6255741 | 4.674023 | 4.6114096 |
| GO:0061308 | cardiac neural crest cell development involved in heart development | 11 | 0.5723520 | 1.594593 | 3.929766e-02 | 0.422543913 | 0.396504073 | 3882 | tags=64%, list=23%, signal=49% | SEMA3C||JAG1||CITED2||MAPK3||HES1||TWIST1||BMP7 | 4.119766 | 4.2972018 | 4.2035867 | 4.258507 | 4.184720 | 4.3615235 | 4.3333092 | 4.3087380 | 4.069814 | 4.109584 | 4.177826 | 4.2855940 | 4.3358225 | 4.2693535 | 4.205937 | 4.23513351 | 4.1689277 | 4.3226894 | 4.2201089 | 4.230498 | 4.194511 | 4.042720 | 4.304976 | 4.332627 | 4.363531 | 4.38788130 | 4.364724 | 4.3055648 | 4.3290229 | 4.2929094 | 4.299292 | 4.3336790 |
| GO:0048640 | negative regulation of developmental growth | 90 | 0.3071079 | 1.447740 | 3.933434e-02 | 0.422543913 | 0.396504073 | 3388 | tags=29%, list=20%, signal=23% | GDF15||MYOZ1||MAP2||SEMA3C||FSTL4||KCNK2||SEMA7A||G6PD||PTEN||CDKN1A||SEMA6B||CAV3||SEMA4G||RGMA||WNT3A||RTN4R||BCL11A||CITED2||RNF6||PTCH1||SEMA6A||NOG||TP73||PTPRS||RAI1||CTDP1 | 3.015774 | 3.7990338 | 3.5383584 | 3.831087 | 3.004285 | 3.4814733 | 3.8056182 | 3.8160415 | 2.951186 | 3.046650 | 3.047380 | 3.7789406 | 3.8205482 | 3.7973109 | 3.669633 | 3.53334490 | 3.3994538 | 3.8151950 | 3.8155195 | 3.862041 | 3.012273 | 2.868167 | 3.121293 | 3.301090 | 3.385443 | 3.72261537 | 3.842698 | 3.7957699 | 3.7776024 | 3.8211467 | 3.838930 | 3.7875775 |
| GO:0001710 | mesodermal cell fate commitment | 13 | 0.5336238 | 1.567064 | 3.994253e-02 | 0.424320941 | 0.398171589 | 1880 | tags=38%, list=11%, signal=34% | MESP1||ETV2||KLF4||WNT3A||EYA2 | 2.700100 | 2.8299631 | 2.6807506 | 2.811879 | 2.688418 | 2.9794270 | 2.8161182 | 2.7825596 | 2.648123 | 2.673785 | 2.775237 | 2.8351749 | 2.8470466 | 2.8073811 | 2.538936 | 2.78305466 | 2.7095108 | 2.8844123 | 2.7300416 | 2.817042 | 2.721937 | 2.486953 | 2.834849 | 2.992187 | 3.083951 | 2.85281030 | 2.797678 | 2.7869949 | 2.8625177 | 2.7799902 | 2.695404 | 2.8671719 |
| GO:0060306 | regulation of membrane repolarization | 21 | 0.4544012 | 1.538006 | 4.008016e-02 | 0.424320941 | 0.398171589 | 3034 | tags=43%, list=18%, signal=35% | AKAP6||KCNQ1||CACNA1D||SNTA1||CAV3||KCNH2||SCN5A||FLNA||SCN4B | 5.157647 | 5.5675253 | 4.7684374 | 5.680573 | 5.057596 | 4.9495794 | 5.5495557 | 5.7058406 | 5.222286 | 5.264967 | 4.968215 | 5.6255451 | 5.5485635 | 5.5265853 | 4.997130 | 4.61503234 | 4.6623451 | 5.6655717 | 5.6428301 | 5.731830 | 5.003634 | 5.182007 | 4.978498 | 5.034196 | 4.975212 | 4.83189792 | 5.552893 | 5.5483342 | 5.5474338 | 5.6984384 | 5.701378 | 5.7176316 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 86 | 0.3107138 | 1.452632 | 4.032258e-02 | 0.424320941 | 0.398171589 | 3255 | tags=35%, list=19%, signal=28% | ENSG00000173867||SH3RF2||HSPA1A||NOP53||HSPA1B||PLK1||AKT1||ARIH1||GABARAP||TRIB3||UBQLN2||HSPBP1||DVL1||SGTA||PIAS1||STUB1||FZR1||ZER1||KEAP1||ELOB||RNF217||DAB2||RAD23A||CBFA2T3||USP5||CDC20||NKD2||DNAJB2||FBXW8||SIRT2 | 7.227376 | 6.9618282 | 7.0386086 | 6.769642 | 7.306276 | 7.2835231 | 6.9321858 | 6.7669969 | 7.188308 | 7.178281 | 7.311676 | 6.9866194 | 6.9606198 | 6.9378322 | 6.885581 | 7.11500554 | 7.1038464 | 6.8231860 | 6.7079763 | 6.775447 | 7.328628 | 7.232477 | 7.354866 | 7.315654 | 7.366991 | 7.15995785 | 6.963261 | 6.9211961 | 6.9115748 | 6.7653970 | 6.777286 | 6.7582441 |
| GO:2001138 | regulation of phospholipid transport | 10 | -0.6771137 | -1.580600 | 4.032896e-02 | 0.424320941 | 0.398171589 | 772 | tags=40%, list=5%, signal=38% | ATP8A2||DBI||ATP8A1||TRIAP1 | 6.541005 | 6.3639805 | 6.4898902 | 6.564579 | 6.528575 | 6.5174628 | 6.3227352 | 6.5513836 | 6.490323 | 6.535298 | 6.595462 | 6.4157658 | 6.3300541 | 6.3446564 | 6.481448 | 6.44760961 | 6.5391254 | 6.5744495 | 6.5310565 | 6.587626 | 6.512901 | 6.504012 | 6.567978 | 6.559008 | 6.596799 | 6.38812715 | 6.353203 | 6.3052628 | 6.3092472 | 6.5638333 | 6.548494 | 6.5417351 |
| GO:0032212 | positive regulation of telomere maintenance via telomerase | 34 | -0.5069822 | -1.555958 | 4.108496e-02 | 0.429067257 | 0.402625407 | 4793 | tags=56%, list=29%, signal=40% | POT1||MAP3K4||CCT7||DKC1||NEK7||MAPK1||CCT5||CCT6A||TNKS||XRCC5||TNKS2||CCT4||ATR||FBXO4||CCT2||MAPK15||CCT3||ATM||CTNNB1 | 7.028298 | 7.9778855 | 7.1886066 | 7.985540 | 6.857717 | 6.9209603 | 8.0281181 | 8.0385963 | 7.062676 | 7.240021 | 6.738144 | 7.9931688 | 7.9573253 | 7.9829266 | 7.511599 | 6.90308643 | 7.0817849 | 7.9514509 | 7.9559925 | 8.047141 | 6.827684 | 7.006843 | 6.724354 | 6.952134 | 6.863453 | 6.94561137 | 8.019309 | 8.0197646 | 8.0451285 | 8.0260059 | 8.041380 | 8.0483127 |
| GO:0098974 | postsynaptic actin cytoskeleton organization | 10 | 0.5895585 | 1.592481 | 4.131557e-02 | 0.430214027 | 0.403701506 | 2814 | tags=80%, list=17%, signal=67% | DBNL||ACTG1||ACTB||INA||WASF2||DBN1||FARP1||ARF1 | 9.611421 | 8.8803594 | 9.0353468 | 8.748667 | 9.650569 | 9.5142658 | 8.8060219 | 8.7501302 | 9.556554 | 9.605662 | 9.669806 | 8.8736579 | 8.8922117 | 8.8751349 | 8.740653 | 9.18176216 | 9.1436222 | 8.8523831 | 8.6305093 | 8.754567 | 9.756036 | 9.510417 | 9.674512 | 9.683485 | 9.483155 | 9.35725809 | 8.805480 | 8.8207896 | 8.7916492 | 8.7631941 | 8.756584 | 8.7304044 |
| GO:0034605 | cellular response to heat | 51 | 0.3421400 | 1.445946 | 4.134763e-02 | 0.430214027 | 0.403701506 | 2080 | tags=25%, list=12%, signal=22% | HMOX1||IRAK1||HSPA1A||HSPA1B||MAPT||EP300||CREBBP||HSF1||CDKN1A||PRKACA||TRPV4||STUB1||BAG3 | 6.647663 | 7.4308014 | 6.4712434 | 7.381524 | 6.674404 | 6.5432630 | 7.4577449 | 7.4213700 | 6.647871 | 6.757094 | 6.529020 | 7.4475939 | 7.4215075 | 7.4231547 | 6.752408 | 6.26979945 | 6.3433156 | 7.3425078 | 7.3449650 | 7.454252 | 6.735094 | 6.651542 | 6.634557 | 6.613015 | 6.572452 | 6.43860478 | 7.455858 | 7.4536540 | 7.4637034 | 7.4199496 | 7.419502 | 7.4246531 |
| GO:0060317 | cardiac epithelial to mesenchymal transition | 26 | 0.4252629 | 1.528295 | 4.151493e-02 | 0.430684780 | 0.404143248 | 3493 | tags=50%, list=21%, signal=40% | FGF8||HEY2||BMP2||SNAI2||JAG1||SMAD4||NOTCH1||NOG||TBX3||SPRY1||HEYL||TWIST1||HEY1 | 3.177750 | 4.3226506 | 3.6229271 | 4.350015 | 3.203631 | 3.5375804 | 4.3053035 | 4.3263499 | 3.181912 | 3.198825 | 3.152126 | 4.2916754 | 4.3733008 | 4.3015929 | 3.884318 | 3.48474920 | 3.4591744 | 4.3088507 | 4.3525090 | 4.387606 | 3.138472 | 3.198711 | 3.270671 | 3.369037 | 3.430983 | 3.77818032 | 4.306568 | 4.3107311 | 4.2985849 | 4.3036954 | 4.376723 | 4.2972737 |
| GO:0043401 | steroid hormone mediated signaling pathway | 115 | 0.2844703 | 1.386093 | 4.154589e-02 | 0.430684780 | 0.404143248 | 3131 | tags=29%, list=19%, signal=23% | JAK2||ESRRG||PMEPA1||NR4A3||RXRG||FOXA1||TRERF1||PADI2||NR1D1||EP300||WBP2||CARM1||GPER1||PKN1||ZBTB7A||VPS18||SMARCA4||RXRA||PAGR1||DDX54||ZMIZ1||NCOR2||ESRRA||ARID1A||LATS1||PAQR7||RNF6||DAB2||SRC||FOXP1||PPARD||TADA3||UBE3A | 5.756047 | 6.2242303 | 6.0117487 | 6.112228 | 5.708559 | 5.8484839 | 6.1946543 | 6.1191765 | 5.746964 | 5.841634 | 5.674672 | 6.2433460 | 6.2340351 | 6.1948522 | 6.142920 | 5.90454294 | 5.9773417 | 6.0994425 | 6.0789327 | 6.157163 | 5.714663 | 5.724809 | 5.685925 | 5.847018 | 5.781068 | 5.91428997 | 6.196941 | 6.1878318 | 6.1991648 | 6.1053771 | 6.143646 | 6.1081902 |
| GO:0061028 | establishment of endothelial barrier | 37 | 0.3777641 | 1.479811 | 4.228330e-02 | 0.435123739 | 0.408308650 | 760 | tags=19%, list=5%, signal=18% | FOXP3||PLCB1||TJP3||SOX18||FASN||RAPGEF3||RAPGEF1 | 3.909685 | 4.6485209 | 4.3032623 | 4.776658 | 3.921715 | 4.4098209 | 4.6915912 | 4.7715677 | 3.834680 | 3.957804 | 3.933643 | 4.5875160 | 4.7021406 | 4.6536165 | 4.440462 | 4.28675675 | 4.1697469 | 4.7384843 | 4.7486537 | 4.840627 | 3.956177 | 3.810052 | 3.992523 | 4.312484 | 4.388397 | 4.52082962 | 4.706143 | 4.6860836 | 4.6824338 | 4.7472174 | 4.800582 | 4.7663968 |
| GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 16 | 0.4998781 | 1.562853 | 4.237805e-02 | 0.435302986 | 0.408476851 | 4718 | tags=56%, list=28%, signal=40% | SOX9||SOX8||NOG||SIX2||MAGED1||HOXB7||WNT2B||PAX2||SMO | 4.481012 | 4.4816184 | 4.6005302 | 4.467058 | 4.500818 | 4.7101279 | 4.5177227 | 4.4634240 | 4.419623 | 4.491523 | 4.529730 | 4.4710818 | 4.5130656 | 4.4601654 | 4.587850 | 4.62515307 | 4.5882684 | 4.4807022 | 4.4111412 | 4.507621 | 4.536059 | 4.396938 | 4.563970 | 4.649966 | 4.633320 | 4.83801193 | 4.539288 | 4.5031830 | 4.5104433 | 4.4560415 | 4.495923 | 4.4376913 |
| GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 83 | 0.3041208 | 1.420842 | 4.274702e-02 | 0.436023217 | 0.409152696 | 2064 | tags=25%, list=12%, signal=22% | HMOX1||MIF||NUPR1||SHISA5||HIPK2||CDIP1||BOK||EP300||PIAS4||NACC2||CDKN1A||PML||DDIT4||BAD||BAK1||TP53||ZNF385A||ABL1||BAG6||SNAI2||BAX | 7.609650 | 7.5976048 | 7.2907634 | 7.518734 | 7.627279 | 7.5633606 | 7.5683618 | 7.5166852 | 7.625864 | 7.519274 | 7.679244 | 7.6438527 | 7.5663570 | 7.5814290 | 7.220351 | 7.33874614 | 7.3105590 | 7.5705876 | 7.4908110 | 7.493372 | 7.628816 | 7.552552 | 7.696862 | 7.612350 | 7.699405 | 7.35675218 | 7.556080 | 7.5805575 | 7.5683437 | 7.5108532 | 7.503009 | 7.5359874 |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | 39 | 0.3707098 | 1.470793 | 4.283860e-02 | 0.436023217 | 0.409152696 | 3959 | tags=41%, list=24%, signal=31% | HMOX1||GPER1||PPARGC1A||PTEN||CDKN1A||BMP2||KLF4||GSTP1||NPR1||PPARD||S1PR2||TNFAIP3||GNA12||IL12A||NDRG2||TRIB1 | 6.341190 | 5.8524163 | 6.2531479 | 5.809365 | 6.411595 | 6.4583874 | 5.8446045 | 5.8911054 | 6.273780 | 6.270758 | 6.469876 | 5.8664301 | 5.8576171 | 5.8329939 | 6.098868 | 6.35903329 | 6.2891441 | 5.8574734 | 5.7375084 | 5.830387 | 6.404625 | 6.303405 | 6.518705 | 6.402333 | 6.569275 | 6.39681023 | 5.850889 | 5.8341257 | 5.8487412 | 5.8845596 | 5.909190 | 5.8793903 |
| GO:0018410 | C-terminal protein amino acid modification | 10 | -0.6744512 | -1.574385 | 4.285940e-02 | 0.436023217 | 0.409152696 | 5180 | tags=90%, list=31%, signal=62% | LCMT1||ATG7||ATG5||AGTPBP1||GPLD1||ATG16L1||TTL||ICMT||ATG12 | 4.578018 | 5.4096511 | 5.0243802 | 5.482174 | 4.489931 | 4.5215406 | 5.3944410 | 5.4802745 | 4.605243 | 4.794255 | 4.290394 | 5.3910110 | 5.4233124 | 5.4144371 | 5.174556 | 4.84336422 | 5.0361817 | 5.3908450 | 5.4802851 | 5.569842 | 4.566949 | 4.608240 | 4.271839 | 4.504909 | 4.180083 | 4.81104312 | 5.392958 | 5.4161402 | 5.3739146 | 5.4795929 | 5.501380 | 5.4595474 |
| GO:0035019 | somatic stem cell population maintenance | 30 | 0.3979800 | 1.489991 | 4.306402e-02 | 0.436023217 | 0.409152696 | 3902 | tags=50%, list=23%, signal=38% | MYC||SOX2||ZFP36L2||SKI||SOX9||LRP5||KLF10||CDX2||HES1||GATA2||SOX4||NOG||SIX2||BMP7||ZHX2 | 4.173648 | 4.5585289 | 4.4704103 | 4.523732 | 4.180579 | 4.5018240 | 4.5628527 | 4.5066494 | 4.135259 | 4.242205 | 4.140953 | 4.5644046 | 4.5661363 | 4.5449501 | 4.488585 | 4.48514799 | 4.4369213 | 4.5815840 | 4.4558000 | 4.531044 | 4.202090 | 4.082159 | 4.252234 | 4.425184 | 4.408868 | 4.65775035 | 4.580626 | 4.5389061 | 4.5687060 | 4.5064294 | 4.509854 | 4.5036578 |
| GO:0046928 | regulation of neurotransmitter secretion | 66 | 0.3304527 | 1.473206 | 4.306785e-02 | 0.436023217 | 0.409152696 | 2893 | tags=30%, list=17%, signal=25% | MCTP1||APBA1||SYT12||PPFIA2||RIMS4||GPER1||CPLX1||SNCAIP||VPS18||ADCY1||NCS1||DVL1||GIPC1||STXBP1||SEPTIN5||SNCG||SYT1||SYN1||CALM1||RAB3A | 4.409249 | 4.7695806 | 4.5916346 | 4.811097 | 4.458599 | 4.7004481 | 4.7660430 | 4.7855095 | 4.346950 | 4.388732 | 4.488383 | 4.7635189 | 4.7861930 | 4.7588816 | 4.562590 | 4.64340844 | 4.5674733 | 4.8206253 | 4.7717720 | 4.840038 | 4.474602 | 4.359890 | 4.535812 | 4.612774 | 4.737126 | 4.74758471 | 4.783559 | 4.7502654 | 4.7641105 | 4.7789284 | 4.798841 | 4.7786665 |
| GO:0070831 | basement membrane assembly | 14 | 0.5172823 | 1.551060 | 4.314646e-02 | 0.436034903 | 0.409163663 | 411 | tags=21%, list=2%, signal=21% | LAMB3||NTNG1||NTN4 | 3.476156 | 4.1734652 | 4.3113074 | 4.172356 | 3.501804 | 4.1039878 | 4.0734782 | 3.9658641 | 3.457023 | 3.391062 | 3.574374 | 4.1572831 | 4.2164917 | 4.1456153 | 4.356462 | 4.40934996 | 4.1559111 | 4.1633909 | 4.2047147 | 4.148372 | 3.457209 | 3.366913 | 3.664915 | 3.804343 | 3.805122 | 4.56305873 | 4.179737 | 4.0415929 | 3.9925141 | 3.9590609 | 4.018489 | 3.9182784 |
| GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 45 | 0.3502535 | 1.447094 | 4.381919e-02 | 0.438839022 | 0.411794974 | 1324 | tags=20%, list=8%, signal=18% | HMOX1||JAK2||NR4A3||MMP2||JUN||GPER1||PTEN||CDKN1A||MEF2D | 6.342760 | 5.9079028 | 6.3029256 | 5.929906 | 6.413908 | 6.4475253 | 5.8991763 | 5.9404081 | 6.288364 | 6.283830 | 6.449815 | 5.9165511 | 5.9131918 | 5.8938617 | 6.212988 | 6.36289898 | 6.3286496 | 5.9674872 | 5.8654204 | 5.954683 | 6.410586 | 6.327920 | 6.498191 | 6.379143 | 6.515700 | 6.44449847 | 5.916116 | 5.8865890 | 5.8946619 | 5.9351090 | 5.953645 | 5.9323773 |
| GO:0050996 | positive regulation of lipid catabolic process | 15 | 0.5046473 | 1.547957 | 4.404762e-02 | 0.438839022 | 0.411794974 | 5903 | tags=73%, list=35%, signal=48% | MTLN||PRKCE||PLIN5||AKT2||TWIST1||PRKCD||IRS2||ADORA1||PNPLA2||ABCD1||DAGLB | 4.134524 | 4.1060112 | 4.4068046 | 3.976068 | 4.163811 | 4.5200043 | 4.0192164 | 3.9523951 | 3.981529 | 4.048396 | 4.346872 | 4.1299853 | 4.1257128 | 4.0613148 | 4.335966 | 4.51103742 | 4.3673061 | 4.0731923 | 3.9113467 | 3.938403 | 4.167150 | 3.954331 | 4.343748 | 4.331539 | 4.559325 | 4.65071225 | 4.084699 | 3.9746829 | 3.9958908 | 3.9319042 | 4.002078 | 3.9218721 |
| GO:0030261 | chromosome condensation | 34 | 0.3848744 | 1.483364 | 4.433696e-02 | 0.440163585 | 0.413037909 | 3850 | tags=32%, list=23%, signal=25% | H1-4||PLK1||H1-10||GPER1||INCENP||NCAPH||CHMP1A||KMT5A||SMC4||BANF1||NCAPH2 | 6.187582 | 6.5143243 | 6.2835150 | 6.488725 | 6.179961 | 6.3950163 | 6.5604507 | 6.6221480 | 6.142787 | 6.186651 | 6.231930 | 6.5530522 | 6.4897605 | 6.4993503 | 6.354187 | 6.26784078 | 6.2255229 | 6.4608239 | 6.4488941 | 6.554138 | 6.179461 | 6.095541 | 6.260187 | 6.381081 | 6.389619 | 6.41414414 | 6.563930 | 6.5474165 | 6.5699110 | 6.5878721 | 6.659994 | 6.6176658 |
| GO:0003148 | outflow tract septum morphogenesis | 23 | 0.4414208 | 1.532595 | 4.465213e-02 | 0.441110155 | 0.413926145 | 2656 | tags=35%, list=16%, signal=29% | SEMA3C||FGF8||PARVA||NKX2-5||TBX2||GATA6||SMAD4||TBX1 | 2.775987 | 3.5832875 | 2.8822998 | 3.599091 | 2.763912 | 3.1234779 | 3.5488523 | 3.5837104 | 2.689948 | 2.736750 | 2.893316 | 3.5440787 | 3.6389842 | 3.5650681 | 2.986549 | 2.88274734 | 2.7694422 | 3.5468640 | 3.6284902 | 3.620522 | 2.802911 | 2.533547 | 2.927641 | 3.060540 | 3.201437 | 3.10483896 | 3.528860 | 3.5921054 | 3.5245960 | 3.5348212 | 3.619146 | 3.5958549 |
| GO:0010830 | regulation of myotube differentiation | 27 | 0.4146058 | 1.502467 | 4.469274e-02 | 0.441110155 | 0.413926145 | 3195 | tags=41%, list=19%, signal=33% | HDAC9||NKX2-5||NIBAN2||CAV3||PLPP7||RBM38||NOTCH1||HDAC5||TBX1||HDAC4||MAMSTR | 4.152368 | 4.4114872 | 4.4306157 | 4.306175 | 4.123688 | 4.3710717 | 4.3908591 | 4.3130239 | 4.108053 | 4.156746 | 4.191100 | 4.4160976 | 4.4325393 | 4.3854290 | 4.492917 | 4.42164282 | 4.3748303 | 4.3226549 | 4.2963999 | 4.299327 | 4.129633 | 3.991617 | 4.239186 | 4.166942 | 4.335085 | 4.58092823 | 4.434895 | 4.3935938 | 4.3426091 | 4.2948027 | 4.347232 | 4.2964187 |
| GO:0003203 | endocardial cushion morphogenesis | 31 | 0.3973022 | 1.495686 | 4.489164e-02 | 0.441473034 | 0.414266661 | 3882 | tags=52%, list=23%, signal=40% | ACVRL1||FGF8||TBX2||HEY2||SOX9||BMP2||SNAI2||SMAD4||NOTCH1||DCHS1||NOG||TBX3||HEYL||TWIST1||HEY1||BMP7 | 2.384188 | 3.6908078 | 2.7155302 | 3.702608 | 2.428304 | 2.6988431 | 3.6728094 | 3.6640593 | 2.372156 | 2.455568 | 2.321658 | 3.6411089 | 3.7386657 | 3.6910000 | 3.029665 | 2.50903430 | 2.5475604 | 3.6267899 | 3.7144489 | 3.763285 | 2.386337 | 2.447721 | 2.449955 | 2.622322 | 2.598767 | 2.86063341 | 3.658017 | 3.6842960 | 3.6759901 | 3.5963073 | 3.727727 | 3.6651507 |
| GO:0032526 | response to retinoic acid | 85 | 0.3024336 | 1.412184 | 4.503217e-02 | 0.441613348 | 0.414398328 | 4281 | tags=35%, list=26%, signal=26% | MEIOSIN||WNT8B||RXRG||PTGES||RARG||WNT10B||COL1A1||TWF2||OVCA2||WNT6||SOX9||RXRA||GATA6||IGFBP2||KLF4||WNT3A||SREBF1||CREB1||PTCH1||TBX1||TMEM161A||SLC10A3||TESC||AQP3||TEAD2||LTK||MYB||HTRA2||DNAAF2||SERPINF1 | 4.910250 | 4.8600729 | 4.8875156 | 4.802467 | 4.951254 | 4.9767523 | 4.7789848 | 4.7665240 | 4.910267 | 4.811037 | 5.003062 | 4.9342106 | 4.8519422 | 4.7904518 | 4.809668 | 4.95850584 | 4.8905309 | 4.8471135 | 4.7531358 | 4.805616 | 4.932286 | 4.871438 | 5.044656 | 4.965463 | 5.020847 | 4.94282574 | 4.812605 | 4.7620400 | 4.7617123 | 4.7588452 | 4.771370 | 4.7693260 |
| GO:0002042 | cell migration involved in sprouting angiogenesis | 39 | 0.3683929 | 1.461601 | 4.586759e-02 | 0.446693188 | 0.419165117 | 4324 | tags=44%, list=26%, signal=32% | HMOX1||HDAC9||AKT1||KLF4||ABL1||SLIT2||SRF||GATA2||CIB1||NOTCH1||HDAC5||HDAC7||CARD10||MAP2K5||EPHB4||MAP3K3||NR4A1 | 4.785927 | 4.8542097 | 4.8396844 | 4.903260 | 4.776082 | 4.9284506 | 4.8646141 | 4.9273115 | 4.729510 | 4.829706 | 4.796766 | 4.8335012 | 4.8761875 | 4.8526233 | 4.876331 | 4.80092353 | 4.8408132 | 4.8858690 | 4.8696830 | 4.952873 | 4.813738 | 4.683877 | 4.826364 | 4.887940 | 4.942626 | 4.95392654 | 4.856402 | 4.8572617 | 4.8800537 | 4.9097888 | 4.950064 | 4.9217850 |
| GO:0042104 | positive regulation of activated T cell proliferation | 14 | -0.6167393 | -1.569777 | 4.609553e-02 | 0.447105486 | 0.419552006 | 1757 | tags=29%, list=10%, signal=26% | CD24||AGER||GPAM||EPO | 8.898680 | 8.4989153 | 8.6176770 | 8.498169 | 8.897371 | 8.6538802 | 8.4597483 | 8.5142729 | 8.922152 | 8.863031 | 8.910182 | 8.5769252 | 8.4848546 | 8.4311850 | 8.468624 | 8.67770056 | 8.6958424 | 8.5666938 | 8.4518396 | 8.473372 | 8.932811 | 8.861404 | 8.897015 | 8.842835 | 8.645512 | 8.44606222 | 8.475569 | 8.4599957 | 8.4435025 | 8.5115459 | 8.484282 | 8.5463207 |
| GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 53 | 0.3350149 | 1.425248 | 4.649948e-02 | 0.447105486 | 0.419552006 | 3727 | tags=34%, list=22%, signal=26% | MVP||GPRC5A||MVB12A||SOS1||GPER1||MVB12B||DAB2IP||CHMP6||RNF126||WDR54||NEURL1||SHKBP1||EGFR||ZGPAT||NUP62||CBL||HIP1||CBLB | 4.490070 | 4.7239863 | 4.6055723 | 4.746347 | 4.511816 | 4.6750253 | 4.7691239 | 4.7759111 | 4.417404 | 4.470474 | 4.577686 | 4.7311354 | 4.7366057 | 4.7040071 | 4.680843 | 4.60347308 | 4.5283721 | 4.7215109 | 4.7243220 | 4.792096 | 4.507516 | 4.408807 | 4.611970 | 4.614496 | 4.697441 | 4.71125282 | 4.788763 | 4.7604612 | 4.7579445 | 4.7700211 | 4.808368 | 4.7487097 |
| GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 14 | 0.5117710 | 1.534534 | 4.696214e-02 | 0.447105486 | 0.419552006 | 2603 | tags=43%, list=16%, signal=36% | JAK2||PRR7||NLE1||BAX||GSN||LGALS9 | 5.973677 | 5.9117182 | 6.2059500 | 5.918359 | 5.898589 | 5.8966222 | 5.9010241 | 5.8695367 | 6.020586 | 6.064017 | 5.825406 | 5.9284334 | 5.8948696 | 5.9116566 | 6.191257 | 6.11927992 | 6.3014547 | 5.9810830 | 5.8367346 | 5.933530 | 5.819099 | 5.994802 | 5.876264 | 5.818229 | 5.888444 | 5.97869864 | 5.952170 | 5.8356731 | 5.9128061 | 5.8606231 | 5.864636 | 5.8832502 |
| GO:0007492 | endoderm development | 64 | 0.3264212 | 1.445264 | 4.703833e-02 | 0.447105486 | 0.419552006 | 2955 | tags=31%, list=18%, signal=26% | LAMB3||MMP2||FGF8||MESP1||SOX2||PAX9||SOX7||ZFP36L1||COL5A2||ARC||GATA6||DUSP2||CDC73||GATA4||SMAD4||BPTF||MED12||NOTCH1||NOG||LEO1 | 3.791565 | 4.6305215 | 4.3617434 | 4.732152 | 3.685761 | 4.0086042 | 4.6294795 | 4.7454182 | 3.800701 | 3.960422 | 3.589789 | 4.6477924 | 4.6462260 | 4.5969694 | 4.535895 | 4.20983771 | 4.3202349 | 4.7204939 | 4.7173425 | 4.758258 | 3.665585 | 3.782935 | 3.602942 | 3.924259 | 3.786945 | 4.27068750 | 4.647414 | 4.6271884 | 4.6136356 | 4.7222147 | 4.775310 | 4.7382148 |
| GO:0060037 | pharyngeal system development | 22 | 0.4395860 | 1.511536 | 4.705484e-02 | 0.447105486 | 0.419552006 | 3909 | tags=45%, list=23%, signal=35% | FGF8||PLXNA2||NKX2-5||GATA3||HES1||PTCH1||TBX1||NOG||BMP7||RIPPLY3 | 3.042231 | 3.5838019 | 3.1760454 | 3.573668 | 3.054499 | 3.3802488 | 3.5256726 | 3.5292626 | 2.989879 | 2.964134 | 3.164349 | 3.5656989 | 3.6234040 | 3.5614691 | 3.160932 | 3.23572712 | 3.1294006 | 3.6041328 | 3.5544292 | 3.561942 | 3.081411 | 2.840837 | 3.216483 | 3.268131 | 3.429798 | 3.43659593 | 3.551686 | 3.5185655 | 3.5063843 | 3.4855493 | 3.598644 | 3.5009679 |
| GO:0050873 | brown fat cell differentiation | 38 | 0.3651890 | 1.439433 | 4.725086e-02 | 0.447105486 | 0.419552006 | 3988 | tags=42%, list=24%, signal=32% | DIO2||LAMB3||CEBPB||PPARGC1A||RREB1||TFE3||TRPV4||SH2B2||FNDC5||ZBTB7B||ZNF516||HNRNPU||CEBPA||BMP7||PRDM16||METRNL | 3.811884 | 4.6016261 | 3.8898298 | 4.664268 | 3.705209 | 3.9504623 | 4.6665514 | 4.6777082 | 3.782021 | 3.930881 | 3.714160 | 4.5541982 | 4.6129262 | 4.6365133 | 4.248448 | 3.67367686 | 3.6671040 | 4.6118254 | 4.6418680 | 4.736173 | 3.726658 | 3.681500 | 3.707113 | 3.991147 | 3.978388 | 3.87927266 | 4.628168 | 4.6798640 | 4.6908491 | 4.6737191 | 4.695708 | 4.6635100 |
| GO:0021953 | central nervous system neuron differentiation | 126 | 0.2740889 | 1.356478 | 4.726891e-02 | 0.447105486 | 0.419552006 | 3234 | tags=28%, list=19%, signal=23% | MAP2||GRID2||FGF8||LBX1||MAPT||EPHB3||RAC3||HES5||PTEN||SUFU||NFIB||EPHB2||ZMIZ1||WNT3A||C12orf57||SPOCK1||TULP3||SLIT2||GLI3||NDNF||HES1||GATA2||PTCH1||DLX2||OGDH||NKX2-2||SHANK3||EMX1||NKX6-1||SOX4||FOXN4||ISL2||NIN||PLXNA4||TSKU | 5.707856 | 5.9123600 | 5.7409727 | 5.963898 | 5.651766 | 5.5424126 | 5.9228385 | 5.9910995 | 5.692403 | 5.760029 | 5.669596 | 5.9655847 | 5.8831371 | 5.8868460 | 5.803557 | 5.65868430 | 5.7569078 | 5.9637085 | 5.9261256 | 6.000891 | 5.698629 | 5.667434 | 5.586944 | 5.634923 | 5.439254 | 5.54642370 | 5.927446 | 5.9171093 | 5.9239412 | 6.0021611 | 5.975830 | 5.9951783 |
| GO:0051974 | negative regulation of telomerase activity | 10 | 0.5819440 | 1.571913 | 4.756727e-02 | 0.447105486 | 0.419552006 | 2749 | tags=60%, list=16%, signal=50% | TEN1||TERF1||TP53||CERS1||SRC||MEN1 | 4.825457 | 5.0552406 | 4.9042272 | 4.973260 | 4.904866 | 5.1394975 | 5.0013761 | 4.9162238 | 4.726479 | 4.787404 | 4.952876 | 5.0546601 | 5.0945759 | 5.0153994 | 4.840072 | 4.99342809 | 4.8746594 | 4.9757100 | 4.9161681 | 5.025815 | 4.905972 | 4.763739 | 5.032397 | 5.111173 | 5.156572 | 5.15032978 | 5.018429 | 4.9891522 | 4.9963857 | 4.8948028 | 4.952609 | 4.9005534 |
| GO:0050830 | defense response to Gram-positive bacterium | 28 | -0.5197415 | -1.537564 | 4.763208e-02 | 0.447105486 | 0.419552006 | 1215 | tags=18%, list=7%, signal=17% | MR1||RPL39||C5AR1||H2BC6||LYG1 | 7.962656 | 7.5357848 | 7.7283106 | 7.545887 | 7.870942 | 7.3083013 | 7.5235272 | 7.2196989 | 8.100271 | 8.075282 | 7.674098 | 7.5899173 | 7.4846997 | 7.5308066 | 7.763851 | 7.54548492 | 7.8580333 | 7.5398303 | 7.5758052 | 7.521494 | 7.799347 | 8.182693 | 7.561943 | 7.595598 | 7.208204 | 7.06818725 | 7.483292 | 7.5526698 | 7.5337309 | 7.5395655 | 6.050367 | 7.6061320 |
| GO:0031110 | regulation of microtubule polymerization or depolymerization | 81 | 0.3020933 | 1.402017 | 4.781705e-02 | 0.447105486 | 0.419552006 | 4051 | tags=38%, list=24%, signal=29% | MAP2||SLAIN2||GAS2L1||HSPA1A||HSPA1B||MAPT||TUBB4A||MAP1A||MAP1S||ARL2||CAV3||TRPV4||CAMSAP1||APC2||ABL1||MAPRE2||MAP1B||MID1||CIB1||ARHGEF2||CKAP2||KATNB1||SPEF1||RANGRF||NIN||MAPRE3||DCTN1||CAMSAP2||GIT1||MECP2||SKA3 | 6.976180 | 6.7944070 | 6.7337797 | 6.622423 | 7.027438 | 6.8246724 | 6.7533541 | 6.6246281 | 6.988510 | 6.975624 | 6.964303 | 6.8496404 | 6.7847168 | 6.7469895 | 6.646873 | 6.75767585 | 6.7928015 | 6.6688865 | 6.5715831 | 6.625155 | 7.084328 | 6.972914 | 7.022910 | 6.958705 | 6.825053 | 6.67647801 | 6.778548 | 6.7436566 | 6.7375177 | 6.6049732 | 6.627496 | 6.6411840 |
| GO:0060009 | Sertoli cell development | 10 | 0.5813838 | 1.570400 | 4.783909e-02 | 0.447105486 | 0.419552006 | 4455 | tags=60%, list=27%, signal=44% | SOX9||FLNA||SOX8||RAB13||NTRK1||SDC1 | 4.727033 | 4.9670327 | 4.9503567 | 5.005851 | 4.734373 | 5.0542333 | 4.9384835 | 4.9080177 | 4.720775 | 4.709141 | 4.750860 | 4.8782746 | 5.0910443 | 4.9229516 | 4.982045 | 4.98077676 | 4.8861673 | 4.9548099 | 5.0353401 | 5.026064 | 4.700390 | 4.665290 | 4.832032 | 4.907523 | 4.968519 | 5.26134497 | 4.932268 | 4.9554581 | 4.9275692 | 4.9254663 | 4.967715 | 4.8272968 |
| GO:0060391 | positive regulation of SMAD protein signal transduction | 17 | 0.4810971 | 1.529580 | 4.797508e-02 | 0.447105486 | 0.419552006 | 2489 | tags=35%, list=15%, signal=30% | JAK2||RBPMS||TGFB1||BMP2||SMAD4||DAB2 | 5.155384 | 5.6588937 | 5.5297829 | 5.717210 | 5.064662 | 5.4222293 | 5.6680395 | 5.7456581 | 5.135345 | 5.360697 | 4.939233 | 5.6303788 | 5.6806547 | 5.6651890 | 5.727526 | 5.34620167 | 5.4896751 | 5.7135641 | 5.6694188 | 5.766989 | 5.109992 | 5.115211 | 4.963718 | 5.556298 | 5.349035 | 5.35139107 | 5.683654 | 5.6469278 | 5.6732891 | 5.7127075 | 5.785918 | 5.7373844 |
| GO:0010942 | positive regulation of cell death | 447 | 0.1926744 | 1.110474 | 4.910035e-02 | 0.456082256 | 0.427975570 | 4324 | tags=33%, list=26%, signal=25% | HMOX1||CCL5||JAK2||UCP2||PRR7||DAPK2||HBA1||EEF1A2||FOXA1||HOXA13||MARK4||ID3||CALHM2||PITX3||NUPR1||HLA-G||KCNK2||MAPT||TPD52L1||RARG||PHLDA2||JUN||NGFR||BOK||AIFM2||SLC9A3R1||WNT10B||GPER1||PIAS4||NACC2||PPARGC1A||AKT1||PTEN||DUSP9||ELK1||MAP3K11||CDKN1A||CDC34||BCL6||PML||TFAP2A||DDIT4||DAB2IP||UBE2M||SLC27A4||BMP2||VDR||TSPO||BAD||H19||BAK1||TP53||TRADD||CTSD||PPP1CA||PIN1||EIF5A||GAL||MTCH1||MYBL2||FZD9||DLC1||MYBBP1A||OSGIN1||DAPK3||TRAF2||ZBTB16||CDKN2A||ENDOG||ABL1||RRP1B||TLE5||EIF4G1||TRAF7||BAX||LATS1||DEDD2||SLIT2||MTCO2P12||PTPA||NEURL1||LATS2||SPHK2||CREB1||ARRB1||GSN||NOTCH1||RIPK2||LGALS9||SOX4||SRC||PRDM11||DNM2||ARRB2||KATNB1||RNF122||CYP1B1||PRELID1||TP73||HTT||MAP3K9||HDAC4||SPRY1||CAPN2||INCA1||SIRT2||PLAGL2||PNMA3||BRMS1||PDCD5||IL12A||SCRIB||APBB1||FIS1||CDK4||RPS6KA2||AKAP12||ITM2C||BIN1||TFPT||RNPS1||MAGED1||TNFRSF1A||CCAR2||CSRNP3||BMP7||TP53BP2||LTK||PEA15||ZC3H12A||PRKCD||MYB||PLEKHF1||RACK1||BBC3||HTRA2||MAP3K10||PHLDA3||FYN||SARM1||E2F1||CDK5||PPP1R13B||DFFA||NTRK1||BACE1||NR4A1 | 7.184863 | 7.1219390 | 7.2356526 | 7.182995 | 7.190797 | 7.2284905 | 7.1435867 | 7.1667140 | 7.142987 | 7.195670 | 7.214974 | 7.0627472 | 7.1444109 | 7.1568589 | 7.165980 | 7.28287423 | 7.2555290 | 7.1682506 | 7.2305155 | 7.148955 | 7.196108 | 7.182235 | 7.194009 | 7.215360 | 7.191064 | 7.27765886 | 7.132172 | 7.1497004 | 7.1488199 | 7.1760502 | 7.153973 | 7.1700290 |
| GO:0051882 | mitochondrial depolarization | 16 | 0.4922651 | 1.539051 | 4.939024e-02 | 0.457118569 | 0.428948019 | 5333 | tags=56%, list=32%, signal=38% | BOK||TSPO||FZD9||ABL1||GCLM||SRC||RACK1||IFI6||ABCD1 | 8.709558 | 8.2623619 | 8.5218492 | 8.179725 | 8.748517 | 8.7588067 | 8.2285001 | 8.1581241 | 8.666478 | 8.661712 | 8.796396 | 8.3038673 | 8.2471990 | 8.2350810 | 8.368807 | 8.57978094 | 8.6054716 | 8.2417952 | 8.1177036 | 8.177004 | 8.750516 | 8.711584 | 8.782576 | 8.827026 | 8.858913 | 8.57393219 | 8.231494 | 8.2249701 | 8.2290283 | 8.1583513 | 8.159701 | 8.1563183 |
| GO:0071371 | cellular response to gonadotropin stimulus | 12 | 0.5460200 | 1.560733 | 4.944270e-02 | 0.457118569 | 0.428948019 | 2604 | tags=50%, list=16%, signal=42% | WT1||PPARGC1A||GATA6||EPHA8||NOTCH1||GCLM | 4.224635 | 4.9689532 | 4.5030997 | 5.205965 | 4.234616 | 4.5337518 | 5.0343360 | 5.2661162 | 4.162101 | 4.169081 | 4.335924 | 4.9870787 | 4.9622544 | 4.9573502 | 4.516201 | 4.55493570 | 4.4356063 | 5.1914649 | 5.1862912 | 5.239538 | 4.209205 | 4.098903 | 4.381583 | 4.383928 | 4.525091 | 4.67730860 | 5.066564 | 5.0157615 | 5.0201310 | 5.2595400 | 5.283650 | 5.2549935 |
| GO:0046635 | positive regulation of alpha-beta T cell activation | 42 | 0.3517196 | 1.422004 | 4.945554e-02 | 0.457118569 | 0.428948019 | 4699 | tags=60%, list=28%, signal=43% | FOXP3||AP3D1||RUNX1||ITPKB||NFKBIZ||ZBTB16||GLI3||RIPK2||LGALS9||CD81||NKAP||HLX||IHH||ZBTB7B||IL12A||PRKCZ||HLA-A||SOCS1||MYB||HLA-E||RARA||HSPH1||RUNX3||IL23A||ADA | 5.360228 | 5.4405358 | 5.4555998 | 5.373249 | 5.363841 | 5.5506390 | 5.3429646 | 5.3243197 | 5.309900 | 5.348992 | 5.419640 | 5.4770086 | 5.4554638 | 5.3876344 | 5.346574 | 5.51640806 | 5.4978742 | 5.3936796 | 5.3450595 | 5.380570 | 5.384325 | 5.278927 | 5.424385 | 5.486110 | 5.561227 | 5.60218704 | 5.354335 | 5.3303470 | 5.3441115 | 5.3157426 | 5.368357 | 5.2876940 |
| GO:0045956 | positive regulation of calcium ion-dependent exocytosis | 14 | 0.5083338 | 1.524228 | 4.960376e-02 | 0.457711165 | 0.429504096 | 5141 | tags=71%, list=31%, signal=50% | CDK5R2||STXBP1||SYT1||ARF1||SYT7||CACNA1G||CACNA1H||CDK5||HYAL3||ZP3 | 6.222425 | 5.8440083 | 6.2539230 | 5.601386 | 6.318570 | 6.5046661 | 5.7555790 | 5.6436023 | 6.149993 | 6.250982 | 6.263633 | 5.8867857 | 5.8486965 | 5.7950738 | 6.053923 | 6.39740044 | 6.2893851 | 5.7055469 | 5.4650238 | 5.623323 | 6.361767 | 6.167848 | 6.414571 | 6.538602 | 6.461302 | 6.51302262 | 5.774766 | 5.7394070 | 5.7523420 | 5.6333937 | 5.673649 | 5.6232708 |
| GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 73 | 0.3071710 | 1.397692 | 4.993679e-02 | 0.457711165 | 0.429504096 | 4683 | tags=45%, list=28%, signal=33% | FOXP3||LRRC32||HLA-G||CEBPB||AKT1||RUNX1||BCL6||KLF4||IL4I1||LAG3||PRDX2||GLI3||LGALS9||SOCS6||HLX||DLG5||IHH||ZBTB7B||ASS1||GNRH1||ERBB2||SCRIB||MAD1L1||WNK1||CBLB||SOCS1||ZC3H12A||MDK||CXCL12||PELI1||RUNX3||LOXL3||PRNP | 4.858572 | 5.2315678 | 4.8727480 | 5.136764 | 4.881437 | 5.0221652 | 5.2385941 | 5.1389396 | 4.833831 | 4.779891 | 4.956288 | 5.2390848 | 5.2335644 | 5.2220016 | 4.901198 | 4.90615527 | 4.8088231 | 5.1264488 | 5.1118319 | 5.171342 | 4.846652 | 4.819173 | 4.973730 | 4.941331 | 5.079173 | 5.04248429 | 5.246365 | 5.2336978 | 5.2356873 | 5.1680708 | 5.117000 | 5.1312653 |
GO: MF
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 345 | 0.2852191 | 1.583622 | 2.694735e-05 | 0.00919803 | 0.008991907 | 4125 | tags=38%, list=25%, signal=29% | MECOM||ESRRG||WT1||NR4A3||FOSL2||MESP1||SOX18||FOXA1||HOXA13||MYC||PITX3||NKX2-5||ETV2||NFIC||DLX3||JUND||JUN||SOX2||ELF4||MEIS2||ETV5||NKX2-8||ZNF574||PAX9||CEBPB||ELF1||DBP||RUNX1||CEBPD||ELK1||NFIB||GLIS2||ZNF580||PITX1||TFAP2A||SOX9||MEF2B||KLF10||MEF2D||RREB1||FOXK2||NKRF||TCF3||TFE3||RELA||ELK3||TP53||ZNF836||MAFB||ZNF668||ALX4||KLF4||HOXA10||GATA4||GLI1||FLI1||MYBL2||SREBF1||FOXC1||ZBTB16||CREB3L1||GLIS1||NFATC1||MAF||GMEB2||SOX12||MEIS3||KLF5||SRF||ZNF319||HOXC10||GATA3||SIX3||CASZ1||SMAD4||ZNF48||IRF2||CSRNP1||ETV4||HOXD3||JUNB||CREB1||GATA2||CREB3L2||ARNT2||DLX2||NKX2-2||STAT3||NFKB2||HAND1||SOX4||MAFA||SALL2||IRX6||ZBED1||FOXO4||RFXANK||ZNF615||TP73||NFATC2||FOSB||TBX3||LHX2||ZBTB7B||ZNF639||HOXB2||SIX2||PLAGL2||MAFG||NR1H3||HEYL||BARHL2||KLF7||ATF5||NFE2L2||ZNF629||SATB2||ESRRB||OSR2||IRF4||MLXIP||CEBPA||HOXB5||CSRNP3||FOXD1||PRDM16||HOXB7||TFDP1||MYB||ZNF395||MAFF||KLF15 | 3.687290 | 3.960871 | 3.902201 | 4.061836 | 3.685663 | 3.993177 | 4.015249 | 4.098594 | 3.642966 | 3.717742 | 3.700106 | 3.954923 | 3.961958 | 3.965712 | 3.988531 | 3.877364 | 3.836383 | 4.067658 | 3.998704 | 4.116717 | 3.683584 | 3.625827 | 3.745110 | 3.926223 | 3.964849 | 4.083749 | 4.029447 | 3.996738 | 4.019367 | 4.092369 | 4.109251 | 4.094103 |
| GO:0042043 | neurexin family protein binding | 12 | -0.8207189 | -2.021894 | 3.853831e-04 | 0.04932904 | 0.048223602 | 633 | tags=25%, list=4%, signal=24% | SYTL2||SYTL4||SYTL5 | 2.286307 | 3.989905 | 2.651083 | 3.965098 | 2.314745 | 2.366000 | 4.025362 | 4.010221 | 2.269519 | 2.436221 | 2.137611 | 3.905062 | 4.056273 | 4.004310 | 3.139721 | 2.387248 | 2.262744 | 3.897274 | 3.967072 | 4.027986 | 2.082798 | 2.573526 | 2.243852 | 2.140495 | 2.279791 | 2.632288 | 4.020314 | 4.082137 | 3.971500 | 4.019795 | 4.050904 | 3.958434 |
| GO:0003714 | transcription corepressor activity | 169 | 0.3282537 | 1.712532 | 5.153235e-04 | 0.05863237 | 0.057318442 | 3725 | tags=39%, list=22%, signal=31% | FOXP3||HDAC9||TRERF1||WTIP||TCP10L||HSPA1A||HIPK2||TLE6||CREBBP||JAZF1||BEND6||PIAS4||SCAI||IRF2BP1||KCTD1||SUFU||RERE||EID1||TFAP2A||TRIB3||SMARCA4||DDX54||HDGF||ZMYND8||PPP1R13L||PIAS1||MYBBP1A||NCOR2||ATN1||RUVBL2||TLE5||CITED2||TRIM28||SPEN||TLE3||DRAP1||MIER2||ARID5A||BCOR||NOC2L||PEX14||TOB2||TLE2||URI1||EID2||CBFA2T3||TOB1||KMT5A||ZFPM1||IRF2BP2||IRF2BPL||PARP10||HDAC4||HIRA||MIDEAS||TLE4||YAF2||HDAC7||TLE1||DPF2||NAB2||HSBP1L1||CTBP1||SFMBT2||HNRNPU||CBX4 | 5.773898 | 5.979136 | 5.856527 | 5.996211 | 5.741232 | 5.846632 | 6.024782 | 6.070197 | 5.746135 | 5.839934 | 5.733258 | 5.994260 | 5.978629 | 5.964363 | 5.905321 | 5.817175 | 5.845678 | 5.993007 | 5.953714 | 6.040599 | 5.733430 | 5.739557 | 5.750657 | 5.799752 | 5.823190 | 5.914393 | 6.026508 | 6.023684 | 6.024152 | 6.053681 | 6.087199 | 6.069516 |
| GO:0003682 | chromatin binding | 491 | 0.2298457 | 1.350349 | 1.008813e-03 | 0.10330243 | 0.100987474 | 4010 | tags=33%, list=24%, signal=26% | JDP2||FOSL2||TRIM24||FOXA1||CDT1||KLF14||NUPR1||ARID1B||ACTL6B||PHC3||NKX2-5||H1-4||PHF19||DLX3||RARG||HMGA1||EP300||THAP7||CREBBP||WBP2||SAMD1||BEND6||GRWD1||HSF1||H1-10||CEBPB||GPER1||PHC2||TAF10||PKN1||PPARGC1A||CITED1||WDR13||HES5||ELK1||RERE||NCOA6||BCL6||TFAP2A||HNRNPD||SMARCA4||SOX9||ZNF431||KDM4D||UPF1||LRWD1||ZIC2||ACTB||GATA6||SBNO2||GTF2F1||RELA||PCGF2||KDM6B||POLR2A||TP53||CENPB||BRD4||L3MBTL3||MBD3||KLF4||GLI1||FLI1||MBD5||SREBF1||FOXC1||NCOR2||SUV39H1||SHMT2||ARID1A||RUVBL2||CREB3L1||POLR1A||TSHZ3||EPOP||MEIS3||PELP1||GLI3||SRF||CITED2||ZNF609||TRIM28||NPM3||SMAD4||MBD6||VAX1||REPIN1||SPHK2||PATZ1||GATA2||KDM8||MED12||CDK9||NELFA||EGFR||DLX2||NOTCH1||NOC2L||CSNK2B||STAT3||NKX6-1||SAMD11||HCFC1||MEN1||PRDM11||FOXN4||EHMT2||TICRR||URI1||NCAPH||FOXO4||EP400||CABIN1||NKAP||SMARCE1||TOX2||THRA||SMC4||NFATC2||PHF21A||DHX30||CRAMP1||LHX2||PYGO2||NUP62||SMARCD2||GATAD2B||HDAC4||HIRA||TLE4||KLHDC3||SIRT2||MLH3||BRD2||HMGN1||USP51||ATF5||APBB1||HDAC7||L3MBTL2||NPM1||CTBP1||SFMBT2||SATB2||FUS||TNRC18||HNRNPU||ARID3C||CBX4||VAX2||BAHD1||CEBPA||NCAPH2||BAP1||RING1||SAFB||ZC3H12A||MECP2||ASF1A||ACTN4||RNF2||CHAF1A | 5.907516 | 6.420548 | 6.049802 | 6.440535 | 5.858539 | 5.982414 | 6.467769 | 6.488990 | 5.902139 | 5.993378 | 5.821926 | 6.422064 | 6.412772 | 6.426772 | 6.208375 | 5.927506 | 5.998542 | 6.418095 | 6.401714 | 6.499868 | 5.841648 | 5.881521 | 5.852151 | 5.948154 | 5.941722 | 6.054559 | 6.454127 | 6.462989 | 6.486002 | 6.490050 | 6.477566 | 6.499272 |
| GO:0017124 | SH3 domain binding | 112 | 0.3511311 | 1.722018 | 1.480174e-03 | 0.11902447 | 0.116357182 | 3892 | tags=40%, list=23%, signal=31% | SH2D2A||ELMO1||MVB12A||MAPT||SYNJ2||GPX1||RAPGEF1||SOS1||SH3BGRL||SH3KBP1||MICAL1||ARHGAP1||USP8||PLSCR3||SH3GL1||DAB2IP||ARHGAP31||CRB3||KHDRBS1||WAS||ENAH||BCAR1||SH3BP1||CTTNBP2||ABL1||WASF2||NCKIPSD||AFAP1||SHANK1||ABI2||SHANK3||ADAM9||CYBA||SIRPA||DNM2||DNAJC6||WIPF1||CBL||PLSCR4||CD2AP||DENND1A||CBLB||ARHGAP17||HCLS1||TP53BP2 | 4.843884 | 4.770646 | 4.800825 | 4.744989 | 4.887548 | 5.044504 | 4.742169 | 4.753456 | 4.811238 | 4.742841 | 4.968231 | 4.806946 | 4.765412 | 4.738760 | 4.770689 | 4.865261 | 4.764295 | 4.768106 | 4.681004 | 4.783748 | 4.864989 | 4.777970 | 5.010095 | 5.005617 | 5.151491 | 4.969923 | 4.769700 | 4.717702 | 4.738631 | 4.759773 | 4.760170 | 4.740335 |
| GO:0005200 | structural constituent of cytoskeleton | 72 | 0.4089320 | 1.853467 | 1.888107e-03 | 0.13810157 | 0.135006778 | 4292 | tags=40%, list=26%, signal=30% | NEFM||NEFH||BICD1||ACTL6B||TUBB4A||SORBS3||VIM||TUBB3||TUBG1||ARPC1B||ACTG1||TUBB4B||ACTB||TUBB2B||NEFL||INA||ANK1||TLN1||TLN2||TUBA1C||ADD2||TUBB2A||PLEC||CD2AP||HIP1||ARPC4||CCDC6||GFAP||TUBB | 7.830722 | 7.331066 | 7.614637 | 7.242591 | 7.896352 | 7.899965 | 7.292249 | 7.250015 | 7.747821 | 7.825591 | 7.913959 | 7.347869 | 7.335458 | 7.309606 | 7.395044 | 7.733382 | 7.692430 | 7.313460 | 7.149301 | 7.260184 | 7.972216 | 7.774297 | 7.934972 | 8.005066 | 7.850733 | 7.838031 | 7.296762 | 7.295155 | 7.284801 | 7.257715 | 7.257798 | 7.234408 |
| GO:0045296 | cadherin binding | 297 | 0.2549076 | 1.409953 | 2.484292e-03 | 0.15899469 | 0.155431688 | 3762 | tags=32%, list=22%, signal=26% | MMP24||CDH22||DBNL||GPRC5A||MICALL1||PARVA||PAK4||CDC42EP1||H3C12||FASN||HSPA1A||CNN2||FXYD5||CDH23||EEF2||SLC9A3R2||EMD||CAPG||EHD1||ALDOA||RPS2||PLIN3||S100A11||H1-10||ARHGAP1||USP8||TWF2||SH3GL1||ASAP1||NOTCH3||DAB2IP||NIBAN2||CORO1B||PFN1||SEPTIN9||JUP||PPP1CA||PUF60||MARK2||PPP1R13L||BAIAP2||CAST||EFHD2||GIPC1||BAG3||PFKP||DIAPH3||EPS15L1||WASF2||DBN1||CHMP2B||TBC1D2||TLN1||PAK6||RSL1D1||MPRIP||PKM||PDLIM1||RAB11B||UNC45A||EGFR||MYH9||TJP1||HCFC1||SRC||EIF2A||DCHS1||FSCN1||FLNA||RPL23A||KRT18||NUDC||TAGLN2||BAIAP2L1||PLEC||TNKS1BP1||KLC2||RANGAP1||CDH4||BSG||CBL||EEF1G||LARP1||LASP1||TBC1D10A||HSPA5||ADD1||SCRIB||STK24||USO1||CD2AP||ERC1||CHMP4B||PTPN1||PPME1||CAPZB | 8.162745 | 7.988948 | 7.856478 | 7.959071 | 8.179302 | 8.106965 | 7.982695 | 7.973273 | 8.148653 | 8.136010 | 8.202698 | 8.034264 | 7.967432 | 7.964056 | 7.789373 | 7.863751 | 7.913610 | 7.998744 | 7.897899 | 7.978608 | 8.176554 | 8.167749 | 8.193484 | 8.186074 | 8.238748 | 7.869080 | 7.977425 | 7.981471 | 7.989164 | 7.971442 | 7.968181 | 7.980170 |
| GO:0008022 | protein C-terminus binding | 178 | 0.2920919 | 1.537913 | 3.481438e-03 | 0.19805515 | 0.193616828 | 3662 | tags=36%, list=22%, signal=28% | JAK2||DBNL||VGLL2||OPRL1||SLC9A3R2||GRIP1||ATXN2||EP300||TAX1BP3||PIAS4||CSK||VPS4A||PABPC1||CITED1||SCAF4||ATP1B1||RABAC1||HRAS||SH3GL1||SDCBP2||CAV3||NEFL||MIF4GD||POLR2A||AP2A1||FIGN||ID1||BAIAP2||PIAS1||DAPK3||ZBTB16||PEX12||CDK7||ABL1||HIC2||SNX17||MYO1C||SHANK1||MED12||CIB1||DAB2||FBLN1||CORO1A||SHANK3||SYT1||SRC||RAB3A||NCL||SREBF2||SASH1||PICK1||CDC20||TNFAIP3||PPP1R9B||SIPA1||BANF1||MKI67||ERBB2||FOXN3||ERCC2||CD2AP||CTBP1||MAPRE3||KCNK3 | 5.554504 | 6.118523 | 5.626989 | 6.128317 | 5.540719 | 5.695126 | 6.140639 | 6.179039 | 5.515181 | 5.587445 | 5.559965 | 6.127328 | 6.115792 | 6.112405 | 5.755312 | 5.541939 | 5.574432 | 6.106231 | 6.096280 | 6.180949 | 5.531487 | 5.503222 | 5.586211 | 5.681126 | 5.726290 | 5.677448 | 6.127916 | 6.147385 | 6.146533 | 6.165373 | 6.200369 | 6.171131 |
| GO:0098918 | structural constituent of synapse | 16 | 0.6548801 | 2.046492 | 5.443000e-03 | 0.27868158 | 0.272436459 | 1639 | tags=56%, list=10%, signal=51% | NEFH||PPFIA2||DNM3||BSN||ACTG1||ACTB||NEFL||INA||ACTN1 | 8.725243 | 7.963423 | 8.057710 | 7.872062 | 8.752271 | 8.565139 | 7.900170 | 7.862679 | 8.676486 | 8.714452 | 8.782773 | 7.948085 | 7.975283 | 7.966767 | 7.754399 | 8.195487 | 8.181247 | 7.968327 | 7.758254 | 7.881913 | 8.864800 | 8.618552 | 8.762908 | 8.759566 | 8.529192 | 8.381270 | 7.893863 | 7.923098 | 7.883254 | 7.879984 | 7.864629 | 7.843186 |
| GO:0008013 | beta-catenin binding | 71 | 0.3841994 | 1.735546 | 6.670342e-03 | 0.32525857 | 0.317969678 | 3526 | tags=42%, list=21%, signal=33% | MED12L||SLC9A3R2||GRIP1||BCL9L||EP300||TAX1BP3||SLC9A3R1||CARM1||SUFU||SOX9||AXIN1||KDM6B||DVL1||PIN1||KLF4||PXN||APC2||RUVBL2||SALL1||SHROOM2||GLI3||MED12||TCF7L1||DVL3||FOXO4||DLG5||PTPRU||TCF7||CD2AP||SETD1A | 4.771901 | 4.989143 | 4.831002 | 5.058934 | 4.791083 | 4.969281 | 4.983631 | 5.058302 | 4.731929 | 4.747833 | 4.833843 | 5.005562 | 4.999787 | 4.961687 | 4.879504 | 4.810053 | 4.802186 | 5.064561 | 5.017164 | 5.094038 | 4.785732 | 4.708023 | 4.874669 | 4.935458 | 5.054644 | 4.913714 | 5.006861 | 4.976743 | 4.966988 | 5.058035 | 5.084059 | 5.032347 |
| GO:0005540 | hyaluronic acid binding | 13 | -0.7281478 | -1.825243 | 7.743534e-03 | 0.36042632 | 0.352349329 | 1205 | tags=31%, list=7%, signal=29% | HAPLN2||C1QBP||ACAN||HAPLN4 | 5.671847 | 6.057610 | 5.779523 | 6.254733 | 5.399473 | 4.921439 | 6.066267 | 6.278316 | 5.791223 | 6.037311 | 4.994789 | 6.158984 | 6.004153 | 6.004088 | 5.836061 | 5.509390 | 5.956853 | 6.232924 | 6.218354 | 6.311182 | 5.228090 | 5.814885 | 5.037926 | 4.901518 | 4.739129 | 5.100851 | 6.026090 | 6.059043 | 6.112351 | 6.243335 | 6.266005 | 6.324391 |
| GO:0031434 | mitogen-activated protein kinase kinase binding | 11 | 0.6724613 | 1.851904 | 8.495478e-03 | 0.37318173 | 0.364818898 | 1690 | tags=55%, list=10%, signal=49% | ACE||MAPK8IP1||MAP3K11||DAB2IP||TRIB3||PIN1 | 5.522235 | 5.022847 | 5.515088 | 4.985044 | 5.582064 | 5.728725 | 5.006487 | 4.939998 | 5.471775 | 5.420152 | 5.663255 | 5.055941 | 5.013341 | 4.998643 | 5.422695 | 5.594740 | 5.522672 | 5.052662 | 4.890469 | 5.007188 | 5.604831 | 5.424112 | 5.703473 | 5.630000 | 5.861942 | 5.683897 | 5.041903 | 4.986421 | 4.990472 | 4.961999 | 4.996936 | 4.857435 |
| GO:0003735 | structural constituent of ribosome | 155 | -0.3945508 | -1.506010 | 9.608118e-03 | 0.37318173 | 0.364818898 | 3780 | tags=35%, list=23%, signal=28% | RPL24||RPS8||MRPS15||RPL7||RPS23||MRPL17||RPL7A||MRPL42||RPL39L||RPS3A||RPS6||MRPS6||MRPS7||MRPL33||MRPL22||MRPL18||MRPL9||RPL22||RPL23||RSL24D1||MRPL51||DAP3||RPS25||MRPL13||RPL7L1||RPS17||RPL37||MRPS17||RPL22L1||RPS4X||RPL10A||RPL21||MRPL21||MRPL36||MRPL35||RPS27A||RPS20||RPL32||RPL27||RPL39||RPL4||RPS12||MRPS18C||RPL34||MRPS14||RPS27L||RPL35A||RPS7||RPL30||MRPL15||MRPS33||MRPL46||RPS15A||MRPS23||RPL9 | 10.935557 | 10.688269 | 10.538882 | 10.566723 | 10.919685 | 10.576383 | 10.663932 | 10.566342 | 10.991702 | 10.927286 | 10.885700 | 10.750258 | 10.641796 | 10.670553 | 10.475583 | 10.494816 | 10.640545 | 10.606965 | 10.536699 | 10.555584 | 10.908976 | 11.004589 | 10.840782 | 10.752806 | 10.664780 | 10.266545 | 10.653919 | 10.668985 | 10.668840 | 10.578059 | 10.496562 | 10.621625 |
| GO:0071889 | 14-3-3 protein binding | 29 | 0.5077757 | 1.882682 | 9.608278e-03 | 0.37318173 | 0.364818898 | 4425 | tags=62%, list=26%, signal=46% | ZFP36L1||AKT1||RPTOR||DDIT4||DAB2IP||BAD||RIPOR1||FOXK1||PRKCE||KLHL22||ARRB2||TBC1D22A||HDAC7||PRKCZ||ZFP36||NEK1||IRS2||SIK1 | 4.663970 | 4.970543 | 4.721096 | 4.968489 | 4.733235 | 4.959346 | 4.972256 | 4.989039 | 4.596010 | 4.624289 | 4.765813 | 4.993658 | 4.980656 | 4.936700 | 4.694168 | 4.776774 | 4.690690 | 5.008045 | 4.890516 | 5.003854 | 4.695128 | 4.619231 | 4.873422 | 4.897307 | 5.036877 | 4.940293 | 4.948363 | 4.986333 | 4.981774 | 4.966434 | 5.020736 | 4.979390 |
| GO:0035256 | G protein-coupled glutamate receptor binding | 12 | 0.6313540 | 1.794337 | 9.839753e-03 | 0.37318173 | 0.364818898 | 4683 | tags=75%, list=28%, signal=54% | HOMER3||DNM3||SLC9A3R2||SLC9A3R1||CALM1||CALM3||FYN||NECAB2||PRNP | 5.890072 | 5.867762 | 5.901243 | 5.905201 | 5.953899 | 6.158080 | 5.898542 | 5.892384 | 5.815215 | 5.866435 | 5.983382 | 5.844882 | 5.872180 | 5.885922 | 5.814670 | 5.971418 | 5.913311 | 5.963252 | 5.848139 | 5.901911 | 5.971544 | 5.845964 | 6.037662 | 6.108276 | 6.234799 | 6.127927 | 5.921830 | 5.869105 | 5.904194 | 5.874149 | 5.908714 | 5.894081 |
| GO:0016922 | nuclear receptor binding | 131 | 0.2941772 | 1.483852 | 1.027221e-02 | 0.37566953 | 0.367250952 | 2943 | tags=31%, list=18%, signal=25% | NR4A3||GAS2L1||TRIM24||FOXP2||TRERF1||PADI2||MED16||GRIP1||RARG||JUND||HMGA1||EP300||WBP2||CEBPB||TAF10||PKN1||PPARGC1A||MED25||NCOA6||ZBTB7A||TRIP6||SMARCA4||RXRA||VDR||PAGR1||BAZ2A||DDX54||TGFB1I1||NCOR2||ARID1A||LATS1||RNF6||ARRB1||MED12||ARID5A||TOB2||STAT3||SRC||FOXP1||SMARCE1 | 5.612464 | 6.117080 | 5.753129 | 6.022628 | 5.573718 | 5.708344 | 6.096415 | 6.034795 | 5.612862 | 5.675409 | 5.546231 | 6.132825 | 6.124317 | 6.093806 | 5.897057 | 5.650700 | 5.699674 | 6.017528 | 5.992120 | 6.057481 | 5.571791 | 5.587606 | 5.561638 | 5.731852 | 5.694582 | 5.698307 | 6.111059 | 6.080597 | 6.097427 | 6.024213 | 6.055867 | 6.024071 |
| GO:0005522 | profilin binding | 10 | 0.6741768 | 1.801025 | 1.125704e-02 | 0.39748981 | 0.388582247 | 5264 | tags=90%, list=31%, signal=62% | ACTG1||ENAH||DBN1||HTT||WIPF1||PCLO||VASP||EVL||RHOQ | 8.706519 | 7.931290 | 8.007100 | 7.865793 | 8.741020 | 8.512024 | 7.889631 | 7.877626 | 8.644297 | 8.692449 | 8.779540 | 7.907343 | 7.942448 | 7.943782 | 7.738144 | 8.127891 | 8.121799 | 7.933927 | 7.764448 | 7.893622 | 8.867346 | 8.581486 | 8.759902 | 8.734262 | 8.475215 | 8.292166 | 7.883770 | 7.906012 | 7.878964 | 7.881715 | 7.879083 | 7.872062 |
| GO:0046332 | SMAD binding | 73 | 0.3693951 | 1.681368 | 1.310542e-02 | 0.42068361 | 0.411256285 | 2880 | tags=34%, list=17%, signal=28% | ACVRL1||PMEPA1||HIPK2||USP15||JUN||ZC3H3||CITED1||COL5A2||SKI||ZBTB7A||PML||AXIN1||BMP2||ZMIZ1||GATA4||TGFB1I1||STUB1||CREB3L1||CITED2||SMAD4||DAB2||MEN1||FLNA||EID2||TOB1 | 4.950818 | 5.488887 | 5.227335 | 5.534876 | 4.931090 | 5.176134 | 5.504252 | 5.536461 | 4.938996 | 5.017139 | 4.893603 | 5.489542 | 5.495525 | 5.481559 | 5.426417 | 5.093160 | 5.139357 | 5.513829 | 5.506970 | 5.582609 | 4.925461 | 4.885062 | 4.981131 | 5.153374 | 5.122293 | 5.249659 | 5.510787 | 5.500948 | 5.500998 | 5.530237 | 5.549818 | 5.529235 |
| GO:0004879 | nuclear receptor activity | 41 | 0.4399311 | 1.766838 | 1.314636e-02 | 0.42068361 | 0.411256285 | 5026 | tags=56%, list=30%, signal=39% | ESRRG||NR4A3||RXRG||NR1D1||NR2F2||RARG||RXRA||VDR||SREBF1||ESRRA||NR2F6||NR2F1||STAT3||THRA||PPARD||NR1H3||AHR||ESRRB||NR4A1||RARA||NR1H2||NR4A2||THRB | 2.909626 | 3.284933 | 3.280782 | 3.345804 | 2.920309 | 3.381359 | 3.341469 | 3.353455 | 2.810122 | 2.896169 | 3.015210 | 3.263794 | 3.318563 | 3.271834 | 3.316380 | 3.338379 | 3.182719 | 3.350973 | 3.316517 | 3.369422 | 2.895404 | 2.745678 | 3.098063 | 3.240250 | 3.390144 | 3.501805 | 3.381518 | 3.339413 | 3.302389 | 3.336092 | 3.399439 | 3.323685 |
| GO:0048156 | tau protein binding | 44 | 0.4132168 | 1.702778 | 1.483813e-02 | 0.46043165 | 0.450113593 | 5650 | tags=55%, list=34%, signal=36% | MAP2||MARK4||MAP1A||EP300||ACTB||TTBK1||MARK2||PIN1||STUB1||MARK1||ROCK1||APBB1||BAG2||BIN1||DCTN1||FYN||CDK5||BRSK2||BRSK1||PPP2R2A||PPP5C||GSK3A||PPP2CA||PICALM | 7.031915 | 7.147887 | 6.884023 | 7.186557 | 7.014063 | 7.067167 | 7.167699 | 7.191382 | 6.994738 | 7.064154 | 7.036008 | 7.154844 | 7.139106 | 7.149666 | 6.911983 | 6.868347 | 6.871327 | 7.199674 | 7.123194 | 7.234565 | 7.070680 | 6.945506 | 7.023244 | 7.132897 | 7.061739 | 7.003971 | 7.163780 | 7.174861 | 7.164429 | 7.200442 | 7.194871 | 7.178745 |
| GO:0008093 | cytoskeletal anchor activity | 18 | 0.5492236 | 1.782661 | 1.529338e-02 | 0.46060072 | 0.450278870 | 2625 | tags=44%, list=16%, signal=38% | GAS2L1||BICD1||MAP1A||JUP||ANK1||BAIAP2||OBSL1||ABI2 | 3.743204 | 4.449683 | 3.841232 | 4.420463 | 3.791075 | 4.003869 | 4.493020 | 4.417719 | 3.660424 | 3.739505 | 3.824987 | 4.425201 | 4.464094 | 4.459442 | 3.969516 | 3.793946 | 3.750814 | 4.423383 | 4.342771 | 4.491405 | 3.744734 | 3.722238 | 3.899707 | 3.917592 | 3.989038 | 4.099153 | 4.545834 | 4.462113 | 4.469616 | 4.418531 | 4.452272 | 4.381486 |
| GO:0003707 | steroid hormone receptor activity | 20 | 0.5150245 | 1.724895 | 1.660281e-02 | 0.47974810 | 0.468997166 | 2187 | tags=40%, list=13%, signal=35% | ESRRG||NR4A3||RXRG||NR1D1||GPER1||RXRA||ESRRA||PAQR7 | 2.935224 | 3.580278 | 3.475244 | 3.736434 | 2.999123 | 3.449331 | 3.619349 | 3.779718 | 2.831893 | 2.964286 | 3.003923 | 3.572835 | 3.614480 | 3.552832 | 3.651527 | 3.449113 | 3.303843 | 3.655581 | 3.748995 | 3.800981 | 2.988174 | 2.823929 | 3.165083 | 3.287974 | 3.442898 | 3.600236 | 3.633893 | 3.634451 | 3.589237 | 3.760931 | 3.802532 | 3.775381 |
| GO:0003713 | transcription coactivator activity | 244 | 0.2360349 | 1.281304 | 1.686614e-02 | 0.47974810 | 0.468997166 | 3177 | tags=27%, list=19%, signal=22% | MED12L||UTF1||MAML2||VGLL2||TRIM24||RBPMS||TRERF1||NUPR1||ARID1B||ACTL6B||TRIM14||MED16||HIPK2||BCL9L||MPND||HMGA1||EP300||CREBBP||WBP2||CARM1||PKN1||MRTFA||PPARGC1A||CITED1||NFKBIB||MAML3||ELK1||RERE||NCOA6||NIBAN2||SMARCA4||JUP||PCBD1||AIP||SRCAP||ACTN1||ZMIZ1||BRD4||WNT3A||SUPT3H||TGFB1I1||ABL1||ARID1A||RRP1B||USP22||UBE2L3||CITED2||TRIM28||TRIM8||DRAP1||ARRB1||PDLIM1||MED12||NOTCH1||TAF6||HCFC1||CITED4||ZBED1||MED26||SMARCE1||DYRK1B||TOX2||TADA3||UBE3A||SMARCD2 | 5.838685 | 6.261872 | 5.950106 | 6.274736 | 5.804228 | 5.876241 | 6.312054 | 6.323572 | 5.843118 | 5.884658 | 5.786604 | 6.265559 | 6.246390 | 6.273534 | 6.092968 | 5.868069 | 5.877902 | 6.266982 | 6.229462 | 6.326115 | 5.807647 | 5.831574 | 5.772858 | 5.894506 | 5.797677 | 5.933184 | 6.310572 | 6.295452 | 6.329931 | 6.317964 | 6.323279 | 6.329448 |
| GO:0015106 | bicarbonate transmembrane transporter activity | 12 | -0.6995033 | -1.723271 | 1.830877e-02 | 0.50670748 | 0.495352402 | 3505 | tags=67%, list=21%, signal=53% | SLC39A8||SLC26A7||BEST1||SLC4A5||SLC4A7||SLC26A2||SLC4A8||SLC26A10 | 1.498586 | 2.913346 | 2.820418 | 3.122480 | 1.543687 | 2.330131 | 2.957148 | 3.060793 | 1.351273 | 1.496778 | 1.633887 | 2.860761 | 2.966393 | 2.910947 | 2.996669 | 2.796857 | 2.646214 | 3.069124 | 3.170219 | 3.126317 | 1.455900 | 1.402701 | 1.747997 | 1.762158 | 2.001519 | 2.942847 | 2.994997 | 2.939561 | 2.936126 | 3.010028 | 3.175476 | 2.989584 |
| GO:0051015 | actin filament binding | 169 | 0.2670676 | 1.393318 | 2.053322e-02 | 0.55331613 | 0.540916574 | 3869 | tags=36%, list=23%, signal=28% | DBNL||GAS2L1||NEBL||FLNC||EEF2||CAPG||TMEM201||ABLIM2||MAP1S||MICAL1||MARCKSL1||TWF2||MACO1||ESPNL||SHROOM1||ARPC1B||MYO1F||CORO1B||COTL1||TRPV4||DMTN||ACTN1||MYO1D||TRIOBP||ABL1||FMNL1||PKNOX2||SHROOM2||DBN1||MYO18A||TLN1||MPRIP||MYO1C||TLN2||CFL1||GSN||EGFR||CORO1A||MYH9||FMNL3||CORO7||FSCN1||FLNA||FERMT1||ADD2||UTRN||PICK1||MYO6||MARCKS||FRG1||PPP1R9B||LASP1||ADD1||HIP1||BIN1||MYH14||CAPZB||ARPC4||HCLS1||WDR1 | 5.676947 | 5.736575 | 5.621480 | 5.704435 | 5.710737 | 5.922433 | 5.726257 | 5.715220 | 5.612048 | 5.671326 | 5.744417 | 5.723185 | 5.757221 | 5.729090 | 5.562741 | 5.687136 | 5.611835 | 5.744171 | 5.641528 | 5.725548 | 5.740445 | 5.593985 | 5.790621 | 5.937300 | 5.991942 | 5.833599 | 5.737878 | 5.720030 | 5.720791 | 5.706954 | 5.736188 | 5.702284 |
| GO:0070742 | C2H2 zinc finger domain binding | 13 | 0.5805407 | 1.706220 | 2.143865e-02 | 0.55423404 | 0.541813907 | 5118 | tags=69%, list=31%, signal=48% | WT1||THAP7||U2AF2||GATA2||EHMT2||ZXDC||EHMT1||MBD2||LEF1 | 6.099409 | 6.063031 | 6.421212 | 6.071550 | 6.134384 | 6.499118 | 6.153853 | 6.152814 | 6.052679 | 5.976892 | 6.254235 | 6.016231 | 6.106443 | 6.065005 | 6.423411 | 6.512609 | 6.321275 | 6.093925 | 6.042966 | 6.077292 | 6.045341 | 5.992214 | 6.340649 | 6.310611 | 6.479369 | 6.683185 | 6.217064 | 6.133926 | 6.108316 | 6.122988 | 6.228607 | 6.103689 |
| GO:0051428 | peptide hormone receptor binding | 11 | 0.6229610 | 1.715584 | 2.164977e-02 | 0.55423404 | 0.541813907 | 4130 | tags=55%, list=25%, signal=41% | JAK2||GPHA2||GNAO1||GNAS||GNRH1||FYN | 6.562776 | 6.892117 | 6.519432 | 6.608199 | 6.552567 | 6.795062 | 6.863396 | 6.607590 | 6.477167 | 6.607435 | 6.600052 | 6.904788 | 6.903185 | 6.868080 | 6.571743 | 6.494381 | 6.490711 | 6.645459 | 6.534259 | 6.642129 | 6.545854 | 6.476921 | 6.630800 | 6.820391 | 6.895935 | 6.658790 | 6.868802 | 6.880838 | 6.840248 | 6.618794 | 6.606404 | 6.597492 |
| GO:0008106 | alcohol dehydrogenase (NADP+) activity | 12 | 0.5861879 | 1.665973 | 2.473995e-02 | 0.55481308 | 0.542379970 | 4901 | tags=58%, list=29%, signal=41% | ALDH3A1||AKR7A2||DHRS13||RDH13||RDH12||DHRS4||AKR1A1 | 6.266329 | 6.100807 | 6.160346 | 5.949283 | 6.299841 | 6.301873 | 6.021519 | 5.885666 | 6.211918 | 6.216178 | 6.365527 | 6.130368 | 6.120002 | 6.050758 | 6.031201 | 6.240712 | 6.200665 | 5.990534 | 5.905673 | 5.950393 | 6.317140 | 6.215032 | 6.363384 | 6.374805 | 6.325199 | 6.200039 | 6.041005 | 6.020358 | 6.002942 | 5.871654 | 5.904327 | 5.880819 |
| GO:0005080 | protein kinase C binding | 45 | 0.3822651 | 1.585758 | 2.489814e-02 | 0.55481308 | 0.542379970 | 3605 | tags=40%, list=22%, signal=31% | HDAC9||PARD6A||HSPB1||PKN1||AKT1||TWF2||KIAA1614||TRPV4||ABL1||HDAC5||ADAM9||SRC||FLNA||PICK1||MARCKS||HDAC7||TIRAP||PRKCSH | 7.614964 | 7.386447 | 7.496521 | 7.332697 | 7.634451 | 7.601159 | 7.366487 | 7.343825 | 7.600100 | 7.605618 | 7.638869 | 7.428252 | 7.379103 | 7.350920 | 7.354965 | 7.532938 | 7.591292 | 7.364476 | 7.283503 | 7.348840 | 7.629971 | 7.633609 | 7.639755 | 7.639371 | 7.644093 | 7.516416 | 7.362898 | 7.368209 | 7.368349 | 7.329836 | 7.348533 | 7.353001 |
| GO:0008138 | protein tyrosine/serine/threonine phosphatase activity | 39 | 0.4101752 | 1.626655 | 2.503237e-02 | 0.55481308 | 0.542379970 | 1900 | tags=26%, list=11%, signal=23% | PTP4A3||DUSP23||PPM1F||SBF1||DUSP15||PTEN||DUSP9||PTP4A1||DUSP2||DUSP8 | 3.984270 | 4.582722 | 4.094752 | 4.601463 | 3.966195 | 4.090847 | 4.646388 | 4.651498 | 4.019387 | 4.007886 | 3.923656 | 4.546367 | 4.609281 | 4.591789 | 4.269374 | 4.005657 | 3.991944 | 4.540545 | 4.608973 | 4.652665 | 3.949961 | 3.941115 | 4.006631 | 4.051291 | 4.063887 | 4.155121 | 4.629638 | 4.635720 | 4.673416 | 4.640418 | 4.654798 | 4.659212 |
| GO:0035497 | cAMP response element binding | 15 | 0.5465071 | 1.670634 | 2.606635e-02 | 0.55481308 | 0.542379970 | 2514 | tags=33%, list=15%, signal=28% | NR4A3||JUN||CREB3L1||CREB1||CREB3L2 | 4.022500 | 4.654279 | 4.515562 | 4.679874 | 4.119378 | 4.579117 | 4.615039 | 4.590156 | 3.950841 | 4.055033 | 4.059040 | 4.623895 | 4.688733 | 4.649470 | 4.672642 | 4.458821 | 4.400806 | 4.682344 | 4.627036 | 4.728457 | 4.152633 | 3.980893 | 4.214572 | 4.414105 | 4.507302 | 4.789055 | 4.628941 | 4.621893 | 4.594049 | 4.595653 | 4.599166 | 4.575537 |
| GO:0035591 | signaling adaptor activity | 69 | 0.3337942 | 1.497813 | 2.654867e-02 | 0.55481308 | 0.542379970 | 3144 | tags=30%, list=19%, signal=25% | GNB2||SHC1||GRIP1||MAPK8IP1||MAP2K2||DAB2IP||AXIN1||KHDRBS1||TRADD||DEDD2||GAB2||SHANK1||MAPK8IP2||ABI2||SHANK3||STAT3||SH2B2||SPATA2||MAVS||DLG5||NUP62 | 6.693436 | 6.372399 | 6.536196 | 6.292966 | 6.747338 | 6.798115 | 6.359034 | 6.292569 | 6.636116 | 6.629741 | 6.807327 | 6.390831 | 6.371904 | 6.354229 | 6.360336 | 6.612926 | 6.620444 | 6.343659 | 6.229456 | 6.303466 | 6.754792 | 6.681485 | 6.803144 | 6.847330 | 6.888958 | 6.646602 | 6.362598 | 6.362677 | 6.351801 | 6.285420 | 6.298771 | 6.293483 |
| GO:0016780 | phosphotransferase activity, for other substituted phosphate groups | 20 | -0.5861077 | -1.619726 | 3.056769e-02 | 0.61011105 | 0.596438741 | 2915 | tags=45%, list=17%, signal=37% | SAMD8||GNPTAB||PIGF||CHPT1||SGMS2||CRLS1||PIGN||SELENOI||CEPT1 | 4.492082 | 5.354306 | 5.169926 | 5.475501 | 4.514812 | 4.921592 | 5.280680 | 5.420943 | 4.440013 | 4.530688 | 4.504045 | 5.360360 | 5.390460 | 5.310985 | 5.212572 | 5.170597 | 5.125289 | 5.435313 | 5.518599 | 5.471380 | 4.507812 | 4.474283 | 4.561012 | 4.685295 | 4.718318 | 5.280552 | 5.302872 | 5.285322 | 5.253412 | 5.421500 | 5.455649 | 5.384810 |
| GO:0001221 | transcription coregulator binding | 77 | 0.3183834 | 1.464128 | 3.098220e-02 | 0.61011105 | 0.596438741 | 4646 | tags=44%, list=28%, signal=32% | TRERF1||NR1D1||EP300||THAP7||CREBBP||WIZ||MED25||ZBTB7A||SMARCA4||RELA||HDGF||KLF4||BAIAP2||ZBTB16||NFATC1||SIX3||SMAD4||CREB1||CDK9||HAND1||EHMT2||ZNF618||PPARD||NEK6||AHR||NFE2L2||CTBP1||TEAD2||MAFK||PHF12||USP11||HIF1A||EHMT1||PER2 | 5.616869 | 5.903016 | 5.680583 | 5.985053 | 5.522391 | 5.528661 | 5.976646 | 6.020160 | 5.647503 | 5.758603 | 5.424733 | 5.878013 | 5.902612 | 5.927992 | 5.784583 | 5.547405 | 5.699830 | 5.959458 | 5.969152 | 6.025659 | 5.427609 | 5.719163 | 5.398099 | 5.458656 | 5.509468 | 5.613517 | 5.973214 | 5.963728 | 5.992843 | 6.017978 | 6.025510 | 6.016977 |
| GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | 255 | 0.2348108 | 1.276392 | 3.183958e-02 | 0.61516470 | 0.601379145 | 3057 | tags=28%, list=18%, signal=23% | FOXP3||MSC||ZBTB2||JDP2||FOXP2||MYC||DACH1||CUX2||NR1D1||ZBTB45||MYPOP||JUN||NFE2L3||KLF16||HSF1||ZNF219||CEBPB||ZNF688||NACC2||DEAF1||NACC1||HES5||ZNF205||DMBX1||TBX2||SKI||HEY2||BCL6||ZBTB7A||ZBTB12||TFAP2A||CDX2||FOXK2||HES6||TCF3||RELA||ELK3||ZFHX3||FOXK1||ZNF668||BHLHE40||ZNF282||GSC||ZBTB16||FOXP4||AEBP2||BCL11A||HIC2||CREB3L1||HOXB13||ESX1||SNAI2||NR2F6||KCNIP3||SALL1||BATF3||ERF||GATA3||VAX1||THAP1||PATZ1||HES1||NR2F1||NKX6-1||HAND1||FOXP1||GTF2IRD1||SREBF2||PPARD||NFATC2||ZGPAT||TBX3 | 4.417988 | 4.354156 | 4.547408 | 4.365057 | 4.444713 | 4.633060 | 4.347157 | 4.366174 | 4.347900 | 4.381820 | 4.518534 | 4.378259 | 4.353024 | 4.330792 | 4.517210 | 4.592762 | 4.531126 | 4.399945 | 4.298589 | 4.394403 | 4.491274 | 4.334005 | 4.502777 | 4.575013 | 4.606362 | 4.714089 | 4.354036 | 4.334668 | 4.352686 | 4.352943 | 4.386231 | 4.359130 |
| GO:0070300 | phosphatidic acid binding | 17 | 0.5047667 | 1.605325 | 3.442976e-02 | 0.64101948 | 0.626654527 | 4063 | tags=47%, list=24%, signal=36% | MICALL1||NRGN||ATP13A2||MARK1||PITPNC1||PITPNM1||PLTP||UQCC3 | 4.203170 | 4.545338 | 4.255715 | 4.646036 | 4.214350 | 4.458214 | 4.484781 | 4.566467 | 4.187770 | 4.132874 | 4.284750 | 4.533295 | 4.564510 | 4.538011 | 4.261531 | 4.284688 | 4.220187 | 4.672116 | 4.599183 | 4.665684 | 4.202505 | 4.146893 | 4.290030 | 4.406186 | 4.530678 | 4.434811 | 4.529214 | 4.447927 | 4.476017 | 4.550458 | 4.600424 | 4.547910 |
| GO:0004865 | protein serine/threonine phosphatase inhibitor activity | 11 | 0.5777139 | 1.590977 | 3.809263e-02 | 0.67290386 | 0.657824393 | 1559 | tags=45%, list=9%, signal=41% | MYOZ1||PPP1R2||PPP1R14A||PPP1R1A||PPP1R14B | 6.100188 | 5.690545 | 5.703406 | 5.560957 | 6.019293 | 6.014542 | 5.686175 | 5.580309 | 6.061414 | 6.083037 | 6.154461 | 5.768544 | 5.619287 | 5.679887 | 5.643877 | 5.737881 | 5.726644 | 5.620434 | 5.484635 | 5.574512 | 5.998668 | 5.910980 | 6.139011 | 5.996220 | 6.217910 | 5.799022 | 5.730170 | 5.642333 | 5.684683 | 5.596975 | 5.606914 | 5.536021 |
| GO:0003678 | DNA helicase activity | 69 | -0.4243151 | -1.473347 | 4.002375e-02 | 0.68037644 | 0.665129515 | 5224 | tags=49%, list=31%, signal=34% | MCM3||DSCC1||RFC3||SUPV3L1||PIF1||DHX36||BRIP1||RUVBL1||MCM9||HFM1||BLM||WRN||DDX11||MCM8||CHD6||MRE11||RFC2||FANCM||ERCC3||CHD1||HELB||XRCC5||POLQ||DHX9||DNA2||RAD51||XRCC6||CHD1L||MCM6||G3BP1||RFC5||RAD54B||SETX||ASCC3 | 5.606440 | 6.153593 | 5.952037 | 6.247922 | 5.523164 | 5.681108 | 6.211893 | 6.338619 | 5.593290 | 5.690022 | 5.531572 | 6.153681 | 6.160431 | 6.146633 | 6.141674 | 5.806698 | 5.886149 | 6.212123 | 6.214604 | 6.314649 | 5.516757 | 5.555384 | 5.496732 | 5.611749 | 5.586286 | 5.832385 | 6.219637 | 6.201356 | 6.214625 | 6.318342 | 6.360093 | 6.337120 |
| GO:0003953 | NAD+ nucleosidase activity | 10 | 0.5956559 | 1.591261 | 4.154382e-02 | 0.68037644 | 0.665129515 | 6614 | tags=90%, list=40%, signal=54% | TLR5||TICAM1||SIGIRR||TIRAP||SARM1||IL18R1||IL1RAP||PIK3AP1||MYD88 | 3.194837 | 2.530099 | 3.070860 | 2.220060 | 3.311292 | 3.298634 | 2.378178 | 2.094408 | 3.162672 | 2.941548 | 3.437367 | 2.554926 | 2.577292 | 2.455171 | 2.846664 | 3.252286 | 3.085105 | 2.395573 | 2.118557 | 2.128600 | 3.297595 | 3.123554 | 3.489486 | 3.160477 | 3.456151 | 3.263521 | 2.415030 | 2.354434 | 2.364336 | 2.090323 | 2.057302 | 2.134557 |
| GO:0005154 | epidermal growth factor receptor binding | 25 | 0.4288501 | 1.525702 | 4.217497e-02 | 0.68037644 | 0.665129515 | 1327 | tags=20%, list=8%, signal=18% | ERBB4||EFEMP1||SHC1||ATXN2||RNF126 | 4.265657 | 5.142523 | 4.457700 | 4.979190 | 4.207554 | 4.352533 | 5.182659 | 5.040409 | 4.271965 | 4.382183 | 4.131967 | 5.157097 | 5.124816 | 5.145470 | 4.618687 | 4.343483 | 4.395895 | 4.933037 | 4.949597 | 5.052034 | 4.199954 | 4.199057 | 4.223518 | 4.327967 | 4.240645 | 4.478854 | 5.176776 | 5.175217 | 5.195891 | 5.045650 | 5.050737 | 5.024708 |
| GO:0030552 | cAMP binding | 18 | 0.4761770 | 1.545567 | 4.431960e-02 | 0.68260093 | 0.667304162 | 2032 | tags=28%, list=12%, signal=24% | RAPGEF3||PRKAR1B||HCN2||POPDC3||HCN4 | 2.411006 | 3.783064 | 2.839651 | 3.686283 | 2.434806 | 2.766468 | 3.797584 | 3.713458 | 2.398082 | 2.490793 | 2.340123 | 3.750074 | 3.792434 | 3.806095 | 3.109223 | 2.681347 | 2.685195 | 3.632419 | 3.675710 | 3.748336 | 2.383512 | 2.454724 | 2.464829 | 2.645987 | 2.619482 | 3.001564 | 3.763192 | 3.807310 | 3.821608 | 3.674741 | 3.742692 | 3.722104 |
| GO:0005338 | nucleotide-sugar transmembrane transporter activity | 11 | -0.6567299 | -1.578015 | 4.611994e-02 | 0.68260093 | 0.667304162 | 3265 | tags=73%, list=20%, signal=59% | SLC35B1||SLC35A1||SLC35E3||SLC35A5||SLC35A3||SLC35B4||SLC35D2||SLC35D1 | 4.354515 | 5.070694 | 4.817417 | 5.035604 | 4.415612 | 4.633562 | 4.992551 | 4.901742 | 4.262632 | 4.440712 | 4.354711 | 5.056634 | 5.127238 | 5.026344 | 4.831559 | 4.871641 | 4.746222 | 5.010397 | 5.033409 | 5.062532 | 4.459630 | 4.337905 | 4.446241 | 4.555426 | 4.439322 | 4.870741 | 4.996914 | 5.009642 | 4.970825 | 4.889651 | 4.935122 | 4.879848 |
| GO:0043425 | bHLH transcription factor binding | 23 | 0.4376439 | 1.523457 | 4.622132e-02 | 0.68260093 | 0.667304162 | 4091 | tags=39%, list=24%, signal=30% | CREBBP||SOX9||TCF3||BHLHE40||USF2||KLF5||HAND1||TWIST1||MAP3K10 | 4.499157 | 4.702176 | 4.662708 | 4.667696 | 4.508109 | 4.720761 | 4.710711 | 4.734687 | 4.479659 | 4.459947 | 4.556069 | 4.684498 | 4.718666 | 4.703160 | 4.744070 | 4.614452 | 4.626003 | 4.710191 | 4.588710 | 4.701043 | 4.511789 | 4.439397 | 4.570172 | 4.715756 | 4.716148 | 4.730331 | 4.730498 | 4.692021 | 4.709357 | 4.723821 | 4.765597 | 4.714124 |
| GO:0098631 | cell adhesion mediator activity | 43 | 0.3583714 | 1.465106 | 4.674978e-02 | 0.68260093 | 0.667304162 | 3218 | tags=42%, list=19%, signal=34% | NTNG1||PAK4||CDC42EP1||EMILIN1||S100A11||IGSF9||JUP||PPP1CA||BAIAP2||VSTM2L||NINJ1||PALLD||PDLIM1||SIRPA||KRT18||BAIAP2L1||MADCAM1||BSG | 7.665182 | 7.105145 | 7.467500 | 7.038711 | 7.741318 | 7.566812 | 7.003833 | 7.043825 | 7.631884 | 7.554782 | 7.798067 | 7.227190 | 7.051539 | 7.028405 | 7.267839 | 7.562872 | 7.552794 | 7.086334 | 6.989580 | 7.038596 | 7.753275 | 7.660180 | 7.806705 | 7.645795 | 7.553566 | 7.497156 | 7.008666 | 6.997436 | 7.005374 | 7.044532 | 7.029778 | 7.057035 |
| GO:0042056 | chemoattractant activity | 15 | 0.5024174 | 1.535854 | 4.709716e-02 | 0.68260093 | 0.667304162 | 853 | tags=27%, list=5%, signal=25% | CCL5||FGF8||MIF||VEGFB | 8.582199 | 8.083155 | 8.135508 | 7.922472 | 8.647181 | 8.719086 | 8.001627 | 7.846047 | 8.562364 | 8.345290 | 8.802645 | 8.212614 | 7.992319 | 8.034949 | 7.936519 | 8.290855 | 8.157200 | 8.080648 | 7.827241 | 7.845433 | 8.649325 | 8.408758 | 8.849869 | 8.615881 | 8.989823 | 8.505872 | 7.994791 | 7.996838 | 8.013183 | 7.876337 | 7.816242 | 7.844934 |
| GO:0016208 | AMP binding | 13 | 0.5325586 | 1.565200 | 4.710860e-02 | 0.68260093 | 0.667304162 | 2411 | tags=46%, list=14%, signal=40% | H1-4||APRT||CYB5R3||PFKL||PFKP||ACSS1 | 6.898484 | 6.568062 | 6.750460 | 6.434695 | 6.987522 | 7.105836 | 6.512773 | 6.430491 | 6.836023 | 6.752831 | 7.085493 | 6.616256 | 6.541067 | 6.545623 | 6.594398 | 6.869678 | 6.773904 | 6.531484 | 6.349840 | 6.416885 | 6.986883 | 6.856422 | 7.108283 | 7.041601 | 7.249981 | 7.014223 | 6.557136 | 6.473166 | 6.506777 | 6.424057 | 6.495364 | 6.369276 |
| GO:0031683 | G-protein beta/gamma-subunit complex binding | 14 | 0.5161813 | 1.548656 | 4.837316e-02 | 0.68260093 | 0.667304162 | 3272 | tags=36%, list=20%, signal=29% | GNAL||GNAO1||GNA11||GNAS||GNA12 | 6.113866 | 6.485529 | 6.056109 | 6.255466 | 6.130329 | 6.357780 | 6.449888 | 6.236429 | 6.023194 | 6.142325 | 6.171825 | 6.490710 | 6.505514 | 6.459989 | 6.070172 | 6.095570 | 6.000929 | 6.277672 | 6.195524 | 6.291351 | 6.128842 | 6.030287 | 6.225273 | 6.371253 | 6.455352 | 6.238500 | 6.451135 | 6.466907 | 6.431402 | 6.227732 | 6.239275 | 6.242238 |
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | 98 | 0.2786077 | 1.342618 | 4.983660e-02 | 0.68260093 | 0.667304162 | 4130 | tags=33%, list=25%, signal=25% | ERBB4||JAK2||EPHA5||EFEMP1||MATK||EPHB3||CSK||MAP2K2||ERBB3||MAP2K7||MAP2K3||EPHB2||ABL1||DDR1||EPHA8||EGFR||RIPK2||IGF1R||SRC||DYRK1B||BTK||TYRO3||EIF2AK2||ERBB2||FLT4||MAP2K5||LTK||EPHB4||NTRK2||PRKCD||EFNB3||FYN | 3.712364 | 4.172040 | 4.060006 | 4.116234 | 3.710856 | 4.094313 | 4.186893 | 4.168269 | 3.628242 | 3.680300 | 3.821552 | 4.137189 | 4.218212 | 4.159500 | 4.133052 | 4.078791 | 3.962982 | 4.103740 | 4.104795 | 4.139873 | 3.716259 | 3.563145 | 3.839898 | 3.938494 | 4.071455 | 4.255359 | 4.219866 | 4.182513 | 4.157620 | 4.143447 | 4.205645 | 4.154954 |
GO: CC
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0005667 | transcription regulator complex | 361 | 0.2660099 | 1.528903 | 4.869429e-05 | 0.005043337 | 0.004810849 | 3562 | tags=32%, list=21%, signal=25% | HDAC9||NR4A3||RXRG||SOX18||TRERF1||WTIP||DACH1||NKX2-5||LBX1||HIPK2||TAF4||BCL9L||RARG||JUND||JUN||HMGA1||EP300||SOX2||TLE6||CREBBP||JAZF1||CEBPB||TAF10||GTF2F2||RUNX1||MED25||DMBX1||TBX2||SKI||HEY2||HOXB9||NCOA6||PITX1||NPAS2||TAF3||SOX9||MEF2B||RXRA||CDX2||VDR||GATAD2A||HES6||GATA6||TCF3||GTF2F1||RELA||HDGF||TP53||ZFHX3||MAFB||MBD3||ALX4||KLF4||HOXA10||GATA4||SUPT3H||GSC||NCOR2||SNAPC3||ZBTB16||TEAD4||CDK7||TAF1B||HOXB13||NFATC1||TLE5||TRRAP||BRF1||GLI3||KLF5||SIX3||TRIM28||SPEN||SMAD4||TLE3||DRAP1||JUNB||CREB1||PDLIM1||GATA2||ARNT2||TCF7L1||STAT3||TAF6||TLE2||HAND1||SOX4||SOX8||MED26||RFXANK||NFATC2||ZFPM1||TADA3||PTOV1||PYGO2||GTF3C6||GATAD2B||HDAC4||SIX2||LIN37||MIDEAS||TLE4||GTF3C5||RBM14||NR1H3||TCF7||SMAD9||ATF5||AHR||TLE1||TEAD3||ERCC2||CDK4||CTBP1 | 5.313596 | 5.668200 | 5.421863 | 5.693322 | 5.289666 | 5.434033 | 5.712764 | 5.749056 | 5.282225 | 5.359858 | 5.297528 | 5.675405 | 5.658081 | 5.671058 | 5.537420 | 5.348763 | 5.371977 | 5.678450 | 5.650532 | 5.749183 | 5.281097 | 5.273198 | 5.314370 | 5.420929 | 5.424798 | 5.456113 | 5.715639 | 5.701416 | 5.721164 | 5.738768 | 5.758863 | 5.749467 |
| GO:0098978 | glutamatergic synapse | 274 | 0.2974865 | 1.641794 | 1.133918e-04 | 0.007749856 | 0.007392603 | 4218 | tags=36%, list=25%, signal=27% | ERBB4||CNKSR2||APBA1||NTNG1||JAK2||GRID2||PRR7||PLCB1||DLGAP4||CACNG7||PPFIA2||DLGAP3||DNM3||CALY||PRKAR1B||BSN||SYN2||SLC1A2||GRIP1||LRFN3||NRGN||EIF4EBP1||CPLX1||USP8||AP3D1||ARC||CRTC1||HRAS||SH3GL1||VPS18||ADCY1||BCAN||PFN1||LRFN4||CNIH2||ACTB||GABRD||NCS1||RELA||PORCN||DVL1||PPP1CA||ACTN1||EPHB2||ARHGAP39||PIN1||ABHD17A||FXYD6||WNT3A||DGKZ||FZD9||BAIAP2||DLGAP1||RTN4R||CTTNBP2||GIPC1||STXBP1||DBN1||CHMP2B||ABHD17B||FARP1||SHANK1||BCR||FLOT2||PCDH17||CORO1A||ARHGEF2||ARHGAP22||STAT3||SYT1||SRC||ARF1||FLNA||DNM2||LRFN5||GRID1||ABR||NAPA||ADRA2C||ELAVL1||PTPRS||RNF216||SCRIB||FLRT3||CTBP1||PRKCZ||HIP1||CPT1C||CACNG4||PCLO||PPP3CA||CLSTN2||CAMK1||GRIK3||EFNB3||FYN||SEMA3F||ADGRL3||CDK5 | 5.882308 | 6.084604 | 5.750214 | 5.990751 | 5.834013 | 5.848802 | 6.066693 | 6.013272 | 5.880433 | 5.952334 | 5.810678 | 6.119734 | 6.074841 | 6.058539 | 5.758868 | 5.715068 | 5.776023 | 5.999423 | 5.940822 | 6.030573 | 5.844530 | 5.836963 | 5.820440 | 5.872991 | 5.854151 | 5.818739 | 6.064944 | 6.066073 | 6.069059 | 6.007752 | 6.021796 | 6.010228 |
| GO:0043197 | dendritic spine | 143 | 0.3362289 | 1.691783 | 4.132207e-04 | 0.024965415 | 0.023814559 | 3163 | tags=32%, list=19%, signal=26% | APBA1||SYNPO||ZNF804A||GRID2||PPFIA2||DNM3||LAMA2||NR1D1||MAPT||SORCS2||NGFR||DDN||NRGN||GPER1||USP8||ARC||PTEN||ASAP1||CNIH2||MPP2||PRKACA||APBA2||DVL1||PPP1CA||ZMYND8||ABHD17A||BAIAP2||CTTNBP2||DAGLA||GIPC1||NEURL1||MAP1B||ABHD17B||FARP1||TANC2||SHANK1||ARRB1||BCR||ABI2||PALM||SHANK3||STRN4||ARRB2||ITPKA||APBA3||ABR | 4.910157 | 5.440384 | 4.996237 | 5.359009 | 4.897267 | 5.057131 | 5.413081 | 5.363712 | 4.885131 | 4.968265 | 4.875254 | 5.450087 | 5.452085 | 5.418738 | 5.101182 | 4.929311 | 4.952111 | 5.340614 | 5.342892 | 5.392915 | 4.919608 | 4.868264 | 4.903451 | 5.046042 | 5.049067 | 5.076095 | 5.420629 | 5.405186 | 5.413387 | 5.351460 | 5.385577 | 5.353847 |
| GO:0005882 | intermediate filament | 52 | 0.4514500 | 1.913113 | 4.970782e-04 | 0.027721671 | 0.026443757 | 4094 | tags=42%, list=24%, signal=32% | NEFM||NEFH||FAM83H||GPER1||MICAL1||VIM||JUP||NEFL||INA||LMNB1||EPPK1||LMNB2||NCKIPSD||KRT18||KRT8||PLEC||EIF6||NES||LMNA||SLC1A4||IFFO2||GFAP | 6.543271 | 6.222835 | 6.446369 | 6.096260 | 6.512482 | 6.297725 | 6.174240 | 6.090384 | 6.586241 | 6.593725 | 6.445038 | 6.266712 | 6.186912 | 6.213735 | 6.386139 | 6.432996 | 6.516917 | 6.145659 | 6.027432 | 6.113119 | 6.574664 | 6.604458 | 6.344566 | 6.461414 | 6.090818 | 6.317013 | 6.186946 | 6.147731 | 6.187680 | 6.072004 | 6.105184 | 6.093767 |
| GO:0005884 | actin filament | 87 | 0.3666011 | 1.709328 | 1.349507e-03 | 0.061149533 | 0.058330662 | 4398 | tags=46%, list=26%, signal=34% | DBNL||GAS2L1||WIPF3||RAC3||TWF2||COBL||ACKR2||VPS18||CORO1B||ACTG1||ACTB||COTL1||WAS||DMTN||ACTN1||MARK2||APC2||PDLIM2||DAPK3||PALLD||KEAP1||DBN1||MYO1C||PDLIM1||CORO1A||SH2B2||SRC||FLNA||MYO6||WIPF1||WIPF2||WASL||CD2AP||HCLS1||PDLIM4||FYN||PDLIM7||TPM4||LMOD1||WHRN | 6.695567 | 6.392682 | 6.238285 | 6.303058 | 6.719366 | 6.622479 | 6.359325 | 6.294126 | 6.650413 | 6.693595 | 6.741261 | 6.382159 | 6.405264 | 6.390527 | 6.092703 | 6.309380 | 6.302439 | 6.349856 | 6.231903 | 6.324757 | 6.803778 | 6.604416 | 6.742736 | 6.753598 | 6.611370 | 6.490420 | 6.361588 | 6.366422 | 6.349915 | 6.301337 | 6.302612 | 6.278299 |
| GO:0030027 | lamellipodium | 179 | 0.2703422 | 1.414568 | 8.679957e-03 | 0.299665185 | 0.285851218 | 4918 | tags=41%, list=29%, signal=29% | DBNL||PARVA||RAPGEF3||PARVB||MTSS2||CAPG||CTTNBP2NL||SRCIN1||FGD1||AKT1||PABPC1||RAC3||TWF2||TUBB3||CORO1B||ACTB||ARHGAP31||WASF1||TRPV4||CSPG4||PXN||APC2||ENAH||DGKZ||BAIAP2||BCAR1||SH3BP1||PALLD||LIMK1||WASF2||DBN1||ROCK1||CIB1||CFL1||FLOT2||CORO1A||CARMIL2||ABI2||FSCN1||PODXL||DNM2||PDXP||SPEF1||RAB3IP||PPP1R9B||FGD3||SCRIB||APBB1||WASL||TESC||CAPZB||GIT1||PLEKHG5||NF2||PDLIM4||ARAP3||RAB13||PLXND1||CDK5||ACTA2||CDC42BPB||FLOT1||VASP||RAPH1||FGD4||PLCE1||ARPIN||DAG1||MCC||EVL||PTPRM||HAX1||IGF2BP1 | 6.224531 | 6.250300 | 6.155270 | 6.188818 | 6.224933 | 6.239616 | 6.219589 | 6.179276 | 6.211661 | 6.247518 | 6.214136 | 6.254999 | 6.256458 | 6.239381 | 6.106699 | 6.174475 | 6.183421 | 6.212155 | 6.140093 | 6.212998 | 6.263159 | 6.201714 | 6.209142 | 6.292298 | 6.198325 | 6.226607 | 6.226953 | 6.213182 | 6.218599 | 6.178590 | 6.193007 | 6.166106 |
| GO:0000137 | Golgi cis cisterna | 16 | -0.6807349 | -1.785696 | 1.000597e-02 | 0.329742316 | 0.314541853 | 3640 | tags=62%, list=22%, signal=49% | ATL1||TMEM59||GOLGA8H||GOLGA8K||GOLGA8A||GOLGA8R||GOLGA8N||GOLGA2P5||GOLGA8T||GOLGA8O | 2.444773 | 3.724498 | 3.293099 | 3.714461 | 2.472183 | 2.970702 | 3.641243 | 3.718105 | 2.367100 | 2.533076 | 2.429247 | 3.782547 | 3.736267 | 3.651639 | 3.707337 | 3.064021 | 2.996598 | 3.657696 | 3.756265 | 3.727647 | 2.388080 | 2.435811 | 2.585250 | 2.533338 | 2.764915 | 3.450400 | 3.680074 | 3.668619 | 3.572642 | 3.668100 | 3.799393 | 3.683215 |
| GO:0071565 | nBAF complex | 15 | 0.5819849 | 1.778293 | 1.364580e-02 | 0.336871828 | 0.321342709 | 5171 | tags=80%, list=31%, signal=55% | ARID1B||ACTL6B||SMARCA4||ARID1A||DPF1||SMARCE1||DPF2||SMARCD1||SMARCA2||SMARCC2||SMARCD3||SMARCB1 | 5.460366 | 5.668966 | 5.609254 | 5.662365 | 5.426045 | 5.774999 | 5.726692 | 5.730822 | 5.355305 | 5.496336 | 5.523852 | 5.660778 | 5.681440 | 5.664597 | 5.603835 | 5.659155 | 5.563160 | 5.697300 | 5.601671 | 5.686240 | 5.506061 | 5.274732 | 5.486148 | 5.792788 | 5.716385 | 5.814007 | 5.727464 | 5.714668 | 5.737849 | 5.726799 | 5.732902 | 5.732756 |
| GO:0044391 | ribosomal subunit | 173 | -0.3838425 | -1.474934 | 1.393952e-02 | 0.336871828 | 0.321342709 | 3267 | tags=35%, list=20%, signal=28% | RPL24||RPS8||MRPS15||RPL7||RPS23||MRPL17||RPL7A||MRPL42||RPL39L||NSUN3||RPS3A||RPS6||MRPS6||MRPL40||MRPS7||MRPL33||MRPL22||MRPL18||MRPL9||RPL22||RPL23||MRPL51||DAP3||RPS25||MRPL13||RPL7L1||RPS17||RPL37||MRPS17||RPS4X||RPL10A||RPL21||MRPL45||MRPL21||MRPS27||MRPL36||MRPL35||RPS27A||RPS20||RPL32||RPL27||MRPL58||RPL39||RPL4||RPS12||MRPS18C||ZCCHC17||RPL34||MRPS14||RPS27L||RPL35A||MRPS28||RPS7||RPL30||MRPL15||MRPS33||MRPL46||RPS15A||RPL9||HBA2 | 10.812276 | 10.576037 | 10.421405 | 10.457971 | 10.797402 | 10.464087 | 10.553284 | 10.458505 | 10.866482 | 10.802817 | 10.765723 | 10.637400 | 10.529809 | 10.558743 | 10.359682 | 10.378633 | 10.520488 | 10.497208 | 10.427157 | 10.448652 | 10.786804 | 10.879599 | 10.721410 | 10.636650 | 10.552373 | 10.159785 | 10.542692 | 10.558128 | 10.558973 | 10.469626 | 10.390654 | 10.512595 |
| GO:0098685 | Schaffer collateral - CA1 synapse | 60 | 0.3747210 | 1.625177 | 1.491742e-02 | 0.348875178 | 0.332792729 | 4218 | tags=52%, list=25%, signal=39% | APBA1||NTNG1||NEFH||PRKAR1B||BSN||SYN2||CPLX1||SH3GL1||ADCY1||ACTG1||ACTB||TUBB2B||NEFL||INA||DVL1||BAIAP2||GIPC1||IQSEC2||SHANK1||BCR||STAT3||SYN1||ABR||PTPRS||RNF216||PRKCZ||AKAP12||CAPZB||PPP3CA||FYN||CDK5 | 7.017200 | 6.545018 | 6.536901 | 6.480494 | 7.040540 | 6.921391 | 6.494004 | 6.468929 | 6.970165 | 7.014665 | 7.065201 | 6.541294 | 6.557670 | 6.536001 | 6.347160 | 6.618510 | 6.627808 | 6.547737 | 6.393707 | 6.495806 | 7.136971 | 6.922458 | 7.054127 | 7.070193 | 6.887637 | 6.792409 | 6.494905 | 6.511291 | 6.475596 | 6.469571 | 6.482053 | 6.455037 |
| GO:0097440 | apical dendrite | 15 | 0.5623185 | 1.718200 | 1.809552e-02 | 0.387125482 | 0.369279770 | 6416 | tags=87%, list=38%, signal=53% | MAP2||PPARGC1A||MYO1D||NEURL1||MAP1B||FLNA||CPEB3||YKT6||NSMF||GSK3A||OSBP2||CLU||PTK2B | 5.064917 | 4.699547 | 5.333213 | 4.610146 | 5.042554 | 5.432146 | 4.749519 | 4.617690 | 4.990274 | 5.046760 | 5.152963 | 4.655994 | 4.706701 | 4.734843 | 5.202820 | 5.457651 | 5.327913 | 4.658497 | 4.552845 | 4.617138 | 5.048620 | 4.876767 | 5.185745 | 5.313080 | 5.410950 | 5.561498 | 4.822017 | 4.725950 | 4.697623 | 4.616353 | 4.648161 | 4.587927 |
| GO:0001725 | stress fiber | 57 | 0.3768358 | 1.615410 | 1.824818e-02 | 0.387125482 | 0.369279770 | 2789 | tags=40%, list=17%, signal=34% | SYNPO||FHL3||GAS2L1||NEBL||CNN2||ZYX||SIPA1L3||TRIP6||CORO1B||SEPTIN9||ACTN1||PXN||TLNRD1||PDLIM2||FBLIM1||BAG3||PALLD||ROR1||PDLIM1||MYH9||SH2B2||CYBA||FSCN1 | 5.084928 | 5.324584 | 5.083249 | 5.254695 | 5.080875 | 5.227353 | 5.277415 | 5.235751 | 5.050068 | 5.062975 | 5.140090 | 5.349754 | 5.325509 | 5.298025 | 5.113171 | 5.104886 | 5.030254 | 5.295909 | 5.188453 | 5.277451 | 5.095083 | 4.991734 | 5.151292 | 5.175310 | 5.262526 | 5.242781 | 5.305800 | 5.262228 | 5.263791 | 5.229716 | 5.243946 | 5.233553 |
| GO:0035102 | PRC1 complex | 15 | 0.5611145 | 1.714522 | 1.868882e-02 | 0.387125482 | 0.369279770 | 6227 | tags=80%, list=37%, signal=50% | PHC3||PHC2||PCGF2||CBX4||RING1||RNF2||CBX8||PCGF5||CBX2||PCGF3||PCGF6||BMI1 | 4.509435 | 4.916814 | 4.747295 | 4.991365 | 4.541368 | 4.868862 | 5.009464 | 5.024041 | 4.421313 | 4.490729 | 4.609924 | 4.861399 | 4.958061 | 4.929283 | 4.750526 | 4.776187 | 4.714507 | 4.979479 | 4.955706 | 5.037672 | 4.558088 | 4.384003 | 4.667922 | 4.726478 | 4.855005 | 5.011000 | 5.001253 | 4.989102 | 5.037595 | 4.990778 | 5.033950 | 5.046800 |
| GO:0030424 | axon | 508 | 0.2034716 | 1.188050 | 2.152742e-02 | 0.409855661 | 0.390962133 | 4628 | tags=37%, list=28%, signal=27% | GAP43||MAP2||ZNF804A||TRPV2||NEFM||EPHA5||RIN3||RASGRF1||NEFH||CDK5R2||CACNG7||PPFIA2||DNM3||CALY||BSN||CA2||SLC1A2||KCNK2||MAPT||TUBB4A||SNCB||MAP7||SEPTIN6||N4BP3||HSPB1||MAPK8IP1||MAP1A||NGFR||SRCIN1||LRFN3||SCN4A||KIF1C||NRGN||GPER1||CPLX1||MICAL1||RAC3||TWF2||AP3D1||MACO1||COBL||PREX1||TUBB3||ELK1||TMEM108||ATP6V0D1||NFIB||IGSF9||DAB2IP||ACTG1||TPGS1||ADGRL1||ACTB||GABRD||LMTK3||NCS1||NEFL||TRPV4||TBC1D24||LLGL1||DVL1||ANK1||EPHB2||KCNQ2||PACSIN1||MAP4||MYO1D||KIF21B||C1QL1||RTN4RL2||BCAR1||VSTM2L||RTN4R||SPOCK1||ADCY9||NECTIN1||DAGLA||TSHZ3||KCNIP3||PALLD||ROR1||KIF1A||STXBP1||RNF6||L1CAM||DBN1||MAP1B||MAF1||TANC2||CREB1||CIB1||PTCH1||BCR||CORO1A||NAV1||IGF1R||SNCG||PALM||SYT1||HCFC1||SYN1||FSCN1||CALM1||FLNA||DNM2||GRM2||KATNB1||RAB3A||SEMA6A||KCNAB2||SIGMAR1||KIF3A||PLEC||MAST1||NIN||HTT||ABR||PINK1||RANGAP1||ADRA2C||BSG||GNRH1||CBL||RAB21||SIRT2||GPRIN1||CALM3||PPP1R9B||CCDC120||IRX3||PTPRS||SLC38A7||CYTH2||GJC2||RAP1GAP||APBB1||ATP1A3||RNF40||CD2AP||FLRT3||PRKCZ||BAG2||GDI1||CPT1C||BIN1||MYH14||DCTN1||ARL8A||MBP||PCLO||KCNC4||GIT1||SH2D3C||NTRK2||PLEKHG5||NDRG2||MGARP||GRIK3||BLVRB||EXOC7||CBARP||SARM1||AZIN2||GAD1||TRAK1||ADGRL3||CDK5||BLOC1S1||SERPINF1||NTRK1||BACE1||BRSK2||NECAB2||BRSK1||SYT2||WHRN||INPP5J||TIMP2||FXR2||POLG||ROGDI||SYAP1||CTSZ||HIF1A||ADORA1||DAG1||CRMP1 | 5.450208 | 5.729239 | 5.403426 | 5.722391 | 5.414049 | 5.461021 | 5.719449 | 5.729881 | 5.432169 | 5.513894 | 5.402234 | 5.739610 | 5.731530 | 5.716482 | 5.454952 | 5.363815 | 5.389980 | 5.718732 | 5.687334 | 5.760179 | 5.450205 | 5.389530 | 5.401695 | 5.488404 | 5.426940 | 5.467045 | 5.725204 | 5.719140 | 5.713979 | 5.721590 | 5.747429 | 5.720462 |
| GO:0043256 | laminin complex | 11 | 0.6100618 | 1.685640 | 2.310867e-02 | 0.409855661 | 0.390962133 | 411 | tags=27%, list=2%, signal=27% | LAMB3||NTNG1||NTN4 | 3.005650 | 3.510428 | 3.984150 | 3.535757 | 2.971468 | 3.521020 | 3.404214 | 3.461136 | 3.052102 | 2.810406 | 3.135069 | 3.387656 | 3.597458 | 3.538144 | 4.185174 | 3.984892 | 3.749636 | 3.482689 | 3.584427 | 3.538359 | 2.883927 | 2.840764 | 3.167431 | 3.086474 | 3.242302 | 4.042825 | 3.432188 | 3.484474 | 3.288949 | 3.514286 | 3.487735 | 3.377786 |
| GO:0022626 | cytosolic ribosome | 95 | -0.4165350 | -1.502331 | 2.317804e-02 | 0.409855661 | 0.390962133 | 3780 | tags=36%, list=23%, signal=28% | RPL24||RPS8||RPL7||RPS23||RPL7A||RPL39L||RPS3A||RPS6||EIF2AK4||RPL22||RPL23||RPS25||RPL7L1||RPS17||RPL37||RPS4X||RPL10A||RPL21||RPS27A||RPS20||RPL32||RPL27||RPL39||RPL4||RPS12||ZCCHC17||RPL34||RPS27L||RPL35A||RPS7||RPL30||RPS15A||RPL9||HBA2 | 11.596661 | 11.350879 | 11.194057 | 11.226321 | 11.580314 | 11.230018 | 11.326906 | 11.226164 | 11.654105 | 11.589969 | 11.543776 | 11.412754 | 11.304510 | 11.333180 | 11.130545 | 11.148735 | 11.297045 | 11.266692 | 11.197505 | 11.213857 | 11.570006 | 11.668751 | 11.497024 | 11.412019 | 11.317549 | 10.913478 | 11.316001 | 11.333187 | 11.331468 | 11.238319 | 11.154159 | 11.283058 |
| GO:0005911 | cell-cell junction | 366 | 0.2063075 | 1.184491 | 2.912496e-02 | 0.459653785 | 0.438464663 | 4339 | tags=33%, list=26%, signal=25% | SYNPO||AKAP6||CDH22||FRMD4A||TJP3||CLMP||WTIP||PARD6A||PAK4||CCDC85B||CDC42EP1||CNN2||ZYX||NGFR||SH3KBP1||S100A11||AMOTL2||CSK||AKT1||MAP2K2||LSR||ATP1B1||SIPA1L3||SHROOM1||JAM2||NIBAN2||ACTG1||KIFC3||JUP||CAV3||ACTB||PLPP3||CCDC85C||CRB3||TRPV4||CLDN9||WAS||PPP1CA||CDC42EP4||ACTN1||PXN||PDLIM2||BAIAP2||SAPCD2||SH3BP1||NECTIN1||EPPK1||OBSL1||JAG1||SCN5A||SHROOM2||POLDIP2||WASF2||DBN1||TLN1||UBN1||PDLIM1||YBX3||PIP5K1C||FLOT2||CORO1A||MYH9||NOTCH1||ARHGEF2||ABI2||TJP1||TMEM47||PDCD6IP||DCHS1||FSCN1||PODXL||FLNA||SYMPK||KRT18||FLRT1||PDXP||KRT8||DLG5||BAIAP2L1||SCN4B||RANGRF||TNKS1BP1||CLDN4||PCDH1||UBA1||CGNL1||LIMS2||PANX2||SIRT2||PTPRU||STARD10||PPP1R9B||CYTH2||GJC2||ADD1||SCRIB||NECTIN2||CD2AP||CDK4||FLRT3||PRKCZ||AQP3||ARHGAP17||PVR||WDR1||DNMBP||PPP3CA||PLEKHG5||PRKCD||NF2||ARVCF||PDLIM4||TRAF4||RAB13||ADGRB1||GAB1||SLC2A1||ADGRL3||PDLIM7||CDC42BPB||FLOT1||VASP | 5.459180 | 5.460447 | 5.339737 | 5.430032 | 5.466570 | 5.522976 | 5.436299 | 5.431324 | 5.407705 | 5.465156 | 5.503085 | 5.458825 | 5.479954 | 5.442315 | 5.287608 | 5.372189 | 5.358000 | 5.448961 | 5.381329 | 5.458583 | 5.521253 | 5.368460 | 5.505164 | 5.561818 | 5.495726 | 5.510548 | 5.447923 | 5.435310 | 5.425576 | 5.425753 | 5.458885 | 5.408885 |
| GO:0005834 | heterotrimeric G-protein complex | 23 | 0.4639106 | 1.609064 | 2.948234e-02 | 0.459653785 | 0.438464663 | 2183 | tags=30%, list=13%, signal=27% | GNAL||GNB2||GNAO1||GNG7||GNA11||GNG5||GNAS | 6.113332 | 6.357188 | 6.010900 | 6.126001 | 6.150595 | 6.354861 | 6.334684 | 6.116183 | 6.029034 | 6.122936 | 6.183826 | 6.346812 | 6.382122 | 6.342299 | 5.971588 | 6.050972 | 6.009045 | 6.163169 | 6.050271 | 6.161682 | 6.147283 | 6.049670 | 6.248015 | 6.367063 | 6.445446 | 6.245044 | 6.339962 | 6.350034 | 6.313813 | 6.116622 | 6.109259 | 6.122637 |
| GO:0043025 | neuronal cell body | 371 | 0.2154216 | 1.233306 | 2.979825e-02 | 0.459653785 | 0.438464663 | 4771 | tags=37%, list=29%, signal=27% | GAP43||MAP2||CNKSR2||ACVRL1||SYNPO||DAB1||ZNF804A||DBNL||KCNN1||EPHA5||RIN3||KCNQ1||IL6ST||EEF1A2||CACNG7||SNPH||KCNK2||MAPT||KLHL14||TUBB4A||SNCB||GRIP1||SORCS2||GRIN3B||MAP1A||NGFR||SRCIN1||DDN||SOS1||MAP1S||NRGN||SLC12A5||CPLX1||ATP13A2||NDUFS7||SNCAIP||PPARGC1A||DMWD||RAC3||DPYSL5||ARC||COBL||RPTOR||TUBB3||ELK1||CRTC1||DAB2IP||CKB||GABRD||NRSN2||TTBK1||DVL1||PPP1CA||EPHB2||MYO1D||GAL||BAIAP2||NCDN||RTN4RL2||ZNF385A||ENDOG||RTN4R||TXN2||ABL1||ITPR3||KIF1A||NEURL1||L1CAM||RBFOX3||MAP1B||CIB1||UBXN1||MAPK8IP2||SNCG||HCFC1||CYBA||FLNA||FLRT1||KATNB1||P2RX2||MAST1||ASS1||ADRA2C||CAPN2||GNRH1||SIRT2||PPP1R9B||DNER||PTPRS||SLC38A7||GJC2||RAP1GAP||APBB1||ATP1A3||ENO2||UNC5A||ASTN2||PRKCZ||GDI1||DENND1A||AKAP12||FUS||FBXO31||DCTN1||MBP||PI4K2A||KCNC4||PTPRF||SEMA4F||MCRS1||TMEM266||GRIK3||RACK1||SLC1A4||AZIN2||CDK5||SERPINF1||NTRK1||BACE1||WHRN||TIMP2||SLC24A1||FLNB||FXR2||NUMA1||ROGDI||SYAP1||GLRX5||ADORA1||CRMP1||ITGA4||ADA||ELOVL5||BRD1||RTN4RL1||YKT6 | 6.088386 | 6.064478 | 6.143699 | 6.064380 | 6.088676 | 6.170687 | 6.061084 | 6.052170 | 6.055216 | 6.072169 | 6.136491 | 6.048338 | 6.082234 | 6.062661 | 6.108822 | 6.167064 | 6.154562 | 6.073336 | 6.054406 | 6.065335 | 6.095114 | 6.050134 | 6.119914 | 6.136132 | 6.150867 | 6.223536 | 6.065745 | 6.063231 | 6.054252 | 6.062128 | 6.050009 | 6.044315 |
| GO:0005901 | caveola | 59 | 0.3452865 | 1.490117 | 3.116746e-02 | 0.461110597 | 0.439854319 | 3228 | tags=34%, list=19%, signal=27% | ASAH2||HMOX1||AKAP6||JAK2||CAVIN1||SCARB1||ATP1B1||EHD2||CAV3||SMPD2||DLC1||SCN5A||GASK1A||F2R||MAPK3||PTCH1||FLOT2||SRC||RANGRF||CBL | 4.344208 | 4.918158 | 4.703642 | 4.977787 | 4.333913 | 4.574809 | 4.848473 | 4.941678 | 4.312115 | 4.424336 | 4.292642 | 4.959774 | 4.943098 | 4.849155 | 4.779204 | 4.652941 | 4.675617 | 4.964942 | 4.986246 | 4.982084 | 4.350337 | 4.305624 | 4.345363 | 4.498396 | 4.446195 | 4.759886 | 4.867887 | 4.837515 | 4.839819 | 4.931210 | 4.974092 | 4.919152 |
| GO:0098686 | hippocampal mossy fiber to CA3 synapse | 28 | 0.4282270 | 1.567693 | 3.142126e-02 | 0.461110597 | 0.439854319 | 4569 | tags=50%, list=27%, signal=36% | SYT12||PRKAR1B||GRIK4||GPER1||SH3GL1||ADCY1||SLC6A9||NECTIN1||SYT1||SYT7||CAPZB||EFNB3||BACE1||STXBP5 | 4.405236 | 5.388755 | 4.376841 | 5.297082 | 4.386720 | 4.537329 | 5.416044 | 5.341099 | 4.376551 | 4.537269 | 4.290965 | 5.401456 | 5.370401 | 5.394225 | 4.517180 | 4.295246 | 4.307122 | 5.272208 | 5.236082 | 5.379096 | 4.361762 | 4.436529 | 4.360547 | 4.557155 | 4.541815 | 4.512664 | 5.402935 | 5.398325 | 5.446384 | 5.322865 | 5.335836 | 5.364285 |
| GO:0000792 | heterochromatin | 71 | 0.3246916 | 1.459650 | 3.307276e-02 | 0.461110597 | 0.439854319 | 4005 | tags=39%, list=24%, signal=30% | IKZF1||H1-4||NOP53||HMGA1||HSF1||PML||KDM4D||LRWD1||BAZ2A||CENPC||PCGF2||CENPB||MBD3||FOXC1||CDKN2A||SUV39H1||INCENP||SALL1||TRIM28||ZNF618||TNKS1BP1||UBA1||SIRT2||RNF40||TCP1||RING1||MECP2||RNF2 | 6.327672 | 6.477028 | 6.443578 | 6.523672 | 6.257339 | 6.369667 | 6.497151 | 6.553231 | 6.312108 | 6.396308 | 6.271790 | 6.483789 | 6.478430 | 6.468827 | 6.484298 | 6.367524 | 6.475994 | 6.527478 | 6.481331 | 6.561098 | 6.234234 | 6.261661 | 6.275813 | 6.307392 | 6.411742 | 6.387808 | 6.496436 | 6.488255 | 6.506705 | 6.542257 | 6.573208 | 6.544019 |
| GO:0005925 | focal adhesion | 371 | 0.2097461 | 1.200813 | 3.875312e-02 | 0.511124474 | 0.487562656 | 3471 | tags=28%, list=21%, signal=23% | CSRP1||JAK2||FHL3||PARVA||PAK4||FLNC||CDC42EP1||GNB2||HSPA1A||CNN2||PARVB||HSPA1B||ZYX||SLC9A3R2||SORBS3||RPS2||HSPB1||HMGA1||SH3KBP1||MRC2||ATP6V0C||VIM||RHOG||PABPC1||MAP2K2||PRAG1||ARL2||ARPC1B||RPLP2||TRIP6||CORO1B||ACTG1||PFN1||JUP||CAV3||ACTB||ARHGAP31||WASF1||TRPV4||CSPG4||RPS9||UBOX5||RPS15||ACTN1||PXN||TRIOBP||ENAH||RPL18||DLC1||FBLIM1||TNS1||TGFB1I1||SCARF2||RPL8||BCAR1||CAPN1||MAPRE2||RRAS||PALLD||LIMK1||L1CAM||TSPAN4||TLN1||AFAP1||MAPK3||IRF2||MPRIP||RPLP1||PDLIM1||DAB2||TLN2||CFL1||PIP5K1C||GSN||EGFR||FLOT2||MYH9||ARHGEF2||ARHGAP22||EHD3||CD81||ADAM9||TLE2||PDCD6IP||SRC||CYBA||CD151||ARF1||FLNA||DNM2||FERMT1||FLRT1||PLEC||MARCKS||LIMS2||CAPN2||BSG||CBL||RAB21||GNA12||LASP1||HSPA5||ADD1||NPM1||NECTIN2 | 9.136977 | 8.970793 | 8.785829 | 8.833651 | 9.143384 | 8.919339 | 8.920220 | 8.847472 | 9.161262 | 9.122729 | 9.126629 | 9.040596 | 8.929816 | 8.939335 | 8.700090 | 8.781031 | 8.871280 | 8.872975 | 8.791723 | 8.835110 | 9.142586 | 9.183388 | 9.103060 | 9.052033 | 9.017650 | 8.655971 | 8.913569 | 8.916987 | 8.930052 | 8.846876 | 8.827840 | 8.867429 |
| GO:0099061 | integral component of postsynaptic density membrane | 30 | 0.4090727 | 1.523217 | 4.060150e-02 | 0.511124474 | 0.487562656 | 4096 | tags=50%, list=24%, signal=38% | ERBB4||GRID2||CACNG7||LRFN3||ADCY1||LRFN4||CNIH2||LRFN1||LRFN5||GRID1||ADRA2C||PTPRS||CACNG4||CLSTN2||EFNB3 | 2.611094 | 3.352667 | 3.200603 | 3.290431 | 2.629267 | 3.200770 | 3.225070 | 3.217013 | 2.548132 | 2.552586 | 2.725390 | 3.313473 | 3.426587 | 3.315001 | 3.182019 | 3.326948 | 3.082306 | 3.277274 | 3.322990 | 3.270460 | 2.651365 | 2.432446 | 2.782544 | 2.971957 | 3.103097 | 3.478306 | 3.264545 | 3.228382 | 3.181070 | 3.204104 | 3.281102 | 3.163343 |
| GO:0098993 | anchored component of synaptic vesicle membrane | 10 | 0.5968951 | 1.597261 | 4.062839e-02 | 0.511124474 | 0.487562656 | 6273 | tags=80%, list=37%, signal=50% | RAB26||RAB11B||SYN1||RAB3A||DNAJC5||RAB4A||RAB35||RAB5A | 4.043962 | 4.362095 | 4.130280 | 4.281540 | 4.074339 | 4.273257 | 4.386651 | 4.288021 | 4.011661 | 3.971732 | 4.142886 | 4.332567 | 4.441864 | 4.308317 | 4.111518 | 4.187067 | 4.090456 | 4.313940 | 4.269006 | 4.261110 | 4.083356 | 3.907346 | 4.215816 | 4.240857 | 4.289778 | 4.288597 | 4.385229 | 4.396701 | 4.377962 | 4.291774 | 4.289091 | 4.283185 |
| GO:0005801 | cis-Golgi network | 56 | -0.4512799 | -1.509376 | 4.165113e-02 | 0.511814674 | 0.488221039 | 2975 | tags=32%, list=18%, signal=27% | RETREG1||GOLGA8H||TRAPPC6B||IFT20||CCN2||TMED10||GOLGA8K||GOLGA8A||KDELR3||RAB30||FKTN||GOLGA8R||ATP2C1||YIPF6||GOLGA8N||GOLGA2P5||GOLGA8T||GOLGA8O | 4.788625 | 5.504775 | 5.197878 | 5.390997 | 4.821075 | 5.100780 | 5.382182 | 5.343580 | 4.736857 | 4.778538 | 4.848277 | 5.499314 | 5.558977 | 5.454114 | 5.213459 | 5.214747 | 5.164871 | 5.394440 | 5.409561 | 5.368696 | 4.833060 | 4.738606 | 4.887636 | 4.934739 | 4.991673 | 5.341465 | 5.416914 | 5.371531 | 5.357429 | 5.351772 | 5.390106 | 5.286985 |
| GO:0030863 | cortical cytoskeleton | 89 | 0.2994238 | 1.397301 | 4.459064e-02 | 0.518272870 | 0.494381525 | 4501 | tags=48%, list=27%, signal=36% | DBNL||RIMS2||RAPGEF3||BSN||MTSS2||GYPC||MYRIP||COBL||SHROOM1||ACTB||LLGL2||COTL1||TRPV4||LLGL1||DMTN||ACTN1||DLC1||SHROOM2||DBN1||CIB2||GSN||FLOT2||CORO1A||MYH9||FLNA||UTRN||CAPN2||PPP1R9B||LASP1||WASL||ERC1||CAPZB||PCLO||HCLS1||WDR1||NF2||ACTN4||CALD1||SLC2A1||TPM4||FLOT1||SHROOM3||NUMA1 | 6.481317 | 7.325804 | 6.387241 | 7.270380 | 6.355019 | 6.230425 | 7.291586 | 7.267764 | 6.537752 | 6.596827 | 6.291552 | 7.429754 | 7.283131 | 7.258522 | 6.579848 | 6.096595 | 6.443322 | 7.305737 | 7.210975 | 7.292612 | 6.361153 | 6.458176 | 6.237260 | 6.275717 | 6.177868 | 6.236016 | 7.244352 | 7.318175 | 7.311088 | 7.312285 | 7.149419 | 7.334599 |
| GO:0098982 | GABA-ergic synapse | 42 | 0.3641806 | 1.471069 | 4.551422e-02 | 0.518272870 | 0.494381525 | 1403 | tags=17%, list=8%, signal=15% | ERBB4||GAP43||PLCB1||GLRB||BSN||LRFN4||GABRD | 5.289682 | 5.535976 | 5.412948 | 5.624199 | 5.166079 | 5.096634 | 5.507755 | 5.628394 | 5.336632 | 5.473698 | 5.022392 | 5.603976 | 5.516468 | 5.484828 | 5.417416 | 5.335934 | 5.481797 | 5.625404 | 5.588567 | 5.657795 | 5.126356 | 5.318769 | 5.038740 | 5.073259 | 4.941023 | 5.257967 | 5.512769 | 5.498284 | 5.512166 | 5.604505 | 5.633418 | 5.646933 |
| GO:0005751 | mitochondrial respiratory chain complex IV | 14 | -0.6202568 | -1.579781 | 4.717840e-02 | 0.518272870 | 0.494381525 | 3970 | tags=57%, list=24%, signal=44% | COX6A1||COX7A2||UQCRFS1||COX7C||COX7A2L||UQCRC2||COX6C||NDUFA4 | 10.314775 | 10.070006 | 10.879264 | 10.286836 | 10.402130 | 10.642883 | 10.135636 | 10.166958 | 10.251250 | 10.315705 | 10.374729 | 9.739911 | 10.202301 | 10.218588 | 10.802131 | 11.014870 | 10.810576 | 10.217302 | 10.478746 | 10.142473 | 10.437682 | 10.334396 | 10.431995 | 10.483636 | 10.426439 | 10.957664 | 10.129553 | 10.156014 | 10.121109 | 10.236378 | 10.111608 | 10.150043 |
| GO:0005940 | septin ring | 12 | 0.5437003 | 1.547337 | 4.865629e-02 | 0.518272870 | 0.494381525 | 2308 | tags=42%, list=14%, signal=36% | SEPTIN6||SEPTIN9||SEPTIN1||TMEM250||SEPTIN5 | 4.516093 | 5.905594 | 4.643834 | 5.816780 | 4.521973 | 4.716752 | 5.970251 | 5.862262 | 4.443374 | 4.571119 | 4.530845 | 5.902275 | 5.893551 | 5.920821 | 4.888127 | 4.563994 | 4.441803 | 5.776783 | 5.750103 | 5.917769 | 4.510117 | 4.442249 | 4.608683 | 4.677190 | 4.738936 | 4.733323 | 5.970545 | 5.937409 | 6.002076 | 5.832414 | 5.860247 | 5.893475 |
| GO:0031105 | septin complex | 12 | 0.5437003 | 1.547337 | 4.865629e-02 | 0.518272870 | 0.494381525 | 2308 | tags=42%, list=14%, signal=36% | SEPTIN6||SEPTIN9||SEPTIN1||TMEM250||SEPTIN5 | 4.516093 | 5.905594 | 4.643834 | 5.816780 | 4.521973 | 4.716752 | 5.970251 | 5.862262 | 4.443374 | 4.571119 | 4.530845 | 5.902275 | 5.893551 | 5.920821 | 4.888127 | 4.563994 | 4.441803 | 5.776783 | 5.750103 | 5.917769 | 4.510117 | 4.442249 | 4.608683 | 4.677190 | 4.738936 | 4.733323 | 5.970545 | 5.937409 | 6.002076 | 5.832414 | 5.860247 | 5.893475 |
| GO:0043198 | dendritic shaft | 29 | 0.4056416 | 1.494949 | 4.947917e-02 | 0.518272870 | 0.494381525 | 1998 | tags=38%, list=12%, signal=33% | MAP2||ZNF804A||MAP1A||GPER1||PREX1||CNIH2||MPP2||ZMYND8||BAIAP2||RTN4R||GIPC1 | 4.504635 | 4.792673 | 4.699407 | 4.739871 | 4.516058 | 4.792480 | 4.686429 | 4.655301 | 4.442568 | 4.491652 | 4.576491 | 4.830459 | 4.836690 | 4.707210 | 4.698192 | 4.729986 | 4.669406 | 4.793415 | 4.729041 | 4.695434 | 4.519652 | 4.413640 | 4.608314 | 4.725899 | 4.768185 | 4.878995 | 4.760611 | 4.663734 | 4.631803 | 4.644037 | 4.699872 | 4.620848 |
KEGG
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa05168 | Herpes simplex virus 1 infection | 386 | -0.4261363 | -1.910536 | 1.774118e-09 | 2.075718e-07 | 1.363269e-07 | 3933 | tags=46%, list=27%, signal=34% | ZNF551||ZNF14||ZNF136||HLA-DMB||EIF2B2||ZNF785||ZNF814||ZNF140||ZNF793||HCFC2||ZNF3||SRSF5||ZNF227||ZNF250||ZNF773||ZNF584||ZNF343||ZNF850||CHUK||ZNF790||PIK3R3||HLA-DPA1||CASP3||ZNF184||ZNF585A||ZNF561||ZNF195||ZNF585B||ZNF527||TSC1||IRF7||ZNF256||ZNF79||ZFP37||ZFP28||ZNF543||ZNF81||ZNF382||ZNF429||ZNF555||ZNF189||AKT3||ZNF468||ZNF514||ZNF623||ZNF25||ZNF606||ZNF567||ZNF670||ZNF302||BIRC2||ZNF568||ZNF45||ZFP30||ZNF582||ZNF419||ZNF317||ZNF235||SOCS3||STAT2||ZNF792||IFNGR1||TBK1||ZNF234||ZNF547||ZNF621||163051||SRSF7||DDX58||ZNF565||ZNF101||RHEB||ZNF273||IFNGR2||ZNF26||EIF2AK3||EIF2AK4||ZNF430||ZNF33B||ZNF595||ZNF583||ZNF440||ZNF669||ZNF112||ZNF888||ZNF571||ZNF552||CASP8||ZNF337||ZNF77||ZNF577||ZNF100||ZNF473||IRAK4||ZNF823||SRPK1||RNASEL||ZNF91||PILRB||ZNF84||ZNF420||ZNF85||POU2F1||ZNF578||ZNF680||ZNF549||ZNF875||ZNF436||ZNF605||ZNF717||ZNF254||ZNF2||ZNF529||ZNF354B||ZNF417||ZNF19||ZNF550||HLA-DOB||ZNF470||ZNF416||ZNF283||ZNF506||ZNF155||ZNF587||ZNF320||ZNF230||ZNF226||ZNF222||ZNF248||ZNF350||ZNF641||ZNF223||APAF1||ZNF268||ZNF562||ZNF557||ZNF799||ZNF224||ZNF891||ZNF44||ZNF30||ZNF559||ZNF546||ZNF528||ZNF169||ZNF726||SRSF3||ZNF433||ZNF736||ZNF700||ZNF304||ZNF682||ZSCAN32||ZNF805||ZNF540||EIF2B3||ZNF519||ZNF426||ZNF404||ZNF649||ZNF214||IRF9||ZNF442||ZNF780B||B2M||ZNF211||ZNF284||C3||ZNF548||ZNF432||CFP||ZNF10||ZNF772||ZNF699||PILRA||ZIK1||ZNF17 | 4.388954 | 5.025723 | 4.546492 | 4.833528 | 4.378123 | 4.492159 | 4.987082 | 4.848860 | 4.372601 | 4.424142 | 4.369463 | 5.044975 | 5.034870 | 4.996879 | 4.667011 | 4.483299 | 4.481168 | 4.815203 | 4.820150 | 4.864713 | 4.366461 | 4.355440 | 4.411848 | 4.427392 | 4.461171 | 4.583216 | 4.994740 | 4.978205 | 4.988253 | 4.835197 | 4.877310 | 4.833647 |
| hsa04926 | Relaxin signaling pathway | 100 | 0.3869252 | 1.910152 | 3.043332e-05 | 1.780349e-03 | 1.169280e-03 | 2530 | tags=36%, list=17%, signal=30% | PLCB1||MMP2||SHC3||GNB2||SHC1||JUN||PLCB2||SOS1||GNAO1||VEGFB||COL1A1||COL4A4||AKT1||MAP2K2||HRAS||TGFB1||ADCY1||MAP2K7||PRKACA||RELA||GNG7||COL4A3||GNG5||ADCY9||CREB3L1||GNAS||MAPK3||SOS2||MAPK12||CREB1||ARRB1||EGFR||CREB3L2||SRC||AKT2||ARRB2 | 5.388034 | 5.430831 | 5.414903 | 5.409789 | 5.406132 | 5.637333 | 5.422060 | 5.418223 | 5.335836 | 5.406474 | 5.420376 | 5.434282 | 5.438369 | 5.419776 | 5.398927 | 5.426962 | 5.418677 | 5.453301 | 5.323546 | 5.448796 | 5.401738 | 5.357043 | 5.457846 | 5.624837 | 5.670315 | 5.616261 | 5.438150 | 5.421370 | 5.406485 | 5.414653 | 5.422515 | 5.417489 |
| hsa01522 | Endocrine resistance | 85 | 0.4069310 | 1.942573 | 5.909188e-05 | 2.304583e-03 | 1.513581e-03 | 2529 | tags=39%, list=17%, signal=32% | MMP2||BIK||SHC3||SHC1||JUN||SOS1||CARM1||DLL3||GPER1||AKT1||MAP2K2||CDKN1A||HRAS||NOTCH3||ADCY1||RPS6KB2||PRKACA||BAD||TP53||CDKN2A||ADCY9||JAG1||BAX||GNAS||MAPK3||SOS2||MAPK12||RPS6KB1||EGFR||NOTCH1||IGF1R||SRC||AKT2 | 5.654703 | 5.638594 | 5.625880 | 5.460227 | 5.687791 | 5.903745 | 5.606608 | 5.447679 | 5.569541 | 5.613897 | 5.772678 | 5.661480 | 5.641043 | 5.612846 | 5.556980 | 5.701108 | 5.615903 | 5.509835 | 5.381360 | 5.486271 | 5.709522 | 5.524970 | 5.814163 | 5.867516 | 5.990136 | 5.849485 | 5.637287 | 5.586396 | 5.595629 | 5.443679 | 5.468727 | 5.430366 |
| hsa04062 | Chemokine signaling pathway | 112 | 0.3508925 | 1.754096 | 1.027656e-04 | 3.005894e-03 | 1.974181e-03 | 2530 | tags=31%, list=17%, signal=26% | CCL5||JAK2||PLCB1||SHC3||ELMO1||GNB2||SHC1||PLCB2||SOS1||AKT1||RAC3||NFKBIB||PREX1||HRAS||GRK2||ADCY1||PRKACA||BAD||RELA||GNG7||WAS||PXN||GNG5||BCAR1||CXCL16||GRK6||ADCY9||MAPK3||ROCK1||SOS2||ARRB1||STAT3||SRC||AKT2||ARRB2 | 5.041486 | 5.181632 | 5.092048 | 5.108712 | 5.057636 | 5.244744 | 5.196751 | 5.132559 | 4.977890 | 5.048646 | 5.095498 | 5.175448 | 5.190905 | 5.178497 | 5.095459 | 5.092803 | 5.087872 | 5.120695 | 5.042287 | 5.160651 | 5.069333 | 4.969094 | 5.129929 | 5.193521 | 5.275796 | 5.263556 | 5.211789 | 5.184706 | 5.193627 | 5.116475 | 5.146370 | 5.134675 |
| hsa05207 | Chemical carcinogenesis - receptor activation | 139 | 0.3345399 | 1.737908 | 1.836054e-04 | 4.296367e-03 | 2.821725e-03 | 2635 | tags=32%, list=18%, signal=26% | JAK2||FGF8||RXRG||MYC||JUN||SOS1||EIF4EBP1||DLL3||CCND3||CACNA1D||AKT1||MAP2K2||HRAS||BCL6||ADCY1||MGST1||RXRA||VDR||RPS6KB2||PRKACA||AIP||BAD||RELA||GSTO2||KLF4||ADCY9||CREB3L1||JAG1||KLF5||GNAS||PAQR7||MAPK3||SOS2||RPS6KB1||CREB1||ARRB1||EGFR||CREB3L2||STAT3||SRC||AKT2||ARRB2||CYP1B1||GSTM4 | 5.659508 | 6.171344 | 5.718537 | 6.144137 | 5.618565 | 5.770986 | 6.174923 | 6.178915 | 5.640350 | 5.727764 | 5.607726 | 6.188402 | 6.165237 | 6.160237 | 5.842413 | 5.633079 | 5.671401 | 6.131545 | 6.093741 | 6.204902 | 5.629789 | 5.625379 | 5.600352 | 5.805908 | 5.777996 | 5.727976 | 6.167106 | 6.173343 | 6.184269 | 6.176708 | 6.187251 | 6.172746 |
| hsa04010 | MAPK signaling pathway | 237 | 0.2644629 | 1.479816 | 5.585412e-04 | 9.562715e-03 | 6.280506e-03 | 2530 | tags=27%, list=17%, signal=22% | ERBB4||MECOM||RASGRF1||FGF8||MYC||CACNG7||FLNC||IRAK1||HSPA1A||HSPA1B||MAPT||HSPB1||MAPK8IP1||JUND||JUN||NGFR||SOS1||VEGFB||CACNA1D||AKT1||RAC3||MAP2K2||DUSP9||ELK1||MAP3K11||HRAS||TGFB1||MAPK7||RASGRF2||ERBB3||CACNG6||ECSIT||TAB1||MAP2K7||PRKACA||MAP2K3||RELA||DUSP2||TP53||TRADD||RPS6KA4||TRAF2||DUSP8||MAP3K12||FLT3LG||RRAS||CDC25B||NFATC1||SRF||MAPK3||SOS2||MAPK12||ARRB1||MAPKAPK2||MAPKAPK3||EGFR||MAPK8IP2||IGF1R||NFKB2||FLNA||RASGRP3||AKT2||ARRB2 | 5.124360 | 5.416650 | 4.999270 | 5.209890 | 5.149400 | 5.126837 | 5.407695 | 5.226722 | 5.125299 | 5.194478 | 5.049670 | 5.454896 | 5.397300 | 5.396981 | 5.050948 | 4.944713 | 5.000194 | 5.208793 | 5.154105 | 5.264654 | 5.176880 | 5.122345 | 5.148459 | 5.127340 | 5.145007 | 5.107926 | 5.431056 | 5.392884 | 5.398853 | 5.212514 | 5.237005 | 5.230535 |
| hsa04935 | Growth hormone synthesis, secretion and action | 93 | 0.3621161 | 1.753721 | 6.000280e-04 | 9.562715e-03 | 6.280506e-03 | 2529 | tags=31%, list=17%, signal=26% | JAK2||PLCB1||SHC3||SHC1||PLCB2||EP300||CREBBP||SOS1||CACNA1D||AKT1||MAP2K2||HRAS||ADCY1||PRKACA||MAP2K3||GNA11||BCAR1||ADCY9||CREB3L1||ITPR3||GNAS||MAPK3||SOS2||MAPK12||JUNB||CREB1||CREB3L2||STAT3||AKT2 | 5.272141 | 5.406182 | 5.381122 | 5.434263 | 5.269300 | 5.540505 | 5.413078 | 5.449958 | 5.217130 | 5.306209 | 5.291513 | 5.412130 | 5.413091 | 5.393237 | 5.409305 | 5.381533 | 5.351959 | 5.457614 | 5.357454 | 5.484634 | 5.271657 | 5.224871 | 5.310110 | 5.524567 | 5.553385 | 5.543416 | 5.435621 | 5.407447 | 5.395876 | 5.437646 | 5.463763 | 5.448346 |
| hsa05224 | Breast cancer | 120 | 0.3229891 | 1.647037 | 6.538609e-04 | 9.562715e-03 | 6.280506e-03 | 3240 | tags=40%, list=22%, signal=31% | FGF8||WNT8B||SHC3||MYC||SHC1||JUN||SOS1||WNT10B||DLL3||AKT1||MAP2K2||HES5||PTEN||CDKN1A||HRAS||HEY2||WNT6||NOTCH3||LRP5||AXIN1||RPS6KB2||BAK1||TP53||DVL1||WNT3A||APC2||FZD9||JAG1||BAX||MAPK3||SOS2||RPS6KB1||HES1||EGFR||TCF7L1||NOTCH1||IGF1R||NFKB2||DVL3||AKT2||ARAF||HEYL||ERBB2||TCF7||HEY1||CDK4||FZD1||FLT4 | 4.666126 | 4.824564 | 4.767687 | 4.773691 | 4.715140 | 4.938707 | 4.799185 | 4.763799 | 4.584171 | 4.635341 | 4.772255 | 4.838010 | 4.834434 | 4.800959 | 4.755714 | 4.798067 | 4.748785 | 4.796897 | 4.716659 | 4.805856 | 4.723243 | 4.584145 | 4.827729 | 4.874774 | 4.977165 | 4.962079 | 4.832240 | 4.772214 | 4.792453 | 4.752922 | 4.785170 | 4.753064 |
| hsa04928 | Parathyroid hormone synthesis, secretion and action | 86 | 0.3672863 | 1.757522 | 7.386566e-04 | 9.602536e-03 | 6.306659e-03 | 2217 | tags=31%, list=15%, signal=27% | MMP24||PLCB1||PTH1R||RXRG||NACA||JUND||PLCB2||SLC9A3R1||CDKN1A||ADCY1||LRP5||RXRA||MEF2D||VDR||PRKACA||MAFB||GNA11||ADCY9||CREB3L1||ITPR3||GNAS||GATA3||MAPK3||CREB1||ARRB1||EGFR||CREB3L2 | 5.776806 | 5.902604 | 5.684113 | 5.928387 | 5.797768 | 5.919981 | 5.914773 | 5.960131 | 5.717281 | 5.812121 | 5.799194 | 5.891986 | 5.895542 | 5.920120 | 5.788497 | 5.638286 | 5.619542 | 5.940828 | 5.886197 | 5.957183 | 5.764727 | 5.792978 | 5.834737 | 5.915992 | 6.028258 | 5.807229 | 5.914373 | 5.902614 | 5.927226 | 5.967003 | 5.963206 | 5.950131 |
| hsa03010 | Ribosome | 132 | -0.4140023 | -1.689454 | 8.302731e-04 | 9.714195e-03 | 6.379993e-03 | 3161 | tags=38%, list=22%, signal=30% | MRPS21||MRPL27||RPL24||RPS8||MRPS15||RPL7||RPS23||MRPL17||RPL7A||RPS3A||RPS6||MRPS6||MRPS7||MRPL33||MRPL22||MRPL18||MRPL9||RPL22||RPL36A-HNRNPH2||RPL23||RSL24D1||RPS25||MRPL13||RPS17||RPL37||MRPS17||RPL22L1||RPS4X||RPL10A||RPL21||MRPL21||MRPL36||MRPL35||RPS27A||RPS20||RPL32||RPL27||RPL39||RPL4||RPS12||MRPS18C||RPL34||MRPS14||RPS27L||RPL35A||RPS7||RPL30||MRPL15||RPS15A||RPL9 | 11.166480 | 10.911299 | 10.766852 | 10.791598 | 11.150579 | 10.807533 | 10.887919 | 10.805717 | 11.222421 | 11.158490 | 11.116552 | 10.970769 | 10.866358 | 10.894739 | 10.704407 | 10.723803 | 10.866831 | 10.830568 | 10.764463 | 10.778921 | 11.140880 | 11.234392 | 11.071840 | 10.983734 | 10.895497 | 10.498576 | 10.877494 | 10.893589 | 10.892618 | 10.802929 | 10.767657 | 10.845510 |
| hsa04072 | Phospholipase D signaling pathway | 105 | 0.3284658 | 1.631587 | 1.140339e-03 | 1.132557e-02 | 7.438294e-03 | 2529 | tags=30%, list=17%, signal=25% | DGKK||PLCB1||SHC3||DNM3||RAPGEF3||SHC1||PLCB2||SOS1||AKT1||MAP2K2||HRAS||AGPAT3||ADCY1||PLPP3||SPHK1||DGKZ||ADCY9||RRAS||GNAS||F2R||AGPAT1||MAPK3||SOS2||GAB2||SPHK2||PIP5K1C||EGFR||AGPAT2||ARF1||DNM2||GRM2||AKT2 | 5.316572 | 5.419658 | 5.430767 | 5.261938 | 5.339656 | 5.589753 | 5.409541 | 5.298150 | 5.247865 | 5.327250 | 5.371877 | 5.413518 | 5.443124 | 5.402021 | 5.402771 | 5.476576 | 5.411826 | 5.291808 | 5.192194 | 5.299354 | 5.359483 | 5.226257 | 5.426107 | 5.551513 | 5.590116 | 5.626651 | 5.422538 | 5.406657 | 5.399331 | 5.287558 | 5.324249 | 5.282281 |
| hsa05202 | Transcriptional misregulation in cancer | 135 | 0.3026471 | 1.563186 | 1.222435e-03 | 1.132557e-02 | 7.438294e-03 | 2429 | tags=29%, list=17%, signal=24% | TLX3||WT1||NR4A3||RXRG||MYC||H3C12||NUPR1||NGFR||ETV5||CEBPB||RUNX1||CDKN1A||BCL6||PML||RXRA||JUP||TCF3||TFE3||BAK1||RELA||DOT1L||TP53||NFKBIZ||HOXA10||SUPT3H||FLI1||ZBTB16||MLLT1||BAX||MAF||TAF15||SLC45A3||ETV4||ASPSCR1||CDK9||ARNT2||IGF1R||H3C15||MEN1 | 5.073516 | 5.562054 | 5.260594 | 5.523933 | 5.059677 | 5.177891 | 5.612324 | 5.523816 | 5.063251 | 5.168363 | 4.982923 | 5.541473 | 5.573462 | 5.571007 | 5.495723 | 5.075245 | 5.176767 | 5.507889 | 5.490751 | 5.571884 | 5.025721 | 5.092227 | 5.060315 | 5.140470 | 5.138578 | 5.251694 | 5.608896 | 5.600479 | 5.627463 | 5.581177 | 5.413334 | 5.570881 |
| hsa05130 | Pathogenic Escherichia coli infection | 149 | 0.2919680 | 1.532449 | 1.369343e-03 | 1.132557e-02 | 7.438294e-03 | 3755 | tags=39%, list=26%, signal=29% | IRAK1||TLR5||WIPF3||TUBB4A||JUN||SLC9A3R1||NFKBIB||TUBB3||ARPC1B||MYO1F||ACTG1||TUBB4B||TAB1||ACTB||TUBB2B||WASF1||BAK1||RELA||CLDN9||TRADD||MYO1D||BAIAP2||TRAF2||ABL1||BAX||WASF2||F2R||MAPK3||ROCK1||MYO1C||MAPK12||MYH9||ARHGEF2||TJP1||TUBA1C||SRC||ARF1||GAPDH||NCL||MYO6||TUBB2A||BAIAP2L1||CLDN4||WIPF1||GNA12||WIPF2||CYTH2||TIRAP||WASL||MYH14||SEC24C||TNFRSF1A||ARPC4||HCLS1||ARHGEF1||IKBKG||FYN||TAB2 | 7.822505 | 7.395991 | 7.653174 | 7.365561 | 7.872882 | 7.875785 | 7.369847 | 7.382459 | 7.763791 | 7.801977 | 7.898391 | 7.420959 | 7.398878 | 7.367639 | 7.479187 | 7.742905 | 7.722792 | 7.423381 | 7.290964 | 7.379204 | 7.922956 | 7.783598 | 7.908074 | 7.973246 | 7.873961 | 7.773218 | 7.372237 | 7.373784 | 7.363498 | 7.381716 | 7.386534 | 7.379116 |
| hsa04919 | Thyroid hormone signaling pathway | 108 | 0.3230867 | 1.611106 | 1.542386e-03 | 1.132557e-02 | 7.438294e-03 | 2755 | tags=31%, list=19%, signal=25% | DIO2||RCAN2||MED12L||PLCB1||RXRG||MYC||MED16||PLCB2||EP300||PLCD3||CREBBP||AKT1||MAP2K2||ATP1B1||HRAS||NOTCH3||ACTG1||RXRA||ACTB||PRKACA||BAD||PFKL||TP53||GATA4||PFKP||MAPK3||MED12||NOTCH1||DIO3||SRC||AKT2||THRA||ATP1B2 | 6.476170 | 6.249961 | 6.191963 | 6.233246 | 6.489857 | 6.470449 | 6.220188 | 6.242962 | 6.430257 | 6.479455 | 6.517473 | 6.241425 | 6.271617 | 6.236589 | 6.088055 | 6.251682 | 6.230720 | 6.279605 | 6.166824 | 6.250941 | 6.575226 | 6.379926 | 6.507649 | 6.567261 | 6.422637 | 6.416339 | 6.227636 | 6.227821 | 6.204988 | 6.240591 | 6.260840 | 6.227257 |
| hsa05225 | Hepatocellular carcinoma | 148 | 0.2964217 | 1.552523 | 1.602678e-03 | 1.132557e-02 | 7.438294e-03 | 2635 | tags=34%, list=18%, signal=29% | HMOX1||TERC||WNT8B||SHC3||MYC||ARID1B||ACTL6B||SHC1||SOS1||WNT10B||AKT1||MAP2K2||PTEN||ELK1||CDKN1A||HRAS||TGFB1||WNT6||MGST1||SMARCA4||LRP5||AXIN1||ACTG1||TXNRD2||ACTB||RPS6KB2||BAD||BAK1||TP53||DVL1||GSTO2||WNT3A||APC2||FZD9||CDKN2A||ARID1A||BAX||KEAP1||GSTP1||SMAD4||MAPK3||SOS2||RPS6KB1||EGFR||TCF7L1||IGF1R||DPF1||DVL3||AKT2||SMARCE1||GSTM4 | 6.520056 | 6.047373 | 6.204462 | 5.986513 | 6.555218 | 6.522314 | 6.008542 | 5.989176 | 6.471510 | 6.479225 | 6.605475 | 6.065013 | 6.048618 | 6.028252 | 6.038778 | 6.301229 | 6.259823 | 6.051075 | 5.901599 | 6.002856 | 6.614600 | 6.431755 | 6.611801 | 6.590629 | 6.553123 | 6.417496 | 6.024769 | 6.001117 | 5.999601 | 5.990542 | 6.001705 | 5.975157 |
| hsa04510 | Focal adhesion | 163 | 0.2845780 | 1.516624 | 1.635848e-03 | 1.132557e-02 | 7.438294e-03 | 2529 | tags=28%, list=17%, signal=23% | LAMB3||RASGRF1||SHC3||PARVA||PAK4||FLNC||TNXB||LAMA2||SHC1||PARVB||ZYX||JUN||RAPGEF1||SOS1||COL9A2||VEGFB||CCND3||COL1A1||COL4A4||AKT1||RAC3||PTEN||ELK1||HRAS||ACTG1||CAV3||ACTB||BAD||COL4A3||PPP1CA||ACTN1||PXN||BCAR1||TLN1||MAPK3||ROCK1||SOS2||PAK6||TLN2||PIP5K1C||EGFR||IGF1R||SRC||FLNA||AKT2 | 6.034361 | 5.763767 | 5.735569 | 5.712333 | 6.053770 | 6.029798 | 5.730961 | 5.720522 | 5.988120 | 6.031457 | 6.081975 | 5.760019 | 5.774157 | 5.757067 | 5.622335 | 5.797246 | 5.780744 | 5.753877 | 5.641618 | 5.738947 | 6.123987 | 5.944166 | 6.086974 | 6.108754 | 6.016775 | 5.959942 | 5.738811 | 5.734458 | 5.719544 | 5.720268 | 5.732465 | 5.708735 |
| hsa05165 | Human papillomavirus infection | 266 | 0.2553652 | 1.445479 | 1.645595e-03 | 1.132557e-02 | 7.438294e-03 | 3239 | tags=33%, list=22%, signal=26% | LAMB3||MAML2||WNT8B||PARD6A||TNXB||PPP2R2C||LAMA2||ATP6V0E2||HLA-G||EP300||TICAM1||CREBBP||SLC9A3R1||SOS1||WNT10B||COL9A2||EIF4EBP1||ATP6V0C||CCND3||COL1A1||COL4A4||AKT1||MAP2K2||HES5||PTEN||MAML3||ATP6V0D1||CDKN1A||HRAS||HEY2||WNT6||TUBG1||NOTCH3||HES4||AXIN1||LLGL2||HES6||RPS6KB2||PRKACA||BAD||CRB3||BAK1||RELA||LLGL1||TP53||COL4A3||DVL1||TRADD||PXN||WNT3A||APC2||FZD9||PPP2R1A||CREB3L1||JAG1||BAX||GNAS||PPP2R3B||MAPK3||SOS2||RPS6KB1||PKM||CREB1||HES1||EGFR||CREB3L2||ATP6V0B||TCF7L1||NOTCH1||TRAF3||DVL3||AKT2||ATP6AP1||TADA3||ATP6V1F||UBE3A||EIF2AK2||HEYL||TCF7||SCRIB||HEY1||PPP2R5D||CDK4||IKBKE||PRKCZ||PPP2R5B||FZD1 | 5.507986 | 5.540893 | 5.591600 | 5.487563 | 5.547183 | 5.742088 | 5.489521 | 5.468480 | 5.443679 | 5.453378 | 5.620006 | 5.579160 | 5.546241 | 5.496067 | 5.530254 | 5.656816 | 5.584932 | 5.517966 | 5.443030 | 5.500630 | 5.553020 | 5.436354 | 5.644645 | 5.689438 | 5.779017 | 5.756314 | 5.513539 | 5.479205 | 5.475513 | 5.465155 | 5.491647 | 5.448307 |
| hsa04915 | Estrogen signaling pathway | 91 | 0.3348526 | 1.616810 | 2.670019e-03 | 1.653388e-02 | 1.085896e-02 | 2536 | tags=33%, list=17%, signal=27% | PLCB1||MMP2||SHC3||HSPA1A||SHC1||HSPA1B||JUN||PLCB2||SOS1||GNAO1||GPER1||AKT1||MAP2K2||HRAS||ADCY1||PRKACA||CTSD||ADCY9||CREB3L1||ITPR3||GNAS||MAPK3||SOS2||CREB1||EGFR||CREB3L2||SRC||CALM1||AKT2||KRT18 | 6.390865 | 7.000558 | 6.264759 | 6.809772 | 6.381271 | 6.343253 | 6.987010 | 6.826563 | 6.392746 | 6.513952 | 6.254220 | 7.033386 | 6.986145 | 6.981571 | 6.404579 | 6.132343 | 6.244315 | 6.793453 | 6.764323 | 6.869486 | 6.422379 | 6.386801 | 6.333242 | 6.388624 | 6.359086 | 6.279870 | 6.990736 | 6.985951 | 6.984336 | 6.820095 | 6.830969 | 6.828602 |
| hsa05226 | Gastric cancer | 117 | 0.3059318 | 1.547512 | 2.684989e-03 | 1.653388e-02 | 1.085896e-02 | 2240 | tags=29%, list=15%, signal=25% | TERC||FGF8||WNT8B||RXRG||SHC3||MYC||SHC1||SOS1||WNT10B||AKT1||MAP2K2||CDKN1A||HRAS||TGFB1||WNT6||LRP5||AXIN1||RXRA||CDX2||JUP||RPS6KB2||BAK1||TP53||DVL1||WNT3A||APC2||FZD9||BAX||SMAD4||MAPK3||SOS2||RPS6KB1||EGFR||TCF7L1 | 4.380070 | 4.635634 | 4.534421 | 4.626403 | 4.410083 | 4.659860 | 4.631026 | 4.648415 | 4.316044 | 4.353424 | 4.466459 | 4.640206 | 4.653558 | 4.612841 | 4.547485 | 4.542474 | 4.513065 | 4.645457 | 4.577595 | 4.654930 | 4.411057 | 4.290316 | 4.519762 | 4.590688 | 4.692997 | 4.693484 | 4.659946 | 4.609245 | 4.623412 | 4.632689 | 4.674570 | 4.637623 |
| hsa05163 | Human cytomegalovirus infection | 172 | 0.2704907 | 1.453431 | 3.168131e-03 | 1.853357e-02 | 1.217229e-02 | 2529 | tags=27%, list=17%, signal=23% | CCL5||PLCB1||MYC||GNB2||HLA-G||PLCB2||SOS1||GNAO1||EIF4EBP1||AKT1||RAC3||MAP2K2||ELK1||CDKN1A||HRAS||ADCY1||RPS6KB2||PRKACA||BAK1||RELA||GNG7||TP53||TRADD||PXN||GNA11||TRAF2||GNG5||CDKN2A||BCAR1||ADCY9||CREB3L1||ITPR3||NFATC1||BAX||GNAS||MAPK3||ROCK1||SOS2||MAPK12||RPS6KB1||CREB1||EGFR||CREB3L2||STAT3||SRC||CALM1||AKT2 | 5.932579 | 6.196340 | 5.974439 | 6.038369 | 5.942837 | 6.093007 | 6.141741 | 6.035940 | 5.887338 | 5.949276 | 5.960061 | 6.217109 | 6.205944 | 6.165455 | 5.951906 | 5.995080 | 5.976007 | 6.063128 | 5.989909 | 6.060875 | 5.951406 | 5.883002 | 5.992005 | 6.067381 | 6.122460 | 6.088645 | 6.154638 | 6.132015 | 6.138477 | 6.036039 | 6.051423 | 6.020189 |
| hsa04725 | Cholinergic synapse | 84 | 0.3325802 | 1.578357 | 4.356306e-03 | 2.427085e-02 | 1.594037e-02 | 2529 | tags=31%, list=17%, signal=26% | JAK2||PLCB1||KCNQ1||GNB2||PLCB2||GNAO1||CACNA1D||AKT1||HRAS||ADCY1||CHRM4||PRKACA||KCNJ4||GNG7||KCNQ2||GNA11||GNG5||ADCY9||CREB3L1||ITPR3||MAPK3||KCNJ12||CREB1||CREB3L2||CHRM3||AKT2 | 4.898453 | 4.818758 | 4.931808 | 4.955285 | 4.915127 | 5.119258 | 4.835006 | 4.983973 | 4.856953 | 4.939996 | 4.897214 | 4.813787 | 4.827704 | 4.814740 | 4.907820 | 4.937064 | 4.950214 | 4.996756 | 4.860379 | 5.004225 | 4.908956 | 4.901372 | 4.934841 | 5.107619 | 5.121924 | 5.128154 | 4.852895 | 4.831052 | 4.820886 | 4.979196 | 4.984557 | 4.988151 |
| hsa05135 | Yersinia infection | 112 | 0.2930307 | 1.464847 | 5.409756e-03 | 2.877007e-02 | 1.889532e-02 | 3168 | tags=33%, list=22%, signal=26% | ELMO1||IRAK1||WIPF3||JUN||TICAM1||PKN1||RHOG||AKT1||RAC3||MAP2K2||ARPC1B||ACTG1||TAB1||ACTB||MAP2K7||MAP2K3||RELA||WAS||PXN||BAIAP2||TRAF2||BCAR1||NFATC1||LIMK1||WASF2||MAPK3||ROCK1||MAPK12||PIP5K1C||SRC||AKT2||NFATC2||WIPF1||WIPF2||RPS6KA1||WASL||RPS6KA2 | 6.576515 | 6.241436 | 6.203325 | 6.173005 | 6.593165 | 6.539123 | 6.218920 | 6.182226 | 6.533226 | 6.571021 | 6.623859 | 6.238322 | 6.245586 | 6.240390 | 6.079108 | 6.264802 | 6.258471 | 6.220458 | 6.089158 | 6.205845 | 6.672138 | 6.480685 | 6.619947 | 6.642114 | 6.540013 | 6.427248 | 6.220911 | 6.217989 | 6.217857 | 6.180259 | 6.193803 | 6.172534 |
| hsa05200 | Pathways in cancer | 411 | 0.2172225 | 1.286420 | 5.757037e-03 | 2.928580e-02 | 1.923404e-02 | 2529 | tags=24%, list=17%, signal=21% | IL12RB2||LAMB3||HMOX1||MECOM||JAK2||DAPK2||PLCB1||MMP2||TERC||FGF8||WNT8B||IL6ST||RXRG||MYC||LAMA2||GNB2||JUN||PLCB2||EP300||CREBBP||SOS1||WNT10B||VEGFB||DLL3||CCND3||COL4A4||AKT1||RAC3||MAP2K2||RUNX1||HES5||PTEN||ELK1||CDKN1A||SUFU||HRAS||TGFB1||HEY2||WNT6||PML||NOTCH3||ADCY1||MGST1||LRP5||AXIN1||RXRA||BMP2||TXNRD2||JUP||RPS6KB2||PRKACA||BAD||BAK1||RELA||GNG7||TP53||COL4A3||DVL1||GSTO2||WNT3A||APC2||GLI1||FZD9||GNA11||DAPK3||TRAF2||GNG5||ZBTB16||CDKN2A||ABL1||ADCY9||FLT3LG||JAG1||BAX||KEAP1||GSTP1||GLI3||GNAS||ELOB||F2R||SMAD4||MAPK3||ROCK1||SOS2||RPS6KB1||HES1||PTCH1||BCR||EGFR||ARNT2||TCF7L1||NOTCH1||IGF1R||STAT3||NFKB2||TRAF3||DVL3||CALM1||RASGRP3||AKT2 | 5.312516 | 5.599183 | 5.345615 | 5.590481 | 5.310491 | 5.415754 | 5.606128 | 5.618291 | 5.288234 | 5.321179 | 5.327825 | 5.612114 | 5.599504 | 5.585809 | 5.403382 | 5.311196 | 5.320470 | 5.589784 | 5.549332 | 5.631166 | 5.306049 | 5.269102 | 5.355033 | 5.390459 | 5.464287 | 5.391264 | 5.609497 | 5.599871 | 5.608995 | 5.611712 | 5.627975 | 5.615135 |
| hsa04934 | Cushing syndrome | 120 | 0.2834812 | 1.445572 | 6.748076e-03 | 3.289687e-02 | 2.160568e-02 | 4601 | tags=46%, list=32%, signal=32% | PLCB1||WNT8B||KCNK2||PLCB2||SCARB1||WNT10B||CACNA1D||USP8||MAP2K2||CDKN1A||WNT6||ADCY1||AXIN1||PRKACA||AIP||DVL1||WNT3A||APC2||FZD9||GNA11||CDKN2A||ADCY9||CREB3L1||ITPR3||GNAS||MAPK3||CREB1||EGFR||CREB3L2||ORAI1||TCF7L1||DVL3||MEN1||CAMK2B||TCF7||AHR||CDK4||FZD1||CACNA1G||KCNK3||DVL2||CACNA1H||E2F1||WNT2B||NR4A1||FH||LDLR||ADCY7||KMT2A||FZD8||ADCY3||PLCB3||ARMC5||FZD2||LEF1 | 5.312905 | 5.396474 | 5.380982 | 5.389708 | 5.313603 | 5.523575 | 5.371101 | 5.401313 | 5.254701 | 5.336738 | 5.345548 | 5.435591 | 5.392724 | 5.360113 | 5.363362 | 5.406736 | 5.372485 | 5.428542 | 5.318274 | 5.419728 | 5.322488 | 5.234340 | 5.380251 | 5.498478 | 5.549633 | 5.522160 | 5.392055 | 5.359797 | 5.361218 | 5.401823 | 5.411286 | 5.390757 |
| hsa04151 | PI3K-Akt signaling pathway | 239 | 0.2413058 | 1.348480 | 7.344046e-03 | 3.437014e-02 | 2.257328e-02 | 2217 | tags=21%, list=15%, signal=18% | ERBB4||LAMB3||JAK2||FGF8||MYC||TNXB||PPP2R2C||LAMA2||GNB2||MLST8||NGFR||SOS1||COL9A2||VEGFB||EIF4EBP1||CCND3||PKN1||COL1A1||COL4A4||AKT1||MAP2K2||PTEN||RPTOR||GYS1||CDKN1A||HRAS||DDIT4||ERBB3||RXRA||RPS6KB2||BAD||RELA||GNG7||TP53||COL4A3||GNG5||PPP2R1A||PCK2||CREB3L1||FLT3LG||CD19||PPP2R3B||F2R||MAPK3||G6PC3||SOS2||RPS6KB1||CREB1||EGFR||CREB3L2 | 5.617384 | 6.313622 | 5.560671 | 6.266753 | 5.564001 | 5.589903 | 6.294716 | 6.314350 | 5.619097 | 5.692520 | 5.536308 | 6.386251 | 6.277671 | 6.274105 | 5.719123 | 5.391307 | 5.552969 | 6.258382 | 6.219673 | 6.320410 | 5.524511 | 5.617894 | 5.547954 | 5.644191 | 5.640354 | 5.479090 | 6.276050 | 6.296343 | 6.311537 | 6.290351 | 6.325640 | 6.326761 |
| hsa05205 | Proteoglycans in cancer | 171 | 0.2566318 | 1.378559 | 7.659322e-03 | 3.446695e-02 | 2.263686e-02 | 3029 | tags=29%, list=21%, signal=23% | ERBB4||MMP2||WNT8B||MYC||FLNC||SOS1||WNT10B||TWIST2||COL1A1||AKT1||MAP2K2||ELK1||CDKN1A||HRAS||TGFB1||WNT6||ERBB3||ACTG1||NUDT16L1||CAV3||ACTB||RPS6KB2||PRKACA||TP53||PPP1CA||ANK1||PXN||WNT3A||FZD9||ITPR3||RRAS||MAPK3||ROCK1||SOS2||MAPK12||RPS6KB1||PTCH1||EGFR||IGF1R||STAT3||SRC||FLNA||AKT2||IHH||CBL||ARAF||CAMK2B||TWIST1||ERBB2 | 6.334007 | 6.545269 | 6.062644 | 6.471930 | 6.312060 | 6.220000 | 6.499938 | 6.505547 | 6.320212 | 6.371106 | 6.309956 | 6.619945 | 6.511124 | 6.501719 | 6.075276 | 5.986814 | 6.122566 | 6.487831 | 6.417904 | 6.508502 | 6.341739 | 6.283580 | 6.310273 | 6.323404 | 6.221208 | 6.107302 | 6.481980 | 6.508369 | 6.509298 | 6.475158 | 6.525925 | 6.515063 |
| hsa05206 | MicroRNAs in cancer | 145 | 0.2730025 | 1.422654 | 9.971547e-03 | 4.321004e-02 | 2.837906e-02 | 2697 | tags=27%, list=19%, signal=22% | HMOX1||MYC||PAK4||TNXB||SHC1||EP300||CREBBP||SOS1||VIM||MAP2K2||PTEN||RPTOR||CDKN1A||HRAS||MAPK7||NOTCH3||DDIT4||ERBB3||BAK1||TP53||WNT3A||APC2||PRKCE||CDKN2A||ABL1||CDC25B||SLC45A3||MAPK3||ROCK1||SOS2||EGFR||NOTCH1||HDAC5||STAT3||SOX4||FSCN1||FOXP1||CYP1B1||MARCKS | 5.043579 | 5.491135 | 5.150761 | 5.487608 | 4.992999 | 5.183911 | 5.524099 | 5.526197 | 5.036860 | 5.113577 | 4.977048 | 5.482871 | 5.490675 | 5.499810 | 5.296489 | 5.057043 | 5.086759 | 5.475534 | 5.445321 | 5.540330 | 5.007241 | 4.957135 | 5.013955 | 5.173010 | 5.172800 | 5.205675 | 5.520363 | 5.509716 | 5.542028 | 5.508173 | 5.543155 | 5.527051 |
| hsa04144 | Endocytosis | 231 | 0.2407968 | 1.342592 | 1.097657e-02 | 4.586639e-02 | 3.012368e-02 | 4051 | tags=37%, list=28%, signal=27% | PSD4||STAM||PARD6A||DNM3||MVB12A||HSPA1A||EPN1||HLA-G||HSPA1B||WIPF3||EHD1||ACAP1||SH3KBP1||USP8||MVB12B||AGAP1||IQSEC1||HRAS||ARF5||GRK2||SH3GL1||PML||ASAP1||VPS37D||ARPC1B||AGAP2||CHMP6||CHMP4C||EHD2||CAV3||AP2A1||GRK6||SNX32||ARF3||AGAP3||EPS15L1||CLTB||CHMP2B||IQSEC2||VPS37B||ARRB1||RAB11B||DAB2||PIP5K1C||EGFR||AP2S1||IGF1R||EHD3||PDCD6IP||SRC||ARF1||DNM2||ARRB2||CHMP1A||DNAJC6||VPS37C||RAB11FIP5||SMAP1||WIPF1||CBL||WIPF2||CYTH2||WASL||PRKCZ||RAB11FIP3||BIN1||HLA-A||CHMP4B||CBLB||CAPZB||VPS28||ARPC4||GIT1||SH3GLB2||ARAP3||HLA-E||SH3GLB1||SNX12||VPS26A||VPS37A||VPS26B||ARF6||LDLR||RNF41||VPS25 | 5.689744 | 6.049107 | 5.666881 | 5.861265 | 5.715640 | 5.732939 | 6.029340 | 5.888571 | 5.676534 | 5.738743 | 5.652579 | 6.084524 | 6.039956 | 6.022123 | 5.705117 | 5.639525 | 5.655183 | 5.860669 | 5.815615 | 5.906092 | 5.744600 | 5.665285 | 5.735734 | 5.714144 | 5.731320 | 5.753088 | 6.038558 | 6.023976 | 6.025440 | 5.873770 | 5.905135 | 5.886637 |
| hsa00190 | Oxidative phosphorylation | 118 | -0.3669673 | -1.475519 | 1.533159e-02 | 5.886554e-02 | 3.866113e-02 | 3621 | tags=36%, list=25%, signal=28% | ATP5F1B||ATP6V1C1||NDUFA2||COX11||NDUFV2||ATP5F1C||COX6A1||UQCRB||ATP5MF||SDHB||COX7A2||UQCRFS1||NDUFB3||ATP5ME||NDUFS1||UQCRH||ATP6V1C2||NDUFB9||NDUFA1||NDUFA8||COX7C||NDUFB1||ATP6V0A2||ATP6V1B1||NDUFA6||NDUFAB1||NDUFB6||COX7A2L||COX7B||UQCRC2||MT-ND6||COX6C||ATP5F1E||NDUFS5||ATP6V1A||ATP6V1G1||ATP5F1A||NDUFA9||ATP5PO||ATP6V1B2||COX6B2||UQCRHL||NDUFA4 | 10.022035 | 9.478658 | 10.462432 | 9.820110 | 10.096000 | 10.376440 | 9.560072 | 9.693546 | 9.946739 | 9.997150 | 10.116885 | 9.094780 | 9.630618 | 9.645434 | 10.349612 | 10.603921 | 10.421726 | 9.740565 | 10.036663 | 9.654724 | 10.136587 | 10.004387 | 10.142833 | 10.242821 | 10.235103 | 10.617399 | 9.543148 | 9.586456 | 9.550236 | 9.777633 | 9.643996 | 9.655161 |
| hsa05012 | Parkinson disease | 233 | -0.3227094 | -1.394711 | 1.540310e-02 | 5.886554e-02 | 3.866113e-02 | 2044 | tags=21%, list=14%, signal=19% | NDUFS1||VDAC2||SOD1||UQCRH||UCHL1||NDUFB9||NDUFA1||NDUFA8||PSMA7||MCU||MFN1||COX7C||NDUFB1||SLC25A5||NDUFA6||PRKN||NDUFAB1||PSMB7||NDUFB6||COX7A2L||COX7B||UQCRC2||MAOA||MT-ND6||COX6C||TUBA8||ATP5F1E||APAF1||NDUFS5||UBA7||PSMD13||UBC||RPS27A||ATP5F1A||DDIT3||NDUFA9||CALM2||MAPK10||PSMB1||SLC39A9||VDAC3||CALML6||ATP5PO||TXN||PSMA6||PARK7||COX6B2||UQCRHL||PSMA5||NDUFA4 | 9.331331 | 8.878639 | 9.705822 | 9.151700 | 9.385400 | 9.617311 | 8.943873 | 9.056675 | 9.268426 | 9.329961 | 9.392920 | 8.597383 | 8.995006 | 9.007098 | 9.602463 | 9.824189 | 9.682013 | 9.094708 | 9.321314 | 9.021929 | 9.422039 | 9.318537 | 9.413358 | 9.511325 | 9.483720 | 9.830530 | 8.930222 | 8.965187 | 8.935967 | 9.124415 | 9.014585 | 9.028529 |
| hsa05131 | Shigellosis | 205 | 0.2432533 | 1.334844 | 1.559685e-02 | 5.886554e-02 | 3.866113e-02 | 2529 | tags=28%, list=17%, signal=23% | CCL5||PLCB1||ELMO1||H3C12||TLR5||SEPTIN6||JUN||PLCB2||PLCD3||AKT1S1||AKT1||RPTOR||NFKBIB||FNBP1||TNIP1||HK2||ARPC1B||ACTG1||CAPNS1||PFN1||SEPTIN9||TAB1||ACTB||RPS6KB2||WASF1||RELA||TP53||TRADD||ACTN1||PXN||TRAF2||PRKCE||CAST||BCAR1||CAPN1||ITPR3||BAX||UBE2V1||WASF2||TLN1||MAPK3||ROCK1||TECPR1||MAPK12||RPS6KB1||TLN2||EGFR||ARHGEF2||RIPK2||FOXO6||HK1||H3C15||SRC||ARF1||U2AF1L4||FOXO4||AKT2 | 6.923646 | 6.846507 | 6.739767 | 6.803847 | 6.888386 | 6.778497 | 6.861113 | 6.819231 | 6.923334 | 6.988665 | 6.855884 | 6.845971 | 6.838498 | 6.855003 | 6.710783 | 6.702012 | 6.804265 | 6.831517 | 6.751738 | 6.826904 | 6.918954 | 6.903807 | 6.841226 | 6.866363 | 6.732918 | 6.732027 | 6.850765 | 6.863540 | 6.868974 | 6.836228 | 6.776484 | 6.844039 |
| hsa04071 | Sphingolipid signaling pathway | 104 | 0.2949001 | 1.463934 | 1.924521e-02 | 7.036528e-02 | 4.621382e-02 | 2138 | tags=26%, list=15%, signal=22% | ASAH2||DEGS2||PLCB1||PPP2R2C||PLCB2||AKT1||RAC3||MAP2K2||PTEN||HRAS||SMPD2||RELA||TP53||TRADD||CTSD||SPHK1||TRAF2||PRKCE||PPP2R1A||BAX||CERS1||PPP2R3B||MAPK3||ROCK1||GAB2||MAPK12||SPHK2 | 5.250577 | 5.429366 | 5.288673 | 5.384295 | 5.270334 | 5.451118 | 5.396560 | 5.390139 | 5.193777 | 5.233537 | 5.321443 | 5.441499 | 5.438643 | 5.407714 | 5.268758 | 5.317219 | 5.279593 | 5.394637 | 5.337201 | 5.419808 | 5.287425 | 5.174888 | 5.343605 | 5.400564 | 5.481898 | 5.469567 | 5.407027 | 5.386731 | 5.395850 | 5.376802 | 5.416771 | 5.376471 |
| hsa05014 | Amyotrophic lateral sclerosis | 306 | -0.3047051 | -1.346122 | 2.033467e-02 | 7.209567e-02 | 4.735028e-02 | 2658 | tags=26%, list=18%, signal=22% | TBK1||DNAH11||SDHB||COX7A2||SRSF7||PSMD1||UQCRFS1||NDUFB3||NUP85||PPP3CB||MAP2K6||NDC1||PSMA4||EIF2AK3||NXT2||RAE1||ANG||SPG11||NDUFS1||GRIN2C||SOD1||ACTR10||UQCRH||BECN1||NDUFB9||NDUFA1||NDUFA8||PIK3R4||PSMA7||RB1CC1||MCU||COX7C||NDUFB1||NDUFA6||DERL1||PRKN||NDUFAB1||PSMB7||NDUFB6||ATG2B||COX7A2L||ALS2||C9orf72||DNAH2||COX7B||NUP58||UQCRC2||DCTN5||MT-ND6||COX6C||TUBA8||ATP5F1E||APAF1||NDUFS5||MAPK14||PSMD13||DNAH10||DNAH17||PFN2||SRSF3||ATP5F1A||DDIT3||NDUFA9||DNAH6||NUP155||PSMB1||NUP160||ATP5PO||PIK3C3||PSMA6||SETX||NUP37||NUP43||NUP205||ATG14||COX6B2||UQCRHL||PSMA5||NDUFA4 | 8.956655 | 8.527291 | 9.311054 | 8.785482 | 9.017930 | 9.259355 | 8.588173 | 8.694237 | 8.885299 | 8.946757 | 9.034026 | 8.259174 | 8.638969 | 8.650444 | 9.210946 | 9.433885 | 9.279207 | 8.729921 | 8.947073 | 8.664026 | 9.058773 | 8.932003 | 9.059321 | 9.157563 | 9.136486 | 9.460695 | 8.575867 | 8.606774 | 8.581690 | 8.759175 | 8.657980 | 8.663293 |
| hsa04530 | Tight junction | 132 | 0.2578675 | 1.327119 | 2.341598e-02 | 8.057851e-02 | 5.292156e-02 | 3580 | tags=36%, list=25%, signal=27% | SYNPO||TJP3||PARD6A||PPP2R2C||JUN||SLC9A3R1||AMOTL2||CACNA1D||RUNX1||JAM2||ARPC1B||ACTG1||ACTB||MAP2K7||LLGL2||PRKACA||CRB3||LLGL1||CLDN9||WAS||ACTN1||GATA4||PRKCE||PPP2R1A||MYL6B||ROCK1||YBX3||MYH9||ARHGEF2||TJP1||TUBA1C||SRC||SYMPK||ARHGEF18||CLDN4||CGNL1||STK11||ERBB2||SCRIB||CDK4||PRKCZ||MYH14||ARPC4||ARHGAP17||HCLS1||NF2||ACTN4 | 7.075219 | 6.732266 | 6.899439 | 6.746924 | 7.123941 | 7.100595 | 6.710945 | 6.777239 | 7.014381 | 7.084438 | 7.124685 | 6.760344 | 6.725514 | 6.710486 | 6.753088 | 6.970803 | 6.963986 | 6.785031 | 6.671753 | 6.781147 | 7.180758 | 7.046177 | 7.141586 | 7.190650 | 7.056406 | 7.050325 | 6.713742 | 6.712552 | 6.706532 | 6.773607 | 6.788751 | 6.769285 |
| hsa04932 | Non-alcoholic fatty liver disease | 134 | -0.3458048 | -1.414323 | 2.635569e-02 | 8.810329e-02 | 5.786361e-02 | 4108 | tags=37%, list=28%, signal=27% | PRKAG2||PIK3R1||PIK3CB||NDUFC2||FOS||MLXIPL||PIK3R3||CASP3||MAPK8||NDUFA2||ADIPOR2||NDUFV2||AKT3||COX6A1||UQCRB||SOCS3||SDHB||COX7A2||UQCRFS1||NDUFB3||PRKAA2||CASP7||EIF2AK3||NDUFS1||CASP8||UQCRH||NDUFB9||NDUFA1||NDUFA8||COX7C||NDUFB1||NDUFA6||NDUFAB1||NDUFB6||COX7A2L||COX7B||UQCRC2||PRKAG1||COX6C||NDUFS5||MAPK14||CDC42||DDIT3||NDUFA9||MAPK10||CYP2E1||PRKAB2||COX6B2||UQCRHL||NDUFA4 | 9.173542 | 8.638620 | 9.522068 | 8.934789 | 9.241107 | 9.436955 | 8.720129 | 8.827533 | 9.107584 | 9.145542 | 9.262919 | 8.304525 | 8.770402 | 8.790573 | 9.391100 | 9.652699 | 9.510493 | 8.878259 | 9.117672 | 8.788029 | 9.265086 | 9.172686 | 9.283132 | 9.319854 | 9.315769 | 9.649251 | 8.708656 | 8.741027 | 8.710472 | 8.886712 | 8.789501 | 8.804474 |
| hsa04024 | cAMP signaling pathway | 134 | 0.2487027 | 1.283688 | 3.293623e-02 | 1.070427e-01 | 7.030248e-02 | 2233 | tags=26%, list=15%, signal=22% | GRIN2D||RAPGEF3||GPHA2||GRIN3B||JUN||EP300||CREBBP||CACNA1D||AKT1||HCN2||RAC3||MAP2K2||ATP1B1||ADCY1||SOX9||PRKACA||BAD||RELA||PPP1CA||GLI1||ADCY9||CREB3L1||RRAS||HCN4||NFATC1||GLI3||GNAS||F2R||NPR1||MAPK3||ROCK1||CREB1||PTCH1||CREB3L2||ORAI1 | 5.146570 | 5.446789 | 5.225692 | 5.388041 | 5.155022 | 5.368533 | 5.440144 | 5.399465 | 5.094942 | 5.163862 | 5.179509 | 5.441578 | 5.460643 | 5.438045 | 5.251942 | 5.230824 | 5.193709 | 5.393890 | 5.348704 | 5.420616 | 5.166154 | 5.078216 | 5.217283 | 5.325020 | 5.401720 | 5.377794 | 5.443694 | 5.438845 | 5.437887 | 5.387051 | 5.417921 | 5.393238 |
| hsa04360 | Axon guidance | 163 | 0.2380976 | 1.268912 | 3.419516e-02 | 1.081306e-01 | 7.101698e-02 | 4887 | tags=47%, list=34%, signal=31% | NTNG1||PLXNC1||SEMA3C||SLIT3||EPHA5||PLXNA2||PARD6A||PAK4||NTN4||SEMA7A||ABLIM2||EPHB3||RAC3||DPYSL5||HRAS||SEMA6B||EPHB2||SEMA4G||RGMA||ENAH||ABL1||RRAS||SLIT2||LIMK1||L1CAM||MAPK3||ROCK1||PAK6||EPHA8||PTCH1||CFL1||SRC||SSH3||SEMA6A||NFATC2||PLXNA4||CAMK2B||UNC5A||SRGAP1||MYL5||PRKCZ||UNC5B||BMP7||SEMA4F||EPHB4||PPP3CA||PRKCA||SRGAP3||EFNB3||SLIT1||FYN||SEMA3F||NTN1||CDK5||CXCL12||DPYSL2||SMO||NGEF||PLXNA1||EFNA2||EFNA4||SEMA4A||RASA1||CFL2||NCK2||PLXNB2||ROBO1||SEMA4B||ROBO2||PPP3CC||SSH2||EPHA3||GNAI2||WNT5B||EPHA2||RAC1 | 4.782213 | 4.803769 | 4.816522 | 4.840771 | 4.808555 | 4.954890 | 4.811167 | 4.863859 | 4.720088 | 4.784887 | 4.839201 | 4.794507 | 4.822989 | 4.793617 | 4.803297 | 4.836322 | 4.809735 | 4.846260 | 4.788513 | 4.885881 | 4.816708 | 4.728058 | 4.877020 | 4.916890 | 4.987283 | 4.959626 | 4.824391 | 4.805834 | 4.803184 | 4.851843 | 4.882331 | 4.857220 |
| hsa04722 | Neurotrophin signaling pathway | 105 | 0.2642910 | 1.312812 | 3.662791e-02 | 1.096218e-01 | 7.199634e-02 | 3850 | tags=40%, list=26%, signal=30% | SHC3||IRAK1||ARHGDIA||SHC1||JUN||NGFR||MATK||RAPGEF1||SOS1||AKT1||MAP2K2||NFKBIB||HRAS||MAPK7||MAP2K7||BAD||RELA||TP53||ARHGDIG||ABL1||BAX||MAPK3||SOS2||MAPK12||MAPKAPK2||RIPK2||SH2B2||CALM1||AKT2||TP73||CALM3||CAMK2B||RPS6KA1||NFKBIE||RPS6KA2||MAGED1||MAP2K5||NTRK2||PRKCD||GAB1||MAP3K3||NTRK1 | 6.016337 | 5.952352 | 5.955489 | 6.002791 | 5.993966 | 6.095876 | 5.974173 | 6.041013 | 6.008955 | 6.067897 | 5.970486 | 5.962448 | 5.944408 | 5.950142 | 5.980340 | 5.913952 | 5.971281 | 6.025701 | 5.926940 | 6.052716 | 5.992483 | 6.006068 | 5.983256 | 6.090956 | 6.135859 | 6.059797 | 5.987265 | 5.956042 | 5.979029 | 6.035523 | 6.045466 | 6.042033 |
| hsa04152 | AMPK signaling pathway | 101 | 0.2686609 | 1.330093 | 3.796762e-02 | 1.096218e-01 | 7.199634e-02 | 3266 | tags=30%, list=22%, signal=23% | PPP2R2C||FASN||EEF2||AKT1S1||EIF4EBP1||PPARGC1A||AKT1||RPTOR||GYS1||RPS6KB2||PFKL||SREBF1||PFKFB4||PPP2R1A||PCK2||CREB3L1||PFKP||PPP2R3B||G6PC3||RPS6KB1||CREB1||RAB11B||CREB3L2||IGF1R||AKT2||STK11||ELAVL1||PPP2R5D||PPP2R5B||CPT1C | 5.780567 | 5.973031 | 5.817950 | 5.929368 | 5.823412 | 6.161538 | 5.957416 | 5.940596 | 5.702210 | 5.765566 | 5.868991 | 5.958782 | 5.999922 | 5.960008 | 5.757743 | 5.898020 | 5.794392 | 5.974479 | 5.861853 | 5.949364 | 5.867556 | 5.672215 | 5.918898 | 6.171375 | 6.230665 | 6.078513 | 5.970387 | 5.964267 | 5.937382 | 5.929570 | 5.972977 | 5.918668 |
| hsa04390 | Hippo signaling pathway | 135 | 0.2455899 | 1.268483 | 3.805497e-02 | 1.096218e-01 | 7.199634e-02 | 3914 | tags=36%, list=27%, signal=26% | WNT8B||WTIP||MYC||PARD6A||PPP2R2C||TPTEP2-CSNK1E||SOX2||WNT10B||CCND3||TGFB1||WNT6||AXIN1||ACTG1||BMP2||ACTB||LLGL2||LLGL1||DVL1||PPP1CA||WNT3A||APC2||ID1||FZD9||TEAD4||PPP2R1A||SNAI2||LATS1||LATS2||SMAD4||TCF7L1||DVL3||TP73||DLG5||NKD2||TCF7||SCRIB||TEAD3||PRKCZ||FZD1||DVL2||TEAD2||BMP7||TP53BP2||NF2||BBC3||WNT2B||NKD1||RASSF1 | 6.225480 | 6.182642 | 5.864648 | 6.146631 | 6.232037 | 6.163088 | 6.165498 | 6.172552 | 6.192335 | 6.245355 | 6.238180 | 6.194859 | 6.180679 | 6.172297 | 5.797434 | 5.895946 | 5.898284 | 6.180768 | 6.074069 | 6.182408 | 6.295670 | 6.153156 | 6.243697 | 6.277992 | 6.141005 | 6.061933 | 6.158025 | 6.172172 | 6.166262 | 6.165842 | 6.186162 | 6.165554 |
| hsa03013 | Nucleocytoplasmic transport | 98 | -0.3622382 | -1.418479 | 3.841448e-02 | 1.096218e-01 | 7.199634e-02 | 3276 | tags=32%, list=23%, signal=25% | THOC2||NCBP1||TMEM33||MAGOH||SAP18||TNPO1||KPNA1||SUMO3||EIF4A3||NUP85||XPO7||NDC1||THOC3||NUP42||THOC1||SNUPN||NXT2||IPO5||IPO8||RAE1||CSE1L||IPO11||XPO4||NUP58||RAN||NUP155||NUP160||NUP37||EEF1A1||NUP43||NUP205 | 6.634100 | 7.685674 | 6.767342 | 7.668316 | 6.465975 | 6.373296 | 7.699191 | 7.710681 | 6.693915 | 6.814767 | 6.355271 | 7.767399 | 7.648530 | 7.637462 | 7.056428 | 6.410580 | 6.763325 | 7.663388 | 7.627110 | 7.713156 | 6.425037 | 6.647286 | 6.304425 | 6.372836 | 6.284857 | 6.457060 | 7.661140 | 7.717789 | 7.717903 | 7.730295 | 7.639917 | 7.759165 |
| hsa05161 | Hepatitis B | 131 | 0.2492888 | 1.284547 | 3.973064e-02 | 1.106782e-01 | 7.269014e-02 | 2661 | tags=27%, list=18%, signal=23% | JAK2||MYC||IRAK1||JUN||EP300||TICAM1||CREBBP||SOS1||AKT1||MAP2K2||ELK1||CDKN1A||HRAS||TGFB1||TAB1||MAP2K7||BAD||MAP2K3||RELA||TP53||CREB3L1||NFATC1||BAX||SMAD4||MAPK3||SOS2||MAPK12||CREB1||CREB3L2||STAT3||TRAF3||SRC||AKT2||ATP6AP1||MAVS||NFATC2 | 5.379189 | 5.699612 | 5.430174 | 5.738766 | 5.383314 | 5.544891 | 5.727114 | 5.789940 | 5.359990 | 5.404002 | 5.373220 | 5.707961 | 5.691975 | 5.698856 | 5.484957 | 5.411215 | 5.392691 | 5.737029 | 5.676689 | 5.799945 | 5.365138 | 5.370306 | 5.413998 | 5.520791 | 5.556304 | 5.557280 | 5.723611 | 5.715621 | 5.741982 | 5.773441 | 5.801432 | 5.794799 |
| hsa04310 | Wnt signaling pathway | 133 | 0.2436478 | 1.257120 | 4.413793e-02 | 1.200962e-01 | 7.887562e-02 | 3883 | tags=38%, list=27%, signal=28% | NOTUM||PLCB1||WNT8B||MYC||TPTEP2-CSNK1E||JUN||PLCB2||EP300||TLE6||CREBBP||WNT10B||CCND3||RAC3||WNT6||LRP5||AXIN1||PRKACA||PORCN||TP53||DVL1||WNT3A||APC2||FZD9||NFATC1||ROR1||SMAD4||SFRP5||TLE3||TCF7L1||CSNK2B||TLE2||DVL3||PPARD||NFATC2||NKD2||TLE4||CAMK2B||TCF7||TLE1||CTBP1||FZD1||DVL2||DAAM2||PPP3CA||PRKCA||ZNRF3||WNT2B||PRICKLE3||SERPINF1||NKD1||LGR5 | 4.760824 | 5.117283 | 4.896897 | 5.146953 | 4.756303 | 4.935633 | 5.113288 | 5.162190 | 4.739385 | 4.809582 | 4.732234 | 5.122814 | 5.122641 | 5.106331 | 4.968346 | 4.853142 | 4.866428 | 5.147554 | 5.107451 | 5.184816 | 4.769056 | 4.718718 | 4.780391 | 4.915511 | 4.917810 | 4.972844 | 5.121873 | 5.105967 | 5.111978 | 5.145604 | 5.177099 | 5.163694 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 80 | 0.2832792 | 1.332979 | 4.729400e-02 | 1.257591e-01 | 8.259479e-02 | 2128 | tags=26%, list=15%, signal=23% | JAK2||PLCB1||MMP2||JUN||PLCB2||PLCD3||VEGFB||COL1A1||COL4A4||AKT1||HRAS||TGFB1||THBD||RELA||COL4A3||PRKCE||NFATC1||BAX||SMAD4||MAPK3||MAPK12 | 4.843154 | 4.971409 | 4.885908 | 4.885794 | 4.883765 | 5.045624 | 4.963974 | 4.895307 | 4.764548 | 4.829839 | 4.930229 | 4.992672 | 4.963566 | 4.957745 | 4.883818 | 4.899980 | 4.873805 | 4.896454 | 4.825712 | 4.933157 | 4.891387 | 4.780923 | 4.972593 | 4.989927 | 5.080244 | 5.065089 | 4.989165 | 4.943488 | 4.958893 | 4.880771 | 4.919981 | 4.884846 |
ORA: All DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: All DEGs” results.
GO: BP
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0010457 | centriole-centriole cohesion | 2/42 | 14/20870 | 0.0003543162 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 4.318051 | 5.487879 | 5.031966 | 5.677370 | 4.225202 | 4.551264 | 5.484452 | 5.695344 | 4.305087 | 4.459975 | 4.174948 | 5.517517 | 5.506111 | 5.438760 | 5.285587 | 4.810304 | 4.958311 | 5.642203 | 5.678232 | 5.710857 | 4.202190 | 4.302561 | 4.167420 | 4.372135 | 4.419951 | 4.818817 | 5.504015 | 5.483828 | 5.465253 | 5.679781 | 5.724709 | 5.681088 |
| GO:0032786 | positive regulation of DNA-templated transcription, elongation | 2/42 | 34/20870 | 0.0021292947 | 0.2820041 | 0.2468512 | CCNT1||CTNNB1 | 2 | 6.286426 | 6.664121 | 6.352703 | 6.732317 | 6.251144 | 6.259029 | 6.722421 | 6.807268 | 6.262236 | 6.357493 | 6.236723 | 6.668241 | 6.667076 | 6.657021 | 6.519235 | 6.257272 | 6.265966 | 6.703654 | 6.701766 | 6.789768 | 6.264429 | 6.234113 | 6.254723 | 6.222563 | 6.203989 | 6.346346 | 6.719482 | 6.711548 | 6.736124 | 6.796412 | 6.828326 | 6.796832 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2/42 | 45/20870 | 0.0037053379 | 0.2820041 | 0.2468512 | NBAS||SMG8 | 2 | 5.826224 | 6.863748 | 5.940690 | 6.831910 | 5.759909 | 5.980144 | 6.907572 | 6.883645 | 5.818207 | 5.897205 | 5.759964 | 6.852338 | 6.864312 | 6.874510 | 6.235001 | 5.767267 | 5.767639 | 6.777477 | 6.807765 | 6.907270 | 5.724801 | 5.748087 | 5.805635 | 5.966783 | 6.038001 | 5.933670 | 6.868892 | 6.922384 | 6.930665 | 6.872288 | 6.897411 | 6.881123 |
| GO:0048489 | synaptic vesicle transport | 2/42 | 45/20870 | 0.0037053379 | 0.2820041 | 0.2468512 | AP3M2||CTNNB1 | 2 | 5.507588 | 5.701124 | 5.650028 | 5.672758 | 5.538711 | 5.676770 | 5.665455 | 5.676478 | 5.484903 | 5.553467 | 5.483276 | 5.729771 | 5.709735 | 5.663056 | 5.657414 | 5.650041 | 5.642590 | 5.688571 | 5.620661 | 5.707601 | 5.559248 | 5.480887 | 5.574266 | 5.691810 | 5.615862 | 5.720619 | 5.667938 | 5.651831 | 5.676485 | 5.673199 | 5.706150 | 5.649523 |
| GO:0031062 | positive regulation of histone methylation | 2/42 | 50/20870 | 0.0045558326 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.750006 | 5.550108 | 5.126810 | 5.630453 | 4.648402 | 4.803877 | 5.590397 | 5.677335 | 4.760291 | 4.892592 | 4.580202 | 5.573517 | 5.561269 | 5.514878 | 5.389858 | 4.900362 | 5.045580 | 5.598566 | 5.627867 | 5.664175 | 4.634212 | 4.729100 | 4.577828 | 4.762938 | 4.695444 | 4.941884 | 5.608590 | 5.580762 | 5.581664 | 5.657744 | 5.698325 | 5.675650 |
| GO:0009101 | glycoprotein biosynthetic process | 4/42 | 336/20870 | 0.0045622098 | 0.2820041 | 0.2468512 | MAGT1||OGT||CTNNB1||OGA | 4 | 4.682544 | 5.249157 | 5.018778 | 5.200676 | 4.723558 | 4.933462 | 5.149768 | 5.143748 | 4.648439 | 4.664582 | 4.733200 | 5.272412 | 5.280474 | 5.192974 | 5.026015 | 5.050159 | 4.979261 | 5.179833 | 5.236821 | 5.184678 | 4.718187 | 4.658518 | 4.790916 | 4.807814 | 4.856109 | 5.116931 | 5.174711 | 5.156777 | 5.117219 | 5.134121 | 5.184441 | 5.111717 |
| GO:0051650 | establishment of vesicle localization | 3/42 | 170/20870 | 0.0048287457 | 0.2820041 | 0.2468512 | AP3M2||SAR1A||CTNNB1 | 3 | 4.980056 | 5.450239 | 5.239036 | 5.422373 | 4.988541 | 5.200836 | 5.424174 | 5.429382 | 4.936233 | 5.027134 | 4.975358 | 5.448124 | 5.475190 | 5.426999 | 5.290537 | 5.220620 | 5.204495 | 5.411573 | 5.401887 | 5.453142 | 4.994078 | 4.933971 | 5.035764 | 5.155852 | 5.141861 | 5.299463 | 5.446825 | 5.417495 | 5.407918 | 5.423481 | 5.456547 | 5.407687 |
| GO:0050982 | detection of mechanical stimulus | 2/42 | 52/20870 | 0.0049189375 | 0.2820041 | 0.2468512 | ADGRV1||CTNNB1 | 2 | 3.499818 | 4.639988 | 4.173924 | 4.654266 | 3.412685 | 3.642595 | 4.617657 | 4.668828 | 3.561730 | 3.584483 | 3.340923 | 4.644063 | 4.671794 | 4.603285 | 4.428225 | 3.960261 | 4.092155 | 4.600020 | 4.672643 | 4.688599 | 3.310536 | 3.459335 | 3.463003 | 3.377338 | 3.439809 | 4.020084 | 4.665643 | 4.643476 | 4.540802 | 4.645476 | 4.718601 | 4.641087 |
| GO:0097479 | synaptic vesicle localization | 2/42 | 58/20870 | 0.0060854215 | 0.2820041 | 0.2468512 | AP3M2||CTNNB1 | 2 | 5.217470 | 5.445932 | 5.382752 | 5.424272 | 5.247815 | 5.406098 | 5.410268 | 5.427370 | 5.192904 | 5.261064 | 5.197430 | 5.468073 | 5.459020 | 5.410029 | 5.402756 | 5.379465 | 5.365793 | 5.434133 | 5.378916 | 5.458615 | 5.268603 | 5.185628 | 5.287199 | 5.409177 | 5.342047 | 5.464470 | 5.414745 | 5.397129 | 5.418840 | 5.423870 | 5.459347 | 5.398240 |
| GO:0051648 | vesicle localization | 3/42 | 188/20870 | 0.0063804352 | 0.2820041 | 0.2468512 | AP3M2||SAR1A||CTNNB1 | 3 | 4.881661 | 5.366256 | 5.142991 | 5.340596 | 4.886856 | 5.106106 | 5.340152 | 5.345965 | 4.839880 | 4.926916 | 4.876863 | 5.362321 | 5.392033 | 5.344007 | 5.200289 | 5.122905 | 5.103965 | 5.326628 | 5.321917 | 5.372697 | 4.893040 | 4.829594 | 4.935954 | 5.057488 | 5.048346 | 5.206927 | 5.362119 | 5.333988 | 5.324078 | 5.339365 | 5.375232 | 5.322800 |
| GO:0032784 | regulation of DNA-templated transcription, elongation | 2/42 | 66/20870 | 0.0078169223 | 0.2820041 | 0.2468512 | CCNT1||CTNNB1 | 2 | 5.958730 | 6.430697 | 6.068448 | 6.480772 | 5.923856 | 6.000263 | 6.475805 | 6.799439 | 5.922412 | 6.044570 | 5.905166 | 6.438347 | 6.433273 | 6.420411 | 6.256668 | 5.950816 | 5.977559 | 6.457350 | 6.443137 | 6.539924 | 5.966957 | 5.873771 | 5.929320 | 5.961885 | 5.960152 | 6.075687 | 6.466964 | 6.468479 | 6.491835 | 6.524698 | 7.229204 | 6.524329 |
| GO:0051568 | histone H3-K4 methylation | 2/42 | 70/20870 | 0.0087563413 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.026450 | 5.608113 | 5.451674 | 5.657117 | 4.988956 | 5.233215 | 5.623612 | 5.664912 | 5.015049 | 5.114676 | 4.944545 | 5.623073 | 5.616994 | 5.583966 | 5.612442 | 5.329157 | 5.398083 | 5.639656 | 5.639943 | 5.691140 | 4.967158 | 5.015707 | 4.983581 | 5.164419 | 5.109939 | 5.407579 | 5.642394 | 5.611257 | 5.616994 | 5.647188 | 5.701963 | 5.644857 |
| GO:0009100 | glycoprotein metabolic process | 4/42 | 415/20870 | 0.0095052649 | 0.2820041 | 0.2468512 | MAGT1||OGT||CTNNB1||OGA | 4 | 4.826752 | 5.336325 | 5.165029 | 5.289770 | 4.852387 | 5.077499 | 5.239369 | 5.236128 | 4.801154 | 4.813325 | 4.864978 | 5.355348 | 5.369110 | 5.283045 | 5.157754 | 5.203645 | 5.132791 | 5.279048 | 5.316361 | 5.273524 | 4.843153 | 4.796539 | 4.914995 | 4.950774 | 5.007408 | 5.255753 | 5.268282 | 5.244688 | 5.204414 | 5.225844 | 5.277689 | 5.203853 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2/42 | 74/20870 | 0.0097437992 | 0.2820041 | 0.2468512 | SELENOK||TXNDC12 | 2 | 5.839547 | 6.100603 | 6.081685 | 6.195694 | 5.787172 | 5.930769 | 6.048638 | 6.176093 | 5.865195 | 5.934880 | 5.709420 | 6.121276 | 6.113808 | 6.066107 | 6.123862 | 6.000933 | 6.116983 | 6.211288 | 6.155738 | 6.219231 | 5.748081 | 5.875035 | 5.734184 | 5.893877 | 5.831452 | 6.057440 | 6.078128 | 6.032101 | 6.035226 | 6.176000 | 6.193362 | 6.158707 |
| GO:0031016 | pancreas development | 2/42 | 79/20870 | 0.0110444676 | 0.2820041 | 0.2468512 | CTNNB1||SELENOT | 2 | 4.010021 | 4.679029 | 4.337938 | 4.680418 | 4.004671 | 4.244189 | 4.644119 | 4.682639 | 3.993476 | 4.036143 | 4.000076 | 4.714118 | 4.695414 | 4.626072 | 4.456530 | 4.245329 | 4.303629 | 4.666934 | 4.659211 | 4.714485 | 3.964888 | 3.962747 | 4.083062 | 4.121147 | 4.238848 | 4.362478 | 4.655243 | 4.648032 | 4.628953 | 4.674603 | 4.712568 | 4.660238 |
| GO:0044242 | cellular lipid catabolic process | 3/42 | 237/20870 | 0.0119836731 | 0.2820041 | 0.2468512 | CPS1||SLC25A17||PPT1 | 3 | 5.005694 | 5.197128 | 5.202633 | 5.215226 | 4.989172 | 5.124164 | 5.146970 | 5.183058 | 4.991541 | 5.020678 | 5.004715 | 5.230945 | 5.201122 | 5.158398 | 5.254230 | 5.180042 | 5.172199 | 5.217895 | 5.190365 | 5.237038 | 4.984216 | 4.962707 | 5.020012 | 5.031598 | 5.098455 | 5.234941 | 5.171214 | 5.138882 | 5.130493 | 5.170218 | 5.216898 | 5.161438 |
| GO:0006486 | protein glycosylation | 3/42 | 240/20870 | 0.0123949928 | 0.2820041 | 0.2468512 | MAGT1||OGT||OGA | 3 | 4.900737 | 5.420245 | 5.220616 | 5.374274 | 4.946433 | 5.145543 | 5.317982 | 5.318482 | 4.867393 | 4.877461 | 4.955726 | 5.446705 | 5.447970 | 5.364483 | 5.214883 | 5.259876 | 5.186132 | 5.358293 | 5.411332 | 5.352465 | 4.943167 | 4.879015 | 5.013957 | 5.027513 | 5.068287 | 5.322753 | 5.342616 | 5.324288 | 5.286474 | 5.306843 | 5.363542 | 5.283891 |
| GO:0043413 | macromolecule glycosylation | 3/42 | 240/20870 | 0.0123949928 | 0.2820041 | 0.2468512 | MAGT1||OGT||OGA | 3 | 4.900737 | 5.420245 | 5.220616 | 5.374274 | 4.946433 | 5.145543 | 5.317982 | 5.318482 | 4.867393 | 4.877461 | 4.955726 | 5.446705 | 5.447970 | 5.364483 | 5.214883 | 5.259876 | 5.186132 | 5.358293 | 5.411332 | 5.352465 | 4.943167 | 4.879015 | 5.013957 | 5.027513 | 5.068287 | 5.322753 | 5.342616 | 5.324288 | 5.286474 | 5.306843 | 5.363542 | 5.283891 |
| GO:0031060 | regulation of histone methylation | 2/42 | 86/20870 | 0.0129865134 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.959030 | 5.537053 | 5.314570 | 5.582833 | 4.894046 | 5.107731 | 5.575414 | 5.617332 | 4.938236 | 5.067591 | 4.863853 | 5.553040 | 5.545649 | 5.512142 | 5.485610 | 5.177762 | 5.262575 | 5.562470 | 5.564283 | 5.620975 | 4.896745 | 4.920251 | 4.864601 | 5.077408 | 4.996953 | 5.238281 | 5.591859 | 5.566242 | 5.567997 | 5.598433 | 5.642485 | 5.610719 |
| GO:1901264 | carbohydrate derivative transport | 2/42 | 86/20870 | 0.0129865134 | 0.2820041 | 0.2468512 | SLC25A17||RSC1A1 | 2 | 6.199161 | 5.964336 | 6.252716 | 5.966508 | 6.217881 | 6.298529 | 5.838792 | 5.913297 | 6.190638 | 6.232173 | 6.174049 | 6.026962 | 5.971292 | 5.891551 | 6.135743 | 6.299603 | 6.316007 | 6.023343 | 5.924545 | 5.949799 | 6.226994 | 6.193210 | 6.233120 | 6.304625 | 6.319828 | 6.270697 | 5.867036 | 5.830690 | 5.818204 | 5.910628 | 5.943493 | 5.885177 |
| GO:0015931 | nucleobase-containing compound transport | 3/42 | 245/20870 | 0.0130984696 | 0.2820041 | 0.2468512 | SLC25A17||NUP205||RSC1A1 | 3 | 6.438449 | 7.082579 | 6.510346 | 7.094881 | 6.373040 | 6.462428 | 7.127235 | 7.150162 | 6.438940 | 6.552925 | 6.313564 | 7.098168 | 7.068785 | 7.080632 | 6.712350 | 6.364320 | 6.430211 | 7.078043 | 7.059849 | 7.145340 | 6.366227 | 6.413224 | 6.338682 | 6.468742 | 6.439786 | 6.478475 | 7.115484 | 7.122134 | 7.143934 | 7.134435 | 7.158441 | 7.157482 |
| GO:0006493 | protein O-linked glycosylation | 2/42 | 89/20870 | 0.0138611687 | 0.2820041 | 0.2468512 | OGT||OGA | 2 | 3.867728 | 4.448050 | 4.315947 | 4.515674 | 3.910499 | 4.214484 | 4.381159 | 4.457858 | 3.822673 | 3.807326 | 3.967696 | 4.439906 | 4.493731 | 4.409241 | 4.373988 | 4.352283 | 4.216580 | 4.468770 | 4.548157 | 4.528912 | 3.896881 | 3.820563 | 4.007881 | 4.030643 | 4.144713 | 4.437110 | 4.413092 | 4.376471 | 4.353283 | 4.445393 | 4.517968 | 4.408040 |
| GO:0070085 | glycosylation | 3/42 | 255/20870 | 0.0145731090 | 0.2820041 | 0.2468512 | MAGT1||OGT||OGA | 3 | 4.910584 | 5.409970 | 5.221002 | 5.358037 | 4.956186 | 5.147244 | 5.308123 | 5.299982 | 4.880487 | 4.887176 | 4.962639 | 5.435063 | 5.438398 | 5.354909 | 5.215864 | 5.259030 | 5.187205 | 5.345321 | 5.392392 | 5.335760 | 4.953172 | 4.891342 | 5.021121 | 5.030589 | 5.069649 | 5.323617 | 5.334816 | 5.312137 | 5.276825 | 5.285607 | 5.347421 | 5.265653 |
| GO:1903312 | negative regulation of mRNA metabolic process | 2/42 | 98/20870 | 0.0166335447 | 0.2820041 | 0.2468512 | CCNT1||NBAS | 2 | 6.953586 | 7.805895 | 7.094086 | 7.788960 | 6.870143 | 7.014798 | 7.845545 | 7.842174 | 6.979546 | 7.091920 | 6.771142 | 7.820955 | 7.779731 | 7.816646 | 7.344902 | 6.906732 | 6.992341 | 7.757249 | 7.749874 | 7.857246 | 6.848988 | 6.941472 | 6.817059 | 7.010416 | 6.951300 | 7.079806 | 7.817907 | 7.835091 | 7.882849 | 7.830747 | 7.847086 | 7.848621 |
| GO:0006354 | DNA-templated transcription, elongation | 2/42 | 103/20870 | 0.0182676376 | 0.2820041 | 0.2468512 | CCNT1||CTNNB1 | 2 | 6.089300 | 6.285959 | 6.096424 | 6.339595 | 6.078955 | 6.157008 | 6.318898 | 6.568793 | 6.056438 | 6.100227 | 6.110664 | 6.299241 | 6.289309 | 6.269163 | 6.187709 | 6.045225 | 6.051809 | 6.341509 | 6.292760 | 6.383102 | 6.091101 | 6.021726 | 6.122212 | 6.128507 | 6.207809 | 6.133331 | 6.327104 | 6.306684 | 6.322824 | 6.379862 | 6.885576 | 6.379760 |
| GO:0031058 | positive regulation of histone modification | 2/42 | 103/20870 | 0.0182676376 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.273117 | 5.635880 | 5.472630 | 5.664516 | 5.245684 | 5.443652 | 5.660638 | 5.705226 | 5.228978 | 5.324696 | 5.264046 | 5.662513 | 5.636461 | 5.608153 | 5.590155 | 5.408622 | 5.411520 | 5.662914 | 5.622010 | 5.707361 | 5.249533 | 5.205437 | 5.281083 | 5.418241 | 5.420622 | 5.490904 | 5.680634 | 5.650586 | 5.650485 | 5.684857 | 5.731238 | 5.699191 |
| GO:0000050 | urea cycle | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.262866 | 4.753968 | 4.216298 | 4.037180 | 4.203726 | 4.138404 | 4.612521 | 3.982695 | 4.248734 | 4.311588 | 4.226930 | 4.830151 | 4.742218 | 4.685856 | 4.216070 | 4.167974 | 4.263276 | 4.070867 | 3.944751 | 4.091589 | 4.188139 | 4.210883 | 4.212030 | 4.153726 | 4.143606 | 4.117641 | 4.628370 | 4.597855 | 4.611176 | 3.965977 | 4.022091 | 3.959187 |
| GO:0002084 | protein depalmitoylation | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.069159 | 5.817692 | 5.362781 | 5.852175 | 5.085838 | 5.187962 | 5.755626 | 5.802579 | 5.066708 | 5.046249 | 5.094122 | 5.920028 | 5.779001 | 5.748167 | 5.455138 | 5.274479 | 5.353025 | 5.830841 | 5.845893 | 5.879361 | 5.037843 | 5.068634 | 5.148754 | 5.067097 | 5.247463 | 5.242129 | 5.751446 | 5.765285 | 5.750099 | 5.780017 | 5.855600 | 5.770607 |
| GO:0007440 | foregut morphogenesis | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.019108 | 5.761821 | 4.903341 | 5.928440 | 3.760901 | 3.779080 | 5.722424 | 5.942940 | 4.141728 | 4.348895 | 3.407489 | 5.820579 | 5.794598 | 5.665556 | 5.265781 | 4.371943 | 4.935465 | 5.899650 | 5.935229 | 5.949979 | 3.619243 | 4.126527 | 3.448109 | 3.659226 | 3.520541 | 4.093817 | 5.741108 | 5.718535 | 5.707423 | 5.934868 | 5.955063 | 5.938808 |
| GO:0044848 | biological phase | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.726582 | 5.458386 | 4.890690 | 5.570369 | 4.575860 | 4.491023 | 5.397307 | 5.528887 | 4.811545 | 4.804027 | 4.549012 | 5.515384 | 5.484355 | 5.371478 | 5.049083 | 4.653473 | 4.941125 | 5.566221 | 5.584105 | 5.560676 | 4.597032 | 4.644232 | 4.481494 | 4.491747 | 4.470625 | 4.510423 | 5.432043 | 5.383960 | 5.375268 | 5.526469 | 5.538622 | 5.521515 |
| GO:0048263 | determination of dorsal identity | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.031406 | 5.863292 | 4.892495 | 6.024686 | 3.706003 | 3.584103 | 5.788910 | 6.009712 | 4.186254 | 4.376437 | 3.333622 | 5.944344 | 5.885735 | 5.753207 | 5.298975 | 4.229623 | 4.953141 | 5.991359 | 6.043379 | 6.038748 | 3.490740 | 4.134779 | 3.370523 | 3.407849 | 3.355850 | 3.919054 | 5.810402 | 5.786083 | 5.769956 | 6.011970 | 6.027201 | 5.989720 |
| GO:0060272 | embryonic skeletal joint morphogenesis | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.782839 | 5.266779 | 4.525953 | 5.417058 | 3.578189 | 3.556549 | 5.198897 | 5.398491 | 3.863101 | 4.058472 | 3.333275 | 5.325153 | 5.315737 | 5.153043 | 4.799533 | 4.101472 | 4.590960 | 5.397542 | 5.433143 | 5.420262 | 3.508772 | 3.818771 | 3.369809 | 3.418839 | 3.310518 | 3.875966 | 5.218642 | 5.191472 | 5.186367 | 5.396558 | 5.408309 | 5.390549 |
| GO:0060439 | trachea morphogenesis | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.776232 | 5.873128 | 5.352962 | 5.973485 | 4.709649 | 4.832058 | 5.861004 | 5.987171 | 4.805518 | 4.939654 | 4.557888 | 5.912865 | 5.901645 | 5.802328 | 5.604790 | 5.059186 | 5.343586 | 5.959827 | 5.974665 | 5.985844 | 4.624959 | 4.831529 | 4.664012 | 4.728300 | 4.708831 | 5.035275 | 5.871724 | 5.855929 | 5.855299 | 5.973311 | 5.993543 | 5.994561 |
| GO:0090037 | positive regulation of protein kinase C signaling | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 3.070775 | 3.346961 | 3.719934 | 3.369778 | 3.157215 | 3.578420 | 3.341566 | 3.414134 | 2.977457 | 3.005810 | 3.216997 | 3.363700 | 3.317371 | 3.359360 | 3.895822 | 3.674089 | 3.570523 | 3.410628 | 3.314521 | 3.382501 | 3.174358 | 2.953274 | 3.320549 | 3.268840 | 3.442379 | 3.938067 | 3.378100 | 3.343178 | 3.302426 | 3.401111 | 3.528955 | 3.303444 |
| GO:0044262 | cellular carbohydrate metabolic process | 3/42 | 292/20870 | 0.0208240440 | 0.2820041 | 0.2468512 | OGT||BPNT1||SLC5A3 | 3 | 5.329203 | 5.412600 | 5.443193 | 5.467164 | 5.367576 | 5.507164 | 5.428294 | 5.478319 | 5.286816 | 5.321192 | 5.378123 | 5.414734 | 5.421972 | 5.401015 | 5.436678 | 5.469923 | 5.422568 | 5.457656 | 5.421542 | 5.520549 | 5.367278 | 5.320691 | 5.413275 | 5.479203 | 5.493094 | 5.548265 | 5.432539 | 5.418411 | 5.433881 | 5.471482 | 5.489537 | 5.473872 |
| GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 2/42 | 111/20870 | 0.0210173142 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 8.364296 | 8.325810 | 8.034230 | 8.265866 | 8.309812 | 7.991268 | 8.324788 | 8.267789 | 8.439584 | 8.419851 | 8.223742 | 8.361510 | 8.292652 | 8.322441 | 8.055229 | 7.938979 | 8.103559 | 8.294871 | 8.241217 | 8.261000 | 8.280245 | 8.426790 | 8.214123 | 8.075687 | 8.098080 | 7.778522 | 8.322109 | 8.307623 | 8.344395 | 8.268677 | 8.224115 | 8.309316 |
| GO:0003263 | cardioblast proliferation | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.974345 | 5.510557 | 4.801961 | 5.709595 | 3.773164 | 3.903646 | 5.468979 | 5.709613 | 4.105747 | 4.240125 | 3.464618 | 5.576149 | 5.533423 | 5.417462 | 5.143816 | 4.351551 | 4.803087 | 5.693283 | 5.715200 | 5.720159 | 3.680403 | 4.048597 | 3.541948 | 3.789858 | 3.601726 | 4.242408 | 5.492024 | 5.457782 | 5.456850 | 5.700744 | 5.727665 | 5.700259 |
| GO:0003264 | regulation of cardioblast proliferation | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.974345 | 5.510557 | 4.801961 | 5.709595 | 3.773164 | 3.903646 | 5.468979 | 5.709613 | 4.105747 | 4.240125 | 3.464618 | 5.576149 | 5.533423 | 5.417462 | 5.143816 | 4.351551 | 4.803087 | 5.693283 | 5.715200 | 5.720159 | 3.680403 | 4.048597 | 3.541948 | 3.789858 | 3.601726 | 4.242408 | 5.492024 | 5.457782 | 5.456850 | 5.700744 | 5.727665 | 5.700259 |
| GO:0033234 | negative regulation of protein sumoylation | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.105625 | 6.624912 | 6.581303 | 6.773236 | 5.899356 | 5.864026 | 6.627485 | 6.774963 | 6.216591 | 6.324840 | 5.701986 | 6.648896 | 6.627838 | 6.597543 | 6.732738 | 6.330967 | 6.649801 | 6.794378 | 6.734884 | 6.789692 | 5.781097 | 6.146362 | 5.734406 | 5.772491 | 5.753933 | 6.046651 | 6.648194 | 6.600379 | 6.633467 | 6.757077 | 6.786055 | 6.781588 |
| GO:0042989 | sequestering of actin monomers | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | TMSB15A | 1 | 7.540491 | 6.485058 | 6.945485 | 6.932227 | 7.489460 | 6.863027 | 6.510972 | 6.993968 | 7.667076 | 7.564411 | 7.374918 | 6.581638 | 6.402066 | 6.465693 | 6.858476 | 6.891828 | 7.076465 | 6.983356 | 6.856507 | 6.953785 | 7.452901 | 7.648256 | 7.351283 | 7.126047 | 6.825142 | 6.587118 | 6.543019 | 6.478360 | 6.510812 | 6.961714 | 6.990878 | 7.028535 |
| GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | RSC1A1 | 1 | 9.073771 | 8.557521 | 8.859841 | 8.503891 | 9.136981 | 9.138581 | 8.522043 | 8.501808 | 9.014612 | 8.995354 | 9.202171 | 8.601061 | 8.551185 | 8.519135 | 8.621458 | 8.960293 | 8.971134 | 8.568863 | 8.447316 | 8.492872 | 9.146143 | 9.078208 | 9.184587 | 9.210067 | 9.246102 | 8.940692 | 8.515292 | 8.535490 | 8.515252 | 8.498172 | 8.502998 | 8.504246 |
| GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | EMC3 | 1 | 6.021302 | 6.326249 | 6.134075 | 6.278797 | 5.877234 | 5.954097 | 6.249406 | 6.234483 | 6.061417 | 6.021047 | 5.980301 | 6.342157 | 6.334987 | 6.301274 | 6.128752 | 6.120721 | 6.152562 | 6.296557 | 6.287626 | 6.251822 | 5.876958 | 5.823458 | 5.929344 | 5.871173 | 5.948511 | 6.037782 | 6.283002 | 6.223818 | 6.240753 | 6.221421 | 6.259323 | 6.222382 |
| GO:0048262 | determination of dorsal/ventral asymmetry | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.949523 | 5.802298 | 4.736327 | 5.942855 | 3.637239 | 3.526067 | 5.737932 | 5.945566 | 4.098650 | 4.283386 | 3.284588 | 5.865177 | 5.826002 | 5.711320 | 5.160109 | 4.075136 | 4.773646 | 5.901551 | 5.959876 | 5.966262 | 3.428077 | 4.041214 | 3.336456 | 3.402649 | 3.354351 | 3.782116 | 5.746838 | 5.740724 | 5.726155 | 5.940482 | 5.964119 | 5.931903 |
| GO:0060742 | epithelial cell differentiation involved in prostate gland development | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.022225 | 6.629284 | 6.595708 | 6.720524 | 6.009211 | 6.320140 | 6.538535 | 6.655493 | 5.960989 | 6.144103 | 5.953390 | 6.697269 | 6.653229 | 6.532343 | 6.594428 | 6.592296 | 6.600389 | 6.720956 | 6.725673 | 6.714922 | 6.031770 | 6.021747 | 5.973443 | 6.212529 | 6.033247 | 6.644619 | 6.553660 | 6.536028 | 6.525779 | 6.671171 | 6.660319 | 6.634748 |
| GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.355605 | 6.050842 | 5.738427 | 6.145062 | 5.315079 | 5.386958 | 6.037946 | 6.156602 | 5.337705 | 5.488764 | 5.228454 | 6.097371 | 6.081003 | 5.970901 | 5.840855 | 5.565269 | 5.794348 | 6.139840 | 6.132648 | 6.162530 | 5.319134 | 5.353807 | 5.271103 | 5.314989 | 5.236217 | 5.585920 | 6.064872 | 6.023481 | 6.025101 | 6.165069 | 6.152262 | 6.152438 |
| GO:0070601 | centromeric sister chromatid cohesion | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.585342 | 6.978778 | 6.580504 | 6.938714 | 6.564130 | 6.641947 | 6.966043 | 6.985288 | 6.560518 | 6.561230 | 6.633066 | 7.023087 | 6.960860 | 6.951329 | 6.688091 | 6.446931 | 6.596293 | 6.928569 | 6.897085 | 6.988973 | 6.544520 | 6.502971 | 6.641383 | 6.625764 | 6.777710 | 6.509815 | 6.984526 | 6.946103 | 6.967243 | 6.978537 | 6.999142 | 6.978085 |
| GO:0044344 | cellular response to fibroblast growth factor stimulus | 2/42 | 115/20870 | 0.0224529946 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.273541 | 4.280472 | 3.783727 | 4.310241 | 3.236434 | 3.514922 | 4.262362 | 4.295562 | 3.262676 | 3.338878 | 3.216409 | 4.283385 | 4.311566 | 4.245710 | 3.953663 | 3.674678 | 3.706369 | 4.276974 | 4.323815 | 4.329362 | 3.205283 | 3.240503 | 3.262931 | 3.381010 | 3.406481 | 3.730355 | 4.285943 | 4.254403 | 4.246436 | 4.284237 | 4.317043 | 4.285163 |
| GO:0019751 | polyol metabolic process | 2/42 | 118/20870 | 0.0235557933 | 0.2820041 | 0.2468512 | BPNT1||SLC5A3 | 2 | 5.642470 | 5.277912 | 5.637300 | 5.334206 | 5.726575 | 5.814037 | 5.263944 | 5.332793 | 5.557484 | 5.569409 | 5.788586 | 5.305675 | 5.287843 | 5.239407 | 5.534141 | 5.699561 | 5.672794 | 5.356616 | 5.270359 | 5.373534 | 5.714491 | 5.651193 | 5.809618 | 5.790942 | 5.897523 | 5.749581 | 5.282758 | 5.253061 | 5.255827 | 5.325077 | 5.345944 | 5.327267 |
| GO:0003306 | Wnt signaling pathway involved in heart development | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.822548 | 5.492218 | 4.547769 | 5.663990 | 3.543254 | 3.390835 | 5.433174 | 5.657189 | 3.989823 | 4.100811 | 3.232523 | 5.551332 | 5.527688 | 5.392606 | 4.934775 | 3.948456 | 4.593570 | 5.622940 | 5.684328 | 5.683843 | 3.392404 | 3.889023 | 3.271745 | 3.376211 | 3.291781 | 3.497104 | 5.447086 | 5.430871 | 5.421448 | 5.659791 | 5.658739 | 5.653029 |
| GO:0006020 | inositol metabolic process | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | SLC5A3 | 1 | 4.720464 | 4.649936 | 5.057921 | 4.793895 | 4.759914 | 5.143587 | 4.696894 | 4.832931 | 4.649341 | 4.627276 | 4.871872 | 4.633840 | 4.691998 | 4.623012 | 5.089160 | 5.097853 | 4.983988 | 4.817245 | 4.709513 | 4.851164 | 4.806135 | 4.524103 | 4.921139 | 4.956594 | 5.077558 | 5.365528 | 4.762570 | 4.688013 | 4.637343 | 4.806723 | 4.910334 | 4.778367 |
| GO:0007042 | lysosomal lumen acidification | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.851089 | 6.350054 | 6.296515 | 6.357931 | 5.906390 | 6.114170 | 6.150319 | 6.224873 | 5.808606 | 5.844606 | 5.898629 | 6.432915 | 6.371834 | 6.238629 | 6.224602 | 6.350590 | 6.311486 | 6.396846 | 6.383081 | 6.291613 | 5.906148 | 5.844348 | 5.966108 | 5.936792 | 5.988694 | 6.375990 | 6.201926 | 6.142437 | 6.104932 | 6.225910 | 6.280729 | 6.165686 |
| GO:0009950 | dorsal/ventral axis specification | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.682251 | 5.440082 | 4.500793 | 5.611397 | 3.391255 | 3.294143 | 5.399783 | 5.607603 | 3.810736 | 3.994129 | 3.094829 | 5.504849 | 5.466615 | 5.343926 | 4.907744 | 3.922414 | 4.507131 | 5.579117 | 5.617519 | 5.636954 | 3.253997 | 3.747087 | 3.089085 | 3.151304 | 3.050920 | 3.615389 | 5.410811 | 5.399987 | 5.388465 | 5.606935 | 5.616809 | 5.599010 |
| GO:0019627 | urea metabolic process | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.262866 | 4.753968 | 4.216298 | 4.037180 | 4.203726 | 4.138404 | 4.612521 | 3.982695 | 4.248734 | 4.311588 | 4.226930 | 4.830151 | 4.742218 | 4.685856 | 4.216070 | 4.167974 | 4.263276 | 4.070867 | 3.944751 | 4.091589 | 4.188139 | 4.210883 | 4.212030 | 4.153726 | 4.143606 | 4.117641 | 4.628370 | 4.597855 | 4.611176 | 3.965977 | 4.022091 | 3.959187 |
| GO:0035112 | genitalia morphogenesis | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.822736 | 5.828054 | 4.788151 | 6.010714 | 3.455638 | 3.109134 | 5.774687 | 6.009180 | 4.027482 | 4.226768 | 2.891245 | 5.899007 | 5.856911 | 5.722405 | 5.233520 | 4.051036 | 4.842512 | 5.964812 | 6.033898 | 6.032359 | 3.247202 | 3.979561 | 2.934178 | 2.994250 | 2.735375 | 3.492935 | 5.785272 | 5.776810 | 5.761883 | 6.001881 | 6.015068 | 6.010561 |
| GO:0048715 | negative regulation of oligodendrocyte differentiation | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.170971 | 5.450019 | 4.800812 | 5.559395 | 4.034610 | 4.092259 | 5.395354 | 5.557069 | 4.239621 | 4.370530 | 3.854295 | 5.511016 | 5.485579 | 5.348203 | 5.069634 | 4.444306 | 4.821043 | 5.546845 | 5.567020 | 5.564238 | 3.989992 | 4.172068 | 3.930669 | 3.997531 | 3.880201 | 4.355589 | 5.423138 | 5.382479 | 5.380036 | 5.548941 | 5.572620 | 5.549519 |
| GO:0060123 | regulation of growth hormone secretion | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | SELENOT | 1 | 3.355548 | 5.031099 | 3.680747 | 5.000469 | 3.111499 | 3.388222 | 5.113121 | 5.086264 | 3.397182 | 3.556729 | 3.071144 | 5.016108 | 5.026565 | 5.050409 | 4.069952 | 3.258634 | 3.597601 | 4.873378 | 4.979491 | 5.136345 | 3.156699 | 3.148149 | 3.025979 | 3.465384 | 3.384796 | 3.310314 | 5.080612 | 5.152742 | 5.105073 | 5.078212 | 5.116917 | 5.063128 |
| GO:0060767 | epithelial cell proliferation involved in prostate gland development | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.210970 | 5.905060 | 5.592999 | 5.999929 | 5.171914 | 5.245224 | 5.891585 | 6.011468 | 5.193842 | 5.343724 | 5.083471 | 5.950172 | 5.936014 | 5.825808 | 5.695415 | 5.420587 | 5.648295 | 5.994629 | 5.987619 | 6.017370 | 5.175552 | 5.210074 | 5.128968 | 5.172828 | 5.094864 | 5.444242 | 5.917723 | 5.877470 | 5.879203 | 6.019934 | 6.007549 | 6.006884 |
| GO:0060788 | ectodermal placode formation | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.110723 | 6.600917 | 5.751653 | 6.709326 | 4.910418 | 5.018709 | 6.616440 | 6.741341 | 5.199344 | 5.372962 | 4.671062 | 6.645857 | 6.605897 | 6.549372 | 6.108657 | 5.366345 | 5.683139 | 6.688013 | 6.695613 | 6.743716 | 4.876271 | 5.145807 | 4.669431 | 4.949772 | 4.686124 | 5.343574 | 6.608273 | 6.607628 | 6.633270 | 6.722135 | 6.758163 | 6.743499 |
| GO:0071377 | cellular response to glucagon stimulus | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.394864 | 5.218736 | 4.043511 | 3.956852 | 4.337042 | 4.213524 | 5.017685 | 3.894576 | 4.404147 | 4.382388 | 4.397969 | 5.326338 | 5.199739 | 5.122769 | 4.009603 | 4.056280 | 4.064053 | 3.940620 | 3.918576 | 4.009782 | 4.328705 | 4.313347 | 4.368511 | 4.362262 | 4.237392 | 4.020416 | 5.018407 | 5.048334 | 4.985634 | 3.910011 | 3.947772 | 3.823130 |
| GO:0071697 | ectodermal placode morphogenesis | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.110723 | 6.600917 | 5.751653 | 6.709326 | 4.910418 | 5.018709 | 6.616440 | 6.741341 | 5.199344 | 5.372962 | 4.671062 | 6.645857 | 6.605897 | 6.549372 | 6.108657 | 5.366345 | 5.683139 | 6.688013 | 6.695613 | 6.743716 | 4.876271 | 5.145807 | 4.669431 | 4.949772 | 4.686124 | 5.343574 | 6.608273 | 6.607628 | 6.633270 | 6.722135 | 6.758163 | 6.743499 |
| GO:0071941 | nitrogen cycle metabolic process | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.262866 | 4.753968 | 4.216298 | 4.037180 | 4.203726 | 4.138404 | 4.612521 | 3.982695 | 4.248734 | 4.311588 | 4.226930 | 4.830151 | 4.742218 | 4.685856 | 4.216070 | 4.167974 | 4.263276 | 4.070867 | 3.944751 | 4.091589 | 4.188139 | 4.210883 | 4.212030 | 4.153726 | 4.143606 | 4.117641 | 4.628370 | 4.597855 | 4.611176 | 3.965977 | 4.022091 | 3.959187 |
| GO:0071774 | response to fibroblast growth factor | 2/42 | 122/20870 | 0.0250603504 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.327762 | 4.253186 | 3.800685 | 4.274480 | 3.296967 | 3.577871 | 4.235277 | 4.260555 | 3.307889 | 3.380179 | 3.293720 | 4.256057 | 4.283337 | 4.219452 | 3.951037 | 3.712244 | 3.726110 | 4.243759 | 4.284661 | 4.294519 | 3.277179 | 3.276558 | 3.336343 | 3.449773 | 3.483435 | 3.777403 | 4.260289 | 4.226032 | 4.219173 | 4.247976 | 4.281908 | 4.251539 |
| GO:0007494 | midgut development | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CPS1 | 1 | 3.366681 | 3.953231 | 3.450559 | 3.246768 | 3.345066 | 3.462190 | 3.831041 | 3.212594 | 3.350536 | 3.352053 | 3.396970 | 4.018595 | 3.986140 | 3.849432 | 3.425912 | 3.458223 | 3.467214 | 3.215793 | 3.233555 | 3.289913 | 3.340172 | 3.297656 | 3.395693 | 3.412552 | 3.418362 | 3.551339 | 3.846067 | 3.840885 | 3.805840 | 3.231117 | 3.259492 | 3.144709 |
| GO:0010739 | positive regulation of protein kinase A signaling | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 9.758394 | 8.851893 | 9.247425 | 8.627674 | 9.839473 | 9.923020 | 8.648873 | 8.460644 | 9.731749 | 9.489519 | 10.007333 | 9.060642 | 8.720571 | 8.749241 | 8.879464 | 9.468512 | 9.330825 | 8.880882 | 8.487597 | 8.476754 | 9.855561 | 9.564954 | 10.056158 | 9.826975 | 10.213026 | 9.674502 | 8.647626 | 8.648984 | 8.650008 | 8.482433 | 8.454293 | 8.444941 |
| GO:0015791 | polyol transport | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | SLC5A3 | 1 | 1.876776 | 2.715215 | 3.217662 | 3.240215 | 1.951667 | 2.800008 | 2.873029 | 3.239115 | 1.689882 | 1.881171 | 2.038281 | 2.617180 | 2.807309 | 2.714897 | 3.313560 | 3.235792 | 3.095206 | 3.194432 | 3.264391 | 3.260753 | 1.871420 | 1.795183 | 2.161925 | 2.238031 | 2.517786 | 3.385262 | 2.975373 | 2.845661 | 2.791835 | 3.190388 | 3.314870 | 3.208934 |
| GO:0021781 | glial cell fate commitment | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.433846 | 5.273683 | 4.333427 | 5.454681 | 3.110581 | 2.923432 | 5.217428 | 5.445408 | 3.596402 | 3.778203 | 2.723437 | 5.343389 | 5.304495 | 5.167294 | 4.747612 | 3.689257 | 4.372698 | 5.425343 | 5.463823 | 5.474415 | 2.936351 | 3.548969 | 2.712127 | 2.755201 | 2.601711 | 3.312946 | 5.226388 | 5.220952 | 5.204859 | 5.438724 | 5.454465 | 5.442988 |
| GO:0032429 | regulation of phospholipase A2 activity | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | PPT1 | 1 | 3.377836 | 4.991773 | 4.292212 | 5.138562 | 3.087942 | 3.149547 | 4.856709 | 5.105324 | 3.498314 | 3.655862 | 2.864029 | 5.109239 | 4.969148 | 4.888212 | 4.634196 | 3.817010 | 4.311038 | 5.084665 | 5.187020 | 5.142179 | 3.039185 | 3.378456 | 2.784212 | 3.009861 | 2.881681 | 3.484956 | 4.842313 | 4.884227 | 4.843189 | 5.084971 | 5.158467 | 5.070996 |
| GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.916178 | 6.184543 | 5.274792 | 6.317106 | 4.754219 | 4.948420 | 6.238215 | 6.361911 | 4.901835 | 5.087523 | 4.737943 | 6.178413 | 6.217741 | 6.156811 | 5.648177 | 4.908925 | 5.167934 | 6.268988 | 6.302193 | 6.377963 | 4.736311 | 4.835977 | 4.686325 | 4.898664 | 4.970406 | 4.974929 | 6.228809 | 6.244169 | 6.241621 | 6.358173 | 6.372078 | 6.355425 |
| GO:0048548 | regulation of pinocytosis | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | PPT1 | 1 | 4.366222 | 5.522390 | 4.941477 | 5.648879 | 4.260752 | 4.335752 | 5.517020 | 5.695505 | 4.386340 | 4.577225 | 4.094736 | 5.583671 | 5.499778 | 5.481656 | 5.242249 | 4.559747 | 4.942456 | 5.587004 | 5.646298 | 5.710683 | 4.211256 | 4.438622 | 4.112771 | 4.279727 | 4.195343 | 4.513130 | 5.505514 | 5.526518 | 5.518950 | 5.669317 | 5.727485 | 5.689104 |
| GO:0048820 | hair follicle maturation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.006386 | 5.709607 | 5.283967 | 5.767784 | 4.883857 | 4.982140 | 5.696175 | 5.773260 | 5.041479 | 5.103736 | 4.863285 | 5.731778 | 5.738599 | 5.657033 | 5.447208 | 5.113869 | 5.271532 | 5.752899 | 5.773354 | 5.776982 | 4.923092 | 4.909888 | 4.816255 | 4.964944 | 4.901754 | 5.074406 | 5.718838 | 5.684935 | 5.684481 | 5.768667 | 5.788331 | 5.762657 |
| GO:0061046 | regulation of branching involved in lung morphogenesis | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.428014 | 6.382084 | 5.395448 | 6.551280 | 4.076479 | 3.761370 | 6.314812 | 6.543815 | 4.617376 | 4.830850 | 3.532719 | 6.450884 | 6.415288 | 6.274086 | 5.810533 | 4.696807 | 5.466483 | 6.520606 | 6.566305 | 6.566448 | 3.884308 | 4.580168 | 3.576011 | 3.623164 | 3.329433 | 4.194422 | 6.326534 | 6.316005 | 6.301791 | 6.534483 | 6.556907 | 6.539960 |
| GO:0061085 | regulation of histone H3-K27 methylation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | OGT | 1 | 5.063009 | 5.638408 | 5.560314 | 5.629488 | 5.038329 | 5.223784 | 5.632937 | 5.503203 | 5.059749 | 5.231469 | 4.875959 | 5.671863 | 5.619298 | 5.623470 | 5.728397 | 5.408940 | 5.525331 | 5.587868 | 5.605939 | 5.692475 | 5.029888 | 5.152388 | 4.923614 | 5.220884 | 4.999323 | 5.420508 | 5.659734 | 5.608375 | 5.630242 | 5.469697 | 5.563310 | 5.474664 |
| GO:0071696 | ectodermal placode development | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.984347 | 6.477260 | 5.620440 | 6.581163 | 4.783979 | 4.891787 | 6.490066 | 6.613027 | 5.071763 | 5.246159 | 4.547116 | 6.519160 | 6.484961 | 6.426128 | 5.976723 | 5.236610 | 5.551708 | 6.561583 | 6.566596 | 6.614712 | 4.749560 | 5.018572 | 4.544412 | 4.825956 | 4.565199 | 5.210462 | 6.481767 | 6.481208 | 6.507073 | 6.594114 | 6.630340 | 6.614398 |
| GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.261654 | 5.229418 | 4.169716 | 5.370776 | 2.917512 | 2.676653 | 5.180140 | 5.383155 | 3.432036 | 3.643548 | 2.454715 | 5.289249 | 5.260675 | 5.133621 | 4.610220 | 3.487255 | 4.198032 | 5.332556 | 5.385385 | 5.393630 | 2.734090 | 3.386735 | 2.473376 | 2.598733 | 2.360469 | 2.997873 | 5.180674 | 5.191412 | 5.168240 | 5.376049 | 5.393594 | 5.379762 |
| GO:0072497 | mesenchymal stem cell differentiation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.063510 | 5.207631 | 4.576614 | 5.278305 | 3.916279 | 3.855521 | 5.135041 | 5.244471 | 4.139131 | 4.210216 | 3.810503 | 5.260201 | 5.239454 | 5.119262 | 4.825911 | 4.211830 | 4.625948 | 5.253766 | 5.291787 | 5.289052 | 3.875432 | 4.056816 | 3.804738 | 3.783198 | 3.723624 | 4.039820 | 5.155388 | 5.130437 | 5.119058 | 5.232454 | 5.266746 | 5.233951 |
| GO:1900364 | negative regulation of mRNA polyadenylation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CCNT1 | 1 | 5.770945 | 6.236909 | 6.026429 | 6.217444 | 5.719692 | 6.033174 | 6.258902 | 6.256497 | 5.689087 | 5.824397 | 5.795855 | 6.237307 | 6.241353 | 6.232050 | 6.076112 | 6.044296 | 5.956221 | 6.182673 | 6.181062 | 6.286069 | 5.761941 | 5.667295 | 5.728248 | 5.912273 | 6.029479 | 6.148134 | 6.299599 | 6.232270 | 6.243936 | 6.223063 | 6.304385 | 6.240771 |
| GO:1990403 | embryonic brain development | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 8.919538 | 8.021670 | 8.670129 | 7.933080 | 9.010001 | 8.886555 | 8.026984 | 8.013662 | 8.939758 | 8.736155 | 9.063912 | 8.018859 | 8.079396 | 7.964462 | 8.441984 | 8.700133 | 8.840378 | 7.971574 | 7.918148 | 7.908719 | 9.054419 | 8.898906 | 9.070529 | 8.991413 | 9.019614 | 8.614251 | 8.073422 | 8.077678 | 7.924649 | 7.963560 | 8.077711 | 7.997319 |
| GO:1904659 | glucose transmembrane transport | 2/42 | 126/20870 | 0.0266033140 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 3.910897 | 4.488307 | 4.284345 | 4.541570 | 3.911282 | 4.250547 | 4.495726 | 4.524655 | 3.848629 | 3.935828 | 3.946255 | 4.469815 | 4.507059 | 4.487807 | 4.391516 | 4.283980 | 4.168965 | 4.503379 | 4.525034 | 4.594712 | 3.890248 | 3.839412 | 3.999518 | 4.103675 | 4.199219 | 4.429097 | 4.509366 | 4.486075 | 4.491633 | 4.519485 | 4.557369 | 4.496454 |
| GO:0048193 | Golgi vesicle transport | 3/42 | 326/20870 | 0.0276833253 | 0.2820041 | 0.2468512 | SAR1A||NBAS||RSC1A1 | 3 | 6.026866 | 6.132262 | 6.118246 | 6.082199 | 6.065056 | 6.198069 | 6.089379 | 6.078730 | 5.974567 | 6.007022 | 6.096241 | 6.151529 | 6.146221 | 6.098444 | 6.057391 | 6.163328 | 6.131976 | 6.093995 | 6.053840 | 6.098344 | 6.070141 | 6.002224 | 6.120372 | 6.181871 | 6.196986 | 6.215158 | 6.099713 | 6.091445 | 6.076888 | 6.071009 | 6.102416 | 6.062457 |
| GO:0000052 | citrulline metabolic process | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CPS1 | 1 | 5.208000 | 5.752459 | 5.585622 | 5.515343 | 5.121159 | 5.392200 | 5.676700 | 5.455829 | 5.235468 | 5.265333 | 5.119097 | 5.745509 | 5.795163 | 5.715582 | 5.615742 | 5.600196 | 5.539817 | 5.462749 | 5.520844 | 5.560755 | 5.127489 | 5.128021 | 5.107876 | 5.282680 | 5.133813 | 5.699139 | 5.707445 | 5.680915 | 5.640964 | 5.447953 | 5.506084 | 5.411880 |
| GO:0006999 | nuclear pore organization | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | NUP205 | 1 | 4.727484 | 6.215271 | 5.318037 | 6.353303 | 4.582429 | 4.961544 | 6.280218 | 6.426828 | 4.712357 | 4.952211 | 4.479159 | 6.194741 | 6.227266 | 6.223587 | 5.733778 | 4.971696 | 5.132694 | 6.268795 | 6.344595 | 6.441324 | 4.573996 | 4.673011 | 4.494738 | 5.046213 | 4.850701 | 4.980902 | 6.265463 | 6.282052 | 6.293005 | 6.396153 | 6.456253 | 6.427453 |
| GO:0031441 | negative regulation of mRNA 3'-end processing | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CCNT1 | 1 | 5.603515 | 6.097142 | 5.862302 | 6.081061 | 5.552510 | 5.865710 | 6.118450 | 6.132236 | 5.521731 | 5.657462 | 5.627849 | 6.092112 | 6.109112 | 6.090126 | 5.916822 | 5.877324 | 5.789849 | 6.044485 | 6.044684 | 6.151358 | 5.593893 | 5.501217 | 5.560895 | 5.744598 | 5.862204 | 5.980675 | 6.155612 | 6.093602 | 6.105380 | 6.088965 | 6.186338 | 6.119680 |
| GO:0034333 | adherens junction assembly | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 8.054318 | 7.668552 | 7.701730 | 7.601310 | 8.063195 | 8.047228 | 7.598870 | 7.577856 | 8.030170 | 8.053189 | 8.079179 | 7.667361 | 7.697642 | 7.640080 | 7.488546 | 7.803250 | 7.791857 | 7.686783 | 7.514023 | 7.597949 | 8.138981 | 7.969526 | 8.076020 | 8.144301 | 8.016510 | 7.975451 | 7.607530 | 7.616015 | 7.572700 | 7.599936 | 7.588731 | 7.544302 |
| GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.202563 | 7.059911 | 6.502002 | 6.975464 | 6.178471 | 6.299322 | 7.009876 | 6.984877 | 6.154535 | 6.296042 | 6.152362 | 7.057858 | 7.094401 | 7.026676 | 6.635525 | 6.376487 | 6.482178 | 6.964278 | 6.976381 | 6.985652 | 6.147944 | 6.196237 | 6.190749 | 6.219211 | 6.220239 | 6.446440 | 7.042116 | 6.998790 | 6.988155 | 6.976897 | 7.017609 | 6.959508 |
| GO:0048208 | COPII vesicle coating | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.202563 | 7.059911 | 6.502002 | 6.975464 | 6.178471 | 6.299322 | 7.009876 | 6.984877 | 6.154535 | 6.296042 | 6.152362 | 7.057858 | 7.094401 | 7.026676 | 6.635525 | 6.376487 | 6.482178 | 6.964278 | 6.976381 | 6.985652 | 6.147944 | 6.196237 | 6.190749 | 6.219211 | 6.220239 | 6.446440 | 7.042116 | 6.998790 | 6.988155 | 6.976897 | 7.017609 | 6.959508 |
| GO:0050667 | homocysteine metabolic process | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CPS1 | 1 | 5.699296 | 5.822978 | 5.854275 | 5.667816 | 5.779181 | 5.986813 | 5.761403 | 5.653922 | 5.651681 | 5.631935 | 5.807764 | 5.865582 | 5.856908 | 5.743245 | 5.805418 | 5.907438 | 5.848148 | 5.708656 | 5.622133 | 5.671356 | 5.796446 | 5.645554 | 5.885427 | 5.858243 | 5.996178 | 6.096174 | 5.809703 | 5.739827 | 5.733432 | 5.649982 | 5.709807 | 5.599877 |
| GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | SAR1A | 1 | 7.117005 | 6.919455 | 7.362000 | 6.925550 | 7.172971 | 7.378534 | 6.749864 | 6.867203 | 7.028314 | 7.002908 | 7.300542 | 6.973389 | 6.964864 | 6.814654 | 7.144745 | 7.503631 | 7.413927 | 6.945211 | 6.978096 | 6.850313 | 7.163357 | 7.059422 | 7.287111 | 7.279743 | 7.357815 | 7.490157 | 6.779679 | 6.762612 | 6.706281 | 6.857446 | 6.927409 | 6.814497 |
| GO:0072498 | embryonic skeletal joint development | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.470475 | 4.875899 | 4.202085 | 5.034418 | 3.288161 | 3.340355 | 4.810159 | 5.009321 | 3.538046 | 3.724248 | 3.074029 | 4.928981 | 4.923531 | 4.769578 | 4.454815 | 3.822575 | 4.258449 | 5.012108 | 5.049655 | 5.041221 | 3.227635 | 3.497417 | 3.111899 | 3.195375 | 3.099424 | 3.661601 | 4.834263 | 4.803901 | 4.791982 | 5.003546 | 5.016351 | 5.008036 |
| GO:0008645 | hexose transmembrane transport | 2/42 | 130/20870 | 0.0281840021 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 4.018011 | 4.530332 | 4.347029 | 4.578882 | 4.030440 | 4.345781 | 4.530434 | 4.551110 | 3.954927 | 4.030586 | 4.066289 | 4.520186 | 4.544127 | 4.526576 | 4.438426 | 4.356249 | 4.239519 | 4.539940 | 4.566264 | 4.628985 | 4.009212 | 3.951865 | 4.124829 | 4.215532 | 4.308877 | 4.498395 | 4.545531 | 4.518466 | 4.527173 | 4.546017 | 4.583752 | 4.522906 |
| GO:0034968 | histone lysine methylation | 2/42 | 131/20870 | 0.0285849894 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.983262 | 5.379789 | 5.275671 | 5.431893 | 4.972092 | 5.199626 | 5.405767 | 5.451188 | 4.943596 | 5.016527 | 4.988726 | 5.395071 | 5.381035 | 5.363081 | 5.395833 | 5.206589 | 5.216624 | 5.412841 | 5.400956 | 5.480595 | 4.979577 | 4.919443 | 5.015623 | 5.142490 | 5.144784 | 5.305479 | 5.425170 | 5.394140 | 5.397792 | 5.435744 | 5.478143 | 5.439292 |
| GO:0015749 | monosaccharide transmembrane transport | 2/42 | 132/20870 | 0.0289882819 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 4.009689 | 4.531466 | 4.346997 | 4.578489 | 4.021472 | 4.342779 | 4.531057 | 4.550399 | 3.946372 | 4.023472 | 4.057002 | 4.520517 | 4.546324 | 4.527433 | 4.439870 | 4.356832 | 4.237121 | 4.538554 | 4.567242 | 4.628212 | 4.001257 | 3.942619 | 4.115174 | 4.209281 | 4.301550 | 4.501833 | 4.545982 | 4.520033 | 4.527032 | 4.544803 | 4.582962 | 4.522789 |
| GO:0002679 | respiratory burst involved in defense response | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | SELENOK | 1 | 9.697135 | 9.008855 | 9.448125 | 8.842761 | 9.692611 | 9.580971 | 8.890680 | 8.796517 | 9.733745 | 9.605015 | 9.748377 | 9.086117 | 8.989835 | 8.947078 | 9.248899 | 9.556756 | 9.519583 | 8.928668 | 8.768104 | 8.826917 | 9.701870 | 9.602748 | 9.768415 | 9.630225 | 9.699079 | 9.396481 | 8.930429 | 8.865373 | 8.875382 | 8.793168 | 8.785013 | 8.811245 |
| GO:0030252 | growth hormone secretion | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | SELENOT | 1 | 5.026271 | 5.908669 | 5.610761 | 5.746560 | 5.053080 | 5.505696 | 5.928146 | 5.828500 | 4.929512 | 5.054772 | 5.089658 | 5.887736 | 5.927985 | 5.910004 | 5.745475 | 5.558380 | 5.518085 | 5.691137 | 5.710031 | 5.834280 | 4.999817 | 4.951396 | 5.196213 | 5.314349 | 5.425982 | 5.741996 | 5.935095 | 5.962810 | 5.885472 | 5.824707 | 5.893083 | 5.764855 |
| GO:0032239 | regulation of nucleobase-containing compound transport | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | RSC1A1 | 1 | 4.791809 | 6.103455 | 5.033763 | 6.109421 | 4.632461 | 4.960208 | 6.198619 | 6.156953 | 4.758135 | 4.957706 | 4.641741 | 6.062079 | 6.112351 | 6.134974 | 5.525185 | 4.659061 | 4.752562 | 6.054491 | 6.068430 | 6.200777 | 4.580923 | 4.668449 | 4.646580 | 4.973383 | 5.016118 | 4.888199 | 6.182533 | 6.223272 | 6.189723 | 6.150770 | 6.171415 | 6.148564 |
| GO:0042159 | lipoprotein catabolic process | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.277006 | 5.843844 | 5.519253 | 5.820153 | 5.319199 | 5.538825 | 5.715315 | 5.730959 | 5.275237 | 5.177100 | 5.372095 | 5.919534 | 5.840335 | 5.767663 | 5.469726 | 5.549192 | 5.537572 | 5.843397 | 5.802951 | 5.813807 | 5.288162 | 5.268699 | 5.397375 | 5.411041 | 5.592985 | 5.604411 | 5.735134 | 5.715411 | 5.695122 | 5.726120 | 5.784753 | 5.680092 |
| GO:0072160 | nephron tubule epithelial cell differentiation | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.031676 | 4.987204 | 3.932166 | 5.137764 | 2.685684 | 2.466261 | 4.941656 | 5.156717 | 3.189773 | 3.422572 | 2.228226 | 5.046532 | 5.019654 | 4.890663 | 4.376446 | 3.252520 | 3.953711 | 5.098517 | 5.153168 | 5.160810 | 2.505531 | 3.147452 | 2.251596 | 2.391907 | 2.153686 | 2.782441 | 4.938351 | 4.956573 | 4.929914 | 5.147879 | 5.170474 | 5.151697 |
| GO:0090110 | COPII-coated vesicle cargo loading | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | SAR1A | 1 | 5.131829 | 5.884984 | 5.471408 | 5.912995 | 5.124147 | 5.364404 | 5.879828 | 5.950802 | 5.126930 | 5.256804 | 5.000358 | 5.883122 | 5.891894 | 5.879908 | 5.649377 | 5.365282 | 5.381573 | 5.899057 | 5.893226 | 5.946118 | 5.110138 | 5.182915 | 5.077357 | 5.349783 | 5.242525 | 5.490179 | 5.883379 | 5.893064 | 5.862876 | 5.950180 | 5.973215 | 5.928666 |
| GO:0034219 | carbohydrate transmembrane transport | 2/42 | 134/20870 | 0.0298017408 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 3.978073 | 4.509439 | 4.318371 | 4.562716 | 3.989838 | 4.309953 | 4.507585 | 4.532671 | 3.915335 | 3.992647 | 4.024078 | 4.498446 | 4.524933 | 4.504804 | 4.414098 | 4.325425 | 4.208226 | 4.522447 | 4.552537 | 4.611728 | 3.968933 | 3.912601 | 4.082740 | 4.176330 | 4.268583 | 4.469232 | 4.522321 | 4.496495 | 4.503817 | 4.527194 | 4.565838 | 4.504311 |
| GO:0043467 | regulation of generation of precursor metabolites and energy | 2/42 | 134/20870 | 0.0298017408 | 0.2820041 | 0.2468512 | OGT||NDUFA4 | 2 | 6.608718 | 6.528474 | 6.577218 | 6.579274 | 6.597014 | 6.532015 | 6.518725 | 6.576511 | 6.614946 | 6.643457 | 6.566711 | 6.570992 | 6.506438 | 6.507035 | 6.549551 | 6.566454 | 6.614849 | 6.606679 | 6.535278 | 6.594855 | 6.609028 | 6.602888 | 6.578951 | 6.564086 | 6.503929 | 6.527392 | 6.532130 | 6.493825 | 6.529901 | 6.582211 | 6.564471 | 6.582776 |
| GO:0009855 | determination of bilateral symmetry | 2/42 | 137/20870 | 0.0310389583 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 3.401227 | 4.373120 | 3.760697 | 4.385691 | 3.360780 | 3.611996 | 4.353899 | 4.369719 | 3.403402 | 3.477703 | 3.318168 | 4.364826 | 4.399866 | 4.354271 | 3.954995 | 3.637988 | 3.667390 | 4.344191 | 4.389736 | 4.422085 | 3.334230 | 3.370911 | 3.376830 | 3.507136 | 3.553315 | 3.762509 | 4.373274 | 4.353909 | 4.334249 | 4.350659 | 4.417113 | 4.340173 |
| GO:0043414 | macromolecule methylation | 3/42 | 341/20870 | 0.0310496467 | 0.2820041 | 0.2468512 | METTL2A||OGT||CTNNB1 | 3 | 5.586565 | 5.733021 | 5.699702 | 5.787615 | 5.589075 | 5.729628 | 5.754018 | 5.810007 | 5.546544 | 5.590010 | 5.622144 | 5.753399 | 5.735313 | 5.710025 | 5.756715 | 5.672381 | 5.668271 | 5.792741 | 5.745501 | 5.823535 | 5.575296 | 5.547233 | 5.643008 | 5.685629 | 5.743527 | 5.758703 | 5.773852 | 5.746003 | 5.741990 | 5.792518 | 5.830659 | 5.806585 |
| GO:0000956 | nuclear-transcribed mRNA catabolic process | 2/42 | 138/20870 | 0.0314558641 | 0.2820041 | 0.2468512 | NBAS||SMG8 | 2 | 5.662942 | 6.375390 | 5.807702 | 6.393633 | 5.619638 | 5.754664 | 6.413931 | 6.445531 | 5.648414 | 5.705622 | 5.633787 | 6.375285 | 6.370738 | 6.380131 | 6.038953 | 5.668446 | 5.684521 | 6.351821 | 6.366227 | 6.460422 | 5.597767 | 5.594101 | 5.665905 | 5.724890 | 5.777431 | 5.761171 | 6.391244 | 6.421997 | 6.428280 | 6.436079 | 6.456582 | 6.443858 |
| GO:0009799 | specification of symmetry | 2/42 | 138/20870 | 0.0314558641 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 3.393418 | 4.363631 | 3.754338 | 4.376096 | 3.352735 | 3.604988 | 4.343977 | 4.360401 | 3.394498 | 3.470736 | 3.310578 | 4.354760 | 4.390994 | 4.344727 | 3.947631 | 3.632126 | 3.661770 | 4.335007 | 4.379736 | 4.412498 | 3.326524 | 3.362505 | 3.368817 | 3.499964 | 3.544896 | 3.756856 | 4.363358 | 4.343831 | 4.324480 | 4.341161 | 4.407839 | 4.330988 |
| GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.550688 | 5.410910 | 4.819770 | 5.465331 | 4.463718 | 4.348379 | 5.363952 | 5.450780 | 4.580634 | 4.606847 | 4.460395 | 5.464439 | 5.447320 | 5.316478 | 5.008776 | 4.526597 | 4.881777 | 5.452085 | 5.455798 | 5.487840 | 4.429122 | 4.512879 | 4.447807 | 4.286981 | 4.305276 | 4.447472 | 5.384866 | 5.361033 | 5.345685 | 5.447661 | 5.457747 | 5.446908 |
| GO:0006206 | pyrimidine nucleobase metabolic process | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.934265 | 5.852420 | 5.348263 | 5.852616 | 4.736181 | 4.895404 | 5.900347 | 5.887289 | 4.993593 | 5.082791 | 4.699093 | 5.859470 | 5.848815 | 5.848950 | 5.614726 | 5.088413 | 5.292327 | 5.792405 | 5.807899 | 5.952108 | 4.688101 | 4.845067 | 4.668819 | 4.743489 | 4.697042 | 5.192192 | 5.886049 | 5.890356 | 5.924330 | 5.836676 | 5.923810 | 5.899982 |
| GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 2.749596 | 3.589305 | 2.959495 | 3.516906 | 2.582547 | 2.457226 | 3.712844 | 3.617395 | 3.019037 | 2.829884 | 2.310172 | 3.503560 | 3.572379 | 3.686061 | 3.190661 | 2.790153 | 2.865568 | 3.312009 | 3.537028 | 3.678282 | 2.498334 | 2.707709 | 2.532749 | 2.360230 | 1.944383 | 2.905263 | 3.815564 | 3.770270 | 3.537567 | 3.592424 | 3.652885 | 3.606178 |
| GO:0060572 | morphogenesis of an epithelial bud | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.386852 | 5.087543 | 4.192199 | 5.250068 | 3.168011 | 3.109333 | 5.015133 | 5.233036 | 3.511079 | 3.662232 | 2.871793 | 5.137808 | 5.144729 | 4.973681 | 4.533120 | 3.695044 | 4.227728 | 5.221340 | 5.284890 | 5.243249 | 3.063951 | 3.471207 | 2.909093 | 2.942116 | 2.822848 | 3.476194 | 5.034064 | 5.011377 | 4.999747 | 5.214250 | 5.265532 | 5.218766 |
| GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.239058 | 6.305491 | 6.282980 | 6.159250 | 6.313135 | 6.466858 | 6.243185 | 6.215928 | 6.167369 | 6.286915 | 6.260182 | 6.338074 | 6.305612 | 6.272033 | 6.203715 | 6.365420 | 6.275246 | 6.193411 | 6.082892 | 6.198515 | 6.333074 | 6.196164 | 6.402581 | 6.500675 | 6.428548 | 6.470442 | 6.238597 | 6.241969 | 6.248968 | 6.208032 | 6.240156 | 6.199274 |
| GO:0061548 | ganglion development | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.473370 | 4.999026 | 4.225322 | 5.198279 | 3.306156 | 3.435391 | 4.946776 | 5.184953 | 3.583890 | 3.697962 | 3.062053 | 5.058124 | 5.027619 | 4.906946 | 4.546874 | 3.774571 | 4.251999 | 5.180050 | 5.202862 | 5.211741 | 3.192665 | 3.558492 | 3.129117 | 3.353792 | 3.283167 | 3.643444 | 4.963257 | 4.946184 | 4.930704 | 5.173278 | 5.199995 | 5.181456 |
| GO:0097091 | synaptic vesicle clustering | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.428653 | 5.042433 | 4.276806 | 5.184572 | 3.239562 | 3.489428 | 4.984551 | 5.173691 | 3.502216 | 3.648454 | 3.075949 | 5.069575 | 5.089633 | 4.965014 | 4.627619 | 3.854770 | 4.245228 | 5.139665 | 5.212711 | 5.200286 | 3.155927 | 3.437734 | 3.102041 | 3.283258 | 3.284630 | 3.829140 | 5.006896 | 4.984953 | 4.961446 | 5.166073 | 5.200376 | 5.154225 |
| GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.579422 | 6.366463 | 6.221458 | 6.415833 | 5.582591 | 5.951472 | 6.372234 | 6.433346 | 5.546448 | 5.630817 | 5.559566 | 6.397133 | 6.391992 | 6.308564 | 6.344020 | 6.119706 | 6.191478 | 6.425288 | 6.406044 | 6.416102 | 5.540048 | 5.521429 | 5.680963 | 5.751997 | 5.812044 | 6.239835 | 6.427097 | 6.357927 | 6.329936 | 6.403739 | 6.480052 | 6.415067 |
| GO:0007098 | centrosome cycle | 2/42 | 140/20870 | 0.0322963640 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 5.165503 | 6.219477 | 5.370243 | 6.282620 | 5.105112 | 5.293807 | 6.282291 | 6.342968 | 5.171460 | 5.211797 | 5.111489 | 6.240009 | 6.186450 | 6.231399 | 5.683592 | 5.194504 | 5.172805 | 6.233205 | 6.256265 | 6.355446 | 5.065257 | 5.116696 | 5.132528 | 5.198054 | 5.252104 | 5.421716 | 6.257726 | 6.273910 | 6.314640 | 6.329398 | 6.347336 | 6.352071 |
| GO:0009581 | detection of external stimulus | 2/42 | 141/20870 | 0.0327199377 | 0.2820041 | 0.2468512 | ADGRV1||CTNNB1 | 2 | 3.350071 | 4.104735 | 3.823986 | 4.115885 | 3.299805 | 3.591122 | 4.094607 | 4.132622 | 3.337849 | 3.393846 | 3.317430 | 4.104460 | 4.130591 | 4.078687 | 3.975044 | 3.735368 | 3.748762 | 4.079464 | 4.110994 | 4.156165 | 3.263889 | 3.245077 | 3.386334 | 3.394095 | 3.483064 | 3.853937 | 4.121198 | 4.112785 | 4.048757 | 4.107397 | 4.172046 | 4.117580 |
| GO:0033865 | nucleoside bisphosphate metabolic process | 2/42 | 142/20870 | 0.0331457138 | 0.2820041 | 0.2468512 | PPT1||BPNT1 | 2 | 4.846220 | 5.246076 | 5.036811 | 5.361007 | 4.861142 | 5.096937 | 5.212573 | 5.321503 | 4.808299 | 4.846371 | 4.883023 | 5.253021 | 5.262983 | 5.221906 | 5.097772 | 5.023157 | 4.987292 | 5.342315 | 5.350938 | 5.389334 | 4.858997 | 4.794579 | 4.926818 | 5.012580 | 5.097391 | 5.176202 | 5.223590 | 5.215749 | 5.198261 | 5.313991 | 5.357139 | 5.292630 |
| GO:0033875 | ribonucleoside bisphosphate metabolic process | 2/42 | 142/20870 | 0.0331457138 | 0.2820041 | 0.2468512 | PPT1||BPNT1 | 2 | 4.846220 | 5.246076 | 5.036811 | 5.361007 | 4.861142 | 5.096937 | 5.212573 | 5.321503 | 4.808299 | 4.846371 | 4.883023 | 5.253021 | 5.262983 | 5.221906 | 5.097772 | 5.023157 | 4.987292 | 5.342315 | 5.350938 | 5.389334 | 4.858997 | 4.794579 | 4.926818 | 5.012580 | 5.097391 | 5.176202 | 5.223590 | 5.215749 | 5.198261 | 5.313991 | 5.357139 | 5.292630 |
| GO:0034032 | purine nucleoside bisphosphate metabolic process | 2/42 | 142/20870 | 0.0331457138 | 0.2820041 | 0.2468512 | PPT1||BPNT1 | 2 | 4.846220 | 5.246076 | 5.036811 | 5.361007 | 4.861142 | 5.096937 | 5.212573 | 5.321503 | 4.808299 | 4.846371 | 4.883023 | 5.253021 | 5.262983 | 5.221906 | 5.097772 | 5.023157 | 4.987292 | 5.342315 | 5.350938 | 5.389334 | 4.858997 | 4.794579 | 4.926818 | 5.012580 | 5.097391 | 5.176202 | 5.223590 | 5.215749 | 5.198261 | 5.313991 | 5.357139 | 5.292630 |
| GO:0007625 | grooming behavior | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | PPT1 | 1 | 7.114518 | 6.908724 | 6.932640 | 6.780642 | 7.208278 | 7.268711 | 6.834968 | 6.748884 | 7.072533 | 6.931522 | 7.313402 | 6.983479 | 6.853543 | 6.885957 | 6.799333 | 7.020083 | 6.969354 | 6.834276 | 6.722540 | 6.782945 | 7.184912 | 7.099816 | 7.330606 | 7.207998 | 7.454914 | 7.122219 | 6.874520 | 6.794500 | 6.834776 | 6.751140 | 6.793700 | 6.700300 |
| GO:0035751 | regulation of lysosomal lumen pH | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.547151 | 6.143089 | 6.012913 | 6.157383 | 5.594329 | 5.812966 | 5.960714 | 6.049120 | 5.507792 | 5.555314 | 5.577469 | 6.220173 | 6.168358 | 6.034425 | 5.983555 | 6.049409 | 6.004990 | 6.185123 | 6.178877 | 6.106844 | 5.588089 | 5.546629 | 5.646520 | 5.629535 | 5.678630 | 6.085951 | 6.010348 | 5.954929 | 5.915282 | 6.043750 | 6.116539 | 5.984015 |
| GO:0043981 | histone H4-K5 acetylation | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | OGT | 1 | 4.971643 | 5.314386 | 5.402001 | 5.392739 | 4.971122 | 5.237293 | 5.388748 | 5.447956 | 4.889608 | 5.000644 | 5.021231 | 5.334475 | 5.313129 | 5.295286 | 5.554904 | 5.304345 | 5.333526 | 5.371212 | 5.348129 | 5.456600 | 4.971169 | 4.897353 | 5.041255 | 5.174966 | 5.200158 | 5.331790 | 5.445765 | 5.352183 | 5.366526 | 5.399964 | 5.511951 | 5.429609 |
| GO:0043982 | histone H4-K8 acetylation | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | OGT | 1 | 4.971643 | 5.314386 | 5.402001 | 5.392739 | 4.971122 | 5.237293 | 5.388748 | 5.447956 | 4.889608 | 5.000644 | 5.021231 | 5.334475 | 5.313129 | 5.295286 | 5.554904 | 5.304345 | 5.333526 | 5.371212 | 5.348129 | 5.456600 | 4.971169 | 4.897353 | 5.041255 | 5.174966 | 5.200158 | 5.331790 | 5.445765 | 5.352183 | 5.366526 | 5.399964 | 5.511951 | 5.429609 |
| GO:0000723 | telomere maintenance | 2/42 | 144/20870 | 0.0340038326 | 0.2820041 | 0.2468512 | RPA2||CTNNB1 | 2 | 6.549257 | 7.378963 | 6.750769 | 7.426868 | 6.436545 | 6.520055 | 7.429793 | 7.483000 | 6.571249 | 6.707475 | 6.346240 | 7.394273 | 7.353542 | 7.388736 | 7.042824 | 6.490400 | 6.662640 | 7.384405 | 7.393826 | 7.499518 | 6.416087 | 6.535723 | 6.351748 | 6.550641 | 6.470744 | 6.537514 | 7.413457 | 7.420806 | 7.454779 | 7.468116 | 7.492484 | 7.488283 |
| GO:0009582 | detection of abiotic stimulus | 2/42 | 144/20870 | 0.0340038326 | 0.2820041 | 0.2468512 | ADGRV1||CTNNB1 | 2 | 3.462736 | 4.177900 | 3.943939 | 4.190665 | 3.414085 | 3.716705 | 4.169095 | 4.203256 | 3.443043 | 3.509949 | 3.434019 | 4.176469 | 4.203565 | 4.153226 | 4.089108 | 3.861953 | 3.869003 | 4.155093 | 4.184674 | 4.231207 | 3.392429 | 3.351136 | 3.494869 | 3.518221 | 3.593903 | 3.991870 | 4.197626 | 4.185306 | 4.123258 | 4.177100 | 4.243575 | 4.188211 |
| GO:0048565 | digestive tract development | 2/42 | 144/20870 | 0.0340038326 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.526943 | 4.420165 | 3.925073 | 4.382544 | 3.463472 | 3.700561 | 4.426743 | 4.426496 | 3.559834 | 3.607378 | 3.406026 | 4.457367 | 4.427743 | 4.374155 | 4.125183 | 3.780195 | 3.846139 | 4.353609 | 4.361842 | 4.430924 | 3.400508 | 3.526340 | 3.460820 | 3.587927 | 3.600922 | 3.891916 | 4.428177 | 4.428898 | 4.423148 | 4.404738 | 4.462168 | 4.411901 |
| GO:0016042 | lipid catabolic process | 3/42 | 354/20870 | 0.0341347463 | 0.2820041 | 0.2468512 | CPS1||SLC25A17||PPT1 | 3 | 4.964218 | 5.134583 | 5.115672 | 5.119145 | 4.966732 | 5.076017 | 5.084825 | 5.088222 | 4.937602 | 4.971876 | 4.982791 | 5.171195 | 5.135511 | 5.096065 | 5.148153 | 5.102639 | 5.095659 | 5.121005 | 5.091167 | 5.144764 | 4.961134 | 4.931475 | 5.006591 | 4.997493 | 5.062472 | 5.163233 | 5.110801 | 5.074962 | 5.068348 | 5.072638 | 5.121803 | 5.069627 |
| GO:0032355 | response to estradiol | 2/42 | 146/20870 | 0.0348706396 | 0.2820041 | 0.2468512 | H2AZ1||CTNNB1 | 2 | 6.705869 | 6.714693 | 6.601020 | 6.561242 | 6.730425 | 6.836428 | 6.649902 | 6.561429 | 6.668979 | 6.699331 | 6.748188 | 6.730504 | 6.723451 | 6.689798 | 6.491638 | 6.642589 | 6.662840 | 6.604171 | 6.499426 | 6.578079 | 6.726367 | 6.667449 | 6.794647 | 6.852974 | 6.966212 | 6.675317 | 6.660034 | 6.647494 | 6.642120 | 6.553081 | 6.583835 | 6.547103 |
| GO:0042157 | lipoprotein metabolic process | 2/42 | 146/20870 | 0.0348706396 | 0.2820041 | 0.2468512 | SELENOK||PPT1 | 2 | 4.699001 | 5.293147 | 5.095771 | 5.322650 | 4.717104 | 4.924364 | 5.225843 | 5.267116 | 4.683838 | 4.706054 | 4.706991 | 5.319957 | 5.304875 | 5.253780 | 5.161892 | 5.069286 | 5.053749 | 5.304604 | 5.321664 | 5.341445 | 4.692560 | 4.714587 | 4.743707 | 4.766219 | 4.817317 | 5.157437 | 5.253284 | 5.215065 | 5.208776 | 5.265082 | 5.304998 | 5.230300 |
| GO:0018022 | peptidyl-lysine methylation | 2/42 | 147/20870 | 0.0353072762 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.896156 | 5.268547 | 5.171412 | 5.316011 | 4.884436 | 5.104911 | 5.292852 | 5.329293 | 4.858990 | 4.920783 | 4.907961 | 5.286036 | 5.267841 | 5.251558 | 5.290027 | 5.105793 | 5.110672 | 5.299049 | 5.284537 | 5.363226 | 4.891414 | 4.828513 | 4.931517 | 5.046205 | 5.056468 | 5.206420 | 5.313729 | 5.281263 | 5.283336 | 5.313586 | 5.358273 | 5.315577 |
| GO:0006703 | estrogen biosynthetic process | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | HSD17B7 | 1 | 3.456693 | 5.113624 | 3.350914 | 4.906031 | 3.440546 | 3.243876 | 4.996812 | 4.901798 | 3.443555 | 3.511312 | 3.413464 | 5.151100 | 5.118942 | 5.069667 | 3.608586 | 3.154021 | 3.249353 | 4.799790 | 4.978254 | 4.934116 | 3.373283 | 3.486259 | 3.459692 | 3.183915 | 3.392489 | 3.142597 | 4.993252 | 5.011944 | 4.985109 | 4.933122 | 4.942235 | 4.827228 |
| GO:0006901 | vesicle coating | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.375713 | 7.039942 | 6.665758 | 6.914431 | 6.362097 | 6.517449 | 6.966981 | 6.914325 | 6.319915 | 6.416088 | 6.389434 | 7.036879 | 7.083706 | 6.997964 | 6.708558 | 6.623968 | 6.663506 | 6.907498 | 6.928985 | 6.906700 | 6.331490 | 6.345095 | 6.408529 | 6.440062 | 6.459905 | 6.643534 | 7.007724 | 6.962568 | 6.929583 | 6.926524 | 6.934904 | 6.880965 |
| GO:0043923 | positive regulation by host of viral transcription | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | CCNT1 | 1 | 5.520201 | 6.352141 | 5.657690 | 6.306795 | 5.487684 | 5.779983 | 6.436201 | 6.375567 | 5.481309 | 5.564440 | 5.513633 | 6.298695 | 6.357884 | 6.398115 | 5.895728 | 5.533036 | 5.511109 | 6.278462 | 6.250333 | 6.387899 | 5.527426 | 5.396491 | 5.534976 | 5.793711 | 5.758767 | 5.787233 | 6.422250 | 6.416735 | 6.469043 | 6.356655 | 6.386815 | 6.383046 |
| GO:0046855 | inositol phosphate dephosphorylation | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.156687 | 4.239506 | 3.708721 | 4.396930 | 3.127006 | 3.434130 | 4.263546 | 4.445901 | 3.152228 | 3.264512 | 3.044967 | 4.226526 | 4.253887 | 4.237973 | 3.995301 | 3.579755 | 3.500773 | 4.308011 | 4.380170 | 4.496325 | 3.098603 | 3.121776 | 3.159972 | 3.302340 | 3.306208 | 3.663291 | 4.283459 | 4.272386 | 4.234335 | 4.425210 | 4.494219 | 4.417022 |
| GO:0070365 | hepatocyte differentiation | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | CPS1 | 1 | 5.035432 | 4.575960 | 4.472858 | 4.131174 | 5.065228 | 4.968086 | 4.481154 | 4.005317 | 5.058295 | 4.866373 | 5.165841 | 4.619095 | 4.609481 | 4.496091 | 4.311927 | 4.575006 | 4.518556 | 4.206733 | 4.032825 | 4.148569 | 5.085259 | 4.951214 | 5.152017 | 5.002515 | 5.194220 | 4.657210 | 4.462300 | 4.488370 | 4.492604 | 4.035131 | 3.983499 | 3.996821 |
| GO:0080182 | histone H3-K4 trimethylation | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | OGT | 1 | 4.169385 | 4.905353 | 4.814708 | 5.088010 | 4.082535 | 4.420420 | 4.985889 | 5.134885 | 4.150066 | 4.315740 | 4.027815 | 4.884448 | 4.926561 | 4.904742 | 5.086125 | 4.651894 | 4.662264 | 4.996613 | 5.095695 | 5.166681 | 4.115634 | 4.102699 | 4.027718 | 4.380702 | 4.217244 | 4.632702 | 4.984148 | 5.010556 | 4.962560 | 5.078557 | 5.225134 | 5.096491 |
| GO:0090036 | regulation of protein kinase C signaling | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 2.976564 | 3.611891 | 3.679205 | 3.591815 | 3.011556 | 3.514147 | 3.590861 | 3.610288 | 2.876324 | 2.959175 | 3.086396 | 3.600954 | 3.612966 | 3.621678 | 3.860031 | 3.629472 | 3.527767 | 3.590995 | 3.589782 | 3.594663 | 3.030531 | 2.850491 | 3.139050 | 3.200127 | 3.357190 | 3.891202 | 3.617194 | 3.610103 | 3.544167 | 3.591716 | 3.691797 | 3.543357 |
| GO:1903830 | magnesium ion transmembrane transport | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 2.902988 | 5.077355 | 3.726273 | 5.014121 | 2.830494 | 3.150318 | 4.996710 | 4.988288 | 2.936599 | 3.159809 | 2.547465 | 5.119175 | 5.078413 | 5.033197 | 4.131741 | 3.451379 | 3.490058 | 4.907910 | 5.087220 | 5.041276 | 2.760378 | 3.022304 | 2.686873 | 3.016643 | 2.886630 | 3.478622 | 4.989658 | 5.031835 | 4.967904 | 4.974955 | 5.043993 | 4.944094 |
| GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.439509 | 5.056724 | 4.255972 | 5.265046 | 3.266567 | 3.484048 | 5.023489 | 5.290780 | 3.546361 | 3.696696 | 2.981473 | 5.104268 | 5.076360 | 4.986957 | 4.609640 | 3.834856 | 4.219961 | 5.225967 | 5.272872 | 5.295430 | 3.184752 | 3.501005 | 3.079984 | 3.381462 | 3.181889 | 3.814723 | 5.038795 | 5.015850 | 5.015700 | 5.272389 | 5.316801 | 5.282774 |
| GO:0006790 | sulfur compound metabolic process | 3/42 | 366/20870 | 0.0371198540 | 0.2820041 | 0.2468512 | CPS1||PPT1||BPNT1 | 3 | 6.095896 | 5.939683 | 5.993266 | 5.923982 | 6.153063 | 6.187191 | 5.894843 | 5.904300 | 6.065748 | 6.037829 | 6.180136 | 5.980998 | 5.934692 | 5.902271 | 5.919585 | 6.027796 | 6.029688 | 5.946167 | 5.876391 | 5.948236 | 6.148859 | 6.091226 | 6.216382 | 6.195865 | 6.281603 | 6.076803 | 5.910465 | 5.882078 | 5.891842 | 5.894964 | 5.924779 | 5.892936 |
| GO:0006044 | N-acetylglucosamine metabolic process | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | OGA | 1 | 3.124689 | 3.954254 | 3.968484 | 4.069396 | 3.074994 | 3.403264 | 4.037562 | 4.284912 | 3.093583 | 3.284847 | 2.978959 | 3.944658 | 3.967801 | 3.950201 | 4.222068 | 3.844017 | 3.801157 | 3.992726 | 4.061697 | 4.149479 | 3.078107 | 3.060668 | 3.086089 | 3.133206 | 3.105046 | 3.844170 | 4.038667 | 4.024647 | 4.049267 | 4.238721 | 4.320770 | 4.294034 |
| GO:0015693 | magnesium ion transport | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 2.810357 | 4.984767 | 3.633543 | 4.921689 | 2.737688 | 3.057520 | 4.904309 | 4.895823 | 2.844132 | 3.067209 | 2.454571 | 5.026472 | 4.986017 | 4.940532 | 4.038924 | 3.358619 | 3.397497 | 4.815426 | 4.994751 | 4.948933 | 2.667529 | 2.929579 | 2.594006 | 2.923923 | 2.793800 | 3.385789 | 4.897205 | 4.939246 | 4.875752 | 4.882485 | 4.951564 | 4.851597 |
| GO:0015867 | ATP transport | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | SLC25A17 | 1 | 7.671523 | 7.164200 | 7.476798 | 7.238043 | 7.627840 | 7.527461 | 7.082855 | 7.265943 | 7.732037 | 7.784559 | 7.479966 | 7.257493 | 7.135802 | 7.094278 | 7.375305 | 7.407617 | 7.633522 | 7.320616 | 7.153340 | 7.235324 | 7.640229 | 7.709368 | 7.528155 | 7.668476 | 7.637660 | 7.237533 | 7.086844 | 7.081727 | 7.079986 | 7.259659 | 7.266909 | 7.271238 |
| GO:0045603 | positive regulation of endothelial cell differentiation | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.627384 | 5.313597 | 4.452743 | 5.487211 | 3.365024 | 3.293362 | 5.256679 | 5.496493 | 3.753341 | 3.961969 | 3.000901 | 5.372157 | 5.350422 | 5.213113 | 4.832143 | 3.873338 | 4.495330 | 5.455923 | 5.491879 | 5.513252 | 3.205442 | 3.746580 | 3.046645 | 3.168071 | 3.001420 | 3.634070 | 5.258107 | 5.263586 | 5.248303 | 5.481915 | 5.508148 | 5.499293 |
| GO:0046838 | phosphorylated carbohydrate dephosphorylation | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.156687 | 4.239506 | 3.708721 | 4.396930 | 3.127006 | 3.434130 | 4.263546 | 4.445901 | 3.152228 | 3.264512 | 3.044967 | 4.226526 | 4.253887 | 4.237973 | 3.995301 | 3.579755 | 3.500773 | 4.308011 | 4.380170 | 4.496325 | 3.098603 | 3.121776 | 3.159972 | 3.302340 | 3.306208 | 3.663291 | 4.283459 | 4.272386 | 4.234335 | 4.425210 | 4.494219 | 4.417022 |
| GO:0060438 | trachea development | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.353478 | 5.247170 | 4.836983 | 5.328615 | 4.302878 | 4.483332 | 5.243716 | 5.351692 | 4.352196 | 4.475705 | 4.221335 | 5.276532 | 5.279546 | 5.183375 | 5.039296 | 4.623441 | 4.818167 | 5.318914 | 5.324260 | 5.342565 | 4.238192 | 4.358600 | 4.309307 | 4.379767 | 4.401652 | 4.652377 | 5.253717 | 5.236220 | 5.241155 | 5.341451 | 5.358409 | 5.355159 |
| GO:0061323 | cell proliferation involved in heart morphogenesis | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.378849 | 4.983953 | 4.192314 | 5.196400 | 3.217407 | 3.437614 | 4.953531 | 5.222285 | 3.480782 | 3.635429 | 2.929052 | 5.032063 | 5.002992 | 4.914209 | 4.549142 | 3.772857 | 4.150878 | 5.157834 | 5.202903 | 5.227599 | 3.134329 | 3.447603 | 3.037834 | 3.329219 | 3.147312 | 3.764898 | 4.968319 | 4.945315 | 4.946842 | 5.204362 | 5.247656 | 5.214480 |
| GO:1903204 | negative regulation of oxidative stress-induced neuron death | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 8.831526 | 8.415356 | 8.724088 | 8.486348 | 8.851282 | 8.847334 | 8.405384 | 8.497900 | 8.801967 | 8.831525 | 8.860493 | 8.460765 | 8.399845 | 8.384323 | 8.586955 | 8.744722 | 8.830128 | 8.541265 | 8.414252 | 8.500627 | 8.852136 | 8.859771 | 8.841884 | 8.930421 | 8.902031 | 8.698656 | 8.408407 | 8.405460 | 8.402281 | 8.494909 | 8.494657 | 8.504114 |
| GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.757745 | 4.926525 | 4.008576 | 4.988606 | 3.609393 | 3.199754 | 4.876304 | 4.989365 | 3.845393 | 3.868099 | 3.536579 | 4.978516 | 4.954779 | 4.842662 | 4.345684 | 3.516433 | 4.045339 | 4.956904 | 4.994425 | 5.013907 | 3.544849 | 3.767725 | 3.501212 | 3.239935 | 3.118745 | 3.237291 | 4.888121 | 4.881944 | 4.858681 | 4.988916 | 4.994725 | 4.984435 |
| GO:0031023 | microtubule organizing center organization | 2/42 | 153/20870 | 0.0379717329 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 5.069343 | 6.120454 | 5.288061 | 6.182125 | 5.011003 | 5.209661 | 6.183373 | 6.243230 | 5.071038 | 5.115869 | 5.019513 | 6.139067 | 6.088790 | 6.132985 | 5.597626 | 5.116000 | 5.092268 | 6.132146 | 6.155772 | 6.255466 | 4.972363 | 5.019740 | 5.040073 | 5.112548 | 5.168375 | 5.338362 | 6.160386 | 6.175615 | 6.213596 | 6.228099 | 6.249667 | 6.251803 |
| GO:0055123 | digestive system development | 2/42 | 154/20870 | 0.0384231564 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.567602 | 4.484743 | 3.976978 | 4.432799 | 3.502208 | 3.736204 | 4.480973 | 4.473696 | 3.595110 | 3.650877 | 3.449390 | 4.523072 | 4.490499 | 4.439427 | 4.176272 | 3.835010 | 3.896252 | 4.400922 | 4.417246 | 4.479047 | 3.447400 | 3.560243 | 3.496760 | 3.618123 | 3.619917 | 3.945380 | 4.481474 | 4.486122 | 4.475301 | 4.451682 | 4.511976 | 4.456651 |
| GO:0001711 | endodermal cell fate commitment | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.262603 | 5.658517 | 4.813982 | 5.832310 | 4.070601 | 4.206884 | 5.654124 | 5.859654 | 4.329451 | 4.483467 | 3.917300 | 5.689963 | 5.678456 | 5.605694 | 5.160114 | 4.408354 | 4.775892 | 5.802419 | 5.833567 | 5.860362 | 3.995812 | 4.285759 | 3.901971 | 4.201360 | 4.117802 | 4.295976 | 5.661698 | 5.644069 | 5.656547 | 5.853598 | 5.868185 | 5.857139 |
| GO:0043586 | tongue development | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.569268 | 5.985129 | 5.168130 | 6.086523 | 4.365965 | 4.500682 | 5.996404 | 6.112092 | 4.646097 | 4.805336 | 4.187551 | 6.022621 | 5.993291 | 5.938204 | 5.508930 | 4.810895 | 5.098501 | 6.067072 | 6.076026 | 6.115999 | 4.329798 | 4.572691 | 4.166200 | 4.419888 | 4.207648 | 4.809018 | 5.988941 | 5.992349 | 6.007851 | 6.096694 | 6.122989 | 6.116464 |
| GO:0046112 | nucleobase biosynthetic process | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CPS1 | 1 | 6.818454 | 6.983781 | 6.840244 | 6.902339 | 6.837005 | 6.927166 | 6.983172 | 6.928267 | 6.793118 | 6.757717 | 6.900659 | 7.015048 | 6.966089 | 6.969688 | 6.835235 | 6.849134 | 6.836322 | 6.900487 | 6.844964 | 6.959301 | 6.829585 | 6.782726 | 6.896436 | 6.861290 | 7.007570 | 6.908757 | 7.003007 | 6.952335 | 6.993672 | 6.911403 | 6.956987 | 6.915974 |
| GO:0048490 | anterograde synaptic vesicle transport | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | AP3M2 | 1 | 4.754411 | 5.012477 | 4.859218 | 5.109481 | 4.744554 | 4.821254 | 5.016193 | 5.081542 | 4.758212 | 4.776416 | 4.728194 | 5.007897 | 5.030347 | 4.999008 | 4.950001 | 4.822136 | 4.800966 | 5.092478 | 5.090698 | 5.144614 | 4.759174 | 4.761990 | 4.711951 | 4.831514 | 4.777981 | 4.853230 | 5.005103 | 4.999482 | 5.043594 | 5.077072 | 5.126039 | 5.040230 |
| GO:0071545 | inositol phosphate catabolic process | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.140300 | 4.213631 | 3.705575 | 4.364860 | 3.103480 | 3.423720 | 4.242233 | 4.411433 | 3.136241 | 3.257736 | 3.016873 | 4.197340 | 4.228810 | 4.214571 | 3.986949 | 3.582919 | 3.498269 | 4.274553 | 4.348726 | 4.464894 | 3.075583 | 3.111606 | 3.122830 | 3.311564 | 3.280943 | 3.649021 | 4.259531 | 4.251708 | 4.215069 | 4.389827 | 4.461457 | 4.381670 |
| GO:0099514 | synaptic vesicle cytoskeletal transport | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | AP3M2 | 1 | 4.754411 | 5.012477 | 4.859218 | 5.109481 | 4.744554 | 4.821254 | 5.016193 | 5.081542 | 4.758212 | 4.776416 | 4.728194 | 5.007897 | 5.030347 | 4.999008 | 4.950001 | 4.822136 | 4.800966 | 5.092478 | 5.090698 | 5.144614 | 4.759174 | 4.761990 | 4.711951 | 4.831514 | 4.777981 | 4.853230 | 5.005103 | 4.999482 | 5.043594 | 5.077072 | 5.126039 | 5.040230 |
| GO:0099517 | synaptic vesicle transport along microtubule | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | AP3M2 | 1 | 4.754411 | 5.012477 | 4.859218 | 5.109481 | 4.744554 | 4.821254 | 5.016193 | 5.081542 | 4.758212 | 4.776416 | 4.728194 | 5.007897 | 5.030347 | 4.999008 | 4.950001 | 4.822136 | 4.800966 | 5.092478 | 5.090698 | 5.144614 | 4.759174 | 4.761990 | 4.711951 | 4.831514 | 4.777981 | 4.853230 | 5.005103 | 4.999482 | 5.043594 | 5.077072 | 5.126039 | 5.040230 |
| GO:1901679 | nucleotide transmembrane transport | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | SLC25A17 | 1 | 7.200952 | 6.769221 | 7.032344 | 6.802130 | 7.161536 | 7.073792 | 6.678675 | 6.821939 | 7.258069 | 7.308064 | 7.020695 | 6.850135 | 6.754867 | 6.698577 | 6.929087 | 6.974691 | 7.180621 | 6.883789 | 6.719048 | 6.798846 | 7.174169 | 7.233518 | 7.072339 | 7.197299 | 7.172773 | 6.821573 | 6.689182 | 6.675873 | 6.670909 | 6.803691 | 6.833819 | 6.828128 |
| GO:1904948 | midbrain dopaminergic neuron differentiation | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.320140 | 6.106337 | 5.961809 | 6.156065 | 5.322954 | 5.692009 | 6.111376 | 6.173298 | 5.287442 | 5.371792 | 5.299740 | 6.136885 | 6.131917 | 6.048512 | 6.084810 | 5.859999 | 5.931396 | 6.165073 | 6.146510 | 6.156552 | 5.280639 | 5.262089 | 5.420854 | 5.492598 | 5.552465 | 5.980413 | 6.166035 | 6.096906 | 6.069463 | 6.144131 | 6.219985 | 6.154600 |
| GO:0016571 | histone methylation | 2/42 | 159/20870 | 0.0407111770 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.607968 | 5.682203 | 5.712590 | 5.726938 | 5.604506 | 5.734573 | 5.702380 | 5.750568 | 5.574012 | 5.618330 | 5.630944 | 5.703772 | 5.685633 | 5.656815 | 5.748105 | 5.693502 | 5.695496 | 5.736963 | 5.687317 | 5.755672 | 5.600790 | 5.564186 | 5.647336 | 5.713425 | 5.724416 | 5.765357 | 5.725735 | 5.692972 | 5.688142 | 5.732111 | 5.770198 | 5.749142 |
| GO:0006066 | alcohol metabolic process | 3/42 | 380/20870 | 0.0407678251 | 0.2820041 | 0.2468512 | HSD17B7||BPNT1||SLC5A3 | 3 | 5.338062 | 5.382816 | 5.505685 | 5.501227 | 5.373726 | 5.517767 | 5.337816 | 5.458979 | 5.289955 | 5.333710 | 5.388817 | 5.398865 | 5.404333 | 5.344496 | 5.489382 | 5.529617 | 5.497743 | 5.503450 | 5.484524 | 5.515537 | 5.371794 | 5.333392 | 5.414839 | 5.453949 | 5.493551 | 5.601717 | 5.361488 | 5.330298 | 5.321353 | 5.450240 | 5.489107 | 5.437083 |
| GO:0007194 | negative regulation of adenylate cyclase activity | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 3.242723 | 3.757289 | 3.539756 | 3.918975 | 3.245422 | 3.466481 | 3.753264 | 3.875733 | 3.179613 | 3.222519 | 3.322301 | 3.719741 | 3.816418 | 3.733804 | 3.574732 | 3.604911 | 3.433916 | 3.922194 | 3.867823 | 3.965258 | 3.272680 | 3.139587 | 3.318089 | 3.343376 | 3.364970 | 3.667781 | 3.814706 | 3.748814 | 3.693725 | 3.875903 | 3.914794 | 3.835410 |
| GO:0010738 | regulation of protein kinase A signaling | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 8.784907 | 7.895652 | 8.278112 | 7.672536 | 8.865779 | 8.948412 | 7.697977 | 7.510711 | 8.757124 | 8.517959 | 9.033440 | 8.099110 | 7.768215 | 7.795740 | 7.914556 | 8.496142 | 8.361645 | 7.919395 | 7.533749 | 7.528679 | 8.881596 | 8.593246 | 9.081286 | 8.852726 | 9.236948 | 8.701627 | 7.696356 | 7.698959 | 7.698614 | 7.531791 | 7.506358 | 7.493723 |
| GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.693665 | 4.105816 | 3.979432 | 4.099248 | 3.710687 | 3.792596 | 4.048815 | 4.008509 | 3.698326 | 3.671414 | 3.710973 | 4.111154 | 4.148149 | 4.056682 | 4.111203 | 3.913643 | 3.903818 | 4.032240 | 4.103322 | 4.159374 | 3.676921 | 3.658376 | 3.793040 | 3.598212 | 3.754153 | 3.997173 | 4.096665 | 4.067908 | 3.979295 | 3.983579 | 4.050033 | 3.990990 |
| GO:0046931 | pore complex assembly | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | NUP205 | 1 | 5.976609 | 6.799557 | 6.329316 | 6.928056 | 5.815089 | 5.820769 | 6.831894 | 7.006158 | 6.057149 | 6.195672 | 5.615579 | 6.844335 | 6.778294 | 6.774976 | 6.575064 | 6.059889 | 6.306990 | 6.886844 | 6.900658 | 6.994281 | 5.801197 | 6.004389 | 5.613135 | 5.951109 | 5.710502 | 5.790198 | 6.828651 | 6.829156 | 6.837856 | 6.975338 | 7.026851 | 7.015776 |
| GO:0048485 | sympathetic nervous system development | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 2.995007 | 4.786575 | 3.879255 | 4.979320 | 2.704889 | 2.686285 | 4.731804 | 4.973569 | 3.153813 | 3.316475 | 2.337646 | 4.839087 | 4.820751 | 4.695710 | 4.290297 | 3.286483 | 3.889729 | 4.953998 | 4.988728 | 4.994897 | 2.534739 | 3.106812 | 2.364781 | 2.599717 | 2.430968 | 2.973681 | 4.739785 | 4.734377 | 4.721187 | 4.964257 | 4.994659 | 4.961556 |
| GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.693665 | 4.105816 | 3.979432 | 4.099248 | 3.710687 | 3.792596 | 4.048815 | 4.008509 | 3.698326 | 3.671414 | 3.710973 | 4.111154 | 4.148149 | 4.056682 | 4.111203 | 3.913643 | 3.903818 | 4.032240 | 4.103322 | 4.159374 | 3.676921 | 3.658376 | 3.793040 | 3.598212 | 3.754153 | 3.997173 | 4.096665 | 4.067908 | 3.979295 | 3.983579 | 4.050033 | 3.990990 |
| GO:0072283 | metanephric renal vesicle morphogenesis | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.315698 | 5.176333 | 4.585195 | 5.227105 | 4.229269 | 4.116094 | 5.128686 | 5.212819 | 4.345130 | 4.372921 | 4.224787 | 5.229257 | 5.213067 | 5.082215 | 4.774436 | 4.290867 | 4.647847 | 5.213514 | 5.217237 | 5.250279 | 4.195775 | 4.276858 | 4.213914 | 4.054289 | 4.073323 | 4.215250 | 5.150482 | 5.125667 | 5.109615 | 5.208869 | 5.220342 | 5.209217 |
| GO:0002053 | positive regulation of mesenchymal cell proliferation | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.569810 | 4.924265 | 4.215580 | 5.080304 | 3.478515 | 3.800405 | 4.872581 | 5.023588 | 3.613724 | 3.729083 | 3.339361 | 4.954052 | 4.972291 | 4.843092 | 4.472609 | 3.939883 | 4.184863 | 5.089167 | 5.070472 | 5.081213 | 3.404233 | 3.585571 | 3.439259 | 3.629592 | 3.683958 | 4.050398 | 4.878464 | 4.873895 | 4.865353 | 5.007600 | 5.047139 | 5.015724 |
| GO:0002089 | lens morphogenesis in camera-type eye | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.954906 | 5.920798 | 6.141591 | 6.396622 | 5.882845 | 6.055203 | 5.940908 | 6.448155 | 5.984288 | 6.122227 | 5.731354 | 5.990893 | 5.889634 | 5.879210 | 6.216924 | 6.002542 | 6.195784 | 6.430677 | 6.303495 | 6.451299 | 5.843072 | 6.060535 | 5.724611 | 6.117773 | 5.948752 | 6.093365 | 5.968146 | 5.919806 | 5.934346 | 6.455572 | 6.433190 | 6.455589 |
| GO:0007063 | regulation of sister chromatid cohesion | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.307569 | 6.811344 | 6.565588 | 6.824220 | 6.284487 | 6.439614 | 6.836784 | 6.868936 | 6.289742 | 6.353699 | 6.278114 | 6.836872 | 6.801055 | 6.795757 | 6.726299 | 6.417107 | 6.536384 | 6.775927 | 6.805020 | 6.889297 | 6.244137 | 6.291599 | 6.316784 | 6.381132 | 6.408507 | 6.525116 | 6.840607 | 6.832830 | 6.836903 | 6.855208 | 6.882464 | 6.869006 |
| GO:0033762 | response to glucagon | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CPS1 | 1 | 8.578909 | 8.200568 | 9.016931 | 8.232266 | 8.667386 | 9.089139 | 8.249581 | 8.066968 | 8.453089 | 8.562287 | 8.709805 | 7.650361 | 8.396300 | 8.429262 | 8.896224 | 9.133824 | 9.010954 | 8.158070 | 8.451542 | 8.057610 | 8.680361 | 8.524208 | 8.785670 | 8.905689 | 8.948501 | 9.366872 | 8.217004 | 8.301671 | 8.228599 | 8.128911 | 8.070154 | 7.998907 |
| GO:0035020 | regulation of Rac protein signal transduction | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | OGT | 1 | 5.013166 | 5.334232 | 5.153032 | 5.211367 | 5.029493 | 5.155393 | 5.325128 | 5.165876 | 4.990107 | 5.033242 | 5.015822 | 5.342166 | 5.318265 | 5.342134 | 5.289150 | 5.069996 | 5.089602 | 5.184994 | 5.162987 | 5.283256 | 5.066433 | 4.954274 | 5.064929 | 5.191598 | 5.118987 | 5.154680 | 5.348269 | 5.308304 | 5.318511 | 5.152910 | 5.182401 | 5.162158 |
| GO:0043576 | regulation of respiratory gaseous exchange | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | SLC5A3 | 1 | 3.912671 | 4.078121 | 4.402305 | 4.279699 | 3.922973 | 4.343005 | 4.127143 | 4.310386 | 3.828573 | 3.924470 | 3.980879 | 4.054395 | 4.096237 | 4.083412 | 4.531435 | 4.362904 | 4.302728 | 4.230157 | 4.291136 | 4.316445 | 3.898070 | 3.823891 | 4.038655 | 4.159110 | 4.291471 | 4.550608 | 4.153966 | 4.170522 | 4.054218 | 4.278814 | 4.342718 | 4.308916 |
| GO:0043984 | histone H4-K16 acetylation | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | OGT | 1 | 4.896010 | 5.377388 | 5.407865 | 5.434215 | 4.905230 | 5.200522 | 5.420465 | 5.470882 | 4.830549 | 4.889957 | 4.964400 | 5.392326 | 5.369116 | 5.370605 | 5.549058 | 5.346758 | 5.316530 | 5.371277 | 5.452029 | 5.477229 | 4.897388 | 4.810365 | 5.001575 | 5.058099 | 5.138210 | 5.384835 | 5.442749 | 5.417507 | 5.400831 | 5.429398 | 5.526935 | 5.454526 |
| GO:0048199 | vesicle targeting, to, from or within Golgi | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.082884 | 6.777208 | 6.375513 | 6.654799 | 6.069498 | 6.225772 | 6.709120 | 6.654195 | 6.026986 | 6.124830 | 6.095101 | 6.772700 | 6.821409 | 6.736249 | 6.423115 | 6.331979 | 6.369990 | 6.644829 | 6.668627 | 6.650835 | 6.039793 | 6.053347 | 6.114258 | 6.147861 | 6.167028 | 6.353369 | 6.748148 | 6.704324 | 6.673920 | 6.664089 | 6.675132 | 6.622838 |
| GO:0071639 | positive regulation of monocyte chemotactic protein-1 production | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | SELENOK | 1 | 4.980524 | 6.573818 | 5.077308 | 6.564822 | 4.722358 | 4.381816 | 6.707812 | 6.586816 | 5.101178 | 5.180731 | 4.591997 | 6.530030 | 6.559523 | 6.630060 | 5.689630 | 4.589825 | 4.677604 | 6.482450 | 6.569703 | 6.638104 | 4.525230 | 5.015313 | 4.574465 | 4.253148 | 4.254017 | 4.608445 | 6.689070 | 6.694257 | 6.739572 | 6.646364 | 6.473729 | 6.634008 |
| GO:0072079 | nephron tubule formation | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 2.924339 | 4.741278 | 3.796253 | 4.911537 | 2.651214 | 2.560772 | 4.692457 | 4.913499 | 3.090569 | 3.244461 | 2.256484 | 4.802690 | 4.768773 | 4.647818 | 4.194777 | 3.186499 | 3.834032 | 4.888465 | 4.919643 | 4.926222 | 2.473201 | 3.052624 | 2.320755 | 2.421208 | 2.286355 | 2.900391 | 4.708279 | 4.691047 | 4.677884 | 4.901626 | 4.928167 | 4.910577 |
| GO:0009150 | purine ribonucleotide metabolic process | 3/42 | 391/20870 | 0.0437579095 | 0.2820041 | 0.2468512 | PPT1||OGT||BPNT1 | 3 | 7.977925 | 7.437593 | 8.162638 | 7.690583 | 8.029391 | 8.191313 | 7.472514 | 7.622055 | 7.905842 | 7.952389 | 8.070514 | 7.270393 | 7.513241 | 7.515653 | 8.016962 | 8.288536 | 8.169630 | 7.664807 | 7.786816 | 7.614653 | 8.069416 | 7.966058 | 8.050612 | 8.152239 | 8.134329 | 8.282774 | 7.464443 | 7.479974 | 7.473083 | 7.663324 | 7.606211 | 7.595710 |
| GO:0032259 | methylation | 3/42 | 391/20870 | 0.0437579095 | 0.2820041 | 0.2468512 | METTL2A||OGT||CTNNB1 | 3 | 5.490085 | 5.652357 | 5.604520 | 5.694933 | 5.489179 | 5.625966 | 5.668069 | 5.714284 | 5.455116 | 5.493634 | 5.520751 | 5.673428 | 5.654162 | 5.629139 | 5.662974 | 5.574896 | 5.573864 | 5.699945 | 5.653094 | 5.730704 | 5.475380 | 5.449544 | 5.541063 | 5.578543 | 5.635919 | 5.662173 | 5.686256 | 5.660276 | 5.657501 | 5.696327 | 5.735655 | 5.710595 |
| GO:0002181 | cytoplasmic translation | 2/42 | 168/20870 | 0.0449563775 | 0.2820041 | 0.2468512 | RPS15A||RPL9 | 2 | 11.087627 | 10.859751 | 10.700510 | 10.744064 | 11.069908 | 10.739194 | 10.838454 | 10.747063 | 11.142651 | 11.083777 | 11.034414 | 10.918261 | 10.816127 | 10.842912 | 10.640620 | 10.656367 | 10.799195 | 10.783099 | 10.713760 | 10.734453 | 11.061758 | 11.155057 | 10.988042 | 10.916088 | 10.822622 | 10.435225 | 10.828015 | 10.844481 | 10.842809 | 10.757254 | 10.680672 | 10.800714 |
| GO:0001759 | organ induction | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 2.876870 | 4.463998 | 3.572536 | 4.676217 | 2.664693 | 2.754658 | 4.436676 | 4.676981 | 2.987352 | 3.116668 | 2.441657 | 4.503138 | 4.511615 | 4.373108 | 3.958598 | 3.028508 | 3.583063 | 4.641752 | 4.688430 | 4.697846 | 2.541967 | 2.915606 | 2.499146 | 2.743088 | 2.631319 | 2.878891 | 4.440305 | 4.440791 | 4.428899 | 4.653232 | 4.684001 | 4.693403 |
| GO:0002052 | positive regulation of neuroblast proliferation | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.015886 | 5.042616 | 4.535878 | 5.215336 | 3.897583 | 4.039361 | 5.043382 | 5.231390 | 4.060572 | 4.134868 | 3.835674 | 5.085494 | 5.058998 | 4.981337 | 4.857027 | 4.174109 | 4.495597 | 5.187179 | 5.202402 | 5.255530 | 3.805248 | 4.046049 | 3.829048 | 3.979735 | 3.948273 | 4.179058 | 5.043593 | 5.049055 | 5.037474 | 5.220309 | 5.255715 | 5.217833 |
| GO:0006907 | pinocytosis | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | PPT1 | 1 | 4.876200 | 5.578026 | 5.241122 | 5.687307 | 4.836261 | 4.997123 | 5.619402 | 5.752191 | 4.831826 | 4.973102 | 4.818530 | 5.618404 | 5.560038 | 5.554766 | 5.441269 | 5.085112 | 5.172701 | 5.629168 | 5.682786 | 5.747529 | 4.832989 | 4.842936 | 4.832836 | 4.950421 | 4.941142 | 5.094616 | 5.614930 | 5.626700 | 5.616548 | 5.732794 | 5.786357 | 5.736802 |
| GO:0021819 | layer formation in cerebral cortex | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.480257 | 5.547444 | 5.046847 | 5.695349 | 4.397219 | 4.613118 | 5.488124 | 5.647027 | 4.486476 | 4.614668 | 4.325100 | 5.614704 | 5.580135 | 5.441747 | 5.214686 | 4.852369 | 5.050788 | 5.671870 | 5.693573 | 5.720197 | 4.403152 | 4.450361 | 4.335855 | 4.502023 | 4.510754 | 4.805450 | 5.515833 | 5.485739 | 5.462301 | 5.640476 | 5.660426 | 5.640086 |
| GO:0060231 | mesenchymal to epithelial transition | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.035807 | 4.862230 | 4.291770 | 4.903576 | 3.949445 | 3.844885 | 4.815852 | 4.893062 | 4.070915 | 4.094461 | 3.937106 | 4.914796 | 4.899049 | 4.768416 | 4.476974 | 4.012437 | 4.347035 | 4.889411 | 4.894861 | 4.926182 | 3.914672 | 3.998823 | 3.933481 | 3.787736 | 3.787785 | 3.952766 | 4.833606 | 4.814158 | 4.799592 | 4.887222 | 4.900684 | 4.891247 |
| GO:0008643 | carbohydrate transport | 2/42 | 170/20870 | 0.0459213317 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 4.067728 | 4.603316 | 4.481574 | 4.654156 | 4.098509 | 4.416416 | 4.573956 | 4.592612 | 4.003992 | 4.084798 | 4.112212 | 4.592728 | 4.632689 | 4.584062 | 4.548696 | 4.501119 | 4.390364 | 4.623737 | 4.643619 | 4.694196 | 4.087781 | 4.017844 | 4.185004 | 4.268240 | 4.334638 | 4.621454 | 4.592995 | 4.566268 | 4.562412 | 4.587029 | 4.626629 | 4.563470 |
| GO:0031056 | regulation of histone modification | 2/42 | 172/20870 | 0.0468939695 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.220205 | 5.708676 | 5.435831 | 5.735372 | 5.173859 | 5.355152 | 5.742145 | 5.766290 | 5.192563 | 5.298517 | 5.166105 | 5.717233 | 5.713263 | 5.695439 | 5.578139 | 5.337440 | 5.380325 | 5.722317 | 5.693881 | 5.788285 | 5.161096 | 5.173874 | 5.186496 | 5.329956 | 5.326631 | 5.407411 | 5.752804 | 5.734507 | 5.739062 | 5.745528 | 5.787200 | 5.765842 |
| GO:0032200 | telomere organization | 2/42 | 172/20870 | 0.0468939695 | 0.2820041 | 0.2468512 | RPA2||CTNNB1 | 2 | 6.527224 | 7.318417 | 6.722691 | 7.351486 | 6.424548 | 6.482130 | 7.369899 | 7.397102 | 6.550322 | 6.687990 | 6.319632 | 7.328375 | 7.296327 | 7.330297 | 7.009867 | 6.456790 | 6.645758 | 7.310837 | 7.318517 | 7.422391 | 6.396083 | 6.530065 | 6.340883 | 6.506180 | 6.430487 | 6.508367 | 7.353876 | 7.359842 | 7.395624 | 7.399303 | 7.377155 | 7.414603 |
| GO:0001702 | gastrulation with mouth forming second | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.233107 | 5.422433 | 4.783825 | 5.527500 | 4.128624 | 4.411596 | 5.407874 | 5.493159 | 4.246409 | 4.376566 | 4.058790 | 5.429362 | 5.481119 | 5.353994 | 5.006921 | 4.564284 | 4.745710 | 5.489367 | 5.544794 | 5.547596 | 4.111046 | 4.200540 | 4.071232 | 4.322602 | 4.344757 | 4.555767 | 5.414145 | 5.422920 | 5.386306 | 5.457333 | 5.539409 | 5.481498 |
| GO:0018279 | protein N-linked glycosylation via asparagine | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 6.611683 | 7.314000 | 6.924215 | 7.178923 | 6.633270 | 6.706728 | 7.177432 | 7.133674 | 6.606879 | 6.644518 | 6.582983 | 7.370854 | 7.323022 | 7.245352 | 6.922732 | 6.932867 | 6.917001 | 7.170953 | 7.219199 | 7.145648 | 6.647122 | 6.616764 | 6.635760 | 6.664916 | 6.533595 | 6.897849 | 7.195621 | 7.190091 | 7.146073 | 7.121780 | 7.171177 | 7.107284 |
| GO:0030539 | male genitalia development | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.185989 | 5.023158 | 4.791298 | 5.281531 | 4.144837 | 4.408482 | 4.963834 | 5.227021 | 4.205925 | 4.316726 | 4.019861 | 5.067848 | 5.057332 | 4.940887 | 4.905699 | 4.660670 | 4.797117 | 5.253182 | 5.297813 | 5.293182 | 4.162048 | 4.196455 | 4.073221 | 4.298571 | 4.131497 | 4.728146 | 4.982185 | 4.968891 | 4.940106 | 5.223126 | 5.251268 | 5.206312 |
| GO:0031280 | negative regulation of cyclase activity | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 3.135829 | 3.645431 | 3.430818 | 3.804674 | 3.136395 | 3.357641 | 3.640858 | 3.762544 | 3.071300 | 3.116350 | 3.216013 | 3.607336 | 3.706078 | 3.620879 | 3.462826 | 3.493845 | 3.330629 | 3.808563 | 3.753030 | 3.850765 | 3.162266 | 3.029940 | 3.210953 | 3.236554 | 3.254794 | 3.558410 | 3.701679 | 3.637046 | 3.581333 | 3.763581 | 3.802010 | 3.720900 |
| GO:0035994 | response to muscle stretch | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.295324 | 5.766372 | 5.482324 | 5.800913 | 5.302583 | 5.534643 | 5.754283 | 5.793782 | 5.206304 | 5.394725 | 5.278651 | 5.759620 | 5.792801 | 5.746296 | 5.541956 | 5.390393 | 5.510235 | 5.797505 | 5.751147 | 5.852309 | 5.319563 | 5.248738 | 5.337920 | 5.481705 | 5.587380 | 5.532909 | 5.774205 | 5.738718 | 5.749695 | 5.798215 | 5.832469 | 5.749458 |
| GO:0051571 | positive regulation of histone H3-K4 methylation | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.933372 | 5.950992 | 5.408666 | 6.057853 | 4.791517 | 4.997187 | 5.967625 | 6.093130 | 4.956718 | 5.132304 | 4.674667 | 5.981102 | 5.970969 | 5.899543 | 5.700721 | 5.124703 | 5.341159 | 6.033299 | 6.050498 | 6.089192 | 4.754633 | 4.920684 | 4.689269 | 4.945504 | 4.827370 | 5.194090 | 5.982310 | 5.958044 | 5.962405 | 6.077487 | 6.110176 | 6.091541 |
| GO:0060479 | lung cell differentiation | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.017536 | 5.185393 | 4.438673 | 5.324341 | 3.879529 | 3.851637 | 5.169273 | 5.307459 | 4.096177 | 4.225878 | 3.674934 | 5.219671 | 5.195400 | 5.139955 | 4.701110 | 4.108405 | 4.446023 | 5.289230 | 5.326481 | 5.356524 | 3.844676 | 4.066458 | 3.704143 | 3.862316 | 3.723575 | 3.959339 | 5.172299 | 5.178618 | 5.156816 | 5.304343 | 5.293869 | 5.324003 |
| GO:0060571 | morphogenesis of an epithelial fold | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.322489 | 4.899118 | 4.005645 | 5.038267 | 3.143350 | 3.210679 | 4.864783 | 5.038273 | 3.426060 | 3.569601 | 2.885632 | 4.927632 | 4.948479 | 4.817857 | 4.339835 | 3.552040 | 4.018737 | 5.004107 | 5.061777 | 5.048290 | 3.067936 | 3.384798 | 2.940366 | 3.129770 | 2.996010 | 3.465025 | 4.877994 | 4.858138 | 4.858124 | 5.020901 | 5.057419 | 5.036266 |
| GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | EMC3 | 1 | 6.347418 | 6.393076 | 6.328499 | 6.314187 | 6.296019 | 6.402076 | 6.323221 | 6.273736 | 6.362783 | 6.259390 | 6.415724 | 6.398470 | 6.416687 | 6.363569 | 6.257137 | 6.366761 | 6.359027 | 6.357352 | 6.285429 | 6.298762 | 6.281865 | 6.205758 | 6.394182 | 6.367062 | 6.450226 | 6.387634 | 6.363332 | 6.289719 | 6.315645 | 6.266644 | 6.302919 | 6.251156 |
| GO:0072077 | renal vesicle morphogenesis | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.056552 | 4.902164 | 4.324336 | 4.946776 | 3.974621 | 3.875965 | 4.852708 | 4.930740 | 4.083091 | 4.110842 | 3.972013 | 4.951501 | 4.940798 | 4.809931 | 4.506500 | 4.042536 | 4.384868 | 4.934744 | 4.936259 | 4.969064 | 3.942896 | 4.016554 | 3.963409 | 3.806471 | 3.830150 | 3.984702 | 4.874389 | 4.849745 | 4.833698 | 4.925829 | 4.939164 | 4.927190 |
| GO:0072530 | purine-containing compound transmembrane transport | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | SLC25A17 | 1 | 7.334416 | 6.876285 | 7.224422 | 6.928702 | 7.302094 | 7.255302 | 6.773800 | 6.936878 | 7.382610 | 7.434372 | 7.173230 | 6.957451 | 6.867393 | 6.799658 | 7.114997 | 7.186200 | 7.360861 | 7.019252 | 6.851025 | 6.910761 | 7.315392 | 7.361404 | 7.226229 | 7.350105 | 7.333013 | 7.065522 | 6.791294 | 6.769064 | 6.760870 | 6.923044 | 6.955512 | 6.931883 |
| GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.050925 | 6.526198 | 6.390253 | 6.595356 | 5.894059 | 5.938846 | 6.512245 | 6.565230 | 6.140803 | 6.261614 | 5.689655 | 6.544052 | 6.530052 | 6.504208 | 6.576084 | 6.138749 | 6.422364 | 6.614854 | 6.548969 | 6.621142 | 5.853012 | 6.106415 | 5.692269 | 6.004148 | 5.848993 | 5.959013 | 6.531359 | 6.485509 | 6.519476 | 6.560244 | 6.567236 | 6.568197 |
| GO:0009259 | ribonucleotide metabolic process | 3/42 | 408/20870 | 0.0485907586 | 0.2820041 | 0.2468512 | PPT1||OGT||BPNT1 | 3 | 7.919737 | 7.388427 | 8.106259 | 7.638884 | 7.970475 | 8.132743 | 7.424156 | 7.571172 | 7.848015 | 7.894685 | 8.011555 | 7.223047 | 7.463410 | 7.465613 | 7.962857 | 8.230679 | 8.112806 | 7.612653 | 7.733855 | 7.564844 | 8.010166 | 7.907618 | 7.991584 | 8.092827 | 8.074938 | 8.225711 | 7.416148 | 7.431574 | 7.424704 | 7.611726 | 7.555722 | 7.545177 |
| GO:0007035 | vacuolar acidification | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.659481 | 5.961256 | 5.974208 | 6.000876 | 5.709970 | 5.929753 | 5.814710 | 5.894791 | 5.612986 | 5.633290 | 5.729466 | 6.014694 | 5.987968 | 5.877468 | 5.886847 | 6.042394 | 5.989076 | 6.033907 | 6.016195 | 5.951220 | 5.713907 | 5.642282 | 5.770851 | 5.800589 | 5.871084 | 6.100306 | 5.852912 | 5.811683 | 5.778573 | 5.890995 | 5.945659 | 5.845990 |
| GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.182462 | 5.257771 | 4.879434 | 5.374684 | 4.081403 | 4.380957 | 5.199590 | 5.361063 | 4.210876 | 4.269704 | 4.058670 | 5.282283 | 5.304603 | 5.183573 | 5.080425 | 4.709979 | 4.822543 | 5.330448 | 5.403935 | 5.388631 | 3.992518 | 4.168673 | 4.077635 | 4.119806 | 4.136483 | 4.783084 | 5.232786 | 5.210340 | 5.154521 | 5.357564 | 5.384399 | 5.340892 |
| GO:0018196 | peptidyl-asparagine modification | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 6.563466 | 7.260196 | 6.878812 | 7.126894 | 6.585306 | 6.664095 | 7.125222 | 7.081463 | 6.559104 | 6.596706 | 6.533894 | 7.316208 | 7.270090 | 7.191547 | 6.878697 | 6.889096 | 6.868571 | 7.119194 | 7.166466 | 7.094084 | 6.600971 | 6.567868 | 6.586887 | 6.622576 | 6.487832 | 6.857394 | 7.143931 | 7.137609 | 7.093605 | 7.069377 | 7.118337 | 7.055924 |
| GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | SELENOK | 1 | 6.344581 | 6.750600 | 6.615334 | 6.718064 | 6.380143 | 6.519095 | 6.625619 | 6.659173 | 6.302951 | 6.305307 | 6.422232 | 6.784635 | 6.787697 | 6.676723 | 6.606354 | 6.616506 | 6.623092 | 6.711851 | 6.750612 | 6.691096 | 6.354220 | 6.333198 | 6.450290 | 6.420558 | 6.484730 | 6.642840 | 6.660838 | 6.628577 | 6.586481 | 6.653228 | 6.705025 | 6.617933 |
| GO:0033233 | regulation of protein sumoylation | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.635760 | 6.419616 | 6.086879 | 6.516595 | 5.495958 | 5.625570 | 6.459275 | 6.571507 | 5.688344 | 5.825571 | 5.353436 | 6.443268 | 6.409013 | 6.406273 | 6.273317 | 5.894603 | 6.067746 | 6.501693 | 6.494084 | 6.553286 | 5.436771 | 5.634912 | 5.405292 | 5.525115 | 5.490884 | 5.835202 | 6.473593 | 6.438171 | 6.465823 | 6.550171 | 6.587489 | 6.576608 |
| GO:0072087 | renal vesicle development | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.969514 | 4.814975 | 4.237995 | 4.859635 | 3.887855 | 3.789980 | 4.765476 | 4.843547 | 3.995731 | 4.023754 | 3.885380 | 4.864444 | 4.853474 | 4.722746 | 4.420112 | 3.956334 | 4.298470 | 4.847517 | 4.848971 | 4.882150 | 3.856042 | 3.929875 | 3.876637 | 3.720590 | 3.743622 | 3.899116 | 4.787014 | 4.762634 | 4.746492 | 4.838785 | 4.851908 | 4.839912 |
| GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | TXNDC12 | 1 | 5.873179 | 6.577389 | 6.359894 | 6.540368 | 5.772846 | 5.844196 | 6.471804 | 6.461404 | 5.939119 | 6.048445 | 5.594002 | 6.608743 | 6.599819 | 6.522040 | 6.435934 | 6.226247 | 6.408625 | 6.546235 | 6.550960 | 6.523764 | 5.713128 | 5.933810 | 5.656488 | 5.692751 | 5.588606 | 6.179917 | 6.497522 | 6.471540 | 6.445888 | 6.457117 | 6.490295 | 6.436286 |
| GO:1903077 | negative regulation of protein localization to plasma membrane | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | RSC1A1 | 1 | 4.927336 | 5.707485 | 5.036068 | 5.711967 | 4.894942 | 5.025592 | 5.692290 | 5.765361 | 4.886543 | 4.969553 | 4.924714 | 5.707900 | 5.731086 | 5.683068 | 5.106005 | 4.969138 | 5.029797 | 5.687209 | 5.704717 | 5.743402 | 4.903275 | 4.897639 | 4.883844 | 5.043208 | 4.991985 | 5.041004 | 5.676750 | 5.723769 | 5.675828 | 5.750329 | 5.775910 | 5.769722 |
GO: MF
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0003735 | structural constituent of ribosome | 5/42 | 193/20678 | 4.322112e-05 | 0.006785716 | 0.005277526 | MRPS33||RPS15A||RPL9||MRPS23||MRPL46 | 5 | 10.9355568 | 10.688269 | 10.5388820 | 10.566723 | 10.9196846 | 10.5763827 | 10.663932 | 10.566342 | 10.9917019 | 10.9272864 | 10.8857001 | 10.750258 | 10.641796 | 10.670553 | 10.4755825 | 10.4948158 | 10.6405452 | 10.606965 | 10.536699 | 10.555584 | 10.9089765 | 11.0045888 | 10.8407820 | 10.7528062 | 10.6647803 | 10.2665451 | 10.653919 | 10.668985 | 10.668840 | 10.578059 | 10.496562 | 10.621625 |
| GO:0015035 | protein-disulfide reductase activity | 2/42 | 39/20678 | 2.845537e-03 | 0.173862098 | 0.135219600 | TXNDC12||SELENOT | 2 | 6.3591793 | 7.129436 | 6.5199649 | 6.998543 | 6.3159593 | 6.3114794 | 7.029613 | 6.943397 | 6.4044616 | 6.4445966 | 6.2185453 | 7.149729 | 7.146930 | 7.090888 | 6.6103900 | 6.4055875 | 6.5365115 | 6.961836 | 7.024432 | 7.008629 | 6.3301616 | 6.3913259 | 6.2212768 | 6.3414663 | 6.1806987 | 6.4032268 | 7.020204 | 7.030988 | 7.037593 | 6.937780 | 6.962004 | 6.930214 |
| GO:0015036 | disulfide oxidoreductase activity | 2/42 | 44/20678 | 3.609513e-03 | 0.173862098 | 0.135219600 | TXNDC12||SELENOT | 2 | 6.3576393 | 7.052684 | 6.5086874 | 6.930216 | 6.2959948 | 6.2409811 | 6.955551 | 6.880823 | 6.4224283 | 6.4515814 | 6.1842966 | 7.079035 | 7.067306 | 7.010792 | 6.5854670 | 6.4045529 | 6.5301328 | 6.891673 | 6.955352 | 6.942836 | 6.3267635 | 6.3993886 | 6.1506224 | 6.2809885 | 6.0747151 | 6.3529936 | 6.947774 | 6.956255 | 6.962584 | 6.875448 | 6.899622 | 6.867201 |
| GO:0016874 | ligase activity | 3/42 | 184/20678 | 6.167614e-03 | 0.173862098 | 0.135219600 | CPS1||YARS2||SUCLG1 | 3 | 7.2088914 | 7.076004 | 7.6262817 | 7.382555 | 7.2240138 | 7.5099627 | 7.117392 | 7.324684 | 7.1497183 | 7.2575240 | 7.2173807 | 6.923483 | 7.144060 | 7.149164 | 7.5855949 | 7.7213818 | 7.5668897 | 7.319226 | 7.496982 | 7.324265 | 7.2745739 | 7.1834315 | 7.2125285 | 7.4502449 | 7.3745850 | 7.6864345 | 7.100005 | 7.124210 | 7.127803 | 7.378091 | 7.303267 | 7.291150 |
| GO:0016597 | amino acid binding | 2/42 | 61/20678 | 6.831887e-03 | 0.173862098 | 0.135219600 | CPS1||YARS2 | 2 | 5.3532404 | 5.622106 | 5.4702382 | 5.577023 | 5.3191520 | 5.4999836 | 5.588206 | 5.559977 | 5.3274449 | 5.4226094 | 5.3070100 | 5.649059 | 5.626284 | 5.590370 | 5.5224862 | 5.4433929 | 5.4433810 | 5.584763 | 5.522128 | 5.622404 | 5.3508084 | 5.2956128 | 5.3104680 | 5.4896058 | 5.4098928 | 5.5944953 | 5.602792 | 5.566174 | 5.595392 | 5.552409 | 5.571974 | 5.555471 |
| GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors | 2/42 | 61/20678 | 6.831887e-03 | 0.173862098 | 0.135219600 | TXNDC12||SELENOT | 2 | 6.0362596 | 6.759133 | 6.1776892 | 6.675592 | 5.9882250 | 5.9438403 | 6.673392 | 6.637094 | 6.0913853 | 6.1267732 | 5.8783152 | 6.789111 | 6.769426 | 6.717929 | 6.2595853 | 6.0773312 | 6.1903157 | 6.639324 | 6.696002 | 6.690773 | 6.0053587 | 6.0891793 | 5.8609675 | 5.9732101 | 5.8006105 | 6.0467591 | 6.661295 | 6.678209 | 6.680596 | 6.632757 | 6.652385 | 6.626010 |
| GO:0016595 | glutamate binding | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | CPS1 | 1 | 4.8812511 | 5.296357 | 5.3460360 | 5.419536 | 4.8029466 | 5.0406410 | 5.294881 | 5.345292 | 4.8528529 | 5.1389030 | 4.6018981 | 5.277354 | 5.342872 | 5.267680 | 5.4672002 | 5.2396336 | 5.3220129 | 5.406858 | 5.364778 | 5.484411 | 4.9177338 | 4.8519293 | 4.6228591 | 4.9962355 | 4.5837323 | 5.4211101 | 5.315271 | 5.270023 | 5.298987 | 5.337181 | 5.345535 | 5.353115 |
| GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | CPS1 | 1 | 4.9987928 | 6.407354 | 5.3754951 | 6.069058 | 4.8944763 | 5.1392240 | 6.405037 | 6.127690 | 5.0509740 | 5.0727487 | 4.8636341 | 6.458865 | 6.359544 | 6.401928 | 5.5928092 | 5.2332673 | 5.2728348 | 6.001681 | 6.052074 | 6.149487 | 4.8225605 | 4.9705534 | 4.8864883 | 4.9873463 | 5.0133116 | 5.3822806 | 6.382541 | 6.399952 | 6.432178 | 6.102092 | 6.145393 | 6.135231 |
| GO:0045294 | alpha-catenin binding | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | CTNNB1 | 1 | 4.3613657 | 5.976656 | 5.0875183 | 6.100524 | 4.1521887 | 4.2589306 | 5.910110 | 6.094956 | 4.4342950 | 4.6308175 | 3.9324116 | 6.015496 | 6.020998 | 5.889689 | 5.4547244 | 4.6319118 | 5.0597610 | 6.054806 | 6.119992 | 6.125704 | 4.1068354 | 4.3874668 | 3.9241824 | 4.2019348 | 4.1354010 | 4.4234558 | 5.933336 | 5.915419 | 5.881088 | 6.090157 | 6.118847 | 6.075529 |
| GO:0098505 | G-rich strand telomeric DNA binding | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | RPA2 | 1 | 7.8639006 | 8.865442 | 7.9019885 | 8.837516 | 7.7471186 | 7.9396794 | 8.933782 | 8.848420 | 7.8381828 | 8.0505253 | 7.6787717 | 8.841937 | 8.849433 | 8.904152 | 8.3449328 | 7.5448896 | 7.6842147 | 8.803391 | 8.817814 | 8.889848 | 7.6985875 | 7.8180269 | 7.7219444 | 7.9734512 | 7.9768184 | 7.8660404 | 8.919179 | 8.919930 | 8.961824 | 8.831142 | 8.862156 | 8.851788 |
| GO:0005347 | ATP transmembrane transporter activity | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | SLC25A17 | 1 | 8.0755980 | 7.506628 | 7.8718292 | 7.567456 | 8.0315895 | 7.9306649 | 7.424731 | 7.596554 | 8.1358878 | 8.1881644 | 7.8848889 | 7.600662 | 7.477620 | 7.436502 | 7.7598370 | 7.8062172 | 8.0342337 | 7.660309 | 7.473092 | 7.562887 | 8.0465041 | 8.1092822 | 7.9335155 | 8.0741904 | 8.0433280 | 7.6340786 | 7.431371 | 7.419349 | 7.423446 | 7.587832 | 7.595954 | 7.605820 |
| GO:0015166 | polyol transmembrane transporter activity | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | SLC5A3 | 1 | -0.5048209 | 1.484559 | 0.4680739 | 1.635668 | -0.4875053 | -0.1353992 | 1.598437 | 1.697323 | -0.5324684 | -0.3243855 | -0.6798407 | 1.409021 | 1.576807 | 1.462728 | 0.8923442 | 0.1181986 | 0.2728783 | 1.367223 | 1.709340 | 1.795805 | -0.4451347 | -0.5983061 | -0.4252156 | -0.4197511 | -0.2662565 | 0.2044235 | 1.687472 | 1.645932 | 1.451032 | 1.607331 | 1.712723 | 1.767354 |
| GO:0043047 | single-stranded telomeric DNA binding | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | RPA2 | 1 | 7.6069557 | 8.614827 | 7.6474848 | 8.588773 | 7.4900772 | 7.6836654 | 8.683166 | 8.597709 | 7.5815007 | 7.7934887 | 7.4216517 | 8.591013 | 8.598829 | 8.653823 | 8.0915533 | 7.2890544 | 7.4291412 | 8.553086 | 8.569484 | 8.642183 | 7.4415228 | 7.5607861 | 7.4651389 | 7.7163974 | 7.7208502 | 7.6110964 | 8.668486 | 8.669000 | 8.711585 | 8.580424 | 8.611864 | 8.600661 |
| GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | CCNT1 | 1 | 5.8361480 | 6.825090 | 5.9379185 | 6.927780 | 5.5786891 | 5.0748901 | 6.910883 | 7.098084 | 6.0036408 | 6.1199833 | 5.2355224 | 6.887575 | 6.775148 | 6.810243 | 6.1727634 | 5.5413501 | 6.0268154 | 6.901721 | 6.919429 | 6.961533 | 5.4733427 | 5.9940951 | 5.1379252 | 5.1409405 | 4.9646409 | 5.1129403 | 6.864738 | 6.937673 | 6.929141 | 7.109520 | 7.061855 | 7.122179 |
| GO:0047134 | protein-disulfide reductase (NAD(P)) activity | 1/42 | 13/20678 | 2.609295e-02 | 0.173862098 | 0.135219600 | SELENOT | 1 | 6.6165227 | 7.008156 | 6.8358345 | 7.257034 | 6.5215753 | 6.1913566 | 7.062189 | 7.255944 | 6.7439812 | 6.7786612 | 6.2740491 | 6.997719 | 6.996135 | 7.030355 | 6.9480716 | 6.6337374 | 6.9059639 | 7.224431 | 7.281470 | 7.264608 | 6.5442389 | 6.7653105 | 6.1998501 | 6.3523581 | 5.8262600 | 6.3360819 | 7.060565 | 7.049792 | 7.076087 | 7.252542 | 7.241333 | 7.273768 |
| GO:1990226 | histone methyltransferase binding | 1/42 | 13/20678 | 2.609295e-02 | 0.173862098 | 0.135219600 | CTNNB1 | 1 | 7.0177141 | 7.894986 | 7.4241274 | 8.123230 | 6.8271261 | 6.9438234 | 7.993981 | 8.174724 | 7.0044536 | 7.2266590 | 6.7888505 | 7.909919 | 7.875652 | 7.899175 | 7.8324877 | 7.0021715 | 7.3138525 | 8.077494 | 8.110965 | 8.179355 | 6.8250696 | 6.9694127 | 6.6715462 | 7.0360965 | 6.8839556 | 6.9067048 | 7.986720 | 7.989305 | 8.005844 | 8.176500 | 8.171574 | 8.176092 |
| GO:0010851 | cyclase regulator activity | 1/42 | 14/20678 | 2.807234e-02 | 0.173862098 | 0.135219600 | ADGRV1 | 1 | 7.8647313 | 8.219995 | 7.8423704 | 8.047909 | 7.8630468 | 8.0440130 | 8.230649 | 8.087167 | 7.8067909 | 7.9229723 | 7.8620888 | 8.215024 | 8.222578 | 8.222371 | 7.8885692 | 7.8251407 | 7.8122387 | 8.081145 | 7.989707 | 8.071143 | 7.8486425 | 7.8174715 | 7.9210602 | 8.0389069 | 8.1317237 | 7.9560504 | 8.220974 | 8.239486 | 8.231427 | 8.080660 | 8.097528 | 8.083256 |
| GO:0072582 | 17-beta-hydroxysteroid dehydrogenase (NADP+) activity | 1/42 | 14/20678 | 2.807234e-02 | 0.173862098 | 0.135219600 | HSD17B7 | 1 | 3.2097645 | 5.015721 | 3.2339704 | 4.905140 | 3.1609219 | 2.9868018 | 4.900907 | 4.897982 | 3.2475434 | 3.3143469 | 3.0550092 | 5.036789 | 5.018901 | 4.991106 | 3.6028599 | 2.9484481 | 3.0629221 | 4.791147 | 4.978592 | 4.938990 | 3.0714517 | 3.2631632 | 3.1415940 | 2.9426533 | 3.0877234 | 2.9244168 | 4.899167 | 4.922718 | 4.880526 | 4.920868 | 4.943932 | 4.826481 |
| GO:0008252 | nucleotidase activity | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | BPNT1 | 1 | 6.2899468 | 6.077989 | 6.2439033 | 6.065577 | 6.3297709 | 6.4343397 | 6.042705 | 6.026160 | 6.2419477 | 6.2360813 | 6.3867097 | 6.097056 | 6.087575 | 6.048886 | 6.1457245 | 6.2868000 | 6.2943817 | 6.062154 | 6.046764 | 6.087518 | 6.3431481 | 6.2781567 | 6.3665602 | 6.3981784 | 6.4879345 | 6.4153240 | 6.050713 | 6.062381 | 6.014592 | 6.007770 | 6.061502 | 6.008547 |
| GO:0015929 | hexosaminidase activity | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | OGA | 1 | 4.6931369 | 5.090033 | 5.3273920 | 5.119205 | 4.7382147 | 5.0766574 | 5.005393 | 5.198033 | 4.5867447 | 4.6959293 | 4.7896074 | 5.136267 | 5.106989 | 5.024528 | 5.3987605 | 5.3536432 | 5.2241282 | 5.063162 | 5.168656 | 5.123857 | 4.7341888 | 4.6221401 | 4.8493686 | 4.8643607 | 4.9025394 | 5.3991956 | 5.048130 | 4.995617 | 4.971361 | 5.188242 | 5.240697 | 5.164093 |
| GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | SELENOT | 1 | 6.5023694 | 7.187224 | 6.6864700 | 7.436011 | 6.4184528 | 6.0585265 | 7.238267 | 7.463466 | 6.6277915 | 6.6769535 | 6.1449031 | 7.198502 | 7.165145 | 7.197775 | 6.8526547 | 6.4535376 | 6.7249193 | 7.399112 | 7.455426 | 7.452800 | 6.4031454 | 6.6902675 | 6.1023356 | 6.2074153 | 5.7496326 | 6.1749237 | 7.218601 | 7.242082 | 7.253893 | 7.461428 | 7.447891 | 7.480887 |
| GO:0032977 | membrane insertase activity | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | EMC3 | 1 | 6.1601178 | 6.274879 | 6.1797816 | 6.241185 | 6.0757611 | 6.0803389 | 6.212230 | 6.186938 | 6.1793181 | 6.1484249 | 6.1524143 | 6.280881 | 6.281896 | 6.261771 | 6.1886570 | 6.1606494 | 6.1898497 | 6.264381 | 6.246935 | 6.211740 | 6.0687282 | 6.0159985 | 6.1398729 | 6.0435058 | 6.0893431 | 6.1074177 | 6.243151 | 6.193962 | 6.199068 | 6.175862 | 6.200969 | 6.183871 |
| GO:0070411 | I-SMAD binding | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | CTNNB1 | 1 | 3.5045979 | 5.252527 | 4.2066234 | 5.366438 | 3.3334134 | 3.4024071 | 5.240125 | 5.405324 | 3.5782712 | 3.7583755 | 3.0996852 | 5.296251 | 5.272257 | 5.186793 | 4.5491324 | 3.7758363 | 4.1923907 | 5.334880 | 5.360564 | 5.403048 | 3.2099192 | 3.5550590 | 3.2067942 | 3.2826035 | 3.2435578 | 3.6460127 | 5.245093 | 5.239893 | 5.235373 | 5.393573 | 5.418427 | 5.403863 |
| GO:0044325 | transmembrane transporter binding | 2/42 | 136/20678 | 3.114688e-02 | 0.173862098 | 0.135219600 | CTNNB1||SLC5A3 | 2 | 5.9095468 | 6.305045 | 5.9339853 | 6.346192 | 5.8727004 | 5.9447286 | 6.310415 | 6.377671 | 5.9104316 | 5.9765472 | 5.8383531 | 6.318074 | 6.305527 | 6.291411 | 6.0455000 | 5.8433793 | 5.9055892 | 6.346504 | 6.302301 | 6.388484 | 5.8595494 | 5.8854946 | 5.8729406 | 5.9564980 | 5.9548215 | 5.9226144 | 6.311496 | 6.304978 | 6.314755 | 6.369875 | 6.390043 | 6.373014 |
| GO:0098847 | sequence-specific single stranded DNA binding | 1/42 | 16/20678 | 3.201934e-02 | 0.173862098 | 0.135219600 | RPA2 | 1 | 7.5404854 | 8.481106 | 7.5888917 | 8.445511 | 7.4396641 | 7.6414837 | 8.546066 | 8.462177 | 7.5114007 | 7.7135784 | 7.3763853 | 8.459732 | 8.464556 | 8.518293 | 7.9740187 | 7.2848387 | 7.4110943 | 8.417424 | 8.422236 | 8.495538 | 7.3961974 | 7.4930833 | 7.4280151 | 7.6752790 | 7.6657888 | 7.5815508 | 8.533020 | 8.532982 | 8.571848 | 8.445639 | 8.477838 | 8.462873 |
| GO:0043177 | organic acid binding | 2/42 | 145/20678 | 3.502018e-02 | 0.173862098 | 0.135219600 | CPS1||YARS2 | 2 | 4.8918083 | 5.296771 | 5.0266792 | 5.230447 | 4.8707182 | 5.0356702 | 5.264285 | 5.194759 | 4.8658025 | 4.9376520 | 4.8708497 | 5.306869 | 5.306377 | 5.276864 | 5.1001442 | 4.9907352 | 4.9862502 | 5.226418 | 5.190996 | 5.272759 | 4.8649033 | 4.8537192 | 4.8932440 | 4.9824659 | 4.9803163 | 5.1384604 | 5.276520 | 5.243545 | 5.272567 | 5.183201 | 5.216687 | 5.184136 |
| GO:0099106 | ion channel regulator activity | 2/42 | 145/20678 | 3.502018e-02 | 0.173862098 | 0.135219600 | SLC5A3||RSC1A1 | 2 | 6.4875454 | 6.249267 | 6.3109826 | 6.219883 | 6.5144603 | 6.5007302 | 6.237431 | 6.231194 | 6.4631031 | 6.4583886 | 6.5396920 | 6.282579 | 6.237837 | 6.226778 | 6.2088125 | 6.3404472 | 6.3782581 | 6.261097 | 6.159472 | 6.237136 | 6.5027427 | 6.5005876 | 6.5397143 | 6.5462467 | 6.5853781 | 6.3607291 | 6.235632 | 6.240891 | 6.235765 | 6.224458 | 6.235980 | 6.233117 |
| GO:0005402 | carbohydrate:cation symporter activity | 1/42 | 18/20678 | 3.595070e-02 | 0.173862098 | 0.135219600 | SLC5A3 | 1 | 1.2994216 | 2.917732 | 2.4149115 | 3.020926 | 1.3251257 | 2.0087806 | 2.869390 | 2.898371 | 1.2451291 | 1.4262759 | 1.2178431 | 2.862021 | 3.007256 | 2.879520 | 2.6857874 | 2.3028151 | 2.2110395 | 2.896213 | 3.100154 | 3.058464 | 1.3447988 | 1.3052264 | 1.3250804 | 1.6917520 | 1.6605450 | 2.5062493 | 2.869477 | 2.916095 | 2.821033 | 2.861849 | 2.949251 | 2.882560 |
| GO:0015095 | magnesium ion transmembrane transporter activity | 1/42 | 18/20678 | 3.595070e-02 | 0.173862098 | 0.135219600 | MAGT1 | 1 | 2.9029881 | 5.077355 | 3.7262729 | 5.014121 | 2.8304944 | 3.1503177 | 4.996710 | 4.988288 | 2.9365992 | 3.1598087 | 2.5474654 | 5.119175 | 5.078413 | 5.033197 | 4.1317413 | 3.4513792 | 3.4900582 | 4.907910 | 5.087220 | 5.041276 | 2.7603776 | 3.0223036 | 2.6868729 | 3.0166431 | 2.8866305 | 3.4786221 | 4.989658 | 5.031835 | 4.967904 | 4.974955 | 5.043993 | 4.944094 |
| GO:0016290 | palmitoyl-CoA hydrolase activity | 1/42 | 18/20678 | 3.595070e-02 | 0.173862098 | 0.135219600 | PPT1 | 1 | 4.8294795 | 5.120267 | 5.0602508 | 5.161528 | 4.7885047 | 4.8861227 | 5.041373 | 5.148558 | 4.8226492 | 4.8583722 | 4.8069345 | 5.222489 | 5.093093 | 5.039012 | 5.1292083 | 4.9594264 | 5.0867559 | 5.169699 | 5.132480 | 5.181945 | 4.8141912 | 4.7425184 | 4.8077206 | 4.8452219 | 4.8948662 | 4.9173389 | 5.047737 | 5.050166 | 5.026093 | 5.115872 | 5.181854 | 5.147193 |
| GO:0016247 | channel regulator activity | 2/42 | 151/20678 | 3.770134e-02 | 0.173862098 | 0.135219600 | SLC5A3||RSC1A1 | 2 | 6.4671635 | 6.225858 | 6.2942541 | 6.196939 | 6.4932338 | 6.4801780 | 6.211999 | 6.206853 | 6.4430616 | 6.4400957 | 6.5170096 | 6.257580 | 6.216457 | 6.202974 | 6.1927860 | 6.3238197 | 6.3608066 | 6.238343 | 6.137018 | 6.213534 | 6.4840260 | 6.4791308 | 6.5162617 | 6.5266167 | 6.5591258 | 6.3457646 | 6.210414 | 6.215494 | 6.210081 | 6.199957 | 6.211835 | 6.208741 |
| GO:0000295 | adenine nucleotide transmembrane transporter activity | 1/42 | 19/20678 | 3.791053e-02 | 0.173862098 | 0.135219600 | SLC25A17 | 1 | 7.3764301 | 6.933433 | 7.2594115 | 6.988879 | 7.3455838 | 7.3057568 | 6.834909 | 6.995078 | 7.4212799 | 7.4732935 | 7.2227886 | 7.011317 | 6.923123 | 6.861940 | 7.1447921 | 7.2274225 | 7.3946877 | 7.074890 | 6.909870 | 6.977090 | 7.3600612 | 7.3994461 | 7.2744268 | 7.4020830 | 7.3879628 | 7.1086807 | 6.851226 | 6.828845 | 6.824511 | 6.986724 | 7.007538 | 6.990886 |
| GO:0005346 | purine ribonucleotide transmembrane transporter activity | 1/42 | 19/20678 | 3.791053e-02 | 0.173862098 | 0.135219600 | SLC25A17 | 1 | 7.3764301 | 6.933433 | 7.2594115 | 6.988879 | 7.3455838 | 7.3057568 | 6.834909 | 6.995078 | 7.4212799 | 7.4732935 | 7.2227886 | 7.011317 | 6.923123 | 6.861940 | 7.1447921 | 7.2274225 | 7.3946877 | 7.074890 | 6.909870 | 6.977090 | 7.3600612 | 7.3994461 | 7.2744268 | 7.4020830 | 7.3879628 | 7.1086807 | 6.851226 | 6.828845 | 6.824511 | 6.986724 | 7.007538 | 6.990886 |
| GO:0140597 | protein carrier activity | 1/42 | 19/20678 | 3.791053e-02 | 0.173862098 | 0.135219600 | EMC3 | 1 | 6.7919224 | 7.575821 | 6.5858978 | 7.342040 | 6.5186937 | 6.2021494 | 7.549400 | 7.349123 | 6.9019202 | 7.0096735 | 6.3919145 | 7.672605 | 7.522322 | 7.527431 | 6.8090106 | 6.2677752 | 6.6294349 | 7.298090 | 7.334717 | 7.391766 | 6.4670455 | 6.7008853 | 6.3675886 | 6.2287711 | 6.1918672 | 6.1854298 | 7.550081 | 7.552320 | 7.545790 | 7.328164 | 7.353442 | 7.365512 |
| GO:0140101 | catalytic activity, acting on a tRNA | 2/42 | 155/20678 | 3.953147e-02 | 0.173862098 | 0.135219600 | METTL2A||YARS2 | 2 | 5.3059236 | 5.742666 | 5.4673500 | 5.797422 | 5.2499815 | 5.3858380 | 5.752520 | 5.808580 | 5.2915743 | 5.3557952 | 5.2689895 | 5.764949 | 5.738389 | 5.724366 | 5.6066706 | 5.3860845 | 5.3984818 | 5.777893 | 5.760628 | 5.852100 | 5.2427907 | 5.2367639 | 5.2701698 | 5.3275825 | 5.3503714 | 5.4751451 | 5.760563 | 5.739297 | 5.757608 | 5.796447 | 5.831111 | 5.797915 |
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 1/42 | 20/20678 | 3.986647e-02 | 0.173862098 | 0.135219600 | H2AZ1 | 1 | 7.3577173 | 7.693184 | 7.5989688 | 7.669032 | 7.2235780 | 6.9983145 | 7.730754 | 7.632202 | 7.4265789 | 7.6057834 | 6.9684120 | 7.692198 | 7.686174 | 7.701140 | 7.7372336 | 7.3198683 | 7.7037279 | 7.648572 | 7.641847 | 7.715522 | 7.1147122 | 7.4708932 | 7.0484470 | 6.9400906 | 6.8646516 | 7.1721325 | 7.717290 | 7.719457 | 7.755200 | 7.746157 | 7.390512 | 7.732582 |
| GO:0015216 | purine nucleotide transmembrane transporter activity | 1/42 | 22/20678 | 4.376671e-02 | 0.176382223 | 0.137179604 | SLC25A17 | 1 | 7.1355776 | 6.716521 | 7.0325027 | 6.773661 | 7.1044585 | 7.0705005 | 6.619053 | 6.778542 | 7.1801002 | 7.2332660 | 6.9813295 | 6.792894 | 6.707558 | 6.645295 | 6.9259988 | 6.9979301 | 7.1632264 | 6.855350 | 6.698049 | 6.763242 | 7.1190568 | 7.1578476 | 7.0336890 | 7.1634563 | 7.1483584 | 6.8827887 | 6.636201 | 6.613710 | 6.607086 | 6.768834 | 6.793324 | 6.773351 |
| GO:0001965 | G-protein alpha-subunit binding | 1/42 | 24/20678 | 4.765148e-02 | 0.176382223 | 0.137179604 | ADGRV1 | 1 | 4.5451885 | 4.952815 | 4.8597640 | 4.897709 | 4.5808287 | 4.8984120 | 4.906254 | 4.840233 | 4.4810233 | 4.4915913 | 4.6561804 | 4.967460 | 4.992218 | 4.897086 | 4.8675584 | 4.9021049 | 4.8080649 | 4.886210 | 4.890219 | 4.916510 | 4.5462766 | 4.4628392 | 4.7212073 | 4.7395118 | 4.8810353 | 5.0570912 | 4.937714 | 4.901734 | 4.878700 | 4.796255 | 4.888172 | 4.834795 |
| GO:0004303 | estradiol 17-beta-dehydrogenase activity | 1/42 | 24/20678 | 4.765148e-02 | 0.176382223 | 0.137179604 | HSD17B7 | 1 | 5.8497982 | 5.892582 | 5.7432939 | 5.874683 | 5.8971862 | 5.9146491 | 5.774762 | 5.854812 | 5.7894273 | 5.7458776 | 6.0009837 | 5.941890 | 5.917151 | 5.815626 | 5.6509454 | 5.7743352 | 5.8002325 | 5.887111 | 5.835413 | 5.900707 | 5.8917791 | 5.8089889 | 5.9853869 | 5.9041476 | 6.0380301 | 5.7911650 | 5.784950 | 5.782315 | 5.756855 | 5.857535 | 5.900151 | 5.805185 |
| GO:0047617 | acyl-CoA hydrolase activity | 1/42 | 24/20678 | 4.765148e-02 | 0.176382223 | 0.137179604 | PPT1 | 1 | 4.6798582 | 5.121637 | 4.9288249 | 5.173884 | 4.6389671 | 4.7444199 | 5.048887 | 5.174105 | 4.6731788 | 4.7060445 | 4.6599603 | 5.214851 | 5.099996 | 5.044816 | 5.0156183 | 4.8189959 | 4.9450289 | 5.157840 | 5.157699 | 5.205583 | 4.6598803 | 4.5939984 | 4.6619920 | 4.6987602 | 4.7518526 | 4.7814339 | 5.044858 | 5.062107 | 5.039600 | 5.138250 | 5.219373 | 5.163492 |
| GO:0019903 | protein phosphatase binding | 2/42 | 176/20678 | 4.967315e-02 | 0.176382223 | 0.137179604 | RPA2||CTNNB1 | 2 | 6.3758622 | 6.385268 | 6.3278158 | 6.306783 | 6.4005539 | 6.4628681 | 6.359854 | 6.320524 | 6.3363370 | 6.3500000 | 6.4390773 | 6.411108 | 6.386076 | 6.358132 | 6.2603575 | 6.3413652 | 6.3791828 | 6.327297 | 6.260612 | 6.331353 | 6.4032105 | 6.3656792 | 6.4320061 | 6.4921351 | 6.5286685 | 6.3625715 | 6.357471 | 6.364424 | 6.357658 | 6.310663 | 6.338630 | 6.312107 |
GO: CC
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0005840 | ribosome | 5/44 | 258/21916 | 0.0001623284 | 0.03068006 | 0.02563080 | MRPS33||RPS15A||RPL9||MRPS23||MRPL46 | 5 | 10.576229 | 10.351360 | 10.189649 | 10.233622 | 10.561440 | 10.239215 | 10.328566 | 10.235635 | 10.628922 | 10.567142 | 10.530915 | 10.411223 | 10.306409 | 10.334396 | 10.129156 | 10.148514 | 10.286146 | 10.272470 | 10.202213 | 10.225292 | 10.551391 | 10.640795 | 10.488041 | 10.408788 | 10.328489 | 9.937772 | 10.318230 | 10.333382 | 10.334031 | 10.246046 | 10.170486 | 10.287928 |
| GO:0044391 | ribosomal subunit | 4/44 | 206/21916 | 0.0007667866 | 0.04918786 | 0.04109262 | MRPS33||RPS15A||RPL9||MRPL46 | 4 | 10.812276 | 10.576037 | 10.421405 | 10.457971 | 10.797402 | 10.464087 | 10.553284 | 10.458505 | 10.866482 | 10.802817 | 10.765723 | 10.637400 | 10.529809 | 10.558743 | 10.359682 | 10.378633 | 10.520488 | 10.497208 | 10.427157 | 10.448652 | 10.786804 | 10.879599 | 10.721410 | 10.636650 | 10.552373 | 10.159785 | 10.542692 | 10.558128 | 10.558973 | 10.469626 | 10.390654 | 10.512595 |
| GO:0032993 | protein-DNA complex | 4/44 | 207/21916 | 0.0007807598 | 0.04918786 | 0.04109262 | RPA2||H2AZ1||CTNNB1||H2BC6 | 4 | 6.015182 | 6.696574 | 6.235620 | 6.698442 | 5.918405 | 5.934967 | 6.735271 | 6.759565 | 6.053975 | 6.150438 | 5.821641 | 6.715227 | 6.669990 | 6.704121 | 6.447156 | 6.022897 | 6.205256 | 6.673546 | 6.659692 | 6.760027 | 5.879047 | 6.016227 | 5.854627 | 5.897595 | 5.860118 | 6.040820 | 6.721054 | 6.719454 | 6.764844 | 6.760027 | 6.743777 | 6.774724 |
| GO:0000791 | euchromatin | 2/44 | 39/21916 | 0.0027844451 | 0.11286067 | 0.09428627 | H2AZ1||CTNNB1 | 2 | 6.780152 | 7.097828 | 6.975175 | 7.136847 | 6.680968 | 6.658912 | 7.116133 | 7.181746 | 6.799782 | 6.918012 | 6.605578 | 7.122914 | 7.086586 | 7.083651 | 7.034150 | 6.819581 | 7.059936 | 7.128353 | 7.110642 | 7.170880 | 6.597744 | 6.807509 | 6.628641 | 6.585276 | 6.649048 | 6.738304 | 7.107403 | 7.115653 | 7.125288 | 7.169972 | 7.195959 | 7.179188 |
| GO:0098798 | mitochondrial protein-containing complex | 4/44 | 302/21916 | 0.0031100405 | 0.11286067 | 0.09428627 | MRPS33||ATP5MK||NDUFA4||MRPL46 | 4 | 8.765320 | 8.343565 | 9.099184 | 8.588167 | 8.819336 | 9.024468 | 8.381052 | 8.489704 | 8.726756 | 8.752249 | 8.815501 | 8.112028 | 8.442274 | 8.451196 | 9.017934 | 9.205907 | 9.067064 | 8.536758 | 8.741074 | 8.472880 | 8.848926 | 8.756129 | 8.850935 | 8.926991 | 8.919554 | 9.207761 | 8.376009 | 8.394005 | 8.373051 | 8.558383 | 8.444859 | 8.463278 |
| GO:0140534 | endoplasmic reticulum protein-containing complex | 3/44 | 153/21916 | 0.0035828784 | 0.11286067 | 0.09428627 | MAGT1||EMC3||NBAS | 3 | 6.827336 | 7.484077 | 7.052437 | 7.263255 | 6.839109 | 6.958832 | 7.349913 | 7.211815 | 6.823243 | 6.849803 | 6.808659 | 7.508672 | 7.515868 | 7.425974 | 7.038785 | 7.068445 | 7.049924 | 7.245957 | 7.297556 | 7.245629 | 6.840203 | 6.826268 | 6.850754 | 6.911243 | 6.885818 | 7.072265 | 7.367800 | 7.352787 | 7.328886 | 7.214265 | 7.245824 | 7.174470 |
| GO:0030117 | membrane coat | 2/44 | 93/21916 | 0.0150113810 | 0.19942106 | 0.16660072 | AP3M2||SAR1A | 2 | 5.761155 | 5.810442 | 5.844522 | 5.805784 | 5.765694 | 5.908312 | 5.800548 | 5.834002 | 5.727401 | 5.752139 | 5.802895 | 5.823064 | 5.815556 | 5.792532 | 5.823519 | 5.858505 | 5.851305 | 5.833517 | 5.743221 | 5.838632 | 5.771333 | 5.725413 | 5.799373 | 5.914741 | 5.912357 | 5.897780 | 5.803623 | 5.800019 | 5.797995 | 5.827896 | 5.843251 | 5.830813 |
| GO:0048475 | coated membrane | 2/44 | 93/21916 | 0.0150113810 | 0.19942106 | 0.16660072 | AP3M2||SAR1A | 2 | 5.761155 | 5.810442 | 5.844522 | 5.805784 | 5.765694 | 5.908312 | 5.800548 | 5.834002 | 5.727401 | 5.752139 | 5.802895 | 5.823064 | 5.815556 | 5.792532 | 5.823519 | 5.858505 | 5.851305 | 5.833517 | 5.743221 | 5.838632 | 5.771333 | 5.725413 | 5.799373 | 5.914741 | 5.912357 | 5.897780 | 5.803623 | 5.800019 | 5.797995 | 5.827896 | 5.843251 | 5.830813 |
| GO:0000313 | organellar ribosome | 2/44 | 94/21916 | 0.0153183474 | 0.19942106 | 0.16660072 | MRPS33||MRPL46 | 2 | 7.099385 | 6.815556 | 6.885067 | 6.783040 | 7.093419 | 6.953522 | 6.780040 | 6.771455 | 7.114499 | 7.048911 | 7.133391 | 6.880959 | 6.768539 | 6.794757 | 6.838523 | 6.868269 | 6.946253 | 6.824405 | 6.715129 | 6.807210 | 7.074105 | 7.068853 | 7.136317 | 6.975376 | 7.060211 | 6.814259 | 6.799303 | 6.747573 | 6.792696 | 6.769105 | 6.767175 | 6.778061 |
| GO:0005761 | mitochondrial ribosome | 2/44 | 94/21916 | 0.0153183474 | 0.19942106 | 0.16660072 | MRPS33||MRPL46 | 2 | 7.099385 | 6.815556 | 6.885067 | 6.783040 | 7.093419 | 6.953522 | 6.780040 | 6.771455 | 7.114499 | 7.048911 | 7.133391 | 6.880959 | 6.768539 | 6.794757 | 6.838523 | 6.868269 | 6.946253 | 6.824405 | 6.715129 | 6.807210 | 7.074105 | 7.068853 | 7.136317 | 6.975376 | 7.060211 | 6.814259 | 6.799303 | 6.747573 | 6.792696 | 6.769105 | 6.767175 | 6.778061 |
| GO:0015935 | small ribosomal subunit | 2/44 | 95/21916 | 0.0156280174 | 0.19942106 | 0.16660072 | MRPS33||RPS15A | 2 | 11.247650 | 10.971873 | 10.846813 | 10.843030 | 11.256433 | 10.926938 | 10.939687 | 10.867337 | 11.287183 | 11.214631 | 11.240195 | 11.044729 | 10.919936 | 10.947961 | 10.755784 | 10.825224 | 10.952492 | 10.878744 | 10.812689 | 10.836881 | 11.254790 | 11.306368 | 11.206408 | 11.121602 | 11.011767 | 10.595932 | 10.931530 | 10.943029 | 10.944468 | 10.861499 | 10.838702 | 10.901116 |
| GO:0005916 | fascia adherens | 1/44 | 10/21916 | 0.0199002911 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.826774 | 6.310780 | 5.413358 | 6.459454 | 4.649234 | 4.818287 | 6.248959 | 6.433961 | 4.850790 | 5.069136 | 4.505361 | 6.334125 | 6.356689 | 6.238832 | 5.721487 | 5.013152 | 5.419331 | 6.409362 | 6.485094 | 6.482628 | 4.657657 | 4.795657 | 4.476765 | 4.804468 | 4.740777 | 4.904854 | 6.270992 | 6.254284 | 6.221157 | 6.441467 | 6.445566 | 6.414654 |
| GO:0000786 | nucleosome | 2/44 | 112/21916 | 0.0212949672 | 0.19942106 | 0.16660072 | H2AZ1||H2BC6 | 2 | 6.040878 | 6.271642 | 6.244724 | 6.193609 | 5.926593 | 5.771782 | 6.283362 | 6.217637 | 6.100219 | 6.237862 | 5.739915 | 6.280463 | 6.256044 | 6.278292 | 6.355001 | 6.018191 | 6.336768 | 6.184069 | 6.161119 | 6.234655 | 5.857554 | 6.091711 | 5.814849 | 5.712095 | 5.666674 | 5.923370 | 6.272912 | 6.267285 | 6.309524 | 6.260776 | 6.157674 | 6.232503 |
| GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex | 1/44 | 11/21916 | 0.0218689019 | 0.19942106 | 0.16660072 | CCNT1 | 1 | 4.287652 | 5.674508 | 4.452047 | 5.779841 | 4.178630 | 4.468446 | 5.788796 | 5.863780 | 4.267631 | 4.375107 | 4.215606 | 5.603559 | 5.691865 | 5.725372 | 4.886521 | 4.254137 | 4.087607 | 5.690652 | 5.762393 | 5.880105 | 4.107616 | 4.172607 | 4.252035 | 4.430845 | 4.514323 | 4.458915 | 5.772502 | 5.808846 | 5.784805 | 5.855136 | 5.897150 | 5.838418 |
| GO:0030877 | beta-catenin destruction complex | 1/44 | 11/21916 | 0.0218689019 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.895837 | 6.228811 | 5.343709 | 6.266643 | 4.722139 | 4.893520 | 6.246604 | 6.302032 | 4.946107 | 5.052677 | 4.660859 | 6.278170 | 6.208876 | 6.198072 | 5.650546 | 5.079174 | 5.239849 | 6.220144 | 6.256106 | 6.321831 | 4.656165 | 4.798321 | 4.708334 | 4.752488 | 4.927941 | 4.989760 | 6.250851 | 6.250119 | 6.238811 | 6.292846 | 6.304294 | 6.308909 |
| GO:0072546 | EMC complex | 1/44 | 11/21916 | 0.0218689019 | 0.19942106 | 0.16660072 | EMC3 | 1 | 6.021302 | 6.326249 | 6.134075 | 6.278797 | 5.877234 | 5.954097 | 6.249406 | 6.234483 | 6.061417 | 6.021047 | 5.980301 | 6.342157 | 6.334987 | 6.301274 | 6.128752 | 6.120721 | 6.152562 | 6.296557 | 6.287626 | 6.251822 | 5.876958 | 5.823458 | 5.929344 | 5.871173 | 5.948511 | 6.037782 | 6.283002 | 6.223818 | 6.240753 | 6.221421 | 6.259323 | 6.222382 |
| GO:0015934 | large ribosomal subunit | 2/44 | 115/21916 | 0.0223715870 | 0.19942106 | 0.16660072 | RPL9||MRPL46 | 2 | 10.413003 | 10.249118 | 10.037763 | 10.148118 | 10.371702 | 10.038477 | 10.239487 | 10.126910 | 10.483518 | 10.428245 | 10.322643 | 10.297769 | 10.208307 | 10.239847 | 10.015651 | 9.969323 | 10.123925 | 10.188113 | 10.116594 | 10.138716 | 10.350078 | 10.489145 | 10.267107 | 10.183035 | 10.128090 | 9.770569 | 10.225378 | 10.245298 | 10.247681 | 10.154486 | 10.019318 | 10.200820 |
| GO:0022626 | cytosolic ribosome | 2/44 | 118/21916 | 0.0234704147 | 0.19942106 | 0.16660072 | RPS15A||RPL9 | 2 | 11.596661 | 11.350879 | 11.194057 | 11.226321 | 11.580314 | 11.230018 | 11.326906 | 11.226164 | 11.654105 | 11.589969 | 11.543776 | 11.412754 | 11.304510 | 11.333180 | 11.130545 | 11.148735 | 11.297045 | 11.266692 | 11.197505 | 11.213857 | 11.570006 | 11.668751 | 11.497024 | 11.412019 | 11.317549 | 10.913478 | 11.316001 | 11.333187 | 11.331468 | 11.238319 | 11.154159 | 11.283058 |
| GO:0044815 | DNA packaging complex | 2/44 | 120/21916 | 0.0242151594 | 0.19942106 | 0.16660072 | H2AZ1||H2BC6 | 2 | 5.954709 | 6.263016 | 6.176568 | 6.211908 | 5.844387 | 5.740177 | 6.285084 | 6.262950 | 6.004341 | 6.142993 | 5.678208 | 6.269988 | 6.249891 | 6.269078 | 6.302815 | 5.955413 | 6.247880 | 6.185480 | 6.185617 | 6.263224 | 5.781094 | 5.992624 | 5.746978 | 5.680456 | 5.644594 | 5.883774 | 6.275823 | 6.270935 | 6.308210 | 6.297365 | 6.212771 | 6.277364 |
| GO:0008250 | oligosaccharyltransferase complex | 1/44 | 13/21916 | 0.0257945375 | 0.19942106 | 0.16660072 | MAGT1 | 1 | 7.768737 | 8.215595 | 8.004530 | 8.102704 | 7.816173 | 7.910804 | 8.091067 | 8.047128 | 7.764994 | 7.765858 | 7.775335 | 8.264308 | 8.230184 | 8.149913 | 7.948329 | 8.048218 | 8.015255 | 8.115981 | 8.142252 | 8.048255 | 7.818305 | 7.785255 | 7.844351 | 7.853867 | 7.816592 | 8.050813 | 8.110839 | 8.106191 | 8.055522 | 8.048960 | 8.072909 | 8.019010 |
| GO:1990907 | beta-catenin-TCF complex | 1/44 | 13/21916 | 0.0257945375 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.394227 | 5.398809 | 5.047398 | 5.554694 | 4.312486 | 4.679961 | 5.388580 | 5.593529 | 4.398681 | 4.538550 | 4.228852 | 5.423650 | 5.437294 | 5.333285 | 5.228209 | 4.861808 | 5.028797 | 5.537110 | 5.562015 | 5.564796 | 4.272584 | 4.352029 | 4.311750 | 4.513853 | 4.567207 | 4.923551 | 5.404935 | 5.382457 | 5.378204 | 5.592157 | 5.605686 | 5.582650 |
| GO:0043601 | nuclear replisome | 1/44 | 14/21916 | 0.0277515772 | 0.19942106 | 0.16660072 | RPA2 | 1 | 6.151814 | 6.337963 | 6.366203 | 6.382033 | 6.078637 | 6.134530 | 6.348359 | 6.391377 | 6.170154 | 6.197162 | 6.085802 | 6.363714 | 6.328202 | 6.321616 | 6.434868 | 6.262834 | 6.395334 | 6.401848 | 6.321022 | 6.421283 | 6.069814 | 6.112455 | 6.052987 | 6.089844 | 6.090340 | 6.219499 | 6.370320 | 6.328926 | 6.345531 | 6.376895 | 6.419214 | 6.377613 |
| GO:0045239 | tricarboxylic acid cycle enzyme complex | 1/44 | 14/21916 | 0.0277515772 | 0.19942106 | 0.16660072 | SUCLG1 | 1 | 5.013463 | 5.741154 | 5.114022 | 5.807362 | 4.931741 | 4.965087 | 5.744561 | 5.843109 | 5.053540 | 5.144023 | 4.824297 | 5.765024 | 5.739200 | 5.718866 | 5.310430 | 4.969059 | 5.039648 | 5.779743 | 5.778671 | 5.862075 | 4.938964 | 4.972808 | 4.881994 | 4.952271 | 4.978319 | 4.964551 | 5.746713 | 5.736804 | 5.750133 | 5.821242 | 5.882646 | 5.824610 |
| GO:0005779 | integral component of peroxisomal membrane | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | SLC25A17 | 1 | 4.724407 | 5.052226 | 4.704894 | 4.914699 | 4.740645 | 4.685002 | 5.015593 | 4.937243 | 4.718907 | 4.669853 | 4.782258 | 5.114933 | 5.052450 | 4.986434 | 4.722691 | 4.663814 | 4.727311 | 4.921234 | 4.887814 | 4.934645 | 4.666373 | 4.735712 | 4.815963 | 4.666586 | 4.786367 | 4.595576 | 5.028921 | 5.012309 | 5.005448 | 4.926072 | 4.985441 | 4.898853 |
| GO:0030127 | COPII vesicle coat | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | SAR1A | 1 | 5.666499 | 6.040382 | 5.869626 | 6.009600 | 5.657401 | 5.724072 | 6.026746 | 6.061777 | 5.641247 | 5.783271 | 5.566517 | 6.069129 | 6.031945 | 6.019610 | 5.928871 | 5.788729 | 5.887704 | 6.026393 | 5.948851 | 6.051579 | 5.640895 | 5.716349 | 5.612965 | 5.728088 | 5.627706 | 5.810619 | 6.034496 | 6.031318 | 6.014342 | 6.045998 | 6.084367 | 6.054684 |
| GO:0031231 | intrinsic component of peroxisomal membrane | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | SLC25A17 | 1 | 4.724407 | 5.052226 | 4.704894 | 4.914699 | 4.740645 | 4.685002 | 5.015593 | 4.937243 | 4.718907 | 4.669853 | 4.782258 | 5.114933 | 5.052450 | 4.986434 | 4.722691 | 4.663814 | 4.727311 | 4.921234 | 4.887814 | 4.934645 | 4.666373 | 4.735712 | 4.815963 | 4.666586 | 4.786367 | 4.595576 | 5.028921 | 5.012309 | 5.005448 | 4.926072 | 4.985441 | 4.898853 |
| GO:0099092 | postsynaptic density, intracellular component | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 5.091148 | 5.580313 | 5.289675 | 5.605286 | 5.062143 | 5.242641 | 5.561879 | 5.609713 | 5.048861 | 5.125067 | 5.098481 | 5.612454 | 5.594366 | 5.532921 | 5.361116 | 5.214170 | 5.289999 | 5.625786 | 5.559093 | 5.629890 | 5.080583 | 4.996744 | 5.106826 | 5.216813 | 5.280042 | 5.230297 | 5.597099 | 5.544250 | 5.543634 | 5.585611 | 5.650397 | 5.592247 |
| GO:1990909 | Wnt signalosome | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 3.996449 | 5.384103 | 4.747863 | 5.545616 | 3.884693 | 4.215555 | 5.331897 | 5.513422 | 4.031314 | 4.151408 | 3.782458 | 5.438268 | 5.421924 | 5.287418 | 4.987654 | 4.446171 | 4.759185 | 5.539094 | 5.544796 | 5.552925 | 3.806818 | 4.000118 | 3.839638 | 4.056167 | 4.160408 | 4.407243 | 5.366752 | 5.315318 | 5.312979 | 5.501291 | 5.538381 | 5.500266 |
| GO:0030894 | replisome | 1/44 | 16/21916 | 0.0316541373 | 0.19942106 | 0.16660072 | RPA2 | 1 | 6.189163 | 6.798801 | 6.347049 | 6.796936 | 6.136233 | 6.153395 | 6.825482 | 6.867803 | 6.226262 | 6.222838 | 6.115672 | 6.817963 | 6.784683 | 6.793553 | 6.500585 | 6.201689 | 6.323126 | 6.785018 | 6.747617 | 6.856062 | 6.085765 | 6.192555 | 6.128371 | 6.117092 | 6.154785 | 6.187451 | 6.829685 | 6.821796 | 6.824954 | 6.836286 | 6.908224 | 6.857951 |
| GO:0098831 | presynaptic active zone cytoplasmic component | 1/44 | 16/21916 | 0.0316541373 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.250594 | 5.269332 | 4.746473 | 5.330539 | 4.128772 | 4.304294 | 5.229451 | 5.326176 | 4.282238 | 4.389956 | 4.060263 | 5.307818 | 5.301699 | 5.195749 | 4.935896 | 4.547610 | 4.729722 | 5.317880 | 5.315665 | 5.357683 | 4.114996 | 4.222738 | 4.042881 | 4.260011 | 4.147506 | 4.484709 | 5.239928 | 5.227754 | 5.220604 | 5.308989 | 5.341887 | 5.327464 |
| GO:0016600 | flotillin complex | 1/44 | 17/21916 | 0.0335996723 | 0.20484962 | 0.17113585 | CTNNB1 | 1 | 5.194723 | 6.253135 | 5.624109 | 6.295464 | 5.126002 | 5.164974 | 6.217457 | 6.280703 | 5.220597 | 5.355720 | 4.983564 | 6.335291 | 6.252033 | 6.167187 | 5.883739 | 5.277872 | 5.647286 | 6.282614 | 6.279140 | 6.324201 | 5.075085 | 5.243121 | 5.052171 | 5.155261 | 5.123861 | 5.214335 | 6.230139 | 6.198447 | 6.223591 | 6.275974 | 6.304235 | 6.261573 |
| GO:0099091 | postsynaptic specialization, intracellular component | 1/44 | 19/21916 | 0.0374792893 | 0.21909206 | 0.18303430 | CTNNB1 | 1 | 4.871904 | 5.402821 | 5.070452 | 5.418462 | 4.845346 | 5.026925 | 5.389288 | 5.420336 | 4.826541 | 4.908510 | 4.879466 | 5.428750 | 5.420661 | 5.358018 | 5.144103 | 4.998267 | 5.065288 | 5.430681 | 5.371050 | 5.452431 | 4.862551 | 4.776792 | 4.894154 | 4.999405 | 5.062876 | 5.017754 | 5.422460 | 5.372637 | 5.372186 | 5.397465 | 5.459088 | 5.403652 |
| GO:0005751 | mitochondrial respiratory chain complex IV | 1/44 | 20/21916 | 0.0394133858 | 0.21909206 | 0.18303430 | NDUFA4 | 1 | 10.314775 | 10.070006 | 10.879264 | 10.286836 | 10.402130 | 10.642883 | 10.135636 | 10.166958 | 10.251250 | 10.315705 | 10.374729 | 9.739911 | 10.202301 | 10.218588 | 10.802131 | 11.014870 | 10.810576 | 10.217302 | 10.478746 | 10.142473 | 10.437682 | 10.334396 | 10.431995 | 10.483636 | 10.426439 | 10.957664 | 10.129553 | 10.156014 | 10.121109 | 10.236378 | 10.111608 | 10.150043 |
| GO:0005753 | mitochondrial proton-transporting ATP synthase complex | 1/44 | 20/21916 | 0.0394133858 | 0.21909206 | 0.18303430 | ATP5MK | 1 | 10.107546 | 9.553595 | 10.485020 | 10.004835 | 10.161447 | 10.417021 | 9.608785 | 9.913688 | 10.037012 | 10.130284 | 10.152751 | 9.318820 | 9.646658 | 9.669360 | 10.384532 | 10.616451 | 10.443903 | 9.935202 | 10.168951 | 9.894915 | 10.215475 | 10.105944 | 10.160843 | 10.371383 | 10.309670 | 10.558254 | 9.586984 | 9.621127 | 9.617997 | 9.995534 | 9.869986 | 9.871923 |
| GO:0045259 | proton-transporting ATP synthase complex | 1/44 | 21/21916 | 0.0413436841 | 0.22325589 | 0.18651286 | ATP5MK | 1 | 10.030221 | 9.477651 | 10.407600 | 9.927974 | 10.084032 | 10.339422 | 9.532764 | 9.836772 | 9.959717 | 10.053019 | 10.075338 | 9.243378 | 9.570723 | 9.593014 | 10.307318 | 10.538912 | 10.366422 | 9.858372 | 10.092116 | 9.817990 | 10.137934 | 10.028672 | 10.083422 | 10.293769 | 10.232011 | 10.480719 | 9.510924 | 9.545093 | 9.542028 | 9.918618 | 9.793010 | 9.795068 |
| GO:0016327 | apicolateral plasma membrane | 1/44 | 22/21916 | 0.0432701914 | 0.22477529 | 0.18778220 | CTNNB1 | 1 | 3.786309 | 5.161394 | 4.342563 | 5.273774 | 3.652876 | 3.702798 | 5.093457 | 5.239956 | 3.839199 | 3.924031 | 3.572892 | 5.194496 | 5.215737 | 5.069675 | 4.635304 | 3.981601 | 4.337344 | 5.233092 | 5.297914 | 5.289458 | 3.581688 | 3.798653 | 3.566398 | 3.625851 | 3.603800 | 3.864049 | 5.113433 | 5.090421 | 5.076273 | 5.222333 | 5.276733 | 5.220086 |
| GO:0098800 | inner mitochondrial membrane protein complex | 2/44 | 168/21916 | 0.0447944167 | 0.22477529 | 0.18778220 | ATP5MK||NDUFA4 | 2 | 9.437482 | 8.978214 | 9.846205 | 9.270005 | 9.504751 | 9.762805 | 9.028746 | 9.153827 | 9.387496 | 9.422663 | 9.499983 | 8.678718 | 9.103199 | 9.111735 | 9.758726 | 9.969995 | 9.801131 | 9.203665 | 9.456050 | 9.129658 | 9.541797 | 9.427655 | 9.541818 | 9.645986 | 9.634636 | 9.980395 | 9.019705 | 9.049535 | 9.016769 | 9.235849 | 9.101725 | 9.120211 |
| GO:0031307 | integral component of mitochondrial outer membrane | 1/44 | 23/21916 | 0.0451929150 | 0.22477529 | 0.18778220 | FUNDC2 | 1 | 5.746309 | 6.029610 | 5.710113 | 5.846095 | 5.750251 | 5.769529 | 5.994107 | 5.874766 | 5.726248 | 5.701205 | 5.809245 | 6.018607 | 6.054383 | 6.015515 | 5.745791 | 5.696369 | 5.687493 | 5.851106 | 5.799906 | 5.885976 | 5.720699 | 5.711620 | 5.816095 | 5.724515 | 5.872407 | 5.705826 | 6.028575 | 5.966725 | 5.986328 | 5.854909 | 5.913791 | 5.854791 |
| GO:0031306 | intrinsic component of mitochondrial outer membrane | 1/44 | 24/21916 | 0.0471118623 | 0.22831133 | 0.19073628 | FUNDC2 | 1 | 5.771639 | 6.042729 | 5.768754 | 5.874368 | 5.775996 | 5.816203 | 6.011752 | 5.900602 | 5.748979 | 5.737767 | 5.826542 | 6.028325 | 6.070324 | 6.029136 | 5.800799 | 5.762210 | 5.742644 | 5.881108 | 5.829611 | 5.911208 | 5.746120 | 5.735739 | 5.843654 | 5.769861 | 5.895530 | 5.779806 | 6.042954 | 5.990956 | 6.000814 | 5.882968 | 5.937615 | 5.880499 |
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
ORA: Down-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Down-regulated DEGs” results.
GO: BP
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0010457 | centriole-centriole cohesion | 2/42 | 14/20870 | 0.0003543162 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 4.318051 | 5.487879 | 5.031966 | 5.677370 | 4.225202 | 4.551264 | 5.484452 | 5.695344 | 4.305087 | 4.459975 | 4.174948 | 5.517517 | 5.506111 | 5.438760 | 5.285587 | 4.810304 | 4.958311 | 5.642203 | 5.678232 | 5.710857 | 4.202190 | 4.302561 | 4.167420 | 4.372135 | 4.419951 | 4.818817 | 5.504015 | 5.483828 | 5.465253 | 5.679781 | 5.724709 | 5.681088 |
| GO:0032786 | positive regulation of DNA-templated transcription, elongation | 2/42 | 34/20870 | 0.0021292947 | 0.2820041 | 0.2468512 | CCNT1||CTNNB1 | 2 | 6.286426 | 6.664121 | 6.352703 | 6.732317 | 6.251144 | 6.259029 | 6.722421 | 6.807268 | 6.262236 | 6.357493 | 6.236723 | 6.668241 | 6.667076 | 6.657021 | 6.519235 | 6.257272 | 6.265966 | 6.703654 | 6.701766 | 6.789768 | 6.264429 | 6.234113 | 6.254723 | 6.222563 | 6.203989 | 6.346346 | 6.719482 | 6.711548 | 6.736124 | 6.796412 | 6.828326 | 6.796832 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2/42 | 45/20870 | 0.0037053379 | 0.2820041 | 0.2468512 | NBAS||SMG8 | 2 | 5.826224 | 6.863748 | 5.940690 | 6.831910 | 5.759909 | 5.980144 | 6.907572 | 6.883645 | 5.818207 | 5.897205 | 5.759964 | 6.852338 | 6.864312 | 6.874510 | 6.235001 | 5.767267 | 5.767639 | 6.777477 | 6.807765 | 6.907270 | 5.724801 | 5.748087 | 5.805635 | 5.966783 | 6.038001 | 5.933670 | 6.868892 | 6.922384 | 6.930665 | 6.872288 | 6.897411 | 6.881123 |
| GO:0048489 | synaptic vesicle transport | 2/42 | 45/20870 | 0.0037053379 | 0.2820041 | 0.2468512 | AP3M2||CTNNB1 | 2 | 5.507588 | 5.701124 | 5.650028 | 5.672758 | 5.538711 | 5.676770 | 5.665455 | 5.676478 | 5.484903 | 5.553467 | 5.483276 | 5.729771 | 5.709735 | 5.663056 | 5.657414 | 5.650041 | 5.642590 | 5.688571 | 5.620661 | 5.707601 | 5.559248 | 5.480887 | 5.574266 | 5.691810 | 5.615862 | 5.720619 | 5.667938 | 5.651831 | 5.676485 | 5.673199 | 5.706150 | 5.649523 |
| GO:0031062 | positive regulation of histone methylation | 2/42 | 50/20870 | 0.0045558326 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.750006 | 5.550108 | 5.126810 | 5.630453 | 4.648402 | 4.803877 | 5.590397 | 5.677335 | 4.760291 | 4.892592 | 4.580202 | 5.573517 | 5.561269 | 5.514878 | 5.389858 | 4.900362 | 5.045580 | 5.598566 | 5.627867 | 5.664175 | 4.634212 | 4.729100 | 4.577828 | 4.762938 | 4.695444 | 4.941884 | 5.608590 | 5.580762 | 5.581664 | 5.657744 | 5.698325 | 5.675650 |
| GO:0009101 | glycoprotein biosynthetic process | 4/42 | 336/20870 | 0.0045622098 | 0.2820041 | 0.2468512 | MAGT1||OGT||CTNNB1||OGA | 4 | 4.682544 | 5.249157 | 5.018778 | 5.200676 | 4.723558 | 4.933462 | 5.149768 | 5.143748 | 4.648439 | 4.664582 | 4.733200 | 5.272412 | 5.280474 | 5.192974 | 5.026015 | 5.050159 | 4.979261 | 5.179833 | 5.236821 | 5.184678 | 4.718187 | 4.658518 | 4.790916 | 4.807814 | 4.856109 | 5.116931 | 5.174711 | 5.156777 | 5.117219 | 5.134121 | 5.184441 | 5.111717 |
| GO:0051650 | establishment of vesicle localization | 3/42 | 170/20870 | 0.0048287457 | 0.2820041 | 0.2468512 | AP3M2||SAR1A||CTNNB1 | 3 | 4.980056 | 5.450239 | 5.239036 | 5.422373 | 4.988541 | 5.200836 | 5.424174 | 5.429382 | 4.936233 | 5.027134 | 4.975358 | 5.448124 | 5.475190 | 5.426999 | 5.290537 | 5.220620 | 5.204495 | 5.411573 | 5.401887 | 5.453142 | 4.994078 | 4.933971 | 5.035764 | 5.155852 | 5.141861 | 5.299463 | 5.446825 | 5.417495 | 5.407918 | 5.423481 | 5.456547 | 5.407687 |
| GO:0050982 | detection of mechanical stimulus | 2/42 | 52/20870 | 0.0049189375 | 0.2820041 | 0.2468512 | ADGRV1||CTNNB1 | 2 | 3.499818 | 4.639988 | 4.173924 | 4.654266 | 3.412685 | 3.642595 | 4.617657 | 4.668828 | 3.561730 | 3.584483 | 3.340923 | 4.644063 | 4.671794 | 4.603285 | 4.428225 | 3.960261 | 4.092155 | 4.600020 | 4.672643 | 4.688599 | 3.310536 | 3.459335 | 3.463003 | 3.377338 | 3.439809 | 4.020084 | 4.665643 | 4.643476 | 4.540802 | 4.645476 | 4.718601 | 4.641087 |
| GO:0097479 | synaptic vesicle localization | 2/42 | 58/20870 | 0.0060854215 | 0.2820041 | 0.2468512 | AP3M2||CTNNB1 | 2 | 5.217470 | 5.445932 | 5.382752 | 5.424272 | 5.247815 | 5.406098 | 5.410268 | 5.427370 | 5.192904 | 5.261064 | 5.197430 | 5.468073 | 5.459020 | 5.410029 | 5.402756 | 5.379465 | 5.365793 | 5.434133 | 5.378916 | 5.458615 | 5.268603 | 5.185628 | 5.287199 | 5.409177 | 5.342047 | 5.464470 | 5.414745 | 5.397129 | 5.418840 | 5.423870 | 5.459347 | 5.398240 |
| GO:0051648 | vesicle localization | 3/42 | 188/20870 | 0.0063804352 | 0.2820041 | 0.2468512 | AP3M2||SAR1A||CTNNB1 | 3 | 4.881661 | 5.366256 | 5.142991 | 5.340596 | 4.886856 | 5.106106 | 5.340152 | 5.345965 | 4.839880 | 4.926916 | 4.876863 | 5.362321 | 5.392033 | 5.344007 | 5.200289 | 5.122905 | 5.103965 | 5.326628 | 5.321917 | 5.372697 | 4.893040 | 4.829594 | 4.935954 | 5.057488 | 5.048346 | 5.206927 | 5.362119 | 5.333988 | 5.324078 | 5.339365 | 5.375232 | 5.322800 |
| GO:0032784 | regulation of DNA-templated transcription, elongation | 2/42 | 66/20870 | 0.0078169223 | 0.2820041 | 0.2468512 | CCNT1||CTNNB1 | 2 | 5.958730 | 6.430697 | 6.068448 | 6.480772 | 5.923856 | 6.000263 | 6.475805 | 6.799439 | 5.922412 | 6.044570 | 5.905166 | 6.438347 | 6.433273 | 6.420411 | 6.256668 | 5.950816 | 5.977559 | 6.457350 | 6.443137 | 6.539924 | 5.966957 | 5.873771 | 5.929320 | 5.961885 | 5.960152 | 6.075687 | 6.466964 | 6.468479 | 6.491835 | 6.524698 | 7.229204 | 6.524329 |
| GO:0051568 | histone H3-K4 methylation | 2/42 | 70/20870 | 0.0087563413 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.026450 | 5.608113 | 5.451674 | 5.657117 | 4.988956 | 5.233215 | 5.623612 | 5.664912 | 5.015049 | 5.114676 | 4.944545 | 5.623073 | 5.616994 | 5.583966 | 5.612442 | 5.329157 | 5.398083 | 5.639656 | 5.639943 | 5.691140 | 4.967158 | 5.015707 | 4.983581 | 5.164419 | 5.109939 | 5.407579 | 5.642394 | 5.611257 | 5.616994 | 5.647188 | 5.701963 | 5.644857 |
| GO:0009100 | glycoprotein metabolic process | 4/42 | 415/20870 | 0.0095052649 | 0.2820041 | 0.2468512 | MAGT1||OGT||CTNNB1||OGA | 4 | 4.826752 | 5.336325 | 5.165029 | 5.289770 | 4.852387 | 5.077499 | 5.239369 | 5.236128 | 4.801154 | 4.813325 | 4.864978 | 5.355348 | 5.369110 | 5.283045 | 5.157754 | 5.203645 | 5.132791 | 5.279048 | 5.316361 | 5.273524 | 4.843153 | 4.796539 | 4.914995 | 4.950774 | 5.007408 | 5.255753 | 5.268282 | 5.244688 | 5.204414 | 5.225844 | 5.277689 | 5.203853 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2/42 | 74/20870 | 0.0097437992 | 0.2820041 | 0.2468512 | SELENOK||TXNDC12 | 2 | 5.839547 | 6.100603 | 6.081685 | 6.195694 | 5.787172 | 5.930769 | 6.048638 | 6.176093 | 5.865195 | 5.934880 | 5.709420 | 6.121276 | 6.113808 | 6.066107 | 6.123862 | 6.000933 | 6.116983 | 6.211288 | 6.155738 | 6.219231 | 5.748081 | 5.875035 | 5.734184 | 5.893877 | 5.831452 | 6.057440 | 6.078128 | 6.032101 | 6.035226 | 6.176000 | 6.193362 | 6.158707 |
| GO:0031016 | pancreas development | 2/42 | 79/20870 | 0.0110444676 | 0.2820041 | 0.2468512 | CTNNB1||SELENOT | 2 | 4.010021 | 4.679029 | 4.337938 | 4.680418 | 4.004671 | 4.244189 | 4.644119 | 4.682639 | 3.993476 | 4.036143 | 4.000076 | 4.714118 | 4.695414 | 4.626072 | 4.456530 | 4.245329 | 4.303629 | 4.666934 | 4.659211 | 4.714485 | 3.964888 | 3.962747 | 4.083062 | 4.121147 | 4.238848 | 4.362478 | 4.655243 | 4.648032 | 4.628953 | 4.674603 | 4.712568 | 4.660238 |
| GO:0044242 | cellular lipid catabolic process | 3/42 | 237/20870 | 0.0119836731 | 0.2820041 | 0.2468512 | CPS1||SLC25A17||PPT1 | 3 | 5.005694 | 5.197128 | 5.202633 | 5.215226 | 4.989172 | 5.124164 | 5.146970 | 5.183058 | 4.991541 | 5.020678 | 5.004715 | 5.230945 | 5.201122 | 5.158398 | 5.254230 | 5.180042 | 5.172199 | 5.217895 | 5.190365 | 5.237038 | 4.984216 | 4.962707 | 5.020012 | 5.031598 | 5.098455 | 5.234941 | 5.171214 | 5.138882 | 5.130493 | 5.170218 | 5.216898 | 5.161438 |
| GO:0006486 | protein glycosylation | 3/42 | 240/20870 | 0.0123949928 | 0.2820041 | 0.2468512 | MAGT1||OGT||OGA | 3 | 4.900737 | 5.420245 | 5.220616 | 5.374274 | 4.946433 | 5.145543 | 5.317982 | 5.318482 | 4.867393 | 4.877461 | 4.955726 | 5.446705 | 5.447970 | 5.364483 | 5.214883 | 5.259876 | 5.186132 | 5.358293 | 5.411332 | 5.352465 | 4.943167 | 4.879015 | 5.013957 | 5.027513 | 5.068287 | 5.322753 | 5.342616 | 5.324288 | 5.286474 | 5.306843 | 5.363542 | 5.283891 |
| GO:0043413 | macromolecule glycosylation | 3/42 | 240/20870 | 0.0123949928 | 0.2820041 | 0.2468512 | MAGT1||OGT||OGA | 3 | 4.900737 | 5.420245 | 5.220616 | 5.374274 | 4.946433 | 5.145543 | 5.317982 | 5.318482 | 4.867393 | 4.877461 | 4.955726 | 5.446705 | 5.447970 | 5.364483 | 5.214883 | 5.259876 | 5.186132 | 5.358293 | 5.411332 | 5.352465 | 4.943167 | 4.879015 | 5.013957 | 5.027513 | 5.068287 | 5.322753 | 5.342616 | 5.324288 | 5.286474 | 5.306843 | 5.363542 | 5.283891 |
| GO:0031060 | regulation of histone methylation | 2/42 | 86/20870 | 0.0129865134 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.959030 | 5.537053 | 5.314570 | 5.582833 | 4.894046 | 5.107731 | 5.575414 | 5.617332 | 4.938236 | 5.067591 | 4.863853 | 5.553040 | 5.545649 | 5.512142 | 5.485610 | 5.177762 | 5.262575 | 5.562470 | 5.564283 | 5.620975 | 4.896745 | 4.920251 | 4.864601 | 5.077408 | 4.996953 | 5.238281 | 5.591859 | 5.566242 | 5.567997 | 5.598433 | 5.642485 | 5.610719 |
| GO:1901264 | carbohydrate derivative transport | 2/42 | 86/20870 | 0.0129865134 | 0.2820041 | 0.2468512 | SLC25A17||RSC1A1 | 2 | 6.199161 | 5.964336 | 6.252716 | 5.966508 | 6.217881 | 6.298529 | 5.838792 | 5.913297 | 6.190638 | 6.232173 | 6.174049 | 6.026962 | 5.971292 | 5.891551 | 6.135743 | 6.299603 | 6.316007 | 6.023343 | 5.924545 | 5.949799 | 6.226994 | 6.193210 | 6.233120 | 6.304625 | 6.319828 | 6.270697 | 5.867036 | 5.830690 | 5.818204 | 5.910628 | 5.943493 | 5.885177 |
| GO:0015931 | nucleobase-containing compound transport | 3/42 | 245/20870 | 0.0130984696 | 0.2820041 | 0.2468512 | SLC25A17||NUP205||RSC1A1 | 3 | 6.438449 | 7.082579 | 6.510346 | 7.094881 | 6.373040 | 6.462428 | 7.127235 | 7.150162 | 6.438940 | 6.552925 | 6.313564 | 7.098168 | 7.068785 | 7.080632 | 6.712350 | 6.364320 | 6.430211 | 7.078043 | 7.059849 | 7.145340 | 6.366227 | 6.413224 | 6.338682 | 6.468742 | 6.439786 | 6.478475 | 7.115484 | 7.122134 | 7.143934 | 7.134435 | 7.158441 | 7.157482 |
| GO:0006493 | protein O-linked glycosylation | 2/42 | 89/20870 | 0.0138611687 | 0.2820041 | 0.2468512 | OGT||OGA | 2 | 3.867728 | 4.448050 | 4.315947 | 4.515674 | 3.910499 | 4.214484 | 4.381159 | 4.457858 | 3.822673 | 3.807326 | 3.967696 | 4.439906 | 4.493731 | 4.409241 | 4.373988 | 4.352283 | 4.216580 | 4.468770 | 4.548157 | 4.528912 | 3.896881 | 3.820563 | 4.007881 | 4.030643 | 4.144713 | 4.437110 | 4.413092 | 4.376471 | 4.353283 | 4.445393 | 4.517968 | 4.408040 |
| GO:0070085 | glycosylation | 3/42 | 255/20870 | 0.0145731090 | 0.2820041 | 0.2468512 | MAGT1||OGT||OGA | 3 | 4.910584 | 5.409970 | 5.221002 | 5.358037 | 4.956186 | 5.147244 | 5.308123 | 5.299982 | 4.880487 | 4.887176 | 4.962639 | 5.435063 | 5.438398 | 5.354909 | 5.215864 | 5.259030 | 5.187205 | 5.345321 | 5.392392 | 5.335760 | 4.953172 | 4.891342 | 5.021121 | 5.030589 | 5.069649 | 5.323617 | 5.334816 | 5.312137 | 5.276825 | 5.285607 | 5.347421 | 5.265653 |
| GO:1903312 | negative regulation of mRNA metabolic process | 2/42 | 98/20870 | 0.0166335447 | 0.2820041 | 0.2468512 | CCNT1||NBAS | 2 | 6.953586 | 7.805895 | 7.094086 | 7.788960 | 6.870143 | 7.014798 | 7.845545 | 7.842174 | 6.979546 | 7.091920 | 6.771142 | 7.820955 | 7.779731 | 7.816646 | 7.344902 | 6.906732 | 6.992341 | 7.757249 | 7.749874 | 7.857246 | 6.848988 | 6.941472 | 6.817059 | 7.010416 | 6.951300 | 7.079806 | 7.817907 | 7.835091 | 7.882849 | 7.830747 | 7.847086 | 7.848621 |
| GO:0006354 | DNA-templated transcription, elongation | 2/42 | 103/20870 | 0.0182676376 | 0.2820041 | 0.2468512 | CCNT1||CTNNB1 | 2 | 6.089300 | 6.285959 | 6.096424 | 6.339595 | 6.078955 | 6.157008 | 6.318898 | 6.568793 | 6.056438 | 6.100227 | 6.110664 | 6.299241 | 6.289309 | 6.269163 | 6.187709 | 6.045225 | 6.051809 | 6.341509 | 6.292760 | 6.383102 | 6.091101 | 6.021726 | 6.122212 | 6.128507 | 6.207809 | 6.133331 | 6.327104 | 6.306684 | 6.322824 | 6.379862 | 6.885576 | 6.379760 |
| GO:0031058 | positive regulation of histone modification | 2/42 | 103/20870 | 0.0182676376 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.273117 | 5.635880 | 5.472630 | 5.664516 | 5.245684 | 5.443652 | 5.660638 | 5.705226 | 5.228978 | 5.324696 | 5.264046 | 5.662513 | 5.636461 | 5.608153 | 5.590155 | 5.408622 | 5.411520 | 5.662914 | 5.622010 | 5.707361 | 5.249533 | 5.205437 | 5.281083 | 5.418241 | 5.420622 | 5.490904 | 5.680634 | 5.650586 | 5.650485 | 5.684857 | 5.731238 | 5.699191 |
| GO:0000050 | urea cycle | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.262866 | 4.753968 | 4.216298 | 4.037180 | 4.203726 | 4.138404 | 4.612521 | 3.982695 | 4.248734 | 4.311588 | 4.226930 | 4.830151 | 4.742218 | 4.685856 | 4.216070 | 4.167974 | 4.263276 | 4.070867 | 3.944751 | 4.091589 | 4.188139 | 4.210883 | 4.212030 | 4.153726 | 4.143606 | 4.117641 | 4.628370 | 4.597855 | 4.611176 | 3.965977 | 4.022091 | 3.959187 |
| GO:0002084 | protein depalmitoylation | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.069159 | 5.817692 | 5.362781 | 5.852175 | 5.085838 | 5.187962 | 5.755626 | 5.802579 | 5.066708 | 5.046249 | 5.094122 | 5.920028 | 5.779001 | 5.748167 | 5.455138 | 5.274479 | 5.353025 | 5.830841 | 5.845893 | 5.879361 | 5.037843 | 5.068634 | 5.148754 | 5.067097 | 5.247463 | 5.242129 | 5.751446 | 5.765285 | 5.750099 | 5.780017 | 5.855600 | 5.770607 |
| GO:0007440 | foregut morphogenesis | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.019108 | 5.761821 | 4.903341 | 5.928440 | 3.760901 | 3.779080 | 5.722424 | 5.942940 | 4.141728 | 4.348895 | 3.407489 | 5.820579 | 5.794598 | 5.665556 | 5.265781 | 4.371943 | 4.935465 | 5.899650 | 5.935229 | 5.949979 | 3.619243 | 4.126527 | 3.448109 | 3.659226 | 3.520541 | 4.093817 | 5.741108 | 5.718535 | 5.707423 | 5.934868 | 5.955063 | 5.938808 |
| GO:0044848 | biological phase | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.726582 | 5.458386 | 4.890690 | 5.570369 | 4.575860 | 4.491023 | 5.397307 | 5.528887 | 4.811545 | 4.804027 | 4.549012 | 5.515384 | 5.484355 | 5.371478 | 5.049083 | 4.653473 | 4.941125 | 5.566221 | 5.584105 | 5.560676 | 4.597032 | 4.644232 | 4.481494 | 4.491747 | 4.470625 | 4.510423 | 5.432043 | 5.383960 | 5.375268 | 5.526469 | 5.538622 | 5.521515 |
| GO:0048263 | determination of dorsal identity | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.031406 | 5.863292 | 4.892495 | 6.024686 | 3.706003 | 3.584103 | 5.788910 | 6.009712 | 4.186254 | 4.376437 | 3.333622 | 5.944344 | 5.885735 | 5.753207 | 5.298975 | 4.229623 | 4.953141 | 5.991359 | 6.043379 | 6.038748 | 3.490740 | 4.134779 | 3.370523 | 3.407849 | 3.355850 | 3.919054 | 5.810402 | 5.786083 | 5.769956 | 6.011970 | 6.027201 | 5.989720 |
| GO:0060272 | embryonic skeletal joint morphogenesis | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.782839 | 5.266779 | 4.525953 | 5.417058 | 3.578189 | 3.556549 | 5.198897 | 5.398491 | 3.863101 | 4.058472 | 3.333275 | 5.325153 | 5.315737 | 5.153043 | 4.799533 | 4.101472 | 4.590960 | 5.397542 | 5.433143 | 5.420262 | 3.508772 | 3.818771 | 3.369809 | 3.418839 | 3.310518 | 3.875966 | 5.218642 | 5.191472 | 5.186367 | 5.396558 | 5.408309 | 5.390549 |
| GO:0060439 | trachea morphogenesis | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.776232 | 5.873128 | 5.352962 | 5.973485 | 4.709649 | 4.832058 | 5.861004 | 5.987171 | 4.805518 | 4.939654 | 4.557888 | 5.912865 | 5.901645 | 5.802328 | 5.604790 | 5.059186 | 5.343586 | 5.959827 | 5.974665 | 5.985844 | 4.624959 | 4.831529 | 4.664012 | 4.728300 | 4.708831 | 5.035275 | 5.871724 | 5.855929 | 5.855299 | 5.973311 | 5.993543 | 5.994561 |
| GO:0090037 | positive regulation of protein kinase C signaling | 1/42 | 10/20870 | 0.0199475685 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 3.070775 | 3.346961 | 3.719934 | 3.369778 | 3.157215 | 3.578420 | 3.341566 | 3.414134 | 2.977457 | 3.005810 | 3.216997 | 3.363700 | 3.317371 | 3.359360 | 3.895822 | 3.674089 | 3.570523 | 3.410628 | 3.314521 | 3.382501 | 3.174358 | 2.953274 | 3.320549 | 3.268840 | 3.442379 | 3.938067 | 3.378100 | 3.343178 | 3.302426 | 3.401111 | 3.528955 | 3.303444 |
| GO:0044262 | cellular carbohydrate metabolic process | 3/42 | 292/20870 | 0.0208240440 | 0.2820041 | 0.2468512 | OGT||BPNT1||SLC5A3 | 3 | 5.329203 | 5.412600 | 5.443193 | 5.467164 | 5.367576 | 5.507164 | 5.428294 | 5.478319 | 5.286816 | 5.321192 | 5.378123 | 5.414734 | 5.421972 | 5.401015 | 5.436678 | 5.469923 | 5.422568 | 5.457656 | 5.421542 | 5.520549 | 5.367278 | 5.320691 | 5.413275 | 5.479203 | 5.493094 | 5.548265 | 5.432539 | 5.418411 | 5.433881 | 5.471482 | 5.489537 | 5.473872 |
| GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 2/42 | 111/20870 | 0.0210173142 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 8.364296 | 8.325810 | 8.034230 | 8.265866 | 8.309812 | 7.991268 | 8.324788 | 8.267789 | 8.439584 | 8.419851 | 8.223742 | 8.361510 | 8.292652 | 8.322441 | 8.055229 | 7.938979 | 8.103559 | 8.294871 | 8.241217 | 8.261000 | 8.280245 | 8.426790 | 8.214123 | 8.075687 | 8.098080 | 7.778522 | 8.322109 | 8.307623 | 8.344395 | 8.268677 | 8.224115 | 8.309316 |
| GO:0003263 | cardioblast proliferation | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.974345 | 5.510557 | 4.801961 | 5.709595 | 3.773164 | 3.903646 | 5.468979 | 5.709613 | 4.105747 | 4.240125 | 3.464618 | 5.576149 | 5.533423 | 5.417462 | 5.143816 | 4.351551 | 4.803087 | 5.693283 | 5.715200 | 5.720159 | 3.680403 | 4.048597 | 3.541948 | 3.789858 | 3.601726 | 4.242408 | 5.492024 | 5.457782 | 5.456850 | 5.700744 | 5.727665 | 5.700259 |
| GO:0003264 | regulation of cardioblast proliferation | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.974345 | 5.510557 | 4.801961 | 5.709595 | 3.773164 | 3.903646 | 5.468979 | 5.709613 | 4.105747 | 4.240125 | 3.464618 | 5.576149 | 5.533423 | 5.417462 | 5.143816 | 4.351551 | 4.803087 | 5.693283 | 5.715200 | 5.720159 | 3.680403 | 4.048597 | 3.541948 | 3.789858 | 3.601726 | 4.242408 | 5.492024 | 5.457782 | 5.456850 | 5.700744 | 5.727665 | 5.700259 |
| GO:0033234 | negative regulation of protein sumoylation | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.105625 | 6.624912 | 6.581303 | 6.773236 | 5.899356 | 5.864026 | 6.627485 | 6.774963 | 6.216591 | 6.324840 | 5.701986 | 6.648896 | 6.627838 | 6.597543 | 6.732738 | 6.330967 | 6.649801 | 6.794378 | 6.734884 | 6.789692 | 5.781097 | 6.146362 | 5.734406 | 5.772491 | 5.753933 | 6.046651 | 6.648194 | 6.600379 | 6.633467 | 6.757077 | 6.786055 | 6.781588 |
| GO:0042989 | sequestering of actin monomers | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | TMSB15A | 1 | 7.540491 | 6.485058 | 6.945485 | 6.932227 | 7.489460 | 6.863027 | 6.510972 | 6.993968 | 7.667076 | 7.564411 | 7.374918 | 6.581638 | 6.402066 | 6.465693 | 6.858476 | 6.891828 | 7.076465 | 6.983356 | 6.856507 | 6.953785 | 7.452901 | 7.648256 | 7.351283 | 7.126047 | 6.825142 | 6.587118 | 6.543019 | 6.478360 | 6.510812 | 6.961714 | 6.990878 | 7.028535 |
| GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | RSC1A1 | 1 | 9.073771 | 8.557521 | 8.859841 | 8.503891 | 9.136981 | 9.138581 | 8.522043 | 8.501808 | 9.014612 | 8.995354 | 9.202171 | 8.601061 | 8.551185 | 8.519135 | 8.621458 | 8.960293 | 8.971134 | 8.568863 | 8.447316 | 8.492872 | 9.146143 | 9.078208 | 9.184587 | 9.210067 | 9.246102 | 8.940692 | 8.515292 | 8.535490 | 8.515252 | 8.498172 | 8.502998 | 8.504246 |
| GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | EMC3 | 1 | 6.021302 | 6.326249 | 6.134075 | 6.278797 | 5.877234 | 5.954097 | 6.249406 | 6.234483 | 6.061417 | 6.021047 | 5.980301 | 6.342157 | 6.334987 | 6.301274 | 6.128752 | 6.120721 | 6.152562 | 6.296557 | 6.287626 | 6.251822 | 5.876958 | 5.823458 | 5.929344 | 5.871173 | 5.948511 | 6.037782 | 6.283002 | 6.223818 | 6.240753 | 6.221421 | 6.259323 | 6.222382 |
| GO:0048262 | determination of dorsal/ventral asymmetry | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.949523 | 5.802298 | 4.736327 | 5.942855 | 3.637239 | 3.526067 | 5.737932 | 5.945566 | 4.098650 | 4.283386 | 3.284588 | 5.865177 | 5.826002 | 5.711320 | 5.160109 | 4.075136 | 4.773646 | 5.901551 | 5.959876 | 5.966262 | 3.428077 | 4.041214 | 3.336456 | 3.402649 | 3.354351 | 3.782116 | 5.746838 | 5.740724 | 5.726155 | 5.940482 | 5.964119 | 5.931903 |
| GO:0060742 | epithelial cell differentiation involved in prostate gland development | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.022225 | 6.629284 | 6.595708 | 6.720524 | 6.009211 | 6.320140 | 6.538535 | 6.655493 | 5.960989 | 6.144103 | 5.953390 | 6.697269 | 6.653229 | 6.532343 | 6.594428 | 6.592296 | 6.600389 | 6.720956 | 6.725673 | 6.714922 | 6.031770 | 6.021747 | 5.973443 | 6.212529 | 6.033247 | 6.644619 | 6.553660 | 6.536028 | 6.525779 | 6.671171 | 6.660319 | 6.634748 |
| GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.355605 | 6.050842 | 5.738427 | 6.145062 | 5.315079 | 5.386958 | 6.037946 | 6.156602 | 5.337705 | 5.488764 | 5.228454 | 6.097371 | 6.081003 | 5.970901 | 5.840855 | 5.565269 | 5.794348 | 6.139840 | 6.132648 | 6.162530 | 5.319134 | 5.353807 | 5.271103 | 5.314989 | 5.236217 | 5.585920 | 6.064872 | 6.023481 | 6.025101 | 6.165069 | 6.152262 | 6.152438 |
| GO:0070601 | centromeric sister chromatid cohesion | 1/42 | 11/20870 | 0.0219208284 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.585342 | 6.978778 | 6.580504 | 6.938714 | 6.564130 | 6.641947 | 6.966043 | 6.985288 | 6.560518 | 6.561230 | 6.633066 | 7.023087 | 6.960860 | 6.951329 | 6.688091 | 6.446931 | 6.596293 | 6.928569 | 6.897085 | 6.988973 | 6.544520 | 6.502971 | 6.641383 | 6.625764 | 6.777710 | 6.509815 | 6.984526 | 6.946103 | 6.967243 | 6.978537 | 6.999142 | 6.978085 |
| GO:0044344 | cellular response to fibroblast growth factor stimulus | 2/42 | 115/20870 | 0.0224529946 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.273541 | 4.280472 | 3.783727 | 4.310241 | 3.236434 | 3.514922 | 4.262362 | 4.295562 | 3.262676 | 3.338878 | 3.216409 | 4.283385 | 4.311566 | 4.245710 | 3.953663 | 3.674678 | 3.706369 | 4.276974 | 4.323815 | 4.329362 | 3.205283 | 3.240503 | 3.262931 | 3.381010 | 3.406481 | 3.730355 | 4.285943 | 4.254403 | 4.246436 | 4.284237 | 4.317043 | 4.285163 |
| GO:0019751 | polyol metabolic process | 2/42 | 118/20870 | 0.0235557933 | 0.2820041 | 0.2468512 | BPNT1||SLC5A3 | 2 | 5.642470 | 5.277912 | 5.637300 | 5.334206 | 5.726575 | 5.814037 | 5.263944 | 5.332793 | 5.557484 | 5.569409 | 5.788586 | 5.305675 | 5.287843 | 5.239407 | 5.534141 | 5.699561 | 5.672794 | 5.356616 | 5.270359 | 5.373534 | 5.714491 | 5.651193 | 5.809618 | 5.790942 | 5.897523 | 5.749581 | 5.282758 | 5.253061 | 5.255827 | 5.325077 | 5.345944 | 5.327267 |
| GO:0003306 | Wnt signaling pathway involved in heart development | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.822548 | 5.492218 | 4.547769 | 5.663990 | 3.543254 | 3.390835 | 5.433174 | 5.657189 | 3.989823 | 4.100811 | 3.232523 | 5.551332 | 5.527688 | 5.392606 | 4.934775 | 3.948456 | 4.593570 | 5.622940 | 5.684328 | 5.683843 | 3.392404 | 3.889023 | 3.271745 | 3.376211 | 3.291781 | 3.497104 | 5.447086 | 5.430871 | 5.421448 | 5.659791 | 5.658739 | 5.653029 |
| GO:0006020 | inositol metabolic process | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | SLC5A3 | 1 | 4.720464 | 4.649936 | 5.057921 | 4.793895 | 4.759914 | 5.143587 | 4.696894 | 4.832931 | 4.649341 | 4.627276 | 4.871872 | 4.633840 | 4.691998 | 4.623012 | 5.089160 | 5.097853 | 4.983988 | 4.817245 | 4.709513 | 4.851164 | 4.806135 | 4.524103 | 4.921139 | 4.956594 | 5.077558 | 5.365528 | 4.762570 | 4.688013 | 4.637343 | 4.806723 | 4.910334 | 4.778367 |
| GO:0007042 | lysosomal lumen acidification | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.851089 | 6.350054 | 6.296515 | 6.357931 | 5.906390 | 6.114170 | 6.150319 | 6.224873 | 5.808606 | 5.844606 | 5.898629 | 6.432915 | 6.371834 | 6.238629 | 6.224602 | 6.350590 | 6.311486 | 6.396846 | 6.383081 | 6.291613 | 5.906148 | 5.844348 | 5.966108 | 5.936792 | 5.988694 | 6.375990 | 6.201926 | 6.142437 | 6.104932 | 6.225910 | 6.280729 | 6.165686 |
| GO:0009950 | dorsal/ventral axis specification | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.682251 | 5.440082 | 4.500793 | 5.611397 | 3.391255 | 3.294143 | 5.399783 | 5.607603 | 3.810736 | 3.994129 | 3.094829 | 5.504849 | 5.466615 | 5.343926 | 4.907744 | 3.922414 | 4.507131 | 5.579117 | 5.617519 | 5.636954 | 3.253997 | 3.747087 | 3.089085 | 3.151304 | 3.050920 | 3.615389 | 5.410811 | 5.399987 | 5.388465 | 5.606935 | 5.616809 | 5.599010 |
| GO:0019627 | urea metabolic process | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.262866 | 4.753968 | 4.216298 | 4.037180 | 4.203726 | 4.138404 | 4.612521 | 3.982695 | 4.248734 | 4.311588 | 4.226930 | 4.830151 | 4.742218 | 4.685856 | 4.216070 | 4.167974 | 4.263276 | 4.070867 | 3.944751 | 4.091589 | 4.188139 | 4.210883 | 4.212030 | 4.153726 | 4.143606 | 4.117641 | 4.628370 | 4.597855 | 4.611176 | 3.965977 | 4.022091 | 3.959187 |
| GO:0035112 | genitalia morphogenesis | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.822736 | 5.828054 | 4.788151 | 6.010714 | 3.455638 | 3.109134 | 5.774687 | 6.009180 | 4.027482 | 4.226768 | 2.891245 | 5.899007 | 5.856911 | 5.722405 | 5.233520 | 4.051036 | 4.842512 | 5.964812 | 6.033898 | 6.032359 | 3.247202 | 3.979561 | 2.934178 | 2.994250 | 2.735375 | 3.492935 | 5.785272 | 5.776810 | 5.761883 | 6.001881 | 6.015068 | 6.010561 |
| GO:0048715 | negative regulation of oligodendrocyte differentiation | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.170971 | 5.450019 | 4.800812 | 5.559395 | 4.034610 | 4.092259 | 5.395354 | 5.557069 | 4.239621 | 4.370530 | 3.854295 | 5.511016 | 5.485579 | 5.348203 | 5.069634 | 4.444306 | 4.821043 | 5.546845 | 5.567020 | 5.564238 | 3.989992 | 4.172068 | 3.930669 | 3.997531 | 3.880201 | 4.355589 | 5.423138 | 5.382479 | 5.380036 | 5.548941 | 5.572620 | 5.549519 |
| GO:0060123 | regulation of growth hormone secretion | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | SELENOT | 1 | 3.355548 | 5.031099 | 3.680747 | 5.000469 | 3.111499 | 3.388222 | 5.113121 | 5.086264 | 3.397182 | 3.556729 | 3.071144 | 5.016108 | 5.026565 | 5.050409 | 4.069952 | 3.258634 | 3.597601 | 4.873378 | 4.979491 | 5.136345 | 3.156699 | 3.148149 | 3.025979 | 3.465384 | 3.384796 | 3.310314 | 5.080612 | 5.152742 | 5.105073 | 5.078212 | 5.116917 | 5.063128 |
| GO:0060767 | epithelial cell proliferation involved in prostate gland development | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.210970 | 5.905060 | 5.592999 | 5.999929 | 5.171914 | 5.245224 | 5.891585 | 6.011468 | 5.193842 | 5.343724 | 5.083471 | 5.950172 | 5.936014 | 5.825808 | 5.695415 | 5.420587 | 5.648295 | 5.994629 | 5.987619 | 6.017370 | 5.175552 | 5.210074 | 5.128968 | 5.172828 | 5.094864 | 5.444242 | 5.917723 | 5.877470 | 5.879203 | 6.019934 | 6.007549 | 6.006884 |
| GO:0060788 | ectodermal placode formation | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.110723 | 6.600917 | 5.751653 | 6.709326 | 4.910418 | 5.018709 | 6.616440 | 6.741341 | 5.199344 | 5.372962 | 4.671062 | 6.645857 | 6.605897 | 6.549372 | 6.108657 | 5.366345 | 5.683139 | 6.688013 | 6.695613 | 6.743716 | 4.876271 | 5.145807 | 4.669431 | 4.949772 | 4.686124 | 5.343574 | 6.608273 | 6.607628 | 6.633270 | 6.722135 | 6.758163 | 6.743499 |
| GO:0071377 | cellular response to glucagon stimulus | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.394864 | 5.218736 | 4.043511 | 3.956852 | 4.337042 | 4.213524 | 5.017685 | 3.894576 | 4.404147 | 4.382388 | 4.397969 | 5.326338 | 5.199739 | 5.122769 | 4.009603 | 4.056280 | 4.064053 | 3.940620 | 3.918576 | 4.009782 | 4.328705 | 4.313347 | 4.368511 | 4.362262 | 4.237392 | 4.020416 | 5.018407 | 5.048334 | 4.985634 | 3.910011 | 3.947772 | 3.823130 |
| GO:0071697 | ectodermal placode morphogenesis | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.110723 | 6.600917 | 5.751653 | 6.709326 | 4.910418 | 5.018709 | 6.616440 | 6.741341 | 5.199344 | 5.372962 | 4.671062 | 6.645857 | 6.605897 | 6.549372 | 6.108657 | 5.366345 | 5.683139 | 6.688013 | 6.695613 | 6.743716 | 4.876271 | 5.145807 | 4.669431 | 4.949772 | 4.686124 | 5.343574 | 6.608273 | 6.607628 | 6.633270 | 6.722135 | 6.758163 | 6.743499 |
| GO:0071941 | nitrogen cycle metabolic process | 1/42 | 12/20870 | 0.0238902098 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.262866 | 4.753968 | 4.216298 | 4.037180 | 4.203726 | 4.138404 | 4.612521 | 3.982695 | 4.248734 | 4.311588 | 4.226930 | 4.830151 | 4.742218 | 4.685856 | 4.216070 | 4.167974 | 4.263276 | 4.070867 | 3.944751 | 4.091589 | 4.188139 | 4.210883 | 4.212030 | 4.153726 | 4.143606 | 4.117641 | 4.628370 | 4.597855 | 4.611176 | 3.965977 | 4.022091 | 3.959187 |
| GO:0071774 | response to fibroblast growth factor | 2/42 | 122/20870 | 0.0250603504 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.327762 | 4.253186 | 3.800685 | 4.274480 | 3.296967 | 3.577871 | 4.235277 | 4.260555 | 3.307889 | 3.380179 | 3.293720 | 4.256057 | 4.283337 | 4.219452 | 3.951037 | 3.712244 | 3.726110 | 4.243759 | 4.284661 | 4.294519 | 3.277179 | 3.276558 | 3.336343 | 3.449773 | 3.483435 | 3.777403 | 4.260289 | 4.226032 | 4.219173 | 4.247976 | 4.281908 | 4.251539 |
| GO:0007494 | midgut development | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CPS1 | 1 | 3.366681 | 3.953231 | 3.450559 | 3.246768 | 3.345066 | 3.462190 | 3.831041 | 3.212594 | 3.350536 | 3.352053 | 3.396970 | 4.018595 | 3.986140 | 3.849432 | 3.425912 | 3.458223 | 3.467214 | 3.215793 | 3.233555 | 3.289913 | 3.340172 | 3.297656 | 3.395693 | 3.412552 | 3.418362 | 3.551339 | 3.846067 | 3.840885 | 3.805840 | 3.231117 | 3.259492 | 3.144709 |
| GO:0010739 | positive regulation of protein kinase A signaling | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 9.758394 | 8.851893 | 9.247425 | 8.627674 | 9.839473 | 9.923020 | 8.648873 | 8.460644 | 9.731749 | 9.489519 | 10.007333 | 9.060642 | 8.720571 | 8.749241 | 8.879464 | 9.468512 | 9.330825 | 8.880882 | 8.487597 | 8.476754 | 9.855561 | 9.564954 | 10.056158 | 9.826975 | 10.213026 | 9.674502 | 8.647626 | 8.648984 | 8.650008 | 8.482433 | 8.454293 | 8.444941 |
| GO:0015791 | polyol transport | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | SLC5A3 | 1 | 1.876776 | 2.715215 | 3.217662 | 3.240215 | 1.951667 | 2.800008 | 2.873029 | 3.239115 | 1.689882 | 1.881171 | 2.038281 | 2.617180 | 2.807309 | 2.714897 | 3.313560 | 3.235792 | 3.095206 | 3.194432 | 3.264391 | 3.260753 | 1.871420 | 1.795183 | 2.161925 | 2.238031 | 2.517786 | 3.385262 | 2.975373 | 2.845661 | 2.791835 | 3.190388 | 3.314870 | 3.208934 |
| GO:0021781 | glial cell fate commitment | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.433846 | 5.273683 | 4.333427 | 5.454681 | 3.110581 | 2.923432 | 5.217428 | 5.445408 | 3.596402 | 3.778203 | 2.723437 | 5.343389 | 5.304495 | 5.167294 | 4.747612 | 3.689257 | 4.372698 | 5.425343 | 5.463823 | 5.474415 | 2.936351 | 3.548969 | 2.712127 | 2.755201 | 2.601711 | 3.312946 | 5.226388 | 5.220952 | 5.204859 | 5.438724 | 5.454465 | 5.442988 |
| GO:0032429 | regulation of phospholipase A2 activity | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | PPT1 | 1 | 3.377836 | 4.991773 | 4.292212 | 5.138562 | 3.087942 | 3.149547 | 4.856709 | 5.105324 | 3.498314 | 3.655862 | 2.864029 | 5.109239 | 4.969148 | 4.888212 | 4.634196 | 3.817010 | 4.311038 | 5.084665 | 5.187020 | 5.142179 | 3.039185 | 3.378456 | 2.784212 | 3.009861 | 2.881681 | 3.484956 | 4.842313 | 4.884227 | 4.843189 | 5.084971 | 5.158467 | 5.070996 |
| GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.916178 | 6.184543 | 5.274792 | 6.317106 | 4.754219 | 4.948420 | 6.238215 | 6.361911 | 4.901835 | 5.087523 | 4.737943 | 6.178413 | 6.217741 | 6.156811 | 5.648177 | 4.908925 | 5.167934 | 6.268988 | 6.302193 | 6.377963 | 4.736311 | 4.835977 | 4.686325 | 4.898664 | 4.970406 | 4.974929 | 6.228809 | 6.244169 | 6.241621 | 6.358173 | 6.372078 | 6.355425 |
| GO:0048548 | regulation of pinocytosis | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | PPT1 | 1 | 4.366222 | 5.522390 | 4.941477 | 5.648879 | 4.260752 | 4.335752 | 5.517020 | 5.695505 | 4.386340 | 4.577225 | 4.094736 | 5.583671 | 5.499778 | 5.481656 | 5.242249 | 4.559747 | 4.942456 | 5.587004 | 5.646298 | 5.710683 | 4.211256 | 4.438622 | 4.112771 | 4.279727 | 4.195343 | 4.513130 | 5.505514 | 5.526518 | 5.518950 | 5.669317 | 5.727485 | 5.689104 |
| GO:0048820 | hair follicle maturation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.006386 | 5.709607 | 5.283967 | 5.767784 | 4.883857 | 4.982140 | 5.696175 | 5.773260 | 5.041479 | 5.103736 | 4.863285 | 5.731778 | 5.738599 | 5.657033 | 5.447208 | 5.113869 | 5.271532 | 5.752899 | 5.773354 | 5.776982 | 4.923092 | 4.909888 | 4.816255 | 4.964944 | 4.901754 | 5.074406 | 5.718838 | 5.684935 | 5.684481 | 5.768667 | 5.788331 | 5.762657 |
| GO:0061046 | regulation of branching involved in lung morphogenesis | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.428014 | 6.382084 | 5.395448 | 6.551280 | 4.076479 | 3.761370 | 6.314812 | 6.543815 | 4.617376 | 4.830850 | 3.532719 | 6.450884 | 6.415288 | 6.274086 | 5.810533 | 4.696807 | 5.466483 | 6.520606 | 6.566305 | 6.566448 | 3.884308 | 4.580168 | 3.576011 | 3.623164 | 3.329433 | 4.194422 | 6.326534 | 6.316005 | 6.301791 | 6.534483 | 6.556907 | 6.539960 |
| GO:0061085 | regulation of histone H3-K27 methylation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | OGT | 1 | 5.063009 | 5.638408 | 5.560314 | 5.629488 | 5.038329 | 5.223784 | 5.632937 | 5.503203 | 5.059749 | 5.231469 | 4.875959 | 5.671863 | 5.619298 | 5.623470 | 5.728397 | 5.408940 | 5.525331 | 5.587868 | 5.605939 | 5.692475 | 5.029888 | 5.152388 | 4.923614 | 5.220884 | 4.999323 | 5.420508 | 5.659734 | 5.608375 | 5.630242 | 5.469697 | 5.563310 | 5.474664 |
| GO:0071696 | ectodermal placode development | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.984347 | 6.477260 | 5.620440 | 6.581163 | 4.783979 | 4.891787 | 6.490066 | 6.613027 | 5.071763 | 5.246159 | 4.547116 | 6.519160 | 6.484961 | 6.426128 | 5.976723 | 5.236610 | 5.551708 | 6.561583 | 6.566596 | 6.614712 | 4.749560 | 5.018572 | 4.544412 | 4.825956 | 4.565199 | 5.210462 | 6.481767 | 6.481208 | 6.507073 | 6.594114 | 6.630340 | 6.614398 |
| GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.261654 | 5.229418 | 4.169716 | 5.370776 | 2.917512 | 2.676653 | 5.180140 | 5.383155 | 3.432036 | 3.643548 | 2.454715 | 5.289249 | 5.260675 | 5.133621 | 4.610220 | 3.487255 | 4.198032 | 5.332556 | 5.385385 | 5.393630 | 2.734090 | 3.386735 | 2.473376 | 2.598733 | 2.360469 | 2.997873 | 5.180674 | 5.191412 | 5.168240 | 5.376049 | 5.393594 | 5.379762 |
| GO:0072497 | mesenchymal stem cell differentiation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.063510 | 5.207631 | 4.576614 | 5.278305 | 3.916279 | 3.855521 | 5.135041 | 5.244471 | 4.139131 | 4.210216 | 3.810503 | 5.260201 | 5.239454 | 5.119262 | 4.825911 | 4.211830 | 4.625948 | 5.253766 | 5.291787 | 5.289052 | 3.875432 | 4.056816 | 3.804738 | 3.783198 | 3.723624 | 4.039820 | 5.155388 | 5.130437 | 5.119058 | 5.232454 | 5.266746 | 5.233951 |
| GO:1900364 | negative regulation of mRNA polyadenylation | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CCNT1 | 1 | 5.770945 | 6.236909 | 6.026429 | 6.217444 | 5.719692 | 6.033174 | 6.258902 | 6.256497 | 5.689087 | 5.824397 | 5.795855 | 6.237307 | 6.241353 | 6.232050 | 6.076112 | 6.044296 | 5.956221 | 6.182673 | 6.181062 | 6.286069 | 5.761941 | 5.667295 | 5.728248 | 5.912273 | 6.029479 | 6.148134 | 6.299599 | 6.232270 | 6.243936 | 6.223063 | 6.304385 | 6.240771 |
| GO:1990403 | embryonic brain development | 1/42 | 13/20870 | 0.0258557199 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 8.919538 | 8.021670 | 8.670129 | 7.933080 | 9.010001 | 8.886555 | 8.026984 | 8.013662 | 8.939758 | 8.736155 | 9.063912 | 8.018859 | 8.079396 | 7.964462 | 8.441984 | 8.700133 | 8.840378 | 7.971574 | 7.918148 | 7.908719 | 9.054419 | 8.898906 | 9.070529 | 8.991413 | 9.019614 | 8.614251 | 8.073422 | 8.077678 | 7.924649 | 7.963560 | 8.077711 | 7.997319 |
| GO:1904659 | glucose transmembrane transport | 2/42 | 126/20870 | 0.0266033140 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 3.910897 | 4.488307 | 4.284345 | 4.541570 | 3.911282 | 4.250547 | 4.495726 | 4.524655 | 3.848629 | 3.935828 | 3.946255 | 4.469815 | 4.507059 | 4.487807 | 4.391516 | 4.283980 | 4.168965 | 4.503379 | 4.525034 | 4.594712 | 3.890248 | 3.839412 | 3.999518 | 4.103675 | 4.199219 | 4.429097 | 4.509366 | 4.486075 | 4.491633 | 4.519485 | 4.557369 | 4.496454 |
| GO:0048193 | Golgi vesicle transport | 3/42 | 326/20870 | 0.0276833253 | 0.2820041 | 0.2468512 | SAR1A||NBAS||RSC1A1 | 3 | 6.026866 | 6.132262 | 6.118246 | 6.082199 | 6.065056 | 6.198069 | 6.089379 | 6.078730 | 5.974567 | 6.007022 | 6.096241 | 6.151529 | 6.146221 | 6.098444 | 6.057391 | 6.163328 | 6.131976 | 6.093995 | 6.053840 | 6.098344 | 6.070141 | 6.002224 | 6.120372 | 6.181871 | 6.196986 | 6.215158 | 6.099713 | 6.091445 | 6.076888 | 6.071009 | 6.102416 | 6.062457 |
| GO:0000052 | citrulline metabolic process | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CPS1 | 1 | 5.208000 | 5.752459 | 5.585622 | 5.515343 | 5.121159 | 5.392200 | 5.676700 | 5.455829 | 5.235468 | 5.265333 | 5.119097 | 5.745509 | 5.795163 | 5.715582 | 5.615742 | 5.600196 | 5.539817 | 5.462749 | 5.520844 | 5.560755 | 5.127489 | 5.128021 | 5.107876 | 5.282680 | 5.133813 | 5.699139 | 5.707445 | 5.680915 | 5.640964 | 5.447953 | 5.506084 | 5.411880 |
| GO:0006999 | nuclear pore organization | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | NUP205 | 1 | 4.727484 | 6.215271 | 5.318037 | 6.353303 | 4.582429 | 4.961544 | 6.280218 | 6.426828 | 4.712357 | 4.952211 | 4.479159 | 6.194741 | 6.227266 | 6.223587 | 5.733778 | 4.971696 | 5.132694 | 6.268795 | 6.344595 | 6.441324 | 4.573996 | 4.673011 | 4.494738 | 5.046213 | 4.850701 | 4.980902 | 6.265463 | 6.282052 | 6.293005 | 6.396153 | 6.456253 | 6.427453 |
| GO:0031441 | negative regulation of mRNA 3'-end processing | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CCNT1 | 1 | 5.603515 | 6.097142 | 5.862302 | 6.081061 | 5.552510 | 5.865710 | 6.118450 | 6.132236 | 5.521731 | 5.657462 | 5.627849 | 6.092112 | 6.109112 | 6.090126 | 5.916822 | 5.877324 | 5.789849 | 6.044485 | 6.044684 | 6.151358 | 5.593893 | 5.501217 | 5.560895 | 5.744598 | 5.862204 | 5.980675 | 6.155612 | 6.093602 | 6.105380 | 6.088965 | 6.186338 | 6.119680 |
| GO:0034333 | adherens junction assembly | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 8.054318 | 7.668552 | 7.701730 | 7.601310 | 8.063195 | 8.047228 | 7.598870 | 7.577856 | 8.030170 | 8.053189 | 8.079179 | 7.667361 | 7.697642 | 7.640080 | 7.488546 | 7.803250 | 7.791857 | 7.686783 | 7.514023 | 7.597949 | 8.138981 | 7.969526 | 8.076020 | 8.144301 | 8.016510 | 7.975451 | 7.607530 | 7.616015 | 7.572700 | 7.599936 | 7.588731 | 7.544302 |
| GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.202563 | 7.059911 | 6.502002 | 6.975464 | 6.178471 | 6.299322 | 7.009876 | 6.984877 | 6.154535 | 6.296042 | 6.152362 | 7.057858 | 7.094401 | 7.026676 | 6.635525 | 6.376487 | 6.482178 | 6.964278 | 6.976381 | 6.985652 | 6.147944 | 6.196237 | 6.190749 | 6.219211 | 6.220239 | 6.446440 | 7.042116 | 6.998790 | 6.988155 | 6.976897 | 7.017609 | 6.959508 |
| GO:0048208 | COPII vesicle coating | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.202563 | 7.059911 | 6.502002 | 6.975464 | 6.178471 | 6.299322 | 7.009876 | 6.984877 | 6.154535 | 6.296042 | 6.152362 | 7.057858 | 7.094401 | 7.026676 | 6.635525 | 6.376487 | 6.482178 | 6.964278 | 6.976381 | 6.985652 | 6.147944 | 6.196237 | 6.190749 | 6.219211 | 6.220239 | 6.446440 | 7.042116 | 6.998790 | 6.988155 | 6.976897 | 7.017609 | 6.959508 |
| GO:0050667 | homocysteine metabolic process | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CPS1 | 1 | 5.699296 | 5.822978 | 5.854275 | 5.667816 | 5.779181 | 5.986813 | 5.761403 | 5.653922 | 5.651681 | 5.631935 | 5.807764 | 5.865582 | 5.856908 | 5.743245 | 5.805418 | 5.907438 | 5.848148 | 5.708656 | 5.622133 | 5.671356 | 5.796446 | 5.645554 | 5.885427 | 5.858243 | 5.996178 | 6.096174 | 5.809703 | 5.739827 | 5.733432 | 5.649982 | 5.709807 | 5.599877 |
| GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | SAR1A | 1 | 7.117005 | 6.919455 | 7.362000 | 6.925550 | 7.172971 | 7.378534 | 6.749864 | 6.867203 | 7.028314 | 7.002908 | 7.300542 | 6.973389 | 6.964864 | 6.814654 | 7.144745 | 7.503631 | 7.413927 | 6.945211 | 6.978096 | 6.850313 | 7.163357 | 7.059422 | 7.287111 | 7.279743 | 7.357815 | 7.490157 | 6.779679 | 6.762612 | 6.706281 | 6.857446 | 6.927409 | 6.814497 |
| GO:0072498 | embryonic skeletal joint development | 1/42 | 14/20870 | 0.0278173664 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.470475 | 4.875899 | 4.202085 | 5.034418 | 3.288161 | 3.340355 | 4.810159 | 5.009321 | 3.538046 | 3.724248 | 3.074029 | 4.928981 | 4.923531 | 4.769578 | 4.454815 | 3.822575 | 4.258449 | 5.012108 | 5.049655 | 5.041221 | 3.227635 | 3.497417 | 3.111899 | 3.195375 | 3.099424 | 3.661601 | 4.834263 | 4.803901 | 4.791982 | 5.003546 | 5.016351 | 5.008036 |
| GO:0008645 | hexose transmembrane transport | 2/42 | 130/20870 | 0.0281840021 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 4.018011 | 4.530332 | 4.347029 | 4.578882 | 4.030440 | 4.345781 | 4.530434 | 4.551110 | 3.954927 | 4.030586 | 4.066289 | 4.520186 | 4.544127 | 4.526576 | 4.438426 | 4.356249 | 4.239519 | 4.539940 | 4.566264 | 4.628985 | 4.009212 | 3.951865 | 4.124829 | 4.215532 | 4.308877 | 4.498395 | 4.545531 | 4.518466 | 4.527173 | 4.546017 | 4.583752 | 4.522906 |
| GO:0034968 | histone lysine methylation | 2/42 | 131/20870 | 0.0285849894 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.983262 | 5.379789 | 5.275671 | 5.431893 | 4.972092 | 5.199626 | 5.405767 | 5.451188 | 4.943596 | 5.016527 | 4.988726 | 5.395071 | 5.381035 | 5.363081 | 5.395833 | 5.206589 | 5.216624 | 5.412841 | 5.400956 | 5.480595 | 4.979577 | 4.919443 | 5.015623 | 5.142490 | 5.144784 | 5.305479 | 5.425170 | 5.394140 | 5.397792 | 5.435744 | 5.478143 | 5.439292 |
| GO:0015749 | monosaccharide transmembrane transport | 2/42 | 132/20870 | 0.0289882819 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 4.009689 | 4.531466 | 4.346997 | 4.578489 | 4.021472 | 4.342779 | 4.531057 | 4.550399 | 3.946372 | 4.023472 | 4.057002 | 4.520517 | 4.546324 | 4.527433 | 4.439870 | 4.356832 | 4.237121 | 4.538554 | 4.567242 | 4.628212 | 4.001257 | 3.942619 | 4.115174 | 4.209281 | 4.301550 | 4.501833 | 4.545982 | 4.520033 | 4.527032 | 4.544803 | 4.582962 | 4.522789 |
| GO:0002679 | respiratory burst involved in defense response | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | SELENOK | 1 | 9.697135 | 9.008855 | 9.448125 | 8.842761 | 9.692611 | 9.580971 | 8.890680 | 8.796517 | 9.733745 | 9.605015 | 9.748377 | 9.086117 | 8.989835 | 8.947078 | 9.248899 | 9.556756 | 9.519583 | 8.928668 | 8.768104 | 8.826917 | 9.701870 | 9.602748 | 9.768415 | 9.630225 | 9.699079 | 9.396481 | 8.930429 | 8.865373 | 8.875382 | 8.793168 | 8.785013 | 8.811245 |
| GO:0030252 | growth hormone secretion | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | SELENOT | 1 | 5.026271 | 5.908669 | 5.610761 | 5.746560 | 5.053080 | 5.505696 | 5.928146 | 5.828500 | 4.929512 | 5.054772 | 5.089658 | 5.887736 | 5.927985 | 5.910004 | 5.745475 | 5.558380 | 5.518085 | 5.691137 | 5.710031 | 5.834280 | 4.999817 | 4.951396 | 5.196213 | 5.314349 | 5.425982 | 5.741996 | 5.935095 | 5.962810 | 5.885472 | 5.824707 | 5.893083 | 5.764855 |
| GO:0032239 | regulation of nucleobase-containing compound transport | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | RSC1A1 | 1 | 4.791809 | 6.103455 | 5.033763 | 6.109421 | 4.632461 | 4.960208 | 6.198619 | 6.156953 | 4.758135 | 4.957706 | 4.641741 | 6.062079 | 6.112351 | 6.134974 | 5.525185 | 4.659061 | 4.752562 | 6.054491 | 6.068430 | 6.200777 | 4.580923 | 4.668449 | 4.646580 | 4.973383 | 5.016118 | 4.888199 | 6.182533 | 6.223272 | 6.189723 | 6.150770 | 6.171415 | 6.148564 |
| GO:0042159 | lipoprotein catabolic process | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.277006 | 5.843844 | 5.519253 | 5.820153 | 5.319199 | 5.538825 | 5.715315 | 5.730959 | 5.275237 | 5.177100 | 5.372095 | 5.919534 | 5.840335 | 5.767663 | 5.469726 | 5.549192 | 5.537572 | 5.843397 | 5.802951 | 5.813807 | 5.288162 | 5.268699 | 5.397375 | 5.411041 | 5.592985 | 5.604411 | 5.735134 | 5.715411 | 5.695122 | 5.726120 | 5.784753 | 5.680092 |
| GO:0072160 | nephron tubule epithelial cell differentiation | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.031676 | 4.987204 | 3.932166 | 5.137764 | 2.685684 | 2.466261 | 4.941656 | 5.156717 | 3.189773 | 3.422572 | 2.228226 | 5.046532 | 5.019654 | 4.890663 | 4.376446 | 3.252520 | 3.953711 | 5.098517 | 5.153168 | 5.160810 | 2.505531 | 3.147452 | 2.251596 | 2.391907 | 2.153686 | 2.782441 | 4.938351 | 4.956573 | 4.929914 | 5.147879 | 5.170474 | 5.151697 |
| GO:0090110 | COPII-coated vesicle cargo loading | 1/42 | 15/20870 | 0.0297751565 | 0.2820041 | 0.2468512 | SAR1A | 1 | 5.131829 | 5.884984 | 5.471408 | 5.912995 | 5.124147 | 5.364404 | 5.879828 | 5.950802 | 5.126930 | 5.256804 | 5.000358 | 5.883122 | 5.891894 | 5.879908 | 5.649377 | 5.365282 | 5.381573 | 5.899057 | 5.893226 | 5.946118 | 5.110138 | 5.182915 | 5.077357 | 5.349783 | 5.242525 | 5.490179 | 5.883379 | 5.893064 | 5.862876 | 5.950180 | 5.973215 | 5.928666 |
| GO:0034219 | carbohydrate transmembrane transport | 2/42 | 134/20870 | 0.0298017408 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 3.978073 | 4.509439 | 4.318371 | 4.562716 | 3.989838 | 4.309953 | 4.507585 | 4.532671 | 3.915335 | 3.992647 | 4.024078 | 4.498446 | 4.524933 | 4.504804 | 4.414098 | 4.325425 | 4.208226 | 4.522447 | 4.552537 | 4.611728 | 3.968933 | 3.912601 | 4.082740 | 4.176330 | 4.268583 | 4.469232 | 4.522321 | 4.496495 | 4.503817 | 4.527194 | 4.565838 | 4.504311 |
| GO:0043467 | regulation of generation of precursor metabolites and energy | 2/42 | 134/20870 | 0.0298017408 | 0.2820041 | 0.2468512 | OGT||NDUFA4 | 2 | 6.608718 | 6.528474 | 6.577218 | 6.579274 | 6.597014 | 6.532015 | 6.518725 | 6.576511 | 6.614946 | 6.643457 | 6.566711 | 6.570992 | 6.506438 | 6.507035 | 6.549551 | 6.566454 | 6.614849 | 6.606679 | 6.535278 | 6.594855 | 6.609028 | 6.602888 | 6.578951 | 6.564086 | 6.503929 | 6.527392 | 6.532130 | 6.493825 | 6.529901 | 6.582211 | 6.564471 | 6.582776 |
| GO:0009855 | determination of bilateral symmetry | 2/42 | 137/20870 | 0.0310389583 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 3.401227 | 4.373120 | 3.760697 | 4.385691 | 3.360780 | 3.611996 | 4.353899 | 4.369719 | 3.403402 | 3.477703 | 3.318168 | 4.364826 | 4.399866 | 4.354271 | 3.954995 | 3.637988 | 3.667390 | 4.344191 | 4.389736 | 4.422085 | 3.334230 | 3.370911 | 3.376830 | 3.507136 | 3.553315 | 3.762509 | 4.373274 | 4.353909 | 4.334249 | 4.350659 | 4.417113 | 4.340173 |
| GO:0043414 | macromolecule methylation | 3/42 | 341/20870 | 0.0310496467 | 0.2820041 | 0.2468512 | METTL2A||OGT||CTNNB1 | 3 | 5.586565 | 5.733021 | 5.699702 | 5.787615 | 5.589075 | 5.729628 | 5.754018 | 5.810007 | 5.546544 | 5.590010 | 5.622144 | 5.753399 | 5.735313 | 5.710025 | 5.756715 | 5.672381 | 5.668271 | 5.792741 | 5.745501 | 5.823535 | 5.575296 | 5.547233 | 5.643008 | 5.685629 | 5.743527 | 5.758703 | 5.773852 | 5.746003 | 5.741990 | 5.792518 | 5.830659 | 5.806585 |
| GO:0000956 | nuclear-transcribed mRNA catabolic process | 2/42 | 138/20870 | 0.0314558641 | 0.2820041 | 0.2468512 | NBAS||SMG8 | 2 | 5.662942 | 6.375390 | 5.807702 | 6.393633 | 5.619638 | 5.754664 | 6.413931 | 6.445531 | 5.648414 | 5.705622 | 5.633787 | 6.375285 | 6.370738 | 6.380131 | 6.038953 | 5.668446 | 5.684521 | 6.351821 | 6.366227 | 6.460422 | 5.597767 | 5.594101 | 5.665905 | 5.724890 | 5.777431 | 5.761171 | 6.391244 | 6.421997 | 6.428280 | 6.436079 | 6.456582 | 6.443858 |
| GO:0009799 | specification of symmetry | 2/42 | 138/20870 | 0.0314558641 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 3.393418 | 4.363631 | 3.754338 | 4.376096 | 3.352735 | 3.604988 | 4.343977 | 4.360401 | 3.394498 | 3.470736 | 3.310578 | 4.354760 | 4.390994 | 4.344727 | 3.947631 | 3.632126 | 3.661770 | 4.335007 | 4.379736 | 4.412498 | 3.326524 | 3.362505 | 3.368817 | 3.499964 | 3.544896 | 3.756856 | 4.363358 | 4.343831 | 4.324480 | 4.341161 | 4.407839 | 4.330988 |
| GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.550688 | 5.410910 | 4.819770 | 5.465331 | 4.463718 | 4.348379 | 5.363952 | 5.450780 | 4.580634 | 4.606847 | 4.460395 | 5.464439 | 5.447320 | 5.316478 | 5.008776 | 4.526597 | 4.881777 | 5.452085 | 5.455798 | 5.487840 | 4.429122 | 4.512879 | 4.447807 | 4.286981 | 4.305276 | 4.447472 | 5.384866 | 5.361033 | 5.345685 | 5.447661 | 5.457747 | 5.446908 |
| GO:0006206 | pyrimidine nucleobase metabolic process | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CPS1 | 1 | 4.934265 | 5.852420 | 5.348263 | 5.852616 | 4.736181 | 4.895404 | 5.900347 | 5.887289 | 4.993593 | 5.082791 | 4.699093 | 5.859470 | 5.848815 | 5.848950 | 5.614726 | 5.088413 | 5.292327 | 5.792405 | 5.807899 | 5.952108 | 4.688101 | 4.845067 | 4.668819 | 4.743489 | 4.697042 | 5.192192 | 5.886049 | 5.890356 | 5.924330 | 5.836676 | 5.923810 | 5.899982 |
| GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 2.749596 | 3.589305 | 2.959495 | 3.516906 | 2.582547 | 2.457226 | 3.712844 | 3.617395 | 3.019037 | 2.829884 | 2.310172 | 3.503560 | 3.572379 | 3.686061 | 3.190661 | 2.790153 | 2.865568 | 3.312009 | 3.537028 | 3.678282 | 2.498334 | 2.707709 | 2.532749 | 2.360230 | 1.944383 | 2.905263 | 3.815564 | 3.770270 | 3.537567 | 3.592424 | 3.652885 | 3.606178 |
| GO:0060572 | morphogenesis of an epithelial bud | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.386852 | 5.087543 | 4.192199 | 5.250068 | 3.168011 | 3.109333 | 5.015133 | 5.233036 | 3.511079 | 3.662232 | 2.871793 | 5.137808 | 5.144729 | 4.973681 | 4.533120 | 3.695044 | 4.227728 | 5.221340 | 5.284890 | 5.243249 | 3.063951 | 3.471207 | 2.909093 | 2.942116 | 2.822848 | 3.476194 | 5.034064 | 5.011377 | 4.999747 | 5.214250 | 5.265532 | 5.218766 |
| GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.239058 | 6.305491 | 6.282980 | 6.159250 | 6.313135 | 6.466858 | 6.243185 | 6.215928 | 6.167369 | 6.286915 | 6.260182 | 6.338074 | 6.305612 | 6.272033 | 6.203715 | 6.365420 | 6.275246 | 6.193411 | 6.082892 | 6.198515 | 6.333074 | 6.196164 | 6.402581 | 6.500675 | 6.428548 | 6.470442 | 6.238597 | 6.241969 | 6.248968 | 6.208032 | 6.240156 | 6.199274 |
| GO:0061548 | ganglion development | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.473370 | 4.999026 | 4.225322 | 5.198279 | 3.306156 | 3.435391 | 4.946776 | 5.184953 | 3.583890 | 3.697962 | 3.062053 | 5.058124 | 5.027619 | 4.906946 | 4.546874 | 3.774571 | 4.251999 | 5.180050 | 5.202862 | 5.211741 | 3.192665 | 3.558492 | 3.129117 | 3.353792 | 3.283167 | 3.643444 | 4.963257 | 4.946184 | 4.930704 | 5.173278 | 5.199995 | 5.181456 |
| GO:0097091 | synaptic vesicle clustering | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.428653 | 5.042433 | 4.276806 | 5.184572 | 3.239562 | 3.489428 | 4.984551 | 5.173691 | 3.502216 | 3.648454 | 3.075949 | 5.069575 | 5.089633 | 4.965014 | 4.627619 | 3.854770 | 4.245228 | 5.139665 | 5.212711 | 5.200286 | 3.155927 | 3.437734 | 3.102041 | 3.283258 | 3.284630 | 3.829140 | 5.006896 | 4.984953 | 4.961446 | 5.166073 | 5.200376 | 5.154225 |
| GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1/42 | 16/20870 | 0.0317290977 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.579422 | 6.366463 | 6.221458 | 6.415833 | 5.582591 | 5.951472 | 6.372234 | 6.433346 | 5.546448 | 5.630817 | 5.559566 | 6.397133 | 6.391992 | 6.308564 | 6.344020 | 6.119706 | 6.191478 | 6.425288 | 6.406044 | 6.416102 | 5.540048 | 5.521429 | 5.680963 | 5.751997 | 5.812044 | 6.239835 | 6.427097 | 6.357927 | 6.329936 | 6.403739 | 6.480052 | 6.415067 |
| GO:0007098 | centrosome cycle | 2/42 | 140/20870 | 0.0322963640 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 5.165503 | 6.219477 | 5.370243 | 6.282620 | 5.105112 | 5.293807 | 6.282291 | 6.342968 | 5.171460 | 5.211797 | 5.111489 | 6.240009 | 6.186450 | 6.231399 | 5.683592 | 5.194504 | 5.172805 | 6.233205 | 6.256265 | 6.355446 | 5.065257 | 5.116696 | 5.132528 | 5.198054 | 5.252104 | 5.421716 | 6.257726 | 6.273910 | 6.314640 | 6.329398 | 6.347336 | 6.352071 |
| GO:0009581 | detection of external stimulus | 2/42 | 141/20870 | 0.0327199377 | 0.2820041 | 0.2468512 | ADGRV1||CTNNB1 | 2 | 3.350071 | 4.104735 | 3.823986 | 4.115885 | 3.299805 | 3.591122 | 4.094607 | 4.132622 | 3.337849 | 3.393846 | 3.317430 | 4.104460 | 4.130591 | 4.078687 | 3.975044 | 3.735368 | 3.748762 | 4.079464 | 4.110994 | 4.156165 | 3.263889 | 3.245077 | 3.386334 | 3.394095 | 3.483064 | 3.853937 | 4.121198 | 4.112785 | 4.048757 | 4.107397 | 4.172046 | 4.117580 |
| GO:0033865 | nucleoside bisphosphate metabolic process | 2/42 | 142/20870 | 0.0331457138 | 0.2820041 | 0.2468512 | PPT1||BPNT1 | 2 | 4.846220 | 5.246076 | 5.036811 | 5.361007 | 4.861142 | 5.096937 | 5.212573 | 5.321503 | 4.808299 | 4.846371 | 4.883023 | 5.253021 | 5.262983 | 5.221906 | 5.097772 | 5.023157 | 4.987292 | 5.342315 | 5.350938 | 5.389334 | 4.858997 | 4.794579 | 4.926818 | 5.012580 | 5.097391 | 5.176202 | 5.223590 | 5.215749 | 5.198261 | 5.313991 | 5.357139 | 5.292630 |
| GO:0033875 | ribonucleoside bisphosphate metabolic process | 2/42 | 142/20870 | 0.0331457138 | 0.2820041 | 0.2468512 | PPT1||BPNT1 | 2 | 4.846220 | 5.246076 | 5.036811 | 5.361007 | 4.861142 | 5.096937 | 5.212573 | 5.321503 | 4.808299 | 4.846371 | 4.883023 | 5.253021 | 5.262983 | 5.221906 | 5.097772 | 5.023157 | 4.987292 | 5.342315 | 5.350938 | 5.389334 | 4.858997 | 4.794579 | 4.926818 | 5.012580 | 5.097391 | 5.176202 | 5.223590 | 5.215749 | 5.198261 | 5.313991 | 5.357139 | 5.292630 |
| GO:0034032 | purine nucleoside bisphosphate metabolic process | 2/42 | 142/20870 | 0.0331457138 | 0.2820041 | 0.2468512 | PPT1||BPNT1 | 2 | 4.846220 | 5.246076 | 5.036811 | 5.361007 | 4.861142 | 5.096937 | 5.212573 | 5.321503 | 4.808299 | 4.846371 | 4.883023 | 5.253021 | 5.262983 | 5.221906 | 5.097772 | 5.023157 | 4.987292 | 5.342315 | 5.350938 | 5.389334 | 4.858997 | 4.794579 | 4.926818 | 5.012580 | 5.097391 | 5.176202 | 5.223590 | 5.215749 | 5.198261 | 5.313991 | 5.357139 | 5.292630 |
| GO:0007625 | grooming behavior | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | PPT1 | 1 | 7.114518 | 6.908724 | 6.932640 | 6.780642 | 7.208278 | 7.268711 | 6.834968 | 6.748884 | 7.072533 | 6.931522 | 7.313402 | 6.983479 | 6.853543 | 6.885957 | 6.799333 | 7.020083 | 6.969354 | 6.834276 | 6.722540 | 6.782945 | 7.184912 | 7.099816 | 7.330606 | 7.207998 | 7.454914 | 7.122219 | 6.874520 | 6.794500 | 6.834776 | 6.751140 | 6.793700 | 6.700300 |
| GO:0035751 | regulation of lysosomal lumen pH | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.547151 | 6.143089 | 6.012913 | 6.157383 | 5.594329 | 5.812966 | 5.960714 | 6.049120 | 5.507792 | 5.555314 | 5.577469 | 6.220173 | 6.168358 | 6.034425 | 5.983555 | 6.049409 | 6.004990 | 6.185123 | 6.178877 | 6.106844 | 5.588089 | 5.546629 | 5.646520 | 5.629535 | 5.678630 | 6.085951 | 6.010348 | 5.954929 | 5.915282 | 6.043750 | 6.116539 | 5.984015 |
| GO:0043981 | histone H4-K5 acetylation | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | OGT | 1 | 4.971643 | 5.314386 | 5.402001 | 5.392739 | 4.971122 | 5.237293 | 5.388748 | 5.447956 | 4.889608 | 5.000644 | 5.021231 | 5.334475 | 5.313129 | 5.295286 | 5.554904 | 5.304345 | 5.333526 | 5.371212 | 5.348129 | 5.456600 | 4.971169 | 4.897353 | 5.041255 | 5.174966 | 5.200158 | 5.331790 | 5.445765 | 5.352183 | 5.366526 | 5.399964 | 5.511951 | 5.429609 |
| GO:0043982 | histone H4-K8 acetylation | 1/42 | 17/20870 | 0.0336791973 | 0.2820041 | 0.2468512 | OGT | 1 | 4.971643 | 5.314386 | 5.402001 | 5.392739 | 4.971122 | 5.237293 | 5.388748 | 5.447956 | 4.889608 | 5.000644 | 5.021231 | 5.334475 | 5.313129 | 5.295286 | 5.554904 | 5.304345 | 5.333526 | 5.371212 | 5.348129 | 5.456600 | 4.971169 | 4.897353 | 5.041255 | 5.174966 | 5.200158 | 5.331790 | 5.445765 | 5.352183 | 5.366526 | 5.399964 | 5.511951 | 5.429609 |
| GO:0000723 | telomere maintenance | 2/42 | 144/20870 | 0.0340038326 | 0.2820041 | 0.2468512 | RPA2||CTNNB1 | 2 | 6.549257 | 7.378963 | 6.750769 | 7.426868 | 6.436545 | 6.520055 | 7.429793 | 7.483000 | 6.571249 | 6.707475 | 6.346240 | 7.394273 | 7.353542 | 7.388736 | 7.042824 | 6.490400 | 6.662640 | 7.384405 | 7.393826 | 7.499518 | 6.416087 | 6.535723 | 6.351748 | 6.550641 | 6.470744 | 6.537514 | 7.413457 | 7.420806 | 7.454779 | 7.468116 | 7.492484 | 7.488283 |
| GO:0009582 | detection of abiotic stimulus | 2/42 | 144/20870 | 0.0340038326 | 0.2820041 | 0.2468512 | ADGRV1||CTNNB1 | 2 | 3.462736 | 4.177900 | 3.943939 | 4.190665 | 3.414085 | 3.716705 | 4.169095 | 4.203256 | 3.443043 | 3.509949 | 3.434019 | 4.176469 | 4.203565 | 4.153226 | 4.089108 | 3.861953 | 3.869003 | 4.155093 | 4.184674 | 4.231207 | 3.392429 | 3.351136 | 3.494869 | 3.518221 | 3.593903 | 3.991870 | 4.197626 | 4.185306 | 4.123258 | 4.177100 | 4.243575 | 4.188211 |
| GO:0048565 | digestive tract development | 2/42 | 144/20870 | 0.0340038326 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.526943 | 4.420165 | 3.925073 | 4.382544 | 3.463472 | 3.700561 | 4.426743 | 4.426496 | 3.559834 | 3.607378 | 3.406026 | 4.457367 | 4.427743 | 4.374155 | 4.125183 | 3.780195 | 3.846139 | 4.353609 | 4.361842 | 4.430924 | 3.400508 | 3.526340 | 3.460820 | 3.587927 | 3.600922 | 3.891916 | 4.428177 | 4.428898 | 4.423148 | 4.404738 | 4.462168 | 4.411901 |
| GO:0016042 | lipid catabolic process | 3/42 | 354/20870 | 0.0341347463 | 0.2820041 | 0.2468512 | CPS1||SLC25A17||PPT1 | 3 | 4.964218 | 5.134583 | 5.115672 | 5.119145 | 4.966732 | 5.076017 | 5.084825 | 5.088222 | 4.937602 | 4.971876 | 4.982791 | 5.171195 | 5.135511 | 5.096065 | 5.148153 | 5.102639 | 5.095659 | 5.121005 | 5.091167 | 5.144764 | 4.961134 | 4.931475 | 5.006591 | 4.997493 | 5.062472 | 5.163233 | 5.110801 | 5.074962 | 5.068348 | 5.072638 | 5.121803 | 5.069627 |
| GO:0032355 | response to estradiol | 2/42 | 146/20870 | 0.0348706396 | 0.2820041 | 0.2468512 | H2AZ1||CTNNB1 | 2 | 6.705869 | 6.714693 | 6.601020 | 6.561242 | 6.730425 | 6.836428 | 6.649902 | 6.561429 | 6.668979 | 6.699331 | 6.748188 | 6.730504 | 6.723451 | 6.689798 | 6.491638 | 6.642589 | 6.662840 | 6.604171 | 6.499426 | 6.578079 | 6.726367 | 6.667449 | 6.794647 | 6.852974 | 6.966212 | 6.675317 | 6.660034 | 6.647494 | 6.642120 | 6.553081 | 6.583835 | 6.547103 |
| GO:0042157 | lipoprotein metabolic process | 2/42 | 146/20870 | 0.0348706396 | 0.2820041 | 0.2468512 | SELENOK||PPT1 | 2 | 4.699001 | 5.293147 | 5.095771 | 5.322650 | 4.717104 | 4.924364 | 5.225843 | 5.267116 | 4.683838 | 4.706054 | 4.706991 | 5.319957 | 5.304875 | 5.253780 | 5.161892 | 5.069286 | 5.053749 | 5.304604 | 5.321664 | 5.341445 | 4.692560 | 4.714587 | 4.743707 | 4.766219 | 4.817317 | 5.157437 | 5.253284 | 5.215065 | 5.208776 | 5.265082 | 5.304998 | 5.230300 |
| GO:0018022 | peptidyl-lysine methylation | 2/42 | 147/20870 | 0.0353072762 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 4.896156 | 5.268547 | 5.171412 | 5.316011 | 4.884436 | 5.104911 | 5.292852 | 5.329293 | 4.858990 | 4.920783 | 4.907961 | 5.286036 | 5.267841 | 5.251558 | 5.290027 | 5.105793 | 5.110672 | 5.299049 | 5.284537 | 5.363226 | 4.891414 | 4.828513 | 4.931517 | 5.046205 | 5.056468 | 5.206420 | 5.313729 | 5.281263 | 5.283336 | 5.313586 | 5.358273 | 5.315577 |
| GO:0006703 | estrogen biosynthetic process | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | HSD17B7 | 1 | 3.456693 | 5.113624 | 3.350914 | 4.906031 | 3.440546 | 3.243876 | 4.996812 | 4.901798 | 3.443555 | 3.511312 | 3.413464 | 5.151100 | 5.118942 | 5.069667 | 3.608586 | 3.154021 | 3.249353 | 4.799790 | 4.978254 | 4.934116 | 3.373283 | 3.486259 | 3.459692 | 3.183915 | 3.392489 | 3.142597 | 4.993252 | 5.011944 | 4.985109 | 4.933122 | 4.942235 | 4.827228 |
| GO:0006901 | vesicle coating | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.375713 | 7.039942 | 6.665758 | 6.914431 | 6.362097 | 6.517449 | 6.966981 | 6.914325 | 6.319915 | 6.416088 | 6.389434 | 7.036879 | 7.083706 | 6.997964 | 6.708558 | 6.623968 | 6.663506 | 6.907498 | 6.928985 | 6.906700 | 6.331490 | 6.345095 | 6.408529 | 6.440062 | 6.459905 | 6.643534 | 7.007724 | 6.962568 | 6.929583 | 6.926524 | 6.934904 | 6.880965 |
| GO:0043923 | positive regulation by host of viral transcription | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | CCNT1 | 1 | 5.520201 | 6.352141 | 5.657690 | 6.306795 | 5.487684 | 5.779983 | 6.436201 | 6.375567 | 5.481309 | 5.564440 | 5.513633 | 6.298695 | 6.357884 | 6.398115 | 5.895728 | 5.533036 | 5.511109 | 6.278462 | 6.250333 | 6.387899 | 5.527426 | 5.396491 | 5.534976 | 5.793711 | 5.758767 | 5.787233 | 6.422250 | 6.416735 | 6.469043 | 6.356655 | 6.386815 | 6.383046 |
| GO:0046855 | inositol phosphate dephosphorylation | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.156687 | 4.239506 | 3.708721 | 4.396930 | 3.127006 | 3.434130 | 4.263546 | 4.445901 | 3.152228 | 3.264512 | 3.044967 | 4.226526 | 4.253887 | 4.237973 | 3.995301 | 3.579755 | 3.500773 | 4.308011 | 4.380170 | 4.496325 | 3.098603 | 3.121776 | 3.159972 | 3.302340 | 3.306208 | 3.663291 | 4.283459 | 4.272386 | 4.234335 | 4.425210 | 4.494219 | 4.417022 |
| GO:0070365 | hepatocyte differentiation | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | CPS1 | 1 | 5.035432 | 4.575960 | 4.472858 | 4.131174 | 5.065228 | 4.968086 | 4.481154 | 4.005317 | 5.058295 | 4.866373 | 5.165841 | 4.619095 | 4.609481 | 4.496091 | 4.311927 | 4.575006 | 4.518556 | 4.206733 | 4.032825 | 4.148569 | 5.085259 | 4.951214 | 5.152017 | 5.002515 | 5.194220 | 4.657210 | 4.462300 | 4.488370 | 4.492604 | 4.035131 | 3.983499 | 3.996821 |
| GO:0080182 | histone H3-K4 trimethylation | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | OGT | 1 | 4.169385 | 4.905353 | 4.814708 | 5.088010 | 4.082535 | 4.420420 | 4.985889 | 5.134885 | 4.150066 | 4.315740 | 4.027815 | 4.884448 | 4.926561 | 4.904742 | 5.086125 | 4.651894 | 4.662264 | 4.996613 | 5.095695 | 5.166681 | 4.115634 | 4.102699 | 4.027718 | 4.380702 | 4.217244 | 4.632702 | 4.984148 | 5.010556 | 4.962560 | 5.078557 | 5.225134 | 5.096491 |
| GO:0090036 | regulation of protein kinase C signaling | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 2.976564 | 3.611891 | 3.679205 | 3.591815 | 3.011556 | 3.514147 | 3.590861 | 3.610288 | 2.876324 | 2.959175 | 3.086396 | 3.600954 | 3.612966 | 3.621678 | 3.860031 | 3.629472 | 3.527767 | 3.590995 | 3.589782 | 3.594663 | 3.030531 | 2.850491 | 3.139050 | 3.200127 | 3.357190 | 3.891202 | 3.617194 | 3.610103 | 3.544167 | 3.591716 | 3.691797 | 3.543357 |
| GO:1903830 | magnesium ion transmembrane transport | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 2.902988 | 5.077355 | 3.726273 | 5.014121 | 2.830494 | 3.150318 | 4.996710 | 4.988288 | 2.936599 | 3.159809 | 2.547465 | 5.119175 | 5.078413 | 5.033197 | 4.131741 | 3.451379 | 3.490058 | 4.907910 | 5.087220 | 5.041276 | 2.760378 | 3.022304 | 2.686873 | 3.016643 | 2.886630 | 3.478622 | 4.989658 | 5.031835 | 4.967904 | 4.974955 | 5.043993 | 4.944094 |
| GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 1/42 | 18/20870 | 0.0356254628 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.439509 | 5.056724 | 4.255972 | 5.265046 | 3.266567 | 3.484048 | 5.023489 | 5.290780 | 3.546361 | 3.696696 | 2.981473 | 5.104268 | 5.076360 | 4.986957 | 4.609640 | 3.834856 | 4.219961 | 5.225967 | 5.272872 | 5.295430 | 3.184752 | 3.501005 | 3.079984 | 3.381462 | 3.181889 | 3.814723 | 5.038795 | 5.015850 | 5.015700 | 5.272389 | 5.316801 | 5.282774 |
| GO:0006790 | sulfur compound metabolic process | 3/42 | 366/20870 | 0.0371198540 | 0.2820041 | 0.2468512 | CPS1||PPT1||BPNT1 | 3 | 6.095896 | 5.939683 | 5.993266 | 5.923982 | 6.153063 | 6.187191 | 5.894843 | 5.904300 | 6.065748 | 6.037829 | 6.180136 | 5.980998 | 5.934692 | 5.902271 | 5.919585 | 6.027796 | 6.029688 | 5.946167 | 5.876391 | 5.948236 | 6.148859 | 6.091226 | 6.216382 | 6.195865 | 6.281603 | 6.076803 | 5.910465 | 5.882078 | 5.891842 | 5.894964 | 5.924779 | 5.892936 |
| GO:0006044 | N-acetylglucosamine metabolic process | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | OGA | 1 | 3.124689 | 3.954254 | 3.968484 | 4.069396 | 3.074994 | 3.403264 | 4.037562 | 4.284912 | 3.093583 | 3.284847 | 2.978959 | 3.944658 | 3.967801 | 3.950201 | 4.222068 | 3.844017 | 3.801157 | 3.992726 | 4.061697 | 4.149479 | 3.078107 | 3.060668 | 3.086089 | 3.133206 | 3.105046 | 3.844170 | 4.038667 | 4.024647 | 4.049267 | 4.238721 | 4.320770 | 4.294034 |
| GO:0015693 | magnesium ion transport | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 2.810357 | 4.984767 | 3.633543 | 4.921689 | 2.737688 | 3.057520 | 4.904309 | 4.895823 | 2.844132 | 3.067209 | 2.454571 | 5.026472 | 4.986017 | 4.940532 | 4.038924 | 3.358619 | 3.397497 | 4.815426 | 4.994751 | 4.948933 | 2.667529 | 2.929579 | 2.594006 | 2.923923 | 2.793800 | 3.385789 | 4.897205 | 4.939246 | 4.875752 | 4.882485 | 4.951564 | 4.851597 |
| GO:0015867 | ATP transport | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | SLC25A17 | 1 | 7.671523 | 7.164200 | 7.476798 | 7.238043 | 7.627840 | 7.527461 | 7.082855 | 7.265943 | 7.732037 | 7.784559 | 7.479966 | 7.257493 | 7.135802 | 7.094278 | 7.375305 | 7.407617 | 7.633522 | 7.320616 | 7.153340 | 7.235324 | 7.640229 | 7.709368 | 7.528155 | 7.668476 | 7.637660 | 7.237533 | 7.086844 | 7.081727 | 7.079986 | 7.259659 | 7.266909 | 7.271238 |
| GO:0045603 | positive regulation of endothelial cell differentiation | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.627384 | 5.313597 | 4.452743 | 5.487211 | 3.365024 | 3.293362 | 5.256679 | 5.496493 | 3.753341 | 3.961969 | 3.000901 | 5.372157 | 5.350422 | 5.213113 | 4.832143 | 3.873338 | 4.495330 | 5.455923 | 5.491879 | 5.513252 | 3.205442 | 3.746580 | 3.046645 | 3.168071 | 3.001420 | 3.634070 | 5.258107 | 5.263586 | 5.248303 | 5.481915 | 5.508148 | 5.499293 |
| GO:0046838 | phosphorylated carbohydrate dephosphorylation | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.156687 | 4.239506 | 3.708721 | 4.396930 | 3.127006 | 3.434130 | 4.263546 | 4.445901 | 3.152228 | 3.264512 | 3.044967 | 4.226526 | 4.253887 | 4.237973 | 3.995301 | 3.579755 | 3.500773 | 4.308011 | 4.380170 | 4.496325 | 3.098603 | 3.121776 | 3.159972 | 3.302340 | 3.306208 | 3.663291 | 4.283459 | 4.272386 | 4.234335 | 4.425210 | 4.494219 | 4.417022 |
| GO:0060438 | trachea development | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.353478 | 5.247170 | 4.836983 | 5.328615 | 4.302878 | 4.483332 | 5.243716 | 5.351692 | 4.352196 | 4.475705 | 4.221335 | 5.276532 | 5.279546 | 5.183375 | 5.039296 | 4.623441 | 4.818167 | 5.318914 | 5.324260 | 5.342565 | 4.238192 | 4.358600 | 4.309307 | 4.379767 | 4.401652 | 4.652377 | 5.253717 | 5.236220 | 5.241155 | 5.341451 | 5.358409 | 5.355159 |
| GO:0061323 | cell proliferation involved in heart morphogenesis | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.378849 | 4.983953 | 4.192314 | 5.196400 | 3.217407 | 3.437614 | 4.953531 | 5.222285 | 3.480782 | 3.635429 | 2.929052 | 5.032063 | 5.002992 | 4.914209 | 4.549142 | 3.772857 | 4.150878 | 5.157834 | 5.202903 | 5.227599 | 3.134329 | 3.447603 | 3.037834 | 3.329219 | 3.147312 | 3.764898 | 4.968319 | 4.945315 | 4.946842 | 5.204362 | 5.247656 | 5.214480 |
| GO:1903204 | negative regulation of oxidative stress-induced neuron death | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 8.831526 | 8.415356 | 8.724088 | 8.486348 | 8.851282 | 8.847334 | 8.405384 | 8.497900 | 8.801967 | 8.831525 | 8.860493 | 8.460765 | 8.399845 | 8.384323 | 8.586955 | 8.744722 | 8.830128 | 8.541265 | 8.414252 | 8.500627 | 8.852136 | 8.859771 | 8.841884 | 8.930421 | 8.902031 | 8.698656 | 8.408407 | 8.405460 | 8.402281 | 8.494909 | 8.494657 | 8.504114 |
| GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1/42 | 19/20870 | 0.0375679014 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.757745 | 4.926525 | 4.008576 | 4.988606 | 3.609393 | 3.199754 | 4.876304 | 4.989365 | 3.845393 | 3.868099 | 3.536579 | 4.978516 | 4.954779 | 4.842662 | 4.345684 | 3.516433 | 4.045339 | 4.956904 | 4.994425 | 5.013907 | 3.544849 | 3.767725 | 3.501212 | 3.239935 | 3.118745 | 3.237291 | 4.888121 | 4.881944 | 4.858681 | 4.988916 | 4.994725 | 4.984435 |
| GO:0031023 | microtubule organizing center organization | 2/42 | 153/20870 | 0.0379717329 | 0.2820041 | 0.2468512 | CTNNB1||RTTN | 2 | 5.069343 | 6.120454 | 5.288061 | 6.182125 | 5.011003 | 5.209661 | 6.183373 | 6.243230 | 5.071038 | 5.115869 | 5.019513 | 6.139067 | 6.088790 | 6.132985 | 5.597626 | 5.116000 | 5.092268 | 6.132146 | 6.155772 | 6.255466 | 4.972363 | 5.019740 | 5.040073 | 5.112548 | 5.168375 | 5.338362 | 6.160386 | 6.175615 | 6.213596 | 6.228099 | 6.249667 | 6.251803 |
| GO:0055123 | digestive system development | 2/42 | 154/20870 | 0.0384231564 | 0.2820041 | 0.2468512 | CPS1||CTNNB1 | 2 | 3.567602 | 4.484743 | 3.976978 | 4.432799 | 3.502208 | 3.736204 | 4.480973 | 4.473696 | 3.595110 | 3.650877 | 3.449390 | 4.523072 | 4.490499 | 4.439427 | 4.176272 | 3.835010 | 3.896252 | 4.400922 | 4.417246 | 4.479047 | 3.447400 | 3.560243 | 3.496760 | 3.618123 | 3.619917 | 3.945380 | 4.481474 | 4.486122 | 4.475301 | 4.451682 | 4.511976 | 4.456651 |
| GO:0001711 | endodermal cell fate commitment | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.262603 | 5.658517 | 4.813982 | 5.832310 | 4.070601 | 4.206884 | 5.654124 | 5.859654 | 4.329451 | 4.483467 | 3.917300 | 5.689963 | 5.678456 | 5.605694 | 5.160114 | 4.408354 | 4.775892 | 5.802419 | 5.833567 | 5.860362 | 3.995812 | 4.285759 | 3.901971 | 4.201360 | 4.117802 | 4.295976 | 5.661698 | 5.644069 | 5.656547 | 5.853598 | 5.868185 | 5.857139 |
| GO:0043586 | tongue development | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.569268 | 5.985129 | 5.168130 | 6.086523 | 4.365965 | 4.500682 | 5.996404 | 6.112092 | 4.646097 | 4.805336 | 4.187551 | 6.022621 | 5.993291 | 5.938204 | 5.508930 | 4.810895 | 5.098501 | 6.067072 | 6.076026 | 6.115999 | 4.329798 | 4.572691 | 4.166200 | 4.419888 | 4.207648 | 4.809018 | 5.988941 | 5.992349 | 6.007851 | 6.096694 | 6.122989 | 6.116464 |
| GO:0046112 | nucleobase biosynthetic process | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CPS1 | 1 | 6.818454 | 6.983781 | 6.840244 | 6.902339 | 6.837005 | 6.927166 | 6.983172 | 6.928267 | 6.793118 | 6.757717 | 6.900659 | 7.015048 | 6.966089 | 6.969688 | 6.835235 | 6.849134 | 6.836322 | 6.900487 | 6.844964 | 6.959301 | 6.829585 | 6.782726 | 6.896436 | 6.861290 | 7.007570 | 6.908757 | 7.003007 | 6.952335 | 6.993672 | 6.911403 | 6.956987 | 6.915974 |
| GO:0048490 | anterograde synaptic vesicle transport | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | AP3M2 | 1 | 4.754411 | 5.012477 | 4.859218 | 5.109481 | 4.744554 | 4.821254 | 5.016193 | 5.081542 | 4.758212 | 4.776416 | 4.728194 | 5.007897 | 5.030347 | 4.999008 | 4.950001 | 4.822136 | 4.800966 | 5.092478 | 5.090698 | 5.144614 | 4.759174 | 4.761990 | 4.711951 | 4.831514 | 4.777981 | 4.853230 | 5.005103 | 4.999482 | 5.043594 | 5.077072 | 5.126039 | 5.040230 |
| GO:0071545 | inositol phosphate catabolic process | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.140300 | 4.213631 | 3.705575 | 4.364860 | 3.103480 | 3.423720 | 4.242233 | 4.411433 | 3.136241 | 3.257736 | 3.016873 | 4.197340 | 4.228810 | 4.214571 | 3.986949 | 3.582919 | 3.498269 | 4.274553 | 4.348726 | 4.464894 | 3.075583 | 3.111606 | 3.122830 | 3.311564 | 3.280943 | 3.649021 | 4.259531 | 4.251708 | 4.215069 | 4.389827 | 4.461457 | 4.381670 |
| GO:0099514 | synaptic vesicle cytoskeletal transport | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | AP3M2 | 1 | 4.754411 | 5.012477 | 4.859218 | 5.109481 | 4.744554 | 4.821254 | 5.016193 | 5.081542 | 4.758212 | 4.776416 | 4.728194 | 5.007897 | 5.030347 | 4.999008 | 4.950001 | 4.822136 | 4.800966 | 5.092478 | 5.090698 | 5.144614 | 4.759174 | 4.761990 | 4.711951 | 4.831514 | 4.777981 | 4.853230 | 5.005103 | 4.999482 | 5.043594 | 5.077072 | 5.126039 | 5.040230 |
| GO:0099517 | synaptic vesicle transport along microtubule | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | AP3M2 | 1 | 4.754411 | 5.012477 | 4.859218 | 5.109481 | 4.744554 | 4.821254 | 5.016193 | 5.081542 | 4.758212 | 4.776416 | 4.728194 | 5.007897 | 5.030347 | 4.999008 | 4.950001 | 4.822136 | 4.800966 | 5.092478 | 5.090698 | 5.144614 | 4.759174 | 4.761990 | 4.711951 | 4.831514 | 4.777981 | 4.853230 | 5.005103 | 4.999482 | 5.043594 | 5.077072 | 5.126039 | 5.040230 |
| GO:1901679 | nucleotide transmembrane transport | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | SLC25A17 | 1 | 7.200952 | 6.769221 | 7.032344 | 6.802130 | 7.161536 | 7.073792 | 6.678675 | 6.821939 | 7.258069 | 7.308064 | 7.020695 | 6.850135 | 6.754867 | 6.698577 | 6.929087 | 6.974691 | 7.180621 | 6.883789 | 6.719048 | 6.798846 | 7.174169 | 7.233518 | 7.072339 | 7.197299 | 7.172773 | 6.821573 | 6.689182 | 6.675873 | 6.670909 | 6.803691 | 6.833819 | 6.828128 |
| GO:1904948 | midbrain dopaminergic neuron differentiation | 1/42 | 20/20870 | 0.0395065206 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.320140 | 6.106337 | 5.961809 | 6.156065 | 5.322954 | 5.692009 | 6.111376 | 6.173298 | 5.287442 | 5.371792 | 5.299740 | 6.136885 | 6.131917 | 6.048512 | 6.084810 | 5.859999 | 5.931396 | 6.165073 | 6.146510 | 6.156552 | 5.280639 | 5.262089 | 5.420854 | 5.492598 | 5.552465 | 5.980413 | 6.166035 | 6.096906 | 6.069463 | 6.144131 | 6.219985 | 6.154600 |
| GO:0016571 | histone methylation | 2/42 | 159/20870 | 0.0407111770 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.607968 | 5.682203 | 5.712590 | 5.726938 | 5.604506 | 5.734573 | 5.702380 | 5.750568 | 5.574012 | 5.618330 | 5.630944 | 5.703772 | 5.685633 | 5.656815 | 5.748105 | 5.693502 | 5.695496 | 5.736963 | 5.687317 | 5.755672 | 5.600790 | 5.564186 | 5.647336 | 5.713425 | 5.724416 | 5.765357 | 5.725735 | 5.692972 | 5.688142 | 5.732111 | 5.770198 | 5.749142 |
| GO:0006066 | alcohol metabolic process | 3/42 | 380/20870 | 0.0407678251 | 0.2820041 | 0.2468512 | HSD17B7||BPNT1||SLC5A3 | 3 | 5.338062 | 5.382816 | 5.505685 | 5.501227 | 5.373726 | 5.517767 | 5.337816 | 5.458979 | 5.289955 | 5.333710 | 5.388817 | 5.398865 | 5.404333 | 5.344496 | 5.489382 | 5.529617 | 5.497743 | 5.503450 | 5.484524 | 5.515537 | 5.371794 | 5.333392 | 5.414839 | 5.453949 | 5.493551 | 5.601717 | 5.361488 | 5.330298 | 5.321353 | 5.450240 | 5.489107 | 5.437083 |
| GO:0007194 | negative regulation of adenylate cyclase activity | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 3.242723 | 3.757289 | 3.539756 | 3.918975 | 3.245422 | 3.466481 | 3.753264 | 3.875733 | 3.179613 | 3.222519 | 3.322301 | 3.719741 | 3.816418 | 3.733804 | 3.574732 | 3.604911 | 3.433916 | 3.922194 | 3.867823 | 3.965258 | 3.272680 | 3.139587 | 3.318089 | 3.343376 | 3.364970 | 3.667781 | 3.814706 | 3.748814 | 3.693725 | 3.875903 | 3.914794 | 3.835410 |
| GO:0010738 | regulation of protein kinase A signaling | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 8.784907 | 7.895652 | 8.278112 | 7.672536 | 8.865779 | 8.948412 | 7.697977 | 7.510711 | 8.757124 | 8.517959 | 9.033440 | 8.099110 | 7.768215 | 7.795740 | 7.914556 | 8.496142 | 8.361645 | 7.919395 | 7.533749 | 7.528679 | 8.881596 | 8.593246 | 9.081286 | 8.852726 | 9.236948 | 8.701627 | 7.696356 | 7.698959 | 7.698614 | 7.531791 | 7.506358 | 7.493723 |
| GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.693665 | 4.105816 | 3.979432 | 4.099248 | 3.710687 | 3.792596 | 4.048815 | 4.008509 | 3.698326 | 3.671414 | 3.710973 | 4.111154 | 4.148149 | 4.056682 | 4.111203 | 3.913643 | 3.903818 | 4.032240 | 4.103322 | 4.159374 | 3.676921 | 3.658376 | 3.793040 | 3.598212 | 3.754153 | 3.997173 | 4.096665 | 4.067908 | 3.979295 | 3.983579 | 4.050033 | 3.990990 |
| GO:0046931 | pore complex assembly | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | NUP205 | 1 | 5.976609 | 6.799557 | 6.329316 | 6.928056 | 5.815089 | 5.820769 | 6.831894 | 7.006158 | 6.057149 | 6.195672 | 5.615579 | 6.844335 | 6.778294 | 6.774976 | 6.575064 | 6.059889 | 6.306990 | 6.886844 | 6.900658 | 6.994281 | 5.801197 | 6.004389 | 5.613135 | 5.951109 | 5.710502 | 5.790198 | 6.828651 | 6.829156 | 6.837856 | 6.975338 | 7.026851 | 7.015776 |
| GO:0048485 | sympathetic nervous system development | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 2.995007 | 4.786575 | 3.879255 | 4.979320 | 2.704889 | 2.686285 | 4.731804 | 4.973569 | 3.153813 | 3.316475 | 2.337646 | 4.839087 | 4.820751 | 4.695710 | 4.290297 | 3.286483 | 3.889729 | 4.953998 | 4.988728 | 4.994897 | 2.534739 | 3.106812 | 2.364781 | 2.599717 | 2.430968 | 2.973681 | 4.739785 | 4.734377 | 4.721187 | 4.964257 | 4.994659 | 4.961556 |
| GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | BPNT1 | 1 | 3.693665 | 4.105816 | 3.979432 | 4.099248 | 3.710687 | 3.792596 | 4.048815 | 4.008509 | 3.698326 | 3.671414 | 3.710973 | 4.111154 | 4.148149 | 4.056682 | 4.111203 | 3.913643 | 3.903818 | 4.032240 | 4.103322 | 4.159374 | 3.676921 | 3.658376 | 3.793040 | 3.598212 | 3.754153 | 3.997173 | 4.096665 | 4.067908 | 3.979295 | 3.983579 | 4.050033 | 3.990990 |
| GO:0072283 | metanephric renal vesicle morphogenesis | 1/42 | 21/20870 | 0.0414413276 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.315698 | 5.176333 | 4.585195 | 5.227105 | 4.229269 | 4.116094 | 5.128686 | 5.212819 | 4.345130 | 4.372921 | 4.224787 | 5.229257 | 5.213067 | 5.082215 | 4.774436 | 4.290867 | 4.647847 | 5.213514 | 5.217237 | 5.250279 | 4.195775 | 4.276858 | 4.213914 | 4.054289 | 4.073323 | 4.215250 | 5.150482 | 5.125667 | 5.109615 | 5.208869 | 5.220342 | 5.209217 |
| GO:0002053 | positive regulation of mesenchymal cell proliferation | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.569810 | 4.924265 | 4.215580 | 5.080304 | 3.478515 | 3.800405 | 4.872581 | 5.023588 | 3.613724 | 3.729083 | 3.339361 | 4.954052 | 4.972291 | 4.843092 | 4.472609 | 3.939883 | 4.184863 | 5.089167 | 5.070472 | 5.081213 | 3.404233 | 3.585571 | 3.439259 | 3.629592 | 3.683958 | 4.050398 | 4.878464 | 4.873895 | 4.865353 | 5.007600 | 5.047139 | 5.015724 |
| GO:0002089 | lens morphogenesis in camera-type eye | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.954906 | 5.920798 | 6.141591 | 6.396622 | 5.882845 | 6.055203 | 5.940908 | 6.448155 | 5.984288 | 6.122227 | 5.731354 | 5.990893 | 5.889634 | 5.879210 | 6.216924 | 6.002542 | 6.195784 | 6.430677 | 6.303495 | 6.451299 | 5.843072 | 6.060535 | 5.724611 | 6.117773 | 5.948752 | 6.093365 | 5.968146 | 5.919806 | 5.934346 | 6.455572 | 6.433190 | 6.455589 |
| GO:0007063 | regulation of sister chromatid cohesion | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.307569 | 6.811344 | 6.565588 | 6.824220 | 6.284487 | 6.439614 | 6.836784 | 6.868936 | 6.289742 | 6.353699 | 6.278114 | 6.836872 | 6.801055 | 6.795757 | 6.726299 | 6.417107 | 6.536384 | 6.775927 | 6.805020 | 6.889297 | 6.244137 | 6.291599 | 6.316784 | 6.381132 | 6.408507 | 6.525116 | 6.840607 | 6.832830 | 6.836903 | 6.855208 | 6.882464 | 6.869006 |
| GO:0033762 | response to glucagon | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CPS1 | 1 | 8.578909 | 8.200568 | 9.016931 | 8.232266 | 8.667386 | 9.089139 | 8.249581 | 8.066968 | 8.453089 | 8.562287 | 8.709805 | 7.650361 | 8.396300 | 8.429262 | 8.896224 | 9.133824 | 9.010954 | 8.158070 | 8.451542 | 8.057610 | 8.680361 | 8.524208 | 8.785670 | 8.905689 | 8.948501 | 9.366872 | 8.217004 | 8.301671 | 8.228599 | 8.128911 | 8.070154 | 7.998907 |
| GO:0035020 | regulation of Rac protein signal transduction | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | OGT | 1 | 5.013166 | 5.334232 | 5.153032 | 5.211367 | 5.029493 | 5.155393 | 5.325128 | 5.165876 | 4.990107 | 5.033242 | 5.015822 | 5.342166 | 5.318265 | 5.342134 | 5.289150 | 5.069996 | 5.089602 | 5.184994 | 5.162987 | 5.283256 | 5.066433 | 4.954274 | 5.064929 | 5.191598 | 5.118987 | 5.154680 | 5.348269 | 5.308304 | 5.318511 | 5.152910 | 5.182401 | 5.162158 |
| GO:0043576 | regulation of respiratory gaseous exchange | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | SLC5A3 | 1 | 3.912671 | 4.078121 | 4.402305 | 4.279699 | 3.922973 | 4.343005 | 4.127143 | 4.310386 | 3.828573 | 3.924470 | 3.980879 | 4.054395 | 4.096237 | 4.083412 | 4.531435 | 4.362904 | 4.302728 | 4.230157 | 4.291136 | 4.316445 | 3.898070 | 3.823891 | 4.038655 | 4.159110 | 4.291471 | 4.550608 | 4.153966 | 4.170522 | 4.054218 | 4.278814 | 4.342718 | 4.308916 |
| GO:0043984 | histone H4-K16 acetylation | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | OGT | 1 | 4.896010 | 5.377388 | 5.407865 | 5.434215 | 4.905230 | 5.200522 | 5.420465 | 5.470882 | 4.830549 | 4.889957 | 4.964400 | 5.392326 | 5.369116 | 5.370605 | 5.549058 | 5.346758 | 5.316530 | 5.371277 | 5.452029 | 5.477229 | 4.897388 | 4.810365 | 5.001575 | 5.058099 | 5.138210 | 5.384835 | 5.442749 | 5.417507 | 5.400831 | 5.429398 | 5.526935 | 5.454526 |
| GO:0048199 | vesicle targeting, to, from or within Golgi | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | SAR1A | 1 | 6.082884 | 6.777208 | 6.375513 | 6.654799 | 6.069498 | 6.225772 | 6.709120 | 6.654195 | 6.026986 | 6.124830 | 6.095101 | 6.772700 | 6.821409 | 6.736249 | 6.423115 | 6.331979 | 6.369990 | 6.644829 | 6.668627 | 6.650835 | 6.039793 | 6.053347 | 6.114258 | 6.147861 | 6.167028 | 6.353369 | 6.748148 | 6.704324 | 6.673920 | 6.664089 | 6.675132 | 6.622838 |
| GO:0071639 | positive regulation of monocyte chemotactic protein-1 production | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | SELENOK | 1 | 4.980524 | 6.573818 | 5.077308 | 6.564822 | 4.722358 | 4.381816 | 6.707812 | 6.586816 | 5.101178 | 5.180731 | 4.591997 | 6.530030 | 6.559523 | 6.630060 | 5.689630 | 4.589825 | 4.677604 | 6.482450 | 6.569703 | 6.638104 | 4.525230 | 5.015313 | 4.574465 | 4.253148 | 4.254017 | 4.608445 | 6.689070 | 6.694257 | 6.739572 | 6.646364 | 6.473729 | 6.634008 |
| GO:0072079 | nephron tubule formation | 1/42 | 22/20870 | 0.0433723298 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 2.924339 | 4.741278 | 3.796253 | 4.911537 | 2.651214 | 2.560772 | 4.692457 | 4.913499 | 3.090569 | 3.244461 | 2.256484 | 4.802690 | 4.768773 | 4.647818 | 4.194777 | 3.186499 | 3.834032 | 4.888465 | 4.919643 | 4.926222 | 2.473201 | 3.052624 | 2.320755 | 2.421208 | 2.286355 | 2.900391 | 4.708279 | 4.691047 | 4.677884 | 4.901626 | 4.928167 | 4.910577 |
| GO:0009150 | purine ribonucleotide metabolic process | 3/42 | 391/20870 | 0.0437579095 | 0.2820041 | 0.2468512 | PPT1||OGT||BPNT1 | 3 | 7.977925 | 7.437593 | 8.162638 | 7.690583 | 8.029391 | 8.191313 | 7.472514 | 7.622055 | 7.905842 | 7.952389 | 8.070514 | 7.270393 | 7.513241 | 7.515653 | 8.016962 | 8.288536 | 8.169630 | 7.664807 | 7.786816 | 7.614653 | 8.069416 | 7.966058 | 8.050612 | 8.152239 | 8.134329 | 8.282774 | 7.464443 | 7.479974 | 7.473083 | 7.663324 | 7.606211 | 7.595710 |
| GO:0032259 | methylation | 3/42 | 391/20870 | 0.0437579095 | 0.2820041 | 0.2468512 | METTL2A||OGT||CTNNB1 | 3 | 5.490085 | 5.652357 | 5.604520 | 5.694933 | 5.489179 | 5.625966 | 5.668069 | 5.714284 | 5.455116 | 5.493634 | 5.520751 | 5.673428 | 5.654162 | 5.629139 | 5.662974 | 5.574896 | 5.573864 | 5.699945 | 5.653094 | 5.730704 | 5.475380 | 5.449544 | 5.541063 | 5.578543 | 5.635919 | 5.662173 | 5.686256 | 5.660276 | 5.657501 | 5.696327 | 5.735655 | 5.710595 |
| GO:0002181 | cytoplasmic translation | 2/42 | 168/20870 | 0.0449563775 | 0.2820041 | 0.2468512 | RPS15A||RPL9 | 2 | 11.087627 | 10.859751 | 10.700510 | 10.744064 | 11.069908 | 10.739194 | 10.838454 | 10.747063 | 11.142651 | 11.083777 | 11.034414 | 10.918261 | 10.816127 | 10.842912 | 10.640620 | 10.656367 | 10.799195 | 10.783099 | 10.713760 | 10.734453 | 11.061758 | 11.155057 | 10.988042 | 10.916088 | 10.822622 | 10.435225 | 10.828015 | 10.844481 | 10.842809 | 10.757254 | 10.680672 | 10.800714 |
| GO:0001759 | organ induction | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 2.876870 | 4.463998 | 3.572536 | 4.676217 | 2.664693 | 2.754658 | 4.436676 | 4.676981 | 2.987352 | 3.116668 | 2.441657 | 4.503138 | 4.511615 | 4.373108 | 3.958598 | 3.028508 | 3.583063 | 4.641752 | 4.688430 | 4.697846 | 2.541967 | 2.915606 | 2.499146 | 2.743088 | 2.631319 | 2.878891 | 4.440305 | 4.440791 | 4.428899 | 4.653232 | 4.684001 | 4.693403 |
| GO:0002052 | positive regulation of neuroblast proliferation | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.015886 | 5.042616 | 4.535878 | 5.215336 | 3.897583 | 4.039361 | 5.043382 | 5.231390 | 4.060572 | 4.134868 | 3.835674 | 5.085494 | 5.058998 | 4.981337 | 4.857027 | 4.174109 | 4.495597 | 5.187179 | 5.202402 | 5.255530 | 3.805248 | 4.046049 | 3.829048 | 3.979735 | 3.948273 | 4.179058 | 5.043593 | 5.049055 | 5.037474 | 5.220309 | 5.255715 | 5.217833 |
| GO:0006907 | pinocytosis | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | PPT1 | 1 | 4.876200 | 5.578026 | 5.241122 | 5.687307 | 4.836261 | 4.997123 | 5.619402 | 5.752191 | 4.831826 | 4.973102 | 4.818530 | 5.618404 | 5.560038 | 5.554766 | 5.441269 | 5.085112 | 5.172701 | 5.629168 | 5.682786 | 5.747529 | 4.832989 | 4.842936 | 4.832836 | 4.950421 | 4.941142 | 5.094616 | 5.614930 | 5.626700 | 5.616548 | 5.732794 | 5.786357 | 5.736802 |
| GO:0021819 | layer formation in cerebral cortex | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.480257 | 5.547444 | 5.046847 | 5.695349 | 4.397219 | 4.613118 | 5.488124 | 5.647027 | 4.486476 | 4.614668 | 4.325100 | 5.614704 | 5.580135 | 5.441747 | 5.214686 | 4.852369 | 5.050788 | 5.671870 | 5.693573 | 5.720197 | 4.403152 | 4.450361 | 4.335855 | 4.502023 | 4.510754 | 4.805450 | 5.515833 | 5.485739 | 5.462301 | 5.640476 | 5.660426 | 5.640086 |
| GO:0060231 | mesenchymal to epithelial transition | 1/42 | 23/20870 | 0.0452995344 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.035807 | 4.862230 | 4.291770 | 4.903576 | 3.949445 | 3.844885 | 4.815852 | 4.893062 | 4.070915 | 4.094461 | 3.937106 | 4.914796 | 4.899049 | 4.768416 | 4.476974 | 4.012437 | 4.347035 | 4.889411 | 4.894861 | 4.926182 | 3.914672 | 3.998823 | 3.933481 | 3.787736 | 3.787785 | 3.952766 | 4.833606 | 4.814158 | 4.799592 | 4.887222 | 4.900684 | 4.891247 |
| GO:0008643 | carbohydrate transport | 2/42 | 170/20870 | 0.0459213317 | 0.2820041 | 0.2468512 | SLC5A3||RSC1A1 | 2 | 4.067728 | 4.603316 | 4.481574 | 4.654156 | 4.098509 | 4.416416 | 4.573956 | 4.592612 | 4.003992 | 4.084798 | 4.112212 | 4.592728 | 4.632689 | 4.584062 | 4.548696 | 4.501119 | 4.390364 | 4.623737 | 4.643619 | 4.694196 | 4.087781 | 4.017844 | 4.185004 | 4.268240 | 4.334638 | 4.621454 | 4.592995 | 4.566268 | 4.562412 | 4.587029 | 4.626629 | 4.563470 |
| GO:0031056 | regulation of histone modification | 2/42 | 172/20870 | 0.0468939695 | 0.2820041 | 0.2468512 | OGT||CTNNB1 | 2 | 5.220205 | 5.708676 | 5.435831 | 5.735372 | 5.173859 | 5.355152 | 5.742145 | 5.766290 | 5.192563 | 5.298517 | 5.166105 | 5.717233 | 5.713263 | 5.695439 | 5.578139 | 5.337440 | 5.380325 | 5.722317 | 5.693881 | 5.788285 | 5.161096 | 5.173874 | 5.186496 | 5.329956 | 5.326631 | 5.407411 | 5.752804 | 5.734507 | 5.739062 | 5.745528 | 5.787200 | 5.765842 |
| GO:0032200 | telomere organization | 2/42 | 172/20870 | 0.0468939695 | 0.2820041 | 0.2468512 | RPA2||CTNNB1 | 2 | 6.527224 | 7.318417 | 6.722691 | 7.351486 | 6.424548 | 6.482130 | 7.369899 | 7.397102 | 6.550322 | 6.687990 | 6.319632 | 7.328375 | 7.296327 | 7.330297 | 7.009867 | 6.456790 | 6.645758 | 7.310837 | 7.318517 | 7.422391 | 6.396083 | 6.530065 | 6.340883 | 6.506180 | 6.430487 | 6.508367 | 7.353876 | 7.359842 | 7.395624 | 7.399303 | 7.377155 | 7.414603 |
| GO:0001702 | gastrulation with mouth forming second | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.233107 | 5.422433 | 4.783825 | 5.527500 | 4.128624 | 4.411596 | 5.407874 | 5.493159 | 4.246409 | 4.376566 | 4.058790 | 5.429362 | 5.481119 | 5.353994 | 5.006921 | 4.564284 | 4.745710 | 5.489367 | 5.544794 | 5.547596 | 4.111046 | 4.200540 | 4.071232 | 4.322602 | 4.344757 | 4.555767 | 5.414145 | 5.422920 | 5.386306 | 5.457333 | 5.539409 | 5.481498 |
| GO:0018279 | protein N-linked glycosylation via asparagine | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 6.611683 | 7.314000 | 6.924215 | 7.178923 | 6.633270 | 6.706728 | 7.177432 | 7.133674 | 6.606879 | 6.644518 | 6.582983 | 7.370854 | 7.323022 | 7.245352 | 6.922732 | 6.932867 | 6.917001 | 7.170953 | 7.219199 | 7.145648 | 6.647122 | 6.616764 | 6.635760 | 6.664916 | 6.533595 | 6.897849 | 7.195621 | 7.190091 | 7.146073 | 7.121780 | 7.171177 | 7.107284 |
| GO:0030539 | male genitalia development | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.185989 | 5.023158 | 4.791298 | 5.281531 | 4.144837 | 4.408482 | 4.963834 | 5.227021 | 4.205925 | 4.316726 | 4.019861 | 5.067848 | 5.057332 | 4.940887 | 4.905699 | 4.660670 | 4.797117 | 5.253182 | 5.297813 | 5.293182 | 4.162048 | 4.196455 | 4.073221 | 4.298571 | 4.131497 | 4.728146 | 4.982185 | 4.968891 | 4.940106 | 5.223126 | 5.251268 | 5.206312 |
| GO:0031280 | negative regulation of cyclase activity | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | ADGRV1 | 1 | 3.135829 | 3.645431 | 3.430818 | 3.804674 | 3.136395 | 3.357641 | 3.640858 | 3.762544 | 3.071300 | 3.116350 | 3.216013 | 3.607336 | 3.706078 | 3.620879 | 3.462826 | 3.493845 | 3.330629 | 3.808563 | 3.753030 | 3.850765 | 3.162266 | 3.029940 | 3.210953 | 3.236554 | 3.254794 | 3.558410 | 3.701679 | 3.637046 | 3.581333 | 3.763581 | 3.802010 | 3.720900 |
| GO:0035994 | response to muscle stretch | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.295324 | 5.766372 | 5.482324 | 5.800913 | 5.302583 | 5.534643 | 5.754283 | 5.793782 | 5.206304 | 5.394725 | 5.278651 | 5.759620 | 5.792801 | 5.746296 | 5.541956 | 5.390393 | 5.510235 | 5.797505 | 5.751147 | 5.852309 | 5.319563 | 5.248738 | 5.337920 | 5.481705 | 5.587380 | 5.532909 | 5.774205 | 5.738718 | 5.749695 | 5.798215 | 5.832469 | 5.749458 |
| GO:0051571 | positive regulation of histone H3-K4 methylation | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.933372 | 5.950992 | 5.408666 | 6.057853 | 4.791517 | 4.997187 | 5.967625 | 6.093130 | 4.956718 | 5.132304 | 4.674667 | 5.981102 | 5.970969 | 5.899543 | 5.700721 | 5.124703 | 5.341159 | 6.033299 | 6.050498 | 6.089192 | 4.754633 | 4.920684 | 4.689269 | 4.945504 | 4.827370 | 5.194090 | 5.982310 | 5.958044 | 5.962405 | 6.077487 | 6.110176 | 6.091541 |
| GO:0060479 | lung cell differentiation | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.017536 | 5.185393 | 4.438673 | 5.324341 | 3.879529 | 3.851637 | 5.169273 | 5.307459 | 4.096177 | 4.225878 | 3.674934 | 5.219671 | 5.195400 | 5.139955 | 4.701110 | 4.108405 | 4.446023 | 5.289230 | 5.326481 | 5.356524 | 3.844676 | 4.066458 | 3.704143 | 3.862316 | 3.723575 | 3.959339 | 5.172299 | 5.178618 | 5.156816 | 5.304343 | 5.293869 | 5.324003 |
| GO:0060571 | morphogenesis of an epithelial fold | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.322489 | 4.899118 | 4.005645 | 5.038267 | 3.143350 | 3.210679 | 4.864783 | 5.038273 | 3.426060 | 3.569601 | 2.885632 | 4.927632 | 4.948479 | 4.817857 | 4.339835 | 3.552040 | 4.018737 | 5.004107 | 5.061777 | 5.048290 | 3.067936 | 3.384798 | 2.940366 | 3.129770 | 2.996010 | 3.465025 | 4.877994 | 4.858138 | 4.858124 | 5.020901 | 5.057419 | 5.036266 |
| GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | EMC3 | 1 | 6.347418 | 6.393076 | 6.328499 | 6.314187 | 6.296019 | 6.402076 | 6.323221 | 6.273736 | 6.362783 | 6.259390 | 6.415724 | 6.398470 | 6.416687 | 6.363569 | 6.257137 | 6.366761 | 6.359027 | 6.357352 | 6.285429 | 6.298762 | 6.281865 | 6.205758 | 6.394182 | 6.367062 | 6.450226 | 6.387634 | 6.363332 | 6.289719 | 6.315645 | 6.266644 | 6.302919 | 6.251156 |
| GO:0072077 | renal vesicle morphogenesis | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.056552 | 4.902164 | 4.324336 | 4.946776 | 3.974621 | 3.875965 | 4.852708 | 4.930740 | 4.083091 | 4.110842 | 3.972013 | 4.951501 | 4.940798 | 4.809931 | 4.506500 | 4.042536 | 4.384868 | 4.934744 | 4.936259 | 4.969064 | 3.942896 | 4.016554 | 3.963409 | 3.806471 | 3.830150 | 3.984702 | 4.874389 | 4.849745 | 4.833698 | 4.925829 | 4.939164 | 4.927190 |
| GO:0072530 | purine-containing compound transmembrane transport | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | SLC25A17 | 1 | 7.334416 | 6.876285 | 7.224422 | 6.928702 | 7.302094 | 7.255302 | 6.773800 | 6.936878 | 7.382610 | 7.434372 | 7.173230 | 6.957451 | 6.867393 | 6.799658 | 7.114997 | 7.186200 | 7.360861 | 7.019252 | 6.851025 | 6.910761 | 7.315392 | 7.361404 | 7.226229 | 7.350105 | 7.333013 | 7.065522 | 6.791294 | 6.769064 | 6.760870 | 6.923044 | 6.955512 | 6.931883 |
| GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1/42 | 24/20870 | 0.0472229488 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 6.050925 | 6.526198 | 6.390253 | 6.595356 | 5.894059 | 5.938846 | 6.512245 | 6.565230 | 6.140803 | 6.261614 | 5.689655 | 6.544052 | 6.530052 | 6.504208 | 6.576084 | 6.138749 | 6.422364 | 6.614854 | 6.548969 | 6.621142 | 5.853012 | 6.106415 | 5.692269 | 6.004148 | 5.848993 | 5.959013 | 6.531359 | 6.485509 | 6.519476 | 6.560244 | 6.567236 | 6.568197 |
| GO:0009259 | ribonucleotide metabolic process | 3/42 | 408/20870 | 0.0485907586 | 0.2820041 | 0.2468512 | PPT1||OGT||BPNT1 | 3 | 7.919737 | 7.388427 | 8.106259 | 7.638884 | 7.970475 | 8.132743 | 7.424156 | 7.571172 | 7.848015 | 7.894685 | 8.011555 | 7.223047 | 7.463410 | 7.465613 | 7.962857 | 8.230679 | 8.112806 | 7.612653 | 7.733855 | 7.564844 | 8.010166 | 7.907618 | 7.991584 | 8.092827 | 8.074938 | 8.225711 | 7.416148 | 7.431574 | 7.424704 | 7.611726 | 7.555722 | 7.545177 |
| GO:0007035 | vacuolar acidification | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | PPT1 | 1 | 5.659481 | 5.961256 | 5.974208 | 6.000876 | 5.709970 | 5.929753 | 5.814710 | 5.894791 | 5.612986 | 5.633290 | 5.729466 | 6.014694 | 5.987968 | 5.877468 | 5.886847 | 6.042394 | 5.989076 | 6.033907 | 6.016195 | 5.951220 | 5.713907 | 5.642282 | 5.770851 | 5.800589 | 5.871084 | 6.100306 | 5.852912 | 5.811683 | 5.778573 | 5.890995 | 5.945659 | 5.845990 |
| GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 4.182462 | 5.257771 | 4.879434 | 5.374684 | 4.081403 | 4.380957 | 5.199590 | 5.361063 | 4.210876 | 4.269704 | 4.058670 | 5.282283 | 5.304603 | 5.183573 | 5.080425 | 4.709979 | 4.822543 | 5.330448 | 5.403935 | 5.388631 | 3.992518 | 4.168673 | 4.077635 | 4.119806 | 4.136483 | 4.783084 | 5.232786 | 5.210340 | 5.154521 | 5.357564 | 5.384399 | 5.340892 |
| GO:0018196 | peptidyl-asparagine modification | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | MAGT1 | 1 | 6.563466 | 7.260196 | 6.878812 | 7.126894 | 6.585306 | 6.664095 | 7.125222 | 7.081463 | 6.559104 | 6.596706 | 6.533894 | 7.316208 | 7.270090 | 7.191547 | 6.878697 | 6.889096 | 6.868571 | 7.119194 | 7.166466 | 7.094084 | 6.600971 | 6.567868 | 6.586887 | 6.622576 | 6.487832 | 6.857394 | 7.143931 | 7.137609 | 7.093605 | 7.069377 | 7.118337 | 7.055924 |
| GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | SELENOK | 1 | 6.344581 | 6.750600 | 6.615334 | 6.718064 | 6.380143 | 6.519095 | 6.625619 | 6.659173 | 6.302951 | 6.305307 | 6.422232 | 6.784635 | 6.787697 | 6.676723 | 6.606354 | 6.616506 | 6.623092 | 6.711851 | 6.750612 | 6.691096 | 6.354220 | 6.333198 | 6.450290 | 6.420558 | 6.484730 | 6.642840 | 6.660838 | 6.628577 | 6.586481 | 6.653228 | 6.705025 | 6.617933 |
| GO:0033233 | regulation of protein sumoylation | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 5.635760 | 6.419616 | 6.086879 | 6.516595 | 5.495958 | 5.625570 | 6.459275 | 6.571507 | 5.688344 | 5.825571 | 5.353436 | 6.443268 | 6.409013 | 6.406273 | 6.273317 | 5.894603 | 6.067746 | 6.501693 | 6.494084 | 6.553286 | 5.436771 | 5.634912 | 5.405292 | 5.525115 | 5.490884 | 5.835202 | 6.473593 | 6.438171 | 6.465823 | 6.550171 | 6.587489 | 6.576608 |
| GO:0072087 | renal vesicle development | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | CTNNB1 | 1 | 3.969514 | 4.814975 | 4.237995 | 4.859635 | 3.887855 | 3.789980 | 4.765476 | 4.843547 | 3.995731 | 4.023754 | 3.885380 | 4.864444 | 4.853474 | 4.722746 | 4.420112 | 3.956334 | 4.298470 | 4.847517 | 4.848971 | 4.882150 | 3.856042 | 3.929875 | 3.876637 | 3.720590 | 3.743622 | 3.899116 | 4.787014 | 4.762634 | 4.746492 | 4.838785 | 4.851908 | 4.839912 |
| GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | TXNDC12 | 1 | 5.873179 | 6.577389 | 6.359894 | 6.540368 | 5.772846 | 5.844196 | 6.471804 | 6.461404 | 5.939119 | 6.048445 | 5.594002 | 6.608743 | 6.599819 | 6.522040 | 6.435934 | 6.226247 | 6.408625 | 6.546235 | 6.550960 | 6.523764 | 5.713128 | 5.933810 | 5.656488 | 5.692751 | 5.588606 | 6.179917 | 6.497522 | 6.471540 | 6.445888 | 6.457117 | 6.490295 | 6.436286 |
| GO:1903077 | negative regulation of protein localization to plasma membrane | 1/42 | 25/20870 | 0.0491425802 | 0.2820041 | 0.2468512 | RSC1A1 | 1 | 4.927336 | 5.707485 | 5.036068 | 5.711967 | 4.894942 | 5.025592 | 5.692290 | 5.765361 | 4.886543 | 4.969553 | 4.924714 | 5.707900 | 5.731086 | 5.683068 | 5.106005 | 4.969138 | 5.029797 | 5.687209 | 5.704717 | 5.743402 | 4.903275 | 4.897639 | 4.883844 | 5.043208 | 4.991985 | 5.041004 | 5.676750 | 5.723769 | 5.675828 | 5.750329 | 5.775910 | 5.769722 |
GO: MF
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0003735 | structural constituent of ribosome | 5/42 | 193/20678 | 4.322112e-05 | 0.006785716 | 0.005277526 | MRPS33||RPS15A||RPL9||MRPS23||MRPL46 | 5 | 10.9355568 | 10.688269 | 10.5388820 | 10.566723 | 10.9196846 | 10.5763827 | 10.663932 | 10.566342 | 10.9917019 | 10.9272864 | 10.8857001 | 10.750258 | 10.641796 | 10.670553 | 10.4755825 | 10.4948158 | 10.6405452 | 10.606965 | 10.536699 | 10.555584 | 10.9089765 | 11.0045888 | 10.8407820 | 10.7528062 | 10.6647803 | 10.2665451 | 10.653919 | 10.668985 | 10.668840 | 10.578059 | 10.496562 | 10.621625 |
| GO:0015035 | protein-disulfide reductase activity | 2/42 | 39/20678 | 2.845537e-03 | 0.173862098 | 0.135219600 | TXNDC12||SELENOT | 2 | 6.3591793 | 7.129436 | 6.5199649 | 6.998543 | 6.3159593 | 6.3114794 | 7.029613 | 6.943397 | 6.4044616 | 6.4445966 | 6.2185453 | 7.149729 | 7.146930 | 7.090888 | 6.6103900 | 6.4055875 | 6.5365115 | 6.961836 | 7.024432 | 7.008629 | 6.3301616 | 6.3913259 | 6.2212768 | 6.3414663 | 6.1806987 | 6.4032268 | 7.020204 | 7.030988 | 7.037593 | 6.937780 | 6.962004 | 6.930214 |
| GO:0015036 | disulfide oxidoreductase activity | 2/42 | 44/20678 | 3.609513e-03 | 0.173862098 | 0.135219600 | TXNDC12||SELENOT | 2 | 6.3576393 | 7.052684 | 6.5086874 | 6.930216 | 6.2959948 | 6.2409811 | 6.955551 | 6.880823 | 6.4224283 | 6.4515814 | 6.1842966 | 7.079035 | 7.067306 | 7.010792 | 6.5854670 | 6.4045529 | 6.5301328 | 6.891673 | 6.955352 | 6.942836 | 6.3267635 | 6.3993886 | 6.1506224 | 6.2809885 | 6.0747151 | 6.3529936 | 6.947774 | 6.956255 | 6.962584 | 6.875448 | 6.899622 | 6.867201 |
| GO:0016874 | ligase activity | 3/42 | 184/20678 | 6.167614e-03 | 0.173862098 | 0.135219600 | CPS1||YARS2||SUCLG1 | 3 | 7.2088914 | 7.076004 | 7.6262817 | 7.382555 | 7.2240138 | 7.5099627 | 7.117392 | 7.324684 | 7.1497183 | 7.2575240 | 7.2173807 | 6.923483 | 7.144060 | 7.149164 | 7.5855949 | 7.7213818 | 7.5668897 | 7.319226 | 7.496982 | 7.324265 | 7.2745739 | 7.1834315 | 7.2125285 | 7.4502449 | 7.3745850 | 7.6864345 | 7.100005 | 7.124210 | 7.127803 | 7.378091 | 7.303267 | 7.291150 |
| GO:0016597 | amino acid binding | 2/42 | 61/20678 | 6.831887e-03 | 0.173862098 | 0.135219600 | CPS1||YARS2 | 2 | 5.3532404 | 5.622106 | 5.4702382 | 5.577023 | 5.3191520 | 5.4999836 | 5.588206 | 5.559977 | 5.3274449 | 5.4226094 | 5.3070100 | 5.649059 | 5.626284 | 5.590370 | 5.5224862 | 5.4433929 | 5.4433810 | 5.584763 | 5.522128 | 5.622404 | 5.3508084 | 5.2956128 | 5.3104680 | 5.4896058 | 5.4098928 | 5.5944953 | 5.602792 | 5.566174 | 5.595392 | 5.552409 | 5.571974 | 5.555471 |
| GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors | 2/42 | 61/20678 | 6.831887e-03 | 0.173862098 | 0.135219600 | TXNDC12||SELENOT | 2 | 6.0362596 | 6.759133 | 6.1776892 | 6.675592 | 5.9882250 | 5.9438403 | 6.673392 | 6.637094 | 6.0913853 | 6.1267732 | 5.8783152 | 6.789111 | 6.769426 | 6.717929 | 6.2595853 | 6.0773312 | 6.1903157 | 6.639324 | 6.696002 | 6.690773 | 6.0053587 | 6.0891793 | 5.8609675 | 5.9732101 | 5.8006105 | 6.0467591 | 6.661295 | 6.678209 | 6.680596 | 6.632757 | 6.652385 | 6.626010 |
| GO:0016595 | glutamate binding | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | CPS1 | 1 | 4.8812511 | 5.296357 | 5.3460360 | 5.419536 | 4.8029466 | 5.0406410 | 5.294881 | 5.345292 | 4.8528529 | 5.1389030 | 4.6018981 | 5.277354 | 5.342872 | 5.267680 | 5.4672002 | 5.2396336 | 5.3220129 | 5.406858 | 5.364778 | 5.484411 | 4.9177338 | 4.8519293 | 4.6228591 | 4.9962355 | 4.5837323 | 5.4211101 | 5.315271 | 5.270023 | 5.298987 | 5.337181 | 5.345535 | 5.353115 |
| GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | CPS1 | 1 | 4.9987928 | 6.407354 | 5.3754951 | 6.069058 | 4.8944763 | 5.1392240 | 6.405037 | 6.127690 | 5.0509740 | 5.0727487 | 4.8636341 | 6.458865 | 6.359544 | 6.401928 | 5.5928092 | 5.2332673 | 5.2728348 | 6.001681 | 6.052074 | 6.149487 | 4.8225605 | 4.9705534 | 4.8864883 | 4.9873463 | 5.0133116 | 5.3822806 | 6.382541 | 6.399952 | 6.432178 | 6.102092 | 6.145393 | 6.135231 |
| GO:0045294 | alpha-catenin binding | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | CTNNB1 | 1 | 4.3613657 | 5.976656 | 5.0875183 | 6.100524 | 4.1521887 | 4.2589306 | 5.910110 | 6.094956 | 4.4342950 | 4.6308175 | 3.9324116 | 6.015496 | 6.020998 | 5.889689 | 5.4547244 | 4.6319118 | 5.0597610 | 6.054806 | 6.119992 | 6.125704 | 4.1068354 | 4.3874668 | 3.9241824 | 4.2019348 | 4.1354010 | 4.4234558 | 5.933336 | 5.915419 | 5.881088 | 6.090157 | 6.118847 | 6.075529 |
| GO:0098505 | G-rich strand telomeric DNA binding | 1/42 | 10/20678 | 2.013114e-02 | 0.173862098 | 0.135219600 | RPA2 | 1 | 7.8639006 | 8.865442 | 7.9019885 | 8.837516 | 7.7471186 | 7.9396794 | 8.933782 | 8.848420 | 7.8381828 | 8.0505253 | 7.6787717 | 8.841937 | 8.849433 | 8.904152 | 8.3449328 | 7.5448896 | 7.6842147 | 8.803391 | 8.817814 | 8.889848 | 7.6985875 | 7.8180269 | 7.7219444 | 7.9734512 | 7.9768184 | 7.8660404 | 8.919179 | 8.919930 | 8.961824 | 8.831142 | 8.862156 | 8.851788 |
| GO:0005347 | ATP transmembrane transporter activity | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | SLC25A17 | 1 | 8.0755980 | 7.506628 | 7.8718292 | 7.567456 | 8.0315895 | 7.9306649 | 7.424731 | 7.596554 | 8.1358878 | 8.1881644 | 7.8848889 | 7.600662 | 7.477620 | 7.436502 | 7.7598370 | 7.8062172 | 8.0342337 | 7.660309 | 7.473092 | 7.562887 | 8.0465041 | 8.1092822 | 7.9335155 | 8.0741904 | 8.0433280 | 7.6340786 | 7.431371 | 7.419349 | 7.423446 | 7.587832 | 7.595954 | 7.605820 |
| GO:0015166 | polyol transmembrane transporter activity | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | SLC5A3 | 1 | -0.5048209 | 1.484559 | 0.4680739 | 1.635668 | -0.4875053 | -0.1353992 | 1.598437 | 1.697323 | -0.5324684 | -0.3243855 | -0.6798407 | 1.409021 | 1.576807 | 1.462728 | 0.8923442 | 0.1181986 | 0.2728783 | 1.367223 | 1.709340 | 1.795805 | -0.4451347 | -0.5983061 | -0.4252156 | -0.4197511 | -0.2662565 | 0.2044235 | 1.687472 | 1.645932 | 1.451032 | 1.607331 | 1.712723 | 1.767354 |
| GO:0043047 | single-stranded telomeric DNA binding | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | RPA2 | 1 | 7.6069557 | 8.614827 | 7.6474848 | 8.588773 | 7.4900772 | 7.6836654 | 8.683166 | 8.597709 | 7.5815007 | 7.7934887 | 7.4216517 | 8.591013 | 8.598829 | 8.653823 | 8.0915533 | 7.2890544 | 7.4291412 | 8.553086 | 8.569484 | 8.642183 | 7.4415228 | 7.5607861 | 7.4651389 | 7.7163974 | 7.7208502 | 7.6110964 | 8.668486 | 8.669000 | 8.711585 | 8.580424 | 8.611864 | 8.600661 |
| GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity | 1/42 | 12/20678 | 2.410962e-02 | 0.173862098 | 0.135219600 | CCNT1 | 1 | 5.8361480 | 6.825090 | 5.9379185 | 6.927780 | 5.5786891 | 5.0748901 | 6.910883 | 7.098084 | 6.0036408 | 6.1199833 | 5.2355224 | 6.887575 | 6.775148 | 6.810243 | 6.1727634 | 5.5413501 | 6.0268154 | 6.901721 | 6.919429 | 6.961533 | 5.4733427 | 5.9940951 | 5.1379252 | 5.1409405 | 4.9646409 | 5.1129403 | 6.864738 | 6.937673 | 6.929141 | 7.109520 | 7.061855 | 7.122179 |
| GO:0047134 | protein-disulfide reductase (NAD(P)) activity | 1/42 | 13/20678 | 2.609295e-02 | 0.173862098 | 0.135219600 | SELENOT | 1 | 6.6165227 | 7.008156 | 6.8358345 | 7.257034 | 6.5215753 | 6.1913566 | 7.062189 | 7.255944 | 6.7439812 | 6.7786612 | 6.2740491 | 6.997719 | 6.996135 | 7.030355 | 6.9480716 | 6.6337374 | 6.9059639 | 7.224431 | 7.281470 | 7.264608 | 6.5442389 | 6.7653105 | 6.1998501 | 6.3523581 | 5.8262600 | 6.3360819 | 7.060565 | 7.049792 | 7.076087 | 7.252542 | 7.241333 | 7.273768 |
| GO:1990226 | histone methyltransferase binding | 1/42 | 13/20678 | 2.609295e-02 | 0.173862098 | 0.135219600 | CTNNB1 | 1 | 7.0177141 | 7.894986 | 7.4241274 | 8.123230 | 6.8271261 | 6.9438234 | 7.993981 | 8.174724 | 7.0044536 | 7.2266590 | 6.7888505 | 7.909919 | 7.875652 | 7.899175 | 7.8324877 | 7.0021715 | 7.3138525 | 8.077494 | 8.110965 | 8.179355 | 6.8250696 | 6.9694127 | 6.6715462 | 7.0360965 | 6.8839556 | 6.9067048 | 7.986720 | 7.989305 | 8.005844 | 8.176500 | 8.171574 | 8.176092 |
| GO:0010851 | cyclase regulator activity | 1/42 | 14/20678 | 2.807234e-02 | 0.173862098 | 0.135219600 | ADGRV1 | 1 | 7.8647313 | 8.219995 | 7.8423704 | 8.047909 | 7.8630468 | 8.0440130 | 8.230649 | 8.087167 | 7.8067909 | 7.9229723 | 7.8620888 | 8.215024 | 8.222578 | 8.222371 | 7.8885692 | 7.8251407 | 7.8122387 | 8.081145 | 7.989707 | 8.071143 | 7.8486425 | 7.8174715 | 7.9210602 | 8.0389069 | 8.1317237 | 7.9560504 | 8.220974 | 8.239486 | 8.231427 | 8.080660 | 8.097528 | 8.083256 |
| GO:0072582 | 17-beta-hydroxysteroid dehydrogenase (NADP+) activity | 1/42 | 14/20678 | 2.807234e-02 | 0.173862098 | 0.135219600 | HSD17B7 | 1 | 3.2097645 | 5.015721 | 3.2339704 | 4.905140 | 3.1609219 | 2.9868018 | 4.900907 | 4.897982 | 3.2475434 | 3.3143469 | 3.0550092 | 5.036789 | 5.018901 | 4.991106 | 3.6028599 | 2.9484481 | 3.0629221 | 4.791147 | 4.978592 | 4.938990 | 3.0714517 | 3.2631632 | 3.1415940 | 2.9426533 | 3.0877234 | 2.9244168 | 4.899167 | 4.922718 | 4.880526 | 4.920868 | 4.943932 | 4.826481 |
| GO:0008252 | nucleotidase activity | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | BPNT1 | 1 | 6.2899468 | 6.077989 | 6.2439033 | 6.065577 | 6.3297709 | 6.4343397 | 6.042705 | 6.026160 | 6.2419477 | 6.2360813 | 6.3867097 | 6.097056 | 6.087575 | 6.048886 | 6.1457245 | 6.2868000 | 6.2943817 | 6.062154 | 6.046764 | 6.087518 | 6.3431481 | 6.2781567 | 6.3665602 | 6.3981784 | 6.4879345 | 6.4153240 | 6.050713 | 6.062381 | 6.014592 | 6.007770 | 6.061502 | 6.008547 |
| GO:0015929 | hexosaminidase activity | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | OGA | 1 | 4.6931369 | 5.090033 | 5.3273920 | 5.119205 | 4.7382147 | 5.0766574 | 5.005393 | 5.198033 | 4.5867447 | 4.6959293 | 4.7896074 | 5.136267 | 5.106989 | 5.024528 | 5.3987605 | 5.3536432 | 5.2241282 | 5.063162 | 5.168656 | 5.123857 | 4.7341888 | 4.6221401 | 4.8493686 | 4.8643607 | 4.9025394 | 5.3991956 | 5.048130 | 4.995617 | 4.971361 | 5.188242 | 5.240697 | 5.164093 |
| GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | SELENOT | 1 | 6.5023694 | 7.187224 | 6.6864700 | 7.436011 | 6.4184528 | 6.0585265 | 7.238267 | 7.463466 | 6.6277915 | 6.6769535 | 6.1449031 | 7.198502 | 7.165145 | 7.197775 | 6.8526547 | 6.4535376 | 6.7249193 | 7.399112 | 7.455426 | 7.452800 | 6.4031454 | 6.6902675 | 6.1023356 | 6.2074153 | 5.7496326 | 6.1749237 | 7.218601 | 7.242082 | 7.253893 | 7.461428 | 7.447891 | 7.480887 |
| GO:0032977 | membrane insertase activity | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | EMC3 | 1 | 6.1601178 | 6.274879 | 6.1797816 | 6.241185 | 6.0757611 | 6.0803389 | 6.212230 | 6.186938 | 6.1793181 | 6.1484249 | 6.1524143 | 6.280881 | 6.281896 | 6.261771 | 6.1886570 | 6.1606494 | 6.1898497 | 6.264381 | 6.246935 | 6.211740 | 6.0687282 | 6.0159985 | 6.1398729 | 6.0435058 | 6.0893431 | 6.1074177 | 6.243151 | 6.193962 | 6.199068 | 6.175862 | 6.200969 | 6.183871 |
| GO:0070411 | I-SMAD binding | 1/42 | 15/20678 | 3.004780e-02 | 0.173862098 | 0.135219600 | CTNNB1 | 1 | 3.5045979 | 5.252527 | 4.2066234 | 5.366438 | 3.3334134 | 3.4024071 | 5.240125 | 5.405324 | 3.5782712 | 3.7583755 | 3.0996852 | 5.296251 | 5.272257 | 5.186793 | 4.5491324 | 3.7758363 | 4.1923907 | 5.334880 | 5.360564 | 5.403048 | 3.2099192 | 3.5550590 | 3.2067942 | 3.2826035 | 3.2435578 | 3.6460127 | 5.245093 | 5.239893 | 5.235373 | 5.393573 | 5.418427 | 5.403863 |
| GO:0044325 | transmembrane transporter binding | 2/42 | 136/20678 | 3.114688e-02 | 0.173862098 | 0.135219600 | CTNNB1||SLC5A3 | 2 | 5.9095468 | 6.305045 | 5.9339853 | 6.346192 | 5.8727004 | 5.9447286 | 6.310415 | 6.377671 | 5.9104316 | 5.9765472 | 5.8383531 | 6.318074 | 6.305527 | 6.291411 | 6.0455000 | 5.8433793 | 5.9055892 | 6.346504 | 6.302301 | 6.388484 | 5.8595494 | 5.8854946 | 5.8729406 | 5.9564980 | 5.9548215 | 5.9226144 | 6.311496 | 6.304978 | 6.314755 | 6.369875 | 6.390043 | 6.373014 |
| GO:0098847 | sequence-specific single stranded DNA binding | 1/42 | 16/20678 | 3.201934e-02 | 0.173862098 | 0.135219600 | RPA2 | 1 | 7.5404854 | 8.481106 | 7.5888917 | 8.445511 | 7.4396641 | 7.6414837 | 8.546066 | 8.462177 | 7.5114007 | 7.7135784 | 7.3763853 | 8.459732 | 8.464556 | 8.518293 | 7.9740187 | 7.2848387 | 7.4110943 | 8.417424 | 8.422236 | 8.495538 | 7.3961974 | 7.4930833 | 7.4280151 | 7.6752790 | 7.6657888 | 7.5815508 | 8.533020 | 8.532982 | 8.571848 | 8.445639 | 8.477838 | 8.462873 |
| GO:0043177 | organic acid binding | 2/42 | 145/20678 | 3.502018e-02 | 0.173862098 | 0.135219600 | CPS1||YARS2 | 2 | 4.8918083 | 5.296771 | 5.0266792 | 5.230447 | 4.8707182 | 5.0356702 | 5.264285 | 5.194759 | 4.8658025 | 4.9376520 | 4.8708497 | 5.306869 | 5.306377 | 5.276864 | 5.1001442 | 4.9907352 | 4.9862502 | 5.226418 | 5.190996 | 5.272759 | 4.8649033 | 4.8537192 | 4.8932440 | 4.9824659 | 4.9803163 | 5.1384604 | 5.276520 | 5.243545 | 5.272567 | 5.183201 | 5.216687 | 5.184136 |
| GO:0099106 | ion channel regulator activity | 2/42 | 145/20678 | 3.502018e-02 | 0.173862098 | 0.135219600 | SLC5A3||RSC1A1 | 2 | 6.4875454 | 6.249267 | 6.3109826 | 6.219883 | 6.5144603 | 6.5007302 | 6.237431 | 6.231194 | 6.4631031 | 6.4583886 | 6.5396920 | 6.282579 | 6.237837 | 6.226778 | 6.2088125 | 6.3404472 | 6.3782581 | 6.261097 | 6.159472 | 6.237136 | 6.5027427 | 6.5005876 | 6.5397143 | 6.5462467 | 6.5853781 | 6.3607291 | 6.235632 | 6.240891 | 6.235765 | 6.224458 | 6.235980 | 6.233117 |
| GO:0005402 | carbohydrate:cation symporter activity | 1/42 | 18/20678 | 3.595070e-02 | 0.173862098 | 0.135219600 | SLC5A3 | 1 | 1.2994216 | 2.917732 | 2.4149115 | 3.020926 | 1.3251257 | 2.0087806 | 2.869390 | 2.898371 | 1.2451291 | 1.4262759 | 1.2178431 | 2.862021 | 3.007256 | 2.879520 | 2.6857874 | 2.3028151 | 2.2110395 | 2.896213 | 3.100154 | 3.058464 | 1.3447988 | 1.3052264 | 1.3250804 | 1.6917520 | 1.6605450 | 2.5062493 | 2.869477 | 2.916095 | 2.821033 | 2.861849 | 2.949251 | 2.882560 |
| GO:0015095 | magnesium ion transmembrane transporter activity | 1/42 | 18/20678 | 3.595070e-02 | 0.173862098 | 0.135219600 | MAGT1 | 1 | 2.9029881 | 5.077355 | 3.7262729 | 5.014121 | 2.8304944 | 3.1503177 | 4.996710 | 4.988288 | 2.9365992 | 3.1598087 | 2.5474654 | 5.119175 | 5.078413 | 5.033197 | 4.1317413 | 3.4513792 | 3.4900582 | 4.907910 | 5.087220 | 5.041276 | 2.7603776 | 3.0223036 | 2.6868729 | 3.0166431 | 2.8866305 | 3.4786221 | 4.989658 | 5.031835 | 4.967904 | 4.974955 | 5.043993 | 4.944094 |
| GO:0016290 | palmitoyl-CoA hydrolase activity | 1/42 | 18/20678 | 3.595070e-02 | 0.173862098 | 0.135219600 | PPT1 | 1 | 4.8294795 | 5.120267 | 5.0602508 | 5.161528 | 4.7885047 | 4.8861227 | 5.041373 | 5.148558 | 4.8226492 | 4.8583722 | 4.8069345 | 5.222489 | 5.093093 | 5.039012 | 5.1292083 | 4.9594264 | 5.0867559 | 5.169699 | 5.132480 | 5.181945 | 4.8141912 | 4.7425184 | 4.8077206 | 4.8452219 | 4.8948662 | 4.9173389 | 5.047737 | 5.050166 | 5.026093 | 5.115872 | 5.181854 | 5.147193 |
| GO:0016247 | channel regulator activity | 2/42 | 151/20678 | 3.770134e-02 | 0.173862098 | 0.135219600 | SLC5A3||RSC1A1 | 2 | 6.4671635 | 6.225858 | 6.2942541 | 6.196939 | 6.4932338 | 6.4801780 | 6.211999 | 6.206853 | 6.4430616 | 6.4400957 | 6.5170096 | 6.257580 | 6.216457 | 6.202974 | 6.1927860 | 6.3238197 | 6.3608066 | 6.238343 | 6.137018 | 6.213534 | 6.4840260 | 6.4791308 | 6.5162617 | 6.5266167 | 6.5591258 | 6.3457646 | 6.210414 | 6.215494 | 6.210081 | 6.199957 | 6.211835 | 6.208741 |
| GO:0000295 | adenine nucleotide transmembrane transporter activity | 1/42 | 19/20678 | 3.791053e-02 | 0.173862098 | 0.135219600 | SLC25A17 | 1 | 7.3764301 | 6.933433 | 7.2594115 | 6.988879 | 7.3455838 | 7.3057568 | 6.834909 | 6.995078 | 7.4212799 | 7.4732935 | 7.2227886 | 7.011317 | 6.923123 | 6.861940 | 7.1447921 | 7.2274225 | 7.3946877 | 7.074890 | 6.909870 | 6.977090 | 7.3600612 | 7.3994461 | 7.2744268 | 7.4020830 | 7.3879628 | 7.1086807 | 6.851226 | 6.828845 | 6.824511 | 6.986724 | 7.007538 | 6.990886 |
| GO:0005346 | purine ribonucleotide transmembrane transporter activity | 1/42 | 19/20678 | 3.791053e-02 | 0.173862098 | 0.135219600 | SLC25A17 | 1 | 7.3764301 | 6.933433 | 7.2594115 | 6.988879 | 7.3455838 | 7.3057568 | 6.834909 | 6.995078 | 7.4212799 | 7.4732935 | 7.2227886 | 7.011317 | 6.923123 | 6.861940 | 7.1447921 | 7.2274225 | 7.3946877 | 7.074890 | 6.909870 | 6.977090 | 7.3600612 | 7.3994461 | 7.2744268 | 7.4020830 | 7.3879628 | 7.1086807 | 6.851226 | 6.828845 | 6.824511 | 6.986724 | 7.007538 | 6.990886 |
| GO:0140597 | protein carrier activity | 1/42 | 19/20678 | 3.791053e-02 | 0.173862098 | 0.135219600 | EMC3 | 1 | 6.7919224 | 7.575821 | 6.5858978 | 7.342040 | 6.5186937 | 6.2021494 | 7.549400 | 7.349123 | 6.9019202 | 7.0096735 | 6.3919145 | 7.672605 | 7.522322 | 7.527431 | 6.8090106 | 6.2677752 | 6.6294349 | 7.298090 | 7.334717 | 7.391766 | 6.4670455 | 6.7008853 | 6.3675886 | 6.2287711 | 6.1918672 | 6.1854298 | 7.550081 | 7.552320 | 7.545790 | 7.328164 | 7.353442 | 7.365512 |
| GO:0140101 | catalytic activity, acting on a tRNA | 2/42 | 155/20678 | 3.953147e-02 | 0.173862098 | 0.135219600 | METTL2A||YARS2 | 2 | 5.3059236 | 5.742666 | 5.4673500 | 5.797422 | 5.2499815 | 5.3858380 | 5.752520 | 5.808580 | 5.2915743 | 5.3557952 | 5.2689895 | 5.764949 | 5.738389 | 5.724366 | 5.6066706 | 5.3860845 | 5.3984818 | 5.777893 | 5.760628 | 5.852100 | 5.2427907 | 5.2367639 | 5.2701698 | 5.3275825 | 5.3503714 | 5.4751451 | 5.760563 | 5.739297 | 5.757608 | 5.796447 | 5.831111 | 5.797915 |
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 1/42 | 20/20678 | 3.986647e-02 | 0.173862098 | 0.135219600 | H2AZ1 | 1 | 7.3577173 | 7.693184 | 7.5989688 | 7.669032 | 7.2235780 | 6.9983145 | 7.730754 | 7.632202 | 7.4265789 | 7.6057834 | 6.9684120 | 7.692198 | 7.686174 | 7.701140 | 7.7372336 | 7.3198683 | 7.7037279 | 7.648572 | 7.641847 | 7.715522 | 7.1147122 | 7.4708932 | 7.0484470 | 6.9400906 | 6.8646516 | 7.1721325 | 7.717290 | 7.719457 | 7.755200 | 7.746157 | 7.390512 | 7.732582 |
| GO:0015216 | purine nucleotide transmembrane transporter activity | 1/42 | 22/20678 | 4.376671e-02 | 0.176382223 | 0.137179604 | SLC25A17 | 1 | 7.1355776 | 6.716521 | 7.0325027 | 6.773661 | 7.1044585 | 7.0705005 | 6.619053 | 6.778542 | 7.1801002 | 7.2332660 | 6.9813295 | 6.792894 | 6.707558 | 6.645295 | 6.9259988 | 6.9979301 | 7.1632264 | 6.855350 | 6.698049 | 6.763242 | 7.1190568 | 7.1578476 | 7.0336890 | 7.1634563 | 7.1483584 | 6.8827887 | 6.636201 | 6.613710 | 6.607086 | 6.768834 | 6.793324 | 6.773351 |
| GO:0001965 | G-protein alpha-subunit binding | 1/42 | 24/20678 | 4.765148e-02 | 0.176382223 | 0.137179604 | ADGRV1 | 1 | 4.5451885 | 4.952815 | 4.8597640 | 4.897709 | 4.5808287 | 4.8984120 | 4.906254 | 4.840233 | 4.4810233 | 4.4915913 | 4.6561804 | 4.967460 | 4.992218 | 4.897086 | 4.8675584 | 4.9021049 | 4.8080649 | 4.886210 | 4.890219 | 4.916510 | 4.5462766 | 4.4628392 | 4.7212073 | 4.7395118 | 4.8810353 | 5.0570912 | 4.937714 | 4.901734 | 4.878700 | 4.796255 | 4.888172 | 4.834795 |
| GO:0004303 | estradiol 17-beta-dehydrogenase activity | 1/42 | 24/20678 | 4.765148e-02 | 0.176382223 | 0.137179604 | HSD17B7 | 1 | 5.8497982 | 5.892582 | 5.7432939 | 5.874683 | 5.8971862 | 5.9146491 | 5.774762 | 5.854812 | 5.7894273 | 5.7458776 | 6.0009837 | 5.941890 | 5.917151 | 5.815626 | 5.6509454 | 5.7743352 | 5.8002325 | 5.887111 | 5.835413 | 5.900707 | 5.8917791 | 5.8089889 | 5.9853869 | 5.9041476 | 6.0380301 | 5.7911650 | 5.784950 | 5.782315 | 5.756855 | 5.857535 | 5.900151 | 5.805185 |
| GO:0047617 | acyl-CoA hydrolase activity | 1/42 | 24/20678 | 4.765148e-02 | 0.176382223 | 0.137179604 | PPT1 | 1 | 4.6798582 | 5.121637 | 4.9288249 | 5.173884 | 4.6389671 | 4.7444199 | 5.048887 | 5.174105 | 4.6731788 | 4.7060445 | 4.6599603 | 5.214851 | 5.099996 | 5.044816 | 5.0156183 | 4.8189959 | 4.9450289 | 5.157840 | 5.157699 | 5.205583 | 4.6598803 | 4.5939984 | 4.6619920 | 4.6987602 | 4.7518526 | 4.7814339 | 5.044858 | 5.062107 | 5.039600 | 5.138250 | 5.219373 | 5.163492 |
| GO:0019903 | protein phosphatase binding | 2/42 | 176/20678 | 4.967315e-02 | 0.176382223 | 0.137179604 | RPA2||CTNNB1 | 2 | 6.3758622 | 6.385268 | 6.3278158 | 6.306783 | 6.4005539 | 6.4628681 | 6.359854 | 6.320524 | 6.3363370 | 6.3500000 | 6.4390773 | 6.411108 | 6.386076 | 6.358132 | 6.2603575 | 6.3413652 | 6.3791828 | 6.327297 | 6.260612 | 6.331353 | 6.4032105 | 6.3656792 | 6.4320061 | 6.4921351 | 6.5286685 | 6.3625715 | 6.357471 | 6.364424 | 6.357658 | 6.310663 | 6.338630 | 6.312107 |
GO: CC
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0005840 | ribosome | 5/44 | 258/21916 | 0.0001623284 | 0.03068006 | 0.02563080 | MRPS33||RPS15A||RPL9||MRPS23||MRPL46 | 5 | 10.576229 | 10.351360 | 10.189649 | 10.233622 | 10.561440 | 10.239215 | 10.328566 | 10.235635 | 10.628922 | 10.567142 | 10.530915 | 10.411223 | 10.306409 | 10.334396 | 10.129156 | 10.148514 | 10.286146 | 10.272470 | 10.202213 | 10.225292 | 10.551391 | 10.640795 | 10.488041 | 10.408788 | 10.328489 | 9.937772 | 10.318230 | 10.333382 | 10.334031 | 10.246046 | 10.170486 | 10.287928 |
| GO:0044391 | ribosomal subunit | 4/44 | 206/21916 | 0.0007667866 | 0.04918786 | 0.04109262 | MRPS33||RPS15A||RPL9||MRPL46 | 4 | 10.812276 | 10.576037 | 10.421405 | 10.457971 | 10.797402 | 10.464087 | 10.553284 | 10.458505 | 10.866482 | 10.802817 | 10.765723 | 10.637400 | 10.529809 | 10.558743 | 10.359682 | 10.378633 | 10.520488 | 10.497208 | 10.427157 | 10.448652 | 10.786804 | 10.879599 | 10.721410 | 10.636650 | 10.552373 | 10.159785 | 10.542692 | 10.558128 | 10.558973 | 10.469626 | 10.390654 | 10.512595 |
| GO:0032993 | protein-DNA complex | 4/44 | 207/21916 | 0.0007807598 | 0.04918786 | 0.04109262 | RPA2||H2AZ1||CTNNB1||H2BC6 | 4 | 6.015182 | 6.696574 | 6.235620 | 6.698442 | 5.918405 | 5.934967 | 6.735271 | 6.759565 | 6.053975 | 6.150438 | 5.821641 | 6.715227 | 6.669990 | 6.704121 | 6.447156 | 6.022897 | 6.205256 | 6.673546 | 6.659692 | 6.760027 | 5.879047 | 6.016227 | 5.854627 | 5.897595 | 5.860118 | 6.040820 | 6.721054 | 6.719454 | 6.764844 | 6.760027 | 6.743777 | 6.774724 |
| GO:0000791 | euchromatin | 2/44 | 39/21916 | 0.0027844451 | 0.11286067 | 0.09428627 | H2AZ1||CTNNB1 | 2 | 6.780152 | 7.097828 | 6.975175 | 7.136847 | 6.680968 | 6.658912 | 7.116133 | 7.181746 | 6.799782 | 6.918012 | 6.605578 | 7.122914 | 7.086586 | 7.083651 | 7.034150 | 6.819581 | 7.059936 | 7.128353 | 7.110642 | 7.170880 | 6.597744 | 6.807509 | 6.628641 | 6.585276 | 6.649048 | 6.738304 | 7.107403 | 7.115653 | 7.125288 | 7.169972 | 7.195959 | 7.179188 |
| GO:0098798 | mitochondrial protein-containing complex | 4/44 | 302/21916 | 0.0031100405 | 0.11286067 | 0.09428627 | MRPS33||ATP5MK||NDUFA4||MRPL46 | 4 | 8.765320 | 8.343565 | 9.099184 | 8.588167 | 8.819336 | 9.024468 | 8.381052 | 8.489704 | 8.726756 | 8.752249 | 8.815501 | 8.112028 | 8.442274 | 8.451196 | 9.017934 | 9.205907 | 9.067064 | 8.536758 | 8.741074 | 8.472880 | 8.848926 | 8.756129 | 8.850935 | 8.926991 | 8.919554 | 9.207761 | 8.376009 | 8.394005 | 8.373051 | 8.558383 | 8.444859 | 8.463278 |
| GO:0140534 | endoplasmic reticulum protein-containing complex | 3/44 | 153/21916 | 0.0035828784 | 0.11286067 | 0.09428627 | MAGT1||EMC3||NBAS | 3 | 6.827336 | 7.484077 | 7.052437 | 7.263255 | 6.839109 | 6.958832 | 7.349913 | 7.211815 | 6.823243 | 6.849803 | 6.808659 | 7.508672 | 7.515868 | 7.425974 | 7.038785 | 7.068445 | 7.049924 | 7.245957 | 7.297556 | 7.245629 | 6.840203 | 6.826268 | 6.850754 | 6.911243 | 6.885818 | 7.072265 | 7.367800 | 7.352787 | 7.328886 | 7.214265 | 7.245824 | 7.174470 |
| GO:0030117 | membrane coat | 2/44 | 93/21916 | 0.0150113810 | 0.19942106 | 0.16660072 | AP3M2||SAR1A | 2 | 5.761155 | 5.810442 | 5.844522 | 5.805784 | 5.765694 | 5.908312 | 5.800548 | 5.834002 | 5.727401 | 5.752139 | 5.802895 | 5.823064 | 5.815556 | 5.792532 | 5.823519 | 5.858505 | 5.851305 | 5.833517 | 5.743221 | 5.838632 | 5.771333 | 5.725413 | 5.799373 | 5.914741 | 5.912357 | 5.897780 | 5.803623 | 5.800019 | 5.797995 | 5.827896 | 5.843251 | 5.830813 |
| GO:0048475 | coated membrane | 2/44 | 93/21916 | 0.0150113810 | 0.19942106 | 0.16660072 | AP3M2||SAR1A | 2 | 5.761155 | 5.810442 | 5.844522 | 5.805784 | 5.765694 | 5.908312 | 5.800548 | 5.834002 | 5.727401 | 5.752139 | 5.802895 | 5.823064 | 5.815556 | 5.792532 | 5.823519 | 5.858505 | 5.851305 | 5.833517 | 5.743221 | 5.838632 | 5.771333 | 5.725413 | 5.799373 | 5.914741 | 5.912357 | 5.897780 | 5.803623 | 5.800019 | 5.797995 | 5.827896 | 5.843251 | 5.830813 |
| GO:0000313 | organellar ribosome | 2/44 | 94/21916 | 0.0153183474 | 0.19942106 | 0.16660072 | MRPS33||MRPL46 | 2 | 7.099385 | 6.815556 | 6.885067 | 6.783040 | 7.093419 | 6.953522 | 6.780040 | 6.771455 | 7.114499 | 7.048911 | 7.133391 | 6.880959 | 6.768539 | 6.794757 | 6.838523 | 6.868269 | 6.946253 | 6.824405 | 6.715129 | 6.807210 | 7.074105 | 7.068853 | 7.136317 | 6.975376 | 7.060211 | 6.814259 | 6.799303 | 6.747573 | 6.792696 | 6.769105 | 6.767175 | 6.778061 |
| GO:0005761 | mitochondrial ribosome | 2/44 | 94/21916 | 0.0153183474 | 0.19942106 | 0.16660072 | MRPS33||MRPL46 | 2 | 7.099385 | 6.815556 | 6.885067 | 6.783040 | 7.093419 | 6.953522 | 6.780040 | 6.771455 | 7.114499 | 7.048911 | 7.133391 | 6.880959 | 6.768539 | 6.794757 | 6.838523 | 6.868269 | 6.946253 | 6.824405 | 6.715129 | 6.807210 | 7.074105 | 7.068853 | 7.136317 | 6.975376 | 7.060211 | 6.814259 | 6.799303 | 6.747573 | 6.792696 | 6.769105 | 6.767175 | 6.778061 |
| GO:0015935 | small ribosomal subunit | 2/44 | 95/21916 | 0.0156280174 | 0.19942106 | 0.16660072 | MRPS33||RPS15A | 2 | 11.247650 | 10.971873 | 10.846813 | 10.843030 | 11.256433 | 10.926938 | 10.939687 | 10.867337 | 11.287183 | 11.214631 | 11.240195 | 11.044729 | 10.919936 | 10.947961 | 10.755784 | 10.825224 | 10.952492 | 10.878744 | 10.812689 | 10.836881 | 11.254790 | 11.306368 | 11.206408 | 11.121602 | 11.011767 | 10.595932 | 10.931530 | 10.943029 | 10.944468 | 10.861499 | 10.838702 | 10.901116 |
| GO:0005916 | fascia adherens | 1/44 | 10/21916 | 0.0199002911 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.826774 | 6.310780 | 5.413358 | 6.459454 | 4.649234 | 4.818287 | 6.248959 | 6.433961 | 4.850790 | 5.069136 | 4.505361 | 6.334125 | 6.356689 | 6.238832 | 5.721487 | 5.013152 | 5.419331 | 6.409362 | 6.485094 | 6.482628 | 4.657657 | 4.795657 | 4.476765 | 4.804468 | 4.740777 | 4.904854 | 6.270992 | 6.254284 | 6.221157 | 6.441467 | 6.445566 | 6.414654 |
| GO:0000786 | nucleosome | 2/44 | 112/21916 | 0.0212949672 | 0.19942106 | 0.16660072 | H2AZ1||H2BC6 | 2 | 6.040878 | 6.271642 | 6.244724 | 6.193609 | 5.926593 | 5.771782 | 6.283362 | 6.217637 | 6.100219 | 6.237862 | 5.739915 | 6.280463 | 6.256044 | 6.278292 | 6.355001 | 6.018191 | 6.336768 | 6.184069 | 6.161119 | 6.234655 | 5.857554 | 6.091711 | 5.814849 | 5.712095 | 5.666674 | 5.923370 | 6.272912 | 6.267285 | 6.309524 | 6.260776 | 6.157674 | 6.232503 |
| GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex | 1/44 | 11/21916 | 0.0218689019 | 0.19942106 | 0.16660072 | CCNT1 | 1 | 4.287652 | 5.674508 | 4.452047 | 5.779841 | 4.178630 | 4.468446 | 5.788796 | 5.863780 | 4.267631 | 4.375107 | 4.215606 | 5.603559 | 5.691865 | 5.725372 | 4.886521 | 4.254137 | 4.087607 | 5.690652 | 5.762393 | 5.880105 | 4.107616 | 4.172607 | 4.252035 | 4.430845 | 4.514323 | 4.458915 | 5.772502 | 5.808846 | 5.784805 | 5.855136 | 5.897150 | 5.838418 |
| GO:0030877 | beta-catenin destruction complex | 1/44 | 11/21916 | 0.0218689019 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.895837 | 6.228811 | 5.343709 | 6.266643 | 4.722139 | 4.893520 | 6.246604 | 6.302032 | 4.946107 | 5.052677 | 4.660859 | 6.278170 | 6.208876 | 6.198072 | 5.650546 | 5.079174 | 5.239849 | 6.220144 | 6.256106 | 6.321831 | 4.656165 | 4.798321 | 4.708334 | 4.752488 | 4.927941 | 4.989760 | 6.250851 | 6.250119 | 6.238811 | 6.292846 | 6.304294 | 6.308909 |
| GO:0072546 | EMC complex | 1/44 | 11/21916 | 0.0218689019 | 0.19942106 | 0.16660072 | EMC3 | 1 | 6.021302 | 6.326249 | 6.134075 | 6.278797 | 5.877234 | 5.954097 | 6.249406 | 6.234483 | 6.061417 | 6.021047 | 5.980301 | 6.342157 | 6.334987 | 6.301274 | 6.128752 | 6.120721 | 6.152562 | 6.296557 | 6.287626 | 6.251822 | 5.876958 | 5.823458 | 5.929344 | 5.871173 | 5.948511 | 6.037782 | 6.283002 | 6.223818 | 6.240753 | 6.221421 | 6.259323 | 6.222382 |
| GO:0015934 | large ribosomal subunit | 2/44 | 115/21916 | 0.0223715870 | 0.19942106 | 0.16660072 | RPL9||MRPL46 | 2 | 10.413003 | 10.249118 | 10.037763 | 10.148118 | 10.371702 | 10.038477 | 10.239487 | 10.126910 | 10.483518 | 10.428245 | 10.322643 | 10.297769 | 10.208307 | 10.239847 | 10.015651 | 9.969323 | 10.123925 | 10.188113 | 10.116594 | 10.138716 | 10.350078 | 10.489145 | 10.267107 | 10.183035 | 10.128090 | 9.770569 | 10.225378 | 10.245298 | 10.247681 | 10.154486 | 10.019318 | 10.200820 |
| GO:0022626 | cytosolic ribosome | 2/44 | 118/21916 | 0.0234704147 | 0.19942106 | 0.16660072 | RPS15A||RPL9 | 2 | 11.596661 | 11.350879 | 11.194057 | 11.226321 | 11.580314 | 11.230018 | 11.326906 | 11.226164 | 11.654105 | 11.589969 | 11.543776 | 11.412754 | 11.304510 | 11.333180 | 11.130545 | 11.148735 | 11.297045 | 11.266692 | 11.197505 | 11.213857 | 11.570006 | 11.668751 | 11.497024 | 11.412019 | 11.317549 | 10.913478 | 11.316001 | 11.333187 | 11.331468 | 11.238319 | 11.154159 | 11.283058 |
| GO:0044815 | DNA packaging complex | 2/44 | 120/21916 | 0.0242151594 | 0.19942106 | 0.16660072 | H2AZ1||H2BC6 | 2 | 5.954709 | 6.263016 | 6.176568 | 6.211908 | 5.844387 | 5.740177 | 6.285084 | 6.262950 | 6.004341 | 6.142993 | 5.678208 | 6.269988 | 6.249891 | 6.269078 | 6.302815 | 5.955413 | 6.247880 | 6.185480 | 6.185617 | 6.263224 | 5.781094 | 5.992624 | 5.746978 | 5.680456 | 5.644594 | 5.883774 | 6.275823 | 6.270935 | 6.308210 | 6.297365 | 6.212771 | 6.277364 |
| GO:0008250 | oligosaccharyltransferase complex | 1/44 | 13/21916 | 0.0257945375 | 0.19942106 | 0.16660072 | MAGT1 | 1 | 7.768737 | 8.215595 | 8.004530 | 8.102704 | 7.816173 | 7.910804 | 8.091067 | 8.047128 | 7.764994 | 7.765858 | 7.775335 | 8.264308 | 8.230184 | 8.149913 | 7.948329 | 8.048218 | 8.015255 | 8.115981 | 8.142252 | 8.048255 | 7.818305 | 7.785255 | 7.844351 | 7.853867 | 7.816592 | 8.050813 | 8.110839 | 8.106191 | 8.055522 | 8.048960 | 8.072909 | 8.019010 |
| GO:1990907 | beta-catenin-TCF complex | 1/44 | 13/21916 | 0.0257945375 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.394227 | 5.398809 | 5.047398 | 5.554694 | 4.312486 | 4.679961 | 5.388580 | 5.593529 | 4.398681 | 4.538550 | 4.228852 | 5.423650 | 5.437294 | 5.333285 | 5.228209 | 4.861808 | 5.028797 | 5.537110 | 5.562015 | 5.564796 | 4.272584 | 4.352029 | 4.311750 | 4.513853 | 4.567207 | 4.923551 | 5.404935 | 5.382457 | 5.378204 | 5.592157 | 5.605686 | 5.582650 |
| GO:0043601 | nuclear replisome | 1/44 | 14/21916 | 0.0277515772 | 0.19942106 | 0.16660072 | RPA2 | 1 | 6.151814 | 6.337963 | 6.366203 | 6.382033 | 6.078637 | 6.134530 | 6.348359 | 6.391377 | 6.170154 | 6.197162 | 6.085802 | 6.363714 | 6.328202 | 6.321616 | 6.434868 | 6.262834 | 6.395334 | 6.401848 | 6.321022 | 6.421283 | 6.069814 | 6.112455 | 6.052987 | 6.089844 | 6.090340 | 6.219499 | 6.370320 | 6.328926 | 6.345531 | 6.376895 | 6.419214 | 6.377613 |
| GO:0045239 | tricarboxylic acid cycle enzyme complex | 1/44 | 14/21916 | 0.0277515772 | 0.19942106 | 0.16660072 | SUCLG1 | 1 | 5.013463 | 5.741154 | 5.114022 | 5.807362 | 4.931741 | 4.965087 | 5.744561 | 5.843109 | 5.053540 | 5.144023 | 4.824297 | 5.765024 | 5.739200 | 5.718866 | 5.310430 | 4.969059 | 5.039648 | 5.779743 | 5.778671 | 5.862075 | 4.938964 | 4.972808 | 4.881994 | 4.952271 | 4.978319 | 4.964551 | 5.746713 | 5.736804 | 5.750133 | 5.821242 | 5.882646 | 5.824610 |
| GO:0005779 | integral component of peroxisomal membrane | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | SLC25A17 | 1 | 4.724407 | 5.052226 | 4.704894 | 4.914699 | 4.740645 | 4.685002 | 5.015593 | 4.937243 | 4.718907 | 4.669853 | 4.782258 | 5.114933 | 5.052450 | 4.986434 | 4.722691 | 4.663814 | 4.727311 | 4.921234 | 4.887814 | 4.934645 | 4.666373 | 4.735712 | 4.815963 | 4.666586 | 4.786367 | 4.595576 | 5.028921 | 5.012309 | 5.005448 | 4.926072 | 4.985441 | 4.898853 |
| GO:0030127 | COPII vesicle coat | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | SAR1A | 1 | 5.666499 | 6.040382 | 5.869626 | 6.009600 | 5.657401 | 5.724072 | 6.026746 | 6.061777 | 5.641247 | 5.783271 | 5.566517 | 6.069129 | 6.031945 | 6.019610 | 5.928871 | 5.788729 | 5.887704 | 6.026393 | 5.948851 | 6.051579 | 5.640895 | 5.716349 | 5.612965 | 5.728088 | 5.627706 | 5.810619 | 6.034496 | 6.031318 | 6.014342 | 6.045998 | 6.084367 | 6.054684 |
| GO:0031231 | intrinsic component of peroxisomal membrane | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | SLC25A17 | 1 | 4.724407 | 5.052226 | 4.704894 | 4.914699 | 4.740645 | 4.685002 | 5.015593 | 4.937243 | 4.718907 | 4.669853 | 4.782258 | 5.114933 | 5.052450 | 4.986434 | 4.722691 | 4.663814 | 4.727311 | 4.921234 | 4.887814 | 4.934645 | 4.666373 | 4.735712 | 4.815963 | 4.666586 | 4.786367 | 4.595576 | 5.028921 | 5.012309 | 5.005448 | 4.926072 | 4.985441 | 4.898853 |
| GO:0099092 | postsynaptic density, intracellular component | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 5.091148 | 5.580313 | 5.289675 | 5.605286 | 5.062143 | 5.242641 | 5.561879 | 5.609713 | 5.048861 | 5.125067 | 5.098481 | 5.612454 | 5.594366 | 5.532921 | 5.361116 | 5.214170 | 5.289999 | 5.625786 | 5.559093 | 5.629890 | 5.080583 | 4.996744 | 5.106826 | 5.216813 | 5.280042 | 5.230297 | 5.597099 | 5.544250 | 5.543634 | 5.585611 | 5.650397 | 5.592247 |
| GO:1990909 | Wnt signalosome | 1/44 | 15/21916 | 0.0297047747 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 3.996449 | 5.384103 | 4.747863 | 5.545616 | 3.884693 | 4.215555 | 5.331897 | 5.513422 | 4.031314 | 4.151408 | 3.782458 | 5.438268 | 5.421924 | 5.287418 | 4.987654 | 4.446171 | 4.759185 | 5.539094 | 5.544796 | 5.552925 | 3.806818 | 4.000118 | 3.839638 | 4.056167 | 4.160408 | 4.407243 | 5.366752 | 5.315318 | 5.312979 | 5.501291 | 5.538381 | 5.500266 |
| GO:0030894 | replisome | 1/44 | 16/21916 | 0.0316541373 | 0.19942106 | 0.16660072 | RPA2 | 1 | 6.189163 | 6.798801 | 6.347049 | 6.796936 | 6.136233 | 6.153395 | 6.825482 | 6.867803 | 6.226262 | 6.222838 | 6.115672 | 6.817963 | 6.784683 | 6.793553 | 6.500585 | 6.201689 | 6.323126 | 6.785018 | 6.747617 | 6.856062 | 6.085765 | 6.192555 | 6.128371 | 6.117092 | 6.154785 | 6.187451 | 6.829685 | 6.821796 | 6.824954 | 6.836286 | 6.908224 | 6.857951 |
| GO:0098831 | presynaptic active zone cytoplasmic component | 1/44 | 16/21916 | 0.0316541373 | 0.19942106 | 0.16660072 | CTNNB1 | 1 | 4.250594 | 5.269332 | 4.746473 | 5.330539 | 4.128772 | 4.304294 | 5.229451 | 5.326176 | 4.282238 | 4.389956 | 4.060263 | 5.307818 | 5.301699 | 5.195749 | 4.935896 | 4.547610 | 4.729722 | 5.317880 | 5.315665 | 5.357683 | 4.114996 | 4.222738 | 4.042881 | 4.260011 | 4.147506 | 4.484709 | 5.239928 | 5.227754 | 5.220604 | 5.308989 | 5.341887 | 5.327464 |
| GO:0016600 | flotillin complex | 1/44 | 17/21916 | 0.0335996723 | 0.20484962 | 0.17113585 | CTNNB1 | 1 | 5.194723 | 6.253135 | 5.624109 | 6.295464 | 5.126002 | 5.164974 | 6.217457 | 6.280703 | 5.220597 | 5.355720 | 4.983564 | 6.335291 | 6.252033 | 6.167187 | 5.883739 | 5.277872 | 5.647286 | 6.282614 | 6.279140 | 6.324201 | 5.075085 | 5.243121 | 5.052171 | 5.155261 | 5.123861 | 5.214335 | 6.230139 | 6.198447 | 6.223591 | 6.275974 | 6.304235 | 6.261573 |
| GO:0099091 | postsynaptic specialization, intracellular component | 1/44 | 19/21916 | 0.0374792893 | 0.21909206 | 0.18303430 | CTNNB1 | 1 | 4.871904 | 5.402821 | 5.070452 | 5.418462 | 4.845346 | 5.026925 | 5.389288 | 5.420336 | 4.826541 | 4.908510 | 4.879466 | 5.428750 | 5.420661 | 5.358018 | 5.144103 | 4.998267 | 5.065288 | 5.430681 | 5.371050 | 5.452431 | 4.862551 | 4.776792 | 4.894154 | 4.999405 | 5.062876 | 5.017754 | 5.422460 | 5.372637 | 5.372186 | 5.397465 | 5.459088 | 5.403652 |
| GO:0005751 | mitochondrial respiratory chain complex IV | 1/44 | 20/21916 | 0.0394133858 | 0.21909206 | 0.18303430 | NDUFA4 | 1 | 10.314775 | 10.070006 | 10.879264 | 10.286836 | 10.402130 | 10.642883 | 10.135636 | 10.166958 | 10.251250 | 10.315705 | 10.374729 | 9.739911 | 10.202301 | 10.218588 | 10.802131 | 11.014870 | 10.810576 | 10.217302 | 10.478746 | 10.142473 | 10.437682 | 10.334396 | 10.431995 | 10.483636 | 10.426439 | 10.957664 | 10.129553 | 10.156014 | 10.121109 | 10.236378 | 10.111608 | 10.150043 |
| GO:0005753 | mitochondrial proton-transporting ATP synthase complex | 1/44 | 20/21916 | 0.0394133858 | 0.21909206 | 0.18303430 | ATP5MK | 1 | 10.107546 | 9.553595 | 10.485020 | 10.004835 | 10.161447 | 10.417021 | 9.608785 | 9.913688 | 10.037012 | 10.130284 | 10.152751 | 9.318820 | 9.646658 | 9.669360 | 10.384532 | 10.616451 | 10.443903 | 9.935202 | 10.168951 | 9.894915 | 10.215475 | 10.105944 | 10.160843 | 10.371383 | 10.309670 | 10.558254 | 9.586984 | 9.621127 | 9.617997 | 9.995534 | 9.869986 | 9.871923 |
| GO:0045259 | proton-transporting ATP synthase complex | 1/44 | 21/21916 | 0.0413436841 | 0.22325589 | 0.18651286 | ATP5MK | 1 | 10.030221 | 9.477651 | 10.407600 | 9.927974 | 10.084032 | 10.339422 | 9.532764 | 9.836772 | 9.959717 | 10.053019 | 10.075338 | 9.243378 | 9.570723 | 9.593014 | 10.307318 | 10.538912 | 10.366422 | 9.858372 | 10.092116 | 9.817990 | 10.137934 | 10.028672 | 10.083422 | 10.293769 | 10.232011 | 10.480719 | 9.510924 | 9.545093 | 9.542028 | 9.918618 | 9.793010 | 9.795068 |
| GO:0016327 | apicolateral plasma membrane | 1/44 | 22/21916 | 0.0432701914 | 0.22477529 | 0.18778220 | CTNNB1 | 1 | 3.786309 | 5.161394 | 4.342563 | 5.273774 | 3.652876 | 3.702798 | 5.093457 | 5.239956 | 3.839199 | 3.924031 | 3.572892 | 5.194496 | 5.215737 | 5.069675 | 4.635304 | 3.981601 | 4.337344 | 5.233092 | 5.297914 | 5.289458 | 3.581688 | 3.798653 | 3.566398 | 3.625851 | 3.603800 | 3.864049 | 5.113433 | 5.090421 | 5.076273 | 5.222333 | 5.276733 | 5.220086 |
| GO:0098800 | inner mitochondrial membrane protein complex | 2/44 | 168/21916 | 0.0447944167 | 0.22477529 | 0.18778220 | ATP5MK||NDUFA4 | 2 | 9.437482 | 8.978214 | 9.846205 | 9.270005 | 9.504751 | 9.762805 | 9.028746 | 9.153827 | 9.387496 | 9.422663 | 9.499983 | 8.678718 | 9.103199 | 9.111735 | 9.758726 | 9.969995 | 9.801131 | 9.203665 | 9.456050 | 9.129658 | 9.541797 | 9.427655 | 9.541818 | 9.645986 | 9.634636 | 9.980395 | 9.019705 | 9.049535 | 9.016769 | 9.235849 | 9.101725 | 9.120211 |
| GO:0031307 | integral component of mitochondrial outer membrane | 1/44 | 23/21916 | 0.0451929150 | 0.22477529 | 0.18778220 | FUNDC2 | 1 | 5.746309 | 6.029610 | 5.710113 | 5.846095 | 5.750251 | 5.769529 | 5.994107 | 5.874766 | 5.726248 | 5.701205 | 5.809245 | 6.018607 | 6.054383 | 6.015515 | 5.745791 | 5.696369 | 5.687493 | 5.851106 | 5.799906 | 5.885976 | 5.720699 | 5.711620 | 5.816095 | 5.724515 | 5.872407 | 5.705826 | 6.028575 | 5.966725 | 5.986328 | 5.854909 | 5.913791 | 5.854791 |
| GO:0031306 | intrinsic component of mitochondrial outer membrane | 1/44 | 24/21916 | 0.0471118623 | 0.22831133 | 0.19073628 | FUNDC2 | 1 | 5.771639 | 6.042729 | 5.768754 | 5.874368 | 5.775996 | 5.816203 | 6.011752 | 5.900602 | 5.748979 | 5.737767 | 5.826542 | 6.028325 | 6.070324 | 6.029136 | 5.800799 | 5.762210 | 5.742644 | 5.881108 | 5.829611 | 5.911208 | 5.746120 | 5.735739 | 5.843654 | 5.769861 | 5.895530 | 5.779806 | 6.042954 | 5.990956 | 6.000814 | 5.882968 | 5.937615 | 5.880499 |
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
ORA: Up-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Up-regulated DEGs” results.
GO: BP
No significcant functional terms were enriched under the threshold of P<0.05
GO: MF
No significcant functional terms were enriched under the threshold of P<0.05
GO: CC
No significcant functional terms were enriched under the threshold of P<0.05
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
Help
still working on
2-2. Summary Plot
GSEA
GO: BP
GO: MF
GO: CC
KEGG
ORA: All DEGs
GO: BP
GO: MF
GO: CC
KEGG
No results under this category
ORA: Down-regulated DEGs
GO: BP
GO: MF
GO: CC
KEGG
No results under this category
ORA: Up-regulated DEGs
GO: BP
No results under this category
GO: MF
No results under this category
GO: CC
No results under this category
KEGG
No results under this category
ORA: Overlapped Terms
GO: BP
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | GO:0010457||GO:0032786||GO:0000184||GO:0048489||GO:0031062||GO:0009101||GO:0051650||GO:0050982||GO:0097479||GO:0051648||GO:0032784||GO:0051568||GO:0009100||GO:0070059||GO:0031016||GO:0044242||GO:0006486||GO:0043413||GO:0031060||GO:1901264||GO:0015931||GO:0006493||GO:0070085||GO:1903312||GO:0006354||GO:0031058||GO:0000050||GO:0002084||GO:0007440||GO:0044848||GO:0048263||GO:0060272||GO:0060439||GO:0090037||GO:0044262||GO:1903321||GO:0003263||GO:0003264||GO:0033234||GO:0042989||GO:0042996||GO:0045050||GO:0048262||GO:0060742||GO:0060768||GO:0070601||GO:0044344||GO:0019751||GO:0003306||GO:0006020||GO:0007042||GO:0009950||GO:0019627||GO:0035112||GO:0048715||GO:0060123||GO:0060767||GO:0060788||GO:0071377||GO:0071697||GO:0071941||GO:0071774||GO:0007494||GO:0010739||GO:0015791||GO:0021781||GO:0032429||GO:0032968||GO:0048548||GO:0048820||GO:0061046||GO:0061085||GO:0071696||GO:0072182||GO:0072497||GO:1900364||GO:1990403||GO:1904659||GO:0048193||GO:0000052||GO:0006999||GO:0031441||GO:0034333||GO:0048207||GO:0048208||GO:0050667||GO:0070863||GO:0072498||GO:0008645||GO:0034968||GO:0015749||GO:0002679||GO:0030252||GO:0032239||GO:0042159||GO:0072160||GO:0090110||GO:0034219||GO:0043467||GO:0009855||GO:0043414||GO:0000956||GO:0009799||GO:0003337||GO:0006206||GO:0050910||GO:0060572||GO:0060628||GO:0061548||GO:0097091||GO:1904953||GO:0007098||GO:0009581||GO:0033865||GO:0033875||GO:0034032||GO:0007625||GO:0035751||GO:0043981||GO:0043982||GO:0000723||GO:0009582||GO:0048565||GO:0016042||GO:0032355||GO:0042157||GO:0018022||GO:0006703||GO:0006901||GO:0043923||GO:0046855||GO:0070365||GO:0080182||GO:0090036||GO:1903830||GO:2000136||GO:0006790||GO:0006044||GO:0015693||GO:0015867||GO:0045603||GO:0046838||GO:0060438||GO:0061323||GO:1903204||GO:2000696||GO:0031023||GO:0055123||GO:0001711||GO:0043586||GO:0046112||GO:0048490||GO:0071545||GO:0099514||GO:0099517||GO:1901679||GO:1904948||GO:0016571||GO:0006066||GO:0007194||GO:0010738||GO:0034035||GO:0046931||GO:0048485||GO:0050427||GO:0072283||GO:0002053||GO:0002089||GO:0007063||GO:0033762||GO:0035020||GO:0043576||GO:0043984||GO:0048199||GO:0071639||GO:0072079||GO:0009150||GO:0032259||GO:0002181||GO:0001759||GO:0002052||GO:0006907||GO:0021819||GO:0060231||GO:0008643||GO:0031056||GO:0032200||GO:0001702||GO:0018279||GO:0030539||GO:0031280||GO:0035994||GO:0051571||GO:0060479||GO:0060571||GO:0071816||GO:0072077||GO:0072530||GO:2000679||GO:0009259||GO:0007035||GO:0010560||GO:0018196||GO:0032469||GO:0033233||GO:0072087||GO:1902236||GO:1903077 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: MF
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | GO:0003735||GO:0015035||GO:0015036||GO:0016874||GO:0016597||GO:0016667||GO:0016595||GO:0016884||GO:0045294||GO:0098505||GO:0005347||GO:0015166||GO:0043047||GO:0061575||GO:0047134||GO:1990226||GO:0010851||GO:0072582||GO:0008252||GO:0015929||GO:0016668||GO:0032977||GO:0070411||GO:0044325||GO:0098847||GO:0043177||GO:0099106||GO:0005402||GO:0015095||GO:0016290||GO:0016247||GO:0000295||GO:0005346||GO:0140597||GO:0140101||GO:0000979||GO:0015216||GO:0001965||GO:0004303||GO:0047617||GO:0019903 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: CC
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | GO:0005840||GO:0044391||GO:0032993||GO:0000791||GO:0098798||GO:0140534||GO:0030117||GO:0048475||GO:0000313||GO:0005761||GO:0015935||GO:0005916||GO:0000786||GO:0019908||GO:0030877||GO:0072546||GO:0015934||GO:0022626||GO:0044815||GO:0008250||GO:1990907||GO:0043601||GO:0045239||GO:0005779||GO:0030127||GO:0031231||GO:0099092||GO:1990909||GO:0030894||GO:0098831||GO:0016600||GO:0099091||GO:0005751||GO:0005753||GO:0045259||GO:0016327||GO:0098800||GO:0031307||GO:0031306 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
KEGG
Summary Plot
No results under this category
Venn Plot
No results under this category
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
Help
still working on
3. Isoform Switch Analysis
Alternative Splicing Events
Amount of Event
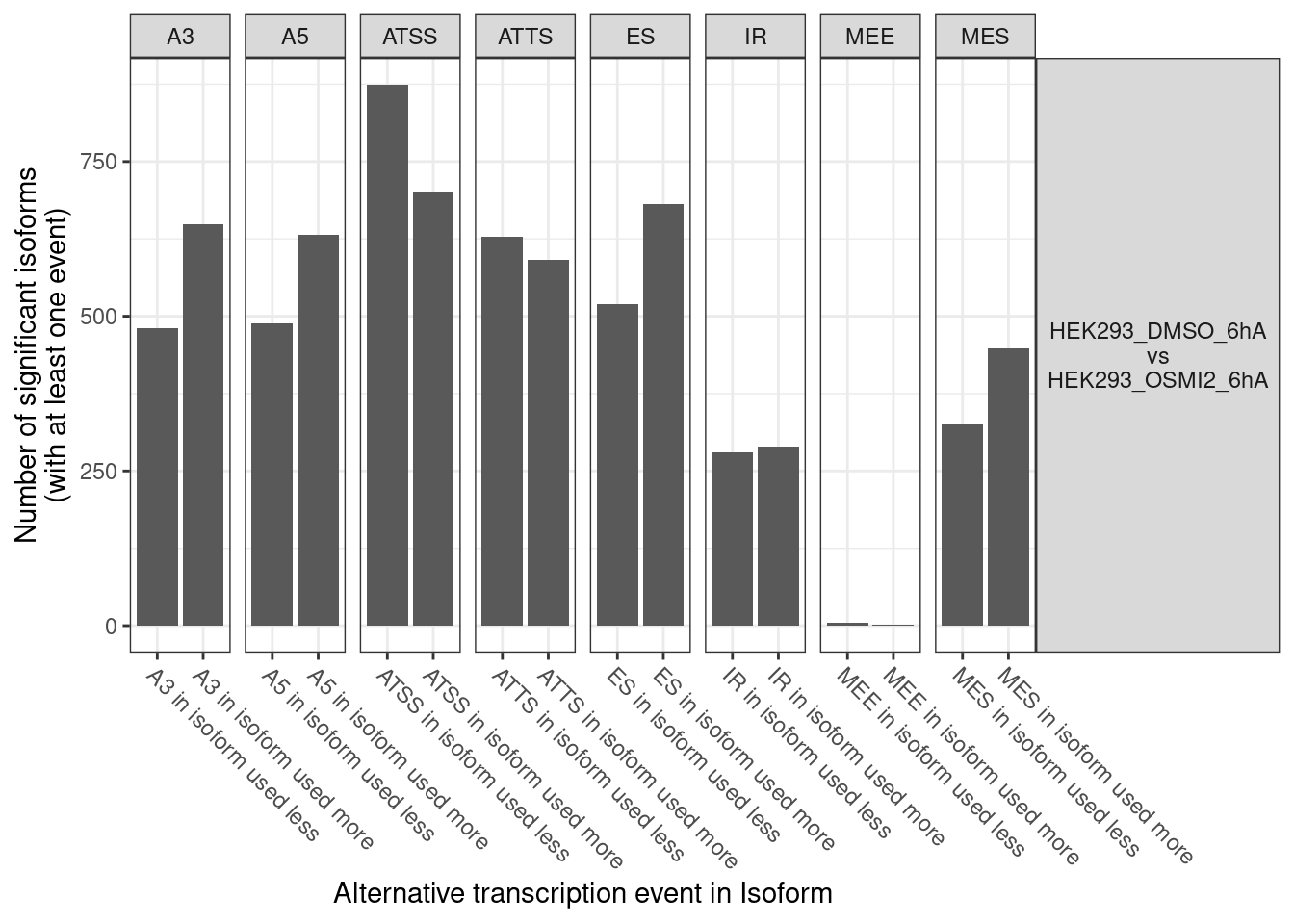
Enrichment Analysis
This below result summarizes the uneven usage within each comparison by for each alternative splicing type calculate the fraction of events being gains (as opposed to loss) and perform a statistical analysis of this fraction.
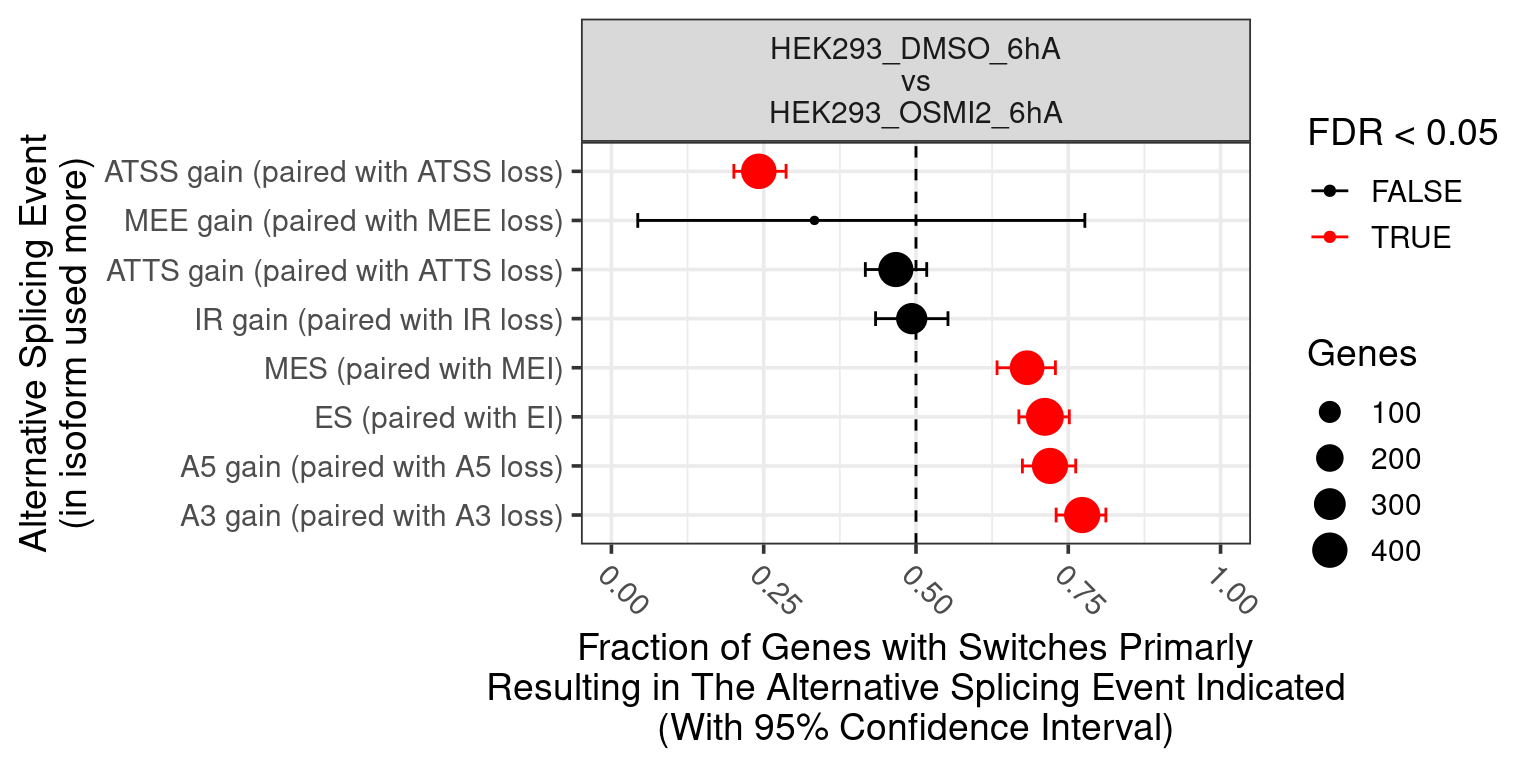
Comparison Analysis
This below result answered the question: How does the isoform usage of all isoforms utilizing a particular splicing type change - in other words is all isoforms or only a subset of isoforms that are affected.
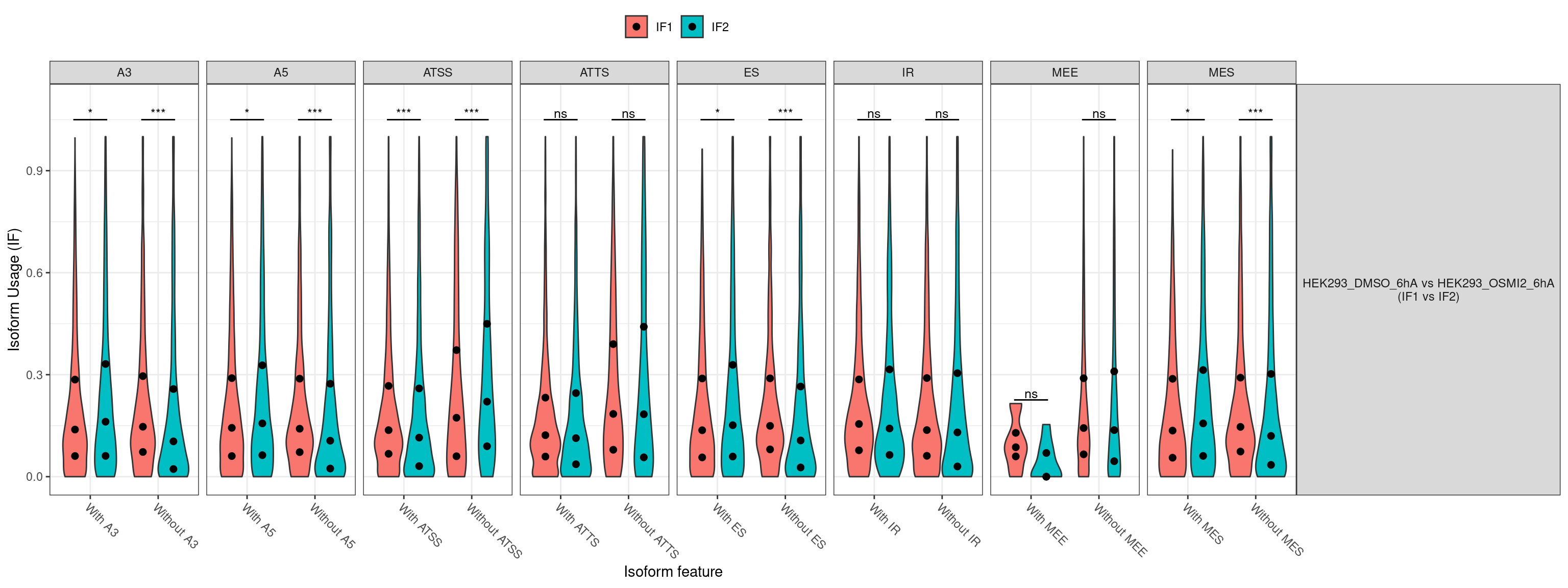
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- IR : Intron Retention.
- A5 : Alternative 5’ donor site (changes in the 5’end of the upstream exon).
- A3 : Alternative 3’ acceptor site (changes in the 3’end of the downstream exon).
- ATSS : Alternative Transcription Start Site.
- ATTS : Alternative Transcription Termination Site.
- ES : Exon Skipping (EI means Exon Inclusion).
- MES : Multiple Exon Skipping. Skipping of >1 consecutive exons. (MEI means Multiple Exon Inclusion).
- MEE : Mutually Exclusive Exons.
Alternative Splicing Consequence
Amount of Event
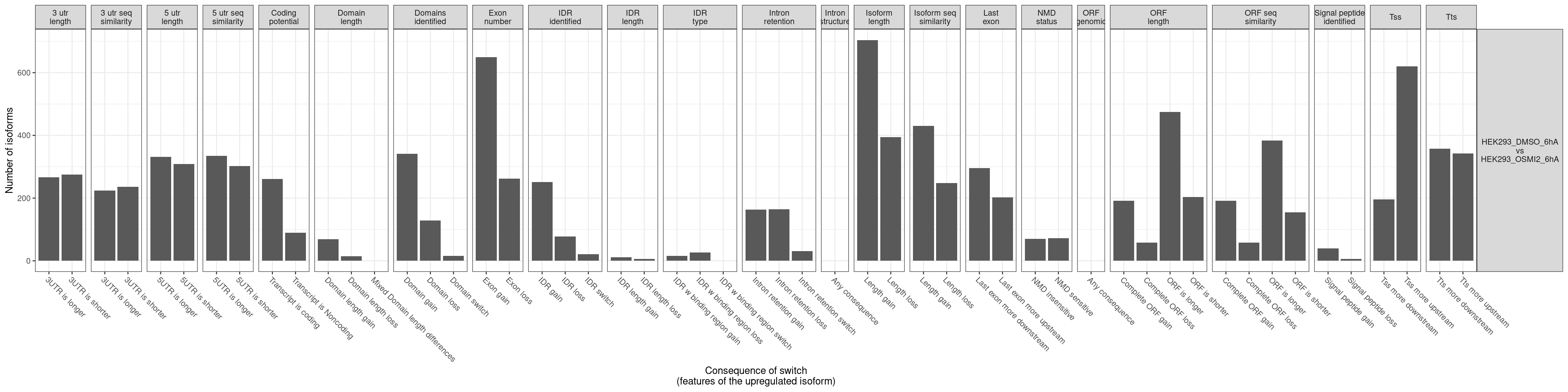
Enrichment Analysis
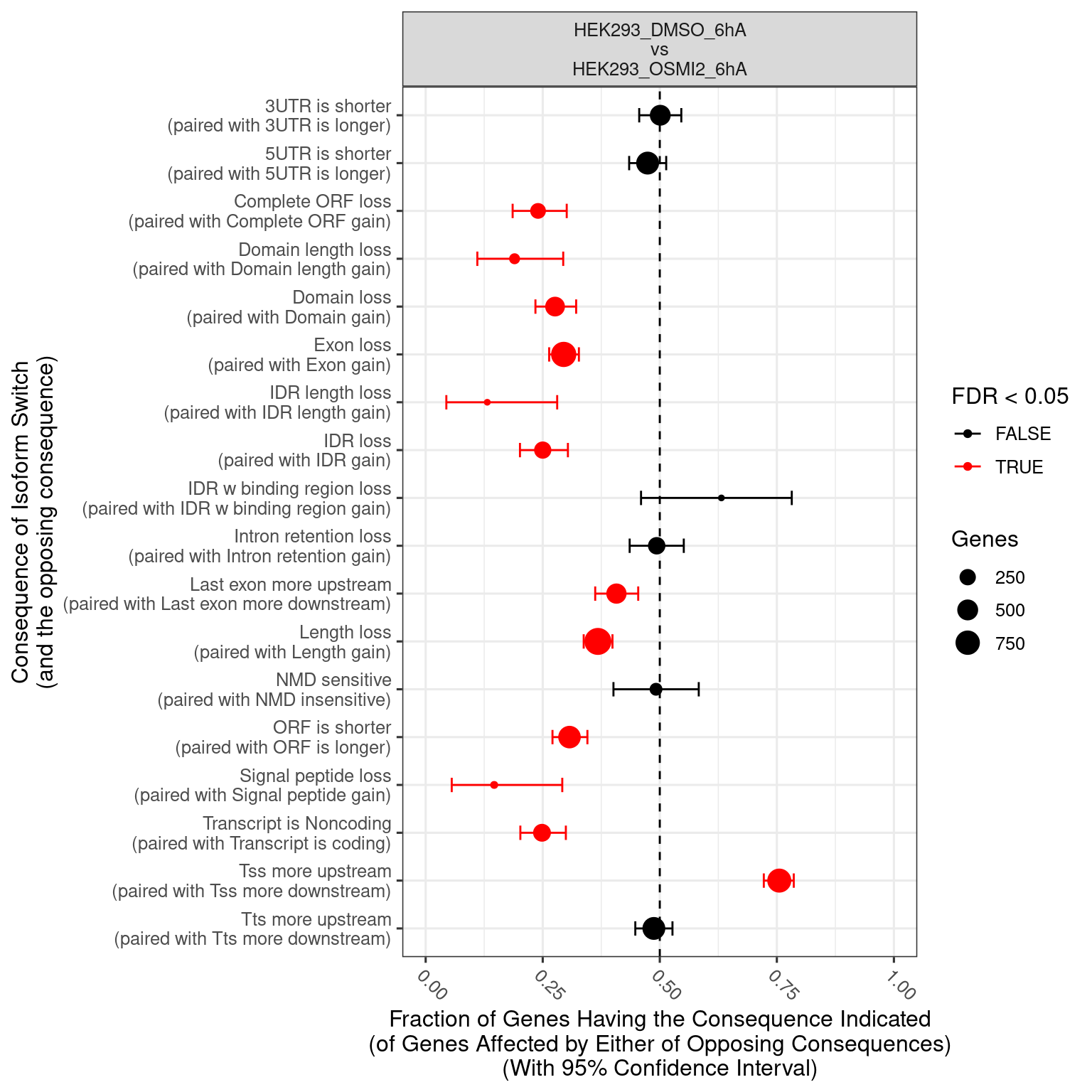
Comparison Analysis
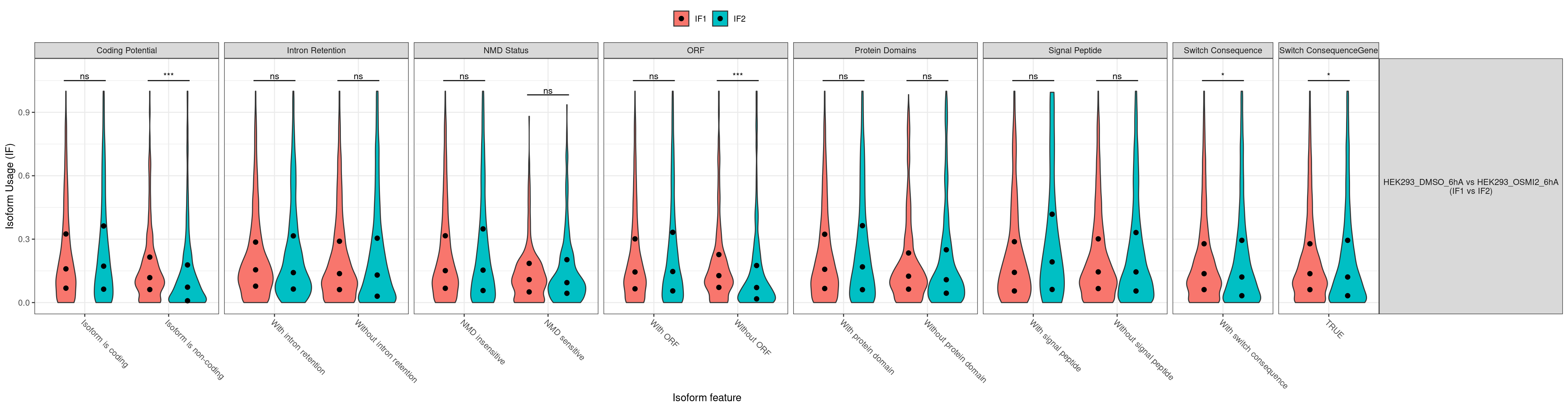
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- tss : Test transcripts for whether they use different Transcription Start Site (TSS) (more than ntCutoff from each other).
- tts : Test transcripts for whether they use different Transcription Termination Site (TTS) (more than ntCutoff from each other).
- last_exon : Test whether transcripts utilizes different last exons (defined as the last exon of each transcript is non-overlapping).
- isoform_seq_similarity : Test whether the isoform nucleotide sequences are different (as described above). Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff.
- isoform_length : Test transcripts for differences in isoform length. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Please * note that this is a less powerful analysis than implemented in ‘isoform_seq_similarity’ as two equally long sequences might be very different.
- exon_number : Test transcripts for differences in exon number.
- intron_structure : Test transcripts for differences in intron structure, e.g. usage of exon-exon junctions. This analysis corresponds to analyzing whether all introns in one isoform is also found in the other isoforms.
- intron_retention : Test for differences in intron retentions (and their genomic positions). Require that analyzeIntronRetention have been run.
- isoform_class_code : Test transcripts for differences in the transcript classification provide by cufflinks. For a updated list of class codes see http://cole-trapnell-lab.github.io/cufflinks/cuffcompare/#transfrag-class-codes.
- coding_potential : Test transcripts for differences in coding potential, as indicated by the CPAT or CPC2 analysis. Requires that analyzeCPAT or analyzeCPC2 have been used to add external conding potential analysis to the switchAnalyzeRlist.
- ORF_seq_similarity : Test whether the amino acid sequences of the ORFs are different (as described above). Reported as different if the measured JCsim is smaller than AaJCsimCutoff and the length difference of the aligned and combined region is larger than AaCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_genomic : Test transcripts for differences in genomic position of the Open Reading Frames (ORF). Requires that least one of the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and settingaddAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_length : Test transcripts for differences in length of Open Reading Frames (ORF). Note that this is a less powerful analysis than implemented in ORF_seq_similarity as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 5’ UnTranslated Region (UTR) are different (as described above). The 5’UTR is defined as the region from the transcript start to the ORF start. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_length : Test transcripts for differences in the length of the 5’ UnTranslated Region (UTR). The 5’UTR is defined as the region from the transcript start to the ORF start. Note that this is a less powerful analysis than implemented in ‘5_utr_seq_similarity’ as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 3’ UnTranslated Region (UTR) are different (as described above). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_length : Test transcripts for differences in the length of the 3’ UnTranslated Regions (UTR). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Note that this is a less powerful analysis than implemented in 3_utr_seq_similarity as two equally long sequences might be very different. Requires that identifyORF have been used to predict NMD sensitivity or that the ORF was imported though one of the dedicated import functions implemented in isoformSwitchAnalyzeR. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- NMD_status : Test transcripts for differences in sensitivity to Nonsense Mediated Decay (NMD). Requires that both the isoforms have been annotated with PTC either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domains_identified : Test transcripts for differences in the name and order of which domains are identified by the Pfam in the transcripts. Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domain_length : Test transcripts for differences in the length of overlapping domains of the same type (same hmm_name) thereby enabling analysis of protein domain truncation. Do however note that a small difference in length is will likely not truncate the protein domain. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff .Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- genomic_domain_position : Test transcripts for differences in the genomic position of the domains identified by the Pfam analysis (Will be different unless the two isoforms have the same domains at the same genomic location). Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
- IDR_identified : Test for differences in isoform IDRs. Specifically the two isoforms are analyzed for whether they contain IDRs which do not overlap in genomic coordinates. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_length : Test for differences in the length of overlapping (in genomic coordinates) IDRs. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_type : Test for differences in IDR type. Specifically the two isoforms are tested for overlapping IDRs (genomic coordinates) and overlapping IDRs are compared with regards to their IDR type (IDR vs IDR w binding site). Only available if analyzeIUPred2A was used to add external IDR analysis to the switchAnalyzeRlist.
- signal_peptide_identified : Test transcripts for differences in whether a signal peptide was identified or not by the SignalP analysis. Requires that analyzeSignalP have been used to add external SignalP analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via analyzeORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
Volcano Plot
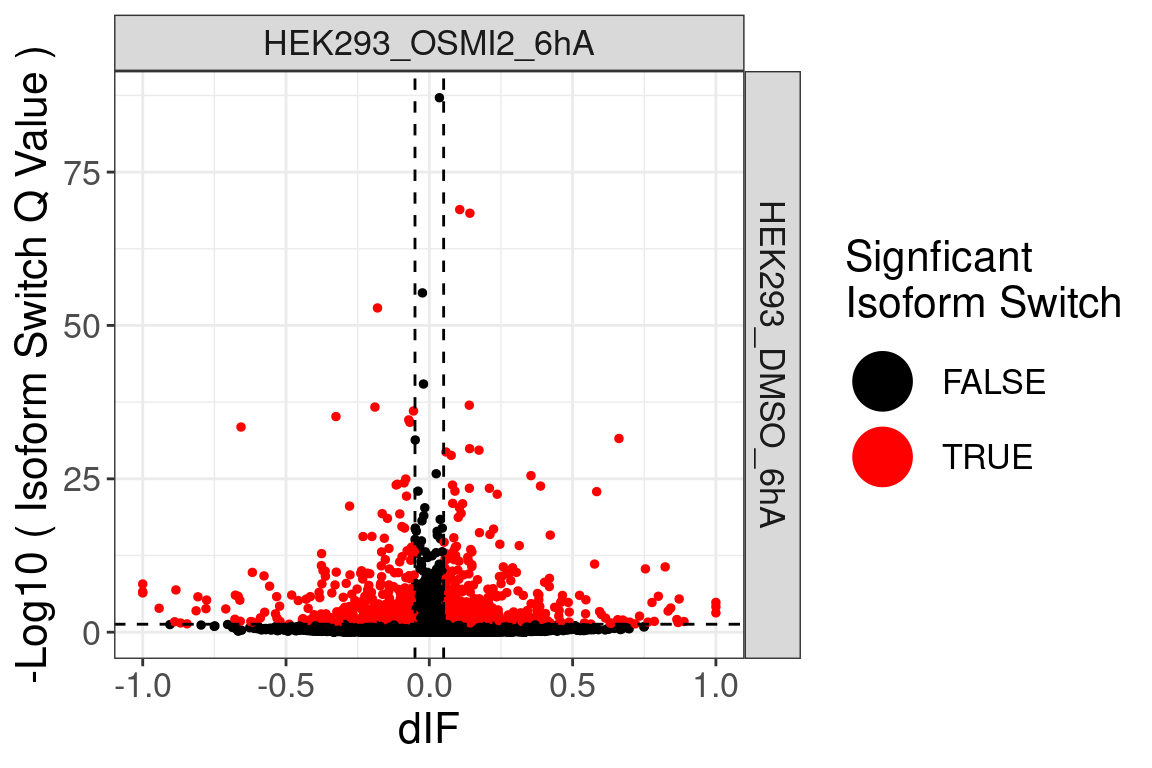
Spreadsheet
| iso_ref | gene_ref | isoform_id | gene_id | gene_name | condition_1 | condition_2 | IF1 | IF2 | dIF | isoform_switch_q_value | switchConsequencesGene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| isoComp_00527574 | geneComp_00101513 | MSTRG.32917.13 | ENSG00000165119 | HNRNPK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.106 | 0.106 | 1.286750e-69 | TRUE |
| isoComp_00438629 | geneComp_00091927 | ENST00000455138 | ENSG00000092203 | TOX4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.142 | 0.142 | 5.107859e-69 | TRUE |
| isoComp_00554458 | geneComp_00105161 | MSTRG.4718.6 | ENSG00000183605 | SFXN4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.181 | 0.000 | -0.181 | 1.403812e-53 | TRUE |
| isoComp_00515150 | geneComp_00100047 | ENST00000672515 | ENSG00000156983 | BRPF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.139 | 0.139 | 9.990394e-38 | TRUE |
| isoComp_00420305 | geneComp_00090303 | ENST00000502499 | ENSG00000015479 | MATR3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.190 | 0.000 | -0.190 | 2.009980e-37 | TRUE |
| isoComp_00535949 | geneComp_00102506 | ENST00000475872 | ENSG00000169221 | TBC1D10B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.055 | 0.000 | -0.055 | 8.838700e-37 | TRUE |
| isoComp_00500344 | geneComp_00098356 | MSTRG.2532.7 | ENSG00000143158 | MPC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.326 | 0.000 | -0.326 | 7.083665e-36 | TRUE |
| isoComp_00521173 | geneComp_00100693 | MSTRG.11864.2 | ENSG00000162062 | TEDC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.072 | 0.000 | -0.072 | 2.473295e-35 | TRUE |
| isoComp_00541107 | geneComp_00103229 | ENST00000537601 | ENSG00000172663 | TMEM134 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.068 | 0.000 | -0.068 | 6.209243e-35 | TRUE |
| isoComp_00494756 | geneComp_00097793 | ENST00000551673 | ENSG00000139131 | YARS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.657 | 0.000 | -0.657 | 3.588834e-34 | TRUE |
| isoComp_00494754 | geneComp_00097793 | ENST00000324868 | ENSG00000139131 | YARS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.338 | 1.000 | 0.662 | 2.666659e-32 | TRUE |
| isoComp_00510723 | geneComp_00099492 | MSTRG.4725.1 | ENSG00000151929 | BAG3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.141 | 0.141 | 1.207607e-30 | TRUE |
| isoComp_00500345 | geneComp_00098357 | ENST00000370509 | ENSG00000143162 | CREG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.822 | 0.995 | 0.173 | 2.147726e-30 | TRUE |
| isoComp_00567206 | geneComp_00107053 | MSTRG.9109.6 | ENSG00000198176 | TFDP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.058 | 0.058 | 4.195464e-30 | TRUE |
| isoComp_00480785 | geneComp_00096351 | MSTRG.16434.6 | ENSG00000130733 | YIPF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.076 | 0.076 | 1.497153e-29 | TRUE |
| isoComp_00438036 | geneComp_00091876 | ENST00000469408 | ENSG00000091127 | PUS7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.355 | 0.355 | 3.022376e-26 | TRUE |
| isoComp_00445483 | geneComp_00092602 | ENST00000683627 | ENSG00000102119 | EMD | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.000 | -0.082 | 1.078791e-25 | TRUE |
| isoComp_00517352 | geneComp_00100294 | MSTRG.706.3 | ENSG00000159023 | EPB41 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.088 | 0.000 | -0.088 | 4.586427e-25 | TRUE |
| isoComp_00448637 | geneComp_00092897 | ENST00000675568 | ENSG00000104419 | NDRG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.111 | 0.000 | -0.111 | 7.655507e-25 | TRUE |
| isoComp_00510722 | geneComp_00099492 | ENST00000450186 | ENSG00000151929 | BAG3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.116 | 0.000 | -0.116 | 1.008425e-24 | TRUE |
| isoComp_00442065 | geneComp_00092260 | ENST00000462640 | ENSG00000100379 | KCTD17 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.081 | 0.081 | 1.041728e-24 | TRUE |
| isoComp_00551027 | geneComp_00104643 | ENST00000303450 | ENSG00000180806 | HOXC9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.388 | 0.388 | 1.510020e-24 | TRUE |
| isoComp_00490244 | geneComp_00097333 | MSTRG.31143.10 | ENSG00000136574 | GATA4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.140 | 0.140 | 3.458525e-24 | TRUE |
| isoComp_00578753 | geneComp_00110045 | MSTRG.29036.4 | ENSG00000225177 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.210 | 0.210 | 3.496360e-24 | TRUE | |
| isoComp_00607004 | geneComp_00122261 | MSTRG.17184.24 | ENSG00000269858 | EGLN2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.089 | 0.089 | 1.017653e-23 | |
| isoComp_00527517 | geneComp_00101507 | ENST00000650836 | ENSG00000165097 | KDM1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.584 | 0.584 | 1.214661e-23 | TRUE |
| isoComp_00474477 | geneComp_00095640 | ENST00000354228 | ENSG00000125354 | SEPTIN6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.237 | 0.237 | 3.243648e-23 | TRUE |
| isoComp_00458755 | geneComp_00093918 | ENST00000534947 | ENSG00000111652 | COPS7A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.000 | -0.080 | 6.695959e-23 | TRUE |
| isoComp_00455934 | geneComp_00093649 | ENST00000511662 | ENSG00000109180 | OCIAD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.082 | 0.082 | 1.001040e-21 | TRUE |
| isoComp_00508836 | geneComp_00099278 | ENST00000394547 | ENSG00000149743 | TRPT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.116 | 0.116 | 1.149984e-21 | TRUE |
| isoComp_00432197 | geneComp_00091357 | ENST00000413543 | ENSG00000077684 | JADE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.278 | 0.000 | -0.278 | 2.800991e-21 | TRUE |
| isoComp_00522464 | geneComp_00100878 | MSTRG.3130.6 | ENSG00000162923 | WDR26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.105 | 0.105 | 4.943738e-21 | TRUE |
| isoComp_00420067 | geneComp_00090281 | ENST00000542838 | ENSG00000013573 | DDX11 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.111 | 0.111 | 4.266770e-20 | |
| isoComp_00500348 | geneComp_00098357 | MSTRG.2527.4 | ENSG00000143162 | CREG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.164 | 0.000 | -0.164 | 4.721177e-20 | TRUE |
| isoComp_00485407 | geneComp_00096819 | ENST00000511186 | ENSG00000133835 | HSD17B4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.000 | -0.103 | 5.318063e-20 | TRUE |
| isoComp_00452191 | geneComp_00093270 | MSTRG.29392.56 | ENSG00000106266 | SNX8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.101 | 0.101 | 1.931478e-19 | TRUE |
| isoComp_00555434 | geneComp_00105322 | ENST00000427528 | ENSG00000184319 | RPL23AP82 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.146 | 0.000 | -0.146 | 2.812975e-19 | TRUE |
| isoComp_00436426 | geneComp_00091733 | MSTRG.20534.4 | ENSG00000088325 | TPX2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.096 | 0.000 | -0.096 | 5.930793e-18 | TRUE |
| isoComp_00432596 | geneComp_00091385 | ENST00000487384 | ENSG00000078399 | HOXA9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.086 | 0.000 | -0.086 | 1.059592e-17 | |
| isoComp_00468553 | geneComp_00094947 | MSTRG.9942.18 | ENSG00000119685 | TTLL5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.224 | 0.224 | 1.559063e-17 | TRUE |
| isoComp_00503083 | geneComp_00098637 | ENST00000650617 | ENSG00000144659 | SLC25A38 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.751 | 0.926 | 0.175 | 5.949315e-17 | TRUE |
| isoComp_00459483 | geneComp_00094008 | MSTRG.28228.4 | ENSG00000112130 | RNF8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.212 | 0.212 | 1.161984e-16 | TRUE |
| isoComp_00608646 | geneComp_00123193 | MSTRG.21751.17 | ENSG00000272779 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.535 | 0.957 | 0.422 | 1.481233e-16 | TRUE | |
| isoComp_00564112 | geneComp_00106645 | ENST00000517766 | ENSG00000196776 | CD47 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.200 | 0.000 | -0.200 | 2.475111e-16 | TRUE |
| isoComp_00606580 | geneComp_00122062 | MSTRG.20996.7 | ENSG00000268858 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.231 | 0.000 | -0.231 | 2.564657e-16 | TRUE | |
| isoComp_00475679 | geneComp_00095786 | ENST00000375379 | ENSG00000126012 | KDM5C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.049 | 0.134 | 0.085 | 4.026481e-16 | |
| isoComp_00549414 | geneComp_00104374 | ENST00000614952 | ENSG00000179094 | PER1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.157 | 0.000 | -0.157 | 4.821420e-16 | TRUE |
| isoComp_00480783 | geneComp_00096351 | MSTRG.16434.11 | ENSG00000130733 | YIPF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.052 | 0.052 | 2.147365e-15 | TRUE |
| isoComp_00481622 | geneComp_00096433 | ENST00000393243 | ENSG00000131149 | GSE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.246 | 0.246 | 4.572006e-15 | TRUE |
| isoComp_00487039 | geneComp_00097002 | ENST00000652225 | ENSG00000134899 | ERCC5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.314 | 0.314 | 7.638572e-15 | TRUE |
| isoComp_00447188 | geneComp_00092765 | ENST00000561664 | ENSG00000103264 | FBXO31 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.052 | 0.000 | -0.052 | 9.290150e-15 | TRUE |
| isoComp_00537155 | geneComp_00102651 | ENST00000568028 | ENSG00000169955 | ZNF747 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.095 | 0.095 | 1.007571e-14 | |
| isoComp_00536590 | geneComp_00102592 | ENST00000581482 | ENSG00000169660 | HEXD | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.062 | 0.000 | -0.062 | 1.342807e-14 | TRUE |
| isoComp_00475975 | geneComp_00095807 | ENST00000592537 | ENSG00000126246 | IGFLR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.090 | 0.090 | 1.532369e-14 | TRUE |
| isoComp_00565308 | geneComp_00106809 | ENST00000419215 | ENSG00000197343 | ZNF655 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.141 | 0.000 | -0.141 | 2.231757e-14 | TRUE |
| isoComp_00519041 | geneComp_00100480 | MSTRG.21539.9 | ENSG00000160298 | C21orf58 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.000 | -0.065 | 2.412472e-14 | TRUE |
| isoComp_00604806 | geneComp_00121282 | MSTRG.15274.5 | ENSG00000266074 | BAHCC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.145 | 0.145 | 3.288702e-14 | TRUE |
| isoComp_00485577 | geneComp_00096834 | ENST00000555276 | ENSG00000133983 | COX16 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.078 | 0.000 | -0.078 | 5.433445e-14 | TRUE |
| isoComp_00554419 | geneComp_00105157 | ENST00000456048 | ENSG00000183597 | TANGO2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.084 | 0.084 | 6.685000e-14 | TRUE |
| isoComp_00559989 | geneComp_00106039 | ENST00000562424 | ENSG00000187741 | FANCA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.052 | 0.000 | -0.052 | 7.279933e-14 | TRUE |
| isoComp_00527577 | geneComp_00101513 | MSTRG.32917.5 | ENSG00000165119 | HNRNPK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.472 | 0.306 | -0.166 | 8.139305e-14 | TRUE |
| isoComp_00565422 | geneComp_00106822 | MSTRG.21480.10 | ENSG00000197381 | ADARB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.148 | 0.148 | 8.178608e-14 | TRUE |
| isoComp_00458441 | geneComp_00093890 | MSTRG.8253.8 | ENSG00000111364 | DDX55 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.057 | 0.057 | 1.452614e-13 | TRUE |
| isoComp_00421336 | geneComp_00090410 | ENST00000696693 | ENSG00000027697 | IFNGR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.376 | 0.000 | -0.376 | 1.553794e-13 | TRUE |
| isoComp_00560590 | geneComp_00106148 | ENST00000430040 | ENSG00000188191 | PRKAR1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.059 | 0.000 | -0.059 | 2.499286e-13 | TRUE |
| isoComp_00516116 | geneComp_00100145 | ENST00000560940 | ENSG00000157823 | AP3S2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.092 | 0.092 | 3.228557e-13 | TRUE |
| isoComp_00425015 | geneComp_00090743 | MSTRG.2880.5 | ENSG00000058673 | ZC3H11A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.077 | 0.077 | 3.245356e-13 | TRUE |
| isoComp_00550544 | geneComp_00104544 | MSTRG.34112.6 | ENSG00000180182 | MED14 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.095 | 0.000 | -0.095 | 4.918148e-13 | TRUE |
| isoComp_00442544 | geneComp_00092303 | ENST00000612399 | ENSG00000100519 | PSMC6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.134 | 0.134 | 6.120838e-13 | TRUE |
| isoComp_00474429 | geneComp_00095635 | MSTRG.14430.1 | ENSG00000125319 | HROB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.088 | 0.088 | 8.177179e-13 | TRUE |
| isoComp_00444272 | geneComp_00092457 | ENST00000642689 | ENSG00000101266 | CSNK2A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.086 | 0.086 | 1.170979e-12 | TRUE |
| isoComp_00443695 | geneComp_00092393 | ENST00000672258 | ENSG00000100997 | ABHD12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.055 | 0.055 | 1.214785e-12 | TRUE |
| isoComp_00571545 | geneComp_00107764 | ENST00000552318 | ENSG00000204852 | TCTN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.061 | 0.061 | 1.375153e-12 | |
| isoComp_00543908 | geneComp_00103569 | ENST00000531856 | ENSG00000174516 | PELI3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.154 | 0.000 | -0.154 | 1.487135e-12 | TRUE |
| isoComp_00472841 | geneComp_00095437 | ENST00000601967 | ENSG00000123815 | COQ8B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.087 | 0.087 | 1.543097e-12 | |
| isoComp_00458439 | geneComp_00093890 | MSTRG.8253.22 | ENSG00000111364 | DDX55 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.000 | -0.065 | 1.884065e-12 | TRUE |
| isoComp_00442604 | geneComp_00092309 | ENST00000554236 | ENSG00000100554 | ATP6V1D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.136 | 0.136 | 2.425779e-12 | TRUE |
| isoComp_00546669 | geneComp_00103969 | ENST00000323274 | ENSG00000176890 | TYMS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.817 | 0.921 | 0.104 | 2.679892e-12 | TRUE |
| isoComp_00461350 | geneComp_00094213 | ENST00000692557 | ENSG00000113719 | ERGIC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.000 | -0.103 | 3.387413e-12 | TRUE |
| isoComp_00590534 | geneComp_00114957 | ENST00000462322 | ENSG00000240204 | SMKR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.423 | 1.000 | 0.577 | 7.871629e-12 | TRUE |
| isoComp_00521187 | geneComp_00100695 | ENST00000643767 | ENSG00000162065 | TBC1D24 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.148 | 0.148 | 9.890239e-12 | TRUE |
| isoComp_00482951 | geneComp_00096563 | ENST00000651735 | ENSG00000132170 | PPARG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.376 | 0.000 | -0.376 | 1.374116e-11 | TRUE |
| isoComp_00423186 | geneComp_00090586 | ENST00000544272 | ENSG00000048028 | USP28 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.168 | 0.000 | -0.168 | 1.504772e-11 | TRUE |
| isoComp_00506395 | geneComp_00099029 | ENST00000522313 | ENSG00000147649 | MTDH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.148 | 0.148 | 2.020477e-11 | TRUE |
| isoComp_00562542 | geneComp_00106475 | MSTRG.4433.11 | ENSG00000196233 | LCOR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.258 | 0.258 | 2.229387e-11 | TRUE |
| isoComp_00595085 | geneComp_00116527 | ENST00000542535 | ENSG00000249196 | TMEM132D-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.823 | 0.823 | 2.256891e-11 | TRUE |
| isoComp_00596916 | geneComp_00117272 | MSTRG.24531.1 | ENSG00000251580 | LINC02482 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.291 | 0.291 | 3.170790e-11 | TRUE |
| isoComp_00439249 | geneComp_00091975 | ENST00000589134 | ENSG00000095066 | HOOK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.055 | 0.055 | 3.587267e-11 | TRUE |
| isoComp_00601176 | geneComp_00119367 | ENST00000620814 | ENSG00000259209 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.754 | 0.754 | 4.783402e-11 | TRUE | |
| isoComp_00422244 | geneComp_00090496 | ENST00000509897 | ENSG00000038219 | BOD1L1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.139 | 0.000 | -0.139 | 4.916764e-11 | TRUE |
| isoComp_00552667 | geneComp_00104896 | ENST00000415337 | ENSG00000182378 | PLCXD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.100 | 0.100 | 7.572195e-11 | TRUE |
| isoComp_00565409 | geneComp_00106821 | ENST00000300875 | ENSG00000197380 | DACT3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.371 | 0.000 | -0.371 | 9.179482e-11 | TRUE |
| isoComp_00605747 | geneComp_00121658 | ENST00000666181 | ENSG00000267374 | MIR924HG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.236 | 0.000 | -0.236 | 9.729488e-11 | TRUE |
| isoComp_00493551 | geneComp_00097686 | MSTRG.19676.10 | ENSG00000138386 | NAB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.363 | 0.000 | -0.363 | 1.098705e-10 | TRUE |
| isoComp_00510955 | geneComp_00099530 | MSTRG.19444.1 | ENSG00000152253 | SPC25 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.080 | 0.080 | 1.177042e-10 | TRUE |
| isoComp_00551502 | geneComp_00104705 | MSTRG.3553.2 | ENSG00000181192 | DHTKD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.081 | 0.081 | 1.177042e-10 | TRUE |
| isoComp_00467593 | geneComp_00094837 | ENST00000537373 | ENSG00000118600 | RXYLT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.286 | 0.286 | 1.455598e-10 | TRUE |
| isoComp_00434632 | geneComp_00091575 | ENST00000536056 | ENSG00000083844 | ZNF264 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.325 | 0.000 | -0.325 | 1.621391e-10 | TRUE |
| isoComp_00568001 | geneComp_00107157 | ENST00000521943 | ENSG00000198586 | TLK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.304 | 0.304 | 1.790655e-10 | TRUE |
| isoComp_00608324 | geneComp_00123041 | MSTRG.51.3 | ENSG00000272438 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.617 | 0.000 | -0.617 | 1.790655e-10 | TRUE | |
| isoComp_00530231 | geneComp_00101849 | ENST00000530175 | ENSG00000166483 | WEE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.000 | -0.108 | 1.918926e-10 | TRUE |
| isoComp_00570026 | geneComp_00107525 | ENST00000678885 | ENSG00000204120 | GIGYF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.121 | 0.000 | -0.121 | 2.068625e-10 | TRUE |
| isoComp_00563491 | geneComp_00106580 | ENST00000431655 | ENSG00000196550 | FAM72A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.217 | 0.000 | -0.217 | 2.407772e-10 | TRUE |
| isoComp_00515334 | geneComp_00100064 | ENST00000519647 | ENSG00000157110 | RBPMS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.239 | 0.000 | -0.239 | 2.461487e-10 | TRUE |
| isoComp_00552915 | geneComp_00104929 | ENST00000329548 | ENSG00000182544 | MFSD5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.628 | 0.898 | 0.270 | 2.655971e-10 | TRUE |
| isoComp_00416161 | geneComp_00089946 | ENST00000413811 | ENSG00000000460 | C1orf112 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.208 | 0.000 | -0.208 | 2.932760e-10 | TRUE |
| isoComp_00555887 | geneComp_00105393 | ENST00000591198 | ENSG00000184640 | SEPTIN9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.113 | 0.250 | 0.138 | 3.087832e-10 | TRUE |
| isoComp_00611097 | geneComp_00124629 | ENST00000689603 | ENSG00000278175 | GLIDR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.073 | 0.073 | 3.865118e-10 | |
| isoComp_00527624 | geneComp_00101520 | MSTRG.33169.7 | ENSG00000165152 | PGAP4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.120 | 0.120 | 4.510663e-10 | TRUE |
| isoComp_00567374 | geneComp_00107073 | ENST00000442123 | ENSG00000198252 | STYX | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.277 | 0.000 | -0.277 | 4.510663e-10 | TRUE |
| isoComp_00423651 | geneComp_00090623 | ENST00000504459 | ENSG00000049860 | HEXB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.053 | 0.000 | -0.053 | 4.776176e-10 | |
| isoComp_00529449 | geneComp_00101767 | ENST00000529334 | ENSG00000166181 | API5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.070 | 0.000 | -0.070 | 6.069028e-10 | TRUE |
| isoComp_00590535 | geneComp_00114957 | ENST00000488846 | ENSG00000240204 | SMKR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.577 | 0.000 | -0.577 | 6.636342e-10 | TRUE |
| isoComp_00424497 | geneComp_00090701 | MSTRG.18218.12 | ENSG00000055332 | EIF2AK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.254 | 0.254 | 7.629295e-10 | TRUE |
| isoComp_00461292 | geneComp_00094210 | ENST00000514777 | ENSG00000113658 | SMAD5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.363 | 0.000 | -0.363 | 7.629295e-10 | TRUE |
| isoComp_00567373 | geneComp_00107073 | ENST00000354586 | ENSG00000198252 | STYX | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.245 | 0.245 | 8.498395e-10 | TRUE |
| isoComp_00525423 | geneComp_00101227 | ENST00000483811 | ENSG00000164076 | CAMKV | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.166 | 0.000 | -0.166 | 8.807201e-10 | TRUE |
| isoComp_00603891 | geneComp_00120780 | ENST00000614048 | ENSG00000263001 | GTF2I | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.109 | 0.027 | -0.082 | 1.278647e-09 | TRUE |
| isoComp_00547445 | geneComp_00104075 | ENST00000395704 | ENSG00000177427 | MIEF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.059 | 0.059 | 1.669662e-09 | |
| isoComp_00565410 | geneComp_00106821 | ENST00000391916 | ENSG00000197380 | DACT3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.554 | 0.973 | 0.419 | 1.681649e-09 | TRUE |
| isoComp_00494700 | geneComp_00097787 | ENST00000535402 | ENSG00000139044 | B4GALNT3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.066 | 0.000 | -0.066 | 1.961853e-09 | TRUE |
| isoComp_00551250 | geneComp_00104681 | ENST00000439109 | ENSG00000181019 | NQO1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.232 | 0.000 | -0.232 | 2.140768e-09 | TRUE |
| isoComp_00464669 | geneComp_00094512 | MSTRG.18597.19 | ENSG00000116001 | TIA1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.103 | 0.103 | 2.317134e-09 | TRUE |
| isoComp_00584201 | geneComp_00112325 | ENST00000686031 | ENSG00000231584 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.077 | 0.077 | 2.317134e-09 | TRUE | |
| isoComp_00438351 | geneComp_00091912 | ENST00000554222 | ENSG00000092020 | PPP2R3C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.168 | 0.168 | 2.751421e-09 | TRUE |
| isoComp_00515357 | geneComp_00100069 | MSTRG.22484.5 | ENSG00000157152 | SYN2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.284 | 0.284 | 3.885268e-09 | TRUE |
| isoComp_00617305 | geneComp_00128605 | ENST00000686810 | ENSG00000289486 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.402 | 0.402 | 7.214247e-09 | TRUE | |
| isoComp_00616489 | geneComp_00128001 | MSTRG.31594.4 | ENSG00000288596 | C8orf44 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.290 | 0.000 | -0.290 | 1.116193e-08 | TRUE |
| isoComp_00566652 | geneComp_00106979 | ENST00000567356 | ENSG00000197943 | PLCG2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.170 | 0.000 | -0.170 | 1.171915e-08 | TRUE |
| isoComp_00481565 | geneComp_00096425 | ENST00000391961 | ENSG00000131115 | ZNF227 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.250 | 0.250 | 1.213856e-08 | TRUE |
| isoComp_00533790 | geneComp_00102249 | ENST00000301897 | ENSG00000168071 | CCDC88B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.375 | 0.000 | -0.375 | 1.328234e-08 | TRUE |
| isoComp_00578511 | geneComp_00109941 | ENST00000427198 | ENSG00000224892 | RPS4XP16 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.000 | -1.000 | 1.393367e-08 | TRUE |
| isoComp_00558051 | geneComp_00105707 | ENST00000684972 | ENSG00000185986 | SDHAP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.057 | 0.057 | 1.939260e-08 | |
| isoComp_00563809 | geneComp_00106624 | ENST00000399759 | ENSG00000196689 | TRPV1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.141 | 0.000 | -0.141 | 1.949394e-08 | TRUE |
| isoComp_00455960 | geneComp_00093651 | ENST00000510808 | ENSG00000109184 | DCUN1D4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.329 | 0.000 | -0.329 | 1.979741e-08 | TRUE |
| isoComp_00537597 | geneComp_00102715 | ENST00000610704 | ENSG00000170265 | ZNF282 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.239 | 0.393 | 0.154 | 2.044630e-08 | TRUE |
| isoComp_00507526 | geneComp_00099146 | MSTRG.4625.7 | ENSG00000148700 | ADD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.211 | 0.000 | -0.211 | 2.340910e-08 | TRUE |
| isoComp_00484505 | geneComp_00096729 | MSTRG.2918.2 | ENSG00000133069 | TMCC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.090 | 0.090 | 2.389419e-08 | TRUE |
| isoComp_00501146 | geneComp_00098441 | ENST00000366992 | ENSG00000143493 | INTS7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.137 | 0.137 | 2.460253e-08 | TRUE |
| isoComp_00495232 | geneComp_00097853 | ENST00000695578 | ENSG00000139496 | NUP58 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.171 | 0.000 | -0.171 | 2.703416e-08 | TRUE |
| isoComp_00611204 | geneComp_00124671 | MSTRG.8638.3 | ENSG00000278390 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.557 | 0.000 | -0.557 | 3.163496e-08 | TRUE | |
| isoComp_00527901 | geneComp_00101556 | MSTRG.32608.4 | ENSG00000165283 | STOML2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.302 | 0.176 | -0.125 | 3.616893e-08 | TRUE |
| isoComp_00555321 | geneComp_00105302 | ENST00000382348 | ENSG00000184221 | OLIG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.581 | 1.000 | 0.419 | 3.659828e-08 | TRUE |
| isoComp_00495697 | geneComp_00097902 | MSTRG.8946.32 | ENSG00000139746 | RBM26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.060 | 0.060 | 4.996702e-08 | TRUE |
| isoComp_00608933 | geneComp_00123382 | MSTRG.21012.29 | ENSG00000273154 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.137 | 0.000 | -0.137 | 5.014274e-08 | TRUE | |
| isoComp_00432516 | geneComp_00091382 | ENST00000675836 | ENSG00000078319 | PMS2P1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.086 | 0.000 | -0.086 | 6.203643e-08 | TRUE |
| isoComp_00419091 | geneComp_00090198 | MSTRG.23042.5 | ENSG00000010318 | PHF7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.083 | 0.000 | -0.083 | 6.768794e-08 | TRUE |
| isoComp_00609894 | geneComp_00123919 | MSTRG.14217.7 | ENSG00000275066 | SYNRG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.223 | 0.223 | 7.975446e-08 | TRUE |
| isoComp_00428543 | geneComp_00091053 | ENST00000676162 | ENSG00000070061 | ELP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.000 | -0.065 | 8.487982e-08 | TRUE |
| isoComp_00575644 | geneComp_00108885 | MSTRG.21586.4 | ENSG00000215193 | PEX26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.062 | 0.000 | -0.062 | 8.487982e-08 | TRUE |
| isoComp_00476293 | geneComp_00095847 | ENST00000644989 | ENSG00000126705 | AHDC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.252 | 0.000 | -0.252 | 9.318378e-08 | TRUE |
| isoComp_00498163 | geneComp_00098128 | ENST00000269159 | ENSG00000141401 | IMPA2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.210 | 0.415 | 0.205 | 9.495220e-08 | TRUE |
| isoComp_00600719 | geneComp_00119112 | MSTRG.16774.10 | ENSG00000258674 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.000 | -0.108 | 1.037805e-07 | TRUE | |
| isoComp_00511969 | geneComp_00099663 | MSTRG.32089.12 | ENSG00000153310 | CYRIB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.116 | 0.116 | 1.039250e-07 | TRUE |
| isoComp_00532835 | geneComp_00102151 | ENST00000422535 | ENSG00000167733 | HSD11B1L | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.082 | 0.082 | 1.039250e-07 | TRUE |
| isoComp_00527575 | geneComp_00101513 | MSTRG.32917.14 | ENSG00000165119 | HNRNPK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.064 | 0.178 | 0.114 | 1.099990e-07 | TRUE |
| isoComp_00599491 | geneComp_00118506 | MSTRG.6794.14 | ENSG00000256673 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.106 | 0.106 | 1.099990e-07 | TRUE | |
| isoComp_00495208 | geneComp_00097850 | ENST00000551627 | ENSG00000139437 | TCHP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.091 | 0.000 | -0.091 | 1.165073e-07 | TRUE |
| isoComp_00460867 | geneComp_00094165 | ENST00000659302 | ENSG00000113318 | MSH3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.215 | 0.000 | -0.215 | 1.171175e-07 | TRUE |
| isoComp_00502137 | geneComp_00098548 | MSTRG.18669.10 | ENSG00000144034 | TPRKB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.068 | 0.068 | 1.230075e-07 | |
| isoComp_00486935 | geneComp_00096992 | ENST00000309964 | ENSG00000134852 | CLOCK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.884 | 0.000 | -0.884 | 1.282177e-07 | TRUE |
| isoComp_00540420 | geneComp_00103122 | MSTRG.18822.3 | ENSG00000172086 | KRCC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.310 | 0.310 | 1.863900e-07 | TRUE |
| isoComp_00514383 | geneComp_00099956 | MSTRG.31849.36 | ENSG00000156170 | NDUFAF6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.072 | 0.000 | -0.072 | 2.202802e-07 | TRUE |
| isoComp_00571720 | geneComp_00107813 | ENST00000466599 | ENSG00000205060 | SLC35B4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.079 | 0.000 | -0.079 | 2.208841e-07 | |
| isoComp_00536560 | geneComp_00102588 | ENST00000565525 | ENSG00000169627 | BOLA2B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.891 | 0.749 | -0.142 | 2.324499e-07 | TRUE |
| isoComp_00500691 | geneComp_00098400 | ENST00000366927 | ENSG00000143353 | LYPLAL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.110 | 0.110 | 2.364243e-07 | TRUE |
| isoComp_00537455 | geneComp_00102690 | MSTRG.6293.5 | ENSG00000170145 | SIK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.115 | 0.115 | 2.364243e-07 | TRUE |
| isoComp_00596007 | geneComp_00116895 | ENST00000503525 | ENSG00000250366 | TUNAR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.000 | -1.000 | 2.457757e-07 | TRUE |
| isoComp_00422518 | geneComp_00090516 | MSTRG.31643.5 | ENSG00000040341 | STAU2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.143 | 0.143 | 2.921550e-07 | TRUE |
| isoComp_00486321 | geneComp_00096924 | ENST00000588446 | ENSG00000134444 | RELCH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.202 | 0.000 | -0.202 | 3.075074e-07 | TRUE |
| isoComp_00534287 | geneComp_00102298 | ENST00000544451 | ENSG00000168300 | PCMTD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.385 | 0.000 | -0.385 | 3.199474e-07 | TRUE |
| isoComp_00481499 | geneComp_00096419 | ENST00000253401 | ENSG00000131089 | ARHGEF9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.101 | 0.101 | 3.260035e-07 | TRUE |
| isoComp_00472288 | geneComp_00095374 | ENST00000509935 | ENSG00000123213 | NLN | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.270 | 0.270 | 3.497818e-07 | TRUE |
| isoComp_00513651 | geneComp_00099871 | ENST00000509008 | ENSG00000155329 | ZCCHC10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.060 | 0.000 | -0.060 | 3.893365e-07 | TRUE |
| isoComp_00604556 | geneComp_00121166 | ENST00000630097 | ENSG00000265366 | GLUD1P2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.000 | -1.000 | 3.955390e-07 | TRUE |
| isoComp_00523960 | geneComp_00101085 | ENST00000495891 | ENSG00000163659 | TIPARP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.340 | 0.000 | -0.340 | 4.024736e-07 | TRUE |
| isoComp_00426462 | geneComp_00090869 | ENST00000468620 | ENSG00000065150 | IPO5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.096 | 0.000 | -0.096 | 4.691554e-07 | TRUE |
| isoComp_00596608 | geneComp_00117133 | MSTRG.34158.2 | ENSG00000251192 | ZNF674 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.378 | 0.378 | 4.848138e-07 | TRUE |
| isoComp_00425456 | geneComp_00090785 | ENST00000536569 | ENSG00000061794 | MRPS35 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.056 | 0.056 | 4.893110e-07 | TRUE |
| isoComp_00487484 | geneComp_00097053 | ENST00000583751 | ENSG00000135164 | DMTF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.101 | 0.000 | -0.101 | 5.139658e-07 | TRUE |
| isoComp_00470526 | geneComp_00095189 | ENST00000480814 | ENSG00000121578 | B4GALT4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.217 | 0.000 | -0.217 | 6.167692e-07 | TRUE |
| isoComp_00509402 | geneComp_00099347 | MSTRG.25929.2 | ENSG00000150756 | ATPSCKMT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.270 | 0.000 | -0.270 | 6.395147e-07 | TRUE |
| isoComp_00424285 | geneComp_00090682 | MSTRG.276.13 | ENSG00000054523 | KIF1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.074 | 0.000 | -0.074 | 6.739663e-07 | TRUE |
| isoComp_00587190 | geneComp_00113575 | ENST00000490451 | ENSG00000235034 | C19orf81 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.075 | 0.075 | 6.739663e-07 | TRUE |
| isoComp_00462541 | geneComp_00094331 | ENST00000505904 | ENSG00000114853 | ZBTB47 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.162 | 0.000 | -0.162 | 6.851236e-07 | TRUE |
| isoComp_00521160 | geneComp_00100693 | ENST00000361837 | ENSG00000162062 | TEDC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.102 | 0.195 | 0.093 | 7.364943e-07 | TRUE |
| isoComp_00571019 | geneComp_00107684 | ENST00000336905 | ENSG00000204588 | LINC01123 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.073 | 0.000 | -0.073 | 7.417888e-07 | TRUE |
| isoComp_00559350 | geneComp_00105931 | MSTRG.5183.6 | ENSG00000187079 | TEAD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.229 | 0.219 | 7.945790e-07 | TRUE |
| isoComp_00530834 | geneComp_00101924 | ENST00000300069 | ENSG00000166831 | RBPMS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.294 | 0.677 | 0.382 | 8.515318e-07 | TRUE |
| isoComp_00424489 | geneComp_00090701 | ENST00000647926 | ENSG00000055332 | EIF2AK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.234 | 0.234 | 8.787667e-07 | TRUE |
| isoComp_00553507 | geneComp_00105019 | ENST00000434675 | ENSG00000182957 | SPATA13 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.480 | 0.000 | -0.480 | 8.907836e-07 | TRUE |
| isoComp_00601175 | geneComp_00119367 | ENST00000558618 | ENSG00000259209 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.677 | 0.000 | -0.677 | 9.162098e-07 | TRUE | |
| isoComp_00602756 | geneComp_00120236 | ENST00000566140 | ENSG00000261040 | WFDC21P | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.524 | 0.524 | 9.887523e-07 | TRUE |
| isoComp_00565574 | geneComp_00106846 | MSTRG.4109.24 | ENSG00000197467 | COL13A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.328 | 0.328 | 9.957997e-07 | TRUE |
| isoComp_00511544 | geneComp_00099616 | ENST00000409831 | ENSG00000152969 | JAKMIP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.465 | 0.465 | 1.004675e-06 | TRUE |
| isoComp_00527562 | geneComp_00101513 | ENST00000360384 | ENSG00000165119 | HNRNPK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.114 | 0.167 | 0.053 | 1.004675e-06 | TRUE |
| isoComp_00600683 | geneComp_00119093 | MSTRG.9229.4 | ENSG00000258643 | BCL2L2-PABPN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.000 | -0.082 | 1.148429e-06 | TRUE |
| isoComp_00517353 | geneComp_00100294 | MSTRG.706.38 | ENSG00000159023 | EPB41 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.059 | 0.059 | 1.217635e-06 | TRUE |
| isoComp_00543987 | geneComp_00103589 | ENST00000564186 | ENSG00000174628 | IQCK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.253 | 0.253 | 1.222151e-06 | TRUE |
| isoComp_00508215 | geneComp_00099211 | ENST00000527446 | ENSG00000149273 | RPS3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.073 | 0.130 | 0.057 | 1.235394e-06 | |
| isoComp_00609526 | geneComp_00123735 | ENST00000634910 | ENSG00000274290 | H2BC6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.201 | 1.000 | 0.799 | 1.384951e-06 | TRUE |
| isoComp_00510174 | geneComp_00099430 | ENST00000644374 | ENSG00000151491 | EPS8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.667 | 0.000 | -0.667 | 1.408615e-06 | TRUE |
| isoComp_00610609 | geneComp_00124361 | ENST00000618412 | ENSG00000277053 | GTF2IP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.332 | 0.107 | -0.225 | 1.442552e-06 | TRUE |
| isoComp_00515146 | geneComp_00100047 | ENST00000433861 | ENSG00000156983 | BRPF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.081 | 0.081 | 1.466240e-06 | TRUE |
| isoComp_00484322 | geneComp_00096705 | ENST00000382907 | ENSG00000132950 | ZMYM5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.533 | 0.000 | -0.533 | 1.574168e-06 | TRUE |
| isoComp_00569056 | geneComp_00107309 | MSTRG.13520.11 | ENSG00000198920 | KIAA0753 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.089 | 0.089 | 1.574168e-06 | TRUE |
| isoComp_00475395 | geneComp_00095750 | ENST00000377868 | ENSG00000125864 | BFSP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.415 | 0.000 | -0.415 | 1.583814e-06 | TRUE |
| isoComp_00592618 | geneComp_00115781 | ENST00000431553 | ENSG00000243970 | PPIEL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.083 | 0.000 | -0.083 | 1.593734e-06 | TRUE |
| isoComp_00429741 | geneComp_00091147 | ENST00000357917 | ENSG00000072422 | RHOBTB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.807 | 0.000 | -0.807 | 1.707029e-06 | TRUE |
| isoComp_00544623 | geneComp_00103668 | ENST00000348124 | ENSG00000175104 | TRAF6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.235 | 0.235 | 1.713706e-06 | TRUE |
| isoComp_00530772 | geneComp_00101911 | ENST00000558779 | ENSG00000166797 | CIAO2A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.058 | 0.000 | -0.058 | 2.010140e-06 | TRUE |
| isoComp_00501908 | geneComp_00098514 | ENST00000594572 | ENSG00000143847 | PPFIA4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.300 | 0.000 | -0.300 | 2.035796e-06 | TRUE |
| isoComp_00554992 | geneComp_00105258 | ENST00000602683 | ENSG00000184007 | PTP4A2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.074 | 0.158 | 0.084 | 2.089506e-06 | TRUE |
| isoComp_00570987 | geneComp_00107680 | ENST00000452441 | ENSG00000204580 | DDR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.205 | 0.205 | 2.223790e-06 | TRUE |
| isoComp_00530835 | geneComp_00101924 | ENST00000560606 | ENSG00000166831 | RBPMS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.706 | 0.323 | -0.382 | 2.320058e-06 | TRUE |
| isoComp_00524876 | geneComp_00101179 | MSTRG.23083.7 | ENSG00000163935 | SFMBT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.073 | 0.073 | 2.588030e-06 | TRUE |
| isoComp_00430215 | geneComp_00091186 | ENST00000442241 | ENSG00000073536 | NLE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.051 | 0.000 | -0.051 | 2.683674e-06 | |
| isoComp_00554236 | geneComp_00105133 | MSTRG.26326.2 | ENSG00000183474 | GTF2H2C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.139 | 0.000 | -0.139 | 2.771810e-06 | TRUE |
| isoComp_00507112 | geneComp_00099104 | ENST00000483302 | ENSG00000148356 | LRSAM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.062 | 0.000 | -0.062 | 2.826033e-06 | TRUE |
| isoComp_00578497 | geneComp_00109934 | ENST00000431388 | ENSG00000224877 | NDUFAF8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.742 | 0.950 | 0.209 | 3.080659e-06 | TRUE |
| isoComp_00546149 | geneComp_00103876 | MSTRG.4402.4 | ENSG00000176273 | SLC35G1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.157 | 0.157 | 3.280308e-06 | TRUE |
| isoComp_00592477 | geneComp_00115722 | ENST00000415654 | ENSG00000243716 | NPIPB5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.174 | 0.174 | 3.538635e-06 | TRUE |
| isoComp_00511754 | geneComp_00099636 | ENST00000674984 | ENSG00000153113 | CAST | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.101 | 0.000 | -0.101 | 3.675726e-06 | TRUE |
| isoComp_00429740 | geneComp_00091147 | ENST00000337910 | ENSG00000072422 | RHOBTB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.107 | 0.978 | 0.872 | 3.842341e-06 | TRUE |
| isoComp_00417433 | geneComp_00090054 | ENST00000514834 | ENSG00000005884 | ITGA3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.198 | 0.000 | -0.198 | 3.961644e-06 | TRUE |
| isoComp_00501187 | geneComp_00098446 | ENST00000366878 | ENSG00000143502 | SUSD4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.430 | 0.000 | -0.430 | 4.107011e-06 | TRUE |
| isoComp_00500257 | geneComp_00098345 | ENST00000369718 | ENSG00000143110 | C1orf162 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.546 | 0.546 | 5.203694e-06 | TRUE |
| isoComp_00526424 | geneComp_00101369 | ENST00000317020 | ENSG00000164542 | KIAA0895 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.382 | 0.382 | 5.259641e-06 | TRUE |
| isoComp_00489188 | geneComp_00097217 | ENST00000438943 | ENSG00000135953 | MFSD9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.064 | 0.064 | 5.289402e-06 | TRUE |
| isoComp_00506101 | geneComp_00098995 | ENST00000276414 | ENSG00000147437 | GNRH1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.223 | -0.777 | 5.794769e-06 | TRUE |
| isoComp_00582327 | geneComp_00111455 | ENST00000664157 | ENSG00000229124 | VIM-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.661 | 0.000 | -0.661 | 6.026694e-06 | TRUE |
| isoComp_00419804 | geneComp_00090255 | MSTRG.34237.3 | ENSG00000012211 | PRICKLE3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.069 | 0.069 | 6.036433e-06 | TRUE |
| isoComp_00432469 | geneComp_00091381 | ENST00000350249 | ENSG00000078304 | PPP2R5C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.083 | 0.022 | -0.061 | 6.420913e-06 | TRUE |
| isoComp_00513370 | geneComp_00099830 | ENST00000493838 | ENSG00000154928 | EPHB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.260 | 0.000 | -0.260 | 6.813204e-06 | TRUE |
| isoComp_00549859 | geneComp_00104440 | ENST00000653319 | ENSG00000179523 | EIF3J-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.457 | 0.000 | -0.457 | 6.914724e-06 | TRUE |
| isoComp_00463799 | geneComp_00094434 | MSTRG.18473.5 | ENSG00000115484 | CCT4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.364 | 0.224 | -0.140 | 6.927759e-06 | TRUE |
| isoComp_00590834 | geneComp_00115083 | ENST00000466399 | ENSG00000240771 | ARHGEF25 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.063 | 0.000 | -0.063 | 7.050141e-06 | TRUE |
| isoComp_00489886 | geneComp_00097294 | ENST00000383029 | ENSG00000136319 | TTC5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.853 | 0.944 | 0.090 | 7.122992e-06 | |
| isoComp_00524449 | geneComp_00101135 | ENST00000466067 | ENSG00000163811 | WDR43 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.059 | 0.000 | -0.059 | 7.186246e-06 | TRUE |
| isoComp_00486047 | geneComp_00096894 | ENST00000541959 | ENSG00000134287 | ARF3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.087 | 0.153 | 0.067 | 7.296032e-06 | TRUE |
| isoComp_00482290 | geneComp_00096495 | ENST00000326266 | ENSG00000131652 | THOC6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.586 | 0.765 | 0.178 | 7.309314e-06 | TRUE |
| isoComp_00451989 | geneComp_00093250 | ENST00000395156 | ENSG00000106089 | STX1A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.052 | 0.052 | 7.623022e-06 | TRUE |
| isoComp_00563617 | geneComp_00106599 | ENST00000586814 | ENSG00000196605 | ZNF846 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.463 | 0.463 | 7.623022e-06 | TRUE |
| isoComp_00473643 | geneComp_00095538 | ENST00000269973 | ENSG00000124459 | ZNF45 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.221 | 0.000 | -0.221 | 7.969943e-06 | TRUE |
| isoComp_00425766 | geneComp_00090811 | ENST00000425675 | ENSG00000063241 | ISOC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.337 | 0.446 | 0.109 | 8.084022e-06 | |
| isoComp_00549517 | geneComp_00104394 | ENST00000538932 | ENSG00000179195 | ZNF664 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.057 | 0.057 | 9.096044e-06 | TRUE |
| isoComp_00582776 | geneComp_00111688 | ENST00000594797 | ENSG00000229833 | PET100 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.653 | 0.816 | 0.163 | 1.053394e-05 | TRUE |
| isoComp_00467443 | geneComp_00094820 | ENST00000437412 | ENSG00000118495 | PLAGL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.098 | 0.098 | 1.094396e-05 | TRUE |
| isoComp_00431447 | geneComp_00091292 | ENST00000265564 | ENSG00000075914 | EXOSC7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.615 | 0.745 | 0.130 | 1.108209e-05 | TRUE |
| isoComp_00431246 | geneComp_00091278 | MSTRG.15705.7 | ENSG00000075643 | MOCOS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.104 | 0.000 | -0.104 | 1.157991e-05 | TRUE |
| isoComp_00521854 | geneComp_00100788 | ENST00000403491 | ENSG00000162599 | NFIA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.239 | 0.000 | -0.239 | 1.161582e-05 | TRUE |
| isoComp_00593695 | geneComp_00116063 | ENST00000533708 | ENSG00000246067 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.074 | 0.074 | 1.161582e-05 | TRUE | |
| isoComp_00551644 | geneComp_00104737 | MSTRG.3174.3 | ENSG00000181450 | ZNF678 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.459 | 0.459 | 1.349564e-05 | TRUE |
| isoComp_00460977 | geneComp_00094178 | ENST00000262462 | ENSG00000113396 | SLC27A6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 1.000 | 1.000 | 1.386438e-05 | TRUE |
| isoComp_00549857 | geneComp_00104440 | ENST00000560049 | ENSG00000179523 | EIF3J-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.100 | 0.585 | 0.485 | 1.396096e-05 | TRUE |
| isoComp_00471019 | geneComp_00095240 | ENST00000381140 | ENSG00000122034 | GTF3A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.789 | 0.896 | 0.108 | 1.435156e-05 | TRUE |
| isoComp_00523784 | geneComp_00101061 | ENST00000469809 | ENSG00000163608 | NEPRO | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.091 | 0.000 | -0.091 | 1.447548e-05 | TRUE |
| isoComp_00506102 | geneComp_00098995 | ENST00000421054 | ENSG00000147437 | GNRH1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.777 | 0.777 | 1.451883e-05 | TRUE |
| isoComp_00424495 | geneComp_00090701 | ENST00000681507 | ENSG00000055332 | EIF2AK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.235 | 0.000 | -0.235 | 1.453785e-05 | TRUE |
| isoComp_00456285 | geneComp_00093681 | ENST00000505308 | ENSG00000109586 | GALNT7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.244 | 0.000 | -0.244 | 1.575514e-05 | TRUE |
| isoComp_00437167 | geneComp_00091803 | ENST00000356638 | ENSG00000089597 | GANAB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.692 | 0.754 | 0.062 | 1.624102e-05 | |
| isoComp_00504178 | geneComp_00098767 | MSTRG.26683.7 | ENSG00000145725 | PPIP5K2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.355 | 0.355 | 2.212857e-05 | TRUE |
| isoComp_00460252 | geneComp_00094102 | ENST00000481109 | ENSG00000112739 | PRPF4B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.062 | 0.000 | -0.062 | 2.331863e-05 | TRUE |
| isoComp_00595018 | geneComp_00116494 | ENST00000364535 | ENSG00000249087 | ZNF436-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.178 | 0.000 | -0.178 | 2.396391e-05 | TRUE |
| isoComp_00462632 | geneComp_00094335 | ENST00000649528 | ENSG00000114861 | FOXP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.097 | 0.097 | 2.447668e-05 | TRUE |
| isoComp_00582290 | geneComp_00111431 | MSTRG.29377.5 | ENSG00000229043 | ZFAND2A-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.207 | 0.207 | 2.481430e-05 | TRUE |
| isoComp_00429620 | geneComp_00091138 | ENST00000431523 | ENSG00000072195 | SPEG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.132 | 0.000 | -0.132 | 2.546781e-05 | TRUE |
| isoComp_00527730 | geneComp_00101541 | MSTRG.33386.11 | ENSG00000165219 | GAPVD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.139 | 0.000 | -0.139 | 2.546781e-05 | TRUE |
| isoComp_00551367 | geneComp_00104692 | MSTRG.32219.5 | ENSG00000181085 | MAPK15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.088 | 0.088 | 2.579502e-05 | TRUE |
| isoComp_00506319 | geneComp_00099019 | ENST00000521605 | ENSG00000147586 | MRPS28 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.224 | 0.102 | -0.122 | 2.673739e-05 | TRUE |
| isoComp_00508327 | geneComp_00099219 | ENST00000675843 | ENSG00000149311 | ATM | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.118 | 0.118 | 2.987868e-05 | TRUE |
| isoComp_00513209 | geneComp_00099815 | ENST00000389169 | ENSG00000154803 | FLCN | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.031 | 0.188 | 0.157 | 2.987868e-05 | TRUE |
| isoComp_00534245 | geneComp_00102292 | MSTRG.3655.24 | ENSG00000168283 | BMI1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.089 | 0.000 | -0.089 | 3.060896e-05 | TRUE |
| isoComp_00557075 | geneComp_00105585 | ENST00000427149 | ENSG00000185485 | SDHAP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.149 | 0.000 | -0.149 | 3.072951e-05 | TRUE |
| isoComp_00604808 | geneComp_00121282 | MSTRG.15274.7 | ENSG00000266074 | BAHCC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.154 | 0.000 | -0.154 | 3.072951e-05 | TRUE |
| isoComp_00480777 | geneComp_00096351 | ENST00000588962 | ENSG00000130733 | YIPF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.092 | 0.039 | -0.053 | 3.221518e-05 | TRUE |
| isoComp_00441826 | geneComp_00092237 | ENST00000397771 | ENSG00000100325 | ASCC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.246 | 0.350 | 0.104 | 3.428958e-05 | |
| isoComp_00426695 | geneComp_00090887 | ENST00000337859 | ENSG00000065548 | ZC3H15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.878 | 0.944 | 0.066 | 3.454816e-05 | |
| isoComp_00527389 | geneComp_00101480 | ENST00000379080 | ENSG00000164970 | FAM219A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.097 | 0.097 | 3.688341e-05 | TRUE |
| isoComp_00421325 | geneComp_00090410 | ENST00000367739 | ENSG00000027697 | IFNGR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.563 | 0.940 | 0.377 | 3.902925e-05 | TRUE |
| isoComp_00496188 | geneComp_00097967 | ENST00000267884 | ENSG00000140319 | SRP14 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.224 | 0.329 | 0.106 | 3.999491e-05 | TRUE |
| isoComp_00490782 | geneComp_00097396 | ENST00000460176 | ENSG00000136895 | GARNL3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.095 | 0.000 | -0.095 | 4.208291e-05 | |
| isoComp_00540857 | geneComp_00103196 | ENST00000641773 | ENSG00000172460 | PRSS30P | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.093 | 0.000 | -0.093 | 4.414586e-05 | TRUE |
| isoComp_00592453 | geneComp_00115717 | ENST00000667634 | ENSG00000243701 | DUBR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.192 | 0.000 | -0.192 | 5.075227e-05 | TRUE |
| isoComp_00502308 | geneComp_00098566 | ENST00000466259 | ENSG00000144161 | ZC3H8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.087 | 0.000 | -0.087 | 5.163765e-05 | TRUE |
| isoComp_00548801 | geneComp_00104269 | ENST00000528666 | ENSG00000178538 | CA8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.057 | 0.000 | -0.057 | 5.296913e-05 | TRUE |
| isoComp_00474535 | geneComp_00095649 | ENST00000398051 | ENSG00000125388 | GRK4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.097 | 0.097 | 5.303806e-05 | TRUE |
| isoComp_00495787 | geneComp_00097921 | ENST00000553556 | ENSG00000139926 | FRMD6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.522 | 0.000 | -0.522 | 5.714057e-05 | TRUE |
| isoComp_00493642 | geneComp_00097695 | ENST00000451836 | ENSG00000138434 | ITPRID2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.146 | 0.000 | -0.146 | 5.740483e-05 | TRUE |
| isoComp_00468259 | geneComp_00094913 | ENST00000398803 | ENSG00000119471 | HSDL2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.113 | 0.000 | -0.113 | 5.763503e-05 | TRUE |
| isoComp_00489865 | geneComp_00097291 | ENST00000258796 | ENSG00000136295 | TTYH3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.407 | 0.555 | 0.148 | 5.857741e-05 | TRUE |
| isoComp_00609547 | geneComp_00123745 | ENST00000624412 | ENSG00000274333 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.109 | 0.109 | 5.915170e-05 | TRUE | |
| isoComp_00487063 | geneComp_00097003 | MSTRG.9039.3 | ENSG00000134900 | TPP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.386 | 0.386 | 6.264062e-05 | TRUE |
| isoComp_00570370 | geneComp_00107583 | ENST00000484849 | ENSG00000204305 | AGER | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.279 | 0.000 | -0.279 | 6.357087e-05 | TRUE |
| isoComp_00574711 | geneComp_00108617 | ENST00000685100 | ENSG00000214174 | AMZ2P1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.067 | 0.067 | 6.551072e-05 | TRUE |
| isoComp_00550272 | geneComp_00104484 | ENST00000686631 | ENSG00000179846 | NKPD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 1.000 | 1.000 | 7.068515e-05 | TRUE |
| isoComp_00538366 | geneComp_00102838 | MSTRG.14818.4 | ENSG00000170832 | USP32 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.116 | 0.116 | 8.106205e-05 | TRUE |
| isoComp_00426712 | geneComp_00090888 | ENST00000602305 | ENSG00000065559 | MAP2K4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.156 | 0.156 | 8.546603e-05 | TRUE |
| isoComp_00453763 | geneComp_00093446 | ENST00000636056 | ENSG00000107779 | BMPR1A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.188 | 0.000 | -0.188 | 8.546603e-05 | TRUE |
| isoComp_00482831 | geneComp_00096557 | MSTRG.1139.3 | ENSG00000132122 | SPATA6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.437 | 0.437 | 8.570507e-05 | TRUE |
| isoComp_00429179 | geneComp_00091109 | ENST00000415597 | ENSG00000071282 | LMCD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.295 | 0.000 | -0.295 | 8.664611e-05 | TRUE |
| isoComp_00557049 | geneComp_00105582 | ENST00000535811 | ENSG00000185480 | PARPBP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.161 | 0.000 | -0.161 | 8.750864e-05 | TRUE |
| isoComp_00482653 | geneComp_00096535 | ENST00000254260 | ENSG00000131941 | RHPN2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.871 | 1.000 | 0.129 | 8.860365e-05 | |
| isoComp_00426548 | geneComp_00090877 | ENST00000548491 | ENSG00000065357 | DGKA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.090 | 0.000 | -0.090 | 9.623567e-05 | TRUE |
| isoComp_00507312 | geneComp_00099121 | ENST00000470978 | ENSG00000148459 | PDSS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.096 | 0.000 | -0.096 | 9.695625e-05 | TRUE |
| isoComp_00500834 | geneComp_00098413 | ENST00000369026 | ENSG00000143384 | MCL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.500 | 0.696 | 0.197 | 1.016039e-04 | TRUE |
| isoComp_00479368 | geneComp_00096205 | ENST00000250930 | ENSG00000129925 | PGAP6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.175 | 0.309 | 0.134 | 1.018355e-04 | TRUE |
| isoComp_00435627 | geneComp_00091669 | ENST00000380122 | ENSG00000086712 | TXLNG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.735 | 0.924 | 0.189 | 1.020299e-04 | TRUE |
| isoComp_00475394 | geneComp_00095749 | MSTRG.20344.8 | ENSG00000125863 | MKKS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.083 | 0.299 | 0.216 | 1.029442e-04 | TRUE |
| isoComp_00511412 | geneComp_00099599 | ENST00000630827 | ENSG00000152795 | HNRNPDL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.283 | 0.503 | 0.220 | 1.057252e-04 | TRUE |
| isoComp_00432425 | geneComp_00091375 | ENST00000590013 | ENSG00000078142 | PIK3C3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.266 | 0.000 | -0.266 | 1.071387e-04 | TRUE |
| isoComp_00465318 | geneComp_00094579 | ENST00000468598 | ENSG00000116560 | SFPQ | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.119 | 0.036 | -0.083 | 1.119327e-04 | TRUE |
| isoComp_00511653 | geneComp_00099628 | ENST00000283025 | ENSG00000153060 | TEKT5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.158 | 1.000 | 0.842 | 1.130555e-04 | TRUE |
| isoComp_00456512 | geneComp_00093700 | ENST00000505478 | ENSG00000109756 | RAPGEF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.136 | 0.000 | -0.136 | 1.158373e-04 | TRUE |
| isoComp_00474032 | geneComp_00095592 | ENST00000379834 | ENSG00000124784 | RIOK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.787 | 0.913 | 0.127 | 1.181009e-04 | TRUE |
| isoComp_00531130 | geneComp_00101967 | ENST00000570004 | ENSG00000166971 | AKTIP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.078 | 0.000 | -0.078 | 1.196455e-04 | TRUE |
| isoComp_00550270 | geneComp_00104484 | ENST00000317951 | ENSG00000179846 | NKPD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.943 | 0.000 | -0.943 | 1.248188e-04 | TRUE |
| isoComp_00497810 | geneComp_00098103 | ENST00000325214 | ENSG00000141179 | PCTP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.056 | 0.000 | -0.056 | 1.282305e-04 | |
| isoComp_00526300 | geneComp_00101345 | MSTRG.28615.13 | ENSG00000164414 | SLC35A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.104 | 0.000 | -0.104 | 1.282305e-04 | TRUE |
| isoComp_00608645 | geneComp_00123193 | MSTRG.21751.15 | ENSG00000272779 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.452 | 0.030 | -0.423 | 1.288617e-04 | TRUE | |
| isoComp_00611178 | geneComp_00124655 | ENST00000620927 | ENSG00000278311 | GGNBP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.146 | 0.015 | -0.131 | 1.306131e-04 | TRUE |
| isoComp_00496566 | geneComp_00097992 | ENST00000561381 | ENSG00000140455 | USP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.100 | 0.000 | -0.100 | 1.318840e-04 | TRUE |
| isoComp_00527894 | geneComp_00101556 | ENST00000356493 | ENSG00000165283 | STOML2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.552 | 0.738 | 0.186 | 1.318840e-04 | TRUE |
| isoComp_00482851 | geneComp_00096560 | ENST00000457607 | ENSG00000132153 | DHX30 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.491 | 0.620 | 0.129 | 1.319213e-04 | TRUE |
| isoComp_00452863 | geneComp_00093343 | ENST00000336180 | ENSG00000106683 | LIMK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.678 | 0.861 | 0.183 | 1.384263e-04 | TRUE |
| isoComp_00555031 | geneComp_00105261 | ENST00000328194 | ENSG00000184014 | DENND5A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.269 | 0.161 | 1.387032e-04 | |
| isoComp_00547913 | geneComp_00104138 | MSTRG.15072.2 | ENSG00000177728 | TMEM94 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.039 | 0.136 | 0.097 | 1.393581e-04 | TRUE |
| isoComp_00435849 | geneComp_00091688 | ENST00000523450 | ENSG00000087116 | ADAMTS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.200 | 0.000 | -0.200 | 1.453436e-04 | TRUE |
| isoComp_00595084 | geneComp_00116527 | ENST00000541320 | ENSG00000249196 | TMEM132D-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.779 | 0.000 | -0.779 | 1.474614e-04 | TRUE |
| isoComp_00610649 | geneComp_00124372 | ENST00000612610 | ENSG00000277117 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.238 | 0.000 | -0.238 | 1.497112e-04 | TRUE | |
| isoComp_00484044 | geneComp_00096675 | ENST00000459879 | ENSG00000132768 | DPH2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.149 | 0.082 | -0.067 | 1.579122e-04 | TRUE |
| isoComp_00437437 | geneComp_00091824 | ENST00000555508 | ENSG00000090060 | PAPOLA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.074 | 0.012 | -0.062 | 1.597681e-04 | TRUE |
| isoComp_00559925 | geneComp_00106026 | ENST00000583241 | ENSG00000187688 | TRPV2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.710 | 0.000 | -0.710 | 1.621410e-04 | TRUE |
| isoComp_00512372 | geneComp_00099712 | MSTRG.1541.2 | ENSG00000153936 | HS2ST1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.132 | 0.132 | 1.646458e-04 | TRUE |
| isoComp_00471025 | geneComp_00095240 | ENST00000482655 | ENSG00000122034 | GTF3A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.066 | 0.009 | -0.057 | 1.706854e-04 | TRUE |
| isoComp_00528913 | geneComp_00101693 | ENST00000370664 | ENSG00000165886 | UBTD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.906 | 0.961 | 0.055 | 1.730439e-04 | TRUE |
| isoComp_00458064 | geneComp_00093849 | ENST00000397196 | ENSG00000111186 | WNT5B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.097 | 0.000 | -0.097 | 1.741706e-04 | TRUE |
| isoComp_00554594 | geneComp_00105190 | ENST00000510796 | ENSG00000183718 | TRIM52 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.065 | 0.065 | 1.764696e-04 | TRUE |
| isoComp_00588049 | geneComp_00113950 | ENST00000623598 | ENSG00000236088 | COX10-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.215 | 0.000 | -0.215 | 2.032502e-04 | TRUE |
| isoComp_00593657 | geneComp_00116058 | ENST00000500559 | ENSG00000245958 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.063 | 0.063 | 2.082224e-04 | TRUE | |
| isoComp_00453438 | geneComp_00093410 | MSTRG.32692.26 | ENSG00000107371 | EXOSC3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.128 | 0.207 | 0.079 | 2.083507e-04 | TRUE |
| isoComp_00535452 | geneComp_00102442 | ENST00000676068 | ENSG00000168944 | CEP120 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.301 | 0.000 | -0.301 | 2.176434e-04 | TRUE |
| isoComp_00448205 | geneComp_00092864 | ENST00000673645 | ENSG00000104213 | PDGFRL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.174 | 0.000 | -0.174 | 2.252866e-04 | TRUE |
| isoComp_00550453 | geneComp_00104521 | ENST00000613509 | ENSG00000180035 | ZNF48 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.583 | 0.789 | 0.206 | 2.272756e-04 | TRUE |
| isoComp_00422524 | geneComp_00090517 | ENST00000432465 | ENSG00000040487 | SLC66A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.190 | 0.096 | -0.094 | 2.323073e-04 | |
| isoComp_00495681 | geneComp_00097902 | ENST00000449987 | ENSG00000139746 | RBM26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.392 | 0.203 | -0.190 | 2.403416e-04 | TRUE |
| isoComp_00596384 | geneComp_00117059 | ENST00000691004 | ENSG00000250903 | GMDS-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.240 | 0.240 | 2.466725e-04 | TRUE |
| isoComp_00525378 | geneComp_00101225 | ENST00000641508 | ENSG00000164073 | MFSD8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.239 | 0.239 | 2.491007e-04 | TRUE |
| isoComp_00468906 | geneComp_00094978 | MSTRG.18522.4 | ENSG00000119844 | AFTPH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.204 | 0.000 | -0.204 | 2.581534e-04 | TRUE |
| isoComp_00525166 | geneComp_00101207 | ENST00000503230 | ENSG00000164038 | SLC9B2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.000 | -0.080 | 2.598344e-04 | TRUE |
| isoComp_00596606 | geneComp_00117133 | ENST00000683375 | ENSG00000251192 | ZNF674 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.176 | 0.000 | -0.176 | 2.706343e-04 | TRUE |
| isoComp_00534952 | geneComp_00102377 | ENST00000517458 | ENSG00000168672 | LRATD2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.064 | 0.000 | -0.064 | 2.724453e-04 | |
| isoComp_00593212 | geneComp_00115987 | ENST00000560359 | ENSG00000244879 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.086 | 0.000 | -0.086 | 2.724453e-04 | TRUE | |
| isoComp_00508265 | geneComp_00099214 | ENST00000533073 | ENSG00000149294 | NCAM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.057 | 0.057 | 2.761074e-04 | |
| isoComp_00472556 | geneComp_00095400 | ENST00000300209 | ENSG00000123427 | EEF1AKMT3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.293 | 0.635 | 0.342 | 2.851079e-04 | TRUE |
| isoComp_00528611 | geneComp_00101661 | MSTRG.4085.7 | ENSG00000165730 | STOX1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.375 | 0.375 | 2.851079e-04 | TRUE |
| isoComp_00529557 | geneComp_00101779 | ENST00000299300 | ENSG00000166226 | CCT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.671 | 0.847 | 0.176 | 2.873513e-04 | TRUE |
| isoComp_00558825 | geneComp_00105836 | ENST00000358777 | ENSG00000186567 | CEACAM19 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.505 | 0.958 | 0.453 | 2.873513e-04 | TRUE |
| isoComp_00551029 | geneComp_00104643 | ENST00000508190 | ENSG00000180806 | HOXC9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.854 | 0.551 | -0.304 | 2.948675e-04 | TRUE |
| isoComp_00520390 | geneComp_00100606 | MSTRG.24337.6 | ENSG00000161217 | PCYT1A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.052 | 0.000 | -0.052 | 3.169477e-04 | TRUE |
| isoComp_00449901 | geneComp_00093030 | ENST00000598785 | ENSG00000105146 | AURKC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.814 | 0.000 | -0.814 | 3.217696e-04 | TRUE |
| isoComp_00526641 | geneComp_00101397 | MSTRG.29491.5 | ENSG00000164654 | MIOS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.151 | 0.000 | -0.151 | 3.241016e-04 | TRUE |
| isoComp_00537972 | geneComp_00102772 | ENST00000408946 | ENSG00000170473 | PYM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.532 | 0.705 | 0.172 | 3.557599e-04 | TRUE |
| isoComp_00552863 | geneComp_00104920 | ENST00000414248 | ENSG00000182511 | FES | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.833 | 0.833 | 3.595120e-04 | TRUE |
| isoComp_00444616 | geneComp_00092495 | ENST00000471511 | ENSG00000101413 | RPRD1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.401 | 0.216 | -0.186 | 3.649716e-04 | TRUE |
| isoComp_00423903 | geneComp_00090644 | ENST00000474243 | ENSG00000051341 | POLQ | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.122 | 0.000 | -0.122 | 3.848681e-04 | TRUE |
| isoComp_00450744 | geneComp_00093111 | ENST00000601104 | ENSG00000105516 | DBP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.051 | 0.051 | 3.852837e-04 | |
| isoComp_00544320 | geneComp_00103630 | ENST00000611884 | ENSG00000174891 | RSRC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.236 | 0.236 | 3.881008e-04 | TRUE |
| isoComp_00454944 | geneComp_00093547 | ENST00000522301 | ENSG00000108515 | ENO3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.000 | -0.080 | 3.975578e-04 | TRUE |
| isoComp_00451478 | geneComp_00093193 | ENST00000222567 | ENSG00000105849 | POLR1F | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.874 | 1.000 | 0.126 | 4.035276e-04 | TRUE |
| isoComp_00519461 | geneComp_00100521 | MSTRG.16248.1 | ENSG00000160633 | SAFB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.025 | 0.089 | 0.064 | 4.095848e-04 | |
| isoComp_00419764 | geneComp_00090251 | MSTRG.17320.19 | ENSG00000012061 | ERCC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.083 | 0.152 | 0.069 | 4.146999e-04 | TRUE |
| isoComp_00453173 | geneComp_00093380 | ENST00000432829 | ENSG00000107099 | DOCK8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.594 | 0.594 | 4.329858e-04 | TRUE |
| isoComp_00467780 | geneComp_00094862 | MSTRG.8895.4 | ENSG00000118939 | UCHL3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.060 | 0.147 | 0.087 | 4.414232e-04 | TRUE |
| isoComp_00589034 | geneComp_00114367 | ENST00000431681 | ENSG00000237296 | SMG1P1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.591 | 0.268 | -0.323 | 4.682545e-04 | TRUE |
| isoComp_00534520 | geneComp_00102322 | ENST00000571750 | ENSG00000168411 | RFWD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.044 | 0.114 | 0.070 | 4.688890e-04 | |
| isoComp_00428569 | geneComp_00091054 | MSTRG.5229.11 | ENSG00000070081 | NUCB2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.209 | 0.063 | -0.146 | 4.928005e-04 | TRUE |
| isoComp_00516555 | geneComp_00100204 | ENST00000336592 | ENSG00000158290 | CUL4B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.161 | 0.455 | 0.294 | 5.002515e-04 | TRUE |
| isoComp_00437195 | geneComp_00091805 | ENST00000588171 | ENSG00000089639 | GMIP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.000 | -0.082 | 5.130999e-04 | TRUE |
| isoComp_00488763 | geneComp_00097174 | ENST00000258243 | ENSG00000135763 | URB2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.852 | 0.949 | 0.097 | 5.130999e-04 | TRUE |
| isoComp_00510832 | geneComp_00099513 | ENST00000409645 | ENSG00000152127 | MGAT5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.812 | 0.904 | 0.092 | 5.233310e-04 | TRUE |
| isoComp_00605821 | geneComp_00121678 | ENST00000587920 | ENSG00000267421 | ZFP28-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.488 | 0.488 | 5.269610e-04 | TRUE |
| isoComp_00534637 | geneComp_00102338 | ENST00000340394 | ENSG00000168488 | ATXN2L | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.105 | 0.184 | 0.079 | 5.288270e-04 | |
| isoComp_00542176 | geneComp_00103342 | ENST00000349820 | ENSG00000173226 | IQCB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.187 | 0.122 | 5.304529e-04 | TRUE |
| isoComp_00481286 | geneComp_00096398 | MSTRG.34178.22 | ENSG00000130985 | UBA1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.089 | 0.010 | -0.079 | 5.336200e-04 | TRUE |
| isoComp_00567643 | geneComp_00107104 | ENST00000482611 | ENSG00000198408 | OGA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.022 | 0.106 | 0.083 | 5.539897e-04 | |
| isoComp_00505082 | geneComp_00098882 | ENST00000485526 | ENSG00000146530 | VWDE | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.575 | 0.000 | -0.575 | 5.542266e-04 | TRUE |
| isoComp_00571745 | geneComp_00107816 | ENST00000692097 | ENSG00000205084 | TMEM231 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.050 | 0.000 | -0.050 | 5.822190e-04 | TRUE |
| isoComp_00491748 | geneComp_00097494 | ENST00000259983 | ENSG00000137434 | C6orf52 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.092 | 0.000 | -0.092 | 5.894031e-04 | TRUE |
| isoComp_00514217 | geneComp_00099939 | ENST00000603118 | ENSG00000156026 | MCU | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.000 | -0.065 | 5.903491e-04 | TRUE |
| isoComp_00446412 | geneComp_00092706 | ENST00000620761 | ENSG00000102977 | ACD | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.390 | 0.594 | 0.203 | 5.915064e-04 | |
| isoComp_00476919 | geneComp_00095924 | ENST00000264771 | ENSG00000127419 | TMEM175 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.124 | 0.247 | 0.123 | 5.981661e-04 | TRUE |
| isoComp_00564566 | geneComp_00106709 | MSTRG.29914.3 | ENSG00000197008 | ZNF138 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.071 | 0.000 | -0.071 | 6.078753e-04 | TRUE |
| isoComp_00417318 | geneComp_00090049 | ENST00000526598 | ENSG00000005801 | ZNF195 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.063 | 0.000 | -0.063 | 6.129286e-04 | TRUE |
| isoComp_00461255 | geneComp_00094208 | ENST00000296702 | ENSG00000113649 | TCERG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.046 | 0.139 | 0.094 | 6.561488e-04 | TRUE |
| isoComp_00469522 | geneComp_00095071 | ENST00000677182 | ENSG00000120526 | NUDCD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.145 | 0.000 | -0.145 | 6.561488e-04 | TRUE |
| isoComp_00458662 | geneComp_00093914 | ENST00000322166 | ENSG00000111641 | NOP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.063 | 0.206 | 0.143 | 6.627493e-04 | TRUE |
| isoComp_00537971 | geneComp_00102772 | ENST00000398213 | ENSG00000170473 | PYM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.336 | 0.186 | -0.150 | 6.735490e-04 | TRUE |
| isoComp_00564901 | geneComp_00106745 | ENST00000489153 | ENSG00000197122 | SRC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.058 | 0.000 | -0.058 | 6.821964e-04 | TRUE |
| isoComp_00525409 | geneComp_00101226 | ENST00000645843 | ENSG00000164074 | ABHD18 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.319 | 0.319 | 6.869296e-04 | TRUE |
| isoComp_00420347 | geneComp_00090306 | ENST00000495473 | ENSG00000015568 | RGPD5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.185 | 0.000 | -0.185 | 6.953104e-04 | TRUE |
| isoComp_00558320 | geneComp_00105751 | ENST00000302979 | ENSG00000186184 | POLR1D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.685 | 0.544 | -0.141 | 7.056126e-04 | TRUE |
| isoComp_00598713 | geneComp_00118162 | ENST00000524824 | ENSG00000255328 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 1.000 | 1.000 | 7.056126e-04 | TRUE | |
| isoComp_00465742 | geneComp_00094625 | MSTRG.366.3 | ENSG00000116786 | PLEKHM2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.270 | 0.120 | -0.149 | 7.181012e-04 | TRUE |
| isoComp_00457723 | geneComp_00093817 | ENST00000228437 | ENSG00000110851 | PRDM4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.858 | 0.754 | -0.104 | 7.247318e-04 | TRUE |
| isoComp_00572447 | geneComp_00107954 | ENST00000416302 | ENSG00000205771 | CATSPER2P1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.335 | 0.335 | 7.425807e-04 | TRUE |
| isoComp_00541961 | geneComp_00103307 | ENST00000581229 | ENSG00000173065 | FAM222B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.067 | 0.067 | 7.469340e-04 | TRUE |
| isoComp_00435994 | geneComp_00091698 | MSTRG.24476.15 | ENSG00000087266 | SH3BP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.109 | 0.316 | 0.207 | 7.556647e-04 | TRUE |
| isoComp_00505110 | geneComp_00098886 | ENST00000389531 | ENSG00000146555 | SDK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.021 | 0.228 | 0.208 | 8.331619e-04 | TRUE |
| isoComp_00559842 | geneComp_00106009 | MSTRG.13312.6 | ENSG00000187624 | C17orf97 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.097 | 0.000 | -0.097 | 8.570240e-04 | TRUE |
| isoComp_00617392 | geneComp_00128682 | ENST00000693024 | ENSG00000289586 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.024 | 0.429 | 0.405 | 8.750109e-04 | TRUE | |
| isoComp_00522983 | geneComp_00100953 | ENST00000457308 | ENSG00000163214 | DHX57 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.442 | 0.741 | 0.299 | 8.899221e-04 | TRUE |
| isoComp_00418982 | geneComp_00090190 | ENST00000394679 | ENSG00000010244 | ZNF207 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.403 | 0.519 | 0.115 | 9.138326e-04 | |
| isoComp_00552487 | geneComp_00104864 | MSTRG.13596.6 | ENSG00000182224 | CYB5D1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.032 | 0.162 | 0.130 | 9.267611e-04 | TRUE |
| isoComp_00502725 | geneComp_00098607 | ENST00000392062 | ENSG00000144468 | RHBDD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.059 | 0.059 | 9.318806e-04 | TRUE |
| isoComp_00466112 | geneComp_00094667 | MSTRG.1489.38 | ENSG00000117114 | ADGRL2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.097 | 0.097 | 9.361085e-04 | TRUE |
| isoComp_00443394 | geneComp_00092372 | ENST00000557969 | ENSG00000100889 | PCK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.137 | 0.030 | -0.107 | 9.379555e-04 | TRUE |
| isoComp_00575164 | geneComp_00108752 | ENST00000398859 | ENSG00000214725 | CDIPTOSP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.168 | 0.000 | -0.168 | 9.649213e-04 | TRUE |
| isoComp_00524031 | geneComp_00101090 | MSTRG.23147.6 | ENSG00000163681 | SLMAP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.060 | 0.060 | 9.685768e-04 | TRUE |
| isoComp_00536563 | geneComp_00102588 | ENST00000651894 | ENSG00000169627 | BOLA2B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.047 | 0.168 | 0.121 | 9.942900e-04 | TRUE |
| isoComp_00452190 | geneComp_00093270 | MSTRG.29392.55 | ENSG00000106266 | SNX8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.078 | 0.016 | -0.062 | 9.994014e-04 | TRUE |
| isoComp_00497214 | geneComp_00098048 | ENST00000299952 | ENSG00000140832 | MARVELD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.545 | 0.545 | 1.001698e-03 | TRUE |
| isoComp_00489628 | geneComp_00097266 | ENST00000317257 | ENSG00000136169 | SETDB2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.267 | 0.000 | -0.267 | 1.005128e-03 | TRUE |
| isoComp_00606116 | geneComp_00121832 | ENST00000655016 | ENSG00000267750 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.537 | 0.000 | -0.537 | 1.005128e-03 | TRUE | |
| isoComp_00441129 | geneComp_00092168 | ENST00000681920 | ENSG00000100139 | MICALL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.180 | 0.297 | 0.117 | 1.008333e-03 | TRUE |
| isoComp_00579378 | geneComp_00110265 | MSTRG.28410.17 | ENSG00000225791 | TRAM2-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.361 | 0.000 | -0.361 | 1.013828e-03 | TRUE |
| isoComp_00566441 | geneComp_00106953 | ENST00000586323 | ENSG00000197863 | ZNF790 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.316 | 0.000 | -0.316 | 1.014887e-03 | TRUE |
| isoComp_00442770 | geneComp_00092324 | MSTRG.9985.5 | ENSG00000100596 | SPTLC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.190 | 0.378 | 0.188 | 1.040301e-03 | TRUE |
| isoComp_00512358 | geneComp_00099711 | ENST00000570738 | ENSG00000153933 | DGKE | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.309 | 0.000 | -0.309 | 1.040301e-03 | TRUE |
| isoComp_00556551 | geneComp_00105507 | MSTRG.8340.5 | ENSG00000185163 | DDX51 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.001 | 0.073 | 0.072 | 1.042456e-03 | TRUE |
| isoComp_00429854 | geneComp_00091157 | ENST00000346183 | ENSG00000072736 | NFATC3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.061 | 0.183 | 0.122 | 1.053760e-03 | TRUE |
| isoComp_00442506 | geneComp_00092301 | ENST00000216392 | ENSG00000100504 | PYGL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.593 | 0.739 | 0.146 | 1.053760e-03 | TRUE |
| isoComp_00486027 | geneComp_00096893 | ENST00000256680 | ENSG00000134285 | FKBP11 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.000 | -0.065 | 1.059417e-03 | TRUE |
| isoComp_00615040 | geneComp_00126931 | ENST00000621411 | ENSG00000286522 | H3C2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.717 | 0.339 | -0.377 | 1.062608e-03 | TRUE |
| isoComp_00520898 | geneComp_00100657 | ENST00000591229 | ENSG00000161860 | SYCE2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.285 | 0.000 | -0.285 | 1.093520e-03 | TRUE |
| isoComp_00441241 | geneComp_00092177 | ENST00000396426 | ENSG00000100167 | SEPTIN3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.091 | 0.091 | 1.095384e-03 | TRUE |
| isoComp_00530220 | geneComp_00101847 | ENST00000578765 | ENSG00000166479 | TMX3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.125 | 0.000 | -0.125 | 1.106985e-03 | TRUE |
| isoComp_00615041 | geneComp_00126931 | MSTRG.27753.1 | ENSG00000286522 | H3C2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.283 | 0.661 | 0.377 | 1.109516e-03 | TRUE |
| isoComp_00498718 | geneComp_00098167 | ENST00000582907 | ENSG00000141562 | NARF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.150 | 0.058 | -0.092 | 1.138348e-03 | TRUE |
| isoComp_00472151 | geneComp_00095364 | ENST00000242783 | ENSG00000123143 | PKN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.219 | 0.302 | 0.083 | 1.176826e-03 | |
| isoComp_00459521 | geneComp_00094012 | ENST00000461222 | ENSG00000112146 | FBXO9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.209 | 0.127 | 1.186698e-03 | TRUE |
| isoComp_00550502 | geneComp_00104533 | MSTRG.692.34 | ENSG00000180098 | TRNAU1AP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.107 | 0.202 | 0.094 | 1.186698e-03 | TRUE |
| isoComp_00545195 | geneComp_00103724 | ENST00000507151 | ENSG00000175414 | ARL10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.137 | 0.263 | 0.126 | 1.192407e-03 | TRUE |
| isoComp_00485971 | geneComp_00096888 | ENST00000460653 | ENSG00000134262 | AP4B1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.098 | 0.026 | -0.071 | 1.198302e-03 | TRUE |
| isoComp_00575541 | geneComp_00108856 | ENST00000571010 | ENSG00000215067 | ALOX12-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.057 | 0.057 | 1.204799e-03 | TRUE |
| isoComp_00561244 | geneComp_00106252 | ENST00000371064 | ENSG00000188706 | ZDHHC9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.163 | 0.093 | -0.071 | 1.260364e-03 | TRUE |
| isoComp_00607390 | geneComp_00122496 | ENST00000603346 | ENSG00000270647 | TAF15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.073 | 0.019 | -0.054 | 1.260364e-03 | TRUE |
| isoComp_00461987 | geneComp_00094274 | ENST00000463304 | ENSG00000114383 | TUSC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.037 | 0.114 | 0.076 | 1.273607e-03 | TRUE |
| isoComp_00427064 | geneComp_00090928 | ENST00000503767 | ENSG00000066427 | ATXN3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.211 | 0.000 | -0.211 | 1.300757e-03 | TRUE |
| isoComp_00455141 | geneComp_00093565 | ENST00000225740 | ENSG00000108602 | ALDH3A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.601 | 0.601 | 1.341354e-03 | TRUE |
| isoComp_00517065 | geneComp_00100270 | ENST00000569901 | ENSG00000158805 | ZNF276 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.128 | 0.073 | -0.055 | 1.365482e-03 | |
| isoComp_00481142 | geneComp_00096379 | MSTRG.35113.6 | ENSG00000130827 | PLXNA3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.071 | 0.207 | 0.136 | 1.395820e-03 | TRUE |
| isoComp_00428757 | geneComp_00091072 | ENST00000521492 | ENSG00000070501 | POLB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.221 | 0.054 | -0.166 | 1.399488e-03 | TRUE |
| isoComp_00452526 | geneComp_00093308 | ENST00000485736 | ENSG00000106477 | CEP41 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.056 | 0.000 | -0.056 | 1.399488e-03 | TRUE |
| isoComp_00470822 | geneComp_00095226 | ENST00000480118 | ENSG00000121905 | HPCA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.402 | 0.000 | -0.402 | 1.414428e-03 | TRUE |
| isoComp_00522743 | geneComp_00100923 | ENST00000295213 | ENSG00000163071 | SPATA18 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.342 | 0.342 | 1.414428e-03 | TRUE |
| isoComp_00422145 | geneComp_00090484 | MSTRG.27322.40 | ENSG00000037241 | RPL26L1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.095 | 0.166 | 0.071 | 1.449212e-03 | TRUE |
| isoComp_00479896 | geneComp_00096268 | ENST00000310326 | ENSG00000130305 | NSUN5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.074 | 0.316 | 0.242 | 1.459813e-03 | TRUE |
| isoComp_00448420 | geneComp_00092884 | ENST00000422906 | ENSG00000104343 | UBE2W | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.236 | 0.236 | 1.474398e-03 | TRUE |
| isoComp_00441741 | geneComp_00092228 | ENST00000475962 | ENSG00000100307 | CBX7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.061 | 0.061 | 1.515133e-03 | |
| isoComp_00437046 | geneComp_00091791 | ENST00000419234 | ENSG00000089234 | BRAP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.760 | 0.530 | -0.230 | 1.515899e-03 | TRUE |
| isoComp_00470959 | geneComp_00095233 | MSTRG.19282.16 | ENSG00000121988 | ZRANB3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.276 | 0.276 | 1.515913e-03 | TRUE |
| isoComp_00511058 | geneComp_00099545 | ENST00000282251 | ENSG00000152404 | CWF19L2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.837 | 1.000 | 0.163 | 1.538678e-03 | TRUE |
| isoComp_00435852 | geneComp_00091689 | ENST00000454077 | ENSG00000087152 | ATXN7L3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.199 | 0.308 | 0.110 | 1.616425e-03 | TRUE |
| isoComp_00454679 | geneComp_00093529 | ENST00000577918 | ENSG00000108424 | KPNB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.068 | 0.158 | 0.091 | 1.631441e-03 | TRUE |
| isoComp_00537595 | geneComp_00102715 | ENST00000470381 | ENSG00000170265 | ZNF282 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.205 | 0.086 | -0.118 | 1.662001e-03 | TRUE |
| isoComp_00606747 | geneComp_00122149 | MSTRG.27872.9 | ENSG00000269293 | ZSCAN16-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.147 | 0.052 | -0.095 | 1.697544e-03 | TRUE |
| isoComp_00485639 | geneComp_00096843 | ENST00000382998 | ENSG00000134030 | CTIF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.243 | 0.178 | 1.736785e-03 | TRUE |
| isoComp_00499680 | geneComp_00098266 | ENST00000576095 | ENSG00000142494 | SLC47A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.004 | 0.063 | 0.060 | 1.737572e-03 | |
| isoComp_00437982 | geneComp_00091870 | ENST00000381295 | ENSG00000090989 | EXOC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.267 | 0.000 | -0.267 | 1.739749e-03 | TRUE |
| isoComp_00430280 | geneComp_00091190 | ENST00000360317 | ENSG00000073605 | GSDMB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.097 | 0.000 | -0.097 | 1.822030e-03 | TRUE |
| isoComp_00509185 | geneComp_00099319 | ENST00000528724 | ENSG00000150433 | TMEM218 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.053 | 0.124 | 0.071 | 1.849350e-03 | TRUE |
| isoComp_00500837 | geneComp_00098413 | ENST00000620947 | ENSG00000143384 | MCL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.428 | 0.240 | -0.188 | 1.856565e-03 | TRUE |
| isoComp_00610650 | geneComp_00124372 | ENST00000620481 | ENSG00000277117 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.213 | 0.213 | 1.856565e-03 | TRUE | |
| isoComp_00533135 | geneComp_00102200 | ENST00000523180 | ENSG00000167904 | TMEM68 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.000 | -0.103 | 1.939998e-03 | TRUE |
| isoComp_00505202 | geneComp_00098894 | MSTRG.5698.4 | ENSG00000146670 | CDCA5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.188 | 0.268 | 0.080 | 2.009346e-03 | TRUE |
| isoComp_00431888 | geneComp_00091331 | ENST00000688892 | ENSG00000077092 | RARB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.280 | 0.280 | 2.018599e-03 | TRUE |
| isoComp_00456212 | geneComp_00093674 | ENST00000394665 | ENSG00000109475 | RPL34 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.192 | 0.123 | -0.069 | 2.018599e-03 | TRUE |
| isoComp_00528626 | geneComp_00101663 | ENST00000354185 | ENSG00000165732 | DDX21 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.807 | 0.884 | 0.077 | 2.083694e-03 | |
| isoComp_00541114 | geneComp_00103229 | ENST00000545682 | ENSG00000172663 | TMEM134 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.133 | 0.050 | 2.098112e-03 | TRUE |
| isoComp_00565104 | geneComp_00106777 | ENST00000409302 | ENSG00000197223 | C1D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.078 | 0.078 | 2.113010e-03 | TRUE |
| isoComp_00438760 | geneComp_00091937 | ENST00000563333 | ENSG00000092531 | SNAP23 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.002 | -0.077 | 2.164324e-03 | TRUE |
| isoComp_00509960 | geneComp_00099408 | ENST00000318473 | ENSG00000151332 | MBIP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.007 | 0.144 | 0.137 | 2.187375e-03 | TRUE |
| isoComp_00494982 | geneComp_00097819 | ENST00000493991 | ENSG00000139219 | COL2A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.050 | 0.000 | -0.050 | 2.200897e-03 | TRUE |
| isoComp_00464492 | geneComp_00094497 | ENST00000395038 | ENSG00000115904 | SOS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.006 | 0.120 | 0.114 | 2.201140e-03 | TRUE |
| isoComp_00442663 | geneComp_00092316 | ENST00000555404 | ENSG00000100575 | TIMM9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.135 | 0.261 | 0.127 | 2.227720e-03 | TRUE |
| isoComp_00526103 | geneComp_00101319 | MSTRG.25816.2 | ENSG00000164323 | CFAP97 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.227 | 0.227 | 2.234285e-03 | TRUE |
| isoComp_00510488 | geneComp_00099464 | ENST00000502461 | ENSG00000151725 | CENPU | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.295 | 0.145 | -0.150 | 2.247794e-03 | TRUE |
| isoComp_00589797 | geneComp_00114652 | ENST00000546413 | ENSG00000238105 | GOLGA2P5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.127 | 0.000 | -0.127 | 2.262697e-03 | TRUE |
| isoComp_00461264 | geneComp_00094208 | ENST00000511077 | ENSG00000113649 | TCERG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.101 | 0.023 | -0.078 | 2.360739e-03 | TRUE |
| isoComp_00456982 | geneComp_00093747 | ENST00000227495 | ENSG00000110080 | ST3GAL4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.070 | 0.156 | 0.086 | 2.366793e-03 | TRUE |
| isoComp_00475396 | geneComp_00095750 | ENST00000377873 | ENSG00000125864 | BFSP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.501 | 0.952 | 0.452 | 2.366793e-03 | TRUE |
| isoComp_00486943 | geneComp_00096992 | ENST00000513440 | ENSG00000134852 | CLOCK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.734 | 0.734 | 2.392412e-03 | TRUE |
| isoComp_00433427 | geneComp_00091457 | ENST00000261263 | ENSG00000080371 | RAB21 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.996 | 0.841 | -0.155 | 2.432693e-03 | TRUE |
| isoComp_00500596 | geneComp_00098385 | ENST00000497515 | ENSG00000143303 | METTL25B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.173 | 0.078 | -0.095 | 2.445171e-03 | TRUE |
| isoComp_00568308 | geneComp_00107198 | ENST00000362007 | ENSG00000198715 | GLMP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.383 | 0.503 | 0.120 | 2.499481e-03 | |
| isoComp_00466537 | geneComp_00094711 | ENST00000434299 | ENSG00000117448 | AKR1A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.071 | 0.123 | 0.052 | 2.522200e-03 | |
| isoComp_00427704 | geneComp_00090978 | MSTRG.15545.2 | ENSG00000067900 | ROCK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.057 | 0.057 | 2.683894e-03 | |
| isoComp_00428802 | geneComp_00091077 | ENST00000521752 | ENSG00000070614 | NDST1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.205 | 0.109 | -0.096 | 2.725462e-03 | TRUE |
| isoComp_00495925 | geneComp_00097942 | ENST00000559060 | ENSG00000140044 | JDP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.398 | 0.000 | -0.398 | 2.849755e-03 | TRUE |
| isoComp_00585402 | geneComp_00112834 | ENST00000667119 | ENSG00000232956 | SNHG15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.232 | 0.066 | -0.166 | 2.963900e-03 | TRUE |
| isoComp_00552367 | geneComp_00104853 | ENST00000488612 | ENSG00000182185 | RAD51B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.119 | 0.119 | 2.993801e-03 | TRUE |
| isoComp_00552302 | geneComp_00104846 | ENST00000333213 | ENSG00000182173 | TSEN54 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.224 | 0.427 | 0.203 | 3.046040e-03 | TRUE |
| isoComp_00573553 | geneComp_00108305 | ENST00000460898 | ENSG00000213397 | HAUS7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.224 | 0.143 | -0.081 | 3.058153e-03 | TRUE |
| isoComp_00457346 | geneComp_00093786 | ENST00000314845 | ENSG00000110497 | AMBRA1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.081 | 0.003 | -0.078 | 3.112576e-03 | TRUE |
| isoComp_00500847 | geneComp_00098414 | ENST00000677611 | ENSG00000143387 | CTSK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.212 | 0.000 | -0.212 | 3.223585e-03 | TRUE |
| isoComp_00554997 | geneComp_00105258 | MSTRG.747.5 | ENSG00000184007 | PTP4A2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.125 | 0.311 | 0.186 | 3.223585e-03 | TRUE |
| isoComp_00461256 | geneComp_00094208 | ENST00000394421 | ENSG00000113649 | TCERG1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.228 | 0.545 | 0.316 | 3.251197e-03 | TRUE |
| isoComp_00507248 | geneComp_00099112 | MSTRG.33713.9 | ENSG00000148399 | DPH7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.052 | 0.173 | 0.121 | 3.289543e-03 | TRUE |
| isoComp_00437229 | geneComp_00091809 | ENST00000203630 | ENSG00000089693 | MLF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.769 | 0.927 | 0.158 | 3.309663e-03 | TRUE |
| isoComp_00520303 | geneComp_00100601 | ENST00000468686 | ENSG00000161179 | YDJC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.280 | 0.206 | -0.073 | 3.311096e-03 | TRUE |
| isoComp_00478170 | geneComp_00096060 | MSTRG.30681.4 | ENSG00000128607 | KLHDC10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.252 | 0.029 | -0.224 | 3.344324e-03 | TRUE |
| isoComp_00526637 | geneComp_00101397 | ENST00000456533 | ENSG00000164654 | MIOS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.007 | 0.114 | 0.106 | 3.361271e-03 | TRUE |
| isoComp_00519286 | geneComp_00100501 | ENST00000602239 | ENSG00000160410 | SHKBP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.087 | 0.035 | -0.052 | 3.441621e-03 | TRUE |
| isoComp_00508928 | geneComp_00099288 | MSTRG.5702.23 | ENSG00000149809 | TM7SF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.039 | 0.089 | 0.050 | 3.516514e-03 | TRUE |
| isoComp_00549104 | geneComp_00104328 | ENST00000314666 | ENSG00000178921 | PFAS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.538 | 0.670 | 0.132 | 3.604681e-03 | |
| isoComp_00510491 | geneComp_00099464 | ENST00000510146 | ENSG00000151725 | CENPU | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.332 | 0.523 | 0.191 | 3.798605e-03 | TRUE |
| isoComp_00488193 | geneComp_00097119 | ENST00000257934 | ENSG00000135476 | ESPL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.399 | 0.561 | 0.162 | 3.817998e-03 | TRUE |
| isoComp_00580245 | geneComp_00110637 | MSTRG.32535.24 | ENSG00000226823 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.652 | 1.000 | 0.348 | 3.842237e-03 | TRUE | |
| isoComp_00433926 | geneComp_00091510 | ENST00000508751 | ENSG00000081791 | DELE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.151 | 0.242 | 0.090 | 3.936662e-03 | TRUE |
| isoComp_00456213 | geneComp_00093674 | ENST00000394667 | ENSG00000109475 | RPL34 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.784 | 0.861 | 0.077 | 3.939965e-03 | TRUE |
| isoComp_00461005 | geneComp_00094181 | MSTRG.26628.3 | ENSG00000113441 | LNPEP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.295 | 0.000 | -0.295 | 3.959454e-03 | TRUE |
| isoComp_00467824 | geneComp_00094868 | ENST00000261254 | ENSG00000118971 | CCND2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.564 | 0.903 | 0.339 | 3.971022e-03 | TRUE |
| isoComp_00440455 | geneComp_00092098 | ENST00000215730 | ENSG00000099940 | SNAP29 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.903 | 0.985 | 0.082 | 3.981257e-03 | |
| isoComp_00512432 | geneComp_00099718 | ENST00000368494 | ENSG00000153989 | NUS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.462 | 0.700 | 0.239 | 3.987201e-03 | TRUE |
| isoComp_00538807 | geneComp_00102891 | ENST00000490390 | ENSG00000171103 | TRMT61B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.064 | 0.000 | -0.064 | 3.987201e-03 | TRUE |
| isoComp_00516020 | geneComp_00100138 | ENST00000642228 | ENSG00000157764 | BRAF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.008 | 0.188 | 0.180 | 3.994510e-03 | TRUE |
| isoComp_00545068 | geneComp_00103711 | MSTRG.15539.2 | ENSG00000175322 | ZNF519 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.121 | 0.000 | -0.121 | 4.095747e-03 | TRUE |
| isoComp_00468504 | geneComp_00094945 | ENST00000356357 | ENSG00000119682 | AREL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.602 | 0.772 | 0.171 | 4.098253e-03 | TRUE |
| isoComp_00513621 | geneComp_00099867 | ENST00000285667 | ENSG00000155304 | HSPA13 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.797 | -0.203 | 4.280653e-03 | TRUE |
| isoComp_00540144 | geneComp_00103092 | ENST00000474627 | ENSG00000171953 | ATPAF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.394 | 0.550 | 0.155 | 4.313051e-03 | TRUE |
| isoComp_00599311 | geneComp_00118423 | ENST00000590612 | ENSG00000256294 | ZNF225 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.344 | 0.000 | -0.344 | 4.343821e-03 | TRUE |
| isoComp_00427903 | geneComp_00090997 | ENST00000376423 | ENSG00000068400 | GRIPAP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.176 | 0.372 | 0.196 | 4.360780e-03 | TRUE |
| isoComp_00582476 | geneComp_00111524 | MSTRG.24004.3 | ENSG00000229320 | KRT8P12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.364 | 0.364 | 4.360780e-03 | TRUE |
| isoComp_00609595 | geneComp_00123771 | ENST00000612689 | ENSG00000274425 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.814 | 0.937 | 0.123 | 4.379379e-03 | TRUE | |
| isoComp_00512995 | geneComp_00099789 | ENST00000524221 | ENSG00000154548 | SRSF12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.139 | 0.000 | -0.139 | 4.430878e-03 | TRUE |
| isoComp_00530563 | geneComp_00101888 | ENST00000540540 | ENSG00000166682 | TMPRSS5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.589 | 0.000 | -0.589 | 4.537392e-03 | TRUE |
| isoComp_00421567 | geneComp_00090429 | ENST00000430118 | ENSG00000029993 | HMGB3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.635 | 0.731 | 0.096 | 4.548632e-03 | TRUE |
| isoComp_00494038 | geneComp_00097737 | ENST00000348405 | ENSG00000138674 | SEC31A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.193 | 0.298 | 0.105 | 4.585101e-03 | TRUE |
| isoComp_00592266 | geneComp_00115638 | ENST00000359751 | ENSG00000243364 | EFNA4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.098 | 0.027 | -0.071 | 4.589140e-03 | TRUE |
| isoComp_00502378 | geneComp_00098579 | ENST00000272645 | ENSG00000144231 | POLR2D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.360 | 0.219 | -0.142 | 4.601418e-03 | TRUE |
| isoComp_00524806 | geneComp_00101173 | ENST00000296277 | ENSG00000163923 | RPL39L | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.943 | 0.885 | -0.057 | 4.696963e-03 | |
| isoComp_00587085 | geneComp_00113525 | ENST00000434411 | ENSG00000234912 | SNHG20 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.336 | 0.528 | 0.192 | 4.713727e-03 | TRUE |
| isoComp_00500211 | geneComp_00098338 | ENST00000446120 | ENSG00000143036 | SLC44A3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.298 | 0.000 | -0.298 | 4.770220e-03 | TRUE |
| isoComp_00502473 | geneComp_00098585 | ENST00000548921 | ENSG00000144306 | SCRN3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.055 | 0.055 | 4.860537e-03 | TRUE |
| isoComp_00509134 | geneComp_00099311 | ENST00000395019 | ENSG00000150281 | CTF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.168 | 0.168 | 4.860537e-03 | TRUE |
| isoComp_00509156 | geneComp_00099317 | ENST00000460318 | ENSG00000150401 | DCUN1D2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.089 | 0.089 | 4.894773e-03 | TRUE |
| isoComp_00599850 | geneComp_00118684 | ENST00000246166 | ENSG00000257365 | FNTB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.061 | 0.676 | 0.615 | 4.968471e-03 | TRUE |
| isoComp_00447565 | geneComp_00092804 | ENST00000448516 | ENSG00000103510 | KAT8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.061 | 0.126 | 0.064 | 5.007696e-03 | TRUE |
| isoComp_00537745 | geneComp_00102736 | ENST00000360871 | ENSG00000170325 | PRDM10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.710 | 0.952 | 0.242 | 5.093207e-03 | TRUE |
| isoComp_00478774 | geneComp_00096132 | ENST00000620009 | ENSG00000129173 | E2F8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.133 | 0.133 | 5.136254e-03 | TRUE |
| isoComp_00495917 | geneComp_00097940 | MSTRG.9885.6 | ENSG00000140043 | PTGR2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.574 | 0.043 | -0.532 | 5.145339e-03 | TRUE |
| isoComp_00606734 | geneComp_00122149 | MSTRG.27872.12 | ENSG00000269293 | ZSCAN16-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.210 | 0.399 | 0.189 | 5.145339e-03 | TRUE |
| isoComp_00505604 | geneComp_00098938 | MSTRG.34148.18 | ENSG00000147050 | KDM6A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.008 | 0.140 | 0.132 | 5.148141e-03 | TRUE |
| isoComp_00422011 | geneComp_00090468 | ENST00000558064 | ENSG00000035664 | DAPK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.399 | 0.399 | 5.510886e-03 | TRUE |
| isoComp_00498011 | geneComp_00098115 | ENST00000269051 | ENSG00000141314 | RHBDL3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.102 | 0.102 | 5.523641e-03 | TRUE |
| isoComp_00530227 | geneComp_00101849 | ENST00000450114 | ENSG00000166483 | WEE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.252 | 0.406 | 0.154 | 5.589391e-03 | TRUE |
| isoComp_00513378 | geneComp_00099832 | ENST00000285243 | ENSG00000154945 | ANKRD40 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.910 | 0.965 | 0.054 | 5.727376e-03 | TRUE |
| isoComp_00588047 | geneComp_00113950 | ENST00000602539 | ENSG00000236088 | COX10-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.094 | 0.094 | 5.761801e-03 | TRUE |
| isoComp_00518573 | geneComp_00100441 | ENST00000454902 | ENSG00000160145 | KALRN | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.223 | 0.000 | -0.223 | 5.838818e-03 | TRUE |
| isoComp_00460797 | geneComp_00094158 | ENST00000231198 | ENSG00000113272 | THG1L | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.505 | 0.225 | -0.280 | 5.931466e-03 | TRUE |
| isoComp_00486041 | geneComp_00096894 | ENST00000256682 | ENSG00000134287 | ARF3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.848 | 0.768 | -0.080 | 5.944937e-03 | TRUE |
| isoComp_00430068 | geneComp_00091171 | ENST00000425690 | ENSG00000073008 | PVR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.236 | 0.152 | -0.084 | 5.964385e-03 | TRUE |
| isoComp_00454440 | geneComp_00093504 | MSTRG.4403.4 | ENSG00000108239 | TBC1D12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.016 | 0.255 | 0.239 | 6.006652e-03 | TRUE |
| isoComp_00423633 | geneComp_00090621 | ENST00000376207 | ENSG00000049768 | FOXP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.963 | 0.522 | -0.441 | 6.110239e-03 | TRUE |
| isoComp_00565078 | geneComp_00106770 | ENST00000475781 | ENSG00000197183 | NOL4L | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.096 | 0.000 | -0.096 | 6.110239e-03 | |
| isoComp_00519956 | geneComp_00100567 | ENST00000292494 | ENSG00000160932 | LY6E | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.470 | 0.401 | -0.069 | 6.189175e-03 | |
| isoComp_00558828 | geneComp_00105836 | ENST00000480278 | ENSG00000186567 | CEACAM19 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.241 | 0.000 | -0.241 | 6.234240e-03 | TRUE |
| isoComp_00440080 | geneComp_00092062 | ENST00000589686 | ENSG00000099622 | CIRBP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.315 | 0.415 | 0.099 | 6.261711e-03 | |
| isoComp_00417285 | geneComp_00090046 | ENST00000006777 | ENSG00000005486 | RHBDD2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.770 | 0.872 | 0.102 | 6.271888e-03 | TRUE |
| isoComp_00514791 | geneComp_00100010 | ENST00000529447 | ENSG00000156599 | ZDHHC5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.054 | 0.002 | -0.052 | 6.271888e-03 | TRUE |
| isoComp_00595002 | geneComp_00116490 | ENST00000512618 | ENSG00000249069 | LINC01033 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.084 | 0.436 | 0.352 | 6.271888e-03 | TRUE |
| isoComp_00507670 | geneComp_00099166 | ENST00000278070 | ENSG00000148840 | PPRC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.331 | 0.423 | 0.093 | 6.379616e-03 | TRUE |
| isoComp_00480396 | geneComp_00096324 | ENST00000493643 | ENSG00000130638 | ATXN10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.162 | 0.076 | -0.086 | 6.448064e-03 | TRUE |
| isoComp_00454678 | geneComp_00093529 | ENST00000577875 | ENSG00000108424 | KPNB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.093 | 0.035 | -0.058 | 6.473780e-03 | TRUE |
| isoComp_00466507 | geneComp_00094708 | MSTRG.1039.13 | ENSG00000117411 | B4GALT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.634 | 0.517 | -0.117 | 6.473780e-03 | TRUE |
| isoComp_00526721 | geneComp_00101412 | ENST00000480376 | ENSG00000164707 | SLC13A4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.119 | 0.000 | -0.119 | 6.681488e-03 | TRUE |
| isoComp_00483980 | geneComp_00096668 | MSTRG.2416.11 | ENSG00000132716 | DCAF8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.015 | 0.090 | 0.076 | 6.827335e-03 | TRUE |
| isoComp_00468862 | geneComp_00094975 | ENST00000495673 | ENSG00000119801 | YPEL5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.004 | -0.103 | 6.859330e-03 | TRUE |
| isoComp_00495007 | geneComp_00097822 | MSTRG.7614.5 | ENSG00000139233 | LLPH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.829 | 0.686 | -0.143 | 6.860590e-03 | TRUE |
| isoComp_00498911 | geneComp_00098185 | ENST00000587697 | ENSG00000141655 | TNFRSF11A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.128 | 0.000 | -0.128 | 6.884645e-03 | TRUE |
| isoComp_00554801 | geneComp_00105221 | ENST00000460719 | ENSG00000183814 | LIN9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.117 | 0.117 | 6.884645e-03 | TRUE |
| isoComp_00557242 | geneComp_00105609 | MSTRG.11686.25 | ENSG00000185596 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.535 | 0.400 | -0.135 | 6.884645e-03 | TRUE | |
| isoComp_00561251 | geneComp_00106252 | MSTRG.34859.15 | ENSG00000188706 | ZDHHC9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.040 | 0.119 | 0.079 | 6.884645e-03 | TRUE |
| isoComp_00480787 | geneComp_00096352 | ENST00000309469 | ENSG00000130734 | ATG4D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.314 | 0.414 | 0.100 | 6.954945e-03 | TRUE |
| isoComp_00505302 | geneComp_00098907 | ENST00000474433 | ENSG00000146776 | ATXN7L1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.000 | -0.080 | 6.996814e-03 | TRUE |
| isoComp_00537450 | geneComp_00102690 | ENST00000304987 | ENSG00000170145 | SIK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.953 | 0.835 | -0.119 | 7.053906e-03 | TRUE |
| isoComp_00513547 | geneComp_00099858 | ENST00000438925 | ENSG00000155229 | MMS19 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.039 | 0.134 | 0.096 | 7.101024e-03 | |
| isoComp_00453163 | geneComp_00093379 | ENST00000480996 | ENSG00000107077 | KDM4C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.106 | 0.106 | 7.175201e-03 | TRUE |
| isoComp_00520953 | geneComp_00100667 | ENST00000293778 | ENSG00000161921 | CXCL16 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.748 | 0.969 | 0.221 | 7.220638e-03 | TRUE |
| isoComp_00571959 | geneComp_00107869 | ENST00000429243 | ENSG00000205352 | PRR13 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.267 | 0.551 | 0.284 | 7.220638e-03 | TRUE |
| isoComp_00478877 | geneComp_00096145 | ENST00000250113 | ENSG00000129245 | FXR2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.835 | 0.917 | 0.082 | 7.256965e-03 | |
| isoComp_00506817 | geneComp_00099072 | ENST00000374686 | ENSG00000148143 | ZNF462 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.329 | 0.329 | 7.492216e-03 | TRUE |
| isoComp_00596190 | geneComp_00116993 | ENST00000504313 | ENSG00000250673 | REELD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.678 | 0.000 | -0.678 | 7.771791e-03 | TRUE |
| isoComp_00422285 | geneComp_00090499 | ENST00000358933 | ENSG00000038358 | EDC4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.116 | 0.333 | 0.217 | 7.807005e-03 | TRUE |
| isoComp_00439903 | geneComp_00092045 | ENST00000282633 | ENSG00000099290 | WASHC2A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.396 | 0.662 | 0.266 | 7.807005e-03 | TRUE |
| isoComp_00486551 | geneComp_00096954 | ENST00000394334 | ENSG00000134602 | STK26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.578 | 0.873 | 0.295 | 7.933157e-03 | TRUE |
| isoComp_00508794 | geneComp_00099272 | MSTRG.20946.1 | ENSG00000149657 | LSM14B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.279 | 0.392 | 0.113 | 7.933157e-03 | |
| isoComp_00448058 | geneComp_00092851 | ENST00000558057 | ENSG00000104081 | BMF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.000 | -0.103 | 8.045211e-03 | TRUE |
| isoComp_00566512 | geneComp_00106966 | ENST00000359864 | ENSG00000197905 | TEAD4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.664 | 0.573 | -0.090 | 8.074932e-03 | |
| isoComp_00450401 | geneComp_00093075 | ENST00000221972 | ENSG00000105369 | CD79A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.634 | -0.366 | 8.124651e-03 | TRUE |
| isoComp_00554535 | geneComp_00105180 | ENST00000508973 | ENSG00000183666 | GUSBP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.054 | 0.054 | 8.124651e-03 | TRUE |
| isoComp_00555106 | geneComp_00105268 | MSTRG.21641.13 | ENSG00000184058 | TBX1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.152 | 0.078 | -0.074 | 8.138234e-03 | TRUE |
| isoComp_00499969 | geneComp_00098312 | MSTRG.627.2 | ENSG00000142751 | GPN2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.186 | 0.301 | 0.115 | 8.152443e-03 | TRUE |
| isoComp_00516684 | geneComp_00100216 | ENST00000289371 | ENSG00000158417 | EIF5B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.735 | 0.874 | 0.139 | 8.152443e-03 | TRUE |
| isoComp_00529169 | geneComp_00101726 | MSTRG.6184.3 | ENSG00000166012 | TAF1D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.093 | 0.214 | 0.121 | 8.169543e-03 | TRUE |
| isoComp_00442070 | geneComp_00092260 | MSTRG.22059.10 | ENSG00000100379 | KCTD17 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.266 | 0.140 | -0.126 | 8.190840e-03 | TRUE |
| isoComp_00435629 | geneComp_00091669 | ENST00000485153 | ENSG00000086712 | TXLNG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.189 | 0.018 | -0.172 | 8.279986e-03 | TRUE |
| isoComp_00509025 | geneComp_00099298 | ENST00000308893 | ENSG00000149930 | TAOK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.418 | 0.504 | 0.086 | 8.475405e-03 | TRUE |
| isoComp_00517641 | geneComp_00100324 | MSTRG.1039.20 | ENSG00000159214 | CCDC24 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.090 | 0.090 | 8.475405e-03 | TRUE |
| isoComp_00566159 | geneComp_00106913 | ENST00000556047 | ENSG00000197734 | C14orf178 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.864 | 0.864 | 8.489150e-03 | TRUE |
| isoComp_00442705 | geneComp_00092318 | ENST00000651759 | ENSG00000100578 | KIAA0586 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.136 | 0.002 | -0.134 | 8.556930e-03 | TRUE |
| isoComp_00575510 | geneComp_00108849 | ENST00000399464 | ENSG00000215041 | NEURL4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.133 | 0.286 | 0.153 | 8.556930e-03 | TRUE |
| isoComp_00480796 | geneComp_00096352 | ENST00000588972 | ENSG00000130734 | ATG4D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.222 | 0.089 | -0.133 | 8.602322e-03 | TRUE |
| isoComp_00548589 | geneComp_00104228 | ENST00000430044 | ENSG00000178234 | GALNT11 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.597 | 0.811 | 0.214 | 8.636143e-03 | TRUE |
| isoComp_00543265 | geneComp_00103479 | MSTRG.18039.5 | ENSG00000173960 | UBXN2A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.102 | 0.102 | 8.759732e-03 | TRUE |
| isoComp_00426945 | geneComp_00090914 | ENST00000394963 | ENSG00000066117 | SMARCD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.474 | 0.577 | 0.103 | 8.764219e-03 | TRUE |
| isoComp_00493256 | geneComp_00097652 | MSTRG.4752.16 | ENSG00000138162 | TACC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.202 | 0.202 | 8.850676e-03 | TRUE |
| isoComp_00449881 | geneComp_00093026 | MSTRG.17796.12 | ENSG00000105136 | ZNF419 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.084 | 0.084 | 8.983839e-03 | TRUE |
| isoComp_00511165 | geneComp_00099565 | ENST00000282356 | ENSG00000152495 | CAMK4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.342 | 1.000 | 0.658 | 9.053502e-03 | TRUE |
| isoComp_00554402 | geneComp_00105155 | ENST00000544604 | ENSG00000183579 | ZNRF3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.254 | 0.418 | 0.164 | 9.080691e-03 | TRUE |
| isoComp_00524445 | geneComp_00101135 | ENST00000407426 | ENSG00000163811 | WDR43 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.830 | 0.982 | 0.153 | 9.176422e-03 | TRUE |
| isoComp_00509758 | geneComp_00099386 | ENST00000551813 | ENSG00000151135 | TMEM263 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.429 | 0.013 | -0.416 | 9.299434e-03 | TRUE |
| isoComp_00539749 | geneComp_00103036 | ENST00000400796 | ENSG00000171729 | TMEM51 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.481 | 0.672 | 0.192 | 9.509020e-03 | TRUE |
| isoComp_00433776 | geneComp_00091493 | ENST00000498274 | ENSG00000081154 | PCNP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.085 | 0.028 | -0.057 | 9.593136e-03 | TRUE |
| isoComp_00433814 | geneComp_00091496 | ENST00000636998 | ENSG00000081189 | MEF2C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.207 | 0.000 | -0.207 | 9.593136e-03 | TRUE |
| isoComp_00527520 | geneComp_00101507 | MSTRG.27699.7 | ENSG00000165097 | KDM1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.468 | 0.025 | -0.443 | 9.618482e-03 | TRUE |
| isoComp_00566451 | geneComp_00106956 | ENST00000361007 | ENSG00000197879 | MYO1C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.489 | 0.672 | 0.182 | 9.640017e-03 | TRUE |
| isoComp_00418067 | geneComp_00090115 | MSTRG.16185.17 | ENSG00000007264 | MATK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.038 | 0.090 | 0.053 | 9.658657e-03 | |
| isoComp_00506280 | geneComp_00099014 | ENST00000316985 | ENSG00000147548 | NSD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.251 | 0.490 | 0.238 | 9.869834e-03 | TRUE |
| isoComp_00603344 | geneComp_00120575 | ENST00000563496 | ENSG00000261787 | TCF24 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.638 | 1.000 | 0.362 | 1.011167e-02 | TRUE |
| isoComp_00464004 | geneComp_00094449 | ENST00000409225 | ENSG00000115561 | CHMP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.124 | 0.209 | 0.085 | 1.029839e-02 | TRUE |
| isoComp_00467237 | geneComp_00094789 | MSTRG.2811.8 | ENSG00000118197 | DDX59 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.182 | 0.000 | -0.182 | 1.029839e-02 | TRUE |
| isoComp_00457308 | geneComp_00093781 | ENST00000532705 | ENSG00000110442 | COMMD9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.048 | 0.110 | 0.061 | 1.033053e-02 | |
| isoComp_00463632 | geneComp_00094421 | ENST00000323926 | ENSG00000115414 | FN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.051 | 0.051 | 1.033053e-02 | TRUE |
| isoComp_00601069 | geneComp_00119303 | ENST00000554824 | ENSG00000259066 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.675 | 0.675 | 1.035983e-02 | TRUE | |
| isoComp_00438644 | geneComp_00091928 | ENST00000524980 | ENSG00000092208 | GEMIN2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.067 | 0.000 | -0.067 | 1.042759e-02 | TRUE |
| isoComp_00513366 | geneComp_00099830 | ENST00000398015 | ENSG00000154928 | EPHB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.718 | 0.967 | 0.249 | 1.061747e-02 | TRUE |
| isoComp_00536624 | geneComp_00102595 | ENST00000302759 | ENSG00000169679 | BUB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.257 | 0.440 | 0.183 | 1.064242e-02 | TRUE |
| isoComp_00454896 | geneComp_00093542 | ENST00000348066 | ENSG00000108509 | CAMTA2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.053 | 0.127 | 0.074 | 1.080053e-02 | TRUE |
| isoComp_00485828 | geneComp_00096868 | MSTRG.1723.4 | ENSG00000134186 | PRPF38B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.008 | 0.074 | 0.067 | 1.086016e-02 | TRUE |
| isoComp_00478234 | geneComp_00096071 | ENST00000409156 | ENSG00000128656 | CHN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.053 | 0.053 | 1.091664e-02 | TRUE |
| isoComp_00455966 | geneComp_00093651 | MSTRG.24868.5 | ENSG00000109184 | DCUN1D4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.074 | 0.074 | 1.099827e-02 | TRUE |
| isoComp_00503158 | geneComp_00098646 | ENST00000295874 | ENSG00000144724 | PTPRG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.223 | 0.621 | 0.398 | 1.111552e-02 | TRUE |
| isoComp_00465955 | geneComp_00094653 | ENST00000366577 | ENSG00000116984 | MTR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.416 | 0.561 | 0.145 | 1.137627e-02 | TRUE |
| isoComp_00530399 | geneComp_00101868 | ENST00000588875 | ENSG00000166562 | SEC11C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.138 | 0.079 | -0.059 | 1.146086e-02 | TRUE |
| isoComp_00467420 | geneComp_00094817 | ENST00000393387 | ENSG00000118482 | PHF3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.206 | 0.000 | -0.206 | 1.165538e-02 | TRUE |
| isoComp_00529249 | geneComp_00101741 | ENST00000467068 | ENSG00000166105 | GLB1L3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.262 | 0.000 | -0.262 | 1.165630e-02 | TRUE |
| isoComp_00425879 | geneComp_00090821 | MSTRG.11800.3 | ENSG00000063854 | HAGH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.177 | 0.295 | 0.119 | 1.171624e-02 | TRUE |
| isoComp_00438581 | geneComp_00091924 | ENST00000554455 | ENSG00000092199 | HNRNPC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.208 | 0.158 | -0.050 | 1.171624e-02 | |
| isoComp_00467644 | geneComp_00094846 | ENST00000373872 | ENSG00000118707 | TGIF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.367 | 0.495 | 0.128 | 1.171624e-02 | TRUE |
| isoComp_00600678 | geneComp_00119093 | ENST00000553781 | ENSG00000258643 | BCL2L2-PABPN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.014 | 0.267 | 0.252 | 1.197284e-02 | TRUE |
| isoComp_00603855 | geneComp_00120771 | ENST00000440428 | ENSG00000262919 | CCNQ | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.057 | 0.149 | 0.092 | 1.200388e-02 | TRUE |
| isoComp_00488589 | geneComp_00097154 | ENST00000258145 | ENSG00000135677 | GNS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.897 | 0.966 | 0.069 | 1.204758e-02 | TRUE |
| isoComp_00545320 | geneComp_00103735 | ENST00000539074 | ENSG00000175482 | POLD4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.107 | 0.243 | 0.136 | 1.207852e-02 | TRUE |
| isoComp_00442555 | geneComp_00092305 | ENST00000553406 | ENSG00000100523 | DDHD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.000 | -0.108 | 1.210658e-02 | TRUE |
| isoComp_00503770 | geneComp_00098720 | ENST00000514689 | ENSG00000145348 | TBCK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.073 | 0.000 | -0.073 | 1.216231e-02 | TRUE |
| isoComp_00504603 | geneComp_00098813 | ENST00000476517 | ENSG00000145996 | CDKAL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.183 | 0.000 | -0.183 | 1.216231e-02 | TRUE |
| isoComp_00500616 | geneComp_00098386 | ENST00000478899 | ENSG00000143314 | MRPL24 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.016 | -0.066 | 1.237760e-02 | |
| isoComp_00581871 | geneComp_00111283 | ENST00000427831 | ENSG00000228643 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.266 | 0.000 | -0.266 | 1.237760e-02 | TRUE | |
| isoComp_00545118 | geneComp_00103718 | ENST00000327283 | ENSG00000175354 | PTPN2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.665 | 0.824 | 0.159 | 1.239566e-02 | TRUE |
| isoComp_00431597 | geneComp_00091307 | ENST00000463510 | ENSG00000076356 | PLXNA2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.122 | 0.122 | 1.242712e-02 | TRUE |
| isoComp_00478309 | geneComp_00096075 | ENST00000496543 | ENSG00000128699 | ORMDL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.305 | 0.206 | -0.098 | 1.242712e-02 | |
| isoComp_00424984 | geneComp_00090742 | MSTRG.2874.5 | ENSG00000058668 | ATP2B4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.004 | 0.095 | 0.091 | 1.243900e-02 | TRUE |
| isoComp_00416406 | geneComp_00089966 | ENST00000381192 | ENSG00000002586 | CD99 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.597 | 0.533 | -0.064 | 1.252704e-02 | TRUE |
| isoComp_00442321 | geneComp_00092284 | ENST00000553935 | ENSG00000100441 | KHNYN | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.334 | 0.580 | 0.245 | 1.252704e-02 | TRUE |
| isoComp_00518735 | geneComp_00100455 | ENST00000478282 | ENSG00000160201 | U2AF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.230 | 0.106 | -0.124 | 1.257099e-02 | TRUE |
| isoComp_00617302 | geneComp_00128603 | MSTRG.1596.1 | ENSG00000289483 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.243 | 0.243 | 1.264390e-02 | TRUE | |
| isoComp_00520259 | geneComp_00100595 | ENST00000586050 | ENSG00000161082 | CELF5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.173 | 0.173 | 1.265037e-02 | TRUE |
| isoComp_00487472 | geneComp_00097053 | ENST00000488352 | ENSG00000135164 | DMTF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.118 | 0.040 | -0.078 | 1.268334e-02 | TRUE |
| isoComp_00590618 | geneComp_00114987 | ENST00000463866 | ENSG00000240344 | PPIL3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.133 | 0.032 | -0.101 | 1.268334e-02 | |
| isoComp_00572033 | geneComp_00107882 | ENST00000567712 | ENSG00000205423 | CNEP1R1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.089 | 0.000 | -0.089 | 1.269165e-02 | TRUE |
| isoComp_00423632 | geneComp_00090621 | ENST00000376199 | ENSG00000049768 | FOXP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.412 | 0.412 | 1.275796e-02 | TRUE |
| isoComp_00467140 | geneComp_00094779 | ENST00000525408 | ENSG00000118058 | KMT2A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.465 | 0.331 | -0.134 | 1.275796e-02 | TRUE |
| isoComp_00565102 | geneComp_00106777 | ENST00000355848 | ENSG00000197223 | C1D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.005 | 0.116 | 0.111 | 1.277945e-02 | TRUE |
| isoComp_00559078 | geneComp_00105878 | ENST00000541517 | ENSG00000186815 | TPCN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.344 | 0.177 | -0.167 | 1.278675e-02 | TRUE |
| isoComp_00451479 | geneComp_00093193 | ENST00000462263 | ENSG00000105849 | POLR1F | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.126 | 0.000 | -0.126 | 1.284821e-02 | TRUE |
| isoComp_00591801 | geneComp_00115451 | ENST00000493287 | ENSG00000242485 | MRPL20 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.153 | 0.074 | -0.078 | 1.305647e-02 | TRUE |
| isoComp_00572156 | geneComp_00107905 | ENST00000380750 | ENSG00000205572 | SERF1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.893 | 0.676 | -0.216 | 1.306750e-02 | TRUE |
| isoComp_00490965 | geneComp_00097419 | MSTRG.32039.10 | ENSG00000136986 | DERL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.059 | 0.122 | 0.064 | 1.311306e-02 | TRUE |
| isoComp_00530850 | geneComp_00101926 | ENST00000496480 | ENSG00000166839 | ANKDD1A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.376 | 0.376 | 1.312051e-02 | TRUE |
| isoComp_00511955 | geneComp_00099663 | ENST00000519540 | ENSG00000153310 | CYRIB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.274 | 0.462 | 0.188 | 1.312734e-02 | TRUE |
| isoComp_00512434 | geneComp_00099718 | MSTRG.28865.3 | ENSG00000153989 | NUS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.527 | 0.298 | -0.228 | 1.325387e-02 | TRUE |
| isoComp_00441796 | geneComp_00092234 | ENST00000695854 | ENSG00000100320 | RBFOX2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.147 | 0.257 | 0.110 | 1.378493e-02 | TRUE |
| isoComp_00446984 | geneComp_00092751 | MSTRG.11985.50 | ENSG00000103199 | ZNF500 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.085 | 0.221 | 0.136 | 1.378493e-02 | TRUE |
| isoComp_00470640 | geneComp_00095204 | ENST00000479971 | ENSG00000121749 | TBC1D15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.055 | 0.000 | -0.055 | 1.387998e-02 | TRUE |
| isoComp_00419600 | geneComp_00090238 | ENST00000491782 | ENSG00000011426 | ANLN | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.190 | 0.035 | -0.155 | 1.396726e-02 | TRUE |
| isoComp_00446358 | geneComp_00092699 | MSTRG.12711.1 | ENSG00000102931 | ARL2BP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.049 | 0.129 | 0.080 | 1.407784e-02 | |
| isoComp_00452864 | geneComp_00093343 | ENST00000435201 | ENSG00000106683 | LIMK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.269 | 0.117 | -0.152 | 1.408939e-02 | TRUE |
| isoComp_00502891 | geneComp_00098619 | ENST00000273047 | ENSG00000144566 | RAB5A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.324 | 0.531 | 0.207 | 1.411962e-02 | TRUE |
| isoComp_00429582 | geneComp_00091135 | ENST00000489215 | ENSG00000072135 | PTPN18 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.083 | 0.172 | 0.089 | 1.415342e-02 | TRUE |
| isoComp_00520983 | geneComp_00100673 | ENST00000581010 | ENSG00000161956 | SENP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.180 | 0.107 | -0.073 | 1.415342e-02 | TRUE |
| isoComp_00570527 | geneComp_00107609 | ENST00000375530 | ENSG00000204371 | EHMT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.118 | 0.200 | 0.082 | 1.415342e-02 | |
| isoComp_00480444 | geneComp_00096327 | ENST00000418356 | ENSG00000130649 | CYP2E1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.442 | 0.000 | -0.442 | 1.419072e-02 | TRUE |
| isoComp_00487549 | geneComp_00097063 | ENST00000482041 | ENSG00000135249 | RINT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.059 | 0.059 | 1.447298e-02 | TRUE |
| isoComp_00518059 | geneComp_00100383 | ENST00000582244 | ENSG00000159640 | ACE | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.125 | 0.000 | -0.125 | 1.463491e-02 | TRUE |
| isoComp_00580370 | geneComp_00110692 | ENST00000411630 | ENSG00000226950 | DANCR | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.717 | 0.808 | 0.091 | 1.468711e-02 | TRUE |
| isoComp_00538368 | geneComp_00102839 | MSTRG.33573.1 | ENSG00000170835 | CEL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.542 | 0.151 | -0.391 | 1.487121e-02 | TRUE |
| isoComp_00495473 | geneComp_00097879 | ENST00000379850 | ENSG00000139631 | CSAD | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.009 | 0.200 | 0.191 | 1.488137e-02 | TRUE |
| isoComp_00539891 | geneComp_00103052 | ENST00000684248 | ENSG00000171798 | KNDC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.095 | 0.000 | -0.095 | 1.488137e-02 | TRUE |
| isoComp_00600092 | geneComp_00118789 | MSTRG.7469.56 | ENSG00000257727 | CNPY2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.041 | 0.115 | 0.074 | 1.497738e-02 | TRUE |
| isoComp_00443630 | geneComp_00092386 | ENST00000555393 | ENSG00000100968 | NFATC4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.073 | 0.000 | -0.073 | 1.499257e-02 | TRUE |
| isoComp_00453469 | geneComp_00093416 | MSTRG.3515.1 | ENSG00000107485 | GATA3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.456 | 0.734 | 0.277 | 1.499257e-02 | TRUE |
| isoComp_00493907 | geneComp_00097724 | ENST00000395002 | ENSG00000138640 | FAM13A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.153 | 0.153 | 1.499257e-02 | TRUE |
| isoComp_00572818 | geneComp_00108054 | ENST00000463533 | ENSG00000206560 | ANKRD28 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.085 | 0.000 | -0.085 | 1.503195e-02 | TRUE |
| isoComp_00432228 | geneComp_00091360 | ENST00000326324 | ENSG00000077782 | FGFR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.027 | 0.136 | 0.109 | 1.507596e-02 | TRUE |
| isoComp_00455083 | geneComp_00093560 | MSTRG.14903.66 | ENSG00000108588 | CCDC47 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.352 | 0.480 | 0.128 | 1.507596e-02 | TRUE |
| isoComp_00567187 | geneComp_00107053 | ENST00000375370 | ENSG00000198176 | TFDP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.193 | 0.089 | -0.104 | 1.515223e-02 | TRUE |
| isoComp_00580139 | geneComp_00110590 | ENST00000429922 | ENSG00000226696 | LENG8-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.091 | 0.091 | 1.524698e-02 | TRUE |
| isoComp_00496988 | geneComp_00098030 | ENST00000321919 | ENSG00000140632 | GLYR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.481 | 0.585 | 0.104 | 1.544741e-02 | TRUE |
| isoComp_00488590 | geneComp_00097154 | ENST00000418919 | ENSG00000135677 | GNS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.088 | 0.017 | -0.072 | 1.555139e-02 | TRUE |
| isoComp_00519704 | geneComp_00100542 | ENST00000368366 | ENSG00000160767 | FAM189B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.164 | 0.066 | -0.099 | 1.572072e-02 | TRUE |
| isoComp_00477020 | geneComp_00095931 | ENST00000375003 | ENSG00000127483 | HP1BP3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.314 | 0.143 | -0.171 | 1.578411e-02 | TRUE |
| isoComp_00505469 | geneComp_00098923 | MSTRG.30987.7 | ENSG00000146909 | NOM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.002 | 0.060 | 0.058 | 1.595342e-02 | |
| isoComp_00501759 | geneComp_00098502 | MSTRG.3131.5 | ENSG00000143786 | CNIH3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.191 | 0.485 | 0.294 | 1.595590e-02 | TRUE |
| isoComp_00500290 | geneComp_00098350 | MSTRG.2538.15 | ENSG00000143147 | GPR161 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.059 | 0.059 | 1.598233e-02 | TRUE |
| isoComp_00476527 | geneComp_00095878 | ENST00000359428 | ENSG00000126883 | NUP214 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.301 | 0.463 | 0.162 | 1.598346e-02 | |
| isoComp_00505822 | geneComp_00098960 | MSTRG.34438.1 | ENSG00000147162 | OGT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.033 | -0.075 | 1.604106e-02 | TRUE |
| isoComp_00564140 | geneComp_00106651 | ENST00000426961 | ENSG00000196793 | ZNF239 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.129 | 0.000 | -0.129 | 1.605596e-02 | TRUE |
| isoComp_00439058 | geneComp_00091960 | ENST00000417572 | ENSG00000093183 | SEC22C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.005 | 0.060 | 0.055 | 1.606031e-02 | TRUE |
| isoComp_00514058 | geneComp_00099920 | ENST00000502783 | ENSG00000155893 | PXYLP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.102 | 0.102 | 1.606031e-02 | TRUE |
| isoComp_00524147 | geneComp_00101100 | ENST00000466712 | ENSG00000163702 | IL17RC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.136 | 0.011 | -0.124 | 1.606031e-02 | TRUE |
| isoComp_00506045 | geneComp_00098989 | ENST00000428169 | ENSG00000147403 | RPL10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.498 | 0.423 | -0.075 | 1.618298e-02 | TRUE |
| isoComp_00519637 | geneComp_00100539 | ENST00000467076 | ENSG00000160752 | FDPS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.224 | 0.135 | -0.089 | 1.618298e-02 | TRUE |
| isoComp_00472953 | geneComp_00095452 | ENST00000521459 | ENSG00000123992 | DNPEP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.086 | 0.170 | 0.084 | 1.620014e-02 | |
| isoComp_00602396 | geneComp_00120046 | ENST00000563716 | ENSG00000260596 | DUX4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.659 | 0.000 | -0.659 | 1.644778e-02 | TRUE |
| isoComp_00465061 | geneComp_00094559 | ENST00000338639 | ENSG00000116288 | PARK7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.155 | 0.206 | 0.051 | 1.656268e-02 | |
| isoComp_00561855 | geneComp_00106361 | ENST00000581874 | ENSG00000189159 | JPT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.133 | 0.242 | 0.109 | 1.682308e-02 | TRUE |
| isoComp_00586823 | geneComp_00113437 | MSTRG.20242.1 | ENSG00000234684 | SDCBP2-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.083 | 0.083 | 1.683890e-02 | TRUE |
| isoComp_00425536 | geneComp_00090792 | ENST00000309096 | ENSG00000062194 | GPBP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.227 | 0.088 | -0.139 | 1.691417e-02 | TRUE |
| isoComp_00504386 | geneComp_00098790 | ENST00000611185 | ENSG00000145860 | RNF145 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.274 | 0.630 | 0.356 | 1.691911e-02 | TRUE |
| isoComp_00435536 | geneComp_00091655 | MSTRG.29457.4 | ENSG00000086232 | EIF2AK1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.122 | 0.201 | 0.079 | 1.698399e-02 | |
| isoComp_00605823 | geneComp_00121678 | ENST00000590613 | ENSG00000267421 | ZFP28-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.946 | 0.425 | -0.521 | 1.698728e-02 | TRUE |
| isoComp_00451108 | geneComp_00093159 | ENST00000222304 | ENSG00000105697 | HAMP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.785 | 0.785 | 1.707305e-02 | TRUE |
| isoComp_00497438 | geneComp_00098072 | ENST00000564659 | ENSG00000140983 | RHOT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.217 | 0.162 | -0.054 | 1.717738e-02 | TRUE |
| isoComp_00538081 | geneComp_00102790 | ENST00000303221 | ENSG00000170571 | EMB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.758 | 0.925 | 0.167 | 1.717738e-02 | TRUE |
| isoComp_00486564 | geneComp_00096957 | ENST00000257075 | ENSG00000134644 | PUM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.246 | 0.446 | 0.200 | 1.735815e-02 | TRUE |
| isoComp_00526994 | geneComp_00101440 | MSTRG.31987.3 | ENSG00000164830 | OXR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.042 | 0.211 | 0.168 | 1.735815e-02 | TRUE |
| isoComp_00504936 | geneComp_00098860 | ENST00000275159 | ENSG00000146350 | TBC1D32 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.152 | 0.000 | -0.152 | 1.756673e-02 | TRUE |
| isoComp_00499372 | geneComp_00098233 | ENST00000300527 | ENSG00000142173 | COL6A2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.695 | 0.781 | 0.085 | 1.758669e-02 | TRUE |
| isoComp_00552232 | geneComp_00104837 | ENST00000566536 | ENSG00000182149 | IST1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.288 | 0.367 | 0.078 | 1.768131e-02 | |
| isoComp_00553724 | geneComp_00105044 | ENST00000350690 | ENSG00000183048 | SLC25A10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.598 | 0.747 | 0.149 | 1.774121e-02 | TRUE |
| isoComp_00438064 | geneComp_00091879 | ENST00000222399 | ENSG00000091136 | LAMB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.514 | 0.735 | 0.221 | 1.776654e-02 | TRUE |
| isoComp_00458645 | geneComp_00093912 | ENST00000229238 | ENSG00000111639 | MRPL51 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.622 | 0.712 | 0.090 | 1.776654e-02 | |
| isoComp_00474203 | geneComp_00095613 | ENST00000413305 | ENSG00000125046 | SSUH2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.280 | 0.280 | 1.798048e-02 | TRUE |
| isoComp_00484072 | geneComp_00096677 | ENST00000350030 | ENSG00000132780 | NASP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.104 | 0.224 | 0.120 | 1.803698e-02 | |
| isoComp_00451551 | geneComp_00093201 | ENST00000682759 | ENSG00000105875 | WDR91 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.093 | 0.030 | -0.064 | 1.807460e-02 | TRUE |
| isoComp_00477647 | geneComp_00095987 | ENST00000598845 | ENSG00000128000 | ZNF780B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.227 | 0.000 | -0.227 | 1.808018e-02 | TRUE |
| isoComp_00495585 | geneComp_00097886 | MSTRG.7349.4 | ENSG00000139651 | ZNF740 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.208 | 0.327 | 0.118 | 1.808087e-02 | TRUE |
| isoComp_00491822 | geneComp_00097507 | ENST00000531815 | ENSG00000137494 | ANKRD42 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.000 | -0.082 | 1.809822e-02 | TRUE |
| isoComp_00615983 | geneComp_00127627 | ENST00000685840 | ENSG00000287642 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.111 | 1.000 | 0.889 | 1.809822e-02 | TRUE | |
| isoComp_00436777 | geneComp_00091767 | ENST00000484809 | ENSG00000089006 | SNX5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.151 | 0.075 | -0.076 | 1.811344e-02 | TRUE |
| isoComp_00421264 | geneComp_00090401 | MSTRG.29303.32 | ENSG00000026297 | RNASET2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.162 | 0.257 | 0.095 | 1.814826e-02 | TRUE |
| isoComp_00450402 | geneComp_00093075 | ENST00000444740 | ENSG00000105369 | CD79A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.366 | 0.366 | 1.817496e-02 | TRUE |
| isoComp_00610122 | geneComp_00124077 | MSTRG.6809.2 | ENSG00000275778 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.623 | 0.000 | -0.623 | 1.822637e-02 | TRUE | |
| isoComp_00436278 | geneComp_00091719 | ENST00000333483 | ENSG00000087995 | METTL2A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.223 | 0.404 | 0.181 | 1.837689e-02 | TRUE |
| isoComp_00600429 | geneComp_00118972 | ENST00000370003 | ENSG00000258366 | RTEL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.107 | 0.050 | -0.057 | 1.852196e-02 | TRUE |
| isoComp_00512156 | geneComp_00099684 | MSTRG.18841.2 | ENSG00000153574 | RPIA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.025 | 0.078 | 0.053 | 1.852668e-02 | |
| isoComp_00477746 | geneComp_00096005 | ENST00000248975 | ENSG00000128245 | YWHAH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.456 | 0.612 | 0.156 | 1.852924e-02 | TRUE |
| isoComp_00442702 | geneComp_00092318 | ENST00000650845 | ENSG00000100578 | KIAA0586 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.089 | 0.089 | 1.858092e-02 | TRUE |
| isoComp_00446313 | geneComp_00092694 | ENST00000562321 | ENSG00000102904 | TSNAXIP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.072 | 0.000 | -0.072 | 1.863741e-02 | TRUE |
| isoComp_00438853 | geneComp_00091943 | ENST00000674426 | ENSG00000092847 | AGO1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.289 | 0.141 | -0.148 | 1.875267e-02 | TRUE |
| isoComp_00444952 | geneComp_00092541 | MSTRG.15575.10 | ENSG00000101773 | RBBP8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.115 | 0.276 | 0.162 | 1.875267e-02 | TRUE |
| isoComp_00529591 | geneComp_00101781 | ENST00000570085 | ENSG00000166233 | ARIH1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.121 | 0.240 | 0.119 | 1.878917e-02 | TRUE |
| isoComp_00438877 | geneComp_00091946 | ENST00000584541 | ENSG00000092871 | RFFL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.062 | 0.062 | 1.894292e-02 | TRUE |
| isoComp_00458908 | geneComp_00093933 | ENST00000399448 | ENSG00000111679 | PTPN6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.341 | 0.341 | 1.894292e-02 | TRUE |
| isoComp_00557113 | geneComp_00105589 | ENST00000443656 | ENSG00000185504 | FAAP100 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.198 | 0.301 | 0.103 | 1.896307e-02 | TRUE |
| isoComp_00575423 | geneComp_00108824 | ENST00000674759 | ENSG00000214960 | CRPPA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.670 | 0.000 | -0.670 | 1.896307e-02 | TRUE |
| isoComp_00442427 | geneComp_00092291 | ENST00000553833 | ENSG00000100473 | COCH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.071 | 0.008 | -0.063 | 1.896384e-02 | TRUE |
| isoComp_00588052 | geneComp_00113950 | ENST00000655970 | ENSG00000236088 | COX10-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.099 | 0.000 | -0.099 | 1.899302e-02 | TRUE |
| isoComp_00446589 | geneComp_00092719 | MSTRG.12767.12 | ENSG00000103037 | SETD6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.002 | 0.096 | 0.094 | 1.901812e-02 | TRUE |
| isoComp_00480999 | geneComp_00096374 | ENST00000340748 | ENSG00000130816 | DNMT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.072 | 0.237 | 0.165 | 1.914277e-02 | TRUE |
| isoComp_00598320 | geneComp_00117977 | MSTRG.12107.13 | ENSG00000254852 | NPIPA2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.940 | 0.740 | -0.200 | 1.914277e-02 | TRUE |
| isoComp_00500896 | geneComp_00098418 | ENST00000409426 | ENSG00000143398 | PIP5K1A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.200 | 0.278 | 0.079 | 1.926698e-02 | TRUE |
| isoComp_00499592 | geneComp_00098255 | ENST00000391984 | ENSG00000142330 | CAPN10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.212 | 0.429 | 0.217 | 1.936846e-02 | |
| isoComp_00499708 | geneComp_00098274 | ENST00000600100 | ENSG00000142530 | FAM71E1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.488 | 0.627 | 0.139 | 1.943543e-02 | TRUE |
| isoComp_00466720 | geneComp_00094732 | ENST00000340385 | ENSG00000117592 | PRDX6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.848 | 0.769 | -0.079 | 1.943593e-02 | TRUE |
| isoComp_00453046 | geneComp_00093362 | ENST00000495170 | ENSG00000106829 | TLE4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.053 | 0.003 | -0.051 | 1.944447e-02 | TRUE |
| isoComp_00429390 | geneComp_00091124 | ENST00000392090 | ENSG00000071994 | PDCD2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.347 | 0.267 | -0.080 | 1.946365e-02 | TRUE |
| isoComp_00499952 | geneComp_00098309 | MSTRG.25442.2 | ENSG00000142731 | PLK4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.207 | 0.196 | 1.946365e-02 | TRUE |
| isoComp_00507844 | geneComp_00099187 | ENST00000278353 | ENSG00000149084 | HSD17B12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.658 | 0.849 | 0.191 | 1.946365e-02 | TRUE |
| isoComp_00530530 | geneComp_00101883 | MSTRG.10369.4 | ENSG00000166664 | CHRFAM7A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.006 | 0.086 | 0.080 | 1.946365e-02 | TRUE |
| isoComp_00574368 | geneComp_00108559 | ENST00000680254 | ENSG00000213995 | NAXD | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.217 | 0.109 | 1.946365e-02 | TRUE |
| isoComp_00604276 | geneComp_00120990 | ENST00000652782 | ENSG00000264247 | ZNF407-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.485 | 0.809 | 0.324 | 1.946365e-02 | TRUE |
| isoComp_00426132 | geneComp_00090842 | ENST00000588233 | ENSG00000064547 | LPAR2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.061 | 0.000 | -0.061 | 1.947132e-02 | TRUE |
| isoComp_00419369 | geneComp_00090223 | ENST00000585134 | ENSG00000011143 | MKS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.088 | 0.151 | 0.063 | 1.948711e-02 | TRUE |
| isoComp_00501166 | geneComp_00098442 | ENST00000493155 | ENSG00000143494 | VASH2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.045 | 0.303 | 0.259 | 1.948711e-02 | TRUE |
| isoComp_00559774 | geneComp_00106002 | ENST00000618554 | ENSG00000187595 | ZNF385C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.458 | 0.458 | 1.952712e-02 | TRUE |
| isoComp_00507328 | geneComp_00099123 | MSTRG.3603.5 | ENSG00000148481 | MINDY3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.169 | 0.159 | 1.956805e-02 | TRUE |
| isoComp_00432121 | geneComp_00091351 | ENST00000599394 | ENSG00000077463 | SIRT6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.132 | 0.037 | -0.095 | 1.958824e-02 | TRUE |
| isoComp_00540264 | geneComp_00103110 | ENST00000305544 | ENSG00000172037 | LAMB2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.225 | 0.356 | 0.131 | 1.958824e-02 | |
| isoComp_00615982 | geneComp_00127627 | ENST00000653733 | ENSG00000287642 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.889 | 0.000 | -0.889 | 1.971497e-02 | TRUE | |
| isoComp_00590864 | geneComp_00115099 | ENST00000371656 | ENSG00000240849 | PEDS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.301 | 0.168 | -0.132 | 1.984221e-02 | TRUE |
| isoComp_00476614 | geneComp_00095888 | MSTRG.34597.8 | ENSG00000126953 | TIMM8A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.092 | 0.197 | 0.105 | 1.989424e-02 | TRUE |
| isoComp_00465089 | geneComp_00094562 | ENST00000342115 | ENSG00000116337 | AMPD2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.310 | 0.207 | 1.989502e-02 | TRUE |
| isoComp_00469579 | geneComp_00095077 | ENST00000319778 | ENSG00000120616 | EPC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.388 | 0.753 | 0.365 | 1.989502e-02 | TRUE |
| isoComp_00552736 | geneComp_00104902 | MSTRG.9437.1 | ENSG00000182400 | TRAPPC6B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.094 | 0.000 | -0.094 | 1.993437e-02 | TRUE |
| isoComp_00423832 | geneComp_00090638 | ENST00000538440 | ENSG00000050820 | BCAR1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.463 | 0.656 | 0.193 | 1.994414e-02 | TRUE |
| isoComp_00606057 | geneComp_00121797 | ENST00000590614 | ENSG00000267680 | ZNF224 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.075 | 0.000 | -0.075 | 1.997814e-02 | TRUE |
| isoComp_00432392 | geneComp_00091373 | ENST00000679754 | ENSG00000078124 | ACER3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.128 | 0.128 | 2.001542e-02 | TRUE |
| isoComp_00503048 | geneComp_00098633 | ENST00000689987 | ENSG00000144647 | POMGNT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.330 | 0.192 | -0.138 | 2.005945e-02 | TRUE |
| isoComp_00500415 | geneComp_00098368 | ENST00000367888 | ENSG00000143198 | MGST3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.180 | 0.256 | 0.076 | 2.006224e-02 | TRUE |
| isoComp_00574792 | geneComp_00108636 | ENST00000592854 | ENSG00000214212 | C19orf38 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.731 | 0.984 | 0.253 | 2.029535e-02 | TRUE |
| isoComp_00418999 | geneComp_00090191 | ENST00000203407 | ENSG00000010256 | UQCRC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.414 | 0.471 | 0.057 | 2.041102e-02 | |
| isoComp_00444263 | geneComp_00092457 | ENST00000400217 | ENSG00000101266 | CSNK2A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.186 | 0.083 | 2.047308e-02 | TRUE |
| isoComp_00472457 | geneComp_00095391 | ENST00000266970 | ENSG00000123374 | CDK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.454 | 0.561 | 0.107 | 2.067963e-02 | TRUE |
| isoComp_00421269 | geneComp_00090402 | ENST00000425428 | ENSG00000026508 | CD44 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.685 | 0.685 | 2.070062e-02 | TRUE |
| isoComp_00445254 | geneComp_00092577 | ENST00000218104 | ENSG00000101986 | ABCD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.579 | 0.752 | 0.173 | 2.075337e-02 | TRUE |
| isoComp_00496368 | geneComp_00097980 | ENST00000558394 | ENSG00000140391 | TSPAN3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.046 | 0.108 | 0.062 | 2.075337e-02 | |
| isoComp_00504356 | geneComp_00098787 | ENST00000452510 | ENSG00000145833 | DDX46 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.016 | 0.292 | 0.276 | 2.075337e-02 | TRUE |
| isoComp_00490887 | geneComp_00097409 | MSTRG.33113.2 | ENSG00000136937 | NCBP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.419 | 0.569 | 0.149 | 2.087903e-02 | TRUE |
| isoComp_00505708 | geneComp_00098949 | MSTRG.34175.3 | ENSG00000147123 | NDUFB11 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.206 | 0.153 | -0.054 | 2.108787e-02 | |
| isoComp_00462346 | geneComp_00094311 | MSTRG.23008.4 | ENSG00000114738 | MAPKAPK3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.119 | 0.175 | 0.056 | 2.114386e-02 | TRUE |
| isoComp_00606575 | geneComp_00122062 | MSTRG.20996.1 | ENSG00000268858 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.008 | 0.142 | 0.134 | 2.123446e-02 | TRUE | |
| isoComp_00546750 | geneComp_00103982 | ENST00000445522 | ENSG00000176945 | MUC20 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.429 | 0.000 | -0.429 | 2.124496e-02 | TRUE |
| isoComp_00434161 | geneComp_00091532 | ENST00000495084 | ENSG00000082438 | COBLL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.201 | 0.000 | -0.201 | 2.133342e-02 | TRUE |
| isoComp_00522134 | geneComp_00100829 | ENST00000314485 | ENSG00000162729 | IGSF8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.303 | 0.418 | 0.115 | 2.136052e-02 | TRUE |
| isoComp_00554802 | geneComp_00105221 | ENST00000481685 | ENSG00000183814 | LIN9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.146 | 0.008 | -0.137 | 2.136052e-02 | TRUE |
| isoComp_00416419 | geneComp_00089966 | MSTRG.33864.2 | ENSG00000002586 | CD99 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.228 | 0.310 | 0.083 | 2.143813e-02 | TRUE |
| isoComp_00584304 | geneComp_00112348 | MSTRG.3303.2 | ENSG00000231663 | COA6-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.109 | 0.028 | -0.081 | 2.146814e-02 | TRUE |
| isoComp_00464907 | geneComp_00094539 | ENST00000371514 | ENSG00000116171 | SCP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.280 | 0.437 | 0.157 | 2.157586e-02 | TRUE |
| isoComp_00517783 | geneComp_00100345 | ENST00000309083 | ENSG00000159335 | PTMS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.656 | 0.745 | 0.089 | 2.158594e-02 | TRUE |
| isoComp_00463545 | geneComp_00094420 | ENST00000402135 | ENSG00000115392 | FANCL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.114 | 0.266 | 0.153 | 2.162126e-02 | TRUE |
| isoComp_00453377 | geneComp_00093402 | ENST00000470082 | ENSG00000107282 | APBA1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.318 | 0.318 | 2.201540e-02 | TRUE |
| isoComp_00491024 | geneComp_00097428 | ENST00000692291 | ENSG00000137070 | IL11RA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.069 | 0.191 | 0.122 | 2.205404e-02 | TRUE |
| isoComp_00570466 | geneComp_00107598 | ENST00000477826 | ENSG00000204348 | DXO | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.120 | 0.060 | -0.060 | 2.207457e-02 | TRUE |
| isoComp_00508550 | geneComp_00099240 | MSTRG.5589.6 | ENSG00000149503 | INCENP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.100 | 0.259 | 0.159 | 2.208118e-02 | TRUE |
| isoComp_00558322 | geneComp_00105751 | ENST00000399697 | ENSG00000186184 | POLR1D | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.118 | 0.245 | 0.127 | 2.212906e-02 | TRUE |
| isoComp_00601074 | geneComp_00119303 | MSTRG.10110.6 | ENSG00000259066 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.941 | 0.325 | -0.616 | 2.227010e-02 | TRUE | |
| isoComp_00606544 | geneComp_00122048 | ENST00000596498 | ENSG00000268750 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.117 | 0.880 | 0.763 | 2.248556e-02 | TRUE | |
| isoComp_00493319 | geneComp_00097662 | ENST00000260818 | ENSG00000138246 | DNAJC13 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.329 | 0.817 | 0.488 | 2.251754e-02 | TRUE |
| isoComp_00514495 | geneComp_00099972 | MSTRG.21196.9 | ENSG00000156273 | BACH1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.123 | 0.123 | 2.258448e-02 | TRUE |
| isoComp_00457049 | geneComp_00093754 | ENST00000529775 | ENSG00000110169 | HPX | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.104 | 0.000 | -0.104 | 2.277357e-02 | TRUE |
| isoComp_00542838 | geneComp_00103430 | ENST00000308696 | ENSG00000173692 | PSMD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.831 | 0.931 | 0.100 | 2.277782e-02 | |
| isoComp_00427494 | geneComp_00090963 | ENST00000467474 | ENSG00000067369 | TP53BP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.172 | 0.050 | -0.122 | 2.278996e-02 | TRUE |
| isoComp_00442754 | geneComp_00092322 | ENST00000557628 | ENSG00000100592 | DAAM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.100 | 0.000 | -0.100 | 2.282668e-02 | TRUE |
| isoComp_00511862 | geneComp_00099650 | ENST00000421804 | ENSG00000153208 | MERTK | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.074 | 0.774 | 0.700 | 2.289277e-02 | TRUE |
| isoComp_00440283 | geneComp_00092084 | ENST00000397582 | ENSG00000099849 | RASSF7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.598 | 0.744 | 0.145 | 2.330811e-02 | TRUE |
| isoComp_00517919 | geneComp_00100363 | MSTRG.2239.3 | ENSG00000159445 | THEM4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.035 | 0.185 | 0.150 | 2.330811e-02 | TRUE |
| isoComp_00461113 | geneComp_00094194 | ENST00000231504 | ENSG00000113575 | PPP2CA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.392 | 0.483 | 0.090 | 2.358171e-02 | |
| isoComp_00433428 | geneComp_00091457 | ENST00000547246 | ENSG00000080371 | RAB21 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.157 | 0.157 | 2.360665e-02 | TRUE |
| isoComp_00486552 | geneComp_00096954 | ENST00000394335 | ENSG00000134602 | STK26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.353 | 0.025 | -0.328 | 2.364612e-02 | TRUE |
| isoComp_00503656 | geneComp_00098702 | ENST00000511055 | ENSG00000145216 | FIP1L1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.145 | 0.024 | -0.121 | 2.364612e-02 | TRUE |
| isoComp_00478213 | geneComp_00096066 | MSTRG.19685.5 | ENSG00000128641 | MYO1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.147 | 0.000 | -0.147 | 2.386414e-02 | TRUE |
| isoComp_00553306 | geneComp_00104997 | ENST00000329236 | ENSG00000182872 | RBM10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.163 | 0.091 | -0.072 | 2.388693e-02 | |
| isoComp_00489238 | geneComp_00097224 | MSTRG.18976.4 | ENSG00000135974 | C2orf49 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.238 | 0.466 | 0.227 | 2.396059e-02 | TRUE |
| isoComp_00454514 | geneComp_00093513 | ENST00000264639 | ENSG00000108344 | PSMD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.892 | 0.963 | 0.071 | 2.401813e-02 | |
| isoComp_00616563 | geneComp_00128034 | ENST00000676884 | ENSG00000288674 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.631 | -0.369 | 2.408814e-02 | TRUE | |
| isoComp_00503478 | geneComp_00098682 | ENST00000381975 | ENSG00000145014 | TMEM44 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.050 | 0.050 | 2.413860e-02 | TRUE |
| isoComp_00524965 | geneComp_00101185 | MSTRG.24444.4 | ENSG00000163950 | SLBP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.499 | 0.353 | -0.146 | 2.413860e-02 | TRUE |
| isoComp_00436768 | geneComp_00091767 | ENST00000377768 | ENSG00000089006 | SNX5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.056 | 0.129 | 0.073 | 2.417387e-02 | TRUE |
| isoComp_00564132 | geneComp_00106650 | ENST00000554124 | ENSG00000196792 | STRN3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.075 | 0.000 | -0.075 | 2.432085e-02 | TRUE |
| isoComp_00577648 | geneComp_00109588 | MSTRG.21545.43 | ENSG00000223875 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.704 | 1.000 | 0.296 | 2.435876e-02 | TRUE | |
| isoComp_00614228 | geneComp_00126446 | MSTRG.30271.38 | ENSG00000285725 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.778 | 0.949 | 0.171 | 2.458245e-02 | TRUE | |
| isoComp_00546108 | geneComp_00103871 | ENST00000483432 | ENSG00000176248 | ANAPC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.116 | 0.062 | -0.054 | 2.460511e-02 | TRUE |
| isoComp_00462181 | geneComp_00094292 | ENST00000489817 | ENSG00000114541 | FRMD4B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.397 | 0.000 | -0.397 | 2.470155e-02 | TRUE |
| isoComp_00590142 | geneComp_00114807 | ENST00000685086 | ENSG00000239523 | MYLK-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.078 | 0.078 | 2.478836e-02 | TRUE |
| isoComp_00580759 | geneComp_00110842 | ENST00000649123 | ENSG00000227372 | TP73-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.125 | 0.038 | -0.087 | 2.484199e-02 | TRUE |
| isoComp_00457966 | geneComp_00093836 | MSTRG.8221.2 | ENSG00000111011 | RSRC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.045 | 0.145 | 0.101 | 2.488703e-02 | |
| isoComp_00548516 | geneComp_00104218 | ENST00000563674 | ENSG00000178188 | SH2B1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.052 | -0.052 | 2.488703e-02 | TRUE |
| isoComp_00495006 | geneComp_00097822 | ENST00000446587 | ENSG00000139233 | LLPH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.135 | 0.279 | 0.143 | 2.514800e-02 | TRUE |
| isoComp_00510840 | geneComp_00099513 | MSTRG.19256.9 | ENSG00000152127 | MGAT5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.146 | 0.064 | -0.082 | 2.526142e-02 | TRUE |
| isoComp_00610034 | geneComp_00124018 | MSTRG.21043.2 | ENSG00000275496 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.316 | 0.000 | -0.316 | 2.526142e-02 | TRUE | |
| isoComp_00599725 | geneComp_00118638 | ENST00000551765 | ENSG00000257218 | GATC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.637 | 0.522 | -0.114 | 2.531112e-02 | TRUE |
| isoComp_00462719 | geneComp_00094341 | ENST00000392222 | ENSG00000114942 | EEF1B2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.822 | 0.740 | -0.082 | 2.558909e-02 | |
| isoComp_00455983 | geneComp_00093656 | ENST00000264218 | ENSG00000109255 | NMU | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.928 | 0.739 | -0.189 | 2.586593e-02 | TRUE |
| isoComp_00532603 | geneComp_00102123 | ENST00000505305 | ENSG00000167653 | PSCA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.311 | 0.000 | -0.311 | 2.604067e-02 | TRUE |
| isoComp_00420439 | geneComp_00090315 | ENST00000494034 | ENSG00000017483 | SLC38A5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.327 | 0.000 | -0.327 | 2.608125e-02 | TRUE |
| isoComp_00501223 | geneComp_00098450 | ENST00000505882 | ENSG00000143515 | ATP8B2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.125 | 0.008 | -0.117 | 2.608125e-02 | TRUE |
| isoComp_00463795 | geneComp_00094434 | ENST00000461370 | ENSG00000115484 | CCT4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.165 | 0.338 | 0.173 | 2.624542e-02 | TRUE |
| isoComp_00507525 | geneComp_00099146 | MSTRG.4625.12 | ENSG00000148700 | ADD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.061 | 0.061 | 2.625810e-02 | TRUE |
| isoComp_00589897 | geneComp_00114707 | ENST00000374971 | ENSG00000238269 | PAGE2B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.132 | 1.000 | 0.868 | 2.654722e-02 | TRUE |
| isoComp_00550511 | geneComp_00104534 | ENST00000512944 | ENSG00000180104 | EXOC3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.173 | 0.418 | 0.245 | 2.674960e-02 | TRUE |
| isoComp_00608451 | geneComp_00123122 | ENST00000608760 | ENSG00000272645 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.442 | 0.219 | -0.223 | 2.678956e-02 | TRUE | |
| isoComp_00478202 | geneComp_00096066 | ENST00000339514 | ENSG00000128641 | MYO1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.157 | 0.157 | 2.703011e-02 | TRUE |
| isoComp_00453432 | geneComp_00093410 | ENST00000489414 | ENSG00000107371 | EXOSC3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.398 | 0.289 | -0.109 | 2.721064e-02 | TRUE |
| isoComp_00457942 | geneComp_00093835 | ENST00000261822 | ENSG00000110987 | BCL7A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.811 | 0.871 | 0.060 | 2.721064e-02 | |
| isoComp_00445992 | geneComp_00092666 | ENST00000384919 | ENSG00000102743 | SLC25A15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.308 | 0.174 | -0.134 | 2.727056e-02 | TRUE |
| isoComp_00459391 | geneComp_00093998 | ENST00000229794 | ENSG00000112062 | MAPK14 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.141 | 0.446 | 0.304 | 2.737810e-02 | TRUE |
| isoComp_00566377 | geneComp_00106944 | ENST00000540565 | ENSG00000197837 | H4-16 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.197 | 0.000 | -0.197 | 2.749007e-02 | TRUE |
| isoComp_00486648 | geneComp_00096962 | ENST00000373062 | ENSG00000134697 | GNL2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.766 | 0.904 | 0.138 | 2.751986e-02 | TRUE |
| isoComp_00443173 | geneComp_00092356 | ENST00000554943 | ENSG00000100796 | PPP4R3A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.062 | 0.157 | 0.094 | 2.758599e-02 | TRUE |
| isoComp_00603345 | geneComp_00120575 | ENST00000566129 | ENSG00000261787 | TCF24 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.362 | 0.000 | -0.362 | 2.769237e-02 | TRUE |
| isoComp_00485742 | geneComp_00096856 | ENST00000476343 | ENSG00000134108 | ARL8B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.139 | 0.005 | -0.134 | 2.779564e-02 | TRUE |
| isoComp_00555961 | geneComp_00105406 | MSTRG.21972.11 | ENSG00000184708 | EIF4ENIF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.140 | 0.130 | 2.815776e-02 | TRUE |
| isoComp_00494060 | geneComp_00097737 | ENST00000509691 | ENSG00000138674 | SEC31A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.082 | 0.027 | -0.056 | 2.913832e-02 | TRUE |
| isoComp_00566714 | geneComp_00106986 | ENST00000498279 | ENSG00000197965 | MPZL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.097 | 0.014 | -0.083 | 2.913832e-02 | TRUE |
| isoComp_00572362 | geneComp_00107943 | ENST00000399352 | ENSG00000205726 | ITSN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.220 | 0.421 | 0.201 | 2.913832e-02 | TRUE |
| isoComp_00513296 | geneComp_00099822 | ENST00000355285 | ENSG00000154856 | APCDD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.557 | 0.978 | 0.421 | 2.933206e-02 | TRUE |
| isoComp_00425427 | geneComp_00090781 | ENST00000434521 | ENSG00000061455 | PRDM6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.075 | 0.075 | 2.936821e-02 | TRUE |
| isoComp_00573525 | geneComp_00108297 | ENST00000562595 | ENSG00000213380 | COG8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.442 | 0.350 | -0.093 | 2.947719e-02 | TRUE |
| isoComp_00555225 | geneComp_00105288 | ENST00000388940 | ENSG00000184178 | SCFD2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.004 | 0.093 | 0.088 | 2.951495e-02 | TRUE |
| isoComp_00439040 | geneComp_00091959 | ENST00000496479 | ENSG00000093167 | LRRFIP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.071 | 0.015 | -0.056 | 2.955274e-02 | |
| isoComp_00501009 | geneComp_00098426 | ENST00000489944 | ENSG00000143434 | SEMA6C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.067 | 0.007 | -0.061 | 2.955274e-02 | TRUE |
| isoComp_00502001 | geneComp_00098531 | ENST00000404735 | ENSG00000143947 | RPS27A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.186 | 0.134 | -0.051 | 2.964547e-02 | TRUE |
| isoComp_00564663 | geneComp_00106722 | MSTRG.17053.7 | ENSG00000197050 | ZNF420 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.221 | 0.000 | -0.221 | 2.965426e-02 | TRUE |
| isoComp_00571176 | geneComp_00107710 | ENST00000434407 | ENSG00000204642 | HLA-F | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.014 | 0.067 | 0.053 | 2.965967e-02 | TRUE |
| isoComp_00591796 | geneComp_00115451 | ENST00000344843 | ENSG00000242485 | MRPL20 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.748 | 0.846 | 0.098 | 2.996135e-02 | TRUE |
| isoComp_00580244 | geneComp_00110637 | ENST00000379514 | ENSG00000226823 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.348 | 0.000 | -0.348 | 2.999493e-02 | TRUE | |
| isoComp_00481910 | geneComp_00096457 | MSTRG.22534.5 | ENSG00000131375 | CAPN7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.069 | 0.000 | -0.069 | 3.016759e-02 | TRUE |
| isoComp_00590097 | geneComp_00114791 | ENST00000426475 | ENSG00000239467 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.164 | 0.000 | -0.164 | 3.017951e-02 | TRUE | |
| isoComp_00457397 | geneComp_00093789 | ENST00000534965 | ENSG00000110583 | NAA40 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.005 | 0.081 | 0.076 | 3.018444e-02 | |
| isoComp_00540539 | geneComp_00103149 | ENST00000506213 | ENSG00000172244 | C5orf34 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.222 | 0.024 | -0.198 | 3.032457e-02 | TRUE |
| isoComp_00590322 | geneComp_00114868 | ENST00000657071 | ENSG00000239791 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.256 | 0.017 | -0.238 | 3.032950e-02 | TRUE | |
| isoComp_00616645 | geneComp_00128070 | ENST00000686621 | ENSG00000288743 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.774 | 0.916 | 0.143 | 3.084988e-02 | TRUE | |
| isoComp_00442052 | geneComp_00092259 | ENST00000462361 | ENSG00000100376 | FAM118A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.173 | 0.061 | -0.111 | 3.096091e-02 | TRUE |
| isoComp_00429585 | geneComp_00091135 | ENST00000495400 | ENSG00000072135 | PTPN18 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.077 | 0.013 | -0.064 | 3.113344e-02 | TRUE |
| isoComp_00589898 | geneComp_00114707 | ENST00000374974 | ENSG00000238269 | PAGE2B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.868 | 0.000 | -0.868 | 3.129718e-02 | TRUE |
| isoComp_00495964 | geneComp_00097948 | ENST00000355338 | ENSG00000140105 | WARS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.163 | 0.221 | 0.058 | 3.155460e-02 | TRUE |
| isoComp_00485974 | geneComp_00096888 | ENST00000479285 | ENSG00000134262 | AP4B1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.106 | 0.030 | -0.076 | 3.158098e-02 | TRUE |
| isoComp_00488477 | geneComp_00097141 | ENST00000381501 | ENSG00000135605 | TEC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.459 | 1.000 | 0.541 | 3.158098e-02 | TRUE |
| isoComp_00440438 | geneComp_00092096 | ENST00000473244 | ENSG00000099917 | MED15 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.226 | 0.130 | -0.096 | 3.167514e-02 | TRUE |
| isoComp_00433109 | geneComp_00091431 | ENST00000600224 | ENSG00000079435 | LIPE | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.060 | 0.000 | -0.060 | 3.176684e-02 | TRUE |
| isoComp_00512507 | geneComp_00099728 | MSTRG.28483.2 | ENSG00000154079 | SDHAF4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.036 | 0.154 | 0.118 | 3.176891e-02 | TRUE |
| isoComp_00447752 | geneComp_00092827 | ENST00000559633 | ENSG00000103707 | MTFMT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.118 | 0.305 | 0.187 | 3.179710e-02 | TRUE |
| isoComp_00608455 | geneComp_00123122 | MSTRG.3119.1 | ENSG00000272645 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.092 | 0.249 | 0.158 | 3.200170e-02 | TRUE | |
| isoComp_00495472 | geneComp_00097879 | ENST00000379846 | ENSG00000139631 | CSAD | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.151 | 0.011 | -0.140 | 3.203893e-02 | TRUE |
| isoComp_00568097 | geneComp_00107168 | ENST00000366574 | ENSG00000198626 | RYR2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.369 | 1.000 | 0.631 | 3.231087e-02 | TRUE |
| isoComp_00596569 | geneComp_00117115 | ENST00000505637 | ENSG00000251141 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.700 | 0.919 | 0.219 | 3.268398e-02 | TRUE | |
| isoComp_00510212 | geneComp_00099433 | ENST00000281187 | ENSG00000151502 | VPS26B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.311 | 0.423 | 0.111 | 3.275683e-02 | |
| isoComp_00475417 | geneComp_00095754 | ENST00000467391 | ENSG00000125871 | MGME1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.172 | 0.082 | -0.090 | 3.320904e-02 | TRUE |
| isoComp_00550023 | geneComp_00104478 | ENST00000419542 | ENSG00000179818 | PCBP1-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.004 | 0.068 | 0.064 | 3.335899e-02 | |
| isoComp_00472399 | geneComp_00095386 | ENST00000547003 | ENSG00000123352 | SPATS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.059 | 0.000 | -0.059 | 3.352304e-02 | |
| isoComp_00612973 | geneComp_00125841 | ENST00000635200 | ENSG00000282988 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.112 | 0.747 | 0.635 | 3.369522e-02 | TRUE | |
| isoComp_00556867 | geneComp_00105560 | ENST00000676462 | ENSG00000185359 | HGS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.182 | 0.278 | 0.096 | 3.371915e-02 | |
| isoComp_00446919 | geneComp_00092750 | ENST00000401874 | ENSG00000103197 | TSC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.124 | 0.202 | 0.078 | 3.394570e-02 | |
| isoComp_00434609 | geneComp_00091569 | ENST00000196482 | ENSG00000083812 | ZNF324 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.430 | 0.213 | -0.217 | 3.399597e-02 | TRUE |
| isoComp_00455006 | geneComp_00093554 | ENST00000574087 | ENSG00000108559 | NUP88 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.166 | 0.098 | -0.069 | 3.415891e-02 | TRUE |
| isoComp_00499201 | geneComp_00098215 | ENST00000524382 | ENSG00000141971 | MVB12A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.125 | 0.044 | -0.081 | 3.419246e-02 | TRUE |
| isoComp_00600889 | geneComp_00119205 | ENST00000556713 | ENSG00000258857 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.101 | 0.000 | -0.101 | 3.426665e-02 | TRUE | |
| isoComp_00545374 | geneComp_00103750 | ENST00000663595 | ENSG00000175567 | UCP2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.882 | 0.949 | 0.067 | 3.431001e-02 | |
| isoComp_00514209 | geneComp_00099937 | ENST00000521027 | ENSG00000156011 | PSD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.194 | 0.194 | 3.442926e-02 | TRUE |
| isoComp_00451347 | geneComp_00093181 | MSTRG.29684.11 | ENSG00000105778 | AVL9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.087 | 0.078 | 3.454445e-02 | TRUE |
| isoComp_00497216 | geneComp_00098048 | ENST00000565261 | ENSG00000140832 | MARVELD3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.361 | 0.000 | -0.361 | 3.454445e-02 | TRUE |
| isoComp_00562353 | geneComp_00106450 | ENST00000379344 | ENSG00000196155 | PLEKHG4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.042 | 0.148 | 0.106 | 3.483255e-02 | TRUE |
| isoComp_00419553 | geneComp_00090234 | ENST00000467936 | ENSG00000011376 | LARS2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.101 | 0.049 | -0.052 | 3.489699e-02 | TRUE |
| isoComp_00474168 | geneComp_00095608 | ENST00000537318 | ENSG00000124920 | MYRF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.068 | 0.000 | -0.068 | 3.497450e-02 | TRUE |
| isoComp_00491847 | geneComp_00097509 | ENST00000540626 | ENSG00000137497 | NUMA1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.243 | 0.143 | -0.100 | 3.512931e-02 | TRUE |
| isoComp_00465328 | geneComp_00094579 | MSTRG.817.19 | ENSG00000116560 | SFPQ | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.114 | 0.188 | 0.074 | 3.520632e-02 | TRUE |
| isoComp_00528121 | geneComp_00101595 | ENST00000528336 | ENSG00000165494 | PCF11 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.075 | 0.022 | -0.053 | 3.547304e-02 | TRUE |
| isoComp_00494390 | geneComp_00097757 | ENST00000395640 | ENSG00000138764 | CCNG2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.339 | 0.000 | -0.339 | 3.578362e-02 | TRUE |
| isoComp_00458176 | geneComp_00093860 | ENST00000549970 | ENSG00000111237 | VPS29 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.389 | 0.287 | -0.102 | 3.589998e-02 | TRUE |
| isoComp_00573531 | geneComp_00108297 | MSTRG.12910.6 | ENSG00000213380 | COG8 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.326 | 0.475 | 0.149 | 3.605027e-02 | TRUE |
| isoComp_00442202 | geneComp_00092271 | ENST00000355209 | ENSG00000100413 | POLR3H | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.072 | 0.125 | 0.053 | 3.627836e-02 | TRUE |
| isoComp_00565032 | geneComp_00106763 | ENST00000356126 | ENSG00000197170 | PSMD12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.202 | 0.087 | -0.115 | 3.631739e-02 | TRUE |
| isoComp_00513341 | geneComp_00099825 | ENST00000593001 | ENSG00000154889 | MPPE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.177 | 0.464 | 0.288 | 3.638835e-02 | TRUE |
| isoComp_00590760 | geneComp_00115040 | ENST00000409611 | ENSG00000240583 | AQP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.372 | 0.020 | -0.352 | 3.638835e-02 | TRUE |
| isoComp_00432685 | geneComp_00091396 | MSTRG.31451.4 | ENSG00000078668 | VDAC3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.034 | 0.087 | 0.053 | 3.641687e-02 | |
| isoComp_00474479 | geneComp_00095640 | ENST00000360156 | ENSG00000125354 | SEPTIN6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.199 | 0.102 | -0.097 | 3.675194e-02 | TRUE |
| isoComp_00459056 | geneComp_00093953 | ENST00000548914 | ENSG00000111785 | RIC8B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.015 | 0.185 | 0.170 | 3.677511e-02 | TRUE |
| isoComp_00496315 | geneComp_00097976 | ENST00000685548 | ENSG00000140374 | ETFA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.191 | 0.096 | -0.096 | 3.686534e-02 | TRUE |
| isoComp_00572992 | geneComp_00108078 | ENST00000551301 | ENSG00000211584 | SLC48A1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.112 | 0.270 | 0.158 | 3.697140e-02 | TRUE |
| isoComp_00423852 | geneComp_00090639 | ENST00000529360 | ENSG00000051009 | FHIP1B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.479 | 0.281 | -0.198 | 3.701889e-02 | TRUE |
| isoComp_00493219 | geneComp_00097648 | MSTRG.4320.2 | ENSG00000138138 | ATAD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.018 | 0.155 | 0.137 | 3.719837e-02 | TRUE |
| isoComp_00422021 | geneComp_00090469 | ENST00000517612 | ENSG00000035681 | NSMAF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.101 | 0.000 | -0.101 | 3.735688e-02 | TRUE |
| isoComp_00515152 | geneComp_00100047 | ENST00000682208 | ENSG00000156983 | BRPF1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.151 | 0.018 | -0.133 | 3.735688e-02 | TRUE |
| isoComp_00480400 | geneComp_00096324 | MSTRG.22323.3 | ENSG00000130638 | ATXN10 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.594 | 0.698 | 0.104 | 3.756351e-02 | TRUE |
| isoComp_00512261 | geneComp_00099699 | MSTRG.20045.18 | ENSG00000153827 | TRIP12 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.059 | 0.185 | 0.126 | 3.762444e-02 | TRUE |
| isoComp_00505951 | geneComp_00098980 | ENST00000320676 | ENSG00000147274 | RBMX | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.674 | 0.794 | 0.120 | 3.792966e-02 | TRUE |
| isoComp_00430501 | geneComp_00091216 | ENST00000327320 | ENSG00000074266 | EED | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.087 | 0.030 | -0.057 | 3.815980e-02 | TRUE |
| isoComp_00474674 | geneComp_00095660 | ENST00000582160 | ENSG00000125458 | NT5C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.119 | 0.051 | -0.068 | 3.829724e-02 | TRUE |
| isoComp_00453846 | geneComp_00093457 | ENST00000489578 | ENSG00000107829 | FBXW4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.087 | 0.022 | -0.065 | 3.838945e-02 | TRUE |
| isoComp_00581217 | geneComp_00111051 | ENST00000358438 | ENSG00000228049 | POLR2J2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.838 | 0.626 | -0.212 | 3.840377e-02 | TRUE |
| isoComp_00465688 | geneComp_00094618 | ENST00000370951 | ENSG00000116754 | SRSF11 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.159 | 0.079 | 3.858823e-02 | TRUE |
| isoComp_00469468 | geneComp_00095063 | ENST00000533318 | ENSG00000120451 | SNX19 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.068 | 0.068 | 3.859541e-02 | TRUE |
| isoComp_00419096 | geneComp_00090199 | ENST00000231721 | ENSG00000010319 | SEMA3G | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.984 | 0.900 | -0.084 | 3.861267e-02 | TRUE |
| isoComp_00466723 | geneComp_00094732 | MSTRG.2603.1 | ENSG00000117592 | PRDX6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.135 | 0.224 | 0.089 | 3.893548e-02 | TRUE |
| isoComp_00490170 | geneComp_00097324 | MSTRG.24221.3 | ENSG00000136527 | TRA2B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.057 | 0.167 | 0.110 | 3.912837e-02 | TRUE |
| isoComp_00510076 | geneComp_00099419 | MSTRG.26704.4 | ENSG00000151422 | FER | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.322 | 0.322 | 3.912837e-02 | TRUE |
| isoComp_00425867 | geneComp_00090821 | ENST00000564445 | ENSG00000063854 | HAGH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.418 | 0.289 | -0.128 | 3.917743e-02 | TRUE |
| isoComp_00579491 | geneComp_00110308 | ENST00000451315 | ENSG00000225921 | NOL7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.559 | 0.708 | 0.149 | 3.933755e-02 | TRUE |
| isoComp_00560151 | geneComp_00106053 | MSTRG.21783.3 | ENSG00000187792 | ZNF70 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.212 | 0.427 | 0.215 | 3.938558e-02 | TRUE |
| isoComp_00465104 | geneComp_00094562 | ENST00000529299 | ENSG00000116337 | AMPD2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.065 | 0.004 | -0.060 | 3.940097e-02 | TRUE |
| isoComp_00499121 | geneComp_00098205 | ENST00000679869 | ENSG00000141867 | BRD4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.117 | 0.252 | 0.135 | 3.959350e-02 | TRUE |
| isoComp_00539013 | geneComp_00102924 | ENST00000342711 | ENSG00000171219 | CDC42BPG | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.850 | -0.150 | 3.987866e-02 | TRUE |
| isoComp_00503044 | geneComp_00098633 | ENST00000344697 | ENSG00000144647 | POMGNT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.650 | 0.774 | 0.124 | 4.027233e-02 | TRUE |
| isoComp_00507936 | geneComp_00099194 | ENST00000278412 | ENSG00000149136 | SSRP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.818 | 0.871 | 0.054 | 4.029238e-02 | |
| isoComp_00522454 | geneComp_00100878 | ENST00000480676 | ENSG00000162923 | WDR26 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.289 | 0.160 | -0.129 | 4.038464e-02 | TRUE |
| isoComp_00517637 | geneComp_00100324 | ENST00000486064 | ENSG00000159214 | CCDC24 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.215 | 0.080 | -0.135 | 4.039985e-02 | TRUE |
| isoComp_00511953 | geneComp_00099663 | ENST00000519110 | ENSG00000153310 | CYRIB | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.243 | 0.017 | -0.226 | 4.074195e-02 | TRUE |
| isoComp_00527706 | geneComp_00101539 | MSTRG.33344.13 | ENSG00000165209 | STRBP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.084 | 0.005 | -0.079 | 4.077105e-02 | TRUE |
| isoComp_00610300 | geneComp_00124181 | ENST00000616284 | ENSG00000276234 | TADA2A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.073 | 0.005 | -0.068 | 4.077105e-02 | TRUE |
| isoComp_00449752 | geneComp_00093010 | MSTRG.17347.1 | ENSG00000104983 | CCDC61 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.064 | 0.005 | -0.059 | 4.086144e-02 | TRUE |
| isoComp_00432419 | geneComp_00091375 | ENST00000587261 | ENSG00000078142 | PIK3C3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.113 | 0.000 | -0.113 | 4.094069e-02 | TRUE |
| isoComp_00434729 | geneComp_00091583 | MSTRG.18873.2 | ENSG00000084090 | STARD7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.128 | 0.039 | -0.089 | 4.104440e-02 | TRUE |
| isoComp_00589252 | geneComp_00114424 | ENST00000497454 | ENSG00000237441 | RGL2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.393 | 0.583 | 0.190 | 4.114907e-02 | TRUE |
| isoComp_00466412 | geneComp_00094698 | ENST00000470824 | ENSG00000117360 | PRPF3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.178 | 0.062 | -0.116 | 4.119399e-02 | TRUE |
| isoComp_00517786 | geneComp_00100345 | ENST00000540667 | ENSG00000159335 | PTMS | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.290 | 0.211 | -0.079 | 4.119399e-02 | TRUE |
| isoComp_00522715 | geneComp_00100918 | ENST00000366777 | ENSG00000163050 | COQ8A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.708 | 0.867 | 0.159 | 4.121375e-02 | TRUE |
| isoComp_00603734 | geneComp_00120719 | MSTRG.13045.9 | ENSG00000262583 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.677 | 0.887 | 0.209 | 4.144870e-02 | TRUE | |
| isoComp_00497461 | geneComp_00098074 | ENST00000422427 | ENSG00000140987 | ZSCAN32 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.056 | 0.000 | -0.056 | 4.161054e-02 | TRUE |
| isoComp_00555871 | geneComp_00105393 | ENST00000541152 | ENSG00000184640 | SEPTIN9 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.163 | 0.078 | -0.085 | 4.161054e-02 | TRUE |
| isoComp_00479374 | geneComp_00096205 | ENST00000475348 | ENSG00000129925 | PGAP6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.213 | 0.145 | -0.068 | 4.172929e-02 | TRUE |
| isoComp_00498411 | geneComp_00098151 | ENST00000570914 | ENSG00000141504 | SAT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.104 | 0.029 | -0.075 | 4.173983e-02 | TRUE |
| isoComp_00500818 | geneComp_00098411 | ENST00000692314 | ENSG00000143379 | SETDB1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.066 | 0.148 | 0.082 | 4.179701e-02 | |
| isoComp_00492802 | geneComp_00097608 | ENST00000681617 | ENSG00000137992 | DBT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.035 | 0.405 | 0.370 | 4.187374e-02 | TRUE |
| isoComp_00494874 | geneComp_00097807 | ENST00000266546 | ENSG00000139182 | CLSTN3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.804 | 0.931 | 0.126 | 4.205907e-02 | TRUE |
| isoComp_00559419 | geneComp_00105938 | MSTRG.7677.2 | ENSG00000187109 | NAP1L1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.208 | 0.144 | -0.063 | 4.205907e-02 | TRUE |
| isoComp_00606795 | geneComp_00122163 | ENST00000597785 | ENSG00000269386 | RAB11B-AS1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.214 | 0.063 | -0.150 | 4.208766e-02 | TRUE |
| isoComp_00543052 | geneComp_00103453 | ENST00000570776 | ENSG00000173821 | RNF213 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.146 | 0.011 | -0.135 | 4.213741e-02 | TRUE |
| isoComp_00434178 | geneComp_00091534 | ENST00000444842 | ENSG00000082482 | KCNK2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 1.000 | 0.398 | -0.602 | 4.221426e-02 | TRUE |
| isoComp_00569332 | geneComp_00107395 | ENST00000398337 | ENSG00000203485 | INF2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.228 | 0.337 | 0.109 | 4.222560e-02 | TRUE |
| isoComp_00590399 | geneComp_00114895 | ENST00000636714 | ENSG00000239900 | ADSL | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.519 | 0.389 | -0.131 | 4.234232e-02 | TRUE |
| isoComp_00558164 | geneComp_00105723 | ENST00000565792 | ENSG00000186073 | CDIN1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.122 | 0.000 | -0.122 | 4.241822e-02 | TRUE |
| isoComp_00564555 | geneComp_00106709 | ENST00000307355 | ENSG00000197008 | ZNF138 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.118 | 0.016 | -0.101 | 4.244926e-02 | TRUE |
| isoComp_00485687 | geneComp_00096849 | ENST00000506789 | ENSG00000134058 | CDK7 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.117 | 0.107 | 4.262976e-02 | TRUE |
| isoComp_00555720 | geneComp_00105366 | ENST00000646620 | ENSG00000184508 | HDDC3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.150 | 0.284 | 0.135 | 4.264275e-02 | TRUE |
| isoComp_00575793 | geneComp_00108916 | ENST00000564861 | ENSG00000215302 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.090 | 0.090 | 4.296402e-02 | TRUE | |
| isoComp_00464197 | geneComp_00094470 | ENST00000401869 | ENSG00000115694 | STK25 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.259 | 0.344 | 0.085 | 4.306596e-02 | |
| isoComp_00482135 | geneComp_00096480 | MSTRG.14372.3 | ENSG00000131480 | AOC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.379 | 0.039 | -0.340 | 4.306596e-02 | TRUE |
| isoComp_00543758 | geneComp_00103549 | ENST00000307897 | ENSG00000174442 | ZWILCH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.628 | 0.819 | 0.191 | 4.315290e-02 | TRUE |
| isoComp_00485736 | geneComp_00096856 | ENST00000256496 | ENSG00000134108 | ARL8B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.720 | 0.929 | 0.209 | 4.318048e-02 | TRUE |
| isoComp_00568337 | geneComp_00107200 | ENST00000361462 | ENSG00000198718 | TOGARAM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.033 | 0.751 | 0.719 | 4.327386e-02 | TRUE |
| isoComp_00556150 | geneComp_00105445 | ENST00000585852 | ENSG00000184922 | FMNL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.234 | 0.000 | -0.234 | 4.334185e-02 | TRUE |
| isoComp_00545427 | geneComp_00103757 | ENST00000531493 | ENSG00000175592 | FOSL1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.845 | 0.000 | -0.845 | 4.358962e-02 | TRUE |
| isoComp_00583502 | geneComp_00111981 | ENST00000620051 | ENSG00000230606 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.073 | 0.073 | 4.358962e-02 | TRUE | |
| isoComp_00518353 | geneComp_00100415 | ENST00000587921 | ENSG00000159917 | ZNF235 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.185 | 0.006 | -0.179 | 4.368914e-02 | TRUE |
| isoComp_00519784 | geneComp_00100547 | ENST00000683032 | ENSG00000160789 | LMNA | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.270 | 0.387 | 0.117 | 4.374200e-02 | TRUE |
| isoComp_00550240 | geneComp_00104481 | ENST00000528664 | ENSG00000179832 | MROH1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.157 | 0.032 | -0.124 | 4.374200e-02 | TRUE |
| isoComp_00424753 | geneComp_00090722 | ENST00000540564 | ENSG00000057252 | SOAT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.006 | 0.076 | 0.070 | 4.389245e-02 | TRUE |
| isoComp_00542037 | geneComp_00103320 | ENST00000308860 | ENSG00000173137 | ADCK5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.155 | 0.294 | 0.139 | 4.404376e-02 | TRUE |
| isoComp_00474175 | geneComp_00095608 | MSTRG.5577.1 | ENSG00000124920 | MYRF | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.017 | 0.298 | 0.281 | 4.424861e-02 | TRUE |
| isoComp_00481345 | geneComp_00096405 | ENST00000537033 | ENSG00000131018 | SYNE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.095 | 0.000 | -0.095 | 4.428241e-02 | TRUE |
| isoComp_00522157 | geneComp_00100832 | ENST00000392220 | ENSG00000162735 | PEX19 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.041 | 0.125 | 0.084 | 4.442924e-02 | TRUE |
| isoComp_00507203 | geneComp_00099111 | ENST00000467838 | ENSG00000148396 | SEC16A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.321 | 0.239 | -0.082 | 4.447758e-02 | TRUE |
| isoComp_00427791 | geneComp_00090987 | ENST00000587003 | ENSG00000068097 | HEATR6 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.106 | 0.182 | 0.076 | 4.490996e-02 | TRUE |
| isoComp_00485419 | geneComp_00096819 | ENST00000646590 | ENSG00000133835 | HSD17B4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.009 | 0.121 | 0.111 | 4.500561e-02 | TRUE |
| isoComp_00493382 | geneComp_00097667 | MSTRG.4144.21 | ENSG00000138303 | ASCC1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.231 | 0.349 | 0.118 | 4.500561e-02 | TRUE |
| isoComp_00441418 | geneComp_00092200 | ENST00000612753 | ENSG00000100239 | PPP6R2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.194 | 0.121 | -0.073 | 4.509974e-02 | TRUE |
| isoComp_00574584 | geneComp_00108590 | ENST00000498056 | ENSG00000214078 | CPNE1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.103 | 0.048 | -0.054 | 4.541499e-02 | TRUE |
| isoComp_00474254 | geneComp_00095617 | ENST00000637247 | ENSG00000125122 | LRRC29 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.015 | 0.207 | 0.191 | 4.544693e-02 | TRUE |
| isoComp_00514804 | geneComp_00100012 | ENST00000287218 | ENSG00000156639 | ZFAND3 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.220 | 0.421 | 0.201 | 4.579964e-02 | TRUE |
| isoComp_00473010 | geneComp_00095458 | ENST00000602942 | ENSG00000124074 | ENKD1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.172 | 0.057 | -0.115 | 4.581132e-02 | |
| isoComp_00504206 | geneComp_00098768 | MSTRG.26679.6 | ENSG00000145730 | PAM | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.010 | 0.074 | 0.064 | 4.590299e-02 | TRUE |
| isoComp_00465864 | geneComp_00094642 | ENST00000354267 | ENSG00000116885 | OSCP1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.206 | 0.362 | 0.156 | 4.594529e-02 | TRUE |
| isoComp_00480301 | geneComp_00096311 | ENST00000371793 | ENSG00000130558 | OLFM1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.059 | 0.623 | 0.564 | 4.632088e-02 | TRUE |
| isoComp_00485940 | geneComp_00096885 | ENST00000357172 | ENSG00000134255 | CEPT1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.497 | 0.764 | 0.267 | 4.646490e-02 | TRUE |
| isoComp_00493805 | geneComp_00097712 | ENST00000559500 | ENSG00000138604 | GLCE | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.053 | 0.000 | -0.053 | 4.646490e-02 | TRUE |
| isoComp_00563243 | geneComp_00106561 | ENST00000354464 | ENSG00000196497 | IPO4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.536 | 0.643 | 0.107 | 4.690213e-02 | |
| isoComp_00578085 | geneComp_00109758 | ENST00000649658 | ENSG00000224389 | C4B | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.000 | 0.227 | 0.227 | 4.690592e-02 | TRUE |
| isoComp_00419469 | geneComp_00090228 | ENST00000225298 | ENSG00000011260 | UTP18 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.858 | 0.938 | 0.079 | 4.743001e-02 | TRUE |
| isoComp_00510310 | geneComp_00099440 | ENST00000483272 | ENSG00000151576 | QTRT2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.170 | 0.020 | -0.150 | 4.759616e-02 | TRUE |
| isoComp_00461982 | geneComp_00094274 | ENST00000232496 | ENSG00000114383 | TUSC2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.928 | 0.863 | -0.065 | 4.781774e-02 | TRUE |
| isoComp_00565344 | geneComp_00106812 | ENST00000409858 | ENSG00000197355 | UAP1L1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.108 | 0.264 | 0.156 | 4.862947e-02 | TRUE |
| isoComp_00427683 | geneComp_00090976 | ENST00000393758 | ENSG00000067840 | PDZD4 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.380 | 0.639 | 0.259 | 4.864593e-02 | TRUE |
| isoComp_00420094 | geneComp_00090283 | ENST00000014914 | ENSG00000013588 | GPRC5A | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.891 | 0.402 | -0.489 | 4.896710e-02 | TRUE |
| isoComp_00477748 | geneComp_00096005 | ENST00000420430 | ENSG00000128245 | YWHAH | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.524 | 0.369 | -0.155 | 4.898263e-02 | TRUE |
| isoComp_00553547 | geneComp_00105025 | ENST00000405646 | ENSG00000182979 | MTA1 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.118 | 0.274 | 0.156 | 4.907708e-02 | TRUE |
| isoComp_00584768 | geneComp_00112524 | MSTRG.17850.4 | ENSG00000232098 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.140 | 0.067 | -0.073 | 4.919408e-02 | ||
| isoComp_00608266 | geneComp_00123022 | ENST00000615331 | ENSG00000272391 | POM121C | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.652 | 0.820 | 0.168 | 4.923968e-02 | TRUE |
| isoComp_00442929 | geneComp_00092340 | MSTRG.9806.4 | ENSG00000100650 | SRSF5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.072 | 0.143 | 0.071 | 4.925368e-02 | TRUE |
| isoComp_00501652 | geneComp_00098495 | ENST00000366862 | ENSG00000143756 | FBXO28 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.805 | 0.598 | -0.207 | 4.929982e-02 | TRUE |
| isoComp_00504102 | geneComp_00098759 | ENST00000380345 | ENSG00000145685 | LHFPL2 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.476 | 0.852 | 0.376 | 4.929982e-02 | TRUE |
| isoComp_00596528 | geneComp_00117113 | ENST00000669938 | ENSG00000251136 | RIPK2-DT | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.063 | 0.000 | -0.063 | 4.935380e-02 | TRUE |
| isoComp_00518991 | geneComp_00100479 | ENST00000397708 | ENSG00000160294 | MCM3AP | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.259 | 0.347 | 0.088 | 4.943979e-02 | TRUE |
| isoComp_00488541 | geneComp_00097148 | ENST00000477038 | ENSG00000135632 | SMYD5 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.080 | 0.026 | -0.054 | 4.971541e-02 | TRUE |
| isoComp_00493441 | geneComp_00097677 | ENST00000236959 | ENSG00000138363 | ATIC | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.654 | 0.743 | 0.090 | 4.971541e-02 | TRUE |
| isoComp_00513380 | geneComp_00099832 | ENST00000513072 | ENSG00000154945 | ANKRD40 | HEK293_DMSO_6hA | HEK293_OSMI2_6hA | 0.085 | 0.027 | -0.058 | 4.984861e-02 | TRUE |
- Note:
- The comparisons made can be identified as “from ‘condition_1’ to ‘condition_2’”, meaning ‘condition_1’ is considered the ground state and ‘condition_2’ the changed state. This also means that a positive dIF value indicates that the isoform usage is increased in ‘condition_2’ compared to ‘condition_1’. Since the ‘isoformFeatures’ entry is the most relevant part of the switchAnalyzeRlist object, the most-used standard methods have also been implemented to work directly on isoformFeatures.
4. Circular RNA Analysis
Volcano Plot
No significantly changed circRNA was detected.
Details of sig. circRNA
No significantly changed circRNA was detected.
Annotation of sig. circRNA
No significantly changed circRNA was detected.